Note: Descriptions are shown in the official language in which they were submitted.
CA 02303086 2000-03-13
v.;
w -1-
Hoechst Aktiengesellschaft
H26102PCT BO/JK/sa
Addressable modular recognition system, its preparation and use
The present invention relates to a recognition system comprising
(a) at least one immobilized binding component A having at least one binding
site
for the recognition species B and
(b) at least one recognition species B which can bind to the binding component
A
and contains at least one binding site for a substrate S, the binding of the
binding
component A to the recognition species B taking place in the form of a
molecular
1 ~ pairing system.
Arrays are arrangements of immobilized recognition species which play an
important role in the simultaneous determination of analytes, especially in
analytical methods and diagnosis. Examples are peptide arrays (Fodor et al.,
Nature 1993, 364, 555) and nucleic acid arrays (Southern et al. Genomics 1992,
13, 1008; U.S. Patent No. 5,632,957).
In experimental analytical systems, arrays permit particularly simple, rapid
and
reproducible data analysis as a result of the localized generation of events.
Examples of this extend from the physical multi-channel detector as far as
microtitre plates in laboratory medicine.
Arrays also serve for the storage and processing of information and are the
fundamental construction element of nanotechnology.
Further important application areas can be found in biology, biochemistry,
medicine and pharmacology. Thus, EP-Al-0 461 462 describes an immunoassay in
which antigens which are positioned and immobilized in a field-like manner are
brought into contact with one or more antibodies. WO 96/01836 describes, for
example, an array of DNA molecules of differing sequence, which was used for
the
detection of gene sections and thus led, for example, to the diagnosis of
pathogenic
bacteria.
CA 02303086 2000-03-13
-2-
Immobilization by means of supramolecular interactions is also known outside
of
the array applications. Thus, excipients having anti-antibodies can be
immobilized
by means of an antigen bonded covalently to the excipient. The analytical
system
of immunoassays is based largely on enzyme immunoassays (EIAs), in which an
S enzymatically catalysed reaction indicates the presence of an antigen-
antibody or
an antigen-antibody-anti-antibody complex. One of the units involved in the
complex is in this case either immobilized on a Garner or itself a Garner,
e.g. in the
form of tissue constituents.
Signal amplification processes of this type, however, have disadvantages, in
particular with respect to the reliability of the qualitative information and
quantification. A particular disadvantage of miniaturized arrays is the outlay
and
the costs in preparation.
The object of the present invention was therefore to find a recognition system
which is simple, reliable, highly selective and moreover inexpensive.
The present invention therefore relates to a recognition system comprising
(a) at least one immobilized binding component A having at least one binding
site
for the recognition species B and
(b) at least one recognition species B which can bind to the binding component
A
and contains at least one binding site for a substrate S, the binding of the
binding
component A to the recognition species B taking place in the form of a
molecular
pairing system.
Such pairing systems are supramolecular systems of non-covalent interaction,
which are distinguished by selectivity, stability and reversibility, and their
properties are preferably influenced thermodynamically, i.e. by temperature,
pH
and concentration. Such pairing systems can also be used, for example, on
account
of their selective properties as "molecular adhesive" for the bringing
together of
different metal clusters to give cluster associates having potentially novel
properties [see, for example, R. L. Letsinger, et al., Nature 1996, 382, 607-
9; P. G.
Schultz et al., Nature 1996, 382, 609-11].
It is therefore particularly advantageous if the pairing system is a complex
which is
formed by association of the binding component A with the recognition species
B
via non-covalent interactions. The non-covalent interactions are, in
particular,
hydrogen bridges, salt bridges, stacking, metal ligands, charge-transfer
complexes
and hydrophobic interactions.
CA 02303086 2000-03-13
_3_
In a particular embodiment, the molecular pairing system according to the
present
invention contains a nucleic acid and its analogues, in particular in the form
of a
pentose, preferably of a pentopyranose or pentofuranose. In general, the
pentose is
selected from a ribose, arabinose, lyxose or xylose. Pyranosyl-RNA (p-RNA),
nucleic acid having one or more aminocyclohexylethanoic acid (CNA) units,
peptide nucleic acid (PNA), or a nucleic acid having one or more [2-amino-
4-(carboxymethyl)cyclohexyl]nucleobases is particularly preferred. Pyranosyl
nucleic acids (p-NAs) and especially p-RNAs are particularly preferred.
p-NAs are in general structural types isomeric to the natural RNA, in which
the
pentose units are present in the pyranose form and are repetitively linked by
phosphodiester groups between the positions C-2' and C-4'. "Nucleobase" is
understood here as meaning the canonical nucleobases A, T, U, C, G, but also
the
pairs isoguanine/isocytosine and 2,6-diaminopurine/xanthine and within the
meaning of the present invention also other purines and pyrimidines such as
purine, 2,6-diaminopurine, 6-purinethiol, pyridine, pyrimidine, isoguanine,
6-thioguanine, xanthine, hypoxanthine, isocytosine, indole, tryptamine,
N-phthaloyltryptamine, caffeine, theobromine, theophylline, benzotriazole or
acridine, and preferably ribopyranosyladenosine, ribopyranosylguanosine,
ribopyranosylthymidine, ribopyranosylcytosine, ribopyranosyltryptamine or
ribopyranosyl-N-phthalotryptamine, ribopyranosyluracil or their [2-amino-
4-(carboxymethyl)ribopyranosyl] derivatives.
p-NAs, namely the p-RNAs derived from ribose, were described for the first
time
by Eschenmoser et al. (see Pitsch, S. et al. Helv. Chim. Acta 1993, 76, 2161;
Pitsch, S. et al. Helv. Chim Acta 1995, 78, 1621; Angew. Chem. 1996, 108, 1619-
1623). They form exclusively so-called Watson-Crick-paired, i.e. purine-
pyrimidine- and purine-purine-paired, antiparallel, reversibly "melting",
quasi-
linear and stable duplices. Homochiral p-RNA strands of the opposite sense of
chirality likewise pair controllably and are strictly non-helical in the
duplex
formed. This specificity, which is valuable for the construction of
supramolecular
units, is connected with the relatively low flexibility of the ribopyranose
phosphate
backbone and with the strong inclination of the base plane to the strand axis
and
the tendency resulting from this for intercatenary base stacking in the
resulting
duplex and can finally be attributed to the participation of a 2',4'-cis-
disubstituted
ribopyranose ring in the construction of the backbone.
CA 02303086 2000-03-13
-4-
These significantly better pairing properties make p-NAs preferred pairing
systems
for use in the construction of supramolecular units compared with DNA and RNA.
They form a pairing system which is orthogonal to natural nucleic acids, i.e.
they
do not pair with DNAs and RNAs occurring in the natural form, which is
advantageous, in particular, in the diagnostic field.
p-NAs are therefore particularly suitable for use in the field of
nanotechnology, for
example for the preparation of novel materials, diagnostics and therapeutics
and
also microelectronic, photonic or optoelectronic components and for the
controlled
bringing together of molecular species to give supramolecular units, such as,
for
example, for the (combinatorial) synthesis of protein assemblies [see, for
example,
A. Lombardi, J. W. Bryson, W. F. DeGrado, Biomolekuls (Pept. Sci.) 1997, 40,
495-504J, as p-NAs, and particularly p-RNAs, form pairing systems which are
strongly and thermodynamically controllable. A further application therefore
1 S results especially in the diagnostic and drug discovery field due to the
possibility of
providing functional, preferably biological, units such as proteins or DNA/RNA
sections, e.g. with a p-RNA code which does not interfere with the natural
nucleic
acids (see, for example, W093/20242).
According to the present invention, the length of the nucleic acid and its
analogues
is at least about 4-50, preferably at least about 4-25, in particular at least
about 4-
15, especially at least about 4-10, nucleotides.
In general, the binding component A is immobilized on a Garner.
The term "immobilized" is understood within the meaning of the present
invention
as meaning the formation of a covalent bond, quasi-covalent bond or
supramolecular bond by association of two or more molecular species such as
molecules having a linear constitution, in particular peptides, peptoids,
proteins,
linear oligo- or polysaccharides, nucleic acids and their analogues, or
monomers
such as heterocycles, in particular nitrogen heterocycles, or molecules having
a
non-linear constitution such as branched oligo- or polysaccharides or
antibodies
and their functional moieties. Functional moieties of antibodies are, for
example,
Fv fragments (Skerra & Pliickthun (1988) Science 240, 1038), single-chain Fv
fragments (scFv; Bird et al. (1988), Science 242, 423; Huston et al. (1988)
Proc.
Natl. Acad. Sci. U.S.A., 85, 5879) or Fab fragments (Better et al. (1988)
Science
240, 1041 ).
CA 02303086 2000-03-13
t
-5-
The attachment to the carrier is thus in general carried out covalently, quasi-
covalently, supramolecularly or physically, such as magnetically (A. R.
Shepard et
al. (1997) Nucleic Acids Res., 25, 3183-3185, No. 15), in an electrical field
or
through a molecular sieve. The binding component A is thereby either
synthesized
directly at the position of the carrier or "linked" to certain positions of
the Garner.
Examples are conjugation and carrier processes via periodote oxidation and
reductive amination of the Schiff's base, N-hydroxysuccinimide esters of,
preferably, dicarboxylic acid linkers, ethylenediaminephosphoamidate linkers,
mercapto-, iodoacetyl or maleimido processes and/or covalent or non-covalent
biotin linker processes.
The term "Garner" is understood within the meaning of the present invention as
meaning a material, in particular chip material, which is present in solid or
alternatively gelatinous form. Suitable Garner materials are, for example,
ceramic,
metal, in particular noble metal, glasses, plastics, crystalline materials or
thin
layers of the Garner, in particular of the materials mentioned, or
(bio)molecular
filaments such as cellulose, structural proteins.
A particular embodiment is therefore a recognition system according to the
invention, in which the binding component A is immobilized on a carrier by
means
of a covalent bond, quasi-covalent bond or supramolecular bond by association
of
two or more molecular species such as molecules of linear constitution, in
particular peptides, peptoids, proteins, linear oligo- or polysaccharides,
nucleic
acids and their analogues, or monomers such as heterocycles, in particular
nitrogen
heterocycles, or molecules of non-linear constitution such as branched oligo-
or
polysaccharides or antibodies and their functional moieties such as Fv
fragments,
single-chain Fv fragments (scFv) or Fab fragments.
In a further embodiment, the binding component A is immobilized at defined
sites
of the Garner, in particular in the form of a matrix, the defined sites of the
Garner
preferably being addressed.
According to the preferred recognition system, a molecule in the mobile
(buffer)
phase having the appropriate complementary sequence will only spontaneously
form a supramolecular complex at the position of the suitable address. If
further
units having particular functions such as, for example, that of an antibody,
are
bonded to these mobile complementary addresses by chemical (conjugates) or
supramolecular compound formation (complexes), depending on the address
CA 02303086 2000-03-13
-6-
pattern used a different functional array will be spread on the same
immobilizate
array.
The great advantages of such a modular system are the identical one-off
provision
of the carrier units for very different applications and the in situ
generation of non-
keepable bioconjugates, for example, from proteins, enzymes or living cells
and
the pairing radical.
A further advantage is the stepwise production of substrate binding event and
the
measurable binding event at the carrier position, i.e. the substrate can form
a first
complex with the soluble, addressed component (recognition species B) in a
completely unhindered manner and then immobilize on the binding component A
in a pairing manner in the space of the Garner position.
1 S It is further particularly preferred if the binding component A is
immobilized on a
carrier electrode of the Garner, since an electronically readable signal is
produced,
for example, by a signal amplification of the impedance behaviour of Garner
electrodes during binding events. Appropriate electrode processes are
described in
R. P. Andres (1996) Science, 272, 1323-1325 and appropriate impedance
measurements are described in M. Stelzle et al. (1993) J. of Physical Chem.,
97,
2974-2981.
A suitable recognition species B is, for example, a biomolecule which, for
example, is selected from a peptide, peptoid, protein, such as receptor or
functional
moieties thereof such as the extracellular domain of a membrane receptor,
antibodies or functional moieties thereof such as Fv fragments, single-chain
Fv
fragments (scFv) or Fab fragments, or cell constituents such as lipids,
glycoproteins, filament constituents, or viruses, viral constituents such as
capsids,
or viroids, or their derivatives such as acetates and their active moieties,
or
substance libraries such as ensembles of structurally differing compounds,
preferably oligomeric or polymeric peptides, peptoids, saccharides, nucleic
acids.
The biomolecule customarily contains a binding region for the binding
component
A, which is preferably one of the nucleic acids described above or their
analogues.
In general, the biomolecule is bonded here to a selected nucleic acid or
analogue
via a linker. For example, a uracil-based linker is suitable, in which the 5-
position
of the uracil has preferably been modified, for example N-
phthaloylaminoethyluracil, but also an indole-based linker, preferably
tryptamine
derivatives, such as, for example, N-phthaloyltryptamine.
CA 02303086 2000-03-13
.,
In a particular embodiment, the immobilized binding component A contains
various binding sites for various recognition species B, by means of which
various
recognition species B can bind to the binding component A.
S
In a further embodiment, at least one further recognition species B is
immobilized
on the binding component A.
Therefore a further recognition system according to the invention is
characterized
in that it comprises
(a) at least one immobilized binding component A having at least 2+n different
binding sites for at least 2+n different recognition species B1, B2 ... Bn and
a
further recognition species B(n+3) different from the recognition species Bl,
B2 ...
Bn, which is immobilized on the immobilized binding component A, and
(b) at least (n+3) different recognition species B1, B2 ... B(n+3),
where n is an integer from 0-20, preferably 0-10, in particular 0-5,
especially 0 or
1.
In a further embodiment, the recognition species Bl, B2 ... Bn originates from
a
substance library.
For the structural analysis of a complex of a substance library, it is
particularly
advantageous if the structure of the recognition species B(n+3) is known,
and/or
the different recognition species B recognize the same substrate S.
The term "substrate" is understood within the meaning of the present invention
as
meaning a non-Garner-bonded substance, which is intended to be recognized by
the
recognition system according to the invention. The substrate S is in general
selected from molecules, preferably pharmaceuticals and plant protection
active
compounds, metabolites, physiological messenger substances, derivatives of
lead
structures, substances which are produced or produced to an increased extent
in the
human or animal body in the case of pathological changes, or transition state
analogues, or peptides, peptoids, proteins such as receptors or functional
moieties
thereof such as the extracellular domain of a membrane receptor, antibodies or
functional moieties thereof such as Fv fragments, single-chain Fv fragments
(scFv)
or Fab fragments, or cell constituents such as lipids, glycoproteins, filament
constituents, or viruses, viral constituents such as capsids, or viroids, or
their
derivatives such as acetates, or monomers such as heterocycles, in particular
nitrogen heterocycles, or molecules of non-linear constitution such as
branched
CA 02303086 2000-03-13
.. _ 8 _
oligo- or polysaccharides, or substance libraries such as ensembles of
structurally
differing compounds, preferably oligomeric or polymeric peptides, peptoids,
saccharides, nucleic acids, esters, acetals or monomers such as heterocycles,
lipids,
steroids, or targets for pharmaceuticals, preferably pharmaceutical receptors,
voltage-dependent ion channels, transporters, enzymes or biosynthesis units of
microorganisms.
Substance libraries are known to the person skilled in the art from the field
of
combinatorial chemistry. Examples are the readily accessible peptide
libraries,
produced by permutation of the peptide sequence. If such libraries pair,
completely
novel supramolecules or complexes result. The appreciable number of possible
complexes possibly includes recognition regions for substrate molecules,
similarly
to the epitope of an antibody. The embodiment then permits screening of such a
stochastic binding event. If one of the conjugate libraries is bonded to the
carrier,
its identity (e.g. the peptide sequence) can be directly fixed by the codon
address
or, if the address is constant, by its mere position. The array produces a so-
called
encoded library for one of the pairing strands and simplifies the complex
analysis
of the supramolecular library.
In a further preferred embodiment, the recognition system according to the
invention is an immunoassay.
Another subject of the present invention is also a process for the
identification of a
substrate S in a sample with the aid of the recognition system according to
the
invention, in which
(a) a recognition species B which recognizes the substrate S is brought into
contact
with the sample,
(b) is simultaneously or successive brought into contact with an immobilized
recognition species B, and
(c) the formation of a complex of immobilized binding component A, recognition
species B and substrate S is detected.
In particular, in the process according to the invention the formation of the
complex is controlled by means of physical parameters such as temperature,
salts,
solvents, electrophoretic processes.
In general, the complex formed is detected by means of labelling such as
radioactive or fluorescent labelling, enzymatic labelling, redox labelling,
spin
labelling of the recognition species B, or by means of the complex itself, for
CA 02303086 2000-03-13
- -9-
example by means of electrode processes such as by means of chemical
processes,
e.g. redox processes in the environment or on the electrode or by means of a
physical parameter such as by means of impedance measurement or direct current
measurement.
Particular amplification or preconcentration steps of the substrates are thus
not
needed for many applications, which is particularly advantageous. The chemical
and physical heterogeneity of the positions before and after the pairing
events can
moreover be eliminated using the direct electronic process, very
advantageously by
parametrization or calibration by means of the software.
The problem that important substrate molecules for such applications can be
molecules of the natural pairing systems DNA and RNA themselves and would
thus interact interferingly with the addressing is solved in that particularly
stable,
selective and non-natural pairing systems, such as, for example, p-NAs, are
used.
The present invention therefore also relates to a process with which
recognition
species, preferably natural DNA or RNA strands and proteins, in this case
preferably antibodies or functional moieties of antibodies, are clearly
encoded by
p-NA sections, preferably p-RNA sections. These can then be hybridized with
the
associated codons on a solid Garner. Thus always novel, diagnostically useful
arrays can be constructed on a solid carrier, which is equipped with codons in
the
form of an array only by adjustment of hybridization conditions with always
novel
combinations of recognition species at the desired positions. If the analyte,
for
example a biological sample such as serum or the like, is then applied, the
species
to be detected are then bonded to the array in a certain pattern, which is
then
recorded indirectly (e.g. by fluorescence labelling of the recognition
species) or
directly (e.g. by impedance measurement at the linkage point of the codons).
The
hybridization is then eliminated by means of suitable conditions (temperature,
salts, solvent, electrophoretic processes), so that again only the Garner with
the
codons remains. This is then again loaded with other recognition species and
is
used, for example, for the same analyte for the determination of another
sample.
The always novel arrangement of recognition species in the array format and
the
use of p-NAs as pairing systems is particularly advantageous compared with
other
systems, see, for example, WO 96/13522.
In the process according to the invention, the complex of recognition species
B and
substrate S can also be isolated in a further step. For this, for example, the
complex
CA 02303086 2000-03-13
_ 10_
is isolated from recognition species B and substrate S after freezing the
binding
equilibrium or covalent cross-linking of recognition species B and substrate
S.
The recognition system according to the invention is consequently particularly
highly suitable for finding a substrate S for diagnosis, for the preparation
of a
catalyst and/or for the preparation of an electronic component, in particular
for the
finding, for the optimization and/or for the preparation of a pharmaceutical
active
compound or plant protection active compound.
Depending on the addresses synthesized, kits which form the test system by
pairing
on the existing codon array in situ can thus be rapidly assembled for
different
questions or diagnostic problems. Biomolecules, for example very generally
cell or
viral constituents, very particularly monoclonal antibodies or their
functional
moieties, are preferred.
The following figures are intended to describe the invention in greater
detail,
without restricting it.
DESCRIPTION OF THE FIGURES
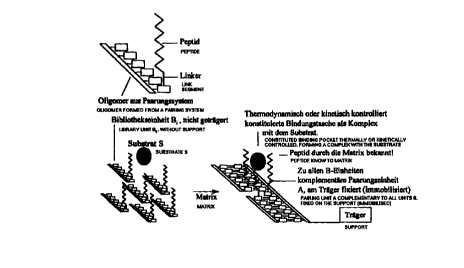
Fig. 1 shows schematically the general principle of a recognition species,
which is produced in situ around a substrate to be recognized. The
complexing unit (peptide) can be known by a carrier matrix. A binding
pocket formed under thermodynamic or kinetic control is formed here as
a complex with the substrate. The pairing unit A complementary to all B
units is immobilized on the carrier.
Fig.2 shows schematically an arrangement of immobilized recognition
structures (arrays) on a solid Garner.
Fig. 3 shows schematically the modular production of a supramolecular array.
Different immunoarrays are constructed on the same anticodon Garner
by addressing with the selective pairing regions.
Fig.4 shows schematically the construction of an array having 4 Garner
positions (electrodes) and the measuring principle.
Fig. 5 shows schematically the detection of the pairing of the anticodon-codon
molecules by UV spectroscopy and impedance spectroscopy. By
CA 02303086 2000-03-13
- -11-
lowering the temperature, the strands pair, the buffer supernatant
weakens, the UV extinction of the supernatant decreases and the change
in the electrode bilayer acts on the impedance measurement.
Fig. 6 shows schematically the functioning of an addressed immunoarray. Only
electrode 3 carries the appropriate address for an antibody-pairing strand
conjugate. If the appropriate antigen is added, the impedance at the
electrode 1 changes other than by mere change of buffer at the other
electrodes.
Fig.7 shows the cooling curves of a temperature-induced UV pairing
experiment with two complementary p-RNA addresses, to which a
histidine peptide is conjugated in each case. The pairing produces a
recognition region for nickel ions as a substrate. The substrate leads to a
clear increase in the transition temperature Tm, which is not observed
without the histidine radicals.
Fig. 8 shows schematically a simple matrix of two vapour-deposited gold
electrodes.
Fig. 9 shows the direct electronic detection of an antigen-antibody complex at
one electrode position of the array by impedance spectroscopy.
Fig. 10 shows an additional detection of the antigen-antibody complex at the
addressed electrode by means of fluorescence.
Examples
Example 1
Synthesis of a p-RNA oligonucleotide containing a linker using linker of the
formula 4' AGGCAIndT 2':
1.1 Solid-phase synthesis of the oligonucleotide
A, G, C, T are the nucleobases adenine, guanine, cytosine and thymine and Ind
is
aminoethylindole (indole CH2-CH2-NH2) as a linker in the form of a nucleobase.
CA 02303086 2000-03-13
-12-
The fully automatic solid-phase synthesis was carried out with 15 pmol in each
case. A synthesis cycle consists of the following steps:
(a) detritylation: 5 minutes with 6% DCA (dichloroacetic acid) in CH2ClZ (79
ml).
(b) washing with CH2C12 (20 ml), acetonitrile (20 ml) and then flushing with
S argon:
(c) coupling: washing of the resin with the activator (0.5 M pyridine.HCl in
CHzCl2 (0.2 ml) and then 30 minutes' treatment with activator (0.76 ml) and
phosphoramidite of the corresponding nucleobase (0.76 ml: 8 eq; 0.1 M in
acetonitrile) in the ratio 1/1;
(d) capping: 2 minutes' treatment with 50% Cap A (10.5 ml) and 50% Cap B
(10.5 ml) from PerSeptive Biosystems, Inc., Texas, USA (Cap A: THF,
lutidine, acetic anhydride; Cap B: 1-methylimidazole, THF, pyridine);
(e) oxidation: 1 minute's treatment with 120 ml of iodine solution (400 mg of
iodine in 100 ml of acetonitrile, 46 ml of Hz0 and 9.2 ml of sym-collidine);
and
(f) washing with acetonitrile (22 ml).
To facilitate the subsequent HPLC purification of the oligonucleotides, the
last
DMT (dimethoxytrityl) group was not removed. To detect the last coupling with
the modified phosphoamidites, after the synthesis with 1% of the resin a
trityl
cation absorption was carried out in UV (503 nm).
1.2 Work-up of the oligonucleotide:
The removal of the allyl ether protective groups was carned out with a
solution of
tetrakis(triphenylphosphine)palladium (272mg), triphenylphosphine (272 mg) and
diethylammonium hydrogencarbonate in CHzCl2 (l5ml) after S hours at RT. The
glass Garners were then washed with CH2C12 (30m1), acetone (30m1) and water
(30m1). In order to remove palladium complex residues, the resin was rinsed
with
an aqueous 0.1 M sodiumdiethyldithiocarbamate hydrate solution. The
abovementioned washing operation was carned out once more in the reverse
order.
The resin was then dried in a high vacuum for 10 minutes. The removal step
from
the glass Garner with simultaneous debenzoylation was carned out in 24%
hydrazine hydrate solution (6m1) at 4°C. After HPLC checking on RP 18
(18-25
hours), the oligonucleotide "Trityl ON" was freed of hydrazine by means of an
activated (acetonitrile, 20 ml) Waters Sep-Pak cartridge. The hydrazine was
washed with TEAB, 0.1 M (30m1). The oligonucleotide was then eluted with
acetonitrile/TEAB, O.1M (lOml). It was then purified by means of HPLC for the
removal of fragment sequences and the DMT deprotection (30 ml of 80% strength
CA 02303086 2000-03-13
~ -13-
aqueous formic acid) was carned out. Final desalting (by means of Sep-Pak
cartridge, with TEAB buffer O.1M/acetonitrile: 111) yielded the pure
oligonucleotide.
Example 2
Iodoacetylation of p-RNA with N-(iodoacetyloxy)succinimide
p-RNA sequence: 4' AGGCAIndT 2' MW = 2266.56 g/mol, prepared according to
Example 1.
1 eq. of the p-RNA was dissolved (1 ml per 350 nmol) in a 0.1 molar sodium
hydrogencarbonate solution (pH 8.4) and treated (40 ul per mg) with a solution
of
N-(iodoacetyloxy)succinimide in DMSO. The batch is blacked out with aluminium
1 S foil and allowed to stand at room temperature for 30-90 minutes.
The progress of the reaction was monitored by means of analytical HPLC. The
standard conditions were:
Buffer A : 0.1 molar triethylammonium acetate buffer in water
Buffer B : 0.1 molar triethylammonium acetate buffer in water:acetonitrile 1:4
Gradient : starting from 10% B to SO% B in 40 minutes
Column material: 10 pM LiChrosphere ~ 100 RP-18 from Merck Darmstadt
GmbH; 250 x 4 mm
Retention time of the starting materials: 18.4 minutes
Retention time of the products in this case: 23.1 minutes
After reaction was complete, the batch was diluted to four times the volume
with
water. A Waters Sep-Pak cartridge RP-18 (from 15 oD 2 g packing) was activated
with 2 x 10 ml of acetonitrile and 2 x 10 ml of water, the oligonucleotide was
applied and allowed to sink in, and the reaction vessel was washed with 2 x 10
ml
of water, rewashed with 3 x 10 ml of water in order to remove salt and
reagent, and
eluted first with 5 x 1 ml of 50:1 water:acetonitrile and then with 1:1. The
product
eluted in the 1:1 fractions in very good purity. The fractions were
concentrated in
the cold and in the dark, combined and concentrated again.
The yields were determined by means of W absorption spectrometry at 260 nm.
Mass spectrometry:
Sequence : 4' AGGCAInd(CH2CHZNHCOCH2-I)T 2'
calculated mass : 2434.50 g/mol
CA 02303086 2000-03-13
' -14-
found mass MHz2+: 1217.9 g/mol = 2433
Example 3
Conjugation ofp-RNA to a peptide of the sequence (His)6:
The iodoacetylated p-RNA (MW = 2434.50 g/mol) was dissolved in a buffer system
(1000p1 per 114 nmol) and then treated with a solution of the peptide in
buffer
(2 mol eq. of (His)6 peptide).
Buffer system : Borax/HCl buffer from Riedel-de Haen, pH 8.0, was mixed in the
ratio 1:1 with a 10 millimolar solution of EDTA disodium salt in water and
adjusted to pH 6.3 with HC1. A solution which contained S mM Na2EDTA was
obtained thereby.
The batch was left at room temperature in the dark until reaction was
complete.
The reaction was monitored by means of HPLC analysis. After reaction was
complete, the batch was purified directly by means of RP-HPLC. The fractions
were concentrated in the cold and in the dark, combined and concentrated
again.
The residue was taken up in water and desalted. A Waters Sep-Pak cartridge of
RP-18 (from 15 oD 2 g packing) was activated with 2 x 10 ml of acetonitrile
and
2 x 10 ml of water, the oligonucleotide was applied and allowed to sink in,
and the
reaction vessel was washed with 2 x 10 ml of water, rewashed with 3 x 10 ml of
water in order to remove the salt, and eluted with water:acetonitrile 1:1. The
product fractions were concentrated, combined and concentrated again.
The yields were determined by means of UV absorption spectrometry at 260 nm.
They reached 70-95% of theory.
HPLC Analysis:
Buffer A : 0.1 molar triethylammonium acetate buffer in water
Buffer B : 0.1 molar triethylammonium acetate buffer in water: acetonitrile
1:4
Gradient : starting from 10% B to 50% B in 40 minutes
Column material : 10 uM LiChrosphere ~ 100 RP-18 from Merck Darmstadt
GmbH; 250 x 4
Retention time of the product: 16.9 minutes
Mass spectrometry:
Sequence : 4' AGGCAInd(CH2CH2NHCOCH2-(His)6T 2'
calculated mass: MH2 2+ : 1626.9 g/mol
found mass MH22+ : 1626.0 g/mol
CA 02303086 2000-03-13
- 1$ -
The complementary sequence 4' Ind(CH2CH2NHCOCHz-(His)6TGCCT 2'
was prepared analogously:
$ calculated mass MH22~: 1436.2 g/mol
found mass MH2z~ : 1436.4 g/mol
Peptide libraries for the formation of recognition regions on the p-RNA were
also
conjugated analogously.
It was possible to demonstrate in the UV solution experiment that the
interaction of
the histidine subunits with a substrate (nickel ions), by itself influences
the pairing
properties. A conjugate solution of in each case $ uM p-RNA, in IOmM Tris HCl
1$OmM ultrapure NaCI showed a Tm of 32°C in the UV pairing experiment,
which
1$ increased by 10° C to 42° after addition of 10 equivalents of
nickel ions per strand.
Thus the detection, i.e. the recognition of a substrate here very
advantageously
accompanies the addressing itself; this corresponds on the Garner matrix to
the
immobilization process.
Example 4
Direct electronic detection of an antibody/antigen recognition on the
addressable
recognition system.
2$ A simple matrix of two vapour-deposited gold electrodes was used as an
example
of an addressable recognition system (see Fig. 8).
A commercially obtainable thiol-reduced antibody unit (Rockland Immuno
chemicals, Pennsylvania, USA) was conjugated to an iodoacetylated p-RNA
sequence as described above.
The complementary p-RNA-unit 4'Ind--TAGGCAAT 2 ' was thiol-activated on the
amino linker by means of 100 equivalents of Traut's reagent in 1mM aqueous
EDTA and borax buffer pH 9.$, purified by reverse-phase HPL chromatography
3$ after 6 hours, and bonded overnight to one of the two gold electrodes which
had
been freshly cleaned by means of UV light.. Only this electrode binds the
antibody-p-RNA conjugate by pairing (see Fig. 9).
CA 02303086 2000-03-13
' -16-
The figure shows the impedance signal (without further wiring; spectrometer
Solarton Instruments 1260 interface; Solarton SI 1287) of the thio-reduced
antibody, which was bonded directly overnight to a freshly cleaned electrode
of the
type described, before and after an antibody-antigen complexation of the
immobilized antibody under the buffer conditions 1/15 moll Na2HP04, KH2PO4,
pH 7.4 and room temperature.
It was possible to check the recognition result in the selected case by means
of
fluorescent labels, as the commercially obtainable antigen (a human IgG-
F(ab')2
fraction of Rockland Immunochemicals) is fluorescein-labelled (see Fig. 10).