Note: Descriptions are shown in the official language in which they were submitted.
CA 02323075 2000-09-14
wo 99/47700 PCT/DE99/00725
- 1 -
Method and device for detecting a nucleotide sequence
The invention relates to a method in accordance with
the preamble of Claim 1. It also relates to a
microtiter plate and a kit for carrying out the method.
US 4,996,143 and DE 195 81 489 Tl disclose methods in
which a first and a second primer are bound to the
nucleotide sequence to be detected at a distance of 2
to 7 nucleotides. The first and the second primer are
each provided with a fluorophoric .molecule. In the
bound state, a radiation-free energy transfer from one
fluorophoric molecule to the other is observed owing to
the Forster effect. This causes a specific
fluorescence. - The known method is not particularly
sensitive.
US 5,607,834 discloses the use of a primer with a hair-
pin loop for detecting a nucleotide sequence. In this
case, fluorophoric molecule and a quencher are provided
opposite each other on the loop sections of the hair-
pin loop. The distance between the fluorophoric
molecule and the quencher allow [sic] a radiation-free
energy transfer which quenches the fluorescence.
However, when the primer hybridizes with a
complementary strand, the hairpin is opened. The
spatial relationship between the fluorophoric molecule
and the quencher, which quenches a fluorescence, is
altered. Thus, a fluorescence is observable.
W093/09250 discloses an amplification method in which a
first primer is bound to a first phase. A second primer
is labeled with a fluorophoric ,dye. When a nucleotide
sequence to be detected is present, the labeled second
primer accumulates on the solid phase. - In order to
recognize a sufficiently discriminating signal on the
solid phase, it is necessary to carry out a washing
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 2 -
step after the PCR. This step requires an additional
effort. Moreover, contaminations may be introduced
while carrying out this step.
The object of the present invention is to eliminate the
disadvantages of the prior art; it is intended in
particular to provide a method with improved
sensitivity, where the possibility of contamination is
reduced and which is simple and inexpensive to carry
out. Moreover, the concentration of the nucleotide
sequence to be detected is to be determined in as
efficient a manner as possible.
This object is achieved by the features of claims 1 and
19. Expedient embodiments result from the features of
claims 2 to 18 and 20 to 34.
In accordance with the invention, at least one of the
fluorophoric molecules is bound to the surface of a
solid phase. The method allows the nucleotide sequence
to be detected to be determined qualitatively and
quantitatively. A simple fluorescence measurement, in
particular an online detection, is possible owing to
the fact that the at least one fluorophoric molecule is
bound to a solid phase. The method can be carried out
in a simple and inexpensive manner since washing steps,
which increase the risk of contamination, can be
dispensed with.
In a particular embodiment, a first primer is bound to
the solid phase. It is possible that the first
fluorophoric molecule is bound to the solid phase via
the first primer. In this case, the first primer
advantageously has a hairpin loop, and the first
fluorophoric molecule is bound to the one loop section
and the second fluorophoric molecule opposite to the
other loop section at a distance which allows the
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 3 -
interaction to take place. The interaction is
eliminated expediently by hybridization with a
complementary strand which is complementary to the
first primer or by a synthesis which takes place on the
first primer. The above-described procedure further
reduces the possibility of contamination.
In a further embodiment of the method, the second
fluorophoric molecule can also be bound to a second
primer. However, it is also possible to incorporate
nucleotides provided with the second fluorophoric
molecule, or a further nucleic acid sequence, into a
synthesis strand. The second primer is in solution.
After amplification and denaturation, the first and the
second primer are advantageously hybridized in such a
manner that the interaction is generated. The distance
between the first and the second fluorophoric molecule
in the hybridized state is preferably 2 to 12
nucleotides. The above variant of the method is
particularly sensitive.
The solid phase can comprise a polymer which is
preferably electroconductive, for example a
polycarbonate, polycarbene, trimethylthiopene and/or
triaminobenzene and/or carbon fibers. It has proved to
be especially advantageous for the solid phase to be a
microtiter plate.
In a further feature of an embodiment, the first
molecule is an acceptor group and the second
fluorophoric molecule a donor group. The acceptor group
can be a 6-carboxytetramethylrhodamine and the donor
group a 6-carboxyfluorescein. Other suitable
donor/acceptor pairs can be seen from the table which
follows:
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 4 -
Donor Acceptor
Fluorescein Fluorescein
Fluorescein Tetramethylrhodamine
IAEDANS (= 5-((((2- Fluorescein
iodacyl) amino) ethyl) amino)
-
naphathalene-lsulon [sic]
acid)
EDANS (=5-((2-aminomethyl)- DABCYL [sic] (4-dimethyl-
amino)naphthalene-1- aminoazo-benzene-4'-
sulfonic acid) sulfoylo chloride) [sic]
BODOPY [sic] FL BODIPY FL
Naturally, it is possible to swap the first and the
second fluorophoric molecule. In a further feature of
an embodiment, the first or second fluorophoric
molecule can be replaced by a quencher, preferably a
quencher formed by 4-[4'-dimethylaminophenylazo]benzoic
acid.
Suitable quencher/fluorophore pairs can be seen from
the table which follows:
Quencher Fluorophore
DABCYL [sic] Coumarin
DABCYL [sic] EDANS
DABCYL [sic] Fluorescein
DABCYL [sic] Lucifer Yellow
DABCYL [sic] Bodipy
DABCYL [sic] Eosin
DABCYL [sic] Tetramethylrhodamine
DABCYL [sic] Texas Red
DABCYL [sic] Erythrosin
To determine the concentration of the nucleotide
sequence to be detected, the fluorescence can be
recorded by means of a fluorometer connected to a data
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 5 -
processing system, the concentration of the nucleotide
sequence to be detected being determined from the
change of the fluorescence intensity over time. The
reference point used is preferably the second
derivative of the fluorescence intensity over the
number of the amplification cycles carried out.
In accordance with the solution with regard to the
device, a microtiter plate is provided for carrying out
the method according to the invention with an upper
face which is provided with a plurality of well-shaped
recesses and to which the first molecule is bound. A
first primer may be bound to the upper face, the first
molecule advantageously being bound to the surface via
the first primer. In a further embodiment, the first
primer has a hairpin loop, and the first molecule is
bound to one loop section and the second molecule
opposite to a second loop section at a distance which
allows the interaction to take place.
In a further embodiment with regard to the device, a
kit is provided with a microtiter plate according to
the invention and with a primer provided with a second
molecule.
The method according to the invention is illustrated in
greater detail with reference to the drawing. In this
drawing,
Fig. 1 shows the pairing of the strand and the
complementary strand of a target DNA with a
first and a second primer,
Fig. 2 the hybridization of the primers synthesized,
Fig. 3 the excitation of the fluorophoric molecules,
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 6 -
Fig. 4 the pairing of the strand and the complementary
strand of a target DNA in a further variant of
the method,
Fig. 5 the excitation of the fluorophoric molecules in
accordance with the variant of the method in
Fig. 4,
Fig. 6 the fluorescence of a detector nucleotide with
and without nucleotide label,
Fig. 7a a particle bound to a second primer following
PCR with template DNA in a dark field image,
Fig. 7b the particle of Fig. 7a in a fluorescence
micrograph,
Fig. 7c a particle bound to a second primer following
PCR without template DNA in a dark field image,
and
Fig. 7d the particle of Fig. 7c in a fluorescence
micrograph.
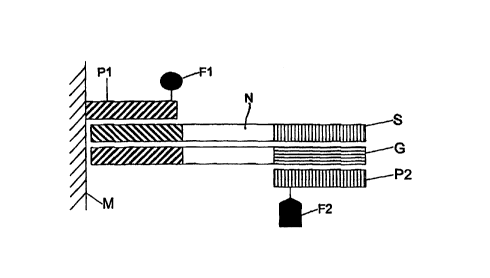
In Fig. l, a first primer Pl is bound to the upper face
within a cavity of a microtiter plate M made of
polycarbonate or polypropylene. The microtiter plate M
may contain a controlled resistance heating. It may
also be an element of a resistance heating itself. A
first fluorophoric molecule Fl is bound to the first
primer P1.
The nucleic acid sequence N to be detected, which is
present in a target DNA, and the further components
required for carrying out a polymerase chain reaction
(PCR) or ligase chain reaction (LCR) are pipetted into
the cavities. The latter comprise in particular a
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
_ 7 _
second primer P2 with a second fluorophoric molecule F2
bound thereto. The target DNA is denatured, i.e.
separated into a strand S and a complementary strand C.
The temperature is then reduced to 50 to 60° [sic]. The
strand S binds with a complementary sequence segment to
the first primer P1. The complementary strand G binds
to the second primer P2 which is present in the fluid.
Then, the sequence segment which is missing in each
case is synthesized by means of a Taq DNA polymerase.
The temperature is then raised to ~94°C, so that the
synthesis strands comprising the fluorophoric molecules
Fl, F2 are present in the fluid as single strands viz
as synthesis strand SSl and as synthesis complementary
strand SC1. The second fluorophoric molecule F2 may
also be incorporated into the synthesis strand SSl in a
form in which it is bound to nucleotides or a further
nucleic acid sequence, instead of via the second primer
P2. The temperature is reduced to 50 to 60° [sic] . The
synthesis strand SS1 and the synthesis complementary
strand SC1 hybridize, so that the first F1 and the
second fluorophoric molecule F2 are present at a
distance of 6 to 12 nucleotides. This is shown
schematically in Fig. 2.
Upon excitation of the first fluorophoric molecule F1,
which is designed as the donor, a radiation-free energy
transfer to the second fluorophoric molecule F2, which
acts as the acceptor, takes place. As a consequence, an
increased fluorescence is observed on the second
fluorophoric molecule F2. The fluorescence is detected
by means of a fluorometer. The readings are transmitted
to a data-processing system.
The first primer Pl may also exhibit a hairpin loop,
the first fluorophoric molecule F1 being bound to a
first loop section and a quencher being bound to a loop
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
_ g _
section opposite at a distance which allows the
interaction to take place. When the hairpin loop is
closed, the interaction causes the fluorescence to be
quenched. As a result of hybridization with a
complementary strand C which is complementary to the
first primer P1 or by a synthesis which takes place on
the first primer P1, the hairpin loop is opened up. The
interaction between the fluorophoric molecule and the
quencher is eliminated. Excitation of the fluorophoric
molecules results in fluorescence.
Then, the next PCR cycle is started by raising the
temperature. The synthesis strand SSl and the synthesis
complementary strand SC1 are multiplied further, and,
as a result, the fluorescence intensity is increased.
The change in fluorescence intensity over the number of
PCR or LCR cycles is a measure for the initial
concentration of the target DNA: the more target DNA a
sample contains, the more rapidly the fluorescence
intensity increases.
To carry out the abovementioned method, a microtiter
plate M made of polycarbonate or polypropylene is used.
The first primer P1 is bound with its 5'-terminus in
the well area to the upper face of the microtiter plate
M to a polypropylene surface via a linker which is
preferably composed of 6 CH2 groups. The first primer
Pl is bound to the polypropylene surface by the method
of Weiler-J. and Hoheisel-JD. (Anal.- [sic] Biochem.,
1996; 243 (2) . 218-27).
Fig. 4 shows a further variant of the method. In this
variant, the first fluorophoric molecule F1 is bound
directly to the solid phase, i.e. the upper face of the
microtiter plate M. The first primer P1 is bound to the
solid phase in the vicinity of the first fluorophoric
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
9
molecule Fl. After hybridization of the synthesis
strands SSl or the synthesis complementary strands SCl,
excitation results in a radiation-free energy transfer
from the first fluorophoric molecule F1 (donor) to the
second fluorophoric molecule F2 (acceptor), where
fluorescence results (Fig. 5).
Fig. 6 shows the fluorescence of PCR products of the
PCR with primers which have 3'-fluorophores attached to
them. The fluorescence of the PCR product has been
measured in relative fluorescence units (RFU) at an
excitation wavelength of 496 nm and an emission
wavelength of 576 nm. The sample ~~PCR without template"
is a PCR preparation without HGH template DNA after
25 cycles . The sample '~PCR with template" is a PCR mix
with HGH template DNA after 25 cycles. The column on
the right shows the PCR mix with template DNA, but
without temperature cycles having been carried out.
It can be seen clearly from Fig. 6 that the template
can be detected readily with the aid of the method
according to the invention, in particular without any
need for washing steps.
Example l:
Fluorescence energy transfer in PCR products of primers
which have 3'-fluorophores attached to them
Two primers labeled with fluorophoric groups in the 3'-
terminus zone are synthesized.
A first primer with a length of 23 bases has the
following sequence:
5'-ACCAGGAGTTTGTAAGCTCTTGG-3'.
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
wo 99/47700 PCT/DE99/00725
- 10 -
The thymidine in position 4 relative to the 3'-terminus
(emboldened in the sequence) is labeled with 6-
carboxyfluorescein (6-FAM). The FAM group is bound via
the amino group of the dT-C2-NH2, which is incorporated
during oligonucleotide synthesis.
A second primer with a length of 19 bases has the
following sequence:
5'-biotin-CCTGATGCGCACCCATTCC-3'
The thymidine in position 3 relative to the 3'-terminus
(emboldened in the sequence) is labeled with
carboxymehtylrhodamine [sic] (TAMRA). The TAMRA group
is bound via the amino group of the dT-C2-NH2, which is
incorporated during oligonucleotide synthesis. The
second primer is labeled at the 5'-terminus with a
biotin group.
The synthesis is carried out on a 0.2~mol scale. The
primers are purified by HPLC. The sequences of the
primers are in immediate vicinity of a sequence segment
of the human growth hormone gene (HGH gene).
First primer: 5'-ACCAGGAGTTTGTAAGCTCTTGG-3'
HGC: 5'-ACCAGGAGTTTGTAAGCTCTTGG-
GGAATGGGTGCGCATCAGG-3'
3'-TGGTCCTCAAACATTCGAGAACC-
CCTTACCCACGCGTAGTCC-5'
Second primer: 3'-CCTTACCCACGCGTAGTCC-biotin-5'
The first and second primers are reacted in a PCR using
a template DNA which covers the sequence segment of the
HGH gene. The PCR is carried out in a total volume of
501 with in each case 0.5~M primer, 2 units of Taq-DNA
polymerase and 1~1 of HGH gene (long) in the relevant
PCR buffers (all solutions and enzymes from Boehringer,
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 11 -
Mannheim). 25 cycles with an annealing temperature of
66°C (45 seconds), elongation temperature of 72°C (45
seconds) and a denaturation temperature of 94°C (30
seconds) are carried out.
As negative control, the same PCRs are carried out, but
without the template DNA. As a further control, the PCR
mix is left at 4°C.
The PCR results in the formation of the PCR product in
which the fluorophores of the first and second primers
are arranged on the strands of opposite polarity at a
distance of a few bases:
FAM
5'-ACCAGGAGTTTGTAAGCTCTTGG-GGAATGGGTGCGCATCAGG-3'
3'-TGGTCCTCAAACATTCGAGAACC-CCTTACCCACGCGTAGTCC-biotin-5'
TAMRA
A suitable excitation of the 5-FAM group at 496nm
results in a fluorescence energy transfer to the TAMRA
group which is located on the complementary strand and
which has an emission maximum at 576nm.
To detect the formation of the PCR product and the
fluorescence energy transfer, the fluorescence is
determined in a fluorescence spectrometer at an
excitation of 496nm(+/- lOnm) and an emission of 576 nm
(+/- lOnm). Owing to the PCR, the fluorescence of the
TAMRA group increases (Fig. 6). This increase in
fluorescence shows the formation of the expected PCR
product.
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 12 -
Example 2:
PCR with 3'-labeled and immobilized primers
The primers which described in Example 1 are also used
for the PCR with 3'-labeled and immobilized primers.
The 5'-biotinylated second primer in accordance with
Example 1 is bound by the PCR to streptavidin-coated,
superparamagnetic particles with a size of approx.
2.8 ~m in diameter (M-280 Dynabeads, Dynal, Hamburg). To
this end, the particles (10~g/~1; 6.7 x 108 particles/ml
suspended in phosphate-buffered saline (PBS) pH 7.4 are
washed with B/W buffer (lOmM Tris-C1, 1mM EDTA, 2M
NaCl) ph [sic] 7.5 and brought to a concentration of
5 ~g/~1 in B/W buffer. 20~t1 of this suspension are
treated with an equal volume of 50~M solution of
primer-2 in distilled water. The suspension is
incubated for 1 hour at room temperature with gentle
shaking. Unbound primer is removed by washing the
particles twice, first with 1001 of B/W buffer and
then by washing with lOmM TrisCl, 0.2 mM EDTA pH8 (TE).
The particles are stored at 4°C in TE in a suspension
of 10~g/~1.
The PCR with the second primer which is bound to the
supermagnetic particle is carried out as in Example 1.
In contrast to Example 1, l~l of the suspension of the
particle-bound primer-2 is employed instead of the free
second primer. After the PCR, the particles are washed
repeatedly with TE and analyzed on the fluorescence
microscope. What is studied is the attachment of the 6-
FAM-labeled first primer to the particles. Fig. 7A
demonstrates the fluorescence of the particles after
conclusion. The PCR mix shown in Fig. 7B shows a
fluorescence of the particles caused by the attachment
of the FAM-labeled first primer to the particles. This
fluorescence is not present in the control without
template DNA (Fig. 7D).
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 13 -
The symbols denote:
S Strand
C Complementary strand
P1 First primer
P2 Second primer
Fl First fluorophoric molecule
F2 Second fluorophoric molecule
SSl Synthesis strand
SCl Synthesis complementary strand
M Microtiter plate
N Nucleotide sequence
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
14
SEQUENCE PROTOCOLS
<110> november AG Novus Medicatus Bertling
Gesellschaft fur Molekolare [sic] Medizin
<120> Method and device for detecting a nucleotide
sequence
<130> 390687ga5
<140>
<141>
<160> 4
<170> PatentIn Ver. 2.1
<210> 1
<211> 23
<212> DNA
<213> human
<400> 1
accaggagtt tgtaagctct tgg 23
<210> 2
<211> 42
<212> DNA
<213> human
<400> 2
accaggagtt tgtaagctct tggggaatgg gtgcgcatca gg 42
<210> 3
<211> 42
<212> DNA
<213> human
REPLACEMENT SHEET (RULE 26)
CA 02323075 2000-09-14
WO 99/47700 PCT/DE99/00725
- 15 -
<400> 3
cctgatgcgc acccattccc caagagctta caaactcctg gt 42
<210> 4
<211> 19
<212> DNA
<213> human
<400> 4
cctgatgcgc acccattcc 19
REPLACEMENT SHEET (RULE 26)