Note: Descriptions are shown in the official language in which they were submitted.
W 093/25684 PC~r/US93/05672
21~77g9
THERAPEUTIC AND DIAGNOSTIC METHODS BASED ON NEUROTROPHIN4
EX~
The present application is a continuation-in-part of copending United
States application Serial No. 07t898,194, filed on June 12, 1992 which is a
continuation in part of copending United States application Serial No.
791,924 filed on November 14, 1991.
1. INTRODUCTION
The present invention relates . to neurotrophin-4 (NT-4), a newly
characterized member of the BDNF/NGF/NT-3 gene family and the
therapeutic and diagnostic methods of utilizing neurotrophin-4 in the
treatment of neurological disorders.
2. BACKGROUNDOFTHEINVENTION
The nerve growth factor family includes nerve growth factor (NGF),
brain-derived neurotrophic factor (BDNF), and neurotrophin-3 (NT-3), also
known as hippocampus-derived neurotrophic factor (HDNF). This family of
proteins plays an important role in both the developing and the adult
verlel~,ale nervous system, where they support neuronal survival.
Based on the amino acid sequence of the mouse NGF protein
(Angeletti, et al., 1973, Biochemistry 12:100-115) DNA sequences coding for
mouse and human NGF have been isolated (Scott et al., 1983, Nature
302:538-540; Ullrich et al., 1983, Nature 303:821-825). Comparison of
mouse and human NGF showed that the protein is conserved within
mammals and in support of this, NGF-like activities have been isolated from
several species (Harper and Thoenen, 1981, Ann. Rev. Pharmacol. Toxicol.
W O 93/25684~ PC~r/US93/05672
21:205-229~. SubserllJently, DNA sequences from bull (Meier et al., 198
EMBO J. 5:1489-1493); chick (Meier et al., 1986, EMBO J. 5:1489-1493;
Ebendal et al., 1986, EMBOJ. 5:1483-1487; Wion et al., 1986, FEBS ~tters
203:82-86; cobra (Selby et al., 1987, J. Neurosci. Res. 18:293-298); rat
(Whittemore et al., 1988, J. Neurosci. Res. 20:403-410); and guinea pig
(Schwarz et al., 1989, Neurochem. 52:1203-1209) NGFs were also
determined. Brain-derived neurotrophic factor (BDNF) was first isolated
from pig brain (Barde et al., 1982, EMBOJ. 1:549-553) and sub.se~uently
cloned as a cDNA from this tissue (Leibrock et al., 1989 Nature 341:149-
1 0 152). The gene for NT-3 has been isolated from mouse (Hohn et al., 1~90,Nature, 344: 339-341), rat (Maisonpierre et al., 1990, Science 247: 1446-
1451; Ernfors et al., 1990, Proc. Natl. Acad. Sci. USA 87: 5454-5458), and
human (Rosenthal et al., 1990, Neuron 4: 767-773), using degenerate
oligonucleotides based on the sequence similarity between the other two
1 5 factors. The three factors show approximately 55% amino acid similarity
to each other, and most sequence differsnces are present in five regions
that contain amino acid motifs characteristic of each protein. The
neurotrophic activity in vitro of two of these proteins have recently been
shown to be acquired by specific combinations of these variable regions.
NGF supports the development and maintenance of peripheral
sympathetic and neural crest-derived sensory neurons (reviewed in Thoenen
and Barde, 1980, Physiol. Rev., 60: 1284-1325; Levi-Montalcini, 1987,
Science, 237: 1154-1162). No activity has been seen for BDNF in peripheral
sympathetic neurons, but this factor supports in vivo the survival of both
placode and neural crest-derived sensory neurons (Hofer and Barde, 1988,
Naturs, 331: 261-262). The neurons sensitive to NT-3 in vivo remain to be
identified. However, in explanted chick ganglia or dissociated neuronal
cultures in vitro, tha three factors support both ov~rlapping and unique sets
of neuronal populations, suggesting that NT-3 exerts both specific and
overlapping neurotrophic activities also in vivo (Hohn et al., 1990, Nature,
WO 93/25684 ~ 7 9 9 PCI`/US93/05672
344: 339-341; Maisonpierr~ et al., 1990, Science, 247: 1446-1451; Ernfors
et al., 1990, Proc. Natl. Acad. Sci. USA, 87: 5454-5458; Rosenthal et al.,
1990, Neuron 4: 767-773). All three factors are ex~,ressed in specific sets
of neurons in the brain, with the highest levels of mRNA for all three factors
in the hippocampus (Ayer-LeLievre et al., 1988, Science 240: 1339-1341;
Ernfors et al., 1990, Proc. Natl. Acad. Sci. USA, 87: 5454-5458; Ernfors et
al., 1990, Neuron 5: 511-526; Watmore et al., 1991, Neurol. 109: 141-152;
Hofer et al., 1990, EMBOJ.,9: 2459-2464; Phillips et al., 1990, Science, 250:
290-294). In th~ brain, NGF has been shown to support basal forebrain
1 0 cholinergic neurons (reviewed in Whittemore and Seiger, 1987, Brain Res.,
434: 439-464; Thoenen et al., 1987, Rev. Physiol. Biochem. Pharmacol., 105:
145-178; Ebendal, 1989, Prog. Growth Factor Res. 1: 143-159) and BDNF
has been shown to stimulate the survival of these neurons in vitro (Alderson
et al., 1990, Neuron 5: 297-306).
1 5 The effects of the three proteins are mediated by their interactionwith specific receptors present on sensitive cells. Molecular clones have
been isolated for the rat, human, and chicken NGF receptor (NGF-R), and
nucleotide sequence analysis of these clones has shown that the NGF-R
contains one plasma membrane-spanning domain, a cytoplasmic region, and
an extr~cellul~r cysteine-rich amino-terminal domain (Johnson et al., 1986,
Cell, 47: 545-554; Radeke et al., 1987, Nature, 325: 593-597; Large et al.,
1989, Neuron 2: 1123-1134). The NGF-Rshows a low but significant
sequence similarity to the receptor for a tumor necrosis factor (Schall et
al., 1990, Cell, 61: 361-370) as well as to the Iymphocyte surface antigens
2 5 CD40 (Stamenkovic et al., 1989, EMBO J., 8: 1403-1410) and OX40 (Mallettet al., 1990, EMBO J., 9: 1063-1068). The NGF-R can occur in two apparent
states, known as the low and high affinity states (Sutter, et al., 1979, J.
Biol. Chem., 254: 5972-5982; Landreth and Shooter, 1980, Proc. Natl. Acad.
Sci. USA, 77: 4751-4755; Schechter and Bothwell, 1991, Cell 24: 867-874).
The gene for the NGF-R appears to encode a prolein that forms part of
WO 93/25684 ~ PCI/US93/05672
both the low and the high affinity states of the receptor (Hempstead et
1989, Science, 243: 373-375), though only the high affinity receptor has
been proposed to mediate the biological activity of NGF. Both BDNF
(Rodriguez-Tebar et al., 1990, Neuron, 4: 487-492) and NT-3 (Ernfors et al.,
1990, Proc. Natl. Acad. Sci. USA, 87: 5454-5458) can interact with the low
affinity NGF-R, suggesting that the low affinity NGF-R may be, in an as yet
unknown way, involved in mediating the biological effects of all three factors.
In the developing nervous system, N,GF and its receptor have been
shown to be synthesized in the target area and in the responsive neurons,
1 0 respectively, at the time when the growing axon reaches its target (Davies
et al., 1987, Nature, 326: 353-358). In agreement with this, the level of NGF
mRNA in the developing chick embryo reaches a maximum at embryonic day
8 (E8) (Ebendal and Persson, 1988, Development, 102: 101 -106), which
coincides with the time of sensory innervation. However, in the chick, NGF-R
1 5 mRNA is maximally expressed at early embryonic stages prior to neuronal
innervation (Ernfors et al., 1988, Neuron, 1, 983-996), and in the E8 chick
embryo high levels of NGF-R mRNA have been detect~d in the mesenchyme,
somites, and neural tube cells (Hallbook et al., 1990, Development, 108: 693-
704; Heuer et al., 1990, Dev. Biol., 137: 287-304; Heuer 1990, Neuron, 5:
283-296). This observation, together with the fact that NGF mRNA is
expressed in th~ E3 chick embryo at relatively high levels (Ebendal and
Bersson, 1988, Development, 102: 101-106), indicates that NGFmay play a
role in early devGlop",ent that is distinct from its function as a neurotrophic
factor. In agreement with this possibility, NGF has rec~ntly been shown to
2 5 control prolif~ration and differentiation of E14 rat embryonic striatal
precursor cells in culture (Cattaneo and McKay, 1990, Nature, 347: 762-
765). In the chick embryo BDNF and NT-3 mRNA are maximally expressed
at E4, 5, and BDNF has been shown to control the differentiation of avian
neural crest cells in vitro (Kalcheim and Gendreau, 1988, Dev. Brain Res., 41:
3 0 79-86).
wo 93/25684 21 ~ 7 7 99 PCI`/US93/05672
Moreover, evidence for a non-neuronal function of NGF has also been
presented. The stili unexplained high levels of NGF found in the male mouse
submandibular gland may indicate other functions for NGF (Levi-Montalcini,
1987, Science, 237: 1154-1162). In the adult rat, NGF has been shown to
5 induce DNA synthesis and to stimulate IgM secretion in B-cells (Otten et al.,
1989, Proc. Natl. Acad. Sci. USA 86: 10059-1006~). Additionally, NGF is
present in sufficient quantity in guinea pig prostate such that Rubin and
Bradshaw (1981, J. Neur. Res. 6: 451-464) were success~ul in isolating and
characterizing substantially pure NGF from this exocrine tissue. The high
1 0 level of NGF in pig prostate support the hypothesis that this neurotrophic
factor functions in a non-neuronal capacity not yet understood (Bradshaw,
1978, Ann. Rev. Biochem. 47:191-216; Harper, et al., 1979, Nature 279:160-
162; Harper and Thoenen, 1980, J. Neurochem. 34:893-903).
Furthermore, NGF mRNA is expressed in spermatocytes and early
1 5 spermatids in the adult rat testis (Ayer-LeLievre et al., 1988, Proc. Natl.
Acad. Sci. USA, 85: 2628-2632), and the NGF protein is present in germ cells
of all stages from spermatocytes to spermatozoa (Olson et al., 1987, Cel
rlssue Res., 248: 275-286; Ayer-LeLievre et al., 1988a, Proc. Natl. Acad. Sci.
USA 85: 2628-2632). NGF-R mRNA has also bsen detected in the adult rat
testis, where it is expressed in Sertoli cells under negative control of
testosterone, and in the testis NGF has been suggested to control meiosis
and spermiation (Persson et al., 1990, Science, 247: 704-707).
3. SUMMARYOFTHEINVENTION
The present invention relates to neurotrophin-4 (NT-4), a newly
characterized member of the BDNF/NGF/NT-3 g2ne family.
The present invention provides for nucleic acid molecules encoding NT-
4. Such molecules may comprise a sequence subslar,lially as set forth for
3 0 NT-4 in Figure 1 [SEQ ID NO:1 (NT-4, viper), SEQ ID NO:2 (NT-4, Xenopus)],
WO 93/25684 7 t~7 ~ 9 ~ i PCI/US93/05672
Figure 4 (SEQ ID NO:43), Figure 8 (SEQ ID NO:49), Figure 14 (SEQ ID NO:~
Figure 15 (SEQ ID NO:63), Figure 17 (SEQ ID NO:69), Figure 18 (SEQ ID
NO:75), Figure 20 (SEQ ID NO:93) and Figure 21 (SEQ ID NO:116) or may
comprise a sequence that is at least about seventy percent homologous to
5 such sequence.
The present invention also provides for protein or peptide molecules
which comprise a sequence sut,st~,lially as set forth for NT-4 in Figure 2
[SEQ ID NO:22 (NT-4, viper), SEQ ID NO:23 (NT-4, Xenopus)], Figure 4 (SEQ
ID NO:44), Figure 8 (SEQ ID NO:50), Figure 14 (SEQ ID NO:62), Figure 15
1 0 (SEQID NO:64), Figure 17 (SEQ ID NO:70), Figure 18 (SEQ ID NO:76), Figure
20 (SEQ ID NO:94), or Figure 21 (SEQ ID NO:117) or may comprise a
sequence that is at least about seventy percent homologous to such
sequence.
The present invention further provides for expression of biologically
1 5 active NT-4 molecules in prokaryotic and eukaryotic systems.
The present invention further provides for the production of NT-4 in
quantities sufficient for therapeutic and diagnostic applications. Likewise,
anti-NT-4 antibodies may be utilized in therapeutic and diagnostic
ar~plic~tions. For most purposes, it is pr~ferable to use NT-4 genes or gene
20 products from the sams species for therapeutic or diagnostic purposes,
although cross-species utility of NT-4 may be useful in specific embodiments
of the invention.
The present invention further provides for therapeutic and diagnostic
applic~tions based on NT-4 expression by ~isc~osing detectable levels of NT-
2 5 4 expression in human skeletal muscle, prostate, thymus and testes.Further therapeutic and diagnostic applications are based on the
demonstration of binding of NT-4 to brain and retina, as well as retrograde
transport of NT~ and on the ability of NT-4 to support the survival of
various neuronal cell popul~tions.
W O 93/25684 . 2 1 3 77 ~ 9 PC~r/US93/05672
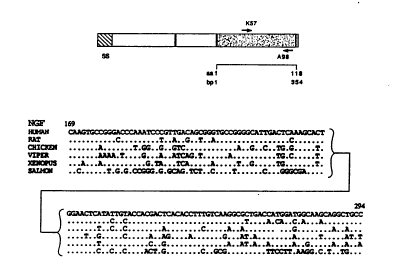
4. DESCRIPTION OF THE FIGURES
FIGURE 1. Alignments of DNA sequ~nces of the isolated
fragments coding for NGF, BDNF, NT-3 and the novel neurotrophic factor
5 NT-4 from different species.
(A) Schematic representation of the mouse preproNGF molecule. The
hatched box indicates the signal sequence (SS), black bars denote
proteolytic cleavage sites and the shaded box represenls the mature NGF.
Regions used for the degenerate primers are indicated by arrows. The
1 0 upstream primer was from the region coding for Iysine 50 to threonine 56
and the downstream primer includes tryptophan 99 to aspartic acid 105.
The amplified region comprises DNA sequencss from base pair (bp) 168 t o
294 in the mature NGF molecules and in all members of the NGF family
desc.ibed so far, this region is loc~ted in one exon.
1 5 (B) Alignment of nucleotide sequences for NGF, BDNF, NT-3 and NT-4isolated from different species. The fragments cGr,espond to amino acids
57 to 98 in the mature mouse NGF. Identical bases are indicated by dots.
The numbering refers to nucleotides in the sequences of mouse mature NGF
(Scott et al., 1983, Nature 300:538-540). SEQ ID NO:1 (NT-4, viper), SEQ
ID NO:2 (NT-4, Xenopus), SEQ ID NO:3 (NGF, human), SEQ ID NO:4 (NGF,
rat), SEQ ID NO:5 (NGF, chicken), SEQ ID NO:6 (NGF, viper), SEQ ID NO:7
(NGF, Xenopus), SEQ ID NO:8 (NGF, salmon), SEQ ID NO:9 (BDNF, human),
SEQ ID NO:10 (BDNF, rat), SEQ ID NO:11 (BDNF, chicken), SEQ ID NO:12
(BDNF, viper), SEQ ID NO:13 (8DNF, Xenopus), SEQ ID NO:14 (BDNF,
2 5 salmon), SEQ ID NO:15 (BDNF, ray), SEQ ID NO:16 (NT-3, human), SEQ ID
NO:17 (NT-3, rat), SEQ ID NO:18 (NT-3, chicken), SEQ ID NO:19 (NT-3,
Xenopus), SEQ ID NO:20 (NT-3, salmon), SEQ ID NO:21 (NT-3, ray).
FIGURE2. Alignment of amino acid sequences deduced for NGF,
BDNF, NT-3 and NT-4 from different sreciQs. The numbering of the amino
3 0 acids (single letter code) is taken from the mature mouse NGF (Scott et al.,
w0 s3/2s684~ Pcr/uss3/os672
1983, Nature 300:538-540). Identical amino acids are inJicaled with do~
Positions that show conservative amino acid replacements in all species
variants of the same factor are underlined. The broken line in~ tes that
the corresponding sequence was not isolated. Bars represent variable
regions in the different molecules~(R59 to S67 and D93 to A98). SEQ ID
NO:22 (NT-4, viper), SEQ ID NO:23 (NT~, Xenopus), SEQ ID NO:24 (NGF,
human), SEQ ID NO:25 (NGF, rat), SEQ ID NO:26 (NGF, chicken), SEQ ID
NO:27 (NGF, viper), SEQ ID NO:28 (NGF, Xenopus), SEQ ID NO:29 (NGF,
salmon), SEQ ID NO:30 (BDNF, human), SEQ ID NO:31 (BDNF, rat), SEQ ID
1 0 NO:32 (BDNF, chicken), SEQ ID NO:33 (BDNF, viper), SEQ ID NO:34 (BDNF,
Xenopus), SEQ ID NO:35 (BDNF, salmon), SEQ ID NO:36 (BDNF, ray), SEQ ID
NO:37 (NT-3, human), SEQ ID NO:38 (NT-3, rat), SEQ ID NO:39 (NT-3,
chicken), SEQ ID NO:40 (NT-3, Xenopus), SEQ ID NO:41 (NT-3, salmon),
ID NO:42 (NT-3, ray).
1 5 FIGURE 3. Deduced phylogeny of members of the NGF family.
Phylogenetic trees showing speciation of NGF (A), BDNF (B), and NT-3 (C)
were constructed using analysis of nucleotide sequences. Human NT-3 was
used as a reference point in (A) and (B), human NGF and human BDNF were
used in (C). The scale bar in (A) represents a branch length corresponding
to a relative difference score of 20. The same scale was used in (B) and
(C). (D) shows a phylogram of the evolutionary rslationship between the
diffarent members of the NGF family. The data ware compiled from
deduced amino acid sequences. The scale bar represents a branch length
of 20. All trses shown are unrooted so that the branches are measured
2 5 relative to one another with no outside reference. Abbreviations: chi,
chicken; hum, human; sal, salmon; vip, viper; xen, Xenopus.
FIGURE 4. Sequence of Xenopus NT-4 and Comparison to NGF, BDNF,
and NT-3.
(A) A potential translation start site is boxed. A putative signal
cleavage site is indicated by the arrow labeled SC. Amino acids within the
WO 93/25684 ~ 1 3 7 7 ~ 9 PCI/US93/05672
signal sequence that are identical between Xenopus NT~ and pig and rat
BDNF are indicated with stars. A consensus sequence for N-glycosylation is
underlined, and the arrow indicates the presumptive start of the mature NT-
4 protein. (SEQ ID NO:43 and SEQ ID NO:44)
(B) Amino acid (singie-letter code) sequence comparison of Xenopus
NT-4 (SEQ ID NO:45) with mouse NGF (Scott et al., 1983, Nature 300: 538-
540) (SEQ ID NO:46), mouse BDNF (Hofer et al., 1990, EMBOJ. 9: 2459-
2464) (SEQ ID NO:47), and mouse NT-3 (Hohn et al., 1990, Nature 344:
339-341) (SEQ ID NO:48). Identical amino acid replacements compared
1 0 with the NT-4 amino acid sequence are shown by dots. Sequences that
differ behr~ocn NGF, BDNF, and NT-3 also differ in the sequence of the NT-4
protein.
FIGURE5. Transient expression of the Xenopus NT-4 protein in COS
cells and its interaction with NGF-Rs on PC12 cells.
1 5 (A) SDS-PAGE of conditioned media from in vivo labeled COS cell
cultures (3 x 104 cpm loaded in each lane) transfected with the rat NGF
gene, a control plasmid without insert, or the Xenopus NT~ gene. Shown is
an autoradiograph of the dried gel after an overnight exposure to X-ray
film.
(B) Serial dilutions of transfected COS cell medium containing equal
amounts of NT-4 (open circles) or NGF (closed circles) protein were
assayed for their ability to ~ispl^^e 1251-NGF from its receptor on PC12 cells.
Binding assays were performed at 37C using 1.5 x 109 M 1251-NGF and 1 x
104 cells per ml. Medium from mock-transfected cells failed to displace
binding of 1251-NGF from PC12 cells. Each point represents the mean + SD
of triplicate determinations.
FK~URE6. Stimulation of neurite outgrowth from chicken embryonic
ganglia.
(A, B, and C) Neurite oulgro~.ll, elicited in dorsal root ganglia with
recombinant NT~ protein (A), recombinant NGF (B), and BDNF protein (C).
W0 93/25684 f~ PCI/US93/05672
(D) The response of dorsal root ganglia to conditioned medi~
from mock-transfected cells.
(E and F) Stimulation of neurite outgrowth from sympathetic ganglia
in response to NT4 (E) or NGF (F).
(G, H, and 1) Nodose ganglia stimulated with recombinant NT-4 (G),
NT-3 (H), and BDNF (I) proteins. All figures are bright-field micrographs of
ganglia after 1.5 days in culture.
FIGURE 7. Detection of NT-4 mRNA in di~erent Xenopus tissues.
(A) Poly(A)+ RNA (10 9 per slot) from the indicated tissues of adult
female Xenopus was electrophoresed in a formaldehyde-containing agarose
gel, blotted onto a nitrocellulose filter, and hybridized to a 500 bp Hindl
fragment from the 3' axon of the Xenopus NT-4 gene. For comparison, the
filter was also hybridized to a 180 bp PCR fragment from the Xenopus NGF
gene (lane marked heart, NGF). Th~ filter hybridized to the NT-4 probe was
exposed for 2 days; the filter hybridized to the NGF probe was exposed for
2 weeks. A prolonged 2 week exposure of the filter hybridized to the NT-4
probe did not reveal NT-4 mRNA in any tissues other than the ovary, which
includes oocytes of different stages. The lane l~heled CNS includes brain
and spinal cord.
(B) Poly(A)+ RNA (10 9) from Xenopus ovary was analyzed for the
e~cpression of the four members of the NGF family. Each filter was
hybridized with the indicated probes obtained by labeling of PCR fragments
from their respective Xenopus genes. The location of the labeled I~R
fragments in the 3' axon of their genes is shown in Figure 1A. The filters
were washed at high stringency and exposed to X-ray films for 5 days.
FIGURE8. Nucleotide sequenca of Xenopus NT-4 with restriction
andonuclease cleavage sites (SEQ ID NO:49 and SEQ ID NO:50).
FIGURE9. NT-4 mRNA expression in the Xenopus laevis ovary. Ovar~
from adult Xenopus laevis was sectionad in a cryostat (14 m thick
sections) and the scctions were then hybridized to the indicated 48-mer
1 0
wo 93/25684 ~ ~ ~ 7 7 ~ ~ PCr/US93/05672
oli~onucleotides labeled with 35S-dATP using termina! deoxynucleotidyl
transferase.
(A) Hybridization using a Xenopus NT-4 m RNA specific
oligonucleotide with the sequence
5 CCCACM(~ I I G I I ~GCATCTATGGTCAGAGCCCTCACATMGA(; ~ G
C3'. (SEQ ID NO:95)
(B) Hybridization using a control oligonucleotide of similar length
and G+C content. After hybridization, sèctions were washed in 1x SSC at
55C followed by exposure to X-ray film for 10 days. Shown in the figure
are photographs of the developed X-ray films. Note the intense labeling
over many small cells with the NT-4 probe and the absence of labeling with
the control probe. Arrows point at large (stage Vl) oocytes which are not
labeled with either of the two probes. Scale bar, 2 mm.
FIGURE 10. Bright-field illumination of emulsionautoradiographs
showing NT~ mRNA expressing oocytes in the Xenopus ovary. Sections
hybridized to the Xenopus NT-4 mRNA specific (A,B) or control (C) probe
as described in FIG. 9 were coated with Kodak NTB2 emulsion, exposed for
5 weeks, developed and lightly counlersldined with cresyl violet This figure
shows bright-field photomicrographs of the developed sections. Note in
panel A the i"lense NT-4 mRNA labeli.19 over small size oocytes (stages I
and ll) and the absence of labeling over large size (stages V and Vl)
oocytes. Panel B shows a higher magnification of the boxed in area in panel
A. Note the intense labeling of the c~iloplas", of the stage ll oocytes shown
in the picture.
2 5 (C) No labeling can be seen using the control probe.
Abbreviations: n, nuc'~us; fc, follicle cells; pl, pigmented layer. Scale bar inA,50 m;inBandC,15 m.
FK3URE 11. Levels of NT-4 mRNA in oocytes at different stages of
oogenesis. Emulsion autoradiographs (shown in figure 10) of sections
hyLridi~ed with the Xenopus NT-4 mRNA specific probe were used to count
WO 93/25684 PCI/US93/05672
e number of grains over an area unit. The area unit chosen was abo~
one hundredth of a stage I oocyte. Fifteen area units were analyzed in 1 0
different oocytes of the il~d;c~ted stages. Error bars show S.D.
FIGURE12. Northern blot analysis of NT~ mRNA expression during
oogenesis in Xenopus laevis. Ovaries from two adult Xenopus laevis were
dissected out and treated with coîlagenase to remove follicle cells and
release the oocytes. The oocytes were then grouped in the indicated
groups following the stages described by Dumont, 1972, J. Morphol. 136:
153-180. Total ovary and the released follicle cells were also included in the
1 0 analysis. Total cellular RNA was then prepared and a 40 g/slot of RNA
was electrophoresed in a formaldehyde-containing 1% agarose gel. This
was blotted onto a nitrocellulose filter and hybridized to a 600bp Hincll
fragment from the 3'exon of the Xenopus NT-4 gene. The filter was washed
at high stringency and exposed for five days to a X-ray film. Note the
1 5 marked decreased in the level of NT-4 mRNA in stages V and Vl oocytes.
FIGURE 13. (A) The xNT-4 partial amino acid sequence (SEQ ID
NO:51) indicating positions where degenerate oligonucleotides were
synthesized and utilized to prime the amplification of human and rat
genomic DNA via the polymerase chain reaction. Arrows indicate
2 0 oligonucleotides representing sense and antisense degenerate
oligonucleotides. A set of deganerate oligonucleotides to primer 2Z
represent amino acids 184-189 of rBDNF (SEQ ID NO:52). The partial
Xenopus NT-4 amino acid sequence represenled is from amino acid 167 -
amino acid 223, as desc~ibed in Figure 4, supra.
(B) Degenerate oligonucleotides used for cloning of human and rat
NT~. Oligonucleotide 3Z in Figure 13 is comprised of a mixtura of 3Z and
3Z' in order to allow for the degeneracy of the sarin~ codon. 2Y (SEQ ID
NO:53), 2Z (SEQ ID NO:54), 3Y (SEQ ID NO:55), 3Z (SEQ ID NO:56), (SEQ ID
NO:57) and 4Z (SEQ ID NO:58). (C) Cloning tails for degenerate
3 0 oligonlJol~otides 3'(SEQ ID NO:59) and 5'(SEQ ID NO:60).
W O 93/25684 ~ 1 ~ 7~ ~ ~ PC~r/US93/05672
FIGURE 14. DNA sequence of the isolated fragment encoding a
portion of rat NT-4(SEQ ID NO:61). The predicted open reading frame for
the peptide encoded by the rNT-4 nucleic acid fragment is represented by
the single letter code (SEQ ID NO:62). Sequence inside brackets is part of
PCR primer.
FIGURE 15. DNA sequence of the isolated fragment encoding a
portion of human NT-4 (SEQ ID NO:63). The predicted open reading frame
for the peptide encoded by the hNT-4 nucleic acid fragment is represented
by the single letter code. (SEQ ID NO:64) Sequence inside brackets is part
1 0 of PCR primer.
FIGURE 16. Alignment of amino acid sequences deduced from
representative neurotrophins. Amino acids are indicated using the single
letter code. Identical amino acids are indicated with dots. Dashed lines
indicate a 7 amino acid insertion within the conserved region of both rNT-4
1 5 (SEQ ID NC):62) and hNT-4 (SEQ ID NO:64). xNT-4 (SEQ ID NO:65), rNGF
(SEQ ID NO:66), rBDNF (SEQ ID NO:67), rNT-3 (SEQ ID NO:68). x=Xenopus,
r=rat, h=human. Sequence inside brackets is part of PCR primer.
Fl(~JRE 17. (A) DNA sequence of an isolated fragment encoding a
portion of human NT~ (SEQ ID NO:69). The predicted peptide encoded by
the 192 bp hNT-4 nucleic acid fragment is represented by the single letter
code (SEQ ID NO:70). Sequence inside brackets is part of PCR primer.
(B) Oligonucleolide sequence of the 5'-end primer, termed hNT4-5"
containing a sequence (SEQ ID NO:71) encodi.-g ETRCKA (SEQ ID NO:72)],
used in the primary amplification of human genomic DNA along with the 3'-
end primer, termed 4Z (SEQ ID NO:58) [containing a nucleotide sequence
encoding WIRIDTl.
(C) Oligonucleotide sequence of the 5'-end primer used to amplify the
primary PCR reaction product. The primer, termed hNT4-5"' [containing a
sequence (SEQ ID NO:73) encoding DNAEEG (SEQ ID NO:74)1 was utilized
3 0 with the 3' primer, 4Z (SEQ ID NO:58), to obtain a fragment of 162 bp (plus
wog3/j~g~ ~ PCl[/US93/05672
bp of cloning tail). The 162 bp PCR fragment was then utilized in a patll
PCR reaction using our previously utilized upstream PCR fragment (termed
2YZ3Z) to generate the single fragment of 192 bp plus cloning tail shown in
(A). Additional 3' extended nucleic acid sequence information was obtained
following the subcloning and sequencing of this fragment.
FIGURE 18. DNA sequence of the portion of the isolated human
genomic phage clone 7-2 encoding human NT-4 (SEQ ID NO:75). The
predicted hNT-4 protein encoded by the genomic clone 7-2 is represented
by the one-letter symbols for amino acids (SEQ ID NO:76). The boxed
region represents the predicted cleavage site of the hNT-4 preprotein.
Arrows indicate conserved residues in the presequence. The underlined
region (N-R-S) represents a consensus sequence for n-glycosylation. The
circled region represents the initiating methionine. The splice acceptor site isIocated at base pair 461-462 (AG) of SEQ ID NO:75, represenLing the 3'-end
of the intron.
FIGURE 19. Alignment of amino acid sequences deduced from
representative neurotrophins (SEQ ID NOS. 77-92) Amino acids are
indicated using the single letter code. Amino acids identical to those
encoded by the human genomic phage clone 7-2 (SEQ ID NO:77) are
indicated with an asterisk. Dashed lines represent breaks in homologous
amino acids as compared to the protein encoded by SEQ ID NO:77.
FIGURE20. DNA sequence of the isolated fragment encoding a
portion of the human genomic phage clone, 2-1 (SEQ ID NO:93). The
predicted open reading frame for the peptide encoded by the isolated
nucleic acid fragment is represented by the single letter code (SEQ ID
NO:94). URE 21. DNA sequence of the isolated fragment encoding a portion
of the human genomic phage clone, 4-2 (SEQ ID NO:116). The predicted
open reading frame for the peptide encoded by the isolated nucleic acid
fragment is represented by the single letter code (SEQ ID NO:117).
1 4
WO 93/25684 21377 Q g PCI'/US93/05672
FIGURE22. Northern blot analysis of human NT4 mRNA ex~.ression.Tissue specific mRNA from human was purchased from Clontech. RNA's (10
g) were fractionated by electrophoresis through a 1 % agarose-
formaldehyde gel followed by capillary transfer to a nylon membrane
5 (MagnaGraph, Micron Separations Inc.) with 10X SSC (pH 7). RNAs were
UV-cross-linked to the membranes by exposure to ultraviolet light
(Stratlinker, Stratagene, Inc.) and hybridized at 6~C with the radiolabeled
probe (a 680bp Xho1-Not1 fragment containing the complete coding region
of HG7-2 NT-4 (see Example Section 9, infra) in the presence of 0.5 M
1 0 NaPO4 (pH 7), 1% bovine serum albumin (Fraction V, Sigma), 7% SDS, 1 mM
EDTA (Mahmoudi and Lin, 1989, Biotechniques 7:331-333), and 100 g/ml
sonicated, denatured salmon sperm DNA. The filter was washed at 65C
with 2X SSC, 0.1% SDS and subjected to autoradiography overnight with
one inlensi~ying screen (Cronex, DuPont) and X-ray film (XAR-5, Kodak) at
1 5 70C. Ethidium bromide staining of the gel demol,slraled that equivalent
levels of total RNA were being assayed for the different samples (as in
Maisonpierre et al., 1990, Science 247:1446-1451).
Lane 1: fetal liver poly(A)+ mRNA; Lane 2: fetal brain poly(A)+
mRNA; Lane 3: proslale poly A+ mRNA; Lane 4: muscle poly(A)+ mRNA;
20 Lane 5: inlesline poly(A)+ mRNA; Lane 6: kidney poly(A)+ mRNA; Lane 7:
liver poly(A)+ mRNA; Lane 8: spleen poly(A)+ mRNA; Lane 9: thymus
poly(A)+ mRNA; Lane 10: ovary poly(A)+ mRNA; Lane 11: testes poly(A)+
mRNA; Lane 12: placenta poly(A)+ mRNA; Lane 13: brain poly(A)+ mRNA;
Lane 14: brain total RNA.
FIGURE23. COSsupernatants from transfected cell lines;Q1 (pCMX-
HG7-2Q), N7 (pCMX-hNT3/hNT4) and X1 (pCMX-xNT4/hNT4) were tested in
volumes of 10 I, 50 1 and 250 I for neurite promoting activity in DRG
ex~lanls. A supernatant from a mock transfected COS oell line was utilized
as a control.
WO 93/25684 ,~ 9~ . ~ PCI/US93/05672
FIGURE 24. COS supernatants from Q1 (pCMX-HG7-2Q). and--
(pCMX-HG7-2M) cell lines were tested for their survival-promoting activity on
DRG associated cells. Volumes tested ranged from 5 I to 250 1 in a total
volume of 2 ml.
FIGURE25. Motor neuron enriched cultures isolated from E14 rat
embryos were treated with two dilutions of COS oell supernatants from the
M cell line (pCMX-HG7-2M). Biological activity was measured by choline
acetyltransferase (CAT) activity as described in Fonnum, 1975, J.
Neurochem. 24:407-409. Both a mock transfected COS cell line (MOC COS)
and an untreated motor neuron (C-NT) are pressnted as controls.
FIGURE26. COS supernatants containing human, rat and xenopus
NT-4 were tested for their ability to induce the tyrosine phosphorylation of
trkA, trkB and trkC.
FIGURE27. (A) Tyrosins phosphorylations of trkB induced by
varying conce"ltdlio,-s of human and xenopus NT-4.
(B) Growth response of NIH3T3 fibroblasts expresssing trkB to
various concentrations of COS supernatants containing human or xenopus
NT-4. A mock transfected COS cell line is presented as a control.
FIGURE28. Tyrosine phosphorylation of trk receptors induced by
varying concenl,alions of purified preparations of NGF, BDNF, NT-3 and NT-
4. (A)Phosphorylation of trkA; (B) Phosphorylation of trkB; (C)
rhosphorylation of trkC.
The effect of varying concentrations of purified pleparalions of NGF,
BDNF, NT-3 and NT-4 on cell growth of NIH3T3 cells expressing (D) trkA;
(E)trkB and (F)trkC.
F~GURE29. The effect of varying concentrations of purified
preparations of NGF, BDNF, NT-3 and NT-4 on parental PC12 cells and PC12
cells expressing trkB. (A)Differentiation as demonstrated by neurite
eAlensioil.
(B) Number of surviving cells; (C) Tyrosine pl,ospho~lation assays.
1 6
W O 93/25684 ~ ~ 3 7 7 9 ~ PC~r/US93/05672
FK3URE 30. (A) Crosslinking of iodinated NGF to PC12 c~lls and rat
postnatal day 7 striatal homogenates and competition with cold
neurotrophins.
(B) Crosslinking of iodinated BDNF to3T3 trkB cells and rat postnatal day 7
5 cortex and competition with cold neurotrophins. (C) Crosslinking of iodinated
NT-4 to 3T3 trkB cells and rat postnatal day 7 cortex and hippocampus
and competition with cold neurotrophins.
FIGURE 31. Cell survival of embryonic E14 rat DRG explants in
response to increasing concentrations of human NT-4.
FIGURE 32. Expl~ssion of trkB and trkC mRNA in ganglionic neurons
surviving in the presence of NT-4.
FIGURE33. (A) Induction of fos mRNA in hippocampi from D18 rat
embryos by purified neurotrophins.
(B) rl,osphorylation of trk in hippocampi from D18 rat embryos by
1 5 purified neu,ultophins.
FIGURE34. (A) The effect of NT-4 on the number of calbindin-
immunopositive cells in l-ippor~."pal cultures.
(B) The effect of NT-4 and BDNF on the number of
acetylcholinesterase-positive cells in h;ppoc~npal cultures.
2 0 FIGURE 35. The dose related effect of purified human NT-4 on
choline acetyltransferase activity in cultures of basal forebrain neurons.
FIGURE 36. The effect of increasing concantrations of purified
human NT-4 on tyrosine hydroxylase positive dopaminergic neurons of the
rat embryonic substantia nigra. (A) NT-4 added on day 1; (B) NT-4 added
on days 1 4 and 7.
FIGURE37. Effect of NT-4 treatment on calbindin-immunoreactive
neurons in D17 striatal cultures at 8 days in vitro.
FIGURE38. Effect of NT-4 on high-affinity GABA uptake in E17
striatal cultures at 8 days in vitro.
W O 93/25684 ~ ~ PC~r/US93/05672
F~URE39. Effect of in~ as;ng concentrations of purified human N--
4 on choline acetyltransferase activity in E14 rat motor neurons.
FIGURE40. Effect of CNTF or NT-3 alone or in combination with NT-
4 on choline acetyltransferase activity in E14 rat motor neurons.
FIGURE41. Immunocytochemical staining of GABAergic neurons.
Cells were prepared as described and plated at 50,000 cells/cm2 in 35 mm
culture dishes. After 3, 6 and 10 days (41A, 41B and 41C), cultures were
processed for immunocytochemical staining using a polyclonal antiserum
directed against recombinant feline GAD (courtesty Dr. A. Tobin (UCLA, CA).
41 D: control indicating lack of staining in 7 day cultures in the absence of
antibody. Scale bar, 10011M.
FIGURE42. Effect of neurotrophins on GAD activity in mesencephalic
cultures. Cells were prepared as described above and plated at ~0,000
cells/cm2 in 35 mm dishes. Upon media change after the initial attachment
period, cultures were exposed to increasing concer,l,dlions of either BDNF,
NT-3, or NT-4 (n=5 per group). All cultures were maintained for 7 days in
vitro, and were than processed for the measurement of GAD as described.
GAD activity is expressed as a percentaga of the control value determined
in untreated cultures. The baseline GAD activity in untreated control
samples was calculated as 6.23 +/-0.39 pmol/120 min/llg protein. ~,
p~0.01; ~p<0.05 compared to control using students t test.
FIGURE43. Effect of neurotrophins on high affinity GABA uptake.
Cells were ~,fepared as described above and plated at 50,000 cells/cm2 in
35 mm dishes. After the initial attachment period, the culture media was
changed to a serum-free formulation, and BDNF, NT-3 or NT-4 were added
in incleasi,)g cGncenl,alions. After 7 days, cultures were processed for the
measurement of high-affinity GABA uptake as described. 43A: dose
response curves for BDNF, NT-3 and NT-4. The GABA uptake activities are
expressed as a percentage of the GABA uptake determined in control
cultures. 43B: GABA uptake activity detcrmined in cultures which were
1 8
WO 93/25684 ~ I ~ 7 7 ~ ! p~/US93/05672
-
mainlained in the presence of either 25 ng/ml BDNF, 10nglml NT-3, 2.5
ng/ml NT~, 50 ng/ml NGF, or combinations of BDNF and NT-3 or NT-4 at
these same concentrations. The baseline GABA uptake activity in untreated
control cultures was calculated as 41,3581/- 2161 cpm/15 min/dish.
FIGURE44. BDNF, NT-3 and NT-4 increase the GABA content of
mesencephalic cultures. Cultures were prepared as describ~d, and plated at
50,000 cells/cm2 into 35 mm dishes. Upon change to serum-free media,
BDNF(50 ng/ml), NT-3 (10 ng/ml), NT-4 (2.5 ng/ml) or NGF(50 ng/ml)
were added (n=6 per group). Cultures were mainained for 7 days before
1 0 being p,ucessed for GABA determination as described. The GABA content isexpressed as pg/~Lg protein. Data are the mean +/- sem ~, p<0.05
compared to control in students t test.
FIGURE45 Localization of trkB and trkC mRNA in adult rat
subsl~nlia nigra. Dark field photomicrographs of coronal sections of adult
1 5 rat brain showing the autoradiographic localization of hybridization signal
to mRNA encoding trkB (45B, 45C) and trkC (45D, 45F). 45A: schematic
illustration of a cross section of the area of midbrain which is representative
of the brain sectiol)s in panels B-F. At low may~ c~lion (45B), trkB mRNA
is detected in the ventral tegmental area (VTA) and medial s. nigra (SN).
45C: higher magnification of the righth hemisphere, ventral aspect of the
tissue section shown in B. 45D: corresponding tissue section to that in C,
hybridized to trkC cRNA. 45E, 45F: high magnification pair of matching
bright field/dark field photomicrographs of the tissue section in D, showing
the autoradiographic localization of trkC hybridization to large perikarya,
presumed to be neurons. Scale bar=3125 ~Lm for B, 2185 llm for C and D,
and 546 llm for E and F.
FIGURE46. Northern blot analysis of TrkB and TrkC mRNA in
cultures derived from E14 ventral mesencepl,alon. Cells were plated at
50,000 cells/cm2 into 60 mm dishes. After maintainence in serum-free
conditions for 72 hours, BDNF (50 ng/ml) or NT-3 (25 ng/ml) was added
wo 93,25684~q~1 g~ PCl/US93/05672
and cultures maintained for 5, 16, 24 or 29 hours. Northern blots prepar~
from the cultures were probed for TrkB or TrkC m RNA. 46A:
autoradiogram of a blot following hybridization with the TrkB probe;
C=control, B=BDNF, TB=total adult rat brain RNA; 46B: the corresponding
(to 46A) ethidium bromide stained gel; 46C: autoradiogram of a blot
folowing hybridization with the TrkC probe; C=control, N=NT-3, TB=total
brain RNA; 46D corresponding (to 46C) ethidium bromide stained gel.
5. DErAILED DESCRIPTION OF lHE INVENTION
1 0
The present invention provides for NT~ genes and proteins. It is
based, at least in part, on the cloning, characterization, and expression of
the NT~ gene.
In particular, the present invention provides for recombinant nudeic
1 5 acid molecules that encode NT-4. Such molecules comprise a sequence
su~st~ntially as sst forth in Figure 1 (SEQ ID NO:1) for viper, Figure 1 (SEQ
ID NO:2), Figure 4 (SEQ ID NO:43) or Figure 8 (SEQ ID NO:49) for Xenopus
NT-4, Figure 14 (SEQ ID NO:61) for rat NT-4, Figure 15 (SEQ ID NO:63),
Figure 17 (SEQ ID NO:69) or Figure 18 (SEQ ID NO:75) for human NT-4,
2 0 Figure 20 (SEQ ID NO:93) and Figure 21 (SEQ ID NO:116) for a human NT-4
like sequence, or a sequence that is at least about seventy percent
homologous to any such sequence, in which homology refers to sequence
identity (e.g. a sequence that is 70 percen~ homologous to a second
sequence shares 70 percent of the same nucleotide residues with the
secol,d sequence).
In a particular aspect the present invention detailed in Example
Section 8 and Figure 15 (SEQ ID NO:63, SEQ ID NO:64) herein, the nucleotide
and amino acid sequence for a portion of a human neurolloph;n molecule is
determin~d. In another aspect of the present inv~ntion detailed in Example
Section 9 and Figure 17 (SEQ ID NO:69, SEQ ID N 0:70) and Figure 18 (SEQ
wo 93/25684 2 ~ ~ 7 ~ ~ 9 PCr/USs3/05672
ID NO:7~, SEQ ID NO:76) herein, the nucleotide and amino acid sequence for
the entire human neurotrophin molecule is determined. In another aspect of
the present invention detailed in Example Section 9 and Figure 20 (SEQ ID
NO:93, SEQ ID NO:94) and Figure 21 (SEQ ID NO:116, SEQ ID NO:117)
herein, the nucleotide and amino acid sequence for a portion of a human
genomic phage clones, 2-1 and 4-2, respectively, which are similar but not
identical to the nucleotide and amino acid sequence described in Figure 18
(SEQ ID NO:75), are detailed. While such human neurotrophin molecule is
referred to herein as human neurotrophin-4, it should be understood that
such a molecule may be the human homologue of the Xenopus neurotrophin-
4 described herein, or alternatively, a distinct yet homologous neurotrophin
molecule. Similarly, the molecula referred to herein as rat NT-4 may be the
rat homologue of NT-4, or alternatively, a distinct yet homologous
neurotrophin molecule. The methods and compositions of the present
invention do not depend on any single nomenclature.
The present invention also providas for subsPntially purified NT-4
protein or peptide molecules Such molecules may comprise a sequence
sui,~ ,lially as set forth in Figure 2, (SEQ ID NO:1 and SEQ ID NO:2), Figure
4 (SEQ ID NO:44) Figure 8 (SEQ ID NO:50), Figure 14 (SEQ ID NO:62), Figure
15 (SEQ ID NO:64) Figure 17 (SEQ ID NO:70), Figure 18 (SEQ ID NO:76),
Figure 20 (SEQ ID NO:94) and Figure 21 (SEQ ID NO:117) for NT-4, or a
sequence that is at least about seventy percent homologous to any such
sequence. In Ad~lilional nonlimiting specific embodiments of the invention, a
substantially purified protein or peptide comprises the sequence
KCN~il IH (SEQ ID NO:96). In another embodiment of the invention, a
subst~ntially purified peptide or protein comprises the sequence H~VD
(SEQ ID NO:97). In yet another embodiment of the invention, a substantially
purified peptids or protein comprises the sequence KQWIS (SEQ ID NO:98).
In a further embodiment of the invention, a slJbslznlially purified peptide or
3 0 protein comprises the sequence KQSWR (SEQ ID NO:99). In yet another
W0 93/25684 ~3~ 9 PCI/US93/05672
em iment of the invention, a substantially purified peptide or prote
comprises the sequenceGK~(GGG (SEQID NO:100), where X represents one
of the set of 20 amino acids. In a related embodiment of the invention, a
subsPntially purified peptide or protein comprises the sequence GPGVGGG
(SEQ ID NO:101) orGPGAGGG (SEQ ID NO:102). In a further embodiment of
the invention, a substantially purified peptide or protein comprises the
sequence ESAGE(SEQ ID NO:103). In yet a further embodiment of the
invention, a substantially purified peptide or protein comprises the sequence
DNAEE(SEQID NO:104).
1 0 The proteins and peptides of the invention may be produced by
chemical synthesis using standard techniques or may be produced using the
NT~-encoding nucleic acid molecules of the invention, using prokaryotic or
eukaryotic ex,uression systems known to one skilled in the art, such as those
described in PCT application PCT/US90/04916, filed August 29, 1990,
published as WO 91/03569, which is incorporated by reference in its entirety
herein, or as exempiified infra (see Section 6.2.4., infra, and Figure 5) for
transient e~cpression in COS cells.
The present invention also provides for the use of NT-4 in promoting
the growth and/or survival of cells of the nervous system, in particular, but
not limited to, dopaminergic neurons, cholinergic neurons, sensory neurons,
striatal cells, cells of the cortex, striatum, hippocampus, cerebellum,
olfactory bulbs, periaqueductal gray, raphe nucle, locus coeruleus, dorsal
root ganglion, neural placode derivatives, sympathetic neurons and upper
and lower motor neurons.
The presenl invention also provides for portions of NT-4 nucleic acid
or amino acid sequence, suL:,lArllially as set forth for NT-4 in Figure 1, 2, 4,8, 14, 15, 17, 18, 20 or 21 (SEQ ID NO's listed, supra) that are not iden~ical
to portions of BDNF, NGF, or NT-3 of sul~st~,lially th2 same size.
The prssent invention further provides for a eukaryotic or
prokaryotic cell that contains r~combinant nucleic acid that encodes NT-4
WO 93/25684 ~1~3 1'19 9 PCr/US93/05672
and that expresses recombinant NT-4 protein. In a specific embodiment the
cell is a eukaryotic cell, such as a COS cell. Accordingly, the present
invention also provides for recombinant NT-4 protein or peptide that is
produced by inserting recombinant nucleic acid encoding NT-4 into a cell
(e.g., by transfection, transduction, electroporation, microinjection, etc.)
under conditions which permit expression of NT-4 and then isolating NT-4
from the cell.
In addition, the present invention provides for molecules produced by
PCR using, for example, the following oligonucleotides as primers:
5'CAGTAI I I I IACGAMCC(SEQ ID NO:105) and3~ilCI 1~31 1 IGGCI I IACA
(SEQ ID NO:106) for human NT-4 and 5'CAGTA I I I I I ACGAGACG (SEQ ID
NO:107)and3'CGATTGlllGGC~llACA(SEQlDNO:108) for rat NT-4, and
using any suitable genomic or cDNA as template. In a specific embodiment
of the invention, these primers may be used in conjunction with human cDNA
as template to produce fragments of the human NT-4 gene that are
suitable for cloning.
The production and use of derivatives, analogues, and peptides
related to NT-4 are also envisioned, and within the scope of the present
invention. Such derivatives, analogues, or peptides which have the desired
2 0 neurotrophic activity, immunogenicity or antigenicity can be used, for
example therapeutically, or in immunoassays, for immunization, etc.
Derivatives, analogues, or peptides related to NT-4 can be tested for the
desired activity by procedures known in the art.
The NT-4 related derivatives, analogues, and peptides of the invention
can be produced by various methods known in the art. The manipulations
which result in their production can occur at the gene or protein level. For
example, the cloned NT-4 gene can be modified by any of numerous
strategies known in the art (Maniatis, T., 1982, Molecul~r Cloning, A
Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor,
New York). The NT-4 sequence can be cleaved at appropriate sites with
WO 93/25684 PCI`/US93/05672
re~tf~ e~ndonuclease(s), followed by further enzymatic modification
desired, isolated, and ligated in vitro. In ths production of the gene
~ncG13ing a derivative, analogue, or peptide related to NT-4, care should be
taken to ensure that the modified gene remains within the same
5 translational reading frame as NT-4, uninterrupted by translational stop
signals, in the gene region where the desired NT-4-specific activity is
encoded.
Additionally, the NT-4 gene can be mutated in vitro or in vivo, to
create and/or destroy translation, initiation, and/or termination sequences,
10 or to create variations in coding regions and/or form new restriction
endonuclease sites or destroy preexisting ones, to facilitate further in vitro
modification. Any technique for mutagenesis known in the art can be used,
including but not limited to, in vitro site-directed mutagenesis (Hutchinson,
C., et al., 1978, J. Biol. Chem. 253:6551), use of TAB~ linkers (Pharmacia),
1 5 etc.
As disa~ssed infra, the prepro or mature coding region of NT-4 may
be utilized to construct neu,Gl,ophin based chimeric genes. For example,
neurotrophin genes, including but not limited to NGF, BDNF and NT-3, can
provide the prepro region for construction of neurotrophin prepro/NT-4
20 mature coding region chimeric genes.
Manir~ tions of the NT-4 sequence may also be made at the protein
level. Any of numerous chemical modifications may be carried out by known
techniques, including but not limited to specific chemical cleavage by
cyanogen bromide, trypsin, chymotrypsin, papain, V8 protease, NaBH4,
25 acetylation, formylation, oxidation, reduction, metabolic synthesis in the
presenc~ of tunicamycin, etc.
In addition, analogues and peptides related to NT-4 can be chemically
synthesi~ed. For 2xample, a peptide corresponding to a portion of NT-4
which me~i~tes ths desired neufol,ophic activity can be synthesized by use
3 0 of a peptide synthesizer.
24
W O 93/25684 ~ 1 ~ 7~ ~ ~ PC~r/US93/05672
O The present invention further provides for a method of treating
fertility disorders related to ovarian/oocyte dysfunction. As shown in the
examples infra, in particular, Section 7,NT-4is involved in the maturation of
oocytss. The discussicn of Section 7.3 demonstrates that NT-4is produced
by oocytes, is concentrated in immature rather than mature oocytes, and
appears to play a roie in oogenesis. The putative function of NT-4 protein in
the ovary appears to be coupled to events occurring in the pre-vitellogenic
and early/mid vitellogenic oocyte.
It has been established that other members of the BDNF/NGF/NT-3
gene family, in particular NGF, are involved in meiotic maturation (Nebrada
et al., 1991, Science, 252:558-563). Rec~lJsQ NT-4 has exhibited properties
similar to NGF (see Section 6, infra) it may be used as a factor involved in
the regulation of oocyte development. These properties of NT-4 can be
exploited to provide a method for treating infartility disorders and/or other
ovarian dysfunctions ~csocieted with oogenesis.
Therefore, in accordance with the invention a method of treating
infertility disorders and/or other ovarian dysfunctions comprising
administering a therapeutically effective amount of NT~ or an NT-4 related
peptide in a pharmaceutically effective carrier is provided. A tharapeutically
effective amount is one which induces proper maturation of an oocyte
and/or ovulation. For example, a therapeutically effective dose may be one
sufficient to maintain circulating serum levels of NT-4 at a concentration of
from about 1 to 100 x 10-10M. Establishing additional effective doses is
within the purview of one skilled in the art.
2 5 In various embodiments of the invention, NT-4 protein, peptide
fragments or derivatives can be administersd to patiants in whom the
nervous system has been damaged by trauma, surgery, ischemia, i"~ection,
metabolic dise~-~e, nutritional dt:fic;G.-cy, malignancy, or toxic agents. The
invention in particular can be used to treat conditions in which damage has
30 occurred to neurons in the basal forebrain, hippocampus or striatum. In
WO 93/25684~ 9~ PCr/US93/05672
addition, it can be used to treat conditions in which damage
degeneration has occurred to spinal sensory neurons, cranial sensory
neurons involved in hearing, taste, vision, balance, etc., motor neurons or
retinal cells, by administering effective~therapeutic amounts of NT~ protein
or peptide fragments or derivatives. Such uses include, but are not limited
to, treatment of retinal detachment, age related or other maculopathies,
photic retinopathy, surgery-induced retinopathy, retinopathy of prematurity,
viral retinopathy, uvetis, ischemic retinopathy due to venous or arterial
occlusion or other vascular disorders, retinopathy due to trauma or
penetrating lesions of the eye, peripheral vitreoretinopathy or inherited
retinal degeneration.
In various specific embodiments of the invention, NT~ can be locally
administered to sensory neurons which have been severed, including, but not
limited to, neurons in dorsal root ganglia or in the retina. It may be
desirable to administer the NT-4-related peptides or NT-4 protein by
adsorption onto a membrane, e.g. a silastic membrane, that could be
implanted in the proximity of the severed nerve. The present invention can
also be used for example in hastening the recovery of patients suffering
from peripheral neu-opall,ies.
In further embodiments of the invention, NT-4 protein or peptide
fragments or derivatives derived therefrom, can be used to treat congenital
conditions or neurodegenerative disorders, including, but not limited to,
Alzheimer's dise~e, Parhil)son's ~ise~ce~ P~hi"son-Plus syndromes (in which
P~klnsonian symptoms result from degeneration of dopaminergic neurons),
such as Progressive Supranuclear Palsy (Steele-Richardson-Olszewski
Syndrome), Olivoponto- cerebellar Atrophy (OPCA), Shy-Drager Syndrome
(multiple systems atrophy), and Guamanian Pa"~ sonism dementia complex,
and Huntington's chorea; in particular, the invention can be used to treat
congenital or neurodegenerative disorders associated with sensory nerve
dysfunction and degenerative dise~ces of the retina. For example, the NT-4
WO 93/25684 2 ~ PCI`/US93/05672
protein, or peptide fragments, or derivatives of the invention can be used in
the treatment of hereditary spastic paraplegia with retinal degeneration
(Kjellin and Barnard-Scholz syndromes), retinitis pigmentosa, Stargardt
dise~-ce~ Usher syndrome (retinitis pigmentosa with congenital hearing loss),
5 and Refsum syndrome (retinitis pigmentosa, hereditary hearing loss, and
polyneuropathy), to name but a few. It is possible that a defect in NT-4
synthesis or responsiveness may be the underlying etiology for syndromes
characterized by a combination of retinal degeneration and other sensory
dysfunction.
In a specific embodiment of the invention, administration of NT-4
protein, or peptide fragments or derivatives derived therefrom, can be used
in conjunction with surgical implantation of tissue in the treatment of
Alzheimer's ~lise~se and/or P~kinson's ~lise~e. As ~iscussed in Section 18
NT4 may be used to promote the survival of dopaminergic neurons
15 of the subst~ntia nigra in a dose-dependen~ manner, supporting the use of
NT-4 in the treatment of disorders of CNS dopaminergic neurons, induding,
but not limited to, P~,hil1son's dice~ce. In addition, NT-4 has been observed
to sustain the survival of CNS cholinergic neurons (Section 17) and, in
particular, basal forebrain cholinergic nsurons, indicating that NT-4 may be
20 useful in the treatment of disorders involving cholinergic neurons, induding, but not limited to Alzheimer's disease. It has been shown that
approximately 35% of patients with Parkinson's disease suffer from
Alzheimer-type dementia; NT-4 produced according to the invention may
prove to be a useful single agent therapy for this disease complex.
2 5 Similarly, NT-4 produced according to tha invention may be used
therapeutically to treat Alzheimer's dise~ce in conjunction with Down's
Syndrome. NT-4 producsd accordin~ to the invention can be used in the
treatment of a variety of dementias as well as congenital learning disorders.
In another specific embodiment of the invention, the administration of
30 NT-4 protein or peptide fragments or derivatives derived therefrom can be
W 0 93/25684 ~ PC~r/US93/05672
used for the treatment of f~ise~es or disorders which involve striatal c~
which include, but are not limited to Huntington~s chorea, striatonigral
degeneration and cerebral palsy. This is based on the disclosure herein
(Section 19) indicating the ability of NT-4 to support striatal cultures, as
5 indicated by an increase in calbindin immunoreactivity and a high affinity
uptake of GABA. A dramatic decrease in calbindin and calbindin mRNA has
bsen detected in the striata of Huntington's chorea patients [Kiyama et al,
Brain Res. 525:209-214 (1990); lacopino et al Proc. Natl. Acad. Sci.
87:4078-4082 (1990)].
1 0 In another embodiment of the invsntion, the administration of NT-4
protein or peptide fragments or derivatives derived therefrom can be used
for the treatment of other diseases or disorders which are related to
damage or degensration of striatal or hippocampal cells. Such dise~es or
disorders may be caused by, for example, stroke, ischemia, hypoglycemia or
1 5 hypoxia.
In yet another embodiment of the invention, NT-4 may be
admin;;,lered in the treatment of epilepsy-related or other seizures. Reduced
Isvels of the inhibitory transmitter GABA are known to be associated with
seizures. For example, high doses of penicillin, which reduce GABA levels, can
20 be used to induce Qxperimsntal focal epilepsy. Adminisration of the GABA
agonist muscimol into the area of the sul,~ rllia nigra has been shown to
markedly suppress motor and limbic seizures (as measured
electrographically) induced by electrical stimulation. McNamara, J. et al.,
1984, J. Neurosci. 4:2410-2417. Accordingly, the present invention
25 contsmplates use of the neurotrophins, including NT-4, to enhance levels of
GABA and thereby prevent the motor mani~e~lalions of seizures.
Effective doses of NT-4 or an NT-4 related peptide formulated ~
suitable pharmacological carriers may be administered by any appropriate
route including but not limited to injection (e.g., intravenous, intraperitoneal,
3 0 intramuscular, subcutaneous, etc.), by absorption through epithelial o r
WO 93/25684 2 1 3 7~ ~ PCI/US93/05672
mucocutaneous linings (e.g., oral mucosa, rectal and intestinal mucosa,
etc.); etc.
In addition, NT-4 or NT-4 peptide may be used in any suitable
pharmacological carrier, linked to a carrier or targeting molecule (e.g.,
5 antibody, hormone, growth factor, etc.) and/or incorporated into
liposomes, microcapsules, and controlled release preparation prior to
adminislfalion in vivo.
Each respective mammalian NT-4 DNA sequence can be utilized as a
32P-labelled probe to isolate a respective genomic and cDNA clone via the
10 procedures outlined in the Materials and Methods portion Section 8, infra.
The rat NT-4 and human NT-4 gene fragments may be utilized directly (as
32P-labelled probes) or indirectly (to deduce a PCR strategy as described
infra) to isolate other mammalian NT-4 genomic and cDNA clones, based on
the unique nature of the 7 amino acid insertion in the rNT-4 and hNT-4
15 coding region, or other unique ~pects of the rat or human NT-4 coding
region.
Any mammalian NT-4 gene isolated via the information disclosed by
the rat and human NT~ sequence may be utilized in, although is not limited
to, the various manipulations rliscussed for Xenopus NT-4. For example, the
20 proteins and peptides of mammalian NT-4, subse~uent to characterization
of the full length gene as rliscussed in Example Section 9, may be produced
using the respective mammalian NT-4 molecules in a prokaryotic or a
eukaryotic ex~.ression system known to one skilled in the art, such as those
described in PCT application PCT/US90/04916, filed August 29, 1990,
published as W091/03569, or as exemplified infra (see Section 6.2.4., supra,
and Figure 5) for transient ex~.ressiol) inCOS cells. Additional functions for
mammalian NT-4, as described infra for Xenopus NT-4, include, but are not
limited to: the promotion of growth and/or survival of cells of the nervous
system, in particular, but not limited to, cells of dorsal root ganglion or
neural placode derivatives (see Section 6.2.4., and Figure 6, for example),
29
WO 93/25684 7~ PCI/US93/05672
treating fertility disorders related to ovarian/oocyte dysfunction (se--
Section 7), the treatment of infertility disorders andtor other ovarian
dysfunction associated with oogenesis (see Section 6), the treatment of
motor neuron diseases (see Section 10), the treatment of an epitheliac
hyperplasia such as benign prostatic hypertrophy (see Section 10), the
treatment of impotence as related to prostate gland function (see Section
10) and, therefore, the therapeutically effective amounts of mammalian NT-
4 for the treatment of said disorders as formulated in suitable
pharmacological carriers to provide a pharmaceutical composition may be
administered by any appropriate route including but not limited to injection
(e.g., intravenous, intraperitoneal, intramusuJl~r, subcutaneous, etc.), by
absorption through epithelial or mucocutaneous linings (e.g., oral mucosa,
rectal and inles~inal mucosa, etc.); etc.
In addition, rat, human or other mammalian NT-4 or NT-4 peptide
may be used in any suitable pharmacological carrier, linked to a carrier or
targeting molecule (e.g., antibody, hormone, growth factor, etc.) and/or
incorporated into liposomes, microcapsules, and controlled release
preparation prior to administration in vivo.
In addition, the present invention, which relates to nucleic acids
encoding NT~ and to proteins, peptide fragments, or derivatives produced
therefrom, as well as antibodies directed against NT-4 protein, peptides, or
derivatives, may be utilized to diagnose or monitor the progression of
dise~ces and disorders of the nervous system which are associated with
alterations in the pattern of NT-4 exl,-ession. Such alterations can be a
decrease or increase relative to that in normal patients, preferably, or in
other samples taken from the patient, or in samples from the same patient
taken at an ~arlier time.
In various embodiments of the invention, NT-4 genes and related
nucleic acid sequences and subsequences, including complementary
sequences, may be used in diagnostic hybridization assays. The NT-4
W O 93/25684 ~ 7 ~ ~ PC~r/US93/05672
nucleic acid sequences, or subsequences thereof comprising about 15
nucleotides, can be used as hybridization probes. Hybridization assays can
be used to detect, prognose, diagnose, or monitor conditions, disorders, or
dise~-se states ~csoci~ted with changes in NT-4 levels. For example, the
5 data presented in Example Section 10 discloses tissue specific expression of
human NT-4 in skeletal muscle as well as the prostate gland, thymus and
testes. The level of expression of human NT4 in the muscle tissue may be
indicative of the presence or absence of neuronal degradation. Therefore,
poly(A)+ mRNA or total RNA from a tissue sample of a patient could be
10 assayed for the presence of human NT-4 mRNA in skeletal muscle tissue.
Additionally, the data presented in Example Section 10 discloses tissue
specific ex,uression of NT-4 in the human proslale gland. DNA sequences
encoding NT-4 or a portion thereof, as well as NT-4 protein or a peptide
may be useful as a therapeutic agent to treat proslate ~lise~e.
In a similar method, diagnostic assays can bs immuno~cs~ys. Thus,
antibodies can be used in immunoassays to quantitate the level of NT-4 in a
sample from a patient, in order to detect, prognose, diagnose, or monitor
conditions, disorders, or dise~ce states associated with changes in NT-4
levels.
The immunoassays which can be used include but are not limited to
competitive and non-competitive assay systems using techniques such as
radioimmunoassays, ELISA (enzyme linked immunosorbent assay),
~sandwich~ immunoassays, precipitin reactions, gel diffusion precipitin
reactions, immunodiffusion assays, agglutination assays, complement-
2 5 fixation assays, immunoradiometric assays, fluorescent immunoassays,
protein A immunoassays, and immunoelectrophoresis assays, to name but a
few.
Anti NT-4 antibody fragments or derivatives containing the binding
domain may also be used in such assays.
W093/25684 ?.,~ 9~ PCr/US93/05672
Antibody fragments which contain the idiotype of the molecule can b~
generated by known techniques. For example, such fragments include but
are not limited to: the F(ab')2 fragment which can be produced by pepsin
digestion of the antibody molecule; the Fab' fragments which can be
generated by reducing the disulfide bridges of and the Fab fragments which
can be generated by treating.the antibody molecule with papain and a
reducing agent.
Diagnostic kits are also provided. For example, such a kit can
comprise in a suitable container an NT-4 specific probe. In one embodiment,
the probe is an antibody specific for NT-4. In another embodiment, the
probe is a nucleic acid (molecular probe) capable of hybridizing to an NT-4
nucleic acid sequence. The probe can be detectably labeled; alternatively,
the kit can further comprise a labeled specific binding partner for the probe.
The above-described hybridization assays and immunoassays can
also be used to quantitate NT-4 levels as an indication of therapeutic
efficacy, by comparing the levels in patient samples before and after
treatment of a disorder, particularly, a motor neuron disease.
In an analogous fashion, tha expression of human NT-4 mRNA in
muscle tissue leads to potential methods of treating motor neuron
disorders comprising administering to a patient in need of such treatment
an effective amount of an NT-4 factor to support the survival, growth,
and/or differentiation of motor neurons. Expression of NT-4 mRNA in
human muscle suggests further avenues for diagnosing and treating neuron
disorder~. Retrograde axonal transport of NT-4 has been demonstrated
in both the central and peripheral nervous system (see Section 14, ~f~.)
The specific retrograde transport of NT-4 can be used to indicate whether
neurons are responsive to NT~ in normal or diss~ed states. Therefore,
the pressnt invention provides for a method of diagnosing NT-4 related
motor neuron, central and peripheral nervous system disorders comprising
injecting a detectably labeled NT-4 protein or peptida into a nerve and
WO 93~25684 2 1 ~ ~ 7 ~ ~ PCI/US93/05672
determining whether the labeled NT-4 protein or peptide is retrogradely
transported, in which a failure to be retrogradely transported positively
correlates with lack of responsiveness to NT-4 and indicates the presence
of a nervous system disorder that is NT-4 related. Evaluation of
5 retrograde transport may be performed by any method known in the art,
including but not limited to MRI, CAT, or scintillation scanning. Such methods
may be used to identify the location of a nervous system lesion, as
retrograde transport should subsl~nlially diminish upon reaching the lesion.
The invention further provides kits for such retrograde evaluation
10 comprising in a container a detectably labeled NT-4 protein, derivative or
fragment. Such a label can be a r~d;Q~r,tive isotope, or other label known in
the art.
The present invention may be utilized to treat diseases and disorders
of the nervous system which may be associated with alterations in the
15 pattern of NT-4 ex~.ression or which may benefit from exposure to NT-4 or
anti-NT-4 antibodies (or fragments thereof containing the binding domain).
We show that human NT-4 is expressed in skeletal muscle (See Example
Section 10, infra). Based on this discovery, the invention provides for the
treatment of motor neuron diseases. A wide array of neurological
20 disorders may affect motor neurons. Upper motor neurons, for example,
are predominantly affected by cerebrovascular accidents, neoplasms,
infections and trauma. Lower motor neurons, or anterior horn cells, are
secondarily affected by these processes, but in addition are subject to a
number of d;sorders in which anterior horn cell loss is the primary feature,
25 including amyotrophic lateral sclerosis, infantile and juvenile spinal muscular
atrophy, poliomyelitis and the post-polio syndrome, hereditary motor and
sensory neuropathies, and toxic motor neuropathies (e.g. vincristine). The
disorders of motor neurons which can be traated according to the present
invention include but are not limited to the foregoing. Methods of
30 formulation and adminislfa~ion of NT-4 protein, derivatives, fragments, or
33
W0 93/2568~'3 PCI/US93/05672
antibodies thereto which can be used include but are not limited to thos--
disclosed supra or known in the art.
The invention may also be utilized to treat benign prostatic
hypertrophy (BPH), a common yet poorly understood condition occurring
mostly in males over 50 years of age. The proliferation of the prostrate
during BPH may be induced by a growth factor such as NT-4 through an
autocrine loop phenomenon. Synthesis and excretion of NT-4 would be
followed by transport of NT-4 back into the prostate cell via a specific
receptor on the proslale cell membrane. Autocrine loops have been defined
for various growth factor molsculQs and tumor cell lines. In some cases,
these autocrine loops have been experimentally defined by the use of
antisense approaches for the disruption of the autocrine loop. Therefore, a
therapeutic application of the present invention includes the use of a nucleic
acid anti-sense to human NT-4 or a portion thereof to inhibit translation of
NT-4 mRNA in the prostate, (for procedures which can be used, see
copending U.S. Application Serial No. 07/728,784 filed July 3, 1991 and
incorporated by reference herein in its entirety). For example, a patient
suffering from a prostate localized disease characterized by increased
transcription in prostate tissus of an NT-4 gene relative to that of
transcription levels of the NT-4 gene in the prostate of normal patients
could be administered an effective amount of an oligonucleotide to treat a
prostate disease, preferably benign prostatic hypertrophy. The
oligonlJc'eotide should be at least 6 nucleotides in length, complementary to
a least a portion of the RNA transcript of the NT~ gene and, hence, being
2 5 capable of hybridizing to the NT-4 transcript. Additionally, anti-NT-4
antibod;Qs may be utilized to inhibit bindil)g of NT-4 to its specific receptor
on the proslale cell membrane. A therapeutically effective amount of either
an NT-4 anlisense nucleic acid or an anti-NT-4 antibody may be delivered in
any fashion deso~ibed supra.
34
WO 93/25684 PCr/US93/05672
Th~ invention may also be u~iz7d to treat other prostate relateddysfunctions, specifically impotence. Such a malady may be the direct or
indirect result of in~de~lu~te levels of NT~ in the prostate. Therefore, both
the detection of the dysfunction as well as treating the patient for
impotence via application of a therapeutically effective amount of NT-4
protein or a functional fragment or derivative of NT-4 may be delivered by
any method desc,ibed supra.
The present invention discloses the detection of NT-4 expression in
human thymus tissue. Therefore, the invention may also be utilized to treat
immunological disorders affecting neuromuscular transmission, including but
not limited to myasthenia gravis, an acquired autoimmune disorder
associated with the acetylcholine receptor (AChR) within the postsynaptic
folds at the neuromuscular junction. The disease manifests itself as
weakness and muscular fatigue due to blockage of post-synaptic AChR or
1 5 muscle membranes by binding of antibodies specific to the AChR. (See, e.g.,
Drachman, 1983, Trends Neurosci. 6:446-451). Treatment of such
immunological mediated neurological disorders may include therapeutic
applications of the NT-4 protein or a functional fragment or derivative of
NT-4, delivered by any of the methods desc,ibed supra.
The present invention provides for a method of treating motor
neuron disorders comprising administering, to a patient in need of such
treatment, an effective amount of an NT-4 protein, derivative or peptide
fragment capable of supporting the survival, growth and/or differentiation
of motor neurons as demonstrated in an in vitro culture system.
2 5 In in vitro embodiments, effective amounts of neurotrophic factor
may desirably be determined on a case by case basis, as motor neurons
from di~rerent tissue sources or from different species may exhibit different
sensitivities to neurotrophic factor. For any particular culture, it may be
desirable to construct a dose response curve that correlates neurotrophic
30 factor concentration and motor neuron response. To evaluate motor
WO 93/25684~ 9 PCI/US93/05672
neuron survival, growth, andlor differentiation, one can compare mot--
neurons exposed to an NT-4 protein, derivative or peptide fragment to
motor neurons not exposed to an NT-4 protein, d~rivative or peptid~
fragments, using, for example, vital dyes to ~valuate survival, phase-
contrast microscopy and/or neurofilament stain to measure neurite
sprouting, or techniques thàt measure the bioactivity of motor neuron-
associated compounds, such as choline acetyltransferase (CAT), or any
other methods known in the art. CAT activity may be measured, for
example, by harvesting and Iysing trea~ed and untreated motor neurons in a
1 0 20 mM Tris-HCI (pH 8.6) solution containing about 0.1% Triton X-100,
removing an aliquot of several microliters, and measuring for CAT activity
using, as a substrate, 0.2 ml ~1 - C] acetyl-CoA, 300 mM NaCI, 8 mM
choline bromide, 20 mM EDTA, and 0.1 mM neostigmine in 50 mM NaH2PO4
(pH 7.4) buffer, using the micro-Fonnum procedure as described in Fonnum,
1 5 1975, J. N~ufoche",. 24:407-409, incorporated by reference in its entirety
herein.
In a specific, non-limiting embodiment of the invention, motor neurons
may be prepared, and cultured in vitro, as follows. At Isast a portion of a
spinal cord, preferably obtained from an embryonic organism such as a rat,
may be ~ceptic~lly obtained and separated from the bulb, sensory ganglia,
and adhering meninges. The ventral segments of the cord may then be
isolated, as motor neurons are localized in the ventral (anterior) horns of
the spinal cord. Ventral cord segments may be diced into small pieces and
incubated in about 0.1%trypsin and 0.01% deoxyribonucleasetype 1 in
calcium and magnesium-free phosphate buffered saline (PBS) at 37C for
about 20 minutes. Thc trypsin solution may then b~ removed, and the cslls
may be rinsed and placed in fresh medium, such as 45% Eagle's minimum
esser,lial (MEM), 45% Ham's nutrient mixture F12, 5% heat inactivated fetal
calf serum, 5% heat inactivated horse serum, glutamine (2 mM), penicillin G
(0.5 U/ml), and streptomycin (0.5 g/ml). The tissue may be m~chanically
36
W O 93/25684 ~ 1 3 7 ~ ~ $ PC~r/US93/05672
dissociated by gentle trituration through a Pasteur pipet, and the
supernatants pooled and filtered through a nylon filter (e.g. Nitex, Tetko; 40
m). The filtered cell suspension may then be fractioned using a modification
of the method set forth in Schnaar and Schaffner (1981, J. Neurosci. 1:204-
5 217). All steps are desirably carried out at 4C. Metrizamide may be
dissolved in F12:MEM medium (1:1) and a discontinuous gradient may be
esPhlished that consists of a 18% mel,i~a",ide cushion (e.g. 0.5 ml), 3 ml
of 17% metrizamide, 3 ml of 12% metrizamide, and 3 ml of 8%
metrizamide. The filtered cell suspension (e.g. 2.5 ml) may be layered over
the step gradient and the tube may be centrifuged at 2500 9 for about 15
minutes using a swing-out rotor (e.g. Sorvall HB4). Centrifugation may be
expected to result in three layers of cells: fraction I (at 0-8% interface),
fraction ll (at 8-12% interface) and fraction lll (at 12-17% interface).
Fraction 1, enriched for motor neurons, may be removed in a small volume
15 (e.g. about 1 ml) and rinsed twice with a serum-free defined medium such
as 50% F12 and 50% MEM s~ppl~mented with glutamine (2 mM), insulin (5
g/ml), transferrin (100 g/ml), progesterone (20 nM), putrescine (100 M),
and sodium selenite (30 nM, see B~llenslein and Sato, 1979, Proc. Natl.
Acad. Sci. U.S.A. 76:514-517). Viable cell count may then be obtained by
20 hemocytometer counting in the presence of trypan blue. The motor neuron
enriched cell suspensiG" may then be plated at a density of about 100,000
cells/cm2 in tissue culture wells (preferably 6 mm) precoated with poly L-
~r"ill,ine (e.g. 10 g/ml) and laminin (e.g. 10 g/ml). An NT-4 protein,
derivative or peptide factor may then be added. For example, in specific
25 embodiments, NT-4 may be added to achieve a final concentration of
between about 0.01 and 100 ng/ml, and preferably about 50 ng/ml. The
motor neuron cultures may then be maintained in serum-free defined medium
at 37C in a 95% air/5% CO2 al",osphere at nearly 100% relative humidity.
In a further embodiment of the invention, the NT-4 related
3 0 recombinant nucleic acid sequence, such as co--lained in bacteriophage HG7-
W093/25684 ~ 9~ ` ~ PCI`/US93/05672
2, 7~G4-2, and/or HG2-1, may be utilized to construct chimel--
prepro/mature NT-4 genes. For example, when it is desired to express a
mature NT~ protein, derivative or peptide fragment in vivo or in vitro, one
can fuse the pre-pro region of a distinct neurotrophic gene to the mature
coding region of the NT4 related sequence. The neurotrophic genes which
can provide the prepro region include but are not limited to NGF, BDNF, and
NT-3. Such a chimeric construct may promote increased stability of the
chimeric mRNA transcript in relation to a wild type NT-4 mRNA transcript,
may increase translational efficiency or may generate a more suitable
1 0 template for proteolytic processing to a mature, biologically active
neurotrophin protein or peptide fragment, thus increasing ex~,ression. One
of ordinary skill in the art possesses the requisite knowledge to construct
such chimeric nucleic acid sequences, given the published DNA sequences of
other neurotrophin genes such as NGF (Scott et al., 1983, Nature 302: 538-
540; Ullrichetal., 1983, Nature 303:821-825), BDNF(Leibrock etal., 1989,
Nature 341:149-152) and NT-3 (Hohn et al., 1990, Nature 344:339-341;
Maisonpierre et al., 1990 Science 247:1446-1451; Ernfors et al., 1990, Proc.
Natl. Acad. Sci. USA 87:5454-5458; Rosenthal et al., 1990, Neuron 4:767-
773), as well as guidance as to strategies for generating a fusion junction
(for example, see Darling et al., 1983, Cold Spring Harbor Symposium
Quantative Biology 48:427-434; Edwards et al., 1988, J. Biol. Chem.
263:6810-6815; Suter et al., 1991, EMBO J. 10:2395-2400). in another
embodiment, chimeric constructions fusing the pre-pro region of an NT-4
related recombinant nucleic acid, such as con~ained in bacteriophage HG7-2,
HG4-2 and HG2-1, to the mature regions of other neurotrophins, can also
be used to promote efficient expression of such other neurotrophins, as
liscu~sed supra.
The present invention also provides methods of detecting or
measuring NT-4 activity. As described in Example 12, we have discovered
that trkB is a functional receptor for NT~. Based on this discoYery, the
38
WO 93/25684 2 1 3 7 7 ~ ~ PCI/US93/0~672
invention provides methods for detecting or measuring NT-4 activity
comprising exposing a cell that expresses trkB to a test agent, and
detec~ing or measuring binding of the test agent to trkB, in which specific
binding to trkB positively correlates with NT~ activity in the test agent. In
5 a specific embodiment, the cell that expresses trkB is a transfected cell such as a 3T3 fibroblast, which expresses recombinant trkB, such that the
survival of the cell is dependent upon exposure to neurotrophin-4 or BDNF.
Thus detecting of binding of the test agent can be carried out by observing
the survival of such transfected cells.
1 0
6. EXAMPLE: EVOLUllONARY STUDIES OF THE
NERVE GROWrH FACTOR FAMILY REVEAL A
NOVEL M~RFP~ ABUNDANTLY EX~ .SS~
IN XENOPUS OVARY
1 5
6. 1. MATERIALS AND METHODS
6.1 .1 . DNA PREPARATION
Genomic DNA was isolated by standard procedures (Davis et al.,
1986, ~Basic Methods In Molecular Biology~, Elsevier, New York)) from
human leukocytes and from liver of Sprague-Dawley rat, frog (Xenopus
laevis) and ray (Raja clavata). Genomic DNA was also obtained from
salmon (Salmon) and from the elephant snake (Vipera lebetina). The DNA
was precipitated with ethanol, collected using a glass hook, washed in 80%
ethanol, dried and dissolved in water to a final concentration of 1 mg/ml.
Salrnon DNA (Sigma, St. Louis, MO) was dissolved in water, extracted twice
with phenol and once with chloroform, and precipit~ted with ethanol.
39
W O 93/25684 PC~r/US93/05672
6.1 ~ RASE CHAIN REACTIONS, MOLECULAR
CLONING AND DNA SEQUENCING
Six separate mixtures of 28-mer oligonucleotides representing all
5 possible codons corresponding to the amino acid sequence KQYFYET(SEQ
ID NO:110) (5'-oligonuclaotide) and WRFIRID (SEQ ID NO:111) (3'-
oligonucleotide) (Fig. 1A) were synthesized on an Applied Biosystem A381
DNA synthesizer. The 5' oligonucleotide contained a synthetic EcoRI site and
the 3'-oligonucleotide contained a synthetic Hindlll site (Knoth et al., 1988,
1 0 Nucl. Acids Res. 16:1093; Nunberg et al., 1989, J. Virology 63:3240-3249).
Each mixture of oligonucleotides was then used to prime the amplification of
0.8 9 of genomic DNA using the polymerase chain reaction (PCR) (Taq DNA
polymerase, Promega) (Saiki et al., 1985, Science 230:1350-1354). The PCR
products were restricted with Hindlll and EcoRI, analyzed on a 2% agarose
1 5 gel and cloned into plasmid Bluescript KS+ (Stratagene, La Jolla,CA). The
size of th2 amplified region plus primers is 179 base pairs (bp) for NGFand
182 bp for BDNF and NT-3. As a result of internal EcoRI sites in some
cases, shorter fragments of 144 bp and 95 bp were also isolated. The
cloned DNA fragments were sequenced using the dideoxy nucleotide chain
2 0 t~rmination method (Sanger st al., 1977, Proc. Natl. Acad. Sci. U.S.A.
74:5463-5467) with T7 DNA polymerase (Pharmacia, Upps~l~). Between 2
and 20 independent clones were sequenced for each gene and species, and
altogether mor~ than 200 independent clones were sequenced.
Approximately 2,000,000 clones from a Xenopus genomic library
prepared by insertion of Mbol-digested genomic DNA in the BamHI site of
phasa ~lEMBL-3 were screened using conventional procedures with a 182 bp
PCR fragment of Xenopus NT-4 labeled with l -32PldCTP by nick translation
to a specific activity of approximately 5 x 108 cpm/ 9. Hybridization was
carried out in 4 x SSC (1 x SSC is 150 mM NaCI, 15 mM sodium citrate (pH
7.0)), 40% formamide, 1 x Denhardts solution, 10% dextran sulfate at 42C.
The filters were washed at 55C in 0.1 x SSC, 0.1% SDS and exposed to
9 q
W O 93/25684 PC~r/US93/05672
Kodak XAR-5 films at -70C. Eig~t~hag~clon~s w~r~ isolated, and a
hyb,idi~ing 1.5 kb Pstl fragment from one of these clones was subcloned in
the plasmid pBS-KS (Stratagene). The nucleotide sequence of the subcloned
fragment was determined by the dideoxy chain termination method (Sanger
et al., 1977, Proc. Natl. Acad. Sci. U.S.A. 74:5463-5467).
6.1.3. COMPUTER ANALYSIS OFTHE SEQUENCE DATA
DNA and amino acid sequence comparisons and alignments shown in
1 0 Table I were performed on a VAX computer using UWGCG software
(Devereux et al., 1984, Nucl. Acids Res. 72:387-395). The results of
comparing amino acid sequences using the UWGCG programs are presented
as percent amino acid similarity or nucleotide identity between the
sequences, taking conservative amino acid changes into consideration
1 5 (Gribskov and Burgess, 1986, Nucl. Acids Res. 14:6745-6763; Schwartz andDayhoff, 1979, ~An Atlas of Protein Sequence and Structure~, ed., Natl.
Biomed. Res. Found., Was~,inylon D. C., pp. 353-358). Phylogenetic Analysis
Using Par:,;",o,ny (PAUPversion 3.0f) was used for the construction of the
phylograms (Felsenslein, 1988; Annu. Rev. Gene. 22:521-555; Swofford and
Olsen, 1990, in ~Molecular Systematics,~ Hills and Morik, Eds., Sunderland,
MA., Sinaver Assoc., Inc. pp. 441-501). Searches for the most probable
trees were run using both exhaustive and heuristic (branch swapping)
algorithms.
2 5 6.1.4. PRODUCTION OF RECOMBINANT PROTEIN, BINDING ASSAY TO PC12
CELLS, AND ASSAYS OF NEUROTROPHIC ACTlVmES
For transient expression of recombinant proteins in COS oells,
appropriate DNA fragments were cloned in the vector pXM (Yang et al.,
1986, Cell 47:3-10). For NT-4 the sequenced 1.5 kb Pstl fragment from
Xenopus was cloned in pXM, and for NGF a 771 bp BstEII-Pstl fragment
w0 93~2~684 3~ PCI/US93/05672
from~t~e 3~ exon of the rat NGFgene was used (Halbook et al., 1988
Development 108:693-704). To express BDNF protein, a PCR-ampiified
fragment containing the prepro-BDNF coding sequence from the mouse
BDNF gene (Hofer et al., l990, EMBO J. 9:2459-2464) was also subcloned in
5 pXM. For NT-3, a 1020 bp rat cDNA cione was inserted in pXM (Ernfors et
al., 1990, Proc. Natl. Acad. Sci. U.S.A. 87:5454-5458).
COS cells (Gluzman, 1981, Cell 3:175-182) grown to about 70%
confluency were transfected with 25 9 of plasmid DNA per 100 mm dish
using the DEAE-dextran-chloroquine protocol (Luthman and Magnusson,
1 0 1983, Nucl. Acids Res. 17:1295-1305). Transfected cells were then grown in
complete medium (DMEM plus 10% FCS), and conditioned medium was
collected 3 days after transfection. Dishes (35 mm) transfected in parallel
were grown over the third night after transfection in the presence of 200
Ci/ml [35S]cysteine (Amersham, UK). Aliquots (10-20 1 each) of the in vivo
1 5 labeled conditioned media were analyzed by SDS-PAGE in 13%
polyacrylamide gels. The gels wer~ treated with EnHance (New England
Nuclear, Boston, MA), dried, and exposed to Kodak XAR5 films with
inlensif~ing screens for 24-48 hr at 80C. Autoradiographs were scanned in
a Shimadzu densitometer, and the relative amounts of the different
recombinant proteins were estimated by Cy~lc~ tirl9 the area corresponding
to each protein rslativ2 to that obtained with rat NGF. The absolute
amount of rat NGF protein was ~-csessed by quanlilali~e immunoblotting of
conditioned media using standards of purified mouse NGF and was used to
determine the protein concentration in the samples containing the other
recombinant proteins.
For binding assay of recombinant proteins to PC12 cells (Greene and
Tischler, 1976, Proc. Natl. Acad. Sci. U.S.A. 73:2424-2428), mouse NGFwas
lab~led wi~h 125I by the chloramine-T method to an average activity of 7 x
107 cpm/ 9. Stsady-state binding was measured in competition assays
performed at 37C or 0C using 1 x 104 c~lls per ml, 1.5 x 10-9 M 125INGF;
42
WO 93/25684 2 1 ~ 7 7 ~ ~ PCr/US93/05672
--and serial dilutions of conditioned media containing equivalent amounts of
NGF or NT-4. All components were added at the same time, and cells were
collected by centrifugation after equilibrium was reached (1-3 hr incubation).
Control experiments using medium from mock-transfected COS cells showed
5 that other proteins present in the conditioned medium had no effect on the
binding of 125I-NGF to PC12 cells. Nonspecific binding was measured in a
parallel incubation to which at least a 1000-fold excess of unlabeled NGF
was added. All results were corrected for this nonspecific binding which
was always less than 10% of the total binding.
10The biological activities of the different proteins were measured by
the ability of transfected COS cell conditioned media, containing equal
amounts of recombinant protein, to stimutate neurite outgrowth from
explanted sympathetic, nodose, and dorsal root ganglia from E9 chicken
embryos (Ebendal, 1984, ~Organizing Principles of Neural Development, S.
1 5Sharms, ed., New York: Plenum Publishing Corp., pp. 93-107; Ebendal, 1~89,
~Use of Collagen Gels to Bioassay Nerve Growth Factor Activity In Nerv~
Growth Factors~, R. A. Rush, ed. (Chichester: John Wiley 8 Sons, pp. 81-93).
Serial dilutions of conditioned medium were assayed and the fiber outgrowth
was scored.
6. 1. 5. RNA PREPARATIONS AND BLOT ANALYSIS
The indicated tissues from adult female Xenopus were dissected and
frozen in liquid nitrogen. The brain and spinal cord were pooled. Several
25lobes of the ovary were dissected out, including oocytes of different stages.
The frozen tissue samples were homogenized in 4 M guanidine
isothiocyanate, 0.1 M û-mercaploethanol, 0.025 M sodium citrate (pH 7.0)
and homogenized three times for 15 s with a Polytron. Each homogenate
was layered ov~r a 4 ml cushion of 5.7 M CsCI in 0.025 M sodium citrate
30(pH 5.5) and centrifuged at 15C in a Beckman SW41 rotor at 35,000 rpm
43
WO 93/256~4~3~ PCI'/US93/05672
for 76 hr (Cl,i,y~:in ot al., 1979, Biochemistry 78:5294-5299). Poly(A)+ RN~
was purified by oligo(dT)-cellulose chromatography (Aviv and Leder, 1972,
Proc. Natl. Acad. Sci. U.S.A. 69:1408-1412), and the recovery of RNA was
quantified spectrophotometrically before use in RNA blot analysis. Poly(A)~
RNA (10 9) from each sampl~ was electrophoresed in a 1% agarose gel
containing 0.7% formaldehyde. UV-transillumination of the stained gel was
used to confirm that all samples contained similar amounts of intact RNA.
The gel was then transferred to a nitrocellulose filter. The filter was
hybridized to the indicated DNA probes. The probes were labeled with [ -
1 0 32P]dCTP by nick translation to a specific activity of around 5 x 105 cpm/ 9,
and the hybridization was carried out as described above. Filters were
washed at high stringency (0.1 x SSC, 0.1% SDS, 54C) and exposed to
Kodak XAR-5 films.
1 5 6.2. RESULTS
DNA fragments coding for NGF, BDNF and NT-3 from human, rat,
snake, frog, and fish were isolated using the PCR technique with degenerate
primars from conserved regions in these three proteins located between
2 0 Iysine 50 and threonine 56 for the upstream primer and between
tryptophan 99 to aspartic acid 105 for the dow"~ am primer (Fig. 1A).
The amplified region contains three of the six cysteine residues and covers
approximately one third of the mature molecules. A comparison of the
amplified region in already characterized NGF molecules from different
2 5 species shows that it contains two variable regions, arginine 59 to serine 67
and aspartic acid 93 to alanine 98. A hydrophilic stretch believed to be
exposed on thQ surface of th~ molecules (Bradshaw, 1978, Ann. Rcv.
Biochem. 47:191-216), as well as the highly conserved regions glycine 68 to
tryptophan 76 and threonine 85 to threonine 91 are also included in the
30 amplified region. The BDNF and NT-3 molecules have an extra amino acid
WO 93/25684 ~ 1 ~ 7 ~ ~ ~ PCr/US93/OS672
between positions 94 and 95 of the mouse NGF protein which is also
included in the ampiified region.
The sequences of the entire mature molecule of mouse NGF, BDNF
and NT-3 proteins were compared in order to calculate how representative
the amplified region is of the complete molecule. The entire mature
molec~ s show 65/57% similarity (amino acid sequence similarity/nucleotide
sequence identity) between NGF and BDNF, 70/61% similarity between NGF
and NT-3 and 68/58% similarity belY:een BDNF and NT-3. When comparing
the region isolated in this study, the similarity between NGF and BDNF is
1 0 62/53%, that bel~:~,on NGF and NT-3 is 67/58%, and that between BDNF
and NT-3 69/60%. This strongly suggests that the region isolated in this
study is representative for the entire molecule and that it can be used to
monitor the evolutionary relationships among the different factors. Pairwise
sequence comparison were performed (Table 1) taking conservative amino
1 5 acid replacements into consideration, using the comparison matrix of
Schwark and Dayhoff (1979, ~In Atlas of Protein Sequence and Structure,
M.O. Dayhoff, ed., Washin~ton, D.C., Natl. Biomed. Res. Found., pp. 353-358).
Therefore, comparisons of amino acid sequences given below and shown in
Table I indicate percent similarity, not identity. Phylogenetic trees wsre
20 constructed using parsimony analysis (Felsenstein, 1988, Ann. Rev. Genet.
22:521-565; Swofford and Olsen, 1990, ~In Molecu~r Systematics, D. M. Hills
and C. Morik, eds., Sunderland, MA:Sinauer Assoc., Inc., pp. 411-501). As
shown below, all isolated DNA fragments with predicted amino acid
sequencss related to those of NGF, BDNF, and NT-3 contained conserved
25 cysteine residues at the correct positions. This was used as an initial
criterion for a sequence to be considered as a member of the nerve growth
factor gene family.
W O 93/25684 ~ ~ ~ PC~r/US93/05672
6. 2. 1. NGF, BDNF AND HDNF/NT-3 ARE HIGHLY
CONSERVED DURING EVOLUllON
6. 2. 1. 1. NERVE GROWrH FQCTOR
The nucleotide sequence (Fig. 1B [human (SEQ ID NO:3), rat (SEQ ID
NO:4), chicken (SEQ ID NO:5), viper (SEQ ID NO:6), Xenopus (SEQ ID NO:7),
salmon (SEQ ID NO:8)] and the predicted amino acid sequence of the
10 isolated fragments coding for NGF are highly conserved from fish to human
(Fig. 2 [human (SEQ ID NO:24), rat (SEQ ID NO:25), chicken (SEQ ID NO:26),
viper (SEQ ID NO:27), Xenopus (SEQ ID NO:28), salmon (SEQ ID NO:29]).
Most of the non-conservative amino acid changes were found in the variable
regions arginine 59 to serine 67 and aspartic acid 93 to alanine 98 (Fig. 2).
1 5 The similarity between the Xenopus and human NGF sequences is 93/79%
(Table 1). Xenopus and chicken NGF are identical excapt for one
conservative change from Iysine 62 to arginine 62 (Fig. 2). The sequences
of viper and salmon NGF contain 11 and 19 amino acid differences (out of
42), respectively, compared with human NGFwhile all other species only
20 showed four dir~erences. None of the NGFamino acid sequences isola~ed
cGnlained the ~xtra amino acid residue present in BDNF and NT-3 between
glutamic acid 94 and Iysine 95 of the human NGF sequences.
The interspecies relationships of the different NGF sequences were
analyzed by the construction of a phylogenetic tree (Figure 3A). The
25 salmon NGF sequence appears to have diverged more than the NGF
sequences isolated from other speçie~. No NGF sequence could be isolated
from ray using the described PCR technique, suggesting that ray NGF
sequences may be above the mismatch tolerance of the primers used in our
PCR protocol. Alternatively, the absence of NGF in cartilaginous fishes would
30 imply that NGF appeared after the splitting of the branch leading to the
evolution of the bony fishes (some 450 million years ago) but before
46
WO 93/25684 2 1 ~ 7 7 ~ J PCI/US93/05672
amphibians and highar vertebrates evolved from this branch (about 400
million years ago).
Nucleotide identities & amino acid similarities were calculated with a
VAX computer (software package from the UW~CG; Devereux et al., 1984,
Nucl. Acids Res. 12: 389-395) according to the comparison matrix of
Schwartz and Dayhoff (1979, Washington, D.C. Nat'l Biomed. Res. Found.
pp. 353-358), taking conservative amino acid changes into consideration.
The figures below the diagonals show percent nucleotide identity. ~he
figures above the diagonals show the percent amino acid similarity. X
indicated that the sequences were not isolated from those species (NGF)
from ray and NT-3 from viper). Hum, human; Chi, chicken; Vip, viper; Sal,
salmon; Xen, Xenopus.
6. 2. 1. 2. BRAIN-DERIVED NEUROTROPHIC FACTOR
1 5
DNA sequences similar to that of human BDNF were found in all
species investigated (Fig. 1B [SEQ ID NOS:1-21, listed supra). The similarity
in amino acid and nucleotide sequences between ray, the most primitive
species investigated, and human are 93/77% (Table 1). Only two non-
20 conservative changes were seen out~ide the variable regions, whereas tensimilar changes were found in the two variable regions (Fig. 2). In Xenopus,
(SEQ ID NO:34) leucine 90 is replaced by a phenylalanine as a result of a
single base pair mutation, C to T in the first position of the codon, and in
salmon (SEQ ID NO:35), tryptophan 77 is repl~^ed by tyrosine as a result of
25 a double mutation, chaoginy the codon from TGG to TAT (Fig. 1B [SEQ ID
NO:14]). All i-~ol~ted sequences contained an extra amino acid residue at
position 96, compared with NGF (Fig. 2 [SEQ ID NO:24-29]). The BDNF
sequences from different species appeared as a homogenous group of
sequences when analyzed by the parsimony method (Figure 3B).
W0 93~25684 ~3~ PCI/US93/05672
6. 2. . . NEUROTROPHIN-3
The nucleotide and predicted amino acid sequences for human (SEQ
ID NO:16 and 37), rat (SEQ ID NO:17 and 38), chicken (SEQ ID NO:18 and
39), Xenopus (SEQ ID NO 19 and 40), salmon (SEQ ID NO:20 and 41), and
ray NT-3 are highly similar (Figs. 1B, Figure 2). Most of the changes ar~
silent mutations resulting from changes in the third position of the codons,
usually transitions that preserve the pyrimidine or purine feature of the base
pair. Only non-conservative amino acid changes were found within the two
variable regions and no amino acid replacements were seen outside the two
variable regions. The salmon sequence lacks Asp-94 which is present in all
other NT-3 molecules (Fig. 2) and has a longer distance from the branching
point in the phylogenetic tree than NT-3 sequences from other species
(Figure 3C).
1 5 6. 2. 1. 4. A NOVEL ~FM~cn OFTHE NERVE
GROWrH FACTOR GENE FAMILY
Additional DNA fragments were isolated from viper (SEQ ID NO:1)
and Xenopus (SEQ ID NO:2), and the predicted amino acid sequences (SEQ
ID NO:22 and SEQ ID NO:23, respectively) revealed that these fragments
contained all three cysteine residues in the same positions as in NGF, BDNF
and NT-3 (Fig. 1B, Fig. 2). A comparison with the sequences of Xenopus
NGF, BDNF and NT-3 indicated that this new sequence is related, but not
identical, to the sequences of the other members of the NGFfamily. The
gene including this sequence was therefore named neurotrophin-4 (NT-4).
Comparison of the nucleotide and amino acid sequences show that Xenopus
and viper NT-4 are 91/73% similar. This similarity is in the same range as
the one seen between Xenopus and viper NGF and BDNF (Table 1). As for
the other members of the NGF family, non-conservative amino acid changes
were only seen in the two variable regions (Fig. 2).
48
Wo 93/25684 ~ ~ ~ 7 7 9 ~ PCI/US93/05672
6. 2. 1. ~. COMPARISONANDPHYLOGENYOFTHEMEMBERS
IN THE NERVE GROWrH FACTOR GENE FAMILY
A comparison of the phylogenetic trees for NGF, BDNF, and NT-3
showed longer branches in the NGF tree, indicating a higher rate of
5 evolutionary change (Figures 3A-3C). The relationship of each member of
the NGFfamily to the other members was studied by the construction of a
phylogram comparing the deduced amino acid sequences for the four
members of the family. The phylogram showed that NGF is more closely
related to NT-3 than to BDNF and NT-4 (Figure 3D). NT-3 is as related to
1 0 NGF as to BDNF. NT-4 is clearly more related to BDNF than to the other
two members.
6.2.2. STRUCTURAL FEATURES OF THE NT-4 PROTEIN
1 5 To enable a more detailed characterization of the NT-4 gene and itsgene product, we screened a Xenopus genomic library with the NT-4 F~R
fragment and isolated a phage clone containing a 16 kb insert. From this
insert, a 1.5 kb Pstl fragment was subcloned and sequenced Figure 4A (SEQ
ID NO:43). The nucleotide sequence contained an open reading frame
encoding a 236 amino acid protein (SEQ ID NO:44) that showed several
structural features characteristic of the other members of the NGF family.
The amino terminus of the predicted NT-4 protein contains an 18 amino acid
putative signal sequence in which a region of 4 amino acids is identical to the
corresponding regions in pig and rat BDNF (Leibrock et al. 1989, Nature
341, 149-152; Maisonpierre, et al., 1990, Science, 247, 1446-1451). A
~oler,lial signal cleavage site, which is also identical to the one proposed forBDNF (Figura 4A), follows. A potential cleavage site for a 123 amino acid
mature NT-4 protein is found after amino acid 113 in the prepro-NT-4
protein. A single predicted N-glycosylation site (Asn-Lys-Thr) is located 8
amino acids before the putative cleavage site.
49
WO 93/25~ 99 PCI/US93/05672
A compa,ison of the mature NT~ protein ~o the mature BDNF, NT~
and NGF proteins from mouse reveaied 60%, 58% and 51% amino acid
identity, respectively. Included in the mature NT-4 protein are all 6 cysteine
residues involved in the formation of disulfide bridges [Figure 4B (SEQ ID
NO:45-48)]. The regions that are identical between NGF, BDNF, and NT-3
are also similar in the NT-4 protein. Most sequence differences between the
NT-4 protein and the other three proteins were found within the same
variable regions previously identified in the other members of the family.
1 0 6. 2. 3. BINDING TO THE NGF-R AND
NEUROTROPHIC ACTIVITY OF NT-4
The 1.5 kb Xenopus Pstl fragment was cloned in the expression
vector pXM (Yang et al., 1986, Cell 47: 3-10) and transiently expressed in
COS cells. SDS-PAGE of conditioned media from transfected cells labeled
with [35S] cysteine sho~NGd an NT-4 protein with an Mr of 14K (Figure 5A).
NGF protein produced and labeled in parallel dishes migrated somewhat
faster than the NT~ protein. This difference in mobility is most likely due to
variations in the charge of the two proteins. Similar mobility differences
have also been observed for NGF prolei.,s with identical sizes from different
2 0 srec;r-s.
Conditioned media from transfected COS cells containing equal
amounts of rat NGF and Xenopus NT-4 protein were tested for their ability
to compet~ for binding of ~25I-labeled NGF to its receptor on PC12 cells.
Binding assays were done at 37C and under conditions in which 80% of the
125I-NGF ~c-soci~ted to the cells is bound to the low affinity NGF-R (Sutter et
al., 1979, J. Biol. Chem. 254, 3972). Similar concentrations of NGF and NT-4
(6x10-lOM) were required to ~ispl~ce 50% of the 125I-NGF from the PC12
cells, indicating that the two proteins bind to the low affinity NGF-R with a
similar affinity (Figure 5B). At higher concer,l~alions, the NT-4 protein was
3 0 less efficient in displ- ,ing 125I-NGF, suggesli,-g that in this case the remaining
WO 93/25684 ~ ~ ~ f ~ ~ ~ PCI/US93/05672
NGF associated with the cells was bound to high affinity or internalized
receptors. The fact that this difference could not be seen in a parallel
assay performed at 0C in which no membrane mobilization or
internalization occurs suggests that the NT-4 protein is not able to
5 compete with NGF for internalization, a process known to be mediated
exclusively through the high affinity receptors (Olsnder and Stach, 1980, J.
Biol. Chem. 255, 9338-9343; Bernd and Greene, 1984; J. Biol. Chem. 259,
15509-15516; Hosang and Shooter, 1987, EMBO ~. 6, 1197-1202).
The NT~ protein t,al)sienlly sx~ressed in COS cells was tested for its
1 0 ability to promote neurite growth from explanted embryonic chick ganglia.
A clear stimulation of neurite outgrowth from explanted chicken dorsal root
ganglia was seen (Figure 6A). Comparison of dose-response curves using
equal amounts of NT~ and NGF protein revealed that the activity obtained
with NT-4 was lower than that seen with NGF (Figures 5A and 5B).
1 5 Recombinant NT-4 and BDNF proteins stimulated neurite outgrowth in the
dorsal root ganglia to a similar extent (Figures 6A and 6C). The NT-4
protein elicited a weak, but consistent, neurite outgrowth from the nodose
ganglia (Figure 6G), whereas no activity could be detected in sympathetic
ganglia (Figure 6E). This is in contrast to NGF,which markedly stimulates
20 neurite outgrowth from sympathetic ganglia (Figure 6F), and NT-3, which
showed a clear activity in the nodose ganglia (Figure 6H). As for NT-4, the
neurite outgrowth-promoting activity of BDNF in the nodose ganglia (Figure
61) was lower than the activity seen with NT-3.
2 5 6. 2. 4. EXPRESSION OF NT-4 mRNA IN
DIFFERENT XENOPUS TIS~SIJFS
Polyadenylated RNAwas prepared from 11 di~erent Xenopus tissues
and used for Northern blot analysis. Hybridization with the Xenopus NT-4
probe revealed high levels of two NT-4 lransc,i~s of 2.3 kb and 6.0 kb in
the ovary (Figure 7A). In contrast, the level of NT~ mRNA was below the
W O 93/2568~ PC~r/US93/05672
detection limit in all other tissues analyzed. Hybridization with the Xenopu~
NGF probe showed a 1.3 kb NGF mRNA in the heart (Figure 7A) and brain.
However, the amount of NGF mRNA in these tissues was on the order of
100 times lower than the level of NT-4 mRNA in the ovary. NGF mRNA was
5 also detected in the ovary, though the amount of NGF mRNA was
approximately 100 times lower than the level of NT-4 mRNA in this tissue
(Figure 7B). The levels of BDNF and NT-3 mRNAs in ovary were both below
the detection limit (Figure 7B).
6. 3. DISCUSSION
We have used the polymerase chain reaction (PCR) in combination
with degenerate oligonucleotide primers to isolate the genes for different
members in the NGF family from different species. A comparison of the
nucleotide and amino acid sequences of the entire mature NGF, BDNF and
NT-3 proteins revealed similarities that are the same as those obtained by
comparing the region of the genes analyzed in this study. Hence, this region
appears to be represenlalive for the rest of the gene and can therefore be
used to study the evolutionary conservation of the entire mature protein.
2 0 The NGF, BDNF and NT-3 genes from different species include regionswhich show complete identity be~wecn fish and mammals, as well as regions
with lower similarity. A comparison of NGF sequences from different species
with the corresponding sequences of BDNF or NT-3 showed that the NGF
gene is less conserved in vertebrates than both BDNF and HDNF/NT-3. The
two latter genes appear to be equally conserved in all species studied,
except in salmon, in which NT-3 is less conserved than BDNF. In this
context, it is interesting to speculate about the fact that the molecular
clock seems sped up in some branches, notably NGF, and not in others. It is
generally be5eved that there is a selective force that preserves the correct
tertiary structure of a protein (Dickerson, 1971, J. Mol. Evol. 1, 26-45;
52
W O 93/25684 ~ 1 ~ 7 7 ~ ~ ; Pc~r/us93/05672
Kimura 8 Ohta, 1974, Proc. Natl. Acad. Sci. USA, 71: 2848-2852). The
dirrer~l,ce in the evolutionary conservation of the three factors suggests
that there has been a higher selective pressure on BDNF and NT-3 than on
the NGF gene. Environmental changes have been proposed to lead to
changes in the selective pressure altering the performance optimum of a
specific gene product (Kimura 1983, in ~Evolution of Genes and Proteins~, pp.
208-233). In this context, it is possib'e that the more extensive evolutionary
changes seen in NGF compared to BDNF and NT-3 reflect the fact that the
function of NGF has changed more during evolution. Structure-function
1 0 studies of NGF have shown that this molecule can tolerate considerable
structural changes without loss or modification of its activity profile,
suggesting that the lower degree of evolutionary conservation of NGFcould
be dua to a more stable structure of this protein, which is therefore less
easily perturbed by substitutions. Another possiblc explanation is that the
1 5 regions of the genome whera the genes for the different factors are located
have dirrerenl general mutation rates. Different mutation rates have been
shown for non-coding ragions of the genome (Wolfe, et al., 1989, Nature,
337: 283-285) but it is less clear if this can lead to an increased number of
changes in coding regions.
Salmon NGF and NT-3 are notably more different when compared
with these molecules in other species. Some amino acids including the
threonine 82 and the histidine-ll,reGni"e-phenylalanine at position 85 to 87 in
NGF, as well as the absence of the amino acid between positions 94 and 95
(compared to the two other proteins), are consistent features of the NGF
protein. The fact that the isolated salmon sequence contains all of these
NGFspecific motifs argues that it is not an additional member of the family,
but rather represents salmon NGF. In cGnlrasl to all other NT-3 sequences
studied, salmon NT-3 lacks the amino acid in position 95. Since the extra
amino acid is present in ray NT-3, it is likely that the common ancestor of
ray and salmon had an ancestral NT-3 sequence which included the extra
WO 93/2~ ~ PCI-/US93/05672
amino acid in position 95. Therefore, the changes in the salmon NT~
molecule must have occurred after this gene split from the common
ancestor. Most of the changes in the amino acids of the salmon sequence
are in the same regions that vary, to a lesser degree, also in ~he other
species, strongly suggesting that the isolated salmon NGF or NT-3
sequences are not pseudogenes. The greater divergence of salmon NGF
and NT-3, compared with the other species, probably reflects the high
degree of evolutionary expansion of the bony fishes.
The results in this study indicate that the NGF family probably existed
1 0 500 million years ago in the primitive fishes, which were the ancestors of
today's higher vertebrates. The gene family could have been formed by
gene duplication, which is believed to be the most common mechanism
whereby new genes evolve (Li, W., 1983, in ~Evolution of Genes and rloteins,
pp. 14-37). Duplications of functional genes could have been facilitated,
1 5 since all information required for the synthesis of a biologically active protein
is cGnlained within a 3' exon (Hallbook, et al., 1988, Mol. Cell. Biol. 8: 452-
456; Leibrock, et al., 1989, Nature, 341: 149-152; Hohn, et al., 1990,
Nature, 344: 339-341). The formation of the family has involved several
gene duplications (Figure 3D).
Since NT~ is more closely related to BDNF than to NT-3 or NGF, it
appears that NT-4 and BDNF were formed from a common ancestral gene.
However, since no progenitor-like molecule for all four factors can be
distinguished from the present data, the evolutionary relation of the
putativs BDNF/NT-4 dnc~slor to the ancestors of NGF and NT-3 cannot be
definil~ly est~hlished. The topology of the phylograms using data from
differant species is in general agreement with the consensus evolutionary
relationship among di~eren~ .spec Qs However, for both NGF and BDNF, the
chicken sequences show an earlier branching in the phylogram than
expected. Comparison of NT-4, NGF, and BDNF from viper and Xenopus
revealed that the NT-4 sequences in these species have 11 amino acid
54
WO 93/25684 2 1~-~?~ PCI`/US93/05672
replacemants, compared with 9 and 8 replacements in NGF and BDNF,
respectively. This suggests that in these species, NT-4 has diverged with a
rate that is comparable to, or faster than, the rate of NGF or BDNF
divergence.
Replacements of highly conserved amino acids in the NGF molecule do
not abolish the biological activity, but in many cases these affect the
amount of protein produced, indicating that there are constraints other
than the biological activity, such as protein stability, which may be
important for the conservation of the NGF protein. In addition, the fact
that all members of the N~iFfamily can interact with the low affinity NGCR
suggests that the complete conservation of certain regions in these factors
may be due to constraints on these genes to retain proteins that can
interact with the NGF-R. The basic mechanisms and strategies for the early
ontogeny of the embryo are similar in all vertebrates and presumably
involve genes that are conserved in all vertebrates. The evolutionary
conservation of the neurotrophic factors is therefore consistent with the
notion that they are important in early embryonic development in many
different species.
The hip~o~."pus contains the highest levels of NGF, BDNF, and NT-3,
mRNA in the brain (Ernfors et al., 1990 J. Dev. Neurosci. 9, ~7-66). It is a
highly specialized structure derived from the archipallium, which first
appeared in the brains of amphibians and reptiles. The mammalian
hippocampus is important for memory, learning and cognitive functions
known to be ~ssoci~ted with high neuronal plasticity (Crutcher and Collins,
1982, Science 277:67-68). These demands may have generated a selective
pressure during phylogeny for pl~cticity-promoting mechanisms, possibly
medicated by neurotrophic factors. However, the results in this study
clearly show that the duplication event of the genes for the neurotrophic
factors preceded by far the formation of the hippocampus. This finding
ind;c~es that the neurotrophic factors did not evolve as a conse~uence of
WO 93/25684 . ' ' - PCI/US93/05672
the formation of the hippocampus and supports the notion that th~
neuronal plasticity in this brain region is at least in part due to these
mlo'~cu'es
The organization of the nervous system of primitive vertebrates, i.e.,
cartilaginous fishes, shows some basic simiiarities to the nervous system of
higher vertebrates. The cranial nerves and the somatic sensory and
autonomic nervous systems in cartilaginous fishes are in general similar to
those of higher vertebrates (Young, J.Z., 1981, The Life of Vertebrates, New
York Oxford University Press). It is therefore likely that the principles of
neurotrophic intsractions are the same in both primitive and higher
vertebrates. The evolutionary conservation of the NGF-like neurotrophic
factors also in primitive vertebrates suggests that these factors first
avolvQd in invertebrates and were later adapted to function in the
development of the vertebrate nervous system.
1 5 Our study of the evolutionary conservation of the NGF farnily led to
the isolation of a novel member of this family, named neurotrophin-4 or NT-
4, PCR fragments from the NT-4 gene ware isolated from Xenopus and
viper, and a genomic clone was subsequently isolated from Xenopus.
Nucleotide sequence analysis of this clone revealed an open reading frame
for a 236 amino acid protein, which showed several structural features
resembling those of the three other members of the NGF family. These
include the presence of a putative amino-terminal signal sequence and a
potential N-glycosylation site close to a proteolytic cleavage site that
predicts a 123 amino acid mature NT-4 protein. The size of the mature NT-
4 protein is 4 amino acids longer than that of BDNF and NT-3 and 5 amino
acids longer than that the mature NGF protein. Within the mature NT-4
protein, all 6 cystein residues involved in the formation of disulfide bridges
are cons~lved. The NT~ protein differs from lhe other members of the
family in the same regions that vary among the sequences of the three
3 0 other family members. As for NGF, BDNF, and NT-3, the entire prepro-NT-4
56
WO 93/25684 ~ 1 ~ 7 7 ~ ~ PCI`/US93/05672
protein is encoded in one singla exon. Hence, both the gene organization
and the structural features of the predicted protein indicate that the NT-4
gene is an additional member of the NGFfamily. The fact that the NT-4
gene was isolated from both reptiles and amphibians suggests that it is
present in several different species.
Both BDNF and NT-3 have been shown to interact with the low
affinity NGF-R (Rodriguez-Tebar et al., 1990, Neuron 4:487-492; Ernfors et
al., 1g90, Proc. Natl. Acad. Sci. USA 87:5454-5458). The Xenopus NT-4
protein displaced 125-NGF from its low affinity recaptor on PC12 cells,
1 0 indicating that the fourth member of this family can also interact with the
low affinity NGF-R. The comparison of displacement curves obtained at
37C and 0C suggesls that the NT-4 protein cannot compete for binding to
the high affinity NGF-R. The protein encoded by the low affinity NGF-R gene
appears to form part of both the low and the high affinity receptors
1 5 (Hempstead et al., 1989, Scienca 243:373-375). The mechanis", by which
two kinetically different receptors are formed from the same receptor gene
is not known, although it has been proposed that the two states can be
generated by the formation of a complex between the cytoplasmic domain
of the receptor and an intr~ellul~r protein (Radeke et al., 1987, Nature
325:593-597; Meakin and Shooter, 1991, Neuron 6:153-163). Alternatively,
a high affinity receptor chain may be encoded by a separate gene and,
similar to the intarleukin-2 receptor (Hatakeyama et al., 1989, Scienoe
744:551-556) and the platelet-derived growth factor receptor (Matsui et
al., 1989, Science 243:800-804), the two receptor chains may form a dimer
that conslilutes the high affinity receptor. The fact that all four members
of the NGF family can interact with the low affinity NGF-R suggests that the
low affinity state of the NGF-R may be, in an as yet ur,hr,o~n way, involved in
mediating the biological effects of all these factGrs. In this context, it is
inlere~ling to note that the low affinity NGF-R gene has been shown to be
expressed in many tissues of both neuronal and nonneuronal origin not
wo 93/~43~ ~ ~9 ~ PCI`/US93/05672
known to respond to NGF. These include mes~ncl~yme, somites and neur~
tube cells in the early chick embryo (Hallbook et al., 1990, Development
108:693-704; Heuer at al., 1990a, Dev. Biol. 137:287-304; Heuer et al.,
1990b, Neuron 5:283-296), as well as developing and regenerating spinal
cord motorneurons (Ernfors et al., 1989, Neuron 2:1605-1613; Ernfors et
al., 1991, J. Dev. Neurosci. 9:57-66). It would therefore be of interest to
investigats whether the NT-4 protein is of functional importance in any of
these tissues or neuronal populations.
The neurotrophic activity of the NT-4 protein was assayed on
1 0 explanted chick embryonic ganglia, and as for the other three members ofthe NGF family, the NT-4 protein showed a clear stimulation of neurite
outgrowth from dorsal root ganglia. However, when compared to NGF,the
NT-4 protein showed lower activity in dorsal root ganglia. Both BDNF and
NT-3 readily elicit neurite outgrowth in explanted nodose ganglia, though the
1 5 response with NT-3 was consistently stronser than that with BDNF. NGF
strongly stimulates neurite outgrowth in sympathetic ganglia, and NT-3 also
has activity in this ganglia, though it is much lower than that of NGF
(Maisonpierre st al., 1990, Science 247:1446-1451; Ernfors et al., 1990,
Proc. Natl. Acad. Sci. USA 87:5454-5458). NT-4 showed weaker activity in
2 0 nodose ganglia compared with NT-3 and no activity in the sympathetic
ganglia. The spectrum of the biological activity of NT-4 on peripheral
explanted ganglia resembles that of BDNF, which is in agreement with the
fact that NT-4 is structurally similar to BDNF.
Northern blot analysis of 11 different tissues from Xenopus showed
high levels of NT-4 in the ovary, whereas the level of NT-4 mRNA was below
the detsction limit in all other tissues examined. Two NT-4 mRNAs of 2.3 kb
and 6.0 kb were seen in the oocytes. The presence of two transcripts from
the same gene has previously been observed for BDNF, in which case two
mRNAs of 1.4 kb and 4.0 kb ar~ presenl in the rat brain (Leibrock et al.,
1989, Nature 341:149-152; Maisonpierre at al., 1990, Science 247:1446-
58
WO 93/25684 ,~ ~ ~ 77 9 ~ PCI/l~S93/05672
1451; Ernfors et al., 1 990a, Proc. Natl. Acad. Sci. USA 87:5454-5458).
Hybridization to a Xenopus NGF probe revealed NGF mRNA in the Xenopus
heart, most likely as a result of NGF mRNA expression in target tissues for
neuronal innervation. The level of NGF mRNA in the heart was, however,
5 more than 100-fold lower than the level of NT-4 mRNA in the ovary. Since
the high level of NT~ mRNA in the ovary does not correlate with neuronal
innervation, it appears unlikely that the NT~ protein has only a neurotrophic
function in this case. Instead, the abundant expression of NT~ mRNA in
Xenopus ovary implies an additional and important nonneurotrophic function
for the NT-4 protein. NGF mRNA was also detected in Xenopus ovary
though at almost 100 times lower levels than those of NT-4 mRNA; BDNF
and NT-3 mRNAs were not detected in this tissue.
mRNAs for two growth factors havs been described as maternal
mRNAs in Xenopus oocytes. One of these mRNAs encodes a protein with
15 strong similarity to basic fibroblast growth factor (Kimelman and Kirschner,
1987, Cell 51:869-877); the other mRNA encodes a protein homologous to
transforming growth factor (Weeks and Melton, 1987, Cell 51:861-867).
These factors have been suggested to function as morphogens for the
formation of mesoderm and the subse~uQnt induction of this tissue into the
20 neural tube. In the rat, in situ hybridization studies have revealed NT-3
mRNA in the epithelium of secondary and tertiary follicles, and a role for NT-
3 in oogenesis has been suggested (Ernfors et al., 1990, Neuron 5:511-
526).
59
WO 93/25684 PCI'/US93/05672
7. E)~AMPLE: IDENT FICAllON OF CEL~S
EXPRESSING M-4 mRNA IN THE XENOPUS
LAEVIS OVARY BY IN SITU HYBRiDlZATlON
7.1. MATERIALS AND METHODS
7.1.1. ISOLATION, HANDLING AND CULTURE OF
XENOPUS OOCYrES, EMBRYOS AND CELLS
1 0
Male and female X. Iaevis frogs were maintained in the laboratory at
19C. After immersion-anesthesia of the animals in 0.25% tricaine methane
sulfonate (Sandoz, Switzerland), ovarian lobes were surgically removed,
washed with modified Barth's saline H~pes (MBSH) (Gurdon and Wickens,
1 5 1983, Methods Enzymol, 101: 370-86) and dissociated by overnight
incubation at 20C in calcium-free MBSH containing 2 mg/ml collagenase.
Crude separation of pre-vitellogenic and vitelloenic oocytes was obtained by
differential sedimentation, and oocytes were further sorted manually under
a dissecting microscope into the developmental classes described by
Dumont (1972, supra).
Sy"chronously cleaving embryos were obtained by in vitro fertilization
essentially as described by Newport and Kirschner (1982).
A6 Xenopus kidney cells were cultured in Leibowitz L15 medium
diluted with distilled water 60:40 (vh) and supp'emented with 10 mM Hepes
pH 7.35, 10 M hypoxanlhine (Sigma), 4 mM glutamine and 10% fetal bovine
serum (Gibco) at 20C. Cultures were equilibrated with air and kept in ~he
dark.
7.1.2. IN SITU HYBRIDIZATION
Fresh-frozen ovaries from adult Xenopus laevis frogs were sectioned
(14) in a cryostat (Leitz, Germany) and the sections were thawed onto
WO 93/25684 ~ 7 ~ ~ ~ PCI'/US93/OS672
poly-L-lysine (50 g/ml) pretreated slides after which they were fixed in 10%
formalin for 30 min and rinsed twice in PBS. Dehydration was carried out in
a graded series of ethanol including a 5 min incubation in chioroform after
which the slides were air dried. Two 53-mer oligonucleotides, one specific
for Xenopus NT-4 mRNA (5Y~CCACMGC I l ~i l I GGCATCTATGGTCAGAGCCCT
CACATMGACIcil I I IGC3' ~SEQ ID NO:109]) and another one, as a control,
specific for chicken BDNFmRNA (corresponding to amino acids 61 to 77 of
the mature chicken BDNF protein (Hallbook et al., 1991, Neuron 6: 845-58
[contained within SEQID NOS.: 11 and 32]), were labeled at the 3' end with
35S-dATP using terminal deoxyribonucleotidyl transferase (IBI, New Haven)
to a specific activity of approximately 1x109 cpm/ . Hybridization was
performed at 42C for 16 hours in 50% formamide, 4x SSC, Ix Denhardts
solution, 1% Sarcosyl, 0.02M NaPO4 (pH 7.0), 10% dextransulphate, 0.5
mglml yeast tRNA, 0.06M DDT,0.1 mg/ml sheared salmon sperm DNA and
1x107 cpmlml of 35S-labeled oligonucleotide probe. Sections were
subse~ ently rinsed, washed 4 times (15 min. each) at 55C in 1 x SSC,
rinsed in water, dehydrated in a graded series of ethanol and air-dried. The
sections were exposed to X-ray film followed by coating in Kodak NTB-3
photo emulsion (diluted 1:1 in water), exposed for 5-6 weeks at -20C,
developed, fixed and counter:jlained with cresyl violet.
7.1.3 RNA BLOT ANALYSIS
The indicated samples were homogenized in 4M guanidine
isothiocyanate, 0.1M l~-mercaploethanol, 0.025M sodium citrate pH 7.0 and
homogenized 3 times for 15 seconds with a Polytrone. Each homogenate
was layered over a 4ml cushion of 5.7M CsCI in 0.025M sodium citrat~ pH
5.5 and centrifuged at 15C in a Beckman SW41 rotor at 35,000 rpm for
16 hrs. (Chirgwin et al., 1979, Biochemistry 78: 5294-5299).
Polyadenylated RNA (Poly(A)+ RNA) was purified by oligo (dT) cellulose
61
Wo 93/25~ Pcr/us93/05672
chromatography (Aviv and Leder, 1972, PNAS 69: 1408-1412) and tt~
recovsry of RNA (40 9) was quantified spectrophotometrically before use
in RNA blot analysis. Total celluiar RNA (40 9) or where indicated
poly(A)+RNA (5 9) from each sample was electrophoresed in a 1% agarose
5 gel conlaining 0.7% formaldehyde. UV-transillumination of the stained gel
was used to confirm that all samples contained similar amounts of intact
RNA. The gel was then transferred to a nitrocellu~ase filter. The filter was
then hybridized to a 350bp Hincll fragment from the 3' exon of the Xenopus
NT-4 gene (Hallbook et al., 1991, Neuron 6: 845-858). The fragment was
1 0 l~heled with -(32p)-dCTP by nick translation to a specific activity of around
5x108 cpm/ 9 and the hybridization was carried out as described (Ernfors
st al., 1988, Neuron 1: 983-96). Filters were washed at high stringency
(0.1xSSC, 0.1% SDS, 54C) and exposed to Kodak AR-5 films at -70C.
1 5 7.2 RESULTS
Tissue sections through the adult Xenopus laevis ovary were
hyLridi~ed to a 3sS-dATP labeled oligonucleotide probe specific for Xenopus
NT-4 mRNA. As a control for the specificity of the hybridization, adjacent
20 sections were hy6ridi~sd to an oligonucleotide probe of the same length and
GC-conlenl complemantary to mRNA for chicken brain-derived neurotrophic
factor (BDNF). The NT-4 mRNA specific probe revealed an intense labeling
over many cslls sodller~d throughout the ovary with a size (50-400 m in
diameter) corresponding to oocytes in early stages of oogenesis (Fig. 9A),
25 No NT~ mRNA could be detected over mature, post-vitellogenic stage Vl
oocytes (arrows in Fig. 9A). The chicken BDNF mRNA specific control probe
did not label any cells in the Xenopus ovary.
Analysis of emulsion autoradiographs from the hybridized sections
revealed an intense lab~ling over the cytoplasm of oocytes with a diameter
3 0 of 50-200 m (Fig. 1 OA and 1 OB) corresponding to stage I oocytes
W O 93/25684 ~ 73~ PC~r/US93/05672
according to Dumont, 1972, supra. The NT-4 mRNA specific probe also
labeled oocytes with a larger diameter corresponding to stages ll to IV,
though the intensity of labeling over these cells was lower than that seen
over stage I oocytes. In agreement with the analysis of low magnification
- 5 dark-field illuminations (Fig. 9), the emulsion autoradiographs did not show
any labeling over more mature oocytes of stages V and Vl. No labeling was
seen over any cells after hybridi~tion with the control BDNF probe (Fig.
10C). To enable a more detailed determination of the level of NT-4 mRNA
during oogenesis, the number of grains per an arbitrarily chosen area unit
was counted. The area unit chosen corresponded to approximately one
hundredth of the cross section area of a stage I oocyte. The result of this
analysis showed that th~ intensity of labeling over stage I oocytes was 1.7
and 4.3 times higher than over stage ll/lll and IV oocytes respectively (Fig.
11). The number of grains per area unit over stage V and Vl oocytes was
not siy"iricanlly above the level of the background labeling.
7.2.1 NORTHERN BLOT ANALYSIS OF NT-4 mRNA EXPRESSION
DURING XENOPUS OX~!-S!S AND EARLY DEVELOPMENT
A fixed amount of total cellular RNA (40 g) prepared from different
stages of oocytes as well as from a fraction enriched from follicle cells was
analyzed by Northern blots using a Xenopus NT~ specific probe (Hallbook
et al., 1991 supra). In agreement with the results of the in situ
hyl~riJi~tion, the highest levels of NT-4 transc,ipls with sizes of 2.3 kb and
6.0 kb was ~rese"l in the smallest oocytes (stages I and ll) (Fig. 12). The
level of NT-4 mRNA declined abruptly in more mature stage V and Vl
oocytes. A weak hyLridi~alion signal was seen in the follicle cell preparation
which was probably due to a contamination with a small number of stage I
and ll oocytes. The same result was obtained when a fixed amount (5 9)
63
w093/256 ~9s~ ;s i~ PCI/US93/05672
~f polyadenylated RNA was analyzed from the differenca samples shown ~
Fig.12.
The results of the analysis of the NT-4 mRNA e~,ression in the ovary
showed that NT-4 mRNA is restricted to immature oocytes. To test the
possibility that expression of NT-4 mRNA is induced after fertilization, the
level of NT-4 mRNA was ~csesssd in developing Xenopus embryos by
Northern blots of polyadenylated RNA. A low level of NT-4 mRNA was
found in Xenopus somatic A6 cultured kidney cells which were also included in
the analysis. How~ver, no NT-4 mRNA could be detected in early embryos
1 0 from the onset of cleavage divisions to the neurula stage.
7.3 DISCUSSION
The abundant expression of NT-4 mRNA in the Xenopus ovary
1 5 (Hallbook et al., 1991 supra) indicates that this membQr of the NGF family
plays a role in oogenesis and/or early embryogenesis. I oc~ tion of oells
expressing NT-4 mRNA in the ovary provided insights into the putative
function of the NT-4 protein in the ovary. In amphibians, as in all other
vertebrates, ferti~ tion of the eg~ triggers a period of rapid cell cleava~e.
This evant is controlled by a class of solublc maternal mRNAs expressad
during oogenesis and stored in the unfertilized egg for subsequent
development (Davidson, 1g86, Gene Activity in Early Development (New
York, Academic Press). This class of maternal mRNAs includes two growth
factors, basic fibroblast growth factor (Kimelman and Kirschner, 1987, Cell,
51: 869-77) and transforming growth factor-~ (Weeks and Melton, 1987,
Cell, 51: 861-67), as wall as several protooncogenes such as c-myc (Godeau
et al., 1986, EMBO J., 5: 3571-77); (Vriz et al., 1989, EMBO J. 8: 4091 -97),
c-fos (Mohun et al., 1989, Development, 107: 835-46), ras (Andeol et al.,
1990, Dev. Biol., 139: 24-34), ets-2 (Chen et al., 1990, Science, 250: 1416- c
18) and c-mos (Sagata et al., 1988, Nature, 335: 519-25). Immature
64
WO 93/25684 ~77~` PCI'/US93/05672
stage Vl Xenopus oocytes are al-e:,led in prophase of meiosis I and both c-
mos (Sagata et al., 1988) and ets-2 (Chen et al., 1990) have been shown
to function during reinitiation of meiotic division. The finding of high levels of
NT-4 mRNA in stage I and ll oocytes but a decreased level below the
detection limit of both Northern blots and in situ hybridization in stage V
and Vl oocytes strongly suggests that the NT-4 mRNA does not belong to
- the class of maternal mRNAs. This result also argues against a role of the
NT-4 protein in the reinitiation of meiotic division or in early embryogenesis.
In agreement with this, addition of recombinant NT-4 protein to immature
1 0 stage Vl oocytes failed to induce germinal vesicle breakdown in vitro and no
NT-4 mRNA was detected in Xenopus early embryos. Instead, the putative
function of the NT-4 protein in the ovary appears to be coupled to events
occurring in the pre-vitellogenic and early mid vitellogenic oocyte. Both N~'
(Ayer-LeLievre et al., 1988, PNAS 85: 2628-2632) the 7~kD low-affinity NGF
1 5 receptor (Persson et al., 1990, Science, 247: 704-707) and the trkA high-
affinity component of the NGF receptor (J.P. Merlo and H. Persson,
unpublished) are expressed in the testes where NGF has recently been
shown to stimulate DNA synthesis at the onset of meiosis (Parvinen et al.,
1991, submitted). Hence, it appears that the neurotrophins do not only
2 0 function as neurotrophic factors but also play an important role in
reproductive tissues
8. E)CAMPLE: ISOLATION AND CHARACTERIZATION
OF NUCLEIC ACID FRAGMENTS ENCODING
2 5 MAMIMALIAN NT4
8.1. MATERIALS AND METHODS
8.1.1. DNA PREPARATION
Genomic DNA was isolated as described in 6.1.1, supra.
WO 93/25684 ~ ` PCI/US93/05672
3~ ` ~
8.1.2. POLYMERASE CHAIN REACTIONS, MOLECULAR CLONING AND DNA
.~yJFNc~lG
Mixtures of 34-mer oligonucleotides (including tail) representing all
possible codons corresponding to the amino acid sequences QYFFET
(contained within SEQ ID NO:51 ) and QYFYET (SEQ ID NO:52) (5'-
oligonucleotide) and, WISECK, CKAKQS and WIRIDT (each contained within
SEQ ID NO:51) (3'-oligonucleotide) (Fig. 13) were synthesized, with linkers,
as described in 6.1.2., supra. Together, 2Y (derived from xNT-4 [SEQ ID
NO:50]) and 2Z (derived from BDNF/NT-3 [SEQ ID NO:51]) represent all
known sequence for neurotrophins from all species in this region. A primary
amplification of both rat and human genomic DNA was carried out with Taq
polymerase (Cetus) with cycles of 1 minute at 95C, 2 minutes at 43C and
2 minutes at 72C. An aliquot from the primary PCR reactions was then
reamplified using either the same primers as in the primary amplification or
with new nested degenerate oligonucleotide primers which would result in an
expected size shift. PCR products from the reamplification procedure were
purified as follows: bands of prospective size were gel purified, reamplified,
and column purified using Stratagene ~primerase~ columns. These were then
digested to completion with EcoRI and Sall, analyzed and re-purified using
Primarase columns (Stratagene) and ligated into EcoRI-Xhol digested
Bluescript KS(-). Transformants were screened for pBS-KS containing an
insert of the approximate predicted size. The cloned fragmants were
subjected to DNA sequence analysis as described in 6.1.2, supra.
8.1.3. ISOLATION OF FULL LENGTH GENOMIC AND cDNA CLONES ENCODING
rNT- 4 AND NT-4
A human ovary cDNA library in ~lGT-10 was obtained from Clontech.
A human hippocampus cDNA library in ~:ZAPII was obtained from
Stratagene. A human genomic DNA library in EMBL3/SP61T7 was obtained
66
wo 93/25684 ~ 1 ~ 7 7 9 9 b PCI /US93/05672
from Clontech. A rat brain cDNA library in ~l-ZAP was obtained from
Stratagene.
Isolation of NT-4 clones can be carried out as follows:
A cloned insert encoding the rNT-4 fragment (Fig. 14 lSEQ ID NO:61])
5 or the hNT~ fragment (Fig. 15 [SEQ ID NO:63]) are labeled by P~P~ to a
specific activity of approximately 5x108 cpmlng. HyL,idi~alion is carried
out in hyl,ridi~alion solution consisting of 0.5 mg/ml salmon sperm DNA at
60C. The filters are washed at 60C in 2 x SSC, 0.1 % SDS and exposed to
Kodak XAR-5 film at -70C. Alternatively, oligonucleotides whose s~quence
10 co,-esponds exactly to the desir~d mammalian neufot,ophin can be used to
generate probes (e.g. kinase labelling) and can be used to screen the sama
libraries by conventional methods. Positive phage are plaque purified and
infected at low multiplicity in an ~pro~.riate E. coli strain in liquid broth asdescribed by Maniatis, et al., Molecular Cloning: A Laboratory Manual, Cold
15 Spring Harbor Laboratory, Cold Spring Harbor, New York). GT-10 and
EMBL3/SP6/T7 phage are prepared as follows: Cultures are inoubated
overnight at 37 with constant shaking. The overnight suspension is
brought to 1M NaCI and 8% PEG, mixed well and inc~ ted over-,iyl,t at 4C
to precipitate the bacteriophage. Tha bacteriophage are pelleted via
centrifugation, resuspended in TM buffer (10mM Tris-HCI, pH 7.~; 10mM
MgCI2), layered upon a CsCI step gradient and centrifuged at appropriate
speed and length of time to band the ba.,1~riophage. The bact~riophage
are removed, transferred to a fresh Eppendorf tube and Iysed by the
~dd~ion of 1 volume of formamide. EMBL-3 DNA is precipilaled by the
addition of 2 volumes of 100% ethanol. The EMBL-3 DNA is recovsred by
microcentrifugation, washed in 70% ethanol and re~uspended in TE buffer
(10mM Tris-HCI, pH 7.5; 1mM Na2-EDTA). The DNA is sxtracted several
times with ph~nol:chloroform:isomyl alcohol (24:24:1), ethanol precipitatad,
resusperided in TE buffer, digested with various restriction enzymes and
electrophoresed through a 1% agarose gel. Subsec~l)ent to electrophoresis,
67
w093/256843~9 PCI/US93/05672
r~strict~d DNA is lr~,~sf~r~ed to nitroc~lluloss and hybridiz~d to the 32p--
l~helled rNT-4 or hNT-4 probe, under conditions described supra. The
hyLridi~ing band is subcloned into pBS-KS plasmid vector and subjected to
DNA sequence analysis by the dideoxy chain termination method (Sanger, et
al., 1977, Proc. Natl. Acad. Sci. U.S.A. 74: 5463-5467).
The ~l-ZAP plasmid preparations are performed as follows: 20011 of
OD600=1.0 XL1-Blue cells, 20011 of the hi-titer phase stock, and 1~L of R408
helper phage (1x10 minutes pfu/ml) are combined. For a negative control,
add no phage stock. Incubate 15 minutes at 37C. Add 5 ml of 2XYT
1 0 media, shake for 3 hours at 37C. At the end of the 3 hours, the nega~ive
control should be cloudy and the samples clear. Samples are heated at
65C for 30 minutes, spun at 40009 for 5 minutes. Supernatant contains
phagemid stock. To rescue the phagemid, add 0.511 of the stock to 200 of
XL1-Blue cells (OD600s1). Incubate 37C for 15 minutes. Plate 1-100
1 5 (preferably 10~L) on LB ampicillin plates. Incubate 37C x overnight andlarge col~n-,es are picked. After plasmid DNA is purified, it is sequenced as
abova.
Lambda phage cDNA libraries are screened according to standard
methods (Mani~lis, at al., supra) as desc,il,ed supra.
2 0 Positiv~ plaques are purified, reisolated and subjected to DNA
sequence analysis as desc,ibed supra.
8.2. RESULTS AND DISCUSSION
A region of the Xenopus NT-4 coding sequence was used as a model
for sy(,ll,ssis of degenerate oligonucleotide primers. Figure 13 d~notes that
the 5'-oligonucleotide primer 2Y [SEQ ID NO:53] (QYFFET) and 3'-
oligonucleotide primers, 3Y [SEQ ID NO:55] (WISECK), 3Z [SEQ ID NO:56]
(CKAKQS) and 4Z [SEQ ID NO:58] (WIRIDT) wer~ derived from the xNT-4
amino acid s~qu~nce. The 5'-oligonucleotide primer 2Z [SEQ ID NO:54]
WO 93/25684 ~ ~ ~ 7 7 9 9 PCI/US93/0~;672
(QYFYET) is derived from the homologous region of rBDNF. All ~ le
combinations of these degenerate oligonuc'~otides were utilized to amplify
DNA from both rat and human genomic DNA libraries. Since the primers
represented by 3Y[SEQIDNO:55] and 3Z[SEQID NO:56~ of xNT~ ar~ not
5 conserved in the NGF/BDNF/NT-3 gene family, and therefore were n~t likely
to amplify NGF,BDNF or NT-3, these two primers were utilized in the
reamplification, or secondary PCR.
DNA fragments of the approximate expected sizes were obtained
from PCR amplification and reamplification of both the rat and human
10 genomic libraries when the following primer combinations were utilized:
(1) 2YI3Z (primary PCR): 2Y, 2Z/3Y (se~nda,~ PCR)
(2) 2Y/3Z (primary PCR): 2Y,2~32(secondafy PCR)
(3) 2Y/4Z (primary PCR): 2Y, 2Z/32 (secGnda,y PCR)
(4) 2~4Z (prima~ PCR): 2Y,2~3Z (secor-Ja~ PCR)
The secondary PCR products of the approximate exl~e~.1e~l size were
electrophoresed through a 2% agarose gel, eluted by standard tec~.ni~ues,
2 0 digested with EcoRI and Sall and ligated in EcoRI-Xhol digested pBS-KS DNA.
Positive transformants were selected, and inserted fragments were
subjected to DNA sequencing by the didsoxy chain termination method
(Sanger, et al., supra).
An open ~eadil-g frame has been ~leduced for a portion of the rat NT-
4 (Fig. 14 [SEQ ID N0:62]) and human NT-4 (Fig. 15 [SEQ ID N0:64]) amino
acid coding sequence. Figure 16 illusliales the homologous region of the
rNT-4 (SEQ ID N0:62) and hNT-4 (SEQ ID N0:64) fragment t o
represent~ti./e members of the NGF/BDNF/NT-3 gene family.
An open reading frame encoding a larger portion of human NT~ than
3 0 that J;sclosefJ in Figure 15 is shown in Figure 17A (SEQ ID N0:69 and SEQ ID
69
PCI~/US93/0~i672
NO:70). Figure 17A presents additional 3~ sequence information for th~
human NT~ coding region. The 192 bp nucleic acid fragment was isolated
as desc,il,ed supra in the Description of the Figures.
The actual size of the PCR products recovered from the
reamplification procedure was larger than predicted due to the ~dditi~.,al 7
amino acids in the rat NT-4 (GPGVGGG) lSEQ ID No:101] and human NT-4
(GPGAGGG) [SEQ ID NO:102] DNA fragment.
The 7 amino acid insertions of rNT-4 and hNT-4 are describ~d as
'GPGXGGG'[SEQID NO:100], where X=V for rNT-4 and X=A for hNT-4.
Valine and alanine possess nonpolar R group. Whether position four is
conserved to contain a nonpolar R group at position 4 in other mammalian
NT~ pro~oins is not preser,lly known, nor wl,ell,er the 7bp insertion itself will
be characteristic of other mammalian NT~ g2nes. It is inlereslins~ to note
that fish NGF has a 22 amino acid b~se,lion in the same region as disclosed
in the presenl invention.
9. EXAMPLE: ISOLATION AND CHARACTERIZATION OF AN NT-4
HUMAN GENOMIC CLONE
We have screened a human placenta genomic library in Ch~RI ~ SP6/T7
(Clontech, K802 as host). A total of 1.25 x 106 pfu were plated on large
NZY plates. Duplicate lifts were made using Schleicher & Schuell
nitrocellulose fil~ers, and were hybridized to a 120 bp probe (from hNT-4
clone 17B, which was obtained from hurnan genomic DNA using primers
2 5 2,4Z followed by 2Z3Z), l~helled by PCR using oligonucleotide primers 2Z/3Z.
The filters were hybridized at 60C with the r~;ol~'~eled probe (10~ cpm/
ml) under the following hybridi7~tion conditions: 0.5 M NaPO", 1% BSA, 7%
SDS, 1 mM EDTA, and 100 g/ml salmon sperm DNA. The filters were then
washed at 60C with 2xSSC and 0.1% SDS, and subjected to
autoradiography. Following four days of exposure, positive signals were
identified on the duplic~te lifts. A total of seven plugs were picked, put into
WO93/25684 ~ 7~i~ PCr/US93/05672
1 ml SM buffer, shaken for 2 hr, and replated as follows: 1) 100 1 of 10-3
dilution (1 1 in 1 ml), mixed with 100 I cells, and plated; an almost confluent
plate was obtained; 2) 200 1 of 10-5 dilution, which gave isolated pl~ es.
Duplicate lifts were made, and screened as described above with the hNT-4
5 120 bp probe. Following a 2 day exposure, many positives were identified
on the confluent plate for plugs HG2, 4, and 7. A well-isolated positive was
identified on both HG4-2 and HG7-2 plates. A single plaque for HG4-2 and
HG7-2 was picked, put into 500 1 of SM buffer, and shaken for 2 hr,
following which 100 1 of eluant was mixed with 100 I cells and plated. The
10 plate was then flooded with 3 ml SM buffer, and supernatant collected as
the first high titer stock. Three plates were then plated using 100 1 of this
first stock mixed with 100 I cells. The plates were flooded with 3 ml SM
and shaken on rotator for 3 hr at room temperature. Supernatant was
removed, spun to remove debris, following which chloroform was added,
15 and this used as the second high titer stock. Two I of HG4-2 and HG7-2
high titer stock was spotted onto Schleicher & Schuell nitrocellulose filter,
and was found to hybridize to the rNT-4 180 bp probe lisolated from the
plasmid cGnlai.-ing an insert obtained by PCR from rat genomic DNA using
primers designed based on our rat NT-4 clone sequence coding for the
20 amino acidGELSVCD (SEQ ID NO:112) (degenerate primer) and KAESAG
(SEQ ID NO:113) (exact primer)]. Plate Iysates and liquid Iysates were
prepared for HG4-2, HG7-2 and HG2-1. Phage DNA was made, an aliquot of
which was run on agarose gel and subjected to Southern analysis. HG4-2,
HG7-2 and HG2-1 were found to hybridize to the rNT-4 180 bp probe
25 (NaPO" hybridization as above, 65C), and a 45mer oligonucleotide probe
(GG~r~~ ~GTGGAcAGGAGG~ CTGGGTATCTGAG) [SEQ ID
NO:114] corresponding to amino acid GGGCRGVDRRHWVSE [SEQ ID
NO:115] coded for by human PCR fragment clone 17B (6xSSC, 45C
hybridization). The size of the insert for these three genomic clones is
30 approximately 9-23 kb. They both contain the coding exon of the gene(s)
WO 93/25684~ 1 3 7 7 9 ~ PCI/US93/05672
that is closely related to the probes ussd for the screening, hNT4 (120 b~
and rNT4 (180 bp). The phage DNA for the genomic clones was digested
with several restriction enzymes and subjected to Southern analysis. The
appropriate fragment that hybridizes to the probe rNT4 (180 bp) can be
subcloned into Bluescript vector. The size of DNA fragments to be
subcloned are as follows: clone 2-1 (1.0 kb Xhol fragment), clone 4-2 (4.0
kb Xhol fragmant) and clone 7-2 (5.0 kb BamHI fragment). Complete
coding sequence can be obtained and this information can be used to
identify the exon boundaries to allow subcloning of this gene into an
appropriate expression vector.
To this end, nucleotide sequence analysis was performsd on human
genomic phage clone 7-2, which had been obtained by screening a human
genomic library with a PCR fragment derived from human genomic DNA
using degenerate oligonucleotides to the DNA sequence of Xenopus NT~
(see ~iscussicn, supra). Sequenca analysis revealed that human phage clone
7-2 contains a sequence identical to the sequence of the PCR fragment used
as a probe to screen the genomic library. This sequence is contained within
what appears to be an exon encoding a novel neulutrophic factor (Figure
18, SEQ ID NO:7~ and SEQ ID NO:76).
Alignment of the protein encoded by this exon (Figure 19, SEQ ID
NO:77) with the known neu,ul,ûph;lls revealed that it shares features found
in all the known neurotrophins (Figure 19, SEQ ID NOS:78-92). It contains a
prepro region in which are conserved many of the identical amino acids
consel.led between the prepro regions of previously defined neurotrophins.
Fullllerl,.ore, this prepro region is preceded by a splice acceptor site
localized in the same region as in other neurotrophin genes. The prepro
region also contains a consensus glycosylation site at the appropriate
position, and terminates at a cleavaga site which was very similar to the
cleavage sites found in the other neurotrophins (Figure 18). The prepro
region of 7-2 is unusual, however, due to its short length as compared to
W O 93/25684 2 ~ ~ 7 ~ ~ ~ PC~r/US93/05672
the prepro region of known neurotrophins. The decrease in length occurs in
the N-terminal portion of the prepro region, which is the least conserved
portion of prepros between family members. The mature region retains all
6 cysteines found in all previously identified neurotrophins. Many of the
5 residues shared between different members of the neurotrophin family are
also conserved. F~ch~ing the exlensive sequence similarity shared by aPCR
fragment derived from rat genomic DNAwhich may correspond to the rat
equivalent of the protein encoded by the human 7-2 clone, computer
alignments revealed that the neurotrophin encoded by the 7-2 phage clone
10 was most similar to that of Xenopus NT-4. This was true for both the
prepro and mature regions. The protein encoded by the 7-2 clone is
unusual, as compared to the known neurotrophins, due to the presence of
an insertion situated between the second and third cysteines in the mature
region.
Ssquence analysis was also performed on two additional human
clones isolated in the same screen;ng procedure that yielded clone 7-2 (see
disc~ssicn, supra). The sequence of these clones was similar to, but not
identical to, that obtained from clone 7-2, raising the possibility that thay
encode novel neulot,ophins more closely related to 7-2 than to the other
20 known neu-ol,ophins. The partial sequence of one of these clones, clone 2-
1, is presented in Figure 20 (SEQ ID NO:93 and SEQ ID NO:94). The
sequence disclosed starts at a position corresponding to amino acid
number 50 in the alignments clepi~1ed in Figure 19. Partial sequence of the
other clone, clone 4-2, is presented in Figure 21 (SEQ ID NO:116 and SEQ ID
NO:117).
10. EXAMPLE: TISSUE SPECIFIC EXPRESSION OF HUMAN NT4
A 680 bp Xhol-Notl fragment, containing the entire coding region of
30 the human genomic NT-4 clone, HG7-2, was radiolabeled and utilized in
W093/~5684 i~,~``f~ PCI/US93/05672
Northern analysis of various human tissue specific PolyA+ RNAs. The huma~
tissue specific mRNAs were fractionated by electrophoresis through a 1%
agarose-formaldehyde gel followed by capillary transfer to a nylon
membrane with 10X SSC. The RNAs were cross-linked to the membranes by
e~posure to ultraviolet light and hybridized at 65C to the 680 bp Xho1-
Not1 r~diol~heled NT-4 probe in the presence of 0.5M NaPO4 (pH 7), 1%
bovine serum albumin (Fraction V, Sigma), 7% SDS, 1 mM EDTA and 100
ng/ml sonicated, denatured salmon sperm DNA. The filter was washed at
65C with 2X SSC, 0.1% SDS and subjected to autoradiography overnight
with one intensifying screen and X-ray film at -70C. Ethidium bromide
staining of the gel demonstrated that equivalent levels of total RNA were
being assayed for the different samples.
The human NT-4 probe hybridized strongly to mRNA from skeletal
muscle, proslale, thymus, testes and place"la (Figure 22). The NT-4 probe
hybridized to a larger transcript in skeletal muscle than prostate mRNA.
This data suggests that a small human NT-4 multigene family, po~sessing
di~erenl e,~,ession levels as well as l,dnsc,ipl sizes, may be pressnl.
The high sx~,ression of human NT-4 in muscle tissue suggests that the
present invention may be utilized to treat disorders of ths nervous system,
2 0 speci~ically the wide array of neurological d;sorders affecting motor neurons
(see ~iscu~sion, supra). Additionally, high expression of human NT~ in
pn.sldle tissue suggests that the pfesenl invention may be utilized to treat
proslate ~ise~se, preferably BPH and impotency (see discussion~ supra).
Finally, ~ression of human NT-4 in thymus tissue suggests that the
presenl invention may be utilized to treat immunological related disorders of
nerve and muscla tissue, including but not limited to myasthenia gravis (see
scussion, supra).
74
WO93/25684 ~ 17~;~ PCr/US93/05672
J~ 11. EXAMPLE: CONSTRUCTION OF HUMAN NT-4 IN EUKARYOTIC
EX~htSSlON VECTORS AND THE MEASU11~1ENT OF BIOLOGICAL
ACTIVITY OF RECOMBINANT HUMAN NT-4
11.1 MATERIALS AND METHODS
11.1.1.CONSTRUCTION OF EUKARYOTIC EXPRESSION VECTORS ENCODING
HUMAN NT-4
Two eukaryotic expression vectors containing the prepro precursor
coding region of the human genomic clone HG7-2 were constructed in pCMX
(NRRLAcce~sion No. B-18790). The first construction utilized the normal
translation initiation site of pCMX (pCMX-HG7-2Q), while the other utilized
the Kozak consensus translation initiation site (pCMX-HG7-2M). A 5 kb
genomic fragment of HG7-2, contaWng the entire coding region cloned in the
BamHI site of Bluescript, was amplified by PCR utilizing the following
oligonucleolides;
hNT4-5XhoM:CGGTACCCTCGAGCCACCATGCTCCCTCTCCCCTCA
lSEQ ID NO:118]
2 0 hNT4-3'N~t~TACM~r~XGC I l l; l 1 GGGCATGGGTCTCAG
[SEQ ID NO:119]
hNT4~XhoQ:CGGTACCC I ~GAGCCACCCA~i l ~; 1 CCGAGAGATG
lSEQ ID NO:1201
Oligonucleotide primer combinations of hNT4-5'XhoM and hNT4-3'Not were
used to construct pCMX-HG7-2M, while oligos hNT4-5'XhoQ and hNT4-3'Not
were used to construct pCMX-HG7-2Q. The PCR fragment was digested
with Xho1/Not1 and subcloned into Xho1/Not1 digested pCMX.
WO 93/25684 . PCr/US93/05672
11.1 ~.CONSTRUCTION OF CHIMERIC GENES FUSING A
NElJ~ ~IIN PREPRO REGION TO THE MATURE CODING REGION OF
HUMAN NT-4
Two additional eukaryotic expression vectors encoding ths mature
portion of human NT-4 were constructed. First, the prepro region of human
NT-4 was replaced with the prepro region of Xenopus NT-4 (pCMX-
xNT4/hNT4). Second, the prepro region of human NT-4 was replaced with
the prepro region of human NT-3 (pCMX-hNT3/hNT4). The following
oligonucleotides were utilized in the construction of pCMX-xNT4/hNT4 and
pCMX-hNT3/hNT4:
(1) 5'CDM8: GAGACCGGMGC I l C l AGAGATC [SEQ ID NO:121]
(2) hNT3/hNT4 fusion (~US~ oligonucleotide):
1 5 TGCAG I I I CGCTCACCCCCCGl~ I CCGCCGTGATGT [SEQ ID NO:122]
(3) hNT3/hNT4 fusion (~DS~ oligonucleotida):
ACATCACGGCGGAAAC(~ r.TGAGCGMACTGCA ~SEQ ID NO:123]
(4) xNT4/hNT3 fusion (~DS~ oligonucleotide):
ACTTCCCGGCTAMACGGt3~t3~`TGAGCGAAACTGCA [SEQ ID NO:124]
(5) xNT4/hNT4 fusion (~US~ oligonucleotide):
TGCAG I I I CGCTCACCCCCCG I I I I AGCCGGGAAGT [SEQ ID NO: 109]
The hNT-3 containing plasmid vector (pC8-hNT3) was amplified by
PCRwith the 5'CDM8 and hNT3/hNT4 fusion oligonucleotides as primers.
The hNT-4 containing plasmid (pCMX-HG7-2Q) was amplified by PCR with the
hNT3/hNT4 fusion ~DS~ oligonucleotide and the hNT4-3'Notl oligonucleotide.
The PCRfragment obtained was excised from the gel, and reamplified by
PCR with the 5'CDM8 and hNT4-3'Notl oligonucleotides. The product was
then digested with Hindlll and Pstl and subcloned into Hindlll/Pstl digested
pCMX-HG7-2Q. Therefore, the expression plasmid pCMX-hNT3/hNT4
contained the hNT3 prepro region fused to the mature coding region of
human NT-4. Similarly, the human NT-4 eA~,r~ssion plasmid (pCMX-HG7-2Q)
WO 93/25684 2 1 ~ 7 7 q 9 PCI`/US93/05672
was amplifi~d by PCR with the 5'CDM8 and xNT4/hNT4/fusion ~US~
oligonucleotides as primers, while pCMX-HG7-2Q was amplified with the
xNT4/hNT4-fusion ~DS~ oligonucleotide and the hNT4-3'Not1 oligonucleotide.
The PCR fragment was excised from the gel, and reamplified with the
5 5'CDM8 and the hNT4-3'Not oligonucleotides. The product was then
digested with Hindlll and Pst1 and subcloned into Hindlll/Pst1 digested
pCMX-HG7-2Q. Therefore, the resulting eukaryotic expression piasmid,
pCMX-xNT4/hNT4 contains the Xenopus NT-4 prepro region fused to the
mature coding region of human NT-4.
11.1.3.EXPRESSION OF RECOMBINANT
HUMAN NT-4 IN COS CELLS
COS M5 cells were set up at a density of 1.5 x 105 cellshNell of a
15 Costar 6 well dish in DMEM media supplemented with 10% FBS, glutamine
and Na pyruvate (all from Irvine Scientific except FBS).
The next day the cells were aspiraled and refed with 2 ml/well of
RPMI media containing 400 g/ml DEAE-Dextran (Pharmacia), 400 M
chloroquine (Sigma), 4 mM glutamine (Irvine), 1 x ITS (insulin, transferrin,
20 selenium, Sigma). To each well 2 9 of the appropriate DNA was added and
mixed by swirling. Three separate constructs were used: pCMX-xNT4,
containing the prepro precursor of Xenopus NT-4; and two human NT-4
constructions, pCMX-HG7-2M and pCMX-HG7-2Q. After the addition of the
DNA the plates were returned to 37C, 5% CO2 incub~tor for 3 hours 15
25 minutes. Ths media/DNA mixture was then aspirated and 2 mlhNell of 10%
DMSO in PBS without Ca2+, Mg2+ was added for 2 minutes. The
DMSO/PBS was aspirated and wells washed once with 10% FBS DMEM, then
- refed with 10% FBS DMEM. The next morning, plates to be l~io~-~s~yed were
washed onc~ with Defined Media (DM) and refed 2 ml/well of DM. Three
30 days post-lra"sfection, supernatants wers removed from cells and debris
WO 9~5~7 7 ~ 9 PCI`/US93/05672
pelleted by microcentrifugation Supernatants were transferred to fres!
tubes and assayed for bioactivity.
11.1.4. PREPARATION OF ENRICHED MOTOR NEURON CULTURES
Embr,vos (E14) from Sprague-Dawley rats (HSD or Zivic-Miller) were
used for all experiments. Pregnant rats were sacrificed by carbon dioxide
asphyxiation, and embryos were rapidly removed and placed in ice-cold
medium for further ~issection. Spinal cords were removed aseptically from
10 rat embryos of 14 days gestation. The spinal cord was severed caudal to
the bulb (at the level of the first dorsal root ganglion), freed of sensory
ganglia and adhering meninges. The cord was then subdivided into ventral
and msdiodorsal segments for separate cultures. The ventral spinal cord
tissues were diced into small pieces and incub~ted in 0.1% trypsin (GIBCO)
and 0.01% deoxyribonuclease type 1 (Sigma) in PBS at 37C for 20 minutes.
Trypsin solution was then removed, rinsed and replaced with medium
consisting of 45% Eagle's minimum essential medium (MEM), 45% Ham's
nutrient mixture F12 (F12), 5% heat inactivated fetal calf serum (GIBCO),
5% heat inactivated horse serum (GIBCO), glutamine (2 mM), penicillin G (0.5
U/ml), and streptomycin (0.5 g/ml). The tissue was then mechanically
dissociated by gentle trituration through a Pasteur pipet, and the
supernatants were pooled and filtered through a nyion fiber (Nitex, Tetko;
40 m). Ths filtered cell suspension were then subjected to a modification of
the fraction procedure described by Schnaar and Schaffner (1981, J.
Neurosci, 1:204-217). All steps were carried out at 4C. Mel(i~cai"ide was
dissolved in F12:MEM (1:1 ) medium, and a discontinuous gradient was
esl~hlished which consialed of a 18% metrizamide cushion (0.5 ml), 3 ml of
17% metrizamide, 3 ml of 12% metrizamide, and 3 ml of a 8% mstrizamide
was prepared. The filtered ventral spinal cord cell suspension (2.~ rnl)
obtained as described above was layered over the step gradient, the tube
78
WO 93/25684 2 1 3 7 7 9 9 PCi'/US93/05672
was centrifuged at 2500 x 9 for 15 minutes using a swing-out rotor (Sorvall
HB4). Centrifugation resulted in three layers of cells: fraction I (at 0-8%
interface), fraction ll (at 8-12% interface), and fraction lll (at 12-17%
interface). The cells from each interface were removed in a small volume
5 (about 1 ml), rinsed twice with serum-free defined medium consisting of
50% F12 and 50% MEM, sllpp!emented with glutamine (2 mM), insulin (5
g/ml), transfe~,in (100 g/ml), progesterone (20 nM), putrescine (100 M),
and sodium selenite (30 nM) (Bollensleil, and Sato, 1979, Proc. Natl. Acad.
Sci. 76:514-517). Viable cell count was obtained by hemocytometer
10 counting in the presence of trypan blue. Fractionated ventral spinal cord
cells (enriched with motor neurons) were then plated at a density of
100,000 cells/cm2 in 6 mm wells precoated with poly-L-ornithine (Sigma: 10
g/ml) and laminin(GlBCO: 10 g/ml). Treatment with COS cell supernatants
conlaining NT-4 was COS cell was given on the day of plating. Cultures were
maintained in serum-free defined medium at 37C in 95% air/ 5% CO2
atmosphere at nearly 100% relative humidity. On day 2 (48 hours), oells
were harvested for measurements of choline acetyltransferase (CAT) as
desc,ibed in Fonnum, 1975, J. Neurochem. 24:407-409.
2 0 11.1.5. PREPARATION OF ENRICHED HUMAN MOTOR NEURON CULTURES
Seven to 9 week old human embryonic material was obtained from
aspiration abortions at the Geneva Cantonal I lospil~i. Appropriate consent
forms for experimental use of embryonic tissues were obtained from the
Ethics Commission for the Department of Gynecology and Obstetrics at the
Hospital. The age of the embryo was estimated according to menstrual
history, foot size (Streeter, 1920 Contri. Embryol. 11, 143) and external
characteristics (Moore, 1982, in UThe Develop;ng Human~, K.L. Moore, ed.,
pp 70-92, W.B. Saunders, Phi'^ lelpl,ia). The material was kept at 4 C for 2
to 6 hours until dicse~tion. The spinal cords were carefully isolated, all the
spinal roots were removed, and the meninges and other adhering tissue
79
W O 93/25684 PC~r/US93/05672
~r~ discarded. The cords were minced and incubated in 0.12% trypsin ir~
Ca2~, Mg2~- free salt solution for 10 minutes at 37C. The cells were
dispersed into a suspension by repeated trituration through a pipette. Cells
were centrifuged and resuspended in a standard culture medium (MEM plus
5 13% decomplemented human serum). The Petri dishes were covered with a
solution of polyornithine at a concentration of 1 mg/ml for 1 hr at 37 C
and rinsed three times with phosphats-buffered saline solution (PBS) before
plating. Cells were plated at a density of 4 X 104 and 10-15 X 104 cells per
6 and 11 mm tissue culture well, respectively. The cultures were maintained
1 0 at 37C in 5% C2/2 air. The medium was changed ev~ry three days and
cytosine arabinoside (ara C) (a0-6M) was added during the last 4 days of
culture. Human neurotrophic factors CNTF, NT-3 and NT-4 were added from
the start of the culture period at a concentration of 1 Ong/ml and lthe
culture medium was changed every 3 days. Choline acetyltransferase
1 5 (ChAT) was determined by measuring the synth~sis of 3H-acdtylcoenzyme
A. The ChAT measurements were done according to the method of
Fonnum, F. J., 1975, Neurochem. 24:407-409 with the modifications of
Raynaud, et al., 1987, Dev. Biol. 119:305-312 and Martinou et al., 1989, J.
Neurosci. 9:3645-3656 (1989).
11.2 RESULTS
11.2.1.EUKARYOTIC EXPRESSION OF BIOLOGICALLY ACTIVE RECOMBII\IANT
HUMAN NT-4
rla~",id DNA from each of the pCMX-based constructions (pCMX-HG7-
2Q, pCMX-HG7-2M, pCMX-hNT3/hNT4 and pCMX-xNT4/hNT4) was prepared
and individually transfected into COS cells. COS supernatants from each
transfectsd cell line were utilized in order to ~Csess the biological activity of
each respective recombinant form of NT-4. The volumes of C06
supernatants tested were 10, 50 and 250 1 in a total volume of 2 ml. Q1
(pCMX-HG7-2Q), N7 (pCMX-hNT3/hNT4 fusion), and X1 (pCMX-xNT4/hNT4)
WO 93/25684 2I 3 7 7 ~ ~ PCr/US93/05672
possessed neurite-promoting activity on DRG explants (Figure 23). In
addition, both Q (pCMX-HG7-2Q) and M (pCMX-HG7-2M) were examined for
their survival-promoting activity on DRG ~issoci~ted cells. Volumes tested
were 5-250 1 in a total volume of 2 ml. When added to cultures of
5 dissociated DRG neurons, COS supernatant containing hNT4 promoted 30%
neuronal survival compared to 10% survival with mock transfected C06
supernatants (Figure 24).
The biological effect of human recombinant protein from
supernatants of COS cells trans~ected with pCMX-HG7-2M was tested on
10 motor neuron enriched cultures prepared as described supra in Example
Section 11.1.4. and on human spinal cord neurons as described supra in
Example Section 11.1.5. Treatment of motor neuron enriched cultures with
pCMX-HG7-2M derived human NT-4 diluted to 1:5 resulted in a 2.9 fold
increase in choline acetyltransferase (CAT) activity after 48 hours as
15 compared to untreated (C-NT) and mock transfected (MOC COS) controls
(Figure 25). The increase in CAT activity dropped to 1.7 fold when a 1:50
dilution was tested, suggesting that it was a dose dependent response
(Figure 25).
The biological effect of the neurotrophins on cultured human spinal
20 cord neurons, as measured by ChAT activity, was as follows. Since there
was a dir~erence in the ChAT values from one experiment to another, the
values in the control wells were normalized to 100% in order to compare
results from dirrerenl experiments. The results have been pooled from 20
dir~erent cultures and are ex,uressed as the mean +/- S.E.M. (n=number of
2 5 wells):
Condition (n) % ChAT Activity
Control (77) 100
NT-3 (24) 231 +/- 23
BDNF (12) 252 +/- 27
NT-4 (25) 318 +/- 38
81
WO 93/2~3 7 7 9 ~ PCI`/US93/05672
11.3 DISCUSSION
The present invention provides for the utilization of an in vitro
eukaryotic expression system to express recombinant human NT-4. The
present invention discloses several strategies to express a biologically active
form of racombinant human NT-4 in COS cells. In one example, the DNA
sequence encoding NT-4 prepro precursor was amplified utilizing two FCR
amplification strategies to yield pCMX-based expression plasmids containing
either the pCMX translation initiation site (pCMX-HG7-2M) or a Kozak
consensus translation site (pCMX-HG7-2Q). In another example, two
chimeric neurotrophin genes fusing either the prepro region of Xenopus N~-4
(pCMX-xNT4/hNT4) or the prepro region of human NT-3 (pCMX-
hNT3/hNT4) to the mature coding region of NT-4 were constructed for
1 5 ex~ ression in COS cells (see Section 5, supra, for a cliscussion of the use of
chimeric constructions to express NT-4 in vitro).
Expression of a biologically active form of human NT-4 in an in vitro
eukaryotic expression system substantially increases the ease at which the
production of human recombinant NT-4, peptides or derivatives thereof may
be scaled up for both therapeutic and diagnostic applications discussed
supra. In view of the instant invention, one of ordinary skill in the art can
readily construct a plasmid containing an identical DNA sequence as
disclQsed or a similar DNA sequence encoding a homologous yet dislin~ NT-
4 like protein or derivative thereof. The skilled artisan can also pick and
2 5 choose between numerous DNA plasmid vectors known in the art to
construct an e~,ression plasmid for use in a eukaryotic expression system.
We have demonsl,aled that recombinant human NT-4, whether produced
as a full prepro precursor or via a neurotrophin-based chimaric
construction, is biologically active as demGns~,dled by the stimulating effect
W0 93/25684 2~ ~ 7 7 n ~ PCr/US93/05672
of recombinant NT-4 COS supernatants on neurite outgrowth in DP~
explants and the bioactivity on rat and human cultured motor neurons.
12. EXAMPLE: TRKB IS A RECEPTOR FOR NEUROTROPHIN-4
12.1 MATERIALS AND METHODS
-
12.1.1 3T3 FIBROBLAST SURVIVAL ASSAYS
C06 cell supernatants were examined in a survival assay utilizing 3T3
fibroblasts. In this assay system, 3T3 fibroblasts, which do not express
neurotrophin receptor proteins, are transfected with mammalian ek~,ression
vectors encoding either trkA, trkB or trkC. 3T3 fibroblast survival is
dependent on the addition and receptor specific binding of the respective
1 5 neurotrophic factor.
COS-M5 cells were cultured and lfansfe~ed with either pCMX-HG7-2Q,
pCMX-HG7-2M or pCMX-HG7-2Q as described in Example Section 11.1.3.
A full-length rat trkA cDNA clone was obtained from Dr. Eric Shooter
of Stanford University. Tha rat trkA cDNA was subcloned into thc
20 mammalian expression vector, pCMX, to generate pCMX-trkA.
Full-length rat trkB and trkC cDNA clones were obtained by screening
a rat brain cDNA library in the lambda ZAP2 vector (Stratagene) with rat
trkB-specific and trkC specific oligonucleotides corresponding to the most 5'
and 3' coding regions of trkB and trkC. Ths rat trkB and trkC cDNAs were
25 subclQned into pCMX to gener;~te pCMX-trkB and pCMX-trkC.
3T3 fibrobl~t-c wera cultured and transfected as described in Glass,
et al., 1991, Cell 66:405-413.
In this survival assay system, 3T3 ribrobl~ , which do not express
neurotrophin receptor proteins, have been l,ansfected with trkA, a proto-
3 0 oncogene encoJ;"g a tyrosine kinase receptor for NGF, with trkB, a tyrosine
WO 9?~?~63~7 7 ~ 9 PCI'/US93/05672
kinase which serves as a functional binding protein for BDNF and NT-3, o~
with trkC, a tyrosine kinase which serves as a functional binding protein for
NT-3. The transfected cells are dependent upon the addition of the
corresponding neurotrophin for survival, and thus may be used to assay for
biological activity of neurotrophins.
12.1.2 TRANSFECTION OF PC12 CELLS
A full length rat trkB cDNA was placed under control of the CMV
promoter in the pcDNA I expression vector (Invitrogen), which also contains
an LTR-promoted neo resistance gene. PC12 cells (a gift of Dr. Eric
Shooter, Stanford University) (107 cells per 10cm dish) were incub~ted for
24 hr in 5 ml OptiMEM medium (GIBCO BRL) conlai,)ing 25 mg DNA and 100
mg Lipofectin (GIBCO BRL), thsn rinsed and placed in frash medium. Five
days after transfection cells were select~d in 0.4 mg/ml G418. Resistant
colonies were assayed for differentiation in the presence of 100ng/ml BDNF
and one clone (PC12/trkB) was selected for further study.
12.1.3 CROSS LINKING STUDIES
Cross-linking studies were performed as follows: Briefly, cell lines and
cell suspensions from cortex, hippocampus and striatum were incub~ted in
PBS-glucose with 1nM of 1251 l~helled NT~, in the presence or absence of
excess cold naurotrophins for 2 hr at 4C. The cross-linking agent (6mM
2 5 EDAC for 1251-BDNF and 1251-NT~, 0.2mM DSS for 1251-NGF) was added, and
rotated at room temperature for 20 minutes. The mixtur~ was washed 3
times with a solution cGnlaining tris/NaCI. The cell pellet was resuspended in
complete RlPAlysis buffer. After centrifugation, the supernatant containing
cross-linked complexes was immunopreci~ led with trk-antibody (RG22),
84
wo 93/25684 ~ I ~ 7 7 9 t~ PCr/US93/05672
and subjected to electrophoresis. The fixed and dried gels were exposed
for autoradiography.
12.2 RESULTS
12.2.1 3T3 trkB CELLS REMAIN VIABLE IN THE PRESENCE OF NT-4
-
Addition of NT-4 containing COS cell supernatants in this bioassay
indicated that viable cslls remain after 48 hours only in 3T3 trkB cultures
10 (Table 2; data not shown indicated inability of NT-4 containing COS
supernatants to support 3T3-trkC cultures). Thus, these results
demonslrate that NT-4 protein has biological activity in this system, and
suggests that trkB, but not trkA or trkC, serves as a functional binding
protein for NT-4.
1 5
TABLE 2
Assay of COS Super"ata,)ts on 3T3
Cell Lines Ex~,essing TrkA and TrkB
DilutionsMock HG7- HG7-hNT3/
2Q 2M hNT4
1:5 _ -- -- --
1:10
2 5 3T3 1:20
1 :50
1:5
1:10
3 0 3T3-trkA 1:20
1 :50
1:5 _ + + +
1:10 _ + + +
3 5 3T3-trkB 1:20 _ + + +
1:50 _ + + +
WO 93/25684 PCI/US93/05672
7 ~ ~ ~
12.2.2 HUMAN AND RAT NT-4 ARE SIMILAR TO XENOPUS NT~ IN THEIR
ABILITY TO SPECIFICALLY ACTIVATE trkB.
To compare human and rat NT-4 to xNT-4 for their abilities t o
activate the various trk receptors, we first expressed all three of these
proteins transiently in COS cells; metabolic labeling was used to
demonstrate that all three proteins were produced in approximately equal
amounts by these cells (data not shown). COScell supsrnatents containing
the three proteins were then assayed for their ability to induce tyrosine
phosphorylation of the three known trks expressed in NIH3T3 fibroblasts.
Human and rat NT-4 were identical to xNT-4 in that they were specifically
able to induce the tyrosine phosphorylation of trkB, but not trkA or trkC
(Figure26). Furthermore, human NT-4 and xNT-4 induced trkB tyrosine
phosphorylation with very similar dose-dependencies (Figure 27A). Human
NT-4 and xNT-4 also displayed sirnilar dose-dependencies in their ability to
slicit grow~h of NIH3T3 cells ex~,lessing trkB receptors (Figure 27B). In
addition, both stimulated neurite outgrowth in PC12 cells expressing
introduced trkB receptors; neither human NT-4 nor xNT-4 could elicit
2 0 phenotypic effects from untransfected versions of these cells, or cells
expressing the other trk receptors (not shown). Thus mammalian NT-4 and
xNT-4 are very similar in their ability to functionally activate trkB but not
trkA or trkC.
2 5 12~.3 TYROSINE PHOSPHORYLATION OF trkB BY NT-4
The assay described above suggested that not only are mammalian
NT-4/5 and xNT-4 similar to BDNF in being spscific ligands for trkB, but they
might also be as potent as BDNF in their ability to activate trkB. To dirsctly
30 determine the specific activities of all the known mammalian neurotrophins
for each of the known trk receptors, we firs~ obtained highly purified
86
-
WO 93/25684 ~ ~ 3 7 ~ ~ PCI/US93/05672
preparations of each of the mammalian neurotrophins. Each of these
purified factors was then tested for its ability to induce tyrosine
phosphorylation of each of the trk receptors expressed in NIH3T3
fibroblasts. NGFwas clearly the preferred ligand for trkA, with very minor
5 inductions by very high concentrations of NT-3 and NT-4, while NT-3 was
clearly the preferred ligand for trkC, with minor induction by very high
concentrations of BDNF (Figure 28A and 28C). BDNF, NT-3 and NT-4 were
all quite effective at inducing tyrosine phosphorylation of trkB, although the
NT-3 induction appeared to raquire higher concentrations to achieve
10 saturation (Figure28B).
To compare specific activities for inducing phosphorylation with those
required for function effects, each of the purified neurotrophins was then
assayed for its ability to promote cell growth of NIH3T3 cells expressing
each of the trk receptors. Strikingly, the dose-responses for cell growth in
15 fibroblasts almost exactly paralleled those for phosphorylation (compare
panels A, B and C with D, E and F in Figure 28); by this assay, BDNF and NT-
4 were essentially indistinguishable in their ability to activate trkB, whereas
NT-3 was about 50-fold less potent. In all cases, the "preferred" ligands for
each trk receptor (i.e. NGF for trkA, BDNF or NT-4 for trkB, and NT-3 for
2 0 trkC) exhibited 50% of their maximum activity (EC50's) of between 1-
1 Ong/ml.
12.2.4 ACTIVATION OF PC12/trkB CELLS BY NT-4
2 5 In addition to the 3T3/trkB system, we hava also compared the
abilities of human NT-4 to activate trkB in PC1 2/trkB cells. Of the 4
- neurollophins tested, parental PC12 cells are only responsive to NGF (Figure
29). In PC12 cells stably l,ansfected with trkB, NT~ increased the survival
-
(Figure 29B) and perce"lage of neurite-bearing PC12/trkB cells (Figure 29A)
30 in a dose-dependent manner, similar to that seen with BDNF. PC12/trkB
87
2137799 PCI/US93/05672
cells responded to NT-3 only at high concer,lrhlions. Results of tyrosine
phosphorylation assays performed with 4 neurotrophins in PC 12 or
PC12trkB cells showed that NGF induced trkA phosphorylation in PC12 cells,
while BDNF and NT-4 and NT-3 (to a lesser extent) induced trkB
phosphorylation in PC12/trkB cells (Figure 29C).
12.2.5 NT~ COMPETES FOR trkB BINDING WITH BDNF and NT-3
Cross-linking experiments were carried out with radiodinated
neurotrophins. Iodinated NGF was found to cross-link to a protein
(presumed trkA) immunoprecipitable with trk antibody in PC12 cells and
striatal homogenates (rat postnatal day 7) (Figure 30A), and 3T3/trkA
cells (data not shown). In all cases, this cross-linked protein could be
competed by excess cold NGF, and not by BDNF, NT-3 or NT~. This data
in~;c~tes that NT-4 did not interact appreciably with the trkA receptor in
cell lines or in vivo. Similarly, iGdi.1dled BDNF could be cross-linked to a trk-
immunoprecipitable protein (presumed trkB) in 3T3/trkB cells, PC12/trkB
cells (data not shown), and cortex (rat postnatal day 7); this cross-linked
protein could be competed by BDNF, and NT-4, to a lesser extent by NT-3,
but not by NGF (Figure 30B). Iodinated NT~ cross-linking experiments gave
similar results. That is, in 3T3nrkB cells, PC12ttrkB cells (data not shown),
cortex and hippocampus (postnatal day 7), NT-4 cross-linked protein could
be competed by BDNF and NT-4, to a lesser extent by NT-3, and not by
NGF (Figurs 30C).
13. E)CAMPLE: NT 4 t~tC 1~ NEURITEOUTGROWlH AND CELL SURVIVAL OF
DORSAL ROOT GANGLION EXPLANTS
Neurotrophin responses in cultured primary neurons were examined.
The survival of sensoly neurons derived from embryonic E14 rat dorsal root
ganglia were supported by all of the known neurotrophins, albeit to differing
WO 93/25684 2 ~ ~ ~ 7 ~ ~ PCr/US93/05672
extents, which suggest that different neuronal subpopulations in the ganglia
are responding to the different neurotrophins (Figure 31). RNA was
prepared from immediately explanted dorsal root ganglia, as well as ganglia
that had been maintained for 24 hours in the presence of individual
5 neurotrophins; after this 24 hour treatment, only neurons responsive to
(and thus presumably expressing the appropriate trk receptor for) the
given neurotrophin survive, while non-responding neurons die. Untreated
ganglia immediately removed from the animals expressed all of the trks
(Figure 32). In contrast, ganglionic neurons surviving in the presence of
10 BDNF or NT-4 showed significant trkB message, but no detectable trkC
message (Figure 32). Ganglionic neurons surviving in the presence of NT-3
expressed trkC but not trkB. These data demonstrated that NT-4 act on
trkB and not on trkC. Similar studies were not possible with trkA because,
in contrast to the mutually exclusive relationship between trkB and trkC
15 ex~ressing neurons, it appears that subsPrltial numbers of trkB and trkC
ex~.ressi,lg neurons co-e,c~ress trkA (data not shown).
14. EXAMPLE: DISTRIBUTION AND RETROGRADE TRANSPORT OF NT-4
2 0 14.1 MATERIALS AND METHODS
NT-4 was iodina~ed by a modification of the lactoperoxidase method.
Briefly, 1 mCi of Na125I (NEN) was added with 1.2 ~lg of lactoperoxidase
(Sigma), 85 ~M H202 and 10 ~9 of NT-4 at pH 6.0 for 12 min. The
25 reaction was stopped by addition of 0.1 M Nal, 0.1 M Na phosphate and
1.0 M NaCI, pH 7.5. The reaction was diluted 1:1 with 2% bovine serum
albumin (Boehringer Mannheim) in PBS. The solution was dialyzed to
eliminate free (unincorporated) 1 25I. Percent incorporation (55%) was
determined by thin layer chromatography. Specific activity (2047
89
W O 93/25684~ PC~r/US93/05672
cpm/fmole) was calculated based on a molecular weight of 26,000 for NT~.
For sciatic nerve studies adult male Sprague-Dawlsy rats (Zivic Miller;
200-220 9; n=30) were anesthetized with a mixture of pentobarbital (35.2
mg/kg) and chloral hydrate (170 mg/kg), and the right sciatic nerve was
exposed. Two ~11 of ~25I-labeled NT-4, containing PBS or a 50-fold excess of
unlabeled neurotrophins, were injected into the nerve at the level of the
tendon of the obturator internus muscle with a Hamilton syringe. Wounds
were sutured and the animals allowed to recover for 18 hr. Rats were
killed, the DRGs were dissected, placed in 4% paraformaldehyde and
1 0 counted in a gamma counter for 1 min. Differences in mean transport
values were analyzed by analysis of variance (ANOVA).
For sympathetic neuron transport studies, 125I-labeled NT-4 was
injected into the anterior eye chamber as described (.lohnson et al., 1978).
After 16 hr SCGswere removed and counted in 4% paraformaldehyde as
1 5 for DRGs.
Fixed DRGs and spinal cords were equilibrated with buffered sucrose,
frozen in methyl butane and se~ioned in a cryostat (10 llm for DRG and
SCG, and 20 ~lm for spinal cord) and then mounted onto microscope slides.
The brains from perfused animals were removed, equilibrated in buffered
sucrose, and 25 ~Lm frozen sections were cut in the coronal plane and
mounted. Slides were then processed for emulsion autoradiography (using
Kodak NTB-2 emulsion) following established procedures (Cowan et al.,
1972). FYposlJre times ranged 1-3 weeks; however, comparable exposure
times were used for any individual region. After being developed, tissues
were counterstained through the emulsion (NTB-2, Kodak) with thionin.
For studies involving transport in the brain, male Sprague-Dawley
rats were anesthetized with chloral hydrate-pentobarbital and fixed in a
stereotaxic apparalus. Small volumes of [~25I]-l~heled trophic factors (0.2-
0.5 ~I) w~re injected slowly into the hippocampus or neostriatum by way of
a borosilic~te glass micropipelle. In other experiments, larger volumes (10
W O 93/25684 ~ 1 3 7 7g~ PC~r/US93/05672
1) were injected into the right lateral cerebral ventricle. The wounds were
closed and the animals allowed to recover. Approximately 24 hours later,
the animals were sacrificed and the brains fixed by transcardial perfusion of
buffered paraformaldehyde. The brains were then removed, sectioned and
5 processed for film and emulsion autoradiography. Hippocampal injections
were centered in the Dentate Gyrus/CA4 - Hilar region. Striatal injections
were located centrally in the rostral caudate-putamen.
Intracerebroventricular (ICV) injections were verified as being made into the
ventricular space. Similar amounts of [125I]-NGF, NT-3 and BDNF have been
10 injected at these sites previous experiments, permitting a clear
determination as to the specificity of the patterns of distribution and
retrograde transport within the CNS.
14.2 RESULTS
1 5
14.2.1 TRANSPORTOF NT-4 IN THE BRAIN
Film and emulsion autoradiographic experiments (Table 3) showed
that, like NGFlabeli"g associated with retrogradely transported NT-4 was
20 well loc~ e~l to magnocellular neurons of the medial septum and diagonal
band, cells which are known to provide the cholinergic input to hippocampus.
In general, more magnocellul~r neurons appear to be labeled in NGF-injected
animals compared to animals injected with NT-4.
Examination of film and emulsion autoradiograms available to date
25 have not provided evidence of transport of NT-4 to any other CNS cell
group following intrastriatal or intrahippocampal injections, in marked
contrast to results obtained with BDNFwhich is widely Iransported within
the CNS (Table 3).
91
WO 93/25684 ~ ~ ` PCI/US93/05672
Table 3: Retrograde Neuronal Labeling rOIIo~:;"y Hippo~mpal
p~37,~9 Injections of Rad;odi,)dled NGF, BDNF and NT~
Brain Area ~ ~E NT-4
Basal Forebrain
Medial Septum ++++ ++ ++
Diagonal Band (v) ++++ ++ ++
Diagonal Band (h) ++ + +
Basal Nuc. Meynert + _ _
1 0
Hippocampus
Hilus/CA 4 _ ++++
Hilus/CA 4 (contra) _ + + +
CA 1 _ +
CA2 _ +
CA3 ++
Brain Area ~3E ~IE NT-4
Dentate Gyrus ? ? ?
2 0 !Subic~ ~'u~n _ +
Par~s~ Ihlicu~um _ + + +
Other
Supramam. NUCIelJS ++ +++
2 5 Reuniens Nucleus _ +
Entorhinal Cortex _ + + +
Data reported above is derived from emulsion autoradiographic
experiments. Pluses represent relative numbers of cells l~hele~ in a given
30 area, from ~++++~ indicating many labeled cells to ~+~ i,-d;caling a few.
92
W O 93/25684 2 1 ~ 7 7 9 ~ PC~r/US93/OS672
Minus signs irl~l;G:~IeS that no l~heled cells were observed. A question mark
indicates areas that were difficult to evaluate given their proximity to the
illjeUliGn site
14.2.2 DISTRIBUTION OF NT~ FOLLOWING ICV ADMINISTRATION
-
In previous experiments, we have found that NGF diffuses widely into
the brain parenchyma following ICV administration, such that the
10 radiolabeled ligand becomes available to and concentrated within NGF-
responsive neuronal populations (eg. the cholinergic neurons of the basal
forebrain). Diffusion of NT-3, and particularly BDNF into the brain subsPnce
is much more limited in the rat, such that at most a few neurons within the
neural parenchyma concentrate sufficient amounts of labeled BDNF to be
1 5 clearly discriminable. Follov :ng ICV adminisl,alion of BDNF (and to a lesser
extent NT-3), labeling of the apical surface of the ventricular ependyma is
particularly prominent.
Following ICV administration of [125I~- NT-4, the pattern of
distribution seen is distinctly different from that described above for the
20 other neurotrophins. As for NGF, label associated with NT-4 is distributed
for some distance into the brain substance bordering the ventricles,
espec;e~ly at the level of the injection site. There is likewise some diffusion
into neural tissues from the extracerebral subarachnoid CSF space. In
co,lttast to NGF, no concentration of NT-4 ~csoci~ted label was apparenl in
25 neurons following ICV admini~lr~lion.
14.2.3 RETROGRADE TRANSPORT OF NT4 BY L4 AND L5 DRG NEURONS
125I-NT-4 was retrogradely transported by L4 and L5 DRG neurons.
30 This accumulation was specific as ~sessed by the fact that few counts
93
WO 93/2~ 8~ t ~ PCI/US93/05672
accumulated in the contralateral (non-injected) L4 or L5 DRGs and that~
transport was blocked by the co-injection of a 16-fold excess of NT-4. NT-
4 transport was blocked to varying degrees by all members of the
neurotrophin family. BDNF and NGF (57-fold) ware approximately equal in
blocking transport, while NT-3 was as effective as NT-4, when injected at a
57-fold excess. NT-4 was transported to SCG neurons when injected into
the anterior eye chamber.
15. EXAMPLE: BINDING OF 125I-NT-4 TO BRAIN AND RETINA
1 0
15.1 MATERIALSANDMETHODS
NT-4 was iodinated by ths lactoperoxidase method to a specific
activity of 2400-3500 cpm/fmol (1211-1789 Ci/mmol of NT-4). [12sI]-NT-4
1 5 was stor~d at 4C and used within 1-3 days after preparation. Biological activity of the radioiiodinated NT-4 was verified by bioassay.
Male Sprague-Dawley rats (200-250 9, Zivic Miller) were maintained
on a 12:12 h li5Jhl.dark cycle and given food and water ad libitum. The
brains and eyes of each rat were frozan in isopentane at -15C within 5 min.
of death. Serial, 12 um thick sections of these tissues were collected on
gelatin coated slides and were used for binding studies.
Sagittal whole body sections of adult male rat were mounted on a
large piece of adhesive tape which was then attached to a plastic frame.
This plastic frame created a rec~ss in which each section was incubated
with the [125I]-NT-4.
Binding assays using [125I]-NT-4 were performed as follows: After
being thawed, adjacent brain and whole body sections were preincubated
for 0.5 h in phosphate buffered saline, pH 7.4 followed by a 3 h incubation
at room temperature in DMEM tissue culture medium containing high
glucose, 10% heat- inactivated fetal calf serum (60C for 0.5 h), 25 mM
94
wo 93/25684 2 1 3 7 7 9 ~ PCI/US93/05672
Hepes buffer, 4 ug/ml leupeptin, 0.5 mM PMSF (BRL, Gaithersburg, MD.,
dissolved to 0.1 mglmg ethanol, 0.5 mM MgCI2 and I nM l125Il-NT-4 with
(non-specific) or without (total) I ~M unlabeled NT-3 or NT-4. Following the
incubation, the sections were washed for 0.5 h in PBS. Eye sections were
5 not preincubated, and were washed for 0.5 h in binding buffer without
labeled or unlabeled neurotrophin, since this procedure maintained the
anatomical integrity of the retina during the binding procedure. After
washing, sections were fixed for lO min in 4% paraformalde-hyde at 22OC,
rinsed for 2 seconds in dH20 and dried with a stream of room temperature
10 air. The labeled sections and 125I-containing radioactivity standards
(Amersham, Inc.) were exposed at room temperature for 2-5 days to 125I-
sensitive film (Hyperfilm, Amersham, Inc.). Slides were then dipped in Kodak
NTB-2 photographic emulsion diluted 1:1 with distilled water and developed
1 to 2 weeks later in Kodak D-19 at 15C.
Slides conl~ning eye sections were stained with hematoxylin-eosin for
histological examination.
15.2 RESULTS
In whole body sections of adult rats, specific binding of [125I]-NT-4
was restricted to the central nervous system, including the brain, spinal
cord and retina as well as to the dorsal root ganglia.
In brain sections, specific [125I]-NT-4 binding was found to be widely
distributed throughout the brain including the cortex, striatum,
hippoc~mpus, cerebellum, olfactory bulbs, periaqueductal gray, and raphe
nuclei.
Results from dry film eYposlJre of [125I]-NT-4 labeled rat eye sections
and human eye sections revealed high levels of displaGe~b'~ binding in the
retina. When examined at the emulsion level, intense, ~ispl~r,e~t~le labeling
WO 93/25684 PCI'/US93/05672
2~3 was found in the inner plexiform and ganglion cell layers of the retina as w~
as in the human optic nerve.
16. EXAMPLE: THE COMPARATIVE EFFECT OF NEUROTROPHINS IN
HIPPOCAMPUS
16.1 MATERIALSANDMETHODS
Hippocampi were dissected from E18 rat embryos of Sprague-
Dawley rats, and collected in F10 medium. The tissues were minced, rinsed
twice with F10 medium (Gibco) and trypsinized with 0.25% trypsin (Gibco)
for 20 minutes at 37C. Trypsin was inactivated by the addition of a serum-
containing medium composed of minimum essential medium (MEM
supplemented with fetal calf serum (FCS, 10%), glutamine (2 mM), penicillin
(25 U/ml) and streptomycin (25 llg/ml) in DME plus 10% fetal calf serum.
After 4 hours of culture, the medium was changed to DME plus I mg/ml BSA
and N2 media supplement [Bottenstein, st al., Methods Enzymol. 58:94-109]
and 1 mM pyruvate, at which time NT-4 was added. The media was
changed every three to four days, with re-addition of the factor.
16.2 RESULTS
Purified recombinant human NT-4 produced an increase in fos mRNA
in these cells similar to that seen with BDNF or NT-3 (Figure 33A). This
increase was followed by an increase in fos protein when examined at 2 hr
after treatment. The three neurotrophins (BDNF, NT-3 and NT-4) were
found to cause tyrosine phosphorylation of proteins immunoprec4)ilabl~ by
a pan trk-specific antibody (Figure 33B). Two cell populations that were
shown to respond to BDNF also responded to NT-4. That is, there was an
increase in the number of acetylcholinesterase-positive cellsand calbindin-
immunoposili~e cells in hippocampal cultures treated with NT-4 (Figure 34).
96
W O 93/25684 PC~r/US93/05672
~1~77~
.
17. EXAMPLE: NT4 INCREASES SURVIVAL AND DIFFERENTIATED FUNCTIONS
OF RAT SEPTAL CHOI INERGIC NEURONS IN CULllJRE
17.1 MATERIALS AND METHODS
17.1.1. PREPARATION OF DISSOCIATED CELLS AND CELL CULTURE
- CONDITIONS
1 0 The septal region from rats (Sprague-Dawley) after 17 days of
gestation was dissected free from the surrounding tissue. Tissue
fragments were pooled, washed three times with Ham's F-10, and then
transferred to a 35mm tissue culture dish and minced. A single cell
suspension was made by incub~ing the tissue with 0.25% trypsin for 20
1 5 minutes at 37~C. Following the inactivation of the trypsin by a five minute
incubation at room temperature in growth medium (in~). containing 100
~lg/ml deoxyribonuclease type 1 (Sigma), the cells were dissociated by
passing the fragments rep~atedly through the constricted tip of a Pasteur
pipet. The dissoc,~led cells were then centrifuged at 500xg for 45 seconds.
The super-,alanl was removed and recentrifuged.
The loose cell pellets were resuspended and combined in normal
growth medium (5% (v/v) horse serum (Gibco), 1 % N3 additives (v/v)
(Romijn et al. 1982, Dev. Brain Res. 2: 583-589), 0.5% (v/v) glutamine
(200mM, Gibco), and 0.25% (v/v) penicillin-sl-eptomycin (10,000 units/ml,
10,000 mcg/ml respectively, Gibco) in Dulbecco's modified Eagle's medium
(DMEM). Three to four hours after plating, sterile coverslips were placed in
each well to cover the cells. Neuronal-enriched cultures were prepared by
replacing 2/3 of the growth medium, 24 and 72 hours after plating with
N3/DMEM: DMEM containing 1% N3 additives, 0.5% glutamine, and 0.25%
~.enicillin and streptomycin. All suhse~luent medium changes, carried out
every other day, were done by removing 1/2 the volume o~ medium and
97
W O 93/25684 9 PC~r/US93/05672
2~ replacing it with an equal volume of fresh N3/DMEM. To limit the growth
astrocytes, the cultures were treated for 24 hours with cytosine
arabinoside at a concenl,dlion of 2~M after 1 week in culture.
17.1 .2 ASSAY OF CHOLINE ACETYLTRANSFERASE (ChAT) ACTIVITY
The growth medium was removed from the cultures and 125 ~l of
the Iysis buffer (50 mM KHaP4 pH 6.7 containing 200 mM NaCI and 25%
(v/v) Triton x-100) was added. With the tissue culture plates on ice, the
10 cells were scraped from the plates and the wells were rinsed with an
additional 125111 of Iysis buffer. The two aliquotes were then combined in
sppindorf tubes and quick frozen in a dry-ice methanol slurry.
17.2 RESULTS
1 5
The culture conditions used in this study limit the proliferatiorl of
astrocytes and allow for the long-term maintenance of basal forebrain
neurons in vitro. E. coli produced NT-4 was added to tha cultures 24 hrs
after plating and was replenished with svery medium change. At the end of
20 the two week treatment period, the cells were harvested. Figure35 depicts
ths dose related effect of NT-4 on ChAT activity. At a saturating
concentration of 25 ng/ml, NT-4 produced an approximate two-fold
il1creass in ChAT activity. This level of enzyme induction was maintained up
to the highest concentration (100 ng/ml) of NT-4 tested. The level of ChAT
25 induction observed with NT-4 is approximately equivalent to that observed
with BDNF. These data thus demonstrat~ that the basal forebrain
cholinersic neurons are a target for NT-4.
98
W 0 93/25684 ~I~77~ PC~r/US93/05672
18 EXAMPLE: NT4 SUSTAINS THE SURVIVAL OF DOPAMINERGIC
NElJPfONS
18.1 MATERIALSANDMElHODS
18.1.1. METHODS FOR CULTURING DOPAMINERGIC
SUBSTANTIA NIGRA NEURONS
-
Ventral mesencephalon was dissected from brains of rat embryos
10 varying in age from embryonic day 13 to embryonic day 15. Typically, two
litters were used in each experiment. The dissection solution had the
f~llowing composition: NaCI, 136.8 mM, KC1, 2.7 mM, Na2HPO4.7H20,
8.0mM, KH2PO4, 1.5 mM, glucose, 6 mg/ml, and BSA, 0.1 mg/ml, pH 7.4.
This solution was prepared and subsequently filter sterilized through a
15 0.211M pore filter. The dissection was performed under non-sterile
conJilions. Once the tissue was dEcsected from all the brains, the rest of
the procedure was carried out under sterile conditions. The tissue
fragments were placed in a 35 mm culture dish and minced using a fine
scissGr~. Two ml of F-12 nutrient media containing 0.125% trypsin was
20 then added to the tissue, snd incub~ted at 37 C. At the end of this
incob~ion period, DNAsel was added to the slurry such that the final
concentration was 80 ng/ml. Another idenlical incub~tion was carried out,
and the tissue slurry was sl~bse~luently added to 8.0 ml of growth medium
consisting of Minimal Csse,)lial Medium (MEM) supplemented with 2mM
25 glutamine, 6 mg/ml glucose, 5 units/ml penicillin, 5mg/ml streptomycin, and
7.5% fetal calf serum (FCS). The sample was centrifuged in a tabletop
centrifuge at room temperature at 500 rpm for a period of 5 minutes. The
medium was aspirated, and 2 ml growth medium was added to the cell
pellet. A fire polished pipette with an Gpening of I mm was used to triturate
3 0 the cells eight times. The remaining tissue fragments were allowed to settleby gravity, and a small aliquot of the supe",atant was taken to ~cse~s cell
99
W O 93/25684 PC~r/US93/05672
number by counting in a hemocytomater. After c~ll density was dstermine~
the cells were plated into tissue culture plates at a density of 50,000/cm2.
The culture plates were prepared on the day prior to dissection.
Tissue plates (24 well, 2 cm2/well) were precoated with polyornithine
(molecular weight 30,000-70,000 g/mol), 0.5 mg/ml, at room temperature
for 3 hours. The plates were extensively washed with water, and
subse~luently treated with mouse laminin, 5 ~lg/ml, at room temperature for
3 hours. The plates were then washed with water as above, and incubated
overnight at 37C in a humidified atmosphere consisling of 5% CO2, 95% air,
in the presence of growth medium. The medium in the plates was removed
the following day and replaced with fresh growth madium.
Once the cells were plated onto the culture plates, the cells were
placed in an incubator set at 37C and 5% CO2/95% air for a period of 24
hours. The culture medium was changed to a serum-free formulation (SFM)
having the following composition: a 1:1 (vol:vol) mixture of Basal Eagle
Medium (BEM) and nutrient mixture F-12 with glucose (33 mM), glutamine
(2mM), NaHCO3 (15 mM)~ HEPES (10mM), supplQmented with insulin (25
~g/ml), putrescine (60 ~M), progesterone (20 nM), sodium selenite (30
nM), penicillin (5 U/ml), strsptomycin (5 mg/ml), and T3 (30 nM). In some
experiments, purified BDNF was added to the cultures after the media
change to SFM on culture day 2.
The solutions used for culturing dopaminergic neurons were
prepared using water taken from a Milli-Q reagent water system. The
tissue culture media formulations were obtained through Gibco Laboratories
(Santa Clara, California), as was the fetal cal serum (lot number 43N1086)
and the mouse laminin. All other media components were purchased fTom
Sigma Chemical (St. Louis, MO), and were cell culture tested grade. The
polyornithine and DNAsel were also obtained from Sigma. Trypsin was
obtained from Worthington (Freehold, NJ), lot number 3667. Commercial
1 00
WO 93/25684 2 ~ ~ 7 7 ~ ~ PCI`/US93/05672
chemicals were of analytical grad~, purchased from Baker Chemical
(Phillipsburg, NJ).
18.1.2. METHODS FOR IMMUNOCYTOCI 1'~11GAL STAINING
OF VENTRAL MESENCEPHALON CULTURES
..
Fixativs solutions were prepared fresh for each experiment. For the
- staining of tyrosine hydroxylase (TH), the fixative was 4.0%
paraformaldehyde in Sorenson's phosphate buffer. The Sorenson buffer
was prepared by adding a 0.2 M solution of KH2PO4 to a stock of 0.2 M
NA2H P04 until tha pH reached 7.3. Ths paraformaldehyde was
subse~uently added to the solution and briefly heated, to allow it to be
dissolved, and cooled to room temperature before use.
To begin the procedure, culture medium was removed from the
15 culture dishes by gentle suction, and the proper fixative solution was gentlyadded to the dish. A room temperature incubation of 20 minutes was
carrisd out. Three washes in Sorenson's phosphate buffer, for 5 minutes
each, with gentle rotation, followed. The cells were then incubated in a
quench solution for 30 minutes at room temperature with gentle rotation.
20 The quench solution for the cultures to be stained for TH consisted of
Sorenson's phosphale buffer containing 2% normal horse serum. Next, the
cultures were incllb~ted in permeabilization buffer at room temperature for
30 minutes with gentle rotation. The solution consisled of Sorenson's buffer
containing 0.2% saponin, and 1.5% of normal horse serum for the cultures
2~ to be stained for TH. Following the perma~hi'i~tion step, the cultures were
incub~ted in the presence of primary antibody overnight at 4C. The
antibody against rat TH was a mouse monoclonal antibody of isotype
IgG2a. It was used at 40 llg/ml in a colution of 20 mM NaPO4, 50mM NaCI,
0.2% saponin pH 7.5. Following the primary antibody incubation, the
30 cultures were washed 5 times for 15 minutes each in the appropriate
permeabilization buffer. Next, the cultures were incub~ted with secondary
1 0 1
WO 93/25684 PCI/US93/05672
anlibody conjugated to biotin, that is biotinylated horse anti-mouse IgO
2 ~ This incubation was carried out at room temperature for two hours with
gentle rotation. Washes identical to those described above followed, and
the cultures were then incubated in the presence of a preformed avidin-
5 biotinylated horseradishperoxidase complex (ABC reagent, Vector
Laboratories, Burlingame, CA) prepared according to manufactur~r's
protocol. After a 30 min. incubation at room temperature with gentle
rotation, the cultures were washed as described above. The cultures were
subse~uently incubated with 55 mM Tris-CI pH 7.3 containing 0.5 mg/ml
10 diaminobenzidine and 0.02% hydrogen peroxide. The development of
reaction product was allowed to proceed for 2-5 min. after which the
solution was removed and the cultures were washed several times with the
ice cold PBS. The number of positive cells/cm2 was then ascertained.
The paraformaldehyde and the glutaraldehyde were obtained from
15 Fluka Chemical. Ve~t~ct~in kits containing normal serum (used as a blocking
agent), biotinylated, affinity-purified anti-immunoglobulin, avidin DH, and
biotinylated HRP-H wera purchased from Vector Laboratories. The
diaminober,~idine was obtained from BRBDNFL (Gaithersberg, MD).
18.2 RESULTS
Two sets of data from experiments in which 2 lots of human NT-
4(made in E. coli) were used are shown in Figures 36A and 36B. Cultures
were prepared as previously described, and were treated with increasing
25 concentrations of NT-4. In ths experiment shown in Figure 36A, the
treatment of cultures with NT-4 was given as a single addition on the day of
plating. In ths experiment shown in Figure 36B, the NT-4 treatment was
given as multiple additions at the day of plating (culture day 1), and
subse~uently at culture days 4 and 7 (CD4, CD7). At culture day 8, the
3 0 cultures were processed for immunocytochemical staining for the
1 0 2
WO 93/25684 2~ ~ 3 7 'i ~ 9 PCI'/US93/05672
dopaminergic marker tyrosine hyroxylase. The number of dopaminergic
neurons presenl in each dish was then determined. Each treatment group
represenls 5 replic~te cultures.
The results from both experiments show that NT-4 treatment leads
5 to an increase in the number of dopaminergic neurons detected by TH
immunocytochemical staining. This increase is dose dependent and
saturable as shown in Figure 36B.
19. EXAMPLE: EFFECTS OF NT-4 ON STRIATAL NEURONS IN VITRO
1 0
19.1 METHODS
19.1.1 DISSOCIATED CULTURE PREPARATION
Striatal neuronal cultures were prspared from E1 7 rat brains as
follows: striatal tissue was minced in calcium- and magnesium-free Hank's
balanced salt solution and dicsoc;~ted by enzymatic treatment with 0.25%
trypsin and DNAase (0.2 mg/ml) followsd by mechanical trituration in
medium composed of D~'bscco'sModified Eagle's Msdium and 10% fetal calf
serum (DME-FCS). Dissociated cells were seeded at a density of 104
cells/well in serum-free N2 medium in 9C woll tissue culture plates that had
been previously coated with polylysine and merosin. Human NT-4 (0.1-50
ng/ml) was added at the time of plating and replenished every other day.
2 5 19.1 ~ IMMUNOHISTOCHEMICAL STAINING FOR CALBINDIN
Striatal cultures at 8 days in vitro (8 DIV) were fixed in 4%
paraformaldehyde for 30 minutes, rinsed with PBS, permeabilized for 15
minutes in 0.1% Triton X-100/PBS, and blocked with 10% horse serum/1%
30 bovine serum albumin/PBS for 90 minutes at room temperature. Cultures
1 0 3
W0~9~84 PCI/US93/05672
were then inu)b~ted with the primary antibody (Mouse-anti-calbindin. Sigma~
1:5000 dilution) plus 5% normal horse serum overnight at 4C prior to
incubation with the secondary antibody (biotinylated horse-anti-mouse
(Vector Labs, 1:400 dilution) for 90 minutes at room temparature.
Call,i".lin immunoreactivity was visu~ ed by using the Vectastatin
ABC kit (Vector Labs). The number of total neurons as well as the numbsr
of neurons immunoreactive for calbindin were counted in approximately
5-10% of the total area of each of four duplicate culture wells.
1 0 19.1.3 MEASUREMENT OF HIGH-AFFINITY GABA UPTAKE
High-affinity GABA uptake was measured according to a modification
of the procedure of Tomozawa and Appel, 1986, Brain Res. 399:111-124.
Cells were washed once in buffer containing 140 mM NaCI, 2.6 mM KCI, 0.75
mM MgCI2, 0.75 mM CaCI2, 1 mM KH2PO4, 1 mM Na2HPO4, 6 mg/ml glucose,
2 mM l~-alanine, and 1 mg/ml BSA. Following one wash, cells were incub~ted
in the uptake buffer for 5 minutes at 37C. 3H-GABA (NEN, NET-191X,100
Ci/mmol) was then added at a final concenlfalion of 17 nM, and cells were
incub~ted for 15 minutes at 37C. In both incubations, neuron-specific
uptaks was verified by the ar~dition of 1 mM nipecotic acid to some wells.
Following the second incub~tion~ cells were washed four times with uptake
buffer at 4C, and then incub~ted with 0.5 M NaOH for two hours at room
tsmperature. The cell extract was then collected and the 3H-GABA in the
extract counted.
19.2 RESULTS
Striatal cultures were treated with human NT-4 purified from E. coli.
In striatal cultures treated with NT-4 for 8 days, the percent of total
neurons that were calbindin-immunoreactive increased by approximately 3
to 4 fold compared to untreated control cultures (Figure 37).
1 04
wo 93/25684 2 ~ ~ 7 7 ~ ~ PCI/US93/05672
NT-4 treatment produced an approximately 3 to 4 fold increase inthe level of high-affinity uptake of GABA as compared to untreated controls
(Figure 38).
20. EXAMPLE: EFFECTOF M~ ON NIGRAL GABAergic NEURONS
20.1 METHODS
20.1.1 PREPARATION OF VENTRAL MESENCEPHALON CULTURES
Cultures were prepared from the ventral mesencephalon of embryonic
day 14 (E14) rat embryos as before (Hyman et al, 1991,Nature, 350:230-
232; Spina et al., 1992, J. Neurochem., 59:99-106). The single cell suspension
obtained following trypsinization and mechanical dissociation of the brain
tissue was seeded at a density of 5 x 104 oell per cm2 onto 35mm dishes
which had been precoated with poly-L-lysine and merosin (Collaborative
Research). After a 4 hour incub~tion in MEM sur)plQmented with glutamine
(2mM), glucose (6 mg/ml), penicillin G (0.5U/ml), streptomycin (5 llg/ml),
and fetal calf serum (FCS,7.5%) to allow for cell attachment to the
subst~atum, cells were cultured in the presence or absence of trophic
factors in a defined medium consisting of F12 and basal Eagle medium
(1:1,vh) with N2 supplements as described by Bottenstein and Sato,1979,
Proc. Natl. Acad. Sci. USA, 64:787-794, except that the insulin concentration
was reduced to 20 ng/ml, and glutathione was included at 2.5 ~g/ml.
2 5 20.1.2 3H~ABA UPTAKE MEASUREMENT
The measurement of high-affinity GABA uptake activity was carried
out accor.ling to the method set forth in Example 19.
20.1.3 GLUTAMIC ACID DECARBOX~LASE (GAD) IMMUNOCYTOCHEMICAL
3 0 STAINING
105
W O 93/25684 PC~r/US93/05672
2,3 3~ 9 Cultures to be stained for GAD were prefixed in a 1 :1 mixture of 4
parafor",aldehyde and F12/BME (1:1, v/v) for 10 minutes, followed by 30
minutes incubation in 4% paraformaldehyde. Cultures were then incub~ted in
the presence of phosphate buffer containing 4% goat serum and 0.02%
5 saponin, after which the cultures were incubated overnight at 4C with goat
anti-feline GAD 67 antiserum (3enerously s~ppi Qd by Dr. A. Tobin, UCLA, CA)
at a dilution of 1/7500 . Cultures were washed, incubated with goat anti-
rabbit IgG at a concenlralion of 1.5 ~Lg/ml, and specifically bound antibody
was detected after binding of avidin-HRP, by development using DAB with
1 0 NiSO4 intensification (Hancock et al, 1982,Neurosci. Lett., 31:247-252).
20.1.4 MEASUREMENT OF GABA CONTENT
Cultures were prepared and maintained in vitro for various times,
either in the presence or absence of trophic factors. At the end of the
1 5 culture period, cells were harvested and immediately acidified ~lvith 0.4 N
perchlorate containing 0.1 mM ascorbate and 2.5 ng/ml
dihydroxybenzylamine (DHBA, an internal standard). Following
homogenization and centrifugation to remove precipitated protein, the
catecholamines in the extract were absorbed onto alumina (ICN). The
2 0 samples were then washed extensively, and the catecholamines recovered
by extraction from the alumina with 174nM acetic acid containing 0.05%
sodium disulfite and 0.025% EDTA. The protein content of the sample was
determined after resuspension of the pelleted precipitate in PBS by the
method of Smith et al, 1985, Anal. Biochem. 76-85. GABA determinations
25 were carried out, after derivatization with ophthalaldehyde, by an HPLC
separation using a reverse phase C18 column in a mobile phase consisting of
25% methanol, 3.1% acetonitrile, 0.1 M Na2PO4, pH 6.8. Quantitation of the
various amino acids was performed after alectrochemical detection (ESA
5500 coulochem electrode array system detector) of the eluted peaks from
3 0 the HPLC, and the data normalized to the protein content of the sample.
1 0 6
WO 93/25684 PCI'/US93/05672
213~7~
20.2 RESULTS
20 ~ 1 EFFECT OF NEUROTROPHINS ON GABAERGIC NEURONS
To examine possi~le effects of the neurotrophins on nigral GABAergic
5 neurons, high-affinity GABA uptake, GABA content, and GAD activity were
measured in control and treated cultures. The presence of GABAergic
neurons in the cultures was first detected by immunocytochemical staining
using an antibody specifically directed against the 67kD form of G~D,
which is found in terminal processes and neuronal perikarya (Gonzales et al,
1991, J. Neurocytol., 20: 953-961; Kaufman et al, 1991, J. Neurochem.
56:720-723). Fig. 4 shows representative GAD staining patterns obtained in
cultures maintained in the absence of any neurotrophic factors for 4, 7, or
11 days (41A-C, respectively). No staining was observed in the absence of
primary antibody (41D). Cell counts indicated that the number of G~D
positive neurons in cultures maintained for one week in vitro in the absence
of neurotrophins represented 3.3+0.7% of the total cell count at that time.
Only NT-3 produced a significant increase (63%) in the number of GAD
positive neurons after 7 days in vitro.
Although BDNF did not increase the number of GAD-positive neurons,
BDNF as well as NT-3 produced dose-dependenl increases in GAD enzymatic
activity in cultures maintained for 7 days, as shown in Fig. 42A and 42B
respectively. NT-3 produced a greater increase than BDNF (3-fold vs 1.8-
fold) whereas NT-4/5 was without effect at any concentration tested (Fig.
42C; and up to 200ng/ml, data not shown).
As another marker of GABAergic neurons, we e,cplored the effects of
each of the neurotrophins on the high-affinity GABA uptake capacity of cells
cultured for 7 days. As shown in Fig. 43A, all three neufol,ophins tested
(BDNF, NT-3 and NT-4/5) elicited increases of 2- to 3-fold in GABA uptake.
The dose responses of BDNF and NT-3 were similar, reaching maxima at
approximately 20 ng/ml. NT-4/5 was effective at lower concenl,dlions than
1 0 7
.:
Wo 93/25684 Pcr/uss3/os672
~,~3~ BDNF or NT-3, saturating at 10 ng/ml with decreased effects at high~
concer,lf~lions. When we assessed possibie additive or synergistic effects
on GABA uptake, no combinatorial effects of any of the neurol~ophins were
observed (Fig. 43B). NGF (50 ng/ml) had no effect on GABA uptake
5 activity.
As a further marker of the GABAergic ph~notype, we measured
GABA conlenl in cultures treated with each of the neurotrophins (Fig. 44).
BDNF, NT-3, and NT4/5 all produced modest but significant increases in the
GABA content of cultures grown with these factors for 7 days. Again,
NGF treatment was without effect. These data are consistent with the
increases observed in the high-affinity GABA uptake activity measurements.
20.2.2 ANALYSIS OF TRK RECEPTOR DISTRIBUTION IN SUBSTANTIA NIGRA
To address the question of the site of action of the neurotrophins in
mediating the above effects on cultured nigral neurons, it was of interest to
ascertain the expression pattern of high-affinity BDNF and NT-3 receptors
(TrkB and TrkC, respectively) in both embryonic cultures and adult rat
brain subsP~ltia nigra. As shown in Fig. 4~, in situ hybridization clearly
indicated high levels of TrkB (45B, 45C) and TrkC (45D-F) mRNA in both the
ventral tegmental area (VTA) and substantia nigra of adult rat brain. The
distribution of TrkC is more widespread than TrkB. Examination of the
TrkC localization pattern at higher magnification (Fig. 45E,45F),
demonstrates that Trk C is expressed in large perikarya, indicative of
neurons.
To ascertain if TrkB and TrkC are expressed in embryonic rat ventral
mesencepl,alic tissue, RNA prepare(J from E14 mesencephalic cultures grown
in the presencs or absencQ of BDNF or NT-3 for various times was probed
by Northern blot analysis. As shown in Fig. 46A, a probe to the kinase
domain of TrkB indic~ted the presence of a 9 kb transcript under all
conditions. F~poslJ~e of cultures to BDNF or NT-3 for up to 29 hours did
108
W O 93/25684 2 ~ ~ 7 7 9 g PC~r/US93/05672
not substantially alter the expression levels of TrkB mRNA. The 9 kb
transcript detected is one of the 2 brain-specific transcripts described by
Klein et al, 1991, Cell 65:189-197 which was shown to correspond to the full
length Trk B tyrosine kinase cell surface receptor. In the blot probed for
5 TrkC (Fig. 46C), a 14kb transcript was detected both in adult brain and
E14VM culture RNA. This species has been identified as the major lransc,ipt
encoding full length TrkC (Valenzuela et al, 1993, in press). The expression
level of this transcript was not strikingly altered in NT-3 treated cultures.
An additional 5kb TrkC transcript was detected. Migrating just above the
1 0 28S ribosomal RNA band, this is one of two known transcripts which encode
truncated forms of TrkC (Valenzuela et al., 1993, in press).
20.2.3 DISCUSSION
The above data provides evidence that the neurotrophins may
1 5 provide a role in supporting GABAergic neurons in the su~ nlia nigra. In
general, altered GABAergic neu~lrans",ission is ~ssoci~ted with generalized
seizures. In particular, generalized seizures can be prevented by increasing
GABAergic neurol(ar,s",ission within the subst~rltia nigra. Gale, K., 1985,
Federation Proceedings 44:2414-2424; Olsen, R.W., et al., 1986, in
20 Neurotransminers, Seizures and Epilepsy lll, Nistico, et al. (eds), Raven
Press, New York. Accordingly, data provided herein suggest that the
neu,ol.ophins may have potential utility for the treatment of seizure related
disorders.
21. EXAMPLE: NT-4 IN COMBINATION WITH OTHERNEUROTROPHIC
FACTORS SUSTAINS THE SURVIVAL OF MOTOR NEURONS
The effect of NT-4 on motor neurons was measured by monitoring
ChAT activity as described above. NT-4 stimulated ChATactivity in motor
3 0 neuron enriched cultures in a dose dependent manner (Figure 39).
Simultaneous treatment of motor neuron enriched cultures with NT-4 and
1 0 9
WO 93/25684 PCI/US93/05672
99 CNTF in combination as well as NT-4 and NT-3 in combination resulted in
mor~ than additive Qffect on ChAT activity. NT-4 (100 ng/ml) alon~
increassd ChAT activity 3.8 foid and CNTF (50ng/ml) alone increase ChAT
activity 4.5 fold. However, when they wer2 added simultaneously, ChAT
activity was elevated 13.4-fold, suggesting synergistic actions of the two
factors.
(Figure 40).
21. DEPOSIT OF MICROORGANISMS
1 0
The following recombinant bacteriophage, containing a human
genomic sequencs related to neurotrophin-4, were deposited on August 22,
1991 (HG4-2 and HG7-2) and September 11, 1991 (HG2-1) with the
Amsrican Type Culture Collection, 12301 Parklawn Drive, Roclcville, Maryland
1 5 20852, and assigned the indicated ~cc~ssicn number. Additionally, the
chimeric gene construction, pCMX-hNT3/hNT4, was deposited on October
30, 1991 with the American Type Culture Collection and assigned the
indicated ~ccession number. A recombinant bacteriophage (hCNTF-G1),
containing a human gsnomic sequence related to ciliary neurotrophic factor
(CNTF), was deposited on September 12, 1989, with the American Type
Culture Collection and assigned accession number ATCC 40657. A
recombinant bacteriophage [phi hN3(G1)], containing a human genomic
sequence related to neurotrophin-3 (NT-3), was deposited on February 28,
1990 with the American Type Culture Collection and assigned accession
2 5 numbsr ATCC 40763.
Bacteriophage ATCC Accession Number
HG4-2 75069
HG7-2 75070
3 0 HG2-1 75098
1 1 0
WO 93/25684 2 ~ ~ 7 7 9 ~ PCI/US93/05672
pCMX-hNT3/hNT4 75 133
The present invention is not to be limited in scope by the deposited
microorganisms or the specific embodiments described herein. indeed,
5 various modifications of the invention in addition to those described herein
will become apparent to those skilled in the art from the foregoing
description and accompanying figures. Such modifications are intended to
fall within the scope of the appended claims.
Various public~l;o,)s have been cited herein which are incorporated by
10 reference in their entireties.
1 1 1
WO 93/25684 PCI'/US93/05672
International ~rpli~ ion No PCT/
MICROORGANISMS
Option~l Sh~ct in with tho ' _ ' rcfened to on pa~p t 10 line~ 9-30 ~md p~oe 1 t 1 lin~t 1 of the
description
A ~uEn N I~lCATION OF DEPOSIT '
Furth~r depo~iti rt~ id~ntifbd on ~n Addition~l ~he~lt '
Name of deposit~try institulion
~mencan Type Culture CoDection
Addrnss of dcpositary institution (includinçl postnl code and country) '
12301 Pnrklnwn Drive
Rockville MD 10582
US
D8te of deposit ' Auaust 22. 1991 Accos~;on Number ' 75069
B ADDITIONAE INDICATIONS ~ b~ U a ~ppl'~tb) Tbi in~otttt l;on i. or~in~d r~ dnl ~et ED
C DESIGNATED STATES FOR WHICH INDlCATtONS ARE MADE ~
D SEPARATE FURNISHING OF INDICATIONS ~'D ~ ur a pp~b)
Tl~- inr~i~ tion h.ted b~low will b~ ubrnitterl to the ~nt~rn tior~ ~u e-u l-ter lsp~city th- oener-l n tur~ Of the i~irJ.~ion. ~ 0
Acre~ion Nurnber of Depo it'l - -
E I~This sneet was received with tne I -' ' application when filed (~o bc chec ed by tne receiving Offlce)
(Aulhorized 0ffil3
O The date of receipt (from tbe applicant) by tbe 1- ~ ' ' 8ureau "
W215 J
(Autnorized OfGcer)
Form PCT/RO/134 ~January 1981 )
112
WO 93/25684 ~ 1 ~ 7 7 ~ ~ PCI/US93/05672
.
I"t~ tiondl A!, ' ~ No: PCT/
Form PCT/RO/134 ~cont.)
Americ~n Type Culture Collection
12301 PArkbwn Drive
Aockville, MD 10582
US
Accession No. D~te of DePosit
75070 Au~ust 22, 1991
75098 Sl."t.,.,.L.~r 11, 1991
75133 October 30, 1991
113
W O 93/25684 P ~ /US93/05672
9 ~h~Uh~r; LISTING
(1) GENP'RAT INFORMATION:
(i) APPLICANT: Ip and Yancopoulo~
(ii) TITLE OF lNvr;h~ION: Therapeutic and DiagnoQtic Method~
Baqed on Neurotrophin-4 Expreusion
(iii) NUMBER OF Sr;yur;N~S: 124
(iv) CORRESPONDENCE ADDRESS:
A~I ADDRESSEE: R~g~neron Pharmaceuticalff, Inc.
BI STREET: 777 Old Saw Mill River Road
C CITY: Tarrytown
D STATE: New York
El COuh~r: U.S.A.
,,F, ZIP: 10591
(v) COMPUTER ~T.~AnARTT.~ FORM:
A'l MEDIUM TYPE: Floppy di~k
Bl COMPUTER: IBM PC compatible
CI OPERATING SYSTEM: PC-DOS/MS-DOS
,D, SOFTWARE: PatentIn Relea~e #1.0, Ver~ion #1.25
(Vi) ~UKR~l APPLICATION DATA:
(A) APPLICATION NUMBER: 07/898,194
(B) FILING DATE: 12-JUN-92
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA
(A) APPLICATION NUMBER: US 07/791,924
(B) FILING DATE: 14-NOV-9l
(viii) A.lORNr;r/AGENT INFORMATION:
(A) NAME: Gail M. Kempler
(B) REGISTRATION NUMBER: 32,143
(C) R~rriK~N~/DOCKET NUMBER: 6526-115/Reg 80
(ix) TELECOMMUNICATION INrORMATION:
(A) TELEPHONE: 914 347-7000
(B) TELEFAX: 914 347-2113
(C) TELEX:
(2) INFORMATION FOR SEQ ID NO:1:
( i ) ~U~I.~ CHARACTERISTICS:
'A' LENGTH: 130 baue pair~
Bl TYPE: nucleic acid
C, STRANDEDNESS: double
~D, TOPOLOGY: unk- ."
(ii) MOLECULE TYPE: DNA ( g~nr ~ C )
(xi) ~riQ~ DESCRIPTION: SEQ ID NO:1:
CAAATGTAAT CCCGCTGGTG GAA~.~lGGG TGGCTGCCGG GG.~.GATC GACGCCATTG 60
GATCTCTGAG TGCAAGGCTA AACAGTCTTA CGTGAGGGCT CTGACTATGG ATTCTGACAA 120
GAl~.. GGC 130
(2) INFORMATION FOR SEQ ID NO:2:
(i) SEQUENCE CHARACTERISTICS:
114
WO 93/25684 ~ 1 ~ 7 7 'I ~ PCr/US93/05672
IA'I LENGTH: 130 base pairs
B TYPE: nucleic acid
,C STRANDEDNESS: double
Dl TOPOLOGY: unknown
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:2:
CAAGTGCAAT CCATCAGGCA GCACCACTAG AGGATGCCGA GGTGTAGACA AAAAGCAATG 60
GATATCTGAG TGCAAAGCAA AACAGTCTTA TGTGAGGGCT CT~.AC~AT~G ATGCCAACAA 120
G~.`~.GG~ 130
(2) INFORMATION FOR SEQ ID NO:3:
(i) ~yu~N~ CHARACTERISTICS:
'A) LENGTH: 127 ba~e pair~
B) TYPE: nucleic acid
C) STRANDEDNESS: double
,D) TOPOLOGY: tlnkn~
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:3:
CAAGTGCCGG r-ACCrAAATC CCGTTGACAG CGGGTGCCGG GGCATTGACT CAAAGCACTG 60
GAACTCATAT TGTACCACGA CTCACACCTT TGTCAAGGCG CTGACCATGG ATGGCAAGCA 120
GGCTGCC 127
(2) INFORMATION FOR SEQ ID NO:4:
(i) S~yu~.._~ CHARACTERISTICS:
'A'l LENGTH: 127 ba~e pair~
,B TYPE: nucleic acid
,C, STRANDEDNESS: double
~Dl TOPOLOGY: I~nk- "
(ii) MOLECULE TYPE: DNA (genomic)
(xi) ~'QU~:N~ DESCRIPTION: SEQ ID NO:4:
CAAGTGCCGG GCCC~AAATC CTGTAr-~r-~G TGGATGCCGG GGCATTGACT CCAAGCACTG 60
GAACTCATAC TGCACCACGA CTCACACCTT TGTCAAGGCG TTGACAACAG ACGA~A.AAr~ 120
GGCTGCC 127
(2) INFORMATION FOR SEQ ID NO:5:
(i) ~Qu~._~ CHARACTERISTICS:
~A~I LENGTH: 127 base pairs
'B, TYPE: nucleic acLd
C, STRANDEDNESS: double
lD, TOPOLOGY: I~nk- ,."
- ( ii ) MOT~CUT~F TYPE: DNA (y~r- ic)
(Xi) ~QU~N~: DESCRIPTION: SEQ ID NO:5:
115
WO 93/25684 PCI'/US93/05672
CAAGTGCAGG GACCCTAGGC CGG~ CCAG CGGGTGCCGA GGGATCGATG CGAAGCATTG 60
GAACTCTTAC TGCACCACGA rACAr~CCTT CGTCAAAGCA CTGACCATGG AGGGCAAGCA 120
AGCAGCC 127
(2) INFORMATION FOR SEQ ID NO:6:
(i) SEQUENCE CHARACTERISTICS:
,~A'I LENGTH: 127 baE~e pair~
B TYPE: nucleic acid
, Cl STRANDEDNESS: double
,,D,I TOPOLOGY: llnknot/"
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:6:
CAAGTGCAAA AATCCAAGTC CAGTATCAGG TGGGTGCAGG GGCATTGATG CCAAGCATTG 60
GAATTCGTAT TGCACCACAA rAt'.A~Ar~TT TGTCAGGGCA TTAACCATGG AAGGCAATCA 120
GGCATCT 127
(2) INFORMATION FOR SEQ ID NO:7:
(i) SEQUENCE CHARACTERISTICS:
,~A~I LENGTH: 127 base pairs
,8, TYPE: nucleic acid
,C, STR~NDEDNESS: double
D TOPOLOGY: , n knl . "
(ii) MOLECULE TYPE: DNA (genomic)
(Xi) ~ iQU~;N~:~; DESCRIPTION: SEQ ID NO:7:
CAAATGCAGG GACCr~AAGC TAGTTTCAAG CGGATGCCGT GGGATTGATG CAAAGCATTG 60
GAACTCTTAT TGTACCACCA CGCACACCTT TGTCAAAGCA TTAAr~ATGG AAGGGAAGCA 120
AGCAGCA 127
(2) INFORMATION FOR SEQ ID NO:8:
(i) SEQUENCE CHARACTERISTICS:
A'l LENGTH: 127 base pair~
, Bl TYPE: nucleic acid
C STRANDEDNESS: double
l,DI TOPOLOGY: 1nlrnot."
(ii) MOLECULE TYPE: DNA (genomic)
(xi) S~:QIJ~ .CE DESCRIPTION: SEQ ID NO:8:
CACGTGCCGT GGCGCCCGGG CGGGCAGCTC TGGCTGCCTG GGCATCGACG GGCGACACTG 60
GAACTCCTAC TGCACCAACT CGCACACCTT CGTGCGGGCG CTGACTTCCT TTAp~r-GAccT 120
GGTGGCC 127
(2) INFORMATION FOR SEQ ID NO:9:
116
W O 93/25684 ~ ~ 3 7 7 9 ~ PC~r/US93/OS672
(i) SEQUENCE CHARACTERISTICS:
/A~I LENGTH: 130 ba~e pairs
'B TYPE: nucleic acid
,C STRANDEDNESS: double
,DI TOPOLOGY l-nknl~."
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE D~Sr~TPTION: SEQ ID NO:9:
CAAGTGCAAT CCCATGGGTT AcAc-AA~An~ AGGCTGCAGG GGCATAGACA AAAGGCATTG 60
GAACTCCCAG TGCCGAACTA CCCAGTCGTA CGTGCGGGCC CTTACCATGG ATAGCAAAAA 120
GAGAATTGGC 130
(2) INFORMATION FOR SEQ ID NO:10:
(i) SEQUENCE CHARACTERrSTICS:
/A'I LENGTH: 130 ba~e pair~
B TYPE: nucleic acid
,C, STRANDEDNESS: double
DJ TOPOLOGY: l~nl-- ~."
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:10:
CAAGTGTAAT CCCATGGGTT ACACr-~A~A AGGCTGCAGG GGCATAGACA AAAGGCACTG 60
GAACTCGCAA TGCCGAACTA CCCAATCGTA ~ CGGGCC CTTACTATGG ATAGCAAAAA 120
GAGAATTGGC 130
(2) lN~O~MATION FOR SEQ ID NO:11:
yDh-.~E CHARACTERISTICS:
'A' LENGTH: 130 base pair~
Bl TYPE: nucleic acid
Cl STRANDEDNESS: double
,Dj TOPOLOGY: nnl--~"
(ii) MOLECULE TYPE: DNA (genomic)
(xi) ~yuhl.~ DESCRIPTION: SEQ ID NO:ll:
CAAATGCAAC CCr-AAGGGGT Ar,ACAAArGA AGGCTGCAGG GGrATAr-~CA AGAGGCACTG 60
GAACTCACAG TGCCGAACTA CCCAGTCTTA CGTGAGAGCT CTCACCATGG ~AAr.,AA~AA 120
GAGAGTTGGC 130
(2) INFORMATION FOR SEQ ID NO:12:
(i) ~yu~_~ CHARACTERISTICS:
~A' LENGTH: 130 ba~e pair~
'B TYPE: nucleic ~cid
,C, STRAN,DEDNESS: double
l,D, TOPOLOGY: l~nl~ , "
(ii) MOLECULE TYPE: DNA (genomic)
117
W O 93/25684 ' P ~ /US93/05672
213~79~ --
(xL) SEQUENCE DESCRIPTION: SEQ ID NO:12:
CAAGTGCAGC ACGAAGGGTT ATG~AAAAr-A AGGCTGTAGA GGCATA~-Ar~ AGAGGTACTG 60
GAATTCCCAG TGCCGAACTA CTCAGTCTTA CGTCCGCGCT CTCACCATGG ATAACAAAAA 120
GAGGATTGGA 130
(2) INFORMATION FOR SEQ ID NO:13:
(i) ~Q~N~L CHARACTERISTICS:
'A' LENGTH: 130 ba~e pair~
,B TYPE: nucleic acLd
C STRANDEDNESS: double
~D, TOPOLOGY: unk- . "
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:13:
CAAATGCAAC CCTATGGGTT ACATGAAAGA AGGCTGCAGA GGCATAGACA AAAGGTACTG 60
GAACTCTCAG TGCCGAACTA CTCAGTCTTA CGTGCGGGCT TTCACCATGG ATAGCAGAAA 120
AAAAGTTGGT 130
(2) INFORMATION FOR SEQ ID NO:14:
(i) S~:yULN~ CHARACTERISTICS:
'A' LENGTH: 130 ba~e pair~
B TYPE: nucleic acLd
C STR~NDEDNESS: double
.,D,I TOPOLOGY: llnkn~tl~l
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:14:
CAAATGTAAC CCTATGGGGT A~-ACAAAr-G~ GGGC~GCCGI GGAATAGACA AGAGGCATTA 60
TAACTCCCAA TGCAGGACAA CCCAGTCCTA CGTGCGAGCG CTCACCATGG ATAGCAAAAA 120
GAAGATTGGC 130
(2) INFORMATION FOR SEQ ID NO:15:
(i) ~QuL.-'CE CHARACTERISTICS:
'A'l LENGTH: 130 baQe pair~
B TYPE: nucleic acLd
C, STRANDEDNESS: double
~D, TOPOLOGY: ~nknl.."
(ii) MOLECULE TYPE: DNA (genomic)
(xi~ ULN~: DESCRIPTION: SEQ ID NO:lS:
CAAATGTAAC CCCAAGGGTT Tr~C~ A AGGCTGCAGA GG~ATAGA~-A A~-AAA-~TTG 60
GAATTCGCAG TGTAGAACCA GCCAATCCTA TGTGCGAGCT CTAAC~ATGG ATAGTAGGAA 120
118
WO 93/25684 2 ~ ;~ 7 ~ ~ ~ PCI/US93/05672
GAAGATTGGG 130
(2) lNrO~MATION FOR SEQ ID NO:16:
(i) ~h~UhN~h CHARACTERISTICS:
'A'l LENGTH: 130 base pairs
B TYPE: nucleic acid
C, STRANDEDNESS: double
,,DI TOPOLOGY: unknown
( ii ) ~OT T'`CUT ~ TYPE: DNA (genomic)
- (xi) ~hyuhN~h DESCRIPTION: SEQ ID NO:16:
GCGATGTAAG GAAGCCAGGC CGGTCAAAAA CGGTTGCAGG GGTATTGATG ATAAAcAcTG 60
GAACTCTCAG TGCAAAACAT CC~AAArCTA CGTCCn~G~A CTGACTTCAG A~-~A-rAATAA 120
A~ GGGC 130
~2) INFORMATION FOR SEQ ID NO:17:
(i) ~hyUb~ : CHARACTERISTICS:
~A~I LENGTH: 130 base pairs
B, TYPE: nucleic acid
,C STRANDEDNESS: double
,D) TOPOLOGY: llnk~
(ii) MOLECULE TYPE: DNA (9~-- ;c)
(xi) ~h~U~ DESCRIPTION: SEQ ID NO:17:
GAGGTGTAAA ~-AAGC~Ar-GC CAGTCA ~AA CGGTTGCAGG GGGATTGATG ACAAACACTG 60
GAACTCTCAG TGrAAAACGT CGCAAACCTA CGTCCGAGCA CTGACTTCAG AAAA~AprAA 120
ACTCGTAGGC 130
(2) INFOR~ATION FOR SEQ ID NO:18:
(i) ~h~U~:N'~: CHARA'CTERISTICS:
'A'l LENGTH: 130 ba~e pair~
,'8, TYPE: nucleic acid
C, STRANDEDNESS: double
~DJ TOPOLOGY: llnknr~,"
( ii ) ~T~T~crJr~ TYPE: DNA (genomic)
(xi) ~r;gDL~ : DESCRIPTION: SEQ ID NO:18:
AAGGTGTAAA GAAGCCAAAC CTGTTAAAAA TGGCTGCCGA GGCATTGACG ACAAGCACTG 60
GAACTCCCAG TGrAA~-ACAT CC~AAACTTA CGTTAGAGCA TTGACTTCAG AAAA~ATAA 120
ACTTGTAGGC 130
= (2) INroR~ATIoN FOR SEQ ID NO:l9:
(i) Shy~hr._r; CHARACTERISTICS:
(A) LENGTH: 66 base pair~
(B) TYPE: nucleic acid
119
W O 93/25684 PC~r/US93/05672
(C) STRANDEDNESS: double
(D) TOPOLOGY: llnkn~"
?, ~ ( ii, MOLECULE TYPE: DNA ~genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:l9:
AAGGTGTAAA GAGGCAAGAC CTGTCAAAAA TGGCTGTCGA GGrAT~r~ArG ACAAACACTG 60
GAATTC 66
(2) INFORMATION FOR SEQ ID NO:20:
(i) SEQUENCE CHARACTERISTICS:
~A' LENGTH: 128 base pairs
B TYPE: nucleic acid
C STRANDEDNESS: lln~
,D, TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(xi) S~QU~:N~: DESCRIPTION: SEQ ID NO:20:
CAAGTGTCGG ACTGCCAAAC CTTTTAAGAG CGGCTGTCGC GGCATCGATG ACAAACACTG 60
GAACTCGCAG TGTAAr-AccT CTCAGACGTA CGTCAGAGTC TCTGACGCAG GACCGTACCT 120
CTGTGGGC 128
(2) INFORMATION FOR SEQ ID NO:21:
Q~ CHARACTERISTICS:
'A' LENGTH: 130 ba~e pair~
Bl TYPE: nucleic acid
C STRANDEDNESS: double
,D,, TOPOLOGY: llnkn~,...
(ii) MOLECULE TYPE: DNA (genomLc)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:21:
CCGCTGCAAG GAGTCGAAGC CGGGCAAGAA CGGGTGCCGG GGCATCGACG ACAAACACTG 6û
GAACTCGCAG TG~Ar-AcrA Gcr~r-AccTA TGTCCGAGCG CTGAGCAAGG Ar-AArAATAA 120
ATATGTGGGC 130
(2) lN~OR~ATION FOR SEQ ID NO:22:
( i ) ~U~N~ CHARACTERISTICS:
,'A', LENGTH: 43 amino acids
IB TYPE: amino acid
,C STRANDEDNESS: single
l,D, TOPOLOGY: 1~ n'~
( i i ) MOT~CUT ~ TYPE: peptide
(xi) ~QU~:N~ DESCRIPTION: SEQ ID NO:22:
Lys Cys Asn Pro Ala Gly Gly Thr Val Gly Gly Cys Arg Gly Val Asp
120
W O 93/25684 ~ 7 ~ ~ PC~r/US93/05672
1 5 10 15
Arg Arg Hi8 Trp Ile Ser Glu Cy~ Ly- Ala Ly~ Gln Ser Tyr Val Arg
20 25 30
Ala Leu Thr Met A~p Ser Asp LYB Ile Val Gly
35 40
(2) INFORMATION FOR SEQ ID NO:23:
(i) SEQUENCE CHARACTERISTICS:
,'A' LENGTH: 43 amino acid~
BI TYPE: amino acid
~ ,C STRANDEDNESS: Hingle
- ~D, TOPOLOGY: unl- - "
(i$) MOLECULE TYPE: peptide
(xi) ~hy~NCE DESCRIPTION: SEQ ID NO:23:
Ly~ Cy~ A~n Pro Ser Gly Ser Thr Thr Arg Gly Cys Arg Gly Val A~p
Ly~ Ly~ Gln Trp Ile Ser Glu Cys Ly~ Ala Ly~ Gln Ser Tyr Val Arg
20 25 30
Ala Leu Thr Ile Asp Ala A~n LyQ Leu Val Gly
(2) INFORMATION FOR SEQ ID NO:24:
(i) SEQUENCE CHARACTERISTICS:
'A' LENGTH: 42 amino acids
BI TYPE: amino acid
,C STRANDEDNESS: single
,D, TOPOLOGY: un~ wn
(ii) MnT.T~CUT.T~ TYPE: peptLde
(Xi) ~Q~N~ DESCRIPTION: SEQ ID NO:24:
Ly~ Cy~ Arg Affp Pro Asn Pro Val A~p Ser Gly Cys Arg Gly Ile A~p
1 5 10 15
Ser Ly~ Hi~ Trp A~n Ser Tyr Cyff Thr Thr Thr His Thr Phe Val Lye
20 25 30
Ala Leu Thr Met A~p Gly Ly~ Gln Ala Ala
(2) lN~0~l5ATION FOR SEQ ID NO:25:
( i ) ~N~: CHARACTERISTICS:
'A) LENGTH: 42 amino acids
B) TYPE: amino acid
C) STRANDEDNESS: single
,D) TOPOLOGY: u nkr "
(ii) MOLECULE TYPE: peptide
-
(Xi) ~y~N~ DESCRIPTION: SEQ ID NO:25:
Lys Cy~ Arg Ala Pro A~n Pro Val Glu Ser Gly Cy3 Arg Gly Ile A~p
1 5 10 15
121
W O 93/25684 ' P ~ /US93/05672
.
~ ~Ser Ly~ Hi~ Trp Ann Ser Tyr Cy~ Thr Thr Thr Hi~ Thr Phe Val Ly~
2~ 9 ~ 20 25 30
Ala Leu Thr Thr Asp A~p Lys Gln Ala Ala
(2) IN~ORMATION FOR SEQ ID NO:26:
(i) SEQUENCE CHARACTERISTICS:
'A' LENGTH: 42 amino acids
~B TYPE: amino acid
C STRANDEDNESS: single
l,D, TOPOLOGY: unknown
(iL) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:26:
Lys Cys Arg Asp Pro Arg Pro Val Ser Ser Gly Cys Arg Gly Ile Asp
1 5 10 15
Ala Ly~ His Trp Asn Ser Tyr Cy~ Thr Thr Thr His Thr Phe Val Ly~
20 25 30
Ala Leu Thr Met Glu Gly Lys Gln Ala Ala
(2) INFORMATION FOR SEQ ID NO:27:
(i) SEQUENCE CHARACTERISTICS:
'A'l LENGTH: 42 amino acids
Bl TYPE: amino acid
,C STRANDEDNESS: ~ingle
l,D~ TOPOLOGY: tlnknl,,"
(ii) M~T.FCrJr.r~ TYPE: peptLde
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:27:
Ly~ Cy8 Ly~ Asn Pro Ser Pro Val Ser Gly Gly Cy8 Arg Gly Ile Asp
1 5 10 15
Ala Ly~ His Trp A~n Ser Tyr Cy~ Thr Thr Thr Asp Thr Phe Val Arg
20 25 30
Ala Leu Thr Met Glu Gly A3n Gln Ala Ser
(2) INFORMATION FOR SEQ ID NO:28:
(i) ~UL.._~ CHARACTERISTICS:
'A'I LENGTH: 42 amino acid~
B TYPE: amino acid
C, STRANDEDNESS: ~ingle
~DJ TOPOLOGY: 1- nk~
( ii ) ~T~T'`CuT~T'' TYPE: peptide
122
W O 93/25684 ~ ~ 3 7 7 9 ~ PC~r/US93/05672
.
(xi) ~P:yu~ DESCRIPTION: SEQ ID NO:28:
Ly~ Cy~ Arg A~p Pro Ly~ Pro Val Ser Ser Gly Cy8 Arg Gly Ile A~p
1 5 10 15
Ala LYB His Trp Asn Ser Tyr Cy~ Thr Thr Thr His Thr Phe Val Lys
20 25 30
Ala Leu Thr Met Glu Gly Ly~ Gln Ala Ala
(2) INFORMATION FOR SEQ ID NO:29:
(i) SEQUENCE CHARACTERISTICS:
'A~ LENGTH: 42 amino acids
B TYPE: amino acid
C STRANDEDNESS: 3ingle
D TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:29:
Thr Cy~ Arg Gly Ala Arg Ala Gly Ser Ser Gly Cy8 Leu Gly Ile Asp
1 5 10 15
Gly Arg Hi~ Trp A~n Ser Tyr Cyq Thr A~n Ser Hi~ Thr Phe Val Arg
20 25 30
Ala Leu Thr Ser Phe Lys Asp Leu Val Ala
(2) lN~uR~ATION FOR SEQ ID NO:30:
(i) ~kyDk~._~ CHARACTERISTICS:
lAI LENGTH: 43 amino acid~
Bl TYPE: amino acid
C, STRANDEDNESS: ~ingle
~D/ TOPOLOGY: nn~
( ii ) MnT~CUT ~ TYPE: peptide
(xi) ~:yuk..~~ DESCRIPTION: SEQ ID NO:30:
Ly~ Cy~ Asn Pro Met Gly Tyr Thr Ly~ Glu Gly CYB Arg Gly Ile Asp
1 5 10 15
Ly~ Arg His Trp A~n Ser Gln Cy~ Arg Thr Thr Gln Ser Tyr Val Arg
20 25 30
Ala Leu Thr Met A~p ser Ly~ Ly~ Arg lle Gly
(2) INFORMATION FOR SEQ ID NO:31:
( i ) x~yu~N~ CHARACTERISTICS:
123
W O 93/25684 PC~r/US93/05672
IA' LENGTH: 43 amino acid~
B TYPE: amino acid
,C, STRANDEDNESS: ~ingle
,DJ TOPOLOGY: I~n~n~.,,
(ii) ~OLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:31:
Ly~ Cys A~n Pro Met Gly Tyr Thr Ly~ Glu Gly Cy~ Arg Gly Ile A~p
1 5 10 15
Ly~ Arg His Trp A~n Ser Gln Cy~ Arg Thr Thr Gln Ser Tyr Val Arg
20 25 30
Ala Leu Thr Met Asp Ser LYB Lys Arg Ile Gly
(2) INFORMATION FOR SEQ ID NO:32:
(i) SEQUENCE CHARACTERISTICS:
~A LENGTH: 43 amino acids
~B TYPE: amino acid
,C STRANDEDNESS: ~ingle
l,D, TOPOLOGY: ~nknr,
(ii) MOLECULE TYPE: peptide
(xi) ~yu~r._~ DESCRIPTION: SEQ ID NO:32:
LYH Cys A~n Pro Ly~ Gly Tyr Thr Ly~ Glu Gly Cy5 Arg Gly Ile A~p
1 5 10 15
Ly~ Arg His Trp A~n Ser Gln Cy~ Arg Thr Thr Gln Ser Tyr Val Arg
20 25 30
Ala Leu Thr Met A~p Asn Lys Lys Arg Val Gly
(2) INFORMATION FOR SEQ ID NO:33:
(i) ~QU~_~ CHARACTERISTICS:
~A' LENGTH: 43 amino acid~
B TYPE: amino acid
C, STRANDEDNESS: ningle
l,DJ TOPOLOGY: l~ n ~n~ .. "
(ii) MOLECULE TYPE: peptide
(xi) ~QD~ DESCRIPTION: SEQ ID NO:33:
Ly~ Cy~ Ser Thr Ly~ Gly Tyr Ala Ly~ Glu Gly Cys Arg Gly Ile Asp
1 5 10 15
Ly~ Arg Tyr Trp Asn Ser Gln Cy~ Arg Thr Thr Gln Ser Tyr Val Arg
124
WO 93/25684 ~ 1 ~ 7 ~ ~ ~ PCI/US93/05672
Ala Leu Thr Met A~p A3n Ly~ Ly~ Arg Ile Gly
(2) lN~u}~ATION FOR SEQ ID NO:34:
( i ) ~Q~N~' CHARACTERISTICS:
'A' LENGTH: 43 amino acid~
B TYPE: amino acid
C, STRANn~nN~CS: ningle
~D, TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:34:
Ly~ Cyu Ann Pro Met Gly Tyr Met Ly~ Glu Gly Cy~ Arg Gly Ile A~p
LYR Arg Tyr Trp Asn Ser Gln Cy~ Arg Thr Thr Gln Ser Tyr Val Arg
20 25 30
Ala Phe Thr Met A~p Ser Arg Ly~ Ly~ Val Gly
(2) INFORMATION FOR SEQ ID NO:35:
(i) S~yU~:N~: CHARACTERISTICS:
(A' LENGTH: 43 amino acid~
(Bl TYPE: amino acid
(C STRANDEDNESS: single
(D, TOPOLOGy: ~In~n~
(ii) MOLECULE TYPE: peptide
(Xi) S~U~N~: DESCRIPTION: SEQ ID NO:35:
Ly~ CYB A~n Pro Met Gly Tyr Thr Ly~ Glu Gly Cy~ Arg Gly Ile A~p
1 5 10 15
Ly~ Arg Hi~ Tyr Asn Ser Gln Cy~ Arg Thr Thr Gln Ser Tyr Val Arg
20 25 30
Ala Leu Thr Met A~p Ser Lys Lyu Lys Ile Gly
(2) INFORMATION FOR SEQ ID NO:36:
- (i) SEQUENCE CHARACTERISTICS:
~A'l LENGTH: 43 amino acid~
B TYPE: amino acid
C STRANDEDNESS: ~ingle
,DJ TOPOLOGY: ~In~- .~"
(ii) MOLECULE TYPE: peptide
125
W O 93/25684 PC~r/US93/05672
) SEQUENCE DESCRIPTION: SEQ ID No:36:
Lys Cy- A~n Pro Ly~ Gly Phe Thr Asn Glu Gly Cyn Arg Gly Ile Asp
Ly~ Lys Hi~ Trp Asn Ser Gln Cys Arg Thr Ser Gln Ser Tyr Val Arg
20 25 30
Ala Leu Thr Met A~p Ser Arg Ly~ Ly~ Ile Gly
(2) INFORMATION FOR SEQ ID NO:37:
(i) SEQUENCE CHARACTERISTICS:
'A' LENGTH: 43 amino acid~
B TYPE: amino acid
C STRANDEDNESS: ~ingle
,D, TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:37:
Arg Cy~ Lys Glu Ala Arg Pro Val Lys A~n Gly Cys Arg Gly Ile Asp
1 5 10 15
A~p Ly~ His Trp Asn Ser Gln Cy~ Ly~ Thr Ser Gln Thr Tyr Val Arg
20 25 30
Ala Leu Thr Ser Glu A~n Asn Ly~ Leu Val Gly
(2) lNrORMATION FOR SEQ ID NO:38:
(i) ~yuh~CE CHARACTERISTICS:
'Al LENGTH: 43 amino acid~
Bl TYPE: amino acid
C, STRANDEDNESS: single
,DI TOPOLOGY: t~nkn~
(ii) MOLECULE TYPE: peptide
(xi) ~u~_~ DESCRIPTION: SEQ ID NO:38:
Arg Cy~ Ly~ Glu Ala Arg Pro Val Lys Asn Gly Cy~ Arg Gly Ile Aap
1 5 10 15
Asp Ly~ Hi~ Trp A~n Ser Gln Cys Lys Thr Ser Gln Thr Tyr Val Arg
20 25 30
Ala Leu Thr Ser Glu A~n A~n Ly~ Leu Val Gly
(2) INFORMATION FOR SEQ ID NO:39:
( i ) ~h~U~ CHARACTERISTICS:
(A) LENGTH: 43 amino acidt
126
WO 93/25684 2 :~ 3 7 7 ~ ~ PCI'/US93/05672
(B) TYPE: amino acid
- (C) STR~ND~n~CS: single
(D) TOPOLOGY un~- "
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:39:
Arg Cys Lys Glu Ala Lys Pro Val Lys Asn Gly Cys Arg Gly Ile Asp
' 1 5 10 15
- Asp Ly~ His Trp Asn Ser Gln Cy~ Ly~ Thr Ser Gln Thr Tyr Val Arg
20 25 30
Ala Leu Thr Ser Glu A~n Asn Lys Leu Val Gly
(2) INFORMATION FOR SEQ ID NO:40:
(i) ~yu~N~: CHARACTERISTICS:
A~l LENGTH: 21 ~mino acids
B TYPE: amino acid
,C, STRANDEDNESS: 8 ingle
l,DI TOPOLOGY: unknown
( ii ) Mor~cuT~F TYPE: peptide
(xi) S~yu~ DESCRIPTION: SEQ ID NO:40:
Arg Cys Lys Glu Ala Arg Pro Val Lys Asn Gly Cys Arg Gly Ile Asp
1 5 10 15
Asp Ly~ His Trp Asn
(2) INFORMATION FOR SEQ ID NO:41:
(i) SEQUENCE CHARACTERISTICS:
Al LENGTH: 42 amino acids
B TYPE: amino acid
C STRANDEDNESS: single
~Dl TOPOLOGY: ~1nl~ ..n
(ii) MOLECULE TYPE: peptide
(xi) ~yD~.._~ DESCRIPTION: SEQ ID NO:41:
Ly~ Cy~ Arg Thr Ala Ly~ Pro Phe Lys Ser Gly Cy~ Arg Gly Ile A~p
1 5 10 15
A~p Lys His Trp Asn Ser Gln Cys Lys Thr Ser Gln Thr Tyr Val Arg
Ala Leu Thr Gln Asp Arg Thr Ser Val Gly
127
W O 93/25684 P ~ /US93/05672
~2~ lNrORhATION FOR SEQ ID NO:42:
~QUL.._~ CHARACTERISTICS:
~A, LENGTH: 43 amino acids
IBI TYPE: amino acid
,C, STRANDEDNESS: ~inqLe
~D, TOPOLOGY: ~lnkn~ "
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:42:
Arg Cys Lys Glu Ser Lys Pro Gly Lys Asn Gly Cys Arg Gly Ile A~p
1 5 10 15
Asp Lys Hi~ Trp Asn Ser Gln Cys Lys Thr Ser Gln Thr Tyr Val Arg
20 25 30
Ala Leu Ser Lys Glu A~n Ann Ly~ Tyr Val Gly
(2) INFORMATION FOR SEQ ID NO:43:
(i) ~u~.~ CHARACTERISTICS:
'A'l LENGTH: 1302 ba~e pairs
,BI TYPE: nucleic acid
C STRANDEDNESS: double
D, TOPOLOGY: unknown
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NANE/REY: CDS
(B) LOCATION: join(529..534, 538..1248)
(xi) ~Qu~-..CE DESCRIPTION: SEQ ID NO:43:
CAATCATACT TATGAACAGC AGGGGGAGCC CTCGCCTTAC TTCCCAGCCA TGCAGAACTC 60
AAGCAGCTTT GTTTATGCCG ATCCCTAAGC AGCC~PGACC ACACTGAGCA TGTGCACAGT 120
CTTAGTCTTG ~AA~-ATGTT TAACAAPGTT ArpAGp~GGT GACCCC~r~ AGCCAACTTT 180
GAAAGCATAA ATCATTTGTT TGATTAGGCT ~rlG~rlGCAG TAAGTTCATG TTTATATTTA 240
G~ATArAAAA TACAGCATTT CTAGCCTTAT TCTATTTTAG ACTTTACCCT TTAATGCCCA 300
GTTCTGCCCA TTGCCTTATA GATGTTAAAG TCCrAATATC ACATTGGCAT CCTCGGCTGT 360
TTA~AA~AAA CATTAAAACT TGTACTTATA TTTAACATTC ~ rl~ 1. C~AATATTCC 420
ATCACACTTA GACCCTAPAA GAATTATATG TA~A~AATTT Gr-ATAAATTA TATA~TGGCA 480
GCCGTATTCT AA..~.~.-. .................. TTTTTGCAGT GGTCTGAG GTG GAT 534
Val A~p
TAA GTA ATG ATC CTC CGC CTT TAT GCC ATG GTG ATC TCA TAC TGT TGT 582
Val Met Ile Leu Arg Leu Tyr Ala Met Val Ile Ser Tyr Cyn Cy~
5 10 15
128
W O 93/25684 ~ ~ 3 7 7 ~ ~ PC~r/US93/05672
GCC ATC TGC GCT GCC CCC TTC CAG AGC CGG ACC ACA GAT TTG GAT TAT 630
Ala Ile Cys Ala Ala Pro Phe Gln Ser Arg Thr Thr Asp Leu Asp Tyr
20 25 30
GGC CCC GAT A-A-A ACA TCA GAA GCC TCA GAC CGG CAA TCA GTT CCC AAC 678
Gly Pro A~p Ly~ Thr Ser Glu Ala Ser ABP Arg Gln Ser Val Pro Asn
35 40 45
AAC TTC AGT CAT GTC CTG CAA AAT GGG TTC TTT CCA GAT TTG TCA TCC 726
Ann Phe Ser His Val Leu Gln Asn Gly Phe Phe Pro Asp Leu Ser Ser
50 55 60 65
ACC TAT TCC AGC ATG GCT GGT AAG GAC TGG AAC CTA TAC TCA CCT AGA 774
Thr Tyr Ser Ser Met Ala Gly Lys Asp Trp Asn Leu Tyr Ser Pro Arg
~ 70 75 80
GTG ACT CTT TCA AGT GAG GAG CCT TCT GGA CCT CCA CTA CTT TTC TTG 822
Val Thr Leu Ser Ser Glu Glu Pro Ser Gly Pro Pro Leu Leu Phe Leu
85 90 95
TCA GAG GAG ACA GTG GTA CAT CCA GAA CCA GCC AAC AAG ACT TCC CGG 870
Ser Glu Glu Thr Val Val His Pro Glu Pro Ala Asn Ly~ Thr Ser Arg
100 105 110
CTA AAA CGG GCA TCA GGA TCT GAT TCG GTC AGC TTG TCC CGT CGG GGA 918
Leu Ly~ Arg Ala Ser Gly Ser Asp Ser Val Ser Leu Ser Arg Arg Gly
115 120 125
GAG CTC TCT GTG TGT GAC AGT GTC AAC GTC TGG GTT ACC GAT AAA CGT 966
Glu Leu Ser Val Cys Asp Ser Val Ann Val Trp Val Thr A~p Lys Arg
130 135 140 145
ACA GCC GTG GAT GAT CGG GGT AAA ATA GTG ACT GTC ATG TCT GAG ATT 1014
Thr Ala Val A~p Asp Arg Gly Lys Ile Val Thr Val Met Ser Glu Ile
150 155 160
CAG ACT CTA ACA GGA CCA CTG AAG CAA TAC TTC TTT GAG ACC AAG TGC 1062
Gln Thr Leu Thr Gly Pro Leu Ly~ Gln Tyr Phe Phe Glu Thr Ly~ Cys
165 170 175
AAT CCA TCA GGC AGC ACC ACT AGA GGA TGC CGA GGT GTA GAC AAA AAG 1110
Asn Pro Ser Gly Ser Thr Thr Arg Gly Cy~ Arg Gly Val Asp Ly~ LYB
180 185 190
CAA TGG ATA TCT GAG TGC AAA GCA AAA CAG TCT TAT GTG AGG GCT CTG 1158
Gln Trp Ile Ser Glu Cys Lys Ala Lys Gln Ser Tyr Val Arg Ala Leu
195 200 205
ACC ATA GAT GCC AAC AAG CTT GTG GGT TGG CGT TGG ATC CGT ATT GAC 1206
Thr Ile A~p Ala Ann Ly~ Leu Val Gly Trp Arg Trp Ile Arg Ile Aap
210 215 220 225
ACA GCG TCT GTC TGT ACC TTG TTG AGT CGG ACA GGA AGG ACG 1248
Thr Ala Cys Val Cy~ Thr Leu Leu Ser Arg Thr Gly Arg Thr
230 235
TAAAAr-Arr-A GGGTTAGCAA AATAr-Ar-AGA AGAGGTTGAT CCGTTGACCT GCAG 1302
-
(2) INFORMATION FOR SEQ ID NO:44:
(L) ~u~._~ CHARACTERISTICS:
A) T- _~n: 239 amino acidn
B) TYPE: amino acid
,D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
129
WO 93/25684 - PCI/US93/05672
(xi) ~k~U~._~ DESCRIPTION: SEQ ID NO:44:
1 Asp Val Net Ile Leu Arg Leu Tyr Ala Met Val Ile Ser Tyr Cy8
Cy~ Ala Ile Cys Ala Ala Pro Phe Gln Ser Arg Thr Thr A~p Leu A~p
Tyr Gly Pro A~p Ly~ Thr Ser Glu Ala Ser A~p Arg Gln Ser Val Pro
Asn A~n Phe Ser Hi~ Val Leu Gln A~n Gly Phe Phe Pro A~p Leu Ser
Ser Thr Tyr Ser Ser Met Ala Gly Ly~ A~p Trp A~n Leu Tyr Ser Pro
Arg Val Thr Leu Ser Ser Glu Glu Pro Ser Gly Pro Pro Leu Leu Phe
Leu Ser Glu Glu Thr Val Val His Pro Glu Pro Ala Asn Lys Thr Ser
100 105 110
Arg Leu Ly~ Arg Ala ser Gly Ser A~p Ser Val Ser Leu Ser Arg Arg
115 120 125
Gly Glu Leu Ser Val Cyn A~p Ser Val A~n Val Trp Val Thr Asp Lys
130 135 140
Arg Thr Ala Val A~p A~p Arg Gly Ly~ Ile Val Thr Val Met Ser Glu
145 150 155 160
Ile Gln Thr Leu Thr Gly Pro Leu Lys Gln Tyr Phe Phe Glu Thr Ly~
165 170 175
Cys Asn Pro Ser Gly Ser Thr Thr Arg Gly Cys Arg Gly Val Asp Lys
180 185 190
Ly~ Gln Trp Ile Ser Glu Cys Ly~ Ala Ly~ Gln Ser Tyr Val Arg Ala
195 200 205
Leu Thr Ile Asp Ala A~n Ly~ Leu Val Gly Trp Arg Trp Ile Arg Ile
210 215 220
Asp Thr Ala Cy~ Val Cyn Thr Leu Leu Ser Arg Thr Gly Arg Thr
225 230 235
(2) INFORMATION FOR SEQ ID NO:45:
(i) ~uk..'E CHARACTERISTICS:
'A LENGTH: 123 amino acids
IBI TYPE: amino acid
,C, STRANDEDNESS: aingle
~D, TOPOLOGY: , n k- - "
(ii) MOLECULE TYPE: peptide
(Xi) ~Qu~N~k DESCRIPTION: SEQ ID NO:45:
Ala Ser Gly Ser A~p Ser Val Ser Leu Ser Arg Arg Gly Glu Leu Ser
1 5 10 15
Val Cy~ Asp Ser Val Affn Val Trp Val Thr Asp Ly~ Arg Thr Ala Val
13û
W O 93/25684 ~ 1 ~ 7 7 ~ ~ PC~r/US93/05672
A~p A~p Arg Gly Lys Ile Val Thr Val Met Ser Glu Ile Gln Thr Leu
Thr Gly Pro Leu Ly~ Gln Tyr Phe Phe Glu Thr Lys Cy~ A~n Pro Ser
Gly Ser Thr Thr Arg Gly Cy~ Arg Gly Val A~p Ly~ Ly~ Gln Trp Ile
Ser Glu Cy~ Ly~ Ala Ly~ Gln Ser Tyr Val Arg Ala Leu Thr Ile A~p
Ala A~n Ly~ Leu Val Gly Trp Arg Trp Ile Arg Ile A~p Thr Ala Cys
100 105 110
Val Cy~ Thr Leu Leu Ser Arg Thr Gly Arg Thr
115 120
(2) INFORMATION FOR SEQ ID NO:46:
(i) SEQUENCE CHURACTERISTICS:
'A, LENGTH: 118 amino acid~
B TYPE: amino acid
C STRANDEDNESS: ~ingle
~D, TOPOLOGY~ knr~."
(ii) MOLECULE TYPE: peptide
(Xi) S~yU~N~: DESCRIPTION: SEQ ID NO:46:
Ser Ser Thr His Pro Val Phe Hi~ Met Gly Glu Phe Ser Val Cy~ Asp
1 5 10 15
Ser Val Ser Val Trp Val Gly Asp Lys Thr Thr Ala Thr Asp Ile Ly~
Gly Ly~ Glu Val Thr Val Leu Ala Glu Val A~n Ile A~n A3n Ser Val
4S
Phe Arg Gln Tyr Phe Phe Glu Thr Lys Cy~ Arg Ala Ser A~n Pro Val
Glu Ser Gly Cy~ Arg Gly Ile A~p Ser Ly~ His Trp Asn Ser Tyr Cy~
Thr Thr Thr His Thr Phe Val Ly~ Ala Leu Thr Thr A~p Glu Ly~ Gln
Ala Ala Trp Arg Phe Ile Arg Ile A~p Thr Ala Cys Val Cy~ Val Leu
100 105 110
Ser Arg Ly Ala Thr Arg
(2) r~-~R~ATION FOR SEQ ID NO:47:
( i ) S~Qu~N~ CH7~RACTERISTICS:
~ lA~I LENGTH: 119 amino acid~
- B, TYPE: amino acid
C, STRANnT~'nNESS: 8ingle
lDI TOPOLOGY: lln~r ~
( ii ) M~T.T~'CUT.T~' TYPE: peptide
131
WO 93/25684 PCr/US93/05672
~X1) 51:,~Uhl~CI!; DESCRIPTION: SEQ ID NO:47:
Hi~ Ser A~p Pro Ala Arg Arg Gly Glu Leu Ser Val Cy~ Asp Ser Ile
Ser Glu Trp Val Thr Ala Ala Anp Ly~ Lys Thr Ala Val Asp Met Ser
Gly Gly Thr Val Thr Val Leu Glu Ly~ Val Pro Val Ser Ly~ Gly Gln
Leu Ly~ Gln Tyr Phe Tyr Glu Thr Ly~ Cy~ Aun Pro Met Gly Tyr Thr
Ly~ Glu Gly CYB Arg Gly Ile A~p Ly~ Arg Hi~ Trp A~n Ser Gln Cy~
Arg Thr Thr Gln Ser Tyr Val Arg Ala Leu Thr Met A~p Ser Ly~ Lys
Arg Ile Gly Trp Arg Phe Ile Arg Ile A~p Thr Ser Cy~ Val Cy~ Thr
100 105 110
Leu Thr I le Ly~ Arg Gly Arg
115
(2) INFORNATION FOR SEQ ID NO: 48:
U~NC;h CHARACTERISTICS:
(A' LENGTH: 119 amino acid~
( B TYPE: amino ac id
(C STRANDEDNESS: ~ingle
( D TOPOLOGY: llnk- "
( i i ) ~tOT.~CUT.~ TYPE: pept ide
(xi) ~guhN~; DESCRIPTION: SEQ ID NO:48:
Tyr Ala Glu Hi~ Ly~ Ser Hil~ Arg Gly Glu Tyr Ser Val CYQ A~p Ser
Glu Ser Leu Trp Val Thr A~p Lys Ser Ser Ala Ile Asp Ile Arg Gly
Hi~ Gln Val Thr Val Leu Gly Glu Ile Lys Thr Gly Asn Ser Pro Val
Ly~ Gln Tyr Phe Tyr Glu Thr Arg Cys Ly~ Glu Ala Arg Pro Val Ly~
Asn Gly Cyn Arg Gly Ile A~p A~p Lys Hi~ Trp Al~n Ser Gln Cy8 Ly~
Thr Ser Gln Thr Tyr Val Arg Ala Leu Thr Ser Glu A~n A~n Ly~ Leu
132
W O 93/25684 ~ ~ 2 7 7 ~ ~ PC~r/US93/05672
Val Gly Trp Arg Trp Ile Arg Ile A~p Thr Ser Cy~ Val Cy~ Ala Leu
100 105 110
Ser Arg Ly~ Ile Gly Arg Thr
115
(2) INFORMATION FOR SEQ ID NO:49:
~ ( i ) ~U~N~ CHARACTERISTICS:
- 'A) LENGTH: 1313 ba~e pair~
B) TYPE: nucleic acid
,C) STRANDEDNESS: double
~D) TOPOLOGY: ~nl~
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: 552 1259
(xi) S~Qu~ DESCRIPTION: SEQ ID NO:49:
CTGCAGGGAA ACAATCATAC TTATGAACAG CAGGGGGAGC CCTCGCCTTA ~l~cc~AGcc 60
ATGCAGAACT CAAGCAGCTT TGTTTATGCC GATCCCTAAG CAGCCrAr-AC CACACTGAGC 120
ATGTGCACAG TCTTAGTCTT GrpAAGATGT TTAAr-AAAGT TArA~r~A-TGG Tr~r,c~c~c~,G 180
TAGCCAACTT TGAPAGCATA AATCATTTGT TTGATTAGGC ..G.G~.GCA GTAAGTTCAT 240
GTTTATATTT AGr-ATAr-APA ATAr,AGrATT TCTAGCCTTA TTCTATTTTA GACTTTACCC 300
TTTAATGCCC AGTTCTGCCC ATTGCCTTAT AGATGTTAAA GTCCCAATAT CACATTGGCA 360
~C~.CGGCTG TTTArAAr-AA ACATTAAAAC TTGTACTTAT ATTTAACATT ~.G..~..~. 420
TC'SAPTATTC CATCACACTT Ar-ACCCTAAA AGAATTATAT GTATATAATT TGrATAAATT 480
ATATAATGGC AGCCGTATTC TAA .~-.G.~ ... .L~..GCAG G~.GAGG 540
TGGATTAAGT A ATG ATC CTC CGC CTT TAT GCC ATG GTG ATC TCA TAC TGT 590
Met Ile Leu Arg Leu Tyr Ala Met Val Ile Ser Tyr Cyn
1 5 10
TGT GCC ATC TGC GCT GCC CCC TTC CAG AGC CGG ACC ACA GAT TTG GAT 638
Cy~ Ala Ile Cyn Ala Ala Pro Phe Gln Ser Arg Thr Thr A~p Leu ABP
15 20 25
TAT GGC CCC GAT AAA ACA TCA GAA GCC TCA GAC CGG CAA TCA GTT CCC 686
Tyr Gly Pro Asp Ly~ Thr Ser Glu Ala Ser A~p Arg Gln Ser Val Pro
30 35 40 45
AAC AAC TTC AGT CAT GTC CTG CAA AAT GGG TTC TTT CCA GAT TTG TCA 734
Asn Asn Phe Ser His Val Leu Gln A~n Gly Phe Phe Pro Asp Leu Ser
50 55 60
TCC ACC TAT TCC AGC ATG GCT GGT AAG GAC TGG AAC CTA TAC TCA CCT 782
Ser Thr Tyr Ser Ser Met Ala Gly Ly~ Asp Trp Asn Leu Tyr Ser Pro
65 70 75
133
~3~ 3 PCr/US93/05672
AGA GTG ACT CTT TCA AGT GAG GAG CCT TCT GGA CCT CCA CTA CTT TTC 830
Arg Val Thr Leu Ser Ser Glu Glu Pro Ser Gly Pro Pro Leu Leu Phe
80 85 90
TTG TCA GAG GAG ACA GTG GTA CAT CCA GAA CCA GCC AAC AAG ACT TCC 878
Leu Ser Glu Glu Thr Val Val Hi~ Pro Glu Pro Ala Asn Lys Thr Ser
95 100 105
CGG CTA AAA CGG GCA TCA GGA TCT GAT TCG GTC AGC TTG TCC CGT CGG 926
Arg Leu Ly~ Arq Ala Ser Gly Ser Aup Ser Val Ser Leu Ser Arg Arg
110 115 120 125
GGA GAG CTC TCT GTG TGT GAC AGT GTC AAC GTC TGG GTT ACC GAT AAA 974
Gly Glu Leu Ser Val Cy~ A~p Ser Val Asn Val Trp Val Thr Asp Ly~
130 135 140
CGT ACA GCC GTG GAT GAT CGG GGT AAA ATA GTG ACT GTC ATG TCT GAG 1022
Arg Thr Ala Val A~p A~p Arg Gly Ly~ Ile Val Thr Val ~et Ser Glu
145 150 lS5
ATT CAG ACT CTA ACA GGA CCA CTG AAG CAA TAC TTC TTT GAG ACC AAG 1070
Ile Gln Thr Leu Thr Gly Pro Leu Lys Gln Tyr Phe Phe Glu Thr Ly~
160 165 170
TGC AAT CCA TCA GGC AGC ACC ACT AGA GGA TGC CGA GGT GTA GAC AAA 1118
Cy~ A~n Pro Ser Gly Ser Thr Thr Arg Gly Cy~ Arg Gly Val Asp Ly~
175 180 185
AAG CAA TGG ATA TCT GAG TGC AAA GCA AAA CAG TCT TAT GTG AGG GCT 1166
Ly3 Gln Trp Ile Ser Glu Cy8 Ly~ Ala Lys Gln Ser Tyr Val Arg Ala
190 195 200 205
CTG ACC ATA GAT GCC AAC AAG CTT GTG GGT TGG CGT TGG ATC CGT ATT 1214
Leu Thr Ile Asp Ala A~n Ly~ Leu Val Gly Trp Arg Trp Ile Arg Ile
210 215 220
GAC ACA GCG TGT GTC TGT ACC TTG TTG AGT CGG ACA GGA AGG ACG 1259
A~p Thr Ala Cy~ Val Cys Thr Leu Leu Ser Arg Thr Gly Arg Thr
225 230 235
TAAAAG~CGA GGGTTAGCAA AATA~.A~.A~.A AGAGGTTGAT CCGTTGACCT GCAG 1313
(2) INFORMATION FOR SEQ ID NO:50:
(i) ~SD~-..CE CHARACTERISTICS:
A) LENGTH: 236 amino acid~
B) TYPE: amino acid
,D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) 5~ ~ DESCRIPTION: SEQ ID NO:50:
Met Ile Leu Arg Leu Tyr Ala Met Val Ile Ser Tyr Cys CYB Ala Ile
1 5 10 15
Cy~ Ala Ala Pro Phe Gln Ser Arg Thr Thr ARP Leu Asp Tyr Gly Pro
Anp Lys Thr Ser Glu Ala Ser Asp Arg Gln Ser Val Pro A~n A~n Phe
Ser Hi~ Val Leu Gln A~n Gly Phe Phe Pro Asp Leu Ser Ser Thr Tyr
Ser Ser Met Ala Gly Ly~ A~p Trp A~n Leu Tyr Ser Pro Arg Val Thr
134
WO 93/25684 Z 1 3 7 7 g ~ PCI'/US93/05672
Leu Ser Ser Glu Glu Pro Ser Gly Pro Pro Leu Leu Phe Leu Ser Glu
Glu Thr Val Val Hi~ Pro Glu Pro Ala A~n Lyu Thr Ser Arg Leu Ly~
100 105 110
Arg Ala Ser Gly Ser A~p Ser Val Ser Leu Ser Arg Arg Gly Glu Leu
115 120 125
Ser Val Cyu Asp Ser Val Aan Val Trp Val Thr Aup Lys Arg Thr Ala
130 135 140
Val Asp Asp Arg Gly Ly~ Ile Val Thr Val Met Ser Glu Ile Gln Thr
145 150 155 160
Leu Thr Gly Pro Leu Ly~ Gln Tyr Phe Phe Glu Thr Ly~ Cy~ Asn Pro
165 170 175
Ser Gly Ser Thr Thr Arg Gly Cy~ Arg Gly Val Anp Ly~ Ly~ Gln Trp
180 185 190
Ile Ser Glu Cy~ Lys Ala Ly~ Gln Ser Tyr Val Arg Ala Leu Thr Ile
195 200 205
Asp Ala Asn Lys Leu Val Gly Trp Arg Trp Ile Arg Ile Asp Thr Ala
210 215 220
Cys Val Cy8 Thr Leu Leu Ser Arg Thr Gly Arg Thr
225 230 235
(2) lNrOR~ATION FOR SEQ ID NO:51:
( i ) ~kQ~k.._~' CHARACTERISTICS:
,~A'I LENGTH: 57 amino acid~
IB TYPE: amino acid
,C, STRANDEDNESS: single
lD, TOPOLOGY -n'~ ,"
(ii) MOLECULE TYPE: peptide
(xi) ~hQur;N~r; DESCRIPTION: SEQ ID NO:51:
Gln Tyr Phe Phe Glu Thr Ly~ Cy~ Asn Pro Ser Gly Ser Thr Thr Arg
1 5 10 , 15
Gly Cy~ Arg Gly Val Asp Lys Lys Gln Trp Ile Ser Glu Cys Lys Ala
Ly~ Gln Ser Tyr Val Arg Ala Leu Thr Ile A~p Ala A~n Ly~ Leu Val
35 40 45
Gly Trp Arg Trp Ile Arg Ile A~p Thr
(2) lNrOR~ATION FOR SEQ ID NO:52:
' ($) SEQUENCE CHARACTERISTICS:
- 'AJ LENGTH: 6 amino acids
B TYPE: amino acid
C STRANDEDNESS: 3ingle
,D~ TOPOLOGY ~nkn~ "
135
W O 93/25684 PC~r/US93/05672
~(Li) MnT~CUT ~ TYPE: pQptide
(xi) S~ur;~ DESCRIPTION: SEQ ID NO:52:
Gln Tyr Phe Tyr Glu Thr
1 5
(2) lNrO~MATION FOR SEQ ID NO:53:
(i) SEQUENCE CHARACTERISTICS:
'A' LENGTH: 17 base pairs
B TYPE: nucleic acid
C, STRANDEDNESS: singlc
,D, TOPOLOGY l~nl-- ,. "
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:53:
CARTAYTTYT TYGARAC 17
(2) INFORMATION FOR SEQ ID NO:54:
(i) ~r;Qur,~ri CHARACTERISTICS:
'A'l LENGTH: 17 base pairs
B TYPE: nucleic acid
C STRANDEDNESS: ~ingle
,,D, TOPOLOGY: I~nkn~,...
(ii) MOLECULE TYPE: DNA (genomic)
(xi) ~bQDr;N~r; DESCRIPTION: SEQ ID NO:54:
CARTAYTTYT AYGARAC 17
(2) INFORMATION FOR SEQ ID NO:55:
(i) ~r;Q~r,N~r; CHARACTERISTICS:
'A' LENGTH: 17 base pairs
~B~ TYPE: nucleic acid
C STRAND~nNESS: ~ingle
,D,I TOPOLOGY: tln~
( ii ) ~r~FCur~ TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/KEY: modified_base
(B) LOCATION: 9
(D) OTHER lNrOh~ATION: /label~ N
/note= "N = I"
(ix) FEATURE:
(A) NAME/KEY: modified base
(B) LOCATION: 12
(D) OTHER INFORMATION: /label~ N
/note= "N = I"
(xi) ~r;QB~N~r; DESCRIPTION: SEQ ID NO:55:
136
W O 93/25684 ~ 1 ~ 7 7 9 9 PC~r/US93/05672
TTRCAYTCNS WNATCCA . 17
(2) lN~Ok~ATION FOR SEQ ID NO:56:
(i) SEQUENCE CHARACTERISTICS:
A~I LENGTH: 17 ba~e pair~
BI TYPE: nucleic acid
C, STRANDEDNESS: ~ingle
,,D~ TOPOLOGY: un~n~..n
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/KEY: modified ba~c
~B) LOCATION: 9
(D) OTHER IhrOR~ATION: /label= N
/note~ ~N ~ I~
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:56:
GAr.~Y~,-.G CYTTRCA 17
(2) INFORMATION FOR SEQ ID NO:57:
(i) SEQUENCE CHARACTERISTICS:
(A~ LENGTH: 17 ba~e pair~
(B TYPE: nucleic acid
(C STRANDEDNESS: single
(D, TOPOLOGy: un~r:....
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/KEY: modified_ba~e
(B) LOCATION: 9
(D) OTHER l~rO~ATION: /label= N
/note~ "N ~ I n
(xi) sriyuri~r; DESCRIPTION: SEQ ID NO:57:
~,Y.~Y,,NG CYTTRCA 17
(2) INFORMATION FOR SEQ ID NO:58:
( i ) sriyDL~-T~ CHARACTERISTICS:
~A' LENGTH: 17 ba~e pair~
Bl TYPE: nucleic acid
C, STRAN~ SS: ~inqle
~D~ TOPOLOGY: 1 l n 1-~ .. "
( ii ) MOT T'~CUT~T~ TYPE: DNA ( gen~ ~ C )
(ix) FEATURE:
(A) NAME/KEY: modified ba~e
(B) LOCATION: 6
(D) OTHER l~rORMATION: /label~ n
/notes ~N ~ I"
(ix) FEATURE:
(A) NAME/KEY: modified_base
(B) LOCATION: 9
(D) OTHER l~rO~ATION: /label~ N
137
W O 93/25684 P ~ /US93/05672
.~ .~ . ,~
i 13 7 7 9 9, F~ATURE ' ~' ~
(A) NAME/KEY: modified ba~e~
(B) LOCATION: 12
(D) OTHER INFORMATION~ ~label= N
/note= "N - I"
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:58:
TNC KNATCCA 17
(2) INFORMATION\FOR SEQ ID NO:59:
( i ) S~'yU~N~ CHARACTERISTICS:
'A' LENGTH: 17 baMe pairs
,B TYPE: nucleic acid
C STR~NDEDNESSs ~inglc
D,l TOPOLOGY: unknown
(ii) MOLECULE TYPE: DNA (genomic)
(xi) ~E~u~n~ DESCRIPTION: SEQ ID NO:59:
CCAAGCTTCT AGAATTC 17
(2) INFORMATION FOR SEQ ID NO:60:
( i ) S~QU~N~ CHARACTERISTICS:
'A'l LENGTH: 17 ba~e pairD
B TYPE: nucleic acid
C, STRANDEDNESS: 3ingle
,D~ TOPOLOGY: llnl-- . "
(ii) MOLECULE TYPE: DNA (genomic)
(xi) ~u~.._~ DESCRIPTION: SEQ ID NO:60:
GACTCGAGTC GACATCG ' 17
(2) INFORMATION FOR SEQ ID NO:61:
QDL..CE CHARACTERISTICS:
IA' LENGTH: 126 ba~e pair~
B TYPE: nucleic acid
C ST~ANn~-nNESS: double
~D, TOPOLOGY: unknown
( ii ) ~r~CuT~F TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: 1..126
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:61:
CAG TAT TTT TAC GAG ACG CGC TGC AAG GCC GAA AGC GCT GGG GAA GGT 48
Gln Tyr Phe Tyr Glu Thr Arg Cy~ Ly~ Ala Glu Ser Ala Gly Glu Gly
1 5 10 15
GGC CCA GGT GTG GGC GGA GGG GGC TGT CGC GGC GTG GAT CGG AGG CAC 96
Gly Pro Gly Val Gly Gly Gly Gly Cy~ Arg Gly Val Asp Arg Arg His
138
W O 93/25684 .' 2 1 37 7 9 g PC~r/US93/0~672
20 ;25 ~ i 30
TGG CTC TCA GAA TGT AAA GCC AA~A CAA TCG 126
Trp Leu S-r Glu Cy~ Ly~ Ala Ly~ Gln Ser
35 40
(2) lN~ORMATION FOR SEQ ID NO:62:
ti) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 42 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: lincar
~ii) MOLECULE TYPE: protein
(xi) SEQUENCE DFC~PTPTION: SEQ ID NOs62:
Gln Tyr Phe Tyr Glu Thr Arg Cy~ Lyff Ala Glu Ser Ala Gly Glu Gly
Gly Pro Gly Val Gly Gly Gly Gly Cys Arg Gly Val A~p Arg Arg Hi~
20 25 30
Trp Leu Ser Glu Cys Lys Ala Lys Gln Ser
(2) INFORMATION FOR SEQ ID NO:63:
( i ) shQUh~h CHARACTERISTICS:
'A~l LENGTH: 126 base pairs
Bl TYPE: nucleic acid
C STRANDEDNESS: double
lD, TOPOLOGy: 1nkn~ ,.,
(ii) MOLECULE TYPE: DNA (g,e~
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: 1..126
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:63:
CAG TAT TTT TAC GAA ACC CGC TGC AAG GCT GAT AAC GCT GAG GAA GGT 48
Gln Tyr Phe Tyr Glu Thr Arg Cys Lys Ala A~p Asn Ala Glu Glu Gly
1 5 10 15
GGC CCG GGG GCA GGT GGA GGG GGC TGC CGG GGA GTG GAC AGG AGG CAC 96
Gly Pro Gly Ala Gly Gly Gly Gly Cy~ Arg Gly Val A~p Arg Arg Hi~
20 25 30
TGG GTA TCT GAG TGT AAA GCC AAA CAA TCG 126
Trp Val Ser Glu Cy~ Ly~ Ala Lys Gln Ser
35 40
(2) lNrOh~ATION FOR SEQ ID NO:64:
(i) SEQUENCE CHA~ACT~TSTICS:
' (A) LENGTH: 42 amino acids
= (B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
139
:
t~
93/25684 ,. -~ ~ P ~ /US93/05672
~ ~ xi) ~r:yu~N~ DESCRIPTION: SEQ ID NO:64:
? ~ ~ 9 Gln Tyr Phe Tyr Glu Thr Ary Cy~ Ly~ Ala Asp A~n Ala Glu Glu Gly 1 5 - 10 15
Gly Pro Gly Ala Gly Gly Gly Gly Cy~l Arg Gly Val A~p Arg Arg Hiu
20 25 30
Trp Val Ser Glu CYR Ly~ Ala Lys Gln Ser
(2) INFORMATION FOR SEQ ID NO:65:
(i) SEQUENCE CHARACTERISTICS:
,~A' LENGTH: 35 amino acid~
B TYPE: amino acid
C, STRANDEDNESS: ~ingle
,DJ TOPOLOGY: llnknrs,,,
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:65:
Gln Tyr Phe Phe Glu Thr Ly~ Cys Asn Pro Ser Gly Ser Thr Thr Arg
1 5 10 15
Gly Cys Arg Gly Val A~p Lys Lys Gln Trp Ile Ser Glu CYR Lys Ala
Lys Gln Ser
(2) INFORMATION FOR SEQ ID NO:66:
(i) SEQUENCE CHARACTERISTICS:
,'A'I LENGTH: 35 amino acid~
IB TYPE: amino acid
,C, STR~NDEDNESS: ~ingle
,,D~ TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) ~yu~CE DESCRIPTION: SEQ ID NO:66:
Gln Tyr Phe Phe Glu Thr LYR Cys Arg Ala Pro Asn Pro Val Glu Ser
1 5 10 15
Gly Cys Arg Gly Ile A~p Ser LYR His Trp ARn Ser Tyr Cy~ Thr Thr
Thr Hi~ Thr
(2) INFORMATION FOR SEQ ID NO:67: -
(i) ~Q~ CE CHARACTERISTICS:
'A'I LENGTH: 35 amino acid~
Bl TYPE: amino acid
C STRANDEDNESS: single
D,I TOPOLOGY: 11nk- ~
( ii ) M~T.T~'.CUT.T~` TYPE: peptide
140
WO 93/25684 2 1 3 7 7 g g PCI`/US93/0567
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:67:
Gln Tyr Phe Tyr Glu Thr Lyn Cyff A~n Pro Met Gly Tyr Thr Lya Glu
1 5 10 15
Gly Cy~ Arg Gly Ile A~p Ly~ Arg Hin Trp Affn Ser Gln Cyff Arg Thr
Thr Gln Ser
(2) INFORMATION FOR SEQ ID NO:68:
(i) SEQUENCE CHARACTERISTICS:
'A' LENGTH: 35 amino acids
B TYPE: amino acid
C STRANDEDNESS: ffingle
,D,I TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:68:
Gln Tyr Phe Tyr Glu Thr Arg Cy~ Ly~ Glu Ala Arg Pro Val Lya A~n
1 5 10 15
Gly Cys Arg Gly Ile Affp Affp Lyff His Trp Asn Ser Gln Cy~ Lys Thr
Ser Gln Thr
(2) INFORMATION FOR SEQ ID NO:69:
(i) SEQUENCE CHARACTERISTICS:
'A' LENGTH: 192 baffe pairs
IB TYPE: nucleic acid
,C, STRANDEDNESS: double
l,DJ TOPOLOGY: unknown
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: 1..192
(xi) ~yu~CE DESCRIPTION: SEQ ID NO:69:
CAA TAT TTT TTC GAG ACC CGC TGC AAG GCT GAT AAC GCT GAG GAA GGT 48
Gln Tyr Phe Phe Glu Thr Arg Cyff Lyff Ala Affp A~n Ala Glu Glu Gly
l 5 10 15
GGT CCG GGG GCA GGT GGA GGG GGC TGC CGG GGA GTG GAC AGG AGG CAC 96
Gly Pro Gly Ala Gly Gly Gly Gly Cyff Arg Gly Val Affp Arg Arg Hiff
20 25 30
TGG GTA TCT GAG TGC AAG GCC AAG CAG TCC TAT GTG CGG GCA TTG ACC 144
Trp Val Ser Glu Cy~ Ly~ Ala Ly~ Gln Ser Tyr Val Arg Ala Leu Thr
35 40 45
GCT GAT GCC CAG GGC CGT GTG GGC TGG CGA TGG ATC CGC ATC GAT ACG 192
Ala Affp Ala Gln Gly Arg Val Gly Trp Arg Trp Ile Arg Ile A~p Thr
50 55 60
141
WO93~256'84 .; ~ PCI`/US93/0567Z
t2) INFORMATION FOR SEQ ID NO:~0~
~ ~ 3 j 7 99 ( i, SEQUENCE CHARACTERISTICS:
(A) LENGTH: 64 amino acLd~
(8) TYPE: amino acid
(D) TOPOLOGY: linear
($i) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:70:
Gln Tyr Phe Phe Glu Thr Arg Cys Lys Ala Asp Asn Ala Glu Glu Gly
1 5 10 15
Gly Pro Gly Ala Gly Gly Gly Gly Cy~ Arg Gly Val Anp Arg Arg His
Trp Val Ser Glu Cys Lys Ala Ly~ Gln Ser Tyr Val Arg Ala Leu Thr
Ala A~p Ala Gln Gly Arg Val Gly Trp Arg Trp Ile Arg Ile AQP Thr
50 55 60
(2) INFORMATION FOR SEQ ID NO:71:
(i) SEQUENCE CHARACTERISTICS:
,~A' LENGTH: 35 base pairs
B TYPE: nucleic acid
C STRANDEDNESS: double
,D TOPOLOGY: ~ n~- - ,, "
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/REY: CDS
(B) LOCATION: 18..35
(xi) ~:yu~N~L DESCRIPTION: SEQ ID NO:71:
GACTCGAGTC GACATCG GAA ACC CGC TGC AAG GCT 35
Glu Thr Arg Cys Ly Ala
(2) INFORMATION FOR SEQ ID NO:72:
( i ) XLS D~N~;~ CHARACTERISTICS:
A) LENGTH: 6 amino acids
,B) TYPE: amino acid
l,D) TOPOLOGY: linear
(ii) ~OLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:72:
Glu Thr Arg Cys Lys Ala
(2) INFORMATION FOR SEQ ID NO:73:
( i ) i~gUL.. - ~ CHARACTERISTICS:
(A) LENGTH: 35 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
142
W O 93/25684 P ~ /US93/05672
2 1 3 7 7 g 9 ~ s ~
(D) TOPOLOGY: un~U"`
(ii) MOLECULE TYPE: DNA (genomic) ; -
. ,; ,
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: 18 35
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:73:
GACTCGAGTC GACATCG GAT AAC GCT GAG GAA 6GT 35
A~p Asn Ala Glu Glu Gly
1 5
(2) INFORMATION FOR SEQ ID NO:74:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 6 amino acid~
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:74:
A~p A~n Ala Glu Glu Gly
1 5
(2) INFORMATION FOR SEQ ID NO:75:
(i) SEQUENCE CHARACTERISTICS:
rA~I LENGTH: 1404 ba~e pairQ
,BI TYPE: nucleic acid
C STRANv~N~SS: double
~,D, TOPOLOGY: 1~ n ~ - - ,, "
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: 460 .1104
(xi) ~yu~N~ DESCRIPTION: SEQ ID NO:75:
~..~.~ACCC A6GT6GCACC CGAGTGGTGC A~ .GCT CACTGCAACC TCGGCCTCCT 60
GGGTTCGAGT GA--~-C~-A CCTCAGCCTA CTGAGTAGCT GGGATTACAG GCGTGCAGCA 120
CTATGCCCGG TTAATTTTGG TA..~..~. AGAGATGAGG TTTCACAATG TTGACCAGCT 180
GCTCTGGAAC TCCTGACCTC AAGTCATCCA CCTGCCTCAG c~.cc~ Ar- TGCTGGGATT 240
AGA6~.~.GG GGCACAGTGC CTGGCCTGTA GTAGTTGAAT ATTTATTATT AATCTACAAG 300
TTGCGCATTA CGCAAGCCCT A~-~TATAGGG ~CCCC-~AAC TTCTA~-AAr~ AG6G~..CCC 360
CACAATCCTG GCAGGCAAGC ~.CCC~.GGG 6..CC~AACT .~..lCCCCA CTGAAGTTTT 420
TA~'CCC~,,C TCTAATCCCA GC~.CC~.~. ....... ~.~.~.C CAG GTG CTC CGA GAG 474
Gln Val Leu Arg Glu
143
WO 93/25684 ' ~ " PCI'/US93/05672
2 13 ~A~G~ CTC CCT CTC CCC TCA TGC TCC CTC CCC ATC CTC CTC CTT TTC CTC 522
- , Me~ Leu Pro Leu Pro Ser Cy~ Ser Leu Pro I le Leu Leu Leu Phe Leu
'' 10 15 20
CTC CCC AGT GTG CCA ATT GAG TCC CAA CCC CCA CCC TCA ACA TTG CCC 570
Leu Pro Ser Val Pro Ile Glu Ser Gln Pro Pro Pro Ser Thr Leu Pro
25 30 35
CCT TTT CTG GCC CCT GAG TGG GAC CTT CTC TCC CCC CGA GTA GTC CTG 618
Pro Phe Leu Ala Pro Glu Trp Asp Leu Leu Ser Pro Arg Val Val Leu
40 45 50
TCT AGG GGT GCC CCT GCT GGG CCC CCT CTG CTC TTC CTG CTG GAG GCT 666
Ser Arg Gly Ala Pro Ala Gly Pro Pro Leu Leu Phe Leu Leu Glu Ala
55 60 65
GGG GCC TTT CGG GAG TCA GCA GGT GCC CCG GCC AAC CGC AGC CGG CGT 714
Gly Ala Phe Arg Glu Ser Ala Gly Ala Pro Ala Asn Arg Ser Arg Arg
70 75 80 85
GGG GTG AGC GAA ACT GCA CCA GCG AGT CGT CGG GGT GAG CTG GCT GTG 762
Gly Val Ser Glu Thr Ala Pro Ala Ser Arg Arg Gly Glu Leu Ala Val
90 95 100
TGC GAT GCA GTC AGT GGC TGG GTG ACA GAC CGC CGG ACC GCT GTG GAC 810
Cy~ Asp Ala Val Ser Gly Trp Val Thr Asp Arg Arg Thr Ala Val Asp
105 110 115
TTG CGT GGG CGC GAG GTG GAG GTG TTG GGC GAG GTG CCT GCA GCT GGC 858
Leu Arg Gly Arg Glu Val Glu Val Leu Gly Glu Val Pro Ala Ala Gly
120 125 130
GGC AGT CCC CTC CGC CAG TAC TTC TTT GAA ACC CGC TGC AAG GCT GAT 906
Gly Ser Pro Leu Arg Gln Tyr Phe Phe Glu Thr Arg Cys Lys Ala Asp
135 140 145
AAC GCT GAG GAA GGT GGT CCG GGG GCA GGT GGA GGG GGC TGC CGG GGA 954
Asn Ala Glu Glu Gly Gly Pro Gly Ala Gly Gly Gly Gly Cys Arg Gly
150 155 160 165
GTG GAC AGG AGG CAC TGG GTA TCT GAG TGC AAG GCC AAG CAG TCC TAT 1002
Val Asp Arg Arg His Trp Val Ser Glu Cys Lys Ala Lys Gln Ser Tyr
170 175 180
GTG CGG GCA TTG ACC GCT GAT GCC CAG GGC CGT GTG GGC TGG CGA TGG 1050
Val Arg Ala Leu Thr Ala A~p Ala Gln Gly Arg Val Gly Trp Arg Trp
185 190 195
ATT CGA ATT GAC ACT GCC TGC GTC TGC ACA CTC CTC AGC CGG ACT GGC 1098
Ile Arg IlQ Asp Thr Ala Cys Val Cys Thr Leu Leu Ser Arq Thr Gly
200 205 210
CGG GCC Tr-AI~-ACc~AT GCC~C~AAA ATAACAGAGC TGGATGCTGA GAGACCTCAG 1154
Arg Ala
215
GGATGGCCCA GCTGATCTAA Gr-ACCCÇ~GT TTGGGAACTC AT~AAATAAT CACAAAATCA 1214
CAA~,. .G ATTTGGAGCT CAATCTCTGC AGGATGGGTG AAACCAr~ATG GGG.~GGA 1274
GGTTGAATAG GA~..~ ~C~;~ GGAGCAACTT GAGGGTAATA ATGATGATGA TATAATAATA 1334
ATAGCCACTA TTTACTGAGT GTTTACTGTT TCTTATCCCT AATAI'ATAA~' TCCTCAGATC 1394
AACTCTCATG 1404
144
W O 93/25684 2 1 ~ ~ i9 9 PC~r/US93/05672
(2) INFORMATION FOR SEQ ID NO:76: ~J~
(i) s~Q~hCE CHARACTERISTICS:
(A) LENGTH: 215 amino acid~
(B) TYPE: amLno acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
r (xi) SEQUENCE DESCRIPTION: SEQ ID NO:76:
Gln Val Leu Arg Glu Met Leu Pro LQU Pro Ser Cys Ser Leu Pro Ile
1 5 10 15
- Leu Leu Leu Phe Leu Leu Pro Ser Val Pro Ile Glu Ser Gln Pro Pro
2S 30
Pro Ser Thr Leu Pro Pro Phe Leu Ala Pro Glu Trp Asp Leu Leu Ser
Pro Arg Val Val Leu Ser Arg Gly Ala Pro Ala Gly Pro Pro Leu Leu
Phe Leu Leu Glu Ala Gly Ala Phe Arg Glu Ser Ala Gly Ala Pro Ala
Asn Arg Ser Arg Arg Gly Val Ser Glu Thr Ala Pro Ala Ser Arg Arg
Gly Glu Leu Ala Val Cy~ A~p Ala Val Ser Gly Trp Val Thr Asp Arg
100 105 110
Arg Thr Ala Val Asp Leu Arg Gly Arg Glu Val Glu Val Leu Gly Glu
115 120 125
Val Pro Ala Ala Gly Gly Ser Pro Leu Arg Gln Tyr Phe Phe Glu Thr
130 135 140
Arg Cy~ Lys Ala Asp Asn Ala Glu Glu Gly Gly Pro Gly Ala Gly Gly
145 150 155 160
Gly Gly Cys Arg G6y Val Asp Arg Arg Hio Trp Val Ser Glu Cy Ly~
Ala Lys Gln Ser Tyr Val Arg Ala Leu Thr Ala Asp Ala Gln Gly Arg
180 185 190
Val Gly Trp Arg Trp Ile Arg Ile A~p Thr Ala Cyn Val Cy~ Thr Leu
195 200 205
Leu Ser Arg Thr Gly Arg Ala
210 215
(2) IN~ORhATION FOR SEQ ID NO:77:
(i) ~Quh~CE CHARACTERISTICS:
'A' LENGTH: 214 amino acid~
B TYPE: amino acid
C STRANDEDNESS: ~ingle
D~ TOPOLOGY: lln~n~
(ii) MOLECULE TYPE: peptide
(xi) X~QD~N~ DESCRIPTION: SEQ ID NO:77:
145
W O 93/25684 '! '~ p ~ /Us93/05672
r ~
21377 9 9 Val Leu Arg Glu Met Leu Pro Leu Pro Ser Cy~ Ser Leu Pro le Leu
Leu Leu Phe Leu Leu Pro Ser Val Pro Ile Glu Ser Gln Pro Pro Pro
Ser Thr Leu Pro Pro Phe Leu Ala Pro Glu Trp A~p Leu Leu Ser Pro
Arg Val Val Leu Ser Arg Gly Ala Pro Ala Gly Pro Pro Leu Leu Phe
Leu Leu Glu Ala Gly Ala Phe Arg Glu Ser Ala Gly Ala Pro Ala A~n
Arg Ser Arg Arg Gly Val Ser Glu Thr Ala Pro Ala Ser Arg Arg Gly
Glu Leu Ala Val Cy~ A~p Ala Val Ser Gly Trp Val Thr Asp Arg Arg
100 105 110
Thr Ala Val A~p Leu Arg Gly Arg Glu Val Glu Val Leu Gly Glu Val
115 120 125
Pro Ala Ala Gly Gly Ser Pro Leu Arg Gln Tyr Phe Phe Glu Thr Arg
130 135 140
Cy~ Lys Ala A~p A~n Ala Glu Glu Gly Gly Pro Gly Ala Gly Gly Gly
145 150 155 160
Gly Cys Arg Gly Val A~p Arg Arg Hi~ Trp Val Ser Glu Cys Lys Ala
165 170 175
Ly~ Gln Ser Tyr Val Arg Ala Leu Thr Ala A~p Ala Gln Gly Arg Val
180 185 190
Gly Trp Arg Trp Ile Arg Ile A~p Thr Ala cys Val Cys Thr Leu Leu
195 200 205
Ser Thr Arg Gly Arg Ala
210
(2) INFORMATION FOR SEQ ID NO:78:
ti) ~yu~:N~ CHARACTERISTICS:
'A' LENGTH: 176 amino acidn
,B TYPE: amino acid
,C STRANDEDNESS: ~ingle
~D TOPOLOGY: lln~ "
t ii ) ~nT~CTJT T~ TYPE: peptide
txi) ~yu~w~E DESCRIPTION: SEQ ID NO:78:
Ser Ser Thr Tyr Ser Ser Met Ala Gly Ly~ ARP Trp Asn Leu Tyr Ser
Pro Arg Val Thr Leu Ser Ser Glu Glu Pro Ser Gly Pro Pro Leu Leu
Phe Leu Ser Glu Glu Thr Val Val Hi~ Pro Glu Pro Ala Asn Ly~ Thr
Ser Arg Leu Ly~ Arg Ala Ser Gly Ser A8p Ser Val Ser Leu Ser Arg
146
WO 93/25684 2 1 3 7 7 9 9 PCI'/US93/05672
50 55 60
Arg Gly Glu Leu Ser Val Cy~ A~p Ser Val A~n Val Trp Val Thr Asp
65 70 75 80
Ly~ Arg Thr Ala Val A~p Asp Arg Gly Lys Ile Val Thr Val Met Ser
85 90 95
Glu Ile Gln Thr Leu Thr Gly Pro Leu Ly~ Gln Tyr Phe Phe Glu Thr
r 100 105 110
Ly~ Cys Asn Pro Ser Gly Ser Thr Thr Arg Gly Cys Arg Gly Val Asp
115 120 125
- Lys Ly~ Gln Trp Ile Ser Glu Cy~ Ly~ Ala Ly~ Gln Ser Tyr Val Arg
130 135 140
Ala Leu Thr Ile Asp Ala Asn Ly~ Leu Yal Gly Trp Arg Trp Ile Arg
145 150 155 160
Ile Asp Thr Ala Cys Val Cy~ Thr Leu Leu Ser Arg Thr Gly Arg Thr
165 170 175
~2) INFORMATION FOR SEQ ID NO:79:
(i) SEQUENCE CHARACTERISTICS:
IA' LENGTH: 177 amino acids
B TYPE: amino acid
C, STRANDEDNESS: ~Lngle
~Dl TOPOLOGY: I~nl-- . "
(ii) MOLECULE TYPE: peptide
(xi) ~Qu~CE DESCRIPTION: SEQ ID NO:79:
Glu Phe Gln Pro Met Ile Ala Thr Asp Thr Glu Leu Leu Arg Gln Gln
1 5 10 15
Arg Arg Tyr A~n Ser Pro Arg Val Leu Leu Ser A~p Ser Thr Pro Leu
Glu Pro Pro Pro Leu Tyr Leu Met Glu Asp Tyr Val Gly Asn Pro Val
Val Ala A~n Arg Thr Ser Pro Arg Arg Ly~ Tyr Ala Glu Hi~ Ly~ Ser
His Arg Gly Glu Tyr Ser Val Cy~ Anp Ser Glu Ser Leu Trp Val Thr
A~p Ly~ Ser Ser Ala Ile Asp Ile Arg Gly His Gln Val Thr Val Leu
Gly Glu Ile Lys Thr Gly Asn Ser Pro Val Lys Gln Tyr Phe Tyr Glu
100 105 110
Thr Arg Cy~ Ly~ Glu Ala Arg Pro Val Ly~ Asn Gly Cys Arg Gly Ile
115 120 125
A~p Asp Ly~ Hi~ Trp A~n Ser Gln Cyn Ly~ Thr Ser Gln Thr Tyr Val
130 135 140
Arg Ala Leu Thr Ser Glu Asn Asn Lys Leu Val Gly Trp Arg Trp Ile
147
WO 93/25684 ~ . P~/US93/05672
2 1 3~ ~95 150 155 160
Arg Ile ARP Thr Ser Cy8 Val Cy8 Ala Leu Ser Arg Ly~ Ile Gly Arg
165 170 175
Thr
(2) INFORMATION FOR SEQ ID NO:80:
(i) SEQUENCE CHARACTERISTICS:
~A LENGTH: 175 amino acid~
IB TYPE: amino acid
,C, STRANDEDNESS: ~ingle
~D, TOPOLOGY: unl~ "
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:80:
Gln Pro Val Ile Ala Met A~p Thr Glu Leu Leu Arg Gln Gln Arg Arg
1 5 lo 15
Tyr Ann Ser Pro Arg Val Leu Leu Ser A~p Thr Thr Pro Leu Glu Pro
Pro Pro Leu Tyr Leu Met Glu A3p Tyr Val Gly Ser Pro Val Val Ala
A~n Arg Thr Ser Arg Arg Lya Arg Tyr Ala Glu His Ly~ Ser Hi~ Arg
Gly Glu Tyr Ser Val Cy~ A~p Ser Glu Ser Leu Trp Val Thr Asp Ly~
Ser Ser Ala Ile A~p Ile Arg Gly Hi~ Gln Val Thr Val Leu Gly Glu
Ile Ly~ Thr Gly Asn Ser Pro Val Lys Gln Tyr Phe Tyr Glu Thr Arg
loo 105 llo
Cys Ly~ Glu Ala Arg Pro Val Ly~ AQn Gly Gly Arg Gly Ile A~p A~p
115 120 125
Ly~ Hi~ Trp ARn Ser Gln Cy~ Ly~ Thr Ser Gln Thr Tyr Val Arg Ala
130 135 140
Leu Thr Ser Glu A~n Asn Ly~ Leu Val Gly Trp Arg Trp Ile Arg Ile
145 150 155 160
A~p Thr Ser Cys Val Cy~ Ala Leu Ser Arg Lys Ile Gly Arg Thr
165 170 175
2) INFORMATION FOR SEQ ID NO:81:
(i) SEQUENCE CHARACTERISTICS:
'A` LENGTH: 178 amino acids
B TYPE: amino acid
C, STRANDEDNESS: ~ingle
,D, TOPOLOGY: l-n~
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:81:
148
WO 93/25684 ~1 ~ 77g9 PCl/US93/05672
Glu Phe Gln Pro Met Ile Ala Thr A~p Thr Glu Leu Leu Arg Gln ~ n~;~ ?
1 5 10 15 ';~
Arg Arg Tyr A~n Ser Pro Arg Val Leu Leu Ser Asp Ser Thr Pro Leu
Glu Pro Pro Pro Leu Tyr Leu Met Glu Asp Tyr Val Gly A~n Pro Val
Val Thr Asn Arg Thr Ser Pro Arg Arg Ly~ Arg Tyr Ala Glu Hi~ Ly~
Ser His Arg Gly Glu Tyr Ser Val Cy~ Asp Ser Glu Ser Leu Trp Val
-
Thr A~p Ly~ Ser Ser Ala Ile Asp Ilo Arg Gly Hi~ Gln Val Thr Val
Leu Gly Glu Ile Lys Thr Gly Asn Ser Pro Val Lys Gln Tyr Phe Tyr
100 105 110
Glu Thr Arg Cys Lys Glu Ala Arg Pro Val Ly~ Atn Gly Gly Arg Gly
Ile A~p A~p Ly~ His Trp Atn Ser Gln Cys Lys Thr Ser Gln Thr Tyr
130 135 140
Val Arg Ala Leu Thr Ser Glu Asn A~n Lys Leu Val Gly Trp Arg Trp
145 150 155 160
Ile Arg Ile Asp Thr Ser Cy~ Val Cys Ala Leu Ser Arg Lyn Ile Gly
165 170 175 .
Arg Thr
~2) INFORMATION FOR SEQ ID NO:82:
(i) SEQUENCE CHARACTERISTICS:
~A' LENGTH: 160 amino acid3
,B TYPE: amino acid
,C, STRANDEDNESS: ~ingle
,D, TOPOLOGY: t,nl~
(ii) MOLECULE TYPE: peptide
(Xi) ~L~UL~ DESCRIPTION: SEQ ID NO:82:
Asp Leu Tyr Thr Ser Arg Val Met Leu Ser Ser Gln Val Pro Leu Glu
1 5 10 15
Pro Pro Leu Leu Phe Leu Leu Glu Glu Tyr Ly~ Asn Tyr Leu A~p Ala
Ala A~n Met Ser Met Arg Val Arg Arg Hi~ Ser Asp Pro Ala Arg Arg
Gly Glu Leu Ser Val Cy~ Asp Ser Ile Ser Glu Trp Val Thr Ala Ala
50 55 60
A~p Lys Lys Thr Ala Val A~p Met Ser Gly Gly Thr Val Thr Val Leu
Glu Ly~ Val Pro Val Ser Lys Gly Gln Leu Lys Gln Tyr Phe Tyr Glu
149
C~
W O 93/25684 PC~r/US93/05672
~ ~ r Ly~ Cys A~n Pro Met Gly Tyr Thr Lys Glu Gly Gly Arg Gly Ile
~1377 loo 10S 110
Aap Ly~ Arg His Trp Asn Ser Gln Cy~ Arg Thr Thr Gln Ser Tyr Val
. ~ 115 120 125
Arg Ala Leu Thr Met Anp Ser Lys Lys Arg Ile Gly Trp Arg Phe Ile
130 135 140
Arg Ile AQp Thr Ser Cys Val Cys Thr Leu Thr Ile Lys Arg Gly Arg
145 150 155 160
(2) INFORMATION FOR SEQ ID NO:83:
(i) SEQUENCE CHARACTERISTICS:
A LENGTH: 160 amino acids
B TYPE: amino acid
C STRANDEDNESS: single
lD TOPOLOGY: llnkn~ .~"
(ii) MOLECULE TYPE: peptide
(xi) ~yu~:N~ DESCRIPTION: SEQ ID NO:83:
Asp Leu Tyr Thr Ser Arg Val Met Leu Ser Ser Gln Val Pro Leu Glu
1 5 10 15
Pro Pro Leu Leu Phe Leu Leu Glu Glu Tyr Lys Asn Tyr Leu A~p Ala
Ala A~n Met Ser Met Arg Val Arg Arg Hi~ Ser Asp Pro Ala Arg Arg
Gly Glu Leu Ser Val Cy~ Asp Ser Ile Ser Glu Trp Val Thr Ala Ala
Anp Ly~ Ly~ Thr Ala Val A~p Met Ser Gly Gly Thr Val Thr Val Leu
Glu Lys Val Pro Val Ser Lys Gly Gln Leu Lys Gln Phe Phe Tyr Glu
Thr Lys Cy~ AQn Pro Met Gly Tyr Thr Lys Glu Gly Gly Arg A~p Ile
100 105 110
A~p Lys Arg Hit Trp A~n Ser Gln Cys Arg Thr Thr Gln Ser Tyr Val
115 120 125
Arg Ala Leu Thr Met A~p Ser Lys Ly~ Arg Ile Gly Trp Arg Phe Ile
130 135 140
Arg Ile A~p Thr Ser Cy8 Val Cys Thr Leu Thr Ile Ly~ Arg Gly Arg
145 150 155 160
(2) INFORMATION FOR SEQ ID NO:84:
(i) ~hQ~:N~ CHARACTERISTICS:
'A'l LENGTH: 160 amino acids
Bl TYPE: amino acid
C, STRANDEDNESS: ~ingle
lD,I TOPOLOGY: tlnkn~
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:84:
150
-
'h.. '
WO93/25684 ~ 77~ PCr/US93/05672
A~p Leu Tyr Thr Ser Arg Val Met Leu Ser Ser Gln Val Pro Leu Glu
1 5 10 15 , ~:
Pro Pro Leu Leu Phe Leu Leu Glu Glu Tyr Ly~ A~n Tyr Leu A~p Ala ~ t
Ala A~n Met Ser Met Arg Val Arg Arg Hi~ Ser A~p Pro Ala Arg Arg
Gly Glu Leu Ser Val Cy~ A~p Ser Ile Ser Glu Trp Val Thr Ala Ala
Asp Ly3 LYB Thr Ala Val Asp Met Ser Gly Gly Thr Val Thr Val Leu
Glu Lys Val Pro Val Ser Lys Gly Gln Leu Lys Gln Tyr Phe Tyr Glu
Thr Lys Cy~ A~n Pro Met Gly Tyr Thr Lys Glu Gly Gly Arg Asp Ile
100 105 110
A~p Ly~ Arg His Trp Asn Ser Gln Cyq Arg Thr Thr Gln Ser Tyr Val
115 120 125
Arg Ala Leu Thr Met A~p Ser Ly~ Ly~ Arg Ile Gly Trp Arg Phe Ile
130 135 140
Arg Ile ABP Thr Ser Cys Val Cy~ Thr Leu Thr Ile Ly~ Arg Gly Arg
145 150 155 160
(2) INFORMATION FOR SEQ ID NO:85:
(i) ~Qu~ CHARACTERISTICS:
,'A' LENGTH: 160 amino acLd~
B TYPE: amino acid
C STRANDEDNESS: single
~DJ TOPOLOGY: unknown
(ii) MOT.T~CU~.E TYPE: pept$de
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:85:
A~p Met Tyr Thr Ser Arg Val Met Leu Ser Ser Gln Val Pro Leu Glu
1 5 10 15
Pro Pro Leu Leu Phe Leu Leu Glu Glu Tyr Ly~ A~n Tyr Leu Asp Ala
Ala A~n Met Ser Met Arg Val Arg Arg Hi~ Ser A~p Pro Ala Arg Arg
Gly Glu Leu Ser Val Cy~ A~p Ser Ile Ser Glu Trp Val Thr Ala Ala
A~p Ly~ Lys Thr Ala Val ARP Met Ser Gly Gly Thr Val Thr Val Leu
Glu Ly~ Val Pro Val Ser Ly~ Gly Gln Leu Ly~ Gln Tyr Phe Tyr Glu
~ Thr Lys Cy5 A~n Pro Met Gly Tyr Thr Ly~ Glu Gly Gly Arg A~p Ile
- 100 105 110
A~p Ly~ Arg HL~ Trp A~n Ser Gln Cy~ Arg Thr Thr Gln Ser Tyr Val
115 120 125
151
W O 93~25684 ,~ P ~ /US93/05672
Arg Ala Leu Thr Met A~p Ser Lys Lys Arg Ile Gly Trp Arg Phe Ile
2~7~ Arg Il~ ABP Thr Ser Cy8 Val Cy8 Thr Leu Thr Ile Ly~ Arg Gly Arg
145 150 155 160
(2) lN~ORhATION FOR SEQ ID NO:86:
(i) SEQUENCE CHARACTERISTICS:
A' LENGTH: 180 amino acids
B TYPE: amino acid
C STRANDEDNESS: ningle
~D TOPOLOGY: I~n~nl "
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:86:
Ala Ala Arg Val Thr Gly Gln Thr Arg Asn Ile Thr Val A~p Pro Arg
1 5 10 15
Leu Phe Lys Lys Arg Arg Leu His Ser Pro Arg Val Leu Phe Ser Thr
Gln Pro Pro Pro Thr Ser Ser A~p Thr Leu A~p Leu Anp Phe Gln Ala
His Gly Thr Ile Pro Phe Asn Arg Thr Hi~ Arg Ser Lys Arg Ser Ser
Ser Hi~ Pro Ile Phe Hi~ Arg Gly Glu Phe Ser Val Cy~ Asp Ser Val
Ser Val Trp Val Gly Asp Ly~ Thr Thr Ala Thr Asp Ile Ly~ Gly Ly~
Glu Val Met Val Leu Gly Glu Val A~n Ile A~n A~n Ser Val Phe Ly~
100 105 110
Gln Tyr Phe Phe Glu Thr Ly~ Cys Arg Asp Pro Asn Pro Val A~p Ser
115 120 125
Gly Gly Arg Asp Ile A~p Ser Ly~ His Trp A~n ser Tyr Cy~ Thr Thr
130 135 140
Thr His Thr Phe Val Lys Ala Leu Thr Thr A~p Glu Lys Gln Ala Ala
145 150 155 160
Trp Arg Phe Ile Arg Ile Asp Thr Ser Cy~ Val Cy~ Val Leu Ser Arg
165 170 175
Ly3 Ala Thr Arg
180
(2) INFORMATION FOR SEQ ID NO:87:
(i) ~Q~ ~ CHARACTERISTICS:
'A'l LENGTH: 180 amino acid~
B TYPE: amino acid
C, STRANDEDNESS: ~ingle
,D TOPOLOGY: unknown
(Li) MOLECULE TYPE: peptide
152
WO 93/25684 2`1 3 7 7 9 g PCI/US93/05672
.
(xi) SEQUENCE DESCRIPTIoN: SEQ ID NO:87:
Ala Ala Arg Val Thr Gly Gln Thr Arg Asn Ile Thr Val Asp Pro Lys
1 5 10 15
Leu Phe LYB Lys Arg Arg Leu Arg Ser Pro Arg Val Leu Phe Ser Thr
Gln Pro Pro Pro Thr Ser Ser Asp Thr Leu Asp Leu Asp Phe Gln Ala
His Gly Thr Ile Ser Phe Asn Arg Thr His Arg Ser Lys Arg Ser Ser
Thr His Pro Val Phe HL~ Met Gly Glu Phe Ser Val Cys Asp Ser Val
Ser Val Trp Val Gly Asp Lys Thr Thr Ala Thr Asp Ile Lys Gly Lys
Glu Val Thr Val Leu Gly Glu Val Asn Ile A~n Asn Ser Val Phe Lys
100 105 110
Gln Tyr Phe Phe Glu Thr Lyn Cys Arg Ala Pro Asn Pro Val Glu Ser
115 120 125
Gly Gly Arg Asp Ile A~p Ser Lys His Trp Asn Ser Tyr Cys Thr Thr
130 135 140
Thr His Thr Phe Val Lys Ala Leu Thr Thr Asp Asp Lys Gln Ala Ala
145 150 155 160
Trp Arg Phe Ile Arg Ile Asp Thr Ser Cys Val Cys Val Leu Ser Arg
165 170 175
Lys Ala Ala Arg
180
(2) lNrORMATION FOR SEQ ID NO:88:
(i) SEQUENCE CHARACTERISTICS:
/A'I LENGTH: 179 amino acids
Bl TYPE: amino acid
,C, STRANDEDNESS: ningle
~D,, TOPOLOGY: In~rlu..
(ii) MOTFCYr~ TYPE: peptide
(xi) sr;QuhL~-r~ DESCRIPTION: SEQ ID NO:88:
Ala Ala Arg Val Ala Gly Gln Thr Arg A~n Ile Thr Val Asp Pro Arg
1 5 10 15
Phe Lys Lys Arg Arg Leu Arg Ser Pro Arg Val Leu Phe Ser Thr Gln
Pro Pro Arg Glu Ala Asp Thr Thr Gln Asp Leu Asp Phe Glu Val Gly
Gly Ala Ala Pro Phe Asn Arg Thr His Arg Ser Lys Arg Ser Ser Ser
His Pro Ile Phe His Arg Gly Glu Phe Ser Val Cy~ Asp Ser Val Ser
153
WO 93/25684 PCI/US93/OS672
~al Trp Val Gly A~p Ly~.~Thr Thr~Ala Thr Asp Ile Ly~ Gly Lys Glu
' 90 95
Val Met Val Leu Gly Glu Val A~n Ile A~n A~n Ser Val Phe Ly~ Gln
2 1 3 7 7 9 9 loo 105 110
Tyr Phe Phe Glu Thr Ly~ Cy~ Arg A~p Pro Asn Pro Val A~p Ser Gly
115 120 125
Gly Arg A~p Ile A~p Ser Lyn Hi~ Trp A~n Ser Tyr Cy~ Thr Thr Thr
130 135 140
Hi~ Thr Phe Val Ly- Ala Leu Thr Met A~p Gly Ly3 Gln Ala Ala Trp
145 150 155 160
Arg Phe Ile Arg Ile Anp Thr Ser Cy~ Val Cy~ Val Leu Ser Arg Ly~
165 170 175
Ala Val Arg
(2) INFORMATION FOR SEQ ID NO:89:
(i) SEQUENCE CHARACTERISTICS:
(A' LENGTH: 163 amino acid~
(B, TYPE: amino acid
(C, STRANDEDNESS: ~ingle
(D,, TOPOLOGY: ~nkn,~
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:89:
Phe Phe Ly~ Ly~ Ly~ Arg Phe Arg Ser Ser Arg Val Leu Phe Ser Thr
1 5 10 15
Gln Pro Pro Pro Glu Ser Arg Lyn Gly Gln Ser Thr Gly Phe Leu Ser
Ser Ala Val Ser Leu A~n Arg Thr Ala Arg Thr Ly~ Arg Thr Ala Hi~
Pro Val Leu His Arg Gly Glu Phe Ser Val Cy~ A~p Ser Val Ser Met
Trp Val Gly A~p Lys Thr Thr Ala Thr Asp Ile Ly~ Gly Ly~ Glu Val
Thr Val Leu Gly Glu Val Ann Ile A~n A~n A~n Val Phe Ly~ Gln Tyr
Phe Phe Glu Thr Ly~ Cy~ Arg A~p Pro Arg Pro Val Ser Ser Gly Gly
100 105 110
Arg Asp Ile A~p Ala Ly~ His Trp A~n Ser Tyr Cy~ Thr Thr Thr Hi~
115 120 125
Thr Phe Val Ly~ Ala Leu Thr Met Glu Gly Ly~ Gln Ala Ala Trp Arg
130 135 140
Phe Ile Arg Ile A~p Thr Ser Cy5 Val Cy~ Val Leu Ser Arg Ly~ Ser
145 150 155 160
Gly Arg Pro
154
WO 93/25684 PCI-/US93/05672
?1~7~
(2) INFORMATION FOR SEQ ID NO:90: ` --
( i ) ~hQ~L~L CHARACTERISTICS: ;
'A) LENGTH: 124 amino acids
l'B) TYPE: amino acid
,C) STRANDEDNESS: single
,D) TOPOLOGY: 1~nkn~ "
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:90:
~ His Arg Ser Ly~ Arg Ser Ser Glu Ser His Pro Val Phe His Arg Gly
- 1 5 10 15
Glu Phe Ser Val Cys Asp Ser Ile Ser Val Trp Val Gly ARP Lys Thr
Thr Ala Thr Asp Tle Lys Gly Lys Glu Val Met Val Leu Gly Glu Val
Asn Ile Asn Asn Ser Val Phe Lys Gln Tyr Phe Phe Glu Thr Ly~ Cys
Arg A~p Pro Asn Pro Val Asp Ser Gly Gly Arg ARP Ile Asp Ala Lys
His Trp Asn Ser Tyr Cys Thr Thr Thr His Thr Phe Val Ly~ Ala Leu
Thr Met A~p Gly Ly~ Gln Ala Ala Trp Arg Phe Ile Arg Ile Asp Thr
100 105 110
Ser Cys Val Cys Val Leu Ser Arg Ly3 Thr Gly Gln
llS 120
(2) INFORMATION FOR SEQ ID NO:91:
(i) SEQUENCE CHARACTERISTICS:
'A'I LENGTH: 164 amino acids
l'B TYPE: amino acid
,C STRhNDEDNESS: 3ingle
~DJ TOPOLOGY: 1-nk- ."
(ii) MOLECULE TYPE: peptide
(Xi) ~h~UL.. - ~ DESCRIPTION: SEQ ID NO:91:
Val A~p Pro Lys Leu Phe Gln Ly3 Arg Gln Phe Gln Ser Pro Ary Val
1 5 10 15
Leu Phe Ser Thr Gln Pro Pro Leu Leu Ser Arg Asp Glu Glu Ser Val
Glu Phe Leu Asp Asn Glu Asp Ser Leu Asn Arg Asn Ile Arg Ala Ly~
Arg Glu Asp Hi~ Pro Val His Asn Leu Gly Glu His Ser Val Cy~ Asp
Ser Val Ser Ala Trp Val Gly Lys Thr Thr Ala Thr Asp Ile Lys Gly
Asn Thr Val Thr Val Met Glu Asn Val Asn Leu Asp Asn Lys Val Tyr
155
2~ 7899 i~ PCI/US93/05672
Ly~ Gln Tyr Phe Pho Glu Thr Lys Cy~ Arg A~n Pro Asn Pro Glu Pro
100 105 110
Ser Gly Gly Arg A~p Ile A~p Ser Ser Hi~ Trp A~n Ser Tyr Cy~ Thr
115 120 125
Glu Thr A~p Gly Phe Ile Ly~ Ala Leu Thr Met Glu Gly Asn Gln Ala
130 135 140
Ser Trp Arg Phe Ile Arg Ile A~p Thr Ser Cy~ Val Cy~ Val Ile Thr
145 150 155 160
Ly~ Ly~ Ly~ Gly
(2) lN~ORMATION FOR SEQ ID NO:92:
~QU~N~ CHARACTERISTICS:
,'A~, LENGTH: 196 amino acid~
IBI TYPE: amino acid
,C STRANDEDNESS: ningle
~D, TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) S~u~_~ DESCRIPTION: SEQ ID NO:92:
Pro Ala Gly Ser Ser Pro Asp Pro Ser Ser Pro Val Val Asp Pro Ly~
1 5 10 15
Leu Phe Ser Ly~ Arg Hi~ Tyr Pro Ser Pro Arg Val Val Phe Ser Glu
2S 30
Val Ile Pro Ser Hi~ A~p Val Leu A~p Gly Glu Gly Tyr Asp Phe Glu
Arg Val Arg Gly Leu Arg Val Arg Arg Ly~ Ala Val Ser Hi~ Thr Met
His Arg Gly Glu Tyr Ser Val Cy~ A~p Ser Ile A~n Thr Trp Val A~n
Thr Ly~ Arg Ala Thr A~p Met Ser Gly A~n Glu Val Thr Val Leu Ser
His Val Thr Val A~n ADn LYB Val Ly~ Lya Gln Leu Phe Tyr Glu Thr
100 105 110
Thr Cys Arg Ser Pro Thr Hi~ Arg Ser Ser Gly Ile Val Ile Gly Gly
115 120 125
Arg Ser Gly Gly Arg Gly Gly Ser Gln Gly Ser Ly~ Thr Gly A~n Ser
130 135 140
Gly Gly Arg A~p Ile Asp Ser Arg Tyr Trp A~n Ser Hi~ Cy~ Thr A~n
145 150 155 160
Thr A~p Ile Tyr Val Ser Ala Leu Thr Val Phe Ly~ Glu Gln Thr Ala
165 170 175
Trp Arg Phe Ile Arg Ile Asn Ala Ser Cyn Val Cy~ Val Ser Arg Thr
180 185 190
156
WO 93/25684 ~ 7 7 9 g PCI`/US93/05672
A~n Ser Trp Ser . ,
(2) INFORMATION FOR SEQ ID NO:93:
(i) SEQUENCE CHARACTERISTICS:
,'A'I LENGTH: 225 ba~e pairn
B TYPE: nucleic acid
,C STRANDEDNESS: double
,,DJ TOPOLOGY: unknown
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/REY: CDS
(8) LOCATION: 1..225
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:93:
CGA GTG GTC CTG TCT AGG GGT GCC GCT GCC GGG CCC CCT CTG GTC TTC 48
Arg Val Val Leu Ser Arg Gly Ala Ala Ala Gly Pro Pro Leu Val Phe
1 5 10 15
CTG CTG GAG ACT GGA GCC TTT CGG GAG TCA GCA GGC GCC CGG GCC AAC 96
Leu Leu Glu Thr Gly Ala Phe Arg Glu Ser Ala Gly Ala Arg Ala Asn
20 25 30
CGC AGC CAG CGA GGG GTG AGC GAT ACT TCA CCG GCG AGT CAT CAG GGT 144
Arg Ser Gln Arg Gly Val Ser Asp Thr Ser Pro Ala Ser His Gln Gly
35 40 45
GAG CTG GCC CTG TGC GAT GCA GTC AGT GTC TGG GTG ACA GAC CCC TGG 192
Glu Leu Ala Leu Cys Asp Ala Val Ser Val Trp Val Thr A~p Pro Trp
50 55 60
ACT GCT GTG GAC TTG GGT GTG CTC GAG GTC GAG 225
Thr Ala Val A~p Leu Gly Val Leu Glu Val Glu
65 70 75
(2) INFORMATION FOR SEQ ID NO:94:
(i) ~QD~CE CHARACTERTSTICS:
A) LENGTH: 75 amino acids
B) TYPE: amino acid
~D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:94:
Arg Val Val Leu Ser Arg Gly Ala Ala Ala Gly Pro Pro Leu Val Phe
1 5 10 15
Leu Leu Glu Thr Gly Ala Phe Arg Glu Ser Ala Gly Ala Arg Ala A~n
Arg Ser Gln Arg Gly Val Ser Asp Thr Ser Pro Ala Ser His Gln Gly
- Glu Leu Ala Leu Cys Asp Ala Val Ser Val Trp Val Thr A~p Pro Trp
Thr Ala Val Asp Leu Gly Val Leu Glu Val Glu
157
W O 93/25684 PC~r/US93/05672
2137~9g ~ ."~
(2) lNrOR~ATION FOR SEQ ID NO:95:
( i ) ~h~U~N~ CHARACTERISTICS:
'A'l LENGTH: 53 ba~e pair~
IB TYPE: nucleic acid
,C STRANDEDNESS: single
~D,l TOPOLOGY: unknown
(ii) MOLECULE TYPE: cDNA
(Xi) ~hyU~Ne~ DESCRIPTION: SEQ ID NO:95:
CCCACAAGCT TGTTGGCATC TATGGTCAGA GCCCTCACAT AAGACTGTTT TGC 53
(2) INFORMATION FOR SEQ ID NO:96:
( i ) ~yU~N~ CHARACTERISTICS:
~'A~ LENGTH: 10 amino acids
B TYPE: amino acid
C STR~NDEDNESS: ~inqle
~D, TOPOLOGY: ~l n ~n~ . "
(ii) ~OLECULE TYPE: peptide
(xi) ~:yu~ DESCRIPTION: SEQ ID NO:96:
Ly~ Cy~ Asn Pro Ser Gly Ser Thr Thr Arg
l 5 lO
(2) INFORMATION FOR SEQ ID NO:97:
yu~.._~ CHARACTERISTICS:
'A', LENGTH: 7 amino acid~
Bl TYPE: amino acid
C STRANDEDNESS: 3ingle
,D, TOPOLOGY: llnl-- ,,"
(ii) MOLECULE TYPE: peptide
(xi) ~gu~N~ DESCRIPTION: SEQ ID NO:97:
Arg Gly Cy~ Arg Gly Val Asp
1 5
(2) INFORMATION FOR SEQ ID NO:98:
( i ) ~Q~N~ CHARACTERISTICS:
~A' LENGTH: 5 amino acid~
B TYPE: ~mino acid
,C, STRANDEDNESS: ~ingle
,D,, TOPOLOGY: llnl~
(ii) MOLECULE TYPE: peptide
(xi) S~:yU~N~ DESCRIPTION: SEQ ID NO:98:
Ly~ Gln Trp Ile Ser
(2) INFORMATION FOR SEQ ID NO:99:
158
W O 93/25684 PC~r/US93/05672
2137799 `; ``
(i) ~yu~nCE CHARACTERISTICS~
'A'l LENGTH: 6 amino acids 7 '
B TYPE: amino acid
C, STRANDEDNESS: ~ingle
~Dl TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:99:
Ly~ Gln Ser Tyr Val Arg
1 5
- (2) INFORMATION FOR SEQ ID NO:100:
(i) SEQUENCE CHARACTERISTICS:
'A'l LENGTH: 7 umino acid~
~B TYPE: amino acid
C STRANDEDNESS: ~ingle
,D, TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:100:
Gly Pro Gly Xaa Gly Gly Gly
1 5
(2) INFORMATION FOR SEQ ID NO:101:
(1) SEQUENCE CHARACTERISTICS:
'A' LENGTH: 7 amino acid~
B TYPE: amino acid
C STRANDEDNESS: ~ingle
,D, TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:101:
Gly Pro Gly Val Gly Gly Gly
1 5
(2) INFORMATION FOR SEQ ID NO:102:
QDriN~ CHARACTERISTICS:
,'A` LENGTH: 7 amino acid~
BI TYPE: amino acid
Cl STRANDEDNESS: ~ingle
~,D~ TOPOLOGY: ~n~n, ...,
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:102:
Gly Pro Gly Ala Gly Gly Gly
(2) lNroR~ATIoN FOR SEQ ID NO:103:
(i) ~r;yu~nCE CHARACTERISTICS:
(A) LENGTH: S amino acid~
(B) TYPE: amino acid
159
W O 93/25684 ,'~ ' , ~ ; P ~ /US93/05672
9 ~c, STRANDEDNESS: single
213 7 7 D) TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) ~Q~hNCE DESCRIPTION: SEQ ID NO:103:
Glu Ser Ala Gly Glu
1 5
(2) INFORMATION FOR SEQ ID NO:104:
(i) s~Qu~ r- CHARACTERISTICS:
'A' LENGTH: 5 amino acid~ -
IB TYPE: amino acLd
,C STRANDEDNESS: ~ingle
~D,l TOPOLOGY: lln~
(ii) MOLECULE TYPE: peptide
(xi) SEQDLN~L DESCRIPTION: SEQ ID NO:104:
A~p A~n Ala Glu Glu
1 5
(2) INFORMATION FOR SEQ ID NO:105:
(i) s~g~:N~ CHARACTERISTICS:
'A' LENGTH: 18 ba~e pair~
BI TYPE: nucleic acid
C, STRANDEDNESS: ~ingle
~D,, TOPOLOGY: t, nkn ~_ ,, ~,
( ii ) M~T.~CUT.T~` TYPE: DNA (genomic)
(Xi) ~:yuL~ : DESCRIPTION: SEQ ID NO:105:
CAGTATTTTT AC~AAACC 18
(2) lN~OR~ATION FOR SEQ ID NO:106:
(i) SEQUENCE CHARACTERISTICS:
'A', LENGTH: 18 ba~e pair~
B TYPE: nucleic acid
C, STRANDEDNESS: ~ingle
,D,I TOPOLOGY: unknown
( ii ) M~T~T~CYT~T~ TYPE: cDNA
(Xi) S~U~N~L DESCRIPTION: SEQ ID NO:106:
ACA~.LCG~ G 18
(2) INFORMATION FOR SEQ ID NO:107:
( i ) SLgUL.. T' CHARACTERISTICS:
'A' LENGTH: 18 ba~e pair~
B TYPE: nucleic ac$d
C, STRANDEDNESS: ~ingle
l,DJ TOPOLOGY: llnkn~ ~"
(ii) MOLECULE TYPE: DNA (genomic)
160
W O 93/25684 2 1 ~ 7 7 9 9 PC~r/US93/05672
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:107: ~,
CAGTATTTTT ~AG~rG 18 .
(2) INFORMATION FOR SEQ ID NO:108:
(i) ~yuh~_E CHARACTERISTICS:
A', LENGTH: 18 ba~e pair~
,BI TYPE: nucleic acid
- C sTRA~n~np~ss: ~inqle
,D, TOPOLOGy llnl-- - ,,"
- (ii) MOLECULE TYPE: cDNA
(xi) ~:QuhNCE DESCRIPTION: SEQ ID NO:108:
ACAlllCGGL TTGTTAGC 18
(2) INFORMATION FOR SEQ ID NO:109:
(i) SEQUENCE CHARACTERISTICS:
'Al LENGTH: 36 base pairs
B TYPE: nucleic acid
C, STRANDEDNESS: ~ingle
~D TOPOLOGY: ~
(ii) MOLECULE TYPE: DNA (genomic)
~xi) SEQUENCE DESCRIPTION: SEQ ID NO:109:
TGCAGTTTCG CTC~CCCCCC GTTTTAGCCG GGAAGT 36
(2) INFORMATION FOR SEQ ID NO:110:
( i ) ~QD~N~ CHARACTERISTICS:
'A' LENGTH: 7 amino acids
BI TYPE: amino acid
,C STRANDEDNESS: single
lDJ TOPOLOGY: ~lnkn~
(ii) MOLECULE TYPE: peptide
(xi) ~:yu~ DESCRIPTION: SEQ ID NO:110:
Lys Gln Tyr Phe Tyr Glu Thr
1 5
(2) INFORMATION FOR SEQ ID NO:lll:
( i ) ~QU~N~ CHARACTERISTICS:
A' LENGTH: 7 amino acids
lB TYPE: amino acid
,C, STRANDEDNESS: single
,D, TOPOLOGY: I-nknot~,,
- (ii) MOLECULE TYPE: pept$de
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:lll:
Trp Arg Phe Ile Arg Ile Asp
161
W O 93/25684 ; ~ t ~ ~ ~ P ~ /Us93/05672
2~3~ 9~ 5
, ~2) INFORMATION FOR SEQ ID NO:112:
Q~N~r; CHARACTERISTICS:
'A'l LENGTH: 7 amino acidn
~B TYPE: amino acid
,C STRANDEDNESS: single
~D,, TOPOLoGy I~nk- ,,"
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:112:
Gly Glu Leu Ser Val Cys Asp
(2) INFORMATION FOR SEQ ID NO:113:
(i) SEQUENCE CHARACTERISTICS:
(A' LENGTH: 6 amino acids
(B TYPE: amino acid
(C STRANDEDNESS: single
(D, TOPOLOGY: llnkn-,~n
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:113:
Lys Ala Glu Ser Ala Gly
1 5
(2) INFORMATION FOR SEQ ID NO:114:
( i ) S~yUh~: CHARACTERISTICS:
'A' LENGTH: 45 ba~e pairs
B' TYPE: nucleic acid
C STRANDEDNESS: ~ingle
~D, TOPOLOGY: ~nkr ....
( ii ) MoT~cuT~ TYPE: peptide
(xi) ~riQuk.._r; DESCRIPTION: SEQ ID NO:114:
GGAGGGGGCT GCCGGGr~ T GGACAGGAGG CACTGGGTAT CTGAG 45
(2) lNru~ATION FOR SEQ ID NO:115:
( i ) ~UL.._~: CHARACTERISTICS:
'A'l LENGTH: 15 amino acids
B TYPE: amino acid
,C STRANDEDNESS: single
~D,l TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
(xi) ~yuh~cE D~Sr~TPTION: SEQ ID NO:115:
Gly Gly Gly Cys Arg Gly Val Asp Arg Arg Hi~ Trp Val Ser Glu
l 5 10 15
(2) INFORMATION FOR SEQ ID NO:116:
162
W O 93/25684 2 1 ~ 7 7 g 9 PC~r/US93/05672
(i) s~:~Dh~_~ CHARACTERISTICS~
fA'I LENGTH: 582 ba~e pairr
~B TYPE: nuclei~ acid
C STRANDEDNESS: double
D,, TOPOLOGY: unknown
(ii) MOLECULE TYPE: DNA (genomic)
(ix) FEATURE:
(A) NAME/KEY: CDS
(B) LOCATION: 1.. 396
.,
- (xi) SEQUENCE DESCRIPTION: SEQ ID NO:116:
GTA GTT TGT CCA ATT ATG TCA CAC CAC AGA AGT AAG GTT CCT TCA CAA 48
Val Val Cy~ Pro Ile Met Ser Hi~ His Arg Ser Lys Val Pro Ser Gln
l 5 10 15
AGA TCC TCT AGA GTC GCG CCC GCG ACC TGC AGG CGC AGA ACT GGT AGG 96
Arg Ser Ser Arg Val Ala Pro Ala Thr Cy~ Arg Arg Arg Thr Gly Arg
TAT GGA AGA TCC CTC GAG GTG GAG GTG TTG GGC GAG GTG CCT CCA GCT 144
Tyr Gly Arg Ser Leu Glu Val Glu Val Leu Gly Glu Val Pro Pro Ala
GTC GGC AGT TCC CTC CGC CAG CAC TTC TTT GTT GCC CGC TTC GAG GCC 192
Val Gly Ser Ser Leu Arg Gln Hi~ Phe Phe Val Ala Arg Phe Glu Ala
GAT AAA TCT GAG GAA GGT GGC CCG GGG GTA GGT GGA GGG GCT GCC GCC 240
A~p Lys Ser Glu Glu Gly Gly Pro Gly Val Gly Gly Gly Ala Ala Ala
GGG GTG TGG ACC GGG GGG CAC TGG GTG TCT GAG TGC AAG GCC AAG CAG 288
Gly Val Trp Thr Gly Gly Hin Trp Val Ser Glu Cyq Lys Ala Ly~ Gln
TCC TAT GTG CGG GCA TTG ACC GCT GAT GCC CAG GGC CGT GTG GAC TGG 336
Ser Tyr Val Arg Ala Leu Thr Ala A~p Ala Gln Gly Arg Val A~p Trp
100 105 110
CGA TGG ATT CAA ACT GGC ACA GCC TGT GTC TGC ACA CTC CTC AGC CGG 384
Arg Trp Ile Gln Thr Gly Thr Ala Cyr Val Cy~ Thr Leu Leu Ser Arg
115 120 125
ACT GGC TGG GCC TGAGACTTAT ACC~PGr7AAC TGGTCAGGCA GAAAPAGAAC 436
Thr Gly Trp Ala
~ 130
AGAGCTGGAT GCTr-AGAGAC CTCAGGGTTG GCCCAGCTGC TCTACGGACG GACCC~GTT 496
- GGGGAACTCA TGAAATCATC A~AAAATCAC AA~-~.GA ATTTGAGCTC AATCTCTGCA 556
GGATGGGTGC CACCACATGT GGTTTT 582
163
W O 93/25684 ~ . P ~ /US93/05672
.. .. . . .. ..
(2) INFORMATION FOR SEQ ID NO:117:
n Q (i) SEQUENCE CH M ACTERISTIC5:
~l~7~ ~ (A) LENGTH: 132 amino acid~
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:117:
Val Val Cyn Pro Ile Met Ser Hi~ Hin Arg Ser LYB Val Pro Ser Gln
1 5 10 15
Arg Ser Ser Arg Val Ala Pro Ala Thr Cyn Arg Arg Arg Thr Gly Arg
Tyr Gly Arg Ser Leu Glu Val Glu Val Leu Gly Glu Val Pro Pro Ala
Val Gly Ser Ser Leu Arg Gln Hi~ Phe Phe Val Ala Arg Phe Glu Ala
Asp Ly~ Ser Glu Glu Gly Gly Pro Gly Val Gly Gly Gly Ala Ala Ala
Gly Val Trp Thr Gly Gly Hi~ Trp Val Ser Glu Cy~ Ly~ Ala Lys Gln
Ser Tyr Val Arg Ala Leu Thr Ala A~p Ala Gln Gly Arg Val A~p Trp
100 105 110
Arg Trp Ile Gln Thr Gly Thr Ala Cys Val CYB Thr Leu Leu Ser Arg
115 120 125
Thr Gly Trp Ala
130
(2) INFORMATION FOR SEQ ID NO:118:
(i) SEQUENCE CHARACTERISTICS:
'A' LENGTH: 36 ba~e pair~
B TYPE: nucleic acid
C STRANDEDNESS: ~ingle
,,DI TOPOLOGY: 1~n1__ ,,"
(ii) MOLECULE TYPE: DNA (genomic)
(xi) ~yur;N~r; DESCRIPTION: SEQ ID NO:118:
CGGTACCCTC GAGCCACCAT G~.CC~.~.C CCCTCA 36
(2) lwrOfi~ATION FOR SEQ ID NO:119:
(i) sriQ~N~r; CHARACTERISTICS:
'A) LENGTH: 36 ba~e pairs
IB) TYPE: nucleic ~cid
,C) STRANDEDNESS: ~ingle
,D) TOPOLOGY: tlnl-~:.l"
(ii) MOLECULE TYPE: DNA (genomic)
(xi) ~r;yur;N~ DESCRIPTION: SEQ ID NO:119:
164
W O 93/25684 21 ~ 7 7 ~ 9 PC~r/US93/05672
CGGTACAAGC GGCCGCTTCT TGGGCATGGG TCTCAG , 36
(2) 1N~fi~ATION FOR SEQ ID NO:120: ,, 1! ,/ ~ ~ "~
(i) ShQDhN~h CHARACTERISTICS
I~A~I LENGTH: 36 ba~e pairs
B, TYPE: nucleic acid
,C, STRANDEDNESS: ~ingle
~D, TOPOLOGY: ~n~n~t,.,
- (ii) MOLECULE TYPE: DNA ( genomic)
Xi) S~YU~ _h DESCRIPTION: SEQ ID NO:120:
CGGTACCCTC GAGC~rC~ GG.~.CCGA GAGATG 36
(2) INFORMATION FOR SEQ ID NO:121:
(i) SEQUENCE CHARACTERISTICS:
~A~, LENGTH: 2 3 base pairs
,B TYPE: nucleic acid
C, STRANDEDNESS: ging1e
~D, TOPOLOGY: unknown
(ii) MOLECULE TYPE: DNA ~ genomic)
(Xi) S~UhN~ DESCRIPTION: SEQ ID NO:121:
~GACCGGAA GCTTCTAGAG ATC 23
(2) INFORMATION FOR SEQ ID NO:122:
(i) ~hYUhhCE CHARACTERISTICS:
,'A' LENGTH: 36 ba~e pairs
,B, TYPE: nucleic acid
C STRANDEDNESS: ffLngle
~D, TOPOLOGY: 11n~
(ii) MOLECULE TYPE: DNA ( genomic)
(Xi) ShgUhh-E DESCRIPTION: SEQ ID NO:122:
TGCAGTTTCG CT~rCCCCC G.-.CCGCCG TGATGT36
(2) INFORMATION FOR SEQ ID NO:123:
(L) X~QUhh-h CHARACTERISTICS:
I~A~I LENGTH: 36 ba~e paLr~
B, TYPE: nucleLc acid
,C, STRANDEDNESS: 8 Lngle
,D, TOPOLOGY lln'~
(LL) MOLECULE TYPE: DNA ( genomLc)
(xL) ~g~hN~ DESCRIPTION: SEQ ID NO:123:
ACATCACGGC GGAAACGGGG GGTrr ,C~-~A ACTGCA 36
(2) INFORMATION FOR SEQ ID NO:124:
(L) SEQUENCE CHARACTERISTICS:
165
WO 93/25684 ,,, ~ j PCI/US93/05672
~ 9 9 I'A'I LENGTH: 36 ba~e pair~
Z~31 B TYPE: nucleic acid
,C, STRANDEDNESS: single
,DJ TOPOLOGY llnkn~
(ii) MOLECULE TYPE: DNA (genomic)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:124:
A~lCCCGGC TAAA~CGGGG GGTGAGCGAA ACTGCA 36
166