Note: Descriptions are shown in the official language in which they were submitted.
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
1
DESCRIPTION
Expression Plasmids For Multiepitope
Nucleic Acid-Based Vaccines
Statement of Related Applications
This application is related to U.S. Patent Application
entitled "IL-12 Gene Expression and Delivery Systems and
Uses", filed October 10, 1997, Serial No. not yet assigned,
by Jeff Nordstrom, Bruce Freimark and Deepa Deshpande, Lyon
& Lyon Docket No. 226/285 and U.S. Patent Application
entitled "Gene Expression and Delivery Systems and Uses",
filed October 10, 1997, Serial No. not yet assigned, by Jeff
Nordstrom, Bruce Freimark and Deepa Deshpande, Lyon & Lyon
Docket No. 226/284, both of which are incorporated herein by
reference in their entirety, including any drawings.
Introduction
The invention relates generally to gene therapy, in
particular, the invention relates in part to improved
plasmids and methods for nucleic acid based vaccines.
Background of the Invention
The following discussion of the background of the
invention is merely provided to aid the reader in
understanding the invention and is not admitted to describe
or constitute prior art to the present invention.
Plasmids are an essential element in genetic engi-
neering and gene therapy. Plasmids are circular DNA
molecules that can be introduced into bacterial cells by
transformation which replicate autonomously in the cell.
Plasmids allow for the amplification of cloned DNA. Some
plasmids are present in 20 to 50 copies during cell growth,
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
2
and after the arrest of protein synthesis, as many as 1000
copies per cell of a plasmid can be generated. (Suzuki et
al. , Genetic Analysis, p. 404, {1989) . )
Current non-viral approaches to human gene therapy
require that a potential therapeutic gene be cloned into
plasmids. Large quantities of a bacterial host harboring
the plasmid may be fermented and the plasmid DNA may be
purified for subsequent use. Current human clinical trials
using plasmids utilize this approach. (Recombinant DNA
Advisory Committee Data Management Report, December, 1994,
Human Gene Therapy 6:535-548). Studies normally focus on
the therapeutic gene and the elements that control its
expression in the patient when designing and constructing
gene therapy plasmids. Generally, therapeutic genes and
regulatory elements are simply inserted into existing
cloning vectors that are convenient and readily available.
Plasmid design and construction utilizes several key
factors. First, plasmid replication origins determine
plasmid copy number, which affects production yields.
Plasmids that replicate to higher copy number can increase
plasmid yield from a given volume of culture, but excessive
copy number can be deleterious to the bacteria and lead to
undesirable effects (Fitzwater, et al., EMBO J. 7:3289-3297
(1988); Uhlin, et al., Mol. Gen. Genet. 165:167-179 (1979)).
Artificially constructed plasmids are sometimes unstably
maintained, leading to accumulation of plasmid-free cells
and reduced production yields.
To overcome this problem of plasmid-free cells, genes
that code for antibiotic resistance phenotype are included
on the plasmid and antibiotics are added to kill or inhibit
plasmid-free cells. Most general purpose cloning vectors
contain ampicillin resistance ((3-lactamase, or b1a) genes.
CA 02309344 2000-OS-11
WO 99/Z4596 PCT/US98/23745
3
Use of ampicillin can be problematic because of the
potential for residual antibiotic in the purified DNA, which
can cause an allergic reaction in a treated patient. In
addition, (3-lactam antibiotics are clinically important for
disease treatment. When plasmids containing antibiotic
resistance genes are used, the potential exists for the
transfer of antibiotic resistance genes to a potential
pathogen.
Other studies have used the neo gene which is derived
from the bacterial transposon Tn5. The neo gene encodes
resistance to kanamycin and neomycin (Smith, Vaccine
12:1515-1519 (1994)). This gene has been used in a number
of gene therapy studies, including several human clinical
trials (Recombinant DNA Advisory Committee Data Management
Report, December, 1994, Human Gene Therapy 6:535-548). Due
to the mechanism by which resistance is imparted, residual
antibiotic and transmission of the gene to potential
pathogens may be less of a problem than for (3-lactams.
In addition to elements that affect the behavior of the
plasmid within the host bacteria, such as E. coli, plasmid
vectors have also been shown to affect transfection and
expression in eukaryotic cells. Certain plasmid sequences
have been shown to reduce expression of eukaryotic genes in
eukaryotic cells when carried in cis (Peterson, et al., Mol.
Cell. Biol. 7:1563-1567 (1987); Yoder and Ganesan, Mol.
Cell. Biol. 3:956-959 (1983); Lusky and Botchan, Nature
293:79-81 (1981); and Leite, et al., Gene 82:351-356
(1989)). Plasmid sequences also have been shown to fortu-
itously contain binding sites for transcriptional control
proteins (Ghersa, et al., Gene 151:331-332 (1994); Tully and
Cidlowski, Biochem. Biophys. Res. Comm. 144:1-10 (1987); and
Kushner, et al., Mol. Endocrinol. 8:405-407 (1994)). This
CA 02309344 2000-OS-11
WO 99/x4596 PCT/US98/23745
4
can cause inappropriate levels of gene expression in treated
patients.
Nucleic acid vaccines, or the use of plasmid encoding
antigens, has become an area of intensive research and
development in the last half decade. Comprehensive reviews
on nucleic acid vaccines have recently been published [M. A.
Liu, et al.(Eds.), 1995, DNA Vaccines: A new era in
vaccinology, Vol. 772, Ann. NY. Acad. Sci., New York;
Kumar, V., and Sercarz, E., 1996, Nat. Med. 2:857-859;
Ulmer, J.B., et al., (Eds.) Current Opinion in Immunology;
8:531-536. Vol. 772, Ann. NY. Acad. Sci., New York].
Protective immunity in an animal model using plasmid
encoding a viral protein was first observed in 1993 by Ulmer
et al. [Ulmer, J.B., et al., 1993, Science 259:1745-1749].
Since then, several studies have demonstrated protective
immunity for several disease targets and human clinical
trials have been started.
Summary
The use of epitopes, small immunologically relevant
protein sequences that are capable of inducing both cellular
and humoral responses that result in a protective or
therapeutic immune response against large and complex
pathogens, is an attractive and amenable strategy provided
by the present invention for incorporation into nucleic
acid-based vaccines. If multiple epitopes are expressed in
the context of a synthetic gene construct, immunity against
many antigenic targets, multiple strain variants or multiple
pathogens is possible. This disclosure describes the
structures and characteristics of gene expression systems
that are capable of expressing multiple epitopes.
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98n3745
Thus, in one aspect the invention provides a method of
genetic immunization comprising the step of presenting
multiple epitopes to an organism in need of said
immunization. '
5 In preferred embodiments, the multiple epitopes are
presented with one or more augmenting cytokines and/or are
presented with a delivery vehicle selected from the group
consisting of cationic lipids, delivery peptides, and
polymer based deliver systems.
In another aspect, the invention features a plasmid for
expression of multiple epitopes comprising a nucleic acid
sequence encoding multiple epitopes, wherein each of said
epitopes is capable of creating an immune response.
In preferred embodiments, the plasmid includes a
promoter, a 5' UTR sequence, and a 3' UTR sequence, a nucleic
acid sequence encoding polyubiquitin, there are spacers
between the nucleic acid regions encoding each of said
epitopes, there are proteolytic cleavage sites between each
of said epitopes, there are ER targeting signals between
each of said epitopes, there are lysosomal and/or endosomal
targeting sequences between each of said epitopes.
In other aspects, the invention provides a multivalent
expression system as shown in Figure 8 and selected from the
group consisting of two plasmids, two genes, IRES, and
alternative splicing and a method of making a plasmid
producing the appropriate nucleic acid sequence.
The summary of the invention described above is non-
limiting and other features and advantages of the invention
will be apparent from the following detailed description of
the preferred embodiments, as well as from the claims.
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
6
Brief Description of The Drawings
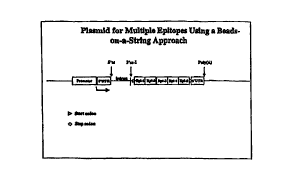
Figure 1 shows a plasmid for multiple epitopes using a
beads on a string approach.
Figure 2 shows a plasmid for multiple epitopes using
beads on a string fused to polyubiquitin.
Figure 3 shows a plasmid for multiple epitopes using
beads on a string with spacers between epitopes.
Figure 4 shows a plasmid for multiple epitopes using
beads on a string with proteolytic cleavage sites between
epitopes.
Figure 5 shows a plasmid for beads on a string epitopes
with ER targeting sequences.
Figure 6 shows a plasmid for multiple epitopes with ER
targeting sequences.
Figure 7 shows a plasmid for multiple epitopes with
lysosomal/endosomal targeting sequences.
Figure 8 shows types of multivalent expression systems.
Figure 9 shows a DNA vaccine expression plasmid with
two genes.
Figure 10 shows a design of a drug-controlled DNA
vaccine expression plasmid.
Detailed Description of the Preferred Embodiments
Various exemplary plasmids and methods for multiepitope
nucleic acid based vaccines are described below.
The following explanation of the invention is to aid in
understanding various aspects of the invention. The
following explanation does not limit the operation of the
invention to any one theory.
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
7
I. Expression Plasmid for Epitopes Arrancted as Beads-on-a-
String
In expression plasmids of this type, the multiple
epitopes are directly linked to each other. No spacer
sequences~between the epitopes are included. The epitope
sequences themselves are sufficient for the formation of a
functional "pseudo" protein that can be processed into
individual peptide epitopes via proteosome cleavage. This
concept, i.e. beads-on-a-string, is supported by data that
shows full CTL responses to numerous epitopes when they are
placed into novel locations within different proteins
(Nomura M, Nakata Y, Inoue T et al., J. Immursol. Methods,
193:41-9 (1996) and Weidt G, Deppert W, Buchhop S, Dralle H,
Lehmanngrube F., J. Virol., 69:2654-8 (1995)).
An and Whitton, J. Virol., 71:2292-302 (1997) have
described that a beads-on-a-string approach is feasible with
a recombinant vaccinia virus vector. They appear to have
demonstrated that a linear array of B-cell, CTL and Th
epitopes was able to induce the corresponding immune
response. Gilbert et al., Nature Bio., 15:1280-84 (1997)
hGs demonstrated that the beads-on-a-string approach is
feasible with a recombinant Ty-VLP vector. They described
that a linear array of 15 defined malaria epitopes induced
protective CTL responses in mice, and that neither epitope
order nor flanking sequences influenced the processing of
the epitopes.
Multiple epitopes expressed from a recombinant vaccinia
virus vector as a string of 10 contiguous minimal CTL
epitopes, which were restricted by five MHC alleles and
derived from five viruses, a parasite, and a tumor model,
induced a primary CTL response in vivo in the appropriate
mouse strain. This illustrates that multiple CTL epitopes
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
8
can be effectively delivered in a beads-on-a-string array
(Thomson et al., J. Immunol., 157:822-6 (1996)).
The present invention provides an exemplary expression
system for beads-on-a-string as shown below (See Figure 1):
Promoter / 5' UTR / intron / AUG / (Epitope)n / stop
codon / 3' UTR / poly(A) signal
Table I below provides a description of each of these
genetic elements.
Table I. Description of genetic elements.
Element Description
Promoter CMV, tissue-specific (e. g. APC-
specific), or synthetic promoter
5' UTR Optimized to assure mRNA stability and
translatability. Current optimal
sequences are UT11 (from human loricrin
gene ) or UT12 ( f rom CMV) .
Intron Synthetic intron that has optimized
5' ss, 3' ss and branch point sequences.
Current optimal sequence is IVS 8.
Initiation codon AUG is placed in the context of the
Kozak sequence to ensure optimal
initiation of translation.
(Epitope)n String of epitopes, each having a length
of 9-10 amino acid residues in length
for class I presentation, or >10 amino
acid residues for class II presentation.
It appears that at least 15 epitopes may
be strung together. One of the main
considerations will be to avoid the
placement of glycine or proline adjacent
to the desired epitope termini.
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
9
Stop codon For termination of translation. To
ensure efficient termination, it is
desirable to string two stop codons in
tandem.
3' To ensure efficient processing of the
UTR/poly(A) mRNA an efficient poly(A) signal, such
signal as from human growth hormone, is
required.
II. Expression Plasmid for Multiple Epitopes as Beads-on-a-
Strincr Linked to a Polyubiquitin Chain
Degradation of many eukaryotic proteins requires their
prior ligation to polyubiquitin (Ub) chains, which target
the substrates to the 26S proteasome, an abundant cellular
protease. Thus, it is advantageous to encode a Ub chain
that is fused to the N-terminus of the multiple epitopes
that are arranged as beads-on-a-string. Expression plasmids
with Ub fused to the antigen have been used to achieve class
I presentation (Gueguen and Long, Proc. Natl. Acad. Sci. USA
93:14692-97 (1996)).
Illustrated below is an expression system for beads-on-
a-string fused to polyubiquitin (Ub) (See Figure 2):
Promoter / 5' UTR / intron / AUG / Ub / (Epitopes-Spacer)n /
stop codon / 3' UTR / poly(A) signal
III. Expression Plasmid for Multiple Epitopes as Beads-on-a-
Strincr with Spacers Between Epitopes
An expression system for beads-on-a-string with spacers
is shown below (See Figure 3):
Promoter / 5' UTR / intron / AUG / (Epitopes-Spacer)n / stop
codon / 3' UTR / poly(A) signal
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
Some investigators have shown that flanking sequence
can profoundly influence the generation of epitopes
(Yellenshaw et al., J. Immunol., 158:1727-33 (1997); Shastri
et al., J. Immunol., 155:4339-46 (1995); i7e1-Val et al.,
5 Cell, 66:1145-93 (1991); Eggers et al., J. Exp. Med.
182:1865-70 (1995); Niedermann et al. Immunity, 2:289-95
(1995); Lippolis et al. J. Virol. 69:3134-46 (1995)). In
particular, glycine or proline residues adjacent to the
minimal epitope should to be avoided, since peptide bonds to
10 these residues are known to be resistant to protease
activity (Niedermann et al., 1995).
Spacers may facilitate the formation of epitopes that
induce immunity. Ideally, the spacer sequence should be one
that does not conform at all to the rules for class I or II
epitope. However, it may be desirable to include a
hydrophobic, basic or acidic residue at the C-terminus of
the spacer to facilitate cleavage between the spacer and the
adjacent epitope. The length of the spacer that would be
optimal is not known and would have to be determined
empirically.
IV. Expression Plasmid for Multiple Epitopes as Beads-on-a-
Strinc~with Proteolytic Cleavage Sites Between Epitopes
An expression system for beads-on-a-string with
proteolytic cleavage sites is diagramed in summary form
below (See Figure 4):
Promoter / 5' UTR / intron / AUG / (Epitopes-Cleavage Site)n
/ stop codon / 3' UTR / poly(A) signal
Many viruses (e. g. retroviruses, flaviviruses) generate
mRNAs that encode polyproteins that must undergo proteolytic
cleavage to form the mature protein products. Cleavage,
which occurs at specific sites, is catalyzed by host
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
11
proteinases or by virally encoded proteinases. For example,
the polyprotein from hepatitis C virus is structured as
follows: H2N-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NSSA-NSSB-COOH.
Host cell signal peptidases cleave the junctions in the
region between C and NS2. The viral proteinase (NS2-3
proteinase)cleaves the junction between NS2 and NS3.
Another viral proteinase (NS3 proteinase) cleaves the
junctions between NS3 and NSSB.
One approach is to insert host cell cleavage sites
between the epitope sequences. This may be achieved by
insertion of the sequences that are located at the junctions
between the C-E1-E2-p7-NS2 proteins of the hepatitis
polyprotein. Other possibilities are to utilize the
recognition site for specific cellular proteases.
A second approach is to insert cleavage sites for the
viral proteinase between the epitope sequences. Thus, the
site for the sequence recognized by the NS3 proteinase,
Asp/Glu-X-X-X-X-Cys/ThrlSer/Ala (Koch and Bartenschlager,
Virology, 237:78-88 (1997)), may be inserted. However, for
this approach to work, the NS3 proteinase must be also
encoded by the expression plasmid. Thus, two transcription
units are required, one for the multiepitopes, one for the
viral proteinase. See Section V. Multivalent expression
plasmids for nucleic acid-based vaccines.
V. Expression Plasmids for Multiple Epitopes With
Tarctet inQ
The classical pathway for antigen presentation in the
context of class I involves the partial degradation of
antigenic proteins into peptides by the proteasome. The
peptides are then transported into the endoplasmic reticulum
by peptide transporters (TAP-1 and TAP-2). It is within the
CA 02309344 2000-OS-11
WO 99/24596 PGT/US98/23745
12
lumen of the ER or cis-Golgi that the peptides are loaded
into the binding pocket of the MHC class I molecules.
Ciernik et al., J. Immunol. 2369-75 (1996) have demonstrated
that the immunogenicity of an epitope may be enhanced if the
epitope sequence is fused in frame with the adenovirus E3
leader sequence and expressed from a plasmid delivered by
particle bombardment. An epitope fused to the E3 leader
yielded greater protection from tumor challenge than an
epitope without the leader.
ER targeting signals have also been used by Overwijk et
al., Identification of a Kb-restricted Ctl Epitope of Beta-
galactosidase: Potential Use in Development of Immunization
Protocols for "Self" Antigens, Methods 12: 117-23 (1997). The
signal sequence allows the epitope to be targeted to the ER
by the standard protein translocation process. The N-
terminal leader sequence targets the peptide to the ER by
first binding, as a nascent sequence, to the 54 kDa subunit
of the SRP particle. The leader sequence subsequently binds
to the b-subunit of the membrane-bound transporter protein,
Sec6lp. Following entry into the lumen of the ER through a
putative channel, a peptidase cleaves the peptide to remove
the leader sequence. This mechanism is independent of the
TAP transporter system. This alternative mechanism may be
advantageous if epitope formation by proteasome cleavage, or
epitope transport by the TAP system, are limiting steps in
antigen presentation.
For ER targeting, a leader sequence preferably needs to
be attached to the N-terminus of each epitope. Adding a
leader sequence to the multiple epitopes that are arranged
as beads-on-a string concept is unlikely to work, since the
leader will be attached only to the first epitope sequence.
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
13
Placing an individual targeting sequence on each of the
epitopes that is arranged in a bead-on-a-string assembly is
a possibility. However, this strategy will depend on
accurate proteolytic cleavage at the N-terminus of the
leader sequence and at the C-terminus of the adjoined
epitope sequence. A gene expression system that utilizes
alternative splicing will yield individual epitopes with
their own leader sequences. The peptide epitopes produced
by this strategy will not depend on random degradation of a
protein precursor.
The only processing that is required is N-terminal
processing that is associated with protein translocation.
The C-terminal ends of the epitopes are defined by the stop
codons that are designed into the system. The preprotein
products may be incompletely synthesized until protein
translocation through the pore into the ER has occurred.
Alternatively, the prepeptides may be synthesized in their
entirety prior to ER translocation. This may expose the
prepeptide to the proteasome and transport of proteins that
transport epitopes to the ER by the standard pathway.
A. Expression Plasmid for Multiple Epitopes with ER
Taraetina Sequences
An expression system for beads-on-a-string epitopes
with targeting sequence is shown below (See Figure 5):
Promoter / 5' UTR / AUG / ER signal sequence / epitope-1 /
stop codon / ER signal sequence / epitope-2 / stop codon /
ER signal sequence / epitope-3 / stop codon / 3' UTR/poly(A)
signal
An alternative splicing system for multiple epitopes
with targeting sequence is shown below (See Figure 6):
Promoter / 5' UTR / AUG / ER signal sequence / 5'ss /
internal intron sequence / 3'ss-1 / epitope-1 / stop codon-1
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
14
/ 3'ss-2 / epitope-2 / stop codon-2 / 3'ss-3 / epitope-3 /
stop codon-3 / 3' UTR/poly (A) signal
Table II below describes the genetic elements used in
the alternative splicing strategy.
Table II. Description of genetic elements for the
alternative splicing strategy.
Element Description
ER Signal The N-terminal leader sequence from
sequence 5' ss adenovirus E3 or preprolactin
Strong 5' splice site, one that exactly
matches the consensus sequence. Such a
sequence is found in the synthetic
intron, IVS8.
Internal This sequence is derived from the
intron synthetic intron, IVS8. It extends from
sequence the 3' end of the 5' splice site to the
5' end of the polypyrimidine tract of
the 3'splice site.
Alternative 3' ss-1, 3' ss-2, 3' ss-3: These splice
3' splice sites sites will be designed to be used
equally. Thus, their relative strengths
need to be mathed. This will be
accomplished by introducing purines
within the polypyrimidine regions of the
splice site sequences.
In the alternative RNA splicing system, the strengths
of the 3' splice sites must be balanced to splicing from the
5'ss to each of the 3' splice sites (3'ss-1, to 3'ss-2, to
3'ss-3, etc.). Balanced splicing will be achieved by
controlling the purine content of the pyrimidine-rich
CA 02309344 2000-OS-11
WO 99/24596 PCTNS98/23745
sequences of the 3' splice sites. In general, the greater
the purine content, the weaker the splice site. There are
model systems to follow. For example, the major late
transcript of adenovirus is alternately spliced into 5
5 families of transcripts that are produced in roughly
equivalent amounts. Thus, one way to design an appropriate
alternatively spliced system for epitopes is to model the 3'
splice sites of adenoviral late transcripts.
Another key feature is that, after splicing, the leader
10 sequence must be fused in frame with the peptide sequence of
each epitope. Also, by altering the strengths of the 3'
splice sites, the relative amounts of the epitopes may be
varied. This may important if certain epitopes are more
dominant than others.
15 Table III below shows an example of a balanced set at 3'
splice sites derived from the adenoviral late transcript.
Table III. Example of a balanced set of 3' splice
sites (derived from the adenoviral late transcript)
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
16
5' ss Source Alternative 3' ssl~ Source2~
CAG~LGTAAGT IVS8 TTTGCTTTTCCCCAG~I~G Ad2 (11039)
Consensus
5' ss)
TTGTATTCCCCTTAGyT Ad2 ( 1419
9 )
GTTGTATGTATCCAG~LC Ad2 (16515)
GTAACTATTTTGTAGJ~A Ad2 (17999)
CCATGTCGCCGCCAGd~A Ad2 (18801)
ATGTTTTGTTTGAAGJ~T Ad2 (21649)
TTCCTTCTCCTATAGJ~G Ad2 (24094)
1/ The consensus sequence for a 3' ss is YYYYYYYYYYYNYAGyG.
Y - C or T, and y = intron/exon junction.
2/ Adenovirus 2 (Ad2) sequences are from the Genbank entry,
ADRCG. Numbers in parentheses indicate the nucleotide position
of each 3' splice site. Note the locations of the purines (A or
G) that interrupt the polypyrimidine (C or T) region.
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/Z3745
17
B. Expression Plasmid for Multiple Epitopes with
Endosomal/Lysosomal Targeting Sequences
An expression system structure is shown below (See
Figure 7):
Promoter / 5' UTR / AUG / ER signal sequence / 5'ss /
internal intron sequence / 3'ss-1 / epitope-1 / LAMP-1
transmembrane-cytoplasmic tail / stop codon-1 / 3'ss-2 /
epitope-2 / LAMP-1 transmbrane-cytoplasmic tail / stop
codon-2 / 3' ss-3 / epitope-3 / LAMP-1 transmbrane-
cytoplasmic tail / stop codon-3 / 3' UTR/poly(A) signal
To target class II antigen presentation, it may be
desirable to directly target the peptide epitope to the
endosomes or lysosomes. One strategy employs the
transmembrane and cytoplasmic tail sequences from a one of
the lysosomal-associated membrane glycoproteins, such as
LAMP-1. Wu et al., (1995) have used such a sequence, in
combination with an N-terminal ER targeting sequence, to
target an antigen to the endosomal and lysosomal
compartments for class II antigen presentation. Thus, each
epitope is preceded by an N-terminal leader sequence (e. g.
adenovirus E3) and followed by the C-terminal
endosomal/lysosomal targeting sequence (e.g. the
transmembrane and cytoplasmic tail region of LAMP-1).
Another sequence that may be employed for endosomal
targeting is the cytoplasmic tail of membrane immunoglobulin
(Weiser et al., Science 276:407-9 (1997); Achatz et al.,
Science 276:409-11 (1997)).
Since the transmembrane/cytoplasmic tail can be added
to some, but not necessarily all epitopes, it would be
possible to target some epitopes to the ER for class I
presentation and others to the endosomes/lysosomes for class
II presentation.
CA 02309344 2000-OS-11
WO 99/24596 PCT/US98/23745
18
VI. Multivalent Expression Plasmids for Nucleic Acid-Based
«____~....
For effective nucleic acid-based vaccines, it may be
important to have the capability of expressing multiple gene
products. For example, expression of multiple intact
antigens or multiepitope gene product may enhance the
potency of the these vaccines. Co-expression of
costimulatory proteins, such as IL-2, IL-6, IL-12, GM-CSF,
B7.1 or B7.2, have been demonstrated to enhance the immune
response to an encoded antigen (Geissler et al., J.
Immunol. 158:1231-1237 (1997), Irvine et al., (1996) Kim et
al., Vaccine 15:879-83 (1997); Okada et al., J. Immunol.
159:3638-47(1997); Barry and Johnston, Scand. J. Immunol.,
45:605-12(1997)). Co-expression of proteins that facilitate
peptide epitope formation, such as proteolytic enzymes (e. g.
the NS3 protease from hepatitis C (Koch and Bartenshlager,
1997)) or chaperone proteins (e. g. heat shock protein Hsp65
(Wells et al., Scand. J. Immunol., 45:605-12 1997)), may
also enhance the response.
The various types of multivalent expression plasmids
are described in Figure 8. They include (1) multiple
complete genes, or transcription units, on a single plasmid,
(2) generation of polycistronic mRNAs using a internal
ribosome entry site (IRES) sequence, and (3) generation of
multiple mRNAs by alternative RNA splicing. The design of
a nucleic acid-based vaccine expression plasmid that has two
genes is shown in Figure 9.
The details of these systems are described in the
related patent applications incorporated herein by reference
on page 1.
CA 02309344 2000-OS-11
WO 99/24596 PGT/US9S/23745
19
Examples
The present invention will be more fully described in
conjunction with the following specific examples which are
not to be construed in any way as limiting the scope of the
invention.
Geneswitch Example
The GeneSwitch is a chimeric protein that consists of
human progesterone receptor with a modified ligand binding
domain, a DNA binding domain from yeast GAL4, and an
activator domain from Herpes Simplex VP16. When a synthetic
steroid, mifepristone, is administered, the GeneSwitch
protein becomes activated (binds the synthetic steroid,
presumably dimerizes, and translocates to the nucleus). The
activated GeneSwitch then binds to a target sequence
(multiple GAL4 binding sites linked to a minimal promoter)
and thereby stimulates the transcription of the desired
transgene (Wang et al., Proc. Natl. Acad. Sci. USA 91:8180-
84 (1994) Wang et al., Nature Biotechnology 15:239-243
(1997a); Wang et al., Gene Therapy 4:432-41 (1997b)).
The GeneSwitch may be used to regulate the expression
of a plasmid for nucleic acid-based vaccines. It is
possible that the timing of expression may influence the
immune response. Thus, with a GeneSwitch regulated system,
the genes that encode the multiepitopes may be turned on at
a defined time after DNA delivery by the administration of
the ligand (mifepristone) to the animal. If the expression
plasmid persists in vivo for a long enough time, the
GeneSwitch system also can be used to provide pulsatile
expression of the multiepitope gene products. An example
of the design of a system that is regulated by the
GeneSwitch is shown in Figure 10.
CA 02309344 2000-OS-11
WO 99/24596 PG"TNS98I23745
One skilled in the art would readily appreciate that
the present invention is well adapted to carry out the
objects and obtain the ends and advantages mentioned, as
well as those inherent therein. The molecular complexes and
5 the methods, procedures, treatments, molecules, specific
compounds described herein are presently representative of
preferred embodiments are exemplary and are not intended as
limitations on the scope of the invention. Changes therein
and other uses will occur to those skilled in the art which
10 are encompassed within the spirit of the invention are
defined by the scope of the claims.
It will be readily apparent to one skilled in the art
that varying substitutions and modifications may be made to
the invention disclosed herein without departing from the
15 scope and spirit of the invention.
All patents and publications mentioned in the speci-
fication are indicative of the levels of those skilled in
the art to which the invention pertains. All patents and
publications are herein incorporated by reference to the
20 same extent as if each individual publication was specific-
ally and individually indicated to be incorporated by
reference.
The invention illustratively described herein suitably
may be practiced in the absence of any element or elements,
limitation or limitations which is not specifically
disclosed herein. Thus, for example, in each instance
herein any of the terms "comprising", "consisting
essentially of" and "consisting of" may be replaced with
either of the other two terms. The terms and expressions
which have been employed are used as terms of description
and not of limitation, and there is no intention that in the
use of such terms and expressions of excluding any
CA 02309344 2000-OS-11
WO 99/24596 PCTNS98/Z3745
21
equivalents of the features shown and described or portions
thereof, but it is recognized that various modifications are
possible within the scope of the invention claimed. Thus,
it should be understood that although the present invention
has been specifically disclosed by preferred embodiments and
optional features, modification and variation of the
concepts herein disclosed may be resorted to by those
skilled in the art, and that such modifications and
variations are considered to be within the scope of this
invention as defined by the appended claims.
In addition, where features or aspects of the invention
are described in terms of Markush groups, those skilled in
the art will recognize that the invention is also thereby
described in terms of any individual member or subgroup of
members of the Markush group. For example, if X is
described as selected from the group consisting of bromine,
chlorine, and iodine, claims for X being bromine and claims
for X being bromine and chlorine are fully described.
Other embodiments are within the following claims.