Note: Descriptions are shown in the official language in which they were submitted.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
SENSE INTRON INHIBITION OF STARCH BRANCHING ENZYME EXPRESSION
The present invention relates to a method of inhibiting gene expression,
particularly
inhibiting gene expression in a plant. The present invention also relates to a
nucleotide
sequence useful in .the method. In addition, the present invention relates to
a promoter
that is useful for expressing the nucleotide sequence.
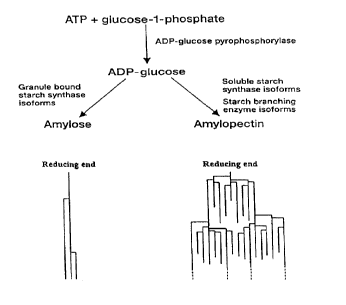
Starch is one of the main storage carbohydrates in plants, especially higher
plants. The
structure of starch consists of amylose and amylopectin. Amylose consists
essentially of
straight chains of a-1-4-linked glycosyl residues. Amylopectin comprises
chains of
a-1-4-linked glycosyl residues with some a-1-6 branches. The branched nature
of
amylopectin is accomplished by the action of inter alia an enzyme commonly
known as
the starch branching enzyme {"SBE"). SBE catalyses the formation of branch
points in
the amylopectin molecule by adding a-1,4 glucans through a-1,6-glucosidic
branching
linkages. The biosynthesis of amylose and amylopectin is schematically shown
in Figure
1, whereas the a-1-4-links and the a-1-6 links are shown in Figure 2.
In Potato, it is known that two classes of SBE exist. In our copending
international patent
applications PCTlEP96103052 and PCT/EP96103053, class B potato SBE and a gene
encoding it are discussed. In international patent application W096/34968,
class A potato
SBE and a cDNA encoding it are disclosed.
It is known that starch is an important raw material. Starch is widely used in
the food,
paper, and chemical industries. However, a large fraction of the starches used
in these
industrial applications are post-harvest modified by chemical, physical or
enzymatic
methods in order to obtain starches with certain required functional
properties.
Within the past few years it has become desirable to make genetically modified
plants
which could be capable of producing modified starches which could be the same
as the
post-harvest modified starches. It is also known that it may be possible to
prepare such
genetically modified plants by expression of antisense nucleotide coding
sequences. In
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
2
this regard, June Bourque provides a detailed summary of antisense strategies
for the
genetic manipulations in plants (Bourque 1995 Plant Science ~5 pp 125-149).
W096/34968 discusses the use of antisense sequences complementary to sequences
which
encode class A and class B potato SBE to downregulate SBE expression in potato
plants.
The sequences used are complementary to SBE coding sequences.
Whilst it is known that enzymatic activity can be affected by expression of
particular
nucleotide sequences (for example see the teachings of Finnegan and McElroy
[1994]
Biotechnology 1? 883-888; and Matzke and Matzke [1995] TIG ~ 1-3) there is
still a
need for a method that can more reliably and/or more efficiently and/or more
specifically
affect enzymatic activity.
According to a first aspect of the present invention there is provided a
method of affecting
I5 enzymatic activity in a plant (or a cell, a tissue or an organ thereof)
comprising
expressing in the plant (or a cell, a tissue or an organ thereof) a nucleotide
sequence
wherein the nucleotide sequence partially or completely codes (is) an intron
of the potato
class A SBE gene in a sense orientation, optionally together with a nucleotide
sequence
which codes, partially or completely, for an intron of a class B starch
branching enzyme
in a sense or antisense orientation; and wherein the nucleotide sequence does
not contain a
sequence that is a sense exon sequence normally associated with the intron.
According to a second aspect of the present invention there is provided a
method of
affecting enzymatic activity in a starch producing organism (or a cell, a
tissue or an organ
thereof) comprising expressing in the starch producing organism (or a cell, a
tissue or an
organ thereof) a nucleotide sequence wherein the nucleotide sequence codes,
partially or
completely, for an intron of the potato class A SBE gene in a sense
orientation optionally
together with a nucleotide sequence which codes, partially or completely, for
an intron of
a class B starch branching enzyme in a sense or antisense orientation; wherein
the
nucleotide sequence does not contain a sequence that is sense to an exon
sequence
normally associated with the intron; and wherein starch branching enzyme
activity is
,r
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
3
affected and/or the levels of amylopectin are affected and/or the composition
of starch is
changed.
Preferably, the class A SBE gene sense intron construct is used in combination
with a
potato class B SBE gene sense intron construct as defined in PCT/EP96/03053.
However, it may also be used independently thereof, to target class A SBE
alone, or in
combination with other transgenes such as other sense and/or antisense
transgenes, for
example antisense intron transgenes such as from SBE genes, to further
manipulate starch
quality in potato plants.
According to a third aspect of the present invention there is provided a
sequence
comprising the nucleotide sequence shown as SEQ. ID. No. 38 or a variant,
derivative or
homologue thereof.
According to a fourth aspect of the present invention there is provided a
promoter
comprising the sequence shown as SEQ.I.D. No. 14 or a variant, derivative or
homologue thereof.
According to a fifth aspect of the present invention there is provided a
construct capable
of comprising or expressing the present invention.
According to a sixth aspect of the present invention there is provided a
vector comprising
or expressing the present invention.
According to a seventh aspect of the present invention there is provided a
cell, tissue or
organ comprising or expressing the present invention.
According to an eighth aspect of the present invention there is provided a
transgenic
starch producing organism comprising or expressing the present invention.
According to
a ninth aspect of the present invention there is provided a starch obtained
from the present
invention.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
4
A key advantage of the present invention is that it provides a method for
preparing
modified starches that is not dependent on the need for post-harvest
modification of
starches. Thus the method of the present invention obviates the need for the
use of
hazardous chemicals that are normally used in the post-harvest modification of
starches.
In addition, the present invention provides inter alia genetically modified
plants which are
capable of producing modified and/or novel and/or improved starches whose
properties
would satisfy various industrial requirements.
Thus. the present invention provides a method of preparing tailor-made
starches in plants
which could replace the post-harvest modified starches.
Also, the present invention provides a method that enables modified starches
to be
prepared by a method that can have a more beneficial effect on the environment
than the
known post-harvest modification methods which are dependent on the use of
hazardous
chemicals and large quantities of energy.
An other key advantage of the present invention is that it provides a method
that may
more reliably and/or more efficiently andlor more specifically affect
enzymatic activity
when compared to the known methods of affecting enzymatic activity. With
regard to
this advantage of the present invention it is to be noted that there is some
degree of
homology between coding regions of SBEs. However, there is little or no
homology with
the intron sequences of SBEs. Thus, sense intron expression provides a
mechanism to
affect selectively the expression of a particular SBE. This advantageous
aspect could be
used, for example, to reduce or eliminate a particular SBE enzyme and replace
that
enzyme with another enzyme which can be another branching enzyme or even a
recombinant version of the affected enzyme or even a hybrid enzyme which could
for
example comprise part of a SBE enzyme from one source and at least a part of
another
SBE enzyme from another source. This particular feature of the present
invention is
1 ~
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
covered by the combination aspect of the present invention which is discussed
in more
detail later.
Thus the present invention provides a mechanism for selectively affecting SBE
activity.
5 This is in contrast to the prior art methods which are dependent on the use
of for example
antisense exon expression whereby it would not be possible to introduce new
SBE activity
without affecting that activity as well.
In the context of the present invention, class B SBE is synonymous with SBE I:
class A
SBE is synonymous with SBE II. Class A SBE is as defined in W096/34968,
incorporated herein by reference. Preferably, the antisense intron construct
used
comprises intron 1 of class A SBE, which is 2.0 kb in length and is located
starting at
residue 45 of the coding sequence of class A SBE. The boundaries of the intron
may be
calculated by searching for consensus intron boundary sequences, and are shown
in
attached figure 11. The sequence of the intron is set forth in SEQ. ID. No.
38. Class B
SBE is substantially as defined in the sequences given herein and in
PCT/EP96/03053.
Preferably with the first aspect of the present invention starch branching
enzyme activity
is affected and/or wherein the levels of amylopectin are affected andlor the
composition
of starch is changed.
Preferably with the first or second aspect of the present invention the
nucleotide sequence
does not contain a sequence that is sense to an exon sequence.
Preferably with the fourth aspect of the present invention the promoter is in
combination
with a gene of interest ("GOI").
Preferably the enzymatic activity is reduced or eliminated.
Preferably the nucleotide sequence codes for at least substantially all of at
least one intron
in a sense orientation.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
6
Preferably the nucleotide sequence codes, partially or completely, for two or
more introns
and wherein each intron is in a sense orientation.
Preferably the nucleotide sequence comprises at least 350 nucleotides (e.g.
350 bp), more
preferably at least 500 nucleotides (e.g. 500 bp).
Preferably the nucleotide sequence comprises the sequence shown as SEQ. ID.
No. 38, or
a fragment thereof.
Preferably the nucleotide sequence is expressed by a promoter having a
sequence shown
as SEQ. LD. No. 14 or a variant, derivative or homologue thereof.
Preferably the transgenic starch producing organism is a plant.
A preferred aspect of the present invention therefore relates to a method of
affecting
enzymatic activity in a plant (or a cell, a tissue or an organ thereof)
comprising
expressing in the plant (or a cell, a tissue or an organ thereof) a nucleotide
sequence
wherein the nucleotide sequence codes, partially or completely, for a class A
SBE intron
in a sense orientation; wherein the nucleotide sequence does not contain a
sequence that is
sense to an exon sequence normally associated with the intron; and wherein
starch
branching enzyme activity is affected and/or the levels of amylopectin are
affected and/or
the composition of starch is changed.
A more preferred aspect of the present invention therefore relates to a method
of affecting
enzymatic activity in a plant (or a cell, a tissue or an organ thereof)
comprising
expressing in the plant (or a cell, a tissue or an organ thereof) a nucleotide
sequence
wherein the nucleotide sequence codes, partially or completely, for an intron
in a sense
orientation; wherein the nucleotide sequence does not contain a sequence that
is sense to
an exon sequence normally associated with the intron; wherein starch branching
enzyme
activity is affected and/or the levels of amylopectin are affected andlor the
composition of
~, _ .
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
7
starch is changed; and wherein the nucleotide sequence comprises the sequence
shown as
SEQ. ID. No. 38, or fragments thereof.
The term "nucleotide" in relation to the present invention includes DNA and
RNA.
Preferably it means DNA, more preferably DNA prepared by use of recombinant
DNA
techniques.
The term "intron" is used in its normal sense as meaning a segment of
nucleotides,
usually DNA, that does not encode part or all of an expressed protein or
enzyme.
The term "exon" is used in its normal sense as meaning a segment of
nucleotides, usually
DNA, encoding part or all of an expressed protein or enzyme.
Thus, the term "intron" refers to gene regions that are transcribed into RNA
molecules,
but which are spliced out of the RNA before the RNA is translated into a
protein. In
contrast, the term "exon" refers to gene regions that are transcribed into RNA
and
subsequently translated into proteins.
The terms "variant" or "homologue" or "fragment" in relation to the nucleotide
sequence
of the present invention include any substitution of, variation of,
modification of,
replacement of, deletion of or addition of one (or more) nucleic acid from or
to the
respective nucleotide sequence providing the resultant nucleotide sequence can
affect
enzyme activity in a plant, or cell or tissue thereof, preferably wherein the
resultant
nucleotide sequence has at least the same effect as the sequence shown in SEQ.
ID. No.
38. In particular, the term "homologue" covers homology with respect to
similarity of
structure and/or similarity of function providing the resultant nucleotide
sequence has the
ability to affect enzymatic activity in accordance with the present invention.
With respect
to sequence homology (i.e. similarity), preferably there is more than 80%
homology,
more preferably at least 85% homology, more preferably at least 90% homology,
even
more preferably at least 95% homology, more preferably at least 98% homology.
The
above terms are also synonymous with allelic variations of the sequences.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
8
Likewise, the terms "variant" or "homologue" or "fragment" in relation to the
promoter
of the present invention include any substitution of, variation of,
modification of,
replacement of, deletion of or addition of one (or more) nucleic acid from or
to the
respective promoter sequence providing the resultant promoter sequence allows
expression of a GOI, preferably wherein the resultant promoter sequence has at
least the
same effect as SEQ.I.D. No. 14. In particular, the term "homologue" covers
homology
with respect to similarity of structure and/or similarity of function
providing the resultant
promoter sequence has the ability to allow for expression of a GOI, such as a
nucleotide
sequence according to the present invention. With respect to sequence homology
(i.e.
similarity), preferably there is more than 80% homology, more preferably at
least 85%
homology, more preferably at least 90% homology, even more preferably at least
95°~°
homology, more preferably at least 98% homology. The above terms are also
synonymous with allelic variations of the sequences.
The intron sequence of the present invention can be any one or all of the
intron sequences
of the present invention, including partial sequences thereof, provided that
if partial sense
sequences are used the partial sequences affect enzymatic activity. Suitable
examples of
partial sequences include sequences that are shorter than any one of the full
sense
sequences shown as SEQ. ID. No. 38 but which comprise nucleotides that are
adjacent
the respective exon or exons.
With regard to the second aspect of the present invention (i.e. specifically
affecting SBE
activity), the nucleotide sequences of the present invention may comprise one
or more
sense or antisense exon sequences of the class A or class B SBE gene (but not
sense exon
sequences naturally associated with the intron sequence), including complete
or partial
sequences thereof, providing the nucleotide sequences can affect SBE activity,
preferably
wherein the nucleotide sequences reduce or eliminate SBE activity. Preferably,
the
nucleotide sequence of the second aspect of the present invention does not
comprise sense
exon sequences.
, . .. .. ~, ~.. . .".... .. . ...... .....
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
9
The term "vector" includes an expression vector and a transformation vector.
The term
"expression vector" means a construct capable of in vivo or in vitro
expression. The term
"transformation vector" means a construct capable of being transferred from
one species
to another - such as from an E. Coli plasmid to a fungus or a plant cell, or
from an
Agrobactericcm to a plant cell.
The term "construct" - which is synonymous with terms such as "conjugate",
"cassette"
and "hybrid" - in relation to the sense nucleotide sequence aspect of the
present invention
includes the nucleotide sequence according to the present invention directly
or indirectly
attached to a promoter. An example of an indirect attachment is the provision
of a
suitable spacer group such as an intron sequence, such as the Shl-intron or
the ADH
intron, intermediate the promoter and the nucleotide sequence of the present
invention.
The same is true for the term "fused" in relation to the present invention
which includes
direct or indirect attachment. The terms do not cover the natural combination
of the wild
type SBE gene when associated with the wild type SBE gene promoter in their
natural
environment.
The construct may even contain or express a marker which allows for the
selection of the
genetic construct in, for example, a plant cell into which it has been
transferred. Various
markers exist which may be used in, for example, plants - such as mannose.
Other
examples of markers include those that provide for antibiotic resistance -
e.g. resistance to
6418, hygromycin, bleomycin, kanamycin and gentamycin.
The construct of the present invention preferably comprises a promoter. The
term
"promoter" is used in the normal sense of the art, e.g. an RNA polymerase
binding site in
the Jacob-Monod theory of gene expression. Examples of suitable promoters are
those
that can direct efficient expression of the nucleotide sequence of the present
invention
and/or in a specific type of cell. Some examples of tissue specific promoters
are
disclosed in WO 92/11375.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
The promoter could additionally include conserved regions such as a Pribnow
Box or a
TATA box. The promoters may even contain other sequences to affect {such as to
maintain, enhance, decrease) the levels of expression of the nucleotide
sequence of the
present invention. Suitable examples of such sequences include the Shl-intron
or an
5 ADH intron. Other sequences include inducible elements - such as
temperature,
chemical, light or stress inducible elements. Also, suitable elements to
enhance
transcription or translation may be present. An example of the latter element
is the TMV
S' leader sequence (see Sleat Gene 217 [1987] 217-225; and Dawson Plant Mol.
Biol. 23
[1993] 97).
As mentioned, the construct and/or the vector of the present invention may
include a
transcriptional initiation region which may provide for regulated or
constitutive
expression. Any suitable promoter may be used for the transcriptional
initiation region,
such as a tissue specific promoter. In one aspect, preferably the promoter is
the patatin
promoter or the E35S promoter. In another aspect, preferably the promoter is
the SBE
promoter.
If, for example, the organism is a plant then the promoter can be one that
affects
expression of the nucleotide sequence in any one or more of seed, tuber, stem,
sprout,
root and leaf tissues, preferably tuber. By way of example, the promoter for
the
nucleotide sequence of the present invention can be the a-Amy 1 promoter
{otherwise
known as the Amy 1 promoter, the Amy 637 promoter or the a-Amy 637 promoter)
as
described in our co-pending UK patent application No. 9421292.5 filed 21
October 1994.
Alternatively, the promoter for the nucleotide sequence of the present
invention can be the
a-Amy 3 promoter (otherwise known as the Amy 3 promoter, the Amy 351 promoter
or
the a-Amy 351 promoter) as described in our co-pending UK patent application
No.
9421286.7 filed 21 October 1994.
The present invention also encompasses the use of a promoter to express a
nucleotide
sequence according to the present invention, wherein a part of the promoter is
inactivated
but wherein the promoter can still function as a promoter. Partial
inactivation of a
t . . r __. _ ._._.~._~.._. _ .. . _ _.....
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98100295
11
promoter in some instances is advantageous. In particular, with the Amy 351
promoter
mentioned earlier it is possible to inactivate a part of it so that the
partially inactivated
promoter expresses the nucleotide sequence of the present invention in a more
specific
manner such as in just one specific tissue type or organ. The term
"inactivated" means
partial inactivation in the sense that the expression pattern of the promoter
is modified but
wherein the partially inactivated promoter still functions as a promoter.
However, as
mentioned above, the modified promoter is capable of expressing a gene coding
for the
enzyme of the present invention in at least one (but not all) specific tissue
of the original
promoter. Examples of partial inactivation include altering the folding
pattern of the
promoter sequence, or binding species to parts of the nucleotide sequence, so
that a part
of the nucleotide sequence is not recognised by, for example, RNA polymerase.
Another, and preferable, way of partially inactivating the promoter is to
truncate it to
form fragments thereof. Another way would be to mutate at least a part of the
sequence
so that the RNA polymerase can not bind to that part or another part. Another
modification is to mutate the bindine sites for regulatory proteins for
example the CreA
protein known from filamentous fungi to exert carbon catabolite repression,
and thus
abolish the cataliolite repression of the native promoter.
The construct and/or the vector of the present invention may include a
transcriptional
termination region.
The nucleotide according to the present invention can be expressed in
combination (but
not necessarily at the same time) with an additional construct. Thus the
present invention
also provides a combination of constructs comprising a first construct
comprising the
nucleotide sequence according to the present invention operatively linked to a
first
promoter; and a second construct comprising a GOI operatively linked to a
second
promoter (which need not be the same as the first promoter). With this aspect
of the
present invention the combination of constructs may be present in the same
vector,
plasmid, cells, tissue, organ or organism. This aspect of the present
invention also covers
methods of expressing the same, preferably in specific cells or tissues, such
as expression
in just a specific cell or tissue, of an organism, typically a plant. With
this aspect of the
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
12
present invention the second construct does not cover the natural combination
of the gene
coding for an enzyme ordinarily associated with the wild type gene promoter
when they
are both in their natural environment.
An example of a suitable combination would be a first construct comprising the
nucleotide
sequence of the present invention and a promoter, such as the promoter of the
present
invention, and a second construct comprising a promoter, such as the promoter
of the
present invention, and a GOI wherein the G01 codes for another starch
branching enzyme
either in sense or antisense orientation.
The above comments relating to the term "construct" for the sense nucleotide
aspect of
the present invention are equally applicable to the term "construct" for the
promoter
aspect of the present invention. In this regard, the term includes the
promoter according
to the present invention directly or indirectly attached to a GOI.
The term "GOI" with reference to the promoter aspect of the present invention
or the
combination aspect of the present invention means any gene of interest, which
need not
necessarily code for a protein or an enzyme - as is explained later. A GOI can
be any
nucleotide sequence that is either foreign or natural to the organism in
question, for
example a plant.
Typical examples of a GOI include genes encoding for other proteins or enzymes
that
modify metabolic and catabolic processes. The GOI may code for an agent for
introducing or increasing pathogen resistance.
The GOI may even be an antisense construct for modifying the expression of
natural
transcripts present in the relevant tissues. An example of such a GOI is the
nucleotide
sequence according to the present invention.
r ~
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
13
The GOI may even code for a protein that is non-natural to the host organism -
e.g. a
plant. The GOI may code for a compound that is of benefit to animals or
humans. For
example, the GOI could code for a pharmaceutically active protein or enzyme
such as any
one of the therapeutic compounds insulin, interferon, human serum albumin,
human
growth factor and blood clotting factors. The GOI may even code for a protein
giving
additional nutritional value to a food or feed or crop. Typical examples
include plant
proteins that can inhibit the formation of anti-nutritive factors and plant
proteins that have
a more desirable amino acid composition (e.g. a higher lysine content than a
non-
transgenic plant). The GOI may even code for an enzyme that can be used in
food
processing such as xylanases and a-galactosidase. The GOI can be a gene
encoding for
any one of a pest toxin, an antisense transcript such as that for a-amylase, a
protease or a
glucanase. Alternatively, the GOI can be a nucleotide sequence according to
the present
invention.
The GOI can be the nucleotide sequence coding for the arabinofuranosidase
enzyme
which is the subject of our co-pending UK patent application 9505479.7. The
GOI can be
the nucleotide sequence coding for the glucanase enzyme which is the subject
of our co-
pending UK patent application 9505475.5. The GOI can be the nucleotide
sequence
coding for the a-amylase enzyme which is the subject of our co-pending UK
patent
application 9413439.2. The GOI can be the nucleotide sequence coding for the a-
amylase enzyme which is the subject of our co-pending UK patent application
9421290.9.
The GOI can be any of the nucleotide sequences coding for the a-glucan lyase
enzyme
which are described in our co-pending PCT patent application PCT/EP94/03397.
In one aspect the GOI can even be a nucleotide sequence according to the
present
invention but when operatively linked to a different promoter.
The GOI could include a sequence that codes for one or more of a xylanase, an
arabinase,
an acetyl esterase, a rhamnogalacturonase, a glucanase, a pectinase, a
branching enzyme
or another carbohydrate modifying enzyme or proteinase. Alternatively, the GOI
may be
a sequence that is antisense to any of those sequences.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
14
As mentioned above, the present invention provides a mechanism for selectively
affecting
a particular enzymatic activity.
In an important application of the present invention it is now possible to
reduce or
eliminate expression of a genomic nucleotide sequence coding for a genomic
protein or
enzyme by expressing a sense intron construct for that particular genomic
protein or
enzyme and (e.g. at the same time) expressing a recombinant version of that
enzyme or
protein - in other words the GOI is a recombinant nucleotide sequence coding
for the
genomic enzyme or protein. This application allows expression of desired
recombinant
enzymes and proteins in the absence of (or reduced levels of) respective
genomic enzymes
and proteins. Thus the desired recombinant enzymes and proteins can be easily
separated
and purified from the host organism. This particular aspect of the present
mventton ~s
very advantageous over the prior art methods which, for example, rely on the
use of anti-
sense exon expression which methods also affect expression of the recombinant
enzyme.
Thus, a further aspect of the present invention relates to a method of
expressing a
recombinant protein or enzyme in a host organism comprising expressing a
nucleotide
sequence coding for the recombinant protein or enzyme; and expressing a
further
nucleotide sequence wherein the further nucleotide sequence codes, partially
or
completely, for an intron in a sense orientation; wherein the intron is an
intron normally
associated with the genomic gene encoding a protein or an enzyme corresponding
to the
recombinant protein or enzyme; and wherein the further nucleotide sequence
does not
contain a sequence that is sense to an exon sequence normally associated with
the intron.
Additional aspects cover the combination of those nucleotide sequences
including their
incorporation in constructs, vectors, cells, tissues and transgenic organisms.
Therefore the present invention also relates to a combination of nucleotide
sequences
comprising a first nucleotide sequence coding for a recombinant enzyme; and a
second
nucleotide sequence which corresponds to an intron in a sense orientation;
wherein the
intron is an intron that is associated with a genomic gene encoding the enzyme
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
corresponding to the recombinant enzyme; and wherein the second nucleotide
sequence
does not contain a sequence that is sense to an exon sequence normally
associated with the
mtron.
5 The GOI may even code for one or more introns but in an antisense
orientation, such as
any one or more of the antisense intron sequences presented in the attached
sequence
listings. For example, the present invention also covers the expression of for
example a
sense intron (e.g. SEQ.I.D.No. 38) in combination with for example an
antisense intron
which preferably is not complementary to the sense intron sequence (e.~.
SEQ.I.D.No.
10 16).
The terms "cell", "tissue" and "organ" include cell, tissue and organ per se
and when
within an organism.
IS The term "organism" in relation to the present invention includes any
organism that could
comprise the nucleotide sequence according to the present invention and/or
wherein the
nucleotide sequence according to the present invention can be expressed when
present in
the organism. Preferably the organism is a starch producing organism such as
any one of
a plant, algae, fungi, yeast and bacteria, as well as cell lines thereof.
Preferably the
organism is a plant.
The term "starch producing organism" includes any organism that can
biosynthesise
starch. Preferably, the starch producing organism is a plant.
The term "plant" as used herein includes any suitable angiosperm, gymnosperm,
monocotyledon and dicotyledon. Typical examples of suitable plants include
vegetables
such as potatoes; cereals such as wheat, maize, and barley; fruit; trees;
flowers; and other
plant crops. Preferably, the term means "potato" .
The term "transgenic organism" in relation to the present invention includes
any organism
that comprises the nucleotide sequence according to the present invention
and/or products
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
16
obtained therefrom, andlor wherein the nucleotide sequence according to the
present
invention can be expressed within the organism. Preferably the nucleotide
sequence of
the present invention is incorporated in the genome of the organism.
Preferably the
transgenic organism is a plant, more preferably a potato.
To prepare the host organism one can use prokaryotic or eukaryotic organisms.
Examples of suitable prokaryotic hosts include E. coli and Bacillces
scebtilis. Teachings on
the transformation of prokaryotic hosts is well documented in the art, for
example see
Sambrook et al (Sambrook et al. in Molecular Cloning: A Laboratory Manual, 2nd
edition, 1989, Cold Spring Harbor Laboratory Press).
Even though the enzyme according to the present invention and the nucleotide
sequence
coding for same are not disclosed in EP-B-0470145 and CA-A-2006454, those two
documents do provide some useful background commentary on the types of
techniques
that may be employed to prepare transgenic plants according to the present
invention.
Some of these background teachings are now included in the following
commentary.
The basic principle in the construction of genetically modified plants is to
insert genetic
information in the plant genome so as to obtain a stable maintenance of the
inserted
~enetic material.
Several techniques exist for inserting the genetic information, the two main
principles
being direct introduction of the genetic information and introduction of the
genetic
information by use of a vector system. A review of the general techniques may
be found
in articles by Potrykus (Annu Rev Plant Physiol Plant Mol Biol [1991] 42:205-
225) and
Christou (Agro-Food-Industry Hi-Tech March/April 1994 17-27).
Thus, in one aspect, the present invention relates to a vector system which
carries a
nucleotide sequence or construct according to the present invention and which
is capable
of introducing the nucleotide sequence or construct into the genome of an
organism, such
as a plant.
rt
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
17
The vector system may comprise one vector, but it can comprise two vectors. In
the case
of two vectors, the vector system is normally referred to as a binary vector
system.
Binary vector systems are described in further detail in Gynheung An et al.
(1980),
Binary Vectors, Plant Molecular Biology Manual A3, 1-19.
One extensively employed system for transformation of plant cells with a given
promoter
or nucleotide sequence or construct is based on the use of a Ti plasmid from
Agrobacterium turnefaciens or a Ri plasmid from Agrobacteritun rhizogenes An
et al.
(1986), Plant Physiol. 81, 301-305 and Butcher D.N. et al. (1980), Tissue
Culture
Methods for Plant Pathologists, eds.: D.S. Ingrams and J.P. Helgeson. 203-208.
Several
different Ti and Ri plasmids have been constructed which are suitable for the
construction
of the plant or plant cell constructs described above. A non-limiting example
of such a Ti
plasmid is pGV3850.
The nucleotide sequence or construct of the present invention should
preferably be
inserted into the Ti-plasmid between the terminal sequences of the T-DNA or
adjacent a
T-DNA sequence so as to avoid disruption of the sequences immediately
surrounding the
T-DNA borders, as at least one of these regions appears to be essential for
insertion of
modified T-DNA into the plant genome.
As will be understood from the above explanation, if the organism is a plant
the vector
system of the present invention is preferably one which contains the sequences
necessary
to infect the plant (e.g. the vir region) and at least one border part of a T-
DNA sequence,
the border part being located on the same vector as the genetic construct.
Furthermore, the vector system is preferably an Agrobacterium turnefaciens Ti-
plasmid or
an Agrobacteritcrn rltizogenes Ri-plasmid or a derivative thereof. As these
plasmids are
well-known and widely employed in the construction of transgenic plants, many
vector
systems exist which are based on these plasmids or derivatives thereof.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98l00295
18
In the construction of a transgenic plant the nucleotide sequence or construct
of the
present invention may be first constructed in a microorganism in which the
vector can
replicate and which is easy to manipulate before insertion into the plant. An
example of a
useful microorganism is E. coli, but other microorganisms having the above
properties
may be used. When a vector of a vector system as defined above has been
constructed in
E. coli, it is transferred, if necessary, into a suitable Agrobactericcm
strain, e.~.
Agrobacterium tcemefaciens. The Ti-plasmid harbouring the nucleotide sequence
or
construct of the present invention is thus preferably transferred into a
suitable
Agrobacteritcm strain, e.g. A. tumefaciens, so as to obtain an Agrobacterium
cel!
harbouring the promoter or nucleotide sequence or construct of the present
invention,
which DNA is subsequently transferred into the plant cell to be modified.
If, for example, for the transformation the Ti- or Ri-plasmid of the plant
cells is used, at
least the right boundary and often however the right and the left boundary of
the Ti- and
Ri-plasmid T-DNA, as flanking areas of the introduced genes, can be connected.
The use
of T-DNA for the transformation of plant cells has been intensively studied
and is
described in EP-A-120516; Hoekema, in: The Binary Plant Vector System Offset-
drukkerij Kanters B.B., Alblasserdam, 1985, Chapter V; Fraiey, et al., Crit.
Rev. Plant
Sci., 4:1-46; and An et al., EMBO J. (1985) 4:277-284.
Direct infection of plant tissues by Agrobacterictm is a simple technique
which has been
widely employed and which is described in Butcher D.N. et al. (1980), Tissue
Culture
Methods for Plant Pathologists, eds.: D.S. Ingrams and J.P. Helgeson, 203-208.
For
further teachings on this topic see Potrykus (Annu Rev Plant Physiol Plant Mol
Biol
[1991] 42:205-225) and Christou (Agro-Food-Industry Hi-Tech March/April 1994
17-
27). With this technique, infection of a plant may be performed in or on a
certain part or
tissue of the plant, i.e. on a part of a leaf, a root, a stem or another pan
of the plant.
Typically, with direct infection of plant tissues by Agrobacterium carrying
the GOI (such
as the nucleotide sequence according to the present invention) and,
optionally, a
promoter, a plant to be infected is wounded, e.g. by cutting the plant with a
razor blade
.. , *_ ,
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
19
or puncturing the plant with a needle or rubbing the plant with an abrasive.
The wound is
then inoculated with the Agrobacterium. The inoculated plant or plant part is
then grown
on a suitable culture medium and allowed to develop into mature plants.
When plant cells are constructed, these cells may be grown and maintained in
accordance
with well-known tissue culturing methods such as by culturing the cells in a
suitable
culture medium supplied with the necessary growth factors such as amino acids,
plant
hormones, vitamins, etc.
Regeneration of the transformed cells into genetically modified plants may be
accomplished using known methods for the regeneration of plants from cell or
tissue
cultures, for example by selecting transformed shoots using an antibiotic and
by
subculturing the shoots on a medium containing the appropriate nutrients,
plant
hormones, etc.
Further teachings on plant transformation may be found in EP-A-0449375.
As reported in CA-A-2006454, a large amount of cloning vectors are available
which
contain a replication system in E. coli and a marker which allows a selection
of the
transformed cells. The vectors contain for example pBR 322, pUC series, M13 mp
series, pACYC 184 etc. In this way, the nucleotide or construct of the present
invention
can be introduced into a suitable restriction position in the vector. The
contained plasmid
is then used for the transformation in E.coli. The E.coli cells are cultivated
in a suitable
nutrient medium and then harvested and lysed. The plasmid is then recovered.
As a
method of analysis there is generally used sequence analysis, restriction
analysis,
electrophoresis and further biochemical-molecular biological methods. After
each
manipulation, the used DNA sequence can be restricted and connected with the
next DNA
sequence. Each sequence can be cloned in the same or different plasmid.
After the introduction of the nucleotide sequence or construct according to
the present
invention in the plants the presence and/or insertion of further DNA sequences
may be
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
necessary - such as to create combination systems as outlined above (e.g. an
organism
comprising a combination of constructs).
The above commentary for the transformation of prokaryotic organisms and
plants with
5 the nucleotide sequence of the present invention is equally applicable for
the
transformation of those organisms with the promoter of the present invention.
In summation, the present invention relates to affecting enzyme activity by
expressing
sense ~ntron sequences.
Also, the present invention relates to a promoter useful for the expression of
those sense
mtron sequences.
The following samples have been deposited in accordance with the Budapest
Treaty at the
recognised depositary The National Collections of Industrial and Marine
Bacteria Limited
(NCIMB) at 23 St Machar Drive, Aberdeen, Scotland, AB2 1RY, United Kingdom, on
13 July 1995:
NCIMB 40754 (which refers to pBEA 11 as described herein);
NCIMB 40751 (which refers to ~.-SBE 3.2 as described herein), and
NCIMB 40752 (which refers to i,.-SBE 3.4 as described herein).
2~ A highly preferred embodiment of the present invention therefore relates to
a method of
affecting enzymatic activity in a plant (or a cell, a tissue or an organ
thereof) comprising
expressing in the plant (or a cell, a tissue or an organ thereof) a nucleotide
sequence
wherein the nucleotide sequence codes, partially or completely, for an intron
in a sense
orientation; wherein the nucleotide sequence does not contain a sequence that
is sense to
an exon sequence normally associated with the intron; wherein starch branching
enzyme
activity is affected and/or the levels of amylopectin are affected and/or the
composition of
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
21
starch is changed; and wherein the intron nucleotide sequence is the sequence
of intron 1
of class A SBE as set forth in SEQ. ID. No. 38, or any other intron of class A
SBE,
including fragments thereof, and including combinations of class A sense
intron sequences
and class B sense or antisense intron sequences. The sequence of introns of
class A SBE
other than intron 1 may be obtained by sequencing of, for example, potato
class A SBE
genomic DNA, isolatable by hybridisation screening of a genomic DNA library
with class
A SBE cDNA obtainable according to W096/34968 according to methods well known
in
the art and set forth, for example, in Sambrook et al. , Molecular Cloning: A
Laboratory
Manual, Cold Spring Harbor, 1989.
The present invention will now be described only by way of example, in which
reference
is made to the following attached Figures:
Figure 1, which is a schematic representation of the biosynthesis of amylose
and
amylopectin;
Figure 2, which is a diagrammatic representation of the a-1-4-links and the a-
1-6 links of
arnylopectin;
Figure 3, which is a diagrammatic representation of the exon-intron structure
of a
genomic SBE clone;
Figure 4, which is a plasmid map of pPATAl, which is 3936 by in size;
Figure 5, which is a plasmid map of pABE7, which is 5106 by in size;
Figure 6, which is a plasmid map of pVictorIV Man, which is 7080 by in size;
Figure 7, which is a plasmid map of pBEAI l, which is 9.54 kb in size;
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
22
Figure 8, which shows the full genomic nucleotide sequence for SBE including
the
promoter, exons and introns;
Figure 9, which is a plasmid map of pVictor5a, which is 9.12 kb in size;
Figure 10, which is a plasmid map of pBEP2, which is 10.32 kb in size;
Figure 11, which shows the positioning of intron 1 in the class A and class B
SBE genes;
Figure 12, which shows the sequence of intron 1 of the potato class A SBE;
Figure 13, which shows pSSlS; and
Figure 14, which shows pSSl6.
Figures 1 and 2 were referred to above in the introductory description
concerning starch
in general. As mentioned, Figure 3 is a diagrammatic representation of the
exon-intron
structure of a genomic SBE clone, the sequence of which is shown in Figure 8.
This
clone, which has about 11.5 k base pairs, comprises 14 exons and 13 introns.
The
introns are numbered in increasing order from the 5' end to the 3' end and
correspond to
SEQ.I.D.No.s 1-13, respectively. Their respective antisense intron sequences
are shown
as SEQ.LD.No.s 15-27.
In more detail, Figures 3 and 8 present information on the 11468 base pairs of
a potato
SBE gene. The 5' region from nucleotides 1 to 2082 contain the promoter region
of the
SBE gene. A TATA box candidate at nucleotide 2048 to 2051 is boxed. The
homology
between a potato SBE cDNA clone (Poulsen & Kreiberg (1993) Plant Physiol 102:
1053-
1054) and the exon DNAs begin at 2083 by and end at 9666 bp. The homology
between
the cDNA and the exon DNA is indicated by nucleotides in upper case letters,
while the
translated amino acid sequences are shown in the single letter code below the
exon DNA.
Intron sequences are indicated by lower case letters.
n ....
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
23
Figure 7 is a plasmid map of pBEA7, which is 9.54 k base pairs in size.
Plasmid pBEA
11 comprises the first intron sequence of the potato SBE gene. This first
intron sequence,
which has 1177 base pairs, is shown in Figure 3 and lies between the first
exon and the
second exon.
These experiments and aspects of the present invention are now discussed in
more detail.
EXPERIMENTAL PROTOCOL
ISOLATION, SUBCLONING IN PLASMIDS, AND SEQUENCING OF GENOMIC
SBE CLONES
Various clones containing the potato SBE gene are isolated from a Desiree
potato
genomic library (Clontech Laboratories Inc., Palo Alto CA, USA) using
radioactively
labelled potato SBE cDNA (Poulsen & Kreiberg (1993) Plant Physiol. 102:1053-
1054) as
probe. The fragments of the isolated ?~-phages containing SBE DNA (~.SBE 3.2 -
NCIMB 40751 - and ~.SBE-3.4 - NCIMB 40752) are identified by Southern analysis
and
then subcloned into pBluescript II vectors (Clontech Laboratories Inc., Palo
Alto CA,
USA). ?~SBE 3.2 contains a 15 kb potato DNA insert and ~.SBE-3.4 contains a 13
kb
potato DNA insert. The resultant plasmids are called pGB3, pGBll, pGBlS, pGBl6
and
pGB25 (see discussion below). The respective inserts are then sequenced using
the
Pharmacia Autoread Sequencing Kit (Pharmacia, Uppsala) and a A.L.F. DNA
sequencer
(Pharmacia, Uppsala).
In total, a stretch of 11.5 kb of the SBE gene is sequenced. The sequence is
deduced from the above-mentioned plasmids, wherein: pGB25 contains the
sequences
from 1 by to 836 bp, pGBlS contains the sequences from 735 by to 2580 bp,
pGBl6
contains the sequences from 2580 by to 5093 bp, pGBll contains the sequences
from
3348 by to 7975 bp, and pGB3 contains the sequences from 7533 by to 11468 bp.
In more detail, pGB3 is constructed by insertion of a 4 kb EcoRI fragment
isolated
from ~.SBE 3.2 into the EcoRI site of pBluescript II SK (+). pGBll is
constructed by
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
24
insertion of a 4.7 kb XhoI fragment isolated from ~.SBE 3.4 into the XhoI site
of
pBluescript II SK (+). pGBIS is constructed by insertion of a 1.7 kb SpeI
fragment
isolated from ~.SBE 3.4 into the SpeI site of pBluescript II SK (+). pGBl6 is
constructed
by insertion of a 2.5 kb SpeI fragment isolated from ~,SBE 3.4 into the SpeI
site of
pBluescript II SK (+). For the construction of pGB25 a PCR fragment is
produced with
the primers
5' GGA ATT CCA GTC GCA GTC TAC ATT AC 3'
(SEQ. ID. No. 30)
and
5' CGG GAT CCA GAG GCA TTA AGA TTT CTG G 3'
(SEQ. ID. No. 31)
and J~SBE 3.4 as a template.
The PCR fragment is digested with BamHI and EcoRI, and inserted in pBluescript
II SK (+) digested with the same restriction enzymes.
CONSTRUCTION OF PLASMID pBEAll
The SBE intron 1 is amplified by PCR using the oligonucleotides
5' CGG GAT CCA AAG AAA TTC TCG AGG TTA CAT GG 3'
(SEQ. ID. No. 32)
and
5' CGG GAT CCG GGG TAA TTT TTA CTA ATT TCA TG 3'
(SEQ. ID. No. 33)
and the ?~SBE 3.4 phage containing the SBE gene as template.
The PCR product is digested with BamHI and inserted in a sense orientation in
the
BamHI site of plasmid pPATAl (described in WO 94/24292) between the patatin
promoter and the 35S terminator. This construction, pABE7, is digested with
KpnI, and
the 2.4 kb "patatin promoter-SBE intron 1- 35S terminator" KpnI fragment is
isolated and
inserted in the KpnI site of the plant transformation vector pVictorIV Man
yielding
plasmid pBEAll.
CONSTRUCTION OF PLASMID pSSlS.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
The 2122 by intron 1 sequence of the potato SBEII gene (see SEQ. ID. No. 38)
is amplified by PCR from a genomic SBEII subclone using the primers 5' - CGG
GAT
CCC GTA TGT CTC ACT GTG TTT GTG GC - 3' (SEQ. ID. No. 34) and 5' - CGG
GAT CCC CCT ACA TAC ATA TAT CAG ATT AG - 3' (SEQ. ID. No. 35). The
5 PCR product is digested with BamHI and inserted in sense orientation after a
patatin
promoter in the BamHI site of a plant transformation vector in which the NPTII
gene is
used as selectable marker (see figure 13).
CONSTRUCTION OF PLASMID pSSlG.
10 The 2122 by intron 1 sequence of the potato SBEII gene (SEQ. ID. No. 38) is
amplified by PCR from a genomic SBEII subclone using the primers ~' - CGG GAT
CCC GTA TGT CTC ACT GTG TTT GTG GC - 3' (SEQ. ID. No. 34) and 5' - CGG
GAT CCC CCT ACA TAC ATA TAT CAG ATT AG - 3' (SEQ. ID. No. 35). The
PCR product is digested with BamHI and inserted in sense orientation after a
patatin
15 promoter in the BamHI site of a plant transformation vector in which the
manA gene is
used as selectable marker {see figure 14).
PRODUCTION OF TRANSGENIC POTATO PLANTS
~xenic stock cultures
20 Shoot cultures of Solanum tLCberosum 'Bintje' and 'Dianelia' are maintained
on a
substrate (LS) of a formula according to Linsmaier, E.U. and Skoog, F. (1965),
Physiol.
Plant. 18: 100-127, in addition containing 2 pM silver thiosulphate at
25°C and 16 h
light/8 h dark.
The cultures are subcultured after approximately 40 days. Leaves are then cut
off
25 the shoots and cut into nodal segments (approximately 0.8 cm) each
containing one node.
Inoculation of o~ tatO~issues
Shoots from approximately 40 days old shoot cultures (height approximately 5-6
cms) are cut into internodal segments (approximately 0.8 cm). The segments are
placed
into liquid LS-substrate containing the transformed Agrobactericcm
tccmefaciens containing
the binary vector of interest. The Agrobacterium are grown overnight in YMB-
substrate
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
26
(di-potassium hydrogen phosphate, trihydrate (0.66 g/1); magnesium sulphate,
heptahydrate (0.20 g/1); sodium chloride (0.10 g/1); mannitol (10.0 g/1); and
yeast extract
(0.40 g/1)) containing appropriate antibiotics (corresponding to the
resistance gene of the
Agrobacterium strain) to an optical density at 660 nm (OD-660) of
approximately 0.8,
centrifuged and resuspended in the LS-substrate to an OD-660 of 0.5.
The segments are left in the suspension of Agrobacterium for 30 minutes and
then
the excess of bacteria are removed by blotting the segments on sterile filter
paper.
Co-cultivation
The shoot segments are co-cultured with bacteria for 48 hours directly on LS-
substrate containing agar (8.0 g/1), 2,4-dichlorophenoxyacetic acid (2.0 mg/i)
and trans-
zeatin (0.5 mg/1). The substrate and also the explants are covered with
sterile filter
papers, and the petri dishes are placed at 25°C and 16 h light/ 8 dark.
"Washing" procedure
After the 48 h on the co-cultivation substrate the segments are transferred to
containers containing liquid LS-substrate containing 800 mg/l carbenicillin.
The
containers are gently shaken and by this procedure the major part of the
Agrobacterium is
either washed off the segments and/or killed.
Selection
After the washing procedure the segments are transferred to plates containing
the
LS-substrate, agar (8 g/1), trans-zeatin (1-5 mgll), gibberellic acid (U.1
mgll),
carbenicillin (800 mg/1), and kanamycin sulphate (SO-100 mg/1) or
phosphinotricin (1-5
mg/1) or mannose (5 g/1) depending on the vector construction used. The
segments are
sub-cultured to fresh substrate each 3-4 weeks. In 3 to 4 weeks, shoots
develop from the
segments and the formation of new shoots continued for 3-4 months.
Rooting of regenerated shoots
The regenerated shoots are transferred to rooting substrate composed of LS-
substrate, agar (8 g/1) and carbenicillin (800 mgll). The transgenic genotype
of the
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
27
regenerated shoot are verified by testing the rooting ability on the above
mentioned
substrates containing kanamycin sulphate (200 mg/I), by performing NPTII
assays
(Radke, S. E. et al, Theor. Appl. Genet. (1988), 75: 685-694) or by performing
PCR
analysis according to Wang et al (1993, NAR 2~ pp 4153-4154). Plants which are
not
positive in any of these assays are discarded or used as controls.
Alternatively, the
transgenic plants could be verified by performing a GUS assay on the co-
introduced (1-
glucuronidase gene according to Hodal, L. et al. (PI. Sci. (1992), 87: 115-
122).
Transfer to soil
The newly rooted plants (height approx. 2-3 cms) are transplanted from rooting
substrate to soil and placed in a growth chamber (21°C. 16 hour light
200-400uE/m~/sec).
When the plants are well established they are transferred to the greenhouse,
where they
are grown until tubers had developed and the upper part of the plants are
senescing.
Harvesting
The potatoes are harvested after about 3 months and then analysed.
BRANCHING ENZYME ANALYSIS
The SBE expression in the transgenic potato lines are measured using the SBE
assays described by Blennow and Johansson (Phytochemistry (1991) 30:437-444)
and by
standard Western procedures using antibodies directed against class A and
class B potato
SBE.
STARCH ANALYSIS
Starch is isolated from potato tubers and analysed for the amylose:amylopectin
ratio {Hovenkamp-Hetmelink et al. (1988) Potato Research 31:241-246). In
addition, the
chain length distribution of amylopectin is determined by analysis of
isoamylase digested
starch on a Dionex HPAEC. The number of reducing ends in isoamylase digested
starch
is determined by the method described by N. Nelson (1944) J. Biol.Chem.
153:375-380.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
28
The results revealed that there is a reduction in the level of synthesis of
SBE
and/or the level of activity of SBE and/or the composition of starch SBE in
the transgenic
plants.
CONSTRUCTION OF SBE PROMOTER CONSTRUCT
An SBE promoter fragment is amplified from ~,-SBE 3.4 using primers:
5 ' CCA TCG ATA CTT TAA GTG ATT TGA TGG C 3'
(SEQ. ID. No. 36)
and
5' CGG GAT CCT GTT CTG ATT CTT GAT TTC C 3' .
(SEQ. ID. No. 37)
The PCR product is digested with CIaI and BamHI. The resultant 1.2 kb
fragment is then inserted in pVictor5a (see Figure 9) linearised with CIaI and
BgIII
yielding pBEP2 (see Figure 10).
STARCH BRANCHING ENZY1~IE MEASUREMENTS OF POTATO TUBERS
Potatoes from potato plants transformed with pBEAI l are cut in small pieces
and
homogenised in extraction buffer (50 mM Tris-HCl pH 7.5, Sodium-dithionite
(0.1 g/1),
and 2 mM DTT) using a Ultra-Turax homogenizer; 1 g of Dowex xl. is added pr.
10 g of
tuber. The crude homogenate is filtered through a miracloth filter and
centrifuged at 4°C
for 10 minutes at 24.700 g. The supernatant is used for starch branching
enzyme assays.
The starch branching enzyme assays are carried out at 25 °C in a volume
of 400 yl
composed of 0.1 M Na citrate buffer pH 7.0, 0.75 mglml amylose, 5 mg/ml bovine
serum albumin and the potato extract. At 0, 15 30 and 60 minutes aliqouts of
50 lZl are
removed from the reaction into 20 pl 3 N HC1. 1 ml of iodine solution is added
and the
decrease in absorbance at 620 nm is measured with an ELISA spectrophotometer.
The starch branching enzyme (SBE) levels in tuber extracts are measured from
24
transgenic Dianella potato plants transformed with plasmid pBEAl 1, pSS 15 and
pSS 16.
The results show that the BEAT l , SS 15 and SS 16 transgenic lines produce
tubers
which have class B and class A SBE levels, respectively, that are only 10 % to
15 % of
the SBE levels found in non transformed Dianella plants.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
29
In a further experiment, plasmids pSSlS and pBEAI l are cotransfected into
potato
plants, as described above. In the cotransfectants, when analysed as set forth
above,
simultaneous reduction of class A and class B SBE levels are observed.
SUMMATION
The above-mentioned examples relate to the isolation and sequencing of a gene
for
potato SBE. The examples further demonstrate that it is possible to prepare
SBE intron
constructs. These SBE intron constructs can be introduced into plants, such as
potato
plants. After introduction, a reduction in the level of synthesis of SBE
and/or the level of
activity of SBE and/or the composition of starch in plants can be achieved.
Without wishing to be bound by theory it is believed that the expressed sense
intron nucleotide sequence according to the present invention affects
enzymatic activity
via co-suppression and/or trans-activation. Reviews of these mechanisms has
been
published by Finnegan and McElroy (1994 Biotechnology 1? pp 883 - 887) and
Matzke
and Matzke (1995 TIG ~1_ No. 1 pp 1 - 3). By these mechanisms, it is believed
that the
sense introns of the present invention reduce the level of plant enzyme
activity (in
particular SBE activity), which in turn for SBE activity is believed to
influence the
amylose:amylopectin ratio and thus the branching pattern of amylopectin.
Thus, the present invention provides a method wherein it is possible to
manipulate
the starch composition in plants, or tissues or cells thereof, such as potato
tubers, by
reducing the level of SBE activity by using sense intron sequences.
The simultaneous reduction or elimination of class A and class B SBE sequences
from the doubly transformed potato plants, moreover, offers the possibility to
transform
such plants with different SBE genes at will, thus allowing the manipulation
of branching
in starch according to the desired result.
In summation the present invention therefore relates to the surprising use of
SBE
class A sense intron sequences in a method to affect class A SBE activity in
plants.
Other modifications of the present invention will be apparent to those skilled
in the
art without departing from the scope of the present invention.
The following pages present a number of sequence listings which have been
consecutively numbered from SEQ.LD. No. 1 - SEQ.LD. No. 38. In brief, SEQ.LD.
No. 1 - SEQ.LD. No. 13 represent sense intron sequences (genomic DNA); SEQ.LD.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
No. 14 represents the SBE promoter sequence (genomic sequence); SEQ.LD. No. 15
-
SEQ.I.D. No. 27 represent antisense intron sequences; and SEQ. I.D. No. 28
represents
the sequence complementary to the SBE promoter sequence - i.e. the SBE
promoter
sequence in antisense orientation. The full genomic nucleotide sequence for
SBE
5 including the promoter, exons and introns is shown as SEQ. LD. No. 29 (see
Figures 3
and 8 which highlight particular gene features). SEQ. ID. No. 30 to 37 show
primers
used in the methods set forth above. SEQ. ID. No. 38 represents the nucleotide
sequence
of intron 1 of the class A potato SBE gene.
...,..... .fi......
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
31
SEQUENCE LISTING
(1) GENERAL INFORMATION:
S
(i) APPLICANT:
(A) NAME: DANISCO A/S
(B) STREET: LANGEBROGADE 1
(C) CITY: COPENHAGEN K
lO (E) COUNTRY: DENMARK
(F) POSTAL CODE (ZIP): DK-1001
(ii) TITLE OF INVENTION: INHIBITION OF GENE EXPRESSION
IS (iii) NUMBER OF SEQUENCES: 38
(iv) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
ZO (C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: PatentIn Release #1.0, Version #1.30 (EPO)
(2) INFORMATION FOR SEQ ID NO: 1:
2S
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1165 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
3O (D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
3S
(iv) ANTI-SENSE: NO
i ~i
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
32
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
NO: 1:
S GTAATTTTTACTAATTTCATGTTAATTTCAATTATTTTTAGCCTTTGCATTTCATTTTCC60
AATATATCTGGATCATCTCCTTAGTTTTTTATTTTATTTTTTATAATATCAAATATGGAA120
GAAAAATGACACTTGTAGAGCCATATGTAAGTATCATGTGACAAATTTGCAAGGTGGTTG180
AGTGTATAAAATTCAAAAATTGAGAGATGGAGGGGGGGTGGGGGAAGACAATATTTAGAA240
AGAGTGTTCTAGGAGGTTATGGAGGACACGGATGAGGGGTAGAAGGTTAGTTAGGTATTT300
LS GAGTGTTGTCTGGCTTATCCTTTCATACTAGTAGTCGTGGAATTATTTGGGTAGTTTCTT360
GTTTTGTTATTTGATCTTTGTTATTCTATTTTCTGTTTCTTGTACTTCGATTATTGTATT420
ATATATCTTGTCGTAGTTATTGTTCCTCGGTAAGAATGCTCTAGCATGCTTCCTTTAGTG480
TTTTATCATGCCTTCTTTATATTCGCGTTGCTTTGAAATGCTTTTACTTTAGCCGAGGGT540
CTATTAGAAACAATCTCTCTATCTCGTAAGGTAGGGGTAAAGTCCTCACCACACTCCACT600
2S TGTGGGATTACATTGTGTTTGTTGTTGTAAATCAATTATGTATACATAATAAGTGGATTT660
TTTACAACACAAATACATGGTCAAGGGCAAAGTTCTGAACACATAAAGGGTTCATTATAT720
GTCCAGGGATATGATAAAAATTGTTTCTTTGTGAAAGTTATATAAGATTTGTTATGGCTT780
TTGCTGGAAACATAATAAGTTATAATGCTGAGATAGCTACTGAAGTTTGTTTTTTCTAGC840
CTTTTAAATGTACCAATA_~TAGATTCCGTATCGAACGAGT TACCTGGTCA900
ATGTTTTGAT
3S TGATGTTTCTATTTTTTACA GTTGTATCCT960
TTTTTTTGGT
GTTGAACTGC
AATTGAAAAT
ATGAGACGGA 1020
TAGTTGAGAA
TGTGTTCTTT
GTATGGACCT
TGAGAAGCTC
AAACGCTACT
~ ......
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
33
CCAATAATTT CTATGAATTC AAATTCAGTT TATGGCTACC AGTCAGTCCA GAAATTAGGA 1080
TATGCTGCAT ATACTTGTTC AATTATACTG TAAAATTTCT TAAGTTCTCA AGATATCCAT 1140
S GTAACCTCGA GAATTTCTTT GACAG 1165
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 317 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
1S (ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
NO: 2:
2S
GTATGTTTGATAATTTATATGGTTGCATGG ATAGTATATAAATAGTTGGAAAACTTCTGG60
ACTGGTGCTCATGGCATATTTGATCTGTGC ACCGTGTGGAGATGTCAAACATGTGTTACT120
3O TCGTTCCGCCAATTTATAATACCTTAACTT GGGAAAGACAGCTCTTTACTCCTGTGGGCA180
TTTGTTATTTGAATTACAATCTTTATGAGC ATGGTGTTTTCACATTATCAACTTCTTTCA240
TGTGGTATATAACAGTTTTTAGCTCCGTTA ATACCTTTCTTCTTTTTGATATAAACTAAC300
3S
TGTGGTGCATTGCTTGC 317
(2) INFORMATION EQ ID NO: 3:
FOR S
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
34
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 504 base pairs
(B) TYPE: nucleic acid
S (C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
IO (iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0: 3:
GTAACAGCCAAAAGTTGTGC TTTAGGCAGT TTGACCTTATTTTGGAAGATGAATTGTTTA 60
TACCTACTTTGACTTTGCTA GAGAATTTTG CATACCGGGGAGTAAGTAGTGGCTCCATTT 120
AGGTGGCACCTGGCCATTTT TTTGATCTTT TAAAAAGCTGTTTGATTGGGTCTTCAAAAA 180
2S AGTAGACAAGGTTTTTGGAG AAGTGACACA CCCCCGGAGTGTCAGTGGCAAAGCAAAGAT 240
TTTCACTAAGGAGATTCAAA ATATAAAAAA AGTATAGACATAAAGAAGCTGAGGGGATTC 300
AACATGTACTATACAAGCAT CAAATATAGT CTTAAAGCAATTTTGTAGAAATAAAGAAAG 360
TCTTCCTTCTGTTGCTTCAC AATTTCCTTC TATTATCATGAGTTACTCTTTCTGTTCGAA 420
ATAGCTTCCTTAATATTAAA TTCATGATAC TTTTGTTGAGATTTAGCAGTTTTTTCTTGT 480
3S GTAAACTGCTCTCTTTTTTT GCAG 504
(2) INFORMATION
FOR SEQ
ID NO:
4:
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
(i} SEQUENCE CHARACTERISTICS:
(A) LENGTH: 146 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
5 (D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
I~
(iv) ANTI-SENSE: NO
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 4:
GTAGGTCCTC GTCTACTACA AAATAGTAGT TTCCATCATC ATAACAGATT TTCCTATTAA 60
ZO AGCATGATGT TGCAGCATCA TTGGCTTTCT TACATGTTCT AATTGCTATT AAGGTTATGC 120
TTCTAATTAA CTCATCCACA ATGCAG 146
(2) INFORMATION FOR SEQ ID NO: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 218 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
3~ (D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
i i
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
36
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5:
S GTTTTGTTAT TCATACCTTG AAGCTGAATT TTGAACACCA TCATCACAGG CATTTCGATT 60
CATGTTCTTA CTAGTCTTGT TATGTAAGAC ATTTTGAAAT GCAAAAGTTA AAATAATTGT 120
GTCTTTACTA ATTTGGACTT GATCCCATAC TCTTTCCCTT AACAAAATGA GTCAATTCTA 180
TAAGTGCTTG AGAACTTACT ACTTCAGCAA TTAAACAG 218
(2) INFORNfATION FOR SEQ ID NO: 6:
IS (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 198 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
2S (iv) ANTI-SENSE: NO
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 6:
GTATTTTAAA TTTATTTCTA CAACTAAATA ATTCTCAGAA CA.~TTGTTAG ATAGAATCCA 60
AATATATACG TCCTGAAAGT ATAAAAGTAC TTATTTTCGC CATGGGCCTT CAGAATATTG 120
GTAGCCGCTG AATATCATGA TAAGTTATTT ATCCAGTGAC ATTTTTATGT TCACTCCTAT 180
TATGTCTGCT GGATACAG 198
~.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
37
(2) INFORMATION FOR SEQ ID NO: 7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 208 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
1$
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 7:
GTTTGTCTGT TTCTATTGCA TTTTAAGGTT CATATAGGTT AGCCACGGAA AATCTCACTC 60
TTTGTGAGGT AACCAGGGTT CTGATGGATT ATTCAATTTT CTCGTTTATC ATTTGTTTAT 120
~S TCTTTTCATG CATTGTGTTT CTTTTTCAAT ATCCCTCTTA TTTGGAGGTA ATTTTTCTCA 180
TCTATTCACT TTTAGCTTCT AACCACAG 208
(2) INFORMATION FOR SEQ ID NO: 8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 293 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
i i
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
38
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 8:
lO GTATGTCTTA CATCTTTAGA TATTTTGTGA TAATTACAAT TAGTTTGGCT TACTTGAACA 60
AGATTCATTC CTCAAAATGA CCTGAACTGT TGAACATCAA AGGGGTTGAA ACATAGAGGA 120
AAACAACATG ATGAATGTTT CCATTGTCTA GGGATTTCTA TTATGTTGCT GAGAACAAAT 180
GTCATCTTAA AAAAAACATT GTTTACTTTT TTGTAGTATA GAAGATTACT GTATAGAGTT 240
TGCAAGTGTG TCTGTTTTGG AGTAATTGTG AAATGTTTGA TGAACTTGTA CAG 293
ZO (2) INFORMATION FOR SEQ ID NO: 9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 376 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
3O (iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 9:
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
39
GTTCAAGTAT TTTGAATCGC AGCTTGTTAA AATTTTTAGA TTGCTTACTT60
ATAATCTAGT
GGAAGTCTAC TTGGTTCTGG GGATGATAGC TTGTTCTACT TATTTTCCAA120
TCATTTCATC
S CCGAATTTCT GATTTTTGTT TCGAGATCCA TCATTTACAC TTATTACCGC180
AGTATTAGAT
CTCATTTCTA CCACTAAGGC CTTGATGAGC GATTCTTTGA AGCTATAGTT240
AGCTTAAGTT
TCAGGCTACC AATCCACAGC CTGCTATATT TTACCTTTTC TTTACAATGA300
TGTTGGATAC
AGTGATACTA ATTGAAATGG TCTAAATCTG TTCTCCGTCT TTCCTCCCCC360
ATATCTATAT
TCATGATGAA ATGCAG 376
IS (2) INFORMATION FOR SEQ ID NO: 10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 172 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
ZS (iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 10:
GTAAAATCAT CTAAAGTTGA AAGTGTTGGG TTTATGAAGT GCTTTAATTC TATCCAAGGA 60
3S
CAAGTAGAAA CCTTTTTACC TTCCATTTCT TGATGATGGA TTTCATATTA TTTAATCCAA 120
TAGCTGGTCA AATTCGGTAA TAGCTGTACT GATTAGTTAC TTCACTTTGC AG 172
CA 02280210II1999-08-10
WO 98/37214 PCT/IB98/00295
(2) INFORMATION FOR SEQ ID NO: 11:
(i) SEQUENCE CHARACTERISTICS:
S (A) LENGTH: 145 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
10 (ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
1S
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 11:
GTATATATGT TTTACTTATC CATGAAATTA TTGCTCTGCT TGTTTTTAAT GTACTGAACA 60
AGTTTTATGG AGAAGTAACT GAAACAAATC ATTTTCACAT TGTCTAATTT AACTCTTTTT 120
2S TCTGATCCTC GCATGACGAA AACAG 145
(2) INFORMATION FOR SEQ ID NO: 12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 242 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
3S (ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
41
(iv) ANTI-SENSE: NO
S
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 12:
GTAAGGATTT GCTTGAATAA CTTTTGATAA TAAGATAACA ACAGTTCTCT 60
GATGTAGGGT
lO CACCAAAAAG AACTGTAATT GTCTCATCCA TCTTTAGTTG CCGACTGTCT 120
TATAAGATAT
GAGTTCGGAA GTGTTTGAGC CTCCTGCCCT CCCCCTGCGT ATTCAAAAAG 180
TGTTTAGCTA
GAGAAAACTG TTTATTGATG ATCTTTGTCT TCATGCTGAC TTCTCATGAC 240
ATACAATCTG
1S
AG 242
(2) INFORMATION FOR SEQ ID NO: 13:
2O (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 797 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
2S
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
3O (iv) ANTI-SENSE: NO
3S (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 13:
GTACAGTTCT TGCCGTGTGA CCTCCCTTTT TATTGTGGTT TTGTTCATAG TTATTTGAAT 60
CA 02280210111999-08-10
WO 98/37214 PCT/IB98/00295
42
GCGATAGAAG TTAACTATTG ATTACCGCCA CAATCGCCAGTTAAGTCCTC TGAACTACTA120
ATTTGAAAGG TAGGAATAGC CGTAATAAGG TCTACTTTTGGCATCTTACT GTTACAAAAC180
S AAAAGGATGC CAAAAAAATT CTTCTCTATC CTCTTTTTCCCTAAACCAGT GCATGTAGCT240
TGCACCTGCA TAAACTTAGG TAAATGATCA AAAATGAAGTTGATGGGAAC TTAAAACCGC300
CCTGAAGTAA AGCTAGGAAT AGTCATATAA TGTCCACCTTTGGTGTCTGC GCTAACATCA360
ACAACAACAT ACCTCGTGTA GTCCCACAAA GTGGTTTCAGGGGGAGGGTA GAGTGTATGC420
AAAACTTACT CCTATCTCAG AGGTAGAGAG GATTTTTTCAATAGACCCTT GGCTCAAGAA480
IS AAAAAGTCCA AAAAGAAGTA ACAGAAGTGA AAGCAACATGTGTAGCTAAA GCGACCCAAC540
TTGTTTGGGA CTGAAGTAGT TGTTGTTGTT GAAACAGTGCATGTAGATGA ACACATGTCA600
GAAAATGGAC AACACAGTTA TTTTGTGCAA GTCAAAAA.AATGTACTACTA TTTCTTTGTG660
CAGCTTTATG TATAGAAAAG TTAAATAACT AATGAATTTTGCTAGCAGAA AAATAGCTTG720
GAGAGAAATT TTTTATATTG AACTAAGCTA ACTATATTCATCTTTCTTTT TGCTTCTTCT780
2S TCTCCTTGTT TGTGAAG 797
(2) INFORMATION FOR SEQ ID NO: 14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2169 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
3S (ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
t n
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
43
(iv) ANTI-SENSE: NO
(xi) SEQUENCE DESCRIPTION:
SEQ ID NO: 14:
ATCATGGCCA ATTACTGGTTCAAATGCATT ACTTCCTTTC AGATTCTTTC GAGTTCTCAT60
IO GACCGGTCCT ACTACAGACGATACTAACCC GTGGAACTGT TGCATCTGCT TCTTAGAACT120
CTATGGCTAT TTTCGTTAGCTTGGCGTCGG TTTGAACATA GTTTTTGTTT TCAAACTCTT180
CATTTACAGT CAAAATGTTGTATGGTTTTT GTTTTCCTCA ATGATGTTTA CAGTGTTGTG240
TTGTCATCTG TACTTTTGCCTATTACTTGT TTTGAGTTAC ATGTTAAAAA AGTGTTTATT300
TTGCCATATT TTGTTCTCTTATTATTATTA TCATACATAC ATTATTACAA GGAAAAGACA360
2O AGTACACAGA TCTTAACGTTTATGTTCAAT CAACTTTTGG AGGCATTGAC AGGTACCACA420
AATTTTGAGT TTATGATTAAGTTCAATCTT AGAATATGAA TTTAACATCT ATTATAGATG480
CATAAAAATA GCTAATGATAGAACATTGAC ATTTGGCAGA GCTTAGGGTA TGGTATATCC540
AACGTTAATT TAGTAATTTTTGTTACGTAC GTATATGAAA TATTGAATTA ATCACATGAA600
CGGTGGATAT TATATTATGAGTTGGCATCA GCAAAATCAT TGGTGTAGTT GACTGTAGTT660
3O GCAGATTTAA TAATAAAATGGTAATTAACG GTCGATATTA AAATAACTCT CATTTCAAGT720
GGGATTAGAA CTAGTTATTA 780
AAAAAATGTA TACTTTAAGT
GATTTGATGG CATATAATTT
AAAGTTTTTC ATTTCATGCTAAAATTGTTA ATTATTGTAA TGTAGACTGC GACTGGAATT840
ATTATAGTGT AAATTTATGC 900
ATTCAGTGTA AAATTAAAGT
ATTGAACTTG TCTGTTTTAG
AAAATACTTT ATACTTTAAT 960
ATAGGATTTT GTCATGCGAA
TTTAAATTAA TCGATATTGA
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
44
ACACGGAATA CCAAAATTAA AAAGGATACA CATGGCCTTC ATATGAACCG 1020
TGAACCTTTG
ATAACGTGGA AGTTCAAAGA AGGTAAAGTT TAAGAATAAA CTGACAAATT 1080
AATTTCTTTT
ATTTGGCCCA CTACTAAATT TGCTTTACTT TCTAACATGT CAAGTTGTGC 1140
CCTCTTAGTT
GAATGATATT CATTTTTCAT CCCATAAGTT CAATTTGATT GTCATACCAC 1200
CCATGATGTT
IO CTGAAAAATG CTTGGCCATT CACAAAGTTT ATCTTAGTTC CTATGAACTT 1260
TATAAGAAGC
TTTAATTTGA CATGTTATTT ATATTAGATG ATATAATCCA TGACCCAATA 1320
GACAAGTGTA
TTAATATTGT AACTTTGTAA TTGAGTGTGT CTACATCTTA TTCAATCATT 1380
TAAGGTCATT
AAAATAAATT ATTTTTTGAC ATTCTAAAAC TTTAAGCAGA ATAAATAGTT 1440
TATCAATTAT
TAAAAACAAA AAACGACTTA TTTATAAATC AACAAACAAT TTTAGATTGC 1500
TCCAACATAT
2O TTTTCCAAAT TAAATGCAGA AAATGCATAA TTTTATACTT GATCTTTATA 1560
GCTTATTTTT
TTTAGCCTAA CCAACGAATA TTTGTAAACT CACAACTTGA TTAAAAGGGA 1620
TTTACAACAA
GATATATATA AGTAGTGACA AATCTTGATT TTAAATATTT TAATTTGGAG 1680
GTCAAAATTT
TACCATAATC ATTTGTATTT ATAATTAAAT TTTAAATATC TTATTTATAC 1740
ATATCTAGTA
AACTTTTAAA TATACGTATA TACAAAATAT AAAATTATTG GCGTTCATAT 1800
TAGGTCAATA
3O AATCCTTAAC TATATCTGCC TTACCACTAG GAGAAAGTAA AAAACTCTTT 1860
ACCAAAAATA
CATGTATTAT GTATACAAAA AGTCGATTAG ATTACCTAAA TAGAAATTGT 1920
ATAACGAGTA
AGTAAGTAGA AATATAAAAA AACTACAATA CTP.AAAAAAA TATGTTTTAC 1980
TTCAATTTCG
AAACTAATGG GGTCTGAGTG AAATATTCAG AAAGGGGAGG ACTAACAAAA 2040
GGGTCATAAT
GTTTTTTTAT AAAAAGCCAC TAAAATGAGG AAATCAAGAA TCAGAACATA 2100
CAAGAAGGCA
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
4S
GCAGCTGAAG CAAAGTACCA TAATTTAATC AATGGAAATT AATTTCAAAG TTTTATCAAA 2160
2169
ACCCATTCG
S
(2) INFORMATION FOR SEQ ID NO: 15:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1165 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
1S
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 15:
2S CTGTCAAAGA AATTCTCGAG GTTACATGGA TATCTTGAGA ACTTAAGAAA TTTTACAGTA 60
TAATTGAACA AGTATATGCA GCATATCCTA ATTTCTGGAC TGACTGGTAG CCATAAACTG 120
AATTTGAATT CATAGAAATT ATTGGAGTAG CGTTTGAGCT TCTCAAGGTC CATACAAAGA 180
ACACATTCTC AACTATCCGT CTCATAGGAT ACAACATTTT CAATTGCAGT TCAACACCAA 240
AAAAATGTAA AAAATAGAAA CATCATGACC AGGTAATCAA AACATACTCG TTCGATACGG 300
3S AATCTATTAT TGGTACATTT AAAAGGCTAG AAAAAACAAA CTTCAGTAGC TATCTCAGCA 360
TTATAACTTA TTATGTTTCC AGCAAAAGCC ATAACAAATC TTATATAACT TTCACAAAGA 420
m
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
46
AACAATTTTT ATCATATCCC TGGACATATA ATGAACCCTTTATGTGTTCA GAACTTTGCC480
CTTGACCATG TATTTGTGTT GTAAAAAATC CACTTATTATGTATACATAA TTGATTTACA540
S ACAACAAACA CAATGTAATC CCACAAGTGG AGTGTGGTGAGGACTTTACC CCTACCTTAC600
GAGATAGAGA GATTGTTTCT AATAGACCCT CGGCTAAAGTAAAAGCATTT CAAAGCAACG660
CGAATATAAA GAAGGCATGA TAAAACACTA AAGGAAGCATGCTAGAGCAT TCTTACCGAG720
GAACAATAAC TACGACAAGA TATATAATAC AATAATCGAAGTACAAGAAA CAGAAAATAG780
AATAACAAAG ATCAAATAAC AAAACAAGAA ACTACCCAAATAATTCCACG ACTACTAGTA840
IS TGAAAGGATA AGCCAGACAA CACTCAAATA CCTAACTAACCTTCTACCCC TCATCCGTGT900
CCTCCATAAC CTCCTAGAAC ACTCTTTCTA AATATTGTCTTCCCCCACCC CCCCTCCATC960
TCTCAATTTT TGAATTTTAT ACACTCAACC ACCTTGCAAATTTGTCACAT GATACTTACA1020
TATGGCTCTA CAAGTGTCAT TTTTCTTCCA TATTTGATATTATAAAAAAT AAAATAAAAA1080
ACTAAGGAGA TGATCCAGAT ATATTGGAAA ATGAAATGCA 1140
AAGGCTAAAA ATAATTGAAA
~S TTAACATGAA ATTAGTAAAA ATTAC 1165
(2) INFORMATION FOR SEQ ID NO: 16:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 317 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
3S (ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
n
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
47
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 16:
GCAAGCAATG CACCACAGTT AGTTTATATC AAAAAGAAGA CGGAGCTAAA 60
AAGGTATTAA
IO AACTGTTATA TACCACATGA AAGAAGTTGA TAATGTGAAA CATAAAGATT 120
ACACCATGCT
GTAATTCAAA TAACAAATGC CCACAGGAGT AAAGAGCTGT TTAAGGTATT 180
CTTTCCCAAG
ATAAATTGGC GGAACGAAGT AACACATGTT TGACATCTCC CAGATCAAAT 240
ACACGGTGCA
1$
ATGCCATGAG CACCAGTCCA GAAGTTTTCC AACTATTTAT TGCAACCATA 300
ATACTATCCA
TAAATTATCA AACATAC 317
ZO (2) INFORMATION FOR SEQ ID NO: 17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 504 base pairs
(B) TYPE: nucleic acid
2$ (C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
3O (iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 17:
I II
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
48
CTGCAAAAAA AGAGAGCAGT TTACACAAGA AAAAACTGCTAAATCTCAAC AAAAGTATCA60
TGAATTTAAT ATTAAGGAAG CTATTTCGAA CAGAAAGAGTAACTCATGAT AATAGAAGGA120
S AATTGTGAAG CAACAGAAGG AAGACTTTCT TTATTTCTACAAAATTGCTT TAAGACTATA180
TTTGATGCTT GTATAGTACA TGTTGAATCC CCTCAGCTTCTTTATGTCTA TACTTTTTTT240
ATATTTTGAA TCTCCTTAGT GAAAATCTTT GCTTTGCCACTGACACTCCG GGGGTGTGTC300
ACTTCTCCAA AAACCTTGTC TACTTTTTTG AAGACCCAATCAAACAGCTT TTTAAAAGAT360
CAAAAAAATG GCCAGGTGCC ACCTAAATGG AGCCACTACTTACTCCCCGG TATGCAAAAT420
IS TCTCTAGCAA AGTCAAAGTA GGTATAAACA ATTCATCTTCCAAAATAAGG TCAAACTGCC480
TAAAGCACAA CTTTTGGCTG TTAC 504
(2) INFORMATION FOR SEQ ID NO: 18:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH. 146 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
2S (D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 18:
CTGCATTGTG GATGAGTTAA TTAGAAGCAT AACCTTAATA GCAATTAGAA CATGTAAGAA 60
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
49
AGCCAATGAT GCTGCAACAT CATGCTTTAA TAGGAAAATC TGTTATGATG ATGGAAACTA 120
CTATTTTGTA GTAGACGAGG ACCTAC 146
S
(2) INFORMATION FOR SEQ ID NO: 19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 218 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
1S
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 19:
2S CTGTTTAATT GCTGAAGTAG TAAGTTCTCA AGCACTTATA GAATTGACTC ATTTTGTTAA 60
GGGAAAGAGT ATGGGATCAA GTCCAAATTA GTAAAGACAC AATTATTTTA ACTTTTGCAT 120
TTCAAAATGT CTTACATAAC AAGACTAGTA AGAACATGAA TCGAAATGCC TGTGATGATG 180
GTGTTCAAAA TTCAGCTTCA AGGTATGAAT AACAAAAC 218
(2) INFORMATION FOR SEQ ID NO: 20:
3S (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 198 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
S (iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 20:
CTGTATCCAG CAGACATAAT AGGAGTGAAC ATAAAAATGT CACTGGATAA ATAACTTATC 60
ATGATATTCA GCGGCTACCA ATATTCTGAA GGCCCATGGC GAAAATAAGT ACTTTTATAC 120
TTTCAGGACG TATATATTTG GATTCTATCT AACAATTGTT CTGAGAATTA TTTAGTTGTA 180
GAAATAAATT TAAAATAC 198
(2) INFORMATION FOR SEQ ID NO: 21:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 208 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
S1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 21:
CTGTGGTTAG AAGCTAAAAG TGAATAGATG AGAAAAATTA CCTCCAAATA AGAGGGATAT 60
S TGAAAAAGAA ACACAATGCA TGAAAAGAAT AAACAAATGA TAAACGAGAA AATTGAATAA 120
TCCATCAGAA CCCTGGTTAC CTCACAAAGA GTGAGATTTT CCGTGGCTAA CCTATATGAA 180
CCTTAAAATG CAATAGAAAC AGACAAAC 208
(2) INFORMATION FOR SEQ ID NO: 22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 293 base pairs
1S (B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(ivy ANTI-SENSE: YES
2S
(xi) SEQUENCE
DESCRIPTION:
SEQ ID NO:
22:
3O CTGTACAAGTTCATCAAACATTTCACAATT ACTCCAAAACAGACACACTTGCAAACTCTA60
TACAGTAATC TTCTATACTACAA.A.AAAGTA AACAATGTTTTTTTTAAGATGACATTTGTT120
CTCAGCAACA TAATAGAAATCCCTAGACAA TGGAAACATTCATCATGTTGTTTTCCTCTA180
35
TGTTTCAACC CCTTTGATGTTCAACAGTTC AGGTCATTTTGAGGAATGAATCTTGTTCAA240
GTAAGCCAAA CTAATTGTAATTATCACAAA ATATCTAAAGATGTAAGACATAC 293
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
52
(2) INFORMATION FOR SEQ ID NO: 23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 376 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
NO: 23:
CTGCATTTCATCATGAGGGGGAGGAAAGAC GGAGAAATATAGATATCAGA TTTAGACCAT60
TTCAATTAGTATCACTTCATTGTAAAGAAA AGGTAAGTATCCAACAAATA TAGCAGGCTG120
2S TGGATTGGTAGCCTGAAACTATAGCTTCAA AGAATCAACTTAAGCTGCTC ATCAAGGCCT180
TAGTGGTAGAAATGAGGCGGTAATAAGTGT AAATGAATCTAATACTTGGA TCTCGAAACA240
AAAATCAGAAATTCGGTTGGAAAATAAGTA GAACAAGATGAAATGAGCTA TCATCCCCAG300
AACCAAGTAGACTTCCAAGTAAGCAATCTA AAAATTACTAGATTATTTAA CAAGCTGCGA360
TTCAAAATACTTGAAC 376
(2) INFORMATION EQ ID NO: 24:
FOR S
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH:172 base pairs
r rt
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
53
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
S (ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
1~
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 24:
CTGCAAAGTG AAGTAACTAA TCAGTACAGC TATTACCGAA TTTGACCAGC TATTGGATTA 60
AATAATATGA AATCCATCAT CAAGAAATGG AAGGTAAAAA GGTTTCTACT TGTCCTTGGA 120
ZO TAGAATTAAA GCACTTCATA AACCCAACAC TTTCAACTTT AGATGATTTT AC 172
(2) INFORMATION FOR SEQ ID N0: 25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 145 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
3~ (ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
i a
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
54
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 25:
CTGTTTTCGT CATGCGAGGA TCAGAAAP.AA GAGTTAAATT AGACAATGTG AAAATGATTT 60
S GTTTCAGTTA CTTCTCCATA AAACTTGTTC AGTACATTAA AAACAAGCAG AGCAATAATT 120
TCATGGATAA GTAAAACATA TATAC 145
(2) INFORMATION FOR SEQ ID NO: 26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 242 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
IS (D) TOPOLOGY. linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
2S
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 26:
CTGTCATGAG AACAGATTGT ATGTCAGCAT GAAGACAAAG ATCATCAATA AACAGTTTTC 60
3O TCCTTTTTGA ATTAGCTAAA CAACGCAGGG GGAGGGCAGG AGGCTCAAAC ACTTCCGAAC 120
TCAGACAGTC GGATATCTTA TACAACTAAA GATGGATGAG ACAATTACAG TTCTTTTTGG 180
TGAGAGAACT GTACCCTACA TCTGTTATCT TATTATCAAA AGTTATTCAA GCAAATCCTT 240
AC 242
(2) INFORMATION FOR SEQ ID NO: 27:
i ~,
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
SS
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 797 base pairs
(B) TYPE: nucleic acid
S (C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(iiy MOLECULE TYPE: DNA (genomic)
lO (iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
NO: 27:
CTTCACAAACAAGGAGAAGAAGAAGCAAAAAGAAAGATGAATATAGTTAGCTTAGTTCAA60
TATAAAAAATTTCTCTCCAAGCTATTTTTCTGCTAGCAAAATTCATTAGTTATTTAACTT120
TTCTATACATAAAGCTGCACAAAGAAATAGTAGTACATTTTTTTGACTTGCACAAAATAA180
2S CTGTGTTGTCCATTTTCTGACATGTGTTCATCTACATGCACTGTTTCAACAACAACAACT240
ACTTCAGTCCCAAACAAGTTGGGTCGCTTTAGCTACACATGTTGCTTTCACTTCTGTTAC300
TTCTTTTTGGACTTTTTTTCTTGAGCCAAGGGTCTATTGAAAAAATCCTCTCTACCTCTG360
AGATAGGAGTAAGTTTTGCATACACTCTACCCTCCCCCTGAAACCACTTTGTGGGACTAC420
ACGAGGTATGTTGTTGTTGATGTTAGCGCAGACACCAAAGGTGGACATTATATGACTATT480
3S CCTAGCTTTACTTCAGGGCGGTTTTAAGTTCCCATCAACTTCATTTTTGATCATTTACCT540
AAGTTTATGCAGGTGCAAGCTACATGCACTGGTTTAGGGAAAAAGAGGATAGAGAAGAAT600
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
56
TTTTTTGGCA TCCTTTTGTT TTGTAACAGT AAGATGCCAA AAGTAGACCT 660
TATTACGGCT
ATTCCTACCT TTCAAATTAG TAGTTCAGAG GACTTAACTG GCGATTGTGG 720
CGGTAATCAA
S TAGTTAACTT CTATCGCATT CAAATAACTA TGAACAAAAC CACAATAAAA 780
AGGGAGGTCA
CACGGCAAGA ACTGTAC 797
(2) INFORMATION FOR SEQ ID NO: 28:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2169 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
15(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE
DESCRIPTION:
SEQ ID NO:
28:
CGAATGGGTT TTGATAAAACTTTGAAATTAATTTCCATTGATTAAATTATGGTACTTTGC60
3O TTCAGCTGCTGCCTTCTTGTATGTTCTGATTCTTGATTTCCTCATTTTAGTGGCTTTTTA120
TAAAAAAACA TTATGACCCTTTTGTTAGTCCTCCCCTTTCTGAATATTTCACTCAGACCC180
CATTAGTTTC GAAATTGAAGTAAAACATATTTTTTTTAGTATTGTAGTTTTTTTATATTT240
CTACTTACTT ACTCGTTATACAATTTCTATTTAGGTAATCTAATCGACTTTTTGTATACA300
TAATACATGT ATTTTTGGTAAAGAGTTTTTTACTTTCTCCTAGTGGTAAGGCAGATATAG360
1 1f
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
$7
TTAAGGATTT ATTGACCTAA TATGAACGCC AATAATTTTA TATTTTGTAT 420
ATACGTATAT
TTAAAAGTTT ACTAGATATG TATAAATAAG ATATTTAAAA TTTAATTATA 480
AATACAAATG
ATTATGGTAA AATTTTGACC TCCAAATTAA AATATTTAAA ATCAAGATTT 540
GTCACTACTT
ATATATATCT TGTTGTAAAT CCCTTTTAAT CAAGTTGTGA GTTTACAAAT 600
ATTCGTTGGT
IO TAGGCTAAAA AAAATAAGCT ATAAAGATCA AGTATAAAAT TATGCATTTT 660
CTGCATTTAA
TTTGGAAAAA TATGTTGGAG CAATCTAAAA TTGTTTGTTG ATTTATAAAT 720
AAGTCGTTTT
TTGTTTTTAA TAATTGATAA ACTATTTATT CTGCTTAAAG TTTTAGAATG 780
TCF~AAAAATA
1$
ATTTATTTTA ATGACCTTAA ATGATTGAAT AAGATGTAGA CACACTCAAT 840
TACAAAGTTA
CAATATTAAT ACACTTGTCT ATTGGGTCAT GGATTATATC ATCTAATATA 900
AATAACATGT
ZO CAAATTAAAG CTTCTTATAA AGTTCATAGG AACTAAGATA AACTTTGTGA 960
ATGGCCAAGC
ATTTTTCAGA ACATCATGGG TGGTATGACA ATCAAATTGA ACTTATGGGA 1020
TGAAAAATGA
ATATCATTCA ACTAAGAGGG CACAACTTGA CATGTTAGAA AGTAAAGCAA 1080
ATTTAGTAGT
GGGCCAAATA AAAGAAATTA ATTTGTCAGT TTATTCTTAA ACTTTACCTT 1140
CTTTGAACTT
CCACGTTATC AAAGGTTCAC GGTTCATATG AAGGCCATGT GTATCCTTTT 1200
TAATTTTGGT
3O ATTCCGTGTT CAATATCGAT TAATTTAAAT TCGCATGACA AAATCCTATA 1260
TTAAAGTATA
AAGTATTTTC TAAAACAGAC AAGTTCAATA CTTTAATTTT ACACTGAATG 1320
CATAAATTTA
CACTATAATA ATTCCAGTCG CAGTCTACAT TACAATAATT AACAATTTTA 1380
GCATGAAATG
3$
AAAAACTTTA AATTATATGC CATCAAATCA CTTAAAGTAT ACATTTTTTT 1440
AATAACTAGT
TCTAATCCCA CTTGAAATGA GAGTTATTTT AATATCGACC GTTAATTACC 1500
ATTTTATTAT
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
58
TAAATCTGCA ACTACAGTCA ACTACACCAA TGATTTTGCTGATGCCAACT CATAATATAA1560
TATCCACCGT TCATGTGATT AATTCAATAT TTCATATACGTACGTAACAA AAATTACTAA1620
ATTAACGTTG GATATACCAT ACCCTAAGCT CTGCCAAATGTCAATGTTCT ATCATTAGCT1680
ATTTTTATGC ATCTATAATA GATGTTAAAT TCATATTCTAAGATTGAACT TAATCATAAA1740
IO CTCAAAATTT GTGGTACCTG TCAATGCCTC CAAAAGTTGATTGAACATAA ACGTTAAGAT1800
CTGTGTACTT GTCTTTTCCT TGTAATAATG TATGTATGATAATAATAATA AGAGAACAAA1860
ATATGGCAAA ATAAACACTT TTTTAACATG TAACTCAAAACAAGTAATAG GCAAAAGTAC1920
AGATGACAAC ACAACACTGT AAACATCATT GAGGAAAACAAAAACCATAC AACATTTTGA1980
CTGTAAATGA AGAGTTTGAA AACAAAAACT ATGTTCAAACCGACGCCAAG CTAACGAAAA2040
2O TAGCCATAGA GTTCTAAGAA GCAGATGCAA CAGTTCCACGGGTTAGTATC GTCTGTAGTA2100
GGACCGGTCA TGAGAACTCG AAAGAATCTG AAAGGAAGTAATGCATTTGA ACCAGTAATT2160
GGCCATGAT 2169
(2) INFORMATION FOR SEQ ID NO: 29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 11469 base pairs
3O (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
1 if
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
S9
S (xi) SEQUENCE DESCRIPTION: SEQ ID N0: 29:
ATCATGGCCA ATTACTGGTT CAAATGCATT ACTTCCTTTC GAGTTCTCAT 60
AGATTCTTTC
GACCGGTCCT ACTACAGACG ATACTAACCC GTGGAACTGT TCTTAGAACT 120
TGCATCTGCT
CTATGGCTAT TTTCGTTAGC TTGGCGTCGG TTTGAACATA TCAAACTCTT 180
GTTTTTGTTT
CATTTACAGT CAAAATGTTG TATGGTTTTT GTTTTCCTCA CAGTGTTGTG 240
ATGATGTTTA
IS TTGTCATCTG TACTTTTGCC TATTACTTGT TTTGAGTTAC AGTGTTTATT 300
ATGTTAAAAA
TTGCCATATT TTGTTCTCTT ATTATTATTA TCATACATAC GGAAAAGACA 360
ATTATTACAA
AGTACACAGA TCTTAACGTT TATGTTCAAT CAACTTTTGG AGGTACCACA 420
AGGCATTGAC
AATTTTGAGT TTATGATTAA GTTCAATCTT AGAATATGAA ATTATAGATG 480
TTTAACATCT
CATAAAAATA GCTAATGATA GAACATTGAC ATTTGGCAGA TGGTATATCC 540
GCTTAGGGTA
2S AACGTTAATT TAGTAATTTT TGTTACGTAC GTATATGAAA ATCACATGAA 600
TATTGAATTA
CGGTGGATAT TATATTATGA GTTGGCATCA GCAAAATCAT GACTGTAGTT 660
TGGTGTAGTT
GCAGATTTAA TAATAAAATG GTAATTAACG GTCGATATTA CATTTCAAGT 720
AAATAACTCT
GGGATTAGAA CTAGTTATTA AAAAAATGTA TACTTTAAGT CATATAATTT 780
GATTTGATGG
AAAGTTTTTC ATTTCATGCT AAAATTGTTA ATTATTGTAA 840
TGTAGACTGC GACTGGAATT
3S ATTATAGTGT AAATTTATGC ATTCAGTGTA AAATTAAAGT 900
ATTGAACTTG TCTGTTTTAG
AAAATACTTT ATACTTTAAT ATAGGATTTT GTCATGCGAA 960
TTTAAATTAA TCGATATTGA
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
ACACGGAATA CCAAAATTAA AAAGGATACA CATGGCCTTC ATATGAACCG TGAACCTTTG 1020
ATAACGTGGA AGTTCAAAGA AGGTAAAGTT TAAGAATAAA CTGACAAATT AATTTCTTTT 1080
S ATTTGGCCCA CTACTAAATT TGCTTTACTT TCTAACATGT CAAGTTGTGC CCTCTTAGTT 1140
GAATGATATT CATTTTTCAT CCCATAAGTT CAATTTGATT GTCATACCAC CCATGATGTT 1200
CTGAAAAATG CTTGGCCATT CACAAAGTTT ATCTTAGTTC CTATGAACTT TATAAGAAGC 1260
TTTAATTTGA CATGTTATTT ATATTAGATG ATATAATCCA TGACCCAATA GACAAGTGTA 1320
TTAATATTGT AACTTTGTAA TTGAGTGTGT CTACATCTTA TTCAATCATT TAAGGTCATT 1380
IS AAAATAAATT ATTTTTTGAC ATTCTAAAAC TTTAAGCAGA ATAAATAGTT TATCAATTAT 1440
TAAAAACAAA AAACGACTTA TTTATAAATC AACAAACAAT TTTAGATTGC TCCAACATAT 1500
TTTTCCAAAT TAAATGCAGA AAATGCATAA TTTTATACTT GATCTTTATA GCTTATTTTT 1560
TTTAGCCTAA CCAACGAATA TTTGTAAACT CACAACTTGA TTAAAAGGGA TTTACAACAA 1620
GATATATATA AGTAGTGACA AATCTTGATT TTAAATATTT TAATTTGGAG GTCAAAATTT 1680
2S TACCATAATC ATTTGTATTT ATAATTAAAT TTTAAATATC TTATTTATAC ATATCTAGTA 1740
AACTTTTAAA TATACGTATA TACAAAATAT AAAATTATTG GCGTTCATAT TAGGTCAATA 1800
AATCCTTAAC TATATCTGCC TTACCACTAG GAGAAAGTAA AAAACTCTTT ACCAAAAATA 1860
CATGTATTAT GTATACAAAA AGTCGATTAG ATTACCTAAA TAGAAATTGT ATAACGAGTA 1920
AGTAAGTAGA AATATAAAAA AACTACAATA CTAAAAAAAA TATGTTTTAC TTCAATTTCG 1980
3S AAACTAATGG GGTCTGAGTG AAATATTCAG AAAGGGGAGG ACTAACAAAA GGGTCATAAT 2040
GTTTTTTTAT AAAAAGCCAC TAAAATGAGG AAATCAAGAA TCAGAACATA CAAGAAGGCA 2100
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/0!0295
61
GCAGCTGAAG CAAAGTACCA TAATTTAATC AATGGAAATT AATTTCAAAG TTTTATCAAA 2160
ACCCATTCGA GGATCTTTTC CATCTTTCTC ACCTAAAGTT TCTTCAGGGG TAATTTTTAC 2220
S TAATTTCATG TTAATTTCAA TTATTTTTAG CCTTTGCATT TCATTTTCCA ATATATCTGG 2280
ATCATCTCCT TAGTTTTTTA TTTTATTTTT TATAATATCA AATATGGAAG AAAAATGACA 2340
CTTGTAGAGC CATATGTAAG TATCATGTGA CAAATTTGCA AGGTGGTTGA GTGTATAAAA 2400
TTCAAAAATT GAGAGATGGA GGGGGGGTGG GGGAAGACAA TATTTAGAAA GAGTGTTCTA 2460
GGAGGTTATG GAGGACACGG ATGAGGGGTA GAAGGTTAGT TAGGTATTTG AGTGTTGTCT 2520
IS GGCTTATCCT TTCATACTAG TAGTCGTGGA ATTATTTGGG TAGTTTCTTG TTTTGTTATT 2580
TGATCTTTGT TATTCTATTT TCTGTTTCTT GTACTTCGAT TATTGTATTA TATATCTTGT 2640
CGTAGTTATT GTTCCTCGGT AAGAATGCTC TAGCATGCTT CCTTTAGTGT TTTATCATGC 2700
CTTCTTTATA TTCGCGTTGC TTTGAAATGC TTTTACTTTA GCCGAGGGTC TATTAGAAAC 2760
AATCTCTCTA TCTCGTAAGG TAGGGGTAAA GTCCTCACCA CACTCCACTT GTGGGATTAC 2820
2S ATTGTGTTTG TTGTTGTAAA TCAATTATGT ATACATAATA AGTGGATTTT TTACAACACA 2880
AATACATGGT CAAGGGCAAA GTTCTGAACA CATAAAGGGT TCATTATATG TCCAGGGATA 2940
TGATAAAAAT TGTTTCTTTG TGAAAGTTAT ATAAGATTTG TTATGGCTTT TGCTGGAAAC 3000
ATAATAAGTT ATAATGCTGA GATAGCTACT GAAGTTTGTT TTTTCTAGCC TTTTAAATGT 3060
ACCAATAATA GATTCCGTAT CGAACGAGTA TGTTTTGATT ACCTGGTCAT GATGTTTCTA 3120
3S TTTTTTACAT TTTTTTGGTG TTGAACTGCA ATTGAAAATG TTGTATCCTA TGAGACGGAT 3180
AGTTGAGAAT GTGTTCTTTG TATGGACCTT GAGAAGCTCA AACGCTACTC CAATAATTTC 3240
i ii
CA 02280210 1999-08-10
WO 98137214 PCT/IB98/00295
62
TATGAATTCA AATTCAGTTT ATGGCTACCA GTCAGTCCAG AAATTAGGAT ATGCTGCATA 3300
TACTTGTTCA ATTATACTGT AAAATTTCTT AAGTTCTCAA GATATCCATG TAACCTCGAG 3360
S AATTTCTTTG ACAGGCTTCT AGAAATAAGA TATGTTTTCC TTCTCAACAT AGTACTGGAC 3420
TGAAGTTTGG ATCTCAGGAA CGGTCTTGGG ATATTTCTTC CACCCCAAAA TCAAGAGTTA 3480
GAAAAGATGA AAGGGTATGT TTGATAATTT ATATGGTTGC ATGGATAGTA TATAAATAGT 3540
TGGAAAACTT CTGGACTGGT GCTCATGGCA TATTTGATCT GTGCACCGTG TGGAGATGTC 3600
AAACATGTGT TACTTCGTTC CGCCAATTTA TAATACCTTA ACTTGGGAAA GACAGCTCTT 3660
IS TACTCCTGTG GGCATTTGTT ATTTGAATTA CAATCTTTAT GAGCATGGTG TTTTCACATT 3720
ATCAACTTCT TTCATGTGGT ATATAACAGT TTTTAGCTCC GTTAATACCT TTCTTCTTTT 3780
TGATATAAAC TAACTGTGGT GCATTGCTTG CATGAAGCAC AGTTCAGCTA TTTCCGCTGT 3840
TTTGACCGAT GACGACAATT CGACAATGGC ACCCCTAGAG GAAGATGTCA AGACTGAAAA 3900
TATTGGCCTC CTAAATTTGG ATCCAACTTT GGAACCTTAT CTAGATCACT TCAGACACAG 3960
2S AATGAAGAGA TATGTGGATC AGAAAATGCT CATTGAAAAA TATGAGGGAC CCCTTGAGGA 4020
ATTTGCTCAA GGTAACAGCC AAAAGTTGTG CTTTAGGCAG TTTGACCTTA TTTTGGAAGA 4080
TGAATTGTTT ATACCTACTT TGACTTTGCT AGAGAATTTT GCATACCGGG GAGTAAGTAG 4140
TGGCTCCATT TAGGTGGCAC CTGGCCATTT TTTTGATCTT TTAAAAAGCT GTTTGATTGG 4200
GTCTTCAAAA AAGTAGACAA GGTTTTTGGA GAAGTGACAC ACCCCCGGAG TGTCAGTGGC 4260
3S AAAGCAAAGA TTTTCACTAA GGAGATTCAA AATATAAAAA AAGTATAGAC ATAAAGAAGC 4320
TGAGGGGATT CAACATGTAC TATACAAGCA TCAAATATAG TCTTAAAGCA ATTTTGTAGA 4380
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
63
AATAAAGAAA GTCTTCCTTC TGTTGCTTCA CAATTTCCTT CTATTATCAT GAGTTACTCT 4440
TTCTGTTCGA AATAGCTTCC TTAATATTAA ATTCATGATA CTTTTGTTGA GATTTAGCAG 4500
S TTTTTTCTTG TGTAAACTGC TCTCTTTTTT TGCAGGTTAT.TTAAAATTTG GATTCAACAG 4560
GGAAGATGGT TGCATAGTCT ATCGTGAATG GGCTCCTGCT GCTCAGTAGG TCCTCGTCTA 4620
CTACAAAATA GTAGTTTCCA TCATCATAAC AGATTTTCCT ATTAAAGCAT GATGTTGCAG 4680
CATCATTGGC TTTCTTACAT GTTCTAATTG CTATTAAGGT TATGCTTCTA ATTAACTCAT 4740
CCACAATGCA GGGAAGCAGA AGTTATTGGC GATTTCAATG GATGGAACGG TTCTAACCAC 4800
IS ATGATGGAGA AGGACCAGTT TGGTGTTTGG AGTATTAGAA TTCCTGATGT TGACAGTAAG 4860
CCAGTCATTC CACACAACTC CAGAGTTAAG TTTCGTTTCA AACATGGTAA TGGAGTGTGG 4920
GTAGATCGTA TCCCTGCTTG GATAAAGTAT GCCACTGCAG ACGCCACAAA GTTTGCAGCA 4980
CCATATGATG GTGTCTACTG GGACCCACCA CCTTCAGAAA GGTTTTGTTA TTCATACCTT 5040
GAAGCTGAAT TTTGAACACC ATCATCACAG GCATTTCGAT TCATGTTCTT ACTAGTCTTG 5100
2S TTATGTAAGA CATTTTGAAA TGCAAAAGTT AAAATAATTG TGTCTTTACT AATTTGGACT 5160
TGATCCCATA CTCTTTCCCT TAACAAAATG AGTCAATTCT ATAAGTGCTT GAGAACTTAC 5220
TACTTCAGCA ATTAAACAGG TACCACTTCA AATACCCTCG CCCTCCCAAA CCCCGAGCCC 5280
CACGAATCTA TGAAGCACAT GTCGGCATGA GCAGCTCTGA GCCACGTGTA AATTCGTATC 5340
GTGAGTTTGC AGATGATGTT TTACCTCGGA TTAAGGCAAA TAACTATAAT ACTGTCCAGT 5400
3S TGATGGCCAT AATGGAACAT TCTTACTATG GATCATTTGG ATATCATGTT ACAAACTTTT 5460
TTGCTGTGAG CAGTAGATAT GGAAACCCGG AGGACCTAAA GTATCTGATA GATAAAGCAC 5520
f II
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
64
ATAGCTTGGG TTTACAGGTT CTGGTGGATG TAGTTCACAG TCATGCAAGC 5580
AATAATGTCA
CTGATGGCCT CAATGGCTTT GATATTGGCC AAGGTTCTCA AGAATCCTAC 5640
TTTCATGCTG
S GAGAGCGAGG GTACCATAAG TTGTGGGATA GCAGGCTGTT CAACTATGCC 5700
AATTGGGAGG
TTCTTCGTTT CCTTCTTTCC AACTTGAGGT GGTGGCTAGA AGAGTATAAC 5760
TTTGACGGAT
TTCGATTTGA TGGAATAACT TCTATGCTGT ATGTTCATCA TGGAATCAAT 5820
ATGGGATTTA
CAGGAAACTA TAATGAGTAT TTCAGCGAGG CTACAGATGT TGATGCTGTG 5880
GTCTATTTAA
TGTTGGCCAA TAATCTGATT CACAAGATTT TCCCAGATGC AACTGTTATT 5940
GCCGAAGATG
LS TTTCTGGTAT GCCGGGCCTT GGCCGGCCTG TTTCTGAGGG AGGAATTGGT 6000
TTTGTTTACC
GCCTGGCAAT GGCAATCCCA GATAAGTGGA TAGATTATTT AAAGAATAAG 6060
AATGATGAAG
ATTGGTCCAT GAAGGAAGTA ACATCGAGTT TGACAAATAG GAGATATACA 6120
GAGAAGTGTA
TAGCATATGC GGAGACCCAT GATCAGGTAT TTTAAATTTA TTTCTACAAC 6180
TAAATAATTC
TCAGAACAAT TGTTAGATAG AATCCAAATA TATACGTCCT GAAAGTATAA 6240
AAGTACTTAT
2S TTTCGCCATG GGCCTTCAGA ATATTGGTAG CCGCTGAATA TCATGATAAG 6300
TTATTTATCC
AGTGACATTT TTATGTTCAC TCCTATTATG TCTGCTGGAT ACAGTCTATT 6360
GTTGGTGACA
AGACCATTGC ATTTCTCCTA ATGGACAAAG AGATGTATTC TGGCATGTCT 6420
TGCTTGACAG
ATGCTTCTCC TGTTGTTGAT CGAGGAATTG CGCTTCACAA GGTTTGTCTG 6480
TTTCTATTGC
ATTTTAAGGT TCATATAGGT TAGCCACGGA AAATCTCACT CTTTGTGAGG 6540
TAACCAGGGT
3S TCTGATGGAT TATTCAATTT TCTCGTTTAT CATTTGTTTA TTCTTTTCAT 6600
GCATTGTGTT
TCTTTTTCAA TATCCCTCTT ATTTGGAGGT AATTTTTCTC ATCTATTCAC 6660
TTTTAGCTTC
~.
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
6S
TAACCACAGA TGATCCATTT TTTCACAATG GCCTTGGGAG GAGAGGGGTA 6720
CCTCAATTTC
ATGGGTAACG AGGTATGTCT TACATCTTTA GATATTTTGT GATAATTACA 6780
ATTAGTTTGG
S CTTACTTGAA CAAGATTCAT TCCTCAAAAT GACCTGAACT GTTGAACATC 6840
AAAGGGGTTG
AAACATAGAG GAAAACAACA TGATGAATGT TTCCATTGTC TAGGGATTTC 6900
TATTATGTTG
CTGAGAACAA ATGTCATCTT AA.AAAP.AACA TTGTTTACTT TTTTGTAGTA6960
TAGAAGATTA
CTGTATAGAG TTTGCAAGTG TGTCTGTTTT GGAGTAATTG TGAAATGTTT 7020
GATGAACTTG
TACAGTTTGG CCATCCTGAG TGGATTGACT TCCCTAGAGA GGGCAATAAT 7080
TGGAGTTATG
IS ACAAATGTAG ACGCCAGTGG AACCTCGCGG ATAGCGAACA CTTGAGATAC 7140
AAGGTTCAAG
TATTTTGAAT CGCAGCTTGT TAAATAATCT AGTAATTTTT AGATTGCTTA 7200
CTTGGAAGTC
TACTTGGTTC TGGGGATGAT AGCTCATTTC ATCTTGTTCT ACTTATTTTC 7260
CAACCGAATT
TCTGATTTTT GTTTCGAGAT CCAAGTATTA GATTCATTTA CACTTATTAC 7320
CGCCTCATTT
CTACCACTAA GGCCTTGATG AGCAGCTTAA GTTGATTCTT TGAAGCTATA 7380
GTTTCAGGCT
ZS ACCAATCCAC AGCCTGCTAT ATTTGTTGGA TACTTACCTT TTCTTTACAA 7440
TGAAGTGATA
CTAATTGAAA TGGTCTAAAT CTGATATCTA TATTTCTCCG TCTTTCCTCC 7500
CCCTCATGAT
GAAATGCAGT TTATGAATGC ATTTGATAGA GCTATGAATT CGCTCGATGA 7560
AAAGTTCTCA
TTCCTCGCAT CAGGAAAACA GATAGTAAGC AGCATGGATG ATGATAATAA 7620
GGTAAAATCA
TCTAAAGTTG AAAGTGTTGG GTTTATGAAG TGCTTTAATT CTATCCAAGG 7680
ACAAGTAGAA
3S ACCTTTTTAC CTTCCATTTC TTGATGATGG ATTTCATATT ATTTAATCCA 7740
ATAGCTGGTC
AAATTCGGTA ATAGCTGTAC TGATTAGTTA CTTCACTTTG CAGGTTGTTG 7800
TGTTTGAACG
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
66
TGGTGACCTG GTATTTGTAT TCAACTTCCA CCCAAAGAAC ACATACGAAG 7860
GGTATATATG
TTTTACTTAT CCATGAAATT ATTGCTCTGC TTGTTTTTAA TGTACTGAAC 7920
AAGTTTTATG
S GAGAAGTAAC TGAAACAAAT CATTTTCACA TTGTCTAATT TAACTCTTTT 7980
TTCTGATCCT
CGCATGACGA AAACAGGTAT AAAGTTGGAT GTGACTTGCC AGGGAAGTAC 8040
AGAGTTGCAC
TGGACAGTGA TGCTTGGGAA TTTGGTGGCC ATGGAAGAGT AAGGATTTGC 8100
TTGAATAACT
TTTGATAATA AGATAACAGA TGTAGGGTAC AGTTCTCTCA CCAAAAAGAA 8160
CTGTAATTGT
CTCATCCATC TTTAGTTGTA TAAGATATCC GACTGTCTGA GTTCGGAAGT 8220
GTTTGAGCCT
IS CCTGCCCTCC CCCTGCGTTG TTTAGCTAAT TCAAAAAGGA GAAAACTGTT 8280
TATTGATGAT
CTTTGTCTTC ATGCTGACAT ACAATCTGTT CTCATGACAG ACTGGTCATG 8340
ATGTTGACCA
TTTCACATCA CCAGAAGGAA TACCTGGAGT TCCAGAAACA AATTTCAATG 8400
GTCGTCCAAA
TTCCTTCAAA GTGCTGTCTC CTGCGCGAAC ATGTGTGGTA CAGTTCTTGC 8460
CGTGTGACCT
CCCTTTTTAT TGTGGTTTTG TTCATAGTTA TTTGAATGCG ATAGAAGTTA 8520
ACTATTGATT
2S ACCGCCACAA TCGCCAGTTA AGTCCTCTGA ACTACTAATT TGAAAGGTAG 8580
GAATAGCCGT
AATAAGGTCT ACTTTTGGCA TCTTACTGTT ACAAAACAAA AGGATGCCAA 8640
AAAAATTCTT
CTCTATCCTC TTTTTCCCTA AACCAGTGCA TGTAGCTTGC ACCTGCATAA 8700
ACTTAGGTAA
ATGATCAAAA ATGAAGTTGA TGGGAACTTA AAACCGCCCT GAAGTAAAGC 8760
TAGGAATAGT
CATATAATGT CCACCTTTGG TGTCTGCGCT AACATCAACA ACAACATACC 8820
TCGTGTAGTC
3S CCACAAAGTG GTTTCAGGGG GAGGGTAGAG TGTATGCAAA ACTTACTCCT 8880
ATCTCAGAGG
TAGAGAGGAT TTTTTCAATA GACCCTTGGC TCAAGAAAAA AAGTCCAAAA 8940
AGAAGTAACA
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
67
GAAGTGAAAG CAACATGTGT AGCTAAAGCG ACCCAACTTG TTTGGGACTG 9000
AAGTAGTTGT
TGTTGTTGAA ACAGTGCATG TAGATGAACA CATGTCAGAA AATGGACAAC 9060
ACAGTTATTT
S TGTGCAAGTC AAAAAAATGT ACTACTATTT CTTTGTGCAG CTTTATGTAT 9120
AGAAAAGTTA
AATAACTAAT GAATTTTGCT AGCAGAAAAA TAGCTTGGAG AGAAATTTTT 9180
TATATTGAAC
TAAGCTAACT ATATTCATCT TTCTTTTTGC TTCTTCTTCT CCTTGTTTGT 9240
GAAGGCTTAT
TACAGAGTTG ATGAACGCAT GTCAGAAACT GAAGATTACC AGACAGACAT 9300
TTGTAGTGAG
CTACTACCAA CAGCCAATAT CGAGGAGAGT GACGAGAAAC TTAAAGATTC 9360
GTTATCTACA
IS AATATCAGTA ACATTGACGA ACGCATGTCA GAAACTGAAG TTTACCAGAC 9420
AGACATTTCT
AGTGAGCTAC TACCAACAGC CAATATTGAG GAGAGTGACG AGAAACTTAA 9480
AGATTCGTTA
TCTACAAATA TCAGTAACAT TGATCAGACT GTTGTAGTTT CTGTTGAGGA 9540
GAGAGACAAG
GAACTTAAAG ATTCACCGTC TGTAAGCATC ATTAGTGATG TTGTTCCAGC 9600
TGAATGGGAT
GATTCAGATG CAAACGTCTG GGGTGAGGAC TAGTCAGATG ATTGATCGAC 9660
CCTTCTACCG
2S ATTGGTGATC GCTATCCTTG CTCTCTGAGA AATAGGTGAG GCGAAACAAA 9720
AAATAATTTG
CATGATAAAA AGTCTGATTT TATGATCGCT ATCCTCGCTC TCTGAGAAAG 9780
AAGCGAAACA
AAGGCGACTC CTGGACTCGA ATCTATAAGA TAACAAAGGC GACTCCTGGG 9840
ACTCGAATCT
ATAAGATAAC AAAGGCAATT CCAAGACTTG AATCTATAAA AAATTTAGTT 9900
AAGAATGATT
AACGTCCGAT CCTAATTCGA ATCGAGGCAT CTTACCACTC CATTGATAAT 9960
TATATAAGTC
3S AATAAGTCAT ATAAAGTATT AAAAACTAAA TTGACTTGAT CGGTCTATCA 10020
AAAATAGATA
AATTGTGTTC ATATGTAACA TTTTTGTTGT CACAATTAGC TTAATTACAT 10080
CTTTCATGTG
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
68
CAATAACAAA GAAATGATAG GAATTTAGAG ATTCCAATTT TTTTGTTGCC ACAATTAACT 10140
TAATTACATC TTTCATTTGC AATAACAAAG AAATGATAGG AATTTAGAGA TCCAGTGTCA 10200
S ATACACAACC TAGGCCAACA TCGAAAGCAT AACTGTAAAC TCATGCATGA AGAAATCAGT 10260
CGTAAAAATG AATAAATGCG ACATAAAAAC AAATTGCATG TATCATTAAT GTGACTTAAC 10320
TACAAGTAAA AATAAATTTA ACAAATGTAA CTTAACTACA AGTAAAAATA AATTGCTTCT 10380
ATCATTAACA AACAAACAGA ATTAAAAAGA AAAAA.ACATA CTAAATCTTA CCGTCATTCG 10440
ATAA.AA.AAAA ATACCAAATT CATAATGCAA GGAAAACGAA ACGCGTCCTG ATCGGGTATC 10500
IS P.ACGATGAAA TGGACCAGTT GGATCGACTG CCTGCACAAC GTTAGGTATG CCAAAAAAA.A 10560
GAACACGATC CTTTGCACCC GTTCGATGAT TATCAGTATG TTCACAAAAA AAACTTAAGT 10620
TCATCCCAGT GTACAACAGC CCCAACATCT GCCCCAAGTA ACAF1A.A.AACA ACCAATTTAT 10680
CTTATTCTTA TCTGCCACAA AATAATCGGT TTCACACTAT TCTCTTGTTA TACAAAATTG 10740
ACAAGTAGGA AGGAGAGGAG TCATCCAAAT AAACGGTGCA CGTTCTTTGA GAAAAGTCTT 10800
2S ATTTTTCGTA AGATCCAATT TCAACAAACT TTTCTTCAAG TCAAAATTCC TGATAGTGTA 10860
TCTCCTCTCG ACGACCTCTT GCATTGAACG ATCTCCGCTT ATCATGAAAA GTTGCTTGGA 10920
TAACAAGTAT TGCAAGGGGG GGACAGTAGC TATTAAGTTA GTCGGCCCAA GGAAATGGAG 10980
GAGTGATAGT CTCGAATATT ATTCACCTCT TTAGCATTAC CCGGTCTGGC TTTAAGGAGT 11040
TACGTCTTTT ACGCTCGCCA ATTTCTTTTT TTAGAATGGT TGGTGTCAAA ATCGCGAGTT 11100
3S GTGGAAGGTT CAAGTTACTC GATTCGTGAT TTTCAAGTAT GAGTGGTGAG AGAGATTCGA 11160
TATTTTCACG AGGTGTATTC GAGGTCTAGT AGAACGAAGG GTGTCACTAA TGAAAGTTTC 11220
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
69
AAGAGTTCAT CATCATCTTC TTCTAGTAGA TTTTCGCTTTCAAATGAGTA TGAAAATTCT11280
TCCTCTTTTC TATTGATTTT CTTCATTGTT TTCTTCATTGTTGTGGTTGT TATTGAAAAG11340
S AAAGAAAATT TATAACAGAA AAAGATGTCA AAAAAAAGGTAAAATGAAAG AGTATCATAT11400
ACTTAAAGAG TTGCGTAGAG ATAAGTCAAA AGAAACAGAATTATAGTAAT TTCAGCTAAG11460
TTAGAATTC 11469
(2) INFORMATION FOR SEQ ID NO: 30:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26 base pairs
1S (B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: other nucleic
acid
(A) DESCRIPTION: /desc = "Synthetic
DNA Primer"
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
2S
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 30:
GGAATTCCAG TCGCAGTCTA CATTAC 26
(2) INFORMATION FOR SEQ ID NO: 31:
3S (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 28 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: other nucleic acid
(A) DESCRIPTION: /desc = "Synthetic DNA Primer"
5
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3I:
IS CGGGATCCAG AGGCATTAAG ATTTCTGG 28
(2) INFORMATION FOR SEQ ID NO: 32:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
ZS (ii) MOLECULE TYPE: other nucleic acid
(A) DESCRIPTION: /desc = "Synthetic DNA Primer"
(iii) HYPOTHETICAL: NO
3O (iv) ANTI-SENSE: YES
3S (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 32:
CGGGATCCAA AGAAATTCTC GAGGTTACAT GG 32
t
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
71
(2) INFORMATION FOR SEQ ID NO: 33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: other nucleic acid
(A) DESCRIPTION: /desc = "Synthetic DNA Primer"
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 33:
CGGGATCCGG GGTAATTTTT ACTAATTTCA TG 32
(2) INFORMATION FOR SEQ ID NO: 34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECUhE TYPE: other nucleic acid
(A) DESCRIPTION: /desc = "Synthetic DNA Primer"
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
i
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
72
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 34:
S CGGGATCCCG TATGTCTCAC TGTGTTTGTG GC 32
(2) INFORMATION FOR SEQ ID NO: 35:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
IS (ii) MOLECULE TYPE: other nucleic acid
(A) DESCRIPTION: /desc = "Synthetic DNA Primer"
(iii) HYPOTHETICAL: NO
2O (iv) ANTI-SENSE: YES
2S (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 35:
CGGGATCCCC CTACATACAT ATATCAGATT AG 32
(2) INFORMATION FOR SEQ ID NO: 36:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 28 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
3S (D) TOPOLOGY: linear
(ii) MOLECULE TYPE: other nucleic acid
(A) DESCRIPTION: /desc = "Synthetic DNA Primer"
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
73
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 36:
CCATCGATAC TTTAAGTGAT TTGATGGC 28
(2) INFORMATION FOR SEQ ID NO: 37:
IS (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 28 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: other nucleic acid
(A) DESCRIPTION: /desc = "Synthetic DNA Primer"
(iii) HYPOTHETICAL: NO
2S
(iv) ANTI-SENSE: YES
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 37:
CGGGATCCTG TTCTGATTCT TGATTTCC 28
3S (2) INFORMATION FOR SEQ ID NO: 3B:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2122 base pairs
i ii
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
74
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (genomic)
(iii) HYPOTHETICAL: NO
(iv) ANTI-SENSE: NO
1~
{xi) SEQUENCE DESCRIPTION: SEQ ID NO: 38:
15
GTATGTCTCA CTGTGTTTGT GGCTGTGTGT GTTTTTTTCT CTGTCTTTTT 60
GTGTTTTGTG
TAATTGGGGC TCTTTAAAGT TGGTATTGTG TATACCCTTT TGAGTATAGT 120
CTTTGAGGAA
2O GCAAAATGAT GAATCTTGAT TGACATTAGT AAGGGTTGTA ACTTTTTGAA 180
GTTTGGTTAG
GTGTAATTGA GTTTGGCTTG TGTGTCTGTG TGTCGAGGTT ATTTTTTTGG 240
TTTGTGTTAT
TGGGGATTCT TAAAAGTTGG TATTGTGTAT ACCCTTTTGA GTATAGTCTT 300
TGAGGAAGCA
25
AAAATGATGA ATCTTGATTG GCATTAGTAA AGGTTGTAGC TTTTTGAAGT 360
GTGGTTAGGT
GTAATTGAGT TTGGCTTGTG TGTCTGTGTG TTTTGGAATC CTGATGTGTG 420
TCAAGTCCTG
3O ATATGGGTCG AGGTTCTTTC TTTGGTTTGT GTAATTGGGG GTTCTTAAAA 480
GTTGGTATTA
TGTACCTTTT TAAGAATAGT GTCTGAGAAA GCAAAATCGA TGAATTTTGA 540
TTGACAGCAT
ATTCTTTGAG AAAGCAAAAA ATGGTGAGTT TTCATGGAGA AACTTGATTG 600
ACATTACTAA
35
AGGTAGCAAC TTTTTCAACT CCTGATATGG GTCAAGGTTC TTTGTTTGGT 660
TTGTGTAATT
TGGGGTTCTT TGAAGTTTTG AGAAAGAAAA ATTATGATTT TTCATGGAGA 720
AATTTGATTT
i ~
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
ACATTAATAA AGGTAGTAGC TTTTTAAAGT GTGGTCAGCT GTAATGAGTT 780
CAGCTTGGTT
TAAAGGGGCC CTACATATGG TGCTTTCTGG TGAGATATTT GTTGCTCCAC 840
CATACGAGTT
5
ATAAGAATCA TAGTGTTAGG ATCTTTTTTC TTTTTTTTTT CATTTTTCAC 900
TTGACTAGCT
ACTAGAGGAG TGATCTTGAC GGCGGAAAAT CTTAGAAAGG GGAAGGTTGT 960
TTGCATCAAC
IO TGGTGTTATA TGTGCAAGGA GACGGGAGAT GATGTAGATC ATCTTCTTCT 1020
TCATTGTGGT
CTTTCCATGA GGTTATGATG TGATATGTTT GAATGGTTTG GTACTTCTTG 1080
GCTATGCCAA
GAACTGTGAA AGAATTGATA TTCAGTTGGA AGTGTGGAGT TGGAAGAGTG 1140
GAAGAATTGA
15
CACTTGGTTC CATTAGCTTT AATGTGGGTG GTGTGGAGAG AGAGAGAAAT 1200
AGGAGAGCTT
TTGAGGGGGT AGAGTTGAGC TTTCCTCAGT TGAGAAGTAG CCTTTGATAT 1260
CTTTTTTTTT
2O TTTTTTTGTA CACCCATAGA ATTCCCAATT GTATAGAAGA TTGGGTGGAG 1320
TTTGTAGAGA
ATCATCTTTT GTAGTAGATT CTTTACCTTT TGGTATATCC ATTGTATACA 1380
GCCAGGCCTT
TGACTATGTT TATGAATGAA TATACATTAC TTGAAAAAAA AAGAAGTGAA 1440
GCCAGTCTGT
25
TGTACCTTTG TAGACAATGT TGTTGCAGCA TCTTGATAAT TCCCTGAAAA 1500
TTGTCTCCCT
GAAGGAATAG TTTGGTTGAT ATTGATTATT TCTTGGTTTG TTTAATTCGG 1560
TGTTCTTGAA
3O GGCCATTTTA AATCCTTTGA CATTGTTAAA GGTGTTTACA AGTGTTGGTC 1620
TGGGTTTAAA
AGCACCTCTT GTATGGTGCT TTCTGGAGTG ATCTTTCTTC CTCCAAAAGA 1680
GAAGTTGCAA
GAATCAGTGT GTGTACTTTT TTCTCTTGTA TGATCAGATC TTTTTTCAAT 1740
TTTTCCGTTT
35
TAGTTGATTT ATCCATATAG TGAAAGTTGG TGTCATAGTT GCTGTTTGTG 1800
GACTTCCTGT
AAAAGTTTTT TGATATACTT AAAAAATTGT CACACAGAAG AAAGAGTTTT 1860
TTACCATTAC
I II
CA 02280210 1999-08-10
WO 98/37214 PCT/IB98/00295
76
TTAAGCTAGA TGGGACTGTT TGATTCTTAG ACCAAATAAT GAACCTTTTT GTTCTCTTAA 1920
CGTGTACTTG AAATAGTTTG GTAAAATTGT GATAGGAAAA AAGATAATTC TTGATTGCTT 1980
TTGGAGCATC ACTTCTAATC ATAAAAGTCT TTGCTCTCTT CAACCATGAA TGATAAATTG 2040
GACACTTATG TGGCCCTAAG TTGCTCTCAG TAGTGGTCTT TAATTGTGGA GATATAACTA 2100
IO ATCTGATATA TGTATGTAGG GA 2122