Note: Descriptions are shown in the official language in which they were submitted.
CA 02210731 1997-07-17
W096/23078 PCT~S95/12429
COMPUTER SYSTEM STORING AND ANALYZING MICROBIOLOGICAL DATA
FIELD OF INVENTION
This invention relates to computer database technology
applied to genetic data and corresponding cell information.
More specifically, a relational database system that stores
DNA sequences, the corresponding source data, and other
related scientific data is disclosed.
BACKGROUND OF THE I~ENTION
Relational databases are generally known in the art. See
for example C.J. Date, "An Introduction To Database Systems"
Addison-Wesley Publishing Company, 1982 (particularly, Part
2).
In general, a relational database can be characterized as
a system for storing data represented as a plurality of
tables. A row of each table, also referred to as a tuple,
represents a record of information. A column is essentially a
collection of values for the same field of the stored records.
Each column is also referred to as an attribute of the stored
records. In other words, each record in a given table of a
relational database includes a set of fields that correspond
to the attributes of the table. A set of all the values from
which the actual values of an attribute can be drawn is
referred to as a domain. As discussed on page 65 of the
above-referenced text, "a crucial feature of relational data
structure is that associations between tuples (rows) are
represented solely by data values in columns drawn from a
common domain."
Previously most of the analysis of genetic information
has been done using chemical methods in a laboratory.
Computerized research tools have been limited essentially to
performing comparisons of sequence information to determine
whether a particular genetic sequence has been previously
identified. Such tools may provide effective searching
techn; ques for genetic sequences; however, they do not store
and manipulate diverse scientific information, such as the
correlation between the cDNA sequences and the types of cells
from which they were derived. Thus, the existing computerized
CA 02210731 1997-07-17
W 096/23078 PCTrUS95/12429
tools have only a very limited use in the field of diagnostics
and drug development research. Presently, there is a pressing
need to develop a computer system which stores genetic data
and related cell information in a well organized form so as to
enable scientists to analyze such data efficiently.
SUMMARY OF THE INVENTION
In accordance with the present lnvention a relational
database for storing biological information is provided. The
relational database is organized as a collection of tables
each of which stores specific records of biological
information. The records are interrelated so that each table
includes a column which is common with at least one other
table. This enables database queries that can search the
database essentially on any attribute of any table.
In a preferred embodiment of the invention, the database
contains cDNA sequencing data and corresponding match logs
indicating the correlation between presently identified cDNA
sequences and previously known sequences. In addition, a
variety of tables of the database store historical data
related to identification of a particular cDNA sequence. Such
tables include the identification of the biological source;
cell culture and treatment data; mRNA preparation data; cDNA
construction data; clone preparation data including tables for
inoculation, preparation, fluorometer data, and excision.
The interrelated information in the database enables the
design of various queries useful in scientific analysis and
other applications. For example, such functions as abundance
analysis which allows one to determine the frequency with
which an RNA transcript appears within a certain source tissue
can be performed using database of the preferred embodiment.
Other analytical results that have previously been obtained
using laboratory chemical techniques can be determined using
database queries. One such application is subtraction
analysis.
BRIEF DESCRIPTION OF THE DRAWINGS
The foregoing features of the present invention may be
fully understood from the following detailed disclosure of a
CA 02210731 1997-07-17
W 096123078 PCTrUS95/12429
specific preferred embodiment in conjunction with the
accompanying drawings in which:
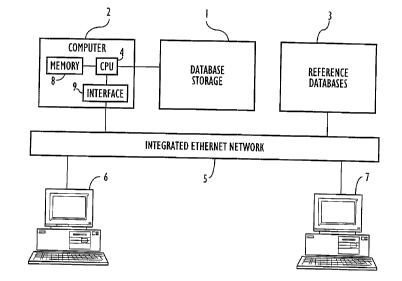
Fig. 1 symbolically depicts an overall architecture of
the system of the preferred embodiment of the present
invention.
Fig. 2 is a flowchart symbolically depicting the process
~ of cloning and sequencing cDNAs.
Figs. 3A, 3B and 4-10 illustrate portions of the
biological relational database of the preferred embodiment of
the present invention.
Fig. 11 illustrates an example of the output of an
abundance analysis query of the relational database of the
preferred embodiment.
Fig. 12 illustrates an example of the output of a
subtraction analysis query using the database of the
preferred embodiment.
DETAILED DESCRIPTION OF THE PREFERRED EMBODIMENT
According to the preferred embodiment of the present
invention, a system for storing, tracking and manipulating
the genetic data is organized as a relational database. As
illustrated in Fig. 1, the users of the system at their
workstations (6 and 7) can access one or more relational
databases via an integrated Ethernet network 5. The
workstations (6, 7) are typically personal computers known in
the art that usually include data entry means, output
devices, display, CPU, memory (RAM and ROM) and interfaces to
network 5. Database storage 1 illustrates the database of
the preferred embodiment of the present invention, which is
stored at a file server connected to network 5. As
illustrated, it is supported by computer 2, which, as known
in the art, usually includes CPU 4, data storage means 8,
interfaces to the network 9, and input and output devices
(not shown). Reference databases 3 illustrate sources of
data which, for example, may be searched as part of the use
of database 1. Such databases may, for example, include
other sequence, nucleic acid, protein, and motif databases.
SUBSTITUTE SHEET (RUEE 26)
CA 02210731 1997-07-17
W O 96/23078 PCT~US95/12429
It is well known that each cell in an organism such as
the human body, contains a complete set of genes or genetic
information. These genes are either active or inactive at
different times in the cell's life cycle. Some genes are
active in all cells and are necessary for normal and common
functions, or housekeeping duties. Gther genes are only
active in a particular cell type, because they specify and
regulate functions peculiar to a tissue or an organ under
normal conditions. Finally, there are genes which are
activated only in response to stress or disease. Some stress
genes, which activate in several cell types, respond to the
general alarm. Other stress genes are very specific and only
activate in a particular cell type. Thus genes can be grouped
into very small and specific subsets or subsets of varying,
larger sizes. The classification and understanding of these
nested sets of genes are important in the diagnosis and
treatment of disease.
Genes, or double-stranded deoxyribonucleic acid (DNA),
are activated by the transcription or copying of the sense
strand of the DNA molecule into single-stranded messenger
ribonucleic acid (mRNA). The message inherent in the mRNA
sequence is subsequently translated into amino acids, the
molecular building blocks of the polypeptides or proteins that
function structurally or enzymatically in the cell.
The activities taking place at any one time and the
relative importance of those activities are reflected in the
numbers of mRNA molecules found in the cell. Some mRNAs
(housekeeping) are always present, and their numbers remain
fairly stable in normal cells of any tissue. These mRNAs (eg.
actin) represent and carry out the constant background
activity essential to most cell types (the exception to this
case is a mature, differentiated red blood cell which lacks
DNA but has a set of mRNAs or enzymes which function for the
remainder of its life). In contrast, the RNAs (routine) which
carry out the duties of a particular cell type are only
activated in that cell type, and the numbers of routine mRNAs
will be stable under normal conditions. If that particular
CA 02210731 1997-07-17
W096/23078 PCT~S95/12429
cell type is stressed or exposed to disease, the numbers of
routine mRNAs fluctuate as genes which respond to the
stress/disease are activated. These stress/disease mRNAs have
priority over other routine or housekeeping mRNAs, and they
quickly increase in number.
For example, the housekeeping genes of brain cells and
liver cells are shared; cel~s from both organs transcribe the
mRNAs that produce the enzymes required to process incoming
molecules of glucose. However, the mRNAs that make proteins
for the normal functions of a pituitary cell are different
from the mRNAs of a liver Kupffer cell although each is
functioning normally. Likewise, the set of mRNAs from a
diseased liver cell differ from those from a normal liver
cell. In each case, a different and diverse subset of mRNAs
characterizes the cell in a particular situation at a
particular time.
The database of the preferred embodiment provides the
storage, manipulation, and retrieval of the information which
relates to the classification and characterization of unique
populations of mRNAs. On the basis of this information,
scientists can diagnose diseases and design specific
treatments. The wealth of detailed information provides clues
to earlier diagnosis and treatment which contribute to rapid
healing and help avoid permanent impairment or death.
The database system of the present invention takes
advantage of the powerful capabilities of modern computers by
storing genetic information in association with a large amount
of related information. More specifically, in the preferred
embodiment, the information on essentially all the steps of
obt~;n;ng tissue, extracting transcripts, cloning, and
identifying cDNA sequences is stored in various relational
tables. Thus, the database of the present invention allows
one to backtrack through the steps performed in the laboratory
in identifying the cDNA sequence. The diverse data stored in
the system of the present invention will in many instances
answer questions frequently asked in molecular biology and
pharmacology without requiring actual experiments, such as:
-
CA 02210731 1997-07-17
W 096/23078 PCTrUS95/12429
What are the most active genes and common functions of a
particular cell type? What happens to housekeeping and cell-
specific routine functions during stress or disease? What
genes are diagnostic of the normal or disease state? Which
gene products are targets for pharmaceutical intervention?
Fig. 2 illustrates the steps of preparing genetic data
stored in the database of the present invention. The
information associated with the steps of Fig. 2 is stored in
the database as tables depicted in Figs. 3A, 3B and 4 through
10. In Fig. 2 the first step 10 is cell preparation. Cell
preparation 10 includes the steps of obtaining and growing
the cells so as to prepare them for ~NA extraction. The
following step 20 indicates the processes associated with
extracting mRNA from the cell. Next, at step 30, the mRNA
becomes cDNA. Alternatively, the cDNA fragment can be
received from an outside source or collaborator without
performing steps 10 and 20. Once the cDN~ molecule is
obtained, it is cloned at step 40 and sequenced at step 50.
The sequence that is obtained at step 50 is then compared at
step 60 to known sequences on the genetic database. Finally,
the function of the DNA sequence is determined at step 70.
Figs. 3A, 3B and 4-10 schematically illustrate the
tables of the database of the preferred embodiment.
Exemplary fields (or attributes) are depicted within each
box, and each table includes an attribute having a domain
which is common to at least one other table. For example,
consider the table indicated as 130 "Biological Source" and
the table indicated as 140 "Cell Culture/Treatment". In
these two tables the common domain is bio_source_ID. Also,
notice that Arrow 135, one end of which is labelled "1" and
the other end is labelled "M", indicates that for each one
tuple in the Biological Source table there may be more than
one tuple in the Cell Culture/Treatment table.
The data received and obtained in steps 10-30 of Fig. 2
is stored in the Library Preparation portion of the database
of the present invention (Figs. 3A and 3B). This data
includes information relating to the biological source of the
cells
SUBSTITUTE SHEET (RULI 26)
-
CA 022l073l l997-07-l7
WO 96/23078 PCTrUS95/12429
used to obtain the cDNA (boxes 130, 110, 120), cell culture
and treatment (boxes 140, 180), mRNA preparation (box 150)
and cDNA construction (boxes 170, 160). More specifically
box 130 depicts the table for storing the biological source
information. The source may be cells grown in tissue culture
or cells obtained during surgery from a single individual or
a pooled sample, e.g., pituitary glands obtained from
patients of both sexes and a range of ages. In the preferred
embodiment the biological source table 130 contains
attributes as depicted in Fig. 3A, such as tissue, organ,
gender, age, pathology, etc. The biological source may
reflect a normal, treated or diseased state. A person
skilled in the art will realize that, if desirable, certain
other biological source information can be stored; and on the
basis of this disclosure, such person will be able to include
other relevant attributes if desired.
The data regarding the collaborators, i.e contributors
of a biological source, is stored in table 110 as depicted in
Fig. 3A, and the information regarding the cell suppliers
contributing to biological sources is stored in table 120.
The source_ID attribute of the biological source table 130
corresponds to either collaborator_ID or supplier_ID of
tables 110 and 120 respectively.
Part of the cell preparation procedure includes the cell
culture and treatment process. Cell culture is carried out
in containers of known size or volume. Density is usually
reported as cells per milliliter (of liquid media) and is
monitored to maintain a healthy cell culture. Density at the
time cells are harvested may be measured either as cell
number or as grams per liter. Treatment may vary. Induction
with a chemical can change a cell from an immature form,
monocyte, to a mature one, macrophage. Stimulation or
activation with a different chemical causes the macrophage to
ingest and digest invading bacteria.
In some cases, a cell culture is split into two or more
parts, with one subsample maintained in its normal growth
mode (as the biological control) and other subsample(s)
subjected
SUBSTITUTE SHEET (RULE 26)
CA 02210731 1997-07-17
W 096/23078 PCTrUS95/12429
to activation and/or stimulation. In a simple scenario, a
subsample of control cells is compared with a subsample of
cells treated with a drug candidate. Drug doses and length
of treatment may vary.
The cell culture and treatment information is stored in
table 140 in Fig. 3A. The attributes of the cell
culture/treatment table 140 of the preferred embodiment are
listed in the table 140. These attributes include such
information as cell density, cell quantity, and treatment.
The cell culture/treatment table 140 has the attribute
bio_source_ID in common with table 130. Specific treatment
information is stored in table 180 which includes the
attributes depicted in Fig. 3A. The culture_ID attribute is
common to both tables 140 and 180.
Step 20 of mRNA preparation begins with the extraction
of total ribonucleic acid (RNA) from cells of a known weight
or volume according to a standard protocol. The protocol and
any modifications are recorded. The extracted RNA is
optionally fractionated to recover the messenger or
transcript RNA (mRNA); if it is fractionated then yield is
calculated as a percent (mRNA/total RNA). The normal
function of mRNAs in the cell is to produce peptides or
proteins.
Spectrophotometry and gel appearance are used to check
the ~uality of the mRNA. In spectrophotometry, an optical
density readout of 1.8, derived from a 260 lambda/280 lambda
ratio, indicates high quality RNA, not unduly contaminated
with DNA or proteins. A subsample of this mRNA is checked
further by moving it via electric current (electrophoresis)
through an agarose gel. The gel is ~min~d visually for
contaminating DNA, which generally moves with higher
molecular weight substances than the RNA, or for degraded
mRNA, which forms a fuzzy rather than a sharp band or signal.
The data related to the mRNA preparation is stored in
table 150 in Fig. 3B. Table 150 has an attribute
mRNA_source_ID, which correlates with either attribute
culture_ID of table 140 or attribute Bio_source_ID of table
130, and an attribute mRNA_source, which identifies the table
SUBSTlTUTE SHEET (RULE 26)
CA 02210731 1997-07-17
W096/23078 PCT~S95112429
with which mRNA_source_ID=correlates. These two attributes
in combination, therefore, link records of table 150 to
tables 140 and table 130.
Next, as shown in step 30 of Fig. 2, a cDNA sequence is
derived from the mRNA. The cDN~ con.struction requires the
conversion of mRNA into complementary DNA (cDNA) preferably
using oligo DT, random priming, reverse transcription or
other protocols, as known in the art. Useful cloning sites
are designed into the bacteriophage into which the DNA is
packaged or incorporated. Packaging or plating efficiency is
determined by ~x~m;n;ng the number of primary plaques, i.e.,
individual bacterial colonies, which resulted from a
particular experiment. Information ls recorded about the
genetic background of host bacterium and the titer of the
bacteriophage, before and after amplification. The quality
of the library is determined by screening for the actin gene,
present in all normal or diseased cell types, and estimation
of the size of the cDNA fragment which has been inserted
(insert size).
The data related to the cDNA construction is stored in
table 170 in Fig. 3B. As apparent to a person skilled in the
art, the attributes of this table depicted in Fig. 3B provide
detailed information about the cDNA construction. Note that
tables 170 and 150 have a common attribute mRNA_prep_ID.
Preprocessed cDNA fragments can be purchased from an
outside supplier or obtained from a collaborator or customer.
In such a case, the relevant data is stored in the cDNA
supplier table 160 is stored in the database. The Table 160
has the attribute supplier_ID which is also a part of the
cDNA construction table 170.
As depicted in Fig. 2, after the cDNA has been
constructed, the cloning process, is performed. The portion
of the database depicted in Fig. 4 relates to the clone
preparation data that is obtained during the cloning process
and includes information relating to excision (box 190),
inoculation (box 200), preparation (box 210), fluorometer
SUBSTITUTE SHEET (RULE 26)
-
CA 022l073l l997-07-l7
W 096/23078 PCTrUS95/12~29
(boxes 220, 230, 240). Cloning includes the steps of
excision, inoculation and preparation.
Excision is the removal of the cDNA fragment from the
vector. This follows an overnight cultivation and induced
amplification of the vector in the SOLR bacterial host cells
which comprise each culture. The plasmid DNA is separated
from the bacterial DNA and quantitated fluorometrically before
sequencing. The table that stores data related to excision is
illustrated as 190 in Fig. 4. The e~cision table 190 has an
attribute cDNA_const_ID in common with cDNA construction table
170.
Inoculation involves growing up or increasing the number
of bacteria in a liquid growth medium. As soon as the
required cell density (optimum growth) is reached, the culture
is plated (streaked or spread thinly) on solid growth media.
Individual colonies which arise on the surface of this solid
media may be subcultured in tubes or microtiter plate wells of
liquid media as pure cultures. The collection of bacterial
cultures corresponds to the numbers and type of genes which
were active in the source tissue. The data that relates to
inoculation is stored in the table illustrated as 200. The
attribute plating_ID of the table 200 is common with the same
attribute in the table 190.
Fluorometers are used to quantitate the cDNA in nanograms
or micrograms per microliter. The total amount of cDNA must
be determined to calculate the amount which will be processed
and separated electrophoretically in any particular lane of a
sequencing gel. The remainder of the sample is stored for
future use. Fluorimetry procedures determine cDNA purity and
help predict performance in subsequent procedures.
The fluorometer information is stored in the tables
illustrated as 220, 230, and 240. More specifically, the data
from the fluorometer analysis is stored as the attributes of
fluorometer log table 220. Table 230 (Fluorometer) stores the
information regarding the instrument and, as illustrated in
Fig. 4, has an attribute fluorometer_~D in common with the
Table 220. The fluorometer calibration table 240 is
-
CA 02210731 1997-07-17
W 096/23078 PCTrUS95/12429
associated with the fluorometer table 230 via a common
calibration ID attribute.
After fluorometry analysis, the cDNAs are prepared for
sequencing. Preparation of the cDNAs for sequencing is
recorded along with the methods (and their modifications) used
at that time. The scientists (SWAT) troubleshoot the
sequencing process and track the results of their custom
protocols. The preparation table is illustrated as 210.
Table 250, clone log, combines the information regarding
the cloning process as illustrated in Fig. 4. In particular,
it contains an attribute Inoculation_ID which is also an
attribute of the inoculation table 200. An attribute clone_ID
is shared with the fluorometer log table 220. An attribute
Preparation_ID is also a part of the preparation table 210.
The dead_or_alive attribute of the clone log table 250, for
example, identifies dead clones in which the plasmid
preparation did not yield enough DNA to sequence.
The data related to the process of sequencing is stored
as depicted in the sequencing portion of the database
illustrated in Fig. 5. This portion includes information
relating to specifications of the sequence and related
information. It includes the sequencing log (box 300) the
sequencing gel (box 280), the reaction set (box 270) and the
sequence archive (box 290). The specification of the sequence
and related information are stored as attributes in sequencing
log table 300. It should be noted that a clone can be
sequenced multiple times. Table 260 (sequencing link) links
the clone log table 250 with the sequencing log table 300.
The sequencing link table 260 contains a clone_ID attribute,
which is in common with the same attribute in the clone log
table 250 and a sequencing_log_ID attribute which is also
included in the table 300.
Sequencing of the cDNAs is performed on an automated ABI
system. The sequencing gel is evaluated for the sharpness and
darkness of the signal which each of the deoxyribonucleotides
or bases (adenine, cytosine, guanine, and thymidine) display,
their physical proximity to one another in the gel, and the
CA 02210731 1997-07-17
WO 96t23078 PCT~US95/12429
clarity of the gel background. These characteristics must
fall within certain parameters for the automatic gel reader to
produce a sequence. An electronic chromatogram, or gel
representation, is stored in the computer system for future
reference.
The tracking of all gel information is reflected by a gel
key. The gel, the conditions under which it was run, the time
required for the gel run, the individual machine/instrument
used, staff and biological preparation are recorded whether or
not a usable sequence is obtained. This data is stored in the
gel key table 280 which has an attribute Gel_key_ID which is
common with the same attribute in the sequence log table 300.
The biological preparation, which runs on the sequencing
gel, is referred to as the reaction set. The Catalyst is the
Model 800 Molecular Biology Station in which robots perform
amplifications, PCRs, dilutions and additions of fluorescent
dyes to the cDNAs. The data related to the reaction set is
stored in table 270. This table has an attribute entitled
Reaction_Set_ID which is also part of the sequence log table
300.
The sequence archive is activated if a sequence is
obtained. The sequence is rated as normal or variant and
evaluated for usefulness and subsequent storage in the
computer system database. Variant sequences identified at
this time may be designated express (see discussion below).
The sequence archive data is stored in the table 290 which has
the sequence_ID attribute in common wlth the Sequence Log
table 300.
Fig. 6 illustrates a portion of the database for storing
information regarding the sequencing equipment. The Sequencer
Maintenance Log table 900 collects information on maintenance
of each DNA sequencing machine, which via the relational .
database can be related back to any DNA sequence. The
Sequencer Maintenance Log table 900 is linked with the Gel Key
table 280 via the common attribute of instrument_number.
Table 900 includes such information as the date service was
requested, the date service/maintenance was performed, the
CA 02210731 1997-07-17
W 096/23078 PCTAUS95/12429
. 13
nature of the problem, staff involved in maintenance and
pertinent comments.
In a preferred embodiment, the Catalyst and Computer
Maintenance Logs tables (905 and 910 respectively) are linked
through the computer_ID attribute and include similar
information to that of the Sequencer Maintenance Log and can
be related to essentially any DNA sequence.
The Equipment Log table 915 connects with Maintenance
tables 900-910 via the instrument_number and computer_ID
attributes and has information on the equipment or instruments
used in the sequencing operation. In a preferred embodiment,
table 915 stores information regarding equipment name and
serial number, vendor identifier, and date installed.
A separate vendor table 920 connects with the Equipment
Log Table 915 via the vendor_identifier attribute, and stores,
for example, the company name, address, phone number, fax
number and contact person. The vendor listing can also have
additional information on the vendor, including E-mail address
and date contract signed.
Fig. 7 illustrates a portion of the database of the
preferred embodiment for storing information regarding the
sequencing reagents. The Gel Link table 925 links to the Gel
Key table 280 via the gel_key_attribute and to the gel
solution table 935 via the gel_solution_ID attribute.
The Gel Solution table 935 includes information on the
gel solution and further includes the date the solution was
made and who prepared the solution. The Gel Solution-lot Link
table 950 links to the gel solution table 935 via the
gel_solution_ID attribute and also includes lot_number, and
reagent_ID attributes which are shared with the Lot table 965.
The Reaction-Cocktail Link table 930 shares the
reaction_set_ID attributes with the reaction set table 270.
The Reaction-Cocktail Link table 930 shares cocktail_ID with
the Cocktail table 940. The cocktail table 940 also has the
date the cocktail was made and staff person who made the
cocktail. The Cocktail-Lot link table 955 has the cocktail_ID
CA 02210731 1997-07-17
W 096/23078 PCTrUS9~/12429
. 14
attribute in common with the Cocktail table 940 and the Lot-
number and Reagent_id in common with the Lot table 965.
The Lot table 965 includes reagent ID and lot number,
vendor identifier, date received and date used. The vendor_ID
attribute is shared with the Vendor table 960. A separate
reagent table 970 shares the Reagent_ID attribute with the Lot
table 965 and also has an expanded reagent name.
Experimental sets of sequences may be stored in the
database in the express sets portion shown in Fig. 8. This
portion includes an express link table 370, a clone variant
table 380, an experimental table 390, a clean up table 400 and
a resequencing table 410. Express Link table 370 stores
sequence sets which have higher priority. They are given
unique identifiers and handled separately from the batch
process materials. Clone Variant table 380 refers to variant
sequences flagged by an individual investigator. The variants
are evaluated by that scientist, collaborator, or customer and
appropriate action is taken. The experimental sequences
stored in Experimental table 390 are similar to the variants
above. They may be homologous, allelic or mutant sequences
which have been flagged by a particular scientist. If only a
fragment has been recovered, a full length expression sequence
is ordered, and investigation continued. Cleanup table 400
stores data reflecting the addition of extra steps to the
protocol. The longer procedure is designed to improve
readability of the sequence. Resequencing is simply repeating
the procedure in order to check a sequence or to obtain more
data. Information regarding resequencing is stored in
Resequencing table 410.
Express Link table 370 contains a clone_ID attribute
which is also included in the Clone Log table 250. Attribute
log_entity_ID of the table 370 provides a correlation with
variant_ID, experimental_set_ID, cleanUp_set_ID, and
resequencing_set_ID of the tables 380, 390, 400, 410
respectively. Log_table_name attribute of the table 370
identifies the table correlated by the Log_entity_ID.
CA 02210731 1997-07-17
W 096/23078 PCT~US95/12429
As illustrated in step 60 of Fig. 2, each cDNA sequence
that has been obtained in step 50 is then compared to the
known sequences in the genetic databases to identify it if
possible. This process involves comparing sequences (a)
within a data set, (b) within the internal database and/or (c)
- with external databases. Since the library represents the
frequency with which an RNA transcript appears within a
certain source tissue, several different clones may contain
all or parts of the same gene or its allele(s). The computer
also analyzes insert size by counting individual nucleotides
in the sequence.
Data relating to sequence comparison is stored in tables
on the sequence comparison portion of the database shown in
Fig. 7. These tables include a first sequence match log table
510 and a second sequence match log table 515.
The database of the present invention may also access
external databases. Genetic databases may have DNA or protein
sequences. Such databases services may also provide searching
or matching tools in addition to named DNAs, proteins or
fragments thereof. As illustrated in Fig. 7, such outside
databases include the GenBank database (box 610), the ProDom
database (box 570), the Blocks database (box 580), the
Pisearch database (box 590) and the Sites database (box 600).
The Genbank database is used as a primary source of known
genes, sequences and other information against which the
sequencing stored in the database are compared. Percent
identity and probability are both considered to determine
whether such fragments may be categorized as "exact"
(apparently identical to a known/named human sequence), or
homologous (partially related) to a gene identified in humans
or another species. Unique and unidentified fragments or
sequences are listed by an identifier.
ProDom, Blocks, and Pisearch databases may be accessed in
order to determine if a particular sequence contains
functional protein domains or motifs. The patterns may
provide important structural information for a peptide or
protein encoded by the sequence.
- - -
CA 02210731 1997-07-17
W 096/23078 PCTrUS95/12429
16
In addition, Vectors database 520 stores the DNA
sequences of the vectors used to clone the cDNAs. By
comparing the identified cDNA sequences to the sequences in
this database, vector sequences or stretches of vector
sequences that show up in a cDNA sequence can be delimited.
Similarly, Repeats database 530 allows repeats which belong to
a multigene family, such as alu, to be identified. Hidden
Markov database 560 contains software which looks at a
nucleotide sequence alignment and computes a predicted peptide
structure from that sequence. As shown in Box 550 of Fig. 9,
other databases which provide additional features can also be
accessed.
When a sequence comparison results in a match, the
information regarding that match is stored in Sequence Match
Log tables 510 and 515. This information generally includes
address information for the matching sequence record in the
external database as well as scores which represent the
quality of the match. In an alternative embodiment it may be
preferable to store the scores in a separate record, since the
scoring methods are not identical for all databases. Sequence
Match Log 510 is linked to sequence archive 290 by the
attribute sequence_ID which they share. It should be noted
that first Sequence_Match_Log 510 contains better matches,
while marginal matches are stored in the second sequence_Match
Log 515. Both tables (510 and 515) have identical attributes.
Function identification, illustrated as step 70 in Fig.
2, is then performed on matches whose quality is above a
specific threshold. The data related to function
identification is stored in the tables as shown in Fig. 10.
These tables include a protein table 720, a protein-sequence
link table 730, a folder table 760 and location table 780.
Protein identification may come from any of the
function/domain databases. The Genbank location or locus and
the international EC number (enzyme or protein classification)
are stored in table 720. Each entry in this table corresponds
to one or more sequences from the sequence archive table which
was conclusively identified with respect to its function.
CA 02210731 1997-07-17
W O 96/23078 PCTrUS95/12429
Protein table 720 is linked to Sequence Archive table 290 via
Protein-Sequence Link table 730. Protein table 720 has the
attribute protein ID in common with Protein-Sequence Link
table 730; and Sequence Archive table 290 has the attribute
sequence_ID in common with Protein-Sequence Link table 730.
Each entry in folder table 760 contains unstructured
annotations for one or more sequences from the archive table
which had interesting but inconclusive matches with the other
databases. Any type of annotation, footnote, or remark can be
recorded in the folder table 760. This permits the researcher
to store desired information without contaminating other
records in the database with information from inconclusive
matches.
Folder table 760 is linked to sequence archive 290 via
function sequence link 750. Function sequence link 750 has an
attribute Folder_ID in common with folder table 760 and an
attribute Sequence_ID in common with sequence archive 290.
The present invention permits a researcher to search the
relational database using keywords and to specify the table(s)
in which the keyword search should be performed. Thus, for
example, a researcher could query the database for all
occurrences of the word "endothelial" in the Biological Source
Table 130.
In addition, the present invention allows the researcher
2S to store queries in Keywords table 790 shown in Fig. 10. Each
query stored in this table is identified by a unique
Keyword_ID. When a researcher wishes to run a particular
stored query, he or she simply enters the keyword_ID for the
query. The computer then pulls up the associated record, and
searches the table(s) identified in the Table_name field for
the keyword(s) stored in the Keyword_text field. The results
of the search can be delivered to the user for example via
E-mail notification as shown in boxes 800-820 of Fig. 10.
Location table 780 stores information regarding the
location within the cell of each identified sequence.
Location table 780 is linked to Protein table 720 by common
attribute Protein_ID, and stores the location information in
CA 02210731 1997-07-17
W O 96/23078 PCTrUS95/12429
. 18
an attribute called "Location." In a preferred embodiment,
the domain for this attribute consists of these categories:
nuclear, cytoplasmic (cytoskeleton), cytoplasmic
(intracellular membranes), cytoplasmic (mitochondria), cell
surface, and secreted.
Also shown in Fig. 10 is GDB links table 770 which
links Protein table 720 to the Human Genome Database. GDB
links table 770 has attribute Protein_ID in common with
Protein table 720 and links to the Human Genome Database via
attribute GDB_ID.
Given the wealth of related information stored in the
database of the preferred embodiment, a user can now perform
new types of data queries not previously available in the
known genetic databases. For example, the relational database
of the preferred embodiment is well suited for performing
abundance analysis. This analysis provides a user with the
relative frequency of mRNAs or transcripts found in a
particular cell in a given state, e.g., normal or activated.
For example, if a researcher were to input a query requesting
the most abundant sequences in an LPS activated THP-1 cell,
the computer system is programmed to search the relational
database and output to the user a display such as, illustrated
in Fig. 11.
In the preferred embodiment, the search is performed as
follows. First, the cell culture/treatment records 140 in
which the cell_line_name field equals "THP-1" (in this
example) are identified. Next, the identified records are
searched for records in which the treatment field equals
"LPS." Then, the sequence match log records 510 correlated in
the database with this subset of identified records are
determined and the number of sequence match lo~ records for
each distinct match ID value is counted to determine the
abundance in the cell of the particular sequence identified by
the match ID number. After the computer has examined all the
biological source records, it sorts the obtained abundance
information in the manner requested in the specific query and
displays it as a chart, as exemplified in Fig. 11.
CA 02210731 1997-07-17
WO 96/23078 PCT~US95/12429
19
Similarly, the database structure described above
provides a convenient way to implement subtraction analysis.
Subtraction analysis determines which sequences are expressed
more commonly in an activated cell compared to a normal cell.
To perform subtraction analysis, abundance analysis is
performed for the normal cell library and the activated cell
library, and when the information is obtained, a ratio of the
values is determined. Fig. 12 exemplifies the output of such
an operation for normal versus LPS activated THP-1.
Location analysis can also be performed. Here, the user
requests, for example, the location of a specific protein
within a particular activated macrophage. The computer
identifies the subset of records associated with the desired
cell in the manner described above, consults the associated
records in Protein table 720 to verify that the protein is
present in the cell, and finally look:s up the location of the
protein in Location table 780 and outputs the location to the
user.
The sequence location table categories in the preferred
embodiment are nuclear, cytoplasmic, cell surface or secreted.
Within the cytoplasm, sequences may be assigned to
cytoskeleton, intracellular membranes, or mitochondria. This
information is provided in the location field of Location
table 780. All of the unidentified sequences, regardless of
their relative abundance, are by default relegated to the
unknown category.
Yet another function supported by the database of the
preferred embodiment is distribution. This function
determines in which tissues or organs for example a given
sequence is found and how frequently. The system steps
through the records in the Sequencing Log 300 and when there
is a match with the desired sequence the system determines the
organ and tissue where the specified sequence was found
through the relational association of the database. After all
the sequences have been ~X~r; ned, an output is prepared
representing the requested distribution statistics.
-
CA 022l073l l997-07-l7
WO 96/23078 PCT~US95/12429
The detailed records and relational structure of the
database allow the researcher to access practically any field
reflecting a step in the mRNA, cDNA sequencing process. Thus,
the database of the present invention provides a powerful tool
for analyzing test results as well as testing procedures. For
example, if a researcher is interested in knowing all the
sequences that resulted from a particular lot or batch of
mRNA, this information can be obtained by stepping through the
mRNA preparation records 150, finding the records with the
desired lot number and outputting the related entries in the
sequencing log.
Given the disclosure above, a person skilled in the art
can design numerous queries to assist the scientist in various
data analysis tasks. From the foregoing description, it is
clear that the present invention may be embodied in other
specific forms without departing from the spirit or essential
characteristics thereof. The presently disclosed embodiments
are therefore to be considered in all respects as illustrative
and not restrictive, the scope of the invention being
indicated by the appended claims rather than the foregoing
description, and all changes which come within the meaning and
range of equivalency of the claims are therefore intended to
be embraced therein.