Note: Descriptions are shown in the official language in which they were submitted.
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
ENZYMATIC SYNTHESIS OF ssDNA
The present invention relates to the production of single stranded DNA {ssDNA)
in
yeast, prokaryotic, and eukaryotic cells from a set of genetic elements
delivered to the cell by a
vector system. The ss DNA is produced in the cell with minimal vector
sequences which could
interfere with the intended function of the ssDNA in the cell.
So far as is known, there is no method for producing single-stranded
deoxyribonucleic
acid (ssDNA) species in eukaryotic cells which do not contain intervening
and/or flanking
vector sequences. The scientific and patent literature does include the
disclosure of cDNA-
producing vectors (see A. Ohshima, et al., 89 Proc. Nati. Aead. Sci. USA 1016-
1020 (1992);
S. Inouye, et al., 3 Current Opir~. Genet. Develop. 713-718 ( 1993); O.
Mirochnitchenko, et al.,
269 J. Biol. Chem. 2380-2383 (;1994); J.-R. Mao, et al., 270 J. Biol. Chem.
19684-19687
( 1995); and U. S. Patent No. 5,436,141 ), but that system does not appear to
have demonstrated
the ability to produce ssDNA in eukaryotic cells without intervening vector
sequences which
can interfere with the intended function of the ssDNA product. It is,
therefore, an object of the
present invention to provide a DANA construct which directs the synthesis of
ssDNA in vitro or
in vivo with reduced or eliminated contiguous and/or intervening nucleotide
vector sequences.
It is also an object of the present invention to provide a method for
producing ssDNA
and/or dsDNA in vivo for use as aptamers to which proteins bind for producing
a therapeutic
effect in a living organism.
It is also an object of the present invention to provide nucleic acid
sequences, and a
method of introducing such sequences into living cells, for producing a
desired effect in a cell,
tissue, or organism.
According to the present invention, there is provided a set of genetic
elements for
delivery into a cell comprising a nucleic acid construct comprising a sequence
of interest, and a
primer binding site for a reverse transcriptase located in a 3' position with
respect to the
sequence of interest.
The set of genetic elements of the present invention provides an efficient
system for
directing the synthesis of a stable, single-stranded nucleic acid sequence,
both in vivo and in
vitro. The single-stranded nucleic acid sequence may be used to provide a
desired effect in a
cell, tissue or organism. Becau:>e production of the single-stranded nucleic
acid sequence of
SUBSTITUTE SHEET (RULE Z6)
CA 02346155 2001-04-09
WO 00lZ2113 PCT/US99/23933
interest takes place within the cell, prior art problems arising from delivery
of the single-
stranded nucleic acid sequence to the cell are overcome, or at least
alleviated.
Because of the arrangement of the nucleic acid construct, with the primer
binding site
in a position which is 3' to the sequence of interest, there is no limit to
the size or type of
sequence of interest that may be produced using the nucleic acid construct of
the present
invention, and the construct may be easily incorporated into a vector for
delivery by any desired
route to a target cell.
Reverse transcription may be earned out by a reverse transcriptase which is
endogenous to the cell (e.g. in the case of infection by human
immunodeficiency virus or simian
immunodeficiency virus) or the set of genetic elements may, preferably,
further comprise a
reverse transcriptase gene.
In the case that the set c>f genetic elements comprises a reverse
transcriptase gene, the
reverse transcriptase gene is, preferably, polycistronically transcribable
with the sequence of
interest and primer binding site.
I 5 Preferably, the reverse transcriptase gene is located on the same nucleic
acid construct
as the sequence of interest and primer binding site and, more preferably, the
reverse
transcriptase gene is located in a 5' position with respect to said sequence
of interest and 3'
primer binding site.
The reverse transcripta.se gene may encode reverse transcriptase or a reverse
transcriptase/RNAse H polyprotein.
The gene encoding reverse transcriptase/RNAse H polyprotein may suitably be
derived
from Moloney murine leukaemia virus, human immunodeficiency virus, or simian
immunodeficiency virus.
Where the set of genetic elements includes a reverse transcr-iptase gene, the
primer
binding site is, preferably, specific for the reverse transcriptase encoded by
the reverse
transcriptase gene. Alternatively, the primer binding site is, preferably,
specific for am
endogenous reverse transcriptase.
Preferably, the primer binding site is complementary to a transfer RNA (tRNA).
Preferably, the set of genetic elements of the present invention also
comprises a
promoter and, optionally, an enhancer for each of said sequence of interest
and/or said reverse
2.
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
transcriptase gene. More preferably, the promoter and/or enhancer is a
eukaryotic promoter
and/or enhancer.
The promoter may be a constitutive, inducible, wide-spectrum or tissue-
specific
promoter.
S Preferably, the set of genetic elements of the present invention further
comprises a
polyadenylation tail sequence located in a 3' position with respect to the
sequence of interest
and 3' primer binding site. The polyA tail provides stability of the mRNA
transcript.
Preferably, the sequence of interest is an antisense sequence. The present
invention,
thus, has far reaching uses in the field of antisense therapy, particularly in
treating pathological
conditions by regulating gene function,
The sequence of interest may also be an aptamer (i.e. an oligonucleotide that
binds to a
non-oligonucleotide target e.g. a protein). Thus, it can, again, be seen that
the present
invention, has far reaching therapeutic uses.
Preferably, the nucleic acid construct is DNA.
1 S Preferably, the set of genetic elements according to any one of the
preceding claims is
incorporated into at least one vector.
For example, the sequence of interest and 3' primer binding site may be
incorporated
into a first vector, with the reverse transcriptase gene incorporated into a
second vector.
Alternatively, the reverse: transcriptase gene, sequence of interest and
primer binding
site may be incorporated into a single vector. In this latter case, the
reverse transcnptase gene
is, preferably, located in a 5' position with respect to the sequence of
interest and 3' primer
binding site.
According to a preferred embodiment of the invention, there is provided a set
of
genetic elements adapted for delivery into a cell comprising
(a) a sequence of interest and a 3' primer binding site; and
(b) a reverse transcriptase gene,
said sequence of interest and 3' primer binding site, and said reverse
transcriptase gene being
incorporated into at last one vector for delivery into the cell.
The nucleic acid constructs of the present invention are such that they may be
incorporated into commercially available delivery vectors for mammalian and
human
3
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCTNS99/23933
therapeutic purposes, and may bE: administered by any feasible route,
depending on the target
cell.
According to the present invention, there is also provided a vector which
comprises:
(a) a primer binding site and as insertion site for a sequence of interest,
the
primer binding site being located in a 3' position with respect to the
insertion site; and
(b) a reverse transcriptase gene.
Preferably, the reverse transcriptase gene is located in a 5' position with
respect to the insertion
site and 3' primer binding site.
According to another aspect of the present invention, there is provided a
vector system
which comprises a first vector, comprising an insertion site for a sequence of
interest and a 3'
primer binding site, and a second vector which comprises a reverse
transcriptase gene.
Preferably, the vector or vector system of the present invention is a plasmid
or modified
viral construct.
Preferably, the reverse transcriptase gene is operably linked to an expression
control
sequence.
The vector or vector systems of the present invention may be advantageously
employed to deliver antisense, sf;nse, triplex, or any other single-stranded
nucleotide sequence
of interest into a cell, using known digestion and ligation techniques to
splice the sequence of
interest into the vector. The vector or vector system described herein
provides all the
necessary signalling instructions and enzymatic functions to allow a host cell
to produce a
single-stranded nucleic acid molecule having a desired sequence.
The vector or vectors systems of the present invention may also be designed to
allow
the primer binding site to be removed and exchanged, so that different primer
binding sites can
be used, depending upon the requirements of the user and the specificity of
the reverse
transcriptase being used.
Also provided by the present invention is a host cell stably transformed or
transfected
with a vector or vector system of the present invention, in particular, a
eukaryotic cell stably
transformed or transfected with a vector or vector system of the present
invention. Eukaryotic
cells include yeast or plant cells, or mammalian cells.
4
SUIBSTITUTE SHEET (R,ULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/IlS99/23933
According to the present invention there is further provided a kit for
producing a single
stranded nucleic acid sequence, which kit comprises a vector or vector system
according to the
present invention, and a restriction endonuclease for the insertion site.
According to another aspect of the present invention, there is provided a kit
for
producing a single-stranded nucleic acid sequence, which kit comprises a
vector or vector
system according to the present invention, a container for the vector/vector
system, and
instructions for use of the vector/vector system.
According to the present invention, there is further provided an iu vivo or iu
vitro
method of producing a single:-stranded nucleic acid sequence of interest,
which method
comprises the steps of introducing a nucleic acid construct into a target
cell, the nucleic acid
construct comprising a sequence of interest and a primer binding site located
in a 3' position
with respect to the sequence of interest, transcribing the nucleic acid
construct into mRNA and
reverse transcribing the mRNA into cDNA.
Preferably, the method further comprises the step of removing the mRNA from an
mRNA/cDNA heteroduplex formed by reverse transcription of the mRNA.
Reverse transcription may be carried out either by a reverse transcriptase
expressed by
a reverse transcriptase gene introduced into the target cell, or by a reverse
transcriptase which
is endogenous to the target cell (e.g. where the target cell has been infected
with human
immunodeficiency virus or simian immunodeficiency virus).
The mRNA transcript may be removed from the mRNA/cDNA heteroduplex by means
of RNAse H. Preferably, the RNAse H is expressed from a gene encoding a
reverse
transcriptase/RNAse H polyprotc:in introduced into the target cell.
Where the single-stranded nucleic acid sequence is prepared by an in vitro
method of
the present invention, the method may comprise the further step of isolating
the mRNA
transcript, mRNA/cDNA heterocluplex and/or single stranded cDNA from the
target cell.
Also provided by the present invention, are a single-stranded cDNA transcript,
an
inhibitory nucleic acid molecule, (e.g. an antisense sequence or an aptamer),
an mRNA
transcript and/or a heteroduplex molecule produced by the iu vivo or in vitro
method of the
present mventron.
An inhibitory nucleic acid: molecule may be single-stranded DNA synthesized
from the
mRNA transcript, or the mRNA transcript itself, which can specifically bind to
a
5
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
complementary nucleic acid sequence. Such inhibitory nucleic acid molecules
are particularly
useful for regulating gene function. An inhibitory nucleic acid molecule may
also be an oligo
nucleotide that specifically binds to an RNA or DNA-binding protein, or an
oligo-nucleotide
that binds to a biomolecule, e.g. thrombin, bradykinin or PGF2oc, which does
not normally bind
to RNA or DNA.
According to the present invention there is further provided a pharmaceutical
composition which comprises a set of genetic elements, a vector or vector
system, or a host
cell according to the present invention, together with a pharmacologically
acceptable adjuvant,
diluent or carrier.
According to the present invention there is also provided a set of genetic
elements, a
vector or vector system, or a host cell according to the present invention for
use in therapy,
especially for use in delivering am inhibitory nucleic acid molecule to a
target cell. The set of
genetic elements, vector and .rector systems, and host cells of the present
invention are
particularly useful for alleviating pathological conditions by regulating gene
expression.
According to a further aspect of the present invention, there is provided the
use of a set
of genetic elements, vector or ve;etor system, or host cell according to the
present invention for
the manufacture of a medicament for alleviating a pathological condition by
regulating gene
expression, especially for alleviating a pathological condition by delivery of
an inhibitory nucleic
acid molecule to a target cell. Other uses are also disclosed.
The sets of genetic elements, vectors, vector systems and host cells of the
present r
invention may be used for the: prophylactic or therapeutic treatment of a wide
range of
conditions or diseases, particularly conditions or diseases which are caused
by abnormal or
altered gene expression, or conditions or diseases which may be alleviated by
regulating gene
expression.
The sets of genetic elements, vectors, host cells, kits and methods of the
present
invention may be used to produce single-stranded nucleic acid molecules or
virtually any
predefined or desired nucleotide base composition in a host cell, and are
adaptable and
applicable to any bmivo or i~r vi~'ro system.
According to a preferred embodiment, the nucleic acid construct of the present
invention is an artificially synthesised, recombinant, chimeric and/or
heterologous product and
the sequence of interest may be foreign to the host cell in which it is
introduced.
6
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00lZ2113 PCTNS99/23933
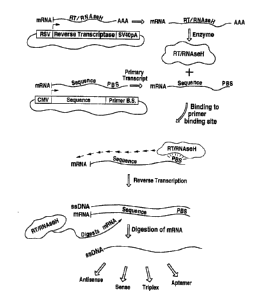
Figure 1 A referenced in the following description is a schematic view of a
plasmid
containing genetic elements encoding the sequence of interest and a primer
binding site for
reverse transcriptase.
Figure 1 B is a schematic view of a plasrnid containing a gene for reverse
transcriptase.
Figure 1 C is a schematic view of a plasmid containing genetic elements
encoding the
sequence of interest, a primer binding site, and a gene for reverse
transcriptase.
Figure 2 is a schematic diagram illustrating one embodiment of the method of
the
present invention.
A vector (as used herein, the term "vector" refers to a plasmid or modified
viral
construct, or any other suitable vehicle, used to deliver and/or manipulate
nucleic acid
sequences of interest) was de<.~igned to produce ssDNA irr nivo. The vector
contains all
necessary signaling instructions and enzymatic functions to allow the host
cell to produce the
ssDNA encoding a desired sequence (a "sequence of interest"). Described are a
set of genetic
elements adapted for delivery into a cell by incorporation into the vector for
synthesizing
I 5 ssDNA in vitro or irr vivo. They include ( 1 ) a reverse transcr-iptase
gene, (2) a genetic element
that supplies the template for the desired ssDNA sequence of interest, and (3)
a second genetic
element located proximal to the genetic element encoding the sequence of
interest that supplies
the primer site for reverse transcription by the reverse transcriptase
molecule. The vector also
contains appropriate promoter(s)/enhancer(s). Also described herein is a
method to construct a
vector into which these genetic f:lements have been incorporated.
Regarding the reverse transcriptase gene which is the first component of the
cassette, the
reverse transcriptase gene from 'the Moloney Murine Leukemia Virus (MoMuLV)
was used to
advantage in the examples described. Many other retroviral reverse
transcriptase genes may be
used to advantage in the cassette of the present invention, it being preferred
that the reverse
transcriptase gene is regulated by an appropriate upstream promoter/enhancer
such as the
Cytomegalovirus (CMV) or Rouse Sarcoma Virus (RSV) promoter for expression in
eukaryotic cells.
The reverse transcriptase gene also preferably includes a downstream
polyadenylation signal sequence so that the mRNA produced from the reverse
transcriptase gene includes a 3' poly(A) tail for mRNA stability. As known to
those skilled
in the art, multiple poly(A) tails are available and are routinely used for
production of
7
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/Z3933
expressed eukaryotic genes. T'he reverse transcriptase produced in the cell
synthesizes a
complementary DNA (cDNA) from the primary mRNA transcript transcribed from the
template encoding the genetic element that includes the sequence of interest
as described
below. The RNase H activity of the reverse transcriptase, along with
endogenous RNase
H activity within the cell, degrades the mRNA component of the heteroduplex
RNA/cDNA hybrid to produce: ssDNA Ill VIVD.
The second component included in the cassette encodes a nucleic acid sequence
that
provides the template for synthesis of ssDNA in target cells. It is this
element that includes the w
sequence of interest. As is the case for the above reverse transcriptase gene,
this genetic
element is preferably regulated by an appropriate wide spectrum or tissue-
specific
promoter(s)/enhancer(s), such as the SV-40 promoter, or combination of
promoter:(s)/enhancer(s), located upstream of the genetic element. Those
skilled in the art
who have the benefit of this disclosure will also recognize that a number of
tissue-specific or
wide spectrum promoters/enhancers, or combinations of promoters/enhancers may
be used to
1 S advantage to regulate the reverse transcriptase gene and sequence of
interest. Although a Iist
of all available promoters/enhancers is not needed to exemplify the invention,
the
promoters/enhancers may be constitutive or inducible and may include the CMV
or RSV (non-
cell type specific) or GFAP (tissue specific) promoters/enhancers listed here
and many other
viral or mammalian promoters. Representative promoters/enhancers that are
appropriate for
use in connection with the prevsent invention may include, but are not limited
to, HSVtk
(McKnight el al., 217 Science 31.6, 1982), human beta-globulin (Breathnach e~
crl., SO Ann.
Rev. of Biochem. 349, I 981 ), beta-actin (Kawamoto et al., 8 Mol. Cell Biol.
267, 1988), rat
growth hormone (Larsen et al, 83 Proc. Natl. Acad. Sci. U.S.A. 8283, 1986),
MMTV
(Huang et al., 27 Celi 245 1981 ), adenovirus 5 E2 (Imperiale, et crl., 4 Mol.
Cell. Biol. 875,
1984), SV40 (Angel e~ al., 49 C'.ell 729, 1987), a-2-macroglobulin (Kunz, et
al., 17 Nucl.
Acids Res. 1121, 1989), MHC class I gene H-2kb (Blanar et al., 8 EMBO J. 1139,
1989), and
thyroid stimulating hormone (Chatterjee et al., 86 Proc. Natl. Acad. Sci.
U.S.A. 9114,
1989).
For expression in eukaryotic cells, the sequence of interest is followed
downstream
by a genetic element encodinf; for a prymer-binding site (PBS) for initiation
of cDNA
synthesis by reverse transcriptiion. The PBS is a sequence that is
complementary to a
8.
SL;rgSTITUTE SHEET (RULE Z6)
CA 02346155 2001-04-09
PCT/US99I23933
W O 00/22113
transfer RNA (tRNA) which resides within the eukaryotic target cell. The PBS
included in
the presently preferred set of gf;netic elements described herein was taken
from the actual
18 nucleotide sequence region of MoMuLV. However, any PBS that is matched to
the
reverse transcriptase that comprises the set of genetic elements may be
utilized for this
purpose. Multiple copies of the sequences of interest, each with its
corresponding PBS,
can be incorporated into the vector for delivery to a cell in accordance with
the method of
the present invention if desired, for example, for use in delivering anti-
sense sequences to
various regions of a gene within the target cell.
The mRNA primary transcript transcribed from the genetic element acts as the
template used by the reverse transcriptase described above to synthesize and
process the
sequence of interest, which as noted above, can be any desired ssDNA. The mRNA
primary transcript contains a primer binding site (PBS) downstream to the
sequence of
interest. The PBS is exclusively recognized by a "primer tRNA." To those
skilled in the
art, tRNAs are endogenous to cells. Each tRNA has the ability to recognize a
unique
sequence (i.e., .codon) on the mRNA transcript coding .for an amino acid, and
has the
ability to covalentiy link to a specific amino acid (i.e., the tRNA becomes
"charged" when
bound to a specific amino acid;). However, a "primer tRNA" when bound to the
mRNA
transcript PBS and not covalently linked (i.e., "uncharged") with an amino
acid, may be
used to initiate ssDNA synthesis by the reverse transcriptase. For example,
the MoMuLV
reverse transcriptase used in the examples described herein, recognizes and
uses an
unchar~~ed lysine tRNA that in turn recognizes and binds to its unique
sequence in the
PBS. Thus, each PBS incorporated into the vector must contain the unique
sequence
recognized by the primer tRNA, and the primer tRNA must be one that is
recognized by
the particular reverse transcriptase utilized.
It is preferred that the vector contain other specialized genetic elements to
facilitate the identification of cells that carry the set of genetic elements
of the present
invention and/or to increase the level of expression of the sequence of
interest. The
specialized genetic elements include selectable marker genes so that the
vector can be
transformed and amplified in a prokaryotic system. For example, the most
commonly used
selectable markers are genes that confer to the bacteria (e.g., E. coli)
resistance to
antibiotics such as ampicillin, c:hloramphenicol, kanamycin (neomycin), or
tetracycline. It
9
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113
PCTNS99/23933
is also preferred that the vector contain specialized genetic elements for
subsequent
transfection, identification and expression in a eukaryotic system. For
expression in
eukaryotic cells, multiple selection strategies (e.g., Chinese Hamster
Ovarian: CHO) may
be used that confer to the cell resistance to an antibiotic or other drug or
alter the
phenotype of the cell such as. morphological changes, loss of contact
inhibition, or
increased growth rate. Selectable markers used in eukaryotic systems include,
but are not
limited to, resistance markers for Zeocin, resistance to 6418, resistance to
aminogIycoside
antibiotics, or phenotypic selection markers such ~3-gal or green fluorescence
protein.
It will also be evident to those skilled in the art from this description that
the linear
ssDNA can be formed into an intact stem-loop ssDNA structure by the addition
of
inverted tandem repeats flanking the sequence of interest that form the "stem"
portion
after duplex formation. The~ stem-loop structure can function similarly in
many
applications as the linear ssDNA form. Such a ssDNA structure may be more
resistant to
intracellular nucleases by retaining the "ends" of a ssDNA in double stranded
form.
It will also be evident to those skilled in the art that the stem (duplex DNA)
can be
designed to contain a predetermined sequence or sequences (i.e., "aptamers")
that are
recognized and bound by specific DNA-binding proteins. Among other uses, such
a stem
structure is used in the cell as a~ competitor to titer out a selected
proteins) that regulate
specific gene expression. For example, a ssDNA stem-loop of the present
invention in a
cell such that the ''stem" contains a binding site for a selected
transcription factor such as
adenovirus E 1 a. Adenovirus F~ 1 a, like other oncogenes, modulates gene
expression of
several adenoviral and cellular genes by affecting the activity of cell-
encoded transcription
factors resulting in changing normal cells to transformed cells. (Jones et
al., Genes Dev.
2, 267-28 I ( 1988)). The duplex structure of the stem thus functions to "bind
up" the
factor, preventing the protein from binding a promoter and thus inhibiting the
expression
of a particular deleterious gene. To those skilled in the art, it will be
clear that the duplex
stem structure may optionally contain multiple binding sites, for example,
sites which are
recognized by various transcription factors that actively regulate expression
of particular
gene. For example, adenovirus E 1 a has been found to repress transcription of
the
collagenase gene via the phorbol ester-responsive element, a promoter element
responsible
for the induction of transcription by 12-O-tetradecanolyphorbol 13-acetate
(TPA), by a
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
W O 00/22113
PCTNS99/23933
number of other mitogens, and by the ras, mos, src, and trk oncogenes. The
mechanism
involves inhibition of the function of the transcription factor family AP-1.
Offringa et al.,
62 Cell 527-538 ( 1990).
In another aspect which will be recognized by those skilled in the art, the
present
invention is used to construct complex secondary ssDNA structures in the loop
portion of
the DNA transcript produced in accordance with the present invention. Such
secondary
structure is engineered to serve any of several functions. For instance, the
sequence of
interest optionally includes {but is not limited to) a sequence which is
incorporated into the
loop portion of the single-stranded cDNA transcript to form so-called "clover
leaf' or
"crucible" like structures such as those found in the long terminal repeats of
adenoassociated virus or in retrotransposons. Under correct circumstances,
such structure
is integrated in site-specific manner into the host genome.
Because a vector incorporating the set of genetic elements of the present
invention
is adaptable for incorporation into multiple commercially available delivery
vectors for
l5 mammalian and human therapeutic purposes, multiple delivery routes are
feasible
depending upon the vector chosen for a particular target cell. For example,
viral vectors
are presently the most frequently used means for transforming the patient's
cells and
introducing DNA into the genome. In an indirect method, viral vectors,
carrying new
genetic information, are used to infect target cells removed from the body,
and these cells
are then re-implanted (i.e., cx rW o). Direct in oivo gene transfer into
postnatal animals has
been reported for formulations of DNA encapsulated in liposomes and DNA
entrapped in
proteoliposomes containing viral envelope receptor proteins (Nicolau et al.,
Proc. Natl.
Acad Sci USA 80:1068-107:? (1983); Kaneda et al., Science 243:375-378 (1989);
Mannino et al., Biotechniques ci:682-690 ( 1988). Positive results have also
been described
with calcium phosphate co-pre:cipitated DNA (Benvenisty and Reshef, Proc.
Natl. Acad
Sci USA 83:9551-9555 (1986.)). Such systems include intravenous,
intramuscular, and
subcutaneous injection, as wel!I as direct intra-tumoral and intra-cavitary
injections. The
set of genetic elements, when incorporated into the vector of choice, can also
be
administered through transmucosal, rectal, oral, or inhalation-type methods of
delivery.
The vector incorporating the set of genetic elements of the present invention
is
advantageously employed to deliver antisense, sense, triplex, or any other
single-stranded
11
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99I23933
nucleotide sequence of interest, using known digestion and ligation techniques
to splice
the particular sequence of interest into the vector in the presence or absence
of inverted
tandem repeats. Those skilled in the art who have the benefit of this
disclosure will also
recognize that the above-described signals used for expression within
eukaryotic cells may
be modified in ways known in the art depending upon the particular sequence of
interest.
The most likely change is to change the promoter so as to confer advantageous
expression
characteristics on the sequence: of interest in the system in which it is
desired to express
the sequence of interest. There are so many possible promoters and other
signals, and
they are so dependent on the particular target cell for which the sequence of
interest has
been selected, that it is impossible to list all the potential enhancers,
inducible and
constitutive promoter systems, and/or poly(A) tailing systems which may be
preferred for
a particular target cell and sequence of interest.
The present invention i s also utilized to produce inhibitory nucleic acids
for use in
therapeutics in ~~iro or in vilrr~. Inhibitory nucleic acids may be ssDNA
synthesized from the
I S mRNA template or the mRNA template itself, which can specifically bind to
a complementary
nucleic acid sequence. By binding to the appropriate target sequence, an RNA--
RNA, a DNA-
-DNA, or RNA-DNA duplex o~r triplex is formed. More commonly, these nucleic
acids are
often termed "antisense" because they are usually complementary to the sense
or coding strand
of the gene, but the "sense'' sequence is also utilized in the cell for
therapeutic purposes. For
example, the identification of oligonucleotides that specifically bind to
biomolecules that do not
normally bind to RNA or DN,A has now been demonstrated for a number of
biomolecules that
vary widely in size, structure <~nd composition. These molecules include: ( 1
) thrombin, a
multifi~nctional regulatory protein that converts fibrinogen to fibrin in the
process of clot
formation; (2) bradykinin, a nonapeptide kinin involved in blood pressure
regulation and
implicated in hypotension; (3) P(JF2.alpha., a prostaglandin or fatty acid
derivative that exhibits
hormonal activity. Additionally, the interaction of oligonucleotides with
biomolecules whose
natural biological function is primarily extracellular has now been
demonstrated. U. S. Pat. No.
5,840,867. The term "inhibitary nucleic acids" as used herein, therefore,
refers to both "sense"
and "antisense" nucleic acids.
By binding to the target nucleic acid, an inhibitory nucleic acid inhibits the
function of
the target nucleic acid. This inhibitory effect results from, for example,
blocking DNA
12
SiJBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99I23933
transcription, processing or poly(A) addition to mRNA, DNA replication,
translation, or
promoting inhibitory mechanisms of the cells, such as promoting RNA
degradation. Inhibitory
nucleic acid methods therefore encompass a number of different approaches to
altering
expression of genes. An example of an antiherpes virus inhibitory nucleic acid
is ISIS 2922
S (ISIS Pharmaceuticals, Carlsbad, CA) which has activity against CMV (see
Biotechnology
News 14:5). These different types of inhibitory nucleic acid technologies are
described in
Helene, C. and Toulme, J. ( 1 ~>90) Biochim. Biophys. Acta. 1049:99-125, which
is referred to
hereinafter as "Helene and Toulme."
In brief, inhibitory nucleic acid therapy approaches can be classified into (
I ) those that
target DNA sequences, (2) those that target RNA sequences (including pre-rnRNA
and
mRNA), (3) those that target proteins (sense strand approaches), and (4) those
that cause
cleavage or chemical modification of the target nucleic acids. The first
approach contemplates
several categories. Nucleic acids are. designed to bind to the major groove of
the duplex DNA
to form a triple helical or "triplex" structure. Alternatively, inhibitory
nucleic acids are desilmed
to bind to regions of single stranded DNA resulting from the opening of the
duplex DNA
during replication or transcription. More commonly, inhibitory nucleic acids
are designed to
bind to mRNA or mRNA precursors. Inhibitory nucleic acids are used to prevent
maturation
of pre-mRNA. Inhibitory nucleic acids may be designed to interfere with RNA
processing,
splicing or translation. In the second approach, the inhibitory nucleic acids
are targeted to
mRNA. In this approach, the inhibitory nucleic acids are designed to
specifically block
translation of the encoded protein. Using this second approach, the inhibitory
nucleic acid can
be used to selectively suppress certain cellular functions by inhibition of
translation of mRNA
encoding critical proteins. For example, an inhibitory nucleic acid
complementary to regions of
c-myc mRNA inhibits c-myc protein expression in a human promyelocytic leukemia
cell line,
HL60, which overexpresses the c-myc proto-oncogene. See Wickstrom E. L., et
al. (1988)
PNAS 85:1028-1032 and Harel-Bellan, A., et al. (1988) Exp. Med. 168:2309-2318.
As
described in Helene and Toulme., inhibitory nucleic acids targeting mRNA have
been shown to
work by several different mechanisms to inhibit translation of the encoded
protein(s).
The inhibitory nucleic acids introduced into the cell can also utilize the
third
approach of designing the "sense' strand of the gene or mRNA to trap or
compete for the
enzymes or binding proteins involved in mRNA translation, as described in
I~elene and
13
SUBSTITUTE SHEET (RULE Z6)
CA 02346155 2001-04-09
WO 00/22113 PCTNS99/23933
Toulme. Lastly, the inhibitory nucleic acids is used to induce chemical
inactivation or
cleavage of the target genes or mRNA. Chemical inactivation occurs by the
induction of
crosslinks between the inhibitory nucleic acid and the target nucleic acid
within the cell.
In another embodiment, the present invention takes the form of a kit comprised
of
a plasmid having the above-described reverse transcriptase gene cloned therein
as well as a
multiple cloning site (MCS) into which the user of the kit inserts a
particular sequence of
interest, which may or may not include the above-described inverted tandem
repeats in
accordance with the user's intended result. The MCS is upstream from l:he
genetic
element encoding the primer binding site. The resulting plasmid is then
purified from the
cell culture in which it is maintained, lyophilized or otherwise preserved for
packaging and
shipping to the user. The kit preferably also includes the restriction
endonuclease(s) for
the MCS into which the sequence of interest is to be cloned, the ligases and
other enzymes
for inserting the sequence of interest into the plasmid, and a map of the
plasmid, along
with suitable reaction buffers.
I S Except where otherwise; indicated, standard techniques are described by
Seabrook,
e~ a!. (1989) (J. Seabrook, er al., Molecular Cloning: A Laboratory Manual
(2nd Ed.),
Cold Spring Harbor Press ( 1989), hereinafter referred to as "Maniatis, et
a~'. ( 1989)")
were utilized in the examples set out below. Several experimental designs are
presented to
illustrate the method of producing ssDNA irr vivo.
EXAMPLES
The following examples are provided for illustrative purposes only and are not
intended
to limit the scope of the invention.
Materials
The plasmid pcDNA3.U2;eo+ was purchased from Invitrogen Corp. (San L>iego, CA)
and piasmid PBK-RSV from Statagene (La Jolla, CA). Oligodeoxyr-ibonucleotides
(ODN)
were synthesized by Midland Certified Reagent Co. (Midland, TX). Polymerase
chain
reactions (PCR) were carried out using Taq DNA polymerase purchased from
Boehringer
Mannheim Corp. (Indianapolis, IN) in a Robo-gradient thermal cycler
(Stratagene {La Jolla,
CA). Restriction endonucleases and T4 DNA lipase were obtained from Boehr-
inger
Mannheim Corp. (Indianapolis, IN). The ODNs used are listed in the attached
Sequence
Listing.
14
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PC'T/U599/23933
Example t
Ifr vivo Synthesis of ssDNA in Eukar r~otic Cells
The following in vivo experiments were designed to deternvne whether ssDNA
could
be produced in intact cells. To control expression of the genetic elements
cloned into the
plasmid in these host cells, the plasmid utilized included the RSV promoter.
However, those
skilled in the art who have the benefit of this disclosure will recoginze that
any of the
eukaryotic promoters listed above can be used for this purpose.
Ptasmid Constructs. The cloning vector pssXB and the plasmids containing the
sequences to be expressed as single-stranded DNA were constructed from a
common
intermediate construct. The host strain for these manipulations was XLl-Blue
MRF'
(Stratagene, La Jolla, CA)
In the first cloning stage, to obtain the common intermediate, the vector
pcDNA3. lZeo (lnvitrogen, San Diego, CA) was digested with the restriction
en:rymes Nhe I
and Apcr I. The double-stranded oligodeoxyribonucleotide having compatible Nhe
I and Apa I
ends, which is formed by annealing the synthetic, single-stranded
oligodeoxynucleotides ODN-
PMMV(+) and ODN-PMMV(-) (see Table I ), was ligated into the digested
pcDNA3.IZeo'
to give pcPMMV. This inserrt contains the Moloney Murine leukemia virus
(MoMuLV)
reverse transcriptase promoter region. It also contains two Not I sites,
unique in pc;PMMV. In
this construct and in the plasmids deriving from this construct, the strands
designated (+) are
positioned to be transcribed into RNA from the cytomegalovirus (CMV) promoter
of
pcDNA3. t/Zeo(+).
The plasmid pssDNA-Express-A (pssXA), containing genes for MoMuLV reverse
transcriptase, was constructed from the vector pBK-RSV (Stratagene, La Jolla,
CA), also
using XL-I Blue MRF' as the host strain. A mouse cell line expressing MoMuLV
was
obtained from the American Type Culture Collection (ATCC #CRL-1858). Virus RNA
was
isolated and reverse transcribed from ODN-RT (-) (Table I). The reverse
transcript was then
PCR amplified according to the manufacturer's intructions using a kit from
Promega (Madison,
WI), primers ODN-RT (+) and ODN-RT (-), and digested with Sac I and Hind lII
(sites for
these restriction endonucleases are present in the S' and 3' primers,
respectively). The 2.4 kb
product obtained includes the sequence of the MoMuLV genome between positions
?546 and
IS
StJBSTITUTE SHEET (RULE Z6)
CA 02346155 2001-04-09
WO OOI22113 PCTNS99/23933
4908. The mature virus reverse transcriptase peptide is encoded by the
sequence between
positions 2337 and 4349 (Petropoulos, C.J. Retroviral taxonomy, protein
structure, sequences
and genetic maps. In: Retroviruses, 757, Appendix 2, Coifrn, J.M. (Ed.). Cold
Spring Harbor
Laboratory Press, Cold Spring :Harbor, New York, USA 1997), but peptides
tnrncated at the
amino terminus retain full activity (Sun, et al. ( 1998)).
The pBRK-RSV vector' was digested with Xba I and Nhe I, which removes the lac
promoter region. The Nhe I end was converted to a Sac I end using the linker
formed by
annealed oligodeoxynucleotides ODN-N>S (+) and ODN-N>S (-). The reverse
transcriptase
amplimers were ligated through the Hind III sites and this construct was
subsequently ligated
I 0 between the Scrc: I and Xba I sites of pBK-RSV to give pBK-RSV-RT.
Those skilled in the an will recognize that the set of genetic elements
comprising the
present invention are also expressed from a single plasmid made by a fusion
of, for instance, the
pc3.lDNA/Zeo-derived plasmids and the pBK-RSV-derived p(asmids such that
fiased plasmids
encode the ss-cDNA-encoding genetic element, the Mo-MuLV-RT gene, and the PBS.
pBK-
RSV-RT/MboL is digested with NsiI to release a 5.3kb fragmrent containing the
Mo-MuLV-
RT gene with an intervening his-pro linker and associated regulatory elements.
The 5.3kb
DNA fragment is ligated to a linker containing an internal EcoRI site and
digested with EcoRI.
The pc3.1/Zeo/N-M and the derivative plasmids containing test sequences are
digested with
BgIII, which recognizes a unique site on pc3.1 DNA/Zeo in the cytomegalovirus
enhancer/promoter (P CMV). The BgIII ends are ligated to Seq. ID I S and Seq.
ID 16, which
contain an internal EcoRI site. After digestion with EcoRl, the 5.3kb fragment
is ligated to
pc3.1/Zeo/N-M and derivatives to generate the plasmid.
Tissue culture studies. Stable and transient transfections are earned out by
using
lipofectant (Boehringer Mannhierrr C;orp.) using the manufacturer's
accompanying instructions.
All plasmid constructs were transfected into Cos-7, U251 and HeLa cell tines.
Assays for
ssDNA were performed by PCR, and by dot-blot analyses 24-48 hours after
transfection.
Reverse transcriptase activity was assayed using the RT-PCR assay developed by
Silver, et al.
(Silver, J., ei al. 21 Nucleic Acids Res. 3593-4 (1993)). The ss-cDNA is
isolated from cells
transfected 48-72-hr earlier using triazol reagent (Gibco Life Technologies,
Gaithersburg,
MD). Assays for specific ss-cDNA species are carried out by both PCR based
assays for
16
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCTNS99/23933
internal fragment and by denatured single stranded gel electrophoresis with
subsequent nylon
blotting and probing with an internal biotin-labeled probe.
The experiments described above demonstrate a method of production of ssDNA
irr
viro by multiple stepwise reactions using eukaryotic reverse transcriptase
reactions and various
cDNA priming reactions. Any nucleotide sequence ~f interest is produced by
this method in a
prokaryotic or eukaryotic cell. The cells were actually co-transfected with
two plasmids, one
plasmid carrying the genetic elements encoding the sequence of interest and
primer binding site
for reverse transcriptase, shown in Fig. 1 A and the other carrying the gene
for reverse
transcriptase shown in Fig. 1 B. Those skilled in the art, however, will
recognize that a single
plasmid including the genetic elements encading the sequence of interest and
PBS for reverse
transcriptase, and the gene for reverse transcriptase also can be used for
this purpose (Fig. 1 C).
Example 2
Reverse Tr:~nscriptase Activity in Transformed Cells
To determine the presence of reverse transcriptase activity in extracts of
cells
containing the pBK-RSV-RT c;onstruct, the following assay is used. This assay
relies upon
reverse transcriptase activity in protein extracts of transfarmed cells to
produce a DNA copy of
the Brome Mosaic Virus RNA l;enome (Silver, el al., 1993). The replication
cycle of this virus
does not involve a DNA intermediate, eliminating the possibility that an
amplification product
could be produced without prior reverse transcription.
Example 3
Demonstration of the presence of single-stranded DNA
in Transformed Mammalian Cells.
A PCR strategy is used to detect single-stranded DNA in transformed cells. The
product obtained from RNA extraction procedures, which presumably includes the
single-
stranded DNA is used as a template in PCR amplifications using primers
specific for the
expected single-stranded DNA molecule, which is not otherwise present in the
cells. A band of
the expected size is produced from untreated RNAIssDNA preparations and from
such
preparations that were treated with RNAase A. Use of preparations treated with
S 1 nuclease,
a highly specific, single-stranded DNA endonuclease, does not result in an
amplified product.
17
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
Example 4
A method and pharmaceutical preparation
for diagnosing and treatin pad , tholo~ical conditions
related to a dopamine rector abnormalitX
S Abnormal activity of the dopaminergic nervous system has been implicated in
a
number of motor and behavioral disorders including Parkinson's disease,
Huntington's disease,
tardive dyskinesia, certain forms of schizophrenia and other dystonias and
dyskinesias.
Dysfunctions of the dopaminergic system may be caused either by a reduced or
increased
activity of the dopaminergic system or by the inability of the systems to be
modulated by a
changing external or internal environment.
For a patient suffering from one of the above mentioned disorders, a plasmid
is
constructed tc»nclude a sequence of interest that generates an antisense
oli,gonucleotide
capable of binding specifically to an expression-controlling sequence of a
nucleic acid encoding
the dopamine receptor. The piasmid is administered under conditions whereby
the plasmid
I S enters cells expressing the dopamine receptor and generates the inhibitory
nucleotide. The
inhibitory nucleotide binds specifically to expression-controlling sequences
of such RNA
molecules, thereby selectively controlling expression of one or more dopamine
receptor
subtypes, and alleviating the pathological conditions related to their
expression. Efficacy is
tested in accordance with the method described in U.S. Patent No. 5,840,708.
Example 5
Inhibitory nucleotides to Kaposi's sarcoma-associated herpesvirus KSHV)
virion protein 2~VP2b~
Kaposi's sarcoma-associated herpes virus (KSHV) is a new human herpes virus
(HHV8) believed to cause Kaposi's sarcoma (KS). Kaposi's sarcoma is the most
common
neoplasm occurring in persons with acquired immunodeficiency syndrome (A1DS).
Approximately I S-20% of AIDS patients develop this neoplasm which rarely
occurs in
immunocompetent individuals. Epidemiologic evidence suggests that AIDS-
associated KS
(AIDS-KS) has an infectious eaiology. Gay and bisexual AIDS patients are
approximately
twenty times more likely than hemophiliac AIDS patients to develop KS, and KS
may be
associated with specific sexual practices among gay men with AIDS. KS is
uncommon among
adult AIDS patients infected tlu-ough heterosexual or parenteral HIV
transmissiorv, or among
18
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
pediatric AIDS patients infected through vertical HIV transmission. Agents
previously
suspected of causing KS include cytomegalovirus, hepatitis B virus, human
papillomavirus,
Epstein-Barr virus (EBV), human herpesvitus 6, human immunodeficiency virus
(HIV), and
Mycoplasma penetrans. Non-infectious environmental agents, such as nitrite
inhalants, also
have been proposed to play a role in KS tumorigenesis. Extensive
investigations, however,
have not demonstrated an etiologic association between any of these agents and
AIDS-KS.
Virion protein 26 (V'P26) is a component of the nucleocapsid structure in most
herpes viruses. This structure serves as a delivery mechanism for the viral
genome as it is
spread from one infected cell to another. As part of the original infecting
virus, it is recognized
as a major antigen by the immune system and can therefore be used to screen
for antibodies to
the herpes virus in patient sera a.nd as a vaccine.
For an infected patient, a plasmid is constructed using the methods described
above
to include a sequence of interest. The sequence of interest is an isolated
nucleic acid molecule
which encodes KSHV virion protein 26 or antisense or triplex oligonucleotide
molecule as
described in L7.S. Patent No. 5,840,708. The plasmid is administered under
conditions
whereby the plasmid enters infected cells and generates the inhibitory
nucleotide. The
inhibitory nucleotide binds specifically to expression-controlling sequences
of such RNA
molecules, or encoding sequences, thereby selectively controlling expression
of KSHV virion
protein 26, and alleviating the pathological conditions related to expression.
Example 6
Inhibitory nucleotides to modulate the expression of IL-8 and/or IL-8 Receptor
to control~rowth. metastasis and/or a~iogenesis in tumors.
Interleukin-8 (IL-8, neutrophil activating protein-I, or NAP-1 ) is a member
of C-X-C
chemokine family of related cytokines having broad involvement in inflammatory
responses,
tissue injury, growth regulation and cellular adhesion. Cerretti, D. P., et
al., Molecular
Characterization of Receptors for Human Interleukin-8, GRO/Melanoma Growth-
Stimulatory
Activity and Neutrophil Activating Peptide-2, Molecular Immunology, 30(4), 359-
367 (1993);
and Koch, A. E;., et al., In situ expression of cytokines and cellular
adhesion molecules in the
skin of patients with systemic: sclerosis, Pathobiology, 61(S-6), 239-46
(1993). 1I,-8 has also
been shown to have a potent stimulatory effect on angiogenesis. See, e.g.,
Koch, A. E.,
19
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PGTNS99l23933
Interleukin-8 as a Macrophage-Derived Mediator of Angiogenesis, Science, 258,
1798-1800
( 1992).
It is known that II,-8 is produced by a variety of normal human samatic cells
including monocytes/macrophages, dermal fibroblasts, vascular endothelial
cells, keratinocytes,
and mesangeal cells. Yasumoto, K., et al., Tumor Necrosis Factor Alpha and
Interferon
Gamma Synergistically Induce Interleukin 8 Production in a Human Gastric
Cancer Cell Line
Though Acting Concurrently on AP-1 and NF-kB-like Binding Sites of the
Interler,ikin 8 Gene,
J. of Biological Chemistry, 267(31 ), 22506-11 ( 1992). Apparently, such cells
produce II,-8
only when stressed, and not under conditions of normal growth and homeostasis.
Factors that
IO induce IL-8 production include inflammation, IL-1, TNF, LPS and thrombin.
It is also known
that II,-8 is commonly secreted by tumor cells. Because of its effects on
growth, it is suspected
that IL-8 has a significant role in the metastatic spread of melanoma and
other cancers.
IL-8 is a ligand for cell-membrane IL-8 Receptor, and it is thought that
interaction
between IL-8 and IL-8 Receptor is reduired for 1L-8 action. Two IL-8 receptor
genes have
been identified so far, IL-8 Receptor type A and type B. Both genes belong to
the so-called
seven transmembrane domain, G protein-coupled receptor family. Receptor A has
been shovm
to be activated by IL-8, and receptor B has been shown to be activated by IL-8
as well as other
cytokines belonging to C-X-C family including Melanoma Growth Stimulatory
Activity
(MGSA).
The role and function of IL-8 Receptor B present in cancer and other tumor
cells is
not fially elucidated. There is, however, evidence that activation of IL,-8R B
( 1 ) is involved in
the mechanism of growth regulation of melanoma and tumorigenic fibroblasts;
(2) is associated
with transformation of lung cells by asbestos, and (3 ) correlates with
metastic potential of
melanoma.
Given the growth stimulatory effect of IL-8 on cells responsive to various
tumor
growth factors., it would be advantageous to provide antisense
oligonucleotides which
modulate expression of either IL8 or IL,-8 Receptor in cancers in vivo. It
would be particularly
advantageous to provide oligonucleotides which are effective against lung
cancer and
melanoma because each of these cancers produce their own growth factors.
There are at least two major types of lung cancer, small cell lung carcinoma
(SCLC)
and non-small cell lung carcinoma (NSCLC). SCLC comprises approximately one-
fourth of
SIJBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
the cases, expresses neuroendocrine markers, and generally metastasizes early
to lymph nodes,
brain, bones, lung and liver. NSCLC comprises the majority of the remaining
lung tumor types,
and includes adeno-carcinoma, squamous cell carcinoma, and large cell
carcinoma. NSCLC is
characterized by epithelial-like growth factors and receptors, and is locally
invasive.
~ Melanoma cells, unlike normal melanocytes, can proliferate in the absence of
exogenous growth factors. 'This independence apparently reflects the
production of growth
factor and cy~tokines for autocrine growth stimulation, including TGF-.ANG.,
TCi<F-, platelet-
derived growth factor A and B chains, basic fibroblast growth factor, II,-8,
IL,-6, IL-1,
granulocyrte macrophage colony stimulating factor, and MGSA. Guo Y, et al.,
Inhibition of
Human Melanoma Growth and Metastasis in Vivo by Anti-CD44 Monolclon<al
Antibody.
Cancer Res., 54, 1561-1565 1;1994).
For a patient suffering from any of the above diseases, a plasmid is
constructed using
the methods described above to include a sequence of interest. The sequence of
interest is an
isolated nucleic acid molecule as described in U S. Patent No. 5,849,903. To
control growth,
metastasis and/'or angiogenesis, the plasmid is administered (e.g., inhalation
or direct injection
into solid tumors) under conditions whereby the plasmid enters cells and
generates the
inhibitory nucleotide. The inhibitory nucleotide binds specifically to
expression-controlling
sequences of such RNA molecules, or encoding sequences, thereby selectively
controlling
expression of IL-8 receptors, and alleviating the pathological conditions
related to expression.
Example 7
Antisense oli~onucleotide inhibition of cytomegalovirus infection.
Cytomegaloviruses (C.'NIVs) are ubiquitous in nature and are the most common
causes of intrauterine infection. Congenital infection is common in newborns
of infected
mothers. In some populations, as much as 10% of children display perinatal
infections. In a
small percentage of newborns, t:he infection is virulent, involving multiple
organs. Pronounced
involvement of the reticuloendothelial and central nervous system is typical;
and the infection is
a major cause of mental retardation. Carefixl testing demonstrates that as
many as 50% of
severely, prenatally infected adults may display neuropsychiatric disease or
deafness. Although
extraneural organs are usually spared chronic morbidity, the virus can be
detected in the kidney
for years.
21
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCTNS99I23933
A plasmid is constructed using the methods describe above to include a
sequence of
interest encoding for an inhibitory nucleotide. Oligonucleotides having a
sequence of
nucleotide bases specifically hybridizable with a selected sequence of a
cytomdgalovirus DNA
or RNA are described in U. S. Patent No. 5,442,049. The plasmid is
administered to the patient
under conditions whereby the plasmid enters cells and generates the inhibitory
nucleotide. The
inhibitory nucleotide binds specifically to expression-controlling sequences
of such RNA
molecules, or encoding sequences, thereby selectively controlling replication
of CMV, and
alleviating the pathological conditions related to CMV infection. This plasmid
is used either
prophylactically or therapeutically to reduce the severity of disease caused
by CMV.
I0 Example 8
Oligonucleotides specifically hybridizable with RNA or DNA
deriving_from a gene corres~ondin~to one of the open readinc~'frames
ULS~ ULB, UL9, U~ 20 UL27, UL29, UL30, UL42. LJL.52 and IE 175
of herpes simplex virus type 1.
I S Oligonucleotides are designed to be specifically hybridizable with DNA or
even more
preferably, RNA from one of the species herpes simplex virus type 1 (HSV-1 ),
herpes simplex
virus type (HSV-2), cytomegalovinrs, human herpes virus 6, Epstein Barr virus
(EBV) or
varicella zoster virus (VZV). Such oligonucleotides are conveniently and
desirably presented as
a pharmaceutical composition in a pharmaceutically acceptable carrier as
described in U. S.
20 Patent No. 5,514,577.
For a patient suffering from any of the above infections, a plasmid is
constructed
using the methods described above to include a sequence of interest. The
sequence of interest
is an isolated nucleic acid molecule as described in U.S. Patent No.
5,514,577. Tthe plasmid is
administered (e.g., inhalation or direct injection into solid tumors) under
conditions whereby
25 the plasmid enters cells and generates the inhibitory nucleotide. The
inhibitory nucleotide binds
specifically to expression-controlling sequences of such RNA molecules, or
encoding
sequences, from one ofthe species herpes simplex virus type I (HSV-1), herpes
simplex virus
type (HSV-2), cytomegalovirus, human herpes virus 6, Epstein Barn virus (EBV)
or varicella
zoster virus (VZV) thereby selectively controlling virus infection, and
alleviating the
30 pathological conditions related to infection.
22
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
Example 9
Antisense olieonucleotides to ~proto oncog_enes and in uarticular to the c-mvb
gene.
and the use of such oli >o~ nucleotides as antineo~lastic and
immunosunpressive agents.
The proto-oncogene c-myb is the normal cellular homologue of the avian
myeloblastosis virus-transforning gene v-myb. The c-myb gene codes for a
nuclear protein
expressed primarily in hematopoietic cells. It is a proto-oncogene, that is,
it codes fi'r a protein
which is required for the survival of normal, non-tumor cells. When the gene
is altered in the
appropriate manner, it has the potential to become an oncogene. Oncogenes are
genes whose
expression within a cell provides some fixnction in the transformation from
normal to tumor
cell. An example is the human c-myb gene which has been isolated, cloned, and
sequenced.
Majello et al, Proc. Natl. Acad. Sci. U.S.A. 83, 9636-9640 (1986).
A plasmid is constructed using the methods describe above to include a
sequence of
interest encoding for an inhibitory nucleotide. Oligonucleotides having a
sequence of
nucleotide bases specifically hybridizable with a selected sequence of the DNA
or RNA as are
described in U.S. Patent No. 5,098,890. The plasmid is administered to the
patient under
conditions whereby the plasmid enters cells and generates the inhibitory
nucleotide thus acting
as an antineoplastic or immunosuppressive agent.
Example 10
Antisense oli~onucleotides Against ICAM-1 Gene Expression
in.Interieukin-1 beta-Stimulated Cells.
It is has been hoped that inhibitors of ICAM-1, VCAM-1 and ELAM-1 expression
would provide a novel therapeutic class of anti-inflammatory agents with
activity towards a
variety of inflammatory diseases or diseases with an inflammatory component
such as asthma,
rheumatoid arthritis, allograft rejections, inflammatory bowel disease,
various dermatological
conditions, and psoriasis. In addition, inhibitors of ICAM-1, VCAM-1, and ELAM-
1 may also
be effective in the treatment of colds due to rhinovirus infection, AIDS,
Kaposi's sarcoma and
some cancers and their metastasis. To date, there are no known therapeutic
agents which
effectively prevent the expression of the cellular adhesion molecules ELAM-l,
VCAM-1 and
ICAM-1. The use of neutralizing monoclonal antibodies against ICAM-1 in animal
models
provide evidence that such inhibitors if identified would have therapeutic
benefit for asthma;
Wegner et al., Science 1990, 247, 456-459, renal allografts; Cosimi et al., J.
Immunol. 1990,
23
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
144, 4604-4612, and cardiac allografts; Isobe et al., Science 1992, 255, 1125-
1127. The use of
a soluble form of ICAM-1 molecule was also effective in preventing rhinovirus
infection of
cells in culture. Marlin et al., 344 Nature 70-72 ( 1990).
Current agents which affect intercellular adhesion molecules include synthetic
peptides, monoclonal antibodies, and soluble foams of the adhesion molecules.
To date,
synthetic peptides which block: the interactions with VCAI.VI-1 or ELAM-1 have
not been
identified. Monoclonal antibodies may prove to be useful for the treatment of
acute
inflammatory response due to expression of ICAM-1, VCAM-1 and SLAM-1. However,
with
chromic treatment, the host animal develops antibodies against the monoclona
antibodies
thereby limiting their usefulness. In addition, monoclonal antibodies are
large proteins which
may have difficulty in gaining access to the inflammatory site. Soluble forms
of the cell
adhesion molecules suffer from many of the same limitations as monoclonal
antibodies in
addition to the expense of their production and their low binding affinity.
Thus, there is a long
felt need for molecules which effectively inhibit intercellular adhesion
molecules. Antisense
oiigonucleotides avoid many of the pitfalls of current agents used to block
the effects of
ICAM-1, VCAM-1 and ELAM-1.
PCT/LJS90/02357 (Hession, et a!.) discloses DNA sequences encoding Endothelial
Adhesion Molecules (ELAMs)., including ELAM-1 and VCAM-1 and VCAM-lb. A number
of uses for these DNA sequences are provided, including ( 1 ) production of
monoclonal
antibody preparations that are reactive for these molecules which may be used
as therapeutic
agents to inhibit leukocyte binding to endothelial cells; (2) production of
ELAM peptides to
bind to the SLAM ligand on leukocytes which, in turn, may bind to SLAM on
endothelial cells,
inhibiting leukocyte binding to endothelial cells; (3) use of molecules
binding to EI~AMS (such
as anti-ELAM antibodies, or markers such as the ligand or fragments of it) to
detect
inflammation; (4) use of ELAM and SLAM ligand DNA sequences to produce nucleic
acid
molecules that intervene in SLAM or SLAM ligand expression at the translations
level using
antisense nucleic acid and riboxymes to block translation of a specific mRNA
either by masking
mRNA with antisense nucleic acid or cleaving it with a ribozyme.
A plasmid is constructed using the methods describe above to include a
sequence of
interest encoding for an inhibitory nucleotide for ICAM-1, VCAM-1 or SLAM-1.
Oligonucleotides having a sequence of nucleotide bases specifically
hybridizable with a selected
24
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
sequence of ICAM-1, VCAM-1 or SLAM-I DNA or RNA are described in U.S. Patent
No.
5,843,738. The plasmid is administered to the patient under conditions whereby
the plasnvd
enters cells and generates the inhibitory nucleotide. The inhibitory
nucleotide binds specifically
to expressiorncontrolling sequences of such RNA molecules, or encoding
sequences, thereby
selectively controlling the expression of ICAM-1, VCAM-1 or ELAM-1, and
alleviating the
pathological conditions related to ICAM-1, VCAM-1 and SLAM-I expression. This
plasmid
is used either prophylactically or therapeutically to reduce the severity of
inflammation caused
by ICAM-1, VC.'AM-1 and ELAM-1.
Example 11
Protein-Binding Oli~onucleotides ~Aptamers) Specifically Bind Target Molecules
The field of rational drub; design using biomolecule targeting and aptamer
development
utilizes oligonucleotides to bind to specific proteins and thus interfere with
their function.
Described in U.S. Pat. No. 5,840,867, are aptamers to biomolecular targets
such as proteins in
general, and thrombin in particular. The novel compounds and methods disclosed
may be
applied broadly to biotechnologry diagnostics and therapeutics.
Conventional methods of detection and isolation of proteins and other
molecules have
employed antibodies and the like which specifically bind such substances.
Recently, however,
the de novo design of specifically binding oligonucleotides for non-
oligonucleotide 'targets that
generally bind nucleic acids has been described. See, e.g., Blackwell, T. K.,
et al., Science
( 1990) 250:1104-1 I 10; Blackwell, T. K., et al., Science ( 1990) 250:1149-
1152; Tuerk, C., and
Gold, L., Science ( 1990) :249:505-510; Joyce, G. F., Gene ( 1989) 82:83-87.
Such
oligonucleotides have been termed "aptamers" herein. Tuerk and Gold describe
the use of a
procedure termed "systematic evolution of ligands by exponential enrichment."
In this method,
a pool of RNAs that are completely randomized at specific positions is
subjected to selection
for binding by a desired nucleic acid-binding protein which has been fixed on
a nitrocellulose
filter. The bound RNAs then are recovered and amplified as double-stranded DNA
that is
competent for subsequent in vitro transcription. 'The newly transcribed RNA
then is recycled
through this procedure to enrich for oligonucleotides that have consensus
sequences for
binding by the cognate protein. 'fhe oligonucleotides so obtained then may be
sequenced for
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99/23933
further study. Tuerk and Gold applied this procedure to identify RNA
oligonucleotides which
are bound by the RNA binding region of T4 DNA polymerase.
The identification of oligonucleotides that specifically bind to biomolecules
that do not
normally bind to RNA or DNA has now been demonstrated for a number of
biomolecules that
vary widely in size, structure and composition. These molecules include: ( 1 )
thrombin, a
multifunctional regulatory protein that converts fibrinogen to fibrin in the
process of clot
formation; (2) bradykinin, a nonapeptide kinin involved in blood pressure
regulation and
implicated in hypotension; (3) PCiF2.alpha., a prostaglandin or fatty acid
derivative that exhibits
hormonal activity. Additionally, the interaction of oligonucleotides with
biomolecules whose
I 0 natural biological fianction is primarily extracellular has now been
demonstrated.
A plasmid is constructed using the methods describe above to include a
sequence of
interest encoding for an aptamer to thrombin. Aptamers having a sequence of
nucleotide bases
specifically binding to thrombin are described in Ll.S. Patent No. 5,840,867.
The plasmid is
administered to the patient under conditions whereby the plasmid enters cells
and generates the
aptamer. Alterntively, an ex vivo administration is performed where cells are
removed from a
patient, the plasmid is transfected into the cells, and the cells are then
placed back into the
patient. The aptamer binds :specifically to thrombin, thereby selectively
controlling the
biological activity of thrombin, and alleviates the pathological conditions
related to thrombin's
presence. This plasmid is used either prophylactically or therapeutically.
Although described with reference to the figures and specific examples set out
herein,
those skilled in the art will recognize that certain changes can be made to
the specific elements
set out herein without changing the manner in which those elements function to
achieve their
intended respective results. 1~or instance, the cassette described herein is
described as being
made up of three primary c.onaponents, genetic elements which comprises a
sequence of
interest and primer binding site, and a reverse transcriptase gene, each of
these components
being provided with appropriate; promoters as described herein. Those skilled
in the art will
recognize that, for instance, the MoMuLV reverse transcriptase gene described
for use as the
reverse transcriptase gene ofthe cassette can be replaced with other reverse
transcriptase genes
and that promoters other than the CMV promoter may be used to advantage. All
such changes
and modifications which do not depart from the spirit of the present invention
are intended to
fall within the scope of the following non-limiting claims.
26
SUBSTITUTE SHEET (RULE 26)
CA 02346155 2001-04-09
WO 00/22113 PCT/US99123933
Table I: Otigodeoxynucleotides
ODN-PMMV(+ 5' -CTACJGTCGGCGGCCGCGAAGATTGGTGCGCAC:ACACACAACGCG
129 bases CACCAATCTTCGCGGCCGCCGACCCGTCAGCGGGGGTCTTTCATTTG
(#23)
TC:GTCCGGGATCGGGAGACCCCTGCCCAGGGCC-3'
GGGGC
ODN-PMMV(-) _
5' -CTGCJGCAGGGGTCTCCC~ATCCCGGACGAGCCCCCAAATGAAAG
121 bases ACCCCCCTCTGACGGGTCGGCGGCCGCGAAGATTGGTGCGCGTTGTGT
(#24)
CGCACCAATCTTCGCGGCCGCCGAC-3'
GTGTG
ODN-Test (+) _
5' -GGCC:GGAAGATTGGGGCGCCAAAGAGTAACTCTCAAAGGCACGC
57 bases (#38GCCCCAATCTTCC-3'
ODN-Test (-) 5' -GGCC:GGAAGATTGGGGCGCGTGCCTTTGAGAGTTACTCTTTGGC
57 bases (#39)GCCCCAATCTTCC-3'
ODN-Telo (+) 5' -GGCC:GGAAGATTGGGGCGTTAGGGTTAGGGTTAGGGTTAGGGTT
92 bases (#40)AGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGCGCCCCAATCTTC
C-3'
_
ODN-Telo (-) 5' -GGCC;GGAAGATTGGGGCGCCCTAACCCTAACCCTAACCCTAACC
92 bases (#41CTAACCC:TAACCCTAACCCTAACCCTAACCCTAACGCCCCAATCTTC
)
C- 3'
__
ODN-XB(+) 5' -GGCC;TTGAAGAGCGGCCGCACTAACACCACCACAGTGCGGCCGC
Sl bases TCTTCAA-3'
ODN-XB(-) 5' -GGCC;TTGAAGAGCGGCCGCACTGTGGTGGTGTTAGTGCGGCCGC
51 bases TCTTCAA-3'
ODN-RT (+) 5' -GGGATCAGGAGCTCAGATCATGGGACCAATGG-3'
32 bases (#13)
ODN-RT (-) 5' -CTTCJTGCACAAGCTTTGCAGGTCT-3'
24 bases (#
12)
_
ODN-N>S (+) 5' -CTAC~CGGCAAGCGTAGCT-3'
18 bases (#25)
_
ODN-N>S (-) 5' -ACGC:TTGCCG-3'
bases (#26)
_
ODN-Mbo (+) 5' -CAATT.AAGGAAAGCTTTGAAAAATTATGTC-3'
30 bases (#
16)
__
ODN-Mbo(-) 5'-TAATGGCCCGGGCATAGTCGGGTAGGG-3'
27 bases #33)__
ODN-HisPro 5' -AGCTGGATCCCCCGCTCCCCACCACCACCACCACCCT'GCCCCT-
(+
43 bases (#36)3'
_
ODN-HisPro 5' -AGCAGGGGCAGGGTGGTGGTGGTGGTGGGGAGCGGGGGATCC-3
(-)
42 bases (#37)
ODN-Rep(+) 5'-ATATCTATTAATTTTGGCAAATCATAGCGGTTATGCTGACTCAG
121 bases GTGAATGCCGCGATAATTTTCAGATTGCAATCTTTCATCAATGAATT
TCAGTGATGAATTGCCAAGATTGATGTTGC-3' _
ODN-Rep(-) 5' -GACGAGATCTCCTCCAGGAATTCTCGAGAATTCGGAT'CCCCCGC
111 bases TCCCCACCACCACCACCACCACCCTGCCCCGCGGATGAAP,AATTATG
TGAGCAACATCAATCTTGGC-3'
Seq )D IS 5' -CT.AGTCGGATGCGGCCGCTGCACAACAACACACAACACAGCGGC
CGCATCCGATCAGCGGGGGTCTTTCATTTGGGGGCTCGTC:CGGATCG
GGAGACCCCTGCCCAGGGCC-3'
SeqID 16 5'-CTGGGGCAGGGGTCTCCCGATCCGGACGAGCCCCCAAATGAAAG
ACCCCCt~CTGATCGGATGCGGCCGCTGTGTTGTTTGTTGTTGTGCAG
CGGCCGCATCCGA-3'
27
SUBSTITUTE SHEET (RULE 26)