Note: Descriptions are shown in the official language in which they were submitted.
" CA 02203801 1999-12-07
NUCLEOTIDE SEQUENCES MEDIATING MALE FERTILITY
AND METHOD OF USING SAME
BACFCGROUND OF 'rHE INVENTION
The goal of plant breeding is to combine in a single
variety/hybrid various desirable traits of the parental
lines. For fiE=ld crops, these traits may include resistance
to diseases a.nd insects, tolerance to heat and drought,
reducing the time to crop maturity, greater yield, and
better agronomic quality. With mechanical harvesting of
many crops, uniformity of plant characteristics such as
germination and stand establishment, growth rate, maturity,
and fruit size, is important.
Field crops are bred through techniques that take
advantage of the plant's method of pollination. A plant is
self-pollinating if pollen from one flower is transferred to
the same or an~~ther_ flower of the same plant. A plant is
cross-pollinate~~ if the pollen comes from a flower on a
different plant.
In Brassica, the plant is normally self sterile and can
only be cross-p~~llinated. In self-pollinating species, such
as soybeans and cotton, the male and female plants are
anatomically juxtaposed. During natural pollination, the
male reproductive organs of a given flower pollinate the
female reproductive organs of the same flower.
Maize plant=s (~ea mays L.) present a unique situation
in that they can be bred by both self-pollination and cross-
1
CA 02203801 1997-04-25
WO 96/13588 PC"T/US94/12444
pollination techniques. Maize has male flowers, located on
the tassel, and female flowers, located on the ear, on the
same plant. It can self or cross pollinate. Natural
pollination occurs in maize when wind blows pollen from the
tassels to the silks that protrude from the tops of the
incipient ears. ,
A reliable method of controlling male fertility in
plants would offer' the opportunity for improved plant
breeding. This is especially true for development of maize
hybrids, which- relies upon some sort of male sterility
system.
The development of maize hybrids requires the
development of homozygous inbred lines, the crossing of
these lines, and the evaluation of the crosses. Pedigree
breeding and recurrent selection are two of the breeding
methods used to develop inbred lines from populations.
Breeding programs combine desirable traits from two or more
inbred lines or various broad-based sources into breeding
pools from which new inbred lines are developed by selfing
and selection of desired phenotypes. A hybrid maize variety
is the cross of two such inbred lines, each of which may
have one or more desirable characteristics lacked by the
other or which complement the other. The new inbreds are
crossed with other inbred lines and the hybrids from these
crosses are evaluated to determine which have commercial
potential. The hybrid progeny of the first generation is
designated Fl. In the development of hybrids only the F1
hybrid plants are sought. The F1 hybrid is more vigorous
than its inbred parents. This hybrid vigor, or heterosis,
can be manifested in many ways, including increased
vegetative growth and increased yield.
0
Hybrid maize seed is typically produced by a male
sterility system incorporating manual detasseling.
Alternate strips of two inbred varieties of maize are
planted in a field, and the pollen-bearing tassels are
removed from one of the inbreds (female). Providing that
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
there is sufficient isolation from sources of foreign maize
pollen, the ears of the detasseled inbred will be fertilized
only with pollen from the other inbred (male), and the
resulting seed is therefore hybrid and will form hybrid
plants. Unfortunately, the manual detasseling process is
not entirely reliable. Occasionally a female plant will be
blown over by a storm and escape detasseling. The natural
variation in plant development can also result in plants
tasseling after manual detassling is completed. Or, a
detasseler will not completely remove the tassel of the
plant. In either event, the female plant will successfully
shed pollen and some female plants will be self-pollinated.
This will result in seed of the female inbred being
harvested along with the hybrid seed which is normally
produced.
Alternatively, the female inbred can be mechanically
detasseled. Mechanical detasseling is approximately as
reliable as manual detasseling, but is faster and less
costly. However, most detasseling machines produce more
damage to the plants than manual detasseling. Thus, no form
of detasseling is presently entirely satisfactory, and a
need continues to exist for alternatives which further
reduce production costs and the eliminate self-pollination
in the production of hybrid seed.
The laborious detasseling process can be avoided by
using cytoplasmic male-sterile (CMS) inbreds. Plants of a
CMS inbred are male sterile as a result of factors resulting
from the cytoplasmic, as opposed to the nuclear, genome.
Thus, this characteristic is inherited exclusively through
the female parent in maize plants, since only the female
provides cytoplasm to the fertilized seed. CMS plants are
fertilized with pollen from another inbred that is not male-
sterile. Pollen from the second inbred may or may not
contribute genes that make the hybrid plants male-fertile.
Usually seed from detasseled normal maize and CMS produced
seed of the same hybrid must be blended to insure that
Z
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
adequate pollen loads are available for fertilization when
the hybrid plants are grown.
There can be other drawbacks to CMS. One is an
historically observed association of a specific variant of .
CMS with susceptibility to certain crop diseases. This
problem has led to virtual abandonment of use of that CMS
variant in producing hybrid maize.
Another form of sterility, genie male sterility, is
disclosed in U.S. Patents 4,654,465 and 4,727,219 to Brar et
al. However, this form of genetic male sterility requires
maintenance of multiple mutant genes at separate locations
within the genome and requires a complex marker system to
track the genes and make use of the system convenient.
Patterson also described a genie system of chromosonal
translocations which are effective, but complicated. U.S.
Patents No. 3,861,709 and 3,710,511.
Many other attempts have been made to improve on these
drawbacks. For example, Fabijanski, et al., developed
several methods of causing male sterility in plants (see EPO
89/3010153.8 publication no. 329,308 and PCT application
PCT/CA90/00037 published as WO 90/08828). One method
includes delivering into the plant a gene encoding a
cytotoxic substance associated with a male tissue specific
promoter. Another involves an antisense system in which a
gene critical to fertility is identified and an antisense to
the gene inserted in the plant. Mariani, et al. also shows
several cytotoxin encoding gene sequences, along with male
tissue specific promoters and mentions an antisense system.
See EP 89/401,194. Still other systems use "repressor"
genes which inhibit the expression of another gene critical
to male sterility. PCT/GB90/00102, published as WO
90/08829.
As noted, an essential aspect of much of the work
underway with male sterility systems is the identification
of genes impacting male fertility.
d
CA 02203801 1997-04-25
WO 96/13588 PG"T/US94112444
Such a gene can be used in a variety of systems to
control male fertility. Previously, a male sterility gene
has been identified in Arabidopis thaliana and used to
produce a male sterile plant. Aarts, et al., "Transposan
Tagging of a Male Sterility Gene in Arabidopsis", Nature,
363:715-717 (June 24, 1993). In the present invention the
inventors provide a novel DNA molecule and the amino acid
sequence it encodes which is critical to male fertility in
plants.
Further, the inventors present a unique variation to
the method of controlling male sterility by using the DNA
molecule to cause a plant to be male sterile after
transformation, with fertility, not sterility, induced.
Thus, one object of the invention is to provide a
nucleic acid sequence, the expression of which is critical
to male fertility in plants.
Another object of the invention is to provide a DNA
molecule encoding an amino acid sequence, the expression of
which is critical to male fertility in plants.
A further object of the invention is to provide a
method of using such DNA molecules to mediate male fertility
in plants.
A still further object is to provide a method of
mediating male fertility in plants by regulating expression
of the DNA molecule naturally occurring in the plant.
Yet another object is to provide a method of mediating
male fertility in plants by delivering the DNA molecule into
a plant such that expression of the DNA molecule may be
controlled.
Another object is to provide plants wherein male
fertility of the plants is mediated by the DNA molecule.
A further object is to use plants having male fertility
mediated by the DNA molecules in a plant breeding system.
Further objects of the invention will become apparent
in the description and claims that follow.
S
r.
- CA 02203801 2001-05-10
J
SUMMARY OF THE INVENTION
This invention relates to nucleic acid sequences, and, specifically, DNA
molecules and the amino acid encoded by the DNA molecules, which are critical
to male
fertility. It also relates to use of such DNA molecules to mediate fertility
in plants. One
such method is to controllably render plants male sterile by using an
inducible promoter
to regulate expression of the DNA molecule such that the gene is normally
"ofi" and the
plant is thus sterile. When the promoter is induced, the plant becomes
fertile.
In one embodiment, the invention provides a nucleotide sequence encoding the
amino acid sequence of SEQ. ID NO. 2. The invention also comprises plasmid
vectors
and transformed plant cells containing this nucleotide sequence.
In another embodiment, the invention provides a DNA molecule that mediates
fertility in plants and encodes the amino acid sequence of SEQ.117 NO. 2.
In a further embodiment, the invention provides a DNA molecule that mediates
fertility in plants comprising SEQ. ID NO. 1.
In another embodiment, the invention provides a method of mediating fertility
of
a plant comprising repressing expression of a nucleotide sequence in the plant
encoding
the amino acid sequence of SEQ. ID NO. 2.
In a further embodiment, the invention provides a method of mediating
fertility of
a plant comprising repressing expression of a DNA molecule in the plant
comprising
SEQ. ID NO. 1.
BRIEF DESCRIPTION OF THE DRAWINGS
Fig. 1 is a restriction map of the transposon Ac.
Fig. 2 is a gel of a Southern Blot analysis of PvuII digested DNA from an Ac
family segregating for sterility and hybridized with an internal 1.6 kb
HindIII from Ac.
Fig. 3 is a schematic representation of inverse polymerase chain reaction.
Fig. 4 is a graphic representation of the l.4kb DNA isolate and its
intervening
sequences.
6
w
CA 02203801 2001-05-10
t
Fig. 5 is a Southern Blot analysis gel of PvuII digested DNA of an Ac family
segregating for sterility and hybridized with the l.4kb DNA isolate.
Fig. 6 is a Northern Blot analysis gel hybridized with the male fertility gene
MS45.
Fig. 7 (SEQ. 1D NOS. 3-6) shows the nucleotide and amino acid sequence of
fertile revenant plant DNA after Ac transposition.
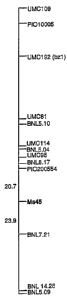
Fig. 8 is an RFLP map of chromosome 9 showing the male fertility gene MS45.
6a
CA 02203801 1999-12-07
~SCLOSURE CF ~:'HE :=NVENTION
Unless aefinecz otherwise, ail technical and scientific
terms used herei;z have the same meaning as commonly
understood by one of ordinary skill in the art to which this
invention belongs. Unless mentioned otherwise, the
techniques employed or contemplated herein are standard
methodologies well anown to one of ordinary skill in the
art. The materials, methods and examples are illustrative
only and not limiting.
MALE FERTILITY DNA MOLECULES
Genetic male sterility results from a mutation in one
of the genes responsible for a specific step in
microsporogenesis, the term applied to the entire process of
pollen formation. These genes can be collectively referred
to as male fertility genes. There are many steps in the
overall pathway where a mutation can lead to male sterility.
This seems aptly supported by the frequency of genetic male
sterility in maize. New alleles of male sterility mutants
are uncovered in materials that range from elite inbreds to
unadapted popu:Lations. To date, published genetic male
sterility research has been mostly descriptive. Some
efforts have been made to establish the mechanism of
sterility in maize, but few have been satisfactory. This
should not be Burp rising given the number of genes that have
been identified as being responsible for male sterility.
One mechanism is unlikely to apply to all mutations.
The invention is of a plant male fertile gene. cDNA's
specific for pollen development and tassel development have
been extensively reported. To date, none of them have led
to cloning a gene that can be referred to as impacting
pollen development.
7
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
The following is presented by way of illustration and
is not intended to limit the scope of the invention.
Tagging
A_c (Activator) is a well known transposable element .
first characterized in 1954 by Barbara McClintock,
(McClintock, B., Cold Spring Harbor Symp. Quant. Biol.
21:197-216 (1956); McClintock, B., Carnegie Inst. Wash.
Yrbook, 53:254-260 (1954) see also Federoff, U.S. Pat. No.
4,732,856 issued March 22, 1988 and Dooner, U.S. Pat. No.
5,013,658 issued May 8, 1991). Ac was used to clone this DNA
molecule. A restriction map of Ac used here is depicted in
Figure 1. Those skilled in the-art are familiar with the
restriction sites of A_c. In sum, The Ac transposon went
from the P-vv locus on chromosome 1 to chromosome 9. The
only currently described male sterility gene on chromosome 9
is ms2, which has never been cloned or sequenced. See
Albertsen, M. and Phillips, R.h, "Developmental cytology of
13 genetic male sterile loci in maize" Canadian Jnl. of
Genetics and Cytology 23:195-208 (Jan, 1981). The only
cloned .fertility gene is the Arabidopsis gene described.
Aarts, et al., s-upra. Test cross progeny have confirmed the
genes are not allelic.
Plant Materials
Three maize lines were used, all of which are widely
available to maize geneticists and regularly used by those
skilled in the art and are described at Chen, et al.,
"Transposition of Ac from the P locus of maize into
unreplicated chromosomal sites" Genetics 117:109-116
(September 1987). Such lines may be obtained, for example,
from the authors of the above article, from Pioneer Hi-Bred
International, Inc., or any one of many public sources such
as the Maize Genetics Stock Cooperation Center, University
H
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
of Illinois, Urbana/Champagne, Department of Agronomy S-123
Turner Hall, 1102 South Goodwin Avenue, Urbana, Illinois,
61801.
The first line is W23P-vv. The P-vv allele is caused
by the insertion of the mobile element Ac into the P locus.
Emerson, R. "The inheritance of a recurring somatic
variation in variegated ears of maize" Am. Nat 48:87-115
(1914); Brink, R. and Nilan, R. "The relation between light
variegated and medium variegated pericarp in maize" Genetics
37:519-544 (1952) and Barclay, P. and Brink, R. "The
relation between modulator and Activator in maize" Proc.
Nat'1. Acad. Sci. USA 40:1118-1126 (1954). The P gene is a
maize gene well characterized and fully detailed in the art.
The P gene induces pigmentation of the pericarp in maize.
Flavanone is reduced to phlobaphenes which cause
pigmentation of the pericarp. One example of the detailed
information on the P gene which is available to one skilled
in the art is the discussion by Lechelt, et al., "Isolation
and molecular analysis of the maize P locus," Mol. Gen.
Genet. 219:225-234 (1989) and Chen, et al., "Molecular
Analysis of Ac transposition and DNA replication" Genetics.
This is an excellent marker gene because of its function in
regulating the color of pericarp, and red striped pericarp
results. The red stripes show the excision of Ac from P,
restoring gene function and providing red pericarp.
The P-gene (P-vv) is on the same chromosome as known
genetic male steriles previously mapped to chromosome 1. It
has been shown that Ac transposes on the same chromosome 67~
of the time. Van Schaik, N.V. and Brink, R.A.,
"Transpositions of modulator, a component of the variegate
- pericarp allele in maize" Genetics 44:725-738 (1959).
However, this did not occur here, as the Ac transposed to
- , chromosome 9. P-vv itself greatly facilitates transposon
tagging because it is possible to visually observe when Ac
has transposed from the P-gene and is elsewhere in the
genome.
4
CA 02203801 1997-04-25
WO 96113588 PC"T/US94/12444
4C063 is a white inbred line that combines well with
W23P-vv to give good hybrid plants with easily scored
kernels. W22r-sc:m3 is a line with the Ds element at the R-
locus. The plant is genetically dominant at all the
anthocyanin pathway genes (A1, A2, Bzl, Bz2, C1, C2, Pr, R) .
Because Ds causes R to become dysfunctional, no anthocyanin ,
are produced in the kernel.
This was coupled with use of W22r-sc:m3 stocks, in
which _Ds is integrated into the R-gene. The Ds element
responds to the presence of Ac, by transposing to another
site on the genome. It is, in fact, a defective Ac. The Ac
transposon can move in and out of a gene on its own, whereas
_Ds cannot move unless A_c is present somewhere on the genome.
The R gene is a gene in maize studied in considerable depth.
It is known to encode enzymes required for synthesis of
anthocyanin pigments. An example of the detailed
information known regarding the R gene is the description
and sequencing information found at Dellaporta, et al.,
Stadler Symposium 18:263 (1988) and Ludwig, et al., "Lc, a
member of the maize R gene family responsible for tissue-
specific anthocyanin production, encodes a protein similar
to transcriptional activators and contains the m~c-homology
region", Proc. Nat. Acad. Sci. 86:7092-7096 (Sept. 1989) and
use of the gene as a visual marker, described at Bowen, et
al., "R Genes as visual markers for corn transformation"
Abstract edit. Gallagher, Academic Press (Oct. 1989) and
Ludwig, et al., "A regulatory gene as a novel visible marker
for maize transformation" Science 247: 449-450 (Jan. 26,
1990 ) .
In the W22 r-sc:m3 stock, all kernel anthocyanin genes
are dominant. The kernel color is yellow, however, because
of _Ds interrupting function of the R-gene. In the presence
. of A_c, however, the Ds element can transpose, resulting in ,
purple-spotted kernels. Therefore, it was possible to 1)
visually determine when Ac transposed away from the P-gene
(red-striped or full red pericarp) and 2) determine whether
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
Ac was still active (purple spots in the aleurone). By
selecting either all red kernels or kernels with red
pericarp stripes over the embryo that also have purple spots
in the aleurone, it was possible to greatly enrich for those
cases where an active Ac has transposed to another location
in the the genome. By selfing plants resulting from these
kernels, one can screen progeny families for any mutations
affecting tassel or anther development. In this case,
selfed families for the segregation of male-sterile plants
were created.
Co-Segregation Analysis
Conducting co-segregation analysis for specific gene
tagging and cloning strictly through a molecular approach
can be tedious and time-consuming. The Ac-system, however,
is well suited to co-segregation analysis at the field
genetics level. Interaction between active Ac and Ds at the
R-gene (r-sc:m3) can be utilized. Plants crossed with A_c
were selfed and grown and those families segregating for
male sterility identified. Once a family was identified that
segregated for male sterility, additional seed was planted
to cross with r-sc:m3 for co-segregation analysis. Each
plant (fertiles and steriles) was crossed with r-sc:m3, the
kernel color segregation observed on each ear and correlated
with whether the plants were male fertile or male sterile.
A family was observed where the plants were mostly male
sterile, with a few extruded abnormal anthers scattered
about the tassel. In most cases, these abnormal anthers did
not have pollen present. When every plant from this family
was crossed with r-sc:m3, co-segregation of Ac with the
male-sterile phenotype was observed as set forth in the
table below.
CA 02203801 1997-04-25
WO 96113588 PCTIUS94/12444
TABLE 1
Segregation of trhn-90-40 crossed with r:m3
Plant Ear Observed Expected
Phenotype Phenotype Number Number ,
Sterile all kernels 8 8.25
purple spotted
Fertile 1/2 kernels 16 16.50
purple spotted
1/2 kernels
no spots
Fertile all kernels 9 8.25
no spot
Male-sterile plants always produced ears with every
kernel purple spotted. Two thirds of the fertile plants had
ears that segregated 50°s spotted kernels and 50~ yellow
kernels. One third of the fertiles produced ears with all
yellow kernels. This showed Ac had transposed into a gene
responsible for male fertility and interrupted its function.
The gene acts as a recessive, and when homozygous, results
in male sterility. This segregation was verified in further
plantings.
Molecular Analysis
Southern analysis was carried out to confirm .
association of A_c with sterility. Southern analysis is a
well known technique to those skilled in the art. This
common procedure involves isolating the plant DNA, cutting
with restriction endonucleases and fractionating the cut DNA
on an agarose and transferring to nitro cellulose membranes
CA 02203801 1999-12-07
~o separate the DNA :,y molecular weight. ~t was then
hybridized with vhe probe fragment which was radioactively
labeled with P32 and washed in an SDS solution. Southern,
E., "Detection o.. a specific sequences among DNA fragments
by gel electrophoresis," J. Mol. Biol. 98:503-517 (1975).
DNA was isolated from sterile-crossed progeny and
fertile-crossed progeny, keeping the purple-spotted kernel
seedlings separate from the yellow kernel seedlings. DNA
was isolated from the t=op two leaves of one month old plants
using an Urea procedure as described at Dellaporta, et al . ,
"A plant DNA minipreparation: version II" Plant Mol. Bio.
Rte. 1:19-21 (1983). '~'he isolated DNA was cut with PvuII in
order to find a 2.5 kb fragment only associated with Ac as
shown in the restriction map (Fig.l). Approximately 8 ug
of DNA was digested with the appropriate enzyme according to
the manufacturer's instructions (Promega). DNA digests were
electrophoresed through a 0.75°s SeaKemT" GTG agarose gel and
transferred to Duralon''"-UV nylon membrane by capillary
blotting and fixE~d to the membrane by baking 1 hour at 85C.
The l.6kb HindIII: fragment of Ac was used as a probe in the
Southern Blot analysis.
The results are shown in the gel at Figure 2. At Figure
2, the male st~=riles are lanes 3-10. Lane 2 is the
heterozygous fertile plant and lane 1 the wild type. As
this gel confirms, a 2.5 kb fragment band appeared in all
sterile (purple spotted kernels) plants and did not appear
in any of the fertile (yellow kernels) plants. This
confirms the Ac was either closely linked to the male
fertility locus ~r inserted into the locus, inhibiting the
function of the gene and resulting in a male sterile
phenotype.
Cloning
The DNA adjacent to the known Ac sequence was cloned
and used in obtaining t:he entire gene
13
CA 02203801 1999-12-07
_..~;a=-__, :._ ___ ____~__ _~nt _:.A _..~ _ne aaie
_ ___~-_ __..= _:;~. ;erg . yes ~..~
_ .. _ _ tr_~.~_..
_.nccnu'__ases __ -, _ _ _ _
_ _ _, ~a~ , sac _, end :~,ba _ _..
_~cate _ __. _~ ..~_::c :~~':: .:.e =:c
_ men.
==a r _ragments were
= _iectrcpr.orese~~, _~et:~:er:: _ransferred, =nd Hybridized with
_~:e =c ~ir.a~:1= _raament. ~ %: kb :st1 =ragment was
=::dentif' as _-.at -... =egregated ~.,rith male-st element. the
-n~~erse _~7R -,F:t::,,c; :~ Daker et ai was ~~sed tc isolate the
DNA associated ~.mth ~c. warp, D.J. owe, 3. and Baker B.,
~0 "Ampiificaticn Ji ~encmic sequences =raking transposable
elements ~.n ;lost and heterologous plants: a tool for
transposon tac~gincr and genomic characterization," Nucleic
Acids Research 1:5271-?279 (1990).
A schematic depicting the well known inverse polymerase
i5 chain reaction ~ro~~edure is shown in Figure 3. After
obtaining the okb _ragment, the ends were religated. A and
B primers wer=_' _dentified readily since the sequence of _Ac
is known. Tr:us she 5' and 3' oligonucleotides could be
identified, and, accordingly to the inverse PCR technique,
20 react to ampl:~fy the intervening sequences. The A and B
primers were run from each side of the religated circle
where the Ac had bf~en. In this way, the DNA between the end
of the Ac was amplified and a l.3kb segment of CNA isolated.
The Known _ . .. ~n =c =ragment plus the amplified i . 3 kb IPCR
~5 product neari,r eau.aled the 6.0 kb Pst I fragment isolated
previously.
Details of this above summarized procedure are as
follows. Genomic DNA was isolated as described above. 20
ug of DNA was digE~sted with 20 units of PstI according to
the manufacturer's instructions (Promega). The digested DNA
was electrophoresed as described above using a preparative
comb. A gel Fragment, which contained DNA with a molecular
weight between 5.5 and 6.5 kilobases, was excised from the
gel. The DNA was electro-eluted from the agarose by using
Spectra/Por~" membrane #2, MWCO 12-1400 (Spectrum Medical
ndustries, -nc.; which contained 0.4 mi sterile water and
14
CA 02203801 1999-12-07
a i eCtrOpnOr°S;.ng ~'lal::S t _.. _ ~ ~:~-a.Cetate- :llL Fer
(TAE). The ;solaced ANA ~Nas Axtracted consecuti~rely with
Tris-equilibrated ~ilenol pH ~.O:chloroform (1.1),
chloroform, '~:len ethanol precipitated, dried and resuspended
in sterile water. LigaLions ~~rere performed according to the
manufacturer's instw.:cticns !Bethesda Researc.'~ Laboratories)
using the PsI diges~ad genomic DNA at a final concentration
of 20 ng/u. Ligations were done 18 hours at 14C.
Oligonucleot:ide primers were synthesized on an Applied
Biosystems model 394 DNA/RNA synthesizer. Primer 85 was
essentially the same as described by Earp et al., su ra,
except for an EcoRI site engineered at the 5' end and an
extra two ba:>es at 3' end. The sequence of both primers
used in the Ac; inverse PCR reaction are as follows:
AS (SEQ. ID NO. 7): 5' GATAGAATTCGGTACGGGATTTTCCCATCCTACTT 3'
85 (SEQ. ID NO. 8): 5' GGTAGAATTCGTTTTCGTT"TCCGTCCCGCAAGTT 3
PCR was ~~arried out using 25ng of circularized genomic
template DNA in a reaction containing 2uM of each primer,
.24mM of each dNTP, 3 units of Hot Tub polymerase (Amersham)
in a 1X reaction buffer supplied by the manufacturer.
Amplification was performed in a MJ Research Inc. model PTC-
100-96 thermocycler under the same conditions as described
by Earp ei. al., supra. Reaction products were
electrophorese~d on I% LMP agarose gels (Bethesda Research
Laboratories). The amplification product was isolated from
the gel using a Magic PCR kit (Promega) and re-amplified
using the above conditions.
cDNA Isolation
cDNA library screenings are commonly known among those
skilled in the art, and are described at Mariatis T. et
al., Molecular Cloning: A Laboratory Manual, Cold Spring
Harbor Laboratory, Cold Spring Harbor, New York). Libraries
CA 02203801 1999-12-07
were _reated as ~oil~ws. ANA
-=om -. :navs =assess was
isolated using a Guanidine ~hiocyanate method °ollowed by
banding in 3 cesium -7loride 3radient. Poll a.+RNA was
selected using oligo ~iT cellulose. Two c~NA libraries were
constructed vn the hectors pCDNAII !invitrogem and Uni-Zao
XR'~"iStratagen~=_) :sing ~ ug of mr-tl~lA for each according to the
manufacturer's instructions.
The 1.3 inverse PCR product was probed onto the arrayed
cDNA tassel library of about 1000 clones and from this a
single homologous clone with an insert size of about l.4kb
obtained. :Ct was 1550 base pairs and is graphically
depicted in figure 4. The genomic piece will, of course,
vary according to the background of the plant ~rom which it
is isolated and the introns may or may not be present.
This, however', shows how the Ac element appeared in this
isolate.
The 1.41cb was hybridized to the PvuII segregation
membrane to insure the 3.4kb co-segregating band found with
the inverse i?CR product was a new genomic region and not
small amounts of Ac DNA contained on the ends of the
fragment. The results are shown in the gel in Figure 5. As
can be seen, the l.4kb from the library hybridized in
sterile planta to the same 3.4kb fragment that co-segregated
with the male sterile phenotype and the purple spotted
kernels planta from the fertile heterozygous.
The l.4kb segment was then used against a second cDNA
tassel library and the full length cDNA was obtained, and
named MS45 (S1:.Q. ID NO. 1) .
Northern Analysis
Tissue from tassels, ears and leaves of sterile and
fertile plants was isolated as described previously, and a
Northern Bloi= analysis run on the extracts. Northern
analysis is also a commonly used technique by those skilled
in the art and is similar to Southern analysis except that
1G
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
RNA is isolated and place on an agarose gel. The RNA is
then hybridized with a labelled probe. Potter, E., et al.,
"Thyrotropin releasing hormone exerts rapid nuclear effects
to increase production of the primary prolactinr mRNA
transcript," Proc. Nat. Acad. 5ci. USA 78:6662-6666 (1981);
Lechelt, et al., supra. Total RNA was isolated from 1)
leaves of plants grown approximately 2 months; 2) tassels at
roughly the mid-vaculate stage; and 3) immature ears between
4.5 - 5.0 cm in length. Tissue was ground in liquid
nitrogen then sequentially treated with a detergent
extraction, a differential LiCl precipitation, and an
ethanol precipitation. The gel was hybridized with the
MS45cDNA isolated as described above. The cDNA hybridized
only with DNA from fertile tassels as can be seen in Fig. 6.
Revertants
To further confirm the gene as one critical to male
fertility, revertants were identified. Since it would not
be possible to distinguish normally fertile plants from
revertants, plants were selected that showed sterility, but
shed some pollen. These were crossed as males to unrelated
lines and no male sterile plants resulted. The MS45 cDNA
was recovered and analyzed to find the A_c had left a
"footprint" when transposing out of the gene of six base
pairs, keeping the sequence in frame. See Figure 7, showing
two amino acids are added, but the frame does not shift.
RFLP Mapping
The IPCR fragment was RFLP-mapped in a B73 X Mol7 F2
population. It mapped to chromosome 9L between probes and
Burr 7.21 as described in Maize Genetics Cooperation
Newsletter, 67:165 (Mar. 15, 1990) and depicted in Figure 8.
l~
CA 02203801 1997-06-26
Sequencing
Sequencing of the MS45 clone was accomplished using the
dideoxy chain termination method of Sanger, et al., Proc.
Nat. Acad. Sci. USA 74:5463-5464 (1977).
By referring to MS45 DNA, it is to be understood that
what is meant is a DNA sequence as set forth below (SEQ. ID
N0. 1) which produces the amino acid sequence also set forth
below (SEQ. ID N0. 2). One skilled in the art readily
appreciates that more than are three member codon may encode
the same amino acid sequence.
METHODS OF CONTROLLING MAhE FERTILITY
WITH MS45 DNA
It is evident to one skilled in the art that the DNA
described herein can be used in any one of a wide variety of
methods to control male fertility in plants. The following
are presented by way of illustrating several of these
methods and are not intended to limit the possible uses of
the DNA molecules herein described, nor the scope of the
invention.
Once one has a DNA molecule that is critical to male
fertility in plants, it is possible to create a sequence
which is in inverse orientation to the 5' to 3' normal
orientation of that DNA sequence. When this antisense
molecule is delivered into the plant, it prevents normal
expression of the male fertile sequence. It is believed the
antisense DNA transcribes to produce an RNA which is
complimentary to and capable of hybridizing to the mRNA
produced by the male fertility gene and thus inhibit
translation. The protein coded for by the mRNA is not
produced and cannot play its role in male fertility. With
the male fertility gene described herein, a construct .is
delivered to the plant having the MS45 DNA therein, the
construct having a transcriptional promoter segment, a
transcriptional termination segment and a DNA segment
18
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
producing an ribonucleotide sequence complimentary to a
ribonucleotide sequence of the MS45 DNA.
This use of antisense to inhibit or control expression
of a gene is known to one skilled in the art and is
described in detail at Inouye, U.S. Patent 5,190,931, issued
March 2, 1993. In one embodiment, the inventors there
describe cutting the DNA with restriction endonucleases, to
result in a relegated plasmid having lost a fragment between
two restriction sites and into which another DNA fragment
may be inserted. The normal DNA is digested, purified and a
fragment inserted in opposite orientation. They thus
inhibited expression of lip, OmpA and OmpC in bacteria and
controlled the development of coliphage SP using such
constructs. An antisense RNA complimentary to the 5' leader
region of the Om~A RNA, but not encompassing the Shine-
Dalgarno sequence was less effective than a transcript
covering the ribosome binding site and initiating codon. An
extensive view of antisense regulation is provided by Claude
Helene and Jean-Jacques Toulme in a review, "Specific
regulation of gene expression by antisense, sense and
antigene nucleic acids," Biochemica et Biophysica Acta
(1990) 99-125.
Another example of antisense and its use in inhibition
or control of a gene include antisense constructs to genes
encoding flavonoid biosynthesis in anthers to provide male
sterility. Vander Meer, et al., "Antisense inhibition of
flavonoid biosynthesis in petunia anthers results in male
sterility," The Plant Cell, 4:253-262 (March 1992).
Antisense chalcone synthesis genes with homologous
sequences to other genes expressed in anthers and a CaMV355
promoter result in male sterile white pollen. As can be
seen, use of antisense to control gene expression is well
- known. See also e.g. Bourque, June E. and Folk, William R.,
"Suppression of gene expression in plant cells utilizing
antisense sequences transcribed by RNA polymerase II", Plant
~9
CA 02203801 1997-04-25
WO 96/13588
PCT/US94/12444
Molecular Biology, 19:641-647 (1992); Weintrab, et al.,
Trends Gen. 1:22-25 (1985).
Another method of controlling gene expression is by
modification of transcriptional activators. During gene .
expression, the double stranded DNA is transcribed to a
corresponding single-stranded messenger RNA. The sense
strand separates from its antisense partner and enzymes
assemble an RNA molecule that compliments the sequence on
the antisense strand. The mRNA migrates to ribosomes which
read the encoded information to produce amino acids.
Transcription of eukaryotic genes is influenced by
various elements, including, transcriptional regulatory
proteins which bind to the DNA in a sequence-specific
manner. These transcriptional activators may be modified so
that they bind to the DNA, but cannot perform their normal
activator function. Transcriptional activators have two
domains, a binding domain, and an activation domain. By
altering the amino acid sequence of the transcriptional
activator proteins for a gene, providing a DNA sequence
which codes for the same, and delivering that DNA into the
plant, expression of the target gene may be blocked. See
Goff, S.A. et al. "Transactivation of the Anthocyanin
pathway structural genes with wild-type and altered cl
proteins" Maize Genetics Cooperation Newsletter 64:6 (March
1, 1990) .
A variation on this method is the isolation of genetic
suppressor elements encoding dominant negative mutant
proteins or inhibitory antisense RNA by random DNA
fragmentation and identified by functional selection for the
phenotype associated with suppression of the target. This
is what Holzmayer, et al.~ described in their article, .
"Isolation of dominant negative mutants and inhibitory
antisense RNA sequences by expression selection of random
DNA fragments" Nucleic Acids Research, Vol. 20, No. 4, 711
717 (Dec. 3, 1991). There they randomly fragmented
bacteriophage lambda DNA to protect E. coli cells from
zo
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
lambda-induced lysis. Multiple genetic suppressor elements
were isolated encoding either protein or antisense RNA
fragments.
Inhibition of normal gene expression has also been
observed when additional or over expression of an endogenous
gene was found to suppress gene expression. This "sense
inhibition", sometimes referred to as "co-suppression" has
been well documented. See e.g. Brussian, et al., "An
Arabidopsis mutant with reduced level of cab 140 RNA is a
result of cosuppression", The Plant Cell, 5:667-677 (June,
1993); Vander Krol et al., "Flavonoid genes in Petunia:
addition of a limited member of gene capus may lead to
suppression of gene expression" The Plant Cell 2:291-299
(April 1990) .
Other means of negative control regulation include
repression of gene transcription. In one system factors
contain DNA binding domains but lack functional activation
domains, competing with activators for binding to the same
sites and blocking activation. Others heterodimerize with
activators reducing either their DNA-binding affinity or
ability to activate transcription. Still other repressors
interact with activator factors when bound to DNA and block
transactivation function. A further type of down-regulators
comprises inhibitory proteins that sequester the activator
in a complex that is unable to bind DNA. See reviews by
Jackson, M.E., J. Cell Sci. 100:1-7 (1991); Jones, N., Curr.
Biol. 1:224-226 (1991); Mitchell. P.J. and Tjian, R.,
Science 245:371-378 (1989).
Direct mutation of the endogenous gene itself will also
change the male fertility gene to a male sterility gene.
Irradiation causes breakage and rearrangement of the
chromosomes and modification of the composition of
- ~ individual genes. Exposure to x-rays is a method of gene
. mutation well known for sometime. See e.g., Stadler, L.J.
"On the genetic nature of induced mutations in plants,"
Reprint, Proceedings of the Sixth International Congress of
z~
CA 02203801 1997-04-25
WO 96113588 PCT/US94/12444
Genetics, Vol. 1, 274-294 (1932). Other techniques include
exposure to chemical mutagens -such - as ethyl
methanesulfonate, and N-methyl-N-nitro-N-nitrosoguanidine,
as was accomplished by Neuffer, M.G., and Coe Jr., E.H. on
pollen grains and described in their early work at "Paraffin
oil technique for treating mature corn pollen with chemical
mutagens" Maydica XXIII (1978) 21-28; also, see Thurling, N.
& Depittayanan, "EMS induction of early flowering mutants in
spring Rape (Brassica na us)" Plant Breeding 108:177-184
(1992). Other methods include treatment with sodium azide
(Rao, B. "A case of genie male sterility induced by sodium
azide in Pearl Millet", Biol. Zentralbl. 104:579-521 (1985);
Conger, B.V. and- Carabia, J.V. "Mutagenic effectiveness and
efficiency of sodium azide versus ethyl methanesulfonate in
maize: induction of somatic mutations at the yg2 locus by
treatment of seeds differing in metabolic state and cell
population" Mutation Research 46:285-296 (1977)) and
exposure to gamma radiation (Filippetti, A. and Deface, C.,
"Improvement of seed yield in Vicia falsa L. by using
experimental mutagenesis II comparisons of gamma-radiation
and ethyl-methanesulfonate (EMS) in production of
morphological mutants" Euphytica 35:49-59x(1986)).
Thus, it is clearly evident to one skilled in the art,
that a male fertility gene, once identified, can be used in
a variety of methods to mediate male fertility in plants.
The foregoing illustrates but a few such methods which can
be used with a novel male fertile gene. Yet one more novel
method is described below created by the inventors of this
application. -
Constitutive Male Sterility Method
This invention differs from conventional approaches to ,
male sterility in plant breeding and seed-production in that
an inducible promoter is used to regulate expression of the
gene which is known to be critical to plant male fertility.
22
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
The first step in the practice of this invention is
therefore the selection of a gene on which fertility is
dependent. One type are the MS45 DNA molecules described,
supra.
The selected gene is cloned, its native promoter
enabled, and the modified gene is inserted into an
expression sequence with an inducible promoter responsive to
external control. Preferably, the promoter is one which
responds to application of a specific non-phytotoxic
chemical to the plant.
Using transformation and gene substitution, the gene is
inactivated in the genome of the plant and replaced by the
genetically-engineered gene incorporated into the expression
sequence with the inducible promoter.
This invention is unique in that the process results in
using the inducible promoter to induce fertility, not
sterility. In this invention, the selected gene's promoter
sequences are removed so that the gene is not transcribed
and the plant is male sterile. When it is desired to
increase the male-sterile plant, male fertility is restored
by inducing expression of the critical gene. In the
preferred embodiment this is accomplished by treating
growing male sterile plants with a specific non-phytotoxic
chemical.
Induction of the inducible promoter by chemical
treatment will be dependent on various factors associated
with the chemical treatment itself and various environmental
conditions at the time of treatment. If the critical gene
were normally "on," to be inactivated by chemical treatment,
a treatment failure would result in self-pollination and
. production and sale of inbred, rather than hybrid seed.
Seed laws that govern the sale of hybrid seed require a high
- degree of seed purity such that percentages of seed that do
not conform to the hybrid specification must be kept very
low. Because one maize plant can produce in excess of six
million pollen granules, even a limited treatment failure
..
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
could result in a high percentage of self-pollination. For
these reasons, the present invention is practiced in such a
manner that the gene is normally "off" and the corresponding
trait is not expressed, so that under normal conditions .
self-pollination cannot occur. In addition, by having the
critical gene normally "off," chemical treatment is not
necessary in the large-scale production of hybrid seed, so
that chemical usage (and associated expense) is minimized
and the risk of treatment failure is present only in the
carefully controlled, limited scale production of parent
seed, where self-pollination is desired. Since treatment
failure in such a case results in underproduction of pollen,
and since pollen is normally overproduced by a wide margin,
the process of this invention for production of parent seed
will tolerate a treatment failure rate as high as 70~ to 80~
with minimal effects on yield of parent seed.
In general, in accordance with the invention described
herein, the DNA molecule herein described is incorporated
into the plant along with a necessary promoter which is
inducible. The plant will be sterile since the DNA molecule
is not expressed-and when the promoter is induced, the plant
will be fertile. The native gene producing the DNA
molecule product is a normally fertile plant which may be
inactivated by any of a variety of methods described below,
such as backcrossing or homologous recombination.
Inducible Promoters
In the practice of this invention the promoter region
is removed from a cloned gene responsible for male fertility
and is replaced with a promoter that only responds to a .
specific external stimulus. Thus, the gene will not be
transcribed except in response to the external stimulus. As ,
long as the gene is not being transcribed, its gene product
-- which is necessary for completion of pollen development -
- is not produced. This causes a breakdown in one or more
24
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
of the biochemical/physiologic pathways of pollen
development, which results in male sterility. The plant can
only become fertile under the specific stimulus that
activates the selected promoter.
An example of a responsive promoter system that can be
used in the practice of this invention is the glutathione-S-
transferase (GST) system in maize. GSTs are a family of
enzymes that can detoxify a number of hydrophobic
electrophilic compounds that often are used as pre-emergent
herbicides (Wiegand, et al., "Messenger RNA Encoding a
Glutathione-S-Transferase Responsible for Herbicide
Tolerance in Maize is Induced in Response to Safener
Treatment", Plant Molecular Biology 7: 235-243, 1986). It
has been discovered that treating maize seed with GSTs
increases the tolerance of the maize to the herbicides.
Studies have shown that the GSTs are directly involved in
causing this enhanced tolerance. This action is primarily
mediated through a specific 1.1 kb mRNA transcription
product. In short, maize has a naturally occurring
quiescent gene already present that can respond to GSTs and
that can be induced to produce a gene product. This gene
has already been identified and cloned. Thus, in one
embodiment of this invention, the promoter is removed from
the GST responsive gene and attached to the male fertility
gene that previously has had its native promoter removed.
This engineered gene is the combination of a promoter that
responds to an external chemical stimulus and a gene
responsible for successful development of fertile pollen.
Gene Introduction
Several methods are known in the art for transferring
cloned DNA into maize. These include electroporation-
facilitated DNA uptake by maize protoplasts (Rhodes et al.,
"Genetically Transformed Maize Plants from Protoplasts",
Science, Vol. 240 (8 April 1988); treatment of maize
CA 02203801 1997-04-25
WO 96/13588 PC"T/US94/12444
protoplasts with polyethylene glycol (Lyznik et al., "Stable
Co-Transformation of Maize Protoplasts with Gus A and Neo
Genes", Plant Molecular Biology 13: 151-161, 1989); and
bombardment of maize cells with DNA laden microprojectiles
(Klein, et al., "Genetic Transformation of Maize Cells by
Particle Bombardment", Plant Physiol. (1989) 91, 440-444) .
and Klein, et al., "Factors Influencing Gene Delivery into
Zea Mays Cells by High-Velocity Microprojectiles",
Bio/Technology Vo_1. 6, May 1988).
Each of -these techniques has ~ advantages and
disadvantages. In each of the techniques, DNA from a
plasmid is genetically engineered such that it contains not
only the gene- of interest, but also s-electable and
screenable marker genes. A selectable marker gene is used
to select only those cells that have integrated copies of
the plasmid (the construction is such that the gene of
interest and the selectable and screenable genes are
transferred as a unit). The screenable gene provides
another check for the successful culturing of only those
cells carrying the genes of interest. A commonly used
selectable marker gene is neomycin phosphotransferase II
(NPTII). This gene conveys resistance to kanamycin, a
compound that- can be added directly to the growth media on
which the cells grow. Plant cells are normally susceptible
to kanamycin and, as a result, die. The presence of the
NPTII gene overcomes the effects of the kanamycin and each
cell with this gene remains viable. Another selectable
marker gene which can be employed in the practice of this
invention is the gene which confers resistance to the
herbicide glufosinate (Basta). A screenable gene commonly
used is the b-glucuronidase gene (GUS). The presence of
this gene~is characterized using a histochemical reaction in
which a sample of putatively transformed cells is treated
with a GUS assay solution. After an appropriate incubation,
the cells containing the GUS gene turn blue. Another
screenable gene is a transcriptional activator for
CA 02203801 1997-04-25
WO 96113588 PCT/US94/12444
anthocyanin biosynthesis, as described in Bowen, et al., "R
Genes as visual markers for corn transformation" Abstract
edit. Gallagher, Academic Press (Oct. 1989); Ludwig, et al.,
"A regulatory gene as a novel visible marker for maize
transformation" Science 247: 449-450 (Jan. 26, 1990). This
gene causes the synthesis of the pigment anthocyanin. Cells
transformed with a plasmid containing this gene turn red.
Preferably, the plasmid will contain both selectable and
screenable marker genes.
The plasmid containing one or more of these genes is
introduced into either maize protoplasts or callus cells by
any of the previously mentioned techniques. If the marker
gene is a selectable gene, only those cells that have
incorporated the DNA package survive under selection with
the appropriate phytotoxic agent. Once the appropriate
cells are identified and propagated, plants are regenerated.
Progeny from the transformed plants must be tested to insure
that the DNA package has been successfully integrated into
the plant genome.
Inactivation of Native Gene
It will be readily appreciated by those skilled in the
art that a wide variety of methods are known to disable the
native gene. Homologous recombination is but one of the
methods known to those skilled in the art for rendering a
native gene inoperative. Thus, when the engineered gene is
homologously recombined into the plant, the native gene will
be rendered inoperative. A good overview of this general
process is provided by Yoder, J. I., and Kmic, Eric, in
"Progress Towards Gene Targeting in Plants", Genetic
Engineering, Vol. 13 (Plenum Press, New York, 1991). At
page 265 of this reference, the authors note "gene targeting
can be used to silence or replace the endogenous gene with
an engineered allele; thus the phenotype of the altered
gene, or its regulatory sequences, can be evaluated in
27
CA 02203801 1997-04-25
WO 96!13588 PG"T/US94/12444
lp anta." It is pointed out that genetic recombination takes
place through breakage and reunion of DNA and the rejoining
mechanism pairs the complimentary DNA sequences. (See, e.g.
271, supra).
S A further discussion of intrachromosomal homologous
recombination in plants is discussed at Peterhans, A.,
Schlupmann, H., Basse, C. and Paszkowski, J.,
"Intrachromosomal Recombination in Plants", The EMBO
Journal, Vol. 9, No. 11, pp. 3437-3445, 1990.
A variety of different means, in addition to these
specific examples, would be available to one skilled in the
art. A still further example includes backcrossing, using
generally accepted plant breeding techniques, to in effect
"delete" the native gene. Backcrossing is often used in
plant breeding to transfer a specific desirable trait from
one inbred or source to an inbred that lacks that trait.
This can be accomplished for example by first crossing a
superior inbred (A) (recurrent parent) to a donor inbred
(non-recurrent parent), which carries the appropriate
genes) for the trait in question. The progeny of this
cross is then mated back to the superior recurrent parent
(A) followed by selection in the resultant progeny for the
desired trait to be transferred from the non-recurrent
parent. After five or more backcross generations with
selection for the desired trait, the progeny will be
heterozygous for loci controlling the characteristic being
transferred, but will be like the superior parent for most
or almost all other genes. The last backcross generation
would be selfed to give pure breeding progeny for the
genes) being transferred. A result of any backcrossing
method is that the "native" gene is replaced by the desired
gene . -
A unique method is discussed in the 1991 Science
magazine, reporting on prior work relating to using
"transgenic scissors". This article describes a method in
which scientists may remove a marker gene which is attached
28
CA 02203801 1997-04-25
WO 96/13588 PCT/US94/12444
to a gene having a desired trait in a plant. The "scissor,"
according to this method, is an enzyme obtained from a
bacterial virus known as "Cre" for control of recombination.
. Science, p. 1457, 6 December 1991. The enzyme is capable of
snipping out any DNA located between a pair of 34-base pair
. sequences, called lox, for locus of crossing over. This is
described in further detail in the patent application filed
by Du Pont, and published at WO 91/09957.
Sterility Selection And Fertility Restoration
After the gene is introduced into a plant, the
appropriate plant types are selected, that is plants that
are male sterile. These plants are male sterile because the
isolated and cloned male fertility gene does not have its
native promoter and, therefore, is not producing its gene
product -that is crucial to successful pollen development.
Therefore, the engineered gene acts as a recessive mutant
allele of -that gene. In normal plant biotechnology, once
the desired genotype is identified following transformation
and regeneration, the plants are selfed to recover that
genotype. However, in the practice of this invention, the
desired genotype cannot be selfed at the first generation
because it is male sterile. To obtain progeny, fertility
must be induced by spraying the plants with a compound which
induces transcription of the gene by activating the altered
promoter. In the case of the GST promoters, the compound is
preferably a GST-inducing compound such as N,N-diallyl-2-2-
dichloroacetanide. The promoter attached to the male
fertility gene responds to this chemical and causes the
. transcription of the gene to begin. Once this occurs, the
normal gene product is produced from the gene and some level
of male fertility is induced. Pollen from this plant is
then used to effect pollination of the original selected
genotype.
29
CA 02203801 1997-04-25
WO 96113588 PCT/US94/12444
Once the initial isolation and propagation of the
desired genotype is completed, the procedure is more
straightforward. Only inbreds that are used as female
parents in hybrid crosses are transformed into male sterile
S variants. Once they are transformed, the amount of male
sterile/female fertile seed must be increased. This is
accomplished by planting in an isolated area (away from
other maize pollen) and spraying with a chemical to which
the promoter responds. Spraying induces the promoter to
start transcription of the gene attached to it. This will
produce some degree of fertility. A particular advantage of
this system in comparison to systems such as that disclosed
in PCT publication W089/10396 of Mariani et al (based on
Intl. Appl. No. PCT/EP89/00495), in which sterility is
induced, is that the treatment does not have to be 100
effective, because normally much more pollen is produced by
a maize plant than is actually needed for fertilization of
all available silks. Therefore; even low fertility
restoration will be effective in obtaining acceptable levels
of seed increase. At the same time, self-pollination does
not occur in hybrid seed production because the plants of
this invention are normally male sterile and must be treated
to become fertile. In systems in which sterility is
induced, induction of sterility must be 100 effective to
avoid self-pollination when hybrid seed is produced.
All the seed harvested continues to be homozygous and
sterile since the fertility is only restored in a single
parent generation by treatment with the fertility inducing
chemical. This seed is then used in a hybrid production
field where it is used as a female parent. Because the
plants are male sterile, they do not have to be detasseled.
All of the hybrid plants produced from such seed are male
fertile because the resulting progeny inherit one modified ,
gene from the female parent and one normal gene from the
male parent. Normal pollen production occurs.
z0
CA 02203801 1997-06-26
SEQUENCE LISTING
(1) GENERAL INf'ORMATIOH:
(i) APPLIG1NT: Pioneer Hi-Bred International, Inc.
(ii) TITLE OF.INVENTION: Nucleotide Sequences Mediating Male
- Fertility and Method of Uaing Same
(iii) NUMBER OF SEQUENCES: 8
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: Pioneer Hi-Bred International, Inc.
(B) STREET: 700 Capital Square, 400 Locust Street
(C) CITY: Des Moines
(D) STATE: Iorua
(E) COUNTRY: U.S.
(F) ZIP: 50309
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTt~IARE: PatentIn Release #1.0, Version X1.25
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER: PCT/US94/12944
(B) FILING DATE: 28 October, 1999 (28.10.94)
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE:
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: SWEENEY, Patricia A.: ROTH, Michael J.:
YATES, Michael J. i SIMON, Soma G.
(B) REGISTRATION NUMBER:
(C) REFEREHCE/DOCKET NUMBER: 12582-PCT
(ix) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: (515) 248 4897
(B) TELEFAX: (515) 248-4894
(2) INFORMATION FOR SEQ ID NO:1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1419 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: cDNA
31
CA 02203801 1997-06-26
(xi) SEQUF.I~ICE DESCRIPTION: SEQ ID NO:1:
GAATTCGGCA CGAGGTCCAC CAGCATGGAG GAGAAGAGGA AGCTGCAGTG60
GCGGCG~F1GGG
CGTGATGGCA TCGTGCAGTA CCCTCACCTG TTCTTCGCGG CCCTGGCCCT120
GGCCCTCCTA
GTCGCGGACC CGTTCGGCCT CAGTCCGCTG GCCGAGGTCG ACTACCGGCC180
GGTGAAGCAC
GAGCTCGCGC CGTACGGGGA GGTCATGGGC AGCTGGCCCA GAGACAATGC240
CAGCCGGCTC
AGGCGCGGGA GGCTGGAGTT CGTCGGCGAG GTGTTCGGGC CGGAGTCCAT300
CGAGTTCGAT
CTCCAGGGCC GCGGGCCGTA CGCCGGCCTC GCCGACGGCC GCGTCGTGCG360
GTGGATGGGC
GAGGAGGCCG GGTGGGAGAC GTTCGCCGTC ATGAATCCTG ACTGGTCAGA420
AGAAGTCTGT
GCCAATGGAG TGAACTCAAC GACGAGGAAG CAGCACGAGA AGGAGGAGTT480
CTfCGGCCGG
CCGCTCGGCC TGAGGTTCCA CGGGGAGACC GGCGAGCTCT ACGTCGCCGA540
CGCGTACTAC
GGTCTCATGG TCGTTGGCCA GAGCGGCGGC GTGGCGTCCT CCGTCGCGa~G600
GGAAGCCGAC
GGGGACCCCA TCCGGTTCGC GAACGACCTC GATGTGCACA GGAATGGATC660
CGTATTCTTC
ACTGACACGA GCATGAGATA CAGCAGRAAG GACCATCTGA ACATCCTGTT720
AGAAGGAGAA
GGCACCGGGA GGCTGCTCAG GTACGATCCA GAAACAAGTG CTGTCCATGT780
CGTGCTCAAG
GGACTGGTGT TCCCAAACGG CGTGCAGATC TCAGe'~AGACC ATCAGTTTCT890
TCTCTTCTCC
GAGACAACAA ACTGCAGGAT AATGAGGTAC TGGCTGGAAG GCCCAAGAGC900
GAGCGAGGTA
GAGGTGTTCG CGAACCTGCC GGGCTTCCCC GACAACGTGC GCTCCAACGG960
CAGGGGCCAG
TTCTGGGTGG CGATCGACTG CTGCCGGACG CCAGCGCAGG AGGTGTTCGC1020
CAAGAGGCCG
TGGCTCCGGA CCCTGTACTT C1~AGTTCCCG CTGTCGCTCA AGGTGCTCAC1080
TTGGAAGGCC
GCCAGGAGGA TGCACACGGT GCTCGCGCTC CTCGACGGCG AAGGGCGCGT1140
CGTGGAGGTG
CTCGAGGACC GGGGCCACGA GGTGATGAAG CTGGTGAGCG AGGTGCGGGA1200
GGTGGGCAGC
AAGCTGTGGA TCGGAACCGT GGCGCACAAC CACATCGCCA CCATCCCCTA1260
CCCTTTAGAG
GACTAACCAT GATCTATGCT GTTTCAATGC CTCCTAATCT GTGTACGTCT1320
ATAAATGTCT
AATGCAGTCA CTGGTTGTAA TC1TGTT'TGT GTTTGGCAAA TTGGCATAAT1380
AATGGACAGA
TTCAATGGGC AAAAAAAP1AA AAP~AAAAAAA AAACTCGAG 1419
(2) INFORMATION FOR SEQ ID N0:2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 473 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: lines=
32
CA 02203801 1997-06-26
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:2:
Glu Phe Gly Thr Azg Ser Thr Ser Met Glu Glu Lys Arg Lys Leu Gln
I 5 10 15
Trp Arg Arg Gly Arg Asp Gly Ile Val Gln Tyr Pro His Leu Phe Phe
ZO 25 30
Ala Ala Leu Ala Leu Ala Leu Leu Val Ala Asp Pro Phe Gly Leu Ser
35 . 40 45
Pro Leu Ala Glu Val Asp Tyr Arg Pro Val Lys His Glu Leu Ala Pro
50 55 60
Tyr Gly Glu Val Met Gly Ser Trp Pro Arg Asp Asn Ala Ser Arg Leu
65 70 75 80
Arg Arg Gly Arg Leu Glu Phe Val Gly Glu Val Phe Gly Pro Glu 3er
85 90 95
Ile Glu Phe Asp Leu Gln Gly Arg Gly Pro Tyr Ala Gly Leu Ala Aap
100 , 105 110
Gly Arg Val Val Arg Trp Het Gly Glu Glu Ala Gly Trp Glu Thr Phe
115 120 I25
Ala Val Met Asn Pro Asp Trp Ser Glu Glu Val Cys Ala Asn Gly Val
I30 I35 140
Asn Set Thr Thr Arg Lys Gln fiis Glu Lys Glu Glu Phe Cys Gly Azg
145 150 155 160
Pro Leu Gly Leu Arg Phe His Gly Glu Thr Gly Glu Leu Tyr Val Ala
165 170 175
Asp Ala Tyr Tyr Gly Leu Met Val Val Gly Gln Ser Gly Gly Val Ala
180 185 190
Ser Ser Val Ala Arg Glu A1a Asp Gly Asp Pro Ile Arg Phe Ala Asn
195 200 205
Asp Leu Asp Val His Arg Asn Gly Ser Val Phe Phe Thr Asp Thr Set
210 2I5 220
Met Arg Tyr Ser Arg Lys Asp His Leu Asn Ile Leu Leu Glu Gly Glu
225 230 235 240
Gly Thr Gly Arg Leu Leu Arg TYr ~P Pro Glu Thr Ser Ala Val His
295 250 255
Val Val Leu Lys Gly Leu Val Phe Pro Asn Gly Val Gln Ile Ser Glu
260 ~ 265 270
Asp iiis Gln Phe Leu Leu Phe Ser Glu Thr Thr Asn Cys Arg Ile Met
275 280 285
Arg Tyr Trp Leu Glu Gly Pro Arg Ala Ser Glu Val Glu Val Phe Ala
290 295 300
Aan Leu Pro Gly Phe Pro Asp Asn Val Arg Ser Asn Gly Arg Gly Gln
305 310 315 320
33
CA 02203801 1997-06-26
Phe Trp Val Ala Zle Asp Cys Cys Arg Thr Pro Ala Gln Glu Val Phe
325 330 335
Ala Lys Arg Pro Tzp Leu Arg Thr Leu Tyr Phe Lys Phe Pro Leu Ser
340 345 350
Leu Lys Val Leu Thr Trp Lys Ala Ala Arg Arg Met His Thr Val Leu
355 - 360 365
Ala Leu Leu Asp Gly Glu Gly Arg Val Val Glu Val Leu Glu Asp Arg
370 375 380
Gly His Glu Val Met Lys Leu Val Ser Glu Val Arg Glu Val Gly Ser
385 390 395 400
Lys Leu Trp Ile Gly Thr Val Ala His Asn iiis Ile Ala Thr Ile Pro
405 410 415
Tyr Pro Leu Glu Asp Xaa Pro Xaa Ser Met Leu Phe Gln Cps Leu Leu
420 425 430
Ile Cys Val Arg Leu Xaa Met Ser Asn Ala Val Thr Gly Cya Asn Leu
435 440 445
Val Cys Val Trp Gln Ile Gly Ile Ile Met Aap Arg Phe Asn Gly Gln
450 955 460
Lys Lys Lys Lys Lys Lys Lys Leu Glu
465 470
(2) INFORMATION FOR SEQ ID N0:3:
ti) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 47 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:3:
GCCCTGGCCC TGGCCCTCCT AGTCGCGGTC GCGACCCGTT CGGCCTC 47
(2) IHE'ORMATIOH FOR SEQ ID N0:4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 16 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID H0:4:
Ala Leu Ala Leu Ala Leu Leu Val Ala Val Ala Aap Pro Phe Gly Leu
1 5 IO 15
34
CA 02203801 1997-06-26
(2) IHFORNATION FOR SEQ ID N0:5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: I1 base pairs
(8) TYPE: nucleic acid
(C) STRAHDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: S:
GCCCTGGCCC TGGCCCTCCT AGTCGCGACC CGTTCGGCCT C 41
(2) INFORMATION FOR SEQ ID N0:6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 14 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:6:
Ala Leu Ala Leu Ala Leu Leu Val Ala Asp Pro Phe Gly Leu
1 5 10
(2) INFORMATION FOR SEQ ID N0:7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 35 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:7:
GATAGAATTC GGTACGGGAT TTTCCCATCC TACTT 35
(2) INFORMATION FOR SEQ ID N0:8:
(i) SEQUENCE CAARF1CTERISTICS:
(A) LENGTH: 34 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:8:
GGTAGAATTC GTrTTCGTTT CCGTCCCGCA AGTT 34
O