Note: Descriptions are shown in the official language in which they were submitted.
CA 022667~0 1999-03-17
W O 98/12352 PCTrUS97/16467
CLEAVED AMPLIFIED RFLP DETECTION METHODS
This invention was made with Government support under Contract No. GM
48707 awarded by the National Institutes of Health and Contract No. MCB-9406240
awarded by the National Science Foundation. The Government has certain rights inthe invention.
Back~round of the Invention
This invention relates to the generation and detection of genetic
polymorphisms.
Genetic maps consisting primarily of restriction fragment length polymorphic
(RFLP) markers are being constructed for many organisms, including man.
Traditional approaches for detecting RFLPs involve Southern blot hybridization.
Recently, techniques based on the polymerase chain reaction (PCR; Mullis et al.,Methods Enzymol. 155:350-355, 1987) have been used in addition to, or in place of,
traditional RFLP markers in genetic analysis (Cox et al., BioEssays 13:193-198,
1991). In contrast to traditional RFLP markers, PCR-generated markers can be scored
using a small sample of DNA, without the use of radioactivity, and without the need
for time-consuming DNA blotting procedures.
One widely used PCR-based approach involves the use of single short PCR
primers of albill~l y sequence called RAPD primers (for Eandom amplified
~2olymorphic DNA; Reiter et al., Proc. Natl. Acad. Sci. USA 89:1477-1481, 1992;
Williams et al., Theoret. Appl. Genet. 82:489-498, 1991). A second category of PCR-
based markers are called SSLPs (for simple sequence length polymorphism). The
method employing SSLPs is based on amplification across tandem repeats of one or a
~ few nucleotides known as "microsatellites." Microsatellites occur frequently and
randomly in most eukaryotic genomes and display a high degree of polymorphism due
to variation in the numbers of repeated units.
A third category of PCR-based markers are called AFLPs (for amplified
fragment length polymorphisms). In the method employing these markers, DNA from
CA 022667~0 1999-03-17
WO 9~/12352 PCT/US97tl6467
two polymorphic strains are cleaved with one or two restriction endonucleases, and
adapters are ligated to the ends of the cleaved fragments. The fragments are then
amplified using primers complementary to the adapter(s). The primers contain short
stretches of random nucleotides at their 3' ends, which result in limiting the number of
5 amplified fragments generated.
Summary of the Invention
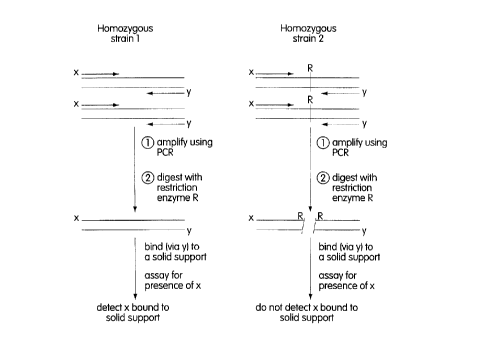
We have developed novel PCR-based methods for detecting the presence or
absence of a polymorphic restriction site in a nucleic acid involving the use of0 differentially labeled PCR primers and oligonucleotides.
Accordingly, in one aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, involving the
steps of (a) amplifying the nucleic acid by PCR using a first and a second primer
fl~nking the polymorphic restriction site, the first primer being tagged with the first
5 member of a specific binding pair, the second primer being tagged with a detectable
label; (b) digesting the PCR product of step (a) with the restriction endonuclease
corresponding to the polymorphic restriction site; (c) contacting the reaction product
of step (b) with the second member of the specific binding pair, immobilized on a
solid support; and (d) measuring the level of the detectable label bound to the solid
2 o support, the presence of the detectable label bound to the solid support being an
indication of the absence of the polymorphic restriction site in the nucleic acid.
In a second aspect, the invention features a method for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, involving the steps
of: (a) amplifying the nucleic acid by PCR using a first and a second primer fl~nking
2 5 the polymorphic restriction site, the first primer being tagged with the first member of
a specific binding pair, the second primer being tagged with a first detectable label; (b)
digesting the PCR product of step (a) with the restriction endonuclease corresponding
to the polymorphic restriction site; (c) annealing and ligating to the single-stranded
CA 022667~0 1999-03-17
W O 98112352 PCTrUS97/16467
ends generated in the reaction of step (b) an oligonucleotide tagged with a second
detectable label; (d) contacting the reaction product of step (c) with the second
member of the specific binding pair, immobilized on a solid support; and (e)
determining the levels of the first and second detectable labels bound to the solid
5 support, the presence of only the first detectable label bound to the solid support being
an indication of a homozygote lacking the polymorphic restriction site, the presence of
only the second detectable label bound to the solid support being an indication of a
homozygote containing the polymorphic restriction site, and the presence of both the
first and second detectable labels bound to the solid support being an indication of a
1 o heterozygote.
In a third aspect, the invention features a method for detecting the presence orabsence of a polymorphic restriction site in a nucleic acid, the method involving the
steps of: (a) amplifying the nucleic acid by PCR using a first and a second primer
fl~nking the polymorphic restriction site, the first primer being tagged with a
5 detectable label, the second primer being unlabeled; (b) digesting a portion of the
reaction of step (a) with the restriction endonuclease corresponding to the
polymorphic restriction site, while leaving another portion of the reaction of step (a)
undigested; (c) denaturing the digested and undigested portions from step (b); (d)
contacting the product of step (c) with an oligonucleotide complementary to a
2 o sequence in the strand of the product of step (c) containing the detectable label, the
sequence being between the polymorphic restriction and the sequence complementary
to the second primer, the oligonucleotide being tagged with a first member of a
specific binding pair; (e) contacting the reaction product of step (d) with the second
member of the specific binding pair, immobilized on a solid support; and (f)
2 5 dele. . " i l~ing the ratio of the levels of the detectable label bound to the solid support
between undigested and digested samples, a ratio of 1:0 between equivalent portions
of the undigested and digested samples being an indication of a homozygote
, .
CA 022667~0 1999-03-17
W O9~/12352 PCTrUS97/16467
containing the polymorphic restriction site, a ratio of 1:1 between equivalent portions
of the undigested and digested samples being an indication of a homozygote lacking
the polymorphic restriction site, and a ratio of 2:1 between equivalent portions of the
undigested and digested samples being an indication of a heterozygote.
In a fourth aspect, the invention features a method for detecting the presence or
absence of a polymorphic restriction site in a nucleic acid, involving the steps of: (a)
amplifying the nucleic acid by PCR using a first and a second primer fl~nking the
polymorphic restriction site, the first primer being tagged with a first detectable label,
the second primer being tagged with a second detectable label; (b) digesting the0 reaction product of step (a) with the restriction endonuclease corresponding to the
polymorphic restriction site; (c) denaturing the reaction product of step (b); (d)
contacting the product of step (c) with a first and a second oligonucleotide, the first
oligonucleotide being complementary to a first sequence in the strand of the product
of step (c) containing the first detectable label, the first sequence being between the
polymorphic restriction site and the sequence corresponding to the first primer, the
first oligonucleotide being tagged with the first member of a first specific binding pair,
the second oligonucleotide being complementary to a second sequence in the strand of
the product of step (c) containing the second detectable label, the second sequence
being on the same side of the polymorphic restriction site as the first sequence, the
2 o second sequence not being contained within or being complementary to either of the
first or second primers, the second oligonucleotide being tagged with the first member
of a second specific binding pair; (e) contacting a first portion of the reaction product
of step (d) with the second member of the first specific binding pair, immobilized on a
first solid support; (f) contacting a second portion of the reaction product of step (d)
2 5 with the second member of the second specific binding pair, immobilized on a second
solid support; and (g) det~ it~ g the ratio of the levels of the first and second
detectable labels bound to the first and second solid supports, a ratio of 1:0 between
equivalent amounts of the first and second portions being an indication of a
CA 022667~0 1999-03-17
WO ~8/12352 PCTAUS97/16467
homozygote containing the polymorphic restriction site, a ratio of 1:1 between
equivalent amounts of the first and second portions being an indication of a
homozygote lacking the polymorphic restriction site, and a ratio of 2 :1 betweenequivalent amounts of the first and second portions being an indication of a
heterozygote.
In a fifth aspect, the invention features a method for detecting the presence orabsence of a polymorphic restriction site in a nucleic acid, involving the steps of: (a)
amplifying the nucleic acid by PCR using a first and a second primer fl~nking the
polymorphic restriction site, the first primer being tagged with a first detectable label,
10 the second primer being tagged with a second detectable label; (b) digesting the
reaction product of step (a) with the restriction endonuclease corresponding to the
polymorphic restriction site; (c) denaturing the reaction product of step (b); (d)
contacting the product of step (c) with a first and a second oligonucleotide, the first
oligonucleotide being complementary to a first sequence in the strand of the product
of step (c) containing the first detectable label, the first sequence being between the
polymorphic restriction site and the sequence complementary to the second primer, the
first oligonucleotide being tagged with the first member of a first specific binding pair,
the second oligonucleotide being complementary to a second sequence in the strand of
the product of step (c) containing the second detectable label, the second sequence
2 o being on the same side of the polymorphic restriction site as the first sequence, the
second sequence not being contained within or being complementary to either of the
first or second primers, the second oligonucleotide being tagged with the first member
of a second specific binding pair; (e) contacting a first portion of the reaction product
of step (d) with the second member of the first specific binding pair, immobilized on a
first solid support; (f) contacting a second portion of the reaction product of step (d)
with the second member of the second specific binding pair, immobilized on a second
solid support; and (g) determining the ratio of the levels of the first and second
detectable labels bound to the first and second solid supports, a ratio of 0:1 between
CA 022667~0 1999-03-17
WO 98112352 PCT/US97/16467
equivalent amounts of the first and second portions being an indication of a
homozygote containing the polymorphic restriction site, a ratio of 1:1 between
equivalent amounts of the first and second portions being an indication of a
homozygote lacking the polymorphic restriction site, and a ratio of 1:2 between
5 equivalent amounts of the first and second portions being an indication of a
heterozygote.
In a sixth aspect, the invention method for detecting the presence or absence ofa polymorphic restriction site in a nucleic acid, involving the steps of: (a) amplifying
the nucleic acid by PCR using a first and a second primer fl~nking the polymorphic
o restriction site, the first primer being tagged with a first detectable label, the second
primer being tagged with a second detectable label; (b) digesting the reaction product
of step (a) with the restriction endonuclease corresponding to the polymorphic
restriction site; (c) denaturing the reaction product of step (b); (d) contacting the
product of step (c) with a first and a second oligonucleotide, the first oligonucleotide
being complementary to a first sequence in the strand of the product of step (c)containing the first detectable label, the first sequence being between the polymorphic
restriction site and the sequence corresponding to the first primer, the first
oligonucleotide being tagged with the first member of a specific binding pair, the
second oligonucleotide being complementary to a second sequence in the strand of the
2 o product of step (c) containing the second detectable label, the second sequence being
on the same side of the polymorphic restriction site as the first sequence, the second
sequence not being contained within or being complementary to either of the first or
second primers, the second oligonucleotide being tagged with the first member of the
specific binding pair; (e) contacting the reaction product of step (d) with the second
member of the specific binding pair, immobilized on a solid support; and
(f) determining the ratio of the levels of the first and second detectable labels bound to
the solid support, a ratio of 1:0 being an indication of a homozygote coll~ahlillg the
polymorphic restriction site, a ratio of 1:1 being an indication of a homozygote lacking
CA 022667~0 1999-03-17
W O9~/12352 PCTrUS97/16467
the polymorphic restriction site, and a ratio of 2:1 being an indication of a
heterozygote.
In a seventh aspect, the invention features a method for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, involving the steps
5 of: (a) amplifying the nucleic acid by PCR using a first and a second primer fl~nking
the polymorphic restriction site, the first primer being tagged with a first detectable
label, the second primer being tagged with a second detectable label; (b) digesting the
reaction product of step (a) with the restriction endonuclease corresponding to the
polymorphic restriction site; (c) denaturing the reaction product of step (b); (d)
0 contacting the product of step (c) with a first and a second oligonucleotide, the first
oligonucleotide being complementary to a first sequence in the strand of the product
of step (c) containing the first detectable label, the first sequence being between the
polymorphic restriction site and the sequence complementary to the second primer, the
first oligonucleotide being tagged with the first member of a specific binding pair, the
second oligonucleotide being complementary to a second sequence in the strand of the
product of step (c) containing the second detectable label, the second sequence being
on the same side of the polymorphic restriction site as the first sequence, the second
sequence not being contained within or being complementary to either of the first or
second primers, the second oligonucleotide being tagged with the first member of the
2 o specific binding pair; (e) contacting the reaction product of step (d) with the second
member of the specific binding pair, immobilized on a solid support; and
(f) determining the ratio of the levels of the first and second detectable labels bound to
the solid support, a ratio of 0:1 being an indication of a homozygote contai~ g the
polymorphic restriction site, a ratio of 1:1 being an indication of a homozygote lacking
2 5 the polymorphic restriction site, and a ratio of 1:2 being an indication of a
heterozygote.
In an eighth aspect, the invention features a method for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, involving the steps
.. ..
CA 022667~0 1999-03-17
W O9~/12352 PCTrUS97/16467
of: (a) amplifying the nucleic acid by PCR using a first and a second primer fl~nking
the polymorphic restriction site, the first primer being tagged with the first member of
a first specific binding pair, the second primer being tagged with a detectable label; (b)
digesting the reaction product of step (a) with the restriction endonuclease
s corresponding to the polymorphic restriction site; (c) contacting the reaction product
of step (b) with the second member of the first specific binding pair, immobilized on a
first solid support; (d) denaturing the reaction product of step (c) not bound to the first
solid support; (e) contacting the product of step (d) with an oligonucleotide
complementary to a sequence in the strand of the product of step (d) containing the
10 detectable label, the sequence being between the polymorphic restriction site and the
sequence corresponding to the second primer, the oligonucleotide being tagged with
the first member of a second specific binding pair; (f) contacting the reaction product
of step (e) with the second member of the second specific binding pair, immobilized
on a second solid support; and (g) detel"lh~ g the ratio of the level of the detectable
5 label bound to the first solid support to the level of the detectable label bound to the
second solid support, a ratio of 0: l being an indication of a homozygote containing the
polymorphic restriction site, in a case where the total amount of the reaction product
from step (c) not bound to the first solid support was used in steps (d), (e), and (f); a
ratio of l :0 being an indication of a homozygote lacking the polymorphic restriction
2 o site, in a case where the total amount of the reaction product from step (c) not bound
to the first solid support was used in steps (d), (e), and (f); and a ratio of 1: l being an
indication of a heterozygote, in a case where the total amount of the reaction product
from step (c) not bound to the first solid support was used in steps (d), (e), and (f).
In a ninth aspect, the invention features a method for detecting the presence or2 5 absence of a polymorphic restriction site in a nucleic acid, involving the steps of: (a)
amplifying the nucleic acid by PCR using a first and a second primer fl~nking the
polymorphic restriction site, the first primer being tagged with a detectable label, the
second primer being unlabeled; (b) digesting the reaction product of step (a) with the
CA 022667~0 1999-03-17
W O98/12352 PCTrUS97116467
restriction endonuclease corresponding to the polymorphic restriction site; (c)
annealing and ligating to the single-stranded ends generated in the reaetion of step (b)
a first oligonucleotide tagged with the first member of a first specific binding pair; (d)
contacting the reaction product of step (c) with the second member of the first specific
5 binding pair, immobilized on a first solid support; (e) denaturing the reaction product
of step (d) not bound to the first solid support; (f) contacting the product of step (e)
with a second oligonucleotide complementary to a sequence in the strand of the
product of step (e) containing the detectable label, the sequence being between the
polymorphic restriction site and either the sequence corresponding to the first primer
0 or the sequence complementary to the second primer, the second oligonucleotidebeing tagged with the first member of a second specific binding pair; (g) contacting
the reaction product of step (f~ with the second member of the second specific binding
pair, immobilized on a second solid support; and (h) determining the ratio of the level
of the detectable label bound to the first solid support to the level of the detectable
label bound to the second solid support, a ratio of l :0 being an indieation of a
homozygote containing the polymorphic restriction site, in a case where the total
amount of the reaction product from step (d) not bound to the first solid support was
used in steps (e), (f), and (g); a ratio of 0: l being an indication of a homozygote
lacking the polymorphic restriction site, in a case where the total amount of the
2 o reaction product from step (d) not bound to the first solid support was used in steps
(e), (f), and (g); and a ratio ofl:l being an indication of a heterozygote; in a ease
where the total amount of the reaetion product from step (d) not bound to the first
solid support was used in steps (e), (f), and (g).
In a tenth aspect, the invention features a method for detecting the presence or2 5 absenee of a polymorphie restriction site in a nucleic acid, involving the steps of: (a)
amplifying the nucleic acid by PCR using a first and a second primer fl~nkin~ the
polymorphic restriction site, the first primer being tagged with the first member of a
first specific binding pair, the second primer being tagged with a deteetable label; (b)
., .~.. .~.. ~ . . .
CA 022667~0 1999-03-17
W O 98/12352 PCTrUS97/16467
digesting the reaction product of step (a) with the restriction endonuclease
corresponding to the polymorphic restriction site; (c) contacting the reaction product
of step (b) with the second member of the first specific binding pair, immobilized on a
first solid support; (d) denaturing the reaction product from step (c) not bound to the
5 first solid support; (e) contacting the product of step (d) with an oligonucleotide
complementary to a sequence in the strand of the product of step (d) containing the
detectable label, the sequence being between the polymorphic restriction site and the
sequence corresponding to the second primer, the oligonucleotide being immobilized
on a second solid support; and (f) determining the ratio of the level of the detectable
o label bound to the first solid support to the level of the detectable label bound to the
second solid support, a ratio of 0:1 being an indication of a homozygote containing the
polymorphic restriction site, in a case where the total amount of the reaction product
from step (c) not bound to the first solid support was used in steps (d) and (e); a ratio
of 1:0 being an indication of a homozygote lacking the polymorphic restriction site, in
15 a case where the total amount of the reaction product from step (c) not bound to the
first solid support was used in steps (d) and (e); and a ratio of 1:1 being an indication
of a heterozygote, in a case where the total amount of the reaction product from step
(c) not bound to the first solid support was used in steps (d) and (e).
In an eleventh aspect, the invention features a method for detecting the
2 o presence or absence of a polymorphic restriction site in a nucleic acid, involving the
steps of: (a) amplifying the nucleic acid by PCR using a first and a second primer
flanking the polymorphic restriction site, the first primer being tagged with a
detectable label, the second primer being unlabeled; (b) digesting the reaction product
of step (a) with the restriction endonuclease corresponding to the polymorphic
2 5 restriction site; (c) annealing and ligating to the single-stranded ends generated in the
reaction of step (b) a first oligonucleotide tagged with the first member of a first
specific binding pair; (d) contacting the reaction product of step (c) with the second
member of the first specific binding pair, immobilized on a first solid support;
CA 022667~0 1999-03-17
W 098/12352 PCTrUS97/16467
11
(e) denaturing the reaction product of step (d) not bound to the first solid support; (f)
contacting the product of step (e) with a second oligonucleotide complementary to a
sequence in the strand of the product of step (e) containing the detectable label, the
sequence being between the polymorphic restriction site and either the sequence
5 corresponding to the first primer or the sequence complementary to the second primer,
the second oligonucleotide being immobilized on a second solid support; and (g)
determining the ratio of the level of the detectable label bound to the first solid support
to the level of the detectable label bound to the second solid support, a ratio of l :O
being an indication of a homozygote containing the polymorphic restriction site, in a
10 case where the total amount of the reaction product from step (d) not bound to the first
solid support was used in steps (e) and (f~; a ratio of 0: l being an indication of a
homozygote lacking the polymorphic restriction site, in a case where the total amount
of the reaction product from step (d) not bound to the first solid support was used in
steps (e) and (f); and a ratio of l: l being an indication of a heterozygote, in a case
5 where the total amount of the reaction product from step (d) not bound to the first
solid support was used in steps (e) and (f).
In a twelfth aspect, the invention features a method for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, involving the steps
of: (a) amplifying the nucleic acid by PCR using a first and a second primer fl~nking
2 o the polymorphic restriction site, the first primer cont~ining a first sequence not
complementary to or present in the nucleic acid, the second primer cont~ining a
second sequence not complementary to or present in the nucleic acid; (b) amplifying
the product of step (a) by PC~ using a third and a fourth primer, the third primer
containing the first sequence or a sequence complementary to the first sequence, the
2 5 third primer being tagged with the first member of a specific binding pair, the fourth
primer containing the second sequence or a sequence complementary to the second
sequence, the fourth primer being tagged with a detectable label; (c) digesting the
product of step (b) with the restriction endonuclease corresponding to the polymorphic
CA 022667~0 1999-03-17
W O98/12352 PCTAUS97/16467
12
restriction site; (d) contacting the reaction product of step (c) with the second member
of the specific binding pair, immobilized on a solid support; and (e) measuring the
level of the detectable label bound to the solid support, the presence of the detectable
label bound to the solid support being an indication of the absence of the polymorphic
5 restriction site in the nucleic acid.
In a thirteenth aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, involving the
steps of: (a) amplifying the nucleic acid by PCR using a first and a second primer
flanking the polymorphic restriction site, the first primer containing a first sequence
0 not complementary to or present in the nucleic acid, the second primer containing a
second sequence not complementary to or present in the nucleic acid; (b) amplifying
the product of step (a) by PCR using a third and a fourth primer, the third primer
containing the first sequence or a sequence complementary to the first sequence, the
third primer being tagged with the first member of a specific binding pair, the fourth
5 primer contai~ g the second sequence or a sequence complementary to the secondsequence, the fourth primer being tagged with a detectable label; (c) digesting the PCR
product of step (b) with the restriction endonuclease corresponding to the polymorphic
restriction site; (d) annealing and ligating to the single-stranded ends generated in the
reaction of step (c) an oligonucleotide tagged with a second detectable label;
2 o (e) contacting the reaction product of step (d) with the second member of the specific
binding pair, immobilized on a solid support; and (f) determining the levels of the first
and second detectable labels bound to the solid support, the presence of only the first
detectable label bound to the solid support being an indication of a homozygote
lacking the polymorphic restriction site, the presence of only the second detectable
2 5 label bound to the solid support being an indication of a homozygote containing the
polymorphic restriction site, and the presence of both the first and second detectable
labels bound to the solid support being an indication of a heterozygote.
CA 022667~0 1999-03-17
W O 98/12352 PCT~US97/16467
13
In a fourteenth aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, involving the
steps of: (a) amplifying the nucleic acid by PCR using a first and a second primer
fl~nking the polymorphic restriction site, the first primer containing a first sequence
5 not complementary to or present in the nucleic acid; (b) amplifying the product of step
(a) by PCR using a third primer and the second primer, the third primer containing the
first sequence, the third primer being tagged with a detectable label; (c) digesting a
portion of the reaction of step (b) with the restriction endonuclease corresponding to
the polymorphic restriction site, while leaving another portion of the reaction of step
10 (b) undigested; (d) denaturing the digested and undigested portions from step (c); (e)
contacting the product of step (d) with an oligonucleotide complementary to a second
sequence in the strand of the product of step (d) containing the detectable label, the
second sequence being between the polymorphic restriction site and the sequence
complementary to the second primer, the oligonucleotide being tagged with a first
5 member of a specific binding pair; (f) contacting the reaction product of step (e) with
the second member of the specific binding pair, immobilized on a solid support; and
(g) determining the ratio of the levels of the detectable label bound to the solid support
between undigested and digested samples, a ratio of l:O between equivalent portions
of the undigested and digested samples being an indication of a homozygote
2 o cont~ining the polymorphic restriction site, a ratio of l: l between equivalent portions
of the undigested and digested samples being an indication of a homozygote lacking
the polymorphic restriction site, and a ratio of 2: l between equivalent portions of the
undigested and digested samples being an indication of a heterozygote.
In a fifteenth aspect, the invention features a method for detecting the presence
2 5 or absence of a polymorphic restriction site in a nucleic acid, involving the steps of:
(a) amplifying the nucleic acid by PCR using a first and a second primer fl~nking the
polymorphic restriction site, the first primer containing a first sequence not
complementary to or present in the nucleic acid, the second primer containing a
CA 022667~0 1999-03-17
WO 98/12352 rCTAUSg7/16467
14
second sequence not complementary to or present in the nucleic acid; (b) amplifying
the product of step (a) by PCR using a third and a fourth primer, the third primer
containing the first sequence or a sequence complementary to the first sequence, the
third primer being tagged with a first detectable label, the fourth primer containing the
5 second sequence or a sequence complementary to the second sequence, the fourthprimer being tagged with a second detectable label; (c) digesting the reaction product
of step (b) with the restriction endonuclease corresponding to the polymorphic
restriction site; (d) denaturing the reaction product of step (c); (e) contacting the
product of step (d) with a first and a second oligonucleotide, the first oligonucleotide
10 being complementary to a third sequence in the strand of the product of step (d)
cont~ining the first detectable label, the third sequence being between the polymorphic
restriction site and the sequence corresponding to or complementary to the firstprimer, the first oligonucleotide being tagged with the first member of a first specific
binding pair, the second oligonucleotide being complementary to a fourth sequence in
5 the strand of the product of step (d) containing the second detectable label, the fourth
sequence being on the same side of the polymorphic restriction site as the thirdsequence, the fourth sequence not being contained within or being complementary to
any of the primers, the second oligonucleotide being tagged with the first member of a
second specific binding pair; (f) contacting a first portion of the reaction product of
2 o step (e) with the second member of the first specific binding pair, immobilized on a
first solid support; (g) contacting a second portion of the reaction product of step (e)
with the second member of the second specific binding pair, immobilized on a second
solid support; and (h) dete~ i"i~-g the ratio of the levels of the first and second
detectable labels bound to the first and second solid supports, a ratio of 1:0 between
2 5 equivalent amounts of the first and second portions being an indication of ahomozygote containing the polymorphic restriction site, a ratio of 1:1 between
equivalent amounts of the first and second portions being an indication of a
homozygote lacking the polymorphic restriction site, and a ratio of 2:1 between
.
CA 022667~0 1999-03-17
W O9B/12352 PCT~US97/16467
equivalent amounts of the first and second portions being an indication of a
heterozygote.
In a sixteenth aspect, the invention features a method for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, involving the steps of:
(a) amplifying the nucleic acid by PCR using a first and a second primer flanking the
polymorphic restriction site, the first primer containing a first sequence not
complementary to or present in the nucleic acid, the second primer containing a
second sequence not complementary to or present in the nucleic acid; (b) amplifying
the product of step (a) by PCR using a third and a fourth primer, the third primer
0 containing the first sequence or a sequence complementary to the first sequence, the
third primer being tagged with a first detectable label, the fourth primer containing the
second sequence or a sequence complementary to the second sequence, the fourth
primer being tagged with a second detectable label; (c) digesting the reaction product
of step (b) with the restriction endonuclease corresponding to the polymorphic
5 restriction site; (d) denaturing the reaction product of step (c); (e) contacting the
product of step (d) with a first and a second oligonucleotide, the first oligonucleotide
being complementary to a third sequence in the strand of the product of step (d)containing the first detectable label, the third sequence being between the polymorphic
restriction site and the sequence corresponding to or complementary to the second
2 o primer, the first oligonucleotide being tagged with the first member of a first specific
binding pair, the second oligonucleotide being complementary to a fourth sequence in
the strand of the product of step (d) containing the second detectable label, the fourth
sequence being on the same side of the polymorphic restriction site as the thirdsequence, the fourth sequence not being contained within or being complementary to
2 5 any of the primers, the second oligonucleotide being tagged with the first member of a
second specific binding pair; (f) contacting a first portion of the reaction product of
step (e) with the second member of the first specific binding pair, immobilized on a
first solid support; (g) contacting a second portion of the reaction product of step (e)
CA 022667~0 1999-03-17
W O 9~/12352 PCTrUS97/16467
16
with the second member of the second specific binding pair, immobilized on a second
solid support; and (h) determining the ratio of the levels of the first and second
detectable labels bound to the first and second solid supports, a ratio of 0: l between
equivalent amounts of the first and second portions being an indication of a
homozygote containing the polymorphic restriction site, a ratio of 1: l between
equivalent amounts of the first and second portions being an indication of a
homozygote lacking the polymorphic restriction site, and a ratio of l :2 betweenequivalent amounts of the first and second portions being an indication of a
heterozygote.
0 In a seventeenth aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, involving the
steps of: (a) amplifying the nucleic acid by PCR using a first and a second primer
fl~nking the polymorphic restriction site, the first primer containing a first sequence
not complementary to or present in the nucleic acid, the second primer containing a
second sequence not complementary to or present in the nucleic acid; (b) amplifying
the product of step (a) by PCR using a third and a fourth primer, the third primer
cont~ining the first sequence or a sequence complementary to the first sequence, the
third primer being tagged with a first detectable label, the fourth primer containing the
second sequence or a sequence complementary to the second sequence, the fourth
2 o primer being tagged with a second detectable label; (c) digesting the reaction product
of step (b) with the restriction endonuclease corresponding to the polymorphic
restriction site; (d) denaturing the reaction product of step (c); (e) contacting the
product of step (d) with a first and a second oli~onucleotide, the first oligonucleotide
being complementary to a third sequence in the strand of the product of step (d)2 5 col~L;~ g the first detectable label, the third sequence being between the polymorphic
restriction site and the sequence corresponding to or complementary to the firstprimer, the first oligonucleotide being tagged with the first member of a specific
binding pair, the second oligonucleotide being complementary to a fourth sequence in
CA 022667~0 1999-03-17
W O 98/12352 PCT~US97/16467
17
the strand of the product of step (d) containing the second detectable label, the fourth
sequence being on the same side of the polymorphic restriction site as the thirdsequence, the fourth sequence not being contained within or being complementary to
any of the primers, the second oligonucleotide being tagged with the first member of
5 the specific binding pair; (~) contacting the reaction product of step (e) with the second
member of the specific binding pair, immobilized on a solid support; and (g)
determining the ratio of the levels of the first and second detectable labels bound to
the solid support, a ratio of 1:0 being an indication of a homozygote containing the
polymorphic restriction site, a ratio of 1:1 being an indication of a homozygote lacking
o the polymorphic restriction site, and a ratio of 2:1 being an indication of a
heterozygote.
In an eighteenth aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, involving (a)
amplifying the nucleic acid by PCR using a first and second primer fl~nking the
polymorphic restriction site, the first primer containing a first sequence not
complementary to or present in the nucleic acid, the second primer colltaillh~g a
second sequence not complementary to or present in the nucleic acid; (b) amplifying
the product of step (a) by PCR using a third and a fourth primer, the third primer
containing the first sequence or a sequence complementary to the first sequence, the
2 o third primer being tagged with a first detectable label, the fourth primer containing the
second sequence or a sequence complementary to the second sequence, the fourth
primer being tagged with a second detectable label; (c) digesting the reaction product
of step (b) with the restriction endonuclease corresponding to the polymorphic
restriction site; (d) den~h-ring the reaction product of step (c); (e) contacting the
2 5 product of step (d) with a first and a second oligonucleotide, the first oligonucleotide
being complementary to a third sequence in the strand of the product of step (d)containing the first detectable label, the third sequence being between the polymorphic
restriction site and the sequence corresponding to or complementary to the second
CA 022667~0 1999-03-17
W O98/12352 PCTrUS97/16467
18
primer, the first oligonucleotide being tagged with the first member of a specific
binding pair, the second oligonucleotide being complementary to a fourth sequence in
the strand of the product of step (d) conlail,ing the second detectable label, the fourth
sequence being on the same side of the polymorphic restriction site as the third5 sequence, the fourth sequence not being contained within or being complementary to
any of the primers, the second oligonucleotide being tagged with the first member of
the specific binding pair; (f) contacting the reaction product of step (e) with the second
member of the specific binding pair, immobilized on a solid support; and (g)
determining the ratio of the levels of the first and second detectable labels bound to
o the solid support, a ratio of 0:1 being an indication of a homozygote cont~ining the
polymorphic restriction site, a ratio of 1:1 being an indication of a homozygote lacking
the polymorphic restriction site, and a ratio of 1:2 being an indication of a
heterozygote.
In a nineteenth aspect, the invention features a method for detecting the
5 presence or absence of a polymorphic restriction site in a nucleic acid, involving the
steps of (a) amplifying the nucleic acid by PCR using a first and a second primer
flanking the polymorphic restriction site, the first primer containing a first sequence
not complementary to or present in the nucleic acid, the second primer containing a
second sequence not complementary to or present in the nucleic acid; (b) amplifying
2 o the product of step (a) by PCR using a third and a fourth primer, the third primer
co~ hlg the first sequence or a sequence complementary to the first sequence, the
third primer being tagged with the first member of a first specific binding pair, the
fourth primer containing the second sequence or a sequence complementary to the
second sequence, the fourth primer being tagged with a detectable label; (c) digesting
2 5 the reaction product of step (b) with the restriction endonuclease corresponding to the
polymorphic restriction site; (d) contacting the reaction product of step (c) with the
second member of the first specific binding pair, immobilized on a first solid support;
(e) denaturing the reaction product of step (d) not bound to the first solid support; (fl
CA 022667~0 1999-03-17
WO 98/12352 PCT/US97/16467
19
contacting the product of step (e) with an oligonucleotide complementary to a third
sequence in the strand of the product of step (e) containing the detectable label, the
third sequence being between the polymorphic restriction site and the sequence
corresponding to or complementary to the second primer, the oligonucleotide being
5 tagged with the first member of a second specific binding pair; (g) contacting the
reaction product of step (f) with the second member of the second specific binding
pair, immobilized on a second solid support; and (h) dete~ lg the ratio of the level
of the detectable label bound to the first solid support to the level of the detectable
label bound to the second solid support, a ratio of 0:1 being an indication of a10 homozygote containing the polymorphic restriction site, in a case where the total
amount of the reaction product from step (d) not bound to the first solid support was
used in steps (e), (f), and (g); a ratio of 1:0 being an indication of a homozygote
lacking the polymorphic restriction site, in a case where the total amount of the
reaction product from step (d) not bound to the first solid support was used in steps
5 (e), (f), and (g); and a ratio of 1:1 being an indication of a heterozygote, in a case
where the total amount of the reaction product from step (d) not bound to the first
solid support was used in steps (e), (f~, and (g).
In a twentieth aspect the invention features a method for detecting the presenceor absence of a polymorphic restriction site in a nucleic acid, involving the steps of:
2 o (a) amplifying the nucleic acid by PCR using a first and a second primer fl~nking the
polymorphic restriction site, the first primer containing a first sequence not
complementary to or present in the nucleic acid; (b) amplifying the product of step (a)
by PCR using a third primer and the second primer, the third primer cont~ining the
first sequence, the third primer being tagged with a detectable label; (c) digesting the
2 5 reaction product of step (b) with the restriction endonuclease corresponding to the
polymorphic restriction site; (d) annealing and ligating to the single-stranded ends
generated in the reaction of step (c) a first oligonucleotide tagged with the f1rst
member of a first specific binding pair; (e) contacting the reaction product of step (d)
CA 022667~0 1999-03-17
W O 9~/12352 PCT~US97116467
with the second member of the first specific binding pair, immobilized on a first solid
support; (f) denaturing the reaction product of step (e) not bound to the first solid
support; (g) contacting the product of step (f) with a second oligonucleotide
complementary to a second sequence in the strand of the product of step (f) containing
5 the detectable label, the second sequence being between the polymorphic restriction
site and either the sequence corresponding to or complementary to the second primer
or the sequence corresponding to or complementary to the first primer, the second
oligonucleotide being tagged with the first member of a second specific binding pair;
(h) contacting the reaction product of step (g) with the second member of the second
10 specific binding pair, immobilized on a second solid support; and (i) determining the
ratio of the level of the detectable label bound to the first solid support to the level of
the detectable label bound to the second solid support, a ratio of 1:0 being an
indication of a homozygote containing the polymorphic restriction site, in a case
where the total amount of the reaction product from step (e) not bound to the first solid
5 support was used in steps (f), (g), and (h); a ratio of 0: l being an indication of a
homozygote lacking the polymorphic restriction site, in a case where the total amount
of the reaction product from step (e) not bound to the first solid support was used in
steps (f), (g), and (h); and a ratio of 1:1 being an indication o~ a heterozygote; in a case
where the total amount of the reaction product from step (e) not bound to the first solid
2 o support was used in steps (f), (g), and (h).
In a twenty-first aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, involving the
steps of: (a) amplifying the nucleic acid by PCR using a first and a second primer
fl~nking the polymorphic restriction site, the first primer cont~ining a first sequence
2 5 not complementary to or present in the nucleic acid, the second primer containing a
second sequence not complementary to or present in the nucleic acid; (b) amplifying
the product of step (a) by PCR using a third and a fourth primer, the third primer
Co~ i t~g the first sequence or a sequence complementary to the first sequence, the
CA 022667~0 1999-03-17
W O 98tl2352 PCT~US97/16467
21
third primer being tagged with the first member of a first specific binding pair, the
fourth primer containing the second sequence or a sequence complementary to the
second sequence, the fourth primer being tagged with a detectable label; (c) digesting
the reaction product of step (b) with the restriction endonuclease corresponding to the
5 polymorphic restriction site; (d) contacting the reaction product of step (c) with the
second member of the first specific binding pair, immobilized on a first solid support;
(e) denaturing the reaction product from step (d) not bound to the first solid support;
(f) contacting the product of step (e) with an oligonucleotide complementary to a third
sequence in the strand of the product of step (e) containing the detectable label, the
10 third sequence being between the polymorphic restriction site and the sequence
corresponding to or complementary to the second primer, the oligonucleotide being
immobilized on a second solid support; and (g) det~ ;t~g the ratio ofthe level of
the detectable label bound to the first solid support to the level of the detectable label
bound to the second solid support, a ratio of 0:1 being an indication of a homozygote
containing the polymorphic restriction site, in a case where the total amount of the
reaction product from step (d) not bound to the first solid support was used in steps (e)
and (f); a ratio of 1:0 being an indication of a homozygote lacking the polymorphic
restriction site, in a case where the total amount of the reaction product from step (d)
not bound to the first solid support was used in steps (e) and (f); and a ratio of 1:1
2 o being an indication of a heterozygote, in a case where the total amount of the reaction
product from step (d) not bound to the first solid support was used in steps (e) and (f).
In a twenty-second aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, the method
involving the steps of: (a) amplifying the nucleic acid by PCR using a first and a
2 5 second primer fl~nking the polymorphic restriction site, the first primer cont~ining a
first sequence not complementary to or present in the nucleic acid; (b) amplifying the
product of step (a) by PCR using a third primer and the second primer, the thirdprimer containing the first sequence, the third primer being tagged with a detectable
CA 022667~0 1999-03-17
WO 98/12352 22 PCTAUS97/16467
label; (c) digesting the reaction product of step (b) with the restriction endonuclease
corresponding to the polymorphic restriction site; (d) annealing and lig;~ting to the
single-stranded ends generated in the reaction of step (c) a first oligonucleotide tagged
with the first member of a first specific binding pair; (e) contacting the reaction
5 product of step (d) with the second member of the first specific binding pair,immobilized on a first solid support; (f) denaturing the reaction product of step (e) not
bound to the first solid support; (g) contacting the product of step (f) with a second
oligonucleotide complementary to a second se4uence in the strand of the product of
step (f) containing the detectable label, the second sequence being between the
0 polymorphic restriction site and either the sequence corresponding to or
complementary to the second primer or the sequence corresponding to or
complementary to the first primer, the second oligonucleotide being immobilized on a
second solid support; and (h) dete~"~ g the ratio of the level of the detectable label
bound to the first solid suppor~ to the level of the detectable label bound to the second
5 solid support, a ratio of 1:0 being an indication of a homozygote cont~ining the
polymorphic restrictlon site, in a case where the total amount of the reaction product
from step (e) not bound to the first solid support was used in steps (f) and (g); a ratio
of 0:1 being an indication of a homozygote lacking the polymorphic restriction site, in
a case where the total amount of the reaction product from step (e) not bound to the
2 o first solid support was used in steps (f) and (g); and a ratio of 1:1 being an indication
of a heterozygote, in a case where the total amount of the reaction product from step
(e) not bound to the first solid support was used in steps (f) and (g).
In a twenty-third aspect, the invention features a kit for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, the kit col-t~ i "g one or
2 5 more sets of a first and a second primer flanking the polymorphic restriction site, the
first primer being tagged with the first member of a specific binding pair, the second
primer being tagged with a detectable label. In a preferred embodiment, the kit further
contains the second member of the specific binding pair, immobilized on a solid
CA 022667~0 1999-03-17
W O 98112352 PCTrUS97/16467
23
support. In another preferred embodiment, the kit further contains an oligonucleotide
complementary to the single-stranded ends generated in the nucleic acid upon
digestion of the nucleic acid with the restriction enzyme corresponding to the
polymorphic restriction site, the oligonucleotide being tagged with a second detectable
label.
In a twenty-fourth aspect, the invention features a kit for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, the kit containing:
(a) a first and a second primer flanking the polymorphic restriction site, the first
primer being tagged with a detectable label, the second primer being unlabeled; (b) an
oligonucleotide complementary to a sequence in the strand of the nucleic acid
complementary to the second primer, the sequence being between the polymorphic
restriction site and the sequence complementary to the second primer, the
oligonucleotide being tagged with a first member of a specific binding pair; and (c) the
second member of the specific binding pair, immobilized on a solid support.
In a twenty-fifth aspect, the invention features a kit for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, the kit contai~ g:
(a) a first and a second primer fl~nking the polymorphic restriction site, the first
primer being tagged with a first detectable label, the second primer being tagged with
a second detectable label; (b) a first oligonucleotide, the first oligonucleotide being
2 o complementary to a first sequence in the strand of the nucleic acid complementary to
the second primer, the first sequence being between the polymorphic restriction site
and either the sequence corresponding to the first primer or the sequence
complementary to the second primer, the first oligonucleotide being tagged with the
first member of a first specific binding pair; (c) a second oligonucleotide, the second
2 5 oligonucleotide being complementary to a second sequence in the strand of the nucleic
acid complementary to the first primer, the second sequence being on the same side of
the polymorphic restriction site as the first sequence, the second sequence not being
contained within or being complementary to either of the first or second primers, the
CA 022667~0 1999-03-17
W O 98/12352 PCT~US97/16467
24
second oligonucleotide being tagged with the first member of a second specific
binding pair; (d) the second member of the first specific binding pair, immobilized on
a first solid support; and (e) the second member of the second specific binding pair,
immobilized on a second solid support. In a preferred embodiment, the first and the
5 second specific binding pairs are identical, and the first and the second solid supports
are identical.
In a twenty-sixth aspect, the invention features a kit for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, the kit containing:
(a) a first and a second primer flanking the polymorphic restriction site, the first
0 primer being tagged with the first member of a first specific binding pair, the second
primer being tagged with a detectable label; (b) the second member of the first specific
binding pair, immobilized on a first solid support; (c) an oligonucleotide
complementary to a first sequence in the strand of the nucleic acid co..~ g the
sequence corresponding to the second primer, the first sequence being between the
15 polymorphic restriction site and the sequence corresponding to the second primer, the
oligonucleotide being tagged with the first member of a second specific binding pair;
and (d) the second member of the second specific binding pair, immobilized on a
second solid support.
In a twenty-seventh aspect, the invention features a kit for detecting the
2 o presence or absence of a polymorphic restriction site in a nucleic acid, the kit
containing: (a) a first and a second primer fl~nking the polymorphic restriction site,
the first primer being tagged with a detectable label, the second primer being
unlabeled; (b) a first oligonucleotide complementary to the single-stranded endsgenerated in the nucleic acid upon digestion of the nucleic acid with the restriction
2 5 enzyme corresponding to the polymorphic restriction site, the oligonucleotide being
tagged with the first member of a first specific binding pair; (c) the second member of
the first specific binding pair, immobilized on a first solid support; (d) a second
oligonucleotide complementary to a sequence in the strand of the nucleic acid
CA 022667~0 1999-03-17
WO 98/123~2 PCTrUS97/16467
complementary to the second primer, the sequence being between the polymorphic
restriction site and either the sequence corresponding to the first primer or the
sequence complementary to the second primer, the second oligonucleotide being
tagged with the first member of a second specific binding pair; and (e) the second
5 member of the second specific binding pair, immobilized on a second solid support.
In a twenty-eighth aspect, the invention features a kit for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, the kit containing:
(a) a first and a second primer fl~nking the polymorphic restriction site, the first
primer being tagged with the first member of a first specific binding pair, the second
0 primer being tagged with a detectable label; (b) the second member of the first specific
binding pair, immobilized on a first solid support; and (c) an oligonucleotide
complementary to a first sequence in the strand of the nucleic acid containillg the
sequence corresponding to the second primer, the first sequence being between the
polymorphic restriction site and the sequence corresponding to the second primer, the
15 oligonucleotide being immobilized on a second solid support.
In a twenty-ninth aspect, the invention features a kit for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, the kit containing:
(a) a first and a second primer fl~nking the polymorphic restriction site, the first
primer being tagged with a detectable label, the second primer being unlabeled; (b) a
2 o first oligonucleotide complementary to the single-stranded ends generated in the
nucleic acid upon digestion of the nucleic acid with the restriction enzyme
corresponding to the polymorphic restriction site, the oligonucleotide being tagged
with the first member of a first specific binding pair; (c) the second member of the
first specific binding pair, immobilized on a first solid support; and (d) a second
2 5 oligonucleotide complementary to a sequence in the strand of the nucleic acid
complementary to the second primer, the sequence being between the polymorphic
restriction site and either the sequence corresponding to the first primer or the
.. . .. , ~
CA 022667~0 1999-03-17
WO 9~1123~2 PCT/US97tl6467
26
sequence complementary to the second primer, the second oligonucleotide being
immobilized on a second solid support.
In a thirtieth aspect, the invention features a kit for detecting the presence or
absence of a polymorphic restriction site in a nucleic acid, the kit containing: (a) a first
5 and a second primer fl~nking the polymorphic restriction site, the first primer
containing a first sequence not complementary to or present in the nucleic acid, the
second primer containing a second sequence not complementary to or present in the
nucleic acid; (b) a third and a fourth primer, the third primer containing the first
sequence or a sequence complementary to the first sequence, the third primer being
0 tagged with the first member of a specific binding pair, the fourth primer containing
the second sequence or a sequence complementary to the second sequence, the fourth
primer being tagged with a detectable label. In a preferred embodiment, the kit further
contains the second member of the specific binding pair, immobilized on a solid
support. In another preferred embodiment, the kit further contains an oligonucleotide
15 complementary to the single-stranded ends generated in the nucleic acid upon
digestion of the nucleic acid with the restriction enzyme corresponding to the
polymorphic restriction site, the oligonucleotide being tagged with a second detectable
label. In a thirty-first aspect, the invention features a kit for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, the kit2 o cont~ining: (a) a first and a second primer fl~nking the polymorphic restriction site,
the first primer containing a first sequence not complementary to or present in the
nucleic acid; (b) a third primer colllailling the first sequence, the third primer being
tagged with a detectable label; (c) an oligonucleotide complementary to a secondsequence in the strand of the nucleic acid contaillillg the sequence complementary to
2 5 the second primer, the second sequence being between the polymorphic restriction site
and the sequence complementary to the second primer, the oligonucleotide being
tagged with a first member of a specific binding pair; and (d) the second member of
the specific binding pair, immobilized on a solid support.
CA 022667~0 1999-03-17
W O 9~/12352 PCTAUS97/16467
27
In a thirty-second aspect, the invention features a kit for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, the kit containing: (a) a
~ first and a second primer flanking the polymorphic restriction site, the first primer
containing a first sequence not complementary to or present in the nucleic acid, the
5 second primer containing a second sequence not complementary to or present in the
nucleic acid; (b) a third and a fourth primer, the third primer col~tai~ g the first
sequence or a sequence complementary to the first sequence, the third primer being
tagged with a first detectable label, the fourth primer containing the second sequence
or a sequence complementary to the second sequence, the fourth primer being tagged
0 with a second detectable label; (c) a first oligonucleotide, the first oligonucleotide
being complementary to a third sequence in the strand of the nucleic acid
complementary to the second primer, the third sequence being between the
polymorphic restriction site and either the sequence complementary to the secondprimer or the sequence corresponding to the first primer, the first oligonucleotide
5 being tagged with the first member of a first specific binding pair, (d) a second
oligonucleotide, the second oligonucleotide being complementary to a fourth sequence
in the strand of the nucleic acid complementary to the first primer, the fourth sequence
being on the same side of the polymorphic restriction site as the third sequence, the
fourth sequence not being contained within or being complementary to any of the
2 o primers, the second oligonucleotide being tagged with the first member of a second
specific binding pair; (e) the second member of the first specific binding pair,immobilized on a first solid support; and (f) the second member of the second specific
binding pair, immobilized on a second solid support. In a preferred embodiment, the
first and the second specific binding pairs are identical, and the first and the second
25 solid supports are identical.
In a thirty-third aspect, the invention features a kit for detecting the presence or
absence of a polymorphic restriction site in a nucleic acid, the kit cont~ining: (a) a first
and a second primer flanking the polymorphic restriction site, the first primer
~.. .... .
CA 022667~0 1999-03-17
W O g8/12352 PCT~US97/16467
28
containing a first sequence not complementary to or present in the nucleic acid, the
second primer containing a second sequence not complementary to or present in the
nucleic acid; (b) a third and a fourth primer, the third primer containing the first
sequence or a sequence complementary to the first sequence, the third primer being
5 tagged with the first member of a first specific binding pair, the fourth primer
containing the second sequence or a sequence complementary to the second sequence,
the fourth primer being tagged with a detectable label; (c) the second member of the
first specific binding pair, immobilized on a first solid support; (d) an oligonucleotide
complementary to a third sequence in the skand of the nucleic acid corresponding to
0 the second primer, the sequence being between the polymorphic restriction site and
the sequence corresponding to the second primer, the oligonucleotide being tagged
with the first member of a second specific binding pair; and (e) the second member of
the second specific binding pair, immobilized on a second solid support.
In a thirty-fourth aspect, the invention features a kit for detecting the presence
5 or absence of a polymorphic restriction site in a nucleic acid, the kit containing: (a) a
first and a second primer fl~nking the polymorphic restriction site, the first primer
containing a first sequence not complementary to or present in the nucleic acid; (b) a
third primer containing the first sequence, the third primer being tagged with adetectable label; (c) a first oligonucleotide complementary to the single-skanded ends
2 o generated in the nucleic acid upon digestion of the nucleic acid with the reskiction
enzyme corresponding to the polymorphic restriction site, the oligonucleotide being
tagged with the first member of a first specific binding pair; (d) the second member of
the first specific binding pair, immobilized on a first solid support; (e) a second
oligonucleotide complementary to a second sequence in the skand of the nucleic acid
2 5 corresponding to the first primer, the second sequence being between the polymorphic
restriction site and either the sequence complementary to the second primer or the
sequence corresponding to the first primer, the second oligonucleotide being tagged
CA 022667~0 1999-03-17
W O 9~/12352 PCTrUS97116467
29
with the first member of a second specific binding pair; and (f) the second member of
the second specific binding pair, immobilized on a second solid support.
In a thirty-fifth aspect, the invention features a kit for detecting the presence or
absence of a polymorphic restriction site in a nucleic acid, the kit containing:5 (a) a first and a second primer flanking the polymorphic restriction site, the first
primer containing a first sequence not complementary to or present in the nucleic acid,
the second primer containing a second sequence not complementary to or present in
the nucleic acid; (b) a third and a fourth primer, the third primer containing the first
sequence or a sequence complementary to the first sequence, the third primer being
0 tagged with the first member of a first specific binding pair, the fourth primer
containing the second sequence or a sequence complementary to the second sequence,
the fourth primer being tagged with a detectable label; (c) the second member of the
first specific binding pair, immobilized on a first solid support; and (d) an
oligonucleotide complementary to a third sequence in the strand of the nucleic acid
5 corresponding to the second primer, the third sequence being between the
polymorphic restriction site and the sequence corresponding to the second primer, the
oligonucleotide being immobilized on a second solid support.
In a thirty-sixth aspect, the invention features a kit for detecting the presence or
absence of a polymorphic restriction site in a nucleic acid, the kit cont~ining:2 o (a) a first and a second primer fl~nking the polymorphic restriction site, the first
primer containing a first sequence not complementary to or present in the nucleic acid;
(b) a third primer containing the first sequence, the third primer being tagged with a
detectable label; (c) a first oligonucleotide complementary to the single-stranded ends
generated in the nucleic acid upon digestion of the nucleic acid with the restriction
2 5 enzyme corresponding to the polymorphic restriction site, the oligonucleotide being
tagged with the first member of a first specific binding pair; (d) the second member of
the first specific binding pair, immobilized on a first solid support; and (e) a second
oligonucleotide complementary to a second sequence in the strand of the nucleic acid
... .
CA 022667~0 1999-03-17
W O 98/12352 rcTrusg7/16467
corresponding to the first primer, the sequence being between the polymorphic
restriction site and either the sequence corresponding to or complementary to the
second primer or the sequence corresponding to or complementary to the first primer,
the second oligonucleotide being immobilized on a second solid support.
In a thirty-seventh aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid, involving the
steps of: (a) amplifying the nucleic acid by PCR using a first and a second primer
flanking the polymorphic restriction site, whereby the resultant PCR product is of a
defined size readily resolved by gel electrophoresis; (b) digesting the PCR product of
step (a) with the restriction endonuclease corresponding to the polymorphic restriction
site, the digestion products being differentially sized; (c) separating the reaction
products of step (b) by gel electrophoresis; and (d) detecting the separated reaction
products, the presence of only uncleaved products being an indication of a
homozygote lacking the polymorphic restriction site, the presence of only cleaved
products being an indication of a homozygote containing the polymorphic restriction
site, and the presence of both cleaved and uncleaved products being an indication of a
heterozygote. In a preferred embodiment, one or both of the first and second primers
are tagged with a detectable label. In another preferred embodiment, the PCR product
is 100-1000 base pairs in length.
2 o In a thirty-eighth aspect, the invention features a kit for detecting the presence
or absence of a polymorphic restriction site in a nucleic acid, the kit containing: a first
and a second primer fl~nking the polymorphic restriction site and capable of
generating a PCR product of a defined size that is readily resolved by gel
electrophoresis. In a preferred embodiment, the first and/or the second primers are
2 5 detectably labeled. In another plefelled embodiment, the PCR product generated is
between 100 and 1000 base pairs in length.
In a thirty-ninth aspect, the invention features a method for identifying a
polymorphic restriction site in a nucleic acid, involving the steps of: (a) digesting
CA 022667~0 1999-03-17
W O98/12352 PCT~US97/16467
31
DNA isolated from a first sample with a first restriction endonuclease; (b) ligating to
each of the ends of the reaction products of step (a) a first adaptor; (c) digesting the
products of step (b) with a second restriction endonuclease; (d) ligating to each of the
ends of the reaction products generated in step (c) a second adaptor; (e) amplifying the
5 reaction products of step (d) by PCR using a first primer complementary to the first
adaptor and a second primer complementary to the second adaptor, the second primer
being tagged with a first member of a specific binding pair (preferably, biotin); (f) in a
separate set of reactions, digesting DNA isolated from a second sample with the first
restriction endonuclease; (g) ligating to each of the ends of the reaction products of
0 step (~) a third adaptor; (h) digesting the products of step (g) with the second
restriction endonuclease; (i) denaturing the products of step (e) and the products of
step (h); (j) combining the denatured products of step (i) under conditions allowing
hybridization; (k) contacting the hybridization products of step (j) with the second
member of the specific binding pair (preferably, avidin), the second member being
5 immobilized on a solid support; (1) recovering the hybridization products captured on
the solid support; and (m) amplifying the products obtained in step (1) by PCR using a
primer complementary to the third adaptor, an amplified product being an indication
of a polymorphic restriction site corresponding to the second restriction endonuclease.
In a fortieth aspect, the invention features a kit for identifying a polymorphic2 o restriction site in a nucleic acid, the kit contAining: (a) a first DNA adaptor, a second
DNA adaptor, and a third DNA adaptor, the first and third DNA adaptors having
regions complementary to the ends generated by a first restriction endonuclease ends
but differing in overall sequence and the second DNA adaptor having a region
complementary to the ends generated by a second restriction endonuclease, the second
2 5 restriction endonuclease site corresponding to the polymorphic restriction site; and (b)
a first primer, a second primer, and a third primer, the first primer being
complementa~y to the first DNA adaptor, the second primer being complementary tothe second DNA adaptor and being tagged with a first member of a specific binding
. ..~,,
CA 022667~0 1999-03-17
W O 9~/12352 PCT~US97/16467
32
pair, and the third primer being complementary to the third DNA adaptor. This kit
may further contain the second member of the specific binding pair immobilized on a
solid support.
In a forty-first aspect, the invention features a method for detecting the
5 presence or absence of a polymorphic restriction site in a nucleic acid. In this method
the nucleic acid is amplified by PCR using a first and a second primer fl~nking the
polymorphic restriction site. The first primer is tagged with a detectable label, and the
amplifying generates a PCR product containing a first strand tagged with the
detectable label and an unlabeled second strand.
The PCR product is then digested with a restriction endonuclease
corresponding to the polymorphic restriction site to generate a digestion product,
which is denatured to generate a denatured product. The denatured product is
contacted with a first probe, which contains a sequence that hybridizes to a first
sequence in the first strand that is between the polymorphic restriction site and the
sequence in the first strand that is complementary to the second primer. The first
probe is also immobilized on a first binding element.
The first binding element is monitored for the presence of the detectable label,and detection of the detectable label on the first binding element indicates the absence
of the polymorphic restriction site in the nucleic acid, and a failure to detect the
2 o detectable label on the first binding element indicates the presence of the polymorphic
restriction site in the nucleic acid. The first binding element is a region on a solid
support, such as a glass plate or a microchip.
This method can also include contacting the denatured product with a second, a
third, or a fourth probe. The second probe, which is immobilized on a second binding
2 5 element, contains a sequence that hybridizes to a second sequence in the first strand
that is between the polymorphic restriction site and the sequence in the first strand that
corresponds to the first primer. The third probe, which is immobilized on a third
binding element, contains a sequence that hybridizes to a third sequence in the second
CA 022667~0 1999-03-17
W O ~B/12352 PCTrUS97/16467
33
strand that is between the polymorphic restriction site and the sequence in the second
strand corresponding to the second primer. The fourth probe, which is immobilized
- on a fourth binding element, contains a sequence that hybridizes to a fourth sequence
in the second strand that is between the polymorphic restriction site and the sequence
5 in the second strand that is complementary to the first primer. ln this method, the
second, third, or fourth binding element can be monitored for the presence of the
detectable label. The first, second, third, and fourth binding elements can each be
distinct regions on a solid support, such as a glass plate or a microchip.
In a forty-second aspect, the invention features a method for detecting the
0 presence or absence of a polymorphic restriction site in a nucleic acid. In this method
the nucleic acid is amplified by PCR using a first and a second primer fl~nking the
polymorphic restriction site. The first primer is tagged with a first detectable label and
the second primer is tagged with a second detectable label. The amplifying generates
a PCR product containing a first strand tagged with the first detectable label and a
5 second strand tagged with the second detectable label. The first and second detectable
labels can be identical or distinct.
The PCR product is treated with a restriction endonuclease corresponding to
the polymorphic restriction site to generate a digestion product, which is denatured to
generate a denatured product. The denatured product is contacted with a first and a
2 o second probe. The first probe, which is immobilized on a first binding element,
contains a sequence that hybridizes to a first sequence in the first strand that is
between the polymorphic restriction site and the sequence in the first strand that is
complementary to the second primer. The second probe, which is immobilized on a
second binding element, contains a sequence that hybridizes to a second sequence in
2 5 the second strand that is between the polymorphic restriction site and the sequence in
the second strand that is complementary to the first primer. The first and second
binding elements can each be distinct regions on a solid support, such as a glass plate
or a microchip.
CA 022667~0 1999-03-17
WO ~8112352 PCT/US97/16467
34
The first binding element is monitored for the presence of the first detectable
label and the second binding element is monitored for the presence of the seconddetectable label. Detection of the first detectable label on the first binding element
and detection of the second detectable label on the second binding element indicates
the absence of the polymorphic restriction site in the nucleic acid, and a failure to
detect the first detectable label on the first binding element and a failure to detect the
second detectable label on the second binding element indicates the presence of the
polymorphic restriction site in the nucleic acid.
This method can also include contacting the denatured product with a third or a
fourth probe. The third probe, which is immobilized on a third binding element,
contains a sequence that hybridizes to a third sequence in the first strand that is
between the polymorphic restriction site and the sequence in the first strand
corresponding to the first primer. The fourth probe, which is immobilized on a fourth
binding element, contains a sequence that hybridizes to a fourth sequence in thesecond strand that is between the polymorphic restriction site and the sequence in the
second strand corresponding to the second primer. The third or fourth binding
element can be monitored for the presence of the first or second detectable label. The
first, second, third, and fourth binding elements can be each distinct regions on a solid
support, such as a glass plate or a microchip.
2 o In a forty-third aspect, the invention features a method for detecting the
presence or absence of a polymorphic restriction site in a nucleic acid. In this method,
the nucleic acid is amplified by PCR using a first and a second primer fl~nking the
polymorphic restriction site. The amplifying generates a PCR product containing a
first strand conlai.~ g a sequence corresponding to the first primer and a second strand
2 5 containing a sequence corresponding to the second primer.
The PCR product is treated with a restriction endonuclease collc~ollding to
the polymorphic restriction site to generate a digestion product, which is denatured to
generate a denatured product. The denatured product is contacted with an
CA 022667~0 1999-03-17
W O 9~/12352 PCT~US97/16467
oligonucleotide to generate a first reaction product. The oligonucleotide contains a 3'
portion that hybridizes to a first region in the first strand that flanks the polymorphic
restriction site on the side of the polymorphic restriction site containing a sequence
corresponding to the first primer. The oligonucleotide is blocked so that it cannot
5 serve as a primer for DNA polymerase. In addition, the oligonucleotide contains a
5' portion that does not hybridize to a second region in the first strand that flanks the
polymorphic restriction site on the side of the polymorphic restriction site cont~illillg a
sequence that is complementary to the second primer.
The first reaction product is treated with a DNA polymerase to extend the
10 unblocked, primed 3' end to generate a second reaction product, which is amplified by
PCR using the first primer, tagged with a first detectable label, and a third primer that
hybridizes to a sequence that is complementary to the S' portion of the oligonucleotide,
to generate a second PCR product. The third primer is tagged with a second
detectable label. The first and the second detectable labels in this method can be
identical or distinct.
The second PCR product is denatured to generate a second denatured product,
which is contacted with a first and a second probe. The first probe, which is
immobilized on a first binding element, contains a sequence that hybridizes to a first
sequence in the second strand that is between the polymorphic restriction site and the
2 o sequence in the second strand that is complementary to the first primer. The second
probe, which is immobilized on a second binding element, contains a sequence that
hybridizes to a second sequence in the first strand that is between the polymorphic
restriction site and the sequence in the first strand that is complementary to the second
primer. The first and second binding elements can each be distinct regions on a solid
2 5 support, such as a glass support or a microchip.
The first binding element is monitored for the presence of the second detectablelabel and the second binding element is monitored for the presence of the first
detectable label. Detection of the second detectable label on the first binding element
CA 022667~0 1999-03-17
WO 9~/12352 PCT/US97/16467
36
and detection of the first detectable label on the second binding element indicates a
heterozygote, detection of the second detectable label on the first binding element and
a failure to detect the first detectable label on the second binding element indicates a
homozygote containing the polymorphic restriction site, and detection of the first
5 detectable label on the second binding element and a failure to detect the second
detectable label on the first binding element indicates a homozygote lacking thepolymorphic restriction site.
This method can also include contacting the second denatured product with a
third or a fourth probe. The third probe, which is immobilized on a third binding
10 element, contains a sequence that hybridizes to a third sequence in the first strand that
is between the polymorphic restriction site and the sequence in the first strandcorresponding to the first primer. The fourth probe, which is immobilized on a fourth
binding element, contains a sequence that hybridizes to a fourth sequence in thesecond strand that is between the polymorphic restriction site and the sequence in the
15 second strand corresponding to the second primer. The third or fourth bindingelement can be monitored for the presence of the first or second detectable label. The
first, second, third, and fourth binding elements can each be distinct regions on a solid
support, such as a glass plate or a microchip.
In a forty-fourth aspect, the invention features a kit for detecting the presence
2 o or absence of a polymorphic restriction site in a nucleic acid. The kit can contain one
or more sets of a first and a second primer fl~nking the polymorphic restriction site.
The first primer is tagged with a detectable label, so that amplifying the nucleic acid
by PCR with the first and second primers generates a PCR product co~ g a first
strand tagged with the detectable label and a second strand. The kit also can include
2 5 one or more first probes, each of which contaillillg a sequence that hybridizes to a first
sequence in the first strand that is between the polymorphic restriction site and the
sequence in the first strand that is complementary to the second primer. Each of the
first probes is immobilized on a first binding element.
CA 022667~0 1999-03-17
W O 9~/12352 PCT~US97/16467
37
This kit can also contain one or more sets of a second, third, or fourth probe.
Each of the second probes, which is immobilized on a second binding element,
contains a sequence that hybridizes to a second sequence in the first strand that is
between the polymorphic restriction site and the sequence in the first strand that
5 corresponds to the first primer. Each of the third probes, which is immobilized on a
third binding element, contains a sequence that hybridizes to a third sequence in the
second strand that is between the polymorphic restriction site and the sequence in the
second strand corresponding to the second primer. Each of the fourth probes, which is
immobilized on a fourth binding element, contains a sequence that hybridizes to a
0 fourth sequence in the second strand that is between the polymorphic restriction site
and the sequence in the second strand that is complementary to the first primer.The first binding element in this kit can be a region on a solid support, such as a
glass plate or a microchip. In addition, in this kit, the first, second, third, and fourth
binding elements can each be distinct regions on a solid support, such as a glass plate
15 or a microchip.
One or more second primers in this kit can each contain a second detectable
label. In addition, the kit can further contain a second probe, which is immobilized on
a second binding element, and contains a sequence that hybridizes to a second
sequence in the second strand that is between the polymorphic restriction site and the
2 o sequence in the second strand that is complementary to the first primer. The kit
can also contain one or more sets of a third or a fourth probe. Each of the third
probes, which are immobilized on a third binding element, contain a sequence that
hybridizes to a third sequence in the first strand that is between the polymorphic
restriction site and the sequence in the first strand corresponding to the first primer.
2 5 Each of the fourth probes, which are immobilized on a fourth binding element, contain
a sequence that hybridizes to a fourth sequence in the second strand that is between
the polymorphic restriction site and the sequence in the second strand corresponding
to the second primer.
CA 022667~0 1999-03-17
W O 9~/12352 rcTrusg7/16467
38
The kit can further contain one or more second probes, which are immobilized
on a second binding element, that each contain a sequence that hybridizes to a second
sequence in the first strand that is between the polymorphic restriction site and the
sequence in the first strand that corresponds to the first primer.
In a forty-fifth aspect, the invention features a kit for detecting the presence or
absence of a polymorphic restriction site in a nucleic acid. This kit can contain one or
more sets of a first and a second PCR primer flanking the polymorphic restriction site.
The first primer can be tagged with a first detectable label, so that amplifying the
nucleic acid by PCR using the first and second primers generates a PCR product
containing a first strand tagged with the first detectable label and a second strand.
Alternatively, the first primer can be unlabeled.
This kit can also include one or more oligonucleotides containing a 3' portion
that hybridizes to a first region in the first strand that flanks the polymorphic
restriction site on the side of the polymorphic restriction site containing a sequence
corresponding to the first primer. The oligonucleotide is blocked so that it cannot
serve as a primer for DNA polymerase. In addition, the oligonucleotide contains a S'
portion that does not hybridize to a second region in the first strand that flanks the
polymorphic restriction site on the side of the polymorphic restriction site cont~ining a
sequence that is complementary to the second primer.
2 o This kit can also include one or more third primers, each of which thathybridizes to a sequence that is complementary to the 5' portion of the oligonucleotide.
The third primer can be tagged with a second detectable label.
Also included in this kit are one or more sets of a first and a second probe.
Each of the first probes, which are immobilized on a first solid support, contain a
2 s sequence that hybridizes to a first sequence in the second strand that is between the
polymorphic restriction site and the sequence in the second strand that is
complementary to the first primer. Each of the second probes, which are immobilized
on a second solid support, contain a sequence that hybridizes to a second sequence in
CA 022667~0 1999-03-17
W O 98/12352 PCTrUS97/16467
39
the first strand that is between the polymorphic restriction site and the se~uence in the
first strand that is complementary to the second primer.
~ The first and the second detectable labels in this kit can be distinct or identical.
In addition, the first and second binding elements can each be distinct regions on a
solid support, such as a glass support or a microchip.
This kit can further contain one or more sets of a third or a fourth probe. Eachof the third probes, which are immobilized on a third binding element, contain asequence that hybridizes to a third sequence in the first strand that is between the
polymorphic restriction site and the sequence in the first strand corresponding to the
first primer. Each of the fourth probes, which are immobilized on a fourth binding
element, contain a sequence that hybridizes to a fourth sequence in the second strand
that is between the polymorphic restriction site and the sequence in the second strand
corresponding to the second primer.
The one or more sets of the first, second, third, and fourth binding elements inthis kit can each be distinct regions on a solid support, such as a glass plate or a
microchip.
In a preferred embodiment of various of the above aspects, multiple
polymorphic restriction sites are detected by the method or kit. In preferred
embodiments of various of the above aspects, the detectable label is selected from the
2 o group consisting of digoxigenin, fluorescent labels (e.g., fluorescein and rhodamine),
enzymes (e.g., horseradish peroxidase and alkaline phosphatase), biotin (which can be
detected by anti-biotin specific antibodies or enzyme-conjugated avidin derivatives),
radioactive labels (e.g., 32p and '2sI), colorimetric reagents, and chemiluminescent
reagents.
2 5 In other preferred embodiments of various of the above aspects, the specific
binding pairs are selected from the group consisting of avidin-biotin, streptavidin-
biotin, hybridizing nucleic acid pairs, interacting protein pairs, antibody-antigen pairs,
reagents containing chemically reactive groups (e.g., reactive amino groups), and
CA 022667~0 1999-03-17
W O98/123S2 PCTrUS97/16467
nucleic acid sequence-nucleic acid binding protein pairs. In related preferred
embodiments of various of the above aspects, the solid supports used in the methods
of the invention are selected from the group consisting of agarose, acrylamide, and
polystyrene beads; polystyrene microtiter plates (for use in, e.g., ELISA); silicon,
gold, or glass chips (e.g., microchips), slides, or plates; and nylon and nitrocellulose
membranes (for use in, e.g., dot or slot blot assays).
The term "heterozygote," as used herein, refers to an individual with different
alleles at corresponding loci on homologous chromosomes. Accordingly, the term
"heterozygous," as used herein, describes an individual or strain having different
0 allelic genes at one or more paired loci on homologous chromosomes.
The term "homozygote," as used herein, refers to an individual with the same
allele at corresponding loci on homologous chromosomes. Accordingly, the term
"homozygous," as used herein, describes an individual or a strain having identical
allelic genes at one or more paired loci on homologous chromosomes.
The term "corresponding" as used herein to describe a nucleic acid strand, e.g.,a nucleic acid strand corresponding to a particular PCR primer, is meant to indicate
that the strand contains the sequence of the particular PCR primer. When used tocompare a polymorphic restriction site to a restriction endonuclease site, the term
again indicates that the two sequences are identical.
2 o An advantage of certain detection methods of the present invention over many
other methods used to detect genetic polymorphisms is that gel electrophoresis is not
required in the analysis. Thus, the methods of the present invention are readilyadaptable for automation, allowing large numbers of samples to be processed in
relatively short periods of time, at lower costs. In certain of the embodiments,2 5 detection of an array of samples is carried out simultaneously on a solid support, such
as a glass slide or a microchip, further reducing processing time and cost. Detection
of signals on arrays can be carried out quantitatively or qualitatively. In addition, in
several variations of the methods of the invention (see, e.g., Examples III and IV
CA 022667~0 1999-03-17
WO 98/12352 PCT/US97/16467
41
below), internal controls are provided, thus controlling for variability detected by
different practitioners. Furthermore, in several of the variations of the methods of the
- invention (see Examples III-VIII and XII-XIV below), an oligonucleotide probe
hybridizing to a sequence in the PCR product internal to the primers is used to purify
the products, thus allowing a reduction in background problems associated with PCR
amplification.
Those detection methods of the invention utilizing gel electrophoresis are also
advantageous because they provide a rapid and inexpensive approach to the
identification of large numbers of PCR-based genetic and RFLP markers.
0 The method of the invention useful for cloning genetic polymorphisms also
represents an improvement over current methods. Because the process of selectingout a tagged (e.g., biotinylated) DNA having a polymorphism involves a specific
hybridization step, candidate DNA from any source may be utilized. For example,
DNA from random clones, CDNA libraries, YAC libraries, or any other DNA
collection may be screened; pure preparations of genomic DNAs are not required.
Moreover, like other methods of the invention, thls cloning procedure is rapid and
mexpenslve.
All methods of the invention are useful in clinical diagnostic testing, genomic
mapping, positional cloning of genes defined by mutation (such as those that cause
2 o inherited disease in humans or resistance to pathogens in crop plants), DNAfingel~ ltillg (e.g., for forensic analysis and paternity testing), crop and livestock
breeding programs, and other related applications.
In one particular example, the detection methods of the invention are useful forbacterial typing utili7ing known conserved polymorphic sequences diagnostic of the
2 5 bacterium. In one application, this approach is useful for distingui.~hing one bacterium
from another (e.g., for the identification of Salmonella in a food sample);
polymorphism-containing sequences preferred for this approach include those present
in conserved ribosomal RNA genes. In another application, this approach is useful for
CA 022667~0 1999-03-17
W O9~/123S2 PCTrUS97116467
42
screening bacteria (e.g., clinical isolates) for antibiotic resistance; in this case, known
polymorphic restriction sites within the antibiotic resistance marker are utilized. The
instant methods of bacterial typing decrease false positive results frequently obtained
using current PCR-based techniques.
Detailed Description
The drawings are first described.
Drawings
Fig. 1 is a schematic of a RFLP detection method involving the use of a first
0 PCR primer tagged with a detcctable label (X) and a second PCR primer tagged with
the first member of a specific binding pair (Y). After amplification by PCR, theproducts are digested with the restriction endonuclease (R) corresponding to thepolymorphic restriction site, contacted with the second member of the specific binding
pair immobilized on a solid support, and the level of the detectable label (X) bound to
the solid support is determined.
Fig. 2 is a schematic of a RFLP detection method involving the use of a first
PCR primer tagged with a first detectable label (X) and a second PCR primer tagged
with the first member of a specific binding pair (Y). After amplification by PCR, the
products are digested with the restriction endonuclease (R) corresponding to the2 o polymorphic restriction site, and an oligonucleotide tagged with a second detectable
label (Z) is annealed and ligated to the single-stranded ends generated in the digestion.
The reaction is then contacted with the second member of the specific binding pair
bound to a solid support, and the levels of the first and second detectable labels (X and
Z) bound to the solid support are determined.
2 5 Fig. 3 is a schematic of a RFLP detection method involving the use of a first
PCR primer tagged with a detectable label (P1) and a second unlabeled PCR primer(P2). After amplification by PCR, half of the reaction (or one of the identical
reactions if carried out in duplicate) is digested with the restriction endonuclease (R)
CA 022667~0 1999-03-17
WO 98/12352 PCT/US97/16467
43
corresponding to the polymorphic restriction site. Both digested and undigested
reactions are then denatured and contacted with an oligonucleotide tagged with the
first member of a specific binding pair, the oligonucleotide being complementary to
the P I strand and located to the right of the restriction site (R) near to, but not
overlapping, primer P2. The reactions are then contacted with the second member of
the specific binding pair immobilized on a solid support, and the levels of P 1 in
digested versus undigested reactions are compared.
Fig. 4 is a schematic of a RFLP detection method involving the use of a first
PCR primer tagged with a first detectable label (P 1) and a second PCR primer tagged
0 with a second detectable label (P2). After amplification by PCR, the products are
digested with the restriction endonuclease (R) corresponding to the polymorphic
restriction site7 denatured, and contacted with a first oligonucleotide complementary to
the P 1 strand and located to the right of the restriction site (R) near to, but not
overlapping primer P2, and a second oligonucleotide complementary to the P2 strand
and located to the right of the restriction site (R) near to, but not overlapping the
sequence complementary to primer P2. Both the first and second oligonucleotides are
tagged with the first member of a specific binding pair (Y). The reactions are then
contacted with the second member of the specific binding pair immobilized on a solid
support, and the ratio of P 1 to P2 bound to the solid support is deterrnined.
2 o Fig. 5 is a schematic of a RFLP detection method involving the use of a first
PCR primer tagged with a detectable label (X) and a second PCR primer tagged with
the first member of a first specific binding pair (Y). After amplification by PCR, the
products are digested with the restriction enzyme (R) corresponding to the
polymorphic restriction site, and are contacted with the second member of the first
2 5 specific binding pair immobilized on a first solid support. The filtrate is then bound to
a solid support with the anchor sequence (or contacted with an oligonucleotide
complementary to the X strand between the restriction site (R) and the label (X), the
oligonucleotide being tagged with the first member of a second specific binding pair,
CA 022667~0 1999-03-17
WO 9~/12352 PCTrUS97/16467
44
and then contacted with the second member of the second specific binding pair
immobilized on a second solid support), and the levels of the detectable label bound to
the first solid support and the anchor sequence (or second solid support) are
determined.
Fig. 6 is a schematic of a RFLP detection method involving the use of a first
unlabeled PCR primer and a second PCR primer tagged with a detectable label (X).After amplification by PCR, the products are digested with the restriction enzyme (R)
corresponding to the polymorphic restriction site, and contacted with an
oligonucleotide complementary to the single-stranded ends generated in the digestion,
the oligonucleotide being tagged with the first member of a specific binding pair. The
products are then contacted with the second member of the first specific binding pair,
bound to a first solid support. The filtrate is then bound to a solid support with the
anchor sequence (or contacted with an oligonucleotide complementary to the X strand,
the oligonucleotide being tagged with the first member of a second specific binding
pair, and then contacted with the second member of the second specific binding pair
immobilized on a second solid support), and the levels of the detectable label bound to
the first solid support and the anchor sequence (or second solid support) are
determined.
Fig. 7 is a schematic of a RFLP detection method involving the use of PCR
2 o primers fl~nking the polymorphic restriction site (the "Alu I" site). Following PCR
amplification, the reaction products are digested with the restriction endonuclease
corresponding to the polymorphic restriction site (Alu I), and the fragments are run on
an agarose gel. The separated fragments are detected as an indication of the presence
or absence of the polymorphic marker.
2 5 Fig. 8 is a schematic of a typical gel analysis according the method described in
Fig. 7.
Figs. 9A-9E are schematics of a method for cloning polymorphic restriction
fragments.
CA 022667~0 1999-03-17
W O 9~/12352 ~CTrUS97/16467
Fig. 10 is a schematic of a non-gel based method for detection of CAPS
markers.
~ Fig. 11 is a schematic of parallel processing of 100 CAPS markers on a
microchip containing an array of oligonucleotide probes.
Fig. 12 is a schematic of a method for detecting a cleaved end of a CAPS
marker.
Fig. 13 is a schematic of a method for distinglli.shing heterozygous CAPS
alleles from homozygous CAPS alleles, involving the use of the method for detecting
a cleaved end of a CAPS marker shown in Fig. 12.
CA 022667~0 1999-03-17
WO g8/12352 PCT/US97/16467
46
Methods for Generatin~ and Detecting Genetic Polymorphisms
The present invention provides several methods for detecting Cleaved
Amplified Polymorphic Sequences (CAPS; Konieczny et al., The Plant Journal
4(2):403-410, 1993). In the CAPS method, a nucleic acid containing apolymorphic
restriction site is amplified using primers fl~nking the restriction site. The resulting
PCR product is digested with the restriction endonuclease corresponding to the
polymorphic restriction site, and the digested products are analyzed by gel
electrophoresis .
The detection methods of the present invention vary greatly from one another
0 in detail, however they share three central features: (1) the nucleic acid cont~ining the
polymorphic restriction site is amplified by PCR using differently labeled primers
flanking the polymorphic restriction site, (2) the resulting PCR product is digested
with the restriction endonuclease corresponding to the polymorphic restriction site
(which will cleave the DNA of some individuals but not cleave the DNA of others,depending on the presence of the polymorphism), and (3) the resulting digestion
products are analyzed by detection of the labels they contain, and/or labels attached to
oligonucleotides complementary to the digestion products, in order to determine the
identity of the polymorphic restriction site. The methods of the invention allow rapid
and efficient analyses of a large number of samples.
2 o The nucleic acid sample containing the polymorphic restriction site being
analyzed can be obtained from any source, e.g., a tissue homogenate, blood, amniotic
fluid, chorionic villus samples, and a bacterial culture; and can be obtained from these
sources using standard methods. Only a minute quantity of nucleic acid is required,
and can be DNA or RNA (in the case of RNA, a reverse transcription step is required
2 5 before the PCR step). The PCR methods used in the methods of the present invention
are carried out using standard methods (see, e.g., Ausubel et al., Current Protocols in
Molecular Biology, John Wiley and Sons, New York, 1989; Erlich, PCR Technology,
Stockton Press, New York, 1989; Innis et al., PCR Protocols: A Guide to Methods and
CA 022667~0 1999-03-17
W O 98/12352 PCT~US97/16467
47
Applications, Academic Press, Harcourt Brace Javanovich, New York, 1990;
Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor
Laboratory Press, Cold Spring Harbor, New York, 1989). Restriction enzyme
digestion is also carried out by standard methods using any of a number of available
5 restriction endonucleases (see, e.g., Ausubel et al., s7lpra; New England Biolabs,
Beverly, MA).
The primers and oligonucleotides used in the methods of the present invention
are preferably DNA, and can be synthesized using standard techniques and, when
~ro~liate, detectably labeled using standard methods (Ausubel et ah, supra).
o Detectable labels that can be used to tag the primers and oligonucleotides used in the
methods of the invention include, but are not limited to, digoxigenin, fluorescent
labels (e.g., fluorescein and rhodamine), enzymes (e.g., horseradish peroxidase and
alkaline phosphatase), biotin (which can be detected by anti-biotin specific antibodies
or enzyme-conjugated avidin derivatives), radioactive labels (e.g., 32p and ~2sI),
5 colorimetric reagents, and chemiluminescent reagents. The labels used in the methods
of the invention are detected using standard methods.
The specific binding pairs useful in the methods of the invention include, but
are not limited to, avidin-biotin, streptavidin-biotin, hybridizing nucleic acid pairs,
interacting protein pairs, antibody-antigen pairs, reagents collt~h~ g chemically
2 o reactive groups (e.g., reactive amino groups), and nucleic acid sequence-nucleic acid
binding protein pairs.
The solid supports useful in the methods of the invention include, but are not
limited to, agarose, acrylamide, and polystyrene beads; polystyrene mic~otiler plates
(for use in, e.g., ELISA); and nylon and nitrocellulose membranes (for use in, e.g., dot
2 5 or slot blot assays).
Some methods of the invention employ solid supports containing arrays of
nucleic acid probes. In these cases, solid supports made of materials such as glass
(e.g., glass plates), silicon or silicon-glass (e.g., microchips), or gold (e.g., gold plates)
CA 022667~0 1999-03-17
WO 98/12352 PCT/US97/16467
48
can be used. Methods for attaching nucleic acid probes to precise regions on such
solid surfaces, e.g., photolithographic methods, are well known in the art, and can be
used to make solid supports for use in the invention. (For example, see, Schena et al.,
Science 270:467-470, 1995; Kozal et al., Nature Medicine 2(7):753-759, 1996; Cheng
et al., Nucleic Acids Research 24(2):380-385, 1996; Lipshutz et al., BioTechniques
19(3):442-447, 1995; Pease et al., Proc. Natl. Acad. Sci. USA 91 :5022-5026, 1994;
Fodor et al., Nature 364:555-556, 1993; Pirrung et al., U.S. Patent No. 5,143,854; and
Fodor et al., WO 92/10092.)
The methods of the invention can be facilitated by the use of kits which contain0 the reagents re4uired for carrying out the assays. The kits can contain reagents for
carrying out the analysis of a single polymorphic restriction site (for use in, e.g.,
diagnostic methods) or multiple polymorphic reskiction sites (for use in, e.g., genomic
mapping). When multiple samples are analyzed, multiple sets of the al~plopliate
primers and oligonucleotides are provided in the kit. In addition to the primers and
oligonucleotides required for carrying out the various methods, the kits may contain
the enzymes used in the methods, and the reagents for detecting the labels, e.g., the
subskates for enzyme labels, etc. The kits can also contain solid substrates for used in
carrying out the method of the invention. For example, the kits can contain solid
substrates, such as glass plates or silicon or glass microchips, containing arrays of
nucleic acid probes.
As discussed above, the invention provides methods and kits for generating and
detecting the presence or absence of a polymorphic reskiction site in a nucleic acid.
Examples I-IX and XII-XIV describe eight variations of the methods of the invention.
Example X describes a preferred use for the methods of the invention. Example XI2 5 describes a preferred method for cloning polymorphic reskiction fragments. The
following examples are meant to illuskate, but not limit, the methods of the present
invention. Other suitable modifications and adaptations of the variety of conditions
... . . ..
CA 022667~0 1999-03-17
WO 98tl2352 PCT/US97/16467
49
and parameters of molecular biology which are obvious to those skilled in the art are
within the spirit and scope of the present invention.
EXAMPT,F.
Example I.
In this method, the nucleic acid containing the polymorphism is amplified by
PCR using a first and a second primer fl~nking the polymorphic restriction site, the
first primer being tagged with the first member of a specific binding pair, the second
primer being tagged with a detectable label. The resulting PCR product is digested
0 with the restriction endonuclease corresponding to the polymorphic restriction site and
the digested products are contacted with the second member of the specific binding
pair, immobilized on a solid support. The level of the detectable label bound to the
solid support is then measured. The presence of the detectable label bound to the solid
support is an indication of the absence of the polymorphic restriction site in the
nucleic acid, while the absence of the detectable label bound to the solid support is an
indication of the presence of the polymorphic restriction site in the nucleic acid. An
embodiment of this method is shown in Fig. l .
Example II.
2 o This method is identical to that described in Example I, with the added step of
annealing and ligating to the single-stranded ends generated in the digestion reaction,
an oligonucleotide tagged with a second detectable label. After applying the reaction
to the second member of the specific binding pair, the levels of both the first and the
second detectable labels bound to the solid support are determined. The presence of
2 5 only the first detectable label bound to the solid support is an indication of a
homozygote lacking the polymorphic restriction site, the presence of only the second
detectable label bound to the solid support is an indication of a homozygote containing
the polymorphic restriction site, and the presence of both the first and the second
., ~...., ~ .
CA 022667~0 l999-03-l7
W O 98/12352 PCTAUS97/16467
detectable labels bound to the solid support is an indication of a heterozygote. An
embodiment of this method is shown in Fig. 2.
In addition to labeling the cleaved ends of the CAPS products by annealing and
ligating an oligonucleotide to the sticky ends generated by the cleavage, as is
5 described above, and, e.g., in Examples VI and VIII, the cleaved ends can be labeled
by using a method described in further detail below in Example XIV. Briefly, in this
method, the denatured product is contacted with an oligonucleotide to generate a first
reaction product. The oligonucleotide contains a 3' portion that hybridizes to a first
region in the first strand that flanks the polymorphic restriction site on the side of the
0 polymorpllic restriction site containing a sequence corresponding to the first primer.
The 3' end of the oligonucleotide is blocked by, e.g., a di-deoxynucleotide, so that it
cannot serve as a primer for DNA polymerase. The oligonucleotide contains a 5'
portion tllat does not hybridize to a second region in the first strand that flanks the
polymorphic restriction site on the side of the polymorphic restriction site conl~ining a
5 sequence that is complementary to the second primer. The use of such an
oligonucleotide to label a cleaved end of a CAPS marker is illustrated in ~ig. 12. This
method can also be applied to the CAPS detection techniques of, for example,
Examples VI and VIII.
20 l~xample III.
In this method, the nucleic acid is amplified using a first and a second primer
fl~nking the polymorphic restriction site, the first primer being tagged with a
detectable label, the second primer being unlabeled. A portion of the PCR reaction is
digested with the restriction endonuclease corresponding to the polymorphic
2 5 restrictiol~ site, while another portion is left undigested. Both the digested and
undigested portions are then denatured, and contacted with an oligonucleotide tagged
with the first member of a specific binding pair. The oligonucleotide is
complementary to a sequence in the strand of the PCR product containing the
CA 022667~0 1999-03-17
WO 98112352 PCTIUS97/16467
51
detectable label, the sequence being between the polymorphic restriction site and the
sequence complementary to the second primer.
The reaction is then contacted with the second member of the specific binding
pair, immobilized on a solid support, and the ratio of the levels of the detectable label
5 bound to the solid support between undigested and digested samples is determined. A
ratio of l :O between equivalent portions of undigested and digested samples is an
indication of a homozygote containing the polymorphic restriction site, a ratio of l: l
between equivalent portions of undigested and digested samples is an indication of a
homozygote lacking the polymorphic restriction site, and a ratio of 2: l between10 equivalent portions of undigested and digested samples is an indication of a
heterozygote. While the sample volumes used for detection and comparison need not
be equivalent, the ~I,rol~liate calculations must be carried out to account for this
adjustment prior to determining the ratio of detectable label in digested and undigested
samples. An embodiment of this method is shown in Fig. 3.
Example IV.
In this method, the nucleic acid is amplified by PCR using a first primer and a
second primer flanking the polymorphic restriction site, the first primer being tagged
with a first detectable label, and the second primer being tagged with a second
2 o detectable label.
The PCR product is digested with the restriction endonuclease corresponding to
the polymorphic restriction site, denatured, and contacting with a first and a second
oligonucleotide. The first oligonucleotide is complementary to a first sequence in the
strand of the PCR product containing the first detectable label, the first sequence being
2 5 between the polymorphic restriction site and the sequence corresponding to the first
primer. The first oligonucleotide is tagged with the first member of a first specific
binding pair. The second oligonucleotide is complementary to a second sequence in
the strand of the PCR product containing the second detectable label. The second
CA 022667~0 1999-03-17
WO 98~12352 PCT/US97/16467
52
sequence is on the same side of the polymorphic restriction site as the ~1rst sequence,
and is not contained within, or complementary to, either the first or the second primer.
The second oligonucleotide is tagged with the first member of a second specific
binding pair.
A first portion of the reaction is then contacted with the second member of the
first specific binding pair, immobilized on a first solid support, while a second portion
of the reaction is contacted with the second member of the second specific binding
pair, immobilized on a second solid support. The ratio of the levels of the first and
second detectable labels bound to the first and second solid supports is then
determined. A ratio of 1:0 between equivalent amounts of the first and second
portions is an indication of a homozygote containing the polymorphic restriction site,
a ratio of 1:1 between equivalent amounts of the first and second portions is anindication of a homozygote lacking the polymorphic restriction site, and a ratio of 2:1
between equivalent amounts of the first and second portions is an indication of a
1 5 heterozygote.
In the case where the first sequence (to which the first oligonucleotide is
complementary) in the strand containing the first detectable label is between the
polymorphic restriction site and the sequence complementary to the second primer, the
ratios differ, as follows. The ratio of the levels of the first and second detectable
2 o labels bound to the first and second solid supports is 0:1 between equivalent amounts
of the first and second portions in the case of a homozygote containing the
polymorphic restriction site. The ratio is 1:1 between equivalent amounts of the first
and second portions in the case of a homozygote lacking the polymorphic restriction
site, and the ratio is 1:2 between equivalent amounts of the first and second portions in
2 5 the case of a heterozygote. An embodiment of this method is shown in Fig. 4.
CA 022667~0 1999-03-17
W O 9g/12352 PCT~US97/16467
53
Example V.
In this method, the nucleic acid is amplified by PCR using a first and a second
primer flanking the polymorphic restriction site, the first primer being tagged with the
first member of a first specific binding pair, the second primer being tagged with a
5 detectable label. The PCR product is digested with the restriction endonuclease
corresponding to the polymorphic restriction site, and the reaction is then contacted
with the second member of the first specific binding pair, immobilized on a first solid
support.
The material not bound to the first solid support is denatured and contacted
0 with an oligonucleotide complementary to a sequence in the strand of the PCR product
containing the detectable label. The sequence is between the polymorphic restriction
site and the sequence corresponding to the second primer, and the oligonucleotide is
tagged with the first member of a second specific binding pair. The reaction is then
contacted with the second member of the second specific binding pair, immobilized on
a second solid support, and the ratio of the level of the detectable label bound to the
first solid support co~ aled to the level of the detectable label bound to the second
solid support is determined. A ratio of 0:1 is an indication of a homozygote
containing the polymorphic restriction site, a ratio of 1:0 is an indication of a
homozygote lacking the polymorphic restriction site, and a ratio of 1:1 is an indication
2 o of a heterozygote. These ratios are correct in cases where the total amount of the
material not bound to the first solid support is used in the following steps, and should
be adjusted accordingly, if a different amount of the material is used. An embodiment
of this method is shown in Fig. 5.
25 Example VI.
In this method, the nucleic acid is amplified by PCR using a first and a second
primer fl~nking the polymorphic restriction site, the first primer being tagged with a
detectable label, the second primer being unlabeled. The PCR product is digested
. .,.~, . .~
CA 022667~0 1999-03-17
W Og8/12352 PCTrUS97/16467
54
with the restriction endonuclease corresponding to the polymorphic restriction site,
and a first oligonucleotide tagged with the first member of a first specific binding pair
is annealed and ligated to the single-stranded ends generated in the digestion reaction.
The reaction is then contacted with the second member of the first specific binding
pair, immobilized on a first solid support.
The material not bound to the first solid support is denatured, and contacted
with a second oligonucleotide complementary to a sequence in the strand of the PCR
product containing the detectable label, the sequence being between the polymorphic
restriction site and either the sequence corresponding to the first primer or the
sequence complementary to the second primer. The second oligonucleotide is tagged
with the first member of a second specific binding pair. The reaction is then contacted
with the second member of the second specific binding pair, immobilized on a second
solid support, and the ratio of the level of the detectable label bound to the first solid
support compared to the level of the detectable label bound to the second solid support
5 is deLe~ ed. A ratio of l :O is an indication of a homozygote containing the
polymorphic restriction site, a ratio of 0: l is an indication of a homozygote lacking the
polymorphic restriction site, and a ratio of l: l is an indication of a heterozygote.
These ratios are correct in cases where the total amount of the material not bound to
the first solid support is used in the following steps, and should be adjusted
2 o accordingly, if a different amount of the material is used. An embodiment of this
method is shown in Fig. 6.
Example VII.
In this method, the nucleic acid is amplified by PCR using a first and a second
2 5 primer flanking the polymorphic restriction site, the first primer being tagged with the
first member of a first specific binding pair, the second primer being tagged with a
detectable label. The PCR product is digested with the restriction endonuclease
CA 022667~0 1999-03-17
W O 98/123S2 PCT~US97/16467
corresponding to the polymorphic restriction site, and contacted with the secondmember of the first specific binding pair, immobilized on a first solid support.~ The material not bound to the first solid support is denatured and contacted
with an oligonucleotide complementary to a sequence in the strand of the PCR product
5 containing the detectable label. The sequence is between the polymorphic restriction
site and the sequence corresponding to the second primer, and the oligonucleotide is
immobilized on a second solid support (e.g., a nylon or nitrocellulose membrane).
The ratio of the level of detectable label bound to the first solid support to the
level of detectable label bound to the second solid support is then determined. A ratio
0 of 0:1 is an indication of a homozygote containing the polymorphic restriction site, a
ratio of 1:0 is an indication of a homozygote lacking the polymorphic restriction site,
and a ratio of 1:1 is an indication of a heterozygote. These ratios are correct in cases
where the total amount of the material not bound to the first solid support is used in
the following steps, and should be adjusted accordingly, if a different amount of the
material is used. An embodiment of this method is shown in Fig. 5.
Example VIII.
In this method, the nucleic acid is amplified by PCR using a first and a second
primer fl~nking the polymorphic restriction site, the first primer being tagged with a
2 o detectable label, the second primer being unlabeled. The PCR product is digested
with the restriction endonuclease corresponding to the polymorphic restriction site,
and a first oligonucleotide tagged with the first member of a first specific binding pair
is annealed and ligated to the single-stranded ends generated in the digestion reaction.
The reaction is contacted with the second member of the first specific binding pair,
2 5 immobilized on a first solid support. The material not bound to the first solid support
is denatured, and contacted with a second oligonucleotide complementary to a
sequence in the strand of the PCR product containing the detectable label. The
sequence is between the polymorphic restriction site and either the sequence
CA 022667~0 1999-03-17
W O 98~12352 PCTrUS97/16467
56
corresponding to the first primer or the sequence complementary to the second primer,
and the second oligonucleotide is immobilized on a second solid support (e.g., a nylon
or nitrocellulose membrane).
The ratio of the level of the detectable label bound to the first solid support to
the level of the detectable label bound to the second solid support is then determined.
A ratio of l :0 is an indication of a homozygote containing the polymorphic restriction
site, a ratio of 0: l is an indication of a homozygote lacking the polymorphic restriction
site, and a ratio of l: l is an indication of a heterozygote. These ratios are correct in
cases where the total amount of the material not bound to the first solid support is used
0 in the following steps, and should be adjusted accordingly, if a different amount of the
material is used. An embodiment of this method is shown in Fig. 6.
PCR primers cont~ining nucleic acid tags on their 5' ends can also be used in
the methods of thc invention. These primers can be used in pairs, or in combination
with un-tagged primers, in the initial cycles of PCR, followed by the addition of a
"universal primer(s)" complementary to the nucleic acid tags in the first primers, and
contain detectable labels (e.g., biotin, fluorescent, or ELISA tags). The use of nucleic
acid tagged primers in the early rounds of PCR is a cost-effective measure, as only one
set of primers, the universal primers, which can be used in the analysis of manydifferent polymorphic sites, need to be detectably }abeled. The sets of primers specific
2 o for individual polymorphic restriction sites do not have to be tagged with detectable
labels, but rather need only to be complementary to the universal primers in their 5'
ends.
Example IX.
2 5 In another method of the invention, the nucleic acid is amplified by PCR using
a first and a second primer flanking the polymorphic restriction site. The PCR product
is digested with the restriction endonuclease corresponding to the polymorphic
restriction site, and, as shown in Fig. 7, the digestion products are run on a gel
CA 022667~0 1999-03-17
W O 9~/12352 PCTrUS97/16467
57
(preferably an agarose gel). To simplify the gel reading, the first and second primers
are preferably designed to generate a PCR product that is easily resolvable on an
agarose gel (e.g., preferably larger than 100 base pairs and smaller than 1000 base
pairs), and the polymorphic restriction site is preferably located at an asymmetric
position within the amplified fragment. Using this technique, short gel runs can be
used for analysis, and the cleaved products are easily detected. In the particular
example shown in Fig. 8, primers are designed to produce PCR amplified products of
300 base pairs, and cleavage at the RFLP site yields products of 200 base pairs and
100 base pairs.
0 In a preferred method of carrying out this method, sets of primer pairs are
provided that detect a number of RFLP markers. Each set of primers may be
provided, for example, in one of the wells of a 96-well microtiter plate, and PCR
reactions run independently. Following restriction digestion, the reaction products are
transferred to an agarose gel and separated by electrophoresis. A typical result of this
method is shown in Fig. 8.
Detection of the amplified and cleaved products after electrophoretic separationcan be carried out by standard methods of DNA staining (e.g., ethidium bromide
staining) or blotting (e.g., Southern blotting). Alternatively, one or both of the PCR
primers can be detectably labeled, and the labels can be detected as described above.
Example X.
A preferred use of the methods of the invention is in conjunction with a method
called RFLP subtraction. RFLP subtraction provides a large number of polymorphicgenetic markers, while the methods of the present invention provide efficient methods
2 5 for their analysis.
Carrying out RFLP subtraction results in the purification of fragments that are
present in one population (the tracer) but absent in another (the driver). Purification is
achieved by removing all of the fragments in the tracer DNA that have counterparts in
CA 022667~0 1999-03-17
W O9~/12352 PCTAUS97/16467 58
the driver DNA using subtractive hybridization (Innis et al., PCR Protocols: A Guide
to Methods and Applications, Academic Press, Harcourt Brace Javanovich, New
York, l 990). ~n RFLP subtraction, the tracer is a size fraction of digested DNA from
one strain and the driver is the same size fraction from a polymorphic strain. The
5 products obtained after removing the common sequences are RFLPs; they are sized
tracer fragments whose driver counterparts are not found in the same size fraction.
There are three steps in RFLP subtraction: preparation of the driver and tracer,subtractive hybridization, and removal of non-hybridizing sequences from the tracer.
To prepare the driver and tracer DNA, genomic DNA from two different strains is
0 digested with a restriction endonuclease, and the ends of the restriction fragents
from each strain are capped with different oligonucleotide adapters. The low
molecular weight fragments are then purified from a slice of an agarose gel and
amplified using one of the adapter strands as a PCR primer. A biotinylated primer can
be used to amplify the driver so that driver DNA can be removed following the
5 subtractive hybridizations by binding to avidin coated beads.
Three rounds of subtractive hybridization are performed to remove tracer
sequences that also occur in the driver. A small amount of tracer is mixed with an
excess of biotinylated driver, the mixture is denatured and allowed to re-anneal. Most
tracer sequences will hybridize to complementary biotinylated driver strands. Some
2 o tracer sequences, however, are not represented in the driver because they reside on
large restriction fragments (i.e., they are RFLPs) or are missing from the driver
genome. These fragments will have no complementary biotinylated strands with
which to anneal. The biotinylated driver DNA, and any tracer that has annealed to it,
is then removed using avidin-coated beads. The unbound fraction is then subjected to
2 5 two more rounds of subtractive hybridization, tracer DNA rem~ining after the third
round is amplified, and poorly hybridizing sequences are removed.
CA 022667~0 1999-03-17
W O98/12352 rcTrusg7/16467
59
F~mrle XI.
Figure 9 shows a preferred method for cloning polymorphic restriction
fragments. The object of this method is to clone restriction fragments from organism
B (generated by restriction endonuclease A) that do not contain cleavage sites for
restriction endonuclease B, and which correspond to restriction fragments in organism
A (generated by restriction endonuclease A) that do contain at least one restriction site
for restriction endonuclease B. These polymorphic restriction fragments are useful as
CAPS markers for the detection methods described above.
Referring to the method outlined in Figs. 9A-9E, in Fig. 9A, genomic DNA
0 isolated from polymorphic individuals A and B is separately digested with restriction
enzyme A, which preferably leaves so-called sticky ends. An oligonucleotide adaptor
(#l), with complementary sticky ends, is ligated to the restriction fragments from
individual A. A different oligonucleotide adaptor (#3) is ligated to the restriction
fragments from individual B.
In Fig. 9B, the restriction fragments from Fig. 9A are cleaved with restriction
endonuclease B, which again preferably leaves sticky ends. In the case of the DNA
fragments from individual A, an oligonucleotide adaptor (#2), with complementarysticky ends for enzyme B, is ligated to the restriction fragments generated by cleavage
with enzyme B.
2 o In Fig. 9C, the DNA fragments from individual A are amplified using the PCR
with an oligonucleotide primer complementary to adaptor #l and with a biotinylated
oligonucleotide primer complementary to adaptor #2.
In Fig. 9D, the amplified products origin~ting from individual A are mixed with
the non-amplified fragments of Fig. 9B from individual B. The mixed DNA
2 5 fragments are then heat denatured, annealed, and adsorbed onto an avidin-coated solid
support (e.g., beads). The avidin coated support containing the adsorbed fragments is
thoroughly washed. If desired, the adsorbed fragments may be eluted, re-amplified
with the same primers as above, adsorbed onto a fresh avidin-cont~ining support, and
CA 022667~0 1999-03-17
WO 9~/12352 PCTNS97/16467
thoroughly washed. This step can be repeated as many times as is necessary or
desired.
In Fig. 9E, the fragments adsorbed to the avidin-coated beads are eluted and
amplified using PCR with primers complementary to adaptor #3. The amplified
products should correspond to the desired restriction fragments described above.These amplified fragments are cloned and then tested individually using the Southern
DNA blot hybridization method for their ability to display the desired RFLP.
Example XII.
0 In this method, the nucleic acid is amplified by PCR using a first and a second
primer fl~nking the polymorphic restriction site, with the first primer being tagged
with a detectable label. The amplification generates a PCR product co~ g a firststrand tagged with the detectable label and a second, unlabeled strand. The PCR
product is digested with the restriction endonuclease corresponding to the
polymorphic restriction site and the digestion product is denatured. The denatured
product is contacted with a first probe that ( 1 ) contains a sequence that hybridizes to a
first sequence in the first strand of the PCR product, and (2) is immobilized on a first
binding element. The first sequence is between the polymorphic restriction site and
the sequence in the first strand that is complementary to the second primer.
2 o The first binding element is monitored for the presence of the detectable label.
Detection of the detectable label on the first binding element indicates the absence of
the polymorphic restriction site in the nucleic acid, and a failure to detect the
detectable label on the first binding element indicates the presence of the polymorphic
restriction site in the nucleic acid.
2 5 In addition to the first probe described above, this method can employ the use
of a second, a third, or a fourth probe. The second probe contains a sequence that
hybridizes to a second sequence which is in the first strand and is between the
polymorphic restriction site and the sequence in the first strand that corresponds to the
CA 022667~0 1999-03-17
W O 98/12352 PCTrUS97/16467
61
first primer. The third probe contains a sequence that hybridizes to a third sequence
which is in the second strand and is between the polymorphic restriction site and the
sequence in the second strand corresponding to the second primer. The fourth probe
contains a sequence that hybridizes to a fourth sequence which is in the second strand
5 and is between the polymorphic restriction site and the sequence in the second strand
that is complementary to the first primer. The second, third, and fourth probes are
immobilized on a second, third, and fourth binding element, respectively. The second
binding element can be monitored for the presence of the detectable label as a positive
control, while the third or fourth binding elements can be monitored for the presence
10 of the detectable label as negative controls. The first, second, third, and fourth binding
elements, in this and in other methods of the invention, can be present on a solid
support having similar sets of binding elements for testing different nucleic acids (see,
for example, Fig. l l ).
The binding elements, for example, the first, second, third, and fourth binding
5 elements, used in this method of the invention can be present as distinct regions on a
single solid support. For example, they can be specific sets of nucleic acids bound to
distinct regions on a glass plate or on a microchip, such as a glass, silicon, or glass-
silicon microchip (see above).
2 o Example XIII.
In this method a nucleic acid is amplified by PCR using a first and a second
primer flanking the polymorphic restriction site. The first primer is tagged with a first
detectable label and the second primer is tagged with a second detectable label. The
amplification thus generates a PCR product containing a first strand tagged with the
2 5 first detectable label and a second strand tagged with the second detectable label. In
this method, the first and second labels can be identical or distinct.
The PCR product is treated with a restriction endonuclease corresponding to
the polymorphic restriction site to generate a digestion product, which is denatured to
CA 022667~0 1999-03-17
W O 98/12352 PCT~US97/16467
62
generate a denatured product. The denatured product is contacted with a first and a
second probe. The first probe, which is immobilized on a first binding element,
contains a sequence that hybridizes to a first sequence in the first strand that is
between the polymorphic restriction site and the sequence in the first strand that is
5 complementary to the second primer. The second probe, which is immobilized on a
second binding element, contains a sequence that hybridizes to a second sequence in
the second strand that is between the polymorphic restriction site and the sequence in
the second strand that is complementary to the first primer.
The first binding element is monitored for the presence of the first detectable
0 label and tlle second binding element is monitored for the presence of the second
detectable label. Detection of the first detectable label on the first binding element
and detection of the second detectable label on the second binding element indicates
the absence of the polymorphic restriction site in the nucleic acid, while a failure to
detect the first detectable label on the first binding element and a failure to detect the
5 second detectable label on the second binding element indicates the presence of the
polymorphic restriction site in the nucleic acid.
In addition to the first and second probes described above, this method can
involve the use of a third and fourth probe. The third probe, which is immobilized on
a third binding element, contains a sequence that hybridizes to a third sequence which
2 o is in the first strand and that is between the polymorphic restriction site and the
sequence in the first strand corresponding to the first primer. The fourth probe, which
is immobilized on a fourth binding element, contains a sequence that hybridizes to a
fourth sequence which is in the second strand and that is between the polymorphic
restriction site and the sequence in the second strand corresponding to the second
2 5 primer.
The third or fourth binding elements can be monitored for the presence of the
first or second detectable labels as controls. For example, the third binding element
CA 022667~0 1999-03-17
WO g8/12352 PCT/US97/16467
63
can be monitored for the presence of the first detectable label and the fourth binding
element can be monitored for the presence of the second detectable label.
The first and second, or the first, second, third, and fourth binding elements can
be present as distinct regions on a solid support, such as glass (e.g., a glass plate) or a
5 microchip (e.g., a silicon or a silicon-glass microchip). Embodiments of this method
are illustrated in Figs. 10 and 11.
Example XIV.
In tllis method, the nucleic acid is amplified by PCR using a first and a second10 primer flanki~1g the polymorphic restriction site. The amplification generates a PCR
product containing a first skand containing a sequence corresponding to the first
primer and a second strand containing a sequence corresponding to the second primer.
The PCR product is treated with a restriction endonuclease corresponding to
the polymolphic restriction site to generate a digestion product, which is denatured to
generate a denatured product. The denatured product is contacted with an
oligonucleotide to generate a first reaction product. The oligonucleotide contains a 3'
portion that hybridizes to a first region in the first strand that flanks the polymorphic
restriction site on the side of the polymorphic restriction site containing a sequence
corresponding to the first primer. The 3' end of the oligonucleotide is blocked by, e.g.,
2 o a di-deoxynucleotide, so that it cannot serve as a primer for DNA polymerase. The
oligonucleotide contains a 5' portion that does not hybridize to a second region in the
first strand that flanks the polymorphic restriction site on the side of the polymorphic
restriction site containing a sequence that is complementary to the second primer. The
use of such an oligonucleotide to label a cleaved end of a CAPS marker is illustrated
25 inFig.12.
As illustrated in Fig. 12., the first reaction product is treated with a DNA
polymerase to extend the unblocked, primed 3' end to generate a second reaction
product, which is amplified by PCR using the first primer, tagged with a first
CA 022667~0 1999-03-17
WO g8/12352 ' rcT/uss7/l6467
64
detectable label, and a third primer, which hybridizes to a sequence that is
complementary to the 5' portion of the oligonucleotide, to generate a second PCRproduct. The third primer is tagged with a second detectable label. In this method, the
first and second detectable labels can be identical or distinct.
The second PCR product is denatured to generate a second denatured product,
which is contacted with a first and a second probe. The first probe, which is
immobilized on a first binding element, contains a sequence that hybridizes to a first
sequence in the second strand that is between the polymorphic restriction site and the
sequence in the second strand that is complementary to the first primer. The second
o probe, which is immobilized on a second binding element, contains a sequence that
hybridizes to a second sequence in the first strand that is between the polymorphic
restriction site and the sequence in the first strand that is complementary to the second
primer.
The first binding element is monitored for the presence of the second detectablelabel and the second binding element is monitored for the presence of the first
detectable label. Detection of the second detectable label on the first binding element
and detection of the first detectable label on the second binding element indicates a
heterozygote, detection of the second detectable label on the first binding element and
a failure to detect the first detectable label on the second binding element indicates a
2 o homozygote containing the polymorphic restriction site, and detection of the first
detectable label on the second binding element and a failure to detect the second
detectable label on the first binding element indicates a homozygote lacking thepolymorphic restriction site.
In addition to the first and second probes described above, this method can
2 5 employ a third or a fourth probe. The third probe, which is immobilized on a third
binding element, contains a sequence that hybridizes to a third sequence in the first
strand that is between the polymorphic restriction site and the sequence in the first
strand corresyonding to the first primer. The fourth probe, which is immobilized on a
CA 022667~0 1999-03-17
W O 98/12352 PCTrUS97/16467
fourth binding element, contains a sequence that hybridizes to a fourth sequence in the
second strand that is between the polymorphic restriction site and the sequence in the
second strand corresponding to the second primer.
The third or fourth binding elements can be monitored for the presence of the
5 first or second detectable labels as controls. For example, the third binding element
can be monitored for the presence of the first detectable label.
The first and second, or the first, second, third, and fourth binding elements can
be present as distinct regions on a solid support, such as a glass (e.g., a glass plate) or
silicon (e.g., ~ microchip) support.
This embodiment is illustrated in Fig. l 3 .
Use of oligonucleotides as described above provides several advantages. For
example, because there can be a significant amount of overlap between the
oligonucleotide and the cleaved product, highly stringent conditions can be used in the
annealing reaction, leading to increased specificity. In addition, the 5' end of the
oligonucleotide can be the same for many CAPS markers, as it is by design not
homologous to any amplified sequences corresponding to a CAPS marker for an
org~ni~m of interest. These advantages also apply to other methods employing such
an oligonucleotide, as are described below.
Other Embodiments
2 o The above examples are, therefore, to be construed as merely illustrative, and
not limitative of the remainder of the disclosure in any way whatsoever.
From the above description, one skilled in the art can easily ascertain the
essential characteristics of the present invention, and without departing form the spirit
and scope thereof, can make various changes and modifications of the invention to
2 5 adapt it to various usages and conditions. All publications cited herein are fully
incorporated by reference herein in their entirety. Other embodiments are in theclaims set forth below.
...... ~, . ....