Note: Descriptions are shown in the official language in which they were submitted.
WO~/19~96 PCT/-S91/04006
7 TITLE OF THE INVENTION:
8 ~ :
x~f~r ~ ~ R~sti~ ~ k~m~
2 a~ P~t ~llc
14
ÇROSS-REFERENCE ~O REL~TED ~PPLIC~TIONS
16
18 This application is a continuation-in-part application
19 of U.S. Patent Application Serial No. 07/537,458, filed on
June 14, 1990.
21
22 FIELD OF THE INVENTION~
23 ::
24 The invention is directed toward recombinant DNA
technology, and more specifically, toward methods for
26 modifying endogenous genes in a chimeric or transgenic
i7 animal or plant. The invention further pertains to the
28 animals/plants produced through application of the method,
29 and to the use of the method in medicine and agriculture.
This invention was supported by Government funds. The
31 Government has certain rights in this invention.
32
33 B~C~RO~D OF T~E INVENT~ON: -
34
I. Chimeric nnd Tran~genic A~imal~
36
37 Recent advances in recombinant DNA and genetic
38 technologies have made it possible to introduce and express
39 a de~ired gene sequence in a recipient animal. Through the
usc o~ such rethods, animals have been engineered to contain
-
`
.... . . . .
:~ ~. : . :. :: : . . .
Wo 9l/19796 PCr/~'S91/0~006
2-
gene sequences that are not normally or naturally present in
2 an unaltered animal. The techniques have also beer~ used to
3 produce animals which exhibit altered expression of
4 naturally present gene sequences.
The animals produced through the use of these me~hods
6 are known as either "chi~neric" or "transger. c" animals. In
7 a "chimeric" animal, only some of the animal's cells contain
8 and express the introduced gene sequence, whereas other
9 cells have been unaltered. The capacity of a chimeric
animal to transmit the introduced gene sequenc~ to its
11 progeny depends upon whether the introduced gene sequences
12 are present in the germ cells of the animal. Thus, only
13 certain chimeric animals can pass along the desired gene
14 sequence to their progeny.
In contrast, all of the cells of a "transgenic" animal
16 contain the introduced gene sequence. Consequently, a
17 transgenic animal is capable of transmitting the introduced
18 gene sequence to its progeny.
19
II. Produ~tiou of TrzL~sgenic A~imal~:
21 Microinjection Method~
22
23 The most widely used method through which transgenic
24 animals have been produced involves in~ecting a DNA molecule
into the male pronucleus of a fertilized egg (Brinster, R.L.
26 et al., Cell 27:223 (1981); Costantini, F. et al., Nature
27 294:92 ~1981); Harbers, K. et al., Nature 293:540 (1981);
28 Wagner, E.F. et al., Proc. Natl. Acad. Sc _LU~s-A-) 78:5016
29 (1981); Gordon, J.W. et al., Proc. Natl. Acad. Sci.~U.S.A.j
3~ 73:1260 (1976)).
31 The gene sequence being introduced need not be incor-
32 porated into any kind of self-replicating plasmid or virus
33 (Jaenisch, R., Science, 240:1468-1474 (1988)). Indeed, the
! ' ; , ,
', ' ' ' , ' ' . " .. ' ~ . , . .
' .~ '
~..
., ., '' ' ~.' `, "' '
' . ' '
' . ' ' '1: '
. ' . , .
WO91/19796 PCT/IS91/04006
--3--
1 presence of vector DNA has been found, in many cases, to be
2 undesirable (Hammer, R.E. et al., Science 235:53 (1987);
3 Chada, X. et_al., Nature 319:685 (1986); Kollias, G. et al.,
4 Cell 46:89 (1936); Shani, M., Molec. Cell. Biol. 6:2624
(1986); Chada, K. et al., Nature 314:377 (1985); Townes, T.
6 et al., EMBO J. 4:171S (1985)).
7 After being injected into the recipient fertilized egg,
8 the DNA molecules are believed to recombine with one another
9 to form extended head-to-tail concatemers. It has been
propo3ed that such concatemers occur at sites of double-
11 stranded DNA breaks at random sites in the egg's
12 chromosomes, and that the concatemers are inserted and
13 integrated into such sites (Brinster, R.L. et_al., Proc.
14 Natl. Acad. Sci. LU.S.A.) 82:4438 (1985)). Although it is,
thus, possible for the injected DNA molecules to be
16 incorporated at se~eral sites within the chromosomes of the
17 fertilized egg, in most instances, only a single $ite of
18 insertion is observed ~Jaenisch, R., Science, 240:1468-1474
19 (1988); Meade, H. et al. (U.S. Patent 4,873,316)).
Once the DNA molecule has been injected into the
21 ~ertilized egg cell, the cell is implanted into the uterus
22 of a recipient female, and a:Llowed to develop into an
23 animal. Since all of the animal's cells are derived from
24 the implanted fertilized egg, all of the cells of ~he
resulting animal (including the germ line cells) shall
26 contain the introduced gene sequence. If, as occurs in
27 about 30% of events, the first cellular division occurs
28 befora the introduced gene sequence has integrated into the
29 cell's genome, the resulting animal will be a chimeric
animal.
31 By breeding and inbreeding such animals, it has been
32 possible to produce heterozygous and homozygous transgenic
33 animals. Despite any unpredicta~ility in the formation of
, . ., , .; . .. . , , , , ~ ..
W~91t19796 PCT/-S91/04006
1 such transgenic animals, the animals have generally been
2 found to be stable, and to be capable of producing offspring
3 which retain and express the introduced gene sequence.
4 Since microinjection causes the injected DNA to be
incorporated into the genome of the fertilized egg through
6 a process involving the disruption and alteration of the
7 nucleotide sequence in the chromosome of the egg at the
8 insertion site, it has been observed to result in the
9 alteration, disruption, or loss of function of the
endo~enous egg gene in which the injected DNA is inserted.
11 Moreover, substantial alterations (deletions, duplications,
12 rearrangements, and translocations) of the endogenous egg
13 sequences flanking the inserted DNA have been obser~ed
14 (Mahon, K.A. et al., Proc. Natl. Acad. Sci. ru.s.A.L-85:ll65
is (1988); Covarrubias, Y. et al., Proc. Natl. Acad. Sci.
16 ~U.S.A.l 83:6020 (1986); Mark, W. et al., Cold S~r. ~arb.
17 Sym~. ~uant. Biol. 50:453 (1985)). Indeed, lethal mutations
18 or gross morphological abnormalities ha~e been observed
19 (Jaenisch, R., Science 240:146~-1474 (1988); First, N.L. et
al., Amer. Meat_Sci. Assn. 39th Reciprocal Meat Conf. 39:41
21 (1986))).
22 Significantly, it has been observed that even if the
23 desired gene sequence of the microinjected DNA molecule is
24 one that is naturally found in the recipient egg's genome,
integration of the desired gene seguence rarely occurs at
26 the site of the natural gene (Brinster, R.L. et al., Proc.
27 ~atl. Acad. Sci. (U.S.A.) 86:7087-7091 (1989)). Moreover,
28 introduction of the desired yene sequence does nat generally
29 alter the sequence of the originally preser.t egg gene.
Although the site in the fertilized egg's geno~e into
31 which the injected DNA ultimately integrates cannot be
32 predetermined, it is possible to control the expression of
33 the desired gene sequence such that, in the animal,
~ .
- ~.... . - . - ~ ~,
'i . ' ~ , ., , , , '
`:~ ' .' ' , : ;' ' " ' ' '': ' ' . ,:'' ~ ,: '' :,. ~:' '
'
WO91/lg796 PCT/~'S91/040~6
-5~ ~ 7~
1 expression of the sequence will occur in an organ or tissue
2 specific manner (reviewed by Westphal, H., FASEB J. 3:117
3 (1989); Jaenisch, X., Science 240:1468-1474 tl988)).
4 The success rate for producing transgenic animals is
greatest in mice. Approximately 25% of fertilized mouse
6 eggs into which DNA has been injected, and which have been
7 implanted in a female, will become transgenic mice. A lower
8 rate has been thus far achieved with rabbits, sheep, cattle,
9 and pigs (Jaenisch, R., Science 240:1468-1474 (1988);
Hammer, R.E. et al., J. Animal. Sci. 63:269 (1986); Hammer,
11 ~.E. et al., Nature 315-680 (19%5); Wagner, T.E. et al.,
12 Therioqenoloav 21:29 (1984)). The lower rate may reflect
13 greater familiarity with the mouse as a genetic system, or
14 may reflect the difficulty of visualizing the male
pronucleus of the fertilized eggs of many farm animals
16 (Wagner, T.E. et_al., Therioaenoloqv 21:29 (1984)).
17 Thus, the productian of transgenic animals by
18 microinjection of DNA su~fers from at least two major
19 drawbacks. First, it can be accomplished only during the
single-cell stage of an animal's life. Second, it requires
21 the disruption of the natural sequence of the DNA, and thus
22 is often mutagenic or teratogenic (Gridley, T. et al.,
23 Trends Genet. 3:162 (1987)3.
24
III. Produetion of Chimeric ,~nd Transgenic Animals:
26 Recombinant Vir~l nnd Retroviral ~ethod~
~7
28 Chimeric and transgenic animals may also, be produced
29 using recombinant viral ar retroviral techniques in which
the gene sequence is introduced into an animal at a multi-
31 cell stage. In such methods, the desired gene sequence is
32 introduced into a virus or retrovirus. Cells which are
33 in~ected with the virus acquire the introduced gene
.. .. . ... ... . . .
~ - . :: . ~ , - . , .
WO91/19796 PCT/~591/04006
s,~ r ~`--~ 1
r t ~*
--6--
l sequence. If the virus or retrovirus infects every cell of
2 the animal, then the method results in the production of a
3 transgenic animal. If, however, the vlrus infects only some
4 of the animal's cells, then a chimeric animal is produced.
S The general advantage of viral or retroviral methods of
6 producing transgenic animals oYer those methods which
7 involve the microinjection of non-replicating DNA, is that
8 it is not necessary to perform the genetic manipulations at
9 a single cell stage. Moreover, infection is a highly
efficient means for introducing the DNA into a desired cell.
11 Recombinant retroviral methods for producing chimeric or
12 transgenic animals ha~e the advantage that retroviruses
13 integrate into a host's genome in a precise manner,
14 resulting generally in the presence of only a single
integrated retrovirus (although multiple insertions may
16 occur). Rearrangements of the host chromosome at the site
17 of integration are, in general, limited to minor deletions
18 (Jaenisch, R., Science 240:1468-1474 tl988); see also,
19 VE.rmUS~ H., In: RNA Tumor Viruses (Weiss, R. et al., Eds.),
Cold Spring Harbor Press, Cold Spring Harbor, NY, pp. 369-
21 Sl2 (1982)). The method is, however, as mutagenic as micro~
22 injection methods.
23 Chimeric animals have, for example, been produced by
24 incorporating a desired gene sequence into a virUs (such as
bovine papilloma virus or polyoma) w~ich is capable of
26 infecting the cells of a host animal. Upon infection, the
27 virus can be maintained in an infected cell as an
28 extrac~romosomal episome (Elbrecht, A. et al., Molec. Cell.
29 iol~ 7:1276 (1987); Lacey, M. et al., Nature 322:609
(1986); Leopold, P. et al., Cell 51:885 (1987)). Although
31 this method decreases the ~utagenic nature uf
32 chimeric/transgenic animal ~ormation, it does so by
33 decreasing germ line stability, and increasing oncogenicity
'~,' '.
.: . . .............................................. . .
:: . ., .. ~ , . . . !
WO91/19796 PCT/~S91/04006
7 ~'T ~ ~ ~
1 Pluripotent embryonic stem cells (referred to as "ES"
2 cells) are cells which may be obtained from embryos until
3 the early post-implantation stage of embryogenesis. The
4 cells may be pr~pagated in culture, and are able to
differentiate either in vitro or in vivo upon implantation
6 into a ~ouse as a tumor. ES cells have a normal Xaryotype
7 ~Evans, M.J. et al., Nature 292:154-156 (1981); Martin, G.R.
8 et al., Proc. Natl. Acad. Sci. (U.S.A.) 78:7634-7638
9 (1981)).
Upon injection into a blastocyst of a developing embryo,
11 ES cells will proliferate and differentiate, thus resulting
12 in the production of a chimeric animal. ES cells are
13 capable of colonizing both the somatic and germ-line
14 lineages of such a chimeric animal (Robertson, E. et al~
15 Cold S~rin~ Harb. Conf. Cell_Prolif. 10:647-663 (1983);
16 Bradley A. et al., Nature 309:255-256 (1984); Bradley, A. et
17 al., Curr. Top. Devel. Biol. 20:357-371 (lg86); Wagner, E.F.
18 et al., Cold SPrina Harb. SYm~. Quant. Biol. 50:691-700
19 (1985); (a:Ll of which references are incorporated herein by
20 reference).
21 In this method, ES cells are cultured in vitro, and
22 infected with a viral or retroviral vector containing the
23 gene sequence of interest. Chimeric animals generated with
24 retroviral vectors have been found to have germ cells which
25 either lack the introduced gene sequence, or contain the
26 introduced sequence but lack the capacity to produce progeny
27 cells capable of expressing the introduced sequence (Evans, -
28 M.J. et al., Cold S~rinq Harb. S~ymP. ~uant Biol. 50:685-689
29 (1985); Stewart, C.L. et al., EMBO J. 4:3701-3709 (1985);
30 Robert~on, L. et al., Nature (1986); which references are
31 incorporated herein by reference).
32 Because ES cells may be propagated in vitro, it is
33 possible to manipulate such cells using the techni~ues of ~
~, '
.
- . : . ,, . , ,, ~ .
.
;
.. . .
, '' ' '~
WO 91/19796 PCr/-S91/04006
l somatic cell genetics. Thus, it is possible to select ES
2 cells which carry mutations (such as in the hPrt gene
3 (encoding hypoxanthine phosphoribosyl transferase) (Hooper,
4 M. et al., Nature 326:292-295 (1987); Kuehn, M.R. et al.,
Nature 326:295-298 (1987)). Such selected cells can then be
6 used to produce chimeric or transgenic mice which fail t~
7 express an active HPRT enzyme, and thus provide animal
8 ~odels for diseases (such as the Lesch-Nyhan syndrome which
9 is characterized by an HPRT deficiency) (Doetschman, T. et
al., Proc. Natl. Acad. Sci. (U.S.A.) 8S 8583-8587 (1988)).
ll As indicated above, it is possible to generate a
12 transgenic animal from a chimeric animal (whose germ line
13 cells con~ain the introduced gene sequence) by inbreeding.
14 The above-described methods permit one to screen for the
desired genetic alteration prior to introducing the trans-
16 fected ~S cells into the blastocyst. One drawback of these
17 methods, however, is the inability to control the site or
18 nature of the integration of the vector.
19 :''
IY. Productio~ of Chimeric ,~nd ~ransgenic Anim~ls:
21 Plasmi~ ~ethods -~
22
23 The inherent drawbacks of t:he above-described methods
24 for producing chimeric and transgenic animals have caused
researchers to attempt to identify additional methods
26 through which such animals could be produced.
27 Gossler, A. et al., ~or example, have described the use
28 of a plasmid vector which had been modified to contain the
29 gene for neomycin phosphotransferase (nPtII gene) to
transfect ES cells in culture. The presence of the nptII
31 gene conferred resistance to the antibiotic G418 to ES cells
32 that had been infected by the plasmi~d (Gossler, A. et_al.,
33 ~roc. Natl. Acad. Sci. (U.S.A.) 83:9065-9069 (1986), which
:
.:.: -. , , ~ : , ~ -
.:: ; , ~ .
.: ., , . - :
WOgl/19~96 PCT/~S91/~4006
_9~ "P 74
1 reference is incorporated herein by reference). The
2 chimeric animals which received the plasmid and which became
3 resistant to G418, were found to have integrated the vector
4 into their chromosomes. Takahashi, Y. et al. have
described the use of a plasmid to produce chimeric mice
6 cells which expressed an avian crystallin gene (DeveloPment
7 102:258-269 (1988), incorporated herein by re~erence). The
8 avian gene was incorporated into a plasmid which contained
9 the nPtII gene. Resulting chimeric animals were found to
express the avian gene.
11
12 V. I~troduction of Ge~e sequenreq anto ~omatic Cells
13
14 DNA has been introduced into somatic cells to produce
variant cell lines. hprt-deficient Chinese hamster ovary
16 ~CHO) cells have been transformed with the CHO hPrt gene in
17 order to produce a prototrophic cell line ~Graf, L.H. et
18 al., Somat. Cell Genet. S:1031-1044 (1979)). Folger et al.
19 examined the fate of a thymidine kinase gene (tk gene) which
had been microinjected into the nuclei of cultured mammalian
21 cells. Recipient cells were found to contain from 1 to lO0
22 copies of the introduced gene sequence integrated as
23 concatemers at one or a few sites in the cellular genome
24 (Folger, K.R. et al., ~olec. Cell. _Biol. 2:1372-1387
(1982)). DNA mediated transfo~mation of an ~NA polymerase
26 II gene into Syrian hamster cells has also been reported
27 (Ingles, C. et al., Molec Cell. Biol. 2:666-673 (1982)).
28 Plasmids conferring host neomycin resistance and
29 guanosine phosphotransferase activity have been transfected
into Chinese hamster ovary cells to generatP novel cell
3~ lines (Robson, C.N. et al., Mutat. Res. 163:201-208 (1986)).
32
.
:: :
'' . . ~
.
,
I
WO91/19796 PCT/~IS91/04006
t )~ 1 0
1 VI. C~imeric or Trnnsge~ic Pl~ntq
3Extensive progress has been made in recent years in the
4fields of plant cell genetics and gene technology. For many
5genera of plants, protoplast regeneration techniques can be
6used to regenerate a plant from a single cell (Friedt, W. et
7al. Proq. Botanv 49:192-215 (1987~; Brunold, C. et al.,
8Molec. Gen. Genet. 208:469 473 (1987); Durand, J. et al.,
9Plant _ Sci. 62:263-272 (1989) which references are
10incorporlted herein by re~erence).
11Several methods can be used to deliver and express a
12foreign gene into a plant cell. The most widely used method
13employs cloning the desired gene sequence into the Ti
14plasmid of the soil bacterium A. tumori~aciens (~omari, T.
15et al., J. Bacteriol. 166:88-94 (1986); Czako, M. et al.,
16Plant Moi. ~3iol. 6:101-109 t1986); Jones, J.D.G. et al., ~ -
17EMBO J. 4:2411-2418 (1985); Shahin, E.A. et al., Theor.
18AP~1. Genet. 73:164-169 (1986)). The frequency of
19transformation may be as high as 70%, depending upon the '~
20type of plant used (Friedt, W. gt al. Pro~_8Otanv 49:192-
21215 (1987)).
22Plant viruses have also been exploited as vectors for
23the delivery and expression of foreign genes in plants. The
24cauliflower mosaic virus (Brisson, N. et al., Nature
25310:511-514 (1984) has been particularly useful for this
26purpose (Shah, D.M. et al., Science 233:478-481 (1986);
27Shew~aker, C.K. et al., Virol. 140:281-288 (1985). Vectors
28have also been prepared from derivatives of RNA viruses
29(French, R. et al., Science 231:1294-1297 ~1986).
30Techniques of microinjection (Crossway, A. et al.,
31Molec. Gen. Genet. 202:179-185 (1986); Potrykus, I. et al.,
32Molec. Gen. &enet. 199:169 177 (1985)), have been ~sed to
33accomplish the direct transfer of gene sequences into plant ;;~
.~ , . . .
~ . ~
.,
~.~
. .
W(~ b 1'(, 1 / l_ ~Y I / U4UUt)
r~ ~,Y~
1 cells. Transformation with a plasmid capable of site
2 specific recombination has bee~ used to introduce gene
3 sequences into As~erqillus (May, G.S., J. Cell Biol.
4 109:2267-2274 (1989); which reference is incorporated herein
by reference).
6 Electroporation has been identified as a method for
7 introducing DNA into plant cells (Fro~m, M.E., et al., Proc.
8 Natl. Acad. sci. (U.S.A.) 8~:5824-5828 (1985); Fromm, M.E.
9 ~t_aL., Nature 319:?91-793 (1986); Morikawa, H. et al., Gene
41:121-124 (1986); Langridge, W.H.R. et al., Theor. Appl.
11 Genet. 67:443-455 (1984)).
12 Gross genetic mutations can be produced in plant cells
13 using transposable elements (Saedler, H. et al., EMBO_J.
14 4:S85-590 (1985); Peterson, P.A., BioEssavs 3:199-204
(1985))- Such elements can initiate chromosomal
16 rearrangements, insertions, duplications, deletions, etc.
17 Chimeric plants can be regenerated from such cells using the
18 procedures described above.
19 A major deficiency of present methods for gene
manipulation in plants is the difficulty of selecting the
21 desired recombinant cell (Bruno:Ld, C. et al., Molec Gen.
22 Genet. 208:469-473 (1987)). In an attempt to address this
23 deficiency, kanamycin resistance and nitrate reductase
24 deficiency have been used as selectable markers (Brunold, C.
et al., Molec. Gen. Genet. 208:469-473 (1987)).
~6
27
.. .. . .. .;
.. . .
WO91/19796 P~T/~S91/04006
~ '~`J'~ 12-
l VII. Conclu~ions
3 The application of the above-described technologies has
4 the potential to produce types of plants and animals which
cannot be produced through classical genetics. For example,
6 animals can be produced which suffer from human diseases
7 (such as AIDS, diabetes, cancer, etc.), and may be valuable
8 in elucidating therapies for such diseases. Chimeric and
9 transgenic plants and animals have substan~i~l use as probes
of natural gene expression. When applied to liv~stock and
ll food crops, the technologies have the potential of yielding
12 improved food, fiber, etc.
13 Despite the successes of the above-described techniques,
14 a method for producing chimeric or transgenic plants and
animals which was less mutagenic, and which would permit
16 defined, specific, and delicate manipulation of the inserted
~7 gene sequence at a specific chromosomal location would be
18 highly desirable. ~
19 .': ,
21 ~R~EF DE~8CRIPTION OF T~IE FIGIJR.E9
2 2 :~
23 Figure l illustrates t~e use of replacement vectors and
24 insertion vectors in gene targeting. Figure lA is a
diagrammatical representation of the use of a replacement ~
26 vector in yene targeting; Figure lB illustrates the use of ~ -
27 an insertion vector to produce subtle mutations in a desired ~ .
28 gene sequence.
29 Figure 2 is a diagra~matical representation of a DNA
molecule which has a region of heterology located at a
31 proposed insertion site. Figure 2A shows a construct with a
32 2 kb region of heterology. Figure 2B shows a construct with
33 a 26 base long region of he~erology which has been
'
;.
';-
: . .. , , : - . . , . : . . ::- . . : : :
. '
: .. . :- ,, .
- , ~
., : . ,
WO91/19796 PCT/US91/04006
r r t ~:
--13 ~
l linearized at the center of the region of heterology.
2 Figure 2C shows a construct with a region of heterology
3 located internal to the re~ion of homology at which
4 recombination is desired. In Figure 2c, the normal BamHI
site of the vector has been changed to an NheI site and the
6 normal EcoRI site of the vector has been changed to a BamHI
7 site. The vector is linearized with XhoI
8 Figure 3 is a diagrammatical representation of the
9 mechanism through which a "humanized" gene may be introduced
into a chromosomal gene sequence in a one step ~ethod.
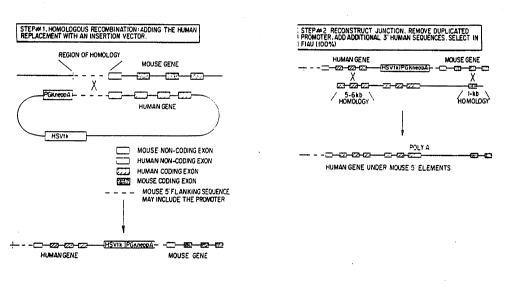
11 Figure 4 is a diagrammatical representation of the
12 mechanism through which a large qene may be introduced into
13 a chromosomal gene sequence so as to place the gene under
14 the transcriptional control of a heterologous promoter (for
example, to place a human gene under the control of a mouse
16 gene). The first step is additive and the !second is a
17 replacement event. Fiyure 4A shows the first step of the
18 process; Figure 4B shows the second step of the process.
l9 The repair recombination event may be configured to remove
all of the mouse coding exons if desired. ~ !`
21 ~ Figure 5 is a diagramatical representation of the use of ~-
22 a positive selection/ negativle selection "cassette" to ;
23 introduce subtle mutations into a chromosome.
24 Figure 6 is a diagrammatical representation of a multi-
step method (~igures 6A-6E) for introducing small or large
26 desired gene sequences into a contiguous region o$ a cellis
27 genome. The figure illustrates a vector capable of
28 facilitating the sequential addition of overlapping clones
29 to construc~ a large locus. Every step i9 selectable. ~;
Subsequent additions may be made by returning to steps 4 and
31 5 as ~any times as required, selecting for insertion in HAT
32 medium, and for repair in media supplemented with 6
: . -: .
~: ~ . . ,: ,. . . .
.: ~ :. .
': . ,.,~ , . ' - , ;: ' ~
wu~ J~ PCT/~S91/0~006
1 thioguanine. This procedure may also be accomplished at the
2 other end of the locus if required.
3 Figure 7 is a diagrammatical reprPsentation of the
4 vectors used in a co~electroporation experiment to mutate
the hprt gene.
6 Figure 8 illustrates the predicted structure ~f the _~E~
7 gene following homologous recombination of the IV6.8 vector.
8 ER is the predicted size fragment indicative of the
9 homologous recombination event. End, ~ is the endogenous
fragment, duplicated by the recombination event. End is the
11 predicted flanking fragment detected by the partial cDNA
12 probe used in these experiments.
13 Figure 9 shows the reversion of homologous recombinants
14 generated with insertion vectors.
Figure 10 illustrates the use of Poly A selection as a
16 means for selecting homologous recombination events. ~
17 Figure 11 illustrates the use of the. invention to -
18 introduce insertions into ~he sequence of a desired gene of
19 a cell. Figure llA is a diagram of the c-src locus showing
relevant restriction sites (E=EcoRI; N~coI; X=XhoI;
21 H=HindIII; ~=BamHI; Nh=NheI). Figure llB illu~trates the ~-
22 src 14 vector used to introduce mutations into the c-src
23 locus; Figure llC illustrates the subtle mutation introduced
24 through the use of this vector. ~-
Figure 12 illustrates the use of the invention to
26 introduce substitutions into the ~equence of a desired gene
27 of a cell. Figure 12A is a diagram of the c-src locus
28 showing relevant restriction sites (E=EcoRI; N=NcoI; X=XhoI;
29 H=HindIII; B=9amHI; Nh-NheI). Figure 12B illustrates the
src 33 vector used to introduce mutations into the c-src
31 locus; Figure 12C illustrates the subtle mutatio~ introduced
32 through the use of this vector.
- .. , . .. , ,. ... , , ... . . ;
.. . . . ~
. . .: . : . . : : . . . : :
. ::. :. , - - ~. . . . - , : .
, . :,- , . .
' : . , : :: . . , -
:: ' :. . - - . .
WO 91/1979~ PCl/l S91/041~06
--15-- ~ r ~r 4
1 Figure 13 illustrates a comparison between targeted and
2 random recombinatlonal events. In a random recombinational
3 event, although concatemers can e~cise duplications, one
4 copy of the vector must remain in the genome. In contrast,
in a targeted recombinational event, all sequences, except
6 the desired sequence is excised from the genome.
8 ~MM~Ry OF T~E INYENTION:
10The present invention provides a method for obtaining a
11desired animal or non-fungal plant cell which contains a
12predefined, specific and desired alteration in its g~nome.
13~he invention further pertains to the non-human animals and
14plants which may be produced from such cells. The invention
15additionally pertains to the use of such non-human animals
16and plants, and their progeny in research, medicine, and
I7agriculture.
~8In detail, the invention provides a method for obtaining
19a desired animal or non-fungal plant cell which contains a
20desired non-selectable gene sequence inserted within a
21predetermined gene sequence of the cell's genome, which
2~method comprises:
23A. incubating a prPcurr,or cell with a DNA molecule
24containing the desired non-selectable gene sequence, wherein
25the DN~ molecule additionally contains two regions of
26homology which flank the desired gene sequence, and which
27are sufficient to permit the desired gene seguence to
28undergo homologous recombination with the predetermined gene
29sequence of the genome of the precursor cell;
3~~. causing the DNA molecule to be introduced into
31the precursor cell;
32C. permitting the introduced DNA molecule to
33undergo homologous recombination with the predetermined gene
:.,: - ,. :
,, . . -. - . . ....
. ~
:,...:
WO91/~9796 PCT/I S91/04006
~_ ~ r ~ ~
--16--
1 sequence of the genome of the precursor cell t3 thereby
2 produce the desired cell wherein the desired non-selectable
~ gene sequence has been inserted into the predeter~ined gene
4 sequence; and
D. recoYering the desired cell.
6 The invention further includes the embodiments of the
7 above-described method wherein the DNA molecule contains a
8 detectable marker gene sequence, and/or wherein the DNA
9 molecule is introduced into the precursor cell by subjecting
the precursor oell and the DNA molecule to electroporation
11 (especially wherein in step ~, the precursor cell is
12 si~ultaneously subjected ~o electroporation with a second
13 DNA molecule, the second DNA molecule containing a
14 detectable marker gene sequence).
The invention further includes the embodiments of the
16 above-described method wherein the desired cell is a non~
17 fungal plant cell, a somatic animal cell (especially one
18 selected from the group consisting of a chicken, a mouse, a
19 rat, a hamster, a rabbit, a sheep, a goat, a fish, a pig, a
cow or bull, a non-human primate and a human), a pluripotent
21 animal cell (especially one selected from the group
22 consisting of a chicken, a mouse, a rat, a hamster, a
23 rabbit, a sheep, a goat, a fish, a pig, a cow or bull, and
24 a non-human primate). The invention includes with the
embodiment wherein the pluripotent cell is an embryonic stem
26 cell.
27 The invention also includes the embodi~ents of the
28 above described methods wherein the desired gene sequence is
29 subst ntially homologous to the predetermined gene ee~uence
of the precursor cell and/or wherein the desired gene
31 sequence is an analog (and especially a human analog) of the
32 predetermined sequence of the precursor cell.
.: . . - - . . , . . ~ ~ :
-;: : :
: : :
- .
. .
WO91/19796 p~r/~s9l/o4oo6
-17- ~ c i ~
1The lnvention also includes the e~bodiment wherein the
2desired gene sequence encodes a protein selected from the
3group consisti~g of: a hormone, an immunoglobulin, a
4receptor molecule, a ligand of a rec2ptor molecule, and an
5enzyme.
6The invention also includes a non-fungal plant cell
7which contains an introduced recombinant DNA molecule
8containing a desired gene sequence, the desired gene
9sequence being flanked by regions of homology which are
10su~fi~ient to permit the desired gene sequence to undergo
11homologous recombination with a predetermined gene sequence
12of the genome of the cell.
13The invention also includes a non-human animal cell
14which contains an introduced recombinant DNA molecule
15containing a desired gene sequence, the desired gene
16sequence being flanked by regions of homolo~y which are
17sufficient to permit the desired gene sequence to u~dergo
18homologous recombination with a predetermined gene sequence
19of the genome of the cell.
20The invention also includes 1:he desired cell produced by
21any of the above-described methods.
22The invention also includes a non-human animal
23containing a cell derived~from the above-described desired -~
24cell, or a descendant thereof, wherein the animal is either
25a chimeric or a transgenic animal, and particularly includes
26the embodiment wherein the non-h~man animal and the desired
27cell are of the same species, and wherein the species is ;
28selected from the group consisting of: a chicken, a mouse,
29a rat, a ha~ster, a rabbit, a sheep, a goat, a fis~, a pig,
30a cow or bull, and a non-human primate.
31The invention also includes a non-fungal plant
32containing a cell derived from the above-described desired
- : . . - .
~ ' ,,
,., . , ' ,'
' ' ~ ' ' :
'', ~ ' .
WOgl/19796 PCT/~591/04~06
-18-
1non-fungal plant cell, wherein said non-fungal plant is
2either a chimeric or a transgenic plant.
~The invention also includes a method of gene therapy
4which comprises introducing to a reclpient in need of such
5therapy, a desired non-selectable gene sequence, the method
6comprising:
7A. providing to the recipient an effective amount
8of a DNA molecule containing the desixed non-selectable gene
9sequence, wherein the D~A molecul~ additionally contains two
10regions of homology which flank the desired gene sequence,
~1and which are sufficient to permit the desired gene seguence
12to undergo homologous recombination with a predetermined
13gene sequence present in a precursor cell of the recipient;
14B. permitting the DNA molecule to be introduced
15into the precursor cell;
16C. permitting the introduced DNA molecule to
17undergo homologous recombination with the predetermined gene
18se~uence of the genome of the precursor cell to thereby
19produce a desired cell wherein the desired non-selectable
20gene sequence has been inserted into the predetermined gene
21sequence; and wherein the presence or expression of the
22introduced gene sequence in the cell of the recipient
23comprises the gene therapy;
24In particular, the invention includes the embodiments of
25the above-stated method wherein the recipient is a non-
26fungal plant, or a human or a non-human animal (particularly
27a non-human animal is selected from the group consisting of~
28a chicken, a mouse, a rat~ a hamster, a rabbit, a sheepj a
29goat, a fish, a pig, a cow or bull, a non-hu~an primate and
30a human).
..... ,..... ..... .,, . ... ,
:: : : .: . ~ .. : :
~ : , . . . ,: , . ~ .
, . . . . .
,: , . ~, . . :, :: . ~.. ; ~ . .,
:. - . , ~: .
WO 91/19796 PCI/~S9~/04006
r r J ~c
1The lnvention also provides a method for obtaining a
2desired animal or non-fungal plant cell which contains a
3desired non-selectable gene sequence inserted within a
4predetermined gene sequence of the cell's geno~e, which
5method comprises:
6A. incubating a precursor cell under non-selective
7culture conditions, or under a first set of selective
8culture conditions, with a DNA molecule containing:
9i) the desired -non-selectable gene seguence,
10wherein the DNA molecule additionally contains
11two regions of homology which flank the desired
12gene sequence, and which are sufficient to
13permit the desired gene sequence to un~ergo
14homologous recombination with the predeterminçd
15gene sequence of the genome of the precursor
16cell; and
17ii~ a selectable gene sequence whose presence or
18expression in the cell can be selected for by
19cuituring the cells under the first set of
20selective culture conditions, and whose
21presence or expression in the cell can be
22selected against by culturing the cells under
23a second set of selective culture conditions;
24B. permitting the DNA molecule to be introduced
25into the precursor cell;
26C. permitting the introduced DNA molecule to
27undergo homologous recombination with the predetermined gene
28sequence of the genome of the precursor cell to thereby
29produce the desired cell wherein the desired non-selectable
30gene sequence has been inserted into the predetermined gene
31sequence; and
32D. recovering the desired cell by culturing the
33cell under the first set of selective culture conditions, by
''~
WO91/1979h PCr/~S91/04006
~ t~ 20-
l then permitting the cell to undergo intrachromosomal
2 recombination under non-selective culture conditions, and by
3 then incubating the cell under the second set of selective
4 culture ~onditions.
The invention also includes the embodiment wherein the
6 cell is deficient in an HPRT, APRT, or TK enzyme, and
7 wherein the sel~ctable gene sequence expresses an active
8 HPRT, APRT, or TX enzyme, and wherein the first set of
9 selective culture conditions comprises incubation of the
cell under conditions in which ~he presence of an active
11 HPRT, APRT, or ~K enzyme in the cell is required for growth,
12 and wherein the second set of selective culture conditions
13 comprises incub~tion of the cell under conditions in which
14 th~ absence of an active HPRT, APRT, or TK enzyme in the ;~
cell is re~uired for growth.
16
17
18 D~8CRIiTION OF THE PREFER~ED EMBODIMENTS:
19 :
The present invention concerns a method for introducing
21 DNA into the genome of a recipi1ent plant or animal cell.
22 The method may be used to introduce such DNA into germ line
23 cells of animals (especially, rodents (i.e. mouse, rat,
24 hamster, etc.), rabbits, sh ep, goats, fish, pigs, cattle
and non-human primates) in order to produce chimeric or
26 transgenic animals. The methods may also be used to
2i introduce DNA into plant cells which can then be manipulated
`28 in order to produce chimeric or transgenic plants.
29 Alternatively, the method may be used to zlter the
s~matic cells of an animal (including humans) or a plant.
31 The plants and plant cells which may be manipulated through
32 application of the disclosed method include all
33 multicellular, higher (i.e. non-fungal or non-yeast) plants.
~.... . ... . . . .
.. . . . .
. .
' ~
W ~ 91/19796 PCT/~591/04006
-2l-
~ ~ ~J~. r
1 I. ~omologou~ Recombination
3The present invention provides a method for introducing
4a desired gene sequence into a plant or animal cell. Thus,
5it is capable of producing chimeric or transgenic plants and
6animals having defined, and specific, gene alterations.
7An understanding of the process of homologous
8recombination (Watson, J.D., In: Molecular ~loloqY of the
9Gene, 3rd Ed., W.A. Benj2min, Inc., Menlo Park, CA ~977),
10which reference is incorporated herein by reference) is
11desirable in order to fully appreciate the present
12invention.
13In brief, homologous recombination is a well-studied
14natural cellular process which results in the scission of
15two nucleic acid molecules having identical or substantially
16similar sequences (i.e. "homologousl'), and the ligation of
17the two molecules such that sne region of each initially
18present molecule is now ligatecl to a region of the other
19initially present molecule (';edivy, J.M., Bio-Technol.
206:1192-1196 (1988), which reference is incorporated herein
21by reference).
22Homologous reco~bination is, thus, a sequence specific
23process by which cèlls can transfer a "region" of DNA from
24one DNA molecule to another. As used herein, a "region" of
25DNA is intended to generally refer to any nucleic acid
26molecule. The region may be of any length from a single
27base to a sub-~tantial fragment of a chromosome.
28For homologous recombination to occur between two DNA
, ~ .
29molecules, the molecules must possess a "region of homology"
30with respect to one another. Such a region of homology must -
31be at least two base pairs long. Two DNA ~olecules possess
32such a "règion of homology" when one contains a region whose
;
'" ',
.
~';
.
WO91/1979~ PCI/I S91/04006
2 2 -
~ ' ' ' .
1 sequence is so similar to a region in the second molecule
2 that homologous recombination can occur.
3 Recombination is catalyzed by enzymes which are
4 naturally present in both prokaryotic and eukaryotic cells.
S The transfer of a region of DNA may be envisioned as
6 occurring through a multi-step process.
7 If either of the two participant molPcules is a circular
8 molecule, then the above recombination event results in the -
9 integration of the circular molecule into the other
participant.
ll Importantly, if a partic~lar region is flanked by
12 regions of homology ~which may be the same, but are
13 preferably different), then two xecombinational events may
14 occur, and result in the exchange of a region of DNA between
two DNA molecules. Recombination may be "reciprocal," and
16 thus results in an exchange of DNA regions between two
17 recombining DNA molecules. Alternatively, it may be "non~
18 reciprocal," (also referred to as "gene conversion") and
19 result in both recombining nucleic acid molecules having the
same nucleotide sequence. There are no constraints
21 regarding the size or sequence of the reyion which is
22 exchanged in a two~event recombinational exchange.
23 The frequency of recombination between two D~A molecules
24 may be enhanced by treating the introduced DNA with agents
which stimulate recombination. Examples of such agents
26 include tximethylpsoralen, W light, etc. ~;
27
'
.: , , - .
: . , - .
~: ' . . - `-, : .
", : ,,
WO 91/t97~6 PCr/l~S91/04006
- 2 3 ~ r
1II. Pro~uction of Chimeric and ~rA~qgenic Animal~:
2 Ge~e Targeting Method3
4 One approach to producing animals having defined and
specific genetic alterations has used homologous
6 recom~ination to control the site of integration of an
7 introduced marker gene sequence in tumor cells and in
8 fusions between diploid human fibroblast and tPtraploid
9 mouse erythroleukemia cells (Smithi~s, O. et al., Nature
317:230-234 (1985)).
11 This approach was further exploited by Thomas, K. R.,
12 and co-workers, who described a general method, known as
13 "gene targeting," for targeting mutations to a preselected,
14 desired gene sequence of an ES cell in order to produce a
transgenic animal (Mansour, S.L. et al., Nature 336:348-352
16 (1988); Capecchi, M.R. Trends Genet. 5:70-76 (1989);
17 Capecchi, M.R. et al., In: Current_ communications in
18 Molecular Biolo~v, Capecchi, M.R. (ed.), Cold Spring Harbor
19 Press, Cold Spring Harbor, NY (1989), pp. 45-52, which
references are incorporated herein by reference).
21 Gene targeting has been used to produce chimeric and
22 transgenic mice in which an nl~tII gene has been inserted
23 into the ~2-microglobulin locus (Xoller, B.H. et al., Proc.
24 Natl. Acad. Sci._(U.S.A.) 86;8932-8935 (1989); Zijlstra, ~.
et al., N_ture 342:435-438 (1989); Zijlstra, M. et al.,
26 Nature 344:742-746 (1989); DeChiaba et al., ature 345:78-80
27 (1990)j. Similar experimen~s have enabled the production of
28 chimeric and transgenic ani~als having a c-abl ~ene which
29 has been disrupted by the insertion of an n~tII gene
(Schwartz~erg, P.L. et al., Scie~ce ~ 799-803 ~19a9)~
31 The technigue has been used to produce chimeric mice in
32 which the en-2 gene has been disrupted by th~ insertion of
.. . .. . . . . .
... ..
~::. .: ::: .:
, ,.:-: . . . ~ . ~. : :
: .: -
- : ~ . . :: .
?~ - ~
W O 91/19796 PCT/~'S91/04006
~t~ -24-
1 an nE~II gene (Joyner, A~L. et al., Nature 338:153-155
2 (1989)).
3 Gene targeting has also been used to correct an hprt
4 deficiency in an hPrt ES cell line. Cells corrected of the
deficiency were used to produce chimeric animals.
6 5ignificantly, all of the corrected cells exhibited gross
7 disruption of the regions flanking the h~rt locus; all of
8 the cells tested were found to contain at least one copy of
9 the vector used to correct the defici~ncy, integrated at the
hDrt locus (Thompson, S. et al., Cell 56:313-321 (1989);
ll Xoller, B.H. et al., Proc. Natl. Acad. sci. (U.S.A.)
12 86:8927-8931 (1989)).
13 In order to utilize the "gene targeting" method, the
14 gene of interest must have been previously cloned, and the
intron-exon boundaries determined. The method results in
16 the insertion of a marker gene (i.e. the n~tII gene) into a
17 translated region of a particular gene of interest. Thus,
~8 use of the gene targeting method results in the gross
l9 destruction of the gene of interest.
Recently, chimeric mice carrying the homeobox hox 1.1
21 allele have been produced using a modification of the gene
22 targeting method (Zimmer, A. et: al., Nature 338:150-154
23 (1989). In this modification, the integration of vector
24 sequences was avoided by microinjecting ES cells with linear
DNA containing only a portion of the hox 1.1 allele, without
26 any accompanying vector sequences. The DNA was found to
27 cause the gene conversion of the cellular hox allele.
28 Setection was not used to facilitate the recovery of the
29 "converted" ES cells, which were identified using the
polymerase chain reaction ("PCR"). Approximately 50~ o~
31 cells which had been clonally purified from "converted"
32 cells were ~ound to contain the introduced hox 1.1 allele,
33 suggesting to Zimmer, A. et al. either chromosomal
i, .
.
: : .
WO91/197g6 PCT/~S9~/04006
-25
1 instability or conta~ination o sa.ple. None of the
2 chimeric mice were found to be able to transmit the
3 "converted" gene to their progeny (Zimmer, A. et al., In:
4 Current Communications in Molecular ~ioloav, Capecchi, M.R.
(ed.), Cold Spring Harbor Press, Cold Spring Harbor, NY
6 (1989), pp. 53-58).
7 The use of the gene targeting method is illustrated in
8 Figure lA. In that figure, a gene construct is produced in
9 which the nptII gene is inserted into an exon (designated
region "3") of a sequence of the hprt gene. The construct
11 is then permitted to undergo recombination with the hPrt
12 gene of a cell. Such reco~bination results in the
13 replacement of the exon 3 sequence of the cell with the
14 disrupted exon 3 - n~tII sequence of the construct.
Significantly, as illustrated in Fiyure lA, the use of gene
16 targeting to alter a gene of a cell results in the formation
17 of a gross alteration in the sequence of that gene. As
18 indicated in Figure lA, the efficiency of gene targeting is
19 approximately 1/300.
~ 0 . .~
21 III. Pro~uction of Chimeric ~nd Tr~n~genic An~m~
22 ~se of Insertion Vectoxs
23
24 In contrast to the above-described methods, the present
invention is capable of producing subtle, precise, and
26 predetermined mutations in the sequence of a desired gene of ~;
27 a cell. The present invention has several embodiments, the
28 simplest of which is illustrated in Figure lB.
29 As shown in Figure lB, an insertion ~ector is used to -~
~utate the nucleotide sequence o~ the hprt gene. The use of ~ `
31 this vector type in combination with a second selectable
32 reversion event prevents the disruption of the chromosome by -
3~ the n~tII gene or by the vector sequences. Thus, gross
~. -.
-., .. , . . .,. .. , .. . . . - .: . :. , .
,. . : -
:: : .. . - -: ,: . : . -
. ::.: : . .
.. . . . ,. .:. ~
WO91/19796 PCT/~'S91/04006
-26-
l distortions of the recipient chromosome are avoided by the
2 present invention. Moreover, the efficiency of the gene
3 targeting was substantially improved (i.e. l/32 as opposed
4 to l/300).
The DNA molecule(s) which are to be introduced into the
6 recipient cell preferably contains a region of homology with
7 a region of the cellular genome. In a preferred embodiment,
8 the DNA molecule will con~ain two regions of homology with
9 the genome (both chromosomal and episomal) of the
pluripotent cell. These regions of homology will preferably
ll flank a "desired gene sequence" whose incorporation into the
12 cellular genome is desired. As stated above, the regions of
13 homology may be of any size greater than two bases long.
14 Most preferably, the regions of homology will be greater
than lO bases long.
16 The DN~ molecule(s) may be single stranded, but are
17 preferably dou~le stranded. The DNA molecule(s~ may be
18 introduced to the cell as one or more RNA molecules which
l9 ~ay be converted to DNA by reverse transcriptase or by other
means. Preferably, the DNA molecule will be double stranded
21 linear molecule. In the best mode fsr conducting the
22 invention, suah a molecule is obtained by cleaving a closed
23 covalent circular molecule to form a linear molecule.
24 Preferably, a restriction endonuclease capable of cleaving
the molecule at a single site to produce either a blunt end
26 or staggered end linear molecule is employed. ~ost
27 preferably, the nucleotides on each side of this restriction
28 site will comprise at least a portion of the preferred two
29 regions of homology betw~en the DNA molecule being
introduced and the cellular genome.
31 The invention thus provides a method for introduciny the
32 "desired gene sequence" into the genome of an animal or
33 plant at a specific chromosomal location. The "desired gene
. .:
:
: : : : : . :: : :
: . . . .: , .~
:: ~ : .
WO91/19796 PCT/~'S91/04006
2r..~
l seque~ce" may be of any length, and have an~ nucleotide
2 sequence. It may comprise one or more qene sequences which
3 encode complete proteins, fragments of such gene sequences,
4 regulatory sequences, etc. Significantly, the desired gene
sequence may differ only slightly from a native yene of the
6 recipient cell (for example, it may contain single, or
multiple base alterations, insertions or deletions relative
8 to the native gene). The use of such desired gene sequences
9 will permit one to create subtle and precise changes in the
genome of the recipient cell. Thus, the present invention
ll provides a means for manipulating and modulatinq gene
12 expression and regulation. ;
13 In particular, the invention provides a mean for
14 manipulating and modulating gene expression and protein
structure through the replacement of a gene sequence with a
16 "non-selectable" "desired gene sequence." A gene sequence
17 is ncn-selectable if its presence or expression in a -;
18 recipient cell provides no survival advantage to the cell
l9 under the culturing conditions employed. Thus, by
definition, one cannot select for cells which have received
21 a "non-selectable" gene sequence. In contrast, a "dominant"
22 gene sequence is one which can under certain circumstances
23 provide a survival advantage to a recipient cell. The
2~ neomycin resistance conferred by the n~tII gene is a
survival advantage to a cell cultured in the presence of
26 neomycin or G418. The n~tII gene is thus a dominant, rather
27 than a non-selectable gene sequence.
28 In particular, the invention permits the replacement of
29 a gene sequence which is present in the recipient cell with
an "analog" sequence. A sequence is said to ~e an analog o~
3l another sequence if the two sequences are su~stantially
32 similar in sequence, but have minor changes in sequence ~;-
33 corresponding to single base substitutions, deletions, or
; '
1'
. . ~ , :... .: .
- ', , , , ' :' : : ~ .,: ' ": . ' :'
' . ' "' ' . : , '', ` " . : ' :, ,
WO 91/19796 PCl/~S91/04006
~r ~ - r ~ 2 8 -
O ~
1 insertions with respect to one anothe-, or if they possess
2 "minor" multiple ~ase alt~rations. Such alterations are
3 intended to exclude insertions of dominant selectable marker
4 genes.
When the desired gene sequence, flanked by regions of
6 homology with the recipient cell, is introduced into the
7 r~cipient cell as a linear double stranded molecule, whose
8 termini correspond to the regions of homology, a single
9 recombination event with the cell's genome will occur in
10 approximately 5% of the transfected cells. Such a single
11 recombinational event will lead to the integration of the
12 entire linear molecule into the genome of the recipient
13 cell.
14 The structure generated by the integration of the lineàr
15 molecule will undergo a subsequent, second recombinational
16 event (approximately 105 - 10-7 per cell generation). This
17 second recombinational event will result in the elimination
18 of all DNA except for the flanking regions of homology, and
19 the desired DNA sequence from the integrated structure.
Thus, the consequence of the second recombinational event
21 is to replace the DNA sequence which is normally present
22 between the flanking regions of homology in the cell's
23 genome, with the desired DNA sequence, and to eliminate the
24 instability of gene replacement.
The DNA molecule containing the desired gene sequence
26 may be introduced into the pluripotent cell by any method
27 which will permit the in~roduced molecule to undergo
28 recombination at its regions of homology. Some methods,
29 suc~ as direct microinjection, or calcium phosphate
30 transformation, may cause the introduced molecule ts form
31 concatemers upon integration. These concatemers may resolve
32 themselves to form non-concatemeric integration structures.
33 Since the presence of concatemers is not desired, methods
: . . ,.................... .. .. . ~ . . :
~ , . . . : . - :
'
. :
. . ... ,.. : ~.: : ... : .. : :.. ~....... :
WO91/1~79~ PCT/~'S91/04006
-29-
1 which produce them are no~ preferred. In a preferred
2 embodiment, the DNA is introduced by electroporation
3 (Toneguzzo, F. et al., Nucleic Acids Res. 16:5515-5532
4 (1988); Quillet, A. et al., J. Immunol. 141:17-20 (1988);
Machy, P. et al., Proc. Natl. Acad. Sci. (U.S.A.) 85:8027-
6 8031 (1988); all of whic~ references are incorporated herein
7 by reference). -
8 After permitting the introduction of the DNA
9 molecule(s), the cells are cultured under conventional
conditions, as are known in the art. -~
ll In order to facilitate the recovPry of those cells which
12 have received the DNA molecule containing the desired g~ne
13 sequence, it is preferable to introduce the DNA containing
14 the desired gene sequence in combination with a second gene ~-
sequence which would contain a detectable marker gene
16 sequence. For the purposes of the present invention, any
17 gene sequence whose presence in a cell permits one to ~;
18 recognize and clonally isolate the cell may be employed as
l9 a detectable marker gene sequenc:e.
In one embodiment, the presence of the detectable marker
21 sequence in a recipient cell is recognized by hybridization,
22 by detection of radiolabelled nucleotides, or by other
23 assays of detection which ~o not require the expression of
24 ~he detectable marker sequence. Preferably, such sequences
are detected using PCR (Mullis, K. et al., Cold S~rinq
26 Harbor SvmP. Ouant. Biol. 51:263-273 (1986); Erlich H. et
27 al., EP 50,424; EP 84,796, EP 258,017, EP 237,362; Mullis,
28 K., EP 201,18~; Mullis X. et al., US 4,683,202; Erlich,
29 ~5 4,582,788; and Saiki, R. et al., US 4,683,194), which
references are incorporated ~erein by reference).
31 PCR achieves the amplification of a specific nucleic
32 acid sequence using two oligonucleotide primers
33 complementary to regions of the sequence to be amplified.
:
' ~ :
::. . :,:
. . ;~ j ..
WO91/1979~ PCT/~'S91/04006
-30-
1 Extensio~ products incorporating the primers then become
2 templates for subsequent repllcation steps. PCR provides a
3 method for selectively increasing the concentration of a
4 nucleic acid molecule having a particular sequence ev~n when
that molecule has not bee~ previously purified and is
6 present only in a single copy in a particular sample. The
7 method can be used to amplify either single or double
8 stranded DNA.
9 Most preferably, however, the detectable marker gene
sequence will be expressed in the recipient cell, and will
11 result in a selectable phenotype. Examples of ~uch
12 preferred detectable gene sequences include the h~rt gene
13 (Littlefield, J.W., Science 145:709-710 (1964), herein
14 incorporated by reference), a xanthine-guanine
phosphoribosyltransferase (q~t) gene, or an adenosine
16 phosphoribosyltransferase (aPrt) ge~e (Sambrook et_al., In:
17 Molecular Cl onlnq A LaboratorY Manual, 2nd. Ed., Cold Spring
18 Harbor Laboratory Press, NY (1989), herein incorporated by
19 reference), a tk gene (i.e. thymidine kinase gene) and
especially the tk gene of herpes simplex virus (Giphart-
21 Gassl2r, M. et al., Mutat. Res 214:223-232 (1989) herein
22 incorporated by reference), the nPtII gene (Thomas, K.R. et
23 al., Cell 51:503-512 (198~); Mansour, S.L. et al., Nature
24 336:348-352 (1988), both references herein incorporated by
reference), or other genes which confer resistance to amino
26 acid or nucleoside analogues, or antibiotics, etc. Examples
27 o~ such genes include gene sequences which encode enzymes
28 such as dihydrofolate reductase (DHFR) enzyme, adenosine
29 deaminase ~ADA), asparagine synthetase (AS), hygromycin B
phosphotransferase, or a CAD enzyme (carbamyl phosphate
31 synthetase, asp rtate transcarbamylase, and dihydroorotase)
32 (Sambrook et al., In: Molecular Cloninq A Laboratorv Manual,
: . , , . :- . . : ~ :
: . , -: : :: . : , .:. , ::: , :". , , ~
, . ' ' ' ~ ' . . . :, :
WO91/l9796 PCT/~S91/04006
-31~
1 2nd. Ed., Cold Spring Harbor Laboratory Press, NY (1989),
2 herein incorporated by reference).
3 Cells that do not contain an active thymidine kinase
4 (TK) enzyme, a hypoxanthine-phophoribosyltransferase (HPRT)
enzyme, a xanthine-guanine phosphoribosyltransferase (XGPRT)
6 enzyme, or an adenosine phosphoribosyltransferase (APRT)
7 enzyme, are unable to grow in medium containing
8 hypoxanthine, aminopterin, and/or mycophenolic acid (and
9 preferably adenine, xanthine, and/or thymidine), and
thymidine, but are able to grow in medium containing
11 nucleoside analogs such as 5-bromodeoxyuridine, 6~
12 thioguanine, 8-azapurine, etc. (Littlefield, J.W., Science
13 145:709-710 (1964); Sambrook et al., In: Molecular Clonina
14 A Laboratorv Manual, 2nd. Ed., Cold Spring Harbor Laboratory
Press, NY (1989)).
16 Conversely, cells that do con~ain such active enzymes
17 are able to grow in such medium, but are unable to grow in
18 medium containing nucleoside analogs such as 5-
19 bromodeoxyuridine, 6-thioguanine, 8-azapurine, etc.
(Sambrook et al., In: Molecular Cloninq A Laporatorv Manual,
21 2nd. Ed., Cold Spring Harbor Laboratory Press, NY (1989)).
22 Cells expressing active thymidine kinase are able to
23 grow in media containing HATG, but are unable to grow in
24 media containing nucleoside analogues such as 5-azacytidine
(Giphart-Gassler, M. et al., Mutat. Bes. 214:223-232
26 (1989)). Cells containing an active HSV-tk gene are
27 incapable of growing in the presence of gangcylovir or
28 similar agents.
29 The detectable marker gene may be any gene which càn
complement for a racognizable cellular deficiency. Thus,
31 for example, the gene for HPRT could be use~ as the
32 detectable marker gene sequence when employing cells lacking
33 HPRT activity. Thus, this gene is an example of a gene
.
.
.
w o 91/19796 PCT/~S91/04006
-32-
1 whose expression product may be used to select mutant cells,
2 or to "negatively select" for cells which express this gene
3 product.
4 The n~tII gene (Southern, P.J., et al., J. Molec. A~l.
S Genet. 1:327-341 (1982); Smithies, O. et al., ~ature
6 317:230-234 (1985), which references are incorporated herein
7 by reference) is the most preferred detectable marker gene
8 sequence. Constructs whi~h contain both an nPtII gene and
9 either a tk gen~ or an hprt gene are especially preferred.
11 A. ~ e of a ~ingle DNA ~olecule cont~i~ing Both the
12 Detectable M~r~r 8equence and the Desired Gene
13 ~eque~ce
14
1~ In a first preferred embodiment, the detectable marker
16 gene sequence, flanked by- the regions of homology, is
17 provided to the recipient cells on the same DNA molecule
18 which contains the desired genP sequence. As discussed
19 previously, it is preferred that this DNA molecule be a
linear molecule.
21 After selection for cells which have incorporated the
22 desired DNA molecule (for example by selection for G418
23 resistant cells when the detectable marker gene sequence is
24 an expressible nptII gene sequence), the cells are cultured,
and the presence of the introduced DNA molecule is confirmed
26 as described above. Approximately 107 cells are cultured and
27 screened for cells which have undergone the second
28 reco~binational event (discussed above) resulting in the
29 replacement of a native se~uence (i.e. a gene sequence
which is normally and naturally present in the recipient
31 cell) with the desired gene se~uence.
32 Any of a variety of methods may be used to identify
33 cells which have undergone the second recombinational event.
.
. ~ , .
wvsl/19796 PCrt~'S9l/04006
~.,1`, , ~ F 7 1
-33-
1 Direct screening of clones, use of PcR, use of hybridization
2 probes, etc., may all be employed for this purpose. In a
3 preferred embodiment, the DNA molecule will, in addition to
4 the desired gene sequence, the flanking regions of homology
and the detectable marker gene sequence, contain an
6 additional gene sequence which will permit the selection or
7 recognition of cells which have undergone the second
8 recombinational event. This additional gene sequence will
9 be excised from the cell's genome as a direct consequence of
the second recombinational event. Thus, gene sequences
11 which are sui~able for this purpose include any gene
1~ sequence whose loss from a cell can be detected or selected
13 for. Examples of such "negative selection" gene sequences
14 include the hPrt gene, and the tk gene (especially the tk
gene of herpes simplex virus).
16 In the first preferred embodiment, the frequency of the
17 second reco~binational event is approximately 105. However,
18 the use of a "negative selection" gene sequence permits one
19 to identify such recombinant cells at a frequency of
approximately 100%.
21 As illustrated in Figure 2, the DNA molecule may have a
22 region of heterology located at the proposed insertion site.
23 Insertion of such a vector permits one to select for
24 recombinants which have recombined at the insertion site
(and not at other potential sites). If recombina~ion occurs
26 at the desired insertion site, it will lead to the loss of
27 the sequence o~ heterology located at the proposed insertion
28 site o~ the DNA molecule (~SVtk, for exam~le, in Figure 2A).
29 Insertions which result from other recombinational events
will retain the sequence of heterology. Thus, by e~ploying
31 a region of heterology which encodes ~n assayable gene
32 product, or which can be used as a "negative selectable"
33 =arker, one oan readily determine thet the loous of
'~
-
WO 91/19796 P~ 'S91/04006
.
~ t~ 34-
l insertlon of the recipient cell contains the precise
2 sequence desired. As indicated in Figure 2A), the
3 efficiency of such a vector is approximately 1/197.
4 The region of heterology which may be introduced at the
insertion site of t~e DNA molecule may be either short tfor
6 example, 26 base pairs, Figure 2B) or of substantial size
7 (or example, 2 kb, Figure 2A). The site of linearization
8 may be 5', 3', or within the region of heterology. When the
9 site of linearization is within the region of heterology,
the efficiency of gene targeting is l/63.
11 As shown in Figure 2c, the region of heterology may be
12 located at a site internal to the region of homology where
13 the desired recombination shall occur. Such a construct can
14 be used when one desires to introduce a subtle mutation into
i5 a locus of the cellular gene at a site other than that of
16 the site of desired recombination~
17
18 B. n3e of a Diff~re~t DNA ~olecules to Pro~i~e the
19 DetectAble Marker 8egue~e and t~e Desired Gene
8equence
21
22 In a second preferred embodiI~ent, the detectable marker
23 gene sequence, flanked by the regions of homology, will be
24 provided to the recipient cell on a different DNA molecule
from that which contains the desired gene sequence. It is
26 preferred that these molecules be linear molecules.
27 When provided on separate DNA molecules, the detectable
~8 marker gene sequence and the desired gene sequence will most
29 preferably be provided to the recipient cell by co~
electroporation, or by other Pqui~alent techniques.
31 After selection of such recipients (preferably through
32 the use of a detectable marker sequence which expresses the
33 nptII gene and thus confers cellular resistance to the
. ., .: :, : . . : : - :- .
, ' ' ' `~ :
WO91/~9796 PCT/~S91/04006
-35~ ~7~ ~
1 antibiotic G418), the cells are gro~n up and screened to
2 confirm the insertion event (preferably using PCR).
3 In the absence of any selection, only one cell in 107
4 would be expected to have the predicted recombinant
structures. If, however, one selects for recipient cells
6 which contain and express a detectable marker sequence (such
7 as the nPtII gene), it is possible to obtain a 103 to 105
8 fold enrichment for cells which have taken up both DNA
9 molecules. Typically, such enrichment enables one to
identify the desired recipient cell (in which the introduced
11 DNA has in~egrated into the cell's genome) by screening only
12 800 -1,500 cells. Such screening is preferably done using
13 PCR, or other equivalent methods. Using such negative
14 selection techniques, one may manipulate the vector copy
number.
16 The two introduced DNA molecules will generally not have
17 integrated into the same site in the genome of the recipient
18 cell. Thus, in some cases, the desired gene sequence will
19 have integrated in a manner so as to replace the native
cellular gene sequence between the flanking regions of
21 homology. The locus of integration of the detectable marker
22 gene is unimportant for the purposes of the present
23 invention, provided it is not genetically linked to the same
24 locus as the desired gene sequence. If desired, however, it
is possible to incorporate a gene sequence capable of
26 negative selection along with the DNA containing the
27 detectable marker sequence. Thus, one can ultimately select
28 for cells which have lost the introduced selectable marker
29 gene sequence DNA.
';" ~
. ~ .
;"` ';" `'
:.. `
.,. ... ~ . : . . . . ~ . .
, ,: .: . .. .
. :, .; : . : ~ : .
WC~I 91/19796 PCr/l ~i91 /0~1006
r~
~ r ~ 3 6--
1C. ~se of Direct Bel~ction to I~entify ~omolog~us
2Recombin~tion Event~
4Although all of the above-described preferred
5embodiments enable the isolation of cells in which one of a
6cell's alleles has been mutated to contain a desired gene
7sequence, each embodiment requires the screening of a
8significant number of candidate cells in order to identify
9the desired recombinant cell. It is, however, possible to
10directly select for the desired recombinant cell by
11employing a variation of the above embodiments. This
12embodiment of the invention is illustrated in Figure 13. In
13the methods illustrated in Figure 13, if the sequence
14located below the asterisk is a neo gene, then only tha
15mutant revertants will be selected if 6-thioguanine and G418
16selection is applied to select for the excision events.
17The method for direct selection of the desired cells
18relies upon the phenotypic difference in targeted and non-
19targeted cells and the use of a single gene which can be
20used for both positive and negative selection.
21Typically, in any homologous recombination experiment
22performed with an insertion vector, three populations o~
23cells will be created. The first class of cells will be
24those which have failed to receive the desired DNA molecule.
25This class will comprise virtually all of the candidate
26cells isolated on completion of the experiment. The second
27cl~ss of cells will be those cells in which the desired gene
28seguence has been incorporated at a random insertion site
29(i.e. a site other than in the gene desired to be mutated).
30Approximately one cell in 103-10~ total cells will be in this
31class. The third class of cells will be those cells in
32which the desired gene sequence has been incorporated by
33homologous recombination into a site in ~he desired gene.
,
' , ~ , . : ,-
. . :
' ..
wo91/l9796 PCT/~S91/04006
g ~
-37-
1 Approximately one cell in l05-106 total cells will be in thls
2 class.
3 In the above-descrlbed embodiments, the cells of the
4 first class (non-transfected cells) can be eliminated by
positive selection, thus necessitating the scxe~ening of only
6 about 1,000 cells in order to identify the desired
7 recombinant oell. In the present embodiment, cells of the
8 third class (homologous recombinants) may be selected from
9 the cells of the second class (random insertions) if a
phenotypic difference exists between the cells of the two
11 classes.
12 Since random integration sites are likely to be
13 concatemeric with few single copy clones (depending upon the
14 DNA concentration with which the cells were transfected?,
such inteyration events are inherently unstable. Thus, such
16 concatemeric constructs will typically undergo intrachromo-
17 somal recombination. Such reco~bination will always leave
18 one intact copy of the vector in the genome. Thus, all
19 random insertion events may be negatively selec~ed from the
population if a negatively selectable marker is included on
21 the vector.
22 In contrast, cells in which the desired gene sequence
23 has been incorporated into the desired gene by homologous
24 recombination will revert with a relatively high frequency
~approximately 1 in 104-105 per cell division (depending upon
26 the size of the duplicated structure) to produce a mutated
27 desired gene that does not contain vector sequences.
28 There~ore, even if the vector contained a negatively
29 selectable gene sequence, such cells will survive negative
selection, and can be recovered. The majority o~ ho~ologous
31 reco~binant cells do not undergo reversion, and will be
32 eliminated by the negative selection. Thus, the sum of th~
~' , .,
.. . .
: : - :
WO 91/19796 PCI/~591/04006
1 selections will result in the isolation of the desired
2 recombinants.
3 The method comprises incubating a "precursor cell" (i.e.
4 a cell which is to be changed by application of the method
S into the "desired" recombinant cell) under non-selective
6 culture conditions, or under a first s~t of selective
7 culture conditions. A culturing condition (i.e. medium,
8 temperature, etc.) is said to be "non-selective" if it is
9 capable of promoting the growth (or sustaining the
viability) of a precursor cell, a desired cell, and an
11 intermediate cell type ti.e. a cell obtained during the
12 progression of a precursor cell into a desired cell). A
13 culturing condition is said to be "selective" if it is
14 capable of promoting the growth (or sustaining the
viability) of only certain cells (i.e. those having a
16 particular genotype and which therefore contain a particular
17 gene product in either an active or an inactive form)~
18 Preferred selective culturing conditions thus depend
l9 upon the genotype of the precursor cell. As stated above,
cells that do not contain an active thymidine kinase (TX)
21 enzyme, a hypoxanthine-phophoribosyltransferase (HPRT)
22 enzyme, a xanthine-guanine phosphoribosyltransferase (XGPR~)
23 enzyme, or an adenosine phosphoribosyltransferase (APRT)
24 enzyme, are unable to grow in medium containing
hypoxanthine, aminopterin, and/or mycophenolic acid (and
26 preferably adenine, xanthine, and/or thymidine), and
27 thymidine, but are able to grow in medium containing
28 nucleoside analogs such as 5-bromodeoxyuridine, 6-
29 thioguanine, 8-azapurine, etc. Conversely, cells that do
contain such active enzymes are able to grow in such medium,
31 but are unable to grow in medium containing nucleoside
32 analogs such as S-bromodeoxyuridine, 6-thioguanine, 8-
33 azapurine, etc.
'
' ' ~ . ' ' : ' ' . ~ ' '
,:, . . . ;
: :
WO 91/19796 PCr/ l 'S9 1 /O~l006
~35~
~ r ~ r ~
1 Such incubation is conducted in the presence of a DMA
2 molecule containing a desired non selectable gene sequence.
3 Preferably, the DNA molecule additionally contains two
4 regions of homology which flank this desired gene sequence,
and which are sufficient to permit the desired gene sequence
6 to undergo homologous recombinatio~ with a predetermined
7 genP sequence of the genome of the precursor cell. The DNA
8 molecule additionally contains a selectable gene sequence
9 whose presence or expression in the cell can be selected for
by culturing the cell under a first set of selective culture
11 conditions, and whose presence or expression in the cell can
12 be selected against ~y culturing the cell under a second set
13 of selective culture conditions.
14 Examples of preferred selectable gene sequences include
gene sequences which encode an active thymidine kinase (TK)
16 enzyme, a hypoxanthine-phophoribosyltransferase (HPRT)
17 enzyme, a xanthine-guanine phosphoribosyltransferase (XGPRT)
18 enzyme, or an adenosine phosphoribosyltransferase (APRT)
19 enzyme. Such gene sequences can be used for both positive
and negative selection.
21 Additional gene sequences which can be used as
22 selectable gene sequences include those which encode enzymes
23 such as dihydrofolate reductase (DHFR) enzyme, adenosine
24 deaminase tADA), asparagine synthetase (AS), hygromycin B
phosphotransferase, or a CAD enzyme (carbamyl phosphate
26 synthetase, aspartate transcarbamylase, and dihydroorotase).
27 Methods for producing cells deficient in expressing these
28 enzymes are described by Sambrook et al. (In: Molecular
29 Cloninq_~ LaboratorY Manual, 2nd. Ed., Cold Spring ~arbor
Laboratory Press, NY (1989), herein incorporated by
31 reference). Such gene sequences can be used only for
32 positive selection.
`:, ..
... . . . . . . .
. : . . .. . . . .
' :' , ~ '' `` ' ,:. ` `' ~.
: ` ' - ~ : .
` ~' ' , .
WO91/19796 PCr/~S91/0~006
~ F
-4c-
l The incubation is performed under conditions sufficient
2 to permit the DNA molecule to be introduc~d into the
3 precursor cell. Such introduced DNA molecules are able to
4 then undergo homologous reco~bination with the predetermined
gene sequence of the genome of the precursor cell to thereby
6 produce the desired cell wherein the desired non-selectable
7 gene sequence has been inserted into the predetermined gene
8 sequence.
9 Such a desired cell can be recovered by culturing the
cell under the first set of selective culture conditions, by
l1 then permitting the cell to undergo intrachromosomal
12 recombination under non-selective culture conditions, and by-
13 then incubating the cell under the second set of selective
14 culture conditions.
Thus, in one preferred embodiment, the precursor cell
16 lacks an active hypoxanthine-phophoribosyltransferase (HPRT)
17 enzyme, a xanthine-guanine phosphoribosyltransferase (XGPRT)
18 enzyme, or an adenosine phosphoribosyltransferase (APRT)
13 enzyme, and the selectable gene sequence expresses an active
HPRT, XGPRT or APRT enzyme. In 1:he first set of selectable
21 culture conditions, medium containing hypoxanthine,
22 aminopterin and/or mycophenolic acid (and preferably
23 adenine, xanthine, and/or thymicline) is employed. In the
24 second set of selectable cult:uring conditions, medium
containing a nucleoside analog such as 5-bromodeoxyuridine,
26 6-thioguanine, 8-azapurine, etc., is employed.
27 In a second preferred embodiment, the precursor cell
28 lacks an active TK enzyme, and the selectable gene sequence
29 expresses an active TK enzyme. In the first et of
selectable culture conditions, medium containing
31 hypoxanthine, aminopterin, and thymidine is employed. In
32 the second set of selectable culturing conditions, medium
33 containing a thymidine analog such as FIAU (Borrelli, Proc.
., . . - . . :.: . : .
- . ... ~ . : ~ -
, . . : , .
. . . . . . . .
- :::
::
WO91/197g6 PCT/~S91/0~00~
rr~.`7~l
1 Nat'l. Acad. Sci. (U.S ~.) 8~:7s72 (1988), or gangcyclovir,
2 etc. is employed (if an HSV t~. gene is used), or 5-
3 bromodeoxyuridine, etc. ( lf a cellular tk gene is employed).
4 A preferred negative selectable marker is the hprt gene
(cells expressing an active HPRT enzyme are unable to gro~
6 in the presence of certain nucleoside analogues such as 6-
7 thioguanine, etc.). When using S-~hioguanine as ~ negative
8 selection agent, a density of 104 cells / cm2 is preferably
9 used since the efficiency of 6-thioguanine selection is cell
density dependent. A typical experiment with 107transfected
11 cells would yield approximately 10 revertant cells after - -
12 successive selection. The relative yield of revertant
13 clones can be substantially increased by using "Poly A -~
14 Selection" for the first round of selection. "Poly A ~
Selection" is discussed in detail in Exa~ple 6 below. - -
16
17 IV. The Pro~uction of Chi~eric and Tr~nsgenic Animsl~
18
l9 The chimeric or transgenic: animals of the present -
invention are prepar~d by int:roducing one or more DNA
21 molecules into a precursor pluripotent cell, most preferably
22 an ES cell, or equivalent (Ro~)ertson, E.J., In: Current
23 Communica~ions in Molecular Bio]oay, Capecchi, M.R. (ed.),
24 Cold Spring Harbor Press, Cold Spring Harbor, NY (1989), pp.
39-44, which reference is incorporated herein by reference).
26 The term ~'precursor" is intended to denote only that the
27 pluripotent cell is a precursor to the desired
28 ("transfected"~ pluripotent cell which i~ prepared in
29 accordance with the teachings of the present invention. The
pluripotent (precursor or transfected) cell may be cultured
31 in VlVo, in a manner known in the art (Evans, ~.J. t al.,
32 ~ature 292:154-156 (19 1)) to form a chi~eric or transgenic
33 ani~al.
:. . . , . - :- : . .. , .- , :-:
- : ., ~, . , - . : , . : : : :
:: . -
.: , : .,' ,.: -: ' :' . : ' ' ,
.: :. . .:: ~ : ' . : : , ; . ..
: :' ~ '' , -' '' '. ' ~.. , ~ :
W~91/~9796 ~T/-S9l/0~006
.
G f ~ r ~
~ -r r f '- --~ 2--
1 Any Es cell may be used ln accordance with the present
2 inve~tion. It is, however, preferred to use primary
3 isolates of ES cells. Such isolates may be obtained
4 directly from embryos such as the CCE cell line disclosed by
Robertson, E.J., In: Current Communications _ln Molecular
6 Bioloqv, Capecchi, M.R. (ed.), Cold Spring Harbor Press,
7 Cold Spring Harbor, NY (1989), pp. 39-44), or from the
8 clonal isolation of ES cells from the CCE cell line
9 (Schwartzberg, P.A. et al., Science 246:799-803 (1989),
which reference is incorporated herein by reference). Such
11 clonal isolation may be accomplished according to the method
12 of E.J. Robertson (In: Teratocarcinomas and Embrvonic Stem
13 Cells: A Practical ADProach, (E.J. Robertson, Ed.), IRL
14 Press, Oxford, 1987) which reference and method are
incorporated herein by reference. The purpose of such
16 clonal propagation is to obtain ES cells which have a
17 greater efficiency Eor differentiating into an animal.
18 Clonally selected ES cells are ~pproximately 10-fold moxe
l9 effective in producing transgenic animals than the
progenitor cell line CCE. For the purposes of the
21 recombination methods of the present invention, clonal
22 selection provides no advantage. An example of ES cell
23 lines which have been clonally derived from embryos are the
24 ES cell lines, ABl (h~rt~) or AB2.1 (hprt).
The ES cells are preferably cultured on stomal cells
26 (such as STO cells (especially SNC4 STO cells) and/or
27 primary embryonic fibroblast cells) as described by E.J.
28 Robertson (In: Teratocarcinomas and Embrvonic Stem Cells: A
29 Practical Ap~roach, (E.J. Robertson, Ed.), IRL Press,
Oxford, 1987, pp 71-112), which reference is incorporated
31 herein by reference. The stomal (and/or fi~roblastj cells
32 serve to eliminate the clonal overgrowth of abnormal ES
33 cells. Most preferably, the cells are cultured in the
:~'
WO 91/19796 PCI`/--IS91/04006
-43- ~ q~i~
1 presence of leukocyte inhibitory factor ("lif") (Gough, N.M.
2 et al., Reprod. Fertil. Dev. 1:281-288 (1989); Yamamori, Y.
3 et al., Science 246:1412-1416 (1989), both of which
4 references are incorporated herein by reference). Since the
gene encoding lif has been cloned (Gough, N.M. et al.,
6 ReProd. Fertil. Dev. 1:281-288 (1989)), it is especially
7 preferred to transform stomal cells with this gene, by means
8 known in the art, and to then culture the ES cells on
9 transformed stomal cells that secrete lif into the culture
medium. -
ll ES cell lines may be derived or isolated from any
12 species (for example, chicken, etc.), although cells derived
13 or isolated from mammals such as rodents (i.e. mouse, rat,
14 hamster, etc.), rabbits, sheep, goats, fish, pigs, cattl~,
primates and humans are preferred.
16
17 V. The Production of ~hi~eric a~d Tr~nsgenic Plant~
18
19 The chimeric or transgenic plants of the invention are
produced through the regeneration of a plant cell which has
21 received a DNA molecule through the use of the methods
22 disclosed herein.
23 All pla~ts from which protc~plasts can be isolated and
24 cultured to give whole regenerated plants can be transformed
by the present invention so that whole plants are recovered
26 which contain the introduced gene sequence. Some suitable
27 plants include, for example, species from the genera
28 Fra~aria, Lotus, Medic~qo, OnobrYchis, Trifolium,
29 Triaonella, Viana, Citrus, Linum, Geranium, Manicot, Daucus,
Arabido~sis, Brassica, Ra~hanus, Sina~is, Atro~a, Cavsicum,
31 Datura, Hyosc~amus, Lvco~ersion, Nisotiana, Solanum,
32 Petunia, Pi~italis, Maiorana, Cich rium, Helianthus,
33 Lactuca, Bromus, AsParaaus, Antirrhinum, Hemer-callis,
,.................. ~ .. ,
. . : , . . .
.. .. ...
: .. . -: . :
:
:: ~'. ': . : :, ,. -
WO 91/19796 PCT/l~S91/n4006
r t ~
~44--
1 Nemesia, Pelaraoniur~, Panicum, Pennisetur~, Ranlln~ulus,
2 Senecio, Salpiqlossis, Cucumis, Browallia, Glycine, Lollum,
3 Zea, Triticum, Sorqhum, Ipomoea, Passiflora, CYclamen,
4 Malus, Prunus, Rosa, Rubus, Populus, Santalum, Allium,
Lilium, Narcissus, Ananas, Ara~his, Phaseolus, Pisum and
6 Datura.
7 There is an increasing body of evidence that practically
8 all plants can be regenerated from cultured cells or
9 tissues, including but not limited to all major cereal crop
species, sugarcane, sugar beet, cot~on, fruit and other
11 trees, legumes and vegetables.
12 Plant regeneration from cultural protoplasts is
13 described in Evans et al., "Protoplast Isolation and
14 Culture," in Handbook of Plant Cell Culture 1:124-176
(MacMillan Publishing Co., New York, 1983); M.R. Davey,
16 "Recent Developments in the Culture and Regeneration of
17 Plant Protoplasts," Proto~lasts, 1983 - Lecture Proceedings,
18 pp. 19-29 (Birkhauser, Basel, 1983); P.J. Dale, "Protoplast
19 Culture and Plant Regeneration of Cereals and Other
Recalcitrant Crops," in P to~lasts 1983 - Lecture
21 Proceedings, pp. 31-41 (Birkhauser, Basel, 1983); and H.
22 Binding, "Regeneration of Plants," in Plant Proto~lasts, pp.
23 21-37 (CRC Press, Boca Raton, 1985).
24 Regeneration varies from sp~ecies to species of plants,
but generally a suspension of transformed protoplasts
26 containing the introduced gene sequence is formed. ~mbryo
27 formation can then be induced from the protoplast
28 ~uspensions, to the stage of ripening and germination as
29 natural embryosO ThP culture media will generally contain
various amino acids and hormones, such as auxin and
31 ~ytokinins. It is also advantageous to add glutamic acid -`~
32 and proline to the medium, especially for such species as
33 corn and alfalfa. Shoots and roots normally develop
:
.~
.
.. .
.; . ~ -
W091/19796 PC~/~591/0~006
_ ~ 5 ~ r ~ g~
1 simultaneously. Efficient regeneration will depend on the
medium, on the genotype, and on the histor~ of the culture.
3 If these three variables ar~ controlled, then regeneration
q is fully reproducible and repeatable.
The mature plants, grown from the transformed plant
6 cells, are selfed to produce an inbred plant. The inbred
7 plant produces seed containing the introduced gene sequence.
8 These seeds can be grown to produce plants that express this
9 desired gene sequence. ~
Parts obtained from the regenerated plant, such as ~ -
11 flowers, seeds, leaves, branches, fruit, and the like are
12 covered by the invent1on. Progeny and variants, and mutants
13 of the regenerated plants are also included within the scope
14 of this invention.
As used herein, variant describes phenotypic changes
16 that are stable and heritable, including heritable variation
17 that is sexually transmitted to progeny of plants.
18 ~ -
19 VI . GENE E~PRESSION
2 0
21 In one embodiment, the DNA molecule(s) which are to be
22 introduced into the recipient cells in accordance with the
23 methods of the present inventioIl will be incorporated into
24 a plasmid or viral vector (or a derivative thereof) capable
of autonomous replication in a host cell.
26 Preferred prokaryotic ~ectors include plasmid6 such as
27 those capable of replication in E. coli such as, for
28 example, pBR322, ColEl, pSC101, pACYC 184, ~X. Such
29 plasmids are, for example, disclosed by Maniatis, T., et al. ~-
tIn Molecular Clonina. A Laboratorv Manual, Cold Spring
31 Harbor Press, Cold Spring Harbor, NY (1982)). Bacillus
32 plasmids include pClg4, pC221, pT127, etc. Such plasmids ~;~
33 are disclosed by Gryczan, T. (In: The Molecular Biolo~v of
;
... . . .. . . . . .
W O 91/19796 P ~ /~S91/04006
-46-
1 the ~acilli, Academic Press, NY (1982), pp. 307-329).
2 Suitable StrePtomvces plasmids include pIJ101 (Kendall,
3 K.J., et al., J. Bacteriol. 169:4177-4183 (1987~), and
4 Stre~tomyces bacteriophages such as ~C31 (Chater, K.F., et
al., In: Sixth International S~m~osium on Actinomvcetales
6 Bioloqv, AXademiai Xaido, Budapest, Hungary (1986), pp. 45-
7 54). Pseudomonas plasmids are reviewed by John, J.F., et
8 al. (Rev. Infect. Dis. 8:693-704 (1986)), and Izaki, K.
9 (Jpn. J. Bacteriol. 33:729-742 (1978)).
Examples of suitable yeast vectors include the yeast 2-
11 micron circle, the expressi~n plasm~ds YE~13, YCP and YRP,
12 etc., or their derivatives. Such plasmids are well known in
13 the art (Botstein, D., et al., Miami wntr. S~mp. 19:265-274
}4 (1982); Broach, J.R., In: The Molecular Bioloqv of the
Yeast Saccharomy~es: Life Cvcle and Inheritance, Cold
16 Spring Harbor Laboratory, Cold Spring Harbor, NY, p. 445-470
17 (1981); Broach, J.R., Cell 28i~i203-204 (1382)).
18 Examples of vectors which may be used to replicate the
19 ~NA molecules in a mammalian host include arlimal viruses
such as bovine papill~ma viruci, polyoma virus, adenovirus,
21 or SV40 virus.
22
23 VII. ~e3 of the PraYent Inventio~
24
The methods of the present invention permit the
26 introduction of a desired ~ene sequence into an animal or
27 plant cell.
28 In a first embodiment, the methods of the present
29 invention may be uied to introduce DNA into germ line cells
of animals in order to produce chimeric or transgenic
31 animals which contain a desired gene sequence. The animals
32 which may be produced through application of the described
33 method include chicken, non-human mammals (especially,
. , . ' .. ' . .
WO9l/19~96 PCT/~'S9lJ0~006
-~7-
1 rodents (i.e. mouse, rat, hamster, etc.), rabbits, sheep,
2 goats, fish, pigs, cattle and non-human primates).
3 As stated above, the desired gene sequence may be of any
4 length, and have any nucleotide sequence. In particular, it
is possible to design the sequence of the desired gene
6 sequence in order to create single, or multiple base
7 alterations, insertions or deletions in any preselected gene
8 of a cell.
9 If such changes are within a translated region of a
native gene sequence, then a nPw protein variant of a native
11 protein can be obtained. Such a procedure can, for example
12 be used to produce animals which produce improved (i.e. more
13 stable, more active, etc.) enzymes, binding proteins,
14 receptors, receptor ligands, etc.
The methods of the present invention may be used to
16 produce cells in which a natural gene has been replaced with
17 a heterologous gene. A gene is said to be heterologous to
7 8 a transgenic cell if it is derivable from a species other
19 than that of the transgenic cell.
In one embodiment, this replacement may be accomplished
21 in a single step (Figure 3). To accomplish such
22 replacement, a DNA molecule containing a desired gene
23 sequence and a region of homology is introduced into a
24 recipient cell. A selectable maeker gene is also introduced
into the cell, and used to select for cells which have
26 underwent recombination. The method results in the
-27 replacement of the normal sequences ad~acent to the region
28 of homology with the heterologous sequences of the desired
29 DNA sequence.
3~ In a second embodiment, this replacement may be
31 accomplished in a two steps (Yigure 4). A~ in the
32 embodiment described above, a cell is provided with a DNA
33 molecule containing a desired gene sequence and a r~gion of
- .
,
~ ~ , ' , ~ ' . .
WO 91/~9796 PCr/-S91/0400S
1 homology. The DNA molecule also contains a selectable
2 marker gene used to select for cells which have undergone a
3 recombinational event that has resulted in the insertion of
4 the introduced DNA molecule into their chromosomes at the
site of homology. The structure of such an insertion site
6 is depicted in Figure 4A.
7 Significantly, in this embodiment, the introduced DNA
8 ~olecule will al50 contain a "negative selectable" marker
9 gene which can be used to select for cells which undergo a
second recombinational event that results in the loss of the
11 inserted DNA.
12 As shown in Figure 4B, a second DNA molecule is employed
13 to complete the gene replacement. This second DNA molecule
14 need not contain any selectable marker gene. ~pon receipt
of the second DNA molecule, a second recombinational event
16 occurs which exchanges the "second" DN~ molecule for the
17 integrated "first" DNA molecule (including the desired DNA
1~ sequence, the selectable marker sequence, and the "negative
19 selectable" marker sequence contained on that molecule).
This aspect of the invention is illustrated in Figure 4B.
21 In another embodiment of the invention, subtle mutations
22 may be introduced into a desired locus using a "cassette"
23 construct containing both a pos:itive selection marker (such
24 as the n~tII gene or the qpt gene~ and a negative selection
marker (such as the tk gene). In this embodiment, one first
26 uses the positive selection capacity of the construct to
27 introduce the two selection markers into a desired ~ocus.
28 One then introduces the desired subtle mutations (substi-
29 tutions, insertions, deletions, etc.) by providing a cell
with a DNA molecule that contains the desired ~utation. By
31 selecting for the loss of the "cassette" (using the negative
32 selection marker), one can select for recombinational events
33 which result in the replacement of the "cassette" sequence
~'
.
.' '' ' , , ' ' :
,
wosl/l979~ PCT/~S91/04006
-49~
l with the DNA sequence containing the desired mutation. This
2 embodiment of the invention is illustrated in Figure 5.
3 The methods of the present invention may also be used tc
4 replace contiguous regions of a chromosome with any desired
gene sequence. Thus, the present invention is not limited
6 in the size of the DNA regions which may ~e altered or
7 replaced. This aspec~ of the present invention is
8 illustrated in Figure 6, as a series of 5 steps (Figures 6A-
9 6E). The method is applicable to any gene sequence. It is
especially useful in producing cells which contain
11 heterologous immunoglobulins (such as the heavy chain locus
12 of an immunoglobulin).
13 The first step in replacing a large region of a
14 chromosome with a desired sequence involves setting up an
initial target. In this step, a recipient cell is provided
16 with a DNA molecule which contains a "first fragment" of the
17 total desired replacement sequence (Figure 6A). This "first
18 ~ragment" of the desired replacement sequence contains a
l9 selectable marker sequence (most preferably the nPtII gene)
at its end.
21 The DNA molecule also contains a "dual selection" gene
22 sequence which encodes a non-functional fra~ment of a gene
23 sequence for which both a positive and a negative selection
24 exists. An example of such a gene is the oDt gene when used
in the context of an h~rt- cell. Cells which express a
26 functional qpt gene can be selected for by their ability to
27 grow in HAT medium; Cells which lack a functional ~t gene
28 can be selected for by their ability to grow in the presence
29 of 6-thioguanine.
Homologous recombination results in the insertion of the
31 DNA molecule into the cell's genome at the region of
32 homology (Figure 6A). Importantly, since this step results
33 in the creation of a cell whose genome contains the
.
; ~ : ., : .i '
, . .::
' ~ '
WO 91/19796 PCT/~'S91/04006
f ~ --50--
1 selectable marker gene, it is possible to sel~ct for the
2 desired reco~binational event.
3 In the second step of the method, a second DNA molecule
4 is provided to the cell. This second DNA molecule contains
a "second fragment" of the desired replacement sequence as
6 well as a sequence of the dual selection gene that, due to
7 an internal deletion, is incapable of encoding a functional
8 gene product. Homologous recombination results in the
9 insertion of the second DNA molecule into the cell's genome
in a manner so as to create a functional dual selection gene
11 (Figure 6B). Recombination also results in the integration
12 of a non-functional fragment of the dual selection gene.
13 Importantly, since this step results in the creation of a
14 cell whose genome contains a functional dual selection gene,
it is possible to select for the desired recombinational
16 event.
17 In the third step of the method, a third DNA molecule is
18 provided to the cell. This third DNA molecule contains both
19 the "first" and l'second" fragments of the desired
replacement sequence. Homologous recombination results in
21 the insertion of the third DN~ molecule into the cell's
22 genome in a manner so as to delete the functional dual
23 selection gene. The non-functional fragment of the dual
24 selection gene (formed in step ~) is not affected by the
recombination, and is retained (Figure 6C). Importantly,
26 since this step results in the creation of a cell whose
27 genome lacks the dual selection gene, it is possible to
28 select for t~e desired recombinational event.
29 In ~he fourth step of the method, a ~ourth DNA molecule
is provided to the cell. This fourth DNA molecule contains
31 a "third fragment" of the desired replacement sequence as
32 well as a seguence of the dual selection gene that, as in
33 step 2, is incapable of encoding a functional gene product
.:: . . . ~
:. : - : .
: . : . ..
. ~ : .. ..
: .
- - .:
Wo 91/1979h PCl/~S91/04006
- 5 1 ~ ~' f~
l due to an internal deletion. Homologous recombinatio~
2 results in the insertion of the fourth DNA molecule into the
3 cell's genome in a manner so as to create a functional dual
4 selection gene (Figure 6D), Recombination also results in
the integration of a non functional fragment of the dual
6 selection gene. Importantly, since this step results in the
7 creation of a cell whose genome contains a functional dual
8 selection gene, it is possible to select for the desired
9 recombinational event.
In the fifth step of the method, a fifth DNA molecule is
11 provided to the cell. This fifth DNA molecule contains both
12 the "second" and "third" fragments of the desired
13 replacement sequence. Homologous recombination results in
14 the insertion of the fifth DNA molecule into the cell's
genome in a manner so as to delete the ~unctional dual
16 selection gene. The non-functional fragment of the dual
17 selection qene (formed in step 4) is not af~ected by the
18 recombination, and is retained (Figure 6C). Importantly,
l9 since this stPp results in the creation of a cell whose
genome lacks the dual selection gene, it is possible to
21 select for the desired recombinational event.
22 As will be appreciated, the net ef~ect of the above-
23 described steps is to produce ,a cell whose genome has been
24 engineered to contain a "first," "second," and "third"
"fragment" of a particular desired gene in a contiguous
26 ~anner. The steps may be repeated as desired in order to
27 introduce additional "fragments" into the cell's genome. In
28 this manner, cells can be constructed which contain
29 heterologous genes, chromosome fragments, or chromosomes,
that could not be introduced using a single vector. As
31 indicated above, each step of the method can be selected
32 for.
~.
. ................................
.
:
.
: '' ' ~ -
,:, ~ ,,. . , - ~ '' '"'
. ~ '
9l/19796 PCT/~591/04006
~ 52--
1 In particular, this aspect of the present invention may
2 be used to produce "humanized" antibodies (i.e. non-human
3 antibodies which are non-immunogenic in a human) (Robinson,
4 R.R. et al., Interna~ional Patent Publication
PCT/US86/02269; AXira, K. et al., European Patent
6 Application 184,187; Taniguchi, M., European Patent
7 Application 171,4g6; Morrison, S.L. et al., European Patent
8 Application 173,494; Neuberger, M.S. et al., PCT Application
9 Wo 86/01533; Cabilly, S. et al., European Patent Application
125,023; Better, M. et al., science 240:1041-1043 (1988);
11 Liu, A.Y. et al., Proc. Natl. Acad. Sci. USA 84:3439-3443
12 ~1987); Liu, A.Y. et al., J. Immunol. 139:3521-3526 (1987);
13 Sun, L.K. et al., Proc. Natl. Acad. Sci. USA 84:214-218
14 (1987~; Nishimura, Y. et_al., Canc. Res. 47:999-1005 (1987);
Wood, C.R. et al., Nature 314:446-449 (1985)); Shaw et al.,
16 J. Natl.Cancer Inst. 80:1553-1559 (1988).
17 The method may also be used to produce animals having
18 superior resistance to disease, animals which constitute or
19 produce improved food sources, animals which provide fibers,
hides, etc. having more desirable characteristics. The
2~ method may also be used to produce new animal models for
22 human genetic diseases. For example, the method may be used
2~ to "humanize" the CD4 analog of an animal, and thus provide
24 an animal model for AIDS. SUCh animal models can be used
for drug testing, and thus hasten the development of new
26 therapies for genetic diseases.
27 In addition, the present invention permits the formation
28 of cells and of transgenic animals which contain mutations
29 in ~edically or clinically significant heterologous genes.
A gene is said to be medically or clinically significant if
31 it e~presses an isotype of a protein associated with a human
32 or animal disease or condition. Examples of such genes
33 include the genes which encode: topoisomerase pl80, 5~
::: :- : ,.: .. : .- : . .. - . -. . .: .. ... - , : :
.:. . : ~ : ,
. :: : . . : ... .
, . . : . . .
~: , ' ' '- . . . 1
Wo91/l9796 PCT/~S91/~4006
-53-
1 reductase, ACAT, 5-lipoxygenase, the insulin receptor, the
2 interleukin-2 receptor, the epidermal growth factor
3 re~eptor, the seratonin receptor, the dopamine receptor, the
4 GABA receptor, the V2 vasopressin receptors, G proteins
(signal transduction), phospholipase C proteins, and
6 insulin. A transgenic mouse produced by microinjection
7 which expresses human insulin was reported by Selden, R.F.
8 et al. (European Patent Publication No. 247,494, which
9 reference is incorporated herein by reference).
The transgenic cells and animals discussed a~ove can be
11 used to study human gene regulation. For example,
12 transgenic animals which express a human isotype of
13 topoisomerase pl80, 5-~ reductase, ACAT,S-lipoxygenase, or
14 hormone or cytokine receptors would have ultility in in vivo
drug screening. The expression of topoisomerase pl80 is
16 associated with resistance to chemotherapeutics. Thus,
17 agents which interfere with this enzyme could be used to
18 enhance the effectiveness of chemotherapy. An animal,
19 es?ecially a rat, capable of expressing a human isotype of
5-~ reductase (especially in the prostate gland) would be
21 highly desirable. ACAT is a key enzyme in lipid metabolism;
22 an animal model for its regulation would be extremely
23 valuableO Animals that express 5-lipoxygenase could be of
2~ interest to many research programs, particularly to screen
isotype selective inhibitors. An animal which expressed
26 human hormone or cytokine receptor proteins would be
27 valuable in identifying agonists and antagonists of receptor
28 action~ Similarly, an animal that expressed components of
29 the human signal transduction system ~i.e. G proteins and
phospholipase Cs, etc.) could be used to study the
31 pathophysiologic consequences of disordered function of
32 these proteins.
:~:
.
. I
.
. . , . :
: . ' . . .. .
WO91/19796 PCT/~591/04006
~? f '' ~
J ~ r 1' ~c
l The present invention can be used to produce cells and
2 animals which express human isotypes of transport proteins
3 (i.e. proteins which facilitate or enable the transport of
4 other molecules or ions across membranes in the gut, blood
brain barrier, kidney, etc.). Such cells or animals can
6 then be used to study the role of such proteins in
7 metabolism. In particular, the extent and patterns of
8 conjugation mediated by such isotypes may be studied in
9 order to investigate the pharmacokinetic consequences of
specifi~ differences in protein structure or sequence.
ll Glucoronide transferase, glycine conjugation and sulfation,
12 methylases, and glu~athione conjugation are examples of
13 enzymes of particular interest in this regard.
14 The clearance of many compounds is mediated by
esterases. Cells or animals which express heterologous
16 isotypes of such esterases may be exploited in investigating
17 such clearance.
18 Cells or animals which express isotypes of proteins
l9 involved in azo or nitro reduction would be desirable for
research on the processes of azo or~nitro reduction.
21 Significantly, potential therapeutic agents are
22 ~requently found to induce toxic~effects in one animal model
23 but not in another animal model. To resolve the potential
24 of such agents, it is often necessary to determine the
2S metabolic patterns in various species, and to then determine
26 the toxicities of the metabolites. The present invention
27 permits one to produce transgenic cells or ~nimals which
28 could facilitate suoh determinations.
29 The methods of the present invention ~ay be used to
produ~e alterations in a regulatory region ~or a native gene
31 sequence. Thus, the invention provides a means for altering
32 the nature or oontrol of transcription or translation of any
33 native gene sequence which is regulated by the regulatory
.' : , ;. ,; , ' ' , .: '............................. ~
, .: : : . .- .
:: ,., :
.: :
W~91/19796 PCT/~591/04006
_55_ ~r~ t~
l region. For example, it is possible to introduce mutations
2 which remove feedback inhibition, and thus result in
3 increased gene expression. Similarly, it is possible to
4 impair the transcriptional capacity of a sequence in order
to decrease gene expression. Such alterations are
6 especially valuable in gene therapy protocols, and in the
7 development of improved animal models of human disease. For
8 example, the capacity to increas~ insulin gene transcription
9 or translation provides a potential genetic therapy for
diabetes. Similarly, the ability to impair the synthesis of
ll beta globln chains provides an animal model for beta-
12 thalassemia.
13 The methods of the present invention, quite apart from
14 their uses in veterinary and human medicine, may be used to
investiyate gene regulation, expression and organization in
16 animals.
l7 Since the methods of the present invention utilize
18 processes of DNA repair and recombination, agents which
l9 inhibit or impair th~ present methods may act by affecting
these processes. Since agents which impair DNA repair and
21 recombination have potential antineoplastic utility, the
22 present invention provides a 1Deans for identifying ~ovel
23 antineoplastic agents.
24 The present invention ~ay additionally be used to
~acilitate both the cloning of gene sequences, and the
26 mapping of chro~osomes or chromosomal abnormalities.
27 Since the desired gene sequence need not be homologous
28 or analogous to any native gene sequence of the recipient
29 c~ll, the methods of the present invention permit one to
produce animals which contain and express foreign gene
31 sequences. I~ the cell expresses an analogous gene, the
32 desired gene sequence may be expressed in addition to such
33 analogous cellular genes (for ex~mple, an animal may express
. , , . . . ~ . . . .
wv 91/1~7~ PCT/~S91/04006
t
--S6--
1 both a "humani~ed" receptor and an analogous native
2 receptor). Thus, for example, t~e invention provides a
3 means for producing animals which express important human
4 proteins (Ruch as human interf~rons, tissue plasminogen
acti~ator, hormones (such as insulin and growth hormone),
6 blood factors (such as Factor VIII), etc.).
7 In a second embodiment, the methods o~ the invention may
8 be used to introduce DNA into plant cells which can then be
9 manipulated in order to produc~ chimeric or transgenic
~0 plants. The plants which may be produced through
11 application of the disclosed method include all
12 multicellular, higher (i.e. non-fungal) plants. A non-
13 fungal plant i5 any plant which is not a fungus or yeast.
14 In a third embodiment, the methods of the invention may
be used to introduce DNA into the somatic cells of an animal
16 (particularly mammals including humans) or plant in order to
17 provide a treatment for genetic dise~se (i.e. "gene
18 therapy"). The principles of gene therapy are disclosed by
19 Oldham, R.X. (In: PrinciPles cf Biotherapy, Raven Press, NY,
1987), and similar texts.
21 In this third embodiment~ the genetic lesion which
22 causes the disease is replaced with a gene sequence encoding
23 a preferred gene product. Examples of such genetic lesions
24 axe those responsible for diseases such as cystic fibrosis,
phenylketonuria, hemophilia, von Willebrand's Disease,
26 sickle cell anemia, thalasse~ia, galactosemia, fructose
27 intolerance, diseases of glycogen storage, hyper-
28 cholesterolemia~ juvenile diabetes, hypothyroidism,
29 Alzheimer's Disease, Huntington's Disease, Gout, Lesch-Nyhan
Syndrome, etc. (Bondy, P.R. et al., In: Disorde~s _of
31 Carbohydrate Metabolism, pp 221-340, Saunders Sl974);
32 Coleman, J. et al., Molecular Mechanisms of Disease, Yale
33 University Press, (1975)~. Disclosures of the methods and
. ~ ~
;:~
~ .
:: . .. .
.,. . . . . . : . . .. . ,,.. , , ,. . , ~:. .
., ,, .:
: , ~. , , ~............... .
WO91/19796 PCT/~591/04006
_57- ~ r~ `5 ~
l uses for gene therapy are provided by Boggs, S.S. (Int. J.
2 Cell Clon. 8:80-96 (1990)); Karson, E.M. (Biol. Re~rod.
3 42:39-49 (l990)); Ledley, F.D., In: BiotechnoloqY A
4 Com~rehensive Treatise volume 7B, Gene Technolo~y, VCH
Publishers, Inc. NY, pp 399-458 (1989)); all of which
6 references are incorporated herein by reference.
7 In a fourth embodiment, the me~hods of the invention may
B be used to provide a treatment to protect recipient animals
9 or plants from exposure to viruses, insects or herbicides
(in the case of plants), insecticides, toxins, etc. In this
11 embodiment, the introduced gene would provide the recipient
1~ with gene sequences capable of mediating either an enhanced
13 or novel expression of an enzyme, or other protein, capable
14 of, for example, degrading an herbicide or toxin. For
example, a plant cell may recaive a gene sequence capable of
16 mediating an enhanced or novel expression of a chitinase,
17 thus conferring increased resistance to insect parasites.
18 When providing the desired gene sequence to the cells of
19 an animal, pharmaceutically acceptable carriers (i.e.
liposomes, etc.) are preferably employed. Such gene
21 sequences can be formulated according to known methods to
22 prepare pharmaceutically useful compositions, whereby these
23 materials, or their functional derivatives, are combined in
24 admixture with a pharmaceutically acceptable carrier
vehicle. Suitable vehicles and their formulation, are
26 described, for example, in Nicolau, C. et al. (Crit. Rev.
27 Ther. Drua Carrier_Sy~t. 6:239-271 (1989)), which reference
28 is incorpor~ted herein by referenoe.
29 In order to form a pharmaceutically acceptable
composition suitable for effective administration, such
31 compositions will contain an effective amount of the desired
32 gene sequence together with a suitable amount of carrier
33 vehicle.
: , ~ - ~ :: ~: . : , :
,
WOs1/19796 PCT/~'S91/04006
~; ^t~J,~ -58-
1 Additional pharmaceutical methods may be employed to
2 control the duration of action. Control release
3 preparations may be ~chieved through the use of polymers to
4 complex or absorb the desired gene sequence (either with or
S without any associated carrier). The controll~d deli~ery
6 may be exercised by selecting appropriate macromolecules
7 (for example polyesters, polyamino acids, polyvinyl,
8 pyrrolidone, ethylenevinylacetate, methylcellulose,
9 carboxymethylcellulose, or protamine, sulfate) and the
concentration of macromolecules as well as the methods of
ll incorporation in order to control relPase. Another possible
12 method to control the duration of action by controlled
13 release preparations is to incorporate the agent into
14 particles of a polymeric material such as polyesters,
polyamino acids, hydrogels, poly(lactic acid) or ethylene
16 vinylacetate copolymers. Alternatively, instead of
17 incorporating these agents into polymeric particles, it is
l8 possible to entrap these materials in microcapsules
19 prepared, ~or example, by coacervation techn~ques or by
interfacial polymerization, for~example, hydroxymethylcellu-
21 lose or gelatine-microcapsules and poly(methylmethacylate)
22 microcapsules, respectively, or in colloidal drug delivery
23 systems, for example, liposomes, albumin ~icrospheres,
24 microemulsions, nanoparticles, and nanocapsules or in
macroemulsions.
26 In a fifth embodiment, the methods of the present
27 invention may be used to improve the food or fiber
28 chara~teristics of plants or non-human animals. For
29 example, the methods can be used to increase the overall
levels of protein synthesis thereby resulting in faster
3l growing plants or non-humah animals, or in the production of
32 plants and non-human animals which have increased food
33 value.
., ; .
:, , , : . . . ~ .............................. ~ , i . .
-: , . :
Wo 91/19796 PCl/~S91/040û6
'7`7~
-5~-
1 Having now generally described the invention, the same
2 will be more readily understood through reference to the
3 following examples which are provided by way of
4 illustration, and are not intended to be limiting of the
presPnt invention, unless specified.
7 E~AMPLE 1
8 ELECT~OPORATION : .
Electroporation was performed as follows: ~
11 . - .
12 DNA Preparation:
13
14 DNA used for electroporation was purified by CsCl
gradient centrifugation. A large-scale digest of this
16 purified DNA was prepared by incubating the DNA with an
17 appropriate restriction enzyme. The large-scale digest was
18 examined for complete digestion by running 500 ~g on a
19 minigel. The DNA concentration of the large-scale digest ~
should be no higher than 1 ~g/~l. ` `
21 The large-scale digest was then extracted once with an
22 equal volume of phenol/chloroform and once with an equal
23 volume o~ chloroform. The DNA was precipitated with 2.4
24 volumes of ethanol, pelleted by centrifugation, and dried
using a Speed-Vac. ;
26 The pelleted DNA was then resuspended at the desired
27 concentration (usually 1 ~g/~l) in a sterile Tris-EDTA
28 bu~fer such as O.lX TE (25 ~1 of DNA per electroporation~
29 The concentration of th~ DNA was then measured with a
fluoro~eter.
31
;,
:~;
: . : , . - .
-,: .~ ' , ' ,'- ., , ; ~ .
:: ~ . - , . . . . .
: .: . ', ' :~ ' ,, ' . ,. . ~ , :
WOgl/19796 PCT/~'S91/0~006
~ F ~ ~ --60--
.
1 Preparation of Cells for_Electroporation:
3~mbryonic stem cells of the AB1 cell line were c~ltured
4to approximately 80% confluence according to the methods of
SE.J. Robertson ~In: Teratocarcinomas and Embryonic Stem
6Cells: A Practical A~pr~ach, ~E.J. Robertson, Ed.), IRL
7Press, Oxford, 1987, pp 71-112). Cells were cultured in the
8presence of stomal cells which expressed lif into the
9culture medium. Cells were passaged 1:2 the day before
10electroporation, and fed 4 hours before harvesting.
11Cells were harvested by trypsinizing the cells, and by
12resuspending in media (cells from 2 x 10 cm plates were
13combined in a total volume of 10 ml in a 15 ml tube).
14The cells were pelleted by centrifugation, and the
15supernatant was removed by aspiration. The cells were then
16resuspended in 10 ml of phosphate buffered saline and the
17total number of cells was determined by counting a 20 ~
18aliquot. The usual yield is 30 x 106 cells per 10 cm plate.
19The cells were then pelleted by centrifugation and the
20supernatant was removed by aspiration. Cells were
21 resuspended at a density of 11 x 106 cells/ml. A 20
22 aliquot was counted to confir~ this cell density.
23
24 Electro~oration
26Cells, prepared as descri~ed above, were incubated in
27the presence of an appropriate amount of DNA in a 15 ml
2tube. 25 ~1 of DNA and 0.9 ml of cells were used for each
29el~ctroporation.
30The mixture was allowed to incubate at room temperature
31for 5 minutes (this step may, however, be omitted).
32The cell/DN~ mixture was then carefully aliquoted into
33electroporation cuvettes (O.~ ml per cuvette; the volume is
,
:`:
';'
U'091/~9796 PCT/~'S91/04006
-61-
1 important). The cuvette was placed in the electroporation
2 holder with the foil electrodes in contact wi~h the metal
3 holding clips.
4 Electroporation was accomplished using a Biorad
GenePulser set at 230V, 500 ~F (this requires a capacitance
6 extender). The time constant should read between 5.6 and
~ 7Ø
8 The cuvette was left at room temperature for 5 minutes
9 and then the cells were plated at an appropriate density (up
to 2 x 1O7 cells/100 mm plate or 6 x lo6 cells/60 mm plate).
11 ~hen G418 was used as a selective agent, this cell density
12 should not be exceeded since G418 takes 3-4 days before
13 killing starts and plates will become over-confluent. When
14 G418 selection was to be applied, it is applied 24 hours
post-electroporation. G418 concentration must be titrated
16 for every batch.
17 The plate(s) were re-fed with fresh media i G418 every
18 day for the first 6-7 days (until colonies are visible and
19 most cell debris has been removed). If usiny FIAU (0.2 ~M)
~election, this may proceed simultaneously.
21 The typical yield for RV4.0 (Thomas, K.R. et al., Cell
22 51:503-512 (1987)) is up to 104 colonies/107 cells/100 mm
23 plate. Although this yield may be significantly ~and
24 unpredictably) different from the yield obtained when other
constructs are used, the use of the meth~d always results in
26 the recovery of some colonies of cells which contain the
27 electroporated DNA.
28 Colonies may be picked as early as 8 days. It is most
29 preferred to pick colonies at around 10-11 days. Colonies
may, however, be recovered up to 18-21 days after t~e
31 electroporation.
32
::
,
; ''-''
: ' "
`
,,
w~ `J~ 7Y~) PCI/I'S~1/0~006
2 - 6 2 -
1 EXAMPL~ 2
2 CO-ELECTROPORATION OF ES CELLS
4 To illustrate the invention, embryonic stem ("~S") cells
were co-electroporated with a 4.5 kb nptII-containing vector
6 (pPGXneobpA) which had been linearized by treatment with
7 XhoI restriction endonuclease, and with the 6.5 kb HPRT
8 vector, AD 8 (linearized with SacI) (Figure 7).
9 Electroporation (230 V, 500 ~F) were done on 0.9 ml aliquot
of CCEp24 cells (7.5 x 106 cells/ml).
11 The electroporation reactions were conducted using molar
12 ratios of 1~ 10, and 1:100 (nptII DNA:HPRT DNA). The
13 total amount of DNA provided was either 25, 50, 100, or 200
14 ~9. The vectors used in this experiment are illustrated in ~',
Figure 7. The results of this experi~ent are shown in Table ~-
16 l. :-
17 . :~
18 ~ TABLE 1
19 CO-ELECTROPORATION OF nptII ~ND hprt GENE SEQUENCES
21 Average of Number of Colonies Formed per 1 x 1o6 Cells ~
22 (~g of DNA (# = Number of trials averaged)) .. :
24 Ratio 200 100 50 25
of :-
26 DNAG418R TGR #G418R TGR # G418R TGQ # G418R TGR # :
28 1:1233 2.7 3101 1.5 3 64 0 3 23 0 5
29 1:1046 0 316 0 5 8.7 0 7 nd nd :~
1:100 8 0.2 5 4.3 0 7 1.6 0 7 nd nd
31
32 This experiment shows that co-electroporation of an h~rt
33 gene sequence with an nptII-containing gene sequence in the
34 presence of selection ~or only the n~tII-containing
sequence, resultad in recombination of both the nPtII and ~ -
36 h~rt DNA molecules. ~
'' ~ ' ~,'' .
, -, , - :. . ., ., , :
!, . ' ' , , . , , ~ . ' , '
' ' ' ' ' ' ' ' : " ~' ' :
WO 91/l!J7Y~ JI/U4UUO
- 6 3 ~
1 The frequencies of recombination are shown in Table 2
2 below.
4 TABLE 2
FREQUENCY OF RECOMBINATION `~
7 ExptRatio [DNA]G418R/10s TGR/107 TGR/G418R
8 Neo:Hprt ~g/ml
A 1:1 20023.3 2.6 1/873
11 B 1:1 10010.1 1.0 1/1010
12 C 1:100 200 0.8 0.2 1/400* :
13 Cont --- 25 10.8 2.7 1/402
14 ~;~
15The reactions were carried out as described above. The
16reproducibility of the experimental results was examined.
17The results of this experiment are shown in Table 3.
1~
19 TABLE 3 ~:
20EFFECT OF MODIFIED CO-ELECTROPORATION PROTOCOL OM ~ :
21RECOMBINATION FREQUENCY
22
23 Molar DNA # G418RIHPRT- HPRT- HPRT- G418R
24 ratios per of colonies G418R (per cell
zap zap (total) transfected)
~ Neo:(~g) (X 10-9)(X 106) ~ .
28 1:1 200 816,150 / 32 1/504 400 202 :
29 100 3 3,030 / 3 1/1,010 100 105
S0 3 1,920 / 0 67 ~
31 25 5 1,150 / 0 24 ~:32 1:10 200 16 608 / 7 1/868 ` 43 47 :. : `
33 100 5 800 / 0 17 ::
34 50 7 609 / 0 9
1:100200 5 400 / 1 1/400 8
36 lO0 7 300 / 0 4.5
37 50 7 ~12 / 0 1.7
38 :::
,,`,~
9.
.. .
, . .
., ~
''`, , ` '
,'','~,'
:
: .
'. :' : ', ` . ~ : ' ` ' ' `
~........... '`: ' , ~
WO 91/19796 PCI`/l,'S91/04006
2 ~ 7 ~-~
--64--
E~lPLE 3
2 HOMOLOGOI~S RECOMBIN~TION
4 In order to investigate the chromosomal structure ;
which is produced by the recombination of the vectors of the
6 above-described vectors into the chromosomes of recipient
7 cells, the following experiments were conducted.
8 For this purpose, a vector was used which contained a
9 6.5 kb region of homology with the cellular h~rt locus. The
vector also contained the n~tII gene, as a selectable ma~er.
11 The vector was linearized with XhoI and provided to ES cells
12 by electroporation, as described above. Cells which became
13 resistant to G418 were selected and their DNA was analyzed --
14 to determine if it contained restriction fragments that were
lS consistent with the predicted integration structure.
16 The vector used, and the predicted integration structure
17 are illustrated in Figure 8. Gel electrophoresis of
18 restriction digests of cellular DNA confirmed that the G418
~9 resistant cells contained the bE~rt structure shown in Figure );
8. This finding confirmed that the vector had integrated
21 into the chromosome of the cell by homologous recombination
22 at the hPrt locus. -~
23
24
~XAMPL~ 4
26 REVERSION OF RECOMaINAN~8 ~-~
27 ~ ~
28 The effect of the size of the region of homology carried ~ -
.
29 by the vector-on the reversion frequency of recombinants was
deter~ined. Recombinants containing a vector having 6.8 kb
31 of homology with the h~rt locus were prepared as described
32 - in Example 3. Using the same method, reco~binants were also
33 prepared which contained a similar vector having only 1.3 kb ;~
.' ,
"
- , . : . . . . . . . .
. . . :. : . . . . ,; . , . . :
~ " . . . ' ., . . , ' ~. ., ' ' I" ',, ,,.. ,., ' , . ,, 1, . " , , " " ,
: . . ; . .
,. , ,.. . . . . : .
WO 91/19796 PCr/l:'S91/04006
-65--
1 of homology with the hprt locus. The structures of the
2 insertion site of the 6.~ kb vector ls illustrated in Figure
3 8. The reversion frequency of the two constructs is shown
4 in Table 4. The structure obtained from the reversion of
the insertion is shown in Figure 9.
7 TABLE 4
8 REVERSION FREQUENCY
Du~lication~ Clones ~ Revertible Freauencv xlO-s
12 6.8 kb l9 19 3.3 to 0.2
13 1.3 kb 2 2 1.2 to 0.3
14
16 E~MPLE 5
17 TARGETING FREO~ENCY OF :~NSERTION AND PcEPL~CEMENT VECTORS ~:
18
19 A series of different Yectors were used to investigate
the targeting freguency achieved through the use of the ~;
21 methods of the invention. These vectors contained 6.8 kb of
22 homology with the murine h~rt gene and had regions of
23 heterology either at the linearization site or internally
24 (Figure 2).
For this pur~ose, 108 cells were electroporated into ES
26 cells, prepared as described above, and plated onto 10 x 90
27 mm plates. After 24 hours G418 (at 350 ~g/ml) was added to
28 the media. After 5 days ~election 105 M 6-thioguanine was
29 added to 9 plates, 1 was retained under G418 selection as
the transfection control. Selection was continued for an
31 additional 7 days. Colo~ies were scored at this time and
32 expanded ~or south~rn analysis as separate clones.
33 Targetiny efficiencies are detailed for each of the vectors
34 IFigur~ 2; Table 5).
.
~ ,
. ~ . .~. . : ,
.: . . - : ~ . . :~ . . - . .. : .. -
. : , , - :
, . : . : .
WO 9 1 / 1 9796 PCT/ l lS9 1 tO4006
r >~
~~ r ~
--66--
1 Southern analysis showed that the majority of the 6-TGR
2 clones had the predicted integration structure depicted for
3 HindIII digestion in Figure 8.
4 Reversion of the hprt clones was done by measuring HATR.
Cells were clonally expanded under 6-TG selection o prevent
6 "jackpot" effects cau~ed by the early recombinational loss
7 of the duplicated element giving rise to a large number of
8 colonies by cell division. When 107 cells were obtained, the
9 cells were reseeded onto 90 mm plates without selection for
48 hours. After 48 hours HAT selection was applied and
11 resistant colonies were scored 10 days later, typically 20
12 to 200 colonies were observed per 107 cells plated (Table 4).
13 Every clone examined reverted at a similar frequency.
14
TABLE 5
16 REPLACEMENT AND INSERTION VECTORS: TARGETING AND FREQUENCY
17
18 Gene Homoloov Vector Frequencv
19 -:
~prt 6.8 kb RV 1/300 ~ 10X
21 ~prt 6.8 kb IV 1/32
22
23 Hprt 1.3 kb RV <1/5000
24 minimum 1 12x :.
Hprt 6.8 kb IV 1/400 -
26
~7 Hox2.6 3.2 kb IV+ 1/33
28 :
29 RV=Replacement Vector; IV=Insertion Vector ~ ~
. ~ :
31
32
33
34 EXA~P~E 6
6ELECTION FOR HO~O~OGO~8 ~ECOM~INATION
36 .
37 It is possible to use "Poly A Selection" in order to ~ -
38 enhance the selection of cells which have integrated the
39 introduced DNA by homologous recombination.
'~
...
~ .
.
.
: :. . : . - - .
: - :. :. . .. : :
,.... . , ~ . .. . . . ..
~.~ :- . . .. : , . : . . ' : .
:
. : . , . : . : .. . .
:
:-: ,. : . . -
:.: . - : - .
WO ~1/19796 S~Cr/-~591/04006
- 6 7 ~, 'C v ~-~ r ~ ~
1 If an introduced DNA molecule were to integrate at
2 random into the host chromosoI~e, it would generally ~ot
3 integrate at a site adjacent to a necessary 3'
4 polyadenylation site. Thus, the mRNA produced by the
transcription of sùch randomly inserted constructs would
6 generally lack polyadenylation. This fact can be exploited
7 by using vectors which permit one to select for a
8 recombinational event that results in integration adjacent
9 to the natural polyadenylation site of the introduced gene
sequence (i.e. by homologous recombination rather than by
11 random insertion).
12 To illustrate this aspect of the invention, three
13 vectors were constructed which contain fragments of the hprt
14 gene (Figure lO). As shown in Fiqure 10, the vectors
contain exons 7, 8, and 9 of the hPrt gene. The
16 polyadenylation site is located in exon 9. A ~inDIII site
17 is present within exon 9, and an EcoRI site is located after
18 the end of the exon.
19 The first vectoE em~)loyed contained a 5.0 kb region, and
thus contained the polyadenylation site of exon 9 (Vector 6,
21 Figure 10). As shown in Table 6, the frequency of insertion
22 was high (i.e. frequency of G418 resistant colonies was 24
23 x 105), but only 1/941 colonies showed the dual thioguanine
24 resistance and G418 resistance which would characterize a
desired recombinant (i.e. a recombinant in which integration
26 had resulted in an intact h~rt gene and an i~tact n~tII
27 gene)0 Thus, some random integration is occurring.
28 Similarly, when a vector of 3.5 kb was employed (Vector
29 lO) which contained DNA from the XbaI site to the coRI site
of Vector 6, the ~reguency of insertion was high (i.e.
31 frequency of G418 resistant colonies was 21 x 105), but only
32 1/770 colonies showed the dual thioguanine resistance and
33 G418 resistance which would characterize a desired
. .
.
., ~ . , . - ,: . .
.~ - ~ . , . -
- ~ .': . ~, :,
W091~19796 PCT/~!S91/04006
-68-
l recombinant (Table 6). This findlng demonstrates that some
2 random integration is occurring.
3 ~f, however, a vector is employed which lacks the
4 polyadenylation site of exon 9 (i.e. Vector 9), random
integration does not result in expression of a functional
6 nptII transcript. Thus, the frequency of G418 resistant
7 colonies is low ( 1 . 4 X lo-5? . Since the number of colonies
8 evidencing random integratio~ is suppressed, the overall
9 frequency of recovery of the desired recombinants is
enhanced (i.e. an overall ef~iciency of l/lOO for the dual
ll resistant colonies (Table 6). Thus, the poly A selection
12 results in an approximate increase of overall efficiency of
13 nearly lO fold. Poly A selection may therefore ~e ;;
14 advantageously used in situations where one desires to
lS minimize or avoid the screening of colonies to identify ~'
16 random versus homologous recombinants.
17 , ;
l8 TABLE 6 ~--
l9 POLY A SELFCTION -
2.1 VECTOR SIZE G418R TGR TGR/G418R
22 # (kb) (x lO-s) (x 107)
23
24 6 5.0 24 2.S l/941 ~ ~ -
9 3.0 l.4 l.4 l/lOO
26 lO 3.5 21 2.7 l/770
27 -~-
28
29
E2A~P~E 7
31 ~NTROD~TION OF 8~T~E ~TATION8 IN ~RE C-8RC ~0~8
32
33 ~he methods of the present invention were further
34 illustrated by their use to produce cells having precise and
subtle mutations in the c-src locus of ES cells. The c-src ~-
36 locus contains several exons, which are designated as
37 ~'boxed" regions 2 and 3' in Figure ll. As shown in Figure
~.
.
.,
.. ... .. , ,. .. ... - ~ ; ... , - :. . . - :
. : .. . , .,, :, :
.
:: :. . . . . .
WO 91/19~96 PCrt~lS91/04006
7~3~
-6~-
1 llA, the natural allele of exon 3' does not contain a
2 HindIII site.
3 The sequence of a portion of exon 3' is shown in Figure
4 llC. As shown in Figure llC, a 9 bp i~sertion into ~his
exon will result in the formation of a HinDI~I site.
6 To accomplish this change in the sequence of exon 3', a
7 vector (src 14) was prepared. As shown in Figure llB, the
8 src 14 vector is homologous to a region of the c-src locus.
9 The exon 3' sequence of the vector, howe~er, has been
altered to contain the 9 base pair insertion needed to
11 creat~ a HindIII site (Figure llC).
12 The src 14 vector was introduced into E5 cells by co-
13 electroporation with a second vector (PGKneo) that oontained
14 the ~E~I gene, at a total DNA concentration of 25 ~g/ml and
a molar ratio of 1:5 (neo vector to targeting vector) in the
16 manner described above.
17 Cells were cultured in the presence of G~18 for 12 days
18 in order to select for recombinant cells in which the nptII
19 gene had integrated. These recom~inant cells were then
screened, using PCR, for cells which had undergone a
21 recombinational event resulting in the replacement of the
22 natural exon 3' locus with the HinDIII site-containing exon
23 3' sequence of the src 14 vector.
24 Southern analysis of the colonies identified by PCR
screening using probes B and C (Figure llB) demonstrated
26 that the natural exon 3' locus had been altered, as de~ired,
27 to contain a HinDIII site. This experiment demonstrated
28 that subtle insertions can be introduced into any`cellular
29 gene.
To further illustrate the capacity o~ the present
31 invention to introduce complex, predetermined mutations into
32 the geno=e of a recipient cell, exon 3" ol the c-sr~ gene of
~ '
WO9l/19796 PCT/~'S91/04006
-70-
1 an ES cell was mutated to contain two different substitution
2 mutations.
3 As shown in Figure 12A, the natural allele of exon 3"
4 does not contain either an NheI site or an EcoRI site. As
shown in Figure 12C, howe~er, thP replacement of the natural
6 sequence ACC TGG TTC of exon 3" with the sequence TAG CTA
7 GCT will result in the formation of an NheI site.
8 Similaxly, replacement of ACA with GAA in exon 3" will
9 create an EcoRI site (Figure 12C).
To accomplish these changes in the sequence of exon ",
11 a vector (src 33) was prepared. As shown in Figure 12B, the
12 src 33 vector is homologous to a region of the c-src locus.
13 The exon 3" sequence of the vector, however, has been
14 altered to c~ntain the substitutions indicated above (Figure
12C).
16 The src 33 vector was introduced into E5 cells by
17 electroporation, in concert with a second vector that
18 contained the nptII gene, in the manner described above.
19 Cells were cultured in the presence of G418 in order to
select for recombinant cells in which the n~tII gene had
21 integrated. These recombinant cells were then screened,
22 using PCR, for cells which had undergone a second
23 recombinational event resulting in the replacement of the
24 natural exon 3" locus with the exon 3" sequence of the src
33 vector.
26 Southern analysis of the colonies identified by PCR
27 screening using probes A and C ~Figure 12C) demonstrated
28 that the natural exon 3" locus had been altered, as desired,
29 to contain both the NheI and the EcoRI sites. This
experiment demonstrated that subtle substitutions can be ~
31 introduced into any cellular gene. -
32 While the invention has been described in connection
33 with specific embodiments thereof, it will be understood
.
~: :: `' ,
W091/19796 PCT/~'S9l/04006
-71-
1 that it is capable of further modifications and this
2 application is intended to cover any variations, uses, or
3 adaptations of the invention following, in general, the
4 principles of the invention and including such departures
from the present disclosure as come within known or
6 customary practice within the art to which the invention
7 pertains and as may be applied to the essential features
8 hereinbefore set forth and as follows in the scope of the
9 appended claims.
',, ,;
.
,`
~',
'
`
, ., : ,: ~.