Note: Descriptions are shown in the official language in which they were submitted.
WO 99/57292 PCT/US99/09925
- 1 -
GENOMIC SEQUENCES UPSTREAM OF THE CODING REGION OF THE IFN-ALPHA2 GENE FOR
PROTEIN
PRODUCTION AND DELIVERY
Field of the Invention
This invention relates to genomic DNA.
Background of the Invention
Current approaches to treating disease with
therapeutic proteins include both administration of
proteins produced in vitro and gene therapy. In vitro
production of a protein generally involves the
introduction of exogenous DNA coding for the protein of
interest into appropriate host cells in culture. Gene
therapy methods, on the other hand, involve' administering
to a patient cells, plasmids,' or viruses that contain a
sequence. encoding the therapeutic protein of interest.
Certain therapeutic proteins may also be produced
by altering the expression of their endogenous genes in a
desired manner with gene targeting techniques. See,
e.g., U.S. Patent Nos. 5,641,670, 5,733,761, and
5,272,071, U.S. Patent Application Serial No. 08/406,030,
WO 91/06666, WO 9I/06667, and WO 90/11354, all of which
are incorporated by reference in their entirety.
Summarv of the Invention
The present invention is based upon the
identification and sequencing of genomic DNA 5' to the
coding sequence of the human interferon-a 2 ("IFNA2")
gene. This DNA can be used, for example, in a DNA
construct that alters (e.g., increases) expression of an
endogenous IFNA2 gene in a mammalian cell upon
integration into the genome of the cell via homologous
recombination. "Endogenous IFNA2 gene" refers to a
genomic (i.e., chromosomal) gene that encodes IFNA2. The
construct contains a targeting sequence including or
CA 02328459 2000-11-07
_ _ , . .
7-UO-GUUV ~ '
" ~~ , ~
! ~ ~ , , ~w . . w v v ~
~ ~ , , ~ w v v v v v .
~ ~ ~ ' ~ ~.~
-~- , .m w ~~ w v w
derived from the newly disclosed 5' noncoding sequence, and a
transcri tional regulatory sequence. The transcriptional
P
regulatory sequence preferably differs in sequence from the
I
transcriptional regulatory sequence of the endogenous IFNA2
gene. The targeting sequence directs the integration of the
I
regulatory sequence into a region upstream of the endogenous
i
IFNA2-coding sequence such that the regulatory sequence becomes
operatively linked to the endogenous coding sequence. By
"operatively linked" is meant that the regulatory sequence can
l0 direct expression of the endogenous IFNA2-coding sequence. The
construct may additionally contain a selectable marker gene to
facilitate selection of cells that have stably integrated the ,
i
construct, and/or another coding sequence linked to a promoter.
In one embodiment, the DNA construct comprises: (a) a
targeting sequence, (b) a regulatory sequence, (c) an exon, (d)
P .
a s lice-donor site, (e) an intron, and (f) a splice-acceptor
i
i.
site, wherein the targeting sequence directs the integration of
f such that elements (b) - (f) are
itself and elements (b) - ( )
within or upstream of the endogenous gene. The regulatory
sequence then directs production of a transcript that includes
not only elements (c) - (f), but also the endogenous IFNA2
coding sequence. Preferably, the intron and the splice-acceptor
site are situated in the construct downstream from the splice-
r..:
donor site.
The targeting sequence is homologous to a pre-
selected target site in the genome with which homologous
recombination is to occur. It contains at least 20 (°.g., at
least 30, 50, 100, or 1000) contiguous nucleotides of SEQ ID
N0:11; and can contain, for instance, at least 20 (e.g., at
least 30, 50, or 100) contiguous nucleotides of SEQ ID N0:7, at
least 20 (e.g., at least 30 or 50) contiguous nucleotides of SEQ
ID N0:8, or at least 20 (e. g., at-'least 30, 50, 100, or 1000)
onnFNnFl7 SHEET
CA 02328459 2000-11-07
15-06-2000 US 009909925
.... ~~ ~~ ~ -, , '.
:. .. .. . . . .
. . . . ... . . ... . . .
. ~ , , ~ , , . . . .
. .. . . ..
_3_ . ... ... .. .. . .. ..
contiguous nucleotides of SEQ ID N0:12. In addition, the
targeting sequence can contain at least 20 (e.g., at least 30,
50, or 100) contiguous nucleotides of SEQ ID N0:15, at least 20
contiguous nucleotides of SEQ ID N0:16, at least 20 (e.g., at
least 30 or 50) contiguous nucleotides of SEQ ID N0:17, or at
least 20 (e. g., at least 30, 50, 100, or 1000) contiguous
nucleotides of SEQ ID N0:18. SEQ ID N0:7 corresponds to
nucleotides 1 to 278 of SEQ ID N0:11; SEQ ID N0:8 corresponds to
nucleotides 3492 to 3564 of SEQ ID N0:11; and SEQ ID N0:12
corresponds to nucleotides 279 to 3491 of SEQ ID N0:11. By
°homologous" is meant that the targeting sequence is identical
or sufficiently similar to its genomic target site so that the
targeting sequence and target site can undergo homologous
recombination within a human cell. A small percentage of
basepair mismatches is acceptable, as long as homologous
recombination can occur at a useful frequency. To facilitate
homologous recombination, the targeting sequence is preferably
at least about 20 (e. g., at least 50, 100, 250, 400, or 1,000)
base pairs ("bp") long. The targeting sequence can also include
genomic sequences from outside the region covered by SEQ ID
N0:12, so long as it includes as least 20 nucleotides from
within this region. For example, additional targeting sequence
could be derived from the sequence lyingwbetween SEQ ID N0:8 and
the endogenous transcription initiation sequence of the IFNA2
gene.
Due to polymorphism that exists at the IFNA2 genetic
locus, minor variations in the nucleotide composition of any
given genomic target site may occur in any given mammalian
species. Targeting sequences that correspond to such
polymorphic variants of SEQ ID NOs:7, 8, 11, 12, 15, 16, 17, and
18 (particularly human
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 4 -
polymorphic variants) are within the scope of this
invention.
Upon homologous recombination, the regulatory
sequence of the construct is integrated into a pre-
y selected region upstream of the coding sequence of an
IFNA2 gene in a chromosome of a cell. The resulting new
transcription unit containing the construct-derived
regulatory sequence alters the expression of the target
IFNA2 gene. The IFNA2 protein so produced may be
identical in sequence to the IFNA2 protein encoded by the
unaltered, endogenous gene, or may contain additional,
substituted, or fewer amino acid residues as compared to
the wild type IFNA2 protein, due to changes introduced as
a result of homologous recombination.
Altering gene expression encompasses activating
(or causing to be expressed) a gene which is normally
silent (i.e, essentially unexpressed) in the cell as
obtained, increasing or decreasing the expression level
of a gene, and changing the regulation pattern of a gene
such that the pattern is different from that in the cell
as obtained. "Cell as obtained" refers to the cell prior
to homologous recombination.
Also within the scope of the invention is a method
of using the present DNA construct to alter expression of
an endogenous IFNA2 gene in a mammalian cell. This
method includes the steps of (i) introducing the DNA
construct into the mammalian cell, (ii) maintaining the
cell under conditions that permit homologous
recombination to occur between the construct and a
genomic target site homologous to the targeting sequence,
to produce a homologously recombinant cell; and (iii)
maintaining the homologously recombinant cell under
conditions that permit expression of the IFNA2 coding
sequence under the control of the construct-derived
regulatory sequence. At least a part of the genomic
CA 02328459 2000-11-07
5-06-200'0 U S 009909925
. , , . .... .. .. _
y .. .. .. . . . .
~ ~ ~ ~ 1 ~ ~ ~ ~ ~ ~ 1 1 1 . f
~ . ~ 1 . 1 ~ . 1 1
~ ~ 1 . . 1 1 . 1 .
-g- . ... ... .. .. . .. ..
target site is 5' to the coding sequence of an endogenous IFNA2
gene. That is, the genomic target site can contain coding
sequence as well as 5' non-coding sequence.
The invention also features transfected or ir_fected
cells in which the construct has undergone homologous
recombination with genomic DNA upstream of the endogenous ATG
initiation codon in one or both alleles of the endogenous IFNA2
gene. Such transfected or infected cells, also called
homologously recombinant cells, have an altered IFNA2 expression
pattern. These cells are particularly useful for in vitro IFNA2
production and for delivering IFNA2 via gene therapy. Methods
of making and using such cells are also embraced by the
invention. The cells can be of vertebrate origin such as
mammalian (e. g., human, non-human primate, cow, pig, horse,
goat, sheep, cat, dog, rabbit, mouse, guinea pig, hamster, or
rat) origin.
The invention further relates to a method of
producing a mammalian IFNA2 protein .in vi tro or in vivo by
introducing the above-described construct into the genome of a
host cell via homologous recombination. The homologously
recombinant cell is then maintained under conditions that allow
transcription, translation, and optionally, secretion of the
IFNA2 protein.
The invention also features isolated nucleic acids
comprising a sequence of at least 20 (e.g., at least 30, 50,
100, 200, or 1000) contiguous nucleotides of SEQ ID N0:11 or its
complement, or of a sequence identical to SEQ ID N0:11 except
for polymorphic variations or other minor variations (e. g., less
than 50 of the sequence) which do not prevent homologous
recombination with the target sequence. Fcr instance, the
isolated DNA can contain at least 20 (e.g., at least 30, 50, or
100)rcontiguous nucleotides of SEQ ID N0:7 or its complement., at
CA 02328459 2000-11-07
5-06-2000 US 009909925
.... .. .. __ __
.. .. .. . . .. . ..
. . . . ... . . ... . . .
. . . .. . . ..
.. . . ..
,6- . ... ... .. .. . .. ..
least 20 {e.g., at least 30 or 50) contiguous nucleotides of SEQ
ID N0:8 or its complement, at least 20 (e.g., at least 30, 50,
100, or 1000) contiguous nucleotides of SEQ ID N0:12 or its
complement, at least 20 (e. g., at least 30, 50, or I00)
contiguous nucleotides of SEQ ID N0:15 or its complement, at
least 20 contiguous nucleotides of SEQ ID N0:16 or its
complement, at least 20 {e. g., at least 30 or 50) contiguous
nucleotides of SEQ ID N0:17 or its complement, or at least 20
(e.g., at least 30, 50, 100, or 1000) contiguous nucleotides of
SEQ ID N0:18 or its complement.
In one embodiment, the isolated nucleic acid of the
invention includes a contiguous 100 by block of SEQ TD NO:11.
For example, the isolated DNA can contain nucleotides 1 to 100,
101 to 200, 201 to 300, 301 to 400, 401 to 500, 501 to 600, 601
to 700, 701 to 800, 801 to 900 ,901 to 1000, 1001 to 1100, 1101
to 1200, 1201 to 1300, 1301 to 1400, 1401 to 1500, 1501 to 1600,
1601 to 1700, 1701 to 1800, 1801 to 1900, 1901 to 2000, 2001 to
2100, 2101 to 2200, 2201 to 2300, 2301 to 2400, 2401 to 2500,
2501 to 2600, 2601 to 2700, 2701 to 2800, 2801 to 2900, 2901 to
3000, 3001 to 3100, 3101 to 3200, 3201 to 3300, 3301 to 3400,
3401 to 3500, or 3465 to 3564 of SEQ ID NO:11 or its complement.
These blocks of SEQ ID N0:11 or its complement are also useful
as targeting sequences in the constructs'of the invention.
In the isolated DNA, the SEQ ID NO:11-derived
sequence is not linked to a sequence encoding intact IFNA2, or
at least is not linked in the same configuration {i.e.,
separated by the same noncoding sequence) as occurs in any wild-
type genome. The term "isolated DNA", as used herein, thus does
not denote a chromosome or a large piece of genomic DNA (as
might be incorporated into a cosmid or yeast artificial
chromosome) that includes not only part or all of SEQ ID NO:11,
but also an intact IFNA2-coding sequence and all of the sequence
CA 02328459 2000-11-07
5-06-2000 , US 009909925
a eeeee. ee
.. .. .e v ~ . v e..
. ..e . . .ee v v a v
. . . . ~ . . . a . . . .
~ . ~ . ~ a a . a
... w .. .e . .e w
which lies between the IFNA2 coding sequence and the sequence
corresponding to SEQ ID NO:11 as it exists in the genome of a
cell. It does include, but is not limited to, a DNA (i) which
is incorporated into a plasmid or virus; or (ii) which exists as
a separate molecule independent of other sequences, e.g., a
fragment produced by polymerase chain reaction ("PCR") or
restriction endonuclease treatment. The isolated DNA preferably
does not contain a sequence which encodes intact IFNA2 precursor
(i.e., IFNA2 complete with its endogenous secretion signal
peptide).
The invention also includes isolated DNA comprising a
strand which contains a sequence that is at least 100 (e.g., at
least 200, 400, or 1000) nucleotides in length and that j
hybridizes under either moderately stringent or highly stringent
i
conditions with SEQ ID N0:7, 8, 11, 12, 15, 16, 17, and/or 18,
or the complement of SEQ ID N0:7, 8, 11, 12, 15, 16, 17, and/or
18. The sequence is not linked to an IFNA2-coding sequence, or
at least is not linked in the same configuration as occurs in
any wild-type genome. By moderately stringent conditions is
meant hybridization at 50°C in Church buffer (7o SDS, 0.50
NaHP04, 1 M EDTA, to bovine serum albumin) and washing at SO°C in
2X SSC. Highly stringent conditions are defined as:
hybridization at 42°C in the presence of.~50% formamide; a first
wash at 65°C with 2X SSC containing 1% SDS; followed by a second
wash at 65°C with O.1X SSC.
Also embraced by the invention is isolated DNA
comprising a strand which contains a sequence that (1) is at I
least 50 (e.g., at least 70 or 100) nucleotides in length and
(2) shares at least 80% (e.g. , at least 85%, 90%, 95 0, or 98%)
sequence identity with a fragment or all of SEQ ID N0:11, or
with the complement of the fragment. This fragment can include,
for instance, a part or all of SEQ ID N0:7, 8, 12, 15, 15, 17,
CA 02328459 2000-11-07
5-06-2000 U S 009909925
.... .. .. __ __
.. .. .. . . . . . . .
. . . ... . . ... . . .
. . . .. . . ..
... .. . . ..
... .. .. . .. ..
or 18. The sequence is not linked to an intact IFNA2-coding
sequence, or at least is not linked in the same configuration as
occurs in any wild-type genome.
Where a particular polypeptide or nucleic acid molecule is said
to have a specific percent identity or conservation to a
reference polypeptide or nucleic acid molecule, the percent
identity or conservation is determined by the algorithm of Myers
;.
;::>
and Miller, CABIOS (1989), which is embodied in the ALIGN
program (version 2.0), or its equivalent, using a gap length
penalty of 12 and a gap penalty of 4 where such parameters are
required. All other parameters are set to their default
positions. Access to ALIGN is readily available. See, e.g.,
http://www2.igh.cnrs.fr/bin/align-guess.cgi on the Internet.
The invention also features a method of delivering IFNA2 to an
animal (e. g., a mammal such as a human, non-human primate, cow,
pig, horse, goat, sheep, cat, dog, rabbit, mouse, guinea pig,
hamster, or rat) by providing a cell whose endogenous IFNA2 gene
has been activated as described herein, and implanting the cell
in the animal, where the cell secretes IFNA2. Also included in
the invention is a method of producing IFNA2 by providing a cell
whose endogenous IFNA2 gene has been activated as described
herein, and culturing the cell in vitro under conditions which
permit the cell to express and secrete IFNA2.
The invention further includes isolated DNA that shares at
least 800 (e. g., at least 850, 900, or 950) sequence identity,
or hybridizes under highly or moderately stringent conditions,
with a portion (e. g., at least about 20, 50, 100, 400, or 1000
by in length) of the HindIII-BamHI insert of plasmid pA2HB
(described below). The 3' end of this portion of the insert is
at
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
g _
least 511 by upstream of the ATG translation initiation
codon of the IFNA2-coding sequence included in the
plasmid insert.
The isolated DNA of the invention can be used, for
example, as a source of an upstream PCR primer for use
(when combined with a suitable downstream primer) in
obtaining the regulatory region and/or complete coding
sequence of an endogenous IFNA2 gene, or as a
hybridization probe for indicating the presence of
chromosome 9 in a preparation of human chromosomes. It
can also be used, as described below, in a method for
altering the expression of an endogenous IFNA2 gene in a
vertebrate cell.
Unless otherwise defined, all technical and
scientific terms used herein.~ave the same meaning as
commonly understood by one of ordinary skill in the art
to which this invention belongs. Exemplary methods and
materials are described below, although methods and
materials similar or equivalent to those described herein
can also be used in the practice or testing of the
present invention. All publications, patent
applications, patents, and other references mentioned
herein are incorporated by reference in their entirety.
In case of conflict, the present specification, including
definitions, will control. The materials, methods, and
examples are illustrative only and not intended to be
limiting.
Other features and advantages of the invention
will be apparent from the following detailed description,
and from the claims.
Brief Description of the Drawings
Fig. 1 is a representation of the published
sequence (SEQ ID NO: l) of a human IFNA2-coding sequence
and some flanking 5' and 3' non-coding sequences (GenBank
CA 02328459 2000-11-07
15-0.6-2000 , U S 009909925
.... .. ..
.. .. .. . . . . . '. ~' .
. . . . ... . . ... . . .
. . . . .. . . ..
. . .. . . ..
-10- ~ ... ... .. .. . .. ..
HUMIFNAA). Sequences of PCR primers IFN1, IFN2, IFN6, and IFN7
are indicated by arrows.
Fig. 2 is a schematic diagram showing the human IFNA2
genomic region encompassed by the insert of plasmid pA2HB.
Fig. 3 is a representation of the nucleotide sequence
(SEQ ID N0:7) of a region upstream of the coding sequence of a
human IFNA2 gene. This nucleotide sequence has not been
reported previously.
Fig. 4 is a representation of a sequence (SEQ ID
N0:9) of a human IFNA2-coding sequence and some flanking 5' and
3' non-coding sequences. The underlined sequence (SEQ ID N0:8)
has not been previously reported. The polypeptide sequence
(SEQ ID N0:2) encoded by this gene is also shown. The N-
terminus of the mature polypeptide is indicated by "Mature."
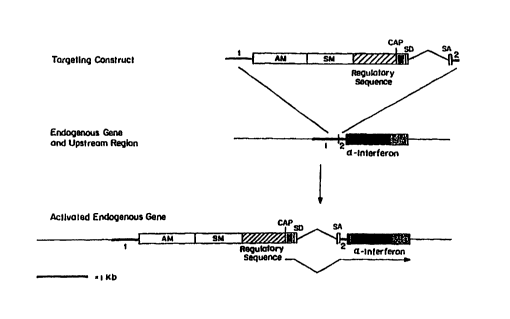
Fig. 5 is a schematic diagram showing a construct of
the invention. The construct contains a first targeting
sequence (1); an amplifiable marker gene (AM); a selectable
marker gene (SM); a regulatory sequence; a CAP site; an exon; a
splice-donor site (SD); an intron; a splice-acceptor site (SA);
and a second targeting sequence (2). The black boxes represent
coding DNA and the stippled boxes represent transcribed but
untranslated sequences.
Fig. 6 is a representation of'a sequence (SEQ ID
N0:10) of a human genomic sequence 5' to the IFNA2 coding
sequence, and including some coding sequence. The underlined
sequence is previously reported while the remainder (-4074 to -
511; SEQ ID NO:11) is new. The framed sequence is SEQ ID N0:12.
The sequence 5' to SEQ ID N0:12 is SEQ ID N0:7. The sequence
between the framed area and the underlined sequence is SEQ ID
N0:8. Nucleotides -4074 to -3270 is SEQ ID N0:15. Nucleotides
-3267 to -3239 is SEQ ID N0:16. Nucleotides -3241 to -3137 is
SEQ ID N0:17_ Nucleotides -3139 to -S11 is SEQ ID N0:18.
CA 02328459 2000-11-07
VJ VVOJVJJLv,J
.... .. ..
15-06-2000
:. .. .. . . . . . ~. . .s
. . . . ... . . ... . . .
. . . . . .
~ ~ ~ ~ . ..
..
-1.1- . ... ... .. .. . .. ~
Fig. 7 is a representation of a first targeting
sequence (SEQ ID N0:13) used in a construct of the invention. i
Fig. 8 is a representation of a second targeting
i
sequence {SEQ ID N0:14) used in a construct of the invention.
Detailed Description
The present invention is based on the discovery of
the nucleotide composition of sequences upstream to the coding
sequence of a human IFNA2 gene.
Interferon-a constitutes a complex gene family with
14 genes clustered on the short arm of chromosome 9. None of
these genes, including the IFNA2 gene, have introns.
Interferon-a is produced by macrophages, T and B cells, and a
variety of many other cells. Interferon-a has considerable
antiviral effects, and has been shown to be efficacious in
treating infections by papilloma virus, hepatitis B and C
viruses, vaccinia, herpes simplex virus, herpes zoster
varicellosus virus, and rhinovirus.
The human IFNA2 gene encodes a 188 amino acid
precursor protein {SEQ ID N0:2) containing a 23 amino acid
signal peptide. The genomic map of the human IFNA2 gene is
shown in Fig. 1. The map is constructed based on 1,733 base
pair ("bp") published sequences (HUMIFNAA, GenEank accession
number J00207 and V00544; SEQ ID NO:1) which begin at position -
510 relative to the translational start site (unless otherwise
specified, all positions referred to herein are relative to the
translational start site), and end at position +1,223. The cap
site is located at position -67.
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
12 -
Specific Secruences 5' to an IFNA2 Gene and Their Use in
Alterinct Endogenous IFNA2 Gene Expression
To obtain genomic DNA containing sequence upstream
to an IFNA2 gene, a human leukocyte genomic library in
lambda EMBL3 (Clontech catalog # HL1006d) was screened
with a 332 by probe generated by PCR. This probe
corresponds to the genomic region between positions -263
and +69, and was amplified from human genomic DNA using
oligonucleotide primers designated IFN7 and IFN6, both of
which were designed from the available IFNA2 genomic DNA
sequence (Fig. 1). The 5' end of primer IFN7 corresponds
to position -263, and the primer's sequence is
5'-AGTTTCTAAAA.AGGCTCTGGGGTA-3' (SEQ ID N0:3). The 5' end
of primer IFN6 corresponds to position +69, and this
primer sequence is 5'-GCCCACAGAGCAGCTTGAC-3' (SEQ ID
N0:4) .
Approximately one million recombinant phage were
screened with the radiolabelled 332 by probe. Sixty
positive plaques were isolated from the primary screening
plates. Lambda phage DNA was isolated from thirty of
these plaques and subjected to PCR assay using
oligonucleotide primers IFN1 and IFN2. Both IFN1 and
IFN2 are derived from the 3' untranslated region of the
IFNA2 gene; their sequences can be found at the website
"http://www.ncbi.nlm.nih.gov/dbSTS," using the
identification code "NCBI ID:42433." The 5' end of
primer IFN1 corresponds to position +639, and the
primer's sequence is 5'-AAAGACTCATGTTTCTGCTATGACC-3' (SEQ
ID N0:5). The 5' end of primer IFN2 corresponds to
position +853, and the primer's sequence is 5'-
GGTGCACATGACATAATATGAACA-3' (SEQ ID N0:6). Of the thirty
phage samples, two generated the expected 215 by PCR
product. One of the two phage plaques was further
purified by two additional rounds of hybridization
screening, yielding phage clone 4-4-1.
CA 02328459 2000-11-07
US 00990992
., , . .,.. .. ..
15-06-2000
.. .. .. . . . .
. ... . . ... . . .
. ~ ' ' ' ' ~ . .. ..
-1'3- ~ ...... .. ..
A 8.3 kb HindIII-BamHI fragment from phage 4-4-1 was
subcloned into pBluescript II SK+ (Stratagene, La Jolla, CA) to
produce pA2HB, which contains approximately 4.3 kb of
untranscribed upstream sequences, the protein-coding region (1.1
kb), and approximately 2.8 kb of downstream sequences of the I,
I
IFNA2 gene. A restriction map of the 8.3 kb HindIII-BamHI
fragment is shown in Fig. 2. '
The pA2HB plasmid was sequenced by the Sanger method.
A 278 by sequence whose 5' terminus is at the 5' end HindIII
site is shown below (see also Fig. 3):
AAGCTTTTATAGGTGTAAATTTTCCACTTAGTACTGCTTTTG
TA.ATGTTGTCTTTTTATTTTCATTTATCTCAAGATGTTTTCT
AATTTCTCTTGACTTCCTTCTTAA.ATTCTTACCTCATGTAGA
CATACATTTTTGGCCCTATGCATTGGGATGCAAAACCAGACT
AATTTACTTTGTACAAAAAGA.AAPATGAGAAAGAAATATATT
TGGTCTTGTGAGCACTATATGGAAATACTTTATATTCCATTT
GTTTCATCATATTCATATATCCCTTT (SEQ ID N0:7)
The HindIII site is located at position -4,073. A
previously unpublished sequence between positions -583 and -511
was also determined, as shown below and as underlined in Fig. 4. '
CATTGGATACTCCATCACCTGCTGTGATATTATGAATGTCTGCCTATATAAATATTCACTAT
TCCATAACACA (SEQ ID N0:8)
The sequence (SEQ ID N0:12) between the regions corresponding to
SEQ ID NOs:7 and 8 was also determined.
The genomic sequence between positions -4,074 and
-511 (SEQ ID NO:11) is the sequence which is not underlined in
Fig. 6. SEQ ID NOs:7 and 8 correspond to nucleotides 1-278 and
nucleotides 3492-3564 of SEQ ID NO:11, respectively.
To alter the expression of an endogenous INFA2 gene,
a DNA fragment containing nucleotides 279-3311 of SEQ ID NO:11
was cloned into a plasmid to produce targeting construct pGA402.
Nucleotides 279-33'!1 of SEQ ID NO:11 was designated SEQ ID
N0:13. The fragment was inserted upstream of a CMV promoter and
a neomycin resistance gene and is schematically represented in
CA 02328459 2000-11-07
VvJ VVJJVJvJLVJ
.... .. .. '. t._, i
.. .. .. . . . ~ tr '
' .. . 1 .. ~
. ~ . . . .
, , . 1 .
-24- ... ... . . . ..
.. ..
Fig. 5. For the second targeting sequence of Fig. 5, a DNA
fragment containing nucleotides -68 to 69 of the IFNA2 gene
sequence shown in Fig. 1 was cloned downstream of the CMV II
promoter and neomycin resistance gene. Nucleotides -68 to 69 of
i
the IFNA2 gene was designated SEQ ID N0:14. The pGA402 plasmid
was introduced into human fibroblast cells exhibiting little or i
no INFA2 gene expression, to allow homologous recombination with
the endogenous INFA2 gene. Cells resistant to 6418 after
plasmid introduction were screened to identify cells with
increased INFA2 gene expression, as would be expected if a
homologous recombination event between pGA402 and the genomic
DNA took place in the vicinity of the endogenous INFA2 gene.
General Methodologies
Alteration of Endogenous IFNA2 Expression
Using the above-described IFNA2 upstream sequences,
one can alter the expression of an endogenous human IFNA2 gene i:
by a method as generally described in U.S. Patent No. 5,641,670.
r
One strategy is shown in Fig. 5. In this strategy, a targeting
construct is designed to include a first targeting sequence
homologous to a first target site upstream of the gene, an
amplifiable marker gene, a selectable marker gene, a regulatory
region, a CAP site, an exon, a splice-donor site, an intron, a
splice-acceptor site, and a second targeting sequence homologous
to a second target site downstream of the first target site and
terminating either within or upstream of the IFNA2-coding ',
sequence. According to this strategy, the 5' end of the second
target site is preferably less than 107 by upstream of the
normal IFNA2 translational initiation site, in order to avoid
undesired ATG start
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 15 -
codons within the transcribed sequence. A transcript
produced from the homologously recombined locus will
include the construct-derived exon, the construct-derived
splice-donor site, the construct-derived intron, the
construct-derived splice-acceptor site, any sequence
between any of those elements, and the sequence from the
construct derived splice acceptor site through the entire
endogenous coding sequence to the transcription
termination site of the IFNA2 gene. Splicing of this
transcript will generate a mRNA which can be translated
to produce a precursor of human IFNA2, having either the
normal IFNA2 secretion signal sequence or a genetically
engineered secretion signal sequence, depending on the
characteristics of the construct-derived exon. The size
of the exogenous intron and thus the position of the
exogenous regulatory region relative to the coding region
of the gene can be varied to optimize the function of the
regulatory region.
In any activation strategy, the first and second
target sites need not be immediately adjacent or even be
near each other. When they are not immediately adjacent
to each other, a portion of the IFNA2 gene's normal
upstream region and/or a portion of the coding region
would be deleted upon homologous recombination.
Mutations that facilitate alteration of endogenous
IFNA2 expression may be introduced into the chromosomal
DNA via homologous recombination. For instance, it may
be desirable to abolish a spurious and undesired ATG
initiation codon upstream of the correct ATG initiation
codon and between the exogenous regulatory region and the
endogenous IFNA2 coding region in the homologously
recombined locus. To do so, one can employ a targeting
construct having a targeting sequence homologous to a
genomic site that spans the undesired ATG initiation
codon. This targeting sequence contains nucleotides that
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 16 -
correspond to the desired mutation, e.g., contains ATT
instead of ATG. The targeting construct optionally
includes one or more selectable markers to facilitate
selection of homologously recombined cells. An exogenous
regulatory region can then be introduced to the
homologously recombined cells upstream of the altered
sites, using the expression alteration method of the
invention.
Alternatively, the exogenous regulatory region and
the desired sequence mutations) may be introduced into
the genomic DNA in a single step. The DNA construct used
in this embodiment may contain both the exogenous
regulatory region and a targeting sequence that contains
nucleotides corresponding to the desired mutation(s).
One may also co-transfect or.co-infect two separate
constructs into target cells, with one construct
containing the regulatory region and the other containing
nucleotides corresponding to the desired mutation.
If desired, a mammalian splice-acceptor site may
be introduced into the genomic DNA, e.g., at a site
between an undesired ATG initiation codon and the correct
ATG initiation codon, in a similar manner. The DNA
construct used for this purpose contains a targeting
sequence homologous to a genomic site upstream of the
correct INFA2 initiation codon, and adjacent to or
embedded within the homologous sequence, a sequence
corresponding to the desired splice-acceptor site. Cells
containing the correctly recombined IFNA2 locus are then
transfected or infected with a second construct
containing an exogenous regulatory region and an exon
with an unpaired splice-donor site at its 3' end,
together with targeting sequences) which target the
second construct to a genomic region upstream of the
inserted splice-acceptor site. A primary transcript
produced under the control of the exogenous regulatory
CA 02328459 2000-11-07
WO 99/57292 PC'T/US99/09925
- 17 -
region will include the exogenous exon, the exogenous
splice-donor site, the exogenous splice-acceptor site,
any sequences between those elements, and the sequence
between the exogenous splice-acceptor site and the
transcriptional termination site of the endogenous IFNA2
gene. Upon splicing, the splice-donor site of the
transcript will be spliced to the splice-acceptor site,
and the intervening intronic RNA, which may contain
undesirable AUG initiation codons, will be removed. Any
problems associated with having a transcript with
undesired AUG translation initiation codons between the
transcription start site and the IFNA2-coding sequence
are thereby avoided. Of course, the regulatory region,
exon, splice donor site, and splice-acceptor site can
instead be introduced in a single step. The DNA
construct used in this embodiment contains a regulatory
region, an exon, a splice-donor site, an intron, a
splice-acceptor site, a targeting sequence homologous to
a genomic site between the correct INFA2 initiation codon
and the undesired ATG codon, and optionally, one or more
selectable markers. Alternatively, two separate
targeting constructs may be useful, with one containing
the regulatory region, the exon, and the splice-donor
site, and the other containing the splice-acceptor site.
The two constructs can be introduced into target cells in
a single step.
The DNA Construct
The DNA construct of the invention includes at
least a targeting sequence and a regulatory sequence. It
can additionally include an exon; or an exon and splice
donor site; or an exon, a splice-donor site, an intron,
and a splice-acceptor site. In the construct, the exon,
if present, is 3' of the regulatory sequence, and the
splice-donor site, if present, is at the 3' end of the
exon. The intron and splice acceptor site, if present,
CA 02328459 2000-11-07
WO 99/57292 PCTIUS99/09925
- 18 -
are 3' of the splice donor site. In addition, there can
be multiple exons and introns (with appropriate splice
donor and acceptor sites) in the construct. The DNA in
the construct is referred to as exogenous, since the DNA
is not an original part of the genome of a host cell.
Exogenous DNA may possess sequences identical to or dif-
ferent from portions of the endogenous genomic DNA
present in the cell prior to transfection or infection by
viral vector. As used herein, "transfection" means
introduction of a plasmid into a cell by nonviral (e. g.,
chemical or physical) means, such as calcium phosphate or
calcium chloride co-precipitation, DEAE-dextran-mediated
transfection, lipofection, electroporation,
microinjection, microprojectiles, or biolistic-mediated
uptake. "Infection" means introduction of a viral vector
into a cell by viral infection. Various elements
included in the DNA construct of the invention are
described in detail below.
The DNA construct can also include cis-acting or
trans-acting viral sequences (e. g., packaging signals),
thereby enabling delivery of the construct into the
nucleus of a cell via infection by a viral vector. Where
necessary, the DNA construct can be disengaged from
various steps of a virus life cycle, such as integrase-
mediated integration in retroviruses or episome
maintenance. Disengagement can be accomplished by
appropriate deletions or mutations of viral sequences,
such as a deletion of the integrase coding region in a
retrovirus vector. Additional details regarding the
construction and use of viral vectors are found in
Robbins et al., Pharmacol. Ther. 80:35-47, 1998; and
Gunzburg et al., Mol. Med. Today 1:410-417, 1995, herein
incorporated by reference.
CA 02328459 2000-11-07
US 009909925
5-06-2000 - -
~ s f ~ f t . . . f
t. .w .. 1 0 . ~ s a s .
w . 1.. . . ... . . . .
~ 1 . ,. . . . . . . . r
~ t t . v . . . ~ . v
-19- ' ~ ~. ... .. .. . .. ..
Taraetina Seauences
Targeting sequences permit homologous recombination
of a desired sequence into a selected site in the host genome.
Targeting sequences are homologous to (i_e., able to
homologously recombine with) their respective target sites in
the host genome.
A circular DNA, construct can employ a single
targeting sequence, or two or more separate targeting sequences.
A linear DNA construct may contain two or more separate target-
ing sequences. The target site to which a given targeting
sequence is homologous can reside within the coding region of
the IFNA2 gene, upstream of and immediately adjacent to the
coding region, or upstream of and at a distance from the coding
region.
The first of the two targeting sequences in the
construct (or the entire targeting sequence, if there is only
one targeting sequence in the construct) is derived from the
newly disclosed genomic regions upstream of the IFNA2-coding
sequence. This targeting sequence contains a portion (e.g:, 20
or more contiguous nucleotides) of SEQ ID NO:11, e.g., a portion
of SEQ ID N0:7, 8, or 12.
The second of the two targeting sequences in the
construct may target a genomic region upstream of the coding
sequence or target part or all of the coding sequence itself.
By way of example, the second targeting sequence may contain, at
its 3' end, an "exogenous" coding region identical to the first
few codons of the IFNA2 coding sequences. Upon homologous
recombination, the exogenous coding region recombines with the
targeted part of the endogenous IFNA2-coding sequence. If
desired, the exogenous coding region may encode a heterologous
amino acid sequence, so long as the exogenous coding region
CA 02328459 2000-11-07
US 00990995
~-06-200.0
1 1 s _ - - '
~
1 1 1 1
1 1 1 ~ 1
1
1 1
remains sufficiently homologous to the endogenous coding region
it replaces to permit homologous recombination.
i-
The targeting sequence may additionally include
sequence derived from a previously disclosed region of the IFNA2
gene, including those described herein, as well as a region
further upstream which is structurally uncharacterized but can
be mapped by one skilled in the art.
Genomic fragments useful as targeting sequences can
be identified by their ability to hybridize to a probe
containing all or a portion of SEQ ID N0:11. Such a probe can
be generated by PCR using primers derived from SEQ ID N0:11.
The ReaulatoY'y Seauence
The regulatory sequence of the DNA construct can
contain one or more promoters (e.g., a constitutive, tissue-
specific, or inducible promoter), enhancers, scaffold-attachment
regions or matrix attachment sites, negative regulatory
i
elements, transcription factor binding sites, or combinations of i
these elements. The regulatory sequence can be derived from
a eukaryotic (e. g., mammalian) or viral genome. Useful
regulatory sequences include, but are not limited to, those that
regulate the expression of SV40 early or late genes,
cytomegalovirus genes, and adenovirus major late genes. They
also include regulatory regions derived from genes encoding
mouse metallothionein-I, elongation factor-la, collagen (e. g.,
collagen Ial, collagen Ia2, and collagen IV), actin (e. g.,
y-actin), immunoglobulin, HMG-CoA reductase, glyceraldehyde
phosphate dehydrogenase, 3-phosphoglyceratekinase, collagenase,
stromelysin, fibronectin, vimentin, plasminogen activator
inhibitor I, thymosin a4, tissue inhibitors of
metalloproteinase, _ribosomal proteins, major histocompatibility
complex molecules, and human leukocyte antigens.
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 21 -
The regulatory sequence preferably contains a
transcription factor binding site, such as a TATA Box,
CCAAT Box, AP1, Spl, or a NF-rcB binding site.
Marker Genes
If desired, the construct can include a sequence
encoding a desired polypeptide, operatively linked to its
own promoter. An example of this would be a selectable
marker gene, which can be used to facilitate the
identification of a targeting event. An amplifiable
marker gene can also be included and used to facilitate
selection of cells having co-amplified flanking DNA
sequences. Cells containing amplified copies of the
amplifiable marker gene can be identified by growth in
the presence of an agent that selects for the expression
of the amplifiable gene. The activated endogenous IFNA2
gene will be amplified in tandem with the amplified
selectable marker gene. Cells containing multiple copies
of the activated endogenous gene may produce very high
levels of IFNA2 and are thus useful for in vitro protein
production and gene therapy.
The selectable and amplifiable marker genes do not
have to lie immediately adjacent to each other. The
amplifiable marker gene and selectable marker gene can be
the same gene. One or both of the marker genes can be
situated in the intron of the DNA construct. Suitable
amplifiable marker genes and selectable marker genes are
described in U.S. Patent No. 5,641,670.
The St~lice-Donor and Splice-Acceptor Sites
The DNA construct may further contain an exon, a
splice-donor site at the 3' end of the exon, an intron,
and a splice-acceptor site.
A splice-donor site is a sequence which directs
the splicing of one exon of an RNA transcript to the
splice-acceptor site of another exon of the transcript,
CA 02328459 2000-11-07
_ US 009909925
5-06-2000
. . . .... .. .. .. ..
.. .. .. . . . . . . .
. 1 f .
. . . . .. . . .t
-22- . .. . . .. .
... ... .. .. . .. ..
resulting in removal of the intron between the two sites.
Typically, the first exon lies 5' of the second exon, and the
splice-donor site located at the 3' end of the first exon is
paired with a splice-acceptor site flanking the second exon on
the 5' side of the second exon. Splice-donor sites have a
characteristic consensus sequence represented as (A/C)AGGUR.AGU
(where R denotes a purine nucleotide), with the GU in the fourth
and fifth positions being required (Jackson, Nucleic Acids
Research 19: 3715-3798, 1991). The first three bases of the
splice-donor consensus site are the last three bases of the
exon: i.e., they are not spliced out. Splice-donor sites are
functionally defined by their ability to effect the appropriate
reaction within the mRNA splicing pathway.
A splice acceptor site in a construct of the
invention directs, in conjunction with a splice donor site, the
splicing of one exon to another exon. Splice-acceptor sites
have a characteristic sequence represented as (Y)loNYAG
(SEQ ID N0:19), where Y denotes any pyrimidine and N denotes any
nucleotide (Jackson, Nucleic Acids Research 19:3715-3798, 1991).
TrP CAP Site
The DNA construct can optionally contain a CAP site.
A CAP site is a specific transcription start site which is
associated with and utilized by the regulatory region. This CAP
site is located at a position relative to the regulatory se-
quence~in the construct such that following homologous
recombination, the regulatory sequence directs synthesis of a
transcript that begins at the CAP site. Alternatively, no CAP
site is included in the construct, and the transcriptional
apparatus will locate by default an appropriate site in the
targeted gene to be utilized as a CAP site.
CA 02328459 2000-11-07
WO 99/57292 PCT/US99109925
- 23 -
Additional DNA elements
The construct may additionally contain sequences
which affect the structure or stability of the RNA or
protein produced by homologous recombination.
Optionally, the DNA construct can include a bacterial
origin of replication and bacterial antibiotic resistance
markers or other selectable markers, which allow for
large-scale plasmid propagation in bacteria or any other
suitable cloning/host system.
All of the above-described elements of the DNA
construct are operatively linked or functionally placed
with respect to each other. That is, upon homologous
recombination between the construct and the targeted
genomic DNA, the regulatory sequence can direct the
production of a primary RNA transcript which initiates at
a CAP site (optionally included in the construct) and
includes the sequence lying between the CAP site and the
endogenous IFNA2 gene's transcription stop site.
Depending on the location of the CAP site, a portion of
this sequence may include the IFNA2 gene's endogenous
regulatory region as well as sequences neighboring that
region that are normally not transcribed. Tf an exon, a
splice-donor site and a splice-acceptor site are present
in the construct, the primary transcript will also
include the exon, the two splice sites, and the intron
between the two sites.
The order of elements in the DNA construct can
vary. Where the construct is a circular plasmid or viral
vector, the relative order of elements in the resulting
structure can be, for example: a targeting sequence,
plasmid DNA (comprised of sequences used for the
selection and/or replication of the targeting plasmid in
a microbial or other suitable host), selectable
marker(s), a regulatory sequence, an exon, a splice-donor
site, an intron, and a splice-acceptor site.
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 24 -
Where the construct is linear, the order can be,
for example: a first targeting sequence, a selectable
marker gene, a regulatory sequence, an exon, a splice-
donor site, an intron, a splice-acceptor site, and a
second targeting sequence; or, in the alternative, a
first targeting sequence, a regulatory sequence, an exon,
a splice-donor site, an intron, a splice-acceptor site, a
selectable marker gene, optionally an internal ribosomal
entry site, and a second targeting sequence.
Alternatively, the order can be . a first
targeting sequence, a first selectable marker gene, a
regulatory sequence, an exon, a splice-donor site, an
intron, a splice-acceptor site, a second targeting
sequence, and a second selectable marker gene; or, a
first targeting sequence, a regulatory sequence, an exon,
a splice-donor site, an intron, a splice-acceptor site, a
first selectable marker gene, a second targeting se-
quence, and a second selectable marker gene.
Recombination between the targeting sequences flanking
the first selectable marker with homologous sequences in
the host genome results in the targeted integration of
the first selectable marker, while the second selectable
marker is not integrated. Desired transfected or
infected cells are those that are stably transfected or
infected with the first, but not second, selectable
marker. Such cells can be selected for by growth in a
medium containing an agent which selects for expression
of the first marker and another agent which selects
against the second marker. Transfected or infected cells
that have improperly integrated the targeting construct
by a mechanism other than homologous recombination would
be expected to express the second marker gene and will
thereby be killed in the selection medium.
A positively selectable marker is sometimes
included in the construct to allow for the selection of
CA 02328459 2000-11-07
WO 99/57292 PG"T/US99/09925
- 25 -
cells containing amplified copies of that marker. In
this embodiment, the order of construct components can
be, for example: a first targeting sequence, an
amplifiable positively selectable marker, a second
selectable marker (optional), a regulatory sequence, an
exon, a splice-donor site, an intron, a splice-acceptor
site, and a second targeting DNA sequence.
The various elements of the construct can be
obtained from natural sources (e.g., genomic DNA), or can
be produced using genetic engineering techniques or
synthetic processes. The regulatory region, CAP site,
splice-donor site, intron, and splice acceptor site of
the construct can be isolated as a complete unit from;
e.g., the human elongation factor-la (Genbank sequence
HUMEF1A) gene or the cytomega7.ovirus (Genbank sequence
HEHCMVPl) immediate early region. These components can
also be isolated from separate genes.
Transfection or Infection and HomoloQOUS Recombination
The DNA construct of the invention can be intro-
duced into the cell, such as a primary, secondary, or
immortalized cell, as a single DNA construct, or as sepa-
rate DNA sequences which become incorporated into the
chromosomal or nuclear DNA of a transfected or infected
cell. By "transfected cell" is meant a cell into which
(or into an ancestor of which) a DNA or RNA molecule has
been introduced by a means other than using a viral
vector. By °infected cell" is meant a cell into which
(or into an ancestor of which) a DNA or RNA molecule has
been introduced using a viral vector. Viruses known to
be useful as vectors include adenovirus, adeno-associated
virus, Herpes virus, mumps virus, poliovirus, lentivirus,
retroviruses, Sindbis virus, and vaccinia viruses such as
canary pox virus. The DNA can be introduced as a linear,
double-stranded (with or without single-stranded regions
at one or both ends), single-stranded, or circular
CA 02328459 2000-11-07
WO 99/57292 PGT/US99/09925
- 26 -
molecule. When the construct is introduced into host
cells in two separate DNA fragments, the two fragments
share DNA sequence homology (overlap) at the 3' end of
one fragment and the 5' end of the other, while one
carries a first targeting sequence and the other carries
a second targeting sequence. Upon introduction into a
cell, the two fragments can undergo homologous
recombination to form a single molecule with the first
and second targeting sequences flanking the region of
overlap between the two original fragments. The product
molecule is then in a form suitable for homologous
recombination with the cellular target sites. More than
two fragments can be used, with each of them designed
such that they will undergo homologous recombination with
each other to ultimately form,'a product suitable for
homologous recombination with the cellular target sites
as described above.
The DNA construct of the invention, if not
containing a selectable marker itself, can be
co-transfected or co-infected with another construct that
contains such a marker. A targeting plasmid may be
cleaved with a restriction enzyme at one or more sites to
create a linear or gapped molecule prior to transfection
or infection. The resulting free DNA ends increase the
frequency of the desired homologous recombination event.
In addition, the free DNA ends may be treated with an
exonuclease to create overhanging 5' or 3' single-
stranded DNA ends (e.g., at least 30 nucleotides in
length, and preferably 100-1000 nucleotides in length) to
increase the frequency of the desired homologous
recombination event. In this embodiment, homologous
recombination between the targeting sequence and the
genomic target will result in two copies of the targeting
sequences, flanking the elements contained within the
introduced plasmid.
CA 02328459 2000-11-07
WO 99/57292 PCT/US99109925
- 27 -
The DNA constructs may be transfected into cells
(preferably in vitro) by a variety of physical or
chemical methods, including electroporation, microin-
jection, microprojectile bombardment, calcium phosphate
precipitation, liposome delivery, or polybrene- or DEAE
dextran-mediated transfection.
The transfected or infected cell is maintained
under conditions which permit homologous recombination,
as described in the art (see, e.g., Capecchi, Science
24:1288-1292, 1989). When the homologously recombinant
cell is maintained under conditions sufficient to permit
transcription of the DNA, the regulatory region
introduced by the DNA construct will alter transcription
of the IFNA2 gene.
Homologously recombinant cells (i.e., cells that
have undergone the desired homologous recombination) can
be identified by phenotypic screening or by analyzing the
culture supernatant in enzyme-linked immunosorbent assays
(ELISA) for IFNA2. Commercial ELISA kits for detecting
IFNA2 are available from Biosource International
(Camarillo, CA). Homologously recombinant cells can also
be identified by Southern and Northern analyses or by
polymerase chain reaction (PCR} screening.
As used herein, the term "primary cells" includes
(i) cells present in a suspension of cells isolated from
a vertebrate tissue source (prior to their being plated,
i.e., attached to a tissue culture substrate such as a
dish or flask), (ii) cells present in an explant derived
from tissue, (iii) cells plated for the first time, and
(iv) cell suspensions derived from these plated cells.
Primary cells can also be cells as they naturally occur
within a human or an animal.
Secondary cells are cells at all subsequent steps
in culturing. That is, the first time that plated
primary cells are removed from the culture substrate and
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 28 -
replated (passaged), they are referred to herein as
secondary cells, as are all cells in subsequent passages.
Secondary cell strains consist of secondary cells which
have been passaged one or more times. Secondary cells
typically exhibit a finite number of mean population
doublings in culture and the property of
contact-inhibited, anchorage-dependent growth
(anchorage-dependence does not apply to cells that are
propagated in suspension culture). Primary and secondary
cells are not immortalized
Immortalized cells are cell lines (as opposed to
cell strains, with the designation "strain" reserved for
primary and secondary cells) that exhibit an apparently
unlimited lifespan in culture.
Cells selected for transfection or infection can
fall into four types or categories: (i) cells which do
not, as obtained, make or contain more than trace amounts
of the IFNA2 protein, (ii) cells which make or contain
the protein but in quantities other than those desired
(such as, in quantities less than the level which is
physiologically normal for the type of cells as
obtained), (iii) cells which make the protein at a level
which is physiologically normal for the type of cells as
obtained, but are to be augmented or enhanced in their
content or production, and (iv) cells in which it is
desirable to change the pattern of regulation or
induction of a gene encoding the protein.
Primary, secondary and immortalized cells to be
transfected or infected by the present method can be
obtained from a variety of tissues and include all
appropriate cell types which can be maintained in
culture. For example, suitable primary and secondary
cells include fibroblasts, keratinocytes, epithelial
cells (e. g., mammary epithelial cells, intestinal
epithelial cells), endothelial cells, glial cells, neural
CA 02328459 2000-11-07
WO 99157292 PGT/US99/09925
- 29 -
cells, formed elements of the blood (e. g., lymphocytes,
bone marrow cells), muscle cells, and precursors of these
somatic cell types. Where the homologously recombinant
cells are to be used in gene therapy, primary cells are
preferably obtained from the individual to whom the
transfected or infected,primary or secondary cells are to
be administered. However, primary cells can be obtained
from a donor (i.e., an individual other than the
recipient) of the same species.
Examples of immortalized human cell lines useful
for protein production or gene therapy include, but are
not limited to, 2780AD ovarian carcinoma cells (Van der
Blick et al., Cancer Res., 48:5927-5932, 1988), A549
(American Type Culture Collection ("ATCC") CCL 185), BeWo
(ATCC CCL 98), Bowes Melanoma cells (ATCC CRL 9607),
CCRF-CEM (ATCC CCL 119), CCRF-HSB-2 (ATCC CCL 120.1),
COL0201 (ATCC CCL 224), COL0205 (ATCC CCL 222), COLD
320DM (ATCC CCL 220), COLD 320HSR (ATCC CCL 220.1), Daudi
cells (ATCC CCL 213), Detroit 562 (ATCC CCL 138), HeLa
cells and derivatives of HeLa cells (ATCC CCL 2, 2.1 and
2.2), HCT116 (ATCC CCL 247), HL-60 cells (ATCC CCL 240),
HT1080 cells (ATCC CCL 121), IMR-32 (ATCC CCL 127),
Jurkat cells (ATCC TIB 152), K-562 leukemia cells (ATCC
CCL 243),~KB carcinoma cells (ATCC CCL 17), KG-1 (ATCC
CCL 246), KG-la (ATCC CCL 246.1), LS123 (ATCC CCL 255),
LS174T (ATCC CCL CL-188), LS180 (ATCC CCL CL-187), MCF-7
breast cancer cells (ATCC BTH 22), MOLT-4 cells (ATCC CRL
1582), Namalwa cells (ATCC CRL 1432), NCI-H498 (ATCC CCL '
254), NCI-H508 (ATCC CCL 253), NCI-H548 (ATCC CCL 249),
NCI-H716 (ATCC CCL 251), NCI-H747 (ATCC CCL 252),
NCI-H1688 (ATCC CCL 257), NCI-H2126 (ATCC CCL 256), Raji
cells (ATCC CCL 86), RD (ATCC CCL 136), RPMI 2650 (ATCC
CCL 30), RPMI 8226 cells (ATCC CCL 155), SNU-C2A (ATCC
CCL 250.1), SNU-C28 (ATCC CCL 250), SW-13 (ATCC CCL 105),
SW48 (ATCC CCL 231), SW403 (ATCC CCL 230), SW480 (ATCC
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 30 -
CCL 227), SW620 (ATCC CCL 227), SW837 (ATCC CCL 235),
SW948 (ATCC CCL 237), SW1116 (ATCC CCL 233), SW1417 (ATCC
CCL 238), SW1463 (ATCC CCL 234), T84 (ATCC CCL 248}, U-
937 cells (ATCC CRL 1593), WiDr (ATCC CCL 218), and WI-
38VA13 subline 2R4 cells (ATCC CLL 75.1), as well as
heterohybridoma cells produced by fusion of human cells
and cells of another species. Secondary human fibroblast
strains, such as WI-38 (ATCC CCL 75) and MRC-5 (ATCC CCL
171), may be used. In addition, primary, secondary, or
immortalized human cells, as well as primary, secondary,
or immortalized cells from other species, can be used for
in vitro protein production or gene therapy.
IFNA2-Expressinct Cells
Homologously recombinant cells of the invention
express IFNA2 at desired levels and are useful for both
in vitro production of IFNA2 and gene therapy.
Protein Production
Homologously recombinant cells according to this
invention can be used for in vitro production of IFNA2.
The cells are maintained under conditions, as described
in the art, which result in expression of proteins. The
IFNA2 protein may be purified from cell lysates or cell
supernatants. A pharmaceutical composition containing
the IFNA2 protein can be delivered to a human or an
animal by conventional pharmaceutical routes known in the
art (e. g., oral, intravenous, intramuscular, intranasal,
pulmonary, transmucosal, intradermal, rectal,
intrathecal, transdermal, subcutaneous, intraperitoneal,
or intralesional}. Oral administration may require use
of a strategy for protecting the protein from degradation
in the gastrointestinal tract: e.g., by encapsulation in
polymeric microcapsules.
Gene Therapy
Homologously recombinant cells of the present
invention are useful as populations of homologously
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 31 -
recombinant cell lines, as populations of homologously
recombinant primary or secondary cells, as homologously
recombinant clonal cell strains or lines, as homologously
recombinant heterogenous cell strains or lines, and as
cell mixtures in which at least one representative cell
of one of the four preceding categories of homologously
recombinant cells is present. Such cells may be used in
a delivery system for treating (i) infections-caused by
such viruses as papilloma virus, hepatitis B and C
viruses, vaccinia, herpes simplex virus, herpes zoster
varicellosus virus, and rhinovirus, and (ii) any other
conditions treatable with IFNA2.
Homologously recombinant primary cells, clonal
cell strains or heterogenous cell strains are
administered to an individual~in whom the abnormal or
undesirable condition is to be treated or prevented, in
sufficient quantity and by an appropriate route, to ex-
press or make available the protein or exogenous DNA at
physiologically relevant levels. A physiologically rele-
vant level is one which either approximates the level at
which the product is normally produced in the body or
results in improvement of the abnormal or undesirable
condition. If the cells are syngeneic with respect to a
immunocompetent recipient, the cells can be administered
or implanted intravenously, intraarterially,
subcutaneously, intraperitoneally, intraomentally,
subrenal capsularly, intrathecally, intracranially, or
intramuscularly.
If the cells are not syngeneic and the recipient
is immunocompetent, the homologously recombinant cells to
be administered can be enclosed in one or more
semipermeable barrier devices. The permeability
properties of the device are such that the cells are
prevented from leaving the device upon implantation into
a subject, but the therapeutic protein is freely
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 32 -
permeable and can leave the barrier device and enter the
local space surrounding the implant or enter the systemic
circulation. See, e.g., U.S. Patent Nos. 5,641,670,
5,470,731, 5,620,883, 5,487,737, and co-owned U.S. Patent
Application entitled "Delivery of Therapeutic Proteins'
(inventors: Justin C. Lamsa and Douglas A. Treco), filed
April 16, 1999, all herein incorporated by reference.
The barrier device can be implanted at any appropriate
site: e.g., intraperitoneally, intrathecally,
subcutaneously, intramuscularly, within the kidney
capsule, or within the omentum.
Barrier devices are particularly useful and allow
homologously recombinant immortalized cells, homologously
recombinant cells from another species (homologously
recombinant xenogeneic cells)., or cells from a nonhisto-
compatibility-matched donor (homologously recombinant
allogeneic cells) to be implanted for treatment of a
subject. The devices retain cells in a fixed position in
vivo, while protecting the cells from the host's immune
system. Barrier devices also allow convenient short-term
(i.e., transient) therapy by allowing ready removal of
the cells when the treatment regimen is to be halted for
any reason. Transfected or infected xenogeneic and
allogeneic cells may also be used in the absence of
barrier devices for short-term gene therapy. In that
case, the IFNA2 produced by the cells will be delivered
in vivo until the cells are rejected by the host's immune
system.
A number of synthetic, semisynthetic, or natural
filtration membranes can be used for this purpose,
including, but not limited to, cellulose, cellulose
acetate, nitrocellulose, polysulfone, polyvinylidene
difluoride, polyvinyl chloride polymers and polymers of
polyvinyl chloride derivatives. Barrier devices can be
utilized to allow primary, secondary, or immortalized
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 33 -
cells from another species to be used for gene therapy in
humans.
Another type of device useful in the gene therapy
of the invention is an implantable collagen matrix in
which the cells are embedded. Such a device, which can
contain beads to which the cells attach, is described in
WO 97/15195, herein incorporated by reference. It can be
implanted as described above.
The number of cells needed for a given dose or
implantation depends on several factors, including the
expression level of the protein, the size and condition
of the host animal, and the limitations associated with
the implantation procedure. Usually the number of cells
implanted in an adult human or other similarly-sized
animal is in the range of 1 X 104 to 5 X 101°, and
preferably 1 X 108 to 1 X 109. If desired, they may be
implanted at multiple sites in the patient, either at one
time or over a period of months or years. The dosage may
be repeated as needed.
Deposit
Under the terms of the Budapest Treaty on the
International Recognition of the Deposit of
Microorganisms for the Purpose of Patent Procedure, the
deposit of plasmid pA2HB was made with the American Type
Culture Collection (ATCC) of Rockville, MD, USA on May
I2, 1998. The deposit was given Accession Number 209872.
Applicants' assignee, Transkaryotic Therapies,
Inc., represents that the ATCC is a depository affording
permanence of the deposit and ready accessibility thereto
by the public if a patent is granted. All restrictions
on the availability to the public of the material so
deposited will be irrevocably removed upon the granting
of the patent. The material will be available during the
pendency of the patent application to.one determined by
the Commissioner to be entitled thereto under 37 CFR 1.14
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 34 -
and 35 U.S.C. X122. The deposited material will be
maintained with all the care necessary to keep it viable
and uncontaminated for a period of at least five years
after the most recent request for the furnishing of a
sample of the deposited material, and in any case, for a
period of at least thirty (30) years after the date of
deposit or for the enforceable life of the patent,
whichever period is longer. Applicants' assignee
acknowledges its duty to replace the deposit should the
depository be unable to furnish a sample when requested
due to the condition of the deposit.
Other Embodiments
It is to be understood that while the invention
has been described in conjunction with the detailed
description thereof, the foregoing description is
intended to illustrate and not to limit the scope of the
invention, which is defined by the scope of the appended
claims.
Other aspects, advantages, and modifications are
within the scope of the following claims.
What is claimed is:
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 1 -
SEQUENCE LISTING
<110> Transkaryotic Therapies Inc.
<120> GENOMIC SEQUENCES FOR PROTEIN PRODUCTION AND DELIVERY
<130> 07236/018W01
<150> US 60/086,555
<151> 1998-05-21
<150> US 60/084,648
<151> 1998-05-07
<160> 19
<170> FastSEQ for Windows Version 3.0
<210> 1
<211> 1733
<212> DNA
<213> Homo sapiens
<400>
1
gcgcctcttatgtacccacaaaaatctattttcaasaaagttgctctaagaatatagtta60
tcaagttaagtaaaatgtcaatagccttttaatttaatttttaattgttttatcattctt120
tgcaataataaaacattaactttatactttttaatttaatgtatagaatagagatataca180
taggatatgtaaatagatacacagtgtatatgtgattaaaatataatgggagattcaatc240
agaaaaaagtttctaaaaaggctctggggtaaaagaggaaggaaacaataatgaaaaaaa300
tgtggtgagaaaaacagctgaaaacccatgtaaagagtgtataaagaaagcaaaaagaga360
agtagaaagtaacacaggggcatttggaaaatgtaaacgagtatgttccctatttaaggc420
taggcacaaagcaaggtcttcagagaacctggagcctaaggtttaggctcacccatttca480
accagtctagcagcatctgcaacatctacaatggccttgacctttgctttactggtggcc540
ctcctggtgctcagctgcaagtcaagctgctctgtgggctgtgatctgcctcaaacccac600
agcctgggtagcaggaggaccttgatgctcctggcacagatgaggagaatctctcttttc660
tcctgcttgaaggacagacatgactttggatttccccaggaggagtttggcaaccagttc720
caaaaggctgaaaccatccctgtcctccatgagatgatccagcagatcttcaatctcttc780
agcacaaaggactcatctgctgcttgggatgagaccctcctagacaaattctacactgaa840
ctctaccagcagctgaatgacctggaagcctgtgtgatacagggggtgggggtgacagag900
actcccctgatgaaggaggactccattctggctgtgaggaaatacttccaaagaatcact960
ctctatctgaaagagaagaaatacagcccttgtgcctgggaggttgtcagagcagaaatc1020
atgagatctttttctttgtcaacaaacttgcaagaaagtttaagaagtaaggaatgaaaa1080
ctggttcaacatggaaatgattttcattgattcgtatgccagctcacctttttatgatct1140
gccatttcaaagactcatgtttctgctatgaccatgacacgatttaaatcttttcaaatg1200
tttttaggagtattaatcaacattgtattcagctcttaaggcactagtcccttacagagg1260
accatgctgactgatccattatctatttaaatatttttaaaatattatttatttaactat1320
ttataaaacaacttatttttgttcatattatgtcatgtgcacctttgcacagtggttaat1380
gtaataaaatgtgttctttgtatttggtaaatttattttgtgttgttcattgaacttttg1440
ctatggaacttttgtacttgtttattctttaaaatgaaattccaagcctaattgtgcaac1500
ctgattacagaataactggtacacttcatttgtccatcaatattatattcaagatataag1560
taaaaataaactttctgtaaaccaagttgtatgttgtactcaagataacagggtgaacct1620
aacaaatacaattctgctctcttgtgtatttgatttttgtatgaaaaaaactaaaaatgg1680
taatcatacttaattatcagttatggtaaatggtatgaagagaagaaggaacg 1733
<210>
2
<211>
188
<212>
PRT
<213>
Homo
sapiens
<400>
2
Met Ala Leu Val
Leu Thr Leu Ser
Phe Ala Cys
Leu Leu
Val Ala
Leu
1 5 10 15
Lys Ser Asp Leu Gln Thr
Ser Cys Pro His Ser
Ser Val Leu
Gly Cys
20 25 30
Gly Ser Leu Ala Met Arg
Arg Arg Gln Arg Ile
Thr Leu Ser
Met Leu
35 40 45
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 2 -
Leu Phe Ser Cys Leu Lys Asp Arg His Asp Phe Gly Phe Pro Gln Glu
50 55 60
Glu Phe Gly Asn Gln Phe Gln Lys Ala Glu Thr Ile Pro Val Leu His
65 70 75 80
Glu Met Ile Gln Gln Ile Phe Asn Leu Phe Ser Thr Lys Asp Ser Ser
85 90 95
Ala Ala Trp Asp Glu Thr Leu Leu Asp Lys Phe Tyr Thr Glu Leu Tyr
100 105 110
Gln Gln Leu Asn Asp Leu Glu Ala Cys Val Ile Gln Gly Val Gly Val
115 120 125
Thr Glu Thr Pro Leu Met Lye Glu Asp Ser Ile Leu Ala Val Arg Lys
130 135 140
Tyr Phe Gln Arg Ile Thr Leu Tyr Leu Lys Glu Lys Lys Tyr Ser Pro
145 150 155 160
Cys Ala Trp Glu Val Val Arg Ala Glu Ile Met Arg Ser Phe Ser Leu
165 170 175
Ser Thr Asn Leu Gln Glu Ser Leu Arg Ser Lys Glu
180 185
<210> 3
<211> 24
<212> DNA
<213> Homo sapiens
<400> 3
agtttctaaa aaggctctgg ggta 24
<210> 4
<211> 19
<212> DNA
<213> Homo sapiens
<400> 4
gcccacagag cagcttgac 19
<210> 5
<211> 25
<212> DNA
<213> Homo sapiens
<400> 5
aaagactcat gtttctgcta tgacc 25
<210> 6
<211> 24
<212> DNA
<213> Homo sapiens
<400> 6
ggtgcacatg acataatatg aaca 24
<210> 7
<211> 278
<212> DNA
<213> Homo sapiens
<400>
7
aagcttttataggtgtaaattttccacttagtactgcttttgtaatgttgtctttttatt 60
ttcatttatctcaagatgttttctaatttctcttgacttccttcttaaattcttacctca 120
tgtagacatacatttttggccctatgcattgggatgcaaaaccagactaatttactttgt 180
acaaaaagaaaaatgagaaagaaatatatttggtcttgtgagcactatatggaaatactt 240
tatattccatttgtttcatcatattcatatatcccttt 278
<210> 8
<211> 73
<212> DNA
<213> Homo sapiens
CA 02328459 2000-11-07
WO 99/57292 PGT/US99/09925
- 3 -
<400> 8
cattggatac tccatcacct gctgtgatat tatgaatgtc tgcctatata aatattcact 60
attccataac aca 73
<210> 9
<211> 1806
<212> DNA
<213> Homo eapiens
<400>
9
cattggatactccatcacctgctgtgatattatgaatgtctgcctatataaatattcact60
attccataacacagcgcctcttatgtacccacaaaaatctattttcaaaaaagttgctct120
aagaatatagttatcaagttaagtaaaatgtcaatagccttttaatttaatttttaattg180
ttttatcattctttgcaataataaaacattaactttatactttttaatttaatgtataga240
atagagatatacataggatatgtaaatagatacacagtgtatatgtgattaaaatataat300
gggagattcaatcagaaaaaagtttctaaaaaggctctggggtaaaagaggaaggaaaca360
ataatgaaaaaaatgtggtgagaaaaacagctgaaaacccatgtaaagagtgtataaaga420
aagcaaaaagagaagtagaaagtaacacaggggcatttggaaaatgtaaacgagtatgtt480
ccctatttaaggctaggcacaaagcaaggtcttcagagaacctggagcctaaggtttagg540
ctcacccatttcaaccagtctagcagcatctgcaacatctacaatggccttgacctttgc600
tttactggtggccctcctggtgctcagctgcaagtcaagctgctctgtgggctgtgatct660
gcctcaaacccacagcctgggtagcaggaggaccttgatgctcctggcacagatgaggag720
aatctctcttttctcctgcttgaaggacagacatgactttggatttccccaggaggagtt780
tggcaaccagttccaaaaggctgaaaccatccctgtcctccatgagatgatccagcagat840
cttcaatctcttcagcacaaaggactcatctgctgcttgggatgagaccctcctagacaa900
attctacactgaactctaccagcagctgaatgacctggaagcctgtgtgatacagggggt960
gggggtgacagagactcccctgatgaaggaggactccattctggctgtgaggaaatactt1020
ccaaagaatcactctctatctgaaagagaagaaatacagcccttgtgcctgggaggttgt1080
cagagcagaaatcatgagatctttttctttgtcaacaaacttgcaagaaagtttaagaag1140
taaggaatgaaaactggttcaacatggaaatgattttcattgattcgtatgccagctcac1200
ctttttatgatctgccatttcaaagactcatgtttctgctatgaccatgacacgatttaa1260
atcttttcaaatgtttttaggagtattaatcaacattgtattcagctcttaaggcactag1320
tcccttacagaggaccatgctgactgatccattatctatttaaatatttttaaaatatta1380
tttatttaactatttataaaacaacttatttttgttcatattatgtcatgtgcacctttg1440
cacagtggttaatgtaataaaatgtgttctttgtatttggtaaatttattttgtgttgtt1500
cattgaacttttgctatggaacttttgtacttgtttattctttaaaatgaaattccaagc1560
ctaattgtgcaacctgattacagaataactggtacacttcatttgtccatcaatattata1620
ttcaagatataagtaaaaataaactttctgtaaaccaagttgtatgttgtactcaagata1680
acagggtgaacctaacaaatacaattctgctctcttgtgtatttgatttttgtatgaaaa1740
aaactaaaaatggtaatcatacttaattatcagttatggtaaatggtatgaagagaagaa1800
ggaacg 1806
<210> 10
<211> 4090
<212> DNA
<213> Homo sapiens
<400> 10
aagcttttataggtgtaaattttccacttagtactgcttttgtaatgttgtctttttatt60
ttcatttatctcaagatgttttctaatttctcttgacttccttcttaaattcttacctca120
tgtagacatacatttttggccctatgcattgggatgcaaaaccagactaatttactttgt180
acaaaaagaaaaatgagaaagaaatatatttggtcttgtgagcactatatggaaatactt240
tatattccatttgtttcatcatattcatatatccctttactaacataaagctgaaggtga300
ataaasaaatcagggttagccaaacaaattttcatggtcaaataccacataaaaagtaaa360
tatacttaagttcccagcaaaatctgaattgaacgtagacaaaatgctcatttctcagtg~
420
tttgacagacttaacagtttgagccaataaaaatgtactgactagataaactactaaaag480
ttgttaatttttgcaatgtatatttctgaaaagaaagtttatctattatagaaattcctg.540
tgcccatttaagaactttgagcattttaattgtttaataatatagtttaattgcatcatg600
aaaataatcaataatacaatttatttggtttatttaaaaaaactgattctttctgctctc660
tctatatatagactgattttatactaatgttgcctaaagatcaccaaattgtttgaagcc720
taggtttctgagggatggaaaatgatgtcacaactatttacagttcacacacacattctg780
gggatttaatacatcctttacaagtgcaggaaaggtggaagattgatgatttgggggaat840
tagagctaccacaccccagagggtggtatggtatgttgtctgttgtgagctgtgtgaatc900
agagagtttgatttagacatatatttagaaagaggaaagatgaaccaatcaaaaataata960
actataatgacttttcaagatatagacaatacagttaagatataaatggaaacaaaaaaa1020
gttaaaagtggggagatgaagtctgattttttggtttttttttttttttgcttttttgtt1080
tgtttatgtaatcagtgttaccagtttaaaataatgggttataagacactatatgcaagc1140
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 4 -
ctcatggtaacctccaatctaaaacatacaacaaatacacacaaaataaaaaggagaaat1200
taaaacacaccaccagagaaaatcacctacattaaaagaaagacaaataggaagaaaata1260
agaaagagaaggccatcaaataatcagaaaatgaataacaaaatgacaggaataagtcct1320
cataaataataacattgaatgtaaatggactaagctctccaatgaaagacagggagtggc1380
tgaatgtattttaaaaaaaatattacaccgagctgtgcgtggtgtctcacacctataatc1440
ccagcattttgggagactgagccgggtggatcacttgagcccaggagttcgagaccagcc1500
tggccaacatggcaaaaccctgtctctactaaaaatacaaaaaattagctgaacatggtg1560
gcacatgcctgtggttccagctactagagaggctgaggcagaagaattgcttgaacttgg1620
gaggtggaggttgcagtgagctaagattgatggagccactgcaccccagcctaggtgaca1680
gaataagactctgcctcaaaaaaaaaaagcaaaacaaaacaaaacaaaaaacccttagac1740
ccaatgattcattgcctacaagaagtatgcttcacctttaaagacacatatagactgaag1800
gtaaagggatggaaaaatattctatgcctatggaaacasacaaaaagaagcagaagctac1860
atttatatcagacaaaatagactgcaagacaaaaactatgaaaagagagaaagaaggtca1920
ttatatagtgatasaggggtccatttagcaagagcatttaacaattctaaatatatattc1980
acccaatactggagtactcaggtatataaagcaaatattattagagccaaagagagagat2040
agacagacccccatacaataataactggagacttcaacaccccactttcagcattggaca2100
gatcatccagacagaaaattaacaaacatcaaatttcatctgcaccataggtcaaatgga2160
cctagtagatatttacagaacatttgatccaacagctgtagaatacacattcttctcctc2220
agcacatggataattctcaaggatataccaaatgctaggtcacaaaacaaatcttaaaat2280
ttagaaaaaaagtgaaataatatcaaacgttttctctcaccacagactaagaaaaaaaga2340
agtcccaaataaatacaatctgagataaaaaaggagacgagacaaccaataccacaaaaa2400
attaaaggatcattagaagatactatgaaactatatgctaataaattggaaaacctgaac2460
aaaatagataattcctagaaacatacaacatactggtctgttcaggttttgtattttttc2520
atagtaccatgaagaaatacaagaattgtttctagaaccattcttgtatttcttcatggt2580
ttttgtatttcttcatggaaccatgaagaaatacaaaatgtgaacaggccaataacaagt2640
aatgagacagaagccatactaaaaagtatcccagaaaagaactcaggatctgatggcttc2700
actgatgaattttgccaaatatttaaaaaactaataccaatccaactcaaattattaaaa2760
aaatagaggtggacagaatctttccaaatgtattctatgaggccagtgttttttctgatt2820
gaatctcccattatattttaatcacatataaaaccagagaaagacacattaaaaagaaag2880
aaaactgtaggccaatatctctgatgaacattgatgcagaaatcctcaacaacaaattag2940
caaactgaattcaagaacacattaaaacaatcattcatcatgaccaagtggaatttgtcc3000
tagagattcaagtgtggttaggtatgtgcagatcaatgggtttaatgttgtccaatgaac3060
ataatgtcctccagctccatccatgttcttgcaaatgacaggatctcattcttttttatg3120
gctaagtagtactccattgtgtataagtgccatattttctttatccattcatctgttaga3180
cacctaagttgcttccaaatcttagctattgtgaatagtgctgcaataaacatgggagtg3240
tasatattttgttgacatactgatttcatttcctttggataaatacccagtagtgggatt3300
gctggatcatatgggggaaaatggagatggctaacgggctcaaaaatatagttagaaaaa3360
atgaatatgatttagtattcgatagcacaataggatgactactgttaatgataatttatt3420
atatattataaaataactaaaatagtataaatgggatgtatgtagcagagagaaatgata3480
aatgtttgaagcattggatactccatcacctgctgtgatattatgaatgtctgcctatat3540
saatattcactattccataacacagcgcctcttatgtacccacaaaaatctattttcaaa3600
aaagttgctctaagaatatagttatcaagttaagtaaaatgtcaatagccttttaattta3660
atttttaattgttttatcattctttgcaataataaaacattaactttatactttttaatt3720
taatgtatagaatagagatatacataggatatgtaaatagatacacagtgtatatgtgat3780
taaaatataatgggagattcaatcagaaaaaagtttctaaaaaggctctggggtaaaaga3840
ggaaggaaacaataatgaaaaaaatgtggtgagaaaaacagctgaaaacccatgtaaaga3900
gtgtataaagaaagcaaaaagagaagtagaaagtaacacaggggcatttggaaaatgtaa3960
acgagtatgttccctatttaaggctaggcacaaagcaaggtcttcagagaacctggagcc4020
taaggtttaggctcacccatttcaaccagtctagcagcatctgcaacatctacaatggcc4080
ttgacctttg 4090
<210> 11
<211> 3564
<212> DNA
<213> Homo Sapiens
<400>
11
aagcttttataggtgtaaattttccacttagtactgcttttgtaatgttgtctttttatt 60
ttcatttatctcaagatgttttctaatttctcttgacttccttcttaaattcttacctca 120
tgtagacatacatttttggccctatgcattgggatgcaaaaccagactaatttactttgt 180
acaaaaagaaaaatgagaaagaaatatatttggtcttgtgagcactatatggaaatactt 240
tatattccatttgtttcatcatattcatatatccctttactaacataaagctgaaggtga 300
ataaaaaaatcagggttagccaaacaaattttcatggtcaaataccacataaaaagtaaa 360
tatacttaagttcccagcaaaatctgaattgaacgtagacaaaatgctcatttctcagtg 420
tttgacagacttaacagtttgagccaataaaaatgtactgactagataaactactaaaag 480
ttgttaatttttgcaatgtatatttctgaaaagaaagtttatctattatagaaattcctg 540
tgcccatttaagaactttgagcattttaattgtttaataatatagtttaattgcatcatg 600
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 5 -
aaaataatcaataatacaatttatttggtttatttaaaaaaactgattctttctgctctc660
tctatatatagactgattttatactaatgttgcctaaagatcaccaaattgtttgaagcc720
taggtttctgagggatggaaaatgatgtcacaactatttacagttcacacacacattctg780
gggatttsatacatcctttacaagtgcaggaaaggtggaagattgatgatttgggggaat840
tagagctaccacaccccagagggtggtatggtatgttgtctgttgtgagctgtgtgaatc900
agagagtttgatttagacatatatttagaaagaggaaagatgaaccaatcaaaaataata960
actataatgacttttcaagatatagacaatacagttaagatataaatggaaacaaaaaaa1020
gttaaaagtggggagatgaagtctgattttttggtttttttttttttttgcttttttgtt1080
tgtttatgtaatcagtgttaccagtttaaaataatgggttataagacactatatgcaagc1140
ctcatggtaacctccaatctaaaacatacaacaaatacacacaaaataaaaaggagaaat1200
taaaacacaccaccagagaaaatcacctacattaaaagaaagacaaataggaagasaata1260
agaaagagaaggccatcaaataatcagaaaatgaataacaaaatgacaggaataagtcct1320
cataaataataacattgaatgtaaatggactaagctctccaatgaaagacagggagtggc1380
tgaatgtattttaaaaaaaatattacaccgagctgtgcgtggtgtctcacacctataatc1440
ccagcattttgggagactgagccgggtggatcacttgagcccaggagttcgagaccagcc1500
tggccaacatggcaaaaccctgtctctactaaaaatacaaaaaattagctgaacatggtg1560
gcacatgcctgtggttccagctactagagaggctgaggcagaagaattgcttgaacttgg1620
gaggtggaggttgcagtgagctaagattgatggagccactgcaccccagcctaggtgaca1680
gaataagactctgcctcaaaaaaaaaaagcaaaacaaaacaaaacaaaaaacccttagac1740
ccaatgattcattgcctacaagaagtatgcttcacctttaaagacacatatagactgaag1800
gtaaagggatggaaaaatattctatgcctatggaaacaaacaaaaagaagcagaagctac1860
atttatatcagacaaaatagactgcaagacaaaaactatgaaaagagagaaagaaggtca1920
ttatatagtgataaaggggtccatttagcaagagcatttaacaattctaaatatatattc1980
acccaatactggagtactcaggtatataaagcaaatattattagagccaaagagagagat2040
agacagacccccatacaataataactggagacttcaacaccccactttcagcattggaca2100
gatcatccagacagaaaattaacaaacatcaaatttcatctgcaccataggtcaaatgga2160
cctagtagatatttacagaacatttgatccaacagctgtagaatacacattcttctcctc2220
agcacatggataattctcaaggatataccaaatgctaggtcacaaaacaaatcttaaaat2280
ttagaaaaaaagtgaaataatatcaaacgttttctctcaccacagactaagaaaaaaaga2340
agtcccaaataaatacaatctgagataaaaaaggagacgagacaaccaataccacaaaaa2400
attaaaggatcattagaagatactatgaaactatatgctaataaattggaaaacctgaac2460
aaaatagataattcctagaaacatacaacatactggtctgttcaggttttgtattttttc2520
atagtaccatgaagaaatacaagaattgtttctagaaccattcttgtatttcttcatggt2580
ttttgtatttcttcatggaaccatgaagaaatacaaaatgtgaacaggccaataacaagt2640
aatgagacagaagccatactaaaaagtatcccagaaaagaactcaggatctgatggcttc2700
actgatgaattttgccaaatatttaaaaaactaataccaatccaactcaaattattaaaa2760
aaatagaggtggacagaatctttccaaatgtattctatgaggccagtgttttttctgatt2820
gaatctcccattatattttaatcacatataaaaccagagaaagacacattaaaaagaaag2880
aaaactgtaggccaatatctctgatgaacattgatgcagaaatcctcaacaacaaattag2940
caaactgaattcaagaacacattaaaacaatcattcatcatgaccaagtggaatttgtcc3000
tagagattcaagtgtggttaggtatgtgcagatcaatgggtttaatgttgtccaatgaac3060
ataatgtcctccagctccatccatgttcttgcaaatgacaggatctcattcttttttatg3120
gctaagtagtactccattgtgtataagtgccatattttctttatccattcatctgttaga3180
cacctaagttgcttccaaatcttagctattgtgaatagtgctgcaataaacatgggagtg3240
taaatattttgttgacatactgatttcatttcctttggataaatacccagtagtgggatt3300
gctggatcatatgggggaaaatggagatggctaacgggctcaaaaatatagttagaaaaa3360
atgaatatgatttagtattcgatagcacaataggatgactactgttaatgataatttatt3420
atatattataaaataactaaaatagtataaatgggatgtatgtagcagagagaaatgata3480
aatgtttgaagcattggatactccatcacctgctgtgatattatgaatgtctgcctatat3540
aaatattcactattccataacaca 3564
<210> 12
<211> 3213
<212> DNA
<213> Homo sapiens
<400> 12
actaacataaagctgaaggtgaataaaaaaatcagggttagccaaacaaattttcatggt 60
caaataccacataaaaagtaaatatacttaagttcccagcaaaatctgaattgaacgtag 120
acaaaatgctcatttctcagtgtttgacagacttaacagtttgagccaataaaaatgtac 180
tgactagataaactactaaaagttgttaatttttgcaatgtatatttctgaaaagaaagt 240
ttatctattatagaaattcctgtgcccatttaagaactttgagcattttaattgtttaat 300
aatatagtttaattgcatcatgaaaataatcaataatacaatttatttggtttatttaaa 360
aaaactgattctttctgctctctctatatatagactgattttatactaatgttgcctaaa 420
gatcaccaaattgtttgaagcctaggtttctgagggatggaaaatgatgtcacaactatt 480
tacagttcacacacacattctggggatttaatacatcctttacaagtgcaggaaaggtgg 540
aagattgatgatttgggggaattagagctaccacaccccagagggtggtatggtatgttg 600
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 6 -
tctgttgtgagctgtgtgaatcagagagtttgatttagacatatatttagaaagaggaaa660
gatgaaccaatcaaaaataataactataatgacttttcaagatatagacaatacagttaa720
gatataaatggaaacaaaaaaagttaaaagtggggagatgaagtctgattttttggtttt780
tttttttttttgcttttttgtttgtttatgtaatcagtgttaccagtttaaaataatggg840
ttataagacactatatgcaagcctcatggtaacctccaatctaaaacatacaacaaatac900
acacaaaataaaaaggagaaattaaaacacaccaccagagaaaatcacctacattaaaag960
asagacaaataggaagaaaataagaaagagaaggccatcaaataatcagaaaatgaataa1020
caaaatgacaggaataagtcctcataaataataacattgaatgtaaatggactaagctct1080
ccaatgaaagacagggagtggctgaatgtattttaaaaaaaatattacaccgagctgtgc1140
gtggtgtctcacacctataatcccagcattttgggagactgagccgggtggatcacttga1200
gcccaggagttcgagaccagcctggccaacatggcaaaaccctgtctctactaaaaatac1260
aaaaaattagctgaacatggtggcacatgcctgtggttccagctactagagaggctgagg1320
cagaagaattgcttgaacttgggaggtggaggttgcagtgagctaagattgatggagcca1380
ctgcaccccagcctaggtgacagaataagactctgcctcaaaaaaaaaaagcaaaacaaa1440
acaaaacaaaaaacccttagacccaatgattcattgcctacaagaagtatgcttcacctt1500
taaagacacatatagactgaaggtaaagggatggaaaaatattctatgcctatggaaaca1560
aacaaaaagaagcagaagctacatttatatcagacaaaatagactgcaagacaaaaacta1620
tgaaaagagagaaagaaggtcattatatagtgataaaggggtccatttagcaagagcatt1680
taacaattctaaatatatattcacccaatactggagtactcaggtatataaagcaaatat1740
tattagagccaaagagagagatagacagacccccatacaataataactggagacttcaac1800
accccactttcagcattggacagatcatccagacagaaaattaacaaacatcaaatttca1860
tctgcaccataggtcaaatggacctagtagatatttacagaacatttgatccaacagctg1920
tagaatacacattcttctcctcagcacatggataattctcaaggatataccaaatgctag1980
gtcacaaaacaaatcttaaaatttagaaaaaaagtgaaataatatcaaacgttttctctc2040
accacagactaagaaaaaaagaagtcccaaataaatacaatctgagataaaaaaggagac2100
gagacaaccaataccacaaaaaattaaaggatcattagaagatactatgaaactatatgc2160
taataaattggaaaacctgaacaaaatagataattcctagaaacatacaacatactggtc2220
tgttcaggttttgtattttttcatagtaccatgaagaaatacaagaattgtttctagaac2280
cattcttgtatttcttcatggtttttgtatttcttcatggaaccatgaagaaatacaaaa2340
tgtgaacaggccaataacaagtaatgagacagaagccatactaaaaagtatcccagaaaa2400
gaactcaggatctgatggcttcactgatgaattttgccaaatatttaaaaaactaatacc2460
aatccaactcaaattattaaaaaaatagaggtggacagaatctttccaaatgtattctat2520
gaggccagtgttttttctgattgaatctcccattatattttaatcacatataaaaccaga2580
gaaagacacattaaaaagaaagaaaactgtaggccaatatctctgatgaacattgatgca2640
gaaatcctcaacaacaaattagcaaactgaattcaagaacacattaaaacaatcattcat2?00
catgaccaagtggaatttgtcctagagattcaagtgtggttaggtatgtgcagatcaatg2760
ggtttaatgttgtccaatgaacataatgtcctccagctccatccatgttcttgcaaatga2820
caggatctcattcttttttatggctaagtagtactccattgtgtataagtgccatatttt2880
ctttatccattcatctgttagacacctaagttgcttccaaatcttagctattgtgaatag2940
tgctgcaataaacatgggagtgtaaatattttgttgacatactgatttcatttcctttgg3000
ataaatacccagtagtgggattgctggatcatatgggggaaaatggagatggctaacggg3060
ctcaaaaatatagttagaaaaaatgaatatgatttagtattcgatagcacaataggatga3120
ctactgttaatgataatttattatatattataaaataactaaaatagtataaatgggatg3180
tatgtagcagagagaaatgataaatgtttgaag 3213
<210> 13
<211> 3033
<212> DNA
<213> Homo eapiens
<400>
13
actaacataaagctgaaggtgaataaaaaaatcagggttagccaaacaaattttcatggt 60
caaataccacataaaaagtaaatatacttaagttcccagcaaaatctgaattgaacgtag 120
acaaaatgctcatttctcagtgtttgacagacttaacagtttgagccaataaaaatgtac 180
tgactagataaactactaaaagttgttaatttttgcaatgtatatttctgaaaagaaagt 2.40
ttatctattatagaaattcctgtgcccatttaagaactttgagcattttaattgtttaat 300
aatatagtttaattgcatcatgaaaataatcaataatacaatttatttggtttatttaaa 360
aaaactgattctttctgctctctctatatatagactgattttatactaatgttgcctaaa 420
gatcaccaaattgtttgaagcctaggtttctgagggatggaaaatgatgtcacaactatt 480
tacagttcacacacacattctggggatttaatacatcctttacaagtgcaggaaaggtgg 540
aagattgatgatttgggggaattagagctaccacaccccagagggtggtatggtatgttg 600
tctgttgtgagctgtgtgaatcagagagtttgatttagacatatatttagaaagaggaaa 660
gatgaaccaatcaaaaataataactataatgacttttcaagatatagacaatacagttaa 720
gatataaatggaaacaaaaaaagttaaaagtggggagatgaagtctgattttttggtttt 780
tttttttttttgcttttttgtttgtttatgtaatcagtgttaccagtttaaaataatggg 840
ttataagacactatatgcaagcctcatggtaacctccaatctaaaacatacaacaaatac 900
acacaaaataaaaaggagaaattaaaacacaccaccagagaaaatcacctacattaaaag 960
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
_ 7 _
aaagacaaataggaagaasataagaaagagaaggccatcaaataatcagaaaatgaataa1020
caaaatgacaggaataagtcctcataaataataacattgaatgtaaatggactaagctct1080
ccaatgaaagacagggagtggctgaatgtattttaaaaaaaatattacaccgagctgtgc1140
gtggtgtctcacacctataatcccagcattttgggagactgagccgggtggatcacttga1200
gcccaggagttcgagaccagcctggccaacatggcaaaaccctgtctctactaaaaatac1260
aaaaaattagctgaacatggtggcacatgcctgtggttccagctactagagaggctgagg1320
cagaagaattgcttgaacttgggaggtggaggttgcagtgagctaagattgatggagcca1380
ctgcaccccagcctaggtgacagaataagactctgcctcaaaaaaaaaaagcaaaacaaa1440
acaaaacaaaaaacccttagacccaatgattcattgcctacaagaagtatgcttcacctt1500
taaagacacatatagactgaaggtaaagggatggaaaaatattctatgcctatggaaaca1560
aacaaaaagaagcagaagctacatttatatcagacaaaatagactgcaagacaaaaacta1620
tgaaaagagagaaagaaggtcattatatagtgataaaggggtccatttagcaagagcatt1680
taacaattctaaatatatattcacccaatactggagtactcaggtatataaagcaaatat1740
tattagagccaaagagagagatagacagacccccatacaataataactggagacttcaac1800
accccactttcagcattggacagatcatccagacagaaaattaacaaacatcaaatttca1860
tctgcaccataggtcaaatggacctagtagatatttacagaacatttgatccaacagctg1920
tagaatacacattcttctcctcagcacatggataattctcaaggatataccaaatgctag1980
gtcacaaaacaaatcttaaaatttagaaaaaaagtgaaataatatcaaacgttttctctc2040
accacagactaagaaaaaaagaagtcccaaataaatacaatctgagataaaaaaggagac2100
gagacaaccaataccacaaaaaattaaaggatcattagaagatactatgaaactatatgc2160
taataaattggaaaacctgaacaaaatagataattcctagaaacatacaacatactggtc2220
tgttcaggttttgtattttttcatagtaccatgaagaaatacaagaattgtttctagaac2280
cattcttgtatttcttcatggtttttgtatttcttcatggaaccatgaagaaatacaaaa2340
tgtgaacaggccaataacaagtaatgagacagaagccatactaaaaagtatcccagaaaa2400
gaactcaggatctgatggcttcactgatgaattttgccaaatatttaaaaaactaatacc2460
aatccaactcaaattattaaaaaaatagaggtggacagaatctttccaaatgtattctat2520
gaggccagtgttttttctgattgaatctcccattatattttaatcacatataaaaccaga2580
gaaagacacattaaaaagaaagaaaactgtaggccaatatctctgatgaacattgatgca2640
gaaatcctcaacaacaaattagcaaactgaattcaagaacacattaaaacaatcattcat2700
catgaccaagtggaatttgtcctagagattcaagtgtggttaggtatgtgcagatcaatg2760
ggtttaatgttgtccaatgaacataatgtcctccagctccatccatgttcttgcaaatga2820
caggatctcattcttttttatggctaagtagtactccattgtgtataagtgccatatttt2880
ctttatccattcatctgttagacacctaagttgcttccaaatcttagctattgtgaatag2940
tgctgcaataaacatgggagtgtaaatattttgttgacatactgatttcatttcctttgg3000
ataaatacccagtagtgggattgctggatcata 3033
<210> 14
<211> 137
<212> DNA
<213> Homo sapiens
<400> 14
gagaacctgg agcctaaggt ttaggctcac ccatttcaac cagtctagca gcatctgcaa 60
catctacaat ggccttgacc tttgctttac tggtggccct cctggtgctc agctgcaagt 120
caagctgctc tgtgggc 137
<210> 15
<211> 805
<212> DNA
<213> Homo sapiens
<400> 15
aagcttttataggtgtaaattttccacttagtactgcttttgtaatgttgtctttttatt 60
ttcatttatctcaagatgttttctaatttctcttgacttccttcttaaattcttacctca I20
tgtagacatacatttttggccctatgcattgggatgcaaaaccagactaatttactttgt 180
acaaaaagaaaaatgagaaagaaatatatttggtcttgtgagcactatatggaaatactt 240
tatattccatttgtttcatcatattcatatatccctttactaacataaagctgaaggtga 300
ataaaaaaatcagggttagccaaacaaattttcatggtcaaataccacataaaaagtaaa 360
tatacttaagttcccagcaaaatctgaattgaacgtagacaaaatgctcatttctcagtg 420
tttgacagacttaacagtttgagccaataaaaatgtactgactagataaactactaaaag 480
ttgttaatttttgcaatgtatatttctgaaaagaaagtttatctattatagaaattcctg 540
tgcccatttaagaactttgagcattttaattgtttaataatatagtttaattgcatcatg 600
aaaataatcaataatacaatttatttggtttatttaaaaaaactgattctttctgctctc 660
tctatatatagactgattttatactaatgttgcctaaagatcaccaaattgtttgaagcc 720
taggtttctgagggatggaaaatgatgtcacaactatttacagttcacacacacattctg 780
gggatttaatacatcctttacaagt 805
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 8 -
<210> 16
<211> 29
<212> DNA
<213> Homo sapiene
<400> 16
aggaaaggtg gaagattgat gatttgggg 29
<210> 17
<211> 105
<212> DNA
<213> Homo sapiens
<400> 17
ggggaattag agctaccaca ccccagaggg tggtatggta tgttgtctgt tgtgagctgt 60
gtgaatcaga gagtttgatt tagacatata tttagaaaga ggaaa 105
<210> 18
<211> 2629
<212> DNA
<213> Homo sapiens
<400> 18
aaagatgaaccaatcaaaaataataactataatgacttttcaagatatagacaatacagt60
taagatataaatggaaacaaaaaaagttaaaagtggggagatgaagtctgattttttggt120
ttttttttttttttgcttttttgtttgtttatgtaatcagtgttaccagtttaaaataat180
gggttataagacactatatgcaagcctcatggtaacctccaatctaaaacatacaacaaa240
tacacacaaaataaaaaggagaaattaaaacacaccaccagagaaaatcacctacattaa300
aagasagacaaataggaagaaaataagaaagagaaggccatcaaataatcagaaaatgaa360
taacaaaatgacaggaataagtcctcataaataataacattgaatgtaaatggactaagc420
tctccaatgaaagacagggagtggctgaatgtattttaaaaaaaatattacaccgagctg480
tgcgtggtgtctcacacctataatcccagcattttgggagactgagccgggtggatcact540
tgagcccaggagttcgagaccagcctggccaacatggcaaaaccctgtctctactaaaaa600
tacaaaaaattagctgaacatggtggcacatgcctgtggttccagctactagagaggctg660
aggcagaagaattgcttgaacttgggaggtggaggttgcagtgagctaagattgatggag720
ccactgcaccccagcctaggtgacagaataagactctgcctcaaaaaaaaaaagcaaaac780
aaaacaaaacaaaaaacccttagacccaatgattcattgcctacaagaagtatgcttcac840
ctttaaagacacatatagactgaaggtaaagggatggaaaaatattctatgcctatggaa900
acaaacaaaaagaagcagaagctacatttatatcagacaaaatagactgcaagacaaaaa960
ctatgaaaagagagaaagaaggtcattatatagtgataaaggggtccatttagcaagagc1020
atttaacaattctaaatatatattcacccaatactggagtactcaggtatataaagcaaa1080
tattattagagccaaagagagagatagacagacccccatacaataataactggagacttc1140
aacaccccactttcagcattggacagatcatccagacagaaaattaacaaacatcaaatt1200
tcatctgcaccataggtcaaatggacctagtagatatttacagaacatttgatccaacag1260
ctgtagaatacacattcttctcctcagcacatggataattctcaaggatataccaaatgc1320
taggtcacaaaacaaatcttaaaatttagaaaaaaagtgaaataatatcaaacgttttct1380
ctcaccacagactaagaaaaaaagaagtcccaaataaatacaatctgagataaaaaagga1440
gacgagacaaccaataccacaaaaaattaaaggatcattagaagatactatgaaactata1500
tgctaataaattggaaaacctgaacaaaatagataattcctagaaacatacaacatactg1560
gtctgttcaggttttgtattttttcatagtaccatgaagaaatacaagaattgtttctag1620
aaccattcttgtatttcttcatggtttttgtatttcttcatggaaccatgaagasataca1680
aaatgtgaacaggccaataacaagtaatgagacagaagccatactaaaaagtatcccaga1740
aaagaactcaggatctgatggcttcactgatgaattttgccaaatatttaaaaaactaat1800
accaatccaactcaaattattaaaaaaatagaggtggacagaatctttccaaatgtattc1860
tatgaggccagtgttttttctgattgaatctcccattatattttaatcacatataaaacc1920
agagaaagacacattaaaaagaaagaaaactgtaggccaatatctctgatgaacattgat1980
gcagaaatcctcaacaacaaattagcaaactgaattcaagaacacattaaaacaatcatt2040
catcatgaccaagtggaatttgtcctagagattcaagtgtggttaggtatgtgcagatca2100
atgggtttaatgttgtccaatgaacataatgtcctccagctccatccatgttcttgcaaa2160
tgacaggatctcattcttttttatggctaagtagtactccattgtgtataagtgccatat2220
tttctttatccattcatctgttagacacctaagttgcttccaaatcttagctattgtgaa2280
tagtgctgcaataaacatgggagtgtaaatattttgttgacatactgatttcatttcctt2340
tggataaatacccagtagtgggattgctggatcatatgggggaaaatggagatggctaac2400
gggctcaaaaatatagttagaaaaaatgaatatgatttagtattcgatagcacaatagga2460
tgactactgttaatgataatttattatatattataaaataactaaaatagtataaatggg2520
atgtatgtagcagagagaaatgataaatgtttgaagcattggatactccatcacctgctg2580
tgatattatgaatgtctgcctatataaatattcactattccataacaca 2629
CA 02328459 2000-11-07
WO 99/57292 PCT/US99/09925
- 9 -
<210> 19
<211> 14
<212> DNA
<213> Homo Sapiens
<220>
<221> misc_feature
<222> (1). .(14)
<223> n = A,T,C or G
<400> 19
YYYYYYYYYY nYag 14
CA 02328459 2000-11-07