Note: Descriptions are shown in the official language in which they were submitted.
CA 02339519 2001-02-02
1
DNA SE UENCE CODING FOR A 1-DEOXY-D-XYLULOSE-5-PHOSPHATE
SYNTHASE AND THE OVERPRODUCTION THEREOF IN PLANTS
The present invention relates to the use of DNA sequences coding
for a 1-deoxy-D-xylulose-5-phosphate synthase (DOXS) for
producing plants with increased tocopherol, vitamin K,
chlorophyll and/or carotenoid contents, specifically to the use
of a DNA sequence SEQ ID No. 1 or SEQ ID No. 3 or of a DNA
sequence hybridizing with the latter, to the use of a DNA
sequence SEQ ID No. 1 or SEQ ID No. 3 and of a DNA sequence
SEQ ID No. 5 or DNA sequences hybridizing with the latter and
coding for a 1-deoxy-D-xylulose-5-phosphate synthase (DOXS) and a
p-dydroxyphenylpyruvate dioxygenase (HPPD) for producing plants
with increased content of tocopherols, vitamin K, chlorophylls
and/or carotenoids, to the use of a DNA sequence SEQ ID No. 1 or
SEQ ID No. 3 and of a DNA sequence SEQ ID No. 7 or DNA sequences
hybridizing with the latter and coding for a
1-deoxy-D-xylulose-5-phosphate synthase (DOXS) and a
geranylgeranyl-pyrophosphate oxidoreductase (GGPPOR) for
producing plants with increased content of tocopherols, vitamin
K, chlorophylls and/or carotenoids, to the use of a DNA sequence
SEQ ID No. 1 or SEQ ID No. 3, of a DNA sequence SEQ ID No. 5 and
of a DNA sequence SEQ ID No. 7 or DNA sequences hybridizing with
the latter and coding for a 1-deoxy-D-xylulose-5-phosphate
synthase (DOXS), a hydroxyphenylpyruvate dioxygenase (HPPD) and a
geranylgeranyl-pyrophosphate oxidoreductase (GGPPOR) for
producing plants with increased content of tocopherols, vitamin
K, chlorophylls and/or carotenoids, to processes for producing
plants with increased tocopherol, vitamin K, chlorophyll and/or
carotenoid contents, comprising a DNA sequence SEQ-ID No. 1 or
SEQ ID No. 3; SEQ ID No. 1 or SEQ ID No. 3 and SEQ ID No. 5; SEQ
ID No. 1 or SEQ ID No. 3 and SEQ ID No. 7 or a DNA sequence SEQ
ID No. 1 or SEQ ID No. 3 and SEQ ID No. 5 and SEQ ID No. 7, to
the plants themselves produced in this way, and to the use of SEQ
ID No. 1 or SEQ ID No. 3 for producing a test system for
identifying DOXS inhibitors.
An important aim of molecular genetic work on plants to date has
been the generation of plants with increased content of sugars,
enzymes and amino acids. However, there is also commercial
interest in the development of plants with increased content of
vitamins, such as increasing the tocopherol content.
The eight compounds with vitamin E activity which occur in nature
are derivatives of 6-chromanol (Ullmann~s Encyclopedia of
Industrial Chemistry, Vol. A 27 (1996), VCH Verlagsgesellschaft,
0817/00006
CA 02339519 2001-02-02
2
Chapter 4, 478-488, Vitamin E). The first group (la-d) is derived
from tocopherol, while the second group consists of derivatives
of tocotrienol (2a- d):
R1
HO
~5 I 1 3
R ~ ~ 4. -
R3
la, a-Tocopherol: R1 = RZ = R3 = CH3
lb, (3-Tocopherol [148-03-8]: R1 = R3 = CH3, Rz = H
lc, y-Tocopherol [54-28-4]: R1 = H, R2 = R3 = CH3
ld, S-Tocopherol [119-13-1]: R1 = RZ = H, R3 = CH3
R1
HO
i i i
R ~ ~ 3. _ 7, 11.
R3
2a, a-Tocotrienol [1721-51-3]: R1 = R2 = R3 = CH3
2b, ~--Tocotrienol [ 490-23-3 ] : R1 = R3 = CH3, RZ = H
2c, y-Tocotrienol [14101-61-2]: R1 = H, RZ = R3 = CH3
2d, b-Tocotrienol [25612-59-3]: R1 = RZ = H, R3 = CH3
The compound of great commercial importance is a-tocopherol.
There are limits on the development of crop plants with increased
tocopherol content through tissue culture or seed mutagenesis and
natural selection. Thus, on the one hand, the tocopherol content
must be measurable even in the tissue culture and, on the other
hand, the only plants which can be manipulated by tissue culture
techniques are those which can be regenerated to whole plants
from cell cultures. In addition, crop plants may, after
mutagenesis and selection, show unwanted properties which must be
eliminated again by backcrossings, several times in some
instances. Moreover increasing the tocopherol content by crossing
would be retricted to plants of the same species.
For these reasons, the genetic engineering procedure of
isolating, and transferring into target crop plants, an essential
biosynthesis gene coding for tocopherol synthesis activity is
0817/00006
CA 02339519 2001-02-02
3
superior to the classical breeding method. The preconditions for
this process are that the biosynthesis and its regulation are
known and that genes which influence the biosynthetic activity
are identified.
Isoprenoids or terpenoids consist of various classes of
lipid-soluble molecules and are formed partly or completely of
CS-isoprene units. Pure prenyl lipids (e. g. carotenoids) consist
of C skeletons derived exclusively from isoprene units, whereas
mixed prenyl lipids (e. g. chlorophyll) have an isoprenoid side
chain connected to an aromatic nucleus.
The biosynthesis of prenyl lipids starts from 3 x acetyl-CoA
units, which are converted via ~-hydroxymethylglutaryl-CoA
(HMG-CoA) and mevalonate into the initial isoprene unit (C5),
isopentenyl pyrophosphate (IPP). It has recently been shown by in
vivo feeding experiments with C13 that a mevalonate-independent
pathway is followed in various eubacteria, green algae and plant
chloroplasts to produce IPP:
25
35
45
0817/00006
CA 02339519 2001-02-02
4
Glyceraldehyde 3-phosphate + pyruvate
DOXS
1-deoxy-D-xylulose 5-phosphate
D P,~ IPP
IPP
GPP
' 2 IPP
GGPP --> > ---~ carotenoids
P~P
tocopherols
This entails hydroxyethylthiamine, which is produced by
decarboxylation of pyruvate, and glyceraldehyde 3-phosphate
(3-GAP) being converted, in a "transketolase" reaction mediated
by 1-deoxy-D-xylulose-5-phosphate synthase, initially into
1-deoxy-D-xylulose-5-phosphate (Schwender et al., FEBS Lett.
414(1),129-134(1997); Arigoni et al., Proc.Natl.Acad.Sci USA
94(2), 10600-10605 (1997); Lange et al., Proc.Natl.Acad.Sci.USA
95(5), 2100-2104(1998); Lichtenthaler et al., FEBS Lett. 400(3),
271-274(1997). The latter is then converted by an intramolecular
rearrangement into IPP (Arigoni et al., 1997). Biochemical data
indicate that the mevalonate pathway operates in the cytosol and
leads to the production of phytosterols. The antibiotic
mevinolin, a specific inhibitor of mevalonate production, leads
only to inhibition of sterol biosynthesis in the cytoplasm,
whereas prenyl lipid production in the plastids is unaffected
(Bach and Lichtenthaler, Physiol. Plant 59(1983), 50-60. By
0817/00006
CA 02339519 2001-02-02
contrast, the mevalonate-independent pathway has a plastidic
localization and leads mainly to the production of carotenoids
and plastidic prenyl lipids (Schwender et al., 1997; Arigoni et
al, 1997).
5
IPP is in equilibrium with its isomer, dimethylallyl pyro-
phosphate (DMAPP). Condensation of IPP with DMAPP in a head-tail
addition affords the monoterpene (Clo) geranyl pyrophosphate
(GPP). Addition of further IPP units results in the sesquiterpene
(C15) farnesy pyrophosphate (FPP) and the diterpene (CZO)
geranyl-geranyl pyrophosphate (GGPP). Linkage of two GGPP
molecules results in the production of the C4p precursors for
carotenoids. GGPP is transformed by a prenyl chain hydrogenase
into phytyl pyrophosphate (PPP), the starting material for
further production of tocopherols.
The ring structures of the mixed prenyl lipids which lead to the
production of vitamins E and K comprise quinones whose initial
metabolites are derived from the shikimate pathway. The aromatic
amino acids phenylalanine and tyrosine are converted into
hydroxyphenylpyruvate, which is transformed by dioxygenation into
homogentisic acid. The latter is bound to PPP in order to produce
2-methyl-6-phytylquinol, the precursor of a-tocopherol and
a-tocoquinone. Methylation steps with S-adenosylmethionine as
methyl group donor result initially in 2,3-dimethyl-6-phytyl-
quinol and then, by cyclization, in y-tocopherol and, by
methylation again, in a-tocopherol (Richter, Biochemie der
Pflanzen, Georg Thieme Verlag Stuttgart, 1996).
Examples are to be found in the literature showing that
manipulation of an enzyme can influence the direction of the
metabolyte flux. It was possible in experiments with modified
expression of phytoene synthase, which links two GGPP molecules
together to give 15-cis-phytoene, to measure a direct effect on
the amounts of carotenoids in these transgenic tomato plants
(Fray and Grierson, Plant Mol.Bio1.22(4),589-602(1993); Fray et
al., Plant J., 8, 693-701(1995). As expected, transgenic tobacco
plants with reduced amounts of phenylalanine-ammonium lyase show
reduced phenylpropanoid amounts. The enzyme
phenylalanine-ammonium lyase catalyzes the breakdown of
phenylalanine and thus removes it from phenylpropanoid
biosynthesis (Bate et al.,Proc. Natl. Acad. Sci USA 91 (16):
7608-7612 (1994); Howles et al., Plant Physiol. 112.
1617-1624(1996).
0817/00006
CA 02339519 2001-02-02
6
To date, little has been disclosed about increasing the
metabolite flux in order to increase the tocopherol content of
plants through overexpression of individual biosynthesis genes.
There is merely a description in w0 97/27285 of modification of
the tocopherol content by increased expression or by
down-regulation of the enzyme p-hydroxyphenylpyruvate dioxygenase
(HPPD).
It is an object of the present invention to develop a transgenic
plant with increased content of tocopherols, vitamin K, chloro-
phylls and carotenoids.
We have found that this object is achieved by overexpression of a
1-deoxy-D-xylulose-5-phosphate sythase (DOXS) gene in the plants.
In order to increase the metabolite flux from primary metabolism
into isoprenoid metabolism, the production of IPP as general
starting substrate for all plastidic isoprenoids was increased.
For this purpose, the DOXS activity in plants was increased by
overexpression of the homologous gene (gene from organism of the
same species). This can also be achieved by expressing a
heterologous gene (gene from remote organisms). Nucleotide
sequences from Arabidopsis thaliana DOXS (Acc. No. U 27099), rice
(Acc. No. AF024512) and peppermint (Acc. No. AF019383) are
described.
In one example 1 there is enhanced expression of the DOXS gene
from Arabidopsis thaliana (SEQ ID No.:l; Mandel et al., Plant J.
9, 649-658(1996); Acc. No. U27099) in transgenic plants.
Plastidic localization is ensured by the transit signal sequence
present in the gene sequence. A suitable expression cassette is
also a DNA sequence which codes for a DOXS gene which hybridizes
with SEQ ID No. 1 and which is derived from other organisms such
as, for example, E. coli (SEQ ID No.3) or, preferably, from other
plants.
The GGPP which is now available in increased quantities is
converted further in the direction of tocopherols and
carotenoids.
Efficient production of carotenoids is essential for photo-
synthesis, where they serve together with chlorophylls as
"light-collecting complexes" for better utilization of the energy
of photons (Heldt, Pflanzenbiochemie. Spektrum Akademischer
Verlag Heidelberg Berlin Oxford, 1996). In addition, carotenoids
carry out important functions protecting from oxygen free
radicals such as singlet oxygen, which they are able to return to
0817/00006
CA 02339519 2001-02-02
7
the ground state (Asada, 1994; Demming-Adams and Adams, Trends in
Plant Sciences 1; 21-26(1996). A 1-deoxy-D-xylulose-5-phosphate
synthase-defective Arabidopsis thaliana mutant showing an "albino
phenotype" has been isolated (Mandel et al., 1996). It can be
inferred from this that a reduced amount of carotenoids in the
plastids has adverse effects on the plant.
We have found that the object is also achieved by overexpression
of a 1-deoxy-D-xylulose-5-phosphate synthase (DOXS) gene and of a
p-hydroxyphenylpyruvate dioxygenase (HPPD) gene in the plants,
see Figure 1.
In order to increase the metabolite flux from primary metabolism
into isoprenoid metabolism, the production of IPP as general
starting substrate for all plastidic isoprenoids was increased.
For this purpose, the DOXS activity in transgenic tobacco and
oilseed rape plants was increased by overexpression of the DOXS
from E. coli. This can be achieved by expression of homologous or
other heterologous genes.
The D-1-deoxy-xylulose 5-phosphate which is now available in
increased quantities is converted further in the direction of
tocopherols and carotenoids.
In addition, the production of homogentisic acid further
intensifies the metabolite flux in the direction of
phytylquinones and thus tocopherol, see Figure 1. Homogentisic
acid is produced from p-hydroxyphenylpyruvate by the enzyme
p-hydroxyphenylpyruvate dioxygenase (HPPD). cDNAs coding for this
enzyme have been described from various organisms such as, for
example, from microorganisms, from plants and from humans.
In Example 11 there was for the first time overexpression of the
HPPD gene from Streptomyces avermitilis (Denoya et al.,
J. Bacteriol. 176(1994), 5312-5319; SEQ ID No. 5) together with
the DOXS from E. coli SEQ ID No. 3 in plants and plant plastids.
The increase in the plastidic IPP production leads to enhanced
production of all plastidic isoprenoids. The increased provision
of homogentisic acid ensures that sufficient substrate is
available for the production of tocopherols in the plastids. This
homogentisate which is now available in increased quantities can
in turn be converted in the transgenic plants with the amount,
which is increased due to the overexpression of DOXS, of phytyl
diphosphate (PPP). PPP occupies a key position, in this
connection, because it serves on the one hand as starting
0817/00006
CA 02339519 2001-02-02
8
substrate for chlorophylls and phylloquinones, and on the other
hand for tocopherols.
The transgenic plants are produced by transformation of the
plants with a construct containing the DOXS and HPPD genes.
Tobacco and oilseed rape were employed as model plants for the
production of tocopherols, vitamin K, chlorophylls and
carotenoids.
The invention also relates to the use of the DNA sequences SEQ ID
No. 1 or SEQ ID No. 3 and SEg ID No. 5, which code for a DOXS or
HPPD or functional equivalents thereof, for producing a plant
with increased tocopherol, vitamin K, chlorophyll and/or
carotenoid contents. The nucleic acid sequences may in these
cases be, for example, DNA or cDNA sequences. Coding sequences
suitable for insertion into an expression cassette are, for
example, those coding for a DOXS or HPPD and conferring on the
host the ability to overproduce tocopherol.
The expression cassettes additionally comprise regulatory nucleic
acid sequences which control the expression of the coding
sequence in the host cell. In a preferred embodiment, an
expression cassette comprises upstream, i.e. at the 5' end of the
coding sequence, a promoter and downstream, i.e. at the 3' end, a
polyadenylation signal and, where appropriate, further regulatory
elements which are operatively linked to the coding sequence for
the DOXS or HPPD gene located in between.
An expression cassette is produced by fusing a suitable promoter
to a suitable DOXS or HPPD DNA sequence and preferably a DNA
which is inserted between promoter and DOXS or HPPD DNA sequence
and codes for a chloroplast-specific transit peptide, and a
polyadenylation signal by conventional recombination and cloning
techniques as described, for example, in T. Maniatis, E.F.
Fritsch and J. Sambrook, Molecular Cloning: A Laboratory Manual,
Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1989), and
in T.J. Silhavy, M.L. Berman and L.W. Enquist, Experiments with
Gene Fusions, Cold Spring Harbor Laboratory, Cold Spring Harbor,
NY (1984) and in Ausubel, F.M. et al., Current Protocols in
Molecular Biology, Greene Publishing Assoc. and
Wiley-Interscience (1987).
It is also possible to use expression cassettes whose DNA
sequence codes for a DOXS or HPPD fusion protein, where part of
the fusion protein is a transit peptide which controls
translocation of the polypeptide. Transit peptides which are
specific for chloroplasts and which are eliminated enzymatically
0817/00006
CA 02339519 2001-02-02
9
from the DOXS or HPPD part after translocation of the DOXS or
HPPD gene into the chloroplasts are preferred. The particularly
preferred transit peptide is derived from the plastidic
transketolase (TK) or a functional equivalent of this transit
peptide (for example the transit peptide of the small subunit of
rubisco or ferredoxin-NADP oxidoreductase).
The fused expression cassette coding for a DOXS gene and an HPPD
gene is preferably cloned into a vector, for example pBinl9,
which is suitable for transformation of Agrobacterium
tumefaciens.
The invention further relates to the use of an expression
cassette comprising DNA sequences SEQ ID No. 1 or SEQ-ID No. 3
and SEQ ID No. 5 or DNA sequences hybridizing with the latter for
the transformation of plants or cells, tissues or parts of
plants. The preferred aim of the use is to increase the
tocopherol, vitamin K, chlorophyll and carotenoid contents of the
plant.
It is moreover possible, depending on the choice of the promoter,
for expression to take place specifically in the leaves, in the
seeds or other parts of the plant. The present invention further
relates to such transgenic plants, propagation material thereof
and cells, tissues or parts of these plants.
The invention additionally relates to transgenic plants
transformed with an expression cassette comprising the sequence
SEQ ID No. 1 or SEQ ID No. 3 and SEQ ID No. 5 or DNA sequences
hybridizing with the latter, and transgenic cells, tissues, parts
and propagation material of such plants. Particular preference is
given in this connection to transgenic crop plants such as, for
example, barley, wheat, rye, corn, oats, soybean, rice, cotton,
sugarbeet, canola, sunflower, flax, hemp, potato, tobacco,
tomato, oilseed rape, alfalfa, lettuce and the various tree, nut
and vine species.
The invention further relates to:
- Process for transforming a plant, which comprises introducing
expression cassettes comprising a DNA sequence SEQ ID No. 1
or SEQ ID No. 3 and a DNA sequence SEQ ID No. 5 or DNA
sequences hybridizing with the latter into a plant cell, into
callus tissue, a whole plant or protoplasts of plants,
0817/00006
CA 02339519 2001-02-02
Use of the DNA sequence SEQ ID No. 1 or SEQ ID No. 3 and SEQ
ID No. 5 or DNA sequences hybridizing with the latter to
produce plants with increased tocopherol, vitamin K,
chlorophyll and/or carotenoid contents by expression of a
5 DOXS and an HPPD DNA sequence in plants.
The object have also been achieved by overexpression of a
1-deoxy-D-xylulose-5-phosphate synthase (DOXS) gene and of a
geranylgeranyl-pyrophosphate oxidoreductase (GGPPOR) gene in the
10 plants, see Figure 1.
In order to increase the metabolite flux from primary metabolism
into isoprenoid metabolism, the production of IPP as general
starting substrate for all plastidic isoprenoids was increased.
For this purpose, the DOXS activity in transgenic tobacco and
oilseed rape plants was increased by overexpression of the DOXS
from E. coli. This can be achieved by expression of homologous or
other heterologous genes.
In order to convert the GGPP which is now available in increased
quantities in the direction of tocopherols and carotenoids, in a
further step essential to the invention in addition the activity
of the enzyme geranylgeranyl-pyrophosphate oxidoreductase is
increased by overexpression of a corresponding gene. This measure
achieves an increased production of phytyl pyrophosphate through
increased conversion of geranylgeranyl pyrophosphate into phytyl
pyrophosphate.
This is done, for example, by enhanced expression of the GGPPOR
gene from Arabidopsis thaliana (SEQ ID No. 7) in transgenic
plants. In order to ensure plastidic localization, a transit
signal sequence is put in front of the Arabidopsis GGPPOR. Also
suitable as expression cassette is a DNA sequence coding for a
GGPPOR gene which hybridizes with SEQ ID No. 7 and which is
derived from other organisms or from other plants.
Example 15 describes the cloning of the GGPPOR gene from
Arabidopsis thaliana.
Increasing the plastidic 1-deoxy-D-xylulose 5-phosphate and
phytyl pyrophosphate production leads to increased production of
all plastidic isoprenoids, so that sufficient substrate for the
production of tocopherols, chlorophylls, vitamin K and
phylloquinones is available in the plastids.
0817/00006
CA 02339519 2001-02-02
11
The transgenic plants are produced by transformation of the
plants with a construct containing the DOXS and GGPPOR genes.
Tobacco and oilseed rape were employed as model plants for the
production of tocopherols, vitamin K, chlorophylls and
carotenoids.
The invention also relates to the use of the DNA sequences SEQ ID
No. 1 or SEQ ID No. 3 and SEQ ID No. 7, which code for a DOXS or
GGPPOR or functional equivalents thereof, for producing plants
with increased tocopherol, vitamin K, chlorophyll and/or
carotenoid contents. The nucleic acid sequences may in these
cases be, for example, DNA or cDNA sequences. Coding sequences
suitable for insertion into an expression cassette are, for
example, those coding for a DOXS or GGPPOR and conferring on the
host the ability to overproduce tocopherol.
The expression cassettes additionally comprise regulatory nucleic
acid sequences which control the expression of the coding
sequence in the host cell. In a preferred embodiment, an
expression cassette comprises upstream, i.e. at the 5' end of the
coding sequence, a promoter and downstream, i.e. at the 3' end, a
polyadenylation signal and, where appropriate, further regulatory
elements which are operatively linked to the coding sequence for
the DOxS or GGPPOR gene located in between. Operative linkage
means sequential arrangement of promoter, coding sequence,
terminator and, where appropriate, further regulatory elements in
such a way that each of the regulatory elements can properly
carry out its function in the expression of the coding sequence.
The sequences which are preferred for the operative linkage, but
are not restricted thereto, are targeting sequences to ensure
subcellular localization in the apoplast, in the vacuole, in
plastids, in the mitochondrion, in the endoplasmic reticulum
(ER), in the cell nucleus, in elaioplasts or other compartments
and translation enhancers such as the 5' leader sequence from
tobacco mosaic virus (Gallie et al., Nucl. Acids Res. 15 (1987),
8693-8711).
For example, the plant expression cassette can be incorporated
into the tobacco transformation vector pBinAR-Hyg. Fig. 1 shows
the tobacco transformation vectors pBinAR-Hyg with the 35S
promoter (A) and pBinAR-Hyg with the seed-specific promoter
phaseolin 796 (B):
- HPT: hygromycin phosphotransferase
- OCS: octopine synthase terminator
- PNOS: nopaline synthase promoter
0817/00006
CA 02339519 2001-02-02
12
- also drawn in are those restriction cleavage sites which cut
the vector only once.
Suitable promoters for the expression cassette are in principle
all promoters able to control expression of foreign genes in
plants. Preferably used is, in particular, a plant promoter or a
promoter derived from a plant virus. The CaMV 35S promoter from
cauliflower mosaic virus (Franck et al., Cell 21 (1980), 285-294)
is particularly preferred. This promoter contains, as is known,
different recognition sequences for transcriptional effectors
which, in their totality, lead to permanent and constitutive
expression of the inserted gene (Benfey et al., EMBO J. 8 (1989),
2195-2202).
The expression cassette may also contain a chemically inducible
promoter by which expression of the exogenous DOXS or GGPPOR gene
in the plant can be controlled at a particular time. Promoters of
this type, such as the PRP1 promoter (Ward et al., Plant. Mol.
Biol. 22 (1993), 361-366), a promoter inducible by salicylic acid
(WO 95/19443), a benzenesulfonamide-inducible (EP-A 388186), a
tetracycline-inducible (Gatz et al., (1992) Plant J. 2, 397-404),
an abscisic acid-inducible (EP-A 335528) and an ethanol- or
cyclohexanone-inducible (WO 93/21334) promoter, inter alia, can
be used.
Further particularly preferred promoters are those which ensure
expression in tissues or parts of plants in which the bio-
synthesis of tocopherol or its precursors takes place. Particular
mention should be made of promoters which ensure leaf-specific
expression. Mention should be made of the promoter of cytosolic
FBPase from potato or the ST-LSI promoter from potato (Stockhaus
et al., EMBO J. 8 (1989) 2445 - 245).
An expression cassette is produced by fusing a suitable promoter
to a suitable DOXS or GGPPOR DNA sequence and, preferably, to a
DNA which is inserted between promoter and DOXS or GGPPOR DNA
sequence and which codes for a chloroplast-specific transit
peptide, and to a polyadenylation signal, by conventional
recombination and cloning techniques as described, for example,
in T. Maniatis, E.F. Fritsch and J. Sambrook, Molecular Cloning:
A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring
Harbor, NY (1989) and in T.J. Silhavy, M.L. Berman and L.W.
Enquist, Experiments with Gene Fusions, Cold Spring Harbor
Laboratory, Cold Spring Harbor, NY (1984) and in Ausubel, F.M. et
al., Current Protocols in Molecular Biology, Greene Publishing
Assoc. and wiley-Interscience (1987).
0817/00006
CA 02339519 2001-02-02
13
It is also possible to use expression cassettes whose DNA
sequence codes for a DOXS or GGPPOR fusion protein, where part of
the fusion protein is a transit peptide which controls
translocation of the polypeptide. Transit peptides specific for
chloroplasts are particularly preferred, and these are eliminated
enzymatically from the DOXS or GGPPOR part after translocation of
the DOXS or GGPPOR gene into the chloroplasts. The particularly
preferred transit peptide is derived from the plastidic
transketolase (TK) or a functional equivalent of this transit
peptide (e. g. the transit peptide of the small subunit of rubisco
or ferredoxin-NADP oxidoreductase).
The fused expression cassette coding for a DOXS gene or a GGPPOR
gene is preferably cloned into a vector, for example pBinl9,
which is suitable for transforming Agrobacterium tumefaciens.
The invention further relates to the use of an expression
cassette comprising DNA sequences SEQ ID No. 1 or SEQ-ID No. 3
and SEQ ID No. 7 or DNA sequences hybridizing with the latter for
the transformation of plants or cells, tissues or parts of
plants. The preferred aim of the use is to increase the
tocopherol, vitamin K, chlorophyll and carotenoid contents of the
plant.
It is moreover possible, depending on the choice of the promoter,
for expression to take place specifically in the leaves, in the
seeds or other parts of the plant. The present invention further
relates to such transgenic plants, propagation material thereof
and cells, tissues or parts of these plants.
The invention additionally relates to transgenic plants
transformed with an expression cassette comprising the sequence
SEQ ID No. 1 or SEQ ID No. 3 and SEQ ID No. 7 or DNA sequences
hybridizing with the latter, and transgenic cells, tissues, parts
and propagation material of such plants. Particular preference is
given in this connection to transgenic crop plants such as, for
example, barley, wheat, rye, corn, oats, soybean, rice, cotton,
sugarbeet, canola, sunflower, flax, hemp, potato, tobacco,
tomato, oilseed rape, alfalfa, lettuce and the various tree, nut
and vine species.
The invention further relates to:
- Process for transforming a plant, which comprises introducing
expression cassettes comprising a DNA sequence SEQ ID No. 1
or a DNA sequence SEQ ID No. 3 and a SEQ ID No. 7 or DNA
0817/00006
CA 02339519 2001-02-02
14
sequences hybridizing with the latter into a plant cell, into
callus tissue, a whole plant or protoplasts of plants,
Use of the DNA sequence SEQ ID No. 1 or SEQ ID No. 3 and SEQ
ID No. 7 or DNA sequences hybridizing with the latter to
produce plants with increased tocopherol, vitamin K,
chlorophyll and/or carotenoid contents by expression of a
DOXS and a GGPPOR DNA sequence in plants.
The object have also been achieved by overexpression of a
1-deoxy-D-xylulose-5-phosphate synthase (DOXS) gene, a
p-hydroxyphenylpyruvate dioxygenase (HPPD) gene and a
geranylgeranyl-pyrophosphate oxidoreductase (GGPPOR) gene in the
plants, see Figure 1.
In order to increase the metabolite flux from primary metabolism
into isoprenoid metabolism, the production of IPP as general
starting substrate for all plastidic isoprenoids was increased.
For this purpose, the DOXS activity was increased by
overexpression of the DOXS from E. coli in transgenic tobacco and
oilseed rape plants. This can also be achieved by expressing
homologous or other heterologous DOXS genes - such as, for
example, a DNA sequence SEQ ID No. 1.
The D-1-deoxy-xylulose 5-phosphate which is now available in
increased quantities is converted further in the direction of
geranylgeranyl pyrophosphate.
In order to convert the GGPP which is now available in increased
quantities in the direction of tocopherols and carotenoids, in a
further step essential to the invention in addition the activity
of the enzyme geranylgeranyl pyrophosphate oxidoreductase is
increased by overexpression of a corresponding homologous or
heterologous gene. This measure achieves an increased production
of phytyl pyrophosphate through increased conversion of
geranylgeranyl pyrophosphate into phytyl pyrophosphate.
This done, for example, by enhanced expression of the GGPPOR gene
from Arabidopsis thaliana (SEQ ID No. 7) in transgenic plants. In
order to ensure plastidic localization, a transit signal sequence
is put in front of the Arabidopsis GGPPOR. Also suitable as
expression cassette is a DNA sequence coding for a GGPPOR gene
which hybridizes with SEQ ID No. 7 and which is derived from
other organisms or from other plants.
CA 02339519 2001-02-02
0817/00006
Example 15 describes the cloning of the GGPPOR gene from
Arabidopsis thaliana.
In order to convert the PPP which is now available in increased
5 quantities in the direction of tocopherol and carotenoids, in a
further step essential to the invention in addition the activity
of the enzyme p-hydroxylphenylpyruvate dioxygenase (HPPD) is
increased by overexpression of a corresponding homologous or
heterologous gene. This measure achieves increased production of
10 homogentisic acid by increased conversion of
hydroxyphenylpyruvate into homogentisic acid.
cDNAs coding for this enzyme have been described from various
organisms such as, for example, from microorganisms, from plants
15 and from humans.
Example 10 describes the cloning of the HPPD gene from
Streptomyces avermitilis (Denoya et al., J. Bacteriol. 176(1994),
5312-5319; SEQ ID No. 5). In order to ensure a plastidic
localization, a transit signal sequence is put in front of the
Streptomyces HPPD. Also suitable as expression cassette is a DNA
sequence which codes for an HPPD gene which hybridizes with SEQ
ID No. 5 and is derived from other organisms or from plants.
The increase in the plastidic D-1-deoxy-xylulose 5-phosphate, the
phytyl pyrophosphate and the homogentisic acid production leads
to increased production of all plastidic isoprenoids. The
increased provision of these precursors ensures that sufficient
substrate is available for the production of tocopherols,
chlorophylls, vitamin K and phylloquinones in the plastids.
The transgenic plants according to the invention are produced by
transforming the plants with a construct containing the DOXS, the
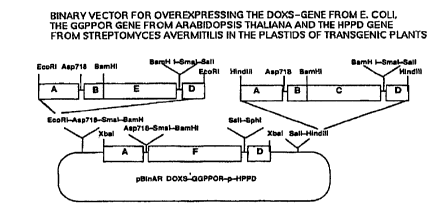
HPPD gene and the GGPPOR gene (Example 17). Tobacco and oilseed
rape were employed as model plants for producing tocopherols,
vitamin K, chlorophylls and carotenoids.
The invention relates to the use of the DNA sequences SEQ ID No.
1 or SEQ ID No. 3, SEQ ID No. 5 and SEQ-ID No. 7, which code for
a DOXS, an HPPD and a GGPPOR or functional equivalents thereof to
produce a plant with increased tocopherol, vitamin K, chlorophyll
and/or carotenoid contents. The nucleic acid sequences may in
these cases be, for example, DNA or cDNA sequences. Coding
sequences suitable for insertion into an expression cassette are,
for example, those coding for a DOXS, an HPPD and a GGPPOR and
conferring on the host the ability to overproduce tocopherol.
0817/00006
CA 02339519 2001-02-02
16
The expression cassettes additionally comprise regulatory nucleic
acid sequences which control the expression of the coding
sequence in the host cell. In a preferred embodiment, an
expression cassette comprises upstream, i.e. at the 5' end of the
coding sequence, a promoter and downstream, i.e. at the 3' end, a
polyadenylation signal and, where appropriate, further regulatory
elements which are operatively linked to the coding sequence for
the DOXS, the HPPD or the GGPPOR gene located in between.
Operative linkage means sequential arrangement of promoter,
coding sequence, terminator and, where appropriate, further
regulatory elements in such a way that each of the regulatory
elements can properly carry out its function in the expression of
the coding sequence. The sequences which are preferred for the
operative linkage, but are not restricted thereto, are targeting
sequences to ensure subcellular localization in the apoplast, in
the vacuole, in plastids, in the mitochondrion, in the
endoplasmic reticulum (ER), in the cell nucleus, in elaioplasts
or other compartments and translation enhancers such as the 5'
leader sequence from tobacco mosaic virus (Gallie et al., Nucl.
Acids Res. 15 (1987), 8693-8711).
For example, the plant expression cassette can be incorporated
into the tobacco transformation vector pBinAR-Hyg. Fig. 2 shows
the tobacco transformation vectors pBinAR-Hyg with the 35S
promoter (A) and pBinAR-Hyg with the seed-specific promoter
phaseolin 796 (B):
- HPT: hygromycin phosphotransferase
- OCS: octopine synthase terminator
- PNOS: nopaline synthase promoter
- also drawn in are those restriction cleavage sites which cut
the vector only once.
Suitable promoters for the expression cassette are in principle
all promoters able to control expression of foreign genes in
plants. Preferably used is, in particular, a plant promoter or a
promoter derived from a plant virus. The CaMV 35S promoter from
cauliflower mosaic virus (Franck et al., Cell 21 (1980), 285-294)
is particularly preferred. This promoter contains, as is known,
different recognition sequences for transcriptional effectors
which, in their totality, lead to permanent and constitutive
expression of the inserted gene (Benfey et al., EMBO J. 8 (1989),
2195-2202).
The expression cassette may also contain a chemically inducible
promoter by which expression of the exogenous DOXS, HPPD and
GGPOR gene in the plant can be controlled at a particular time.
0817/00006
CA 02339519 2001-02-02
17
Promoters of this type, such as the PRP1 promoter (Ward et al.,
Plant. Mol. Biol. 22 (1993), 361-366), a promoter inducible by
salicylic acid (WO 95/19443), a benzenesulfonamide-inducible
(EP-A 388186), a tetracycline-inducible (Gatz et al., (1992)
Plant J. 2, 397-404), an abscisic acid-inducible (EP-A 335528)
and an ethanol- or cyclohexanone-inducible (WO 93/21334)
promoter, inter alia, can be used.
Further particularly preferred promoters are those which ensure
expression in tissues or parts of plants in which the bio-
synthesis of tocopherol or its precursors takes place. Particular
mention should be made of promoters which ensure leaf-specific
expression. Mention should be made of the promoter of cytosolic
FBPase from potato or the ST-LSI promoter from potato (Stockhaus
et al., EMBO J. 8 (1989) 2445 - 245).
An expression cassette is produced by fusing a suitable promoter
to a suitable DOXS, HPPD and GGPPOR DNA sequence and, preferably,
to a DNA which is inserted between promoter and DOXS, HPPD and
GGPOR DNA sequence and which codes for a chloroplast-specific
transit peptide, and to a polyadenylation signal, by conventional
recombination and cloning techniques as described, for example,
in T. Maniatis, E.F. Fritsch and J. Sambrook, Molecular Cloning:
A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring
Harbor, NY (1989) and in T.J. Silhavy, M.L. Berman and L.W.
Enquist, Experiments with Gene Fusions, Cold Spring Harbor
Laboratory, Cold Spring Harbor, NY (1984) and in Ausubel, F.M. et
al., Current Protocols in Molecular Biology, Greene Publishing
Assoc. and Wiley-Interscience (1987).
It is also possible to use expression cassettes whose DNA
sequence codes for a DOXS, HPPD and GGPOR fusion protein, where
part of the fusion protein is a transit peptide which controls
translocation of the polypeptide. Transit peptides specific for
chloroplasts are particularly preferred, and these are eliminated
enzymatically from the DOXS, HPPD and GGPPOR part after
translocation of the DOXS, HPPD and GGPOR gene into the
chloroplasts. The particularly preferred transit peptide is
derived from the plastidic transketolase (TK) or a functional
equivalent of this transit peptide (e.g. the transit peptide of
the small subunit of rubisco or ferredoxin-NADP oxidoreductase).
The fused expression cassette coding for a DOXS gene, an HPPD
gene or a GGPPOR gene is preferably cloned into a vector, for
example p8in19, which is suitable for transforming Agrobacterium
tumefaciens.
0817/00006
CA 02339519 2001-02-02
18
The invention further relates to the use of an expression
cassette comprising DNA sequences SEQ ID No. 1 or SEQ-ID No. 3,
SEQ ID No. 5 and SEQ ID No. 7 or DNA sequences hybridizing with
the latter for the transformation of plants or cells, tissues or
parts of plants. The preferred aim of the use is to increase the
tocopherol, vitamin K, chlorophyll and carotenoid contents of the
plant.
It is moreover possible, depending on the choice of the promoter,
for expression to take place specifically in the leaves, in the
seeds or other parts of the plant. The present invention further
relates to such transgenic plants, propagation material thereof
and cells, tissues or parts of these plants.
The invention additionally relates to transgenic plants
transformed with an expression cassette comprising the sequence
SEQ ID No. 1 or SEQ ID No. 3, SEQ ID No. 5 and SEQ ID No. 7 or
DNA sequences hybridizing with the latter, and transgenic cells,
tissues, parts and propagation material of such plants.
Particular preference is given in this connection to transgenic
crop plants such as, for example, barley, wheat, rye, corn, oats,
soybean, rice, cotton, sugarbeet, canola, sunflower, flax, hemp,
potato, tobacco, tomato, oilseed rape, alfalfa, lettuce and the
various tree, nut and vine species.
The invention further relates to:
- Processes for transforming a plant, which comprises
introducing expression cassettes comprising a DNA sequence
SEQ ID No. 1 or SEQ ID No. 3, a DNA sequence SEQ ID No. 5 and
a DNA sequence SEQ ID No. 7 or DNA sequences hybridizing with
the latter into a plant cell, into callus tissue, a whole
plant or protoplasts of plants,
- Use of the DNA sequence SEQ ID No. 1 or SEQ ID No. 3, SEQ ID
No. 5 and SEQ ID No. 7 or DNA sequences hybridizing with the
latter to produce plants with increased tocopherol, vitamin
K, chlorophyll and/or carotenoid contents by expression of a
DOXS, an HPPD and an GGPPOR DNA sequence in plants.
It was therefore an additional object of the present invention to
develop a test system for identifying DOXS inhibitors.
0817/00006
CA 02339519 2001-02-02
19
This object has been achieved by expressing a DOXS gene from
Arabidopsis or E. coli, or DNA sequences hybridizing therewith,
and subsequently testing chemicals for inhibition of the DOXS
enzyme activity.
The transgenic plants are produced by transforming the plants
with a construct containing the DOXS gene. Arabidopsis and
oilseed rape were employed as model plants for the production of
tocopherols, vitamin K, chlorophylls and carotenoids.
Cloning of the complete DOXS gene from Arabidopsis takes place by
isolating the cDNA (SEQ ID No. 1) specific for the DOXS gene.
The invention relates to the use of the DNA sequence SEQ ID No. 1
or SEQ ID No. 3 which codes for a DOXS or functional equivalent
thereof for producing a plant with increased tocopherol, vitamin
K, chlorophyll and/or carotenoid content. The nucleic acid
sequence can moreover be, for example, a DNA or cDNA sequence.
Examples of coding sequences suitable for insertion into an
expression cassette are those which code for a DOXS and which
confer on. the host the ability to overproduce tocopherol.
The expression cassettes additionally comprise regulatory nucleic
acid sequences which control the expression of the coding
sequence in the host cell. In a preferred embodiment, an
expression cassette comprises a promoter upstream, i.e. at the 5'
end of the coding sequence, and a polyadenylation signal
downstream, i.e. at the 3' end, and, where appropriate, further
regulatory elements which are operatively linked to the coding
sequence for the DOXS gene located in between. Operative linkage
means sequential arrangement of promoter, coding sequence,
terminator and, where appropriate, further regulatory elements in
such a way that each of the regulatory elements can properly
carry out its function in the expression of the coding sequence.
The sequences which are preferred for the operative linkage, but
are not restricted thereto, are targeting sequences to ensure
subcellular localization in the apoplast, in the vacuole, in
plastids, in the mitochondrion, in the endoplasmic reticulum
(ER), in the cell nucleus, in elaioplasts or other compartments
and translation enhancers such as the 5' leader sequence from
tobacco mosaic virus (Gallie et al., Nucl. Acids Res. 15 (1987)
8693 - 8711).
For example, the plant expression cassette can be incorporated
into the tobacco transformation vector pBinAR-Hyg. Fig. [lacuna]
shows the tobacco transformation vectors pBinAR-Hyg with the 35S
0817/00006
CA 02339519 2001-02-02
promoter (A) and pBinAR-Hyg with the seed-specific promoter
phaseolin 796 (B):
- HPT: hygromycin phosphotransferase
5 - OCS: octopine synthase terminator
- PNOS: nopaline synthase promoter
- also drawn in are those restriction cleavage sites which cut
the vector only once.
10 Suitable promoters for the expression cassette are in principle
all promoters able to control expression of foreign genes in
plants. Preferably used is, in particular, a plant promoter or a
promoter derived from a plant virus. The CaMV 35S promoter from
cauliflower mosaic virus (Franck et al., Cell 21 (1980), 285-294)
15 is particularly preferred. This promoter contains, as is known,
different recognition sequences for transcriptional effectors
which, in their totality, lead to permanent and constitutive
expression of the inserted gene (Benfey et al., EMBO J. 8 (1989),
2195-2202).
The expression cassette may also contain a chemically inducible
promoter by which expression of the exogenous DOXS gene in the
plant can be controlled at a particular time. Promoters of this
type, such as the PRP1 promoter (Ward et al., Plant. Mol. Biol.
22 (1993), 361-366), a promoter inducible by salicylic acid
(WO 95/19443), a benzenesulfonamide-inducible (EP A 388186), a
tetracycline-inducible (Gatz et al., (1992) Plant J. 2, 397-404),
an abscisic acid-inducible (EP-A 335528) and an ethanol- or
cyclohexanone-inducible (WO 93/21334) promoter, inter alia, can
be used.
Further particularly preferred promoters are those which ensure
expression in tissues or parts of plants in which the bio-
synthesis of tocopherol or its precursors takes place. Particular
mention should be made of promoters which ensure leaf-specific
expression. Mention should be made of the promoter of cytosolic
FBPase from potato or the ST-LSI promoter from potato (Stockhaus
et al., EMBO J. 8 (1989) 2445 - 245).
It has been possible with the aid of a seed-specific promoter to
express a foreign protein stably up to a content of 0.67 of the
total soluble seed protein in the seeds of transgenic tobacco
plants (Fiedler and Conrad, Bio/Technology 10 (1995), 1090-1094).
The expression cassette can therefore contain, for example, a
seed-specific promoter (preferably the phaseolin promoter
(US 5504200), the USP (Baumlein, H. et al. Mol. Gen. Genet.
(1991) 225 (3), 459 - 467) or LEB4 promoter (Fiedler and Conrad,
0817/00006
CA 02339519 2001-02-02
21
1995)), the LEB4 signal peptide, the gene to be expressed, and an
ER retention signal. The construction of a cassette of this type
is depicted diagrammatically by way of example in Figure 2.
An expression cassette is produced by fusing a suitable promoter
to a suitable DOXS DNA sequence and, preferably, to a DNA which
is inserted between promoter and DOXS DNA sequence and which
codes for a chloroplast-specific transit peptide, and to a
polyadenylation signal, by conventional recombination and cloning
techniques as described, for example, in T. Maniatis,
E.F. Fritsch and J. Sambrook, Molecular Cloning: A Laboratory
Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
(1989) and in T.J. Silhavy, M.L. Berman and L.W. Enquist,
Experiments with Gene Fusions, Cold Spring Harbor Laboratory,
Cold Spring Harbor, NY (1984) and in Ausubel, F.M. et al.,
Current Protocols in Molecular Biology, Greene Publishing Assoc.
and Wiley-Interscience (1987).
Particularly preferred sequences are those which ensure targeting
in the apoplast, in plastids, in the vacuole, in the
mitochondrion, in the endoplasrnic reticulum (ER) or, due to
absence of appropriate operative sequences, ensure retention in
the compartment of production, the cytosol (Kermode, Crit. Rev.
Plant Sci. 15, 4 (1996), 285 - 423). Localization in the ER has
proved particularly beneficial for the amount of protein
accumulated in transgenic plants (Schouten et al., Plant Mol.
Biol. 30 (1996), 781 - 792).
It is also possible to use expression cassettes whose DNA
sequence codes for a DOXS fusion protein, where part of the
fusion protein is a transit peptide which controls translocation
of the polypeptide. Transit peptides specific for chloroplasts
are particularly preferred, and these are eliminated enzymati-
cally from the DOXS part after translocation of the DOXS gene
into the chloroplasts. The particularly preferred transit peptide
is derived from the plastidic transketolase (TK) or a functional
equivalent of this transit peptide (e.g. the transit peptide of
the small subunit of rubisco or ferredoxin-NADP oxidoreductase).
The inserted nucleotide sequence coding for a DOXS can be
prepared by synthesis or be obtained naturally or comprise a
mixture of synthetic and natural DNA constituents, and may
consist of different heterologous DOXS gene sections from
different organisms. In general, synthetic nucleotide sequences
are produced with codons preferred by plants. These codons
preferred by plants can be identified from codons with the
highest protein frequency which are expressed in most plant
0817/00006
CA 02339519 2001-02-02
22
species of interest. For preparing an expression cassette it is
possible to manipulate various DNA fragments in order to obtain a
nucleotide sequence which expediently reads in the correct
direction and is equipped with a correct reading frame. Adapters
or linkers can be attached to the fragments for connecting the
DNA fragments to one another.
It is possible and expedient for the promoter and terminator
regions to be provided in the direction of transcription with a
linker or polylinker which contains one or more restriction sites
for inserting this sequence. As a rule, the linker has 1 to 10,
usually 1 to 8, preferably 2 to 6, restriction sites. The linker
generally has a size of less than 100 bp, frequently less than
60 bp, but at least 5 bp, inside the regulatory regions. The
promoter may be both native or homologous and foreign or
heterologous to the host plant. The expression cassette comprises
in the 5'-3' direction of transcription the promoter, a DNA
sequence which codes for a DOXS gene, and a region for
termination of transcription. Various termination regions are
interchangeable as desired.
It is furthermore possible to employ manipulations which provide
appropriate restriction cleavage sites or delete the redundant
DNA or restriction cleavage sites. It is possible in relation to
insertions, deletions or substitutions, e.g. transitions and
transversions, to use in vitro mutagenesis, primer repair,
restriction or ligation. It is possible with suitable mani-
pulations, e.g. restriction, chewing back or filling in overhangs
for blunt ends, to provide complementary ends of the fragments
for ligation.
It may be important for success according to the invention inter
alia to attach the specific ER retention signal SEKDEL (Schouten,
A. et al. Plant Mol. Biol. 30 (1996), 781 - 792), whereby the
average level of expression is tripled or quadrupled. It is also
possible to employ other retention signals which naturally occur
with plant and animal proteins which are localized within the ER
for constructing the cassette.
Preferred polyadenylation signals are plant polyadenylation
signals, preferably those which essentially correspond to T-DNA
polyadenylation signals from Agrobacterium tumefaciens,
especially of gene 3 of the T-DNA (octopine synthase) of the Ti
plasmid pTiACH5 (Gielen et al., EMBO J. 3 (1984) 835 ff) or
functional equivalents.
0817/00006
CA 02339519 2001-02-02
23
An expression cassette may comprise, for example, a constitutive
promoter (preferably the CaMV 35 S promoter), the LeB4 signal
peptide, the gene to be expressed, and the ER retention signal.
The ER retention signal preferably used is the amino acid
sequence KDEL (lysine, aspartic acid, glutamic acid, leucine).
The fused expression cassette which codes for a DOXS gene is
preferably cloned into a vector, for example pBinl9, which is
suitable for transforming Agrobacterium tumefaciens. Agrobacteria
transformed with such a vector can then be used in a known manner
for transforming plants, in particular crop plants, e.g. tobacco
plants, by, for example, bathing wounded leaves or pieces of leaf
in a solution of agrobacteria and then cultivating in suitable
media. The transformation of plants by agrobacteria is dislosed
inter alia by F.F. White, Vectors for Gene Transfer in Higher
Plants; in Transgenic Plants, Vol. 1, Engineering and
Utilization, edited by S.D. Kung and R. Wu, Academic Press, 1993,
pp. 15-38. Transgenic plants containing a gene, integrated in the
expression cassette, for expression of a DOXS gene can be
regenerated in a known manner from the transformed cells of the
wounded leaves or pieces of leaf.
For transformation of a host plant with a DNA coding for a DOXS,
an expression cassette is incorporated as insertion into a
recombinant vector whose vector DNA comprises additional
functional regulatory signals, for example sequences for
replication or integration. Suitable vectors are described inter
alia in "Methods in Plant Molecular Biology and Biotechnology"
(CRC Press), Chap. 6/7, pp. 71-119 (1993).
It is possible by using the recombination and cloning techniques
cited above to clone the expression cassettes into suitable
vectors which make their replication possible, for example in E.
coli. Suitable cloning vectors are, inter alia, pBR332, pUC
series, Ml3mp series and pACYC184. Binary vectors able to
replicate both in E. coli and in agrobacteria are particularly
suitable.
The invention further relates to the use of an expression
cassette comprising a DNA sequence SEQ No. 1 or SEQ ID No. 3; SEQ
ID No. 1 or SEQ ID No. 3 and SEQ ID No. 5; SEQ ID No. 1 or SEQ-ID
No. 3 and SEQ-ID No. 7 or a DNA sequence SEQ ID No. 1 or SEQ ID
No. 3 and SEQ ID No. 5 and SEQ ID No. 7, or DNA sequences
hybridizing with the latter for transforming plants, or cells,
tissues or parts of plants. The aim of the use is preferably to
0817/00006
CA 02339519 2001-02-02
24
increase the tocopherol, vitamin K, chlorophyll and carotenoid
contents of the plant.
It is moreover possible, depending on the choice of the promoter,
for expression to take place specifically in the leaves, in the
seeds, or other parts of the plant. The present invention further
relates to such transgenic plants, to propagation material
thereof and to cells, tissues or parts of the plants.
The expression cassette can in addition be employed for
transforming bacteria, cyanobacteria, yeasts, filamentous fungi
and algae with the aim of increasing tocopherol, vitamin K,
chlorophyll and/or carotenoid production.
The transfer of foreign genes into the genome of a plant is
referred to as transformation. In this connection, the described
methods for transforming and regenerating plants from plant
tissues or plant cells are utilized for transient or stable
transformation. Suitable methods are protoplast transformation by
polyethylene glycol-induced DNA uptake, the biolistic method
using the gene gun - called the particle bombardment method,
electroporation, incubation of dry embryos in DNA-containing
solution, microinjection and gene transfer mediated by
Agrobacterium. Said processes are described, for example, in
B. Jenes et al., Techniques for Gene Transfer, in: Transgenic
Plants, Vol. 1, Engineering and Utilization, edited by S.D. Kung
and R. Wu, Academic Press (1993) 128-143 and in Potrykus Annu.
Rev. Plant Physiol. Plant Molec. Biol. 42 (1991) 205-225). The
construct to be expressed is preferably cloned into a vector
which is suitable for transforming Agrobacterium tumefaciens, for
example pBinl9 (Bevan et al., Nucl. Acids Res. 12 (1984) 8711).
Agrobacteria transformed with an expression cassette can likewise
be used in a known manner for transforming plants, in particular
crop plants such as cereals, corn, oats, soybean, rice, cotton,
sugarbeet, canola, sunflower, flax, hemp, potato, tobacco,
tomato, oilseed rape, alfalfa, lettuce and the various tree, nut
and vine species, e.g. by bathing wounded leaves or pieces of
leaf in a solution of agrobacteria and subsequently cultivating
in suitable media.
Functionally equivalent sequences which code for a DOXS gene are
sequences which, despite differing in nucleotide sequence, still
have the required functions. Functional equivalents thus comprise
naturally occurring variants of the sequences described herein,
and artificial artificial nucleotide sequences obtained, for
0817/00006
CA 02339519 2001-02-02
example, by chemical synthesis and adapted to the codon usage of
a plant.
A functional equivalent also means in particular natural or
5 artificial mutations of an originally isolated sequence coding
for a DOXS, which still show the required funtion. Mutations
comprise substitutions, additions, deletions, transpositions or
insertions of one or more nucleotide residues. Thus, the present
invention also includes, for example, nucleotide sequences which
10 are obtained by modifying the DOXS nucleotide sequence. The aim
of such a modification may be, for example, to localize further
the coding sequence present therein or else, for example, to
insert further restriction enzyme cleavage sites.
15 Functional equivalents are also variants whose function is
attenuated or enhanced by comparison with the initial gene or
gene fragment.
Artificial DNA sequences are also suitable as long as they
20 confer, as described above, the required property, for example of
increasing the tocopherol content in the plant by overexpression
of the DOXS gene in crop plants. Such artificial DNA sequences
can be identified, for example, by back-translation of proteins
which have been constructed by molecular modelling and have DOXS
25 activity, or by in vitro selection. Particularly suitable coding
DNA sequences are those which have been obtained by back-
translation of a polypeptide sequence in accordance with the
codon usage specific for the host plant. The specific codon usage
can easily be established by a skilled worker familiar with plant
genetic methods through computer analyses of other, known genes
in the plant to be transformed.
Further suitable equivalent nucleic acid sequences which should
be mentioned are sequences which code for fusion proteins, in
which case a plant DOXS polypeptide or a functionally equivalent
part thereof is a constituent of the fusion protein. The second
part of the fusion protein can be, for example, another poly-
peptide with enzymatic activity, or an antigenic polypeptide
sequence with whose aid it is possible to detect DOXS expression
(e.g, myc tag or his tag). However, this is preferably a
regulatory protein sequence, e.g. a signal or transit peptide
which guides the DOXS protein to the required site of action.
However, the invention also relates to the expression products
generated according to the invention, and to fusion proteins
composed of a transit peptide and a polypeptide with DOXS
0817/00006
CA 02339519 2001-02-02
26
activity.
Increasing the tocopherol, vitamin K, chlorophyll and/or
carotenoid content means for the purpose of the present invention
the artificially acquired capability of increased activity in the
biosynthesis of these compounds through functional overexpression
of the DOXS gene in the plant compared with the plant which has
not been genetically modified, for the duration of at least one
plant generation.
The site of tocopherol biosynthesis is generally the leaf tissue
so that leaf-specific expression of the DOXS gene is sensible.
However, it is obvious that tocopherol biosynthesis need not be
confined to the leaf tissue, but may also take place
tissue-specifically in all other parts of the plant - for example
in oilbearing seeds.
Constitutive expression of the exogenous DOXS gene is an
additional advantage. However, on the other hand, inducible
expression may also appear to be desirable.
The effectiveness of expression of the transgenically expressed
DOXS gene can be determined, for example, in vitro by shoot
meristem propagation. In addition, an alteration in the nature
and level of the expression of the DOXS gene and its effect on
tocopherol biosynthesis activity can be tested in glasshouse
experiments on test plants.
The invention additionally relates to transgenic plants
transformed with an expression cassette comprising the sequence
SEQ ID No.l or SEQ ID No. 3; SEQ ID No. 1 or SEQ ID No. 3 and SEQ
No. 5; SEQ ID No. 1 or SEQ ID No. 3 and SEQ ID No. 7 or a DNA
sequence SEQ ID No. 1 or SEQ ID No. 3 and SEQ ID No. 5 and SEQ ID
No. 7, or DNA sequences hybridizing with the latter, and
transgenic cells, tissues, parts and propagation material of such
plants. Particularly preferred in this connection are transgenic
crop plants such as, for example, barley, wheat, rye, corn, oats,
soybean, rice, cotton, sugarbeet, canola, sunflower, flax, hemp,
potato, tobacco, tomato, oilseed rape, alfalfa, lettuce and the
various tree, nut and vine species.
Plants for the purpose of the invention are mono- and
dicotyledonous plants or algae.
In order to be able to find efficient DOXS inhibitors, it is
necessary to provide suitable test systems with which inhibitor/
enzyme binding studies can be carried out. For this purpose, for
0817/00006
CA 02339519 2001-02-02
27
example, the complete cDNA sequence of DOXS from Arabidopsis is
cloned into an expression vector (pQE, Qiagen) and overexpressed
in E. coli.
The DOXS protein expressed using the expression cassette is
particularly suitable for finding inhibitors specific for DOXS.
For this purpose, DOXS can be employed, for example, in an enzyme
assay in which the activity of DOXS is determined in the presence
and absence of the active substance to be tested. Comparison of
the two activity determinations allows qualitative and
quantitative information to be obtained about the inhibitory
behavior of the active substance to be tested. Methods for DOXS
activity determination are described (Putra et. al., Tetrahedron
Letters 39 (1998), 23-26; Sprenger et al., PNAS 94 (1997),
12857-12862).
The test system according to the invention can be used to examine
rapidly and simply a large number of chemical compounds for
inhibitory properties. The method allows reproducible selection,
from a large number of substances, specifically of those with
high activity in order subsequently to carry out with these
substances further, more intensive tests familiar to the skilled
worker.
It is possible in principle by overexpression of the gene
sequence SEQ ID NO: 1 or SEQ ID NO: 3 coding for a DOXS in a
plant to achieve increased resistance to DOXS inhibitors. The
invention likewise relates to transgenic plants produced in this
way.
The invention further relates to:
- A process for transforming a plant, which comprises
introducing an expression cassette comprising a DNA sequence
SEQ ID No. 1 or SEQ ID No. 3 or a DNA sequence hybridizing
with the latter into a plant cell, into callus tissue, a
whole plant or protoplasts of plants.
- The use of a plant for producing plant DOXS.
- The use of the expression cassette comprising a DNA sequence
SEQ ID No. 1 or SEQ ID NO. 3 or a DNA sequence hybridizing
with the latter for producing plants with increased
resistance to DOXS inhibitors by enhanced expression of a DNA
0817/00006
CA 02339519 2001-02-02
28
sequence SEQ ID No. 1 or SEQ ID NO: 3 or a DNA sequence
hybridizing with the latter.
The use of the DNA sequence SEQ ID No. 1 or SEQ ID NO. 3 or
of a DNA sequence hybridizing with the latter for producing
plants with increased tocopherol, vitamin K, chlorophyll
and/or carotenoid content by expression of a DOXS DNA
sequence in plants.
- The use of the expression cassette comprising a DNA sequence
SEQ ID No. 1 or SEQ ID NO: 3 or a DNA sequence hybridizing
with the latter for producing a test system for identifying
DOXS inhibitors.
The invention is illustrated by the examples which now follow,
but is not confined to these:
General cloning methods
The cloning steps carried out for the purpose of the present
invention, such as restriction cleavages, agarose gel electro-
phoresis, purification of DNA fragments, transfer of nucleic
acids to nitrocellulose and nylon membranes, linkage of DNA
fragments, transformation of E. coli cells, cultivation of
bacteria, replication of phages and recombinant DNA sequence
analysis were carried out as described in Sambrook et al. (1989)
Cold Spring Harbor Laboratory Press; ISBN 0-87969-309-6).
The bacterial strains (E. coli, XL-I Blue) used below were
purchased from Stratagene. The agrobacterium strain used for
plant transformation (Agrobacterium tumefaciens, C58C1 with the
plasmid pGV2260 or pGV3850kann) has been described by Deblaere et
al. in (Nucl. Acids Res. 13 (1985) 4777). Alternative
possibilities are also to employ the agrobacterium strain LBA4404
(Clontech) or other suitable strains. Vectors which can be used
for cloning are pUCl9 (Vanish-Perron, Gene 33 (1985), 103-119)
pBluescript SK- (Stratagene), pGEM-T (Promega), pZerO
(Invitrogen), pBinl9 (Bevan et al., Nucl. Acids Res. 12 (1984),
8711-8720) and pBinAR (Hofgen and Willmitzer, Plant Science 66
(1990), 221-230).
Recombinant DNA sequence analysis
Recombinant DNA molecules were sequenced using a Licor laser
fluorescence DNA sequencer (marketed by MWG Biotech, Ebersbach)
CA 02339519 2001-02-02
. 0817/00006
29
using the Sanger method (Sanger et al., Proc. Natl. Acad. Sci.
USA 74 (1977), 5463-5467).
Example 1
Production of the Arabidopsis thaliana DOXS transformation
constructs
The Arabidopsis thaliana DOXS gene was cloned as described in
Mandel et al. (1996) as complete cDNA into the vector pBluescript
KS- (Stratagene).
To produce overexpression constructs, a 2.3 kb fragment
(designated F-23-C) was isolated via the pBluescript KS- Hincll
(blunt-end) and Sacl cleavage sites. This sequence contains the
complete DOXS cDNA from the ATG start codon to the EcoRlI
cleavage site located 80 by downstream of the stop codon. This
fragment was cloned via the Smal (blunt-end) and Sacl cleavage
sites into the pBINl9 vector (Figure 3) (Bevan et al., (1980)
which contains the 35S promoter of cauliflower mosaic virus
(Franck et al., Cell 21(1), 285-294 (1980)) arranged three times
in sequence.
To produce antisense constructs, a region of the 3' end of the
cDNA (called F-23-C antisense) was cloned into the abovementioned
pBINl9-3X35S vector. Part of the 5' region of the DOXS cDNA in
pBluescript KS- was digested via Hincll and the DOXS-internal
BglII cleavage site, and the resulting fragment was removed.
(Figure 4). The BglII cleavage site was filled in by the Klenow
fill-in reaction (Klenow polymerase; Roche; after reaction
according to manufacturer's protocol) so that a blunt end was
produced. The ends which were now compatible (BglII blunt end and
HinclIl were ligated. The 3' region of the DOXS cDNA was then
cloned via KpnI and Xbal (both cleavage sites are located in the
polylinker of pBluescript KS-5' and 3' of the DOXS cDNA) in
antisense orientation into the pBINl9 vector described above in
antisense orientation.
Transformations of Arabidopsis thaliana plants with the
constructs described above using Agrobacterium tumefaciens took
place by the vacuum infiltration method (Bent et al., Science 265
(1994), 1856-1860). Several independent transformands were
isolated for each construct. Each letter (see Table 1) denotes an
independent transformed line. Plants from the T1 generation
obtained therefrom were examined for homo- or heterozygosity.
Several plants from each line were crossed in order to carry out
a segregation analysis. The number in Table 1 corresponds to the
0817/00006
CA 02339519 2001-02-02
individual plant selected for further analyses. Both homo- and
heterozygous lines were obtained. The segregation analysis of the
resulting lines is shown in Table 1 below:
5 Table 1. Segregation analysis of the transgenic DOXS T2 plants
'LINES SEGREGATION
~A9 175%
10 IA19 X100%
IB11 175%
IB4 1100%
IC2 1100%
D3 75%
15 ID17 (100%
E9 175%
E14 1100%
F9 ~ 75%
20 F14 100%
Example 2
Isolation of genomic DNA of the bacterium Escherichia coli XL1
25 Blue
A culture of Escherichia coli XL1 Blue was grown in 300 ml of
Luria broth medium at 37°C for 12 hours. The genomic DNA of the
bacterium was isolated from this culture by first pelleting it at
30 5 000 revolutions in a Sorvall RC50 fuge. The pellet was then
resuspended in 1/30 of the volume of the original culture of
lysis buffer (25 mM EDTA, 0.5% SDS; 50 mM Tris HC1, pH 8.0). An
equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) was
added and incubated at 70 degrees for 10 minutes. The aqueous
phase was then separated from the phenolic in a Heraeus floor
centrifuge at 3 500 rev for 15 minutes. The aqueous supernatant
was mixed with 2.5 volumes of ethanol and 1/10 volume of 8 M
lithium chloride, and the nucleic acids were precipitated at room
temperature for 10 minutes. The pellet was then taken up in
400 ~1 of TE/RNAse and incubated at 37 degrees for 10 minutes.
The solution was again shaken with one volume of phenol/
chloroform/isoamyl alcohol (25:24:1), and the supernatant was
precipitated with 2.5 volumes of ethanol and 1/10 volume of 8 M
lithium chloride. The pellet was then washed with 80% ethanol and
taken up in 400 ~1 of TE/RNAse.
0817/00006
CA 02339519 2001-02-02
31
Example 3
Isolation of the DOXS from E. coli
Oligonucleotides for a PCR were derived from the DOXS DNA
sequence (Acc. Number AF035440), and a BamHI restriction cleavage
site was attached to them at the 5' end, and an Xbal or another
BamHI restriction cleavage site was attached to them at the 3'
end. The oligonucleotide at the 5' end comprises the sequence
5'-ATGGATCCATGAGTTTT-GATATTGCCAAATAC-3' (nucleotides 1-24 of the
DNA sequence; in italics) starting with the ATG start codon of
the gene, and the oligonucleotide at the 3' end comprises the
sequence 5'-ATTCTAGATTATGCCAGCCAGGCCTTG-3' or
5'-ATGGATCCTTATGCCAGCCAGGCCTTG-3' (nucleotides 1$45-1863 of the
reverse complementary DNA sequence; in italics) starting with the
stop codon of the gene. The PCR reaction with the two
BamHI-containing oligonucleotides was carried out with Pfu
polymerase (Stratagene GmbH, Heidelberg) in accordance with the
manufacturer's information. 500 ng of the genomic DNA from E.
coli were employed as template. The PCR program was as follows:
5 cycles: 4 sec 94°C, 30 sec 52°C, 2 min 72°C;
5 cycles: 4 sec 94°C, 30 sec 48°C, 2 min 72°C;
cycles: 4 sec 94°C, 30 sec 44°C, 2 min 72°C
The fragment was purified using a Gene-Clean kit (Dianova GmbH,
Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script (Stratagene GmbH,
Heidelberg). The correctness of the sequence was established by
sequencing. The fragment was BamHI isolated from the PCR-Script
vector and ligated into a correspondingly cut Binl9 vector which
additionally contains the transit peptide of potato transketalase
downstream of the CaMV 35S as promoter. The transit peptide
ensures plastidic localization. The constructs are depicted in
Figure 5 and 6, and the fragments have the following
significance:
Fragment A (529 bp) comprises the 35S promoter of cauliflower
mosaic virus (nucleotides 6909 to 7437 of cauliflower mosaic
virus). Fragment B (259 bp) comprises the transit peptide of
transketolase. Fragment E comprises the DOXS gene. Fragment D
(192 bp) comprises the polyadenylation signal of gene 3 of the
T DNA of the Ti plasmid pTIACHS (Gielen et al., 1984) to
terminate transcription.
0817/00006
CA 02339519 2001-02-02
32
The PCR reaction with the 5'-BamHI and 3'-XbaI-containing
oligonucleotides was carried out with Taq polymerase (Takara,
Sosei Co., Ltd.) in accordance with the manufacturer's
information. 500 ng of the genomic DNA from E. coli were employed
as template. The PCR program was as follows:
5 cycles: 4 sec 94°C, 4 sec 50°C, 2 min 30°C
5 cycles: 4 sec 94°C, 30 sec 46°C, 2 min 68°C
25 cycles: 4 sec 94°C, 30 sec 42°C, 2 min 68°C
The fragment was purified using a Gene-Clean kit and ligated into
the vector pGemT (Promega GmbH, Mannheim). It was cloned as
BamHI/XbaI fragment into a correspondingly cut pBinl9AR vector
downstream of the CaMV 35S promoter. The sequence was checked by
sequencing (SEQ-ID No. 3). This revealed two non-conservative
base exchanges which, compared with the published sequence, lead
to a change in amino acid 152 (asparagine) to valine and amino
acid 330 (cysteine) to tryptophan.
Example 4
Detection of increased amounts of DOXS RNA in transgenic plants
Total RNA from 15-day old seedlings of various transgenic lines
possessing the DOXS overexpression construct was extracted by the
method of Logeman et al., Anal. Biochem. 163, 16-20 (1987),
fractionated in a 1.2~ agarose gel, transferred to filters and
hybridized with a 2.1 kb long DOXS fragment as probe (Figure 7).
Example 5
Detection of increased amounts of DOXS protein in transgenic
plants
Total protein (Figure 8) from 15-day old seedlings of various
independent transgenic plants possessing the DOXS overexpression
construct was isolated and detected in a Western analysis using a
polyclonal anti-DOXS antibody (IgG) (Figure 9).
Example 6
Measurement of the carotenoid and chlorophyll contents
The total amounts of carotenoids and chlorophylls were determined
as described by Lichtenthaler and Wellburn (1983) using 1000
acetone extracts. The results of the multiple measurements of the
CA 02339519 2001-02-02
0817/00006
33
transgenic lines possessing the DOXS overexpression construct are
shown in Table 2 below.
Table 2: Total carotenoid and chlorophyll contents of transgenic
DOXS lines
LINE ~ TOTAL CHLOROPHYLLS~ TOTAL CAROTENOIDS
~
clal mutant 5 5
10Wild type 100 100
B-4 86 89
B-11 84 90
C-2 98 107
D-3 128 135
D-17 136 149
E-14 121 139
F-7 80 90
F-14 85 107
Example 7
Transformation of oilseed rape
The production of transgenic oilseed rape plants is based on a
protocol of Bade, JB and Damm, B (in Gene, Transfer to Plants,
Potrykus, I. and Spangenberg, G., eds, Springer Lab Manual,
Springer Verlag, 1995, 30-38), in which the composition of the
media used are also stated. The transformations took place with
the agrobacterium strain LBA4404 (Clontech). The binary vectors
used were the pBINl9 constructs with the complete DOXS cDNA
already described above. The NOS terminator sequence in these
pBIN vectors was replaced by the OCR terminator sequence.
Brassica napus seeds were surface-sterilized with 70~ (v/v)
ethanol, washed in H20 at 55~C for 10 min, incubated in 1~
strength hypochlorite solution (25~ v/v Teepol, 0.1~ v/v Twenn
20) for 20 min and washed six times with sterile H20 for 20 min
each time. The seeds were dried on filter paper for three days
and 10-15 seeds were induced to germinate in a glass flask with
15 ml of germination medium. The roots and apices were removed
from several seedlings (about 10 cm in size), and the remaining
hypocotyls were cut into pieces about 6 mm long. The approx. 600
explants obtained in this way are washed with 50 ml of basal
medium for 30 min and transferred into a 300 ml flask. After
addition of 100 ml of callus induction medium, the cultures were
incubated at 100 rpm for 24 h.
0$17/00006
CA 02339519 2001-02-02
34
An overnight culture of the agrobacterium strain was set up in LB
with kanamycin (20 mg/1) at 29~C, and 2 ml of this were incubated
in 50 ml of LB without kanamycin at 29~C for 4 h until the OD6oo
was 0.4-0.5. After pelleting of the culture at 2 000 rpm for
25 min, the cell pellet was resuspended in 25 ml of basal medium.
The concentration of the bacteria in the solution was adjusted to
an OD600 of 0.3 by adding further basal medium.
The callus induction medium was removed from the oilseed rape
explants using sterile pipettes, 50 ml of agrobacterium solution
were added and, after cautious mixing, incubated for 20 min. The
agrobacteria suspension was removed, the oilseed rape explants
were washed with 50 ml of callus induction medium for 1 min and
then 100 ml of callus induction medium were added. The
cocultivation was carried out on a rotary shaker at 100 rpm for
24 h. The cocultivation was stopped by removing the callus
induction medium, and the explants were washed twice for 1 min
each time with 25 ml and twice for 60 min with 100 ml each time
of washing medium at 100 rpm. The washing medium with the
explants was transferred into 15 cm Petri dishes, and the medium
was removed using sterile pipettes. For regeneration, in each
case 20-30 explants were transferred into 90 mm Petri dishes
which contained 25 ml of shoot-induction medium with kanamycin.
The Petri dishes were sealed with 2 layers of Leukopor and
incubated at 25~C and with 2000 lux in 16/8 H photoperiods. The
calli which developed was transferred every 12 days to fresh
Petri dishes with shoot-induction medium. All further steps for
regenerating whole plants',-were-'carried out as described by
Bade, J.B. and Damm, B. (in Gene Transfer to Plants, Potrykus, I.
and Spangenberg, G.,eds, Springer Lab Manual, Springer Verlag,
1995, 30-38).
Example 8
Increasing tocopherol biosynthesis in oilseed rape
The DOXS cDNA (SEQ-ID No. 1) was provided with a CaMV 35S
promoter and over-expressed in oilseed rape using the 35S
promoter. In parallel with this, the seed-specific promoter of
the phaseolin gene was used in order specifically to increase the
tocopherol content in the rapeseed. Oilseed rape plants
transformed with the corresponding constructs were grown in a
glasshouse. The a-tocopherol content of the whole plant and of
the seeds of the plant was then determined. In all cases, the
a-tocopherol concentration was increased by comparison with the
untransformed plant.
0817/00006
Example 9
CA 02339519 2001-02-02
Detection of the expression of DOXS from E. coli in transgenic
tobacco plants
5
Leaf disks with a diameter of 0.9 cm were taken from completely
unfolded leaves of plants containing the construct pBinAR
HPPD-DOXS, and were frozen in liquid nitrogen. The leaf material
was homogenized in an HEPES-KOH buffer containing proteinase
10 inhibitors, and the protein concentration was determined from the
extract using the Bio-Rad protein assay in accordance with the
manufacturer's information. 45 ~g of protein from each extract
were mixed with one volume of loading buffer (Laemmli, 1970) and
incubated at 95°C for 5 min. The proteins were then fractionated
15 on a 12.5 percent SDS-PAGE gel. The proteins were then
transferred by means of semi-dry electroblots to Porablot
membrane (Machery and Nagel). Detection of the DOXS protein took
place using a rabbit antibody against E. coli DOXS. The color
reaction is based on the binding of a secondary antibody and of
20 an alkaline phosphatase which converts NBT/BCIP into a dye.
Secondary antibody and alkaline phosphatase were obtained from
Pierce, and the procedure was in accordance with the
manufacturer's information.
25 Figure 10 shows the detection of the DOXS protein in leaves of
transgenic plants. 1: marker; 2: plant 10; 3:62; 4: 63; 5: 69;
7:71; 8:112; 9:113; 10:116; 11:WT1; 12:WT2; 13:100 ng of
recombinant protein; 14:50 ng of recombinant protein; 15: 10 ng
of recombinant protein.
Example 10
Cloning of an HPPD gene from Streptomyces avermitilis U11864
Isolation of genomic DNA of the bacterium Streptomyces
avermitilis U11864:
A culture of Streptomyces avermitilis U11864 was grown in 300 ml
of YEME medium (5 g of malt extract, 2 g of yeast extract, 2 g of
glucose) at 28~C for 96 h. The genomic DNA of the bacterium was
isolated from this culture by pelleting it initially at 5000 rev
in a Sorvall RCSC fuge. The pellet was then resuspended in 1/30
of the volume of lysis buffer (25 mM EDTA, 0.5~ SDS, 50 mM
Tris-HC1, pH 8.0). An equal volume of phenol/chloroform/isoamyl
alcohol (25:24:1) was added and incubated at 70oC for 10 minutes.
The aqueous phase was then separated from the phenolic in a
Heraeus floor centrifuge at 3 500 rev for 15 minutes. The aqueous
0817/00006
CA 02339519 2001-02-02
36
supernatant was mixed with 2.5 volumes of ethanol in 1/10 volume
of 8 M lithium chloride, and the nucleic acids were precipitated
at room temperature for 10 minutes. The pellet was then taken up
in 400 ~1 of TE/RNAse and incubated at 37 degrees for 10 minutes.
The solution was again shaken with one volume of
phenol/chloroform/isoamyl alcohol (25:24:1), and the supernatant
was precipitated with 2.5 volumes of ethanol and 1/10 volume of
8 M lithium chloride. The pellet was then washed with 80~ ethanol
and taken up in 400 ~1 of TE/RNAse.
Oligonucleotides were derived for a PCR from the DNA sequence of
the HPPD from Streptomyces avermitilis (Denoya et al., 1944; Acc.
Number U11864), and a BamHI restriction cleavage site was
attached to the 5' end of them and an XbaI restriction cleavage
site was attached at the 3' end of them. The oligonucleotide at
the 5' end comprises the sequence 5'-GGATCCAGCGGACAAGCCAAC-3'
(37 tv 55 bases distant from the ATG in the 5' direction; in
italics), and the oligonucleotide at the 3' end comprises the
sequence 5'-TCTAGATTATGCCAGCCAGGCCTTG-3' (nucleotides 1845-1863
of the reverse complementary DNA sequence; in italics).
The PCR reaction was carried out with Pfu polymerase (Stratagene
GmbH, Heidelberg) in accordance with the manufacturer's
information. 400 ng of the genomic DNA was employed as pattern.
The PCR program was as follows:
5 cycles: 4 sec 94°C, 30 sec 54°C, 2 min 72°C
5 cycles: 4 sec 94°C, 30 sec 52°C, 2 min 72°C
25 cycles: 4 sec 94°C, 30 sec 50°C, 2 min 72°C
The fragment was purified by means of a Gene-Clean kit (Dianova
GmbH, Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script (Stratagene GmbH,
Heidelberg). The correctness of the sequence was checked by
sequencing. This revealed that the isolated gene codes for an
additional amino acid. It contains the three bases TAC (coding
for tyrosine) in front of nucleotide N429 in the quoted sequence
(Denoya et al., 1994).
The fragment was isolated by a BamHI and XbaI digestion from the
vector and ligated into a correspondingly cut Binl9AR vector
downstream of the CaMV 35S promoter for expression of the gene in
the cytosol. The gene was isolated as BamHI fragment from the
same PCR-Script vector and was ligated into a correspondingly cut
pBinl9 vector which additionally comprises the transit peptide of
the potato plastidic transketolase downstream of the CaMV 35S
promoter. The transit peptide ensures the plastidic localization.
0817/00006
CA 02339519 2001-02-02
37
The constructs are depicted in Figure 11 and 12, and the
fragments have the following significance:
Fragment A (529 bp) comprises the 35S promoter of cauliflower
mosaic virus (nucleotides 6909 to 7437 of cauliflower mosaic
virus). Fragment B (259 bp) comprises the transit peptide of
transketolase. Fragment C comprises the HPPD gene. Fragment D
(192 bp) comprises the polyadenylation signal of gene 3 of the
T DNA of the Ti plasmid pTIACHS (Gielen, J. et al., EMBO J. 3
(1984), 835-846) to terminate transcription.
Example 11
Production of constructs for transformation of plants with DOXS
and HPPD DNA sequences
To produce plants which are transgenic for DOXS and HPPD, a
binary vector which contains both gene sequences was manufactured
(Figure 13). The DOXS and HPPD gene sequences were each cloned as
BamHI fragments as described in Example 3 and 10. The vector
pBinAR-Hyg contains the 35S promoter of cauliflower mosaic virus
and the polyadenylation signal of gene 3 of the T DNA of the Ti
plasmid pTIACHS (Gielen et al., 1984) for termination of
transcription. The pBinAR-Hyg vector confers on plants resistance
to the antibiotic hygromycin and is therefore suitable for
superinfection of plants with kanamycin resistance.
To clone the HPPD in vectors which additionally contain another
cDNA, oligonucleotides were derived for a PCR and had a BamHI
restriction cleavage site attached at the 5' end and at the 3'
end. The oligonucleotide at the 5' end comprises the sequence
5'-GGATCCTCCAGCGGACAAGCCAAC-3' (nucleotides 37 to 55 distant from
the ATG in the 5' direction; in italics), and the oligonucleotide
at the 3' end comprises the sequence
5'-ATGGATCCCGCGCCGCCTACAGGTTG-3' (terminating at base pair 1140
of the coding sequence, starting 8 base pairs 3' from the TAG
stop codon; in italics). The PCR reaction was carried out with
Tli polymerase (Promega GmbH, Mannheim) in accordance with the
manufacturer's information. 10 ng of the plasmid pBinAR-HPPD were
employed as template. The PCR program was as follows:
5 cycles: 94°C 4 sec, 68°C 30 sec, 72°C 2 min
5 cycles: 94°C 4 sec, 64°C 30 sec, 72°C 2 min
25 cycles: 94°C 4 sec, 60°C 30 sec, 72°C 2 min
0817/00006
CA 02339519 2001-02-02
38
The fragment was purified using a Gene-Clean kit (Dianova GmbH,
Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script (Stratagene GmbH,
Heidelberg). The correctness of the sequence was checked by
sequencing. It was cut as BamHI fragment out of the vector
PCR-Script and ligated into a correspondingly cut pBinAR vector
which additionally contains the transit peptide of transketolase
for introducing the gene product into the plastids. The result
was the plasmid pBinAR-TP-HPPD (Figure 12).
For the cloning, the 35S promoter, the transketolase transit
peptide, the HPPD gene and the polyadenylation signal of gene 3
of the T DNA of the Ti plasmid pTIACHS (Gielen et al. 1984) for
termination of transcription was isolated from the plasmid
pBinAR-TP-HPPD by PCR. A HindIII cleavage site was attached in
each case to the oligonucleotides for the promoter and the
terminator. The sequence of the oligonucleotide which anneals
onto the 5' region of the promoter (in italics) is
5'-ATAAGCTTCATGGAGTCAAA-GATTCAAATAGA-3', and that of the
oligonucleotide which anneals onto the termination sequence (in
italics) is 5'-ATAAGCTTGGACAATCAGTAAATTGAACGGAG-3'. The resulting
fragment was purified using a Gene-Clean kit (Dianova GmbH,
Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script from Stratagene GmbH,
Heidelberg. The correctness of the sequence was checked by
sequencing. It was transferred as HindIII fragment from this
PCR-Script vector into the correspondingly cut vector pBinl9
(Bevan, 1984, Nucleic Acids Res. 12, 8711-8721).
The 35S promoter, the transketolase transit peptide, the DOXS
gene and the polyadenylation signal of gene 3 of the T DNA of the
Ti plasmid pTIACH5 (Gielen et al., 1984) for termination of
transcription was isolated by PCR from the plasmid
pBinAR-TP-DOXS. An EcoRI cleavage site was attached to each of
the oligonucleotides for the promoter and terminator sequence.
The sequence of the oligonucleotide which anneals onto the
promoter (in italics) is 5'-ATGAATTCCATGGAGTCAAAGATTCAAATAGA-3',
and that of the oligonucleotide which anneals onto the terminator
sequence (in italics) is 5'-ATGAATTCGGACAATCAGTAAATTGAA-CGGAG-3'.
The fragment was purified using a Gene-Clean kit (Dianova GmbH,
Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script (Stratagene GmbH,
Heidelberg). The correctness of the sequence was checked by
sequencing (SEQ ID No. 3). It was transferred as EcoRI fragment
from the PCR-Script vector into the correspondingly cut vector
pBinl9 (Bevan, 1984).
0817/00006
CA 02339519 2001-02-02
39
It was transferred as XbaI fragment from the PCR-Script vector
into the correspondingly cut vector which, as described above,
already contains the HPPD sequence. The result was the construct
pBinAR-HPPD-DOXS (Figure 13), whose fragments have the following
significance:
Fragment A (529 bp) comprises the 35S promoter of the cauliflower
mosaic virus (nucleotides 6909 to 7437). Fragment B comprises the
transit peptide of plastidic transketolase. Fragment C comprises
the HPPD gene. Fragment D comprises the polyadenylation signal of
gene 3 of the T DNA of the Ti plasmid pTIACH5 (Gielen et al.,
1984) for termination of transcription. Fragment E comprises the
DOXS gene.
Example 12
Production of transgenic tobacco plants
(Nicotiana tabacum L. cv. Samsun NN)
Transgenic tobacco plants having an altered prenyl lipid content
were produced by transforming tobacco leaf disks with DOXS and
HPPD sequences. To transform tobacco plants, 10 ml of an
Agrobacterium tumefaciens overnight culture which had grown under
selection were centrifuged, the supernatant was discarded and the
bacteria were resuspended in the same volume of antibiotic-free
medium. Leaf disks from sterile plants (diameter about 1 cm) were
bathed in this bacterial suspension in a sterile Petri dish. The
leaf disks were then placed on MS medium (Murashige and Skoog,
Physiol. Plant (1962) 15, 473) with 2% sucrose and 0.8% Bacto
agar in Petri dishes. After incubation in the dark at 25~C for
2 days, they were transferred to MS medium with 100 mg/1
kanamycin, 500 mg/1 Claforan, 1 mg/1 benzylaminopurine (BAP),
0.2 mg/1 naphthylacetic acid (NAA), 1.6% glucose and 0.8% Bacto
agar, and the cultivation was continued (16 hours of light/
8 hours of dark). Growing shoots were transferred to hormone-free
MS medium with 2% sucrose, 250 mg/1 Claforan and 0.8% Bacto agar.
Example 13
Production of transgenic oilseed rape plants (Brassica napus)
The production of transgenic oilseed rape plants having an
altered prenyl lipid content was based on a protocol by
Bade, J.B. and Damm, B. (in Gene Transfer to Plants, Potrykus, I.
and Spangenberg, G., eds, Springer Lab Manual, Springer Verlag,
0817/00006
CA 02339519 2001-02-02
1995, 30-38), in which the compositions of the media and buffers
used are also indicated.
The transformations took place with the Agrobacterium tumefaciens
5 strain LBA4404 (Clontech GmbH, Heidelberg). The binary vectors
used were the binary constructs already described above with the
total cDNAs of DOXS and HPPD. In all the binary vectors used
here, the NOS terminator sequence was replaced by the
polyadenylation signal of gene 3 of the T DNA of the Ti plasmid
10 pTIACH5 (Gielen et al., 1984) for termination of transcription.
Brassica napus seeds were surface-sterilized with 70~ (v/v)
ethanol, washed in H20 at 55~C for 10 min, incubated in 1~
strength hypochlorite solution (25~ v/v Teepol, 0.1~ v/v
Tween 20) for 20 min and washed six times with sterile H20 for
15 20 min each time. The seeds were dried on filter paper for
three days and 10-15 seeds were induced to germinate in a glass
flask with 15 ml of germination medium. The roots and apices were
removed from several seedlings (about 10 cm in size), and the
remaining hypocotyls were cut into pieces about 6 mm long. The
20 approx. 600 explants obtained in this way are washed with 50 ml
of basal medium for 30 min and transferred into a 300 ml flask.
After addition of 100 ml of callus induction medium, the cultures
were incubated at 100 rpm for 24 h.
25 An overnight culture of the agrobacterium strain was set up in
Luria Broth medium with kanamycin (20 mg/1) at 29~C, and 2 ml of
this were incubated in 50 ml of Luria Broth medium without
kanamycin at 29~C for 4 h until the OD6oo was 0.4-0.5. After
pelleting of the culture at 2 000 rpm for 25 min, the cell pellet
30 was resuspended in 25 ml of basal medium. The concentration of
the bacteria in the solution was adjusted to an OD6oo of 0.3 by
adding further basal medium.
The callus induction medium was removed from the oilseed rape
35 explants using sterile pipettes, 50 ml of agrobacterium solution
were added and, after cautious mixing, incubated for 20 min. The
agrobacteria suspension was removed, the oilseed rape explants
were washed with 50 ml of callus induction medium for 1 min and
then 100 ml of callus induction medium were added. The
40 cocultivation was carried out on a rotary shaker at 100 rpm for
24 h. The cocultivation was stopped by removing the callus
induction medium, and the explants were washed twice for 1 min
each time with 25 ml and twice for 60 min with 100 ml each time
of washing medium at 100 rpm. The washing medium with the
explants was transferred into 15 cm Petri dishes, and the medium
was removed using sterile pipettes.
0817/00006
CA 02339519 2001-02-02
41
For regeneration, in each case 20-30 explants were transferred
into 90 mm Petri dishes which contained 25 ml of shoot-induction
medium with kanamycin. The Petri dishes were sealed with 2 layers
of Leukopor and incubated at 25~C and with 2 000 lux in
photoperiods of 16 hours of light/8 hours of dark. The calli
which developed were transferred every 12 days to fresh Petri
dishes with shoot-induction medium. All further steps for
regenerating whole plants carried out as described by Bade,
J.B. and Damm, B. (in Gene Transfer to Plants, Potrykus, I. and
Spangenberg, G.,eds, Springer Lab Manual, Springer Verlag, 1995,
30-38).
Example 14
Increasing tocopherol biosynthesis in oilseed rape
The cDNA of DOXS (SEQ-ID No. 3) and of HPPD (SEQ-ID No. 5) was
provided with a CaMV35S promoter and overexpressed in oilseed
rape using the 35S promoter. In parallel with this, the
seed-specific promoter of the phaseolin gene was used in order
specifically to increase the tocopherol content in the rape seed.
Oilseed rape plants transformed with the corresponding constructs
were grown in a glasshouse. The a-tocopherol content of the whole
plant and of the seeds of the plant was then determined. In all
cases, the a-tocopherol concentration was increased by comparison
with the untransformed plant.
Example 15
Cloning of a GGPPOR gene from Arabidopsis thaliana
Isolation of total RNA from completely unfolded leaves of
Arabidopsis thaliana:
Completely unfolded leaves of Arabidopsis thaliana were harvested
and frozen in liquid nitrogen. The material was then powdered in
a mortar and taken up in Z6 buffer (8 M guanidium hydrochloride,
20 mM MES, 20 mM EDTA pH 7.0). The suspension was transferred
into reaction vessels and shaken with one volume of phenol/
chloroform/isoamyl alcohol 25:24:1). After centrifugation at
15 000 rpm for 10 minutes, the supernatant was transferred into a
new reaction vessel, and the RNA was precipitated with
1/20 volumes of 1N acetic acid and 0.7 volume of ethanol
(absolute). After renewed centrifugation, the pellet was first
washed with 3M sodium acetate solution and, after a further
centrifugation, in 70~ ethanol. The pellet was then dissolved in
0817/00006
CA 02339519 2001-02-02
42
DEPC water, and the RNA concentration was determined by
photometry.
Production of cDNA from total RNA from completely unfolded leaves
of A. thaliana:
20 ~g of total RNA were initially mixed with 3.3 ~1 of 3M sodium
acetate solution and 2 ~1 of 1M magnesium sulfate solution and
made up to a final volume of 100 ~1 with DEPC water. 1 ~1 of
RNase-free DNase (Boehringer Mannheim) was added to this and
incubated at 37°C for 45 min. After removal of the enzyme by
shaking with phenol/chloroform/isoamyl alcohol, the RNA was
precipitated with ethanol, and the pellet was taken up in 100 ~1
of DEPC water. 2.5 ~g of RNA from this solution were transcribed
into cDNA by means of a cDNA kit (Gibco, Zife Technologies).
Oligonucleotides were derived for a PCR from the geranylgeranyl-
pyrophosphate oxidoreductase DNA sequence (Keller et al., Eur. J.
Biochem. (1998)251(1-2), 413-417); Accession Number Y14044), and
a BamHI restriction cleavage site was attached at the 5' end of
these and a Sall restriction cleavage site was attached at the 3'
end. The oligonucleotide at the 5' end comprises the sequence
5'-ATGGATCCATGGCGACGACGGTTACACTC-3' starting with the first codon
of the cDNA (in italics), and the oligonucleotide at the 3' end
comprises the sequence 5'-ATGTCGACGTGATGATAGATTACTAACAGAC-3'
starting with base pair 1494 of the cDNA sequence (in italics).
The PCR reaction was carried out with Pfu polymerase from
Stratagene GmbH, Heidelberg in accordance with the manufacturer's
information. 1/8 of a volume of the cDNA was employed as template
(equivalent to 0.3 ~g of RNA). The PCR program was as follows:
5 cycles: 94°C for 4 sec, 48°C for 30 sec, 72°C for 2 min
5 cycles: 94°C for 4 sec, 46°C for 30 sec, 72°C for 2 min
25 cycles: 94°C for 4 sec, 44°C for 30 sec, 72°C for 2
min
The fragment was purified using a Gene-Clean kit (Dianova GmbH,
Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script from Stratagene GmbH,
Heidelberg. The correctness of the fragment was checked by
sequencing (SEQ ID No. 7). The gene was cloned by means of the
restriction cleavage sites attached to the sequence by the
primers as BamHI/SalI fragment into the correspondingly cut
vector BinAR-Hyg. The latter contains the 35S promoter of
cauliflower mosaic virus and the polyadenylation signal of gene 3
of the T DNA of the Ti plasmid pTIACH5 (Gielen et al., EMBO J. 3
(1984), 835-846) for termination of transcription. The plasmid
0817/00006
CA 02339519 2001-02-02
43
confers on plants resistance to the antibiotic hygromycin and is
thus suitable for superinfection of plants with kanamycin
resistance. Since the plastid transit peptide of GGPPOR was also
cloned, the protein ought to be transported into the plastids in
transgenic plants. The construct is depicted in Figure 14. The
fragments have the following significance:
Fragment A (529 bp) comprises the 35S promoter of cauliflower
mosaic virus (nucleotides 6909 to 7437 of cauliflower mosaic
virus). Fragment D comprises the polyadenylation signal of gene 3
of the T DNA of Ti plasmid pTIACHS (Gielen et al., 1984) for
termination of transcription. Fragment F comprises the gene of
GGPPOR including the intrinsic plastid transit sequence.
Example 16
Production of constructs for transformation of plants with DOXS
and GGPPOR sequences
To produce plants which are transgenic for DOXS and GGPPOR, a
binary vector comprising both gene sequences was manufactured
(Figure 15). The GGPPOR gene with the intrinsic plastidic
localization sequence was cloned (as described in Example 15) as
BamHI/Sall fragment into the correspondingly cut vector
pBinAR-Hyg. The DOXS gene was cloned as BamHI fragment as
described in Example 3. The vector pBinAR-Hyg contains the 35S
promoter of cauliflower mosaic virus and the polyadenylation
signal of gene 3 of the T DNA of the Ti plasmid pTIACHS (Gielen
et al., 1984) for termination of transcription. This plasmid
confers on plants resistance to the antibody hygromycin and is
thus suitable for superinfection of plants with kanamycin
resistance.
The 35S promoter, the transketolase transit peptide, the DOXS
gene and the polyadenylation signal of gene 3 of the T DNA of the
Ti plasmid pTIACH5 (Gielen et al., 1984) for termination of
transcription was isolated from the plasmid pBinAR-TP-DOXS by
PCR. An EcoRI cleavage site was attached in each case to the
oligonucleotides for the promoter and the terminator sequence.
The sequence of the oligonucleotide which anneals onto the
promoter (in italics) is 5'-ATGAATTCCATGGAGTCAAAGATTCAAATAGA-3',
and that of the oligonucleotide which anneals onto the terminator
sequence (in italics) is 5'-ATGAATTCGGACAATCAGTAAATTGAACGGAG-3'.
The fragment was purified using a Gene-Clean kit (Dianova GmbH,
Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script from Stratagene GmbH,
Heidelberg. The correctness of the sequence was checked by
0817/00006
CA 02339519 2001-02-02
44
sequencing. It was transferred from the PCR-Script vector as
EcoRI fragment into the correspondingly cut vector pBinl9 (Bevan,
Nucleic Acids Res. 12 (1984), 8711-8721).
The 35S promoter, the GGPPOR gene and the polyadenylation signal
of gene 3 of the T DNA of the Ti plasmid pTIACHS (Gielen et al.,
1984) for termination of transcription was isolated from the
plasmid pBinARHyg-GGPPOR by PCR. An Xbal cleavage site was
attached in each case to the oligonucleotides for the promoter
and the terminator. The sequence of the oligonucleotide which
anneals onto the promoter (in italics) is
5'-ATTCTAGACATGGAGTCAAA-GATTCAA.ATAGA-3', and that of the
oligonucleotide which anneals onto the terminator sequence (in
italics) is 5'-ATTCTAGAGGACAA-TCAGTAAATTGA,ACGGAG-3'. The fragment
was purified using a Gene-Clean kit (Dianova GmbH, Hilden) and
cloned in accordance with the manufacturer's information onto the
vector PCR-Script from Stratagene GmbH, Heidelberg. The
correctness of the sequence was checked by sequencing. It was
transferred from the PCR-Script vector as XbaI fragment into the
correspondingly cut vector which already contained, as described
above, the DOXS sequence. The result was the construct
pBinAR-DOXS-GGPPOR (Figure 15), whose fragments have the
following significance:
Fragment A (529 bp) comprises the 35S promoter of cauliflower
mosaic virus (nucleotides 6909 to 7437 of cauliflower mosaic
virus). Fragment B comprises the transit peptide of the plastidic
transketolase. Fragment E comprises the DOXS gene. Fragment D
comprises the polyadenylation signal of gene 3 of the T DNA of
the Ti plasmid pTIACHS (Gielen et al., 1984) for termination of
transcription. Fragment F comprises the GGPPOR gene including the
intrinsic plastid transit sequence.
Example 17
Production of constructs for transformation of plants with DOXS,
GGPPOR and HPPD DNA sequences
To produce plants which are transgenic for DOXS, GGPPOR and HPPD,
a binary vector containing all three gene sequences was
manufactured (Figure 16). The GGPPOR gene was provided with the
intrinsic plastidic localization sequence (as described in
Example 15). The pBinAR-Hyg vector used confers on plants
resistance to the antibiotic hygromycin and is thus suitable for
superinfection of plants with kanamycin resistance.
0817/00006
CA 02339519 2001-02-02
To clone HPPD into vectors which additionally contain another
cDNA, oligonucleotides were derived for a PCR, and a BamHi
restriction cleavage site was attached to them at the 5' end and
3' end. The oligonucleotide at the 5' end comprises the sequence
5 5'-GGATCCTCCAGCGGACAAGCCAAC-3' (nucleotides 37 to 55 distant from
ATG in the 5' direction; in italics), and the oligonucleotide at
the 3' end comprises the sequence
5'-ATGGATCCCGCGCCGCCTACAGGTTG-3' (ending with base pair 1140 of
the coding sequence, starting 8 base pairs 3' of the TAG stop
10 codon; in italics). The PCR reaction was carried out with Tli
polymerase from Promega GmbH, Mannheim in accordance with the
manufacturer's information. 10 ng of the plasmid pBinAR-HPPD were
employed as template. The PCR program was as follows:
15 5 cycles: 94°C 4 sec, 68°C 30 sec, 72°C 2 min
5 cycles: 94°C 4 sec, 64°C 30 sec, 72°C 2 min
25 cycles: 94°C 4 sec, 60°C 30 sec, 72°C 2 min
The fragment was purified using a Gene-Clean kit (Dianova GmbH,
20 Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script from Stratagene GmbH,
Heidelberg. The correctness of the sequence was checked by
sequencing. It was cut out of the vector PCR-Script as BamHI
fragment and ligated into a correspondingly cut pBinAR vector
25 which additionally contains the transit peptide of transketolase
for introducing the gene product into plastids. The result was
the plasmid pBinAR-TP-p-HPPD.
For the cloning, the 35S promoter, the transketolase transit
30 peptide, the p-HPPD gene and the polyadenylation signal of gene 3
of the T DNA of the Ti plasmid pTIACH5 (Gielen et al. 1984) for
termination of transcription was isolated from the plasmid
pBinAR-TP-p-HPPD by PCR. A HindIII cleavage site was attached in
each case to the oligonucleotides for the promoter and the
35 terminator. The sequence of the oligonucleotide which anneals
onto the 5' region of the promoter (in italics) is
5'-ATAAGCTTCATGGAGTCAAA-GATTCAAATAGA -3', and that of the
oligonucleotide which anneals onto the termination sequence (in
italics) is 5'-ATAAGCTTGGAC-AATCAGTAAATTGAACGGAG-3'. The
40 resulting fragment was purified using a Gene-Clean kit (Dianova
GmbH, Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script from Stratagene GmbH,
Heidelberg. The correctness of the sequence was checked by
sequencing. It was transferred as HindIII fragment from this
45 PCR-Script vector into the correspondingly cut vector pHinl9
(Bevan, 1984, Nucleic Acids Res. 12, 8711-8721).
0817/00006
CA 02339519 2001-02-02
46
The 35S promoter, the transketolase transit peptide, the DOXS
gene and the polyadenylation signal of gene 3 of the T DNA of the
Ti plasmid pTIACH5 (Gielen et al., 1984) for termination of
transcription was isolated from the plasmid pBinAR-TP-DOXS by
PCR. An EcoRI cleavage site was attached in each case to the
oligonucleotides for the promoter and the terminator sequence.
The sequence of the oligonucleotide which anneals onto the
promoter (in italics) is 5'-ATGAATTCCATGGAGTCAAAGATTCAAATAGA-3',
and that of the oligonucleotide which anneals onto the terminator
sequence (in italics) is 5'-ATGAATTCGGACAATCAGTAAATTGAACGGAG-3'.
The fragment was purified using a Gene-Clean kit (Dianova GmbH,
Hilden) and cloned in accordance with the manufacturer's
information into the vector PCR-Script from Stratagene GmbH,
Heidelberg. The correctness of the sequence was checked by
sequencing. It was transferred from the PCR-Script vector as
EcoRI fragment into the correspondingly cut vector which already
contained the HPPD sequence as described above.
The 35S promoter, the GGPPOR gene and the polyadenylation signal
of gene 3 of the T DNA of the Ti plasmid pTIACHS (Gielen et al.,
1984) for termination of transcription was isolated from the
plasmid pBinARHyg-GGPPOR by PCR. An XbaI cleavage site was
attached in each case to the oligonucleotides for the promoter
and the terminator. The sequence of the oligonucleotide which
anneals onto the promoter (in italics) is
5'-ATTCTAGACATGGAGTCAAA-GATTCAAATAGA-3', and that of the
oligonucleotide which anneals onto the terminator sequence (in
italics) is 5'-ATTCTAGAGGACAA-TCAGTAAATTGAACGGAG-3'. The fragment
was purified using a Gene-Clean kit (Dianova GmbH, Hilden) and
cloned in accordance with the manufacturer's information into the
vector PCR-Script from Stratagene GmbH, Heidelberg. The
correctness of the sequence was checked by sequencing. It was
transferred from the PCR-Script vector as XbaI fragment into the
correspondingly cut vector which already contained the HPPD and
DOXS sequences as described above. The result was the construct
pBinAR-DOXS-GGPPOR-HPPD (Figure 16), whose fragments have the
following significance:
Fragment A (529 bp) comprises the 35S promoter of cauliflower
mosaic virus (nucleotides 6909 to 7437 of cauliflower mosaic
virus). Fragment B comprises the transit peptide of the plastidic
transketolase. Fragment C comprises the HPPD gene. Fragment D
comprises the polyadenylation signal of gene 3 of the T DNA of
the Ti plasmid pTIACH5 (Gielen et al., 1984) for termination of
transcription. Fragment E comprises the DOXS gene. Fragment F
0$17/00006
CA 02339519 2001-02-02
47
comprises the GGPPOR gene including the intrinsic plastid transit
sequence.
Example 18
Increasing tocopherol biosynthesis in oilseed rape
The cDNA of DOXS (SEQ ID No. 3) and of GGPOR (SEQ ID No. 7) was
provided with a CaMV35S promoter and overexpressed in rape using
the 35S promoter. In parallel with this, the seed-specific
promoter of the phaseolin gene was used in order specifically to
increase the tocopherol content in the rapeseed. Oilseed rape
plants transformed with the corresponding constructs were grown
in a glasshouse. The a-tocopherol content of the whole plant and
of the seeds of the plant was then determined. In all cases, the
a-tocopherol concentration was increased by comparison with the
untransformed plant.
Example 19
Increasing the tocopherol biosynthesis in oilseed rape
The cDNA of DOXS (SEQ ID No. 3), of HPPD (SEQ ID No. 5) and of
GGPPOR (SEQ-ID No. 7) was provided with a CaMV35S promoter and
overexpressed in rape using the 35S promoter. In parallel with
this, the seed-specific promoter of the phaseolin gene was used
in order specifically to increase the tocopherol content in the
rapeseed. Oilseed rape plants transformed with the corresponding
constructs were grown in a glasshouse. The a-tocopherol content
of the whole plant and of the seeds of the plant was then
determined. In all cases, the a-tocopherol concentration was
increased by comparison with the untransformed plant.
40
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
SEQUENZPROTOKOLL
<110> SunGene GmbH & Co.KGaA
<120> DNA-Sequenz kodierend fuer eine
1-Deoxy-D-xylulose-5-phosphat Synthase
<130> 0050/49249
<140> 0817 - 00006
<141> 1999-08-04
<160> 8
<170> PatentIn Vers. 2.0
<210> 1
<211> 2458
<212> DNA
<213> Arabidopsis thaliana
<220>
<221> CDS
<222> (1)..(2154)
<400> 1
atg get tct tct gca ttt get ttt cct tct tac ata ata acc aaa gga 4B
Met Ala Ser Ser Ala Phe Ala Phe Pro Ser Tyr Ile Ile Thr Lys Gly
1 5 10 15
gga ctt tca act gat tct tgt aaa tca act tct ttg tct tct tct aga 96
Gly Leu Ser Thr Asp Ser Cys Lys Ser Thr 5er Leu Ser 5er Ser Arg
20 25 30
tct ttg gtt aca gat ctt cca tca cca tgt ctg aaa ccc aac aac aat 144
Ser Leu Val Thr Asp Leu Pro Ser Pro Cys Leu Lys Pro Asn Asn Asn
35 40 45
tcc cat tca aac aga aga gca aaa gtg tgt get tca ctt gca gag aag 192
Ser His Ser Asn Arg Arg Ala Lys Val Cys Ala Ser Leu Ala Glu Lys
50 55 60
ggt gaa tat tat tca aac aga cca cca act cca tta ctt gac act att 240
Gly Glu Tyr Tyr Ser Asn Arg Pro Pro Thr Pro Leu Leu Asp Thr Ile
65 70 75 80
aac tac cca atc cac atg aaa aat ctt tct gtc aag gaa ctg aaa caa 288
Asn Tyr Pro Ile His Met Lys Asn Leu Ser Val Lys Glu Leu Lys Gln
1
CA 02339519 2001-02-02
WO 00/08169 PCTlEP99/05467
85 90 95
ctt tct gat gag ctg aga tca gac gtg atc ttt aat gtg tcg aaa acc 336
Leu Ser Asp Glu Leu Arg Ser Asp Val Ile Phe Asn Val Ser Lys Thr
100 105 110
ggt gga cat ttg ggg tca agt ctt ggt gtt gtg gag ctt act gtg get 3B4
Gly Gly His Leu Gly Ser Ser Leu Gly Val Val Glu Leu Thr Val Ala
115 120 125
ctt cat tac att ttc aat act cca caa gac aag att ctt tgg gat gtt 432
Leu His Tyr Ile Phe Asn Thr Pro Gln Asp Lys Ile Leu Trp Asp Val
130 135 140
ggt cat cag tct tat cct cat aag att ctt act ggg aga aga gga aag 480
Gly His Gln Ser Tyr Pro His Lys Ile Leu Thr Gly Arg Arg Gly Lys
145 150 155 160
atg cct aca atg agg caa acc aat ggt ctc tct ggt ttc acc aaa cga 528
Met Pro Thr Met Arg Gln Thr Asn Gly Leu Ser Gly Phe Thr Lys Arg
165 170 175
gga gag agt gaa cat gat tgc ttt ggt act gga cac agc tca acc aca 576
Gly Glu Ser Glu His Asp Cys Phe Gly Thr Gly His Ser Ser Thr Thr
180 185 190
ata tct get ggt tta gga atg gcg gta gga agg gat ttg aag ggg aag 624
Ile Ser Ala Gly Leu Gly Met A1a Val Gly Arg Asp Leu Lys Gly Lys
195 200 205
aac aac aat gtg gtt get gtg att ggt gat ggt gcg atg acg gca gga 672
Asn Asn Asn Val Val Ala Val Ile Gly Asp Gly Ala Met Thr Ala Gly
210 215 220
cag get tat gaa gcc atg aac aac gcc gga tat cta gac tct gat atg 720
Gln Ala Tyr Glu Ala Met Asn Asn Ala Gly Tyr Leu Asp Ser Asp Met
225 230 235 240
att gtg att ctt aat gac aac aag caa gtc tca tta cct aca get act 768
Ile Val Ile Leu Asn Asp Asn Lys Gln Val Ser Leu Pro Thr Ala Thr
245 250 255
ttg gat gga cca agt cca cct gtt ggt gca ttg agc agt get ctt agt 816
Leu Asp Gly Pro Ser Pro Pro Val Gly A1a Leu Ser Ser Ala Leu Ser
260 265 270
cgg tta cag tct aac ccg get ctc aga gag ttg aga gaa gtc gca aag 864
Arg Leu Gln Ser Asn Pro A1a Leu Arg Glu Leu Arg Glu Val Ala Lys
2
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
275 280 285
ggt atg aca aag caa ata ggc gga cca atg cat cag ttg gcg get aag 912
Gly Met Thr Lys Gln Ile Gly Gly Pro Met His Gln Leu Ala Ala Lys
290 295 300
gta gat gtg tat get cga gga atg ata agc ggt act gga tcg tca ctg 960
Val Asp Val Tyr Ala Arg Gly Met Ile Ser Gly Thr Gly Ser Ser Leu
305 310 315 320
ttt gaa gaa ctc ggt ctt tac tat att ggt cca gtt gat ggg cac aac 1008
Phe Glu Glu Leu Gly Leu Tyr Tyr Ile Gly Pro Val Asp Gly His Asn
325 330 335
ata gat gat ttg gta gcc att ctt aaa gaa gtt aag agt acc aga acc 1056
Ile Asp Asp Leu Val Ala Ile Leu Lys Glu Val Lys Ser Thr Arg Thr
340 345 350
aca gga cct gta ctt att cat gtg gtg acg gag aaa ggt cgt ggt tat 1104
Thr Gly Pro Val Leu Ile His Val Val Thr Glu Lys Gly Arg Gly Tyr
355 360 365
cct tac gcg gag aga get gat gac aaa tac cat ggt gtt gtg aaa ttt 1152
Pro Tyr Ala Glu Arg Ala Asp Asp Lys Tyr His Gly Val Val Lys Phe
370 375 380
gat cca gca acg ggt aga cag ttc aaa act act aat gag act caa tct 1200
Asp Pro Ala Thr Gly Arg Gln Phe Lys Thr Thr Asn Glu Thr Gln Ser
385 390 395 400
tac aca act tac ttt gcg gag gca tta gtc gca gaa gca gag gta gac 1248
Tyr Thr Thr Tyr Phe Ala Glu Ala Leu Val Ala Glu Ala Glu Val Asp
405 410 415
aaa gat gtg gtt gcg att cat gca gcc atg gga ggt gga acc ggg tta 1296
Lys Asp Val Val Ala Ile His Ala A1a Met Gly Gly Gly Thr Gly Leu
420 425 430
aat ctc ttt caa cgt cgc ttc cca aca aga tgt ttc gat gta gga ata 1344
Asn Leu Phe Gln Arg Arg Phe Pro Thr Arg Cys Phe Asp Val Gly Ile
435 440 445
gcg gaa caa cac gca gtt act ttt get gcg ggt tta gcc tgt gaa ggc 1392
A1a Glu Gln His Ala Val Thr Phe Ala Ala Gly Leu Ala Cys Glu Gly
450 455 460
ctt aaa ccc ttc tgt gca atc tat tcg tct ttc atg cag cgt get tat 1440
Leu Lys Pro Phe Cys Ala Ile Tyr Ser Ser Phe Met Gln Arg Ala Tyr
3
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
465 470 475 4g0
gac cag gtt gtc cat gat gtt gat ttg caa aaa tta ccg gtg aga ttt 1488
Asp Gln Val Val His Asp Val Asp Leu Gln Lys Leu Pro Val Arg Phe
485 490 495
gca atg gat aga get gga ctc gtt gga get gat ggt ccg aca cat tgt 1536
Ala Met Asp Arg Ala Gly Leu Val Gly Ala Asp Gly Pro Thr His Cys
500 505 510
gga get ttc gat gtg aca ttt atg get tgt ctt cct aac atg ata gtg 1584
Gly Ala Phe Asp Val Thr Phe Met Ala Cys Leu Pro Asn Met Ile Val
515 520 525
atg get cca tca gat gaa gca gat ctc ttt aac atg gtt gca act get 1632
Met Ala Pro Ser Asp Glu A1a Asp Leu Phe Asn Met Val Ala Thr Ala
' 530 535 540
gtt gcg att gat gat cgt cct tct tgt ttc cgt tac cct aga ggt aac 1680
Val Ala Ile Asp Asp Arg Pro Ser Cys Phe Arg Tyr Pro Arg Gly Asn
545 550 555 560
ggt att gga gtt gca tta cct ccc gga aac aaa ggt gtt cca att gag 1728
Gly Ile Gly Val Ala Leu Pro Pro Gly Asn Lys Gly Val Pro Ile Glu
565 570 575
att ggg aaa ggt aga att tta aag gaa gga gag aga gtt gcg ttg ttg 1776
Ile Gly Lys Gly Arg Ile Leu Lys Glu Gly Glu Arg Val Ala Leu Leu
580 585 590
ggt tat ggc tca gca gtt cag agc tgt tta gga gcg get gta atg ctc 1824
Gly Tyr Gly Ser Ala Val Gln Ser Cys Leu Gly Ala Ala Val Met Leu
595 600 605
gaa gaa cgc gga tta aac gta act gta gcg gat gca cgg ttt tgc aag 1872
Glu Glu Arg Gly Leu Asn Val Thr Val Ala Asp Ala Arg Phe Cys Lys
610 615 620
cca ttg gac cgt get ctc att cgc agc tta get aag tcg cac gag gtt 1920
Pro Leu Asp Arg Ala Leu Ile Arg Ser Leu Ala Lys Ser His Glu Val
__ 625 630 635 640
ctg atc acg gtt gaa gaa ggt tcc att gga ggt ttt ggc tcg cac gtt 1968
Leu Ile Thr Val Glu Glu Gly Ser Ile Gly Gly Phe Gly Ser His Val
645 650 655
gtt cag ttt ctt get ctc gat ggt ctt ctt gat ggc aaa ctc aag tgg 2016
Val Gln Phe Leu Ala Leu Asp Gly Leu Leu Asp Gly Lys Leu Lys Trp
4
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
660 665 670
aga cca atg gta ctg cct gat cga tac att gat cac ggt gca cca get 2064
Arg ProMetVal LeuPro AspArgTyr IleAsp HisGly Ala Pro Ala
675 680 685
gat caactaget gaaget ggactcatg ccatct cacatc gca gca acc
2112
Asp GlnLeuAla GluAla GlyLeuMet ProSer HisIle Ala Rla Thr
690 695 700
gca cttaactta atcggt gcaccaagg gaaget ctgttt tga 2154
Ala LeuAsnLeu IleGly AlaProArg GluAla LeuPhe
705 ?10 715
gagtaagaat ctgttggcta aaacatatgt atacaaacac tctaaatgca acccaaggtt 2219
1:
tcttctaagt actgatcaga attcccgccc gagaagtcct ttggcaacag ctatatatat 2274
ttactaagat tgtgaagaga aaggcaaagg caaaggttgt gcaaagatta gtattataga 2334
taaaactggt atttgttttg taattttagg attgtgatga gatcgtgttg taccaataac 2394
taacatcttg taaaatcaat tactctcttg tgatcttcaa taagcttgag tgacaaaaaa 2454
aaaa
2458
<210> 2
<211> 717
<212> PRT
<213> Arabidopsis thaliana
(,
<400> 2
Met Ala Ser Ser A1a Phe Ala Phe Pro Ser Tyr Ile Ile Thr Lys Gly
1 5 10 15
Gly Leu Ser Thr Asp Ser Cys Lys Ser Thr Ser Leu Ser Ser Ser Arg
20 25 30
Ser Leu Val Thr Asp Leu Pro Ser Pro Cys Leu Lys Pro Asn Asn Asn
35 40 95
Ser His Ser Asn Arg Arg Ala Lys Val Cys Ala Ser Leu Ala Glu Lys
50 55 60
Gly Glu Tyr Tyr Ser Asn Arg Pro Pro Thr Pro Leu Leu Asp Thr Ile
65 70 75 80
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
Asn Tyr Pro Ile His Met Lys Asn Leu Ser Va1 Lys Glu Leu Lys Gln
85 90 95
Leu Ser Asp Glu Leu Arg Ser Asp Val Ile Phe Asn Val Ser Lys Thr
100 105 110
Gly Gly His Leu Gly Ser Ser Leu Gly Val Val Glu Leu Thr Val Ala
115 120 125
Leu His Tyr Ile Phe Asn Thr Pro Gln Asp Lys Ile Leu Trp Asp Val
130 135 140
Gly His Gln Ser Tyr Pro His Lys Ile Leu Thr Gly Arg Arg Gly Lys
145 150 155 160
Met Pro Thr Met Arg Gln Thr Asn Gly Leu Ser Gly Phe Thr Lys Arg
165 170 175
Gly Glu Ser Glu His Asp Cys Phe Gly Thr Gly His Ser Ser Thr Thr
180 185 190
Ile Ser Ala Gly Leu Gly Met Ala val Gly Arg Asp Leu Lys Gly Lys
195 200 205
Asn Asn Asn Val Val Ala Val Ile Gly Asp Gly Ala Met Thr Ala Gly
210 215 220
Gln Ala Tyr Glu Ala Met Asn Asn Ala Gly Tyr Leu Asp 5er Asp Met
225 230 235
240
Ile Val Ile Leu Asn Asp Asn Lys Gln Val Ser Leu Pro Thr Ala Thr
245 250 255
Leu Asp Gly Pro 5er Pro Pro Val Gly Rla Leu Ser Ser Ala Leu Ser
260 265 270
Arg Leu Gln Ser Asn Pro Ala Leu Arg Glu Leu Arg Glu Val Ala Lys
275 280 285
Gly Met Thr Lys Gln Ile Gly Gly Pro Met His Gln Leu Ala A1a Lys
290 295 300
Val Asp Val Tyr Ala Arg Gly Met Ile Ser Gly Thr Gly Ser Ser Leu
305 310 315 320
Phe Glu Glu Leu Gly Leu Tyr Tyr Ile Gly Pro Val Asp Gly His Asn
325 330 335
6
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
Ile Asp Asp Leu Val Ala Ile Leu Lys Glu Val Lys Ser Thr Arg Thr
340 345 350
Thr Gly Pro Val Leu Ile His Val Val Thr Glu Lys Gly Arg Gly Tyr
355 360 365
Pro Tyr Ala Glu Arg Ala Asp Asp Lys Tyr His Gly Val Val Lys Phe
370 375 380
Asp Pro Ala Thr Gly Arg Gln Phe Lys Thr Thr Asn Glu Thr Gln Ser
385 390 395 400
Tyr Thr Thr Tyr Phe Ala Glu Ala Leu Val Ala Glu Ala Glu Val Asp
405 410 415
Lys Asp Val Val Rla Ile His Ala Ala Met Gly Gly Gly Thr Gly Leu
420 425 430
Asn Leu Phe Gln Arg Arg Phe Pro Thr Arg Cys Phe Asp Val Gly Ile
435 440 445
Ala Glu Gln His Ala Val Thr Phe Ala Ala Gly Leu Ala Cys Glu Gly
450 455 460
Leu Lys Pro Phe Cys Ala Ile Tyr Ser Ser Phe Met Gln Arg Ala Tyr
465 470 475 480
Asp Gln Val Val His Asp Val Asp Leu Gln Lys Leu Pro Val Arg Phe
985 490 495
Ala Met Asp Arg Ala Gly Leu Val Gly Ala Asp Gly Pro Thr His Cys
500 505 510
Gly Ala Phe Asp Val Thr Phe Met Ala Cys Leu Pro Asn Met Ile Val
515 520 525
Met Ala Pro Ser Asp Glu Ala Asp Leu Phe Asn Met Val Ala Thr Ala
530 535 540
Val Ala Ile Asp Asp Arg Pro Ser Cys Phe Arg Tyr Pro Arg Gly Asn
545 550 555 560
Gly Ile Gly Va1 Ala Leu Pro Pro Gly Asn Lys Gly Val Pro Ile Glu
565 570 575
Ile Gly Lys Gly Arg Ile Leu Lys Glu Gly Glu Arg Val Ala Leu Leu
580 585 590
7
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
Gly Tyr Gly Ser Ala Val Gln Ser Cys Leu Gly A1a Ala Val Met Leu
595 600 605
Glu Glu Arg Gly Leu Asn Val Thr Val Ala Asp Ala Arg Phe Cys Lys
610 615 620
Pro Leu Asp Arg Ala Leu Ile Arg Ser Leu Ala Lys Ser His Glu Val
625 630 635 640
Leu Ile Thr Val Glu Glu Gly Ser Ile Gly Gly Phe Gly Ser His Val
645 650 655
Val Gln Phe Leu Ala Leu Asp Gly Leu Leu Asp Gly Lys Leu Lys Trp
660 665 670
Arg Pro Met Val Leu Pro Asp Arg Tyr Ile Asp His Gly Ala Pro A1a
w 675 680 685
Asp Gln Leu Ala Glu Ala Gly Leu Met Pro 5er His Ile Ala Ala Thr
690 695 700
Ala Leu Asn Leu Ile Gly A1a Pro Arg Glu Ala Leu Phe
705 710 715
<210> 3
<211> 1863
<212> DNA
<213> Escherichia coli
<220>
<221> CDS
<222> (1)..(1863)
<400> 3
atg agt ttt gat att gcc aaa tac ccg acc ctg gca ctg gtc gac tcc 4B
Met Ser Phe Asp Ile Rla Lys Tyr Pro Thr Leu Ala Leu Val Asp Ser
1 5 10 15
acc cag gag tta cga ctg ttg ccg aaa gag agt tta ccg aaa ctc tgc 96
Thr Gln Glu Leu Arg Leu Leu Pro Lys Glu Ser Leu Pro Lys Leu Cys
20 25 30
gac gaa ctg cgc cgc tat tta ctc gac agc gtg agc cgt tcc agc ggg 149
Asp Glu Leu Arg Arg Tyr Leu Leu Asp Ser Val Ser Arg Ser Ser Gly
35 40 45
cac ttc gcc tcc ggg ctg ggc acg gtc gaa ctg acc gtg gcg ctg cac 192
8
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
His Phe Ala Ser Gly Leu Gly Thr Val Glu Leu Thr Val Ala Leu His
50 55 60
tat gtc tac aac acc ccg ttt gac caa ttg att tgg gat gtg ggg cat 240
Tyr Val Tyr Asn Thr Pro Phe Asp Gln Leu Ile Trp Asp Val Gly His
65 70 75 BO
cag get tat ccg cat aaa att ttg acc gga cgc egc gae aaa atc ggc 288
Gln Ala Tyr Pro His Lys Ile Leu Thr Gly Arg Arg Asp Lys Ile Gly
85 90 95
acc atc cgt cag aaa ggc ggt ctg cac ccg ttc ccg tgg cgc ggc gaa 336
Thr Ile Arg Gln Lys Gly Gly Leu His Pro Phe Pro Trp Arg Gly Glu
100 105 110
agc gaa tat gac gta tta agc gtc ggg cat tca tca acc tcc atc agt 384
Ser Glu Tyr Asp Val Leu Ser Val Gly His Ser Ser Thr Ser Ile Ser
115 120 125
gcc gga att ggt att gcg gtt get gcc gaa aaa gaa ggc aaa aat cgc 432
Ala Gly Ile Gly Ile Ala Va1 Ala Ala Glu Lys Glu Gly Lys Asn Arg
130 135 140
cgc acc gtc tgt gtc att ggc gat ggc gcg att acc gca ggc atg gcg 480
Arg Thr Val Cys Val Ile Gly Asp Gly Ala Ile Thr Ala Gly Met A1a
145 150 155 160
ttt gaa gcg atg aat cac gcg ggc gat atc cgt cct gat atg ctg gtg 528
Phe Glu Ala Met Asn His Ala Gly Asp Ile Arg Pro Asp Met Leu Val
165 170 175
att ctc aac gac aat gaa atg tcg att tcc gaa aat gtc ggc gcg ctc 576
Ile Leu Asn Asp Asn Glu Met Ser Ile Ser Glu Asn Val Gly Ala Leu
180 185 190
aac aac cat ctg gca cag ctg ctt tcc ggt aag ctt tac tct tca ctg 624
Asn Asn His Leu Ala Gln Leu Leu Ser Gly Lys Leu Tyr Ser Ser Leu
195 200 205
cgc gaa ggc ggg aaa aaa gtt ttc tct ggc gtg ccg cca att aaa gag 672
Arg Glu Gly Gly Lys Lys Val Phe Ser Gly Val Pro Pro Ile Lys Glu
210 215 220
ctg ctc aaa cgc acc gaa gaa cat att aaa ggc atg gta gtg cct ggc 720
Leu Leu Lys Arg Thr Glu Glu His Ile Lys Gly Met Val Val Pro Gly
225 230 235 240
acg ttg ttt gaa gag ctg ggc ttt aac tac atc ggc ccg gtg gac ggt 768
9
CA 02339519 2001-02-02
- WO 00/08169 PCT/EP99/05467
Thr Leu Phe Glu Glu Leu Gly Phe Asn Tyr Ile Gly Pro Val Asp Gly
245 250 255
cac gat gtg ctg ggg ctt atc acc acg cta aag aac atg cgc gac ctg 816
His Asp Val Leu Gly Leu Ile Thr Thr Leu Lys Asn Met Arg Asp Leu
260 265 270
aaa ggc ccg cag ttc ctg cat atc atg acc aaa aaa ggt cgt ggt tat 864
Lys Gly Pro Gln Phe Leu His Ile Met Thr Lys Lys Gly Arg Gly Tyr
275 280 285
gaa ccg gca gaa aaa gac ccg atc act ttc cac gcc gtg cct aaa ttt 912
Glu Pro Ala Glu Lys Asp Pro Ile Thr Phe His Ala Val Pro Lys Phe
290 295 300
gat ccc tcc agc ggt tgt ttg ccg aaa agt agc ggc ggt ttg ccg agc 960
Asp Pro Ser Ser Gly Cys Leu Pro Lys Ser Ser Gly Gly Leu Pro Ser
305 310 315 320
tat tca aaa atc ttt ggc gac tgg ttg tgc gaa acg gca gcg aaa gac 1008
Tyr Ser Lys Ile Phe Gly Asp Trp Leu Cys Glu Thr Ala Ala Lys Asp
325 330 335
aac aag ctg atg gcg att act ccg gcg atg cgt gaa ggt tcc ggc atg 1056
Asn Lys Leu Met Ala Ile Thr Pro Ala Met Arg Glu Gly Ser Gly Met
340 345 350
gtc gag ttt tca cgt aaa ttc ccg gat cgc tac ttc gac gtg gca att 1104
Val Glu Phe Ser Arg Lys Phe Fro Asp Arg Tyr Phe Asp Val Ala Ile
355 360 365
gcc gag caa cac gcg gtg acc ttt get gcg ggt ctg gcg att ggt ggg 1152
A1a Glu Gln His Ala Val Thr Phe Ala Ala Gly Leu Ala Ile Gly Gly
370 375 380
tac aaa ccc att gtc gcg att tac tcc act ttc ctg caa cgc gcc tat 1200
Tyr Lys Pro Ile Val Ala Ile Tyr Ser Thr Phe Leu Gln Arg Ala Tyr
385 390 395 400
gat cag gtg ctg cat gac gtg gcg att caa aag ctt ccg gtc ctg ttc 1248
Asp Gln Val Leu His Asp Val Ala Ile Gln Lys Leu Pro Val Leu Phe
405 410 415
gcc atc gac cgc gcg ggc att gtt ggt get gac ggt caa acc cat cag 1296
Ala Ile Asp Arg Ala Gly Ile Val Gly Ala Asp Gly Gln Thr His Gln
420 425 430
ggt get ttt gat ctc tct tac ctg cgc tgc ata ccg gaa atg gtc att 1344
CA 02339519 2001-02-02
WO 00108169 PCT1EP99/05467
Gly Ala Phe Asp Leu Ser Tyr Leu Arg Cys Ile Pro Glu Met Val Ile
435 440 445
atg acc ccg agc gat gaa aac gaa tgt cgc cag atg ctc tat acc ggc 1392
Met Thr Pro Ser Asp Glu Asn Glu Cys Arg Gln Met Leu Tyr Thr Gly
450 455 460
tat cac tat aac gat ggc ccg tca gcg gtg cgc tac ccg cgt ggc aac 1440
Tyr His Tyr Asn Asp Gly Pro Ser A1a Val Arg Tyr Pro Arg Gly Asn
465 470 475 480
gcg gtc ggc gtg gaa ctg acg ccg ctg gaa aaa cta cca att ggc aaa 1488
Ala Val Gly Val Glu Leu Thr Pro Leu Glu Lys Leu Pro Ile Gly Lys
485 490 495
ggc att gtg aag cgt cgt ggc gag aaa ctg gcg atc ctt aac ttt ggt 1536
Gly Ile Val Lys Arg Arg Gly Glu Lys Leu Ala Ile Leu Asn Phe Gly
500 505 510
acg ctg atg cca gaa gcg gcg aaa gtc gcc gaa tcg ctg aac gcc acg 1584
Thr Leu Met Pro Glu Ala Ala Lys Val Ala Glu Ser Leu Asn Ala Thr
515 520 525
ctg gtc gat atg cgt ttt gtg aaa ccg ctt gat gaa gcg tta att ctg 1632
Leu Val Asp Met Arg Phe Val Lys Pro Leu Asp Glu Ala Leu Ile Leu
530 535 540
gaa atg gcc gcc agc cat gaa gcg ctg gtc acc gta gaa gaa aac gcc 1680
Glu Met Ala Ala Ser His Glu Ala Leu Val Thr Val Glu Glu Asn A1a
545 550 555 560
att atg ggc ggc gca ggc agc ggc gtg aac gaa gtg ctg atg gcc cat 1728
Ile Met Gly Gly Ala Gly Ser Gly Val Asn Glu Val Leu Met Ala His
565 570 575
cgt aaa cca gta ccc gtg ctg aac att ggc ctg ccg gac ttc ttt att 1776
Arg Lys Pro Val Pro Val Leu Asn Ile Gly Leu Pro Asp Phe Phe Ile
5B0 585 590
ccg caa gga act cag gaa gaa atg cgc gcc gaa ctc ggc ctc gat gcc 1824
Pro Gln Gly Thr Gln Glu Glu Met Arg Ala Glu Leu Gly Leu Asp Ala
595 600 605
get ggt atg gaa gcc aaa atc aag gcc tgg ctg gca taa 1863
Ala Gly Met Glu Ala Lys Ile Lys Ala Trp Leu Ala
610 615 620
11
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
<210> 4
<211> 620
<212> PRT
<213> Escherichia coli
<400> 4
Met Ser Phe Asp Ile Ala Lys Tyr Pro Thr Leu Ala Leu Val Asp Ser
1 5 10 15
Thr Gln Glu Leu Arg Leu Leu Pro Lys Glu Ser Leu Pro Lys Leu Cys
20 25 30
Asp Glu Leu Arg Arg Tyr Leu Leu Asp Ser Val Ser Arg Ser 5er Gly
35 40 95
(.
His Phe Ala Ser Gly Leu Gly Thr Val Glu Leu Thr Val Ala Leu His
50 55 60
Tyr Val Tyr Asn Thr Pro Phe Asp Gln Leu Ile Trp Asp Val Gly His
65 70 75 BO
Gln Ala Tyr Pro His Lys Ile Leu Thr Gly Arg Arg Asp Lys Ile Gly
85 90 95
Thr Ile Arg Gln Lys Gly Gly Leu His Pro Phe Pro Trp Arg Gly Glu
100 105 110
Ser Glu Tyr Asp Val Leu Ser Val Gly His Ser Ser Thr Ser Ile Ser
115 120 125
Ala Gly Ile Gly I1e Ala Val Ala Ala Glu Lys Glu Gly Lys Asn Arg
t 130 135 140
Arg Thr Val Cys Val Ile Gly Asp Gly Ala Ile Thr Ala Gly Met Ala
145 150 155 160
Phe Glu Ala Met Asn His Ala Gly Asp Ile Arg Pro Asp Met Leu Val
165 170 175
Ile Leu Asn Asp Asn Glu Met Ser I1e Ser Glu Asn Val Gly Ala Leu
180 185 190
Asn Asn His Leu Ala Gln Leu Leu Ser Gly Lys Leu Tyr Ser Ser Leu
195 200 205
Arg Glu Gly Gly Lys Lys Val Phe Ser Gly Val Pro Pro Ile Lys Glu
210 215 220
12
CA 02339519 2001-02-02
_ WO 00/OSi69 PCT/EP99/05467
Leu Leu Lys Arg Thr Glu Glu His Ile Lys Gly Met Val Val Pro Gly
225 230 235 240
Thr Leu Phe Glu Glu Leu Gly Phe Asn Tyr Ile Gly Pro Val Asp Gly
245 250 255
His Asp Val Leu Gly Leu Ile Thr Thr Leu Lys Asn Met Arg Asp Leu
260 265 270
Lys Gly Pro Gln Phe Leu His Ile Met Thr Lys Lys Gly Arg Gly Tyr
275 280 285
Glu Pro Ala Glu Lys Asp Pro Ile Thr Phe His Ala Val Pro Lys Phe
290 295 300
Asp Pro Ser Ser Gly Cys Leu Pro Lys Ser Ser Gly Gly Leu Pro Ser
305 310 315 320
Tyr Ser Lys Ile Phe Gly Asp Trp Leu Cys Glu Thr A1a Ala Lys Asp
325 330 335
Asn Lys Leu Met Ala Ile Thr Pro Ala Met Arg Glu Gly Ser Gly Met
340 345 350
Val Glu Phe Ser Arg Lys Phe Pro Asp Arg Tyr Phe Asp Val Ala Ile
355 360 365
A1a Glu Gln His Ala Val Thr Phe A1a Ala Gly Leu Ala Ile Gly Gly
370 375 380
Tyr Lys Pro Ile Val Ala Ile Tyr Ser Thr Phe Leu Gln Arg Ala Tyr
385 390 395 400
Asp Gln Val Leu His Asp Val Ala Ile Gln Lys Leu Pro Val Leu Phe
405 410 415
Ala Ile Asp Arg Ala Gly Ile Val Gly Ala Asp Gly Gln Thr His Gln
920 425 430
Gly A1a Phe Asp Leu Ser Tyr Leu Arg Cys Ile Pro Glu Met Val Ile
435 440 4q5
Met Thr Pro Ser Asp Glu Asn Glu Cys Arg Gln Met Leu Tyr Thr Gly
450 455 460
Tyr His Tyr Asn Asp Gly Pro Ser Ala Val Arg Tyr Pro Arg Gly Asn
465 470 475 480
13
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
Ala Val Gly Val Glu Leu Thr Pro Leu Glu Lys Leu Pro Ile Gly Lys
485 490 495
Gly Ile Val Lys Arg Arg Gly Glu Lys Leu Ala Ile Leu Asn Phe Gly
500 505 510
Thr Leu Met Pro Glu A1a Ala Lys Val Ala Glu Ser Leu Asn Ala Thr
515 520 525
Leu Val Asp Met Arg Phe Val Lys Pro Leu Asp Glu Ala Leu Ile Leu
530 535 540
Glu Met Ala Ala Ser His G1u Ala Leu Val Thr Val Glu Glu Asn Ala
545 550 555 560
I;,
Ile Met Gly Gly Ala Gly Ser Gly Val Asn Glu Val Leu Met Ala His
565 570 575
Arg Lys Pro Val Pro Val Leu Asn Ile Gly Leu Pro Asp Phe Phe Ile
580 5B5 590
Pro Gln Gly Thr Gln Glu Glu Met Arg Ala Glu Leu Gly Leu Asp Ala
595 600 605
Ala Gly Met Glu Ala Lys Ile Lys Ala Trp Leu Ala
610 615 620
<210> 5
<211> 1469
<212> DNA
i- <213> Streptomyces avermitilis
<220>
<221> CDS
<222> (218)..(1138)
<400> 5
gatatccgag cgccgccggg tccactgcgg tccgaagccg cggatgactc cattcgactg 60
aagccggtcg agccgcgcct gcacggtgcc gcgcgcgacc ccgagccgcc gggacatctc 120
gagcactccg atgcgcggct cccgcgccag cagcaccagg agccggccgt ccagatgatc 180
gatcgccacg gcagcccctc cagtggtcat cctgtac atg cag ccc cac gcc atg 235
Met Gln Pro His Ala Met
1 5
14
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
ggc ggt gca ctg aac aca ttg tcc agc gga caa gcc aac tat tgc gca 2B3
Gly Gly Ala Leu Asn Thr Leu Ser Ser Gly Gln Ala Asn Tyr Cys Ala
15 20
cct tgc gga acg gag cga ccc tgc cgc cat gac gca gac cac aca cca 331
' Pro Cys Gly Thr Glu Arg Pro Cys Arg His Asp Ala Asp His Thr Pro
25 30 35
cac tcc cga cac cgc ccg gca ggc cga ccc ctt ccc ggt gaa ggg aat 379
His Ser Arg His Arg Pro Ala Gly Arg Pro Leu Pro Gly Glu Gly Asn
40 45 50
gga cgc ggt cgt ctt cgc cgt agg caa cgc caa gca ggc cgc gca cta 427
Gly Arg Gly Arg Leu Arg Arg Arg Gln Arg Gln Ala Gly Arg Ala Leu
55 60 65 70
ctc cac cgc ctt cgg cat gca get tgt ggc gta ctc cgg acc gga gaa 475
Leu His Arg Leu Arg His Ala Ala Cys Gly Val Leu Arg Thr Gly Glu
75 80 85
cgg cag ccg cga gac cgc ttc gta cgt cct cac caa cgg ctc ggc acg 523
Arg Gln Pro Arg Rsp Arg Phe Val Arg Pro His Gln Arg Leu Gly Thr
90 95 100
ctt cgt cct cac ctc cgt cat caa gcc cgc cac ccc ctg ggg cca ctt 571
Leu Arg Pro His Leu Arg His Gln Ala Arg His Pro Leu Gly Pro Leu
105 110 115
cct cgc cga cca tgt ggc cga gca cgg cga cgg cgt cgt cga cct cgc 619
Pro Arg Arg Pro Cys Gly Arg Ala Arg Arg Arg Arg Arg Arg Pro Arg
120 125 130
~i_ .:.
cat cga ggt ccc gga cgc ccg cgc cgc cca cgc gta cgc gat cga gca 667
His Arg Gly Pro Gly Arg Pro Arg Arg Pro Arg Val Arg Asp Arg Rla
135 140 145 150
cgg cgc ccg ctc ggt cgc cga gcc gta cga get gaa gga cga gca cgg 715
Arg Arg Pro Leu Gly Arg Arg Ala Val Arg Ala Glu Gly Arg Ala Arg
155 160 165
cac ggt cgt cct cgc cgc gat cgc cac cta cgg caa gac ccg cca cac 763
His Gly Arg Pro Arg Arg Asp Arg His Leu Arg Gln Asp Pro Pro His
170 175 180
cct cgt cga ccg gac cgg cta cga cgg ccc cta cct ccc cgg cta cgt 811
Pro Arg Rrg Pro Asp Arg Leu Arg Arg Pro Leu Pro Pro Arg Leu Arg
185 190 195
CA 02339519 2001-02-02
a
WO 00/08169 PCT/EP99J05467
ggc cgc cgc ccc gat cgt cga acc gcc cgc cca ccg cac ctt cca ggc 859
Gly Arg Arg Pro Asp Arg Arg Thr Ala Arg Pro Pro His Leu Pro Gly
200 205 210
cat cga cca ctg cgt cgg caa cgt cga get cgg ccg gat gaa cga atg 907
' His Arg Pro Leu Arg Arg Gln Arg Arg Ala Arg Pro Asp Glu Arg Met
215 220 225 230
ggt cgg ctt cta caa caa ggt cat ggg ctt cac gaa cat gaa gga gtt 955
Gly Arg Leu Leu G1n Gln Gly His Gly Leu His Glu His Glu Gly Val
235 240 245
cgt ggg cga cga cat cgc gac cga gta ctc ggc get gat gtc gaa ggt 1003
Arg Gly Arg Arg His Arg Asp Arg Val Leu Gly Ala Asp Val Glu Gly
250 255 260
-- cgt ggc cga cgg cac get caa ggt caa gtt ccc gat caa cga gcc cgc 1051
Arg Gly Arg Arg His Ala Gln Gly Gln Val Pro Asp Gln Arg Ala Arg
265 270 275
cct cgc caa gaa gaa gtc cca gat cga cga gta cct gga gtt cta cgg 1099
Pro Arg Gln Glu Glu Val Pro Asp Arg Arg Val Pro Gly Val Leu Arg
280 285 290
cgg cgc ggg cgt cca gca cat cgc get gaa cac ggg tga catcgtcgag 1148
Arg Arg Gly Arg Pro A1a His Arg Ala Glu His Gly
295 300 305
acggtacgca cgatgcgcgc cgccggcgtc cagttcctgg acacgcccga ctcgtactac 1208
gacaccctcg gggagtgggt gggcgacacc cgcgtccccg tcgacaccct gcgcgagctg 1268
aagatcctcg cggaccgcga cgaggacggc tatctgctcc agatcttcac caagccggtc 1328
caggaccgcc cgacggtctt cttcgagatc atcgaacgcc acggctcgat gggattcggc 1388
aagggcaact tcaaggccct gttcgaggcg atcgagcggg agcaggagaa gcggggcaac 1448
ctgtaggcgg cgcggcccgg g 1469
<210> 6
<211> 306
<212> PRT
<213> Streptomyces avermitilis
<400> 6
Met Gln Pro His Ala Met Gly Gly Ala Leu Asn Thr Leu Ser Ser Gly
16
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
1 5 10 15
Gln Ala Asn Tyr Cys Ala Pro Cys Gly Thr Glu Arg Pro Cys Arg His
20 25 30
Asp Ala Asp His Thr Pro His Ser Arg His Arg Pro Ala Gly Arg Pro
35 40 45
Leu Pro Gly Glu Gly Asn Gly Arg Gly Arg Leu Arg Arg Arg Gln Arg
50 55 60
Gln Ala Gly Arg Ala Leu Leu His Arg Leu Arg His Ala Ala Cys Gly
65 70 75 g0
Val Leu Arg Thr Gly Glu Arg Gln Pro Arg Asp Arg Phe Val Arg Pro
85 90 95
(;
His Gln Arg Leu Gly Thr Leu Arg Pro His Leu Arg His Gln Ala Arg
100 105 110
His Pro Leu Gly Pro Leu Pro Arg Arg Pro Cys Gly Arg Ala Arg Arg
115 120 125
Arg Arg Arg Arg Pro Arg His Arg Gly Pro Gly Arg Pro Arg Arg Pro
130 135 140
Arg Val Arg Asp Arg Ala Arg Arg Pro Leu Gly Arg Arg Ala Val Arg
145 150 155 160
Ala Glu Gly Arg Ala Arg His Gly Arg Pro Arg Arg Asp Arg His Leu
165 170 175
..
Arg Gln Asp Pro Pro His Pro Arg Arg Pro Asp Arg Leu Arg Arg Pro
180 185 190
Leu Pro Pro Arg Leu Arg Gly Arg Arg Pro Asp Arg Arg Thr Ala Arg
195 200 205
Pro Pro His Leu Pro Gly His Arg Pro Leu Arg Arg Gln Arg Arg Ala
210 215 220
Arg Pro Asp Glu Arg Met Gly Arg Leu Leu Gln Gln Gly His Gly Leu
225 230 235 290
His Glu His Glu Gly Val Arg Gly Arg Arg His Arg Asp Arg VaI Leu
245 250 255
Gly Ala Asp Val Glu Gly Arg Gly Arg Arg His A1a Gln Gly Gln Val
17
CA 02339519 2001-02-02
. WO 00/08169 PCT/EP99/05467
260 265 270
Pro Asp Gln Arg Ala Arg Pro Arg Gln Glu Glu Val Pro Asp Arg Arg
275 280 285
Val Pro Gly Val Leu Arg Arg Arg Gly Arg Pro Ala His Arg A1a Glu
290 295 300
His Gly
305
<210> 7
<211> 1479
<212> DNA
<213> Arabidopsis thaliana
<220>
<221> CDS
<222> (1)..(1401)
<400> 7
atg gcg acg acg gtt aca ctc aaa tcc ttc acc gga ctt cgt caa tca 48
Met Ala Thr Thr Val Thr Leu Lys Ser Phe Thr Gly Leu Arg Gln Ser
1 5 10 15
tca acg gag caa aca aac ttc gtc tct cat gta ccg tca tca ctt tct 96
Ser Thr Glu Gln Thr Asn Phe Val Ser His Val Pro Ser Ser Leu Ser
20 25 30
ctc cct caa cga cgg acc tct ctc cga gta acc gca gcc agg gcc act 194
Leu Pro Gln Arg Arg Thr Ser Leu Arg Val Thr Ala Ala Arg Ala Thr
35 40 45
ccc aaa ctc tcc aac cgt aaa ctc cgt gtc gcc gtc atc ggt ggt gga 192
Pro Lys Leu Ser Asn Arg Lys Leu Arg Val Ala Val Ile Gly Gly Gly
50 55 60
cca gca ggc ggg gca get gca gag act cta gca caa gga gga atc gag 290
Pro Ala Gly Gly Ala A1a Ala Glu Thr Leu Ala Gln Gly Gly Ile Glu
65 70 75 80
acgatt ctc atc cgtaag atggac tgcaag ccttgc ggtggc 288
gag aat
ThrIle Leu Ile ArgLys MetAsp CysLys ProCys GlyGly
Glu Asn
85 90 95
gcgatt cct ctc atggtc ggagaa aacttg ccgttg gatatt 336
tgt ttc
AlaIle Pro Leu MetVal GlyGlu AsnLeu ProLeu AspIle
Cys Phe
18
CA 02339519 2001-02-02
WO 00/08169 PCTJEP99/05467
100 105 110
att gat cgg aga gtg acg aag atg aag atg att tcg ccg tcg aac att 384
Ile Asp Arg Arg Val Thr Lys Met Lys Met Ile Ser Pro 5er Asn Ile
115 120 125
get gtt gat att ggt cgt acg ctt aag gag cat gag tat ata ggt atg 432
A1a Val Asp Ile Gly Arg Thr Leu Lys Glu His Glu Tyr Ile Gly Met
130 135 140
gtg aga aga gaa gtt ctt gat get tat ctg aga gag aga get gag aag 480
Val Arg Arg Glu Val Leu Asp Ala Tyr Leu Arg Glu Arg Ala Glu Lys
145 150 155 160
agt gga gcc act gtg att aac ggt ctc ttc ctt aag atg gat cat ccg 528
Ser Gly Ala Thr Val Ile Asn Gly Leu Phe Leu Lys Met Asp His Pro
165 170 175
gag aat tgg gac tcg ccg tac act ttg cat tac act gag tac gat ggt 576
Glu Asn Trp Asp Ser Pro Tyr Thr Leu His Tyr Thr Glu Tyr Asp Gly
180 185 190
aaa act gga get aca ggg acg aag aaa aca atg gag gtt gat get gtc 624
Lys Thr Gly Ala Thr Gly Thr Lys Lys Thr Met Glu Val Asp Ala Val
195 200 205
att gga get gat gga get aac tct agg gtt get aaa tct att gat get 672
Ile Gly Ala Asp Gly Ala Asn Ser Arg Val A1a Lys Ser Ile Asp Ala
210 215 220
ggt gat tac gac tac gca att gca ttt cag gag agg att agg att cct 720
Gly Asp Tyr Asp Tyr Ala Ile Ala Phe Gln Glu Arg Ile Arg Ile Pro
225 230 235 2q0
gat gag aaa atg act tac tat gag gat tta get gag atg tat gtt gga 768
Asp Glu Lys Met Thr Tyr Tyr Glu Asp Leu Ala Glu Met Tyr Val Gly
245 250 255
gat gat gtg tcg ccg gat ttc tat ggt tgg gtg ttc cct aag tgc gac 816
Asp Asp Val Ser Pro Asp Phe Tyr Gly Trp Val Phe pro Lys Cys Asp
. 260 265 270
cat gta get gtt gga aca ggt act gtg act cac aaa ggt gac atc aag 864
His Val Ala Val Gly Thr Gly Thr Val Thr His Lys Gly Asp Ile Lys
275 280 285
aag ttc cag ctc gcg acc aga aac aga get aag gac aag att ctt gga 912
Lys Phe Gln Leu Ala Thr Arg Asn Arg A1a Lys Asp Lys Ile Leu Gly
19
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
290 295 300
ggg aag atc atc cgt gtg gag get cat ccg att cct gaa cat ccg aga 960
Gly Lys Ile Ile Arg Val Glu Ala His Pro Ile Pro Glu His Pro Arg
305 310 315 320
cca cgt agg ctc tcg aaa cgt gtg get ctt gta ggt gat get gca ggg 1008
Pro Arg Arg Leu Ser Lys Arg Val Ala Leu Val Gly Asp Ala Ala Gly
325 330 335
tat gtg act aaa tgc tct ggt gaa ggg atc tac ttt get get aag agt 1056
Tyr Val Thr Lys Cys Ser Gly Glu Gly Ile Tyr Phe Ala Ala Lys Ser
340 345 350
gga aga atg tgt get gaa gcc att gtc gaa ggt tca cag aat ggt aag 1104
Gly Arg Met Cys Aia Glu Rla Ile Val Glu Gly Ser Gln Asn Gly Lys
355 360 365
aag atg att gac gaa ggg gac ttg agg aag tac ttg gag aaa tgg gat 1152
Lys Met Ile Asp Glu Gly Asp Leu Arg Lys Tyr Leu Glu Lys Trp Asp
370 375 380
aag aca tac ttg cct acc tac agg gta ctt gat gtg ttg cag aaa gtg 1200
Lys Thr Tyr Leu Pro Thr Tyr Arg Val Leu Asp Val Leu Gln Lys Val
385 390 395 400
ttt tac aga tca aat ccg get aga gaa gcg ttt gtg gag atg tgt aat 1248
Phe Tyr Arg Ser Asn Pro Ala Arg Glu Ala Phe Val Glu Met Cys Asn
405 410 415
gat gag tat gtt cag aag atg aca ttc gat agc tat ctg tac aag cgg 1296
t' Asp Glu Tyr Val Gln Lys Met Thr Phe Asp Ser Tyr Leu Tyr Lys Arg
420 425 430
gtt gcg ccg ggt agt cct ttg gag gat atc aag ttg get gtg aac acc 1349
Val Ala Pro Gly Ser Pro Leu Glu Asp Ile Lys Leu Ala Val Asn Thr
435 440 445
att gga agt ttg gtt agg get aat get cta agg aga gag att gag aag 1392
Ile Gly Ser Leu Val Arg Ala Asn Ala Leu Arg Arg Glu Ile Glu Lys
450 455 460
ctt agt gtt taagaaacaa ataatgaggt ctatctcctt tcttcatctc 1491
Leu Ser Val
465
tatctctctt tttttgtctg ttagtaatct atctacac 1479
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
<210> B
<211> 467
<212> PRT
<213> Arabidopsis thaliana
<400> 8
Met A1a Thr Thr Val Thr Leu Lys Ser Phe Thr Gly Leu Arg Gln Ser
1 5 10 15
Ser Thr Glu Gln Thr Asn Phe Val Ser His Val Pro Ser Ser Leu Ser
20 25 30
Leu Pro Gln Arg Arg Thr Ser Leu Arg Val Thr Ala A1a Arg Ala Thr
35 40 45
(.
Pro Lys Leu Ser Asn Arg Lys Leu Arg Val Ala Val Ile Gly Gly Gly
50 55 60
Pro A1a Gly Gly Ala Ala Ala Glu Thr Leu Ala Gln Gly Gly Ile Glu
65 70 75 80
Thr Ile Leu Ile Glu Arg Lys Met Asp Asn Cys Lys Pro Cys Gly Gly
85 90 95
Ala Ile Pro Leu Cys Met Val Gly Glu Phe Asn Leu Pro Leu Asp Ile
100 105 110
Ile Asp Arg Arg Val Thr Lys Met Lys Met Ile Ser Pro Ser Asn Ile
115 120 125
Ala Val Asp Ile Gly Arg Thr Leu Lys Glu His Glu Tyr Ile Gly Met
130 135 140
Val Arg Arg Glu Val Leu Asp A1a Tyr Leu Arg Glu Arg Ala Glu Lys
145 150 155 160
Ser Gly Ala Thr Val Ile Asn Gly Leu Phe Leu Lys Met Asp His Pro
165 170 175
Glu Asn Trp Asp Ser Pro Tyr Thr Leu His Tyr Thr Glu Tyr Asp Gly
180 185 190
. Lys Thr Gly Ala Thr Gly Thr Lys Lys Thr Met Glu Val Asp Ala Val
195 200 205
Ile Gly Ala Asp Gly A1a Asn Ser Arg Val A1a Lys Ser Ile Asp Ala
210 215 220
21
CA 02339519 2001-02-02
WO 00/08169 PCT/EP99/05467
Gly Asp Tyr Asp Tyr Ala Ile Ala Phe Gln Glu Arg Ile Arg Ile Pro
225 230 235 240
Asp Glu Lys Met Thr Tyr Tyr Glu Asp Leu Ala Glu Met Tyr Val Gly
245 250 255
Asp Asp Val Ser Pro Asp Phe Tyr Gly Trp Val Phe Pro Lys Cys Asp
260 265 270
His Val Ala Val Gly Thr Gly Thr Val Thr His Lys Gly Asp Ile Lys
275 280 285
Lys PheGlnLeu AlaThr Arg Arg Lys AspLys LeuGly
Asn Ala Ile
290 295 300
Gly LysIleIle ArgVal GluAlaHis ProIle proGlu ProArg
His
305 310 315 320
Pro ArgArgLeu SerLys ArgValAla LeuVal GlyAsp AlaGly
Ala
325 330 335
Tyr ValThrLys CysSer GlyGluGly IleTyr PheAla LysSer
Ala
340 345 350
Gly ArgMetCys AlaGlu AlaIleVal GluGly SerGln GlyLys
Asn
355 360 365
Lys Met IleAsp GluGly LeuArg LysTyr LeuGluLys TrpAsp
Asp
370 375 380
Lys Thr TyrLeu ProThr TyrArgVal LeuAsp ValLeuGln LysVal
385 390 395 400
Phe Tyr ArgSer AsnPro AlaArgGlu A1aPhe ValGluMet CysAsn
405 410 415
Asp Glu TyrVal GlnLys MetThrPhe AspSer TyrLeuTyr LysArg
420 425 430
Val Ala ProGly SerPro LeuGluAsp IleLys LeuAlaVal AsnThr
435 440 qq5
Ile Gly SerLeu ValArg AlaAsnAla LeuArg ArgGluIle GluLys
450 455 460
Leu Ser Val
465
22