Note: Descriptions are shown in the official language in which they were submitted.
CA 02283497 1999-09-03
WO 98/39425 PCT/LTS98/04155
:L
NOVEL VECTORS AND EXPRESSION METHODS
FOR PRODUCING MiJTANT PROTEINS
BACKGROUND OF THE INVENTION
Field of the Invention
The invention relates to novel expression systems and
vectors for engineered immunot.oxins. More specifically
the invention relates to a shuatle vector for E. coli and
Corynebacteria. The invention. also relates to a method of
expressing engineered toxin mutants and toxin fusion
proteins in a mutant form of Pichia pastoris, a mutant
form Chinese hamster ovary cells, and mutant insect cells
and a methods for producing these mutants.
Background Art
U.S. Patent No. 5,167,956 describes in vivo T cell
killing of 3 logs by immunotoxin anti-CD3-CRM9 or
derivatives. This patent also describes the treatment of
graft versus host disease, autoimmune disease and T cell
leukemia.
A shuttle vector construct=ed for use in
Corynebacterium and E. coli mu;~t contain a replication
region (oriR) and a selectable marker that function in
both host bacteria, or contain an oriR functional in E.
coli and an chromosomal integrative mechanism functional
in Corynebacterium. A natural7_y occurring plasmid, pNG2,
isolated from an erythromycin-resistant Corynebacterium
strain fulfills the former criteria (3). However this
vector is large (14.4 kb), which reduces its
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
2
transformation frequency, which is additionally severely
compromised by restriction incompatibilities between
Corynebacterium and E. coli DNA (4). In addition multiple
cloning sites are not present in pNG2 which are required
to facilitate splicing inserts of toxin and fusion protein
toxin genes into the vector.
Thus, the invention meets an important need for an E.
coli and Corynebacteria shuttle vector with multiple
cloning sites.
SU1~1ARY OF THE INVENTION
The present invention describes a new shuttle vector
permitting the expression of engineered toxin mutants and
toxin fusion proteins in Corynebacterium. In addition the
invention provides a mutant Pichia pastoris, a method for
producing this mutant and a method of expressing
engineered toxin mutants and toxin fusion proteins in the
mutant form of Pichia pastoris. The invention further
provides a mutant Chinese hamster ovary (CHO) cell, a
method for producing this mutant and a method of
expressing engineered toxin mutants and toxin fusion
proteins in the mutant CHO cells. These three systems
have distinct advantages over E. coli expression systems
because of their higher yields of secretion into the media
compared to E. coli (1) thus eliminating the need for
refolding procedures from insoluble aggregates (2).
Refolding procedures employing denaturing agents although
somewhat successful for single chain fusion proteins will
not correctly refold divalent single chain fusion proteins
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
3
due to their greater complexity. Because divalent
immunotoxins are necessary far successful in vivo clinical
application, the present expression system for engineered
immunotoxins is the key to producing a successful
immunotoxin.
BRIEF DESCRIPTION OF THE FIGURES
Fig. 1 depict=s the method for producing an E.
coli/Corynebacterium shuttle vector from the plasmid pNG2.
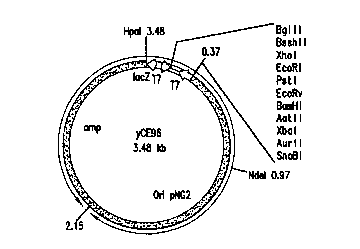
Fig. 2 depict=s the new E. coli/ Corynebacterium
shuttle vector, yCE96, cotaining nucleotides 1-3476 as
shown in SEQ ID NC>:1. Residues from positions 1 to 373
and 2153 to 3476 are from the vector LITMUS 29 and contain
the polycloning linker sites ;end the ampicillin resistance
marker respectively. Residue; from positions 374 to 2152
were the origin sequences from the plasmid pNG2.
Fig. 3a depicts a single chain divalent
antibody-mutant-toxin fusion protein produced in
Corynebacterium. The toxin i;; CRM9 and is preceded by the
CRM9 promoter and signal seque=nce. VL and VH linked by a
spacer described in U.S. Serial No. 08/739,703, herein
incorporated by reference, are from UCHT1. ~.CH3 and ~,CH2
are from human IgM. The fusion protein forms the
disulfide dimer from the cystE:ine between the CH2 and CH3
domains during or shortly after secretion. The gene for
this fusion protein is constructed by PCR overlap
extension to avoid cloning CRNI9 until its toxicity is
further reduced by a second genetic event, in this case an
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
4
additional carboxy terminal protein domain (NIH
guidelines, see reference 20).
Fig. 3b shows a double mutant of DT containing the
S525F mutation of CRM9 plus an additional replacement
within the 514-525 exposed binding site loop to introduce
a cysteine coupling site for example T521C can be produced
in Corynebacterium ulcerans preceded by the CRM9 promoter
and signal sequence. Residue numbering is based on the
sequence provided by Shen et al. (17). The double mutant
is made in Corynebacterium ulcerans by a recombination
event between the plasmid producing CRM9-antibody fusion
protein and PCR generated mutant DNA with a stop codon at
526. This CRM9-C's can be used to form specific thioether
mutant toxin divalent antibody constructs by adding excess
bismaleimidohexane to CRM9-C's and coupling to single
chain divalent antibody containing a free cysteine at
either the end of the ~CH4 domain or the ~.CH3 domain.
DETAILED DESCRIPTION OF THE INVENTION
Provided is an E. coli/Corynebacterium shuttle vector
comprising the origin of replication of shuttle vector
pNG2, polycloning linker sites, and an antibiotic
resistance marker with a size less than 4 kb. There are
multiple possibilities for the polylinker cloning site and
antibiotic resistance marker, which can be selected from
those known or later developed. An example of the vector
described above is yCE96. The sequence of yCE96 is given
below.
CA 02283497 1999-09-03
WO 98/39425
PCT/US98/04155
The vector can further comprise an insert consisting
of a protein-encoding nucleic acid. The encoded protein
can contain a disulfide bond. For example, the protein-
encoding nucleic acid can encode the binding site mutant
DT toxin, CRM9. Alternatively, the protein-encoding
nucleic acid can encode a CRMS~ further comprising a second
attenuating mutation. The second attenuating mutation can
be the insertion of a COOH terminal protein domain. The
second attenuating mutation can be a COOH terminal
IO mutation that reduces binding activity, but not
translocating activity. The second attenuating mutation
can be selected from the group consisting of S508F,
Y514A/C , K516A/C, V523A/C, N524A/C, K526A/C and F530A/C.
A protein encoding nucleic acid construct of the
invention can include a mutation that introduces a
cysteine residue into CRM9 or :its derivatives. For
example, the mutation can be selected from the group
consisting of K530C, K516C, D5:L9C and S535C.
A protein encoding nucleic: acid construct of the
invention can include a mutation that introduces residues
or the replacement of residues in CRM9 or its derivatives,
to attenuate the blocking effects of anti-diphtheria toxin
antibodies. Examples of these mutations are described in
the related applications.
A vector of the invention can further comprise the
CRM9 iron-independent promoter, wherein the protein-
encoding nucleic acid encodes a binding site mutant of
diphtheria toxin, and the protein-encoding nucleic acid is
CA 02283497 1999-09-03
WO 98/39425 PCT/US98l04155
6
under the control of the CRM9 iron-independent promoter
and is preceded by the CRM9 signal sequence.
A method of expressing a diphtheria toxin moiety or
other protein is provided. The expression method
comprises transfecting a Corynebacterium ulcerans or a
Corynebacterium diphtheriae cell with a vector of the
invention under conditions that permit expression of the
protein-encoding nucleic acid. The conditions required
for expression are the same as those previously used or
described herein, including limited iron in the medium.
A method of making a vector of the invention is
provided. For example the method comprises a) deleting
COOH terminal base pairs of the attenuated CRM9 toxin-
encoding nucleic acid using the restriction site Sph I at
the toxin nucleotide position 1523 and a restriction site
used to clone the COON terminal part of the toxin into the
polylinker cloning sites of yCE96, to produce a gapped,
linear, plasmid deleted in the COOH terminal coding
region; b) amplifying a product that corresponds to the
COON terminal region of CRM9 deleted in step a), with a
PCR primer that includes the desired mutation and 30-40
base pairs homologous to the down stream and upstream
regions adjacent to the deletion; c) purifying the
amplified product of step b) on an electrophoretic gel;
and d) electroporating the product of step c) into a
Corynebacterium along with the gapped plasmid of step a),
under conditions which permit homologous recombination to
occur intracellularly.
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
.7
A method of mutating a protein-encoding nucleic acid
in a vector of the invention :is provided. The method can
comprise a) deleting a region of the COOH terminus-
encoding nucleotides of the protein-encoding nucleic acid
using a unique restriction site and a restriction site
used to clone the COOH terminal part of the toxin into the
polylinker cloning sites of tree shuttle vector, to produce
a gapped, linear plasmid deleted in the COOH terminal
coding region is produced; b) amplifying a product that
corresponds to the COOH terminus-encoding region deleted
in step a), using a PCR primer that includes the desired
mutation and 30-40 base pairs homologous to the downstream
and upstream regions adjacent to the deletion; c)
purifying the amplified product of step b) on an
electrophoretic ge:l; and d) electroporating the purified
product of step c) into a Cor~.nebacterium along with the
gapped plasmid of step a), under conditions which permit
homologous recombination to occur intracellularly.
The Corynebacterium used in the methods of making the
present vectors or mutating proteins can be
Corynebacterium ulcerans. Alternatively, the
Corynebacterium can be a Corynebacterium diphtheriae,
which has been mutated by chemical mutagenesis to exhibit
less DNA restrictian. Any number of mutations can
accomplish this. The presence of the desired mutation is
measured by a reduction in restriction in the organism.
For example, this can be determined by measuring the
efficiency of transformation, i.e., if number of
transformants per /.cg of vector DNA increases over wild
type or other mutants.
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
8
A mutant strain of Pichia pastoris is provided. The
mutant strain comprises a mutation in at least one gene
encoding elongation factor 2 (EF2). This the mutation
comprises a Gly>Arg replacement at a position two residues
to the carboxyl side of the modified histidine residue
diphthamide. In this manner, the strain is made resistant
to the toxic ADP-ribosylating activity of diphtheria and
pseudomonas toxins.
l0 A method of expressing a diphtheria toxin protein
moiety or a pseudomonas exotoxin A toxin protein moiety is
provided. Such a method of the invention comprises
transfecting a mutated Pichia cell of the invention with a
vector comprising a toxin protein-encoding nucleic acid
under conditions that permit expression of the protein-
encoding nucleic acid in the cell. The conditions are
those used for Pichia cells and can be optimized for the
particular system.
In an expression method using a mutant Pichia strain,
the encoded protein is glycosylated in the cell to produce
of immunotoxins that are resistant to the blocking effects
of anti-diphtheria toxin antibodies on the T cell
depleting function of CRM9-containing immunotoxins in
human patients in vivo. There is a consensus sequence for
glycosylation (NXS/T), which may be removed or inserted to
control glycosylation which occurs in all eukaryotes,
e.g., Pichia.
A method of expressing a diphtheria toxin or a
pseudomonas exotoxin A toxin in Chinese hamster ovary
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
9
(CHO) cells, insect cells or ether eukaryote cells is
provided. The method can comprise first making a mutant
strain of cells, comprising a mutation in at least one
gene encoding elongation factor 2 (EF2), wherein.the
mutation comprises a Gly>Arg replacement at a position two
residues to the carboxyl side of the modified histidine
residue diphthamide, and the :train is resistant to the
toxic ADP-ribosylating activity of diphtheria and
pseudomonas toxins. This homologous recombination method
can be used in any eukaryote, due to the high conservation
of diphthamide in ekaryotes. Then the mutant cells are
transfected with a vector of a type appropriate for the
particular cell used, under conditions that permit
expression of protein-encoding nucleic acid in the cells.
The conditions can be those u~:ed for CHO cells and can be
routinely optimized for the particular system and cells
used.
As with the Pichia-expressed nucleic acids, the CHO
method produces a glycosylated protein. This can produce
immunotoxins that are resistant the blocking effects of
anti-diphtheria toxin antibodies on the T cell depleting
function of CRM9-containing immunotoxins in human patients
in vivo.
30
CA 02283497 1999-09-03
WO 98!39425 PCT/US98104155
EXAMPLES
Example 1
A Corynebacterium/Escherichia coli Shuttle Vector
5 for Gene Expression
In brief, a new shuttle vector is constructed using
the oriR of pNG2 and the antibiotic resistance marker and
multiple cloning sites of the vector Litmus p29 (New
10 England Bio Labs, Inc. Figure 1 depicts a method of
making of a vector according to the invention. The new
vector, yCE96, is only 3.4 kb in size and can transform
both E. coli and Corynebacteriurn ulcerans and, thus, can
be used to produce toxins and mutant toxins (5).
Cloning OriR from pNG2 plasmid:
pNG2 plasmid DNA was purified from E. coli JM109
strain. The 2.6 kb fragment containing oriR was released
from pNG2 plasmid by restriction digestion with EcoRI and
ClaI endonuclease. After separation and isolation, the
2.6 kb fragment was cloned into pBR322 vector (4361 bp) by
the same two restriction sites EcoRI and ClaI, to form a
construct of pBRNG with a size of 6.96 kb. This new
vector is of limited use, because of its relatively large
size and lack of multiple cloning sites.
Sub cloning pNG2 plasmid OriR to Generate a New Vector:
To generate a functional expression vector in
Corynebacterium with a small size and a multiple cloning
sites, the oriR DNA fragment, released from pBRNG
constructs, is combined with a DNA fragment borrowed from
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
L1
vector litmus p29. The DNA fragment containing the pNG2
oriR was released from pBRNG 'vector by endonucleases EcoRI
and SnaBI, and decreased in size from 2.6 kb to 1.77 kb by
cleavage with these enzymes. It is noted that the
sequence of the oriR is published, such that it could be
constructed without the pNG2 plasmid. Its nt numbers are
374-2152 in the sequence disc:Losed for yCE96. The DNA
fragment borrowed from litmus 29 vector was released by
restriction enzyme SnaBI and I)raIII, and contains an
ampicillin resistant gene and a multiple cloning sites.
The two DNA pieces could not be ligated together, because
the sticky end of the EcoRI site was not compatible with
the sticky end of DraIII. To turn the sticky end into a
blunted end, these DNA fragments were treated with T4
polymerase in the presence of 0.1 mM dNTP.
After digestion with restriction enzymes SnaBI and
DraIII, and treatment with T4 polymerase in the presence
of 0.1 mM dNTP, the litmus 29 vector DNA was
dephosphorylated with alkaline phosphatase to remove the
5' phosphate group. The recorr~bined vector was selected by
transformation of the ligation mixture into Novablue E.
coli cells. Five colonies were picked up. All of them
contained a plasmid vector. The restriction digestion
patterns of purified DNA from these five colonies
demonstrated that four colonies contained the correct
vector size of 3.4 kb.
The selection of endonuclease to release the oriR
from pBRNG was based on the nucleotide sequence of plasmid
pNG2 oriR identified by Messerotti et al.(6). The
CA 02283497 1999-09-03
WO 98/39425 PCT/LTS98/04155
12
replication region of pNG2 is 1854 bp, consists of a
single oriR ans one major open reading frame. When
digested with SnaBI, 75 nucleotides were deleted from 5'
end of the identified 1.85 kb oriR sequence. Therefore,
the oriR cloned into the new vector, yCE96, was 75
nucleotides smaller than the previously identified 1.85 kb
oriR sequence of plasmid pNG2, that is 1779 bp.
Transformation of Corynebacterium and E. coli by yCE96
E. coli cells were rendered competent by overnight
growth followed by resuspension in LB medium containing
10% PEG 8000, 5o DMSO and 50 mM MgCL2, pH 6.5. The cells
were heat shocked in the presence of 20 ng of vector DNA
per 106 cells. C. ulcerans were converted to protoplasts
as described (4) and transformed by electroporation of
40/.cg of vector DNA (4) .
Unless they have undergone transformation by yCE96,
no colonies of Corynebacterium ulcerans or E. coli grow up
under the selection pressure of the presence of
carbenicillin. Many colonies are formed after
transformation of E. coli cells and C. ulcerans with the
yCE96 vector on the LB plate with 0.1 mg/ml of
carbenicillin. The results demonstrate that yCE96 can
stably transform E. coli and C. ulcerans; thus it is an
effective shuttle vector between E. coli and C. ulcerans.
yCE96 is used to introduce foreign proteins into C.
ulcerans, for example, diphtheria toxin mutants made by
PCR site directed mutagenesis. To this end the CRM9
promoter carrying the iron-insensitive mutation (see U.S.
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
13
Serial No. 08/739,703, hereby incorporated by reference),
as well as the toxin signal sequence have been cloned from
CRM9 chromosomal DNA and precede mutant toxin constructs
and mutant toxin single chain antibody fusion proteins.
Thus, high level synthesis of these engineered proteins,
and their secretion into the medium, is expected.
Insertion of tandem repeats of these protein-encoding
constructs into yCE96 can also be made to achieve even
higher production levels. Several constructs and their
uses are detailed in Fig. 3 a~.ld Fig. 3 legend.
Example 2
Corynebacteriophage-based Vector
An alternate type of shuttle vector containing the
integrative mechanism of Corynebacteriophages can be
constructed by cloning the corynephage attachment site
(attP) and the integrase gene (int) sites from beta
Corynebacteriophages into small E. coli vectors such as
pUCl9) not capable of replicating in Corynebacteriae.
Addition of another protein coding sequence such as
modified CRM sequences (see below) will permit the
integration of these sequence: into tox-Corynebacteriae.
These sequences can be further modified by excision
followed by a gapped plasmid methodology described below
using a PCR product to achieve any desired mutation or
combination of mutations. An advantage of the integrative
vector is that it recombines at high efficiency.
CA 02283497 1999-09-03
WO 98139425 PCT/US98/04155
14
Example 3
Using Shuttle Vector yCE96 to Perform Site-Specific
Mutagenesis On Diphtheria Toxin Binding Site
Mutants in Corynebacteria
A binding site mutant of full length diphtheria toxin
residues 1-535 (16) S525F (17) is further modified for
chemical coupling by changing a residue in the binding
domain (residues 379-535) to cysteine. Preferred residues
are those with exposed solvent areas greater than 380.
These residues are K516, V518, D519, H520, T521, V523,
K526, F530, E532, K534 and 5535 (17). Of these, K516 and
F530 are presently preferred, since they are likely to
block any residual binding activity (17). However,
maximal coupling of the new cysteine residue will be
enhanced by the highest exposed solvent surface and
proximity to a positively charged residue (which has the
effect of lowering cysteine -SH pKa). These residues are
at D519 and S535, so that these are also preferred from
the above list of possibilities.
These mutations are accomplished by gapped plasmid
PCR mutagenesis (18) using the newly designed E. coli/C.
ulcerans shuttle vector yCE96 containing either the double
mutant DT S508F S525F or a CRM9 COOH terminus fusion
protein construct having reduced toxicity due to the COON
terminal added protein domain (19). Both of these
constructs follow current NIH guidelines for cloning DT
derivatives into E. coli (Federal Register, Notices,
May 7, 1986), Appendix F-II-B, p. 16971) in that they
contain two mutations which both individually diminish
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
toxicity and, therefore, greatly reduce the chance of
introducing a wild. type toxin into E. coli by a single
base pair reversion. This mui:ation is made by deleting
the COOH terminal 52 base pairs of the toxin construct
5 using the restriction site Sph I at the toxin nucleotide
position 1523 (16) and the re:~triction site used to clone
the COOH terminal part of the toxin into the polylinker
cloning sites of yCE96 (Xba or BamHI for example). Since
Sph I, Xba, and BamHI only occur singly within vector
10 yCE96 containing the inserted toxin construct, a gapped,
linear plasmid, deleted in thE~ COOH terminal coding region
is the result. Using PCR the COOH terminal region of CRM9
is rebuilt introducing the de~;ired mutation and including
30-40 base pairs homologous to the down stream and
15 upstream regions adjacent to the gap. The amplified
product is gel purified and electroporated into C.
ulcerans along with the gapped plasmid (18).
Recombination at the homologous regions occurs
intracellularly, accomplishing site specific mutagenesis
of DT products within Corynebacteriae which are not
specifically subject to NIH toxin cloning restrictions
(20) .
Using methods analogous to those described above, but
with different restriction enzymes, mutagenesis can be
performed anywhere within the toxin molecule. This would
be highly useful for the construction of toxin B chain
mutations having full translocating activity that are
relatively free fram the antitoxin blocking activity
variably present in the sera of patient populations
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
16
resulting from prior immunizations with diphtheria toxoid.
Because of the unique features of the present vector, a
virtually unlimited number of mutated proteins can be
expressed.
Example 4
Expression of mutant ADP-ribosylating toxins and toxin
fusion proteins in an EF2 mutant of Pichia pastoris
The invention provides a system for expressing mutant
ADP-ribosylating toxins and toxin fusion proteins in a
Pichia pastoris mutant. The presently preferred mutant is
one that has been rendered insensitive to these toxins by
substituting arginine for glycine at a position two amino
acids carboxyl to the modified histidine residue
diphthamide (position 701 in the EF2 gene based on the
numbering system in S. cerevisiae).
This mutation has been performed in S. cerevisiae and
prevents toxin induced ADP-ribosylation rendering the
toxin inactive (7). However, the literature has not
described or proposed the use of this mutation to generate
mutant cells for toxin production. To date, all DT based
mutant toxins generated by site directed mutagenesis have
been expressed in E coli. To date, toxin mutants
generated by the application of mutating reagents which
are not site specific have only been produced in C.
diphtheriae and E. coli.
CA 02283497 1999-09-03
WO 98/39425 PCT/LTS98/04155
1'7
Among the advantages of using this mutant P.ichia are
that for antibody toxin fusion. proteins, which must have
appropriate folding of critical antibody disulfide bonds,
the endoplasmic ret:iculum compartment of the eukaryote
yeast offers a similar oxidizing environment to natural
eukaryote antibody producing cells such as hybridomas.
Pichia pastoris has numerous advantages over S. cerevisiae
(8), including the observation that, for therapeutic
proteins, the glycosylation pattern of Pichia is much more
similar to that of humans compared to S. cerevisiae.
The Pichia mutant can be constructed by direct
transformation of yeast spherohlasts with a mutating
oligonucleotide complimentary to the sense strand of S.
cerevisiae EF2 in the region o7= the desired mutation but
not at the desired mutation (9;. Since homology in this
area is very high across phyla (10), this sequence will
very likely undergo homologous recombination in P.
pastoris. One example of such a mutating oligonucleotide
has the following sequence:
5-GTT ACT TTA CAT GCC GAT GCT ATC CAC AGA B,Qg GGT GGT CAA
ATC ATC CCA-3 (SEQ ID N0:2)
The arginine substitution has been underlined. This
nucleic acid can be modified, for example, by shortening
on both ends, but this may result in reduced recombination
frequency.
The transformation is done in the presence of 10 ~.M
wild type diphtheria toxin or a binding site mutant DT
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
18
toxin, such as CRM9, as a selecting agent (11). Several
repeat rounds of selection can be utilized. The final
selection is performed by transforming Pichia with a
toxin-containing construct, which will inhibit growth in
any cells which lack toxin-resistance mutation in EF2.
The invention also provides a Pichia EF2 toxin-
resistant clone, which is free of toxin-encoding
constructs, to be used for to express alternative
constructs. This cell can be generated by popping out the
construct via homologous recombination (12).
Many Pichia pastoris strains and E. coli/Pichia
shuttle vectors are available (Invitrogen Corporation),
and can be used in an expression system as described
above. This technology will be useful for diphtheria
toxin and pseudomonas exotoxin A constructs and fusion
immunotoxins based on these toxins. The same mutant
toxins and toxin-antibody fusion proteins produced in
Corynebacterium shown in Fig 2 can also be produced in P.
pastoris. The genetic constructs differ in that in P.
pastoris the promotor is AOXI and the secretion or signal
sequence is either PHO1 or @-factor. In addition certain
codons unique to Corynebacterium may require changing in
P. pastoris, as will some codons specifying potential
glycosylation sites which would be active in Pichia but
inactive in Corynebacterium. Alternatively, the
introduction of glycosylation sites within the CRM9 gene
could be used as a method to block antibodies directed at
the toxin in patients with high antitoxin titers secondary
to recent immunization with diphtheria toxoid.
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
19
Example 5
Expression of mutant ADP-rilbosylating toxins and toxin
fusion proteins in an E:F2 mutant of CHO cells.
CHO cells have certain advantages for the production
of fusion proteins used as therapeutic reagents: a)
Hamster cells are relatively free from retrovirus
contamination, b) Protein glycosylation patterns are
relatively similar to that seen in humans, and c) CHO
cells generally, respond well to gene amplification
systems that increase the yield of proteins introduced
through DNA transfection. Although it has been reported
that mammalian cells can secrete an ETA based fusion
protein without succumbing to ETA induced cytotoxicity via
ADP-ribosylation (13), this situation is likely due to the
resistance of ETA to proteolytic processing at pH >6.0, a
feature not shared by DT. Therefore the production of DT
based fusion immunotoxins must utilize a line of CHO cells
that has been rendered DT insensitive by mutating EF2 by
substituting arginine for glyc.ine at a position two amino
acids carboxyl to the modified histidine residue
diphthamide (position 717 in the EF2 gene based on the
numbering system in CHO cell E1~2). Two such cell lines
have been reported. RE1.22c (:L4) and KEE1 (15) were
isolated by double rounds of chemical mutagenesis and
selection with DT or ETA. There mutants were used to
determine the physiologic role of dipthamide, the modified
histidine residue whose format_Lon is blocked by this
mutation. No mention is made un the art of using these
cells to make toxins or immunot:oxins. The same procedure
can be applied to other eukaryote cell lines expressing
CA 02283497 1999-09-03
WO 98/39425 PCTIUS98/04155
toxin sensitivity, such as insect cell lines. These lines
have the advantage of secreting large amounts of foreign
proteins introduced through baculovirus (21).
5 Throughout this application various publications are
referenced by numbers within parentheses. Full citations
for these publications are as follows. The disclosures of
these publications in their entireties are hereby
incorporated by reference into this application in order
10 to more fully describe the state of the art to which this
invention pertains.
REFERENCES
1. Greenfield, L., Targeted Diagnosis and Therapy,
Chapter 16, p. 307; 1992.
2. vanderSpek, J. C., Mindell, J. A., et al. J. Biol.
Chem. 268:12077-82, 1993.
3. Serwold-Davis, T. M., Groman, N. B., et al. FEMS
Microbiology Letters. 66:119-124, 1990.
4. Serwold-David, T. M., Groman, N. B., et al. Proc.
Natl. Acad. Sci. 84:4964-4968, 1987.
5. Groman, N., Schiller, J., et al. Infection and
Immunity 45;511-517, 1984.
6. Messerotti, L. J., Radford, A. J. et al. Gene
96:147-8, 1990.
CA 02283497 1999-09-03
WO 98/39425 PCT/LTS98/04155
21
7. Kimata, Y., Harashima, S., et al. Biochemical and
Biophysical Research Communications 191:1145-1151, 1993.
8. Buckholz, R. G., and Gleeson, M. A. G.
Bio/Technology. 9:1067, 1991..
9. Moerschell, R. P., Das, G., et al. Methods in
Enzymology. 194:362, 1991.
10. Perenetesis, .J. P., Phan, L. D., et al. J. Biol.
Chem. 267:1190-1197, 1992.
11. Murakami, S., Bodley, J. W., et al. Molecular and
Cellular Biology :?:588-592, 1982.
12. Rothstein, R. Methods in Enzymology 194:281, 1991.
13. Chen, S. Y. et al. Nature 385(2):78, January 2, 1997.
14. Foley, B.T. et al. J. Biolog. Chem. 370(39}:23218-
23225, September 29, 1995.
15. Kohno, K. and Uchida, T. J. Biolog. Chem.
262(25):12298-12305, 1987.
16. Kaczorek M, Delpeyroux F, Chenciner N, Streeck R
(1983) Science; 221:855.
17. Shen WH, Choe S, Eisenberg D, Collier RJ (1994) .
Biol. Chem.; 469(46):29077-29084.
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
22
18. Muhlrad D, Hunter R, Parker R (1992) Yeast; 8:79-82.
19. Madshus IH, Stenmark H, Snadvig K, Olsnes S (1991) J.
Biol. Chem.; 266(26):17446-53.
20. Federal Register, Notices, May 7, 1986), Appendix
F-II-B, p. 16971.
21. Bei, R, Schlom, J and Kashmiri, SVS (1995) J.
Immunol. Meth.; 186:245-255.
CA 02283497 1999-09-03
WO 98/39425 PCT/US98/04155
23
SEQUENCE LISTING
(1) GENERAL INFORMATION
(i) APPLICANT: The Government of the United States of America, as
represented by the Secretary, Department of Health
and Human Services
(ii) TITLE OF THE INVENTION: NOVEL VECTORS AND EXPRESSION
METHODS FOR PRODUCING MUTANT PROTEINS
(iii) NUMBER OF SEQUENCES: 2
(iv) CORRESPONDENCE ADDRESS:
{A) ADDRESSEE: NEEDLE & ROSENEiERG, P.C.
(B) STREET: 127 Peachtree Street, N.E., Suite 1200
(C) CITY: Atlanta
(D) STATE: GA
(E) COUNTRY: USA
(F) ZIP: 30303-1811
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Diskette
(B) COMPUTER: IBM Compatible
(C) OPERATING SYSTEM: DOS
(D) SOFTWARE: FastSEQ for Windows Version 2.0
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE: 05-MAR-1998
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA:
{A) APPLICATION NUMBER: 60/037,196
(B) FILING DATE: 05-MAR-1997
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: Spratt, Gwendolyn D.
(B) REGISTRATION NUMBER: 36,016
(C) REFERENCE/DOCKET NUMBER: 14014.0286/P
(ix) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: 4U4 688 0770
(B) TELEFAX: 404 688 9880
(C) TELEX:
(2) INFORMATION FOR SEQ ID NO::1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3476 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
CA 02283497 1999-09-03
WO 98/39425 PCT/LTS98104155
24
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:1:
AAAAAAAAGCCCGCCGAAGCGGGCTTTATTACCAAGCGAAGCGCCATTCGCCATTCAGGC60
TGCGCAACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGA120
AAGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGAC180
GTTGTAAAACGACGGCCAGTCCGTAATACGACTCACTTAAGGCCTTGACTAGAGGGAAGA240
TCTGGATGCATTCGCGCGCACGTACGGTCTCGAGGAATTCCTGCAGGATATCGTGGATCC300
AAGCTTCACCATGGGAGACGTCACCGGTTCTAGAACCTAGGGAGCTCTGGTACCCACTAG360
TGAGTCGTATTACGTAACCGCAGGTAAAAGGCATATTTTTCGCGTGTCATGGCTAGTAAA420
TAACACCGGTGTCATTTAGAGTCAGGGAAAGACAATGAAAAACGAAGAAAGCCACCGGGC480
GGCAACCCGATGACTTTCGCTTATCACCCAGCACACACCTGGGAGAAATCACGGTCATGA540
GTTTACAGACTCATGCGCAGAATGCGCACACTAAAACACCTACCCGCGTCGAGCGCGACC600
GTGGTGGACTGGACAACACCCCAGCATCTGCCAGTGACCGCGACCTTTTACGCGATCATC660
TAGGCCGCGATGTACTCCACGGTTCAGTCACACGAGACTTTAAAAAGGCCTATCGACGCA720
ACGCTGACGGCACGAACTCGCCGCGTATGTATCGCTTCGAGACTGATGCTTTAGGACGGT780
GCGAGTACGCCATGCTCACCACCAAGCAGTACGCCGCCGTCCTGGTCGTAGACGTTGACC840
AAGTAGGTACCGCAGGCGGTGACCCCGCAGACTTAAACCCGTACGTCCGCGACGTGGTGC900
GCTCACTGATTACTCATAGCGTCGGGCCAGCCTGGGTGGGTATTAACCCAACTAACGGCA960
AAGCCCAGTTCATATGGCTTATTGACCCTGTCTACGCTGACCGTAACGGTAAATCTGCGC1020
AGATGAAGCTTCTTGCAGCAACCACGCGTGTGCTGGGTGAGCTTTTAGACCATGACCCGC1080
ACTTTTCCCACCGCTTTAGCCGCAACCCGTTCTACACAGGCAAAGCCCCTACCGCTTATC1140
GTTGGTATAGGCAGCACAACCGGGTGATGCGCCTTGGAGACTTGATAAAGCAGGTAAGGG1200
ATATGGCAGGACACGACCAGTTCAACCCCACCCCACGCCAGCAATTCAGCTCTGGCCGCG1260
AACTTATCAACGCGGTCAAGACCCGCCGTGAAGAAGCCCAAGCATTCAAAGCACTCGCCC1320
AGGACGTAGACGCGGAAATCGCCGGTGGTCTCGACCAGTATGACCCGGAACTTATCGACG1380
GTGTGCGTGTGCTCTGGATTGTCCAAGGAACCGCAGCACGCGACGAAACAGCCTTTAGAC1440
ATGCGCTTAAGACTGGCCACCGCTTGCGCCAGCAAGGCCAACGCCTGACAGACGCAGCAA1500
TCATCGACGCCTATGAGCACGCCTACAACGTCGCACACACCCACGGCGGTGCAGGCCGCG1560
ACAACGAGATGCCACCCATGCGCGACCGCCAAACCATGGCAAGGCGCGTGCGCGGGTATG1620
TCGCCCAATCCAAGAGCGAGACCTACAGCGGCTCTAACGCACCAGGTAAAGCCACCAGCA1680
GCGAGCGGAAAGCCTTGGCCACGATGGGACGCAGAGGCGGACAAAAAGCCGCACAACGCT1740
GGAAAACAGACCCCGAGGGCAAATATGCGCAAGCACAAAGGTCGAAGCTTGAAAAGACGC1800
ACCGTAAGAAAAAGGCTCAAGGACGATCTACGAAGTCCCGTATTAGCCAAATGGTGAACG1860
ATCAGTATTTCCAGACAGGGACAGTTCCCACGTGGGCTGAAATAGGGGCAGAGGTAGGAG1920
TCTCTCGCGCCACGGTTGCTAGGCATGTCGCGGAGCTAAAGAAGAGCGGTGACTATCCGG1980
ACGTTTAAGGGGTCTCATACCGTAAGCAATATACGGTTCCCCTGCCGTTAGGCAGTTAGA2040
TAAAACCTCACTTGAAGAAAACCTTGAGGGGCAGGGCAGCTTATATGCTTCAAAGCATGA2100
CTTCCTCTGTTCTCCTAGACCTCGCAACCCTCCGCCATAACCTCACCGAATTGTGGGCCA2160
TCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGA2220
CTCTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGGCTATTCTTTTGATTTATAA2280
GGGATTTTGCCGATTTCGGCCTATTGGTTAAAAAATGAGCTGATTTAACAAAAATTTAAC2340
GCGAATTTTAACAAAATATTAACGTTTACAATTTAAATATTTGCTTATACAATCTTCCTG2400
TTTTTGGGGCTTTTCTGATTATCAACCGGGGTAAATCAATCTAAAGTATATATGAGTAAA2460
CTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTAT2520
TTCGTTCATCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCT2580
TACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGACCCACGCTCACCGGCTCCAGATT2640
TATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAACTTTAT2700
CCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTA2760
ATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTG2820
GTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCCCCATGT2880
TGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCCG2940
CAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCG3000
CA 02283497 1999-09-03
WO 98139425 PCT/US98/04155
TAAGATGCTT TTCTGTGACT GGTGAGTACT CAACCP,AGTC ATTCTGAGAA TAGTGTATGC 3060
GGCGACCGAG TTGCTCTTGC CCGGCGTCAA CACGGG%ATAA TACCGCGCCA CATAGCAGAA 3120
CTTTAAAAGT GCTCATCATT GGAGAACGTT CTTCGGGGCG AAAACTCTCA AGGATCTTAC 3180
CGCTGTTGAG ATCCAGTTCG ATGTAACCCA CTCGTGCACC CAACTGATCT TCAGCATCTT 3240
TTACTTTCAC CAGCGTTTCT GGGTGAGCAA AAACAGGAAG GCAAAATGCC GCAAAAAAGG 3300
GAATAAGGGC GACACGGAAA TGTTGAATAC TCATACTCTT CCTTTTTCAA TATTATTGAA 3360
GCATTTATCA GGGTTATTGT CTCATGAGCG GATACATATT TGAATGTATT TAGAAAAATA 3420
AACAAATAGG GGTTCCGCGC ACATTTCCCC GAAAAGTGCC ACCTGACGTA GTTAAC 3476
(2) INFORMATION FOR SEQ ID N0:2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 51 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:2:
GTTACTTTAC ATGCCGATGC TATCCACAGA AGAGGTGGTC AAATCATCCC A 51