Note: Descriptions are shown in the official language in which they were submitted.
~,.,1 e~ ~~ ~ ~. ~t
WO 93!20216 PCT/CA93/00141
OII~IiODY PROTEIN CIS-ELEMENTS
AS REGULATORY SIGNALS
INTRODUCTION
Technical ~'~eld_
This invention relates to upstream DNA sequences and their use to control
expression of genes in developing plant seeds and their use.
Studies in plant gene expression have yielded a number of general
conclusions concerning the elements that control expression. PDants, like
other
organisms both prokaryotic and eukaryotic, contain conserved or consensus
sequences upstream (5') of the transcriptional start site of genes which
appear
capable of regulating transcriptional rates. In eukaryotes, these sequences
include
a motif found typically about 25 by 5' to the transcriptions! initiation site
which
has the, sequence TATAAITAA/T and is referred to as a TATA box. The role of
this TATA box appears to be to define the transcriptions! start for RNA
polymerase II. A second upstream sequence is referred to as a CART box.
Typically, this is found about.75 bases upstream of the transcriptions! start
and is
associated with regulating the frequency of transcriptions! initiation. In
plants the
consensus sequence may be either CCAAT or sometimes AGGA. However,
neither of these alternative consensus sequences need be present in all plant
genes.
These sequence motifs and their DNA context within 70-90 bases upstream of the
transcriptions! start are often referred to as promoters. In general, 5' of
the
promoter region and most frequently within 200 bases of it are cis-acting
elements which confer a variety of properties on the promoter and which can
modulate transcrip6onal activity in either a constitutive or a non-
constitutive
manner. These cis-acting sequences may be referred to as enhancers (if they
are
responsible for increases in transcription) or silencers (if they are
responsible for
~t~BS'Ti'T13TE S~~ET
'~1~~~1'l
WO-93/20216 PCT/CA93/00141
2
decreases or suppression of transcription). Enhancers and silencers are
frequently
the sites at which nuclear proteins bind or interact. 'The modulating nuclear
proteins are called traps-acting factors. They are considered to be very
important
for non-constitutive or regulated expression as they may be the major
determinant
of the activity of a gene in a particular tissue or organ or in response to an
euternal stimulus. The relationship between this protein binding and the
enhancer/silencer element may determine the transcriptional activity. The
isolation
of genes which are activated by heat, light or chemicals such as endogenous
hormones or are activated in specific organs such as seeds, leaves or flowers
has
permitted analysis of factors which may determine how expression is regulated.
In
numerous, but not all, cases, it has been shown that the construction of
chimeric
genes which contain the promoter and optionally cis-elements from a given
regulated gee and a coding sequence of a reporter protein not normally
associated
with that promoter gives rise to regulated expression of the reporter. The use
of
promoters firom seed-specific genes for the expression of sequences in seed of
gages that are either not normally expressed in a seed-specific manner or
those
that require an altered pattern of expression has been attempted on only a few
occasions. In all cases to date, chimeric genes designed for seed-specific
~. ~,e used seed-storage protein regulatory signals and promoters.
However, it is evident from work on storage protein gene expression that
expression commences at a fairly late stage in embryogenesis, namely once the
embryo has reached (in the case of divots) the classical torpedo shape. Thus,
although storage proteins express at high levels and their regulation is often
transcriptional, the timing and level of expression may not be ideal for all
seed-
specific applications. It is, therefore, of interest to identify other seed-
specific
promoters and enhancers with temporal or cellular specificity different from
that of
seed storage proteins, such as those from oleosins.
3(f The following disclose organ or tissue specific regulatory sequences
used to produce tissue or organ-specific expression in transformed plants.
There
are several by now "classical" examples of regulated gene expression in non-
seed
I ~~.~~i ~~1'~
WO X93/30216 ~ PCT/CA93/00141
3
protein chloramphenicol acetyl transferase could be expressed in a light-
regulated
and organ-specific manner in transgenic plants if the coding sequence for the
reporter protein was fused with the promoter and upstream sequences from a pea
gene encoding ribulose bisphosphate carboxylase (Fluhr, Science (1986),
x:1106-1112).
Sengupta-Gopaian et al. Proc. Natl. Acad. Sci. USA, (1985)
$x:3320-3324 reportod e~cpression of a major storage protein of french beans,
called B-phaseolin, in tobacco plants. The gene expressed correctly in the
seeds
and only at very low levels elsewhere in the plant. However, the constructs
used
by Sengupta-Gopalan were not chimeric. The entire &phaseolin gene including
the native 5'-flanking sequences were used. Subsequent experiments with other
species (Radke et al. (1988) Theor. App. Genet. 75:685-694) or other genes
(Perez-Grau, L., Goldberg, R.B., 1989, Plant Cell, ,x:1095-1109) showed the
fidelity of expression in a seed-specific manner in both Arabidopsis and
Brassica.
Itadke et al. (1988), vide supra, used a "tagged" gene i.e., one containing
the
entire napin gene plus a non-translated "tag".
In tissue and organ specific expression there have been several
examples showing that sequences upstream of the transcriptional start may be
used
to confer tissuelorgan specificity to a gene introduced into plants by genetic
engineering. Examples include engineering seed-specific gene regulation (Radke
a
et al. (1988) vide supra; Bustos et al. (1989), Plant Cell, x:839-853). In
both
examples, sequences upstream of the coding sequ~ces of seed proteins were
linked to a reporter tag (either as RNA or protein) and seed specificity was
conferred on expression of the reporter. These were all storage protein genes
rather than oleosins. Seed storage proteins have different temporal expression
patterns from oleosins.
The DNA motifs that might give rise to seed-specific expression are
now the subject of many studies. Marcotte et al. (Marcotte, W.R., Russel, LS.,
Quantrano, R.S., 1989, Plant Cell, x,:969-97~ studied the Em gene of wheat and
proposed two motifs called "Em-boxes" ~rhich might be consensus sequences for
seed-specific expression. Interestingly, one of these boxes called EM-2 is
similar
to that found in other storage protein genes from monocots (triticin-wheat)
and
PGT/CA93/00141
WO.93/20216
4
even divots (B-conglycinin-soybean). Hatzopoulos et al. (1990, Plant Cell,
2_:457-
467) investigated the sequences directing embryo-specific expression of a
carrot
lipid-body protein gene. A number of AT rich motifs were identified, being
protected from digestion during DNAse treatment presumably by trans-acting
proteins. The motifs identified, however, were not shown to be consensus
motifs
for other seed-specific genes.
DeClercq et al. Plant Physiol., (1990), 24:970-979 used the
promoter of the Arabidopsis 2S albumin and combined coding sequences from both
the Arabidopsis and Brazil nut 2S albumins. Fusions were made in regions
showing low conserdation. Transformation of both tobacco and Brassica napes
y~_c ~pression and correct accumulation of the modified storage
proteins. Levels of expression were between 0.05 % and 0.3 % of total cellular
protein.
Another example of this form of seed-specific expression of foreign
sequences was the expression of lee-enkephalins in seeds. To obtain seed
specific
expression, a chimeric DNA sequence encoding a 2S albumin and a short
oligonucleotide encoding leuphalin (a pentapeptide) was included in the
albumin coding sequences between the 6th and 7th cysteines of the native
protein
(yanderkerhove et al. Bio/Technology, (1989) x:929-932). Again this gene
e~cpressed in a seed-specific manner allo~g ~e accumulation of up to 50 nmol
lee-enlcaphalin per g of seed.
Genomic clones encoding oil-body proteins with their associated
upstream regions have been reported for two species, maize (Zea ways, Bowman-
yance and Huang, (1987) J. Biol. Chem., X2_:11275-11279; and Qu and Huang,
(1990) J. Biol. Chem., x:2238-2243) and carrot (Hatzopoulos et al. (1990)
Plant
Cell, x:457-467). cDNAs and genomic clones have also been reported for one
cultivated oilseed, Brassaca napes (Murphy, et al. (1991), Biochem. Biophys.
Acts, .1$$:86-94; and Lee and Huang (1991) Plant Physico 96:1395-1397.) .
Reports on the expression of these oil-body protein genes in developing seeds
have
3Q varied. In the case of Zea mays, transcription of genes encoding oil-body
protein
isoforms began quite early in seed development and were easily detected 18
days
after pollination. Tn non-endospermic seeds such as the dicotyledonous plant
WO 93/20216 ~ ~ ~ ~ ~ ~ ~ PCT/CA93/00141
after pollination. In non-endospermic seeds such as the dicotyledonous plant
Brassica napes (Canola), expression of oil-body protein genes seems to occur
much later in seed development (Murphy, et al. (1989), Biochem. J., ~$:285-
293) than with corn.
5
Methods and compositions are described for the exploitation of an
oil-body protein transcriptional regulatory sequence and optionally its
accompanying 5' untranslated leader sequence for the expression of
heterologous
genes in a seed-specific manner. The method includes the steps of transforming
a
plant cell with a DNA construct comprising the regulatory sequence and a DNA
sequence other than the open reading frame native to the regulatory sequence,
generating a plant from the transformed cell and growing it under conditions
whereby seed is produced and the DNA sequence is expressed under the
transcriptional control of the regulatory region. These sequences will be
valuable
in applications where expression of a seed-borne product neais to be modified,
enhanced or suppressed. They could also be used to produce modified seeds
containing foreign proteins to increase the intrinsic value of the seed.
BRIEF DESCRIPTION OF THE DRAWINGS
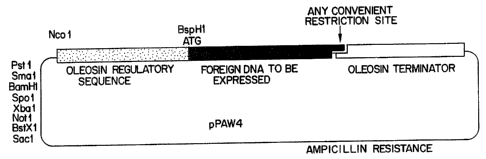
Fig. 1 shows a schematic diagram of vector pPAW4 enclosing an
oleosin regulatory sequence, an initiation colon, foreign DNA to be expressed,
an
oleosin terminator s~uence and an ampicillin resistance gene.
Fig. 2A shows the DNA sequence of an Arabifopsis genomic clone
encoding a l8KDa oil-body protein. The open reading frame is interrupted by a
short intron (which is marked) and the two axons are translated and indicated
in
IUPAC single letter amino-acid codes.
Fig. 2B shows the restriction fragment from an Agrobacterium
EMBL3 genomic library which encloses the Arabidopsis l8KDa oil-body protein
coding sequence. The approximate position of the coding region is highlighted.
Fig. 3 shows the effects of lO~cM ABA on the developmental
expression of oleosin mRNA using a Northern blot analysis of total RNA. A)
W4 93/Z0216 ' PCT/CA93/00141
6
(?O~eg per lane) using 50 ng32P dCTP labelled OB990 as a probe (spec. act. 109
dpm/~cg DNA). Heart-(H): (13-day), torpedo-('I~ (17-day), and cotyledonary-(C)
(21-25 day) stage microsponr-derived embryos with (+) and without (-)
treatment
for 48 h with 10~,M ABA. The blot was exposed to Kodak XARS film at
70°C
for 20 minutes. The apparent size difference of the mRNAs in the different
lanes
is due to interfering quantities of starch in the different mRNA preparations.
All
tlu lanes wart equally loaded as judged by OD260 measurements and EtBr-
staining. B) A 4.5 hour exposure of Fig. 3-A C) Relative intensity of the mRNA
accumulation as determined by scanning densitometry.
Fig. 4 shows the tissue specificity of oleosin. 50 ~cg of poly (A)+
RNA of mots (R), callus (Ca), Cotyledons (Co), leaves (L), and 24-day post-
anthesis zygotic embryos (E) was probed with 50 ng of 3zP dCTP labelled OB990
(spec. act. 1f' dpml~cg DNA).
Fig. 5 shows tl~ developmental sensitivity of oil body protein
synthesis to applied ABA. An estimated 10,000 dpm were loaded per well for
paged samples of controls (lanes A,C,E,G) and ABA treated (lanes B,D,F,H).
All samples were for 2d with ABA, the labeled for 4 h with 1.85
MBq/mL ~S]methionine. Lanes A and B, 10-d-old cultures, sieved on 62~cm
s to obtain globular embryos. Lanes C and D, 13-d-old ~culturPs sieved on
125~m screens to obtain heart stage embryos. Lanes E and F, 17-d-old cultures
sieved on 250~cm screens to obtain torpedo to early cotyledonary embryos.
Lanes
'G and H, 25-d-old cultures, sieved on 500~cm screens to obtain cotyledonary
stage
embryos.
~JfEF DESCRIPTION OF THE SPECIFIC EMBODIMENTS
In accordance with the subject invention, DNA constructs are
provided which allow for modulation of plant phenotype in seed, particularly
during early phases of embryogenesis. The DNA constructs provide for
regulation
of transcription in sped; using 5' untranslated sequences from genes active
from
the late globular stage through to embryo maturity (cotyledonary stage).
Dawhstream from and under t<anscriptional initiation-regulation of an oil body
protein gene initiation region will be a DNA sequence of interest which will
v ,',. v.~ ~ )
f.r..~c)J~~,~
WO 93/20216 PCT/CA93/00141
7
be prepared which allow for integration of the transcription cassette into the
genome of a plant cell. Conveniently, a multiple cloning site downstream from
the seed specific transcriptional initiation region may be included so that
the
integration construct may be employed for a variety of DNA sequences in an
S efficient manner.
Of particular interest is a regulatory sequence from an oil body
protein gene, preferably an oil body protein gene expressed in dicotyledonous
oil
seeds. It has been reported that oil-body proteins accumulate considerably
later
than either oils (triacylglycerides) or storage proteins. This later
expression would
limit the value of any promoters associated with these genes for seed-specific
a~pressio~n as they could not be used for modification of expression of genes
during early phases of embryogenesis. Surprisingly, however, expression of
these
genes in dicotyledonous oilseeds was found to occur much earlier than had
hitherto
been believed. Thus, the promoters and upstream elements of these genes are
valuable for a variety of uses invol ring the modification of metabolism
during
phases of embryogwhich precede the accumulation of stoiage proteins.
Oil~ody proteins have been identified in a wide range of
ta~wnomically diverse spocies (see, for example, Moreau et al: Plant Physiol.
(1980), ~:1I76-1180; Qu ~ al. Biochem. J:, (1986) x:57-6~). These proteins
are uniquely localized in oil-bodies and are not found in organelles of
vegetative
f
tissues. In Brassica »apus (rapeseed) there are at least three polypeptides
associated with the oil-bodies of developing seeds (Taylor et al. (1990),
Planter,
1$x,:18-26). The numbers and sizes of oil-body associated proteins may vary
from
species to species. In corn, for example, there are four immunologically
distinct
~ polypeptides found in oil-bodies (Bowman-Vance and liuang, 1988, J. Biol.
Chem., ~:1476-1481). Oleosins have been shown to comprise regions of
alternate hydrophilicity, hydrophobicity and hydrophilicity (Bowman-Vance and
Huang, 1987, J. Biol. Chem., X2_:11275-11279). The amino acid sequences of
oleosins from corn, rid and carrot have been obtained. See Qu and Huang,
1990, J. Biol. Cfum., x:2238-2243; Hatzopoulos ex al. 1990, Plant Cell; ~:457-
467; respectively. In an oilseed such as rapeseed, oleosin may comprise
between
896 (Taylor et al. 1990, Planter, x$1":18-26) and 20% (Murphy et al. 1989,
W093/20216 ~ ~ ~ ~ ~ ~ ' PCT/CA93/OOldl
8
Biochem. 1., ?x$:285-293) of total seed protein. Such a level is comparable to
that found for many seed storage proteins.
Of particular interest is a transcriptional initiation region associated
with early embryogenesis, particularly the period preceding expression of
storage
proteins, so that in the early development of seed, it provides the desired
level of
transcription of the DNA sequence of interest. Normal plant embryogenesis
typically goes through a series of defined phases. For dicotyledonous seeds,
embryogenesis includes the following phases: globular stage, heart stage,
torpedo
stage, and cotyledonary stage. For the purposes of this application, the
definition
of these terms is provided by Ray, Steves, and Fultz in Botany, (Saunders
College
Publishing), Chapter 17, page 294. Normally, the transcriptional initiation
region
will be obtainable from a gene which is expressed in the early formation of
seed.
Desirably the transcriptional initiation region maintains its activity from
the late
globular through cotyledonary stage, more desirably continues active from the
globular stage through the heart, torpedo and cotyledonary stages of
embryog~esis. By obtainable is intended a transcriptional initiation region
having
a nucleotide sequence-sufficie~ntly similar to that of a natural oil body
protein gene
transcriptional initiation region sequence to provide for transcription in the
early
formation of seed. The sequence may be naturally occurring, sjmthetic or
partially
ZO synthetic.
The transcriptional initiation region from the oil body protein
generally will be provided in a which will include in the 5'-3' direction of
transcription, a transcriptional initiation region, a DNA sequence of interest
and a
transcriptional termination region, wherein the transcriptional regulatory
regions
are operably joined and functional in plant cells. One or more introns may
also be
present. After each manipulation, a DNA to be used in the final construct may
be
restricted and operably joined to other DNA to be used in the final construct,
where each of the partial constructs may be cloned in the same or different
plasmids. In a preferred embodiment, a coding sequ~ce with a compatible
restziction site may be ligated at the position con~esponding to colon ~1 of
the oil-
body protein gene. A schematic diagram of this substitution is shown in figure
1.
The recombinant coding sequence may be inserted in such a way that it
completely
<~~~~4i~
WO 93/20216 PCT/CA93/00141
9
replaces the coding sequence of the oil-body protein gene and is thus flanked
at its
3' end by the oil-body protein gene terminator and polyadenylation signal.
Alternatively, polymerise chain reaction amplification may be carried out to
produce DNA fragments containing the transcriptional initiation region
conveniently flanked by restriction sites. The amplified fragments can be
joined to
the casing sequence for a polypeptide of interest, in a transcriptional or
translational fusion, for example, to produce a chimeric gene in which the
coding
sequence of the polypeptide of interest is transcribed under the control of
the
transcription initiation region on the PCR amplified fragment.
The transcriptional initiation region may be native to or homologous
to the host cell, or foreign or heterologous to the host cell. By foreign is
intended
that the transcriptional initiation region is not found in the wild-type host
into
which the construct comprising the transcriptional initiation region is
inserted.
Generally, the regulatory .sequence comprises DNA of up to 1.5 Kb 5' of the
translational start of an oil-body protein gene. lfiis sequence may be
modified at
the position corresponding to the first colon of the desired protein by sito-
directed
mutagenesis (Kunkel TA, 1985, Proc. Natl. Acid. Sci. USA, $x:488-492) or by
introduction of a convenient linker oligonucleotide by ligation if a suitable
resuiction site is found near the N-terminal codan.
In some cases it will be desirable to express the DNA sequence of
interest as a fusion protein, particularly as a fusion protein with the oil
body
protein. The DNA sequence of interest can be insert~l by routine techniques
into
the oil body protein coding sequence, in frdrne with the oil body protein
coding
sequence, such that transcription of the chimeric gene will produce a fusion
protein. The fusion protein will preferably contain the coding region for
amino
acids number 44 through 122 in the Arabidopsis oil body protein as shown an
Figure 2A, or the ~uivalent region from an oil body protein of a skies other
than Ar~abadopsis, to provide for transport to the oil body in cases where
this is
desirable.
In order to isolate oil body protein coding sequences from other
species, at least two approaches may be used. The first is to use the
Arabidopsis
clone described in the Examples as a probe in genomic libraries of other plant
' , ., ,
WO 93/20216 ~' ~ '~ '~ ~ ~ ~ PCT/CA93/00141
species. This clone will hybridize well with oleosin clones from closely
related
species, in particular, essentially all cruciferous plants. For species which
are
evolutionarily divergent from Arabidopsis, for example, solanaceae,
leguminaceae
and all monocotyledons, an alternative method involves the use of an antibody
5 raised against the gene product of an oleosin clone such as the Arabidopsis
clone.
This antibody may be used to scan a seed-derived cDNA expression library, for
example using lambda gtll; Huynh et al. (1985) in cDNA Cloning, Vol. 1, A
Practical Approach, Ed. Grover IRI. Press, pp. 49-78. This approach yields a
cDNA clone of the oleosin for the new species which may then be used to
isolate
10 the genomic clone from a genomic library of that species by standard DNA
hybridization techniques.
The DNA sequence of interest may be any open reading frame
encoding a peptide of interest, for example, an enzyme, or a sequence
complementary to a genomic sequence, where the genomic sequence may be at
least one of an open reading frame, an intron, a nun-coding leader sequence,
or
any other sequence where the complementary sequence will inhibit
transcription,
messenger RNA processing, for example splicing or translation. The DNA
sequence of interest may be synthetic, naturally derived or a combination
thereof.
Depending upon the nature of the DNA sequence of interest, it~may be desirable
to synthesize the sequence with plant preferr~ colons. The plant preferred
a
colons may be determined from the colons of highest frequency in the proteins
expre$sed in the largest amount in the particular plant species of interest.
The DNA sequence of interest may encode any of a variety of
recombinant proteins. Examples of recombinant proteins which might be
expressed by this procedure include anticoagulants, such as Hirudin,
lympholdnes
such as those of the interleukin family, peptide hormones such as
gonadotrophin
releasing hormone, immunologic~l reagents such as multi or single-chain
antibodies and a variety of industrial valuable enzymes such as proteases,
lipases
and polyglucan hydrolases.
3Q The termination region which is employed will be primarily one of
convenience, since the termination regions appear to be relatively
interchangeable.
The termination region may be native with the DNA sequence of interest, or may
~~.~~~~1'l
WO 93/20216 PCT/CA93/00141
11
be derived from another source. Convenient termination regions are available
and
include the 3' end of the oil body protein gene terminator and polyadenylation
signal from the same gene from which the 5' regulatory region is obtained.
Alternatively, a different terminator and polyadenylation signal may be
employed
with similar results, for example, the terminator of the nopaline synthase
gene of
~lgrobacterium.
The expression cassette may additionally contain a means for
identifying transformed cells and/or selecting for transformed cells. For
example
the recombinant gene may be linked with a constitutively expressed selectable
marlxr such as a gene for antibiotic resistance or herbicide resistance or a
screarable marker, such as a gene conferring bioluminescence or colored
properties to transformed cells.
The DNA sequence of interest flanked at its 5' end by the oil-body
protein promoter and regulatory sequences and at its 3' end by a terminator
may
be introduced into a suitable transformation vector including Agrobacterium Ti
or
binary plasmids, or a simple cloning plasmid (e.g., pUCl9, pBR322) for use in
diroct DNA uptake to plant cells via microinjection, electroporation, PEG-
mediated uptake or a biolistic method. These methods are well known to those
stilled in the art of plant transformation. See, for example, I~oisch et al.
(1985),
ZO Science, 227:1229-1231; Newhaus and Spangenberg (1990). Physiol. Plant,
79:213 217; and Sandford et al. (1990), Physiol. Plant, 79:206-209.
Transforms plants may be obtained from the transformed cells
using standard regeneration protocols (see for example: Moloney et al. (1989),
Plant Cell Rep., $:238-242) compatible with the transfornation method.
The expression cassette, constructed as described above, expresses
essentially preferentially in developing seeds. The plant cells which have
been
transformed with an appropriate fusion peptide therefore are grown into plants
in
accordance with conv~tional ways and allowed to set seed. See, for example,
McCormick et al., Plant Cell Rep. (1986) 5:81-84. Two or more generations may
be groom and either pollinated with the same transformed strain or different
strains, identifying the resulting hybrid having the desired phenotypic
characteristic, to ensure that the subject phenotypic characteristic is stably
CA 02133417 2002-07-15
12
maintained and inherited and then seeds harvested for isolation of the peptide
of
interest or for use to provide seeds with the new phenotypic property. The
regenerated plants are then cultivated identically to non-recombinant plants
in
growth chambers, greenhouses or in the field and will show seed-specific
S expression of the recombinant gene at the ml2NA level and often at the level
of
polypeptide or protein.
It is possible that the polypeptide/protein will itself be valuable and
could be extracted and, if desired, further purified. Alternatively the
polypeptidelprotein or even the mRNA itself may be used to confer a new
biochemical phenotype upon the developing seed. New phenotypes could include
such modiFtcations as altered seed-protein or seed oil composition, enhanced
production of pre-existing desirable products or properties and the reduction
or
even suppression of an undesirable gene product using antisense, ribozyme or
co-suppression technologies (Izant and Weintraub (1984), Cell 36: 1007-1015,
IS antisense; Hazeihoff and Gerlach (1988), Nature 334:585-591, ribozyme;
Napoli,
et al. {1990), Plant Cell, 2:279-289, co-suppression).
If the transformation has been performed to produce a new seed
protein or peptide which requires extraction, this can be done using aqueous
extraction with or without low concentrations of detergents, such as non-
denaturing amounts of sodium dodecyl sulphate (;SDS), TritonTM-X-100, TweenTM
20,
iviEGA-8 or any other detergent known not co irreversibly inactivate the
desired
protein. To extract the protein or polypeptide, dry seeds are ground by hand
or in
a mechanical grinder to produce an aqueous slurry or suspension. This can be
resolved into three phases (particulate, aqueous soluble, and hydrophobic) by
centrifugation, such as at 50,000 x g. Depending upon the nature of the
product,
it may be further purified in each of these phases and after solublization,
may be
selectively precipitated by the use of ammonium sulfate or puriF~ed using
column
chromatography, for example, using ion exchange, gel filtrates or affinity
matrices.
While the ideal host for the regulatory sequence reported here would
be a cruciferous plant, it is possible to use these promoters in a wide
variety of
plant species given the relatively high conservation oleosin of genes. The
major
b,
~r ~.~~~lv
W0.93/20216 PCT/CA93/00141
13
barrier to the use of these promoters is between monocotyledonous and
dicotyledonous species. For transformations involving this specific expression
on
a monocot, a monocot olesin regulatory sequence should be used. For divot seed-
specific expression, a divot oleosin regulatory sequence should be employed.
The
reported sequence can be used in a wide variety of dicotyledonous plants,
including all members of the Brassica genus and crucifers in general.
Solanaceous
plants, such as tobacco and tomato, also recognize the sequences and show
correct
regulation of expression in developing seeds.
It is expected that the desired proteins would be expressed in all
embryonic tissue, although different cellular e~cpression can be detected in
different
tissues of the embryonic auis and cotyledons. This invention has a variety of
uses
which include improving the intrinsic value of plant seeds by their
accumulation of
altered polypeptides or novel recombinant peptides or by the incorporation or
elimination of a metabolic step. In its simplest embodiment, use of this
invention
IS may result in improved protein quality (for example, increased
concentrations of
essential or rare amino acids), improved liquid quality by a modifs~.~.tion of
fatty
acid composition, or improved or elevated carbohydrate composition. P~camples
include the expression of sulfur-rich proteins, such as those found in lupins
or
brazil nuts in a seed deficient in sulphurous amino acid residues'
Alternatively, a
fatty aryl coenzyme A (COA) a transferase enzyme capable of modifying fatty
Los in triglycerides (storage lipid) could be expressed. In cases where a
recombinant pmtein is allows to acxumulate in the seed, the protein could also
be
a peptide which has pharmaceutical, industrial or nutritional value. In this
rise,
the peptide could be extracts from the seed and used in crude or purified
form,
as appropriate for the intended use. The protein could be one truly foreign to
the
plant kingdom, such as an animal hormone, enzyme, lymphokine, anticoagulant,
or the like could be expressed in seed. The heterologous protein could then be
extracted from the seeds and used for experimental, nutritional or
pharmaceutical
purposes after partial or complete purification.
The following examples are offered by way of illustration and not
by limitation.
W0.93/20216 ~ ~ ~ ~ ~~ 1 rt PCT/CA93/0(1141
14
SAMPLES
The oil body protein gene from Arabidopsis was isolated on a lSkb
insert present in a clone from an Arabidopsis thaliana v. Columbia genomic
library in phage ~ EMBL3A by hybridization to a B. napes oleosin clone. A
l.8kb fragment containing approximately 868 base pairs 5' of the oleosin
protein
translational start was subcloned into a plasmid vector. The Arabidopsis ~ 8
KDa
oleosin gene is conveniently cloned as a 1803 by fragment flanked by Ncol and
Kpnl sites in a vector called pPAVV4 (see Figure 1). In order to convert the
fragment into an expression cassette for general use with a variety of
foreign/alternative genes, two modifications must be made. Firstly, using the
technique of site-directed mutagenesis (Kunkel, supra) mutations at positions -
2, -1
and +4 are introduced using a mis-matched oligonucleotide. The mutations
required are A to T (-2), A to C (-1) and G to A (+4). These mutations have
the
. effect of creating a BspHl site at positions -2 to +4. The BspHl site
(T/CATGA)
encloses the ATG initiation colon and gives a recessed end compatible with an
Ncol cut. A second modification involves digestion with EcoRV and P~iscl which
releases a 658 by fragment containing most of the coding sequence of the
native
oleosin. This leaves blunt ends at the cut sites which on separation of the
vector
and an ancillary sequence from the EcoRV-Idfscl fragment, permits
recircularization of the vector-promoter terminator combination. This
recircularization is performed in the presence of an oligonucleotide linker
containing restriction sites not found in the original 1803 Kb fragment.
Gn recircularization, a plasmid containing all the upstream
sequences of the oleosin gene, a transcriptional start site and an initiation
colon
embedded in a BspHl site is obtained. Thirty-one bases downstream of this is a
short polylinker containing one or more unique restriction sites. To introduce
any
DNA sequence into this cassette the foreign sequence should have, or should be
modified to contain, a BspHl or Ncol site at the initial ATG position. For
sequences to be expmessed as proteins this will assure conservation of the
distance
between the "cap" site and the initiator colon.
iw ~. ~) tl ~ .,~. '~
W0.93/20216 PCT/CA93/00141
The DNA sequence to be inserted should terminate with a cohesive
~d of a restriction site not found on the plasmid. The polylinker interposed
into
the e~cpression cassette may be chosen with this site in mind. Digesting the
plasmid with BspHl and the appropriate restriction enzyme for the 3' end of
the
5 foreign sequence will ensure that a directional cloning of the desired DNA
fragment may be effected. Using appropriate ligation conditions, the plasmid
acpre~on cassette with BspHl and a site compatible with the desired DNA,
fragment are incubated together to produce a ligated product as shown in
Figure 1.
The complete construct from Ncol-Kpnl is now excised and
10 introduood into an appropriate plant transformation vector such as an
Agritan plasmid. In order to introduce the construct into common
Agrobacteriwn plasmids such as Bin 19 (Bevan, Nucl. Acid Research (1984)
x:8711-8721) it may be necessary to use one of the additional restriction
sites in
p>asmid pPAW4. In one scenario the plasmid could be cut with Smal and Kpnl.
15 The rr,~ulting purified fragment then is ligated to a Kpnl oligonucleotide
linker
and digested withy Kpnl. This provides a non-directional Kpnl fragment for
i~oduction into Bin 19. Alternatively, the construct may be excised with Kpnl
and BamHl and ligated directionally into pBIN 19 pnwiously cut with the same
re~riction enzymes. The resulting Agrobacterirrm binary plasmid is mobilized
into
a disarmed Agrobacterium strain by tripartite mating (Ditta, et al. (1980),
PNAS
77: 7347 7351) or DNA transformation of competent Agrobacteriwn (An, (1988),
Plant Mol. Biology Manual, A3 1-19, Kluwer Academic, Dordrecht, Netherlands).
The Agnvbacterium harboring the recombinant Bin 19 is used to
transform any susceptible plant, e.g., Brassica sp. by standard explant co-
cultivation (Horsch et al. (198, supra). The transformed cells are selected in
culture with kanamycin using the co-transferral antibiotic resistance genes
(neomycin phosphotransferase) also contained between the T-DNA borders of pain
19. These transformed calls are induced to regenerate whole plants by standard
procedures (e.g. for an oilseed such as rapeseed: ~, Molameyr et al. Plant
Cell
Rep., (1989), $: 238-242). Tht regenerated plants are permitted to flower and
are
self fertilized (or may be cross-fertilized). In cases wrhere the foreign DNA
in the
construct encodes a translatable product, this product may be isolated from
WO 93/20216 ~~ ~ ~ j ,~ {~ ~ ~ ~ PGT/GA93/00141
16
aqueous extractions of the mature seed and subsequent fractionation of the
slurry
by centrifugation (30 min at 100,000 xg). Depending on the desired product it
may partition with any one of the three phases obtained. It may be localized
in
the pellet, aqueous soluble phase or in the lipid film on the surface of the
centrifuged sample.
Alternatively, it may not be necessary to extract the product as the
purpose of the expression may be to divert metabolism in the seed thus
changing
the phenotype of the seed (e.g. by altering size or colour of the seed,
changing the
ratio of fatty acid residues in the seed or interdicting a particular
metabolic step
considered to render the seed less useful or valuable. Such metabolic steps
might
ir~lude the production of antinutritional secondary products which reduce the
value
or desirability of the seed when present. In such cases, the seed, per se, is
simply
harvested and used in accordance with usual procedures.
A number of constructs containing varying amounts of the DNA
from the 5' transcriptional initiation region of the Arabidopsis oleosin
gee joined operably to the coding region for ~-glucuronidase (GUS) were
prepared using PCR. The constructs are designated according ~'o the amount of
the oleosin 5' region contained, for example, the 25~ construct has
approximately
f
2500 base pairs of the.oleosin 5' region. The constructs were introduced into
Brassica napes and tobacco and the expression of the ~-glucuronidase gene was
measured as described in detail below. The GUS expression results of five
constructs, the 2500, the 1200, the 800, the 600 and the 200 constructs in
transformed Brassica napes plants are shown in Table I. A negative control
(untransformed plant) is also shown. The GUS expression results of two
constructs, the 2500 and the 800 constructs, in transformed tobacco plants are
shown in Table II. Table III shows the developmental timing of the expression
of
the oleosin promoter in transgenic embryos.
3Q The constructs were made using standard molecular biology
techniques, including restriction enzyme digestion, ligation and polymerase
chain
W0.93/20216 PCT/CA93/00141
17
reaction (PCR). As an illustration of the techniques employed, the
construction of
the 800 construct is described in detail.
In order to obtain a DNA fragment containing approximately 800
base pairs from the 5' transcriptional initiation region of the Arabidopsis
oleosin
gene in a configuration suitable for ligation to a GUS coding sequence, a PCR
based approach was used. This involves the use of the polymerise chain
reaction
to amplify the procise sequence desired for the expression analysis. To
perform
the necessary PCR amplification, two oligonucleotide primers were synthesized
(Milligen-Biosearch, Cyclone DNA synthesizer) having the following sequences:
Pst 1 oltosin seq
5' primer: 5'CACTG(~IGGAAGTGTGTGGTAA 3'
(GVR10)
IS The italicized bases correspond to nucleotide positions -833 to -817
in the sequence reported in Fig. 2A. The additional nucleotides 5' of this
sequence in the primer are not identical to the oleosin gene, but were
included in
order to place a Pstl site at the 5' end of the amplification product. The
Pstl site
~,
is tmderlined.
A second (3') primer was synthesized which had the following sequence:
3' primer (Aa,P 1)
oleosin seq
5-CTA~~~ATCCTGT?TAGTAGAGAGAAT~3
Smal
This primer contains the precise complement (shown in italics) to
the sequence reported in Fig. 2A from base -13 to -30. In addition, it
contains a
13 bases at the 5' end. lfiis sequence is not complementary to the oleosin
gene, but was added to provide two (overlapping) restriction sites, Smal and
BamHl, at the 3' end of the amplification pmduct to facilitate cloning of the
PCR
fragment.
;~~. -~'~'~r7 !'..;.~' 1 .. .2JW a'.o ~ ~~~ 7
x -r.::- .l ., t... . k..'
.,. c, : rrr-.. , . :,~ > , s . "r.5~ , _ . .. . ~. ... ..~....s ..
r..,..~.~........m:~r. T...,rw,%'r.~_~:/~?F71~83~ ~m , ....".,~,...,a , ,__
... ,.,... .:.., i'. -x ...... .... ~,.....~~1.. ... ..... _, , .. , r , ~..
~:.. a , . ". .. . v. , ... , . , . .. . ~ . .. ... . .r. ... , ..
CA 02133417 2002-07-15
18
These two primers were used in a PCR amplification reaction to
produce DNA fragment containing the sequence between nucleotides -833 and -13
of the oleosin gene with a Pstl site at the 5' end and Smal and BamHl sites at
the
3' end. PCR amplification ,uas performed using the enzyme TaqTM enzyme
(Perkin-Elmer-Cetus) using the conditions recommended by the enzyme
manufacturer and a temperature program of 92°C (denaturation) 1 min,
55°C
(annealing) 1 min and 72°C (elongation) 1 min. The template was the
oleosin
genomic clone shown in Figure 28, top panel, which in the original 7. library
isolate contained approximately I5 kilobases of Arabidop.sis DNA.
The amplification product (OLEO p800) was gel purified on 0.7%
agarose, recovered using the glass bead method of Vogelstein and Gillespie
(Preparative and analytical purification of DNA from agarose. Proc. Natl.
Acad.
5ci. USA 1979 '76:615-619) and digested with Pstl. The digestion product was
gel purified and end filled using DNA polymerase Klenow fragment then cut with
Smal to produce a blunt ended fragment. This was cloned into the Smal site of
pUC 19 to yield the plasmid pUC OLEOp800. Using the asymmetric positioning
of the Accl site in the insert (at the position corresponding to -649 in the
oleosin
gene as shown in Figure 2B) it was possible to select both orientations of
insertion
into pUC vector. The clone having the insert oriented sur.l~ that the 5' most
end
of the amplified fragment (in the direction of transcription) is proximal to
the
unique Hind III site in the puCl9 cloning vector and the 3' most end of the
amplified fragment is proximal to the unique I;~:o RI site in the pUCl9
closing
vector.
The resulting plasmid was then cut with BarnHl to yield the
fragment OLEOp800 flanked by BamH 1 sites. 'This fragment, BamH 1-OLE0800,
was cloned into the BamHl sites of a BamHl digested plasmid designated
HspGUS 1559. HspGUS 1559 is a plasmid used as a binary vector in
Agrobacterium, derived from the vector pCGN 1559 (htacl3ride and Summerfeldt,
1990, Plant Molecular Biology, 14, 269-2?6) with an insert containing heat
shock
promoter (flanked by BamHl sites), the ~i-glucuranidase open reading frame and
a
nopaline synthase terminator (derived from pB1221, Jefferson RA in Cloning
Vectors 1988, Eds. Pouwels P., Enger-Valk BE, Brammer WJ., Elsevier Science
WO 93/20216 .. i. t ~ ~~ PCT/CA93/00141
Pub BV, Amsterdam section VII, Ail l). BamHl digestion of HspGUS1559
results in the release of the heat shock promoter and permits the insertion of
any
other BamHl fragment in its place. The BamHl-OLEOp800 fragment was ligated
into this site to yield the Agrobacterium pOLEOp800GUS1559. This plasmid was
used to transform E. coli and the amplified plasmid was introduced into
Agrobacterium (strain EHA101) by electroporation as described above (Rogers et
al., 1988, Plant Molecular Biology Manual, A2: 1-12, Eds. Gelvin S. and
Schilperoort, R. Kluwer Academic, Dordrecht, Netherlands).
The resultant Agrobacterium strain (EHA 101 x
pOLF.Op800GUS1559) was used to transform Brassica napes plants by the method
of Moloney et al. (Moloney, M.M., Walker, J.M., Sharma, K.K. (1989) Plant
Cell Reports 8:238-242) or tobacco plants by the method of Horsch et al.
(Horsch
et al. Science (1985) 227:1299-1302). The resultant transgenic plants were
allowed to set seed, and GUS expression assays (Jefferson R.A. (1987), Plant
Mol. Biol. Rep. 5 387-405) were performed on the developing seeds and also on
non-reproductive plant parts as controls. GUS expression reported is an
average
obtained from approximately five seeds from each of approximately five
different
transgenic plants.
The other constructs were prepared by the same SCR method
described above using the appropriate primers for amplifying the -2500
fragment,
the -1200 fragment, the -600 fragment or the -200 fragment. The results in
Brassica napes expressed as specific activity of GUS enzyme are shown in Table
I. The results in tobacco are shown in Table II.
These results demonstrate that the oleosin fragment from -833 to -
813 used in the 800 construct contains sufficient information to direct seed-
spa;ific
expression of a reporter gene in transgenic Brassica napes embryos as early as
heart stage and that the Arobidopsis oleosin promoter is capable of directing
transcription in plants other than Arabidopsis. These experiments also show
that
the sequences present in this promoter construct contain the cis elements
required
34 for an increase in transcription in response to the addition of abscisic
acid, a
characteristic of the native oleosin promoter.
WO 93/20216 ~ ~_ ~ ~ ~ ~, ~ PCT/CA93/00141
It should be noted that the seed-specific expression demonstrated
here does not depend on interactions with the native terminator of an oleosin
gene
3' end. In this example, the 3' oleosin terminator was replaced by a
terminator
derived from the nopaline synthase gene of Agrobacteriarm. Thus, the sequence
in
5 the 800 construct is suf~~cient to drive the desired expression profile
independent
of ancillary sequences.
Y
1 r r' r,~'~k'~
~t~.:... . . '~'~'J a ... .Vine., ~. h.1 -~4.~.._. '~. ~..v.. .-t.W... :'"v~ ~
., m
fi r
WU~ 93/Z0216 PCT/CA93/00141
21
Table I
~~p~~~~pressiQn in .8rassica napes .
$ GUS Activity (in pmol product/min/mg protein)
Promoter/GUS
Construct
ABA* -ABA
2500 10,185 7,709 444 46.9 88.2 11,607
1200 18,298 1,795 8,980
800 2,256 475 285 277 650 7,130
600 1,506 144 1,365
200 18.1 64.8 260 5.9 26 11
Negative 18.4 13.9 300 6.1 30 14
r
Control-Non-
transform~l
Plant
*ABA is treatment for abscisicacid
24 hours with 10'sM prior
to
GUS
activity
measurement
Seed Root ~f ~gm Seed (Late-
(Toraedo Stage) ~Q~yledon~
._::: . ~ . ..._. .: ,.... , . :.
W0.93/20216 PCT/CA93/00141
22
Table II
S~~ific Expression in Tobacco
S Promoter/GUS GUS Activity (in pmol productlmin/mg protein)
Constcvcts Mature Seeds
2500 11,330
800 10,970
Table III
~eve~mental Expression in Br~~sica na~us
GUS ACTIVITY (in pmol product/min/mg protein)'
Promoter/ Early Mid- Late
r
GUS Heart Torpedo CotyledonaryCotyledonaryCotyleaonary
2500 272 1207 2541 1819 11,607
1200 124 262 388 5094 8,980
800 149 260 962 2617 7,128
600 59 41 29 38 1,365
200 30 25 15 20 11
Negative 11 14 14
Comrol
CA 02133417 2002-07-15
23
The invention now being fully described, it will be apparent to one of
ordinary skill in the art that many changes and modifications can be made
thereto
without departing from tine spirit or scope of the appended claims.