Note: Descriptions are shown in the official language in which they were submitted.
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99100453
Process to collect metabolites from modified nectar by
insects.
Field of the invention
The present invention relates to isolated, purified DNA
sequences which can act as promoters in eukaryotic cells.
More specifically, the present invention is related to such
DNA sequences which act as promoters to express genes in
nectaries of plants. The present invention also relates to
chimerical gene constructs comprising a structural or a
synthetic gene under the control of a promoter that effects
expression of said genes in nectaries. This invention also
relates to a process for producing metabolites in honey by
allowing insects, preferably bees, to collect and process
nectar from plants that excrete said metabolites in nectar
or other exudates. Further, this invention relates to plant
cells, plants or derivatives therefrom, that express the
said chimerical gene.
Background of the invention
Nectaries are nectar secreting organs or tissues that can
be located inside (floral) or outside (extrafloral) the
flower. The main component of nectar is sugar, the variati-
on between nectars of flowers from different species mainly
being the concentration and ratio of glucose, fructose and
sucrose (Baker a:nd Baker, 1982). In addition, depending on
the plant species, varying amounts of polysaccharides,
lipids, organic acids, volatiles, minerals, phosphates,
alkaloids, amino acids and proteins have been detected
SUBSTITUTE SHEET (RUE.E 2S~
CA 02333897 2001-O1-16
WO 00/04176 PCT/Nh99/00453
_ Z _
(Baker and Baker, 1982). Being a specialised sink organ,
the nectaries are supplied with sucrose by phloem unloading
(Davis et al., 1985, Hagitzer and Fahn, 1992).
The mechanisms of sugar accumulation and nectar secretion
have been described for several plant species (Fahn et al.,
1979). Sugar transport to the nectaries is achieved by
active transport mechanisms and/or osmotic and chemical
gradients. In the nectaries of many plants sucrose is
to converted to glucose and fructose, resulting in a hexose
dominant nectar. Part of the hexoses are converted to
starch, which is hydrolysed prior to anthesis and nectar
secretion. Cell to cell transport of nectar in the nectary
parenchyma tissue is mainly symplastic, as demonstrated by
the presence of many plasmodesmata between these cells
(Fahn et al., 1979). Nectar is secreted from secretory
cells via the cell membrane (eccrine secretion) or via the
Golgi and endoplasmatic reticulum vesicles (granulocrine
secretion). Research on the molecular regulation of nectary
development and nectary biochemistry has not been reported.
The main function of floral nectar is to reward pollinating
insects. Insects collect nectar to meet-their short-term
energy requirements. Colony-living honeybees process large
quantities of nectar into honey, which is stored in honey-
cambs of the beehive and is used as food supply during the
winter period. Within the bee colony different classes of
worker bees cooperate in the honey production process.
Foraging bees collect pollen and nectar from the flowers
and bring it to the hive. On returning to the hive they
give most of it u.p to household bees . Pollen is used as a
protein source, especially to feed the brood. Adult nurse
and worker bees use little protein, their capacity to
digest proteins being very low (Crailsheim et al., 1993).
Honey processing takes place by repeated swallowing and
bringing up of the nectar from the honey stomach. In the
first process 15% of the water content is lost. This semi-
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 3 -
processed nectar is temporarily stored in a honeycomb cell
and taken out later far further processing. The final
process includes filtering the honey to discard small
particles like pollen grains. Sugar metabolising enzymes
(invertase, amylase) are added and the honey is concentra-
ted to an average water content of 20%. Most nectars and
honeys only contain traces of protein (<0.2%). However,
CaZZuna vulgaris (heather) honey can contain up to 1.8%
protein, giving it thixotropic properties (Butler, 1962).
It is known that bees add enzymes like invertase to nectar
during the honey processing. Therefore, the probability
that proteases are also added is very low. Protein digesti-
on does not take place in the honey stomach but in the
intestine of the honeybee. However, the ability of adult
worker bees to digest proteins is very low, their main
requirement being energy which they obtain from nectar.
Until now, it was not established which proteins are
present in heather honey and whether these originate from
floral heather nectar or are added to honey by honeybees.
In the present invention it was established that heather
honey contains two unique proteins that originate from
floral nectar of heather. Based on these results a produc
tion system for proteins in nectar and honey was estab
fished.
It is an object of the present invention to show that
recombinant proteins can be secreted in nectar of transge-
nic plants, that this nectar is collected by honeybees and
that the bees process this nectar into honey that contains
the unaltered protein in a concentrated form.
Defi,niti.ons
Honev: A substance that contains approximately 80% sugar
and varying amounts of other components and that is produ-
ced by insects, preferably bees, that collect and process
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/4176 PCT/NL99/00453
_ g _
nectar from floral or extrafloral nectaries, from honeydew,
other plant exudates or artificial sugar solutions.
MADS box crepe: a gene coding for a transcription factor
having a region of 5& amino acids which is homologous to a
similar region a_n the Arabidopsis AGAMOUS protein and
Antirrhinum DEFICIENS protein. This region is called the
'MADS box'. At least 500 of the amino acids in this region
should be identical to the amino acid composition in the
MADS boxes of AGA1!~OUS and/or DEFICIENS.
Nectarv: secretory organ or secretory tissue of plants,
located in the flowers (floral nectaries) or outside the
flower (extrafloral nectaries) that excrete nectar.
Nectar: sugar containing fluid that is secreted by necta-
ries. Nectar can also contain substances like minerals,
amino acids, proteins, organic acids, volatiles, alkaloids
etc.
Recombinant urotein: the gene product of a recombinant DNA
molecule.
Recombinant DNA molecule: A DNA molecule in which sequences
which are not naturally contiguous have been placed next to
each other by in vitro manipulations.
Promoter: The DNA region, usually upstream to the coding
sequence of a gene, which binds RNA polymerase and directs
the enzyme to the correct transcriptional start site.
Summary of the invention
The production of: recombinant proteins for pharmaceutical
purposes is a growing market. Until now, mainly bacterial
and yeast systems have been used for bulk production of
proteins. Recently animal production systems have also been
SUBSTITUTE 5HEET (RULE 26)
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99/00453
- 5 -
developed. With the availability of efficient transformati-
on techniques fox plants, procedures to use plants for the
production of proteins are now in progress. In plants, the
recombinant proteins are targeted to sink organs like
tubers and seeds. A serious draw-back of these production
methods is that the recombinant protein can only be obtai-
ned after extended, and therefore expensive, purification
steps.
l0 The present invention provides a method to produce metab-
olites, preferably recombinant proteins in honey, which is
manufactured by insects, preferably honeybees; that collect
floral nectar of transgenic plants. Harvesting of honey is
very simple and purification of the protein is very
straight forward and requires no advanced purification
steps. To give an estimation of the protein yield in a crop
like rapeseed, we suggest an average protein production of
2% in honey, as has been found in honey of heather. Tf one
hectare of rapeseed yields 100-500 kilo honey in one
season, a yield of 2 to l0 kilo protein can be obtained. Tn
addition, the present invention provides a method to
collect metabolites from honey that is derived from non-
transgenic plants that secrete these metabolites in nectar.
An example are secondary metabolites like acetylandromedol,
a diterpine compound, that is excreted in nectar of
Rhododendron arboreum and Rhododendron barbatum and of
Piptanthus nepalens.is (Martini et al., 1990).
This invention provides a gene from petunia, NEC1, that is
highly expressed in the nectaries of petunia and weakly
expressed in the stamens. It also provides another gene
from petunia, FBP15, that encodes a MARS box protein and
which is specifically expressed in the nectaries of petu-
nia. Further, it provides the isolated DNA sequences of the
promoters of the NEC1 and the FBPlS genes. Furthermore,
this invention provides an isolated DNA sequence expressed
in nectaries encoding a signal peptide that is translation-
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
W~ 00/04176 PCT/NL99/00453
-
ally fused to a germin-like protein (Lane et al., 1993,
Dumas et al., 1995), having the function to target the
mature germin-Like protein to nectar of heather (Calluna
vulgar.is). This invention gives proof that protein-contai-
ning sugar solution is collected by honeybees to produce
honey that has a higher protein content than the sugar
solution itself, the protein having undergone no qualitati-
ve alterations. Tlzis invention also proofs that a recombi-
nant protein can be produced in nectar of transgenic plants
to and that this protein is present in honey produced by
honeybees that collected this nectar.
Accordingly, this invention provides an isolated DNA
sequence which encodes a protein indicated NEC1 and having
the amino acid sequence given in SEQ ID NO:1 of the sequen-
ce listing hereafter or homologs of NEC1. A homolog of NEC1
is predominantly expressed in nectaries and/or has at least
60% homology with the amino acid sequence given in SEQ ID
NO:1. Further this invention provides an isolated DNA
2o sequence which encodes a protein indicated FBP15 and having
the amino acid sequence given in SEQ ID N0:2 of the sequen-
ce listing hereafter or a homolog of FBP15. A homolog of
FBP15 is specifically expressed in nectaries and belonging
to the MARS box family. Furthermore, a homolog is also a
gene sequence that has at least 80% homology within the
MARS box region and a 60 0 overall homology with the amino
acid sequence given in SEQ ID N0:2. Further this invention
provides the characterisation and the isolation of a DNA
sequence which encodes a signal peptide indicated "CVSP"
(Calluna v_ulgaris signal peptide), wherein the information
contained in the DNA sequence permits, upon translational
fusion with a DNA sequence encoding a protein that is
expressed in nectaries, targeting of the protein to nectar.
The DNA sequences of the invention can also be characteri-
sed in that they comprise the NEC1 gene and the FBPI5 gene
having the nucleotide sequences given in SEQ ID N0:4 and
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/1YL99/00453
sEQ ID N0:5 respectively, or a functionally homologous gene
or an essentially identical nucleotide sequence or part
thereof or derivatives thereof which are derived from said
sequences by insertion, deletion or substitution of one or
more nucleotides, said derived nucleotide sequences being
obtainable by hybridisation with the nucleotide sequences
given in SEQ TD NO:4 and 5 respectively.
Furthermore, the DNA sequences of the invention can also be
characterised in that they comprise signal sequence CUSP
having the nucleotide sequence given in SEQ ID N0:6, or an
essentially identical nucleotide sequence or part thereof
or derivatives thereof which are derived from said sequen
ces by insertion, deletion or substitution of one or more
nucleotides.
Further, this invention provides an isolated DNA sequence
from the promoter region upstream of a nectary-specific
expressed sequence, which nectary-specific expressed
sequence encodes a protein comprising the amino acid
sequence given in SEQ ID NO: I, or a homologous protein.
Furthermore, this invention provides an isolated DNA
sequence from the promoter region upstream of a nectary
specific expressed sequence, which nectary-specific expres
sed sequence encodes a protein comprising the amino acid
sequence given in SEQ ID N0:2, or a homologous protein.
In a further aspect, the invention provides a protein
encoded by any of the above defined DNA sequences. Further,
3o the invention provides processes of producing transgenic
plants exhibiting excretion of recombinant proteins in
nectar, the expression of the chimerical genes and the
targeting of the recombinant proteins being under the
control of promoter sequences anal a signal sequence as
described in this invention. Still further, the invention
provides processes of producing transgenic plants that
produce recombinant proteins in nectar, the expression of
SUBSTITUTE SHEET (RULE 2B)
CA 02333897 2001-O1-16
WO 00/04176 PCTINL99/00453
_ g -
the chimerical genes being under the control of promoter
regions upstream of other genes that are expressed in
nectaries. Still further, the invention provides processes
of producing transgenic plants that produce recombinant
proteins in nectar, the expression of these proteins being
under the control of any signal peptide that affects
targeting of a protein in nectar.
Also; the invention provides recombinant double stranded
1o DNA molecules comprising expression cassettes to be used in
the above process. Further, the invention provides transge-
nic bacteria, transgenic plants producing recombinant
proteins in nectar, and also plant cells, tissue culture,
plant parts or prcogeny plants derived from said transgenic
plants. Finally, the invention provides a process to
produce recombinant gene products in honey, produced by
bees that collect nectar from transgenic plants and process
this nectar into honey.
Brief description of the figures:
Figure 1 shows a polyacrylamide gel with PCR products after
Differential Display mRNA amplification. PCR reactions were
performed with the oligo-dT primer T12MG in combination
with 5 different random primers Apl1-AP15 on cDNA samples
of pistils without nectaries (two independent samples),
nectaries (two independent samples), leaves and a mixture
of sepals (s), petals (p) and stamens (a). Bold arrow
depicts the cloned fragment DD18.
Figure 2 is the DNA sequence of the Differential Display
RT-PCR clone DDl8a. The primers prat 122 and prat lI9 that
were used for 5' RACE PCR reactions are underlined.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCTINL99/00453
_ g _
Figure 3 is the DIVA sequence of clone RC8, obtained by RACE
PCR with gene specific primers prat 122 and prat 119 (Fig.
2) in combination with adapter primers. Primex prat 129
(underlined) is used in the next step together with primer
prat 122 to amplify the coding region of the NECI cDNA.
Figure 4 is the full length sequence of NECI cDNA. The
translation start (ATG) and translation stop (TAA} are
depicted bold.
Figure 5 shows the expression of NEC1 (A) and FBP.I5 (B) in
wild type petunia plants (line W115) as: determined by
Northern blot analysis. Blot A contains total RNA, while
blot B is enriched for mRNA. The tissues are indicated as:
1= leaf, 2= sepal, 3= petal, 4= stamen, 5= pistil, 6=
nectary. For blot A the HindIII/EcoRI fragment of pDDlBa
was used as a probe. For blot B the full length cDNA of
FBP15 was used as a probe.
2o Figure 6 Expression of NEC1 by in situ localisation of
NEC1 transcripts (A) and activity of the NECI promoter in
the nectaries (B) and the stamen (C) as shown by GUS
expression driven by the NECI promoter. The GUS assay used
for the stamens was incubated overnight without modificati-
ons to prevent di:Efusion (example 8}. The GUS assay for the
nectaries was incubated far 5 hrs, using an assay mixture
to prevent diffusion (example 8). For in situ localisation
longitudinal sections of flowers of Petunia hybrida were
hybridised with digoxigenin-labeled antisense NEC1 RNA
Figure 7 is the DNA sequence from the promoter region
upstream of a sequence encoding the NEC1 protein. Underli-
ned is the translation start of NEC1 cDNA.
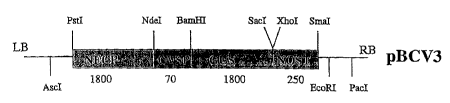
Figure 8 depicts a schematic presentation of the T-DNA
region between the borders of the binary vector pBNEPI,
containing the NECI promoter (Figure 7), the GUS reporter
SUBSTITUTE SHEET {RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCTINL99/00453
- 10 --
gene and the nos terminator in pBINPLUS. This vector was
used to generate transgenic plants to study the expression
of the NEC1 promoter.
Figure 9 shows the SDS-PAGE separation of proteins that are
present in commercial honey samples from different flowers.
M= marker, lane :I: wattle bark, lane 2: flower mixture,
lane 3: heather, lane 4: clover, lane 5: rapeseed.
Figure 10 shows the SDS-PAGE separation of proteins that
are present in commercial honey samples of rapeseed (RH2x,
RHlOx) and heather (HH2x, HHlOx) and of nectar samples of
rapeseed (RN2x, RNlOx) and heather (HN2x, HNlOx). M
molecular weight marker. Two (2x) or ten (10x) fold diluti-
ons were used.
Figure 11 shows the SDS-PAGE separation of proteins present
in dilutions of the sugar/BSA feeding solution (A) and of
honey from bees that had collected the sugar/BSA solution
(B). The dilutions of the sugar/BSA and honey/BSA solution
was the same far both gels: 1~ 15x, 2= 30x, 3= &Ox, 4= 75x,
5= 75x, 6= 90x, 7w 105x, 8= 120x, 9= 135x. M= marker
Figure 12 shows the sequence homology of the N-terminal
protein sequence of CVH29, a unique protein present in
heather honey and nectar, with a germin-like protein GER1
from a gene bank homology search (BLAST).
Figure 13 shows the deduced DNA sequence of the N-terminal
3o protein sequence of CVH29. The degenerated primers prat 176
and prat 177 are underlined (A). The DNA sequence of the
PCR product obtained with prat 176 and prat 177 performed
on genomic DNA of heather is shown in B. The gene-specific
primers prat 207 and prat 206 used to perform 5'RACE PCR
reactions on cDNA from heather flowers axe underlined.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 11 -
Figure 14 shows the DNA sequence of four independent clones
obtained by 5' RACE PCR with prat 207 and prat 206 on cDNA
of heather flowers. The ATG translation start of the
putative signal sequence is boxed. The end of the putative
signal sequence and the start of the mature protein are
indicated by arrows.
Figure I5 is the sequence of the synthetically produced
DNA molecule encoding the signal sequence CUSP (boxed} with
l0 linkers.
Figure 16 is the schematic representation of the plasmid
pCVI. Not all restriction sites are indicated.
Figure 17 is the schematic representation of the plasmid
pCV2. Not all restriction sites are indicated.
Figure 18 is the schematic representation of the plasmid
pCV3. Not all restriction sites are indicated.
Figure 19 is the DNA sequence of the full length cDNA of
FBP15. The translation start (ATG) arid translation stop
(TAA) are boxed. The MAD-box and K-box region are underli-
ned.
Detailed description of the invention
This invention provides processes of producing transgenic
plants that produce recombinant proteins in nectaries and
nectar that is collected by foraging honeybees. This
invention gives evidence that honeybees process protein
containing nectar into honey that contains the unaltered
protein in a concentrated form. Subsequently, the desired
protein can be purified from the honey.
SUBSTITUTE SHEET (RULE 26~
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99100453
- 12 -
To express recombinant proteins in nectaries of transgenic
plants, a translational fusion of an isolated DNA sequence
from a promoter region upstream of a sequence encoding a
protein that is expressed in nectaries with a sequence
encoding the recombinant protein has to be carried out.
Preferably, the isolated DNA sequence from a promoter
region is upstream of a sequence that is specifically or
highly expressed in nectaries.
The invention relates to a DNA sequence isolated from
Petunia hybrida that encodes a protein indicated NEC1 or a
homologous protein or part thereof. A homologous protein
has at least 65~ homology with the amino acid sequence
given in SEQ ID NO:1. The cDNA sequence of the NEC1 gene is
given in Fig. 4 and in SEQ ID N0:4. The deduced amino acid
sequence of the NEC.I gene is given in SEQ TD NO:1. The NEC1
gene shows strong expression in the nectaries and in a very
localised region of the anther filaments of Petunia hybri-
da. The deduced amino acid sequence of NEC1 predicts a
membrane bound protein. The precise function of the gene
has not been elucidated yet, but considering the phenotype
of transgenic plants that ectopically express NEC1 in the
leaves, a role in sugar metabolism of NECI is apparent.
The present invention also relates to homologous DNA
sequences that can be isolated from other organisms,
preferably plants, using standard methods and the already
known DNA sequence of the NEC1 gene. More precisely, it is
also possible to use DNA sequences which have a high degree
of homology to the DNA sequence of the NEC1 gene, but which
are not completely identical, in the process according to
the invention. The use of sequences having homologies
between 85 and 100 % is to be preferred. DNA sequences can
also be used which result from the sequence shown in SEQ ID
N0:4 by insertion, deletion or substitution of one or more
nucleotides. This includes naturally Occurring variations
or variations introduced through targeted mutagenesis or
SU8ST1TUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 13 -
recombination. The DNA sequence shown in SEQ ID N0:4 can
also be produced by using DNA synthesis techniques.
The invention also relates to a DNA sequence isolated from
Petunia hybrida that encodes a MADS box protein indicated
FBP15 or a homo7.ogous protein or part thereof. The cDNA
sequence of FBP1S is given in SEQ ID N0:5. FBP15 shows
exclusively expression in the nectaries of Petuzxia hybrida.
The function of F'BP15 is unknown.
The present invention also relates to homologous DNA
sequences that can be isolated from other organisms,
preferably plants, using standard methods and the already
known DNA sequence of FBP1S. More precisely, it is also
possible to use DNA sequences which have a high degree of
homology to the DNA 'sequence of FBP15, but which are not
completely identical, in the process according to the
invention. The use of sequences having homologies between
85 and 100 °s is t:o be preferred. DNA sequences can also be
used which result from the sequence shown in SEQ ID N0:5 by
insertion, deletion or substitution of one or more nucleo-
tides. This includes naturally occurring variations or
variations introduced through targeted mutagenesis or
recombination. The DNA sequence shown in SEQ ID N0:5 can
also be produced by using current DNA synthesis techniques.
Further, this invention provides an isolated DNA sequence
from the promoter region upstream of a nectary-specific
expressed sequence, which nectary-specific expressed
sequence encodes a protein comprising the amino acid
sequence given in SEQ ID N0:1, or a homologous protein that
is expressed in nectaries. Furthermore, this invention
provides an isolated DNA sequence from the promoter region
upstream of an isolated DNA sequence from the promoter
region upstream of a nectary-specific expressed sequence,
which nectary specific sequence encodes a protein compri-
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 14 -
sing the amino acid sequence given in SEQ ID N0:2, or a
homologous protein that is expressed in nectaries.
More specifically this invention provides an isolated DNA
sequence from the promoter region upstream of a nectary-
specific expressed sequence, which nectary-specific expres-
sed sequence has:
a) a nucleotide sequence given in SEQ ID N0:4, or
b) a nucleotide sequence obtainable by hybridisation with
l0 the nucleotide sequence of (a) or with a fragment of (a).
In a more specific embodiment this invention provides an
isolated DNA sequence from the promoter region upstream of
a nectary-specific expressed sequence, obtained from a
plant of Petunia hybrids, the sequence consisting essenti-
ally of the sequence given in SEQ ID N0:7, or a functional
fragment thereof having promoter activity.
In a further aspect, the invention provides an isolated DNA
2o sequence from the promoter region upstream of a nectary-
specific expressed sequence, which nectary-specific expres-
sed sequence has:
a) a nucleotide sequence given in SEQ ID N0:5, or
b) a nucleotide sequence obtainable by hybridisation with
the nucleotide sequence of (a) or with a fragment of (a).
In a more specific embodiment this invention provides an
isolated DNA sequence from the promoter region upstream of
a nectary-specific expressed sequence, obtained from a
plant of Petunia hybrids, the sequence consisting essenti-
ally of the sequence given in SEQ ID N0:8 , or a functional
fragment thereof having promoter activity.
Further, this invention provides an isolated DNA sequence
comprising the coding region for a signal peptide, wherein
the information contained in the DNA sequence permits, upon
translational fusion with a DNA sequence encoding a protein
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99100453
_ ~5
that is expressed. in nectaries, targeting of the protein to
nectar. More specifically, the DNA sequence comprises the
nucleotide sequence given in SEQ ID N0:6 obtained from a
plant of Calluna vulgaris, or a nucleotide sequence obtai-
noble by hybridisation with the nucleotide sequence given
in SEQ ID N0:6. The use of sequences having homologies
between 95 and 100 % is to be preferred. DNA sequences can
also be used which result from the sequence shown in SEQ ID
N0:6 by insertion,-deletion or substitution of one or more
nucleotides. This includes naturally occurring variations
or variations introduced through targeted mutagenesis or
recombination.:'The DNA sequence shown in SEQ ID N0:6 can
also be produced by using DNA synthesis techniques. The
signal peptide CVSP was isolated tram nectar of Calluna
vulgaris flowers and from honey processed by honeybees that
collected the nectar. The function of CVSP in heather
nectaries is to target the germin-like protein to nectar.
The DNA sequence CVSP can also be used to target other
proteins to nectar in plant species.
A subject of the present invention is the use of
DNA sequences for- producing recombinant proteins in nectar
of plants, wherein the protein is produced in nectaries and
targeted to nectar, and wherein expression in nectaries is
achieved by using a DNA sequence consisting of the promoter
region upstream of a DNA sequence that is expressed in
nectaries, and wherein secretion in nectar is achieved by
using a DNA sequence that encodes a signal sequence that
targets the recombinant protein to nectar. In a further
aspect the present invention relates to processes wherein a
recombinant protein is expressed in other plant tissues
than the nectaries and wherein the biochemical. composition
of nectar is changed as a consequence of the recombinant
gene expression. The present invention also relates to
processes wherein a recombinant protein is expressed in
nectaries of a transgenic plant, wherein the biochemical
composition of nectar or the nectar secretion is changed as
SUHSTiTUTE SHEET (RULE 2f)
CA 02333897 2001-O1-16
WO 00/04176 PCT/1YL99100453
- I6 -
a consequence of this pratein expression. In particular, it
relates to processes where the recombinant protein is an
enzyme that interferes with the sugar metabolism in necta-
ries.
The production of a recombinant protein in nectaries and
nectar is achieved by integrating into the genome of the
plants a recombinant double-stranded DNA molecule compri
sing an expression cassette having the following constitu
ents and expressing it:
i) a promoter functional in nectaries of plants,
ii) a DNA sequence encoding a protein which is fused
to the promoter,
iii) a DNA sequence encoding a signal peptide that
targets the recombinant protein to nectar, which
is translationally fused to the DNA sequence enco
ding the recombinant protein, and optionally
iv) a signal sequence functional in plants for the
transcription termination and polyadenylation of
ari RNA molecule.
Such DNA molecules are also subject of the invention. The
present invention provides an example of such a DNA molecu-
le that contains the described expression cassettes in the
form of plasmid pCV3 (Fig. 18), which comprises the promo-
ter region of the NEC.Z gene from petunia, the signal
sequence CVSP from heather, the coding region of the
reporter gene GUS and the NOS terminator. In principle, any
promoter that is active in the nectaries of plants can be
used as promoter. The promoter is to ensure that the chosen
gene is expressed in nectaries. Also, in principle, any
signal sequence that targets the expressed protein to
nectar can be used as a signal sequence. The signal sequen-
ce is to ensure that the protein is excreted in nectar.
Furthermore, any sequence that encodes a recombinant
protein in nectaries can be used in the present invention.
Preferably, the subject of this invention relates to DNA
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 17 --
sequences that encode proteins to be used for pharmaceuti-
cal purposes . It is also possible to use the invention to
produce proteins for other purposes, e.g. enzymes for
biotests or antioxidants for food additives. Furthermore,
it is possible to use the invention to produce metabolites
in nectar that attract predators of pest insects or that
kill or repel pest insects. In another aspect it is possi-
ble to use the invention to produce metabolites in nectar
that modify the attractiveness of the plant for pollinating
insects or improve the health of pollinating insects.
It is also possible to use DNA sequences that encode
proteins that modify the nectar composition or the sink
strength of nectaries. This means that the recombinant
protein interferes with metabolic pathways in the necta-
ries, resulting in changed levels of compounds that are
already present in nectar, or the formation of new com-
pounds in nectar.
In addition, the present invention also relates to expres
sion cassettes that contain the above mentioned DNA sequen
ces, except for a signal sequence. The recombinant protein
is then only expressed in the nectaries, but not targeted
to the nectar. Consequently, the expression of the recombi-
nant protein in the nectaries can still affect nectar
composition.
In a further aspect, the present invention also relates to
expression cassettes that contain DNA sequences coding for
a protein that is expressed in other tissues than the
nectaries. The expression of the recombinant protein
affects changes in the biochemical composition of nectar or
in nectar secretion.
Finally, the present invention also relates to non-transge-
nic plants that produce metabolites in nectar that can be
harvested and purified from honey that is produced by
honeybees that collect this nectar. Examples for these
metabolites are alkaloids, terpines, amino acids, proteins,
pigments and volatiles.
SUBSTITUTE SHEET {RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99J00453
- 18 -
A preferred embodiment of the process discussed above
provides that the expression cassette is transformed to a
plant species that produces nectar. Preferably, the recom-
binant protein is produced in nectar of plants that are
visited by honeybees that collect the nectar. Honeybees
collect floral as well as extrafloral nectar. The present
invention relates to plants that produce recombinant
proteins in floral or extrafloral nectar. In addition, the
present invention also relates to plants that produce
recombinant proteins in other plant organs, said plant
organs producing an exudate that is collected by insects,
preferably bees, and processed into honey. A particularly
preferred embodiment of the present invention are plants
that can be grown under controlled conditions. Controlled
Z5 conditions are greenhouses or field facilities where
transgenic plants can be grown according to the safety
rules that are required. Preferably, the controlled condi-
tions are such that bee colonies that perform normal
foraging behaviour can be maintained in the same compart-
2o ment during the flowering period. Preferred plants origina-
te from the Brassicaceae family, in particular Brassica
napus.
Examples
Example l:
Cloning of NEC1
The NEC.I cDNA was isolated using the mRNA Differential
3o Display system (Genhunter Corporation, Brookline USA). The
isolation of total RNA from nectaries, sepals, petals,
stamens and pistils from open flowers and fram young leaves
of Petunza hybrids was done according to Verwoerd et al.
(1989). Two independent RNA isolations were performed on
nectaries as well as on pistils. A DNase treatment was
carried out on each RNA sample, using the RNA MessageCleanTM
Kit (Genhunter Corporation Brookline USA, cat. No. M6o1). A
SUBSTITUTE SHEET (RULE 2fi)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99I00453
- 19 -
reverse transcription reaction was carried out on 0.1 ~,g
RNA of each samp7.e, using the oligo-dT primer T12MG from
the Genhunter Kit.. Following the protocol, PCR reactions
were carried out using the arbritary primers AP11-AP15 in
combination with primer T12MG from the Kit. The PCR pro-
ducts were loaded on a sequencing gel and after electropho-
resis the gel was blotted on 3M paper, dried and exposed to
x-ray film (Figure 1). Two adjacent nectary-specific bands
were cut out from the blot and the DNA was purified accor-
1o ding to the manual. Reamplification of the fragment was
carried out using the oligo-dT primer T12MG and the arbri-
tary primer AP15. After electrophoresis, the PCR product
was extracted from the agarose gel by freezing the isolated
fragment in liquid nitrogen, followed by centrifugation.
DNA was precipitated by adding 1/10 volume 1% HAc, 0.1M
MgCl2 and 2.5 volume of 96% ethanol to the supernatant. The
pellet was dissolved in 10 ~.1 TE buffer. The fragment, now
called DDlBa, was cloned into a PMOSBlue T-vector {RPN
1719, Amersham Little Chalfont UK) giving the vector
pDDlBa.
The nucleotide sequence of this 3' cDNA clone was determi-
ned by the dideoxynucleot,ide chain termination method (ABI
PRISMT'" Ready Reaction DyeDeoxy~M Terminator Cycle Sequen-
cing Kit, P/N 402078, Perkin Elmer) and is shown in Figure
2. The DNA fragment has a length of 460 nucleotides. The
missing 5' part of the cDNA was isolated using the Marathon
TM cDNA Amplification Kit of Clontech (catalog K1802-1) and
following the procedure as described in the manual. Brief-
ly, Poly A~- RNA was isolated from nectaries of Petunia
hyhrida flowers. After double stranded cDNA synthesis,
adapters were ligated and a 5'RACE reaction was carried out
using the adapter primer API supplied in the kit and a
gene-specific primer prat 122. The nucleotide sequence of
prat 122 is: 5'-gtgggaaggctatgctacaagc-3' (Figure 2). The
PCR product was diluted lOx and 1 ~.1 was used in a second
5' RACE reaction with the nested adapter primer supplied by
the kit {AP2) and the nested gene-specific primer prat 119
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO OOI04176 PCTINL99l00453
- 20 -
(Figure 2). The nucleotide sequence of prat 119 is: 5'-
ccttctccatggactgcaatgcg-'3 . After gel electrophoreses a
fragment of ~850 by was obtained that hybridised with
clone DDl8a. The fragment, now called RC8, was extracted
from the gel, purified and cloned into a PMOSBlue T-vector
as described above. The sequence is shown in Figure 3. The
combined (overlapping} sequences of clones DDl8a and RC8
are shown in Figure 4, comprising the full length cDNA of a
gene called NEC.2 :hereafter. The NEC1 clone has a length of
1205 nucleotides and encodes for a polypeptide of 265 amino
acid residues. Based an the deduced amino acid sequence,
high homology was found with a cDNA that is associated with
Rhizobium-induced nodule development in the legume Med.icago
trunculata (MtN3, gene bank number: gnl/PID/e274341). The
I5 percentages of identity and similarity are 47% and 72%
respectively. Analysis of the predicted protein, using the
CAOS/CAMM programme (Protein analysis 1991, Genetics
Computer Group inc., Wisconsin USA), shows that the putati-
ve protein structure resembles membrane proteins, having
six evenly spaced hydrophobic loops that traverse the cell
membrane. In addition, a signal sequence is predicted at
the N-terminus, while the C-terminus is highly hydrophilic.
Highest homology with MtN3 is found in the N-terminal
signal sequence, the first two membrane-spanning loops and
the last two membrane-spanning loops. The C-terminal
hydrophilic part shows the lowest homology (28% identity,
30% similarity). The function of NEC1 has not yet been
determined.
Example 2:
Cloning of FBP15
Petunia MADS box cDNA clones were isolated from a cDNA
library made from nectaries of Petunia hybrida flowers. The
cDNA library was constructed using the lambda ZAP cloning
vector (Stratagene, La Jolla USA, catalog nr. 200400-
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCTINL99/00453
-21--
200402). The library was screened under low stringency
hybridisation conditions with a mixed probe comprising the
MADS box regions of Floral binding protein gene FBP2, FBP6
and pMADS3 (Angensnt et al., 1993, 1994, Tsuchimoto 1993).
The hybridizing phage plaques were purified using standard
techniques. Using the in vivo excision method, E.coli
clones which contain a double-stranded Bluescript SK-
plasmid with the cDNA insertion between the EcoRI and Xhol
cleavage site of he polylinker were generated. Cross-
to hybridisation of the purified clones revealed 3 independent
clones that did not cross hybridise with previously isola-
ted FBP cDNA's and which were designated FBP.IS, FBP16 and
FBP17. The nucleotide sequence of FBP15 was determined by
the dideoxynucleotide-mediated chain termination method and
is depicted in SEQ ID N0:5. The FBP15 cDNA clone has a
length of 1157 nucleotides and encodes a peptide of 222
amino acid residues. All characteristics of a MADS box
protein axe present in FBP15: a N-terminal located MADS box
region which shows a high degree of similarity with other
MARS box proteins, and a K-box in the middle of the protein
with an alpha helical structure, FBPIS is most similar to
the tobacco MADS box protein NAGl, which is an Agamous
homolog and expressed in whorl 3 and 4 (Huang et al., 1996,
Mizukami et al., 1996).
Example 3:
Expression of FBP:~5
Expression of FBP15 was determined by standard Northern
blot hybridisation experiments. A DNA fragment comprising
the complete cDNA of FBP15 was used as a probe. High
stringency hybridisation and washing conditions were used.
Using 10 ~g of total RNA from various petunia tissues,
expression of FBPaS was only detectable in nectaries. Using
10 ~.g of mRNA from various tissues, prepared by using the
kit and protocol of the Quickprep Micro mRNA Purification
SU6STiTUTE SHEET tRULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 22 -
Kit (Pharmacia Biotech), expression of FBP15 was only
detectable in nectaries as shown in Figure 5B.
The expression in the ovary and nectaries was determined by
in situ hybridisation using a DIG labelled antisense RNA
probe corresponding to the full length cDNA of FBP15. In
vitro antisense RNA transcripts were made using T7 RNA
polymerase. A standard protocol for in situ hybridisation
was used as described by Canas et al., 3.994. A hybridizing
signal was observed evenly strong in all cells of the
l0 nectary tissue.
Example 4:
Expression of NECI
The RNA expression of NEC1 was determined by standard
Northern blot hybridisation experiments. A DNA fragment
comprising the complete sequence of the Differential
2o Display clone DD18 (Figure 2) was used as a probe. Using
10 ~g of total RNA from various petunia tissues, strong
expression of NEC.1 was detectable in necta.ries and weak
expression in anthers. No expression was detectable in
other floral organs, in leaves or in roots (Figure 5A).
The expression in the ovary and nectaries was determined by
in situ hybridisation using a DIG labelled antisense RNA
probe corresponding to the nucleotides 79 to 1036 of NEC1
cDNA, comprising the coding region and part of the 3'
untranslated region. A clone containing this sequence was
obtained by PCR on adapter-ligated cDNA, using two gene-
specific primers prat 122 and prat 129 (Figure 4). The
nucleotide sequence of prat 122 is: 5'-gtgggaaggctatgctaca-
agc-3"', comprising the nucleotides 1015 to 1036 of the
NEC1 cDNA. The nucleotide sequence of prat 129 is: 5'-
gggatccatgctcgcaattacQtqctgatg-3°, comprising the nucleoti-
des 79 to 100 of i:he NEC1 cDNA. The gene-specific region of
the primers is underlined. The primer contains an extra
SU8ST1TUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCTINL99/00453
- 23 -
BamHI and Ncol site at the 5'end. A PCR fragment of 958
nucleotides was obtained and cloned into a PMOSBIue vector.
The fragment was subcloned in a vector containing the T7
promoter and in vitro antisense RNA transcripts were made
using T7 RNA polymerase. A standard protocol for in situ
hybridisation was used as described by Canas et al., 1994.
Strong hybridizing signals were observed in the outer cell
layers of the nectaries (Figure 5A)
Example 5:
Isolation of NEC1. promoter fragment
The promoter fragment of NEC.I was cloned using the genome
walker protocol (PT3042-1) and kit as provided by Clontech
Laboraties. Briefly, genomic DNA from Petunia hybrids was
digested with 5 different blunt cutting restriction enzy-
mes. GenomeWalker adapters were ligated and PCR reactions
2o were carried out on each GenomeWalker "library" with a gene
specific, reversed primer prat 148 and the adapter primer
from the kit (AP1). The nucleotide sequence of prat 148 is:
5'-ccaagaaggccaaatatgaaagac-3' comprising the nucleotides
105 to 128 of the NEC1 cDNA (Figure 4 ) . PCR products were
subjected to a second round of PCR, using the nested
adapter primer AP2 and the nested gene specific, reversed
primer prat 149. The nucleotide sequence of prat 149 is:
5'-aagtcatcagcacgtaattgcgcc-3', comprising the nucleotides
81 to 104 of the NECZ cDNA. From the second PCR a 2 kb
fragment was isolated from the StuI library, which was
cloned in the PMOSBlue T-vector, yielding the construct
pMAS -10 . Figure 'I ( SEQ ID NO : 7 ) shows the DNA sequence of
the NECZ promoter in the construct pMA5-10, including the
translation start. of NECI cDNA.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 24 -
Example 6:
Construction of ,NEC1 promoter-GUS
A PCR reaction was performed on pMA5-10 (example 5), using
the forward vector primer U19 of pMOSBIue and the gene-
specific primer prat 169. The nucleotide sequence of prat
169 is:
5'-cgctgcagcgccataattttttttagtgaa~ccccc-3' The gene-speci
fic region is underlined. The primer contains an Ncol and
BgIII restriction site at the 3' end. The PCR product was
digested with Kpnl and Ncol and ligated into a pBluescript--
derived vector (pM04) that contains the NTM19 promoter
(Custers et al., 1997), the reporter gene GUS and the nos
terminator. The KpnT/NcoI NTM19 promoter fragment was
replaced, resulting in a .NECI-promoter/GUS translational
fusion. The resulting plasmid pNEPI was digested with Smal
to release the NEC1 promoter/GUS/nos fragment and this
fragment was ligated into a derivative of the binary
plasmid pBIN (Bevan, 1984) yielding the binary plasmid
pBNEPl (Figure 8). pBNEPI was introduced,into Agrobacterium
tumefaciens strain LBA4404 or C58pMP90 by electroporation.
Plasmid DNA from the Agrobacterium transformants was
isolated and the structure of the binary vector was veri-
fied by restriction analysis and PCR.
Example 7:
Generation of transgenic Petunia plants
Agrobacterium strain LBA4404 transformants were used to
transform Petunia hybrida using leaf discs as described by
Horsch et al. (1985). After shoot and root induction on
kanamycin selection media, plants were transferred to soil
in the greenhouse.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00104176 PCTlNL99100453
- 25 -
Example 8:
Histochemical GUS assay
Different plant parts of Kanamycin-resistant plants trans-
formed with the pBNEPI construct were analysed for the
distribution of a-glucuronidase activity (GUS) using the
method described by (Jefferson et al., 1987). In transgenic
plants with high expression levels diffusion of reaction
products to other tissues was observed. To avoid this
spreading a modified GUS assay was used. Briefly, tissues
were pre-treated with 90% cold acetone at -20°C for 1 h,
then rinsed three times 20' with 100 mM phosphate buffer
containing 1 mM potassium ferricyanide. After this treat-
went the standard GUS assay was performed with the modifi-
cation that ferricyanide was excluded from the reaction
mixture.
Example 9:
Results histochemical GUS assay
In very young flowers (~1,4 cm) na blue staining was
observed, in flowers of 2-4 cm weak blue staining of the
nectaries was observed. In flowers of (4-6 cm) strong blue
staining was observed in the nectaries (figure 6B) and in a
very restricted region of the upper part of the anther
filaments (Figure 6C). GUS expression was highest in the
outer cell layers of the ne~tary parenchyma. In cross
sections of the anther filaments GUS expression was obser
ved in all cells except in the xylem of the inner vascular
bundle.
Example 10:
Protein analysis of heather honey and nectar
Samples of pure heather honey, together with samples of
rapeseed, clover, wattle bark and lavender honey were di-
luted, dialysed and loaded on a 12% SDS page gel (Laemmli,
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99/00453
- 26 -
1970). All honey samples showed several identical high
molecular weight protein bands. Heather honey contained 2
unique protein bands of 29 and 50 kDa (Figure 9). The
proteins were named CVH29 and CVH50 (CVH stands for Calluna
vu.Igaris honey). To determine the origin of the proteins,
nectar and honey samples of rapeseed and heather were
prepared and loaded on a 12% SDS page gel. The high molecu-
lar weight protein bands of around 70 kDa that are present
in all honey samples were not observed in rapeseed or
heather nectar (Figure 10). These proteins are added by
honeybees during honey processing. Proteins CVH29 and CVH50
are present in.heather honey and heather nectar, but not in
nectar of rapeseed. Therefore, it was concluded that CVH29
and CVH50 are secreted in nectar of heather and can be
recovered from honey derived from this nectar. The protein
concentration in the heather honey we tested was around
0.5°s.
Example 11:
N-terminal sequence analysis of CVH29 and CVH50
Honey samples were loaded on an SDS PAGE gel and after
electrophoreses the gel was blotted on a PVDF membrane.
After staining the CVH29 and CVH50 bands were cut out from
the blot and N-terminal sequencing was performed on both
proteins. The N-terminal sequence of CVH50 is: SVLDFCVADPS-
LPDGPAGYSCTEPSTVTSQDF. The N--terminal sequence of CVH29 is:
SVLDFCVADPSLPDGPAGYSCKEPAKVTVDDFVFHGLGTA. A gene bank
homology search (BLAST) showed high amino acid sequence
homology (630) with germin-like proteins isolated from
Arabidopsis (Figure 12).
SUBSTITUTE SHEET (RULE 2fi)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99100453
- 27 -
Example 12:
Identification signal sequence of CVH29
Because the gerrnin-like protein CVH29 is excreted in
heather nectar it was expected that part of the cDNA
encodes a signal sequence. Based on the N-terminal amino
acid sequence, degenerated primers were designed. The
sequence of the forward primer prat 176 is: 5'-gayttyt-
gygtngcngaycc-3' (y= c or t, n= c, t, a or g). The sequence
of the reversed primer prat 177 is: ccrtgraanacraartcrtc
(r= g or a). A PCR reaction performed on genomic DNA of
heather yielded a 99 by DNA fragment. The fragment was
sequenced and two reversed, gene-specific 5' primers were
designed to clone the 5' cDNA by "Marathon cDNA racing"
using the kit and protocol of Clontech laboratories (proto-
col PT1115-1, Clontech Palo Alto USA). The sequence of
gene-specific primer prat 207 that was used is: 5'-
ggtgactttagagggctccttgc-3', the sequence of gene-specific
nested primer prat 206 is:
5'-gctccttgcaggagtagcctgc-3' (Figure 13). RNA was isolated
from open flowers of heather and mRNA was prepared using
the Pharmacia quickprep micro mRNA kit. After cDNA synthe-
sis and adapter ligation a PCR reaction was performed,
using the adapter primer AP1 and the gene-specific primer
prat 207. The PCR product was used for a second PCR, using
adapter primer AP2 and the nested gene-specific primer prat
206. A single fragment of around 300 nucleotides was
obtained and cloned in a PMOSBlue T-vector. Four clones
were sequenced. Figure 14 shows that three clones were
identical and one clone had two different nucleotides in
the untranslated S' region. A putative signal sequence of
17 amino acids was identified between the ATG start codon
and the first codon of the mature protein CVH29 that was
identical in all four clones. The nucleotide sequence of
the putative signal sequence (SEg ID N0:6) is:
5'-atgtttcttccaattctcttcaccatttccctcctcttctcctcctcccatgct-
3'.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 28 -
Example 13:
Construction of an expression cassette for excretion of
proteins in nectar
To clone the NECI promoter into a PMOSBlue vector a PCR
reaction was carried out on pMAS-10 (example 5) using the
forward primer prat 247 and the reversed primer prat 248
(Fig. 7). Prat 247 contains an extra Pst1 restriction site.
The Ndel restriction site of prat 248 coincides with the
ATG translation start of NEC.I. The nucleotide sequence of
prat 247 is: 5'-ggctgcaggaqtgttctttgataqaatg-3', the
nucleotide sequence of prat 248 is: 5'-cgcca-
tatgtttttttatggaaqcccc-3'. Gene-specific regions are
underlined. A 1,8 kb promoter fragment was obtained and
3.5 cloned into a pMOSBlue vector, yielding the plasmid pNECP.
A DNA molecule encoding the signal sequence CVSP as depic-
ted in SEQ ID N0:6 was produced by synthesis and subsequent
annealing of two oligo molecules prat 245 and prat 246. The
sequence of prat 245 is: 5'tatattccttccaattcttttcactatttct-
cttcttttctcttcttctcatgcttctgttcttgatttc'3, the sequence of
prat 246 is: 5'gatccgaaatcaagaacagaagcatclaqaagaagagaaaacxaa-
ctagaaataat aaaagaattggaacLqaaca'3. The region encoding the
signal sequence CVSP is underlined. To ensure correct
cleavage of the signal peptide, the linkers were extended
with the coding region for the first five amino acids of
the mature germin-like protein (Fig. 13). The codon usage
of the signal peptide sequence was optimised for Arabidop-
sis. By addition of a BamHI restriction site at the 3' end,
2 extra amino acids were linked in frame to the mature
protein. The resulting DNA molecule is shown in Figure 15.
The fragment was ligated into a Nde1/BamH1 cut PMOSBlue
vector, yielding the plasmid pCVSP.
pNECP was digested with Ndel and PstI to release the NEC1
promoter fragment which was cloned into the Pstl/Nde1 cut
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 29 -
pCVSP, yielding the plasmid pCVI. A schematic representati-
on of pCV1 is given in Figure 16.
A 2S0 by long fragment containing the NOS terminator
sequence (NOST} was obtained by PCR, using the forward
primer prat 251 and the reversed primer prat 2S2 on DNA of
pRAP 33, which is a pUC 19 derived plasmid. Prat 251 adds a
SacI and Xhol site, prat 252 adds a Smal and EcoRI site.
The sequence of prat 251 is: 5'-gggagctcgagtcgttcaaa-
catttctacaataaaQ-3'. The sequence of prat 252 is: 5'-cgaatt-
cccgggatctaQtaacataQatgacac-3' The HOST-specific regions
are underlined. The PCR product was cloned into pCR-ScriptTM
Amp SK(+) Cloning Kit (Catalog 21188-21190, Stratagene La
Jolla USA}, yielding the plasmid pCR-NOST. pCR-NOST was
digested with Sacl and EcoRl and the resulting fragment was
cloned into the pUC 19 (ClonTech), derived plasmid pUCAP
yielding the plasmid pCVNOS.
The plasmid pGUSN358 was purchased from Clontech (catalog
6030-1) containing the reporter gene GUS in pUC 119,
modified to destroy the N-linked glycosylation site within
the 1.814 Kb GUS coding sequence. A PCR reaction was
carried out with gene-specific primers prat 249 and prat
250, yielding a fragment that contains the GUS gene coding
region and a BamHl restriction site at the 5'end and a Sacl
restriction site at the 3'end. The sequence of prat 249 is:
5'-ccggatccatctttaca~tcctcrtagaaacc-3'. The sequence of prat
250 is: 5'-gggagctcccacccLaggctqtaq=3'. The GUS specific
regions are underlined. Subsequently, the PCR fragment was
digested with BamHI and Sacl and ligated into the BamHI/Sa-
cI cut plasmid pCVNOS, yielding the plasmid pCV2. A schema-
tic representation of pCV2 is given in Figure 17.
pCVI is digested with PstI and BamHI and the resulting
fragment is cloned into the PstI/BamHI cut plasmid pCV2,
yielding the plasmid pCV3. A schematic representation of
pCV3 is given in Figure 18. pCV3 is digested with Ascl and
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
_ 30
Smal and the resulting fragment is cloned into a derivati-
ve of the binary plasmid pBIN, yielding the binary plasmid
pBCV3. pBCV3 was transferred from Eschexich.ia coli to the
Agxobactexium tumefaciens strain LBA4404 and C58pMP90 by
electroporation. The transformed Agrobacterium strain was
used to transform Arabidopsis and petunia.
Example l4:
Protein production in nectar
Using the GUS reporter gene, GUS activity in nectar of
transgenicplants was measured according to the method as
described by Jefferson et al., (19871. Briefly, the assay
was carried out by measuring the amount of methyl umbelli-
ferone (MU) produced by GUS fluorometrically by emission of
light of 455 nm. The absolute emission was corrected for
artificial quenching using an internal standard of 1nM MU
(Angenent et al., 1993).
Example 15:
Feeding experiments with honeybees
In September 1996 a beehive located outside was supplied
with a 25o sucrose solution supplemented with 2% BSA
(bovine serum albumin). After 3 weeks the bees had consumed
15 litters of the feeding solution and honey was harvested
from the hive. Although the flowering season had mostly
past, bees still foraged on flowers to collect nectar
outside. Therefore, the honey produced during this period
is derived from a mixture of the feeding solution and
nectar from flowers. An SDS page protein gel was loaded
with dialysed honey samples and sugar/BSA solutions. Figure
11 shows that the protein band of BSA was present in all
the samples tested. and no qualitative changes were observed
in the honey samples compared to the sugar/BSA solutions.
The BSA concentration in honey was 1.5 times higher than in
the feeding samples, demonstrating that protein is concen-
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
W4 00/04176 PCT/NL99/00453
- 31 -
trated in honey, Honeybees that foraged on the sugar/BSA
solution did not show any aberrant behaviour and the colony
developed normally.
Example 16:
Process of honey production from transgenic plants
Twohundred and fifty transgenic plants that each produce
recombinant protein in nectar were grown in a greenhouse of
25 square meters. The facilities were adjusted according to
the safety rules according to European law, including
safety measures to prevent in- or outflow of insects. A
beehive adjusted for small populations, containing around
200 worker honeybees and a queen, was placed in the green-
house when the plants were flowering. When a queen is
present, she will start laying eggs and larvae will come
out . The presence of brood stimulates the bees to collect
nectar and process it into honey. After 2-3 weeks bees
processed the nectar into honey and stored in sealed cells
of the honeycomb. Under the described conditions the amount
of honey that can be harvested is 250-1000 grams.
Example 17:
Ablation of nectaries
By introducing the highly sensitive Rnase BARNASE in plant
cells, under the control of a tissue-specific promoter,
cell ablation can be achieved in very specific tissues or
organs. Ablation of nectaries can be applied to decrease
the attractiveness of plants for pest insects that forage
on the nectar that is secreted by nectaries . In addition,
plants without nectaries can be obtained that are more
resistant to bacterial and fungal infections. An example is
given for the ablation of nectary tissue by expressing
bacterial BARNASE in nectaries, using the NEC1 promoter.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/0417b PCT/NL99/00453
- 32 -
Plasmid DNA of pNEPI (example 6) was digested with Kpnl and
NcoI to release the 1800 by NECK promoter fragment. The
purified promoter fragment was ligated into a pWP90 derived
vector, upstream of the BARNASE-BARSTAR bacterial operon
construct (Hartley, 1988). The construct contains a 35SCaMV
terminator of pol.yA signal cauliflower mosaic virus termi-
nator sequence downstream of the BARNASE-BARSTAR operon.
The resulting plasmid pWP126 was digested with Kpnl/ XhoI
to release the NEC1-promoter/BARNASE-BARSTAR/CamVpolyA
fragment and this fragment was ligated into a pBIN-derived
vector pBIN Plus. The recombinant vector was transferred
via Agrobacterium tumefaciens (LBA4404) to petunia variety
W115. Transgenic petunia plants were selected with flowers
without nectaries or underdeveloped nectaries.
Many promoters a.re less specific than can be concluded
based on promoter/GUS expression is concluded. Because the
bacterial BARNASE is highly cytotoxic at very low concen-
trations it can be preferred to protect other plant tissues
by expression of a ribonuclease inhibitor gene under the
control of a weak, constitutive promoter (e. g. NOS promo-
ter) or a tissue-specific promoter that is not active in
the tissues where cell ablation is to be achieved (Mariani
et al., 1992, Beals et al., 1997).
30
Example 18:
Ectopic nectary development
MADS box genes regulate floral meristem and floral organ
identity. Ectopic expression of MARS box genes can change
the developmental fate of floral organs or cells. Transge-
nic petunia plants ectopically expressing FBP11, an ovule-
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 33 -
specific MADS box gene, develop ovule-like structures on
sepals and petals ( Colombo et al., 1995). FBP15 is a
nectary-specific MARS box gene, involved in the molecular
regulation of nectary development. In petunia nectaries
develop at the base of the carpel. Ectapic expression of
FBP15-in petunia may result in the development of nectaries
on other organs of the flower ar on vegetative parts of the
plant . An example is given of a gene construct that , when
transformed to a plant, results in ectapic expression of
FBP15.
FBP3.5 was amplified using:a 5' primer that hybridises with .
FBP15 sequences just upstream of the ATG translation start
site and a 3' primer that hybridises with FBP15 sequences
just downstream the translation stop site. The 5'primer
contains a NcoI recognition site, the 3'primer contains a
BamHI recognition site. After the sequence was confirmed,
the amplified FBP15 fragment was inserted as a BamHI/NcoI
fragment into the binary vector pCP03~. This binary vector
was derived from pPCV708, as described by Florack et al.
(1994), and contains three expression cassettes with a
multiple cloning site between the left and right T-DNA
borders. The cDNA was cloned in sense orientation between a
modified CaMV 35S promoter and the nopaline synthase
terminator sequence. The chimerical gene construct was
transferred via Agrobacteriurn GV31o1 to petunia variety
W115, using the transformation method as described in
example 7. Transgenic petunia plants were selected that
show ectopic nectary development.
Example 1.9:
Modification of sugar composition and nectar secretion
Although sugar content of nectar from different petunia
W115 flowers shows some variation, the ratio between
hexoses and sucrose is very stable. Down-regulation or up-
regulation of genes involved in the establishment of the
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 34 -
ratio between hexoses and sucrose in nectar will therefore
modify nectar composition. An example is given for anti-
sense expression of a petunia-derived invertase gene.
PCR primers were designed that hybridise with the cDNA of
an invertase gene cloned from Solanum tuberoaum. The 5'
primer 5'-AAGGACTTTAGAGAGACCCGACCACTGCTGG-3'and the 3'
primer 5'-AAATGTC:TTTGATGCATAATATTTCCCATAATC-3' were used
for a PCR reaction on genomic DNA of petunia to yield a
l0 fragment of around 420 bp. The fragment was sequenced and
cloned into a pM4SBlue vector to used as a probe to screen
a petunia nectary-specific cDNA library. Hybridizing phage
plaques were purified and cDNAs were retrieved by in vivo
excision as described in example 2. The expression of the
1.5 cDNA's was determined by Northern blotting as described in
example 3 and the sequence of a nectary-specific invertase
was determined as described in example 2. The invertase
gene was amplified using a 5' primer that hybridises with
sequences just upstream of the ATG translation start site
20 and a 3' primer that hybridises with sequences just down-
stream of the translation stop site. Extra restriction
enzyme recognition sites were generated to allow cloning of
the cDNA in sense (overexpression) or antisense direction
into the binary vector pCP031 as described in example 18.
25 The chimerical gene constructs are transferred via Agrobac-
terium Gv31o2 to petunia variety W115, using the transfor-
mation method as described in example 7. Transgenic petunia
plants were selected that exhibit modified sugar compositi-
on in nectar.
Example 20
Modification of plant development
A DNA which is the IiTECI gene or a homologous gene is
introduced into a plant cell, the said DNA being induced
by promoter elements controlling the expression of the
introduced DNA in such a way that transcription produces
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO OOI04176 PCT/NL99/00453
- 35 -
sense RNA. Plants were regenerated from the transgenic
cells as described in example 7. Plants that ectopically
express the NEC1 gene exhibited modified leaf morphology
and modified sugar composition. Furthermore, plants that
ectopically express the NEC.I gene showed a delay in flowe-
ring time.
REFERENCES:
Angenent G.C., Franken J., Busscher M., Colombo L. and van
Tunen A.J., 1993. Petal and stamen formation in petunia
is regulated by the homeotic gene fbpl. Plant J. 4, 101
112.
Angenent G.C., Franken J., Busscher M., Weiss D. and van
Tuners A.J., 1994. Co-suppression of the petunia homeotic
gene fbp2 affects the identity of the generative meri
stem. Plant J. 5, 33-44.
Baker H.G. and Baker I., 1982. Floral nectar constituents
in relation to pollinator type. In: Handbook of experi-
mental pollination biology (ED. by CE Jones and RJ
Little), pp 117-141. Scientific and Academic Edition,
Van Nostrand Reinhold, New York.
Seals T.P. and Goldberg R.B. (1997). A novel cell ablation
strategy blocks tobacco anther dehiscence. The Plant
Cell 9, 1527--1545.
Bevan M. (1984). Binary Agrobacterium vectors for plant
transformatian. Nucl. Acids Res. 12, 8711-8721
Butler C.G. (1962). The world of the honeybee. London:
Collins.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99100453
- 36 -
Canas L.A., Busscher M., Angenent G.C., Beltran J-P and van
Tunen A.J. (1994). Nuclear localisation of the petunia
MADS box gene protein FBP1. Plant J. 6, 597-604.
Colombo L., Franken J., Koetje E., van Went J., Dons H.J.M,
Angenent G.C. and van Tunen A.J. (1995). The petunia
MARS Box gene FBP1I determines ovule identity. The Plant
Cell 7, 1859-1868.
Crailsheim k., Hrassnigg N., Lorenz W and Lass A. Protein
consumption and distribution in a honeybee colony (Apis
mellifera carnica Pollm). Apidologie 24 (5), 510-511.
Ousters J.B.M., Oldenhof, Schrauwen J.A.M., Cordewener
J.H.G., Wullems G.J. and Lookeren Campagne M.M. (1997).
Analysis of microspore-specific promoters in transgenic
tobacco. Plant molecular biology 35, 689-699.
Davis A.R., Peterson R.L. and Shuel R.W. (1986). Anatomy
and vasculature of the floral nectaries of Brassica
napes (Brassicaceae). Can. J. Bot. vol. 64, 2508-2516.
Dumas B., Freyssinet G. and Pallett E. (1995). Tissue-
specific expression of germin-like oxalate oxidase
during development and fungal infection of barley seed-
lings. Plant Physiol. 107: 1091-1096.
Fahn A. (1979). Ultrastructure of nectaries in relation to
nectar secretion. Amer. J. Bat. 66(8), 977-985.
Florack D.E.A, Dirkse W.G., Visser B., F. Heidekamp and
Stiekema W.J. (1994). Expression of biologically
active hordothionins in tobacco: Effects of pre- and
pro-sequences at the amino and carboxyl termini of the
hordothionin precursor on mature protein expression
and sorting. Plant Mol. Biol. 24. 83-96.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 37 -
Hartley R.W. (1988). Barnase and Barstar: Expression of its
cloned inhibitor permits expression of a cloed ribonu-
clease. J. Mol. Biol. 202, 913-915
Horsch R.B., Fry J.E., Hofman N.L., Eichholz D., Rogers
S.G. and Fraley R.T. (1985). A simple and general method
for transferring genes into plants. Science 227, 1229-
1231.
Huang H., Tudor M., Su T., Hu Y. and Ma H. (1996). DNA
binding properties of two Arabidopsis MARS domain pro-
teins: Binding consensus and dimer formation. The Plant
Cell 8, 81-94.
Jefferson R.A., Kavanagh T.A. and Bevan M. (1987). GUS
fusion: J-glucuronidase as a sensitive and versatile
gene fusion marker in higher plants. EMBO J. 6, 3901-
3907.
Laemmli U.K. (1970). Cleavage of structural proteins during
the assembly of the head bacteriophage T4. nature 227,
680-685.
Lane B.G., Dunwell J.M., Ray J.A., Schmitt M.R. and Cuming
A.C. (1993). Germin, a protein marker of early plant
development, is an oxalate oxidase. The Journal of
Biological Chemistry 268 (17), 12239-12242.
Mariani C., Gossele V., De Beuckeleer M., De Block M.,
Goldberg R.B., De Greef W. and Leemans J. (1992). a
chimaeric ribonuclease-inhibitor gene restores fertility
to male sterile plants. Nature 357, 384-387.
Martini M., Schmid A. and Hess D. (1990). Antibiotics, and
amino acids in nectar of Rhododendron and Piptanthus
species from Nepal. Bot. Acta 103, 343-348.
SUBSTITUTE SHEET (RULE 26)
~ 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 38 -
Mizukami Y., Huang H., Tudor M. and Ma H. (1996). Functio-
nal domains of the floral regulator AGAMOUS: Characteri-
sation of the DNA binding domain and analysis of domi-
nant negative ~tnutations. The Plant Cell 8, 831-845.
Tsuchimoto S., Van der Kro1 A.R. and Chua N.-H, 1993.
Ectopic expression of pMA:DS3 in transgenic petunia
phenocopies the petunia blind mutant. Plant Cell 5, 843-
853.
Verwoerd T.C., Dekker, B.M.M, and Hoekema A. (1989}. A
small-scale procedure fox the rapid isolation of plant
RNAs. Nucl. Acids Res. 17, 2362.
Zer H. and Fahn A. (1992). Floral nectaries of Rosmarinus
officinalis L. Structure, Ultrastructure and Nectar
Secretion. Annals of Botany 70, 391-397.
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 39 -
Sequences:
SEQ ID N0:1 amino acid sequence NEC1
1 MAQLRADDLS FIFGLLGNIV SFMVFLAPVP TFYKIYKRKS SEGYQAIPYM
51 VALFSAGLLL YYAYLRKNAY LIVSINGFGC AIELTYISLF LFYAPRKSKI
101 FTGWLMLLEL GALGMVMPIT YLLAEGSHRV MIVGWTCAAI NVAVFAAPLS
151 TMRQVTKTKS VEFMPFTLSL FLTLCATMWF FYGFFKKDFY IAFPNILGFL
201 FGIVQMLLYF VYKDSKRIDD EKSDPVREAT KSKEGVETIT NIEDDNSDNA
251 LQSMEKDFSR LRTSK
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
-40-
SEQ ID No: 2 amino acid sequence of FBP15
Met G1y Arg Gly Lys Ile Glu Ile Lys Arg Ile Glu Asn Thr Thr Asn
1 5 10 15
Arg Gln Val Thr Phe Cys Lys Arg Arg Asn Gly Leu Leu Lys Lys Ala
20 25 30
Tyr Glu Leu Ser Val Leu Cys Asp Ala Glu val Ala Leu Ile Val Phe
35 40 45
Ser Ser Arg Gly Arg Leu Tyr Glu Tyr Ala Asn Asn Ser Val Lys Ala
50 55 60
Thr Ile Asp Arg Tyr Lys Lys Ala Sex Ser Asp Ser Ser Asn Thr Gly
65 70 ?5 80
Ser Thr Ser Glu Ala Asn Thr Gln Phe Tyr Gln Gin Glu Ala Ala Lys
85 90 95
Leu Arg Val Gln Ile Gly Asn Leu Gln Asn Ser Asn Arg Asn Met Leu
100 105 110
Gly Glu Ser Leu Ser Ser Leu Thr Ala Lys Asp Leu Lys Gly Leu Glu
115 120 125
Thr Lys Leu Glu Lys Gly Ile Ser Arg Ile Arg Ser Lys Lys Asn Glu
130 135 140
Leu Leu Phe Ala Glu Ile G1u Tyr Met Arg Lys Arg Glu Ile Asp Leu
145 150 155 150
His Asn Asn Asn Gln Met Leu Arg Ala Lys Ile Ala Glu Ser Glu Arg
165 170 175
Asn Val Asn Met Met Gly Gly Glu Phe Glu Leu Met Gln Ser His Pro
180 185 190
Tyr Asp Pro Arg Asp Phe Phe Gln Val Asn Gly Leu Gln His Asn His
195 200 205
Gln Tyr Pro Arg Gln Asp Asn Met Ala Leu Gln Leu Val
210 27.5 220
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99100453
- 41 -
SEQ ID N0:3 amino acid sequence CUSP
MFLPILFTISLLFSSSH.A
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99/00453
- 42 -
SEQ ID N0:4 Nucleotide sequence NEC1
1 TCGAGCGGCCGCCCGGGCAGGTATTCAACAAGAGTATTCACCACTTGAAC
51 TCAAAAGGGGCTTCACTAAAAAAAAATCATGGCGCAATTACGTGCTGATG
101 ACTTGTCTTTCATATTTGGCCTTCTTGGTAATATTGTATCATTCATGGTC
151 TTCCTAGCACCCGTGCCAACATTTTACAAAATATATAAAAGGAAATCATC
201 AGAAGGATATCAAGCAATACCATATATGGTAGCACTGTTCAGCGCCGGAC
251 TATTGCTATATTATGCTTATCTCAGGAAGAATGCCTATCTTATCGTCAGC
301 ATTAATGGCTTTGGATGTGCCATTGAATTAACATATATCTCTCTGTTTCT
351 CTTTTACGCGCCCAGAAAGTCTAAGATTTTCACAGGGTGGCTGATGCTCT
401 TAGAATTGGGAGCCCTAGGAATGGTGATGCCAATTACTTATTTATTAGCA
451 GAAGGCTCACATAGAGTGATGATAGTGGGATGGATTTGTGCAGCTATCAA
2 501 TGTTGCTGTCTTTGCTGCTCCTTTAAGCATCATGAGGCAAGTAATAAAAA
5
551 CAAAGAGTGTAGAGTTCATGCCCTTCACTTTATCTTTGTTCCTCACTCTC
601 TGTGCCACTATGTGGTTTTTCTATGGGTTTTTCAAGAAGGACTTTTACAT
651 TGCGTTTCCAAATATACTGGGCTTTCTATTCGGAATCGTTCAAATGCTAT
701 TATATTTTGTTTACAAGGATTCAAAGAGAATAGATGATGAAAAATCTGAT
3 751 CCTGTTCGAGAAGCTACAAAATCAAAAGAAGGTGTAGAAATCATTATCAA
5
801 CATTGAAGATGATAATTCTGATAACGCATTGCAGTCCATGGAGAAGGATT
851 TTTCCAGACTGCGGACATCAAAATAAGCAAGAAGATGATCAAAAAATGAC
901 AAAGCTAAGGAGTTTGAAGTAAGGCAAGGAACTTGACACTGAATATCTAA
951 GCTAATTAGCAAGACTTTAGCAGCTTGTAATATTTAGTGTTTGTGAGGTG
4 1001 TTACCTTATAATT.AGCTTGTAGCATAGCCTTCCCACTAATAATTCTGCTT
5
1051 AGCGAATCTTATATATGGGAAATACTTACACTAGTATGCATCTTCTATAT
1101 ACATGTTTGGCACTTGACTATACATAGAAAAATTAACAAGCATTTCTCAC
1151 CTCAATTTGTCACTTACTTATAAGTAGCTGAATAATATAATGCAATTTTC
1201 ACCCC
60
SUBSTtTUTE SHEET (RULE 26~
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 43 -
SEQ ID N0:5 Nucleotide sequence FBP15
1 TCTGAATACAAGCTGTGTGTGTAGAGAGATTTCATAAAGACAGCAAACAT
51 CCCTTCTTTTTGTTCTGTTTTAAAAGTTCCCTTCTTCAACCAGCTCTTTT
101 CCTCATCAGGGTAAGTTGCAAATAAAGGGGATGTTCCAGAATCAAGAAGA
151 GAAGATGTCAGACT(~GCCTCAGAGGAAGATGGGAAGAGGAAAGATTGAGA
l0
201 TTAAGAGGATTGAAAATACAACAAATCGTCAAGTCACTTTCTGTAAGAGA
251 AGAAATGGGTTGCTTAAAAAAGCTTATGAACTTTCTGTTCTTTGTGATGC
i5 301 TGAAGTTGCTCTCATCGTTTTCTCAAGCCGTGGCCGCCTCTATGAATATG
351 CTAACAACAGTGTGAAGGCAACAATTGATAGATATAAGAAAGCATCCTCA
401 GATTCCTCCAACACTGGATCTACTTCTGAAGCTAACACTCAGTTTTATCA
451 ACAAGAAGCTGCCAAACTCCGAGTTCAGATTGGTAACTTACAGAACTCAA
501 ACAGGAACATGCTAGGCGAGTCTCTAAGTTCTCTGACTGCAAAAGATCTG
2 5 551 AAAGGCCTGGAGACCAAACTTGAGAAAGGAATTAGTAGAATTAGGTCCAA
601 AAAGAATGAACTCCTGTTTGCTGAGATTGAGTATATGCGAAAAAGGGAAA
651 TTGATTTGCACAACAACAATCAGATGCTTCGGGCAAAGATAGCTGAGAGT
701 GAAAGAAATGTGAACATGATGGGAGGAGAATTTGAGCTGATGCAATCTCA
751 TCCGTACGATCCAAGAGACTTCTTCCAAGTGAACGGCTTACAGCATAATC
3 5 801 ATCAATATCCACGCCAAGACAACATGGCTCTTCAATTAGTATAAGTTTAT
851 AATAAAATGCATGGTTTGAAGCACTCTGATTGTGGTGGATTTGGATTATG
901 TATAAGGGAGTGCAGGCCATTTGCCAATTATTGAAAGGTACTCAAACAGG
951 AAGTTGAAGAAGTTCATCATCTCTCTCATCTATATGTCTTAACAAAAGTC
1001 TTAGCTTATGGACTCTAAAACAAAGACTTAATTTAACATATAAATATAAT
4 5 1051 TGTGTAATGCTGTTGTATTGTATGGTATGTATCCAAAAACATTAATAACC
1101 TATCTTTTTCTTCAAATTATGTCTCCTTTGATACAAACTACTAACATATT
1151 TTCTTAT
SUBSTITUTE SHEET (RULE 261
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99100453
- 44 -
SEQ ID N0:6 Nucleotide sequence CVSP
ATGTTTCTTCCAATTCTCTTCACCATTTCCCTCCTCTTCTCCTCCTCCCATGCT
SUBSTITUTE SHEET (RULE 2~j
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
- 45 -
SEQ TD N0:7 Nucleotide sequence NECI promoter
1 CCTAGGAGAAATCAAGCCTACTCTTAAGATGGATGACTCACTTGCCCCGA .
51 TGGTAAGGT(3AAGGATCTGTTGATTAGAGTTGGGAAGTTCATGTTCTCTG
101 CTGATTTTATTATTCTAGACTATGAAGAGGACCAAGAAGCTCCAATAATT
151 TTGGGAAGAGCATTCTTAATCACATCGATGGCAATTATTGACATGGAACT
201 TGGGGAGATGACTGTGAGAGCGCATGGAGAAAAGGTTACTTTCAAGGTTT
251 ATAATAAAAAGGATCATATGGCTAAGTTTGAAGAGTGTTCTTTGATAGAA
301 TGTGTCAGA(~GAGAACATGAAAGTAAACCGAAAGAGGTGTTTGAGCGGAA
351 TGTAGAACAAAGTGACCACGGCACAATAATTGACAAGTTGAAGGAAAATT
401 CACCTAAAGGAAGGAAGAAGACAAAAGTTCGTCGTAACAAGAGGAGACGT
451 AAATGCTGGA.AGTGAGCTTAAAGGTGTTGTCGTACTACGACGTTAACTAA
501 GGCGCTTGTCGGGAGGCAACCCTAGCTTTGTATGTAAATGTAAA.AGTAAA
551 AAATATATATATAGAAAAAGGAAAATACAAAAAGAGTCGTGCCGCGACGT
601 TAAATCAAGCGCTTGTTGGAAGGCAACCCAATTTTTATTGTTTTAGTTGT
651 TTTACTTATTTAGTATTACGTAGTTTCTTGTTGTTTTTGTAGGGCTCGGG
7a1 ACTTTCGGAAGGTGAGGTAATTTCAAGGCATCGCGGTGTGTATTGCAGCG
751 AGGTAAGTGTAAGAGTTGAGTTGGAAGCGTTTGGCCAAGTGTTGCACCGT
801 GAGAGGCTTTCAACCTGTTGCGACACGTGAAA.AATTAAGAGCCAGATCTG
851 CTACATTAGCACTGAAGCATCGCTTGGCCAATAGCTTGGAATGGAAGCAA
901 GAATTCAAACCAAA.ATCAGAAACGCCACAAGAGATGTGTCGCACACTGCA
951 AAGCTTTGTGCAAACTAGTGAACGCAGAAATAGAAATGCTACAGCCCATG
1001 CGTCGCTTGGCTTATGGCAGGCAGCAAAAATTCAGCAGCAAAACAGAAAC
1051 GCTGCGAGAAACGCGTCGCATACGCCATAGCTTTGTGTCAAACAGAACGT
1101 CCAGAAATTGAAAAGCTATAAGCCTGCGTCGCTTGGCTCATGGCGTGCAG
1151 ACTAGAAAAGCTCTAGCAGATGCGTCGCGTATTGTATAGCTTGGTGTGAA
1201 ACAGAAAGTTCGAAACTTGGAAAACGATAACCCAGCGTCGCCTCTTCAAC
1251 CGCGTCCAGGTAAGTTCAAGATTCTTACGGGTTGACCCATTAACCCATTG
1301 ATCGGCTGATTATAAACAATAAAACATCACCTTCAACTATCACATGATTT
1351 CATAAGTTTGACCTAGGATATTTTATATATATATATATATATATACACAC
1401 ACACACCATTTCCAGCGATCTTACCTCATTTTTATTCAAACCATTTTTCT
SUBSTtTUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99100453
- 4fi
1451 GCTTCAAA,AGTTTAAATTATTAATATGATAAGTCATCCATAGTCAAACAA
1501 GATTTTCTATACTATTTTGTCCCTTGTAATTTTAAAAAAAAAATGAGCGA
1551 TGGTAAGATAAACATTGTTTGCAAGTGTACAATTTTAGTATATGCAAACC
1601 AACGCTTCTTCTTCCAACTATCACCTAAAACTACATCATTTATGGCGGGC
1651 GGACTAGACGTAGCCAAATATAAAAACGCAATGGCCATTCAGTTCATGTC
1701 ATTTTTATATCCTTCATCCAATAATATTACTCAAAATTGATGTACAGTTT
1751 GGTCTCTGATGTGCACTTTACTATACGTAATACGGAATTTACATTATAAT
1802 TAAAGAGAACTGTTCCACTAAATTTTAATGATTTAATTAATTTAACTCGG
1851 TTACTTGTATTATTATTATTGCTGTATTTGTTTGTCATTTGAATTTGGCA
1901 CCGCAGATTTTTGTATGCAATTAACCCTCATATATCTTTTGGCCAAATAA
1951 AGAAAAAGTCTGCATATTTCTTGCCAAACATTTATCATACTTTACCGAAT
2001 TCTTGTTTTTTGTTTCTCTGTTGTTGTTCTCCACTATAAATAACATTTGC
2051 AGTGAGTAAAGTTTCTTCAGGTCTCTTTTGTAGATTCAACAAGAGTATTC
2101 AGCACTTGAACTCAAAAGGGGCTTCACTAAAAAAAATCATG
SUBSTITUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99/00453
- 47 -
SEQ ID N0:8 Nucleotide sequence FBP15 promoter
SU9ST~TUTE SHEET (RULE 26)
CA 02333897 2001-O1-16
WO 00104176 PCTIlVL99/00453
SE(ZUENCE LISTING
<110> CPRO-DLO
<120> Process to collect metabolites from modified nectar by
insects
<130> 159782
<190> pct/n199/00953
<141> 1999-07-15
<160> 10
<170> PatentIn Vex. 2.7.
<210> 1
<211> 265
<212> PRT
<213> Petunia x hybrids
.. <220>
<223> strain: W115
<220>
<223> tissue type: nectar gland
<220>
<223> NEC1 amino acid sequence
<900> 1
Met Ala Gln Leu Arg Ala Asp Asp Leu Ser Phe Ile Phe Gly Leu Leu
1 5 10 15
Gly Asn Ile Val Ser Phe Met Val Phe Leu Ala Pro Val Pro Thr Phe
20 25 30
Tyr Lys Ile Tyr Lys Arg Lys Ser Ser Glu Gly Tyr Gln Ala Ile Pro
35 40 45
Tyr Met Val Ala Leu Phe Ser Ala Gly Leu Leu Leu Tyr Tyr Ala Tyr
50 55 60
Leu Arg Lys Asn Ala Tyr Leu Ile Val Sex Ile Asn Gly Phe Gly Cys
65 70 75 80
Ala Tle Glu Leu Thr Tyr Ile Ser Leu Phe Leu Phe Tyr Ala Pro Arg
85 90 95
1
CA 02333897 2001-O1-16
WO 00/04176 . PCT/NL99/00453
Lys Ser Lys Ile Phe Thr Gly Trp Leu Met Leu Leu Glu Leu Gly Ala
100 105 110
Leu Gly Met Val Met Pro Ile Thr Tyr Leu Leu Ala Glu Gly Ser fiis
115 120 125
Arg Val Met Ile Val Gly Trp Ile Cys Ala Ala Ile Asn Val Ala Val
130 135 140
Phe Ala Ala Pro Leu Ser Ile Met Arg Gln Val Ile Lys Thr Lys Ser
145 150 155 160
Val Glu Phe Met Pro Phe Thr Leu Ser Leu Phe Leu Thr Leu Cys Ala
165 170 175
Thr Met Trp Phe Phe Tyr Gly Phe Phe Lys Lys Asp Phe Tyr Ile Ala
180 185 190
Phe Pro Asn Ile Leu GIy Phe Leu Phe Gly Ile Val Gln Met Leu Leu
195 200 205
Tyr Phe Val Tyr Lys Asp Ser Lys Arg Ile Asp Asp Glu Lys Ser Asp
210 215 220
Pro Val Arg Glu Ala Thr Lys Ser Lys Glu Gly Val Glu Ile Ile Ile
225 230 235 240
Asn Ile Glu Asp Asp Asn Ser Asp Asn Ala Leu Gln Ser Met Glu Lys
245 250 255
Asp Phe Ser Arg Leu Arg Thr Sex Lys
250 265
<210> 2
<211> 221
<212> PRT
<213> Petunia x hybrida
<220>
<223> strain: W115
<220>
<223> tissue type: nectar gland, secretory cell
<220>
2
CA 02333897 2001-O1-16
WO 00104176 PCTINL99100453
<223> FBP15 amino acid sequence
<900> 2
Met Gly Arg Gly Lys Ile Glu I1e Lys Arg Ile Glu Asn Thr Thr Asn
1 5 10. 15
Arg Gln Val Thr Phe Cys Lys Arg Arg Asn Gly Leu Leu Lys Lys Ala
20 25 30
Tyr Glu Leu Ser Vai Leu Cys Asp Ala Glu Val Ala Leu Ile Val Phe
35 40 45
Ser Ser Arg Gly Arg Leu Tyr Glu Tyr Ala Asn Asn Ser Val Lys Ala
50 55 60
Thr Ile Asp Arg Tyr Lys Lys Ala Ser Ser Asp Ser Ser Asn Thr Gly
65 70 75 80
Ser Thr Ser Glu Ala Asn Thr Gln Phe Tyr Gln Gln Glu Ala Ala Lys
85 90 95
Leu Arg Val Gln Ile Gly Asn Leu Gln Asn Ser Asn Arg Asn Met Leu
100 105 110
Gly Glu Ser Leu Ser Sex Leu Thr Ala Lys Asp Leu Lys Gly Leu Glu
115 120 125
Thr Lys Leu Glu Lys Gly Ile Ser Arg Ile Arg Ser Lys Lys Asn Glu
130 135 140
Leu Leu Phe Ala Glu Iie Glu Tyr Met Arg Lys Arg Glu Ile Asp Leu
145 150 155 160
His Asn Asn Asn Gln Met Leu Arg Ala Lys Ile Ala Glu Ser Glu Arg
165 170 175
Asn Val Asn Met Met Gly Gly Glu Phe Glu Leu Met Gln Ser His Pro
180 185 190
Tyr Asp Pro Arg Asp Phe Phe Gln Val Asn Gly Leu Gln His Asn His
195 200 205
Gln Tyr Pro Arg Gln Asp Asn Met Ala Leu Gln Leu Val
210 215 220
<220> 3
3
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
<211> 18.
<212> PRT
<213> Calluna vulgaris
<220>
<223> tissue type: flower
<220>
<223> Calluna vulgaris signal peptide
<A00> 3
Met Phe Leu Pro Ile Leu Phe Thr Ile Ser Leu Leu Phe Ser Ser Ser
1 5 10 15
His Ala
<210> 4
<211> 1205
<212> DNA
<213> Petunia x hybrida
<220>
<221> CDS
<222> (79)..(873)
<220>
<223> strain: W115
<220>
<223> tissue type: nectar gland
<220>
<223> NEC1
<400> 9
tcgagcggcc gcccgggcag gtattcaaca agagtattca ccacttgaac tcaaaagggg 60
cttcactaaa aaaaaatc atg gcg caa tta egt get gat gac ttg tct ttc 111
Met Ala Gln Leu Arg Ala Asp Asp Leu Ser Phe
1 5 10
ata ttt ggc ctt ctt ggt aat att gta tca ttc atg gtc ttc cta gca 159
Ile Phe Gly Leu Leu Gly Asn Ile Val Ser Phe Met Val Phe Leu Ala
15 20 25
4
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
ccc gtg cca aca ttt tac; aaa ata tat aaa agg aaa tca tca gaa gga 207
Pro Val Pro Thr Phe Tyr Lys Ile Tyr Lys Arg Lys Ser Ser Glu Gly
30 35 40
tat caa gca ata cca tat. atg gta gca ctg ttc agc gcc gga cta ttg 255
Tyr Gln Ala Ile Pro Tyr Met Val Ala Leu Phe Ser Ala Gly Leu Leu
45 50 55
eta tat tat get tat etc agg aag aat gee tat ett atc gtc age att 303
Leu Tyr Tyr Ala Tyr Leu Arg Lys Asn Ala Tyr Leu Ile Val Ser Ile
60 65 70 75
aat ggc ttt gga tgt gcc att gaa tta aca tat atc tct ctg ttt ctc 351
Asn Gly Phe Gly Cys Ala Ile Glu Leu Thr Tyr Ile Ser Leu Phe Leu
80 85 ~ 90
ttt tac gcg ccc aga aag tct aag att ttc aca ggg tgg ctg atg ctc 399
Phe Tyr Ala Pro Arg Lys Ser Lys Ile Phe Thr Gly Trp Leu Met Leu
95 100 105
tta gaa ttg gga gcc cta gga atg gtg atg cca att act tat tta tta 447
Leu Glu Leu Gly Ala Leu Gly Met Val Met Pro Ile Thr Tyr Leu Leu
110 115 120
gca gaa ggc tca cat aga gtg atg ata gtg gga tgg att tgt gca get 495
Ala Glu Gly Ser His Arg Val Met Ile Val Gly Trp Ile Cys Ala Ala
125 130 135
ate aat gtt get gtc ttt get get cct tta agc ate atg agg caa gta 543
Ile Asn Val Ala Val Phe Ala Ala Pro Leu Ser Ile Met Arg Gln VaI
140 145 I50 155
ata aaa aca aag agt gta gag ttc atg ccc ttc act tta tct ttg ttc 591
Ile Lys Thr Lys Ser Val Glu Phe Met Pro Phe Thr Leu Ser Leu Phe
160 155 170
ctc act ctc tgt gcc act atg tgg ttt ttc tat ggg ttt ttc aag aag 639
Leu Thr Leu Cys Ala Thr Met Trp Phe Phe Tyr Gly Phe Phe Lys Lys
175 180 185
gac ttt tac att gcg ttt cca aat ata ctg ggc ttt cta ttc gga atc 687
Asp Phe Tyr Ile Ala Phe Pro Asn Ile Leu Gly Phe Leu Phe Gly Ile
190 195 200
gtt caa atg cta tta tat ttt gtt tac aag gat tca aag aga ata gat 735
Val Gln Met Leu Leu Tyr Phe Val Tyr Lys Asp Ser Lys Arg Ile Asp
205 210 215
CA 02333897 2001-O1-16
WO 00!04176 PCT/NL99100453
gat gaa aaa tct gat cct: gtt cga gaa get aca aaa tca aaa gaa ggt 783
Asp Glu Lys Ser Asp Pra Val Arg Glu Ala Thr Lys Ser Lys Glu Gly
220 225 230 235
gta gaa atc att atc aac: att gaa gat gat aat tct gat aac gca ttg 831
Val Glu Ile Ile Ile Asn Ile Glu Asp Asp Asn Ser Asp Asn Ala Leu
290 295 250
cag tcc atg gag aag gat ttt tcc aga ctg cgg aca tca aaa g73
Gln Ser Met Glu Lys Asp Phe Ser Arg Leu Arg Thr Ser Lys
255 260 265
taagcaagaa gatgatcaaa aaatgacaaa gctaaggagt ttgaagtaag gcaaggaact 933
tgacactgaa tatctaagct aattagcaag actttagcag cttgtaatat ttagtgtttg 993
tgaggtgtta ccttataatt agcttgtagc atagccttcc cactaataat tctgcttagc 1053
gaatcttata tatgggaaat acttacacta gtatgcatct tctatataca tgtttggcac 1113
ttgactatac atagaaaaat taacaagcat ttctcacctc aatttgtcac ttacttataa 1173
gtagctgaat aatataatgc aattttcacc cc 1205
<210> 5
<211> 265
<2I2> PRT
<213> Petunia x hybrida
<223> NEC1
<400> 5
Met Ala Gln Leu Arg Ala Asp Asp Leu Ser Phe Ile Phe Gly Leu Leu
1 S 10 15
Gly Asn Ile Val Ser Phe Met Val Phe Leu Ala Pro Val Pro Thr Phe
20 25 30
Tyr Lys Ile Tyr Lys Arg Lys Ser Ser Glu Gly Tyr Gln Ala Ile Pro
35 40 45
Tyr Met Val Ala Leu Phe Ser Ala Gly Leu Leu Leu Tyr Tyr Ala Tyr
50 55 60
Leu Arg Lys Asn Ala Tyr Leu Ile Val Ser Ile Asn Gly E'he Gly Cys
65 70 75 80
Ala Ile Glu Leu Thr Tyr Ile Ser Leu Phe Leu Phe Tyr Ala Pro Arg
6
CA 02333897 2001-O1-16
WO 00/04176 PCTINL99100453
85 90 g5
Lys Sex Lys Ile Phe Thr Gly Trp Leu Met Leu Leu Glu Leu Gly Ala
100 105 110
Leu Gly Met Val Met Pro Ile Thr Tyr Leu Leu Ala Glu Gly Ser His
I15 120 125
Arg Val Met Ile Val Gly Trp Ile Cys Ala Ala Ile Asn Val Ala Val
130 135 190
Phe Ala Ala Pro Leu Ser Ile Met Arg Gln Val Ile Lys Thr Lys Ser
195 , i50 155 160
Val Glu Phe Met Pro Phe Thr Leu Ser Leu Phe Leu Thr Leu Cys Ala
165 170 175
Thr Met Trp Phe Phe Tyr Gly Phe Phe Lys Lys Asp Phe Tyr Ile Ala
180 185 190
Phe Pro Asn Ile Leu Gly Phe Leu Phe Gly Iie Val Gln Met Leu Leu
195 200 205
Tyr Phe Val Tyr Lys Asp 5er Lys Arg Iie Asp Asp Glu Lys Ser Asp
zlo 215 220
Pro Val Arg Glu Ala Thr Lys Ser Lys Glu Gly Val Glu Ile Ile Ile
225 230 235 2qp
Asn Ile Glu Asp Asp Asn Ser Asp Asn Ala Leu Gln Ser Met Glu Lys
245 250 255
Asp Phe Ser Arg Leu Arg Thr Ser Lys
260 265
<zlo> 6
<211> 1157
<212> DNA
<213> Petunia x hybrida
<220>
<221> CDS
<222> (I79} " (841)
<220>
<223> strain: WlIS
7
CA 02333897 2001-O1-16
WO 00/04176 PCTlNL99/00453
<220>
<223> tissue type: nectar gland
<220>
<223> cDNA library of nectaries from Petunia hybrida
flowers
<220>
<223> FBP15
<400> 6
tctgaataca agctgtgtgt gtagagagat ttcataaaga cagcaaacat cccttctttt 60
tgttctgttt t~aaagttcc cttcttcaac cagctctttt cctcatcagg gtaagttgca 120
aataaagggg atgttccaga atcaagaaga gaagatgtca gactcgcctc agaggaag 178
atg gga aga gga aag att gag att aag agg att gaa aat aca aca aat 226
Met Gly Arg Gly Lys Ile Glu Ile Lys Arg Ile Glu Asn Thr Thr Asn
1 5 10 15
cgt caa gtc act ttc tgt aag aga aga aat ggg ttg ctt aaa aaa get 274
Arg Gln Val Thr Phe Cys Lys Arg Arg Asn Gly Leu Leu Lys Lys Ala
20 25 30
tat gaa ctt tct gtt ctt tgt gat get gaa gtt get ctc atc gtt ttc 322
Tyr Glu Leu Ser Val Leu Cys Asp Ala Glu Val Ala Leu Ile Val Phe
35 90 45
tca agc cgt ggc egc ctc tat gaa tat get aac aac agt gtg aag gca 370
Ser Ser Arg Gly Arg Leu Tyr Glu Tyr Ala Asn Asn Ser Val Lys Ala
50 55 60
aca att gat aga tat aag aaa gca tcc tca gat tcc tcc aac act gga 418
Thr Ile Asp Arg Tyr Lys Lys Ala Ser Ser Asp Ser Ser Asn Thr Gly
65 70 75 80
tct act tet gaa get aac act cag ttt tat caa caa gaa get gec aaa 466
Ser Thr Ser Glu Ala Asn Thr Gln Phe Tyr Gln Gln Glu Ala Ala Lys
85 90 95
ctc cga gtt cag att ggt aac tta cag aac tca aac agg aac atg cta 519
Leu Arg Val Gln Ile Gly Asn Leu Gln Asn Ser Asn Arg Asn Met Leu
100 105 110
ggc gag tct cta agt tct ctg act gca aaa gat ctg aaa ggc ctg gag 562
Gly Glu Ser Leu Ser Ser Leu Thr Ala Lys Asp Leu Lys Gly Leu Glu
8
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99/00453
115 120 125
acc aaa ctt gag aaa gga att agt aga att agg tcc aaa aag aat gaa 610
Thr Lys Leu Glu Lys Gly Ile Ser Arg Ile Arg Ser Lys Lys Asn Glu
i30 135 140
ctc ctg ttt get gag att gag tat atg cga aaa agg gaa att gat ttg 658
Leu Leu Phe Ala Glu Ile Glu Tyr Met Arg Lys Arg Glu Ile Asp Leu
145 150 155 160
cac aac aac aat cag atg ctt cgg gca aag ata get gag agt gaa aga 706
His Asn Asn Asn Gln Met Leu Arg Ala Lys Ile Ala Glu Ser Glu Arg
165 170 175
aat gtg aac atg atg.gga gga gaa ttt gag ctg atg caa tct cat ccg 754
Asn Val Asn Met Met Gly Gly Glu Phe Glu Leu Met Gln Ser His Pro
180 185 190
tac gat cca aga gac ttc ttc caa gtg aac ggc tta cag cat aat cat 802
Tyr Asp Pro Arg Asp Phe Phe Gln Val Asn Gly Leu Gln His Asn His
195 200 205
caa tat cca cgc caa gac aac atg get ctt caa tta gta taagtttata 851
Gln Tyr Pro Arg Gln Asp Asn Met Ala Leu Gln Leu Val
210 215 220
ataaaatgca tggtttgaag cactctgatt gtggtggatt tggattatgt ataagggagt 911
gcaggccatt tgccaattat tgaaaggtac tcaaacagga agttgaagaa gttcatcatc 971
tctctcatct atatgtctta acaaaagtct tagcttatgg actctaaaac aaagacttaa 1031
tttaacatat aaatataatt gtgtaatgct gttgtattgt atggtatgta tccaaaaaca 1091
ttaataacct atctttttct tcaaattatg tctcctttga tacaaactac taacatattt 1151
tcttat 1157
<210> 7
<21i> 221
<212> PRT
<213> Petunia x hybrida
<223> FBP15
<900> 7
Met Gly Arg Gly Lys Ile Glu Ile Lys Arg Ile Glu Asn Thr Thr Asn
1 5 10 15
9
CA 02333897 2001-O1-16
WO 00104176 PCT/NL99100453
Arg Gln Val Thr Phe Cys Lys Arg Arg Asn Gly Leu Leu Lys Lys Ala
20 25 30
Tyr Glu Leu Ser Val Leu Cys Asp Ala Glu Val Ala Leu Ile Val Phe
35 40 45
Ser Ser Arg Gly Arg Leu Tyr Glu Tyr Ala Asn Asn Ser Val Lys Ala
50 55 60
Thr Ile Asp Arg Tyr Lys Lys Ala Sex Ser Asp Ser Ser Asn Thr Gly
65 70 75 80
Ser Thr Ser Glu Ala Asn Thr Gln Phe Tyr Gln Gln Glu Ala Ala Lys
85 90 95
Leu Arg Val Gln Ile Gly Asn Leu Gln Asn Ser Asn Arg Asn Met Leu
100 105 110
Gly Glu 5er Leu Ser Ser Leu Thr Ala Lys Asp Leu Lys Gly Leu Glu
115 120 125
Thr Lys Leu Glu Lys Gly Ile Ser Arg Ile Arg Ser Lys Lys Asn Glu
130 135 140
Leu Leu Phe Ala Glu Ile Glu Tyr Met Arg Lys Arg Glu Ile Asp Leu
145 150 155 160
His Asn Asn Asn Gln Met Leu Arg Ala Lys Ile Ala Glu Ser Glu Arg
165 170 175
Asn Val Asn Met Met Gly Gly Glu Phe Glu Leu Met Gln Ser His Pro
180 185 190
Tyr Asp Pro Arg Asp Phe Phe Gln Val Asn Gly Leu Gln His Asn His
195 200 205
Gln Tyr Pro Arg Gln Asp Asn Met Ala Leu Gln Leu Val
210 215 220
<210> 8
<211> 59
<212> DNA
<213> Calluna vulgaris
<220>
to
CA 02333897 2001-O1-16
WO 00/04176 PCT/NL99/00453
<221> CDS
<222> (1)..(54)
<220>
<221> sig_peptide
<222> (1)..(54)
<400> 8
atg ttt ctt cca att ctc ttc acc att tcc ctc ctc ttc tcc tcc tcc 48
Met Phe Leu Pro Ile Leu Phe Thr Ile Ser Leu Leu Phe Ser Ser Ser
1 5 10 15
cat get 54
His Ala
<210> 9
<211> 18
<212> PRT
<213> Calluna vulgaris
<400> 9
Met Phe Leu Pro Ile Leu Phe Thr Ile Ser Leu Leu Phe Ser Ser Sex
1 5 10 15
His Ala
<210> 10
<211> 2141
<212> DNA
<213> Petunia x hybrida
<220>
<223> strain: W115
<220>
<223> NEC1 promoter
<400> 10
cctaggagaa atcaagccta ctcttaagat ggatgactca cttgccccga tggtaaggtg 60
aaggatctgt tgattagagt tgggaagttc atgttctctg ctgattttat tattctagac 120
tatgaagagg accaagaagc tccaataatt ttgggaagag cattcttaat cacatcgatg 180
gcaattattg acatggaact tggggagatg actgtgagag cgcatggaga aaaggttact 240
ttcaaggttt ataataaaaa ggatcatatg gctaagtttg aagagtgttc tttgatagaa 300
tgtgtcagac gagaacatga aagtaaaccg aaagaggtgt ttgagcggaa tgtagaacaa 360
11
CA 02333897 2001-O1-16
WO 00/04176 PCTINL99100453
agtgaccacg gcacaataat_tgacaagttg aaggaaaatt cacctaaagg aaggaagaag 420
acaaaagttc gtcgtaacaa gaggagacgt aaatgctgga agtgagctta aaggtgttgt 480
cgtactacga cgttaactaa ggcgcttgtc gggaggcaac cctagctttg tatgtaaatg 540
taaaagtaaa aaatatatat atagaaaaag gaaaatacaa aaagagtcgt gccgcgacgt 600
taaatcaagc gcttgttgga aggcaaccca atttttattg ttttagttgt tttacttatt 660
tagtattacg tagtttcttg ttgtttttgt agggctcggg actttcggaa ggtgaggtaa 720
tttcaaggca tcgcggtgtg tattgcagcg aggtaagtgt aagagttgag ttggaagcgt 780
ttggccaagt gttgcaccgt gagaggcttt caacctgttg cgacacgtga aaaattaaga 840
gccagatctg ctacattagc actgaagcat cgcttggcca atagcttgga atggaagcaa 900
gaattcaaac caaaatcaga a.acgccacaa gagatgtgtc gcacactgca aagctttgtg 960
caaactagtg aacgcagaaa tagaaatgct acagcccatg cgtcgcttgg cttatggcag 1020
gcagcaaaaa ttcagcagca aaacagaaac gctgcgagaa acgcgtcgca tacgccatag 1080
ctttgtgtca aacagaacgt ccagaaattg aaaagctata agcctgcgtc gcttggctca 1190
tggcgtgcag actagaaaag ctctagcaga tgcgtcgcgt attgtatagc ttggtgtgaa 1200
acagaaagtt cgaaacttgg aaaacgataa cccagcgtcg cctcttcaac cgcgtccagg 1260
taagttcaag attcttacgg g~ttgacccat taacccattg atcggctgat tataaacaat 1320
aaaacatcac cttcaactat cacatgattt cataagtttg acctaggata ttttatatat 1380
atatatatat atatacacac acacaccatt tccagcgatc ttacctcatt tttattcaaa 1440
ccatttttct gcttcaaaag tttaaattat taatatgata agtcatccat agtcaaacaa 1500
gattttctat actattttgt cccttgtaat tttaaaaaaa aaatgagcga tggtaagata 1560
aacattgttt gcaagtgtac aattttagta tatgcaaacc aacgcttctt cttccaacta 1620
tcacctaaaa ctacatcatt tatggcgggc ggactagacg tagccaaata taaaaacgca 16$0
atggccattc agttcatgtc atttttatat ccttcatcca ataatattac tcaaaattga 1790
tgtacagttt ggtctctgat gtgcacttta ctatacgtaa tacggaattt acattataat 1800
taaagagaac tgttccacta aattttaatg atttaattaa tttaactcgg ttacttgtat 1860
tattattatt gctgtatttg tttgtcattt gaatttggca ccgcagattt ttgtatgcaa 1920
ttaaccctca tatatctttt ggccaaataa agaaaaagtc tgcatatttc ttgccaaaca 1980
tttatcatac tttaccgaat tcttgttttt tgtttctctg ttgttgttct ccactataaa 2040
taacatttgc agtgagtaaa gtttcttcag gtctcttttg tagattcaac aagagtattc 2100
agcacttgaa ctcaaaaggg gcttcactaa aaaaaatcat g 2141
12