Note: Descriptions are shown in the official language in which they were submitted.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
1 STEM-LOOP AND CIRCULAR OLIGONUCLEOTIDES
This invention was made with United States
government support ~nder grant GM-46625 awarded by the
National Institutes of Health and grant N00014-92-J-1740
awarded by the Office of Naval Research. The United
States government has certain rights in the invention.
FIE~D OF THE INVENTION
The present ~nvention provides stem-loop and
circular oligonucleotides that are capable of binding to
nucleic acid targets with high affinity. The
oligonucleotides of the present invention are useful in
detection and isolation of target nucleic acids, and in
15 inhibiting the biological function of DNA and RNA.
BACKGROUND OF THE INVENTION
An oligonucleotide binds to a target nucleic
acid by forming hydrogen bonds between bases in the
20 target and the oligonucleotide. Common ~ DNA has
conventional adenine-thymine (A-T) and guanine-cytosine
(G-C) Watson and Crick base pairs with two and three
hydrogen bonds, respectively. An understanding of base
pairing motifs and duplex and triplex formation has
25 allowed the development of oligonucleotides for a
variety of utilities. For example, oligonucleotides can
be used as probes for targeting nucleic acids that are
immobilized onto a filter or membrane, or are present in
tissues. Sambrook _ al. (1989, Molecular Cloninq: A
3O Laboratory Manual, Vols. 1-3, Cold Spring Harbor ~ress,
,.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95103602
1 NY) provide a detailed review of hybridization
techniques.
Conventional hybridization technology is based
upon the capability of sequence-specific DNA or RNA
5 probes to bind to a target nucleic acid via Watson-Crick
hydrogen bonds. Other types of hydrogen bonding
patterns are known wherein some atoms of a base that are
not involved in Watson-Crick base pairing can form
hydrogen bonds to another nucleotide. For example,
10 thymine (T) can bind to an A-T Watson-Crick base pair
via hydrogen bonds to the adenine, thereby forming a T-
AT base triad. Hoogsteen (1959, Acta Crystalloqraphy
12:822) first described the alternate hydrogen bonds
present in T AT and C-GC base triads. G-TA base triads,
15 wherein guanine can hydrogen bond with a central
thymine, have also been observed (Griffin et al., 1989,
Science 245:967-971). Beal et al. (1991) Science
251:1360 propose models for hydrogen bonding in G-GC,
A-AT and T-AT base triplets. Such non-Watson-Crick
20 hydrogen bonding is generally referred to as Hoogsteen
bonding.
Oligonucleotides have been observed to bind to
duplex DNA by non-Watson-Crick hydrogen bonding in
vitro. For example, Cooney et al., 1988, Science
25 241:456 disclose a 27-base single-stranded
oligonucleotide which bound to a double-stranded nucleic
acid via Hoogsteen base pairing. However, triple-
stranded complexes of this type are not very stable,
because the oligonucleotide is bound to its target only
3o with less stable alternate hydrogen bonds, i.e., without
any Watson-Crick bonds. Further, as disclosed by Beal
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
3--
1 et al., oligonucleotide directed recognition of double
stranded DNA as described by Cooney et al. is limited to
binding of purine-rich tracts of DNA.
Giovannangeli et al. (1991, J. Am. Chem. Soc.
113:7775 and 1993, Proc. Natl. Acad. Sci. 90:10013)
disclose oligonucleotides capable of binding to single
stranded homopurine DNA targets. The oligonucleotides
contain one domain capable of binding to the target by
Watson-Crick binding, and a second domain capable of
lO binding to the target strand by parallel Hoogsteen
binding. A significant limitation of these
oligonucleotides is the requirement for homopurine
targets. In addition, linear oligonucleotides are
nuclease sensitive, thus limiting their use for many
15 biological applications.
Kool (1991, J. Am. Chem. Soc. 113:7775) and
Prakash et al. (1992, J. Am. Chem. Soc. 114:3523) report
circular oligonucleotides capable of binding to single
stranded nucleic acids by complexing the target strand
20 on both sides to form a pyrimidine/purine/pyrimidine
(pyr/pur/pyr) triplex. In particular, one side of the
circular compound is complementary to the target in the
antiparallel Watson-Crick sense, whereas the other side
of the circle is complementary to the target in the
25 parallel Hoogsteen sense. The use of the circular
oligonucleotides is limited to the recognition of purine
sequences in the target nucleic acid.
Samadashwily et al. (1993, EMBO Journal
12:4975) report that purine-rich linear
3o oligonucleotides, when annealed to a single-stranded
template, resulted in termination of polymerization by
CA 022l~82~ l997-09-l8
W096/29097 PCT~S95/03602
l DNA polymerase in vitro. The linear oligonucleotides
were characterized as having triplex forming potential
when annealed to a DNA template, but neither actual
triplex formation nor binding affinities nor binding to
5 RNA templates were investigated.
In vitro use of oligonucleotides by
hybridization technology thus suffers from problems
including suboptimal binding affinity. Hybridization
technology based upon triplex formation suffers from
lO limitations to particular target sequences such as
purines. The development of oligonucleotides for in
vivo regulation of biological processes has been
hampered by still further problems, including the
nuclease sensitivity of linear oligonucleotides.
For example, transcription of the human c-mvc
gene has been inhibited in a cell free, in vitro assay
system by a 27-base linear oligonucleotide designed to
bind to the c-myc promoter. Inhibition was only
observed using a carefully controlled in vitro assay
20 system wherein lower than physiological temperatures
were employed, and many cellular enzymes had been
removed or inactivated. These conditions were necessary
because linear oligonucleotides bind with low affinity
and are highly susceptible to enzymes which degrade
25 linear pieces of DNA (Cooney et al.). Splicing of a
pre-mRNA transcript essential for Herpes Simplex virus
replication has also been inhibited with a linear
oligonucleotide that was complementary to an acceptor
splice junc~ion. In this instance, a methylphosphonate
3o linkage was employed in the linear oligonucleotide to
increase its nuclease resistance. Addition of this
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95103602
5--
1 chemically-modified oligonucleotide to the growth medium
caused reduction in protein synthesis and growth of
uninfected cells, most likely because of toxicity
problems at high concentrations (Smith et al., 1986,
Proc. Natl. Acad. Sci. USA 83:2787-2791).
Linear oligonucleotides have been examined for
the ability to inhibit human immunodeficiency virus
replication in cultured cells (Goodchild et al., 1988,
Proc. Natl. Acad. Sci. USA 85:5507-5511). Linear
lO oligonucleotides complementary to sites within or near
the terminal repeats of the retrovirus genome and within
sites complementary to certain splice junctions were
most effective in blocking viral replication. However,
these experi~ents required large amounts of the linear
15 oligonucleotides before an effect was obtained,
presumably because of the low binding stability and
vulnerability of these linear oligonucleotides to
nucleases.
Accordingly, there is a need in the art for
20 stable oligonucleGtide compounds capable of strong and
specific binding to nucleic acids, and in particular to
pyrimidine rich nucleic acids. The present invention
represents an innovation characterized by a substantial
improvement relative to the prior art since the subject
25 stem-loop and circular oligonucleotides exhibit high
specificity, low or no toxicity, and more resistance to
nucleases than linear oligonucleotides, while binding to
single-stranded pyrimidine rich target nucleic acids
more strongly than conventional linear oligonucleotides.
3o
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-6-
1 SUMMARY OF THE INVENTION:
The present invention provides circular
oligonucleotides having at least one first binding
domain capable of detectably ~inding to a de~ined
nucleic acid target by antiparallel Watson-Crick base
pairing (hereinafter the WC domain), and at least one
second binding domain capable of binding to the first
binding domain by antiparallel Hoogsteen base pairing
(hereinafter the H domain). The circular nucleotides of
the invention further comprise a linker domain
separating each binding domain.
The present invention is further directed to
stem-loop oligonucleotides comprising a double-stranded
stem domain of at least about two base pairs and a loop
domain having at least one first binding domain capable
of binding to a defined nucleic acid target by
antiparallel Watson-Crick base pairing, and at least one
second binding domain capable of binding to the first
binding domain by antiparallel Hoogsteen base pairing.
The binding domains are separated by a linker domain.
In the case where multiple binding domains are
included in the stem-loop and circular oligonucleotides
of the present invention, the linker ~o~in~ separating
the binding domains can constitute, in whole or in part,
another binding domain that functions as a binding
domain in an alternate conformation. In other words,
depending upon the particular target, a binding domain
can also function as a linker domain for another binding
domain and vice versa.
Another aspect of the present invention
provides the subject stem-loop and circular
.
CA 022l~82~ l997-09-l8
W096/29097 PCT~S95/03602
l oligonucleotides derivatized with a reporter group to
provide a probe for a target nucleic acid, or with a
drug or other pharmaceutical agent to provide cell
specific drug delivery, or with agents which can cleave
or otherwise modify the target nucleic acid or,
furthermore, with agents that can facilitate cellular
uptake or target binding of the oligonucleotide.
An additional aspect of the present invention
provides stem-loop or circular oligonucleotides linked
10 to a solid support for isolation of a nucleic acid
complementary to the oligonucleotide.
Another aspect of the present invention
provides a compartmentalized kit for detection,
diagnosis or isolation of a target nucleic acid
15 including at least one first container providing at
least one of the present stem-loop or circular
oligonucleotides.
A further aspect of the present invention
provides a method of detecting a target nucleic acid
20 which involves contacting a stem-loop or circular
oligonucleotide of the present invention with a sample
containing the target nucleic acid, for a time and under
conditions sufficient to form an oligonucleotide-target
complex, and detecting the complex.
A still further aspect of the present
invention provides a method of regulating biosynthesis
of a DNA, an RNA or a protein. This method includes
contacting at least one of the subject stem-loop or
circular oligonucleotides with a nucleic acid template
3o for the DNA, the RNA or the protein under conditions
.
CA 0221~82~ 1997-09-18
WO96/290s7 PCT~S95/03602
-8-
1 sufficient to permit binding of the oligonucleotide to a
target sequence contained in the template.
The present invention further provides a
method of cell specific drug delivery comprising
5 administering to a mammal a drug covalently linked to a
stem-loop or circular oligonucleotide of the present
invention.
An additional aspect of the present invention
provides pharmaceutical compositions for regulating
lO biosynthesis of a nucleic acid or protein containing a
biosynthesis regulating amount of at least one of the
subject oligonucleotides and a pharmaceutically
acceptable carrier.
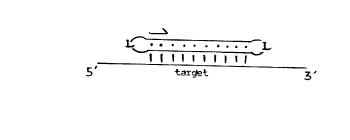
15 BRIEF DESCRIPTION OF THE D~A~INGS:
Figures lA and lB present schematic
illustrations of the relative orientations of
representative circular and stem-loop oligonucleotides,
respectively. Arrows indicate 5' to 3' directionality,
20 linker domains are indicated by L, Watson Crick base
pairs are indicated by lines and Hoogsteen base pairs
are indicated by dots. Stem-loop oligonucleotides may
also be in the opposite orientation, i.e. the stem may
be at the 5' or 3' end.
DETAILED DESCRIPTION OF THE INVENTION:
The present invention relates to stem-loop and
circular oligonucleotides that can bind to nucleic acid
targets with higher affinity, selectivity and stability
3o than a corresponding linear oligonucleotide.
CA 0221~82~ 1997-09-18
Wos6/2sos7 PCT~S95103602
_g _
1 Furthermore, the nuclease resistance and the
strong, selective binding of the subject stem-loop and
circular oligonucleotides to nucleic acid targets
provides a variety of uses, including methods of
regulating such biological processes as DNA replication,
RNA transcription, RNA splicing and processing, protein
translation and the like. Additionally, the present
oligonucleotides are useful for isolation of
complementary nucleic acids or for sequence-specific
lO delivery of drugs or other molecules into cells.
The circular oligonucleotides of the present
invention have at least one first binding domain capable
of detectably binding to a defined nucleic acid target
by antiparallel Watson-Crick base pairing (the WC
15 domain), and at least one second binding domain capable
of binding to the first binding domain by antiparallel
Hoogsteen base pairing (the H domain). The circular
nucleotides of the invention further comprise a linker
domain separating each binding domain. In a preferred
20 embodiment, the clrcular oligonucleotide has one WC
domain and one corresponding H domain separated by
linker domains of two or three oligonucleotides.
The stem-loop oligonucleotides of the present
invention comprise a double-stranded stem domain of at
25 least about two base pairs and a loop domain having at
least one first binding domain capable of binding to a
defined nucleic acid target by antiparallel Watson-Crick
base pairing (the WC domain), and at least one second
binding domain capable of binding to the first binding
3o domain by antiparallel Hoogsteen base pairing (the H
domain). The binding domains are separated by a linker
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-10-
1 domain. In a preferred embodiment, the stem-loop
oligonucleotide has one WC domain and one corresponding
H domain separated by linker domains of two or three
nucleotides.
As defined herein, antiparallel binding means
that the 5' to 3' orientations of two strands are in
opposite directions, i.e. the strands are aligned as
found in the typical Watson-Crick base pairing
arrangement of double helical DNA. Schematic
10 illustrations of the relative orientations of
representative circular and stem-loop oligonucleotides
are set forth in Figures lA and lB, respectively.
Arrows indicate 5' to 3' directionality, linker domains
are indicated by L, Watson Crick base pairs are
15 indicated by lines and Hoogsteen base pairs are
indicated by dots. The stem of the stem-loop
oligonucleotides may be at the 5' or 3' end.
When more than one WC and H binding domains
are present, such binding domains are separated from
20 other binding domains by linker domains of sufficient
length to permit binding to multiple targets. Moreover,
when a stem-loop or circular oligonucleotide has
multiple WC and H domains, a linker domain for one pair
of corresponding WC and H binding domains can constitute
25 a WC or H domain for binding to another target. When an
oligonucleotide of the present invention includes, e.g.,
two pairs of corresponding binding domains, these pairs
of corresponding binding domains can also bind separate
target sites. Moreover, when an oligonucleotide has
3o multiple WC and H domains, the corresponding targets
need not be present on one nucleic acid strand.
CA 0221~82~ 1997-09-18
Wos6J290s7 PCT~S95/03602
--11--
1 Furthermore, a linker domain of a stem-loop or circular
oligonucleotide bound to a given target can be a WC or H
- domain for binding to a second target when the
oligonucleotide is released from the first target.
The nucleotide sequences of the WC and H
domains of the subject stem-loop and circular
oligonucleotides are determined with reference to a
defined nucleic acid target. The base pairing rules
provided hereinbelow define what is referred to herein
10 as Watson Crick and Hoogsteen base pairing.
The defined nucleic acid target may be DNA or
RNA and is preferably single-stranded. Double-stranded
targets are also contemplated, and in a preferred
embodiment a double-stranded target is subjected to
15 denaturating conditions, ionic strength conditions or
other conditions that provide strand opening or strand
displacement such that a single strand is available for
binding to the oligonucleotide of the present invention.
A target may be selected by its known
20 functional and structural characteristics. For example,
some preferred targets can be coding regions, origins of
replication, reverse transcriptase binding sites,
transcription regulatory elements, RNA splicing
junctions, or ribosome binding sites, among others.
25 Messenger RNA and viral DNA and RNA are particularly
preferred targets. A target can also be selected to
effect the detection or isolation of a DNA or RNA
template. Double-stranded DNA and RNA that have been
subjected to denaturation are preferred targets for in
3o vitro and in situ applications. Preferred targets are
..
CA 022l~82~ l997-09-l8
W096l29097 PCT~S95/03602
-12-
l rich in pyrimidines, i.e. in cytosine, uracil and
thymine.
The nucleotide sequence of the target DNA or
RNA may be known in full or in part. When the target
5 nucleotide sequence is completely known, the sequence of
the WC domain is designed with the necessary degree of
complementarity to achieve binding, as detected by known
procedures, for example by a change in light absorption
or fluorescence. In some instances, the target seguence
lO may be represented by a consensus sequence or may be
only partially known. In this instance a target may
match the consensus sequence exactly or may have some
mismatched bases, but not enough mismatch to prevent
binding. Likewise, if a portion of a target sequence is
15 known, one skilled in the art can refer to the base
pairing rules provided hereinbelow to design
oligonucleotides that detectably bind to the target with
higher affinity than a corresponding linear
oligonucleotide.
The sequence of the WC domain of the subject
stem-loop and circular oligonucleotides is determined by
the following base pairing rules with reference to a
defined nucleic acid target.
The general rules for determining the sequence
25 of a sufficient number of nucleotides of the WC domain
are as follows: when a base for a position in the
target is guanine, or a guanine analog, then WC has
cytosine or uracil or pseudouracil, or suitable analogs
thereof, in a corresponding position;
3o when a base for a position in the target is
adenine, or an adenine analog, then WC has thymine or
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-13-
1 uracil or pseudouracil, or suitable analogs thereof, in
a corresponding position;
when a base for a position in the target is
thymine, or a thymine analog, then WC has adenine, or a
suitable analog thereof, in a corresponding position;
and
when a base for a position in the target is
cytosine, or a cytosine analog, then WC has a guanine,
or a suitable analog thereof, in corresponding position;
when a base for a position in the target is
uracil, or a uracil analog, then WC has adenine or
guanine, or suitable analogs thereof, in a corresponding
position.
In particular, a sufficient number of
nucleotide positions of the WC domain is determined
according to the foregoing rules such that detectable
binding is achieved. Detectable binding is defined
herein as binding of the oligonucleotide to the target
such that the formation of a resulting complex can be
detected by methods well-known in the art, including for
example the determination of a change in light
absorption upon binding or melting. Accordingly, the
base pairing rules must be satisfied to the extent
needed to achieve detectable binding of a stem-loop or
circular oligonucleotide to its nucleic acid target.
The degree of complementarity, i.e. adherence to the
base-pairing rules, need not be 100% so long as binding
can be detected. For the present invention sufficient
complementarity means that a sufficient number of base
pairs exist between a target nucleic acid and the WC
domain of the circular or stem-loop oligonucleotide to
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-14-
1 achieve detectable binding. When expressed or measured
by percentage of base pairs formed, the degree of
complementarity can range from as little as about 30-40%
complementarity to full, i.e. 100%, complementarity. In
5 general, the overall degree of complementarity between
the WC domain and the target is preferably at least
about S0%, and more preferably about 90%. In a most
preferred embodiment, the degree of complementarity
between the target and the WC domain is 100%.
Moreover, the degree of complementarity that
provides detectable binding between the subject circular
oligonucleotides and the respective targets is dependent
upon the conditions under which that binding occurs. It
is well know~ that binding, i.e. hybridization, between
15 nucleic acid strands depends on factors besides the
degree of mismatch between two sequences. Such factors
include the GC content of the region, temperature, ionic
strength, the presence of formamide and types of counter
ions present. The effect of these conditions upon
20 binding is known to one skilled in the art.
Furthermore, conditions are frequently determined by the
circumstances of use. For example, when a circular
oligonucleotide is made for use in vivo, no formamide
will be present and the ionic strength, types of counter
25 ions, and temperature correspond to physiological
conditions. Binding conditions can be manipulated ln
vitro to optimize the utility of the present
oligonucleotides. A thorough treatment of the
qualitative and quantitative considerations involved in
3o establishing binding conditions that allow one skilled
in the art to design appropriate oligonucleotides for
CA 0221~82~ 1997-09-18
wos6l2sos7 PCT~S95/03602
-15-
1 use under the desired conditions is provided by Beltz et
al., 1983, Methods Enzymol. 100:266-285 and by Sambrook
et al.
As used herein "binding" or "stable binding"
means that a sufficient amount of the oligonucleotide is
bound or hybridized to its target to permit detection of
that binding. Binding can be detected by either
physical or functional properties of the target:circular
oligonucleotide complex. Binding between a target and
10 an oligonucleotide can be detected by any procedure
known to one skilled in the art, including both
functional or physical binding assays. Binding may be
detected functionally by determining whether binding has
an observable effect upon a biosynthetic process such as
15 DNA replication, RNA transcription, protein translation
and the like.
Physical methods of detecting the binding of
complementary strands of DNA or RNA are well known in
the art, and include such methods as DNase I or chemical
20 footprinting, gel shift and affinity cleavage assays,
Northern blotting, dot blotting and light absorption
detection procedures. For example, a method that is
widely used, because it is simple and reliable, involves
observing a change in light absorption of a solution
25 containing an oligonucleotide and a target nucleic acid
at 220 to 300 nm as the temperature is slowly increased.
If the oligonucleotide has bound to its target, there is
a sudden increase in absorption at a characteristic
temperature as the oligonucleotide and target dissociate
3O or melt.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-16-
1 Having determined the sequence of the WC
domain with reference to a defined nucleic acid target
and the base pairing rules described hereinabove, the
sequence of the H domain is determined with reference to
the corresponding WC domain. The sequence of the H
domain is determined such that the H domain binds in
antiparallel manner to the corresponding WC domain by
Hoogsteen base pairing. Antiparallel Hoogsteen base
pairing is also referred to herein as reverse Hoogsteen
binding. Hoogsteen binding or base pairing is defined
herein as binding in accordance with the following base
pairing rules. These general rules are used for
determining the sequence of the H domain:
when a base for a position in the WC domain is
guanine or a guanine analog, then H has guanine or a
suitable analog thereof, in a corresponding position;
when a base for a position in the WC domain is
adenine or an adenine analog, then H has adenine or
thymine or uracil, or suitable analogs thereof, in a
corresponding position;
when a base for a position in the WC domain is
thymine, cytosine, uracil, or analogs thereof, then H
has adenine, cytosine, guanine, thymine, uracil,
imidazole, or suitable analogs thereof, in a
corresponding position.
The presence of the H domain in the subject
stem-loop and circular oligonucleotides serves to
increase the binding affinity of the present
oligonucleotides relative to corresponding linear
3o oligonucleotides. While not being limited to a
particular mechanism, the subject oligonucleotides
CA 0221~82~ 1997-09-18
wos6l29o97 PCT~S95103602
-17-
l presumably bind directly to the target only by the WC
domain, and the Hoogsteen interactions between the H and
WC domain serve to rigidify the oligonucleotide and
provide a benefit in complexation. Accordingly, the
binding affinity of the present oligonucleotides is
optimal when 100% of the nucleotides in the H domain are
determined by the above base pairing rules relative to
the WC domain. However, a certain amount of mismatch in
the H domain can be tolerated with retention of a gain
in binding affinity relative to Watson-Crick binding
alone. The ordinarily skilled artisan can determine the
tolerable amount of mismatch under given conditions. In
accordance with the present invention, the se~uence of
the H domain must be determined in accordance with the
above base pairing rules only to the degree that the
resulting oligonucleotide exhibits an increased binding
affinity for a particular target relative to a
corresponding linear oligonucleotide. A corresponding
linear oligonucleotide is one that contains only the
sequence of the WC domain, i.e. is complementary to the
target in a Watson-Crick sense. In a preferred
embodiment, 50-100% of the nucleotides in the H domain
are determined in accordance with the base pairing
rules. In a more preferred embodiment, 80-100% of the
nucleotides in the H domain are determined in accordance
with the base pairing rules. In a most preferred
embodiment, 100% of the nucleotides in the H domain are
determined in accordance with the base pairing rules.
In a preferred embodiment of the present
invention, the nucleic acid target is pyrimidine rich;
RNA targets are UC rich, and DNA targets are TC rich.
CA 022l~82~ l997-09-l8
W096/29097 PCT~S95/03602
-18-
l In accordance with the above base pairing rules, the WC
domains of the subject stem-loop and circular
oligonucleotides are thus AG rich and the H domains are
thus TG rich. In a preferred embodiment, the potential
triplexes formed by binding of nucleotides from the
H/WC/target domains are GGC, GGU, AAT, TAT, UAT, AAU,
UAU and TAU.
Each WC and H domain can independently have
about 2 to about 200 nucleotides with preferred lengths
being about 6 to about 80 nucleotides. The most
preferred lengths are 8 to 36 nucleotides.
The double stranded stems of the stem-loop
oligonucleotides of the present invention are formed by
base pairing of complementary bases in the 5' and 3'
ends of the oligonucleotide and are at least about two
base pairs in length. A preferred length is from about
three base pairs to about ten base pairs. The stem is
preferably kept as short as possible without adversely
affecting the stability of the stem-loop structure,
although longer stems are also contemplated. Short
stems are less likely to cause steric hindrance of
binding of oligonucleotide to target. The stem may
further comprise an overhanging single-stranded region,
i.e. the strand may be a partial duplex. The two
strands of the stem can be covalently cross-linked such
that the stability of the stem structure is independent
of its length. Thus the stem length need only be long
enough to permit cross linking, i.e. about two base
pairs. Cross-linking may be performed by procedures
known to one of ordinary skill in the art, e.g. as
described by Calabresi et al. in Gilman et al., eds. The
CA 022l~82~ l997-09-l8
W096/29097 PCT~S95/03602
-19-
l Pharmacoloqical Basis of Therapeutics, 1980, MacMillan
Publishing Co., Inc., pp. 1256-1272 and Glick et al.
(1992) J. Am. Chem. Soc. 114:5447. Further, a
nucleotide linker as described herein may be used to
covalently link the base of the stem.
The stem-loop and circular oligonucleotides
are DNA or RNA, or hybrids of DNA and RNA. For example,
the subject oligonucleotides can comprise: a DNA
binding domain and an RNA binding domain; RNA binding
domains and DNA linkers; all RNA; all DNA; RNA
interspersed with 2'-O-methyl RNA, and so on. The
oligonucleotides comprise the bases guanine (G), adenine
(A), thymine (T), cytosine (C) or uracil (U) in the
nucleotides, or any nucleotide analog that is capable of
hydrogen bonding in an anti-parallel manner. Nucleotide
analogs include pseudocytidine, isopseudocytidine,
imidazole, 3-aminophenyl-imidazole, 2'-O-methyl-
adenosine, 7-deazadenosine, 7-deazaguanosine, 7-
deazaxanthosine, 4-acetylcytidine, 5-(carboxy-
hydroxylmethyl)-uridine, 2'-O-methylcytidine, 5-
carboxymethylaminomethyl-2-thioridine, 5-
carboxymethylamino-methyluridine, dihydrouridine, 2'-O-
methyluridine, pseudouridine, 2'-O-methyl-pseudouridine,
beta,D-galactosylqueosine, 2'-O-methylguanosine,
inosine, N6-isopentenyladenosine, l-methyladenosine, 1-
methyl-pseudouridine, l-methylguanosine, 1-
methylinosine, 2,2-dimethylguanosine, 2-methyladenosine,
2-methylguanosine, 3-methylcytidine, 5-methylcytidine,
5-methyluridine, N6-methyl-adenosine, 7-methylguanosine,
5-methylamino-methyluridine, 5-methoxyaminomethyl-2-
thiouridine, ~-D-mannosylqueosine, 5-
CA 0221~82~ 1997-09-18
W O 96/29097 PC~rrUS95/03602
-20 -
1 methoxycarbonylmethyluridine, 5-methoxyuridine, 2-
methyl-thio-N6-isopentenyladenosine, N-(9-beta-D-
ribofuranosyl-2-methylthiopurine-6-yl)-
carbamoyl)threonine, N-(9-beta-D-ribofuranosylpurine-6-
yl)-N-methylcarbamoyl)threonine and thioguanosine.
Either ribose or deoxyribose sugars can be used with
these analogs. Modified sugars, such as 2'-O-methyl
ribose, are also contemplated. Nucleotides bases in an
a-anomeric conformation can also be used in the stem-
loop and circular oligonucleotides of the presentinvention.
Preferred nucleotide analogs are unmodified G,
A, T, C and U nucleotides; pyrimidine analogs with lower
alkyl, alkyn~l or alkenyl groups in the 5 position of
the base and purine analogs with similar groups in the 7
or 8 position of the base. Especially preferred
nucleotide analogs are 5-methylcytosine, 5-methyluracil,
diaminopurine, and nucleotides with a 2'-0-methylribose
moiety in place of ribose or deoxyribose. As used
herein lower alkyl, lower alkynyl and lower alkenyl
contain from 1 to 6 carbon atoms and can be straight
chain or branched. These groups include methyl, ethyl,
propyl, isopropyl, butyl, isobutyl, tertiary butyl,
amyl, hexyl and the like. A preferred alkyl group is
methyl.
The WC and H domains are separated by linker
domains which can independently have from about 1 to
about 2000 nucleotides. A preferred linker length is
from about 1 to about 10 nucleotides with an especially
preferred length being about 2 or 3 nucleotides. In a
preferred embodiment, the linker is C-C or T-T. The
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-21-
1 stem domain is considered to contribute two nucleotides
to the linker domain separating the WC and H domains of
the subject stem-loop oligonucleotides.
According to the present invention, the linker
domains may be non-nucleotide linkers. Non-nucleotide
linkers can make the present stem-loop and circular
oligonucleotides less expensive to produce. More
significantly, oligonucleotides with non-nucleotide
loops are more resistant to nucleases (Rumney and Kool,
1992, Anqewandte Chemie, Intl. Ed., 31:1617) and
therefore have a longer biological half-life than linear
oligonucleotides. Furthermore, linkers having no
charge, or a positive charge, can be used to promote
binding by eliminating negative charge repulsions
between the linker and target. In addition,
oligonucleotides having uncharged or hydrophobic non-
nucleotide linkers can penetrate cellular membranes
better than oligonucleotides with nucleotide loops.
As contemplated herein, non-nucleotide linker
domains can be composed of alkyl chains, polyethylene
glycol or oligoethylene glycol chains or other chains
providing the necessary steric or flexibility properties
which are compatible with oligonucleotide synthesis.
The length of these ch~in~ is equivalent to about 2 to
about 2000 nucleotides, with preferred lengths
equivalent to about 2 to about 8 nucleotides. The most
preferred length for these chains is equivalent to about
2 to 4 nucleotides.
Preferred chains for non-nucleotide linker
3o domains are polyethylene glycol or oligoethylene glycol
chains. In particular, oligoethylene glycol chains
CA 0221~82~ 1997-09-18
Wos6/2sos7 PCT~S95103602
-22-
1 having a length similar to a 2 to 4 nucleotide chain,
e.g. a di-, tri-, tetra-, penta- or hexaethylene glycol
chain, are preferred but longer oligoethylene glycol
chains are contemplated. Covalent bonds such as
disulfide bonds can also function as linker domains.
The circular oligonucleotides of the present
invention can be made first as linear oligonucleotides
and then circularized. Linear oligonucleotides can be
made by any of a myriad of procedures known for making
DNA or RNA oligonucleotides. For example, such
procedures include enzymatic synthesis and chemical
synthesis.
Enzymatic methods of DNA oligonucleotide
synthesis frequently employ Xlenow, T7, T4, ~g or E.
coli DNA polymerases as described in Sambrook et al.
Enzymatic methods of RNA oligonucleotide synthesis
frequently-employ SP6, T3 or T7 RNA polymerase as
described in Sambrook et al. Reverse transcriptase can
also be used to synthesize DNA from RNA (Sambrook
et al.). To prepare oligonucleotides enzymatically
requires a template nucleic acid which can either be
synthesized chemically, or be obtained as mRNA, genomic
DNA, cloned genomic DNA, cloned cDNA or other
recombinant DNA. Some enzymatic methods of DNA
oligonucleotide synthesis can require an additional
primer oligonucleotide which can be synthesized
chemically. Finally, linear oligonucleotides can be
prepared by polymerase chain reaction (PCR) techniques
as described, for example, by Saiki et al., 1988,
3o Science 239:487.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-23-
1 Chemical synthesis of linear oligonucleotides
is well known in the art and can be achieved by solution
or solid phase techniques. Moreover, linear
oligonucleotides of defined sequence can be purchased
commercially or can be made by any of several different
synthetic procedures including the phosphoramidite,
phosphite triester, H-phosphonate and phosphotriester
methods, typically by automated synthesis methods. The
synthesis method selected can depend on the length of
the desired oligonucleotide and such choice is within
the skill of the ordinary artisan. For example, the
phosphoramidite and phosphite triester method produce
oligonucleotides having 175 or more nucleotides while
the H-phosphonate method works well for oligonucleotides
~f less than 100 nucleotides. If modified bases are
incorporated into the oligonucleotide, and particularly
if modified phosphodiester linkages are used, then the
synthetic procedures are altered as needed according to
known procedures. In this regard, Uhlm~nn et al. (1990,
Chemical Reviews 90:543-584) provide references and
outline procedures for making oligonucleotides with
modified bases and modified phosphodiester linkages.
Synthetic linear oligonucleotides may be
purified by polyacrylamide gel electrophoresis, or by
any of a number of chromatographic methods, including
gel chromatography and high pressure liquid
chromatography. To confirm a nucleotide sequence,
oligonucleotides may be subjected to DNA sequencing by
any of the known procedures, including Maxam and Gilbert
3o sequencing, Sanger sequencing, capillary electrophoresis
sequencing, the wandering spot sequencing procedure or
CA 022l~82~ l997-09-l8
wo96l2so97 PCT~S95/03602
-24-
l by using selective chemical degradation of
oligonucleotides bound to Hybond paper. Se~uences of
short oligonucleotides can also be analyzed by laser
desorption mass spectroscopy or by fast atom bombardment
(McNeal, et al., 1982, J. Am. Chem. Soc. 104:976; Viari,
et al., 1987, Biomed. Environ. Mass Spectrom. 14:83;
Grotjahn et al., 1982, Nuc. Acid Res. 10:4671).
Sequencing methods are also available for RNA
oligonucleotides.
The present invention provides several methods
of preparing circular oligonucleotides from linear
precursors (i.e. precircles), including a method wherein
a precircle is synthesized and bound to an end-joining-
oligonucleotide and the two ends of the precircle are
joined. Any method of joining two ends of an
oligonucleotide is contemplated by the present
invention, including chemical methods employing, for
example, known coupling agents like BrCN, N-
cyanoimidazole ZnCl2, 1-ethyl-3-(3-dimethylaminopropyl)-
carbodiimide and other carbodiimides and carbonyldiimidazoles. Furthermore, the ends of a precircle can
be joined by condensing a 5' phosphate and a 3' hydroxy,
or a 5' hydroxy and a 3' phosphate.
A simple one-step chemical method to construct
the subject circular oligonucleotides from precircles is
provided in U.S. Application Serial No. 08/044,800, now
U.S. Patent No. . An oligonucleotide is
constructed which has the same sequence as the target
nucleic acid; this is the end-joining oligonucleotide.
3o A DNA or RNA linear precircle is chemically or
enzymatically synthesized and phosphorylated on its 5'
CA 022l~82~ l997-09-l8
W096/29097 PCT~S95/03602
-25-
l or 3' end, again by either chemical or enzymatic means.
The precircle and the end-joining oligonucleotide are
mixed and annealed, thereby forming a complex in which
the 5' and 3' ends of the precircle are adjacent. It is
preferred that the ends of the precircle fall within a
binding domain, not within a linker. Moreover, it is
preferred that a precircle have a 3'-phosphate rather
than a 5'-phosphate. After complex formation, the ends
undergo a condensation reaction in a buffered aqueous
solution containing divalent metal ions and BrCN at
about pH 7Ø In a preferred embodiment the buffer is
imidazole-Cl at pH 7.0 with a divalent metal such as Ni,
Zn, Mn, or Co. Ni is the most preferred divalent metal.
Condensation occurs after about 6-48 hr. of incubation
at 4-37~C. Other divalent metals, such as Cu, Pb, Ca
and Mg, can also be used.
One method for RNA circularization
incorporates the appropriate nucleotide sequences,
preferably in a linker domain, into an RNA
oligonucleotide to promote self splicing, since a
circular product is formed under the appropriate
conditions (Sugimoto et al., 1988, Biochemistry 27:6384-
6392).
Enzymatic circle closure is also possible
using DNA ligase or RNA ligase under conditions
appropriate for these enzymes.
Circular oligonucleotides can be separated
from the end joining oligonucleotide by denaturing gel
electrophoresis or melting followed by gel
3o electrophoresis, size selective chromatography, or other
appropriate chromatographic or electrophoretic methods.
CA 0221~82~ 1997-09-18
WO96/290s7 PCT~S95/03602
-26-
1 The recovered circular oligonucleotide can be further
purified by standard techniques as needed for its use in
the methods of the present invention. Alternately, the
end joining oligonucleotide may be attached to a solid
support and recovered by filtration.
The subject stem-loop oligonucleotides can be
made by the procedures described hereinabove for the
synthesis of linear oligonucleotides. The present stem-
loop oligonucleotides can be also made recombinantly by
placing a nucleic acid having a sequence which is
complementary to the desired stem-loop oligonucleotide
into an expression vector. Such an expression vector
minimally encodes a segment which can effect expression
of the stem-loop oligonucleotide when the segment is
operably linked to the nucleic acid encoding the stem-
loop oligonucleotide. However, such an expression
vector can-also encode additional elements such as
origins of replication, selectable markers,
transcription termination signals, centromeres,
autonomous replication sequences.
As used herein, an expression vector can be a
replicable or a non-replicable expression vector.
Further, the expression vectors of the present invention
can be chromosomally integrating or chromosomally
nonintegrating expression vectors, and may be designed
to function in prokaryotic, yeast, insect or m~mm~l ian
cells.
The present invention also contemplates
derivatization or chemical modification of the subject
oligonucleotides with chemical groups to facilitate
cellular uptake. For example, covalent linkage of a
CA 0221~82~ 1997-09-18
w096/29097 PCT~S95/03602
-27-
1 cholesterol moiety to an oligonucleotide can improve
cellular uptake by 5- to 10- fold which in turn improves
DNA binding by about 10- fold (Boutorin et al., 1989,
FEBS Letters 254:129-132). Other ligands for cellular
receptors may also have utility for improving cellular
uptake, including, e.g. insulin, transferrin and others.
Similarly, derivatization of oligonucleotides with poly-
L-lysine can aid oligonucleotide uptake by cells
- (Schell, 1974, Biochem. Biophys. Acta 340:323, and
Lemaitre et al., 1987, Proc. Natl. Acad. Sci. USA
84:648). Certain protein carriers can also facilitate
cellular uptake of oligonucleotides, including, for
example, serum albumin, nuclear proteins possessing
signals for transport to the nucleus, and viral or
bacterial proteins capable of cell membrane penetration.
Therefore, protein carriers are useful when associated
with or linked to the circular oligonucleotides of this
invention. Accordingly, the present invention
contemplates derivatization of the subject stem-loop and
circular oligonucleotides with groups capable of
facilitating cellular uptake, including hydrocarbons and
non-polar groups, cholesterol, poly-L-lysine and
proteins, as well as other aryl or steroid groups and
polycations having analogous beneficial effects, such as
phenyl or naphthyl groups, quinoline, anthracene or
phenanthracene groups, fatty acids, fatty alcohols and
sesquiterpenes, diterpenes and steroids.
The present invention further contemplates
derivatization of the subject oligonucleotides with
3o agents that can cleave or modify the target nucleic acid
or other nucleic acid strands associated with or in the
CA 0221~82~ 1997-09-18
Wo96/2sos7 PCT~S95/03602
-28-
1 vicinity of the target. For example, viral DNA or RNA
can be targeted for destruction without harming cellular
nucleic acids by administering a stem-loop or circular
oligonucleotide having a WC domain that is complementary
to the targeted nucleic acid which is linked to an agent
that, upon binding, can cut or render the viral DNA or
RNA inactive. Nucleic acid destroying agents that are
contemplated by the present invention as having cleavage
or modifying activities include, for example, RNA and
DNA nucleases, ribozymes that can cleave RNA,
azidoproflavine, acridine, EDTA/Fe, chloroethylamine,
azidophenacyl and phenanthroline/Cu. Uhlmann et al.
(1990, Chemical Reviews 90:543-584) provide further
information On the use of such agents and methods of
derivatizing oligonucleotides that can be adapted for
use with the subject oligonucleotides.
~ erivatization of the subject stem-loop and
circular oligonucleotides with groups that facilitate
cellular uptake or target binding, as well as
2Q derivatization wi'h nucleic acid destroying agents or
drugs, can be done by any of the procedures known to one
skilled in the art. Moreover, the desired groups can be
added to nucleotides before synthesis of the
oligonucleotide. For example, these groups can be
linked to the 5-position of T or C and these modified T
and C nucleotides can be used for synthesis of the
present oligonucleotides. In addition, derivatization
of selected nucleotides permits incorporation of the
group into selected domains of the subject
oligonucleotides. For example, in some instances it is
preferable to incorporate certain groups into a linker
CA 022l~82~ l997-09-l8
W096/29097 PCT~S95/03602
-29-
1 where that group will not interfere with binding, or
into a WC domain to facilitate cleavage or modification
of the target nucleic acid.
In accordance with the present invention,
modification in the phosphodiester backbone of the
subject oligonucleotides is also contemplated. Such
modifications can aid uptake of the oligonucleotide by
cells or can extend the biological half-life of such
nucleotides. For example, the subject oligonucleotides
may penetrate the cell membrane more readily if the
negative charge on the internucleotide phosphate is
eliminated. This can be done by replacing the
negatively charged phosphate oxygen with a methyl group,
an amine or by changing the phosphodiester linkage into
a phosphotriester linkage by addition of an alkyl group
to the negatively charged phosphate oxygen.
Alternatively, one or more of the phosphate atoms that
are part of the normal phosphodiester linkage can be
replaced. For example, NH-P, CH2-P or S-P linkages can
be formed. Accordingly, the present invention
contemplates using methylphosphonates,
phosphorothioates, phosphorodithioates, phosphotriesters
and phosphorus-boron (Sood et al., 1990, J. Am. Chem.
Soc. 112:9000) linkages. The phosphodiester group can
be replaced with siloxane, carbonate, acetamidate or
thioether groups. These modifications can also increase
the resistance of the subject oligonucleotides to
nucleases. Methods for synthesis of oligonucleotides
with modified phosphodiester linkages are reviewed by
3o Uhl m~nn et al.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S9S/03602
-30-
1 Stem-loop and circular oligonucleotides with
non-nucleotide linkers can be prepared by any ~nown
procedure. For example, Durand et al. (1990, Nucleic
Acids Res. 18:6353-6359) provide synthetic procedures
for linking non-nucleotide chains to DNA. Such
procedures can generally be adapted to permit an
automated synthesis of a linear oligonucleotide
precursor which is then used to make a stem-loop or
circular oligonucleotide of the present invention. In
general, groups reactive with nucleotides in standard
DNA synthesis, e.g. phosphoramidite, H-phosphonate,
dimethoxytrityl, monomethoxytrityl and the like, can be
placed at the ends of non-nucleotide chains and
nucleotides corresponding to the ends of WC and H
1~ domains can be linked thereto.
Additionally, different nucleotide sugars can
be incorporated into the oligonucleotides of this
invention. Additional binding stability can be provided
by using 2'-0-methyl ribose in the present
oligonucleotides. Phosphoramidite chemistry can be used
to synthesize RNA oligonucleotides as described (Reese,
C. B. in Nucleic Acids & Molecular Bioloqy; Springer-
Verlag: Berlin, 1989; Vol. 3, p. 164; and Rao et al.,
1987, Tetrahedron Lett. 28:4897).
The synthesis of RNA 2'-0-methyl-
oligoribonucleotides and DNA oligonucleotides differ
only slightly. RNA 2'-0-methyloligonucleotides can be
prepared with minor modifications of the amidite, H-
phosphonate or phosphotriester methods (Shibahara et al,
3o 1987, Nucleic Acids Res. 15:4403; Shibahara et al.,
CA 0221~82~ 1997-09-18
Wos6/2sos7 PCT~S95/03602
-31-
1 1989, Nucleic Acids Res. 17:239; Anoue et al., 1987,
Nucleic Acids Res. 15:6131).
- The present invention contemplates a variety
of utilities for the subject stem-loop and circular
oligonucleotides which are made possible by their
selective and stable binding properties with nucleic
acid targets, nuclease resistance, and lack of toxicity.
Some utilities include, but are not limited to: use of
the subject oligonucleotides of defined sequence, bound
to a solid support, for affinity isolation of
complementary nucleic acids; use of the subject
oligonucleotides to provide sequence specific stop
signals during polymerase chain reaction (PCR); covalent
attachment of a drug, drug analog or other therapeutic
agent to the subject oligonucleotides to allow cell type
specific drug delivery; labeling the stem-loop or
circular oligonucleotides with a detectable reporter
molecule for localizing, quantitating or identifying
complementary target nucleic acids; and binding the
subject oligonucleotides to a cellular or viral nucleic
acid template and regulating biosynthesis directed by
that template.
The nucleic acid templates useful in the
methods of the present invention can be RNA or DNA and
can be single-stranded or double-stranded. In a
preferred embodiment the nucleic acid template is
single-stranded DNA or RNA. In another preferred
embodiment the nucleic acid target is mRNA or viral DNA
or RNA. In a most preferred embodiment the nucleic acid
target is pryimidine rich, single-stranded DNA or RNA.
In accordance with the present invention, pyrimidine
CA 0221~82~ 1997-09-18
W096l29097 PCT~S95/03602
-32-
1 rich refers to targets containing at least 50~
pyrimidines. In a preferred embodiment the pyrimidine
rich targets contain at least 70% pyrimidines. In a
most preferred embodiment, the pyrimidine rich targets
contain from 90-100~ pyrimidines. Double-stranded
templates that are opened during biosynthetic processes
or that can be denatured are also contemplated as
targets.
The skilled artisan can determine the
appropriate composition of the subject oligonucleotides
by the base pairing rules and further considerations
addressed herein for particular diagnostic and
therapeutic applications, and with reference to the
desired target nucleic acid. It has been discovered in
accordance with the present invention that DNA
oligonucleotides are useful for targeting both DNA and
RNA templates. Further RNA oligonucleotides are useful
for targeting both DNA and RNA templates. DNA
oligonucleotides are preferred for binding to RNA
targets.
In a method of isolation of a target nucleic
acid, the subject oligonucleotides can be attached to a
solid support such as silica, cellulose, nylon,
polyacrylamide, polystyrene, agarose and other natural
or synthetic materials that are used to make beads,
filters, and column chromatography resins. Attachment
procedures for nucleic acids to solid supports of these
types are well known; any known attachment procedure is
contemplated by the present invention. A stem-loop or
3o circular oligonucleotide attached to a solid support can
then be used to isolate a complementary nucleic acid.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-33-
1 Isolation of the complementary nucleic acid can be done
by incorporating the oligonucleotide:solid support into
a column for chromatographic procedures. Other
isolation methods can be done without incorporation of
the oligonucleotide:solid support into a column, e.g. by
utilization of filtration procedures. The subject stem-
loop and circular oligonucleotides are ideally suited to
applications of this type because they are nuclease
resistant and bind target nucleic acids so strongly.
Further utilities are available for the
subject oligonucleotides in the field of polymerase
chain reaction (PCR) technology. PCR technology
provides methods of synthesizing a double-standard DNA
fragment encoded in a nucleic acid template between two
known nucleic acid sequences which are employed as
primer binding sites. The subject oligonucleotides can
be used to selectively prevent amplification of a
particular species, e.g. a mutant or allelic variant of
known sequence. This can be done by, for example,
binding a circular oligonucleotide of the present
invention to one of the primer binding sites or to a
site lying between the primer binding sites.
The present invention also contemplates using
the subject oligonucleotides for targeting drugs to
specific cell types. Such targeting can allow selective
destruction or enhancement of particular cell types,
e.g. inhibition of tumor cell growth can be attained.
Different cell types express different genes, so that
the concentration of a particular mRNA can be greater in
one cell type relative to another cell type. Such an
mRNA is a target mRNA for cell type specific drug
_
CA 0221~82~ 1997-09-18
WO 96/29097 PCT/US95N3602
--34--
1 delivery by stem-loop or circular oligonucleotides
linked to drugs or drug analogs. Cells with high
concentrations of target mRNA are targeted for drug
delivery by administering to the cell an drug-conjugated
oligonucleotide of the present invention having a WC
domain that is complementary to the target mRNA.
The present invention also contemplates
labeling the subject oligonucleotides for use as probes
to detect a target nucleic acid. Labelled circular
1~ oligonucleotide probes have utility in diagnostic and
analytical hybridization procedures for localizing,
quantitating or detecting a target nucleic acid in
tissues, chromosomes or in mixtures of nucleic acids.
Labeling of a stem-loop or circular oligonucleotide can
be accomplished by incorporating nucleotides linked to a
reporter group into the subject oligonucleotides. A
reporter group, as defined herein, is a molecule or
group which, by its chemical nature, provides an
identifiable signal allowing detection of the circular
oligonucleotide. Detection can be either qualitative or
quantitative. The present invention contemplates using
any commonly used reporter molecule including
radionuclides, enzymes, biotins, psoralens,
fluorophores, chelated heavy metals, and luciferin. The
most commonly used reporter molecules are either
enzymes, fluorophores or radionuclides linked to the
nucleotides which are used in circular oligonucleotide
synthesis. Commonly used enzymes include horseradish
peroxidase, alkaline phosphatase, glucose oxidase and ~-
galactosidase, among others. The substrates to be usedwith the specific enzymes are generally chosen because a
-
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-35-
1 detectably colored product is formed by the enzyme
acting upon the substrate. For example, p-nitrophenyl
phosphate is suitable for use with alkaline phosphatase
conjugates; for horseradish peroxidase, 1,2-
phenylenediamine, 5-aminosalicyclic acid or toluidine
are commonly used. Fluorophores may be detected for
example by microscopy or digital imaging. Similarly,
methods for detecting radionucleotides are well-known in
the art. The probes so generated have utility in the
1~ detection of a specific DNA or RNA target in, for
example, Southern analysis, Northern analysis, in situ
hybridization to tissue sections or chromosomal squashes
and other analytical and diagnostic procedures. The
methods of using such hybridization probes are well
known and examples of such methodology are provided by
Sambrook et al.
The present stem-loop and circular
oligonucleotides can be used in conjunction with any
known detection or diagnostic procedure which is based
upon hybridization of a probe to a target nucleic acid.
Moreover, the present oligonucleotides can be used in
any hybridization procedure which quantitates a target
nucleic acid, e.g., by competitive hybridization between
a target nucleic acid present in a sample and a labeled
tracer target for one of the present oligonucleotides.
Furthermore, the reagents needed for making a stem-loop
or circular oligonucleotide probe and for utilizing such
a probe in a hybridization procedure can be provided in
a kit.
3o The kit can be compartmentalized for ease of
utility and can contain at least one first container
CA 0221~82~ 1997-09-18
wos6l2sos7 PCT~S95103602
-36-
1 providing reagents for making a precircle precursor for
a circular oligonucleotide, at least one second
container providing reagents for labeling the precircle
with a reporter molecule, at least one third container
providing regents for circularizing the precircle, and
at least one fourth container providing reagents ~or
isolating the labeled circular oligonucleotide.
Moreover the present invention provides a kit
for isolation of a template nucleic acid. Such a kit
has at least one first container providing a stem-loop
or circular oligonucleotide having a WC domain that is
complementary to a target contained within the template.
Further, a kit for the detection of any target
nucleic acid is provided which contains an
oligonucleotide of the present invention linked to a
reporter group. Additional containers providing
reagents for detecting a linked reporter group can also
be provided in the kit.
Furthermore, the present invention provides
kits useful when diagnosis of a disease depends upon
detection of a specific, known target nucleic acid.
Such nucleic acid targets can be, for example, a viral
nucleic acid, an extra or missing chromosome or gene, a
mutant cellular gene or chromosome, an aberrantly
expressed RNA and others. The kits can be
compartmentalized to contain at least one first
container providing a stem-loop or circular
oligonucleotide linked to a reporter molecule and at
least one second container providing reagents for
3o detection of the reporter molecule.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95103602
-37-
1 Therefore, as contemplated by the present
invention, the kits disclosed herein can include any
elements recognized or conventionally used by the
skilled artisan for constructing, purifying and using
oligonucleotides. Moreover, the present kits can
include specific chemical reagents or end-joining-
oligonucleotides for making the present circular
oligonucleotides.
One aspect of the present invention provides a
method of regulating biosynthesis of a DNA, an RNA or a
protein by contacting at least one of the subject stem-
loop or circular oligonucleotides with a nucleic acid
template for that DNA, that RNA or that protein in an
amount and under conditions sufficient to permit the
binding of the oligonucleotide to a target sequence
contained in the template. The binding between the
oligonucleotide and the target blocks access to the
template, and thereby regulates biosynthesis of the
nucleic acid or the protein. Blocking access to the
template prevents proteins and nucleic acids involved in
the biosynthetic process from binding to the template,
from moving along the template, or from recognizing
signals encoded within the template. Alternatively,
when the template is RNA, regulation can be accomplished
by allowing selective degradation of the template. For
example, RNA templates bound by the subject
oligonucleotides are susceptible to degradation by RNase
H and RNase H. Degradation of a selected RNA template
can thereby regulate use of the template in biosynthetic
processe
CA 0221~82~ 1997-09-18
WO 96/29097 P~,'l/u595/03602
--38--
1 As used herein, biosynthesis of a nucleic acid
or a protein includes cellular and viral processes such
as DNA replication, DNA reverse transcription, RNA
transcription, RNA splicing, RNA polyadenylation, RNA
translocation and protein translation, all of which can
lead to production of DNA, RNA or protein, and involve a
nucleic acid template at some stage of the biosynthetic
process.
As used herein, regulating biosynthesis
includes inhibiting, stopping, increasing, accelerating
or delaying biosynthesis. Regulation may be direct or
indirect, i.e. biosynthesis of a DNA, RNA or protein may
be regulated directly by binding a stem-loop or circular
oligonucleotide to the template for that DNA, RNA or
protein; alternatively, biosynthesis may be regulated
indirectly by oligonucleotide binding to a second
template encoding a protein that plays a role in
regulating the biosynthesis of the first DNA, RNA or
protein. In a preferred embodiment, the subject
oligonucleotides are used to arrest translation, reverse
transcription, or replication.
DNA replication from a DNA template is
mediated by proteins which bind to an origin of
replication where they open the DNA and initiate DNA
synthesis along the DNA template. To inhibit DNA
replica~ion in accordance with the present invention,
oligonucleotides are selected which bind to one or more
targets in an origin of replication. Such binding
blocks template access to proteins involved in DNA
3o replication. Therefore initiation and procession of DNA
replication is inhibited. As an alternative method of
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-39-
1 inhibiting DNA replication, expression of the proteins
which mediate DNA replication can be inhibited at, for
example, the transcriptional or translational level. As
one skilled in the art recognizes, DNA replication can
also be increased, e.g. by inhibiting expression of a
protein repressor of DNA replication.
DNA replication from an RNA template is
mediated by reverse transcriptase binding to a region of
RNA also bound by a nucleic acid primer. To inhibit DNA
replication from an RNA template, reverse transcriptase
or primer binding can be blocked by binding a stem-loop
or circular oligonucleotide to the primer binding site,
and thereby blocking access to that site. Moreover,
inhibition of DNA replication can occur by binding a
stem-loop or circular oligonucleotide to a site residing
in the RNA template since such binding can block access
to that site and ~o downstream sites, i.e. sites on the
3' side of the target site.
To initiate RNA transcription, RNA polymerase
recognizes and binds to specific start sequences, or
promoters, on a DNA template. Binding of RNA polymerase
opens the DNA template. There are also additional
transcriptional regulatory elements that play a role in
transcription and are located on the DNA template.
These transcriptional regulatory elements include
enhancer sequences, upstream activating sequences,
repressor binding sites and others. All such promoter
and transcriptional regulatory elements, singly or in
combination, are targets for the subject circular
oligonucleotides. Oligonucleotide binding to these
sites can block RNA polymerase and transcription factors
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-40-
1 from gaining access to the template and thereby
regulating, e.g., increasing or decreasing, the
production of RNA, especially mRNA and tRNA.
Additionally, the subject oligonucleotides can be
targeted to the coding region or 3'-untranslated region
of the DNA template to cause premature termination of
transcription. One skilled in the art can readily
design oligonucleotides for the above target sequences
from the known sequence of these regulatory elements,
from coding region sequences, and from consensus
sequences.
Protein biosynthesis begins with the binding
of ribosomes to an mRNA template, followed by initiation
and elongation of the amino acid chain via translational
1~ "reading" of the mRNA. Protein biosynthesis, or
translation; can thus be blocked or inhibited by
blocking access to the template using the subject
oligonucleotides to bind to targets in the template
mRNA. Such targets contemplated by this invention
include the ribosome binding site (Shine-Delgarno
sequence), the 5' mRNA cap site, the initiation codon,
and sites in the protein coding sequence. There are
also classes of protein which share domains of
nucleotide sequence homology. Thus, inhibition of
protein biosynthesis for such a class can be
accomplished by targeting the homologous protein domains
(via the coding sequence) with the subject circular
oligonucleotides.
Regulation of biosynthesis by any of the
3o aforementioned procedures has utility for many
applications. For example, genetic disorders can be
3~
.
CA 022l~82~ l997-09-l8
W096/29097 PCT~S95/03602
-41-
l corrected by inhibiting the production of mutant or
over-produced proteins, or by increasing production of
under-expressed proteins; the expression of genes
encoding factors that regulate cell proliferation can be
~ 5 inhibited to control the spread of cancer; and virally
encoded functions can be inhibited to combat viral
infection.
In accordance with the present invention, it
has been determined that in some instances the
biosynthesis of a DNA, RNA or protein is more
effectively regulated by binding the template at more
than one target site. The present stem-loop and
circular oligonucleotides which are prepared to bind to
multiple target sites, e.g. by having more than one WC
and H domain, can also be more effective at regulating
the biosynthesis of a DNA, RNA or protein than
oligonucleotides that can bind only one target site.
For example, the binding of two sites within a gene can
provide greater inhibition than achieved with single-
site binding (Lisziewicz et al., 1992, Proc. Natl. Acad.Sci. USA 89:11209; Maher et al., 1987, J. Arch. Biochem.
Biophys. 253:214-220; Tannock, I.F. in "The Basic
Science of Oncology" 2nd ed.; Tannock, I.F. and Hill, R.
P., eds. McGraw-Hill, New York, 348-349). In targeting
viral sequences, the binding of two genes in a virus can
inhibit viral replication more effectively than binding
a single target. It has been shown, for example, that
the use of multiple probes against a virus reduces the
ability of the virus to escape inhibition by mutation
3o (Kern et al., 1991, Science 252:17~8-1711). A broader
spectrum o~ inhibition by targeting two mutants of one
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95103602
-42-
l virus or two viruses which are commonly found together,
such as HIV-l and cytomegalovirus (CMV) can also be
achieved in accordance with the present invention.
Therefore, the present methods of regulating
the biosynthesis of a DNA, RNA or protein can also
include binding to more than one target within a
template, whether the targets are bound by separate
stem-loop and circular oligonucleotides or by the same
oligonucleotide which includes multiple WC and H
domains.
Some types of genetic disorders that can be
treated by the circular oligonucleotides of the present
invention include Alzheimer's disease, beta-thalassemia,
some types of arthritis, sickle cell anemia,
osteogenesis imperfecta and others. Many types of viral
infections can be treated by utilizing the
oligonucleotides of the present invention, including
infections caused by influenza, hepatitis, rhinovirus,
HIV, herpes simplex, papilloma virus, cytomegalovirus,
Epstein-Barr virus, adenovirus, vesticular stomatitus
virus, rotavirus and respiratory syncytial virus among
others. According to the present invention, ~nim~ and
plant viral infections may also be treated by
administering the subject oligonucleotides.
Human immunodeficiency virus (HIV) is a
retrovirus causing acquired immunodeficiency syndrome
(AIDS). The stem-loop and circular oligonucleotides of
this invention provide a means of blocking the
replication of the virus without deleteriously affecting
3o normal cellular replication in humans infected with HIV.
Inhibition of HIV infection can be accomplished by
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-43-
1 designing oligonucleotides to bind to a number of
regions within the HIV genome, including coding regions
~ for functions that replicate the genome (i.e., tha pol
or reverse transcriptase function) or functions that
~ 5 control gene expression (e.g. the tat, rev or other
functions). Previous work with linear oligonucleotides
has suggested that splice sites, poly(A) addition
signals, cap or initiator codon sites, and sites
implicated in ribosome assembly can be particularly
effective for inhibiting eucaryotic protein expression.
Furthermore, the terminal structures of the retroviral
genome are also excellent targets for inhibiting
retrovirus production not only because these structures
encode control regions which mediate the rate of
transcription and replication, but also because these
structures are repeated, allowing an oligonucleotide to
bind and block access to each repeat.
In vitro screening for stem-loop or circular
oligonucleotide effectiveness against HIV infection
permits one skilled in the art to judge the stability of
oligonucleotide: target binding and to assess in vivo
efficacy and binding stability. To observe in vitro
inhibition, stem-loop or circular oligonucleotides can
be added to the growth medium of an appropriate cell
line infected with HIV. Cells can be pretreated with
the subject oligonucleotides or the oligonucleotides can
be added at the time of infection or after HIV
infection. Addition before or after infection allows
assessment of whether the subject oligonucleotide can
3o prevent or simply inhibit HIV infection respectively.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-44-
l The extent of inhibition of HIV infection or
replication can be judged by any of several assay
systems, including assessment of the proportion of
oligonucleotide-treated cells surviving after infection
relative to survival of untreated cells, assessment of
the number of syncytia formed in treated and untreated
HIV infected cells and det~rmin~tion of the amount of
viral antigen produced in treated and untreated cells.
In vivo studies of the efficacy of subject
oligonucleotides can be done in a suitable animal host,
such as transgenic mice, immune deficient mice or
chimpanzees. Levels of HIV antigens can be monitored to
assess the effect of subject oligonucleotides on HIV
replication and thereby to follow the course of the
disease state. Alternatively, human volunteers with
AIDS or ARC can be administered with the subject
circular oligonucleotides since the oligonucleotides do
not appear to be cytotoxic. The disease status of these
volunteers can then be assessed to determine the
efficacy of the subject oligonucleotides in treating and
preventing AIDS infection.
A further aspect of this invention provides
pharmaceutical compositions containing the subject stem-
loop or circular oligonucleotides with a
pharmaceutically acceptable carrier. In particular, the
subject oligonucleotides are provided in a
therapeutically effective amount of about O.l ~g to
about lO0 mg per kg of body weight per day, and
preferably of about O.l ~g to about lO mg per kg of body
3o weight per day, to bind to a nucleic acid in accordance
with the methods of this invention. Dosages can be
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-45-
1 readily determined by one of ordinary skill in the art
and formulated into the subject pharmaceutical
compositions.
As used herein, "pharmaceutically acceptable
carrier" includes any and all solvents, dispersion
media, coatings, antibacterial and antifungal agents,
isotonic and absorption delaying agents, and the like.
The use of such media and agents for pharmaceutical
active substances is well known in the art. Except
insofar as any conventional media or agent is
incompatible with the active ingredient, its use in the
therapeutic compositions is contemplated. Supplementary
active ingredients can also be incorporated into the
compositions:
1~ The subject oligonucleotides may be
administered topically or parenterally by, for example,
by osmotic pump, intravenous, intramuscular,
intraperitoneal subcutaneous or intradermal route, or
when suitably protected, the subject oligonucleotides
may be orally administered. The subject
oligonucleotides may be incorporated into a cream,
solution or suspension for topical administration. For
oral administration, oligonucleotides may be protected
by enclosure in a gelatin capsule. Oligonucleotides may
be incorporated into liposomes or liposomes modified
with polyethylene glycol or admixed with cationic lipids
for parenteral administration. Incorporation of
additional substances into the liposome, for example,
antibodies reactive against membrane proteins found on
3o specific target cells, can help target the
oligonucleotides to specific cell types. Stem-loop
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-46-
1 oligonucleotides may be delivered as an expression
vector.
Moreover, the present invention contemplates
administering the subject oligonucleotides with an
osmotic pump providing continuous infusion of such
oligonucleotides, for example, as described in Rataiczak
et al. (1992, Proc. Natl. Acad. Sci. USA 89:11823-
11827). Such osmotic pumps are commercially available,
e.g., from Alzet Inc (Palo Alto, CA). Topical
administration and parenteral administration in a
cationic lipid carrier are preferred modes of
administration.
The following examples further illustrate the
invention.
3o
CA 0221~82~ 1997-09-18
wos6l2sos7 PCT~S95/03602
-47-
1 EXAMPLE 1
Design and Synthesis of Oligonucleotides
r
Four deoxynucleotides were designed to bind
the 12-mer target sequences 5'-dCTCCTCCCTCCT (SEQ ID
NO:l) and 5'-rCUCCUCCCUCCU (SEQ ID NO:2) by conventional
Watson-Crick base pairing or by the formation of
pur-pur-pyr triplexes. The oligonucleotides are as
follows:
Oligonucleotide 1: 5'-AGGAGGGAGGAG (SEQ ID
No:3)
Oligonucleotide 2: 5'-AGGAGGGAGGAGCACACGTGG
TGGGTGGT (SEQ ID NO:4)
Oligonucleotide 3:
1 ~
CGTGGTGGGTGGTC
C AC
A A
CGAGGAGGGAGGAC (SEQ ID
NO:5)
Oligonucleotide 4:
5'-AGGAGGGAGGAGCACACGTGGTGTGTGGT (SEQ ID NO:6)
Oligonucleotide 1 is the Watson-Crick
complement of the target sequence. Linear
oligonucleotide 2 and circular oligonucleotide 3 each
contain a 12-nucleotide domain that is complementary to
the target in antiparallel Watson-Crick sense, and an
opposing 12-nucleotide domain that is complementary to
the first domain in antiparallel reverse Hoogsteen
sense. Oligonucleotide 4 is similar to
3~ oligodeoxynucleotide 2 but contains a single mismatch in
- the Hoogsteen domain.
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95/03602
-48-
1 The strategy for binding of oligonucleotides
1-4 to the target strand is illustrated in Table 1
wherein short lines denote Watson-Crick hydrogen bonds
and dots indicate Hoogsteen bonds.
DNA oligomers were synthesized on an Applied
Biosystems 392 automated synthesizer using the st~n~d
phosphoramidite method described by Beaucage et al.
(1981) Tetrahedron Lett. 22:1859. RNA oligonucleotides
were prepared using t-butyl-dimethylsilyl-protected
phosphoramidites (Applied Biosystems), and following the
oligoribonucleotide synthesis procedure of Scaringe et
al. (1990) Nucleic Acids Res. 18:5433.
Tetrabutylammonium fluoride in THF (Aldrich) was dried
over molecular sieves prior to use in the desilylation
step as described by Horn et al. (1986) Tetrahedron
Lett. 27:4705. For synthesis of circular DNAs the
linear precursors were 5'-phosphorylated on the
synthesizer using a commercially available reagent
purchased from Cruachem as described by Hogrefe et al.
(1993) Nucleic Acids Res. 21:4739. Oligonucleotides
were purified by preparative 20% denaturing
polyacrylamide gel electrophoresis and quantitated by
absorbance at 260 nm. Extinction coefficients for the
oligomers were calculated by the nearest neighbor method
Of Borer (1985) in G. D. Fasman (ed.) Handbook of
Biochemistry and Molecular Bioloqy, 3rd Ed. CRC Press,
Cleveland, Vol. I, p. 589.
The cyclizations of the 5'-phosphorylated
precursor of circular llgand 3 was carried out
3o essentially as described by Prakash et al. (1992) J. Am.
Chem. Soc. 114:3523 and Wang et al. (1994) Nucleic Acids
CA 0221~82~ 1997-09-18
W096l29097 PCT~S95/03602
-49-
1 Res. _:2326. The precursor was reacted at 50 ~M
concentration; the cyclization of 3 was aided by the
template dCTCCTCCCTCCT (SEQ ID NO:1) (55 ~M). The
ligation was carried out in a buffer containing
imidazole-HCl (200 mM, from a 0.5 M pH 7.0 stock), and
NiCl2 (100 mM). Solid BrCN was added with vortex mixing
to give a final calculated concentration of 125 mM, and
the reaction was allowed to proceed at 25~C for 12 hr.
As the reaction proceeded a light tan precipitate was
observed, and previous studies have shown that this
precipitate contains the majority of the nucleic acids.
The solution (including solids) was dialyzed against
water and lyophilized. The resulting solid was loaded
onto a preparative 20~ denaturing polyacrylamide gel for
separation. The circular product was isolated by
excision from the gel after visualization by W
shadowing; the circle 3 migrated at a rate -0.9 times
that of the linear 34-mer precursor.
The circularity of 3 was confirmed by partial
digestion by S1 nuclease. The reaction was carried out
using 1 nmol DNA in 5.1 ~L of a buffer containing 50 mM
NaOAc, 50 mM NaCl, and 5 mM ZnCl2. Then 0.4 units
nuclease S1 (0.9 ~L, Pharmacia) was added, and the
mixture incubated at 37~C for 10 min. The reaction was
stopped by addition of 6 ~L of an 8 M urea, 30 mM EDTA
solution, and the mixture was loaded onto a 20~
denaturing analytical polyacrylamide gel. Products were
visualized for photography with Stains-all dye (Sigma).
The compound showed a single initial product which
migrates with the mobility of the 34-mer precursor.
CA 0221~82~ 1997-09-18
W096l29097 PCT~S95/03602
-50-
1 EXAMPLE 2
Binding Properties of Linear
and C~rcular Oligonucleotides
The binding properties of oligonucleotides 1-4
were characterized by thermal denaturation and gel
titration studies with DNA and RNA target strands
monitored at 260 nm.
Solutions for the thermal denaturation studies
contained a 1:1 molar ratio of oligonucleotide ligand
and complementary 12-nt pyrimidine target (1 ~M each).
Solutions were buffered with 10 mM Na-PIPES (1,4-
piperazine-bis(ethanesulfonate), Sigma) at pH 7.0 or at
pH 5.5. The buffer pH is that of a 500 mM stock
solution at 25~C; after dilution the final solution pH
was shown to be within 0.1 unit of the buffer stock.
Also present in the denaturation solutions were 100 mM
NaCl and 10 mM MgCl2. After preparation the solutions
were heated to 90~C and allowed to cool slowly to room
temperature prior to the melting experiments.
The melting studies were carried out in
teflon-stoppered 1 cm pathlength quartz cells under
nitrogen atmosphere on a Varian ~ary 1 W-vis
spectrophotometer equipped with thermoprogrammer.
Absorbance (260 nm) was monitored while temperature was
raised from 5.0 to 95~C at a rate of 0.5~C/min. Melting
temperatures (Tm) were determined by computer fit of the
first derivative of absorbance with respect to 1/T.
Uncertainty in Tm is estimated at + 0.5~C ~ased on
repetitions of experiments.
Free energies were determined by curve fitting
as described by Petersheim et al. (lg82) Biochemistry
CA 022l~82~ l997-09-l8
W O 96/29097 PC~rAUS95/03602
-5 1-
1 _:256. Values at 60~C are more accurate than those at
37~C because of smaller extrapolation from the Tm
r valueS.
For gel titration studies; pH 7.0 solutions of
ratio 2:1, 1:1 and 1:2 circle:substrate oligomer (0.25
or 0.5 nmol each) were prepared at 4~C in 5 ~L of a
buffer containing 70 mM Tris-borate, 10 mM MgCl2, and 6%
glycerol and incubated for 4 hr prior to loading on a
20% nondenaturing PAGE gel. The gels were
electrophoresed at 2.5 mW at 4~C using the same buffer
as the electrophoresis buffer, and the resulting bands
were visualized with Stains-all dye.
Results of binding studies of oligomers 1-4
with complementary single stranded DNA targets are shown
in Table 1.
3o
CA 02215825 1997-09-18
W 096/29097 PCTrUS95103602
-52-
1 Table 1. Melting Transition Temperatures (T (~C)) and Free Energies
(-~G~ (kcal/mol)) for Complexes of Linear and Circular Purine-Rich DNAs
with Complementary Pyrimidine DNA Single Strands at pH 7.0 (Lines
Indicate Watson-Crick Complementarity, Dots Hoogsteen Complementarity,
and Arrows 5' to 3' Directionality )
OLIGONU- -~G~ ' -AG~6
5 CLEOTIDE COMPLEX T (~C)- (k 1/3~ 1) (kcal/mol)
3'-GAGGAGGGAGGA
56.914.2 8.4
5'-CTCCTCCCTCCT
C ~l~lGGGTGGT
A
C ~----.-.....
2 A 68.4 16.9 11.6
C GAGGAGGGAGGA-5'
5'-~1C~l~CLl~CT
C ~l~ G~l~l C
A A
3 C ~----------- C 71.0 17.3 12.0
A A
C-,GAGGAGGGAGGA C
l l l l l l l l l l l l
5'-~lC~lC~lCCT
C ~~ ~l~lG~
C X
4 A 61.9 12.4 9.4
C GAGGAGGGAGGA-5'
111111111111
5'-CTCCTCCCTCCT
Uncertainties in T values and in free energies are estimated at
+ 1.0~C and + 15%, respectively.
3o
CA 0221~82~ 1997-09-18
wos6l2sos7 PCT~S95/03602
53
1 The results in Table I show that
oligonucleotides 2 and 3 do in fact bind the 12-mer DNA
target, and with considerably higher affinity than does
the Watson-Crick 12-mer complement (oligonucleotide 1).
Specifically, oligonucleotide 2 binds
dCTCCTCCCTCCT with a T~ of 68.4~C (conditions: pH 7.0
(10 mM Na-PIPES buffer), 100 mM Na , 10 mM Mg2) and a
free energy estimated at -16.9 kcal/mol at 37~C. This
is in contrast to the simple 12-mer Watson-Crick
1~ complement oligonucleotide 1, which binds with a T~ of
56.9~C and a free energy of -14.2 kcal/mol under
identical conditions. The closed circular
oligonucleotide 3 binds the target with the highest
thermal stability, with a Tm of 71.0~C and a free energy
15 ~f -17.3 kcal/mol. This represents an advantage of 2
orders of magnitude in association constant over simple
Watson-Crick recognition. A similar advantage is seen
in free energies calculated at 60~C.
The stoichiometry of the complex of 3 was
20 measured by titration of the target with the ligand,
monitored by denaturing gel electrophoresis. This
confirmed 1:1 stoichiometry as predicted for the
expected triple-helical complex. The single-mismatched
oligonucleotide 4 was also hybridized to the pyrimidine
25 complement- Results show that it binds the DNA target
with significantly lower affinity; this confirms the
importance of the Hoogsteen strand in increasing the
affinity of binding, even though it presumably does not
come in direct contact with the target.
The binding of oligonucleotide 3 to the single
stranded DNA target was not significantly pH dependent,
CA 02215825 1997-09-18
W096/29097 PCT~S95103602
-54-
1 giving the same Tm, within experimental error, at pH 5.5
as that measured at pH 7Ø
Results of binding studies of oligonucleotides
1-4 with complementary single stranded RNA targets are
5 shown in Table 2.
3G
CA 0221~82~ 1997-09-18
WO 96/29097 PCT/US95/03602
--55--
1 Table 2. Melting Transition Temperatures (T (~C)) and Free Energies.
(-~G~ (kcal/mol)) for Complexes of Linear and Circular Purine-Rich DNAs
with Complementary Pyrimidine RNA Single Strands at pH 7.0 (Lines
r Indicate Watson-Crick Complementarity, Dots Hoogsteen Complementarity,
and Arrows 5' to 3' Directionality
OLIGONU- -~G~ ~ -~G~ A
5 CLEOTIDE COMPLEX T (~C)' (kcal/mol) (kcal/mol)
3'-dGAGGAGGGAGGG
58.9 15.6 8.3
5'-rCUCCUCCCUCCU
C GTGGTGGGTGGT
C ~--
2 A 65.3 14.5 10.4
C GAGGAGGGAGGA-5'
l l l l l ~ l l l l l l
5'-rCUCCUCCCUCCU
C GTGGTGGGTGGT C
A A
3 C ~----------- C 69.2 15.4 11.2 A A
C GAGGAGGGAGGA C
5'-rCUCCUCCCUCCU
C ~~ G~
cA X
4 A 64.3 15.3 10.3
C GAGGAGGGAGGA-5'
llllllllllll
5'-rCUCCUCCCUCCU
Uncertainties in T values and in free energies are estimated at
+ 1.0~C and + 15%, respectively.
3o
CA 0221~82~ 1997-09-18
W096/29097 PCT~S95103602
-56-
1 As shown in Table 2, oligonucleotides 2 and 3
also bind single-stranded RNA with high affinity.
~airpin-shaped oligonucleotide 2 binds rCUCCUCCCUCCU
with a T~ of 65.3~C, for an advantage of 5.4~C over the
5 Watson-Crick complement, and circular oligonucleotide 3
binds with the highest thermal stability, with a Tm
advantage of 10.3~C over simple Watson-Crick binding.
The estimated free energies at 37~C do not reflect this
difference; however, more accurate values calculated for
10 60~C do mirror the advantage seen in the melting
temperatures. Mismatched oligonucleotide 4 binds the
RNA target somewhat less strongly than the fully
complementary hairpin. There is a clear binding
advantage for circular ligands over the simple Watson-
15 Crick complement. The data in Tables 1 and 2demonstrate that both DNA and RNA strands can be
strongly bound using the same DNA ligands.
3o
CA 0221~82~ 1997-09-18
W096129097 PCT~S95103602
-57-
1 EXAMPLE 3
Circular DNA Oligonucleotides Bind
with High Affinity to DNA and RNA Targets
A circular DNA (oligonucleotide 5) having the .
sequence:
1 ~
GTGGTGGGTGGT
C CC
GAGGAGGGAGGA (SEQ ID NO:7)
was synthesized by the method described in Example l.
The following targets were also synthesized:
5'.rCUCCUCCCUCCU (SEQ ID NO:2)
5'-dCTCCTCCCTCCT (SEQ ID NO:l)
5'-dCCCCCACTCCTCCCTCCTACCCCC (SEQ ID NO:8)
SEQ ID NO:8 represents a "long" target in which the
sequence complementary to the WC domain is embedded in a
longer template.~0
Binding properties of oligonucleotide 8 to the
RNA target (SEQ ID NO:2), short DNA target (SEQ ID NO:l)
and long DNA target (SEQ ID NO:8) were assessed by the
method described in Example 2. The results in Table 3
demonstrate that the circular DNA binds with high
affinity to DNA and RNA targets.
3o
CA 022l5825 l997-09-l8
W096l29097 PCT~S95/03602
-58-
l Table 3. Melting transition temperature (Tm(~C)) and
Free Energies (-~G~37 (kcal/mol) for complexes of
Oligonucleotide 5 wlth Complementary Pyrimidine DNA.and
RNA single Strands at pH 7.0
A- ~G~ ~
TARGET T~(~C) (kcal/mol)
RNA (SEQ ID NO:2) 71.7 12.5
short DNA (SEQ ID NO:l) 70.5 12.3
long DNA (SEQ ID NO:8) 69.6 11.9
Error in Tm values and in free energies are
estimated at + 1.0~C and + 15%, respectively.
3o
-
CA 02215825 1997-09-18
Wo 96/29097 PCT/US95/03602
--59--
SEQUENCE LISTING
r ( 1 ) ~.~N~R~T. INFORMATION:
(i) APPLICANT: Research Corporation Technologies, Inc.
~ 5 (ii) TITLE OF lNv~hllON: STEM-LOOP AND frR~TTr~AR OLIGONUCLEOll~:S
(iii) NUMBER OF ~yu~ :S: 8
(iv) CORRE~nr"~.r~ An~
fA'I Al-l)R~.~ :: Scully, Scott, Murphy & Presser
~Bl STREET: 400 Garden City Plaza
C~ CITY: Garden City
~Dj STATE: New York
(El COu,~-~Y: U.S.A.
o ~Fj ZIP: 11530-0299
(v) CO.I~U1~K R~AnART-~ FORM:
A' MEDIUM TYPE: Floppy disk
Bj CU. ~U~K: IBM PC compatible
fC, OPERATING SYSTEM: PC-DOS/MS-DOS
~Dj SOFTWARE: PatentIn Release #1.0, Version #1.25
(vi) ~uKR~:~L APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE: 21-MAR-1995
(C).CLASSIFICATION:
(viii) All~RN~:Y/AGENT INFORMATION:
(A) NAME: DiGiglio, Frank S.
(B) REGISTRATION NUMBER: 31,346
(C) K~N~:/DOCRET NUMBER: 9373
(ix) TELEC~.~.JNlCATION INFORMATION:
(A) TELEPHONE: (516) 742-4343
(B) TELEFAX: (516) 742-4366
(C) TELEX: 230 901 SANS UR
(2) INFORMATION FOR SEQ ID NO:l:
:yU~NC~ CHARACTERISTICS:
Aj LENGTH: 12 base pairs
~B) TYPE: nucleic acid
fCI STRANu~N~SS: single
~Dj TOPOLOGY: linear
(xi) ~yu~Nc~ DESCRIPTION: SEQ ID NO:l:
~ ~C~-C CT 12
3o
CA 02215825 1997-09-18
WO 96/29097 PCT/US9S/03602
--60--
1 (2) INFORMATION FOR SEQ ID NO:2:
~N~ CHARACTERISTICS:
(A) LENGTH: 12 base pairs
( B ) TYPE: nucleic acid
(C) STRAN~N~SS: single
(D) TOPOLOGY: linear
(xi) ~UU~N~ DESCRIPTION: SEQ ID NO:2:
L:U~ UI :C~.:UL CU 12
(2) INFORMATION FOR SEQ ID NO:3:
(i) ~QU~N_~: CHARACTERISTICS:
(A) LENGTH: 12 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(Xi) ~ U~I.C~ DESCRIPTION: SEQ ID NO:3:
AG&AGGGAGG AG 12
(2) INFORMATION FOR SEQ ID NO:4:
(i) ~UU~L~C~ CHARACTERISTICS:
(A)-LENGTH: 29 base pairs
( B ) TYPE: nucleic acid
(C) STR~Nn~nN~S: single
(D) TOPOLOGY: linear
(xi~ yu~--N~ DESCRIPTION: SEQ ID NO:4:
A~r.ArGr.Ar,G Ar.rArArGTG ~lG~l~ 29
(2) INFORMATION FOR SEQ ID NO:5:
(i) ~r:UUr1._~-' CHARACTERISTICS:
(A' LENGTH: 34 base pairs
~Bl TYPE: nucleic acid
~C~ STRAN~ ~N~:SS: single
~D, TOPOLOGY: circular
(xi) ~Uu~-~ DESCRIPTION: SEQ ID NO:5:
~L~LG~ GTrArArAGG ArGr.ArrArC ACAC 34
3o
CA 02215825 1997-09-18
WO 96/29097 PCT/US95/03602
--61--
1 (2) INFORMATION FOR SEQ ID NO:6:
(i) s-~Q~N~ CHARACTERISTICS:
A LENGTH: 29 base pairs
B'I TYPE: nucleic acid
~C STRAN~E~N~-SS: single
~D TOPOLOGY: linear
(xi) SEQurN~ DESCRIPTION: SEQ ID NO:6:
AGGAGGGAGG AGCACACGTG ~l~I~ 29
(2) INFORMATION FOR SEQ ID NO:7:
(i) sE~u~N~ CHARACTERISTICS:
(A) LENGTH: 28 base pairs
tB) TYPE: nucleic acid
0 C) STRANDEDNESS: single
'D) TOPOLOGY: circular
(xi) s~:~U N~ DESCRIPTION: SEQ ID NO:7:
G~l~G~l~ GTCCAGGAGG GAGGAGCC 28
(2) INFORMATION FOR SEQ ID NO:8:
(i) ~Qu~r~c~ CHARACTERISTICS:
(A) LENGTH: 24 base pairs
(8) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:8:
CrCCrACTCC ~ AC CCCC 24
3o