Note: Descriptions are shown in the official language in which they were submitted.
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
IDENTIFICATION OF A cDNA ASSOCIATED WITH ISCHEMIA IN HUMAN
HEART TISSUE
FIELD OF THE INVENTION
The invention relates generally to the changes in gene expression in ischemic
heart
tissue compared to normal human heart tissue. The invention relates
specifically to a
novel human gene which is up-regulated in ischemic human heart tissue. This
application
is related to U.S. Provisional Application Serial No. 60/079,377, filed March
26, 1998,
which is incorporated herein by reference.
BACKGROUND OF THE INVENTION
Cardiovascular disease is a general diagnostic category consisting of several
separate diseases. Coronary heart disease and cerebrovascular disease are
major
components of cardiovascular disease with 478,530 dying of coronary heart
disease and
144,070 dying of cerebrovascular disease in the U.S. in 1991. See Cecil
Textbook of
Medicine, Bennet and Plum Eds., W.B. Saunders Co., 1996. Many of the acute
fon~ns of
coronary heart disease are caused by coronary artery abnormalities such as
coronary
atherosclerosis. Among the more common causes and contributing factors in
sudden
cardiac death are chmnic ischemic heart disease and ischemic cardiomyopathy.
Chronic ischemic heart disease and ischemic cardiomyopathy are caused in part
by
episodes of insufficient myocardial oxygen supply. Myocardial oxygen supply is
governed by coronary blood flow and the ability of the myocardium to extract
oxygen
from the blood delivered to it. Unlike other organs, the heart always extracts
oxygen with
near maximal efficiency from the blood. Even under situations of minimal
demand, there
is little potential for enhanced oxygen extraction to counter increased oxygen
demands.
Coronary blood flow, on the other hand, can increase several-fold in normal
subjects as a
result of coronary arterial vasodiladon. Coronary arterial vasodilation is
regulated by the
coronary endothelium which releases vasodilatory substances, most importantly
nitric
oxide.
In atherosclerotic coronary heart disease, endothelial dysfunction may
diminish
CA 02323574 2000-09-22
WO 99/49062 PCTNS99106662
-2-
production of vasodilatory substances, such as nitric oxide. Myocardial
ischemia results
when autoregulatory vasodilation is prevented, whether by flow-limiting
coronary arterial
stenosis or by endothelial dysfunction. In both cases, arterial blood flow can
no longer
increase proportional to rising oxygen demands. In other situations,
myocardial ischemia
may occur when oxygen demands are constant but there is a primacy decrease in
coronary
blood flow mediated via coronary artery spasm, rapid evolution of the
underlying
atherosclerotic plaque leading to a reduced coronary arterial lumen caliber,
andlor
intermittent microvascular plugging by platelet aggregates.
At the molecular level, ischemia is characterized by the differential
expression of
numerous genes compared to normal heart tissue. For instance, in human heart
failure
caused by ischemic cardiomyopathy, expression of the Vii,- and (32-adrenergic
receptors of
the adenyl cyclase signal transduction system is impaired by reductions in the
expression
of mRNA for each receptor (Ihl-Vahl et al., 1996, J. Mol. Cell. Cardiol. 28:1-
10).
Ischemic injury is also known to lead to the differential expression of heat
shock and
immediate early genes such as hsp70, c fos, c jun, jun-B as well the genes
encoding
angiotensin receptor subtypes (Plumier et al., 1996, J. Mol. Cell. Cardiol.
28:1251-1260;
Wharton et al., 1998, J. Pharmoc. Experiment. Therap. 284(1) 323-336; and
Heads et al.
1995, J. Mol. Cell. Cardiol. 27:2133-2148).
The identification of new genes that are differentially expressed in ischemic
heart
tissue will allow for the development of numerous diagnostic and therapeutic
applications
such as molecular probes and new agents which modulate the activity or
expression of
these genes.
SUMMARY OF THE INVENTION
The present invention is based on our discovery of a new gene which is up-
regulated in ischemic heart tissue. The invention includes isolated nucleic
acid molecules
selected from the group consisting of an isolated nucleic acid molecule that
encodes the
amino acid sequence of SEQ ID No.2, an isolated nucleic acid molecule that
encodes a
fragment of at least 10 amino acids of SEQ ID No.2, an isolated nucleic acid
molecule
which hybridizes to a nucleic acid molecule comprising SEQ ID No. 1 under
conditions of
sufficient stringency to produce a clear signal and an isolated nucleic acid
molecule which
hybridizes to a nucleic acid molecule that encodes the amino acid sequence of
SEQ ID No.
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
-3-
2 under conditions of sufficient stringency to produce a clear signal.
The present invention further includes the nucleic acid molecules operably
linked
to one or more expression control elements, including a vector comprising the
isolated
nucleic acid molecules. The invention further includes host cells transformed
to contain
the nucleic acid molecules of the invention and methods for producing a
protein
comprising the step of culturing a host cell transformed with the nucleic acid
molecule of
the invention under conditions in which the protein is expressed.
The invention further provides an isolated polypeptide selected from the group
consisting of an isolated polypeptide comprising the amino acid sequence of
SEQ ID No.2,
an isolated polypeptide comprising a fragment of at least 10 amino acids of
SEQ ID No.2,
an isolated polypeptide comprising conservative amino acid substitutions of
SEQ ID No.2
and naturally occurring amino acid sequence variants of SEQ ID No.2.
The invention further provides an isolated antibody that binds to a
polypeptide of
the invention, including monoconal and polyclonal antibodies.
The invention further provides methods of identifying an agent which modulates
the expression of a nucleic acid encoding the protein having the sequence of
SEQ ID No.2
comprising the steps of exposing cells which express the nucleic acid to the
agent and
determining whether the agent modulates expression of said nucleic acid,
thereby
identifying an agent which modulates the expression of a nucleic acid encoding
the protein
having the sequence of SEQ ID No.2.
The invention further provides methods of identifying an agent which modulates
at
least one activity of a protein comprising the sequence of SEQ ID No.2
comprising the
steps of exposing cells which express the protein to the agent and determining
whether the
agent modulates at least on activity of said protein, thereby identifying an
agent which
modulates at least one activity of a protein comprising the sequence of SEQ TD
No.2.
The invention further provides methods of identifying binding partners for a
protein comprising the sequence of SEQ ID No. 2, comprising the steps of
exposing said
protein to a potential binding partner and determining if the potential
binding partner binds
to said protein, thereby identifying binding partners for a protein comprising
the sequence
of SEQ ID No. 2
The present invention further provides methods of modulating the expression of
a
nucleic acid encoding the protein having the sequence of SEQ ID No.2
comprising the step
CA 02323574 2000-09-22
WO 99/49061 PCTIUS99/06662
-4-
of administering an effective amount of an agent which modulates the
expression of a
nucleic acid encoding the protein having the sequence of SEQ ll~ No.2. The
invention
also provides methods of modulating at least one activity of a protein
comprising the
sequence of SEQ ID No.2 comprising the step of administering an effective
amount of an
agent which modulates at least one activity of a protein comprising the
sequence of SEQ
)D No.2.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 is a section of an autoradiograph of the expression profile
generated from cDNAs made with RNA isolated from ischemic and control, non-
ischemic
heart tissue.
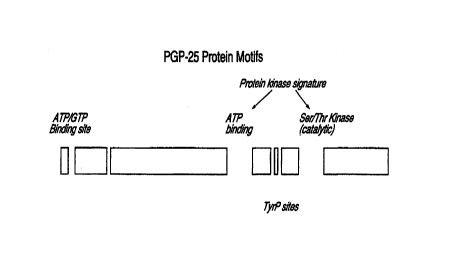
Figure 2 Figure 2 shows the potential binding, phosphorylation and
enzymatic sites of the protein of SEQ ID No.2.
Figure 3 is a Northern blot of RNA isolated from various tissues.
Figure 4 Figure 4 is a PCR-expression analysis of RNA isolated from various
tissues.
DETAILED DESCRIPTION OF THE PREFERRED EMBODIMENT
I. General Description
The present invention is based in part on identifying a new gene that is
differentially regulated or expressed in human ischemic heart tissue compared
to normal
human heart.tissue. This gene, which may be distantly related to human myosin
light
chain kinases, encodes a protein predicted to consist of 819 amino acids.
The protein can serve as a target for agents that can be used to modulate the
expression or activity of the protein. For example, agents may be identified
which
modulate biological processes associated with ischemic injury to the heart
such as chronic
ischemic heart disease and ischemic cardiomyopathy. Agents may also be
identified
which modulate the biological processes associated with recovery to ischemic
injury to the
heart.
The present invention is further based on the development of methods for
isolating
binding partners that bind to the protein. Probes based on the protein are
used as capture
probes to isolate potential binding partners, such as other proteins. Dominant
negative
CA 02323574 2000-09-22
WO 99/49062 PCT/US99l06662
-5-
proteins, DNAs encoding these proteins, antibodies to these proteins, peptide
fragments of
these proteins or mimics of these proteins may be introduced into cells to
affect function.
Additionally, these proteins provide a novel target for screening of synthetic
small
molecules and combinatorial or naturally occurring compound libraries to
discover novel
therapeutics to regulate heart function.
II. Specific Embodiments
A. The Protein Associated with Ischemic Heart Tissue
The present invention provides isolated protein, allelic variants of the
protein, and
conservative amino acid substitutions of the protein. As used herein, the
protein or
polypeptide refers to a protein that has the human amino acid sequence of that
depicted in
SEQ ID No.2. The invention includes naturally occurring allelic variants and
proteins that
have a slightly different amino acid sequence than that specifically recited
above. Allelic
variants, though possessing a slightly different amino acid sequence than
those recited
above, will still have the same or similar biological functions associated
with the 819
amino acid protein.
As used herein, the family of proteins related to the 819 amino acid pmtein
refer to
proteins that have been isolated from organisms in addition to humans. The
methods used
to identify and isolate other members of the family of proteins related to the
819 amino
acid protein are described below.
The proteins of the present invention are preferably in isolated form. As used
herein, a protein is said to be isolated when physical, mechanical or chemical
methods are
employed to remove the protein from cellular constituents that are normally
associated
with the protein. A skilled artisan can readily employ standard purification
methods to
obtain an isolated protein.
The proteins of the present invention further include conservative variants of
the
proteins herein described. As used herein, a conservative variant refers to
alterations in the
amino acid sequence that do not adversely affect the biological functions of
the protein. A
substitution, insertion or deletion is said to adversely affect the protein
when the altered
sequence prevents or disrupts a biological function associated with the
protein. For
example, the overall charge, structure or hydrophobic/hydrophilic properties
of the protein
can be altered without adversely affecting a biological activity. Accordingly,
the amino
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/0666Z
-6-
acid sequence can be altered, for example, to render the peptide more
hydrophobic or more
hydrophilic, without adversely affecting the biological activities of the
protein.
Ordinarily, the allelic variants, the conservative substitution variants, the
members
of the protein family, will have an amino acid sequence having at least 75%
amino acid
sequence identity with the human sequence set forth in SEQ ID No.2, more
preferably at
least 80%, even more preferably at least 90%, and most preferably at least
95%. Identity
or homology with respect to such sequences is defined herein as the percentage
of amino
acid residues in the candidate sequence that are identical with the known
peptides, after
aligning the sequences and introducing gaps, if necessary, to achieve the
rnaximum~percent
homology, and not considering any conservative substitutions as part of the
sequence
identity. N-terminal, C-terminal or internal extensions, deletions, or
insertions into the
peptide sequence shall not be construed as affecting homology.
Thus, the proteins of the present invention include molecules having the amino
acid sequence disclosed in SEQ ID No.2; fragments thereof having a consecutive
sequence
of at least about 3, 5, 10 or 15 amino acid residues of the 819 amino acid
protein; amino
acid sequence variants of such sequence wherein an amino acid residue has been
inserted
N- or C-terminal to, or within, the disclosed sequence; and amino acid
sequence variants
of the disclosed sequence, or their fragments as defined above, that have been
substituted
by another residue. Contemplated variants further include those containing
predetermined
mutations by, e.g., homologous recombination, site-directed or PCR
mutagenesis, and the
corresponding proteins of other animal species, including but not limited to
rabbit, rat,
marine, porcine, bovine, ovine, equine and non-human primate species, and the
alleles or
other naturally occurring variants of the family of proteins; and derivatives
wherein the
protein has been covalently modified by substitution, chemical, enzymatic, or
other
appropriate means with a moiety other than a naturally occurring amino acid
{for example
a detectable moiety such as an enzyme or radioisotope).
As described below, members of the family of proteins can be used: 1 ) to
identify
agents which modulate at least one activity of the protein, including agents
which may
modulate phosphorylation mediated by the protein, 2) in methods of identifying
binding
partners for the protein, 3) as an antigen to raise polyclonal or monoclonal
antibodies, and
4) as a therapeutic agent.
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
_7_
B. Nucleic Acid Molecules
The present invention further provides nucleic acid molecules that encode the
protein having SEQ ID No.2 and the related proteins herein described,
preferably in
isolated form. As used herein, "nucleic acid" is defined as RNA or DNA that
encodes a
peptide as defined above, or is complementary to nucleic acid sequence
encoding such
peptides, or hybridizes to such nucleic acid and remains stably bound to it
under
appropriate stringency conditions, or encodes a polypeptide sharing at least
75% sequence
identity, preferably at least 80%, and more preferably at least 85%, with the
peptide
sequences. Specifically contemplated are genomic DNA, cDNA, mRNA and antisense
molecules, as well as nucleic acids based on alternative backbone or including
alternative
bases whether derived from natural sources or synthesized. Such hybridizing or
complementary nucleic acids, however, are defined further as being novel and
unobvious
over any prior art nucleic acid including that which encodes, hybridizes under
appropriate
stringency conditions, or is complementary to nucleic acid encoding a protein
according to
the present invention.
"Stringent conditions" are those that (1) employ low ionic strength and high
temperature for washing, for example, 0.01 SM NaC1/0.0015M sodium titrate/0.1
% SDS at
50°C., or {2) employ during hybridization a denaturing agent such as
formamide, for
example, 50% {vol/vol) formamide with 0.1 % bovine serum albumin/0.1 %
Fico11/0. l
polyvinylpyrrolidone/50 mM sodium phosphate buffer at pH 6.5 with 750 mM NaCI,
75
mM sodium citrate at 42°C. Another example is use of 50% formatnide, 5
x SSC (0.75M
NaCI, 0.075 M sodium citrate), 50 mM sodium phosphate (pH fi.8), 0.1 % sodium
pyrophosphate, 5 x Denhardt's solution, sonicated salmon sperm DNA (50 pg/ml),
0.1%
SDS, and 10% dextran sulfate at 42°C., with washes at 42°C. in
0.2 x SSC and 0.1% SDS.
A skilled artisan can readily determine and vary the stringency conditions
appropriately to
obtain a clear and detectable hybridization signal.
As used herein, a nucleic acid molecule is said to be "isolated" when the
nucleic
acid molecule is substantially separated from contaminant nucleic acid
encoding other
polypeptides from the source of nucleic acid.
The present invention further provides fragments of the encoding nucleic acid
molecule. As used herein, a fragment of an encoding nucleic acid molecule
refers to a small
portion of the entire protein encoding sequence. The size of the fragment will
be determined
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
_g_
by the intended use. For example, if the fragment is chosen so as to encode an
active portion
of the protein, the fragment will need to be large enough to encode the
functional regions) of
the protein. If the fiagment is to be used as a nucleic acid probe or PCR
primer, then the
fragment length is chosen so as to obtain a relatively small .number of false
positives during
probing/priming.
Fragments of the encoding nucleic acid molecules of the present invention
(i.e.,
synthetic oligonucleotides) that are used as probes or specific primers for
the polymerase chain
reaction (PCR), or to synthesize gene sequences encoding proteins of the
invention can easily
be synthesized by chemical techniques, for example, the phosphotriester method
of Matteucci
et al., (J. Am. Chem. Soc. 103:3185-3191, 1981) or using automated synthesis
methods. 1n
addition, larger DNA segments can readily be prepared by well known methods,
such as
synthesis of a group of oligonucleotides that define various modular segments
of the gene,
followed by ligation of oligonucleotides to build the complete modified gene.
The encoding nucleic acid molecules of the present invention may further be
1 S modified so as to contain a detectable label for diagnostic and probe
purposes. A variety
of such labels are known in the art and can readily be employed with the
encoding
molecules herein described. Suitable labels include, but are not limited to,
biotin,
radiolabeled nucleotides and the like. A skilled artisan can employ any of the
art known
labels to obtain a labeled encoding nucleic acid molecule.
Modifications to the primary structure itself by deletion, addition, or
alteration of the
amino acids incorporated into the protein sequence during translation can be
made without
destroying the activity of the protein. Such substitutions or other
alterations result in proteins
having an amino acid sequence encoded by a nucleic acid falling within the
contemplated
scope of the present invention.
C. Isolation of Other Related Nucleic Acid Molecules
As described above, the identification of the human nucleic acid molecule
having SEQ
lD No. l allows a skilled artisan to isolate nucleic acid molecules that
encode other members
of the protein family in addition to the human sequence herein described.
Further, the
presently disclosed nucleic acid molecules allow a skills artisan to isolate
nucleic acid
molecules that encode other members of the family of proteins in addition to
the 819 amino
acid protein having SEQ ID No.2.
CA 02323574 2000-09-22
WO 99/49062 PCT/US99I06662
_9_
Essentially, a skilled artisan can readily use the amino acid sequence of SEQ
ID No.2
to generate antibody probes to screen expression libraries prepared from
appropriate cells.
Typically, polyclonal antisenun from mammals such as rabbits immunized with
the purified
protein (as described below) or monoclonal antibodies can be used to probe a
mammalian
cDNA or genomic expression library, such as lambda gtll library, to obtain the
appropriate
coding sequence for other members of the protein family. The cloned cDNA
sequence can be
expressed as a fusion protein, expressed directly using its own control
sequences, or expressed
by constructions using control sequences appropriate to the particular host
used for expression
of the enzyme.
Alternatively, a portion of the coding sequence herein described can be
synthesized and
used as a probe to retrieve DNA encoding a member of the protein family from
any
mammalian organism. Uligomers containing approximately 18-20 nucleotides
(encoding
about a 6-7 amino acid stretch) are prepared and used to screen genomic DNA or
cDNA
libraries to obtain hybridization under stringent conditions or conditions of
sufficient stringency
to eliminate an undue level of false positives.
Additionally, pairs of oligonucleotide primers can be prepared for use in a
polymerase
chain reaction (PCR) to selectively clone an encoding nucleic acid molecule. A
PCR
denature/anneaUextend cycle for using such PCR primers is well known in the
art and can
readily be adapted for use in isolating other encoding nucleic acid molecules.
D. rDNA molecules Containing a Nucleic Acid Molecule
The present invention further provides recombinant DNA molecules (rDNAs) that
contain a coding sequence. As used herein; a rDNA molecule is a DNA molecule
that has
been subjected to molecular manipulation in situ. Methods for generating rDNA
molecules are
well known in the art. See for example, Sambrook et al., ~I~folecular Cloning
(1989). In the
preferred rDNA molecules, a coding DNA sequence is operably linked to
expression control
sequences and/or vector sequences.
The choice of vector and/or expression control sequences to which one of the
protein
family encoding sequences of the present invention is operably linked depends
directly, as is
well known in the art, on the functional properties desired, e.g., protein
expression, and the host
cell to be transformed. A vector contemplated by the present invention is at
least capable of
directing the replication or insertion into the host chromosome, and
preferably also expression,
CA 02323574 2000-09-22
WO 99!49062 PCT/US99/06662
-10-
of the structural gene included in the rDNA molecule.
Expression control elements that are used for regulating the expression of an
operably
linked protein encoding sequence are known in the art and include, but are not
limited to,
inducible promoters, constitutive promoters, secretion signals, and other
regulatory elements.
Preferably, the inducible promoter is readily controlled, such as being
responsive to a nutrient
in the host cell's m~ium.
In one embodiment, the vector containing a coding nucleic acid molecule will
include a
prokaryotic replicon, i.e., a DNA sequence having the ability to direct
autonomous replication
and maintenance of the recombinant DNA molecule extrachromosomally in a
prokaryotic host
cell, such as a bacterial host cell, transformed therewith. Such replicons are
well known in the
art. In addition, vectors that include a prokaryotic replicon may also include
a gene whose
expression confers a detectable marker such as a drug resistance. Typical
bacterial drug
resistance genes are those that confer resistance to ampicillin or
tetracycline.
Vectors that include a prokaryotic replicon can further include a prokaryotic
or
bacteriophage promoter capable of directing the expression (transcription and
translation) of
the coding gene sequences in a bacterial host cell, such as E. coli. A
promoter is an expression
control element formed by a DNA sequence that permits binding of RNA
polymerise and
transcription to occur. Promoter sequences compatible with bacterial hosts are
typically
provided in plasmid vectors containing convenient restriction sites for
insertion of a DNA
segment of the present invention. Typical of such vector plasrnids are pUCB,
pUC9, pBR322
and pBR329 available from Biorrd Laboratories, (Richmond; CA), pPL and pKK223
available
from Pharmacia, Piscataway, N.J.
Expression vectors compatible with eukaryotic cells, preferably those
compatible with
vertebrate cells, can also be used to form a rDNA molecules the contains a
coding sequence.
Eukaryotic cell expression vectors are well known in the art and are available
from several
commercial sources. Typically, such vectors are provided containing convenient
restriction
sites for insertion of the desired DNA segment. Typical of such vectors are
pSVL and pKSV-
10 (Pharmacia), pBPV-1/pML2d (International Biotechnologies, Inc.), pTDTl
(ATCC,
#31255), the vector pCDM8 described herein, and the like eukaryotic expression
vectors.
Eukaryotic cell expression vectors used to construct the rDNA molecules of the
present
invention may further include a selectable marker that is effective in an
eukaryotic cell,
preferably a drug resistance selection marker. A preferred drug resistance
marker is the gene
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
-11-
whose expression results in neomycin resistance, i.e., the neomycin
phosphotransferase (neo)
gene. (Southern et al., J. Mol. Anal. Genet. 1:327-341, 1982.) Alternatively,
the selectable
marker can be present on a separate plasmid, and the two vectors are
introduced by co-
transfection of the host cell, and selected by culturing in the appropriate
drug for the selectable
marker.
E. Host Cells Containing an Exogenously SuppUed Coding Nucleic Acid
Molecule
The present invention further provides host cells transformed with a nucleic
acid
molecule that encodes a protein of the present invention. The host cell can be
either
prokaryotic or eukaryotic. Eukaryotic cells useful for expression of a protein
of the invention
are not limited, so long as the cell line is compatible with cell culture
methods and compatible
with the propagation of the expression vector and expression of the gene
product. Preferred
eukaryotic host cells include, but are not limited to, yeast, insect and
mammalian cells,
preferably vertebrate cells such as those from a mouse, rat, monkey or human
cell line.
Preferred eukaryotic host cells include Chinese hamster ovary (CHO) cells
available from the
ATCC as CCL61, N»i Swiss mouse embryo cells NIH/3T3 available from the ATCC as
CRL
1658, baby hamster kidney cells (BHK), and the like eukaryotic tissue culture
cell lines.
Any prokaryotic host can be used to express a rDNA molecule encoding a protein
of
the invention. The prefen~ed prokaryotic host is E. coli.
Transformation of appropriate cell hosts with a rDNA molecule of the present
invention is accomplished by well known methods that typically depend on the
type of vector
used and host system employed. With regard to transformation of prokaryotic
host cells,
electroporation and salt treatment methods are typically employed; see, for
example, Cohen et
al., Proc. Natl. Acad. Sci. USA 69:2110, 1972; and Maniatis et al.,
IVIolecular Cloniy,g. A
Laboratory M, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1982).
With
regard to transformation of vertebrate cells with vectors containing rDNAs,
electroporation,
cationic lipid or salt treatment methods are typically employed; see, for
example, Graham et
al., Virol. 52:456,1973; Wigler et al., Proc. Natl. Acad. Sci. USA 76:1373-76,
1979.
Successfully transformed cells, i.e., cells that contain a rDNA molecule of
the present
invention, can be identified by well known techniques including the selection
for a selectable
marker. For example, cells resulting from the introduction of an rDNA of the
present invention
CA 02323574 2000-09-22
WO 99/49062 PCTIUS99/06662
-12-
can be cloned to produce single colonies. Cells from those colonies can be
harvested, lysed
and their DNA content examined for the presence of the rDNA using a method
such as that
described by Southern, J. Mol. Biol. 98:503, 1975, or Berent et al., Biotech.
3:208, 1985 or the
proteins produced from the cell assayed via an imrnunological method.
F. Production of Recombinant Proteins using a rDNA Molecule
The present invention further provides methods for producing a protein of the
invention
using nucleic acid molecules herein described. In general terms, the
production of a
recombinant form of a protein typically involves the following steps:
First, a nucleic acid molecule is obtained that encodes a protein of the
invention, such
as the nucleic acid molecule depicted in SEQ 1D No. 1, or nucleotides 136-2592
of SEQ ID
No.l. If the encoding sequence is uninterrupted by introns, it is directly
suitable for expression
in any host.
The nucleic acid molecule is then preferably placed in operable linkage with
suitable
1 S control sequences, as described above, to form an expression unit
containing the protein open
reading frame. The expression unit is used to transform a suitable host and
the transformed
host is cultured under conditions that allow the production of the recombinant
protein.
Optionally the recombinant protein is isolated from the medium or from the
cells; recovery and
purification of the protein may not be necessary in some instances where some
impurities may
be tolerated.
Each of the foregoing steps can be done in a variety of ways. For example, the
desired
coding s~uences may be obtained from genomic fragments and used directly in
appropriate
hosts. The construction of expression vectors that are operable in a variety
of hosts is
accomplished using appropriate replicons and control sequences, as set forth
above. The
control sequences, expression vectors, and transformation methods are
dependent on the type
of host cell used to express the gene and were discussed in detail earlier.
Suitable restriction
sites can, if not normally available, be added to the ends of the coding
sequence so as to
provide an excisable gene to insert into these vectors. A skilled artisan can
readily adapt any
host/expression system known in the art for use with the nucleic acid
molecules of the
invention to produce recombinant protein.
CA 02323574 2000-09-22
WO 99/49062 PCTNS99106662
-13-
G. Methods to Identify Binding Partners
Another embodiment of the present invention provides methods for use in
isolating
and identifying binding partners of proteins of the invention. In detail, a
protein of the
invention is mixed with a potential binding partner or an extract or fraction
of a cell under
conditions that allow the association of potential binding partners with the
protein of the
invention. After mixing, peptides, polypeptides, proteins or other molecules
that have
become associated with a protein of the invention are separated from the
mixture. The
binding partner that bound to the protein of the invention can then be removed
and further
analyzed. To identify and isolate a binding partner, the entire protein, for
instance the
entire 819 amino acid protein of SEQ ID No.2 can be used. Alternatively, a
fragment of
the protein can be used.
As used herein, a cellular extract refers to a preparation or fraction which
is made
from a lysed or disrupted cell. The preferred source of cellular extracts will
be cells
derived from human heart tissue, for instance, ischemic human heart tissue.
Alternatively,
cellular extracts may be prepared from normal human heart tissue or available
cell lines,
particularly heart or muscle derived cell lines.
A variety of methods can be used to obtain an extract of a cell. Cells can be
disrupted using either physical or chemical disruption methods. Examples of
physical
disruption methods include, but are not limited to, sonication and mechanical
shearing.
Examples of chemical lysis methods include, but are not limited to, detergent
lysis and
enzyme lysis. A skilled artisan can readily adapt methods for preparing
cellular extracts in
order to obtain extracts for use in the present methods.
Once an extract of a cell is prepared, the extract is mixed with the protein
of the
invention under conditions in which association of the protein with the
binding partner can
occur. A variety of conditions can be used, the most preferred being
conditions that
closely resemble conditions found in the cytoplasm of a human cell. Features
such as
osmolarity, pH, temperature, and the concentration of cellular extract used,
can be varied
to optimize the association of the protein with the binding partner.
After mixing under appropriate conditions, the bound complex is separated from
the mixture. A variety of techniques can be utilized to separate the mixture.
For example,
antibodies specific to a protein of the invention can be used to
immunoprecipitate the
binding partner complex. Alternatively, standard chemical separation
techniques such as
CA 02323574 2000-09-22
WO 99149062 PCT/US99/06662
-14-
chromatography and density/sediment centrifugation can be used.
After removal of nonassociated cellular constituents found in the extract, the
binding partner can be dissociated from the complex using conventional
methods. For
example, dissociation can be accomplished by altering the salt concentration
or pH of the
mixture.
To aid in separating associated binding partner pairs from the mixed extract,
the
protein of the invention can be immobilized on a solid support. For example,
the pmtein
can be attached to a nitrocellulose matrix or acrylic beads. Attachment of the
protein to a
solid support aids in separating peptide/binding partner pairs from other
constituents found
in the extract. The identified binding partners can be either a single protein
or a complex
made up of two or more proteins.
Alternatively, the nucleic acid molecules of the invention can be used in a
yeast
two-hybrid system. The yeast two-hybrid system has been used to identify other
protein
partner pairs and can readily be adapted to employ the nucleic acid molecules
herein
described.
H. Methods to Identify Agents that Modulate the Expression a Nucleic Acid
Encoding the Ischemic Heart Associated Protein
Another embodiment of the present invention provides methods for identifying
agents that modulate the expression of a nucleic acid encoding a protein of
the invention
such as a protein having the amino acid sequence of SEQ ID No. 2. Such assays
may
utilize any available means of monitoring for changes in the expression level
of the nucleic
acids of the invention. As used herein, an agent is said to modulate the
expression of a
nucleic acid of the invention, for instance a nucleic acid encoding the
protein having the
sequence of SEQ 117 No.2, if it is capable of up- or down-regulating
expression of the
nucleic acid in a cell.
In one assay format, cell lines that contain reporter gene fusions between the
open
reading frame defined by nucleotides 136-2592 of SEQ ID No. l and any
assayable fusion
partner may be prepared. Numerous assayable fusion partners are known and
readily
available including the firefly luciferase gene and the gene encoding
chloramphenicol
acetyltransferase (Alam et al. (1990) Anal. Biochem. 188:245-254). Cell lines
containing
the reporter gene fusions are then exposed to the agent to be tested under
appropriate
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
-15 -
conditions and time. Differential expression of the reporter gene between
samples exposed
to the agent and contml samples identifies agents which modulate the
expression of a
nucleic acid encoding the protein having the sequence of SEQ ID No.2.
Additional assay formats may be used to monitor the ability of the agent to
modulate the expression of a nucleic acid encoding a protein of the invention
such as the
protein having SEQ ID No.2. For instance, mRNA expression may be monitored
directly
by hybridization to the nucleic acids of the invention. Cell lines are exposed
to the agent
to be tested under appropriate conditions and time and total RNA or mRNA is
isolated by
standard procedures such those disclosed in Sambmok et al. (Molecular Cloning:
A
Laboratory Manual, 2nd Ed. Clod Spring Harbor Laboratory Press, 1989).
Probes to detect differences in RNA expression levels between cells exposed to
the
agent and control cells may be prepared from the nucleic acids of the
invention. It is
preferable, but not necessary, to design probes which hybridize only with
target nucleic
acids under conditions of high stringency. Only highly complementary nucleic-
acid
hybrids form under conditions of high stringency. Accordingly, the stringency
of the
assay conditions determines the amount of complementarity which should exist
between
two nucleic acid strands in order to form a hybrid. Stringency should be
chosen to
maximize the difference instability between the probeaarget hybrid and
potential
probe:non-target hybrids.
Probes may be designed from the nucleic acids of the invention through methods
known in the art. For instance, the G+C content of the probe and the probe
length can
affect probe binding to its target sequence. Methods to optimize probe
specificity are
commonly available in Sambrook et al. (Molecular Cloning: A Laboratory
Approach,
Cold Spring Harbor Press, NY, 1989) or Ausubel et al. (Current Protocols in
Molecular
Biology, Greene Publishing Co., NY, 1995).
Hybridization conditions are modified using known methods, such as those
described by Sambrook et al. and Ausubel et al. as required for each probe.
Hybridization
of total cellular RNA or RNA enriched for polyA RNA can be accomplished in any
available format. For instance, total cellular RNA or RNA enriched for polyA
RNA can
be affixed to a solid support and the solid support exposed to at least one
probe comprising
at least one, or part of one of the sequences of the invention under
conditions in which the
probe will specifically hybridize. Alternatively, nucleic acid fragments
comprising at least
CA 02323574 2000-09-22
WO 99149062 PCT/US991066b2
-16-
one, or part of one of the sequences of the invention can be affixed to a
solid support, such
as a porous glass wafer. The glass wafer can then be exposed to total cellular
RNA or
polyA RNA from a sample under conditions in which the affixed sequences will
specifically hybridize. Such glass wafers and hybridization methods, such as
those
disclosed by Beattie (WO 95/11755), are widely available. By examining for the
ability of
a given probe to specifically hybridize to an RNA sample from an untreated
cell
population and from a cell population exposed to the agent, agents which up or
down
regulate the expression of a nucleic acid encoding the protein having the
sequence of SEQ
ID No.2 are identified.
I. Methods to Identify Agents that Modulate at Least One Activity of the
Ischemic Heart Associated Protein
Another embodiment of the present invention provides methods for identifying
agents that modulate at least one activity of a protein of the invention such
as the protein
having the amino acid sequence of SEQ 1D No.2. Such methods or assays may
utilize any
means of monitoring or detecting the desired activity.
In one format, the relative amounts of a protein of the invention between a
cell
population that has been exposed to the agent to be tested compared to an un-
exposed
control cell population may be assayed. In this format, probes such as
specific antibodies
are used to monitor the differential expression of the protein in the
different cell
populations. Cell lines or populations are exposed to the agent to be tested
under
appropriate conditions and time. Cellular lysates may be prepared from the
exposed cell
line or population and a control, unexposed cell line or population. The
cellular lysates are
then analyzed with the probe.
Antibody pmbes are prepared by immunizing suitable mammalian hosts in
appropriate immunization protocols using the peptides, polypeptides or
proteins of the
invention if they are of sufficient length, or, if desired, or if required to
enhance
immunogenicity, conjugated to suitable carriers. Methods for preparing
immunogenic
conjugates with carners such as BSA, KLH, or other carrier proteins are well
known in the
art. In some circumstances, direct conjugation using, for example,
carbodiimide reagents
may be effective; in other instances linking reagents such as those supplied
by Pierce
Chemical Co., Rockford, IL, may be desirable to provide accessibility to the
hapten. The
CA 02323574 2000-09-22
WO 99!49062 PCTlUS99/06662
-17-
hapten peptides can be extended at either the amino or carboxy terminus with a
Cys
residue or interspersed with cysteine residues, for example, to facilitate
linking to a carrier.
Administration of the immunogens is conducted generally by injection over a
suitable time
period and with use of suitable adjuvants, as is generally understood in the
art. During the
immunization schedule, titers of antibodies are taken to determine adequacy of
antibody
formation.
While the polyclonal antisera produced in this way may be satisfactory for
some
applications, for pharmaceutical compositions, use of monoclonal preparations
is
preferred. Immortalized cell lines which secrete the desired monoclonal
antibodies may be
prepared using the standard method of Kohler and Milstein or modifications
which effect
immortalization of lymphocytes or spleen cells, as is generally known. The
immortalized
cell lines secreting the desired antibodies are screened by immunoassay in
which the
antigen is the peptide hapten, polypeptide or protein. When the appropriate
immortalized
cell culture secreting the desired antibody is identified, the cells can be
cultured either in
1 S vitro or by production in ascites fluid.
The desired monoclonal antibodies are then recovered from the culture
supernatant
or from the ascites supernatant. Fragments of the monoclonals or the
polyclonal antisera
which contain the immunologically significant portion can be used as
antagonists, as well
as the intact antibodies. Use of immunologically reactive fragments, such as
the Fab, Fab',
of F(ab')i fragments is often preferable, especially in a therapeutic context,
as these
fragments are generally less immunogenic than the whole immunoglobulin.
The antibodies or fragments may also be produced, using current technology, by
recombinant means. Regions that bind specifically to the desired regions of
receptor can
also be produced in the context of chimeras with multiple species origin.
In an alternative format, a specific activity of a protein of the invention
may be
assayed, such as the ability of the protein to phosphorylate a substrate such
as myosin.
Cell lines or populations are exposed under appropriate conditions to the
agent to be
tested. Agents which modulate the kinase activity of the protein of the
invention are
identified by assaying the kinase activity of the protein from the exposed
cell line or
population and a control, unexposed cell line or population, thereby
identifying agents
which modulate the kinase activity of the protein.
Kinase assays to measure the ability of the agent to modulate the kinase
activity of
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
-18-
a protein of the invention are widely available such as the assays disclosed
by Mishima et
al. (1996) .1. Biochem. 119:906-913) and Michnoff et al. (1986) J. Biol. Chem.
261:8320-
8326. Alternative assay formats include actin-myosin motility assays such as
those
disclosed by Kohama et al. (1996) TIPS 17:284-287 or Warrick et al. (1987)
Ann. Rev.
S Cell. Biol. 3:379-421.
Agents that are assayed in the above method can be randomly selected or
rationally
selected or designed. As used herein, an agent is said to be randomly selected
when the
agent is chosen randomly without considering the specific sequences involved
in the
association of the a protein of the invention alone or with its associated
substrates, binding
partners, etc. An example of randomly selected agents is the use a chemical
library or a
peptide combinatorial library, or a growth broth of an organism.
As used herein, an agent is said to be rationally selected or designed when
the
agent is chosen on a nonrandom basis which takes into account the sequence of
the target
site and/or its conformation in connection with the agent's action. As
described in the
Examples, there are proposed binding sites for ATP/GTP and calmodulin as well
as
cAMP/cGMP kinase sites, Tyre sites and Ser/Thr kinase (catalytic) sites in the
protein
having SEQ ID No.2. Agents can be rationally selected or rationally designed
by utilizing
the peptide sequences that make up these sites. For example, a rationally
selected peptide
agent can be a peptide whose amino acid sequence is identical to the ATP or
calmodulin
binding sites or domains.
The agents of the present invention can be, as examples, peptides, small
molecules,
vitamin derivatives, as well as carbohydrates. A skilled artisan can readily
recognize that
there is no limit as to the structural nature of the agents of the present
invention.
The peptide agents of the invention can be prepared using standard solid phase
(or
solution phase) peptide synthesis methods, as is known in the art. In
addition, the DNA
encoding these peptides may be synthesized using commercially available
oligonucleotide
synthesis instrumentation and produced recombinantly using standard
recombinant
production systems. The production using solid phase peptide synthesis is
necessitated if
non-gene-encoded amino acids are to be included.
Another class of agents of the present invention are antibodies immunoreactive
with critical positions of proteins of the invention. Antibody agents are
obtained by
immunization of suitable mammalian subjects with peptides that contain as
antigenic
CA 02323574 2000-09-22
WO 99/49062 PCTIUS99106662
-19-
regions those portions of the protein intended to be targeted by the
antibodies.
J. Uses for Agents that Modulate at Least One Activity of the Ischemic Heart
Associated Protein
As provided in the Examples, the proteins and nucleic acids of the invention,
such
as the protein having the amino acid sequence of SEQ ID No.l, are up-regulated
in
ischemic heart tissue. Agents that modulate or down-regulate the expression of
the protein
or agents such as agonists or antagonists of at least one activity of the
protein may be used
to modulate biological and pathologic processes associated with the protein's
function and
activity.
As used herein, a subject can be any mammal, so long as the mammal is in need
of
modulation of a pathological or biological process mediated by a protein of
the invention.
The term "mammal" is meant an individual belonging to the class Mammalia. The
invention is particularly useful in the treatment of human subjects.
As used herein, a biological or pathological process mediated by a protein of
the
invention may include binding of substrates such as ATP, GTP or calmodulin or
phosphorylation of a substrate such as skeletal myosin.
Pathological processes refer to a category of biological processes which
produce a
deleterious effect. For example, expression or up-regulation of expression of
a protein of
the invention is associated with chronic ischemic heart disease and ischemic
cardiomyopathy. As used herein, an agent is said to modulate a pathological
process when
the agent reduces the degree or severity of the process. For instance, chronic
ischemic
heart disease or ischemic cardiomyopathy may be prevented or disease
progression
modulated after an ischemic event by the administration of agents which reduce
or
modulate in some way the expression or at least one activity of a protein of
the invention.
The agents of the present invention can be provided alone, or in combination
with
other agents that modulate a particular pathological process. For example, an
agent of the
present invention can be administered in combination with anti-thrombotic
agents. As
used herein, two agents are said to be administered in combination when the
two agents
are administered simultaneously or are administered independently in a fashion
such that
the agents will act at the same time.
The agents of the present invention can be administered via parenteral,
CA 02323574 2000-09-22
WO 99149062 PCT/US99106662
-20-
subcutaneous, intravenous, intramuscular, intraperitoneal, transdermal, or
buccal routes.
Alternatively, or concurrently, administration may be by the oral route. The
dosage
administered will be dependent upon the age, health, and weight of the
recipient, kind of
concurrent treatment, if any, frequency of treatment, and the nature of the
effect desired.
The present invention further provides compositions containing one or more
agents
which modulate expression or at least one activity of a protein of the
invention. While
individual needs vary, determination of optimal ranges of effective amounts of
each
component is within the skill of the art. Typical dosages comprise 0.1 to 100
pg/kg body
wt. The preferred dosages comprise 0.1 to 10 ~,g/kg body wt. The most
preferred dosages
comprise 0.1 to 1 pg/kg body wt.
In addition to the pharmacologically active agent, the compositions of the
present
invention may contain suitable pharmaceutically acceptable carriers comprising
excipients
and auxiliaries which facilitate processing of the active compounds into
preparations
which can be used pharmaceutically for delivery to the site of action.
Suitable
formulations for parenteral administration include aqueous solutions of the
active
compounds in water-soluble form, for example, water-soluble salts. In
addition,
suspensions of the active compounds as appropriate oily injection suspensions
may be
administered. Suitable lipophilic solvents or vehicles include fatty oils, for
example,
sesame oil, or synthetic fatty acid esters, for example, ethyl oleate or
triglycerides.
Aqueous injection suspensions may contain substances which increase the
viscosity of the
suspension include, for example, sodium carboxymethyl cellulose, sorbitol,
andlor dextran.
Optionally, the suspension may also contain stabilizers. Liposomes can also be
used to
encapsulate the agent for delivery into the cell.
The pharmaceutical formulation for systemic administration according to the
invention may be formulated for enterai, parenteral or topical administration.
Indeed, all
three types of formulations may be used simultaneously to achieve systemic
administration
of the active ingredient.
Suitable formulations for oral administration include hard or soft gelatin
capsules,
pills, tablets, including coated tablets, elixirs, suspensions, syrups or
inhalations and
controlled release forms thereof.
In practicing the methods of this invention, the compounds of this invention
may
be used alone or in combination, or in combination with other therapeutic or
diagnostic
CA 02323574 2000-09-22
WO 99149062 PCT/US99/Obbb2
-21-
agents. In certain preferred embodiments, the compounds of this invention may
be
coadministered along with other compounds typically prescribed for these
conditions
according to generally accepted medical practice, such as anticoagulant
agents,
thrombolytic agents, or other antithrombotics, including platelet aggregation
inhibitors,
tissue plasminogen activators, urokinase, prourokinase, streptokinase,
heparin, aspirin, or
warfarin. The compounds of this invention can be utilized in vivo, ordinarily
in mammals,
such as humans, sheep, horses, cattle, pigs, dogs, cats, rats and mice, or in
vitro.
K. Transgenic Animals
Transgenic animals containing a mutant, knock-out or modified gene
corresponding to the cDNA sequence of SEQ ID No. l or a construct to modify
the
expression level of the gene, for instance for up-regulating the expression,
are also
included in the invention. Transgenie animals are genetically modified animals
into which
recombinant, exogenous or cloned genetic material has been experimentally
transferred.
Such genetic material is often referred to as a "transgene". The nucleic acid
sequence of
the transgene, in this case a form of SEQ ID No.l, may be integrated either at
a locus of a
genome where that particular nucleic acid sequence is not otherwise normally
found or at
the normal locus for the transgene. The transgene may consist of nucleic acid
sequences
derived from the genome of the same species or of a different species than the
species of
the target animal.
The term "germ cell line transgenic animal" refers to a transgenic animal in
which
the genetic alteration or genetic information was introduced into a germ line
cell, thereby
conferring the ability of the transgenic animal to transfer the genetic
information to
offspring. If such offspring in fact possess some or all of that alteration or
genetic
information, then they too are transgenic animals.
The alteration or genetic information may be foreign to the species of animal
to
which the recipient belongs, foreign only to the particular individual
recipient, or may be
genetic information already possessed by the recipient. In the last case, the
altered or
introduced gene may be expressed differently than the native gene.
Transgenic animals can be produced by a variety of different methods including
transfection, electroporation, microinjection, gene targeting in embryonic
stem cells and
recombinant viral and retroviral infection (see, e.g., U.S. Patent No.
4,736,866; U.S. Patent
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06bb2
-22-
No. 5,602,307; Mullins et al. (1993) Hypertension 22(4):630-633; Brenin et al.
(1997)
Surg. Oncol. 6(2)99-110; Tuan (ed.), Recombinant Gene Expression Protocols,
Methods
in Molecular Biology No. 62, Humana Press (1997)).
A number of recombinant or transgenic mice have been produced, including those
which express an activated oncogene sequence {U.S. Patent No. 4,736,866);
express
simian SV 40 T-antigen (U.S. Patent No. 5,728,915); lack the expression of
interferon
regulatory factor 1 (IRF-1) (U.S. Patent No. 5,731,490); exhibit dopaminergic
dysfunction
(U.S. Patent No. 5,723,719); express at least one human gene which
participates in blood
pressure control (U.S. Patent No. 5,731,489); display greater similarity to
the conditions
existing in naturally occurnng Alzheimer's disease (U.S. Patent No.
5,720,936); have a
reduced capacity to mediate cellular adhesion (U.S. Patent No. 5,602,307);
possess a
bovine growth hormone gene (Clutter et al. (1996) Genetics 143{4):1753-1760);
or, are
capable of generating a fully human antibody response (McCarthy (1997) The
Lancet
349(9049):405).
I S While mice and rats remain the animals of choice for most transgenic
experimentation, in some instances it is preferable or even necessary to use
alternative
animal species. Transgenic procedures have been successfully utilized in a
variety of non-
murine animals, including sheep, goats, pigs, dogs, cats, monkeys,
chimpanzees, hamsters,
rabbits, cows and guinea pigs (see, e.g., Kim et al. (1997) Mol. Reprod. Dev.
46(4):515-
526; Houdebine (1995) Reprod. Nutr. Dev. 35(6):609-617; Petters (1994) Reprod.
Fertil.
Dev. 6(5):643-645; Schnieke et al. (1997) Science 278(5346):2130-2133; and
Amoah
(1997) J. Animal Science 75{2):578-585).
The method of introduction of nucleic acid fragments into recombination
competent mammalian cells can be by any method which favors co-transformation
of
multiple nucleic acid molecules. Detailed procedures for producing transgenic
animals are
readily available to one skilled in the art, including the disclosures in U.S.
Patent No.
5,489,743 and U.S. Patent No. 5,602,307.
Without further description, it is believed that one of ordinary skill in the
art can,
using the preceding description and the following illustrative examples, make
and utilize
the compounds of the present invention and practice the claimed methods. The
following
working examples therefore, specifically point out preferred embodiments of
the present
invention, and are not to be construed as limiting in any way the remainder of
the
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
-23-
disclosure.
EXAMPLES
Identi~,cadon of Differentially Expressed Ischemic Heart mRNA
Heart tissue was obtained from five male patients with inotrope-dependent post-
ischemic cardiomyopathy exhibiting severe myocyte and or cardiac hypertrophy
with at
least three years since their first myocardial infarction. Heart tissue was
also obtained
from 5 female patients with idiopathic dilated cardiomyopathy exhibiting
severe myocyte
and/or cardiac hypertrophy and CHF duration of at least 2 years.
Total cellular RNA was prepared from the heart tissue described above as well
as from control, non-ischemic heart tissue using the procedure of Newburger et
al. (1981)
J. Biol. Chem. 266(24): 16171-7 and Newburger et al. (1988) Proc. Natl. Acad.
Sci. USA
85:5215-5219.
Synthesis of cDNA was performed as previously described by Prashar et al. in
WO 97/05286 and in Prashar et al. (1996) Proc. Natl. Acad. Sci. USA 93:659-
663.
Briefly, cDNA was synthesized according to the protocol described in the
GIBCOBRL
kit for cDNA synthesis. The reaction mixture for first-strand synthesis
included 6 pg of
total RNA, and 200 ng of a mixture of 1-base anchored oligo(dT) primers with
all three
possible anchored bases
(ACGTAATACGACTCACTATAGGGCGAATTGGGTCGACTTTTTTTTTTTTTTTTT
nl wherein nl=A/C or G) (SEQ ID No.3) along with other components for first-
strand
synthesis reaction except reverse transcriptase. This mixture was incubated at
65°C for
Sm, chilled on ice and the process repeated. Alternatively, the reaction
mixture may
include l0pg of total RNA, and 2 pmol of 1 of the 2-base anchored oligo(dT)
primers a
heel such as RP5.0 {CTCTCAAGGATCTTACCGCTT,BAT) (SEQ ID No.4), or RP6.0
(TAATACCGCGCCACATAGCAT,BCG) (SEQ ID No.S), or RP9.2
{CAGGGTAGACGACGCTACGCT,BGA) (SEQ ID No.6) along with other components
for first-strand synthesis reaction except reverse transcriptase. This mixture
was then
layered with mineral oil and incubated at 65 °C for 7 min followed by
50°C for another 7
min. At this stage, 2~c1 of Superscript reverse transcriptase (200 units/~cl;
GIBCOBRL)
was added quickly and mixed, and the reaction continued for 1 hr at 45-
50°C. Second-
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
-24-
strand synthesis was performed at 16 °C for 2 hr. At the end of the
reaction, the cDNAs
were precipitated with ethanol and the yield of cDNA was calculated. In our
experiments,
200 ng of cDNA was obtained from l0~cg of total RNA.
The adapter oligonucleotide sequences were
A1 (TAGCGTCCGGCGCAGCGACGGCCAG) (SEQ ID No.7) and
A2 (GATCCTGGCCGTCGGCTGTCTGTCGGCGC) (SEQ ID No.B). One
microgram of oligonucleotide A2 was first phosphorylated at the 5' end using
T4
polynucleotide kinase (PNK). After phosphorylation, PNK was heated denatured,
and 1/.cg
of the oligonucleotide A1 was added along with lOX annealing buffer (1 M
NaCI/100
mM Tris-HCI, pH8.0/10 mM EDTA, pH8.0} in a final vol of 20 ~cl. This mixture
was then
heated at 65 °C for 10 min followed by slow cooling to room temperature
for 30 min,
resulting in formation of the Y adapter at a final concentration of 100 ngl~l.
About 20 ng
of the cDNA was digested with 4 units of Bgl II in a final vol of l0,ul for 30
min at 37°C.
Two microliters (~4 ng of digested cDNA) of this reaction mixture was then
used for
ligation to 100 ng (~50-fold) of the Y-shaped adapter in a final vol of S,ul
for 16 hr at
15 °C. After ligation, the reaction mixture was diluted with water to a
final vol of 80 ~1
(adapter ligated cDNA concentration, ~ 50 pg/,ul) and heated at 65 °C
for 10 min to
denature T4 DNA ligase, and 2-,ul aliquots (with ~ 100 pg of cDNA) were used
for PCR.
The following sets of primers were used for PCR amplification of the adapter
ligated 3' -end cDNAs:
TGAAGCCGAGACGTCGGTCG(T),$ nl, n2 (wherein nl, n2 = AA, AC, AG
AT CA CC CG CT GA GC GG and GT) (SEQ 117 No.9) as the 3' primer with A1 as the
5'
primer or alternatively RP 5.0, RP 6.0, or RP 9.2 used as 3' primers with
primer Ai.l
serving as the S' primer. To detect the PCR products on the display gel, 24
pmol of
oligonucleotide A1 or Al.l was 5' -end-labeled using 15 ~ul of [y 32 P]ATP
(Amersham;
3000 Ci/mmol) and PNK in a final volume of 20 ~1 for 30 min at 37°C.
After heat
denaturing PNK at 65 °C for 20 min, the labeled oligonucleotide was
diluted to a final
concentration of 2 ~cM in 80 ~1 with unlabeled oligonucleotide Al .l. The PCR
mixture
{201) consisted of 2 /.d (~ 100 pg) of the template, 2~c1 of lOx PCR buffer
(100 mM
Tris~HCl, pH 8.3/500 mM KCl), 2 ~cl of 15 mM MgCl2 to yield 1.5 mM final Mgz+
concentration optimum in the reaction mixture, 200 ~cM dN'TPs, 200 nM each 5 '
and 3'
PCR primers, and 1 unit of Amplitaq Gold. Primers and dNTPs were added after
CA 02323574 2000-09-22
WO 99149062 PCT/US99/06662
- 25 -
preheating the reaction mixture containing the rest of the components at 85
°C. This "hot
start" PCR was done to avoid artefactual amplification arising out of
arbitrary annealing of
PCR primers at lower temperature during transition from room temperature to 94
°C in the
first PCR cycle. PCR consisted of 5 cycles of 94°C for 30 sec,
55°C for 2 min, and 72°C
for 60 sec followed by 25 cycles of 94°C for 30 sec, 60°C for 2
min, and 72°C for 60 sec.
A higher number of cycles resulted in smeary gel patterns. PCR products (2.51)
were
analyzed on 6% polyacrylamide sequencing gel. For double ox multiple digestion
following adapter Iigation, 13.2 ~cl of the ligated cDNA sample was digested
with a
secondary restriction enzymes) in a final vol of 20 ~cl. From this solution,
3~c1 was used as
template for PCR. This template vol of 3 ,ul carried = 100 pg of the cDNA and
10 mM
MgClz (from the lOX enzyme buffer), which diluted to the optimum of 1.5 mM in
the final
PCR vol of 20 ~cl. Since Mg2+ comes from the restriction enzyme buffer, it was
not
included in the reaction mixture when amplifying secondarily cut cDNA.
Individual
cDNA fragments corresponding to mRNA species were separated by denaturing
polyacrylamide gel electrophoresis and visualized by autoradiography. Bands
were
extracted from the display gels as described by Liang et al. (1995 Curr. Opin.
Immunol.
7:274-280), reamplified using the 5' and 3' primers, and subcloned into pCR-
Script with
high efficiency using the PCR-Script cloning kit from Stratagene. Plasmids
were
sequenced by cycle sequencing on an ABI automated sequencer. Alternatively,
bands
were extracted (cored) from the display gels, PCR amplified and sequenced
directly
without subcloning.
Figure 1 presents a section of an autoradiogram of the expression profile
generated from cDNAs made from RNA isolated from control and ischemic heart
tissue.
Ci-25 is a band that corresponds to a cDNA derived from a mRNA species that is
up-
regulated in ischemic heart tissue in a female patient with idiopathic dilated
cardiomyopathy exhibiting severe myocyte and/or cardiac hypertrophy and CHF
duration
of at least 2 years. The band corresponding to Ci-25 was sequenced. The
sequence of CI-
25 is:
GGCTCACATCTGTAATCCCAGCACTTTGGGAGGCCAAGGTGGGCAGATTGCTG
GCCAACATGGTAAAACCCCATCTCTAAAGATATAAAAATTAGCTGGGCGTGGT
GGCGCATACCTGTAATCCCAGCTACTTGGGAGGCTAAGGCACAAGAATCACTT
AAACAGGAGGCGGGGGTTGCAGTGAGCTGAGATCACACCACTGCACTCCAGC
CA 02323574 2000-09-22
WO 99/49062 PGTlUS99/06662
-26-
CTGGGTGGCAGAGCAAAACTTTGTCCCCACCCCTGACAAAAAACAAACAAAC
AAACAAAACAAAAAAAAACCTGTCAATTCA (SEQ ID No.lO).
Cloning of a Full Length cDNA Corresponding to Ci-25
The full length cDNA corresponding to Ci-25 band was obtained by the
oligo-pulling method. Briefly, a gene-specific oligo was designed based on
cDNA
fragment Ci-25. The oligo was labeled with biotin and used to hybridize with 2
ug of
single strand plasmid DNA (cDNA recombinants) from a human heart cDNA library
following the procedures of Sambrook et al.. The hybridized cDNAs were
separated by
streptavidin-conjugated beads and eluted by heating. The eluted cDNA was
converted to
double strand plasmid DNA and used to transform E. toll cells (DH10B) and the
longest
cDNA was screened. After confirmed by PCR using gene-specific primers, the
cDNA
clone was subjected to DNA sequencing.
The nucleotide sequence of the full-length cDNA corresponding to the
differentially regulated Ci-25 band is set forth in SEQ ID No.l. The cDNA
comprises
5532 base pairs with an open reading frame encoding a protein predicted to
contain 819
amino acids. The predicted amino acid sequence is presented in SEQ ID Nos. 1
and 2.
Comparison of the open reading to the sequences of known proteins and genes
indicates
that the gene may be distantly related to a known myosin light chain kinase
gene as it
exhibits 61% identity to a myosin light chain kinase at the nucleotide
sequence level.
The tissue distribution of RNA encoding the differentially regulated gene
encoding
the protein of SEQ ID N0.2 was analyzed by Northern Blot as well as ,PCR
expression
analysis of RNA isalated from various tissues. RNA was isolated from human
heart,
brain, placenta, lung, liver, skeletal muscle, kidney, leukocytes, testis and
pancreas using
standard procedures. Northern blots were prepared using a probe derived from
SEQ ID
No.l with hybridization conditions as described by Sambrook et al. PCR
expression
analysis was also performed using primers derived from SEQ ID No.l using
AmpliTaq
Gold PCR amplification kits (Perkin Elmer). Figure 3 is a Northern blot
demonstrating the
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
- 27 -
presence of specific RNA in heart and skeletal muscle. Figure 4 is a PCR
expression
analysis demonstrating the presence of specific RNA in heart and testis.
Example 4
G_ enera_~tj.on ojf TT~~genic Mice
Transgenic mice were generated using a gene corresponding to the cDNA sequence
of SEQ >D No.l . Briefly, a cDNA fragment encoding the protein of SEQ >D No.2
was
cloned into an alpha-MHC vector using standard techniques. The vector was then
used to
produce transgenic mice in accordance with standard techniques. Of the 22
resulting mice,
six were confirmed by Southern blot to be transgene positive.
Although the present invention has been described in detail with reference to
the
examples above, it is understood that various modifications can be made
without departing
from the spirit of the invention. Accordingly, the invention is limited only
by the
following claims. All cited patents and publications referred to in this
application are
herein incorporated by reference in their entirety.
CA 02323574 2000-09-22
WO 99/49062 PGTIUS99/06662
SEQUENCE LISTING
<110> Gene Logic, Inc., Larry Tiffany
<120> Identification of a cDNA.Associated with Ischemia in
Human Heart Disease, Yatindra PRASHAR, Inventor
<130> genelogic5005heartdiseasegene
<140> 09/
<141> 1999-03-26
<150> 60/079,377
<151> 1998-03-26
<160> 10
<170> PatentIn Ver. 2.0
<210> 1
<211> 5532
<212> DNA
<213> human gene expressed in ischemic heart disease
<220>
<221> CDS
<222> (136)..(2592)
<223> SEQ. ID. N0. 1
<400> 1
cgtggaggtt ggtgctgcca ggagtctgtc agctacggag gacaatgacc ttgcagacac 60
caccgcctga gtgagaacca ggggtctgtg cctctcctca ttccccgctc ttgcccttgt 120
caagcctgca ccagc atg tca gga acc tcc aag gag agt ctg ggg cat ggg 171
Met Ser Gly Thr Ser Lys Glu Ser Leu Gly His Gly
1 5 10
ggg ctg cca ggg ttg ggc aag acc tgc tta aca acc atg gac aca aag 219
Gly Leu Pro Gly Leu Gly Lys Thr Cys Leu Thr Thr Met Asp Thr Lys
15 20 25
ctg aac atg ctg aac gag aag gtg gac cag ctc ctg cac ttc caa gaa 267
Leu Asn Met Leu Asn Glu Lys Val Asp Gln Leu Leu His Phe Gln Glu
30 35 40
gat gtc aca gag aag ttg cag agc atg tgc cga gac atg ggc cac ctg 315
Aep Val Thr Glu Lya Leu Gla 8er Met Cye Arg Aep Met Gly Hia Leu
1
CA 02323574 2000-09-22
WO 99/49062 PCTIUS99I06662
45 50 55 60
gag cgg ggc ctg cac agg ctg gag gcc tcc cgg gca ccg ggc ccg ggc 363
Glu Arg Gly Leu His Arg Leu Glu Ala Ser Arg Ala Pro Gly Pro Gly
65 70 75
ggg get gat ggg gtt ccc cac att gac acc cag get ggg tgg ccc gag 911
Gly Ala Asp Gly Val Pro His Ile Asp Thr Gln Ala Gly Trp Pro Glu
80 85 90
gtc ctg gag ctg gtg agg gcc atg cag cag gat gcg gcc cag cac ggt 459
Val Leu Glu Leu Val Arg Ala Met Gln Gln Asp Ala Ala Gln His Gly
95 100 105
gcc agg ctg gag gcc ctc ttc agg atg gtg get gcg gtg gac agg gcc 507
Ala Arg Leu Glu Ala Leu Phe Arg Met Val Ala Ala Val Asp Arg Ala
110 115 120
atc get ttg gtg ggg qcc acg ttc cag aaa tca aag gtg gcg gat ttc 555
Ile Ala Leu Val Gly Ala Thr Phe Gln Lys Ser Lys Val Ala Asp Phe
125 130 135 190
ctc atg cag ggg cgt gtg ccc tgg agg aga ggc agc cca ggt gac agc 603
Leu Met Gln Gly Arg Val Pro Trp Arg Arg Gly Ser Pro Gly Asp Ser
145 150 155
cct gag gag aat aaa gag cga gtg gaa gaa gag gga gga aaa cca aag 651
Pro Glu Glu Asn Lys Glu Arg Val Glu Glu Glu Gly Gly Lys Pro Lys
160 165 170
cat gtg ctg agc acc agt ggg gtg cag tct gat gcc agg gag cct ggg 699
His Val Leu Ser Thr Ser Gly Val Gln Ser Asp Ala Arg Glu Pro Gly
175 180 185
gaa gag agc cag aag gcg gac gtg ctg gag ggg aca gcg gag agg ctg 747
Glu Glu Ser Gln Lys Ala Asp Val Leu Glu Gly Thr Ala Glu Arg Leu
190 195 200
ccc ccc atc aga gcg tca ggg ctg gga get gac ccc gcc cag gca gtg 795
Pro Pro Ile Arg Ala Ser Gly Leu Gly Ala Asp Pro Ala Gln Ala Val
205 210 215 220
gtc tca ccg ggc cag gga gat ggt gtt cct ggc cca gcc cag gca ttc 893
Val Ser Pro Gly Gln Gly Asp Gly Val Pro Gly Pro Ala Gln Ala Phe
225 230 235
cct ggc cac ctg ccc ctg ccc aca aag gtg gaa gcc aag get cct gag 891
Pro Gly His Leu Pro Leu Pro Thr Lys Val Glu Ala Lys Ala Pro Glu
2
CA 02323574 2000-09-22
WO 99/49062 PCT/US99106662
240 245 250
aca ccc agc gag aac ctc agg act ggc ctg gaa ttg get cca gca ccc 939
Thr Pro Ser Glu Asn Leu Arg Thr Gly Leu Glu Leu Ala Pro Ala Pro
255 260 265
ggc agg gtc aat gtg gtc tcc ccg agc ctg gag gtt gca cca ggt gca 987
Gly Arg Val Asn Val Val Ser Pro Ser Leu Glu Val Ala Pro Gly Ala
270 275 280
gga caa gga gca tcg tcc agc agg cct gac cct,gag ccc tta gag gaa 1035
Gly Gln Gly Ala Ser Ser Ser Arg Pro Asp Pro Glu Pro Leu Glu Glu
285 2.90 295 300
ggc acg agg ctg act cca ggg cct ggc cct cag tgc cca ggg cct cca 1083
Gly Thr Arg Leu Thr Pro Gly Pro Gly Pro Gln Cys Pro Gly Pro Pro
305 310 315
ggg ctg cca gcc cag gcc agg gca acc cac agt ggt gga gaa aca cct 1131
Gly Leu Pro Ala Gln Ala Arg Ala Thr His Ser Gly Gly Glu Thr Pro
320 325 330
cca agg atc tcc atc cac ata caa gag atg gat act cct ggg gag atg 1179
Pro Arg Ile Ser Ile His Ile Gln Glu Met Asp Thr Pro Gly Glu Met
335 340 345
ctg atg aca ggc agg ggc agc ctt gga ccc acc ctc acc aca gag get 1227
Leu Met Thr Gly Arg Gly 5er Leu Gly Pro Thr Leu Thr Thr Glu Ala
350 355 360
cca gca get gcc cag cca ggc aag cag ggc cca cct ggg acc ggg cgc 1275
Pro Ala Ala Ala Gln Pro Gly Lys Gln Gly Pro Pro Gly Thr Gly Arg
365 370 375 380
tgc ctc caa gcc cct ggg act gag ccc gga gaa cag acc cct gaa gga 1323
Cys Leu Gln Ala Pro Gly Thr Glu Pro Gly Glu Gln Thr Pro Glu Gly
385 390 395
gcc aga gag ctc tcc ccg ctg cag gag agc agc agc ccc ggg gga gtg 1371
Ala Arg Glu Leu Ser Pro Leu Gln Glu Ser Ser Ser Pro Gly Gly Val
900 405 410
aag gca gag gag gag caa agg get ggg gcc gag cct ggc acg aga cca 1919
Lys Ala Glu Glu Glu Gln Arg Ala Gly Ala Glu Pro Gly Thr Arg Pro
415 420 425
agc ttg gcc agg agt gac gac aat gac cac gag gtt ggg gcc ctg ggc 1467
Ser Leu Ala Arg Ser Asp Asp Asn Asp His Glu Val Gly Ala Leu Gly
3
CA 02323574 2000-09-22
WO 99149062 PCTIUS99106662
930 435 440
ctg cag cag ggc aaa agc cca ggg gcg gga aac cct gag cct gag cag 1515
Leu Gln Gln Gly Lys Ser Pro Gly Ala Gly Asn Pro Glu Pro Glu Gln
945 950 455 960
gac tgt gca gcc agg get ccg gtg aga get gaa gca gta agg agg atg 1563
Asp Cys Ala Ala Arg Ala Pro Val Arg Ala Glu Ala Val Arg Arg Met
465 470 475
ccc cca ggc gcc gag get ggc agc gtg gtt ctg gat gac agt ccg gcc 1611
Pro Pro Gly Ala Glu Ala Gly Ser Val Val Leu Asp Asp Ser Pro Ala
480 985 490
cca cca get cct ttt gaa cac cgg gta gtg agc gtc aag gag acc tcc 1659
Pro Pro Ala Pro Phe Glu His Arg Val Val Ser Val Lys Glu Thr Ser
995 500 505
atc tct gcg ggt tac gag gtg tgc cag cac gaa gtc ttg gga ggg ggt 170%
Ile Ser Ala Gly Tyr Glu Val Cys Gln His Glu Val Leu Gly Gly Gly
510 515 520
cgg ttt ggc cag gtc cac agg tgc aca gag aag tcc aca ggc ctc cca 1755
Arg Phe Gly Gln Val His Arg Cys Thr Glu Lys Ser Thr Gly Leu Pro
525 530 535 590
ctg get gcc aag atc atc aaa gtg aag agc gcc aag gac cgg gag gac 1803
Leu Ala Ala Lys Ile Ile Lys Val Lys Ser Ala Lys Asp Arg Glu Asp
595 550 555
gtg aag aac gag atc aac atc atg aac cag ctc agc cac gtg aac ctg 1851
Val Lys Asn Glu Ile Asn Ile Met Asn Gln Leu Ser His Val Asn Leu
560 565 570
atc cag ctc tat gac gcc ttc gag agc aag cac agc tgc acc ctt gtc 1899
Ile Gln Leu Tyr Asp Ala Phe Glu Ser Lys His Ser Cys Thr Leu Val
575 580 585
atg gag tac gtg gac ggg ggt gag ctc ttc gac cgg atc aca gat gag 1997
Met G1u Tyr Val Asp Gly Gly Glu Leu Phe Asp Arg Ile Thr Asp Glu
590 595 600
aag tac cac ctg act gag ctg gat gtg gtc ctg ttc acc agg cag atc 1995
Lys Tyr His Leu Thr Glu Leu Asp Val Val Leu Phe Thr Arg Gln Ile
605 610 615 620
tgt gag ggt gtg cat tac ctg cac cag cac tac atc ctg cac ctg gac 2043
Cys Glu Gly Val His Tyr Leu His Gln His Tyr Ile Leu His Leu Asp
4
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
625 630 635
ctc aag ccg gag aac ata ttg tgc gtc aat cag aca gga cat caa att 2091
Leu Lys Pro Glu Asn Ile Leu Cys Val Asn Gln Thr Gly His Gln Ile
690 645 650
aag atc att gac ttt ggg ctg gcc aga agg tac aag cct cga gag aag 2139
Lys Ile Ile Asp Phe Gly Leu Ala Arg Arg Tyr Lys Pro Arg Glu Lys
655 660 665
ctg aag gtg aac ttc ggc act cct gag ttc ctg gcc cca gaa gtc gtc 2187
Leu Lys Val Asn Phe Gly Thr Pro Glu Phe Leu Ala Pro Glu Val Val
670 675 680
aat tat gag ttt gtc tca ttc ccc aca gac atg tgg agt gtg gga gtc 2235
Asn Tyr Glu Phe Val Ser Phe Pro Thr Asp Met Trp Ser Val Gly Va=
685 690 695 700
atc acc tac atg cta ctc agt ggc ttg tcc cca ttt cta ggg gaa aca 2283
Ile Thr Tyr Met Leu Leu Ser Gly Leu Ser Pro Phe Leu Gly Glu Ti:r
705 710 ~ 715
gat gca gag acc.atg aat ttc att gta aac tgt agc tgg gat ttt gay 2331
Asp Ala Glu Thr Met Asn Phe Ile Val Asn Cys Ser Trp Asp Phe Asp
720 725 730
get gac acc ttt gaa ggg ctc tcg gag gag gcc aag gac ttt gtt tcc 2379
Ala Asp Thr Phe Glu Gly Leu Ser Glu Glu Ala Lys Asp Phe Val Ser
735 740 795
cgg ttg ctg gtc aaa gag aag agc tgc aga atg agt gcc aca cag tgc 2427
Arg Leu Leu Val Lys Glu Lys Ser Cys Arg Met Ser Ala Thr Gln Cys
750 755 760
ctg aaa cac gag tgg ctg aat aat ttg cct gcc aaa get tca aga tcc 2975
Leu Lys His Glu Trp Leu Asn Asn Leu Pro Ala Lys Ala Ser Arg Ser
765 770 775 780
aaa act cgt ctc aaa tcc caa cta ctg ctg cag aaa tac ata get caa 2523
Lys Thr Arg Leu Lys Ser Gln Leu Leu Leu Gln Lys Tyr Ile Ala Gln
785 790 795
aga aaa tgg aag aaa cat ttc tat gtg gtg act get gcc aac agg tt~ 2571
Arg Lys Trp Lys Lys His Phe Tyr Val Val Thr Ala Ala Asn Arg Lei
800 805 810
agg aaa ttt cca act tct ccc taatcttcaa ctctgctgct ccaatgggtc 2622
Arg Lys Phe Pro Thr Ser Pro
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
815
cagaaattac tgaggccagt ggtgaagtga agagatgact caaacattta aataatttgg 2682
ctttttggta ttattgattc cacttatttt gtaaaaatgg ttatggctgc tgccttcctt 2792
gtggatgaaa agtggctgta aagaagcttc ctaagaacgt ttttttctgc cttgtaagat 2802
cactacgtgt gaaatgctct gagtaccttt caaatatacc tacttttggt ggtaagtgta 2862
gggatgcttt aggtaggtac tttgcatctg tcgaatttaa attctaaact cacactgatt 2922
aaggaactca gtagactact ttgcaggggc catgttattc agtgttatct cctccagtac 2982
aaagaattcc tagaattttg atttgctcag gtgtgagctg acattttatt gtactacccc 3042
attcttgtgt taagccatgt ggatttagga cagtgatctt caaacttgct ttaacttatg 3102
ctcccttttg agaatcagca tcacttgaaa tgtcaaaata tgtcaactct catagccaaa 3162
tcagagaagt gatcattttg actgtgcttt ttaaatctcc acacacctcc acctctctcc 3222
taattctgcc tgtcttaacc cctctgtctt agttataaat ttctggtctt gtaagtctgg 3282
aagctgatag gcaatttatg aaagagataa gaatgtaatg aggttcagct ttctgagaaa 3392
cacagaaatg atacatcctg agacataaag gaaagctgct cttctgctgc ctcaggctgt 3902
agcactctca atgttgtcac tctacacata cactttctat atacatgtac agttgaccct 3462
tgaacagggt ttgaattgca gtcaacttaa atgtggattt tctttcacct ttgtcacccc 3522
tgagacagca acaccatgct ctcctcttca tcccactctg cagcctactc aacaggaaga 3582
tgatgaagat gaacaccttt atgatgatac actttcactt aatgaatagt aaacatatgt 3692
tttcctcctt atgattttag taacttttct ctagcttact ttattgtaag aatacagcta 3702
atttttgtat ttttagtgga gatagggttt tgccatgttg cccaacctgg tcttgagctc 3762
aagcgatcca cctgccccag cctcccaaag tgctgggatt ataggtgtga gccaccacac 3822
ccagcttcaa ctaacacatt tacgaacttg tatacatgta tatttagata ctttaccaac 3882
ttgtaaaatg gttaaaggag tactttatta tgaaaaaata tacaatcttt aaaatttcct 3942
tacttctaca tgatttttgt gctattccca tttttttcct caggtgagca gctttagtca 4002
6
CA 02323574 2000-09-22
WO 99149062 PCTIUS99106662
atgttgtgaa ttttagattt taattagaca tgcaacagtt tcactacctt tcaggatttt 9062
tgtcctgtaa cagaggctct tgctttttga cagagaggta ggcaggtgga gaggttatcc 4122
tgctgctgca gttctcaagt tgttaagttt cctctggaag gctaaccctt gttgggaact 9182
aacagtttca ataccagcaa gtctaggcct gctccaagtt ggtcagctga agaatgaaca 9242
tcagaagaca cagctgctga aagttgtcct ttgatgagac agtgatagtg atttggtaaa 9302
atgtcttatt ttttaaatgt cagttatctt tctttaaaag gttttttgag ggcagcctcc 4362
agaaggagct agagagtata ttttatagtt ctattgtggt tcataccctg ttttcgactt 9422
aagattctgg agaatgctat gaaacatctc cccagaaaaa gacagttaat taccatatct 9482
agagcagcac tgcccaacaa aaatatagta caggctatac acataattaa aacatttcta 9542
gtagcttctc taacaaaacc cattgaaagt caattttaat aatttatata acttagtgta 9602
tcaaaaatat ttcaatatgt aatcaacata aaattgagat actttaccag ctactaggga 4662
ggctgaggca caagaatcac ttaaacagga ggcaggggtt gcagtgagct gagatcacac 9722
cactgcactc cagcctgggt ggcagagcaa aactttgtcc ccacccctga caaaaaacaa 9782
acaaacaaac aaaacaaaaa aaaacctgtc aattcagatg ctaggttttc atcagacgta 4842
cttaatctgt atttagattt cttaaaactt actgtggaaa atgtatttac atactcaagt 4902
tgtttgaaac ataactcact gttttccaat aactgaagta tccactttta catgtattaa 9962
aattaaataa aattagaaat tcagttctgc agttgcacta gccacatttt aagtgtttaa 5022
tagccacacg tggttagtgg catctatatt ggacagggca gatctagaga gaatcctgta 5082
tctaacaatt ttaatttttt tccctttatg ctgttattcc ttacctagag aaacaatttc 5142
cctccaaagt tcctttgagg ggtctgttta ggccaggcca acacaagtga cctatgtgga 5202
ttttagcatc ctttttttga aatttgaggt tttatgaagc ttgagttttt ctggatattt 5262
ttagtaattt gctggtgtgt acttagctca gatacttgat tgcaactgtg ttgggtcaac 5322
tatttctaat gggacttttc catttgcatg tacagtcact ggaaactgct gggcagagaa 5382
actctaaaag gtagttgggg cacacttttt ccacctgtca gattggtgaa gaattggtga 5442
7
CA 02323574 2000-09-22
WO 99149062 PCTNS99/Obbb2
ggctgtgggg aaaatggcat tctcccactt ttgatggata tgtatccaaa taaaagtcat 5502
tcccatgaaa aaaaaaaaaa aaaaaaaaaa 5532
<210> 2
<211> B19
<212> PRT
<213> human gene expressed in ischemic heart disease
<400> 2
Met Ser Gly Thr Ser Lys Glu Ser Leu Gly His Gly Gly Leu Pro Gly
1 5 10 15
Leu Gly Lys Thr Cys Leu Thr Thr Met Asp Thr Lys Leu Asn Met Leu
20 25 30
Asn Glu Lys Val Asp Gln Leu Leu His Phe Gln Glu Asp Val Thr Glu
35 90 95
Lys Leu Gln Ser Met Cys Arg Asp Met Gly His Leu Glu Arg Gly Leu
50 55 60
His Arg Leu Glu Ala Ser Arg Ala Pro Gly Pro Gly Gly Ala Asp Gly
65 70 75 80
Val Pro His Ile Asp Thr Gln Ala Gly Trp Pro Glu Val Leu Glu Leu
g5 90 95
Val Arg Ala Met Gln Gln Asp Ala Ala Gln His Gly Ala Arg Leu Glu
100 105 110
Ala Leu Phe Arg Met Val Ala Ala Val Asp Arg Ala Ile Ala Leu Va1
115 120 125
Gly Ala Thr Phe Gln Lys Ser Lys Val Ala Asp Phe Leu Met Gln Gly
130 135 190
Arg Val Pro Trp Arg Arg Gly Ser Pro Gly Asp Ser Pro Glu Glu Asn
145 150 155 160
Lys Glu Arg Val Glu Glu Glu Gly Gly Lys Pro Lys His Val Leu Ser
165 170 175
Thr Ser Gly Val Gln Ser Asp Ala Arg Glu Pro Gly Glu Glu Ser Gln
180 185 190
Lys Ala Asp Val Leu Glu Gly Thr Ala Glu Arg Leu Pro Pro Ile Arg
8
CA 02323574 2000-09-22
WO 99/49062 PGT/US99/06662
195 200 205
Ala Ser Gly Leu Gly Ala Asp Pro Ala Gln Ala Val Val Ser Pro Gly
210 215 220
Gln Gly Asp Gly Val Pro Gly Pro Ala Gln Ala Phe Pro Gly His Leu
225 230 235 240
Pro Leu Pro Thr Lys Val Glu Ala Lys Ala Pro Glu Thr Pro Ser Glu
295 250 255
Asn Leu Arg Thr Gly Leu Glu Leu Ala Pro Ala Pro Gly Arg Val Asn
260 265 270
Val Val Ser Pro Ser Leu Glu Val Ala Pro Gly Ala Gly Gln Gly Ala
275 280 285
Ser Ser Ser Arg Pro Asp Pro Glu Pro Leu Glu Glu Gly Thr Arg Leu
290 295 300
Thr Pro Gly Pro Gly Pro Gln Cys Pro Gly Pro Pro Gly Leu Pro Ala
305 310 315 320
Gln Ala Arg Ala Thr His Ser Gly Gly Glu Thr Pro Pro Arg Ile Ser
325 330 335
Ile His Ile Gln Glu Met Asp Thr Pro Gly Glu Met Leu Met Thr Gly
340 345 350
Arg Gly Ser Leu Gly Pro Thr Leu Thr Thr Glu Ala Pro Ala Ala Ala
355 360 365
Gln Pro Gly Lys Gln Gly Pro Pro Gly Thr Gly Arg Cys Leu Gln Ala
370 375 380
Pro Gly Thr Glu Pro Gly Glu Gln Thr Pro Glu Gly Ala Arg Glu Leu
385 390 395 400
Ser Pro Leu Gln Glu Ser Ser Ser Pro Gly Gly Val Lys Ala Glu Glu
405 410 415
Glu Gln Arg Ala Gly Ala Glu Pro Gly Thr Arg Pro Ser Leu Ala Arg
920 925 430
Ser Asp Asp Asn Asp His Glu Val Gly Ala Leu Gly Leu Gln Gln Gly
435 440 495
Lys Ser Pro Gly Ala Gly Asn Pro Glu Pro Glu Gln Asp Cys Ala Ala
9
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
950 955 460
Arg Ala Pro Val Arg Ala Glu Ala Val Arg Arg Met Pro Pro Gly Ala
965 470 975 980
Glu Ala Gly Ser Val Val Leu Asp Asp Ser Pro Ala Pro Pro Ala Pro
485 490 995
Phe Glu His Arg Val Val Ser Val Lys Glu Thr Ser Ile Ser Ala Gly
500 505 510
Tyr Glu Val Cys Gln His Glu Val Leu Gly Gly Gly Arg Phe Gly Gln
515 520 525
Val His Arg Cys Thr Glu Lys Ser Thr Gly Leu Pro Leu Ala Ala Lys
530 535 540
Ile Ile Lys Val Lys Ser Ala Lys Asp Arg Glu Asp Val Lys Asn Glu
595 550 555 560
Ile Asn Ile Met Asn Gln Leu Ser His Val Asn Leu Ile Gln Leu Tyr
565 570 575
Asp Ala Phe Glu Ser Lys His 5er Cys Thr Leu Val Met Glu Tyr Val
580 585 590
Asp Gly Gly Glu Leu Phe Asp Arg Ile Thr Asp Glu Lys Tyr His Leu
595 600 605
Thr Glu Leu Asp Val Val Leu Phe Thr Arg Gln Ile Cys Glu Gly Val
610 615 620
His Tyr Leu His Gln His Tyr Ile Leu His Leu Asp Leu Lys Pro Glu
625 630 635 640
Asn Ile Leu Cys Val Asn Gln Thr Gly His Gln Ile Lys Ile Ile Asp
645 650 655
Phe Gly Leu Ala Arg Arg Tyr Lys Pro Arg Glu Lys Leu Lys Val Asn
660 665 670
Phe Gly Thr Pro Glu Phe Leu Ala Pro Glu Val Val Asn Tyr Glu Phe
675 680 685
Val Ser Phe Pro Thr Asp Met Trp Ser Val Gly Val Ile Thr Tyr Met
690 695 700
Leu Leu Ser Gly Leu Ser Pro Phe Leu Gly Glu Thr Asp Ala Glu Thr
CA 02323574 2000-09-22
WO 99149062 PCTIUS99/06662
705 710 715 720
Met Asn Phe Ile Val Asn Cys Ser Trp Asp Phe Asp Ala Asp Thr Phe
725 730 735
Glu Gly Leu Ser Glu Glu Ala Lys Asp Phe Val Ser Arg Leu. Leu Val
790 795 750
Lys Glu Lys Ser Cys Arg Met Ser Ala Thr Gln Cys Leu Lys, His Glu
755 760 765
Trp Leu Asn Asn Leu Pro Ala Lys Ala Ser Arg Ser Lys Thr Arg Leu
770 775 780
Lys Ser Gln Leu Leu Leu Gln Lys Tyr Ile Ala Gln Arg Lys Trp Lys
785 790 795 800
Lys His Phe Tyr Val Val Thr Ala Ala Asn Arg Leu Arg Lys Phe Pro
805 810 815
Thr Ser Pro
<210> 3
<211> 55
<212> DNA
<213> oligo(dT) primer, 1-base anchored
<400> 3
acgtaatacg actcactata gggcgaattg ggtcgacttt tttttttttt ttttv 55
<210> 4
<211> 40
<212> DNA
<213> oligo(dT) primer, 2-base anchored
<400> 4
ctctcaagga tcttaccgct tttttttttt ttttttttat 40
<210> 5
<211> 40
<212> DNA
<213> oligo(dT) primer, 2-base anchored
<400> 5
11
CA 02323574 2000-09-22
WO 99/49062 PGT/US99/06662
taataccgcg ccacatagca tttttttttt ttttttttcg 90
<210> 6
<211> 40
<212> DNA
<213> oligo(dT) primer, 2-base anchored
<900> 6
cagggtagac gacgctacgc tttttttttt ttttttttga 90
<210> 7
<211> 25
<212> DNA
<213> adapter oligonucleotide sequence
<900> 7
tagcgtccgg cgcagcgacg gccag 25
<210> 8
<211> 29
<212> DNA
<213> adapter oligonucleotide sequence
<900> 8
gatcctggcc gtcggctgtc tgtcggcgc 29
<210> 9
<211> 40
<212> DNA
<213> pcr primer, adapter-ligated 3' end
<400> 9
tgaagccgag acgtcggtcg tttttttttt ttttttttvn 90
<210> 10
<211> 293
<212> DNA
<213> sequence of Ci-25 band
<400> 10
ggctcacatc tgtaatccca gcactttggg aggccaaggt gggcagattg ctggccaaca 60
tggtaaaacc ccatctctaa agatataaaa attagctggg cgtggtggcg catacctgta 120
12
CA 02323574 2000-09-22
WO 99/49062 PCT/US99/06662
atcccagcta cttgggaggc taaggcacaa gaatcactta aacaggaggc gggggttgca 180
gtgagctgag atcacaccac tgcactccag cctgggtggc agagcaaaac tttgtcccca 290
cccctgacaa aaaacaaaca aacaaacaaa acaaaaaaaa acctgtcaat tca 293
13