Note: Descriptions are shown in the official language in which they were submitted.
2198209
'`
METHOD AND APPARATUS FOR CELL COUNTiNG
AND CELL CLASSIFICATION
~ ;l
.,
I llMlTFn COPYRIGHT WAIVFR
A portion of the d;sclosllre of this patent document contains
material to which the claim of copyright protection is made. The
copyright owner has no objection to the facsimile reproduction by any
person of the patent document or the patent ~isclos~ ~re, as it appears in
the U.S. Patent and Trademark Office file or records, but reserves atl
other rights whatsoever.
CONTINUING APPl ICATION QATA
15
The present application is a continuation-in-part of Serial Number
08/018,762 filed February 17, 1993 en~itled "Method and Apparatus for
Volumetric Capillary Cytometry", assigned to the same assignee of the
present application.
CROSS-RFFFRFNCF TO RELATFn APPLI(`~
The present application is related to U.S. patent applicalion
entitled "Method and Apparatus for Volumetric Capiliary Cytometry",
invented by Baer et al, filed on the same day as the present application
and owned by the same Assignee. The related application is
incorporated by reference as if fully set forth herein.
AKornoy Dock-t No.: E' "~ ' ~H
mah/blT il2X2.X2
- 2148204
`_
BACKGROUND OF THF INVFI~ITION
Field of the Invention
The present invention relates generally to the processing of
volumetric capilla, y cytometry data for the purpose of counting and
characterizing cells or cell constituents in a volume of material.
Desc, ;~tio~ of Related Art
The optical analysis of biological specimens, such as blood, has
widespread applicalions. There are many computer controlled
instruments in the market for providing such analysis, including flow
cytometers, automated blood cell analyzers and blood cell classifiers.
As described in the above cross-referenced application, a
volumetric cytometry instrument has many advantages. In particular,
the amount of blood being analyzed is controllable, the handling of the
blood is reduced, and analyzed samples of blood can be stored for
further processing.
However, the processing of blood samples in a volumetric system
raises a number of problems. Particularly if counting of blood cells is
required, it is necess~ry to analyze the entire volume in the capillary or
other container which holds the material to be analyzed. Blood cells on
the side of a container may be difficult to detect, if the ar!alysis
instrument is not calibrated for the precise dimensions of the container.
For instance, in Kamentsky, U.S. Patent No. 5,072,382, samples of
blood were a, F l ~ to a slide. A region to be analyzed was defined by
sy"ci~ror,ization pulses in the scanr.ing apparalus. (See column 14,
lines 49~3 of Kamentsky). Synchronization pulses in the scanning
mechanism cannot be precisely aligned with a container such as a
capillary or cuvette for a blood sample because of the variations in the
shapes of such con~;ners, and va, ia~ions in the alignment of mounts for
Anomey Oock-~ No.: B~ H
mah/bmi/2002.002
- 21g8204
the containers in the scanning mechanism. It will be appreciated that
the ability to precisely mount and manufacture containers is quite
advanced. However, the scanning of containers for the,purposes of
processing cells may require resolution on the micron scale.
An additional difficulty arises because of the charac~erislics of
dyes used to mark target cells. For instance, when analyzing cells for
the presence of specific antibodies, it is common to tag the cells with
dyes which fluoresce with a particular spectrum in response to an
excitation beam. If more than one antibody is to be detected, more than
one dye is used. However, the fluorescence spectrums of various dyes
may overlap. Thus, it is difficult to fully process the informatioll in
detected fluorescence generated by plural dyes with overlapping
spectra.
Furthermore, when it is necessary to count a particular number of
target cells within a volume, to achieve a slatistically valid count, a
relatively large sample must be used. A large sample of blood, when it
is scanned on the micron scale, can generate very large amounts of
data. It is important for practical analysis machines that the data be
processed in a reasonable amount of time. For instance, as described
in the above cross-referenced application, the sample can scan for the
presence of two dyes with overlapping spectra, with two,channels of
data. Each channel of data includes information relevant to both dyes.
Further, the scan involves about 10,000 lines of 200 pixels each,
resulting in 2 million samples per channel, which for 2 b~tes per sample
in 2 channels amounts to a total of 8 megabytes of raw data.
Furthermore, the fluorescence monitoring techniques are
susceptible to a low signal-to-noise ratio. Thus, it is important to be able
to process these large amounts of data with high bacl~ground noise to
Atlorn-y Dockot No.: e~ H
mah/bmi/2002.002
214820~
accurately characteri a and identify target cells within the volume,
particularly when unbound antil)~y is present.
Accordingly, it is desirable to provide a method a~nd apparatus for
processing data from a volumetric cytometry system which is robust and
accurate. Further, the system should be relatively fast and operate in a
system having a relatively low memory requirement.
SUMMARY OF THF INVFI~ITION
The present invention provides a method and an apparatus for
analyzing a sample of cells or cell constituents, including but not limited
to blood, within a capillary in a volumetric cytometry system. According
to the invention, the sample cells or cell constituents have one or more
detec~ characterislics. The system provides for detecting the edges
of the container, counting the cells or cell constituents within the
container, charac~eri~ing the cells or cell constituents within the
container, and evaluating channels of data which contain information
relevant to more than one of the detect~hle characteristics of the cells or
cell constituents.
Accordingly, the present invention can be characterized as an
apparatus which comprises a scanner for scanning a container of
sample including target cells or cell constituents. Data sampling circuitry
is coupled to the scanner to genera~e scanned images of the sample in
the container. According to one aspect, the scanner and data sampling
circuitry produce a plurality of channels of data, and a corresponding
plurality of scanned images. A processing system is coupled to the
sampling circuitry, and includes resources to count and/or charac~eri~e
the target cells or cell constituents in response to the scanned images.
These processing resources, according to one aspect, are
capable of processing the scanned image, which includes information
At~om y Oock~t No.: BM12002MAH
mahhmU2002.002
2148201
,
relevant to more than one of the dete.,~hlo characleris~ics of the cells or
cell constituents, to distinguish such detectable cl~ara.,terislics. Such
resources may include software for ~e, ror"~ing a correlation analysis
between the scanned image having information relevant to more than
one chara~terislic and anoll ,er scanned image in the plurality of
scanned images. In one prefer,ed system, two scanned images are
generated based on fluorescel~ce data from dyes that have overlapping
spectra. The two scanned images are processed using a linear
regression analysis among corlesponding pixels in the scanned images
near target cells to characterize relative cGntents of two fluorescing dyes
in a target cell or cell constituents.
AccGr~ing to another aspect of the present invention, target cells
or cell constituents are identified from the scanned images using
processing resources which identify a peak pixel within a neighborhood,
and compare the amplitude of the peak with the amplitude of pixels on
the perimeter of the neigl,borhood. If the peak pixel value exceeds the
perimeter pixel values by more than a predetermined threshold, then the
resources characterize the neighborhood as containing a target cell.
Upon identifying a target cell in this manner, segments of data from the
plurality of scanned images corresponding to the identified cell can be
saved for further analysis, such as the linear regression analysis
discussed above.
f urther, according to another aspect of the present invention, in
addition to determining a relative con~, ibution from more than one dye in
a scanned image, a parameter indica~ing the in~ensity of the
fluorescence of a target cell or cell constituents is determined by filtering
the identified segments of data from the plurality of scanned images
based upon the expe~ted cl)aracteristics of target cells or cell
constituents. For example, in one novel species of the invention, the
Anomey Oocket No.: E'"~'~H
mal ~bmU2002 002
214820~
segmer,ts are fiKered by defining a neiyhbo, hood of pixels for each
identified segment in the scanned images wherein the neigl ,bo, hood is
larger than the e~p6~ted size of the target cell. The pixels within the
neighbGrl,ood are processed to compensate for background noise and
generate an intensity value for the target cell within the neighborhood
based solely on pixel values within the nei!Jhborl ,ood. For instance this
processing may involve a matched filter multiplying the intensity values
of the neiyhborhood of a cell by a set of values which reflects the
expected i"tensi~y profile of a typical cell. The resulting products are
summed to yield an amplitude estimate which optimizes signal to noise
ratio for that cell. The perimeter of the neighborl,ood is determined
based on the expected shape of the target cells or cell constituents
within the nei~ orhood.
According to yet another aspect of the present invention the
processing resources pei~o"n edge detection and ignore contributions
to the scanned images which fall outside of the deteàed edges of the
container.
In the prefel,e~ system the cells or cell constituents are
characterized using a slope value deterrrlined from the linear regression
analysis over a neiyhbo, i ,ood defined for a target cell between two
scanned images of the cell generated from overlapping fluorescence
spectra of two dyes. Using the linear regression analysis a "slope
value" is determined for each target cell. This slope value is then
multiplied by the intensity value of the neighborhood in one of the
scanned images to produce an analysis coordinate. The cell or cell
constituents is characteri~ed based upon the position of the analysis
coordinate on a chara~:teri~dlion graph. The characte, i~alion graph is
defined with a first region within which target cells or cell constituents
having one dye should fall a second region within which target cells or
Attorney Dockd No.~ H
mah~i~7 n~
21~8204
cell constituents having the second dye should fall, and a third region
within which target cells or cell constituents stained with both dyes
s~ould fall. The regions are defi- ,ed based on the background signal
cl ,aract6ris~ics of the scanned images for the purposes of signal
immunity and more accurate characteri~ations.
The present invention can also be characl~ri~6d as a method for
analyzing the sample within such a container. The method includes the
following:
scanning the material with a detector to generate a plurality of
channels of data, in which at least one of the channels may contain
inforrrlation relevant to more than one of a plurality of detectable
characteristics of the target cells or cell constituents;
sampling the plurality of channels of data to produce a plurality of
scanned images of the sample; and
15 analyzing the plurality of scanned images to characterize the
target cells or cell constituents in response to the plurality of channels of
data, including processing the scanned image corresponding to the one
channel which includes information relevant to more than one
characteristic to distinguish such detectable characleristics, processing
at least one of the plurality of scanned images to identify segments in
the plurality of scanned images con~aWng target cells oç cell
constituents, filtering identified segments of data based upon expected
cllar~c~erislies of target cells or cell constituents to generate respective
intensity values for the iden~iried segments, and characterizing the
target cells or cell constituents based on the intensity value in at least
one of the plurality of scanned images for a particular segment, and a
value based on correlation analysis (such as the slope in a linear
regression analysis) between two scanned images of a segment of data.
Attom~y Dodt-t No.: Q'"~'~H
,.~h~ ~ ~
214820~ 1
`._
The system may also mclude analyzing at least one of the
scanned images to detect the edges of the container and ignoring data
found outside of the det~ d edges. Furthermore, th~ process of
characleri~ing the in~e"sit~ value for a particular segment of data may
include defining a nei!JhLo, hood of pixels for each identified segment
the neighborhood being larger than the ~Ypected size of the target cell
and processing the pixels within the neighbo, hood to compensate for
background signal and generale an intensit~ value for the target cell or
cell constituents within the neighborhood.
Other aspects and advantages of the present invention can be
seen upon review of the figures, the detailed desc, i,l~tion and the claims
which follow.
BRIFF IlFSCRlPTlON OF THF FIGUF?FS
Fig. 1 is a schematic block d;agra,., of a scanner apparatus with a
data processing system according to the present invention.
Fig. 2 is a schematic block diagra,., of the data processing
system used in combinalion with the system of Fig. 1.
Fig. 3 is a schematic diayrar~, illuslrating the scanning process
used in the scanner of Fig. 1.
Fig. 4 schematically illustrates an organization of data in the
scanned images generated by sampling the output of the scanner of
Fig. 1.
Fig. 5 is a plot of a representative scanned image from the
system according to Fig. 1.
Figs. 6A and 6B together make up a flow chart for the basic data
processing loop for the system according to the p,esent invention.
Fig. 7 illust. ates the overlapping spectra of two dyes which are
analyzed according to the present invention.
Attome~ odcd No.: F'~
mah/bmUX~002
- 2148204
-
Fig. 8 is a graph illust~aling the linear regression analysis used in
the characte, i alion of cells according to the pres~nt invention.
Fig. 9 illustrates a cell classilioalio" graph used for classifying
cells based on chann~ls of data which include ove, lapFing information,
S accordi"g to the presenl invention.
Fig. 10 is a flow chart illuslfaling the generation of background
noise indices according ta the p~5~3nt invention.
Fig. 11 is a flow chart illuslr~ling the edge detection process
according to the present invention.
Fig. 12 is a flow chart of the process used for detecting cells
according to the present invention.
Fig. 1~ IS a flow chart illustrating the ~.rocess for linear regression
analysis accord;,)g to the present invention.
DFTAII Fn DESCRIPTION
A detailed description of prefer,ed embodiments of the present
invention is provided with respec~ to the figures. Figs. 1 and 2 illustrate
a hardware environment for the present invention. Figs. 3-13 provide
an explanation of the processing resources used to count and
characterize target cells or cell constituents according to the present
invention.
As can be seen in Fig.1, an apparatus for volumetric capillary
cytometry is provided. The machine is designed to process material
within a capilla~y 10, which has a known volume. In the prefer,ed
system, the capillary may be rectangular in cross-section, having a
width of about 0.4 millimeters to 1.5 millimeters, a length of about 40
millimeters, and a depth of about 25 to 225 microns, and in one
embodiment, 100 microns. This capillary is suitable for detecting or
characterizing a variety of cells or cell constituents. In one embodiment.
Attomey Dockd No.: BM1200~MH
" ,dl Ll ~ ~
219820~
.._
it is used for the eharacte~ i~ation of a CD3/CD4 assay, in which
concent~tion of CD3 and CD4 anti~edies are to be determined. In this
type of assay, there are typically three populations present. CD3
positive cells stained with a CyS l~hs"~ antibody only; CD4 cells
S stained with both a Cy5 l~h~ d CD3 antibody and a CyFr labelled CD4
anli~Jo-~y. Monocytes are stained with the CyFr labelled CD4 antibody
only. It is found that the cells of int~resl in this assay are about 10
microns in diameter and are well classified using a capillary of the
dimensions outlined above. It will be appreciated that the present
invention is not limited to the c~,araderi~dlion of a CD3/CD4 assay.
According to one method useful with the invention, a biological
fluid, such as~whole unco~ lated blood, can be reacted with an excess
amount of a binding agent that contains a fluorophone excitable at a
given wavelength. The fluorescently-l~helsd binding agent is selected
to react with binding sites present within the sample. For example, a
fluorescently-labeled antibody directed to the CD4 cell surface marker
present on some leukocyte blood subcl~sses is reacted with a sample of
whole blood. The lah~le~ binding agents and the binding sites, i.e.
Iabeled anti-CD4 antibodies and the surfaces of CD4-bearing leukocytes
in the example, form fluorescent complexes that will emit a signal when
used with the apparatus of the present invention.
After the fluidic sample is reacted with the labeled binding agent,
it is diluted and the into capillary 10. Minimal processing of components
of the biological fluid nor separation of bound and unbound binding
agent is required at any point in the praclice of the method of the
present invention. An optical scan is made of the sample in a volumetric
manner and fluorescence emission is sequentially recorded from each
illuminated columnar region.
- 10 -
AKomey Docket No.~ H
mah/bmi/2002.002
21~8204
Fluorescence emission occurs from both the binding agent-
binding site complexes and from the free binding agent but a more
if~tense signal relative to background level comes from areas where the
binding agent is clustered, i.e. cells or cell constituents exhibiting binding
sites to which the binding agent is d;rected. Therefore, a signal of
heightened fluorescence cGr,aspGnds to a cell or cell constituents, and
is recorded as such. When the fluorophores with which the anti-CD4
antibodies are lAheled and excited in the given example, the
fluorescence emitted and recorded as an event sign;~ies the presence of
a leukocyte that expresses the CD4 antigen.
The enumeration may occur in an absolute volume, depending
on a desired application, by noting the beginning and ending points of
the lengthwise scan of the capillary tube and measuring incremental
steps therebetween. This quanUtalion of all of the fluorescent targets in
a fixed, precise volume is a powerful method of quickly obtaining
detailed population data.
Fluorophores that activate at dilrerenl wavele.-~ths can be
combined with binding agents directed to dilrerent binding sites, so that
the presence of multiple reaction moieties in the sample can be
detected. From the precise known volume of the capillary tube that has
been scanned, a quick reading will identify the number o~f cells or cell
constituents of a particular subclass per unit volume that are present in
the sample. To illustrate, this method can quickly distinguish and
enumerate the monocyte granulocyte or Iymphocyte subsets of a given
volume of a blood sample through reaction of the whole sample with
differentially-excitable fluorescently-labeled antibodies d.recled to the
cell surface an~igens. The T-cell leukocytes can be directed to CD3,
CD4, and CD8. The optical system is simply set to excite each
fluorophore at its crucial wavelength and a detection channel is created
1 1
Attorney Dock~t No.: 8Mi200~AH
mahlbmU2002.002
2I4820~
_
to correspond to the emission wavelen~ll, of each fluoropl,Gre.
Al~e")dlively, a ratio can be obtained without counting a precise volume,
e.g. this is a rapid technique for obtaining CD4/CD8 T-c.ell ratios,
important in determining the progression of AIDS.
S When an assay is l~e,ror",ed to determine leukocyte subclasses
in whole unco~gu'ated blood using the lechnique of the present
invention, a two or three minute wait between placement of the reacted
sample into the capillary tube and the optical scan allows for the natural
density of the numerous red blood cells present in the sample to cause
settling of the red blood cells to the bottom of the capillary tube and the
sl ~hse~uent displacement of the white blood cells. This natural buoyant
effect causes a resultant localion of the white blood cells near the upper
portion of the capi'l2~ tube and assists in fluorescence detection
l~ecAIJse of the top-down scan geometry of the presenl invention.
Because of this effect, coincidence of targets is also negligible.
The scanner is based on use of a laser 11, such as a helium -
neon laser (as shown), an ion laser, a semiconductor laser, or the like,
which generates a laser beam along path 12. Laser 11 preferably emits
in the 600 to 1000 nm range. The laser beam along path 12 passes
through a beam splitter 13, such that a portion of the beam is diverted to
a power meter 14 for monitoring the laser output power., The ;nain
beam passes through the beam splitter 13 to a narrow line filter 15,
which selects the wavelength of interest generated by the laser 11.
Next, a dichr~ic beam splitter 16 receives the beam which passes
through the filter 15. The dichr~.~ beam splitter diverts the selected
output from the laser toward a steering mirror 17. The steering mirror
17 diverts the beam to a folding mirror or prism 18. From the folding
mirror or prism 18, the beam is d;re~ed to a scanner 19.
Attorney Docket No.: e'"~'~H
, n..h/~ ~ ~
2I 1820~
The scanner 19 allows transverse and longitudinal scanning of
the laser beam aaoss the sample capillary 10. The scanner assembly
includes a galvanometer mounted mirror 25 which rotat~es a few degrees
back and forth in a rapid fashio n at about 20-200 Hz (peak-to-peak 6
12). The beam is defleded by the galvanometer mounted mirror 25 to
a first lens 22, through a second lens 23 and through an objective lens
24 to the capillary 10. Two lenses 22 and 23 are desig"~d to be
confocal, that is, they are sepa~at~d along the optical path by their focal
length, and they have equal focal lengths. It is not necessAry that the
lenses be confocal, but they must have ove, lap,~i.)g focal planes.
Similarly, the distance between lens 23 and the objective lens 25 of the
microscope rnust be precisely conlfolled so that the beam, as it is
rotated by the galvanometer mounted mirror 25, appears to be rotating
about a virtual point directly in front of the micr~scope objective.
As schematically illustrated by ghosted outline 39, the scanning
assembly 19 is designed to move longitudinaily along the capillary 10 for
a distance of about 40 millimeters.
A variety of other scar-ner mechanisms can be used as suits a
particular application of the invention.
The whole scanner assembly 19 is con~ by computer 30, as
schematically illustrated by line 26.
The beam impinging upon the outer wall of capillary 10 traverses
the wall and illuminates a columnar region of the sample causing
fluorescent emission from the sample. Light collection occurs in an epi-
illumination manner. The emitted fluorescence is collected by
microscop~ objective 24 and ~i,ec~ed back, as a retrobeam.
Microscope objective 24 has a central portion for p~ssage of incident
beam and uniform depth of focus of the inc;dent beam through capillary
- 13-
Atlorney Qockot No.: 3M1200~UAH
malVbmU2002.0~2
- 214820~
10. BecalJse flolJrescenl emission is over a very wide angle, flourescent
~ol'ec-tion occurs over a wider portion of microscope objective 24.
Fluor~sc6"ce given off by the dyes in re3pollse t~o the exGita~ion
beam generated by the laser 11 retl aces the optical patn through the
scanning mechanism 19, the prism 18, the steering mirror 17, to the
dichroic beam splitter 16. The fluorescence comes from the focal point
of the microscope objective 24 and is collimated with a diameter of
about 8 millimeters as it comes out of the microscope objective. The
d;ch,a9c beam splitter 16 allows the fluorescing wavelengths to proceed
along path 26.
The beam on path 26 enters a bandpass filter 27, or a series of
the same, whlch are designed to filter the backcc~lering of the laser
beam itself which are due to weak reflections from the optical elements
and surfaces of the capillary. These r~le~lions may be much stronger
than the actual fluorescence that is detecled from the sample.
From the bandpass filters 27, a folding mirror 28 directs the beam
through a focusing lens 29. The focusing lens brings the colimated light
from the sample into a focus, and through a pinhole filter 30. The light
from the capillary, which arises outside the focal point of the microscope
objective 25 will not be columnated when it enters the lens 29. Thus,
the pinhole filter 30 rejects fluorescence from regions that are not of
interest. The pinhole size is chosen so as to define a volume from
which to collect fluo, escence intensity from the sample. Typically, this
volume is ss'~te~ to be about five or ten times the expected volume of
the target cells.
Through the pinhole 30, the beam enters a pl ,olo" ,ultiplier box
31. The photomultiplier box includes a dichroic beam splitter 32 which
separates the detected fluorescence into two basic components. The
first component, channel 0 is light having wavelengths below about 680
- 14 -
Attom-y Dockd No.: B~"m~'~H
mahlbmU2002 002
21~8204
_
nanometers in the pr~sent embodiment. The second component,
channel 1, is light having wavelengths above about 680 nanometers.
Cl,snnel 0 is directed to a first ~hotG."ultiplier 33. Channel 1 is directed
to a second photomultiplier 34. rl ,otomulli~ rs are connected across
line 35 to the computer 40.
Also incl~ed in the mechanism, but not shown, is an autofocus
mechanism. The autofocusing system uses an algor~thn, which involves
measuring the fluorescence in a preliminary scan. At a first position in
the capillary, the microscope objective focus is scanned to find the
position of maximum fluorescence. This value is stored, and the
objective is moved to a second position in the capillary. Again, focus in
this position is scanned for maximum fluorescence. That value is
stored. Using the two values, as a beginning and end point for the scan,
the microscope focus is linearly exl,apolated between the two to
optimize the fluorescence reading along the length. In addition, the
length of each scan line is set to be slightly larger than the width of the
interior of the capillary. This is done to insure that every cell is detected
by overscanning capillary dimensions, and later detecting the edges of
the capillary to filter irrelevant information.
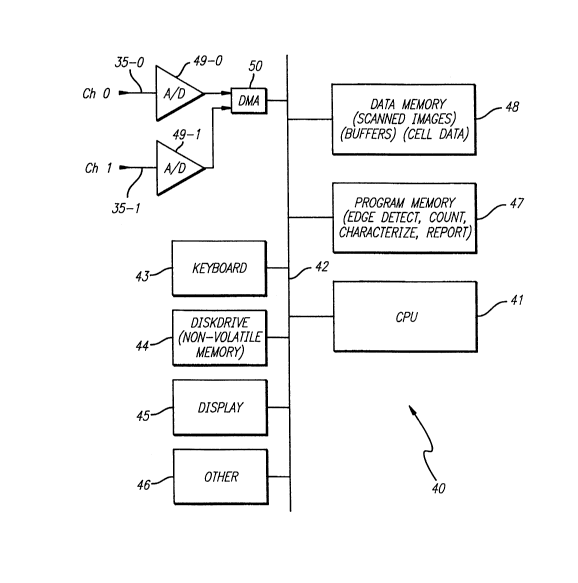
Fig. 2 schematically illustrates processing resources in the
computer 40. The computer 40 includes a CPU 41 cou~led through a
system bus 42, as schematically ill-,sl,aled. On the system bus 42 are a
keyboard 43, a disk drive 44, or other non-volatile memory system, a
display 45, and other peripherals 46, as known in the art. Also coupled
to the bus 42 are a program memory 47 and a data memory 48. The
output of the ~I,olo",ultipliers, channel 0 and channel one, are supplied
on lines 35~ and 35-1, respectively, through analog to digital converters
49-0 and 49-1. The outputs of the analog to digital converters are 16 bit
pixel values of the analog signals from channels 0 and 1. These values
- 1 5 -
At~om-y Oockct No.: ?''~'t.H
n~ t 7nno r~
2148204
are supplied through a direct memory access (DMA) circuit 50 which
~r~nsrers the pixel values into the data memory 48.
The processing system 40 includes resources that store scanned
images of the data for channel 0 and 1, buffers used during the
processing of the data, and memory for storing cell data once the cells
or cell constituents are Iscate~l cl ,aracteri~ed and/or classified.
Similarly, the progra", memoly 47 includes resources for detecting the
edges of the capillary, counting and locdling target cells or cell
constituents within the scanned images, cl~aracleri~ing the target cells
or cell constituents, and repo, (ing results. More details concerning the
processing resources in the computer 40 are provided below with
respect to the flow charts and graphs of Figs. 3-13. As will be
appreciated, processing resources may be implemented with hardware,
software, or a combination of both, as suits a particular use of the
1 5 invention.
Fig. 3 illustrates the scanning technique used for gathering data
from the capillary. The capillary 10, as illu .ll.ated in Fig. 3, has a width
of about 0.667 millimeters, and a length of about 40 millimeters. A
galvanometer scanning system scans the laser beam along a track 1 on
a line longer than the capillary 10 is wide. At the end of line 1, the beam
snaps back to the begin"ing of line 2 and scans line 2. The distance
between the center of lines 1 and 2 is about 4 microns in the present
embodiment.
For a capillary of about 0.667 millimeters in width, the scan lines
are about 0.8 millimeters long. This provides 200 four micron samples
along each scan line. For a 40 millimeter long capillary, with scan lines
separated by 4 microns, about 10,000 scan lines are collected for each
blood sample.
- 16 -
Attorn-y Docl~-t No.~ H
mah/bmil20~..002
21~820~
The analog to digital converters sample the scanned data at a
rate which creates a pixel value representing fluor~scence in a spot
having dimensioos of about 4 microns by 4 microns. Fig. 4 illus~rales a
7X7 neighborl,ood of pixels. Thus, in the upper left hand corner, pixel in
row 1, column 1, is found. In the upper right hand comer, pixel in row 1,
column 7, is found. In the center of the n~ llbo-i,ood of pixels, pixel
row 4, column 4, is found. Similarly, in the lower right hand corner, pixel
in row 7, column 7, is found. Fig. 4 also illustrates the size of the laser
spot relative to the sample dimensions. In the prefer, ed embodiment,
the laser spot (e.g., 50) has a diameter of about 10 microns. Thus,
oversampling occurs. That is, the laser spot 50 excites a region of 10
microns in diameter for the pixel at row 7, column 1. At row 6, column 1,
a second spot 51 illuminates a region 10 microns in diameter which
substantially overlaps with the spot 50 for row 7, column 1. Similarly,
the spot 52 for row 7, column 2, substan~ia:ly overlaps with the spot 50
and the spot 51 in column 1.
Fig. 5 illustrates a portion of a scanned image generated with one
of the channels of the present invention. The graph of Fig. 5 is a
baseline sub~racted represenldtion, where the baseline is illustrated at
line 60. The baseline is essentially the average height of all the lines in
the scan region. With this value subtracted, a number of peaks, e.g.,
peak 61, can be seen in the scanned image. These peaks typically
correspond to target cells and are processed as described below. Also,
each scan includes a region, generally 62, and a region, generally 63,
which lie out~ide the capillary. The baseline 60 can be used to define
the edges of the capillary becsu-se of the rapid falloff at 64 and 65
co" esponding with the edges of the capillary. The processing
resources characterizing the cells ignore pixel values outside the
detected edges.
AnorTley Dockot No.: BM1200~UAH
m-h/bmi/~02.002
214820~
.~
As mentioned above, there are two c~annels detected according
to the pr~s~al invention. Fig. 5 illusb ales a single cnannel. There will
be a cGr, espGn~ing scanned image from the second channel having a
similar profile, however, the amplitudes of the peaks wili dffler
depending on the magnitude of the fluorescence delec~6d in the second
channel. Also, some peaks may be found in one image but not the
other.
The basic data processil-g steps ex~ted by the processing
resources are illusll aled in Figs. 6A and 6B. The first step in Fig. 6A is
to receive the data from channel O and ~ ,anl ,el 1 (block 100). As the
data is received, the DMA circuitry loads it into a buffer in the data
memory (block 101). The buffer may be a circular buffer or other data
structure used to keep track of the amount of data being received. The
algorill"n then determines whether a block of data having a prespecified
size has been received (block 102). For the purposes of the Rresent
example, about 100 to 150 scan lines may comprise a suitable block
size. If a block has not yet been completely receivcd, then the algorithm
determines whether the last block from a blood sample has been
received (block 103). If it has, then the alg~ritl,tl, is done (block 104). If
it has not, the algoriUI", loops into block 101 to continue loading data in
the buffer.
When it is detecled that a complete block has been received at
block 102, then the algo,itl"" parses the data into a plurality of scanned
images by dividing the data into a raster image file ImO for the first
channel, and a raster image file Im1 for the second chanl~el (block 105).
Data pre-processing involves reading in and processing the data
in blocks. The signal processing requires signed numbers, thus, the
unsigned 16 bit data is converted into 15 bit signed data.
- 18 -
Altorn-y Dock-t No.~ H
mah/bmU2002.002
` _ 2148204
Next the two scanned images ImO and Im1 are summed or
averagecJ to g~erale a composite image Im2 and the composite
irnage Im2 is stored (block 106).
Next an edge dete~tion algorithm such as described below with
respect to Fig. 11 is executed (block 107). The edge deteclion
algo,itl"" may be supplemented with an algo,ilh,n for evaluating the
results to ensure that no false edges such as might be detected by a
bubble in a capillary are found.
The edge detection is done using a baseline profile where the
baseline is the average of all the scans in the block ignoring the scans
that have peaks higher than a certain threshold
After btock 107 the algo,iU"~, p,oceeds to determine thresholds
to be used for pa, tic e detection in the composite image Im2. These
thresholds may be prespec;fied empirically determined values or they
may be adaptively computed for each buffer or each sample. One
algo,ill"" for determining the threshold may involve determining the
maximum and minimum values for each 7X7 pixel neighbo,l,ood in the
block being processed The maximum minus minimum value with the
highest frequency of occurrence is used to estimate the threshold for
particle deteclion. Distribution of the maximum and minimum for peaks
of neighborhoods in the buffer are determined and the thresholds are
set so that peaks are detected if the maximum and minimum values in
the nei!Jhbo, h ood differ by amounts smaller than a threshold 3 standard
deviations below the average peak height.
After determining the lhreshclds for particle detection in block
108 the algorithn, proceeds to compute the background indices for the
scanned images ImO and Im1 using an algorit~",~ such as described with
respect to Fig. 10 (block 109). Afler computing the background indices
the algori~l"" may then proceed to do a baseline su6sl,action step for
- 19-
AttomeyDock-tNo.: EIM1200~AH
~ 7~ ~?
2148204
-
the scanned images ImO and Im1. This is an optional step, depending
on the tech,)iques used for particle deteetiGn and cell cl~ar~cteri~dlion
set out below.
One baseline removal technique involves finding the minimum, or
average of the N minimum lowest points along a given scan (N equal
about 10). The above minimum, or average minimum, is then
subt(acted from all pixels in the block, and negative values are clipped
to 0. Baseline removal is optional. In particular, if cell detection uses
the peak slope criteria, as described below, it is not necess~ry to do
baseline removal. However, it may be desirable to have baseline
subtraction to eliminate edge effects in an overscanning situation.
After baseline sublraclion for images ImO and Im1, the algorithm
does a p81licle detection routine using image Im2 (block 1 1 1). The
particle detection process is illustrated below with respect to Fig. 12.
The next block determines whether a particle is detected (block
112). If a particle is detected, then the neigllbGrl,oods of pixels from
images ImO, Im1 and optionally Im2 are saved, and cell parameters are
computed using data in the nei~l,bG,l,ood of the pixel maps, e.g. in
respGnse to the saved neighbo,l,oods. If a particle is not detected, then
the algo,itl"" determines whether the Im2 buffer has been completely
processed for particle detection (block 1 14). If not, the algorithm loops
to block 111 to continue the particle detection routine. If the buffer has
been finished, then the algorithm loops to block 102 in Fig. 6A, as
illus~a~ed, to begin processing a next block of data.
Thus, for example, the raw data consists of two 200xN (where
is less than or equal to 10,000) raster image files for each channel O and
channel 1, where each pixel is a 16 bit unsiy, 16d integer, generaled by
the output of the analog to digital converters. The image data may be
processed in blocks. Data for scan lines at the boundaries of the image
- 20 -
At~om-y Dock-t No.: '`'"~'~H
21~8204
-
block may be buffered to deal with cells crossing the image block
boundaries.
The data block size is 200 pixeis high by N scans wide, where N
is about 128. The buffered block size may be 200 X 16 pixels on the
S block bound~. ies. 8y image processing in blocks, a smaller arnount of
memory resources are used for the image processing.
The technique of saving the nei,Jhbo,l,ood values from the
scanned images for each block, as p~, licles are detected, and then
continuing to process ~itional blocks allows real time gathering of
data and cell or cell constituent deledion, with ability to compute cell
parameters and characte- i~a the cell later, or with a time shared
processing technique. This greatly enhances the efficiency of use of the
computation resources to allow sampling of very large amounts of
scanned data in substantially real time.
Cell and cell constKuent cl~arac~eri~dtion, according to a
preferred embodiment of the presen~ invention, involves utilizing
information from the two cl ,an,lals. The two ~;hannels in the present
system include data which is relevant to both of the dyes which are to
be detected. Thus, a technique must be used to discriminate
information from the two dyes in the two channels that is efficient and
noise immune. Accordingly, one species of the presenl invention
applies a linear ~gfessiGn technique over the neighbo,hoods of pixels
saved for detec~ pa, t: ~' s.
The problem to be solved can be appreciated with respect to Fig.
7, which schematically illustrates the spectfa of fluorescence for the two
dyes detected by cha"nel 0 and channel 1, respectively. Thus, a cell
containing an~igens stained by the first dye will fluoresce with a
spectrum suc'n as spectrum 120. Similarly, the cell with a dye attached
to an a.ltigen of the second type will fluoresce with the spectrum 121.
Anomey Oocket No.: E~
mah/bmi/20~2.002
21~820-~
As can be seen, the two spectra sul,slanli~lly overla,o. The dichroic
beam splitter 32 in the phot~multiplier mechanism 31, as shown in Fig.
~, splits the fluore-~cen~ beam along the 680 nanometer line 122 to
generate two signals. This line has been empirically determined for the
presently described system to provide good separation. Thus, the two
signals both contain information which is generaled in response to both
dyes.
The linear regression technique utilized to discriminate the
information in the two channels is described with reference to Fig. 8. In
particular, a 7X7 neighborhood of pixels is saved from each of image
ImO and Im1, cenlered on each target cell which is detected using the
algorithm of F~lgs. 6A and 6B. The 49 coordinates may be plotted as
shown in Fig. 8, where each dot is positioned with the magnitude of the
first channel on axis X and the magnitude of the second channel along
15 axis Y, such that sample at row 1, column 1, and sample at row 7,
column 7, may appear at the points (1, 1 ) and (7, 7). Similarly, the
sample at row 4, column 4, may appear at the point (4, 4). As can be
seen, the sample from row 4, column 4, will have the highest average of
amplitude from both channels because of the cell classification
technique. The dots are then a, pl'8d to a linear regression algorithm,
as well known in the art, to find the best fit line 123 to the dot plot, Pres
it al., Numerir~l RP~.U; Pe in C. 1988, p. 523. This produces the slope
"m" and the offset "a" for each target cell. Thus, the magnitude of the
cont, ibution from channel O can be expressed as the slope "m" times
the magnitude of the contribution of channel 1, plus the offset value "a".
Fig. 9 is a graph which illusl~ales the technique used for
characterizing the detec~ed cells. In particular, the intensity value for a
detected cell is de~ined as [CH1 ] for the scanned image Im1 and [ChO]
for the sc~nned image ImO in the respecti~/e 7X7 nei~JhbGrhoods. This
Attomey Docke~ No.~ H
mah/bmU2002.002
21~820~
.
intensily value is determined by a matched filtering technique based on
the expected cl lafac~l isli~s of target cells over the 7X7 nei~l ;bo, hoods.
A matched filter can bQ constructed from e~pected or typical pixel
values in the neighborl-~od of a cell. The matched filter is a matrixl of
coefficient which will be multiplied with the conesponding pixels from the
neighborhood of the Jete~led cell. The 49 resulting products can be
summed to yield a single intensity value (~CH0] or [CH1]) for the
detected cell. The cG~ff~-~nts can be chosen to sum to zero, in which
case the conslant background signal is cancelled out.
The inlensity value for the cell is then plotted on the graph of Fig.
9 by multiplying the slope determined using the linear regression
analysis above by the inlensity value of one of the channels.
As can be seen, the graph of Fig. 9 is divided into five regions.
The first region, 130, is for target cells which are dyed substantially only
with the first dye which is centered on channel 1. The second region,
131, is defined for target cells which are dyed substantially only with the
second dye. The third region 132 is defined for cells which are believed
to be stained with both dyes. The fourth region 133 and the fifth region
135 are a "no call" region to ensure that bad data is ignored.
The first region 130 is defined during calil,ralion of the device by
doing a scan of a sample dyed only with the first dye. Linear regression
analysis is applied to create a line 136 based on the one dye detection
from channel 1. A similar technique is used to define line 137 for the
second dye which has most of its spectrum det6.,~ed by channel 0. A
background index is c~l~J~ated for the buffer in question to define
regions indicated by dotted lines 138 and 139 below which for channel 1
and above which for channel 0, samples are charac~ei i ed as having
only one dye. The samples which fall in the region 132 are
characterized as having both. Samples which fall in the regions 133
- 23 -
AnomeyDocketNo.: 9~'12~'~H
rn~h/bmu2w2~oo2
21~820~
and 135, or which have a magnitude value which is too low, are
~;har~eri~e~ as no calls.
Ap,vendi~ A provides a source code for the cell c~!dr~ctefi2ation
routine according to one embodiment of the pr~sent invention as a
means of providing an example of pfocassing resources which might be
used to accomplish this sort of classification.
The goal of cell or cell constituent classifi~3~io,) is to determine
the decisiQn boundaries based on pop!Jl~tion~ dependent parameters.
If necessary, the boundaries determined can be validated with
population ~dtislics.
In the CD3/CD4 assay, there are 3 cell popul~tions present. The
CD3 positive cells are stained with the Cy5 l~hel~d antibody only. The
CD4 cells are stained with both the CyS lal-ele~ CD3 antibody and the
CyFr l~hele~ CD4 antibody. The monocytes are stained with the CyFr
labeled CD4 antil~o~y only.
Since the CD3 positive cells and the monocytes are stained with
one dye only, their slope distributions should cluster around the Cy5 and
CyFr slopes respectively. The Cy5 and CyFr slopes are determined in
the compensation matrix calibralion. The spread or distribution of the
clusters can be determined from the background noise estimates.
Similarly, in the CD3/CD8 assay, there are 3 cell populations
present. The CD3 positive cells are stained with the Cy5 labeled
anli~o~Jy only. The CD8 cells are stained with both the Cy5 labeled CD3
antiL~ody and the CyFr labeled CD8 antibody. The NK cells are stained
with the CyFr l~heled CD8 antibody only.
Since the CD3 positive cells and the NK cells are stained with
one dye only, their slope distributions should cluster around the Cy5 and
CyFr slopes respectively. The Cy5 and CyFr slopes are determined in
- 24 -
Attomey Dock-t No.: B"'~'~H
1 Idl Ll : ~}2.002
21~820~
` .
the compensation matrix calibration. The spread or distribution of the
clusters can be determined from the background noise estimates.
The following classi~ioa~ion rules may be applie~
Non-Cells
S A particle is classified as non-cell, if any of the following criteria is
met:
1. Particles with cc ~rela~iGn coefficient (for the regression fit
for planes O and 1 ) less than the threshold value (0.8).
2. Channel O value less than background noise threshold.
3. Channel 1 value less than background noise threshold.
Monocytes and NK Cells
1. Cells with channel 1 value grealer than the CyFr slope line
minus a constant background noise offset for channel 1.
. 15
CD3 Cells
1. Cells with channel 1 value less than the CyS slope line
plus a constant background noise offset for d ,annel 1.
C D 4/8 C ells
Any cell that does not satisfy the above criteria are potentially a
C D 4/8 cell. Cells that lie too close to the Cy5 or CyFr slopes are labeled
as "no-calls". The slopes di~rence between the CyFr and CyS slope
are divided into slope regions. Cells that are below the 10% or above
the 90% region boundaries are classified as no~alls.
The use of the slope value from the linear regression a ,nalysis
provides a noise immunity, and improves the robustness of the system.
This analysis may be replaced by solving two equ~tions with two
- 25 -
Atton~y ~OCk-t No.: F~ H
mah~bmi~2002.002
2148204
-
unknowns based on the intensity values for channel O and channel 1,
respectively.
The cross co" ~,lation COerriC;en~ betweG,- col I dsQond;n9 pixels
from image O and image 1 is determined. H the signal is dominated
S random noise, one would expect a poor correlation coefric;enl. A cell
with good signal gives a co"~lation that ranges from 0.9 to 1Ø
The cross CGI I elatiGn technique can be e~ten~ed to correlate
individual cêlls with thQ average cell profile. A con,posite average cell
profile can be generated by averaging the cell profile of all the cells
detecte-J. A good cor, ela~iGn coefficient indicates that the cell shape is
similar to the average cell shape. An artifact peak usually has a
~lirreren~ cell shape profile and thus gives a poorer cross correlation
coefficient.
Fig. 10 illusllatês an algoriU,I,~ for computation of the background
noise indices, used as described above, with respect to Fig. 9, to define
the regions around the line for the two dyes within which a cell will be
characterized as containing only that dye.
This background index can be computed for each buffer, or it can
be computed across the entire image.
The technique involves defining a plurality of bins for N
background baseline noise levels, where N is about 2,000, and each bin
16 bits wide, in one embodiment (block 200). for each buffer, a V\IXW
pixel map surrounding each pixel is derined, such that WXW is large
enough to contain almost all the signal from the exrected size of the
cell. This may be about 5X5 in the present example (block 201).
For each V\~N pixel map, the maximum and minimum pixel
values are determined for each pixel in images Im1 and ImO (block 202).
Each pixel is then assigned an integer bin number based on the
dirrerence between the maximum and minimum values within the pixel
AKorr~y Dock t No.: BM1200~AH
rn~hhm~O02
21~820~
-
map divided by the size of the allocated bins (block 203). As mentioned
above, there may be 2,000 bins, 16 bits wide each for a range of about
32,000 values.
Wlth the integer bin number, the count for the co,.~,sponding bin
is incremented for that pixel (block 204). After incrementing the bin
number, the algori~l)." determines whetl,er there are more pixels to
process for the buffer (block 205). If there are, it loops back to block
201. If not, the algGl ith m determines the bin number with the highest
count (block 206). The background noise index for this buffer is set to
the bin number with the highest occurrence times the bin size (block
207). These indices are then stored for both channels (block 208). As
mentioned above, the process is carried out for both channels to
achieve two separate background noise indices.
Fig. 11 illustrates an algoritl "" for performing edge detection, as
menlio"ed above. Rec~use the blood sample in this example will
contain free dye labelled antibody, a distinct background signal results.
This signal is used to locate the boundary of the lumen of the capillary.
The average image Im2 from chal)nel 0 and channel 1 is gene~led
(block 300). A specified number N of scans are averaged pixel by pixel
to produce an average scan (block 301). The ma~timum and minimum
values are determined from the average scan (block 302). Next, a
threshold is set based on the difrerence between the maximum and the
minimum times a factor which ranges from 0.1 to 0.8 (block 303). This
est~l shes a ll,reshald which is a percenlage of the average amplitude
of fluorescence from a given scan lins. The scan lines are then
searched from center of the scan to the left until the baseline value is
less than the threshold for left edge delec~ion. The left edge is then set
as the lo~a~io~ where the threshcl~ is crossed (block 304). Similarly,
the algorith,n searches from the center of the scan to the right until the
Altomoy Dockot No.: E~ H
m~h~bmV2002 002
21~820~
threshold for right edge detec~ion is pAsse~l The lo~a~ion of the right
edge is then defined as the localion where the threshold is crossed
(block 305). Using edge detec~ion, sc~nned pixels out.s,ide of the
detec~ed edges are ignored in the additional processing des~ ibed
above.
Fig. 12 illusbdt~s the algo.iU,.~, for detec(ing a cell or cell
constituent. The cell dele~ion algorithm uses the image Im2, which is
based on the average (or sum which for this purpose is substantially the
same thing) of images ImO and Im1 (block 400). A 5X5 pixel map
surrounding each pixel is then detined in sequence (block 401). This
pixel map is tested. Ihe test is a five step test whidl involves
determining whether the center pixel is the pixel having the highest
value of all pixels in the pixel map. If it is, then the test takes the
difrarence bet Necn the center pixel and the top left comer pixel and
determines whether this di~ference is greater than a threshold which is
assigned for cell or cell constituent detection. Next, the center pixel and
the top right comer are used to make the ll,resho'i determination. Next,
the center pixel and the bottom left corner pixel are used to make the
threshold determination. Next, the center pixel and the bottom right
corner pixel are used to maker the tl,resho'd determination. If the
center pixel has the highest value, and anotl,er pixel in the map has an
equal value, then the algorithm passes the test. In order to avoid
counting a cell twice, in this inslance, the center pixel value is
incremented by one, so that when the other pixel value having the high
value is encountered, It will be determined that it is not the highest value
pixel (block 402).
If all five conditions have been satistied (block 403), the x and y
coordinate for thé center pixel is saved, and a 7X7 pixel map, or
neighbo, hood, surrounding the center pixel is saved from each image
AKom-y Dockd No.: B"'~'~l
..: ~ ~qO2
21~820~
ImO and Im1 in the cell parameter list (block 404). These values can be
used for later processing of the data as des~ il~ed above.
Using the averaged (or summed) image for cell detection
provides better cell resolution, because of magnitude of the signal from
any one of the two dyes may be very low on a given cell. This cell or
cell constituent det~ion is based on the maximum an minimum values
in the neighbG- hood around the detected cell, rather than an absolute
peak value. This provides immunity from background levels may vary
over the scanned region.
Fig. 13 illustrates the basic algoritl "" for linear regression
analysis of the 7X7 pixel map. The linear reg~ssion analysis begins by
taking the 7Xrpixel map of a detected cell from both images lO and 11
(block 500). A linear regression line is fitted for corresponding pixel
points from images ImO and Im1 over the indices for the row i and the
column j, such that Im1 (i,j) approximates to the slope times ImO (i,j)
plus an offset (block ~01). After fitting the regression line, the slope,
offset and CGI, elation coefficient r2 (goodness of ft) are stored in the cell
parameter list (block 502).
Thus, a linear regression line fit is computed between the
corresponding pixels from image O and image 1 and the slope and
goodness of fit values are obtained. The slope determin,ed indicates
whether the cell is stained with 1 of 2 antibGJies. Using a 7X7 pixel map
to determine the slope gives a better estimate of the slope of the cell.
The goodl ,ess of fit indicates the gooJI ,ess of the data. The advantages
of the linear regression analysis include noise reduction, a good
estimate of signal quality, a system insens~ti~/e to baseline subtraction,
and the well defined oi U~ogonal coordinates.
In sum, a method and apparat~ls is provided for processing data
generated by one or more ;hannels of data, where the channels include
- 29 -
Anom y Dock-t No.: g~ H
mahhni~.O02
21 48204
information relevant to more than one characlerislic to be determined,
and are taken from a container. The technique allows for counting and
ct)atact~ ing the cells or cell constituents within the contained region
with minimum ope,alor handling of the samples, repe~t~l~i'ity, and
efficient utili~dlion of processing resources.
The data processing resources accomplish data collection, image
averaging for capillary edge and cell detection opera~i~ns, background
noise determination, baseline removal, and cell charac~e, i~ation.
The data from a detected cell or cell constituent is then extracted
by saving it into a cell parameter list in memory. This allows continuous
scans of large volumes of data, with processing of thè data saved for
later steps when more processi"g resources may be available. Also, it
allows reanalysis of data for detected cells in the future based on much
reduced file sizes, as compared to what would result if the entire 200 by
10,000 pixel file must be saved for later analysis.
The present invention provides a system for processing scanned
data which is very robust and accurate. It allows concurrent data
ccl'~ction and analysis, as well as analysis of data after collection.
Further, it allows for completion of data analysis very rapidly, shortly
after completion of the data collection systems.
- The for2~ing desuiplion of a prere"ed embodiment of the
invention has been presenled for purposes of illualldliG,i and
description. It is not inle,)-led to be exhaustive or to limit the invention to
the prec;3e forms disclose~ Obviously, many mod~ficalio"s and
variatiol,s will be apparenl to pra~,1ilioners skilled in this art. It is
intended that the scope of the invention be defined by the following
claims and their equivalents.
- 30 -
Attomey Dock-t No.: ~'"~'Ul
. . ,; ~ .:~2.002
21~820~
._
APPFI~IDIX A
Copyright Biometric Imaging, Inc., 1994
SOURCF CODF FOR CFI I CHARACTFRI7~TIQN
const float kCellCo.,~' YcnCoefThresha~ 0.8;
CELL-TYPE CCell::CellC-b ~ni' ~ ~ffc n( )
{
float rnaxSlope - CyFrSlope - ((CyFrSlope-CySSlope) 0.1);
float minSlope ~ CySSlope + ((CyFrSlope-CySSlope) 0.1);
if ( (cellCo m 'a' nCoef ~ kCellCo, . ~ 'aflc nCoefThreshokl) l l
(peakValueO ~ noiseTh.~73h~'~0) ll
(peakValue1 ~ noisemr~h ~
retum (NON_CELL); ll noise peak
else if ( (peakValue1 ~ (CyfrSlope peakValueO ~ noiseLever) )
retum (CYFR_POSlTlVE); //~.~no. ytl~ or NK cells
else if ( peakValue1 ~ (Cy5Slope peakValueO ~ n - ~LAvel) )
return(CYS_POSlTlVE); /ICD3 celk
else if ( (cellSlope ~ rnaxSlope) l l (cellSlope ~ rninSlope) )
retum(NO CALL); llNocall
else
retum (CYFR_CYF_POSITIVE); ll CD418 cell
)
-31 -
AKor~ k t No.~ H