Note: Descriptions are shown in the official language in which they were submitted.
CA 02297616 2000-O1-31
1
VIRAL VECTORS
TECHNICAL FIELD
The present invention relates generally to recombinant plant viral
nucleic acids, and methods of use thereof.
PRIOR ART
Recombinant plant viral nucleic acids are of interest generally for
their utility as expression vectors in plants.
Additionally, such nucleic acids can be used to initiate virus
induced gene silencing (VIGS). This phenomenon is based on the
observation that virus infection in plants can initiate sequence-
specific nucleic-acid based defence mechanisms that resemble either
transcriptional, or post-transcriptional gene silencing (PTGS)
(Covey, Al-Kaff 1997; Ratcliff, Harrison et al. 1997; A1-Kaff, Covey
et al. 1998). PTGS is also manifest as an inhibition of nuclear gene
expression when a virus is modified to carry sequence from a nuclear
expressed gene (Kumagai, Donson et al. 1995; Kjemtrup, Sampson et al.
1998; Ruiz, Voinnet et al. 1998). PTGS can also cause recovery from
viral infection when a plant expressing a transgene derived from
viral cDNA is infected by a homologous virus (Lindbo, Silva-Rosales
et al. 1993; Guo and Garcia 1997). Both the inhibition of nuclear
gene expression, and recovery from viral infection are caused by
sequence-specific RNA degradation.
Because modified viruses inhibit the expression of homologous plant
genes, VIGS can be used to induce an apparent null-phenotype or a
loss of function and therefore identify the function of any gene.
Viruses that have been modified in this manner include tobacco mosaic
virus (TMV) (Kumagai, Donson et al. 1995) potato virus X (PVX) (Ruiz,
Voinnet et al. 1998), and tomato golden mosaic virus (Kjemtrup,
Sampson et al. 1998).
DISCLOSURE OF THE INVENTION
The present invention is concerned with novel recombinant plant viral
nucleic acids.
In preferred forms the present invention is concerned with providing
VIGS-based methods and materials which may be more suitable as a tool
for functional genomics than those which have been used in the past.
For instance TMV, PVX and TGMV infections cause significant symptoms,
such as a chlorosis, leaf-distortion and necrosis. Phenotypes caused
by VIGS of a plant gene can therefore be hard to differentiate from
these viral symptoms. Secondly, like most viruses, TMV, PVX and TGMV
form mosaic, vein-based infections, and therefore do not cause
confluent VIGS across the whole leaf. Leaves may therefore contain a
mixture of cells with and without VIGS, complicating interpretation
of any phenotype. Thirdly, TMV, PVX and TGMV do not infect
meristems (Matthews 1991) and can not therefore inhibit expression of
genes that determine the identity and development of plant tissue.
Finally, although the first plant genome to be fully sequenced will
CA 02297616 2000-O1-31
2
be that of Arabidopsis thaliana, TMV, PVX and TGMV vectors do not
infect this plant. Therefore the potential of VIGS to identify gene
function in Arabidopsis is limited with available technology. A
VIGS vector which overcame one or more of these drawbacks would
therefore represent a contribution to the art.
The present inventors have developed novel recombinant cDNA viral
constructs based on tobacco rattle virus (TRV) which in preferred
forms are particularly adapted for use with VIGS. Such vectors may
induce few or no symptoms, cause confluent VIGS across the leaf,
operate in Nicotiana species and in Arabidopsis, and inhibit gene
expression in meristems.
A viral expression vector based on TRV has previously been described
in which non-viral proteins were expressed from a sub-genomic
promoter (Ratcliff, MacFarlane et al. 1999). The viral RNA was
synthesised in vitro and then inoculated into the plant. The TRV
vectors of the present invention include inter alia modifications to
facilitate both the insertion of plant gene sequences and the
subsequent infection of plants. Other TRV based vectors are
disclosed by Hamilton & Baulcombe (1989) J. Gen. Virol 70: 963-968
and Mueller et al (1997) J. Gen. Virol 78: 2085-2088.
Thus in a first aspect of the present invention there is disclosed a
nucleic acid vector which comprises:
(a) a transfer nucleotide sequence comprising (i) a plant active
promoter, operably linked to (ii) a recombinant tobacco rattle virus
(TRV) cDNA which includes at least cis acting elements permitting
replication of said cDNA; a subgenomic promoter operably linked to a
sequence encoding a TRV coat protein; and a heterologous nucleotide
sequence which is foreign to said virus;
(b) border sequences which permit the transfer of the transfer
nucleotide sequence into a plant cell nucleus.
The transfer nucleotide sequence is situated between the border
sequences and is capable of being inserted into a plant genome under
appropriate conditions. Generally this may be achieved by use of so
called "agro-infiltration" which uses Agrobacterium-mediated
transient transformation. Briefly, this technique is based on the
property of Agrobacterium tumafaciens to transfer a portion of its
DNA ("T-DNA") into a host cell where it may become integrated into
nuclear DNA. The T-DNA is defined by left and right border sequences
which are around 25 nucleotides in length. In the present invention
the border sequences are included around the transfer nucleotide
sequence (the T-DNA) with the whole vector being introduced into the
plant by agro-infiltration, optionally in the form of a binary-
transformation vector.
By "plant active promoter" is meant a sequence of nucleotides from
which transcription may be initiated of DNA operably linked
downstream (i.e. in the 3' direction on the sense strand of double-
stranded DNA). "Operably linked" means joined as part of the same
nucleic acid molecule, suitably positioned and oriented for
transcription to be initiated from the promoter. Nucleic acid
operably linked to a promoter is "under transcriptional initiation
CA 02297616 2000-O1-31
3
regulation" of the promoter.
The cDNA includes cis acting elements permitting replication of said
cDNA. However the vector need not include all of the sequence
required to replicate and move within the plant. The vectors of the
present invention will generally require supplementary proteins
and/or nucleic acids from TRV in order to achieve this. Thus the
cDNA may correspond to part of TRV RNA 2, and will thus require
proteins encoded by TRV RNA1 for replication.
The TRV coat protein (as with other defined or recited sequences
herein) need not be 'wild-type', but may optionally be a variant
(e. g. mutant, or other variant, or a substantially homologous
derivative) provided that its function (to encapsulate and permit
movement of the TRV genome) is not negated. By "Substantially
homologous" is meant that the sequence in question shares at least
about 70%, or 80% identity, most preferably at least about 90%, 95%,
96%, 97%, 98% or 99% identity with the reference sequence. Identity
may be at the nucleotide sequence and/or encoded amino acid sequence
level. Homology may be over the full-length of the relevant sequence
shown herein (e.g. in the sequence Annex) or may be over a part of
it. Identity may be determined by the TBLASTN program, of Altschul
et al. (1990) J. Mol. Biol. 215: 403-10, or BestFit, which is part
of the Wisconsin Package, Version 8, September 1994, (Genetics
Computer Group, 575 Science Drive, Madison, Wisconsin, USA, Wisconsin
53711). Preferably sequence comparisons are made using FASTA and
FASTP (see Pearson & Lipman, 1988. Methods in Enzymology 183: 63-98).
Parameters are preferably set, using the default matrix, as follows:
Gapopen (penalty for the first residue in a gap): -12 for proteins /
-16 for DNA; Gapext (penalty for additional residues in a gap): -2
for proteins /-4 for DNA; KTUP word length: 2 for proteins / 6 for
DNA.
The heterologous nucleotide sequence is foreign (non-native) to TRV,
which is to say that it does not occur naturally in the TRV viral
genome at the position in which it is present in the VIGS vector.
The sequence will generally be either a cloning site (to permit the
insertion of a desired sequence) or a desired sequence itself.
Some preferred embodiments of the invention will now be discussed.
Vector
This is preferably based on plant binary transformation vector pGreen
(see Materials and Methods below). The vector may be an expression
vector (for transcription of a desired sequence, which may then be
translated). Alternatively (and preferably) the vector is a "VIGS
vector", by which is meant one which is adapted to cause or permit
virus induced gene silencing of a desired target nucleotide sequence
corresponding to a sequence included in the vector.
Nucleic acid vectors according to the present invention may be
provided isolated and/or purified, in substantially pure or
homogeneous form, or free or substantially free of other nucleic
acid. The term "isolated" encompasses all these possibilities.
CA 02297616 2000-O1-31
4
Generally speaking, in the light of the present disclosure, those
skilled in the art will be able to construct vectors according to the
present invention. Such vectors may include, in addition to the
promoter, a suitable terminator or other regulatory sequence such as
to define an expression cassette consisting of the recombinant TRV
cDNA and the heterologous nucleotide sequence. For further details
see, for example, Molecular Cloning: a Laboratory Manual: 2nd
edition, Sambrook et al, 1989, Cold Spring Harbor Laboratory Press.
Many known techniques and protocols for manipulation of nucleic acid,
for example in preparation of nucleic acid constructs, mutagenesis,
sequencing, introduction of DNA into cells and gene expression, and
analysis of proteins, are described in detail in Protocols in
Molecular Biology, Second Edition, Ausubel et al. eds., John Wiley &
Sons, 1992. Specific procedures and vectors previously used with
wide success upon plants are described by Bevan, Nucl. Acids Res.
(1984) 12, 8711-8721), and Guerineau and Mullineaux, (1993) Plant
transformation and expression vectors. In: Plant Molecular Biology
Labfax (Croy RRD ed) Oxford, BIOS Scientific Publishers, pp 121-148.
Plant promoter
Suitable promoters will be well known to those skilled in the art and
include the Cauliflower Mosaic Virus 35S (CaMV 35S) gene promoter
that is expressed at a high level in virtually all plant tissues. The
promoter may in principle be an inducible promoter such as the maize
glutathione-S-transferase isoform II (GST-II-27) gene promoter which
is activated in response to application of exogenous safener
(W093/01294, ICI Ltd). The GST-II-27 gene promoter has been shown to
be induced by certain chemical compounds which can be applied to
growing plants. Another suitable promoter may be the DEX promoter
(Plant Journal (1997) 11: 605-612).
Recombinant TRY cDNA
This is preferably based on a modified, reduced, cDNA clone of TRV
RNA2. In the Examples herein the strain used is ppk20. However any
appropriate strain, which can give rise to replicating, infectious
viral transcripts, could be used (see e.g. Macfarlane, 1999 for
further examples).
Within the cDNA it is preferable that non-essential ORFs or other
sequences are deleted, provided that the cDNA can still be used to
generate replicating, infectious transcripts. Preferably, where the
cDNA is based on TRV RNA2 of ppk20, two open reading frames (37K and
32.8K) are deleted to leave only the 5' and 3' untranslated regions
and the viral gene encoding the coat-protein. The deleted ORFs are
replaced by a heterologous nucleotide sequence between the coat
protein and the untranslated region (UTR). The sequence is shown in
the Sequence appendix (No. 1). Naturally substantially homologous
variants of the sequence are also included within the scope of the
invention. In particular, vectors derived from pTV00 and having the
characteristics (described herein) of that vector, are also embraced.
Vectors based on TRV RNA2 require proteins encoded by TRV RNA1 for
CA 02297616 2000-O1-31
replication, which can be achieved as described below.
Heterologous nucleotide sequence.
5 This can in principle be a single or multiple cloning site (i.e. a
sequence encoding two or more restriction endonuclease target sites)
to facilitate the incorporation of a desired nucleotide sequence.
For expression vectors according to the present invention, the
sequence will generally include or be operably linked to a subgenomic
promoter which is recognised by a TRV-effective replicase (e.g. the
PEBV CP subgenomic promoter) and an ORF sequence which it is desired
to express and which is therefore transcribed as a subgenomic RNA.
For VIGS vectors the sequence will be a "targeting sequence" which
corresponds to a sequence in a target gene, either in the sense or
anti-sense orientation, or a sequence which has sufficient homology
to a target sequence for down-regulation of expression of the target
gene to occur. Such a targetting sequence may be included in the
vector anywhere in the viral cDNA irrespective of the location of any
subgenomic promoter (provided it does not interfere with the cis-
acting replication elements or the coat protein). Generally speaking
it will be preferable for VIGS vectors according to the present
invention not to include a subgenomic promoter within or operably
linked to the heterologous gene sequence. Such preferred vectors
have the advantage that they are more stable (reduced likelihood of
self-recombination) that those of the prior art such as those
described by Ratcliff, MacFarlane et al. (1999) supra which had more
than one subgenomic promoter.
In general the targeting sequence may be derived from a plant nuclear
gene or transgene, or a gene on an extrachromosomal element such as a
plastid.
VIGS is particularly preferred for investigating gene function in
that it can be used to impose an intermediate or a null phenotype for
a particular gene, which can provide information about the function
of that gene in vivo. In such cases the targeting sequence may not be
known, but the methods of the present invention may be used to
identify it with a particular phenotype.
The complete sequence corresponding to the coding sequence (in
reverse orientation for anti-sense) need not be used. For example
fragments of sufficient length may be used. It is a routine matter
for the person skilled in the art to screen fragments of various
sizes and from various parts of the coding sequence to optimise the
relationship between target and targeting sequence. It may be
preferable that there is complete sequence identity between the
targeting sequence in the vector and the target sequence in the
plant, although total complementarity or similarity of sequence is
not essential. One or more nucleotides may differ in the targeting
sequence from the target gene. Thus, a targeting sequence employed
in a construct in accordance with the present invention may be a
wild-type sequence (e.g. gene) selected from those available, or a
substantially homologous mutant, derivative, variant or allele, by
CA 02297616 2000-O1-31
6
way of insertion, addition, deletion or substitution of one or more
nucleotides, of such a sequence. Such a sequence need not include an
open reading frame or specify an RNA that would be translatable.
A further possibility is to target a conserved sequence of a gene,
e.g. a sequence that is characteristic of one or more genes in one or
more pathogens against which resistance is desired, such as a
regulatory sequence.
Other aspects of the invention will now be discussed.
One aspect of the present invention is a process for producing a
vector as described above, the process being substantially as set out
in the Examples hereinafter. A further aspect is a process for
producing a vector as described above, which process comprises the
step of cloning a heterologous nucleotide sequence which is a
targeting sequence into the vector.
Thus one aspect of the present invention includes a method of
silencing a target gene in a plant tissue using VIGS which method
comprises the steps of introducing a vector as described above into
the plant, wherein said vector includes a heterologous nucleotide
sequence which is a targeting sequence.
"Plant tissue" is any tissue of a plant in planta or in culture,
including the whole plant an organ thereof, a cutting, or any group
of plant cells organised into a structural and functional unit.
"Silencing" is a term generally used to refer to suppression of
expression of a gene. The degree of reduction may be so as to
totally abolish production of the encoded gene product, but more
usually the abolition of expression is partial, with some degree of
expression remaining. The term should not therefore be taken to
require complete "silencing" of expression. It is used herein where
convenient because those skilled in the art well understand this.
The method may be preferably used to cause confluent VIGS of the
target gene across a whole leaf and/or to silence a target gene in
meristematic tissue.
As discussed above, for introduction into the plant, the vector may
be in the form of an Agrobacterium binary vector. The vector is
introduced into the plant cell by Agrobacterium-mediated T-DNA
transfer, the transfer sequence may be integrated transiently into
the plant (cell) genome, and is then transcribed to RNA from the
plant promoter. In the published vector of Ratcliff, MacFarlane et
al. (1999), the viral cDNA and any cDNA inserted after the sub-
genomic promoter was transcribed to infectious RNA in vitro by T7 RNA
polymerase and subsequently introduced into the plant.
TRV RNA 2 and all derived constructs require proteins encoded by TRV
RNA1 for replication within and movement though out the plant. TRV
RNA1 infections can be initiated either by rub-inoculating the plant
with purified RNA 1 (Matthews 1991), or by transient Agrobacterium
mediated expression in the plant of the plasmid pBINTRA6, which
CA 02297616 2000-O1-31
7
contains a CaMV 35S driven infectious clone of TRV PPK20 RNA 1 (see
Materials and Methods).
The present invention may particularly be applied in plants which are
natural hosts (compatible with) TRV. By "compatible" is meant
capable of operating with the other components of a system, in this
case TRV must be capable of replicating in the plant in question.
These include Arabidopsis thaliana. Others include (but are not
limited to) Allium cepa; Amaranthus caudatus; Amaranthus retroflexus;
Antirrhinum majus; snap-dragon; Arachis hypogaea; Avena sativa;
Bellis perennis; Beta vulgaris; Brassica campestris; Brassica
campestris ssp. napus; Brassica campestris ssp. pekinensis;
Brassica juncea; Calendula officinalis; Capsella bursa-pastoris;
Capsicum annuum; Catharanthus roseus; Cheiranthus cheiri;
Chenopodium album; Chenopodium amaranticolor; Chenopodium
foetidum; Chenopodium quinoa; Coriandrum sativum; Cucumis melo;
Cucumis sativus; Glycine max; Gomphrena globosa; Gypsophila
elegans; Helianthus annuus; Hyacinthus; Hyoscyamus niger;
Lactuca sativa; Lathyrus odoratus; Linum usitatissimum; Lobelia
erinus; Lupinus mutabilis; Lycopersicon esculentum;
Lycopersicon pimpinellifolium; Melilotus albus; Momordica
balsamina; Myosotis sylvatica; Narcissus pseudonarcissus;
Nicandra physalodes; Nicotiana benthamiana; Nicotiana
clevelandii; Nicotiana glutinosa; Nicotiana rustica; Nicotiana
sylvestris; Nicotiana tabacum; Nicotiana edwardsonii; Ocimum
basilicum; Petunia hybrida; Phaseolus vulgaris; Phytolacca
americana; Pisum sativum; Raphanus sativus; Ricinus communis;
Salvia splendens; Senecio vulgaris; Solarium melongena; Solarium
nigrum; Solarium tuberosum; Spinacia oleracea; Stellaria media;
Trifolium pratense; Trifolium repens; Tropaeolum majus; Tulipa;
Vicia faba; Vicia villosa; Viola arvensis.
Target genes include those which confer 'unwanted' traits in the
plant and which it may therefore be desired to silence using VIGS.
Examples include ripening specific genes in tomato to improve
processing and handling characteristics of the harvested fruit; genes
involved in pollen formation so that breeders can reproducibly
generate male sterile plants for the production of F1 hybrids; genes
involved in lignin biosynthesis to improve the quality of paper pulp
made from vegetative tissue of the plant; gene silencing of genes
involved in flower pigment production to produce novel flower
colours; gene silencing of genes involved in regulatory pathways
controlling development or environmental responses to produce plants
with novel growth habit or (for example) disease resistance;
elimination of toxic secondary metabolites by gene silencing of genes
required for toxin production.
A further aspect provides a process which includes introducing the
vector into a plant, optionally including the further step of
introducing a source of proteins encoded by TRV RNA1 into the plant.
A further aspect of the present invention provides a method which
includes causing or allowing transcription from a construct as
disclosed within the genome of a plant cell to produce a
cytoplasmically-replicating RNA.
CA 02297616 2000-O1-31
8
A further aspect of the present invention provides a method of
reducing or suppressing or lowering the level of a target gene in a
plant cell, the method including causing or allowing transcription
from a vector as disclosed above.
In preferred forms the present invention is concerned with providing
VIGS-based methods are useful in functional genomics. Thus in one
aspect of the present invention, the target gene may be of unknown
phenotype, in which case the VIGS system may be employed to analyse
the phenotype by generating a widespread null (or nearly null)
phenotype. The target gene may be essential, which is to say that
the null phenotype is lethal to the cell or tissue in question.
This aspect of the invention may comprise a method of characterising
a target gene comprising the steps of:
(a) silencing the target gene in a part or at a certain development
stage of the plant using the TRV VIGS system described above,
(b) observing the phenotype of the part of the plant in which or when
the target gene has been silenced.
Generally the observation will be contrasted with a plant wherein the
target gene is being expressed in order to characterise (i.e.
establish one or more phenotypic characteristics of) the gene.
The advantage of the TRV system over certain prior art constructs is
discussed above. There are also several advantages of the current
method over alternative methods in which the targeted gene is
inactivated by insertional or other mutagenic procedures. The
advantage over mutagenic procedures applies when there is more than
one homologous gene carrying out the role of the target gene.
Mutagenic procedures will not normally reveal a phenotype in that
situation. A second situation where the current invention has
advantage over both mutagenic and unregulated gene silencing
procedures applies when the target gene has a lethal phenotype. The
controllable attribute of the gene silencing will allow the phenotype
of such genes to be investigated and exploited more efficiently than
using the alternative methods available prior to the disclosure of
the current invention.
Nor, for the identification of endogenous genes, would it be
necessary to try and generate a transgenic plant in which gene
silencing is already activated to observe the effect.
In a further aspect there is disclosed a method of altering the
phenotype of a plant comprising use of the silencing method discussed
above. Traits for which it may be desirable to change the phenotype
include the following: colour; disease or pest resistance; ripening
potential; male sterility.
In a further aspect of the present invention there is disclosed a
virus or viral particle including, preferably encapsulating, a
vector (or transcript from the expression cassette in the vector)
according to the present invention.
a
CA 02297616 2000-O1-31
9
In a further aspect of the present invention there is disclosed a kit
comprising a vector as described above, plus a source of TRV RNA1
polypeptide or vector encoding the same (e. g. pBINTRA6).
In a further aspect of the present invention there is disclosed a
host cell including a vector according to the present invention.
These may be plant cells, or may be microbial (particularly bacterial
and especially Agrobacterium) cells.
In a further aspect there is disclosed a plant, or plant tissue,
including, or transiently transformed by, a vector of the present
invention.
The invention will now be further described with reference to the
following non-limiting Figures and Examples. Other embodiments of
the invention will occur to those skilled in the art in the light of
these.
FIGURES
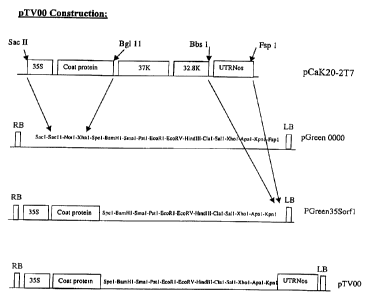
Figure 1; Schematic illustration of the cloning steps to produce
pTV00. 35S is the CaMV 35S promoter; 37K and 32.8K are the open
reading frames for the 37K and 32.8K proteins; UTR is the
untranslated region; Nos is the nopaline synthase transcriptional
terminator; RB is the right border; LB is the left border.
Figure 2.
A; Schematic drawing of TRV RNA1; 5'UTR and 3'UTR are the 5' and 3'
untranslated regions respectively; Rep 134 K is the 134KDa replicase
protein; Rep 194 K is the 194 KDa read-through replicase protein; MP
is the movement protein; 16K is the 16 KDa protein. B; The relative
positions of the PCR1 and PCR2 cDNA fragments.
Figure 3.
Schematic illustration of the cloning strategy for pBSTRF16.
Figure 4.
Schematic representation of the cloning strategy for the introduction
of intron 3 from A. thaliana NIA1 gene to TRV RNA 1, to obtain
pBSTRA3. INT is NIA1 gene intron 3.
Figure 5.
Schematic representation of pBINTRA6. LB and RB respectively are the
left border and right border of pBINTRA6 T-DNA.
SSQU$NCS APPBNDIC$S
1. The full sequence of pTV00.
2. PEBV promoter sequence.
3. N. benthamiana pds cDNA fragment.
4. A. thaliana pds cDNA fragment.
CA 02297616 2001-04-25
5. N. benthamiana rubisco cDNA fragment.
6. GFP (,PCR amplified) .
5 7. N. benthamiana NFL cDNA fragment.
8' The partial sequence of pBINTRA6.
EXAMPLES
General materials and methods
All DNA modifications and digestions were performed using enzymes
according to the manufacturers' instructs a.nd following protocols
described by Sambrook et aI. (Sambrook, Fritsch et al. 1989).
The TRV vector pTV00 was derived from pCaK20-2T7, a previously
described clone of TRV RNA2 (Hernandez, Mathis et al. 1995). pCaK20-
2T7 contains a full-length cDNA of TRV strain PPK20 RNA2 cloned
between the cauliflower mosaic virus (CaMV) 35S promoter and the Nos
terminator.
For the construction of pTV00 two fragments of pCaK20-2T7 were cloned
into the binary plant transformation vector pGreen0000 (described
below). The first fragment, of 1536bp, included the CaMV 35S
promoter, and cDNA sequence encoding the TR.V RNA2 5' untranslated
region and coat protein. This fragment was released by digesting
pCaK20-2T7 with Bgl II, treating with T4 DNA polymerase , and
subsequently digesting with Sac II. The resulting fragment was
ligated into the binary plasmid pGreen 0000 that had been cut with
Xba I, blunted with T4 DNA polymerase and digested with Sac II, to
form the plasmid pGreen35Sorfl.
The second fragment of pCaK20-2T7, containing the 3' untranslated
region (UTR) of TRV RNA2 and the Nos terminator, was released by
digestion with BbsI and FspI and then blunted with T4 DNA polymerase.
This 835bp fragment was ligated into pGreen.35Sorf1 at an Fsp I site,
to form pTV00. The full sequence of pTV00 is given in Appendix 1. A
schematic illustration of the cloning procedure is shown in Figure 1.
These cloning steps had three purposes. Firstly, the TRV RNA 2 cDNA
clone was introduced to the T-DNA of a plant binary-transformation
vector. This allows Agrobacterium mediated-infection of TRV RNA 2
without the need for in vitro transcription. as previously described
for another TRV RNA2 clone (Ratcliff, MacFarlane et al. 1999). The
second effect of these cloning steps was to remove the cDNA sequences
that encode the 37K and 32.8K proteins. These proteins are
dispensable for TRV infection, and are required for transmission of
TRV by nematode vectors (MacFarlane 1999). The third effect was to
introduce 12 unique restriction enzyme sites into the genome of the
TRV RNA 2. This multiple cloning site (MCS) is more convenient for
the insertion of novel sequences to TRV RNA. 2 than the single
restriction enzyme site present in the previously described TRV RNA 2
vector (Ratcliff, MacFarlane et al. 1999).
CA 02297616 2000-O1-31
11
A 409bp cDNA fragment of pds was PCR amplified from N. benthamiana
cDNA using Taq DNA polymerase and the primers 5'ggcactcaactttataaacc
and 5'cttcagttttctgtcaaacc. This pds cDNA fragment (Appendix 3) was
cloned into the Sma 1 sites of pTV00 and into pGR107, to form pTV.pds
and pGR107.pds respectively.
For construction of pTV.apds a l.7kb fragment of the pds gene was PCR
amplified from A. thaliana cDNA using Taq DNA polymerase and the
primers 5'cccctcgagagatgtcaaatc and 5'cccctcgaggcactttcatctgg. This
A. thaliana pds cDNA fragment (Appendix 4) was cloned into pTV00 at
the Sma 1 site.
A 500bp cDNA fragment of the rubisco small sub-unit was PCR amplified
from N. benthamiana cDNA using Taq DNA polymerase and the primers
5'cagtctagatggcttcctcagttctttcc and 5'cagggatcccacttgacgcacgttgtc.
This rubisco cDNA fragment (Appendix 5) was cloned into the Sma 1
sites of pTV00 and pGR107 to form pTV.rubisco and pGR107.rubisco
respectively.
A 321bp fragment corresponding to the 3' end of GFP (designated P)
was PCR amplified using Taq DNA polymerase and the primers
5'aacatcctcggcccacaagtt and 5'gagctcttagagttcgtcatg. This fragment
(Appendix 6) was cloned into the Sma 1 sites of pGR107 and pTV00 to
form pGR107.P and pTV.P respectively.
A 421bp cDNA fragment of the NFL gene was PCR amplified from N.
benthamiana using Taq DNA polymerase and the primers
5'tggacccagaggctttctc and 5'cttcttgtgagagagcgtca. This NFL cDNA
fragment (Appendix 7) was cloned into pGR107 and pTV00 at the Sma 1
sites to form pGR107.NFL and pTV.NFL respectively.
pBINTRA6, a full length infectious clone of TRV (strain PPK20) RNA1,
was constructed as follows.
Total RNA was prepared from TRV (strain ppk20) infected N.
benthamiana plants as previously described (Devic, Jaegle et al.
1989). Full length cDNA corresponding to TRV RNA1 was prepared from
this RNA using Superscript Reverse Transcriptase (Gibco) and the
primer TRV2 5'ggggggatccgggcgtaataacgcttacg3' which anneals to the 3'
end of TRV RNAl. A schematic drawing of TRV RNA1 is shown in Figure
2. All primers in this work were derived from the sequence of a
closely related TRV strain SYM (Hamilton, Boccara et al. 1987) The
full-length cDNA was used as a template for PCR amplification of two
overlapping fragments, PCR1 and PCR2, which together cover all of TRV
4 5 RNA1.
PCR1, a 3.2 kb fragment, was amplified using Expand HiFi polymerase
(Roche). The primers were: TRV1
~ggggggatccataaaacatttcaatcctttg3'(which anneals to positions 1-21 of
TRV) and TRV4U 5'ttagcaccagctatctgagcgc3' (positions 3168-3189).
PCR2, a 4.1 kb product, was also amplified using Expand HiFi
polymerase (Roche) and the primers TRV4D 5'gttccaaccagacaaacgtatgg3'
(positions 2698-2720) and TRV2 (see above).
PCR1 and PCR2 share a 491nt overlap in the replicase open reading
CA 02297616 2000-O1-31
12
frame (ORF). The primers TRV1 and TRV2 contain BamHI sites to allow
cloning of the full-length product (Figure 2). PCR2 was blunt-ended
using T4 DNA polymerase, digested with BamHI, and cloned into the
plasmid pBAC/SacBl (Bendahmane, Kanyuka et al. 1999) which had
previously digested with BamHI and EheI to form pBSTR3'C. The PCR1
fragment was blunted-ended with T4 DNA polymerase and ligated into
HpaI digested-pBSTR3'C, to form pBSTRF16 (Figure 3). pBSTRF16
therefore contains 302bp that are duplicated within the replicase
ORF.
The 302bp of duplicated replicase sequence was replaced with a 438bp
intron. Intron 3 of Arabidopsis thaliana Col-0 nitrate reductase NIA1
gene (Wilkinson and Crawford 1993) was amplified using the primers
AraF and Aran. AraF is 5'CGTATCTTTGCAA
TAACAGgtaataatcctctctcttgatatt3', where the sequence in upper case
corresponds to positions 2826-2845 of TRV RNA1 and the sequence in
lower case corresponds to positions 1-24 of the intron. Similarly,
Aran is 5'TTAAATTGTCCAAGATCAACct gtttaacacaagtcaacgtc3' where the
sequence in upper case corresponds to positions 2846-2864 of TRV RNA
1 and the sequence in lower case corresponds to positions 416-438 of
the intron. The PCR amplified intron 3 fragment was therefore flanked
by the AGGT intron splice-sites, and l9bp of TRV (exon) sequence
(Figure 4). Two TRV-exons (exon 1 and exon 2) that flank the intron
insertion site were then PCR amplified. For exon 1 the primers were
TRV2D 5'tcgcacaaaaccaaggtgatag3' (positions 1772-1793) and AraS'R
5'ggattatt acCTGTTATTGCAAAGATACGTCTG3' where the sequence in lower
case corresponds to positions 1-10 of the intron and sequence in
upper case corresponds to positions 2822-2845 of TRV RNA1. Exon 1 was
amplified as a 1.07kb fragment from pBSTRI6. For exon 2 the primers
were Ara3'F 5'tgttaaacagGTTGATC TTGGACAATTTAAGTGC3', where the
sequence in upper case corresponds to positions 2846-2868 of TRV RNA1
and the sequence in lower case corresponds to positions 428-438 of
the intron, and TRV4U (see above). Exon 2 was amplified as a 0.35kb
fragment from PCR 1 (see above). Exon l, intron3 and exon 2 were all
amplifed using Pfu polymerase (Promega). To introduce intron 3 to the
TRV RNA 1 genome, chimeric PCR was performed with Pfu polymerase and
the primers TRV2D and TRV4U using a mixture of exon 1, intron 3 and
exon 2 as template to give a l.8kb fragment.
This l.8kb intron-containing-fragment was digested with ApaI and SalI
and cloned in pBSTRF16 using ApaI-partial digestion and SalI, thus
replacing the region that included duplicated sequence, and forming
pBSTRA3 (Figure 4).
To transfer the cloned RNA1 to a binary transformation vector, the
7.2kb fragment corresponding to TRV RNA 1 was released from pBSTRA3
with BamHI and cloned into the BamHI site between the CaMV 35s
promoter and the CaMV terminator on the plasmid pBIN61 to form
pBINTRA6 (Figure 5). pBIN61 is a modified version of the pBINl9
(Frisch, Harris-Haller et al. 1995) binary vector that carries a
transcription cassette comprising the CaMV 35S promoter and
terminator. To construct the pBIN61 binary vector, the transcription
cassette containing the CaMV 35S promoter and terminator was released
by digestion with KpnI and XhoI from the plasmid pJIT61 (kindly
provided by P. Mullineaux, JIC, Norwich, UK). The transcription
CA 02297616 2001-04-25
13
cassette was then ligated to the pBINl9 plasmid vector digested with
KpnI and SalI to create pBIN6l.
Agrobacterium strain GV3101 containing pBINTRA6 was infiltrated into
N. benthamiana leaves causing a TRV RNA 1 infection. The partial
sequence of pBINTRA6 is given in Appendix 8.
The kanamycin resistance gene (NptI) from pACYCI~7 (Chang and Cohen
1978) was cloned as a NheI-NcoI fragment into the SpeI-BspHI sites of
pBluescript SKII+, creating intermediate L. The NcoI site was
introduced and restriction sites that would have been duplicated in
the pGreen polylinker were removed by site-directed mutagenesis
(mutagenic oligos: Xhol (5' cgtcttgctcaaggccgcgat 3'), ClaI (5'
cgacaatcta ccgattgtatg 3'), SmaI (5' ctgcgatcccagggaaaacag 3'),
HindIII (5'aaatgcataaagtttt gccat 3') and NcoI (5'
tggttgtaaccatggcagagca 3'). Two complementary oligos
(5'gaattcagatcta3' and 5'acatgtagatctg3' respecively ) were annealed
and inserted between the EcoRI and AfIIII sites, to introduce a
unique BglII site, this was intermediate II. The pSa-on sequence
from pJIT134Sa-Bam was inserted a a BamHI-SmaI fragment into the
BamHI-SmaI sites (remaining from the original pBluescript plasmid) of
intermediate II. These sites, along with intervening PstI and NotI
sites were removed by successive rounds of treatment with T4 DNA
polymerase I (T4 poll) and re-ligation. The StuI site in the NptI
promoter and ClaI sites, introduced when the pSa-on was inserted,
were removed by transformation into E.coli strain SCS110 (dam;dcm),
digestion with T4 poll and re-ligation. This produced the pGreen
backbone that was ready to receive the T-DN.A cassette.
The complementary olignucleotides (5'-
catgaaggccttgacaggatatattggcgggtaaactaa gtcgctgtatgtgtttgtttgagatct-
3' and 5'-catgagatctcaaacaaacacatacagcgacttagtttacccg
ccaatatatcctgtcaaggcctt -3') were annealed to produce a DNA fragment
consisting of a StuI site, the RB sequence, the RB "overdrive"
sequence and a BglII site. This RB DNA fragment was inserted into the
AfIIII site of pBluescript SKII- and its orientation determined by
sequencing. A recombinant plasmid (intermediate A) was selected which
had the orientation of the RB fragment such that the StuI site was
nearest to the SK multiple cloning site.
Two further oligonucleotides (5'-
tccacacattatacgagccgatgattaattgtcaacagatcttggcag
gatatattgtggtgtaaacgttaac-3' and 5'-
ggtaacgtttacaccacaatatatcctgccaagatctgttgacaatta
atcatcggctcgtataatgtgtgga-3') were then annealed to produce an LB DNA
fragment consisting of HpaI and BglII site and LB sequence. This
fragment was inserted between the two SspI .sites of intermediate A,
simultaneously deleting the pBluescript SKII- fl ori. This 815bp
BglII fragment was cloned into the pGreen backbone to produce
pGreen0000.
All work involving virus infected material was carried out in
containment glasshouses under MAFF license :PHF 1420c/1773(12/1996).
N, benthamiana and A. thaliana plants were germinated on a 1:1
mixture of JIC compost and peat, then grown individually in pots at
CA 02297616 2000-O1-31
14
25°c during the day and 20°c during the night. Supplementary
winter
lighting from halogen quartz iodide lamps provided a 16 hour day
length.
Virus infections on N. benthamiana were achieved by Agrobacterium-
mediated transient gene expression of infectious constructs from the
T-DNA of a binary plasmid (e. g. pGR107, pTV00 or pBINTRA6).
Agrobacterium was grown to an OD of 600 in L broth. The culture was
then centrifuged and re-suspended in lOmM MgCl2, lOmM MES and 150mM
acetosyringone, and kept at room temperature for 2 hours. The culture
was then infiltrated to the underside of a leaf using a 2m1 syringe
without a needle.
For virus infections on A.thaliana, Agrobacterium cultures carrying
pBINTRA6 and pTV.apds were first infiltrated to N. benthamiana, as
described above. A week later, systemically infected leaves were
ground in 50mM phosphate buffer pH7 using a pestle and mortar. The
solution was then centrifuged for 1 minute at 3000 rpm and the
supernatant was rubbed onto the leaves of carborundum-dusted plants.
For RNA infection, total RNA was purified from infected plants as
previously described (Devic, Jaegle et al. 1989). 5m of this RNA was
rubbed onto the leaves of carborundum dusted plants.
Total RNA from N. benthamiana was prepared using Tri-reagent (Sigma).
Genomic DNA was removed from this RNA by incubation with DNase
(Sigma) at 37°c for 2 hours followed by phenol extraction. This RNA
was used as a template for first-strand cDNA synthesis using random
hexanucleotide primers and Super-Script Reverse Transcriptase (Roche)
according to the manufacture's instructions. This cDNA was diluted
2000 fold in sterile distilled water, and used as a template for
quantitative PCR in an ABI Prism 7700 Sequence Detection System (PE
Applied Biosystems) with 2X Taqman Universal PCR Mastermix (PE
Applied Biosystems). For rubisco amplification, the primer sequences
were cgtcaagtgcagtgcatcagt and gacaatagggtaagttgtcctaatatgaaa and the
probe sequence was cattgcctccaagcctgacgga. Each sample was
quantitatively standardised by amplification of 25S ribosomal cDNA
using the primers accacagggataactggcttgt and ccgacatcgaaggatcaaaaa
with the probe cagccaagcgtcatagcgacgttg. The probe DNA was modified
to contain 5' FAM fluorescent reporter and 3' Tamra quencher (MGW).
To confirm that PCR amplification corresponded to mRNA levels and not
from genomic or contaminating DNA, additional reactions were
performed without template cDNA and with RNA that had not been
treated with reverse-transcriptase.
Example 1 - comparison of silencing of pds Qene by TRV and PVX VIGS
vectors
To assess whether constructs derived from TRV have advantages over
previously described virus vectors, we compared symptoms and VIGS
caused by TRV and another virus. PVX was used for this comparison,
because it inhibits expression of a wide range of plant genes (Ruiz,
CA 02297616 2000-O1-31
Voinnet et al. 1998; Jones, Hamilton et al. 1999). Furthermore, as
with TRV, an infectious PVX cDNA construct is available under control
of the CaMV 35s promoter on the binary transformation plasmid
"pGreen" (pGR107, (Jones, Hamilton et al. 1999)). As both TRV and PVX
5 derivatives are therefore introduced to plants in the same way, any
differences can be attributed to the virus itself, rather than the
means of inoculation.
We next compared the ability of TRV and PVX derived constructs to
10 inhibit plant gene expression in N. benthamiana. Identical plant cDNA
sequences were inserted into pTV00 and pGR107 to compare the
efficiency of TRV and PVX at silencing the same plant gene. cDNA
sequences from two plant genes were used for this comparison.
15 The first cDNA we used was part of the phytoene desaturase (pds)
gene. Pds is essential for production of carotenoids that protect
plants from photobleaching (Demmig-Adams and Adams 1992). Plant
tissue in which pds is inhibited therefore turns white due to
photobleaching. This whitening phenotype provides a useful visual
marker for assessing the extent and severity of VIGS (Kumagai, Donson
et al. 1995; Ruiz, Voinnet et al. 1998). A further rationale for
choosing pds as a target of VIGS is that the copy number and
expression level of the pds gene is low (Ruiz, Voinnet et al. 1998).
VIGS of pds therefore indicates the potential of a virus to inhibit
the expression of a low abundance mRNA. For these experiments a 409bp
cDNA fragment of pds (Appendix 3) was PCR amplified from N.
benthamiana cDNA and cloned in the sense orientation into pTV00 and
pGR107, to form pTV.pds and pGR107.pds respectively.
Leaves of 3 week old N. benthamiana plants were infiltrated with
Agrobacterium containing either pGR107.pds, to cause PVX.pds
infections, or with Agrobacterium containing pBINTRA6 and pTV.pds, to
cause TRV.pds infections. As previously reported (Ruiz, Voinnet et
al. 1998), after 3 weeks plants infected with PVXpds developed
symptoms of photobleaching that are typical of pds inhibition. Stems,
axillary shoots, and sepals were affected. In leaves the
photobleaching was initiated and predominantly maintained around the
veins. However, a large proportion of the leaf surface remained
green. Plants infected with TRV.pds also developed photobleaching
symptoms. Again, stem, axillary shoots, sepals and leaves were
affected. However, unlike leaves infected with PVX.pds, leaves
infected with TRV.pds became predominantly white, indicating
widespread silencing of pds gene by the TRV based construct.
These data are typical of 10 repeats, and show that when TRV and PVX
derivatives carry the same fragment of the pds gene, the TRV
construct inhibits pds activity in a higher proportion of leaf tissue
than the PVX vector.
Example 2 - comparison of silencing of rubisco by TRV and PVX VIGS
vectors
A second comparison of VIGS caused by TRV and PVX was made using the
small sub-unit of ribulose -1,5-bisphosphate carobxylase oxygenase
(rubisco). Antisense or PVX.rubisco induced inhibition of rubisco
CA 02297616 2000-O1-31
16
causes chlorotic and stunting symptoms (Rodermel, Abbott et al. 1988;
Jones, Hamilton et al. 1999). A further rationale for choosing
rubisco as a target of VIGS is that the copy number and expression
level of the rubisco genes is very high. In contrast to pds, VIGS of
rubisco would therefore indicate the potential of a virus to inhibit
the expression of a high abundance endogenous mRNA. A 500bp cDNA
fragment of the rubisco small sub-unit (Appendix 4) was PCR amplified
from N. benthamiana cDNA and cloned in the sense orientation into
pTV00 and pGR107 to form pTV.rubisco and pGR107.rubisco respectively.
Leaves of 3 week old N. benthamiana plants were infiltrated either
with Agrobacterium containing pGR107.rubisco, to cause a PVX.rubisco
infection, or with Agrobacterium containing pBINTRA6 and pTV.rubisco,
to cause a TRV.rubisco infection.
Three weeks after infiltration, the upper leaves of plants that were
infected with PVX.rubisco developed very pale green or yellow
patches. However, as with PVX induced inhibition of pds, leaves were
not uniformly affected. Similar areas of pale green developed on
stems, axillary shoots and sepals. These pale green areas were
distinct and different from the mosaic normally associated with PVX
infection. Plants that were infected with TRV.rubisco also developed
pale green or yellow tissue in systemically infected leaves, stems,
axillary shoots and sepals. However, in contrast to PVX.rubisco
infected plants, in TRV.rubisco infected leaves there was no mosaic,
and the pale green or yellow symptoms appeared uniform across the
leaf. This experiment was repeated more than 10 times, with the same
result.
To confirm that the symptoms associated with PVX.rubisco and
TRV.rubisco infection were caused by inhibition of rubisco, and to
quantify any reduction in rubisco mRNA caused by each construct,
rubisco mRNA levels were measured by Taqman PCR (PE Applied
Biosystems). In these experiments, total RNA from infected plants was
used as a template for first-strand cDNA synthesis. Quantitative PCR
was then used to assess the ratio of rubisco cDNA to that of
ribosomal cDNA in PVX.rubisco or TRV.rubisco infected plants, and in
uninfected plants. The rubisco sequence carried by the PVX and TRV
constructs was not PCR amplified in these experiments as the primers
used were outside the region carried by in the virus vectors (see
Materials and Methods). Nine separate plants were used for each
treatment, and each plant was analysed three times.
In leaves infected with PVX.rubisco there was a 37 fold reduction in
rubisco mRNA compared to the-~evels in mock inoculated plants
(Student's T-Test; P=1.96 10 ). However, in leaves infected with
TRV.rubisco there was an even greater reduction in rub~sco levels of
200 fold compared to mock inoculated plants (P=2.1 10 ). These data
show that the TRV based construct is 5.4 (P=0.02) fold more efficient
at suppressing rubisco accumulation than the PVX based construct
carrying an identical rubisco sequence.
Example 3 - comparison of silencing in A thaliana by TRV and PVX
VIGS vectors
CA 02297616 2000-O1-31
17
One disadvantage of currently available virus vectors is that they
have a limited host range. For example, neither PVX nor TMV vectors
infect the model plant A. thaliana. In an attempt to overcome this
limited host range, some authors have made transgenic A. thaliana
plants that express a full-length infectious PVX cDNA carrying
endogenous plant gene sequence. This strategy was pursued because PVX
replicates in A. thaliana protoplasts, but does not infect whole
plants. However, even when PVX is expressed from a transgene and
replicating in every cell, there was little or no inhibition of
homologous Arabidopsis gene expression (unpublished data).
We tested whether constructs based on TRV would inhibit gene
expression in A. thaliana. A fragment of the pds gene was PCR
amplified from A. thaliana cDNA (Appendix 5) and cloned the sense
orientation into pTV00 to form pTV.apds. No TRV infections could be
established by infiltrating A. thaliana with Agrobacterium carrying
pBINTRA6 and pTV.pds or pTV00. Therefore, TRV.apds infections were
established by infiltrating Agrobacterium that contained pBINTRA6 and
pTV.apds to N. benthamiana. Infectious sap from these N. benthamiana
plants was subsequently used to inoculate A. thaliana ecotype Col-0
(see Materials and Methods). As with N. benthamiana, A. thaliana
plants infected with TRV developed no symptoms. However, A. thaliana
plants infected with TRV.apds developed confluent photobleaching in
systemic leaves that is typical of pds inhibition. These data suggest
that unlike PVX, TRV can inhibit gene expression in a range of plant
species including A. thaliana.
Example 4 - comparison of silencing of GFP transaene in meristematic
tissue by TRV and PVX VIGS vectors
A sub-set of genes that control identity and development of newly
forming plant tissues are expressed in meristematic regions.
Understanding the function of these genes is of particular interest
as they determine characteristics such as fruit and flower
production. However, PVX, TMV and TGMV do not infect meristems
(Matthews 1991), and therefore cannot be used to inhibit meristem-
expressed genes.
We assessed whether our TRV based vector would overcome this
limitation by comparing the ability of the PVX and TRV derived
constructs to inhibit the expression of two genes in meristems. The
first of these genes encodes the green fluorescent protein, GFP
(Chalfie, Tu et al. 1994). A 321bp fragment corresponding to the 3'
end of GFP (designated P; Appendix 6) was PCR amplified and cloned in
the sense orientation into pGR107 and pTV00 to form pGR107.P and
pTV.P respectively. For these experiments N. benthamiana plants were
used that express GFP from a CaMV 35S driven transgene (line 16c,
(Ruiz, Voinnet et al. 1998)). In UV light these plants fluoresce
green due to expression of the GFP transgene. In contrast, non-
transgenic plants, and 16c plants in which GFP expression is
inhibited, are red under UV illumination, due to fluorescence from
chlorophyll.
Three week old 16c N. benthamiana plants were infiltrated with
Agrobacterium containing either pGR107.P, to cause a PVX.P infection,
CA 02297616 2000-O1-31
18
or pBINTRA6 and pTV.P to cause a TRV.P infection, or with water. As
previously reported, 3 weeks after infiltration the leaves, stems and
axillary shoots of plants infected with PVX.P had lost all green
fluorescence, and appeared red under UV light (Figure 5A) (Ruiz,
Voinnet et al. 1998). Similarly, 16c plants that were infected with
TRV.P also lost GFP expression in leaves, stems and axillary shoots.
We then assessed the level of GFP expression in vegitative
meristematic regions using confocal laser-scanning microscopy. As
previously reported (Ruiz, Voinnet et al. 1998) PVX.P did not inhibit
GFP expression in this region, as the meristem and surrounding leaf
primordia in these plants remained green-fluorescent. In contrast,
TRV.P inhibited GFP expression in the meristematic dome and
surrounding leaf primordia.
Example 5 - comparison of silencing of LFY endocrenous crene iw
meristematic tissue by TRV and PVX VIGS vectors
We next compared the ability of PVX and TRV derived constructs to
inhibit the expression of an endogenous gene in meristems. The gene
we chose was NFL, the Nicotiana floricola homologue of the A.
thaliana gene leafy (LFY). LFY is important for determination of
floral organs. A. thaliana lfy mutants have two characteristics;
additional inflorescence shoots are formed in the place of early
flowers, and flowers that develop later are abnormal and contain
shoot-inflorescence tissue (Weigel, Alvarez et al. 1992). We
therefore predicted that inhibiting NFL expression would have similar
effects on flower formation in N. benthamiana. A 421bp cDNA fragment
of the NFL gene was PCR amplified from N. benthamiana cDNA (Appendix
7) and cloned into in the sense orientation into pGR107 and pTV00 to
form pGR107.NFL and pTV.NFL respectively.
Three week-old N. benthamiana plants were infiltrated with water
(mock), or with Agrobacterium containing pGR107 or pGR107.NFL to
cause PVX or PVX.NFL infections, or pBINTRA6 and pTV00 or pTV.NFL to
cause a TRV or TRV.NFL infections respectively. Six plants were used
for each treatment. The arrangement and formation of leaves, flowers
and branches was recorded after 12 weeks. In N. benthamiana, organs
are formed at nodes around the main stem. Each node typically
consists of a leaf, a primary branch, and a flower. The branches vary
in size and complexity, and may give rise to secondary, or
occasionally tertiary branches, before producing flowers. Floral
organs such as petals, sigma and stamen are formed after a whorl of
leaves known as sepals. On each plant, the number of primary,
secondary and tertiary branches was recorded, as well as the number
of correct and incorrectly formed flowers. Although there was
structural variation between plants, there were no consistent changes
between mock inoculated plants, and plants that were infected with
PVX, PVX.NFL or TRV. All plants produced secondary branches. One TRV
and one PVX.NFL infected plant each produced a single tertiary
branch. All flowers on these plants were normal compared to mock
inoculated plants. In contrast, plants infected with TRV.NFL had a
more complex and branched structure. Each of these plants produced
secondary and tertiary branches. Five of the six plants also produced
quartenary branches. These additional branches were formed in place
of flowers. Many of the flowers that were formed on TRV.NFL infected
CA 02297616 2000-O1-31
19
plants were abnormal. Flower defects ranged from repeated whorls of
sepals, to floral structures containing single or multiple
inflorescence branches which themselves gave rise to more floral
structures. These data are reminiscent of floral abnormalities in
Arabidopsis lfy mutants, and are entirely consistent with an
inhibition of NFL expression in the meristematic regions of N.
benthamiana.
Taken together, these two experiments with GFP and NFL show that
unlike other viral vectors, constructs based on TRV can inhibit gene
expression in meristems.
Example 6 - TRV as an expression vector
The TRV derived virus vector pTV00 has been specifically designed to
inhibit rather than direct protein synthesis. For certain purposes it
may be advantageous to modify pTV00 to allow protein expression from
this vector. If the multiple cloning site of pTV00 was immediately
preceded by a sub-genomic promoter that was recognised by the
replicase protein of TRV RNA 1, then proteins would be translated
from a sub-genomic RNA.
For example, total RNA could be prepared from PEBV infected N.
benthamiana plants as previously described (Devic, Jaegle et al.
1989). cDNA corresponding to the coat protein sub-genomic promoter of
PEBV could be prepared and then PCR amplified from this total RNA
using Superscript Reverse Transcriptase (Gibco) followed by Pfu
polymerase (Promega) with the primers ggatccgcacacaaggtta and
gggcgcgccctcgttaac. This PEBV sub-genomic promoter cDNA fragment
(Appendix 2) could be cloned into pTV00 that was previously digested
with Spe 1 and blunted with T4 DNA polymerase, to form pTVl.O. Open
reading frames cloned in sense and in frame in the remainder of the
multiple cloning site would then be expressed. For example, GFP cDNA
sequence (Chalfie, Tu et al. 1994), could be inserted into the BamHi
site, to achieve GFP expression in any host plant such as N.
benthamiana.
References
Inasmuch as they may be required by the person skilled in the art to
practice the present invention, all citations are specifically
included herein by cross-reference.
A1-Kaff, N. S., S. N. Covey, et al. (1998). "Transcriptional and
post-transcriptional plant gene silencing in response to a pathogen."
Science 279: 2113-2115.
Bendahmane, A., K. Kanyuka, et al. (1999). "The Rx gene from potato
controls separate virus resistance and cell death responses." Plant
Cell 11: 781-791.
Chalfie, M., Y. Tu, et al. (1994). "Green fluorescent protein as a
marker for gene expression." Science 263: 802-805.
Chang, A. C. Y. and S. N. Cohen (1978). "Construction and
CA 02297616 2000-O1-31
characterisation of amplifiable multicopy DNA cloning vehicles
derived from the P15A cryptic miniplasmid." Journal of Bacterioloay
134: 1141-1156.
5 Covey, S. N., N. S. A1-Kaff, et al. (1997). "Plants combat infection
by gene silencing." Nature 385: 781-782.
Demmig-Adams, B. and W. W. Adams (1992). "Photoprotection and other
responses of plants to high light stress." Annu.Rev.Plant
10 Phvsiol.Plant Mol.Biol. 43: 599-626.
Devic, M., M. Jaegle, et al. (1989). "Symptom production on tobacco
and tomato is determined by two distinct domains of the satellite RNA
of cucumber mosaic virus (strain Y)." J.Gen.Virol. 70: 2765-2774.
Frisch, D. A., L. W. Harris-Haller, et al. (1995). "Complete Sequence
of the binary vector Bin 19." Plant Molecular Biolocrv 27: 405-409.
Guo, H. S. and J. A. Garcia (1997). "Delayed resistance to plum pox
potyvirus mediated by a mutated RNA replicase gene: involvement of a
gene-silencing mechanism." Molecular Plant-Microbe Interactions 10:
160-170.
Hamilton, W. D. O., M. Boccara, et al. (1987). "The complete
nucleotide sequence of tobacco rattle virus RNA-1." J.Gen.Virol. 68:
2563-2575.
Hernandez, C., A. Mathis, et al. (1995). "Sequence of RNA 2 of a
nematode transmissible isolate of tobacco rattle virus." Journal Of
General Virolocrv 76: 2847-2851.
Jones, L., A. J. Hamilton, et al. (1999). "RNA-DNA interactions and
DNA methylation in post-transcriptional gene silencing." Plant Cell
11: 2291-2302.
Kjemtrup, S., K. S. Sampson, et al. (1998). "Gene silencing from
plant DNA carried by a geminivirus." Plant Journal 14(1): 91-100.
Kumagai, M. H., J. Donson, et al. (1995). "Cytoplasmic inhibition of
carotenoid biosynthesis with virus-derived RNA." Proceedings Of The
National Academy Of Sciences Of The United States Of America 92:
1679-1683.
Lindbo, J. A., L. Silva-Rosales, et al. (1993). "Induction of a
highly specific antiviral state in transgenic plants: implications
for regulation of gene expression and virus resistance." Plant Cell
5: 1749-1759.
MacFarlane, S. A. (1999). "Molecular biology of the tobraviruses."
Journal Of General Viroloav 80: 2799-2807.
Matthews, R. E. F. (1991). Plant Viroloav. San Diego, CA, Academic
Press.
Ratcliff, F., B. D. Harrison, et al. (1997). "A similarity between
CA 02297616 2000-O1-31
21
viral defense and gene silencing in plants." Science 276: 1558-1560.
Ratcliff, F., S. MacFarlane, et al. (1999). "Gene silencing without
DNA: RNA-mediated cross protection between viruses." Plant Cell 11:
1207-1216.
Rodermel, S. R., M. S. Abbott, et al. (1988). "Nuclear-organelle
interactions: Nuclear antisense gene inhibits ribulose bisphosphate
carboxylase enzyme levels in transformed tobacco plants." Cell 55:
673-681.
Ruiz, M. T., O. Voinnet, et al. (1998). "Initiation and maintenance
of virus-induced gene silencing." Plant Cell 10: 937-946.
Sambrook, J., E. F. Fritsch, et al. (1989). Molecular Cloning: A
Laboratory Manual. New York, Cold Spring Harbor Laboratory Press.
Weigel, D., J. Alvarez, et al. (1992). "LEAFY Controls Floral
Meristem Identity in Arabidopsis." Cell 69: 843-859.
Wilkinson, J. Q. and N. M. Crawford (1993). "Identification and
characterisation of a chorate-resistant mutant of Arabidopsis
thaliana with mutations in both nitrate reductase structural genes
NIA1 and NIA2." Molecular & General Genetics 239: 289-297.
CA 02297616 2001-04-25
- 22 -
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT:
(A) NAME: Plant Bioscience Limited
(B) STREET: Norwich Research Park, Colney Lane
(C) CITY: Norwich
(D) STATE: Norfolk
(E) COUNTRY: United Kingdom
(F) POSTAL CODE (ZIP): NR4 7UH
(ii) TITLE OF INVENTION: Viral vectors
(iii) NUMBER OF SEQUENCES: 46
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: Fetherstonhaugh & Co
(B) STREET: 438 University Avenue, Suite 100, Box 111
(C) CITY: Toronto
(D) STATE: Ontario
(E) COUNTRY: Canada
(F) POSTAL CODE (ZIP): M5G 2K8
(v} COMPUTER READABLE FORM:
(A) COMPUTER: IBM PC compatible
(B) OPERATING SYSTEM: PC-DOS/MS-DOS
(C) SOFTWARE: PatentIn Release #1.0, 'Version #1.25 (EPO)
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER: CA 2,297,616
(B) FILING DATE: 31-JAN-2000
(viii) PATENT AGENT INFORMATION:
(A} NAME: Ms Yoon Kang
(B) REFERENCE NUMBER: 04900-36
(2) INFORMATION FOR SEQ ID N0: l:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: l:
GGCACTCAAC TTTATAAACC 20
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 base pairs
CA 02297616 2001-04-25
- 23 -
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ LD N0: 2:
CTTCAGTTTT CTGTCAAACC 20
(2) INFORMATION FOR SEQ ID N0: 3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 3:
CCCCTCGAGA GATGTCAAAT C 21
(2) INFORMATION FOR SEQ ID N0: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 23 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 4:
CCCCTCGAGG CACTTTCATC TGG 23
(2) INFORMATION FOR SEQ ID N0: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5:
CAGTCTAGAT GGCTTCCTCA GTTCTTTCC 29
(2) INFORMATION FOR SEQ ID NO: 6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
CA 02297616 2001-04-25
- 24 -
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 6:
CAGGGATCCC ACTTGACGCA CGTTGTC 27
(2) INFORMATION FOR SEQ ID NO: 7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 7:
AACATCCTCG GCCCACAAGT T 21
(2) INFORMATION FOR SEQ ID N0: 8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 8:
GAGCTCTTAG AGTTCGTCAT G 21
(2) INFORMATION FOR SEQ ID NO: 9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 9:
TGGACCCAGA GGCTTTCTC 19
(2) INFORMATION FOR SEQ ID NO: 10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
CA 02297616 2001-04-25
- 25 -
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 10:
CTTCTTGTGA GAGAGCGTCA 20
(2) INFORMATION FOR SEQ ID N0: 11:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 11:
GGGGGGATCC GGGCGTAATA ACGCTTACG 29
(2) INFORMATION FOR SEQ ID N0: 12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 31 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 12:
GGGGGGATCC ATAAAACATT TCAATCCTTT G 31
(2) INFORMATION FOR SEQ ID N0: 13:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 13:
TTAGCACCAG CTATCTGAGC GC 22
(2) INFORMATION FOR SEQ ID NO: 14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 23 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
CA 02297616 2001-04-25
- 26 -
(xi} SEQUENCE DESCRIPTION: SEQ ID N0: 14:
GTTCCAACCA GACAAACGTA TGG 23
(2) INFORMATION FOR SEQ ID N0: 15:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 43 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 15:
CGTATCTTTG CAATAACAGG TAATAATCCT CTCTCTTGAT ATT 43
(2) INFORMATION FOR SEQ ID N0: 16:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 42 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 16:
TTAAATTGTC CAAGATCAAC CTGTTTAACA CAAGTCAACG TC 42
(2) INFORMATION FOR SEQ ID N0: 17:
(i) SEQUENCE CHARACTERISTICS:
(A} LENGTH: 22 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 17:
TCGCACAAAA CCAAGGTGAT AG 22
(2) INFORMATION FOR SEQ ID N0: 18:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 18:
ii
CA 02297616 2001-04-25
- 27 -
GGATTATTAC CTGTTATTGC AAAGATACGT CTG 33
(2) INFORMATION FOR SEQ ID N0: 19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 34 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 19:
TGTTAAACAG GTTGATCTTG GACAATTTAA GTGC 34
(2) INFORMATION FOR SEQ ID N0: 20:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 20:
CGTCTTGCTC AAGGCCGCGA T 21
(2) INFORMATION FOR SEQ ID N0: 21:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 21:
CGACAATCTA CCGATTGTAT G 21
(2) INFORMATION FOR SEQ ID N0: 22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 22:
CTGCGATCCC AGGGAAAACA G 21
CA 02297616 2001-04-25
- 28 -
(2) INFORMATION FOR SEQ ID NO: 23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 23:
AAATGCATAA AGTTTTGCCA T 21
(2) INFORMATION FOR SEQ ID N0: 24:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 24:
TGGTTGTAAC CATGGCAGAG CA 22
(2) INFORMATION FOR SEQ ID N0: 25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 25:
GAATTCAGAT CTA 13
(2) INFORMATION FOR SEQ ID NO: 26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 26:
ACATGTAGAT CTG 13
(2) INFORMATION FOR SEQ ID N0: 27:
~ii
CA 02297616 2001-04-25
- 29 -
(i} SEQUENCE CHARACTERISTICS:
(A) LENGTH: 66 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 27:
CATGAAGGCC TTGACAGGAT ATATTGGCGG GTAAACTAAG TCGCTGTATG TGTTTGTTTG 60
AGATCT 66
(2) INFORMATION FOR SEQ ID NO: 28:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 66 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single.
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 28:
CATGAGATCT CAAACAAACA CATACAGCGA CTTAGTTTAC CCGCCAATAT ATCCTGTCAA 60
GGCCTT 66
(2) INFORMATION FOR SEQ ID N0: 29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 73 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 29:
TCCACACATT ATACGAGCCG ATGATTAATT GTCAACAGAT CTTGGCAGGA TATATTGTGG 60
TGTAAACGTT AAC 73
(2) INFORMATION FOR SEQ ID N0: 30:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 73 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 30:
ii
CA 02297616 2001-04-25
GGTAACGTTT ACACCACAAT ATATCCTGCC AAGATCTGTT GAC.AATTAAT CATCGGCTCG 60
TATAATGTGT GGA 73
(2) INFORMATION FOR SEQ ID N0: 31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 31:
CGTCAAGTGC AGTGCATCAG T 21
(2) INFORMATION FOR SEQ ID NO: 32:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 32:
GACAATAGGG TAAGTTGTCC TAATATGAAA 30
(2) INFORMATION FOR SEQ ID N0: 33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 33:
CATTGCCTCC AAGCCTGACG GA 22
(2) INFORMATION FOR SEQ ID N0: 34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 34:
~ii
CA 02297616 2001-04-25
- 31 -
ACCACAGGGA TAACTGGCTT GT 22
(2) INFORMATION FOR SEQ ID N0: 35:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 35:
CCGACATCGA AGGATCAAAA A 21
(2) INFORMATION FOR SEQ ID N0: 36:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
NO: 36:
CAGCCAAGCGTCATAGCGAC GTTG 24
(2) INFORMATION
FOR SEQ
ID N0:
37:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 5592 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: circular
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0: 37:
TTTTTATCCCCGGAAGCCTG TGGATAGAGG GTAGTTATCC ACGTGAAACCGCTAATGCCC 60
CGCAAAGCCTTGATTCACGG GGCTTTCCGG CCCGCTCCAA AAACTATCCACGTGAAATCG 120
CTAATCAGGGTACGTGAAAT CGCTAATCGG AGTACGTGAA ATCGCTAATAAGGTCACGTG 180
AAATCGCTAATCAAAAAGGC ACGTGAGAAC GCTAATAGCC CTT'TCAGATCAACAGCTTGC 240
AAACACCCCTCGCTCCGGCA AGTAGTTACA GCAAGTAGTA TGTTCAATTAGCTTTTCAAT 300
TATGAATATATATATCAATT ATTGGTCGCC CTTGGCTTGT GGACAATGCGCTACGCGCAC 360
CGGCTCCGCCCGTGGACAAC CGCAAGCGGT TGCCCACCGT CGAGCGCCAGCGCCTTTGCC 420
CACAACCCGGCGGCCGGCCG CAACAGATCG TTTTATAAAT TTTTTTTTTTGAAAAAGAAA 480
ii
CA 02297616 2001-04-25
- 32 -
AAGCCCGAAAGGCGGCAACCTCTCGGGCTTCTGGATTTCCGATCCCCGGAATTAGATCTC540
AAACAAACACATACAGCGACTTAGTTTACCCGCCAATATATCC'CGTCAAGGCCTTCATGT600
TCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGTGAGCTG660
ATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGAAGCGGAAG720
AGCGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGAT'.CCATTAATGCAGCTGGC780
ACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGAGCGCAACGCAATTAATGTGAGTTAGC840
TCACTCATTAGGCACCCCAGGCTTTACACTTTATGCTTCCGGCTCGTATGTTGTGTGGAA900
TTGTGAGCGGATAACAATTTCACACAGGAAACAGCTATGACCA'.LGATTACGCCAAGCTCG960
GAATTAACCCTCACTAAAGGGAACAAAAGCTGGAGCTCCACCGCGGTGGAGCTCCACCGG1020
GGAAACCTCCTCGGGATTCCATTGCCCAGCTATCTGTCACTTTATTGAGAAGATAGTGGA1080
AAAGGAAGGTGGCTCCTACAAATGCCATCATTGCGATAAAGGAAAGGCCATCGTTGAAGA1140
TGCCTCTGCCGACAGTGGTCCCAAAGATGGACCCCCACCCACGAGGAGCATCGTGGAAAA1200
AGAAGACGTTCCAACCACGTCTTCAAAGCAAGTGGATTGATGTCiATATCTCCACTGACGT1260
AAGGGATGACGCACAATCCCACTATCCTTCGCAAGACCCTTCCTCTATATAAGGAAGTTC1320
ATTTCATTTGGAGAGGCTAGATAAAACATTGCACCTATGGTGT7.'GCCCTGGCTGGGGTAT1380
GTCAGTGATCGCAGTAGAATGTACTAATTGACAAGTTGGAGAA7.'ACGGTAGAACGTCCTT1440
ATCCAACACAGCCTTTATCCCTCTCCCTGACGAGGTTTTTGTCAGTGTAATATTTCTTTT1500
TGAACTATCCAGCTTAGTACCGTACGGGAAAGTGACTGGTGTGC:TTATCTTTGAAATGTT1560
ACTTTGGGTTTCGGTTCTTTAGGTTAGTAAGAAAGCACTTGTC7.'TCTCATACAAAGGAAA1620
ACCTGACGTATCGCTTACGAAAGTAGCAATGAAAGAAAGGTGGTGGTTTTAATCGTACCG1680
CAAAAAACGATGGGGTCGTTTTAATTAACTTCTCCTACAAGCG9'CTAAACGGACGTTGGG1740
GTTTTGCTAGTTTCTTTAGAGAAAACTAGCTAAGTCTTTAATGTTATCATTAGAGATGGC1800
ATAAATATAATACTTGTGTCTGCTGATAAGATCATTTTAATTTCTGACGATTAGACTTGTT1860
GAACTACAGGTTACTGAATCACTTGCGCTAATCAACATGGGAGATATGTACGATGAATCA1920
TTTGACAAGTCGGGCGGTCCTGCTGACTTGATGGACGATTCTTC7GGTGGAATCAGTTTCG1980
TGGAAAGATTTGTTGAAGAAGTTACACAGCATAAAATTTGCACTACAGTCTGGTAGAGAT2040
GAGATCACTGGGTTACTAGCGGCACTGAATAGACAGTGTCCTTATTCACCATATGAGCAG2100
TTTCCAGATAAGAAGGTGTATTTCCTTTTAGACTCACGGGCTAACAGTGCTCTTGGTGTG2160
ATTCAGAACGCTTCAGCGTTCAAGAGACGAGCTGATGAGAAGAATGCAGTGGCGGGTGTT2220
i,,
CA 02297616 2001-04-25
- 33 -
ACAAATATTCCTGCGAATCCAAACACAACGGTTACGACGAACCAAGGGAGTACTACTACT 2280
ACCAAGGCGAACACTGGCTCGACTTTGGAAGAAGACTTGTACACTTATTACAAATTCGAT 2340
GATGCCTCTACAGCTTTCCACAAATCTCTAACTTCGTTAGAGAACATGGAGTTGAAGAGT 2400
TATTACCGAAGGAACTTTGAGAAAGTATTCGGGATTAAGTTTGGTGGAGCAGCTGCTAGT 2460
TCATCTGCACCGCCTCCAGCGAGTGGAGGTCCGATACGTCCTAATCCCTAGGGATTTAAG 2520
GACGTGAACTCTGTTGAGATCCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATA 2580
TCAAGCTTATCGATACCGTCGACCTCGAGGGGGGGCCCGGTACCCAATTCGCCCTATAGT 2640
GAGTCGTATTACAATTCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGC 2700
GTTACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAA 2760
GAGGCCCGCACCGATCGCCCTTCCCAACAGTTGCGAAGACATTAAACTACGGTTCTTTAA 2820
GTAGATCCGTGCCTGAAGTTTTAGGTTCAATTTAAACCTACGAGATTGACATTCTCGACT 2880
GATCTTGATTGATCGGTAAGTCTTTTGTAATTTAATTTTCTTT'rTGATTTTATTTTAAAT 2940
TGTTATCTGTTTCTGTGTATAGACTGTTTGAGATCGGCGTTTGGCCGACTCATTGTCTTA 3000
CCATAGGGGAACGGACTTTGTTTGTGTTGTTATTTTATTTGTA'PTTTATTAAAATTCTCA 3060
ACGATCTGAAAAAGCCTCGCGGCTAAGAGATTGTTGGGGGGTGAGTAAGTACTTTTAAAG 3120
TGATGATGGTTACAAAGGCAAAAGGGGTAAAACCCCTCGCCTACGTAAGCGTTATTACGC 3180
CCTCGAGTATCGAATTGCTGGAGGCATGCAAGCGATCCCCGATCGTTCAAACATTTGGCA 3240
ATAAAGTTTCTTAAGATTGAATCCTGTTGCCGGTCTTGCGATGATTATCATATAATTTCT 3300
GTTGAATTACGTTAAGCATGTAATAATTAACATGTAATGCATGACGTTATTTATGAGATG 3360
GGTTTTTATGATTAGAGTCCCGCAATTATACATTTAATACGCGATAGAAAACAAAATATA 3420
GCGCGCAAACTAGGATAAATTATCGCGCGCGGTGTCATCTATGTTACTAGATCGGGAATT 3480
GCCAAGCTGCTTGGCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGCGT 3540
TACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGA 3600
GGCCCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAATGGCGCGAAAT 3660
TGTAAACGTTAATGTTAACGTTACACCACAATATATCCTGCCAAGATCTCATGTGAGCAA 3720
AAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGC 3780
TCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGA 3840
CAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTC 3900
CGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTT 3960
ii
CA 02297616 2001-04-25
- 34 -
CTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGT'TCGCTCCAAGCTGGGCT 4020
GTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTG 4080
AGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTA 4140
GCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCT 4200
ACACTAGAAGGACAGTATTTGGTATCTGCGCTCTGCTGAAGCC.AGTTACCTTCGGAAAAA 4260
GAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTT 4320
GCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTA 4380
CGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGGTTACAA 4440
CCAATTAACCAATTCTGATTAGAAAAACTCATCGAGCATCAAA'rGAAACTGCAATTTATT 4500
CATATCAGGATTATCAATACCATATTTTTGAAAAAGCCGTTTC'TGTAATGAAGGAGAAAA 4560
CTCACCGAGGCAGTTCCATAGGATGGCAAGATCCTGGTATCGG'rCTGCGATTCCGACTCG 4620
TCCAACATCAATACAACCTATTAATTTCCCCTCGTCAAAAATAAGGTTATCAAGTGAGAA 4680
ATCACCATGAGTGACGACTGAATCCGGTGAGAATGGCAAAAGT'PTATGCATTTCTTTCCA 4740
GACTTGTTCAACAGGCCAGCCATTACGCTCGTCATCAAAATCACTCGCATCAACCAAACC 4800
GTTATTCATTCGTGATTGCGCCTGAGCGAGACGAAATACGCGA'PCGCTGTTAAAAGGACA 4860
ATTACAAACAGGAATCGAATGCAACCGGCGCAGGAACACTGCCAGCGCATCAACAATATT 4920
TTCACCTGAATCAGGATATTCTTCTAATACCTGGAATGCTGTT'rTCCCTGGGATCGCAGT 4980
GGTGAGTAACCATGCATCATCAGGAGTACGGATAAAATGCTTGATGGTCGGAAGAGGCAT 5040
AAATTCCGTCAGCCAGTTTAGTCTGACCATCTCATCTGTAACA'PCATTGGCAACGCTACC 5100
TTTGCCATGTTTCAGAAACAACTCTGGCGCATCGGGCTTCCCA'rACAATCCATAGATTGT 5160
CGCACCTGATTGCCCGACATTATCGCGAGCCCATTTATACCCATATAAATCAGCATCCAT 5220
GTTGGAATTTAATCGCGGCCTGGAGCAAGACGTTTCCCGTTGAATATGGCTCATAACACC 5280
CCTTGTATTACTGTTTATGTAAGCAGACAGTTTTATTGTTCATGATGATATATTTTTATC 5340
TTGTGCAATGTAACATCAGAGATTTTGAGACACAACGTGGCTTTGTTGAATAAATCGAAC 5400
TTTTGCTGAGTTGAAGGATCAGATCACGCATCTTCCCGACAACGCAGACCGTTCCGTGGC 5460
AAAGCAAAAGTTCAAAATCACCAACTGGTCCACCTACAACAAAGCTCTCATCAACCGTGC 5520
TCCCTCACTTTCTGGCTGGATGATGGGGCGATTCAGGCGATCCCCATCCAACAGCCCGCC 5580
GTCGAGCGGGCT 5592
(2) INFORMATION
FOR SEQ
ID NO:
38:
I~I
CA 02297616 2001-04-25
- 35 -
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 249 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ LD N0: 38:
GGATCCGCAC ACAAGGTTAA AAACGCTGTA GTAATACATG CGCAAGAACA GGCTGAGCAT 60
CTTGTTCTGG GGTTTCACAC TATCTTTAGA GAAAGTGTTA AGTTAATTAA GTTATCTTAA 120
TTAAGAGCAT AATTATACTG ATTTGTCTCT CGTTGATAGA GTC'TATCATT CTGTTCTAAA 180
AATTTGACAA CTCGGTTTGC TGACCTACTG GTTACTGTAT CACTTACCCG AGTTAACGAG 240
GGCGCGCCC 249
(2) INFORMATION FOR SEQ ID NO: 39:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 409 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID
N0: 39:
TCTGTCAAAC CATATATGGA CATTTATCAC AGGAACTCCCACTAGCTTCTCCAACTTTTG 60
GAAATATGGG ATCTCTTTCC AGTCTTCAGG CAAAAGAAGCTTCAAGATATCCACTGGAGT 120
GGCAAACACA AAAGCATCTC CTTTAATTGT ACTGCCATTATTCTGTATAAAACATTTGAC 180
ACTTCCATCC TCATTCAGCT CGATCTTTTT TATTCGTGAGTTTAGTCTGACTTGGCCACC 240
TTTTGACTCA ATATGTTCCA CAATCGGCAT GCAAAGTCTCTCAGGAGGGTTACCATCTAA 300
AAAGGCCATT TTTGAACCAT GTTTCTCCTG AAGAAATCTGTTCAAAGCAATCAAGATGCA 360
CTGCATCGAA AGCTCGTCAG GGTTTATAAA GTTGAGTGCCTCTAGACTG 409
(2) INFORMATION FOR SEQ ID N0: 40:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1773 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 40:
!ii
CA 02297616 2001-04-25
- 36 -
CTCGAGAGATGTCAAATCTGTGAGATTCATTTACTGAAGAGAGTAAGGTTAAGTTTGGAT 60
TGCAATGAGATTTATACACAAATAATAATGCTTACAAGCAAATCATCTTTAAGTTTTGTC 120
CTCTTCTCATGATGATGATACTGTTGCCTCCGACAACTTTCTTGGTCCAGACGCAGCCAG 180
TAGCTCGTAATCCTGAACAATAGACTGAGAGCAGAATTTGCCAGAGAGGACAGCGCCTTC 240
CATGGAAGCTAAGTACTTCTGTTTTGTGTAATCTCCAGCTAAGTAGAATCCTTCAATAGG 300
TGATCTTTGTAGAGGACGACATGGTTCACAGTTTGGGATGGTCTTGTACACAGATCTTGG 360
AGTCTTAACGACATGGTACTTCAGAATTTTAGCTTTGCTTTGGTCAGCTGAGATTTCATC 420
AGGGAAGAGTTTCTCAAGTTCTTTCATTGTTGCATCTATGATGTCAGAATCAGTCCGTGA 480
TATCCATTCCTCTGCTGGTGCAAATACTAGCTCCAGCATTGACCGGTTAGGATCGTAATA 540
TTCCTTACAAGTTAAGGACATGTCGGCATACACGCTCAGAAGGTTACTTCTGCTAAAGAG 600
TAGGTGATCATATGTGTTCTTCAGTTTTCGATCAAACCATATATGAACATTAATAACTGG 660
TACTCCAACTAATTTATCCAATTTCTTGAAGTACGGTATTTCTTTCCAGGGATCTGGTAA 720
AAGGAGCTTCAGGATATCGACTGGAGCGGCAAACACATAAGCG'rCTCCTTCGACAGTGCT 780
TCCATTAGTGAGTAAGAAACTCTTAACCGTGCCATCGTCATTGAGCTCAATTTTCTTTAT 840
CCTAGAATTAAGTTGCACTTCCCCACCTAGTGATCGAATATGA'PCCACTACTGGCATACA 900
AAGCCTTTCCGGAGGATTACCATCCAAGAATGCCATCTTGGAACCATGTTTTTCCTGAAG 960
AAACCGGTTCAAAGCTATCAAAATGCATTGCATTGACAGTTCA'rCAGGGTTTATAAAGTT 1020
TAGCGCCTTTGACATGGCAATAAACACCTCGTCGGTCACGCGC'PCAGGTACTCCCTGCTT 1080
TTCCATCCATTCTTTGACTGATAAACCATCTTGGGCCTCAACA'rAAGCCTGACCGCCGAC 1140
CATGGCTGGCAAAAGTCCAATAGCAAACTTTATTTTCTCTGGCCATGTCAGCATCTCGTT 1200
GTTCCGCAAAATAGCCCAAATACCATTTAAGGGTGCTGGTAGGACATCTGGGAAGTCAAA 1260
TCTACTAAATTCTCCAGGTTTACTTGGCATAGCAAAAATCATGGAGTGTTCCTTCCACTG 1320
CAACCGATCATTGATCCCAAGTTCTCCAAATAAATTCTGCACATTCGGATAAGCACCGAA 1380
GAAAATATGTAAACCAGTCTCATACCAGTCCCCATCTTCATCC'PTCCATGCAGCTATCTT 1440
TCCACCAAGAACATCTCTTGCTTCAAGCAACAGAGGTTTGTGGCCTGCATCAGCCAGGTA 1500
CTTTGCAGTTGACAATCCAGCCAATCCAGCACCAGCAATTACAACTTTCAAAGGCTTAGC 1560
AGGACGAGGAGCACTACGGAAGGATGCAGATAAACTAGCAGCTTCCAAGAAATTGACAGT 1620
GTTCTCTAGCTCTGGCCTTGGAATATCCACACAAACTACCTGCAAAGGACCAGCAGTACT 1680
CCTCCTCCTTGTTCTTGTCTTAAGCGCTTGAGAAGTGGGAACCCTAAAGCTATGTCCCAT 1740
;ii
CA 02297616 2001-04-25
_ 37 _
TAGTTCACAA CCTCCAGATG AAAGTGCCTC GAG 1773
(2) INFORMATION FOR SEQ ID N0: 41:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 499 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID
NO: 41:
AGTCTAGATG GCTTCCTCAG TTCTTTCCTC TGCAGCAGTTGCCACCCGCA GCAATGTTGC60
TCAAGCTAAC ATGGTTGCAC CTTTCACTGG CCTTAAGTCAGCTGCCTCAT TCCCTGTTTC120
AAGGAAGCAA AACCTTGACA TCACTTCCAT TGCCAGCAACGGCGGAAGAG TGCAATGCAT180
GCAGGTGTGG CCACCAATTA ACAAGAAGAA GTACGAGACTCTC'rCATACC TTCCTGATTT240
GAGCCAGGAG CAATTGCTTA GTGAAGTTGA GTACCTTTTGAAAAATGGAT GGGTTCCTTG300
CTTGGAATTC GAGACTGAGC ACGGATTTGT CTACCGTGAAAACAACAAGT CACCAGGATA360
CTATGATGGC AGATACTGGA CCATGTGGAA GCTACCTATTTCGGATGCAC TGATGCCACC420
CAAGTGTTGG CTGAGGTGGA AGAGGCGAAG AAGGCATACCCACAGGCCTG GATCCGTATC480
ATTGGATTCG ACAACGTGG 499
(2) INFORMATION FOR SEQ ID N0: 42:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 321 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION:
SEQ ID NO: 42:
GATGGAAACA TTCTTGGACACAAATTGGAATACAACTATA ACTCACACAATGTATACATC 60
ATGGCAGACA AACAAAAGAATGGAATCAAAGTTAACTTCA AAA'PTAGACACAACATTGAA 120
GATGGAAGCG TTCAACTAGCAGACCATTATCAACAAAATA CTCCAATTGGCGATGGCCCT 180
GTCCTTTTAC CAGACAACCATTACCTGTCCACACAATCTG CCC'CTTCGAAAGATCCCAAC 240
GAAAAGAGAG ACCACATGGTCCTTCTTGAGTTTGTAACAG CTG(:TGGGATTACACATGGC 300
ATGGATGAAC TATACAAATAA 321
(2) INFORMATION
FOR SEQ ID NO:
43:
!II
CA 02297616 2001-04-25
- 38 -
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 421 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID
N0: 43:
TGGACCCAGA GGCTTTCTCA GCGAGTTTGT TCAAATGGGACCCTAGAGGTGCAATGCCAC 60
CGCCAACCCG GCTGTTGGAA GCCGCGGTGG CGCCTCCTCCTCCACCACCAGTTCTGCCAC 120
CGCCGCAGCC TCTATCGGCG GCCTATTCCA TTAGGACAAGGGAGTTAGGAGGGCTAGAGG 180
AGTTGTTTCA AGCTTACGGT ATACGTTATT ACACTGCTGCTAAAATAGCGGAGCTAGGTT 240
TTACGGTGAA TACTCTATTG GACATGAAAG ATGAGGAACTTGATGATATGATGAATAGCC 300
TTTCACAGAT TTTCAGATGG GAACTCCTCG TCGGAGAAAGGTACGGTATCAAAGCTGCAA 360
TCAGGGCGGA ACGGCGGAGG CTTGAGGAGG AAGAACTACGGCGGCGCAGCCACCTTCTGT 420
C 421
(2) INFORMATION FOR SEQ ID N0: 44:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 8976 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: circular
(xi) SEQUENCE
DESCRIPTION:
SEQ ID
N0: 44:
TACTCCAAAAATGTCAAAGATACAGTCTCAGAAGACCAAAGGGCTATTGAGACTTTTCAA 60
CAAAGGGTAATTTCGGGAAACCTCCTCGGATTCCATTGCCCAGCTATCTGTCACTTCATC 120
GAAAGGACAGTAGAAAAGGAAGGTGGCTCCTACAAATGCCATCATTGCGATAAAGGAAAG 180
GCTATCATTCAAGATGCCTCTGCCGACAGTGGTCCCAAAGATGGACCCCCACCCACGAGG 240
AGCATCGTGGAAAAAGAAGACGTCCCAACCACGTCTTCAAAGCAAGTGGATTGATGTGAC 300
ATCTCCACTGACGTAAGGGATGACGCACAATCCCACTATCCTTCGCAAGACCCTTCTTCT 360
ATATAAGGAAGTTCATTTCATTTGGAGAGGACAGCCCAAGCTTTCTAGAGGATCCATAAA 420
ACATTTCAATCCTTTGAACGCGGTAGAACGTGCTAATTGGATT'.CTGGTGAGAACGCGGTA 480
GAACGTACTTATCACCTACAGTTTTATTTTGTTTTTCTTTTTGGTTTAATCTATCCAGCT 540
TAGTACCGAGTGGGGGAAAGTGACTGGTGTGCCTAAAACCTTT'.PCTTTGATACTTTGTAA 600
I~I
CA 02297616 2001-04-25
- 39 -
AAATACATACAGATACAATGGCGAACGGTAACTTCAAGTTGTCTCAATTGCTCAATGTGG 660
ACGAGATGTCTGCTGAGCAGAGGAGTCATTTCTTTGACTTGATGCTGACTAAACCTGATT 720
GTGAGATCGGGCAAATGATGCAAAGAGTTGTTGTTGATAAAGTCGATGACATGATTAGAG 780
AAAGAAAGACTAAAGATCCAGTGATTGTTCATGAAGTTCTTTCTCAGAAGGAACAGAACA 840
AGTTGATGGAAATTTATCCTGAATTCAATATCGTGTTTAAAGACGACAAAAACATGGTTC 900
ATGGGTTTGCGGCTGCTGAGCGAAAACTACAAGCTTTATTGCTTTTAGATAGAGTTCCTG 960
CTCTGCAAGAGGTGGATGACATCGGTGGTCAATGGTCGTTTTGGGTAACTAGAGGTGAGA 1020
AAAGGATTCATTCCTGTTGTCCAAATCTAGATATTCGGGATGATCAGAGAGAAATTTCTC 1080
GACAGATATTTCTTACTGCTATTGGTGATCAAGCTAGAAGTGGTAAGAGACAGATGTCGG 1140
AGAATGAGCTGTGGATGTATGACCAATTTCGTGAAAATATTGCTGCGCCTAACGCGGTTA 1200
GGTGCAATAATACATATCAGGGTTGTACATGTAGGGGTTTTTCTGATGGTAAGAAGAAAG 1260
GCGCGCAGTATGCGATAGCTCTTCACAGCCTGTATGACTTCAAGTTGAAAGACTTGATGG 1320
CTACTATGGTTGAGAAGAAAACTAAAGTGGTTCATGCTGCTATGCTTTTTGCTCCTGAAA 1380
GTATGTTAGTGGACGAAGGTCCATTACCTTCTGTTGACGGTTACTACATGAAGAAGAACG 1440
GGAAGATCTATTTCGGTTTTGAGAAAGATCCTTCCTTTTCTTACATTCATGACTGGGAAG 1500
AGTACAAGAAGTATCTACTGGGGAAGCCAGTGAGTTACCAAGGGAATGTGTTCTACTTCG 1560
AACCGTGGCAGGTGAGAGGAGACACAATGCTTTTTTCGATCTACAGGATAGCTGGAGTTC 1620
CGAGGAGGTCTCTATCATCGCAAGAGTACTACCGAAGAATATA'PATCAGTAGATGGGAAA 1680
GCATGGTTGTTGTCCCAATTTTCGATCTGGTCGAATCAACGCGAGAGTTGGTCAAGAAAG 1740
ACCTGTTTGTAGAGAAACAATTCATGGACAAGTGTTTGGATTACATAGCTAGGTTATCTG 1800
ACCAGCAGCTGACCATAAGCAATGTTAAATCATACTTGAGTTCAAATAATTGGGTCTTAT 1860
TCATAAACGGGGCGGCCGTGAAGAACAAGCAAAGTGTAGATTC'PCGAGATTTACAGTTGT 1920
TGGCTCAAACTTTGCTAGTGAAGGAACAAGTGGCGAGACCTGTCATGAGGGAGTTGCGTG 1980
AAGCAATTCTGACTGAGACGAAACCTATCACGTCATTGACTGATGTGCTGGGTTTAATAT 2040
CAAGAAAACTGTGGAAGCAGTTTGCTAACAAGATCGCAGTCGGCGGATTCGTTGGCATGG 2100
TTGGTACTCTAATTGGATTCTATCCAAAGAAGGTACTAACCTGGGCGAAGGACACACCAA 2160
ATGGTCCAGAACTATGTTACGAGAACTCGCACAAAACCAAGGTGATAGTATTTCTGAGTG 2220
TTGTGTATGCCATTGGAGGAATCACGCTTATGCGTCGAGACATCCGAGATGGACTGGTGA 2280
AAAAACTATGTGATATGTTTGATATCAAACGGGGGGCCCATGTCTTAGACGTTGAGAATC 2340
CA 02297616 2001-04-25
- 40 -
CGTGCCGCTATTATGAAATCAACGATTTCTTTAGCAGTCTGTATTCGGCATCTGAGTCCG 2400
GTGAGACCGTTTTACCAGATTTATCCGAGGTAAAAGCCAAGTCTGATAAGCTATTGCAGC 2460
AGAAGAAAGAAATCGCTGACGAGTTTCTAAGTGCAAAATTCTCTAACTATTCTGGCAGTT 2520
CGGTGAGAACTTCTCCACCATCGGTGGTCGGTTCATCTCGAAGCGGACTGGGTCTGTTGT 2580
TGGAAGACAGTAACGTGCTGACCCAAGCTAGAGTTGGAGTTTC.AAGAAAGGTAGACGATG 2640
AGGAGATCATGGAGCAGTTTCTGAGTGGTCTTATTGACACTGAAGCAGAAATTGACGAGG 2700
TTGTTTCAGCCTTTTCAGCTGAATGTGAAAGAGGGGAAACAAGCGGTACAAAGGTGTTGT 2760
GTAAACCTTTAACGCCACCAGGATTTGAGAACGTGTTGCCAGCTGTCAAACCTTTGGTCA 2820
GCAAAGGAAAAACGGTCAAACGTGTCGATTACTTCCAAGTGATGGGAGGTGAGAGATTAC 2880
CAAAAAGGCCGGTTGTCAGTGGAGACGATTCTGTGGACGCTAGAAGAGAGTTTCTGTACT 2940
ACTTAGATGCGGAGAGAGTCGCTCAAAATGATGAAATTATGTCTCTGTATCGTGACTATT 3000
CGAGAGGAGTTATTCGAACTGGAGGTCAGAATTACCCGCACGGACTGGGAGTGTGGGATG 3060
TGGAGATGAAGAACTGGTGCATACGTCCAGTGGTCACTGAACATGCTTATGTGTTCCAAC 3120
CAGACAAACGTATGGATGATTGGTCGGGATACTTAGAAGTGGC'TGTTTGGGAACGAGGTA 3180
TGTTGGTCAACGACTTCGCGGTCGAAAGGATGAGTGATTATGTCATAGTTTGCGATCAGA 3240
CGTATCTTTGCAATAACAGGTAATAATCCTCTCTCTTGATATT'rTTAAATTATAGAATTA 3300
ATTAGTTTACTTTATTCTTTACTATATGATTTAAATAGTTTAA'rCTTGTTTTTGAGTAAA 3360
CTATTCGATTTTGATATTTGTATTCGTCCTACAAAGTTGGAAA'rACTGATGATATTTTCT 3420
TTTGAACGTGATACCTACCAATACTAATCTTACGGAATCTTTTAATAGAGCACTAATCAA 3480
CATGGAACTAAAGACCAATTCTTAAGTGTCTCTGTTGTACAGT'rCATTTTAGTAGTGCGT 3540
TTAAGTATTATTATCTCCCTTCATGCGGGGCAATTATGTAGAT'rAAAATCGAAATTATAT 3600
AAAATTTACATAAGTCTAAGTCTAGGGTCTCCAGCTAATTGTTATTTTTTTAACGATGTT 3660
GACTAAAGCAATAACGACGTTGACTTGTGTTAAACAGGTTGATCTTGGACAATTTAAGTG 3720
CCCTGGATCTAGGACCAGTTAACTGTTCTTTTGAATTAGTTGACGGTGTACCTGGTTGTG 3780
GTAAGTCGACAATGATTGTCAACTCAGCTAATCCTTGTGTCGA'PGTGGTTCTCTCTACTG 3840
GGAGAGCAGCAACCGACGACTTGATCGAGAGATTCGCGAGCAAAGGTTTTCCATGCAAAT 3900
TGAAAAGGAGAGTGAAGACGGTTGATTCTTTTTTGATGCATTG'PGTCGATGGTTCTTTAA 3960
CCGGAGACGTGTTGCATTTCGACGAAGCTCTCATGGCCCATGC'CGGTATGGTGTACTTTT 4020
GCGCTCAGATAGCTGGTGCTAAACGATGTATCTGTCAAGGAGATCAGAATCAAATTTCTT 4080
!ii
CA 02297616 2001-04-25
- 41 -
TCAAGCCTAGGGTATCTCAAGTTGATTTGAGGTTTTCTAGTCTGGTCGGA 4140
AAGTTTGACA
TTGTTACAGAAAAAAGAGAAACTTACAGAAGTCCAGCAGATGTGGCTGCCGTATTGAACA 4200
AGTACTATACTGGAGATGTCAGAACACATAACGCGACTGCTAATTCGATGACGGTGAGGA 4260
AGATTGTGTCTAAAGAACAGGTTTCTTTGAAGCCTGGTGCTCAGTACATAACTTTCCTTC 4320
AGTCTGAGAAGAAGGAGTTGGTAAATTTGTTGGCATTGAGGAAAGTGGCAGCTAAAGTGA 4380
GTACAGTACACGAGTCGCAAGGAGAGACATTCAAAGATGTAGTCCTAGTCAGGACGAAAC 4440
CTACGGATGACTCAATCGCTAGAGGTCGGGAGTACTTAATCGTGGCATTGTCGCGTCACA 4500
CACAATCACTTGTGTATGAAACTGTGAAAGAGGACGATGTAAGCAAAGAGATCAGGGAAA 4560
GTGCCGCGCTTACGAAGGCGGCTTTGGCAAGATTTTTTGTTACTGAGACCGTCTTATGAC 4620
GGTTTCGGTCTAGGTTTGATGTCTTTAGACATCATGAAGGGCCTTGCGCCGTTCCAGATT 4680
CAGGTACGATTACGGACTTGGAGATGTGGTACGACGCTTTGTTTCCGGGAAATTCGTTAA 4740
GAGACTCAAGCCTAGACGGGTATTTGGTGGCAACGACTGATTGCAATTTGCGATTAGACA 4800
ATGTTACGATCAAAAGTGGAAACTGGAAAGACAAGTTTGCTGAAAAAGAAACGTTTCTGA 4860
AACCGGTTATTCGTACTGCTATGCCTGACAAAAGGAAGACTACTCAGTTGGAGAGTTTGT 4920
TAGCATTGCAGAAAAGGAACCAAGCGGCACCCGATCTACAAGA.F1AATGTGCACGCGACAG 4980
TTCTAATCGAAGAGACGATGAAGAAGCTGAAATCTGTTGTCTACGATGTGGGAAAAATTC 5040
GGGCTGATCCTATTGTCAATAGAGCTCAAATGGAGAGATGGTGGAGAAATCAAAGCACAG 5100
CGGTACAGGCTAAGGTAGTAGCAGATGTGAGAGAGTTACATGAAATAGACTATTCGTCTT 5160
ACATGTATATGATCAAATCTGACGTGAAACCTAAGACTGATTTAACACCGCAATTTGAAT 5220
ACTCAGCTCTACAGACTGTTGTGTATCACGAGAAGTTGATCAACTCGTTGTTCGGTCCAA 5280
TTTTCAAAGAAATTAATGAACGCAAGTTGGATGCTATGCAACCACATTTTGTGTTCAACA 5340
CGAGAATGACATCGAGTGATTTAAACGATCGAGTGAAGTTCTTAAATACGGAAGCGGCTT 5400
ACGACTTTGTTGAGATAGACATGTCTAAATTCGACAAGTCGGCAAATCGCTTCCATTTAC 5460
AACTGCAGCTGGAGATTTACAGGTTATTTGGGCTGGATGAGTGGGCGGCCTTCCTTTGGG 5520
AGGTGTCGCACACTCAAACTACTGTGAGAGATATTCAAAATGGTATGATGGCGCATATTT 5580
GGTACCAACAAAAGAGTGGAGATGCTGATACTTATAATGCAAATTCAGATAGAACACTGT 5640
GTGCACTCTTGTCTGAATTACCATTGGAGAAAGCAGTCATGGT'tACATATGGAGGAGATG 5700
ACTCACTGATTGCGTTTCCTAGAGGAACGCAGTTTGTTGATCCGTGTCCAAAGTTGGCTA 5760
CTAAGTGGAATTTCGAGTGCAAGATTTTTAAGTACGATGTCCCAATGTTTTGTGGGAAGT 5820
~ii
CA 02297616 2001-04-25
42 -
TCTTGCTTAAGACGTCATCGTGTTACGAGTTCGTGCCAGATCCGGTAAAAGTTCTGACGA 5880
AGTTGGGGAAAAAGAGTATAAAGGATGTGCAACATTTAGCCGAGATCTACATCTCGCTGA 5940
ATGATTCCAATAGAGCTCTTGGGAACTACATGGTGGTATCCAAACTGTCCGAGTCTGTTT 6000
CAGACCGGTATTTGTACAAAGGTGATTCTGTTCATGCGCTTTGTGCGCTATGGAAGCATA 6060
TTAAGAGTTTTACAGCTCTGTGTACATTATTCCGAGACGAAAACGATAAGGAATTGAACC 6120
CGGCTAAGGTTGATTGGAAGAAGGCACAGAGAGCTGTGTCAAACTTTTACGACTGGTAAT 6180
ATGGAAGACAAGTCATTGGTCACCTTGAAGAAGAAGACTTTCG.AAGTCTCAAAATTCTCA 6240
AATCTAGGGGCCATTGAATTGTTTGTGGACGGTAGGAGGAAGAGACCGAAGTATTTTCAC 6300
AGAAGAAGAGAAACTGTCCTAAATCATGTTGGTGGGAAGAAGAGTGAACACAAGTTAGAC 6360
GTTTTTGACCAAAGGGATTACAAAATGATTAAATCTTACGCGTTTCTAAAGGTAGTAGGT 6420
GTACAACTAGTTGTAACATCACATCTACCTGCAGATACGCCTGGGTTCATTCAAATCGAT 6480
CTGTTGGATTCGAGACTTACTGAGAAAAGAAAGAGAGGAAAGACTATTCAGAGATTCAAA 6540
GCTCGAGCTTGCGATAACTGTTCAGTTGCGCAGTACAAGGTTG.?~ATACAGTATTTCCACA 6600
CAGGAGAACGTACTTGATGTCTGGAAGGTGGGTTGTATTTCTGAGGGCGTTCCGGTCTGT 6660
GACGGTACATACCCTTTCAGTATCGAAGTGTCGCTAATATGGG'PTGCTACTGATTCGACT 6720
AGGCGCCTCAATGTGGAAGAACTGAACAGTTCGGATTACATTGAAGGCGATTTTACCGAT 6780
CAAGAGGTTTTCGGTGAGTTCATGTCTTTGAAACAAGTGGAGA'rGAAGACGATTGAGGCG 6840
AAGTACGATGGTCCTTACAGACCAGCTACTACTAGACCTAAGTCATTATTGTCAAGTGAA 6900
GATGTTAAGAGAGCGTCTAATAAGAAAAACTCGTCTTAATGCA'TAAAGAAATTTATTGTC 6960
AATATGACGTGTGTACTCAAGGGTTGTGTGAATGAAGTCACTG'rTCTTGGTCACGAGACG 7020
TGTAGTATCGGTCATGCTAACAAATTGCGAAAGCAAGTTGCTGACATGGTTGGTGTCACA 7080
CGTAGGTGTGCGGAAAATAATTGTGGATGGTTTGTCTGTGTTG'PTATCAATGATTTTACT 7140
TTTGATGTGTATAATTGTTGTGGCCGTAGTCACCTTGAAAAGTGTCGTAAACGTGTTGAA 7200
ACAAGAAATCGAGAAATTTGGAAACAAATTCGACGAAATCAAGCTGAAAACATGTCTGCG 7260
ACAGCTAAAAAGTCTCATAATTCGAAGACCTCTAAGAAGAAATTCAAAGAGGACAGAGAA 7320
TTTGGGACACCAAAAAGATTTTTAAGAGATGATGTTCCTTTCGGGATTGATCGTTTGTTT 7380
GCTTTTTGATTTTATTTTATATTGTTATCTGTTTCTGTGTATAGACTGTTTGAGATTGGC 7440
GCTTGGCCGACTCATTGTCTTACCATAGGGGAACGGACTTTGT'TTGTGTTGTTATTTTAT 7500
TTGTATTTTATTAAAATTCTCAATGATCTGAAAAGGCCTCGAGGCTAAGAGATTATTGGG 7560
i
CA 02297616 2001-04-25
- 43 -
GGGTGAGTAAGTACTTTTAA AGTGATGATGGTTACAAAGGCAAAAGGGGT AAAACCCCTC7620
GCCTACGTAAGCGTTATTAC GCCCGGATCCCCCGGGGAGCTCGAATTCGC TGAAATCACC7680
AGTCTCTCTCTACAAATCTA TCTCTCTCTATTTTTTCCATAAATAATGTG TGAGTAGTTT7740
CCCGATAAGGGAAATTAGGG TTCTTATAGGGTTTCGCTCATGTGTTGAGC ATATAAGAAA7800
CCCTTAGTATGTATTTGTAT TTGTAAAATACTTCTATTATCAATAAAATT TCTAATTCCT7860
AAAACCAAAATCCAGTACTA AAATCCAGATCTCCTAAAGTCCCTATAGAT CTTTGTCGTG7920
AATATAAACCAGACACGAGA CGACTAAACCTGGAGCCCAGACGCCGTTCG AAGCTAGAAG7980
TACCGCTTAGGCAGGAGGCC GTTAGGGAAAAGATGCTAAGGCAGGGTTGG TTACGTTGAC8040
TCCCCCGTAGGTTTGGTTTA AATATGATGAAGTGGACGGAAGG.AAGGAGG AAGACAAGGA8100
AGGATAAGGTTGCAGGCCCT GTGCAAGGTAAGAAGATGGAAATTTGATAG AGGTACGCTA8160
CTATACTTATACTATACGCT AAGGGAATGCTTGTATTTATACCCTATACC CCCTAATAAC8220
CCCTTATCAATTTAAGAAAT AATCCGCATAAGCCCCCGCTTAA.AAATTGG TATCAGAGCC8280
ATGAATAGGTCTATGACCAA AACTCAAGAGGATAAAACCTCACCAAAATA CGAAAGAGTT8340
CTTAACTCTAAAGATAAAAG ATCTTTCAAGATCAAAACTAGTTCCCTCAC ACCGGAGCAT8400
GCGATATCCTCGACCTGCAG GCATGCAAGCTTGGCGTAATCATGGTCATA GCTGTTTCCT8460
GTGTGAAATTGTTATCCGCT CACAATTCCACACAACATACGAGCCGGAAG CATAAAGTGT8520
AAAGCCTGGGGTGCCTAATG AGTGAGCTAACTCACATTAATTGCGTTGCG CTCACTGCCC8580
GCTTTCCAGTCGGGAAACCT GTCGTGCCAGCTGCATTAATGAA'PCGGCCA ACGCGCGGGG8640
AGAGGCGGTTTGCGTATTGG GCCAAAGACAAAAGGGCGACATTCAACCGA TTGAGGGAGG8700
GAAGGTAAATATTGACGGAA ATTATTCATTAAAGGTGAATTATCACCGTC ACCGACTTGA8760
GCCATTTGGGAATTAGAGCC AGCAAAATCACCAGTAGCACCAT'PACCATT AGCAAGGCCG8820
GAAACGTCACCAATGAAACC ATCGATAGCAGCACCGTAATCAG'PAGCGAC AGAATCAAGT8880
TTGCCTTTAGCGTCAGACTG TAGCGCGTTTTCATCGGCATTTTCGGTCAT AGCCCCCTTA8940
TTAGCGTTTGCCATCTTTTC ATAATCAAAATCACCG 8976
(2) INFORMATION
FOR SEQ
ID N0:
45:
(i) SEQUENCE
CHARACTERISTICS:
(A) LENGTH: 19 base
pairs
(B) TYPE: nucleic
acid
(C) STRANDEDNESS:
single
(D) TOPOLOGY: linear
CA 02297616 2001-04-25
44 -
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 45:
GGATCCGCAC ACAAGGTTA 19
(2) INFORMATION FOR SEQ ID NO: 46:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 18 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 46:
GGGCGCGCCC TCGTTAAC 18