Note: Descriptions are shown in the official language in which they were submitted.
W'O 91/07660 ~ ~ ~ ~ ~ ~ ~23
r'
;;:,~<
-1-
NON-INVASIVE METHOD FOR ISOLATION AND DETECTION
OF FETAL DNA
Description
Funding
05 Work described herein was supported by the
National Institutes of Health and Children's Hospital
Medical Center.
Background
A variety of fetal cell types--platelets,
trophoblasts, erythrocytes and leucocytes--cross the
placenta and circulate transiently within maternal
blood (Schroder, J., J. Med. Genet., 12:230-242
(1975); Douglas G.W. et al., Am. J. Obstet. Gynec.,
78:960-973 (1959)). There have been numeraus reports
of efforts to separate fetal cells from maternal cells
present in maternal blood, but none has been
successful in isolating cells subsequently shown to
contain fetal DNA. Distinguishing fetal cells from
maternal cells has not been successful for several
reasons, including the small number of fetal cells in
a maternal blood sample and the fact that
morphological differences are slight (e. g.,
trophoblasts are the only fetal cells Which can be
distinguished from maternal cells by morphology
alone).
Others report screening the peripheral blood of
pregnant women for cells of fetal origin. Fetal
identification relied on the presence of a single
cytogenetic marker, the Y chromosame. Lymphocytes
WO 91/07660 PCT/US90/06623
~',?3
2
with a putative "XY" karyotype were found in the
maternal circulation as early as 14 weeks gestation
(Walknowska, J., et al., The Lancet, 1119-1122
(1979)).
05 The availability of flow cytometry has led many
to suggest that fetal cells could be obtained through
the use of a flow cytometer and that such cells could
be exploited for prenatal genetic diagnosis. However,
although cells sorted in this manner have been said to
be of fetal origin, based on analysis of cell surface
antigens, morphology, or cytogenetic criteria, there
has not been confirmation that the cells contain fetal
DNA. A method by which fetal DNA could be obtained
from maternal blood during pregnancy would be
valuable, particularly if it made it possible to carry
out prenatal diagnosis by a noninvasive technique.
Disclosure of the Invention
The present invention is an in vitro method of
separating or isolating fetal DNA present in the blood
of a pregnant woman from maternal DNA, as well as an
in vitro method of detecting the presence of and/or
quantitating selected fetal DNA in fetal DNA, which is
useful as a noninvasive prenatal diagnostic or
analytical method.
In the present method, fetal nucleated cells are
isolated from a maternal blood sample by means of a
detectable material which binds to the fetal nucleated
cells but not to maternal cells and is. then separated
from the maternal sample, resulting in separation of
the fetal nucleated cells from the sample. The fetal
nucleated cells can be any undifferentiated
hematopoietic cell and, particularly, fetal nucleated
WO 91/07660 PCT/US90/06623
206860
-3-
erythrocytes. In one embodiment of the present method
of isolation, at least one detectably labelled
monoclonal antibody specific for an antigen present on
fetal nucleated cells, but not for an antigen present
OS on maternal cells, is combined with a maternal blood
sample and, once bound to fetal nucleated cells, is
separated from the maternal sample. Alternatively, at
least one detectably labelled monoclonal antibody
specific for an antigen present on maternal cells, but
not for an antigen present on fetal nucleated cells is
used. In a further embodiment, the two types of
monoclonal antibodies are used.
In the case in which the detectable label is a
fluorescent molecule, separation is carried out by
means of flow cytometry, in which fluorescently-
labelled molecules are separated from unlabelled
molecules. This results in separation of fetal
nucleated cells, such as fetal nucleated erythrocytes,
from maternal cells and, thus, of fetal DNA from
maternal DNA. That this separation has occurred can
be verified using known techniques, such as microscopy
or detection of fetal hemoglobin.
In one embodiment of the method of the present
invention by which the occurrence of a selected DNA
sequence or sequences (gene(s) or gene portion(s)) in
fetal DNA is determined (detected and/or quantitated),
the isolated fetal nucleated cells, such as fetal
nucleated erythrocytes, are treated to render DNA
present in them available for amplification.
Amplification of DNA from fetal nucleates cells (fetal
DNA) is carried out using a known amplification
technique, such as the polymerase chain reaction
WO 91/07660 PCT/US90/06623
J
'.YY
(PCR). Amplified fetal nucleated cell DNA is
subsequently separated on the basis of size (e.g., by
gel electrophoresis) and contacted with a selected
labelled probe, such as labelled DNA complementary to
05 a selected DNA sequence (e.g., complementary to an
abnormal gene or gene portion, or Y-specific DNA).
Detection of the labelled probe after it has
hybridized to fetal DNA results in detection of the
sequence of interest in the fetal DNA. Quantitation
of the hybridized labelled probe results in
quantitation of the fetal DNA.
In a second embodiment of the present method of
determining the occurrence of a selected DNA sequence
(or sequences), cells isolated as described above are
sorted onto a solid support, such as a slide, and
screened for chromosomal abnormalities using in situ
hybridization. In this embodiment, a selected nucleic
acid probe, such as a labelled DNA probe for
chromosomal DNA associated with a congenital
abnormality, is combined With the fetal DNA, under
conditions appropriate for hybridization of
complementary sequences to occur. Detection and/or
quantitation of the labelled probe after hybridization
results in detection and/or quantitation of the fetal
DNA to which the probe has hybridized.
The present method of detecting the occurrence of
selected fetal DNA is useful for prenatal evaluation
or diagnostic purposes, such as determination of the
sex of the fetus, assessment of chromosomal
abnormalities and determination of the presence of
abnormal genes associated with human disease.
WO 91/07660 PCT/US90/06623
_ ,_
A particular advantage of the method of the
present invention by which fetal nucleated
erythrocytes are isolated is that such cells can be
reliably separated from cells of maternal origin. In
OS addition, because such cells are nucleated and, thus,
contain a full complement of fetal genes, the present
method makes available complete fetal DNA. The
present method of detecting and/or quantitating a
selected fetal DNA sequence is particularly valuable
not only because of the advantages associated with the
present method of isolating fetal cells, but also
because it is a noninvasive technique which can be
applied early in gestation.
Brief Description of the Drawings
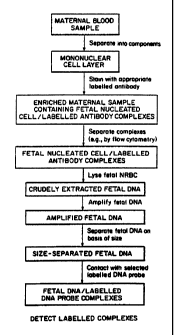
Figure 1 is a schematic representation of the
method of the present invention by which fetal
nucleated cells are isolated from maternal cells and
DNA within the fetal cells is assessed for the
occurrence of a particular fetal DNA sequence.
2O Figure 2 is an autoradiograph of diluted male DNA
amplified for 222 by sequence. Lane 1: reagent
control; lane 2: øX174 molecular weight standard;
lane 3: 100 ng; lane 4: 10 ng; lane 5: 1 ng; lane
6: 200 pcg.; lane 7: l0.pcg; lane 8: 1 pcg.
Figure 3 is a composite autoradiograph of
amplified patient DNA. Lane 1: 10 ng normal male;
lane 2: 10 ng normal female; lane 3: reagent
control; lane 4: øXI74; land 5: sorted cells from
patient 1 (male fetus); lane 6: sorted cells from
patient 2 (male fetus); lane 7: sorted cells from
patient 3 (female fetus); lane 8: sorted cells from
WO 91 /07660 PCT/US90/06623
-6-
patient 6 (female fetus); lane 9: sorted cells from
patient 7 (male fetus); lane 10: sorted cells from
patient 8 (male fetus); lane 11: sorted cells from
patient 9 (female fetus); lane 12: cord blood from female
05 infant whose cells were prenatally sorted in lane 8.
Figure 4 is a diagram demonstrating the detection of
Y chromosomal DNA sequences at various points of
gestation in women bearing male pregnancies.
Figure 5 is a series of histograms (A through F)
obtained when FITC-anti transferrin receptor was used to
determine the presence of mononuclear cells in samples
from non-pregnant females to which male cells have been
added.
Figure 6 is a composite autoradiograph of amplified
male DNA detected in TfR+ cells when 102-106 male cells
are added to samples from non-pregnant females and in
TfR cells when 105-106 male cells are added to samples
from non-pregnant females.
Figure 7 is a series of histograms (A through H)
obtained when anti HFCA-1 antibody was used to determine
the presence of mononuclear cells in samples from
non-pregnant females to which male cells have been added.
Figure 8 is a photograph illustrating a fluorescent
cell due to the positive results of in situ hybridization
of the pDP97 probe for the Y chromosome to a fetal
nucleated red blood cell.
Detailed Description of the Invention
The present invention relates to an in vitro
method of separating or isolating fetal nucleated
cells present in the blood of a pregnant woman (a
maternal blood sample) from the pregnant woman's cells
and of separating or isolating fetal DNA from-maternal
DNA. It further relates to an in vitro method of
WO 91/07660 PCT/US90/06623
'.:~;;
prenatal detection and/or quantitation of selected
fetal DNA in fetal DNA isolated from the maternal
blood sample. The method provides a noninvasive
approach to detect and/or quantitate fetal DNA, such
05 as that associated with a disease or a condition whose
assessment during gestation is desired. It also _
provides a noninvasive means by which the sex of a
fetus can be determined.
The following is a description of the basis for
the subject method; of the present method of isolating
nucleated fetal cells present in the blood of a
pregnant woman from maternal cells and, subsequently,
separating fetal DNA from maternal DNA; and of the
present method of prenatal determination of the
occurrence (presence/absence or quantitation) of
selected DNA in fetal cells.
Nucleated erythrocyte as a potential source of fetal
eg nes
It has now been determined that fetal nucleated
cells, present in the blood of a pregnant woman are a
source of fetal genes. That is, it has been shown
that fetal nucleated erythrocytes (also referred to as
fetal NRBC) can be isolated or separated from maternal
blood and that DNA present in the isolated fetal cells
can be used to assess fetal characteristics (e. g.,
sex, presence or absence of chromosomal
abnormalities).
Fetal nucleated erythrocytes were selected for
sorting based on the following rationale:
1. In any given fetomaternal hemorrhage, no matter
how small, the ratio of fetal erythrocytes to
WO 91/07660 PCT/US90/06623
f-r..
.,:. ;:':
206~66U
_8_
fetal lymphocytes should remain the same as in
whole fetal blood; thus, there would be 1,000
times as many red cells as white cells available
for analysis.
05 2. Normal pregnant females do not usually have
circulating NRBC; therefore, an isolated NRBC
would a priori have a greater chance of being
fetal in origin.
3. The majority of pregnancies are blood group
compatible, which means that the "transfused"
NRBC would probably be tolerated by the mother
and remain in her circulation.
4. Because they are nucleated, the NRBC contain a
full complement of fetal genes.
Advances in molecular biology applied to fetal cell
sorting
Recent advances in molecular biology have had an
enormous impact on the feasibility of fetal cell
identification. For example, fluorescent in situ
hybridization can be used for this purpose.
The development of the polymerase chain reaction
(PCR) (Mullis, K., et al., Cold Spring Harb. Symp.
Quant. Biol.,.51:263-272 (1986)), with its capacity
for DNA analysis from a single cell (Li, H., et al.,
Nature, 355:414-417 (1988); Handyside, A.H., et al.,
Lancet _1:347-349 (1989)), has eliminated the technical
problems associated with the small number of fetal
BYO 91/07660 PCT/US90/06623
4ts
._ 2468660
,:,
;; ..:
_9_
cells in maternal blood. It makes DNA diagnosis from
a single cell possible.
As described below, fetal nucleated erythroblasts
have been shown to be present in blood obtained from
OS pregnant women, thus making maternal blood a
useful/reliable potential source of fetal DNA; fatal
nucleated cells have been distinguished from maternal
cells on the basis of surface antigenic
characteristics, thus making it possible to separate
the two cell types from one another; and fetal DNA
present in the separated fetal nucleated cells has
been analyzed and characterized.
Detection of fetal gene sequences in maternal blood
One of the first steps in developing the present
method of isolating fetal nucleated cells from the
maternal blood supply was identification of monoclonal
antibodies that permit identification and separation
of fetal cells from maternal cells present in blood
obtained from a pregnant woman. This has been done,
as described in detail in the Examples. As a result,
it has been determined that monoclonal antibodies
which recognize maternal leucocytes and monoclonal
antibodies which recognize fetal cell surface antigens
are useful in separating maternal and fetal cells.
The following is a brief description of monoclonal
antibodies which have been shown to be useful in
separating fetal nucleated cells from maternal cells
present in a maternal blood sample. However, other
monoclonal antibodies which distinguish between fetal
and maternal cells on the basis of surface antigenic
differences, can also be used in the present method.
WO 91/07660 PCT/US90/06623
,~ .
2U6~U~U'Uv ~. ,
The present method requires the use of at least one
type of antibody which is specific for (or recognizes)
a surface antigen present on fetal nucleated cells,
for a surface antigen present on maternal cells, but
05 not specific for both. That is, the present method
can be carried out using one or more antibody which
distinguishes fetal nucleated cells from maternal
cells. The present method can be carried out using
whole blood or blood treated or processed to enrich
for (increase the concentration of) fetal nucleated
cells.
Described below is the selection and successful
use of monoclonal antibodies which distinguish fetal
nucleated erythrocytes from maternal cells. It is to
be understood, however, that in a similar manner,
monoclonal antibodies which make it possible to select
for another fetal nucleated cell type (or types) can
be identified and used in the present method to
separate fetal nucleated cells from maternal sells
(and, thus, fetal DNA sources from maternal DNA).
Initial efforts focused on the elimination of
contaminating maternal leucocytes in the mononuclear
cell layer and identification of monoclonal antibodies
effective in carrying out this separation, which
results in production of a maternal sample enriched in
fetal nucleated cells.
Hhe-1 (Becton-Dickinson Monoclonal center,
Mountain View, CA, catalog X7463) is a monoclonal
antibody available as a direct fluorescein isothi-
ocyanate (FITC) conjugate. It recognizes an antigen
present on mature human leucocytes and on very
immature erythrocyte precursors, but not on mature
WO 91/07660 PCT/US90/06623
2 6 6 8 6 6,-~.v: ~~
-11-
nucleated erythrocytes (token, M.E., et al., Blood,
_69:255-263 (1987)). Thus, maternal leucocytes are
recognized and bound, but fetal nucleated erythrocytes
are not, making separation of the two possible. As
OS described in detail in Example 1, this labelled
antibody was used to eliminate maternal leucocytes in
the mononuclear cell layer.
As is also described (Example 1), a combination
of monoclonal antibadies has been used for the same
purpose (i.e., elimination of maternal cells from the
blood sample). As described, anti-monocyte antibody
(M3) and anti-lymphocytes antibody (L4) have been used
to remove maternal cells from the mononuclear cell
layer resulting from density gradient centrifugation.
1S Monoclonal antibodies which recognize fetal
nucleated cells but do not recognize maternal cells
were also identified. As described in detail in
Example 1, a monoclonal antibody which recognizes the
transferrin receptor was identified. Erythroblasts
have been shown to express the transferrin receptor
(token, M.R:, et al., Blood, 69:255-263 (1987))
antigen on their cell surfaces from the BFU-E stage
until nuclear extrusion (token, M.R. et al., Blood,
_69:255-263 (1987)). The transferrin receptor is also
present on activated lymphocytes (Trowbridge, I.S. and
M.B. Omary, Proc. Natl. Acad. Sci. USA, 78:3039-3043
(1981)), certain tumor cells (Greaves, M. et al., Int.
J. Immunopharmac., _3:283-300 (1981)), and trophoblast
cells (Galbraith, G.M.P. et al., Blood, 55:240-242
(1980)). Thus, such an antibody is specific for or
recognizes (binds to) fetal nucleated cells, but not
maternal leucocytes. As described in Example 1,
CA 02068660 2000-11-14
V4'O 91 /07660 PCT/US90/4 '?3
-12-
commercially available fluorescein-conjugated
monoclonal antibodies against the transferrin receptor
(TfR) were used to separate fetal nucleated
erythrocytes from maternal cells. although the
05 antibody is not specific for fetal nucleated
erythrocytes, it facilitated their enrichment in the
flow-sorted samples. Other monoclonal antibodies
which are able to distinguish between fetal nucleated
cells and maternal cells present in a blood sample can
also be used. Such antibodies include commercially
available monoclonal antibodies and those which can be
produced using known techniques.
Separation of fetal nucleated cells from a
maternal blood sample using antibodies described above
can be carried out with samples of whole blood or a
fraction of whole blood (i.e., one resulting from
treatment or processing of whole blood to increase the
proportion of fetal nucleated cells present, referred
to as an enriched maternal sample. An enriched
maternal sample is produced, for example, in a
two-step process. The maternal sample is subjected to
initial separation on the basis of size, such as by
Ficoll-HypaqueTM density gradient centrifugation. This
results in production of a supernatant layer, which
contains platelets; a mononucluear cell layer; and an
agglutinated pellet which contains non-nucleated
erythrocytes and granulocytes. The mononuclear layer
is separated from the other layers, to produce a
maternal sample which is enriched in fetal nucleated
cells.
The maternal sample, whether maternal whole blood
or an enriched maternal sample, is subjected to
WO 91/07660 PCT/US90/06623
t;;..
~ss~sss
-13_ f. .. . .
separation, based on surface antigenic differences
between fetal nucleated cells and maternal cells using
antibodies described above. The maternal sample is
contacted with at least one monoclonal antibody which
05 is specific for either fetal nucleated cells or
maternal cells, but not for both and, thus, makes it
possible to separate the two types of cells. The
maternal sample can be combined with a set of two or
more monoclonal antibodies, each of which is specific
for either fetal or maternal cells, but not for both.
The combination of monoclonal antibodies can be
designed to enhance separation of the two types of
cells (e.g., the combination of anti-TfR antibody and
HLe-1 antibody described previously) beyond that
Possible with a single monoclonal antibody.
Separation of the fetal cells is carried out using
known techniques, such as flow cytometry, use of
immunomagnetic beads and cell panning. In general,
the monoclonal antibodies have a detectable label
(e~g-, radioactive material, fluorophore).
An embodiment of the method of the present
invention by which fetal cells are isolated and fetal
DNA is detected is represented schematically in Figure
1. A maternal blood sample (typically 20 ml.) is ob-
tained, using known techniques. The sample is
separated into component layers on the basis of size
and the mononuclear cell layer, referred to as, the
maternal sample enriched in nucleated cells (or
enriched maternal sample), is removed for further
processing. The enriched maternal sample is contacted
with at least one monoclonal antibody, as described
above, and the resulting fetal nucleated cell/antibody
WO 91/07660 PCT/US90/06623
~j
-14-
complexes are separated using known methods (e. g.,
flow cytometry, immunomagnetic beads, cell panning).
Fetal DNA is crudely extracted from the resulting
complexes (e. g., by heat), thus rendering it available
05 for hybridization with nucleic acid probes. Fetal DNA
can be analyzed for a selected DNA sequence or DNA
sequences, using known techniques. Prior to analysis,
fetal DNA can be amplified, as needed, using known
methods (e. g., PCR).
If amplification is to be carried out, the sorted
samples are amplified for an appropriate number of
cycles of denaturation and annealing (e. g.,
approximately 25-60). Control samples include a tube
without added DNA to monitor for false positive
amplification. With proper modification of PCR
conditions, more than one separate fetal gene can be
amplified simultaneously. This technique, known as
"multiplex" amplification, has been used with six sets
of primers in the diagnosis of DMD (Chamberlain, J.S.,
et al., Prenat. Diagnosis, 9:349-355 (1989)). When
amplification is carried out, the resulting
amplification product is a mixture which contains
amplified fetal DNA of interest (i.e., the DNA whose
occurrence is to be detected and/or quantitated) and
other DNA sequences. The amplified fetal DNA of
interest and other DNA sequences are separated, using
known techniques. Subsequent analysis of amplified
DNA can be carried out using known techniques, such
as: digestion with restriction endonuclease,
ultraviolet light visualization of ethidium bromide
stained agarose gels, DNA sequencing, or hybridization
with allele specific oligonucleotide probes (Saiki,
WO 91/07660 PCT/US90/06623
~~,.sai:;5
-15- ,
R.K., et al., Am. J. Hum. Genet., 43 (Suppl):A35
(1988)). Such analysis will determine whether
polymorphic differences exist between the amplified
"maternal" and "fetal" samples. In one embodiment,
05 the amplification mixture is separated on the basis of
size and the resulting size-separated fetal DNA is
contacted with an appropriate selected DNA probe or
probes (DNA sufficiently complementary to the fetal
DNA of interest that it hybridizes to the fetal DNA of
interest under the conditions used). Generally, the
DNA probes are labelled (e. g., with a radioactive
material, a fluorophore or other detectable material).
After the size-separated fetal DNA and the selected
DNA probes have been maintained for sufficient time
under appropriate conditions for hybridization of
complementary DNA sequences to occur, resulting in
production of fetal DNA/DNA probe complexes, detection
of the complexes is carried out using known methods.
For example, if the probe is labelled, fetal
DNA/labelled DNA probe complex is detected and/or
quantitated (e.g., by autoradiography, detection of
the fluorescent label). The quantity of labelled
complex (and, thus, of fetal DNA) can be determined by
comparison with a standard curve (i.e., a
predetermined relationship between quantity of label
detected and a given reading).
The present method has been used to identify
Y-specific DNA in nucleated erythrocytes obtained from
peripheral blood of pregnant women. This. is described
in Example 3. Briefly, candidate fetal cells from
blood samples obtained from 19 pregnant women were
isolated by flow sorting. The DNA in these cells was
WO 91/07660 PCT/US90/06623
z.~
20686fiU -16-
amplified for a 222 base pair (bp) sequence present on
the short arm of the Y chromosome as proof that the
cells were derived from the fetus. The amplified DNA
was compared with standardized DNA concentrations; 0.1
05 to 1 ng fetal DNA was obtained in the 20 ml maternal
samples. In 7/19 cases, a 222 by band of amplified
DNA was detected, consistent with the presence of male
DNA in the isolated cells; 6/7 of these were confirmed
as male pregnancies by karyotyping amniocytes. In the
case of the female fetus, DNA prepared from cord blood
at delivery also showed the presence of the Y chromo-
somal sequence. In 10/12 cases where the 222 by band
was absent, the fetuses were female. Thus, the Y
chromosomal sequence was successfully detected in 750
of the male-bearing pregnancies, demonstrating for the
first time that it is possible to isolate fetal gene
sequences from maternal blood.
As described in Example 6, male (Y-specific) DNA
has been detected in cells sorted from pregnant women
at various points in gestation. Briefly, the
mononuclear cell layer was isolated from venous blood
samples obtained from women between 11 and 16 weeks
gestation. Separation was carried out using
Ficoll/Hypaque density centrifugation, followed by
incubation with monoclonal antibodes (Anti-TfR,
anti-Leu 4 and anti-LeuM3) conjugated with a
fluorescent marker or compound (fluorescein,
phycoerythrin) and dual color analysis and flow
sorting on a fluorescence-activated cell sorter. The
cells that displayed green fluorescence, but not red
fluorescence (TfR positive, Leu 4 negative, Leu M3
negative), were fetal nucleated cells and were
WO 91/07660 PCT/US90/06523
~~;:rc.
h:~ .,a
_17_
separated from the remainder of the sample. These
cells were lysed, after which the DNA was amplified
and probed for the presence of a 397 by sequence of
the Y chromosome.
05 The results presented in Example 6 indicate the
procedure allows the detection of the 397 by sequence
present in as little as 5 pg of male DNA. In
addition, they suggest that there is a relationship
between gestational age and detection of male DNA, as
illustrated in Figure 4. This data suggests there may
be a biologic "window" for transfer of fetal nucleated
erithrocytes into maternal circulation.
The present method also has been used to
distinguish female fetal DNA from maternal DNA. The
two types of female DNA were distinguished using
amplification of paternal polymorphism, as described
in detail in Example 7. Briefly, venous blood samples
were collected from women with uncomplicated
pregnancies. Separation of fetal nucleated cells was
conducted using Ficoll/Hypaque density centrifugation,
followed by incubation with monoclonal antibodies
(anti-TfR, anti-Leu 4 and anti-Leu M3) conjugated with
a fluorescent marker (fluorescein, phycoerythrin) and
dual color analysis and flow sorting on a
fluorescence-activated cell sorter. Fetal nucleated
cells identified by displaying green fluorescence (TfR
positive), but not red fluorescence (Leu-4, Leu-3
negative), were collected and lysed. The DNA from the
cells was amplified and probed for paternal sequences
of the highly polymorphic region of chromosome 17,
which allows the distinction of female fetal DNA from
maternal DNA.
WO 91/07660 PCT/US90/06623
~:<
2 py~gy ~ ~ -18-
The results demonstrated that DNA sequences from
the father can be identified in the autosomal
chromosomes of the fetus. Consequently, the method of
the present invention can be used to separate female
05 fetal nucleated cells, as well as male fetal nucleated
cells, from maternal blood. Thus, the method can be
used for all DNA-based diagnostic procedures currently
being used in other methods, such as amniocentisis.
Further support for of the present method's
capability to identify Y-specific DNA in nucleated
erthyroctyes obtained from peripheral blood of
pregnant women is given by reconstruction experiments.
As described in Example 8, male cord blood was added
to blood obtained from non-pregnant females to
simulate the presence of fetal cells in maternal
blood. Briefly, venous blood samples were collected
from healthy, non-pregnant women and the mononuclear
cell layers isolated by Ficoll/Hypaque density
centrifugation. Mononuclear cells from the umbilical
cords of male infants (ranging from 102 to 106 cells)
were added to the mononuclear cell layers of the blood
of non-pregnant women. The cord blood contains a
large percentage of nucleated erythrocytes. The
results obtained from these experiments were
substantially similar to those obtained from pregnant
women at various stages in gestation. Amplified
sequences from the Y chromosome, consistent with the
presence of male DNA, were detected when 102 male
cells were added to the female cells.
The results of the work described above and in
the Examples demonstrate that nucleated fetal cells
have been isolated from maternal blood; genomic DNA
WO 91/07660 PCT/US90/06623
r:.;;
l:rNi i~
20~8~5Q
-19-
has been extracted from the fetal cells and identified
as being of fetal origin; fetal genes have been
amplified using PCR; and selected DNA sequences have
been identified in the fetal DNA. They demonstrate
05 that for the first time, fetal DNA has been detected
in sells isolated from maternal blood.
Uses of the Present Method of Fetal Nucleated Cell
Isolation and Fetal DNA Characterization
Thus, it has been demonstrated that fetal DNA can
be obtained from fetal nucleated cells present in a
maternal blood sample. The method of detecting and/or
quantitating fetal DNA which is represented in Figure
1 is useful as a tool for prenatal assessment (e. g.,
as a means for assessing chromosomal abnormalities,
for determining whether DNA associated with a disease
is present, or for detecting Y-specific DNA). It is
particularly useful because it is noninvasive and
requires only a small samples of blood.
Fetal DNA sequences in fetal nucleated erythro-
cytes, isolated as described herein or by other means
by which fetal nucleated cells can be separated from a
maternal blood sample, can be analyzed or assessed for
the occurrence of a DNA sequence or DNA sequences
(gene(s) or gene portion(s)) which are of interest for
diagnostic or other purposes. The DNA sequences) or
gene(s)/gene portions) present in fetal cells are
referred to herein as fetal DNA of interest. For
example, the selected DNA whose presence or absence is
to be determined and whose quantity can also be
determined is the gene for a disease, such as cystic
WO 91 /07660 PCT/US90/06623
~~=a,
..:j'
2~6~8660
-20-
fibrosis, where the causative gene or gene portion has
been cloned and sequenced; alternatively, it is a
probe for X- or Y- specific DNA. The same procedure
can also be used, with appropriate modifications
OS (e~g.. an appropriate DNA probe, time, temperature),
to detect other genes or gene portions.
As used in a diagnostic context, such as to
detect the gene known to cause cystic fibrosis, the
present method is carried out as follows: Initially,
a maternal blood sample (typically 20 ml.) is obtained
and separated into component layers based on relative
weights (e. g., by Ficoll-Hypaque density gradient
centrifugation) to remove non-nucleated erythrocytes
and produce a mononuclear cell layer. This results in
production of a maternal blood sample enriched in
fetal nucleated erythrocytes. The mononuclear cell
layer is stained with at least one appropriate
monoclonal antibody (e.g., one which is specific for
the type of fetal nucleated cell to be separated from
the sample). For example, a monoclonal antibody
specific for fetal nucleated cells, such as anti-TfR
antibody, described above, can be used. In general,
the monoclonal antibody used bears a detectable label.
Alternatively, a combination of selected labelled
monoclonal antibodies, such as monoclonal antibodies
specific for fetal nucleated cells (e.g., anti-TfR
antibody) and monoclonal antibodies specific for
maternal leucocytes (HLe-1 or L4 and M3), each
labelled with a different fluorescent compound, can be
used to remove essentially all maternal cells.
Labelled cells are subsequently separated from one
another using a known method, such as flow cytometry.
WO 91 /07660 PCT/US90/06623
,v
3"'
1,~'
206~6f~U
_21-
Binding of the monoclonal antibodies to cells for
which they are specific results in production of
labelled monoclonal antibody-cell complexes. For
example, in the case in which anti-TfR antibodies and
05 HLe-1 are used, fetal nucleated erythrocytes are bound
by anti-TfR antibody, to produce fetal nucleated
erythrocytes/anti-TfR antibody complexes, and maternal
leucocytes are bound by HLe-1 antibodies, to produce
maternal leucocyte/HLe-1 antibody complexes. The
fetal nucleated erythrocyte/anti-TfR antibody
complexes are separated from maternal cell/HLe-1
antibody complexes, using, for example, flow
cytometry. The fetal cells are lysed, to produce
crudely extracted fetal DNA which is subsequently
amplified, using, for example, PCR. This results in
production of amplified fetal DNA, which is
subsequently separated on the basis of size.
Size-separated fetal DNA is contacted with labelled
DNA probes (i.e., in prenatal detection of cystic
fibrosis, a labelled DNA probe complementary to the
gene associated with cystic fibrosis). If the fetal
DNA contains DNA of interest (in this case, the gene
associated with cystic fibrosis), fetal DNA of
interest/labelled probe complexes are formed.
Fetal DNA of interest/labelled probe complexes
are subsequently detected, using a known technique,
such as autoradiography. Simple presence or absence
of labelled fetal DNA of interest can be determined or
the quantity of fetal DNA of interest present can be
determined. In either case, the result is assessment
of fetal DNA obtained from a maternal blood sample for
selected DNA.
WO 91/07660 PCT/LS90/06623
zo~es~so
-22-
The occurrence of fetal DNA associated with
diseases or conditions other than cystic fibrosis can
also be detected and/or quantitated by the present
method. In each case, an appropriate probe is used to
05 detect the sequence of interest. For example,
sequences from probes Stl4 (Oberle, I., et al., New
Engl. J. Med., 312:682-686 (1985)), 49a (Guerin, P.,
et al., Nucleic Acids Res., 16:7759 (1988)), KM-19
(Gasparini, P., et al., Prenat. Diagnosis, 9:349-355
(1989)), or the deletion-prone exons for the Duchenne
muscular dystrophy (DMD) gene (Chamberlain, J.S., et
al., Nucleic Acids Res., 16:11141-11156 (1988)) are
used as probes. Stl4 is a highly polymorphic sequence
isolated from the long arm of the X chromosome that
has potential usefulness in distinguishing female DNA
from maternal DNA. It maps near the gene for Factor
VIII:C and, thus, may also be utilized for prenatal
diagnosis of Hemophilia A. Primers corresponding to
sequences flanking the six most commonly deleted exons
in the DMD gene, which have been successfully used to
diagnose DMD by PCR, can also be used (Chamberlain,
J.S., et al., Nucleic Acids Res., 16:11141-11156
(1988)). Other conditions which can be diagnosed by
the present method include ~-thalassemia (Cai, S-P.,
et al., Blood, 73:372-374 (1.989); Cai, S-P., et al.,
Am. J. Hum. Genet., 45:112-114 (1989); Saiki, R.K., et
al., New Engl. J. Med., 319:537-541 (1988)), sickle
cell anemia (Saiki, R.K., et al., New Engl. J. Med.,
319:537-541 (1988)), phenylketonuria (DiLella, A.G.,
et al., Lancet, 1:497-499 (1988)) and Gaucher disease
(Theophilus, B., et al., Am. J. Hum. Genet., 45:212°
215 (1989)). An appropriate probe (or probes) is
CA 02068660 2000-11-14
W(. . /07660 PCT/US90/06623
-23-
available for use in the present method for assessing
each condition.
It is also possible to separate fetal cells from
maternal cells by means other than flow cytometry, as
05 mentioned previously, and to analyze fetal nucleated
erythrocyte DNA obtained in this way. Such separation
procedures may be used in conjunction with or
independent of flow cytometry. This is advantageous
because lack of access to a flow cytometer, as well as
expense, could limit potential applications of this
technique. Thus, other methods of fetal cell
separation can be used. The separation method used
can result in elimination of unwanted cells ("negative
selection") or isolation of rare but desirable cells
("positive selection").
For example, separation by immunomagnetic beads
or by cell panning can be used. In this embodiment,
the mononuclear cell layer is isolated, as described
previously. This layer is then mixed with antibody-
coated polymer particles containing magnetic cores
(e.g., "DynabeadsTM"). These immunomagnetic beads are
available coated with a variety of antibodies. For
example, immunomagnetic beads coated with antibody to
leucocyte antigens and antibody to mouse immuno-
globulins, which can be subsequently conjugated to
mouse monoclonal antibody against the human trans-
ferrin receptor, can be used. After mixing, the
rosetted cells are isolated with a magnetic particle
concentrator. In one embodiment, two sets of
antibody-coated immunomagnetic beads are used in
succession. First, the maternal leucocytes are
depleted and then the remaining TfR positive cells are
WO 91/07660 PCT/US90/06623
;, ~.
Y;
'Y:-'i~
-24-
206~86,~6.0
collected. Subsequent steps in the method
(amplification, separation, contact with an appro-
priate DNA probe or probe set) are as described for
cells separated by flow cytometry.
OS Mueller et al. Lancet, 336: 197-200 (1990)) have
described a method of isolating placenta-derived
trophoblast cells in the blood of pregnant women using
magnetic beads. This method included mixing 1 ml of
monoclonal antibody hybridoma culture supernatant with
2 x 107 magnetic beads precoated with sheep antibody
to mouse IgG (Fc fragment) (Dynabeads M-450, Dynal AS,
Oslo, Norway) and incubated overnight at room
temperature. The coated beads were stored at 4°C and
washed three times in ice-cold RPMI 1640 medium
containing lithium heparin (10 IU/ml). The blood from
the pregnant women was collected into tubes containing
10 IU of lithium per ml of whole blood, diluted 1:10
with RPMI containing lithium, and incubated with the
antibody coated beads at 4°C overnight. The desired
cells were bound to the antibody on the bead; the
beads collected by means of a cobalt-samarium magnet.
Although in this case the antibody was directed
against trophoblast antigens, a similar technique
can be utilized with, for example, antibody to cell
surface aritigens present on fetal nucleated
erythrocytes and not present on maternal cells. An
advantage to this particular technique is that an
initial step which results in mononuclear cell
isolation is not added. Additionally, the magnetic
beads can be used for both positive (fetal cells) and
negative (maternal cells) selection.
An alternative method of isolation can be a
modificatian of the method described by R.J. Berenson
et al. (J. of Immunol. Methods, 91: 11-19 (1986))
WO 91 /07660 PCf/US90/06623
~_y:.i
2flfl86flfl
-25-
in which the high affinity between the protein avidin
and the vitamin biotin was exploited to create an
indirect immunoadsorptive procedure. In this
technique, avidin was linked to cyanogen bromide
O5, activated sepharose 6MB beads and washed in an
alternating fashion with coupling buffer (0.1 M NaHCO3
in 0.5 M NaCl at pH 8.3) and washing buffer (0.1 M
sodium acetate in 0.5 M NaCl at pH 4.5) and stored at
4°C. The blood cells were incubated with 1) murine
monoclonal antibody, and 2) biotinylated goat
anti-mouse immunoglobulin. A 3 ml column of gel was
.packed in a Pharmacia K 19/15 column. The treated
cells were passed through the column in phosphate
buffered saline containing 2% bovine serum albumin.
Adherent cells were dislodged by mechanical agitation.
This technique can be applied ~o fetal cell separation
if the antibodies used recognize fetal cell surface
antigens or maternal cell surface antigens, but not
both. Variations in methods for conjugating
antibodies to beads exist; examples include those
described by Thomas and co-workers (Thomas, T.E., et
al. (3. of Immuno. Methods, 120: 221-131 (1989)) and
by deKretser and co-workers (deKretser, T.A., et al.
(Tissue Antigens, 16: 317-325 (1980)). The use of an
antibody-bound column does not require the preliminary
isolation of the mononuclear cell fraction from whole
blood.
Once the fetal cells, are isolated from maternal
blood, they may be cultured to increase the numbers of
cells available for diagnosis, if desired. E. Fibach
et al. Blood, _73: 100-103 (1989)) have described a
method that supports the growth of human hematopoietic
WO 91/07660 PCT/US90/06523
yD,
.y.?
r'.:;I~
2~6~650
-26-
progenitor cells. This step-wise method involves 1)
initial culture in the presence of conditioned medium
from human bladder carcinoma cells, 2) removal of
leucocytes by harvest of non-adherent cells and lysis
05 with monoclonal antibodies, and 3) reculture of cells
in medium supplemented by recombinant erythropoietin.
Other methods of separating fetal nucleated cells
from maternal cells can also be used, provided that
they make it possible to differentiate between fetal
cells and maternal cells, and to isolate one from the
other.
A kit for use in carrying out the present method-
of isolating and detecting fetal DNA of interest, such
as a chromosomal abnormality associated with a disease
or other condition, in a maternal blood sample can be
produced. It includes, for example, a container for
holding the reagents needed; the reagents and,
optionally, a solid support for use in separating
fetal nucleated cell/specific antibody complexes from
other sample components or for removing maternal cells
complexed with specific antibody. For example,
reagents in a kit to be used in detecting fetal DNA of
interest after amplification of fetal DNA by PCR can
include: 1) at least one antibody specific for a
surface antigen characteristic of fetal nucleated
cells but not specific for a surface antigen
characteristic of maternal leucocytes; selected DNA
primers for use in amplifying fetal DNA by PCR; and at
least one DNA probe complementary to the fetal DNA to
be detected (fetal DNA of interest).. The kit, as
indicated, can also include a solid support to be used
in separating complexes formed from other samples
WO 91 /07660 PCT/US90/06623
.vf\\:...p
r.
-27-
components. Such solid support can be, for example, a
glass slide, nitrocellulose filter, or immunomagnetic
beads and can have affixed thereto an antibody
selective for the antibody present in the fetal
05 nucleated cell/specific antibody complexes.
The present invention will now be illustrated by
the following examples, which are not intended to be
limiting in any way.
EXAMPLE 1 Antibody selection for isolation and
sorting of fetal nucleated erythrocytes
NRBCs
Removal of maternal leucocytes from maternal
blood using human leucocyte antigen (HLe-1)
The technique of fetal NRBC isolation began with
an initial Ficoll-Hypaque density gradient
centrifugation to remove the tremendously high number
of non-nucleated erythrocytes in maternal blood.
Peripheral blood was centrifuged and separated into a
supernatant layer containing platelets, a mononuclear
cell layer, and an agglutinated pellet consisting of
non-nucleated erythrocytes and granulocytes. The
mononuclear cell layer consisted of lymphocytes,
monocytes, possible trophoblasts, and, due to their
increased size and density, NRBCs and some
reticulocytes. While the Ficoll-Hypaque
centrifugation represented an initial enrichment in
the proportion of fetal NRBCs present in the maternal
sample, flow cytometry and cell sorting was used to
improve the purity of the isolated cell population.
WO 91/07660 PCT/US90/06623
~osssso
-28-
The mononuclear cell layer from peripheral blood
samples in 63 pregnant women, 15 nonpregnant adults,
and 39 umbilical cords, was stained with FITC-HLe-1
for flow cytometric analysis. Umbilical cord samples
05 were used as a substitute for whole fetal blood.
Representative histograms displaying fluorescence
versus low-angle light scatter (an approximation of
cell size) for each of the three groups were
generated. Histogram peaks were identified that
corresponded to leucocytes, erythrocytes and
platelets. In 9 pregnant women, 7 nonpregnant adults
and 12 umbilical cord samples, fluorescent (HLe-1
positive) and non-fluorescent (HLe-1 negative) cell
populations were sorted for detailed microscopy after
Wright-Giemsa staining. While the HLe-1 positive
populations were always composed of leucocytes
independent of the sample source, the HLe-1 negative
populations differed.
In cord blood, the HLe-1 negative cells were
non-nucleated and nucleated erythrocytes with
occasional platelets. In the pregnant women,
there were platelets, non-nucleated erythrocytes, and
a very rare NRBC. In non-pregnant adults, only
platelets and debris were seen. Thus, cord blood,
with its high percentage of NRBCs, was used as a
reference to establish cell sorting parameters.
Microscopy confirmed the specificity of the
antibody-antigen binding and that the sorted HLe-1
negative cells were relatively free from leucocyte
contamination. These sorting parameters were utilized
to isolate potential fetal NRBC on 40 pregnancies.
WO 91/07660 PCT/US90/06623
~i Ar,
-29-
Enrichment of fetal NRBC in maternal blood using
transferrin receptor antigen (TfR)
The transferrin receptor (Newman, R., et al.,
Trends Biochem. Sci. 1:397-399 (1982)) is a surface
05 glycoprotein important in cellular iron transport.
The TfR is present on activated lymphocytes
(Trowbridge, I.S., et al., Proc. Natl. Aced. Sci. USA,
_78:3039-3043 (1981)), certain tumor cells (Greaves,
M., et al., Int. J. Immunopharmac., 3:283-300 (1981)),
and trophoblast cells (Galbraith, G.M.P., et al.,
Blood, _55:240-242 (1980)). Erythroblasts express the
TfR on their cell surfaces from the BFU-E stage until
nuclear extrusion (Loken, M.R., et al., Blood,
_69:255-263 (1987)). Thus, TfR is an excellent
"candidate antigen" for enrichment of fetal NRBCs
found in maternal blood. Monoclonal antibody against
TfR is available as both a fluorescein conjugate
(Becton-Dickinson catalog X7513) and a phycoerythrin
(PR) conjugate (gift of Dr. Michael Loken,
Becton-Dickinson). The mononuclear cell layer was
isolated from peripheral blood samples in 6 pregnant
women, 4 non-pregnant adults, and 3 newborn umbilical
cords for TfR analysis and microscopy. Representative
histograms of fluorescence versus light scatter from
these three groups were generated.
Whereas umbilical cord samples had a large
population of fluorescent (TfR positive cells) that
were heterogenous in size, non-pregnant adults and
pregnant adults had smaller percentages of fluorescent
cells that clustered in discrete groups. In addition,
there were slight differences in the percentages of
WO 91/07660 PCT/US90/06623
AJ-'a
2068u60 -30-
TfR positive cells in the pregnant (mean = 0.83)
versus non-pregnant (mean = 0.32) samples studies.
Micrascope studies of the TfR positive cells Were
performed using Wright-Giemsa stain for morphology and
03 Kleihauer-Betke technique for the detection of fetal
hemoglobin (Kleihauer, E., et al., Klin Wochenschr.,
_35:637-638 (1957)). In the umbilical cord samples,
large numbers of nucleated and non-nucleated
erythrocytes containing fetal hemoglobin and
occasional leucocytes were identified visually. In
the pregnant women, the predominant cell types were
nucleated and non-nucleated erythrocytes containing
fetal hemoglobin, although leucocytes were
infrequently observed. In contrast, the samples from
the non-pregnant controls consisted almost exclusively
of lymphocytes and monocytes. Because trophoblast
cells express TfR, it was postulated that they might
be present in the sorted population from the pregnant
women; none was detected.
Dual antibody analysis
Because both antibodies enriched the proportion
of NRBCs present, but did not completely exclude other
cell types in the sorted samples, combinations of
antibodies were used to isolate pure populations of
fetal NRBCs. Preliminary dual antibody studies were
performed using PE-conjugated TfR and FITC-conjugated
IiLe-1. NRBCs are TfR positive and HLe-1 negative,
whereas maternal leucocytes are HLe-1 positive. These
experiments worked well and resulted in separation of
maternal leucocytes.
VfO 91/07660 PCT/US90/06623
,.y
20~8~~~
-31-
Thus, the work described above defined flow
cytometric parameters for enrichment and sorting of
NRBCs in peripheral blood from pregnant women. In
addition, microscopic studies revealed that
05 morphologic differences occur in mononuclear cell
populations derived from venous blood samples in
pregnant versus non-pregnant adults.
EXAMPLE 2 DNA hybridization studies in HLe-1
.necLative cells sorted from maternal blood
To confirm fetal origin of the cells sorted as
described in Example 1, Y chromosomal probes were used
because it is the Y chromosome that is unquestionably
fetal in origin. The assessments were designed to
study whether the presence of Y chromosomal DNA in
maternal blood as detected on autoradiographs
performed antenatally correlated with the subsequent
birth of a male infant.
DNA isolation
HLe-1 negative cells from cord blood and pregnant
women were sorted into test tubes. Conventional
methods of DNA isolation as well as modification of
cruder methods (Lau, Y-F., et al. Lancet, 1:14-16
(1984); McCabe, E.R.B., et al., Hum Genet., 75:213-216
(1987)) were attempted without success in detecting Y
chromosome derived bands on Southern Blots. All were
limited by the small numbers of cells present.
WO 91/07660 PCf/US90/06623
L'fi.::Y
.
°32-
EXAMPLE 3 Direct hybridization to cells deposited
on filters
In order to circumvent technical problems
associated with DNA isolation, a method of direct DNA
05 hybridization to cells flow sorted onto nitrocellulose
filters was developed (Bianchi, D.W., et al.,
Cytometr~, 8:197-202 (1987)). In control experiments,
the sex of a newborn was determined from as few as 50
sorted cord blood leucocytes or 5,000 HLe-1 negative
cells (a mixture of nucleated and non-nucleated
cells).
The methodology was then applied to detection of
Y chromosomal sequences in HLe-1 negative cells sorted
from peripheral blood samples in 40 women between 8~
and 38 weeks gestation. Results were the following:
Dot Blot Hybridization with Delivered Delivered Lost to
Y Chromosomal Probe Male Infant Female Infant Follow-up
3 2 0
- 21 12 2
It was concluded that hybridization with this probe
was not predictive of male pregnancy. The possibility
exists that there was fetal DNA present on the filters
where DNA hybridization occurred, but that this DNA
bound to the Y probe nonspecifically. Thus, the
filters interpreted as "positive" for male DNA might
actually have been "positive" for fetomaternal
hemorrhage.
WO 91/07660 PCT/US90/06623
~~~~a,
-...~.
-33-
EXAMPLE 4 Use of the polymerise chain reaction (PCR)
to amplify gene sequences in sorted fetal
cells
PCR, which has a capacity for making 106 copies
05 of rare target gene sequences, was used to amplify
gene sequences in sorted fetal cells. Optimum
conditions for PCR, given the minute amounts of DNA
expected after a fetal cell sort (approximately 1 pg
to 100 ng), were determined. Experimental conditions
were modified as new information became available.
For example, Taq polymerise was used instead of Klenow
fragment of E. Coli DNA polymerise (Kogan, S.C. et
al., New England J. Med. 317:990 (1987)) because of
its increased specificity in DNA replication.
Initially, studies were performed on repeated
sequences from the long arm of the Y chromosome, probe
Y431-Hinfa (given by Dr. Kirby Smith, Johns Hopkins
University, Baltimore, MD) and the short arm of the Y
chromosome, probe Y411 (Given by Dr. Ulrich Mullet,
Children's Hospital, Boston, MA). Repeated sequences
were selected because they would create a stronger
amplification signal from a rare male fetal cell.
Y411 is identical to Y156 (Mullet, U., et al.,Nucleic
Acids Res., 14:1325-1329 (1986)), is repeated 10-60
fold, and is absolutely Y specific on Southern blots.
Sequence Y431 has autosomal homology in females that
limited its usefulness in sex determination.
PCR standardization
To define the minimum amount of DNA detectable in
maternal blood, a series of standardization
experiments were done. DNA from male and female
individuals was prepared in tenfold dilutions (1 pg to
1 mcg) and amplified using the standard reagents in
CA 02068660 2000-11-14
WO 91/07660 PCT/US90i _ 623 .
-34-
the GeneAmpTM kit (Perkin-Elmer Cetus cat #N801-0055) on
a Perkin-Elmer DNA Thermal Cycler. Primers 411-01 and
411-03 were designed to amplify a 222 base pair (bp)
sequence within probe Y411. The number of
05 amplification cycles varied between 18 and 30.
Amplified DNA samples were electrophoresed on agarose
gels, transferred to nylon filters, and hybridized to
32P-labeled Y411 probe. While it appeared possible to
detect Y specific bands on autoradiographs in lanes
containing as little as 10 pg of male DNA, results
were often muddled by the presence of amplified DNA in
female lanes or control lanes containing no added DNA.
The phenomenon of "false positive amplification" has
now received universal recognition (Lo, Y-M.D., _et
al., Lancet, 2:697 (1988); Kwok, S., et al., Nature,
339:237-238 (1989)).
Elimination of "false positive" amplification
Due to the limited amount of starting material in
a fetal cell sort, every effort was made to eliminate
background amplification in order to determine which
fetuses truly possess Y chromosomal DNA. Thus,
measures were taken to prevent aerosol contamination
of male DNA. All PCRs were performed under sterile
conditions, wearing gloves, and using positive
displacement pipettes. All reagents were prepared in
a sterile manner and incubated overnight prior to PCR
with a restriction endonuclease having a digestion
site within the target sequence. These precautions
resulted in a significant decrease and virtual absence
of false positive amplification, as monitored by
running control reactions with all reagents but no
DNA.
WO 91/07660 PCT/US90/06623
:;
-35-
Successful isolation and amplification of fetal
gene seguences from NRBCs in maternal blood
After eliminating sources of DNA contamination
and determining that as little as 10 pg of male DNA (1
05. cell = 7 pg of DNA) could be detected after PCR
amplification, candidate fetal cells from the
peripheral blood of 19 women at 12'~ to 17 weeks
gestation were sorted. Monoclonal antibody against
TfR was used to identify the presumed NRBC. The DNA
in the sorted cells was amplified for the 222 by
sequence in probe Y411 as proof that the cells were
derived from the fetus in male pregnancies. In 7/19
cases the 222 by band of amplified DNA was detected on
autoradiographs, consistent with the presence of male
DNA in the isolated cells; 6/7 of these were confirmed
as male pregnancies by karyotyping amniocytes. In the
case of one female fetus, repeat studies at 32 weeks
gestation and cord blood at delivery also showed the
presence of the Y chromosomal sequence. This result
might be explained by a low level of sex chromosome
mosaicism, XX/XY chimerism (Farber, C.M., et al., Hum.
Genet., 82:197-198 (1989)), or the presence of the
Y411 sequence in single copy on the X chromosome or
autosomes. 'In 10/12 cases where the 222 by was
absent, the fetuses were female. Therefore, detection
of the Y chromosomal sequence was successful in 6/8 or
75% of the male-bearing pregnancies. In the two
pregnancies where male DNA was not detected, there may
have been fetomaternal blood group incompatibility.
Alternatively, there may not have been fetomaternal
hemorrhage or the number of NRBCs present may have
been below the limit of sensitivity for detection of
WO 91/07660 PCT/US90/06623
36
DNA. The conditions used made it possible to detect a
minimum of 100 pg of fetal DNA, or the equivalent of
15 fetal cells. The limit of sensitivity can be
improved by extending the number of cycles used in
OS PCR. This work demonstrated that for the first time,
fetal DNA was detected in cells isolated from maternal
blood.
To further decrease false positive amplification
and permit detection of fetal DNA at the single cell
level on agarose gels, PCR is being carried out using
primers derived from a single copy of sequence
specific for the long arm of the Y chromosome, pY49a
(Guerin, P., et al.,Nucleic Acids Res., 16:7759
(1988)). In preliminary experiments using 6o cycles
of PCR, Y chromosomal DNA is visible on ethidium~
bromide stained agarose gels. This extraordinary
degree of sensitivity will now be applied to DNA from
sorted fetal cells.
EXAMPLE 5 Determination of the Volume, Morphology
and Universality of Fetomaternal
Hemarrhage
a. General Strategy
It is also possible, because of the availability
of the present method of isolating fetal nucleated
cells from blood obtained from a pregnant woman, to
determine whether fetal cells can be found in the
maternal blood in all pregnancies. A data base can be
created that can provide information on the number and
type of fetal cells circulating in maternal blood as
pregnancy progresses. Based on previous work, it is
WO 91 /07660 PCT/US90/06623
-37_
anticipated that there will be a normal range of
values that is dependent on gestational age; deviation
from these values will be studied as a potential
indication of a pregnancy at risk. Specifically,
05 large amounts of fetal blood in the maternal
circulation may be correlated with placental
abnormalities, threatened miscarriage and intrauterine
growth retardation.
Maternal venous blood samples are collected from
pregnant women, generally prior to any invasive
procedures. In general, a single 20 ml. venous blood
sample will be obtained. In a subgroup of patients,
permission will be sought to draw blood samples every
4 weeks to follow changes in numbers of fetal cells
Present. Blood is collected in EDTA, diluted 1:1 with
Hanks Balanced Salt Solution (HBSS), layered over a
Ficoll-Hypaque column (Pharmacia) and spun at 1400 rpm
for 40 minutes at roam temperature. The mononuclear
cell layer will be isolated, washed twice with HBSS,
and stained with fluorescent monoclonal antibodies.
For example, this can be a combination of fluorescein
isothiocyanate-conjugated antitransferrin receptor
(TfR) and phycoerythrin-conjugated anti-monocyte
antibodies (M3, Becton-Dickinson catalog #7497) and
anti-lymphocyte antibodies (L4,, Becton-Dickinson
catalog #7347). The staining occurs on ice, in
phosphate buffered saline (PBS) containing 2% fetal
calf serum and 0.1% sodium azide. The cells are
washed in PBS prior to flow cytometry. Analysis and
sorting are performed on a Becton-Dickinson FAGS-IV
interfaced with a Consort 40 program. Data will be
acquired on the relative size and fluorescence (in two
WO 91/07660 PCT/US90/06623
-38-
colors) of the analyzed cells. Cells that are
fluorescent in the green wavelength (TfR positive) and
not fluorescent in the red wavelength (L4 and M3
negative) will contain the presumed fetal NRBCs. The
OS percentage of these cells in the mononuclear cell
layer arerecorded and analyzed as a function of
gestational age. These cells are sorted for
microscopy and FCR amplification. In addition, cells
that are not fluorescent in the green wavelength (TfR
negative) but are fluorescent in the red wavelength
(L4 and/or M3 positive) are sorted as a presumed
maternal leucocyte population and source of materna l
DNA polymorphisms.
An additional benefit of studying nucleated fetal
cells in maternal blood is that the amount of fetal
DNA present can be extrapolated to determine the
extent of fetomaternal hemorrhage in normal and
unusual pregnancies. In the pregnancies studied, an
average amount of 1 ng of fetal DNA (corresponding to
2p 150 NBCRs) was present. Using published values of the
number of NRBCs per liter of fetal blood at 16 weeks
(3.6 x 109) (Millar, D.S., et al., Prenat. Diagnosis,
5:367-373 (1985); (Forestier, F., et al., Pediatr.
Res., 20:342-346 (1986)) and doing simple algebra,
these results were calculated.to be consistent with
2-20 ~cl hemorrhage of fetal blood into maternal
circulation. This is a trivial amount when compared
with the fetoplacental blood volume at 16 weeks, about
20 ml. It is important to validate and extend these
results to generate normative data regarding
fetomaternal transfusion in early pregnancies. It
WO 91/07660 PC'f/US90/06623
2g~8~~0
-39-
will be equally important to correlate deviations from
the expected results with pregnancy complications.
Example 6 Detection of Male DNA in Cells Sorted from
Pregnant Women at Different Points in
05 Gestation
Venous blood samples (20 ml) were collected in
EDTA from healthy women with uncomplicated
pregnancies, prior to invasive diagnostic procedures,
at different points in gestation. The mononuclear
cell layer was isolated by Ficoll/Hypaque density
centrifugation and incubated with the monoclonal
antibodies fluorescein (FITC)-conjugated anti-TfR,
phycoerythrin (PE)-conjugated anti-Leu 4 and
PE-conjugated anti-Leu M3 (Becton-Dickinson). Dual
color analysis and flow sorting were performed on a
fluorescence-activated cell sorter.
Cells that display green fluorescence but not red
fluorescence (TfR positive, Leu 4 negative, Leu M3
negative) were collected into sterile micro test tubes
and frozen at -20°C. Prior to polymerase chain
reaction amplification, the cells were lysed by
boiling. The polymerase chain reaction (PCR) was
performed under standard conditions using standard
reagents as described in Example 4. The primers used
to amplify material from the Y chromosome define a 397
base pair (bp) sequence. After PCR, the patient
samples were analyzed with conventional Southern blots
using 32P labelled probe. Ethidium bromide stained
agarose gels and autoradiographs Were examined for the
presence of the 397 by band, which is considered
WO 91/07660 PCT/U590/06623
2Y~68660 -40-
significant only if reagent controls do not reveal
false positive amplification.
Under the reaction conditions described above, it
was possible to detect the 397 by male specific band
05 if 5 pg of male DNA was present. This is
approximately the amount of DNA present in one cell.
When excess female DNA (500 ng) was added to the
reaction mixture, the male specific band was
consistently detectable at 100 pg.
Figure 4 represents a summation of samples
obtained from twelve women bearing male fetuses.
These samples were taken at different times in
pregnancy, and one woman was sampled twice. The data
indicates that there is a relationship between
gestational age and the detection of male DNA. This
implies a potential biologic "window" for the transfer
of fetal nucleated erythrocytes into the maternal
circulation.
Example 7 Detection of Female Fetal DNA by
Amplification of Paternal Polymorphisms
Venous blood samples (20 ml) were collected in
EDTA from healthy women with uncomplicated
pregnancies. The mononuclear cell layer Was isolated
by Ficoll/Hypaque density centrifugation and incubated
with the monoclonal antibodies fluorescein (FZTC)-
conjugated anti-TfR, phycoerythrin (PE)-conjugated
anti-Leu 4 and PE-conjugated anti-Leu M3
(gecton-Dickinson). Dual color analysis and flow
sorting were performed on a fluorescence-activated
cell sorter.
WO 91/07660 PCTlUS90106623
C,:;:'J
~06866D
-41-
Cells that display green fluorescence but not red
fluorescence (TfR positive, Leu 4 negative, Leu M3
negative) were collected into sterile micro test tubes
and frozen at -20°C. Additionally, cells that
05 displayed red fluorescence but not green fluorescence
(TfR negative, Leu 4 positive, Leu M3 positive) were
collected in an identical manner. Prior to polymerase
chain reaction (PCR) amplification, the cells were
lysed by boiling. PCR was performed using buffers
containing 1 mM MgCl2. The primers used in PCR
amplify a highly polymorphic region of chromosome 17.
Amplified DNA sequences correspond to blocks of genes
transmitted directly from parent to child. As a
result of the high degree of individual variation in
these sequences, it is uncommon for two parents to
manifest identical DNA patterns. Thus, it is possible
to demonstrate inheritance of the paternal sequences
in the sorted fetal cells. Since these sequences are
from chromosome 17, they are independent of fetal sex,
and may be used to distinguish female fetal DNA from
maternal DNA. Amplified DNA was separated by
electrophoresis through ethidium bromide stained
agarose gels. The DNA was transferred to nylon
filters and probed using 32P labeled sequence. The
maternal DNA, paternal DNA, TfR+ cells, and TfR cells
were then compared.
In 5 of 10 pregnant women, it was possible to
show the presence of paternal sequences in the sorted
candidate fetal cell population. In the other 5
women, no differences were seen between the maternal
DNA and the DNA obtained from the candidate fetal
cells.
WO 91/07660 PCT/US90/06623
'~'~te'.,~.
~'.) . ;.
-42-
Example 8 Reconstruction Experiments Using
Non-Pregnant Female Blood and Added Male
Cord Blood to Simulate the Presence of
Fetal Cells in Maternal Blood
05 Venous blood samples (20 ml) were collected in
EDTA from healthy non-pregnant women. Umbilical cord
blood samples (10 ml) were Collected in EDTA from
normal newborns. The mononuclear cell layer was
isolated by Ficoll/Hypaque density centrifugation.
Cell counts were performed with a hemocytometer.
Separate aliquots of cells were made containing: 1)
female cells alone; 2) female cells plus 102 added
male cord blood cells; 3) female cells plus 103 added
male cord blood cells; 4) female cells plus 10'1 added
male cord blood cells; 5) female cells plus 105 added
male cord blood cells; 6) female cells plus 106 added
male cord blood cells; 7) male cord blood cells alone.
The separate aliquots were then incubated with the
individual monoclonal antibodies being tested.
Analysis and sorting were performed using a flow
cytometer. For each aliquot, a bivariate histogram
was obtained, and gating parameters were established
for antibody positive and antibody negative cells.
The sorted cells were collected into sterile micro
test tubes and frozen at -20°C. PCR amplification was
performed with primers that detect a 397 by sequence
unique to the Y chromosome. The presence of a band at
397 by in autoxadiographs was used to confirm the
presence of male unbilical cord blood cells in sorted
samples.
Figure 5 shows the histograms obtained when
FITC-anti transferrin receptor is used. In the
WO 91/07660 PCT/US90/06623
2~f ~~5~
-43-
non-pregnant female, 0.1% of the mononuclear cells
react with the antibody. In male cord blood, 24.9% of
the mononuclear cells react with the antibody. With
the addition of more and more umbilical cord cells to
05 the non-pregnant female cells, an increased percentage
of cells that react with the antibody is seen.
Figure 6 shows that male DNA is detected in the
TfR+ cells when 102°106 male cells are added. Male
DNA is detected in the TfR cells when 105-106 male
cells are added. This results from the presence of
male white blood cells in the TfR population.
Figure 7 shows the histograms obtained when anti
HPCA-1 antibody is used. In the non-pregnant female,
0.9% of the mononuclear cells react with antibody. In
umbilical cord blood, a well-defined population of
cells is seen, but the percentage is only 1.1%. Thus,
the addition of umbilical cord blood cells to the
non-pregnant female calls is not seen on the
histograms as clearly as with the transferrin receptor
2p antibody. An increased number of HPCA+ cells were
collected as the amounts of added cord blood cells
increased.
In agarose gels, the 397 by band consistent with
DNA was detected in the HPCA+ cells when 103-105 male
cells were added to the female cells. Male DNA was
detected in agarose gels in the HPCA cells when 106
male cells were added to the female cells.
WO 91 /07660 PCT/US90/06623
~~n~'
20~~fi5~
-44-
Example 9 In situ Hybridization Using Molecular Probes
Recognizing Individual Chromosomes in Flow
Sorted Nucleated Erythrocytes
To demonstrate diagnostic utility of the present
05, invention, a DNA probe set was constructed of chromosome
specific probes that provided both good signal to noise
ratios and good spatial resolution of the fluorescent
signals. Accordingly, specific probes were developed for
five chromosomes frequently seer. as liveborn
aneuploidies; chromosomes 13, 18, 21, X and Y. A probe
for chromosome 1 was used as a control. In constructing
the probes, the general strategy was to identify a
starting clone that mapped to the desired chromosomal
region by multiple genetic and physical methods, and then
to use that clone to identify a matching cosmid "contig"
which was then used as a hybridization probe.
Hybridization of the high copy number repeat
sequences was suppressed by inclusion of total genomic
human DNA, and the chromosomal specificity verified by
hybridization to metaphase spreads. The probes gave
sharp, punctate fluorescent signals in interphase cells
that was easily discriminated and enumerated. The Y
probe used in this study was pDP97, a repetitive clone (a
5.3 kb EcoRI Y fragment from cosmid Y97 subcloned into
EcoRI site of pUC-13). All probes were labeled with
biotin, hybridized under suppression conditions, and
specific hybridization detected by conjugated
streptoavidin-FITC, which showed as a single "dot" in the
FITC image. As illustrated in Figure 8, the Y chromosome
was detected by in situ hybridization of the pDP97 probe
for the Y chromosome in a fetal nucleated red blood cell.
Thus, prenatal diagnosis for chromosomal abnormalities
could be performed on fetal cells isolated from maternal
blond.