Note: Descriptions are shown in the official language in which they were submitted.
~13g623
WO 94/26934 ^ PCT/US94/0~085
HUMAN PAPILLOMAVIRUS DETECTION ASSAY
BACKGROUND OF THE INVENTION
Human papillomaviruses (HPVs) are a heterogeneous group
of DNA viruses associated with a variety of proliferative lesions of the
epithelium. Many of these lesions are benign such as those associated
with HPV 6 and HPV 11, and are considered causative of such conditions
as warts, and condylomas (see C.i~sm~n, Canc. Surv., 3: 161 (1984)).
However, epidemiological and molecular studies implicate several high
risk types that infect the genital tract associated with dysplasia and
sometimes progress to cervical cancer (see, for example, Durst. et al.,
PNAS, 80: 3812 (1983)). High risk HPV types are predominately HPV 16
and HPV 18, with HPV 31, HPV 33, and HPV 35 being of lesser significance.
More recently, another HPV type associated with malignancy, HPV 44, has
been identified (Lorincz, U.S. Patent No. 4,849,331).
HPV of any type is generally found in extremely low numbers
in biological specimens. Therefore, molecular techniques must be
performed for amplifying nucleic acid viral markers from very low copy
number in a specimen to detectable levels. Polymerase chain reaction
20 (PCR) has been utilized to amplify HPV viral DNA in this manner, as
disclosed in WO 90/02821, and Shibata, et al., J. Exp. Med., 167: 225 (DATE).
Other applications of PCR to HPV diagnostics are Maitland, et al., May
WO 94l2cg34 PCT/US94/0~085
2~396~3 2
1988. Seventh International Papillomavirus Workshop, Abstract, p. 5 and
Campione-Piccardo, et al., May 1988, $eventh International
Papillomavirus Workshop. One major problen~ with PCR amplification
of HPV is that these viruses are detectable as fortuitous passengers in a
5 significant percentage of healthy women showing no evidence of any
benign of malignant pathology. Percentage estimates of such passenger
presence range 10% (see U.S. Patent No. 4,983,728) to as high as 60%.
Detection of HPV per se is thus of limited diagnostic value.
Many nucleic acid-based assays utilize the well-known
10 sandwich configuration in a heterogeneous format. In this format a
capture oligonucleotide is chPmicAlly conjugated to a solid support such as
a micloliler well or bead, the sample is added, and the target nucleic acid
having base homology to capture oligonucleotide is allowed to hybridize.
After a wash (phase separation), a detection oligonucleotide hybridizes,
15 and after a second wash to remove unhybridized detection
oligonucleotide, the amount of tracer or reporter is measured, or the signal
generating means produces a signal. For the details of such assays, refer to
Ranki, U.S. Patent No. 4,486,539 and U.S. Patent No. 4,731,325. The basic
problem with such sandwich assays is relatively low capture efficiency on
20 the solid support, which may p.o~o~,dly reduce sensiLivily of the assay.
SUMMARY OF TE~F. INVENTION
It is an object of this invention to provide a specific assay for
HPV infections associated with cervical dysplasia and cellular
25 transformation to malignancy. In achieving this object, it is essential to
first amplify to detectable levels only the messenger RNA (mRNA)
expressed from oncogene regions (genes E6/E7) of HPV types implicated in
malignant or pre-malignant cervical lesions. This not only restricts
detection to malignant and pre-malignant HPV types, but also
30 distinguishes actual oncogene expression from mere passenger presence of
virus.
. 213g623
W094/~934 PCT~S94/05085
3 . `--
It is a further object to provide a highly sensitive assay for
HPV having a high capture efficiency in the initial capture hybridization
step. This is important because in situations in which the patient
specimen contains very low copy number of viral mRNA, amplification
may not occur to a level high enough for detection unless the assay itself is
sensitive.
It is a still further aspect of the invention to provide reagents
such as primer f~mili~ for optimally efficient amplification, and probes
which anneal to their targets under stringent conditions to give high
selectivity and specificity. Finally, the invention contemplates a kit
comprising these reagents, buffers, sample preparation solutions, solid
supports, and reaction vessels.
In accordance with the assay of the present invention, a
patient specimen suspected of containing messenger RNA encoded by at
least one type of HPV associated with cervical dysplasia, malignant cells, or
pre-malignant cells is
(1) subjected to nucleic acid amplification by self sustained
sequence replication utilizing two primers separated by at least ten
nucleotides, at least one such primer containing a transcriptional
promoter,
annealing the first such primer to its complementary
sequence on the target region messenger RNA, extending the 3' end of the
primer by action of a strand-extending polymerase in the presence of
cofactors and nucleotide triphosphates,
digesting the RNA strand of the nascent RNA/DNA duplex
with an enzyme having exogenous or endogenous RNAse H activity,
annealing the second such primer to its complementary
sequence on the resultant single stranded cDNA, primer extending the 3'
end of the primer by action of a strand-extending polymerase,
transcribing the double stranded DNA with a transcriptase in
the presence of nucleoside triphosphates, and
;~ *
WO 94t26934 PCT/US94/05085
2~3~3 4
repeating the amplification utilizing the newly synthesized
transcripts as new targets,
(2) hybridizing in solution amplified messenger RNA to a
free biotinylated reagent capture probe having a sequence complementary
5 to a first segment of the amplified RNA to form a reagent capture complex,
(3) attachment of the capture complex to a solid phase by
reaction of the biotin residue of the~ c~pture probe with streptavidin bound
to the surface of the solid phase,
t4) washing the bound complex to remove unbound and
10 unreacted reagents,
(5) hybridizing a virus type-specific enzyme-conjugated
detection probe having a sequence complementary to a second segment of
the amplified RNA not overlapping the sequence of the first such RNA
segment to form a solid phase-bound capture probe-target sequence-
15 detection probe complex,
(6) washing the complex to remove unhybridized
detection probe, and
(7) adding a fluorogenic or chromogenic enzyme substrate
and reacting the conjugated enzyme to produce a detectable fluorophor or
20 chromogen.
The present invention is also directed to certain primer
families and selected probes for use in the HPV detection assay, and to kits
for conveniently providing reagents to users.
25 BRTFF DESCRIPTION OF THE DRAWINGS
Figure 1: HPV 16 genome organization. Transcription
proceeds clockwise from the P97 promotor. AE and AL are the
polyadenylation sites for the early and late transcripts.
Figure 2: Sequence of HPV 16. The primers are indicated by
30 underlines. Boxes indicate splice donor and acce~lor sequences.
2139623
wo 94n6934 - PCT/US94/05085
~ ;
Figure 3: Sequence of HPV 18. Sequences of HPV 18 primers
are indicated by underlines. Boxes indicate splice donors and acceptor
sequences.
Figure 4: HPV 16 primer famili~s. A variety of primers were
5 tested by the ability to amplify total RNA from SiHa cells (infected with
HPV 16). The reactions contained 10% DMSO and 15% sorbitol. The
primers are indicated on the autoradiogram.
Figure 5: The effect of increasing the RNAse H concentration
using HPV 16 primer families.
Figure 6: HPV 16 primer sensitivity. Total RNA is titrated
from 1, 0.1, 0.01, 0.001 attomoles of specific E6-7 RNA isolated from SiHa
RNA. p. 32. N5.
Figure 7: Primer sensitivity using cells which contain HPV 18
DNA. From right to left is 104 to 10 cells. p34 N4.
Figure 8: An autoradiogram slotting 3SR reaction products.
A RNAse titration was performed using primers 32-54 which amplified
HPV 18 RNA.
Figure 9: Autoradiogram of a 3SR reaction using primers 32-
54 containing different additives. The additives (left to right) were 10%
20 DMSO, 10% polyethylene glycol and 10% glycerol. The cross reactivity
using primers 29-15 using SiHa cell using these additives were included to
determine if there was any cross reactivities of the reactions.
Figure 10: Autoradiogram of a 3SR reaction comparing
primers 32-54 and 69-54. The 3SR reaction using primers 69-54 contained
25 either no additives (column 1) or 15% sorbitol (column 2). The reactions
using Prirners 32-54 contained 10% polyethylene glycol (column 3). From
top to bottom was a titration of RNAse H, 1-3 units per reaction.
Figure 11: Co-amplification. Lane A used primers 136-73
(HPV 16), Lane B used primers 136-91 (HPV 16) amplifying 5 amol of SiHa
30 RNA using decreasing amounts of DMSO/sorbitol mixture. Lane C from
top to bottom: 136-73 (HPV 16) and 54-69 (HPV 18), 136-91 and 54-69, and
5~69 amplifying a mixture of 5 amol of SiHa cell (infected with HPV 16)
WO 94/2693,~ ' PCT/US94/0~085
a~39623 6
and HeLa cell (infected with HPV 18) RNA. Duplicate blots were prepared
and probed with an HPV 18 specific probe (59) and an HPV 16 specific
probe (98).
Figure 12: HPV 16 plate optimization. Capture 245
temperature optimum. Absorbance values using CAP245 at different
temperature ranges: 30C, 40C, 50C, 60C and 70C. Each line represents
a different detectors; DET 251, DET 252, and DET 254.
Figure 13: HPV 16 plate optimization. Capture 250
temperature optimum. Absorbance values using CAP250 at different
temperature ranges: 30C, 40C, 50C, 60C and 70C. Each line represents
a different detectors; DET 251, DET 252, and DET 254.
Figure 14: Detector hybridization optimum using CAP 245.
Detectors were hybrirli7e.1 using different temperature ranges: 30C, 40C,
50C, 60C and 70C. Each line represents different detectors: DET 98, DET
251, DET 252, and DET 254.
Figure 15: Detector hybridization optimum using CAP 250.
Detectors were hybridized using different temperature ranges: 30C, 40C,
50C, 60C and 70C. Each line represents different detectors: DET 98, DET
251, DET 252, and DET 254.
Figure 16: HPV 16 plate assay. A comp~ri~on of captures 245,
250, and 253 using DET 98, DET 251, DET 252, and DET 254. Each capture
was hybridized to the 3SR product at 50C. The detectors were hybridized
at room temperature.
Figure 17: HPV 16 detector performance. A comparison of all
the detector oligos for HPV 16 using CAP 250. The detector names are
listed in the bottom of each figure.
Figure 18: A comparison of detector lengths using CAP 250 in
the enzyme probe assay. DET 256 is a 17mer oligo and DET 257 is a 15mer
oligo. The sequence was identical except that 2 bases were omitted for DET
257.
Figure 19: A comparison in absorbance values using different
additives in the capture buffer. From left to right are duplicate wells using
wo 9412C934 2 1 3 9 6 2 3 PCT~US94/05085
DET 255, DET 98 and DET 256. Columns 1-6 are 3SR products using
primers 96-91. Columns 7-12 are 3SR products using primer 137-91 using
different detectors. The additives are indicated on the left of the
absorbance values. Rows 1 and 2 are plus and minus templates using 5%
polyethylene glycol. Rows 3 and 4 are plus and minus templates using 1%
BSA. Rows 5 and 6 are plus and minus templates using 5% PEG, 1% BSA.
Rows 7 and 8 are the standard hybridization buffer using 0.1%
polyvinylpyrrolidone, 5X SSC.
Figure 20: A comparison in absorbance values using different
additives in the detection buffer. From left to right using different
detectors: DET 256, DET 98, and DET 255. Columns 1, 5, and 9 contained
the standard hybridization buffer 30% glycerol, 0.1% PVP, 1% BSA and 5X
SSC. Columns 2, 6, and 10 contained 5% PEG, 0.1% PVP, and 5X SSC as the
hybridization buffer. Columns 3, 7, and 11 contained 1% BSA, 0.1% PVP
and 5X SSC as the hybridization buffer. Columns 4, 8, and 12 contained 5%
PEG, 1% BSA, 0.1% PVP, and 5X SSC as the hybridization buffer. Rows A
and B are plus and minus templates using primers 96-91 which amplify
SiHa RNA. Rows C and D is plus and minus template using primers 136-
91 which amplify SiHa RNA.
Figure 21: Different primers sets which amplify HeLa RNA
(HPV 18). Primers are noted on the autoradiogram.
Figure 22: Comparison of capture oligos for HPV 18 using the
enzyme probe assay. The 3SR product was amplified from HeLa RNA
using primer 54-69. Column 1 is substrate only. Columns 2 and 3 are plus
and minus templates using capture 56. Columns 4 and 5 is plus and
minus templates using capture 267. Rows indicate different detectors.
Row A DET 59, Row B DET 260, Row C DET 262, Row D DET 268, Row E
DET 269, and Row F DET 270.
Figure 23: Comparison of ca~ re oligos for HPV 16 and HPV
18 using the enzyme probe assay. The 3SR product was a co-amplification
from HeLa and SiHa RNA using primers 136-91 (HPV 16) and 54-69 (HPV
18).
WO 94/26934 PCT/US94/05085
2139623 8
Figure 24: HPV 16 and HPV 18 EPA. The absorbance levels of
a typical specimen. HPV 16 and HPV 18 were co-amplified using primers
136-91 and 54-69. CAP 265 and CAP 267 were added and allowed to
hybridize. The reaction was added to two microwells and detected using a
5 type specific oligo DET 256 and HPV 16 and DET 260 for HPV 18.
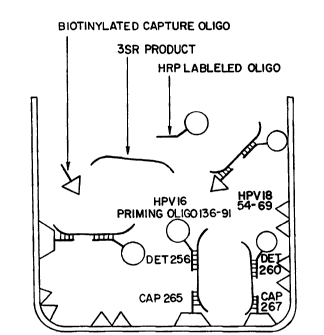
Figure 25: Schematic of the Enzyme Probe Assay. The
capture oligo hybridizes to the amplified 3SR product either HPV 16 or
HPV 18. The complex is detected using HRP labeled oligonucleotide.
Figures 26 and 27: Autoradiographs of amplific~tion products
10 comparing yields of reaction performed at 50C and at 42C.
DETAILED DESCRIPTION OF THE PREFERRED EMBODIMENT
Figure 1 is a schematic drawing showing a generalized HPV
16 genome. The heavy concentric lines indicate open reading frames.
15 Figures 2 and 3 locate the splice donor and acce~lor~ for HPV 16 and 18
genes (indicated by boxes around the terminal two bases involved in the
splice in the E6/E7 region). The portion of the HPV 16 and 18 viral
genomes coding for E6/E7 polypeptides are identified in the Sequence
Listing as SEQ. ID. Nos. 1 and 2 respectively. This is a significant region of
20 the genome since the ~roleins encoded are thought to be involved in
degradation of the p53 su~ressor ~roLeill, which regulates cell growth.
Loss of p53 function is associated with malignancy. Thus, expression of
E6/E7 is diagnostic for cervical cancer or pre-malignant states.
In the ex~ression of the E6/E7 region, splicing at the positions
25 indicated in the figures occurs at substantial but unknown frequency. In
designing primers for amplification of mRNA targets transcribed from this
region, it is therefore important to make certain that all primer pairs lie
outside the portion of the transcript from which the splice leads to excision
of an mRNA fragment. Typical primers selected are illustrated in figures 2
30 and 3.
Since the rationale of the assay of the present invention is to
detect only gene products produced in cells actually expressing genes
wo 94/26934 213 9 6 2 3 PCT/US94/05085
E6/E7, self-sustained sequence replication (3SR) is the amplification
method of choice. Polymerase chain reaction amplifies DNA, and while it
may detect the presence of virus with great sensitivity, it is unsuitable for
detecting gene expression. The method of 3SR is fully described in
Gingeras, et al., Ann. Biol. Clin., 48: 498 (1990), Guatelli, et al., PNAS,
87: 1874 (1990), and WO 90/06995. The methods described therein are
followed herein except as noted, and define the procedure to be followed
in the practice of the present invention. The general 3SR amplification
procedure as set forth in Gingeras et al. and Guatelli et al. involves the
following steps: One hundred-microliter 3SR amplification reactions
contained the target RNA, 40 mM Tris-HCl at pH 8.1, 20 mM MgCl2, 25
mM NaCl, 2 mM spermidine hydrochloride, 5 mM dithiothreitol, 80
~g/ml bovine serum albumin, 1 mM dATP, 1 mM dCTP, 1 mM dGTP, 1
mM dTTP, 4 mM ATP, 4 mM CTP, 4 mM GTP, 4 mM UTP, and 250 ng of
each selected oligonucleotide primer. After heating at 65C for 1 minute
and cooling at 37C for 2 minutes, 30 units of AMV reverse transcriptase,
100 units of T7 RNA polymerase, and 4 units of E. coli RNase H were
added to each reaction. All reactions were incubated at 37C for 1 hour
and stopped by placing the reaction on ice.
In general, 3SR is carried out as follows on HPV specimens:
samples are obtained by vaginal lavage or cervical scrape. Messenger RNA
is released by treatment with chaotrophic/phenol reagents and precipitated
conventionally with ethanol. A prefel,ed one step extraction utilizes
RNAzol B (Cinna/Tiotecx Laboratories, Inc.) according to the
manufacturer's instructions. The RNA is then dissolved in 3SR buffer,
together with nucleotide and nucleoside triphosphates, primers, enzymes,
and cofactors to carry out 3SR amplification. Reagents were obtained as
follows:
Primer Oligonucleotides
All oligonucleotides may be synthesized on a commercially
available synthesizer such as a Milligen 8700 DNA synthesizer.
Wo 94t26934 PCT/US94/05085
2~39623 10
Oligonucleotides which contained a 5' biotin may be synthesized using a
biotin phosphoramidite (Glenn Research). Oligonucleotides which
contain a 3' biotin may be synthesized using control pore glass containing a
protected biotin (Glenn Research?. Oligonucleotides which contain a 3'
amine are conveniently synthesized using a amino-on control pore glass
column (Glenn Research). Below is~ a list of oligonucleotides used in the
development of HPV16/18 enzyrnë`probe assay of the present invention.
All of the sequences are from left to right 5' to 3'. The oligonucleotide
primers are also listed in the Sequence Listing as SEQ. ID. Nos. 3-31.
SEO. ID. No. Primer Probes
3 HPV15: AAT TTA ATA CGA CTC ACT ATA GGG
AGC l'l l TCT TCA GGA CAC AGT GGC
T
4 HPVl9: AAT GTT TCA GGA CCC ACA GGA GC
HPV20: GAA TGT GTG TAC TGC AAG CAA
CAG
6 HPV29: ATG CAC AGA GCT GCA AAC AAC TA
7 HPV32: CAC TTC ACT GCA AGA CAT AGA A
8 HPV48: AAT TTA ATA CGA CTC ACT ATA GGG
ATG TGT CTC CAT ACA CAG AGT C
9 HPV53: GAA TGT GTG TAC TGCC AAG CAA
CAG
HPV54: AAT TTA ATA CGA CTC ACT ATA GGG
AAA GGT GTC TAA GTT TTT CTG CTG
G
11 HPV69: CTG AAC ACT TCA CTG CAA GAC
12 HPV73: CAG TTA TGC ACA GAG CTG CAA AC
13 HPV74: GTT ATG CAC AGA GCT GCA AAC AA
14 HPV77: CAA GCA ACA GTT ACT GCG AC
15 . HPV89: AGC AAC AGT TAC TGC GAC GT
16 HPV90: GCA CAG AGC TGC AAA CAA CTA TA
WO 94/26g34 213 9 6 2 3 PCT/US94/05085
11 -` ~i. ~.
17 HPV91: ACA GAG CTG CAA ACA ACT ATA CA
18 HPV92: AAT TTA ATA CGA CTC ACT ATA GGG
ACT TTT CTT CAG GAC ACA GTG GCT
TTT
- 5 19 HPV93: AAT TTA ATA CGA CTC ACT ATA GGG
ATT TGC'lTl TCT TCA GGA CAC AGT
GG
HPV94: AAT TTA ATA CGA CTC ACT ATA GGG
ATC ll-l GCT TTT CTT CAG GAC ACA
GT
21 HPV95: AAT TTA ATA CGA CTC ACT ATA GGG
ATG TCT TTG CTT TTC TTC AGG ACA
CA
22 HPV96: AAT TTA ATA CGA CTC ACT ATA GGG
AGA TGT CTT TGC TTT TCT TCA GGA
CA
23 HPV101: AGA GCT GCA AAC AAC TAT ACA TG
24 HPV106: AAT TTA ATA CGA CTC ACT ATA GGG
ATT CAT GCA ATG TAG GTG TAT CTC
C
HPV107: AAT TTA ATA CGA CTC ACT ATA GGG
ATA TTC ATG CAA TGT AGG TGT ATC
T
26 HPV118: AGC TGC AAA CAA CTA TAC ATG AT
27 HPV120: AAT TTA ATA CGA CTC ACT ATA GGG
ATG CAA TGT AGG TGT ATC TCC ATG
C
28 HPV129: AAT TTA ATA CGA CTC ACT ATA GGG
AAA TGT AGG TGT ATC TCC ATG CAG
29 HPV131: AAA CAA CTA TAC ATG ATA TAA TA
Wo 94/26934 PCT/US94/05085
2~39623 12
HPV136: AAT TTA ATA CGA CTC ACT ATA GGG
AAT GTA GGT GTA TCT CCA TGC ATG
A
31 HPV137: AAT TTA ATA CGA CTC ACT ATA GGG
ATG TAG GTG TAT CTC CAT GCA TGA
T
Primer selection for high level amplification is basically a
directed trial and error process. To define a first set of primers a span of
400 bases (with beginning and ending sites outside the spliced region) was
selected by designating the first 10-30 nucleotides at the 5' end of the E6
gene beginning with the ATG codon and counting off 400 bases, then
selecting as primers the next 10-30 bases. Note that for each pair, at least
one of the primers must contain a promoter for transcription. The
bacteriophage T7 RNA polymerase binding site (SEQ. ID. No. 44), AAT
TTA ATA CGA CTC ACT ATA GGG A, is ~rerelled because of its strength
and specificity.
The primer pairs are tested for their amplification efficiency.
To optimize, the second primer position is held stationary and the first
primer is moved arbitrarily 20 bases towards the second (thereby
decreasing the inlelplilller span, e.g. the bases between the position of the
3' end of the first primer and the 5' end of the second primer, by 20 bases to
380 bases). Fine tuning is accomplished by walking the primers from the
best pairings by 2-5 base jumps.
Primer f~mili~. Figure 4 gives primer f~milies that amplify
the HPV 16 E6-7. All primers amplified total RNA isolated from the SiHa
cell line which contain the HPV 16 transcripts. The reaction conditions
include 7mM rNTPs, lmM dNTPs, 40mM Tris pH 8.1, 30mM MgCl2 20mM
KCl, 50mM dithiothreitol, 20 mM spermidine, 10% DMSO, 15% sorbitol,
and 15pmol each priming oligonucleotide. After pre-warming each tube at
42C for 5 minutes 30 units of AMV-RT, 2 units RNAse H, and 250 units of
T7 RNA polymerase were added as a cocktail to each reaction. The
213g623
-- WO 94/26934 PCT/US94/05085
13
reaction was allowed to proceed for one hour at 42C. A sample of the 3Sl~
reaction was slotted onto nitrocellulose. The nitrocellulose was baked for
45 minutes and then hybridized for 45 minutes using a type specific
detection oligo. An autoradiogram was generated by exposing the
5 nitrocellulose to film for 45 minutes at -70C. The primer family for 120 is
29 and 90. The primer family for 15 is 19, 20, 77, 53, and 89. The primer
family for primer 129 is 29, 74, 73, 118, 130, and 131. The prime~ family for
primer 136 is 91, 29, 90, 74, 73, 130, 131, and 118. The primer family for
primer 137 is 29, 90, 74, 73, 131, and 118.
Figure 5 illustrates the effect of titrating the RNAse H HPV 16
primer families. The 3SR reaction conditions are identical as described in
figure 4 except the DMSO and sorbitol were omitted from the reaction.
Ten microliters were slotted onto nitrocellulose then baked and probed
with a type specific detection oligo (HPV55). The primer family for primer
15 93 is 73 and 91. The optimal RNAse H needed for the reaction using these
two primer pairs is between 1 and 2 units. The primer family 95 is 101 and
91. These primer sets do not appear to be sensitive to different RNAse H
concentrations. A single primer set was defined for primer 92; 92-91,
primer 94; 94-91, and primer 85; 85-77. The primer family for primer 96 is
20 73 and 91. All of these primer sets amplify optimally using between 2 and
3 units of RNAse H. The sensilivil~ of primers 96-73, 96-91, and 94-91 were
tested using a titration of E6-7 isolated from SiHa cells. Once each primer
set has been defined and optimized the sensitivity can be measured by
amplifying decreasing amounts of RNA from control cells (figure 6). The
25 3SR reaction conditions are identical to those described in figure 4 except,
using primers 96-73 the DMSO was included and the sorbitol was omitted,
and using primers 94-91 only 10% sorbitol was included.
Figures 7-10 describe the primers used to amplify HPV 18 E6-
7. The primer family for primer 54 is 32, 69, and 70. Primers 48 and 32 also
30 amplify HeLa RNA. Primers 54-32 and 54-48 both require the addition of
additives 10% polyethylene glycol or DMSO and sorbitol to the 3SR
reaction. Primers 54-69 do not require the addition of additives for
WO 94/26934 . ~ PCT/US94/05085
14
.396~ successful amplification. Additional primer families for primer 214 is 69,
244, 214, and 70 all which require additives to the amplification reaction.
Co-amplification. Once primers have been selected for both
HPV 16 and HPV 18 a co-amplification of both targets is required for
clinical use. Co-amplification is required because only a single specimen
is obtained. This can be done not only for HPV 16 or HPV 18, but also can
be applied to a plurality of HPV types including but not limited to HPV 31,
33, and 35, as well as any other types that prove to be oncogenic. It is not
practical to split a single specimen for two.independent reactions. Figure
11 is a duplicate blot which is probed with a 16 and 18 type specific
detection probe. Lane C demonstrates the cross reactivity of amplifying
two independent targets.
Capture and Detection Probes. Because it is impractical to
incubate the plate in elevated temperatures the detector should produce
maximum signal at room temperature. Many times uneven temperatures
across a microwell can cause differences in hybridization thereby causing
variability of absorbance values. The format of the plate affects the
performance of the assay. Incubating both capture and detector probes
simultaneously rather than capturing the 3SR product first and detecting
in a separate incubation step affects the relative OD values. There are
disadvantages of co-incubation of both capture and detection probes. In
high template concentration, the 3SR reaction produces very high product
concentrations. When the capture is incubated to the target in one step
then applied to the microwell and allowed to bind, excess target is
subsequently washed away. The detection probe is then applied which
only hybridizes to the capture 3SR target.
When designing capture oligonucleotide sequences, defining
the hybridization temperatures is critical to the performance of the assay.
Figures 12 and 13 define the optimum temperature of hybridization for
HPV 16 capture oligonucleotide. The 3SR product is diluted 1:10,000 to
reduce the absorbance levels thereby allowing differences of different
detection probes to become more pronounced. The hybridization reaction
WO 94/26934 213 9 6 2 3 PCTIUS94/0508
~-
..
contain 50 ~ll of the diluted 3SR product in 0.1% PVP, 2X SSC, and 4 pmol
capture oligonucleotide. The reaction was incubated at different
temperatures ranging from room temperature to 70C. The reactionproceeded in the microwell for 20 minutes and the well washed 3 times
with 2X SSC (0.6 M NaCl, 0.06 M Na citrate pH 7.0), 0.05% Tween 20'3', and
0.01% ThimersolTM. The detection probe was added and incubated for 30
minutes at room temperature. The microwell was again washed 3 times
with 2X SSC, 0.5% Tween 20, and 0.01% Thimersol. Substrate for the
horseradish peroxidase enzyme, 3', 3', 5', 5', tetra methyl benzidine and
hydrogen peroxide was added to each well and allowed to develop for 15
minutes at room temperature. The reaction was stopped by ~he addition of
1 M phosphoric acid and read at 450 nm.
The optimum temperature of hybridization for capture 245 is
between 50C and 60C. The signal remains relatively constant at 70C but
thermal degradation of the RNA is a concern at this temperature. Capture
250 hybridization optimum is between 50C and 60C. A variety of
detection probes should be tested because the optimum temperatures for
hybridization of the detection probes must be empirically determined.
Once the capture oligo temperature optimum has been defined, the same
experiments must be repeated using different probes.
Best Mode. Figures 14 and 15 define the detector optimum.
CAP 250 and CAP 245 produced the highest absorbance values when
hybridizing DET 251 at room temperature. The reaction was performed as
described in figure 13. The following is a list of useful detection, capture
probes, and positive hybridization control probes. The detection, capture
and positive hybridization control probes are also listed in the Sequence
Listing as SEQ. ID. Nos. 32-43.
SEO. ID. No. Capture Probes:
32 CAP235: TGT ATT AAC TGT CAA AAG CCA BIOTIN
33 CAP250: TGT ATT AAC TGT CAA AAG CCA AAA AAA
BIOTIN
WO 94n6934 - ` PCT/US94105085 _,-
~396~3 16
34 CAP 253: TGT ATT AAC TGT CAA AAG CCA AAA AAA
AAA A BIOTIN
CAP265: GTA GAG AAA CCC AGC TGT AAA AAA
BIOTIN
36 CAP267~: GTG CCT GCG GTG CCA GAA AAA AAA
BIOTIN
SEO. ID. No. Detection Probes:
37 DET59: GAC AGT ATT GGA ACT TAC AG
38 DET98: TTA GAA TGT GTG TAC TGC AAG NH2
39 DET255: CAA CAG TTA CTG CGA CGT GAG NH2
DET256: TTA CTG CGA CGT GAG GT NH2
41 DET260: GTA TAT TGC AAG ACA GTA NH2
15 SEO. ID. No. Positive Hybridization Control Probes:
42 PHC271: TGT CTT GCA ATA TAC AAA AA BIOTIN
43 PHC272: CTC ACG TCG CAG TAA AAA AAA BIOTIN
Figure 16 is a comparison of all the best performing capture
20 probes using 4 different detection probes. The capture probes were
hybridized to the 3SR product at the temperature optima for 30 minutes in
0.1% PVP, 2X SSC and 8 pmol capture probe. The reaction was applied to
the microwell and allowed to incubate at room temperature for 20
minutes. The microwell was washed 3 times in 2X SSC, 0.05% Tween 20
25 and 0.01% Thimersol. The detection probe was added to the microwell
and hybridized at room temperature for 30 minutes. The well was again
washed 3 times and developed for 15 minutes. The reaction was stopped
and read at A450. The pelLollnance of the capture probes on the plate assay
could be increased by the addition of adenine residues on the end of the
30 oligos closest to the well (data not shown). Different bases were targeted
(G, C, A, and T). T was not chosen because most mRNA's are
polyadenylated which would cause end hybridization. CAP 250 produces
WO 94/26g34 213 9 6 2 3 PCT/US94/05085
17 ~ ~
the highest signal when amplifying SiHa cells; however, CAP 250 only can
capture two of the three spliced E6 RNA's. Several other capture probes
were investigated and CAP 265 captures all three E6 transcripts. Each cell
line splices E6 at different rates. CAP 265 was chosen because clinical
specimens may be heterogenous in splicing E6.
Once the capture probe has been defined, selecting an
enzyme-conjugated detection probe is undertaken. Figure 17 is a
comparison of all the detection probes for HPV 16. DET 256 produces the
highest absorbance values in the present assay. Two detection probes were
synthesized for illustration. The first a 17mer and the second a 15mer to
define the minimum number of bases needed for efficient hybridization.
The minimum length a detector oligo can be is about 17 bases (figure 18).
Please note that best results are achieved when the signal enzyme is
conjugated to the oligonucleotide at the 3' end.
Various additives in the capture buffer were performed with
little increase in the relative absorbance in the plate assay (figure 19).
When these same additives were added to the detection buffer the signal
was more than doubled (figure 20). This effect appears to be related to the
length of the 3SR product. The longer the product the more pronounced
- 20 the effect. Primers 9~91 produce a shorter 3SR product than 13~91 (figure
20). Including additives in the detection buffer increases background
levels. A titration using glycerol reduces background levels. Figure 21 is
an autoradiogram of additional primer set that amplify HPV 18 using
HeLa RNA. Figure 22 demonstrates the performance of HPV 18 capture
probes using a variety of detection probes. Figure 23 demonstrates the
absorbance values of a co-amplification and co-capture of HPV 16 and HPV
18 using type specific detection probes. Best results were achieved in co-
amplification for HPV 16 and HPV 18 simultaneously utilizing primers
136-91 (HPV 16), 54-69 (HPV 18), CAP 265 (HPV 16), CAP 267 (HPV 18), and
DET 256 (HPV 16), DET 60 (HPV 18) as shown in figure 24. The
configuration of this assay is shown in figure 25.
WO 94/26g34 : PCT/US94/05085
18
~,~ 396 The Assay Format. Utilizing the reagents describedhereinabove, the assay format of the present invention was devised to
optimize the signal obtainable from specimens having low viral mRNA
copy number. A fluid phase capture of sample target sequence
5 complementary to a capture prove sequence is much more efficient than
adsorbing directly onto;a~ solid phase. In fact, in a typical sandwich
configuration, it is not uncommon to capture only 1-3% of total available
nucleic acid in the sample. This reduces sensitivity correspondingly by
two orders of magnitude.
Since it is still rlecess~ry to separate nucleic acid complexes on
a solid phase, the "capture" sample must be immobilized onto the solid
phase before the detection probe is added. The present assay takes
advantage of the extremely high binding constant for the interaction
between biotin and streptavidin. The capture oligonucleotide is
15 biotinylated through 3' or 5' terminal labeling by conventional techniques.
It has been empirically determined for the probes studied to date that
biotinylating the capture probe at the 3' terminus is more efficient in
immobilizing the probe hybridized to sample target sequence.
The solid phase is coated with streptavidin, so that when the
20 hybri~i7e.1 capture-sample sequence complex is brought into contact with
it, the reaction between slre~lavidin and biotin takes place. The solid
phase is preferably the inner surface of microtiter tray wells, but any solid
phase separation system known to the art is satisfactory including but not
limited to poly~lyl~lle beads, magnetic microparticles, test strips of plastic
25 or metal, dipsticks, columns packed with a variety of materials, etc. The
fluid phase capture method of the present invention is expected to give
enhanced results with solid supports made of plastic because of the
especially low capture efficiencies with plastic supports in conventional
assays.
Any signal-generating enzyme or other reporter or tracer
~ysLell~ capable of being conjugated covalently or electrostatically to a
oligonucleotide without hindering its hybridizing to a complementary
WO 94/26g34 213 9 6 2 3 PCTIUS94/05085
19 ` 'f
sequence is contemplated in the present assay. Horseradish peroxidase^is
preferred, but alkaline phosphatase and synthetic fluorogenic and
- chromogenic molecule hydrolyzing enzymes may also be employed. Non-
isotopic reporter/tracer ~ystems are preferred over radioactive tracers
5 because of environmental and stability considerations.
The kinetics of hybridization of various capture and detection
probes will differ according to their thermodynamic characteristics, and
some relatively insignificant amount of experimentation may be required
to optimize the assay for probes of similar but not identical sequence
10 disclosed herein for illustrative purposes.
Alternative Amplification Reaction Conditions
Figure 26 compares amplification reactions performed using
the standard 3SR reaction conditions (42C) with amplification reactions
15 performed at an elevated temperature (50C). The assays used the primer
sets 136-91 (HPV 16) and 54-69 (HPV 18) together and separately. The
standard 3SR reaction conditions were 40 mM Tris-HCl, pH 8.1; 30 mM
MgCl2; 20 mM KCl; 10 mM dithiothreitol; 4 mM spermidine; 15 pmole
each priming oligonucleotide; 1 mM dNTP's; 7 mM rNTP's; 30 units AMV
20 reverse transcriptase; 2 units RNAse H; and 1000 units T7 RNA
polymerase. The reaction was incubated for 1 hour at 42C. The elevated
temperature reaction conditions were 40 mM Tris acetate, pH 8.1; 30 mM
Mg acetate; 10 mM dithiothreitol; 100 mM potassium glutamate, pH 8.1; 1
mM dNTP's; 6 mM rNTP's; 15% sorbitol; 30 units AMV reverse
25 transcriptase; 2 units RNAse H; and 1000 units T7 RNA polymerase. The
reaction was incubated for 1 hour at 50C.
After incubating the amplification reactions, l/lOth of the
amplification products were denatured in 90 ~11 of 7.4% formaldehyde and
lOX SSC in a 65C water bath for 10 minutes and quick-chilled on ice for at
30 least 1 minute. BA-85 nitrocellulose was pre-wetted with water and then
with lOX SSC. The denatured amplification samples were applied to a slot
blot apparatus containing the pre-wetted nitrocellulose and the samples
Wo 94l26934 PCTJUS94/0508~ -
2~39623 20
were drawn onto the nitrocellulose using a vacuum. The filter was then
baked for 45 minutes at 80C a~nd hybridized with a type-specific
oligonucleotide specific for HPV;18 (DET59) or HPV 16 (DET98). The
hybridization solution cor~tains 6X SSC; 10X Denhardts; 10 mM Tris, pH
7.4; 0.2 mg/ml sheared salmon sperm DNA; and 1% SDS.
Figures 26 and 27 depict a comparison of the amplification
yields of reactions performed at 50C and at 42C. In both figures, the
amplification reactions in column 1 used the HPV 16 primers 136-91, the
reactions in column 2 used the HPV 18 primers 54-69, and the reactions in
column 3 used a combination of the HPV 16 and HPV 18 primers 136-91
and 54-69. The target sequence was a mixture of 5 amol each of SiHa cell
(infected with HPV 16) and HeLa cell (infected with HPV 18) RNA. Rows
1-4 contained sorbitol concentrations of 15%, 10%, 5% and 0% respectively;
row 5 was a minus template reaction using 15% sorbitol; row 6 was blank;
and rows 7-11 contained sorbitol concentrations of 15%, 10%, 5% and 0%
respectively. Rows 1-5 were incubated at 50C and rows 7-11 were
incubated at 42C. The amplification products in figure 26 were probed
with DET 98 which is specific for HPV 16. The amplification products in
figure 27 were probed with DET 59 which is specific for HPV 18.
Figure 26 depicts that the bands were much stronger at the
15% and 15% sorbitol levels than at the 5% or 0% levels. These results
demonstrate that the increased sorbitol concentrations protect the enzymes
so that the reaction can be incubated at 50C rather than 42C. When the
sorbitol concentration was dropped below 10% the enzymes were not
thermally protected and denatured at elevated temperatures, resulting in
the decreased level of amplification. Figures 26 and 27 demonstrate that
the elevated temperature increased the level of amplification when
compared to the 42C reaction conditions. This was particularly evident
when the target sequence was co-amplified using the mixed primer set,
136-91 (HPV 16) and 5~69 (HPV 18). The estimated level of amplification
using the elevated temperature was 10 fold higher than the level of
arnplification using the 42C reaction conditions.
2139623
`_ WO 94/26934 PCT/US94/05085
21
The foregoing detailed description has been provided for a
better understanding of the invention only and no unnecessary limitation
should be understood thereLloln as some modifications will be apparent to
those skilled in the art without deviating from the spirit and scope of the
5 appended claims.
WOg4/~934 ; PCT~S94/05085
a,,396~3 22
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: Jani~ T. Brown
(ii) TITLE OF INVENTION'HUMAN PAPILLOMAVIRUS DETECTION
ASSAY
(iii) NUMBER OF SEQUENCES:44
(iv) CORRESPONDENCE ADDRESS
(A) ADDRESSEE: Baxter Diagnostics Inc.
(B) STREET: One Baxter Parkway, Building DP-3E
(C) CITY: Deerfield
(D) STATE: Illinois
(E) COUNTRY: USA
(F) ZIP: 60015
(v)COMPUTER READABLE FORM
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: Apple Macintosh
(C) OPERATING SYSTEM: Apple Macintosh System 7.0
(D) SOFTWARE: Macintosh Text File
(vi)CURRENT APPLICATION DATA
(A) APPLICATION NUMBER: N/A
(B) FILING DATE: N/A
(C) CLASSIFICATION: N/A
(vii)PRIOR APPLICATION DATA
(A) APPLICATION NUMBER: US 08/058,920
(B) FILING DATE: May 6, 1993
(viii)ATTORNEY/AGENT INFORMATION
(A) NAME: Mark Buonaiuto
(B) REGISTRATION NUMBER: 31,593
(C) REFERENCE/DOCKET NUMBER: BA-4448
Og4/26934 213 9 6 2 3 PCT~S94/05085
(ix)TELECOMMUNICATION INFORMATION
(A) TELEPHONE: 708/948-2537
(B) TELEFAX: 708/948-2642
W094/~934 PCT~S94/05085
2 ~3 9 6 2 3 24
- (2) INFORMATION FOR SEQ ID NO: 1
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 570
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Papaoviridae, Human papilloma
virus
(B) STRAIN: 16
(ix) FEATURE:
(A) NAME/KEY: Portion of viral genome coding for
E6/E7 polypeptides.
(x) PUBLICATION INFORMATION:
(A) AUTHORS: Seedorf, K., Krammer, G., Durst,
M.,
Suhai, S., and Rowekamp, W.
(B) TITLE: Human Papillomavirus Type 16 DNA
Sequence
(C) JOURNAL: Virology
(D) VOLUME: 145
(E) ISSUE:
(F) PAGES: 181-185
(G) DATE: 1985
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:
T ATG CAC CAA AAG AGA ACT GCA ATG TTT CAG GAC CCA CAG GAG
CGA 46
Met His Gln Lys Arg Thr Ala Met Phe Gln Asp Pro Gln Glu
Arg
WO 94/26934 213 9 6 2 3 PCT/US94/05085
CCC AGA AAG TTA CCA CAG TTA TGC ACA GAG CTG CAA ACA ACT
ATA 91
Pro Arg Lys Leu Pro Gln Leu Cys Thr Glu Leu Gln Thr Thr
Ile
20 25
CAT GAT ATA ATA TTA GAA TGT GTG TAC TGC AAG CAA CAG TTA
CTG 136
His Asp Ile Ile Leu Glu Cys Val Tyr Cys Lys Gln Gln Leu
Leu
35 40
CGA CGT GAG GTA TAT GAC TTT GCT TTT CGG GAT TTA TGC ATA
GTA 181
Arg Arg Glu Val Tyr Asp Phe Ala Phe Arg Asp Leu Cys Ile
Val
50 55
TAT AGA GAT GGG AAT CCA TAT GCT GTA TGT GAT AAA TGT TTA
AAG 226
Tyr Arg Asp Gly Asn Pro Tyr Ala Val Cys Asp Lys Cys Leu
Lys
65 70
TTT TAT TCT AAA ATT AGT GAG TAT AGA CAT TAT TGT TAT AGT
TTG 271
Phe Tyr Ser Lys Ile Ser Glu Tyr Arg His Tyr Cys Tyr Ser
Leu
80 85
TAT GGA ACA ACA TTA GAA CAG CAA TAC AAC AAA CCG TTG TGT
GAT 316
Tyr Gly Thr Thr Leu Glu Gln Gln Tyr Asn Lys Pro Leu Cys
Asp
95 100
105
TTG TTA ATT AGG TGT ATT AAC TGT CAA AAG CCA CTG TGT CCT
GAA 361
Leu Leu Ile Arg Cys Ile Asn Cys Gln Lys Pro Leu Cys Pro
Glu
110 115
120
GAA AAG CAA AGA CAT CTG GAC AAA AAG CAA AGA TTC CAT AAT
ATA 406
Glu Lys Gln Arg His Leu Asp Lys Lys Gln Arg Phe His Asn
Ile
125 130
135
W094/26934 PCT~S94/05085
2 ~ 3 9 6~ 3 26
AGG GGT CGG TGG ACC GGT CGA TGT ATG TCT TGT TGC AGA TCA
TCA 451
Arg Gly Arg Trp Thr Gly Arg Cys Met Ser Cys Cys Arg Ser
Ser
140 145
150
AGA ACA CGT AGA GAA ACC CAG CTG TAATC ATG CAT GGA GAT ACA
495
Arg Thr Arg Arg Glu Thr Gln Leu Met His Gly Asp Thr
155 5
CCT ACA TTG CAT GAA TAT ATG TTA GAT TTG CAA CCA GAG ACA
ACT 540
Pro Thr Leu His Glu Tyr Met Leu Asp Leu Gln Pro Glu Thr
Thr
GAT CTC TAC TGT TAT GAG CAA TTA AAT GAC
570
Asp Leu Tyr Cys Tyr Glu Gln Leu Asn Asp
(2) INFORMATION FOR SEQ ID NO: 2
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH:483
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Papovaviridae, Human papilloma
45 virus
(B) STRAIN: 18
(viii) POSITION IN GENOME
(A) CHROMOSOME/SEGMENT
(ix) FEATURE:
(A) NAME/KEY: Portion of viral genome coding for
E6/E7 polypeptides.
_ W094/~934 213 9 6 2 3 PCT~S94/05085
. : , : ''~ ,~. .
27
(x) PUBLICATION INFORMATION:
(A) AUTHORS: Cole, S., and Danos, O.
(B) TITLE: Nucleotide Sequence and
Comparative
Analysis of the Human
Papillomavirus
Type 18 Genome.
(C) JOURNAL: Journal of Molecular Biology
(D) VOLUME: 193
(E) ISSUE:
(F) PAGES: 599-608
(G) DATE: 1987
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2
ATG GCG CGC TTT GAG GAT CCA ACA CGG CGA CCC TAC AAG CTA CCT
Met Ala Arg Phe Glu Asp Pro Thr Arg Arg Pro Tyr Lys Leu Pro
GAT CTG TGC ACG GAA CTG AAC ACT TCA CTG CAA GAC ATA GAA ATA
Asp Leu Cys Thr Glu Leu Asn Thr Ser Leu Gln Asp Ile Glu Ile
ACC TGT GTA TAT TGC AAG ACA GTA TTG GAA CTT ACA GAG GTA TTT
135
Thr Cys Val Tyr Cys Lys Thr Val Leu Glu Leu Thr Glu Val Phe
GAA TTT GCA TTT AAA GAT TTA TTT GTG GTG TAT AGA GAC AGT ATA
180
Glu Phe Ala Phe Lys Asp Leu Phe Val Val Tyr Arg Asp Ser Ile
CCG CAT GCT GCA TGC CAT AAA TGT ATA GAT TTT TAT TCT AGA ATT
225
Pro His Ala Ala Cys His Lys Cys Ile Asp Phe Tyr Ser Arg Ile
AGA GAA TTA AGA CAT TAT TCA GAC TCT GTG TAT GGA GAC ACA TTG
270
Arg Glu Leu Arg His Tyr Ser Asp Ser Val Tyr Gly Asp Thr Leu
GAA AAA CTA ACT AAC ACT GGG TTA TAC AAT TTA TTA ATA AGG TGC
315
Glu Lys Leu Thr Asn Thr Gly Leu Tyr Asn Leu Leu Ile Arg Cys
~ r
W094/26934 PCT~S94/05085
?,~39623 28
100 105
CTG CGG TGC CAG AAA CCG TTG AAT CCA GCA GAA AAA CTT AGA CAC
360
Leu Arg Cys Gln Lys Pro Leu Asn Pro Ala Glu Lys Leu Arg His
110 115 120
CTT AAT GAA AAA CGA CGA TTT CAC AAC ATA GCT GGG CAC TAT AGA
405
Leu Asn Glu Lys Arg Arg Phe His Asn Ile Ala Gly His Tyr Arg
125 130 135
GGC CAG TGC CAT TCG TGC TGC AAC CGA GCA CGA CAG GAA CGA CTC
450
Gly Gln Cys His Ser Cys Cys Asn Arg Ala Arg Gln Glu Arg Leu
140 145 150
CAA CGA CGC AGA GAA ACA CAA GTA TAATATTAA
483 Gln Arg Arg Arg Glu Thr Gln Val
155
(2) INFORMATION FOR SEQ ID NO: 3
25 (i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 49
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV15. Phage T7 RNA polymerase
binding site at 5'end, followed by HPV-16/18 sequence.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3
AATTTAATAC GACTCACTAT AGGGAGCTTT TCTTCAGGAC ACAGTGGCT
49
~W094/~934 213 9 6 2 3 PCT~S94/0508~
29
(2) INFORMATION FOR SEQ ID NO: 4
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 23
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV19.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 4
AATGTTTCAG GACCCACAGG AGC 23
(2) INFORMATION FOR SEQ ID NO: 5
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 24
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
W094/269~4 PCT~S94/05085
2~39623
(A) NAME/KEY: HPV20.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5
5 GAATGTGTGT ACTGCAAGCA ACAG 24
(2) INFORMATION FOR SEQ ID NO:6
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 23
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(ix) FEATURE:
(A) NAME/KEY: HPV29.
_ W094/~g34 21 3 9 6 2 3 PCT~S94/05085
31
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:6
ATGCACAGAG CTGCAAACAA CTA 23
(2) INFORMATION FOR SEQ ID NO:7
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 22
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV32.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:7
CACTTCACTG CAAGACATAG AA 22
(2) INFORMATION FOR SEQ ID NO:8
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 46
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
W094/26934 PCT~S94105085 _-
?,~396~3 32
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE: .~ ~
(A) NAME/KEY: HPV48.
~xi) SEQUENCE DESCRIPTION: SEQ ID NO:8
AATTTAATAG CACTCACTAT AGGGATGTGT CTCCATACAC AGAGTC
46
(2) INFORMATION FOR SEQ ID NO:9
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 25
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV53.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:9
GAATGTGTGT ACTGCCAAGC AACAG 25
(2) INFORMATION FOR SEQ ID NO:l0
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 49
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
W094/~934 213 9 6 2 3PCT~S94/05085
33 `'t ~ ' ~
,
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV54.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:10
AATTTAATAC GACTCACTAT AGGGAAAGGT GTCTAAGTTT TTCTGCTGG
49
(2) INFORMATION FOR SEQ ID NO:11
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 21
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV69.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:11
CTGAACACTT CACTGCAAGA C 21
(2) INFORMATION FOR SEQ ID NO:12
WOg4/~934 PCT~S94/05085
2 ~3 9 6~ ~ 34
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 23
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV73.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:12
CAGTTATGCA CAGAGCTGCA AAC 23
(2) INFORMATION FOR SEQ ID NO:13
(i) SEQUENCE CHARACTERISTICS
(A) . LENGTH: 23
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV74.
_ W094/26934 213 96 23 PCT~S94/05085
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:13
GTTATGCACA GAGCTGCAAA CAA 23
(2) INFORMATION FOR SEQ ID NO:14
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 20
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV77.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:14
CAAGCAACAG TTACTGCGAC 20
(2) INFORMATION FOR SEQ ID NO:15
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 20
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
W094t26934 PCT~S94/05085
2 ~3 9 6 ~ 3 36
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV89.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:15
10 AGCAACAGTT ACTGCGACGT 20
(2) INFORMATION FOR SEQ ID NO:16
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 23
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV90.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:16
GCACAGAGCT GCAAACAACT ATA 23
(2) INFORMATION FOR SEQ ID NO:17
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 23
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
213g623
_W094/~934 . ~ PCT~S94/05085
37
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV91.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 17
ACAGAGCTGC AAACAACTAT ACA 23
(2) INFORMATION FOR SEQ ID NO:18
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 51
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV92.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:18
AATTTAATAC GACTCACTAT AGGGACTTTT CTTCAGGACA CAGTGGCTTT T
(2) INFORMATION FOR SEQ ID NO:19
(i) SEQUENCE CHARACTERISTICS
W094/26934 PCT~S94/05085
2 ~ 3 9 6~ 3 38
(A) LENGTH: 50
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV93.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:19
AATTTAATAC GACTCACTAT AGGGATTTGC TTTTCTTCAG GACACAGTGG
(2) INFORMATION FOR SEQ ID NO:20
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 50
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV94.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:20
- W094/~g34 213962 3 PCT~S94/05085
39
AATTTAATAC GACTCACTAT AGGGATCTTT GCTTTTCTTC AGGACACAGT
(2) INFORMATION FOR SEQ ID NO:21
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 50
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV95.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:21
AATTTAATAC GACTCACTAT AGGGATGTCT TTGCTTTTCT TCAGGACACA
(2) INFORMATION FOR SEQ ID NO:22
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 50
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
W094~6934 PCT~S94/05085
2 ~3 9 6~ 3 40
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV96.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:22
AATTTAATAC GACTCACTAT AGGGAGATGT CTTTGCTTTT CTTCAGGACA
(2) INFORMATION FOR SEQ ID NO:23
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 23
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV101.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:23
AGAGCTGCAA ACAACTATAC ATG 23
(2) INFORMATION FOR SEQ ID NO:24
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 49
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
213962~
~ W094/26934 PCT~S94/05085
41
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV106.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:24
AATTTAATAC GACTCACTAT AGGGATTCAT GCAATGTAGG TGTATCTCC
49
(2) INFORMATION FOR SEQ ID NO:25
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 49
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV107.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:25
AATTTAATAC GACTCACTAT AGGGATATTC ATGCAATGTA GGTGTATCT
49
(2) INFORMATION FOR SEQ ID NO:26
W094t~934 PCT~S94/05085
42
~396~3
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 23
~;~
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV118.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:26
AGCTGCAAAC AACTATACAT GAT 23
(2) INFORMATION FOR SEQ ID NO:27
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 49
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
213g623
~- W094/~g34 PCT~S94/05085
43
(A) NAME/KEY: HPV120. Phage T7 RNA polymerase
binding site at 5'end, followed by HPV-16/18 sequence.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:27
AATTTAATAC GACTCACTAT AGGGATGCAA TGTAGGTGTA TCTCCATGC
49
(2) INFORMATION FOR SEQ ID NO:28
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 48
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
30(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV129.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:28
AATTTAATAC GACTCACTAT AGGGAAATGT AGGTGTATCT GGATGCAT 48
(2) INFORMATION FOR SEQ ID NO: 29
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 23
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
~ D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
W094l26934 PCT~S94/05085
44
2~39623
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV131.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 29
AAACAACTAT ACATGATATA ATA 23
(2) INFORMATION FOR SEQ ID NO:30
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 49
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV136. Phage T7 RNA polymerase
binding site at 5'end, followed by HPV-16/18 sequence.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:30
AATTTAATAC GACTCACTAT AGGGAATGTA GGTGTATCTC CATGCATGA
49
(2) INFORMATION FOR SEQ ID NO:31
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 49
2139623
~ WOg4/26934 PCT~S94/05085
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: HPV137.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:31
AATTTAATAC GACTCACTAT AGGGATGTAG GTGTATCTCC ATGCATGAT
49
(2) INFORMATION FOR SEQ ID NO:32
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 21
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
4S (iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: CAP245.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:32
55 TGTATTAACT GTCAAAAGCC A 21
W094/26934 ` PCT~S94/05085
~3 9 62 3 46
(2) INFORMATION FOR SEQ ID NO:33
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 27
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: CAP250.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:33
TGTATTAACT GTCAAAAGCC AAAAAAA 27
(2) INFORMATION FOR SEQ ID NO:34
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 31
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
~_wog4/~934 21~ 9 6 2 3 PCT~S94/05085
47
(ix) FEATURE:
(A) NAME/KEY: CAP253.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:34
TGTATTAACT GTCAAAAGCC AAAAAAAAAA A 31
(2) INFORMATION FOR SEQ ID NO:35
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 24
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: CAP265.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:35
GTAGAGAAAC CCAGCTGTAA AAAA 24
(2) INFORMATION FOR SEQ ID NO:36
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 24
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
W094/26934 ~CT~S94/05085
48
2~396~3
- (iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: CAP267.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:36
GTGCCTGCGG TGCCAGAAAA AAAA 24
(2) INFORMATION FOR SEQ ID NO:37
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 20
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: DET59.
~ W094/26g34 213 9 6 2 3 PCT~S94/05085
49 - .
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:37
GACAGTATTG GAACTTACAG 20
(2) INFORMATION FOR SEQ ID NO:38
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 21
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
20 DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: DET98.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:38
TTAGAATGTG TGTACTGCAA G 21
(2) INFORMATION FOR SEQ ID NO:39
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 21
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
W094/~934 PCT~S94/05085
2 ~39623 50
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: DET255.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:39
CAACAGTTAC TGCGACGTGA G 21
(2) INFORMATION FOR SEQ ID NO:40
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 17
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME~KEY: DET 256.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:40
TTACTGCGAC GTGAGGT 17
(2) INFORMATION FOR SEQ ID NO:41
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 18
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
2139623
_W094/26934 PCT~S94/0508
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: DET260.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:41
GTATATTGCA AGACAGTA 18
(2) INFORMATION FOR SEQ ID NO:42
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 20
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: PHC271.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:42
TGTCTTGCAA TATACAAAAA 20
(2) INFORMATION FOR SEQ ID NO:43
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 21
W094/26g34 . PCT~S94/05085
2 ~3 9 6 ~ 3 52
tB) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE:
(A) DESCRIPTION: Other nucleic acid, synthetic
10 DNA
(iii) HYPOTHETICAL: no
(iv) ANTI-SENSE: no
(vii) IMMEDIATE SOURCE:
(A) LIBRARY: DNA synthesizer
(ix) FEATURE:
(A) NAME/KEY: PHC272.
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:43
CTCACGTCGC AGTAAAAAAA A 21
(2) INFORMATION FOR SEQ ID NO:44
(i) SEQUENCE CHARACTERISTICS
(A) LENGTH: 25
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:44
AATTTAATAC GACTCACTAT AGGGA 25