Note: Descriptions are shown in the official language in which they were submitted.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98114215
Modified Lepidopteran Receptors and Hvbrid
Multifunctional Proteins for Use in Regulation of
Transaene Expression
ACKNOWLEDGMENT
This invention was made with Government support under
. Grant No. AG 10435, awarded by the National Institutes of
Health. The Government has certain rights in the
invention.
RELATED APPLICATIONS
This application claims priority from Provisional
Application, U.S.S.N. 60/ , filed July 7, 1998, and
also claims priority from United States Serial No.
08/891,298, filed July 10, 1997, now pending, the entire
contents of both of which are hereby incorporated by
reference herein.
FIELD OF THE INVENTION
The present invention relates to methods in the field
of recombinant DNA technology, and products related
thereto. In a particular aspect, the invention relates to
methods for modulating the expression of exogenous genes in
mammalian systems, and products useful therefor.
BACKGROUND OF THE INVENTION
In the field of genetic engineering, precise control
of gene expression is an invaluable tool for studying,
manipulating and controlling development and other
_ physiological processes. For example applications for
regulated gene expression in mammalian systems include
inducible gene targeting, overexpression of toxic and
teratogenic genes, anti-sense RNA expression, and gene
CA 02296093 2000-O1-10
WO 99102683 PCTIUS98I14215
2
therapy (see, for example, Jaenisch, R. (1988) Science 240,
1468-1474). For cultured cells, glucocorticoids and other
steroids have been used to induce the expression of a
desired gene.
As another means for controlling gene expression in
mammalian systems, an inducible tetracycline regulated
system has been devised and utilized in transgenic mice,
whereby gene activity is induced in the absence of the
antibiotic and repressed in its presence (see, e.g, Gossen
et al. (1992) PNAS 89, 5547-5551; Gossen et al.(1993) TIBS
18, 471-475; Furth et al. (1994) PNAS 91, 9302-9306; and
Shockett et al. (1995) PNAS 92, 6522-6526). However,
disadvantages of the inducible tetracycline system include
the requirement for continuous administration of
tetracycline to repress expression and the slow clearance
of antibiotic from bone, which interferes with regulation
of gene expression. While this system has been improved by
the recent identification of a mutant tetracycline
repressor which acts conversely as an inducible activator,
the pharmacokinetics of tetracycline may hinder its use
during development when a precise and efficient "on-off"
switch is essential (see, e.g., Gossen et al. (1995)
Science 268, 1766-1769).
Historically, the expression of transgenes with
potential pathogenic and cytotoxic properties has been
difficult in the context of stable cell lines since cells
chronically producing significant levels of the toxic
transgene generally die from the cytopathic effects. For
example, CAG repeats and concomitant polyglutaminel(polyQ)
expression has been linked to a variety of
neurodegenerative conditions, including Hungtington's
disease (Koide et al. (1994) Nat Genet 6:9-13),
dentatorubro-pallidoluysian atrophy (Ranum et al. (1994)
CA 02296093 2000-O1-10
WO 99/02683 PCTJUS98/14215
3
Nat Genet 8:280-284), spinocerebellar ataxias (Kawaguch et
al. (1994) Nat Genet 8:221-228, Imbert et al. (1993) Nat
Genet 4:72-76, Sanpei et al. (1996) Nat Genet 14:277-284,
and Zhuchenko et al. (1997) Nat Genet 15:62-69), and
~ 5 spinobulbar muscular atrophy (David et al. (1997) Nat Genet
17:65-70). PolyQ expansion has also been linked to the
' formation of intracellular protein aggregates in vivo in
transgenic mice expressing human huntingtin fragments with
expanded repeats (Ordway et al (1997) Cell 91:753-763),
brain tissue from Huntington's patients (DiFiglia et al.
(1997) Science 277:1990-1993) and transiently transfected
cultured cells (Martindale et al (1998) Nat Genet 18:150-
154 and Cooper et al. (1998) Hum Mol Genet 7:783-790).
In transient transfection models, a 79-Q expanded
repeat from the ataxia 3 gene has been shown to form a
"punctate pattern" within the cytoplasm, and to result in
the apopotic cell death of 70-80% of transfected COS-7
cells within 48 hours of transfection. Paulson et al. has
reported that 293 cells transfected with a separate 78-Q
ataxia-3 derived construct acquired intranuclear inclusion
bodies and also underwent apoptotic cell death in a subset
(300) of the transfected population with 72 hours of
transfection. Neuroblastoma cells transiently transfected
with an 82-Q construct derived from an expanded huntingtin
gene exon 1 were found to be 2-3 times more susceptible to
staurosporine induced apoptosis than cells transfected with
a LacZ control construct (18), however, spontaneous cell
death was not described. Variations in culture techniques,
transfection efficiencies, and cell lines, however, make it
difficult to rigorously characterize the progression of
polyQ expression, intracellular aggregate (IA) formation,
- and cell death. One impediment to the comparative study of
IAs in vitro is that transient transfection studies predict
~ that toxicity from polyQ over-expression will prevent the
isolation of stable cultured cells.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
4
Accordingly, there is a need in the art for improved
systems to precisely modulate the expression of exogenous
genes in mammalian subjects. For example, a non-
mammalian-based transcription regulating system would be
desirable for general application to transgene regulation
in in vitro, ex vivo, and in vivo applications, as well as
transgenic animals. Such systems would be extremely
desirable to conditionally express pathogenic or toxic
compounds in order to produce stable cells. A system that
is simple, compact and dependent on ligands which are
relatively inexpensive, readily available and of low
toxicity in animals would prove useful for stimulation of
regulated systems.
BRIEF DESCRIPTION OF THE INVENTION
In accordance with the present invention, it has been
discovered that nuclear receptor proteins isolated from the
silk moth bombyx mori (bR) are useful for the regulation of
transgene expression. bR is generally thought to be a
strong transcriptional regulator within cells of the silk
moth. In accordance with the present invention, it has
been discovered that bR is also functional in mammalian
cells without the addition of exogenous dimer partners
therefor. It has further been discovered that the addition
of activation domains to the bR open-reading frame (VbR)
enhances the activity of the ligand modulated regulator to
afford high-level transcriptional induction (see, e.g.,
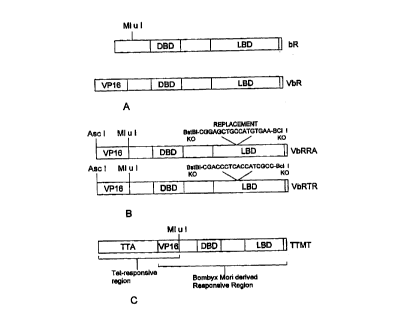
Figure lA) . Further modifications to the bR ligand binding
domain result in receptors with unique transactivation
characteristics (see, e.g., Figure 1B).
In accordance with another aspect of the present
invention, hybrid proteins produced by fusion of modified
bRs with other ligand-regulated proteins have been found to
be capable of high level, regulated transactivation of
CA 02296093 2000-O1-10
WO 99102683 PCTIUS98I14215
sequences controlled by both hormone response elements and
tetracycline operators. VbR variants and hybrid proteins
(see, e.g., Figure 1C), in combination with the appropriate
promoters and transgenes, can be introduced into target
5 cells by common methods such as transfection of plasmids or
by virus mediated gene transfer. The small size and
simplicity of these proteins makes them particularly
attractive for use in retroviral vectors.
Invention receptor peptides have proven capable of
providing a high degree (>100 fold) of ligand-dependent
transgene expression in a variety of infected and
transfected cell types including primary cell types. In
addition, several transgenes including luciferase, LacZ,
human GH, GFP, tyrosine hydroxylase, and the low-affinity
NGF receptor, have all been shown to function in a
ligand-dependent fashion in cells infected with invention
constructs.
Invention methods provide for regulated gene
expression by exogenous non-mammalian inducers, and
therefor can be advantageously employed in a variety of in
vivo and in vitro mammalian expression systems. For
example, inducible expression of flp recombinase in
transgenic mammals, in accordance with invention methods,
would enable those of skill in the art to accomplish
temporally specific inducible gene targeting of the adult
or the developing embryo (See, e.g., U.S. Patent Nos.
5,654,182 and 5,677,177). Stable cell lines conditionally
expressing desired genes provide a valuable tool for the
study and treatment of diseases. In order to maximize the
chance of obtaining stably transfected/infected cultured
" cells expressing a desired transgene, such as polyglutamine
expression cassette (PQEC), a retroviral construct is
' provided in which expression of the transgene is controlled
by an exogenous ligand.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
6
For example, in order to study possible mechanisms
underlying Huntington's disease, cultured cell types that
conditionally express the glutamine (CAG) repeats
characteristic of the pathological form of the huntingtin
protein are produced. Two aspects of polyQ-mediated
cytotoxicity are well-suited to characterization in vitro;
the expression, distribution, and aggregation of proteins
with expanded polyQ tracts within individual cells, and the
progression of polyQ expression to death of the cell. The
huntingtin protein is expressed predominantly in the
cytoplasm of both neuronal and non-neuronal cell types.
The expression of polyglutamine tracts separately from the
huntingtin protein will allow one to determine what toxic
properties, if any, polyglutamine tracts themselves may
harbor. A variable length polyglutamine fused to cytosolic
reporter proteins such as green fluorescent protein (GFP)
facilitates the selection and characterization of PQEC
ligand-responsive cell populations and clonal lines. The
use of recombinant retroviruses as PQEC transfer vectors
will allow the stable integration of the transgene into a
wide variety of cultured cell types.
BRIEF DESCRIPTION OF THE FIGURES
Figure lA provides a schematic drawing of the
nuclear receptor protein isolated from the silk moth
bombyx rnori (bR), as well as a construct containing a
VP16 activation domain fused at an internal MluI site
near the N-terminus of the bombyx mori nuclear receptor,
referred to as VbR.
Figure 1B depicts the replacement of several amino
acids of the ligand binding domain of the bombyx mori
nuclear receptor with sequences from the retinoic acid
CA 02296093 2000-O1-10
WO 99/02683 PCTIUS98114215
7
receptor or the thyroid hormone receptor (VbRRA and
VbRTR, respectively).
Figure 1C provides a schematic of a hybrid
transactivating construct, designated TTMT, containing a
tetracycline-responsive transactivator (Tet-responsive
region), a VP16 activation domain and a peptide derived
from bombyx mori.
Figure 2A shows the induction of fusion proteins
(VP16 activation domain fused to either Drosophila
ecdysone receptor (dVEcR) or bombyx mori nuclear receptor
derived(VbR)) with 5 ~g/ml tebufenozide in the absence of
exogenous dimer partners.
Figure 2B shows that a peptide derived from bombyx
mori responds to tebufenozide (teb) with greater than
200-fold induction, while Drosophila EcR responds weakly,
it at all.
Figure 2C illustrates the action of tebufenozide
(teb) on the expression of EIRE-Luc reporter when co-
transfected with the following receptors:
LINX: which encodes the tet-transactivator (TTA),
VbR: which encodes VbR,
TTMT: which encodes the TTA-VbR fusion protein, and
TTMT-2V: similar to TTMT but with two VP16 t-domains.
Figure 2D depicts the same experiment illustrated in
Figure 2C, utilizing a TetO-luc reporter, which responds
only to TTA or fusion proteins in the absence of
doxycycline.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
8
Figure 2E shows the results of an experiment
employing CV-1 cells transfected with TTMT and two
reporters: 6Tet0-luc and 4-EIRE-~3-galactosidase.
Figure 3 shows that lower levels of teb are as
effective as higher levels for stimulating full activity.
Figure 4 is a schematic drawing of representative
single-plasmid constructs involving VbR use.
Figure 5 illustrates the construction of the pBW
plasmid.
Figure 6A provides a comparison of Bombyx mori
ecdysone receptor (BE) and Drosophila melanogaster ecdysone
receptor (DE) by transient transfection with both receptors
and the dimer partners Usp or RXR in 293 cells.
Figure 6B provides a similar comparison as in Figure
&A, however, in CV-1 cells.
Figure 7A is a schematic drawing of native and
chimeric ecdysone receptors.
Figure 7B is a graph depicting the transient
transfection of VEI-i into CV-1 cells.
Figure 8 is a schematic drawing comparing several
different chimeric receptors.
Figure 9A provides a schematic of receptor variant
viral vectors and an ecdysone response element responsive
reporter virus (MS).
CA 02296093 2000-O1-10
WO 99102683 PCT/US98114215
9
Figure 9B illustrates induction levels of luciferase
in 293, CV-1 and primary fibroblast cells infected with MS-
luc virus.
Figure 10 illustrates the construction of the Boris I
plasmid.
Figure 11 illustrates the construction of the Boris
III plasmid.
Figure 12 illustrates the construction of the Boris IV
plasmid.
Figure 13 illustrates the construction of the pBW-6E-
GFP-CB-VBE plasmid.
Figure 14 illustrates the retroviral construct, BORIS
III transcribes a transgene in the presence and absence of
a ligand.
Figures 15A and B illustrate the construction of a
PolyQ expression construct.
Figure 16 provides a comparison of various polyQ-GFP
vectors constructed.
DETAILED DESCRIPTION OF THE INVENTION
In accordance with the present invention, there are
provided nucleic acid constructs encoding a receptor
peptide comprising:
(i) a ligand binding domain and hinge region of
a non-mammalian member of the nuclear
receptor superfamily,
CA 02296093 2000-O1-10
WO 99102683 i'CT/US98114215
(ii) a DNA binding domain, and
(iii) an activation domain,
wherein the receptor peptide activates a regulatory element
in the absence of an exogenous dimer partner therefor and
5 in the presence of a ligand for said ligand binding domain.
Alternatively, there are provided nucleic acid constructs
encoding a receptor peptide comprising:
(i) a ligand binding domain and hinge region of a
non-mammalian member of the nuclear receptor
10 superfamily, and
(ii) a DNA binding domain, wherein the DNA
binding domain is not obtained from the same
member as the ligand binding domain and
hinge region,
wherein the receptor peptide activates a regulatory
element in the absence of an exogenous dimer partner
therefor and in the presence of a ligand for said ligand
binding domain.
In accordance with another embodiment of the present
invention, there are provided methods) employing such
constructs. For example, there are provided methods) for
modulating the transcription of exogenous nucleic acids)
in a host containing:
(i) a nucleic acid construct comprising a promoter
and said exogenous nucleic acids) under the
control of a regulatory element; and
(ii) a receptor peptide comprising a DNA binding
domain, and the ligand binding domain and hinge
region of a non-mammalian member of the nuclear
receptor superfamily which is not normally
present in the cells of said host, wherein said
receptor peptide activates said regulatory
element in the absence of an exogenous dirner
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
11
partner therefor and in the presence of a ligand
for said ligand binding domain,
said method comprising administering to said host an
amount of ligand effective to modulate the transcription of
said exogenous nucleic acid(s); wherein said ligand is not
normally present in the cells of said host.
As employed herein, the terms "modulate" and
"modulating" refer to the ability of a given
ligand/receptor peptide complex to activate/deactivate
and/or up-regulate/down-regulate transcription of exogenous
nucleic acids, relative to the transactivation activity of
said receptor peptide in the absence of ligand. The actual
effect of complex formation on the transactivation activity
of a receptor peptide will vary depending on the specific
ligand and DNA binding domains employed in the receptor
peptide and on the regulatory element with which the
ligand/receptor peptide complex interacts.
As employed herein, the term "host" refers to the
cell, tissue, organ or organism in need of transcriptional
regulation of exogenous or endogenous nucleic acids.
Preferably, hosts are mammalian or mammalian derived cells
or tissue. Exemplary mammals include: humans; domesticated
animals, e.g., rat, mouse, rabbit, canine, and feline; farm
animals, e.g., chicken, bovine, porcine and ovine; and
animals of zoological interest, e.g., monkey and baboon;
and the like.
As employed herein, a "dimer partner" refers to
members of the nuclear receptor superfamily to which other
members preferentially bind to form heterodimeric species.
For example, members of the nuclear receptor superfamily
preferentially form heterodimers with a common partner, the
retinoid X (or 9-cis retinoic acid) receptor (RXR, see, for
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
12
example, Yu et al. (1991) Cell 67:1251-1266; Bugge et al.
(1992) EMBO J. 11:1409-1418; Kliewer et al. (1992) Nature
355:446-449; Leid et al, (1992) Cell 68:377-395; Marks et
al. (1992) EMBO J. 11:1419-1435; Zhang et al. (1992) Nature
355:441-446; Issemann et al. (1993) Biochimie. 75:251-256).
Additional dimer partners for members of the nuclear
receptor superfamily include ultraspiracle (Usp), farnesoid
X receptor (FXR), and the like.
Receptor peptides utilized in the present invention
are characterized as being fully functional in mammalian
cells without the addition of any exogenous dimer partners
therefor. For such receptor peptides, the presence of
native level or concentration of endogenous dimer partner
is suf f icient to promote transcription . As employed herein,
the phrase "exogenous dimer partner" refers to a dimer
partner for the receptor peptide that is introduced into
the host. Exogenous dimer partner may be employed because
the dimer partner is not present in the host cell (i.e.,
Usp) or because the dimer partner is required at elevated
levels in the host (i.e., higher levels of RXR).
Oppositely, the phrase "endogenous dimer partner(s)" refers
to dimer partners which are native to the unmodified
mammalian host cell. Invention receptors are advantageous
because dimer partner is not necessary at elevated levels
in order for formation of heterodimers to occur, thereby
promoting transcription.
Members of the nuclear receptor superfamily are
characterized by the presence of five domains: A/B, C, D,
E and F (Evans, R. Science 240:889-895 (1988)), wherein "C"
corresponds to the DNA binding domain, "D" corresponds to
the hinge region, and "E" corresponds to the ligand binding
domain. In accordance with a particular aspect of the
invention, it has been discovered that the hinge region
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
13
(D), in coordination with the ligand binding domain (E), of
certain non-mammalian members of the nuclear receptor
superfamily enable such members to interact with dimer
partners endogenous to mammalian cells, i.e., in the
absence of and without requiring introduction of exogenous
dimer partners. Specifically, it has been identified that
regions within the hinge region ("D") and the ligand
binding domain ("E") of the bombyx mori receptor function
in concert to produce high affinity, ligand-dependent
heterodimerization between bR and mammalian dimer partners,
without requiring the introduction of exogenous dimer
partners such as Usp.
Thus, in accordance with a preferred embodiment of the
present invention, the invention receptor peptide comprises
the hinge region and ligand binding domain derived from
insect receptors of lepidopteran species such as bombyx
mori, (Swevers et al. Insect Biochem. Moles. Biol.
(7) :857-866 (1995) ) , Choristoneura fumiferana (Palli et
al. Insect Biochem. Moles. Biol. 26(5):485-499 (1996)),
20 Manduca sexta (Fujiwara et al. Insect Biochem. Moles. Biol.
25(7):845-856 (1995)), Aedes aegypti (Cho et al. Insect
Biochem Moles. Biol. 25:19-27 (1995), Chorinomus tentans
(Imhof et al. Insect Biochem. Moles. Biol. 25:115-124
(1993), and the like. Presently preferred insect receptors
25 from which the hinge region and/or ligand binding domain is
derived substantially lack the C-terminal "F" domain. In
addition, presently preferred insect receptors share less
than 50% sequence similarity with Drosophila ecdysone
receptors, comparisons performed by Higgins and Sharp
sequence alignment.
As employed herein, the term "hinge region" or "D
domain" refers to the domain located between the DNA
binding domain and the ligand binding domain of intact
CA 02296093 2000-O1-10
WO 99/02683 PCT1US98/14215
14
members of the nuclear receptor superfamily. In accordance
with a preferred embodiment of the present invention, the
hinge region facilitates high affinity interaction with
dimer partners endogenous to mammalian cells, also referred
herein as "endogenous dimer partners." Preferably, the
hinge region of a non-mammalian member of the nuclear
receptor superfamily can be characterized as any sequence
having substantial sequence identity with amino acid
residues 273-362 set forth in SEQ ID N0:3, or substantial
portions thereof which confer sufficiently high affinity
for endogenous dimer partner.
The hinge region can be functionally located in either
orientation and at various positions within the receptor
peptide. For example, the hinge region can be positioned
at either the amino or carboxy terminus of the receptor
peptide, or therebetween. In a preferred embodiment of the
present invention, the hinge region is positioned
internally between the ligand binding and DNA binding
domains of the receptor peptide (see Figure lA).
As employed herein, the phrase "ligand binding domain
of a non-mammalian member of the nuclear receptor
superfamily" refers to ligand binding domains derived from
receptors which are not endogenous to a mammalian host.
Ligand binding domains which are not endogenous to a
mammalian host include ligand binding domains which are
modified to be non-responsive to ligands endogenous or
native to the host. Ligand binding domains contemplated
for use according to the present invention can be derived
from non-mammalian members) of the nuclear receptor
superfamily which members are not normally present in the
cells of a host. Ligand binding domains are preferably
derived from the carboxy-terminal portion of non-mammalian
receptors which are capable of activating transcription of
regulatory elements in the absence of exogenous dimer
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
partner therefore. Exemplary receptors which are not
normally present in mammalian cells include insect
receptors, plant receptors, and the like.
Ligand binding domains can be functionally located in
5 either orientation and at various positions within the
receptor peptide. For example, the ligand binding domain
can be positioned at either the amino or carboxy terminus
of the receptor peptide, or therebetween. In a preferred
embodiment of the present invention, the ligand binding
10 domain is positioned at the carboxy terminus of the
receptor peptide (see Figure lA).
Exemplary ligand binding domains can alternatively be
characterized as comprising substantial sequence identity
with amino acid residues 393-508 ( "Ez" ) , preferably amino
15 acid residues 393-586 (EZ and E3, wherein "E3" is amino acid
residues 509-586), as set forth in SEQ ID N0:3, or
substantial portions thereof (i.e., typically at least 46
or more contiguous nucleotides thereof). It has been
determined that the specific ligand response determinant of
bombyx lies within the Ez region of the native bR, whereas
both EZ and E3 regions facilitate heterodimer formation with
endogenous dimer partners. Modifications of this sequence
contemplated for use in the practice of the present
invention include replacing several amino acids of the
ligand binding domain with sequences from ligand binding
domains of other members of the nuclear receptor
superfamily, such as retinoic acid receptor and/or thyroid
hormone receptor (Figure 1B). These modifications provide
unique transactivating characteristics and/or eliminate
restriction sites, which facilitate the construction of
useful peptide-based viral constructs.
- DNA-binding domains contemplated for use in the
preparation of invention receptor peptides are typically
CA 02296093 2000-O1-10
WO_ 99/02683 PCT/US98/14215
16
obtained from DNA-binding proteins (e. g., transcription
factors). The term "DNA-binding domain" is understood in
the art to refer to an amino acid sequence that is able to
bind to DNA. As used herein, the term "DNA-binding domain"
encompasses a minimal peptide sequence of a DNA-binding
protein up to the entire length of a DNA-binding protein,
so long as the DNA-binding domain functions to associate
with a particular regulatory element.
DNA-binding domains are known to function
heterologously in combination with other functional domains
by maintaining the ability to bind DNA recognition sequence
(see, e.g., Brent and Ptashne, (1985) Cell, 43:729-736).
For example, with respect to hormone receptors, DNA-binding
domains are interchangeable, thereby providing numerous
chimeric receptor proteins (see, e.g., U.S. Patent
4,981,784; and Evans, R., (1988) Science, 240:889-895).
Similar to the ligand binding domain, the DNA-binding
domain can be positioned at either the carboxy terminus or
the amino terminus, or the DNA-binding domain can be
positioned between the ligand binding domain and the
activation domain. In preferred embodiments of the present
invention, the DNA-binding domain is positioned internally
between the ligand binding domain and the activation
domain.
"DNA-binding protein(s)" contemplated for use herein
belong to the well-known class of proteins that are able to
directly bind DNA and facilitate initiation or repression
of transcription. Exemplary DNA-binding proteins
contemplated for use herein include transcription control
proteins (e. g., transcription factors and the like; Conaway
and Conaway, 1994, "Transcription Mechanisms and
Regulation", Raven Press Series on Molecular and Cellular
Biology, Vol. 3, Raven Press, Ltd., New York, NY).
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98114215
17
Transcription factors contemplated for use herein as
a source of such DNA binding domains include, e.g.,
homeobox proteins, zinc finger proteins, hormone receptors,
helix-turn-helix proteins, helix-loop-helix proteins,
basic-Zip proteins (blip), ~3-ribbon factors, and the like.
See, for example, Harrison, S., "A Structural Taxonomy of
DNA-binding Domains," Nature, 353:715-719. Homeobox DNA-
binding proteins suitable for use herein include, for
example, HOX, STF-1 (Leonard et al., 1993, Mol. Endo.,
7:1275-1283), Antp, Mat a-2, INV, and the like. See, also,
Scott et al. (1989), Biochem. Biophys. Acta, 989:25-48. It
has been found that a fragment of 76 amino acids
(corresponding to amino acids 140-215 described in Leonard
et al., (1993) MoI. Endo., 7:1275-1283) containing the STF-
1 homeodomain binds DNA as tightly as wild-type STF-1.
Suitable zinc finger DNA-binding proteins for use herein
include Zif268, GLI, XFin, and the like. See also, Klug
and Rhodes (1987) Trends Biochem. Sci., 12:464; Jacobs and
Michaels (1990? New Biol., 2:583; and Jacobs (1992), EMBG
J., 11:4507-4517.
An additional DNA binding domain contemplated for use
in the practice of the present invention is the GAL4 DNA
binding domain. The DNA binding domain of the yeast GAL4
protein comprises at least the first 74 amino terminal
amino acids thereof (see, for example, Keegan et al.,
Science 231:699-704 (1986)). Preferably, the first 90 or
more amino terminal amino acids of the GAL4 protein will be
used, with the 147 amino terminal amino acid residues of
yeast GAL4 being presently most preferred.
Preferably, DNA-binding domains) used herein is(are)
obtained from a member of the nuclear receptor superfamily.
As used herein, the phrase "member(s) of the nuclear
receptor superfamily" (also known as "intracellular
CA 02296093 2000-O1-10
WO 99102683 PCTIUS98/14215
18
receptors" or "steroid/thyroid hormone superfamily of
receptors") refers to hormone binding proteins that operate
as ligand-dependent transcription factors, including
identified members of the nuclear receptor superfamily for
which specific ligands have not yet been identified
(referred to hereinafter as "orphan receptors").
Exemplary members of the steroid/thyroid hormone
superfamily of receptors (including the various isoforms
thereof) include steroid receptors such as glucocorticoid
receptor (GR}, rnineralocorticoid receptor (MR), estrogen
receptor (ER), progesterone receptor (PR), androgen
receptor (AR), vitamin D3 receptor (VDR), various isoforms
of peraxisome proliferator-activated receptors (PPARs), and
the like; plus retinoid receptors, such as the various
isoforrns of retinoic acid receptor (e.g., RARa, RAR(3, or
RARY}, the various isoforms of retinoid X receptor (e. g.,
RXRa, RXR(3, or RXRY), and the like (see, e.g., U.S. Patent
Nos. 4,981,784; 5,171,671; and 5,071,773); thyroid
receptors (TR), such as TRa, TRH, and the like; insect
derived receptors such as the ecdysone receptor, and the
like; as well as other gene products which, by their
structure and properties, are considered to be members of
the superfamily, as defined hereinabove, including the
various isoforms thereof. Examples of orphan receptors
contemplated for use herein as a source of DNA binding
domain include HNF4 (see, for example, Sladek et al., in
Genes & Development 4: 2353-2365 (1990)), the COUP family
of receptors (see, for example, Miyajima et al., in Nucleic
Acids Research 16: 11057-11074 (1988), and Wang et al., in
Nature 340: 163-166 (1989)), COUP-like receptors and COUP
homologs, such as those described by Mlodzik et al., in
Cell 60: 211-224 (1990) and Ladias et al., in Science 251:
561-565 (1991), the insect derived knirps and knirps-
related receptors, and the like.
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
19
The DNA-binding domains of all members of the nuclear
receptor superfamily are related. Such domains consist of
66-68 amino acid residues, and possess about 20 invariant
amino acid residues, including nine cysteines. Members of
the superfamily are characterized as proteins which contain
these 20 invariant amino acid residues. The highly
conserved amino acids of the DNA-binding domain of members
of the superfamily are as follows:
Cys - X - X - Cys - - X - Asp*- X - Ala* -
X X
-
Gly* - X - Tyr* - X X - X - - Cys - -
- X X X
-
Cys - Lys* - X - Phe - Phe - Arg* - X -
X - X
-
X - X - X - X - X - - X - (X X - ) Cys X
X - - -X
- X - X - X - (X - X X -) Cys - X -X - -
- X Lys
- X - X - Arg - X - - Cys - - X - Cys Arg
X X -
- X - X - Lys* - Cys - X - X X - Gly* -
- Met
(SEQ ID NO:1);
wherein X designates non-conserved amino acids within the
DNA-binding domain; an asterisk denotes the amino acid
residues which are almost universally conserved, but for
which variations have been found in some identified hormone
receptors; and the residues enclosed in parenthesis are
optional residues (thus, the DNA-binding domain is a
minimum of 66 amino acids in length, but can contain
several additional residues).
It has also been found that in vitro evolution methods
can be applied to modify and improve existing DNA-binding
domains (see, e.g., Devlin et al., 1990, Science, 249:404-
406; and Scott and Smith, 1990, Science, 249:386-390).
Alternatively, DNA-binding domains which are engineered
with novel DNA-recognition specificity (see, e.g.,
Pomerantz et al. Science 267:93-96, 1995, ZFHD1, an
engineered transcription factor with a composite DNA-
binding domain) are also contemplated.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
Two polynucleotides or polypeptides are said to be
"identical" if the sequence of nucleotides or amino acid
residues in the two sequences is the same when aligned for
maximum correspondence. Optimal alignment of sequences for
5 comparison may be conducted by the local homology algorithm
of Srnith and Waterman, Adv. Appl. Math., 2:482 (1981), by
the homology alignment algorithm of Needleman and Wunsch,
J. Mol. Biol., 48:443 {1970), by the search for similarity
method of Pearson and Lipman, Proc. Natl. Acad. Sci.
10 {U.S.A.), 85:2444 (1988), by computerized implementations
of these algorithms (GAP, BESTFIT, FASTA, and TFASTA in the
Wisconsin Genetics Software Package, Genetics Computer
Group, 575 Science Dr., Madison, Wis.), or by inspection.
These references are incorporated herein by reference.
15 The percentage of sequence identity between two
sequences is determined by comparing two optimally aligned
sequences over a window of comparison of at least 20
positions. The percentage is calculated by determining the
number of positions at which the identical nucleic acid
20 base or amino acid residue occurs in both sequences to
yield the number of matched positions, dividing the number
of matched positions by the total number of positions in
the window of comparison (i.e., the window size), and
multiplying the result by 100 to yield the percentage of
sequence identity.
For instance, a preferred method for comparing
sequences uses the GAP program based on the algorithm of
Needleman, et at., supra. Typically, the default values for
all parameters are selected. These are gap weight: 5.0,
length weight: 0.30, average match: 1.0, and average
mismatch: 0Ø
CA 02296093 2000-O1-10
WO 99/02683 PCTIUS98/14215
21
As used herein, the phrase "substantial sequence
identity" refers to nucleotide or amino acid sequences
which share at least 80% sequence identity, preferably 90%,
more preferably 95 % or more, regardless of the algorithm
used to determine sequence identity, compared to a
reference sequence over a comparison window of about 20 by
to about 2000 bp, typically about 50 to about 1500 bp,
usually about 350 by to about 1200. The values of percent
identity are preferably determined using the GAP program,
referred to above. Another indication that nucleotide
sequences are substantially identical is if two molecules
hybridize to each other under stringent conditions. It is
recognized, however, that proteins (and DNA or mRNA
encoding such proteins) containing less than the above-
described level of homology produced as splice variants or
as a result of conservative amino acid substitutions (or
substitution of degenerate codons) are contemplated to be
within the scope of the present invention.
The phrase "substantially the same, " used in reference
to a DNA nucleotide sequence, an RNA ribonucleotide
sequence, or an amino acid sequence, refers to sequences
that have slight and non-consequential sequence variations
from the actual sequences disclosed herein. Species that
are substantially the same are considered to be equivalent
to the disclosed sequences. In this regard, "slight and
non-consequential sequence variations" mean that sequences
that are substantially the same as the DNA, RNA, or
proteins disclosed and claimed herein are functionally
equivalent to the sequences disclosed and claimed herein.
Functionally equivalent sequences will function in
substantially the same manner to produce substantially the
same effect as the sequences disclosed.
Receptor peptides employed in the present invention
can be modified by the introduction of activation domains.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
22
Activation domains contemplated for use in the practice of
the present invention are well known in the art and can
readily be identified by those of skill in the art.
Activation domains contemplated for use herein are
typically derived from transcription factors and comprise
a contiguous sequence that functions to activate gene
expression when associated with a suitable DNA-binding
domain and a suitable ligand binding domain. An activation
domain can be positioned at any convenient site within the
receptor peptide, i.e., at the carboxy terminus, the amino
terminus or between the ligand binding domain and the DNA
binding domain. In preferred embodiments of the present
invention, the activation domain is positioned at the amino
terminus of the receptor peptide.
Suitable activation domains can be obtained from a
variety of sources, e.g., from the N-terminal region of a
member of the steroid/thyroid hormone superfamily of
receptors, from a transcription factor activation domain,
such as, for example, VP16, GAL4, NF-KB or BP64 activation
domains, and the like. The presently most preferred
activation domain contemplated for use in the practice of
the present invention is obtained from the N-terminal
region of the VP16 protein.
In a particular embodiment of the present invention,
the invention nucleic acid construct further comprises
promoters and regulatory elements operatively associated
with exogenous nucleic acids. In a preferred embodiment of
the present invention, receptor peptide, in the presence of
a ligand therefor, binds the regulatory element and
activates transcription of the exogenous nucleic acids.
Regulatory elements contemplated for use in the
practice of the present invention include elements
responsive to the invention receptor peptide. In a
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98I14215
23
preferred embodiment of the present invention, such
elements are exogenous regulatory elements not normally
present in the cells of the host. One class of exogenous
regulatory elements contemplated for use herein includes
hormone response elements which modulate transcription of
exogenous nucleic acid when bound to the DNA binding domain
of an invention receptor peptide.
Additional regulatory elements that may be utilized in
the practice of the present invention include exogenous
l0 regulatory elements responsive to a non-mammalian
transactivator. One such transactivator-responsive
regulatory element is an operator which confers
responsiveness to antibiotics. Exemplary operators
contemplated for use in this aspect of the invention
include the tetracycline-analog regulated operator, the TET
operator, the Lac operator, and the like.
Exogenous response elements contemplated for use
herein are short cis-acting sequences (i.e., having about
12-20 bp) that are required for activation of transcription
in response to association with a suitable ligand, such as
diacyl hydrazines, and an invention receptor peptide.
Response element sequences contemplated for use herein
function in a position- and orientation-independent
fashion. Exemplary response elements include hormone
response elements, GAL4 response elements and the like.
Hormone response elements contemplated for use in the
practice of the present invention are response elements
which are responsive to members of the nuclear receptor
superfamily. These response elements comprise at least two
half-sites (in either direct repeat or inverted repeat
orientation to one another), separated by a spacer of 0-5
- nucleotides. As used herein, the term "half-site" refers
to a contiguous 6 nucleotide sequence that is bound by a
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
24
particular member of the nuclear receptor superfamily.
Typically, two half-sites with a corresponding spacer make
up a hormone response element. Hormone response elements
can be incorporated in multiple copies into various
transcription regulatory regions.
Preferred hormone response elements employed in the
practice of the present invention comprise a first half-
site and a second half-site, separated by a spacer of 0-5
nucleotides;
wherein each half-site has the sequence:
-RGBNNM-,
(or complements thereof) wherein
each R is independently selected from A or G;
each B is independently selected from G, C, or T;
each N is independently selected from A, T, C, or G;
and
each M is independently selected from A or C;
with the proviso that at least 4 nucleotides of each
-RGBNNM- group of nucleotides are identical with the
nucleotides at comparable positions of the sequence
-AGGTCA-.
Exemplary half-sites having the -RGBNNM- motif for use
in preparing response elements useful in the practice of
the present invention include, for example, half-sites
selected from -AGGGCA-, -AGTTCA-, -AGGTAA-, -AGGTCA-,
-GGTTCA-, -GGGTTA-, -GGGTGA-, -AGGTGA-, -GGGTCA-, and the
like. A particularly preferred first half-site is
-AGTGCA-.
Additional response elements included the GAL4
response element. Exemplary GAL4 response elements are
those containing the palindromic 17-mer:
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
5'-CGGAGGACTGTCCTCCG-3' (SEQ ID N0:4),
such as, for example, 17MX, as described by Webster et al.,
in Cell 52:169-178 (1988), as well as derivatives thereof.
Additional examples of suitable response elements include
5 those described by Hollenberg and Evans in CeI3 55:899-906
(1988); or Webster et al. in CeI1 54:199-207 {1988).
Regulatory elements employed in the practice of the
present invention are operably linked to a suitable
promoter for transcription of exogenous nucleic acids)
10 product(s). When exogenous nucleic acid(s), operatively
linked to a suitable promoter, is(are) introduced into the
cells of a suitable host, expression of the exogenous
nucleic acids) is(are) controlled by the presence of
ligands, which are not normally present in the host cells.
15 As used herein, when referring to nucleic acids, the
phrase "exogenous to said mammalian host" or simply
"exogenous" refers to nucleic acids not naturally found at
levels sufficient to provide a function in the particular
cell where transcription is desired. For example,
20 exogenous nucleic acids can be either natural or synthetic
nucleic acids, which are introduced into the host in the
form of DNA or RNA. The nucleic acids of interest can be
introduced into target cells (for in vitro applications),
or the nucleic acids of interest can be introduced directly
25 or indirectly into a host by the transfer of transformed
cells into a host.
In contrast to exogenous nucleic acids, the phrase
"endogenous nucleic acids" or "endogenous genes" refers to
nucleic acids naturally found at levels sufficient to
provide a function in the particular cell where
transcription is desired.
CA 02296093 2000-O1-10
WO 99!02683 PCT/US98/14215
26
Exogenous nucleic acids contemplated for use in the
practice of the present invention include wild type and/or
therapeutic nucleic acids.
"Wild type" genes are those that are native to cells
of a particular type. Exemplary wild type nucleic acids
are genes which encode products:
the substantial absence of which leads to the
occurrence of a non-normal state in a host; or
a substantial excess of which leads to the
occurrence of a non-normal state in a host.
Such genes may not be expressed in biologically significant
levels or may be undesirably overexpressed, respectively.
Thus, for example, while a synthetic or natural gene coding
for human insulin would be exogenous genetic material to a
yeast cell (since yeast cells do not naturally contain
insulin genes), a human insulin gene inserted into a human
skin fibroblast cell would be a wild type gene with respect
to the fibroblast since human skin fibroblasts contain
genetic material encoding human insulin, although human
skin fibroblasts do not express human insulin in
biologically significant levels.
Therapeutic nucleic acids contemplated for use in the
practice of the present invention include those which:
encode products which are toxic to the cells in
which they are expressed; or
encode products which impart a beneficial
property to a host; or
those which transcribe nucleic acids which modulate
transcription and/or translation of endogenous genes.
As employed herein, the phrase "therapeutic nucleic
acids" refers to nucleic acids which impart a beneficial
function to the host in which such nucleic acids are
transcribed. Therapeutic nucleic acids are those that are
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
27
not naturally found in host cells. For example, synthetic
or natural nucleic acids coding for wild type human insulin
would be therapeutic when inserted into a skin fibroblast
cell so as to be expressed in a human host, where the human
host is not otherwise capable of expressing functionally
active human insulin in biologically significant levels.
' Further examples of therapeutic nucleic acids include
nucleic acids which transcribe antisense constructs used to
suppress the expression of endogenous genes. Such
antisense transcripts bind endogenous nucleic acid (mRNA or
DNA) and effectively cancel out the expression of the gene.
In accordance with the methods described herein,
therapeutic nucleic acids are expressed at a level that
provides a therapeutically effective amount of the
corresponding therapeutic protein.
Exogenous nucleic acids useful in the practice of the
present invention include genes that encode biologically
active proteins of interest, such as, e.g., secretory
proteins that can be released from said cell; enzymes that
can metabolize a toxic substance to produce a non-toxic
substance, or that metabolize an inactive substance to
produce a useful substance; regulatory proteins; cell
surface receptors; and the like. Useful genes include
genes that encode blood clotting factors such as human
factors VIII and IX; genes that encode hormones such as
insulin, parathyroid hormone, luteinizing hormone releasing
factor (LHRH), alpha and beta seminal inhibins, and human
growth hormone; genes that encode proteins such as enzymes,
the absence of which leads to the occurrence of an abnormal
state; genes encoding cytokines or lymphokines such as
interferons, granulocytic macrophage colony stimulating
- factor (GM-CSF), colony stimulating factor-1 (CSF-1), tumor
necrosis factor (TNF), and erythropoietin (EPO); genes
- encoding inhibitor substances such as alphas-antitrypsin;
genes or nucleotides which characterize specific conditions
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
28
or diseases (i.e., CAG repeat expansion and concomitant
polyglutamine (polyQ) expression has been linked to a
variety of neurodegenerative conditions including
Huntington's disease, dentatorubro-pallidoluysian atrophy,
spinocerebellar ataxias, and spiriobulbar muscular atrophy,
and the like); genes encoding substances that function as
drugs, e.g., genes encoding the diphtheria and cholera
toxins; and the like.
Additional nucleic acids contemplated for use in
accordance with the present invention include genes which
encode proteins present in dopaminergic neurons (useful,
for example, for the treatment of Parkinson's disease),
cholinergic neurons (useful, for example, for the treatment
of Alzheimer's disease), hippocampal pyramidal neurons
(also useful for the treatment of Alzheimer's disease),
norepinephrine neurons (useful, for example, for the
treatment of epilepsy), spinal neurons (useful, for
example, for the treatment of spinal injury), glutamatergic
neurons (useful, for example, for the treatment of
schizophrenia), cortical neurons (useful, for example, for
the treatment of stroke and brain injury), motor and
sensory neurons (useful, for example, for the treatment of
amyotrophic lateral sclerosis), and the like.
Typically, nucleic acid sequence information for
proteins encoded by exogenous nucleic acids) contemplated
for use employed herein can be located in one of many
public access databases, e.g., GENBANK, EMBL, Swiss-Prot,
and PIR, or in related journal publications. Thus, those
of skill in the art have access to sequence information for
virtually all known genes. Those of skill in the art can
either obtain the corresponding nucleic acid molecule
directly from a public depository or the institution that
published the sequence. Optionally, once the nucleic acid
sequence encoding a desired protein has been ascertained,
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
29
the skilled artisan can employ routine methods, e.g.,
polymerase chain reaction (PCR) amplification, to isolate
the desired nucleic acid molecule from the appropriate
nucleic acid library. Thus, all known nucleic acids
encoding proteins of interest are available for use in the
methods and products described herein.
Additional components which can optionally be
incorporated into invention constructs include selectable
markers. Selectable markers contemplated for use in the
practice of the present invention include radiolabeled
molecules, fluorescent molecules, ligands, enzymes, and the
like. Preferable selectable markers are enzymes such as
antibiotic resistance genes, genes which enable cells to
process metabolic intermediaries, and the like. Exemplary
antibiotic resistance genes include genes which impart
tetracycline resistance, genes which impart ampicillin
resistance, zeomycin resistance, neomycin resistance,
hygromycin resistance, puromycin resistance, and the like.
Genes which enable cells to process metabolic
intermediaries include genes which permit cells to
incorporate L-histidinol, genes encoding thymidine kinase,
genes encoding xanthine-guanine phosphoribosyl transferase
(gpt), genes encoding dihydrofolate reductase, genes
encoding asparagine synthetase, and the like.
In accordance with a preferred embodiment of the
present invention, the invention nucleic acid construct
further comprises, in addition to the receptor peptide, a
non-mammalian transactivator, not a member of the nuclear
receptor superfarnily and not normally present in the cells
of said host, and a compatible transactivator responsive
regulatory element not normally present in cells of said
host. The transactivator responsive regulatory element
- controls transcription of the exogenous nucleic acids) or
a second nucleic acid construct comprising a second
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
exogenous nucleic acid(s). Examples of transactivator
responsive regulatory elements include operators which are
responsive to non-mammalian transactivators which confer
responsiveness to antibiotics. Exemplary operators
5 contemplated for use in this aspect of the invention
include the tetracycline-analog regulated operator, the TET
operator, the Lac operator, and the like. Preferably, the
transactivator responsive regulatory elements employed in
the practice of the present invention are operably linked
10 to a suitable promoter for transcription of exogenous
nucleic acids) proteins.
Non-mammalian transactivators, other than members of
the nuclear receptor superfamily, contemplated for use in
the practice of the present invention function in the
15 absence of exogenous dimer partner. Examples of
transactivators that typically function in the absence of
exogenous dimer partner are tetracycline-controlled
transactivators, Vpl6-Lac fusion transactivators, and the
like. When contained as part of a transactivating
20 construct, the transactivator can be positioned at any
convenient site within the construct, i.e., at the carboxy
terminus or the amino terminus of the transactivating
construct. In preferred embodiments of present invention,
the transactivator is positioned at the amino terminus of
25 the transactivating construct (Figure 1C).
Preferably, the non-mammalian transactivator confers
responsiveness to antibiotics. An example of a non-
mammalian transactivator which confers responsiveness to
antibiotics, as contemplated for use in the practice of the
30 present invention is the tetracycline-controlled
transactivator. The tetracycline inducible system is well-
known in the art (see, e.g, Gossen et al. (1992) PNAS 89,
5547-5551; Gossen et al. (1993) TIBS 18, 471-475; Furth et
al. (1994) PNAS 91, 9302-9306; Shockett et al. (1995) PNAS
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
31
92, 6522-6526; and Hoshimaru et al. (1996) PNAS 93(4):1518-
1523). Other examples of non-mammalian transactivators
well known in the art include the IPTG inducible system
based on a VP16-Lac repressor fusion which functions
through lac operator sequences inserted into heterologous
promoters (see, e.g., Baim et al. (1991) PNAS 88:5072-
5076 ) .
In addition to the receptors/transactivators set forth
above, those of skill in the art recognize that other
transactivators can be used herein, e.g., homeobox
proteins, zinc finger proteins, hormone receptors, helix-
turn-helix proteins, helix-loop-helix proteins, basic-Zip
proteins (blip), (3-ribbon factors, and the like. See, for
example, Harrison, S., "A Structural Taxonomy of DNA-
binding Domains," Nature, 353:715-719. Homeobox DNA-
binding proteins suitable for use herein include, for
example, HOX, STF-1 (Leonard et al., (1993) MoI. Endo.,
7:1275-1283), Antp, Mat a-2, INV, and the like. See, also,
Scott et al. (1989) Biochem. Biophys. Acta, 989:25-48. It
has been found that a fragment of 76 amino acids
(corresponding to amino acids 140-215 described in Leonard
et al., (1993) MoI. Endo., 7:1275-1283) containing the STF-
1 homeodomain binds DNA as tightly as wild-type STF-1.
Suitable zinc finger DNA-binding proteins for use herein
include Zif268, GLI, XFin, and the like. See also, Klug
and Rhodes (1987) Trends Biochem. Sci., 12:464; Jacobs and
Michaels (1990) New Biol., 2:583; and Jacobs (1992) EMBO
J., 11:4507-4517.
In a preferred embodiment of the present invention,
expression cassettes are prepared by operably linking
invention nucleic acid constructs to a suitable promoter
for expression of the encoded receptor peptide. As used
herein, the term "promoter" refers to a specific nucleotide
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
32
sequence recognized by RNA polymerase, the enzyme that
initiates RNA synthesis. The promoter sequence is the site
at which transcription can be specifically initiated under
proper conditions. When nucleic acid constructs,
operatively linked to a suitable promoter, is introduced
into the cells of a suitable host, expression of the
nucleic acid construct is conditionally or perpetually
initiated.
Promoters contemplated for use in the practice of the
present invention include inducible (e.g., minimal CMV
promoter, minimal TK promoter, modified MMLV LTR),
constitutive (e.g., chicken b-actin promoter, MMLV LTR
(non-modified), DHFR), and/or tissue specific promoters.
Inducible promoters contemplated for use in the
practice of the present invention comprise transcription
regulatory regions that function maximally to promote
transcription of mRNA under inducing conditions. Examples
of suitable inducible promoters include DNA sequences
corresponding to: the E. coli lac operator responsive to
IPTG (see Nakamura et al., Cell, 18:1109-1117, 1979); the
metallothionein promoter metal-regulatory-elements
responsive t.o heavy-metal (e. g., zinc) induction (see Evans
et al., U.S. Patent No. 4,870,009), the phage T7lac
promoter responsive to IPTG (see Studier et al., Meth.
Enzymol., 185: 60-89, 1990; and U.S. #4,952,496), the heat-
shock promoter; the TK minimal promoter; the CMV minimal
promoter; a synthetic promoter; and the like.
Exemplary constitutive promoters contemplated for use
in the practice of the present invention include the CMV
promoter, the SV40 promoter, the DHFR promoter, the mouse
mammary tumor virus (MMTV) steroid-inducible promoter,
Moloney murine leukemia virus (MMLV) promoter, elongation
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
33
factor la (EFla) promoter, albumin promoter, APO A1
promoter, cyclic AMP dependent kinase II (CaMKII) promoter,
keratin promoter, CD3 promoter, immunoglobulin light or
heavy chain promoters, neurofiliment promoter, neuron
specific enolase promoter, L7 promoter, CD2 promoter,
myosin light chain kinase promoter, HOX gene promoter,
thymidine kinase (TK) promoter, RNA Pol II promoter, MYOD
promoter, MYFS promoter, phophoglycerokinase (PGK)
promoter, Stfl promoter, Low Density Lipoprotein (LDL)
promoter, chicken b-actin promoter (used in conjunction
with ecdysone response element) and the like.
As readily understood by those of skill in the art,
the term "tissue specific" refers to the substantially
exclusive initiation of transcription in the tissue from
which a particular promoter, which drives expression of a
given gene, is derived (e. g., expressed only in T-cells,
endothelial cells, smooth muscle cells, and the like).
Exemplary tissue specific promoters contemplated for use in
the practice of the present invention include the GH
promoter, the NSE promoter, the GFAP promoter,
neurotransmitter promoters (e.g., tyrosine hydroxylase, TH,
choline acetyltransferase, ChAT, and the like), promoters
for neurotropic factors (e. g., a nerve growth factor
promoter, NT-3, BDNF promoters, and the like), and so on.
In yet another embodiment of the present invention,
there are provided constructs comprising a promoter, a
tetracycline-controlled transactivator, a VP16 activation
domain, a DNA binding domain and bombyx mori-derived hinge
region and ligand binding domain encoding sequence, wherein
the components of the construct are operatively associated
' with the other components of the construct.
As used herein, the phrase "operatively associated
with" refers to the functional relationship of DNA with
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
34
regulatory and effector sequences of nucleotides, such as
promoters, enhancers, transcriptional and translational
stop sites, and other signal sequences. For example,
operative linkage of DNA to a promoter refers to the
physical and functional relationship between the DNA and
promoter such that the transcription of such DNA is
initiated from the promoter by an RNA polymerase that
specifically recognizes, binds to and transcribes the DNA.
In yet another embodiment of the present invention,
there are provided constructs comprising nucleic acids)
encoding a a VP16 activation domain operatively associated
with nucleic acid encoding a DNA binding domain and bombyx
mori-derived hinge region and ligand binding domain.
In accordance with another embodiment of the present
invention, there are provided gene transfer vectors useful
for the introduction of invention constructs into suitable
host cells. Such gene transfer vectors preferably comprise
a first reporter under the control of a regulatory element,
a second reporter under the control of an operator which is
responsive to a ligand-mediated receptor which confers
responsiveness to antibiotics, and a construct comprising
a promoter and nucleic acid encoding a VP16 activation
domain, a DNA binding domain and bombyx mori-derived hinge
region and ligand binding domain, optionally, a
tetracycline-controlled transactivator. The number of
copies of regulatory elements can readily be varied by
those of skill in the art. For example, transcription
regulatory regions can contain from 1 up to about 50 copies
of a particular regulatory element, preferably 2 up to
about 25 copies, more preferably 3 up to about 10-15
copies, with about 4-6 copies being especially preferred.
Gene transfer vectors (also referred to as "expression
vectors") contemplated for use herein are recombinant
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
nucleic acid molecules that are used to transport invention
nucleic acid constructs into host cells for expression
and/or replication thereof. Expression vectors may be
either circular or linear, and are capable of incorporating
5 a variety of nucleic acid constructs therein. Expression
vectors typically come in the form of a plasmid that, upon
introduction into an appropriate host cell, results in
expression of the inserted nucleic acid.
Suitable expression vectors for use herein include a
10 recombinant DNA or RNA construct(s), such as plasmids,
phage, recombinant virus or other vectors that, upon
introduction into an appropriate host cell, results) in
expression of the inserted DNA. Appropriate expression
vectors are well known to those of skill in the art and
15 include those that are replicable in eukaryotic cells
and/or prokaryotic cells and those that remain episomal or
those which integrate into the host cell genome.
As used herein, the phrase "transcription regulatory
region" refers to that portion of a nucleic acid or gene
20 construct that controls the initiation of mRNA
transcription. Regulatory regions contemplated for use
herein, in the absence of the non-mammalian transactivator,
typically comprise at least a minimal promoter in
combination with a regulatory element responsive to the
25 ligand/receptor peptide complex. A minimal promoter, when
combined with a regulatory element functions to initiate
mRNA transcription in response to a ligand/receptor peptide
complex. However, transcription typicallywill not occur
unless the required inducer (ligand therefor) is present.
30 Conversely, when the non-mammalian transactivator, other
- than members of the nuclear receptor superfamily, is
present in the host, the transactivator-responsive
regulatory element will typically only induce transcription
in the absence of ligand therefor.
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
36
Preferably, the transcription regulatory region
further comprises a binding site for ubiquitous
transcription factor(s). Such binding sites are preferably
positioned between the promoter and the regulatory element.
Suitable ubiquitous transcription factors for use herein
are well-known in the art and include, for example, Spl.
Expression vectors suitable for use in the practice of
the present invention are well known to those of skill in
the art and include those that are replicable in eukaryotic
cells and/or prokaryotic cells as well as those that remain
episomal and those that integrate into the host cell
genome. Expression vectors typically further contain other
functionally important nucleic acid sequences encoding
antibiotic resistance proteins, and the like.
Exemplary eukaryotic expression vectors include
eukaryotic constructs, such as the pSV-2 gpt system
(Mulligan et al., (1979) Nature, 277:108-114); pBlueSkript
(Stratagene, La Jolla, CA), the expression cloning vector
described by Genetics Institute (Science, (1985) 228:810-
815), and the like. Each of these plasmid vectors are
capable of promoting expression of the receptor peptide of
interest.
In a specific embodiment, a gene transfer vector
contemplated for use herein is a viral vector, such as
Adenovirus, adeno-associated virus, or herpes-simplex virus
based vectors, and synthetic vectors for gene therapy, and
the like (see, e.g., Suhr et al. (1993} Arch. of Neurol.
50:1252-1268). Adenovirus and adeno-associated virus are
extremely suitable as gene transfer vectors to produce
receptor peptides. Alternatively, a gene transfer vector
employed herein is a retroviral vector. Retroviral vectors
are gene transfer plasmids that have an expression
construct containing an exogenous nucleic acid residing
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
37
between two retroviral LTRs. Retroviral vectors typically
contain appropriate packaging signals (e. g., a retroviral
psi (~Y) packaging signal), elements which enable invention
construct integration into a host cell (e. g., a 5' and/or
a 3' long terminal repeat (LTR)), and/or genes encoding
proteins required for retroviral packaging (e.g., the pot
gene, the gag gene and the env gene), that enable the
retroviral vector, RNA transcribed using the retroviral
vector as a template, to be packaged into a viral virion in
an appropriate packaging cell line (see, e.g., U.S. Patent
4,650,764).
Suitable retroviral vectors for use herein are
described, for example, in U.S. Patents 5,399,346 and
5,252,479; and in WIPO publications WO 92/07573, WO
90/06997, WO 89/05345, WO 92/05266 and WO 92/14829, each of
which is hereby incorporated herein by reference, in their
entirety. These documents provide a description of methods
for efficiently introducing nucleic acids into human cells
using such retroviral vectors. Other retroviral vectors
include, for example, mouse mammary tumor virus vectors
(e.g. , Shackleford et al. , (1988) PNAS, FJSA, 85:9655-9659) ,
human immunodeficiency virus (e. g., Naldini et al. (1996)
Science 272:165-320), and the like.
Various procedures are also well-known in the art for
providing helper cells which produce retroviral vector
particles which are essentially free of replicating virus.
See, for example, U.S. Patent 4,650,764; Miller, Human Gene
Therapy, 1:5-14 (1990); Markowitz, et al., Journal of
Virology, 61(4):1120-1124 (1988); Watanabe, et al.,
Molecular and Cellular Biology, 3(12):2241-2249 (1983);
Danos, et al., PNAS, 85:6460-6464 (1988); and Bosselman, et
al., Molecular and Cellular Biology, 7(5):1797-1806 (1987),
which disclose procedures for producing viral vectors and
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
38
helper cells which minimize the chances for producing a
viral vector which includes a replicating virus.
Recombinant retroviruses suitable for carrying out the
invention methods are produced employing well-known methods
for producing retroviral virions. See, for example, U.S.
Patent 4,650,764; Miller, Human Gene Therapy, 1:5-14
(1990); Markowitz, et al., Journal of Virology, 51(4):1120-
1124 (1988); Watanabe, et al., Molecular and Cellular
Biology, 3(12):2241-2249 (1983); Danos, et al., PNAS,
85:6460-6464 (1988); and Bosselrnan, et al., Molecular and
Cellular Biology, 7 (5) :1797-1806 (1987) .
Thus, in one embodiment, a recombinant retroviral
vector can be utilized to express receptor peptide.
Preferably, the retroviral vector will further comprise a
I5 regulatory element and exogenous nucleic acid under the
control of the regulatory element. Optionally, the
retroviral vector can express an antibiotic resistance gene
(see Figure 1C). A "covector" can also be utilized to
provide a nucleic acid construct comprising the promoter,
the regulatory element and exogenous nucleic acid and a
second antibiotic resistance gene. The co-vector carrying
exogenous nucleic acid also has LTRs modified to promote
high-level expression of exogenous nucleic acid (s) only in
the presence of the receptor peptide encoded by the
recombinant retrovirus and exogenous ligand therefor. Co-
infected primary mammalian cells can then be selected using
both antibiotics, resulting in a cell population that is
dependent on ligand for high-level expression of the
exogenous nucleic acid.
The use of invention nucleic acid constructs allows
the stable integration and expression of an exogenous
nucleic acids) or transgene into a wide variety of
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98114215
39
cultured cell types. In accordance with another embodiment
of the present invention, there are provided recombinant
cells containing invention nucleic acid constructs encoding
receptor peptides as described herein, optionally further
containing regulatory elements operatively associated with
exogenous nucleic acid(s). Alternatively, recombinant
cells constructs comprising a regulatory element, such as
the bombyx mori receptor response element, operatively
associated with exogenous nucleic acids, optionally further
containing invention receptor peptides.
The amount of exogenous nucleic acid introduced into
a host can be varied by those of skill in the art. For
example, when a viral vector is employed to achieve gene
transfer, the amount of nucleic acid introduced can be
increased by increasing the amount of plaque forming units
(PFU) of the viral vector.
Suitable means for introducing (transducing)
expression vectors containing invention nucleic acid
constructs into host cells to produce transduced
recombinant cells (i.e., cells containing recombinant
heterologous nucleic acid) are well-known in the art (see,
for review, Friedmann, (1989) Science, 244:1275-1281;
Mulligan, (1993) Science, 260:926-932, each of which are
incorporated herein by reference in their entirety).
Exemplary methods of transduction include, e.g., infection
employing viral vectors (see, e.g., U.S. Patent 4,405,712
and 4,650,764), calcium phosphate transfection (U. S.
Patents 4,399,216 and 4,634,665), dextran sulfate
transfection, electroporation, lipofection (see, e.g., U.S.
Patents 4,394,448 and 4,619,794), cytofection, particle
" bead bombardment, and the like. The transduced nucleic
acid can optionally include sequences which allow for its
' extrachromosomal (i.e., episomal) maintenance, or the
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
transduced nucleic acid can be donor nucleic acid that
integrates into the genome of the host.
Exemplary eukaryotic cells suitable for introducing
invention expression vectors include, e.g., CV-1 cells, P19
5 cells and NT2/D1 cells (which are derived from human embryo
carcinomas), ES cells (embryonic stem cells), COS cells,
mouse L cells, Chinese hamster ovary (CHO) cells, primary
human fibroblast cells, human embryonic kidney cells,
African green monkey cells, HEK 293 (ATCC accession #CRL
10 1573; U.S. Patent No. 5,024,939}, Ltk- cells (ATCC accession
#CCL1.3), COS-7 cells (ATCC under accession #CRL 1651),
DG44 cells (dhfr- CHO cells; see, e.g. , Urlaub et al. (1986)
Cell. Molec. Genet. 12: 555), cultured primary tissues,
cultured tumor cells, neuronal progenitor or precursor
15 cells, such as hcn/v-myc (a tetracycline sensitive
retroviral vector selected by 6418 resistance), neuronal
cells lines such as cerebellum derived neuronal precursors
and PC12 cells, neurons, primary astrocytes,
oligodendrocytes, and the like. Presently preferred cells
20 include CV-1 and 293 cells.
Invention nucleic acid constructs may be stably
incorporated into cells or may be transiently expressed
using methods known in the art. Cells are cultivated under
growth conditions (as opposed to protein expression
25 conditions) until a desired density is achieved. Stably
transfected mammalian cells may be prepared by transfecting
cells with an expression vector having a selectable marker
gene (such as, for example, the gene for thymidine kinase,
dihydrofolate reductase, neomycin resistance, and the
30 like), and growing the transfected cells under conditions
selective for cells expressing the marker gene. To prepare
transient transfectants, mammalian cells are transfected
with a reporter gene (such as the E. coli f3-galactosidase
gene) to monitor transfection efficiency. Selectable
CA 02296093 2000-O1-10
WO 99/U2683 PCT/US98114215
41
marker genes are typically not included in the transient
transfections because the transfectants are typically not
grown under selective conditions, and are usually analyzed
within a few days after transfection.
In accordance with a still further embodiment of the
present invention, there are provided transgenic animals
and methods for producing transgenic animals capable of
prolonged and regulated expression of exogenous nucleic
acid(s), said method comprising introducing into early-
stage embryos or stem cells:
(i) a nucleic acid construct comprising a promoter
and said exogenous nucleic acids) under the
control of a regulatory element;
(ii) nucleic acid encoding a receptor peptide
comprising a DNA binding domain, and the ligand
binding domain and hinge region of a non-
mammalian member of the nuclear receptor
superfamily which is not normally present in the
cells of said host, wherein said receptor peptide
activates said regulatory element in the absence
of an exogenous dimer partner therefor and in the
presence of a ligand for said ligand binding
domain.
As used herein, the phrase "transgenic animal" refers
to an animal that contains one or more inheritable
expression constructs containing one or more exogenous
nucleic acids) under the transcription control of an
operator and/or hormone response element as described
herein.
Methods of making transgenic animals using a
particular nucleic acid construct are well-known in the
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
42
art. When preparing invention transgenic animals, it is
preferred that two transgenic lines are generated. The
first line will express, for example, a receptor peptide as
described above (e.g., VbR). Tissue specificity is
conferred by the selection of a tissue-specific promoter
(e.g., T-cell specific) that will direct expression of the
receptor peptide to appropriate tissue. A second line
contains a nucleic acid construct comprising a promoter and
exogenous nucleic acid under the control of a regulatory
element. Preferably, transgenic animals are produced by
the transfection/infection of embryonic stem cells which
are employed to produce invention transgenic animals
employing methods known to those of skill in art.
In a presently preferred embodiment, an invention
transgenic animal contains one or more expression
constructs containing nucleic acid encoding receptor
peptide and exogenous nucleic acid under the transcription
control of a regulatory element. Thus, with tissue
specific expression of the receptor peptide as described
above and timely hormone treatment, inducible gene
expression can be achieved with spatial, dosage, and
temporal specificity.
In yet another embodiment of the present invention,
there are provided methods of inducing the transcription of
an exogenous nucleic acids) in a host containing:
(i) a nucleic acid construct comprising a promoter
and said exogenous nucleic acids) under the
control of a regulatory element;
(ii) nucleic acid encoding a receptor peptide
comprising a DNA binding domain, and the ligand
binding domain and hinge region of a non-
mammalian member of the nuclear receptor
CA 02296093 2000-O1-10
WO 99/02683 PCTIUS98114215
43
superfamily which is not normally present in the
cells of said host, wherein expression of said
receptor peptide is under the control of an
inducible promoter, wherein said receptor peptide
activates said regulatory element in the absence
of an exogenous dimer partner therefor and in the
presence of a ligand for said ligand binding
domain; and
(iii) said ligand for said ligand binding domain,
wherein said ligand is not normally present
in the cells of said host;
said method comprising subjecting said host to
conditions suitable to induce expression of said receptor
peptide.
In accordance with yet another embodiment of the
present invention, there are provided methods for the
expression of recombinant products detrimental to a host
organism, said method comprising:
transforming suitable host cells with:
(i) a nucleic acid construct comprising a promoter
and exogenous nucleic acids) which express said
recombinant product under the control of a
regulatory element; wherein said regulatory
element is not normally present in the cells of
said host, and
(ii) nucleic acid encoding a receptor peptide
comprising a DNA binding domain, and the ligand
- binding domain and hinge region of a non
mammalian member of the nuclear receptor
~ 30 superfamily which is not normally present in the
cells of said host, wherein said receptor peptide
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
44
activates said regulatory element in the absence
of an exogenous dimer partner therefor and in the
presence of a ligand for said ligand binding
domain, and
growing said host cells to the desired level in the
substantial absence of ligand for said receptor peptide;
and
inducing expression of said recombinant product by
introducing into said host cells a ligand, which, in
combination with said receptor peptide, binds to said
regulatory element and activates transcription therefrom.
Recombinant products detrimental to a host organism
contemplated for expression in accordance with the present
invention include any gene product that functions to confer
a toxic effect on the organism. For example, inducible
expression of a toxin such as the diptheroid toxin would
allow for specific ablation of tissue (Ross et al. Genes
and Development 7:1318-1324 (1993)). Moreover, the
numerous gene products that are known to induce apoptosis
in cells expressing such products are contemplated for use
herein (see, e.g, Apoptosis, The Molecular Basis of Cell
Death, Current Communications In Cell & Molecular Biology,
Cold Spring Harbor Laboratory Press, 1991). For example,
high level expression of 79-amino acid polyglutamine tracts
in cells results in apoptosis and rapid cell death.
In accordance with still another embodiment of the
present invention, there are provided nucleic acid
constructs and cell lines which express a polyglutamine
expression cassette (PQEC) under the control of an
exogenous ligand to maximize obtaining stable
transfected/infected cultured cells which do not undergo
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
undesired apoptosis. Expression of polyglutamine tracts
separately from the huntingtin protein facilitates
identification of the toxic properties associated with the
polyglutamine tracts themselves. In another embodiment of
5 the present invention, there are provided methods for
employing these cell lines to identify novel proteins that
could target developing intracellular aggregates (IAs) and
either block subsequent growth or facilitate or mark the
IAs for degradation.
10 In accordance with still another embodiment of the
present invention, there are provided methods for
modulating the transcription of nucleic acids) in an in
vitro system, said method comprising administering to said
system an amount of a ligand effective to modulate the
15 transcription of said nucleic acid(s); wherein said ligand
is not normally present in said cellular system; wherein
said system comprises:
(i) a nucleic acid construct comprising a promoter
and said nucleic acids) under the control of a
20 regulatory element; and
(ii) nucleic acid encoding a receptor peptide
comprising a DNA binding domain, and the ligand
binding domain and hinge region of a non-
mammalian member of the nuclear receptor
25 superfamily which is not normally present in the
cells of said host, wherein said receptor peptide
activates said regulatory element in the absence
of an exogenous dimer partner therefor and in the
presence of a ligand for said ligand binding
3 0 domain,
- In accordance with yet another embodiment of the
present invention, there are provided methods for the
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
46
treatment of a host in need of gene therapy, said method
comprising:
introducing into cells of said host:
(i) a nucleic acid construct comprising a promoter
and said exogenous nucleic acids) under the
control of a regulatory element;
(ii) nucleic acid encoding a receptor peptide
comprising a DNA binding domain, and the ligand
binding domain and hinge region of a non-
mammalian member of the nuclear receptor
superfamily which is not normally present in the
cells of said host, wherein expression of said
receptor peptide is under the control of an
inducible promoter, wherein said receptor peptide
activates said regulatory element in the absence
of an exogenous dimer partner therefor and in the
presence of a ligand for said ligand binding
domain.
administering, to said host, ligand for said ligand
binding domain.
As used herein, the term "in vivo delivery" refers to
delivery of biological materials by such routes of
administration as oral, intravenous, subcutaneous,
intraperitoneal, intrathecal, intramuscular, intracranial,
inhalational, topical, transdermal, suppository (rectal),
pessary (vaginal), and the like.
As employed herein, the term "ligand" (or ligand
precursor) refers to a non-steroidal substance or compound
which, in its native state (or after conversion to its
"active" form), binds to the receptor peptide, thereby
creating a ligand/receptor peptide complex, which in turn
can bind an appropriate response element and activate
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98114215
47
transcription therefrom. Ligands function to modulate
transcription of nucleic acids) maintained under the
control of a response element. In accordance with one
aspect of the present invention, unless and until a
suitable ligand is administered to the host, substantially
no transcription of the desired exogenous nucleic acids
occurs.
Preferred ligands contemplated for use in the practice
of the present invention are ligands characterized as not
normally present in the cells of the host to be treated.
Such ligands are referred to as being exogenous to the
host. An example of a class of ligands not naturally
present in mammalian systems are compounds referred to as
hydrazines, preferably diacyl hydrazines.
Hydrazines contemplated for use in the present
invention include compounds which are readily available and
are/or relatively inexpensive to manufacture. One such
compound, tebufenozide, is a non-steroidal ecdysone agonist
which is used commercially as an insecticide. This
compound specifically targets lepidopteran species,
including bombyx mori. Tebufenozide has undergone
extensive testing in animal hosts and has proved to be of
very low toxicity to mammals and other non-insect species.
Exemplary hydrazines contemplated for use herein are
mimics of the naturally occurring ecdysones, i.e.,
synthetic organic compounds which have transactivation
activity characteristic of the naturally occurring
ecdysones. Examples of such compounds, referred to herein
as ecdysone mimics, include 1,2-diacyl hydrazines (e. g.,
tebufenozide and others described in U.S. Patent Nos.
5,424,333 and 5,354,762, the entire contents of each of
' which are hereby incorporated by reference herein?,
N'-substituted-N, N'-di-substituted hydrazines (e. g., those
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
48
described in U:S. Patent No. 5,117,057, the entire contents
of which are hereby incorporated by reference herein),
dibenzoylalkyl cyanohydrazines (e.g., those described in
European Application No. 461,809, the entire contents of
which are hereby incorporated by reference herein),
N-substituted-N-alkyl-N, N'-diaroyl hydrazines (e. g., those
described in U.S. Patent No. 5,225,443, the entire contents
of which are hereby incorporated by reference herein),
N-substituted-N-acyl-N-alkyl, carbonyl hydrazines (e. g.,
those described in European Application No. 234,944, the
entire contents of which are hereby incorporated by
reference herein), N-aroyl-N'-alkyl-N'-aroyl hydrazines
(e.g., those described in U.S. Patent No. 4,985,451, the
entire contents of which are hereby incorporated by
reference herein?, and the like.
Since it has been previously reported that the above-
described diacyl hydrazines are neither toxic, teratogenic,
nor known to affect mammalian physiology, they are ideal
candidates for use as inducers in cultured cells and
transgenic mammals according to invention methods.
Ligands are administered in a manner compatible with
the route of administration, the dosage formulation, and in
a therapeutically effective amount. The required dosage
will vary with the particular treatment desired, the degree
and duration of therapeutic effect desired, the judgment of
the practitioner, as well as properties peculiar to each
individual. Moreover, suitable dosage ranges for systemic
application depend on the route of administration. It is
anticipated that dosages between about 10 micrograms and
about 1 milligram per kilogram of body weight per day will
be used for therapeutic treatment.
An effective amount of ligand contemplated for use in
the practice of the present invention is the amount of
CA 02296093 2000-O1-10
WO 99/02683 PCTIUS98/I4215
49
ligand (e.g., diacyl hydrazine(s)) required to achieve the
desired level of transcription and/or translation of
exogenous nucleic acid. A therapeutically effective amount
is typically an amount of a ligand or ligand precursor
that, when administered in a physiologically acceptable
composition, is sufficient to achieve a plasma
concentration of the transcribed or expressed nucleic acid
product from about 0.1 ~.g/ml to about 100 ~Cg/ml, preferably
from about 1.0 ~.g/ml to about 50 ~g/ml, more preferably at
least about 2 ~g/ml and usually 5 to 10 ~.g/ml.
Ligand can be administered in a variety of ways, as
are well-known in the art, i.e., by any means that produces
contact between ligand and receptor peptide. For example,
such ligands can be administered topically, orally,
intravenously, intraperitoneally, intravascularly, and the
like. The administration can be by any conventional means
available for use in conjunction with pharmaceuticals,
e.g., by intravenous injection; either as individual
therapeutically active ingredients or in a combination with
other therapeutically active ingredients. Ligands
contemplated for use in the practice of the present
invention can be administered alone, but are generally
administered with a pharmaceutical carrier selected on the
basis of the chosen route of administration and standard
pharmaceutical practice.
Therapeutic compositions containing suitable ligand
are preferably administered intravenously, as by injection
of a unit dose, for example. The term "unit dose," when
used in reference to a therapeutic composition of the
present invention, refers to a quantity of ligand suitable
' as unitary dosage for the subject, each unit containing a
predetermined quantity of active material calculated to
' produce the desired therapeutic effect in association with
the required diluent, i.e., carrier, or vehicle. It may be
CA 02296093 2000-O1-10
WO 99102683 PCT/US98114215
particularly advantageous to administer such compounds in
depot or long-lasting form as discussed hereinafter.
Suitable regimes for initial administration and
booster shots are variable, but are typified by an initial
5 administration followed by repeated doses at one or more
intervals by a subsequent injection or other
administration. Alternatively, continuous intravenous
infusion sufficient to maintain concentrations in the blood
in the ranges specified for in vivo therapies are
10 contemplated.
In accordance with still another embodiment of the
present invention, there are provided methods for the
treatment of a host in need of gene therapy, said method
comprising:
15 introducing into cells obtained from said host:
(i) a nucleic acid construct comprising a promoter
and exogenous nucleic acids) under the control
of a regulatory element; and
(ii) nucleic acid encoding a receptor peptide
20 comprising a DNA binding domain, and the ligand
binding domain and hinge region of a non-
mammalian member of the nuclear receptor
superfamily which is not normally present in the
cells of said host, wherein said receptor peptide
25 activates said regulatory element in the absence
of an exogenous dimer partner therefor and in the
presence of a ligand for said ligand binding
domain; and
to provide modified cells;
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
51
reintroducing the modified cells into said host; and
administering, to said host, ligand for said ligand
binding domain.
In accordance with this embodiment of the present
invention, the exogenous nucleic acid is introduced
' directly into cells obtained from a donor (host or separate
donor). Such cells are then implanted within the host. In
a presently preferred embodiment, the transplanted cells
are autologous with respect to the host. Autologous means
that the donor and recipient of the cells are one and the
same.
The concept of gene replacement therapy for humans
involves the introduction of functionally active "wild
type" or "therapeutic" nucleic acids into the somatic cells
of an affected host to correct a gene defect or deficiency.
However, in order for gene replacement therapy to be
effective, it must be possible to control the time and
location at which gene expression occurs.
Genes that encode useful "gene therapy" proteins that
2D are not normally transported outside the cell can be used
in the invention if such genes are "functionally appended"
to a signal sequence that can "transport" the encoded
product across the cell membrane. A variety of such signal
sequences are known and can be used by those skilled in the
art without undue experimentation.
The invention will now be described in greater detail
by reference to the following non-limiting examples.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
52
Example 1
Construction of Lepidopteran-derived fusion receptor
peptide
The VP16 t-domain (activation domain) is fused in
frame onto bombyx mori-derived nuclear receptor (bR) at an
internal MluI site near the N-terminus {VbR, Figure lA).
An AscI site immediately downstream of the ATG start codon
allows multimerization of VP16 domains for VbRs with
multiple VP16 activation domains. Note that unlike
Drosophila ecdysone receptor (dEcR), bR has virtually no
C-terminal domain downstream of the LBD. Several amino
acids of the hormone binding domain are replaced with
sequences from the retinoic acid receptor or the thyroid
hormone receptor (VbRRA and VbRTR, respectively) (Figure
1B). These mutant variations have properties slightly
different from VbR, and in addition have silent mutations
that eliminate BclI and BstBl sites in VbR. These are
useful in the construction of many VbR-based retroviral
constructs.
Example 2
Construction of multifunctional regulatory proteins
The small size and simplicity of the VbR system lends
itself to the development of multifunctional regulatory
proteins which are functional on both hormone receptor and
TTA responsive promoters. VbR fusion to the tetracycline
transactivator results in a hybrid protein called TTMT (for
Tebufenozide/Tetracycline Modulated Transactivator; see
Figure 1C). This protein functions jointly as a
ligand-modulated regulator of gene expression from both
tet0 and EIRE (ecdysone response element)-containing
promoters either separately or simultaneously. This
compact protein, encoded in approximately 3 kb, confers
CA 02296093 2000-O1-10
WO 99/02683 PCTIUS98/14215
53
constitutive activation of promoters containing
tetracycline operators (tetOs) in the absence of
tetracycline/analogs. In the presence of tet analogs, the
tet0-binding function of TTMT is blocked, deactivating
responsive promoters. The VbR half of the chimeric protein
is constitutively inactive on EIRE-containing promoters,
but may be stimulated to transactivate responsive promoters
to a high level by the addition of muristerone A (murA) or
tebufenozide (teb). Through such a dual system, two
separate responsive promoters may be simultaneously
regulated by two separate ligands through a single protein.
Additional variants using the reverse-Tet-transactivator,
other VbR variants, novel response elements, and other
regulatory proteins, will presumably result in further
customized variations on this same theme.
A single transgenic animal generated with the TTMT
protein would be simultaneously responsive to transgene
regulation of two separate transgenes by two separate
ligands. Further, individual promoters can be produced
that respond to both halves of the protein, allowing the
transgene to be regulated by both ligands, at the
discretion of the investigator.
Example 3
Transient transfection assay
Transient transfection experiments are carried out in
CV-1 cells or 293 cells using modified drosophila ecdysone
receptor (VdEcR) or modified bombyx hormone receptor (VbR)
with or without dirner partners. Luc activity is examined
40 hours after transfection and stimulation with ligand
- 30 (Figure 2A). Both receptors are assayed in CV-1 cells with
1 ~.M MurA and no exogenous dimer partner . Note that bR
responds, while dEcR does not. Both receptors are also
assayed in the presence of 5 ~g/ml tebufenozide (Figure
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
54
2B). Note that VbR responds with greater than 200-fold
induction while drosophila EcR does not respond at all, and
in fact, decreases slightly. Both receptors are also
assayed with and without dimer partners. VbR is stimulated
with 5 ~.g/ml tebufenozide and VdEcR is stimulated with 1 ~M
MurA. Note that RXR and Usp are necessary for VdEcR while
VbR is constitutively activated by the presence of
exogenous dimer partner and is inhibited by ligand
addition. VbR, with no exogenous dimer partner, has an
extremely low base-line which is stimulated 200-300 fold by
ligand. Note also from this experiment that VdEcR tends to
have a much higher baseline than VbR without dimer partner.
A side-by side comparison of 2.5 ~.g/ml teb and 1 ~M
MurA on VbR show both ligands are effective at stimulating
VbR. Tebufenozide is administered to cells co-transfected
with nucleic acids encoding receptors: LINX (which encodes
the tet-transactivator (TTA)), VbR, TTMT (which encodes the
TTA-VbR fusion protein, and TTMT-2V which is like TTMT, but
with two VP16 t-domains) to assay for EIRE-Luc activity
(Figure 2C) . Note that teb stimulates VbR, and the TTMT
fusions, but has no action on TTA (LINX). The same
experiment as that described for 2C, only with a TetO-Luc
reporter, which responds only to TTA or fusion proteins is
summarized in Figure 2D. Doxycycline (dox) acts to block
constitutive activity of TTA. Note that in this
experiment, TTMT and TTMT-2V work better than TTA only
(LINX) to stimulate the TetO-luc promoter/reporter. They
are also efficiently blocked by dox. An experiment on CV-1
cells transfected with TTMT and two reporters (6Tet0-luc
and 4-EIRE-~igalactosidase) shows both promoters could be
simultaneously influenced by the presence or absence of
both ligands (Figure 2E).
CA 02296093 2000-O1-10
WO 99102b83 PCT/US98/14215
Example 4
Liaand mediated transaene reaulation
Transgene regulation by teb or MurA in stably infected
rat fibroblasts is assayed utilizing recombinant
5 retroviral constructs as the reporter, and VbR encoding
retroviral vectors to provide the receptor. Bulk infected
selected populations stimulated with 2.5 ~,g/ml teb or 1 ~.M
MurA indicate b-gal or tyrosine hydroxylase infected cells.
Quantitative analysis of an individual VbR-luc clonal
10 fibroblast line, and a VbR-GH clonal line show that 0.25
~.g/ml teb is as effective at stimulating full activity as
higher levels (Figure 3).
Example 5
Single-~lasmid retroviral constructs
15 The plasmid, pBO, utilizes a responsive internal
promoter to regulate transgene expression and also contains
a VbR-responsive 3' LTR to autoregulate the VbR receptor
itself (Figure 4). The plasmid, pNA, is constructing using
a 3'-responsive LTR to regulate transgene expression. pB0
20 utilizes an EcR responsive internal promoter and a TTA
responsive 3' LTR. Both are host to regulation by the TTMT
protein, simultaneously. The purpose of this design is to
provide chronic stimulation of the duplicated 5' LTR along
with regulated expression of the internal EcR responsive
25 promoter. By removing all of the native promoting
sequences from the 6To LTR with the exception of the
TetO's, variants of this construct are also inactivatable.
Example 6
Construction of p,lasmid vectors
30 VbR and TTMT systems including constitutive and
inducible promoters are integrated into a modular plasmid
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
56
vector known as "pBW" (Figure 5) . The purpose of pBW is to
integrate all of the elements of bombyx receptor- or
tetracycline-analog responsiveness into a single plasmid
vector destined for use in producing transgenic animals by
methods such as pronuclear injection. pBW is to facilitate
construction of transgenic animals or stably transfected
cells using VbR variants. The pBW design simplifies the
insertion of responsive promoters, transgenes, constitutive
promoters, and the various VbRs, TTMTs, and related
regulatory proteins. With pBW, the transgene regulatory
properties of generated animals should be more predictable
and reproducible from founder to founder than multi-vector
systems.
There are two major components of this system: the
first is a tissue-specific expression cassette to produce
the receptor and/or antibiotic resistance genes. The
second is an expression cassette with a responsive promoter
and a transgene. Polyadenylation signals flank all of the
expression cassettes to provide efficient polyadenylation
(p(A)) of transcribed RNAs. The use of rare restriction
endonuclease sites within strategic locations and
polylinkers of pBW will ease cloning of transgenes. An
adjunct shuttle vector derived from pBSK (Stratagene)
called SKSP contains rare sites flanking the polylinker and
compatible with pBW. pBW also shares a number of sites
with pB0 to further simplify construction. pBW is
constructed within the plasmid pcDNA3 (Invitrogen).
Example 7
Regulating transaene
expression in target cells in vitro
Individual desired elements can be inserted
sequentially into a recombinant retrovirus. Recombinant
retrovirus starts as a polylinker composed of 5' NotI-MIuI-
CA 02296093 2000-O1-10
WO 99/02683 PCT/1JS98/14215
57
NruI-EcoRI-AscI-PmlI-BstBI-BamHI-HindIII-HpaI-CIaI-NsiI-
KpnI 3' sites. The retroviral ~Y gln (see Adam and Miller
in J. Virol. 62:3802 (1988) and Barklis et al., in CeI1
47:391 (1986)) is inserted into sites of the recombinant
retroviral polylinker. The internal CMV promoter is
inserted BamHI-CZaI into the BamHI-ClaI sites of the
evolving recombinant retroviral vector. All LTRs destined
for insertion into the 5' location are produced by low-
cycle/high-fidelity PCR production and end primers with
compatible NotI-MluI sites for insertion into the NotI-MZuI
sites. 3' LTRs are generated with CZaI-NsiI compatible
ends for insertion into these sites. The R region of the
3' LTR is included in the downstream primer sequence.
Transgenes, receptor cDNAs, or selectable marker genes are
inserted into the remaining polylinker from the EcoRI site
through the BamHI site, or the remaining polylinker after
the internal promoters (HindIII-CIaI).
The resulting vectors are characterized by restriction
digests and mapping, amplified by large-scale plasmid
preparation, and prepared for transfection into packaging
cells by standard methods. Transient retroviral production
in 293 and 293T cells has been previously described (see
Pear et al., in PNAS 90:8392 (1993)). Forty-eight hours
after transient retroviral production, the conditioned
media is removed, filtered through 0.45 mm filters, and
frozen at -70°C until use. 10-cm dishes of primary-cultured
Fisher rat abdominal fibroblasts (Rosenberg et al., Science
242:1575 (1988)) at approximately 50o density are infected
with 1/10 volume of virus-containing media and 8 ~g
polybrene/ml for 48 hours. Infected cells are then
selected under the appropriate antibiotic until non-
infected cells are cleared from the population
(approximately 10 days). Resistant colonies are
trypsinized, pooled and passaged. The resulting population
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
58
is then infected with the second virus, and reselected
again in media containing both antibiotics. The infection
and selection process for the second round of viral
infection proceeds as described for the first round. The
final population of doubly resistant primary fibroblasts
are then pooled and passaged to 6 or 24-well Costar plates
for assay in vitro.
The short (24 hour) course is performed in 24-well
Costar dishes with cells at approximately 80% confluency.
5 ~,1 of ligand (final concentration of 1 ACM) or vehicle
(20o EtOH) is added to the 1 ml of media in each well.
Eight hours after first ligand administration, the cells
are all washed in PBS and the media is replaced.
galactosidase histochemical reaction is performed on 1.50
glutaraldehyde-fixed cells essentially as described by
Shimohama et al. in Brain Res Mo1 Brain Res 5:271 (1989) in
a 37°C environment for 2 hours. The 4-day time course is
performed in an identical fashion, except that the cell
population is plated at only 20 o confluency at the start of
the experiments (and grow to near-total confluency by the
end of the 4 day period) and neither ligand nor media is
changed or replaced.
For long-term culture experiments, MMBG fibroblacts
are plated in triplicate at high density in 24-well plates.
Within 48-72 hours of plating, when the cells reach 100%
confluency within the wells, 1 ~M ligand or vehicle is
added to one group of plates, while others are left
completely untreated. The wells are then placed in a 37°C-
l0o C02 environment for the next 25 days without any media
changes. On day 23, the plates that had received neither
vehicle nor ligand on day 1 are removed and half of the
wells are given vehicle and the other half are given 1 ~,M
ligand for 40 hours.
CA 02296093 2000-O1-10
WO 99102683 PCT/US98I14215
59
After 25 days of nutrient deprivation and contact
inhibition, all of the plates are processed for (3gal
histochemical staining as above.
MMGH fibroblacts are produced by infecting a
fibroblast line with the recombinant retroviral vectors
containing desired elements. Cells are selected under both
antibiotics as described previously. After preliminary
examination of ligand-induced hGH production in the bulk
population, the MMGH population is plated at high density
l0 in 6-well Costar plates. Forty-eight hours after the
initial plating and when the cells are essentially
completely growth inhibited by contact, the medium of all
plates is replaced with DME containing 2o FBS. Under these
conditions, primary rat fibroblacts stop dividing even if
they are not confluent, and settle out into a distended
morphology with prominent nuclei characteristic of severely
growth-arrested fibroblacts. Six days after
acclimatization to this culture environment, 1 ml of medium
is harvested from each well and plate of the experiment and
stored frozen at -20°C. The remaining unused media is
discarded at the same time of harvest and replaced with
fresh DME-2o FBS. Groups of wells are treated or not with
1 ~M ligand. Hormonally treated wells are washed with
medium to remove as much residual ligand as possible prior
to medium replacement when ligand treatment is
discontinued. This routine is repeated daily for the 21-
day extent of the experiment.
After all time-point samples have been collected and
frozen at -20°C, they are simultaneously processed for the
presence of the hGH protein by ELISA (Boehringer Mannheim)
following the protocol recommended by the manufacturer.
ELISA data are quantitated on an MR700 microplate reader
(Dynatech Laboratories, Chantilly, VA) and compared to a
standard curve generated using purified hGH to determine
CA 02296093 2000-O1-10
WO 99102683 PCTlUS98/14215
picogram GH amounts. Control wells are TH (tyrosine
hydroxylase) producing fibroblasts, but display all of the
characteristics of the recombinant retrovirus except for
the hGH transgene and secreted protein.
5 Example 8
Expression Vectors and Transient Transfection Assays
All receptors were subcloned into vector LNCX (A. D.
Miller, GenBank accession no. M28247, with an extended
polylinker) for use in transient transfection assays. VBR
10 (or CVBR) when referring to the retrovirus) was produced by
insertion of VP16 (14) sequences with a synthetic ATG start
codon into the amino-terminal region of BE up to, and
in-frame with, the Mlu1 site (corresponding to amino acid
26). VP16 primers: (5') 5'-GAGAGAAGCTTATGGCGCGCCCGACCGATG
15 (SEQ ID N0:5) and (3') 5'-CACACACGCGTGTACTCGTCAATTCCAAG
(SEQ ID N0:6) . VDE (or CVDE) was produced in a similar
fashion by fusing VP16 sequences in-frame at a novel NcoI
site corresponding to amino acid 68. The downstream primer
was made Ncol compatible. Plasmid template (100 ng), each
20 primer (500 ng), and reaction conditions outlined by the
manufacturer for Pwo (Boehringer Mannheim) high fidelity
polymerase, were used with a program of 1 min at 94°C,
1 min at 45°C, and 1 min at 72°C for 20 cycles for
production of all PCR products used herein. LNCX-Usp and
25 RXR were produced from EcoRI fragments encoding this
complete cDNAs, filling with the Klenow fragment of DNA
polymerase, and inserting the fragments into the HpaI site
of LNCX. Orientation was determined by restriction
endonuclease digestion. Transient transfection in CV-1 and
30 293 cell was performed by using standard methods (Sambrook
et al (1989) Molecular Cloning: A Laboratory Manual 2na
edition) in triplicate in 24-well plates by calcium
phosphate coprecipitation of 100-ng receptor(s), reporter
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
61
plasmid E4-luc, and pCH110 as an internal control.
Briefly, E4-luc is four tandem EcREs inserted upstream of
a thymidine kinase gene minimal promoter directing
luciferase expression EIRE oligonucleotides were as
described (Thomas et al. (1993) Nature 362:471-475) with
BamHI-Bg/II compatible ends. Ligand (1 ~.M) was added at
the time of transfection, and 40 h later cells were lysed
and extracts were used for [3-galactosidase assay and
measurement of luciferase activity in an analystical
bioluminescence photometer.
CVDEiR and MS vectors used only as retroviruses in
production of stable cell lines were produced as follows:
The VDEiR expression cassette was constructed by modifying
the ATG start codon of human RXRa to contain an overlapping
BstXI site for fusion into the ATG start codon of the
0.8-kb EMCV IRES (fang et al (1989) J. Virol 63:1651-1660
and Hoshimaru et ai (1996) PNAS 93:1518-1523) sequence.
The IRES-RXR cassette was then subcloned downstream of VDE
in LNCX-ofVDE (CofVDE), a VDE variant vector with lower
basal transactivation levels in superphysiological RXR
environments, to produce CVDEiR. The MS retroviral
reporter was produced by removing the Moloney murine
leukemia virus enhancer core from the 3' long terminal
repeat (LTR) with NheI-XbaI digestion and replacing this
sequence with four tandem EcREs, resulting in a reporter
analogous to the E4-luc reporter plasmid.
Example 9
Gel Mobility Shift Assays
Gel mobility shift analysis was performed by in vitro
translation of receptor ORFs subcloned into pBSK
(Stratagene), pGEM-3 (Promega), and PSL301 (Invitrogen) in
the presence of [35S]Met by using T3/T7 TNT (Promega)
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
62
transcription/translation and following the manufacturer's
protocol. In vitro translated proteins were qualitatively
examined by 5o SDS/PAGE with protocols as described
elsewhere (Sambroak et al (1989) Molecular Cloning: A
Laboratory Manual 2nd edition) and quantified by
PhosphorImager (Molecular Dynamics) exposures of dried
gels. Amounts of proteins used in gel shift assays were
normalized by using quantitative data and correction for
predicted Met residues in individual constructs.
Double-stranded EIRE probes corresponding to response
elements described above were labeled by filling of the
Klenow fragment of DNA polymerase with [32P] dCTP and
unlabeled dGAT by standard methods. Reaction conditions
for protein-probe interaction and gel electrophoresis were
essentially as described by Yao et a1. ((1992) Cell 71:63-
72) except, to facilitate comparison between samples,
reaction mixtures (including dimer partners) and probe)
were prepared as a mixture and distributed equally to
individual tubes with receptor proteins. The reactions
were allowed to proceed at 23°C for 5 min. at which time
ligand or vehicle was added and the reaction allowed to
continue for an additional 20 min.
Example 10
Construction of bombyx rnori receptor (BE)- drosoph.ila
mel.anocraster ecdysone receptor (DE) Chimeric Receptors
Chimeric receptors were produced by taking advantage
of conserved DNA sequences and sites within the receptors
and by introducing compatible sites by low cycle, high
fidelity PCR (described above) of bombyx mori receptor
(BE) and drosophila melanogaster ecdysone receptor (DE)
templates to produce fusion proteins. DEBE-A/B was
produced by PCR and replaced the BE /AB domain (amino acids
1-197, as set forth in SEQ ID N0:3) with amino acids 1-255
CA 02296093 2000-O1-10
WO 99/02683 PCTIUS98/14215
63
of DE introducing a novel Apal restriction site. DEBE-C
replaced the A/B and C domains (amino acids 1-273, as set
forth in SEQ ID N0:3) of BE with amino acids 1-331 of the
DE protein by introduction of a compatible Eagl site into
BE at the fusion junction. DEBE-D replaced BE from amino
acids 1-363, as set forth in SEQ ID N0:3, with amino acids
1-430 of DE by KpnI digest. BEDE was the reverse of this
construct and replaced amino acids 1-430 of DE with amino
acids 1-363 (as set forth in SEQ ID N0:3 ) of BE by using
KpnI partial digest. DEBH was produced from DEBE-C by
removing all BE sequences except the D domain by KpnI
digestion and inserting sequences encoding the intact E and
F domains (amino acids 430-878) of the original DE after
partial KpnI digestion. VEH was produced by excision of
the central region of DEBH, including the heterologous
hinge domain, and inserted into VDE, replacing the
analogous region.
Example 11
Construction of E Domain Chimeric Receptors
The chimera, BEDB, was produced by insertion of the
654-by KpnI-Bg/II fragment encoding precisely the DE
hormone-binding domain into the identical analogous sites
in BE. Two unique sites within the hormone-binding domain
of BEDB were used in the production of the chimeras. A
unique AatII site lies approximately 120 by downstream of
the 5' KpnI site, and a unique EagI sites lies
approximately 440 by downstream. PCR was used as described
above, and corresponding fragments of BE with appropriate
compatible ends were generated and subcloned into BEDB
digested with KpnI + AatII (BKE) , AatII + Eagl (BAE) , AatII
+ Bg/II (BAB), and EagI-Bg/II for BEB. The resulting
constructs were translated, examined by SDS/PAGE,
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
64
quantified, and normalized as described above. Conditions
for gel mobility shift assay were also as described above.
Example 12
Infection of Cells with Recombinant Retroviruses
Transient restroviral production in 293 cell types has
been described (Pear et al. ((1993) PNAS 90:8392-8396).
Forty-eight hours after transient retroviral production,
the conditioned medium was removed, filtered through
0.45-~m filters, and frozen at - 70°C until use.
Ten-centimeter dishes of primary-cultured Fischer rat
abdominal fibroblasts (Schinstine et al. (1992) Neurochem
58:2019-2029) and 293 and CV-1 cells at approximately 500
density were infected at a multiplicity of infection of
approximately 0.05 with virus-containing media and 8 rng/ml
Polybrene for 48 h. Infected cells were then selected with
4-6 mM z-histidinol until noninfected cells were cleared
from the population: approximately 10 d for FFl2s, 21
dafor 293s, and >30 d for CV-is (CV-1 cell populations were
resistant to rigorous selection with i.-histidinol).
Resistant colonies from multiple plates were trypsinized,
pooled, and passaged. The resulting bulk population was
then split and infected with receptor-bearing virus and
reselected again in medium containing both z-histidinol and
400-800 ~.g/ml 6418. The infection and selection process
for the second round of viral infection proceeded as
described for the first round. The final population of
doubly resistant primary fibroblasts was then pooled and
passaged in triplicate in 6- or 24-well Costar plates for
~-galactosidase staining by standard methods or luciferase
assay.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98114215
Example 13
CV-1 Cells Support BE-Mediated Transactivation Without
Exogenous Dimer Partner
Both receptors were tested to determine the effect
5 that differential endogenous dimer partner availability had
on transactivation of responsive promoters. Relative
luciferase activity employing the reporter plasmid, E4-luc,
were performed from VDE and VBR transiently cotransfected
into 293 or CV-1 cells with either Usp, RXR, or no added
10 heterodimer partner. Luciferase activity was assayed in
the presence of either vehicle, 1 ~.M murA, or 1 ~.M
tebufenozide.
In the presence of an equimolar exogenously added
dimer partner, VDE and VBR transactivation was similar in
15 both cell types. With Usp, both proteins were induced less
than 2-fold by ligand and displayed a high level of basal
transactivation. With RXR, VDE displayed an average
relative 5.35-fold induction across both cell types,
whereas VBR was induced only 2.35-fold, even though the
20 absolute level of induction matched or exceeded the
expression with VDE. The decreased relative induction of
VBR + RXR resulted from approximately doubled basal
activity levels compared with VDE + RXR. With no exogenous
dimer partner, both receptors in both cell types exhibited
25 dramatic 15- to 80-fold decreases in basal transactivation.
With no exogenous dimer partner, VDE failed to respond
to tebufenozide, whereas VBR continued to respond well to
both murA and tebufenozide. The addition of RXR to
VDE-transfected 293 cells increased the maximum
30 murA-treated expression level by only 20%, indicating that
the high level of endogenous RXR in 293 cells is near
saturation for heterodimerization with DE. By comparison,
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
66
CV-1 cells supported murA stimulation of VDE at only 130 of
the expression level of VDE with added RXR (Fig. IB). VBR
was active at levels similar to VDE in the high RXR
background of the 293 cells; however, in the CV-1 cells,
tebufenozide-stimulated VBR exhibited both the highest
absolute level of transactivation and the greatest relative
induction (160.2-fold) of any of the other combinations of
receptors and ligands tested. In the CV-1 cells,
tebufenozide-stimulated VBR displayed 21-fold greater
relative induction and an absolute expression level 9.25
times the level of VDE treated with mutA.
Example 14
BE Dist~lays Facilitated Heterodimer Formation with Usp
and RXR
The high level function of VBR in the low heterodimer
partner environment of the CV-1 cells suggested that BE
might be capable of efficient function with lower levels of
dimer partner than DE. To explore this possibility,
cell-free experiments with gel mobility shift analysis and
equivalent amount of in vitro translated BE and DE in
combination with Usp or RXR dimer partners and both ligands
were performed. Neither protein alone formed a dimeric
complex with the EIRE probe unless either the Usp or RXR
dimer partner proteins were added. In the presence of Usp,
DE formed a comparatively weak interaction with the EcREs
that was increased approximately 4-fold by murA and 2-fold
by tebufenozide. In contrast, BE produced a strong shift
with Usp that was only slightly influenced by either
ligand. In the absence of any ligand, BE bound probe 5
times more efficiently than DE. With ligand, the BE probe
shift was observed to still exceed the DE shift and was
double the DE + Usp band volume. Both receptors combined
with RXR displayed an absolute dependence on ligand for
heterodimer formation; however, whereas BE + RXR with
CA 02296093 2000-O1-10
WO 99102683 PCTIUS98l14215
67
either ligand displayed a prominent shifted band, an
equivalent amount of DE resulted in a barely detectable
shifted band with murA and no detectable band shift with
tebufenozide. The BE + RXR tebufenozide- and murA-induced
shifts were determined to be >15 times the DE + RXR shift
with murA.
Example 15
BE-DE Chimeras Reveal That the D Domain Is Critical to
High Affinitv Heterodimerization
Gel mobility shift assays indicated that BE had higher
affinity heterodimer formation than DE. To further define
the subdomains and molecular determinants resulting in the
BE high affinity phenotype, PCR mutagenesis and internal
shared restriction endonuclease sites were used to produce
BE-DE chimeric receptors that were assayed for their
ability to bind the EIRE probe with either Usp or RXR and
with or without ligand. Because previous results indicated
that murA stimulated both EcRs, for purposes of direct
comparison only murA was used for gel shift analysis of the
chimeras. Fusion constructs began at the N terminus of BE
and sequentially replaced the A/B (DEBE-A), C (DEBE-C),
and D (DEBE-D) domains with DE sequences. BEDE was the
reverse, encoding BE up to the D-E domain boundary with the
DE E and F domains. DEBE-A and DEBE-C with Usp shifted
nearly as efficiently as native BE. The absence of shift
for DEBE-D, however, and the substantial shift seen for
BEDS + Usp suggested that the high affinity determinant for
BE-Usp heterodimerization resided in the D, or hinge
domain. The same constructs in combination with RXR
revealed a different shift pattern. BE and DEBE-A both
responded strongly to ligand to bind and shift the probe.
DEBE-C with ligand was approximately 40% decreased relative
to native BE; however, it still displayed 16-fold greater
binding than native DE. DEBE-D had lost all high affinity
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
68
DNA-binding complex formation and functioned
indistinguishably from the native DE with or without
ligand. MurA-stimulated BEDS + RXR produced a band shift
4 times the intensity of the native DE + RXR stimulated
with murA, but BEDS was clearly significantly impaired to
heterodimerization with RXR compared with native BE and
other higher affinity chimeras like DEBE-A and DEBE-C.
For further confirmation of the role of the BE D
domain in dimer partner affinity, the D region of DE was
replaced with the BE D domain to produce DEBH. Native DE
and DEBH were compared side by side for binding to EcREs
with both Usp and RXR dimer partners. Gel mobility shift
of in vitro translated and normalized DE and
hinge-substituted DEBH was performed, in the presence of
either Usp or RXR, and further in the presence of vehicle
or 1 ~,M murA. Compared with DE + Usp, DEBH + Usp averaged
5-fold greater probe binding with murA and 9-fold greater
binding without ligand. DEBH + RXR exhibited both
decreased probe shift in the absence of ligand and
increased probe binding with murA (approximately 3-fold
over native DE) for a 10-fold relative induction by ligand
compared with 2.5-fold for DE. Transient transfection
analysis of a VP16-DEBH construct (VEH) in CV-1 cells
revealed that VEH shared characteristics of both DE and BE.
VEH displayed the high basal transactivation level and
low relative induction (average of 2.5 fold) characteristic
of BE with superphysiological RXR. VEH displayed 5- to
8-fold greater basal and murA-activated expression than
native DE in the absence of any exogenous dimer partner.
MurA-stimulated VEH expression exceeded even the expression
level for murA-stimulated BE by nearly 2-fold. As
predicted, without the BE hormone-binding domain, DEBH did
not to respond to tebufenozide. Additional chimeric
proteins with a subset for BED-region sequences did not
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
69
function as well as DEBH with replacement of the entire D
domain, suggesting that multiple discrete determinants
presumably over much of the hinge region contribute to the
high affinity phenotype.
Example 16
Discrete Functional Regions Within the E Domain Revealed
by BE-DE Chimeras
Although the preceding data reveal that the BED domain
alone was sufficient for high affinity heterodimerization
with Usp, a comparison of the relative shift of the DEBE-D
and BEDS chimeras with both partners suggested that
additional determinants within the BE hormone-binding E
domain contributed to a high affinity interaction with RXR.
To further study the BE E domain, chimeric proteins were
produced by using BEDE as a template. Five E-domain
chimeras named BEDB, BKE, BAE, BAB, and BEB were prepared.
BEDB is identical with BE with the exception of complete
replacement of the E domain with corresponding sequences
from DE, BEDB was identical with BEDE with the exception of
replacement of the large C-terminal DE F domain for the
20-amino acid F domain of BE. BKE, BAE, BAB, and BEB are
derived from BEDB and are sequential, C-terminal moving
replacements of three regions within the DE-derived E
domain. Gel mobility shift assays with BE, BEDE, and BEDB
revealed that BEDE and BEDB are essentially identical in
their properties of complex formation, suggesting that the
F domain does not play a significant role in
heterodimerization and DNA binding. In addition, BEDB
recapitulates the high affinity USP binding and the low
affinity RXR binding of BEDE.
The BEDB-derived chimeric receptors reveal several
functional subdomains over most of the BE E domain. By
using unique internal sites, the E domain was subdivided
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
into approximately thirds called E1 E2 and E3. Treatments
were with vehicle (-) , 1 ~.M murA (M) , or 1 ~.M tebufenozide
(T) . Chimera BKE with replacement of the DE E1 and E2,
regions and chimera BAE with replacement of only the E2
5 region displayed very similar patterns of shift. Both
chimeras were significantly impaired in complex formation
with Usp and RXR relative to the original BEDB chimera,
suggesting that there were fundamental incompatibilities
between subdomains of the Bombyx and Drosophila E domains.
10 The similarity of these two constructs suggested that DE
and BE E1 regions were likely to be functionally similar,
but that the EZ regions had very different properties.
Curiously, although both basal and murA-stimulated
heterodirnerization were inhibited, BKE/BAE-Usp
15 heterodimerization was significantly stimulated by
tebufenozide, indicating that the tebufenozide response
determinant is within EZ but that the high affinity RXR-
binding determinant lies elsewhere.
When Ez was combined in tandem with E3 in the chimeric
20 receptor BAB, high affinity heterodimer formation
indistinguishable from native BE heterodimerization was
observed with both Usp and RXR. To determine whether the
E3 region alone could confer high affinity
heterodimerization with RXR, the chimera BEB, with
25 substitution of only the E3 region, was produced and tested.
BEB displayed a pattern of response essentially identical
with the original BEDB with low-level interaction with RXR
and only upon stimulation with murA. BEB, which lacked the
E2 region of native BE, was not responsive to tebufenozide.
30 Taken together, these results indicate that the BE D domain
and E3 region, in combination with ligand-binding
determinants within the Ez region, function in concert to
produce high affinity, ligand-dependent heterodimerization
between BE and the RXR protein.
CA 02296093 2000-O1-10
WO 99/02683 PCTIUS98/14215
71
Example 17
Comparison of VBR and VDE Function in Stablv Transduced
Cells
Building on transient transfection and gel shift data,
DE an BE variants were directly compared for their ability
to function at low or single copy in stably transduced cell
types. In addition to direct comparison of CVDE and CVBR
retroviruses, an RXR-bearing VDE variant called CVDEiR was
also tested that previous studies had determined to
provide the best combination of low basal expression and
relatively high induction for VDE cotransferred with
supplemental RXR. These three receptor-bearing vectors and
an empty LNCX control were used to infect 293, CV-1, and
primary fibroblast cells harboring an EIRE-responsive
retroviruses termed MS. MS encoded a 3' LTR with the core
enhancer elements replaced with four tandem EcREs and an
internal simian virus 40 promoter vector directing
resistance to the drug L-histidinol. Parallel MS vectors
encoding either the LacZ gene (MS-Z) or a luciferase
transgene (MS-luc) were used in this comparative study, the
former as an indication of the number of responding cells
and the latter for quantitative purposes.
Three cell types, 293s, CV-Is, and rat primary
fibroblasts (line FF12), were selected for assay of
reporter/receptor virus combinations based on their
relative capacities to supports high level DE-mediated
transactivation. The human 293 cell line support
essentially full DE activation with no requirement for
added RXR, whereas CV-is require the contransfection of
exogenous dimer partner for DE function. FFl2s were
selected as the third cell type because they are refractory
to chemical methods of transduction and reliant on the use
of biological vectors such as recombinant retroviruses for
efficient gene transfer. Furthermore, primary fibroblasts
CA 02296093 2000-O1-10
WO 99/02683 PCT/U898/14215
72
are frequently used as autologous donor cells for
transplantation approaches to somatic gene therapy.
All three cell types were infected with both the
reporter and receptor-bearing vectors and treated for 72 h
with either vehicle, 1 ~.M murA or 1 ~M tebufenozide.
Histochemical staining of MS-Z infected cells indicated
that all three receptor types were capable of high level
ligand-responsive transactivation in coinfected 293 cells.
DE-derived constructs responded well to murA but were
unable to respond strongly to tebufenozide. CVBR, on the
other hand, responded moderately to murA but strongly to
tebufenozide. MS-LacZ cell types infected with an empty
LNCX vector displayed no staining when stimulated with any
ligand and further confirmed observations from
vehicle-treated cells that the MS vector permitted only
relatively low basal levels of transgene expression.
Approximately 30% of CVDE-, CV-DEiR-, and CVBR-infected
MS-Z 293 cells responded to murA. Tebufenozide stimulated
<lo of CVDE-infected cells and <4% of CVDEiR-infected MS-Z
293s. More than 500 of CVBR-infected MS-Z 293s responded
to tebufenozide, however, exceeding all DE-based vectors
with murA. More dramatic differences were seen in CV-1 and
FF12 MS-Z infected cells. CVDE alone was not inducible in
either cell type, indicating that the endogenous level of
RXR in either CV-is or primary fibroblasts is not
sufficient to support VDE-mediated transactivation. When
the endogenous level of RXR is supplemented, as in CVDEiR,
3-10% of the infected population was observed to strongly
respond to murA. CVBR, however, was clearly the most
potent transactivator in the CV-1 and FF12 cell types with
200 of CV-1 and 550 of CVBR-infected FF12 cells
histochemically positive.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
73
Luciferase activity in MS-Luc cells infected in
parallel with the same receptor viruses confirmed that CVBR
was not the most potent transactivator in the CV-1 and FF12
cell types and that all receptor types were functional in
293 cells. The MS-Z data suggest that the low absolute
level of CVDEiR induction in CV-1 and FF12 cells with murA
most likely reflects a small number of robustly responding
cells in a largely nonresponsive population as opposed to
low level induction by most MS-luc-infected cells.
Examgle 18
Transient transfections
Transient transfections are performed by calcium-
phosphate coprecipitation employing standard methods (see
Sambrook et al., in Molecular Cloning: A Laboratory Manual.
Cold Spring Harbor Press, New York, New York (1989)). All
tissue culture experiments are performed using DMEM 10 o FBS
in a 10% COz incubator unless otherwise specified. All
transfections are performed in triplicate in 24-well Costar
plates using CV-1 cells at an approximately 5X104 plating
density. Immediately following transfection, 1 ~,m ligand
in 20% EtOH/PBS is added to wells. After 40 additional
hours of incubation, cells are harvested and luciferase
activity measured in an analytical bioluminescence
photometer. Cell extracts are simultaneously examined for
LacZ activity of the internal control by standard methods.
Bar graph levels represent relative luciferase activity
after correction using internal control values. All
molecular biology enzymes and reagents used in this study
are provided by either NEB (Beverly, MA) or Strategene (La
Jolla, CA).
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
74
Example 19
Prolonged transaene expression for
ex vivo gene therapy applications
Autologous, explanted skin fibroblasts genetically
modified to express tyrosine hydroxylase (TH), the enzyme
responsible for synthesis of L-dopa and the precursor for
the neurotransmitter dopamine, have proven to ameliorate a
loss of local dopamine in animal models of Parkinson's
disease. Although quite effective in providing dopamine to
the area of neural damage, this technique is only
therapeutically useful for 2-3 weeks following
transplantation. The decrease in effectiveness has been
traced back to a dramatic loss of TH transgene expression
in transplanted, post-mitotic cells. The use of invention
constructs allows one to overcome the loss of transgene
expression by providing stimulation of the retroviral LTR
promoter through either ligand-activated transactivating
complexes or through constitutive transactivating receptor
variants. In this way, transgene expression may be
maintained for longer periods of time, even indefinitely if
desired.
Stem-type cells, such as stem cells of the
hematopoetic system, nervous system, or embryo, could be
infected or transfected with VbR regulated transgenes and
subsequently implanted into adult, fetus, or early
embryonic animals for either therapeutic or research
purposes. Cells of the hematopoetic system could
conditionally express proteins producing blood clotting
factors such as factor IX, metabolic factors such as
glucocerebrosidase, or protective factors including
anti-HIV proteins.
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
Examgle 20
Prolon~c~ed transcrene expression for
in vivo gene therapy applications
The small size and regulatory capacity of VbRs lend
5 themselves to use in recombinant retroviruses as a method
of gene transfer. VbRs have been introduced into both MLV
and lentiviral-based retroviral systems. When these
viruses are introduced directly into target cells of either
a mature or developing organism, expression of the virally
10 encoded transgenes may be regulated by systemic addition of
ligands such as tebufenozide and derivatives. An example
of a disease that could be theoretically ameliorated by
application of in vivo VbR encoding retroviruses is
Parkinson's disease, described above.
15 Another application would be to use VbR encoding
retroviruses as an anti-viral agent. Lentiviral vectors
with regulated properties and harboring suicide genes or
"protective" proteins could be used as a means of
conditionally depleting or destroying HIV-positive cells.
20 Example 21
Modulated transaene expression for either
in vivo or ex vivo gene therapy at~plication
Treatment of Parkinson's disease with the chemical
precursor of dopamine, L-dopa, has proven effective in
25 ameliorating many of the deficits of Parkinsonism. With
time, however, patients become refractory to L-dopa
therapy, with the deleterious effects of chronic treatment
' outweighing even the serious symptoms of the disease
itself. Eventually, patients are left with few therapeutic
30 options. While the transplantation of TH expressing cells
may be effective when constantly producing low-levels of L-
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98114215
76
dopa, a potentially far more beneficial approach would be
to allow the physician some degree of control over L-dopa
production in the patient. This would allow sufficient
control to ensure that the transgenic factor is expressed
at appropriate therapeutic levels. At times when
endogenous systems are capable of providing full function,
the transgene may be allowed to become quiescent and
transcriptionally inactive until needed again. Because the
transcriptional induction of the invention retroviral
constructs a.s dependent on an exogenous ligand, expression
of an integrated therapeutic transgene can be placed under
the control of the physician and patient.
Example 22
Construction of a BORIS vector expressing PolyO-GFP
PolyQ-GFP are constructed from an existing GFP clone,
GFP-SKSP. PCR primers encoding a novel restriction site
and ATG start codon at the 5' end, and an Age I site
in-frame with the GFP ORF at the 3' end are obtained for
PCR amplification of huntingtin polyQ regions. These PCR
products are inserted in-frame into GFP-SKSP to produce the
fusion constructs of pQ-GFP. The clones are analyzed for
mutations and function by sequencing and in vitro
translation to confirm the creation of a pQ-GFP ORF.
Alternate antibiotic resistance BORIS regulatable
retroviral vectors are produced by removing the
L-Histidinol gene from plasmid LSHL, by PCR, and inserting
it in place on the neon into a BORIS vector for production
of BORIS-LHis. The plasmid are large-scale prepped. A
preliminary test for function is transient transfection
experiments into transfectable cell types such as 293 cells
and examination for the expression of beta-galactosidase or
GFP reporter genes. Retrovirus are transiently produced
and used for infection of target cells . Infected cells are
selected in L-Histidinol containing media to ensure proper
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
77
selection. polyQ-GFP is then inserted into regulated
retroviral vectors. pQ-GFPs are excised from SKSP and
inserted into the regulatable BORIS and BORIS-LHis vectors.
The plasmids are large-scale prepped. Function is
determined by examination for green fluorescence and
altered phenotype. Additional plasmid are used for
transient production of retroviruses.
Primary rat fibroblasts, and the hcnlv-myc are selected for
initial testing of BPQGS. Both are infected with the
appropriate vectors) place under selection, and individual
resulting colonies subcloned. The subclones are expanded
and passaged for analysis. Infected cells are treated or
not with their respective ligands and the resulting
phenotypes analyzed. Preliminary analysis will examine GFP
fluorescence and distribution within the cells. At timed
intervals following ligand induced expression, the pattern
of fluorescence are recorded to determine if the fusion
protein changes in distribution. Morphological
characteristics of stimulated cells their growth rates, and
cell death will also be examined at intervals following
induction. For the hcn/v-myc cells, the consequences of
polyQ-GFP expression in the presence and absence of
tetracycline are compared.
Examgle 23
Aaareaation of goly0-fusion proteins
GFP fusion proteins with long (97Q), short (13Q), or
no N-terminal polyQ tracts were constructed. Unique
restriction endonuclease sites were designed into the
regions flanking the polyQ tracts to allow insertion of an
SV40 nuclear localization signal (NLS) or other sequences
either C-terminal to the polyQ tract. Transient
transfection analysis of these constructs in the NIT
expression vector revealed that cells transduced with GFP
CA 02296093 2000-O1-10
WO 99/02683 PCT1US98/14215
78
only or 13Q-GFP expressed GFP uniformly throughout their
cytoplasm and that the NLS variant 13QN-GFP localized
predominantly in the nucleus with a uniform distribution.
97Q- and 97QN-GFP constructs, on the other hand, initially
manifested as uniform GFP positive fluorescence throughout
the cytoplasm or nucleus, but within 24-72 hours after
transfection condensed into bright granules indicative of
intracellular aggregates (IAs). IAs were localized within
the cytoplasm, surrounding the nuclear envelope, and within
the nucleus itself (Figure 1B). The presence of the SV40
nuclear localization signal (NLS), though observed to
efficiently direct localization of either 13Q- or 97Q-GFP
variants to the nucleus, resulted in a quantifiable
difference in the rate of IA formation after transfection.
Example 24
GFP-fusion proteins with short ~olvglutamine tracts are
recruited into IAs
It was sought to determine whether overexpression of
13Q-GFP constructs would inhibit or interfere with the
formation of IAs from 97Q-fusion constructs. 13Q-GFP was
not observed to exert any inhibitory influence on 97Q-GFP
mediated IA formation, and contrary to expectations,
appeared to result in a qualitative difference in the IAs
that formed. To test whether the 13Q-GFP fusion proteins
were participating in the formation of IAs, the
GFP-encoding portion of 97Q-GFP (97Q~) was deleted and this
construct was cotransfected with 13Q-GFP into cultured
293 cells. Fluorescent IAs were readily observed in a
large population of the GFP-positive cells within the 24-72
hr. time course observed for 97Q-GFP alone. 13Q-GFP/97Q~
doubly infected cells were distinctive from 97Q-GFP cells
because they continued to display faint uniform cytoplasmic
GFP positivity in addition to the formation of
predominantly nuclear IAs, whereas IA formation within
CA 02296093 2000-O1-10
WO 99102683 PCT/US98114215
79
97Q-GFP cells generally results in condensation of all GFP
fluorescence into IAs leaving other areas of the cell
nonfluorescent and dark. Control cotransfection of
NIT-97Qo with NIT-GFP constructs with no N-terminal
polyglutamine tracts did not result in the formation of IAs
under any conditions tested, indicating that the short
polyQ tract of 13Q-GFP is necessary for participation in IA
formation.
Preliminary studies of 13Q-GFP recruitment were
performed with a molar excess of 97Q~ plasmid. To further
determine the molar ratio of 97Q to 13Q required for
participation of 13Q in IA formation, 13Q-GFP was titrated
with 97Q~ in a cotransfection assay and the percent of IA
containing cells quantified relative to the total number of
visibly GFP positive cells. No IAs were observed with non
cotransfected 97Q~ or at a 0.1 molar ratio of 97Q~ plasmid;
however, at a 0 . 5 molar ratio of 97Q~ to 13Q-GFP, 10 0 of
GFP positive cells contained GFP positive IAs. At a 1:1
ratio, nearly 40% of GFP positive cells also displayed IAs.
At 10:1 ratio of NIT-97Q~ to NIT-13Q-GFP, over 750 of
positive cells displayed IAs 60 hours post-transfection.
Example 25
Characterization of IAs formed from a combination of long
and short polvQ-reporters
To better characterize the formation of IAs in the
presence of both long and short polyQ tracts, 97Qo was
modified to contain an in-frame fusion of the LacZ gene
encoding (3-galactosidase (97QZ). Preliminary transient
transfection studies of 97QZ revealed that like 97Q-GFP,
97QZ condensed within transfected cells to form both
immunopositive and histochemically reactive IAs. NIT-97QZ
and NIT-13Q-GFP were cotransfected into cells and 60 hours
later, performed irnmunohistochemistry and histochemical
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98114215
reactions on the transfected cells to simultaneously
examine both reporter constructs. Although the
~-galactosidase (~i-gal) histochemical reaction obscures GFP
fluorescence, through slight modification of standard
5 staining techniques and limiting the time of exposure to
reagents, a low level of GFP fluorescence could be
preserved and imaged along with the characteristic blue
stain of the (3-galactosidase reaction. A population of IAs
are observed that are both GFP and (3-galactosidase
10 positive. Numerous large (3-gal only positive IAs are also
present, presumably from the aggregation of 97QZ only (or
predominantly). Curiously, several IAs that appear only
positive for GFP were also observed using the histochemical
stain and native GFP fluorescence imaging.
15 Further exploration of this observation was performed
using confocal microscopy and immunohistochemistry to
identify reporters within IAs. As in the above study,
individual IAs were clearly observed to be positive for
both reporters while other IAs were positive only for (3-gal
20 for GFP. Taken together, these results clearly indicate
that long polyglutamine tract-reporter fusion constructs
form IAs in vitro, and that the forming aggregates can
either recruit short polyglutamine-reporters into the
aggregate, induce them to aggregate on their own, or both.
25 Example 26
Construction of cell lines with regulated IA formation
and 97Q-GFP expression
The study of IA formation has thus far relied heavily
on transient transfection analysis and has been hampered by
30 a lack of stable IA-forming cultured cell populations.
Through the use of retroviral vectors, a variety of stably
cultured cell lines conditionally or chronically expressing
the polyQ-GFP constructs were constructed. Two regulatable
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
81
vector systems, an induced expression system based on a
modified ecdysone receptor variant (VbR) and a repressed
expression system based on the tetracycline transactivator
(TTA), were used to create cell lines with inducible IA
formation. These two vectors are best suited to different
ranges of induction in cultured cell types. VBR-based B3
vectors are particularly suited to applications where very
low basal expression levels that can be rapidly stimulated
to moderately high levels are desired. The TTA-based NIT
1Q vector tends not have the broad-based low level of
"uninduced" expression characteristic of the BORIS vectors
(identified in SEQ ID N0:6); however, because
transactivator expression and transactivation is continuous
in the absence of ligand, very high transgene expression
levels may be achieved through the use of this vector.
97Q-GFP was cloned via a custom shuttle vector into
both retroviral vectors. Transiently produced B3-polyQ-GFP
retroviruses (B3-13Q or B3-97Q) and TTA-based NIT polyQ-GFP
retroviruses (NIT-13Q or NIT-97Q) were used at low MOI to
infect several well-characterized cell lines used
previously in transduction studies of IA formation. COS-7
cells were selected because several studies had examined IA
formation and IA-induced apoptosis in this cell type.
293 cells were utilized for the same reason and also
because they are amenable to efficient transfection and
would have significant utility in the development of
gene-based factors that interact with or interrupt IA
formation. Primary rat fibroblasts were selected because
they are non-transformed and readily enter a prolonged
post-mitotic state following contact inhibition at high
culture densities.
Pilot studies with reporter constructs revealed that
the B3 vector functioned most efficiently in the 293 and
COS cell lines and that NIT functioned at highest
CA 02296093 2000-O1-10
WO 99102683 PCT/US98/14215
82
efficiency in the primary fibroblast line. Individual cell
lines infected with the appropriate vector were placed
under 6418 selection to remove uninfected cells. For
B3-polyQ infected 293 and COS-7 cells, the resulting
resistant population was pooled and passaged without
ligand. For NIT-polyQ infected primary fibroblasts, cells
were cultured with the tetracycline analog doxycycline
prior to, during, and after infection to prevent chronic
polyQ overexpression to disrupt growth of resistant
colonies. 293 and COS-7 cells were also infected with NIT
constructs and cultured without added doxycycline to
determine the effects of chronic overexpression of 97Q-GFP
on these cell types as described below.
Example 27
Stable IA-forming' 293 cells
6418-resistant B3-97Q 293s displayed little to no
visible GFP fluorescence during culture in the absence of
ligand; however, introduction of 1 ~.M tebufenozide into the
culture medium resulted in a rapid increase of cytoplasmic
fluorescence within 24 hours post-induction. Nearly 1000
of cells would display GFP-positive fluoresence within
24-72 hours of induction. After 24 hours, only widely
scattered cells would contain small, detectable IAs. By
48 hours, >8% of the cell population would contain one or
more IAs increasing to >150 of the population between 48
and 72 hours. Occasional IA containing cells would be
observed floating in the culture medium; however, there was
no evident "wave" of cell death at the 72 hour time point
or later times. Passage of B3-97Q 293s in the presence of
ligand for three passages resulted in only a small but
detectable decrease in the percentage of cells containing
IAs (15% to llo) .
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
83
NIT-97Q infection sheds further light on the toxic
potential of long-polyglutamine repeats on 293 cells.
Surprisingly, colonies of 100% IA-positive 293 cells
survived for over two weeks and apparently divided and
grew, in some cases, to colonies of hundreds of cells
(Figure 4D). By the end of 21 days of culture; however,
the majority of IA positive colonies had disappeared
leaving only scattered IA positive cells and numerous
colonies of GFP positive cells without IAs that eventually
grew to fill the plates. The expression time-course and
level of B3- AND NIT-13Q cells paralleled the 97Q cells
although no IAs were formed.
Example 28
Stable IA-forming COS-7 cells
6418-resistant B3-97Q COS-7s displayed no visible GFP
fluoresence during culture in the absence of ligand in a
manner almost identical to the 293 B3-97Q population.
Addition of 1 ~.M tebufenozide into the culture medium
resulted in a rapid increase of cytoplasmic fluorescence
within 24-48 hours post-induction. IA formation peaked at
4-50 of total cells (at any given moment) 72-96 hours
post-induction. Unlike the B3-97Q 293 cells, a continuous
population of 6418-resistant cells did not display even GFP
fluoresence suggesting that one or more components of the
regulatable system were lost from the COS population. Like
B3-97Q 293 cells, IA containing COS-7 cells were also
observed floating in the culture medium; however a
depletion of the expressing population was not observed
presumably because the continuous formation of IAs in other
cells.
The effects of chronic 97Q-GFP overexpression in
infected COS-7 cells was examined using the NIT vector.
Like NIT-97Q infected 293s, colonies in which the majority
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98/14215
84
of cells contained large and numerous IAs were readily
observed. Unlike the overexpressing 293 cells; however,
several selected colonies continued to grow and express IAs
in a continual percentage of the population even after
passage. Three lines, COSN97Q-4, 6 and 7, continually
express IAs after continuous growth for over four weeks.
COSN97Q-7 displays the highest level of spontaneous IA
formation. At any given time, approximately 60 of the
total population contains readily observable IAs. Cv-1
cells, a predecessor of the COS-7 cells were also infected
with NIT-97Q and formed IA containing colonies in much the
same manner as COS cell populations.
Example 29
Stable IA forming ~rimarv fibroblasts
B3-97Q infected primary fibroblasts (FFl2s) became
faintly GFP fluorescent within 72 hours of tebufenozide
treatment but did not achieve a maximum level of 97Q-GFP
expression sufficient to result in IA formation in a
significant population of cells. The NIT vectors were
employed to generate regulated 97Q-GFP expression.
Infected NIT-97Q FFl2s were infected and cultured
continuously in the presence of doxycycline (dox) to
prevent IA formation. Twelve individual colonies of
NIT-97Q FFl2s were selected in order to screen for
individual cell populations with low dox inhibited
expression and high 97Q expression in the absence of
ligand. One line, FFN97Q-5 displayed GFP fluoresence
within 900 of the confluent cell population within 5 days
of ligand removal. IAs began forming on day 5 and
continued to increase through day 8. Approximately 15% of
the total cell population displayed IAs 10 days post ligand
removal. IA formation in the confluent FFN97Q-5 population
was distinctive from other cell types examined in that it
was occasionally exclusively nuclear and at other times
CA 02296093 2000-O1-10
WO 99102683 PCTlUS98/14215
predominantly cytoplasmic in an unusual pattern within the
cell body. IAs with a distinctive stellate or tri-partite
appearance were often observed. Large cytoplasmic
aggregates of multiple star-like IAs were also occasionally
5 seen. Curiously, confluent FFN97Q-5 cells with multiple
cytoplasmic or nuclear IAs appeared visually normal.
Floating dead cells with or without IAs were rarely
observed and in numbers no different from control wells.
FF970Q-%s cultured in the presence of dox neither expressed
10 visible 97Q-GFP nor formed IAs.
Example 30
Use of invention for efficient
production of transaenic animals
Transgenic animals are generally produced by either
15 pronuclear injection of DNA or by transfection of embryonic
stem (ES) cells followed by selection and injection of the
stem cell into the inner cell mass of very early embryos.
Pronuclear injection results in approximately 5-10% stable
gene transfer in the production of transgenic mice. The
20 use of ES cells in producing transgenics is likewise
inefficient in generating mosaics with germ-line
transmission of the transgene. It was proposed in the mid-
1980's to use retroviruses to transfer transgenes with high
efficiency into early embryos or ES cells to dramatically
25 enhance the odds of producing transgenic animals. All
attempts at this failed, not because the virus was
incapable of stably integrating into the target cell
genome, but because the integrated provirus did not express
any of the genes encoded within the viral transcriptional
30 cassette.
The present invention is capable of overcoming the
transcriptional block to result in germ-line transgenic
animals with full expression from the integrated transgene.
CA 02296093 2000-O1-10
WO 99/02683 PCTIUS98114215
86
In addition to expressing the transgene, the level of
transcription may still be regulated by controlling the
supply of Iigand to the transgenic animal. The increased
efficiency of producing transgenic animals by retroviral
infection should open up the way to producing mutant
animals of a variety of species previously impractical for
genetic modification because of the potential cost of
producing a large number of non-positive animals by
classical methods.
To produce transgenic mice, the following nucleic acid
constructs are prepared and subsequently injected into
fertilized eggs: CD3-VbR and a ligand inducible f3-gal
reporter. Two separate lines of transgenic mice are
generated harboring either a ligand inducible reporter, or
a T-cell specific expression construct of VbR,
respectively. The former are referred to as reporter mice,
the latter are referred to as receptor mice, and double
transgenic mice are referred to as receptor/reporter mice.
Constructs CD3-VbR are injected, while the reporter is
injected alone. Primary genotyping is performed by
Southern blot analysis and the transmission of transgenic
mice is monitored by dot blot analysis. Receptor mice are
analyzed for VbR expression by Northern blot analysis of
RNA collected from these mice. For Northern blot analysis,
l5,ug of total RNA obtained from the thymus, and various
tissues as a control, is run on a denaturing gel and
blotted onto a nitrocellulose membrane. The blot is probed
with a radiolabeled f3-gal-specific probe and exposed on
film for 2 days. In addition, the transgene can be
transferred to the offspring as expected by Mendelian
genetics.
CA 02296093 2000-O1-10
WO 99/02683 PCT/US98114215
87
Example 31
Use of retroviral constructs in the invention for
efficient gene transfer to develot~ina embryos
Since the present invention can effectively overcome
the block of viral expression in embryonic cells, invention
constructs are a potent tool in the delivery of transgenes
to somatic tissues of a developing embryo. With many
diseases, considerable damage is done during embryonic
development so that therapies applied after birth are
essentially ineffective to ameliorate the disease
phenotype.
The present invention provides methodology where one
can infect cells of the embryo and can provide therapeutic
factors to the developing fetus either constitutively, or
under the regulation of exogenously produced ligand.
Example 32
Use of vector constructs in invention
with inducible hiah titers
One obstacle to the use of retroviruses as gene
transfer agents is that titers of retroviruses from
existing producer cell lines are only on the order of 1X104
or 1X105. By using a retroviral construct of the invention
having intact enhancers and regulatory elements, expression
of the retrovirus may be induced by greater than ten-fold,
resulting in correspondingly higher titers of infectious
virus.
While the invention has been described in detail with
reference to certain preferred embodiments thereof, it will
be understood that modifications and variations are within
the spirit and scope of that which is described and
claimed.