Note: Descriptions are shown in the official language in which they were submitted.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
MUTANTS OF YEAST Cdc24p, DEFECTIVE IN BINDING OF THE G-PROTEIN BETA SUBUNIT
The present invention relates to nucleotide sequences and protein sequences.
In particular, the
present invention relates to nucleotide sequences and protein sequences that
affect interactions
of cellular components.
According to Cerione and Zheng (The Dbl family of oncogenes Current Opinion In
Cell
Biology $, 216-222 ( 1996)), genetic screening and biochemical studies during
the past years
have led to the discovery of a certain family of cell growth regulatory
proteins and oncogene
1 o products for which the Dbl oncoprotein is the prototype. Another review on
Dbl is presented by
Machesky and Hall (1996 Trends In CeII Biology ~ pp 3-4-310).
Cerione and Zheng {ibicT) say that proto-Dbl is a 1 I S kDa cytoskeleton-
associated protein that is
found in tissues such as brain. ovary, testis and adrenal glands. Oncogenic
activation of proto-
I5 Dbl occurs as a result of an amino-terminal truncation of proto-Dbl which
leaves residues 498-
925 fused with the product of an as yet unidentified gene which is localised
on chromosome 3.
Cerione and Zheng also say that a region located between residues 498 and 674
of proto-Dbl -
which is retained by oncogenic Dbl - has significant similarities with the
Saccharomyces
2o cerevisiae cell division cycle molecule Cdc24p and the breakpoint cluster
gene product Bcr (see
also Hart et al 1991 Nature ~ 3I I-314; Miyamoto et al 1991 Biochem Biophys
Res Commun
1$1604-610; Ron et al 1991 New Biol ~ 372-379). This region - which is
referred to as being
the DH domain - was later shown to be responsible for the GEF (SDP-GTP
exchange )_actor -
otherwise known as a guanine nucleotide e:cchange factor) activity of the Dbl
oncoprotein and to
25 be critical for its transforming function (see also Hart et al J Biol Chem
~ 62-65).
Cerione and Zheng also report that since the initial identification of Dbl as
a GEF for Rho-type
GTP binding proteins, a number of oncogene products and growth regulatory
molecules have
been shown to contain a DH domain in tandem with another region designated PH
(i.e. a
3o pleckstrin homology domain which is found between residues 703-812 in of
proto-Dbl). Many
of these products and molecules, such as Bcr, Cdc24, Sos, Vav, ect-2, Ost,
Tim, Lbc, Lfc and
Dbc, form a family of GEFs which have been implicated in cell growth
regulation. Cerione and
CA 02305707 2000-04-07
WO 99/I8213 PCT/GB98/03033
2
Zheng provide details on each of these products and molecules. In addition,
these and other
products and molecules are discussed below.
Cerione and Zheng (ibid) end their Abstract by saying:
''Despite the increasing interest in the Dbl family of proteins, there is
still a good
deal to learn regarding the biochemical mechanisms that underlie their diverse
biological functions."
1o As mentioned above, it is known that proto-Dbl has significant similarities
with the S cerevisiae
cell division cycle molecule Cdc24p which is a GEF for the Rho-family GTPase
molecule
Cdc42p (see again Hart et al 1991 Nature ~ 311-314: Miyamoto et al 1991
Biochem Biophys
Res Commun ].$L 604-610; Ron et al 1991 New BioI ~ 372-379; Zheng et al 1994 J
Biol Chem
~Q 2369-2372). However, whilst it is known that the Rho-family GTPases and
their regulators
are essential for cytoskeletal reorganisation and transcriptional activation
in response to
extracellular signals~~2, little is known about what links these molecules to
membrane receptors.
For example, in the budding yeast S. cerevisiae, haploid cells respond to
mating pheromone
through a G-protein coupled receptor (Ste2p/Ste3p) via G~iy (Ste4p/Stel8p)
resulting in cell
cycle arrest, transcriptional activation, and polarised growth towards a
mating partner4~s.
2o Recently, the Rho-family GTPase Cdc42p and its exchange factor Cdc24p have
been implicated
in the mating processb'~ but their specific role is unknown.
However, in our studies (which are presented below) on S. cerevisiae we have
been able to
identify hitherto unrecognised regions that play a key role in the interaction
of cellular
components. This finding has broad implications - not only for the design of
anti-fungal drugs
(such as those that could be directed against the yeast Candida) but also in
the screening and
design of agents that can affect oncogenes such as Dbl, in particular proto-
Dbl.
Moreover, in our studies (which are presented below), we have identified novel
cdc24 alleles
3o which do not affect vegetative growth but drastically reduce the ability of
yeast cells to mate.
When exposed to mating pheromone these mutants arrest growth, activate
transcription, and
undergo characteristic morphological and actin cytoskeleton polarisation.
However, the mutants
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
3
are unable to orient towards a pheromone gradient and instead position their
mating projection
adjacent to their previous bud site. Strikingly, these mutants are
specifically defective in the
binding of Cdc24p to G~3y. This work demonstrates that the association of a
GEF and the (3y-
subunit of a hetero-trimeric G-protein (G(3y) links receptor-mediated
activation to oriented cell
growth.
The present invention also demonstrates that Farl, a cyclic dependent kinase
inhibitor (CDKI)
may also be implicated as being important for orientated cell growth.
to Thus. according to one broad aspect of the present invention there is
provided a GEF capable
of interacting with a G/i such that the interaction provides a connection
between G protein
coupled receptor activation and polarised cell growth.
According to another broad aspect of the present invention there is also
provided an agent
capable of affecting a GEF/G(3 interaction, which interaction provides a
connection between
G protein coupled receptor activation and polarised cell growth.
These and other aspects of the present invention are set out in the claims.
3o By way of example, in a broad aspect, the present invention provides a
nucleotide sequence
shown as SEQ LD. No. 1 or a derivative. fragment, variant or homologue
thereof, wherein the
expression product of the nucleotide sequence has the capability of not
substantially affecting
the interaction of G~3 with GEF or a homologue thereof that is usually capable
of being
associated therewith.
The term "expression product of the nucleotide sequence has the capability of
not substantially
affecting the interaction of G(3 with GEF or a homologue thereof that is
usually capable of
being associated therewith" means that if the expression product were to be
present within
GEF and the GEF were to be contacted with G~i then the expression product
would not
3o substantially affect the interaction of G(3 with GEF.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
4
Thus, alternatively expressed, the present invention covers a nucleotide
sequence shown as
SEQ LD. No. 1 or a derivative, fragment, variant or homologue thereof, wherein
the
expression product of the nucleotide sequence has the capability of not
substantially affecting
the interaction of G(3 with GEF or a homologue thereof that is usually capable
of being
associated therewith if the expression product were to be present within GEF
and the GEF
were to be contacted with G(3.
With this aspect of the present invention, the expression product need not
necessarily be
present within GEF and/or the GEF need not necessarily be contacted with G(3.
By way of
t o example, the expression product can be part of a truncated GEF and/or part
of a fused protein.
However, if the expression product were present within GEF, then preferably
the GEF is not
in its natural environment. By way of example, the GEF can be in an isolated
form - such as
in an assay device. Likewise, if the expression product were contacted with
G~3 then
preferably the G~i is not in its natural environment. By way of example, the
G(3 can be in an
l5 isolated form - such as in an assay device.
The present invention also covers a mutant of the nucleotide sequence shown as
SEQ LD. No. 1
or a derivative, fragment, variant or homologue thereof, wherein the
expression product of the
mutant nucleotide sequence has the capability of substantially affecting the
interaction of G(3
2o with GEF or a homologue thereof that is usually capable of being associated
therewith.
The term ''expression product of the mutant nucleotide sequence has the
capability of
substantially affecting the interaction of G~i with GEF or a homologue thereof
that is usually
capable of being associated therewith" means that if the expression product
were to be present
25 within a GEF like entity (such as GEF bearing that mutation) and that GEF
like entity were to
be contacted with G(3 then the expression product would substantially affect
the interaction of
G(3 with that GEF like entity.
Thus, alternatively expressed, the present invention also covers a mutant of
the nucleotide
30 sequence shown as SEQ LD. No. 1 or a derivative, fragment, variant or
homologue thereof,
wherein the expression product of the mutant nucleotide sequence has the
capability of
substantially affecting the interaction of G(3 with GEF or a homologue thereof
that is usually
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
capable of being associated therewith if the expression product were to be
present within GEF
and the GEF were to be contacted with G~i.
With this aspect of the present invention, the expression product need not
necessarily be
5 present within the GEF like entity and/or the GEF like entity need not
necessarily be
contacted with G(3. By way of example, the expression product can be part of a
truncated
GEF and/or part of a fused protein. The GEF like entity may be in an isolated
form - such as
in an assay device. Likewise, if the expression product were contacted with
G(3 then
preferably the G~i is not in its natural environment. By way of example, the
G(3 can be in an
to isolated form - such as in an assay device.
In one preferred aspect, the GEF is Cdc24p. Other suitable GEFs have been
mentioned
above.
Thus, the present invention also covers in a broad aspect a nucleotide
sequence shown as SEQ
LD. No. 1 or a derivative, fragment, variant or homologue thereof, wherein the
expression
product of the nucleotide sequence has the capability of not substantially
affecting the
interaction of G~i with Cdc2:Ip or a homologue thereof that is usually capable
of being
associated therewith.
The term ''expression product of the nucleotide sequence has the capability of
not substantially
affecting the interaction of G~3 with Cdc24p or a homologue thereof that is
usually capable of
being associated therewith" means that if the expression product were to be
present within
Cdc24p and the Cdc24p were to be contacted with G(3 then the expression
product would not
substantially affecting the interaction of G(3 with Cdc24p.
Thus, alternatively expressed, the present invention covers in a broad aspect
a nucleotide
sequence shown as SEQ LD. No. 1 or a derivative, fragment, variant or
homologue thereof,
wherein the expression product of the nucleotide sequence has the capability
of not
3o substantially affecting the interaction of G(3 with Cdc24p or a homologue
thereof that is usually
capable of being associated therewith if the expression product were to be
present within
Cdc24p and the Cdc24p were to be contacted with G(3.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
6
With this aspect of the present invention, the expression product need not
necessarily be
present within Cdc24p and/or the Cdc24p need not necessarily be contacted with
G(3. By way
of example, the expression product can be part of a truncated Cdc24p and/or
part of a fused
protein. However, if the expression product is present within Cdc24p, then
preferably the
Cdc24p is not in its natural environment. By way of example, the Cdc24p can be
in an
isolated form - such as in an assay device. Likewise, if the expression
product were contacted
with G(3 then preferably the G~i is not in its natural environment. By way of
example, the G(3
can be in an isolated form - such as in an assay device.
t o By way of further example, the present invention also covers a mutant ~af
the nucleotide
sequence shown as SEQ LD. No. 1 or a derivative, fragment, variant or
homologue thereof,
wherein the expression product of the mutant nucleotide sequence has the
capability of
substantially affecting the interaction of G~i with Cdc24p or a homologue
thereof that is usually
capable of being associated therewith.
t5
The term "expression product of the mutant nucleotide sequence has the
capability of
substantially affecting the interaction of G~ with Cdc24p or a homologue
thereof that is usually
capable of being associated therewith" means that if the expression product
were to be present
within a Cdc24p like entity (such as Cdc24p bearing that mutation) and that
Cdc24p like
2o entity were to be contacted with G~3 then the expression product would
substantially affect the
interaction of G(3 with that Cdc24p like entity.
With this aspect of the present invention, the expression product need not
necessarily be
present within the Cdc24p like entity and/or the Cdc24p like entity need not
necessarily be
25 contacted with G~i. By way of example, the expression product can be part
of a truncated
Cdc24p and/or part of a fused protein. The Cdc24p like entity may be in an
isolated form -
such as in an assay device. Likewise, if the expression product were contacted
with G(3 then
preferably the G(3 is not in its natural environment. By way of example, the
G~i can be in an
isolated form - such as in an assay device.
In a preferred aspect, the present invention covers the sequences of the
present invention in
isolated form - in other words the sequences are not in their natural
environment and when
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
7
they have been expressed by their natural coding sequences which are under the
control of
their natural expression regulatory elements (such as the natural promoter
etc.). By way of
example the sequences may be in an assay device.
s It is to be noted that the nucleotide sequence presented as SEQ ID No. 1 is
quite different to the
DH domain and the PH domain discussed by Cerione and Zheng (ibic~. It is also
to be noted
that the nucleotide sequence presented as SEQ ID No. 1 is in a region quite
different to the DH
domain and the PH domain.
i o One important aspect of the present invention is that we have found it is
possible to affect the
interaction of Cdc24p with a ~3 subunit (such as Ste4p) or even a (3y subunit
(such as
Ste4p/StelBp) of a hetero-trimeric G-protein (hereinafter collectively
referred to as ''G(3"). If the
interaction is detrimentally affected (such as lost) then this may in turn
prevent (or at least
reduce) signalling (possibly GEF activity) being passed to the the Rho-family
GTPase
15 (Cdc42p). Hence, the present invention also covers the use of any one or
more of the
aforementioned aspects of the present invention to have an effect on a signal
being passed to
the Rho-family GTPases.
The term "derivative, fragment, variant or homologue" in relation to the
nucleotide Sequence
2o ID No. 1 of the present invention includes any substitution of modification
of, replacement of,
deletion of or addition of one (or more) nucleic acid from or to the sequence
providing the
resultant nucleotide sequence or the expression product thereof has the
capability of not
substantially affecting the interaction of G~i with Cdc24p or a homologue
thereof that is usually
capable of being associated with the Cdc24p or the homologue thereof. In
particular, the term
25 "homologue" covers homology with respect to function. With respect to
sequence homology
(i.e. similarity), preferably there is at least 75%, more preferably at least
85%, more preferably at
least 90% homology to the sequence shown as SEQ ID No.l in the attached
sequence listings.
More preferably there is at least 95%, such as at least 98%, homology to the
sequence shown as
SEQ ID No. 1 in the attached sequence listings.
The term "derivative, fragment, variant or homologue" in relation to the
protein Sequence ID
No. 2 of the present invention includes any substitution of, modification of,
replacement of,
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
8
deletion of or addition of one (or more) amino acid from or to the sequence
providing the
resultant amino acid sequence has the capability of not substantially
affecting the interaction of
G~3 with Cdc24p or a homologue thereof that is'usually capable of being
associated with the
Cdc24p or the homologue thereof. In particular, the term "homologue" covers
homology with
s respect to function. With respect to sequence homology (i.e. similarity),
preferably there is at
least 75%, more preferably at least 8~%, more preferably at least 90% homology
to the sequence
shown as SEQ ID No.2 in the attached sequence listings. More preferably there
is at least 95%,
such as at least 98%, homology to the sequence shown as SEQ ID No. 2 in the
attached
sequence listings.
l0
An example of a fragment of the expression product of SEQ ID No. 1 that has
the capability of
not substantially affecting the interaction of G(3 with Cdc24p or a homologue
thereof that is
usually capable of being associated with the Cdc24p or the homologue thereof
is the amino
acid sequence presented as SEQ ID No. 15 or SEQ ID No. 16. The present
invention also
15 covers nucleotide sequences coding for such sequences.
With respect to the mutated sequences then, in a preferred aspect, the mutated
sequence
comprises one or more mutations in the region presented as SEQ ID No. 1 ~ or
SEQ ID No. 16.
2o An example of a fra~ment of the expression product of a mutant SEQ ID No. 1
that has the
capaloilit~~ of substantially affecting the interaction of G(3 with Cdc24p or
a homologue thereof
that is usually capable of being associated with the Cdc24p or the homologue
thereof is the
amino acid sequence presented as SEQ ID No. 17 or SEQ ID No. 18 or SEQ ID No.
19. The
present invention also covers nucleotide sequences coding for such sequences.
In particular, the term "homology" as used herein may be equated with the term
"identity".
Relative sequence homology (i.e. sequence identity) can be determined by
commercially
available computer programs that can calculate % homology between two or more
sequences.
Typical examples of such computer programs are BLAST and CLUSTAL.
Sequence homology (or identity) may moreover be determined using any suitable
homology
algorithm, using for example default parameters. Advantageously, the BLAST
algorithm is
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
9
employed, with parameters set to default values. The BLAST algorithm is
described in
detail at http://www.ncbi.nih.govlBLAST/biast help.html, which is incorporated
herein by
reference. The search parameters are defined as follows, and are
advantageously set to the
defined default parameters.
Advantageously, "substantial homology" when assessed by BLAST equates to
sequences
which match with an EXPECT value of at least about 7, preferably at least
about 9 and
most preferably 10 or more. The default threshold for EXPECT in BLAST
searching is
usually 10.
~o
BLAST (Basic Local Alignment Search Tool) is the heuristic search algorithm
employed by
the programs blastp, blastn, blastx, tblastn, and tblastx; these programs
ascribe significance
to their findings using the statistical methods of Karlin and Altschul (see
http://www.ncbi.nih.gov/BLAST/blast help.html) with a few enhancements. The
BLAST
~5 programs were tailored for sequence similarity searching, for example to
identify
homologues to a query sequence. The programs are not generally useful for
motif-style
searching. For a discussion of basic issues in similarity searching of
sequence databases,
see Altschul et al (1994) Nature Genetics 6:119-129.
3o The five BLAST programs available at http://www.ncbi.nlm.nih.gov perform
the following
tasks:
blastp compares an amino acid query sequence against a protein sequence
database;
25 blastn compares a nucleotide query sequence against a nucleotide sequence
database;
blastx compares the six-frame conceptual translation products of a nucleotide
query
sequence (both strands) against a protein sequence database;
3o tblastn compares a protein query sequence against a nucleotide sequence
database
dynamically translated in all six reading frames (both strands).
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
tblastx compares the six-frame translations of a nucleotide query sequence
against the six-
frame translations of a nucleotide sequence database.
BLAST uses the following search parameters:
s
HISTOGRAM Display a histogram of scores for each search; default is yes. (See
parameter
H in the BLAST Manual).
DESCRIPTIOnucleotide sequence Restricts the number of short descriptions of
matching
to sequences reported to the number specified; default limit is 100
descriptions. (See
parameter V in the manual page). See also EXPECT and CUTOFF.
ALIGNMENTS Restricts database sequences to the number specified for which high-
scoring segment pairs (HSPs) are reported; the default limit is 50. If more
database
I S sequences than this happen to satisfy the statistical significance
threshold for reporting (see
EXPECT and CUTOFF below), only the matches ascribed the greatest statistical
significance are reported. (See parameter B in the BLAST Manual).
EXPECT The statistical significance threshold for reporting matches against
database
?o sequences; the default value is 10, such that 10 matches are expected to be
found merely by
chance, according to the stochastic model of Karlin and Altschul (1990). If
the statistical
significance ascribed to a match is greater than the EXPECT threshold, the
match will not
be reported. Lower EXPECT thresholds are more stringent, leading to fewer
chance
matches being reported. Fractional values are acceptable. (See parameter E in
the BLAST
25 Manual).
CUTOFF Cutoff score for reporting high-scoring segment pairs. The default
value is
calculated from the EXPECT value (see above). HSPs are reported for a database
sequence
only if the statistical significance ascribed to them is at least as high as
would be ascribed to
3o a lone HSP having a score equal to the CUTOFF value. Higher CUTOFF values
are more
stringent, leading to fewer chance matches being reported. (See parameter S in
the BLAST
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
11
Manual). Typically, significance thresholds can be more intuitively managed
using
EXPECT.
MATRIX Specify an alternate scoring matrix for BLASTP, BLASTX, TBLASTN and
s TBLASTX. The default matrix is BLOSUM62 {Henikoff & Henikoff, 1992). The
valid
alternative choices include: PAM40, PAM120, PAM250 and IDENTITY. No alternate
scoring matrices are available for BLASTN; specifying the MATRIX directive in
BLASTN
requests returns an error response.
t o STRAND Restrict a TBLASTN search to just the top or bottom strand of the
database
sequences; or restrict a BLASTN, BLASTX or TBLASTX search to just reading
frames on
the top or bottom strand of the query sequence.
FILTER Mask off segments of the query sequence that have low compositional
15 complexity, as determined by the SEG program of Wootton & Federhen (1993)
Computers
and Chemistry 17:149-163, or segments consisting of short-periodicity internal
repeats, as
determined by the XNU program of Claverie & States (1993) Computers and
Chemistry
17:191-201, or, for BLASTN, by the DUST program of Tatusov and Lipman (see
http://www.ncbi.nlm.nih.gov). Filtering can eliminate statistically
significant but
2o biologically uninteresting reports from the blast output (e.g., hits
against common acidic-,
basic- or proline-rich regions), leaving the more biologically interesting
regions of the
query sequence available for specific matching against database sequences.
Low complexity sequence found by a filter program is substituted using the
letter "N" in
25 nucleotide sequence (e.g., "NNNNNNNNNNNNN") and the letter "X" in protein
sequences (e.g., "XXXXXXx:XX").
Filtering is only applied to the query sequence (or its translation products),
not to database
sequences. Default filtering is DUST for BLASTN, SEG for other programs.
It is not unusual for nothing at all to be masked by SEG, XNU, or both, when
applied to
sequences in SWISS-PROT, so filtering should not be expected to always yield
an effect.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
12
Furthermore, in some cases, sequences are masked in their entirety, indicating
that the
statistical significance of any matches reported against the unfiltered query
sequence should
be suspect.
s NCBI-gi Causes NCBI gi identifiers to be shown in the output, in addition to
the accession
and/or locus name.
Most preferably, sequence comparisons are conducted using the simple BLAST
search
algorithm provided at http://www.ncbi.nlm.nih.gov/BLAST.
Other computer program methods to determine identify and similarity between
the two
sequences include but are not limited to the GCG program package (Devereux et
al 1984
Nucleic Acids Research 12: 387and FASTA (Atschul et al 1990 J Molec Biol 403-
410).
t 5 The term "variant" also encompasses sequences that are complementary to
sequences that are
capable of hydridising to the nucleotide sequences presented herein.
Preferably, the term "variant" encompasses sequences that are complementary to
sequences
that are capable of hydridising under stringent conditions (eg. 65°C
and O.IxSSC {lxSSC =
20 0.15 M NaCI, 0.015 Na3 citrate pH 7.0}) to the nucleotide sequences
presented herein.
The present invention also relates to nucleotide sequences that can hybridise
to the nucleotide
sequences of the present invention (including complementary sequences of those
presented
herein).
The present invention also relates to nucleotide sequences that are
complementary to
sequences that can hybridise to the nucleotide sequences of the present
invention (including
complementary sequences of those presented herein).
3o The term "hybridization" as used herein shall include "the process by which
a strand of
nucleic acid joins with a complementary strand through base pairing" (Coombs J
(1994)
Dictionary of Biotechnology, Stockton Press, New York NY) as well as the
process of
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
13
amplification as carried out in polymerase chain reaction technologies as
described in
Dieffenbach CW and GS Dveksler (1995, PCR Primer, a Laboratory Manual, Cold
Spring
Harbor Press, Plainview NY).
Also included within the scope of the present invention are polynucleotide
sequences that
are capable of hybridizing to the nucleotide sequence of the present invention
or other
nucleotide sequences coding for the protein sequence of the present invention
under
conditions of intermediate to maximal stringency. Hybridization conditions are
based on
the melting temperature (Tm) of the nucleic acid binding complex, as taught in
Berger and
t o Kimmel ( 1987, Guide to Molecular Cloning Techniques, Methods in
Enzymology, Vol
152, Academic Press, San Diego CA), and confer a defined "stringency" as
explained
below.
Maximum stringency typically occurs at about Tm-5°C (5°C below
the Tm of the probe);
high stringency at about 5°C to 10°C below Tm; intermediate
stringency at about 10°C to
20°C below Tm; and low stringency at about 20°C to 25°C
below Tm. As will be
understood by those of skill in the art, a maximum stringency hybridization
can be used to
identify or detect identical polynucleotide sequences while an intermediate
(or low)
stringency hybridization can be used to identify or detect similar or related
polynucleotide
2o sequences.
In a preferred aspect, the present invention covers nucleotide sequences that
can hybridise to
the nucleotide sequence of the present invention under stringent conditions
(e.g. 65°C and
0. lxSSC).
Examples of homologues of Cdc24p include but are not limited to any one or
more of the
homologues listed above or below, such as proto-Dbl, Bcr, Sos, Vav, ect-2,
Ost, Tim, Lbc, Lfc
and Dbc.
3o The term ''mutant" in relation to the nucleotide sequence of the present
invention means a
variant of SEQ ID No. 1 but wherein that variant or the expression product
thereof has the
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
14
capability of substantially affecting the interaction of G~3 with Cdc24p or a
homologue thereof
that is usually capable of being associated with the Cdc24p or the homologue
thereof.
Preferred mutants of the nucleotide sequence of the present invention include
any one or more
of the nucleotide sequences presented as SEQ ID No. 3, SEQ ID No. 5 or SEQ ID
No. 7.
The term "mutant" in relation to the protein sequence of the present invention
means a variant
of SEQ ID No. 2 but wherein that variant has the capability of substantially
affecting the
interaction of G(3 with Cdc24p or a homologue thereof that is usually capable
of being
to associated with the Cdc24p or the homologue thereof.
Preferred mutants of the protein sequence of the present invention include any
one or more of
the protein sequences presented as SEQ ID No. 4, SEQ ID No. 6 or SEQ ID No. 8.
The term "growth behaviour" includes growth per se (but not vegetative growth
of yeast),
growth control and growth orientation of cells. In some aspects, it includes
at least growth
orientation of cells. The term may also include the mating pattern (e.g.
mating per se or
mating behaviour) of cells.
For a preferred aspect of the present invention, any one or more of the
nucleotide sequence of
the present invention or the expression product thereof, or the mutant
nucleotide sequence of
the present invention or the expression product thereof, or the protein of the
present invention,
or the mutant protein of the present invention may be within a transgenic
organism or cell
(such as being an integral part thereof) - that is an organism or cell that is
not a naturally
occurring organism or cell and wherein the organism or cell has been prepared
by use of
recombinant DNA techniques. The transgenic cell may be part of or contained
within tissue.
Preferably, the transgenic organism or cell is a yeast, an animal (such as a
mammal) or an
animal cell (such as a mammalian cell).
In preferred embodiments, the transgenic organism is a transgenic yeast or a
transgenic
mouse.
CA 02305707 2000-04-07
WO 99/I8213 PCTlGB98/03033
Transgenic yeast may be prepared by appropriately adapting the teachings of
Ito et al Journal
of Bacteriology ~ 163-168; Rose et al 1991 Methods in yeast genetics: a
laboratory course
manual Cold Spring Harbor, N.Y.: Cold Spring Harbor Press) .
5
Transgenic mammals or mammalian cells may be prepared by appropriately
adapting the
teachings of Ausubel et al 1992 Short Protocols in Molecular Biology 2nd Ed.
New York:
John Wiley and Sons) .
to The transgenic organism or transgenic cell of the present invention
therefore provides a
simple assay system that can be used to determine whether one or more agents
(e.g.
compounds or compositions) have one or more beneficial properties. By way of
example, the
assay system of the present invention may utilise a mating phenotype and/or
the assay system
may be a two-hybrid interaction assay.
By way of example, if the transgenic organism is a transgenic yeast which
comprises the
nucleotide sequence presented as SEQ ID No. 1 or the expression product
thereof (namely the
protein sequence presented as SEQ ID No. 2) then the yeast could be used to
screen for agents
that bind to this nucleotide sequence or the expression product thereof and in
doing so affect
the growth behaviour of the yeast. If an agent produces such a detrimental
effect (such as
drastically reducing the ability of the yeast to mate), then that agent may
also affect the
interaction of Gp with Cdc24p or another Cdc24p entity that is usually capable
of being
associated therewith. This aspect of the present invention could allow workers
to screen for
anti-fungal agents, such as agents that could be used to treat or combat
Candida.
By way of further example, if the transgenic organism is a transgenic yeast
which comprises
the nucleotide sequence presented as SEQ ID No. 1 or the expression product
thereof then the
yeast could be used to screen for agents that bind to this nucleotide sequence
or expression
product thereof and in doing so affect the growth behaviour of the yeast. If
an agent produces
3o a detrimental affect (such as drastically reducing the ability of the yeast
to mate), then that
agent is likely to detrimentally affect the interaction of G(3 with a
homologue of Cdc24p with
which it is usually capable of being associated. This could allow workers to
screen for
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
16
compounds or compositions that could for example influence the in vivo
expression or
behaviour of effect of proto-oncogenes and the like - such as proto-Dbl.
By way of further example, if the transgenic organism is a transgenic yeast
which comprises a
mutant of the nucleotide sequence in accordance with the present invention
then the yeast
could be used to screen for agents that affect the growth behaviour of the
yeast. If an agent
produces a marked affect - such as restoration to a normal growth behaviour or
a further
detrimental growth behaviour - then workers could screen for compounds or
compositions
that could for example influence the in vivo expression or behaviour or effect
or activity of a
to Cdc24 homologue, such as, but not limited to proto-oncogenes such as Dbl
and/or Vav.
By way of further example, if the transgenic organism is a transgenic yeast
which comprises a
homologue (e.g. Dbl) of the nucleotide sequence shown as SEQ ID No. 1 or an
expression
product thereof then workers could see if that homologue or the expression
product thereof
t5 had an effect on the growth behaviour of yeast, and thus also to see if it
had an effect on the
interaction of G~3 with a homologue of Cdc24p. In addition, workers could use
those
transgenic yeast to screen for agents that modified the effect - such as
enhance the growth
behaviour or detrimentally affect the growth behaviour. In this aspect, agents
that affect the
growth behaviour may also influence the activity of oncogenes (or even parts
thereof) and
2o therefore have potential as therapeutic agents.
The assays of the present invention may also be used to screen for agents that
affect the
interaction of Cdc24p or a Cdc24p homologue with G~i to determine whether that
effect has a
downstream effect on a Rho-family GTPase.
For example, with the present invention - such as by use of the assays of the
present invention -
it is possible to devise and/or to screen for peptide inhibitors which block
GEF/G~i interaction.
In this regard, peptides and peptidyl derivatives based regions encompassing
mutants may be
used to block and/or antagonise GEF (such as the proto-oncogenes Dbl or Vav)
G(3 interaction.
3o Derivatives of these peptides (including peptide mimics) which bind with
higher affinity may
also be used. The perturbation of these interactions may be of therapeutic
value for example in
treatment of cancers.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
17
In addition, by use of the present invention it is possible to devise simple
yeast based assay
systems (utilising mating function and interaction reporters). These assay
systems will be
extremely useful for high through-put screening to identify molecules
perturbing the GEF/G~i
interaction.
In addition, it is possible to devise and/or screen for agents that can
modulate (e.g. interact),
preferably selectively modulate (interact), with and affect Cdc24p/G~i
interactions. Hence, it
would be possible to devise and/or to screen for anti-fungal agents directed
at invasive and/or
l0 pathogenic yeasts such as. but not limited to Candida albicans and/or
Cryptococcus
neoformans.
If the assay of the present invention utilises a transgenic organism according
to the present
invention then transgenic organism may comprise nucleotide sequences etc. that
are additional
to the nucleotide sequences of the present invention in order to maintain the
viability of the
transgenic organism.
In the assays of the present invention, the agent can be any suitable
compound, compostion as
well as being (or even including) a nucleotide sequence of interest or the
expression product
2o thereof. Hence. if any one of the nucleotide sequences of the present
invention are contained
within a transgenic organism - such as a transgenic yeast - then that
transgenic organism may
also contain that nucleotide sequence of interest. If the agent is a
nucleotide sequence, then the
agent may be, for example, nucleotide sequences from organisms (e.g. higher
organisms - such
as eukaryotes) that restore or increase the growth behaviour. Agents which
affect the growth
behaviour may also influence the activity of homologous oncogenes and may
therefore be
potential therapeutic agents.
CA 02305707 2000-04-07
WO 99/18213 PC'T/GB98/03033
18
The following samples were deposited in accordance with the Budapest Treaty at
the recognised
depositary of The National Collections of Industrial and Marine Bacteria
Limited (NCIMB) at
23 St. Machar Drive, Aberdeen, Scotland, United Kingdom, AB2 1 RY on 3 October
1997:
E.coli CMK603 PRS414CDC24 (WT) - Deposit Number NCIMB 40898
E.coli CMK603 PRS414CDC24 (M1) - Deposit Number NCIMB 40899
E. coli CMK603 PRS414CDC24 (M2) - Deposit Number NCIMB 40900
E. coli CMK603 PRS414CDC24 (M3) - Deposit Number NCIMB 40901
Deposit NCIMB 40898 is in respect of cdc2=l (wt); Deposit NCIMB 40899 is in
respect of
cdc2=l-ml; Deposit NCIMB 40900 is in respect of cdc24-m2; Deposit NCIMB 40901
is in
respect of cdc2:l-m3.
In accordance with a preferred aspect of the present invention, the nucleotide
sequence is
obtainable from, or the protein is expressable from the nucleotide sequence
contained within, the
respective deposit. By way of example, the respective nucleotide sequence may
be isolated from
2o the respective deposit by use of appropriate restriction enzymes or by use
of PCR techniques.
The present invention will now be described only by way of example, in which
reference is
made to the following Figures:
Figure 1 which presents some photographs and a graph;
Figure 2 which presents some images and graphs;
Figure 3 which presents some photographs, a sequence, and a pictorial
representation of Cdc24
3o and DBD Cdc24; and
Figure 4 which presents a pictorial representation of a cellular interaction.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
19
The Figures are discussed in more detail later on.
Materials and Methods
General techniques
Strains were constructed using standard techniques2~. All constructs were
verified by DNA dye
terminator cycle sequencing (ABI377 sequencer).
Strains
pRS414CDC24 contains the CDC24 ORF including 258 by upstream of ATG.
Oligonucleotide-directed mutagenesis was used to introduce silent base changes
that resulted in
the following ten new restriction sites in CDC24: NheI (bp -12), KasI (bp
283), AatII (bp 681),
PstI (bp 1207), RsrII (bp 1369), BstEII (bp 1426), XhoI (bp 1758), NIIuI (bp
1963), SaII (bp
2061 ), BamHi {bp 2485). RAY410 (MATa, leu2, CDC24::LEU2, ade2, lys2, his3,
trp 1, ura3,
pEG(KT)CDC24) was derived from the diploid YOC380z2 which was transformed with
2o pEG{KT)CDC242' and sporulated. RAY9~0 is isogenic to RAY410 but has
pRS416Ga1His6CDC24 as a rescuing plasmid. RAY928 (MATa, leu2-3, 112, ura3-52,
his3-
D200, trill-D901, lys2-801, suc2-D9, CDC24::HISS pEG[KT]CDC24) and RAY931
(same as
RAY928 but ~LlATa, ade2, LYS2) were made by transformation of SEY6210 and 6211
with
pEG(KT)CDC24 followed by PCR-based gene disruption of CDC24. The CDC24 ORF was
replaced with S. pombe HIS.i2'~, flanked by LoxP sites. Replacement of CDC24
in SEY6211
with a PCR-generated integration cassette consisting of TRPI fused to 343 by
of CDC24
promoter followed by 1704 by of CDC24 or cdc24-ml ORF was used to construct
RAY1034 or
RAY1035, respectively.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
IDENTIFICATION OF cdc24 MUTANTS WITH SPECIFIC DEFECTS IN CELL
MATING:
s A) Construction of a library of cdc24 random mutants
Error-prone PCR was used to generate a library of cdc2=1 mutants in a plasmid
vector suitable for
phenotypic screening in yeast.
1 ) Plasmid:
pRS414 CDC2~ with upstream region and new restriction sites (referred to as
pRS414CDC24).
2) Mutagenic PCRs:
is
Conditions from Fromant, M., Blanquet, S. & Plateau, P. Direct random
mutagenesis of gene-
sized DNA fragments using polymerase chain-reaction. Analytical Biochemistry
224, 347-353
(1995).
2o Different PCR-conditions were tested and the error-rate was determined by
DNA sequencing.
The following conditions were used for constructing the library used in the
screen.
Composition of PCR-reactions (25 pl each):
DNA pRS414CDC24 600pM
dATP 0.23 mM
dCTP 0.20 mM
dTTP 2.9 mM
3o dGTP 0.42 mM
Buffer PCR Buffer supplied with Taq-polymerase
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
21
MgCl2 4 mM
MnCl2 O.S mM
Taq (Ampli-Taq) 2 U per reaction
Primer: ~ 0.5 mM
PCR-cycles:
1 o step 1 94 C S min
step 2 91 C 1 min
step 3 S 1 C 1 min
step 4 72 C 3 min
step S 72 C 5 min
step 6 4 C pause
16 cycles (steps 2-4)
3) Library construction:
The PCR products were digested with AatII and NheI (680 by corresponding to
amino acid 1 -
227) were mutagenised and the resulting fragment ligated into pRS414CDC24 (cut
with the
same enzymes). Ligations were transformed into E. coli by electroporation and
> 50,000
transformants pooled for plasmid isolation.
B) Phenotypic screening for cell-mating specific cdc24 alleles
Rationale:
To identify mutant cdc2~ alleles which cause defects in cell mating but allow
vegetative growth.
Yeast strain RAY9S0, in which expression of CDC24 is repressed in glucose
medium, was used.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
22
1 ) Library plasmids were transformed into RAY950 and transformants selected
on SC -trp
plates which contained 2% glucose. As RAY950 does not grow on glucose plates
this procedure
eliminated all non-functional cdc24 mutants.
2) Transformants were replica-plated onto a lawn of WT (screen 1 ) or
OfuslOfus2 (screen 2)
tester cells, incubated at 30°C for 3 hrs and replica-plated onto
plates selecting for diploids or
RAY950 derived haploids. Mating defective mutants were identified by comparing
the pattern
of colonies on the two sets of plates and candidate mutants were picked from
the original
transformation plates for retesting.
1o
3) Plasmids from mutants were isolated by transformation into E. coli.
Isolated plasmids were
retransformed into RAY950, RAY928 and RAY931 for independent confirmation of
phenotype
and retested for defects in cell mating.
4) Mutations of confirmed mutants were identified by DNA sequencing. Multiple
mutations
were separated by subcloning and the mutation responsible for the phenotype
identified by
mating tests in RAY950.
5) A total of ~ x,000 yeast transformants were tested in each screen.
- Screen 1 identified two mutants {cdc24-ml, cdc24-m2).
- Screen 2 identified one mutant (cdc24-m3).
Phenotypic analyses
Quantitative matings~°, matings in the presence of saturating
pheromonel3, halo-assays26 using
sstl::URA3 strains, and FusllacZ measurements with pSG23111 were carried out
as described.
Halo assays showed MATa and MATa cdc24-ml cells secreted a-factor and a-
factor,
respectively. Actin was visualised with rhodamine phalloidin2~ on a Biorad-MRC-
600 confocal
microscope and pictures are projections of 4-6 0.5 mm z-series steps. For a-
factor treatment,
cells were incubated with 5 mM a-factor for 2 hr. RAY1034 and RAY1035 cells
were used to
determine bud scar positions on zygotes~4 visualised with Calcoflour28.
Similar results were
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
23
observed with the position of the bud scar on shmoos. Direct measurement of
cell orientation in
a pheromone gradient was carried out essentially as describedl2. A pheromone
gradient was
generated using a micropipet filled with 80 mM a-factor injected at 105 kPa
into lml of YEPD
media layered on top of cells embedded in 2% Low Melting Point (LMP) agarose.
Cells shape
was recorded by video microscopy on a heated stage at 35° for 4 - 7 hr
and data analysis was
from traced cell outlines~4. Mating projections were formed at the same
pheromone
concentrations and budding, that is non-responding cells were seen at similar
distances from the
micropipet in both strains.
t o Two-Hybrid methods
STE=l, BEM/ (372 - 5~1 aa), CDC-t2[C178S], and CDC2-l Icdc2=l-ml (1-288, 1-
160, and 170-
245 aa) were cloned by PCR into pGAD424 (AD, GAL; activation domain) or pAS 1
(DBD,
GAL:/ DNA binding domain). Plasmids were transformed into HF7c. For
determination of
t5 STE18 requirement, PCR-based gene disruption was carned out in PJ69-4A
(MATa, trill-901,
leu2-3,112, ura3-52, his3-200, gal4D, ga180D, GAL2-ADE2, LYS2::GAL1-HIS3,
met2::GAL7-
lacZ)29, replacing the entire STEl8 ORF with K. Lactis URA33°. For all
two-hybrid
experiments, equal amounts of transformants were spotted on SC-leu-trp and SC-
leu-trp-his
plates, identical results were obtained with at least four transformants, and
for Dstel8 two
2o independent deletion strains. All strains for two-hybrid analyses expressed
similar amounts of
AD- and DBD- fusion proteins of the expected sizes, as determined by SDS-PAGE
and
immuno-blotting. None of the DBD fizsions showed any self activation using two
different non-
interacting AD fusions.
25 In vitro binding studies
A fragment of CDC24 (1-472 aa) in pGEX-2T (Pharmacia) and His6Ste4p (pTrcSte4)
were
expressed in E. coli. Cells were resuspended in buffer A (PBS, 0.1% TX-100,
Phenyl Methyl
Sulfonyl Fluoride (PMSF), leupeptin, chymostatin, pepstatin, aprotinin) and
lysed by snap
3o freezing in liquid nitrogen followed by sonication. Insoluble material was
removed by
centrifugation ( 10,000g). Mixed supernatants (denoted cell extracts)
containing His6Ste4 and
GSTCdc24 fusions were incubated with GSH-agarose (Sigma Chemical Co.) at
4° for 1 hr.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
24
Resin was washed 3 times with buffer A. Resin samples (referred to as eluates)
and extracts
were analyzed by SDS-PAGE, immuno-blotting probed with Omni-probe anti-sera
(Santa
Cruz), and visualised with enhanced chemiluminescence (Amersham). GSTCdc24p (1-
127 aa),
similar to GST, did not bind His6Ste4p. Similar results were observed in 5
independent
experiments.
C) Ste4p mutants
Ste4p is the ~i-subunit of the heterodimeric G protein that can usually
associate with Cdc24p
exemplified by nucleotide SEQ ID No. 9 and amino acid SEQ ID No. 10. A
mutation in
STE4 exemplified by nucleotide SEQ ID No. 11 and SEQ ID No. 13 and amino acid
SEQ ID
No. 12 and SEQ ID No. 14 prevented the interaction of the mutant G protein (3
subuttit with
Cdc24p. Thus, it is possible to devise assays based on this mutation to screen
for agents
capable of modifying the non-interactive behaviour of the mutant G protein (3
subunit with
t5 Cdc24p. In addition, the assay could be used to study Cdc24p homologues or
even Cdc24p
derivatives or homologues to see if those derivatives or homologues affect the
non-interactive
behaviour of the mutant G protein ~i subunit.
The Ste4p mutants are also aspects of the present invention.
In this regard, the present invention also covers an STE4 mutant.
The present invention also covers a mutation of the ~3-subunit of the
heterodimeric G protein
that can usually associate with GEF (preferably Cdc24p) that is capable of
preventing the
interaction of the mutant G protein subunit with GEF (preferably Cdc24p).
Hence, a further aspect of the present invention is a mutation in STE4 - i.e.
on the (3-subunit
of the heterodimeric G protein that can usually associate with Cdc24p. This
mutation
prevents the interaction of the mutant G protein subunit with Cdc24p. Thus,
likewise, it is
3o possible to devise similar assays based on this mutation to screen for
agents that modify the
non-interactive behaviour of the mutant G protein with Cdc24p. In addition,
the assay could
be used to study Cdc24p homologues or even Cdc24p derivatives or variants to
see if those
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
derivatives or variants affect the non-interactive behaviour of the mutant G
protein. The
sequences associated with this aspect of the present invention are shown as
SEQ ID No. 9 etc.
The present invention also covers variants or derivatives of such sequences -
wherein the
variants or derivatives of the wildtype sequences do not substantially affect
Cdc24 interaction;
5 and wherein the variants or derivatives of the mutant sequences do
substantially affect Cdc24
interaction.
D) Assay system to monitor the effects of two human oncogenic agents on an S.
cerevisiae yeast mutant with a mating defect.
l0
An assay system was devised to establish whether two different proto-oncogenes
could
complement the S. cerevisiae yeast phenotype (cdc24-ml ) mating defect as
described above
and in Nern and Arkowitz (Nature ( 1998) 391: 195-198). The two oncogenic
agents used
were the human proto-oncogene, proto-Dbl and the mouse C4 protein which is
almost
1s identical to the human sequence, C5 Vav, and which is referred to hereafter
as Vav. The S.
cerevisiae cell division cycle molecule, Cdc24p, which is a protein with
similiarities to proto-
Dbl was used as a positive control in addition to the Cdc24p of the related
yeast K. lactis.
Transgenic yeast organisms which co-expressed the nucleotide sequence (SEQ ID
No. 3) for
3o the cdc2:l-ml mating defect and the nucleotide sequence of interest (NOI)
encoding either
proto-Dbl, Vav or two related Cdc24p's were used.
The expression levels of the proto-oncogene, proto-Dbl, in S. cerevisiae were
relatively low
compared with the expression levels of the Cde24p protein from either S.
cerevisiae or K
25 lactis.
Qualitatively, both proto-Dbl and K. lactis Cdc24 proteins partially
complemented the mating
defect in the cdc2~t-ml mutant. This result is in contrast to that obtained
with the oncogenic
form of Dbl alone which, although expressed, did not complement the cdc24-ml
mating
defect. The Vav protein, did not display any effect on the mating defect. This
lack of effect
may be due to either insufficient expression of the Vav protein or to the fact
that Vav function
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
26
requires a phosphorylation of the Lck kinase which must be co-expressed with
the Vav
protein before an effect can be observed.
E) Assays to determine FART interaction with Cdc24p and G(3
Studies have shown that FARI may play an important role both for pheromone
mediated
growth arrest and growth orientation during mating (Valtz, N., Peter, M. &
Herskowitz, I. J.
Cell Biol. 131, 863-73 ( 1995); Chang, F. & Herskowitz, I. Cell 63, 999-1011 (
1990); Peter,
M., Gartner, A., Horecka, J., Ammerer, G. & Herskowitz, I. Cell 73, 747-60 (
1993)). The
to orientation function, which is specifically disrupted in a farl-H7 mutant,
is required for the
Cdc24 G~3 interaction suggesting that Farl might interact with Cdc24. Two-
hybrid analyses
show that indeed Farl interacts with Cdc24.
While the Cdc24 G~3 interaction requires the presence of FARI, the Farl Cdc24
interaction is
independent of G~i, suggesting that Farl might bind Cdc24 directly whereas
Cdc24 G(3 are
part of a complex which include Farl. Farl also interacts by two-hybrid assays
with G(3,
consistent with the notion that Cdc24, Farl, and G(3 form a complex. In a
diploid two-hybrid
strain, in which a number of pheromone response genes are not expressed, we
are unable to
detect the Cdc24 G(i interaction. However, overexpression of Farl results in
an interaction
2o and further overexpression of Cry results in a maximal interaction,
indicating that a complex
comprised of Cdc24, G~3~r, and Farl forms even in diploid cells.
Although cdc2=t-m and farl-s mutants result in similar defects in growth
orientation during
mating, we set out to determine if these genes function in the same
orientation process.
Generation of a cdc24-ml mutation in a Marl strain did not result in a
substantial decrease in
mating efficiency, suggesting these two genes function in the same process. In
contrast,
results from double mutants of cdc24-ml with ~,rpa2, ~.ste20, or ~beml suggest
that these
three genes do not function in the same orientation process as Cdc24 and Farl.
Cdc24 and
Farl were epitope tagged in order to determine whether these proteins interact
in yeast cells.
3o The chromosomal copy of Cdc24 was replaced with a 3xmyc tagged Cdc24 and
the
chromosomal copy of Farl was replaced with Farl protein A fusion. Both of
these fusion
proteins are fully functional. Isolation of Farl-protein A from yeast extracts
using IgG-
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
27
Sepharose co-precipitated 3xmyc-Cdc24. In contrast, the 3xmyc-Cdc24-ml mutant
was
defective in binding Farl in similar immunoprecipitation assays. These results
indicate that
Cdc24 and Farl bind one-another and this interaction may be essential for
growth orientation
during mating.
Farl binds Cdc24 and G[3
The binding relationships between Cdc24, Farl, and G~i were examined in vitro
using
proteins purified from bacteria and yeast. G(3y was purified from yeast cells
using a
to chromosomal copy of the gene which has HA epitope (Tyr-Pro-Tyr-Asp-Val-Pro-
Asp-Tyr-
Ala) fused to the amino-terminus and protein A fused to the carboxyl-terminus.
A Iobacco
etch virus (TEV) protease cleavage site (recognition site Glu-Asn-Leu-Tyr-Phe-
Gln-Gly with
cleavage occurring between Gln and Gly) was placed between G(iand the protein
A domain so
that material isolated from yeast using IgG-Sepharose can be specifically
eluted with
commercially available recombinant TEV protease. Maltose binding protein (MBP)
Farl
fusions have been expressed and purified from E. coli. Similarly, a
glutathione-S-transferase
(GST) Cdc24 fusion (residues 1 - 472) has been expressed and purified from E.
coli. MBP-
Farl binds GST-Cdc24 specifically. The removal of the 7~ carboxyl-terminal
residues of
Farl (H7) prevents Cdc24 binding. Furthermore GST alone is unable to bind MBP-
Farl.
These results show that Cdc24 can directly bind Farl in the absence of any
other yeast
proteins. Farl fragments containing either the amino-terminal Lim domain (a
domain
implicated in protein-protein interactions) or the carboxyl-terminus were
tested for their
ability to bind GST-Cdc24. Both fragments showed very little binding to GST-
Cdc24
indicating that although the Farl carboxyl-terminus is necessary, it is not
sufficient for Cdc24
binding. Using MBP-Farl we have been able to observe binding to G(3 purified
from yeast.
Binding of G~i is reduced using amino-terminal or carboxy-terminal MBP-Farl
fragments, yet
G~i binds FarlH7 as well as Farl.
3o In one preferred asepct of the present invention the assay also includes
the presence of Farl.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
28
Table 1 cdc24-ml is defective in cell mating
Strain Tester % Mating efficiency
1 o CDC24 MATa MATa WT 100 (21 )
cdc2-~-ml MATa lLIATa WT 0.5 (0.2)
CDC24 MATa MATa WT 100 (20)
cdc24-ml MATa MATa WT 3.8 (1.6)
CDC2-I MATa MATa Ofusl Ofus2 100{17)
cdcZ4-ml MATa MATa ~fusl ~fus2 <_ 0.02
CDC24 ~t~lATa CDC24 MATa 100( 18)
2o cdc24-ml MATa cdc24-ml ~LIATa <_ 0.0006
Mating efficiencies are the number of diploid cells divided by the total cells
with CDC24 WT
set to 100%. The values are means of 4 determinations with standard deviation
n. Absolute
mating e~ciency was 14-15% with MATa and MATa testers, 1.8% with ~fusl Ofus2
tester, and
3.4% with CDC24 tester.
Some of the results are also shown in the accompanying Figures. These Figures
are now
discussed in more detail.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
29
FIGURE. 1
cdc2=!-ml phenotypes. a, Actin cytoskeleton of cdc2;t-ml cells shows polarised
distribution.
Bar equals 5 mm. b, Pheromone-induced growth arrest is similar in cdc2=l-ml
with WT cells.
Sterile filter disks spotted with a-factor (1, 0.5, 0.2, 0.1, 0.05, and 0.012
mg) were placed onto
cells in agarose. c, MAP-kinase pathway signalling is unaffected in cdc2.~-ml.
LacZ activities
are the average of 2 experiments (2-3 determinations per experiment) with
standard deviation.
WT maximum (29.6 Miller Units) was set to 100%.
t o FIGURE. 2
cdc2:l-ml cells are unable to orient in a pheromone gradient. a, Excess
pheromone has a
negligible effect on cdc2-~-ml mating. :LIATa cells were mated with a WT
tester and mating
efficiency for CDC2;l (2.8%) was set to 100%. Values are means (n=2). b,
cdc2:l-ml cells are
1 s unable to orient in a pheromone gradient. A trace of cell shapes after 6-7
hr in a pheromone
gradient is shown with arrowheads indicating orientation. Quantitation of cell
projection angle
relative to the micropipet (needle) from 4-7 separate experiments (n=112 CDC24
and 167
cdc24-ml cells). The average cosine of the angle of cell projection relative
to the micropipet
was 0.52 for CDC2=l and -0.02 for cdc2-!-ml cells (a cosine of 1 represents
perfect orientation
2o and 0. random orientation). c, cdc2=l-ml cells position their shmoos
adjacent their bud scar. The
position of the bud scar on zygotes was determined for approximately 120
cells.
FIGURE. 3
25 cdc24-m mutants are defective in mating and Ste4p (G~i) binding. a,
Location of Cdc24p mating
mutations. Mating patches show diploids from mating with MATa WT tester. Ste4
2-H patch
growth on -leu-trp-his indicates an interaction of Cdc24p (I-288 aa) with
Ste4p. Similar results
were obtained using a LacZ reporter in strain YI87 (relative Miller Units I00
for Cdc24/Ste4
and 3 for Cdc24-ml/Ste4). b, Two hybrid interactions of Cdc24p. For
interactions with Ste4p,
30 a fragment of Cdc24p (1-288 aa) was used, however, full length Cdc24p also
interacts with
Ste4p. c, Region of Cdc24p necessary for Ste4p interaction. Numbers refer to
Cdc24p as fused
to DBD. d, Cdc24p binds to Ste4p in the absence of other yeast proteins. Mixed
bacterial cell
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
extracts (1 eq) containing either His6Ste4p and GST or GSTCdc24p (1-472 aa),
and GSH-
agarose eluates (800 eq) were separated by SDS-PAGE, immuno-blotted and probed
with anti-
sera to His6Ste4p. Anti-GST sera showed similar amounts of GST and GSTCdc24p
in eluates.
Due to proteolysis, His6Ste4p migrates as a doublet.
5
FIGURE. 4
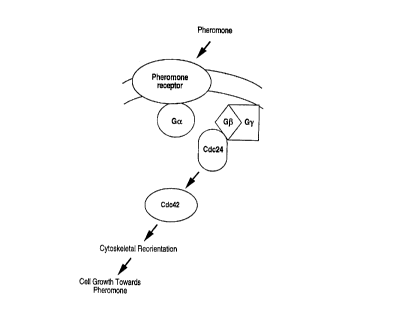
Model for signal transduction pathway required for cell orientation. For
clarity we have omitted
components of MAP-kinase cascade. The role of Cdc42p (a Rho-family GTPase) in
cell
10 orientation is speculative. Pheromone binds the pheromone receptor (Ste2p
on Ste3p) resulting
in the dissociation of Ga (Gpalp) from G(3y (Ste4p/Stel8p). Direct binding of
Cdc24p to G(3y
(in the vicinity of the receptor) activates or recruits Cdc42p which is
necessary for oriented
growth towards a mating partner.
15 .~F.~LJFNCE ANALYS.I~
The DH and PH sequences were analysed by a Blast homology search. In addition,
an analysis
of the amino acid identity over the entire protein to S. cerevisiae Cdc24p was
conducted. DH
refers to the Dbl homology region (GEF region) - see Hart et al 1991 Nature ,~
311-314;
2o Miyamoto et al 1991 Biochem Biophys Res Commun ~ 604-610; Ron et al 1991
New Biol ~
372-379. PH refers to the Pleckstrin homology region - see Musacchio et al
Trends Biochem
Sci 1$ 343-348.
The results are as follows:
A. Blast homology search using Cdc24 DH and PH region TBLASTN 1.4.9 MP
Query= yeast Cdc24p DH PH (392 aa):
3o K,IIKEFVATERKYVHDLEILDKYRQQLLDSNLITSEELYMLFPNLGDAIDFQRRFLISLEI
NALVEPSKQRIGALFMHSKHFFKLYEPWSIGQNAAIEFLSSTLHKMRVDESQRFIINNKL
ELQSFLYKPVQRLCRYPLLVKELLAESSDDNNTKELEAALDISKNIARSINENQRRTEN
CA 02305707 2000-04-07
WO 99/18213 PCTlGB98/03033
31
HQVVKKLYGRVVNWKGYRISKFGELLYFDKVFISTTNSSSEPEREFEVYLFEKIIILFSE
VVT'KKSASSLILKKKSSTSASISASNITDNNGSPHHSYHKRHSNSSSSNNIHLSSSSAAAII
HSSTNSSDNNSNNSSSSSLFKLSANEPKLDLRGRIMIMNLNQIIPQNNRSLNITWESIKEQ
GNFLLKFKNEETRDNWSSCLQQLIHDLKN
Database: Non-redundant Genbank+EMBL+DDBJ+PDB sequences
349,525 sequences; 540,957,745 total letters
Reference: Altschul, Stephen F., Warren Gish, Webb Miller, Eugene W. Myers,
and David J.
to Lipman (1990). Basic local alignment search tool. J. Mol.Biol. 215:403-410.
Reading High SmallestSmallest
Frame ScoreSum ProbSum Prob
ability ability
P(N) N
gb~U12538~SPU12538 Schizosaccharomyces+3 171 l.Oe-51 6
pombe scdl
embiX57298~MM1VICF2P0Ivt.musculus +1 128 8.3e-10 3
Mcf2
proto-oncogene
(Mc~ is Dbl)
gb~U 16296~HSU 16296Human T-lymphoma+3 88 2.3e-09 3
invasion and
metastasis
inducing TIAM1
gb~U05245~MMU0.i245Mus musculus +3 88 S.Se-09 3
BALB/c
invasion inducing
protein (Tiam-1)
gbJ036391HUMDBLTP Human DBL oncogene+2 121 2.1e-07 3
encoding a
transforming
protein
gb~S76992~576992 VAV2=VAV oncogene+3 125 2.6e-07 2
homolog human
dbjjD86547~D86547 Fruitfly still+2 76 5.4e-07 5
life type
1
gb~U37017~MMU37017 Mus musculus +1 126 6.4e-07 2
Vav2
oncogene
dbj~D86546~D86546 Fruitfly still+I 76 l.Oe-06 5
life type
2
gb[U39476~RNU39476 Rattus norvegicus+3 116 6.3e-06 1
p95
Vav proto-oncogene
gb~S76838~S76838 Dbs (Dbl guanine+3 112 4.4e-OS 2
nucleotide
exchange
factor homology
marine
dbjjAB002360~AB002360Human ICIAA0362+2 I 4.5e-05 2
13
emb~Z35654~RNOSTOG Rnorvegicus +I 112 4.9e-OS 2
Ost -
oncogene
embIX83931~HSVAVONCOH.sapiens VAV +1 109 S.Se-05 1
oncogene
gb~AF0U3147~CELC11D9Caenorhabditis+3 81 0.0070 3
elegans
C11D9
gb~U96634~MMU96634 Mus musculus +2 62 0.016 3
p85SPR
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
32
embjY10159jDDY10159D.discoideum +1 71 0.025 3
racGAP
gb~U58203~MMU58203htus musculus +2 75 0.044 2
Lsc
oncogene
embjY09160~HSSUB15H.sapiens Subl.S+1 80 0.063 2
gb~AF003740~CELC41Caenorhabditis +2 81 0.064 4
D 11 elegans
C41D11
gbjU02081~HSU02081Human guanine +1 77 0.12 2
nucleotide regulatory
protein (NET1)
gb~U00055~CELR02F2Caenorhabditis +1 85 O.13 1
elegans
R02F2
gbjU64lOS~HSU64105Human guanine +I 77 0.14 1
nucleotide exchange
factor p I 15-lthoGEF
gbjU42390jHSU42390Homo sapiens +I 74 0.33 3
Trio
gbjM24603/HUMBCRD Human bcr protein+1 58 0.91 3
~
amino end
embiX02596~HSBCRR Human bcr (breakpoint+3 58 0.996 3
cluster region)
in
Philadelphia
chromosome
gbjU 11690~I-iSU Human faciogenitat+2 73 0.999 1
11690
dvsplasia (FGD1)
gbjU22325jMMU22325Mus musculus +3 73 0.9997 2
faciogenital
dysplasia
(Fgdl)
gbjM1~025jHUMBCRABLHuman BCR/ABL +3 58 0.999955
product of the
translocation
of
t(222q11; 9q34)
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
33
B. Amino acid identity over entire protein to S. cerevisiae Cdc24p
Organism gene protein % identity
size (aa) (aa)
______~____~___~____~~_~_______~____~__~__~___~________
Schizosaccharomyces Scdl 834 21.9
pombe
Mouse Fgdl 960 16.7
Human Fgd 1 961 16.5
t Mouse Vav2 868 16.5
o
Mouse Ect2 768 16.2
Human Vav2 878 15.8
Worm Q I 8479 860 15.4
Mouse V av 844 14.6
t Rat Vav 843 14.5
Human V av 846 14.4
Mouse Dbs 1150 14.3
Human Tim 519 14.0
Human proto-Dbl 925 13.4
2o Human p 11 ~RhoGEF 912 13.4
Mouse Lfc 572 13.4
Rat Ost 872 12.9
Worm Q22354 862 12.9
Mouse Lsc 919 12.5
2s Human Lbc 424 12.4
Human Net 1 460 12.3
Human BCR 1271 11.9
Mouse Tiam 1 1591 11.2
Human Tiaml 1591 10.9
3o Mouse proto-Dbl 320 (partial)9.7
Drosophila Still Life 1 2064 9.0
Drosophila Still Life 2 2044 8.4
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
34
Scdl: Schizosaccharomyce pombe Cdc24plo~.
Fgdi Faciogenital Dysplasia Protein. FGD also known as Aarskog-Scott syndrome,
is
an X-linked developmental disorders°2.
Vav/Vav2 A oncogene derived from hematopoietic cells'°3.
Q I 8479 (similar to Vav)
Q22354 (similar to Vav)
Ect2 Oncogene e~cpressed in epithelial cells and possessing transforming
potential~°~.
to Tim Mammary epithelial oncogene~°s.
Dbl/Dbs Diffuse b-cell lymphoma (dbl) oncogenelo6, ioy
pl ISRhoGEF Regulates cell proliferation. induces the transformation of
cells'°g.
Lfc Hematopoietic oncogene~°9.
Ost Osteosarcoma derived proto-oncogene. Truncation is oncogenic and highly
tumorigenic in micel~o.
Lsc Oncoprotein~ ~ ~.
Lbc Oncogene involved in chronic myeloid leukemias~~2.
Netl Neuroepithelioma transforming oncogene~ 13.
BCR bcr (breakpoint cluster region), an oncogene which is the translocation
2o breakpoint in chronic myeloid leukemias (CML) ~ ~4~ ~ ~ s.
Tiam 1 Human invasion- and metastasis-inducing tiam 1 gene and is e~cpressed
in tumor-
cell lines of different tissue origin"6.
Still Life I/2 A synaptic terminal protein~~~.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
CDC42 and its GDP/GTP exchange factor (GEF) CDC24 are required for vegetative
growthg'9
and cell mating6'~'l0. The precise function of these proteins in cell mating
has been difficult to
5 study because they are essential for viability. In accordance with the
present invention, we
reasoned that if CDC24 has a specific function in the mating pathway, cdc24
alleles should exist
which affect cell mating but not vegetative growth. To identify such alleles,
a collection of
CDC24 random mutants was screened and three recessive mating mutants, cdc24-ml-
3 were
isolated (Figure 3A). This screen required isolated cdc24 mutants to be able
to support
t o vegetative growth. Further characterisation of cdc24-m cells revealed
normal growth between
18° and 37° and cell morphology, bud site selection, and actin
distribution were similar to WT
cells (see below and Figure 1 A). The specificity of the cdc24-m phenotype is
in contrast to that
of all other described cdc24 mutants which have strong defects in vegetative
growth8'~°.
is To elucidate the role of CDC24 in mating, we examined cdc24-ml cells for
defects in the mating
pathway. The mating e~ciency of cdc24-ml cells with a WT partner was reduced
approximately 100-fold compared to WT (Table 1 ), and this effect was
essentially independent
of mating type. When cdc24-ml or an enfeebled mater defective in cell fusion
were used as
mating partners. significantly stronger defects were observed. Such bilateral
mating defects
3o suggest impairment in a process such as shmoo (mating projection)
formation. orientation. or
fusion in which a WT mating partner can partially compensate for the mutant
strain.
Pheromone activation results in a number of responses including cell cycle
arrest, MAP-kinase
cascade mediated induction of mating specific genes, and changes in cell
morphology '~'S.
25 Pheromone-induced growth arrest determined by halo-assays showed both cdc24-
ml and WT
cells responded similarly (Figure 1 B). Furthermore, overexpression of the ~i-
subunit of the yeast
hetero-trimeric G-protein, Ste4p, from an inducible promoter arrested growth
of both cdc24-ml
and WT cells (data not shown). Microscopic examination revealed identical
numbers of WT
and cdc24-ml cells (78%, n=1600) formed shmoos after 4 hr exposure to 10 mM
pheromone.
3o The actin distribution of cdc24-ml budding and shmooing cells was also
similar to that of WT
cells (Figure 1 A), demonstrating that the mating defect was not due to an
inability to polarise the
actin cytoskeleton. The level of pheromone induced FUS1-lacZ expression, a
reporter used to
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
36
measure induction of mating specific genes~~, was similar in cdc2:l-ml and WT
cells (Figure
1 C). However, examination of mating mixtures of cdc2=1-ml and WT tester cells
showed a
greater than ten-fold decrease in the number of zygotes, indicating that the
cdc2=~-ml defect
occurs prior to cell fusion. Thus cdcZ.l-m cells appear normal for cell cycle
arrest, shmoo
formation, actin cytoskeleton polarisation, and MAP-kinase signalling, yet are
defective at a step
prior to cell fusion.
During mating, polarised growth towards a mating partner requires a pheromone
gradient''' and
saturation with pheromone during mating results in random orientation of
growth and mating
1o partner selection, and hence a decrease in mating efficiency~3'~4. WT cells
showed a 16-fold
decrease in mating e~ciency in the presence of saturating pheromone (20 rtWt),
whereas only
10% reduction was observed with cdc2,l-ml cells (Figure 2A), suggesting that
this mutant is
unable to orientate towards a pheromone gradient during mating. Similar
results were observed
with cdc2~t-m2 and cdc2~-m3 cells. To test directly whether cdc2~-ml cells are
defective in
mating projection orientation their response to an artificial pheromone
gradient created by a
micropipet was examined. While CDC2;~ cells oriented growth towards the
pheromone source
(greater than 70% of cells oriented within 60° angle of micropipet),
cdc24-ml cells did not show
a preferred orientation (Figure 2B). No difference in the sensitivity of WT or
mutant cells to
pheromone was observed.
Although cdc2~-ml cells oriented randomly in a pheromone gradient, the choice
of shmoo site
could be dictated by an internal cue, such as the previous bud site. To
examine this possibility,
the location of the bud scar (in cells with a single bud scar) relative to the
neck of the zygote was
determined. While WT cells showed a random position of their bud scar on the
zygotes, 86% of
cdc2.l-ml zygotes had formed a shmoo adjacent to their previous bud site
(Figure 2C). Together
these results establish a specific role for Cdc24p in orientation towards a
mating partner.
Sequencing of cdc2~-m alleles revealed mutations that changed one of two
adjacent amino acid
residues (Figure 3A). cdc24-ml and cdcl=l-m3 both have a single amino acid
change from Ser
189 to either a Phe or Pro. cdc24-m2 had two amino acid substitutions and
subcloning
demonstrated that the mutation responsible for the mating defect is Asp to Gly
at residue 190.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98103033
37
The grouping of these mutations suggests that this region of Cdc24p is
important for an
interaction required for oriented growth.
Previous two-hybrid studies have suggested that the amino-terminus of Cdc24p
might interact
with Ste4p', however, the in vivo significance of this association was
unclear. We determined
whether Cdc24p mating mutants could interact with Ste4p (Figure 3A). In
contrast to the wild-
rype Cdc24p, the mutants did not show a detectable interaction with Ste4p. In
agreement with
the clustering of the cdc2.l-m mutations, amino acid residues 170 to 245 of
Cdc24p were
sufficient for the Ste4p interaction (Figure 3C), while an amino-terminal
fragment consisting of
to the first 160 amino acid residues. although expressed, failed to interact.
Consistent with a
functional significance of the Cdc24p Ste4p interaction. we have isolated
mutants in STE=t,
(exemplified by SEQ ID No. 9 and SEQ ID No. 10), using a two-hybrid screen,
which are
unable to interact with Cdc24p and are phenorypically similar to cdc2-~-m
mutants.
To assess the specificity of the defect in the interaction between Ste4p and
Cdc24-mlp,
interactions with Cdc42p and Bemlp, two proteins known to bind to Cdc24p~''16
were
investigated. Bemlp is an SH3 domain protein involved in bud formation and
mating".
Cde24-mlp was able to interact with both Cdc42p and Bemlp (Figure 3B)
consistent with the
absence of an effect of cdc?.l-ml on vegetative growth.
While the cdc2-~-ml phenotype along with the two-hybrid results indicates that
the interaction
between Cdc2=lp and G(3 is central to cell orientation, these results do not
address whether this
interaction is direct or indirect. G~i typically functions as a complex with
the third subunit of a
hetero-trimeric G-protein, Cry. We therefore determined whether the yeast Gy,
Stel8p, was
required for the Cdc24p Ste4p interaction. Deletion of STE18 abolished the
Cdc24p Ste4p two-
hybrid interaction (data not shown), suggesting that Cdc24p interacts with the
G[3~y-complex. To
determine if Cdc24p could directly bind Ste4p, these proteins were expressed
in bacteria.
Hexahistidine-tagged Ste4p specifically bound to GSTCdc24p (Figure 3D). These
results
demonstrate that Cdc24p can directly bind G(3 in the absence of any other
yeast proteins. We
3o attribute the requirement for Gy in the two-hybrid assays to its
stabilisation of G(3~8.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
38
Pheromone receptor activation results in dissociation of G(iy from Ga at the
receptor. Our
results indicate that the orientation defect in cdc2=~-m cells is due to a
specific defect in the
Cdc24p G~iy interaction. This suggests a model in which direct binding of
Cdc24p to G(3y
results in recruitment (to the vicinity of the receptor) or activation of
Cdc42p and that this local
concentration of activated Cdc42p is responsible for oriented growth towards a
pheromone
gradient (Figure 4). In the absence of this recruitment or activation a site
adjacent to the
previous bud site appears to function as a default site for shmoo formation.
Our results together
with previous studies implicating Cdc24p in bud site selections, suggest that
Cdc24p acts as a
crucial component required both for bud and shmoo site selection, perhaps
functioning as a kind
of molecular selector switch between internal signals for bud site selection
and external signals
for shmoo site selection. It is likely that local activation of Cdc24p
recruits and activates the
Rho GTPase Cdc42p, which could then interact with downstream targets required
for orientation
of the cytoskeleton. Cdc42p interactions with the protein kinase
Ste20p~9'2° are not necessary for
cell orientation2°, suggesting that novel targets of Cdc42p are
required for oriented growth
towards a mating partner.
Cdc24p belongs to a diverse family of GEFs which include many mammalian proto-
oncogenes2.
This group of proteins shares a conserved region consisting of a Dbl-domain
(named after the
human proto-oncogene Dbl) followed by a plecktstrin-homology domain (PH).
Sequence
2o comparison revealed similarity between a small stretch of amino acids
flanking the cdc24
mating mutations and Dbl (Figure 3A). Our results indicate that an association
between
Cdc24p and G~3y links pheromone receptor activation to shmoo orientation. We
propose that
other GEFs, such as the proto-oncogene Dbl, provide a similar connection
between G-protein
coupled receptor activation and polarised cell growth.
Hence, in accordance with the present invention there are provided the
following uses and
utilities of Cdc24p/Ste4 interaction and cdc24-m mutants
1 ) Peptide inhibitors which block GEF/G~3 interaction. Peptides and peptidyl
derivatives based
regions encompassing mutants will be used to block and/or antagonise GEF (such
as the proto-
oncogenes Dbl or Vav) G(3 interaction. Derivatives of these peptides
(including peptide mimics)
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
39
which bind with higher affinity will also be used. The perturbation of these
interactions will be
of therapeutic value for example in treatment of cancers.
2) Simple yeast based assays systems (utilising mating function and
interaction reporters) will
be extremely useful for high through-put screening to identify molecules
perturbing this
GEF/G(3 interaction. In particular, the qualitative effect on mating observed
with the proto-
oncogene, proto-Dbl, even at low levels of expression, indicates that this
type of assay is
amenable to large scale screening for the effect of agents, such as proto-
oncogenes, on
induced defects in yeast and other host cells.
t0
3) Similar Cdc24p/G~3 interactions will be ideal targets for anti-fungal drugs
directed at the
pathogenic yeast Candida.
1) We have identified an important interaction between two general cellular
components,
Cdc24p and G(3 which provides a connection between G protein coupled receptor
activation and
polarised cell growth. This work has been exemplified by work done with yeast
genes/proteins,
however, both cellular components involved have homologues in humans.
2) We show the physiological consequence of this interaction and from these
data
extrapolate to the general role of this interaction in human cells.
3) In addition, we have identified sequences required for this interaction.
Specifically, we
have identified a short stretch of one protein (Cdc24p) encompassing 7~ as
sufficient for this
interaction and three amino acid changes (within this stretch) which block the
interaction and
have physiological consequences. These amino acid changes fall within a I9
amino acid piece
with similarity to the human proto-oncogene Dbl. Indeed, removal of this
region from proto-
Dbl (when the amino terminus is removed) results in oncogenicity in tissue
culture cells.
4) We have also identified specific mutants in the (3-subunit of the
heterodimeric G protein
(Ste4p) which appear to block its interaction with Cdc24p. We believe that
several of these
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
mutations will fall in conserved retions of G~3. Thus, it is possible to
devise assays based on
this mutation to screen for agents capable of modifying the non-interactive
behaviour of the
mutant G protein (3 subunit with Cdc24p. In addition, the assay could be used
to study
Cdc24p homologues or even Cdc24p derivatives or homologues to see if those
derivatives or
s homologues affect the non-interactive behaviour of the mutant G protein.
5) There is a wealth of information on human G(3's, human GEF's (~DP/GTP
exchange
Factors), such as Cdc24p homologues and the rho family of GTP-binding-proteins
(such as rho
like Cdc42p) which the GEFs work on. Most human GEF's are oncogenes such as
Dbl, Vav,
1o and Ect and are involved in some way in growth control. Furthermore G(3's
are involved in
linking signals from receptors to intracellular responses. The present
invention has shown that
that a GEF from yeast, Cdc24p, can directly bind G(3 in the absence of any
other yeast proteins.
Although unproven, it is likely that interactions between human GEF's and
G~3's are also crucial
in erowth control and chemotaxis.
l5
6) We propose the interaction we have identified will have broad cellular
ramifications and
manipulation of these interactions (such as peptidic inhibitors and peptides
mimicking activated
species) will be of therapeutic value.
20 7) In addition, simple yeast based assays systems could be extremely useful
for high
through-put screening to identify molecules perturbing this interaction. In
particular, a
qualitative assay using a yeast mutant with a mating defect could prove useful
in the design of
agents, such as anti-cancer agents, that can affect the function of oncogenes
such as proto-Dbl,
in terms of its ability to complement a yeast mutant mating defect and/or its
function in
25 mammalian tissue culture cells.
8) We also believe similar interactions will be ideal targets for anti-fungal
drugs directed at
invasive and pathogenic yeasts such as Candida albicans and Cryptococcus
neoformans.
3o All publications mentioned in the above specification are herein
incorporated by reference.
Various modifications and variations of the described methods and system of
the invention
will be apparent to those skilled in the art without departing from the scope
and spirit of the
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
41
invention. Although the invention has been described in connection with
specific preferred
embodiments, it should be understood that the invention as claimed should not
be unduly
limited to such specific embodiments. Indeed, various modifications of the
described
modes for carrying out the invention which are obvious to those skilled in
molecular
biology or related fields are intended to be within the scope of the following
claims.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
42
RFERRFNCES
1. Machesky, L.M. & Hall, A. Rho: a connection between membrane receptor
signaling and the
cytoskeleton. Trends In Cell Biology ~, 304-310 {1996).
2. Cerione, R.A. & Zheng, Y. The Dbl family of oncogenes. Ca~rrent Opinion In
Cell Biology $,
216-222 ( 1996).
3. Whiteway, M., et al. The STEM and STE18 genes of yeast encode potential b-
subunits and y-
to subunits of the mating factor receptor-coupled G protein. Cell $~, 467-477
(1989).
4. Sprague, G.F.J. & Thorner, J.W. Pheromone response and signal transduction
during the
mating process of Saccharomyces cerevisiae. In The molecular and cellular
biology of the yeast
Saccharomyces (eds. Jones, E.W., Pringle, J.R. & Broach, J.R.) 657-744 (Cold
Spring Harbor
t5 Press, Cold Spring Harbor, N.Y., 1992).
5. Leberer, E., Thomas, D.Y. & Whiteway, M. Pheromone signalling and polarized
morphogenesis in yeast. Current Opinion in Genetics & Development Z, 59-66
(1997).
20 6. Simon, M.-N., et al. Role for the Rho-family GTPase Cdc42 in yeast
mating pheromone
signal pathway. Nature ~, 702-705 (1995).
7. Zhao, Z.S., Leung, T., Manser, E. & Lim, L. Pheromone signalling in
Saccharomyces
cerevisiae requires the small GTP-binding protein Cdc42p and its activator
CDC24. Nlol Cell
25 Biol ~, 5246-57 (1995).
8. Sloat, B.F., Adams, A. & Pringle, J.R. Roles of the CDC24 gene product in
ceiluiar
morphogenesis during the Saccharomyces cerevisiae cell cycle. JCell Biol $~,
395-405 (1981).
30 9. Adams, A.E., Johnson, D.L, Longnecker, R.M., Sloat, B.F. & Pringle, J.R.
CDC42 and
CDC43, two additional genes involved in budding and the establishment of cell
polarity in the
yeast Saccharomyces cerevisiae. JCell Biol llh 131-42 (1990).
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
43
10. Chenevert, J., Valtz, N. & Herskowitz, I. Identification of genes required
for normal
pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics ~,
1287-96
( i 994).
11. Trueheart, J., Boeke, J.D. & Fink, G.R. Two genes required for cell fusion
during yeast
conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol
Z, 2316-2328
(1987).
12. Segall, J.E. Polarization of yeast cells in spatial gradients of a mating
factor. Proc Natl Acad
Sci USA ~( , 8332-6 (1993).
13. Dorer, R., Pryciak, P.M. & Hartwell, L.H. Saccharomyces cerevisiae cells
execute a default
pathway to select a mate in the absence of pheromone gradients. JCell Biol ]~,
845-61 (1995).
14. Valtz, N., Peter, M. & Herskowitz, I. FARI is required for oriented
polarization of yeast
cells in response to mating pheromones. J Cell Biol ~, 863-73 ( I 995).
15. Zheng, Y., Cerione, R. & Bender, A. Control of the yeast bud-site assembly
GTPase Cdc42.
Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase
activity by
Bem3. JBiol Chem ~Q, 2369-72 (1994).
16. Zheng, Y., Bender, A. & Cerione, R.A. Interactions among proteins involved
in bud-site
selection and bud-site assembly in Saccharomyces cerevisiae. JBiol Chem Z7~,
626-30 (1995).
17. Chenevert, J., Corrado, K., Bender, A., Pringle, J. & . Herskowitz, I. A
yeast gene (BEMl )
necessary for cell polarization whose product contains two SH3 domains. Nature
~, 77-9
( 1992).
18. Hirschman, J.E., DeZutter, G.S., Simonds, W.F. & Jenness, D.D. The G~3~y
complex of the
yeast pheromone response pathway - subcellular fractionation and protein-
protein interactions. J
Biol Chem ~, 240-248 (1997).
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
44
19. Peter, M., Neiman, A.M., Park, H.O., Van Lohuizen, M. & Herskowitz, I.
Functional
analysis of the interaction between the small GTP binding protein Cdc42 and
the Ste20 protein
kinase in yeast. Embo Journal ~, 7046-7059 (1996).
20. Leberer, E., et al. Functional characterization of the Cdc42p binding
domain of yeast Ste20p
protein kinase. Embo Journal j,~, 83-97 (1997).
21. Rose, M.D., Winston, F. & Hieter, P. Methods in yeast genetics: a
laboratory course
manual (Cold Spring Harbor Press, Cold Spring Harbor, N.Y., 1991).
to
22. Miyamoto, S., et al. A DBL-homologous region of the yeast CLS4/CDC2;t gene
product is
important for Ca(2+)-modulated bud assembly. Biochem Biophys Res Common ~, _
604-10
(1991).
23. Mitchell, D.A., Marshall, T.K. & Deschenes, R.J. Vectors for the inducible
overexpression
of glutathione-S-transferase fusion proteins in yeast. Yeast ~, 715-722
(1993).
24. Wach, A., Brachat, A., Alberti-Segui, C., Rebischung, C. & Philippsen, P.
Heterologous
HISS marker and GFP reporter modules for PCR-targeting in Saccharomyces
cerevisiae. Yeast
~, 1065-1075 (1997).
25. Fromant, M., Blanquet, S. & Plateau, P. Direct random mutagenesis of gene-
sized DNA
fragments using polymerise chain-reaction. Analytical Biochemistry ~, 347-353
(1995).
26. Sprague, G.F. Assay of yeast mating reaction. Methods In Enzymol ~, 77-93
( 1991 ).
27. Adams, A.E. & Pringle, J.R. Staining of actin with fluorochrome-conjugated
phalloidin.
Methods Enrymol ,1~, 729-31 ( 1991 ).
28. Pringle, J.R. Staining of bud scars and other cell wall chitin with
calcofluor. Methods
Enrymol ~, 732-5 (1991).
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
29. Langle-Ronault, F. & Jacobs, E. A method for perfoming precise alterations
in the yeast
genome using a recyclable selectable marker. Nucleic Acids Research ~, 3079-
3081 (1995).
30. James, P., Halladay, J. & Craig, E.A. Genomic libraries and a host strain
designed for
5 highly efficient two-hybrid selection in yeast. Genetics ~, 1425-1436
(1996).
101. Chang, E.C., Barr, M., Wang, Y., Jung, V., Xu, H.P., and Wigler, M.H.
1994. Cooperative
interaction of S. pombe proteins required for mating and morphogenesis. Cell.
~: 131-41.
l0 102. Pasteris, N.G., Cadle, A., Logie, L.J., Porteous, M., Schwartz, C.E.,
Stevenson, R.E.,
Glover, T.W., Wilroy, R.S., and Gorski, J.L. 1994. Isolation and
characterization of the
faciogenital dysplasia (aarskog-scoff syndrome) gene - a putative Rho/rac
guanine-nucleotide
exchange factor. Cell. 7Q: 669-678.
is 103. Katzav, S., Martinzanca, D., and Barbacid, M. 1989. Vav, a novel human
oncogene
derived from a locus ubiquitously expressed in hematopoietic-cells. Embo
Journal. $: 2283-
2290.
104. Miki, T., Smith, C.L., Long, J.E., Eva, A., and Fleming, T.P. 1993.
Oncogene ect2 is
2o related to regulators of small GTP-binding proteins. Nature. 462-465.
105. Chan, A.M., McGovern, E.S., Catalano, G., Fleming, T.P., and Miki, T.
1994. Expression
cDNA cloning of a novel oncogene with sequence similarity to regulators of
small GTP-binding
proteins. Oncogene. Q: 1057-63.
106. Eva, A. and Aaronson, S.A. 1985. Isolation of a new human oncogene from a
diffuse B-
cell lymphoma. :Vature. 3~: 273-275.
107. Ron, D., Tronick, S.R., Aaronson, S.A., and Eva, A. 1988. Molecular-
cloning and
3o characterization of the human dbl proto- oncogene - evidence that its
overexpression is sufficient
to transform nih/3t3 cells. Embo Journal. Z: 2465-2473.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
46
108. Hart, M.J., Sharma, S., Elmasry, N., Qiu, R.G., Mccabe, P., Polakis, P.,
and Bollag, G.
1996. identification of a novel guanine-nucleotide exchange factor for the Rho-
gtpase. Journal
Of Biological Chemistry. ?~7~.: 25452-25458.
109. Whitehead, L, Kirk, H., Tognon, C., Trigo-Gonzalez, G., and Kay, R. 1995.
Expression
cloning of lfc, a novel oncogene with structural similarities to guanine
nucleotide exchange
factors and to the regulatory region of protein kinase C. J Biol Chem. ~Q:
18388-95.
110. Horii, Y., Beeler, J.F., Sakaguchi, K., Tachibana, M., and Miki, T. 1994.
A novel
oncogene, ost, encodes a guanine nucleotide exchange factor that potentially
links Rho and Rac
signaling pathways. Embo J. .~.,~: 4776-86.
111. Glaven, J.A., Whitehead, LP., Nomanbhoy, T., Kay, R., and Cerione, R.A.
1996. Lfc and
lsc oncoproteins represent 2 new guanine-nucleotide exchange factors for the
Rho-gtp-binding
protein. Journal Of Biological Chemistry. ~: 27374-27381.
112. Toksoz, D. and Williams, D.A. 1994. Novel human oncogene ibc detected by
transfection
with distinct homology regions to signal-transduction products. Oncogene. 621-
628.
113. Chan, A., Takai, S., Yamada, K., and Miki, T. 1996. Isolation of a novel
oncogene, netl,
from neuroepithelioma cells by expression cdna cloning. Oncogene. ~: 1259-
1266.
114. Hariharan, LK. and Adams, J.M. 1987. Cdna sequence for human-bcr, the
gene that
translocates to the abl- oncogene in chronic myeloid-leukemia. Embo Journal.
~: 115-119.
115. Diekmann, D., Brill, S., Garrett, M.D., Totty, N., Hsuan, J., Monfries,
C., Hall, C., Lim, L.,
and Hall, A. 1991. Bcr encodes a gtpase-activating protein for p2lrac. Nature.
651: 400-402.
116. Habets, G., Vanderkammen, R.A., Stam, J.C., Michiels, F., and Collard,
J.G. 1995.
Sequence of the human invasion-inducing tiaml gene, its conservation in
evolution and its
3o expression in tumor-cell lines of different tissue origin. Oncogene. 1371-
1376.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
47
117. Sone, M., Hoshino, M., Suzuki, E., Kuroda, S., Kaibuchi, K., Nakagoshi,
H., Saigo, K.,
Nabeshima, Y., and Hama, C. 1997. Still life, a protein in synaptic terminals
of Drosophila
homologous to GDP-GTP exchangers. Science. ~: 543-5~7.
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
1
~'Qt 1RNCE LISTINGS
A. cdc24 (wt)
SEQ ID NO. 1
s DNA:
cccctctgtatacmtcaactctgtgaagccgcaatttaaattaccggtaatagcatctgacgamgaaagtctgtaaaaa
atccatttatgactt
tatattgggctgcaagaaacacmgcamaacgatgaggagctmcactatatccgacgtttttgccaactcgacgtcccag
ctggtcaaag
tgctagaagtagtagaaacgctaatgaattccagc
io
SEQ ID NO. 2
Protein:
PLCILFNSVKPQFKLPVIASDDLKVCKKSIYDFILGCKKHFAFNDEELFTISDVFA~'~'STSQ
LVKVLEVVETLivINSS
B. cdc24-ml
SEQ ID NO. 3
DNA:
2o
cccctctgtatactmcaactctgtgaagccgcaatttaaattaccggtaatagcamgacgamgaaagtctgtaaaaaat
ccatttatgactt
tatattgggctgcaagaaacacmgcamaacgatgaggagcmtcactatatccgacgtrittgccaactcgacgtcccag
ctggtcaaag
tgctagaagtagtagaaacgctaatgaattccagc
SEQ ID NO. 4
Protein:
PLCILFNSVKPQFKLPVIAFDDLKVCKKSIYDFILGCKKHFAFNDEELFTISDVFANSTSQ
LVKVLEWETLMNSS
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
2
C. cdc24-m2
SEQ ID NO. S
DNA:
S
cccctctgtatacttttcaactctgtgaagccgcaatttaaattaccggtaatagcatctggcgatttgaaagtctgta
aaaaatccatttatgactt
tatattgggctgcaagaaacactttgcatttaacgatgaggagcttttcactatatccgacgtttttgccaactcgacg
tcccagctggtcaaag
tgctagaagtagtagaaacgctaatgaattccagc
1o SEQ ID NO. 6
Protein:
PLCILFNSVKPQFKLPVLASGDLKVCKKSIYDFILGCKKHFAFiV'DEELFTISDVF A.~1STSQ
LVKVLEV~'ETLivINSS
is
D. cdc24-m3
SEQ ID NO. 7
DNA:
cccctctgtatacttttcaactctgtgaagccgcaatttaaattaccggtaatagcacctgacgatttgaaagtctgta
aaaaatccatttatgact
ttatattaggctgcaagaaacactttgcatttaacgatgaggagcttttcactatatccgacgtttttgccaactcgac
gtcccagctggtcaaa
gtgctagaagtagtagaaacgctaatgaattccagc
2, SEQ ID NO. 8
Protein:
PLCILFNSVKPQFKLPVIAPDDLKVCKKSIYDFILGCKKHFAFNDEELFTISDVFA.~1STSQ
LVKVLEWETLMNSS
SEQ ID NO. 9
STE4 DNA sequence (wild-type)
SUBSTITUTE SHEET (RULE 26~
CA 02305707 2000-04-07
WO 99/18213 PCT/GB98/03033
AT~~-.GCACATCnG=s':GG:CTCGATnACGTnTTCT.'-.i:Tr'~='.'::~TCCCCnACr'1GTATe-
._'=:CA.'ACC.'-..~_==.AGTCi.'-:CAGGi-.
TATCTCTGCA~'GGAGGi~~GP.AATTCAP.AATAF_=.::TAGi:GGCCGCCAGACAAG=G=GTA.3A'CnGC'~'TC
A?'GC"_'CP.ai TP_3
Ai=.nAGCP_~IAi:CnCAeyG=.TACPAGATGCAAGCT='ATTCCAGATGGCCP.ACP.AAG='°_nC':'
:Ca_C CCe"in= =-.':A~:Gi:TC
P.F:CTTAAAGCC:.e~.ATATCG'i'GTTGAAAGGCCAT~-
.ATAn:e'~Ar~.P.TCTCAGATTTTC:~GTGGAGTC =:~GATTCisi,AACGTAT
Tm_'"GAGTGCAniCAAGATGGCTTTATGCTTnT=.TGGGACAGTGCTTCAGGTTTr~
=u:CAGP.FC=CTATTCCP.TTP-GnTT
CTCAATGGGTTCT.TTCCTGCGCTATTTCGCCATCGAGTnCTTTGGTAGCAAGCGCnGGATTP.P.ACe~ATAACTGTA
CCATT
TnCAGAGTTT W: P ~.AGi~.AP.e'~CAGAGTAGCGCAAt-.ACGTT GCGT. CAAT. TTTCAAAGGACATAC
T'i G CT. r'1T. ATT : CTGACAT
TG~-.nTTTACAGi==~-.nCt:CACi:Ti-~T_F~TTGACAGC.r.GTGGGGAT.ATGi-
.CATGTGCC::'GiGGGn- :P:CCGi:.z=.GCr'1AAG:y
GGGTGAGAGir='.CAT'T_'CiGr~CCi:TTTAGGTGATG'='TTTGGCATTnGCT:,TT_CCTG
~~GF.GCCAr=.C'_"TAGr".F ~ '"TC'FTCG
P.~.~.nCATTCGC'_:1GCLVTGGH~il~.A~GACGGGTATACTTnCA'iATGS.-
.GnT~.GCi:Gi=TCTCCGiC:..CTGTnCi:== GCTi"~...
'"L '"" _._..C=.C,TTC~~:mT._-_...__._..____.._____.._
__._.._.__._._._....._._~___......
..__. r~..~Jr:_...._...._.._...
T ~~.-11~TVTC:.J_.. _T:1.".VVTVVVC:CTVTTCTI:.TGV.T:1W~~........r v . T..v:-
:VGTT.__ _.~.Vtl.'...VT :..w.V.T~\.~.r.
ym ~ r..I=~~n~n~r.", Tm T /"' .....ACrW ~~'''.t' Tl.:~'v~GI.TV.:IV~.TATCT
:._? LT..TA_...:..r~.,...~ .W':_GGAGTACAA.=CCGC::..r.:_'=~..»-.-
IS .._ :v.til'.L.l. JJ..VT':v'/aTT~.T1~i~GATITmP.VTVC :T...TG'.:!"_'~'-7~
Vt:l~%TY-~..~'v~': ~.:.._.._~vAGr.L:lT~V'J~~~1V
.__TGTGGG=C__.._.. ~~.nGGAGAGATT.GiTG~AA~nTTi-i=='..W:'_"CA'-'GG'-GG~nGe:G'-C-
_~:'~GTG:va.~CTCGi:~'-
C~., AT.- ~~T°-_G~TG""T '"" A
GTTCATGGGACi:.i'==_C_..CGri'~,.~"'P.':'G.:T..T~~:'.V4'T-.:._"CAAT=:~
.._ J 1VVV ._ .r-V:l'1C V
2o SEQ ID No. 10
Ste4 Protein sequence (wild-type)
h~ _::QMDSITY S:WVTQQYIQPQSLQDISAVEEc.IQNKIEAi-iRQES:CQLiii-~QINiC~
K:'.KT_QDe:S=°QMANK'.'=SLTK:iKIh1
L K?h1I'v i,KGH":~;KIS'JF RWSRDSKRILSASQDG F MLIidCSi:SGLKQNA = cLDSQ:~J~i
LSCe:I S?SSTLVAS.'-:G=.NhINC i I'''
2~ i<..'SKEhiRVAQ:~-
~=.SI:~CGii"'~YISDIEFTDNA::ILTASGDN'TCALWDiPKr'1KR'iREYSDH:.G~'.~n~AT:._._?N=.V
SS::
_ ~' SCG S CG'_ _ _ - ~: uS=cS?S~:VQS :'YVNDS DI:vnLRF : KDG:~iS -VAGS
Dt3Ga.T_ cl~'.': DLRS D C.. _. . _' c S :. F.... _ .._.. _ - -
. ::~:PAhIh!EY\:
P.QSPQ='_::~jTSSSYLDNQGVVSLDFSASGRLMYSCYTDiGCV'v:°:DVLKGe.T_'i
G=CT~EGHG~:~ ITGVRSS°
.,GLAVCTGS::DSTMKI'tISPGYQ
SEQ ID No. 11
sre4-o1.5 DI~~ sequence (mutant)
. -GGChCAT.C.'-.GATGG C'ICGATAACGTATTC'_'AATe"-sr'?TS:TCF-
~CCCP.ACAGTA':i:'FACAFCC=...P.iz.~GTC_.-'-.Ci-sGG=.
". ,. ,., r. _. .-
3~ ..._..T..iV~. _.:':'VV~-xGVrie:Gr-~A': TCAAAATri-r.ATi:VrVGv,CGCCAGAC?.e-
:GnVr?VTrsAr:Cr.:..: m.n_.:..-.-._....._.
~'v~1_ ~~~w - wT ~.~ I' ~~ Tmr~r._i.., y ~.I~m-T m /~~~' .._..... ....
.._. ~.V ...~~...tl.:V.'y.I.:a.Cl.'.J :'GCCY1V',..-:C1
1..V.~V:SLVVC'v.A!'ICCIt~':J m :C:1 _ :.:1'..'..'~ :~'~~'-
._:CTTe~GCC:.i'ii'_'=s':CC'-
.T.GTTGAAAGGCCATAATF~ITt'1~'~P_~,'TCTCAGi,T':TTCGGTGGAG'~ CGAG::T'-'C=W
~CGT~:C
SUBSTITUTE SHEET (RULE 26~
CA 02305707 2000-04-07
WO 99118213 " PCT/GB98/03033
TTTGAGTGC.~AGTCAAGATGGCTTTATGCTTATATGGGACAGTGCTTCAGG'=TTAAAACAGArCG::ATTCCATTAG
ATT
CTCAATGGG':T.CTTTCCTGCGCTATTTCGCCATCGAGTACTTTGGTAGCAr~iCGCAGGAT. TFF.F.i:~~-
.nTAACTGTACCATT
TATAGAG""_"T~GAAAGAP.AACAGAGTAGCGCAAAACGTTGCGTCAATTTTCF.AAGGACATri~'""'G~T~TATT
TCTGACAT
TGAA T TTe=.Ci:GA TAACGCACA TATATTGACAGCAAGTGGGGA T ATGA CATG : GCCTTGTGGG~:Te
:T ~:CCGAAnG CAAAG'?.
GGGTGAGAGGATATTCTGACCATTTAGGTGATGTTTTGGCATTAGCTATTCCTGAAGAGCCt~.AC'TAGAAAAT.TCT
TCG
AACACATTCGCTAGCTG T GGATCAGACGGGTATACTTFiCA TiiTGGGiITAGC~GA'=C T CCC:TCCGC':
G iACAAAGC':'T. T'TA
CGTTAACGATAGTGATATTAATGCACTTCGTTTTTTCAAAGACGGGATGTCGATTGTTGCAGGArGTGACAATGGTGCG
A
TAAATATGTATGATTTAAGGTCGGACTGTTCTATTGCTr'1CTTTTTCTCTTTI'TCGAGGTTATGAA~vAiaCGTACC
CCTACC
CCTACTTATATGGCAGCTAACATGGAGTACAATACCGCGCAATCGCCACAArCTTTAAA~'.TCnF.CF~
GCTCAAGCTATCT
AGACAACC:ie=:GGCGTTGTTTCTTTAGATTTTAGTGCATCTGv~AAGATTGATGTACTCATGCTnTi:~.nGACATT
GGTTGTG
TTGTGTGGG:.TGTATTAAAAGGAGAGATTGTTGGAAAAT
TnGAAGGTCATGGTGGCr~.GAGTCnC'='G('.~TGTGCGCTCGAGT
..v.:lv.~.T'v'v.:.:~Tf=VU.~VTI:T'.:':W.~GVTmV..I:~'JGG~.v.ll.C :v..... '.:r'-
."-.......7G_~..M.~.v. .-..__...-:'~~1~:..
SEQ ID No. 12
Ste4-o15 Protein sequence {mutant)
MA:IQMDST_ T YSNNVTQQYIQPQSLQDISAVEEEIQNKIEAARQESKQLHAQ;NKAKHKIQDi":ST_=
,~~:~IANKVTSLTKNKIN
LKPNIVLKG.'~'.NNKISDFRWSRDSKRILSASQDGFMLIWDSASGLKQNAIPLDSQWVLSCA_S?SS'"LVASAGLN
NNCTIY
RVSKENR6'AQNVASI FKGHTCYISDIEF
TDNAHIi~TASGDMTCALTnTDIPK.:CRVRGYSDH:..G:~V=:~LAIPEEPNLENSSN
TFASCGSDGYTYIWDSRSPSAVQSFYVNDSDINALRF FKDG:~.SIVAGSDh~.INMYDLRSD'.:.'ST_A':'F
SLFRGYF.ERTPTP
TYMAANM~YNTAQSPQTLKSTSSSYLDNQGVVSLDFSASGRi.MYSCYTD=G:VVWDVLKGF.IVG:CEGHGGRVTGVR
SSP
DGLAVCTGSWDSTMKIWSPGYQ
SEQ ID NO. 13
ste4-ol7 DNA sequence (mutant)
ATGGCACF:T CAGATGGACT. CGATAACGTi-s'-'TCTAATAATGTCACCCe':ACi'iG :
ATATACAACCACAnAGTCTACAGu~a
TA?'CTCTWAGTGGAGGAAGAAATTCAA.~r1' TAAAATAGAGGCCGCCnGAC.~'-
u=~GAGAGTAAACAGC'ITCATGCTC~r=~ATAA
ATAAAGCA.=AP.CACAAGATACAAGATGCAAGCTTAT T C CAGATGGC C ~ri :CF .nAGTTACTTCG':'
T anC CAAAn.= :'f.AGATC
AACT TAP.F-.GCCAAATATCGTGTTGAAAGGCCATAATAATAAAATCTCAG."-.':
TTTCGGTGGAGTCGe,GATTCAF.AACGTAT
TTTGAGTGCAAGTCAAGATGGCTTTATGCTTATATGGGACAGTGCTTCAG~'i'TTAAP.ACAGAF.CGCTATTCC~:T
.TAGATT
CTCP.ATGGGTTCTTTCCTGCGCTATTTCGCCATCGAGTACTTTGGTAGCF.l:GCGCAGGATTAAF.CrrITAACTGT
ACCATT
. C. _-. ~ ~~i. ~,GACr.Te":.. _ : C a A T. . .. T ~:nCe°:~:
_. ::.Gn-="_'.~.v~"'i."-..'~G«isil?CAGF,GT=.G:GCAA.-~u°-.CGTTGCG'"
~"_°.-_ ~~ " "'"'"G
TGF~':'""' :C=.G~T~CGCACATATATTG:~CF:GCAAGTGGGGATA
TGF,CA'_"GTGCCTTGTGGGATFTACCG:-..?.AGCP.AAGA
GGG'"Ge'~GnGP.ATATTCTGACCATTTAGGTGATGTTTTGGCATTAGCTAT_"_'CCTGAAGAGCCAF.AC'_'TAG
P.AAATTCTTCG
AACACAT : ~GCTAGCTGTGGATCAGACGGGTATACTTACATATGGGF TAG~:AGATC T CCG : CCGC
T'GT:'iW-.AnGC': TTTA
CGTTAACGaTAGTGaTATTAATGCACTTCGTTTTTTCAAAGACGGGATGTCGATTGTTGCAGGAF:GTGACAnTC:"TG
CGA
SUBSTITUTE SHEET tR(lLE 2B1
CA 02305707 2000-04-07
WO 99/18213 5 PCT/GB98/03033
TAAATATGTATGATTTAAGGTCGGACTGTTCTATTGCTACTTTTTCTCTTTTTCGAGGTTATGAAG?.ACGTACCCC':
'rCC
CCTACTTATATGGCAGCTAACATGGAGTACAATACCGCGCAATCGCCACAAACTTTAAAATCAACAAGCTCAAGCTATC
T
AGACP.ACCAAGGCGCTGTTTCTTTAGATTTTAGTGCATCTGGAAGATTGATGTACTCATGCTATACF:GACATTGGiT
GTG
TTGTGTGGGATGTATTAAAAGGAGAGATTGTTGGAAAATTAGAAGG~_'CATGGTGGCAGAGTCACT.GGTGTGCGCTC
GAGT
CCAGATGGGTTAGCTGTATGTACAGGTTCATGGGACTCAACCATGAe'~AATATGGTCTCCAGGTTnTCAATAG
SEQ ID No. 14
Ste4-ol7 Protein sequence (mutant)
MAHQMDSITYSNNVTQQYIQPQSLQDISAVEEEIQNitIEAARQESKQLHAQINKAKHKIQDASLFQi~IANKVTSLTK
NKIN
LKPNIVLKGHNNKISDFRWSRDSKRILSASQDGFMLIWDSASGLKQNAIPLDSQWVLSCAISPSS':LVASAGLNNNCT
IY
RVSK= NRVAQNJASIF KGHTCYISDIEF
TDNAHILTASGDMTCrI~.::_=P:~~:CRVREYSDHLGDV_r:.nI?F.EP:i:..F:;SS'i
TFASCGSDGYTYIWDSRSPSAVQSFYVNDSDIIVALRFFKDGMSIVi-
nGSDN~VAINMYDLRSDCST_A=:jLFRGYE'.~TPTP
TYMAANMEYNTAQSPQTLKSTSSSYLDNQGA'!
'SLDFSASGRLMYS~'_'TDIVCVVWDVLhGtIVG:C:.~GHGGRV~GVR.SSn
1~ DGLAVCTGSWDSTMKIWSPGYQ
SEQ ID No. 15 is presentedFigure 3A as
in Dbl Hu.
SEQ ID No. 16 is presentedFigure 3A as
in Cdc24 Sc.
SEQ ID No. 17 is presentedFigure 3A as
in Cdc24-ml .
2o SEQ ID No. 18 is Figure 3A as
presented in Cdc24-m2.
SEQ ID No. 19 is presentedFigure 3A as
in Cdc24-m3.
SUBSTITUTE SHEET (RULE 261