Note: Descriptions are shown in the official language in which they were submitted.
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
ANTISENSE OLIGONUCLEOTIDE CONSTRUCTS BASED ON
f3-ARABINOFURANOSE AND ITS ANALOGUES
BACKGROUND OF THE INVENTION
(a) Field of the Invention
It is the primary objective of this invention
to provide modified oligonucleotide therapeutic agents
to selectively prevent gene transcription and expres-
sion in a sequence-specific manner. In particular,
this invention is directed to the selective inhibition
of protein biosynthesis via antisense strategy using
oligonucleotides constructed from arabinonucleotide or
modified arabinonucleotide residues. More particularly
this invention relates to the use of antisense oligonu-
cleotides having arabinose sugars to hybridize to com-
plementary RNA such as cellular messenger RNA, viral
RNA, etc. More particularly this invention relates to
the use of arabinonucleic acid or modified arabinonu-
cleic acid strands to hybridize to and induce cleavage
of (via RNaseH activation) the complementary RNA.
Other applications of this invention relates to the use
of antisense oligonucleotides based on arabinonucleo-
- tides or modified arabinonucleotides in combination
with RNaseH as laboratory reagents for the sequence
specific cleavage and mapping of RNA. This invention
also relates to the use of oligonucleotides based on
arabinonucleotides or modified arabinonucleotides, par-
ticularly those comprised of 2'F-arabinonucleic acid
strands to hybridize duplex DNA to form a triple heli-
cal complex and thereby block DNA transcription.
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 2 -
(b) Description of Prior Art
The Antisense Strategy
Antisense oligonucleotides (AON) are novel
therapeutic agents which can inhibit specific gene
expression in a sequence-specific manner. Many AON are
currently in clinical-trial for the treatment of cancer
and viral diseases (for reviews see: (i) Uhlmann, E.;
Peyman, A. Chem. Rev. 1990, 90, 543. (ii) Cook, P.D.
Anti-Cancer Drug Design 1991, 6, 585. (iii) Crooke,
S.T. Annu. Rev. Pharmacol. Toxicol. 1992, 32, 329.
(iv) Crooke, S.T.; Lebleu, B. Antisense Research and
Applications; 1993, pp.579, CRC Press, Boca Raton, FL.
(v) Agrawal; S.; Iyer, R.P. Cur. Op. Biotech. 1995, 6,
12.). (vi) DeMesmaeker, A.; Haner, R.; Martin, P.;
Moser, H. Acc. Chem. Res. 1995, 28, 366. (vii) Crooke,
S.T.; Bennett, C.F. Annu. Rev. Pharmacol. Toxicol.
1996, 36, 107). For potential clinical utility, AON
should exhibit stability against degradation by serum
and cellular nucleases, show low non-specific binding
to serum and cell proteins (this binding would diminish
the amount of antisense oligonucleotide available to
base-pair with the target RNA), exhibit enhanced recog-
nition of the target RNA sequence (in other words, pro-
vide increased stability of the antisense-target RNA
duplex at physiological temperature), and to some
extent, demonstrate cell-membrane permeability. The
formation of a duplex between the antisense oligomer
and its target RNA blocks the translation of that RNA,
by a mechanism termed "translation arrest". This mecha-
nism may however be a minor contributor to the overall
antisense effect. More important is the ability of the
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 3 -
antisense oligonucleotide to induce the activation of
ribonuclease H(RNaseH), an endogenous enzyme that spe-
cifically degrades RNA when duplexed with a complemen-
tary DNA oligonucleotide (or antisense oligonucleotide)
component (Walder, R.T.; Walder, J.A. Proc. Natl. Acad.
Sci. USA 1988, 85, 5011). For example, when an
antisense DNA oligonucleotide hybridizes to a mRNA
transcript, RNase H then cuts the mRNA at that site.
Antisense oligomers that modulate gene expression by
more than one mechanism of action are highly desirable
as this increases the potential efficacy of the
antisense compound in vivo.
Oligonucleotide Analogs
Oligonucleotides containing natural sugars (D-
ribose and D-2-deoxyribose) and phosphodiester (PO)
linkages are rapidly degraded by serum and intracellu-
lar nucleases, which limits their utility as effective
therapeutic agents. Chemical strategies to improve
nuclease stability include modification of the sugar
moiety, the base moiety, and/or modification or
replacement of the internucleotide phosphodiester link-
age. To date, the most widely studied analogues are
the phosphorothioate (PS) oligodeoxynucleotides, in
which one of the non-bridging oxygen atoms in the phos-
phodiester backbone is replaced with a sulfur
(Eckstein, F. Ann. Rev. Biochem. 1985, 54, 367).
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 4 -
Oligonucleotides that activate RNaseH
t Base ase t p/Base t O Base
H ~.--~~
0=P-O O4-O
S=~-O O~P-O
O
St ase A= ase IH3 Base O- Base
' li~~ X
H O\ O
O4-O g 4-O O_P-O 04-0
St AH3- I
0.
PS_DNA PS2-DNA Borano.DNA ANA X= OH, F
(presentinvention)
Several phosphorothioate oligonucleotide ana-
logues are undergoing clinical trial evaluation in the
treatment of cancer and viral diseases, and some are
moving rapidly towards New Drug Application (NDA) fil-
ings (Akhtar, S.; Agrawal, S. "In vivo studies with
antisense oligonucleotides" TiPS 1997, 18, 12). Phos-
phorothioates retain the ability to induce RNaseH deg-
radation of the target RNA and exhibit good stability
to degradation by nucleases. However, the PS oligode-
oxynucleotides form less stable duplexes with the tar-
get nucleic acid than do PO oligodeoxynucleotides, and
also exhibit significant nonspecific binding to cellu-
lar proteins, which can reduce the probability of find-
ing and interacting with the target nucleic acid; these
characteristics can limit the therapeutic utility of
PS-AON (for a review see: Brach, A.D.; "A good
antisense molecule is hard to find", TIBS, 1998, 23,
45). Furthermore, PS-AONs are less efficient at induc-
ing RNaseH degradation of the target RNA than are the
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 5 -
corresponding PO-AONs (Agrawal, S.; Mayrand, S.H.;
Zamenick, P.; Pederson, T. Proc. Natl. Acad. Sci. USA
1990, 87, 1401).
Specificity of action may be improved by devel-
oping novel oligonucleotide analogues. Current strate-
gies to generate novel oligonucleotides are to alter
the internucleotide phosphate backbone, the heterocy-
clic base, and the sugar ring, or a combination of
these. Alteration or complete replacement of the
internucleotide linkage has been the most popular
approach, with over 60 types of modified phosphate
backbones studied since 1994 (Sanghvi, Y. DNA in
"Altered Backbones in Antisense Applications", in Com-
prehensive Natural Product Chemistry, Barton, D.H.R.;
Nakanishi, K.; Meth-Coth, O. (eds), 1998, Elsevier Sci-
ence, Oxford, UK). Apart from the phosphorothioate
backbone, only two others have been reported to acti-
vate RNaseH activity, i.e., the phosphorodithioate (PSZ)
(Seeberger, P.H.; Yen, E.; Caruthers, M.H. J. Am. Chem.
Soc. 1995, 117, 1472) and the boranophosphonate back-
bones (Sergueev, D. et al., Poster 269, XIII Interna-
tional Round Table, Montpellier, France, Sept. 6-10,
1998; Higson, A.P. et al. Tetrahedron Letters 1998, 39,
3899). Because of the higher sulfur content of phos-
phorodithioate-linked (PS2) oligodeoxynucleotides, they
appear to bind proteins tighter than the phosphorothio-
ate (PS) oligomers, and to activate RNaseH mediated
cleavage with reduced efficiency compared to the PS
analogue. Boranophosphonate-linked oligodeoxynucleo-
tides activate RNaseH mediated cleavage of RNA targets,
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 6 -
but less well than PO- or PS-linked oligodeoxynucleo-
tides.
Among the reported sugar-modified oligonucleo-
tides most of them contain a five-membered ring,
closely resembling the sugar of DNA (D-2-deoxyribose)
and RNA (D-ribose). Example of these are a-oligodeoxy-
nucleotide analogs, wherein the configuration of the 1'
(or anomeric) carbon has been inverted as shown below
(Morvan, F.; Rayner, B.; Imbach, J.-L., Chang, D.K.;
Lown, J.W. Nucleic Acids Res. 1987, 15, 7027).
These analogues are nuclease resistant, form
stable duplexes with DNA and RNA sequences, and are
capable of inhibiting 0-globin mRNA translation via an
RNaseH-independent antisense mechanism (Boiziau, C;
Kurfurst, R.; Cazanave, C; Roig, V.; Thuong, N.T.
Nucleic Acids Res. 1991, 19, 1113) . Other examples
shown also below are xylo-DNA, 2'-O-Me RNA and 2'-F RNA
(reviewed in Sanghvi, Y.S.; Cook, P.D. in "Carbohydrate
Modifications in Antisense Research", Sanghvi, Y.S.;
Cook, P.D. (eds), ACS Symposium Series, vol. 580, pp.
1, American Chemical Society, Washington DC, 1994).
Sugar modified oligonucleotide analogs that do not activate RNaseH
0 1 1 t 0 Base Base ~ O Base
ase
O\P-O CH
OP-O O P-O O~-O
O1~ ase I
o O ase 6- 0 Base
Base 0-P-0
_ GH3 O
\
O-P-O O_\P-O paP-O
3# O- 0-
a.-DNA xylo-DNA 2'_ M RN 2'-F RNA
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 7 -
These analogues form stable duplexes with RNA
targets, however, these duplexes are not substrates for
RNaseH. To overcome this limitation, mixed-backbone
oligonucleotides ("MBO") composed of either
phosphodiester (PO) and phosphorothioate (PS)
oligodeoxynucleotide segments flanked on both sides by
sugar-modified oligonucleotide segments have been
synthesized (Zhao, G. et al., Biochem. Pharmacol. 1996,
51, 173; Crooke, S.T. et al. J. Pharmcol. Exp. Ther.
1996, 277, 923). Among the MBOs most studied to date
is the [2' -OMe RNA] -[PS DNA] -[2'OMe RNA] chimera. The
PS segment in the middle of the chain serves as the
RNaseH activation domain, whereas the flanking 2'-OMe
RNA regions increase affinity of the MBO strand for the
target RNA. MBOs have increased stability in vivo, and
appear to be more effective than phosphorothioate
analogues in their biological activity both in vitro
and in vivo. Examples of this approach incorporating
2'-OMe and other alkoxy substituents in the flanking
regions of an oligonucleotide have been demonstrated by
Monia et al. by enhanced antitumor activity in vivo
(Monia, P.B.; Johnston, J.F.; Geiger, T.; Muller, M.;
Fabbro, D. Nature Med. 1996, 2, 668). Several pre-
clinical trials with these analogues are ongoing
(Akhtar, S.; Agrawal, S. "In vivo studies with
antisense oligonucleotides" TiPS 1997, 18, 12).
The synthesis of oligonucleotides containing
hexopyranoses instead of pentofuranose sugars has also
been reported (Herdewijn, P. et al., in "Carbohydrate
Modifications in Antisense Research", Sanghvi, Y.S.;
Cook, P.D. (eds), ACS Symposium Series, vol. 580, pp.
80, American Chemical Society, Washington DC, 1994). A
--- - ----- ----
CA 02331333 2000-12-19
WO 99/67378 - PCT/CA99/00571
- 8 -
few of these analogues have increased enzymatic stabil-
ity but generally suffer from a reduced duplex forming
capability with the target sequence. A notable excep-
tion are 6'->4' linked oligomers constructed from 1,5-
anhydrohexitol units which, due to their highly pre-
organized sugar structure, form very stable complexes
with RNA (van Aeroschot, A.C. et al., Nucleosides &
Nucleotides 1997, 16, 973). However, none of these
hexopyranose oligonucleotide analogues have been shown
to elicit RNaseH activity. Recently, oligonucleotides
containing completely altered backbones have been syn-
thesized. Notable examples are the peptide nucleic
acids ("PNA") with an acyclic backbone (Nielsen, P.E.
in "Perspectives in Drug Discovery and Design", vol. 4,
pp. 76, Trainor, G.L. (ed.), ESCOM, Leiden, 1996).
These compounds have exceptional hybridization proper-
ties, and stability towards nucleases and proteases.
However, efforts to use PNA oligomers as antisense con-
structs have been hampered by poor water solubility,
self-aggregation properties, poor cellular uptake, and
inability to activate RNaseH. Very recently, PNA-[PS-
DNA]-PNA chimeras have been designed to maintain RNaseH
mediated cleavage via the PS-DNA portion of the chimera
(Bergman, F; Bannworth, W.; Tam, S. Tetrahedron Lett.
1995, 36, 6823; Van der Laan, A.C. et al. Trav. Chim
Pays-Bas 1995, 114, 295).
Arabinonucleosides and Arabinonucleic Acids (ANA)
Arabinonucleosides are stereoisomers of ribonu-
cleosides, differing only in the configuration at the
2'-position of the sugar ring. They have had a sub-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 9 -
stantial impact on chemotherapy and as such they have
been extensively used as antiviral and anticancer drugs
(for a review, see: Wright, G.E.; Brown, N.C. Pharma-
col. Ther. 1990, 47, 447) . P-D-Arabinofuranosylcyto-
sine (ara-C) is the most successful nucleoside antileu-
kemic agent and is widely used in combination therapy
or at high doses as a single agent to treat patients
with acute lymphoblastic and myeloblastic leukemias
(Kufe, D.W.; Spriggs, D.R. Semin. Oncol. 1985, 12, 34;
Lauer et al. Cancer 1987, 60, 2366).
Oligonucleotides constructed from arabinonu-
cleotides ("arabinonucleic acids" or ANA, have been
under investigation from various different aspects.
ANA oligomers have been synthesized as pro-drugs in an
attempt to improve the solubility of arabinonucleoside
therapeutics. Incorporation of ara-C into DNA strands
has also been the focus of research to understand the
mechanism of action of this anticancer drug (Mikita,
T.; Beardsley, G.P. Biochemistry 1988, 27, 4698;
Mikita, T.; Beardsley, G.P. Biochemistry 1994, 33,
9195).
DNA strands containing arabinonucleosides have
also been a subject of a number of structural studies.
In the crystal, DNA duplexes containing araC adopt a
normal B-type double helix with only small conforma-
tional perturbations at the araC-dG base pair (Chwang,
A.K.; Sundaralingam, M. Nature 1973, 243, 78; Teng, M.
et al. Biochemistry 1989, 28, 4923; Gao, Y.-G. et al.,
Biochemistry 1991, 30, 9922). Mikita and Beardsley
prepared DNA/DNA and DNA/RNA duplexes containing a sin-
gle araC-G base pair to investigate the structural dis-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 10 -
tortions caused by arabinonucleotides. They found that
both the DNA duplex and the DNA/RNA hybrid can accommo-
date araC-dG(rG) base pair with only a moderate and
equivalent loss of stability (Mikita, T.; Beardsley,
G.P. Biochemistry 1994, 33, 9195). Pfleiderer and co-
workers synthesized an all-arabinose oligonucleotide
mimicking a transfer RNA molecule (Resmini, M;
Pfleiderer, W. Helv. Chim. Acta 1993, 76, 158).
The association properties of uniformly modi-
fied oligoarabinonucleotides (ANA) were investigated by
Giannaris and Damha and independently by Watanabe and
co-workers (Giannaris, P.A.; Damha, M.J. Can. J. Chem.
1994, 72, 909; Kois, P.; Watanabe, K.A. Nucleic Acids
Symposium Series 1993, 29, 215; Kois, P. et al. Nucleo-
sides & Nucleotides 1993, 12, 1093) . Giannaris and
Damha showed that oligomers of either purine or pyrimi-
dine 0-arabinonucleosides generally associate with com-
plementary DNA and RNA with thermal stabilities compa-
rable with those of the corresponding DNA strands
(Giannaris, P.A.; Damha, M.J. Can. J. Chem. 1994, 72,
909). For example, they showed that (a) an octaarabino-
adenylate, ara-Ae associated with poly ribo-U and poly
deoxy-T; the melting temperature of the resulting com-
plex was slightly higher than the corresponding com-
plexes formed by the normal ribo-A8 and deoxy-A8
strands; (b) ara-C8 and ara(UCU UCC CUC UCC C) associ-
ated with their complementary RNA strand, albeit with
lower affinity relative to the corresponding unmodified
strands; (c) ara-Ua did not bind with poly rA under con-
ditions where ribo-U8 and deoxy-U8 formed a complex with
poly rA. Giannaris and Damha also reported that
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 11 -
replacement of the normal phosphodiester (PO) linkage
in ANA oligomers with phosphorothioate (PS) linkages
had a severe destabilizing effect; the destabilization
was greater than that observed when the PO linkages of
a normal DNA strand were replaced with PS internucleo-
tide linkages (Giannaris, P.A.; Damha, M.J. Can. J.
Chem. 1994, 72, 909). ANA oligomers displayed some
stability against cleavage by snake-venom phosphodi-
esterase; however, they were rapidly degraded by nucle-
ase Pl, ribonuclease S1 and spleen-phosphodiesterase
(Giannaris, P.A.; Damha, M.J. Can. J. Chem. 1994, 72,
909).
Watanabe and co-workers incorporated 2'-deoxy-
2'-fluoro-(3-D-arabinofuranosylpyrimidine nucleosides
(2'F-ara-N, where N=C, U and T) at multiple positions
within a normal DNA chain and evaluated the hybridiza-
tion properties of such (2'-F)ANA-DNA "chimeras"
towards complementary DNA (Kois, P. et al. Nucleosides
& Nucleotides 1993, 12, 1093). They found that substi-
tutions with 2'F-araU and 2'F-araC had a destabilizing
effect on duplex stability, whereas substitution with
2'F-araT was stabilizing compared to unmodified oligo-
deoxynucleotide strands. The authors also reported
that 2'F-araT11 and 2'F-araU11 oligomers were able to
bind to the complementary DNA with equal or slightly
better affinity compared to the control dTll (DNA) oli-
gomer. Marquez and co-workers recently evaluated the
self-association of a DNA strand in which two internal
thymidines were replaced by 2'F-araT's (Ikeda et al.
Nucleic Acids Res. 1998, 26, 2237). They confirmed the
findings of Watanabe and co-workers that internal 2'F-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 12 -
araT residues stabilize significantly the DNA double
helix. The association of these (2'-F)ANA-DNA "chime-
ras" with complementary RNA (the typical antisense tar-
get) was not reported.
Recently, Noronha and Damha tested oligonucleo-
tides based on (3-D-arabinose for their ability to rec-
ognize duplex DNA, duplex RNA and DNA/RNA hybrids
(Noronha, A.; Damha, M.J. Nucleic Acids Res. 1998, 26,
2665). A pyrimidine oligoarabinonucleotide was shown
to form triple-helical complexes with duplex DNA and
hybrid DNA(purine)/RNA(pyrimidine). However, this oli-
goarabinonucleotide was found to bind with an affinity
that was lower relative to the natural pyrimidine oli-
godeoxynucleotide or oligoribonucleotide controls.
Oligomers constructed from a-arabinofurano-
sylthymine (a-ara-T) exhibited a large decrease in
melting temperature towards complement DNA when com-
pared to the control DNA ((3-dT) strand (Adams, A.D.;
Petrie, C.R.; Meyer Jr., R.B. Nucleic Acids Res. 1991,
19, 3647). On the other hand, the duplexes formed
between either a-ara-T15 or dTls and complementary RNA
(poly-rA) were of similar strength. More recently,
Wengel and co-workers reported the synthesis and asso-
ciation properties of DNA oligomers containing one and
two (3-2'-OMe-araT inserts (Gotfredsen, C.H.; Spielmann,
P.; Wengel, J.; Jacobsen, J.P. Bioconjugate Chem. 1996,
7, 680). These oligomers showed moderately lowered
thermal stabilities towards both single stranded DNA
and RNA, compared to unmodified DNA controls. The same
authors reported that oligomers constructed from a-2'-
OMe-araT units exhibited increased affinity towards the
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 13 -
riboadenylate (RNA) target compared to normal DNA con-
trols; however, a-2'-OMe araT strands did not display
any advantage relative to the known a-dT oligomers. The
susceptibility of the above duplexes to RNase H-medi-
ated cleavage was not investigated.
Activation of RNase H by Antisense Oligonucleotides
As described above, an important mechanism of
action of antisense oligonucleotides is the induction
of cellular enzymes such as RNaseH to degrade the tar-
get RNA (Walder, R.T.; Walder, J.A. Proc. Natl. Acad.
Sci. USA 1988, 85, 5o11; Chiang et al. J. Biol Chem.
1991, 266, 18162; Monia et al. J. Biol. Chem. 1993,
268, 14514; Giles, R.V.; Spiller, D.G.; Tidd, D.M.
Antisense Res. & Devel. 1994, 5, 23; Giles et al.
Nucleic Acids Res. 1995, 23, 954). RNase H selectively
hydrolyzes the RNA strand of a DNA/RNA heteroduplex
(Hausen, P.; Stein, H. Eur. J. Biochem. 1970, 14, 279).
RNase Hl from the bacterium Escherichia coli is the
most readily available and the best characterized
enzyme. Studies with eukaryotic cell extracts contain-
ing RNase H suggest that both prokaryotic and eukary-
otic enzymes exhibit similar cleavage properties (Monia
et al. J. Biol. Chem. 1993, 268, 14514; Crooke et al.
Biochem J. 1995, 312, 599; Lima, W.F.; Crooke, S.T.
Biochemistry 1997, 36, 390). Escherichia coli RNase H
is thought to bind in the minor groove of the DNA/RNA
double helix and to cleave the RNA by both endonuclease
and processive 3'-to-5' exonuclease activities
(Nakamura, H. et al. Proc. Natl. Acad. Sci. USA 1991,
88, 11535; Federoff, O.Y.; Salazar, M.; Reid, B.R. J.
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 14 -
Mol. Biol. 1993, 233, 509; Daniher, A.T. et al. Bioorg.
& Med. Chem. 1997; 5, 1037). The efficiency of RNase H
degradation displays minimal sequence dependence and is
quite sensitive to chemical changes in the antisense
oligonucleotide. For example, RNaseH degrades RNA in
PS-DNA/RNA hybrids (Gao et al. Mol. Pharmacol. 1991,
41, 223), but not in hybrids containing methylphospho-
nate-DNA, a-DNA, or 2'-OMe RNA antisense strands (For a
review, see: Sanghvi, Y.S.; Cook, P.D. (eds), ACS Sym-
posium Series, vol. 580, pp. 1, American Chemical Soci-
ety, Washington DC, 1994). Furthermore, while E. coli
RNaseH binds to RNA/RNA duplexes, it cannot cleave
them, despite the fact that the global helical confor-
mation of RNA/RNA duplexes is similar to that of
DNA/RNA substrate duplexes ("A"-form helices)(Oda et
al. Nucleic Acids Res. 1993, 21, 4690). These results
suggest that local structural differences between
DNA/RNA (substrate) and RNA/RNA duplexes is responsi-
ble, at least in part, for substrate discrimination
(Oda et al. Nucleic Acids Res. 1993, 21, 4690; Lima,
W.F.; Crooke, S.T. Biochemistry 1997, 36, 390). In
this regard it is interesting to note that HIV-1
reverse transcriptase (RT)-associated RNaseH cleaves
both DNA/RNA and RNA/RNA duplexes; however, cleavage of
the latter is at least 30-fold slower and occurs only
when RT is artificially arrested (Gotte et al., EMBO J.
1995, 14, 838).
Arabinonucleic Acids as Activators of RNaseH Activity
An essential requirement in the antisense
approach is that an oligonucleotide or its analogue
recognize and bind tightly to its complementary target
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 15 -
RNA. The ability of the resulting antisense oli-
gomer/RNA hybrid to serve as a substrate of RNaseH is
likely to have therapeutic value by enhancing the
antisense effect relative to oligomers that are unable
to activate this enzyme. Apart from PS-DNA (phosphoro-
thioates), PS2-DNA (phosphorodithioates), boranophospho-
nate-linked DNA, and MBO oligos containing an internal
PS-DNA segment, there are no other examples of fully
modified oligonucleotides that elicit RNaseH activity.
For this reason, and because of the problems encoun-
tered with PS-oligonucleotides (e.g., non-antisense
effects and potential risk of toxicity), we have
designed alternative oligonucleotide analogues that
selectively block gene expression through the activa-
tion of RNaseH activity. As a starting point, we felt
that such analogues should (a) retain the natural (3-D-
furanose configuration, (b) possess the unmodified
phosphate groups for solubility purposes, and (c) be
able to mimic the conformation of DNA strands (e.g.,
with sugars puckered in the C2' -endo conformation).
The latter requirement stems from the fact that the
antisense strand of natural substrates is DNA, and as
indicated above, its primary structure (and/or confor-
mation) appears to be essential for RNaseH/substrate
cleavage. Since the DNA sugars of DNA/RNA hybrids
adopt primarily the C2'-endo conformation (Salazar, M.;
Champoux, J.J.; Reid, B.R. Biochemistry 1993, 32, 739;
Salazar, M.; Federoff, O.Y.; Reid, B.R. Biochemistry
1996, 35, 8126), we were interested in an oligonucleo-
tide analog that favored this conformation. Analogues
mimicking the RNA structure (i.e., those that adopt the
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 16 -
C3'-endo rather than the C2'-endo conformation) would
not be suitable for evoking RNase H activity since it
is known that RNA/RNA duplexes are generally not sub-
strates of RNaseH. This prompted us to consider oli-
gomers constructed from arabinonucleotides (i.e., the
arabinonucleic acids or ANA). ANA is an stereoisomer
of RNA differing only in the stereochemistry at the 2'-
position of the sugar ring. ANA/RNA duplexes adopt a
helical structure that is very similar to that of
DNA/RNA substrates ("A"-form), as shown by similar cir-
cular dichroism spectra of these complexes. In addi-
tion, X-ray crystallographic studies on ara-C nucleo-
sides and on DNA duplexes containing ara-C indicated
that the arabinose sugar adopts the C1'-exo or the C2'-
endo conformation; the latter conformation is found in
the DNA sugars of DNA/RNA substrates. Furthermore,
examination of molecular models of an A-type ANA/RNA
duplex suggested that the P-2'-OH group of the arabi-
nose strand is positioned within the major groove of
the hybrid and thus should not interfere with RNase H's
binding and catalytic processes. We also considered
replacing the (3-2'-OH by other electronegative sub-
stituents, e.g., 0-2'-fluorine, since strong stereoe-
lectronic effects are expected to stabilize the C2'-
endo form (Saenger, W. Principles of Nucleic Acids
Structure, Cantor, C.R. (ed.), Springer-Verlag, N.Y.,
1984; Marquez, V.E.; Lim, B.B.; Barchi, J.J., Jr.;
Nicklaus, M.C., "Conformational studies of anti-HIV
activity of mono- and difluorodideoxynucleosides", in
Nucleosides and Nucleotides as Antitumor and Antiviral
Agents, Chu, C.K.; Baker, D.C. (eds.), pp. 265-284,
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 17 -
Plenum Press, N.Y., 1993). The possibility of an ANA
oligomer activating RNaseH has not been reported.
It would be highly desirable to be provided
with ANA oligomers and their analogues for sequence
specific inhibition of gene expression via association
to (and RNaseH mediated cleavage of) complementary mes-
senger RNA.
It would be highly desirable to be provided
with ANA oligomers and their analogues that modulate
gene expression by binding directly to gene sequences
(duplex DNA).
SUbIIKARY OF THE INVENTION
It is the purpose of this invention to provide
ANA oligomers and their analogues for sequence specific
inhibition of gene expression via association to (and
RNaseH mediated cleavage of) complementary messenger
RNA. It is also the purpose of this invention to pro-
vide ANA oligomers and their analogues that modulate
gene expression by binding directly to gene sequences
(duplex DNA).
In one aspect, the present invention provides
sugar-modified oligonucleotides that form a duplex with
its target RNA sequence. The resulting duplex is a
substrate for RNaseH, an enzyme that recognizes this
duplex and degrades the RNA target portion. RNaseH
mediated cleavage of RNA targets is considered to be a
major mechanism of action of antisense oligonucleo-
tides. The sugar-modified oligomers are composed of P-
D-arabinonucleotides (i.e., ANA oligomers) and 2'-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 18 -
deoxy-2'-fluoro-p-D-arabinonucleosides (i.e., 2'F-ANA
oligomers).
B B
O O
OH F
O H O H
B I B
O~-O O O F5-O O
O- OH O_ F
O H
O-d-O_ 0-d-O-
I I
(ANA) O O_ (2'F-ANA)
Prior to this invention, only the natural DNA
(deoxyribonucleic acid phosphodiester) or deoxyribonu-
cleic acid oligonucleotides based on phosphorothioate
(PS-DNA), dithioate, and boranophosphonate backbones,
had been reported to elicit RNaseH degradation of the
target RNA. The present invention relates to the dis-
covery that certain uniformly sugar-modified oligonu-
cleotides, namely those based on P -D-arabinonucleotides
(i.e., ANA oligomers) and 2'-deoxy-2'-fluoro-(3-D-arabi-
nonucleotides (i.e., 2'F-ANA oligomers), can activate
RNaseH activity when duplexed to the target RNA
sequences.
Also provided are oligonucleotides based on 2'-
deoxy-2'-fluoro-(3-D-arabinonucleosides (i.e., 2'F-ANA
oligomers) that bind to duplex DNA with higher affinity
relative to unmodified oligodeoxynucleotides.
CA 02331333 2008-01-03
- 19 -
In another aspect of the invention, defined sequence
oligoarabinonucleotides (ANA and 2'F-ANA) were prepared and
found to inhibit the expression of a specific targetmRNA
that codes for the expression of a specific protein
(luciferase). This inhibition was noted both in experiments
that assessed inhibition of target protein expression in in
vitro transcription/translation of the target (in the
presence of a large excess of non-specific exogenous
protein contributed by the in vitro
transcription/translation system) and in experiments
assessing target protein expression in intact cells.
In summary, our experiments establish that ANA
oligomers serve as excellent models of antisense agents
that have enhanced resistance to the action of degradative
nucleases, bind to RNA through duplex formation, elicit
RNase H activity, and inhibit in vitro and intracellular
specific gene expression. Accordingly, ANA and its
analogues have potential utility as therapeutics agents
and/or tools for the study and control of specific gene
expression in cells and organisms.
In a further aspect, there is provided an
oligonucleotide for selectively preventing or modulating
gene expression in a sequence-specific manner in a host;
wherein said oligonucleotide is a uniformly sugar-modified
oligonucleotide consisting of 2'-deoxy-2'-fluoro-p-D-
arabinonucleotides.
In a further aspect, there is provided a therapeutic
composition for selectively preventing or modulating gene
expression in a sequence-specific manner in a host, wherein
said composition comprises an effective amount of at least
one oligonucleotide described herein and a pharmaceutically
acceptable carrier.
CA 02331333 2008-01-03
- 19a -
In a further aspect, there is provided use of the
oligonucleotide described herein for selectively preventing
or modulating gene expression in a sequence-specific manner
in a host.
BRIEF DESCRIPTION OF THE DRAWINGS
Figs. lA-1B illustrate thermal melting curves of 18-
bp heteroduplexes;
Figs. 2A-2C illustrate circular dichroic (CD)
spectra of duplexes;
Fig. 3 illustrates thermal melting curves of triple
helical complexes formed by the association of
oligoarabinonucleotide SEQ ID NO:13 with DNA/DNA ( DD") and
DNA/RNA ("DR") hairpin duplexes;
CA 02331333 2006-02-27
- 20 -
Fig. 4 illustrates gel mobility shift triplex
assay under non-denaturing conditions;
Fig. 5 illustrates oligonucleotides with (3-D-
arabinose as sugar component elicit RNaseH degradation
of complementary target RNA;
Fig. 6 illustrates homopolymeric oligonucleo-
tides with 2'-F-(3-D-arabinose as sugar component elicit
RNaseH degradation of complementary target RNA;
Fig. 7 illustrates heteropolymeric oligonucleo-
tides with 2'-F-(3-D-arabinose as sugar component elicit
RNaseH degradation of complementary target RNA;
Fig. 8 illustrates stability of oligonucleo-
tides with 2'-F-(3-D-arabinose as sugar component to
degradation by serum nucleases;
Fig. 9 illustrates stability of oligonucleo-
tides with 2'-F-(3-D-arabinose as sugar component to
degradation by snake venom phosphodiesterase I;
Fig. 10 illustrates oligonucleotides based on
(3-D-arabinose and 2'-deoxy-2'-fluoro-p-D-arabinose show
low nonspecific binding to cellular proteins;
Figs. 11A-11D illustrate oligonucleotide
inhibition of specific gene expression in an in vitro
protein translation system; and
Figs. 12A and 12B illustrate oligonucleotide
inhibition of luciferase gene expression in Hela X1/5
cells.
DETAILED DESCRIPTION OF THE INVENTION
The present invention relates to oligonucleo-
tides based on P-D-arabinose and its derivatives and
the therapeutic use of such compounds. It is the
object of the present invention to provide a new oligo-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 21 -
nucleotide analogue that hybridizes to complementary
nucleic acids which may be mRNA, viral RNA (including
retroviral RNA), or duplex DNA for the purpose of
inhibiting gene transcription and expression. More
particularly this invention relates to the use of ara-
binonucleic acid strands and their analogues to cleave
complementary RNA via RNaseH activation. Other appli-
cations of this invention relates to the use of
antisense oligonucleotides based on arabinonucleotides
in combination with RNaseH as laboratory reagents for
the sequence specific cleavage and mapping of RNA.
The oligonucleotides of this invention may be
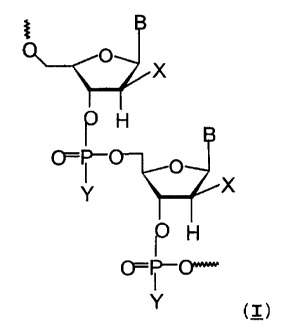
represented by the following Formula (I):
X
~;O
r H
O
CF=~ '~~
Y X
p H
0=~0~'
Y (I)
where B includes but it is not necessarily limited to a
common purine or pyrimidine base such as adenine, gua-
nine, cytosine, thymine, and uracil. The sugar is P-D-
arabinofuranose, its mirror image enantiomer P-L-arabi-
nofuranose, and the corresponding carbocyclic sugars
(i.e., in which the ring oxygen at position 4' is
replaced by a methylene or CHZ group). The substituent
at the 2' position of the sugar ring includes but it is
not necessarily limited to a halogen (fluorine, chlo-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 22 -
rine, bromine, iodine), hydroxy, alkyl, alkylhalide
(e.g. CHzF) , alkylsulfhydryl (e.g. -SCH3) , allyl, amino,
aryl, alkoxy, and azido. Y at the internucleotide
phosphate linkage includes but it is not necessarily
limited to oxygen, sulfur, methyl, amino, alkylamino,
dialkylamino, methoxy, and ethoxy. The ANA oligomers
may also include modified sugars in part of the oli-
gomer. The oligonucleotides may also include the 2'-
deoxy-2',2"-difluoro-o-ribofuranose sugar (D or L con-
figuration) in part or all of the oligomer (this struc-
ture is obtained by replacing the 2'-H atom in formula
I with a fluorine atom, thus providing an oligonucleo-
tide containing two fluorine atoms at carbon 2'). The
oligonucleotides may also include stretches of ssDNA
flanked by ANA segments (e.g., ANA-DNA-ANA, 2'F-ANA-
DNA-2'F-ANA chimeras), or combination of ANA and 2'F-
ANA segments (e.g., ANA-2'F-ANA chimeras). The ANA
oligomers of this invention contains a sequence that is
complementary to a specific sequence of a mRNA, or
genomic viral RNA, such that the oligonucleotide can
specifically inhibit protein biosynthesis, or virus
replication (reverse transcription), respectively. A
complementary target may also be duplex or single
stranded DNA, such that the arabinonucleotide strand
can specifically inhibit DNA replication and/or tran-
scription. Partial modifications to the oligonucleo-
tide directed to the 5' and/or 3'-terminus, or the
phosphate backbone or sugar residues to enhance their
antisense properties (e.g. nuclease resistance) are
within the scope of the invention.
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 23 -
A preferred group of oligonucleotides useful in
this invention, are those wherein B is a natural base
(adenine, guanine, cytosine, thymine, uracil); the
sugar moiety is (3-D-arabinofuranose; X is fluorine; Y
is oxygen since these modifications give rise to oli-
gomers that exhibit high affinity for single stranded
RNA, single stranded DNA, and duplex DNA. In addition,
these oligomers have been shown to meet the require-
ments necessary for antisense therapeutics. For exam-
ple, they activate RNaseH activity, show resistance to
cellular and serum nucleases, inhibit the expression of
a specific target mRNA that codes for the expression of
a specific protein in intact cells; and exhibit little
(if any) nonspecific binding to cellular proteins; as
such they may be potentially more effective in vivo.
The free P-D-arabinose pyrimidine (araU, araC)
nucleoside monomers may be prepared from the corre-
sponding ribonucleosides in good yields, and can be
further elaborated to the corresponding 5'-O-mono-
methoxytrityl-2'-O-acetyl-3'-0-(P-cyanoethylphospho-
ramidite) derivatives suitable for solid-phase oligonu-
cleotide synthesis (Giannaris, P.A.; Damha, M.J. Can.
J. Chem. 1994, 72, 909). The corresponding araA nucleo-
side is commercially available, and can be prepared
readily from riboadenosine (via oxidation of the 2'-OH
group and reduction of the 2'-keto group with a hydride
source, e.g., Robins, M.J. et al. in "Nucleosides,
Nucleotides and their Biological Applications",
Rideaut, J.L.; Henry, D.W.; Beacham III, L.M. (eds.),
pp. 279, Academic Press, Inc., 1993). The correspond-
ing ara-G monomer can be prepared by the method of
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 24 -
Pfleiderer and co-workers (Resmini, M.; Pfleiderer, W.
Helv. Chim. Acta 1994, 77, 429) . The 3' -O- (P-cyano-
ethyl-N,N-diisopropylphosphoramidite) derivatives of
5'-MMT-2'-deoxy-2'-fluoro-(3-D-arabinonucleosides (2'F-
ara-C, 2'F-ara-A, 2'F-ara-G and 2'F-ara-T) may be syn-
thesized following published procedures (Tann, C.H.;
Brodfuehrer, P.R.; Brundidge, S.P.; Sapino, C. Jr.;
Howell, H.G. J. Org. Chem. 1985, 50, 3644; Howell;
H.G.; Brodfuehrer, P.R.; Brundidge, S.P.; Benigni,
D.A.; Sapino, C., Jr. J. Org. Chem. 1988, 53, 85; Kois,
P.; Tocik, Z.; Spassova, M.; Ren, W.-Y.; Rosenberg, I.;
Farras Soler, J.; Watanabe, K.A. Nucleosides & Nucleo-
tides 1993, 12, 1093; Chou, T.-C.; Burchenal, J.H.;
Fox, J.J.; Watanabe, K.A. Chem. Pharm. Bull. 1989, 37,
336).
The protected arabinonucleoside monomers can be
attached to the solid support by known methods. In a
preferred embodiment, the solid support is long-chain
alkylamine controlled pore glass, and the procedure of
Damha et al. is used for its derivatization (Damha et
al. Nucleic Acids Res. 1990, 18, 3813).
The oligomers of this invention (constructed
from either P-D-arabinose or 2-deoxy-2-fluoro-(3-D-ara-
binose) exhibit a number of desirable properties:
(1) They were found to bind to and cleave sin-
gle stranded RNA by activating RNaseH. Circular
dichroism studies in solution showed that DNA/RNA
hybrids (the natural substrate of RNase H) and ANA/RNA
duplexes adopt a very similar helical structure that
falls within the "A" -conformational family. The abil-
ity of RNaseH to degrade RNA in the ANA:RNA duplexes
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 25 -
may be due, at least in part, to (a) the similarity of
the structure of ANA/RNA to that of DNA/RNA duplexes,
and (b) the fact that the 2'-substituent of the sugar
ring is located in the major groove, where it does not
interfere in RNase H's binding and catalytic processes.
The 2'-fluorinated ANA derivatives in particular were
found to have excellent affinity towards RNA targets,
compared to normal DNA and phosphorothioate oligodeoxy-
nucleotide strands.
(2) An oligonucleotides based on P-D-arabinose
and containing four nucleobases (U, C, A and G) was
found to hybridize to complementary RNA but not comple-
mentary single stranded DNA. This property suggests
that these oligomers may be useful for targeting retro-
viral genomic RNA to inhibit early stages of virus rep-
lication, including reverse transcription. This high
level of RNA specificity has previously been reported
for other types of oligonucleotide analogs (e.g.,
2',5'-linked RNA and 2',5'-linked DNA; Giannaris, P.A.;
Damha, M.J., Nucleic Acids Res. 1993, 20, 4742; Alul,
R.; Hoke, G.D. Antisense Res. Dev. 1995, 5, 3), how-
ever, none of these oligonucleotides elicit RNaseH
activity.
(3) Pyrimidine oligonucleotides constructed
from 2'-deoxy-2'-fluoro-(3-D-arabinonucleoside units
were also found to hybridize to duplex DNA and DNA/RNA
hybrids via triplex helix formation. The thermal sta-
bility of these triplexes are significantly higher than
those formed by normal oligodeoxynucleotides. These
results were unexpected given that the (3-D-arabinose
series produces triplexes with only modest stability
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 26 -
(Noronha, A.; Damha, M.J. Nucleic Acids Res. 1998, 26,
2665) .
(4) Results from metabolic stability studies
indicate that the arabinose modification, particularly
the (3-D-arabinose (2'-OH) derivatives, confers greater
resistance to degradation by both serum and cellular
nucleases compared with natural strands (PO-DNA),
although less than to phosphorothioate (PS-DNA) deriva-
tives. Partial modifications to the ANA or 2'F-ANA
oligonucleotide directed to the 5' and/or 3'-terminus,
or the phosphate backbone or sugar residues to further
enhance nuclease resistance are within the scope of the
invention.
(5) ANA and 2'F-ANA show little (if any) non-
specific binding to cellular proteins and serum pro-
teins. This property results in a significantly
improved interaction of arabinooligonucleotides with
target RNA in the presence of cell proteins compared to
the phosphorothioate analogs.
These properties combined establish that ANA
and 2'F-ANA oligomers serve as excellent models of
antisense agents that have resistance to the action of
degradative nucleases, bind to RNA and single stranded
DNA through duplex formation, bind to duplex DNA
through triplex formation, and elicit RNase H activity.
Consequently, antisense oligonucleotide constructs con-
taining arabinose and their analogues should serve as
therapeutics and/or valuable tools for studying and
controlling gene expression in cells and organisms.
The following examples are given by way of
illustration of the present invention. The examples
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 27 -
are not intended in any way to limit the scope of the
invention.
EXAMPLE 1
Preparation of Oligonucleotides containing P-D-
Arabinofuranoses
Oligoarabinonucleotides (Formula I; X= OH, Y=
O') were synthesized using standard phosphoramidite
chemistry and 3'-ara-C(Bz)-long-chain alkylamine con-
trolled pore glass solid support (lcaa-CPG; 500 A; 1
mol scale). The required monomers, namely 5'-MMT-2'-
OAc-3'-O-((3-cyanoethyl-N,N-diisopropylphosphoramidite)
derivatives of ara-A(Bz), ara-C(Bz) and ara-U were syn-
thesized by the method of Damha et al. (Damha, M.J.;
Usman, N.; Ogilvie, K.K., Can. J. Chem. 1988, 67, 831;
Giannaris, P.A.; Damha, M.J.; Can. J. Chem. 1994, 72,
909). The corresponding ara-G (N2-i-Bu, 06-NPE) mono-
mer was prepared by a modification of the procedure of
Resmini et al. (Resmini, M.; Pfleiderer, W. Helv. Chim.
Acta 1994, 77, 429). Thus, the monomers were dissolved
to 0.12 M in anhydrous acetonitrile. Prior to chain
assembly, the support (1 mol) was treated with the
capping reagents, acetic anhydride/N-methylimidazole/4-
dimethylaminopyridine (Damha, M.J.; Ogilvie, K.K. in
Methods in Molecular Biology, 20, Protocols for Oligo-
nucleotides and Analogs: Synthesis and Properties,
Agrawal, S. (ed.), pp. 81, The Humana Press, Inc.
Totawa, N.J., 1993). Chain assembly of sequences was
carried out using an Applied Biosystem DNA synthesizer
(Model 381A) as follows: (i) detritylation: 3% trichlo-
roacetic acid in dichloroethane delivered in 100 s (+
s'burst') steps. The eluate from this step was col-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 28 -
lected and the absorbance at 478 nm (MMT+, arabino
sequences) measured to determine the average coupling
reaction yield (ca. 60-90%) ; (b) nucleoside phospho-
ramidite coupling time of 7.5 min; (c) capping: 1:1
(v/v) of acetic anhydride/collidine/THF 1:1:8 (solution
A) and 1-methyl-lH-imidazole/THF 16:84 (solution B)
delivered in 15 s + 35 s"wait" steps; (d) oxidation:
0.1M iodine in THF/water/pyridine 7:2:1, delivered in
20 s + 35 s "wait" step. The 5'-terminal trityl group
was removed by the synthesizer and the oligomers were
then removed from the support and deprotected by treat-
ment of the CPG with a solution containing concentrated
ammonium hydroxide/ethanol (3:1 v/v, 1 mL) for two days
at room temperature. The ammonium hydroxide/ethanol
solution was evaporated and the crude product purified
by preparative polyacrylamide gel electrophoresis
(PAGE) followed by gel filtration (desalting) on a
Sephadex G-25 column. For sequences containing ara-G
units, it was necessary to subject the partially pro-
tected oligomer to an additional step; that is, follow-
ing the ammonia treatment and evaporation step, the
oligomer was treated with a solution of 1M tetra-n-
butylammonium fluoride (50 L, r.t., 16 h) in THF.
This step cleaves the p-nitrophenylethyl protecting
group at the 06-position of guanine residues. This
solution is then quenched with water (1 mL) and
desalted via size exclusion chromatography (Sephadex G-
25 column). Purification is then carried out by gel
electrophoresis as described above, and its molecular
weight confirmed by MALDI-TOF mass spectrometry. The
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 29 -
yield, base sequence, hybridization properties of the
oligomers synthesized are given in Table 1.
TABLE 1
Base composition, yield and properties of
oligoarabinonucleotides (ANA)
Melting Temperature
oC Y
Base sequence of Oligonucieotide' SEOID Yieldb RNA target DNA target
NO:
ara(AGC UCC CAG GCU CAG AUC) 1 5 44 26 d
ara(AAA AAA AAA AAA AAA AAA) 2 10 26 45
ara(UUU UUU UUU UUU UUU UUU) 3 9 n.o.e n.o.e
ara(UUA UAU UUU UUC UUU CCC) 4 10 32 <15d
ara(AUA UCC UUG UCG UAU CCC) 5 8 47 n.m.f
a Sequence is written in the 5' -* 3' direction;
b Optical density units (AZ60 nm);
Buffer containing 140 mM KCI, 1 mM MgC12, 5 mM Na2HPO4 (pH 7.2);
d weak and broad transition
e no melt curve or sharp transition observed;
f not measured.
Presently only the 5'-DMT, 2'-OAc, ara-C (Bz)
3'-phosphoramidite derivative and 3'-ara-C (Bz) long-
chain alkylamino controlled pore glass (lcaa-CPG) are
commercially available. Of the free (unprotected)
nucleosides, only ara-C, ara-U and ara-A are commer-
cially available.
EXAMPLE 2
Preparation of Oligonucleotides containing 2-Deoxy-2-
Fluoro-p-D-Arabinose sugars
Oligoarabinonucleotide synthesis (Formula I; X=
F, Y = O-) was performed on an Applied Biosystem DNA
synthesizer (model 381A) using the phosphoramidite
approach. Oligomers were prepared on a 1.0 mol scale
using lcaa-CPG solid support bearing 3'-terminal 2'-
deoxy-2'-fluoro-(3-D-arabinonucleosides. Coupling
yields ranged from 60 to 100% (average ca. 80%) as
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 30 -
monitored by the release of the MMT cation. The
required 3'-O-((3-cyanoethyl-N,N-diisopropylphospho-
ramidite) derivatives of 5'-MMT-2'-deoxy-2'-fluoro-(3-D-
arabinonucleosides (2'F-ara-C, 2'F-ara-A, 2'F-ara-G and
2'F-ara-T) were synthesized by published procedures
(Tann, C.H.; Brodfuehrer, P.R.; Brundidge, S.P.;
Sapino, C. Jr.; Howell, H.G. J. Org. Chem. 1985, 50,
3644; Howell; H.G.; Brodfuehrer, P.R.; Brundidge, S.P.;
Benigni, D.A.; Sapino, C., Jr. J. Org. Chem. 1988, 53,
85; Kois, P.; Tocik, Z.; Spassova, M.; Ren, W.-Y.;
Rosenberg, I.; Farras Soler, J.; Watanabe, K.A. Nucleo-
sides & Nucleotides 1993, 12, 1093; Chu, C.K.; Matulic-
Adamic, J.; Huang, J.-T.; Chou, T.-C.; Burchenal, J.H.;
Fox, J.J.; Watanable, K.A. Chem. Pharm. Bull. 1989, 37,
336). Thus, the monomers were dissolved to 0.10 M in
anhydrous acetonitrile. Prior to chain assembly, the
support (1 mol) was treated with the capping reagents,
acetic anhydride/N-methylimidazole/4-dimethylamino
pyridine (Damha, M.J.; Ogilvie, K.K. in Methods in
Molecular Biology, 20, Protocols for Oligonucleotides
and Analogs: Synthesis and Properties, Agrawal, S.
(ed.), pp. 81, The Humana Press, Inc. Totawa, N.J.,
1993). Chain assembly of sequences was carried out as
follows: (i) detritylation: 3% trichloroacetic acid in
dichloroethane delivered in 140 s (+ 80 s 'burst')
steps. (b) nucleoside phosphoramidite coupling time of
10 min; (c) capping of 5'-hydroxyl groups: 1:1 (v/v) of
acetic anhydride/collidine/THF 1:1:8 (solution A) and
1-methyl-lH-imidazole/THF 16:84 (solution B) delivered
in 15 s + 35 s"wait" steps; (d) oxidation of phosphite
triester linkage: 0.1M iodine in THF/water/pyridine
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571 -
- 31 -
7:2:1, delivered in 20 s + 35 s "wait" step. The 5'-
terminal trityl group was removed by the synthesizer
and the oligomers were then removed from the support
and deprotected by treatment of the CPG with a solution
containing concentrated ammonium hydroxide/ethanol (3:1
v/v, 1 mL) for two days at room temperature. The ammo-
nium hydroxide/ethanol solution was evaporated and the
crude product purified by preparative polyacrylamide
gel electrophoresis (PAGE) followed by gel filtration
(desalting) on a Sephadex G-25 column. Molecular weight
of oligomers were confirmed by MALDI-TOF mass spec-
trometry. The yield, base sequence, and hybridization
of the oligomers synthesized are given in TABLE 2.
TABLE 2
Base composition, yield and properties of 2'-F-
oligoarabinonucleotides (2'F-ANA)
Melting Temperature
( C)'
Base sequence of Oiigonucieotidee SEGl ID Yieidb RNA target DNA target
NO:
2'F-ara(AGC TCC CAG GCT CAG 6 11 86 68
ATC)
2'F-ara(TTT TTT TTT TTT TTT TTT) 7 15 52 55
2'F-ara(AAA AAA AAA AAA AAA AAA) 8 27 30 63
2'F-ara(TTA TAT TTT TTC TTT CCC) 9 15 64 54
2'F-ara(ATA TCC TTG TCG TAT CCC) 10 21 76 n.m.d
a Sequence is written in the 5' -a 3' direction;
b Optical density units (A260 nm);
c Buffer containing 140 mM KCI, 1 mM MgCi2, 5 mM Na2HPO4 (pH 7.2);
Not measured.
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 32 -
EXAMPLE 3
Association Properties of Uniformly Modified
Oligonucleotides possessing j3-D-Arabinose and P-D-2-
Fluoro-2-Deoxyarabinose Sugars
Binding to single stranded DNA and RNA Targets
The ability of oligonucleotides to hybridize to
single-stranded nucleic acids to give a double- helical
complex is crucial for their use as antisense therapeu-
tic agents. The formation of such a complex involves
stacking and hydrogen bonding interactions between the
base chromophores, a process which is accompanied by a
reduction in UV absorption ("hypochromicity"),. When
the temperature of the solution containing the double-
helical complex is gradually raised, the hydrogen bonds
break and the duplex dissociates into single strands.
This reduces the amount of base-base interactions and
hence leads to a sudden increase of the UV absorbance.
The temperature at which the double-helical complex
dissociates, or more precisely, the point at which half
the population exists as complex and the remaining half
as single strands, is termed the "melting temperature"
(Tm). Thus a common technique used in nucleic acid
chemistry to investigate duplex formation (and its
strength) involves mixing equimolar amounts of the
strand of interest together, incubating at low tempera-
ture to allow strands to anneal, and then observing the
UV-absorption at 260 nm (absorption maxima) as a func-
tion of temperature. The result is an absorbance ver-
sus temperature plot, or sigmoidal "melt profile" or
"melting curve" from which the T. (the midpoint of the
raise of the melt curve) is calculated (Wickstrom, E.;
Tinoco, I. Jr. Biopolymers 1974, 13, 2367). Circular
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 33 -
dichroism (CD) is another powerful optical technique
for the study of nucleic acid structure and conforma-
tion. The CD spectrum usually includes a region of
rapid change (Cotton effects) with respect to wave-
length (200-350 nm region). The signs, absolute inten-
sity and position of the Cotton effects are particu-
larly sensitive to chemical composition and three-
dimensional structure of the nucleic acid complexes.
CD measurements can therefore be applied to determine
global helical conformation (or helix type) as well as
to investigate structural changes (e.g., helix-to-coil
transitions) as a function of temperature (Bloomfield,
V.A.; Crothers, D.; Tinoco, Jr., I. "Physical Chemistry
of Nucleic Acids", Harper and Row, N.Y., 1974; Ts'o,
P.O.P. (ed.), "Basic Principles in Nucleic Acid Chemis-
try", vol. 1 and 2, Academic Press, N.Y., 1974).
The binding properties of oligonucleotides (SEQ
ID NO:1, and SEQ ID NO:6) (for base sequences see
Tables 1 and 2) with complementary DNA and RNA single
strands were evaluated in a buffer containing 140 mM
KC1, 1 mM MgC1Z1 5 mM Na2HPO4 (pH 7.2), which is repre-
sentative of intracellular conditions (Alberts, B.
Molecular Biology of the Cell, pp. 304, Garland, N.Y.,
1989). Molar extinction coefficients for oligoarabino-
nucleotide strands were calculated using the nearest-
neighbor approximation, and were assumed to be the same
as those of normal strands (Puglisi, J.D.; Tinoco, I.
Jr. Methods in Enzymology, Dahlberg, J.E.; Abelson,
J.N. (eds.), 180, pp. 304, Academic Press, S.D., 1989).
Thermal denaturation curves were acquired at 260 nm
from 5 C to 90 C (rate of heating: 0.5 C/min), at a con-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 34 -
centration of approximately 2.8 M of each strand.
Melting temperatures (Tm) were calculated from first-
derivative plots of absorbance versus temperature.
Thermal denaturation curves of the heteroduplexes are
shown in Figs. 1A & 1B. Oligoarabinonucleotides ANA
(SEQ ID NO:1), 2'F-ANA (SEQ ID N0:6), and control DNA,
PS-DNA and RNA oligonucleotides were hybridized to
(Fig. 1A) complementary single-stranded RNA, and (Fig.
1B) complementary single-stranded DNA.
The results (Fig. 1A) show that the arabinonu-
cleotide of mixed base sequence, ANA (SEQ ID NO:1), has
the ability to form a stable heteroduplex with its RNA
complement, exhibiting a T, of 44 C, compared to 72 C
for the corresponding natural DNA/RNA heteroduplex. A
1:1 mixture of ANA (SEQ ID NO:1) and its DNA complement
showed a much weaker and broader transition suggesting
that, under the conditions used, SEQ ID NO:1 does not
bind to single stranded DNA (Fig. 1B). The data shown
in Fig. lA and Table 2 also show that interaction of
2'F-oligoarabinonucleotides with complement RNA results
in the formation of heteroduplexes that are of superior
thermal stability relative to the complexes formed by
the natural (PO-DNA) and thioate (PS-DNA) strands. For
example, the T,n of 2' F-araA18 (SEQ ID NO: 8) / rU18 hetero-
duplex was 30.2 C, compared to 25.4 C for the natural
dA1e/ rU18 heteroduplex. Similarly, the Tm of the 2' F-
araT1e (SEQ ID N0: 7) /rA18 heteroduplex was 43.90C, which
represents an increase in Tm of ca. 5 C relative to the
natural dT1B/rA18 heteroduplex (Tm 39 C) . Also, the mixed
base heteroduplex formed by the association of 2'F-ANA
(SEQ ID N0:6) and its target RNA sequence is thermally
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 35 -
more stable (T,n 86 C) than the corresponding of PO-
DNA/RNA and PS-DNA/RNA duplexes (Fig. 1A). In contrast
to the behavior observed for ANA sequence (SEQ ID NO:1)
(which exhibited selective binding to RNA), the 2'F-ANA
oligonucleotides (e.g. SEQ ID N0:6) bind strongly to
both single stranded complementary DNA and RNA. In
fact, 2'F-araA18 (SEQ ID N0:8) formed a more stable het-
eroduplex with single stranded complementary DNA (dTle)
than with RNA (rU1e) , i.e., T. 63 . 3 C and 30.20C, respec-
tively (Table 2). This amounts to a binding selectiv-
ity of OTm =+33 C . The selective binding to single
stranded PO-DNA (over RNA) was also observed for the
natural dA18 strand, although in this case the selectiv-
ity observed was less (OTm = +20 C) .
As shown in Fig. 2A, the CD spectra of the
heteroduplexes ANA (SEQ ID NO:l)/RNA and 2'F-ANA (SEQ
ID N0:6)/RNA, closely resembled those of the corre-
sponding DNA/RNA control duplexes (the normal substrate
of RNaseH), suggesting that all of these complexes
share the same helical conformation. The spectral fea-
tures observed are characteristic of a "A"-type helix,
a structure that appears to be important in the recog-
nition of DNA/RNA substrates by RNase H (Lima, W.F.,
Crooke, S.T. Biochemistry 1997, 36, 390). The fact
that ANA/RNA and 2'F-ANA/RNA heteroduplexes are sub-
strates of RNaseH (see Examples 5 and 6) is fully con-
sistent with this notion. The CD spectra of the
DNA/DNA duplex (of the same base sequence) is very dif-
ferent, and is characteristic of the "B-form" helical
conformation. The CD spectra of the A-form RNA/RNA
control duplex is also shown.
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 36 -
Fig. 2B shows the CD spectra of 2' F-araA18 (SEQ
ID NO: 8)/rU18 and dA1e/rU18 duplexes, as well as the cor-
responding dA18/ dTl$ duplex. The first two duplexes
exhibit a similar CD profiles (i.e., peak pattern and
peak position) that is characteristic of the A-helix
conformation, whereas the spectrum of the DNA/DNA
duplex (dA18/ dT18) falls into a pattern that is -typical
of "B-form" helices.
The same conclusions can be reached from the
spectra shown in Fig. 2C. The CD spectra of the 2'F-
araT18 (SEQ ID NO:7)/ rA1e duplex displays very similar
spectral features of the normal dT18/rAl8 hybrid, also
typical of the A-form conformation. The spectra of the
DNA/DNA duplex, dA18/dTle (B-form), is also shown for
comparison.
EXAMPLE 4
Association Properties of Oligonucleotides possessing
2-Deoxy-2-Fluoro-(3-D-Arabinose Sugars
Binding to DNA Duplexes and DNA/RNA Hybrids
To study the interaction between oligomers of
2'-deoxy-2'-fluoroarabinonucleotides and DNA/DNA and
DNA/RNA duplexes, the experimental design of Roberts and
Crothers was adopted (Roberts, R.W.; Crothers, D.M.
Science 1992, 258, 1463). The target duplexes are the
following purine-pyrimidine hairpins:
5'-GGAGAGGAGGGA T
DNA/DNA T
(SEQ ID NO:11) T
3'-CCTCTCCTCCCT T
5'-GGAGAGGAGGGA T
DNA/ RNA T
(SEQ ID NO:12) T
3 ' - CCUCUCCUCCCU T
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571 -
- 37 -
Triplex-helix formation can occur when an oli-
gonucleotide binds in the major groove of the targeted
duplexes (Le Doan, T. et al. Nucleic Acids Res. 1987,
238, 645; Moser; H.E.; Dervan, P.B. Science 1987, 238,
645). The oligopyrimidine strand containing 2'-deoxy-
2'-fluoro-arabinose sugars, i.e., 2'F-ara(CCT CTC CTC
CCT) (SEQ ID NO:13) was used to assess triple helix
formation with the above hairpin duplexes. For the pur-
pose of comparisons, the association of the correspond-
ing oligodeoxyribopyrimidine ("DNA", 2'-deoxy-(3-D-
ribose) and oligoarabinopyrimidine ("ANA", P-D-arabi-
nose) sequences were also examined. The ability of
these oligomers to form triple helices was determined
from ultraviolet spectroscopic melting experiments (as
described in Example 3) and native gel electrophoresis,
in a solution containing 100 mM sodium acetate and 1 mM
ethylenediamine tetraacetate (EDTA), pH 5.5. Molar
extinction coefficients for oligonucleotides were cal-
culated from those of the mononucleotides and dinucleo-
tides according to nearest-neighbouring approximations
(Puglisi, J.D.; Tinoco, I. Jr. Methods in Enzymology,
Dahlberg, J.E.; Abelson, J.N. (eds.), 180, pp. 304,
Academic Press, S.D., 1989). The values for the hybrid
hairpin was assumed to be the sum of their DNA plus RNA
components: DNA/DNA, 26.5; DNA/RNA, 27.1; (units= 104 M-
1 cm-1). The molar extinction coefficient for the 2'-
fluoroarabinonucleotide strand SEQ ID NO:13 was assumed
to be the same as a normal DNA strand (9.6 x 104 M-1 cm-1
units). Complexes were prepared by mixing equimolar
amounts of interacting strands, e.g., 2'F-ANA (SEQ ID
NO:13) + hairpin DNA/DNA or DNA/RNA, and lyophilizing
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 38 -
the resulting mixture to dryness. The resulting pellet
was then re-dissolved in 100 mM NaOAc/ 1 mM EDTA buffer
(pH 5.5). The final concentration was 2 M in each
strand. The solutions were then heated to 80 C for 15
min, cooled slowly to r.t., and stored at 4 C overnight
before measurements. Prior to the thermal run, samples
were degassed by placing them in a speed-vac concentra-
tor (2 min). Denaturation curves were acquired at 260
nm at a rate of heating of 0.5 C/min. Melting tempera-
tures (Tm) were calculated from the first derivative of
the melting curves. The results of the melting experi-
ments are shown in Fig. 3.
The results show that when 2'F-ANA (SEQ ID
NO:13) was mixed with an equimolar concentration of
hairpin DNA/DNA (SEQ ID NO:11) or DNA/RNA (SEQ ID
NO:12), a biphasic transition was observed upon heating
the solution from 10 C to 90 C. The low temperature
transition is assigned to the dissociation of 2'F-ANA
(SEQ ID NO:13) from the target hairpins (Roberts, R.W.;
Crothers, D.M. Science 1992, 258, 1463). The high tem-
perature transition corresponds to the melting of the
hairpin duplexes, since it was also observed when a
solution of hairpin duplex alone was heated under iden-
tical conditions. The data show that Tm values for low
temperature transitions resulting from mixtures of SEQ
ID NO:13 (2'-deoxy-2'-fluoro-(3-D-arabinose oligomer) +
hairpins are considerable higher than T. values for tran-
sitions from the corresponding ANA ((3-D-arabinose oli-
gomer) + hairpin, or DNA (2'-deoxy-(3-D-ribose oligomer)
+ hairpin mixtures. For example, as can be seen from
the melting curves shown in Fig. 3, the Tm for the disso-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 39 -
ciation of 2' F-ANA strand SEQ ID NO:13 from the DNA/DNA
hairpin (DD, SEQ ID NO:11) is 49 C, compared to 34 C and
40 C, for the dissociation of the ANA ((3-D-arabinose; not
shown) and DNA (2'-deoxy-(3-D-ribose) oligonucleotides,
respectively. Similarly, the first transition for the
triplex formed by 2'F-ANA (SEQ ID NO:13) and DNA/RNA
hairpin (SEQ ID NO:12) was 53 C, compared to 43 C and
45 C, for the corresponding triplexes formed by the ANA
(P-D-arabinose; not shown) and DNA (2'-deoxy-P-D-ribose)
strands, respectively.
The equilibrium between single-, double-, and
triple-stranded species of 2'F-ANA (SEQ ID NO:13) +
hairpin mixtures was also directly monitored by poly-
acrylamide gel electrophoresis (Fig. 4). Fig. 4 shows a
photograph of a polyacrylamide gel of single stranded
oligo-2'F-arabinopyrimidine SEQ ID NO:13 and target
hairpins DNA/DNA (SEQUENCE ID NO:11) ("DD") and DNA/RNA
(SEQUENCE ID NO:12) ("DR"), and 1:1 ratios of SEQ ID
NO:13:hairpins. The first lane shows marker dyes
xylene cyanol (XC) and bromophenol blue (BPB). The
next lane shows the SEQ ID NO:13 strand, whereas the
"DD (-)" lane shows the DNA/DNA hairpin (SEQ ID NO:11).
The SEQ ID NO:13:DD triplex is clearly seen in the "DD
(+)" lane, which contains a 1:1 molar mixture of 2'F-
ANA (SEQ ID NO:13) and "DD". The DNA/RNA hairpin (SEQ
ID NO:12) is visible in the "DR (-)" lane. The triplex
SEQ ID NO:13:DR triplex is clearly visible in the "DR
(+)" lane, which consists of a 1:1 mixture of 2'F-ANA
(SEQ ID NO:13) and "DR". Gels were visualized by UV-
shadowing. Base sequence of single strand 2'F-ANA (SEQ
ID NO:13) and hairpins DD (SEQ ID NO:11) and DR (SEQ ID
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 40 -
NO:12) and experimental conditions are given above in
Example 4.
This method provides a convenient way to monitor
triplex formation and to qualitative check on the stoi-
chiometry of interaction the strands (Kibler-Herzog, L.
et al. Nucleic Acids Res. 1990, 18, 3545). The results
in Fig. 4 show that the 2'F-ANA strand (SEQ ID NO:13),
hairpins, and triple-helical complexes can be separated
with excellent resolution on a polyacrylamide gel at low
temperature. As can be seen from the gel results, the
triple-helical complex is nearly quantitatively formed
at a 1:1 molar ratio of 2'F-ANA (SEQ ID NO:13):hairpin.
This is in contrast to the incubation of ANA (D-arabi-
nose strand) and hairpin (1:1 molar ratio) which, under
the same conditions, gave a mixture of ANA + hairpin +
triplex (see Noronha, A.; Damha, M.J. Nucleic Acids Res.
1998, 26, 2665). This is in complete agreement with the
T. results which indicates that the SEQ ID NO:13 (2'-
deoxy-2'-fluoro-(3-D-arabinose) strand has a signifi-
cantly higher affinity for the DNA/DNA and DNA/RNA hair-
pin duplexes relative to the ANA (P-D-arabinose) strand.
EXAMPLE 5
Induction of Ribonuclease H (RNaseH) Activity by
Oligonucleotides possessing P-D-Arabinose as Sugar
Component
Defined-sequence oligonucleotides, 18-units in
length, were used in these experiments, i.e.,
5'-d(AGCTCCCAGGCTCAGATC) -3' "DNA" (SEQ ID NO:14)
5'-ara(AGCUCCCAGGCUCAGAUC)-3' "ARA" (SEQ ID N0:1)
5'-ribo(AGCUCCCAGGCUCAGAUC)-3' "3',5'-RNA" (SEQ ID NO:15)
5'-ribo(AGCUCCCAGGCUCAGAUC)-3' "2',5'-RNA" (SEQ ID NO:16)
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 41 -
These oligomers are complementary to a sequence
within the R region of HIV-1 genomic RNA. The target
RNA used was a synthetic 18 nt 3',5'-RNA oligonucleo-
tide, identical to the sequence within the HIV R
region, and exactly complementary to the sequence of
the above oligonucleotides.
The ability of the above oligonucleotides to
elicit RNaseH degradation of target RNA was determined
in assays (10 L final volume) that comprised 5 pmol of
5' -[3zP] - target RNA and 15 pmol of test oligonucleotide
in 60 mM Tris-HC1 (pH 7.8, 25 C) containing 2mM dithio-
threitol, 60 mM KC1, and either 10 mM MgC12 or 0.1 mM
MnC12. Reactions were started by the addition of HIV-RT
or E. coli RNaseH. Incubations were carried out at 25 C
for varying times (generally 20 to 30 minutes). Reac-
tions were quenched by the addition of loading buffer
(98% deionized formamide containing 10 mM EDTA and
lmg/mL each of bromophenol blue and xylene cyanol), and
heating at 100 C for 5 minutes. The reaction products
were resolved by electrophoresis using a 16% polyacry-
lamide sequencing gel containing 7 M urea, and visual-
ized by autoradiography. The result of such experiments
is shown in Fig. 5.
The results show that the oligonucleotide based
on 2'-deoxyribose ("DNA" (SEQ ID NO:14)) or P-D-arabi-
nose ("ARA" (SEQ ID NO:1)) are able to form duplexes
with target RNA that serve as substrates for the RNase
H activity of either HIV-1 RT or E. coli RNase H, as
indicated by the numerous smaller sized degradation
products of the target RNA in Fig. 5. This RNase H deg-
radation was noted with either Mn2+ (illustrated) or Mg2'
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 42 -
(not shown) as metal. In contrast, oligonucleotides
based on D-ribose, either in 3',5'-linkages (3',5'-RNA
(SEQ ID NO:15)), or in 2',5'-linkages (2',5'-RNA (SEQ
ID NO:16)) were unable to elicit this RNase H degrada-
tion of target RNA, even though these test oligonucleo-
tides were competent to form duplexes with the target
RNA. Similarly, an oligonucleotide based on (3-D-21-
deoxyribose, but of a random base sequence (DNA random)
not complementary to the target RNA (and therefore
unable to form duplexes), was also unable to elicit
RNase H activity.
EXAMPLES 6
Induction of Ribonuclease H (RNAseH) Activity by
Oligonucleotides possessing 2-Deoxy-2-Fluoro-p-D-
Arabinose as Sugar Component
One set of experiments (A) made use of test
homopolymeric octadecanucleotides with either thymine
(T) or uracil (U) as base component, namely PO-araUla
(SEQ ID NO:3), PO-2'F-araT18 (SEQ ID NO:7), PO-dT18, PS-
dT18, PO-2'F-riboTle, PO-riboU18. The target RNA in
experiment set A was a synthetic 3',5'-phosphodiester-
linked rA1e oligonucleotide, exactly complementary to
the sequence of the test oligonucleotides.
Another set of experiments (B) made use of test
heteropolymeric octadecanucleotides of the following
sequence:
5'-TTA TAT TTT TTC TTT CCC-3' (SEQ ID NO:9) for
PO-DNA, PS-DNA and 2'F-ANA oligonucleotides
5'-UUA UAU UUU UUC UUU CCC-3' for ANA (SEQ ID
N0:4) and RNA oligonucleotides
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571 - 43 -
The target RNA in experiment set B was a het-
eropolymeric octadecaribonucleotide exactly complemen-
tary to the sequence of the test oligonucleotides.
Experiments set (A):
The ability of homopolymeric oligonucleotides
with 2'-deoxy-2'-fluoro-p-D-arabinose as sugar compo-
nent, and other oligonucleotides, to elicit RNaseH deg-
radation of target RNA was determined in assays (10 L
final volume) that comprised 1 pmol of 5' -[32P] - target
RNA and 8 pmol of test oligonucleotide in 60 mM Tris-
HC1 (pH 7.8, 25 C) containing 2mM dithiothreitol, 60 mM
KC1, and 2.5 mM MgC12. Reactions were started by the
addition of E. coli RNaseH. Incubations were at 22 C for
0, 5, 10 and 20 minutes. Reactions were quenched by the
addition of loading buffer (98% deionized formamide
containing 10 mM EDTA and lmg/mL each of bromophenol
blue and xylene cyanol, and heating at 100 C for 5 min-
utes. The reaction products were resolved by electro-
phoresis using a 16% polyacrylamide sequencing gel con-
taining 7 M urea, and visualized by autoradiography.
The result of such an experiment is shown in Fig. 6.
Each of the oligonucleotides based on (3-D-2.'-
deoxyribose with phosphodiester bonds (i.e., PO-dT18,
abbreviated as "dT18"), P-D-2'-deoxyribose with phos-
phorothioate bonds (PS-dT16, or "dTlBthioate"), 2'-deoxy-
2'-fluoro-(3-D-arabinose (PO-2'F-araTla SEQ ID NO:7, or
"aFT18" ) , (3-D-ribose (PO-rU18, or "rUle" ) and 2' -deoxy-2' -
fluoro-p-D-ribose (PO-2' F-rT18, or "rFTle" ) were able to
form duplexes with target RNA (rA1e). Only duplexes
formed with oligonucleotides "dT18", "dT18thioate" or
"aFT18" served as substrates for the RNase H activity of
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571 - 44 -
either HIV-1 RT or E. coli RNase H, as indicated by the
numerous smaller sized degradation products of the tar-
get RNA in Fig. 6. Duplexes formed with rFT18 or rUle
could not serve as substrates for the RNase H activity
of E. coli RNase H. Under these conditions an octade-
canucleotide based on P-D-arabinosyluracil (p0-araU1e
SEQ ID N0:3, or "aU1e") was unable to form a duplex with
target rA, and accordingly was unable to elicit RNase H
activity (these findings are consistent with those
reported by Giannaris and Damha, who found that araUB
was unable to form a duplex with poly rA; Giannaris,
P.A.; Damha, M.J., Can.J.Chem. 1994, 74, 909).
Experiments set (B):
Heteropolymeric octadecanucleotides of the
sequence 5'-TTA TAT TTT TTC TTT CCC-3' (SEQ ID N0:9)
for PO-DNA, PS-DNA and 2'F-ANA oligonucleotides, and
5'-UUA UAU UUU UUC UUU CCC-3' (SEQ ID N0:4) for ANA and
RNA oligonucleotides were annealed to 5'-[32P]-labeled
target RNA exactly complementary to the test AON
sequence. The ability of heteropolymeric oligonucleo-
tides with 2'-deoxy-2'-fluoro-(3-D-arabinose as sugar
component, and other oligonucleotides, to elicit RNaseH
degradation of target RNA was determined in assays (50
L final volume) that comprised 100 nM 5' -[32P] - target
RNA and 500 nM test oligonucleotide in 60 mM Tris-HC1
(pH 7.8, 25 C) containing 2mM dithiothreitol, 60 mM KC1,
and 2.5 mM MgC12. Reactions were started by the addition
of E. coli RNaseH (final activity 25U/ml). RNaseH
digestions were carried out at either 25 C or 37 C. At
various times, 10 L aliquots were removed and quenched
by the addition of loading buffer (98% deionized form-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 45 -
amide containing 10 mM EDTA and lmg/mL each of bromo-
phenol blue and xylene cyanol) followed by heating at
100 C for 5 minutes. The reaction products were resolved
by electrophoresis using a 16% polyacrylamide sequenc-
ing gel containing 7 M urea, and visualized by autora-
diography. The result of such an experiment is shown in
Fig. 7. The susceptibility of pre-formed oligonucleo-
tide/RNA duplexes to degradation by E. coli RNaseH was
assessed at 37 C (left panel) and 25 C (Fig. 7, right
panel). For each test compound, the lanes correspond to
the absence (-) or the presence (+) of added E. coli
RNaseH.
All heteropolymeric oligonucleotides were able
to form duplexes with target RNA (UV melting experi-
ments). Oligonucleotides based on P-D-2'-deoxyribose
with phosphodiester bonds (PO-DNA), (3-D-2'-deoxyribose
with phosphorothioate bonds (PS-DNA), P-D-arabinose
(ANA, SEQ ID N0:4) and 2'-deoxy-2'-fluoro-(3-D-arabinose
(2'F-ANA, SEQ ID N0:9) were able to elicit degradation
of the complementary target RNA in the presence of E.
coli RNaseH, as indicated by the numerous smaller sized
degradation products of the target RNA in Fig. 7. Oli-
gonucleotides based on (3-D-ribose (RNA) could not
elicit RNaseH degradation of the target RNA.
EXAMPLE 7
Nuclease Stability of Oligoarabinonucleotides
Thymine octadecanucleotides based on 2'-deoxy-
ribose with phosphodiester bonds (PO-DNA dT1e, abbrevi-
ated as "dT") and 2'-deoxy-2'-F-P-D-arabinose (2'F-
araT1e SEQ ID NO : 7, or "aFT18" ) were compared for stabil -
ity against degradation by serum nucleases and cellular
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 46 -
nucleases. The antisense oligonucleotides were radioac-
tively labeled at the 5' -terminus using [y-32P] -ATP and
and T4 polynucleotide kinase according to standard pro-
tocols (Ausubel, F.M. et al., Current Protocols in
Molecular Biology, John Wiley & Sons, Inc., 1994).
Stability against serum nucleases was assessed
by adding 1 pmol of 5'-[32P]-AON to a reaction assay (10
L final volume) containing 90% horse serum. Reactions
were incubated at 20 C for varying times (0, 5, 10, 15,
20 and 30 min.), then aliquots were removed and diluted
with gel loading buffer (98% deionized formamide con-
taining 10 mM EDTA and 1 mg/mL each of bromophenol blue
and xylene cyanol), boiled for 5 minutes then resolved
by electrophoresis on a 16% polyacrylamide sequencing
gel containing 7 M urea. Separated products were visu-
alized by autoradiography. The results of such an
experiment is shown in Fig. 8.
The "aFT1B" oligonucleotide (SEQ ID NO:7) was
substantially more resistant to degradation by serum
nucleases, as indicated by the decreased number of
smaller molecular size degradation products compared to
that noted with normal "dT1e" oligonucleotide. Qualita-
tively similar results were obtained in similar experi-
ments in which human serum was substituted for horse
serum (data not presented). Under these same condi-
tions, oligonucleotides based on P-D-2'-deoxyribose
with phosphorothioate bonds (PS-dT1e) were virtually
unaffected by serum nucleases (data not shown), as pre-
viously established by many investigators.
Unfractionated mouse liver crude homogenates
(prepared by homogenizing mouse livers in an equal vol-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 47 -
ume of 20 mM Tris-HC1 (pH 7.9, 20 C) containing 60 mM
KC1, imM dithiothreitol and 12% (v/v) glycerol) were
used as a source of cellular nucleases. Stability
against cellular nucleases was assessed by adding 1
pmol of 5' -[32P] -ODN to a reaction assay (10 L final
volume) containing 90% unfractionated mouse liver crude
homogenate. After varying times (0, 10, 20, 30 and 60
min.) of incubation at 20 C, aliquots were removed,
diluted with gel loading buffer (98% deionized formam-
ide containing 10 mM EDTA, lmg/mL each of bromophenol
blue and xylene cyanol), boiled for 5 minutes then
resolved by electrophoresis on a 16% polyacrylamide
sequencing gel containing 7 M urea. Separated products
were visualized by autoradiography. The results estab-
lished that the "aFT18" oligonucleotide was signifi-
cantly more resistant than the corresponding "dT18" oli-
gonucleotide to degradation by cellular nucleases (data
not shown).
Snake venom phosphodiesterase I is an aggres-
sive enzyme that rapidly degrades single strand nucleic
acids. Each of the ol igonucleot ides based on P-D-21-
deoxyribose with phosphodiester bonds (PO-DNA dTle,
abbreviated as "dT"), (3-D-2'-deoxyribose with phos-
phorothioate bonds (PS-DNA dT16, or "dTle thioate"), P-D-
arabinosyluracil (PO-araU18 SEQ ID NO: 3, or "aU18" ), 2' -
deoxy-2'-fluoro-p-D-arabinose (PO-2'-F-araT18 SEQ ID
NO:7, or "aFT1e"), and 2'-deoxy-2'-fluoro-p-D-ribose
(PO-2'-F-rT1e, or "rFT18") was examined for stability
against degradation by snake venom phosphodiesterase I
in assays (50 L final volume) that comprised 100 nM
5'-[32P]-oligonucleotide in 100 mM Tris-HC1 (pH 8.9,
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 48 -
37 C) containing 100 mM NaCl and 10 mM MgC12. Reactions
were started by the addition of snake venom phosphodi-
esterase I (final activity 0.4U/ml). Digestions were
carried out at or 37 C. At various times, 10 l aliquots
were removed and quenched by the addition of loading
buffer (98% deionized formamide containing 10 mM EDTA
and lmg/mL each of bromophenol blue and xylene cyanol)
followed by heating at 100 C for 5 minutes. The reaction
products were resolved by electrophoresis using a 16%
polyacrylamide sequencing gel containing 7 M urea, and
visualized by autoradiography. The results of such an
experiment is shown in Fig. 9. They established that
the order of nuclease stability is "dT1e thioate" >
"aU18 11 N "aFT1e 11 >> "rFT18 11 > 11 dTig It
EXAMPLE 8
Nonspecific Interaction of Oligoarabinonucleotides with
Cellular Proteins
The ability of thymine octadecanucleotides
based on (3-D-2'-deoxyribose with phosphodiester bonds
(PO-DNA dT18, abbreviated as "dTle" ) , (3-D-2' -deoxyribose
with phosphorothioate bonds (PS-DNA dT18, or "dT18 thio-
ate"), J3-D-arabinosyluracil (PO-araU16 SEQ ID N0:3, or
"aU18"), 2'-deoxy-2'-fluoro-(3-D-arabinose (PO-2'-F-araT18
SEQ ID N0:7, or "aFT18"), and 2'-deoxy-2'-fluoro-(3-D-
ribose (PO-2' -F-rT1B, or "rFT18" ) to bind nonspecifically
to proteins in a Hela cell crude extract was analyzed
by a gel shift assay procedure. The antisense oligonu-
cleotides were radioactively labeled at the 5'-terminus
using [y-32P]-ATP and and T4 polynucleotide kinase
according to standard protocols (Ausubel, F.M. et al.,
Current Protocols in Molecular Biology, John Wiley &
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 49 -
Sons, Inc., 1994). Hela cell crude extracts were pre-
pared by homogenizing cells in buffer (20 mM Tris-HC1
(pH 7.9, 20 C) containing 60 mM KC1, imM dithiothreitol
and 12% (v/v) glycerol), followed by centrifugation to
remove membrane particles and cell debris.
The binding experiments comprised 1 pmol of 5'-
32P-labeled AON and 5 g Hela cell extract protein in a
total volume of 20 l comprising 10 mM Tris-HC1 (pH
7.5, 25 C) containing 100 mM KC1, 1 mM MgClZ1 1 mM EDTA
and 5 % glycerol. AON and Hela cell proteins were incu-
bated for 10 min at 25 C, then electrophoresed on 6%
non-denaturing gels. Following completion of the elec-
trophoresis, the gels were dried and the positions of
the free and protein-bound oligonucleotides visualized
by autoradiography. The results of such an experiment
are shown in Fig. 10.
Essentially all of the phosphorothioate "dTle
thioate" was bound to the extract proteins, as indi-
cated by the "smear" of radioactive material throughout
the extent of electrophoresis. None of the other
antisense oligonucleotides showed any appreciable
interaction with Hela cell proteins under the same con-
ditions.
EXAMPLE 9
Antisense oligonucleotide inhibition of specific gene
expression
Antisense oligonucleotides have the potential
to inhibit expression of virtually any gene, based on
the specific base sequence of the chosen target mRNA.
We examined the ability of antisense oligonucleotides
based on P-D-arabinose and 2'-deoxy-2'-fluoro-p-D-ara-
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 50 -
binose to interfere with the expression of a well-char-
acterized marker model, namely expression of the enzyme
luciferase, in both in vitro cell-free translation
experiments (Fig. 11), and in cells stably transfected
with the luciferase gene (Fig. 12).
The ability of oligonucleotides complementary
to a specific region of mRNA coding for luciferase was
tested for inhibition of luciferase protein expression
in an in vitro protein translation system. The specific
antisense oligonucleotide sequences were 5'-ATA TCC TTG
TCG TAT CCC-3' (SEQ ID NO:10) for 2'F-ANA, ANA (SEQ ID
N0:5) and the corresponding PO-DNA and PS-DNA strands,
which are complementary to bases 1511-1528 of the cod-
ing region of the luciferase gene (M. Gossen, H. Bujard
1992, Proc. Natl. Acad. Sci. USA. 89, 5547-5551; (M.
Gossen, H. Bujard 1992, Proc. Natl. Acad. Sci. USA. 89,
5547-5551; W.M. Flanagan et al. 1996, Nucl. Acids Res.
24, 2936-2941). As a control, randomized oligonucleo-
tide sequences (5'-TAA TCC CTA TCG TCG CTT-3' (SEQ ID
N0:17) for 2'F-ANA, ANA, PO-DNA and PS-DNA were used;
these are of the same base composition as the specific
AON sequence, but have no complementarity to any por-
tion of the luciferase gene. Translation reaction
assays (15 l total volume) comprised 0.15 pmol lucif-
erase mRNA, 10 l commercial rabbit reticulocyte lysate
supplemented with complete amino acids mixture and
excess single strand RNA ribonuclease inhibitor. To
this mixture was added varying amounts (0.1 - 5 pmol)
of specific or random oligonucleotide, followed by
addition of E. coli RNaseH (20U/ml final concentra-
tion). Translation reactions were carried out for 60
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 51 -
min at 37 C, then the amount of full-length active
luciferase produced was assayed by luminometry (W.M.
Flanagan et al 1996, Nucl. Acids Res. 24, 2936-2941).
The results of such an experiment are shown in Fig. 11.
Panel (A) shows the inhibitory activity of oligonucleo-
tides based on (3-D-2'-deoxyribose with phosphodiester
bonds (PO-DNA). Panel (B) shows the inhibitory activity
of oligonucleotides based on (3-D-2'-deoxyribose with
phosphorothioate bonds (PS-DNA) Panel (C) shows the
inhibitory activity of oligonucleotides based on 2'-
deoxy-2'-fluoro-(3-D-arabinose with phosphodiester bonds
(2'F-ANA SEQ ID NO:10); Panel (D) shows the inhibitory
activity of oligonucleotides based on (3-D-arabinose
with phosphodiester bonds (ANA, SEQ ID N0:5).
The results (Fig. 11) show that 2'F-ANA and PO-
DNA oligonucleotides are potent and specific inhibitors
of luciferase gene expression in the in vitro protein
expression model, at low AON:mRNA ratios. In these
experiments, the in vitro inhibitory potency (IC50) of
oligonucleotides based on 2'-deoxy-2'-fluoro-(3-D-arabi-
nose with phosphodiester bonds (2'F-ANA SEQ ID NO:10)
is at least four-fold greater than that of the same-
sequence oligonucleotide based on P-D-2'-deoxyribose
with phosphorothioate bonds (PS-DNA).
Hela X1/5 cells stably express the luciferase
gene (W.M. Flanagan et al 1996, Nucl. Acids Res. 24,
2936-2941). Cells were treated with Lipofectin and o1i-
gonucleotide (1 M final concentration) for 24h, then
cells were harvested and cell extracts were assayed for
luciferase activity (Fig. 12, panel A) and toxicity
(residual cell protein, Fig. 12, panel B) . The chemical
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571 - 52 -
character of the oligonucleotides is indicated on Fig.
12. Specific sequences used are as those described
above, namely, 2'F-ANA (SEQ ID NO:10), ANA (SEQ ID
N0:5) and their corresponding PO-DNA and PS-DNA
sequences. Random sequences were 2'F-ANA (SEQ ID
N0:17, ANA and corresponding randomized PO-DNA and PS-
DNA sequences..
X1/5 Hela cells were plated in 96-well plates
and allowed to grow, in DMEM/10% FBS, to 80% conflu-
ence, as assessed by microscopy. The medium was
removed, and the cells washed several times with phos-
phate-buffered saline. The cells were overlaid with
serum-free DMEM medium containing 20 g/ml Lipofectin,
then a small volume aliquot of concentrated test AON
stock solution was added with mixing to ensure good
distribution. After 24h incubation, the Hela cells were
harvested, homogenized and assayed for luciferase.
Panel A of Fig. 12 shows that PS-DNA (sequence
specific) completely eliminated intracellular lucifer-
ase gene expression. The 2'F-ANA (SEQ ID NO:10)and ANA
(SEQ ID No:5) sequence specific oligomers were also
potent inhibitors decreasing intracellular luciferase
gene expression by about 40-50%. These data were sig-
nificantly different from the PO-DNA control as indi-
cated in the Panel A (statistical significance is indi-
cated as well). Panel B shows that neither 2'F-ANA or
ANA were toxic to Hela cells as determined by effect on
cell number (i.e., residual cell protein after 24-h
exposure). In contrast, PS-DNA (both random and spe-
cific sequences) exhibited significant toxicity reduc-
ing cell number by ca. 40% after 24-h exposure.
CA 02331333 2000-12-19
WO 99/67378 PCT/CA99/00571
- 53 -
While the invention has been described in con-
nection with specific embodiments thereof, it will be
understood that it is capable of further modifications
and this application is intended to cover any varia-
tions, uses, or adaptations of the invention following,
in general, the principles of the invention and
including such departures from the present disclosure
as come within known or customary practice within the
art to which the invention pertains and as may be
applied to the essential features set forth hereinbe-
fore, and as follows in the scope of the appended
claims.
CA 02331333 2000-12-19
- 53a -
SEQUENCE LISTING
<110> McGILL UNIVERSITY
<120> ANTISENSE OLIGONUCLEOTIDE CONSTRUCTS BASED ON BETA-
ARABINOFURANOSE AND ITS ANALOGUES
<130> 1770-206"CA" FC/gc
<150> PCT/CA99/00571
<151> 1999-06-17
<150> CA 2,241,361
<151> 1998-06-19
<160> 17
<170> FastSEQ for Windows Version 3.0
<210> 1
<211> 18
<212> RNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 1
agcucccagg cucagauc 18
<210> 2
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 2
aaaaaaaaaa aaaaaaaa 18
<210> 3
<211> 18
<212> RNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
CA 02331333 2000-12-19
- 53b -
<400> 3
uuuuuuuuuu uuuuuuuu 18
<210> 4
<211> 18
<212> RNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 4
uuauauuuuu ucuuuccc 18
<210> 5
<211> 18
<212> RNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 5
auauccuugu cguauccc 18
<210> 6
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 6
agctcccagg ctcagatc 18
<210> 7
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 7
tttttttttt tttttttt 18
CA 02331333 2000-12-19
- 53c -
<210> 8
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 8
aaaaaaaaaa aaaaaaaa 18
<210> 9
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 9
ttatattttt tctttccc 18
<210> 10
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 10
atatccttgt cgtatccc 18
<210> 11
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 11
ggagaggagg gatttttccc tcctctcc 28
<210> 12
<211> 28
<212> RNA
<213> Artificial Sequence
CA 02331333 2000-12-19
- 53d -
<220>
<223> Use as an oligomer
<400> 12
ggagaggagg gattttuccc uccucucc 28
<210> 13
<211> 12
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 13
cctctcctcc ct 12
<210> 14
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 14
agctcccagg ctcagatc 18
<210> 15
<211> 18
<212> RNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 15
agcucccagg cucagauc 18
<210> 16
<211> 18
<212> RNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
CA 02331333 2000-12-19
- 53e -
<400> 16
agcucccagg cucagauc 18
<210> 17
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Use as an oligomer
<400> 17
taatccctat cgtcgctt 18