Note: Descriptions are shown in the official language in which they were submitted.
W0 94/01560 2 1 2 ~ 9 ~ 4 PCI/US93/00139
ALLERGENIC PROTEI NS ~ND PEPTIDES FROM JAPANESE CE:DAEt
POLLEN
S
Back~round of the Invention
Genetically predisposed individuals, who make up about 10% of
the population, become hypersensitized (aller~ic) to antigens from a variety of
environmental sources to which ~hey are exposed. Those antigens that can
induce immediate and/or delayed types of hypersensitivity are known as
allergens. (King,T.P.,Adv. Immunol. 23: 77-105, (1976))~ Anaphylaxis or
atopy, which includes the symptoms of hay fever, asthma, and hives. is one forrnof immediate allergy. It can be caused by a variety of atopic aller~ens, such asproducts of grasses, trees, weeds, animal dander. insects. food, dru~s. and
chemicals.
- The antibodies involved in atopic allergy belong pnmarily to the
I~E class of immunoglobulins. IgE binds to mast cells and basophils. Upon
combination of a specific allergen with lCE bound to mast cells or basophils, ~eIgE may be cross-linked on the cell surface. resultin~ in the physiological effects
~0 of I~E-anti~en interaction. These physiological effects include the release of.
among other substances~ histamine. serotonin, heparin. a chemotactic factor for
eosinophilic leukocytes and/or the leukotrienes, C4. D4, and E4~ which cause
prolon~ed constriction of bronchial smooth muscle cells (Hood. L.E. et al. `
ImJmmology (2nd ed.), The Benjamin/Cumming Publishing Co.~ Inc. (1984)).
1~ These released substances are the mediators which result in allergic symptoms
- caused by a combination of I~E with a specific aller~en. Throu~h them. the
effects of an allergen are manifested. Such effects may be systemic or local in
nature. depending on the route by which the ~nti~en entered the body and the
pattern of deposition of IgE on mast cells or basophils. Local manif~stations
~0 generally occur on epithelial surfaces at the k)cation at which the aller~en
entered the body. Systemic effects can include anaphylaxis (anaphylactic
shock), which is the result of an I~E-basophil response to circulatin~
(intravascular) antigen.
I
j r
WO 94/01560 ~ ' PCr/US93/00139
~1~095~
Japanese cedar (Suoi: Cr~,utol71eria japonica) pollinosis is one of
the most important allergic diseases in Japan. The number of patients sufferin~
t`rom this disease is on the increase an(i in some areas, more than 1()~ of the
population are affected. Treatment of Japanese cedar pollinosis by ~ ¦
administration of Japanese cedar pollen extract to eft`ect hyposensitization to the
aller~en has been attempted. Hyposensitization using Japanese cedar pollen
extract, however, has drawbacks in that it can elicit anaphylaxis if hi~h doses are
used, whereas when low doses are used to avoid anaphylaxis, treatm~nt must be
continued for several years to build up a tolerance for the extract.
The major aller~en from Japanese cedar pollen has been purified
and desi~nated as Su~i basic protein (SBP) Ol' Cr,~j I. This protein is reported to
be a basic protein with a molecular weioht of 41-50 kDa and a pI of 8.8. There
appear to be multiple isoforrns of the alleroen. app~rently due in part to
differential glycosylation (Yasueda et al. (1983) J. Allerg~ Clin. Immunol. 71:
77-86; and Taniai et al. (1988) FEBS Letters 239: 329-332. The sequence of the
first twenty amino acids at the N-telminal end of C ~! j I and a sixteen amino acid
internal sequence have been determined (Taniai supra). - :A second allergen from Japanese cedar pollen having a molecular
- wei~ht of about 37 kDa known as Cr.~ j II has also been reported (Saka~uchi et
al. ( 1990) Allerg~ 45: 309-312). This aller~en was t`ound to have no
immunological cross-reactivity with G~ j 1. Most patients with Japanese cedar
pollinosis were found to haYe IgE antibodies to both Ct~ j I and C~
howe~er. sera from some patients reacted with only Ct~' j I or Cr~ j 11.
In addition to hyposensitization of Japanese cedar pollinosis
2s patients with low doses of Japanese cedar pollen extract. U.S. patent 4~939,239
- issued July 3, 1990 to Matsuhashi et al. discloses a hyposensitization agent i~
comprisino a saccharide covalently linked to a Japanese cedar pollen aller~en for
hyposensitization of persons sensitive to Japanese cedar pollen. This
hyposensitization a~ent is reported to enhance the production of I G and I~M
antibndies. but reduce production of I~E antibodies which are specific to the
alleroen and responsible t`or anaphyla.Yis and aller~y. The aller,~ens use~l in the
hyposensitization a~oent preferably have an NH~-terrninal amino acid sequence of
wos4/01s6n ' 2~.æo~4 PCr/US93/0ol39
Asp-Asn-Pro-Ile-Asp-Sçr-X-Trp-Ar~-Gly-Asp-Ser-Asn-Trp-Ala-Gln-Asn-Arg- s
Met-Lys-, wherein X is Ser, Cys. Thr~ or His (SEQ ID NO: 18). Additionally.
Usui et al. (1990) Int. A-ch. Allerg~ Appl. In1tnlll701. ~: 74-7~ reported ~hat the
ability of a Su~i basic protein (i.e., C~ j l)-pullulan conju~ate to elicit the Asthus
reaction was markedly reduced, about l.()()0 times lower than that of native Su~i !
basic protein and sug~ested that the Su~i basic protein-pullulan conju~ate wouldbe a good candidate for desensitization therapy acainst cedar pollinosis.
The C~y j I aller~en found in C~ ptnm~ria japot7ica has also been
found to be cross-reactive with allergens in the pollen flom other species of
trees, including Cllpressl~s semp~l~i)el?s~ Panzani et al. (Annals of Allelg! ~7:
26-30 (1986)) reported that cross reactivity was detected between allergens in the
pollens of cupressLls semp~rvi)ens and C~ om~ria juponica in skin testin~.
RAST and RAST inhibition. A 50 kDa allerl~en isolated from Mountain Cedar
(Jurliperus sabinoides, also known as Junipe)us ashei) has the NH2-terminal
sequence AspAsnProIleAsp (SEQ ID NO: 25) (Gross et al, (19 î 8) Scand. J.
Immunol. 8: 437-441) which is the same sequence as the first five amino acids ofthe NH-2 terminal end of the Cry j I aller~en. The C~? j I aller~en has also been
found to be allergenically cross-reactive with the following species of trees:
Cupressus a~i~onica, CI~presslls macrocaspa, Jllniperus virgiMiana, Jl~nipe)us
communis, 7`huYa orientalis, and Cham~c~paris o~tusa.
Despite the attention Japanese cedar pollinosis aller ens have
received. definition or characterization of the aller~ens responsible for i~s
adverse effects on people is far ~'rom complete. Current desensitization therap~involves treatment with pollen extract with its attendant risks of anaphyl~xis if
high doses of pollen extract are administered. or lon; desensitizatit)n times when
low doses of pollen extract are administered.
Summarv of the Invention
The present invention provides nucleic acid sequences codin~ for
the Cl~ ptomeria japonica major pollen alles ~en Cr! j I and ~;aoments thereof.
The present invention also provides isolated Cr! j I or at least one fr;~ment
thereof produced in a host cell transformed with a nucleic acid sequence codin~
WO ~4/01560 ` PCr/US93/00139
U~4
for Cry j I or at least one fragment thereof and fragments of ~rYj I prepared
synthetically.
The present invention also provides Jun l~ l and Jun s I protein
allergens which are immunologically cross-reactive with Cr~ j I and fragmen~s ofS J~n v 1 and Jun s I produced in a host cell transformed with a nucleic acid
sequence coding for Jun s I or JuJt v I respectively and fra~ments of Jult s I and
Jun v I prepared synthetically. The present invention further provides nucleic
acid sequences coding for Jun v I and Jlm s I and fia~ments thereof. As used
herein, a fragment of the nucleic acid sequence coding for the entire amino acidsequence of Cry j I, Jun s I or Jun v I refers to a nucleotide sequence having
fewer ~ases than the nucleotide sequence codin~ for the entire amino acid
sequence of CrYj I, J~m s I or Jun v I andlor rnature Cr~ j I. Jult s I or Jl/l? v I.
C~ ~ j I, Jun s I or Jun v I and fra~ments thereof are useful for dia~nosing.
treatin~, and preventint, Japanese cedar pollinosis as well as pollinosis caused by
pollen from other species of trees wherein such pollen is immunolo; ically cross-
reactive with Japenese cedar pollen allergen.
Peptides within the scope of the invention preferably complise at
Ieast one T cell epitope, and more preferably at least two T cell epitopes of Cry j
I. The invention further provides peptides comprisin~ at least two rec~ions~ each
~0 region comprising at least one T cell epitope of a Japanese cedar pollen protein
allergen. The invention also provides modified peptides having similar or
enhanced therapeutic properties as the correspondin~ naturally-occurrin~ ^
allergen or portion thereof, but havin~ reduced side effects. as well as modified `
peptides having improved properties such as increased solubility and stability.
2S Peptides of the invention are capable of modifying~ in a Japanese cedar pollen- ¦
sensitive individual or in an individual who is sensitive to an allergen cross-
reactive with Japanese cedar pollen. to whom they are administered. the aller~icresponse of the individual to a Japanese cedar pollen aller~en or an aller~en
cross-reactive with Japanese cedar pollen such dS JUI? S I or Jl~n ~ I. Methods of
treatment or diagnosis of sensitivity to Japanese cedar pollen or a cross-reactive
aller~en in an individual and therapeutic compositions complising one or more
peptides of the invention are also provided. This in~ention is more particularly
WO 94/01560 2 1 ~ 1~ 9 ~ 4 PCl /US93/00139
described in the appended claims and is described in its preferred embodiments
in the following description.
Brief Description of the Drawin~s
s Fi~. la is a graphic representation of affinity purified Cn~ j I on
Superdex 75 (2.6 by 60 cm) equilibrated with 10 mM sodium acetate (pH 5.0)
and 0.15 M Na~1;
Fi~. lb shows an SDS-PAGE (1~.5%) analysis of the fractions
from the major peak shown in Fig 1 a;
Fig. 2 shows a Western blot of isoforms of purified native Cr~
proteins separated by SDS-PAGE and probed with mAB CBF2;
Fig. 3 is a graphic representation of allergic sera titration ot
different purified fractions of purified native Cr~! j I usin~ plasma from a pool of
fifteen allergic patients;
Figs. 4a-b show the composite nucleic acid sequence trom the two
overlapping clones JC 71.6 and pUC19JC9 la coding for Cry j I. The complete
- cDNA sequence for Cr~ j I is composed of 1312 nucleotides~ including 66
nucleotides of 5' untranslated sequence~ an open readin; frame starting with thecodon for an initiatin~ methionine of 1122 nucleotides~ and a 3' untranslated
re~ion. Figs. 4a-b also show the deduced amino acid sequence of C~
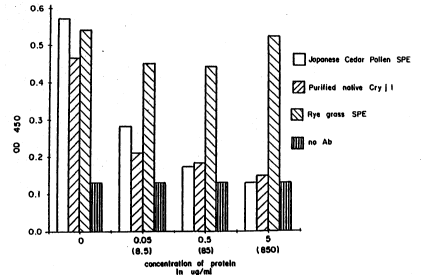
Fig. 5a is a graphic representation of the results of IgE bindino
reactivity wherein the coatin~ antigen is soluble pollen extract (SPE) trom
Japanese cedar pollen;
Fig. Sb is a graphic representation of the results of IC~E bindino
2S reactivity wherein the coating antigen is purified native C~
Fig. 6 is a graphic representation of the results of a competition ~.
ELISA with pooled human plasma (PHP) t`rom 15 patients wherein Ihe coatino
anti~en is soluble pollen extract (SPE) ~om Japanese cedar pollen:
Fig. 7 is a graphic representation of the results ot a c~mpetition
ELISA using plasma from individual patients (indicated by patient numbers!
wherein the coatin~ antigen is soluble pollen extract (SPE) t`rom Japallese cedapollen and the competing antigen is puritied native Cr~
WO 94/01560 , ~ PCr/US93/00139
ZlC~ o(~4
Fi~. 8a is a ~raphic representation of the results from a direct
binding ELISA using plasma frc)m seven individual patients (indicated by
patient numbers) wherein the coating antioen is soluble pollen extract (SPE)
fi om Japanese cedar pollen;
Fig. 8b is a ~raphic representation ot' the results from a direct
binding ELISA using plasma from seven individual patients (indicated by patient
nurnbers) wherein the coating antigen is denatured soluble pollen extrdct which
has been denatured by boiliri~ in the presence of a reducin~ agent, DTI`:
Fig. 9 is a graphic representation of a direct ELISA where the
wells were coated with recombinant C~ j I (rC~ j I) and IgE bindin~ was
assayed on individual patients;
Fig. lOa is a graphic representation of the results of a capture
ELlSA usin~ pooled human plasma from fifteen patients wherein the wells were
coated with CBF2 (IgG) mAb, PBS was used as a negative anti~en control. and `
the anti~en was purified recombinant C~ j I;
Fig~. lOb is a graphic representation of the results of a capture
ELISA using rabbit anti-Amb aI and II. wherein the wells were coated with 20
- ~glml CBF2 (I~G), PBS was used as a ne~ative antigen eontrol and the anti,~en
was purified recombinant Cr~
'0 - Fig. 11 is a graphic representation of a histamine release assay
perforrned on one Japanese cedar pollen aller~ic patient using SPE from
Japanese cedar pollen, purified native C~ j l and recombinant C~ j I as the
added anti~ens; and
Fig. 12 is a graphic representation of the results of a T cell
~s proli~ration assay usin~ blood from patient #999 wherein the antioen is
recombinant G~ j 1 protein, puri~led native C1~ j I protein, or selected Cr~
peptides recombinant Amb a 1.1.
Fig. 13 shows valious peptides of desired lengths derived trom
C~
~0 Fig. 14 is a graphic representation depictino responses of T cell
lines from twenty-five patients primed in vitro with purif~ed native Cr! j I and ~;-
analyzed for response to various G~ j I peptides by percent o f respons~s ,'
WO 94/01~60 P~/US93/00139
2120954
(positive) with an S.I of at least two (shown over each b~r)~ the mean stimulation
index of positive response for the peptide (shown over each bar in parenthesis)
and the positivity index (Y ~xis).
Fig. 15 is a graphic representation of the results of a direct
s binding assay of IgE to certain Cr~ j I peptides. purified native C~,~ j I and rC)~ j ;
I.
Figs. 16 shows the nucleotide sequence of Jlln s l: this sequence is
a composite from the two overlapping cDNA clones pUC19JS42e and
pUC19JS45a as well as the full-length clone JS53iib codin~ for Jltn s I; the
complete cDNA sequence ~or Jun s I is composed of l 17() nucleotides, includin~
25 nucleotides of 5' untranslated sequence. an open reading frame of l.101
nucleotides. and a 3' untranslated region~ Fig. 16 also shows the deduced amino
acid sequence of Jun s I.
Fig. i7 shows the nucleotide sequence of Jun v I: this sequence is a
composite from the two overlapping cDNA clones pUCl9JV46a and
pUC19JV49iia coding for Jun v I: the complete cDNA sequence for Ju11 v I is
composed of 1278 nucleotides. including 35 nucleotides of 5' untranslated
sequence, an open reading frame of 1,110 nucleotides. and a 3' untranslated
region; Fi~. 17 also show the deduced amino acid sequence of J1~n
Fi~. 18 shows various peptides of desired lengths derived t`rom C~
Figs. 19a and 19b show Northern blots of pollen-derived RNA probed
with C)~ j cDNA for identification of mRNA capable of encodino Cr~ j I or a
C~ j I homologue: Fig. 19a shows RNA from C. japnnica (U.S. and Japanese
sources)~ J. sabinoides and J. virginiana probed with C)~ j I cDNA: Fio. l 9b
shows RNA from J. sabinoides and C. ari_onica probed with the same cDNA:
the position of molecular weight standards are shown in each pal~ ot` the Fi~ure.
Detailed Description of the Invention
The present invention provides nucleic acid sequenc~s coding ~or
Cn j I. the major aller~en found in Japanese cedar pollen as well as nucleic acid
sequences coding for Jlm 1,~ 1 and Jun s I. The nucleic acid sequence coding ~or , r
Cr! j I preferably has the sequence shown in Figs. 4a and 4b (SEQ ID NO: I ).
WO 94/01560 - PCI/US93/00139
2 ~ 4
The nucleic acid sequence coding for c~ j 1 shown in Fi~s. 4a and 4b (SE(;~ ID
NO: 1) contains a 21 iamino acid leader sequence from base 66 throu~h base 128.
This leader se~uence is cleaved from the mature protein which is encoded by
basies 129 through 1187. The deduced amino acid sequence of Cr~ j I is also
shown in Figs. 4a and 4b (SEQ ID NO: 2). The nucleic acid sequence of the
invention codes for a protein having a predicted molecular weight of 3g.5 kDa~
with a pI of 7.8. and five potential N-linked glycosylation sites~ Utilization of
these glycosylation sites will increase the molecular wei~ht ~nd affect the pI of
the mature protein. The deduced amino acid sequence for the mature protein
encoded by the nucleic acid sequence of the invention is identical with the
known NH2-tenninal and internal amino acid sequences reported by Taniai et
al.. supra. The NH2-terrninal end of C ~ j I reported by Taniai et al.. sl/pr~ has
the sequence shown in SEQ ID NO: 18. The internal sequence repor~ed by
Taniai et al.~ supra has the sequence
lS GluAlaPheAsnValGluAsnGlyAsnAlaThrProGln`LeuThrLys (SEQ ID NO: 19).
There are sequence polymorphisms observed in the nucleic acid sequence of the
invention. For example. single independent nucleotide substitutions at the
codons encoding amino acids 38, 51 and 74 (GGA vs. GAA. GTG vs. ~CG. and
GGG vs. CiAG. respectively) of SEQ ID #l may result in arnino acid
~0 polymorphisms (G vs. E, V vs. A. and G vs. E. respectiYely) at these sites. In
addition. a single nucleotide substitution has been detected in one cDI~A clone
derived from Cr~ptomeriajaponica pollen collected in Japan. This su~Stitution
in the codon for amino acid 60 (TAT vs. CAT) of SEQ ID #l may result in an `
amino acid polymorphism (Y vs. H) at this site. Additional silent nucleotide
~5 substitutions have been detected. It is expected that there are additional sequence
polymorphisms. and it will be appreciated by one slcilled in the art tha[ o ne or
more nucleotides (up to about 1~ of the nucleotides) in the nucleic acid
sequence coding for Cl,~ j I may vary amon~ individual C)~l7tom~ia j(~ nica
plants due to natural allelic variation. Any and all such nucleotide ~ tions andO resultinD amino acid polymorphisms are within the scope of the in~ention.Furthermore. there may be one or more ~mily members of C~ j I. Sueh ~`amily
members are defined as proteins related in function and amino acid sequence to
WO 94/01560 ' 2 ~ 2 0 ~ S ~ PCI/U~93/~013~ '
C y j I but encoded by genes at separate ~enetic loci.
Fragments of the nucleic acid sequence codin~ for fragments of
Cr~ j I or a cross-reactive allergen are also within the scope of the invention.Fra~ments within the scope of the invention include those coding for palts of Cnj I or a cross-reactive allergen such as Jun v I or Jun s l which induce an immune ?
response in mammals, preferably humans~ such as stimulation of minimal
amounts of IgE: binding of I~E; eliciting the production of IgG and IgM
antibodies: or the elicitin~ of a T cell response such as proliferation andlor
lymphokine secretion and/or the induction of T cell anergy. The foregoing ~~
fragments of C)y j I are referred to herein as antigenic fra~ments. Fra,~,ments
within the scope of the invention also include those capable of hybridizinc~ with
nucleic acid from other plant species for use in screening protocols to detect
alleroens that are cross-reactive with C~ j I. As used herein~ a fragment of thenucleic acid sequence coding for C)y j I refers to a nucleotide sequence having '
fewer bases than the nucleotide sequence coding for the entire amino acid
sequence of C~! j I and/or mature Cr~, i I. Generally. the nucleic acid sequencecoding for the fragment or fragments of C~ j I will be selected from the bases
coding for the mature protein, however, in some instarlces it may be desirable to
select all sr a part of a fragment or fra~ments from the leader sequence portionof the nucleic acid sequence of the invention. The nucleic acid sequence of the
invention may also contain linker sequences. modified restliction endonuclease
- sites and other sequences useful for clonin~ expression or purification of C~
or fra~ments thereof.
A nucleic acid sequence codin~ for Cl~ j I may be obtained from
'5 C yptomeria japonica plants. However. Applicants have found that mRNA
codin~ for Cn~ j I could not be obtained from commercially available
Cr~ptomeria juponica pollen. This inability to obtain mRNA from the pollen
may be due to problems with stora~e or transportation of commercially a~ ailablepollen. Applicants have found that fresh pollen and staminate cones are a oood
source of Cry j I mRNA. It may also be possible to obtain the nucleic acid
sequence codin~ for Cl~ j I from ~enomic DN A. Cr~pt )n7~-ia japol7ic~ is a
well-known species of cedar. and plant materiul may be obtained from w ild.
.
~ .... ~ ...... .
WO 94/01~60 PCI/USg3/00139
'
~20~4
cultivated, or ornamental plants. The nucleic acid sequence codi~ r G~ j I
may be obtaine~ using the method disclosed herein or any other sui~ble
techniques for isolation and cloning of genes. The nucleic acid sequence of the
invention may be DNA or RNA. .
The present invention provides expression vectors and host cells
transforrned to express the nucleic acid sequences of the invention. A nucleic
acid sequence codin;~ for C~ j L Jurt v I or Jull s I or at least one fra~ment
thereof may be expressed in bacterial cells such as E. coti, insect cells
(baculovirus). yeast, or mammalian cells such as Chinese hamster ovary cells
(CHO). Suitable expression vectors. promoters, enhancers. and other expression
control elements may be found in Sambrook et al. Molecular Cloning: A
LaboJa~o~y Manual. second edition. Cold Sprin~ Harbor Laboratory Press. Cold
Spring Harbor. New York (1989). Other suitable expression vectors. promoters,
enhancers, and other expression elements are known to those skilled in the art.
Expression in mammalian, yeast or insect cells leads tO partial or complete
glycosylation of the recombinant material and fo~nation ot any inter- or intra-
chain disulfide bonds. Suitable vectors for expression in yeast include YepSec 1(Baldari et al. (1987) Embo J. 6: 229-234); pMFa (Kurjan and Herskowitz
(1982) Cell 3n 933-943~; JRY88 (Schultz et al. (1987) Gene 54: 113-123) and
pYES2 (Invitrogen Corporation~ San Diego, CA). These vectors are freely
available. Baculovirus and mammalian expression systems are also available.
For example, a baculovirus system is commercially available (PharMingen. San - ~-
Diego. CA) for expression in insect cells while the pMSG vector is commercially
- available ~Pharmacia. Piscataway, NJ) for expression in mammalian cells.
~5 For expression in E. coli. suitable expression vectors include. among
others. pTRC (Amann et al. (1988) Gene 69: 301-315): pGEX (Amrad Corp
Melbourne. Australia); pMAL (N.E. Biolabs. Beverly. MA): pRlT~ (Pharrnacia.
Piscataway. NJ); pET- 1 ld (Nova~en. Madison. WI) Jameel et al.. ( l 990) J.
Vi)ol. 64:3963-3966; and pSEM (Knapp et al. (1990) BioTechrliqlt~s ~: 280-
281) The use of pTRC. and pET-I ld. tor example. will lead to the expression ,
of unfused protein. The use of pMAL. pRIT5 pSEM and pGEX will lead to the ~ c
expression of aller~en fused to maltose E binding protein (pMAL)~ protein A
10.
i
WO 94/01560 PCr/US93/00139
2 1 2 0 9 ~ 4
(pRIT5). truncated B-galactosidase (PSEM), or glutathione S-transferase
(pGEX). When Ct~ j I. fra~ment~ or fragments thereot i~ expressed as a fusion
protein, it is particularly advantageous to introduce an enzymatic cleavage site at
the fusion junction between the carrier protein and C~ j I or fra~ment thereof.
s C~ j I or fragment thereof may then be recovered from the fusion plotein
through enzymatic cleavage at the enzymatic site and biochemical purification
using conventional techniques for purification of proteins and peptides. Suitable
enzymatic cleavage sites include those for blood clotting Factor Xa or thrombin
for which the appropriate enzymes and protocols for cleava~e are commercially
available from, for example. Sigma Chemical Company~ St. Louis. MO and N.E.
Biolabs, Beverly, MA. The different vectors also have different promoter
re. ions allowing constitutive or inducible expression with~ for example. IPTG
induction (PRTC, Amann et al.~ (1988) supru: ~ET-1 ld, Nova~en~ Madison.
WI) or temperature induction ~pRIT5. Pharmacia, Piscataway, NJ) . It may also
be appropriate to express recombinant C~ j I in different E. coli hosts that have
an altered capacity to degrade recombinantly expressed proteins (e.g. U.S. patent
4,758.512). Alternatively, it may be advantageous to alter the nucleic acid
sequence to use codons preferentially utilized by E. coli. where such nucleic acid
alteration would not affect the arnino acid sequence of the expressed protein.
Host cells can be transformed to express the nucleic acid
sequences of the invention using conventional techniques such as calcium
phosphate or calcium chloride co-precipitation. DEAE-dextran-media~d -
transfection. or electroporation. Suitable methods t`or transforming the host cells
may be found in Sambrook et al. supra. and other laboratory textbool;s.
The nucleic acid sequences of the invention may also be
synthesized using standard techniques.
The present invention also provides a method of producill~
isolated Japanese cedar pollen aller~en C~, ,j I or at least one fra~ment thereof
cornprising the steps of culturing a host cell transformed with a nucleic acid
sequence encodin~ Japanese cedar pollen aller~en C~ j I or at least on~ t`raoment
thereof in an appropriate medium to produce a mixture of cells and medium
conr~tining snid Japmese cednr pollen rller~on C~ j I or at least one trlgment
!
WO 94J01560 PCr/US93/00139
, 4
thereof; and purifying the mixture to produce substantially pure Japanese cedar I ;
pollen allergen Gy j I or at least one fra~ment thereof. Host cells transformed
with an expression vector containing DNA coding for C~' j I or at least one
fragment thereof are cultured in a suitable mediurn for the host cell. C~
protein and peptides can be purified from cell culture medium. host cells. or both
usin~ techniques known in the art for purifyin~ peptides and proteins including ~ ;
ion-exchan~e chromatography. ~el filtration chromatography~ ultrafiltratiom
electrophoresis and irnmunopurification with antibodies specific for C~ j I or
fragments thereof. The terms isolated and purified are used interchan~eably
0 herein and refer to peptides. protein, protein fragments~ and nucleic acid
sequences substantially free of cellular material or culture medium when
produced by recombinant DNA techniques~ or chemical precursors or other
chemicals when synthesized chemically.
Another aspect of the invention provides preparations comprising
Japanese cedar pollen aller~en G~ j I or a cross-reactive aller~en such as Ju~ v I
or Jun s I or at least one fragment thereof synthesized in a host cell transformed
with a nucleic acid sequence encoding all or a portion of Japanese cedar pollen
aller~en Cry j I or such cross-reactive allergen, or chemically synthesized. andisolated Japanese cedar pollen allergen C~ j I protein or a cross-reactive allergen
such as Jlm v I or Jl~n s I, or at least one antigenic fragment thereof produced in a
host cell transformed with a nucleic acid sequence of the invention. or
chemically synthesiæd. ln preferred embodiments of the invention th~ Cr~ j I i;
protein is produced in a host cell transformed with the nucleic acid sequence
coding for at least the mature G~! j I protein.
~5 Antigenic fragments as defined herein refer to any protein
fra~ment or peptide which induces an immune response. Unique antigenic
fra~ments as defined herein refer to any antioenic fraoment derived t`rom Cry j 1.
with the exception of the fragments consistino of amino acids 1-2() or 3 ~ .40 as
sho~vn in Figs. 4a-4b. Antigenic ~;agments of an allergen from Japanese ceda
pollen. or a cross-reactive allergen such as J~n v I or Ju11 s I may be obt;lined.
for example. by screening peptides recombinantly produced from the
corresponding fragment of the nucleic acid sequence of the invention codin~ for
WO94~01~60 '~ 2~20954 PCr/VS93/00139
such peptides or synthesized chemically using techniques known in the art. The
aller~en may be arbitrarily divided into fra~ments of a desired len~th with no
overlap of the peptides, or pret`erably divided into overlapping fragments ot a
desired !en~th. The fra~ments are tested to deterrnine their antigenicity (e.~. the
ability of the fragment to induce an immune response). If fragments of Cr~ j I
are to be used for therapeutic purposes~ then the fra~ments of C~ j 1 aller~en
which are capable of elicitin~ a T cell response such as stimulation (i.e.,
proliferation or Iymphokine secretion) and/or are capable of inducing T cell
anervy are particularly desirable and fragments of Japanese cedar pollen which
0 have minimal IgE stirnulating activity are also desirable. Minimal IgE
stimulating activity refers to IgE stimulating activity that is less than the amount
of IgE production stimulated by the native Ct~ j I protein. Additionally. for
therapeutic pulposes, it is preferable to use isolated Japanese cedar pollen
aller~ens, e.~. Cty j I, or fra~ments thereof which are capable of elicitin~ T cell -
responses and which do not bind I~E specific for Japanese cedar pollen or bind
such I~E to a substantially lesser extent than the purified native Japanese cedar
pollen allergen binds such IgE. If the isolated Japanese cedar pollen a}ler~en or
fragment or fragments thereof bind IgE~ it is preferable that such bindin~ does
not result in the release of mediators (e.g. histamines) from mast cells or
basophils. Furthermore, if Jun v I or Jun s I are to be used for therapeutic
purposes~ it is preferable to use Juniperl-s pollen aller~ens~ e.g. J~J7 v I or Jln7 s I
or a t`ra~ment thereof which are capable of elicitin~ T cell responses and which ~.``':3
do not bind IgE specific for pollen from the species Juniperlls or bind such I~Eto a substantially lesser extent than the purified native Juniperlls pollen aller~en
'5 binds such IgE. If the isolated Jl~n v I or J~n s l or &agment or fra2ments
thereof bind I~E, it is preferable that such binding does not result in the release
of mediators (e.g. histarnines) from mast cells or basophils.
Isolated protein allergens from Japanese cedar pollen or pret`ened
antigenic fra~ments thereof, when administered to a Japanese cedar pollell-
sensitive individual, or an individual aller~ic to an aller~en cross-reactive ~vith
Japanese cedar pollen allergen, such as aller~en from the pollen ot Jllni~7e) l~S
virginiana or J~mipetus sabinoides etc. (discussed previously) are capa~le o t
WO 94/01~60 PC~/US93/00139
21209~4 i -
~
modifying the allergic response of the individual to Japanese cedar pollen or
such cross-reactive aller~en of the individual. and preferably are capable of
modifying the B-cell response. T-cell response or both the B-cell and the T-cellresponse of the individual to the allergen. As used herein. modi~lcation of the
allergic response of an individual sensitive to a Japanese cedar pollen aller~en or 1~ -
cross-reactive aller~en can be de~lned as non-responsiveness or diminution in
symptoms to the allergen. as determined by standard clinical procedures (See e.~.
Varney et al, British Medical Journal, 3()2:265-269 (1990~) includin~
dimunition in Japanese cedar pollen induced asthmatic symptoms. As refe-Ted to
herein, a dimunition in symptoms includes any reduction in aller~ic response of
an individual to the allergen after the individual has completed a treatment
regimen with a peptide or protein of the invention. This dimunition may be
subjective (i.e. the patient feels more comfortable in the presence of the
allergen). Dimunition in symptoms can be determined clinically as well~ using
standard skin tests as is known in the art.
The isolated C~ j I protein or fra, ments thereof are preferably
tested in mammalian models of Japanese cedar pollinosis such as the mouse
model disclosed in Tarnura et al. (1986) Microbiol. ~mm~moL 30: 883-89~ or
U.S. patent 4,939,239: or the primate model disclosed in Chiba et al. ( 1990) Int.
Arch. Allergy Imm41nol. ~: 83-88. Initial screenin~ for I~E binding to the
protein or fragments thereof may be performed by scratch tests or intradelmal
skin tests on laboratory animals or human volunteers~ or in in vitro systems such
as RAST (radioallergosorbent test)~ RAST inhibition~ ELISA assay~
radioimmunoassay (RlA)~ or histamine release (see Examples 7 and 8).
2s Antigenic fra~ments of the present invention which have T cell
stimulating activity, and thus comprise at least one T cell epitope are palticularly
desirable. T cell epitopes are believed to be involved in initiation and
perpetuation of the immune response to a protein aller~en which is respol1sible
for the clinical symptoms of allergy. These T cell epitopes are thou; ht to tri~er
early events at the level of the T helper cell by bindin~ to an appropriate HLA
molecule on the sultace ot an anti~en presentin~ cell and stimulatin~ the relevant i-~
T cell subpopulation. These events lead to T cell prolit`eration~ lymphol;ine
WO 94/01~60 2 1 2 0 9 ~ 4 PCI`/US93/00139
secretion, local inflammatory reactions, recruitment of additional immune cells
to the site, and activation of the B cell cascade leading to production of
antibodies. One isotype of these antibodies. IgE, is fundamentally important to
the development of allergic symptoms and its production is influenced early in
S the cascade of events, at the level of the T helper cell. by the nature of the
Iymphokines secreted. A T cell epitope is the basic element or smallest unit of
recognition by a T cell receptor, where the epitope comprises amino acids
essential to receptor recognition. Amino acid sequences which mimic those of
the T cell epitopes and which modify the allergic response to protein alleroens
are within the scope of this invention.
Exposure of cedar pollen patients to isolated protein allergens of
the present invention or to the antigenic fragments of the present invention which
comprise at least one T cell epitope and are derived from protein allergens may
tolerize or anergize appropriate T cell subpopulations such that they become
unresponsive to the protein allergen and do not participate in stimulating an
immune response upon such exposure. ln addition. administration of a protein
aller~en of the invention or an antigenic fragment of the present invention which
comprises at least one T cell epitope may modify the lymphokine secreuon
profile as compared with exposure to the naturally-occurring protein alleroen orportion thereof (e.g. result in a decrease of IL-4 and/or an increase in IL-2).
Furthermore, exposure to such protein allergen or anti~enic fra~ment of such
protein allergen may influence T cell subpopulations which normally participate
in the response to the allergen such that these T cells are drawn away from the
site(s) of normal exposure to the allergen ~e.g., nasal mucosa, skin~ and lun~)
'5 towards the site(s) of therapeutic administration of the fragment or protein
allergen. This redistribution of T cell subpopulations may ameliorate or reduce
the ability of an individual's immune systern to stimulate the usual immune
response at the site of normal exposure to the allerPen~ resulting in a dimunution
inl aller~ic symptoms.
The isolated C~ j 1. JUJ1 ~ I or Jun s 1 protein~ and tra~m~nts or
portions derived therefrom (peptides) can be used in methods of diagnosino.
treating and preventing aller~ic reactions to Japanese cedar pollen aller en or a
WO 94~01560 PCr/US93/00139
21209S4
cross reactive protein aller~en. Thus the present invention provides therapeuticcompositions comprising isolated Japanese cedar pollen allergen C~ j I, JIJn v Ior Jun s I or at least one fragment thereof produced in a host cell transt'ormed to
express Cr ~ j I. Jun v I or Jun s I or at least one fragment thereof, and a
s pharmaceutically acceptable carrier or diluent. The therapeutic compositions of
the invention may also comprise synthetically prepared CrY j I. J~n v I or Jun s I
or at least one fragment thereof and a pharmaceutically acceptable carrier or
diluent. Administration of the therapeutic compositions of the present inventionto an individual to be desensitized can be carried out using known techniques.
lo C~y j I. Jun v I or Jl~n s I protein or at least one fra~ment thereof may be
administered to an individual in combination with, for example, an appropriate
diluent~ a carrier and/or an adjuvant. Pharrnaceutically acceptable diluents
include saline and aqueous buffer solutions. Pharrnaceutically acceptable
carriers include polyethylene glycol (Wie et al. (1981) Int. Arch. Alle~g! Appl.
..
Immlmol. 64:84-99) and liposomes (Strejan et al. (1984) J. Neu-oimml010l 7:
27). For purposes of inducing T cell anergy, the therapeutic composition is
preferably administered in nonimmuno~enic forrn, e.g. it does not contain
~` adjuvant. The therapeutic compositions of the invention are administered to
Japanese cedar pollen-sensitive individuals or individuals sensitive to an allergen
which is immunolo~ically cross-reactive with Japanese cedar pollen allergen (i.e.
J~miperus virginiana, or Juniperus sabinoides. etc.).
Administration of the therapeutic compositions of the present , .
invention to an individual to'be desensitiæd can be carried out using known
procedures at dosa~es and for periods of time effective to reduce sensitivity (i.e
reduce~ the ~allerg* response) of the individual to the allergen. Effécti~e amounts
of the therapeutic compositions will vary according to t'actors such as the degree
; ~ of sensitivity of the individual to Japanese cedar pollen. the age~ sex. and ~vei~ht
of the individual. and the ability of the protein or fra~ment thereof to elicit an
' ~ i antigenic response in the individual.
The active compound (i.e.~ protein or fragment thereof~ mav be
administered in a convenient manner su'ch asi by injection (subcutaneous.
intravenous, etc.), oral administ~ation, inhalation~ transdermal application. or
,~
~
.
,~
,
' 16
:~:
WO 94~01560 Pcr/us93JO0139
21209~5~ `
rectal administration. Depending on the route of administration, the active
compound may be coated within a material to protect the compound from the
action of enzymes, acids and other natural conditions which may inactivate the
compound.
For example, preferably about 1 ,ug- 3 mg and more preferably
from about 20-500 ~Lg of active compound (i.e., protein or fragment thereof) perdosace unit may be administered by injection. Dosage regimen may be adjusted
to provide the optimurii therapeutic response. For example, several divided
doses may be administered daily or the dose may be propoltionally reduced as
indicated by the exigencies of the therapeutic situation.
To administer protein or peptide by other than parenteral
administration, it may be necessary to coat the protein with~ or co-administer the
protein with~ a material to prevent its inactivation. For example~ protein or
fragment thereof may be administered in an adjuvant~ co-administered with
enzyme hibitors or in liposomes. Enzyme inhibitors include pancreatic trypsin
inhibitol~ diisopropylfluorophosphate (DEP) and trasylol. Liposomes include
water-in-oil-in-water CGF emulsions as well as conventional liposomes (Strejan
e~ al.~ (1984) J. Ne~immunol. 7:27).
The active compound may also be administered parenterally or
~0 intraperitoneally. Dispersions can also be prepared in glycerol~ liquid
polyethyline glycols~ and mixtures thereof and in oils. Under ordinary
conditions of storage and use, these preparations may contain a preservative to .
prevent the growth of microorganisms.
Pharmaceutical compositions suitable for injectable use includ~
~5 sterile aqueous solutions (where water soluble) or dispersions and sterile powders
for the extemporaneous preparation of sterile injectable solutions of dispersion.
In all cases~ the composition must be sterile and must be fluid to the extent that
easy syringability exists. It must be stable under the conditions of manutactureand storage and must be preserved against the contaminating action of
~0 microorganisms such as bacteria and fungi. The currier can be a solvent or ~i
dispersion medium containing, for example, wuter, ethanol~ polyol (t`or example, ~,
glyceral, propylene glycol, and liquid polyetheylene glycol, and the like). ~j
,
WO 94/01560 PCr/US93/00139
.
?,~.209~ :
suitable mixtures thereof, and vegetable oils. The proper fluidity can be
maintained. for example~ by the use of a coating such as licithin. by the
maintenance of the required particle size in ehe case of dispersion and by the use
of surfactants. Prevention of the action ot microor~anisms can be achieved by
various antibacterial and antifun~al agent~s~ for example~ parabens~ chlorobutanok
phenol, ascorbic acid, thirmerosal, and the like. In many cases, it will be
preferable eo include isoeonic agents, for example~ su~ars. polyalcohols such asmanieol and sorbitol or sodium chloride in the composition. Prolonged
absorption of the injectable compositions can be brought about, includin~ in thecomposition, an agent which delays absorption, for example~ aluminum
monostearate and gelatin.
Sterile injectable solutions can be prepared by incorporatino
active compound (i.e., protein or peptide) in the required amount in an
appropriate solvent with one or a combination of inoredients enumerated above.
as required, followed by filtered sterilization. Generally, dispersions are
prepared by incorporating the active compound into a sterile vehicle which
contains a basic dispersion medium and the required other ingredients from thoseenumerated above. In the case of sterile powders for the preparation of sterile
indectable solutions, the preferred methods of preparation are vacuum dryino and20 freeze-drying which yields a powder of the active ingredient ~i.eprotein or
peptide) plus any additional desired ingredient from a previously sterile-filtered
solution thereof.
When protein or peptide thereof is suitably protected. as described
above. the protein may be orally administered. for example. with an inelt diluent
~3 or an assimilable edible carrier. The protein and other in~redients may ~Iso be
enclosed in a hard or soft shell gelatin capsule. compressed into tablets. or
incorporated directly into the individual's diet. For oral therapeutic
administration, the active compound may be incorporated with excipients and
used in the forrn of in~estible tablets, buccal tablets. troches~ capsules~ elixirs.
~0 suspensions. syrups. wafers~ and the like. Such compositions and preparali( ns
should contain at least 15~ by wei~ht of active compound. The percenta_e o t` the ~.;
composition and preparations may~ of course. be v alied and may conveniently be
~.
WO 94/~1560 2 1 2 ~ 9 5 ~ PCT'/US93/00~39
between about 5 to 80% of the weight of the unit. The amount of active
compound in such therapeutically useful compositions is such that a suitable
dosa~e will be obtained. Preferred compositions or preparations accordin~ to thepresent invention are prepared so that an oral dosage unit contains between fromabout lO ~lg to about 200 mg of active compound.
The tablets, troches, pills~ capsules and the like may also contain
the following: a binder such as gum gragacanth, acacia, corn starch or gelatin;
excipients such as dicalcium phosphate; a disinte~rating a~en~ such as corn
starch, potato starch, alginic acid and the like; a lubricant such as magnesium
stearate; and a sweetening agent such as sucrose. Iactose or saccharin or a
flavoring agent such as peppermint, oil of winter~reen, or cherry flavoring.
When the dosage unit forrn is a capsule~ it may contain~ in addition to materials
of the above type, a liquid carrier. Various other materials may be present as
coatings or to otherwise modify the physical form of the dosage unit. For
instance, tablets, pills, or capsules may be coated with shellac. sugar or both. A
syrup or elixir may contain the active compound. sucrose as a sweetenino agent,
methyl and propylparabens as preservative, a dye and flavoring such as cherry ororange flavor. Of course, any material used in preparing any dosage unit form
should be pharmaceutically pure and substantially non^toxic in the amounts
~0 employed. In addition. the active compound may be incorporated into sustained-
release preparations and formulations.
As used herein "pharmaceutically acceptable carrier" includes any
and all solvents. dispersion media. coatings. antibactelial and antifun~al a~ents.
isotonic and absorption delaying agents. and the like. The use of such media and- ~5 a~ents for phannaceutically active substances is well known in the a~t. Except
insofar as any conventional media or a~ent is incompatible with the active il
compound. use thereof in the therapeutic compositions is contemplated
Supplementary active compounds can also be incorporated into the
compositions. ~.
~0 It is especially advanta~eous to formulate parenteral compositions
in dosa~e unit form for ease of administration and uniformi~y of dosa~e. Dnsa~e
unit from as used herein refers to physically discrete units suited as unitary
WO 94/01560 PCI`/lJS93/00139
9S 4
dosa~es for the mammalian subjects to be treated; èach unit containin~ a
predetetmined quantity of active compound calculated to produce the desired I -
therapeutic effect in association with the required pharmaceutical carrier. The
specification for the novel dosa~e unit forms of the invention are dictated by and
directly dependent on (a) the unique characteristics of the active compound and
the particular therapeutic effect to be achieved, and (b) the limitations inherent in
the art of compounding such an active compound for the treatment of sensitivity
in individuals. I ~
The C~ j I cDNA (or the mRNA trom which it was transcribed)
or a portion thereof can be used to identify similar sequences in any variety ortype of plant and thus. to identify or "pull out" sequences which have sufficient
homology to hybridize to the Cry j I cDNA or mRNA or portion thereof. for
example, DNA from allergens of Juniperlls vi-ginlana, Juniperus sabinoides
etc., under conditions of low stringency. Those sequences which have sufficient
homology ~generally greater than 40%) can be selected for further assessment
usi~ng the method de~scribed herein. Alternatively~ hi~h stringency conditions can
be~used. In th~s manner, DNA of the present invention can be used to identify~ in
other~types ~of plants, preferably related families, genera~ or species such as
Junipcrus,~ or ~ (~préssus, sequences encoding polypeptides having amino acid
20 ~ ~ ; sequences~slmilar~to ~at of~Japanese cedar pollen allergen Cry; I~ and thus to
identify allergens in other species. Thus~ the present invention includes not only `-
G-~ j I, but also other aller~ens encoded by DNA which hybridizes to DNA of
the present~lnvention. ~The invention further includes isolated alkrgenic proteins
or fragments therèof that are if nmunolo~ically related to G~ j I or fragments
2~ ~thereof. such~as by antibody cross-reactivity wherein the isolated aller~enic
proteins or~fragments thereof are capable of binding to antibodies specific for the
protein~ and~;peptides~ of the invention, or by T cell cross-reactivity wherein the
isolated aller~enic proteins or fragments thereof are capable ot stimulatin~ T
cells specific for the protein and peptides of this invention.
30 ~ Proteins or peptides encoded by the cDNA of the present ~ ~
- invention can be used, forexample as "pulifted" alleloens~ Such purified ~;
aller~ens are useful in the standardizadon of alleroen extracts which ale l;ey
, ~ .
~o
WO 94/01560 2 1 2 0 9 ~ ~ PCI`/US93/00139
reagents for the diagnosis and treatment of Japanese cedar pollinosis.
Furthermore, by usin~ peptides based on the nucleic acid sequence of Cr~
anti-peptide antisera or monoclonal antibodies can be made using standard
methods. For example. an animal such as a mouse or rabbit can be immunized
with an irnmunogenic from of the isolated c,~ j I protein (e.g., C~ ~ j I protein or
anti~Tenic fragment which is capable of elicitin~ an antibody response).
Techniques for conferring immuno~enicity on a protein or peptide subunit
include conju~ation to carriers or other techniques well-known in the art. The ',
C~ j I protein or peptide therof can be administered in the presence of adjuvant.
lo The progress of immunization can be monitored by detection of antibody titers in
plasma or serum standard ELISA or other immunoassay can be used with the
immuno~en as antigen to assess the levels of antibodies.
Following immunization, anti-Cr~ j I antisera can be obtained
and, if desired. polyclonal anti-Cry j I antibodies t`rom the serum. To produce
1~ monoclonal antibodies, antibody producing cells (lymphocytes) can be harvested
from an immunized animal and fused by standard somatic cell fusion procedures
with immortalizing cells such as myeloma cells to yield hybridoma cells.
Hybridoma cells can be screened immunochemically for production of antibodies
reactive with the Cry j I protein or peptide thereof. These sera or monoclonal
~0 antibodies can be used to stanaaldize aller~en extracts.
Through use of the peptides and protein of the present invention.
preparations of consistent, well-defilned composition and biolo~ical activity can ;~
be made and administered for therapeutic pulposes ~e.g. to modify the aller~ic
response of a Japanese cedar sensitive individual to pollen of such trees).
Administration of such peptides or protein may, for example, modify B-cell
response to C~ Y j I allergen, T-cell response to G~ j I allergen or both responses.
lsolated peptides can also be used to study the mechanism ot immunotherapy of
C)~ptomeria japonica aller~y and to design modified derivatives or analooues
useful in immunotherapy.
Work by others has shown that hi~h doses of aller~ens generally
produce the best results (i.e., best symFtom relie~). However~ many people are
unable to tolerate large doses of aller~ens because of aller~ic reactions to the
WO 94/01560 PCr/US93/00139
212~95~
allergens. Modification of naturally-occurring allergens can be desi~ned in such '
a rnanner that modified peptides or modified aller~ens which have the same or
enhanced therapeutic properties as the corresponding naturally-occurrin~
allergen but have reduced side effects (especially anaphyl~ctic reactions) can be
s produced. These can be~ for example, a protein or peptide of the present .
invention (e.~., one having all or a portion of the amino acid sequence of C~ j
I), or a modified proteill or peptide, or protein or peptide analogue.
It is also possible to modify the structure of a peptide ()f the
invention for such purposes as increasing solubility, enhancin~ therapeutic or
preventive efficacy, or stability (e.g., shelf life ex vivo, and resistance to
proteolytic degradation in vivo). A modified peptide can be produced in which
the amino acid sequence has been altered. such as by amino acid substitution~
deletion~ or addition. to modify immunogenicity andlor reduce aller~enicity~ or
to which a component has been added for the same purpose.
For example, a peptide can be modified so that it maintains the
ability to induce T cell anergy and bind MHC proteins without the ability to
induce a stron~ proliferative response or possibly. and proliferative response
when administered in immuno~enic form. In this instance, critical bindin~ -
residues for the T cell receptor can be determined using known techniques (e.~
substitution of each residue and determination of the presence or absence of T
cell reactivity). Those residues shown to be essential to interact with the T cell
receptor can be modified by replacing the essential amino acid with another~ !',~;~;,
preferably similar amino acid residue (a conservative substitution) whos~
presence is shown to enhance, diminish but not eliminate, or not affect T cell
~5 reactivity. In addition, those amino acid residues which are not essential ~r T
cell receptor interaction can be modified by bein~ replaced by another amino
acid whose incorporation may enhance. diminish or not aft`ect T cell reacti~ it~but does not eliminate bindin~ to relevant MHC.
Additionally, peptides of the invention can be modified by
replacinu an amino acid shown to be essential to interact with the MHC protein
complex with another. preferably similar amino acid residue (conser~ ~ti\ e
substitution) whose presence is shown to enhance. diminish but not elimin;l~e ol .
I
WO 94/01560 2 1 ~ Q ~ 3 4 PCI /US93/00139
not affec$ T cell activity. In addition, arnino acid residues which are not essential ~.
for interaction with the MHC protein complex but which still bind the MHC
protein complex can be modified by being replaced by another amino acid whose
incorporation may enhance. not affect, or diminish but not eliminate T cell
reactivity. Preferred amino acid substitutions for non-essential amino acids
include, but are not limited to substitutions with alanine, ~lutamic acid, or a
methyl amino acid.
In order to enhance stability and/or reactivity, the protein or
peptides of the invention can also be modified to incorporate one or more
polymorphisms in the amino acid sequence of the protein allergen resulting from
natural allelic variation. Additionally, D-amino acids, non-natural amino acids
or non-amino acid analogues can be substituted or added to produce a modified
protein or peptide within the scope of this invention. Furthermore. proteins or
peptides of the present invention can be modified using the polyethylene glycol
(PEG) method of A. Sehon and co-workers (Wie et al. sup?a) tO produce a
protein or peptide conjugated with PEG. In addition, PEG can be added during
chemical synthesis of a protein or peptide of the invention. Modifications of
proteins or peptides or portions thereof can also include reductionl alyklation
~Tarr in: Met~ods of Protein Microcharacteri~ation, J.E. Silver ed. Humana
~o Press~ Clifton, NJ, pp 155-194 (1986)); acylation (Tarr, sl~p-a); chemical
coupling to an appropriate carrier (Mishell and Shiigi~ eds. Sel~cted M~t~10~s il7
Celfular Imn~nology, WH Freeman~ San Francisco. CA (1980): U~S. Patent
4.939.239; or mild formalin treatment (Marsh International Archi~es of AII~Jg~
and Applied Immunology, 41:199-215 (1971)).
To facilitate purification and potentially increase solubility ot
proteins or peptides of the invention, it is possible to add reporter group(s) to the
peptide backbone. For example, poly-histidine can be added to a peptide IO
purify the peptide on immobilized metal ion affinity chromatography (Hochuli~
E. et al.. Bio/Technolog~, 6:1321-1325 (1988)). In addition, specific
~0 endoprotease cleava~e sites can be introduced~ if desired~ between a repoltergroup and amino acid sequences of a peptide to facilitate isolation ot` peplidesfree of irrelevant sequences. In order to successfully desensitize an indi~ idual to
~ !
- .
WO 94/0156~ PCI`/US93/00139
212Q9~4
a protein antigen, it may be necessary to increase the solubility of a protein or
peptide by adding functional groups to the peptide or by not including
hydrophobic T cell epitopes or regions containing hydrophobic epitopes in the
peptides or hydrophobic regions of the protein or peptide.
To potentially aid proper antigen processing of T cell epitopes
within a peptide, canonical protease sensitive sites can be recombinantly or
synthetically engineered between regions. each comprising at least one T cell
epitope. For example. charged amino acid pairs, such as KK or RR, can be
introduced between regions within a peptide durin~ recombinant construction of
0 the peptide. The resulting peptide can be rendered sensitive to cathepsin and~or
other trypsin-like enzymes cleavage to generate portions of the peptide
containing one or more T cell epitopes. In addition, such charged amino acid
residues can result in an increase in solubility of a peptide.
Site-directed mutagenesis of DNA encoding a peptide or protein
of the invention (e~g. Gy j I or a fragment thereof) can be used to modify the
structure of the peptide or protein by methods known in the art. Such methods
may~ among others, include PCR with de~enerate oligonucleotides (Ho et al
Gene, 77:51-59 (1989)) or total synthesis of mutated genes (Hostomsky, Z. et al.,
Biochem. Biophys, Res. Comm., 161:1056-1063 (1989)). To enhance bacterial
_0 expression, the aforementioned methods can be used in conjunction with other
procedures to chan~e the eucaryotic codons in DNA constructs encoding protein
or peptides of the invention to ones preferentially used iR E. coli. yeast~ ~`
mammalian cells, or other eukaryotic cells.
Usin~ the structural information now available. it is possible to
~5 design Cr~ j I peptides which, when administered to a Japanese cedar pollen
sensitive individual in sufficient quantities, will modify the individual's alleroic
response to Japanese cedar pollen. This can be done, for example. by examinin~ '
the structure of Cr~ j I, producing peptides (via an expression system~
synthetically or othen,vise) to be examined for their ability to influence B-cell
~0 and/or T-cell responses in Japanese cedar pollen sensitive individuals and
selecting appropriate peptides which contain epitopes reco; nized by the c~lls. In
referrin~ to an epitope~ the epitope will be the basic element or smallest unit ot
24
WO 94/01560 2 1 2 Q 9 ~ 4 PCr~US93/00139
recognition by a receptor. particularly immuno~lobulins, histocompatibility
antigens and T cell receptors where the epitope comprises amino acids essential
to receptor recognition. Amino acid sequences wbich mimic those of the
epitopes and which are capable of down regulating allergic response to C~
can alsobe used.
It is now also possible to design an agent or a drug capable of
blocking or inhibiting the ability of Japanese cedar pollen allergen to induce an
allergic reaction in Japanese cedar pollen sensitive individuals. Such a~ents
could be designed, for exarnple, in such a manner that they would bind to
relevant anti-Cry j I IgEs, thus preventing IgE-aller~en bindin~ and subsequent
mast cell degranula~ion. Alternatively~ such a~ents could bind to cellular
cornponents of the immune system, resulting in suppression or desensiti-zation of
the allergic response to C~yptomeria japonica pollen allergens. A non-
restrictive example of this is the use of appropriate B- and T-cell epitope
1~ peptides. or modifications thereof, based on the cDNA/protein structures of the
present invention to suppress the aller~ic response to Japanese cedar pollen. This
can be carried out by defining the structures of B- and T-cell epitope peptides
which affect B- and T-cell function in in vitro studies with blood components
from Japanese cedar pollen sensitive individuals.
- 'O Protein, peptides or antibodies of the present invention can also be
used for detecting and diagnosing Japanese cedar pollinosis. For example. this
could be dorie by combining blood or blood products obtained from an
individual to be assessed for sensitivity to Japanese cedar pollen with an isolated
antigenic peptide or peptides of Cr~ j I. or isolated C~y j I protein. under
~5 conditions appropriate for binding of components in the blood (e.g.. antibodies.
T-cells~ B- cells) with the peptide~s) or protein and determining the extent to
which such binding occurs.
The present invention also provides ~ method ot producinc~ C)~ j I
or fra~ment thereof comprising culturing a host cell containing an expression
vector which contains a nucleic acid sequence e.g. DNA. encodin all or at least
one fra~ment of Cr \~ j I under conditions appropriate tor expressi~ of C/~ j I Ol
at least one fnagment. The expressed product is then lecovered. using kno~n
WO 94/01560 PCI/US93/00139
2120954
techniques. Altematively, Cr~ j I or fragment thereof can be synthesized usin~ '
known mechanical or chemical techniques.
The DNA used in any embodiment of this invention can be cDNA
obtained as described herein. or alternatively. can be any oligodeoxynucleotide
sequence having all or a portion of a sequence represented herein. or their
functional equivalents. Such oligodeoxynucleotide sequences can be produced
chemically or enzymatically. using known techniques. A functional equivalent of
an oligonucleotide sequence is one which is 1) a sequence capable of hybridizingtO a complementary oligonucleotide to which the sequence (or corresponding ~~
sequence portions) of SEQ ID NO: I or fragments thereof hybridizes~ or ~) the
sequence (or correspondin~ sequence portion) complementary to SEQ ID NO: 1.
and/or 3) a sequence which encodes a product (e~g., a polypeptide or peptide)
having the same functional characteristics of the product encoded by the
sequence (or corresponding sequence portion) of SEQ ID NO: 1. Whether a
functional equivalent must mèet one or both critena will depend on its use (e.~Jif it is to be used only as an oligoprobe, it need m`eet only the first or second
critelia and if it is to be used to produce a Cr~ j I aller~en~ it need only meet the
third cnterion).
The present invention also provides isolated peptides derived from
~0 Japanese cedar pollen protein. As used herein, a peptide or fragment of a protein
refers to an amino acid sequence having fewer amino acid residues than the
entire amino acid sequence of the protein from which it is derived. Peptides of
the invention include peptides derived from Cr~ j 1 which comprise at least one T
cell epitope of the allergen.
~s Peptides comprising at least two regions, each re~ion comprising at
least one T cell epitope of Japanese cedar pollen are also within the scope ot the
invention. Isolated peptides or regions of isolated peptides~ each complisino atleast two T cell epitopes of a Japanese cedar pollen protein aller~en are
particularly desirable for increased therapeutic et`tectiveness. Peptides which are
immunologically related (e.~.. by antibody or T cell cross-reactivity~ to peptides
of the present invention are also within the scope of the invention. ~,
Isolated peptides of the invention can be produced by recombinant
'6
. ~
WO 94/01560 2 ~ ~ Q 9 ~ '~ PCI/US93/00139
.
DNA techniques in a host cell transformed with a nucleic acid having a sequence
encoding such peptide as discussed above. The isolated peptides of the inventioncan also be produced by chemical synthesis. With re~ard to isolated Jun 1~ I
protein or peptides. such protein or peptides may be produced by biochemically
S purifying the native Jun v I proteins from Juniperus ~irginiana pollen as is
known in the art. When a peptide is produced by recombinant techniques. host
cells transformed with a nucleic acid having a sequence encoding the peptide or
the functional equivalent of the nucleic acid sequence are cultured in a medium
suitable for the cells and peptides can be purified from cell culture medium. host
cells, or both using techniques known in the art for purifying peptides and
proteins including ion-exchange chromatography, gel filtration chromatography,
- _ ultrafiltration, electrophoresis or immunopurification with antibodies specific for
the peptide, the protein allergen Japanese cedar pollen from which the peptide is
derived~ or a portion thereof. lsolated peptides of the invention are substantially
fr~ee of celIular material or culture medium when produced by recombinant DNA
techniques, or substandally free of chemical precursors or other chemicals when
synthesized chemically.
~,
To obtain isolated peptides of the present invention, C~ j I is
divided into non-overlapping peptides of desired length or overlapping peptides
20 of desired lengths as discussed in Example 6 which can be produced
recombinantly. or synthetically. Peptides comprising at least one T cell epitopeare capable of eliciting a T cell response. such as T cell proliferation or ~i .
Iympho~ine secretion, andlor are capable of inducing T cell anergy ~1.e.~
t olerization). T o determine peptides comprising at least one T cell epitope.
2~ ~ isolated peptides are tested by, tor example. T cell biology techniques~ todetermino whether the peptides elicit a T cell response or induce T cell anergy.Those peptides found to elicit a T cell response or induce T cell anergy are
defined as having T cell stimulating activity.
As discussed in Example 6~ human T cell stimulating activity can be tested
30 by culturing T cells obtained from an individual sensitive to Japanese cedar
pollen allergen~ (i.e.. an individual who has an 1E mediateti immune response to
lapanese cedar pollen allergen) with a peptide derived t`rom the allergen and
~7
WO 94/01560 PCI /USg3/00~39
2~209S4
deterrnining whether proliferation of T cells occurs in response to the peptide as
measured, e.g., by cellular uptake of tritiated thymidine. Stimulation indices for
responses by T cells to peptides can be calculated as the maximum CPM in
response to a peptide divided by the cc)ntrol CPM. A stimulation index (S.I.)
equal to or greater than two times the background level is considered "positive".
Positive results are used to calculate the mean stimulation index for each peptide
for the group of patients tested. Preferred peptides of this invention comprise at
-
least one T cell epitope and have a mean T cell stimulation index of 8reater than
or equal to 2Ø A peptide having a mean T cell stimulation index of greater than
or equal to 2.0 is considered useful as a therapeutic agent. Preferred peptides
have a mean T cell stimulation index of at least 2.5~ more preferably at least 3.5.
more preferably at least 4.0, more preferably at least 5, even more preferably at
least 7 and and most preferably at least about 9. For example, peptides of the
invention having a mean T cell stimulation index of at least 5, as shown in Fi~.1~ 14, include CJ1-2, CJ1-7, CJ1-10, CJl-15. CJl-17, CJl-20, CJl-22. CJl-23.
CJl-24, CJ1-27, CJ1-31, CJl-32 and CJl-35. For example, peptides of the
invention having a mean T cell stimulation index of at least 7, as shown in Fi~.14. include CJI-16, CJl-20, CJl-22, and CJl-32.
- In addition, preferred peptides have a positivity index (P.I.) of at least
~0 about 100, more preferably at least about 250 and most preferably at least about
350. The positivity index for a peptide is deterrnined by multiplyin~ the mean Tcell stimulation index by the percent of individuals. in a population of ,~
individuals sensitive to Japanese cedar pollen (e.g.. pret`erably at least 15
~- individuals, more preferably at least 30 individuals or more), who have a T cell
stimulation index to such peptide of at least 2Ø Thus, the positivity index
represents both the strength of a T cell response to a peptide (S.I.) and the
frequency of a T cell response to a peptide in a population of individuals s
sensiti-e to Japanese cedar pollen. For example. as shown in Fi~. 14~ peptid~
; i CJi-22 has a mean S.I. of 14.5 and 60.0% of positive responses in the ~roup ot`
individuals tested resultin~ in a positivity index of X70.0(). Peptides ot` C~
havin~ a positivity index of at least about l()l) and u mean T cell stimulation
index of at least about 4 include: CJ l - l fi~ CJ l- 17~ CJ I-20~ CJ I -22~ CJ l-~. CJ I -
,
,~
Wo 94/01560 '- 2 1 2 0 ~ j ~ PCI/US93/00139
24, CJ 1-26, CJ 1-27, CJ 1-32 and CJ 1-35.
In order to determine precise T cell epitopes by, for example. fine
mapping techniques, a peptide having T cell stimul~ ting activity and thus
comprising at least one T cell epitope as determined by T cell biolooy cechniques
is modified by addition or deletion of amino acid residues at either the amino or
carboxy terrninus of the peptide and tested to determine a change in T cell
reactivity to the modified peptide. If two or more peptides which share an area
of overlap in the native protein sequence are found to have human T cell
stimulatin~ activity, as determined by T cell biology techniques. additional
peptides can be produced comprising all or a portion of such peptides and these
additional peptides can be tested by a similar procedure. Followin~ this
technique, peptides are selected and produced recombinantly or synthetically.
Peptides are selected based on various factors~ including the strength of the T cell
response to the peptide (e.g., stimulation index). the frequency of the T cell
1~ response to the peptide in a popula~ion of individuals sensitive to Japanese cedar
pollen, and the potential cross-reactivity of the peptide with other allergens from
other species of trees as discussed earlier (e.g. Cupressus sempervirens,
Cupressus arizonica, Juniperus virginiana, Juniper~s sabinoides, etc.) or
ragweed (Amb a I.1) . The physical and chemical properties of these selected
'0 peptides (e.g., solubility, stability) are examined to determine whether the
- peptides are suitable for use in therapeutic compositions or whether the peptides
require modi~lcation as described herein. The ability of the selected peptides or
selected modified peptides to stimulate human T cells (e.~., induce proliferation.
lymphokine secretion) is determined.
Additionally, preferred T cell epitope-containing peptides of th~
invention do not bind immunoglobulin E (I~E) or bind IgE to a substantially
lesser extent than the protein allergen trom which the peptide is derived binds
IgE. The major complications of standard immunotherapy are I~E-media~d
responses such as anaphylaxis. Immunoglobulin E is a mediator o~ anaphyl;lctic
reactions which result from the binding and cross-linking of antigen to IgE on
mast cells or basophils and the release ot mediators (e.~.~ histamine~ serotonin~
eosinophil chemotacic factors). Thus. anaphylnxis in a substantial percen~ of
WO 94~01~60 PCI/USg3100139
2~2~95~
a population of individuals sensitive to Cr~ j I could be avoided by the use in
immunotherapy of a peptide or peptides which do not bind IgE in a substantial
percentage (e.g., at least about 75%) of a population of individuals sensitive to
C~ j I allergen. or if the peptide binds lgE. such binding does not result in the
release of mediators from rnast cells or basophils. The risk of anaphylaxis could
be reduced by the use in immunotherapy o~ a peptide or peptides which have
reduced IgE binding. Moreover, peptides which have minimal IgE stimulatin~
activity are desirable for therapeutic effectiveness. Minimal IgE stimulating
activity refers to IgE production that is less than the amount of IgE productionandlor IL-4 production stimulated by the native C)~ j I protein allergen.
A T cell epitope containing peptide of the invention, when administered to
a Japanese cedar pollen-sensitive individual~ is capable of modifying the aller~ic
response of the individual to the allergen. Particularly, peptides of the invention
comprising at least one T cell epitope of Cr~ j I or at least two regions derived
from Cr~ j I, e~ch comprisins at least one T cell epitope. when administered to
an individual sensitive to Japanese cedar pollen are capable of modifying T cellresponse of the individual to the allergen.
A preferred isolated peptide of the invention comprises at least one T cell
epitope of the Japanese cedar pollen allergen. G~ j I and accordingly the
peptide comprises at least approximately seven amino acid residues. For
purposes of therapeutic effectiveness. preferred therapeutic compositions of theinvention preferably comprise at least two T cell epitopes of C~ ~ j I. and
accordingly, the peptide comprises at least approximately eight amino acid
residues and preferably at least fifteen amino acid residues. Additionally.
therapeutic compositions comprising preferred isolated peptides of the inventinnpreferably comprise a sufficient percentage of the T cell epitopes of the entireprotein allergen such that a therapeutic regimen of administration of the
composition to an individual sensitive to Japanese cedar pollen~ results in T cells
of the individual being tolerized to the protein allergen. Synthetically produced
peptides of the invention comprising up to approximately forty-five amino acid
residues in length, and most preferably up to approximately thirty amino acid ë .
residues in length are particularly desirable as increases in length may result in
,
~0
WO 94/01560 21 2 0 9 5 4 PCr/US93/00139
dif~lculty in peptide synthesis. Peptides of the invention may also be produced
recombinantly as described above, and it is preferable that peptides of 45 amino s
acids or longer be produced recombinantly.
Preferred peptides comprise all or a portion of the areas of major T cell
s reactivity within the Cry; I protein allergen designated herein as, Region 1.
Region 2,~Region 3, Region 4 and Region 5. Each major area of T cell activity
is de~med as follows and is shown in Fig. 4 a-b. Region 1 comprises amino acid
residues 1-50 of Cry; I; Region 2 comprises amino acid residues 61-120 of C
j I; Region 3 comprises amino acid residues 131-180 of Cry j I; Region 4
comprises amino acid residues 191-280 of C~ j 1: Region 5 comprises amino
acid residues 291-353 of the C~ j I. Preferred areas of major T cell reactivity
within each Region as shown in Fig 4 a-b and comprise: amino ~cid residues 1-
40; amino acid residues 81-110; amino acid residues 15 l -180; amino acid
residues 191-260; and amino acid residues 291-330.
IS Peptides derived from the Cr~ j I protein allergen which can be
used for therapeutic purposes comprise all or a portion of the following peptides:
CJ 1- 1, CJl-2, C~J1-3, CJl-4, CJl-7, CJl-8, CJ 1-9~ CJl- 10, CJ1- 1 1, CJl- 12. CJ I -
14,CJl-15,CJI-16,CJl-17.CJl-18,CJl-l9.CJl-20.CJl-21,CJl-22,CJl-23~
CJl-2A,CJl-25,C~1-26,CJl-27,CJl-28~CJI-30.CJl-31.CJl-32~ CJl-33, CJ l-
34 and CJl-35 wherein the portion of the peptide preferably has a mean T cell
stimulation index equivalent to, or greaur than the mean T cell stimulation index
of the peptide from which it is derived as shown in Fi~. 14. Even more
preferably peptides derived from the Cr~ j I protein aller~en which can be used
for therapeutic purposes comprise all or a portion of the~followin~ peptides: CJl-
2.CJl-9,CJl-10,CJl-16.CJl-17,CJl-20.CJ1-22.CJl-23.CJ1-24,CJI-25.
CJl-26.CJl-27,CJl-30,CJl-31.CJl-32 and CJl-35 as shown in Fig. 14.
Additionally, other preferred peptides derived from the Cry j I protein comprisethe following peptides: CJ1-41.CJl-41.1.CJI-41.2.CJ1-41.3.CJI-4~.CJI-4
i CJ~-42.2.CJl-43.CJl-43.1.CJI-43.6.CJ1-43.7.CJl-43.~,CJl-43.9.CJI-43.1~.CJl-43.11,CJl-43.12,CJl-45.CJl-45.1.CJl-45.2.CJl-44.CJI-44.1.CJI-44.2
and CJl-44.3. all as shown in Fi~, 18. Another preferted antigenic peptide ot`
the invonoon may comprise more than one Reoion, i.e.~ rll or a portion o~ amino
~ .
i
I
WO 94/01560 PCr/US93/00139
?,120954 ~'"
acids 151-352 of the amino acid sequence of Cry; 1~ as shown in Fig. 4a-b.
One embodiment of the present invention features a peptide or
portion thereof of Cr~ j I which comprises at least one T cell epitope of the
protein allergen and has a formula Xn-Y-Zm. Accordin! to the formula, Y is an
amino acid sequence selected from the ~roup consisting of CJI-I.CJl-2,CJl-3.
CJI-4.C11-7.CJI-8.CJl-9.CJl-lO.CJI-ll.CJI-12.CJI-14.CJI-lS~CJl-16.
CJI-17~CJI-18,CJI-l9,CJl-20.CJI-21,CJl-22,CJl-23.CJl-24,CJl-25.CJl-
- 26.CJl-27,CJl-28 CJI-30,CJl-3I,CJl-32.CJl-33~ CJ1-34 and CJ 1-35~ and
preferably selected from the group consisting of CJ1-2.CJI-9~CJl-lO,CJl-16.
0 CJl-17,CJl-20,CJl-22,CJl-23,CJl-24,CJl-25~CJl-26~CJl-27,CJl-30~CJl-
31~CJl-32 and CJI-35. In addition, Xn are amino acid residues contiguous to
the amino terminus of Y in the amino acid sequence of the protein allergen and
Zm are amino acid residues contiguous to the carboxy terminus of Y in the
amino acid sequence of the protein allergen. In the forrnula. n is 0-30 and m is ;
0-30. Preferably, the peptide or portion thereof has a mean T cell stimulation
index equivalent to greater than the mean T cell stimulation index of Y as shown
n Fig. 14.
Another embodiment of the present invention provides peptides
compnsing at least two regions, each region comprisin~ at least one T cell
20 ~ e pitope of`C)yj I~ and accordingly each re~ion comprises at least approximately
seven amino acid resldues. These peptides comprisin~ at least two regions can
- comprise as many arnino acid residues as desired and preferably comprise at least
about 14. even more preferably about 30. and most preferably at least about 40
amino acid residues of a Cr~ j I allergen. If desired. the amino acid sequences of
2~ ~ the regions can be produced and joined by a linker to increase sensitivity to
processmg by antigen-presenting cells. Such linker can be any non-epitope
amino acid sequence or other appropriate linking or joining agent. To obtain
preferred peptides comprising at least two regions. each complising at least oneT cell epitope. the regions are arranged in a conficuration dit'ferent from a
naturally-occurring configuradon of the regions in the allergen. For example~ ;
the regions containing T cell epitope(s) can be arranged in a noncontiguous
confïguration and can preferably be derived t;om the s~me protein allercen.
:
~ ~ r
' ~ '
WO 94/01560 2 I 2 D 9 ~5 ~ PCI/US93/00139
Noncontiguous is defined as an arrangement of regions con~ining T cell
epitope(s) which is different than that of an amino acid sequence present in theprotein aller~en from which the regions are derived. Furtherm()re. the
noncontiguous re~ions containing T cell epitopes can be arranged in a
nonsequential order (e.g., in an order different from the order of the amino acids
of the native protein aller~en from which the re~ion containino T cell epitope(s~
are derived in which amino acids are arran~ed from an amino terminus to a
carboxy terminus). A peptide can comprise at least 15%~ at least 30~, at least
50% or up tO 100% of the T cell epitopes of Cry j I.
The individual peptide regions can be produced and tested to detennine
which regions bind immunoglobulin E specific for Cry j 1 and which of such
regions would cause the release of mediators (e.g.. histamine) from mast cells or
basophils. Those peptide regions found to bind immunoglobulin E and cause the
release of mediators from mast cçlls or basophils in greater than approximatel~
10-15% of the allergic sera tested are preferably not included in the peptide
regions arranged to form preferred pep~ides of the invention.
Additionally, regions of a peptide of the invention preferably
comprise all or a portion of the above discussed preferred areas of major T cellreactivity within Cr y j I ~i.e.. Regions 1-S) or the above discussed preferred areas
20 of major T cell activity within each Re~ion (i.e. amino acids from residues 1-40.
81-110~ 151-180, 191-260 and 291-330). For example. one region can comprise
all or a portion of Region 1 (amino acid residues 1-5 l ) and one region can
comprise all or a portion of Re~ion 2 (amino acid residues 61-120). Peptides of
the~ invention can comprise all or a por~ion ot` two or more of these Rè~ions (i.e..
2~ Regions 1-5) and preferred resulting peptides do not bind I~E and cause the
release of mediators from most cells or basophils. PrefelTed peptides derived
from Cr~ j I comprise all or a portion of Re~ion 3~ Region 4 and Re~ion 5. and.
optionally. Region l or Region 2. Further. if one of these Regions is foulld t~) ;
bind IgE and cause the release of mediators from mast cells or basophils. then it
30 is preferred that the peptide not comprise such Re; ion. but rather comprisesvarious regions delived from such Region which d~) not bind IgE or cause rele;~
of mediators from mast cells ot basophils.
Wo 94/01~0 PCI'/US93/00139
~1~0954
Examples of preferred regions include: CJl-l,CJl-2.CJl-3.CJl-
4,CJl-7.CJI-8.CJl-9,CJl-lO,C~l-ll,CJI-12,CJl-14,CJl-lS,CJl-16,CJl-
17,CJl-18.CJl-l9.CJl-20,CJl-21,CJl-22,CJl-23~CJl-24,CJl-25.CJl-2S,
CJl-27,CJl-28,CJl-30,CJl-31,CJl-32,CJI-33,CJl-34.CJl-35. CJI-41,
CJl-41.1.CJ1-41.2,C~1-41.3,CJl-42,CJl-42.1,CJl-42.2.CJI-43, CJ1-43.1. i`
CJ1-43.6,CJ1-43.7.CJ1-43.8,CJI-43.9.CJI-43.10.CJ1-43.11~CJ1-43.12.CJI-
45,CJl-45.1,CJl-45.2,C31-44,CJl-44.1,CJl-44.2 and CJl-44.3. the amino
acid sequences of such regions being shown in Fi~. 13 and Fi~. 18. or portions of
said regions comprising at least one T cell epitope.
Preferred pep~ides comprise various combinations of two or more
regions, each region comprising all or a portion of the above-discussed preferred
areas of major T cell reactivity. Preferred peptides comprise a combination of
two or more regions (each region having an amino acid sequence as shown in
Fi~. 13), including: ~ -
CJ1-1, CJ1-2 and CJ1-3;
CJl-l and CJl-2;
CJ1-9 and CJl-10;
CJ1-14, CJ1-15, CJ1-16 and CJ1-17;
CJ1-20, CJ1-21, CJ1-22, CJ1-23;
CJ1-20, CJ1-22 and CJ1-23;
CJ1-22 and CJ1-23;
CJ1-22, CJ1-23 and CJ1-24: i` ~
CJ1-24 and CJ1-25;
CJ1-30, CJ1-31 and CJ1-32:
2~ CJ1-31 andCJ1-32;
CJl-22. CJl-23. CJl-16 and CJl-17:
CJ1-22. CJ1-23. CJ1-31 and CJ1-32:
CJ1-16, CJ1-17. CJl-31 and CJI-32;
CJl-9, CJ1-10 and CJI-16;
CJ 1 - 16 and CJ 1- 17:
CJl-17. CJ1-22 and CJI-23;
CJ1-16, CJl-17 and CJ1-20;
:
W o 94/01560 2 1 2 0 3 S ~ P ~ /~S93/00139
CJl-31,CJl-32and CJl-20;
CJl-22. CJ1-23, CJ1-1. CJl- and CJl-3;
CJI-16.CJl-17.CJl-22 and CJl-23.CJl-31 and CJI-32:
CJl-9.CJI-lO.CJl-16.CJl-17. CJ1-22 and CJl-23;
CJI-9,CJl-lO,CJl-16, CJl-17. CJl-31 and CJl-32;
CJI-9,CJl-lO.CJl-22,CJl-23,CJl-31 and CJl-32;
CJl-9,CJl-lO,CJl-16,CJl-17.CJl-22,CJl-23.CJl-31 and CJ1-
32;
CJl-l.CJl-2,CJl-16,CJl-17,CJl-22 and CJl-23;
lQ CJl-2~,CJl-23,CJl-24,CJl-9, and CJl-10;
CJl-22.CJl-23,CJl-24,CJl-9.CJl-lO.CJl-16. and CJl-17;
CJl-22,CJl-23,CJl-24,CJl-16,CJl-17.CJl-31 and CJl-32;
CJl-22,CJl-23,CJl-24.CJl-16. and CJl-17;
CJl-22, CJl-23,CJl-24,CJl-9.CJl-lO,CJl-31 and CJl-32;
CJl-22,CJl-23.CJl-24.CJl-9,CJl-lO,CJl-16.CJl-17.CJl-31
and CJl-32; and
CJl-22,CJl-23,CJl-24,CJl-31, and CJ1-32.
lsolated C yj I protein or peptides of Cryj I within the scope of
the invention can be used in methods of treating and preventing allergic reactions
to Japanese cedar pollen. Thus, one aspect of the present invention provides
therapeutic compositions cornprisin~ a peptide of Cr j 1 including at least one T ~,
cell epitope. or preferably at least two T cell epitopes. and a pharrnaceutically
acceptable carrier or diluent. ln another aspect. the therapeutic composition
comprises a pharmaceutically acceptable carrier or diluent and a peptide
comprising at least two regions. each region comprisino at least one T cell 1
epitope of Cry j I.
Preferred therapeutic compositions comprise a suft'icient percentage of the
T'cell epitopes of Cr~ j I such that a therapeutic reoimen of administration of the
composition to an individual sensitive to Japanese cedar pollen aller2en. resul~s
in T cells of the individual being tolerized to the protein aller~en. More
preferably, the composition comprises a suft'icient percenta e of the T cell
WO 94/01560 PCr/US93/00139
~12095~
epitopes such that at least abnut 40~. and more preferably at least about 60% ofthe T cell reactivity of Cr~; I is included in the composition. Such compositions
can be administered to an individual to treat or prevent sensitivity to Japanesecedar pollen or to an allergen which is immunolo~ically cross-reactive with
Japanese cedar pollen allergen.
In yet another aspect of the present inven~ion. a compositinn is provided
comprisin~ at least two peptides (e.g., a physical mixture of at least two
pep~ides). each complising at least one T cell epitope of C~j I. Such
compositions can be administered in the forrn of a therapeutic composition with
0 a pharmaceutically acceptable camer of diluent. A therapeutically effective
amollnt of one or more of such compositions can be administered simultaneously
or sequentially to an indiv;dual sensitive to Japanese cedar pollen.
Preferred compositions and preferred combinations of peptides which can
be administered simultaneously or sequentially (comprising peptides havina~ -
amino acid sequences shown in Fig. 13) include the ~llowing combinations:
CJI-l,CJl-2 and CJ1-3:
CJI-1 and CJ1-2;
CJl-9 and CJl-10;
CJl-14,CJl-l5,CJl-16 and CJl-17;
CJl-20,CJl-21,CJl-22 and CJl-23;
CJl-20,CJl-22 and CJl-23;
CJl-22 and CJl-23;
CJ1-22~CJl-23 and CJl-24;
CJl-24 and CJl-25;
CJl-30. CJl-31 and CJl-32;
CJl-31 and CJl-32;
CJl-22,CJl-23.CJl-16 and CJI-17;
CJl-22.CJl-23.CJl-31 and CJl-32:
CJl-16.CJl-17. CJl-31 and CJl-32;
CJl-9.CJl-10 and CJl-16;
CJI-16 and CJl-17; i '
CJ1-17.CJl-22 and CJl-23;
36
W O 94/01560 2 1 ~ ~ 9 .S ~ PCT/U~93/00139
CJl-16.CJl-17and CJl-20:
CJl-31.CJl-32and CJl-20;
CJl-22.CJl-23.CJl-l~CJl-2 and CJl-3;
CJI-16,CJl-17,CJl-22.CJI-23.CJl-31and CJl-32;
CJl-9,CJl-lO,CJl-16,CJl-17.CJl-22and CJl-23; ;
CJl-9.CJl-lO.CJI-16.CJl-1'.7. CJl-31 and CJl-32:
CJl-9,CJI-lO.CJI-22, CJ1-23, CJl-31 and CJl-32;
CJl-9.CJl-lO,CJl-16.CJl-17.CJl-22,CJl-23.CJl-31 and CJI-
32;
lo CJl-l,CJl-2.CJl-16,CJl-17.CJl-22 and CJl-23.
C31-22,CJl-23.CJl-24,CJl-9. and CJl-10,
CJ1-22, CJl-23. CJl-24, CJl-9,CJl-10. CJ1-16. and CJl-17;
CJl-22,CJl-23,CJl-24,CJl-16~CJl-17~CJl-31 and CJl-3~;
CJl-22,CJl-23,CJl-24.CJl-16~ and CJl-17; ~;
CJl-22.CJl-23,CJl-24, U l-9.CJl-lO,CJl-31 and CJl-32;
CJl-22,CJl-23,CJl-24,CJl-9,CJl-lO.CJl-16.CJl-17,CJl-~l
and CJl-32; and
CJl-22,CJl-23,CJl-24,CJl-31,and CJl-32.
The invention is further illustrated by the followino non-limi~in~
examples. -~
Example I
25 Purification of Native .~apanese ~ar Po~len Aller~en (Crv i I )
The following is a description of the work done to bi~)chemicall~
purify the major allergen, C~ j I in the native form. The purification was
modified from published procedures (Yasueda et al.. J. Allerg! Clin. Imm~n
7 1 :17, 1983).
100~ of Japanese cedar pollen obtained from Japan (Hollister- t
Stier, Spokane, WA) was defatted in I L diethyl ether three times, the pollen ~ as
collccted after filtration and the ether was dried off in a vacuum.
~7
WO 94/01~60 PCI/US93/00139
21~Q9~
The defatted pollen was extracted at 4C overnight in 2 L
extraction buffer containin~ 50 mM tris-HCL, pH 7.8, 0.2 M NaCI and protease
inhibitors in final concentrations: soybean trypsin inhibitor (2 ,u~/ml). leupeptin
g/ml), pepstatin A (1 llg/ml) and phenyl methyl sulfonyl fluoride (0.17
mg/ml). The insoluble material was reextracted with 1.2 L extraction buffer at
4C overnight and both extracts were combined to~ether and depigmented by
batch absorption with Whatman DE-52 DEAE cellulose (200 ~ dry weight)
equilibrated with the extraction buffer.
The depigmented rnaterial was then fractionated by arnmonium
sulfate precipitation at 80% saturation (4C), which removed much of the lower
molecular weight material. The resultant partially purifled G~ j I was either
dialyzed in PBS buffer and used in T cell studies (see Example 6) or subjected to
further purification (biochemically or by monoclonal affinity chromatography)
as described below.
The enriched C)y j I material was then dialyzed against S0 mM Na-
acetate, pH 5.0 at 4C with 50 mM Na-acetate, pH 5.0 with protease inhibitors. The
sample was next applied to a 100 ml DEAE cellulose column (Whatman DE-52)
- equilibrated at 4C with 50 mM Na-acetate pH 5.0 with protease inhibitors. Th~unbound material (basic proteins) was then applied to a 50 ml cation exchan~e
column i~Whatman CM-52) which was equilibrated at 4C with 10 mM Na-ace~ate~
pH 5.0 with protease inhibitors. C~y j I was eluted in the early fractions of a lin~ar
gradient 0.3 M NaCl. The enriched C~ j 1 material was lyophilized and was th~n
purified by FPLC over a 300 ml Superdex 75 column (Pharmacia) at a flow rate o~`30 ml/h in 10 mM Na-acetate, pH 5.0 at 25C.
The purified Cr~ j I was further applied to FPLC S-Sepharose 1611(:)
column chromatography (Pharmacia) with a linear gradient of 0 - 1 M NaCI at ~S~C.
Cr~! j I, eluted as the major peak, was subjected to a second gel filtration
chromatography. FPLC Superdex 75 column (2.6 by 60 cm)(Pharmacia~
Piscataway, NJ) was eluted with a downward tlow of 10 mM Na-acetate, pH 5.()
with 0.15 M NaCl at a flow rate of 30 mUh at 25C. Fi~. la shows the
chromatography on ~el filtration. Only C~ j I was detected (Fig. Ib. Iane 2 to Iane
S). CJ~ j I was fractionated into 3 bands us analyzed by SDS-PAGE using sil~er
~X
WO 94/01560 2 1 2 0 9 j ~ PCI/US93/00139
staining (Fig. Ib) As shown in Fig. lb. SDS PAGE (12.5%) analysis of the fractions
from the major pealc shown in Fi~. la was performed under reducing conditions.
The gel was silver stained using the silver staining kit from Bio-Rad. The samples
in each lane were as follows: Lane 1, prestained standard proteins (Gibco BRL)
including ovalbumin (43,000 kD). carbonic anhydrase (29,000 kD). anà cx-
lactoglobulin (18,400 kD); lane 2, ~raction 36; lane 3 fraction 37; lane 4 fraction
38; lane 5 fraction 39; lane 6 fraction 41. Iane 7 fraction 43: and lane 8 fraction 44.
All fractions are shown in Fig. la.
These proteins were also analyzed by Western blo~ting usin~ mouse
monoclonal antibody CBF2 (Fig. 2). As shown in Fig. 2. an aliquot of fraction 36(lane 1), fraction 39, (lane 2) and fraction 43 (lane 3) purified from the Superdex 7
- as shown in Fig. 1 was separated by SDS-PAGE. electroblot~ed onto nitrocelluslose
and probed with mAB CBF2. Biotinlylated goat anti-mouse I~ was used for the
second antibody and bound antibody was revealed by 125I-streptavidin. The
monoclonal CBF2 was raised ag,ainst ragweed allergen Amb a I by Dr. D. Klapper
(Chapel Hill, NC~. Because of the homology between the Amb ~ I and C~
sequences, a number of antibodies raised against Amb a I were tested for reactivity
with Cry j I. The results showed that CBF2 recognized denatured C~ J I as detected
by ELISA and Western blotting. In addition, Western blotting also demonstrated
that no other bands were detected by CBF2. other than Cn j I in the expected
molecular weight range (Fig. 2). These results were consistent with the findin~sfrom protein sequencing. When fraction 44 and fraction 39 (Fig I b) were subjected
to N-terrninal sequencing, only C~ j I sequence was detected.
In summary, three C~yj I isoforms of different molecular wei~ht were
purified from pollen extract. The molecular weights estimated by SDS-PAGE
ranged from 40-35 kD under both reducing and non-reducing conditions. The
isoelectric point of these isoforms is approximately 9.5-8.6~ with an avera~ge pl ot
9Ø The N-terrninal 2() amino acid sequence was the same in these 3 bands and was
identical to previously published C ~ j I sequence (Taniai et al~ suprw). The 3
isoforrns are all recognized by monoclonal antibody CBF2 as shown in the allervi~
sera titration of dit`ferent purified subfractions of Cry j I usin~ a pool of fiftee
allergic patient plasma~ They all bind aller~gic patient IgE (Fi~ 3)~ The difteren~e in
WO 94/01560 PCr/US93/00139
21209~
`
molecular wei~ht and isoelectric point in these isotorms might in part be due to post-
translational modifica~ion. e.g. glycosylation~ phosphorylation or lipid content. The
possibility that t~ese different isoforms mioht be due to protease de~radation ~:annot
be ruled out at present even though it is unlikely due to the fact that ~our different
protease inhibitors were used durin~ extraction and purification. The other
possibility could be due tO polymorphism in the cene or altemate splicing in themRNA though only one major form of C ~ j I protein has been detected in cDNA
cloning studies (see Example 4).
Another approach which may be used to purify native CJY j I or
recombinant C ~ j I is immunoaf~lnity chromatography. This technique provides
a very selective protein purification due to the specificity of the interaction
between monoclonal antibodies and antigen. For the purpose of producino C~
I-reactive monoclonal antibodies. female BalbVc mice were obtained from
Jackson Labs. Each mouse was initially immunized intraperitoneally with 70-10C)
llg purified native C~y j I. (>99% purity lower band. as shown in Fig. 1 b)~
emulsified in Freund's complete adjuvant. One further intravenous injection of
10 ~g purified native Cr~j I in PBS was given 54 days after the initial injection.
The spleen was removed 3 days later and myeloma t`usion was conducted as
described (Current Protocols in Immunoloj~y. 1991. Coligan et al. eds.) using the
myeloma line SP2Ø The cells were cultured in 105~ fetal calf serum
tHybrimax), hypoxanthine and azaserine and wells containing colonies of
hybridoma cells were screened for antibody production using anti~en-binding
ELISA.
Cells from positive wells were cloned at threè-tenths cell/well in 10'~
2~ fetal calf serum (Hybrimax). hypoxanthine and positive clones were subcloned one
more time in hypoxanthine medium . Capture ELISA (see Example 7) was used ~;)r
secondary and tertiary screening This assay oft`ers the advantage that a clone th;lt
reco~nizes the native protein may be selected and thus may be useful tor
imrnunoaffinity purification. For example. two monoclonal antibodies (4B 11. ~B I I )
were ~enerated. These antibodies were pulified by Gammabind G. Sepharose
(Pharmacia~ Piscataway. NJ) accordinn to rnanutacturer's procedures and were
immobilized to cyanogen bromide - activated Sepharose 4B (Pharmacia. Piscat~ a! .
I
~ 21209~4
WO 94/01560 PCl /US93/00139
NJ) according to the procedures describe~ ~y Pharmacia. The ammonium sulphate
prepara~ion containing C~ j I was applied to the resin and unbound material was
washed extensively with PBS. C~y j I was eluted with 2 column volumes of 0.1 M
glycine, pH 2.7. Silver staining of the eluate fractions run on SDS PAGE showed
that Cryj I was purified almost to homogeneity. These fractions did not contain
detectable levels of Cry j Jl. Other methods to immobilize MAb 8B l l were also
tested. Similar results were obtained usin~ puri~led MAb 8B l l covalently cross-
linked to Gammabînd G Sepharose by dimethylpimelimidate (Schneider C., et al. J.Biol. Che~ (1982) volume 257:10766-10769). However, experiments using
purified MAb 8B11 covalently cross-linked to Af~l-gel 10 (Biorad. Richmond. CA)
showed that althou~h greater than 90% of the monoclonal antibody was covalently
coupled to Affi-gel 10, the yield of Cr.~ j I purified over this resin was si~nificantly
less than that purified from MAb 8B11 cross-linked to cyanogen bromide-acti~atedSepharose 4B ~data not shown). Nevertheless, the puri~led Cr~ j I from these
monoclonal antibodies immobilized on dit`ferent resins is still intact and can be
recognized by MAb 8B l l and 4B 11 by capture ELISA. Thus, these MAbs will
provide a useful tool in purification of Cr~ j I from pollen extracts. Similarly~
monoclonal antibodies that bind to recombinant C~y j I can also be used for
immunoaf~lnity chromatography. In addition. the monoclonal antibodies ~enerated
may be useful for diagnostic pUlpOSeS. It may also be possible to raise diff`erent
MAbs that show some specificity towards these different isoforms of C-y j I and thus
would provide a useful tool to characterize these isoforms.
Example 2
Attempted Extraction of RNA From Japanese Cedar Pollen
Multiple attempts were made to obtain RNA from commerciall~-
available. non-defatted. Cryptome-ia japol7ica (Japanese cedar) pollen (Hollister
Stier. Seattle. WA). Initially, the method of Sambrook et al.. Molecul~r Cl0ni
A Laborator~ Manual, Cold Sprin~ Harbor Laboratory Press. Cold Sprin~
1 1
WO 94/0156û PCr/US93/00139
209~4
Harbor. New York (1989) was used in which the sample was suspended and
lysed in 4 M guanidine buffer. ~round under liquid nitro~en, and pelleted
throu~h 5.7 M cesium chloride by ultracentrifu~ation. Various arnounts (3. 5
and 10 g) of pollen in varying arnounts of ~uanidine lysis buffer (10 and 25 ml)s were tned. Centrifugation through cesium resulted in viscous matenal in the
bottom of the tube. from which it was not possible to recover an RNA pellet.
Although it was possible to obtain RNA from defatted Ambrosia artemisi~olia
(ragweed) pollen (Greer Laboratories. Lenior. NC) using this protocol. defattingthe Cr~ptomeria japonica pollen with acetone before guanidine extraction also
did not yield any RNA, as determined by absorbance at A~60.
An acid phenol extraction of RNA according to the method in
Sambrool. et aL, supra was attempted from C .~ ptomeria japonica pollen. The
pollen was ground and sheared in 4.5 M guanidine solution. acidified by additionof 2 M sodium acetate, and extracted with water-saturated phenol plus
ls chloroform. After precipitation, the pellet was washed with 4 M lithium
chloride,~redissolved in 10 mM TrislS mM EDTA/1% SDS, chloroform
extracted, and re-precipitated with NaCI and absolute ethanol. It was possible to
extract Ambrosia artemisiifolia but not G~ptomeria japonica RNA with this
~- procedure.
~ Next, 4 g of Cryp~ome~ia japonica pollen was suspended in 10 ml
extraction buffer (50 mM Tris, pH 9.0~ 0.2 M NaCl. 10 mM Mg acetate and
diethylpyrocarbonate (DEPC) to 0.1%), ground in a mortar and pestle on dry ice
transferred to a centrifuge tube with l~o SDS. 10 mM EDTA and 0.5% N-
lauroyl sarcosine, and the mixture was extracted five times with war~m phenol.
23 The aqueous phase was recovered after the final centrifu~ation. 2.5 vol. absolute
ethanol was added, and the mixture was incubated overnight at 4C. The pellet
was recovered by centrifugation. resuspended in 1 ml dH20 by heatin~ to 65C.
and reprecipitated by the addition of 0.1 vol. 3 M Na acetate and 2.0 vol. of
ethanol. No detectable RNA was recovered in the pellet as judged by absorbance
at A260 and ~el electrophoresis.
Finally, 500 mg of Gyptont~ia jap->ni~a pollen was ~round by
mortar and pestle on dry ice and suspended in S ml of 5~) mM Tris pH 9.0 with
: 1,
-
~ 212û95~
WO 94/01560 PCr/US93/00139
0.2 M NaCI, 1 mM EDTA~ lYo SDS that had been treated overni~ht with 0.1%
DEPC, as previously described in Frankis and Mascarhenas (1980) Ann. Bot. 4~:
595- 599. After five extractions with phenoVchloroform/isoamyl alcohol (mixed
at 25:24:1), material was precipitated from the aqueous phase with 0.1 volume 3
s M sodium acetate and 2 volumes ethanol. The pellet was recovered by
centrifu~ation, resuspended in dH20 and heated to 65C to solubilize the
precipitated material. Further precipitations with lithium chloride were not done.
There was no detectable RNA recovered. as determined by absorbance at A260
and gel electrophoresis.
In summary. it has not been possible to recover RNA from the
commercial pollen. It is not known whether the RNA has been de~raded durin~
stora~e or shipment~ or whether the protocols used in this example did not allowrecovery of extant RNA. However. RNA was recovered from fresh C)Yptom~ia
japonica pollen and staminate cone samples. (See Example 3.)
Example 3
Extraction of RNA From Japanese Cedar Pollen and Staminat~ Cones and
Clor~ing of C y j I
Fresh pollen and staminate cone samples. collected from a sin~le
~yptomeria japonica (Japanese cedar) tree at the Arnold Arboretum (Boston.
MA), were frozen immediately on dry ice. RNA was prepared from 500 m~ of
each sample~ essentially as described by Frankis and Mascarenhas. supra. The
- ~ samples were ~round by mortar and pestle on dry ice and suspended in S ml of
2s ~ 50 mM Tris pH 9.0 with 0.2 M NaCl. 1 mM EDTA. 1~ SDS that had been
treated overnight with 0.1% DEPC. After five extractions with
phenoVchloroform/ isoamyl alcohol (mixed at 25:24~ the RNA was
precipitated from the aqueous phase with 0. l volume 2 M sodium acetate and 2
~, volumes ethanol. The pellets were recnvered by centrifugation. resuspended in
dH20 and heated to 65C for 5 min. Two ml ot 4 M lithium chloride were added
to the RNA preparations and they were incubated overni~ht at ()C. The RNA
pellets were recovered by centrifugation. resuspended in l ml dH20. and a~ain
1 ~s
WO 94/01~60 PCr/US93/00139
21~09S~ . .
precipitated with 3 M sodium acetate and ethanol overni~ht. The final pellets
were resuspended in 100 ,ul dH20 and stored at -~0C.
First strand cDNA was synthesized from 8 ~1~ flowerhead and 4
~ pollen RNA usin~ a commercially available kit (cDNA synthesis systems kit.
s BRL, Gaithersburg, MD) with oligo dT primino accordin~ to the method of
5ubler and Hoffman (1983) Cene 25: 263-2~9. An attempt was made to amplify
cDNA encoding C)y j I using the degenerate oligonucleotide CP- I (which has
the sequence 5'-GATAATCCGATAGATAG-3'. wherein T at p~sition 3 can also
be C; T at position 6 can also be C; G at position 9 can also be A,T. or C; A atposition 12 can also be T, or C; T at position 15 can also be C; A at position 16
can also be T; and G at position 17 can also be C: SEQ ID NO: 3) and primers
EDT and ED. Primer EDT has the sequence 5'-GGAAl~CTCTAGACTGCA-
GGlmm~l m-3'(SEQ ID NO: 24). Primer ED has the sequence ~'-
GGAATTCTCTAGACTGCAGGT-3' (SEQ ID NO: 23). CP-l is the de~enera~e
oli~onucleotide sequence encoding the first six amino acids of the amino
terminus (AspAsnProIleAspSer, amino acids 1-6 of SEQ ID NO: 1) of C~yj I.
EDT will hybridiæ with the poly A tail of the gene. All oligonucleotides were
synthesized by Research Genetics, Inc. Huntsville. AL. Polymerase chain
reactions (PCR) were carried out using a commercially available kit (GeneAmp
DNA Amplification kit, Perkin Elmer Cetus. Norwal}c, CT) whereby 10 ,ul lOx
buffer containing dNTPs was mixed with I ~1 of CP- 1 and 1 ,u~ of EDtEDT
pnmers (ED:EDT in a 3:1 M ratio), cDNA (3-5 111 of a 20 ~1 first strand cDNA -
reaction mix), 0.5 ~1 Amplitaq DNA polymerase, and distilled water to 100 ~1.
The samples were amplified with a pro~rammable thermal
2S controller (MJ Research. Inc., Carnbrid~e. MA). The first 5 rounds ofampli~1cation consisted of denaturation at 94C for I minute. annealing of
primers to the template at 45C for 1.5 minutes, and chain elonoation at 70C for
2 minutes. The final 20 rounds of amplification consisted of denaturation as
above. annealing at 55C for 1.5 minutes, and elon~ation as above. Five percen
(5 ~11) of this initial amplification was then used in a secondary amplificationwith 1 llg each of CP-2 (which has the sequence 5'- GGGAATlCAAl-rGGGC- ~ `
GCAGAATGG-3' wherein T at position I I can also be C; G at position 17 can
WO 94/OlS60 21 2 D 9 ~ ~ Pcr/US93/ool39
also be A, T, or C; G at position 20 can also be A; T at position 23 can also be C;
and G at position 24 can also be C) (SEQ ID NO: 4). a nested primer, and ED. as
above. The se~uence 5'-GGGAAl-rC-3' (bases 1 through ~ of SEQ ID NO: 4)
in primer CP-2 represents an Eco Rl site added for cloning purposes: the
s remaining degenerate oligonucleotide sequence encodes amino acids 13-18 of
Ctyj I (AsnTrpAlaGlnAsnArg, amino acids 13 through 18 of SEQ ID NO: 1).
Multiple DNA bands were resolved on a 1% GTG agarose gel (FMC, Rockport.
ME), none of which hybridized with 32p end- labeled probe CP-3 (SEQ ID NO:
S) in a Southern blot performed according to the method in Sambrook et al.
supra. Therefore, it was not possible to select a specffic CJY j I DNA band and
this approach was not pursued. CP-3 has the sequence 5'- CTGCAGCCATT-
TTCIACATTAAA-3' wherein A at position 9 can also be G; T at position 12 can
also be C; A at position 18 can also be G; and A at position 21 can also be G)
(SEQ ID NO: 5). Inosine (I) is used at position 15 in place of G or A or T or C
to reduce degeneracy (Knoth et al. (1988 j Nucleic Acids Res. 1~: 10932). The
sequence 5'-CTGCAG-3' (bases 1 through 6 of SEQ ID NO: 5) in primer CP-3
represent a Pst I site added for cloning purposes; the remaining degenerate
oligonucleotide seguence is the non-coding strand sequence corresponding to
codin_ strand sequence encoding amino acids PheAsnValGluAsnGly (amino
acids 327 through 332 of SEQ rD NO: 1) from the internal sequence of Cr~ j I.
A primary PCR was also performed on first-strand cDNA usino
CP-1 (SEQ ID NO: 3) and CP-3 (SEQ ID NO: 5). as above. A secondary PCR -
was perforrned using 5% of the primary reaction using CP-2 (SEQ ID NO: 4)
and CP-3 (SEQ ID NO: 5). Again, multiple bands were observed, none of which
could be specifically identified in a Southern blot as Cr~ j I, and this approach
was also not pursued.
Double-stranded cDNA was then synthesized from approximatel
4 llo (pollen) or 8 11~ (flowerhead) RNA using a commercially available kit
(cDNA Synthesis System kit, BRL. Gaithersbur . MD)~ After a phenol
extraction and ethanol precipitation, the cDNA was blunted with T4 DNA
polymerase (Promega, Madison, WI)~ and li~ated to ethanol precipitated, self-
annealed, AT ~SEQ ID NO: 20) and AL (SEQ ID NO: 22) oli~onucleotides fo
l`"i
L~,~
- .
WO 94/01560 PCr/U~93/00139
9S~
use in a modified Anchored PCR reaction. according to the method in Rafnar et
al. (1991) J. Biol. C~e~ 266: 1229-1236; Frohman et al. (1990) Proc. Na~l.
Acad. Sci. U~A 85: 89g8-9002; and Roux et al. (1990) BioTech. 8: 48-57.
Oligonucleotide AT has the sequence 5'^ GGGTCTAGAGGTACCGTC-
CGATCGATCATT-3'(SEQIDNO:20) (Rafnar et al. sup~a ). Oligonucleotide
AL has the sequence 5'-AATGATCGATGCT-3'(SEQIDNO:22)(Rafnar et al.
Supra. The amino terminus of C)y j I was amplified from the linkered cDNA ~3
ul from a 20 111 reaction~ with 1 llg each of oligonucleotides AP (SEQ ID NO:
21 ) and degenerate Cry j I primer CP-7 (which has the sequence 5'-
0 TI~CATICGAl'rCTGGGCCCA-3' wherein G at position 8 can also be T; A at
position 9 can also be G; C at position 12 can also be T; and G at position 15 can
also be A, T, or C)(SEQ ID NO: 6). Inosine (I) is used at position 6 in place ofG or A or T or C to reduce degeneracy (Knoth et al. supra). The de,~enerate
oligonucleotide CP-7 (SEQ ID NO: 6) is the non-coding strand sequence
corTesponding to coding strand sequence encoding amino acids 14-20
(TrpAlaGlnAsnAr~MetLys) from the amino terminus of C~ j I (amino acids 14-
20 of SEQ ID NO: 1). Oligonucleotide AP has the sequence 5'-GGGTCTA-
GAGGTACCGTCCG-3' (SEQ ID NO: 21).
The primary PCR reac~ion was carried out as described herein.
Five percent (5 111) of this initial amplification was then used in a secondary
amplification with 1 ,ug each of AP (SEQ ID NO: 21 ) and degenerate C)y j I
primer CP-8 (SEQ ID NO: 7) an internally nested C~ j I oligonucleotide primer,
as described herein. Primer CP-8 has the sequence 5'-CCTGCAGCGATTCT-
GGGCCCAAAl-r-3' wherein G at position 9 can also be T: A at position 10 can
also be G; C at position 13 can also be T; G at position 16 can also be A, T~ or C;
and A at position 23 can also be G)(SEQ ID NO: 7). The nucleotides 5'-
CCTGCAG-3' (bases 1 through 7 of SEQ ID NO: 7) represent a Pst I restriction
site added for cloning purposes. The remaining de~enerate oligonucleotide
sequence is the non-coding strand sequence colTesponding to codin~ strand
sequence encoding amino acids 13-18 of Ct~; I (AsnTrpAlaGlnAsnArg, amino
acids 13-18 of SEQ ID NO: 1) from the amino terminus of C~ j 1. The ~.
dominant amplified product was a DNA band of approximately 193 base pairs.
4~
WO 94/01560 21 2 0 9 ~ 4 PCI`/US93/00139
as visualized on an ethidium bromide (EtBr)-stained 3~ GTG a~arose ~el.
Amplified DNA was recovered by sequential chloroform, phenol,
and chloroform extractions. followed by precipitation at -20C with 0.5 volumes
of 7.5 ammonium acetate and 1.5 volumes of isopropanol. After precipitation
s and washing with 70% ethanol, the DNA was simultaneously digested with Xba
I and Pst I in a 15 !ll reaction and electrophoresed through a preparative 3%
GTG NuSieve low melt gel (FMC, Rockport, ME). The appropliate sized DNA
band was visualized by EtBr staining, excised, and ligated into appropriately
di~ested M 1 3mp 18 for sequencing by the dideoxy chain termination method ~-
(Sanger et al. (1977) Proc. Natl Acad Sci. USA 74: 5463-5476) usin~ a
commercially available sequencing kit (Sequenase kit, U.S. Biochemicals,
Cleveland, OH). It was initially thought that li~atable material could only be
derived from starninate cone-derived RNA. However, upon subsequent
examination, it was shown that li&atable material could be recovered from PCR
product oenerated from pollen-derived RNA, and from staminate cone-derived
RNA.
The clone desi~nated JC7 1.6 was found to contain a partial
sequence of C~y j I. This was confirrned as an authentic clone of C~y j I by
having complete identity to the disclosed NH2-terminal sequence of C7.~' j I
(Taniai et al. sup)a). The amino acid at position 7 was determined to be cysteine
(Cys) in a~reemen~ with the sequence disclosed in U.S. patent 4, 939,239.
Amino acid numberinc is based on the sequence of the mature protein: amino
acid 1 corresponds to the aspartic acid (Asp) disclosed as the NH2-terminus of
C~ j I (Taniai et al. sup)a) The initiating methionine was found to be amino
acid -21 relative to the first amino acid of the mature protein. The position ofthe initiating methionine was supported by the presence of upstream in-frame- -
stop codons and by 78% homology of the surrounding nucleotide sequence with
the planl consensus sequence that encompasses the initiating methionine, as
reported by Lutcke et al. (1987) EMBO J. 6:43-48.
The cDNA encodin~ the remainder of C~ ene was cloned
~;orn the linkered cDNA by usin~ oligonucleotides CP-9 (which has the se4uenc~
5'-ATGGATTCCCCTTGCTTA-3')(SEQ ID NO: 8) and AP (SEQ ID NO: 21) in
~7
z~ ~
WO 94~01560 PCr/US93/00139
~,l2,09S~ '' '
the primary PCR reaction. Oligonucleotide CP-9 (SEQ ID NO: 8) encodes amino
acids MetAspSerProCysLeu of Cr~ j I (amino acids -21 throu~h -16 ot SEQ ID
NO: 1) from the leader sequence of C~; I. and is based c)n the nllcleotide
sequence determined for the partial Cr~ j I clone JC76. 1.
s A secondary PCR reaction was performe~i on 55~ of the initial
amplification mixture~ with 1 ~Lg each of AP (SEQ ID NO: 21) and CP-I()
~which has the sequence 5'-GGGAAl-rCGATAATCCCATAGACAGC-3')(SEQ
ID NO: 9), the nested primer. The nucleotide sequence 5'-GGGAAl~C-3' ot
primer CP- 10 (bases 1 throu~h 8 of SEQ ID NO: 9) represent an Eco RI
restriction site added for cloning purposes. The remainino oli_onucleotide
sequence encodes amino acids 1-6 of C~y j I ~AspAsnProlleAspSer) (amino acids
1 through 6 of SEQ ID NO: 1). and is based on the nucleotide sequence
determined for the partial Cr~ j I clone JC76. 1. The amplified DNA product was
purified and precipitated as above, followed by digestion with Eco RI and Xba 1
and electrophoresis through a preparative lq~ low melt ~el. The dominant DNA
band was excised and ligated into M13mpl9 and pUCl9 for sequencing. A~ain~
ligatable material was recovered from cDNA generated from pollen-derived
RNA. and from staminate cone-derived RNA. Two clones. desi~nated
pUC19JC9la and pUC19JC9ld, were selected for full-len~th sequencing. They
were subsequently found to have identical sequences.
DNA was sequenced by the dideoxy chain termination method
(Sanger et al. supra) usin~ a commercially available kit (sequenase kit (U.S.
Biochemicals. Cleveland. OH). Both strands were completely sequenceà usin~
M13 forward and reverse primers (N.E. Biolabs. Beverly. MA) and internal
sequencing primers CP-13 (SEQ ID NO: 10). CP-14 (SEQ ID NO~ CP-15
(SEQ ID NO: 12). CP-16 (SEQ ID NO: 13)~ CP-18 (SEQ ID NO: 1~). CP-19
(SEQ ID NO: 16). and CP-20 (SEQ ID NO: 17). CP-13 has the sequence 5'-
ATGCCTATGTACATTGC-3' (SEQ ID NO: 10). CP-13 (SEQ ID NO: 1~))
encodes amino acids 82-87 of C~,~ j I (MetProMetTyrlleAI~ amino acids 82
throu~h 87 of SEQ ID NC): 1). CP-14 has the sequence 5'-
GCAATGTACATAGGCAT-3' (SEQ ID NO: 11 ) and ct)rresponds to the non-
coding strand sequence of CP- 13 SEQ ID NO: 1()). CP- l~ h~s the sequence ~'-
4~
WO 94/01~60 2 1 2 ~ 9 ~ ~ PCI/US93/00139
TCCAAl~CTI`CTGATGGT-3' ((SEQ ID NO: 12) which encodes amino acids
169-174 of C~ I (SerAsnSerSerAspGly. amino acids 169 through 174 of SEQ
ID NO: 1). CP-16 has the sequence 5'- ~GTCAAl~GAGGAGT-3' (SEQ
ID NO: 13) which is the non-coding strand sequence which corresponds to
S coding strand sequence encoding amino acids 335-340 of C~
(ThrProGlnLeuThrLys, amino acids 335 through 340 of SEQ ID NO 1). CP- 18
has the sequence 5'-TAGCAACTCCAGTCGAAGT-3' (SEQ ID NO: 15) which
is the non-coding strand sequence which substantially corresponds to codin(r
strand sequence encoding amino acids 181 through 186 of Cr~ j I
o (ThrSerThrGlyValThr, amino acids 181 through 186 of SEQ ID NO: 1) except
that the fourth nucleotide of CP-18 (SEQ ID NO: 15) was synthesized as a C
rather than the correct nucleotide~ T. CP- 19 which has the sequence 5'-
TAGCTCTCATITGGTGC-3' (SEQ ID NO: 1~) is the non-coding strand
sequence which corresponds to coding strand sequence encodin~ amino acids
270 through 275 of C7y j I (AlaProAsnGluSerTyr, amino acids 270 throuoh 275
of SEQ ID NO: 1). CP-20 has the sequence 5'- TATGCAAl-rGGTGGGAGT-3'
(SEQ ID NO: 17) which is the coding strand sequence for arnino acids 251-256
of Cr,~ j I (TyrAlaIleGlyGlySer~ amino acids 251 through 256 of SEQ ID NO:
1). The sequenced DNA was found to have the sequence shown in Fics. 4a and
4b (SEQ ID NO: 1). This is a composite sequence from the two overlappin~
clones JC 71.6 and pUC19J9la. The complete cDNA sequence for Cn j I is
composed of 1312 nucleotides~ including 66 nucleotides of 5' untranslated
sequence~ an open reading frame startin~ with the codon for an initiatin~
methionine~ of 1122 nucleotides~ and a 3' untranslated re~ion. There is a
consensus polyadenylation signal sequence in the 3' untranslated region 25
nucleotides 5' to the poly A tail (nucleotides 1279-1283 of Fig 4 and SEQ. ID
NO: 1). Nucleotides 1313-1337 of Fi; . 4 and SEQ. ID NO: 1 represent vector
sequences. The position of the initiatin~ methionine is confirmed by the
presence of in-frame upstream stop codons and by 785~ homolo~y with the plant
consensus sequence that encompasses the initiatino methionine (AAAAAUGGA
(bases 62 throu~h 71) of SEQ ID NO~ ound in Cr.~ j I compared with the
AACA~!~iGC consensus sequence fol plzmts, ~tcke et al. (19~7) EMBO J. ~: ¦
4,~ .
WO 94/01~60 PCr/US93/00139
~;J,.2~95i~ '
43-48). The open reading f~ame encodes a protein of 374 amino acids of which
the first 21 amino acids comprise a leader sequence that is cleaved from the
mature protein. The amino terrninus of the mature protein was identified by
comparison with the published NH2-terminal sequence (Taniai et al. (1988)
sl~pra) and with sequence deterrnined by direct amino acid analysis of purified
native Cr~; I (Example l ). The deduced amino acid sequence of the mature
protein, comprised of 353 arnino acids has complete sequence identity with the
published protein sequence for C~ j I (Taniai et al. s~lpra), includin~ the first
twenty amino acids for the NH2-terrninal and sixteen contiguous internal amino
acids. The mature protein also contains five potential N-linked ~lycosylation
sites corresponding to the consensus sequence N-X-S/T.
Example 4
i
Extraction of RNA from Japanese Cedar Pollen Collected in Japan
Fresh pollen collected from a pool of Cryptomeria japonica
(Japanese cedar) trees in Japan was frozen immediately on dry ice. RNA was
prepared from 500 m~ ~f the pollen, essentially as described by Frankis and
Mascarenhas Ann. Bot. 45:595-599. The samples were ~round by mortar and
pestle on dry ice and suspended in S ml of 50 mM Tris pH 9.0 with 0.2 M NaCl.
1 mM EDTA. 1% SDS that had been treated overnight with 0.1% DEPC. After
five extractions with phenol/chloroforrn/isoamyl alcohol ~mixed at 25:24:1), theRNA was precipitated from the aqueous phase with O. l volume 3 M sodium
acetate and 2 volumes ethanol. The pellets were recovered by centrifu~ation.
resuspended in dH~0 and heated to 65C for 5 minutes. Two ml of 4 M lithium
chloride were added to the RNA preparations and they were incubated overnioht
at 9C. The RNA pellets were recovered by centritugation~ resuspended in l ml
dH20. and again precipitated with 3 M sodium acetate and ethanol overnight.
The final pellets were resuspended in 100 ~l dH20 and stored at -81)"C.
Double stranded cDNA was synthesized trom 8 ug pollen RNA
using the cDNA Synthesis Systems kit (BRL) with oligo dT priming according
~()
Wo 94/01560 2 12 D 9 3 4 PCr/US93/00~39
to the method of Gubler and Hoffman (1983) Gene 25:2~3-269. Polymerase
chain reactions ~PCR) were carlied out usin~ the GeneAmp DNA Amplification
kit (Perkin Elmer Cetu~) whereby 1(3 ~l lOx buffer containing dNTPs was
mixed with 100 pmol each of a sense oli~onucleotide and an anti-sense
S oli~onucleotide, ( 10 ~11 ot` a 40() ~I double stranded cDNA reaction mix)~ ().5
Amplitaq DNA polymerase. and distilled water to 100 ~
The samples were amplified with a pro~rammable therrnal
controller from MJ Research~ lnc. (Camblidge. MA). The filrst 5 rounds of
amplification consisted of denaturation at 94C for 1 minute, annealing of
lo primers to the template at 45C for 1 minute~ and chain elongation at 72C for I
- minute. The final 20 rounds of amplification consisted of denaturation as above~
annealing at 55C for 1 minute, and elongation as above~
Seven different Cr!~j I primer pairs were used to amplify the double
stranded cDNA as follows: CP-9 (SEQ. ID #8) and CP-17 (SEQ. ID #14). CP-
lO(SEQ.ID#9) and CP-17 (SEQ. ID #14). CP-lO(SEQ.ID#g) and CP-16
(SEQ.ID#13),CP-lO(SEQ.ID~9) and CP-19 (SEQ.ID #16). CP-10 (SEQ.ID
#9) and CP-18 (SEQ. ID #15). CP-13(SEQ.ID#10) and CP-17 (SEQ. ID#14).
dCP-13(SEQ.ID#10) and CP-19 (SEQ.ID#16). CP-17 (SEQ. ID#14)has
the sequence~5'- CCTGCAGAAGCTTCATCAACAACG~AGA-3' and
corresponds to non-coding strand sequence that corresponds to codin~ strand
sequence encoding amino acids SKRC* (amino acids 350-353 and the stop
codon o~ SEQ.ID#1). The nucleotide sequence 5'-CCTGCAGAAGCTI-3'
(bases 1 throu~h 13 of SEQ.ID#14) represents Pst I and Hin dIlI restriction
sites added for clonin~ purposes. The nucleotide sequence 5'-TCA-3' (bases 13
2s ~ through 15 of SEQ.ID#14) correspond to the non-coding strand sequence of a
stop codon. All of the amplifications yielded p[oducts of the expected size
when viewed on ethidium bromide (EtBr)-stained a~arose gels. Two of these
primer pairs were used in ampli~lcations whose products were cloned into
pUCl9 for full-length sequencin~n The PCR reaction with CP~10 (SEQ. ID #9)
and CP-16 (SEQ. ID #13) on the double stranded cDNA yielded a band of
approximately 1.1 kb, and was ~:alled JC13û. A separate t`irst strand cDNA
reaction was done with 8 llg pc)llen RNA as described above and amplit`ied with
WO 94/01~60 PCr/US93/0013~
?.,~æo9~4
oli~onucleotide primers CP-10 (SEQ. ID #9) and CP-17 (SEQ. ID ~14). This
amplification yielded a full-length cDNA. named ~Cl35~ from ~he arnino
terrninus of the mature protein to the stop codon.
Amplified DNA was recovered by sequential chloroform. phenol, and
s chloroform extractions, followed by precipitation at -20C with 0.5 volumes of
7.5 ammonium acetate and l.5 volumes of isopropanol. After precipitation and
washing with 70% ethanol. the DNA was blunted with T4 polymerase followed
by digestion with Eco RI, in the case of JC 130 . or simultaneously digested with
Eco RI and Ps~ I, in the case of JC135, in a 15 111 reaction and electrophoresedthrou~h a preparative 1% SeaPlaque low melt gel (FMC). Appropriate sized
DNA bands were visualized by EtBr staining. excised. and ligated into
appropriately digested pUCl9 for dideoxy DNA sequencin~ by the dideoxy
chain termination method (Sanger et al. (l977? P-oc. Na~l. Acad. Sci. USA
74:~463-5476) using a commercially available sequencing kit (Sequenase kit~
U.S. Biochemicals, Cleveland, OH).
Both strands were sequenced using M13 forward and reverse primers (N.E.
Biolabs, Beverly, MA) and internal sequencing primers CP-13 (SEQ. ID #10).
CP-1~ (SEQ. ID #12), CP-16 (SEQ ID #13), CP- 18 (SEQ. ID #15), CP- 19
(SEQ. ID #16) and CP-20 (SEQ. ID #17). Two clones from amplification JC 130
(JC130a and JC130b) and one clone from amplification JC135 (JC135g) were
found to be C--.Yj I clones upon sequencing. The nucleotide and deduced amino
acid sequences of clones JC130a and JCl35g were identical to previously l;nown .:;
C)~j I sequence (SEQ. ID ~1). Clone JC130b was found to contain a sin~le
nucleotide difference from the previously known Cr~j I sequence (SEQ. ID #l).
Clone JC130b had a C at nucleotide position 306 of Seq. ID #l. This nucleotide
change results in a predicted amino acid change from a Tyr to a His at amino
acid 60 of the mature Cr~ j I protein. This polymorphism has not yet been
confirmed in an independently-derived PCR clone or by direct amino acid
sequencing. However~ such polymorphisms in primary nucleotide and amino
acid sequences are expected.
5~
WO 94/01560 ~ 1 2~ Pcr/US~3/00139
Example 5
Expression of C y j I
Expression of C yj I was performed as follows. Ten ~1 of pUC 19JC9 l a
was digested with Xba I. precipitated, then blunted with T4 polymerase. BclmH I
linkers (N.E. Biolabs. Beverly, MA) were blunt^end ligated to pUC19JC9la
overnight and excess linkers were removed by filltration throu,o~h a NACS ion
exchange minicolumn (BRL, Gaithersbur~. MD). The linkered cDNA was then
digested simultaneously with EcoR I and BamH I. The Gy j I insert (extendin~
from the nucleotides encoding the amino terminus of the mature protein throu~h
the stop codon) was isolated by electrophoresis of this digest throuoh a 1%
SeaPlaque low melt agarose gel. The insert was then ligated into the
appropriately digested expression vector pET-l ld (Nova~en, Madison. WI:
Jameel et al. (1990) J. Virol. 64:3963-3966) modified to contain a sequence
lS encoding 6 histidines (His 6) immediately 3' of the ATG initiation codon
followed by a unique EcoR I endonuclease restriction site. A second EcoR I
endonuclease restriction site in the vector, along with neighboring Cla I and
Hind III endonuclease restriction sites. had previously been removed by
digestion with EcoR I and Hind ~I. blunting and religation. The histidine (HiS6)sequence was added for affinity purification of the recombinant protein (Cr~ j I)
on a Ni2+ chelatinp, column (Hochuli et al. ( 1987) J. Chrornatog. 41 1:177-184:Hochuli et al. (198&) Bio~rech. 6:1321-1325.). A recombinant clone was used to
transforrn Escherichia coli strain BL2 1-DE3 which harbors a plasmid that has anisopropyl-~-D-thiogalactopyranoside (IPI G3-inducible promoter precedin~ the
~ene encoding 17 polymerase. Induction with IPTG leads to high levels of T7
polymerase expression. which is necessary for expression of the recombinant
protein in pET-lld~ which has al7 promoter. Clone pET-lld~ HRhis6JC9la.d
was confirmed by dideoxy sequencino (Sanoer et al. Supra) with CP-14 (SEQ.
ID #l 1) to be a Cryj I clone in the correct readino t`rame for expression.
' Expression of the recombinant protein was confirmed in an initial small
culture (50 ml). An overni&ht culture of clone pET- I ld~HRhis6JC9 la.d was
used to inoculate 50 ml of media (Brain Heart Infusion Media. Difco) containino
ampicillin (200 ,ug/ml). ~rown to an A6 ~ = l.0 Imd then induced with IPTG (l
WO 94/01560 PCr/US93/00139
2~,09S~ ~ '
mM, final concentration) for 2 hrs. One ml aliquots of the bacteria were
collected before and after induction. pelleted by centrifugation, and crude cellIysates prepared by boiling the pellets ~or S minutes in 50 mM Tris HCh pH 6.8,
2 mM EDTA, 1% SDS. 1% ~-mercaptoethanol, 10~h glycerol. 0.25%
bromophenol blue (Studier et al., (1990) Methods in En~.Ymology I 85:60-89).
Recombinant protein expression was visualized as a band with the predicted
molecular weight of approximately 38 kDa on a Coomassie blue-stained SDS- ~
PAGE gel, according to the method in Sambrook et al.~ sl~pra, on which 40 ~1 of
the crude Iysate was loaded. A negative control consisted of crude lysates from
uninduced bacteria containing the plasmid with C~ j I and an induced Iysate
from bacteria carrying no plasmid.
The pET-l ld~ HRhis6JC91a.d clone was then grown on a large scale for
,recombinant protein expression and purification. A 2 ml culture bacteria
containing the recombinant plasmid was grown for 8 hr, then streaked onto solid
media (e.g. 6 petri plates (100 x 15 mm) with 1.5% agarose in LB medium
(Gibco-BRL, Gaithersburg, MD) containing 200 llg/ml ampicillin), grown to
confluence overnight, then scraped into 9 L of liquid media (Brain Heart
Infuslon media. Difco) containing ampicillin (200 ~g/ml). The culture was
grown until the A600 was 1.0, IPTG added ( 1 mM final concentration)~ and the
2 o culture grown for an additional 2 hours.
Bacteria were recovered by centrifugation (7.930 x g~ 10 min), and Iysed
- - ~; in 90 ml of 6M Guanidine-HCI. 0.1M Na2HPO4, pH 8.0 for I hour with
vigorous shaking. Insoluble material was removed by centrifugation ~11,0()() x
g, 10 min, 4 C). The pH of the lysate was adjusted to pH 8Ø and the Iysate
~ ~ '5 applied to an 80 ml Nickel NTA a~arose column (Qiagen) that had been
- - equilibrated with 6 M Guanidine HCI, 100 mM Na2HP04. pH 8Ø The column
was sequentially washed with 6 M Guanidine HCI, 100 mM Na2HP04 10 mM
Tris-HCl, pH 8.0, then 8 M urea, 100 mM Na2HPO4. pH 8Ø and finally 8 M
urea. 100 mM sodium acetate, 10 mM Tris-HCI. pH 6.3. The column was
washcd with each buffer until the flow through had an A280~ 1).05.
The recombinant protein, C~ was eluted with 8 M urea~ 1()(1 mM
- sodium acetate, 10 mM Tris-HCI, pH 4.5. and collected in 1() ml aliquots. The
WO 94/01~60 212 D !3 ~ 4 PCI~/US93/00139
protein concentration of each fraction was deterrnined by absorbance at A280
and the peak fractions pooled. An aliquot of the collected recombinant protein
was analyzed on SDS-PAGE according to the method in Sarnbrook et al.. Sllpl W.
The first 9 L prep. JCpET- I. yielded 30 mg of Cr~ j I with approximately
s 78~o purity, as determined by densitometry (Shimadzu Flying Spot Scanner.Shimadzu Scientific Instruments. Inc.. Braintree. MA) of the Coomassie-blue
stained SDS-PAGE gel. A second 9 E prep prepared the same way. JCpET-2
yielded 41 mg of Cr~j I with approximatel~7 77~ purity.
Example 6
lapanese Cedar Pollen Aller~ic Patient T Ce~l Studies with Crv j I - the
Primarv Cedar Pollen Anti~en.
Synthesis of Overlapping Peptides
Japanese cedar pollen Cr~ j I overlapping peptides were synthesized
using standard Fmoc/tBoc synthetic chemistly and purified by Reverse Phase
HPLC. Figure 13 shows Cryj I peptides used in these studies. The peptide
names are consistent throughout.
T Cell Responses to Cedar Pollen Antigen Peptides
Peripheral blood mononuclear cells (PBMC) were purified by
Iymphocyte separation medium (LSM) centrifugation of 60 ml of heparinized
2~ blood from Japanese cedar pollen-aller~ic patients who exhibited clinicalsymptoms of seasonal rhinitis and were MAST andJor skin test positive for
Japanese cedar pollen. Long term T cell lines were established by stimulation of? X 106 PBL/ml in bulk cultures of complete medium (RPMI-1640~ 2 mM L-
glutamine~ 100 U/ml penicillin/streptomycim Sx10-5M 2-mercaptoethanol. and
10 mM HEPES supplemented with 5~ heat inacti-ated human AB serum) with
20 ,ug/ml of partially purified native G~ j I (755~ purity containing three bands
similar to the three bands in Fig. 2) for 7 days at 37C in a humidified Sq~ CO~incubator to select for G~ j I reactive T cells. This amount of priming antigen
WO 94tO1560 PCI/US93/00139
21209~4
was determined to be optimal for the activation of T cells frorn most cedar pollen
allergic patients. Viable cells were purified by LSM centrifugation and culturedin complete medium supplemented with 5 units recombinant human IL-2tml and
5 units recombinant human IL-4/ml for up to three weeks until the cells no
5 lon~er responded to lymphokines and were considered "rested". The ability of
the T cells to proliferate to selected peptides. recombinant C-~ j I (rCr~ j I),purified native Cryj I, or recombinantAmb a 1.1 (rAmb aI.1) was then assessed.
For assay, 2 X 104 rested cells were restimulated in the presence of 2 X 104
autolo~ous Epstein-Barr virus (EBV)-transformed B cells (prepared as described
below) (gamma-irradiated with 25P00 RADS) with 2-50 ~lg/ml of rGy j 1.
purified native Cry j I or rAmb a I. 1. in a volume of 200 ,ul complete medium in
duplicate or triplicate wells in 96-well round bottom plates for 2-4 days. The
optimal incubation was found to be 3 days. Each well then received 1 ~lCi
tritiated thymidine for 16-20 hours. The counts incorporated were collected onto~glass fiber filter mats and processed for liquid scintillation counting. Fi~. 12
shows the effect of va ying antigen dose in assays with recombinant C ~
purified~nat`ive Cry j I, and recombinant Amb a I. 1 and several anti~enic peptides
synthesized~as~described above. Some peptides were found to be inhibitory at
hi~h~concentrations~in thes,e assays. The titrations were used to optimize the
20 ~ dose~of peptides in T cell assays. The maximum response in a titration~of each
peptidc;is expressed æ the stimulation index (S.I.). The S.I. is thè counts per
minute (CPM) incorporated by cells in response to peptide. divided by the CPM
incorporated by cells in medium only. An S.I. value equal to or greater than 2
times the back~round level is considered "positive" and indicates that the peptide
25 ~ contains a T cell epitope. The positive results were used in calculating mean
stimulation indices for each peptide for the, roup of patients tested. The results
-" ~ shown In Flg. 12~demonstrate that patient ~999 responds well to recombinant
Cr~ j I. and purified native Cr~ j I. as well as to peptides CJ 1-2~ 3. 2(1~ and 22
but not to recombinant Amb a I. l . This indicates that Cr.~ j I T cell epitopes are
recognized by T cells from this particular allergic patient and that rCn j I andpeptides CJ l -2. 3. 20 and 22 contain such T cell epitopes. Fulthermc)re. the 5
epitopes were often not detected with the adjacen~ overlappin~ peptides. and
:
, ~ ,
~ Sfi
;-
WO 94/015~0 2 1 2 0 9 5 4 PCI`/US93/0013~
therefore probably span the non-overlappin~ central residues of the reactive
peptides. No si~nificant cross-reactivity was found in T cell assays using T cells
primed with control antigens or with C~ j I primed T cells against other
antigens.
The above procedure was followed with a number of other patients.
Individual patient results were used in calculating the mean S.I. for each peptide
if the patient responded to the Cr~j I protein at an ~.I. of 2.0 or ~reater and the
patient responded to at least one peptide derived from C/y j I at an S.I. of 2.0 or
greater. A summary of positiYe experiments from twenty-five patients is shown
in Fi~ure 14. The bars represent the positivity index. Above each bar is the
percent of positive responses with an S.I. of at least two to the peptide or protein
in the group of patients tested. In parenthesis above each bar are the mean
stimulation indices for each peptide or protein for the group of patients tested.
All twenty-five T cell lines responded to purified native Cry j I and 68.0% of the
T cell lines responded to rC)y j I. These twenty-five T cell lines also responded
at a significantly lower level to rAmb a I. 1 indicating that the Amb a I allergens
share a de~ree of homology with Cryj I and that "shared" T cell epitopes might
exist between Gy j I and Am~ a I. This panel of Japanese cedar allergic patientsresponded to peptides CJl-l,CJl-2,CJl-3,CJl-4,CJl-7.CJl-8.CJl-9,CJl-10.
CJl-ll,CJl-12,CJl-14,CJl-lS.CJl-16,CJl-17.CJl-18,CJI-l9.CJl-20.CJl-
~l.CJl-22.CJl-23,CJl-24~CJl-25.CJl-26.CJl-27.CJl-28.CJl-30,CJI-31.
CJl-32.CJl-33,CJl-34 and CJl-35indicating that these peptides contain T cell
epitopes.
2~ Preparation of (EBV)-transformed B Cells for Use as Antigen Presenting
Cells
Autolo~ous EBV-transformed cell lines were ~y-irradiated with 25.()0()
Rad'and used as antig~en presenting cells in secondary proliferation assays and
secondary bulk stimulations. These cell lines were also used as a control in theimmuno-fluorescence llow cytometry analysis. These EBV-transt`ormed eell ;~
lines weR made by incubatino S X 106 PBL with I ml of B-59/8 Marmoset cell '-
WO 94/01560 PCr/US93/0û139
,o95~
line (ATCC CRL1612, American Type Culture Collection~ Rockville, MD)
conditioned medium in the presence of I ~lg/ml phorbol 12-myristate 13-acetate
(PMA) at 37C for 60 minutes in 12 X 75 mm polypropylene round-bottom
Falcon snap cap tubes (Becton Dickinson Labware. Lincoln Park, NJ). These
cells were then diluted to 1.25 X 106 cells/ml in RPMI-1640 as described above
except supplemented with 10% heat-inactivated fetal bovine sen~m and cultured
in 200 ~1 aliquots in flat bottom culture plates until visible colonies were
detected. They were then transferred to larger wells until the cell lines were
established.
Example 7
-
Cry j I as the Major Cedar Pollen Allergen
To examine the importance of Cr~ j I, reported as the major allergen of
Japanese cedar pollen, both direct and competition ELISA assays were
performed. For the direct ELISA assays~ wells were coated with either soluble
pollen extract (SPE) of Japanese cedar pollen or purifled native C~ j I (assayedat 905~ purity by protein sequencing) and human IgE antibody binding to these
20~ntigens was analyzed. Pooled human plasma. consisting of an equal volume of
plasma from 15 patients with a Japanese cedar pollen MAST score of 2.5 or
grzater, and two individual patient plasma samples were compared in this assay.
Fig. 5 shows the results of the binding reac.ivity with these two antigens. The
overall pattern of binding is very similar whether the coating antigen is SPE
(Fig. 5a~ or purified native Cr~ j I (Fig. Sb).
In the competition assay, ELISA wells were coated with Japanese cedar
pollen SPE and then allergic patient IgE binding was measured ili the presence ot`
competing purified native Cryj I in solution. The source of al~ergic IgE in these
assays was either the pool of plasma from 15 patients (denoted PHP) or seven
individual plasma samples from patients with a Japanese cedar MAST score of
'.~ or greater. The competition assay usin the pooled human plasma samples
compares the competitive binding capacity of purified native C~ j I to Japanese
WO 9~/01~60 21 2 ~ 9 .S 4 PCr/US93/00139
cedar pollen SPE and an irrelevant allergen source, rye ~rass SPE. Fi8. 6 shows
the graphed results of the competition ELISA with pooled human plasma. The
concentration of protein present in the Japanese cedar pollen SPE is
approximately 170 tirmes greater at each competin~ point than is the purified
native Cryj I . From this analysis it is clear that the purified native C/~ j I
competes very well for IgE binding to the whole range of proteins present in theJapanese cedar pollen soluble pollen extract. This implies that most of the anti-
C)y j IgE reactivity is directed against native C)y j I . The negative control
shows no specific competitive activity and the competing SPE in solution can
completely remove binding to the coated wells. This assay was repeated with
individual patients as a measure of the ran~e of the IgE response within the
- allergic population. Fig. 7 shows this result where the competition of binding to
SPE was performed with purified native C)y j I . The results demonstrate that
although the patients show different dose response to Japanese cedar pollen SPE.each of the seven patients' IgE binding to Japanese cedar pollen SPE could be
competed with pùrified native Cry j I. The implications of these data are that
for each patient the IgE reactivity directed against C~ j I is predominant but
that there is variation in this reactivity between patients. The overall conclusion
is that these data support the previous findings (Yasueda et al., (1988) ~L ) that
Cr~ j I is the major allergen of Japanese cedar pollen.
The reactivity of IgE from cedar pollen allergic patients to the pollen
proteins is dramatically reduced when these proteins are denatured. One method -
of analyzing this property is through direct binding ELISA where the coatin_
antigen is the Japanese cedar pollen SPE or denatured Japanese cedar pollen SPE
2s which has been denatured by boiling in the presence of a reducing a~ent DTT.
This is then examined with allergic patient plasma for lgE binding reactivity.
Fig. 8a. shows the direct binding assay to the SPE with seven individual plasm~
samples. In Fig. 8b. the binding results with the denatured SPE demonstrates themarked decrease in reactivity following this treatment. To determine the extent
of Cr~ j I binding to the ELISA wells. C)~ j I was detected with a rabbit
polyclonal antisera against the Amb a 1 & Il protein family. These ragweed
proteins have high sequence identity (465~) with C-! j I and this antisera can be -~
7~ .
WO 94/01560 PCI/US93/00139
2.~.?.1~9S~ ,
used as a cross reactive antibody detection system. In conclusion, these data
demonstrate a marked loss in IgE reactivity following denaturation of the
Japanese cedar pollen SPE.
s Example 8
IgE Reac~vity and Histamine Release Analysis
The recombinant Cryj I protein (rC~j I), expressed in bacteria and then
purified (as described in Example 5), has been examined for IgE reactivity. The
first method applied to this exarnination was direct ELISA where wells were
coated with the recornbinant Cn~ j I and IgE binding was assayed on individual
- patients. Fig. 9 is the graphic representation of this direct ELISA. The only
positive signals on this data set are from the two control antisera rabbit
polyclonal anti-Amb a I & II prepared by conventional means (Rabbit anti-Am~ a
I & II~ and CBF2, a monoclonal antibody raised against Amb a I that cross
reacts with Gyj I . By this method all patients tested showed no IgE reactivity
with the recombinant Gyj I .
Another method of analysis that was applied to the examination of IgE
reactivity to the recombinant Cry j I was a capture ELISA. This analysis relies
on the use of a defined andbody, in this case CBF2 to bind the antigen and allowfor the binding of antibodies to other epitope sites. The format of this captureELISA is 1) wells are coated with MAb CBF2, 2) antigen or PBS (as one type of
negative control) is added and captured by specific interaction with the coated
MAb~ 3) either the control antibody anti-Amb a I & II (Fi;,. lOb) or human
allergic plasma (Fig. lOa) is added as the detecting antibody, and 4) detection of
antibody binding is assayed. Figs. lOa and lOb are the graphed results of these
assays. For the I~E analysis, the pooled human plasma (PHP) (15 patients) was
used. The conclusion from these results is that there is no indication of any
specit`ic binding of human allergic I~E to rC) ~ j I by this method ot' analysis.
However, the capture of rCrYj I works as evidenced by the control antibody
bindin~ curve, shown in Fi~. lOb. The lacl; ot' I~E bindin~ to E. ~Q~ expressed
rC~ j I may be due to absence of carbohydrate or any other post-translational
~' 212~3,;1
WO 94/01560 PCr/US93/00139
modification and/or that the majority of IgE cannot react with denatured C~ j I.RAST. competition EUSA and Western blotting data also demonstrates no
specific IgE reactivity to ~he rCr~ j I (data not shown).
A histamine release assay was performed on one Japanese cedar pollen
allergic patient using Japanese cedar pollen SPE. purified native C~; I and
rCr~ j I as the added antigens. This assay is a measure of IgE reactivity through
human basophil mediator release. The results of this assay. shown in Fig. 11.
demonstrate strong his~nine release with both purified native Cr~ j I and the
Japanese cedar pollen SPE over a wide concentration range. The only point
where there is any measurable histarnine release with the Cr~ j I is at the highest
concentration, 50 ~/ml. Two possible explanations for this release by the rC~ j
- I are: 1) specific reactivity with a very low proportion of the anti-Cr,~; I IgE
capable of recognizing the recombinant form of Cr! j I . or 2) non-specific
release caused by low abundance of bacterial contaminants observed only at the
hi~hest antigen concentration. Thus far, this result has only been shown in a
single patient. In addition, the data shown are from single data points at each
protein concentration.
It may be possible to use this recombinantly expressed Cry j I protein for
irnmunotherapy as E. coli expressed material has T cell reactivity (Example 6)~
but does not appear to bind IgE from C ytpomeria japonica atopes nor cause
~-~ histamine release from the mast cells and basophils of such atopes i~i vitro.
E:~pression of rG~ j I which is capable of binding IgE could possibly be
achieved in yeast, insect (baculovirus) or mammalian cells (e.g. CHO. human
and mouse). A specific example of mammalian cell expression could be the use
of the pcDNA VAmp mammalian expression vector (Invitro~en. San Diego. CA)
e~pressing recombinant Cr~ j I in COS cells. A rC~ ~ j I capable of actively
binding IgE may be important for the use of recombinant material for diagnostic
purposes.
To analyze IgE reactivity to selected C~ j I peptides a direct ELISA
fo~nat was used. ELSIA wells were coated with 2~ peptides derived trom C~ j
I lnd assayed for IgE binding. Fig. l5a and l5b are graphs ot these bindin~
r~sults using PHP (15 patients) as the cedar pollen aller~ic I E source. This pool
fil
WO 94/01~60 PCr/US93/00139
2~2~95~ -
of plasma was forrnulated for enrichment of IgE that could bind to denatured
SPE (as detennined by direct ELISA) and therefore increase the chance of
reactivity toward the peptides. In this assay~ the peptide I~E binding capacity
was compared to that of purified native Cn j I and to rCr~ j 1. The only specific
IoE detected in this assay was to purified native Cr~j 1 which supports the
~inding that Japanese cedar allergic patient IgE does not bind to recombinant C~j I or the recombinant Cr}~j I peptides tested (Fig. 15).
Although the invention has been described with reference to its preferred
embodiments, other embodiments, can achieve the same results. Variations and
modifications to the present invention will be obvious to those slcilled in the art
and it is intended to cover in the appended claims all such modification and
equivalents and follow in the true spirit and scope of this invention.
Example 9
- Extraction of RNA from Juniperus sabinoides, Juniperus virgini~na andCl~pressus arizonica pollens and the cloning of Jun s I and Jun v I, homologs
of GyjI.
Fresh pollen was collected from a single J~miperus virgil1iana tree at the
Arnold Arboretum (Boston, MA), and was frozen immediately on dry ice;
J~ iperus sabinoides and Cupressus ari~onica pollens were purchased from
Greer Laboratories, Inc. (Lenoir, NC). Total RNA was prepared from J.
~irginiana, J. sabinoides. and C arizonica pollens as described in Example 3.
Single stranded cDNA was synthesized from S llg total pollen RNA from J~
lir~iniana and 5 ~g total pollen RNA from 1. sabinoides usino the cDNA
S~ nthesis System Icit (BRL, Gaithersburg. MD). as described in Example 3.
The initial attempt at cloning C ~ j I homologue from the tw() juniper
speci~s was made using various pairs of C~ specific olioonucleotides in PCR
amplifications on both juniper cDNAs. PCRs were carried out as described in
E.~ample 3 The oligonucleotide primer pairs used were: CP-9JCP-17. CPl~)/CP-
6~
WO94/01~60 ' 212D9.~-~ PCr/USs3/00139 ~'
17, CP-10/CP-16, CP-10/CP-l9. CP-lWCP-18, CP-13/CP-17. and CP-13/CP-19.
CP-10 was used in the majority of the reactions as the ~' primer since it has been
reported by Gross et. al. (1978) Scand. 1. Immunol. 8: 437-441 that the first 5
amino-terminal amino acids of J. sabinoid~s are identical to those of C~ j I.
S These oli~onucleotides and oligonucleotide primers pairs are described in ~-
Example 3.
None of the primer pairs cited above resulted in a PCR product for either
juniper species when viewed on an EtBr-stained 1% a~arose (FMC Bioproducts.
Rockland, ME) minigel.
The next series of PCR amplifications attempting to clone the C~
homologues from J. sabinoides and J. virginiaMa from were made on double
stranded linkered cDNA synthesized from RNA from each species. Oouble
stranded cDNA was synthesized from 5 ,u~ of J. virginiana and S llg J.
sabinoides pollen RNA as described in Example 3. The double-stranded cDNA .
was li~ated to ethanol precipitated, self annealed. AT and AL olioonucleotides
for use in a modified Anchored PCR as described in Example 3. A number of
C~ j I primers were then used in combination with AP in an attempt to isolate
the Cr~ j I homologues from the two juniper species. The sequences of AT. AL
and AP are given in Example 3. First, a primary PCR was carried out with 100
pmol each of the oligonucleotides CP-10 and AP. Three percent (3 ,ul) ot' this
initial ampli~lcation was then used in a secondary PCR with 100 pmoles each of
CP- 10 and 'APA, which has the sequence ~'-GGGCTCGAGCTGCAG'l l~ l~
ImTG-3'. where nucleotides 1-15 represent Pst I and Xho I
endonuclease restriction sites added for clonin~ purposes. and nucleotide 33 canalso be an A or C. A broad smear, with no discreet band. was revealed upon
examination of the secondary PCR reactions on an EtBr-stained agarose gel.
Attempts to clone C~ j I homolo~ues from these PCR products were not
successful. This approach would have cloned a carboxyl portion of these genes.
The~ degenerate Cn~ j I primers CP-l, CP-4. and CP-7 as described in Example 3
were then each used in primary PCRs with AP on the double stranded linl;ered J.
~ ir,~iniana and J. sabinoides cDNAs. Various plimer pair combinations were
used in secondary PCRs as follows: CP-2/AP and CP-4/AP on the CP- I/AP
~,Y~ . ~
Wo 94/01560 PCr/USs3/00139
2~20954
primary PCR arnplification mixture, CP-2/AP and CP-S/AP on the CP-4/AP
primary PCR amplification mixture, and CP-8/AP on the CP-7/AP primary PCR
arnplification mixture. Only the last amplification, the CP-8/AP seeondary PCR
amplification, yielded a band upon examination on an EtBr-stained minigel; the
others gave smears that could not be cloned into pUC19. Both the J. virginiana ~ t
and J. sabinoides secondary PCRs with CP-8 and AP, described in Example 3
called JV21 and JS17, respectively, resulted in amplified products that were
approximately 200 base pairs long. The amplified DNA was recovered as
described in Example 3 and simultaneously digested with Xba I and Pst I in a 50
~11 reaction, precipitated to reduce the volume to 10 ~1. and electrophoresed
through a preparative 2% GTG NuSeive low melt gel (FMC. Rockport, ME).
The appropriate sized DNA band was visualized by EtBr staining~ excised~ and
ligated into appropriately digested pUCl9 for sequencing by the dideoxy chain
termination method of Sanger et al. (~) using a commercially available
sequencing ki~ (Sequenase kit, U.S. Biochemicals, Cleveland, OH). Two JS17
clones (pUC19JS17d and pUC19JS17f) and one JV21 clone (pUC19JV21g)
were sequenced, and found tO contain sequences homologous to the Cry j I
nucleotide and deduced amino acid sequences. The Cr~ j I homologues isolated
from J. sabinoides and J. virginiana RNA were desi~nated Jun s I and Jun
respectively.
The C~ j 1 primers CP-9 and CP-10 should worl; in primary and
secondary PCRs, respectively, with AP to amplify the carboxyl portion of the
Jlm s I and Jun v T cDNAs. The sequence of these primers are essentially
identical to the sequences of Jun s I and Jlm v I, with the exception of 2
nucleotides in CP-9 (T instead of A in position 5 of CP-9. C instead of A in
position 123, and 1 in CP-10 (C instead of A in position 12 for Jun s I only)~
However~ primary PCRs with CP-9 and AP and secondary PCRs with CP-10 and
AP did not yield identifiable Jun s I nor Jun v I product when viewed on an
EtBr-stained agarose gel.
Oligonucleotide J 1 was synthesized~ J 1 and all subsequent
oligonucleotides were synthesized on an ABI 394 DNA/RNA synthesizer
(Applied Biosystems, Foster City. CA)~ Primary PCRs were carried out usino
~4
WO94/01560 2 1 2 0 9 ~ ~ PCT/US93/00139
APandJl with J. virginiana and J. sabinoides cDNAs. J1 has the sequence 5'-
CTAAAAATGGCl~CCCCA-3'~ which corresponds to nucleotides 20-37OfJun
s I (Fig. 16) and nucleotides 30-47OfJunvI(Fig.17). A secondary PCR
amplification was performed on the pnmary Jl/AP~mplificationofJ. sabinoides
S cDNA using primers J2 and AP.J2 has the sequence 5'-
CGGGAATTCTAGATGTGCAATTGTATCTTGTTA-3'. whereby nucleotides
1-13 represent EcoR land Xba I endonuclease restriction sites added for cloning
purposes. and the remaining nucleotides correspond to nucleotides 65-84 in the
Jl~n s I sequence (Fi~. 16). The secondary arnplification from J. virginiana
io cDNA was perforrned with AP and J3, which has sequence 5'-
CGGGAATTCTAGATGTGCAATAGTATCTTGTTG-3' whereby nucleotides
1-13 represent EcoR I and Xba I endonuclease restriction sites added for clonin~purposes and tbe remainin~ nucleotides correspond to nucleotides 75-94 in the
Jl/n 1~ I sequence ~Fig. 17). Nospecificamplified product was observed in eithersecondary reaction. The primers desi~nated ED and EDT were used at a molar
ratio of 3:1 (ED:EDT) in conjunction with primers Jl, J2 and J3, as described
below. EDT has the sequence 5'-GGAATICTCTAGACTGCAGG-
lmmm~-3'. The nucleotides 1 throu~h 20 of EDT were added to
- the poly-T track to create EcoR I. Xba I, and Pst I endonuclease restriction sites
for cloning putposes. ED has the sequence 5'-GGAAl-rCTCTAGACTGCA-
GGT-3'. correspondin~ to nucleotides 1 to 21 of EDT. These oli~onucleotides
,and their use have been previously described (Mor~enstern et al. ( I g9 1 ) Proc.
Natnl. Acad. Sci. USA 88:9690-9694). EDtEDT were used in primary PCRs
with oli~onucleotide Jl for amplifications ~rom J. sabinoides and J. virgil7ianacDNAs, followed by secondary PCRs with oligonucleotides J2 and APA (for J.
sabinoides) or J3 and APA (for J. virginiana). No speci~lc product was
identified from these amplifications. A final set of PCRs with J 1. J2 and J3 was
tried with oli~onucleotide APA. APA was used in a primary PCR reaction with
J l for J. sabinoides and J. vi--giniana. followed by secondary amplifications with
J~ (for J. sa~inoides) or J3 (for J. virginiana) and APA. No specific product was
identified from these amplifications. The dec~enerate primer CP-57 was then
s~nthesized. CP-57 has the sequence 5'-GGCCTGCAGTTAACAGCG-
t
6s
WO 94/01560 PCI/US93/00~39
9 r 4
l~GCAGAAGGTGCA-3'. wherein T at position 10 can also be C~ T at
position 11 can also be C. A at position 13 can also be G.G at position 16 can
also be A.T. or C. G at position 18 can also be T, T at position 19 can also be C.
G at position 22 can also be A. T or C. C at position 23 can also be G. A at
position 24 can also be C. G at position 25 can also be A. T. or C. A at position
27 can also be G. G at position 28 can also be A, T. or C. G at position 29 can
also be C. T at position 30 can also be A, and G at position 31 can also be A.
The nucleotides 1 through 9 of CP-57 were added to create a Pst I site for
cloning purposes, the nucleotides 10 through 12 are complementary to a stop
Io codon and nucleotides 13 through 33 are complementary to coding strand
sequence essentially encoding the amino acids CysSerLeuSerLysArgCys (amino
acids 347 through 353 of Figure 4b, corresponding to nucleotides 1167 through
1187 of Figure 4b). This was used in a primary PCR with Jl on both J.
sabinoides and J. virginiana double stranded linkered cDNA, followed by a
secondary PCRs with CP-57 and J2 for J. sabinoides and CP-57 and J3 for 3.
virginiana. No PCR products were recovered. Three additional degenerate C~ j
- ~ I oligonuc~leotides were synthesized. CP-62 has sequence 5'-
CCACTAAATAl-rATCCA-3', wherein A at position 3 can also be G, A at
position 6 can also be G, T at position 9 can also be A or G. and T at position 12
can also be A or G: this degenerate oligonucleodde sequence is complementary
to the coding strand sequence essentially encoding the amino acids
~- TrpIleIlePheSerGly (amino acids 69 through 74 of Figure 4a, corresponding to
nucleotides 333 throuch 349 of Figure 4a). CP-63 has sequence 5'-
GCATCCCCATCTTGGGGATG-3'. wherein A at position 3 can also be G. A at
position 9 can also be G. T at position 12 can also be C G at position 1~ can also
; be A. T. or C. and A at position 18 can also be G; this degenerate
oligonucleotide sequence is complementary to the sequence capable of encodin~
the arnino acids HisProGlnAspGlyAspAla (amino acids 146-152 of Fi~ure ~a.
~; corresponding to nucleotides 564 tO 583 of Figure 4a). CP-64 has the sequen~:e
5'-GTCCATGGATCATAAl~ATT-3'. wherein T at position 6 can also be C A
at-position 9 can also be G. A at position 12 can also be G~ A at position 15 can
also be G. and A at position 18 can also be G: this de~enerate oli~onucleotide
66
WO 94/01560 2 1 ~ 0 ~ ~ 4 Pcr/usg3/00l39
sequence is complementary to the coding strand sequence capable of encoding
the amino acids AsnAsnTyrAspProTrpThr (amino acids 243-249 of Figure 4b.
corresponding to nucleotides 855 through 874 of Figure 4b). AP was used in a
primary PCR amplification with CP-62. CP-63. CP-64 and CP-3 (described in
Example 3) for both J. sabinoides and J. v;rginiana double-stranded linkered
cDNA. A diagnostic PCR was performed on each primary reaction mixture. In
this diagnostic PCR. 3~Q of the primary reaction was amplified as described
above using AP and CP-8. For both J. sabinoides and J. virginia~la, the expectedbands of approximately 200 base pairs were observed in diagnostic PCRs from
the primary PCR with AP and CP-63.
The degenerate primer CP-65 was then synthesized. CP-65 has the
sequence 5'-GCCCTGCAGTCCCCATCTTGGGGATGGAC-3'. wherein A at
position 15 can also be G7 T at position 18 can also be C. G at position 2 l canalso be G, A, T, or C, A at position 24 can also be G. and G at position 27 can
1S also be A, T, or C. Nucleotides 1-9 of CP-65 were added to create a Pst I
restriction site for cloning purposes, while the remaining de~enerate
o ligonucleodde sequence is complementary to coding strand sequence essentially
7', ~ capable o~ encoding the amino acids ValHisProGlnAspGlyAsp (amino acids
145-151 of Figure 4a corresponding to nucleotides 561 through 580 of Fi~ure
20 ~ ~ 4a). AP was used in conjunction with CP-65 in a secondary PCR of the plimary
AP/CP-63 àmplifications of J. sabinoid~s and J. virginiana desclibed above.
These reactions were designated JS42 for J. sabinoides and JV46 for J.
irgini~na. Both secondary PCRs gave bands of approximately 6~10 base pairs
hen examined on 1% agarose minigels stained with EtBr. The DNA from the
2s JS42 and JV46 PCRs was recovered as described in Example 3, simultaneouslydi~ested with Xba l and Pst I in 15 ,ul reactions then electrophoresed throuoh apr~parative 2% GTG SeaPlaque low melt ~el tFMC Rockport. ME). The
appropriate sized DNA bands were visualized by EtBr staining. excised. and
ligated into appropriately digested pUCI9 for sequencing by the dideoxy chain
t~rmination method (Sanger et al., ~) usin~ a commercially available
sequencing kit (Sequenase l;it. U.S. Biochemicals. Cleveland. OH) Clones were
s~quenced using M13 forward and reverse primers (N.E. Biolabs. Beverly. MA) t
67
Wo 94/01~6~ PCr/US93/00139
2~2U95'~
and internal sequencing primer J4 for both Jun s I and Jun v I. J4 has the
sequence 5'-GCTC(: ACCATGGGAGGCA-3' (nucleotides 177-194 of Fig. 16
and nucleotides 187-204 of Fig. 17), which is the coding strand sequence that
essentially encodes amino acids SerSerThrMetGlyGly (amino acids 30 through
35 of Jun s I and Jun v I as shown in Figs. 16 and 17, respectively).
The sequence of the J~n s I clone designated pUC19JS42e was found to
be identical to that of clones pUC19JS17d and pUC19JS17f in their regions of
overlap, although they had different lengths in ~he 5' untranslated region. Clone
pUC19JS17d had the longest 5' untranslated sequence. Nucleotides 1 through
141 of Fig. 16correspondtosequenceofclonepUC19JS17d. Clone
pUC19JS42e corresponds to nucleotides 1 through 538 of Fig. 16.
The sequences of the Jun v I clones designa~ed pUC19JV46a and
pUC19JV46b were identical to the sequence of clone pUC19JV21g in their
regions of overlap, with the exception that nucleotide 83 of Figure 17 was A in
clone pUC19JV21~ rather than the T shown. This nucleotide difference does not
result in a predicted amino acid change. Clones pUC19JV46a, pUC19JV46b and
pUC19JV21g correspond to nucleotides 1 throu~h 548. 1 through 548 and 2
through 151 of Figure 17, respectively.
The cDNAs encoding the remainder of the Jun s I and Jur1 l~ I genes were
cloned from the respective linkered cDNAs by using degenerate oligonucleotide
CP-66. which has the sequence 5'-CATCCGCAAGATGGGGATGC-3'~ wherein
T at position 3 can also be C. G at- position 6 can also be A, T, or C, A at
position 9 can also be G. T at position 12 can also be C. and T at position 18 can
also be C. and AP in a primary PCR. The sequence of CP-66 is complementary
2s ~o that of CP-63. A secondary PCR was perforrned on 3~G of the initial
arnplification mixture, with 100 pmoles each of AP and CP-67. which has the
sequence 5'-CGGGAAl~CCCTCAAGAl'GGGGATGCGCT-3'. wherein A at
position 15 can also be G, T at position 18 can also be C. T at position 24 can
also be C. G ~at position 27 can also be A, T. or C, and C at position 28 can be T.
The nucleotide sequence 5'-CGGGAATTC-3' ot` primer CP-67 (bases I throu~h
9) were added to create an EcoR I restriction site t'or cloning pulposes. The
- remaining oligonucleotide sequence essentially encodes amino acids ~,
6~
WO 94/01560 2 1 2 0 9 S 1 PCr/US93/00139
ProGlnAspGlyAspAlaLeu (amino acids 147 through 153 of Figure 4a,
corresponding to nucleotides 567 throu~h 5~ of Fi~ure 4a). The amplified
DNA products~ designated JS45 from the J. sabi~loides amplification and JV49ii
from the J. virginiana amplification, were purified as described in Example 3,
digested with EcoR I and Xba I (JS45) or EcoR I and Asp7 18 I (JV49ii) and
electrophoresed through a preparative 1% IQW melt gel. The dominant DNA
bands, which were approximately 650 bp in length, were excised and ligated into
pUC19 for sequencing. DNA was sequenced by the dideoxy chain terrnination
method (Sanger et al. ~upra) using a commercially available kit ~sequenase kit.
U.S. ~iochemicals, Cleveland, OH).
Two clones, designated pUC19JS45a and pUC19JV49iia for Jun s I and
Jl~n v I. respectively. were sequenced using M13 forward and reverse primers
(N.E. BioLabs, Beverly, MA) and internal sequencing primers J8, J9, and J12 for
Jlln s I. and J6 and J 11 for Jlm v I . J8 has the sequence 5'- ,
TAGGACATGATGATACAT-3' (nucleotides 690-707 of Fig. 16), which is the
coding st~and sequence essentially encoding arnino acids LeuGlyHisAspAspThr
o~` Jun s I (amino acids 201-206 of Fig. 16). J9 has the sequence 5'-
GAGATCTACACGAGATGC-3' (nucleotides 976-993 of Fig. 16) which is the
coding strand sequence essentially encoding amino acids Ar~SerThrArgAspAla
of Jlm s I (amino acids 297-302 of Fig. 16). J12 has the sequence 5'-
AAAACTATTCCCTTCACT-3', wherein A at position 1 can also be G~ and A at
position 4 can also be T: This is the non-coding strand sequence that
corresponds to coding strand sequence (nucleotides 875-892 of Fig. 16) encodin~
arnino acids SerGluGlyAsnSerPhe of Jun s I (amino acids 263-268 of Fig. 16).
J~; has the sequence 5'-l`AG(iACATAGTGATTCAT-3' (nucleotides 700-717 of
Fig. 17), which is the coding strand sequence essentially encoding amino acids
LeuGlyHisSerAspSer of Jun v I ~amino acids 201-206 of Fi~.17). Jl l has the
sequence 5'-CCGGGATCCTTACAAATAACACA ITAT-3'. where nucleotides
1-~ encode a BamH 1 restriction site for cloning purposes and nucletides 1(~-27
correspond to noncoding strand sequence complemental~ to nucleotides 1165-
1182 of Fig. 17 in the 3' untranslated region of J~J7 v I. The sequence of clonep~ C19JS45a corresponds to nucleotides 527 throuoh 117() of Fig. 16. The `~
WO 94/01560 P~/US93/00139
'' 2~,209~
sequence of clone pUC29JV49iia corresponds to nucleotides 537 through 1278
of Fig. 17.
Afull len~th clone of Jun s I was ampli~led usinC~ PCR. Oligonucleotides
J7 and J10 were used in a PCR reaction as above with J. sabinoid~s double
stranded, linkered cDNA. 17 has the sequence S'-CCCGAA~CATGGCrrCC-
CCATGCTrA-3', where nucleotides 1-9 encode an &oR I restriction site added
for cloning purposes and nucleotides 10-27 (correspondin~ to nucleotides 26-43
of Fig. 16) are the coding strand sequence that encode arnino acids
MetAlaSerProCysLeu of Jl(n S I (amino acids -21 to - 16, Fig. 16). ~ 10 has the
0 sequence 5~-ccGGGATcccGlTrcATAAGcAAGArr-3~ where nucleotides
1-9 encode a BamH I restriction site dded for cloning purposes and nucleotides
10-27 are the non-codinc~ strand sequence complementary to nucleotides 1140-
1157 from the 3' untranslated re~ion of JIIJ7 S I (Fi~. 16). The PCR product.
designated JS53ii. gave a band of approximately 1200 bp when examined on a
1% aga~rose minigel stained with EtBr. The DNA from the JS53ii PCR was
Rcovered as described in Example 3. After precipitation and washing with 70q~
-~ EtOH, the DNA was simultaneously dioested with EcoR I and BamH I in a 15 ~11
reaction ~and electrophoresed through a preparative 1% GTG SeaPlaque low melt
gel (FMC. Rockport. ME). The appropriate sized DNA band was vlsualized by
~- 20 EtBr staining. excised. and ligated into appropriately digested pUCl9 for
s~quencing by the dideoxy chain termination method (Sanger et al. (1977) ~!
using ~a ~commercially available sequencing kit (Sequenase kit. U.S. q~
Biochemicals. Cleveland. OH). The resultant clone. pUC19JS53iib was partiall~
sequenced using~MI3 forward and reverse primers (N.E. Biolabs Beverly~ MA)
2~ and internal sequencing primer J4. The sequence of pUC19JS53iib that ~as
- ~ determincd was identical to that obtained from clones pUC19JS17d~
pUC~19JS42e.and~pUC19JS45a. Thenucleotidesequenceofclone
p~rC19JS53iib corresponds to nucleotides 26 through 1157 of Ficn 16.
The nucleotide and predicted amino acid sequences of Jut1 s I are shown
in Ficn 16. Jun s I has an open reading *ame c)f 11()1 nucleotides~ correspondin~
~ - to nucleotides 26 throu~h 1126 of Fig. 16~ that can encode a protein of 367 ~;
- ~ arnino acids. Nucleotides 1-25 and 1130-117n of Fio. 16 are untranslated S' and
, ~
,
WO 94/01560 '- 21 2 0 9 ï ~ PCI`/US93/00~39
3' regions, respectively. The initiatin~ Met, encoded by nucleotides 26-28 of
Fig. 16~ has been identified through the 89% identity of nucleotides 23 throu~h
30 (AAAAATGGC) of Fig. 16 with the consensus sequence encompassin~ the
initiating Met in plants ~AACAATGGC; Lutcke, supra). There is also an in-
frame stop codon just 5' of the codon encodin~ the initiatin~ Met. Amino acids
-21 to -1 of Fi~. 16 correspond to a predicted leader sequence. The amino
terminus of the mature form of Jun s I was identified as amino acid 1 of Fi~. 16through direct protein sequence analysis of purified Ju11 s I (Gross et al supra).
The mature fonn of Jun s I. corresponding to amino acids 1 throu; h 346 of Fig.
16. has a predicted molecular weight of 37.7 kDa. Jun s I has three potential N-linked glycosylation sites with the consensus sequence of Asn-Xxx-Ser/Thr.
The nucleic and predicted amino acid sequences of Jun v I are shown in
Fig. 17. Nucleotides 1-35and 1130-1170areuntranslatedS'and3'regions.
respectively. The initiatin~ Met, encoded by nucleotides 36-38 of Fi~. 17~ was
identified throu~h the 89% identity of nucleotides 23 through 30
(AAAAATGGC) of Fig. 17 with the consensus sequence encompassing the
initiatin~ Met in plants (AACAATGGC: Lutcke, supra). The nucleic acids of
Jlm s I (Fig~. 16) and Jlm v I (Fig. 17) are identical in this re~ion surrounding the
initiating Met. There are also 2 in-frame stop codons in the 5' untranslated
region of Fig. 17. Jun v I has an open readin~ frame of 1.110 nucleotides.
corresponding to nucleotides 36 through 1145 of Fig 17, that can encode a
protein of 370 amino acids. Nucleotides 1146-1148 of Fi~. 17 encode a StOp
codon. Amino acids -21 to -1 of Jun v I (Fig. 17) correspond to a predicted
leader sequence. The amino terminus of the mature form of Jlm v I was
identified as amino acid 1 of Fi; . 17 by comparison with the sequences of C~
(Fio. 4a) and Jun s I ~Fig. 16). The mature forrn of Jun v I, correspondin~ to
amino acids 1 through 349 of Fig. 17 has a predicted molecular wei~ht of 3~.()
~Da. Jl/n v I has four potential N-linked glycosylation sites with the consensussequence of Asn-Xxx-Ser/Thr.
As shown in Table I. the amino acid sequences of the mature forms of
Ju~7 s I and Jun v I are 8().9~ homolo~ous t75.4~ identity and 5.5~ similarity)
with each other. The amino acid sequences ot the mature forms of Jun s I and
7~ 3
WO 94/01~60 PCr/US93/00139
21~095~ `
-
Gyj I are 87% homologous (80.1% identity. 6.9% similarity) and the sequences
of the mature fonTls of Jl~n v I and ~j I are 80.5% homolo~ous (72.5%
identity, 8% similarity). The homologies between Cn~; I peptide sequences
identified in Example 6 as containing T cell epitopes and the colTespondin~ Jun s
s I and Jun v I sequences are also very hi~h. For example, peptide CJ 1-22.
corresponding to amino acids 21 1-230 of Cr~ j I (Fig. 13), contains a major T
cell epitope ~Fig. 14). CJ1-22 has 95~o identity (19/20 identical amino acids)
and 85~ homolo~y (16/20 identical amino acids, 1/20 similar amino acid) with
the corresponding regions of Jun s I and Jun v I. respectively (see Table I). This
high degree of sequence homology suggests that an immunotherapy effective in
treating aller~ic disease caused by Cr~ j I may also be effective in treatin~
aller~ic diseases caused by Cr~ j I homolo~ues. All nucleic and amino acid
analyses were perforrned using software contained in PCGENE (lntelligenetics.
Mountain View, CA).
Table I
ProteinlPeptide Total
Comparisons Identity Similaritv Hnmol(~v
Jun s I vs. Jun v I 75.4% 5.5% 80.9%
J~nsIvs. CryjI 80.1% 6.9% 87.9~
Jl~n v I vs. Cryj I 72.5% 8.0% 80.5~7c
CJl-22vs.JunsI ~ 30 95.0~ O.Oqo 95.
CJ1-22 vs. Jun v I,ll 230 80.0% 5.0% 85.0
O
Example 10
~orthern blot analysis of C. japonica, J. sabinoides, J. virginiana
and C. arizon&a RNA.
!
.~ Northern blot analysis was performed on RNA isolated flom C. jclponicu. J.
sal~inoid~s and J. virginiarla pollens. RNA frt)m C. japonica pollens colleeted in
both the l;nited States tExample 3) and Japan (Example 4) were examined.
WO 94/~1560 2 1 2 0 9 ~ ~ Pcr/US93/00l39
Using essentially the method of Sambrook, supra, 15 llg of each RNA were run
on a 1.2% agarose gel containing 38~o formaldehyde and lX MOPS (20X =
0.4M MOPS, 0.02M EDTA, 0. lM NaOAc. pH 7.0~ solution. The RNA samples
(first precipitated with 1/10 volume sodium acetate~ 2 volumes ethanol to reducevolume and resuspended in 5.5 ~11 dH20) were run with 10 1l1
formaldehyde/formamide buffer containing loading dyes with 15.5%
t`ormaldehyde. 42% formamide, and 1.3X MOPS solution~ final concentration.
The samples were transferred to Genescreen Plus (NEN Research Products,
Boston, MA) by capillary transfer in 10X SSC (20X = 3M NaCI, 0.3M Sodium
Citrate)~ after which the membrane was baked 2 hr. at 80C and UV irradiated
for 3 minutes. Prehybridization of the membrane was at 60C for I hour in 4 ml
0.5M NaPO4 (pH 7.2), lmM EDTA, 1% BSA. and 7% SDS. The antisense
probe was synthesized by asymmetric PCR (McCabe, P.C.. in: PCR Protocols.
A Guide to Methods and Applications, Innis. M.. et al., eds. Academic Press~
Boston, (1990), pp 76-83) on the JC9la amplification in low melt agarose
(described in Example 3), where 2 ,ul DNA is amplified with 2 111 dNTP mix
(0.167 mM dATP, 0.167mM dl~P, 0.167mM dGTP, and 0.033mM dCTP). 2 1ll
10X PCR buffer, 10 ~ul 32P-dCTP (100 IlCi; Amersham, Arlington Hei~hts, Il)~ -
1 111(100 pmoles) antisense primer CP-17, 0.5 ,ul Taq polymerase, and dH2O to
''0 ,ul; the 10X PCR buffer, dNTPs and Taq polymerase were from Perkin Elmer
Cetus (Norwalk, CT). Amplification consisted of 30 rounds of denaturation at
94C for 45 sec, annealing of primer to the template at 60C for 45 sec. and
chain elongation at 72C for 1 min. The reaction was stopped by addition of I()~)
1 TE, and the probe recovered over a 3cc G-50 spin column (2 ml G-50
Sephadex [Pharmacia, Uppsala, Sweden] in a 3cc syrin~e plugged with glass
wool. equilibrated with TE) and counted on a 1500 TriCarb Liquid Scintillation
Counter (Packard, Downers Grove, L). The probe was added to the
prehybridizin~ buffer at 10~ cpm/ml and hybridization was canied out at 6()C
~; r 16 hrs. The blot was washed in high stringency conditions: 3x15 min at
65C with 0.2xSSCtl~ SDS, followed by wl~ppin~ in plastic wr~p ~nd
.cposure to film at -80C. A seven hour exposure of this Nolthem blot reveale~
single thick band at approximately 1.2 kb for C. japc nica (United States) (Fi~
WO 94/OlS60 PCr/US93/00139
2~2~9~
19a. lane 1), C. japonica (Japan) ~Fi~. 19a, lane 2), J. sa~inoides (Fi8. 19a. Iane
3) and ~. Yirginiana (Fig.- l9a, lane 4) RNAs. This band is the expected size for
Cry j 1. Jun s I and Jlln v I as predicted by PCR analysis of the cDNA. The
different band intensities in each lane may reflect differences in the amount ofS RNA loaded on the gel. The position ot 1.6 and 1.0 kb molecular weight
standards are shown on the Fi~s. I9a and l9b.
RNA isolated from ~1. sabinoides and C. ari~onica were analyzed in a
separate Northern blot. Five ,ug of total RNA from 3. sabinoides and 5 ~ of
total RNA from C. arizonica were probed as described. The 1.2 kb band was
observed in this blot for both J. sabinoides (Fi~ure l9b, lane 1) and C. ari70nica
(Figure l9b~ lane 2), indicating that C. ari~onica has a C~y j I homolo~ue.
Other, related, trees are also expected to have a C~y j I homologue.
Although this invention has been described with reference to its preferred
embodiments, other embodiments can achieve the s~ne results. Variations and .
modifications to the present invention will be obvious to those skilled in the art
and it is intended to cover in the appended claims all such modifications and
equlvalents that follow in the true spirit and scope of the invention.
WO 94~01~60 2 1 2 0 9 ~ ~ PCr/US93/00139
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: Grif~lth. lrwin J.
Pollock, Joanne. Bond Julian
(ii) TITLE OF INVENTION: Allergenic Proteins And Peptides From
Japanese Cedar Pollen
(iii) NUMBER OF SEQUENCES: 25
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: ImmuLogic Pharmaceutical Corporation
- ~B) STREET: 610 Lincoln Street
(C) CITY: Waltham
(D) STATE: MA
(E) COUNTRY: USA
~o (F) ZIP: 02154
(v~ COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERAllNG SYSTEM: PC-DOStMS-DOS
(D) SOFTWARE: PatentIn Release #l.0, Version #1.25
~i) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE:
: (C) CLASSIFICATION:
iii) ATTORNEY/AGENT INFORMATION:
(A) NAME: Stacey L. Channin~
: 35 (B) REGISTRATION NUMBER: 31.095
(C) REFERENCE/DOCKET NUMBER: IPC~ 5CCC PCT
) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: (617) 466-fi()0()
(B) TELEFAX: ~617) 466-604()
7~
WO 94/01560 PCr/US93/00139
21209~ `
(2) INFORMATION FOR SEQ ID NO~
(i) SEQUENCE CHARACTERISTICS:
S (A) LENGTH: 1337 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA to mRNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Crytpomeria japonica
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: 66..1187
.
(ix) FEATURE:
(A) NAME/KEY: mat_peptide
: (B) LOCATION: 129..1187
(xi~ SEQUENCE DESCRIPTION: SEQ ID NO:l:
.GTCAATCTG CTCATAATCA TAGCATAGCC GTATAG~iAG AAATTCTACA CTCTGCTACC 6 0
.``~iA ATG GAT TCC CCT TGC TTA GTA GCA TT.. CTG GTT TTC TCT TTT 107
~- ~ Met Asp Ser Pro Cys Leu Val Ala Leu Leu Val Phe Ser Phe
-2 1 -20~ -15 -10
GT.`. ,.TT GGA TCT TGC m TCT GAT AAT CCC ATA GAC AGC TGC TGG AGA 15 5
- Val Ile Gly Ser Cys Phe Ser Asp Asn Pro Ile Asp Ser Cys Trp Arg
-5 1 S
~ ~ ~
- C-^-.-. GAC TCA AAC TG G GCC CAA AAT AGA ATG .'- ~G CTC GCA GAT TGT GCA 2 0 3
G:. Asp Ser Asn Trp Ala Gln Asn Arg Met Lys Leu Ala Asp Cys .3,la
lS 20 25
40: ~ C, GGC TTC GGA AGC TCC ACC ATG GGA GGC .'- `.G GGA GGA GAT CTT TAT 2 5 1
2' Gly Phe Gly Ser Ser Thr Met Gly Gly L ~s Gly Gly Asp Leu Tyr
30 3S 40
- ~ `._ - GTC ACG AAC TCA ,GAT GAC GAC CCT GTG .`- `.T CCT GCA CCA GGA P,CT 2 ~ c
4~ ~.r Val Thr Asn Ser Asp Asp Asp Pro Val .isn Pro Ala Pro Gly Thr
45 S0 SS
..
CG' TAT GGA GCA ACC CGA GAT AGG CCC _TG TGG ATA ATT TTC AGT 3 4,
.3.rg Tyr Gly Ala Thr Arg Asp Arg Pro Leu Trp Ile Ile Phe Ser
::
, . ~
"', ~
~ ~ 76
WO 94/01560 2 1 2 0 9 5 ~ PCI/US93/00139
GGG AAT ATG AAT ATA AAG CTC AAA ATG CCT ATG TAC ATT GCT GGG TAT 395
Gly Asn Met Asn Ile Lys Leu Lys Mee Pro Met Tyr Ile Ala Gly q~r
5 75 80 85
AAG ACT TTT GAT GGC AGG GGA GCA CAA GTT TAT ATT GGC AAT GGC GGT 443
Lys Thr Phe Asp Gly Arg Gly Ala Gln Val Tyr Ile Gly Asn Gly Gly
90 95 100 105 `''
CCC TGT GTG TTT ATC AAG AGA GTT AGC AAT GTT ATC ATA CAC GGT TTG 491
Pro Cys Val Phe Ile Lys Arg Val Ser Asn Val Ile Ile His Gly Leu
110 115 120
TAT CTG TAC GGC TGT AGT ACT AGT GTT TTG GGG AAT GTT TTG ATA AAC 539
r Leu Tyr Gly Cys Ser Thr Ser Val Leu Gly Asn Val Leu Ile Asn
125 130 135
GAG AGT TTT GGG GTG GAG CCT GTT CAT CCT CAG GAT GGC GAT GCT CTT 58
Glu Ser Phe Gly Val Glu Pro Val His Pro Gln Asp Gly Asp Ala Leu
140 145 150
ACT CTG CGC ACT GCT ACA AAT ATT TGG ATT GAT CAT AAT TCT TTC TCC 635
Thr Leu Arg Thr Ala Thr Asn Ile Trp Ile Asp His Asn Ser Phe Ser
155 160 165
T TCT TCT GAT GGT CTG GTC GAT GTC ACT CTT ACT TCG ACT GGA GTT o 8 3
sn Ser Ser Asp Gly Leu Val Asp Val Thr Leu Thr Ser Thr Gly Val
170 ~ 175 180 185
CT ATT TCA~AAC AAT CTT TTT TTC AAC CAT CAT AAA GTG ATG TTG TTA ~81
;Thr~Ile~ Ser~Asn~Asn Leu Phe Phe Asn His His Lys Val Met Leu Leu
190 195 200
35~ GGG~CAT GAT GAT GCA TAT AGT GAT GAC AAA TCC ATG AAG GTG ACA GTG 779
Gly His Asp Asp Ala Tyr Ser Asp Asp Lys Ser Met Lys Val Thr Val
205 210 215
GCG TTC AAT CAA TTT GGA CCT AAC TGT GGA CAA AGA ATG CCC AGG GCA 82
.'.la Phe Asn Gln Phe Gly Pro Asn Cys Gly Gln Arg Met Pro Arg Ala
220 225 230
;CG.~ TAT GG~ CTT GTA CAT GTT GCA AAC AAT AAT TAT GAC CCA TGG ACT 8~5
A-~Tyr~Gly Leu Val His Val Ala Asn Asn ~sn Tyr Asp Pro Trp Thr
~235~ 240 ~ 2g5
'`. TAT GCA~ATT GGT GGG AGT TCA A~T CCA .. CC ATT CTA AGT G.'A GGG 92
l8 Tvr Ala Ile Gly GIy Ser Ser Asn Pro Thr Ile Leu Ser Glu Gly
255 260 265
.:'.T AGT TTC ACT GCA CCA AAT GAG AGC TAC .~G AAG CAA GTA ACC AT.; ^,1
;i `.'.sn Ser Phe Thr Ala Pro Asn Glu Ser Tfr Lj~s Lys Gln Val Thr Ile
270 275 280
~55_GT .3.TT GGA TGC AAA ACA TCA TCA TCT TGT T^A A~T TGG GTG TG~ CA~ 1~19
Ile Gly Cys Lys Thr Ser Ser Ser C~s Ser Asn Trp Val Trp Gln
285 290 295
. . ,
:~ ~
~ ~ 77
:: ~
WO 94/01560 PCr/US93/00139
~20954
TCT ACA CAA GAT GTT TTT TAT AAT GGA GCT TAT TTT GTA TCA TCA GGG 1067
Ser Thr Gln Asp Val Phe Tyr Asn Gly Ala Tyr Phe Val Ser ser Gly
300 305 310 ,
A~A TAT GAA GGG GGT AAT ATA TAC ACA AAG A~A GAA GCT TTC AAT GTT 1115
Lys Tyr Glu Gly Gly Asn Ile Tyr Thr Lys Lys Glu Ala Phe Asn Val
315 320 325
GAG AAT GGG AAT GCA ACT CCT CAA TTG ACA ~A AAT GCT GGG GTT TTA 1163
Glu Asn Gly Asn Ala Thr Pro Gln Leu Thr Lys Asn Ala Gly Val Leu
; 330 335 340 345
ACA TGC TCT CTC TCT AAA CGT TGT TGATGATGCA TATATTCTAG CATGTTGTAC 1217
Thr Cys Ser Leu SeS0r Lys Arg Cys
TATCTAA~TT AACATCAACA AGAAAATATA TCATGATGTA TATTGTTGTA TTGATGTCAA 1277
.3TAAAAATG TATCTTTTAC TATTAAAAAA .~AAAATGATC GATCGGACGG TACCTCTAGA 1337
- 20 ~ ~ ~
2~ INFORMATION FOR SEQ ID NO:2:
25 ~ SEQUENCE CHARACTERISTICS:
- ~A) LENGTH-: 374 amino acids
(B~ TYPE: amino acid
D) TOPOLOGY: linear
30~ MOLECULE TYPE: protein
(xi) SEQUENCE~DESCRIPTION: SEQ ID NO:2:
Asp Ser Pro~Cys Leu Yai Ala Leu Leu Val Phe Ser Phe Val Ile
G!. Ser~Cys Phe Ser Asp~Asn Pro Ile Asp Ser Cys Trp Arg Gly Asp
40~ S-- Asn Trp~Ala Gln Asn Arg Met Lys Leu Ala Asp Cys Ala Val Gly
5~ 20 25
P.~.e Gly~Ser~5er Thr Met Gly Gly Lys Gly Gly Asp Leu Tyr Thr Val
30~ ~ 35 40
sn S~er Asp Asp Asp Pro Val Asn Pro .`.la Pro Gly Thr Leu Arg
45~ 50 55
Gly ~3.1a~Thr~Arg Asp Arg Pro Leu Trp Ile Ile Phe Ser Glv .~sn
-:~ 65 70 /5
- M- .isn Ile Lys Leu Lys Met Pro Met Tyr Ile Ala Gly Tyr Lys Thr
55~ ~~-.- .3Sp Gly Arg Gly Ala Gln ~al T~r Ile Gly ~sn Gly Gly Pro Cys
gs lOO lO5 !~`;
~ Phe Ile Lys Arg Val Ser Asn Val Ile Ile His Gly Leu Tyr Leu
:. :
.
~ ~ 7~;
~': ': ,
-:
;~
WO94/01560 212 0 ~ 5 ~ PCT/US93/00139
110 115 120
Tyr Gly Cys Ser Thr Ser Val Leu Gly Asn Val Leu Ile Asn Glu Ser
125 130 135
Phe Gly Val Glu Pro Val His Pro Gln Asp Gly Asp Ala Leu Thr Leu
140 145 150 155
Arg Thr Ala Thr Asn Ile Trp Ile Asp His Asn Ser Phe Ser Asn Ser
0160 ` 165 170
::
:~ ~ Ser Asp Gly Leu Val Asp Val Thr Leu Thr Ser Thr Gly Val Thr Ile
~: 175 180 185
15Ser Asn Asn Leu Phe Phe Asn His His Lys Val Met Leu Leu Gly His
: ~ 190 195 200
Asp Asp Ala Tyr Ser Asp Asp Lys Ser Met Lys Val Thr Val Ala Phe
`~ : 205 210 215
~: Asn Gln Phe Gly Pro Asn Cys Gly Gln Arg Met Pro Arg Ala Arg Tyr
` 22a ~ : 225 230 235
~; Gly Leu Val His Val Ala Asn Asn Asn Tyr Asp Pro Trp Thr Ile Tyr
25`240 245 250
3.1a Ile Gly Gly Ser Ser Asn Pro Thr Ile Leu Ser Glu Gly Asn Ser
255 260 : 265
30 ~Phe Thr Ala Pro Asn Glu Ser Tyr Lys Lys Gln Val Thr Ile Arg Ile
270~ 275 280
Glv ;~ys Lys Thr Ser Ser Ser Cys Ser Asn Trp Val Trp Gln Ser Thr
G_r ~sp~ V~l Phe ~Yr Asn Gly Ala Tyr Phe Val Ser Ser Gly Lys Tyr
30`~ 305 310 315
G' ~ ly Gly Asn Ile- Tyr Thr Lys Lvs Glu Ala Phe Asn Val Glu Asn
20~ ~ ' 325: 330 ~` `
: G~ ..sn Ala Thr Pro Gln Leu Thr Lys Asn:Ala Gly Val Leu Thr Cys
;335. 340 345
~ _r _eu~Ser Lys Arg Cys:
! ` INFO~MATION ~VOR SEQ ID NO:3:
~50 :(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17 base pairs
::(B) TYPE: nucleic acid
C) STRANDEDNESS: single
(D) TOPOLOGY: linear
, :: ~
~ 7~
.
.
- :-
~ ::
WO94/01560 PCT/US93/00139
2120954
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:3:
GAYAAYCCNA THGAYWS
17
(2) INFORMATION FOR SEQ ID NO:4: :
(i) SEQUENCE CHARACTERISTICS: ~`
: l0 (A) LENGTH: 25 base pairs
-(B) TYPE: nucleic acid
: (C) STRANDEDNESS: single
(D) TOPOLOGY: linear
~ -
l:5
xi) SEQUENCE DESCRIPTION: SEQ ID NO:4:
~ GGG.~ATTCAA~YTGGGCNCAR~AAYSG
-~ 20 25 :
: - (2) INFORMATION FOR SEQ ID NO:5:
SEQUENCE CHARACTERISTICS:
25~ (A) LENGTH: 23 base pairs
(3) TYPE: nucleic acid
(C)~STRANDEDNESS: single
:D~TOPOLOGY: linear
(ix:)~:FEATURE:
(A) NAME/KEY: modified_base
(:B) LOCATION::: 15
D)~-OTHER~:INFORMATION: /mod_base= i `-~
(Xl~ SEQUENCE DESCRIPTION: SEQ ID NO:5:
-GCAGCCRT~TYTCNACRTT RAA
: ') INFORMATION FOR~SEQ ID NO:6:
: ::
SEQUENCE CHARACTERISTICS:
.: 4~ :~ (A) LENGTH: 20 base pairs
: (B) TYPE: ~ucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
;" 80
...,.., :~ ~
WO94/01560 2 1 2 0 9 .~ ,~ PCT/US93/00139
(ix) FEA~TURE:
(A) NAME/KEY: modified_base
(B) LOCATION: 6
(D) OTHER INFORMATION: /mod_base= i
.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:6:
TTCATNCKRT TYTGNGCCCA
~) INFORMATION FOR SEQ ID NO:7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 25 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
~ ~D) TOPOLOGY: linear
(Xl) SEQUENCE DESCRIPTION: SEQ ID NO:7:
CCTGCAGCKR TTYTGNGCCC AARTT
2~
(~) INFQR~TION FOR SEQ ID NO:8:
~i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 18 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:8:
~ GATTCCC CTTGCTTA
(^ INFORMATION FOR SEQ ID NO:9: '.
( i ) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26 base ir~
`(B) TYPE: nucleic ac
(C) STRANDEDNESS: si~A~,le
(D) TOPOLOGY: linear
Xl
~ .
WO94/01560 PCT/US93/~013~
21~l~95~
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:9:
GGGAATTCGA TAATCCCATA GACAGC
26
(2) INFORMATION FOR SEQ ID NO:10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17 base pairs
~B) TYPE: nucleic acid --
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:10:
ATGCCTATGT ACATTGC
17
(2~ INFORMATION FOR SEQ ID NO:11:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
~D) TOPOLOGY: linear
.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:11:
C~ TGTACA TAGGCAT
_,
. ~
'_! INFORMATION FOR SEQ ID NO:12:
( ) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 18 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
~.~
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:12:
~'
~ 3~
W~94/~1560 P~T/US93/00139
TCCAATTCTT CTGATGGT
18
(2) INFORMATION FOR SEQ ID NO:13:
(i) SEQUENCE CHARACTERISTICS.
tA) LENGTH: 18 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:13:
TTTTC-TCAAT TGAGGAGT
18
(2) INFORMATION FOR SEQ ID NO:14:
.
(i) SEQUENCE C~ARACTERISTICS:
(A) LENGTH: 30 base ~ rs
: (B) TYPE: nucleic aci
(C) STRANDEDNESS: singLe
(D) TOPOLOGY: linear
.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:14:
CCTGCAGAAG CTTCATCAAC AACGTTTAGA
3C
~ 35 (2) INFORMATION FOR SEQ ID NO:15:
: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
~ Xl ) SEQUENCE DESCRIPTION: SEQ ID NO:15:
T-.^-CP~CTCC AGTCGAAGT
1 . ,
'
WO94/01560 PCT/US93/00139
(2) INFORMATION FOR SEQ ID NO:16:
- (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17 base pairs
(Ei) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:16:
TAGCTCTCAT TTGGTGC
17
(2) INFORMATION FOR SEQ ID NO:17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 18 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
2S
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:17:
TGCAATTG GTGGGAGT
1 ~i
- ~ 30
) INFORMATION FOR SEQ ID NO:18:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 amino acids
(B) TYPE: amino acid
~; (D) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
(v) FRAGMENT TYPE: N-terminal
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Cryptomeria japonica
(i~) FEATURE:
(A~ NAME/KEY: Modified-site
(B) LOCATION: 7
(D) OTHER INFOR~TION: inote= l~the amino acid a.
~-sltion
81
WO94/01560 ~ PCT/US93/00139
7 is Ser, Cys, Thr, or His~
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:18:
Asp Asn Pro Ile Asp Ser Xaa Trp Arg Gly Asp Ser Asn Trp
Ala Gln
l 5 l0
Asn Arg Met Lys
(2) INFORMATION FOR SEQ ID NO:l9:
~i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 16 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
(v) FRAGMENT TYPE: internal
25 (vi) ORIGINAL SOURCE:
(A) ORGANISM: Cryptomeria japonica
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:l9:
- :Glu Ala Phe Asn Val Glu Asn Gly Asn Ala Thr Pro Gln Leu
Thr Lys
l 5 l0
(2) INFORMATION FOR SEQ ID NO:20:
- (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:20: .
GGC-TCTAGAG GTACCGTCCG ATCGATCATT
WO94/01560 PCT/US93/00139
: .
2120~
(2) INFORMATION FOR SEQ ID NO:21:
(i) SEQUENCE CHAR-ACTERISTICS:
~A) LENGTH: 20 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:21:
GGGTCTAGAG GTACCGTCCG
(2) INFORMATION FOR SEQ ID NO:22:
(i) SEQUENCE CHARACTERISTICS:
tA) LENGTH: 13 base pairs -
(B) TYPE: nucleic acid
(C) STRANDEDNESS. single
(D) TOPOLOGY: linear
2~
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:22:
30 ~TGATCGAT GCT
13
(~) _NFORMATION ~OR SEQ ID NO:23:
( i ) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
~xi) SEQUENCE DESCRIPTION: SEQ ID NO:23:
45 GG:-.TTCTCT AGACTGCAGG T
2: .
(~. _NFORMATION FOR SEQ ID NO:24:
WO94/01560 2 1 2 0 ~ ~ 4 PCT/US93/00139
ti) SEQUENCE CHARACTERISTICS:- j
~A) LENGTH. 35 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:24:
GGAATTCTCT AGACTGCAGG TTTTTTTTTT TTTTT 35
(2) INFORMATION FOR SEQ ID NO:25:
li) SEQUENCE CHARACTERISTICS:
~A) LENGTH: 5 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
(v) FRAGMENT TYPE: N-terminal
- (vi) ORIGINAL SOURCE:
(A) ORGANISM: Juniperus sabinoides
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:25:
Asp Asn Pro Ile Asp
l 5
~7