Note: Descriptions are shown in the official language in which they were submitted.
~o ~t~ 79~ 2 ~ 9 ~ ~ 3 ~ PC~ S9~/OX2:~
. . .. .
Chinese Hamster Ovary Cell retrov~rus-l~ke C particles
Backaround of ~he Invention
This invenlion relates to recombinant cell culture lines and methods, particularly the
identification and use therein of nucleic acid sequences encoding retrovirus-like particles.
Retrovirus-like particles are observed in ail Chinese hamster ovary (CHOI cells, when
thin-sections of cells are viewed by transmission electron microscopy. Two types of particles
are consistently observed: Intracytoplasmic A-type particles frequently associated with
centrioles, and budding C-type particles {Donahue, P. R. et~l., J. Viro/. 62: 722-731 (19881;
Lieber, M.M. Science 182:56-58 [1973]; Lubiniecki, Develp.~iol.St~ndards 70:187-191
[1989]; Lubiniecki et ~1., Develp.Blol.St~ 60:141-146 [1985]; Manley, I<.F. J.Gen.Vlrol.
39:505 517 [1978]; Tihon, C., Nat.New Biol. 244:227-231 [1973]}. Infection or
transmission to other cells has never been detected ~Ibid., and Hojman, F.R., Develop. Biol.
70:195-202 [1990}~. Despite their apparently non-infectious nature, the origin of the
observed retrovirus-like particles is of interest since recombinant CH0 cells are used as
substrates for the production of biopharrnaceuticals intended for human use {Collen, D.,
J.Ph~rm.~xp. 231:146-152 [19841; Patzer, E.J., BioTechnology 4:630-636 [1986l; Egrie,
J.C., Prog. Clin.Biol. 191 :339-350 [1985]}. To the extent that reverse transcriptase activity,
indicative of potential biological activity, is shown to be associated with retrovirus-like
particles in recombinant cell lines, concern is heightened regarding potential transmission of
virus to recipients of biopharmaceutical products. ~r
Antisense technology has previously been used to inhibit expression of specific gene
products in mammalian cell lines (Kasid et a/., Science 243:1354-1356 [19891; Khoka et a/.,
Science 243:947-950 [19891: Izant et a/., Science 229:345-352 [1985]) including some
retroviruses: (von Ruden et al., J. Virol. 63:677-682 [1989l: Chang et ~1., J. Virol.
61 :921-924 [19871), but not all attempts have been successful ~Kerr et a/., Eur. J. Biochem.
175:65-73 [1988]). The secondary structure of the target RNA may influence susceptibility
to antisense inhibition. Antisense inhibition is currently believed to require an excess of
antisense RNA relative to coding mRNA in order to be effective.
Ribozymes are small RNA molecules that can cleave substrate RNAs in a sequence
specific manner. For example, "hammerhead" ribozymes are described by Haseloff et al.,
Nature 334:585-591 [1988] which cleave mRNAs at specific sites in vitro. With these
hammerhead ribozymes, target sequences need only contain a GUA or GUC sequence at
which cleavage occurs. Specificity of cleavage is determined by flanking sequences which
hybridize to the ribozyme sequence. Thus, ribozymes can be engineered to cleave at any 18
bp RNA sequence flanking a GUA or GUC triplet, thus generating very precise specificity.
Published literature also describes RNA enzymes with highly specific endoribonuclease
activities, such as those disclosed by Koizumi et a/., FEBS Lett. 239:285-288 [19881.
~ . .
- ~:
~O 9~/0~,9~ .
2 ~ ~9 ;~ ~ 3 5 PCI /I~S91/08~
-2-
Ribozymes have also been used with in vivo applications; for example, in unpublished
data, it has been demonstrated that insertion of a ribozyme sequence into a tRNA gene
improvss stability in v;vo, and that tRNA/ribozyme constructs specific for the tat gene of
HIV-I may inhibit transactivation of the HIV-I LTR promoter by the HIV-I tat product
Because of the low numbers of retrovirus-like particles heretofore shown to be present
in CH0 cells, recognition and characterization has been difficult. It is therefore an object of
this invention to identify and characterize novel extracellular C-type retrovirus-like particles
in culture fluid from a recombinant CH0 cell line. Methods for the detection of these particles
are also objects of this invention.
A further object of this invention is to provide methods for increased or decreased
expression of these retrovirus-like particles in cultured cells.
Methods for the isolation of cell lines with reduced or undetectable levels of C-type
particles, or reverse transcriptase activity associated with those particles, are provided. Such
cell lines provide an ideal parental line for subsequent genetic modification and use as a
t5 substrate for production of biopharmaceuticals, because regulatory requirements for viral
clearance and inactivation studies during manufacturing processes would be minimized and
concerns regarding transmissibility of retrovirus particles would be obviated.
Summar~ the !nvention
The objects of this invention are accomplished through novel nucleic acid sequences
which encode extracellular retrovirus-like particles and provirus sequences. Further objects
have been accomplished by a method comprising providing cells transformed with such
nucleic acid, or cells containing such nucleic acid operably linked to exogenous control
sequences, culturing the host cell to allow the retrovirus-like particles to accumulate in the
culture, and recovering them from the culture.
Surprisingly, a full-length clone encoding these retrovirus-like particles has been
identified and recovered. These sequences have been found to be present as conserved,
repetitivej provirus sequences in genomic DNA of CH0 cell lines and in Chinese hamster liver
DNA,
Polynucleotide probes can be made for determining the level of transcriPtion of said
DNA in cells and receptors produced capable of specifically recognizing determinant sites of
peptide products of said DNA sequences~; The probes and receptors may be labeled with a
wide variety of labels for a variety of applications.
Yet another object is to provide molecules with novel characteristics for use asdiagnostic reagents for the in vitro assay of the retroviruses. The sequences provided by this
invention have homology to the conserved endGnuclease domain of a murine leukemia virus,
but the present sequences contain multiple interruptions of the endonuclease-encoding
reading frame. Because the ciaimed sequences do not effectively code for an endonuclease
. :.
,
2 1~ g ~ PCT/~'S91/o~
-3-
which is required for virus integration and replication, the retrovirus-like particles of this
invention are presently believed to be non-infectious.
Methods for increasing or decreasing expression of these particles in cell culture are
provided, utilizing complementary (antisense) nucleic acid sequences, ribozymes, RNA
nucleases, DNA control sequences and exogenbus translation modulatory elements. Cells
having increased or decreased expression of these particles are provided, as weli as methods
for the detection of expression levels.
The compositions and methods of this invention find application to the detection of
contaminants in recombinant cell culture production, for example through the use of PCR,
immunoassay or other means of identifying these retrovirus-like particles in a product sample.
Brief Description of the Draw!ncls
Figure 1~ Purification of particles from CHO 3-3000-44 cells using a double sucrose
gradient protocol. 4000-fold roncentrated culture fluid from CHO 3-3000-44 cells was
adjusted to 50% sucrose by addition of powdered sucrose and overlayed with successive
layers of 40%, 30% and 20% sucrose. After centrifuging to equilibrium RT containing
fractions from the main peak, shown in panel A, were pooled, dialyzed, and layered on top
of a preformed 10-60% sucrose gradient and again centrifuged ~o equilibrium. RT activity
in the second gradient is shown in panel B.
Figure 2. Electron micrographs of double-gradient purified particles from CHO
3-3000-44 cells. RT containing fractions from the second gradient of a double-gradient
purification were pooled, concentrated, and dialyzed. After spotting onto EM grids, particles
were visualized by negative staining using uranyl acetate. Shown are three examples of the
particles observed. The size and morphology are consistent with previous descriptions of
retrovirus particles visualized by this technique.
Figure 3. Retrovirus-related proteins and nucleic acids in gradient puri~ied particles.
Pools of 4 fractions each from the second gradient of a double-gradient particle purification
~shown in Fig. 1B) were evaluated for the presence of C-type structural proteins and
retrovirus nucleic acid sequences. (A) Western blot analysis using goat anti-FeLV p27 serum
as probe for C-type retrovirus structural proteins. The p31 band represents the major core
protein and the p64 band represents unprocessed gag precursor. (B) Nucleic acids were
extracted from samples of each pool, applied to membranes and hybridized to the indicated
probes. Cytoplasmic RNAs from CHO cells and MRC5 cells (human diploid fibroblasts) were
used as controls.
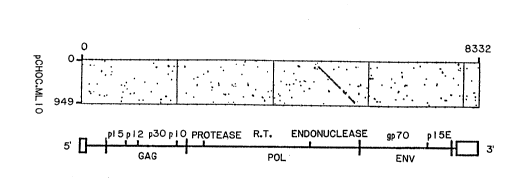
Figure ~. Homology of pCHOC.ML'iO cDNA with Moloney MLV genome. The
35 nucleotide sequence of the Moloney MLV genome {Shinnick, T., N~ture 293:543-548
11981 ]} is plotted on the horizontal axis, and the pCHOC.ML10 nucleotide sequence is plotted
on the venical axis. Each do~ represent 75%: identity over a stretch of 16 nucleotides.
. : .
.
~\'0'~0f~9~ 2 ~ 3~ PCT/-S'~I/OX~
~4-
Regions of significant homology appear as diagonal lines. Organization of the Moloney'MLV
RNA genome is shown for orientation.
Figure 5. Initiation and stop codons of the pCHOC.ML10 sequence. Methionine
codons ~M) and stop codons (O) in the three reading frames of the strand collinear with the
Moloney MLV genome are shown. Multiple interruptions of coding sequences in all three
reading frames are present. No significant open reading frames were observed on the
opposite strand.
Figure 6. Blot analysis of C-type provirus sequences in senomic DNA of CHO cellsand Chinese Hamster liver. Hindlll digested DNAs (2 ~9 per lane) isolated from parental and
10 recombinant CHO cell lines and from Chinese hamster liver were fractionated on agarose gels,
blotted onto membranes and hybridized with retrovirus probes. CHO-1<1 is the ancestral cell
line from which all others shown are derived. CHO-dhfr' is the parent line, CHO-DUX~11,
from which the recombinant line, CHO 3-3000-44 (designated here as CHO-44) was derived
following transfection with an expression vector. Copy number controls are restriction
15 enzyme cut plasmid loaded onto the gel so that the amount of insert is equivalent to the
indicated number of copies per Chinese hamster haploid genome. Labeled Hindlll fragments
of lambda DNA were used as molecular weight markers (MW). Sizes in kilobase pairs of
molecular weight markers are indicated to the right of each panel. Deduced sizes of major
genomic Hindlll fragments hybridizing with each probe are shown to the left of each panel.
20 (A) C-type related sequences were detected by hybridization with the pCHOC.ML10 partial
cDNA clone representing purified particle RNA tsee text). Copy number controls are EcoR1
digested pCHOC.ML10 plasmid. The 0.6 kb genomic Hindlll fragment was predicted from
the sequence of the pCHOC.ML10 clone used as probe. (B) CHO IAP family ll related
sequences were detected by hybridization with the pCHlAP.YL9 cellular cDNA clone25 {Anderson, K. P. et al., J. Viro/. 64:2021-2032 11990]~. Related sequences were observed
in purified extracellular particles of CHO cells (see Figure 3). Copy number controls are
Hindlll digested pCHlAP.YL9 plasmid.
Detailed Descrirtion
C-type retrovirus-like particles are budding particles with retrovirus-like morphology and
30 immunological characteristics, described generally in the literature cited above.
Intracytoplasrnic A-type particles frequently associated with centrioles are specifically
excluded from this definition. C-type retrovirus nucleic acid sequences are defined herein as
sequences encoding such particles, allowing for multiple interruptions of Potential coding
sequences.
Homology with respect to a C-type re;rovirus sequence js defined herein as the
percentage of residues in the candidate sequence that are identical with the residues in the
sequences identified below as Seq. !D No. 1 and Seq. ID No 2., after aligning the sequences
and introducing gaps, if necessary, to achieve the maximum percent homology. Homology
::
:
2 B 9 ~ 6 3 ~ Pcr/~ s9l/ox~ ~
5-
with respect to a C-type retrovirus particle is definecl similarly with respect to the particles
encoded by such sequences. Where the candidate sequence is a fragment of a the particles
encoded by Seq. ID No 1, it preferably but not necessarily has at least ~75,c homology to
the C-type retrovirus particle as defined herein.
Included within the scope of the C-type retrovirus particle as that term is used herein
are particles having the amino acid sequences encoded by Seq. ID No. 1, glycosylated or
unglycosylated derivatives of the particle, homologous amino acid sequence variants of the
sequences encoded by Seq. ID No. 1, and homologous in vit~o-gener3ted variants and
derivatives of the amino acid sequences encoded by Seq. ID No. 1, which are capable of :
exhibiting a biologicai activity in common with the particles encoded by Seq. ID No. 1.
C-type particle biological activity is defined as immunological cross-reactivity with at
least one epitope of a C-type retrov;rus-like particle encoded by Seq. ID No. 1.
:
Immunologically cross-reactive as used herein means that the candidate polypeptide
is capable of competitively inhibiting the qualitative biological activity of the C-type retrovirus-
like particle having this activity with polyclonal antisera raised against the known active
analogue. Such antisera are prepared in conventional fashion by injecting goats or rabbits, : .
for example, subcutaneously with the known active analogue in complete Freund's adjuvant,
followed by booster intraperitoneal or subcutaneous injection in incomplete Freunds.
An "exogenous" element is defined herein to mean foreign to the cell, or homologous
to the cell but in a position within the host cell in which the element is ordinarily not found. .
Novel nucleic acid compositions are provided herewith. This invention encompasses
the following DNA sequence (Seq. ID No. t ) which encodes an RNA transcript of a C-type
retrovirus-like particle endogenous in CH0 cells.
~ :
Sequence ID No. 1: 15' to 3') . `
GAATTCAATG TCCTTGTGGG GAAGAACGAT ATTCTAAAAC AGGTAACTGA 50 :
GCAATGTGAT GCGTGCGCCC GAGTCAACGC ATCCAGACTG AAGCTTCCTC 100
CCGGGAACCG GTCAGAGGCT ACCGGCCCGG AACACATTGG GAGATAGATT 150
TCACTGAGAT TAAACCAGGA AAATATGGAT ACAAGTATCT ATTAATTTTT 200 ~ :
GTAGACACCT TTTCAGGATG GGTTGAAGCC TTCCCTACTA AACATGAAAC 250 .
AGCCAGATCG TTACTAAGAA ATTGCTTGAA GAAATCTTTC CCCGTTATCG 300
: :GATGCCTCAG GTATTGGGAA CAGACAATGG GCCCGCCTTC GTCTCCAGGT 350 ..
: AAGTCAGTCA GTGGCCACCT~TATTGGGGAT TGATTGGAAA TTACATTGTG 400
CTTATAGACC CCAAAGTTCA GGACAGGTAG AAAGGATGAA TAGAACAATC 450
AAGGAGACTT TAACAAAATT GTCGCTTGCA ACTGGCACTA GAGAGCTGGG 500
TCCTCCTACT CCCCCTAGCA CTCTACCGCG CTCGTAATAC CCCTGGACCA 550
CATGGGCTCA CACCCTTTGA GATCCTGTAT GGAGTACCTA GCTCCTATCA 600
TTAACTTTCT TGATCAAGAT GTCTCAGTTT TGCTAACTCC CCTTCTCTCC 650
AAGCTCATTT ACAGGCCCTC CAACTAGTAC AACGGGAGGT CTGGAAACCC 700
CTTGCTCAAG CTTATAAAGA CCAGAGGGAC CATCCCACCA TCCCCCATTC 750
CTACCAGATC GGGGACACTG TTTGGGTCCG GCGTCACCAG GCCAAGAACC 800
' ~
~ : -
~\ 9--/0~ 2 ~ ~ ~ 6 3 ~ Pcr/~ S91 /OX_~ !
-6- f
TTGAACCCCG ~TGGAAGGGA CCCTACATCG TTTTGCTTAC CACTCCCACC 850
GCACTC~AGG TAGACGGCAT TGCAGCTTGG ATACATGCTT CACATGTAAA 900
GCCAGCCCAA CCCACCGATT CAGCCACTGC ATCAGAATGG AGCCACAC~ 949
This invention further encompasses fragments of such DNA and RNA sequences which retain
hybridization specificity.
Novel nucleic acid compositions which are complementary lantisense) to the
sequences just described are encompassed herewith. The following DNA sequence (Seq. ID
No. 2) is complementary to Seq. ID No. 1, and encodes an RNA transcript complementary to
the RNA transcript of the C-type retrovirus-like particle endogenous in CHO cells described
1 0 above.
Sequence ID No. 2: (5' to 3'):
GGTGTGGCTC CATTCTGATG CAGTGGCTGA ATCGGTGGGT TGGGCTGGCT 50
TTACATGTGA AGCATGTATC CAAGCTGCAA TGCCGTCTAC CTT~AGTGCG 100
GTGGGAG~GG TAAGCAAAAC GATGTAGGGT CCCTTCCAGC GGGGTTCAAG 150
GTTCTTGGCC TGGTGACGCC GGACCCAAAC AGTGTCCCCG ATCTGGTAGG 200
AATGGGGGAT GGTGGGATGG TCCCTCTGGT CTTTATAAGC TTGAGCAAGG 250
GGTTTCCAGA CCTCCCGTTG TACTAGTTGG AGGGCCTGTA AATGAGCTTG 300
GAGAGAAGGG GAGTTAGCAA AACTGAGACA TCTTGATCAA GAAAGTTA~T 350
GATAGGAGCT AGGTACTCCA TACAGGATCT CAAAGGGTGT GAGCCCATGT 400
GGTCCAGGGG TATTACGAGC GCGGTAGAGT GCTAGGGGGA GTAGGAGGAC 450
CCAGCTCTCT AGTGCCAGTT GCAAGCGACA ATTTTGTTAA AGTCTCCTTG 500
ATTGTTCTAT TCATCCTTTC TACCTGTCCT GAACTTTGGG GTCTAT~AGC 550
ACAATGTAAT TTCCAATCAA TCCCCAATAA GGTGGCCACT GACTGACTTA 600
CCTGGAGACG AAGGCGGGCC CATTGTCTGT TCCCAATACC TGAGGCATCC 650
GATAACGGGG AAAGATTTCT TCAAGCAATT TCTTAGTAAC GATCTGGC~G 700
TTTCATGTTT AGTAGGGAAG GCTTCAACCC ATCCTGAAAA GGTGTCTACA 750
AAAATTAATA GATACTTGTA TCCATAT~TT CCTGGTTTAA TCTCAGTGAA 800
ATCTATCTCC CAATGTGTTC CGGGCCGGTA GCCTCTGACC GGTTCCCGGG 850
AGGAAGCTTC AGTCTGGATG CGTTGACTCG GGCGCACGCA TCACATTGCT 900
CAGTTACCTG TTTTAGAATA TCGTTCTTCC CCACAAGGAC ATTGAATTC 949
Within the scope of this invention is this complementary DNA, as well as complementary RNA
transcripts, and fragments of such complementary DNA and RNA sequences which retain
hybridization specificitY~
This invention also contemplates amino acid sequence variants of the C-type particles
encoded by Seq. ID No. 1. Amino acid sequence variants of the particle are prepared with
various objectives in mind, including increasing the binding affinity of the particle for its
ligand, modulating its infectivity/transmissibility, and facilitating its stability, purification and
preparation .
Amino acid sequence variants of the retrovirus-like particles of this invention fall into
one or more of three classes: Insertional, substitutional, or deletional variants. These
variants ordinarily are prepared by site specific mutagenesis of nuclsotides in the DNA
.
,
,' '
: ~.
- :.
~ 2 0 9 ~
encoding the particle, by which DNA encoding the variant is obtained, and thereafter
expressing the DNA in recombinant cell culture. However, fragments having up to about
100-150 amino acid residues are prepared conveniently by in vitro synthesis.
The amino acid sequence variants of the C-type retrovirus-like particle of this invention
are predetermined variants not found in nature or naturally occurring alleles. The variants
typically exhibit the same qualitative biological--for example, receptor binding--activity as the
naturally occurring analogue. However, the variants and derivatives that are not capable of
binding to a receptor are useful nonetheless ~a) as a reagent in diagnostic assays for the C-
type retrovirus-like particle of this invention or antibodies thereto, ~b) whan insolubilized in
accord with known methods, as agents for purifying anti-retroviral antibodies from antisera
or hybridoma culture supernatants, and (c) as imrnunogens for raising antibodies to the C-type
retrovirus of this invention or as immunoassay kit components llabelled. as a competitive
reagent for the native particle or unlabelled as a standard for the C-tyPe particle assay) so
long as at least one C-type retrovirus-like epitope remains active.
While the site for introducing an amino acid sequence variation is predetermined, the
mutation per se need not be predetermined. For example, in order to optimize theperformance of a mutation at a given site, random or saturation mutagenesis Iwhere all 20
possible residues are inserted) is conducted at the target codon and the expressed particle
variant is screened for the optinnal combination of desired activities. Such screening is within
the ordinary skill in the art. .
Amino acid insertions usually will be on the order of about from 1 to 10 amino acid
residues; substitutions ara typically introduced for single residues; and deletions will range
about from 1 to 30 residues. Deletions or insertions preferably are made in adjacent pairs,
i.e. a deletion of 2 residues or insertion of 2 residues. It will be amply apparent from the
following discussion that substitutions, deletions, insertions or any combination thereof are
introduced or combined to arrive at a final construct.
Insertional amino acid sequence variants are those in which one or more amino acid
residues extraneous to the C type retrovirus-like particle are intrr duced into a predetermined
site in the target particle and which displace the preexisting residuesi
Commonly, insertional variants are fusions of heteroloqous proteins or polypeptides to
the amino or carboxyl terminus of the particle. Such variants are referred to as fusions of the
retrovirus-like sequence and a polypeptide containing a sequence which is other than that
which is normally found in the particle at the inserted position. Several groups of fusions are
contemplated herein.
The novel polypeptides of this invention are useful in diagnostics or in purification of ~ -
the C-type particle by immunoaffinity techniques known per se.
: .:
........................................................................................................ .: :: -::
:, .
,: , . :.
::
~\ o 9~0~7~ PCr/~ S9 1 /OX~
2~5~3~ -8- ~ .
Desirable fusions of the C-type particle of this invention which may or may not also be
retrovirally active, include fusions of the mature particle sequence with a signal sequence
heterologous to the binding partner.
Signal sequence fusions are employed in order to more expeditiously direct the
5 secretion of the particle. The heterologous signal replaces the native particle signal, and
when the resulting fusion is recognized, i.e. processed and cleaved by the host cell, the
retrovirus-like particle is secreted. Signals are selected based on the intended host cell, and
may include bacterial yeast, mammalian and viral sequences. The native LHR signal or the
herpes gD glycoprotein signal is suitable for use in mammalian expression systems.
Substitutional variants are those in which at least one residue in the sequencesencoded by the Seq. ID No. 1 sequence has been removed and a different residue inserted
in its place. Such substitutions generally are made in accordance with the common practice
when it is desired to finely modulate the characteristics of the particle.
Novel amino acid sequences, as well as isosteric analogs lamino acid or otherwise), as
1 S included within the scùpe of this invention. :
Substantial changes in furlction or immunological identity are made by selecting residue
substitutions that differ significantly in their effect on maintaining (a) the structure of the
polypeptide backbone in the area of the substitution, for example as a sheet or helical
conformation, ~b) the charge or hydrophobicity of the molecule at the target site or (c) the
20 bulk of the side chain.
Some deletions, insertions, and substitutions will not produce radical changes in the
characteristics of the C-type particle.` However, when it is difficult to predict the exact effect
of the substitution, deletion, or insertion in advance of doing so, however one skilled in the
art will appreciate that the effect will be evaluated by routine screening assays. For example,
25 a variant typicallY is made by site specific mutagenesis of the particle-encoding nucleic acid,
expression of the variant nucleic acid in recombinant cell culture and, optionally, purification
from the cell culture for example by immunoaffinity adsorption on a polyclonal anti-C-type
retrovirus column ~in order to adsorb the variant by at least one remaining immune epitopel.
The activity of the cell Iysate or purified particle variant is then screened in a suitable
30 screening assay for the desired characteristic. For example, a change in the character of the
particle such as retroviral activity, is measured by standard assays. As more becomes known
about the functions ln vivo of the particles of this invention other assays will become useful
in such screening. Modifications of such protein properties as redox or therrnal stability,
hydrophobicity, susceptibility to proteolytic degradation, or the tendency to aggregate wi1h
35 carriers or into multimers are assayed by methods weil known to ~he artisan.
Excluded from the scope of deletional variants are the digestion fragments heretofore
obtained in the course of elucida~ing amlno:acid sequences of C-type particles, and protein
.: :..
' ' :: -
, ~
: : .
2 ~ PCr/~'S91/~X~ ~
fragments haYing less than ~ 75% sequence homology to any particle encoded by Seq~ ID
No 1 or 2.
Covalent modifications of the retrovirus-like particle are included within the scope
hereof. Such modifications are introduced by reacting targeted amino acid residues of the
recovered protein with an organic derivatizing agent that is capable of reacting with selected
side chains or terminal residues, or by harnessing mechanisms of post-translational
modification that function in selected recombinant host cells. The resulting covalent
derivatives are useful in programs directed at identifying residues important for biological
activity, for immunoassays or for the preparation of anti-particle antibodies for immunoaffinity
10 purification of the recombinant retrovirus-like particle. Such modifications are within the
ordinary skill in the art and are performed without undue experimentation.
Nucleic acid encoding the C-type retrovirus-like particles of this invention is synthesized
by in vitro methods or is obtained readily from cDNA libraries. The means for synthetic
creation of the DNA encoding the particle, either by hand or with an automated apparatus,
15 ate generally known to one of ordinary skill in the art, particularly in light of the teachings
contained herein. As examples of the current state of the art relating to polynucleotide
synthesis, one is directed to Maniatis et a/" Molecular Cloning--A Laboratory Manual, Cold
Spring Harbor Laboratory (1984), and Horvath et ~ An Automated DNA Synthesizer
Employing Deoxynucleoside 3'-Phosphoramidites, Methods in Enzvmology 154: 313-326,
20 1 987.
Alternatively, to obtain DNA encoding the Particle from sources other than hamster,
since the entire DNA sequence for the preferred embodiment of the C-type retrovirus-like
particle ~Seq. ID No. 1) is given, one needs only to conduct hybridization screening with
labelled DNA encoding the particle or fragments thereof (usually, greater than about 20, and
25 ordinariiy about 50bp) in order to detect clones which contain homologous sequences in the
cDNA libraries of the particular anirnal, followéd by analyzing the clones by restriction enzyme
analysis and nucleic acid sequencing to identify full-length clones. If full length clones are
not present in the library, then appropriate fragments are recovered from the various clones
and ligated at restriction sites common to the fragments to assemble a full-length clone. DNA
30 encoding retrovirus-like particles from other animal species is obtained by probing libraries
from such species with the hamster sequences, or by synthesizing the genes ~n v/tro. DNA
for other retrovirus-like particles having known sequence may be obtained with the use of
analogous routine hybridization procedures.
Provided herein are nucleic acid sequences that hybridize under stringent conditions to
35 a fragment of the DNA sequence of Seq. ID No.1, which fragment is greater than about 10
bp, preferably 20-50 bp,;and even greater than 100 bp. Also included within the scope
hererf are nucleic acid sequences that hybridize under stringent conditlons to a fragment of
~O g~ 79:~ PCT/~iS91/OX~
~9S~3~ -10- ~'
Seq. ID No. 1 or Seq. ID No. 2, and which retain such hybridization specificity to these
sequences.
Included also within the scope hereof are nucleic acid probes which are capable of
hybridizing under stringent conditions to the DNA of Seq. ID No. 1 or 2, or to the genomic
gene for the C-type retrovirus-like particle (including introns and 5' or 3' flanking regions
extending to the adjacent genes or about 5,000 bp, whichever is greater).
Identification of the genomic DNA for the C-type particle is a straight-forward matter
of probing a particular genomic library with the cDNA or its fragments which have been
labelled with a detectable group, e.g. radiophosphorus, and recovering clonels) containing the
gene. The complete gene is pieced together by "walking" if necessary. Typically, such
probes do not encode sequences with less than 75% homology to the particles of this
invention, and they range from about from 10 to 100 bp in length. Homologies and sizes
with respect to other retrovirus-like particles may be determined without undue
experimentation .
~Iybrid DNA technology may be employed for obtaining expression. The DNA sequence
may be restriction mapped and appropriate sites for cleavage defined. In this way, the
sequence may be excised and introduced into a vector having the appropriate control,
transcription modulatory regions and regulatory signals. After obtaining expression of the
DNA sequence, particles may be purified and antibodies can be made to them.
In general, prokaryotes are used for cloning of DNA sequences in constructing the
vectors useful in the invention. For example, E. coli K12 strain 294 IATCC No. 31446) is
particularly useful. Other microbial strains which may be used include ~. co/i B and E. coli
X1776 IATCC No. 31537j. These examples are illustrative rather than limiting.
Alternatively, in vitro methods of cloning, e.g. polymerase chain reaction, are suitable.
The polypeptides of this invention are expressed directly in recombinant cell culture as
an N-terminal methionyl analogue, or as a fusion with a heterologous polypeptide, preferably
a signal sequence or other polypeptide having a specific cleavage site at the N-terminus of
the particle/portion. For example, in constructing a prokaryotic secretory expression vector
for the particle, the nat;ve C-type particle signal is employed with hosts that recognize that
signal. When the secretory leader is "recognized" by the host, the host signal peptidase is
capable of cleaving a fusion of the leader polypeptide fused at its C-terminus to the desired
mature retrovirus-like particle. For host prokaryotes that do not process the retrovirus-like
particle signal, the signal is substituted by a prokaryotic signal selected for example from the
group of the alkaline phosphatase, penicillinase, Ipp or heat stable enterotoxin ll leaders. For
yeast secretion the native signal may be substituted by the yeast invertase, alpha factor or
acid phosphatase léaders. In mammalian cell expression the native signal is satisfactory,
although other mammalian secretory protein signals are suitable, as are viral secretory
leaders, for example the herpes simplex gD signal.
',
~ ~ ,
~ 9~~79`` PCI/~'S91/OX'~
-11- 2~95i63~3
The novel particles of this invention may be expressed in any host cell, but preferably
are synth2sized in mammalian hosts. However, host cells from prolcaryotes, fungi, yeast,
insects and the like are also are used for expression. Exemplary prokaryotes are the strains
suitable for cloning as well as E. coli W3110 (F ,l prototrophic, ATTC No. 27325), other
5 enterobacteriaceae such as Serratia marcescans, bacilli and various pseudomonads.
Preferably the host cell should secrete minimal amounts of proteolytic en~ymes.
Expression hosts typically are transformed with DNA encoding the hybrid which has
been ligated into an expression vector. Such vectors ordinarily carry a replication site
~although this is not necessary where chromosomal integration will occur). Expression vectors
10 also include marker sequences which are caPable of providing phenotypic selection in
transformed cells, as will be discussed further below. For example, E. coli is typically
transformed using p~R322, a plasmid derived from an E. coli species (Bolivar, e~ al.., Gene
2: 95 [1977]). pBR322 contains genes for ampicillin and tetracycline resistance and thus
provides easy means for identifying transformed cells, whether for purposes of cloning or
15 expression. Expression vectors also optimally will contain sequences which are useful for the
control of transcription and translation, e.g., promoters and Shine-Dalgarno sequences (for
prokaryotes) or promoters and enhancers (for mammalian cells). The promoters may be, but
need not be, inducible. While it is conceivable that expression vectors need not contain any
expression control, replicative sequences or selection genes, their absence may hamper the
20 identification of transformants and the achievement of high level particle expression.
Promoters suitable for use with prokaryotic hosts illustratively include the ,B-lactamase
and lactose promoter systems (Chang et al., "Nature", 275: 615 119781; and Goeddel et al.,
"Nature" 281: 544 [1979]), alkaline phosphatase, the tryptophan ItrP) promoter system
(Goeddel "Nucleic Acids Res." 8: 4057 ~1980] and EP0 Appln. Publ. No. 36,776) and hybrid
25 promoters such as the tac promoter IH. de Boer et al., "Proc. Natl. Acad. Sci. USA" 8Q: 21-
25 [1983]). However, other functional bacterial promoters are suitable. Their nucleotide
sequences are generally known, thereby enabling a skilled worker operably to ligate them to
DNA 0ncoding the LHR (Siebenlist et a/., "Cell" 20: 269 [1980]) using linkers or adaptors to
supply any required restriction sites. Promoters for use in bacterial systems also will contain
: 30 ; a Shine-Dalgarno ~S.D.) sequence operably linked to the DNA encoding the C-type retrovirus-
like particle.
In addition to prokaryotes, eukaryotic microbes such as yeast or filamentous fungi are
satisfactory. Saccharom. vces cerevisiae is the most commonly used eukaryotic
microorganism, although a number of other strains are commonly available. The plasmid YRp7
35 is a satisfactory expression vector in yeast ~Stinchcomb, et al.. , Nature 282: 39 [19791;
Kingsmanetal.,Gene~J:141 [1979];Tschemperetal.,Gene10:157[19801). Thisplasmid
already contains the trp1 gene which provides a selection marker for a mutant strain of yeast
- lacking the ability to grow in tryPtophan~ for exarnple ATCC no. 44076 or PEP4-1 (Jones
~0 9'/0~791 PC'r/~S"l/OX'~
3~ -12- ~
Genetics 85: 12 11977]). The presence of the trp1 lesion as a characteristic of the yeast
host cell genome then provides an effective environment for detecting transformation by
growth in the absence of tryptophan.
Suitable promoting sequences for use with yeas~ hosts include the promoters for 3-
phosphoglycerate kinase (Hitzeman et a/., "J. Biol. Chem." 255: 2073 11980~) or other
glycolytic enzymes (Hess et a/., "J. Adv. En~yme Reg." 7: 149 11968]; anr~ Holland,
"Biochemistry" 17: 4900 [1978]), such as enolase, glyceraldehyde-3-phosphate
dehydrogenase, hexokinase, pyruvate decarboxylase, phosphofructokinase, glucose-6-
phosphate isomerase, 3-phosphoglycerate mutase, pyruvate kinase, triosephosphateisomerase, phosphoglucose isomerase, and glucokinase.
Other yeast promoters, which are inducible promoters having the additional advantage
of transcription controlled by growth conditions, are the promoter regions for alcohol
dehydrogenase 2, isocytochrome C, acid phosphatase, degradative enzymes associated with
nitrogen metabolism, metallothionein, glyceraldehyde-3-phosphate dehydrogenase, and
enzymes responsible for maltose and galactose utilization. Suitable vectors and promoters
for use in yeast expression are further described in R. Hitzeman et ~/., European Patent
Publication No. 73,657A.
Expression control sequences are known for eucaryotes. Virtually all eùkaryotic genes
have an AT-rich region located approximately 25 to 30 bases upstream from the site where
transcription is initiated. Another sequence found 70 to 80 bases upstream from the start
of transcription of many genes is a CXCAAT region where X may be any nucleotide. At the
3' end of most eukaryotic genes is an AATAAA sequence which may be the signal for
addition of the poly A tail to the 3' end of the coding sequence. All of these sequences are
inserted into mammalian expression vectors. :
Suitable promoters for controlling transcription from vectors in mammalian host cells
are readily obtained from various sources, for example, the genomes of viruses such as ~: i
polyoma virus, SV40, adenovirus, MMV (steroid inducible), retroviruses (e.g. the LTR of HIV),
hepatitis-B virus and most preferably cytomegalovirus, or from heterologous mammalian
promoters, e.g. the beta actin promoter. The early and late promoters of SV40 are .
conveniently obtained as an SV40 restriction fragment which also contains the SV40 viral
origin of replication. Fiers et a/., Nature, 273: 113 11978~. The immediate early promoter
of the human cytomegalovirus is conveniently obtained as a Hindlll E restriction fragment.
Greenaway, P.J. et a/., Gene 18: 355-360 11982).
Transcription of a DNA encoding the retrovirus-like particles of this invention by higher
eukaryotes is increased by inserting an enhancer sequence into the vector. Enhancers are cis- .
acting elements ùf DNA, usually about from 1 0-300bp, that act on a promoler to increase
its transcription. Enhancers are relatively orientation and position independent having been
found 5' (Laimins, L. et ~1., PNAS 78: 993 11981 ]) and 3' ILuskY, M.L., er al., Mol. Cell Bio.
.
'
' ,
~O ~2/0~79?i PCI/US91/OX~
~ 13 ,2,095,~3,~
3: 1108 11983]1 to the transcription unit, within an intron IBanerji, J.L. et a/., Cell 33: 729
I1983]) as well as within tl1e coding sequence itself (Osborne, T.F., et a/., Mol. Cell Bio. 4
1293 [1984]). Many enhancer sequences are now known from mammalian genes (globin,
elastase, albumin, a-fetoprotein and insulin). Typically, however, one will use an enhancer
5 from a eukaryotic cell virus. Examples include the SV40 enhancer on the late side of ~he
replication origin Ibp 100-270), the cytomegalovirus early promoter enhancer, the polyoma
enhancer on the late side of the replication origin, and aclenovirus enhancers.
Expression vectors used in eukaryotic host cells iyeast, fungi, insect, plant, animal,
human or nucleated cells from other multicellular organisms) will also contain sequences
10 necessary for the termination of transcription which may affect mRNA expression. These
regions are transcribed as polyadenylated segments in the untranslated por~ion of the mRNA
encoding the particle. The 3' untranslated regions also include transcription termination sites.
Expression vectors may contain a selection gene, also termed a selectable marker.
Examples of suitable selectable markers for mammalian cells are dihydrofolate reductase
15 IDHFR), thymidine kinase (TK) or neomycin. When such selectable markers are successfully
transferred into a mammaiian host cell, the transformed mammalian host cell is able to survive
if placed under selective pressure. There are two widely used distinct categories of selective
regimes. The first category is based on a cell's metabolism and the use of a mutant cell line
which lacks the ability to grow independent of a supplemented media. Two examples are
20 CHO DHFR cells and mouse LTK cells. These cells lack the ability to grow without the
addition of such nutrients as thymidine or hypoxanthine. Because these cells lack certain
genes necessary for a complete nucleotide synthesis pathway, they cannot survive unless the
missing nucleotides are provided in a supplemented media. An alternative to supplementing
the media is to introduce an intact DHFR or TK gene into cells lacking the respective genes,
25 thus altering their growth requirements. Individual cells which were not transformed with the
DHFR or TK gene will not be capable of survival in non supplemented media.
The second category of selective regimes is dominant selection which refers to aselection scheme used in anv cell type and does not require the use of a mutant cell line.
These schemes typically use a drug to arrest growth of a host cell. Those cells which are
30 successfully transformed with a heterologous gene express a protein conferring drug
resistance and thus survive the selection regimen. Exarnples of such dominant selec.ion use
the drugs neomycin (Southern et a/., J. Molec. Appl. Genet. 1: 327 (1982)1, mycophenolic
acid IMulligan et a/., Science 209: 1422 l1980)) or hygromycin ISugden et a/., Mol. Cell. Biol.
5: 410-413 11985)). The three examples given above employ bacterial genes under
35 eukaryotic control to convey resistance to the appropriate drug G418 or neomycin ~geneticin),
xgpt Imycophenolic acid) or hygromycin, respectively.
Suitable eukarvotic host cells for expressing the C type retrovirus-like particle include
monkey kidney CV1 line transformed by SV40 icos-7, ATCC CRL 1651 ); human embrvonic
. ..-
~ ~9~/OX79~ PCT/~'S91/OX':~
-20~9~ 14- ~
kidney line 1293 or 293 cells subcloned for growth in suspension culture, Graham, F.L. et al.,
J. Gen Virol. 36: 59 [1977]); baby hamster kidney cells (BHK, ATCC CCL 10); chinese
hamster ovary-cells-DHFR (CHO, Urlaub and Chasin, PNAS (USA) 77: 4216, (1980]); mouse
sertoli cells (TM4, Mather, J.P., Biol. Reprod. ~3: 243-251 [1980]); monkey kidney cells
(CV1 ATCC CCL 70); african green monkey kidney cells (VERO-76, ATCC CRL-1587);
human cervical carcinoma cells IHELA, ATCC CCL 2); canine kidney cells (MDCK, ATCC CCL
34); buffalo rat liver cells IBRL 3A, ATCC CRL 1442): human lung cells IW138, ATCC CCL
75); human liver cells (Hep G2, HB 8065); mouse mammary tumor IMMT 060562, ATCC
CCL51); and, TRI cells IMather, J.P. et a/., Annals N.Y. Acad. Sci. 383: 44-68 [1982])
Construction of suitable vectors containing the desired coding and control sequences
employ standard ligation techniques. Isolated plasmids or DNA fragments are cleaved,
tailored, and religated in the form desired to form the plasmids required. ..
For analysis to confirm correct sequences in plasmids constructed, the ligation mixtures
are used to transform E. coli K12 strain 294 (ATCC 31446) and successful transformants
selected by ampicillin or tetracycline resistance where appropriate. Plasmids from the
transformants are prepared, analyzed by restriction and/or sequenced by the method of ::
Messing et a/., Nucleic Acids Res. 9: 309 11981) or by the method of Maxam et a/., Methods . .
in Enzymology 65: 499 (1980~.
Host cells are transformed with the expression vectors of this invention and cultured
in conventional nutrient media modified as appropriate for inducing promoters, selecting . .
transformants or amplifying the genes encoding the desired sequences.
The host cells used to practice this invention may be cultured in a variety of media.
Commercially available media such as Ham's F10 ISigma), Minimal Essential Medium (~MEM~,
Sigma), RPMI-1640 (Sigma), and Dulbecco's Modified Eagle's Medium ((DME], Sigma) may
be suitable for culturing the host cells. In addition, any of the media described in Ham and
Wallace IMeth. Enz., 58: 44 I1979]), Barnes and Sato (Anal. Biochem., 102:255 11980J),
U.S. Patent 4,767,704, U.S. Patent 4,657,866, WO 90/03430, WO 87/00195, U.S. Patent
~e. 30,985, U.S. Patent 4,927,762, or U~S. Patent 4,560,655 may be used as culture media
for the host cells. Any of these media may be supplemented as necessary with hormones
and/or other ~rowth factors Isuch as insulin, transferrin, or epidermal growth factor), salts
Isuch as sodium chloride, calcium, magnesium and phosphate), buffers (such as HEPES),
nucleosides ~such as adenosine and thvmidine), antibiotics Isuch as Gentamycin), trace
elements Idefined as inorganic compounds~ usually presen~ at final concentrations in the
: micromolar range), and glucose or an equivalent energy source. Any other necessary .
35 supplements may also be included at appropriate concentrations. The culture conditions,
such as temPerature, pH and the like, are those previously used with the~ host cell selected .~ :~
for expression, and will be apparent to the ordinarily skilled artisan. :
~ .
~ 0 9~/0~79:~ PCI /~ S9 1 /I)X~
f~ 15 23~3~
The host cells referred to in this disclosure encompass cells in in vitro culture as well
as cells which are within a host animal.
"Transformation" means introducing DNA into an organism so that the DNA is
replicable, either as an extrachromosomal element or by chromosomal in.egration. Unless
5 indicated otherwise, the method used herein for transformation of the host cells is the
method of Graham, F. and van der Eb, A., Virology 52: 456-457 (1973). However, other
methods for introducing DNA into cells such as by nuclear injection or by protoplast fusion
may also be used. If prokaryotic cells or cells which contain substantial cell wall
constructions are used, the preferred method of transfection is calcium treatment using
10 calcium chloride as described by Cohen, F.N. et ~/.. , Proc. Natl. Acad. Sci. (USA), 69: 2110
1 1 972) .
"Transfection" refers to the introduction of DNA into a host cell whether or not anv
coding sequences are ultimately expressed. Numerous methods of transfection are known
to the ordinarily skilled artisan, for example, CaPO4 and electroporation. Transformation of
15 the host cell is the indicia of successful transfection.
"PCR" (polymerase chain reaction) refers to a technique whereby a piece of DNA is
amplified. Oligonucleotide primers which correspond to the 3' and S' ends (sense or
antisense strand-check) of the segment of the DNA to be amplified are hybridized under
appropria~e conditions and the enzyme Taq polymerase, or equivalent enzyme, is used to
20 synthesize copies of the DNA located between the primers.
It is ~urther envisioned that the particles of this invention may be produced by. homologous recombination, or with recombinant production methods utilizing control
elements introduced into celis already containing DNA encoding the desired product currently
in use in the field. For example, a powerful promoter/enhancer element, a suppressor, or an
25 exogenous transcription modulatory element, is inserted in the genome of the intended host
cell in proximity and orientation sufficient to influence the transcription of DNA encoding the
desired particle; the element does not encode the particle of this inventian, but the DNA is
present in the host cell genome. One next screens for cells making the particles of this
invention, or increased or decreased levels of expression, as desired.
The novel polypeptide is recovered and purified from recombinant cell cultures by
known methods, including ammonium sulfate or ethanol precipitation, acid extraction, anion
or cation exchange chromatography, phosphocellulose chromatography, irnmunoaffinity
chromatography, hydroxyapatite chromatography and lectin chromatography.
Gene amplification and/or expression may be measured in a sample either directly, for
35 example, by conventional Southern blotting or dot blot ~DNA analysis) using an appropriately
labelled probe, based on the sequences provided herein. Various labels may be employed,
most commonly radionuclides, particularly 32p, However, other techniques may also be
employed, such as using biotin modified nucleotides for introduction into a polynucleotide.
:
9?~0~79~ PC~ S91/OX':~
2~9$6~ 16- ~'
The biotin then serves as the site for binding to avidin or antibodies, which may be labeled
with a wide variaty of labels, such as radionuclides, fluorescers, enzymes, or the like.
Alternatively, antibodies may be employed which can recognize specific duplexes, including
DNA duplexes, RNA duplexes and DNA-RNA hybrid duplexes or DNA-protein duplexes. The
5 antibodies in turn may be labeled and the assay may be carried out where the duplex is bound
to a surface, so that upon the foTmation of duplex on the surface, the presence of antibody
bound to the duplex can be detected.
Gene expression, alternatively, rnay be measured either by Northern blotting to
quantitate the transcription of mRNA (Thomas t1980) Proc. Natl. Acad. Sci`. US~
10 77:5201-5205), dot biots, and in situ hybridization, or by immunological methods, such as
immunohistochemical staining of. tissue sections and assay of cell culture or body fluids, to
directly quantitate the expression of gene product.
With immunohistochemical staining techniques, a cell sample is prepared, tyPically by
dehydration and fixation, followed by reaction with labeled antibodies specific for the gene .
15 product.coupled, where the labels are usually visually detectable, such as enzymatic labels,
fluorescent labels, luminescent labels, and the like. A particularly sensitive staining technique
suitable for use in the present invention is described by Hsu et al. (1980) Am. J. Clin. Path.
75:734-738.
Antibodies useful for immunohistochemical staining andlor assay of sample fluids may
20 be either monoclonal or polyclonal. Conveniently, the antibodies may be prepared against a
synthetic peptide based on the DNA sequences provided herein. Such synthetic peptides may
then be used as an immunogen in preparing antibodies by well-known techniques.
Alternatively, the natural gene product and/or portions thereof may be isolated and used as
the immunogen.
In certain embodiments of this invention, cell lines with reduced expression of
endogenous C-type retrovirus-like sequences are isolated by introduction of anti-sense or
ribozyme genes, or by screening and selection procedures. While CH0 cell lines are
particularly appropriate for the application of these methods, these methods are used :
~: routinely with any host cell described above.
Two approaches are illustrated for the isolation of a cell line with reduced or
undetectable endogenous, C-type retrovirus gene expression. The first approach involves the
introduction of an~i-sense RNA genes or ribozyme genes into parental cell lines in order to
specifically inhibit expression of endogenous C-type retrovirus genes. An alternate approach
is the isolation of low-expressor subclones using screening and selection techniques. Both
35 approaches are encompassed by this invention and are discussed in more detail below.
The nucleic acid sequences of this invention permit the routine obtaining of cell lines
;: with reduced C-type particle expression by enabling the introduction of specific antisense .
RNA or ribozyme genes. Inhibition of a~ specific gene~ product using antisense technology
. ,
~0 92/0~79:~ 2 ~ 9 5 6 3 S PCr/~l~i91/0~
-17-
involves introduction of a gene into target cells which codes for an RNA transcript which is
complementary lantisense) to messenger RNA of the gene product in question. Antisense
RNA in ~he cell presumably base-pairs with the complementary mRNA and thereby inhibits
translation or processing of message. The end result is a reduction in expression of a specific
gene product. High-level expression of antisense RNA is achievable using current technology,
as shown in Kasid et a/., Science 243: 1354-1356 11983]; Khoka et a/., Science 243:947-950
[1989]; Izant et a/., Science 229:345-352 119851; von Ruden et a/., J. Virol. 63:677~682
[1989]; and Chang et a/., J. Virol. 61 :921 -924 l1987~). Optimization of levels of expression
of antisense sequences is within the routine skili in the art, considering the expression levels
of the "sense" sequence in the target cell and the potential adverse consequences of the
production capabilities of the transformed cells which may result. For example, since nucleic
acid blot snalysis has revealed hundreds of copies of C-type related DNA sequences and
moderately abundant C type RNA transcripts in CHO cells, this approach may require high
level expression of antisense RNA in order to be effective.
~Iternately, RNA may be introduced into the host cell which is complementary to the
messenger RNA of the C-type retrovirus-like particle to prevent its translation. .
An alternative to the introduction of antisense RNA genes is the use of ribozymes. As
described above, ribozymes are small RNA molecules that can cleave substrate RNAs in a
sequence specific manner. Particularly preferred ribozymes for the practice of this invention
are the "hammerhead" ribozymes described by Haseloff et al., Nature 334:585-591 [1988];
this reference discloses the use of hammerhead ribozymes to cleave mRNAs at specific sites .
in vitro. With these hammerhead ribozymes, target sequences need only contain a GUA or
GUC sequence at which cleavage occurs. Specificity of cleavage is determined by flanking :
sequences which hybridize to the ribozyme sequence. Thus, ribozymes can be engineered
to cieave at any 18 bp RNA sequence flanking a GUA or GUC triplet, thus generating very
precise specificity. Alternatively, RNA enzymes with highly specific endoribonuclease
activities may be utilized in the practice of this invention, such as those disclosed by Koizumi
et a/., FEBS Lett. 239:285-288 11988I. .. .
Utilizing the sequences of this invention, antisense or ribozyme genes specific for
several regions of coding C-type mRNA are constructed and introduced into host cells using
standard transfection protocols. Parental cell lines suitable for construction of recombinant
clones are isolated using unique selectable markers so as not to interfere with the utility of
these cells for subsequent genetic manipulation. It may be desirable to use amplification
methods to facilitate isolation of clones and allow for amplification of antisense or ribozyme
genes if desired.
Cell lines are then evaluated for expression of endogenous C-type products. Detection .
of expression may be made by immunoassay for structural proteins, enzymatic assay for
_ ' ' , .. .
' ~ ' ''
9~/0~79~'~ PCT/~'S91/08~
2 0 9 ~ ~ 3 ~ -1 8-
reverse transcriptase activity, blot analysis for RNA transcripts, and electron microscope
examination of cell sections for the presence of C-type particles.
In other embodiments, low expressor subclones are isolated through routine screening
as described above. For example, subclones of parental cells are evaluated for expression
5 of C-type RNA sequences by blot analysis using probes consisting of fragments of the
sequences claimed herein. Low expressing subclones are subjected to additional rounds of
screening until RNA levels are undetectable or not reduced further. Different transfected cell
lines may vary significantly in their level of C-type mRNA expression and it is anticipated that
variation will occur in non-transfected subclones as well.
This invention also contemplates more powerful selection schemes. Genes encoding . .
envelope proteins for C-type particles are isolated from cDNA or genomic libraries using the
cDNA provided herein as probe. An expression vector is then directly linked to envelope
coding sequences at the DNA level and so as to partially overlap at the mRNA level.
Following provision of sequence from a substantial open reading frame, a protein construct
15 is made and expressed in a suitable recombinant host such as E. coli according to known
techniques. Next, anti-envelope protein antibodies are then generated by immunization with
ths protein.
Since retrovirus envelope proteins are expressed on the surface of cells secreting
C-type particles, many direct selection schemes employing C-type envelope antibodies can
20 be employed. One embodiment involves incubation of a host cell in the presence of
anti-envelope protein antibodies and complement. The antibody and complement Iyse
expressor cell lines and leave only non-expressor cells as survivors. Some routine
experimentation with antibody concentrations may be necessary in order to select for
low-expressor cells in the event that non-expressor cells do not exist in the initial population.
A second approach using the fluorescence activated cell sorter (FACS) allows flexibility
in the selection of low-expressor cells. Host cells are stained using anti-envelope primary
antibody and a fluorescein conjugated second antibody. High expressor cells show high
- fluorescence and low expressors low fluorescence. Physical sorting of individual calls by
FACS according to fluorescence intensity is used to:enrich for a population of cells which
30 express low levels of C-type envelope protein. Multiple rounds of FACS sorting enhances the
selection procedure. Once`a population of low-expressing cells is obtained, subclones are
propagated and analyzed more rigorously for C-type expression as outlined for the screening
protocol above.
The nucleic acid sequences encoding the retrovirus-like particles of this invention are
35 desirably incorporated into a host cell in more than one copy.
It is understood that the application of ~he teachings of the present invention to a
specific problem or si~uation will be within the capabilities of one having ordinary skill in the .
art in light of the teachings contained herein. Examples of the oroducts of the present
,.
:
~ O 9~0~79~ P~r/~'S91 /082:!~
~ ,9 2~9~3~` ~
invention and representative processes for their is~lation, use, and manufacture appear
below, but should not be construed to limit the invention.
Example: Preparation and analYsis of C-tvPe particles
MATERIALS AND METHODS
5 Cells. Extracellular particles were prepared from culture fluid of the recombinant CHO cell
subclone, 3-3000-44 {Lasky, L.A. et al., Science 233:209-212 [1986]}. This subclone was
derived from dihydrofolate reductase (dhfr) deficient CHO-DUXB11 cells {Lubiniecki, A. S. et
al., Develop. ~iol. Stanr~l~rd. 70:187-191 [19891; Heine, U. I. et al., J. Gen. Virol. 44:45-55
l19791} following transfection with an expression vector containing the genes for murine dhfr
10 and recombinant envelope glycoprotein (gpl2C) of human immunodeficiency virus type 1
(HIV-1). The CHO-K1 cell line (progenitor of the CH0-DUXB11 line) was originally derived
from an ovarian biopsy of an adult Chinese hamster (Cricetulus griseus) {Lieber, M. M. et a/.,
Science 182:56-58 l1973]~ and was obtained from the American Type Culture Collection
(ATCC CCL 61). The mouse myeloma cell line, X63-Ag8.653 ~Lubiniecki, A. S. and L. H
May, Develop. Biol. Standard. 60:141-146 [19851), and the human diploid fibroblast cell
line, MRC5 {Hojman, F. et al., Develop. Biol. S~and~rd. 70:195-202 119891 ~ATCC CC-
171)}, were also obtained from ATCC.
Centrifugal concentrates of celi culture fluid. Cell culture fluid used for the preparation of
particles ~,vas obtained from cells grown under serum-free manufacturing conditions.
Typically 20-30 liters of clarified cell culture fluid was passed through a high-capacity,
continuous-flow ultracentrifuge rotor (Beckman model CF32) at 100,000 x 9. Pelleted
particles were harvested by scraping from the walls of the rotor in 150 mM NaCI, 10 mM Tris
(pH 7.5) containing 0.1% BSA ~TN-BSA~. The harvested particles were then pelleted in a
Beckman Ti45 rotor at 100,000 x 9 for 90 minutes, resuspended in a final volume of 4 5 mls
of TN-BSA, and stored at -80C. This procedure results in a 4000-6000 fold concentration
of particles present in starting cell culture fluid.
Reverse transcriptase IRT~ assays. Reverse transcriptase assays were per~ormed in a total
volume of 0.1 mls using 0.05 mls of sample per assay. Samples were treated with 0.2%
triton-X100 for 30 minutes on ice to solubilize RT activity prior to a 1 hour assay incubation
at 37C~ Incorporation of [3H]-TTP into DNA using a poly(rAj:oligo(dT) template in the
presence of 0.2 mM manganese was monitored by counting radioactivity bound to DE81
filters. Polymerase activity in the presence of poly(dA):oligo~dT) template was monitored as
a control for the presence of non-specific polymerase activity. RT activity in the presence
of manganese or 5 mM magnesium was compared for some samples. A modified assay using
15 ~l of sample in a total volume of 65 ~JI, and [32Pl-TTP as label was used in some
experiments.
Sucrose gradient fractionation. For purification of particles, a double sucrose densitv
. gradient procedure was used. Concentrated culture fluid was adjusted to 50% Iw/w) sucrose
-
~o s~/n~7s~ PCT/~iS91/OX~
20~$3~ -20- ~
by addition of solid sucrose, and overlayed with equal volumes of 40%, 30%, and 20%
sucrose in TN buffer (10 mM Tris-HCI pH 7.5, 150 mM NaCI). After centrifuging overnight
at 25K rpm in an SW27 rotor, RT containing peak fractions were pooled, dialyzed vs. TN
buffer, and layered on top of a linear, preformed 10-60% (w/w) continuous sucrose gradient
in TN buffer. Centrifugation for 2 hours at 39K rpm in an SW41 rotor was sufficient tO bar)d
particles at their appropriate density.
Electron microscope analysis of sucrose gradient fractions. Samples of sucrose
gradient fractions (10-50,ul) were dialyzed vs. TN buffer and spotted onto grids. Particles
were allowed to settle for at least 30 min., after which they were negatively stained using
uranyl acetate and visualized in a transmission electron microscope.
Detection of C-type immunoreactive capsid core polypeptides using prot~in blot
analyses. Protein samples were fractionated on reducing SDS-polyacrylamide gels and
transferred electrophoretically to Immobilon filters (Pharmacia). Primary antisera (goat
anti-feline leukemia .virus p27 sera) was used at a 1 :400 dilution. A secondary detector
antibody (donkey anti-goat IgG coupled to horssradish peroxidase) was used at a dilution of
1: 1000. The signal was developed using 4-chloro,1 -naphthol/hydrogen peroxide as substrate.
Characterization of nucleic acids present in retrovirus-like particles. Nucleic acids were
extracted from sucrose gradient fractions by phenol-chloroform extraction. Samples were
applied to membranes (Genescreen plus; NEN Research Products) using a slot-blot manifold
(Schleicher and Schuell mini-fold). Hybridizations were performed at 42C in buffer
containing 0.5 M NaCI, 0.1 M HEPES (pH 7.2~, 5mM EDTA, 0.1% sodium pyrophosphate,
5X Denhardt's solution, 100 ,ugsiml denatured salmon sperm DNA and either 50,0
formamide (high stringency) or 25% formamide (low stringency). Cloned probe for the
endogenous, polytropic murine leukemia virus (MLV) isolate, MX27 {Stoye, J.P., J. Virol.
61 :2659-2669 t19S7]} was provided by J. M. Coffin and J. P. Stoye. Probes for
endogenous CH0 intracisternal A-particle sequences (pCHlAP.SW2 for family I and
pCHlAP.YL9 for farnily ll) have been described elsewhere {Anderson, 1990, supra). Probe
for murine~B-actin was synthesized based on published sequence {Tokunaga, K., Nucleic
:: Acids Res. 14:28-29~11986]} and consisted of 45mer oligodeoxynucleotides of opposite
polarity which contained complementary sequences at their 3' ends. All probes were
radioactively labeled by incorporation of a-[32Pl-dCTP using nick translation for plasmid
probes or Klenow fragment extension for overlapping oligodeoxynucleotide probes.Isolation and characterization of C-type cDNA clones. C type related sequences were
isolated from a randomly primed cDNA library of particle RNA in ~Igt10. Low stringency
hybridization using the MLV-MX27~ probe was used to identify;plaques of interest. DNA
sequencing was performed using the dideoxy chain termination method on subcloned:: single-stranded temPlate {Sanger, F., Proc. Nat/. Acad. Sci. USA 74:34~63-3467 [19771}.
-:
: ~
.
_., , .. ., .. . ., ., : .. , ~ .,,: ., , .. , , ,,:, , : .
~\'0 9~/0~79~ PC'r/~'S91/0~
~ 2~9~
Genomic DNA blots of Chinese hamster DNA. Standard procedures were used for
preparation of genomic DNA from CH0 cells {Maniatis, T. et al., 1982. Moleculat cloning.
a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.}. High
molecular weight Chinese hamster liver DNA was kindly supplied by K. K. Lueders. DNA was
5 digested to completion using the restriction enzyme Hindlll, and 2 1l9 samples were
fractionated by electrophoresis on 0.7% agarose gels. After capillary transfer of DNA to
charged nylon membranes (Genescreen plus, NEN Research Products), hybridizations were
performed using conditions and probes described above for slot blots.
RESULTS
Purification of RT-containing particles ftom culture fluid of CH0 ceils. A protocol was
devised for purification of extracellular, retrovirus-like particles from CH0 cells which involved
centrifugal concentration of large volumes of culture fluid, fo!lowed by fractionation on 2 . .
sequential sucrose density gradients Isee Methods and Materialsl. Cell culture fluid which
had been concentrated 4000-fold by ultracentrifugation was initially fractionated by
15 centrifugation to equilibrium in a sucrose step gradient. Peak reverse transcriptase
lRT)-containing fractions at the appropriate density j1.12-1.14 g/ml) were pooled, dialyzed,
and layered on top of a continuous 10-60% preformed sucrose gradient and again centrifuged
to equilibrium. RT profiles of the first and second gradients in a typical purification protocol
are shown in Fig. 1. A single peak of RT activity with only background activity in flanking
20 fractions was observed on the second sucrose gradient. Fractions comprising the RT peak
of the second gradient were pooled for subsequent characterization. RT activity of purified
Rauscher murine leukemia virus banded at a similar density in identical gradients (not shown).
Preferential polymerase activity in the presence of polylrA) template compared to
poly~dAI template confirmed that the polymerase activity of peak fractions represented
25 reverse transcriptase, and preferential activity in the presence of manganese compared to
magnesium suggested that the particles were related to mammalian C-type retroviruses (not
shown). :
- Electron microscope (EM) evaluation of purified particles from CH0 3-3000-44 cells.
Samples of peak RT-containing fractions were examined in the electron microscope for the
30 presence of retrovirus-like particles. After negative staining with uranyl acetate, pleiomorphic
particles comparable in size and morphology to retroviruses visualized by others using this
methodology were observed (Fig. 2).
Protein and nucleic acid content of purified retrovirus-like particles. Pools of 4 fractions
each from the second sucrose gradient of a particle purification Ishown in Fig. l BI were
35 combined and tested for the presence of retrovirus structural antigens and nucleic acid
sequences. Western blot analysis of gradient pools using antisera to purified p27 core protein ;
of feiine leukemia virus ~FeLV, a mammalian C-type~ retrovirus) revealed the presence of a 31
'~
- .:
', ',
,:
~'O 9'/0~79`~ 2 ~3 9 ~ 6 3 ~ Pcr/~i~9l/nx2 ~
-22- ~t
kd core protein (p31), as well as a 64 kd gag precursor (p64~ in the gradient pool containing
fractions with peak RT activity (Fig~ 3A).
Nucleic acids were isolated from each pool, applied to membranes using a slot-blot
manifold, and hybridized with retrovirus and control probes (Fig. 3B). Nucleic acids
5 specifically hybridizing to the murine leukemia virus (C-type) probe (MLV-MX27, see Materials
and Methods) were observed only in the pool comprised. of fractions with RT activity and
containing detectable p31 and p64 polypeptides. Nucleic acids from RT and p31 containing
fractions also hybridized to pooled probe for family I and family ll CHO cell intracisternal
A-particle (IAP) sequences (pCHlAP.SW2 and pCHlAP.YL9, see Materials and Methods).
10 Subsequent experiments revealed homology to family ll (pCHlAP.YL9), but not to family I
(pCHlAP.SW2) IAP sequences (not shown). Nucleic acids with homology to a major cellular
mRNA l,B-actin) were not detected in any gradient frac~ions.
Hybridization signals for retrovirus probes IMLV-MX27 and CHIAP-YL9) were reduced
significantly when nucleic acids were treated with RNAse or alkali prior to blottin~, but no
15 reduction in signal intensity was observed when samples were treated with RNAse free
DNAse prior to blotting. These findings indicate that the nucleic acids detected were RNA
and not DNA. The presence of RNA in particles is consistent with the morphological and
immunological characterization of these particles as retrovirus related.
Characterkation of particle cDNA sequences homologous to MLV. Several cDNA
20 clones of purified particle RNA which hybridized to MLV probe were isolated and
characterized. Three of the clones were characterized in detail. All showed homology to the
endonuclease domain of MLV. The largest clone IPCHOC.ML10, 949 bp) exhibited 73/o
nucleotide iden~ity with the published sequence of Moloney MLV ~Shinnick, T. supra Nature .:
1981 ) (Fig. 4, Table 1). Reduced homology was found to endonuclease domains of C-type
25 retroviruses of evolutionarily divergent species suggesting that the pCHOC.ML10 clone is
most closely related to rodent C-type retroviruses ITable 1). The pCHOC.ML10 sequence :
contained multiple interruptions of potential coding sequences in all 3 reading frames and
therefore was not capable of encoding an intact endonuclease (Fig. 5).
.
- .
,
\~'09~/0~79~ PC~ i91/OX~
23
Table 1. Nucleotide homology of pCHOC.ML10cDNA to
mamrnalian C-type retrovirus genomes.
% nucleotide homology
c-type retrovirus identity score'
~ . , .
Mo10ney murine leukemia virus 74 2008
t Shinnick et al ., 1981)
{supra}
Fe1ine 1eukemia virus 70 1910
(Donahue et al., 1988
{J.Virol. 62:722-73176
sabOOn endogenous virus 64 171
(~Cato et al ., 1987)
{Jpn . J. Genetics 62: 127-137 ~
* ~ * * *
Presence of homologous DNA sequences in the Chinese hams~er genome. Southern
blot analysis of Chinese hamster DNA using the pCHOC.ML10 clone as probe revealed that
conserved, homologous sequences were present at 100 to 300 copies per genome in DNA
25 of parental and recombinant CHO cell lines, as well as in DNA isolated from a Chinese
hamster liver (Fig. 6A). Homologous sequences were not detected in DNA isolated from
human MRC5 cells or mouse myeloma cells using the same high stringency hybridization
conditions ~data not shown). .
CHO IAP family ll RNA sequences were also detected in gradient purified particles lsee
30 above). Related DNA sequences have previously been demonstrated to exist as a moderately
repetitive family in the genomes of recombinant and parental CHO cell lines ~Anderson e~ al..
supra 1990). Comparison of DNA from several CHO cell lines with DNA from Chinesehamster liver by Southern blot analysis using CHO IAP family ll probe (Fig. 6BI revealed that
this conserved family of repetitive sequences is also present in the Chinese hamster germline.
35 DISCUSSION
Particles morphologically similar to retroviruses have previously been observed in CHO
cells, but detailed characterization ~has not been reported. The use of high-capacity,
flow-through ultracentrifugation to concentrate extracellular, retrovirus-like particles from
large quantities of culture fluid of a recombinant CWO cell line has facilitated purification and
40 biochemical characterization of these particles.
Extracellular particles of CHO cells are similar to those of characterized mammalian
C-type retroviruses by a number of criteria. They band in a sucrose gradient at an appropriate
density, and exhibit RT activity with a preference for manganese. Polypeptides of the
particles are immunologically related to those of mammalian C-type retroviruses, and RNA
- : : ~ . . ..
:
~ Based on algorithm parameeers descrLbed by Fitch and Smith (19Q3)~81}. ~ ~
'' .
~\ 0 9~0~S79 PCr/~'S91/ûX~
209~3~ -24~
from purified particles specifically hybridizes tO a cloned murine C-type genome under low
stringency conditions.
Purified particles also contained RNA which specifically hybridized to a cloned probe
representing one of two characteri~ed families of CHO cell intracisternal A-particle ICHlAP)
5 related sequences {Anderson et a/., supr~ 1990). One possible explanation for this result is
that 2 types of particles with similar buoyant densities in sucrose gradients are present in
supernatants of CHO cells. At least one of these particles must contain structural antigens
which cross-react with C-type antisera. Alternatively, a single type of particle containing
C-type structural proteins may be present which encapsidates the two different RNAs.
10 Examples of retrovirus particles packaging heterologous retrovirus RNA species have
previously been described ~Scolnick, E., J. Virol. 29:964-972 ~19901; Sherwin, S.A, J. Virol.
26:257-264 ~1978]}. Another possibility which should be considered is that the particle RNA
represents the product of a recombinant provirus gene composed of sequences from both
unrelated retrovirus classes. The available data do not allow differentiation between these
1 5 alternatives.
Sequence analysis of a cDNA clone representing purified particle RNA revealed
significant nucleotide homology with endonuclease encoding genes of several mammalian
C-type retroviruses (Table 1). However, the cloned cDNA sequence contained no open
reading frames capable of encoding an intact endonuclease. Since a functional retrovirus
20 endonuclease is required for provirus integration, an obligatory step in the retrovirus
replication cycle, these findings provide one possible explanation for the non-infectious nature
of the particles observed in CHO cells.
Nucleotide homology of particle cDNA to murine C-type virus genomes was greater
than that observed to C-type retrovirus genomes of evolutionarily more distant mammalian
25 species suggesting that the particles are of rodent origin. Furthermore, comparison of DNA
from several CHO cell lines and Chinese hamster liver using Southern b!ot analysis revealed
that both types of retrovirus-like sequences identified in purified extracellular particles ~CHO
IAP family 11 and C-type) are present in the germline of Chinese hamsters. It is therefore likely
that the extracellular retrovirus-like particles of CHO cells are the products of endogenous
30 provirus elements present in the. germline of Chinese hamsters.
,'
' " ',
' ' :';
,
: . :
~,~ o 9_~0~79~ PC~I~ S91/08"~
-25- 2 0 9 -5 ~ 3 ~
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: GENENTECH, INC.
~ ii) TITLE OF INVENTION: Chinese Hamster Ovary Cell Retrovirus~like
Particles
(iii) NUMBER OF SEQUENCES: 2
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: Genentech, In~.
(B) STREET: 460 Point San Bruno Blvd
(C) CITY: South San Francisco
(D) STATE: California
(E) COUNTRY: USA
(F) ZIP: 94080
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: 5.25 inch, 360 Kb floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS .
(D) SOFTWARE: patin (Genentech)
(vi) CURRENT APPLICATION DATA:
tA) APPLICATION NUMBER:
(B~ FILING DATE: 07-NOV-1991
(C) CI,ASSIFICATION:
(~ii) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER: 07/611,529
(B) FILING DATE: 09-NOV-1990
35 (viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: Adler, Carolyn R.
(B) REGISTRATION NUMBER: 32,324
(C) REFERENCE/DOCKET NUMBER: 678 -
(ix) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: 415/266-2614
(8) TELEFAX: 415/952-9881 `
(C) TELEX: 910/371-7168
45 (2) INFORMATION FOR SEQ ID NO:1: ~ ~-
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 949 bases
(8) TYPE: nueleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:1:
GAATTCAATG TCCTTGTGGG GAAGAACGAT ATTCTAAAAC AGGTAACTGA 50
.
GCAATGTGAT GCGTGCGCCC GAGTCAACGC ATCCAGACTG AAGCTTCCTC 100
CCGGGAACCG GTCAGAGGCT ACCGGCCCGG AACACATTGG GAGATAGATT 150
TCACTGAGAT TAAACCAGGA AAATATGGAT ACAAGTATCT ATTAATTTTT 200
GTAGACACCT TTTCAGGATG GGTTGAAGCC TTCCCTACTA AACATGAAAC 250
.
PCT~S91/0X~
2~5635 -26- ~ !
AGCCAGATCG TTACTAAGAA ATTGCTTGAA GAAATCTTTC CCCGTTATCG 300
GATGCCTCAG GTATTGGGAA CAGACAATGG GCCCGCCTTC GTCTCCAGGT 350
AAGTCAGTCA GTGGCCACCT TATTGGGGAT TGATTGGAAA TTACATTGTG 400
CTTATAGACC CCAAAGTTCA GGACAGGTAG AAAGGATGAA TAGAACAATC 450
AAGGAGACTT TAACAAAATT GTCGCTTGCA ACTGGCACTA GAGAGCTGGG 500
TCCTCCTACT CCCCCTAGCA CTCTACCGCG CTCGTAATAC CCCTGGACCA 550
CATGGGCTCA CACCCTTTGA GATCCTGTAT GGAGTACCTA GCTCCTATCA 600
TTAACTTTCT TGATCAAGAT GTCTCAGTTT TGCTAACTCC CCTTCTCTCC 650
AAGCTCATTT ACAGGCCCTC CAACTAGTAC AACGGGAGGT CTGGAAACCC 700
CTTGCTCAAG CTTATAAAGA CCAGAGGGAC CATCCCACCA TCCCCCATTC 750
CTACCAGATC GGGGACACTG TTTGGGTCCG GCGTCACCAG GCCAAGAACC 800
TTGAACCCCG CTGGAAGGGA CCCTACATCG TTTTGCTTAC CACTCCCACC 850
GCACTCAAGG TAGACGGCAT TGCAGCT'~GG ATACATGCTT CACATGT~AA 900
GCCAGCCCAA CCCACCGATT CAGCCACTGC ATCAGAATGG AGCCACACC 949
. . .
(2) INFORMATION FOR SEQ ID NO:2:
(i) SEQUENCE CHARACTERISTICS:
~A) LENGTH: 949 bases
:~ : : (8) TYPE: nucleic acid
~50 (C) STRANDEDNESS: single : :
~D) TOPOLOGY: linear
~xi) SEQUENCE DESCRIPTION: SEQ ID NO:2:
GGTGTGGCTC CATTCTGATG CAGTGGCTGA ATCGGTGGGT TGGGCTGGCT 50
TTACATGTGA AGCATGTATC CAAGCTGCAA TGCCGTCTAC CTTGAGTGCG l00
: GTGGGAGTGG TAAGCAAAAC GATGTAGGGT CCCTTCCAGC GGGGTTCAAG 150
GTTCTTGGCC TGGTGACGCC GGACCCAAAC AGTGTCCCCG ATCTGGTAGG 200
AATGGGGGAT:GGTGGGATGG TCCCTCTGGT.CTTTATAAGC TTGAGCAAGG 250 :
.
~ 9~/~79~ PCT/~S91/0
-27- 2 ~ g ~ ~ 3 ~
GGTTTCCAGA CCTCCCGTTG TACTAGTTGG AGGGCCTGTA AATGAGCTTG 300
GAGAGAAGGG GAGTTAGCAA AACTGAGACA TCTTGATCAA GAAAGTTAAT 350
GATAGGAGCT AGGTACTCCA TACAGGATCT CAAAGGGTGT GAGCCCATGT 400
GGTCCAGGGG TATTACGAGC GCGGTAGAGT.GCTAGGGGGA GTAGGAGGAC 450
CCAGCTCTCT AGTGCCAGTT GCAAGCGACA ATTTTGTTAA AGTCTCCTTG 500
ATTGTTCTAT TCATCCTTTC TACCTGTCCT GAACTTTGGG GTCTATAAGC 550
ACAATGTAAT TTCCAATCAA TCCCCAATAA GGTGGCCACT GACTGACTTA 600
CCTGGAGACG AAGGCGGGCC CATTGTCTGT TCCCAATACC TGAGGCATCC 650
GATAACGGGG AAAGATTTCT TCAAGCAATT TCTTAGTAAC GATCTGGCTG 700
TTTCATGTTT AGTAGGGAAG GCTTCAACCC ATCCTGAAAA GGTGTCTACA 750
AAAATTAATA GATACTTGTA TCCATATTTT CCTGGTTTAA TCTCAGTGAA 800
ATCTATCTCC CAATGTGTTC CGGGCCGGTA GCCTCTGACC GGTTCCCGGG 850
AGGAAGCTTC AGTCTGGATG CGTTGACTCG GGCGCACGCA TCACATTGCT 900
CAGTTACCTG TTTTAGAATA TCGTTCTTCC CCACAAGGAC ATTGAATTC 949 `:
,
.