Note: Descriptions are shown in the official language in which they were submitted.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
De novo DNA Cytosine Methyltransferase Genes,
Polypeptides and Uses Thereof
Background of the Invention
Field of the Invention
The present invention relates generally to the fields of molecular biology,
developmental biology, cancer biology and medical therapeutics. Specifically,
the present invention relates to novel DNA cytosine methyltransferases. More
specifically, isolated nucleic acid molecules are provided encoding mouse
Dnmt3a and Dnmt3b and human DNMT3A and DNMT3B de novo DNA
cytosine methyltransferase genes. Dnmt3a and Dnmt3b mouse and DNMT3A
and DNMT3B human polypeptides are also provided, as are vectors, host cells
and recombinant methods for producing the same. The invention further relates
to an in vitro method for cytosine C5 methylation. Also provided is a
diagnostic
method for neoplastic disorders, and methods of gene therapy using the
polynucleotides of the invention.
Related Art
Methylation at the C-5 position of cytosine predominantly in CpG
dinucleotides is the major form of DNA modification in vertebrate and
invertebrate animals, plants, and fungi. Two distinctive enzymatic activities
have
been shown to be present in these organisms. The de novo DNA cytosine
methyltransferase, whose expression is tightly regulated in development,
methylates unmodified CpG sites to establish tissue or gene-specific
methylation
patterns. The maintenance methyltransferase transfers a methyl group to
cytosine
in hemi-methylated CpG sites in newly replicated DNA, thus functioning to
maintain clonal inheritance of the existing methylation patterns.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-2-
De novo methylation of genomic DNA is a developmentally regulated
process (Jahaner, D. and Jaenish, R., "DNA Methylation in Early Mammalian
Development," In DNA Methylation: Biochemistry and Biological Significance,
Razin, A. et al., eds., Springer-Verlag (1984) pp. 189-219 and Razin, A., and
Cedar, H., "DNA Methylation and Embryogenesis," in DNA Methylation:
Molecular Biology and Biological Significance, Jost., J. P. et al., eds.,
Birkhauser
Verlag, Basel, Switzerland (1993) pp. 343-357). It plays a pivotal role in the
establishment of parental-specific methylation patterns of imprinted genes
(Chaillet, J. R. et al., Cell 66:77-83 (1991); Stoger., R. et al., Cell 73:61-
71
(1993); Brandeis, M. et al., EMBOJ. 12:3669-3677 (1993); Tremblay, K. D. et
al., Nature Genet. 9:407-413 (1995); and Tucker, K. L. et al., Genes Dev.
10:1008-1020 (1996)), and in the regulation of X chromosome inactivation in
mammals (Brockdoff, N. "Convergent Themes in X Chromosome Inactivation
and Autosomal Imprinting," in Genomic Imprinting: Frontiers in Molecular
Biology, Reik, W. and Sorani, A. eds., IRL Press Oxford (1997) pp. 191-210;
Ariel, M. et al., Nature Genet. 9:312-315 (1995); and Zucotti, M. and Monk, M.
Nature Genet. 9:316-320 (1995)).
Thus, C5 methylation is a tightly regulated biological process important
in the control of gene regulation. Additionally, aberrant de novo methylation
can
lead to undesirable consequences. For example, de novo methylation of growth
regulatory genes in somatic tissues is associated with tumorigenesis in humans
(Laird, P. W. and Jaenisch, R. Ann. Rev. Genet. 30:441-464 (1996); Baylin, S.
B.
et al., Adv. Cancer. Res. 72:141-196 (1998); and Jones, P. A. and Gonzalgo, M.
L. Proc. Natl. Acad. Sci. USA 94:2103-2105 (1997)).
The gene encoding the major maintenance methyltransferase, Dnmtl, was
first cloned in mice (Bestor, T. H. et al., J. Mol. Biol. 203:971-983 (1988),
and
the homologous genes were subsequently cloned from a number of organisms,
including Arabidoposis, sea urchin, chick, and human. Dnmtl is expressed
ubiquitously in human and mouse tissues. Targeted disruption of Dnmtl results
in a genome-wide loss of cytosine methylation and embryonic lethality (Li
etal.,
CA 02331781 2001-03-20
-3-
1992). Interestingly, Dnmtl is dispensable for the survival and growth of the
embryonic stem cells, but appears to be required for the proliferation of
differentiated somatic cells (Lei et al., 1996). Although it has been shown
that
the enzyme encoded by Dnmtl can nlethylate DNA de novo in vitro (Bestor,
1992). there is no evidence that Dnmt 1 is directly involved in de novo
methylation
in normal development. Dnmtl appears to function primarily as a maintenance
methyltransferase because of its strong preference for hemi-methylated DNA and
direct association with newly replicated DNA (Leonhardt, H. et al., Cell
71:865-
873 (1992)). Additionally, ES cells homozygous for a null mutation of Dnmtl
can methylate newly integrated retroviral DNA, suggesting that Dnmtl is not
required for de novo methylation and an independently encoded de novo DNA
cytosine methyltransferase is present in mammalian cells (Lei et al., 1996).
Various methods of disrupting Dnmtl protein activity are known to those
skilled in the art. For example, see PCT Publication No. W092/06985, wherein
mechanism based inhibitors are discussed. Applications involving antisense
technology are also known; U.S. Patent No. 5578716 discloses the use of
antisense oligonucleotides to inhibit Dnmtl activity, and Szyf et al., J.
Biol.
Chem. 267: 12831-12836, 1992, demonstrates that myogenic differentiation can
be affected through the antisense inhibition of Dnmtl protein activity.
Thus, while there is a significant amount of knowledge in the art regarding
the maintenance C5 methyltransferase (Dnmtl ), there is no information
regarding
nucleic acid or protein structure and expression or enzymatic properties of
the de
novo C5 methyltransferase in mammals.
Summary of the Invention
An object of the present invention is to provide de novo DNA cytosine
methyltransferase genes, polypeptides and uses thereof. In accordance with an
aspect of the present invention, there is provided an isolated nucleic acid
molecule
comprising a polynucleotide selected from the group consisting of:
CA 02331781 2001-03-20
-3a-
(a) a polynucleotide sequence encoding a polypeptide
comprising amino acids from about 1 to about 908 in SEQ ID NO:5;
(b) a polynucleotide sequence encoding a polypeptide
comprising amino acids from about 1 to about 859 in SEQ ID NO:6;
(c) a polynucleotide sequence encoding a polypeptide
comprising amino acids from about I to about 912 in SEQ ID NO:7;
(d) a polynucleotide sequence encoding a polypeptide
comprising amino acids from about 1 to about 853 in SEQ ID NO:8; and
(e) a polynucleotide sequence that is at least 90% identical to
the polynucleotide sequence of (a), (b), (c) or (d).
In accordance with another aspect of the invention, there is provided an
isolated nucleic acid molecule comprising polynucleotides selected from the
group
consisting of:
(a) at least 20 contiguous nucleotides of SEQ ID NO:I,
provided that said nucleotides are not AA052791(SEQ ID NO: 9);
AA111043(SEQ ID NO:10); AA154890(SEQ ID NO:11); AA240794(SEQ ID
NO:12); AA756653(SEQ ID NO:13); W58898(SEQ ID NO:14); W59299(SEQ
ID NO:15); W91664(SEQ ID NO:16); W91665(SEQ ID NO:17); or any
subfragment thereof; and
(b) a nucleotide sequence complementary to a nucleotide
sequence in (a).
In accordance with another aspect of the invention, there is provided an
isolated nucleic acid molecule comprising polynucleotides selected from the
group
consisting of:
(a) at least 20 contiguous nucleotides of SEQ ID NO:2,
provided that said nucleotides are not AA116694 (SEQ ID NO: 18); AA119979
(SEQ ID NO:19); AA177277 (SEQ ID NO:20); AA210568 (SEQ ID NO:21);
AA399749 (SEQ ID NO:22); AA407106 (SEQ ID NO:23); AA575617 (SEQ ID
NO:24); or any subfragment thereof; and
CA 02331781 2001-03-20
-3b-
(b) a nucleotide sequence complementary to a nucleotide
sequence in (a).
In accordance with another aspect of the invention, there is provided an
isolated nucleic acid molecule comprising polynucleotides selected from the
group
consisting of:
(a) at least 20 contiguous nucleotides of SEQ ID NO:3,
provided that said nucleotides are not AA004310 (SEQ ID NO:25); AA004399
(SEQ ID NO:26); AA312013 (SEQ ID NO:27); AA355824 (SEQ ID NO:28);
AA533619 (SEQ ID NO:29); AA361360 (SEQ ID NO:30); AA364876 (SEQ ID
NO:31); AA503090 (SEQ ID NO:32); AA533619 (SEQ ID NO:33); AA706672
(SEQ ID NO:34); AA774277 (SEQ ID NO:35); AA780277 (SEQ ID NO:36);
H03349 (SEQ ID NO:37); H04031 (SEQ ID NO:38); H53133 (SEQ ID NO:39);
H53239 (SEQ ID NO:40); H64669 (SEQ ID NO:41); N26002 (SEQ ID NO:42);
N52936 (SEQ ID NO:43); N88352 (SEQ ID NO:44); N89594 (SEQ ID NO:45);
R19795 (SEQ ID NO:46); R47511 (SEQ ID NO:47); T50235 (SEQ ID NO:48);
T78023 (SEQ ID NO:49); T78186 (SEQ ID NO:50); W22886 (SEQ ID NO:51);
W67657 (SEQ ID NO:52); W68094 (SEQ ID NO:53); W76111 (SEQ ID NO:54);
Z38299 (SEQ ID NO:55); Z42012 (SEQ ID NO:56); G06200(SEQ ID NO:74);
or any subfragment thereof; and
(b) a nucleotide sequence complementary to a nucleotide
sequence in (a).
In accordance with another aspect of the invention, there is provided an
isolated polypeptide molecule comprising an amino acid sequence selected from
the
group consisting of:
(a) amino acids from about 1 to about 908 in SEQ ID NO:5;
(b) amino acids from about 1 to about 859 in SEQ ID NO:6;
(c) amino acids from about 1 to about 912 in SEQ ID NO:7;
CA 02331781 2001-03-20
-3c-
(d) amino acids fronl about I to about 853 in SEQ ID NO:8;
and
(e) a polypeptide sequence at least about 90% identical to the
amino acid sequence of (a), (b), (c) or (d).
In accordance with another aspect of the invention, there is provided an
isolated polypeptide molecule, wherein except for at least one conservative
amino acid
substitution said polypeptide has a sequence selected from the group
consisting of:
(a) amino acids from about I to about 908 in SEQ ID NO:5;
(b) amino acids from about I to about 859 in SEQ ID NO:6;
(c) amino acids from about I to about 912 in SEQ ID NO:7;
(d) amino acids from about I to about 853 in SEQ ID NO:8;
and
(e) a polypeptide sequence at least about 90% identical to the
amino acid sequence of (a), (b), (c) or (d).
In accordance with another aspect of the invention, there is provided a method
for in vitro de novo methylation of DNA, comprising:
(a) contacting said DNA with an effective amount of a de novo
DNA cytosine methyltransferase polypeptide;
(b) providing an appropriately buffered solution with substrate
and cofactors; and
(c) purifying said DNA.
In accordance with another aspect of the invention, there is provided a method
for diagnosing or determining a susceptibility to neoplastic disorders,
comprising:
(a) assaying a de novo DNA cytosine methyltransferase
expression level in mammalian cells or body fluid; and
(b) comparing said de novo DNA cytosine methyltransferase
expression level with a standard de novo DNA cytosine methyltransferase
expression level whereby an increase or decrease in said de novo DNA cytosine
CA 02331781 2005-07-20
-3d-
methyltransferase expression level over said staridard is indicative of an
increased
or decreased susceptibility to a neoplastic disorder.
In accordance with another aspect of the invention, there is provided an
isolated de no vo DNA cytosine methyltransferasepolypeptide having the amino
acid
sequence encoded by the cDNA clone contained in ATCC Deposit No. 209933.
In accordance with another aspect of the invention, there is provided an
isolated de novo DNA cytosine methyltransferase polypeptide having the amino
acid
sequence encoded by the cDNA clone contained in ATCC Deposit No. 209934.
In accordance with another aspect of the invention, there is provided an
isolated de novo DNA cytosine methyltransferase polypeptide having the amino
acid
sequence encoded by the. cDNA clone contained in ATCC Deposit No. 98809.
In accordance with another aspect"of the invention, there is provided an
isolated de novo DNA cytosine methyltransferase polypeptide having the amino
acid
sequence encoded by the eDNA clone contained in ATCC Deposit No. 326637.
In accordance with another aspect of the invention, there is provided an
isolated de novo DNA cytosine methyltransferase Dnmt3b polypeptide wherein,
except for at least one conservative amino acid substitution, said polypeptide
has a
sequence selected from the group consisting of:
(a) aniino acid residues I to 362 and 383 to 859 from SEQ ID
NO:6; and
(b) amino acid residues I to 362 and 383 to 749 and 813 to
859 from SEQ ID NO:6
In accordance with another aspect of the invention, there is provided an
isolated de novo DNA cytosine methyltransferase D1TMT3B polypeptide wherein,
except for at least one conservative amino acid substitution, said polypeptide
has a
sequence selected from the group consisting of:
CA 02331781 2005-07-20
-3e-
(a) amino acid residues I to 355 and 376 to 853 from SEQ ID
NO:8; and
(b) amino acid residues 1 to 355 and 376 to 743 and 807 to
853 from SEQ ID N0:8
In accordance with another aspect of the invention, there is provided a method
of screening for an agonist or antagonist of DNMT3 DNA cytosine
methyltransferase
activity comprising:
(a) contacting a substrate to a DNMT3 DNA cytosine
methyltransferase protein or polypeptide in the presence of a putative agonist
or
antagonist; and
(b) assaying the activity of said agonist or said antagonist by
determining at least one of the following:
(i) binding of said agonist or said antagonist to said
DNMT3 DNA cytosine methyltransferase protein or polypeptide; and
(ii) determining the activity of said to said DNMT3
DNA cytosine methyltransferase protein or polypeptide in the presence of said
agonist or said antagonist.
A first aspect of the invention provides novel de novo DNA cytosine
methyltransferase nucleic acids and polypeptides that are not available in the
art.
A second aspect of the invention relates to de novo DNA cytosine
methyltransferase recombinant materials and methods for their production. A
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-4-
third aspect of the invention relates to the production of recombinant de novo
DNA cytosine methyltransferase polypeptides. A fourth aspect of the invention
relates to methods for using such de novo DNA cytosine methyltransferase
polypeptides and polynucleotides. Such uses include the treatment of
neoplastic
disorders, among others. Yet another aspect of the invention relates to
diagnostic
assays for the detection of diseases associated with inappropriate de novo DNA
cytosine methyltransferase activity or levels and mutations in de novo DNA
cytosine methyltransferases that might lead to neoplastic disorders.
Brief Description of the Figures
Figure lA-1D shows the nucleotide sequences of mouse Dnmt3a and
Dnmt3b and human DNMT3A and DNMT3B genes, respectively.
Figure 2A-2D shows the deduced amino acid sequence of mouse Dnmt3a
and Dnmt3b and human DNMT3A and DNMT3B genes, respectively. Sequences
are presented in single letter amino acid code.
Figure 3A shows a comparison of mouse Dnmt3a and Dnmt3b amino acid
sequences, and Figure 3B presents a comparison oi' the protein sequences of
human DNMT3A and DNMT3B1.
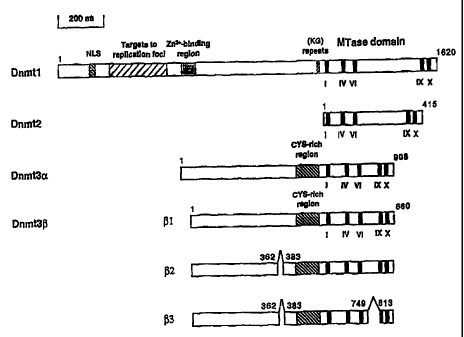
Figure 4A presents a schematic comparison of mouse Dnmtl, Dnmt2,
Dnmt3a and Dnmt3b protein structures. Figure 4B presents a schematic of the
DNMT3A, DNMT3B and zebrafish Zmt3 proteins. Figure 4C and 4D present a
schematic of the human DNMT3B gene organization and exon/intron junction
sequences.
Figure 5A presents a comparison of highly conserved protein structural
motifs for eukaryotic and prokaryotic C5 methyltransferase. Figure 5B presents
a sequence alignment of the C-rich domain of vertebrate DNMT3 proteins and the
X-lined ATRX gene. Figure 5C presents a non-rooted phylogenic tree of
methyltransferase proteins.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373 -
-5-
Figure 6A-6C demonstrates the expression of Dnmt3a and Dnmt3b in
mouse adult tissues, embryos, and ES cells by northern blot.
Figure 7A-7D demonstrates in vitro methyltransferase activities ofmouse
Dnmt3a and Dnmt3b proteins.
Figure 8 demonstrates in vitro analysis of de novo and maintenance
activities of Dnmt3a, Dnmt3bl and Dnmt3b2 proteins.
Figure 9 presents Northem blot expression analysis of DNMT3A and
DNMT3B.
Figure 10 presents DNMT3 Northern Blot expression analysis of
DNMT3A and DNMT3B in human tumor cell lines.
Detailed Description of the Preferred Embodiments
Definitions
In the description that follows, a number of terms used in recombinant
DNA technology are utilized extensively. In order to provide a clear and
consistent understanding of the specification and claims, including the scope
to be
given such terms, the following definitions are provided.
Cloning vector: A plasmid or phage DNA or other DNA sequence which
is able to replicate autonomously in a host cell, and which is characterized
by one
or a small number ofrestriction endonuclease recognition sites at which such
DNA
sequences may be cut in a determinable fashion without loss of an essential
biological function of the vector, and into which a DNA fragment may be
spliced
in order to bring about its replication and cloning. The cloning vector may
further
contain a marker suitable for use in the identification of cells transformed
with the
cloning vector. Markers, for example, provide tetracycline resistance or
ampicillin
resistance.
Expression vector: A vector similar to a cloning vector but which is
capable of enhancing the expression of a gene which has been cloned into it,
after
RECTIFIED SHEET (RULE 91)
ISA/EP
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-6-
transformation into a host. The cloned gene is usually placed under the
control
of (i.e., operably linked to) certain control sequences such as promoter
sequences.
Promoter sequences may be either constitutive or inducible.
Recombinant Host: According to the invention, a recombinant host may
be any prokaryotic or eukaryotic host cell which contains the desired cloned
genes
on an expression vector or cloning vector. This term is also meant to include
those prokaryotic or eukaryotic cells that have been genetically engineered to
contain the desired gene(s) in the chromosome or genome of that organism. For
examples of such hosts, see Sambrook et al., Molecular Cloning: A Laboratory
Manual, Second Edition, Cold Spring Harbor Laboratory, Cold Spring Harbor,
New York (1989). Preferred recombinant hosts are eukaryotic cells transformed
with the DNA construct ofthe invention. More specifically, mammalian cells are
preferred.
Recombinant vector: Any cloning vector or expression vector which
contains the desired cloned gene(s).
HostAnimal: Transgenic animals, all of whose germ and somatic cells
contain the DNA construct of the invention. Such transgenic animals are in
general vertebrates. Preferred Host Animals are mammals such as non-human
primates, mice, sheep, pigs, cattle, goats, guinea pigs, rodents, e.g. rats,
and the
like. The term Host Animal also includes animals in all stages of development,
including embryonic and fetal stages.
Promoter: A DNA sequence generally described as the 5' region of a
gene, located proximal to the start codon. The transcription of an adjacent
gene(s) is initiated at the promoter region. If a promoter is an inducible
promoter,
then the rate of transcription increases in response to an inducing agent. In
contrast, the rate of transcription is not regulated by an inducing agent if
the
promoter is a constitutive promoter. According to the invention, preferred
promoters are heterologous to the de novo DNA cytosine methyltransferase
genes,
that is, the promoters do not drive expression of the gene in a mouse or
human.
Such promoters include the CMV promoter (InVitrogen, San Diego, CA), the
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-7-
SV40, MMTV, and hMTlIapromoters (U.S. 5,457,034), the HSV-14/5 promoter
(U.S. 5,501,979), and the early intermediate HCMV promoter (W092/17581).
In one emdodiment, it is preferred that the promoter is tissue-specific, that
is, it
is induced selectively in a specific tissue. Also, tissue-specific enhancer
elements
may be employed. Additionally, such promoters may include tissue and cell-
specific promoters of an organism.
Gene: A DNA sequence that contains information needed for expressing
a polypeptide or protein.
Structural gene: A DNA sequence that is transcribed into messenger
RNA (mRNA) that is then translated into a sequence of amino acids
characteristic
of a specific polypeptide.
Complementary DNA (cDNA): A "complementary DNA," or "cDNA"
gene includes recombinant genes synthesized by reverse transcription of mRNA
and from which intervening sequences (introns) have been removed.
Expression: Expression is the process by which a polypeptide is
produced from a structural gene. The process involves transcription of the
gene
into mRNA and the translation of such mRNA into polypeptide(s).
Homologous/Nonhomologous: Two nucleic acid molecules are
considered to be "homologous" if their nucleotide sequences share a similarity
of
greater than 40%, as determined by HASH-coding algorithms (Wilber, W.J. and
Lipman, D.J., Proc. Natl. Acad. Sci. 80:726-730 (1983)). Two nucleic acid
molecules are considered to be "nonhomologous" if their nucleotide sequences
share a similarity of less than 40%.
Polynucleotide: This term generally refers to any polyribonucleotide or
polydeoxyribonucleotide, which may be unmodified RNA or DNA or modified
RNA or DNA. "Polynucleotides" include, without limitation single- and double-
stranded DNA, DNA that is a mixture of single- and double-stranded regions,
single- and double-stranded RNA, and RNA that is mixture of single- and double-
stranded regions, hybrid molecules comprising DNA and RNA that may be
single-stranded or, more typically, double-stranded or a mixture of single-
and
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-8-
double-stranded regions. In addition, "polynucleotide" refers to triple-
stranded
regions comprising RNA or DNA or both RNA and DNA. The term
polynucleotide also includes DNAs or RNAs containing one or more modified
bases and DNAs or RNAs with backbones modified for stability or for other
reasons. "Modified" bases include, for example, tritylated bases and unusual
bases such as inosine. A variety of modifications have been made to DNA and
RNA; thus, "polynucleotide" embraces chemically, enzymatically or
metabolically modified forms of polynucleotides as typically found in nature,
as
well as the chemical forms of DNA and RNA characteristic of viruses and cells.
"Polynucleotide" also embraces relatively short polynucleotides, often
referred to
as oligonucleotides.
Polypeptide: This term refers to any peptide or protein comprising two or
more amino acids joined to each other by peptide bonds or modified peptide
bonds, i.e., peptide isosteres. "Polypeptide" refers to both short chains,
commonly referred to as peptides, oligopeptides or oligomers, and to longer
chains, generally referred to as proteins. Polypeptides may contain amino
acids
other than the 20 gene-encoded amino acids. "Polypeptides" include amino acid
sequences modified either by natural processes, such as post-translational
processing, or by chemical modification techniques which are well known in the
art. Such modifications are well described in basic texts and in more detailed
monographs, as well as in a voluminous research literature. Modifications can
occur anywhere in a polypeptide, including the peptide backbone, the amino
acid
side-chains and the amino or carboxyl termini. It will be appreciated that the
same type of modification may be present in the same or varying degrees at
several sites in a given polypeptide. Also, a given polypeptide may contain
many
types of modifications. Polypeptides may be branched as a result of
ubiquitination, and they may be cyclic, with or without branching. Cyclic,
branched and branched cyclic polypeptides may result from post-translation
natural processes or may be made by synthetic methods. Modifications include
acetylation, acylation, ADP-ribosylation, amidation, covalent attachment of
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-9-
flavin, covalent attachment of a heme moiety, covalent attachment of a
nucleotide
or nucleotide derivative, covalent attachment of a lipid or lipid derivative,
covalent attachment of phosphotidylinositol, cross-linking, cyclization,
disulfide
bond formation, demethylation, formation of covalent. cross-links, formation
of
cystine, formation of pyroglutamate, formylatiori, gamma-carboxylation,
glycosylation, GPI anchor formation, hydroxylation, iodination, methylation,
myristoylation, oxidation, proteolytic processing, phosphorylation,
prenylation,
racemization, selenoylation, sulfation, transfer-RNA mediated addition of
amino
acids to proteins such as arginylation, and ubiquitination. See, for instance,
Proteins-Structure and Molecular Properties, 2nd Ecl., T. E. Creighton, W. H.
Freeman and Company, New York, 1993 and Wold, F.., Posttranslational Protein
Modifications: Perspectives and Prospects, pgs. 1-12 in Posttranslational
Covalent Modification of Proteins, B. C. Johnson, Ed., Academic Press, New
York, 1983; Seifter et al., "Analysis for protein modifications and nonprotein
cofactors", Methods in Enzymol. 182:626-646 (1990) and Rattan et al., "Protein
Synthesis: Posttranslational Modifications and Aging", Ann NYAcad Sci 663:48-
62 (1992).
Variant: The term used herein is a polynucleotide or polypeptide that
differs from a reference polynucleotide or polypeptide respectively, but
retains
essential properties. A typical variant of a polynucleotide differs in
nucleotide
sequence from another, reference polynucleotide. Changes in the nucleotide
sequence of the variant may or may not alter the amino acid sequence of a
polypeptide encoded by the reference polynucleotide. Nucleotide changes may
result in amino acid substitutions, additions, deletions, fusions and
truncations in
the polypeptide encoded by the reference sequence, as discussed below. A
typical
variant of a polypeptide differs in amino acid sequence from another,
reference
polypeptide. Generally, differences are limited so that the sequences of the
reference polypeptide and the variant are closely similar overall and, in many
regions, identical. A variant and reference polypeptide may differ in amino
acid
sequence by one or more substitutions, additions, deletions in any
combination.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-10-
A substituted or inserted amino acid residue may or may not be one encoded by
the genetic code. A variant of a polynucleotide or polypeptide may be a
naturally
occurring such as an allelic variant, or it may be a variant that is not known
to
occur naturally. Non-naturally occurring variants of polynucleotides and
polypeptides may be made by mutagenesis techniques or by direct synthesis.
Identity: This term refers to a measure of the identity of nucleotide
sequences or amino acid sequences. In general, the sequences are aligned so
that
the highest order match is obtained. "Identity" per- se has an art-recognized
meaning and can be calculated using published techniques. (See, e.g.:
Computational Molecular Biology, Lesk, A.M., ed., Oxford University Press,
New York, 1988; Biocomputing: Informatics and Genome Projects, Smith, D.W.,
ed., Academic Press, New York, 1993; Computer Analysis of Sequence Data,
Part 1, Griffin, A.M., and Griffin, H.G., eds., Humana Press, New Jersey,
1994;
Sequence Analysis in Molecular Biology, von Heinje, G., Academic Press, 1987;
and Sequence Analysis Primer, Gribskov, M. and Devereux, J., eds., M Stockton
Press, New York, 1991). While there exist a number of methods to measure
identity between two polynucleotide or polypeptide sequences, the term
"identity"
is well known to skilled artisans (Carillo, H. & Lipton, D., SIAMJApplied Math
48:1073 (1988)). Methods commonly employed to determine identity or
similarity between two sequences include, but are not limited to, those
disclosed
in Guide to Huge Computers, Martin J. Bishop, ed., Academic Press, San Diego,
1994, and Carillo, H. & Lipton, D., SIAM J Applied Math 48:1073 (1988).
Methods to determine identity and similarity are codified in computer
programs.
Preferred computer program methods to determine identity and similarity
between
two sequences include, but are not limited to, GCS program package (Devereux,
J., et al., NucleicAcids Research 12(1):387 (1984)), BLASTP, BLASTN, FASTA
(Atschul, S.F., et al., J Mol. Biol 215:403 (1990)).
Therefore, as used herein, the term "identity" represents a comparison
between a test and reference polynucleotide. More specifically, reference
polynucleotides are identified in this invention as SEQ ID Nos: 1, 2, 3 and 4,
and
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-11-
a test polynucleotide is defined as any polynucleotide that is 90% or more
identical to a reference polynucleotide. As used herein, the term "90% or
more"
refers to percent identities from 90 to 99.99 relative to the reference
polynucleotide. Identity at a level of 90% or more is indicative of the fact
that,
assuming for exemplification purposes a test and reference polynucleotide
length
of 100 nucleotides, that no more than 10% (i.e., 10 out of 100) nucleotides in
the
test polynucleotide differ from that of the reference polynucleotide. Such
differences may be represented as point mutations randomly distributed over
the
entire length of the sequence or they may be clustered in one or more
locations
of varying length up to the maximum allowable 10 nucleotide difference.
Differences are defined as nucleotide substitutions, deletions or additions of
sequence. These differences may be located at any position in the sequence,
including but not limited to the 5' end, 3' end, coding and non coding
sequences.
Fragment: A "fragment" of a molecule such as de novo DNA cytosine
methyltransferases is meant to refer to any polypeptide subset of that
molecule.
Functional Derivative: The term "functional derivatives" is intended to
include the "variants," "analogues," or "chemical derivatives" of the
molecule.
A "variant" of a molecule such as de novo DNA cytosine methyltransferases is
meant to refer to a naturally occurring molecule substantially similar to
either the
entire molecule, or a fragment thereof. An "analogue" of a molecule such as de
novo DNA cytosine methyltransferases is meant to refer to a non-natural
molecule
substantially similar to either the entire molecule or a fragment thereof.
A molecule is said to be "substantially similar" to another molecule if the
sequence of amino acids in both molecules is substantially the same, and if
both
molecules possess a similar biological activity. Thus, provided that two
molecules possess a similar activity, they are considered variants as that
term is
used herein even if one of the molecules contains additional amino acid
residues
not found in the other, or if the sequence of amino acid residues is not
identical.
As used herein, a molecule is said to be a "chemical derivative" of another
molecule when it contains additional chemical moieties not normally a part of
the
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-12-
molecule. Such moieties may improve the molecule's solubility, absorption,
biological half-life, etc. The moieties may alternatively decrease the
toxicity of
the molecule, eliminate or attenuate any undesirable side effect of the
molecule,
etc. Examples of moieties capable of mediating such effects are disclosed in
Remington's Pharmaceutical Sciences (1980) and will be apparent to those of
ordinary skill in the art.
Protein Activity or Biological Activity of the Protein: These expressions
refer to the metabolic or physiologic function of de novo DNA cytosine
methyltransferase protein including similar activities or improved activities
or
these activities with decreased undesirable side-effects. Also included are
antigenic and immunogenic activities of said de novo DNA cytosine
methyltransferase protein. Among the physiological or metabolic activities of
said protein is the transfer of a methyl group to the cytosine C5 position of
duplex
DNA. Such DNA may completely lack any methylation of may be
hemimethylated. As demonstrated in Example 8, de novo DNA cytosine
methyltransferases methylate C5 in cytosine moieties in nonmethylated DNA.
De novo DNA Cytosine Methyltransferases Polynucleotides: This term
refers to a polynucleotide containing a nucleotide sequence which encodes a de
novo DNA cytosine methyltransferase polypeptide or fragment thereof or that
encodes a de novo DNA cytosine methyltransferase polypeptide or fragment
wherein said nucleotide sequence has at least 90% identity to a nucleotide
sequence encoding the polypeptide of SEQ ID Nos: 5, 6, 7 or 8, or a
corresponding fragment thereof, or which has sufficient identity to a
nucleotide
sequence contained in SEQ ID NO: 1, 2, 3 or 4.
De novo DNA Cytosine Metltyltransferases Polypeptides: This term
refers to polypeptides with amino acid sequences sufficiently similar to the
de
novo DNA cytosine methyltransferase protein sequence in SEQ ID NO:5, 6, 7 or
8 and that at least one biological activity of the protein is exhibited.
Antibodies: As used herein includes polyclonal and monoclonal
antibodies, chimeric, single chain, and humanized antibodies, as well as Fab
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-13-
fragments, including the products of an Fab or other immunoglobulin expression
library.
Substantially pure: As used herein means that the desired purified
protein is essentially free from contaminating cellular components, said
components being associated with the desired protein in nature, as evidenced
by
a single band following polyacrylamide-sodium dodecyl sulfate gel
electrophoresis. Contaminating cellular components may include, but are not
limited to, proteinaceous, carbohydrate, or lipid impurities.
The term "substantially pure" is further meant to describe a molecule
which is homogeneous by one or more purity or homogeneity characteristics used
by those of skill in the art. For example, a substantially pure de novo DNA
cytosine methyltransferases will show constant and reproducible
characteristics
within standard experimental deviations for parameters such as the following:
molecular weight, chromatographic migration, amino acid composition, amino
acid sequence, blocked or unblocked N-terminus, HPLC elution profile,
biological activity, and other such parameters. The term, however, is not
meant
to exclude artificial or synthetic mixtures of the factor with other
compounds. In
addition, the term is not meant to exclude de novo DNA cytosine
methyltransferase fusion proteins isolated from a recombinant host.
Isolated: A term meaning altered "by the hand of man" from the natural
state. If an "isolated" composition or substance occurs in nature, it has been
changed or removed from its original environment, or both. For example, a
polynucleotide or a polypeptide naturally present in a living animal is not
"isolated," but the same polynucleotide or polypeptide separated from the
coexisting materials of its natural state is "isolated", as the term is
employed
herein. Thus, a polypeptide or polynucleotide produced and/or contained within
a recombinant host cell is considered isolated for purposes of the present
invention. Also intended as an "isolated polypeptide" or an "isolated
polynucleotide" are polypeptides or polynucleotides that have been purified,
partially or substantially, from a recombinant host cell or from a native
source.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-14-
For example, a recombinantly produced version of a de novo DNA cytosine
methyltransferase polypeptide can be substantially purified by the one-step
method described in Smith and Johnson, Gene 67:31-40 (1988).
Neoplastic disorder: This term refers to a disease state which is related
to the hyperproliferation of cells. Neoplastic disorders include, but are not
limited to, carcinomas, sarcomas and leukemias.
Gene Tlzerapy: A means of therapy directed to altering the normal pattern
of gene expression of an organism. Generally, a recombinant polynucleotide is
introduced into cells or tissues of the organism to effect a change in gene
expression.
Antisense RNA gene/Antisense RNA. In eukaryotes, mRNA is
transcribed by RNA polymerase II. However, it is also known that one may
construct a gene containing a RNA polymerase II template wherein a RNA
sequence is transcribed which has a sequence complementary to that of a
specific
mRNA but is not normally translated. Such a gene construct is herein termed an
"antisense RNA gene" and such a RNA transcript is termed an "antisense RNA."
Antisense RNAs are not normally translatable due to the presence of
translation
stop codons in the antisense RNA sequence.
Antisense oligonucleotide: A DNA or RNA molecule or a derivative
of a DNA or RNA molecule containing a nucleotide sequence which is
complementary to that of a specific mRNA. An antisense oligonucleotide binds
to the complementary sequence in a specific mRNA and inhibits translation of
the
mRNA. There are many known derivatives of such DNA and RNA molecules.
See, for example, U.S. Patent Nos. 5,602,240, 5,596,091, 5,506,212, 5,521,302,
5,541,307, 5,510,476, 5,514,787, 5,543,507, 5,512,438, 5,510,239, 5,514,577,
5,519,134, 5,554,746, 5,276,019, 5,286,717, 5,264,423, as well as W096/35706,
W096/32474, W096/29337 (thiono triester modified antisense
oligodeoxynucleotide phosphorothioates), W094/17093 (oligonucleotide
alkylphosphonates and alkylphosphothioates), W094/08004 (oligonucleotide
phosphothioates, methyl phosphates, phosphoramidates, dithioates, bridged
CA 02331781 2005-07-20
-15-
pliosphorothioates, bridge phosphoramidates, sulfones, sulfates, ketos,
phosphate
esters and phosphorobutylamines (van der Krol et al., Biotech. 6:958-976
(1988);
Uhlmann et aL, Chem. Rev. 90:542-585 (1990)), W094/02499 (oligonucleotide
alkylphosphonothioates and arylphosphonothioates), and W092/20697 (3'-end
capped oligonucleotides). Particular de novo DNA cytosine methyltransferase
antisense oligonucleotides ofthe present invention include derivatives such as
S-
oligonucleotides (phosphorothioate derivatives or S-oligos, see, Jack Cohen,
Oligodeoxynucleotides, Antisense Inhibitors of Gene Expression, CRC Press
(1989)). S-oligos (nucleoside phosphorothioates) are isoelectronic analogs of
an
oligonucleotide (0-oligo) in which a nonbridging oxygen atom of the phosphate
group is replaced by a sulfur atom. The S-oligos of the present invention may
be
prepared by treatment ofthe corresponding 0-oligos with 3h1-1,2-benzodithiol-3-
one-1,1-dioxide which is a sulfur transfer reagent. See Iyer et al., J. Org.
Chem.
55:4693-4698 (1990); and Iyer et al., J. Am. Chem. Soc. 112:1253-1254 (1990).
Antisense Tl:erapy: A method of treatment wherein. antisense
oligonucleotides are adniinistered to a patient in order to inhibit the
expression
of the corresponding protein.
L Deposited Material
The invention relates to polynucleotides encoding and polypeptides of
novel de novo DNA cytosine methyltransferase proteins. The invention relates
especially to de novo DNA cytosine methyltransferase mouse Dnmt3a and
Dnmt3b cDNAs and the human DNMT3A and DNMT3B cDNAs set out in SEQ
ID NOs: l. 2, 3 and 4, respectively. The invention also relates to mouse
Dnmt3a
and Dnint3b and human DNMT3A and DNMTB de novo DNA cytosine
methyltransferase polypeptides set out in SEQ ID NOs:5, 6, 7, and 8,
respectively:
The invention further relates to the de novo DNA cytosine methyltransferase -
nucleotide sequences ofthe,mouse Dnmt3a cDNA (plastnid pMT3a) and Dnmt3b
cDNA (plasmid pMT3b) and the human DNMTa cDNA (plasmid pMT3A) in
CA 02331781 2004-08-12
-16-
ATCC Deposit Nos.209933, 209934, and 98809, respectively, and the amino acid
sequences encoded therein. '
The nucleotide sequence of the human DNMT3B cDNA identified in SEQ
ID NO:4 is available in a clone (ATCC Deposit No. 326637) independently
deposited by the I.M.A.G.E. Consortium. The invention relates to the de novo
DNA cytosine methyltransferase polypeptide encoded therein.
Clones containing mouse Dnmt3a and Dnmt3b cDNAs were deposited
with the American Type Culture Collection (ATCC), Manassas, Virginia 20110-
2209, USA, on June 16, 1998, and assigned ATCC Deposit Nos. 209933 and
209934, respectively. The human DNMT3A cDNA was deposited witll the
ATCC on July 10, 1998, and assigned ATCC Deposit No. 98809.
While the ATCC deposits are believed to contain the de novo DNA
cytosine methyltransferase cDNA sequences shown in SEQ ID NOs: 1, 2, 3, and
4, the nucleotide sequences of the polynucleotide contained in the deposited
material, as well as the amino acid sequence of the polypeptide encoded
thereby,
are controlling in the event of any conflict with any description of sequences
herein.
The deposits for mouse Dnmt3a and Dnmt3b cDNAs and the human
DNMT3A cDNA were made under the terms of the Budapest Treaty on the
international recognition of the deposit ofmicro-organisms for purposes of
patent
procedure. The deposits are provided merely as a convenience for those of
skill
in the art and are not an admission that a deposit is required for enablement.
II. Polynucleotides of the Invention
Another aspect o,f the invention relates to isolated polynucleotides, and
polynucleotides closely related thereto, which encode the de novo DNA cytosine
methyltransferase polypeptides. As shown by the results presented in Figure 5,
sequencing of the cDNAs contained in the deposited clones encoding mouse and
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-17-
human de novo DNA cytosine methyltransferases confirms that the de novo DNA
cytosine methyltransferase proteins of the invention are structurally related
to
other proteins of the DNA methyltransferase family.
The polynucleotides of the present invention encoding de novo DNA
cytosine methyltransferase proteins may be obtained using standard cloning and
screening procedures as described in Example 1. Polynucleotides ofthe
invention
can also be obtained from natural sources such as genomic DNA libraries or can
be synthesized using well known and commercially available techniques.
Among particularly preferred embodiments of the invention are
polynucleotides encoding de novo DNA cytosine methyltransferase polypeptides
having the amino acid sequence set out in SEQ ID NO:5, SEQ ID NO:6, SEQ ID
NO:7, or SEQ ID NO:8, and variants thereof.
A particular nucleotide sequence encoding a de novo DNA cytosine
methyltransferase polypeptide may be identical over its entire length to the
coding
sequence in SEQ ID NOs:l, 2, or 3. Alternatively, a particular nucleotide
sequence encoding a de novo DNA cytosine methyltransferase polypeptide may
be an alternate form of SEQ ID NOs: 1, 2, 3 and 4 due to degeneracy in the
genetic code or variation in codon usage encoding the polypeptides of SEQ ID
NOs:5, 6, 7. or 8. Preferably, the polynucleotides of the invention contain a
nucleotide sequence that is highly identical, at least 90% identical, with a
nucleotide sequence encoding a de novo DNA cytosine methyltransferase
polypeptide or at least 90% identical with the encoding nucleotide sequence
set
forth in SEQ ID NOs:l, 2, or 3. Polynucleotides of the invention may be 90 to
99% identical to the nucleotides sequence set forth in SEQ ID NO:4.
When a polynucleotide of the invention is used for the recombinant
production of a de novo DNA cytosine methyltransferase polypeptide, the
polynucleotide may include the coding sequence for the full-length polypeptide
or a fragment thereof, by itself; the coding sequence for the full-length
polypeptide or fragment in reading frame with other coding sequences, such as
those encoding a leader or secretory sequence, a pre-, or pro or prepro-
protein
CA 02331781 2005-07-20
-18-
sequence, or other fusion peptide portions. For example, a marker sequence
that
facilitates purification of the fused polypeptide can be encoded. In certain
preferred embodiments of this aspect of the invention, the marker sequence is
a
hexa-histidine peptide, as provided in the pQE vector (Qiagen, Inc.) and
described
in Gentz et al., Proc Natl Acad Sci USA 86:821-824 (1989), or it may be the HA
tag, which corresponds to an epitope derived from the influenza hemagglutinin
protein (Wilson, I., et al., Cell 37:767, 1984). The polynucleotide may also
contain non-coding 5' and 3' sequences, such as transcribed, non-translated.
sequences, splicing and polyadenylation signals, ribosome binding sites and
sequences that stabilize mRNA.
Embodiments of the invention include isolated nucleic acid molecules
comprising a polynucleotide having a nucleotide sequence at least 90%
identical,
and more preferably at least 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99%
identical to (a) a nucleotide sequence encoding a de novo DNA cytosine
methyltransferase polypeptide having the amino acid sequence in SEQ ID NO:5,
SEQ ID NO:6, or SEQ ID NO:7; (b) a nucleotide sequence encoding a de novo
DNA cytosine methyltransferase polypeptide having the amino acid sequence
encoded by the cDNA clone contained in ATCC Deposit No. 209933, ATCC
Deposit No. 209934, or ATCC Deposit No. 98809; or (c) a nucleotide sequence
complementary to any of the nucleotide sequences in (a) or (b). Additionally,
an
isolated nucleic acid of the invention may be a polynucleotide at least 90%
but
not more than 99% identical to (a) a nucleotide sequence encoding a de novo
DNA cytosine methyltransferase polypeptide having the amino acid sequence in
SEQ ID NO:4; (b) a nucleotide sequence encoding a de novo DNA cytosine
methyltransferase polypeptide having the amino acid sequence encoded by the
cDNA clone contained in ATCC Deposit No.326637; or (c) a nucleotide
sequence complementary to any of the nucleotide sequences in (a) or (b).
Conventional means utilizing known computer programs such as the
BestFit program (Wisconsin Sequence Analysis Package, Version 10 for Unix,
Genetics Computer Group, University Research Park, 575 Science Drive,
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-19-
Madison, WI 53711) may be utilized to determine if a particular nucleic acid
molecule is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99%
identical to any one of the nucleotide sequences shown in SEQ ID NO: 1, SEQ ID
NO:2, SEQ ID NO:3, or SEQ ID NO:4 or to any one of the nucleotide sequences
of the deposited cDNA clones contained in ATCC Deposit No. 209933, ATCC
Deposit No. 209934, ATCC Deposit No. 98809, or ATCC Deposit No. 326637.
Further preferred embodiments are polynucleotides encoding de novo
DNA cytosine methyltransferases and de novo DNA cytosine methyltransferase
variants that have an amino acid sequence of the de novo DNA cytosine
methyltransferase protein of SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:7, or SEQ
ID NO:8 in which several, 1, 1-2, 1-3, 1-5 or 5-10 amino acid residues are
substituted, deleted or added, in any combination.
Further preferred embodiments of the invention are polynucleotides that
are at least 90% identical over their entire length to a polynucleotide
encoding a
de novo DNA cytosine methyltransferase polypeptide having the amino acid
sequence set out in SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:7, or SEQ ID
NO:8, and polynucleotides which are complementary to such polynucleotides.
Most highly preferred are polynucleotides that comprise regions that are at
least
90% identical over their entire length to a polynucleotide encoding the de
novo
DNA cytosine methyltransferase polypeptides of the ATCC deposited human
DNMT3A cDNA clone and polynucleotides complementary thereto, and 90% to
99% identical over their entire length to a polynucleotide encoding the de
novo
DNA cytosine methyltransferase polypeptides of the ATCC deposited human
DNMT3B cDNA clone and polynucleotides complementary thereto. In this
regard, polynucleotides at least 95% identical over their entire length to the
same
are particularly preferred, and those with at least 97% identity are
especially
preferred. Furthermore, those with at least 98% identity are highly preferred
and
with at least 99% identity being the most preferred.
In a more specific embodiment, the nucleic acid molecules of the present
invention, e.g., isolated nucleic acids comprising a polynucleotide having a
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-20-
nucleotide sequence encoding a de novo DNA cytosine methyltransferase
polypeptide or fragment thereof, are not the sequence of nucleotides, the
nucleic
acid molecules (e.g., clones), or the nucleic acid inserts identified in one
or more
of the below cited public EST or STS GenBank Accession Reports.
The following public ESTs were identified that relate to portions of SEQ
ID NO:1: AA052791(SEQ ID NO:9); AA111043(SEQ ID NO:10);
AA154890(SEQ ID NO:11); AA240794(SEQ ID NO:12); AA756653(SEQ ID
NO:13); W58898(SEQ ID NO:14); W59299(SEQ ID NO:15); W91664(SEQ ID
NO:16); W91665(SEQ ID NO: 17); to portions of SEQ IDNO:2: AA116694 (SEQ
ID NO:18); AA119979 (SEQ ID NO:19); AA177277 (SEQ ID NO:20);
AA210568 (SEQ ID NO:21); AA399749 (SEQ ID NO:22); AA407106 (SEQ ID
NO:23); AA575617 (SEQ ID NO:24); to portions of SEQ ID NO:3: AA004310
(SEQ ID NO:25); AA004399 (SEQ ID NO:26); AA312013 (SEQ ID NO:27);
AA355824 (SEQ ID NO:28); AA533619 (SEQ ID NO:29); AA361360 (SEQ ID
NO:30); AA364876 (SEQ ID NO:3 1); AA503090 (SEQ ID NO:32); AA533619
(SEQ ID NO:33); AA706672 (SEQ ID NO:34); AA774277 (SEQ ID NO:35);
AA780277 (SEQ ID NO:36); H03349 (SEQ ID NO:37); H04031 (SEQ ID
NO:38); H53133 (SEQ ID NO:39); H53239 (SEQ ID NO:40); H64669 (SEQ ID
NO:41); N26002 (SEQ ID NO:42); N52936 (SEQ ID NO:43); N88352 (SEQ ID
NO:44); N89594 (SEQ ID NO:45); R19795 (SEQ ID NO:46); R47511 (SEQ ID
NO:47); T50235 (SEQ ID NO:48); T78023 (SEQ ID NO:49); T78186 (SEQ ID
NO:50); W22886 (SEQ ID NO:5 1); W67657 (SEQ ID NO:52); W68094 (SEQ ID
NO:53); W76111 (SEQ ID NO:54); Z38299 (SEQ ID NO:55); Z42012 (SEQ ID
NO:56); and that relate to SEQ ID NO:4: AA206103(SEQ ID NO:57);
AA206264(SEQ ID NO:58); AA216527(SEQ ID NO:59); AA216697(SEQ ID
NO:60); AA305044(SEQ ID NO:61); AA477705(SEQ ID NO:62);
AA477706(SEQ ID NO:63); AA565566(SEQ ID NO:64); AA599893(SEQ ID
NO:65); AA729418(SEQ ID NO:66); AA887508(SEQ ID NO:67); F09856(SEQ
ID NO:68); F12227(SEQ ID NO:69); N39452(SEQ ID NO:70); N48564(SEQ ID
NO:71); T66304(SEQ ID NO:72); and T66356(SEQ ID NO:73); AA736582(SEQ
RECTIFIED SHEET (RULE 91)
ISA/EP
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
21-
ID NO:77); AA748883(SEQ ID NO:78); AA923295(SEQ ID NO:79);
AAI000396(SEQ ID NO:80); A1332472(SEQ ID NO:81); W22473(SEQ ID
NO:82) and the I.M.A.G.E. Consortium clone ID 22089 (ATCC Deposit No.
326637)(SEQ ID NO:76). Additionally, STSs G06200(SEQ ID NO:74) and
G15302(SEQ ID NO:75) were identified in a search with SEQ ID NOS.:3 and 4,
respectively.
The present invention is further directed to fragments of SEQ ID NO: 1, 2
or 3, or to fragrnents of the cDNA nucleotide sequence found in ATCC Deposit
Nos. 209933, 209934, or 98809. A fragment may be defined to be at least about
15 nt, and more preferably at least about 20 nt, still more preferably at
least about
30 nt, and even more preferably, at least about 40 nt in length. Such
fragments are
useful as diagnostic probes and primers as discussed herein. Ofcourse larger
DNA
fragments are also useful according to the present invention, as are fragments
corresponding to most, if not all, of the nucleotide sequence of the cDNA
clones
contained in the plasmids deposited as ATCC DepositNo. 209933, ATCC Deposit
No. 209934 or ATCC Deposit No. 98809,or as shown in SEQ ID NO:1, SEQ ID
NO:2, or SEQ ID NO:3. Generally, polynucleotide fragments of the invention may
be defined algebraically in the following way: (a) for SEQ ID NO:1, as 15 + N,
wherein N equals zero or any positive integer up to 4176; (b) for SEQ ID NO:2,
as 15 + N, wherein N equals zero or any positive integer up to 4180; and (c)
for
SEQ ID NO:3, as 15 + N, wherein N equals zero or any positive integer up to
4401. By a fragment at least 20 nt in length, for example, is intended
fragments
which include 20 or more contiguous bases from a nucleotide sequence of the
ATCC deposited cDNAs or the nucleotide sequence as shown in SEQ ID NO:1,
SEQ ID NO:2 or SEQ ID NO:3.
In another embodiment, the invention is directed to fragments of SEQ ID
NO:4. Such fragments are defined as comprising the nucleotide sequence
encoding the specific amino acid residues integral and immediately adj acent
to the
site where DNMT3B exons are spliced together. The DNMT3B sequence of
SEQ ID NO:4 consists of 23 exon sequences defined accordingly: Exon 1 consists
RECTIFIED SHEET (RULE 91)
ISA/EP
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-22-
of nucleotides 1-108 of SEQ ID NO:4; Exon 2 consists of nucleotides 109-256
of SEQ ID NO:4; Exon 3 consists of nucleotides 257-318 of SEQ ID NO:4;
Exon 4 consists of nucleotides 319-420 of SEQ ID NO:4; Exon 5 consists of
nucleotides 421-546 of SEQ ID NO:4; Exon 6 consists of nucleotides 547-768 of
SEQ ID NO:4; Exon 7 consists of nucleotides 769-927 of SEQ ID NO:4; Exon
8 consists of nucleotides 928-1035 of SEQ ID NO:4; Exon 9 consists of
nucleotides 1036-1180 of SEQ ID NO:4; Exon 10 consists of nucleotides 1181-
1240 of SEQ ID NO:4; Exon I 1 consists of nucleotides 1241-1366 of SEQ ID
NO:4; Exon 12 consists of nucleotides 1367-1411 of' SEQ ID NO:4; Exon 13
consists of nucleotide 1412-1491 of SEQ ID NO:4; Exon 14 consists of
nucleotides 1492-1604 of SEQ ID NO:4; Exon 15 consists of nucleotides 1605-
1788 of SEQ ID NO:4; Exon 16 consists of nucleotides 1789-1873 of SEQ ID
NO:4; Exon 17 consists of nucleotides 1874-2019 of SEQ ID NO:4; Exon 18
consists of nucleotides 2020-2110 of SEQ ID NO:4; Exon 19 consists of
nucleotides 2111-2259 of SEQ ID NO:4; Exon 20 consists of nucleotides 2260-
2345 of SEQ ID NO:4; Exon 21 consists of nucleotides 2346-2415 of SEQ ID
NO:4; Exon 22 consists of nucleotides 2416-2534 of SEQ ID NO:4; and Exon 23
consists of nucleotides 2535-4145 of SEQ ID NO:4.
It should be understood by those skilled in the art that with regards to SEQ
ID NO:4, Exon I and Exon 23 are herein defined for the purposes of the
invention. The first nucleotide of Exon 1 may or may not be the
transcriptional
start site for the DNMT3B genomic locus, and the last nucleotide identified
for
Exon 23 may or may not reflect the last nucleotide transcribed in vivo.
Thus, by way of example, fragments of SEQ ID NO:4 comprise the
following exon-exon junctions of 20 nucleotides in length: the exonl/exon 2
junction of nucleotides 98-118 of SEQ ID NO:4; the exon 2/exon 3 junction of
nucleotides 246-266 of SEQ ID NO:4; the exon 3/exon 4 junction of nucleotides
308-328 of SEQ ID NO:4; the exon 4/exon 5 junction of nucleotides 410-430 of
SEQ ID NO:4; the exon 5/exon 6 junction of nucleotides 536-556 of SEQ ID
NO:4; the exon 6/exon 7 junction of nucleotides 758-778 of SEQ ID NO:4; the
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-23-
exon 7/exon 8 junction of nucleotides 917-937 of SEQ ID NO:4; the exon 8/exon
9 junction of nucleotides 1025-1045 of SEQ ID NO:4; the exon 9/exon 10
junction of nucleotides 1170-1190 of SEQ ID NO:4; the exon 10/exon 11
junction of nucleotides 1230-1250 of SEQ ID NO:4; the exon 11/exon 12
junction of nucleotides 1356-1376 of SEQ ID NO:4; the exon 12/exon 13
junction of nucleotides 1401-1421 of SEQ ID NO:4; the exon 13/exon 14
junction of nucleotides 1481-1501 of SEQ ID NO:4; the exon 14/exon 15
junction of nucleotides 1594-1614 of SEQ ID NO:4; the exon 15/exon 16
junction of nucleotides 1778-1798 of SEQ ID NO:4; the exon 16/exon 17
junction of nucleotides 1863-1883 of SEQ ID NO:4; the exon 17/exon 18
junction of nucleotides 2009-2029 of SEQ ID NO:4; the exon 18/exon 19
junction of nucleotides 2100-2120 of SEQ ID NO:4; the exon 19/exon 20
junction of nucleotides 2249-2269 of SEQ ID NO:4; the exon 20/exon 21
junction of nucleotides 2335-2355 of SEQ ID NO:4; the exon 21/exon 22
junction of nucleotides 2405-2425 of SEQ ID NO:4; and the exon 22/exon 23
junction of nucleotides 2524-2544 of SEQ ID NO:4.
As will be clear to those skilled in the art, other exon-exon junction
fragments of SEQ ID NO:4 are possible which comprise 30, 40, 50, 60, 70, 80,
90, 100, 200, 300, 400, 500, etc., nucleotides of SEQ II) NO:4. For the
purposes
of constructing such fragments, the following exon-exonjunctions are
identified:
the exon 1/exon 2 junction of nucleotides 108 and 109 of SEQ ID NO:4; the exon
2/exon 3 junction of nucleotides 256 and 257 of SEQ ID NO:4; the exon 3/exon
4 junction of nucleotides 318 and 319 of SEQ ID NO:4; the exon 4/exon 5
junction of nucleotides 420 and 421 of SEQ ID NO:4; the exon 5/exon 6 junction
of nucleotides 546 and 547 of SEQ ID NO:4; the exon 6/exon 7 junction of
nucleotides 768 and 769 of SEQ ID NO:4; the exon 7/exon 8 junction of
nucleotides 927 and 928 of SEQ ID NO:4; the exon 8/exon 9 junction of
nucleotides 1035 and 1036 of SEQ ID NO:4; the exon 9/exon 10 junction of
nucleotides 1180 and 1181 of SEQ ID NO:4; the exon 10/exon 11 junction of
nucleotides 1240 and 1241 of SEQ ID NO:4; the exon 11 /exon 12 junction of
CA 02331781 2004-08-12
-24-
nucleotides 1366 and 1367 of SEQ ID NO:4; the exon 12/exon 13 junction of
nucleotides 1411 and 1412 of SEQ ID NO:4; the exon 13/exon 14 junction of
nucleotides 1491 and 1492 of SEQ ID NO:4; the exon 14/exon 15 junction of
nucleotides 1604 and 1605 of SEQ ID NO:4; the exon 15/exon 16 junction of
nucleotides 1788 and 1789 of SEQ ID NO:4; the exon 16/exon 17 junction of
nucleotides 1873 and 1874 of SEQ ID NO:4; the exon 17/exon 18 junction of
nucleotides 2019 and 2020 of SEQ ID NO:4; the exon 18/exon 19 junction of
nucleotides 2110 and 2111 of SEQ ID NO:4; the exon 19/exon 20 junction of
nucleotides 2259 and 2260 of SEQ ID NO:4; the exon 20/exon 21 junction of
nucleotides 2345 and 2346 of SEQ ID NO:4; the exon 21/exon 22 junction of
nucleotides 2415 and 2416 of SEQ ID NO:4; and the exon 22/exon 23 junction
of nucleotides 2534 and 2535 of SEQ ID NO:4. Junction nucleotides may be
located at any position of the selected SEQ ID NO:4 fragment.
The present invention further relates to polynucleotides that hybridize to
the above-described sequences. In this regard, the present invention
especially
relates to polynucleotides that hybridize under stringent conditions to the
above-
described polynucleotides. As herein used, the term "stringent conditions"
means
hybridization will occur only if there is at least 90% and preferably at least
95%
identity and more preferably at least 97% identity between the sequences.
Furthermore, a major consideration associated with hybridization analysis
of DNA or RNA sequences is the degree of relatedness the probe has with the
sequences present in the specimen under study. This is important with a
blotting
technique (e.g., Southern or Northern Blot), since a moderate degree of
sequence
homology under nonstringent conditions of hybridization can yield a strong
signal
even though the probe and sequences in the sample represent non-homologous
genes.
The particular hybridization technique is not essential to the invention,
any technique commonly used in the art is within the scope of the present
invention. Typical probe technology is described in United States Patent
4,358,535 to Falkow et al. e For example,
CA 02331781 2004-08-12
-25-
hybridization can be carried out in a solution containing 6 x SSC (10 x SSC:
1.5
M sodium chloride, 0.15 M sodium citrate, pH 7.0). 5 x Denhardt's (1 x
Denhardt's: 0.2% bovine serum albumin, 0.2% polyvinylpyrrolidone, 0.02%
Ficoll 400), 10 mM EDTA, 0.5% SDS and about l 0' cpm of nick-translated DNA
for 16 hours at 65 C. Additionally, if hybridization is to an immobilized
nucleic
acid, a washing step may be utilized wherein probe binding to polynucleotides
of
low homology, or nonspecific binding of the probe, may be removed. For
example, a stringent wash step may invol ve a buffer of 0.2 x SSC and 0.5% SDS
at a temperature of 65 C.
Additional information related to hybridization technology and, more
particularly, the stringency ofhybridization and washing conditions may be
found
in Sambrook et al., Molecular Cloning: A Laboratory Manual, Second Edition,
Cold Spring Harbor Laboratory, Cold Spring Harbor, New York (1989),
Polynucleotides of the invention which are sufficiently identical to a
nucleotide sequences contained in SEQ ID NO: 1, SEQ ID NO:2, SEQ ID NO:3
or SEQ ID NO:4, or in the cDNA inserts of ATCC Deposit No. 209933, ATCC
Deposit No. 209934, ATCC Deposit No. 98809 or ATCC Deposit No. 326637,
may be used as hybridization probes for cDNA and genomic DNA, to isolate full-
length cDNAs and genomic clones encoding de novo DNA cytosine
methyltransferase proteins and to isolate cDNA and genomic clones of other
genes that have a high sequence similarity to the de novo DNA cytosine
methyltransferase genes. Such hybridization techniques are known to those of
skill in the art. Typically, these nucleotide sequences are at least about 90%
identical, preferably at least about 95% identical, more preferably at least
about
97%, 98% or 99% identical to that of the reference. The probes generally will
comprise at least 15 nucleotides. Preferably, such probes will have at least
30
nucleotides and may have at least 50 nucleotides. Particularly preferred
probes
will range between 30 and 50 nucleotides.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373 -
-26-
The polynucleotides and polypeptides of the present invention may be
employed as research reagents and materials for discovery of treatments and
diagnostics to animal and human disease.
III. Vectors, Host Cells, and Recombinant Expression
The present invention also relates to vectors that comprise a
polynucleotide of the present invention, host cells which are genetically
engineered with vectors of the invention and the production of polypeptides of
the
invention by recombinant techniques. Cell-free translation systems can also be
employed to produce such proteins using RNAs derived from the DNA constructs
of the invention.
For recombinant production, host cells can be genetically engineered to
incorporate expression systems for polynucleotides of the invention.
Introduction
of polynucleotides into host cells can be effected by methods described in
many
standard laboratory manuals, such as Sambrook et al., Molecular Cloning:
A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold
Spring Harbor, N.Y. (1989). For example, calcium phosphate transfection,
DEAE-dextran mediated transfection, transvection, microinjection, cationic
lipid-
mediated transfection, electroporation, transduction, scrape loading,
ballistic
introduction, infection or any other means known in the art may be utilized.
Representative examples of appropriate hosts include bacterial cells, such
as streptococci, staphylococci, E. coli, Streptomyces and Bacillus subtilis
cells;
fungal cells, such as yeast cells and Aspergillus cells; inse_L cells such as
Drosophila S2 and Spodoptera Sf9 cells; animal cells such as CHO, COS, HeLa,
C127, 3T3, BHK, 293 and Bowes melanoma cells; and plant cells.
A great variety of expression systems can be used. Such systems include,
among others, chromosomal, episomal and virus-derived systems, e.g., vectors
derived from bacterial plasmids, from bacteriophages, from transposons, from
yeast episomes, from insertion elements, from yeast chromosomal elements, from
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-27-
viruses such as baculoviruses, papova viruses, such as SV40, vaccinia viruses,
adenoviruses, fowl pox viruses, pseudorabies viruses, and retroviruses, and
vectors derived from combinations thereof, such as those derived from plasmid
and bacteriophage genetic elements, such as cosmids and phagemids. The
expression systems may contain control regions that regulate as well as
engender
expression. Generally, any system or vector suitable to maintain, propagate or
express polynucleotides to produce a polypeptide in a host may be used. The
appropriate nucleotide sequence may be inserted into an expression system by
any
of a variety of well-known and routine techniques, such as, for example, those
set
forth in Sambrook et al., Molecular Cloning: A Laboratory Manual (supra).
RNA vectors may also be utilized for the expression of the de novo DNA
cytosine methyltransferases disclosed in this invention. These vectors are
based
on positive or negative strand RNA viruses that naturally replicate in a wide
variety of eukaryotic cells (Bredenbeek, P.J. and Rice, C.M., Virology 3: 297-
310, (1992)). Unlike retroviruses, these viruses lack an intermediate DNA life-
cycle phase, existing entirely in RNA form. For example, alpha viruses are
used
as expression vectors for foreign proteins because they can be utilized in a
broad
range of host cells and provide a high level of expression; examples of
viruses of
this type include the Sindbis virus and Semliki Forest virus (Schlesinger, S.,
TIBTECH 11: 18-22, (1993); Frolov, I., et al., Proc. Natl. Acad. Sci. (USA)
93:
11371-11377, (1996)). As exemplified by Invitrogen's Sinbis expression system,
the investigator may conveniently maintain the recombinant molecule in DNA
form (pSinrep5 plasmid) in the laboratory, but propagation in RNA form is
feasible as well. In the host cell used for expression, the vector containing
the
gene of interest exists completely in RNA form and may be continuously
propagated in that state if desired.
For secretion of the translated protein into the lumen of the endoplasmic
reticulum, into the periplasmic space or into the extracellular environment
appropriate secretion signals may be incorporated into the desired
polypeptide.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373 -
-28-
These signals may be endogenous to the polypeptide or they may be heterologous
signals.
As used herein, the term "operably linked," when used in the context of
a linkage between a structural gene and an expression control sequence, e.g.,
a
promoter, refers to the position and orientation of the expression control
sequence
relative to the structural gene so as to permit expression of the structural
gene in
any host cell. For example, an operable linkage would maintain proper reading
frame and would not introduce any in frame stop codons.
As used herein, the term "heterologous promoter," refers to a promoter not
normally and naturally associated with the structural gene to be expressed.
For
example, in the context of expression of a de novo DNA cytosine
methyltransferase polypeptide, a heterologous promoter would be any promoter
other than an endogenous promoter associated with the de novo DNA cytosine
methyltransferase gene in non-recombinant mouse or human chromosomes. In
specific embodiments of this invention, the heterologous promoter is a
prokaryotic or bacteriophage promoter, such as the lac promoter, T3 promoter,
or
T7 promoter. In other embodiments, the heterologous promoter is a eukaryotic
promoter.
In other embodiments, this invention provides an isolated nucleic acid
molecule comprising a de novo DNA cytosine methyltransferase structural gene
operably linked to a heterologous promoter. As used herein, the term "a de
novo
DNA cytosine methyltransferase structural gene" refers to a nucleotide
sequence
at least about 90% identical to one of the following nucleotide sequences:
(a) a nucleotide sequence encoding the de novo DNA cytosine
methyltransferase polypeptide having the complete amino acid sequence in SEQ
ID N0:5, SEQ ID NO:6, or SEQ ID NO:7;
(b) a nucleotide sequence encoding the de novo DNA cytosine
methyltransferase polypeptide having the complete amino acid sequence encoded
by the cDNA insert of ATCC Deposit No. 209933, ATCC Deposit No. 209934,
or ATCC Deposit No. 98809; or
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373-
-29-
(c) a nucleotide sequence complementary to any of the nucleotide
sequences in (a) or (b).
In preferred embodiments, the de novo DNA cytosine methyltransferase
structural gene is 90%, and more preferably 91%, 92%, 93%, 94%, 95%, 97%,
98%, 99%, or 100% identical to one or more of nucleotide sequences (a), (b),
or
(c) supra.
In another embodiment the term "a de novo DNA cytosine
methyltransferase structural gene" refers to a nucleotide sequence about 90%
to
99% identical to one of the following nucleotide sequences:
(a) a nucleotide sequence encoding the de novo DNA cytosine
methyltransferase polypeptide having the complete amino acid sequence in SEQ
ID NO:8;
(b) a nucleotide sequence encoding the de novo DNA cytosine
methyltransferase polypeptide having the complete amino acid sequence encoded
by the cDNA insert of ATCC Deposit No. 326637; or
(c) a nucleotide sequence complementary to any of the nucleotide
sequences in (a) or (b).
In preferred embodiments, the de novo DNA cytosine methyltransferase
structural gene is 90%, and more preferably 91%, 92%, 93%, 94%, 95%, 97%,
98%, or 99% identical to SEQ ID NO:8, ATCC Deposit No. 326637 or
polynucleotides complementary thereto.
This invention also provides an isolated nucleic acid molecule comprising
a de novo DNA cytosine methyltransferase structural gene operably linked to a
heterologous promoter, wherein said isolated nucleic acid molecule does not
encode a fusion protein comprising the de novo DNA cytosine methyltransferase
structural gene or a fragment thereof.
This invention further provides an isolated nucleic acid molecule
comprising a de novo DNA cytosine methyltransferase structural gene operably
linked to a heterologous promoter, wherein said isolated nucleic acid molecule
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-30-
is capable of expressing a de novo DNA cytosine methyltransferase polypeptide
when used to transform an appropriate host cell.
This invention also provides an isolated nucleic acid molecule comprising
a polynucleotide having a nucleotide sequence at least 90%, 91%, 92%, 93%,
94%, 95%, 96%, 97%, 98%, 99%, or 100% identical to a sequence encoding a de
novo DNA cytosine methyltransferase polypeptide having the amino acid
sequence of SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:7 or SEQ ID NO:8,
wherein said isolated nucleic acid molecule does not contain a nucleotide
sequence at least 90% identical to the 3' untranslated region of SEQ ID NO:1
(nucleotides 2942-4191), SEQ ID NO:2 (nucleotides 2847-4174), SEQ ID NO:3
(nucleotides 3090-4397) or SEQ ID NO:4 (nucleotides 2677-4127), or a fragment
of the 3' untranslated region greater than 25, 50, 75, 100, or 125 bp in
length.
This invention further provides an isolated nucleic acid molecule
comprising a polynucleotide having a nucleotide sequence at least 90%, 91%.
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% identical to a sequence
encoding a de novo DNA cytosine methyltransferase polypeptide having the
amino acid sequence of SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:7 or SEQ ID
NO:8, wherein said isolated nucleic acid molecule does not contain a
nucleotide
sequence at least 90% identical to the 5' untranslated region of SEQ ID NO:1
(nucleotides 1-216), SEQ ID NO:2 (nucleotides 1-268), SEQ ID NO:3
(nucleotides 1-352) or SEQ ID NO:4 (nucleotides 1-114), or a fragment of the
5'
untranslated region greater than 25, 35, 45, 55, 65, 75, 85, or 90 bp.
Suitable known prokaryotic promoters for use in the production of
proteins of the present invention include the E. coli lacl and lacZ promoters,
the
T3 and T7 promoters, the gpt promoter, the lambda PR and PL promoters and the
trp promoter. Suitable eukaryotic promoters include the CMV immediate earl}r
promoter, the HSV thymidine kinase promoter, the early and late SV40
promoters, the promoters of retroviral LTRs, such as those of the Rous Sarcoma
Virus (RSV), adenovirus promoter, Herpes virus promoter, and metallothionein
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373 -
-31-
promoters, such as the mouse metallothionein-I promoter and tissue and organ-
specific promoters known in the art.
If the de novo DNA cytosine methyltransferase polypeptide is to be
expressed for use in screening assays, generally, it is preferred that the
polypeptide be produced at the surface of the cell. In this event, the cells
may be
harvested prior to use in the screening assay. If de novo DNA cytosine
methyltransferase polypeptide is secreted into the medium, the medium can be
recovered in order to recover and purify the polypeptide; if produced
intracellularly, the cells must first be lysed before the polypeptide is
recovered.
De novo DNA cytosine methyltransferase polypeptides can be recovered
and purified from recombinant cell cultures by well-known methods including
ammonium sulfate or ethanol precipitation, acid extraction, anion or cation
exchange chromatography, phosphocellulose chromatography, hydrophobic
interaction chromatography, affinity chromatography, hydroxylapatite
chromatography and lectin chromatography. Most preferably, high performance
liquid chromatography is employed for purification. Well known techniques for
refolding proteins may be employed to regenerate active conformation when the
polypeptide is denatured during isolation and or purification.
IV. Polypeptides of the Invention
The de novo DNA cytosine methyltransferase polypeptides of the present
invention include the polypeptide of SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:7
or SEQ ID NO:8, as well as polypeptides and fragments which have activity and
have at least 90% identity to the polypeptide of SEQ ID NO:5, SEQ ID NO:6,
SEQ ID NO:7 or SEQ ID NO:8, or the relevant portion and more preferably at
least 96%. 97% or 98% identity to the polypeptide of SEQ ID NO:5, SEQ ID
NO:6, SEQ ID NO:7 or SEQ ID NO:8, and still more preferably at least 91%,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% identity to the polypeptide
of SEQ ID NO:5. SEQ ID NO:6. SEQ ID NO:7 or SEQ ID NO:8.
CA 02331781 2005-07-20
-32-
The polypeptides of the present invention are preferably provided in an
isolated form.
The polypeptides ofthe present invention include the polypeptide encoded
by the deposited cDNAs; a polypeptide comprising amino acids from about I to
about 908 in SEQ ID NO:5; a polypeptide comprising amino acids from about I
to about 859 in SEQ ID NO:6; a polypeptide comprising amino acids from about
I to about 912 in SEQ ID NO:7 and a polypeptide comprising amino acids from
about I to about 853 in SEQ ID NO:8; as well as polypeptides which are at
least
about 90% identical, and more preferably at least about 91%, 92%, 93%, 94%,
95%, 96%, 97%, 98%; 99%, or 100% identical to the polypeptides described
above and also include portions of such polypeptides with at least 30 amino
acids
and more preferably at least 50 amino acids.
Polypeptides of the invention also include alternative splicing variants of
the Dnmt3 sequences disclosed herein. For example, alternative variant spliced
proteins of mouse Dnmt3 b include but are not limited to a polypeptide
wherein,
except for at least one conservative amino acid substitution, said polypeptide
has
a sequence selected from the group consisting of: (1) amino acid residues l to
362
and 383 to 859 from SEQ ID NO:6; and (2) amino acid residues I to 362 and 383
to 749 and 813 to 859 from SEQ ID NO:6; and alternative variant
splicedproteins
of hunian DNMT3B include but are not limited to a polypeptide wherein, except
for at least one conservative amino acid substitution, said polypeptide has a
sequence selected from the group consisting of: (1) amino acid residues l to
355
and 376 to 853 from SEQ ID NO:8; and (2) amino acid residues 1 to 355 and 376
to 743 and 807 to 853 from SEQ ID NO:8.
The de novo DNA cytosine methyltransferase polypeptides may be a part
of a larger protein such as a fusion protein. It is often advantageous to
include
additional amino acid sequence which contains secretory or leader sequences,
pro-sequences, sequences which aid in purification such as multiple histidine
residues, or additional sequence for stability during recombinant production.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-33-
Biologically active fragments of the de novo DNA cytosine
methyltransferase polypeptides are also included in the invention. A fragment
is
a polypeptide having an amino acid sequence that entirely is the same as part
but
not all of the amino acid sequence of one of the aforementioned de novo DNA
cytosine methyltransferase polypeptides. As with de novo DNA cytosine
methyltransferase polypeptides, fragments may be "free-standing," or comprised
within a larger polypeptide of which they form a part or region, most
preferably
as a single continuous region. In the context of this invention, a fragment
may
constitute from about 10 contiguous amino acids identified in SEQ ID NO:5,
SEQ ID NO:6, SEQ ID NO:7 or SEQ ID NO:8. More specifically, polypeptide
fragment lengths may be defined algebraically as follows: (a) for SEQ ID NO:5,
as 10 + N, wherein N equals zero or any positive integer up to 898; (b) for
SEQ
ID NO:6, as 10 + N, wherein N equals zero or any positive integer up to 849;
(c)
for SEQ ID NO:7, as 10 + N, wherein N equals zero or any positive integer up
to
902; and (d) for SEQ ID NO:8, as 10 + N, wherein N equals zero or any positive
integer up to 843.
Preferred fragments include, for example, truncation polypeptides having
the amino acid sequence of de novo DNA cytosine methyltransferase
polypeptides, except for deletion of a continuous series of residues that
includes
the amino terminus, or a continuous series of residues that includes the
carboxyl
terminus or deletion of two continuous series ofresidues, one including the
amino
terminus and one including the carboxyl terminus. Also preferred are fragments
characterized by structural or functional attributes such as fragments that
comprise alpha-helix and alpha-helix forming regions, beta-sheet and beta-
sheet-
forming regions, turn and turn-forming regions, coil and coil-forming regions,
hydrophilic regions, hydrophobic regions, alpha amphipathic regions, beta
amphipathic regions, flexible regions, surface-forming regions, substrate
binding
region, and high antigenic index regions. Biologically active fragments are
those
that mediate protein activity, including those witli a similar activity or an
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-34-
improved activity, or with a decreased undesirable activity. Also included are
those that are antigenic or immunogenic in an animal, especially in a human.
Thus, the polypeptides of the invention include polypeptides having an
amino acid sequence at least 90% identical to that of SEQ ID NO:5, SEQ ID
NO:6, SEQ ID NO:7 or SEQ ID NO:8, or fragments thereof with at least 90%
identity to the corresponding fragment of SEQ ID NO:5, SEQ ID NO:6, SEQ ID
NO:7 or SEQ ID NO:8, all of which retain the biological activity of the de
novo
DNA cytosine methyltransferase protein, including antigenic activity. Included
in this group are variants of the defined sequence and fragment. Preferred
variants are those that vary from the reference by conservative amino acid
substitutions, i.e., those that substitute a residue with another of like
characteristics. Typical substitutions are among Ala, Val, Leu and Ile; among
Ser
and Thr; among the acidic residues Asp and Glu; among Asn and Gln; and among
the basic residues Lys and Arg, or aromatic residues Phe and Tyr. Particularly
preferred are variants in which several, 5 to 10, 1 to 5, or I to 2 amino
acids are
substituted, deleted, or added in any combination.
The de novo DNA cytosine methyltransferase polypeptides of the
invention can be prepared in any suitable manner. Such polypeptides include
isolated naturally occurring polypeptides, recombinantly produced
polypeptides,
synthetically produced polypeptides, or polypeptides produced by a combination
of these methods. Means for preparing such polypeptides are well understood in
the art.
V. In Vitro DNA Metl:ylation
One preferred embodiment of the invention enables the in vitro
methylation at the C5 position of cytosine in DNA. The starting substrate DNA
may be hemimethylated (i.e., one strand of the duplex DNA is methylated) or
may
lack methylation completely. The polypeptides of the invention, being de novo
DNA cytosine methyltransferases. are uniquely suited to the latter function,
owing
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-35-
to the fact that, unlike maintenance methyltransferases, their preferred
substrate
is not hemimethylated DNA.
As exemplified in Examples 7 and 8, isolated polypeptides of the
invention function as in vitro DNA methyltransferases when combined in an
appropriately buffered solution with the appropriate cofactors and a substrate
DNA. The substrate DNA may be selected from any natural source, e.g., genomic
DNA, or a recombinant source such as a DNA fragment amplified by the
polymerase chain reaction. The substrate DNA may be prokaryotic or eukaryotic
DNA. In a preferred embodiment, the substrate DNA is mammalian DNA, and
most preferredly, the substrate DNA is human DNA.
It will be well appreciated by those in the art that in vitro methylation of
DNA may be used to direct or regulate the expression of said DNA in a
biological
system. For example, over-expression, under-expression or lack of expression
of
a particular native DNA sequence in a host cell or organism may be attributed
to
the fact that the DNA is under-methylated (hypomethylated) or not methylated.
Thus, in vitro methylation of a recombinant form of said DNA, and the
subsequent introduction of the methylated, recombinant DNA into the cell or
organism, may effect an increase or decrease in the expression of the encoded
polypeptide.
Also, it will be readily apparent to the skilled artisan that the in vitro
methylation pattern will be maintained after introduction into a biological
system
by the action of maintenance methyltransferase polypeptides in said system.
In one embodiment of the invention, the biological system selected for the
introduction of in vitro methylated DNA may be prokaryotic or eukaryotic. In a
preferred embodiment, the biological system is mammalian, and the most
preferred embodiment is when the biological system is human.
Methods for introducing the in vitro methylated DNA into the biological
system are well known in the art, and the skilled artisan will recognize that
the in
vitro methylation of DNA may be a preliminary step to any system of gene
therapy detailed herein.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-36-
VI. Genetic Screening and Diagnostic Assays
To map the human chromosome locations, the GenBank STS database
was searched using Dnmt3a and Dnmt3b sequences as queries. The search
identified markers WI-6283 (GenBank Accession number G06200) and SHGC-
15969 (GenBank Accession number G15302) as matching the cDNA sequence
of Dnmt3a and Dnmt3b, respectively. WI-6283 has been mapped to 2p23
between D2S 171 and D2S 174 (48-50 cM) on the radiation hybrid map by
Whitehead Institute/MIT Center for Genome Research. The corresponding mouse
chromosome location is at 4.0 cM on chromosome 12. SHGC-15969 has been
mapped to 20pl 1.2 between D20S184 and D20S106 (48-50 cM) by Stanford
Human Genome Center. The corresponding mouse chromosome locus is at 84.0
cM on chromosome 2.
These data are valuable as markers to be correlated with genetic map data.
Such data are found, for example, in V. McKusick, Mendelian Inheritance in Man
(available on-line through Johns Hopkins, University Welch Medical Library).
The relationship between genes and diseases that have been mapped to the same
chromosomal region are then identified through linkage analysis (coinheritence
of physically adjacent genes).
The differences in the cDNA or genomic sequence between affected and
unaffected individuals can also be determined. If a mutation is observed in
some
or all of the affected individuals but not in any normal individuals, then the
mutation is likely to be the causative agent of the disease.
This invention also relates to the use of de novo DNA cytosine
methyltransferase polynucleotides for use as diagnostic reagents. Detection of
a
mutated form of a de novo DNA cytosine methyltransferase gene associated with
a dysfunction will provide a diagnostic tool that can add to or define a
diagnosis
of a disease or susceptibility to a disease which results from under-
expression,
over-expression or altered expression of the mutated de novo DNA cytosine
methyltransferase. Individuals canying mutations in one or more de novo DNA
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373-
-37-
cytosine methyltransferase genes may be detected at the DNA level by a variety
of techniques.
Nucleic acids for diagnosis may be obtained from a subject's cells, such
as from blood, urine, saliva, tissue biopsy or autopsy material. The genomic
DNA may be used directly for detection or may be amplified enzymatically by
using PCR or other amplification techniques prior to analysis. RNA or cDNA
may also be used in similar fashion. Deletions and insertions can be detected
by
a change in size of the amplified product in comparison to the normal
genotype.
Point mutations can be identified by hybridizing amplified DNA to labeled de
novo DNA cytosine methyltransferase nucleotide sequences. Perfectly matched
sequences can be distinguished from mismatched duplexes by RNase digestion
or by differences in melting temperatures. DNA sequence differences may also
be detected by alterations in electrophoretic mobility of DNA fragments in
gels,
with or without denaturing agents, or by direct DNA sequencing (see, e.g.,
Myers,
et al., Science 230:1242 (1985)). Sequence changes at specific locations may
also
be revealed by nuclease protection assays, such as RNase and S 1 protection or
the
chemical cleavage method (see Cotton, et al., Proc. Natl. Acad. Sci. USA
85:4397-4401 (1985)).
The diagnostic assays offer a process for diagnosing or determining a
susceptibility to neoplastic disorders through detection of mutations in one
or
more de novo DNA cytosine methyltransferase genes by the methods described.
In addition, neoplastic disorders may be diagnosed by methods that
determine an abnormally decreased or increased level of de novo DNA cytosine
methyltransferase polypeptide or de novo DNA cytosine methyltransferase
mRNA in a sample derived from a subject. Decreased or increased expression
may be measured at the RNA level using any of the tnethods well known in the
art for the quantitation of polynucleotides; for example, RT-PCR, RNase
protection, Northern blotting and other hybridization methods may be utilized.
Assay techniques that may be used to determine the level of a protein, such as
an
de novo DNA cytosine methyltransferase protein, in a sample derived from a
host
CA 02331781 2000-12-21
WO 99/67397 PCTNS99/14373
-38-
are well known to those of skill in the art. Such assay methods include
radioimmunoassays, competitive-binding assays, Western blot analysis and
ELISA assays.
Additionally, methods are provided for diagnosing or determining a
susceptibility of an individual to neoplastic disorders, comprising (a)
assaying the
de novo DNA cytosine methyltransferase protein gene expression level in
mammalian cells or body fluid; and (b) comparing said de novo DNA cytosine
methyltransferase protein gene expression level with a standard de novo DNA
cytosine methyltransferase protein gene expression level whereby an increase
or
decrease in said de novo DNA cytosine methyltransferase gene expression level
over said standard is indicative of an increased or decreased susceptibility
to a
neoplastic disorder.
VII. De novo DNA Cytosine Methyltransferase Antibodies
The polypeptides of the invention or their fragments or analogs thereof,
or cells expressing them may also be used as immunogens to produce antibodies
immunospecific for the de novo DNA cytosine methyltransferase polypeptides.
By "immunospecific" is meant that the antibodies have affinities for the
polypeptides of the invention that are substantially greater in their
affinities for
related polypeptides such as the analogous proteins of the prior art.
Antibodies generated against the de novo DNA cytosine methyltransferase
polypeptides can be obtained by administering the polypeptides or epitope-
bearing fragments, analogs or cells to an animal, preferably a nonhuman, using
routine protocols. For preparation of monoclonal antibodies, any technique
which
provides antibodies produced by continuous cell line cultures can be used.
Examples include the hybridoma technique (Kohler, G. and Milstein, C., Nature
256:495-497 (1975)), the trioma technique, the human B-cell hybridoma
technique (Kozbor, et al., Immunology Today 4:72 (1983)) and the EBV-
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-39-
hybridoma technique (Cole, et al., Monoclonal Antibodies and Cancer Therapy,
pp. 77-96, Alan R. Liss, Inc., (1985)).
Techniques for the production of single chain antibodies (U.S. Patent No.
4,946,778) may also be adapted to produce single chain antibodies to
polypeptides of this invention. Also, transgenic mice, or other organisms
including other mammals, may be used to express humanized antibodies.
The above-described antibodies may be employed to isolate or to identify
clones expressing the polypeptide or to purify the polypeptides by affinity
chromatography.
Antibodies against de novo DNA cytosine methyltransferase polypeptides
may also be employed to treat neoplastic disorders, among others.
VIII. Agonist and Antagonist Screening
The de novo DNA cytosine methyltransferase polypeptides of the present
invention may be employed in a screening process for compounds which bind one
of the proteins and which activate (agonists) or inhibit activation of
(antagonists)
one of the polypeptides of the present invention. Thus, polypeptides of the
invention may also be used to assess the binding of small molecule substrates
and
ligands in, for example, cells, cell-free preparations, chemical libraries,
and
natural product mixtures. These substrates and ligands may be natural
substrates
and ligands or may be structural or functional mimetics (see Coligan, et al.,
Current Protocols in Immunology 1(2):Chapter 5 (1991)).
By "agonist" is intended naturally occurring and synthetic compounds
capable of enhancing a de novo DNA cytosine methyltransferase activity (e.g.,
increasing the rate of DNA methylation). By "antagonist" is intended naturally
occurring and synthetic compounds capable of inhibiting a de novo DNA cytosine
methyltransferase activity.
DNA methylation is an important, fundamental regulatory mechanism for
gene expression, and, therefore, the methylated state of a particular DNA
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-40-
sequence may be associated with many pathologies. Accordingly, it is desirous
to find both compounds and drugs which stimulate de novo DNA cytosine
methyltransferase activity and which can inhibit the function of de novo DNA
cytosine methyltransferase protein. In general, agonists are employed for
therapeutic and prophylactic purposes including the treatment of ceratin types
of
neoplastic disorders. For example, de novo methylation of growth regulatory
genes in somatic tissues is associated with tumorigenesis in humans (Laird, P.
W.
and Jaenisch, R. Ann. Rev. Genet. 30:441-464 (1996); Baylin, S. B. et al.,
Adv.
Cancer. Res. 72:141-196 (1998); and Jones, P. A. and Gonzalgo, M. L. Proc.
Natl. Acad. Sci. USA 94:2103-2105 (1997)).
In general, such screening procedures involve producing appropriate cells
which express the polypeptide of the present invention. Such cells include
cells
from mammals, yeast, Drosophila or E. coli. Cells expressing the protein (or
cell
membrane containing the expressed protein) are then contacted with a test
compound to observe binding, stimulation or inhibition of a functional
response.
Alternatively, the screening procedure may be an in vitro procedure in
which the activity of isolated DNMT3 protein is tested in the presence of a
potential agonist or antagonist of DNMT3 de novo DNA cytosine
methyltransferase activity. Such in vitro assays are known to those skilled in
the
art, and by way of example are demonstrated in Example 4.
The assays may simply test binding of a candidate compound wherein
adherence to the cells bearing the protein is detected by means of a label
directly
or indirectly associated with the candidate compound or in an assay involving
competition with a labeled competitor. Further, these assays may test whether
the
candidate compound affects activity of the protein, using detection systems
appropriate to the cells bearing the protein at their surfaces. Inhibitors of
activation are generally assayed in the presence of a known agonist and the
effect
on activation by the agonist in the presence of the candidate compound is
observed. Standard methods for conducting such screening assays are well
understood in the art.
CA 02331781 2000-12-21
WO 99/67397 PCTNS99/14373
-41-
Examples of potential de novo DNA cytosine methyltransferase protein
antagonists include antibodies or, in some cases, oligonucleotides or proteins
which are closely related to the substrate of the de novo DNA cytosine
methyltransferase protein, e.g., small molecules which bind to the protein so
that
the activity of the protein is prevented.
IX. Gene Therapy Applications
For overview of gene therapy, see Strachan, T. & Read A.P., Chapter 20,
"Gene Therapy and Other Molecular Genetic-based T'herapeutic Approaches,"
(and references cited therein) in Human Molecular Genetics, BIOS Scientific
Publishers Ltd. (1996).
Initial research in the area of gene therapy focused on a few well-
characterized and highly publicized disorders: cystic fibrosis (Drumm, M.L. et
al.,
Cell 62:1227-1233 (1990); Gregory, R.J. et al., Nature 347:358-363 (1990);
Rich,
D.P. et al., Nature 347:358-363 (1990)); and Gaucher disease (Sorge, J. et
al.,
Proc. Natl. Acad. Sci. (USA) 84:906-909 (1987); Fink, J.K. et al., Proc. Natl.
Acad. Sci. (USA) 87:2334-2338 (1990)); and certain forms of hemophilia-
Bontempo, F.A. et al., Blood 69:1721-1724 (1987); Palmer, T.D. et al., Blood
73:438-445 (1989); Axelrod, J.H. et al., Proc. Natl. Acad. Sci. (USA) 87:5173-
5177 (1990); Armentano, D. et al., Proc. Natl. Acad. Sci. (USA) 87:6141-6145
(1990)); and muscular dystrophy (Partridge, T.A. et al., Nature 337:176-179
(1989); Law, P.K. et al., Lancet 336:114-115 (1990); Morgan, J.E. et al., J.
Cell
Biol. 111:2437-2449 (1990)).
More recently, the application of gene therapy in the treatment of a wider
variety of disorders is progressing, for example: cancer (Runnebaum, I.B.,
Anticancer Res. 17(4B): 2887-2890, (1997)), heart disease (Rader, D.J., Int.
J.
Clin. Lab. Res. 27(1): 35-43, (1997); Malosky, S., Curr. Opin. Cardiol. 11(4):
361-368, (1996)), central nervous system disorders and injuries (Yang, K., et
al.,
Neurotrauma J. 14(5): 281-297, (1997); Zlokovic, B.V.. et al., Neurosurgery
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373-
-42-
40(4): 789-803, (1997); Zlokovic, B.V., et al., Neurosurgery 40(4): 805-812,
41997)), vascular diseases (Clowes, A.W., Thromb. 1-laemost. 78(1): 605-610,
1997), muscle disorders (Douglas, J.T., et al., Neuromuscul. Disord. 7(5): 284-
298, (1997); Huard, J., et al., Neuromuscul. Disord. 7(5): 299-313, (1997)),
rheumatoid arthritis (Evans, C.H., et al., Curr. Opin. Rheumatol. 8(3): 230-
234,
(1996)) and epithelial tissue disorders (Greenhaigh, D.A., et al., lnvest
Dermatol.
J. 103(5 Suppl.): 63S-93S, (1994)).
In a preferred approach, one or more isolated nucleic acid molecules of
the invention are introduced into or administered to the animal. Such isolated
nucleic acid molecules may be incorporated into a vector or virion suitable
for
introducing the nucleic acid molecules into the cells or tissues of the animal
to be
treated, to form a transfection vector. Techniques for the formation of
vectors or
virions comprising the de novo DNA cytosine methyltransferase-encoding nucleic
acid molecules are well known in the art and are generally described in
"Working
Toward Human Gene Therapy," Chapter 28 in Recombinant DNA, 2nd Ed.,
Watson, J.D. et al., eds., New York: Scientific American Books, pp. 567-581
(1992). An overview of suitable vectors or virions is provided in an article
by
Wilson, J.M. (Clin. Exp. Immunol. 107(Suppl. 1): 31-32, (1997)). Such vectors
are derived from viruses that contain RNA (Vile, R.G., et al., Br. Med Bull.
51(1): 12-30, (1995)) or DNA (Ali M., et al., Gene Ther. 1(6): 367-384,
(1994)).
Example vector systems utilized in the art include the following: retroviruses
(Vile, R.G., supra.), adenoviruses (Brody, S.L. et al., Ann. N. Y. Acad. Sci.
716:
90-101, (1994)), adenoviral/retroviral chimeras (Bilbao, G., et al., FASEB J.
11(8): 624-634, (1997)), adeno-associated viruses (Flotte, T.R. and Carter,
B.J.,
Gene Ther. 2(6): 357-362, (1995)), herpes simplex virus (Latchman, D.S., Mol.
Biotechnol. 2(2): 179-195, (1994)), Parvovirus (Shaughnessy, E., et al., Semin
Oncol. 23(1): 159-171, (1996)) and reticuloendotheliosis virus (Donburg, R.,
Gene Therap. 2(5): 301-310, (1995)). Also of interest in the art, the
development
of extrachromosomal replicating vectors for gene therapy (Calos, M.P., Trends
Genet. 12(11): 463-466, (1996)).
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-43-
Other, nonviral methods for gene transfer known in the art (Abdallah, B.
et al., Biol. Cel185(1): 1-7, (1995)) might be utilized for the introduction
of de
novo DNA cytosine methyltransferase polynucleotides into target cells; for
example, receptor-mediated DNA delivery (Philips, S.C., Biologicals 23(1): 13-
16, (1995)) and lipidic vector systems (Lee, R.J. and Huang, L., Crit. Rev.
Ther.
Drug Carrier Syst. 14(2): 173-206, (1997)) are promising alternatives to viral-
based delivery systems.
General methods for construction of gene therapy vectors and the
introduction thereof into affected animals for therapeutic purposes may be
obtained in the above-referenced publications, the disclosures of which are
specifically incorporated herein by reference in their entirety. In one such
general
method, vectors comprising the isolated polynucleotides of the present
invention
are directly introduced into target cells or tissues of the affected animal,
preferably by injection, inhalation, ingestion or introduction into a mucous
membrane via solution; such an approach is generally referred to as "in vivo"
gene
therapy. Alternatively, cells, tissues or organs may be removed from the
affected
animal and placed into culture according to methods that are well-known to one
of ordinary skill in the art; the vectors comprising the de novo DNA cytosine
methyltransferase polynucleotides may then be introduced into these cells or
tissues by any of the methods described generally above for introducing
isolated
polynucleotides into a cell or tissue, and, after a sufficient amount of time
to
allow incorporation of the de novo DNA cytosine methyltransferase
polynucleotides, the cells or tissues may then be re-inserted into the
affected
animal. Since the introduction of a de novo DNA cytosine methyltransferase
gene
is performed outside of the body of the affected animal, this approach is
generally
referred to as "ex vivo" gene therapy.
For both in vivo and ex vivo gene therapy, the isolated de novo DNA
cytosine methyltransferase polynucleotides of the invention may altematively
be
operatively linked to a regulatory DNA sequence, which may be a de novo DNA
cytosine methyltransferase promoter or an enhancer, or a heterologous
regulatory
CA 02331781 2004-08-12
-44-
DNA sequence such as a promoter or enhancer derived from a different gene,
cell
or organism, to form a genetic construct as described above. This genetic
construct may then be inserted into a vector, which is then used in a gene
therapy
protocol. The need for transcriptionally targeted and regulatable vectors
providing cell-type specific and inducible promoters is well recognized in the
art
(Miller, N. and Whelan, J., Hum. Gene Therap. 8(7): 803-815, (1997); and
Walther, W. and Stein, U., Mol. Med. J., 74(7): 379-392, (1996)), and for the
purposes of de novo DNA cytosine methyltransferase gene therapy.
The construct/vector may be introduced into the animal by an in vivo gene
therapy approach, e.g., by direct injection into the target tissue, or into
the cells
or tissues of the affected animal in an ex vivo approach. In another preferred
embodiment, the genetic construct of the invention may be introduced into the
cells or tissues of the animal, either in vivo or ex vivo, in a molecular
conjugate
with a virus (e.g., an adenovirus or an adeno-associated virus) or viral
components (e.g., viral capsid proteins; see WO 93/07283). Alternatively,
transfected host cells, which may be homologous or heterologous, may be
encapsulated within a semi-permeable barrier device and implanted into the
affected animal, allowing passage of de novo DNA cytosine methyltransferase
polypeptides into the tissues and circulation of the animal but preventing
contact
between the animal's immune system and the transfected cells (see
WO 93109222). These approaches result in increased production of de novo DNA
cytosine methyltransferase by the treated animal via (a) random insertion of
the
de novo DNA cytosine methyltransferase gene into the host cell genome; or (b)
incorporation of the de novo DNA cytosine methyltransferase gene into the
nucleus of the cells where it may exist as an extrachromosomal genetic
element.
General descriptions of such methods and approaches to gene therapy may be
found, for example, in U.S. Patent No. 5,578,461, WO 94/12650 and WO
93/09222.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-45-
Antisense oligonucleotides have been described as naturally occurring
biological inhibitors of gene expression in both prokaryotes (Mizuno et al.,
Proc.
Natl. Acad. Sci. USA 81:1966-1970 (1984)) and eukaryotes (Heywood, Nucleic
Acids Res. 14:6771-6772 (1986)), and these sequences presumably function by
hybridizing to complementary mRNA sequences, resulting in hybridization arrest
of translation (Paterson, et al., Proc. Natl.Acad. Sci. USA, 74:4370-
4374(1987)).
Thus, another gene therapy approach utilizes antisense technology.
Antisense oligonucleotides are short synthetic DNA or RNA nucleotide
molecules formulated to be complementary to a specific gene or RNA message.
Through the binding of these oligomers to a target DNA or mRNA sequence,
transcription or translation of the gene can be selectively blocked and the
disease
process generated by that gene can be halted (see, for example, Jack Cohen,
Oligodeoxynucleotides, Antisense Inhibitors of Gene Expression, CRC Press
(1989)). The cytoplasmic location of mRNA provides a target considered to be
readily accessible to antisense oligodeoxynucleotides entering the cell; hence
much of the work in the field has focused on RNA as a target. Currently, the
use
of antisense oligodeoxynucleotides provides a useful tool for exploring
regulation
of gene expression in vitro and in tissue culture (Rothenberg, et al., J.
Natl.
Cancer Inst. 81:1539-1544 (1989)).
Antisense therapy is the administration of exogenous oligonucleotides
which bind to a target polynucleotide located within the cells. For example,
antisense oligonucleotides may be administered systemically for anticancer
therapy (Smith, International Application Publication No. WO 90/09180).
The antisense oligonucleotides of the present invention include derivatives
such as S-oligonucleotides (phosphorothioate derivatives or S-oligos, see,
Jack
Cohen, supra). S-oligos (nucleoside phosphorothioates) are isoelectronic
analogs
of an oligonucleotide (0-oligo) in which a nonbridging oxygen atom of the
phosphate group is replaced by a sulfur atom. The S-oligos of the present
invention may be prepared by treatment of the corresponding 0-oligos with 3H-
1,2-benzodithiol-3-one-1,1-dioxide which is a sulfur transfer reagent. See
lyer et
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-46-
al., J. Org. Chem. 55:4693-4698 (1990); and Iyer et al., J. Am. Chem. Soc.
112:1253-1254 (1990), the disclosures of which are fully incorporated by
reference herein.
As described herein, sequence analysis of SEQ ID NO: 1, SEQ ID NO:2,
SEQ ID NO:3 or the SEQ ID NO:4 cDNA clone shows that sequence that is
nonhomologous to known DNA methyltransferase sequences may be identified
(see Figures 1 and 4). Thus, the antisense oligonucleotides of the present
invention may be RNA or DNA that is complementary to and stably hybridize
with such sequences that are specific for a de novo DNA cytosine
methyltransferase gene of the invention. Use of an oligonucleotide
complementary to such regions allows for selective hybridization to a de novo
DNA cytosine methyltransferase mRNA and not to an mRNA encoding a
maintenance methyltransferase protein.
Preferably, the antisense oligonucleotides of the present invention are a
15 to 30-mer fragment of the antisense DNA molecule coding for unique
sequences of the de novo DNA cytosine methyltransferase cDNAs. Preferred
antisense oligonucleotides bind to the 5'-end of the de novo DNA cytosine
methyltransferase mRNAs. Such antisense oligonucleotides may be used to down
regulate or inhibit expression of the gene.
Other criteria that are known in the art may be used to select the antisense
oligonucleotides, varying the length or the annealing position in the targeted
sequence.
Included as well in the present invention are pharmaceutical compositions
comprising an effective amount of at least one of the antisense
oligonucleotides
of the invention in combination with a pharmaceutically acceptable carrier. In
one
embodiment, a single antisense oligonucleotide is utilized.
In another embodiment, two antisense oligonucleotides are utilized which
are complementary to adjacent regions of the genome. Administration of two
antisense oligonucleotides that are complementary to adjacent regions of the
genome or corresponding mRNA may allow for more efficient inhibition of
CA 02331781 2004-08-12
-4 7-
Qenomic transcription ormRNA translation, resulting in more effective
inhibition
of protein or mRNA production.
Preferably, the antisense oligonucleotide is coadministered with an agent
which enhances the uptake of the antisense molecule by the cells. For example,
the antisense oligonucleotide may be combined with a lipophilic cationic
compound which may be in the form of liposomes. The use of liposomes to
introduce nucleotides into cells is taught, for example, in U.S. Patent Nos.
4,897,355 and 4,394,448,
(see also U.S. Patent Nos. 4,235,871, 4,231,877, 4,224,179,
4,753,788, 4,673,567, 4,247,411, and 4, 814,270 for general methods of
preparing
liposomes comprising biological materials).
Alternatively, the antisense oligonucleotide may be combined with a
lipophilic carrier such as any one of a number of sterols including
cholesterol,
cholate and deoxycholic acid. A preferred sterol is cholesterol.
In addition, the antisense oligonucleotide may be conjugated to a peptide
that is ingested by cells. Examples of useful peptides include peptide
hormones,
antigens or antibodies, and peptide toxins. By choosing a peptide that is
selectively taken up by the targeted tissue or cells, specific delivery of the
antisense agent may be effected. The antisense oligonucleotide may be
covalently
bound via the 5'OH group by formation of an activated aminoalkyl derivative.
The peptide of choice may then be covalently attached to the activated
antisense
oligonucleotide via an amino and sulfhydryl reactive hetero bifunctional
reagent.
The latter is bound to a cysteine residue present in the peptide. Upon
exposure
of cells to the antisense oligonucleotide bound to the peptide, the peptidyl
antisense agent is endocytosed and the antisense oligonucleotide binds to the
target mRNA to inhibit translation (Haralambid et al., WO 8903849 and Lebleu
et al., EP 0263740).
The antisense oligonucleotides and the pharmaceutical compositions of
the present invention may be administered by any means that achieve their
intended purpose. For example, administration may be by parenteral,
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-48-
subcutaneous, intravenous, intramuscular, intraperitoneal, or transdermal
routes.
The dosage administered will be dependent upon the age, health, and weight of
the recipient, kind of concurrent treatment, if any, frequency of treatment,
and the
nature of the effect desired.
Compositions within the scope of this invention include all compositions
wherein the antisense oligonucleotide is contained in an amount effective to
achieve the desired effect, for example, inhibition of proliferation and/or
stimulation of differentiation of the subject cancer cells. While individual
needs
vary, determination of optimal ranges of effective amounts of each component
is
with the skill of the art.
Alternatively, antisense oligonucleotides can be prepared which are
designed to interfere with transcription of the gene by binding transcribed
regions
of duplex DNA (including introns, exons, or both) and forming triple helices
(e.g., see Froehler et al., WO 91/06626 or Toole, WO 92/10590). Preferred
oligonucleotides for triple helix formation are oligonucleotides which have
inverted polarities for at least two regions of the oligonucleotide (Id.).
Such
oligonucleotides comprise tandem sequences of opposite polarity such as 3'---
5'-
L-5------ ', or 5'---3'-L-3'---5', wherein L represents a 0-10 base
oligonucleotide
linkage between oligonucleotides. The inverted polarity form stabilizes single-
stranded oligonucleotides to exonuclease degradation (Froehler et al., supra).
The criteria for selecting such inverted polarity oligonucleotides is known in
the
art, and such preferred triple helix-forming oligonucleotides of the invention
are
based upon SEQ ID NO:1, SEQ ID NO:2, SEQ ID NO:3 or SEQ ID NO:4.
In therapeutic application, the triple helix-forming oligonucleotides can
be formulated in pharmaceutical preparations for a variety of modes of
administration, including systemic or localized administration, as described
above.
The antisense oligonucleotides of the present invention may be prepared
according to any of the methods that are well known to those of ordinary skill
in
the art, as described above.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-49-
Ribozymes provide an alternative method to inhibit mRNA function.
Ribozymes may be RNA enzymes, self-splicing RNAs, and self-cleaving RNAs
(Cech et al., Journal of Biological Chemistry 267:17479-17482 (1992)). It is
possible to construct de novo ribozymes which have an endonuclease activity
directed in trans to a certain target sequence. Since these ribozymes can act
on
various sequences, ribozymes can be designed for virtually any RNA substrate.
Thus, ribozymes are very flexible tools for inhibiting the expression of
specific
genes and provide an alternative to antisense constructs.
A ribozyme against chloramphenicol acetyltransferase mRNA has been
successfully constructed (Haseloff et al., Nature 334:585-591 (1988);
Uhlenbeck
et al., Nature 328:596-600 (1987)). The ribozyme contains three structural
domains: 1) a highly conserved region of nucleotides which flank the cleavage
site in the 5' direction; 2) the highly conserved sequences contained in
naturally
occurring cleavage domains of ribozymes, forming a base-paired stem; and 3)
the
regions which flank the cleavage site on both sides and ensure the exact
arrangement of the ribozyme in relation to the cleavage site and the cohesion
of
the substrate and enzyme. RNA enzymes constructed according to this model
have already proved suitable in vitro for the specific cleaving of RNA
sequences
(Haseloff et al., supra).
Alternatively, hairpin ribozymes may be used in which the active site is
derived from the minus strand of the satellite RNA of tobacco ring spot virus
(Hampel et al., Biochemistry 28:4929-4933 (1989)). Recently, a hairpin
ribozyme was designed which cleaves human immunodeficiency virus type 1
RNA (Ojwang et al., Proc. Natl. Acad. Sci. USA 89:10802-10806 (1992)). Other
self-cleaving RNA activities are associated with hepatitis delta virus (Kuo et
al.,
J. Virol. 62:4429-4444 (1988)).
As discussed above, preferred targets for ribozymes are the de novo DNA
cytosine methyltransferase nucleotide sequences that are not homologous with
maintenance methyltransferase sequences such as Dnmt 1 or Dnmt 2. Preferably,
the ribozyme molecule of the present invention is designed based upon the
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-50-
chloramphenicol acetyltransferase ribozyme or hairpin ribozymes, described
above. Alternatively, ribozyme molecules are designed as described by Eckstein
et al. (International Publication No. WO 92/07065) who disclose catalytically
active ribozyme constructions which have increased stability against chemical
and
enzymatic degradation, and thus are useful as therapeutic agents.
In an alternative approach, an external guide sequence (EGS) can be
constructed for directing the endogenous ribozyme, RNase P, to intracellular
mRNA, which is subsequently cleaved by the cellular ribozyme (Altman et al.,
U.S. Patent No. 5,168,053). Preferably, the EGS comprises a ten to fifteen
nucleotide sequence complementary to an mRNA and a 3'-NCCA nucleotide
sequence, wherein N is preferably a purine (Id.). After EGS molecules are
delivered to cells, as described below, the molecules bind to the targeted
mRNA
species by forming base pairs between the mRNA and the complementary EGS
sequences, thus promoting cleavage of mRNA by RNase P at the nucleotide at
the 5'side of the base-paired region (Id. ).
Included as well in the present invention are pharmaceutical compositions
comprising an effective amount of at least one ribozyme or EGS of the
invention
in combination with a pharmaceutically acceptable carrier. Preferably, the
ribozyme or EGS is coadministered with an agent which enhances the uptake of
the ribozyme or EGS molecule by the cells. For example, the ribozyme or EGS
may be combined with a lipophilic cationic compound which may be in the fozm
of liposomes, as described above. Alternatively, the ribozyme or EGS may be
combined with a lipophilic carrier such as any one of a number of sterols
including cholesterol, cholate and deoxycholic acid. A preferred sterol is
cholesterol.
The ribozyme or EGS, and the pharmaceutical compositions of the
present invention may be administered by any means that achieve their intended
purpose. For example, administration may be by parenteral, subcutaneous.
intravenous, intramuscular, intra-peritoneal, or transdermal routes. The
dosage
administered will be dependent upon the age, health, and weight of the
recipient,
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-51-
kind of concurrent treatment, if any, frequency of treatment, and the nature
of the
effect desired. For example, as much as 700 milligrams of antisense
oligodeoxynucleotide has been administered intravenously to a patient over a
course of 10 days (i.e., 0.05 mg/kg/hour) without signs of toxicity (Sterling,
"Systemic Antisense Treatment Reported," Genetic Engineering News 12(12):1,
28 (1992)).
Compositions within the scope of this invention include all compositions
wherein the ribozyme or EGS is contained in an amount which is effective to
achieve inhibition of proliferation and/or stimulate differentiation of the
subject
cancer cells, or alleviate AD. While individual needs vary, determination of
optimal ranges of effective amounts of each component is with the skill of the
art.
In addition to administering the antisense oligonucleotides, ribozymes, or
EGS as a raw chemical in solution, the therapeutic molecules may be
administered as part of a pharmaceutical preparation containing suitable
pharmaceutically acceptable carriers comprising excipients and auxiliaries
which
facilitate processing of the antisense oligonucleotide, ribozyme, or EGS into
preparations which can be used pharmaceutically.
Suitable formulations for parenteral administration include aqueous
solutions of the antisense oligonucleotides, ribozymes, EGS in water-soluble
form, for example, water-soluble salts. In addition, suspensions of the active
compounds as appropriate oily injection suspensions may be administered.
Suitable lipophilic solvents or vehicles include fatty oils, for example,
sesame oil,
or synthetic fatty acid esters, for example, ethyl oleate or triglycerides.
Aqueous
injection suspensions may contain substances which increase the viscosity of
the
suspension include, for example, sodium carboxymethyl cellulose, sorbitol,
and/or dextran. Optionally, the suspension may also contain stabilizers.
Alternatively, antisense RNA molecules, ribozymes, and EGS can be
coded by DNA constructs which are administered in the form of virions, which
are preferably incapable of replicating in vivo (see, for example, Taylor, WO
92/06693). For example, such DNA constructs may be administered using
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-52-
herpes-based viruses (Gage et al., U.S. Patent No. 5,082,670). Alternatively,
antisense RNA sequences, ribozymes, and EGS can be coded by RNA constructs
which are administered in the form of virions, such as retroviruses. The
preparation of retroviral vectors is well known in the art (see, for example,
Brown
et al., "Retroviral Vectors," in DNA Cloning: A Practical Approach, Volume 3,
IRL Press, Washington, D.C. (1987)).
Specificity for gene expression may be conferred by using appropriate
cell-specific regulatory sequences, such as cell-specific enhancers and
promoters.
Such regulatory elements are known in the art, and their use enables therapies
designed to target specific tissues, such as liver, lung, prostate, kidney,
pancreas,
etc., or cell populations, such as lymphocytes, neurons, mesenchymal,
epithelial,
muscle, etc.
In addition to the above noted methods for inhibiting the expression of the
de novo methyltransferase genes of the invention, gene therapeutic
applications
may be employed to provide expression of the polypeptides of the invention.
Examples
Example 1: Cloning and Sequence Analysis of tlie Mouse Dnmt3a and
Dnmt3b and the Human DNMT3A and DNMT3B Genes and
Polypeptides
In search of a mammalian de novo DNA methyltransferase, two
independent approaches were undertaken, based on the assumption that an
unknown mammalian DNA methyltransferase must contain the highly conserved
cytosine methyltransferase motifs in the catalytic domain of known
methyltransferases (Lauster, R. et al., J. Mol. Biol. 206:305-312 (1989) and
Kumar, S. et al., Nucl. Acids Res. 22:1-10 (1994)). Our first approach, an
RT/PCR-based screening using oligonucleotide primers corresponding to the
conserved motifs of the known cytosine DNA methyltransferases, failed to
detect
any novel methyltransferase gene from Dnmtl null ES cells (data not shown).
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-53-
The second approach was a tblastn search of the dbEST database using full
length
bacterial cytosine methyltransferase sequences as queries.
A search of the dbEST database was performed with the tblastn program
(Altschul, S. F. et al., J. Mol. Biol. 215:403-410 (1990)) using bacterial
cytosine
methyltransferases as queries. Candidate EST sequences were used one by one
as queries to search the non-redundant protein sequence database in GenBank
with the blastx program. This process would eliminate EST clones corresponding
to known genes (including known DNA methyltransferases) and those which
show a higher similarity to other sequences than to DNA methyltransferases.
Two EST clones (GenBank numbers W7611 l and N88352) were found after the
initial search. Two more EST sequences (fl2227 and T66356) were later found
after a blastn search of dbEST with the EST sequence of W76111 as a query.
Two of the EST clones (W761 11 and T66356) were deposited by the I.M.A.G.E.
Consortium (Lawrence Livermore National Laboratory, Livermore, CA) and
obtained from American Type Culture Collection (Manassas, VA). Sequencing
of these two cDNA clones revealed that they were partial cDNA clones with
large
open reading frames corresponding to two related genes. The translated amino
acid sequences revealed the presence of the highly conserved motifs
characteristic
of DNA cytosine methyltransferases. The EST sequences were then used as
probes for screening mouse E7.5 embryo and ES cell cDNA libraries and a
human heart cDNA library (Clontech, CA).
In a screening of the dbEST database using 35 bacterial cytosine-5 DNA
methyltransferase sequences as queries, eight EST clones were found to have
the
highest similarity but not to be identical to the known cytosine-5-DNA
methyltransferase genes. Six of the eight EST sequences were deposited by the
I.M.A.G.E. Consortium (Lawrence Livermore National Laboratory, Livermore,
CA) and obtained from TIGR/ATCC (American "Type Culture Collection,
Manassas, VA). Sequencing of these 6 cDNA clones revealed that they were
partial cDNA clones with large open reading frames corresponding to three
novel
genes. The translated amino acid sequences revealed the presence of the highly
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-54-
conserved motifs characteristic of DNA cytosine methyltransferases. The EST
sequences were then used as probes for screening a mouse ES cell cDNA library,
a mouse E 11.5 embryonic cDNA library (Clontech, CA) and human heart cDNA
library.
Human and mouse cDNA libraries were screened using EST sequences
as probes. Sequencing analysis of several independent cDNA clones revealed
that two homologous genes were present in both human and mouse. This was
further confirmed by Southern analysis of genomic DNA, intron/exon mapping
and sequencing of genomic DNA (data not shown). The full length mouse
cDNAs for each gene were assembled and complete sequencing revealed that
both genes contained the highly conserved cytosine methyltransferase motifs
and
shared overall 51 % of amino acid identity (76% identity in the catalytic
domain)
(Fig. 3). Since these two genes showed little sequence similarities to
Dnmtl(Bestor, T. H. et al., J. Mol. Biol. 203:971-983 (1988) and Yen, R-W. C.
et al., Nucleic Acids Res. 20:2287-2291 (1992)) and a recently cloned putative
DNA methyltransferase gene, Dnmt2 (see Yoder, J. A. and Bestor, T. H. Hum.
Mol. Genet. 7:279-284 (1998)) and Okano, M., Xie, S. and Li, E., (submitted)),
beyond the conserved methyltransferase motifs in the catalytic domain, they
were
named Dnmt3a and Dnmt3b.
The full length Dnmt3a and Dnmt3b genes encode 908 and 859 amino
acid polypeptides, termed Dnmt3a and Dnmt3bl, respectively. Nucleotide and
amino acid sequences of each are presented in Figures 1 A, 1 B, 2A, and 2B.
The
Dnmt3b gene also produces through alternative splicing at least two shorter
isoforms of 840 and 777 amino acid residues, termed Dnmt3b2 and Dnmt3b3,
respectively, (Fig. 4).
To obtain full length human cDNA, fetal heart and fetal testis cDNA
libraries were screened using EST clones as probes. Sequencing analysis of
several overlapping DNMT3A cDNA clones indicates that the DNMT3A gene
encodes a polypeptide of 912 amino acid residues. DNMT3B cDNA clones were
not detected in the fetal heart library, but several DNMT3B cDNA clones were
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-55-
obtained after screening the fetal testis library. PCR screening of large cDNA
clones from 24 human tissues was also performed using the Human Rapid-
ScreenTM cDNA Library Panels (OriGene Technologies, MD). The largest cDNA
clone contained a 4.2 kb insert from a small intestine cDNA library.
Sequencing
analysis of overlapping cDNA clones indicated that the deduced full length
DMNT3B consists of 853 amino acid residues. Since in-frame stop codons are
found upstream of the ATG of both DNMT3A and DNMT3B, it is concluded that
these cDNA clones encode full-length DNMT3A and DNMT3B proteins.
The full length human DNMT3A and DNMT3B cDNAs encode 912 and
853 amino acid polypeptides, termed DNMT3A and DNMT3B1, respectively.
Nucleotide and polypeptide sequences are presented in Figures 1C, 1D, 2C and
2D, respectively. The DNMT3B gene also produces through alternative splicing
at least two shorter isoforms, termed DNMT3B2 and DNMT3B3, respectively.
DNMT3B2 comprises amino acid residues 1 to 355 and 376 to 853 of SEQ ID
NO:4; and DNMT3B3 comprises amino acid residues 1 to 355 and 376 to 743
and 807 to 853 of SEQ ID NO:4.
Also identified through screening was a related zebrafish gene, termed
Zmt-3, which from the EST database (GenBank number AF135438).
The GenBank STS database was used to map chromosome localization by
using DNMT3A and DNMT3B sequences as queries. The results identified
markers WI-6283 (GenBank Accession number G06200) and SHGC-15969
(GenBank Accession number G15302), which matched the cDNA sequence of
DNMT3A and DNMT3B, respectively. WI-6283 has been mapped to 2p23
between D2S171 and D2S 174 (48-50 cM) on the radiation hybrid map by
Whitehead Institute/MIT Center for Genome Research. The corresponding mouse
chromosome location is at 4.0 cM on chromosome 12. SHGC-15969 has been
mapped to 20pl 1.2 between D20S 184 and D20S 106 (48-50 cM) by Stanford
Human Genome Center. The corresponding mouse chromosome locus is at 84.0
cM on chromosome 2.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-56-
Taking the advantage of the newly identified DNMT3A and DNMT3B
cDNA sequences, the human genomic sequence database was searched by BLAST.
While human DNMT3A cDNA did not match any related genornic sequences in
the database, a DNMT3B genomic YAC clone from GenBank (AL035071) was
identified when DNMT3B cDNA sequences were used as queries.
The DNMT3B cDNA and the genomic DNA GenBank (AL035071) clone
were used to map all exons using BESTFIT of the GCG program. As shown in
Figure 4C, there are total 23 exons, spanning some 48 kb genomic DNA. The
putative first exon is located within a CpG island where the promoter is
probably
located as predicted by the GENSCAN program (Whitehead/MIT Center for
Genome Research).
Sequencing of various cDNA clones indicates that the human DNMT3B
gene contains three alternatively spliced exons, exons 10, 21 and 22. Similar
to
the mouse gene, DNMT3B I contains a1123 exons, whereas DNMT3B2 lacks exon
10 and DNMT3B3 lacks exons 10, 21 and 22. The nucleotide sequences at the
exon/intron boundaries are shown in Figure 4D. T'he elucidation of human
DNMT3B gene structure may facilitate analysis of DNMT3B mutations in certain
cancers with characteristic hypomethylation of genomic: DNA (Narayan, A., et
al.,
Int. J. Cancer 77:833-838 (1998); Qu, G., et al., Mutan. Res. 423:91-101
(1999)).
Figure 3A presents an alignment of mouse Dnmt3a and Dnmt3b
polypeptide sequences that was accomplished using the GCG program. The
vertical lines indicate amino acid identity, while the dots and the colons
indicate
similarities. Dots in amino acid sequences indicate gaps introduced to
maximize
alignment. The conserved Cys-rich region is shaded. The full length mouse
Dnmt3a and Dnmt3b genes encode 908 and 859 amino acid polypeptides.
Furthermore, the analysis reveals that both genes contained the highly
conserved
cytosine methyltransferase motifs and share overall 51 % of amino acid
identity
(76% identity in the catalytic domain). The Dnmt3b gene also produces at least
two shorter isoforms of 840 and 777 amino acid residues, termed Dnmt3b2 and
Dnmt3b3, respectively, through alternative splicing (Fig. 4).
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373-
-57-
Figure 3B presents a GCG program alignment using the of the protein
sequences of human DNMT3A and DNMT3B 1. Vertical lines represent identical
amino acid residues, whereas dots represent conserved changes. Dots in amino
acid sequences indicate gaps introduced to maximize alignment.
In Figure 4A, presents a schematic diagram of the overall protein
structures for mouse Dnmtl, mouse Dnmt2, a putative methyltransferase, and the
family of Dnmt3a and Dnmt3b(1-3) methyltransferases. Dnmtl, Dnmt3a and
Dnmt3bs all have a putative N-terminal regulatory domain. The filled bars
represent the five conserved methyltransferase motifs (I, IV, VI, IX, and X).
The
shaded boxes in Dnmt3a and Dnmt3bs represent the Cys-rich region that shows
no sequence homology to the Cys-rich, Zn2+-binding region of Dnmtl
polypeptide. Sites of alternative splicing at amino acid residues 362-383 and
749-
813 in Dnmt3bs are indicated.
An analysis of the human DNMT3 proteins provides similar results as
with the mouse Dnmt proteins. Figure 4B presents a similar schematic of the
human DNMT3 proteins and zebrafish Znmt3 protein. The homology between
differences between these DNMT3 proteins is indicated by the percentage of
sequence identity when compared to DNMT3A.
In addition, the genomic organization of the human DNMT3B 1 locus is
presented in Figure 4C as possessing 23 exons (filled rectangles), a CpG
island
(dotted rectangle),a translation initiation codon (ATG) and a stop codon (TAG)
in exons 2 and 23, respectively. Figure 4D presents the size of the exons and
introns as well as sequences (uppercase for exons and lowercase for introns)
at
exon/intron boundaries.
In Figure 5, sequence analysis of the catalytic domain indicates that this
new family of DNA methyltransferases contains conserved amino acid residues
in each of the five highly conserved motifs, but significant differences are
discernible when compared to the known consensus sequences.
Figure 5A presents an alignment by ClustalW 1.7 of the amino acid
sequences of the five highly conserved motifs in eukaryotic methyltransferase
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-58-
genes. Amino acid residues which are conserved in five or more genes are
highlighted. The Dnmt3 family methyltransferases ai=e most closely related to
a
bacterial DNA methyltransferase (M. Spr.). Sequence comparison of the
catalytic
domain of all known eukaryotic DNA methyltransferases and most of the
bacterial cytosine methyltransferases used in the tblastn search indicates
that this
family of methyltransferases are distantly related to all the known eukaryotic
DNA methyltransferases, including the Dnmt 1 polypeptide from vertebrate and
plant (Bestor, T. H. et al., J. Mol. Biol. 203:971-983 (1988), Yen, R-W. C. et
al.,
Nucleic Acids Res. 20:2287-2291 (1992) and Finnegan, E. J. and Dennis, E. S.
Nucleic Acids Res. 21:2383-2388 (1993)); the human and mouse Dnmt 2
polypeptides (Yoder, J. A. and Bestor, T. H. Hum. Mol. Genet. 7:279-284
(1998),
Okano, M., Xie, S. & Li, E., (submitted)); and masc 1 from Ascobolus
(Malagnac,
F. et al., Cell 91:281-290 (1997)), indicating that the Dnmt3 gene family
originated from a unique prokaryotic prototype DNA methyltransferase during
evolution.
The cysteine-rich region located upstream of the catalytic domain was
found to be conserved among all of the DNMT3 proteins (Fig. 5B). This
Cysteine-rich region, however, is unrelated to the Cysteine-rich (or Zn'-, -
binding)
region of DNMT l(Bestor, T.H., et al., J. Mo. Biol. 203:971-983 (1998);
Bestor,
T.H., EMBO J. 11:2611-2617 (1992)). Interestingly, the Cysteine-rich domain
of DNMT3 proteins shares homology with a similar domain found in the X-
linked ATRX gene of the SNF2/SWI family (Picketts, D.J., et al., Hum. Mol.
Genet. 5:1899-1907 (1996)), raising the interesting possibility that this
domain
may mediate protein-protein or protein-DNA interactions.
The evolutionary relatedness of cytosine-5 methyltransferases as shown
by a non-rooted phylogenic tree is presented in Figure 5C. Amino acid
sequences
from motif I to motif VI of bacterial and eukaryotic cytosine-5
methyltransferases
were used for sequence alignment, and the alignment data was analyzed by
ClustalW 1.7 under conditions excluding positions with gaps. Results were
visualized utilizing Phlip version 3.3. Amino acid sequences from motif IX to
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-59-
motif X were also analyzed and provided similar results (data not shown).
(Abbreviation Ath; Arabidopsis thaliana, Urc; sea urchin, Xen; Xenopus
laevis).
Example 2: Baculovirus-mediated Expression of Dnmt3a andDnmt3b
To test whether the newly cloned Dnmt3 genes encode active DNA
methyltransferases, the cDNAs ofDnmt3a, Dnmt3bl, Dnmt3b2, and Dnmtl were
overexpressed in insect cells using the baculovirus-mediated expression system
(Clontech, CA).
To construct the Dnmt3a expression vector, pSX134, the Xma I/Eco RI
fragment of Dnmt3a cDNA was first cloned into the Nco I/Eco RI sites of pET2
ld with the addition of an Xma I/Nco I adapter (SX165: 5'-
CATGGGCAGCAGCCATCATCATCATCATCATGGGAATTCCATGCCC
TCCAGCGGCC and SX166: 5'-CCGGGGCCGCTGGAGGGCATGGA
ATTCCCATGATGATGATGATGATGGCTGCTGCC) that produced
pSX 132His. pSX 134 was obtained by cloning the EcoR I/Xba 1 fragment of pSX
132His into the EcoR I/Xba I sites of pBacPAK9. The Dnmt3b 1 and Dnmt3b2
expression vectors, pSX153 and pSX154, were constructed by cloning Eco RI
fragments of Dnmt3bl and Dnmt3b2 cDNA into the Eco RI site of pBacPAK9,
respectively. The Dnmtl expression vector pSX 148 was constructed by cloning
the Bgl I/Sac I fragment of Dnmtl cDNA into the Bgl II/Sac 1 sites of pBacPAK-
His2 with the addition of a Bgl 1/Bgl II adapter (SX180: 5'-
GATCTATGCCAGCGCGAACAGCTCCAGCCCGAGTGCCTGCGCTTGC
CTCCC and SX 181: 5'- AGGCAAGCGCAGGCACTCGGGCTGGAGCTGTT
CGCGCTGGCATA).
pSX 134 (Dnmt3a), pSX 153 (Dnmt3b 1), pSX 153 (Dnmt3b2) and pSXl48
(Dnmtl) were used to make the recombinant baculoviruses according to the
procedures recommended by the manufacturer. T175 flasks were used for cell
culture and virus infection. SfZI host cells were grown in the SF-900 II SFM
medium with 10% of the certified FBS (both from GIBCO, MD) and infected
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373 -
-60-
with the recombinant viruses 12-24 hours after the cells were split when they
reached 90-95% affluence. After 3 days, the infected insect cells were
harvested
and frozen in the liquid nitrogen for future use.
Example 3: RNA Expression Analysis
ES cells were routinely cultured on a feeder layer of mouse embryonic
fibroblasts in DMEM medium containing LIF (500 units/ml) and were
differentiated as embryoid bodies in suspension culture as described (Lei, H.,
et
al., Development 122:3195-3205 (1996)). Ten days after seeding, embryoid
bodies were harvested for RNA preparation.
Total RNA was prepared from ES cells, ovary and testis tissue using the
GTC-CsCl centrifugation method, fractionated on a formaldehyde denaturing 1%
agarose gel by electrophoresis and transferred to a nylon membrane. PolyA+
RNA blots (2 g per lane) of mouse and human tissues were obtained from
Clontech, CA. All blots were hybridized to randoni-primed cDNA probes in
hybridization solution containing 50% formamide at 42 C and washed with 0.2
X
SSC, 0.1 % SDS at 65 C and exposed to X-ray film (Kodak).
Fig. 6A presents mouse polyA+ RNA blots of adult tissues (left) and
embryos (right) probed with full length Dnmt3a, Dnmt3b and a control (3-actin
cDNA probe. Each lane contains 2 g of polyA+ RNA. (Ht, Heart; Br, Brain; Sp,
Spleen; Lu, Lung; Li, Liver; Mu, Skeletal Muscle; Ki, Kidney; Te, Testis; and
embryos at gestation days 7 (E7), 11 (E11), 15 (E15). and 17 (E17). Fig. 6B is
a mouse total RNA blot (10 g per lane) of ES cell and adult organ RNA samples
and Fig. 6C shows a mouse total RNA blot (20 g per lane) of undifferentiated
(Undiff.) and differentiated (Diff.) ES cells RNA hybridized to Dnmt3a, Dnmt3b
or P-actin probes.
It has been shown that the maintenance methylation activity is
constitutively present in proliferating cells, whereas the de novo methylation
activity is highly regulated. Active de novo methylation has been shown to
occur
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-61-
primarily in ES cells (or embryonic carcinoma cells), early postimplantation
embryos and primordial germ cells (Jahaner, D. and Jaenish, R., "DNA
Methylation in Early Mammalian Development," In DNA Methylation:
Biochemistry and Biological Significance, Razin, A. et al., eds., Springer-
Verlag
(1984) pp. 189-219; Razin, A., and Cedar, H., "DNA Methylation and
Embryogenesis," in DNA Methylation: Molecular Biology and Biological
Significance, Jost., J. P. etal., eds., Birkhauser Verlag, Basel, Switzerland
(1993)
pp. 343-3 57; Chaillet, J. R. et al., Ce1166:77-83 (1991;); and Li, E. "Role
of DNA
Methylation in Development," in Genomic Imprinting: Frontiers in Molecular
Biology, Reik, W. and Sorani, A. eds., IRL Press, Oxford (1997) pp. 1-20). The
expression of both Dnmt3a and Dnmt3b in mouse embryos, adult tissues and ES
cells was examined. The results indicate that two Dnmt3a transcripts, 9.5 kb
and
4.2kb, are present in embryonic and adult tissue RNA. The 4.2 kb transcript,
corresponding to the size of the full length cDNA, was expressed at very low
levels in most tissues, except for the E11.5 embryo sample (Fig. 6A). A single
4.4 kb Dnmt3b transcript is detected in embryo and adult organ RNAs, with
relatively high levels in testes and E 11.5 embryo samples (Fig. 6A).
Interestingly,
both genes are expressed at much higher levels in ES cells than in adult
tissues
(Fig. 6B), and their expression decreased dramatically upon differentiation of
ES
cells in culture (Fig. 6C). In addition, Dnmt3a and Dnmt3b expression levels
are
unaltered in Dnmtl-deficient ES cells (Fig. 6C), suggesting that regulation of
Dnmt3a and Dnmt3b expression is independent of Dnmtl.
These results suggest that both Dnmt3a and Dnmt3b are expressed
specifically in ES cells and El 1.5 embryo and/or testes. The expression in
the
E11.5 embryo and testes may correlate with the presence of developing or
mature
germ cells in these tissues. Therefore, the expression pattern of Dnmt3a and
Dnmt3b appears to correlate well with de novo methylation activities in
development.
For the RNA expression analysis of human DNMT3 genes, polyA+ RNA
blots were hybridized using DNMT3A and DNMT3B cDNA fragments as probes.
CA 02331781 2004-08-12
-62-
Results indicate that DNMT3A RNA was expressed ubiquitously and was readily
detected in most tissues examined at levels slightly lower than DNMTI RNA
(Fig. 9). Three major DNMT3A transcripts, approximately 4.0, 4.4, and 9.5 kb,
were detected. The relative expression level of the transcripts appeared to
vary
from tissue to tissue. Transcripts of similar sizes were also detected in
mouse
tissues. Results utilizing DNMT3B cDNA probes indicate that transcripts of
about 4.2 kb were expressed at much lower levels in most tissues, but could be
readily detected in the testis, thyroid and bone marrow (Fig. 9). Sequence
analyses of different cDNA clones indicate the presence of alternatively
spliced
transcripts, although the size differences between these transcripts are too
small
to be detected by Northern analysis.
Hypermethylation of tumor suppressor genes is a common epigenetic
lesion found in tumor cells (Laird, P. W. & Jaenisch, R., Ann. Rev. Genet.
30:441-
464 (1996); Baylin, S.B., Adv. Cancer Res. 72:141-196 (1998)). To investigate
whether DNMT3A and DNMT38 ann abnormally activated in tumor cells,
DNMT3 RNA expression was analyzed in several tumor cell lines by Northern
blot hybridization. Results demonstrated that DNMT3A was expressed at higher
levels in most tumor cell lines examined. (Figure 10). As in the normal
tissues,
three different size transcripts were also detected in tumor cells. The ratio
of
these transcripts appeared to be variable in different tumor cell lines.
DNMT3B
expression was dramatically elevated in most tumor cell lines examined though
it was expressed at very low levels in normal adult tissues (Figure 10). The
expression levels of both DNMT3A and DNMT3B appear to be comparable and
proportional to that of DNMTI.
The murine Dnmt3a and Dnmt3b genes are highly expressed in
undifferentiated ES cells, consistent with their potential role in de novo
methylation during early embryonic development. Additionally, both genes are
highly expressed in early embryos. Differences in their expression patterns in
adult tissues in both human and mice suggest that each gene may have a
distinct
function in somatic tissues and may methylate different genes or genomic
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-63-
sequences. The elevated expression of DNMT3 genes in human tumor cell lines
suggests that the DNMT3 enzyme may be responsible for de novo methylation of
CpG islands in tumor suppressor genes during tumor formation.
Example 4: Methyltransferase Activity Assay
In order to demonstrate DNA cytosine methyltransferase activity, the
polypeptides of the invention were expressed and purified from recombinant
host
cells for use in in vitro assays.
Infected insect Sf21 cells and NIH3T3 cells were homogenized by
ultrasonication in lysis solution (20 mM Tris-HCI, p1-17.4, 10 mM EDTA, 500
mM NaC1, 10% glycerol, 1mM DTT, IrnM PMSF, 1 ug/ml leupeptin, 10 ug/ml
TPCK, 10 ug/ml TLCK) and cleared by centrifugation at 100,000 g for 20 min.
The methyltransferase enzyme assay was carried out as described
previously (Lei, H. et al., Development 122:3195-3205 (1996)). DNA substrates
used in the assays include: poly (dI-dC), poly (dG-dC) (Pharmacia Biotech),
lambda phage DNA (Sigma), pBluescriptIlSK (Stratagene, CA), pMu3 plasmid,
which contains tandem repeats of 535bp RsaI-Rsal fcagment of MMLV LTR
region in pUC9, and oligonucleotides. The oligonucleotide sequences utilized
include:
#1, 5'-AGACMGGTGCCAGMGCAGCTGAGCMGGATC-3',
#2, 5'-GATCMGGCTCAGCTGMGCTGGCACMGGTCT-3',
#3, 5'-AGACCGGTGCCAGCGCAGCTGAGCCGGATC-3', and
#4, 5'-GATCCGGCTCAGCTGCGCTGGCACCGGTCT-3' (M represents 5-
methylcytosine).
These sequences are the same as described in a previous study (Pradhan,
S. et al., Nucleic Acids Res. 25:4666-4673 (1997)). Oligonucleotides were
synthesized and purified by polyacrylamide gel electrophoresis (PAGE). To
make double strand oligonucleotides, equimolar amounts of the two
complimentary oligonucleotides were heated at 94"C for 10 min., mixed,
CA 02331781 2004-08-12
-64-
incubated at 78 C for 1 hr and cooled down slowly at room temperature. The
annealing products were quantified for the yield of double-stranded
oligonucleotides (dsDNA) by PAGE and methylene blue staining. In all cases,
the yield of dsDNA was higher than 95%. The dsDNA of #1 and #2 were used
as 'fully' methylated substrates, dsDNA of #1 and #4 as the hemi-methylated
substrates, and dsDNA of #3 and #4 as unmethylated substrates.
For Southern analysis of the methylation of retrovirus DNA, 2 ug of
pMMLV8.3, an 8.3kb Hind III fragment of Moloney murine leukemia virus
cDNA in pBluescriptlISK, was methylated in vitro for 15 hrs under the same
reaction conditions described above except that 160 uM of cold SAM
(S-adenosyl-L-Met) was used instead of 3H-methyl SAM. Then, an equal
volume of the solution containing 1% SDS, 400 mM NaC1, and 0.2 mg/ml
Proteinase K was added, and the sample was incubated at 37 C for 1 hr. After
phenol/chloroform extraction, DNA was precipitated with ethanol, dried and
dissolved in TE buffer. This procedure was repeated 5 times. An aliquot of
DNA was purified after the first, third and fifth reaction, digested with Hpa
II
or Msp I in combination with Kpn I for 16 hrs, separated on 1% agarose gels,
blotted and hybridized to the pMu3 probe.
In a standard methyltransferase assay, enzyme activity was detected
with protein extracts from Sf21 cells overexpressing Dnmt3a and Dnmt3b
polypeptides. Similar to the results obtained with the Dnmtl polypeptide, the
overexpressed Dnmt3 proteins were able to methylate various native and
synthetic DNA substrates, among which poly(dI-dC) consistently gave rise to
the highest initial velocity (Fig. 7a). An analysis of the methylation of Hpa
II
sites in retroviral DNA by these enzymes was also performed. An MMLV full
length cDNA was methylated for 1-5 times by incubation with protein extract
from control Sf21 cells or Sf21 cells infected with baculoviruses expressing
Dnmtl, Dnmt3a or Dnmt3b polypeptides. The Hpa II/Msp I target sequence,
CCGG, is resistant to the Hpa II restriction enzyme, but sensitive to Msp I
digestion when the internal C is methylated, and the restriction site becomes
resistant to Msp I digestion when the external C is methylated (Jentsch, S. et
al.,
Nucleic Acids Res.
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
-65-
9:2753-2759 (1981)). Both Dnmt3a and Dnmt3b polypeptides could methylate
multiple Hpa II sites in the 3' LTR regions of the MMLV DNA, as indicated by
the presence of Hpa II-resistant fragments, though less efficiently than Dnmtl
polypeptide (Fig. 7b). Significantly, even after five consecutive rounds of in
vitro
methylation, the viral DNA was completely digested by Msp I. This result
indicates that both Dnmt3a and Dnmt3b polypeptides methylate predominantly
the internal cytosine residues, therefore, CpGs. Previously it was shown that
the
same region of the proviral DNA was efficiently methylated in Dnmtl null ES
cells infected by the MMLV virus (Lei, H. et al., Development 122:3195-3205
(1996)).
Fig. 7A shows 'H-methyl incorporation into different DNA substrates
(poly (dI-dC), poly (dG-dC) (squares), lambda phage DNA (circles),
pBluescriptIISK (triangles), and pMu3 (diamonds)) when incubated with protein
extracts of Sf21 cells expressing Dnmtl, Dnmt3a, or Dnmt3bl. Fig. 7B shows
Southern blot analysis of the in vitro methylation of untreated pMMLV DNA
(lanes 1-3) and pMMLV DNA incubated with MT1 (lane 4-10), MT3a (lanes 11-
15), MT3(3 (lanes 16-20) or control Sf21 (lanes 21-25) extracts that were
digested
with Kpn I(K), Kpn I and Msp I(K/M) or Kpn I and Hpa II (K/H). Restriction
enzyme digested samples were then subjected to Southern blot analysis using
the
pMu3 probe.
Dnmtl protein appears to function primarily as a maintenance
methyltransferase because of its strong preference for hemimethylated DNA and
direct association with newly replicated DNA (Leonhardt, H. et al., Cell
71:865-
873 (1992)). To determine whether Dnmt3a and Dnmt3b polypeptides show any
preference for hemimethylated DNA over unmethylated DNA, a comparison was
done to examine the methylation rate of unmethylated versus hemimethylated
oligonucleotides. Gel-purified double stranded oligonucleotides were incubated
with protein extracts of Sf21 cells expressing Dnmtl, Dnmt3a, Dnmt3bl,
Dnmt3b2 or NIH3T3 cell extract (unmethylated substrates (open circles), hemi-
methylated substrates (half black diamonds) or completely methylated
substrates
CA 02331781 2000-12-21
WO 99/67397 PCTIUS99/14373
-66-
(closed squares)). While baculovirus-expressed Dnmt 1 polypeptide or 3T3 cell
extract showed much higher activities when hemimetliylated DNA was used as
a substrate, Dnmt3a, Dnmt3bl and Dnmt3b2 polypeptides showed no detectable
preference for hemimethylated DNA (Fig. 8).
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373 -
66-1
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule 13bis)
A. The indications made below relate to the microorganism referred to in the
description on page 16, line 1.
B. IDENTIFICATION OF DEPOSIT Further deposits are id~ntified on an additional
sheet
Name of depositary institution
American Type Culture Collection (ATCC )
Address of depositary institution (including postal code and country)
10801 University Boulevard
Manassas, Virginia 20110-2209
United States of America
Date of deposit June 16. 1998 Accession Number 209933
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is
continued on an additional sheet
pMT3a cDNA clone in bacterial cell DH5alpha
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (i>'the indications are
notjor all designated States)
E. SEPARATE FURNISHING OF INDICATIONS tlemY b1ank lnnropplieabte~
The indications listed below will be submitted to the intemational Bureau
later (specifv the general narure of the indications, e.g.,
"Accession Number ojDeposiJ %
For receiving Office use only For Intemational Bureau use only
YZ This sheet was received with the internationai application ~ This sheet was
received by the lntemational Bureau on:
Authorized officer Authorized officer
SGlljlt n - I
PCT Inlemadpnsl piyision
Form PCT/RO/134 (July 1992)
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
66-2
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule 13bis)
A. The indications made below relate to the microorganism referred to in the
description on page 16, line I.
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional
sheet
Name of depositary institution
American Type Culture Collection (ATCC )
Address of depositary institution (including postal code and country)
10801 University Boulevard
Manassas, Virginia 20110-2209
United States of America
Date of deposit June 16. 1998 j Accession Number 209934
C. ADDITIONAL INDICATIONS (leave blank iJnot applicable) This infortnation is
continued on an additional sheet 0
pMT3b cDNA clone in bacterial cell DH5alpha
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (ijthe indications are nor
jor all designated States)
E. SEPARATE FURNISHING OF INDICATIONS neave btank ynosappdcabrei
The indications listed below will be submitted to the intemational Bureau
later (specijv the generai nature of the indications, e.g.,
"Accession Number ojDeposit')
For receivine Office use only For international Bureau use only
)14-This sheet was received with the intemational application ~ This sheet was
received by the intemational Bureau on:
Authorize officerem mm Authorized officer
PCT lntematlorral Dlvlai=
Fonn PCT/RO/134 (July 1992)
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373 "
66-3
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule 13bis)
A. The indications made below relate to the microorganism referred to in the
description on page 16, line 1.
B. IDENTIFICATION OF DEPOSIT Furthcr deposits are identified on an additional
sheet ~
Name of depositary institution
American Type Culture Collection (ATCC )
Address of depositary institution (including postal code and country)
10801 University Boulevard
Manassas, Virginia 20110-2209
United States of America
Date of deposit July 10, 1998 Accession Number 98809
C. ADDITIONAL INDICATIONS (leave blank ijnot applicable) This information is
continued on an additional sheet ~
pMT3A (human) cDNA clone in bacterial cell DH5alpha
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (f the indications are
notjor all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leare blank Inot applicabrel
The indications listed below will be submitted to the intemational Bureau
later (specifv the general nature ojthe indications, e.g.,
"Accession iti'umber ojDeposil')
For receiving Office use only For Intemational Bureau use onh)~1 This sheet
was reccived with the intemational application ~ This sheet was received by
the Intemational Bureau on:
Authorized officer Authorized officer
SCt" seMM
PCT rrnemadcnw olyisim
Fotm PCT/RO/l34 (July 1992)
CA 02331781 2000-12-21
WO 99/67397 PCTIUS99/14373
66-4
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule 13bis)
A. The indications made below relate to the microorganism referred to in the
description on page 16, line l.
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional
sheet ~
Name of depositary institution
American Type Culture Collection (ATCC)
Address of depositary institution (including postal code and country)
10801 University Boulevard
Manassas, Virginia 20 1 1 0-2209
United States of America
Date of deposit July 10, 1998 Accession Number 98809
C. ADDITIONAL INDICATIONS (leave blank ijnot applicable) This information is
continued on an additional sheet ~
pMT3A (human) cDNA clone in bacterial cell DH5alpha
In respect of those designations in which a European Patent is sought a sample
of the deposited microorganism will be made available
until the publication of the mention of the grant of the European patent or
until the date on which the application has been refused or
withdrawn or is deemed to be withdrawn, only by the issue of such a sample to
an expert nominated by the person requesting the
sample (Rule 28(4) EPC).
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (iJthe indications are nar
jor all designared States)
E. SEPARATE FURNISHING OF INDICATIONS Ilea+-e blank inoa applieab/e)
The indications listed below will be submitted to the intemational Bureau
later (specify the general nature ojthe indications, e.g.,
"Accession Number ojDeposit')
For receiving Office use only For International Bureau use only
This sheet was received with the international application ~ This sheet was
received by the international Bureau on:
Authorizcd o~ ~ Authorized officer
=PCf1T tntematloRW DlyeSbg
Fortn PCT/RO/134 (July 1992)
CA 02331781 2000-12-21
WO 99/67397 PCT/US99/14373
66-5
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM.
(PCT Rule 13bis)
A. The indications made below relate to the microorganism referred to in the
description on page 16, line 1.
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional
sheet
Name of depositary institution
American Type Culture Collection (ATCC)
Address of depositary institution (including postal code and country)
10801 University Boulevard
Manassas, Virginia 20 1 1 0-2209
United States of America
Date of deposit June 16, 1998 Accession Number 209933
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This infotmation is
continued on an additional sheet ~
pMT3a cDNA clone in bacterial cell DH5alpha
In respect of those designations in which a European Patent is sought a sample
of the deposited microorganism will be made available
until the publication of the mention of the grant of the European patent or
until the date on which the application has been refused or
withdrawn or is deemed to be withdrawn, only by the issue of such a sampie to
an expert nominated by the person requesting the
sample (Rule 28(4) EPC).
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (itthe indications are not
for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS tteavebtank;nm appi;eabie/
The indications listed below will be submitted to the intemational Bureau
later (specify the general nature of the indications, e.g.,
"Accession Number of Deposit")
For recciving Office use only For International Bureau use only
IK This sheet was received with the intemational application ~ This sheet was
received by the lnternational Bureau on:
Authorized ofticer Authorized officer
80f" ftf=
PCT tiftmaduua! OK41ple
Form PCTIRO/134 (July 1992)
CA 02331781 2000-12-21
WO 99/67397. PCT/US99/14373
66-6
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule 13bis)
A. The indications made below relate to the microorganism referred to in the
description on page line
B. IDENTIFICATION OF DEPOSIT Funher deposits are identified on an additional
sheet
Name of depositary institution
American Type Culture Collection (ATCC)
Address of depositary institution (including postal code and country)
10801 University Boulevard
Manassas, Virginia 20 1 1 0-2209
United States of America
Date of deposit June 16, 1998 Accession Number 209934
C. ADDITIONAL INDICATIONS /leave blank ijnot applicable) This information is
continued on an additional sheet 0
pMT3b cDNA clone in bacterial cell DH5alpha
In respect of those designations in which a European Patent is sought a sample
of the deposited microorganism will be made available
until the publication of the mention of the grant of the European patent or
until the date on which the application has been refused or
withdrawn or is deemed to be withdrawn, only by the issue of such a sample to
an expert nominated by the person requesting the
sample (Rule 28(4) EPC).
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (f the indications are not
jor all designated States)
E. SEPARATE FURNISHING OF INDICATIONS tteme btmik t/naappiscab/e/
The indications listed below will be submitted to the intemational Bureau
later (specify the general nature of the indications, e.g..
"Accession Number ojDeposit')
For receiving Office use only For International Bureau use only
This sheet was received with the intemational application ~ This sheet was
received by the Intemational Bureau on:
Authorized officer Authorized officer
~ mmbpw omaim
Form PCT/RO/134 (July 1992)
CA 02331781 2005-04-22
-~.-
SEQUENCE LISTING
<110> The General Hospital Corporation
<120> De Novo DNA Cytosine Methyltransferase Genes,
Polypeptides & Uses Thereof
<130> 184-336
<140> CA 2,331,781
<141> 1999-06-25
<150> PCT/US99/14373
<151> 1999-06-25
<150> 60/090,906
<151> 1998-06-25
<150> 60/093,993
<151> 1998-07-24
<160> 82
<170> PatentIn Ver. 2.0
<210> 1
<211> 4192
<212> DNA
<213> Mus musculus
<220>
<221> Unsure
<222> (4161) .. (4161)
<223> May be any nucleic acid
<400> 1
gaattccggc ctgctgccgg gccgcccgac ccgccgggcc acacggcaga gccgcctgaa 60
gcccagcgct gaggctgcac ttttccgagg gcttgacatc agggtctatg tttaagtctt 120
agctcttgct tacaaagacc acggcaattc cttctctgaa gccctcgcag ccccacagcg 180
ccctcgcagc cccagcctgc cgcctactgc ccagcaatgc.cctccagcgg ccccggggac 240
accagcagct cctctctgga gcgggaggat gatcgaaagg aaggagagga acaggaggag 300
CA 02331781 2005-04-22
-2-
aaccgtggca aggaagagcg ccaggagccc agcgccacgg cccggaaggt ggggaggcct 360
ggccggaagc gcaagcaccc accggtggaa agcagtgaca cccccaagga cccagcagtg 420
accaccaagt ctcagcccat ggcccaggac tctggcccct cagatctgct acccaatgga 480
gacttggaga agcggagtga accccaacct gaggaaggga gcccagctgc agggcagaag 540
ggtggggccc cagctgaagg agagggaact gagaccccac cagaagcctc cagagctgtg 600
gagaatggct gctgtgtgac caaggaaggc cgtggagcct ctgcaggaga gggcaaagaa 660
cagaagcaga ccaacatcga atccatgaaa atggagggct cccggggccg actgcgaggt 720
ggcttgggct gggagtccag cctccgtcag cgacccatgc caagactcac cttccaggca 780
ggggacccct actacatcag caaacggaaa cgggatgagt ggctggcacg ttggaaaagg 840
gatgctgaga agaaagccaa ggtaattgca gtaatgaatg ctgtggaaga gaaccaggcc 900
tctggagagt ctcagaaggt ggaggaggcc agccctcctg ctgtgcagca gcccacggac 960
cctgcttctc cgactgtggc caccacccct gagccagtag gaggggatgc tggggacaag 1020
aatgctacca aagcacccga cgatgagcct gagtatgagg atggccgggg ctttggcatt 1080
ggagagctgg tgtgggggaa acttcggggt ttctcttggt ggccaggccg aattgtgtct 1140
tggtggatga caggccggag ccgagcagct gaaggcactc gctgggtcat gtggttcgga 1200
gatggcaagt tctcagtggt gtgtgtggag aagctcatgc cgctgagctc cttctgcagt 1260
gcattccacc aggccaccta caacaagcag cccatgtacc gcaaagccat ctacgaagtc 1320
ctccaggtgg ccagcagccg tgccgggaag ctgtttccag cttgccatga cagtgatgaa 1380
agtgacagtg gcaaggctgt ggaagtgcag aacaagcaga tgattgaatg ggccctcggt 1440
ggcttccagc cctcgggtcc taagggcctg gagccaccag aagaagagaa gaatccttac 1500
aaggaagttt acaccgacat gtgggtggag cctgaagcag ctgcttacgc cccaccccca 1560
ccagccaaga aacccagaaa gagcacaaca gagaaaccta aggtcaagga gatcattgat 1620
gagcgcacaa gggagcggct ggtgtatgag gtgcgccaga agtgcagaaa catcgaggac 1680
atttgtatct catgtgggag cctcaatgtc accctggagc acccattctt cattggaggc 1740
atgtgccaga actgtaagaa ctgcttcttg gagtgtgctt accagtatga cgacgatggg 1800
taccagtcct attgcaccat ctgctgtggg gggcgtgaag tgctcatgtg tgggaacaac 1860
aactgctgca ggtgcttttg tgtcgagtgt gtggatctct tggtggggcc aggagctgct 1920
caggcagcca ttaaggaaga cccctggaac tgctacatgt gcgggcataa gggcacctat 1980
gggctgctgc gaagacggga agactggcct tctcgactcc agatgttctt tgccaataac 2040
catgaccagg aatttgaccc cccaaaggtt tacccacctg tgccagctga gaagaggaag 2100
cccatccgcg tgctgtctct ctttgatggg attgctacag ggctcctggt gctgaaggac 2160
ctgggcatcc aagtggaccg ctacattgcc tccgaggtgt gtgaggactc catcacggtg 2220
ggcatggtgc ggcaccaggg aaagatcatg tacgtcgggg acgtccgcag cgtcacacag 2280
aagcatatcc aggagtgggg cccattcgac ctggtgattg gaggcagtcc ctgcaatgac 2340
ctctccattg tcaaccctgc ccgcaaggga ctttatgagg gtactggccg cctcttcttt 2400
gagttctacc gcctcctgca tgatgcgcgg cccaaggagg gagatgatcg ccccttcttc 2460
tggctctttg agaatgtggt ggccatgggc gttagtgaca agagggacat ctcgcgattt 2520
cttgagtcta accccgtgat gattgacgcc aaagaagtgt ctgctgcaca cagggcccgt 2580
tacttctggg gtaaccttcc tggcatgaac aggcctttgg catccactgt gaatgataag 2640
CA 02331781 2005-04-22
-3 -
ctggagctgc aagagtgtct ggagcacggc agaatagcca agttcagcaa agtgaggacc 2700
attaccacca ggtcaaactc tataaagcag ggcaaagacc agcatttccc cgtcttcatg 2760
aacgagaagg aggacatcct gtggtgcact gaaatggaaa gggtgtttgg cttccccgtc 2820
cactacacag acgtctccaa catgagccgc ttggcgaggc agagactgct gggccgatcg 2880
tggagcgtgc cggtcatccg ccacctcttc gctccgctga aggaatattt tgcttgtgtg 2940
taagggacat gggggcaaac tgaagtagtg atgataaaaa agttaaacaa acaaacaaac 3000
aaaaaacaaa acaaaacaat aaaacaccaa gaacgagagg acggagaaaa gttcagcacc 3060
cagaagagaa aaaggaattt aaagcaaacc acagaggagg aaaacgccgg agggcttggc 3120
cttgcaaaag ggttggacat catctcctga gttttcaatg ttaaccttca gtcctatcta 3180
aaaagcaaaa taggcccctc cccttcttcc cctccggtcc taggaggcga actttttgtt 3240
ttctactctt tttcagaggg gttttctgtt tgtttgggtt tttgtttctt gctgtgactg 3300
aaacaagaga gttattgcag caaaatcagt aacaacaaaa agtagaaatg ccttggagag 3360
gaaagggaga gagggaaaat tctataaaaa cttaaaatat tggttttttt tttttttcct 3420
tttctatata tctctttggt tgtctctagc ctgatcagat aggagcacaa acaggaagag 3480
aatagagacc ctcggaggca gagtctcctc tcccaccccc cgagcagtct caacagcacc 3540
attcctggtc atgcaaaaca gaacccaact agcagcaggg cgctgagaga acaccacacc 3600
agacactttc tacagtattt caggtgccta ccacacagga aaccttgaag aaaaccagtt 3660
tctagaagcc gctgttacct cttgtttaca gtttatatat atatgataga tatgagatat 3720
atatatataa aaggtactgt taactactgt acatcccgac ttcataatgg tgctttcaaa 3780
acagcgagat gagcaaagac atcagcttcc gcctggccct ctgtgcaaag ggtttcagcc 3840
caggatgggg agaggggagc agctggaggg ggttttaaca aactgaagga tgacccatat 3900
caccccccac ccctgcccca tgcctagctt cacctgccaa aaaggggctc agctgaggtg 3960
gtcggaccct ggggaagctg agtgtggaat ttatccagac tcgcgtgcaa taaccttaga 4020
atatgaatct aaaatgactg cctcagaaaa atggcttgag aaaacattgt ccctgatttt 4080
gaattcgtca gccacgttga aggccccttg tgggatcaga aatattccag agtgagggaa 4140
agtgacccgc cattaacccc ncctggagca aataaaaaaa catacaaaat gt 4192
<210> 2
<211> 4195
<212> DNA
<213> Mus musculus
<400> 2
gaattccggg cgccggggtt aagcggccca agtaaacgta gcgcagcgat cggcgccgga 60
gattcgcgaa cccgacactc cgcgccgccc gccggccagg acccgcggcg cgatcgcggc 120
gccgcgctac agccagcctc acgacaggcc cgctgaggct tgtgccagac cttggaaacc 180
tcaggtatat acctttccag acgcgggatc tcccctcccc catccatagt gccttgggac 240
caaatccagg gccttctttc aggaaacaat gaagggagac agcagacatc tgaatgaaga 300
agagggtgcc agcgggtatg aggagtgcat tatcgttaat gggaacttca gtgaccagtc 360
CA 02331781 2005-04-22
-4-
ctcagacacg aaggatgctc cctcaccccc agtcttggag gcaatctgca cagagccagt 420
ctgcacacca gagaccagag gccgcaggtc aagctcccgg ctgtctaaga gggaggtctc 480
cagccttctg aattacacgc aggacatgac aggagatgga gacagagatg atgaagtaga 540
tgatgggaat ggctctgata ttctaatgcc aaagctcacc cgtgagacca aggacaccag 600
gacgcgctct gaaagcccgg ctgtccgaac ccgacatagc aatgggacct ccagcttgga 660
gaggcaaaga gcctccccca gaatcacccg aggtcggcag ggccgccacc atgtgcagga 720
gtaccctgtg gagtttccgg ctaccaggtc tcggagacgt cgagcatcgt cttcagcaag 780
cacgccatgg tcatcccctg ccagcgtcga cttcatggaa gaagtgacac ctaagagcgt 840
cagtacccca tcagttgact tgagccagga tggagatcag gagggtatgg ataccacaca 900
ggtggatgca gagagcatat atggagacag cacagagtat caggatgata aagagtttgg 960
aataggtgac ctcgtgtggg gaaagatcaa gggcttctcc tggtggcctg ccatggtggt 1020
gtcctggaaa gccacctcca agcgacaggc catgcccgga atgcgctggg tacagtggtt 1080
tggtgatggc aagttttctg agatctctgc tgacaaactg gtggctctgg ggctgttcag 1140
ccagcacttt aatctggcta ccttcaataa gctggtttct tataggaagg ccatgtacca 1200
cactctggag aaagccaggg ttcgagctgg caagaccttc tccagcagtc ctggagagtc 1260
actggaggac cagctgaagc ccatgctgga gtgggcccac ggtggcttca agcctactgg 1320
gatcgagggc ctcaaaccca acaagaagca accagtggtt aataagtcga aggtgcgtcg 1380
ttcagacagt aggaacttag aacccaggag acgcgagaac aaaagtcgaa gacgcacaac 1440
caatgactct gctgcttctg agtccccccc acccaagcgc ctcaagacaa atagctatgg 1500
cgggaaggac cgaggggagg atgaggagag ccgagaacgg atggcttctg aagtcaccaa 1560
caacaagggc aatctggaag accgctgttt gtcctgtgga aagaagaacc ctgtgtcctt 1620
ccaccccctc tttgagggtg ggctctgtca gagttgccgg gatcgcttcc tagagctctt 1680
ctacatgtat gatgaggacg gctatcagtc ctactgcacc gtgtgctgtg agggccgtga 1740
actgctgctg tgcagtaaca caagctgctg cagatgcttc tgtgtggagt gtctggaggt 1800
gctggtgggc gcaggcacag ctgaggatgc caagctgcag gaaccctgga gctgctatat 1860
gtgcctccct cagcgctgcc atggggtcct ccgacgcagg aaagattgga acatgcgcct 1920
gcaagacttc ttcactactg atcctgacct ggaagaattt gagccaccca agttgtaccc 1980
agcaattcct gcagccaaaa ggaggcccat tagagtcctg tctctgtttg atggaattgc 2040
aacggggtac ttggtgctca aggagttggg tattaaagtg gaaaagtaca ttgcctccga 2100
agtctgtgca gagtccatcg ctgtgggaac tgttaagcat gaaggccaga tcaaatatgt 2160
caatgacgtc cggaaaatca ccaagaaaaa tattgaagag tggggcccgt tcgacttggt 2220
gattggtgga agcccatgca atgatctctc taacgtcaat cctgcccgca aaggtttata 2280
tgagggcaca ggaaggctct tcttcgagtt ttaccacttg ctgaattata cccgccccaa 2340
ggagggcgac aaccgtccat tcttctggat gttcgagaat gttgtggcca tgaaagtgaa 2400
tgacaagaaa gacatctcaa gattcctggc atgtaaccca gtgatgatcg atgccatcaa 2460
ggtgtctgct gctcacaggg cccggtactt ctggggtaac ctacccggaa tgaacaggcc 2520
cgtgatggct tcaaagaatg ataagctcga gctgcaggac tgcctggagt tcagtaggac 2580
agcaaagtta aagaaagtgc agacaataac caccaagtcg aactccatca gacagggcaa 2640
aaaccagctt ttccctgtag tcatgaatgg caaggacgac gttttgtggt gcactgagct 2700
CA 02331781 2005-04-22
-5-
cgaaaggatc ttcggcttcc ctgctcacta cacggacgtg tccaacatgg gccgcggcgc 2760
ccgtcagaag ctgctgggca ggtcctggag tgtaccggtc atcagacacc tgtttgcccc 2820
cttgaaggac tactttgcct gtgaatagtt ctacccagga ctggggagct ctcggtcaga 2880
gccagtgccc agagtcaccc ctccctgaag gcacctcacc tgtccccttt ttagctcacc 2940
tgtgtggggc ctcacatcac tgtacctcag ctttctcctg ctcagtggga gcagagcctc 3000
ctggcccttg caggggagcc ccggtgctcc ctccgtgtgc acagctcaga cctggctgct 3060
tagagtagcc cggcatggtg ctcatgttct cttaccctga aactttaaaa cttgaagtag 3120
gtagtaagat ggctttcttt taccctcctg agtttatcac tcagaagtga tggctaagat 3180
accaaaaaaa caaacaaaaa cagaaacaaa aaacaaaaaa aaacctcaac agctctctta 3240
gtactcaggt tcatgctgca aaatcacttg agattttgtt tttaagtaac ccgtgctcca 3300
catttgctgg aggatgctat tgtgaatgtg ggctcagatg agcaaggtca aggggccaaa 3360
aaaaattccc cctctccccc caggagtatt tgaagatgat gtttatggtt taagtcttcc 3420
tggcaccttc cccttgcttt ggtacaaggg ctgaagtcct gttggtcttg tagcatttcc 3480
caggatgatg atgtcagcag ggatgacatc accaccttta gggcttttcc ctggcagggg 3540
cccatgtggc tagtcctcac gaagactgga gtagaatgtt tggagctcag gaagggtggg 3600
tggagtggcc ctcttccagg tgtgagggat acgaaggagg aagcttaggg aaatccattc 3660
cccactccct cttgccaaat gaggggccca gtccccaaca gctcaggtcc ccagaacccc 3720
ctagttcctc atgagaagct aggaccagaa gcacatcgtt ccccttatct gagcagtgtt 3780
tggggaacta cagtgaaaac cttctggaga tgttaaaagc tttttacccc acgatagatt 3840
gtgtttttaa ggggtgcttt ttttaggggc atcactggag ataagaaagc tgcatttcag 3900
aaatgccatc gtaatggttt ttaaacacct tttacctaat tacaggtgct attttataga 3960
agcagacaac acttcttttt atgactctca gacttctatt ttcatgttac catttttttt 4020
gtaactcgca aggtgtgggc ttttgtaact tcacaggtgt ggggagagac tgccttgttt 4080
caacagtttg tctccactgg tttctaattt ttaggtgcaa agatgacaga tgcccagagt 4140
ttacctttct ggttgattaa agttgtattt ctctaaaaaa aaaaaaaaaa aaaaa 4195
<210> 3
<211> 4416
<212> DNA
<213> Homo sapiens
<400> 3
ggccggcgtc gaccgacagc gagcggaggg agggagcgag cgagcgagca gcagcggccg 60
ggagggaggg agggcgcgcg ggcggcggcg gcggcgagag cagaggacga gccgggacgc 120
ggcgccgcgg caccagggcg cgcagccggg ccggcccgac cccaccggcc atacggtgga 180
gccatcgaag cccccaccca caggctgaca gaggcaccgt tcaccagagg gctcaacacc 240
gggatctatg tttaagtttt aactctcgcc tccaaagacc acgataattc cttccccaaa 300
gcccagcagc cccccagccc cgcgcagccc cagcctgcct cccggcgccc agatgcccgc 360
catgccctcc agcggccccg gggacaccag cagctctgct gcggagcggg aggaggaccg 420
CA 02331781 2005-04-22
-6-
aaaggacgga gaggagcagg aggagccgcg tggcaaggag gagcgccaag agcccagcac 480
cacggcacgg aaggtggggc ggcctgggag gaagcgcaag caccccccgg tggaaagcgg 540
tgacacgcca aaggaccctg cggtgatctc caagtcccca tccatggccc aggactcagg 600
cgcctcagag ctattaccca atggggactt ggagaagcgg agtgagcccc agccagagga 660
ggggagccct gctggggggc agaagggcgg ggccccagca gagggagagg gtgcagctga 720
gaccctgcct gaagcctcaa gagcagtgga aaatggctgc tgcaccccca aggagggccg 780
aggagcccct gcagaagcgg gcaaagaaca gaaggagacc aacatcgaat ccatgaaaat 840
ggagggctcc cggggccggc tgcggggtgg cttgggctgg gagtccagcc tccgtcagcg 900
gcccatgccg aggctcacct tccaggcggg ggacccctac tacatcagca agcgcaagcg 960
ggacgagtgg ctggcacgct ggaaaaggga ggctgagaag aaagccaagg tcattgcagg 1020
aatgaatgct gtggaagaaa accaggggcc cggggagtct cacaaggtgg aggaggccag 1080
ccctcctgct gtgcagcagc ccactgaccc cgcatccccc actgtggcta ccacgcctga 1140
gcccgtgggg tccgatgctg gggacaagaa tgccaccaaa gcaggcgatg acgagccaga 1200
gtacgaggac ggccggggct ttggcattgg ggagctggtg tgggggaaac tgcggggctt 1260
ctcctggtgg ccaggccgca ttgtgtcttg gtggatgacg ggccggagcc gagcagctga 1320
aggcacccgc tgggtcatgt ggttcggaga cggcaaattc tcagtggtgt gtgttgagaa 1380
gctgatgccg ctgagctcgt tttgcagtgc gttccaccag gccacgtaca acaagcagcc 1440
catgtaccgc aaagccatct acgaggtcct gcaggtggcc agcagccgcg cggggaagct 1500
gttcccggtg tgccacgaca gcgatgagag tgacactgcc aaggccgtgg aggtgcagaa 1560
caagcccatg attgaatggg ccctgggggg cttccagcat tatggcccta agggcctgga 1620
gccaccagaa gaagagaaga atccctacaa agaagtgtac acggacatgt gggtggaacc 1680
tgaggcagct gcatacgcac cacctccacc agccaaaaag ccccggaaga gcacagcgga 1740
gaagcccaag gtcaaggaga ttattgatga gcgcacaaga gagcggctgg tgtacgaggt 1800
gcggcagaag tgccggaaca ttgaggacat ctgcatctcc tgtgggagcc tcaatgttac 1860
cctggaacac cccctcttcg ttggaggaat gtgccaaaac tgcaagaact gctttctgga 1920
gtgtgcgtac cagtacgacg acgacggcta ccagtcctac tgcaccatct gctgtggggg 1980
ccgtgaggtg ctcatgtgcg gaaacaacaa ctgctgcagg tgcttttgcg tggagtgtgt 2040
ggacctcttg gtggggccgg gggctgccca ggcagccatt aaggaagacc cctggaactg 2100
ctacatgtgc gggcacaagg gtacctacgg gctgctgcgg cggcgaaagg actggccctc 2160
ccggctccag atgttcttcg ctaataacca cgaccaggaa tttgaccctc caaaggttta 2220
cccacctgtc ccagctgaga aaaggaagcc catccgggtg ctgtctctct ttgatggaat 2280
cgctacaggg ctcctggtgc tgaaggactt gggcattcag gtggaccgct acattgcctc 2340
ggaggtgtgt gaggactcca tcacggtggg catggtgcgg caccagggga agatcatgta 2400
cgtcggggac gtccgcagcg tcacacagaa gcatatccag gagtggggcc cattcgatct 2460
ggtgattggg ggcagtccct gcaatgacct ctccatcgtc aaccctgctc gcaagggcct 2520
ctacgagggc actggccggc tcttctttga gttctaccgc ctcctgcatg atgcgcggcc 2580
caaggaggga gatgatcgcc ccttcttctg gctctttgag aatgtggtgg ccatgggcgt 2640
tagtgacaag agggacatct cgcgatttct cgagtccaac cctgtgatga ttgatgccaa 2700
agaagtgtca gctgcacaca gggcccgcta cttctggggt aaccttcccg gtatgaacag 2760
CA 02331781 2005-04-22
-7-
gccgttggca tccactgtga atgataagct ggagctgcag gagtgtctgg agcatggcag 2820
gatagccaag ttcagcaaag tgaggaccat tactacgagg tcaaactcca taaagcaggg 2880
caaagaccag cattttcctg tcttcatgaa tgagaaagag gacatcttat ggtgcactga 2940
aatggaaagg gtatttggtt tcccagtcca ctatactgac gtctccaaca tgagccgctt 3000
ggcgaggcag agactgctgg gccggtcatg gagcgtgcca gtcatccgcc acctcttcgc 3060
tccgctgaag gagtattttg cgtgtgtgta agggacatgg gggcaaactg aggtagcgac 3120
acaaagttaa acaaacaaac aaaaaacaca aaacataata aaacaccaag aacatgagga 3180
tggagagaag tatcagcacc cagaagagaa aaaggaattt aaaacaaaaa ccacagaggc 3240
ggaaataccg gagggctttg ccttgcgaaa agggttggac atcatctcct gatttttcaa 3300
tgttattctt cagtcctatt taaaaacaaa accaagctcc cttcccttcc tcccccttcc 3360
cttttttttc ggtcagacct tttattttct actcttttca gaggggtttt ctgtttgttt 3420
gggttttgtt tcttgctgtg actgaaacaa gaaggttatt gcagcaaaaa tcagtaacaa 3480
aaaatagtaa caataccttg cagaggaaag gtgggaggag aggaaaaaag ggaaattttt 3540
aaagaaatct atatattggg ttgttttttt ttttgttttt tgtttttttt ttttgggttt 3600
ttttttttta ctatatatct tttttttgtt gtctctagcc tgatcagata ggagcacaag 3660
caggggacgg aaagagagag acactcaggc ggcagcattc cctcccagcc actgagctgt 3720
cgtgccagca ccattcctgg tcacgcaaaa cagaacccag ttagcagcag ggagacgaga 3780
acaccacaca agacattttt ctacagtatt tcaggtgcct accacacagg aaaccttgaa 3840
gaaaatcagt ttctagaagc cgctgttacc tcttgtttac agtttatata tatatgatag 3900
atatgagata tatatataaa aggtactgtt aactactgta caacccgact tcataatggt 3960
gctttcaaac agcgagatga gtaaaaacat cagcttccac gttgccttct gcgcaaaggg 4020
tttcaccaag gatggagaaa gggagacagc ttgcagatgg,cgcgttctca cggtgggctc 4080
ttccccttgg tttgtaacga agtgaaggag gagaacttgg gagccaggtt ctccctgcca 4140
aaaagggggc tagatgaggt ggtcgggccc gtggacagct gagagtggga ttcatccaga 4200
ctcatgcaat aaccctttga ttgttttcta aaaggagact ccctcggcaa gatggcagag 4260
ggtacggagt cttcaggccc agtttctcac tttagccaat tcgagggctc cttgtggtgg 4320
gatcagaact aatccagagt gtgggaaagt gacagtcaaa accccacctg gagcaaataa 4380
aaaaacatac aaaacgtaaa aaaaaaaaaa aaaaaa 4416
<210> 4
<211> 4145
<212> DNA
<213> Homo sapiens
<400> 4
ggccgcgaat tcggcacgag ccctgcacgg ccgccagccg gcctcccgcc agccagcccc 60
gacccgcggc tccgccgccc agccgcgccc cagccagccc tgcggcagga aagcatgaag 120
ggagacacca ggcatctcaa tggagaggag gacgccggcg ggagggaaga ctcgatcctc 180
gtcaacgggg cctgcagcga ccagtcctcc gactcgcccc caatcctgga ggctatccgc 240
CA 02331781 2005-04-22
-8-
accccggaga tcagaggccg aagatcaagc tcgcgactct ccaagaggga ggtgtccagt 300
ctgctaagct acacacagga cttgacaggc gatggcgacg gggaagatgg ggatggctct 360
gacaccccag tcatgccaaa gctcttccgg gaaaccagga ctcgttcaga aagcccagct 420
gtccgaactc gaaataacaa cagtgtctcc agccgggaga ggcacaggcc ttccccacgt 480
tccacccgag gccggcaggg ccgcaaccat gtggacgagt cccccgtgga gttcccggct 540
accaggtccc tgagacggcg ggcaacagca tcggcaggaa cgccatggcc gtcccctccc 600
agctcttacc ttaccatcga cctcacagac gacacagagg acacacatgg gacgccccag 660
agcagcagta ccccctacgc ccgcctagcc caggacagcc agcagggggg catggagtcc 720
ccgcaggtgg aggcagacag tggagatgga gacagttcag agtatcagga tgggaaggag 780
tttggaatag gggacctcgt gtggggaaag atcaagggct tctcctggtg gcccgccatg 840
gtggtgtctt ggaaggccac ctccaagcga caggctatgt ctggcatgcg gtgggtccag 900
tggtttggcg atggcaagtt ctccgaggtc tctgcagaca aactggtggc actggggctg 960
ttcagccagc actttaattt ggccaccttc aataagctcg tctcctatcg aaaagccatg 1020
taccatgctc tggagaaagc tagggtgcga gctggcaaga ccttccccag cagccctgga 1080
gactcattgg aggaccagct gaagcccatg ttggagtggg cccacggggg cttcaagccc 1140
actgggatcg agggcctcaa acccaacaac acgcaaccag tggttaataa gtcgaaggtg 1200
cgtcgtgcag gcagtaggaa attagaatca aggaaatacg agaacaagac tcgaagacgc 1260
acagctgacg actcagccac ctctgactac tgccccgcac ccaagcgcct caagacaaat 1320
tgctataaca acggcaaaga ccgaggggat gaagatcaga gccgagaaca aatggcttca 1380
gatgttgcca acaacaagag cagcctggaa gatggctgtt tgtcttgtgg caggaaaaac 1440
cccgtgtcct tccaccctct ctttgagggg gggctctgtc agacatgccg ggatcgcttc 1500
cttgagctgt tttacatgta tgatgacgat ggctatcagt cttactgcac tgtgtgctgc 1560
gagggccgag agctgctgct ttgcagcaac acgagctgct gccggtgttt ctgtgtggag 1620
tgcctggagg tgctggtggg cacaggcaca gcggccgagg ccaagcttca ggagccctgg 1680
agctgctaca tgtgtctccc gcagcgctgt catggcgtcc tgcggcgccg gaaggactgg 1740
aacgtgcgcc tgcaggcctt cttcaccagt gacacggggc ttgaatacga agcccccaag 1800
ctgtaccctg ccattcccgc agcccgaagg cggcccattc gagtcctgtc attgtttgat 1860
ggcatcgcga caggctacct agtcctcaaa gagttgggca taaaggtagg aaagtacgtc 1920
gcttctgaag tgtgtgagga gtccattgct gttggaaccg tgaagcacga ggggaatatc 1980
aaatacgtga acgacgtgag gaacatcaca aagaaaaata ttgaagaatg gggcccattt 2040
gacttggtga ttggcggaag cccatgcaac gatctctcaa atgtgaatcc agccaggaaa 2100
ggcctgtatg agggtacagg ccggctcttc ttcgaatttt accacctgct gaattactca 2160
cgccccaagg agggtgatga ccggccgttc ttctggatgt ttgagaatgt tgtagccatg 2220
aaggttggcg acaagaggga catctcacgg ttcctggagt gtaatccagt gatgattgat 2280
gccatcaaag tttctgctgc tcacagggcc cgatacttct ggggcaacct acccgggatg 2340
aacaggcccg tgatagcatc aaagaatgat aaactcgagc tgcaggactg cttggaatac 2400
aataggatag ccaagttaaa gaaagtacag acaataacca ccaagtcgaa ctcgatcaaa 2460
caggggaaaa accaactttt ccctgttgtc atgaatggca aagaagatgt tttgtggtgc 2520
actgagctcg aaaggatctt tggctttcct gtgcactaca cagacgtgtc caacatgggc 2580
CA 02331781 2005-04-22
-9-
cgtggtgccc gccagaagct gctgggaagg tcctggagcg tgcctgtcat ccgacacctc 2640
ttcgcccctc tgaaggacta ctttgcatgt gaatagttcc agccaggccc caagcccact 2700
ggggtgtgtg gcagagccag gacccaggag gtgtgattcc tgaaggcatc cccaggccct 2760
gctcttcctc agctgtgtgg gtcataccgt gtacctcagt tccctcttgc tcagtggggg 2820
cagagccacc tgactcttgc aggggtagcc tgaggtgccg cctccttgtg cacaaatcag 2880
acctggctgc ttggagcagc ctaacacggt gctcattttt tcttctccta aaactttaaa 2940
acttgaagta ggtagcaacg tggctttttt tttttccctt cctgggtcta ccactcagag 3000
aaacaatggc taagatacca aaaccacagt gccgacagct ctccaatact caggttaatg 3060
ctgaaaaatc atccaagaca gttattgcaa gagtttaatt tttgaaaact gggtactgct 3120
atgtgtttac agacgtgtgc agttgtaggc atgtagctac aggacatttt taagggccca 3180
ggatcgtttt ttcccagggc aagcagaaga gaaaatgttg tatatgtctt ttacccggca 3240
cattcccctt gcctaaatac aagggctgga gtctgcacgg gacctattag agtattttcc 3300
acaatgatga tgatttcagc agggatgacg tcatcatcac attcagggct attttttccc 3360
ccacaaaccc aagggcaggg gccactctta gctaaatccc tccccgtgac tgcaatagaa 3420
ccctctgggg agctcaggaa ggggtgtgct gagttctata atataagctg ccatatattt 3480
tgtagacaag tatggctcct ccatatctcc ctcttcccta ggagaggagt gtgaagcaag 3540
gagcttagat aagacacccc ctcaaaccca ttccctctcc aggagaccta ccctccacag 3600
gcacaggtcc ccagatgaga agtctgctac cctcatttct catcttttta ctaaactcag 3660
aggcagtgac agcagtcagg gacagacata catttctcat accttcccca catctgagag 3720
atgacaggga aaactgcaaa gctcggtgct ccctttggag attttttaat ccttttttat 3780
tccataagaa gtcgttttta gggagaacgg gaattcagac aagctgcatt tcagaaatgc 3840
tgtcataatg gtttttaaca ccttttactc ttcttactgg tgctattttg tagaataagg 3900
aacaacgttg acaagttttg tggggctttt tatacacttt ttaaaatctc aaacttctat 3960
ttttatgttt aacgttttca ttaaaatttt tttgtaactg gagccacgac gtaacaaata 4020
tggggaaaaa actgtgcctt gtttcaacag tttttgctaa tttttaggct gaaagatgac 4080
ggatgcctag agtttacctt atgtttaatt aaaatcagta tttgtctaaa aaaaaaaaaa 4140
aaaaa 4145
<210> 5
<211> 908
<212> PRT
<213> Mus musculus
<400> 5
Met Pro Ser Ser Gly Pro Gly Asp Thr Ser Ser Ser Ser Leu Glu Arg
1 5 10 15
Glu Asp Asp Arg Lys Glu Gly Glu Glu Gln Glu Glu Asn Arg Gly Lys
20 25 30
CA 02331781 2005-04-22
-10-
Glu Glu Arg Gln Glu Pro Ser Ala Thr Ala Arg Lys Val Gly Arg Pro
35 40 45
Gly Arg Lys Arg Lys His Pro Pro Val Glu Ser Ser Asp Thr Pro Lys
50 55 60
Asp Pro Ala Val Thr Thr Lys Ser Gln Pro Met Ala Gln Asp Ser Gly
65 70 75 80
Pro Ser Asp Leu Leu Pro Asn Gly Asp Leu Glu Lys Arg Ser Glu Pro
85 90 95
Gln Pro Glu Glu Gly Ser Pro Ala Ala Gly Gln Lys Gly Gly Ala Pro
100 105 110
Ala Glu Gly Glu Gly Thr Glu Thr Pro Pro Glu Ala Ser Arg Ala Val
115 120 125
Glu Asn Gly Cys Cys Val Thr Lys Glu Gly Arg Gly Ala Ser Ala Gly
130 135 140
Glu Gly Lys Glu Gln Lys Gln Thr Asn Ile Glu Ser Met Lys Met Glu
145 150 155 160
Gly Ser Arg Gly Arg Leu Arg Gly Gly Leu Gly Trp Glu Ser Ser Leu
165 170 175
Arg Gln Arg Pro Met Pro Arg Leu Thr Phe Gln Ala Gly Asp Pro Tyr
180 185 190
Tyr Ile Ser Lys Arg Lys Arg Asp Glu Trp Leu Ala Arg Trp Lys Arg
195 200 205
Asp Ala Glu Lys Lys Ala Lys Val Ile Ala Val Met Asn Ala Val Glu
210 215 220
Glu Asn Gln Ala Ser Gly Glu Ser Gln Lys Val Glu Glu Ala Ser Pro
225 230 235 240
CA 02331781 2005-04-22
-11-
Pro Ala Val Gin G1n Pro Thr Asp Pro Ala Ser Pro Thr Val Ala Thr
245 250 255
Thr Pro Glu Pro Val Gly Gly Asp Ala Gly Asp Lys Asn Ala Thr Lys
260 265 270
Ala Pro Asp Asp Glu Pro Glu Tyr Glu Asp Gly Arg Gly Phe Gly Ile
275 280 285
Gly Glu Leu Val Trp Gly Lys Leu Arg Gly Phe Ser Trp Trp Pro Gly
290 295 300
Arg Ile Val Ser Trp Trp Met Thr Gly Arg Ser Arg Ala Ala Glu Gly
305 310 315 320
Thr Arg Trp Val Met Trp Phe Gly Asp Gly Lys Phe Ser Val Val Cys
325 330 335
Val Glu Lys Leu Met Pro Leu Ser Ser Phe Cys Ser Ala Phe His Gln
340 345 350
Ala Thr Tyr Asn Lys Gln Pro Met Tyr Arg Lys Ala Ile Tyr Glu Val
355 360 365
Leu Gln Val Ala Ser Ser Arg Ala Gly Lys Leu Phe Pro Ala Cys His
370 375 380
Asp Ser Asp Glu Ser Asp Ser Gly Lys Ala Val Glu Val Gln Asn Lys
385 390 395 400
Gln Met Ile Glu Trp Ala Leu Gly Gly Phe Gln Pro Ser Gly Pro Lys
405 410 415
Gly Leu Glu Pro Pro Glu Glu Glu Lys Asn Pro Tyr Lys Glu Val Tyr
420 425 430
Thr Asp Met Trp Val Glu Pro Glu Ala Ala Ala Tyr Ala Pro Pro Pro
435 440 445
CA 02331781 2005-04-22
-12-
Pro Ala Lys Lys Pro Arg Lys Ser Thr Thr Glu Lys Pro Lys Val Lys
450 455 460
Glu Ile Ile Asp Glu Arg Thr Arg Glu Arg Leu Val Tyr Glu Val Arg
465 470 475 480
Gln Lys Cys Arg Asn Ile Glu Asp Ile Cys Ile Ser Cys Gly Ser Leu
485 490 495
Asn Val Thr Leu Glu His Pro Phe Phe Ile Gly Gly Met Cys Gln Asn
500 505 510
Cys Lys Asn Cys Phe Leu Glu Cys Ala Tyr Gln Tyr Asp Asp Asp Gly
515 520 525
Tyr Gin Ser Tyr Cys Thr Ile Cys Cys Gly Gly Arg Glu Val Leu Met
530 535 540
Cys Gly Asn Asn Asn Cys Cys Arg Cys Phe Cys Val Glu Cys Val Asp
545 550 555 560
Leu Leu Val Gly Pro Gly Ala Ala Gln Ala Ala Ile Lys Glu Asp Pro
565 570 575
Trp Asn Cys Tyr Met Cys Gly His Lys Gly Thr Tyr Gly Leu Leu Arg
580 585 590
Arg Arg Glu Asp Trp Pro Ser Arg Leu Gln Met Phe Phe Ala Asn Asn
595 600 605
His Asp Gln Glu Phe Asp Pro Pro Lys Val Tyr Pro Pro Val Pro Ala
610 615 620
Glu Lys Arg Lys Pro Ile Arg Val Leu Ser Leu Phe Asp Gly Ile Ala
625 630 635 640
Thr Gly Leu Leu Val Leu Lys Asp Leu Gly Ile Gln Val Asp Arg Tyr
645 650 655
CA 02331781 2005-04-22
-13-
Ile Ala Ser Glu Val Cys Glu Asp Ser Ile Thr Val Gly Met Val Arg
660 665 670
His Gln Gly Lys Ile Met Tyr Val Gly Asp Val Arg Ser Val Thr Gln
675 680 685
Lys His Ile Gln Glu Trp Gly Pro Phe Asp Leu Val Ile Gly Gly Ser
690 695 700
Pro Cys Asn Asp Leu Ser Ile Val Asn Pro Ala Arg Lys Gly Leu Tyr
705 710 715 720
Glu Gly Thr Gly Arg Leu Phe Phe Glu Phe Tyr Arg Leu Leu His Asp
725 730 735
Ala Arg Pro Lys Glu Gly Asp Asp Arg Pro Phe Phe Trp Leu Phe Glu
740 745 750
Asn Val Val Ala Met Gly Val Ser Asp Lys Arg Asp Ile Ser Arg Phe
755 760 765
Leu Glu Ser Asn Pro Val Met Ile Asp Ala Lys Glu Val Ser Ala Ala
770 775 780
His Arg Ala Arg Tyr Phe Trp Gly Asn Leu Pro Gly Met Asn Arg Pro
785 790 795 800
Leu Ala Ser Thr Val Asn Asp Lys Leu Glu Leu Gln Glu Cys Leu Glu
805 810 815
His Gly Arg Ile Ala Lys Phe Ser Lys Val Arg Thr Ile Thr Thr Arg
820 825 830
Ser Asn Ser Ile Lys Gln Gly Lys Asp Gin His Phe Pro Val Phe Met
835 840 845
Asn Glu Lys Glu Asp Ile Leu Trp Cys Thr Glu Met Glu Arg Val Phe
850 855 860
CA 02331781 2005-04-22
-14-
Gly Phe Pro Val His Tyr Thr Asp Val Ser Asn Met Ser Arg Leu Ala
865 870 875 880
Arg Gln Arg Leu Leu Gly Arg Ser Trp Ser Val Pro Val Ile Arg His
885 890 895
Leu Phe Ala Pro Leu Lys Glu Tyr Phe Ala Cys Val
900 905
<210> 6
<211> 859
<212> PRT
<213> Mus musculus
<400> 6
Met Lys Gly Asp Ser Arg His Leu Asn Glu Glu Glu Gly Ala Ser Gly
1 5 10 15
Tyr Glu Glu Cys Ile Ile Val Asn Gly Asn Phe Ser Asp Gln Ser Ser
20 25 30
Asp Thr Lys Asp Ala Pro Ser Pro Pro Val Leu Glu Ala Ile Cys Thr
35 40 45
Glu Pro Val Cys Thr Pro Glu Thr Arg Gly Arg Arg Ser Ser Ser Arg
50 55 60
Leu Ser Lys Arg Glu Val Ser Ser Leu Leu Asn Tyr Thr Gin Asp Met
65 70 75 80
Thr Gly Asp Gly Asp Arg Asp Asp Glu Val Asp Asp Gly Asn Gly Ser
85 90 95
Asp Ile Leu Met Pro Lys Leu Thr Arg Glu Thr Lys Asp Thr Arg Thr
100 105 110
Arg Ser Glu Ser Pro Ala Val Arg Thr Arg His Ser Asn Gly Thr Ser
CA 02331781 2005-04-22
-15-
115 120 125
Ser Leu Glu Arg Gln Arg Ala Ser Pro Arg Ile Thr Arg Gly Arg Gin
130 135 140
Gly Arg His His Val Gln Glu Tyr Pro Val Glu Phe Pro Ala Thr Arg
145 150 155 160
Ser Arg Arg Arg Arg Ala Ser Ser Ser Ala Ser Thr Pro Trp Ser Ser
165 170 175
Pro Ala Ser Val Asp Phe Met Glu Glu Val Thr Pro Lys Ser Val Ser
180 185 190
Thr Pro Ser Val Asp Leu Ser Gln Asp Gly Asp Gln Glu Gly Met Asp
195 200 205
Thr Thr Gln Val Asp Ala Glu Ser Ile Tyr Gly Asp Ser Thr Glu Tyr
210 215 220
Gln Asp Asp Lys Glu Phe Gly Ile Gly Asp Leu Val Trp Gly Lys Ile
225 230 235 240
Lys Gly Phe Ser Trp Trp Pro Ala Met Val Val Ser Trp Lys Ala Thr
245 250 255
Ser Lys Arg Gln Ala Met Pro Gly Met Arg Trp Val Gln Trp Phe Gly
260 265 270
Asp Gly Lys Phe Ser Glu Ile Ser Ala Asp Lys Leu Val Ala Leu Gly
275 280 285
Leu Phe Ser Gln His Phe Asn Leu Ala Thr Phe Asn Lys Leu Val Ser
290 295 300
Tyr Arg Lys Ala Met Tyr His Thr Leu Glu Lys Ala Arg Val Arg Ala
305 310 315 320
Gly Lys Thr Phe Ser Ser Ser Pro Gly Glu Ser Leu Glu Asp Gln Leu
CA 02331781 2005-04-22
-16-
325 330 335
Lys Pro Met Leu Glu Trp Ala His Gly Gly Phe Lys Pro Thr Gly Ile
340 345 350
Glu Gly Leu Lys Pro Asn Lys Lys Gln Pro Val Val Asn Lys Ser Lys
355 360 365
Val Arg Arg Ser Asp Ser Arg Asn Leu Glu Pro Arg Arg Arg Glu Asn
370 375 380
Lys Ser Arg Arg Arg Thr Thr Asn Asp Ser Ala Ala Ser Glu Ser Pro
385 390 395 400
Pro Pro Lys Arg Leu Lys Thr Asn Ser Tyr Gly Gly Lys Asp Arg Gly
405 410 415
Glu Asp Glu Glu Ser Arg Glu Arg Met Ala Ser Glu Val Thr Asn Asn
420 425 430
Lys Gly Asn Leu Glu Asp Arg Cys Leu Ser Cys Gly Lys Lys Asn Pro
435 440 445
Val Ser Phe His Pro Leu Phe Glu Gly Gly Leu Cys Gln Ser Cys Arg
450 455 460
Asp Arg Phe Leu Glu Leu Phe Tyr Met Tyr Asp Glu Asp Gly Tyr Gln
465 470 475 480
Ser Tyr Cys Thr Val Cys Cys Glu Gly Arg Glu Leu Leu Leu Cys Ser
485 490 495
Asn Thr Ser Cys Cys Arg Cys Phe Cys Val Glu Cys Leu Glu Val Leu
500 505 510
Val Gly Ala Gly Thr Ala Glu Asp Ala Lys Leu Gln Glu Pro Trp Ser
515 520 525
Cys Tyr Met Cys Leu Pro Gln Arg Cys His Gly Val Leu Arg Arg Arg
CA 02331781 2005-04-22
-17-
530 535 540
Lys Asp Trp Asn Met Arg Leu Gln Asp Phe Phe Thr Thr Asp Pro Asp
545 550 555 560
Leu Glu Glu Phe Glu Pro Pro Lys Leu Tyr Pro Ala Ile Pro Ala Ala
565 570 575
Lys Arg Arg Pro Ile Arg Val Leu Ser Leu Phe Asp Gly Ile Ala Thr
580 585 590
Gly Tyr Leu Val Leu Lys Glu Leu Gly Ile Lys Val Glu Lys Tyr Ile
595 600 605
Ala Ser Glu Val Cys Ala Glu Ser Ile Ala Val Gly Thr Val Lys His
610 615 620
Glu Gly Gln Ile Lys Tyr Val Asn Asp Val Arg Lys Ile Thr Lys Lys
625 630 635 640
Asn Ile Glu Glu Trp Gly Pro Phe Asp Leu Val Ile Gly Gly Ser Pro
645 650 655
Cys Asn Asp Leu Ser Asn Val Asn Pro Ala Arg Lys Gly Leu Tyr Glu
660 665 670
Gly Thr Gly Arg Leu Phe Phe Glu Phe Tyr His Leu Leu Asn Tyr Thr
675 680 685
Arg Pro Lys Glu Gly Asp Asn Arg Pro Phe Phe Trp Met Phe Glu Asn
690 695 700
Val Val Ala Met Lys Val Asn Asp Lys Lys Asp Ile Ser Arg Phe Leu
705 710 715 720
Ala Cys Asn Pro Val Met Ile Asp Ala Ile Lys Val Ser Ala Ala His
725 730 735
Arg Ala Arg Tyr Phe Trp Gly Asn Leu Pro Gly Met Asn Arg Pro Val
CA 02331781 2005-04-22
-18-
740 745 750
Met Ala Ser Lys Asn Asp Lys Leu Glu Leu Gln Asp Cys Leu Glu Phe
755 760 765
Ser Arg Thr Ala Lys Leu Lys Lys Val Gln Thr Ile Thr Thr Lys Ser
770 775 780
Asn Ser Ile Arg Gln Giy Lys Asn Gln Leu Phe Pro Val Val Met Asn
785 790 795 800
Gly Lys Asp Asp Val Leu Trp Cys Thr Glu Leu Glu Arg Ile Phe Gly
805 810 815
Phe Pro Ala His Tyr Thr Asp Val Ser Asn Met Gly Arg Gly Ala Arg
820 825 830
Gln Lys Leu Leu Gly Arg Ser Trp Ser Val Pro Val Ile Arg His Leu
835 840 845
Phe Ala Pro Leu Lys Asp Tyr Phe Ala Cys Glu
850 855
<210> 7
<211> 912
<212> PRT
<213> Homo sapiens
<400> 7
Met Pro Ala Met Pro Ser Ser Gly Pro Gly Asp Thr Ser Ser Ser Ala
1 5 10 15
Ala Glu Arg Glu Glu Asp Arg Lys Asp Gly Glu Glu Gln Glu Glu Pro
20 25 30
Arg Gly Lys Glu Glu Arg Gln Glu Pro Ser Thr Thr Ala Arg Lys Val
35 40 45
CA 02331781 2005-04-22
-19-
Gly Arg Pro Gly Arg Lys Arg Lys His Pro Pro Val Glu Ser Gly Asp
50 55 60
Thr Pro Lys Asp Pro Ala Val Ile Ser Lys Ser Pro Ser Met Ala Gln
65 70 75 80
Asp Ser Gly Ala Ser Glu Leu Leu Pro Asn Gly Asp Leu Glu Lys Arg
85 90 95
Ser Glu Pro Gln Pro Glu Glu Gly Ser Pro Ala Gly Gly Gln Lys Gly
100 105 110
Gly Ala Pro Ala Glu Gly Glu Gly Ala Ala Glu Thr Leu Pro Glu Ala
115 120 125
Ser Arg Ala Val Glu Asn Gly Cys Cys Thr Pro Lys Glu Gly Arg Gly
130 135 140
Ala Pro Ala Glu Ala Gly Lys Glu Gln Lys Glu Thr Asn Ile Glu Ser
145 150 155 160
Met Lys Met Glu Gly Ser Arg Gly Arg Leu Arg Gly Gly Leu Gly Trp
165 170 175
Glu Ser Ser Leu Arg Gln Arg Pro Met Pro Arg Leu Thr Phe Gln Ala
180 185 190
Gly Asp Pro Tyr Tyr Ile Ser Lys Arg Lys Arg Asp Glu Trp Leu Ala
195 200 205
Arg Trp Lys Arg Glu Ala Glu Lys Lys Ala Lys Val Ile Ala Gly Met
210 215 220
Asn Ala Val Glu Glu Asn Gln Gly Pro Gly Glu Ser His Lys Val Glu
225 230 235 240
Glu Ala Ser Pro Pro Ala Val Gln Gln Pro Thr Asp Pro Ala Ser Pro
245 250 255
CA 02331781 2005-04-22
-20-
Thr Val Ala Thr Thr Pro Glu Pro Val Gly Ser Asp Ala Gly Asp Lys
260 265 270
Asn Ala Thr Lys Ala Gly Asp Asp Glu Pro Glu Tyr Glu Asp Gly Arg
275 280 285
Gly Phe Gly Ile Gly Glu Leu Val Trp Gly Lys Leu Arg Gly Phe Ser
290 295 300
Trp Trp Pro Gly Arg Ile Val Ser Trp Trp Met Thr Gly Arg Ser Arg
305 310 315 320
Ala Ala Glu Gly Thr Arg Trp Val Met Trp Phe Gly Asp Gly Lys Phe
325 330 335
Ser Val Val Cys Val Glu Lys Leu Met Pro Leu Ser Ser Phe Cys Ser
340 345 350
Ala Phe His Gln Ala Thr Tyr Asn Lys Gln Pro Met Tyr Arg Lys Ala
355 360 365
Ile Tyr Glu Val Leu Gln Val Ala Ser Ser Arg Ala Gly Lys Leu Phe
370 375 380
Pro Val Cys His Asp Ser Asp Glu Ser Asp Thr Ala Lys Ala Val Glu
385 390 395 400
Val Gln Asn Lys Pro Met Ile Glu Trp Ala Leu Gly Gly Phe Gln His
405 410 415
Tyr Gly Pro Lys Gly Leu Glu Pro Pro Glu Glu Glu Lys Asn Pro Tyr
420 425 430
Lys Glu Val Tyr Thr Asp Met Trp Val Glu Pro Glu Ala Ala Ala Tyr
435 440 445
Ala Pro Pro Pro Pro Ala Lys Lys Pro Arg Lys Ser Thr Ala Glu Lys
450 455 460
CA 02331781 2005-04-22
-21-
Pro Lys Val Lys Glu Ile Ile Asp Glu Arg Thr Arg Glu Arg Leu Val
465 470 475 480
Tyr Glu Val Arg Gln Lys Cys Arg Asn Ile Glu Asp Ile Cys Ile Ser
485 490 495
Cys Gly Ser Leu Asn Val Thr Leu Glu His Pro Leu Phe Val Gly Gly
500 505 510
Met Cys Gln Asn Cys Lys Asn Cys Phe Leu Glu Cys Ala Tyr Gln Tyr
515 520 525
Asp Asp Asp Gly Tyr Gln Ser Tyr Cys Thr Ile Cys Cys Gly Gly Arg
530 535 540
Glu Val Leu Met Cys Gly Asn Asn Asn Cys Cys Arg Cys Phe Cys Val
545 550 555 560
Glu Cys Val Asp Leu Leu Val Gly Pro Gly Ala Ala Gln Ala Ala Ile
565 570 575
Lys Glu Asp Pro Trp Asn Cys Tyr Met Cys Gly His Lys Gly Thr Tyr
580 585 590
Gly Leu Leu Arg Arg Arg Lys Asp Trp Pro Ser Arg Leu Gln Met Phe
595 600 605
Phe Ala Asn Asn His Asp Gln Glu Phe Asp Pro Pro Lys Val Tyr Pro
610 615 620
Pro Val Pro Ala Glu Lys Arg Lys Pro Ile Arg Val Leu Ser Leu Phe
625 630 635 640
Asp Gly Ile Ala Thr Gly Leu Leu Val Leu Lys Asp Leu Gly Ile Gln
645 650 655
Val Asp Arg Tyr Ile Ala Ser Glu Val Cys Glu Asp Ser Ile Thr Val
660 665 670
CA 02331781 2005-04-22
-22-
Gly Met Val Arg His Gln Gly Lys Ile Met Tyr Val Gly Asp Val Arg
675 680 685
Ser Val Thr Gln Lys His Ile Gin Glu Trp Gly Pro Phe Asp Leu Val
690 695 700
Ile Gly Gly Ser Pro Cys Asn Asp Leu Ser Ile Val Asn Pro Ala Arg
705 710 715 720
Lys Gly Leu Tyr Glu Gly Thr Gly Arg Leu Phe Phe Glu Phe Tyr Arg
725 730 735
Leu Leu His Asp Ala Arg Pro Lys Glu Gly Asp Asp Arg Pro Phe Phe
740 745 750
Trp Leu Phe Glu Asn Val Val Ala Met Gly Val Ser Asp Lys Arg Asp
755 760 765
Ile Ser Arg Phe Leu Glu Ser Asn Pro Val Met Ile Asp Ala Lys Glu
770 775 780
Val Ser Ala Ala His Arg Ala Arg Tyr Phe Trp Gly Asn Leu Pro Gly
785 790 795 800
Met Asn Arg Pro Leu Ala Ser Thr Val Asn Asp Lys Leu Glu Leu Gln
805 810 815
Glu Cys Leu Glu His Gly Arg Ile Ala Lys Phe Ser Lys Val Arg Thr
820 825 830
Ile Thr Thr Arg Ser Asn Ser Ile Lys Gln Gly Lys Asp Gln His Phe
835 840 845
Pro Val Phe Met Asn Glu Lys Glu Asp Ile Leu Trp Cys Thr Glu Met
850 855 860
Glu Arg Val Phe Gly Phe Pro Val His Tyr Thr Asp Val Ser Asn Met
865 870 875 880
CA 02331781 2005-04-22
-23-
Ser Arg Leu Ala Arg Gln Arg Leu Leu Gly Arg Ser Trp Ser Val Pro
885 890 895
Val Ile Arg His Leu Phe Ala Pro Leu Lys Glu Tyr Phe Ala Cys Val
900 905 910
<210> 8
<211> 853
<212> PRT
<213> Homo sapiens
<400> 8
Met Lys Gly Asp Thr Arg His Leu Asn Gly Glu Glu Asp Ala Gly Gly
1 5 10 15
Arg Glu Asp Ser Ile Leu Val Asn Gly Ala Cys Ser Asp Gin Ser Ser
20 25 30
Asp Ser Pro Pro Ile Leu Glu Ala Ile Arg Thr Pro Glu Ile Arg Gly
35 40 45
Arg Arg Ser Ser Ser Arg Leu Ser Lys Arg Glu Val Ser Ser Leu Leu
50 55 60
Ser Tyr Thr Gln Asp Leu Thr Gly Asp Gly Asp Gly Glu Asp Gly Asp
65 70 75 80
Gly Ser Asp Thr Pro Val Met Pro Lys Leu Phe Arg Glu Thr Arg Thr
85 90 95
Arg Ser Glu Ser Pro Ala Val Arg Thr Arg Asn Asn Asn Ser Val Ser
100 105 110
Ser Arg Glu Arg His Arg Pro Ser Pro Arg Ser Thr Arg Gly Arg Gln
115 120 125
CA 02331781 2005-04-22
-24-
Gly Arg Asn His Val Asp Glu Ser Pro Val Glu Phe Pro Ala Thr Arg
130 135 140
Ser Leu Arg Arg Arg Ala Thr Ala Ser Ala Gly Thr Pro Trp Pro Ser
145 150 155 160
Pro Pro Ser Ser Tyr Leu Thr Ile Asp Leu Thr Asp Asp Thr Glu Asp
165 170 175
Thr His Gly Thr Pro Gln Ser Ser Ser Thr Pro Tyr Ala Arg Leu Ala
180 185 190
Gln Asp Ser Gln Gln Gly Gly Met Glu Ser Pro Gln Val Glu Ala Asp
195 200 205
Ser Gly Asp Gly Asp Ser Ser Glu Tyr Gln Asp Gly Lys Glu Phe Gly
210 215 220
Ile Gly Asp Leu Val Trp Gly Lys Ile Lys Gly Phe Ser Trp Trp Pro
225 230 235 240
Ala Met Val Val Ser Trp Lys Ala Thr Ser Lys Arg Gin Ala Met Ser
245 250 255
Gly Met Arg Trp Val Gln Trp Phe Gly Asp Gly Lys Phe Ser Glu Val
260 265 270
Ser Ala Asp Lys Leu Val Ala Leu Gly Leu Phe Ser Gln His Phe Asn
275 280 285
Leu Ala Thr Phe Asn Lys Leu Val Ser Tyr Arg Lys Ala Met Tyr His
290 295 300
Ala Leu Glu Lys Ala Arg Val Arg Ala Gly Lys Thr Phe Pro Ser Ser
305 310 315 320
Pro Gly Asp Ser Leu Glu Asp Gln Leu Lys Pro Met Leu Glu Trp Ala
325 330 335
CA 02331781 2005-04-22
-25-
His Gly Gly Phe Lys Pro Thr Gly Ile Glu Gly Leu Lys Pro Asn Asn
340 345 350
Thr Gln Pro Val Val Asn Lys Ser Lys Val Arg Arg Ala Gly Ser Arg
355 360 365
Lys Leu Glu Ser Arg Lys Tyr Glu Asn Lys Thr Arg Arg Arg Thr Ala
370 375 380
Asp Asp Ser Ala Thr Ser Asp Tyr Cys Pro Ala Pro Lys Arg Leu Lys
385 390 395 400
Thr Asn Cys Tyr Asn Asn Gly Lys Asp Arg Gly Asp Glu Asp Gln Ser
405 410 415
Arg Glu Gln Met Ala Ser Asp Val Ala Asn Asn Lys Ser Ser Leu Glu
420 425 430
Asp Gly Cys Leu Ser Cys Gly Arg Lys Asn Pro Val Ser Phe His Pro
435 440 445
Leu Phe Glu Gly Gly Leu Cys Gln Thr Cys Arg Asp Arg Phe Leu Glu
450 455 460
Leu Phe Tyr Met Tyr Asp Asp Asp Gly Tyr Gln Ser Tyr Cys Thr Val
465 470 475 480
Cys Cys Glu Gly Arg Glu Leu Leu Leu Cys Ser Asn Thr Ser Cys Cys
485 490 495
Arg Cys Phe Cys Val Glu Cys Leu Glu Val Leu Val Gly Thr Gly Thr
500 505 510
Ala Ala Glu Ala Lys Leu Gln Glu Pro Trp Ser Cys Tyr Met Cys Leu
515 520 525
Pro Gln Arg Cys His Gly Val Leu Arg Arg Arg Lys Asp Trp Asn Val
530 535 540
CA 02331781 2005-04-22
-26-
Arg Leu Gln Ala Phe Phe Thr Ser Asp Thr Gly Leu Glu Tyr Glu Ala
545 550 555 560
Pro Lys Leu Tyr Pro Ala Ile Pro Ala Ala Arg Arg Arg Pro Ile Arg
565 570 575
Val Leu Ser Leu Phe Asp Gly Ile Ala Thr Gly Tyr Leu Val Leu Lys
580 585 590
Glu Leu Gly Ile Lys Val Gly Lys Tyr Val Ala Ser Glu Val Cys Glu
595 600 605
Glu Ser Ile Ala Val Gly Thr Val Lys His Glu Gly Asn Ile Lys Tyr
610 615 620
Val Asn Asp Val Arg Asn Ile Thr Lys Lys Asn Ile Glu Glu Trp Gly
625 630 635 640
Pro Phe Asp Leu Val Ile Gly Gly Ser Pro Cys Asn Asp Leu Ser Asn
645 650 655
Val Asn Pro Ala Arg Lys Gly Leu Tyr Glu Gly Thr Gly Arg Leu Phe
660 665 670
Phe Glu Phe Tyr His Leu Leu Asn Tyr Ser Arg Pro Lys Glu Gly Asp
675 680 685
Asp Arg Pro Phe Phe Trp Met Phe Glu Asn Val Val Ala Met Lys Val
690 695 700
Gly Asp Lys Arg Asp Ile Ser Arg Phe Leu Glu Cys Asn Pro Val Met
705 710 715 720
Ile Asp Ala Ile Lys Val Ser Ala Ala His Arg Ala Arg Tyr Phe Trp
725 730 735
Gly Asn Leu Pro Gly Met Asn Arg Pro Val Ile Ala Ser Lys Asn Asp
740 745 750
CA 02331781 2005-04-22
-27-
Lys Leu Glu Leu Gln Asp Cys Leu Glu Tyr Asn Arg Ile Ala Lys Leu
755 760 765
Lys Lys Val Gln Thr Ile Thr Thr Lys Ser Asn Ser Ile Lys Gln Gly
770 775 780
Lys Asn Gln Leu Phe Pro Val Val Met Asn Gly Lys Glu Asp Val Leu
785 790 795 800
Trp Cys Thr Glu Leu Glu Arg Ile Phe Gly Phe Pro Val His Tyr Thr
805 810 815
Asp Val Ser Asn Met Gly Arg Gly Ala Arg Gln Lys Leu Leu Gly Arg
820 825 830
Ser Trp Ser Val Pro Val Ile Arg His Leu Phe Ala Pro Leu Lys Asp
835 840 845
Tyr Phe Ala Cys Glu
850
<210> 9
<211> 393
<212> DNA
<213> Mus musculus
<400> 9
tttctacagt atttcaggtg cctaccacac aggaaacctt gaagaaaacc agtttctaga 60
agccgctgtt acctcttgtt tacagtttat atatatatga tagatatgag atatatatat 120
ataaaaggta ctgttaacta ctgtacatcc cgacttcata atggtgcttt caaaacagcg 180
agatgagcaa agacatcagc ttccgcctgg ccctcgtgtg caaatggcgt ttcatgccca 240
tggatggtgt agaggggagc agctggaggg ggtttcacaa actgaaggat gacccatatc 300
accccccacc cctgccccat gcctagcttc acctgccaaa aaggggctca gctgaggtgg 360
tcggaccctg gggaagctga gtgtggaatt tat 393
<210> 10
<211> 424
CA 02331781 2005-04-22
-28-
<212> DNA
<213> Mus musculus
<400> 10
gaagaaaacc agtttctaga agccgctgtt acctcttgtt tacagtttat atatatatga 60
tagatatgag atatatatat ataaaaggta ctgttaacta ctgtacatcc cgacttcata 120
atggtgcttt caaaacagcg agatgagcaa agacatcagc ttccgcctgg ccctctgtgc 180
aaagggtttc agcccaggat ggtgagaggg gagcatctgg agggggtttt aacaaactga 240
aggatgaccc atatcacccc ccacccctgc cccatgccta gcttcacctg ccaaaaaggg 300
gctcagctga ggtggtcgga ccctggggaa gctgagtgtg gaatttatcc agactcgcgt 360
gcaataacct tagaatatga atctaaaatg actgcctcag aaaaatggct tgagaaaaca 420
ttgt 424
<210> 11
<211> 461
<212> DNA
<213> Mus musculus
<400> 11
,tttaaagcaa accacagagg aggaaaacgc cggaggcttg gccttgcaaa agggttggac 60
atcatctcct gagttttcaa tgttaacctt cagtcctatc taaaaagcaa aataggcccc 120
tccccttcgt tcccctccgg tcctaggagg cgaacttttt gttttctact ctttttcaga 180
ggggttttct gtttgtttgg gtttttgttt cttgctgtga ctgaaacaag agagttattg 240
cagcaaaatc agtaacaaca aaaagtagaa atgccttgga gcggaaaggg agagagggaa 300
aattctataa aaacttaaaa tattggtttt tttttttttc cttttctata tatctctttg 360
gttgtctcta gcctgatcag ataggagcac aaacaggaag agaatagaga ccctcggagg 420
cagagtctcc tctcccaccc cccgagcagt ctcaacagca c 461
<210> 12
<211> 465
<212> DNA
<213> Mus musculus
<400> 12
tcagaggggt tttctgtttg tttgggtttt tgtttcttgc tgtgactgaa acaagagagt 60
tattgcagca aaatcagtaa caacaaaaag tagaaatgcc ttggagagga aagggagaga 120
gggaaaattc tataaaaact taaaatattg gttttttttt tttttccttt tctatatatc 180
tctttggttg tctctagcct gatcagatag gagcacaaac aggaagagaa tagagaccct 240
cggaggcaga gtctcctctc ccaccccccg agcagtctca acagcaccat tcctggtcat 300
CA 02331781 2005-04-22
-29-
gcaaaacaga acccaactag cagcagggcg ctgagagaac accacaccag acacttttct 360
acagtatttc aggtgcctac cacacaggaa accttgaaga aaaccagttt ctagaagccg 420
ctgttacctc ttgtttacag tttatatata tatgatagat atgag 465
<210> 13
<211> 393
<212> DNA
<213> Mus musculus
<400> 13
aaaacgccgg aggcctttgc cttgcacaag ggttggacat catctcctga gttttcaatg 60
ttaaccttca gtcctatcta aaaagcaaaa taggcccctc cccttcttcc cctccggtcc 120
taggaggcga actttttgtt ttctactctt tttcagaggg gttttctgtt tgtttgggtt 180
tttgtttctt gctgtgactg aaacaagaga gttattgcag caaaatcagt aacaacaaaa 240
agtagaaatg ccttggagag gaaagggaga gagggaaaat tctataaaaa cttaaaatat 300
tggttttttt ttttttcctt ttctatatat cgctttggtt gtctctagcc tgatcagata 360
ggagcacaaa caggaagaga atagagaccc tcg 393
<210> 14
<211> 309
<212> DNA
<213> Mus musculus
<400> 14
gtgatgattg acgccaaaga agtgtctgct gcacacaggg cccgttactt ctaggggtaa 60
ccttcctggc atgaacaggc ctttggatcc actgtgaatg ataagctgga gctgcaagag 120
tgtctggagc acggcagaat agccaagttc agcaaagtga ggaccattac caccaggtca 180
aactctataa agcagggcaa agaccagcat ttccccgtct tcatgaacga gaaggaggac 240
atcctgtggt gcactgaaat ggaaagggtc tttggcttcc ccgtccacta cacagacgtc 300
tccaacatg 309
<210> 15
<211> 341
<212> DNA
<213> Mus musculus
<400> 15
tgttaacctt cagtcctatc taaaaagcaa aataggcccc tccccttctt cccctccggt 60
cctaggaggc gaactttttg ttttctactc tttttcagag gggttttctg tttgtttggg 120
CA 02331781 2005-04-22
-30-
tttttgtttc ttgctgtgac tgaaacaaga gagttattgc agcaaaatca gtaacaacaa 180
aaagtagaaa tgccttggag aggaaaggga gagagggaaa attctataaa aacttaaaat 240
attggttttt ttttttttcc ttttctatat atctctttgg ttgtctctag cctgatcaga 300
taggagcaca aacaggaaga gaatagagac cctcggaggc a 341
<210> 16
<211> 240
<212> DNA
<213> Mus musculus
<220>
<221> Unsure
<222> (32)..(32)
<223> May be any nucleic acid
<400> 16
acattttgta tgttttttta tttgctccag gnggggttaa tggcgggtca ctttccctca 60
ctctggaata tttctgatcc cacaaggggc cttcaacgtg gctgacgaat tcaaaatcag 120
ggacaatgtt ttctcaagcc atttttctga ggcagtcatt ttagattcat attctaaggt 180
tattgcacgc gagtctggat aaattccaca ctcagcttcc ccagggtccg accacctcag 240
<210> 17
<211> 256
<212> DNA
<213> Mus musculus
<220>
<221> Unsure
<222> (75)..(75)
<223> May be any nucleic acid
<400> 17
atcagcttcc gcctggccct ctgtgcaaag ggtttcagcc caggatgggg agaggggagc 60
agctggaggg ggttntaaca aactgaagga tgacccatat caccccccac ccctgcccca 120
tgcctagctt cacctgccaa aaaggggctc agctgaggtg gtcggaccct ggggaagctg 180
agtgtggaat ttatccagac tcgcgtgcaa taaccttaga atatgaatct aaaatgactg 240
cctcagaaaa atggct 256
<210> 18
<211> 435
<212> DNA
<213> Mus musculus
CA 02331781 2005-04-22
-31-
<400> 18
gtggaagccc atgcaatgat ctctctaacg tcaatcctgc ccgcaaaggt ttatatgagg 60
gcacaggaag gctcttcttc gagttttacc acttgctgaa ttatacccgc cccaaggagg 120
gcgacaaccg tccattcttc tggatgttcg agaatgttgt ggccatgaaa gtgaatgaca 180
agaaagacat ctcaagattc ctggcatgta acccagtgat gatcgatgcc atcaaggtgt 240
ctgctgctca cagggcccgg tacttctggg gtaacctacc cggaatgaac aggcccgtga 300
tggcttcaaa gaatgataag ctcgagctgc aggactgcct ggagttcagt aggacagcaa 360
agttaaagaa agtgcagaca ataaccacca agtcgaactc catcagacag ggcaaaaacc 420
agcttttccc tgtag 435
<210> 19
<211> 522
<212> DNA
<213> Mus musculus
<400> 19
gatgatgtca gcagggatga catcaccacc tttagggctt ttccctggca ggggcccatg 60
tggctagtcc tcacgaagac tggagtagaa tgtttggagc tcaggaaggg tgggtggagt 120
ggagtctctt ccaggtgtga gggatacgaa ggaggaagct tagggaaatc cattccccac 180
tccctcttgc caaatgaggg gcccagtccc caacagctca ggtccccaga accccctagt 240
tcctcatgag aagctaggac cagaagcaca tcgttcccct tatctgagca gtgtttgggg 300
aactacagtg aaaaccttct ggagatgtta aaagcttttt accccacgat agattgtgtt 360
tttaaggggt gcttttttta ggggcatcac tggagataag aaagctgcat ttcagaaatg 420
ccatcgtaat ggtttttaaa caccttttac ctaattacag gtgctatttt atagaagcag 480
acaacacttc tttttatgac tctcagactt ctattttcat gt 522
<210> 20
<211> 348
<212> DNA
<213> Mus musculus
<400> 20
aaaggaggcc cattagagtc ctgtctctgt ttgatggaat tgcaacgggg tacttggtgc 60
tcaaggagtt gggtattaaa gtggaaaagt acattgcctc cgaagtctgt gcagagtcca 120
tcgctgtggg aactgttaag catgaaggcc agatcaaata tgtcaatgac gtccggaaaa 180
tcaccaagaa aaatattgaa gagtggggcc cgttcgactt ggtgattggt ggaagcccat 240
gcaatgatct ctctaacgtc aatcctgccc gcaaaggttt atatgagggc acaggaaggc 300
tcttcttcga gttttaccac ttgctgaatt atacccgccc caaggagg 348
CA 02331781 2005-04-22
-32-
<210> 21
<211> 258
<212> DNA
<213> Mus musculus
<400> 21
gtttatggtt taagtcttcc tggcaccttc cccttgcttt ggtacaaggg ctgaagtcct 60
gttggtcttg tagcatttcc caggatgatg atgtcagcag ggatgacatc atcaccttta 120
gggcttttcc ctggcagggg cccatgtggc tagtcctcac gaagactgga gtagaatgtt 180
tggagctcag gaagggtggg tggagtgtgc ctcttccagg tgtgagggat acgaaggagg 240
aagcttaggg aaatccat 258
<210> 22
<211> 334
<212> DNA
<213> Mus musculus
<400> 22
tggggtaacc tacccggaat gaacagttaa agaaagtgca gacaataacc accaagtcga 60
actccatcag acagggcaaa aaccagcttt tccctgtagt catgaatggc aaggacgacg 120
ttttgtggtg cactgagctc gaaaggatct tcggcttccc tgctcactac acggacgtgt 180
ccaacatggg ccgcggcgcc cgtcagaagc tgctgggcag gtcctggagt gtaccggtca 240
tcagacacct gtttgccccc ttgaaggact actttgcctg tgaatagttc tacccaggac 300
tggggagctc tcggtcagag ccagtgccca gagt 334
<210> 23
<211> 299
<212> DNA
<213> Mus musculus
<220>
<221> Unsure
<222> (59)..(59)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (173)..(173)
<223> May be any nucleic acid
<400> 23
CA 02331781 2005-04-22
-33-
ctgtttttgt ttgttttttt ggtatcttag ccatcacttc tgagtgataa actcaggang 60
gtaaaagaaa gccatcttac tacctacttc aagttttaaa gtttcagggt aagagaacat 120
gagcaccatg ccgggctact ctaagcagcc aggtctgagc tgtgcacacg ganggagcac 180
cggggctccc ctgcaaggcc aggaggctct gctcccactg agcaggagaa agctgaggta 240
cagtgatgtg aggccccaca caggtgagct aaaaagggga caggtgaggt gccttcagg 299
<210> 24
<211> 455
<212> DNA
<213> Mus musculus
<400> 24
gatcgcttcc tagagctctt ctacatgtat gatgaggacg gctatcagtc ctactgcacc 60
gtgtctgtga gggccgtgaa ctgctgctgt gcagtaacac aagctgctgc agatgcttct 120
gtgtggagtg tctggaggtg ctggtgggcg caggacagct gaggatgcca agctgcagga 180
accctggagc tgctatatgt gcctccctca gcgctgccat ggggtcctcc gacgcaggaa 240
agattggaac atgcgcctgc aagacttctt cactactgat cctgacctgg aagaatttca 300
ggagccaccc aagttgtacc cagcaattcc tgcagccaaa aggaggccca ttagagtcct 360
gtctctgttt gatggaattg caacggggta cttggtgctc aaggagttgg gtattaaagt 420
ggaaaagtac attgcctccg aagtctgtgc agagt 455
<210> 25
<211> 368
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (307)..(307)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (335)..(335)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (353)..(353)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (360)..(360)
CA 02331781 2005-04-22
-34-
<223> May be any nucleic acid
<400> 25
acgttttgta tgttttttta tttgctccag gtggggtttt gactgtcact ttcccacact 60
ctggattagt tctgatccca ccacaaggag ccctcgaatt ggctaaagtg agaaactggg 120
cctgaagact ccgtaccctc tgccatcttg ccgagggagt ctccttttag aaaacaatca 180
aagggttatt gcatgagtct ggatgaatcc cactctcagc ttgtccacgg gcccgaccac 240
ctcatctagc cccctttttg gcaagggaga acctggctcc caagttctcc tccttcactt 300
tcgttancaa accaaggggg aagaagccca ccgtngagaa cgcgccatct tgnaaagctn 360
ggtcttcc 368
<210> 26
<211> 399
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (87) .. (87)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (314)..(314)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (318)..(318)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (370)..(370)
<223> May be any nucleic acid
<400> 26
gaacatgagg atggagagaa gtatcagcac ccagaagaga aaaaggaatt taaaacaaaa 60
accacagagg cggaaatacc ggaggcnttt gcttgcgaaa agggttggac atcatctcct 120
gatttttcaa tgttattctt cagtcctatt taaaaacaaa accaagctcc cttcccttcc 180
tcccccttcc cttttttttc ggtcagacct tttattttct actcttttca gaggggtttt 240
ctgtttgttt gggttttgtt tcttgctgtg actgaaacaa gaaggttatt gcagcaaaaa 300
tcaggtaaca aaanatangt aacaatacct tgcagaggaa aggtgggagg agaggaaaaa 360
agggaaattn ctatagaaat ctatatattg gggttggtt 399
CA 02331781 2005-04-22
-35-
<210> 27
<211> 318
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (205)..(205)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (275)..(275)
<223> May be any nucleic acid
<400> 27
gtacgaggtg cggcagaagt gccggaacat tgaggacatc tgcatctcct gtgggagcct 60
caatgttacc ctggaacacc ccctcttcgt tggaggaatg tgccaaaact gcaagaactg 120
ctttctggag tgtgcgtacc agtacgacga cgacggctac cagtcctact gcaccatctg 180
ctgtgggggc cgtgaggtgc tcatntgcgg aaacaacaac tgctgcaggt gcttttgcgt 240
ggagtgtgtg gacctcttgg tggggccggg ggctncccag gcagcagtta aggaagatca 300
tgtacgtcgg ggacgtcc 318
<210> 28
<211> 259
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (227)..(227)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (234)..(234)
<223> May be any nucleic acid
<400> 28
gagccgagca gctgaaggca cccgctgggt catgtggttc ggagacggca aattctcagt 60
ggtgtgtgtt gagaagctga tgccgctgag ctcgttttgc agtgcgttcc accaggccac 120
gtacaacaag cagcccatgt accgcaaagc catctacgag gtcctgcagg tggccagcag 180
ccgcgcgggg aagctgttcc cggtgtgcca cgacagcgat gagagtnaca ctgncaaggc 240
cgtgggaggt gcagaacaa 259
CA 02331781 2005-04-22
-36-
<210> 29
<211> 483
<212> DNA
<213> Homo sapiens
<400> 29
tttttttttt ttgtatgttt ttttatttgc tccaggtggg gttttgactg tcactttccc 60
acactctgga ttagttctga tcccaccaca aggagccctc gaattggcta aagtgagaaa 120
ctgggcctga agactccgta ccctctgcca tcttgccgag ggagtctcct tttagaaaac 180
aatcaaaggg ttattgcatg agtctggatg aatcccactc tcagctgtcc acggggccga 240
ccacctcatc taggcccctt tttggcaagg agaacccggg tcccaagttc tcctccttca 300
cttcgttaca aaccaggggg aaaaagccca cgtgaaaacg cggcatctgc aaaatggttc 360
cctttcttca tccctgggga aacctttgcg ccaaggcaac gtggaaactg atggttttac 420
tcaactcgct gttttgaagc gccattatga aatcggggtt gtacgtaggt aaagtcccgt 480
gcc 483
<210> 30
<211> 337
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (41) . . (41)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (45).. (45)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (176)..(176)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (190)..(190)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (207)..(207)
<223> May be any nucleic acid
= CA 02331781 2005-04-22
-37-
<220>
<221> Unsure
<222> (265)..(265)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (290)..(290)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (317)..(317)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (322)..(322)
<223> May be any nucleic acid
<400> 30
gggcattcag gtggaccgct acattgcctc ggaggtgtgt naggnctcca tcacggtggg 60
catggtgcgg caccagggga agatcatgta cgtcggggac gtccgcagcg tcacacagaa 120
gcatatccag gagtggggcc cattcgatct ggtgattggg ggcagtccct gcaatnacct 180
ctccatcgtn aaccctgctc gcaaggncct ctacgagggc actggccggc tcttctttaa 240
gttctaccgc ctcctgcatg atgcncggcc caaggagggg agatgatcgn cccttcttct 300
ggctctttaa gaatgtngtg gnccatgggc gtttagt 337
<210> 31
<211> 271
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (234)..(234)
<223> May be any nucleic acid
<400> 31
cttgtttaca gtttatatat atatgataga tatgagatat atatataaaa ggtactgtta 60
actactgtac aacccgactt cataatggtg ctttcaaaca gcgagatgag taaaaacatc 120
agcttccacg ttgccttctg cgcaaagggt ttcaccaagg atggagaaag ggagacagct 180
tgcagatggc gcgttctcac ggtgggctct tccccttggt ttgtaacgaa gtgnaggagg 240
agaacttggg agccaggttc tccctgccaa a 271
<210> 32
CA 02331781 2005-04-22
-38-
<211> 430
<212> DNA
<213> Homo sapiens
<400> 32
acgttttgta tgttttttta tttgctccag gtggggtttt gactgtcact ttcccacact 60
ctggattagt tctgatccca ccacaaggag ccctcgaatt ggctaaagtg agaaactggg 120
cctgaagact ccgtaccctc tgccatcttg ccgagggagt ctcctttaga aaacaatcaa 180
agggttattg catgagtctg gatgaatccc actctcagct gtccacgggc ccgaccacct 240
catctagccc cctttttggc agggagaacc tggctcccaa gttctcctcc ttcacttcgt 300
tacaaaccaa ggggaagagc ccaccgtgag aacgcgccat ctgcaagctg tctccctttc 360
tccatccttg gtgaaacccc tttgcgcaga aggcaacgtg gaagctgatg tttttactca 420
tctcgctgtt 430
<210> 33
<211> 483
<212> DNA
<213> Homo sapiens
<400> 33
tttttttttt ttgtatgttt ttttatttgc tccaggtggg gttttgactg tcactttccc 60
acactctgga ttagttctga tcccaccaca aggagccctc gaattggcta aagtgagaaa 120
ctgggcctga agactccgta ccctctgcca tcttgccgag ggagtctcct tttagaaaac 180
aatcaaaggg ttattgcatg agtctggatg aatcccactc tcagctgtcc acggggccga 240
ccacctcatc taggcccctt tttggcaagg agaacccggg tcccaagttc tcctccttca 300
cttcgttaca aaccaggggg aaaaagccca cgtgaaaacg cggcatctgc aaaatggttc 360
cctttcttca tccctgggga aacctttgcg ccaaggcaac gtggaaactg atggttttac 420
tcaactcgct gttttgaagc gccattatga aatcggggtt gtacgtaggt aaagtcccgt 480
gcc 483
<210> 34
<211> 411
<212> DNA
<213> Homo sapiens
<400> 34
ttttttttta cgttttgtat gtttttttat ttgctccagg tggggttttg actgtcactt 60
tcccacactc tggattagtt ctgatcccac cacaaggagc cctcgaattg gctaaagtga 120
gaaactgggc ctgaagactc cgtaccctct gccatcttgc cgagggagtc tccttttaga 180
CA 02331781 2005-04-22
-3 9-
aaacaatcaa agggttattg catgagtctg gatgaatccc actctcagct gtccacgggc 240
ccgaccacct catctagccc ccttttggca gggagaacct ggctcccaag ttctcctcct 300
tcacttcgtt acaaaccaag gggaagagcc caccgtgaga acgcgccatc tgcaagctgt 360
ctccctttct ccatccttgg tgaaaccctt tgcgcagaag gcaacgtgga a 411
<210> 35
<211> 530
<212> DNA
<213> Homo sapiens
<400> 35
cgcctggacg agcccagact gctgggccgg tcatggagcg cgccagtcat ccgccacctc 60
ttcgctccgc tgaaggcgta ttttgcgtgt gtctaaggga catgggggca aactgaggta 120
gcgacacaaa gttaaacaca caaacacccc acacacaaca taatacaaca ccaagaacat 180
gaggatggag agaagtatca gccacccaga agagaacaag gaatttaaaa ccaaaaccac 240
agaggcggaa ataccggagg actttgcctt gcgaccaggg ttggacatca tctcctgatt 300
tttcaatgtt attcttcagt cctatttaaa aacaaaacca agctcccttc ccttcctgcg 360
gcttcccttt tttttcggtc agacctttta ttttctactc ttttcagagg ggttttctgt 420
ttgtttgggt tttgtttctt gctgtgactg aaacaagaag gttattgcag caaaaatcag 480
taacaaaaaa tagtaacaat accttgcaga ggaaaggtgg gagagaggaa 530
<210> 36
<211> 535
<212> DNA
<213> Homo sapiens
<400> 36
tttacgtttt gtatgttttt ttatttgctc caggtggggt tttgactgtc actttcccac 60
actctggatt agttctgatc ccaccacaag gagccctcga attggctaaa gtgagaaact 120
gggcctgaag actccgtacc ctctgccatc ttgccgaggg agtctccttt tagaaaacaa 180
tcaaagggtt attgcatgag tctggatgaa tcccactctc agctgtccac gggcccgacc 240
acctcatcta gccccctttt tggcagggag aacctggctc ccaagttctc ctccttcact 300
tcgttacaaa ccacggggaa gagcccaccg tgagaacgcg ccatctgcaa gctgtctccc 360
tttctccatc cttggtgaaa ccctttgcgc agaaggcaac gtggaagctg atgtttttac 420
tcatctcgct gtttgaaagc accattatga agtcgggttg tacagtagtt aacagtacct 480
tttatatata tatctcatat ctatcatata tatataaact gtaaacaaga ggtaa 535
<210> 37
<211> 428
CA 02331781 2005-04-22
-40-
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (12) .. (12)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (15) .. (15)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (415)..(415)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (424)..(424)
<223> May be any nucleic acid
<400> 37
acgttttgta tntantttta tttgctccag gtggggtttt gactgtcact ttcccacact 60
ctggattagt tctgatccca ccacaaggag ccctcgaatt ggctaaagtg agaaactggg 120
cctgaagact ccgtaccctc tgccatcttg ccgagggagt ctccttttag aaaacaatca 180
aagggttatt gcatgagtct ggatgaatcc cactctcagc tgtccacggg cccgaccacc 240
tcatctagcc ccctttttgg cagggagaac ctgggctccc aagttctcct ccttcacttc 300
gttacaaacc aaggggaagg agcccaccgt gagaacggcg ccatcttgca agctgtctcc 360
ctttctccat ccttgggtga aacccttttg cgcagaaggg caacgtggga agctngatgt 420
tttntaac 428
<210> 38
<211> 419
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (306)..(306)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (325)..(325)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-4 l -
<220>
<221> Unsure
<222> (341)..(341)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (367)..(367)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (385)..(385)
<223> May be any nucleic acid
<400> 38
atgggcgtta gtgacaagag ggacatctcg cgatttctcg agtccaaccc tgtgatgatt 60
gatgccaaag aagtgtcagc tgcacacagg gcccgctact tctggggtaa ccttcccggt 120
atgaacaggc cgttggatcc actgtgaatg ataagctgga gctgcaggag tgtctggagc 180
atggcaggat agccaagttc agcaaagtga ggaccattac tacgaggtca aactccataa 240
agcagggcaa agaccagcat tttcctgtct tcatgaatga gaaagaggac atcttatggt 300
gcactnaaat tggaaagggt atttngggtt tcccagtcca ntatactgac gtctccaaca 360
tgagccnctt tgggagggca gagantgctg gggccggttc atgggagcgt gcccagttc 419
<210> 39
<211> 437
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (2)..(2)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (11) . . (11)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (23) . . (23)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (76) .. (76)
<223> May be any nucleic acid
<220>
CA 02331781 2005-04-22
-42-
<221> Unsure
<222> (224)..(224)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (290)..(290)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (362)..(362)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (376)..(376)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (386)..(386)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (426)..(426)
<223> May be any nucleic acid
<400> 39
tnttttgttg nctctagcct gancagatag gagcacaagc aggggacgga aagagagaga 60
cactcaggcg gcacanttcc ctcccagcca ctgagctgtc gtgccagcac cattcctggt 120
cacgcaaaac agaacccagt tagcagcagg gagacgagaa caccacacaa gacatttttc 180
tacagtattt caggtgccta ccacacagga aaccttgaag aaantcagtt tctaggaagc 240
cgctgttacc tcttgtttac agtttatata tatatgatag atatgagatn tatatataaa 300
aggtactgtt aactactgta caacccgact tcataatggg tgctttcaaa caggcgaggt 360
gngtaaaaac atcagnttcc acgttngcct tttgcgcaaa gggtttcacc aggttgggga 420
aagggngaca gcttttt 437
<210> 40
<211> 385
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (340)..(340)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-43-
<220>
<221> Unsure
<222> (365)..(365)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (376)..(376)
<223> May be any nucleic acid
<400> 40
tacgttttgt atgttttttt atttgctcca ggtggggttt tgactgtcac tttcccacac 60
tctggattag ttctgatccc accacaagga gccctcgaat tggctaaagt gagaaactgg 120
gcctgaagac tccgtaccct ctgccatctt gccgagggag tctcctttta gaaaacaatc 180
aaagggttat tgcatgagtc tggatgaatc ccactctcag ctgtccacgg gcccgaccac 240
ctcatctagc cccctttttg gcagggagaa cctgggctcc caagttctcc tccttcactt 300
cgttacaaac caaggggaag agcccaccgt gagaacgcgn catctgcaag ctgtctccct 360
ttttncatcc ttggtngaaa ccctt 385
<210> 41
<211> 294
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (66) .. (66)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (73) . . (73)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (267)..(267)
<223> May be any nucleic acid
<400> 41
aaaggtggga gagaggaaaa aaggaaattc tatagaaatc tatatattgg gttgtttttt 60
tttttntttt ttnttttttt ttttttgggt tttttttttt tactatatat cttttttttg 120
ttgtctctag cctgatcaga taggagcaca agcaggggac ggaaagagag agacactcag 180
gcggcacatt tgccctccca gccactgagc tgtcgtgcca gcaccattcc tgggtcacgc 240
aaaacagaac ccagttagca gcagggnaga cgagaacacc acacaagaca tttt 294
CA 02331781 2005-04-22
-44-
<210> 42
<211> 610
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (576)..(576)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (590)..(590)
<223> May be any nucleic acid
<400> 42
tacgttttgt atgttttttt atttgctcca ggtggggttt tgactgtcac tttcccacac 60
tctggattag ttctgatccc accacaagga gccctcgaat tggctaaagt gagaaactgg 120
gcctgaagac tccgtaccct ctgccatctt gccgagggag tctcctttta gaaaacaatc 180
aaagggttat tgcatgagtc tggatgaatc ccactctcag ctgtccacgg gcccgaccac 240
ctcatctagc cccctttttg gcagggagaa cctggctccc aagttctcct ccttcacttc 300
gttacaaacc aaggggaaga gcccaccgtg agaacgcgcc atctgcaagc tgtctccctt 360
tctccatcct ttggtggaaa cccttttgcg cagaaggcaa cgtggaagct gatgttttta 420
ctcatctcgc tgtttgaaag caccattatg aagtcgggtt gtacagtagt taacagtacc 480
ttttatatat atatctcata tctatcatat atatataaac tggtaaacaa gaggtaacag 540
cgggcttcta gaaactgatt ttcttcaagg tttccngtgt ggtaggcacn tgaaatactg 600
gtagaaaatg 610
<210> 43
<211> 283
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (72)..(72)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (272)..(272)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-45-
<400> 43
taactttgtg tcgctacctc agtttgcccc catgtccctt acacacacgc aaaatactcc 60
ttcagcggag anacgaggtg gcggatgact ggcacgctcc atgaccggcc cagcagtctc 120
tgcctcgcca agcggctcat gttggagacg tcagtatagt ggactgggaa accaaatacc 180
ctttccattt cagtgcacca taagatgtcc tctttctcat tcatgaagac aggaaaaatg 240
ctggtctttg gcctgcttta tggagttttg anctcgtaag taa 283
<210> 44
<211> 383
<212> DNA
<213> Homo sapiens
<400> 44
gcggggacgt ccgcagcgtc acacagaagc atatccagga gtggggccca ttcgatctgg 60
tgattggggg cagtccctgc aatgacctct ccatcgtcaa ccctgctcgc aagggcctct 120
acgagggcac tggccggctc ttctttgagt tctaccgcct cctgcatgat gcgcggccca 180
aggagggaga tgatcgcccc ttctctggct ctttgagaat ttggtggcca tggcgttagt 240
acacagagag gacacatctc gcgatttctc gagtccaacc ctgtatatga ttgatgccaa 300
agaagtctca tctgcacaga ggcccctcta cttctggggt cacctccccg tattaacagg 360
ccgtaggatc cactgttatt ata 383
<210> 45
<211> 447
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (445) .. (445)
<223> May be any nucleic acid
<400> 45
acgttttgta tgttttttta tttgctccag gtggggtttt gactgtcact ttcccacact 60
ctggattagt tctgatccca ccacaaggag ccctcgaatt ggctaaagtg agaaactggg 120
cctgaagact ccgtaccctc tgccatcttg ccgagggagt ctccttttag aaaacaatca 180
aagggttatt gcatgagtct ggatgaatcc cactctcagc tgtccacggg cccgaccacc 240
tcatctaagc cccctttttg gcagggagaa cctggctccc aagttctcct ccttcacttc 300
gttacaaacc aaggggaaga gcccaccgtg agaacgcgcc atctgcaagc tgtctccctt 360
tctccatcct tggtgaaacc tttgcgcaga aggcaacgtg gaaagctgaa ggtttttact 420
catctcgctg tttgaaaagc accanta 447
<210> 46
CA 02331781 2005-04-22
-46-
<211> 100
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (96) . . (96)
<223> May be any nucleic acid
<400> 46
acaccaagaa catgagggat ggagagaagt atcagcaccc agaagagaaa aaggaattta 60
aaacaaaaac cacagaggcg gaaataccgg tgactnttct 100
<210> 47
<211> 150
<212> DNA
<213> Homo sapiens
<400> 47
tactccttca gcgggtagga ggtggcggat gactggcacg ctccatgacc ggcccagcag 60
tctctgcctc gccaagcgct catgttggag aggtcagtat agtggactgg gaaaccaaat 120
accctttcca tttcagtgca ccataagatg 150
<210> 48
<211> 237
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (7) . . (7)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (42)..(42)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (45)..(45)
<223> May be any nucleic acid
<400> 48
gctgtcncag gggtgtgtgg gtctaggagc ctggctggag gncancgctg ggtgggagct 60
CA 02331781 2005-04-22
-47-
tgggacaccg atgggcctgc atctgacctg ttgtgctcac tgcttaggac cctccaaagg 120
tttacccacc tgtcccagct gagaagagga agcccatccg ggtgctgtct ctctttgatg 180
gaatcgctac aggtgagggg tgcaggccca agaggtgctg gcctcgtgcg aattcct 237
<210> 49
<211> 442
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (19)..(19)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (91)..(91)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (137)..(137)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (388)..(388)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (397)..(397)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (428)..(428)
<223> May be any nucleic acid
<400> 49
ttttttacta tatatcttnt ttttgttgtc tctagcctga tcagatagga gcacaagcag 60
gggacggaaa gagagagaca ctcaggcggc natttccctc ccagccactg agctgtcgtg 120
ccagcaccat tcctggncac gcaaaacaga acccagttag cagcagggag acgagaacac 180
cacacaagac atttttctac agtatttcag gtgcctacca cacaggaaac cttgaagaaa 240
atcagtttct aggaagccgc tgttacctct tgtttacagt ttatatatat atggatagga 300
tatgaggata tatatataaa agggtactgt ttaactactg taccaacccg actttcataa 360
tgggtgcttt tcaaacagcc gaggatgngg ttaaaancat cagcttccac gttgccttct 420
gcggcaangg gtttcaccag gg 442
CA 02331781 2005-04-22
-48-
<210> 50
<211> 395
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (343)..(343)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (372)..(372)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (379)..(379)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (384)..(384)
<223> May be any nucleic acid
<400> 50
tacgttttgt atgttttttt atttgctcca ggtggggttt tgactgtcac tttcccacac 60
tctggattag ttctgatccc accacaagga gccctcgaat tggctaaagt gagaaactgg 120
gcctgaagac tccgtaccct ctgccatctt gccgagggag tctcctttta gaaaacaatc 180
aaagggttat tgcatgagtc tggatgaatc ccactctcag ctgtccacgg gcccgaccac 240
ctcatctagc cccctttttg ggcagggaga aacctgggct cccaagttct cctccttcac 300
ttcgttaaca aaccaagggg aagagcccac cgtgaggaac ggngccatct ggcaaggttg 360
ttctcccttt tnttccatnc cttnggtgaa aaccc 395
<210> 51
<211> 835
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (2) . . (9)
<223> May be any nucleic acid
<220>
<221> Unsure
CA 02331781 2005-04-22
-49-
<222> (11)(16)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (19)..(21)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (32)..(32)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (37)..(37)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (46)..(46)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (48)..(49)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (62)..(63)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (75)..(76)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (120)..(120)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (140)..(140)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (146)(146)
<223> May be any nucleic acid
<220>
<221> Unsure
CA 02331781 2005-04-22
-50-
<222> (199)..(199)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (300)..(300)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (365)..(365)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (388)..(388)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (397)..(397)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (403)..(403)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (421)..(421)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (441)..(441)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (461)..(461)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (475)..(475)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (494)..(494)
<223> May be any nucleic acid
<220>
<221> Unsure
CA 02331781 2005-04-22
-51-
<222> (514)..(514)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (536)..(537)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (545)..(545)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (550)..(550)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (554)..(554)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (562)..(562)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (565)..(565)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (569)..(569)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (580)..(580)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (584)..(584)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (587)..(587)
<223> May be any nucleic acid
<220>
CA 02331781 2005-04-22
-52-
<221> Unsure
<222> (595)..(595)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (599)..(599)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (617)..(617)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (629)..(629)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (639)..(639)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (658)..(658)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (660)..(660)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (663) .. (663)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (695)..(696)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (699)..(701)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (706)..(706)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-53-
<220>
<221> Unsure
<222> (710)..(710)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (716)..(719)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (727)..(727)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (731)..(731)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (735)..(737)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (739)..(739)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (743)..(745)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (754)..(755)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (781)..(781)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (787)..(787)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (790)..(790)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-54-
<220>
<221> Unsure
<222> (800)..(801)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (805)..(805)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (809)..(809)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (820)..(820)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (827)..(830)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (832)..(832)
<223> May be any nucleic acid
<400> 51
cnnnnnnnng nnnnnnttnn nctgccttta tnctcgntgc cgatantnnt atccatcatc 60
annttcttgg tgttnnatta tgttttgtgt tttttgtttg tttgtttaac tttgtgtcgn 120
tacctcagtt tgcccccatn tccctnacac acacgcaaaa tactccttca gcggagcgaa 180
gaggtggcgg atgactggna cgctccatga ccggcccagc agtctctgcc tcgccaagcg 240
gatcatgttg gagacgtcag tatagtggac tgggaaacca aatacccttt ccatttcagn 300
gcaccataag atgtcctctt tctcattcat gaagacaggg aaaatgctgg tctttggcct 360
gctcnatgga gtttgactcc gtagtaangg ccctcanttt ggntgacttg ggctatcctg 420
ncatgctcca gacacttccg nagggtcaca acagaagcat nttccagggg gtggnggcca 480
ttccgacctt tggnggattg ggggggaagc cccnaaaaat aaccccttca aacggnnaaa 540
ccctngttcn gaangggccc cnttncgang ggaaactggn ccgnttnttt ctttngggnt 600
tcctcccccc ccccccnaaa ataatgggng gccccaagna ggggaattac cccccccncn 660
ttnttttttt tttggaaatt tgggggcccg ggggnnaann naaaanggcn acttcnnnnt 720
ttttggnccc ncccnnnant ttnnncccaa aaannttaat taaaaaggcc cttttctggg 780
ncccccnttn aaccgccccn ngatnggtnc ttggttcccn aacacannnn cncaa 835
<210> 52
CA 02331781 2005-04-22
-55-
<211> 479
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (364)..(364)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (416)..(416)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (464)..(464)
<223> May be any nucleic acid
<400> 52
tacgttttgt atgttttttt atttgctcca ggtggggttt tgactgtcac tttcccacac 60
tctggattag ttctgatccc accacaagga gccctcgaat tggctaaagt gagaaactgg 120
gcctgaagac tccgtaccct ctgccatctt gccgagggag tctcctttta gaaaacaatc 180
aaagggttat tgcatgagtc tggatgaatc ccactctcag ctgtccacgg gcccgaccac 240
ctcatctagc cccctttttg gcagggagaa cctggctccc aagttctcct ccttcacttc 300
gttacaaacc aaggggaaga gcccaccatg agaacgcgcc atctgcaagc tgtctccctt 360
tctncatcct tggtgaaacc tttgcgcaga aggcaacgtg gaagctgatg tttttntcat 420
ctcgctgttt gaaagcacca ttatgaagtc gggttgtaca gtantaacag tacttttag 479
<210> 53
<211> 521
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (327)..(327)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (507)..(507)
<223> May be any nucleic acid
<400> 53
agaacaccac acaagacatt tttctacagt atttcaggtg cctaccacac aggaaacctt 60
CA 02331781 2005-04-22
-56-
gaagaaaatc agtttctaga agccgctgtt acctcttgtt tacagtttat atatatatga 120
tagatatgag atatatatat aaaaggtact gttaactact gtacaacccg acttcataat 180
ggtgctttca aacagcgaga tgagtaaaaa catcagcttc cacgttgcct tctgcgcaaa 240
gggtttcacc aaggatggag aaagggagac agcttgcaga tggcgcgttc tcatggtggg 300
ctcttcccct tggtttgtaa cgaagtntag gaggagaact tgggagccag gttctccctg 360
ccaaaaaggg ggctagatga ggtggtcggg cccgtggaca gctgagagtg ggattcatcc 420
agactcatgc aataaccctt tgattgtttc taaaaggaga ctccctcggc aagatggcag 480
agggtacgga gtcttcaggc ccagttntca ctttagccaa t 521
<210> 54
<211> 440
<212> DNA
<213> Homo sapiens
<400> 54
ctctctttga tggaatcgct acagggctcc tggtgctgaa ggacttgggc attcaggtgg 60
accgctacat tgcctcggag gtgtgtgagg actccatcac ggtgggcatg gtgcggcacc 120
aggggaagat catgtacgtc ggggacgtcc gcagcgtcac acagaagcat atccaggagt 180
ggggcccatt cgatctggtg attgggggca gtccctgcaa tgacctctcc atcgtcaacc 240
ctgctcgcaa gggcctctac gagggcactg gccggctctt ctttgagttc taccgcctcc 300
tgcatgatgc gcggcccaag gagggagatg atcgcccctt cttctggctc tttgagaatg 360
tggtggccat gggcgtttag tgacaagagg gacatctcgc gatttctcga gtccaaccct 420
gtgatgattg atgccaaaga 440
<210> 55
<211> 273
<212> DNA
<213> Homo sapiens
<400> 55
acgttttgta tgttttttta tttgctccag gtggggtttt gactgtcact ttcccacact 60
ctggattagt tctgatccca ccacaaggag ccctcgaatt ggctaaagtg agaaactggg 120
cctgaagact ccgtaccctc tgccatcttg ccgagggagt ctccttttag aaaacaatca 180
aagggttatt gcatgagtct ggatgaatcc cactctcagc tgtccacggg cccgaccacc 240
tcatctagcc ccctttttgg cagggagaac ctg 273
<210> 56
<211> 190
<212> DNA
CA 02331781 2005-04-22
-57-
<213> Homo sapiens
<220>
<221> Unsure
<222> (39)..(39)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (83)..(83)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (181)..(181)
<223> May be any nucleic acid
<400> 56
aaaaacacaa aacataataa aacaccaaga acatgaggnt ggagagaagt atcagcaccc 60
agaagagaaa aaggaattta aancaaaaac cacagaggcg gaaataccgg agggctttgc 120
cttgcgaaaa gggttggaca tcatctcctg atttttcaat gttattcttc agtcctattt 180
naaaacaaag 190
<210> 57
<211> 445
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (167)..(167)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (353)..(353)
<223> May be any nucleic acid
<400> 57
ttagacaaat actgatttta attaaacata aggtaaactc taggcatccg tcatctttca 60
gcctaaaaat tagcaaaaac tgttgaaaca aggcacagtt ttttccccat atttgttacg 120
tcgtggctcc agttacaaaa aaattttaat gaaaacgtta aacatanaaa tagaagtttg 180
agattttaaa aagtgtataa aaagccccac aaaacttgtc aacggttgtt ccttattcta 240
caaaatagca ccagtaagaa gagtaaaagg tgttaaaaac catttatgac agcatttctg 300
aaatgcagct tgtctgaatt cccggttctc cctaaaaacg acttctttat ggnattaaaa 360
CA 02331781 2005-04-22
-58-
aagggtttaa aaaaatctcc aaaggggagc accgagcttt gcaggttttc cctgtcatct 420
ctcagatgtg ggggaagctc gtggc 445
<210> 58
<211> 287
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (38)..(38)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (171)..(171)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (204)..(204)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (274)..(274)
<223> May be any nucleic acid
<400> 58
ttccccacat ctgagagatg acagggaaaa ctgcaaanct cggtgctccc tttggagatt 60
ttttaatcct tttttattcc ataagaagtc gtttttaggg agaacgggaa ttcagacaag 120
ctgcatttca gaaatgctgt cataatggtt tttaacacct tttactcctc nttactggtg 180
ctatttttgt agaataaggg aacnacgttg acaagttttg gtgggggcct ttttatacac 240
cttttttaaa atctccaact tcctaatttt taanggttta accgttt 287
<210> 59
<211> 535
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (452)..(452)
<223> May be any nucleic acid
<220>
<221> Unsure
CA 02331781 2005-04-22
-59-
<222> (526)..(526)
<223> May be any nucleic acid
<400> 59
tagacaaata ctgattttaa ttaaacataa ggtaaactct aggcatccgt catctttcag 60
cctaaaaatt agcaaaaact gttgaaacaa ggcacagttt tttccccata tttgttacgt 120
cgtggctcca gttacaaaaa aattttaatg aaaacgttaa acataaaaat agaagtttga 180
gattttaaaa agtgtataaa aagccccaca aaacttgtca acgttgttcc ttattctaca 240
aaatagcacc agtaagaaga gtaaaaggtg ttaaaaacca ttatgacagc atttctgaaa 300
tgcagcttgt ctgaattccc gttctcccta aaaacgactt cttatggaat aaaaaaggat 360
taaaaaatct ccaaagggag caccgagctt tgcagttttc cctgtccgtc tctcagatgt 420
ggggaaggta tgagaaatgt atgtctgtcc cngactgctg tcactgcctc tgagttagta 480
aaaggtgaga atgagggtag cagcttccca tctggggcct gtgccngtgg agggt 535
<210> 60
<211> 449
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (7) . . (7)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (200) . . (200)
<223> May be any nucleic acid
<400> 60
atcgcancag gctacctagt cctcaaagag ttgggcataa aggtaggaaa gtacgtcgct 60
tctgaagtgt gtgaggagtc cattgctgtt ggaaccgtga agcacgaggg gaatatcaaa 120
tacgtgaacg acgtgaggaa catcacaaag aaaaatattg aagaatgggg cccatttgac 180
ttggtgattg gcggaaccan tgcaacgatc tctcaaatgt gaatccagcc aggaaaggcc 240
tgtatgaggg tacaggccgg ctcttcttcg aattttacca cctgctgaat tactcacgcc 300
ccaaggaggg tgatgaccgg ccgttcttct ggatgtttga gaatgttgta gccatgaagg 360
ttggcgacaa gagggacatc tcacggttcc tggagtgtaa tccagtgatg attgatgcca 420
tccaaagttt ctgctgctca cagggcccg 449
<210> 61
<211> 522
<212> DNA
CA 02331781 2005-04-22
-60-
<213> Homo sapiens
<220>
<221> Unsure
<222> (146)..(146)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (281)..(281)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (304) . . (304)
<223> May be any nucleic acid
<400> 61
aagagggaca tctcacggtt cctggagtgt aatccagtga tgattgatgc catcaaagtt 60
tctgctgctc acagggcccg atacttctgg ggcaacctac ccgggatgaa caggcccgtg 120
atagcatcaa agaatgataa actcgngctg caggactgct tggaatacaa taggatagcc 180
aagttaaaga aagtacagac aataaccacc aagtcgaact cgatcaaaca ggggaaaaac 240
caacttttcc ctgttgtcat gaatggcaaa gaagatgttt ngtggtgcac tgagctcgaa 300
aggntctttg gctttcctgt gcactacaca gacgtgtcca acatgggccg tggtgcccgc 360
cagaagctgc tgggaaggtc ctggagcgtg cctgtcatcc gacacctctt cgcccctctg 420
aaggactact ttgcatgtga atagttccag ccagggccca agcccactgg ggtgtgtggc 480
agagcaggac ccaggaggtg tgattctgaa ggcatcccca gg 522
<210> 62
<211> 573
<212> DNA
<213> Homo sapiens
<400> 62
ctaagatcca ttttctaaac tccaattgag cattctctgt atctgggtgg tttttacttt 60
tttacttaat cttgcttgat caggaactct ggtgtcttct tggcccccca cgtgatctcg 120
ttcatggtca cttttttgtt tatctcattt tctctgaggc tggtccttcc tgttaacgtc 180
ttggcatttg tgggaagcac aaaatgttct tgtccctcca actctgcttt tcgctccctg 240
ccctgccatt cctctcccgc gcctgccctc tcccttccat ctttcccagg tacttttctc 300
tcccagccct gccactcttc tgccgcacct gcgctctccc ctccatcttt cccaggtact 360
tttgagcctt gactccccag gtcccttcat tctgtgctca ctccatgatg tcattttgtt 420
ctccagttaa agaaagtaca gacaataacc accaagtcga actcgatcaa acaggggaaa 480
aaccaacttt tccctgttgt catgaatggc aaagaagatg ttttgtggtg cactgagctc 540
CA 02331781 2005-04-22
-61-
gaaaggatct ttggctttcc tgtgcactac aca 573
<210> 63
<211> 559
<212> DNA
<213> Homo sapiens
<400> 63
agacaaatac tgattttaat taaacataag gtaaactcta ggcatccgtc atctttcagc 60
ctaaaaatta gcaaaaactg ttgaaacaag gcacagtttt ttccccatat ttgttacgtc 120
gtggctccag ttacaaaaaa attttaatga aaacgttaaa cataaaaata gaagtttgag 180
attttaaaaa gtgtataaaa agccccacaa aacttgtcaa cgttgttcct tattctacaa 240
aatagcacca gtaagaagag taaaaggtgt taaaaaccat tatgacagca tttctgaaat 300
gcagcttgtc tgaattcccg ttctccctaa aaacgacttc ttatggaata aaaaaggatt 360
aaaaaatctc caaagggagc accgagcttt gcagttttcc ctgtcatcta tcagatgtgg 420
ggaaggtatg agaaatgtat gtctgtccct gactgctgtc actgcctctg agtttagtaa 480
aaagatgaga aatgagggta gcagacttct catctgggga cctgtgcctg tggagggtag 540
gtctcctgga gagggaatg 559
<210> 64
<211> 391
<212> DNA
<213> Homo sapiens
<400> 64
ttttttttta gacaaatact gattttaatt aaacataagg taaactctag gcatccgtca 60
tctttcagcc taaaaattag caaaaactgt tgaaacaagg cacagttttt tccccatatt 120
tgttacgtcg tggctccagt tacaaaaaaa attttaatga aaacgttaaa cataaaaata 180
gaagtttgag attttaaaaa gtgtataaaa agccccacaa aacttgtcaa cgttgttcct 240
tattctacaa aatagcacca gtaagaagag taaaaggtgt taaaaaccat tatgacagca 300
tttctgaaat gcagcttgtc tgaattcccg ttctccctaa aaacgacttc ttatggaata 360
aaaaaggatt aaaaaatctc caaagggagc a 391
<210> 65
<211> 517
<212> DNA
<213> Homo sapiens
<400> 65
CA 02331781 2005-04-22
-62-
acaaatactg attttaatta aacataaggt aaactctagg caggggcatc tttcagccta 60
aaaattagca aaaactgttg aaacaaggca cagttttttc cccatatttg ttacgtcgtg 120
gctccagtta cggaaaaatt ttaatgaaaa cgttaaacat aaaaatagaa gtttgagatt 180
ttaaaaagtg tataaaaagc cccacaaaac ttgtcaacgt tgttccttat tctacaaaat 240
agcaccagta agaagagtaa aaggtgttaa aaaccattat gacagcattt ctgaaatgca 300
gcttgtctga attcccgttc tccctaaaaa cgacttctta tggaataaaa aaggattaaa 360
aaatctccaa agggagcacc gagctttgca gttttccctg tcatctctca gatgtgggga 420
aggtatgaga aatgtatgtc tgtccctgac tgctgtcact gcctctgagt ttagtaaaaa 480
gatgagaaat gagggtagca gacttctcat ctgggga 517
<210> 66
<211> 442
<212> DNA
<213> Homo sapiens
<400> 66
gacaaatact gattttaatt aaacataagg taaactctag gcatccgtca tctttcagcc 60
taaaaattag caaaaactgt tgaaacaagg cacagttttt tccccatatt tgttacgtcg 120
tggctccagt tacaaaaaaa attttaatga aaacgttaaa cataaaaata gaagtttgag 180
attttaaaaa gtgtataaaa agccccacaa aacttgtcaa cgttgttcct tattctacaa 240
aatagcacca gtaagaagag taaaaggtgt taaaaaccat tatgacagca tttctgaaat 300
gcagcttgtc tgaattcccg ttctccctaa aaacgacttc ttatggaata aaaaaggatt 360
aaaaaatctc caaagggagc accgagcttt gcagttttcc ctgtcatctc gcagatgtgg 420
ggaaggtatg agaaatgtat gt 442
<210> 67
<211> 396
<212> DNA
<213> Homo sapiens
<400> 67
gcagtcaggg acagacatac atttctcata ccttccccac atctgagaga tgacagggaa 60
aactgcaaag ctcggtgctc cctttggaga ttttttaatc cttttttttt ccataagaag 120
tcgtttttag ggagaacggg aattcagaca agctgcattt cagaaatgct gtcataatgg 180
tttttaacac cttttactct tcttactggt gctattttgt agaataagga acaacgttga 240
caagttttgt ggggcttttt atacactttt taaaatctca aacttctatt tttatgttta 300
acgttttcat taaaattttt ttgtaactgg agccacgacg taacaaatat ggggaaaaaa 360
ctgtgccttg tttcaacagt ttttgctaat ttttag 396
CA 02331781 2005-04-22
-63-
<210> 68
<211> 287
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (7) . . (7)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (169)..(169)
<223> May be any nucleic acid
<400> 68
agacaantac tgattttaat taaacataag gtaaactcta ggcatccgtc atctttcagc 60
ctaaaaatta gcaaaaactg ttgaaacaag gcacagtttt tcccccatat ttgttacgtc 120
gtggctccag ttacaaaaaa aattttaatg aaaacgttaa acataaaant agaagtttga 180
gattttaaaa agtgtataaa aagccccaca aaacttgtca acgttgttcc ttattctaca 240
aaatagcacc agtaagaaga gtaaaaggtg ttaaaaacca ttatgac 287
<210> 69
<211> 356
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (193)..(193)
<223> May be any nucleic acid
<400> 69
attgaagaat ggggcccatt tgacttggtg attggcggaa ccgatgcaac gatctctcaa 60
atgtgaatcc agccaggaaa ggcctgtatg agggtacagg ccggctcttc ttcgaatttt 120
accacctgct gaattactca cgccccaagg agggtgatga ccggccgttc ttctggatgt 180
ttgagaatgt tgnagccatg aaggttggcg acaagaggga catctcacgg ttcctggagt 240
gtaatccagt gatgattgat gccatcaaag tttctgctgc tcacagggcc cgatacttct 300
ggggcaacct acccgggatg aacaggatct ttggctttcc tgtgcactac acagac 356
<210> 70
<211> 408
CA 02331781 2005-04-22
-64-
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (408)..(408)
<223> May be any nucleic acid
<400> 70
tttagacaaa tactgatttt aattaaacat aaggtaaact ctaggcatcc gtcatctttc 60
agcctaaaaa ttagcaaaaa ctgttgaaac aaggcacagt tttttcccca tatttgttac 120
gtcgtggctc cagttacaaa aaaaatttta atgaaaacgt taaacataaa aatagaagtt 180
tgagatttta aaaagtgtat aaaaagcccc ac.aaaacttg tcaacgttgt tccttattct 240
acaaaatagc accagtaaga agagtaaaag gtgttaaaaa ccattatgac agcatttctg 300
aaatgcagct tgtctgaatt cccgttctcc ctaaaaacga cttcttatgg aataaaaaag 360
gattaaaaaa tctccaaagg gagcaccgag ctttgcagtt ttccctgn 408
<210> 71
<211> 439
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (50) . . (50)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (85)..(85)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (405)..(405)
<223> May be any nucleic acid
<400> 71
gcatgtagct acaggacatt tttaagggcc caggatcgtt ttttcccagn tgcaagcaga 60
agagaaaatg ttgtatatgt ctttnacccg gcacattccc cttgcctaaa tacaagggct 120
ggagtctgca cgggacctat tagagtattt tccacaatga tgatgatttc agcagggatg 180
acgtcatcat cacattcagg gctatttttt cccccacaaa cccaagggca ggggccactc 240
ttagctaaat ccctccccgt gactgcaata gaaccctctg gggagctcag gaaagggggt 300
CA 02331781 2005-04-22
-65-
gtgctgagtt ctataatata agctgccata tattttgtag acaagtatgg ctcctcccat 360
atctccctct tccctaggag aggagtgtga aagcaaggga gcttngataa gacaccccct 420
caaacccatt ccctctcca 439
<210> 72
<211> 491
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (26) . . (27)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (33) . . (33)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (188)..(188)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (301)..(301)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (339)..(339)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (360)..(360)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (379)..(379)
<223> May be any nucleic acid
<400> 72
ttaattaaac ataaggtaaa ctctanngca tcngtcatct ttcagcctaa aaattagcaa 60
aaactgttga aacaaggcac agttttttcc ccatatttgt tacgtcgtgg ctccagttac 120
aaaaaaaatt ttaatgaaaa cgttaaacat aaaaatagaa gtttgagatt ttaaaaagtg 180
tataaaangc cccacaaaac ttgtcaacgt tgttccttat tctacaaaat agcaccagta 240
CA 02331781 2005-04-22
-66-
agaagagtaa aaggtgttaa aaaccattat gacagcattt ctgaaatgca gcttgtctga 300
nttcccgttc tccctaaaaa cgacttctta tgggataana aagggattaa aaaatctccn 360
aaagggaggc accgagcttt gcaggttttc cctggtcatc tctcaggatg tggggggagg 420
gtatggggaa atggtatggt ctggtccctg gactggctgg tcactgcctc tggggtttng 480
gtaaaagggt g 491
<210> 73
<211> 443
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (9) . . (9)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (11)..(11)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (23) .. (24)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (126)..(126)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (157)..(157)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (170)..(170)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (341)..(341)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (347)..(347)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-67-
<220>
<221> Unsure
<222> (371)..(371)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (405)..(405)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (412)..(412)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (430)..(430)
<223> May be any nucleic acid
<400> 73
ttggcggcna ntgcaacgat ctnnaaatgt gaatcagcca ggaaaggctg tatgagggac 60
aggcggctct tcttcgaatt ttccacctgc tgaattactc acgccccaag gagggtgatg 120
accggncgtt cttctggatg tttgagaatg ttgtagncat gaaggttggn gacaagaggg 180
acatctcacg gttcctggag tgtaatccag tgatgattga tgccatcaaa gtttctgctg 240
ctcacagggc ccgatacttc tggggcaacc tacccgggat gaacaggatc tttggctttc 300
ctgtgcacta cacagacgtg tcccaacatg gggccgtggg ngccgcncca ggaagcttgc 360
tggggaaggt nctggggagc gttgccttgt tcatcccgac acctntttcg gnccctattg 420
gaagggattn atttttgcca tgt 443
<210> 74
<211> 273
<212> DNA
<213> Homo sapiens
<400> 74
acgttttgta tgttttttta tttgctccag gtggggtttt gactgtcact ttcccacact 60
ctggattagt tctgatccca ccacaaggag ccctcgaatt ggctaaagtg agaaactggg 120
cctgaagact ccgtaccctc tgccatcttg ccgagggagt ctccttttag aaaacaatca 180
aagggttatt gcatgagtct ggatgaatcc cactctcagc tgtccacggg cccgaccacc 240
tcatctagcc ccctttttgg cagggagaac ctg 273
<210> 75
<211> 250
CA 02331781 2005-04-22
-68-
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (26)..(27)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (33)..(33)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (188)..(188)
<223> May be any nucleic acid
<400> 75
ttaattaaac ataaggtaaa ctctanngca tcngtcatct ttcagcctaa aaattagcaa 60
aaactgttga aacaaggcac agttttttcc ccatatttgt tacgtcgtgg ctccagttac 120
aaaaaaaatt ttaatgaaaa cgttaaacat aaaaatagaa gtttgagatt ttaaaaagtg 180
tataaaangc cccacaaaac ttgtcaacgt tgttccttat tctacaaaat agcaccagta 240
agaagagtaa 250
<210> 76
<211> 443
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (9) . . (9)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (11)..(11)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (23) . . (24)
<223> May be any nucleic acid
<220>
<221> Unsure
CA 02331781 2005-04-22
-69-
<222> (126)..(126)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (157)..(157)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (170)..(170)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (341)..(341)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (347)..(347)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (371)..(371)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (405). .(405)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (412)..(412)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (430)..(430)
<223> May be any nucleic acid
<400> 76
ttggcggcna ntgcaacgat ctnnaaatgt gaatcagcca ggaaaggctg tatgagggac 60
aggcggctct tcttcgaatt ttccacctgc tgaattactc acgccccaag gagggtgatg 120
accggncgtt cttctggatg tttgagaatg ttgtagncat gaaggttggn gacaagaggg 180
acatctcacg gttcctggag tgtaatccag tgatgattga tgccatcaaa gtttctgctg 240
ctcacagggc ccgatacttc tggggcaacc tacccgggat gaacaggatc tttggctttc 300
ctgtgcacta cacagacgtg tcccaacatg gggccgtggg ngccgcncca ggaagcttgc 360
tggggaaggt nctggggagc gttgccttgt tcatcccgac acctntttcg gnccctattg 420
CA 02331781 2005-04-22
-70-
gaagggattn atttttgcca tgt 443
<210> 77
<211> 394
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (1) . (1)
<223> May be any nucleic acid
<400> 77
nttttttttt ttttgaaaaa attgtgaaaa aatttaaacc ccaggggact atccaagggg 60
aaaagtgaaa tatggaaaaa ttggcggtat gaccaatttg ggcattgcaa agagccttgc 120
agaattatga agcataaaag gaaattattg gcttttggag agttttcttt tctctcttct 180
ttttttgtaa tttcaatcta tatcagtagt ggaaaggtca tagcaaaata tggagaatcc 240
aaatggtaga tacaacctga tatcttgtgg aacaaggcat acaacagcaa agcaacacca 300
gtgaaaccaa ggacaccaaa cagtccccag agaactccag ctgtcatgag gtctcttcta 360
tagccatcag gtcctgagat ggagactggc actg 394
<210> 78
<211> 277
<212> DNA
<213> Homo sapiens
<400> 78
gtcatctttc agcctaaaaa ttagcaaaaa ctgttgaaac aaggcacagt tttttcccca 60
tatttgttac gtcgtggctc cagttaccaa aaaattttaa tgaaaacgtt aaacataaaa 120
atagaagttt gagattttaa aaagtgtata aaaagcccca caaaacttgt caacgttgtt 180
ccttattcta caaaatagca ccagtaagaa gagtaaaagg tgttaaaaac cattatgaca 240
gcatttctga aatgcagctt gtctgaattc ccgttct 277
<210> 79
<211> 469
<212> DNA
<213> Homo sapiens
<400> 79
ttttagacaa atactgattt taattaaaca taaggtaaac tctaggcatc cgtcatcttt 60
CA 02331781 2005-04-22
-71-
cagcctaaaa attagcaaaa actgttgaaa catggcacag ttttttcccc atatttgtta 120
cgtcgtggct ccagttacaa aaaaatttta atgaaaacgt taaacataaa aatagaagtt 180
tgagatttta aaaagtgtat aaaaagcccc acaaaacttg tcaacgttgt tccttattct 240
acaaaatagc accagtaaga agagtaaaag gtgttaaaaa ccattatgac agcatttctg 300
aaatgcagct tgtctgaatt cccgttctcc ctaaaaacga cttcttatgg aataaaaaag 360
gattaaaaaa tctccaaagg gagcaccgag ctttgcagtt ttccctgtca tctctcagat 420
gtggggaagg tatgagaaat gtatgtctgt ccctgactgc tgtcactgc 469
<210> 80
<211> 206
<212> DNA
<213> Homo sapiens
<400> 80
gacaaatact gatcccccct acacataagg taaactctag gcatccgtca tctttcagcc 60
taaaaattag caaaaactgt tgaaacaagg cacagttttt tccccatatt tgttacgtcg 120
tggctccagt tacgaaaaaa attttaatga aaacgttaaa cataaaaata gaagtttgag 180
attttaaaaa gtgtataaaa agcccc 206
<210> 81
<211> 391
<212> DNA
<213> Homo sapiens
<400> 81
ttttagacaa atactgattt taattaaaca taaggtaaac tctaggcatc cgtcatcttt 60
cagcctaaaa attagcaaaa actgttgaaa caaggcacag ttttttcccc atatttgtta 120
cgtcgtggct ccagttacaa aaaaaatttt aatgaaaacg ttaaacataa aaatagaagt 180
ttgagatttt aaaaagtgta taaaaagccc cacaaaactt gtcaacgttg ttccttattc 240
tacaaaatag caccagtaag aagagtaaaa ggtgttaaaa accattatga cagcatttct 300
gaaatgcagc ttgtctgaat tcccgttctc cctaaaaacg acttcttatg gaataaaaaa 360
ggattaaaaa atctccaaag ggagcaccga g 391
<210> 82
<211> 755
<212> DNA
<213> Homo sapiens
<220>
<221> Unsure
<222> (10) .. (10)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-72-
<220>
<221> Unsure
<222> (19)..(19)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (47) . . (47)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (117)..(117)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (119)..(119)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (134)..(134)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (136)..(136)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (138)..(139)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (146)..(147)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (149)..(149)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (157)..(158)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (160)..(160)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-73-
<220>
<221> Unsure
<222> (162)..(162)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (164)..(164)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (170)..(172)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (176)..(178)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (180)..(181)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (186)..(186)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (191)..(194)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (199)..(199)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (205)..(205)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (215)..(215)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (217)..(217)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-74-
<220>
<221> Unsure
<222> (219)..(220)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (226)..(226)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (231)..(231)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (234)..(234)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (237)..(237)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (243)..(244)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (257)..(257)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (259)..(259)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (275)..(275)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (298)..(298)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (301)..(301)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-75-
<220>
<221> Unsure
<222> (338)..(338)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (374)..(374)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (382)..(832)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (416). .(416)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (436)..(436)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (481)..(481)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (486)..(486)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (498) . . (498)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (502)..(502)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (504)..(504)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (524)..(524)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-76-
<220>
<221> Unsure
<222> (528)..(528)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (568)..(568)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (572)..(572)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (577)..(577)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (579)..(579)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (587)..(587)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (596)..(596)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (598)..(599)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (615)..(615)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (619)..(619)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (624)..(624)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-77-
<220>
<221> Unsure
<222> (626)..(626)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (628)..(628)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (631)..(631)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (638)..(638)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (643)..(643)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (655)..(655)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (663)..(663)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (666)..(666)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (668)..(668)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (701)..(701)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (716)..(716)
<223> May be any nucleic acid
CA 02331781 2005-04-22
-78-
<220>
<221> Unsure
<222> (739)..(739)
<223> May be any nucleic acid
<220>
<221> Unsure
<222> (747)..(747)
<223> May be any nucleic acid
<400> 82
tcttcgaagn cgagtcggnc tgtaccctca tacaggcctt tcctggntgg attcacattt 60
gagagatcgt tgcatgggct tccgccaatc accaagtcaa atgggcccca ttcttcnana 120
tttttctttg gggngngnnc cccccnngnc ccccccnngn tntntttttn nntttnnncn 180
ngtccncccg nnnngggtnc tcacncactt cagangngnn gggctntcct nccnttntgg 240
ccnnctcttt gcggatngnt aggctgtcgc gatgncatca aacaatgaca ggactcgnct 300
nggcgccttc gggctgcggg aatgggagga tctttggntt tcctgtgcac tacacagacg 360
tgtccaacat gggncgtggt gnccgccaga agcttgctgg ggaaggtcct tggagnggtg 420
tcttgtcaat cccganaacc tctttccggc cccccttgga aggggcttac ttctgggaat 480
ngttgnattt ggtcccangc cnangggccc caaaaggccc ccantttngg gggttgtttt 540
ttggaaagga ggcccaaggg accccccngg gnggggngnt tgtttcnccc ctgggnanng 600
ggaattcccc cccangggnc cccngntntt nttccccncc aantttttgg ggttnggggt 660
tanaanancc cgggggtttc ccccccaagg ccccccctct ntttgggttc aaaaangggg 720
gggggggaag gggcccccnc cctgaanttt ttttc 755