Note: Descriptions are shown in the official language in which they were submitted.
CA 02350776 2001-05-08
WO 00/28031 PCTIUS99/26592
1
NOVEL MITOGENIC REGULATORS
The U.S. Government has a paid-up license in this
invention and the right in limited circumstances to require the
patent owner to license others on reasonable terms as provided
for by the terms of National Institutes of Health grants HL38206
and HL58000.
TECHNICAL FIELD
The present invention relates to the field of normal
and abnormal cell growth, in particular mitogenic regulation.
The present invention provides the following: nucleotide
sequences encoding for the production of enzymes that are
mitogenic regulators; amino acid sequences of these enzymes;
vectors containing these nucleotide sequences; methods for
transfecting cells with vectors that produce these enzymes;
transfected cells; methods for administering these transfected
cells to animals to induce tumor formation; and antibodies to
these enzymes that are useful for detecting and measuring levels
of these enzymes, and for binding to cells possessing
extracellular epitopes of these enzymes.
BACKGROUND OF THE INVENTION
Reactive oxygen intermediates (ROI) are partial
reduction products of oxygen: 1 electron reduces 02 to form
superoxide (02-), and 2 electrons reduce 02 to form hydrogen
peroxide (H202). ROI are generated as a byproduct of aerobic
metabolism and by toxicological mechanisms. There is growing
evidence for regulated enzymatic generation of 02 and its
conversion to H202 in a variety of cells. The conversion of 02-
to H202 occurs spontaneously, but is markedly accelerated by
superoxide dismutase (SOD). High levels of ROI are associated
with damage to biomolecules such as DNA, biomembranes and
proteins. Recent evidence indicates generation of ROI under
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
2
normal cellular conditions and points to signaling roles for 02
and H202.
Several biological systems generate reactive
oxygen. Phagocytic cells such as neutrophils generate large
quantities of ROI as part of their battery of bactericidal
mechanisms. Exposure of neutrophils to bacteria or to various
soluble mediators such as formyl-Met-Leu-Phe or phorbol
esters activates a massive consumption of oxygen, termed the
respiratory burst, to initially generate superoxide, with
secondary generation of H202, HOCI and hydroxyl radical. The
enzyme responsible for this oxygen consumption is the
respiratory burst oxidase (nicotinamide adenine dinucleotide
phosphate-reduced form (NADPH) oxidase).
There is growing evidence for the generation of
ROI by non-phagocytic cells, particularly in situations related to
cell proliferation. Significant generation of H2021 02_, or both
have been noted in some cell types. Fibroblasts and human
endothelial cells show increased release of superoxide in
response to cytokines such as interleukin-1 or tumor necrosis
factor (TNF) (Meier et al. (1989) Biochem J. 263, 539-545.;
Matsubara et al. (1986) J. Immun. 137, 3295-3298). Ras-
transformed fibroblasts show increased superoxide release
compared with control fibroblasts (Irani, et al. (1997) Science
275, 1649-1652). Rat vascular smooth muscle cells show
increased H202 release in response to PDGF (Sundaresan et al.
(1995) Science 270, 296-299) and angiotensin II (Griendling et
al. (1994) Circ. Res. 74, 1141-1148; Fukui et al. (1997) Circ.
Res. 80, 45-51; Ushio-Fukai et al. (1996) J. Biol. Chem. 271,
23317-23321), and H202 in these cells is associated with
increased proliferation rate. The occurrence of ROI in a
variety of cell types is summarized in Table 1 (adapted from
Burdon, R. (1995) Free Radical Biol. Med. 18, 775-794).
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
3
Table 1
Superoxide Hydrogen Peroxide
human fibroblasts Balb/3T3 cells
human endothelial cells rat pancreatic islet cells
human/rat smooth muscle cells murine keratinocytes
human fat cells rabbit chondrocytes
human osteocytes human tumor cells
BHK-21 cells fat cells, 3T3 L1 cells
human colonic epithelial cells
ROI generated by the neutrophil have a cytotoxic
function. While ROI are normally directed at the invading
microbe, ROI can also induce tissue damage (e.g., in
inflammatory conditions such as arthritis, shock, lung disease,
and inflammatory bowel disease) or may be involved in tumor
initiation or promotion, due to damaging effects on DNA.
Nathan (Szatrowski et al. (1991) Canc. Res. 51, 794-798)
proposed that the generation of ROI in tumor cells may
contribute to the hypermutability seen in tumors, and may
therefore contribute to tumor heterogeneity, invasion and
metastasis.
In addition to cytotoxic and mutagenic roles, ROI
have ideal properties as signal molecules: 1) they are generated
in a controlled manner in response to upstream signals; 2) the
signal can be terminated by rapid metabolism of 02 and H202 by
SOD and catalase/peroxidases; 3) they elicit downstream effects
on target molecules, e.g., redox-sensitive regulatory proteins
such as NF kappa B and AP-1 (S chreck et al. (1991) EMBO J.
10, 2247-2258; Schmidt et al. (1995) Chemistry & Biology 2,
13-22). Oxidants such as 02 and H202 have a relatively well
defined signaling role in bacteria, operating via the Soxl/II
regulon to regulate transcription.
ROI appear to have a direct role in regulating cell
division, and may function as mitogenic signals in pathological
conditions related to growth. These conditions include cancer
and cardiovascular disease. 02 is generated in endothelial cells
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
4
in response to cytokines, and might play a role in angiogenesis
(Matsubara et al. (1986) J. Immun. 137, 3295-3298). 02- and
H202 are also proposed to function as "life-signals", preventing
cells from undergoing apoptosis (Matsubara et al. (1986) J.
Immun. 137, 3295-3298). As discussed above, many cells
respond to growth factors (e.g., platelet derived growth factor
(PDGF), epidermal derived growth factor (EGF), angiotensin
II, and various cytokines) with both increased production of 02
/H202 and increased proliferation. Inhibition of ROI generation
prevents the mitogenic response. Exposure to exogenously
generated 02" and H202 results in an increase in cell
proliferation. A partial list of responsive cell types is shown
below in Table 2 (adapted from Burdon, R. (1995) Free
Radical Biol. Med. 18, 775-794).
Table 2
Superoxide Hydrogen peroxide
human, hamster fibroblasts mouse osteoblastic cells
Balb/3T3 cells Balb/3T3 cells
human histiocytic leukemia rat, hamster fibroblasts
mouse epidermal cells human smooth muscle cells
rat colonic epithelial cells rat vascular smooth muscle cells
rat vascular smooth muscle cells
While non-transformed cells can respond to growth
factors and cytokines with the production of ROI, tumor cells
appear to produce ROI in an uncontrolled manner. A series of
human tumor cells produced large amounts of hydrogen
peroxide compared with non-tumor cells (Szatrowski et al.
(1991) Canc. Res. 51, 794-798). Ras-transformed NIH 3T3
cells generated elevated amounts of superoxide, and inhibition
of superoxide generation by several mechanisms resulted in a
reversion to a "normal" growth phenotype.
02" has been implicated in maintenance of the
transformed phenotype in cancer cells including melanoma,
breast carcinoma, fibrosarcoma, and virally transformed tumor
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
cells. Decreased levels of the manganese form of SOD
(MnSOD) have been measured in cancer cells and in vitro-
transformed cell lines, predicting increased 02" levels (Burdon,
R. (1995) Free Radical Biol. Med. 18, 775-794). MnSOD is
5 encoded on chromosome 6q25 which is very often lost in
melanoma. Overexpression of MnSOD in melanoma and other
cancer cells (Church et al. (1993) Proc. of Natl. Acad. Sci. 90,
3113-3117; Fernandez-Pol et al. (1982) Canc. Res. 42, 609-617;
Yan et al. (1996) Canc. Res. 56, 2864-2871) resulted in
suppression of the transformed phenotype.
ROI are implicated in growth of vascular smooth
muscle associated with hypertension, atherosclerosis, and
restenosis after angioplasty. 02. generation is seen in rabbit
aortic adventitia (Pagano et al. (1997) Proc. Natl. Acad. Sci.
94, 14483-14488). Vascular endothelial cells release 02 in
response to cytokines (Matsubara et al. (1986) J. Immun. 137,
3295-3298). 02 is generated by aortic smooth muscle cells in
culture, and increased 02 generation is stimulated by
angiotensin II which also induces cell hypertrophy. In a rat
model system, infusion of angiotensin II leads to hypertension as
well as increased 02. generation in subsequently isolated aortic
tissue (Ushio-Fukai et al. (1996) J. Biol. Chem. 271, 23317-
23321.; Yu et al. (1997) J. Biol. Chem. 272, 27288-27294).
Intravenous infusion of a form of SOD that localizes to the
vasculature or an infusion of an 02" scavenger prevented
angiotensin II induced hypertension and inhibited ROI
generation (Fukui et al. (1997) Circ. Res. 80, 45-51).
The neutrophil NADPH oxidase, also known as
phagocyte respiratory burst oxidase, provides a paradigm for
the study of the specialized enzymatic ROI-generating system.
This extensively studied enzyme oxidizes NADPH and reduces
oxygen to form 0,-. NADPH oxidase consists of multiple
proteins and is regulated by assembly of cytosolic and
membrane components. The catalytic moiety consists of
flavocytochrome bS58, an integral plasma membrane enzyme
comprised of two components: gp91 phox (gp refers to
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
6
glycoprotein; phox is an abbreviation of the words phagocyte
and oxidase) and p22phox (p refers to protein). gp9lphox
contains I flavin adenine dinucleotide (FAD) and 2 hemes as
well as the NADPH binding site. p22phox has a C-terminal
proline-rich sequence which serves as a binding site for
cytosolic regulatory proteins. The two cytochrome subunits,
gp9lphox and p22phox appear to stabilize one another, since the
genetic absence of either subunit, as in the inherited disorder
chronic granulomatous disease (CGD), results in the absence of
the partner subunit (Yu et al. (1997) J. Biol. Chem. 272,
27288-27294). Essential cytosolic proteins include p47phox,
p67phox and the small GTPase Rac, of which there are two
isoforms. p47phox and p67phox both contain SH3 regions and
proline-rich regions which participate in protein interactions
governing assembly of the oxidase components during
activation. The neutrophil enzyme is regulated in response to
bacterial phagocytosis or chemotactic signals by
phosphorylation of p47phox, and perhaps other components, as
well as by guanine nucleotide exchange to activate the GTP-
binding protein Rac.
The origin of ROI in non-phagocytic tissues is
unproven, but the occurrence of phagocyte oxidase components
has been evaluated in several systems by immunochemical
methods, Northern blots and reverse transcriptase-polymerase
chain reaction (RT-PCR). The message for p22phox is
expressed widely, as is that for Racl. Several cell types that are
capable of 02 generation have been demonstrated to contain all
of the phox components including gp91 phox, as summarized
below in Table 3. These cell types include endothelial cells,
aortic adventitia and lymphocytes.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
7
Table 3
Tissue gp9lphox p22phox p47phox p67phox
neutrophil +1.2 +1j2 +1.2 +1.2
aortic adventitia +1 +1 +1 +1
lymphocytes +2 +2 +1.2 +1.2
endothelial cells +22 +2 +1.2 +1.2
glomerular mesangial +1.2 +1.2 +1.2
cells
fibroblasts - +2 +1.2 +2
aortic sm. muscle - +1.2 ? ?
1= protein expression shown. 2= mRNA expression shown.
However, a distinctly different pattern is seen in
several other cell types shown in Table 3 including glomerular
mesangial cells, rat aortic smooth muscle and fibroblasts. In
these cells, expression of gp9lphox is absent while p22phox and
in some cases cytosolic phox components have been
demonstrated to be present. Since gp9lphox and p22phox
stabilize one another in the neutrophil, there has been much
speculation that some molecule, possibly related to gp91 phox,
accounts for ROI generation in glomerular mesangial cells, rat
aortic smooth muscle and fibroblasts (Ushio-Fukai et al. (1996)
J. Biol. Chem. 271, 23317-23321). Investigation of fibroblasts
from a patient with a genetic absence of gp9lphox provides
proof that the gp91 phox subunit is not involved in ROI
generation in these cells (Emmendorffer et al. (1993) Eur. J.
Haematol. 51, 223-227). Depletion of p22phox from vascular
smooth muscle using an antisense approach indicated that this
subunit participates in ROI generation in these cells, despite the
absence of detectable gp9lphox (Ushio-Fukai et al. (1996) J.
Biol. Chem. 271, 23317-23321). At this time the molecular
candidates possibly related to gp9lphox and involved in ROI
generation in these cells are unknown.
Accordingly, what is needed is the identity of the
proteins involved in ROI generation, especially in non-
phagocytic tissues and cells. What is also needed are the
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
8
nucleotide sequences encoding for these proteins, and the
primary sequences of the proteins themselves. Also needed are
vectors designed to include nucleotides encoding for these
proteins. Probes and PCR primers derived from the nucleotide
sequence are needed to detect, localize and measure nucleotide
sequences, including mRNA, involved in the synthesis of these
proteins. In addition, what is needed is a means to transfect
cells with these vectors. What is also needed are expression
systems for production of these molecules. Also needed are
antibodies directed against these molecules for a variety of uses
including localization, detection, measurement and passive
immunization.
SUMMARY OF THE INVENTION
The present invention , solves the problems
described above by providing a novel family of nucleotide
sequences and proteins encoded by these nucleotide sequences
termed mox proteins and duox proteins. In particular the
present invention provides compositions comprising the
nucleotide sequences SEQ ID NO: 1, SEQ ID NO:3, SEQ ID
NO:22, SEQ ID NO:41, SEQ ID NO:45, SEQ ID NO:47, and
fragments thereof, which encode for the expression of proteins
comprising SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ
ID NO:42, SEQ ID NO:46, SEQ ID NO:48, respectively, and
fragments thereof. While not wanting to be bound by the
following statement, it is believed that these proteins are
involved in ROI production. The present invention also
provides vectors containing these nucleotide sequences, cells
transfected with these vectors which produce the proteins
comprising SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ
ID NO:42, SEQ ID NO:46, SEQ ID NO:48, and fragments
thereof, and antibodies to these proteins and fragments thereof.
The present invention also provides methods for stimulating
cellular proliferation by administering vectors encoded for
production of the proteins comprising SEQ ID NO:2, SEQ ID
NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ
ID NO:48 and fragments thereof. The present invention also
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
9
provides methods for stimulating cellular proliferation by
administering the proteins comprising SEQ ID NO:2, SEQ ID
NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ
ID NO:48 and fragments thereof. The nucleotides and
antibodies of the present invention are useful for the detection,
localization and measurement of the nucleic acids encoding for
the production of the proteins of the present invention, and also
for the detection, localization and measurement of the proteins
of the present invention. These nucleotides and antibodies can
be combined with other reagents in kits for the purposes of
detection, localization and measurement.
Most particularly, the present invention involves a
method for regulation of cell division or cell proliferation by
modifying the activity or expression of the proteins described as
SEQ ID NO:2, SEQ ID NO:4, SEQ ID -NO:21, SEQ ID NO:42,
SEQ ID NO:46, SEQ ID NO:48 or fragments thereof. These
proteins, in their naturally occurring or expressed forms, are
expected to be useful in drug development, for example for
screening of chemical and drug libraries by observing inhibition
of the activity of these enzymes. Such chemicals and drugs
would likely be useful as treatments for cancer, prostatic
hypertrophy, benign prostatic hypertrophy, hypertension,
atherosclerosis and many other disorders involving abnormal
cell growth or proliferation as described below. The entire
expressed protein may be useful in these assays. Portions of the
molecule which may be targets for inhibition or modification
include but are not limited to the binding site for pyridine
nucleotides (NADPH or NADH), the flavoprotein domain
(approximately the C-terminal 265 amino acids), and/or the
binding or catalytic site for flavin adenine dinucleotide (FAD).
The method of the present invention may be used
for the development of drugs or other therapies for the
treatment of conditions associated with abnormal growth
including, but not limited to the following: cancer, psoriasis,
prostatic hypertrophy, benign prostatic hypertrophy,
cardiovascular disease, proliferation of vessels, including but
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
not limited to blood vessels and lymphatic vessels, arteriovenous
malformation, vascular problems associated with the eye,
atherosclerosis, hypertension, and restenosis following
angioplasty. The enzymes of the present invention are excellent
5 targets for the development of drugs and other agents which
may modulate the activity of these enzymes. It is to be
understood that modulation of activity may result in enhanced,
diminished or absence of enzymatic activity. Modulation of the
activity of these enzymes may be useful in treatment of
10 conditions associated with abnormal growth.
Drugs which affect the activity of the enzymes
represented in SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21,
SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48, or fragments
thereof, may also be combined with other therapeutics in the
treatment of specific conditions. For example, these drugs may
be combined with angiogenesis inhibitors in the treatment of
cancer, with antihypertensives for the treatment of
hypertension, and with cholesterol lowering drugs for the
treatment of atherosclerosis.
Accordingly, an object of the present invention is to
provide nucleotide sequences, or fragments thereof, encoding
for the production of proteins, or fragments thereof, that are
involved in ROI production.
Another object of the present invention is to
provide vectors containing these nucleotide sequences, or
fragments thereof.
Yet another object of the present invention is to
provide cells transfected with these vectors.
Still another object of the present invention is to
administer cells transfected with these vectors to animals and
humans.
Another object of the present invention is to
provide proteins, or fragments thereof, that are involved in ROI
production.
Still another object of the present invention is to
provide antibodies, including monoclonal and polyclonal
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
11
antibodies, or fragments thereof, raised against proteins, or
fragments thereof, that are involved in ROI production.
Another object of the present invention is to
administer genes containing nucleotide sequences, or fragments
thereof, encoding for the production of proteins, or fragments
thereof, that are involved in ROI production, to animals and
humans and also to cells obtained from animals and humans.
Another object of the present invention is to
administer antisense complimentary sequences of genes
containing nucleotide sequences, or fragments thereof, encoding
for the production of proteins, or fragments thereof, that are
involved in ROI production, to animals and humans and also to
cells obtained from animals and humans.
Yet another object of the present invention is to
provide a method for stimulating or inhibiting cellular
proliferation by administering vectors containing nucleotide
sequences, or fragments thereof, encoding for the production of
proteins, or fragments thereof, that are involved in ROI
production, to animals and humans. It is also an object of the
present invention to provide a method for stimulating or
inhibiting cellular proliferation by administering vectors
containing antisense complimentary sequences of nucleotide
sequences, or fragments thereof, encoding for the production of
proteins, or fragments thereof, that are involved in ROI
production, to animals and humans. These methods of
stimulating cellular proliferation are useful for a variety of
purposes, including but not limited to, developing animal
models of tumor formation, stimulating cellular proliferation of
blood marrow cells following chemotherapy or radiation, or in
cases of anemia.
Still another object of the present invention is to
provide antibodies useful in immunotherapy against cancers
expressing the proteins represented in SEQ ID NO:2, SEQ ID
NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ
ID NO:48 or fragments thereof.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
12
Yet another object of the present invention is to
provide nucleotide probes useful for the detection, localization
and measurement of nucleotide sequences, or fragments thereof,
encoding for the production of proteins, or fragments thereof,
that are involved in ROI production.
Another object of the present invention is to
provide antibodies useful for the detection, localization and
measurement of nucleotide sequences, or fragments thereof,
encoding for the production of proteins, or fragments thereof,
that are involved in ROI production.
Another object of the present invention is to
provide kits useful for detection of nucleic acids including the
nucleic acids represented in SEQ ID NO: 1, SEQ ID NO:3, SEQ
ID NO:22, SEQ ID NO:41, SEQ ID NO:45, SEQ ID NO:47, or
fragments thereof, that encode for proteins, or fragments
thereof, that are involved in ROI production.
Yet another object of the present invention is to
provide kits useful for detection and measurement of nucleic
acids including the nucleic acids represented in SEQ ID NO:1,
SEQ ID NO:3, SEQ ID NO:22, SEQ ID NO:41, SEQ ID
NO:45, SEQ ID NO:47, or fragments thereof, that encode for
proteins, or fragments thereof, that are involved in ROI
production.
Still another object of the present invention is to
provide kits useful for the localization of nucleic acids including
the nucleic acids represented in SEQ ID NO: 1, SEQ ID NO:3,
SEQ ID NO:22, SEQ ID NO:41, SEQ ID NO:45, SEQ ID
NO:47, or fragments thereof, that encode for proteins, or
fragments thereof that are involved in ROI production.
Another object of the present invention is to
provide kits useful for detection of proteins, including the
proteins represented in SEQ ID NO:2, SEQ ID NO:4, SEQ ID
NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48, or
fragments thereof, that are involved in ROI production.
Yet another object of the present invention is to
provide kits useful for detection and measurement of proteins,
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
13
including the proteins represented in SEQ ID NO:2, SEQ ID
NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ
ID NO:48, or fragments thereof, that are involved in ROI
production.
Still another object of the present invention is to
provide kits useful for localization of proteins, including the
proteins represented in SEQ ID NO:2, SEQ ID NO:4, SEQ ID
NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48, or
fragments thereof, that are involved in ROI production.
Yet another object of the present invention is to
provides kits useful for the detection, measurement or
localization of nucleic acids, or fragments thereof, encoding for
proteins, or fragments thereof, that are involved in ROI
production, for use in diagnosis and prognosis of abnormal
cellular proliferation related to ROI production.
Another object of the present invention is to
provides kits useful for the detection, measurement or
localization of proteins, or fragments thereof, that are involved
in ROI production, for use in diagnosis and prognosis of
abnormal cellular proliferation related to ROI production.
These and other objects, features and advantages of
the present invention will become apparent after a review of the
following detailed description of the disclosed embodiments and
the appended drawings.
BRIEF DESCRIPTION OF THE FIGURES
Fig. 1(a-d). Comparison of amino acid sequences
of the human mox 1 protein (labeled mox l .human, SEQ ID
NO:2), rat mox 1 protein (labeled mox l .rat, SEQ ID NO:21),
human mox2 protein (labeled mox2.human., SEQ ID NO:4) of
the present invention to human (gp 91 phox/human.pep, SEQ ID
NO:12) bovine (gp 91 phox/bovine.pep, SEQ ID NO:37), and
murine (gp 91 phox/mouse.pep, SEQ ID NO:38) proteins. Also
included are related plant enzyme proteins cytb
558.arabidopsis.pep (SEQ ID NO:39) and cytb558.rice.pep,
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
14
(SEQ ID NO:40). Enclosed in boxes are similar amino acid
residues.
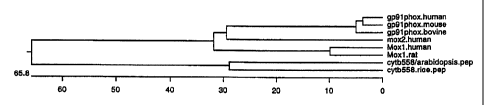
Fig. 2. Sequence similarities among proteins
related to gp9lphox including human mox 1 (SEQ ID NO:2),
human mox2 (SEQ ID NO:4), and rat mox 1 (SEQ ID NO:21).
The dendrogram indicates the degree of similarity among this
family of proteins, and also includes the related plant enzymes.
Fig. 3. Cell free assay for mox-l activity.
Superoxide generation was measured using the
chemiluminescent reaction between lucigenin and superoxide in
cell lysates from vector control NEF2 and mox 1 transfected
NIH3T3 cells.
Fig. 4. Superoxide generation by human mox 1.
Reduction of NBT in moxl transfected and control fibroblasts
was measured in the absence (filled bars) or presence (open
bars) or superoxide dismutase.
Fig. 5. Aconitase (filled bars), lactate
dehydrogenase (narrow hatching) and fumarase (broad
hatching) were determined in lysates of cells transfected with
vector alone (NEF2) or with moxl (YA26, YA28 and YA212).
DETAILED DESCRIPTION OF THE INVENTION
The present invention solves the problems
described above by providing a novel family of nucleotide
sequences and proteins, encoded by these nucleotide sequences,
termed mox proteins and duox proteins. The term "mox"
refers to "mitogenic oxidase" while the term "duox" refers to
"dual oxidase". In particular, the present invention provides
novel compositions comprising the nucleotide sequences SEQ ID
NO:1, SEQ ID NO:3, SEQ ID NO:22, SEQ ID NO:41, SEQ ID
NO:45, SEQ ID NO:47, and fragments thereof, which encode,
respectively, for the expression of proteins comprising SEQ ID
NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID
NO:46, SEQ ID NO:48 and fragments thereof.
Both the mox and duox proteins described herein
have homology to the gp9 l phox protein involved in ROI
CA 02350776 2001-05-08
WO 00/28031 PCTIUS99/26592
generation, however, the mox and duox proteins comprise a
novel and distinct family of proteins. The mox proteins
included in the present invention have a molecular weight of
approximately 65 kDa as determined by reducing gel
5 electrophoresis and are capable of inducing ROI generation in
cells. As described in more detail below, the mox proteins of
the present invention also function in the regulation of cell
growth, and are therefore implicated in diseases involving
abnormal cell growth such as cancer. The present invention
10 describes mox proteins found in human and rat, however, it is
likely that the mox family of genes/proteins is widely
distributed among multicellular organisms.
The duox proteins described herein are larger than
the mox proteins and have three distinct regions: the amino
15 terminal region having homology to peroxidase proteins, the
internal region having homology to calmodulin (CAM) proteins
and the carboxy-terminal region having homology to mox
proteins. Human duoxl is shown in SEQ ID NO:46 and a
portion of human duox2 is shown in SEQ ID NO:48.
Nucleotides encoding duox 1 and duox2 proteins are also shown
in SEQ ID NO: 45 and SEQ ID NO:47, respectively. In
addition to the human duox proteins, comparison of the
sequence of human duox 1 and human duox2 with genomic
databases using BLAST searching resulted in the identification
of two homologs of duox in C. elegans (Ce-duoxl and Ce-
duox2). Drosophila also appears to have at least one duox
homolog. Thus, the duox family of genes/proteins is widely
distributed among multicellular organisms.
Although not wanting to be bound by the following
statement, it is believed that duox l and duox2 have dual
enzymatic functions, catalyzing both the generation of
superoxide and peroxidative type reactions. The latter class of
reactions utilize hydrogen peroxide as a substrate (and in some
cases have been proposed to utilize superoxide as a substrate).
Since hydrogen peroxide is generated spontaneously from the
dismutation of superoxide, it is believed that the NAD(P)H
CA 02350776 2001-05-08
WO 00/28031 PCT/US"/26592
16
oxidase domain generates the superoxide and/or hydrogen
peroxide which can then be used as a substrate for the
peroxidase domain. In support of this hypothesis, a model for
the duox 1 protein in C. elegans has been developed that has an
extracellular N-terminal peroxidase domain, a transmembrane
region and a NADPH binding site located on the cytosolic face
of the plasma membrane. By analogy with the neutrophil
NADPH-oxidase which generates extracellular superoxide,
human duox 1 is predicted to generate superoxide and its
byproduct hydrogen peroxide extracellularly where it can be
utilized by the peroxidase domain.
While the ROI generated by duox 1 and duox2 may
function as does mox 1 in regulation of cell growth, the presence
of the peroxidase domain is likely to confer additional
biological functions. Depending upon the co-substrate,
peroxidases can participate in a variety of reactions including
halogenation such as the generation of hypochlorous acid
(HOCI) by myeloperoxidase and the iodination of tyrosine to
form thyroxin by thyroid peroxidase. Peroxidases have also
been documented to participate in the metabolism of
polyunsaturated fatty acids, and in the chemical modification of
tyrosine in collagen (by sea urchin ovoperoxidase). Although
not wanting to be bound by this statement, it is believed that the
predicted transmembrane nature of duox 1 facilitates its function
in the formation or modification of extracellular matrix or
basement membrane. Since the extracellular matrix plays an
important role in tumor cell growth, invasion and metastasis, it
is believed that the duox type enzymes play a pathogenic role in
such conditions.
In addition to the nucleotide sequences described
above, the present invention also provides vectors containing
these nucleotide sequences and fragments thereof, cells
transfected with these vectors which produce the proteins
comprising SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ
ID NO:42, SEQ ID NO:46, SEQ ID NO:48 and fragments
thereof, and antibodies to these proteins and fragments thereof.
CA 02350776 2001-05-08
WO 00/28031 PCTIUS99/26592
17
The present invention also provides methods for stimulating
cellular proliferation by administering vectors, or cells
containing vectors, encoded for production of the proteins
comprising SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ
ID NO:42, SEQ ID NO:46, SEQ ID NO:48 and fragments
thereof. The nucleotides and antibodies of the present invention
are useful for the detection, localization and measurement of the
nucleic acids encoding for the production of the proteins of the
present invention, and also for the detection, localization and
measurement of the proteins of the present invention. These
nucleotides and antibodies can be combined with other reagents
in kits for the purposes of detection, localization and
measurement. These kits are useful for diagnosis and prognosis
of conditions involving cellular proliferation associated with
production of reactive oxygen intermediates.
The present invention solves the problems
described above by providing a composition comprising the
nucleotide sequence SEQ ID NO:1 and fragments thereof. The
present invention also provides a composition comprising the
nucleotide sequence SEQ ID NO:3 and fragments thereof. The
present invention also provides a composition comprising the
nucleotide sequence SEQ ID NO:22 and fragments thereof. The
present invention also provides a composition comprising the
nucleotide sequence SEQ ID NO:41 and fragments thereof. The
present invention also provides a composition comprising the
nucleotide sequence SEQ ID NO:45 and fragments thereof. The
present invention also provides a composition comprising the
nucleotide sequence SEQ ID NO:47 and fragments thereof.
The present invention provides a composition
comprising the protein SEQ ID NO:2 encoded by the nucleotide
sequence SEQ ID NO:1. The present invention provides a
composition comprising the protein SEQ ID NO:4 encoded by
the nucleotide sequence SEQ ID NO:3. The present invention
provides a composition comprising the protein SEQ ID NO:21
encoded by the nucleotide sequence SEQ ID NO:22. The
present invention provides a composition comprising the protein
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
18
SEQ ID NO:42 encoded by the nucleotide sequence SEQ ID
NO:41. The present invention provides a composition
comprising the protein SEQ ID NO:46 encoded by the
nucleotide sequence SEQ ID NO:45. The present invention
provides a composition comprising the protein SEQ ID NO:48
encoded by the nucleotide sequence SEQ ID NO:47.
The present invention provides a composition
comprising the protein SEQ ID NO:2 or fragments thereof,
encoded by the nucleotide sequence SEQ ID NO:1 or fragments
thereof. The present invention also provides a composition
comprising the protein SEQ ID NO:4 or fragments thereof,
encoded by the nucleotide sequence SEQ ID NO:3 or fragments
thereof. The present invention also provides a composition
comprising the protein SEQ ID NO:21 or fragments thereof,
encoded by the nucleotide sequence SEQ ID NO:22 or
fragments thereof. The present invention also provides a
composition comprising the protein SEQ ID NO:42 or
fragments thereof, encoded by the nucleotide sequence SEQ ID
NO:41 or fragments thereof. The present invention also
provides a composition comprising the protein SEQ ID NO:46
or fragments thereof, encoded by the nucleotide sequence SEQ
ID NO:45 or fragments thereof. The present invention also
provides a composition comprising the protein SEQ ID NO:48
or fragments thereof, encoded by the nucleotide sequence SEQ
ID NO:47 or fragments thereof.
The present invention also provides vectors
containing the nucleotide sequences SEQ ID NO:l, SEQ ID
NO:3, SEQ ID NO:22, SEQ ID NO:41, SEQ ID NO:45, SEQ
ID NO:47 or fragments thereof. The present invention also
provides cells transfected with these vectors. In addition, the
present invention provides cells stably transfected with the
nucleotide sequence SEQ ID NO:1 or fragments thereof. The
present invention also provides cells stably transfected with the
nucleotide sequence SEQ ID NO:3 or fragments thereof. The
present invention also provides cells stably transfected with the
nucleotide sequence SEQ ID NO:22 or fragments thereof. The
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
19
present invention also provides cells stably transfected with the
nucleotide sequence SEQ ID NO:41 or fragments thereof. The
present invention also provides cells stably transfected with the
nucleotide sequence SEQ ID NO:45 or fragments thereof. The
present invention also provides cells stably transfected with the
nucleotide sequence SEQ ID NO:47 or fragments thereof.
The present invention provides cells stably
transfected with the nucleotide sequence SEQ ID NO:1 or
fragments thereof, which produce the protein SEQ ID NO:2 or
fragments thereof. In addition, the present invention provides
cells stably transfected with the nucleotide sequence SEQ ID
NO:3 or fragments thereof which produce the protein SEQ ID
NO:4 or fragments thereof. In addition, the present invention
provides cells stably transfected with the nucleotide sequence
SEQ ID NO:22 or fragments thereof which produce the protein
SEQ ID NO:21 or fragments thereof. The present invention
also provides cells stably transfected with the nucleotide
sequence SEQ ID NO:41 or fragments thereof which produce
the protein SEQ ID NO:42 or fragments thereof. The present
invention also provides cells stably transfected with the
nucleotide sequence SEQ ID NO:45 or fragments thereof which
produce the protein SEQ ID NO:46 or fragments thereof. The
present invention also provides cells stably transfected with the
nucleotide sequence SEQ ID NO:47 or fragments thereof which
produce the protein SEQ ID NO:48 or fragments thereof.
The present invention provides a method for
stimulating growth by administering cells stably transfected
with the nucleotide sequence SEQ ID NO:1 which produce the
protein SEQ ID NO:2 or fragments thereof. The present
invention also provides a method for stimulating growth by
administering cells stably transfected with the nucleotide
sequence SEQ ID NO:3 or fragments thereof, which produce
the protein SEQ ID NO:4 or fragments thereof. The present
invention also provides a method for stimulating growth by
administering cells stably transfected with the nucleotide
sequence SEQ ID NO:22 or fragments thereof, which produce
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
the protein SEQ ID NO:21 or fragments thereof. The present
invention also provides a method for stimulating growth by
administering cells stably transfected with the nucleotide
sequence SEQ ID NO:41 or fragments thereof, which produce
5 the protein SEQ ID NO:42 or fragments thereof. The present
invention also provides a method for stimulating growth by
administering cells stably transfected with the nucleotide
sequence SEQ ID NO:45 or fragments thereof, which produce
the protein SEQ ID NO:46 or fragments thereof. The present
10 invention also provides a method for stimulating growth by
administering cells stably transfected with the nucleotide
sequence SEQ ID NO:47 or fragments thereof, which produce
the protein SEQ ID NO:48 or fragments thereof.
Specifically, the present invention provides a
15 method for stimulating tumor formation by administering cells
stably transfected with the nucleotide sequence SEQ ID NO: 1 or
fragments thereof, which produce the protein SEQ ID NO:2 or
fragments thereof. The present invention also provides a
method for stimulating tumor formation by administering cells
20 stably transfected with the nucleotide sequence SEQ ID NO:3 or
fragments thereof, which produce the protein SEQ ID NO:4 or
fragments thereof. The present invention also provides a
method for stimulating tumor formation by administering cells
stably transfected with the nucleotide sequence SEQ ID NO:22
or fragments thereof, which produce the protein SEQ ID NO:21
or fragments thereof. The present invention also provides a
method for stimulating tumor formation by administering cells
stably transfected with the nucleotide sequence SEQ ID NO:41
or fragments thereof, which produce the protein SEQ ID NO:42
or fragments thereof. The present invention also provides a
method for stimulating tumor formation by administering cells
stably transfected with the nucleotide sequence SEQ ID NO:45
or fragments thereof, which produce the protein SEQ ID NO:46
or fragments thereof. The present invention also provides a
method for stimulating tumor formation by administering cells
stably transfected with the nucleotide sequence SEQ ID NO:47
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
21
or fragments thereof, which produce the protein SEQ ID NO:48
or fragments thereof.
The present invention may also be used to develop
anti-sense nucleotide sequences to SEQ ID NO: 1, SEQ ID NO:3,
SEQ ID NO:22, SEQ ID NO:41, SEQ ID NO:45, SEQ ID
NO:47 or fragments thereof. These anti-sense molecules may
be used to interfere with translation of nucleotide sequences,
such as SEQ ID NO:1, SEQ ID NO:3, SEQ ID NO:22, SEQ ID
NO:41, SEQ ID NO:45, SEQ ID NO:47, or fragments thereof,
that encode for proteins such as SEQ ID NO:2, SEQ ID NO:4,
SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ ID
NO:48 or fragments thereof. Administration of these anti-sense
molecules, or vectors encoding for anti sense molecules, to
humans and animals, would interfere with production of
proteins such as SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21,
SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48, or fragments
thereof, thereby decreasing production of ROIs and inhibiting,
cellular proliferation. These methods are useful in producing
animal models for use in study of tumor development and
vascular growth, and for study of the efficacy of treatments for
affecting tumor and vascular growth in vivo.
The present invention also provides a method for
high throughput screening of drugs and chemicals which
modulate the proliferative activity of the enzymes of the present
invention, thereby affecting cell division. Combinatorial
chemical libraries may be screened for chemicals which
modulate the proliferative activity of these enzymes. Drugs and
chemicals may be evaluated based on their ability to modulate
the enzymatic activity of the expressed or endogenous proteins,
including those represented by SEQ ID NO:2, SEQ ID NO:4,
SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ ID
NO:48 or fragments thereof. Endogenous proteins may be
obtained from many different tissues or cells, such as colon
cells. Drugs may also be evaluated based on their ability to bind
to the expressed or endogenous proteins represented by SEQ ID
NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID
CA 02350776 2001-05-08
WO 00/28031 PCTIUS99/26592
22
NO:46, SEQ ID NO:48 or fragments thereof. Enzymatic
activity may be NADPH- or NADH-dependent superoxide
generation catalyzed by the holoprotein. Enzymatic activity
may also be NADPH- or NADH-dependent diaphorase activity
catalyzed by either the holoprotein or the flavoprotein domain.
By flavoprotein domain, is meant approximately
the C-terminal half of the enzymes shown in SEQ ID NO:2,
SEQ ID NO:4, SEQ ID NO:21, SEQ ID NO:42, or fragments
thereof, and the C-terminal end of the enzymes shown in SEQ
ID NO:46 and SEQ ID NO:48 (approximately the C-terminal
265 amino acids). This fragment of gp9lphox has NADPH-
dependent reductase activity towards cytochrome c,
nitrobluetetrazolium and other dyes. Expressed proteins or
fragments thereof can be used for robotic screens of existing
combinatorial chemical libraries. While not wanting to be
bound by the following statement, it is believed that the NADPH
or NADH binding site and the FAD binding site are useful for
evaluating the ability of drugs and other compositions to bind to
the mox and duox enzymes or to modulate their enzymatic
activity. The use of the holoprotein or the C-terminal half or
end regions are preferred for developing a high throughput
drug screen. Additionally, the N-terminal one-third of the duox
domain (the peroxidase domain) may also be used to evaluate
the ability of drugs and other compositions to inhibit the
peroxidase activity, and for further development of a high
throughput drug screen.
The present invention also provides antibodies
directed to the proteins SEQ ID NO:2, SEQ ID NO:4, SEQ ID
NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48 and
fragments thereof. The antibodies of the present invention are
useful for a variety of purposes including localization, detection
and measurement of the proteins SEQ ID NO:2, SEQ ID NO:4,
SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ ID
NO:48 and fragments thereof. The antibodies may be employed
in kits to accomplish these purposes. These antibodies may also
be linked to cytotoxic agents for selected killing of cells. The
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
23
term antibody is meant to include any class of antibody such as
IgG, IgM and other classes. The term antibody also includes a
completely intact antibody and also fragments thereof, including
but not limited to Fab fragments and Fab + Fc fragments.
The present invention also provides the nucleotide
sequences SEQ ID NO:1, SEQ ID NO:3, SEQ ID NO:22, SEQ
ID NO:41, SEQ ID NO:45, SEQ ID NO:47 and fragments
thereof. These nucleotides are useful for a variety of purposes
including localization, detection, and measurement of messenger
RNA involved in synthesis of the proteins represented as SEQ
ID NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ
ID NO:46, SEQ ID NO:48 and fragments thereof. These
nucleotides may also be used in the construction of labeled
probes for the localization, detection, and measurement of
nucleic acids such as messenger RNA or alternatively for the
isolation of larger nucleotide sequences containing the
nucleotide sequences shown in SEQ ID NO:1, SEQ ID NO:3,
SEQ ID NO:22, SEQ ID NO:41, SEQ ID NO:45, SEQ ID
NO:47 or fragments thereof. These nucleotide sequences may
be used to isolate homologous strands from other species using
techniques known to one of ordinary skill in the art. These
nucleotide sequences may also be used to make probes and
complementary strands. In particular, the nucleotide sequence
shown in SEQ ID NO:47 may be used to isolate the complete
coding sequence for duox2. The nucleotides may be employed
in kits to accomplish these purposes.
Most particularly, the present invention involves a
method for modulation of growth by modifying the proteins
represented as SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21,
SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48 or fragments
thereof.
The term "mitogenic regulators" is used herein to
mean any molecule that acts to affect cell division.
The term "animal" is used herein to mean humans
and non-human animals of both sexes.
CA 02350776 2001-05-08
WO 00/28031 PCTIUS99/26592
24
The terms "a", "an" and "the" as used herein are
defined to mean one or more and include the plural unless the
context is inappropriate.
"Proteins", "peptides," "polypeptides" and
"oligopeptides" are chains of amino acids (typically L-amino
acids) whose alpha carbons are linked through peptide bonds
formed by a condensation reaction between the carboxyl group
of the alpha carbon of one amino acid and the amino group of
the alpha carbon of another amino acid. The terminal amino
acid at one end of the chain (i.e., the amino terminal) has a free
amino group, while the terminal amino acid at the other end of
the chain (i.e., the carboxy terminal) has a free carboxyl group.
As such, the term "amino terminus" (abbreviated N-terminus)
refers to the free alpha-amino group on the amino acid at the
amino terminal of the protein, or to the alpha-amino group
(imino group when participating in a peptide bond) of an amino
acid at any other location within the protein. Similarly, the
term "carboxy terminus" (abbreviated C-terminus) refers to the
free carboxyl group on the amino acid at the carboxy terminus
of a protein, or to the carboxyl group of an amino acid at any
other location within the protein.
Typically, the amino acids making up a protein are
numbered in order, starting at the amino terminal and
increasing in the direction toward the carboxy terminal of the
protein. Thus, when one amino acid is said to "follow" another,
that amino acid is positioned closer to the carboxy terminal of
the protein than the preceding amino acid.
The term "residue" is used herein to refer to an
amino acid (D or L) or an amino acid mimetic that is
incorporated into a protein by an amide bond. As such, the
amino acid may be a naturally occurring amino acid or, unless
otherwise limited, may encompass known analogs of natural
amino acids that function in a manner similar to the naturally
occurring amino acids (i.e., amino acid mimetics). Moreover,
an amide bond mimetic includes peptide backbone modifications
well known to those skilled in the art.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
Furthermore, one of skill will recognize that, as
mentioned above, individual substitutions, deletions or additions
which alter, add or delete a single amino acid or a small
percentage of amino acids (typically less than 5%, more
5 typically less than 1 %) in an encoded sequence are
conservatively modified variations where the alterations result
in the substitution of an amino acid with a chemically similar
amino acid. Conservative substitution tables providing
functionally similar amino acids are well known in the art. The
10 following six groups each contain amino acids that are
conservative substitutions for one another:
1) Alanine (A), Serine (S), Threonine (T);
2) Aspartic acid (D), Glutamic acid (E);
3) Asparagine (N), Glutamine (Q);
15 4) Arginine (R), Lysine (K);
5) Isoleucine (I), Leucine (L), Methionine (M), Valine
(V); and
6) Phenylalanine (F), Tyrosine (Y), Tryptophan (W).
20 When the peptides are relatively short in length
(i.e., less than about 50 amino acids), they are often synthesized
using standard chemical peptide synthesis techniques. Solid
phase synthesis in which the C-terminal amino acid of the
sequence is attached to an insoluble support followed by
25 sequential addition of the remaining amino acids in the sequence
is a preferred method for the chemical synthesis of the antigenic
epitopes described herein. Techniques for solid phase synthesis
are known to those skilled in the art.
Alternatively, the antigenic epitopes described
herein are synthesized using recombinant nucleic acid
methodology. Generally, this involves creating a nucleic acid
sequence that encodes the peptide or protein, placing the nucleic
acid in an expression cassette under the control of a particular
promoter, expressing the peptide or protein in a host, isolating
the expressed peptide or protein and, if required, renaturing the
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
26
peptide or protein. Techniques sufficient to guide one of skill
through such procedures are found in the literature.
When several desired protein fragments or peptides
are encoded in the nucleotide sequence incorporated into a
vector, one of skill in the art will appreciate that the protein
fragments or peptides may be separated by a spacer molecule
such as, for example, a peptide, consisting of one or more
amino acids. Generally, the spacer will have no specific
biological activity other than to join the desired protein
fragments or peptides together, or to preserve some minimum
distance or other spatial relationship between them. However,
the constituent amino acids of the spacer may be selected to
influence some property of the molecule such as the folding, net
charge, or hydrophobicity. Nucleotide sequences encoding for
the production of residues which may be useful in purification
of the expressed recombinant protein may be built into the
vector. Such sequences are known in the art. For example, a
nucleotide sequence encoding for a poly histidine sequence may
be added to a vector to facilitate purification of the expressed
recombinant protein on a nickel column.
Once expressed, recombinant peptides, polypeptides
and proteins can be purified according to standard procedures
known to one of ordinary skill in the art, including ammonium
sulfate precipitation, affinity columns, column chromatography,
gel electrophoresis and the like. Substantially pure
compositions of about 50 to 99% homogeneity are preferred,
and 80 to 95% or greater homogeneity are most preferred for
use as therapeutic agents.
One of skill in the art will recognize that after
chemical synthesis, biological expression or purification, the
desired proteins, fragments thereof and peptides may possess a
conformation substantially different than the native
conformations of the proteins, fragments thereof and peptides.
In this case, it is often necessary to denature and reduce protein
and then to cause the protein to re-fold into the preferred
conformation. Methods of reducing and denaturing proteins
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
27
and inducing re-folding are well known to those of skill in the
art.
The genetic constructs of the present invention
include coding sequences for different proteins, fragments
thereof, and peptides. The genetic constructs also` include
epitopes or domains chosen to permit purification or detection
of the expressed protein. Such epitopes or domains include
DNA sequences encoding the glutathione binding domain from
glutathione S-transferase, hexa-histidine, thioredoxin,
hemagglutinin antigen, maltose binding protein, and others
commonly known to one of skill in the art. The preferred
genetic construct includes the nucleotide sequences of SEQ ID
NO: I, SEQ ID NO:3, SEQ ID NO:22, SEQ ID NO:41, SEQ ID
NO:45, SEQ ID NO:47 or fragments thereof. It is to be
understood that additional or alternative nucleotide sequences
may be included in the genetic constructs in order to encode for
the following: a) multiple copies of the desired proteins,
fragments thereof, or peptides; b) various combinations of the
desired proteins, fragments thereof, or peptides; and c)
conservative modifications of the desired proteins, fragments
thereof, or peptides, and combinations thereof. Preferred
proteins include the human mox 1 protein and human mox2
protein shown as SEQ ID NO:2 and SEQ ID NO:4, respectively,
and fragments thereof. Some preferred fragments of the human
mox 1 protein (SEQ ID NO:2) include but are not limited to the
proteins shown as SEQ ID NO:23, SEQ ID NO:24, and SEQ ID
NO:25. The protein moil is also called p65mox in this
application. Another preferred protein of the present invention
is rat mox 1 protein shown as SEQ ID NO:21 and fragments
thereof. Another preferred protein of the present invention is
rat mox 1 B protein shown as SEQ ID NO:42 and fragments
thereof. Yet another preferred protein of the present invention
is duox 1 protein shown as SEQ ID NO:46 and fragments
thereof. Still another preferred protein of the present invention
is duox2 protein. A partial amino acid sequence of the duox2
protein is shown as SEQ ID NO:48.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
28
The nucleotide sequences of the present invention
may also be employed to hybridize to nucleic acids such as DNA
or RNA nucleotide sequences under high stringency conditions
which permit detection, for example, of alternately spliced
messages.
The genetic construct is expressed in an expression
system such as in NIH 3T3 cells using recombinant sequences in
a pcDNA-3 vector (Invitrogen, Carlsbad, CA) to produce a
recombinant protein. Preferred expression systems include but
are not limited to Cos-7 cells, insect cells using recombinant
baculovirus, and yeast. It is to be understood that other
expression systems known to one of skill in the art may be used
for expression of the genetic constructs of the present invention.
The preferred proteins of the present invention are the proteins
referred to herein as human moxi. and human mox2 or
fragments thereof which have the amino acid sequences set forth
in SEQ ID NO:3 and SEQ ID NO:4, respectively, or an amino
acid sequence having amino acid substitutions as defined in the
definitions that do not significantly alter the function of the
recombinant protein in an adverse manner. Another preferred
protein of the present invention is referred to herein as rat
moxl and has the amino acid sequence set forth in SEQ ID
NO:21. Yet another preferred protein of the present invention
is referred to herein as rat mox 1 B and has the amino acid
sequence set forth in SEQ ID NO:42. Two other preferred
proteins of the present invention are referred to herein as
human duox 1 and human duox2, or fragments thereof, which
have the amino acid sequences set forth in SEQ ID NO:46 and
SEQ ID NO:48, respectively, or an amino acid sequence having
amino acid substitutions as defined in the definitions that do not
significantly alter the function of the recombinant protein in an
adverse manner.
Terminology
It should be understood that some of the
terminology used to describe the novel mox and duox proteins
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
29
contained herein is different from the terminology in U.S.
Provisional Application Serial No. 60/107,911 and U.S.
Provisional Application Serial No. 60/149,332 upon which this
application claims priority in part. As described herein, the
term "human moxl" refers to a protein comprising an amino
acid sequence as set forth in SEQ ID NO:2, or a fragment
thereof, and encoded by the nucleotide sequence as set forth in
SEQ ID NO:1, or a fragment thereof. As described herein, the
term "human mox2" refers to a protein comprising an amino
acid sequence as set forth in SEQ ID NO:4, or a fragment
thereof, and encoded by the nucleotide sequence as set forth in
SEQ ID NO:3, or a fragment thereof. As described herein, the
term "human duox 1" refers to a protein comprising an amino
acid sequence as set forth in SEQ ID NO:46, or a fragment
thereof, and encoded by the nucleotide -sequence as set forth in
SEQ ID NO:45, or a fragment thereof. As described herein,
the term "human duox2" refers to a protein comprising an
amino, acid sequence as set forth in SEQ ID NO:48, or a
fragment thereof, and encoded by the nucleotide sequence as set
forth in SEQ ID NO:47, or a fragment thereof.
Construction of the Recombinant Gene
The desired gene is ligated into a transfer vector,
such as pcDNA3, and the recombinants are used to transform
host cells such as Cos-7 cells. It is to be understood that
different transfer vectors, host cells, and transfection methods
may be employed as commonly known to one of ordinary skill
in the art. Six desired genes for use in transfection are shown
in SEQ ID NO:1, SEQ ID NO:3, SEQ ID NO:22, SEQ ID
NO:41 SEQ ID NO:45 and SEQ ID NO:47. For example,
lipofectamine-mediated transfection and in vivo homologous
recombination was used to introduce the moxl gene into NIH
3T3 cells.
The synthetic gene is cloned and the recombinant
construct containing mox or duox gene is produced and grown
in confluent monolayer cultures of a Cos-7 cell line. The
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
expressed recombinant protein is then purified, preferably using
affinity chromatography techniques, and its purity and
specificity determined by known methods.
A variety of expression systems may be employed
5 for expression of the recombinant protein. Such expression
methods include, but are not limited to the following: bacterial
expression systems, including those utilizing E. coli and Bacillus
subtilis; virus systems; yeast expression systems; cultured insect
and mammalian cells; and other expression systems known to
10 one of ordinary skill in the art.
Transfection of Cells
It is to be understood that the vectors of the present
invention may be transfected into any desired cell or cell line.
15 Both in vivo and in vitro transfection of cells are contemplated
as part of the present invention. Preferred cells for transfection
include but are not limited to the following: fibroblasts
(possibly to enhance wound healing and skin formation),
granulocytes (possible benefit to increase function in a
20 compromised immune system as seen in AIDS, and aplastic
anemia), muscle cells, neuroblasts, stem cells, bone marrow
cells, osteoblasts, B lymphocytes, and T lymphocytes.
Cells may be transfected with a variety of methods
known to one of ordinary skill in the art and include but are not
25 limited to the following: electroporation, gene gun, calcium
phosphate, lipofectamine, and fugene, as well as adenoviral
transfection systems.
Host cells transfected with the nucleic acids
represented in SEQ ID NO: I, SEQ ID NO:3, SEQ ID NO:22,
30 SEQ ID NO:41 SEQ ID NO:45 and SEQ ID NO:47, or
fragments thereof, are used to express the proteins SEQ ID
NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID
NO:46 and SEQ ID NO:48, respectively, or fragments thereof.
These expressed proteins are used to raise
antibodies. These antibodies may be used for a variety of
applications including but not limited to immunotherapy against
CA 02350776 2001-05-08
WO 00/28031 PCTIUS99/26592
31
cancers expressing one of the mox or duox proteins, and for
detection, localization and measurement of the proteins shown
in SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ ID
NO:42, SEQ ID NO:46 or SEQ ID NO:48 or fragments thereof.
Purification and Characterization of the Expressed Protein
The proteins of the present invention can be
expressed as a fusion protein with a poly histidine component,
such as a hexa histidine, and purified by binding to a metal
affinity column using nickel or cobalt affinity matrices. The
protein can also be expressed as a fusion protein with
glutathione S-transferase and purified by affinity
chromatography using a glutathione agarose matrix. The
protein can also be purified by immunoaffinity chromatography
by expressing it as a fusion protein, for example with
hemagglutinin antigen. The expressed or naturally occurring
protein can also be purified by conventional chromatographic
and purification methods which include anion and cation
exchange chromatography, gel exclusion chromatography,
hydroxylapatite chromatography, dye binding chromatography,
ammonium sulfate precipitation, precipitation in organic
solvents or other techniques commonly known to one of skill in
the art.
Methods of Assessing Activity of Expressed Proteins
Different methods are available for assessing the
activity of the expressed proteins of the present invention,
including but not limited to the proteins represented as SEQ ID
NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID
NO:46 or SEQ ID NO:48 substituted analogs thereof, and
fragments thereof.
1. Assays of the holoprotein and fragments
thereof for superoxide generation:
A. General considerations.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
32
These assays are useful in assessing efficacy of
drugs designed to modulate the activity of the enzymes of the
present invention. The holoprotein may be expressed in COS-7
cells, NIH 3T3 cells, insect cells (using baculoviral technology)
or other cells using methods known to one of skill in the art.
Membrane fractions or purified protein are used for the assay.
The assay may require or be augmented by other cellular
proteins such as p47phox, p67phox, and Racl, as well as
potentially other unidentified factors (e.g., kinases or other
regulatory proteins).
B. Cytochrome c reduction.
NADPH or NADH is used as the reducing substrate,
in a concentration of about 100 M. Reduction of cytochrome c
is monitored spectrophotometrically - by the increase in
absorbance at 550 nm, assuming an extinction coefficient of 21
mM-1 cm-1. The assay is performed in the absence and
presence of about 10 g superoxide dismutase. The superoxide-
dependent reduction is defined as cytochrome c reduction in the
absence of superoxide dismutase minus that in the presence of
superoxide dismutase (Uhlinger et al. (1991) J. Biol. Chem.
266, 20990-20997). Acetylated cytochrome c may also be used,
since the reduction of acetylated cytochrome c is thought to be
exclusively via superoxide.
C. Nitroblue tetrazolium reduction.
For nitroblue tetrazolium (NBT) reduction, the
same general protocol is used, except that NBT is used in place
of cytochrome c. In general, about 1 mL of filtered 0.25 %
nitrotetrazolium blue (Sigma, St. Louis, MO) is added in Hanks
buffer without or with about 600 Units of superoxide dismutase
(Sigma) and samples are incubated at approximately 37 C. The
oxidized NBT is clear, while the reduced NBT is blue and
insoluble. The insoluble product is collected by centrifugation,
and the pellet is re-suspended in about 1 mL of pyridine
(Sigma) and heated for about 10 minutes at 100 C to solubilize
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
33
the reduced NBT. The concentration of reduced NBT is
determined by measuring the absorbance at 510 nm, using an
extinction coefficient of 11,000 M-1 cm-1. Untreated wells are
used to determine cell number.
D. Luminescence.
Superoxide generation may also be monitored with
a chemiluminescence detection system utilizing lucigenin (bis-N-
methylacridinium nitrate, Sigma, St. Louis, MO). The sample
is mixed with about 100 .tM NADPH (Sigma, St. Louis, MO)
and 10 M lucigenin (Sigma, St. Louis, MO) in a volume of
about 150 L Hanks solution. Luminescence is monitored in a
96-well plate using a LumiCounter (Packard, Downers Grove,
IL) for 0.5 second per reading at approximately 1 minute
intervals for a total of about 5 minutes; the highest stable value
in each data set is used for comparisons. As above, superoxide
dismutase is added to some samples to prove that the
luminescence arises from superoxide. A buffer blank is
subtracted from each reading (Ushio-Fukai et al. (1996) J. Biol.
Chem. 271, 23317-23321).
E. Assays in intact cells.
Assays for superoxide generation may be
performed using intact cells, for example, the mox-transfected
NIH 3T3 cells. In principle, any of the above assays can be used
to evaluate superoxide generation using intact cells, for
example, the mox-transfected NIH 3T3 cells. NBT reduction is
a preferred assay method.
2. Assays of truncated proteins comprised of
approximately the C-terminal 265 amino acid residues -
While not wanting to be bound by the following
statement, the truncated protein comprised of approximately the
C-terminal 265 amino acid residues is not expected to generate
superoxide, and therefore, superoxide dismutase is not added in
assays of the truncated protein. Basically, a similar assay is
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
34
established and the superoxide-independent reduction of NBT,
cytochrome c, dichlorophenolindophenol, ferricyanide, or
another redox-active dye is examined.
Nucleotides and Nucleic Acid Probes
The nucleotide sequences SEQ ID NO: 1, SEQ ID
NO:3, SEQ ID NO:22, SEQ ID NO:41 SEQ ID NO:45 and SEQ
ID NO:47, as well as fragments thereof and PCR primers
therefor, may be used, respectively, for localization, detection
and measurement of nucleic acids related to SEQ ID NO: 1, SEQ
ID NO:3, SEQ ID NO:22, SEQ ID NO:41 SEQ ID NO:45 and
SEQ ID NO:47, as well as fragments thereof. The nucleotide
sequences SEQ ID NO:1 and SEQ ID NO:3 are also called the
human moxl gene and the human mox2 gene in this application.
SEQ ID NO:22 is also known as the -rat moxl gene in this
application. SEQ ID NO:41 is also known as the rat mox1B
gene in this application. SEQ ID NO:45 is also known as the
human duoxl gene in this application. SEQ ID NO:47 is also
known as the human duox2 gene in this application.
The nucleotide sequences SEQ ID NO:1, SEQ ID
NO:3, SEQ ID NO:22, SEQ ID NO:41 SEQ ID NO:45 and SEQ
ID NO:47, as well as fragments thereof, may be used to create
probes to isolate larger nucleotide sequences containing the
nucleotide sequences SEQ ID NO:1, SEQ ID NO:3, SEQ ID
NO:22, SEQ ID NO:41 SEQ ID NO:45 and SEQ ID NO:47,
respectively. The nucleotide sequences SEQ ID NO:1, SEQ ID
NO:3, SEQ ID NO:22, SEQ ID NO:41 SEQ ID NO:45 and SEQ
ID NO:47, as well as fragments thereof, may also be used to
create probes to identify and isolate mox and duox proteins in
other species.
The nucleic acids described herein include
messenger RNA coding for production of SEQ ID NO:2, SEQ
ID NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46,
SEQ ID NO:48 and fragments thereof. Such nucleic acids
include but are not limited to cDNA probes. These probes may
be labeled in a variety of ways known to one of ordinary skill in
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
the art. Such methods include but are not limited to isotopic
and non-isotopic labeling. These probes may be used for in situ
hybridization for localization of nucleic acids such as mRNA
encoding for SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21,
5 SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48 and fragments
thereof. Localization may be performed using in situ
hybridization at both ultrastructural and light microscopic levels
of resolution using techniques known to one of ordinary skill in
the art.
10 These probes may also be employed to detect and
quantitate nucleic acids and mRNA levels using techniques
known to one of ordinary skill in the art including but not
limited to solution hybridization.
15 Antibody Production
The proteins shown in SEQ ID NO:2, SEQ ID
NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ
ID NO:48, or fragments thereof, are combined with a
pharmaceutically acceptable carrier or vehicle to produce a
20 pharmaceutical composition and administered to animals for the
production of polyclonal antibodies using methods known to one
of ordinary skill in the art. The preferred animals for antibody
production are rabbits and mice. Other animals may be
employed for immunization with these proteins or fragments
25 thereof. Such animals include, but are not limited to the
following; sheep, horses, pigs, donkeys, cows, monkeys and
rodents such as guinea pigs and rats.
The terms "pharmaceutically acceptable carrier or
pharmaceutically acceptable vehicle" are used herein to mean
30 any liquid including but not limited to water or saline, oil, gel,
salve, solvent, diluent, fluid ointment base, liposome, micelle,
giant micelle, and the like, which is suitable for use in contact
with living animal or human tissue without causing adverse
physiological responses, and which does not interact with the
35 other components of the composition in a deleterious manner.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
36
The pharmaceutical compositions may conveniently
be presented in unit dosage form and may be prepared by
conventional pharmaceutical techniques. Such techniques
include the step of bringing into association the active ingredient
and the pharmaceutical carrier(s) or excipient(s). In general,
the formulations are prepared by uniformly and intimately
bringing into association the active ingredient with liquid
carriers.
Formulations suitable for parenteral administration
include aqueous and non-aqueous sterile injection solutions
which may contain anti-oxidants, buffers, bacteriostats and
solutes which render the formulation isotonic with the blood of
the intended recipient; and aqueous and non-aqueous sterile
suspensions which may include suspending agents and thickening
agents. The formulations may be presented in unit-dose or
multi-dose containers, for example, sealed ampules and vials,
and may be stored in a freeze-dried (lyophilized) condition
requiring only the addition of the sterile liquid carrier, for
example, water for injections, immediately prior to use.
Extemporaneous injection solutions and suspensions may be
prepared from sterile powders, granules and tablets commonly
used by one of ordinary skill in the art.
Preferred unit dosage formulations are those
containing a dose or unit, or an appropriate fraction thereof, of
the administered ingredient. It should be understood that in
addition to the ingredients, particularly mentioned above, the
formulations of the present invention may include other agents
commonly used by one of ordinary skill in the art.
The pharmaceutical composition may be
administered through different routes, such as oral, including
buccal and sublingual, rectal, parenteral, aerosol, nasal,
intramuscular, subcutaneous, intradermal, and topical. The
pharmaceutical composition of the present invention may be
administered in different forms, including but not limited to
solutions, emulsions and suspensions, microspheres, particles,
microparticles, nanoparticles, and liposomes. It is expected that
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
37
from about 1 to 7 dosages may be required per immunization
regimen. Initial injections may range from about 0.1 g to 1
mg, with a preferred range of about 1 jig to 800 g, and a more
preferred range of from approximately 25 g to 500 g.
Booster injections may range from 0.1 g to 1 mg, with a
preferred range of approximately 1 g to 800 g, and a more
preferred range of about 10 pg to 500 g.
The volume of administration will vary depending
on the route of administration and the size of the recipient. For
example, intramuscular injections may range from about 0.1 ml
to 1.0 ml.
The pharmaceutical composition may be stored at
temperatures of from about 4 C to -100 C. The pharmaceutical
composition may also be stored in a lyophilized state at different
temperatures including room temperature. The pharmaceutical
composition may be sterilized through conventional means
known to one of ordinary skill in the art. Such means include,
but are not limited to filtration, radiation and heat. The
pharmaceutical composition of the present invention may also
be combined with bacteriostatic agents, such as thimerosal, to
inhibit bacterial growth.
Adjuvants
A variety of adjuvants known to one of ordinary
skill in the art may be administered in conjunction with the
protein in the pharmaceutical composition. Such adjuvants
include, but are not limited to the following: polymers, co-
polymers such as polyoxyethylene-polyoxypropylene
copolymers, including block co-polymers; polymer P1005;
Freund's complete adjuvant (for animals); Freund's incomplete
adjuvant; sorbitan monooleate; squalene; CRL-8300 adjuvant;
alum; QS 21, muramyl dipeptide; trehalose; bacterial extracts,
including mycobacterial extracts; detoxified endotoxins;
membrane lipids; or combinations thereof.
Monoclonal antibodies can be produced using
hybridoma technology in accordance with methods well known
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
38
to those skilled in the art. The antibodies are useful as research
or diagnostic reagents or can be used for passive immunization.
The composition may optionally contain an adjuvant.
The polyclonal and monoclonal antibodies useful as
research or diagnostic reagents may be employed for detection
and measurement of SEQ ID NO:2, SEQ ID NO:4, SEQ ID
NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48 and
fragments thereof. Such antibodies may be used to detect these
proteins in a biological sample, including but not limited to
samples such as cells, cellular extracts, tissues, tissue extracts,
biopsies, tumors, and biological fluids. Such detection
capability is useful for detection of disease related to these
proteins to facilitate diagnosis and prognosis and to suggest
possible treatment alternatives.
Detection may be achieved through the use of
immunocytochemistry, ELISA, radioimmunoassay or other
assays as commonly known to one of ordinary skill in the art.
The mox 1, mox2, duox 1 and duox2 proteins, or fragments
thereof, may be labeled through commonly known approaches,
including but not limited to the following: radiolabeling, dyes,
magnetic particles, biotin-avidin, fluorescent molecules,
chemiluminescent molecules and systems, ferritin, colloidal
gold, and other methods known to one of skill in the art of
labeling proteins.
Administration of Antibodies
The antibodies directed to the proteins shown as
SEQ ID NO:2, SEQ ID NO:4, SEQ ID NO:21, SEQ ID NO:42,
SEQ ID NO:46 or SEQ ID NO:48, or directed to fragments
thereof, may also be administered directly to humans and
animals in a passive immunization paradigm. Antibodies
directed to extracellular portions of SEQ ID NO:2, SEQ ID
NO:4, SEQ ID NO:21, SEQ ID NO:42, SEQ ID NO:46 or SEQ
ID NO:48 bind to these extracellular epitopes. Attachment of
labels to these antibodies facilitates localization and visualization
of sites of binding. Attachment of molecules such as ricin or
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
39
other cytotoxins to these antibodies helps to selectively damage
or kill cells expressing SEQ ID NO:2, SEQ ID NO:4, SEQ ID
NO:21, SEQ ID NO:42, SEQ ID NO:46, SEQ ID NO:48 or
fragments thereof.
Kits
The present invention includes kits useful with the
antibodies, nucleic acids, nucleic acid probes, labeled antibodies,
labeled proteins or fragments thereof for detection, localization
and measurement of SEQ ID NO: 1, SEQ ID NO:2, SEQ ID
NO:3, SEQ ID NO:4, SEQ ID NO:21, SEQ ID NO:22, SEQ ID
NO:41, SEQ ID NO:42, SEQ ID NO:45, SEQ ID NO:46, SEQ
ID NO:47, SEQ ID NO:48 or combinations and fragments
thereof.
Kits may be used for immunocytochemistry, in situ
hybridization, solution hybridization, radioimmunoassay,
ELISA, Western blots, quantitative PCR, and other assays for
the detection, localization and measurement of these nucleic
acids, proteins or fragments thereof using techniques known to
one of skill in the art.
The nucleotide sequences shown in SEQ ID NO:1,
SEQ ID NO:3, SEQ ID NO:22, SEQ ID NO:41 SEQ ID NO:45,
SEQ ID NO:47, or fragments thereof, may also be used under
high stringency conditions to detect alternately spliced messages
related to SEQ ID NO: I, SEQ ID NO:3, SEQ ID NO:22, SEQ
ID NO:41 SEQ ID NO:45, SEQ ID NO:47 or fragments
thereof, respectively.
As discussed in one of the Examples, rat moxl
protein (SEQ ID NO: 21) is similar to mouse gp9l protein
(SEQ ID NO: 38), whereas rat mox1B protein (SEQ ID NO:42)
is similar to human gp9l protein (SEQ ID NO:12). This
observation suggests that other isoforms of mouse and human
gp91 may exist. In addition, another subtype of human moxl,
similar to rat mox1B (SEQ ID NO:42), also exists. The
presence of two isoforms of rat moxl protein in vascular
smooth muscle may have important physiological consequences
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
and biomedical applications. For example, the two isoforms
may have different biological activities, different tissue
distributions and may be regulated differently in physiological
and/or pathological conditions. The fact that moxiB (SEQ ID
5 NO:42) was isolated from cells exposed to angiotensin II,
known to promote oxidative stress and vascular growth,
suggests that it may be upregulated by this hormone and may be
overexpressed in disease. Therefore, the diagnostic kits of the
present invention can measure the relative expression of the two
10 mox isoforms. The diagnostic kits may also measure or detect
the relative expression of the mox proteins described herein
(i.e. human moxi and/or human mox2) and duox proteins
described herein (i.e. human duox 1 and/or human duox2).
Fragments of SEQ ID NO: 1, SEQ ID NO:3, SEQ
15 ID NO:22, SEQ ID NO:41 SEQ ID NO:45 and SEQ ID NO:47
containing the relevant hybridizing sequence can be synthesized
onto the surface of a chip array. RNA samples, e.g., from
tumors, are then fluorescently tagged and hybridized onto the
chip for detection. This approach may be used diagnostically to
20 characterize tumor types and to tailor treatments and/or provide
prognostic information. Such prognostic information may have
predictive value concerning disease progression and life span,
and may also affect choice of therapy.
The present invention is further illustrated by the
25 following examples, which are not to be construed in any way
as imposing limitations upon the scope thereof. On the
contrary, it is to be clearly understood that resort may be had to
various other embodiments, modifications, and equivalents
thereof, which, after reading the description herein, may
30 suggest themselves to those skilled in the art without departing
from the spirit of the present invention.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
41
EXAMPLE 1
Sequence Analysis and Cloning of the Human moxl cDNA
(SEQ ID NO:1) Encoding for Production of the Human moxl
Protein p65mox (SEQ ID NO:2)
Using gp9lphox as a query sequence, a 334 base
sequenced portion of expressed sequence tag (EST) 176696
(GenBank Accession number AA305700) showed 68.8%
sequence identity at the predicted amino acid level with human
(h) gp9 l phox. The bacterial strain number 129134 containing
the EST sequence in the pBluescript SK- vector, was purchased
from American Tissue Type Culture Collection (ATCC,
Rockville, MD). The EST sequence was originally cloned from
a Caco-2 human colon carcinoma cell line. The EST176696
DNA was further sequenced using the T7 and T3 vector
promoters and primers designed to match the known 3'
sequence. Internal primers used for sequencing were as
follows: 5'-AAC AAG CGT GGC TTC AGC ATG-3' SEQ ID
NO:5 (251S, numbering is based on the nucleotides from the 5'
end of EST 176696, and S indicates the sense direction), 5'-AGC
AAT ATT GTT GGT CAT-3' SEQ ID NO:6 (336S), 5'-GAC
TTG ACA GAA AAT CTA TAA GGG-3' SEQ ID NO:7
(393S), 5'-TTG TAC CAG ATG GAT TTC AA-3' SEQ ID
NO:8 (673A, A indicates the antisense direction), 5'-CAG GTC
TGA AAC AGA AAA CCT-3' SEQ ID NO:9 (829S), 5'-ATG
AAT TCT CAT TAA TTA TTC AAT AAA-3' SEQ ID NO:10
(1455A). The coding sequence in EST176696 showed
homology to a 250 amino acid stretch corresponding to the N-
terminal 44% of human gp9 l phox, and contained a stop codon
corresponding to the location in human gp9 l phox. 5' Rapid
amplification of cDNA ends (RACE) was carried out using a
human colon cDNA library and Marathon cDNA Amplification
Kit (ClonTech) using 5'-ATC TCA AAA GAC TCT GCA CA-
3' SEQ ID NO:11 (41A) as an internal gene-specific primer
(Frohman et al. (1988) Proc. Natl. Acad. Sci. USA 85, 8998-
9002). 5' RACE resulted in a 1.1 kb fragment representing the
complete 5' sequence, based on homology with gp9 l phox.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
42
Reamplification was performed with primers spanning the
putative start and stop codons, using the 1.1 kb 5' RACE
product and pSK-EST176696 for primer design. The amplified
1.7kb fragment was TA cloned into the PCR2.1 vector
(Invitrogen, Carlsbad, CA). This recombinant vector is
referred to as PCR-mox.
Figure 1(a-d) presents a comparison of the present
amino acid sequences of human, bovine and murine gp91 phox
with the human and rat mox 1 proteins of the present invention
and the human duox2 protein of the present invention. Also
shown are the amino acid sequences for related plant enzyme
proteins.
The encoded hp65mox ("mox" referring to
mitogenic oxidase and "65" referring to its predicted molecular
weight) is listed as SEQ ID NO:2. h-gp9lphox (SEQ ID
NO: 12) and SEQ ID NO:2 differ in length by 3 residues and are
70% identical in their amino acid sequence. h-gp9lphox and
SEQ ID NO:2 show a greater percentage identity in the C-
terminal half of the molecule which contains the putative
NADPH and FAD binding sites, and there are several relatively
long stretches of complete identity within this region.
A dendrogram (Figure 2) comparing the amino
acid sequences of mouse and human gp9lphox with that of
moxl SEQ ID NO:2 shows that the latter probably represents a
distinct isoform of gp9lphox. Two plant homologs of
cytochrome b558 large subunit are also indicated and represent
more distant relatives of the human sequences. Human (and rat
moxl described more fully below) lack asparagine-linked
glycosylation sites, which are seen in the highly glycosylated
human and mouse gp9lphox. Additionally, the hydropathy
profiles of human gp9lphox and moxl are nearly identical and
include five very hydrophobic stretches in the amino-terminal
half of the molecules which are predicted to be membrane-
spanning regions.
CA 02350776 2007-02-06
WO 00/28031 PCT/US99/26592
43
EXAMPLE 2
Expression of Mox1
Human multiple tissue northern (MTN) Blot I and
Human MTN Blot IV (ClonTech) membranes were hybridized
with the putative coding region of the PCR-mox vector at 68 C
for several hours. The mox coding region was labeled by
random priming with [a-32P]dCTP (10 tCi) using the Prime-It
II kit (Stratagene). For analysis of mox 1 expression in cell
lines, total RNA was prepared from 106 cells using the High
Pure RNA Isolation Kit (Boehringer Mannheim) or RNeasy kit
(Quiagen). Total RNA (10-20 g) was separated on a 1%
agarose formaldehyde mini-gel and transferred to a Nytran
filter (Biorad) and immobilized by ultraviolet cross-linking.
Northern blotting revealed that the major location
of mRNA coding for the mox 1 protein was colon. The message
was also detected in prostate and uterus. The human colon-
carcinoma cell line, Caco-2, also expressed large quantities of
mox 1 message. Northern blotting of mRNA from rat aortic
smooth muscle cells also showed strong hybridization, which
increased roughly two-fold within 12 hours after treatment with
platelet-derived growth factor. This increase in the expression
of rat mox 1 is consistent with the idea that mox 1 contributes to
the growth-stimulatory effects of PDGF.
EXAMPLE 3
Transfection of NIH3T3 Cells with SEQ ID NO:1
The nucleotide sequence (SEQ ID NO:1) encoding
for production of the mox 1 protein (SEQ ID NO:2) was
subcloned into the Notl site of the pEF-PAC vector (obtained
from Mary Dinauer, Indiana University Medical School,
Indianapolis, IN) which has a puromycin resistance gene.
Transfection was carried out as described in Sambrook et al.,
Molecular Cloning, A Laboratory Manual, Volumes 1-3, 2nd
edition, Cold Spring Harbor Laboratory Press, N.Y., 1989.
The SEQ ID NO:1 in pEF-PAC and the empty vector were
separately transfected into NIH 3T3 cells using FugeneTM 6
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
44
(Boeringer Mannheim). About 2 x 106 cells maintained in
DMEM containing 10% calf serum were transfected with 10 g
of DNA. After 2 days, cells were split and selected in the same
medium containing 1 mg/mi puromycin. Colonies that survived
in selection media for 10 to 14 days were subcultured
continuously in the presence of puromycin.
Transfected cells exhibited a "transformed"-like
morphology, similar to that seen with (V 12)Ras-transfected
cells, characterized by long spindle-like cells. The parent NIH
3T3 cells or cells transfected with the empty vector showed a
normal fibroblast-like morphology.
EXAMPLE 4
Expression of Moxl (SEQ ID NO:1) in Transfected NIH3T3
Cells
To verify the expression of moxl mRNA after
transfection, RT-PCR and Northern blotting were performed.
Total RNAs were prepared from 106 cells using the High Pure
RNA Isolation Kit (Boeringer Mannheim) or RNeasy kit
(Qiagen). cDNAs for each colony were prepared from 1-2 .tg
of total RNA using Advantage RT-PCR Kit (ClonTech). PCR
amplification was performed using primers, 5'-TTG GCT AAA
TCC CAT CCA-3' SEQ ID NO:13 (NN459S, numbering
containing NN indicates numbering from the start codon of
moxl) and 5'-TGC ATG ACC AAC AAT ATT GCT G-3' SEQ
ID NO:14 (NN1435A). For Northern blotting, 10-20 g of
total RNA was separated on a 1% agarose formaldehyde gel and
transferred to a nylon filter. After ultraviolet (UV) cross-
linking, filters were used for Northern blotting assay as
described in Example 2.
Colonies expressing large amounts of moxl mRNA
were chosen for further analysis. The expression of mRNA for
glyceraldehyde 3 phosphate dehydrogenase in the various cell
lines was normal.
EXAMPLE 5
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
Colony Formation on Soft Agar
105 to 103 cells stably transfected with human
mox 1 gene SEQ ID NO: 1 and with empty vector were prepared
in 0.3% warm (40 C) agar solution containing DMEM and 10%
5 calf serum. Cells were distributed onto a hardened 0.6% agar
plate prepared with DMEM and 10% calf serum. After three
weeks in culture (370C, 5% CO2) colony formation was
observed by microscopy.
Cells which were stably transfected with the empty
10 vector and cultured in soft agar for 3 weeks as above did not
display anchorage independent growth. In contrast, NIH 3T3
cells which had been stably transfected with the moxl (SEQ ID
NO:1) and cultured for 3 weeks in soft agar demonstrated
anchorage independent growth of colonies.
EXAMPLE 6
NADPH-Dependent Superoxide Generation Assay
In one embodiment of the present invention, NIH
3T3 cells stably transfected with the human moxl gene (SEQ ID
NO: 1) were analyzed for superoxide generation using the
lucigenin (Bis-N-methylacridinium luminescence assay (Sigma,
St. Louis, MO, Li et al. (1998) J. Biol. Chem. 273, 2015-
2023). Cells were washed with cold HANKS' solution and
homogenized on ice in HANKS' buffer containing 15% sucrose
using a Dounce homogenizer. Cell lysates were frozen
immediately in a dry ice/ethanol bath. For the assay, 30 p.g of
cell lysate was mixed with 200 p.M NADPH and 500 M
lucigenin. Luminescence was monitored using a LumiCounter
(Packard) at three successive one minute intervals and the
highest value was used for comparison. Protein concentration
was determined by the Bradford method.
Superoxide generation was monitored in lysates
from some of the stably transfected cell lines and was compared
with superoxide generation by the untransfected NIH 3T3 cell
lysates. The results are shown in Table 4. Cell lines 26, 27,
and 28 gave the highest degree of morphological changes by
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
46
microscopic examination corresponding to the highest degree of
superoxide generation. The luminescent signal was inhibited by
superoxide dismutase and the general flavoprotein inhibitor
diphenylene iodonium, but was unaffected by added
recombinant human p47phox, p67phox and Rac 1(GTP-yS),
which are essential cytosolic factors for the phagocyte
respiratory-burst oxidase.
Table 4
Cell Line Name Superoxide Generation
(RLU)
Control (untransfected) 6045
mox 1-26 17027
mox l -27 14670
mox l -28 18411
mox l -65 5431
mox l -615 11331
moxl-+3 8645
moxl-+10 5425
mox 1-pcc 16 8050
In an alternate and preferred embodiment of the present
invention, cells that had been stably transfected with mox 1
(YA28) or with empty vector (NEF2) were grown in 10 cm
tissue culture plates in medium containing DMEM, 10% calf
serum, 100 units/ml penicillin, 100 p g/ml streptomycin, and 1
g/ml puromycin to approximately 80% confluency. Cells (five
tissue culture plates of each cell type) were washed briefly with
5 ml phosphate buffered saline (PBS) then dissociated from the
plates with PBS containing 5 mM EDTA. Cells were pelleted
by centrifuging briefly at 1000 x g.
To permeabilize the cells, freeze thaw lysis was carried
out and this was followed by passage of the cell material
through a small bore needle. The supernatant was removed and
the cells were frozen on dry ice for 15 minutes. After cells
were thawed, 200 l lysis buffer (HANKS' Buffered Salt
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
47
Solution - HBBS) containing a mixture of protease inhibitors
from Sigma (Catalog # P2714) was added. Cells on ice were
passed through an 18 guage needle 10 times and 200 l of HBSS
buffer containing 34% sucrose was added to yield a final
concentration of 17% sucrose. Sucrose appeared to enhance
stability upon storage. The combination of freeze-thawing and
passage through a needle results in lysis of essentially all of the
cells, and this material is referred to as the "cell lysate."
The cell lysates were assayed for protein concentration
using the BioRad protein assay system. Cell lysates were
assayed for NADPH-dependent chemiluminescence by
combining HBSS buffer, arachidonic acid, and 0.01 - 1 g
protein in assay plates (96 well plastic plates). The reaction was
initiated by adding 1.5 mM NADPH and 75 M lucigenin to the
assay mix to give a final concentration of 200 M NADPH and
10 M lucigenin, and the chemiluminescence was monitored
immediately. The final assay volume as 150 l. The optimal
arachidonic acid concentration was between 50-100 M. A
Packard Lumicount luminometer was used to measure
chemiluminescence of the reaction between lucigenin and
superoxide at 37 C. The plate was monitored continuously for
60 minutes and the maximal relative luminescence unit (RLU)
value for each sample was used for the graph.
Figure 3 shows the RLU at various concentrations of cell
lysates from moxl-transfected (YA28) and vector control
(NEF2) cells. The presence of NaCl or KC1 within a
concentration range of 50-150 M is important for optimal
activity. MgCl, (1-5 mM) further enhanced activity by about 2-
fold. This cell-free assay for moxl NADPH-oxidase activity is
useful for screening modulators (inhibitors or stimulators) of
the mox 1 enzyme. The assay may also be used to detect mox
and duox NADPH-oxidase activity in general and to screen for
modulators (inhibitors or stimulators) of the mox and duox
family of enzymes.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
48
EXAMPLE 7
Nitro Blue Tetrazolium Reduction by Superoxide Generated by
NIH 3T3 cells Transfected with the Moxl cDNA (SEQ ID
NO:1)
Superoxide generation by intact cells was
monitored by using superoxide dismutase-sensitive reduction of
nitroblue tetrazolium. NEF2 (vector alone control), YA26
(mox 1 (SEQ ID NO: 1)-transfected) and YA28 (mox 1 (SEQ ID
NO:1)-transfected) cells were plated in six well plates at
500,000 cells per well. About 24 hours later, medium was
removed from cells and the cells were washed once with 1 mL
Hanks solution (Sigma, St. Louis, MO). About 1 mL of filtered
0.25% Nitro blue tetrazolium (NBT, Sigma) was added in
Hanks without or with 600 units of superoxide dismutase
(Sigma) and cells were incubated at 37 C in the presence of 5%
CO2. After 8 minutes the cells were scraped and pelleted at
more than 10,000g. The pellet was re-suspended in 1 mL of
pyridine (Sigma) and heated for 10 minutes at 100 C to
solubilize the reduced NBT. The concentration of reduced NBT
was determined by measuring the absorbance at 510 nm, using
an extinction coefficient of 11,000 M-1 cm-1. Some wells were
untreated and used to determine cell number.
The data are presented in Table 5 and Figure 4 and
indicate that the moxl (SEQ ID NO:1)-transfected cells
generated significant quantities of superoxide.
Table 5
NBT Reduction (nmols/106 cells) - SOD + SOD -
vector control cells 2.5 0.5 2.1 0.5
YA26 (mox 1) cells 6.4 0.2 3.4 0.1
YA28 (mox 1) cells 5.2 0.6 3.4 0.3
-SOD, and +SOD mean in the absence or presence of added
superoxide dismutase, respectively.
Because superoxide dismutase is not likely to penetrate cells,
superoxide must be generated extracellularly. The amount of
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
49
superoxide generated by these cells is about 5-10% of that
generated by activated human neutrophils.
EXAMPLE 8
Modification of Intracellular Components in Moxl Transfected
Cells
To test whether superoxide generated by mox 1 can affect
intracellular "targets," aconitase activity in control and mox-
transfected cell lines was monitored as described in Suh et al.
(1999) Nature 401, 79-82. Aconitase contains a four-iron-
sulphur cluster that is highly susceptible to modification by
superoxide, resulting in a loss of activity, and has been used as a
reporter of intra-cellular superoxide generation . Acotinase
activity was determined as described in Gardner et al. (1995) J.
Biol. Chem. 270, 13399-13405. Acotinase activity was
significantly diminished in all three mox-transfected cell lines
designated YA26, YA28 and YA212 as compared to the
transfected control (Figure 5). Approximately 50% of the
aconitase in these cells is mitochondrial, based on differential
centrifugation, and the cytosolic and mitochondrial forms were
both affected. Control cytosolic and mitochondrial enzymes
that do not contain iron-sulfur centres were not affected.
Superoxide generated in moxl-transfected cells is therefore
capable of reacting with and modifying intracellular
components.
EXAMPLE 9
Tumor Generation in Nude Mice Receiving Cells Transfected
with the Human moxl cDNA (SEQ ID NO: I)
About 2 x 106 NIH 3T3 cells (either mox 1-
transfected with SEQ ID NO: 1 or cells transfected using empty
vector) were injected subdermally into the lateral aspect of the
neck of 4-5 week old nude mice. Three to six mice were
injected for each of three mox 1-transfected cell lines, and 3
mice were injected with the cells transfected with empty vector
(control). After 2 to 3 weeks, mice were sacrificed. The
tumors were fixed in 10% formalin and characterized by
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
histological analysis. Tumors averaged 1.5 x 1 x 1 cm in size
and showed histology typical of sarcoma type tumors. In
addition, tumors appeared to be highly vascularized with
superficial capillaries. Eleven of twelve mice injected with
5 mox 1 gene-transfected cells developed tumors, while none of
the three control animals developed tumors.
In another study, 15 mice were injected with mox l -
transfected NIH 3T3 cells. Of the 15 mice injected, 14 showed
large tumors within 17 days of injection, and tumors showed
10 expression of mox 1 mRNA. Histologically, the tumors
resembled fibrosarcomas and were similar to ras-induced
tumors. Thus, ras and mox 1 were similarly potent in their
ability to induce tumorigenicity of NIH 3T3 cells in athymic
mice.
EXAMPLE 10
Demonstration of the Role of Mox1 in Non-Cancerous Growth
A role in normal growth was demonstrated in rat
aortic vascular smooth-muscle cells by using antisense to rat
mox 1. Transfection with the antisense DNA resulted in a
decrease in both superoxide generation and serum-dependent
growth. Mox 1 is therefore implicated in normal growth in this
cell type.
EXAMPLE 11
Expression of Human Mox1 Protein (SEQ ID NO:2) in a
Baculovirus Expression System
SEQ ID NO:2 was also expressed in insect cells
using recombinant baculovirus. To establish the p65mox1
expressing virus system, the moxl gene (SEQ ID NO:l) was
initially cloned into the pBacPAK8 vector (Clontech, Palo Alto,
CA) and recombinant baculovirus was constructed using
standard methods according to manufacturer's protocols.
Briefly, PCR amplified moxl DNA was cloned into the KpnI
and EcoRI site of the vector. Primers used for PCR
amplification were: 5'-CAA GGT ACC TCT TGA CCA TGG
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
51
GAA ACT-3' , SEQ ID NO:15, and 5'-ACG AAT TCA AGT
AAA TTA CTG AAG ATA C-3' , SEQ ID NO:16. Sf9 insect
cells (2 x 106 cells) were infected with 0.5 mg of linearized
baculovirus DNA sold under the trademark BACULOGOLD
(PharMingen, San Diego, CA) and 5 mg pBacPAC8-p65moxl
using Transfection Buffers A and B (PharMingen, San Diego,
CA). After 5 days, the supernatants containing recombinant
viruses were harvested and amplified by infecting fresh sf9 cells
for 7 days. Amplification was carried out three times and the
presence of the recombinant virus containing moxl DNA was
confirmed by PCR using the same primers. After three times
amplification of viruses, plaque purification was carried out to
obtain the high titer viruses. Approximately 2 x 108 sf9 cells in
agar plates were infected for 5 days with serial dilutions of
virus and were dyed with neutral red for easy detection of virus
plaques. Selected virus plaques were extracted and the presence
of the human moxl DNA was confirmed again by PCR.
EXAMPLE 12
Cloning of a Rat Homolog of p65mox (SEQ ID NO:2)
cDNA clones of p65mox from a rat aortic smooth
muscle cell have been obtained. RT-PCR (reverse transcription
polymerase chain reaction) was carried out as follows: first
strand cDNA synthesis was performed using total RNA from rat
aortic vascular smooth muscle cells, oligo dT primer and
superscript II reverse transcriptase, and followed by incubation
with RNase H. Degenerate PCR primers were designed to
anneal to conserved areas in the coding regions of h-mox 1 and
gp9lphox of human (X04011), mouse (U43384) and porcine
(SSU02476) origin. Primers were: sense 5'-
CCIGTITGTCGIAATCTGCTSTCCTT-3', SEQ ID NO:17 and
antisense 5'-TCCCIGCAIAICCAGTAGAARTAGATCTT-3',
SEQ ID NO: 18. A major PCR product of the expected 1.1 kb
size was purified by agarose electrophoresis and used as
template in a second PCR amplification reaction.
An aliquot of the RT-PCR product was blunt-
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
52
ended, ligated into a modified Litmus 29 vector and used to
transform XL10 competent E. coli. Approximately 120
bacterial colonies were screened for the presence of a full-
length insert by direct PCR using vector primers and Taq
polymerase. Plasmids were purified from 25 positive colonies
and mapped by digestion with Bam HI. Representative plasmids
from each digestion pattern were partially sequenced. Five out
of 25 clones contained non-specific amplification products and
20 contained identical inserts similar to human (h)-moxl. One
of the latter clones was fully sequenced and found to be 83%
identical to h-mox 1 over 1060 nucleotides. A 1.1 kb probe was
generated by PCR amplification of the insert of a rat mox 1
clone with the degenerate primers described above and used to
hybridize to a Northern blot of rat vascular smooth muscle cell
RNA. A single band, migrating between 28S rRNA and 18S
rRNA, indicated the presence of a message with a size
compatible to that of human mox-1 (2.6 kb).
To obtain full-length rat mox 1, 3' and 5' rapid
amplification of cDNA ends (RACE) reactions were performed
as describe above, using the gene-specific primers 5' -
TTGGCACAGTCAGTGAGGATGTCTTC-3', SEQ ID NO:19
and 5'-CTGTTGGCTTCTACTGTAGCGTTCAAAGTT-3',
SEQ ID NO:20 for 3' and 5' RACE, respectively. Single major
1.5 kb and 850 bp products were obtained for 3' and 5' RACE,
respectively. These products were purified by agarose gel
eletrophoresis and reamplified with Taq polymerase. Both
products were cloned into the pCR 2.1 vector and used to
transform electrocompetent XL1 blue E. coli. The RACE
products were sequenced and new terminal primers were
designed: sense 5'-
TTCTGAGTAGGTGTGCATTTGAGTGTCATAAAGAC-3'
(SEQ ID NO:43), and antisense 5'-
TTTTCCGTCAAAATTATAACTTTTTATTTTCTTTTTATA
ACACAT-3' (SEQ ID NO:44). PCR amplification of rat
VSMC cDNA was performed using these primers.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
53
A single 2.6 kb product was obtained, ligated into
pCR 2.1 and used to transform electrocompetent XL 1 blue E.
coli. The insert was sequenced with 12 sense and 14 antisense
primers. Its length is 2577 bp (including primer sequences),
comprising a 1692 bp open reading frame, 127 bp 5' and 758
bp 3' untranslated regions. The presence of six in-frame stop
codons in the 5' untranslated region suggests that the full length
coding region has been obtained. Consensus polyadenylation
sequences are present at nucleotides 2201 and 2550. Conceptual
translation yields a 563 amino acid peptide, one residue shorter
than the human deduced sequence. This new amino acid
sequence is more similar to human moxl SEQ ID NO:3 (82%
identity) than to mouse gp9lphox SEQ ID NO:38 (55%
identity), suggesting that it is indeed rat mox 1 (SEQ ID NO:21).
This rat (r) homolog of p65mox protein is called r-p65mox or
p65mox/rat.pep and is shown as SEQ ID NO:21. The
nucleotide sequence encoding for r-p65mox is shown as SEQ ID
NO:22.
EXAMPLE 13
Expression of rat (r)-p65mox mRNA in Vascular Smooth
Muscle and Induction by Angiotensin 11, Platelet-Derived
Growth Factor (PDGF), and Phorbol Myristic Acid (PMA)
Using the partial cDNA clone from rat, we have
examined cultured rat aortic smooth muscle cells for expression
of message for r-p65mox. We have observed the mRNA for r-
p65mox in these cells. It has been previously reported
(Griendling et al. (1994) Circ. Res. 74, 1141-1148; Fukui et al.
(1997) Circ. Res. 80, 45-51; Ushio-Fukai et al. (1996) J. Biol.
Chem. 271, 23317-23321) that in vitro or in vivo treatment
with angiotensin II (AII) is a growth stimulus for vascular
smooth muscle cells, and that All induces increased superoxide
generation in these cells. Platelet-derived growth factor
(PDGF) and PMA are proliferative signals for vascular smooth
muscle cells. We observed that the mRNA for r-p65mox was
induced approximately 2-3 fold by angiotensin II (100 nM),
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
54
corresponding to the increased level of superoxide generation.
Thus, the increased superoxide generation in these cells
correlates with increased expression of the mRNA for this
enzyme. The mRNA for r-p65mox also increased 2 or more
fold in response to the growth stimulus PDGF (20 ng/ml), and
2-3 fold in response to PMA. Quantitation by densitometry
revealed that rat mox 1 message was induced nearly 4-fold at the
6 and 12 hour time points in response to PDGF, and about 2-
fold at the 12 hour time point in response to All. 28S RNA was
used as a control for RNA recovery.
EXAMPLE 14
Antibodies to Fragments of Human (h) p65mox (SEQ ID NO:2)
Polyclonal antibodies were raised in rabbits against
the C-terminal half of h-p65mox (residues 233 through 564,
SEQ ID NO:23) which is predicted to fold into a cytosolic
domain containing FAD and the NADPH or NADH binding
site. This domain was expressed in E. coli as an N-terminal
GST-fusion protein and was purified on glutathione agarose by
standard methods. Two antipeptide antibodies were also made
against h-p65mox (residues 243-256, referred to as Peptide A,
SEQ ID NO:24) and h-65mox (residues 538-551, referred to as
Peptide B, SEQ ID NO:25). Peptides were conjugated to
keyhole limpet hemocyanin (KLH) using glutaraldehyde.
Antigens were injected into different rabbits
initially in complete Freund's adjuvant, and were boosted 4
times with antigen in incomplete Freund's adjuvant at intervals
of every three weeks. Approximately 0.5 mg to 1 mg of
peptide was administered at each injection. Blood was drawn 1
week after each boost and a terminal bleed was carried out 2
weeks after the final boost. Antibodies to Peptide A and Peptide
B were affinity purified by column chromatography through
peptide A or peptide B conjugated to Affigel 15 (Bio-Rad,
Richmond, CA). 10 mg of peptide was covalently crosslinked
to 2 ml of Affigel 15 resin and the gel was washed with 20 ml
of binding buffer (20 mM Hepes/NaOH, pH 7.0, 200 mM NaCl,
CA 02350776 2007-02-06
WO eau MO31 PCT/US"/26592
and ().5 re Triton X-100TH). The remaining functional N-
hydrosuccinimide was blocked with 100 l of 1 M
ethanolamine. After washing with 20 ml of binding buffer, 5
n -d of the antiserum was incubated with the pep A-conjugated
5 Affigel 15 resin overnight at 4 C. Unbound protein was washed
away with 20 ml of binding buffer. Elution of the antibodies
from the gel was performed with 6 ml of elution buffer (100
mM glycine/HCI, pH 2.5, 200 mM NaCl, and 0.5% Triton X-
10OT"").The eluate was then neutralized by adding 0.9 ml of I M
10 rris/HCI. pH 8Ø The GST-fusion form of truncated pb5mox l
protein (residues 233-566, SEQ ID NO:23) was expressed in E.
coll. Samples (20 g each) were run on 12 % SDS-PAGE
either before or 1 or 4 hours after induction with 100 M IPTG
(isopropyl 0-thiogalactoside).
15 The extracted proteins were subjected to
immunoprobing with affinity purified antiserum to peptide A at
a 1:1000 dilution. The detection of antigens was performed
using an enhanced chemiluminescence kit (Amersham,
Buckinghamshire, UK). The affinity purified antibody to mox 1
20 (243-256, SEQ ID NO:24) was used at a dilution of 1:1000 in a
Western blot in which a total of 10 g of protein was added to
each lane. The major band observed at 4 hours after IPTG
induction corresponded to the size of the GST-moxl expressed
in bacteria containing the pGEX-2T vector encoding the GST-
25 mox 1 fusion protein.
Example 15
Presence of an NAD(P)H Oxidase in Ras-Transformed
Fibroblasts
30 A superoxide-generating NADPH oxidase activity
was detected in homogenates from NIH 3T3 cells, and this
activity increased about 10-15 fold in Ras-transformed NIH 3T3
cells (Table 6). To establish the stable Ras-transformed cell
lines, the DNA for human Ras encoding an activating mutation
35 at amino acid number 12 (Valine, referred to as V12-Ras) was
subcloned into BamH 1 and EcoRl sites of pCDNA3 vector
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
56
which has a neomycin resistance gene. V12-Ras in pCDNA3
and empty vector were transfected into NIH 3T3 cells using
Lipofectamine Plus (Gibco). 2 X 106 cells were maintained
with DMEM containing 10% calf serum and transfected with 1
mg of DNA. After 2-days, cells were split and selected with the
same medium but containing 1 mg/ml neomycin. Colonies
surviving in selection media for 10 to 14 days were sub-
cultured and characterized by immunoblot analysis using
antibody against human H-Ras.
The expression of Ras in cells transfected with
pcDNA-3 vector alone or in three cell lines transfected with
V12-Ras in the same vector was analyzed on a Western blot.
The three cell lines were named V12-Ras-7, V12-Ras-4, and
V12-Ras-8. The expression of V12-Ras varied widely among
the three cell lines tested. The V12-Ras-4 cell line expressed the
highest level of Ras followed by the V12-Ras-8 cell line. The
V12-Ras-7 cell line expressed the lowest level of Ras.
Lysates from each of these lines were then prepared
and tested for their ability to generate superoxide. For each cell
line, cells were washed with cold HANKS' balanced salt solution
(HBSS), collected by centrifugation, kept on dry-ice for more
than 30 min, and disrupted by suspending in low salt buffer
(LSB; 50 mM Tris/HC1, pH 7.5, 1 mM PMSF, and protease
cocktail from Sigma) and passing through a syringe needle (18
gauge) ten times. Cell lysates were frozen in dry-ice
immediately after determining the protein concentration.
Table 6 shows superoxide generation in the
transfected cells measured using the lucigenin luminescence
assay. For the assay, 5 g of cell lysates were incubated with
the reaction mixture containing 10 M lucigenin (luminescent
probe) and 100 p.M NADPH (substrate) in the presence or
absence of 100 M arachidonate in the absence or presence of
100 U of superoxide dismutase (SOD) or 1 M
diphenyleneiodonium (DPI). Luminescence of the reaction
mixture was monitored for 0.5 second by LumiCounter
(Packard) for four times at 3 second intervals. RLU in Table 5
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
57
refers to relative luminescence units.
As shown in Table 6, the luminescence was
partially inhibited by superoxide dismutase indicating that the
signal was due at least in part to the generation of superoxide.
DPI, a known inhibitor of both neutrophil and non-neutrophil
NADPH oxidase activities, completely inhibited activity. The
generation of superoxide correlated with the expression of Ras
in the three cell lines. Thus, oncogenic Ras appears to induce an
NADPH-dependent superoxide generating activity that is similar
to the activity catalyzed by p65mox 1.
Table 6
RLU/5 g protein
no additions plus SOD plus DPI
Vector Control (1) 465 154 48
V12-Ras-7 (2) 1680 578 39
V12-Ras-4 (3) 5975 2128 36
V12-Ras-8 (4) 4883 2000 35
EXAMPLE 16
Molecular Cloning of Another Rat moxl cDNA Called Rat
moxl B
A rat cDNA library was screened in an effort to
identify new rat mox sequences. The library was constructed in
a ZAP express lambda phage vector (Stratagene, La Jolla, CA)
using RNA isolated from rat vascular smooth muscle cells
which had been exposed to 100 nM angiotensin II for 4 hours.
The library was screened using standard blot hybridization
techniques with the rat mox 1 probe described previously.
Fifteen individual clones were obtained that were characterized
by PCR and restriction mapping. Two different types of clones
were thus identified and representatives of each type were
sequenced. A clone of the first type (representative of 13) was
found to be similar to the previously identified rat mox 1 and
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
58
was thus named rat mox1B. Clones of the second type
(representative of 2) were incomplete rat mox sequences.
The length of the rat mox 1 B nucleotide sequence is
2619 bp and is listed as SEQ ID NO:41. The single longest
1497 bp open reading frame encompasses nucleotides 362 to
1858. The presence of two in-frame stop codons in the 5'
untranslated region at nucleotides 74 and 257 indicates that the
full-length coding region has been isolated. Two putative
polyadenylation sites are present at positions 2243 and 2592.
Alignment of the rat moxl nucleotide sequence (SEQ ID
NO:22) and the rat mox1B nucleotide sequence (SEQ ID
NO:41) shows that the two nucleotides sequences are identical
except at their 5' ends, suggesting that they may represent two
alternatively spliced messages from the same gene. Sequence
identity starts at nucleotides 269 and 311, for rat mox 1 and rat
mox1B, respectively.
Conceptual translation of the rat mox 1 B nucleotide
sequence (SEQ ID NO:41) yields a 499 amino acid sequence
with a predicted molecular weight of 58 kDa. This amino acid
sequence for rat moxiB protein is shown in SEQ ID NO:42.
Alignment of the deduced amino acid sequences for rat mox 1
(SEQ ID NO:21) and rat mox 1 B (SEQ ID NO:42) indicates that
rat mox 1 B is identical to rat mox 1 A, except for a missing
stretch of 64 residues at the N-terminus. Therefore, rat mox1B
appears to be a splicing variant derived from the same gene as
rat moxl.
EXAMPLE 17
Sequence Analysis and Cloning of the Human Mox2 cDNA
(SEQ ID NO:3) Encoding for Production of the Human Mox2
Protein (SEQ ID NO:4)
Note that the mox2 protein as described herein, was
described in U.S. Provisional Application Serial No. 60/149,332
as mox3.
A blast search was carried out using the sequence of
mox 1 as a query sequence. The sequence identified by this
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
59
search was a sequence present in the GenBank database that
contains regions of homology with moxl and gp9lphox. The
GenBank sequence located in the search was a 90.6 kb
sequenced region of human chromosome 6 (6g25.1-26) that was
reported as a GenBank direct submission dated February 9,
1999 and given the Accession No. AL031773. Sequencing was
carried out as part of the human genome sequencing project by
S. Palmer, at Sanger Centre, in Hinxton, Cambridgeshire, UK.
The GenBank sequence was reported as being similar to
"Cytochrome B" and was not reported as having any homology
or relation to a mox protein. The sequence contained a
theoretical amino acid sequence that was derived by computer
using an algorithm that predicted intron/exon boundaries and
coding regions. This predicted region contained a 545 amino
acid sequence that was 56% identical to -mox 1 and 58% identical
to gp9lphox.
In the present invention, based on the GenBank
genomic sequence and the homologies described above, several
specific primers were designed and used to determine the tissue
expression patterns of a novel mox protein, mox2, using Human
Multiple Tissue PCR Panels (Clontech, Palo Alto, CA). The
primers were as follows: Primer 1: 5'-
CCTGACAGATGTATTTCACTACCCAG-3' (SEQ ID NO:49);
Primer 2: 5' -GGATCGGAGTCACTCCCTTCGCTG-3' (SEQ
ID NO:50); Primer 3: 5'-
CTAGAAGCTCTCCTTGTTGTAATAGA-3' (SEQ ID NO:51);
Primer 4: 5'-ATGAACACCTCTGGGGTCAGCTGA-3' (SEQ
ID NO:52). It was determined that mox2 is expressed primarily
in fetal tissues, with highest expression in fetal kidney, with
expression also seen in fetal liver, fetal lung, fetal brain, fetal
spleen and fetal thymus. Among 16 adult tissues tested, mox2
expression was seen in brain, kidney, colon and lung, although
levels of expression appeared to be very low.
Additionally, the 5' RACE (RACE = Rapid
Amplification of cDNA Ends) and 3' RACE techniques were
used to complete the sequence of the 5' and 3' regions of mox2.
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
(5' RACE kit and 3' RACE kit were from Clontech, Palo Alto,
CA and are more fully described in Frohman et al. (1988)
Proc. Natl. Acad. Sci. USA 85, 8998-9002. The 5' RACE and
3'-RACE techniques were carried out using a human fetal
5 kidney library (Marathon-Ready cDNA library, Cat. #7423-1),
using the following specific primers: 5'-RACE: Primer 4: 5'-
ATGAACACCTCTGGGGTCAGCTGA-3' (SEQ ID NO:53);
Primer 5: 5'-GTCCTCTGCAGCATTGTTCCTCTTA-3' (SEQ
ID NO:54); 3' -RACE: Primer 1: 59-
10 CCTGACAGATGTATTTCACTACCCAG-3' (SEQ ID NO:55);
Primer 2: 5' -GGATCGGAGTCACTCCCTTCGCTG-3' (SEQ
ID NO:56). The RACE procedures were successful in
completing the 5' sequence and in confirming the 3' sequence.
The complete coding sequence of mox2 is shown in SEQ ID
15 NO:2, while the predicted amino acid sequence of mox2 is
shown in SEQ ID NO:4.
In comparing the sequences of the present invention
to the, predicted coding regions of the GenBank sequence, the
GenBank sequence did not contain a start codon, appeared to be
20 missing approximately 45 base pairs at the N-terminus, and
contained one other major difference in the predicted coding
region which could have been due to inaccurate computer
prediction of intron/exon boundaries.
25 EXAMPLE 18
Sequence Analysis and Partial Cloning of the Human Duox2
cDNA (SEQ ID NO:47) Encoding for Production of the Human
Duox2 Protein (SEQ ID NO:48)
A partial cDNA clone of duox2 was obtained as follows.
30 A 535-base portion of an expressed sequence tag (EST
zc92h03.rl; Genbank accession no. W52750) from human
pancreatic islet was identified using the human gp9lphox
amino-acid sequence as a query in a Blast search. The bacterial
strain #595758 containing the EST sequence zc92h03.rl in the
35 pBluescript SK-vector was purchased from ATCC (Rockville,
MD). The DNA inserted into the pBluescript SK-vector was
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
61
further sequenced using T7 and T3 vector promoters as well as
sequence specific internal primers. The EST encoded 440
amino acids showing a 24.4% identity to gp9 l phox, including a
stop codon corresponding to the C-terminus of gp9lphox. 5'-
RACE was carried out using mRNA obtained from human
colon carcinoma cells (CaCo2) and the Marathon cDNA
Amplification Kit (ClonTech, Palo Alto). The following gene-
specific primers were used for this procedure: 5'-
GAAGTGGTGGGAGGCGAAGACATA-3' (SEQ ID NO:26)
and 5'-CCTGTCATACCTGGGACGGTCTGG-3' (SEQ ID
NO:27).
The results of the 5'-RACE yielded an additional 2
kilobase of sequenced DNA but this region did not contain the
start codon. To complete the sequence of the 5' and 3' regions
of duox2, 5'- RACE and 3' -RACE were carried out using a
human adult pancreas mRNA (Clontech, Palo Alto, CA) with
the kit of 5' RACE System for Rapid Amplification of cDNA
Ends version 2.0 (Gibco BRL, Gaithersburg, MD). PCR done
using the following specific primers resulted in a total predicted
amino acid sequence of about 1000 residues: 5'-RACE: Primer
3: 5' -GAGCACAGTGAGATGCCTGTTCAG-3' (SEQ ID
NO:28); Primer 4: 5' -
GGAAGGCAGCAGAGAGCAATGATG-3' (SEQ ID NO:29)
(for nested PCR); 3'-RACE Primer 5: 5'-
ACATCTGCGAGCGGCACTTCCAGA-3' (SEQ ID NO:30)
Primer 6: 5' -AGCTCGTCAACAGGCAGGACCGAGC-3'
(SEQ ID NO:31) (for nested PCR).
EXAMPLE 19
Sequence Analysis and Cloning of the Human Duox1 cDNA
(SEQ ID NO:45) Encoding for Production of the Human Duox1
Protein (SEQ ID NO:46)
A cDNA clone of duox 1 was obtained as follows. A
homologous 357-base portion of an expressed sequence tag
(EST nr80dl2.sl; Genbank accession no. AA641653) from an
invasive human prostate was identified by using the partial
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
62
duox2 predicted amino-acid sequence described above as a
query in a Blast search. The bacterial strain #1441736
containing the EST sequence nr80dl2.sl in the pBluescript SK-
vector was purchased from ATCC (Rockville, MD). The DNA
inserted into the pBluescript SK-vector was further sequenced
using T7 and T3 vector promoters as well as sequence specific
internal primers. The EST insert encoded 673 amino acids with
no start or stop codons present. Northern Blot analysis of
duox 1 indicated the gene was about 5.5 kilobase pairs. To
complete the sequence of 5' and 3' regions of duoxl, 5' RACE
and 3'-RACE were carried out using a human adult lung mRNA
(Clontech, Palo Alto, CA) with the kit of 5' RACE System for
Rapid Amplification of cDNA Ends version 2.0 (Gibco BRL,
Gaithersburg, MD). The RACE procedure was carried out
using the following specific primers: 5'-RACE: Primer 5: 5'-
GCAGTGCATCCACATCTTCAGCAC-3' (SEQ ID NO:32);
Primer 6: 5' -GAGAGCTCTGGAGACACTTGAGTTC-3'
(SEQ ID NO:33) (for nested PCR); 3'-RACE Primer 7: 5'-
CATGTTCTCTCTGGCTGACAAG-3' (SEQ ID NO:34);
Primer 8: 5'-CACAATAGCGAGCTCCGCTTCACGC-3' (SEQ
ID NO:35) (for nested PCR). RACE procedures were
successful in completing the 5' sequence and the 3' sequence of
duox 1. The open reading frame is approximately 4563 base
pairs.
EXAMPLE 20
Tissue Expression of Duoxl and Duox2
Based on the duoxl sequence data, several specific
primers were designed (Primer la: 5'-
GCAGGACATCAACCCTGCACTCTC-3' (SEQ ID NO:36);
Primer 2a: 5' -AATGACACTGTACTGGAGGCCACAG-3'
(SEQ ID NO:57); Primer 3a: 5'-
CTGCCATCTACCACACGGATCTGC-3' (SEQ ID NO:58);
Primer 4a: 5'-CTTGCCATTCCAAAGCTTCCATGC-3' (SEQ
ID NO:59) and used these to determine the tissue expression
patterns of duox 1 using Human Multiple Tissue PCR Panels
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
63
(Clontech, Palo Alto, CA). It was determined that duox 1 is
expressed primarily in lung, testis, placenta, prostate, pancreas,
fetal heart, fetal kidney, fetal liver, fetal lung, fetal skeletal
muscle and thymus, with highest expression in adult and fetal
lung. Among 16 adult tissues and 8 fetal tissues tested, duoxl
expression in brain, heart, kidney, colon, ovary, thymus, fetal
brain and fetal spleen appeared to be low.
Two duox2 specific primers were also used to
determine the tissue expression patterns of duox2 using Human
Multiple Tissue PCR (polymerise chain reaction) Panels
(Clontech, Palo Alto, CA). (Primer lb: 5' -
GTACAAGTCAGGACAGTGGGTGCG-3' (SEQ ID NO:60);
Primer 2b: 5'-TGGATGATGTCAGCCAGCCACTCA-3' (SEQ
ID NO:61)). Duox2 is expressed primarily in lung, pancreas,
placenta, colon, prostate, testis and fetal tissues, with highest
expression in adult lung and fetal tissues. Among 16 adult
tissues and 8 fetal tissues tested, duox2 expression in brain,
heart,. kidney, liver, skeletal muscle, thymus and fetal brain
appeared to be low.
EXAMPLE 21
Role of Duoxl and Duox2 in Collagen Crosslinking
To investigate a possible role for the human duoxl
and duox2, the model organism Caenorhabditis elegans and a
new reverse genetic tool, RNA interference (RNAi), were used
to "knock out" the homologues of duox in this organism (Fire et
al. (1998) Nature 391, 806-811). This technique involved
injection of double stranded RNA encoding a segment of Ce-
duox 1 or Ce-duox2 into gonads of C. elegans N2
hermaphrodites. Injected worms were then allowed to lay eggs,
and the harvested eggs were allowed to develop and the Fl
progeny were scored for phenotypes. This procedure has been
documented to "knock-out" the expression of the gene of
interest (Fire et al. (1998) Nature 391, 806-811).
In the case of Ce-duoxl and Ce-duox2, the
knockout animals resulted in a complex phenotype including
CA 02350776 2007-02-06
WO 00/28031 PCT/US99/26592
64
worms with large superficial blisters, short or "dumpy" worms,
worms with locomotion disorders, and worms with retained
eggs and/or larvae. Because of the high identity between Ce-
duox 1 and Ce-duox2, three different RNA constructs were
predicted to knock out either both genes or Ce-duox2 alone. In
all cases, essentially the same group of phenotypes was obtained.
Most or all of these phenotypes had been described previously
in C. elegans mutated in the collagen biosynthetic pathway. C.
elegans has an extracellular structure known as the cuticle, a
complex sheath composed largely of cross-linked collagen,
which functions as the exoskeleton of the nematode. Cross-
linking of collagen in nematodes occurs in part by cross-linking
tyrosine residues, and peroxidases such as sea urchin
ovoperoxidase and human myeloperoxidase have previously
been shown to be capable of carrying out this reaction.
Based upon the similarities of the phenotypes
obtained, the Ce-duoxl/2 knockout worms were examined for
the presence of dityrosine linkages, using an HPLC
methodology (Andersen, S.O. (1966) Acta Physiol. Scand. 66,
Suppl. 263-265; Abdelrahim et al. (1997) J. Chromatogr. B
Biomed. Sci. Appl. 696, 175-182). It was determined that
dityrosine linkages, while easily detected in the wild type
worms, were almost completely lacking in the knockout worms.
Thus, an inability to catalyze dityrosine cross-linking accounts
for the phenotype of C. elegans failing to express Ce-duox 1/2.
These data support the concept that the duox enzymes in higher
organisms can probably function in a similar manner to
modulate the extracellular milieu, possibly the extracellular
matrix and/or the basement membrane.
It should
be understood that the foregoing relates only to preferred
embodiments of the present invention and that numerous
modifications or alterations may be made therein without
CA 02350776 2001-05-08
WO 00/28031 PCT/US99/26592
departing from the spirit and the scope of the present invention
as defined in the follo\ving claims.
CA 02350776 2001-11-09
66
SEQUENCE LISTING
<110> Emory University
<120> Novel Mitogenic Regulators
<130> 6857-25
<140> CA 2,350,766
<141> 1999-11-10
<150> US 60/151,242
<151> 1999-08-27
<150> US 60/149,332
<151> 1999-08-17
<150> US 60/107,911
<151> 1998-11-10
<160> 61
<170> Patentln Ver. 2.0
<210> 1
<211> 2609
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (207)..(1901)
<400> 1
gctgatagca cagttctgtc cagagaagga aggcggaata aacttattca ttcccaggaa 60
ctcttggggt aggtgtgtgt ttttcacatc :taaaggctc acagaccctg cgctggacaa 120
atgttccatt cctgaaggac ctctccagaa tccggattgc tgaatcttcc ctgttgccta 180
gaagggctcc aaaccacctc ttgaca atg gga aac tgg gtg gtt aac cac tgg 233
Met Gly Asn Trp Val Val Asn His Trp
1 5
ttt tca gtt ttg tt:t ctg gtt gtt tgg t:t:a ggg ctg aat gtt ttc ctg 281
Phe Ser Val Leu Phe Leu Val Val Trp Leu Gly Leu Asn Val Phe Leu
15 20 25
ttt gtg gat gcc ttc ctg aaa tat gag aag gcc gac aaa tac tac tac 329
Phe Val Asp Ala Phe Leu Lys Tyr Glu Lys Ala Asp Lys Tyr Tyr Tyr
30 35 40
aca aga aaa atc ctt ggg tca aca ttg gcc tgt gcc cga gcg tct get 377
Thr Arg Lys Ile Leu Gly Ser Thr Leu Ala Cys Ala Arg Ala Ser Ala
45 50 55
ctc tgc ttg aat ttt aac agc acg ctg atc ctg ctt cct gtg tgt cgc 425
Leu Cys Leu Asn Phe Asn Ser Thr Leu Ile Leu Leu Pro Val Cys Arg
60 65 70
aat ctg ctg tcc ttc ctg agg ggc acc tgc tca ttt tgc agc cgc aca 473
Asn Leu Leu Ser Phe Leu Arg Gly Thr Cys Ser Phe Cys Ser Arg Thr
75 80 85
ctg aga aag caa tog gat cac aa.c ctc acc ttc cac aag ctg gtg gcc 521
Leu Arg Lys Gln Leu Asp His Asn Leu Thr Phe His Lys Leu Val Ala
90 95 100 105
tat atg atc tgc cta cat aca get att cac atc att gca cac ctg ttt 569
Tyr Met Ile Cys Lau His Thr Ala Ile His Ile Ile Ala His Leu Phe
CA 02350776 2001-11-09
67
110 115 120
aac ttt gac tgc tat agc aga ago cga cag gcc aca gat ggc tcc ctt 617
Asn Phe Asp Cys Tyr Ser Arg Ser Arg Gln Ala Thr Asp Gly Ser Leu
125 130 135
gcc tcc att ctc tcc agc cta tct cat gat gag aaa aag ggg ggt tct 665
Ala Ser Ile Leu Ser Ser Leu Ser His Asp Glu Lys Lys Gly Gly Ser
140 145 150
tgg cta aat ccc atc cag too cga ,ac acg aca gtg gag tat gtg aca 713
Trp Leu Asn Pro Ile Gln Ser Arg_ Len Thr Thr Val Glu Tyr Val Thr
155 160 165
ttc acc agc gtt gc: ggt ctc act gga gtg atc atg aca ata gcc ttg 761
Phe Thr Ser Val Ala Gly Leu Thr GG;ly Val Ile Met Thr Ile Ala Leu
170 175 180 185
att ctc atg gta act tca get act crag ttc atc cgg agg agt tat ttt 809
Ile Leu Met Val Thr Ser Ala Thr Glu Phe Ile Arg Arg Ser Tyr Phe
190 195 200
gaa gtc ttc tgg tat act cac cac ctt ttt atc ttc tat atc ctt ggc 857
Glu Val Phe Trp Tyr Thr His His Leu Phe Ile Phe Tyr Ile Leu Gly
205 x'.10 215
tta ggg att cac ggc att ggt gga art gtc cgg ggt caa aca gag gag 905
Leu Gly Ile His Gly Ile Gly Gly __1e Val Arg Gly Gln Thr Glu Glu
220 225 230
agc atg eat gag agt cat cct cg(-,- aag tgt gca gag tct ttt gag atg 953
Ser Met Asn Glu Ser His Pro Arg Lys Cys Ala Glu Ser Phe Glu Met
235 240 245
tgg gat gat cgt gac tcc cac tgt agg cgc cct aaq ttt gaa ggg cat 1001
Trp Asp Asp Arg Asp Ser His Cys Lg Arg Pro Lys Phe Glu Gly His
250 255 260 265
ccc cct gag tct tgg aag tgg atc Ott gca ccg gtc att ctt tat atc 1049
Pro Pro Glu Ser Trp Lys Trp Ile Leu Ala Pro Val Ile Leu Tyr Ile
270 275 280
tgt gaa agg atc ctc cgg ttt tat cgc tcc cag cag aag gtt gtg att 1097
Cys Glu Arg Ile Leu Arg Phe Tyr Arg Ser Gln Gin Lys Val Val Ile
285 290 295
acc aag gtt gtt atg cac cca tcc aaa gtt ttg gaa ttg cag atg aac 1145
Thr Lys Val Val Met His Pro Ser Lys Val Leu Glu Leu Gln Met Asn
300 305 310
aag cgt ggc ttc agc atg gaa gtg ggg cag tat atc ttt gtt eat tgc 1193
Lys Arg Gly Phe Ser Met Glu Val Gly Gln Tyr Ile Phe Val Asn Cys
315 320 325
ccc tca atc tct ctc ctg gaa tgg cat cct ttt act ttg acc tct get 1241
Pro Ser Ile Ser Leu Leu Glu Trp His Pro Phe Thr Leu Thr Ser Ala
330 335 340 345
cca gag gaa gat ttc ttc tcc att cat at:c cga gca gca ggg gac tgg 1289
Pro Glu Glu Asp Phe Phe Ser Ile His Ile Arg Ala Ala Gly Asp Trp
350 355 360
aca gaa aat ctc eta agg get ttc gaa caa caa tat tca cca att ccc 1337
Thr Glu Asn Leu Ile Arg Ala Phe Glu Gin Gln Tyr Ser Pro Ile Pro
365 370 375
agg att gaa gtg gat ggt ccc ttt ggc aca gcc agt gag gat gtt ttc 1385
Arg Ile Glu Val Asp Gly Pro Phe Gly Thr Ala Ser Glu Asp Val Phe
380 385 390
CA 02350776 2001-11-09
68
cag tat gaa gtg get gtg ctg gtt gca gca gga att. ggg gtc acc ccc 1433
Gln Tyr Glu Val Ala Val Leu Val. G y Ala Gly Iles Gly Val Thr Pro
395 400 405
ttt get tct atc ttg aaa tcc atc t;gq tac aaa ttc cag tgt gca gac 1481
Phe Ala Ser Ile Leu Lys Ser Ile Vrp Tyr Lys Phe Gln Cys Ala Asp
410 415 420 425
cac aac ctc aaa aca aaa aag atc tat ttc tac tgg atc tgc agg gag 1529
His Asn Leu Lys Thr Lys Lys Ile Tyr Phe Tyr Trp Ile Cys Arg Glu
430 435 440
aca ggt gcc ttt tcc tgg ttc aac aac ctg ttg act; tcc ctg gaa cag 1577
Thr Gly Ala Phe Ser Trp Phe Asn Asn Leu Leu Thr Ser Leu Glu Gln
445 X150 455
gag atg gag gaa tta ggc aaa gtg qqt ttt cta aac tac cgt ctc ttc 1625
Glu Met Glu Glu Leu Gly Lys Va'_. Gly Phe Leu Asn Tyr Arg Leu Phe
460 465 470
ctc acc gga tgg gac agc aat att got ggt cat gca gca tta aac ttt 1673
Leu Thr Gly Trp Asp Ser Asn Ile Val Gly His Ala Ala Leu Asn Phe
475 480 485
gac aag gcc act gac atc gtg ac,-) ggt ctg aaa cag aaa acc tcc ttt 1721
Asp Lys Ala Thr Asp Ile Val Thr G-_y Leu Lys Gin Lys Thr Ser Phe
490 495 500 505
ggg aga cca atg tgg gac aat gag ttt tct aca ate get acc tcc cac 1769
Gly Arg Pro Met Trp Asp Asn Glu he Ser Thr Ile Ala Thr Ser His
510 515 520
ccc aag tct gta gtg gga gtt ttc tta tgt ggc cc,,. cgg act ttg gca 1817
Pro Lys Ser Val Val Gly Val Phe Leu Cys Gly Pro Arg Thr Leu Ala
525 330 535
aag agc ctg cgc aaa tgc tgt cac cga tat tcc ago ctg gat cct aga 1865
Lys Ser Leu Arg Lys Cys Cys His Arg Tyr Ser Ser Leu Asp Pro Arg
540 545 550
aag gtt caa ttc ta.c ttc aac aaa gaa aat ttt tga gttataggaa 1911
Lys Val Gln Phe Tyr Phe Asn Lys G3Lu Asn Phe
555 560 565
taaggacggt aatctgcatt ttgtctcttt gtatcttcag taattgagtt ataggaataa 1971
ggacggtaat ctgcattttg tctctttgta octtcagtaa tttacttggt ctcntcaggt 2031
ttgancagtc actttaggat aagaatgtgc ctctcaagcc ttgactccct ggtattcttt 2091
ttttgattgc attcaacttc gttacttgag cttcagcaac ttaagaactt ctgaagttct 2151
taaagttctg aanttcttaa agcccatgga ccctttctca gaaaaataac tgtaaatctt 2211
tctggacagc catgactgta gcaaggcttg atagcagaag tttggtggtt canaattata 2271
caactaatcc caggtgattt tatcaattcc agtgttacca tctcctgagt tttggtttgt 2331
aatcttttgt ccctcccacc cccacagaag attttaagta gggtgacttt ttaaataaaa 2391
atttattgaa taattaatga taaaacataa taat;aaacat aaataataaa caaaattacc 2451
gagaacccca tccccatata acaccaacag tgtacatgtt tactgtcact tttgatatgg 2511
tttatccagt gtgaacagca atttattatt tttgctcatc aaaaaataaa ggattttttt 2571
tcacttgaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaa 2609
<210> 2
CA 02350776 2001-11-09
69
<211> 564
<212> PRT
<213> Homo sapiens
<400> 2
Met Gly Asn Trp Val Val Asn His ?rp Phe Ser Val Leu Phe Leu Val
1 5 10 15
Val Trp Leu Gly Leu Asn Val Phe Leu Phe Val Asp Ala Phe Leu Lys
20 25 30
Tyr Glu Lys Ala Asp Lys Tyr Tyr P'yr Thr Arg Lys Ile Leu Gly Ser
35 4(1) 45
Thr Leu Ala Cys Ala Arg Ala Se--,. 'Ala Leu Cys Leu Asn Phe Asn Ser
50 55 60
Thr Leu Ile Leu Leu Pro Val Cys Arg Asn Leu Leu Ser Phe Leu Arg
65 70 75 80
Gly Thr Cys Ser Phe Cys Ser Arg_ Thr Leu Arg Lye; Gln Leu Asp His
85 90 95
Asn Leu Thr Phe His Lys Leu Val Ala Tyr Met Ile Cys Leu His Thr
100 L05 110
Ala Ile His Ile Ile Ala His Leu Phe Asn Phe Asp Cys Tyr Ser Arg
115 120 125
Ser Arg Gin Ala Thr Asp Gly Ser. Leu Ala Ser Ile Leu Ser Ser Leu
130 135 140
Ser His Asp Glu Lys Lys Gly Gly Ser Trp Leu Asn Pro Ile Gln Ser
145 150 155 160
Arg Asn Thr Thr Val Glu Tyr Val "fir Phe Thr Ser Val Ala Gly Leu
165 ].70 175
Thr Gly Val Ile Met Thr Ile Ala Leu Ile Leu Met Val Thr Ser Ala
180 185 190
Thr Glu Phe Ile Arg Arg Ser Tyr ?he Glu Val Phe Trp Tyr Thr His
195 200 205
His Leu Phe Ile Phe Tyr Ile Leu sly Leu Gly Ile His Gly Ile Gly
210 215 220
Gly Ile Val Arg Gly Gln Thr Glu Glu Ser Met Asn Glu Ser His Pro
225 230 235 240
Arg Lys Cys Ala Glu Ser Phe Glu Met Trp Asp Asp Arg Asp Ser His
245 250 255
Cys Arg Arg Pro Lys Phe Glu Gly 3iss Pro Pro Glu Ser Trp Lys Trp
260 265 270
Ile Leu Ala Pro Val Ile Leu Tyr Ile Cys Glu Arg Ile Leu Arg Phe
275 280 285
Tyr Arg Ser Gln Gln Lys Val Val Ile Thr Lys Val Val Met His Pro
290 295 300
Ser Lys Val Leu Glu Leu Gln Met Asn Lys Arg Gly Phe Ser Met Glu
305 310 315 320
Val Gly Gln Tyr Ile Phe Val Asn '-ys Pro Ser Ile Ser Leu Leu Glu
325 330 335
Trp His Pro Phe Thr Leu Thr Ser Ala Pro Glu Glu Asp Phe Phe Ser
340 345 350
CA 02350776 2001-11-09
Ile His Ile Arg Ala Ala Gly Asp Trp Thr Glu Asn Leu Ile Arg Ala
355 360 365
Phe Glu Gln Gln Tyr Ser Pro Ii Pro Arg Ile Glu Val Asp Gly Pro
370 375 380
Phe Gly Thr Ala Ser Glu Asp Val Phe Gln Tyr Glu Val Ala Val Leu
385 390 395 400
Val Gly Ala Gly Ile Gly Val Thr. Pro Phe Ala Ser Ile Leu Lys Ser
405 410 415
Ile Trp Tyr Lys Phe Gln Cys Ala Asp His Asn Leu Lys Thr Lys Lys
420 425 430
Ile Tyr Phe Tyr Trp Ile Cys Arg Glu Thr Gly Ala Phe Ser Trp Phe
435 440 445
Asn Asn Leu Leu Thr Ser Leu Glu Gin Glu Met Glu Glu Leu Gly Lys
450 455 460
Val Gly Phe Leu Asn Tyr Arg Leu Phe Leu Thr Gly Trp Asp Ser Asn
465 470 475 480
Ile Val Gly His Ala Ala Leu Asri Phe Asp Lys Ala Thr Asp Ile Val
485 490 495
Thr Gly Leu Lys Gln Lys Thr Sec Phe Gly Arg Pro Met Trp Asp Asn
500 505 510
Glu Phe Ser Thr Ile Ala Thr Sear. His Pro Lys Ser Val Val Gly Val
515 520 525
Phe Leu Cys Gly Pro Arg Thr Leu Ala Lys Ser Leu Arg Lys Cys Cys
530 535 540
His Arg Tyr Ser Ser Leu Asp Pro Ar:g Lys Val Gln Phe Tyr Phe Asn
545 550 555 560
Lys Glu Asn Phe
<210> 3
<211> 2044
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (104)..(1810)
<400> 3
caaagacaaa ataatttact agggaagccc ttactaacga cccaacatcc agacacaggt 60
gagggagaag aaatttcctg acagccgaag agccacaagt atc atg atg ggg tgc 115
Met Met Gly Cys
1
tgg att ttg aat gag ggt ctc too acc ata tta gte ctc tca tgg ctg 163
Trp Ile Leu Asn Glu Gly Leu Ser Thr Ile Leu Val- Leu Ser Trp Leu
5 10 15 20
gga ate aat ttt tat ctg ttt att: gac acg ttc tac tgg tat gaa gag 211
Gly Ile Asn Phe Tyr Leu Phe Ile Asp Thr Phe Tyr. Trp Tyr Glu Glu
25 30 35
gag gag tct ttc cat tac aca cga gt;t ett ttg ggt tca aca ctg get 259
Glu Glu Ser Phe His Tyr Thr Arg Val Ile Leu Gly Ser Thr Leu Ala
40 45 50
CA 02350776 2001-11-09
71
tgg gca cga gca tcc gca ctg tgc cog aat ttt aac tgc atg cta att 307
Trp Ala Arg Ala Ser Ala Leu Cys Leu Asn Phe Asn Cys Met Leu Ile
55 60 65
cta ata cct gtc agt cga aac ctt att tca ttc ata aga gga aca agt 355
Leu Ile Pro Val Ser Arg Asn Leu ---le Ser Phe Ile Arg Gly Thr Ser
70 75 80
att tgc tgc aga gga ccg tgg agg agg caa tta gac aaa aac ctc aga 403
Ile Cys Cys Arg Gly Pro Trp Arg Arg Gln Leu Asp Lys Asn Leu Arg
85 90 95 100
ttt cac aaa ctg gtc gcc tat ggg ate get gtt aat gca acc atc cac 451
Phe His Lys Leu Val Ala Tyr Gly --le Ala Val Asn Ala Thr Ile His
105 110 115
atc gtg gcg cat ttc ttc aac ctg gaa cgc tac cac tgg agc cag tcc 499
Ile Val Ala His Phe Phe Asn Leu Gl.u Arg Tyr His Trp Ser Gln Ser
120 0:5 130
gag gag gcc cag gga ctt ctg gcc gca ctt tcc aag ctg ggc aac acc 547
Glu Glu Ala Gln Gly Leu Leu Ala Ala Leu Ser Lys Leu Gly Asn Thr
135 140 145
cct aac gag agc tac ctc aac cc: gt:c cgg acc ttc ccc aca aac aca 595
Pro Asn Glu Ser Tyr Leu Asn Pro Val Arg Thr Phe Pro Thr Asn Thr
150 155 160
acc act gaa ttg cta agg aca ate qca ggc gtc acc ggt ctg gtg atc 643
Thr Thr Glu Leu Leu Arg Thr Ile Ala Gly Val Thr Gly Leu Val Ile
165 170 175 180
tct ctg get tta gtc ttg atc atg acc tcg tca act gag ttc atc aga 691
Ser Leu Ala Leu Val Leu Ile Met Thr Ser Ser Thr Glu Phe Ile Arg
185 190 195
cag gcc tcc tat gag ttg ttc tgg tac aca cac cat gtt ttc atc gtc 739
Gln Ala Ser Tyr Glu Leu Phe Trp Tyr Thr His His Val Phe Ile Val
200 :220%5 210
ttc ttt ctc agc ctg gcc atc ca, qqg acg ggt cgq att gtt cga ggc 787
Phe Phe Leu Ser Leu Ala Ile His Gl.y Thr Gly Arq Ile Val Arg Gly
215 220 225
caa acc caa gac agt ctc tct ctg cac aac atc acc ttc tgt aga gac 835
Gln Thr Gln Asp Ser Leu Ser Leu His Asn Ile Thr Phe Cys Arg Asp
230 235 240
cgc tat gca gaa tgg cag aca gtg gcc caa tgc ccc gtg cct caa ttt 883
Arg Tyr Ala Glu Trp Gln Thr Vat Ala Gln Cys Pro Val Pro Gln Phe
245 250 255 260
tct ggc aag gaa ccc tcg get tgcj aaa tgg att tta ggc cct gtg gtc 931
Ser Gly Lys Glu Pro Ser Ala Trp Lys Trp Ile Leu Gly Pro Val Val
265 270 275
ttg tat gca tgt gaa aga ata att agg ttc tgg cga ttt caa caa gaa 979
Leu Tyr Ala Cys Glu Arg Ile Ile Arg Phe Trp Arg Phe Gln Gln Glu
280 285 290
gtt gtc att acc aag gtg gte agc cac ccc tct gga gtc ctg gaa ctt 1027
Val Val Ile Thr Lys Val Val Ser His Pro Ser Gly Val Leu Glu Leu
295 300 305
cac atg aaa aag cgt ggc ttt aaa atg gcg cca ggg cag tac atc ttg 1075
His Met Lys Lys Arg Gly Phe Lys Met Ala Pro Gly Gln Tyr Ile Leu
310 315 320
gtg cag tgc cca gcc ata tct tog cog gag tgg cac ccc ttc acc ctt 1123
CA 02350776 2001-11-09
72
Val Gln Cys Pro Ala Ile Ser Ser Leu Glu Trp His Pro Phe Thr Leu
325 330 335 340
acc tct gcc ccc cag gaa gac ttt t:t.c agc gtg cac atc cgg gca gca 1171
Thr Ser Ala Pro Gln Glu Asp Phe Phe Ser Val His Ile Arg Ala Ala
345 350 355
gga gac tgg aca gca gcg cta ctg gag gcc ttt ggg gca gag gga cag 1219
Gly Asp Trp Thr Ala Ala Leu Leo Lb Ala Phe Gly Ala Glu Gly Gln
360 365 370
gcc ctc cag gag ccc tgg agc ctq cca agg ctg gca gtg gac ggg ccc 1267
Ala Leu Gln Glu Pro Trp Ser Leu Pro Arg Leu Ala Val Asp Gly Pro
375 380 385
ttt gga act gcc ctg aca gat gta ttt cac tac cca gtg tgt gtg tgc 1315
Phe Gly Thr Ala Leu Thr Asp Val PLe His Tyr Pro Val Cys Val Cys
390 395 400
gtt gcc gcg ggg atc gga gtc act: ccc ttc get got, ctt ctg aaa tct 1363
Val Ala Ala Gly Ile Gly Val Thr Pro Phe Ala Ala Leu Leu Lys Ser
405 410 415 420
ata tgg tac aaa tgc agt gag gca cag acc cca ctg aag ctg agc aag 1411
Ile Trp Tyr Lys Cys Ser Glu Ala Gin Thr Pro Leo Lys Leu Ser Lys
425 430 435
gtg tat ttc tac tgg att tgc cgg gat gca aga get ttt gag tgg ttt 1459
Val Tyr Phe Tyr Trp Ile Cys Arg Asp Ala Arg Ala Phe Glu Trp Phe
440 4445 450
get gat ctc tta ctc tcc ctg gaa aca cgg atg agt gag cag ggg aaa 1507
Ala Asp Leu Leu Leu Ser Leu Glu "hi. Arg Met Ser Glu Gln Gly Lys
455 461) 465
act cac ttt ctg agt tat cat ata t:tt ctt acc ggc tgg gat gaa aat 1555
Thr His Phe Leu Ser Tyr His Ile Phe Leu Thr Gly Trp Asp Glu Asn
470 475 480
cag get ctt cac ata get tta cac t:gg gac gaa aat. act gac gtg att 1603
Gin Ala Leu His Ile Ala Leu His Yrp Asp Glu Asn Thr Asp Val Ile
485 490 495 500
aca ggc tta aag cag aag acc ttc t:at ggg agg ccc aac tgg aac aat 1651
Thr Gly Leu Lys Gln Lys Thr Phe ~'yr Gly Arg Pro Asn Trp Asn Asn
505 510 515
gag ttc aag cag att gcc tac aat cac ccc ago agc agt att ggc gtg 1699
Glu Phe Lys Gln Ile Ala Tyr Asn Hi.s Pro Ser Ser Ser Ile Gly Val
520 555 530
ttc ttc tgt gga cct aaa got cto- tog agg aca ctt caa aag atg tgc 1747
Phe Phe Cys Gly Pro Lys Ala Leu Ser Arg Thr Leu Gln Lys Met Cys
535 540 545
cac ttg tat tca tca got gac ccc aga ggt gtt cat. ttc tat tac aac 1795
His Leu Tyr Ser Ser Ala Asp Pro Arg Gly Val His Phe Tyr Tyr Asn
550 555 560
aag gag agc ttc tag actttggagg tcaagtccag gcatt:gtgtt ttcaatcaag 1850
Lys Glu Ser Phe
565
ttattgattc caaagaactc caccaggaat tcct:gtgacg gcctgttgat atgagctccc 1910
agttgggaac tggtgaataa taattaac:a ttgtgaacag tacactatac catacttcct 1970
tagcttataa ataacatgtc at:atacaaca qaacaaaaac atttactgaa attaaaatat 2030
attatgtttc tcca 2044
CA 02350776 2001-11-09
73
<210> 4
<211> 568
<212> PRT
<213> Homo sapiens
<400> 4
Met Met Gly Cys Trp Ile Leu As. Glu Gly Leu Ses: Thr Ile Leu Val
1 5 10 15
Leu Ser Trp Leu Gly Ile Asn Phe ^yr Leu Phe Ile Asp Thr Phe Tyr
20 25 30
Trp Tyr Glu Glu Glu Glu Ser Phe His Tyr Thr Arg Val Ile Leu Gly
35 40 45
Ser Thr Leu Ala Trp Ala Arg Ala Oar Ala Leu Cys Leu Asn Phe Asn
50 55 60
Cys Met Leu Ile Leu Ile Pro Val Ser Arg Asn Leu Ile Ser Phe Ile
65 70 75 80
Arg Gly Thr Ser Ile Cys Cys Arg Gly Pro Trp Arg Arg Gln Leu Asp
85 90 95
Lys Asn Leu Arg Phe His Lys Len Val. Ala Tyr Gly Ile Ala Val Asn
100 105 110
Ala Thr Ile His Ile Val Ala His Phe Phe Asn Leu Glu Arg Tyr His
115 120 125
Trp Ser Gln Ser Glu Glu Ala Gln Gly Leu Leu Ala Ala Leu Ser Lys
130 135 140
Leu Gly Asn Thr Pro Asn Glu Ser.-'Pyr Leu Asn Pro Val Arg Thr Phe
145 150 155 160
Pro Thr Asn Thr Thr Thr Glu Len Leu Arg Thr Ile Ala Gly Val Thr
165 170 175
Gly Leu Val Ile Ser Leu Ala Len Val Leu Ile Met Thr Ser Ser Thr
180 -85 190
Glu Phe Ile Arg Gln Ala Ser Tyr Gin Leu Phe Trp Tyr Thr His His
195 202 205
Val Phe Ile Val Phe Phe Leu Ser Leu Ala Ile His Gly Thr Gly Arg
210 215 220
Ile Val Arg Gly Gln Thr Gln Asp Ser Leu Ser Leu His Asn Ile Thr
225 230 235 240
Phe Cys Arg Asp Arg Tyr Ala Gin Top Gln Thr Val Ala Gln Cys Pro
245 250 255
Val Pro Gln Phe Ser Gly Lys Gin Pro Ser Ala Trp Lys Trp Ile Leu
260 265 270
Gly Pro Val Val Leu Tyr Ala Cys Glu Arg Ile Ile Arg Phe Trp Arg
275 282 285
Phe Gln Gln Glu Val Val Ile Thr Lys Val Val Ser His Pro Ser Gly
290 295 300
Val Leu Glu Leu His Met Lys Lys Arg Gly Phe Lys Met Ala Pro Gly
305 310 315 320
Gln Tyr Ile Leu Val Gln Cys Pro Ala Ile Ser Ser Leu Glu Trp His
325 330 335
CA 02350776 2001-11-09
74
Pro Phe Thr Leu Thr Ser Ala Pro Sin Glu Asp Phe Phe Ser Val His
340 345 350
Ile Arg Ala Ala Gly Asp Trp Thr. A.La Ala Leu Leu Glu Ala Phe Gly
355 360 365
Ala Glu Gly Gln Ala Leu Gln Glu Pro Trp Ser Leu Pro Arg Leu Ala
370 375 380
Val Asp Gly Pro Phe Gly Thr Ala Leu Thr Asp Val Phe His Tyr Pro
385 390 395 400
Val Cys Val Cys Val Ala Ala Gly Ile Gly Val Thr Pro Phe Ala Ala
405 410 415
Leu Leu Lys Ser Ile Trp Tyr Lys Cys Ser Glu Ala Gln Thr Pro Leu
420 425 430
Lys Leu Ser Lys Val Tyr Phe Tyr Trp :Ile Cys Arg Asp Ala Arg Ala
435 440 445
Phe Glu Trp Phe Ala Asp Leu Leu Leu Ser Leu Glu Thr Arg Met Ser
450 455 460
Glu Gln Gly Lys Thr His Phe Leu Ser Tyr His Ile Phe Leu Thr Gly
465 470 475 480
Trp Asp Glu Asn Gin Ala Leu His Ile Ala Leu His Trp Asp Glu Asn
485 490 495
Thr Asp Val Ile Thr Gly Leu Lys Gin Lys Thr Phe Tyr Gly Arg Pro
500 505 510
Asn Trp Asn Asn Glu Phe Lys Gln Ile Ala Tyr Asn His Pro Ser Ser
515 520 525
Ser Ile Gly Val Phe Phe Cys Gly ?ro Lys Ala Leu Ser Arg Thr Leu
530 535 540
Gln Lys Met Cys His Leu Tyr Ser Ser Ala Asp Pro Arg Gly Val His
545 550 555 560
Phe Tyr Tyr Asn Lys Glu Ser Phe
565
<210> 5
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 5
aacaagcgtg gcttcagcat g 21
<210> 6
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 6
agcaatattg ttggtcat 18
CA 02350776 2001-11-09
<210> 7
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 7
gacttgacag aaaatctata aggg 24
<210> 8
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 8
ttgtaccaga tggatttcaa 20
<210> 9
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 9
caggtctgaa acagaaaacc t 21
<210> 10
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 10
atgaattctc attaattatt caataaa 27
<210> 11
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 11
atctcaaaag actctgcaca 20
<210> 12
<211> 569
<212> PRT
<213> Homo sapiens
<400> 12
Gly Asn Trp Ala Val Asn Glu Gly Leu Ser Ile Phe Ala Ile Leu Val
1 5 10 15
CA 02350776 2001-11-09
76
Trp Leu Gly Leu Asn Val Phe Leu Phe Val Trp Tyr Tyr Arg Val Tyr
20 25 30
Asp Ile Pro Pro Lys Phe Phe Tyr Thr Arg Lys Leu Leu Gly Ser Ala
35 40 45
Leu Ala Leu Ala Arg Ala Pro Ala --',La Cys Leu Asn Phe Asn Cys Met
50 55 60
Leu Ile Leu Leu Pro Val Cys Arg Asn Leu Leu Ser Phe Leu Arg Gly
65 70 75 80
Ser Ser Ala Cys Cys Ser Thr Arg Jal Arg Arg Gln Leu Asp Arg Asn
85 90 95
Leu Thr Phe His Lys Met Val Ala Top Met Ile Ala Leu His Ser Ala
100 L05 110
Ile His Thr Ile Ala His Leu Phe Asn Val G1u Trp Cys Val Asn Ala
115 120 125
Arg Val Asn Asn Ser Asp Pro Tyr Oer Val Ala Leo Ser Glu Leu Gly
130 135 140
Asp Arg Gln Asn Glu Ser Tyr Leu Asn Phe Ala Arq Lys Arg Ile Lys
145 150 155 160
Asn Pro Glu Gly Gly Leu Tyr Lea Ala Val Thr Leu Leu Ala Gly Ile
165 170 175
Thr Gly Val Val Ile Thr Leu Cys eu :[le Leu Ile Ile Thr Ser Ser
180 i85 190
Thr Lys Thr Ile Arg Arg Ser Tyr he Glu Val Phe Trp Tyr Thr His
195 200 205
His Leu Phe Val Ile Phe Phe Ile G1y Leu Ala Ile His Gly Ala Glu
210 215 220
Arg Ile Val Arg Gly Gln Thr Ala G1u Ser Leu Ala Val His Asn Ile
225 230 235 240
Thr Val Cys Glu Gln Lys Ile Ser Glu Trp Gly Lys Ile Lys Glu Cys
245 250 255
Pro Ile Pro Gln Phe Ala Gly Asn Pro Pro Met Thr Trp Lys Trp Ile
260 265 270
Val Gly Pro Met Phe Leu Tyr Leo Cys Glu Arg Leu Val Arg Phe Trp
275 280 285
Arg Ser Gln Gln Lys Val Val Ile Thr Lys Val Val. Thr His Pro Phe
290 295 300
Lys Thr Ile Glu Leu Gln Met Lys Lys Lys Gly Phe Lys Met Glu Val
305 310 315 320
Gly Gln Tyr Ile Phe Val Lys Cys Pro Lys Val Set Lys Leu Glu Trp
325 330 335
His Pro Phe Thr Leu Thr Ser Alai Pro Glu Glu Asp Phe Phe Ser Ile
340 -~45 350
His Ile Arg Ile Val Gly Asp Trp Thr Glu Gly Leu Phe Asn Ala Cys
355 360 365
Gly Cys Asp Lys Gln Glu Phe Gln Asp Ala Trp Lys Leu Pro Lys Ile
370 375 380
Ala Val Asp Gly Pro Phe Gly Thr Lla Ser Glu Asp Val Phe Ser Tyr
CA 02350776 2001-11-09
77
385 390 395 400
Glu Val Val Met Leu Val Gly Ala Gly lie Gly Val Thr Pro Phe Ala
405 410 415
Ser Ile Leu Lys Ser Val Trp Tyr Lys Tyr Cys Asn Asn Ala Thr Asn
420 425 430
Leu Lys Leu Lys Lys Ile Tyr Phe lyr Trp Leu Cys Arg Asp Thr His
435 440 445
Ala Phe Glu Trp Phe Ala Asp Leu Lou Gln Leu Leu Glu Ser Gln Met
450 455 460
Gln Glu Arg Asn Asn Ala Gly Phe leu Ser Tyr Asn Ile Tyr Leu Thr
465 470 475 480
Gly Trp Asp Glu Ser Gln Ala Asn 3is Phe Ala Val. His His Asp Glu
485 490 495
Glu Lys Asp Val Ile Thr Gly Leu Lvs Gln Lys Thr Leu Tyr Gly Arg
500 305 510
Pro Asn Trp Asp Asn Glu Phe Lys Thr Ile Ala Ser Gln His Pro Asn
515 520 525
Thr Arg Ile Gly Val Phe Leu Cys (3-y Pro Glu Ala Leu Ala Glu Thr
530 535 540
Leu Ser Lys Gln Ser Ile Ser Asn Ser Glu Ser Gly Pro Arg Gly Val
545 550 555 560
His Phe Ile Phe Asn Lys Glu Asn The
565
<210> 13
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial sequence: primer
<400> 13
ttggctaaat cccatcca 18
<210> 14
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 14
tgcatgacca acaatattgc t 21
<210> 15
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 15
caaggtacct cttgaccatg ggaaact 27
CA 02350776 2001-11-09
78
<210> 16
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 16
acgaattcaa gtaaattact gaa(:iata 27
<210> 17
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<221> modified-base
<222> O .. )
<223> n at position 3 = inosine
<220>
<221> modified-base
<222> ()
<223> n at position 6 = inosine
<220>
<221> modified-base
<222> ()
<223> n at position 12 = inosine
<220>
<223> Description of Artificial Sequence: primer
<400> 17
ccngtntgtc gnaatctgct st:cctt. 26
<210> 18
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<221> modified-base
<222> (5)
<223> n at position 5 inosine
<220>
<221> modified-base
<222> (9)
<223> n at position 9 = inosine
<220>
<221> modified-base
<222> (11) <223> n at position 11 == inosine
<220>
<223> Description of Artificial Sequence: primer
<400> 18
tcccngcana nccagtagaa rtagatct?:: 29
<210> 19
<211> 26
CA 02350776 2001-11-09
79
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial sequence: primer
<400> 19
ttggcacagt cagtgaggat gtcttc 26
<210> 20
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 20
ctgttggctt ctactgtagc gttcaaaatt 30
<210> 21
<211> 563
<212> PRT
<213> Rattus sp.
<400> 21
Met Gly Asn Trp Leu Val Asn His Trp Lou Ser Val Leu Phe Leu Val
1 5 10 15
Ser Trp Leu Gly Leu Asn Ile Phe Leu Phe Val Tyr Val Phe Leu Asn
20 25 30
Tyr Glu Lys Ser Asp Lys Tyr Tyr Tyr Thr Arg Gl,i Ile Leu Gly Thr
35 40 45
Ala Leu Ala Leu Ala Arg Ala Se-- Ala Leu Cys Lea Asn Phe Asn Ser
50 55 60
Met Val Ile Leu Ile Pro Val Cys Arg Asn Leu Leu Ser Phe Leu Arg
65 70 75 80
Gly Thr Cys Ser Phe Cys Asn His Thr Leu Arg Lys Pro Leu Asp His
85 90 95
Asn Leu Thr Phe His Lys Leu Val Ala Tyr Met Ile Cys Ile Phe Thr
100 105 110
Ala Ile His Ile Ile Ala His Leu Phe Asn Phe Glu Arg Tyr Ser Arg
115 120 125
Ser Gln Gln Ala Met Asp Gly Ser ~,eu Ala Ser Val. Leu Ser Ser Leu
130 135 140
Phe His Pro Glu Lys Glu Asp Ser. Tap Leu Asn Pro Ile Gln Ser Pro
145 150 155 160
Asn Val Thr Val Met Tyr Ala Ala Phe Thr Ser Ile Ala Gly Leu Thr
165 170 175
Gly Val Val Ala Thr Val. Ala Lea Val lieu Met Va.. Thr Ser Ala Met
180 L85 190
Glu Phe Ile Arg Arg Asn Tyr Phe GGu Leu Phe Trp Tyr Thr His His
195 200 205
Leu Phe Ile Ile Tyr Ile Ile Cys :leu Gly Ile His Gly Leu Gly Gly
210 215 220
CA 02350776 2001-11-09
Ile Val Arg Gly Gln Thr Glu Glu Ser Met Ser Glu Ser His Pro Arg
225 230 235 240
Asn Cys Ser Tyr Ser Phe His Glu Trp Asp Lys Tyr Glu Arg Ser Cys
245 250 255
Arg Ser Pro His Phe Val Gly G_n Pro Pro Glu Ser Trp Lys Trp Ile
260 265 270
Leu Ala Pro Ile Ala Phe Tyr Ile Phe Glu Arg Ile Leu Arg Phe Tyr
275 280 285
Arg Ser Arg Gln Lys Val Val Ile Thr Lys Val Val Met His Pro Cys
290 295 300
Lys Val Leu Glu Leu Gln Met Arg Lys Arg Gly Phe Thr Met Gly Ile
305 310 315 320
Gly Gln Tyr Ile Phe Val Asn Cys Pro Ser Ile Ser Phe Lou Glu Trp
325 330 335
His Pro Phe Thr Leu Thr Ser Ala Pro Glu Glu Glu Phe Phe Ser Ile
340 345 350
His Ile Arg Ala Ala Gly Asp Trp Thr Glu Asn Leu Ile Arg Thr Phe
355 360 365
Glu Gln Gln His Ser Pro Met Pro Arg Ile Glu Val Asp Gly Pro Phe
370 375 380
Gly Thr Val Ser Glu Asp Val Pre Gln Tyr Glu Val Ala Val Leu Val
385 390 395 400
Gly Ala Gly Ile Gly Val Thr Pro Phe Ala Ser Phe Leu Lys Ser Ile
405 410 415
Trp Tyr Lys Phe Gin Arg Ala His Asn Lys Leu Lys Thr Gln Lys Ile
420 425 430
Tyr Phe Tyr Trp Ile Cys Arg Glu Thr Gly Ala Phe Ala Trp Phe Asn
435 440 445
Asn Leu Leu Asn Ser Leu Glu Gin Glu Met Asp Glu Leu Gly Lys Pro
450 455 460
Asp Phe Leu Asn Tyr Arg Leu Phe Leu Thr Gly Trp Asp Ser Asn Ile
465 470 475 480
Ala Gly His Ala Ala Leu Asn Phe Asp Arg Ala Thr Asp Val Leu Thr
485 490 495
Gly Leu Lys Gln Lys Thr Ser Phe Sly Arg Pro Met- Trp Asp Asn Glu
500 505 510
Phe Ser Arg Ile Ala Thr Ala His Pro Lys Ser Val Val Gly Val Phe
515 520 525
Leu Cys Gly Pro Pro Thr Leu Ala Lys Ser Leu Arg Lys Cys Cys Arg
530 535 540
Arg Tyr Ser Ser Leu Asp Pro Arg_ Lvs Val Gln Phe Tyr Phe Asn Lys
545 550 555 560
Glu Thr Phe
<210> 22
<211> 2577
<212> DNA
CA 02350776 2001-11-09
81
<213> Rattus sp.
<400> 22
ttctgagtag gtgtgcattt gagtgtcata aagacatata tcttgagcta gacagaagtt 60
cctatcctga aggatcccat cagagaaacc agattgctcc taagaggctc cagacctcca 120
tttgacaatg ggaaactggc tggttaacca ctggctctca at.tttgtttc tggtttcttg 180
gttggggctg aacatttttc tgtttgtgta cgtcttcctg aattatgaga agtctgacaa 240
gtactattac acgagagaaa ttctc_ggaac tgccttggcc ttggccagag catctgcttt 300
gtgcctgaat tttaacagca tggtgatcct gattcctgtg tgtcgaaatc tgctctcctt 360
cctgaggggc acctgctcat tttgcaacca cacgctgaga aagccattgg atcacaacct 420
caccttccat aagctggtgg catatatgat ctgcatattc acagctattc atatcattgc 480
acatctattt aactttgaac gctacagtac! aagccaacag gccatggatg gatctcttgc 540
ctctgttctc tccagcctat tccctcccge gaaagaagat tcttggctaa atcccatcca 600
gtctccaaac gtgacagtga tgtatgcagc atttaccagt attgctggcc ttactggagt 660
ggtcgccact gtggctttgg ttct:catcrgt aact_tcagct atggagttta tccgcaggaa 720
ttattttgag ctcttctggt atacacat:ca ccttttcatc atctatatca tctgcttagg 780
gatccatggc ctggggggga ttgtccgggg tcaaacagaa gagagcatga gtgaaagtca 840
tccccgcaac tgttcatact ctttccacga gtgggataag tatgaaagga gttgcaggag 900
tcctcatttt gtgggacaac cccctcagtc ttggaagtgg atcctcgcgc cgattgcttt 960
ttatatcttt gaaaggatcc ttcgctttta tcgctcccgg cagaaggtcg tgattaccaa 1020
ggttgtcatg cacccatgta aatttttgga attgcagatg aggaagcggg gctttactat 1080
gggaatagga cagtatatat tcgtaaattg cccctcgatt tccttcctgg aatggcatcc 1140
ctttactctg acctctgctc cagaggaaga atttttctcc attcatattc gagcagcagg 1200
ggactggaca gaaaatctca taaggacatt: tgaacaacag cactcaccaa tgcccaggat 1260
cgaggtggat ggt:ccctttg gcacagtcag tgaggatgtc ttccagtacg aagtggctgt 1320
actggttggg gcagggattg gcgtcactcc ctttgcttcc ttcttgaaat ctatctggta 1380
caaattccag cgtgcacaca acaagctgaa aacacaaaag atctatttct actggatttg 1440
tagagagacg ggtgcctttg cctgggtcaa caacttattg aattccctgg aacaagagat 1500
ggacgaatta ggcaaaccgg at:ttcctaaa ctaccgactc ttc_ctcactg gctgggatag 1560
caacattgct ggtcatgcag cattaaactt tgacagagcc actgacgtcc tgacaggtct 1620
gaaacagaaa acctcctttg gtggaccaat. gtgggacaat gagttttcta gaatagctac 1680
tgcccacccc aagtctgtgg tgggggtttt cttatgcggc cctccgactt tggcaaaaag 1740
cctgcgcaaa tgctgtcggc ggtactcaag tctggatcct aggaaggttc aattctactt 1800
caacaaagaa acgttctgaa tt:ggaggaag ccgcacagta gtacttctcc atcttccttt 1860
tcactaacgt gtgggtcagc tactagatag tccgttgtcg cacaaggact tcactcccat 1920
cttaaagttg actcaactcc at.cattcttg ggctttggca acatgagagc tgcataactc 1980
acaattgcaa aacacatgaa ttattattgg ggggattgta aat.ccttctg ggaaacctgc 2040
CA 02350776 2001-11-09
82
ctttagctga atcttgctgg ttgacacttg cacaatttaa cctcaggtgt cttggttgat 2100
acctgataat cttccctccc acctgtccct cacagaagat ttctaagtag ggtgatttta 2160
aaatatttat tgaatccacg acaaaacaat. aatcataaat aa.taaacata aaattaccaa 2220
gattcccact cccatatcat acccactaaq aacatcgtta tacatgagct tatcatccag 2280
tgtgaccaac aatttatact ttactgtgcc= aaaataatct tcatctttgc_ ttattgaaca 2340
attttgctga ctttccctag taatatct_ta agtatattaa ctggaatcaa atttgtatta 2400
tagttagaag ccaactatat tgccagtttcr ttttgtttga aataactgga aaggcctgac 2460
ctacatcgtg gggtaattta acagaagctc tttccatttt ttgtrgttgt tgttaaagag 2520
ttttgtttat gaatgtgtta taaaaagaaa ataaaaagtt ataattttga cggaaaa 2577
<210> 23
<211> 332
<212> PRT
<213> Homo sapiens
<400> 23
Glu Ser Met Asn Giu Ser His Pro Asp Lys Cys Ala Glu Ser Phe Glu
1 5 10 15
Met Trp Asp Asp Arg Asp Ser His Cys Arg Arg Pro Lys Phe Glu Gly
20 25 30
His Pro Pro Glu Ser Trp Lys Trp Ile Leu Ala Pro Val Ile Leu Tyr
35 40 45
Ile Cys Glu Arg Ile Leu Arg Phe Tyr Arg Ser Gln Gln Lys Val Val
50 55 60
Ile Thr Lys Val Val Met His Pro Ser Lys Val Led Glu Leu Gln Met
65 70 75 80
Asn Lys Arg Gly Phe Ser Met Glu Val :ply Gln Tyr Ile Phe Val Asn
85 90 95
Cys Pro Ser Ile Ser Leu Leu Glu Trp His Pro Phe Thr Leu Thr Ser
100 105 110
Ala Pro Glu Glu Asp Phe Phe Ser 11e His Ile Arg Ala Ala Gly Asp
115 120 125
Trp Thr Glu Asn Leu Ile Arg Ala Phe Glu Gln Gln Tyr Ser Pro Ile
130 135 140
Pro Arg Ile Glu Val Asp Gly Pro Phe Gly Thr Ala Ser Glu Asp Val
145 150 155 160
Phe Gln Tyr Glu Val Ala Val Leu Val Gly Ala Gly Ile Gly Val Thr
165 170 175
Pro Phe Ala Ser Ile Leu Lys Se--- Ile Trp Tyr Lys Phe Gln Cys Ala
180 185 190
Asp His Asn Leu Lys Thr Lys Lys LLe Tyr Phe Tyr Trp Ile Cys Arg
195 200 205
Glu Thr Gly Ala Phe Ser Trp Phe Asn Asn Leu Leu Thr Ser Leu Glu
210 215 220
Gln Glu Met Glu Glu Leu Gly Lys Val G1y Phe Leu Asn Tyr Arg Leu
225 230 235 240
CA 02350776 2001-11-09
83
Phe Leu Thr Gly Trp Asp Ser Asn Ile Val Gly His Ala Ala Leu Asn
245 250 255
Phe Asp Lys Ala Thr Asp Ile Vol .Thr Gly Leu Lys Gln Lys Thr Ser
260 265 270
Phe Gly Arg Pro Met Trp Asp Asn Glu Phe Ser Thr Ile Ala Thr Ser
275 280 285
His Pro Lys Ser Val Val Gly Vol Phe Leu Cys Gly Pro Arg Thr Leu
290 295 300
Ala Lys Ser Leu Arg Lys Cys Cys His Arg Tyr Ser Ser Leu Asp Pro
305 310 315 320
Arg Lys Val Gln Phe Tyr Phe Asn Lys Glu Asn Phe
325 330
<210> 24
<211> 14
<212> PRT
<213> Homo sapiens
<400> 24
Cys Ala Glu Ser Phe Glu Met Trp Asp Asp Arg Asp Ser His
1 5 10
<210> 25
<211> 14
<212> PRT
<213> Homo sapiens
<400> 25
Lys Ser Leu Arg Lys Cys Cys His Arg Tyr Ser Ser Leu Asp
1 5 10
<210> 26
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 26
gaagtggtgg gaggcgaaga cata 24
<210> 27
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 27
cctgtcatac ctgggacggt ctgg 24
<210> 28
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
CA 02350776 2001-11-09
84
<223> Description of Artificial Sequence: primer
<400> 28
gagcacagtg agatgcctgt tcag 24
<210> 29
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 29
ggaaggcagc agagagcaat gatg 24
<210> 30
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 30
acatctgcga gcggcacttc caga 24
<210> 31
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 31
agctcgtcaa caggcaggac cgagc 25
<210> 32
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 32
gcagtgcatc cacatcttca gcac 24
<210> 33
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 33
gagagctctg gagacacttg agttc 25
<210> 34
<211> 22
<212> DNA
<213> Artificial Sequence
CA 02350776 2001-11-09
<220>
<223> Description of Artificial Sequence: primer
<400> 34
catgttctct ctggctgaca ag 22
<210> 35
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 35
cacaatagcg agctccgctt cacgc 25
<210> 36
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 36
gcaggacatc aaccctgcac tctc 24
<210> 37
<211> 570
<212> PRT
<213> Bovine sp.
<400> 37
Met Gly Asn Trp Val Val Asn Glu Gly Ile Ser Ile Phe Val Ile Leu
1 5 10 15
Val Trp Leu Gly Met Asn Val Phe Leu Phe Val Trp Tyr Tyr Arg Val
20 25 30
Tyr Asp Ile Pro Asp Lys Phe Phe ryr Thr Arg Lys Leu Leu Gly Ser
35 40 45
Ala Leu Ala Leu Ala Arg Ala Pro %La Ala Cys Leu Asn Phe Asn Cys
50 55 60
Met Leu Ile Leu Leu Pro Val Cys Arg Asn Leu Leu Ser Phe Leu Arg
65 70 75 80
Gly Ser Ser Ala Cys Cys Ser Thr Arg Ile Arg Arg Gln Leu Asp Arg
85 90 95
Asn Leu Thr Phe His Lys Met Val Ala Trp Met Ile Ala Leu His Thr
100 L05 110
Ala Ile His Thr Ile Ala His Leu The Asn Val Glu Trp Cys Val Asn
115 120 125
Ala Arg Val Asn Asn Ser Asp Pro Tyr Ser Ile Ala Leu Ser Asp Ile
130 135 1.40
Gly Asp Lys Pro Asn Glu Thr Tyr Thu Asn The Val Arg Gln Arg Ile
145 150 155 160
Lys Asn Pro Glu Gly Gly Leu Tyr Val Ala Val Thr Arg Leu Ala Gly
165 170 1.75
CA 02350776 2001-11-09
86
Ile Thr Gly Val Val Ile Thr Leu _ys Leu Ile Leu Ile Ile Thr Ser
180 185 190
Ser Thr Lys Thr Ile Arg Arg Ser Tyr Phe Glu Val Phe Trp Tyr Thr
195 200 205
His His Leu Phe Val Ile Phe Phe Ile Gly Leu Ala Ile His Gly Ala
210 215 220
Gln Arg Ile Val Arg Gly Gln Thr Ala Glu Ser Len Leu Lys His Gln
225 230 235 240
Pro Arg Asn Cys Tyr Gin Asn Ile Ser Gln Trp Gly Lys Ile Glu Asn
245 250 255
Cys Pro Ile Pro Glu Phe Ser Glv Asn Pro Pro Met Thr Trp Lys Trp
260 265 270
Ile Val Gly Pro Met Phe Leu Tye- Leu Cys Glu Arg Leu Val Arg Phe
275 283 285
Trp Arg Ser Gln Gln Lys Val Val I].e Thr Lys Val Val Thr His Pro
290 295 300
Phe Lys Thr Ile Glu Leu Gln Me;_ Lys Lys Lys Gly Phe Lys Met Glu
305 310 315 320
Val Gly Gln Tyr Ile Phe Val Lys Cys Pro Val Val Ser Lys Leu Glu
325 330 335
Trp His Pro Phe Thr Leu Thr Ser Ala Pro Glu Glu. Asp Phe Phe Ser
340 345 350
Ile His Ile Arg Ile Val Gly Asp Trp Thr Glu Gly Leu Phe Lys Ala
355 360 365
Cys Gly Cys Asp Lys Gln Glu Phe Gln Asp Ala Trp Lys Leu Pro Lys
370 375 380
Ile Ala Val Asp Gly Pro Phe Gly Thr Ala Ser Glu Asp Val Phe Ser
385 390 395 400
Tyr Glu Val Val Met Leu Val Gly Ala Gly Ile Gly Val Thr Pro Phe
405 410 415
Ala Ser Ile Leu Lys Ser Val Trp Tyr Lys Tyr Cys Asn Lys Ala Pro
420 425 430
Asn Leu Arg Leu Lys Lys Ile Tyr She Tyr Trp Leu Cys Arg Asp Thr
435 440 445
His Ala Phe Glu Trp Phe Ala Asp Leu Leu Gin Leu Leu Glu Thr Gln
450 455 460
Met Gln Glu Lys Asn Asn Thr Asp Phe Leu Ser Tyr Asn Ile Cys Leu
465 470 4'75 480
Thr Gly Trp Asp Glu Ser Gin Ala Ser His Phe Ala Met His His Asp
485 490 495
Glu Glu Lys Asp Val Ile Thr Gly Leu Lys Gln Lys Thr Leu Tyr Gly
500 505 510
Arg Pro Asn Trp Asp Asn Glu Phe Lys Thr Ile Gly Ser Gln His Pro
515 520 525
Asn Thr Arg Ile Gly Val Phe Leu Cys Gly Pro Glu Ala Leu Ala Asp
530 !535 540
CA 02350776 2001-11-09
87
Thr Leu Asn Lys Gin Cys Ile Ser Asn Ser Asp Ser Gly Pro Arg Gly
545 550 555 560
Val His Phe Ile Phe Asn Lys Glu Asn Phe
565 570
<210> 38
<211> 570
<212> PRT
<213> Murinae gen. sp.
<400> 38
Met Gly Asn Trp Ala Val Asn Glu Gly Leu Ser Ile Phe Val Ile Leu
1 5 10 15
Val Trp Leu Gly Leu Asn Val Prig Leu Phe Ile Asn Tyr Tyr Lys Val
20 25 30
Tyr Asp Asp Gly Pro Lys Tyr Asn Tyr Thr Arg Lys Leu Leu Gly Ser
35 40 45
Ala Leu Ala Leu Ala Arg Ala Pro Ala Ala Cys Leo Asn Phe Asn Cys
50 55 60
Met Leu Ile Leu Leu Pro Val Cys Arg Asn Leu Leu Ser Phe Leu Arg
65 70 75 80
Gly Ser Ser Ala Cys Cys Ser Thr Arg Ile Arg Arg Gin Leu Asp Arg
85 90 95
Asn Leu Thr Phe His Lys Met Val Ala Trp Met Ile Ala Leu His Thr
100 L05 110
Ala Ile His Thr Ile Ala His Leu Phe Asn Val Glu Trp Cys Val Asn
115 120 125
Ala Arg Val Gly Ile Ser Asp Arg Tyr Ser Ile Ala Leu Ser Asp Ile
130 135 140
Gly Asp Asn Glu Asn Glu Glu Tyr :~eu Asn Phe Ala Arg Glu Lys Ile
145 150 155 160
Lys Asn Pro Glu Gly Gly Leu Tyr Val Ala Val Thr Arg Leu Ala Gly
165 170 175
Ile Thr Gly Ile Val Ile Thr Leu Cys Leu Ile Leu Ile Ile Thr Ser
180 185 190
Ser Thr Lys Thr Ile Arg Arg Sep- Vvr Phe Glu Val Phe Trp Tyr Thr
195 200 205
His His Leu Phe Val Ile Phe Phe -le Gly Leu Ala Ile His Sly Ala
210 215 220
Glu Arg Ile Val Arg Gly Gln Thr Ala Glu Ser Leu. Glu Glu His Asn
225 230 235 240
Leu Asp Ile Cys Ala Asp Lys Ile Glu Glu Trp Gly Lys Ile Lys Glu
245 250 255
Cys Pro Val Pro Lys Phe Ala Gly Asn Pro Pro Met Thr Trp Lys Trp
260 =65 270
Ile Val Gly Pro Met Phe Leu Tyr Leu Cys Glu Arg Leu Val Arg Phe
275 280 285
Trp Arg Ser Gln Gln Lys Val Val ]le Thr Lys Val Val Thr His Pro
290 295 30C
CA 02350776 2001-11-09
88
Phe Lys Thr Ile Glu Leu Gln Met Lys Lys Lys Gly Phe Lys Met Glu
305 310 315 320
Val Gly Gln Tyr Ile Phe Val Lys Cys Pro Lys Val Ser Lys Leu Glu
325 330 335
Trp His Pro Phe Thr Leu Thr Ser Ala Pro Glu Glu Asp Phe Phe Ser
340 345 350
Ile His Ile Arg Ile Val Gly Asp Trp Thr Glu Gly Leu Phe Asn Ala
355 360 365
Cys Gly Cys Asp Lys Gln Glu Phe Gin Asp Ala Trp Lys Leu Pro Lys
370 375 380
Ile Ala Val Asp Gly Pro Phe Gly Thr Ala Ser Glu Asp Val Phe Ser
385 390 395 400
Tyr Glu Val Val Met Leu Val Gly Ala Gly Ile Gly Val Thr Pro Phe
4C5 410 415
Ala Ser Ile Leu Lys Ser Val Trj_> Tyr Lys Tyr Cys Asp Asn Ala Thr
420 425 430
Ser Leu Lys Leu Lys Lys Ile Tyr Phe 'I'yr Trp Lei). Cys Arg Asp Thr
435 440 445
His Ala Phe Glu Trp Phe Ala Asp Lou Leu Gln Leu Leu Glu Thr Gln
450 455 460
Met Gln Glu Arg Asn Asn Ala Asn _?he Leu Ser Tyr. Asn Ile Tyr Leu
465 470 475 480
Thr Gly Trp Asp Glu Ser Gln Ala Asn His Phe Ala Val His His Asp
485 490 495
Glu Glu Lys Asp Val Ile Thr Gly Leu Lys Gln Lys Thr Leu Tyr Gly
500 505 510
Arg Pro Asn Trp Asp Asn Glu Phe Lys Thr Ile Ala Ser Glu His Pro
515 520 525
Asn Thr Thr Ile Gly Val Phe Leu Cys Gly Pro Glu Ala Leu Ala Glu
530 535 540
Thr Leu Ser Lys Gln Ser Ile Ser sn Ser Glu Ser Gly Pro Arg Gly
545 550 555 560
Val His Phe Ile Phe Asn Lys Glu Asn Phe
565 570
<210> 39
<211> 944
<212> PRT
<213> Arabidopsis sp.
<400> 39
Met Lys Pro Phe Ser Lys Asn Asp Arg Arg Arg Trp Ser Ph.e Asp Ser
1 5 10 15
Val Ser Ala Gly Lys Thr Ala Val. G1y Ser Ala Ser Thr Ser Pro Gly
20 25 30
Thr Glu Tyr Ser Ile Asn Gly Asp Gl.n Glu Phe Val Glu Val Thr Ile
35 40 45
Asp Leu Gln Asp Asp Asp Thr Ile Val Leu Arg Ser Val Glu Pro Ala
50 55 60
CA 02350776 2001-11-09
89
Thr Ala Ile Asn Val Ile Gly Asp Ile Ser Asp Asp Asn Thr Gly Ile
65 70 75 80
Met Thr Pro Val Ser Ile Ser Arg Ser Pro Thr Me- Lys Arg Thr Ser
85 90 95
Ser Asn Arg Phe Arg Gln Phe Ser Gln Glu Leu Lys Ala Glu Ala Val
100 105 110
Ala Lys Ala Lys Gln Leu Ser Gln Glu Leu Lys Arg Phe Ser Trp Ser
115 120 125
Arg Ser Phe Ser Gly Asn Leu The Thr Thr Ser Thr Ala Ala Asn Gln
130 135 140
Ser Gly Gly Ala Gly Gly Gly Lea Val Asn Ser Ala Leu Glu Ala Arg
145 150 155 160
Ala Leu Arg Lys Gln Arg Ala G1-i Leu Asp Arg The Arg Ser Ser Ala
165 170 175
Gln Arg Ala Leu Arg Gly Leu Arg Phe Ile Ser Asn Lys Gln Lys Asn
180 L85 190
Val Asp Gly Trp Asn Asp Val Gln Per Asn Phe Glu Lys Phe Glu Lys
195 200 205
Asn Gly Tyr Ile Tyr Arg Ser Asp Phe Ala Gln Cys Ile Gly Met Lys
210 215 220
Asp Ser Lys Glu Phe Ala Leu Glu Leu Phe Asp Ala Leu Ser Arg Arg
225 230 235 240
Arg Arg Leu Lys Val Glu Lys Ile 1ksn His Asp Glu. Leu Tyr Glu Tyr
245 250 255
Trp Ser Gin Ile Asn Asp Glu Ser Phe Asp Ser Arg Leu Gln Ile Phe
260 065 270
Phe Asp Ile Val Asp Lys Asn Glu Asp Giy Arg Ile Thr Glu Glu Glu
275 280 285
Val Lys Glu Ile Ile Met Leu Ser Ala Ser Ala Asn Lys Leu Ser Arg
290 295 300
Leu Lys Glu Gln Ala Glu Glu Tyr Ala Ala Leu Ile Met Glu Glu Leu
305 310 315 320
Asp Pro Glu Arg Leu Gly Tyr Ile Glu Leu Trp Gln Leu Glu Thr Leu
325 330 335
Leu Leu Gln Lys Asp Thr Tyr Leu Asn Tyr Ser Gln Ala Leu Ser Tyr
340 345 350
Thr Ser Gin Ala Leu Ser Gln Asn Leu Gin Gly Leu Arg Gly Lys Ser
355 360 365
Arg Ile His Arg Met Ser Ser Asp the Val Tyr Ile Met Gln Glu Asn
370 375 380
Trp Lys Arg Ile Trp Val Leu Ser Leu Trp Ile Met Ile Met Ile Gly
385 390 395 400
Leu Phe Leu Trp Lys Phe Phe Gln Tyr Lys Gln Lys Asp Ala Phe His
405 410 415
Val Met Gly Tyr Cys Leu Leu Thr Ala Lys Gly Ala Ala Glu Thr Leu
420 425 430
Lys Phe Asn Met Ala Leu Ile Lou Phe Pro Val Cys Arg Asn Thr Ile
CA 02350776 2001-11-09
435 440 445
Thr Trp Leu Arg Ser Thr Arg Leu Ser Tyr Phe Val Pro Phe Asp Asp
450 455 460
Asn Ile Asn Phe His Lys Thr Ile Ala Gly Ala Ile Val Val Ala Val
465 470 475 480
Ile Leu His Ile Gly Asp His Leu Ala Cys Asp Phe Pro Arg Ile Val
485 490 495
Arg Ala Thr Glu Tyr Asp Tyr Asn Arg Tyr Leu Phe His Tyr Phe Gin
500 505 510
Thr Lys Gln Pro Thr Tyr Phe Asp Leu Val Lys Gly Pro Glu Gly Ile
515 520 525
Thr Gly Ile Leu Met Val Ile Leu Met Ile Ile Ser Phe Thr Leu Ala
530 535 540
Thr Arg Trp Phe Arg Arg Asn Leu Val Lys Leu Pro Lys Pro Phe Asp
545 550 555 560
Arg Leu Thr Gly Phe Asn Ala Phe Tr_p Tyr Ser His His Leu Phe Val
565 570 575
Ile Val Tyr Ile Leu Leu Ile Leu His Gly Ile Phe Leu Tyr Phe Ala
580 535 590
Lys Pro Trp Tyr Val Arg Thr Thr Trp Met Tyr Leu Ala Val Pro Val
595 600 605
Leu Leu Tyr Gly Gly Glu Arg Thr Leu Arg Tyr Phe Arg Ser Gly Ser
610 615 620
Tyr Ser Val Arg Leu Leu Lys Val Ala Ile Tyr Pro Gly Asn Val Leu
625 630 635 640
Thr Leu Gln Met Ser Lys Pro Thr Gin Phe Arg Tyr Lys Ser Gly Gln
645 650 655
Tyr Met Phe Val Gln Cys Pro Ala Val Ser Pro Phe Glu Trp His Pro
660 665 670
Phe Ser Ile Thr Ser Ala Pro Glu Asp Asp Tyr Ile Ser Ile His Ile
675 680 685
Arg Gln Leu Gly Asp Trp Thr Gln Glu Leu Lys Arg Val Phe Ser Glu
690 695 700
Val Cys Glu Pro Pro Val Gly Gly Lys Ser Gly Leu Leu Arg Ala Asp
705 710 715 720
Glu Thr Thr Lys Lys Ser Leu Pro Lys Leu Leu Ile Asp Gly Pro Tyr
725 730 735
Gly Ala Pro Ala Gln Asp Tyr Arg Lys Tyr Asp Val Leu Leu Leu Val
740 '745 750
Gly Leu Gly Ile Gly Ala Thr Pro the Ile Ser Ile Leu Lys Asp Leu
755 760 765
Leu Asn Asn Ile Val Lys Met Glu Olin His Ala Asp Ser Ile Ser Asp
770 '775 780
Phe Ser Arg Ser Ser Glu Tyr Ser Thr Gly Ser Asn Gly Asp Thr Pro
785 790 795 800
Arg Arg Lys Arg Ile Leu Lys Thr Thr Asn Ala Tyr Phe Tyr Trp Val
805 810 815
CA 02350776 2001-11-09
91
Thr Arg Glu Gln Gly Ser Phe Asp Trp Phe Lys Gly Val Met Asn Glu
820 325 830
Val Ala Glu Leu Asp Gln Arg Gly 'Jal lie Glu Met His Asn Tyr Leu
835 840 845
Thr Ser Val Tyr Glu Glu Gly Asp AOa Arg Ser Ala Leu Ile Thr Met
850 855 860
Val Gln Ala Leu Asn His Ala Lys Asn Gly Val Asp Ile Val Ser Gly
865 870 875 880
Thr Arg Val Arg Thr His Phe Ala Arg Pro Asn Trp; Lys Lys Val Leu
885 890 895
Thr Lys Leu Ser Ser Lys His Cys Asn Ala Arg Ile Gly Val Phe Tyr
900 905 910
Cys Gly Val Pro Val Leu Gly Lys Gl.u Leu Ser Lys Leu Cys Asn Thr
915 920 925
Phe Asn Gin Lys Gly Ser Thr Lys Phe Glu Phe His Lys Glu His Phe
930 935 940
<210> 40
<211> 590
<212> PRT
<213> Oryza sp.
<400> 40
Asn Leu Ala Gly Leu Arg Lys Lys Per Ser Ile Arg Lys Ile Ser Thr
1 5 10 15
Ser Leu Ser Tyr Tyr Phe Glu Asp Asn Trp Lys Arg Leu Trp Val Leu
20 25 30
Ala Leu Trp Ile Gly Ile Met Ala Gly Leu Phe Thr Trp Lys Phe Met
35 40 45
Gln Tyr Arg Asn Arg Tyr Val. Phe Asp Val Met Gly Tyr Cys Val Thr
50 55 60
Thr Ala Lys Gly Ala Ala Glu Thr Leu Lys Leu Asn Met Ala Ile Ile
65 70 75 80
Leu Leu Pro Val Cys Arg Asn Thr Ile Thr Trp Leu Arg Ser Thr Arg
85 90 95
Ala Ala Arg Ala Leu Pro Phe Asp Asp Asn Ile Asn Phe His Lys Thr
100 105 110
Ile Ala Ala Ala Ile Val Val Gly Ile Ile Leu His Ala Gly Asn His
115 12C 125
Leu Val Cys Asp Phe Pro Arg Leu Ile Lys Ser Ser Asp Glu Lys Tyr
130 135 140
Ala Pro Leu Gly Gln Tyr Phe Gly Glu Ile Lys Pro Thr Tyr Phe Thr
145 150 155 160
Leu Val Lys Gly Val. Glu Gly Ile Thr Gly Val Ile Met Val Val Cys
165 170 175
Met Ile Ile Ala Phe Thr Leu Ala Thr Arg Trp Phe Arg Arg Ser Leu
180 185 190
CA 02350776 2001-11-09
92
Val Lys Leu Pro Arg Pro Phe Asp 'Lys Leu Thr Gly Phe Asn Ala Phe
195 200 205
Trp Tyr Ser His His Leu Phe Ile Ile Val Tyr Ile Ala Leu Ile Val
210 215 220
His Gly Glu Cys Leu Tyr Leu Ile 1i-Is Val Trp Ty: Arg Arg Thr Thr
225 230 235 240
Trp Met Tyr Leu Ser Val Pro Val Cys Leu Tyr Val Gly Glu Arg Ile
245 250 255
Leu Arg Phe Phe Arg Ser Gly Ser Tyr Ser Val Arg Leu Leu Lys Val.
260 265 270
Ala Ile Tyr Pro Gly Asn Val Le,:: '"hr Leu Gln Met: Ser Lys Pro Pro
275 28,0 285
Thr Phe Arg Tyr Lys Ser Gly Gln Tyr Met Phe Val. Gln Cys Pro Ala
290 295 300
Val Ser Pro Phe Glu Trp His Pro Phe Ser Ile The Ser Ala Pro Gly
305 310 315 320
Asp Asp Tyr Leu Ser Ile His Val Arg Gln Leu Gly Asp Trp Thr Arg
325 330 335
Glu Leu Lys Arg Val Phe Ala Ala Ala Cys Glu Pro Pro Ala Gly Gly
340 345 350
Lys Ser Gly Leu Leu Arg Ala Asp Glu Thr Thr Lys Lys Ile Leu Pro
355 360 365
Lys Leu Leu Ile Asp Gly Pro Tyr Gly Ser Pro Ala Gln Asp Tyr Ser
370 375 380
Lys Tyr Asp Val Leu Leu Leu Val Gly Leu Gly Ile Gly Ala Thr Pro
385 390 395 400
Phe Ile Ser Ile Leu Lys Asp Leu Leu Asn Asn Ile Ile Lys Met Glu
405 410 415
Glu Glu Glu Asp Ala Ser The Asp Leu Tyr Pro Pro Met Gly Arg Asn
420 425 430
Asn Pro His Val Asp Leu Gly Thr Leu Met Thr Ile Thr Ser Arg Pro
435 440 445
Lys Lys Ile Leu Lys Thr Thr. Asn 1,la Tyr Phe Tyr Trp Val Thr Arg
450 455 460
Glu Gln Gly Ser Phe Asp Trp Phe Lys Gly Val Met Asn Glu Ile Ala
465 470 475 480
Asp Leu Asp Gln Arg Asn Ile Ile Glu Met His Asn Tyr Leu Thr Ser
485 490 495
Val Tyr Glu Glu Gly Asp Ala Arg Ser Ala Leu Ile Thr Met Leu Gln
500 505 510
Ala Leu Asn His Ala Lys Asn Gly Val Asp Ile Val Ser G117 Thr Lys
515 520 525
Val Arg Thr His Phe Ala Arg Pro Asn Trp Arg Lys Val Leu Ser Lys
530 535 540
Ile Ser Ser Lys His Pro Tyr Ala Lys Ile Gly Val Phe Tyr Cys Gly
545 550 555 560
CA 02350776 2001-11-09
93
Ala Pro Val Leu Ala Gln Glu Leu Ser Lys Leu Cys His Glu Phe Asn
565 570 575
Gly Lys Cys Thr Thr Lys Phe Asp PLe His Lys Glu His Phe
580 585 590
<210> 41
<211> 2619
<212> DNA
<213> Rattus sp.
<400> 41
gtgctgtcag agctttacag agcctctggg catgcgcatg gctacccatt tcattgattt 60
acagaagtca tgctaaaatc tctttcatcrc atgtcttcct ttttcagtct ctcctttccc 120
aaagcttttc agtttgccct ttgcttgtac caactgctat ccctcctcaa aggctgctgc 180
aaaaggtatg cctttttctt ggaggcttt.c agcaaatact acctgggaac ctgcttcagc 240
tcttggaata tttaagtgaa gagaacatt.t catagcattt gaatctttct ttgaaggagc 300
caccagacag actgccttgg cct_tggccag agcatctgct ttgtgcctga attttaacag 360
catggtgatc ctgattcctg tgtgtcgaaa tctgctctcc ttcctgaggg gcacctgctc 420
attttgcaac cacacgctga gaaagccatt ggatcacaac ctcaccttcc ataagctggt 480
ggcatatatg atctgcatat tcacagctat tcatatcatt gcacagctat ttaactttga 540
acgctacagt agaagccaac aggccatgga tcigat:ctctt gcctctgt.tc tctccagcct 600
attccatccc gagaaagaag attcttggct aaatcccatc cagtctccaa acgtgacagt 660
gatgtatgca gcatttacca gtattgctgg ccttactgga gtggtcgcca ctgtggcttt 720
ggttctcatg gtaacttcag ctatggagtL tatccgcagg aattattttg agctcttctg 780
gtatacacat caccttttca tcatctatat catctgctta gggatccatg gcctgggggg 840
gattgtccgg ggtcaaacag aagagagcat gagtgaaagt catccccgca actgttcata 900
ctctttccac gagtgggata agtatgaaag gagttgcagg agtcctcatt ttgtggggca 960
accccctgag tcttggaagt ggatcctcgc gccgattgct ttttatatct ttgaaaggat 1020
ccttcgcttt tatcgctccc ggcagaaggt cgtgattacc aaggttgtca tgcacccatg 1080
taaagttttg gaattgcaga tgaggaagcg gggctt:tact atgggaatag gacagtatat 1140
attcgtaaat tgcccctcga tttccttcct ggaatggcat ccctttactc tgacctctgc 1200
tccagaggaa gaatttttct ccattcatat tcgagcagca ggggactgga cagaaaatct 1260
cataaggaca tttgaacaac agcactcacc aatgcccagg atcgaggtgg atggtccctt 1320
tggcacagtc agtgaggatg tcttccagta cgaagtggct gtactggttg gggcagggat 1380
tggcgtcact ccttttgctt ccttcttgga atctatctgg tacaaattcc agcgtgcaca 1440
caacaagctg aaaacacaaa agatctattt ctact:ggatt tgtagagaga cgggtgcctt 1500
tgcctggttc aacaacttat tgaattccct ggaacaagag atggacgaat taggcaaacc 1560
ggatttccta aactaccgac tcttcctcac tggctgggat agcaacattg ctggtcatgc 1620
agcattaaac tttgacagag ccactgacgt cctgacaggt ctgaaacaga aaacctcctt 1680
tgggagacca atgtgggaca atgagttttc tagaatagct actgcccacc ccaagtctgt 1740
CA 02350776 2001-11-09
94
ggtgggggtt ttcttatgcg gccctccgac tttggcaaaa agcctgcgca aatgctgtcg 1800
gcggtactca agtctggatc ctaggaaggt tc:aattctac ttcaacaaag aaacgttctg 1860
aattggagga agccgcacag tagtacttct ccatcttcct tttcactaac gtgtgggtca 1920
gctactagat agtccgttgt cgcacaggaa cttatctccc atcttaaagt tgactcaact 1980
ccatcattct tgggctttgg caacatgaga gctgcataac tcacaattgc aaaacacatg 2040
aattattatt ggggggattg taaatccttc tgggaaacct gcctttagct gaatcttgct 2100
ggttgacact tgcacaattt aacctcagcrt gtcttggttg atacctgata atcttccctc 2160
ccacctgtcc ctcacagaag atctttaagt agggtgattt taaaatattt attgaatcca 2220
cgacaaaaca ataatcataa ataataaaca taaaattacc aagattccca ctcccatatc 2280
atacccacta agaacatcgt tatacatgag cttatcatcc agtgtgacca acaatttata 2340
ctttactgtg ccaaaataat cttcatctt.t gcttattgaa caatcttgct gactttccct 2400
agtaatatct taagtatatt aactggaatc aaatttgtat tatagttaga agccaactat 2460
attgccagtt tgtattgttt gaaataactg gaaaggcctg acctacatcg tggggtaatt 2520
taacagaagc tctttccatt ttttgttgt:t gttgttaaag agttttgttt atgaatgtgt 2580
tataaaaaga aaataaaaag ttataattt.t gacggaaaa 2619
<210> 42
<211> 499
<212> PRT
<213> Rattus sp.
<400> 42
Met Val Ile Leu Ile Pro Val Cys Arg Asn Leu Leu Ser Phe Leu Arg
1 5 10 15
Gly Thr Cys Ser Phe Cys Asn His Thr Leu Arg Lys Pro Leu Asp His
20 25 30
Asn Leu Thr Phe His Lys Leu Val Pla Tyr Met Ile Cys Ile Phe Thr
35 40 45
Ala Ile His Ile Ile Ala His Leu Phe Asn Phe Glu Arg Tyr Ser Arg
50 55 60
Ser Gln Gln Ala Met Asp Gly Ser Leu Ala Ser Val Leu Ser Ser Leu
65 70 75 80
Phe His Pro Glu Lys Glu Asp Ser Trp Leu Asn Pro Ile Gln Ser Pro
85 90 95
Asn Val Thr Val Met Tyr Ala Ala Phe Thr Ser Ile Ala Gly Leu Thr
100 005 110
Gly Val Val Ala Thr Val Ala Leu Val Leu Met Val Thr Ser Ala Met
115 120 125
Glu Phe Ile Arg Arg Asn Tyr Phe (1u Leu Phe Trp Tyr Thr His His
130 135 140
Leu Phe Ile Ile Tyr Ile Ile Cys Leu Gly Ile His Gly Leu Gly Gly
145 150 155 160
Ile Val Arg Gly Gln Thr Glu Glu Ser Met Ser Glu Ser His Pro Arg
165 170 175
CA 02350776 2001-11-09
Asn Cys Ser Tyr Ser Phe His Gin 'Irp Asp Lys Tyr Glu Arg Ser Cys
180 185 190
Arg Ser Pro His Phe Val Gly Gln Pro Pro Glu Ser Trp Lys Trp Ile
195 200 205
Leu Ala Pro Ile Ala Phe Tyr Ile Phe Glu Arg Ile Leu Arg Phe Tyr
210 215 220
Arg Ser Arg Gln Lys Val Val Ile T.Chr Lys Val Val Met His Pro Cys
225 230 235 240
Lys Val Leu Glu Leu Gin Met Ara Lys Arg Gly Phe Thr Met Gly Ile
245 250 255
Gly Gln Tyr Ile Phe Val Asn Cys Pro Ser Ile Ser Phe Leu Glu Trp
260 265 270
His Pro Phe Thr Leu Thr Ser Ala Fro Glu Glu Glu Phe Phe Ser Ile
275 280 285
His Ile Arg Ala Ala Gly Asp Trp Thr Glu Asn Leu Ile Arg Thr Phe
290 295 300
Glu Gln Gln His Ser Pro Met Pro i?rg lie Glu Val Asp Gly Pro Phe
305 310 315 320
Gly Thr Val Ser Glu Asp Val Phe Gln Tyr Glu Val Ala Val Leu Val
325 330 335
Gly Ala Gly Ile Gly Val Thr Pro The Ala Ser Phe Leu Lys Ser Ile
340 345 350
Trp Tyr Lys Phe Gln Arg Ala His Psn Lys Leu Lys Thr Gin Lys Ile
355 360 365
Tyr Phe Tyr Trp Ile Cys Arg Gin Thi Gly Ala Phe Ala Trp Phe Asn
370 375 380
Asn Leu Leu Asn Ser Leu Glu Gln Clu Met Asp Glu Leu Gly Lys Pro
385 390 395 400
Asp Phe Leu Asn Tyr Arg Leu Phe Leu Thr Gly Trp Asp Ser Asn Ile
405 4:L0 415
Ala Gly His Ala Ala Leu Asn Phe Asp Arg Ala Thr Asp Val Leu Thr
420 425 430
Gly Leu Lys Gln Lys Thr Ser Phe Cly Arg Pro Met Trp Asp Asn Glu
435 440 445
Phe Ser Arg Ile Ala Thr Ala His Pro Lys Ser Val Val Gly Val Phe
450 455 460
Leu Cys Gly Pro Pro Thr Leu Ala Lys Ser Leu Arg Lys Cys Cys Arg
465 470 475 480
Arg Tyr Ser Ser Leu Asp Pro Arg Lys Val Gln Phe Tyr Phe Asn Lys
485 490 495
Glu Thr Phe
<210> 43
<211> 35
<212> DNA
<213> Artificial Sequence
CA 02350776 2001-11-09
96
<220>
<223> Description of Artificial Sequence: synthetic
primer
<400> 43
ttctgagtag gtgtgcattt gagtgtcata aagac 35
<210> 44
<211> 45
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial. Sequence: synthetic
primer
<400> 44
ttttccgtca aaattataac tttttattt.t ctttttataa cacat 45
<210> 45
<211> 5494
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (155)..(4810)
<400> 45
gcagagctgc agaggcaccg gacgagagag ggctccgcgg gcccagctgg cagccaggcc 60
ggagacaagt tgcagtcccg ggctctggtg acgccgtggc cgcagggtct ccattttggg 120
acattctaat ccctgagccc ctattattt.t Tate atg ggc too tgc ctg get cta 175
Met Gly Phe Cys Leu Ala Leu
1 5
gca tgg aca ctt ctg gtt ggg gca tgg ar_c cct ctg gga get cag aac 223
Ala Trp Thr Leu Leu Val Gly Ala Trp Thr Pro Leu Gly Ala Gln Asn
15 20
ccc att tcg tgg gag gtg cag cga ttt gat ggg tgg tac aac aac ctc 271
Pro Ile Ser Trp Glu Val Gln Arcr Phe Asp Gly Trp Tyr Asn Asn Leu
25 30 35
atg gag cac aga tgg ggc age aaa ggc tcc cgg ctg cag cgc ctg gtc 319
Met Glu His Arg Trp Gly Ser Lys Gly Ser Arg Leu Gln Arg Leu Val
40 45 50 55
cca gcc agc tat gca gat ggc gtg tae cag ccc ttg gga gaa ccc cac 367
Pro Ala Ser Tyr Ala Asp Gly Val Tyr Gin Pro Leu Gly Glu Pro His
60 65 70
ctg ccc aac ccc cga gac ctt ago aac ace ate tca agg ggc cot gca 415
Leu Pro Asn Pro Arg Asp Leu Ser Asn Thr Ile Ser Arg Gly Pro Ala
75 80 85
ggg ctg gcc tcc ctg aga aac cgc aca gtg ttg ggg gtc ttc ttt ggc 463
Gly Leu Ala Ser Leu Arg Asn Ara Thr Val Leu Gly Val Phe Phe Gly
90 95 100
tat cac gtg ctt tea gac ctg gtcr ago gtg gaa act ccc ggc tgc ccc 511
Tyr His Val Leu Ser Asp Leu Val Ser Val Glu Thr Pro Gly Cys Pro
105 110 115
gcc gag ttc ctc sac att cgc atc ccg ccc gga gac ccc atg ttc gac 559
Ala Glu Phe Leu Asn Ile Arg Ile Pro Pro Gly Asp Pro Met Phe Asp
120 125 130 135
CA 02350776 2001-11-09
97
ccc gac cag cgc ggg gac gtg gtg ctg ccc ttc cag aga agc cgc tgg 607
Pro Asp Gln Arg Giy Asp Val Val Lee Pro Phe Gln Arg Ser Arg Trp
140 145 150
gac ccc gag acc gga cgg agt ccc agc aat ccc cgg gac ccg gcc aac 655
Asp Pro Glu Thr Gly Arg Ser Pro Ser Asn Pro Arg Asp Pro Ala Asn
155 160 165
cag gtg acg ggc tgg ctg gac ggc aqc gcc atc tat ggt tcc tcg cat 703
Gln Val Thr Gly Trp Leu Asp Gly ;;er Ala Ile Tyr Gly Ser Ser His
170 175 180
tcc tgg agc gac gcg ctg cgg age t.tc tcc agg gga cag ctg gcg tcg 751
Ser Trp Ser Asp Ala Leu Arg Ser Phe Ser Arg Gly Gin Leu Ala Ser
185 190 195
ggg ccc gac ccc get ttt ccc cga qa.c tcg cag aac ccc ctg ctc atg 799
Gly Pro Asp Pro Ala Phe Pro Arg Asp Ser Gln Asn Pro Leu Leu Met
200 205 210 215
tgg gcg gcg ccc gac ccc gcc ace qgg cag aac ggg ccc cgg ggg ctg 847
Trp Ala Ala Pro Asp Pro Ala The Gly Gin Asn Gly Pro Arg Gly Leu
220 225 230
tac gcc ttc ggg gca gag aga ggg aac cgg gaa ccc ttc ctg cag gcg 895
Tyr Ala Phe Gly Ala Glu Arg Gly Asn Arg Glu Prc Phe Leu Gln Ala
235 240 245
ctg ggc ctg ctc tgg ttc cgc tac cac aac ctg tgg gcg cag agg ctg 943
Leu Gly Leu Leu Trp Phe Arg Tyr His Asn Leu Trp Ala Gln Arg Leu
250 25` 260
gcc cgc cag cac cca gac tgg gag qac gag gag ctg ttc cag cac gca 991
Ala Arg Gln His Pro Asp Trp Glu t.sp Glu Glu Leu Phe Gln His Ala
265 270 275
cgc aag agg gtc atc gcc acc tar, cag aac atc get gtg tat gag tgg 1039
Arg Lys Arg Val Ile Ala The Tyr Gln Asn Ile Ala Val Tyr Glu Trp
280 285 290 295
ctg ccc agc ttc ctg cag aaa aca etc ccg gag tat aca gga tac cgg 1087
Leu Pro Ser Phe Leu Gln Lys The Leu Pro Glu Tyr Thr Gly Tyr Arg
300 305 310
cca ttt ctg gac ccc agc atc toe tea gag ttc gtg gcg gcc tct gag 1135
Pro Phe Leu Asp Pro Ser Ile Ser Ser Glu Phe Val Ala Ala Ser Glu
315 _'20 325
cag ttc ctg tcc acc atg gtg ccc cct ggc gtc tac atg aga aat gcc 1183
Gln Phe Leu Ser The Met Val Pro Pro Gly Val Tyr Met Arg Asn Ala
330 335 340
agc tgc cac ttc cag ggg gtc ate aat cgg aac tca agt gtc tcc aga 1231
Ser Cys His Phe Gln Gly Val Ile Asn Arg Asn Ser Ser Val Ser Arg
345 350 355
get ctc cgg gtc tgc aac agc tac lgg agc cgt gag cac cca agc cta 1279
Ala Leu Arg Val Cys Asn Ser Tyr Trp Ser Arg Glu His Pro Ser Leu
360 365 370 375
caa agt get gaa gat gtg gat gca. ctg ctg ctg ggc atg gcc tcc cag 1327
Gln Ser Ala Glu Asp Val Asp Ala. Leu Leu Leu Gly Met Ala Ser Gln
380 38.5 390
atc gca gag cga gag gac cat gtg ttg gtt gaa gat gtg cgg gat ttc 1375
Ile Ala Glu Arg Glu Asp His Val Leu Val Glu Asp Val Arg Asp Phe
395 900 405
tgg cct ggg cca ctg aag ttt tcc cgc aca gac cac ctg gcc agc tgc 1423
CA 02350776 2001-11-09
98
Trp Pro Gly Pro Lea Lys Phe Ser Arg Thr Asp His Leu Ala Ser Cys
410 41`5 420
ctg cag cgg ggc cgg gat ctg ggc ctg ccc tct tac acc aag gcc agg 1471
Leu Gln Arg Gly Arg Asp Leu Gly Leu Pro Ser Tyr Thr Lys Ala Arg
425 430 435
gca gca ctg ggc ttg tct ccc att acc cgc tgg cag gac atc aac cct 1519
Ala Ala Leu Gly Leu Per Pro Ile PLo Arg Trp Gin Asp Ile Asn Pro
440 445 450 455
gca ctc tee cgg agc aat gac act_ qta ctg gag gcc aca get gcc ctg 1567
Ala Leu Ser Arg Ser Asn Asp Thr_ iaJL Leu Glu Ala Thr Ala Ala Leu
460 465 470
tac aac cag gac tta tcc tgg cta gag ctg etc cct ggg gga ctc ctg 1615
Tyr Asn Gln Asp Leu Ser Trp Let. Glu Leu Leu Prc Gly Gly Leu Leu
475 X80 485
gag agc cac cgg gac cct gga cct:. ctg ttc agc acc atc gtc ctt gaa 1663
Glu Ser His Arg Asp Pro Giy Pro Leu Phe Per Thi Ile Val Leu Glu
490 495 500
caa ttt gtg cgg cta cgg gat ggt ([ac cgc tac tgg ttt gag aac acc 1711
Gln Phe Val Arg Leu Arg Asp Gly Asp Arg Tyr Trp Phe Glu Asn Thr
505 510 515
agg aat ggg ctg ttc tcc aag aag gag att gaa gaa etc cga aat acc 1759
Arg Asn Gly Leu Phe Ser Lys Lys Glu Ile Glu Glu Ile Arg Asn Thr
520 525 530 535
acc ctg cag gac gtg ctg gtc gct- gtt a.tc aac att gac ccc agt get 1807
Thr Leu Gln Asp Val Leu Val Ala Val Ile Asn Ile Asp Pro Per Ala
540 545 550
ctg cag ccc eat gtc ttt gtc tgg cat aaa gga gac ccc tgt ccg cag 1855
Leu Gln Pro Asn Val Phe Gal Trp His Lys Gly Asp Pro Cys Pro Gln
555 560 565
ccg aga cag ctc agc act gaa ggc ctg cca gcg tgt get ccc tct gtt 1903
Pro Arg Gln Leu Per Thr Glu Gly Leu Pro Ala Cys Ala Pro Per Val
570 571 580
gtt cgt gac tat ttt gag ggc agt gga ttt ggc ttc ggg gtc acc etc 1951
Val Arg Asp Tyr Phe Glu Gly Ser Gly Phe Gly Phe Gly Val Thr Ile
585 590 595
ggg acc etc tgt tgc ttc cct ttg ctg agc ctg etc agt gcc tgg att 1999
Gly Thr Leu Cys Cys Phe Pro Leu Val Per Leu Leu Per Ala Trp Ile
600 605 610 615
gtt gcc cgg ctc cgg atg aga eat; ttc aag agg etc cag ggc cag gac 2047
Val Ala Arg Leu Arg Met Arg Asn the Lys Arg Leu Gln Gly Gln Asp
620 625 630
cgc cag agc ate gtg tct gag aaq etc gtg gga ggc atg gaa get ttg 2095
Arg Gin Per Ile Val Per Glu Lys Lieu Val Gly Gly Met Glu Ala Leu
635 Ã45 645
gaa tgg caa ggc cac aag gag ccc tgc cgg ccc gtg ctt gtg tac ctg 2143
Glu Trp Gln Gly His Lys Glu Pro Cys Arg Pro Val Leu Val Tyr Leu
650 655 660
cag ccc ggg cag etc cgt gtg gte cat ggc agg etc acc gtg ctc cgc 2191
Gln Pro Gly Gln Ile Arg Val Val Asp Gly Arg Leu Thr Val Leu Arg
665 670 675
acc etc cag ctg cag cct cca cag a.ag gtc aac ttc gtc ctg tcc agc 2239
Thr Ile Gln Leu Gln Pro Pro Gln Lys Val Asn Phe Val Leu Per Per
680 685 690 695
CA 02350776 2001-11-09
99
aac cgt gga cgc cgc act ctg ctg ct:c aag atc ccc aag gag tat gac 2287
Asn Arg Gly Arg Arg Thr Leu Leu Leu Lys Ile Pro Lys Glu Tyr Asp
700 705 710
ctg gtg ctg ctg ttt aac ttg gag gaa gag cgg cag gcg ctg gtg gaa 2335
Leu Val Leu Leu Phe Asn Leu Glu Glu Glu Arg Gln Ala Leu Val Glu
715 720 725
aat ctc cgg gga get ctg aag gag agc ggg ttg agc atc cag gag tgg 2383
Asn Leu Arg Gly Ala Leu Lys Glu tar Gly Leu Ser Ile Gln Glu Trp
730 73`5 '740
gag ctg cgg gag cag gag ctg atg aga gca get gtg aca cgg gag cag 2431
Glu Leu Arg Glu Gln Glu Leu Met Arg Ala Ala Val Thr Arg Glu Gln
745 750 755
cgg agg cac ctc ctg gag acc ttt t:tc agg cac ctt ttc tcc cag gtg 2479
Arg Arg His Leu Leu Glu Thr Phe Phe Arg His Leu Phe Ser Gln Val
760 765 770 775
ctg gac atc aac cag gcc gac gca cgg acc ctg ccc ctg gac tcc tcc 2527
Leu Asp Ile Asn Gin Ala Asp Ala Gly Thr Leu Prc Leu Asp Ser Ser
780 785 790
cag aag gtg cgg gag gcc ctg acc tgt gag ctg agc agg gcc gag ttt 2575
Gin Lys Val Arg Glu Ala Leu Thr Cys Glu Leu Ser Arg Ala Glu Phe
795 8(11 0 805
gcc gag tcc ctg ggc ctc aag ccc cag gac atg ttt gtg gag tcc atg 2623
Ala Glu Ser Leu Gly Leu Lys Pro Gin Asp Met Phe Val Glu Ser Met
810 81`i 820
ttc tct ctg get gac aag gat ggc at ggc tac ctg tcc ttc cga gag 2671
Phe Ser Leu Ala Asp Lys Asp Gly FAsn Gly Tyr Leu Ser Phe Arg Glu
825 830 835
ttc ctg gac atc ctg gtg gtc ttc at_g aaa ggc tct cct gag gaa aag 2719
Phe Leu Asp Ile Leu Val Val Phe Met Lys Gly Ser Pro Glu Glu Lys
840 845 850 855
tct cgc ctt atg ttc cgc atg tac gac ttt gat ggg aat ggc ctc att 2767
Ser Arg Leu Met Phe Arg Met Tyr Asp Phe Asp Gly Asn Gly Leu Ile
860 865 870
tcc aag gat gag ttc atc agg atg ctg aga tcc ttc atc gag atc tcc 2815
Ser Lys Asp Glu Phe Ile Arg Met: Leu Arg Ser Phe Ile Glu Ile Ser
875 X80 885
aac aac tgc ctg tcc aag gcc caq ct:a get gag gtg gtg gag tcc atg 2863
Asn Asn Cys Leu Ser Lys Ala Gln. Leu Ala Glu Val Val Glu Ser Met
890 89_`) 900
ttc cgg gag tcg gga ttc cag gac rag gag gaa ctg aca tgg gaa gat 2911
Phe Arg Glu Ser Gly Phe Gln Asp Lys Glu Glu Leu Thr Trp Glu Asp
905 910 915
ttt cac ttc atg ctg cgg gac cac rat agc gag ctc cgc ttc acg cag 2959
Phe His Phe Met Leu Arg Asp His Asn Ser Glu Leu Arg Phe Thr Gln
920 925 930 935
ctc tgt gtc aaa ggg gtg gag gtg cct gaa gtc atc aag gac ctc tgc 3007
Leu Cys Val Lys Gly Val Glu Val Pro Glu Val Ile Lys Asp Leu Cys
940 945 950
cgg cga gcc tcc tar_ atc agc cag cat atg atc tgt ccc tot ccc aga 3055
Arg Arg Ala Ser Tyr Ile Ser Gln. Asp Met Ile Cys Pro Ser Pro Arg
955 960 965
gtg agt gcc cgc tgt tcc cgc agc cac att gag act gag ttg aca cct 3103
CA 02350776 2001-11-09
100
Val Ser Ala Arg Cys Ser Arg Ser Asp Ile Glu Thr Glu Leu Thr Pro
970 975 980
cag aga ctg cag tgc ccc atg gac aca gac cct ccc cag gag att cgg 3151
Gln Arg Leu Gln Cys Pro Met Asp Thr Asp Pro Pro Gln Glu Ile Arg
985 990 995
cgg agg ttt ggc aag aag gta acg t:ca ttc cag ccc ttg ctg ttc act 3199
Arg Arg Phe Gly Lys Lys Val Thr Ser. Phe Gln Pro Leu Leu Phe Thr
1000 1005 1010 1015
gag gcg cac cga gag aag ttc caa cgc agc tgt ctc cac cag acg gtg 3247
Glu Ala His Arg Glu Lys Phe Gln Arg Ser Cys Leu His Gln Thr Val
1020 1025 1030
caa cag ttc aag cgc ttc att gag aac tac cgg cgc cac atc ggc tgc 3295
Gln Gln Phe Lys Arg Phe Ile Glu Asn Tyr Arg Arg His Ile Gly Cys
1035 1040 1045
gtg gcc gtg ttc tac gcc atc gct: ggg ggg ctt ttc ctg gag agg gcc 3343
Val Ala Val Phe Tyr Ala Ile Ala Gly Gly Leu Phe Leu Glu Arg Ala
1050 1051; 1060
tac tac tac gcc ttt gcc gca cat cac acg ggc atc acg gac acc acc 3391
Tyr Tyr Tyr Ala Phe Ala Ala His His Thr Gly Ile Thr Asp Thr Thr
1065 1070 1075
cgc gtg gga atc atc ctg tcg cgg ggc aca gca gcc agc atc tct ttc 3439
Arg Val Gly Ile Ile Leu Ser Arg Gly Thr Ala Ala Ser Ile Ser Phe
1080 1085 1090 1095
atg ttc tcc tac ate ttg ctc acc atg tgc cgc aac ctc ate acc ttc 3487
Met Phe Ser Tyr Ile Leu Leu Thr Net Cys Arg Asn Leu Ile Thr Phe
1100 1105 1110
ctg cga gaa acc ttc ctc aac cg(-, t.a.c gtg ccc ttc gac gcc gcc gtg 3535
Leu Arg Glu Thr Phe Leu Asn Arg Tyr Val Pro Phe Asp Ala Ala Val
1115 1730 1125
gac ttc cat cgc ctc att gcc too acc gcc atc gtc ctc aca gtc tta 3583
Asp Phe His Arg Leu Ile Ala Ser Thr Ala Ile Val Leu Thr Val Leu
1130 1135 1140
cac agt gtg ggc cat gtg gtg aat qtg tac ctg ttc tcc ate agc ccc 3631
His Ser Val Gly His Val Val Asn Val Tyr Leu Phe Ser Ile Ser Pro
1145 1150 1155
ctc agc gtc ctc tct tgc ctc ttt cct ggc ctc ttc cat gat gat ggg 3679
Leu Ser Val Leu Sear Cys Leu Phe Pro Gly Leu Phe His Asp Asp Gly
1160 1165 1170 1175
tct gag ttc ccc cag aag tat tac. t.gg tgg ttc ttc cag acc gta cca 3727
Ser Glu Phe Pro Gln Lys Tyr Tyr Tip Trp Phe Phe G1n Thr Val Pro
1180 1185 1190
ggc ctc acg ggg gtt gtg ctg ctc ctg atc ctg gcc atc atg tat gtc 3775
Gly Leu Thr Gly Val Val Leu Leu Leu Ile Leu Ala Ile Met Tyr Val
1195 1100 1205
ttt gcc tcc cac cac ttc cgc cgc cgc agt ttc cgg ggc ttc tgg ctg 3823
Phe Ala Ser His His Phe Arq Arc] Arg Ser Phe Arg Gly Phe Trp Leu
1210 1215 1220
acc cac cac ctc tac atc ctg ctc tat gtc ctg ctc atc atc cat ggt 3871
Thr His His Leu Tyr Ile Leu Lei. Tyr Val Leu Lei, Ile Ile His Gly
1225 1230 1235
agc ttt gcc ctg atc cag ctg ccc cgt ttc cac atc ttc ttc ctg gtc 3919
Ser Phe Ala Leu Ile Gln Leu Pro Arg Phe His Ile Phe Phe Leu Val
1240 1245 1250 1255
CA 02350776 2001-11-09
101
cca gca atc atc tat ggg ggc gac aag ctg gtg agc ctg agc cgg aag 3967
Pro Ala Ile Ile Tyr Gly Gly Asp lays Leu Val Ser Leu Ser Arg Lys
1260 1265 1270
aag gtg gag atc agc gtg gtg aag gcg gag ctg ctg ccc tca gga gtg 4015
Lys Val Glu Ile Ser Val Val Lys Ala Glu Leu Leu Pro Ser Gly Val
1275 1280 1285
acc cac ctg cgg ttc cag cgg ccc cog ggc ttt gag tac aag tca ggg 4063
Thr His Leu Arg Phe Gln Arg Pro Gin Gly Phe Glu Tyr Lys Ser Gly
1290 1295 1300
cag tgg gtg cgg ate get tgc etq get ctg ggg acc acc gag tac cac 4111
Gln Trp Val Arg Ile Ala Cys Le,.. Ala Leu Gly Thr Thr Glu Tyr His
1305 1.310 1315
ccc ttc aca ctg acc tct gcg ccc cat gag gac acg ctt agc ctg cac 4159
Pro Phe Thr Leu Thr Ser Ala Pro His Glu Asp Thr Leu Ser Leu His
1320 1325 1330 1335
atc cgg gca gca ggg ccc tgg acc act cgc ctc agg gag atc tac tca 4207
Ile Arg Ala Ala Gly Pro Trp Thr -hr Arg Leu Arg_ Glu Ile Tyr Ser
1340 1345 1350
gcc ccg acg ggt gac aga tgt gcc aga tac cca aag ctg tac ctt gat 4255
Ala Pro Thr Gly Asp Arg Cys Ala Arg Tyr Pro Lys Leu Tyr Leu Asp
1355 1360 1365
gga cca ttt gga gag ggc cac cag gag tgg cat aag ttt gag gtg tca 4303
Gly Pro Phe Gly Glu Gly His Gln Glu Trp His Lys Phe Glu Val Ser
1370 1375 1380
gtg tta gtg gga ggg ggc att ggg gtc acc cct ttt gcc tcc atc ctc 4351
Val Leu Val Gly Gly Gly Ile Gly Val Thr Pro Phe Ala Ser Ile Leu
1385 1.390 1395
aaa gac ctg gtc ttc aag tca too gtc agc tgc caa gtg ttc tgt aag 4399
Lys Asp Leu Val Phe Lys Ser Set Val Ser Cys Gln Val Phe Cys Lys
1400 1405 1410 1415
aag atc tac ttc atc tgg gtg acg cgg acc cag cgt cag ttt gag tgg 4447
Lys Ile Tyr Phe Ile Trp Val Thr Arg Thr Gin Arg Gln Phe Glu Trp
1420 1425 1430
ctg get gac atc atc cga gag gtg gag gag aat gac cac cag gac ctg 4495
Leu Ala Asp Ile Ile Arg Glu Val Glu Glu Asn Asp His Gln Asp Leu
1435 1440 1445
gtg tct gtg cac atc tac atc acc cag ctg get gag aag ttc gac ctc 4543
Val Ser Val His Ile Tyr Ile Thr Gin Leu Ala Glu Lys Phe Asp Leu
1450 1455 1460
agg acc act atg ctg tac atc tgt_ gag cgg cac ttc cag aag gtt ctg 4591
Arg Thr Thr Met Leu Tyr Ile Cys Glu Arg His Phe Gln Lys Val Leu
1465 1470 1475
aac cgg agt cta ttc aca ggc ctg cqc tcc atc acc cac ttt ggc cgt 4639
Asn Arg Ser Leu Phe Thr Gly Leu Arg Ser Ile Thr His Phe Gly Arg
1480 1485 1490 1495
ccc ccc ttt gag ccc ttc ttc aac '-cc ctg cag gag gtc cac ccc cag 4687
Pro Pro Phe Glu Pro Phe Phe Asn Ser Lou Gln Glu Val His Pro Gln
1500 1505 1510
gtc cgg aag atc ggg gtg ttt ago Sgt ggc ccc cct ggc atg acc aag 4735
Val Arg Lys Ile Gly Val Phe Ser Cys Gly Pro Pro Gly Met Thr Lys
1515 1520 1525
aat gtg gaa aag gcc tgt cag ctc atc aac agg cag gac cgg act cac 4783
CA 02350776 2001-11-09
102
Asn Val Glu Lys Ala Cys Gin Leo Ile Asn Arg Gln Asp Arg Thr His
1530 1535 1540
ttc tcc cac cat tat gag aac ttc tag gcccctgccc gggggttctg 4830
Phe Ser His His Tyr Glu Asn Phe
1545 1550
cccactgtcc agttgagcag aggtttgagc ccacacctca cctctgttct tcctatttct 4890
ggctgcctca gccttctctg atttcccacc tcccaacctt gttccaggtg gccatagtca 4950
gtcaccatgt gtgggctcag ggacccccag gaccaggatg tgtctcagcc tggagaaatg 5010
gtgggggggc agtgtctagg gactagagtg agaagtaggg gagctactga tttggggcaa 5070
agtgaaacct ctgcttcaga cttcagaaac aaatctcaga agacaagctg acctgacaag 5130
tactatgtgt gtgcatgtct gtatgtgtgt tggggcggtg agtgtaagga tgcagtggga 5190
gcatggatgc tggcatctta gaaccctccc tactcccata cctcctcctc ttctgggctc 5250
cccactgtca gacgggctgg caaatgcctt gcaggaggt_a gaggctggac ccatggcaag 5310
ccatttacag aaacccactc ggaaccccag tctaacacca caactaattt cacccaaggt 5370
tttaagcacg ttctttcatc agaccctggc ocaataccta tgtatgcaat gctcctcagc 5430
cctcttctcc ctgctccagt agtctccctt ccaaataaat cacttttctg ccaaaaaaaa 5490
aaaa 5494
<210> 46
<211> 1551
<212> PRT
<213> Homo sapiens
<400> 46
Met Gly Phe Cys Leu Ala Leu Ala Trp Thr Leu Leu Val Gly Ala Trp
1 5 10 15
Thr Pro Leu Gly Ala Gln Asn Pro Ile Ser. Trp Glu Val Gln Arg Phe
20 25 30
Asp Gly Trp Tyr Asn Asn Leu Met Glu His Arg Trp Gly Ser Lys Gly
35 40 45
Ser Arg Leu Gln Arg Leu Val Pro Ala Ser Tyr Ala Asp Gly Val Tyr
50 55 60
Gln Pro Leu Gly Glu Pro His Leu Pro Asn Pro Arg Asp Leu Ser Asn
65 70 75 80
Thr Ile Ser Arg Gly Pro Ala Gly Leu Ala Ser Leu Arg Asn Arg Thr
85 90 95
Val Leu Gly Val Phe Phe Gly Tyr His Val Leu Ser Asp Leu Val Ser
100 105 110
Val Glu Thr Pro Gly Cys Pro Ala Gle Phe Leu Asn Ile Arg Ile Pro
115 120 125
Pro Gly Asp Pro Met Phe Asp Pro Asp Gin Arg Gly Asp Val Val Leu
130 135 140
Pro Phe Gln Arg Sec Arg Trp Asp Pro Glu Thr Gly Arg Ser Pro Ser
145 150 155 160
Asn Pro Arg Asp Pro Ala Asn Gln Val Thr Gly Trp Leu Asp Gly Ser
165 170 175
CA 02350776 2001-11-09
103
Ala Ile Tyr Gly Ser Ser His Ser Trp Ser Asp Ala Leu Arg Ser Phe
180 185 190
Ser Arg Gly Gin Leu Ala Ser Gly Fro Asp Pro Ala Phe Pro Arg Asp
195 200 205
Ser Gln Asn Pro Leu Leu Met Trp Ala Ala Pro Asp Pro Ala Thr Gly
210 215 220
Gin Asn Gly Pro Arg Gly Leu Tyr Ala Phe Gly Ala Glu Arg Gly Asn
225 230 235 240
Arg Glu Pro Phe Leu Gin Ala Leu Gly Leu Leu Trp Phe Arg Tyr His
245 250 255
Asn Leu Trp Ala Gin Arg Leu Ala Arg Gln His Pro Asp Trp Glu Asp
260 265 270
Glu Glu Leu Phe Gin His Ala Arc Lys Arg Val Ile Ala Thr Tyr. Gin
275 280 285
Asn Ile Ala Val Tyr Glu Trp Leu. Pro Ser Phe Leu Gin Lys Thr Leu
290 295 300
Pro Glu Tyr Thr Gly Tyr Arg Pro Phe Leu Asp Pro Ser Ile Ser Ser
305 310 315 320
Glu Phe Val Ala Ala Ser Glu Gin Phe Leu Ser Thr Met Val Pro Pro
325 330 335
Gly Val Tyr Met Arg Asn Ala Ser Cys His Phe Gin Gly Val Ile Asn
340 345 350
Arg Asn Ser Ser Val Ser Arg Ala Leu Arg Val Cys Asn Ser Tyr Trp
355 36C 365
Ser Arg Glu His Pro Ser Leu Gin Ser Ala Glu Asp Val Asp Ala Leu
370 375 380
Leu Leu Gly Met Ala Ser Gin Ile Ala Glu Arg Glu Asp His Val Leu
385 390 395 400
Val Glu Asp Val Arg Asp Phe Trp Pro Gly Pro Leu Lys Phe Ser Arg
405 410 415
Thr Asp His Leu Ala Ser Cys Leu Gln Arg Gly Arg Asp Leu Gly Leu
420 425 430
Pro Ser Tyr Thr Lys Ala Arg Ala Ala Leu Gly Leu Ser Pro Ile Thr
435 440 445
Arg Trp Gin Asp Ile Asn Pro Ala Leu Ser Arg Ser Asn Asp Thr Val
450 455 460
Leu Glu Ala Thr Ala Ala Leu Tyr Asn Gin Asp Leu Ser Trp Leu Glu
465 470 475 480
Leu Leu Pro Gly Gly Leu Leu Glu Per His Arg Asp Pro Gly Pro Leu
485 490 495
Phe Ser Thr Ile Val Leu Glu Gin Phe Val Arg Leu Arg Asp Gly Asp
500 505 510
Arg Tyr Trp Phe Glu Asn Thr Arg Asn Gly Leu Phe Ser Lys Lys Glu
515 520 525
Ile Glu Glu Ile Arg Asn Thr Thr Leu Gln Asp Val Leu Val Ala Val
530 535 540
CA 02350776 2001-11-09
104
Ile Asn Ile Asp Prc Ser Ala Leu Gin Pic Asn Val Phe Val Trp His
545 550 555 560
Lys Gly Asp Pro Cys Pro Gln Pro Arg G.1n Leu Ser Thr Glu Gly Leu
565 570 575
Pro Ala Cys Ala Pro Ser Val Val Arg Asp Tyr Phe Glu Gly Ser Gly
580 535 590
Phe Gly Phe Gly Val Thr 11e Gly Thr Leu Cys Cys Phe Prc Leu Val
595 600 605
Ser Leu Leu Ser Ala Trp Isle Val Ala Arg Leu Arg Met Arg Asn Phe
610 615 620
Lys Arg Leu Gln Gly Gin Asp Arg Gin Ser Ile Val Ser Glu Lys Leu
625 630 635 640
Val Gly Gly Met Glu Ala Leu Glu Trp Gin Gly His Lys Glu Pro Cys
645 650 655
Arg Pro Val Leu Val Tyr Leu Gln Prc Gly Gln Ile Arg Val Val Asp
660 665 670
Gly Arg Leu Thr Val Leu Arg Thr Ile GIn Leu Gin Pro Pro Gin Lys
675 680 685
Val Asn Phe Val Leu Ser Ser Asn Arg Gly Arg Arg Thr Leu Leu Leu
690 695 700
Lys Ile Pro Lys Glu Tyr Asp Leu Val Leu Leu Phe Asn Leu Glu Glu
705 710 715 '720
Glu Arg Gln Ala Leu Val Glu Asn Leu Arg Gly Ala Leu Lys Glu Ser
725 730 735
Gly Leu Ser Ile Gln Glu Trp Glu Leu Arg Gbu Gln Glu Leu Met Arg
740 745 750
Ala Ala Val Thr Arg Glu Gln Arg Arg His Leu Leu Glu Thr Phe Phe
755 760 765
Arg His Leu Phe Ser Gln Val Leu Asp Ile Asn Gin Ala Asp Ala Gly
770 775 780
Thr Leu Pro Leu Asp Ser Ser Gln Lys Val Arg Glu Ala Leu Thr Cys
785 790 795 800
Glu Leu Ser Arg Ala Glu Phe Ala Glu Ser Leu Gly Leu Lys Pro Gln
805 810 815
Asp Met Phe Val Glu Ser Met Phe Se:r Leu Ala Asp Lys Asp Gly Asn
820 825 830
Gly Tyr Leu Ser Phe Arg Glu Phe Leu Asp Ile Leu Val Val Phe Met
835 840 845
Lys Gly Ser Pro Glu Glu Lys Ser Arg Leu Met Phe Arg Met Tyr Asp
850 855 860
Phe Asp Gly Asn Gly Leu Ile Ser Lys Asp G1u Phe Ile Arg Met Leu
865 870 875 880
Arg Ser Phe Ile Glu Ile Ser Asn Asn Cys Leu Ser Lys Ala Gln Leu
885 890 895
Ala Glu Val Val Glu Ser Met Phe Arg Glu Ser Gly Phe Gln Asp Lys
900 905 910
Glu Glu Leu Thr Trp Glu Asp Phe His Phe Met Leu Arg Asp His Asn
CA 02350776 2001-11-09
105
915 920 925
Ser Glu Leu Arg Phe Thr Gln Leu Cys Val. Lys Gly Val Glu Val Pro
930 935 940
Glu Val Ile Lys Asp Leu Cys Arg Arg Ala Ser Tyr Ile Ser Gln Asp
945 950 955 960
Met Ile Cys Pro Ser Pro Arg Val Ser Ala Arg Cys Ser Arg Ser Asp
965 970 975
Ile Glu Thr Glu Leu Thr Pro Gln Arg Leu Gln Cys Pro Met Asp Thr
980 985 99C
Asp Pro Pro Gln Glu Ile Arg Arg Arg Phe Gly Lys Lys Val Thr Ser
995 1000 1005
Phe Gln Pro Leu Leu Phe Thr Glu Ala His Arg Glu Lys Phe Gln Arg
1010 1015 1020
Ser Cys Leu His Gln Thr Val Gln Gln Phe Lys Arg Phe Ile Glu Asn
025 1030 1035 1040
Tyr Arg Arg His Ile Gly Cys Val Ala Val Phe Tyr Ala Ile Ala Gly
1045 1050 1055
Gly Leu Phe Leu Glu Arg Ala Tyr Tyr Tyr Ala Phe Ala Ala His His
1060 1065 1070
Thr Gly Ile Thr Asp Thr Thr Arg Val Gly Ile Ile Leu Ser Arg Gly
1075 1080 1085
Thr Ala Ala Ser Ile Ser Phe Met Phe Ser Tyr Ile Leu Leu Thr Met
1090 1095 1100
Cys Arg Asn Leu Ile Thr Phe Leu Arg Glu Thr Phe Leu Asn Arg Tyr
105 1110 1115 1120
Val Pro Phe Asp Ala Ala Val Asp Phe His Arg Leu Ile Ala Ser Thr
1125 1130 1135
Ala Ile Val Leu Thr. Val Leu His Ser: Val Gly His Val Val Asn Val
1140 114.5 1150
Tyr Leu Phe Ser Ile Ser Pro Leu Ser Val Leu Ser Cys Leu Phe Pro
1155 116C 1165
Gly Leu Phe His Asp Asp Gly Ser Glu Phe Pro Gln Lys Tyr Tyr Trp
1170 1175 1180
Trp Phe Phe Gln Thr Val Pro Gly Leu Thr Gly Val Val Leu Leu Leu
185 1190 1195 1200
Ile Leu Ala Ile Met Tyr Val Phe Ala Ser His His Phe Arg Arg Arg
1205 1210 1215
Ser Phe Arg Gly Phe Trp Leu Thr His His Leu Tyr Ile Leu Leu Tyr
1220 1225 1230
Val Leu Leu Ile Ile His Gly Ser She Ala Leu Ile Gln Leu Pro Arg
1235 1240 1245
Phe His Ile Phe Phe Leu Val Pro Ala Ile Ile Tyr Gly Gly Asp Lys
1250 1255 1260
Leu Val Ser Leu Ser Arg Lys Lys Val Glu Ile Ser Val Val Lys Ala
265 1270 1275 1280
Glu Leu Leu Pro Ser Gly Val Thr Lis Lou Arg Phe Gln Arg Pro Gin
1285 1290 1295
CA 02350776 2001-11-09
106
Gly Phe Glu Tyr Lys Ser Gly Gln Trp Val Arg Ile Ala Cys Leu Ala
1300 1305 1310
Leu Gly Thr Thr Glu Tyr His Pro Phe Thr Leu Thr Ser Ala Pro His
1315 1320 L325
Glu Asp Thr Leu Ser Leu His Ile Arg Ala Ala Gly Pro Trp Thr Thr
1330 1335 1340
Arg Leu Arg Glu Ile Tyr Ser Ala Pro Thr Gly Asp Arg Cys Ala Arg
345 1350 1355 1360
Tyr Pro Lys Leu Tyr Leu Asp Gly Pro Phe Gly Glu Gly His Gln Glu
1365 1370 1375
Trp His Lys Phe Glu Val Ser Val Leu Val Gly Gly Gly Ile Gly Val
1380 1385 1390
Thr Pro Phe Ala Ser Ile Leu Lys Asp Leu Val Phe Lys Ser Ser Val
1395 1400 L405
Ser Cys Gln Val Phe Cys Lys Lys Ile Tyr Phe Ile Trp Val Thr Arg
1410 1415 1420
Thr Gln Arg Gln Phe Glu '."rp Leu Ala Asp Ile Ile Arg Glu Val Glu
425 1430 1435 1440
Glu Asn Asp His Gin Asp Leu Val Ser Val His Ile Tyr Ile Thr Gln
1445 1450 1455
Leu Ala Glu Lys Phe Asp Leu Arg Thr Thr Met Leu Tyr Ile Cys Glu
1460 1465 1470
Arg His Phe Gln Lys Val Leu Asn Arg Ser Leu Phe Thr Gly Leu Arg
1475 1480 1485
Ser Ile Thr His Phe Gly Arg Pro Pro Phe Glu Pro Phe Phe Asn Ser
1490 1495 1500
Leu Gln Glu Val His Pro Gln Val Arg Lys Ile Gly Val Phe Ser Cys
505 1510 1515 1520
Gly Pro Pro Gly Met: Thr Lys Asn Val Glu Lys Ala Cys Gin Leu Ile
1525 1530 1535
Asn Arg Gln Asp Arc{ Thr His Phe Ser His His Tyr Glu Asn Phe
1540 L545 1550
<210> 47
<211> 3453
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (438)..(3134)
<400> 47
gtcctcgacc agtttgtacg gctgcgggat ggtgaccgct actggtttga gaacaccagg 60
aatgggctgt tctccaagaa ggagattgag acatccgaaa taccaccgtg cgggacgtgc 120
tggtcgctgt tatcaacatt gaccccagtg ccctgcagcc caatgtcttt gtctggcata 180
aaggtgcacc ctgccctcaa cctaagcagc tcacaactga cggcctgccc cagtgtgcac 240
ccctgactgt gcttgacttc tttgaaggca gcagccctgg ttttgccatc accatcattg 300
CA 02350776 2001-11-09
107
ctctctgctg ccttccctta gtgagtctgc ttctct:ctgg agtggtggcc tatttccggg 360
gccgagaaca caagaagcta caaaagaaac tcaaagagag cgtgaagaag gaagcagcca 420
aagatggagt gccagcg atg gag tgg cca ggc ccc aag gag agg agc agt 470
Met Glu Trp Pro Sly Pro Lys Glu Arg Ser Ser
1 `i 10
ccc atc atc atc cag ctg ctg tca gac agg tgt ctg cag gtc ctg aac 518
Pro Ile Ile Ile Gln Leu Leu Ser Asp Arg Cys Leu Gln Val. Leu Asn
15 20 25
agg cat ctc act gtg ctc cgt gtg gtc cag ctg cag cct ctg cag cag 566
Arg His Leu Thr. Val Leu Arg Val Val Gin Leu Gln Pro Leu Gln Gln
30 35 40
gtc aac ctc atc ctg tcc aac aac cga gga tgc cgc acc ctg ctg ctc 614
Val Asn Leu Ile Leu Ser Asn Asn Arg Sly Cys Arg Thr Leu Leu Leu
45 50 55
aag atc cct aag gag tat gac ctg gtg ctg ctg ttt agt tct gaa gag 662
Lys Ile Pro Lys Glu Tyr Asp Leu Val Leu Leu Phe Ser Ser Glu Glu
60 65 70 75
gaa cgg ggc gcc ttt gtg cag cag cta tgg gac ttc tgc gtg cgc tgg 710
Glu Arg Sly Ala Phe Val Gln Gln Leo Trp Asp Phe Cys Val. Arg Trp
80 85 90
get ctg ggc ctc cat gtg get gag atg agc gag aag gag cta ttt agg 758
Ala Leu Sly Leu His Val Ala Glu Met Ser Glu Lys Glu Leu Phe Arg
95 100 105
aag get gtg aca aag cag cag cgg gaa cgc atc ctg gag atc ttc ttc 806
Lys Ala Val Thr Lys Gln Gln Arg Glu Arg Ile Leu Glu Ile Phe Phe
110 115 120
aga cac ctt ttt gct: cag gtg ctg gac atc aac cag gcc gac gca ggg 854
Arg His Leu Phe Ala Gln Val Leu Asp Ile Asn Gln Ala Asp Ala Sly
125 130 135
acc ctg ccc ctg gac tcc 1-cc cag aag gtg cgg gag gcc ctg acc t:gc 902
Thr Leu Pro Leu Asp Ser Ser Gin Lys Val Arg Glu Ala Leu Thr Cys
140 145 150 155
gag ctg agc agg gcc gag ttt gcc gag tcc ctg ggc ctc aag ccc cag 950
Glu Leu Ser Arg Ala Glu Phe Ala Gb Ser Leu Sly Leu Lys Pro Gln
160 165 170
gac atg ttt gtg gag tcc atg ttc tact: ctg get gac aag gat ggc aat 998
Asp Met Phe Val Glu Ser Met Phe Ser Leu Ala Asp Lys Asp Sly Asn
175 1.80 185
ggc tac ctg tcc ttc cga gag ttc ctg gac atc ctg gtg gtc ttc atg 1046
Sly Tyr Leu Ser Phe Arg Glu Phe Leu Asp Ile Leu Val Val Phe Met
190 195 200
aaa ggc tcc cca gag gat aag tcc cgt cta atg ttt acc atg tat gac 1094
Lys Sly Ser Pro Glu Asp Lys Ser Arg Leu Met Phe Thr Met Tyr Asp
205 210 215
ctg gat gag aat ggc ttc ctc tcc aag gac gaa ttc ttc acc atg atg 1142
Leu Asp Glu Asn Sly Phe Leu Ser Lys Asp Glu Phe Phe Thr Met Met
220 225 230 235
cga tcc ttc atc gag atc tcc aac aac tgc ctg tcc aag gcc cag ctg 1190
Arg Ser Phe Ile Glu Ile Ser Asn Asn Cys Leu Ser Lys Ala Gln Leu
240 245 250
gcc gag gtg gtg gag tct atg ttc cgg gag tcg gga ttc cag gac aag 1238
Ala Glu Val Val Glu Ser Met Phe Arg Giu Ser Sly Phe Gln Asp Lys
CA 02350776 2001-11-09
108
255 260 265
gag gag ctg aca tgg gag gat ttt cac ttc atg ctg cgg gac cat gac 1286
Glu Glu Leu Thr Trp Glu Asp Phe His Phe Met Leu Arg Asp His Asp
270 275 280
agc gag ctc cgc ttc acg cag ctc tg: gtc aaa ggt gga ggt gga ggt 1334
Ser Glu Leu Arg Phe Thr Gin Leu Cys Val Lys Gly Gly Gly Gly Gly
285 290 295
gga aat ggt att aga gat atc ttt aaa caa aac atc agc tgt cga gtc 1382
Gly Asn Gly Ile Arg Asp Ile Phe Lys Gin Asn Ile Sex: Cys Arg Val
300 305 310 315
tcg ttc atc act cgg aca cct ggg gag cgc tcc cac ccc cag gga ctg 1430
Ser Phe Ile Thr Arg Thr Pro Gly Glu Arg Ser His Pro Gln Gly Leu
32C 325 330
ggg ccc cct gtc cca gaa gcc cca gag ctg gga ggc cct gga ctg aag 1478
Gly Pro Pro Val Pro Glu Ala Pro Glu Leu Gly Gly Pro Gly Leu Lys
335 340 345
aag agg ttt ggc aaa aag gca gca gtg ccc act ccc cgg ctg tac aca 1526
Lys Arg Phe Gly Lys Lys Ala Ala Val Pro Thr Pro Arg Leu Tyr Thr
350 355 360
gag gcg ctg caa gag aag atg cag cga ggc ttc cta gcc caa aag ctg 1574
Glu Ala Leu Gln Glu Lys Met Gln Arg Gly Phe Leu Ala Gln Lys Leu
365 370 375
cag cag tac aag cgc ttc gtg gag aac tac cgg agg cac atc gtg tgt 1622
Gln Gln Tyr Lys Arg Phe Val Glu Asn Tyr Arg Arg His Ile Val Cys
380 385 390 395
gtg gca atc ttc tcg gcc atc tgt gtt ggc gtg ttt gca gat cgt get 1670
Val Ala Ile Phe Ser Ala Ile Cys Val Gly Val Phe Ala Asp Arg Ala
400 405 410
tac tac tat ggc ttt gcc ttg cca ccc tcg gac att gca cag acc acc 1718
Tyr Tyr Tyr Gly Phe Ala Leu Pro Pro Ser Asp Ile Ala Gln Thr Thr
415 420 425
ctc gtg ggc atc atc ctg tca cga ggc acg gcg gcc agc gtc t--cc ttc 1766
Leu Val Gly Ile Ile Leu Ser Arg Gty Thr Ala Ala Ser Val Ser Phe
430 435 440
atg ttc tct tat atc ttg ctc acc a:g tgc cgc aac ctc ata acc ttc 1814
Met Phe Ser Tyr Ile Leu Leu Thr Met_ Cys Arg Asn Leu Ile Thr Phe
445 450 455
ctg cga gag act ttc ctc aac cgc tat gtg cct ttt gat gcc gca gtg 1862
Leu Arg Glu Thr Phe Leu Asn Arg Tyr. Val Pro Phe Asp Ala Ala Val
460 465 470 475
gac ttc cac cgc tgg atc gcc atg got get gtt gtc ctg gcc att ttg 1910
Asp Phe His Arg Trp Ile Ala Met Ala Ala Val Val Leu Ala Ile Leu
480 485 490
cac agt get ggc cac gca gtc aat g.c tac atc ttc tca gtc agc cca 1958
His Ser Ala Gly His Ala Val Asn Val Tyr Ile Phe Ser Val Ser Pro
495 500 505
ctc agc ctg ctg gcc tgc ata ttc ccc aac gtc ttt gtg aat gat ggg 2006
Leu Ser Leu Leu Ala Cys Ile Phe Pro Asn Val Phe Val Asn Asp Gly
510 515 520
tcc aag ctt ccc cag aag ttc tat tgg tgg ttc ttc cag acc gtc cca 2054
Ser Lys Leu Pro Gln Lys Phe Tyr Trp Trp Phe Phe Gln Thr Val Pro
525 530 535
CA 02350776 2001-11-09
109
ggt atg aca ggt gtg ctt ctg ctc ctg gtc ctg gcc atc atg tat gtc 2102
Gly Met Thr Gly Val Leu Leu Leu Leu Val. Leu Ala Ile Met Tyr Val
540 545 550 555
ttc gcc tcc cac cac ttc cgc cgc cgc agc ttc cgg ggc ttc tgg ctg 2150
Phe Ala Ser His His Phe Arg Arg Arg Ser Phe Arg Gly Phe Trp Leu
560 565 570
acc cac cac etc tae ate ctg etc tai:. gcc ctg etc ate atc cat ggc 2198
Thr His His Leu Tyr Ile Leu Leu Tye Ala Leu Leu Ile Ile His Gly
575 580 585
agc tat get ctg ate cag ctg ccc act ttc cac ate tae ttc ctg gtc 2246
Ser Tyr Ala Leu Ile Gln Leu Pro Thr Phe His Ile Tyr Phe Leu Val
590 595 600
ccg gca ate ate tat gga ggt gac asp ctg gtg agc ctg agc cgg aag 2294
Pro Ala Ile Ile Tyr Gly Gly Asp Lis Leu Val Ser Leu Ser Arg Lys
605 610 615
aag gtg gag ate agc gtg gtg aag gop gag ctg ctg ccc tea gga gtg 2342
Lys Val Glu Ile Ser Val Val Lys Ala Glu Leu Leu Pro Ser Gly Val
620 625 630 635
acc tac ctg caa ttc cag agg ccc caa ggc ttt gag tae aag tea gga 2390
Thr Tyr Leu Gln Phe Gln Arg Pro Gin Gly Phe Glu Tyr Lys Ser Gly
640 645 650
cag tgg gtg cgg ate gcc t::gc ctg get cog ggg acc acc gag tac cac 2438
Gln Trp Val Arg Ile Ala Cys Leu Ala Leu Gly Thr Thr Glu Tyr His
655 650 665
ccc ttc aca ctg acc tee gcg ccc cat gag gac aca etc agc ctg cac 2486
Pro Phe Thr Leu Thr Ser Ala Pro His Glu Asp Thr Leu Ser Leu His
670 675 680
ate cgg gca gtg ggg ccc tgg acc act_ cgc etc agg gag ate tac tea 2534
Ile Arg Ala Val Gly Pro Trp Thr The Arg Leu Arg Glu Ile Tyr Ser
685 690 695
tee cea aag ggc aat ggc tgt get gga tac cca aag etg tae ctt gat 2582
Ser Pro Lys Gly Asn Gly Cys Ala Gly Tyr Pro Lys Leu Tyr Leu Asp
700 705 710 715
gga ccg ttt gga gag ggc cat cag gag tgg cat aaa ttt gag gtg tea 2630
Gly Pro Phe Gly Glu Gly His Gln Glu Trp His Lys Phe Glu Val Ser
720 725 730
gtg ttg gtg gga ggg ggc att ggg g,_c acc ccc ttt gcc tee ate etc 2678
Val Leu Val Gly Gly Gly Ile Gly Val Thr Pro Phe Ala Ser Ile Leu
735 7,10 745
aaa gac ctg gtc ttc aag tea tee t_g ggc agc caa atg ctg tgt aag 2726
Lys Asp Leu Val Phe Lys Ser Ser Liu Gly Ser Gln Met Leu Cys Lys
750 755 760
aag ate tac ttc ate tgg gtg aca egg acc cag cgt cag ttt gag tgg 2774
Lys Ile Tyr Phe Ile Trp Val Thr A.rg Thr Gln Arg Gln Phe Glu Trp
765 770 775
etg get gac ate ate caa gag gtg gag gag aac gac cac cag gac etg 2822
Leu Ala Asp Ile Ile Gln Glu Val GLu Glu Asn Asp His Gln Asp Leu
780 785 790 795
gtg tct gtg cac att tat gtc acc cag ctg get gag aag ttc gac etc 2870
Val Ser Val His Ile Tyr Val Thr GLn Leu Ala Glu Lys Phe Asp Leu
800 805 810
agg acc acc atg eta tac ate tgc gag cgg cac ttc cag aaa gtg ctg 2918
Arg Thr Thr Met Leu Tyr Ile Cys Glu Arg His Phe Gln Lys Val Leu
CA 02350776 2001-11-09
110
815 820 825
aac cgg agt ctg ttc: acg ggc ctg cgc tcc atc acc cac ttt. ggc cgt 2966
Asn Arg Ser Leu Phe Thr Gly Leu Arg Ser Ile Thr His Phe Gly Arg
830 835 840
ccc ccc ttc gag ccc ttc *,-tc aac too ctg cag gag gtc cac cca cag 3014
Pro Pro Phe Glu Pro Phe Phe Asn Ser Leu Gln Glu Val His Pro Gln
845 850 855
gtg cgc aag atc ggg gtg ttc agc tgc ggc cct cca gga atg acc aag 3062
Val Arg Lys Ile Gly Val Phe Ser Cys Gly Pro Pro Gly Met Thr Lys
860 865 870 875
aat gta gag aag gcc tgt cag ctc gtc aac agg cag gac cga gcc cac 3110
Asn Val Glu Lys Ala Cys Gln Leu Val Asn Arg Gln Asp Arg Ala His
880 885 890
ttc atg cac cac tat. gag aac ttc tgagcctgtc ctccctggct gctgcttcca 3164
Phe Met His His Tyr Glu Asn Phe
895
gtatcctgcc ttctcttctg tgcacctaag ttgcccagcc ctgctggcaa tctctccatc 3224
agaatccacc ttaggcctca gctggagggc Lgcagagccc ctcccaatat tgggagaata 3284
ttgacccaga caattataca aatgagaaaa ggcattaaaa tttacgtttc tgatgatggc 3344
aaagctcatt tttctatt.ag taactctgct 3aagatccat ttattgcaat tcatgctgaa 3404
tctaaattgt aaaatttaaa attaaatgca tgtcctcaaa aaaaaaaaa 3453
<210> 48
<211> 899
<212> PRT
<213> Homo sapiens
<400> 48
Met Glu Trp Pro Gly Pro Lys Glu Arg Ser Ser Pro Ile Ile Ile Gin
1 5 10 15
Leu Leu Ser Asp Arg Cys Leu Gln Val Leu Asn Arg His Leu Thr Val
20 25 30
Leu Arg Val Val Gln Leu Gln Pro Lou Gin Gln Val Asn Leu Ile Leu
35 40 45
Ser Asn Asn Arg Gly Cys Arg Thr Lou Leu Leu Lys Ile Pro Lys Glu
50 55 60
Tyr Asp Leu Val Leu Leu Phe Ser Ser Glu Glu Glu Arg Gly Ala Phe
65 70 75 80
Val Gln Gln Leu Trp Asp Phe Cys Val Arg Trp Ala Leu Gly Leu His
85 90 95
Val Ala Glu Met Ser Glu Lys Glu Lou Phe Arg Lys Ala Val Thr Lys
100 1.35 110
Gln Gln Arg Glu Arg Ile Leu Glu 1Ie Phe Phe Arg His Leu Phe Ala
115 120 125
Gln Val Leu Asp Ile Asn Gln Ala Asp Ala Gly Thr Leu Pro Leu Asp
130 135 140
Ser Ser Gln Lys Val Arg Glu Ala Leu Thr Cys Glu Leu Ser Arg Ala
145 150 155 160
Glu Phe Ala Glu Ser Leu Gly Leu Lys Pro Gln Asp Met Phe Val Glu
CA 02350776 2001-11-09
111
165 170 175
Ser Met Phe Ser Leu Ala Asp Lys Asp Gly Asn Gly Tyr Leu Ser Phe
180 185 190
Arg Glu Phe Leu Asp Ile Leu Val Val Phe Met Lys Gly Ser Pro Glu
195 200 205
Asp Lys Ser Arg Leu Met Phe Thr Met Tyr Asp Leu Asp Glu Asn Gly
210 215 220
Phe Leu Ser Lys Asp Glu Phe Phe Thr Met Met Arg Ser Phe Ile Glu
225 230 235 240
Ile Ser Asn Asn Cys Leu Ser Lys Ala Gin Leu Ala Glu Val. Val Glu
245 250 255
Ser Met Phe Arg Glu Ser Gly Phe Gln Asp Lys Glu Glu Leu Thr Trp
260 265 270
Glu Asp Phe His Phe Met Leu Arg Asp His Asp Ser Glu Leu Arg Phe
275 280 285
Thr Gln Leu Cys Val Lys Gly Gly Gly Gly Gly Gly Asn Gly Ile Arg
290 295 300
Asp Ile Phe Lys Gln Asn Ile Ser Cys Arg Val Ser Phe Ile Thr Arg
305 310 315 320
Thr Pro Gly Glu Arg Ser His Pro Gln Gly Leu Gly Pro Pro Val Pro
325 330 335
Glu Ala Pro Glu Leu Gly Gly Pro Gly Leu Lys Lys Arg Phe Gly Lys
340 345 350
Lys Ala Ala Val Pro Thr Pro Arg Leo Tyr Thr Glu Ala Leu Gln Glu
355 360 365
Lys Met Gln Arg Gly Phe Leu Ala Gln Lys Leu Gln Gln Tyr Lys Arg
370 375 380
Phe Val Glu Asn Tyr Arg Arg His Ile Val Cys Val Ala Ile Phe Ser
385 390 395 400
Ala Ile Cys Val Gly Val Phe Ala Asp Arg Ala Tyr Tyr Tyr Gly Phe
405 410 415
Ala Leu Pro Pro Ser Asp Ile Ala Gln Thr Thr Leu Val Gly Ile Ile
420 425 43C
Leu Ser Arg Gly Thr Ala Ala Ser Val Ser Phe Met Phe Ser Tyr Ile
435 440 445
Leu Leu Thr Met Cys Arg Asn Leu Ile Thr Phe Leu Arg Glu Thr Phe
450 455 460
Leu Asn Arg Tyr Val Pro Phe Asp Ala Ala Val Asp Phe His Arg Trp
465 470 475 480
Ile Ala Met Ala Ala Val Val Leu Ala Ile Leu His Ser Ala Gly His
485 490 495
Ala Val Asn Val Tyr Ile Phe Ser Val Ser Pro Leu Ser Leu Leu Ala
500 535 510
Cys Ile Phe Pro Asn Val Phe Val Asn Asp Gly Ser Lys Leu Pro Gln
515 520 525
Lys Phe Tyr Trp Trp Phe Phe Gln Thr Val Pro Gly Met Thr Gly Val
530 535 540
CA 02350776 2001-11-09
112
Leu Leu Leu Leu Val Leu Ala Ile Pet Tyr Val Phe Ala Sex.- His His
545 550 555 560
Phe Arg Arg Arg Ser Phe Arg Gly She Trp Leu Thr His His Leu Tyr
565 5'70 575
Ile Leu Leu Tyr Ala Leu Leu Ile Ile His Gly Ser Tyr Ala Leu Ile
580 585 590
Gln Leu Pro Thr Phe His Ile Tyr Phe Leu Val Pro Ala Ile Ile Tyr
595 60C 605
Gly Gly Asp Lys Leu Val Ser Leu Ser Arg Lys Lys Val Glu Ile Ser
610 615 620
Val Val Lys Ala Glu Leu Leu Pro Ser Gly Val Thr Tyr Leu Gln Phe
625 630 635 640
Gln Arg Pro Gln Gly Phe Glu Tyr Lys Ser Gly Gin Trp Val Arg Ile
645 650 655
Ala Cys Leu Ala Leu Gly Thr Thr Glu Tyr. His Pro Phe Thr Leu Thr
660 665 670
Ser Ala Pro His Glu Asp Thr Leu Ser Leu His Ile Arg Ala Val Gly
675 680 685
Pro Trp Thr Thr Arg Leu Arg Glu Ile Tyr Ser Ser Pro Lys Gly Asn
690 695 700
Gly Cys Ala Gly Tyr Pro Lys Leu Tyr Leu Asp Gly Pro Phe Gly Glu
705 710 715 720
Gly His Gln Glu Trp His Lys Phe Glu Val Ser Val Leu Val Gly Gly
725 730 735
Gly Ile Gly Val Thr Pro Phe Ala Ser Ile Leu Lys Asp Leu Val Phe
740 745 750
Lys Ser Ser Leu Gly Ser Gln Met Leu Cys Lys Lys Ile Tyr Phe Ile
755 760 765
Trp Val Thr Arg Thr Gln Arg Gln Phe Glu Trp Leu Ala Asp Ile Ile
770 775 780
Gln Glu Val Glu Glu Asn Asp His Gin Asp Leu Val Ser Val His Ile
785 790 795 800
Tyr Val Thr Gln Leu Ala Glu Lys Phe Asp Leu Arg Thr Thr Met Leu
805 810 815
Tyr Ile Cys Glu Arg His Phe Gin Lys Val Leu Asn Arg Ser Leu Phe
820 825 830
Thr Gly Leu Arg Ser Ile Thr His Phe Gly Arg Pro Pro Phe Glu Pro
835 840 845
Phe Phe Asn Ser Leu Gln Glu Val His Pro Gln Val Arg Lys Ile Gly
850 855 860
Val Phe Ser Cys Gly Pro Pro Gly Met Thr Lys Asn Val Glu Lys Ala
865 870 875 880
Cys Gln Leu Val Asn Arg Gln Asp Arg Ala His Phe Met His His Tyr
885 890 895
Glu Asn Phe
CA 02350776 2001-11-09
113
<210> 49
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 49
cctgacagat gtatttcact acccag 26
<210> 50
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 50
ggatcggagt cactcccttc gctg 24
<210> 51
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 51
ctagaagctc tccttgttgt aataga 26
<210> 52
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 52
atgaacacct ctggggtcag ctga 24
<210> 53
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 53
atgaacacct ctggggtcag ctga 24
<210> 54
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 54
CA 02350776 2001-11-09
114
gtcctctgca gcattgttcc tctta 25
<210> 55
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 55
cctgacagat gtatttcact acccag 26
<210> 56
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 56
ggatcggagt cactcccttc gctg 24
<210> 57
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 57
aatgacactg tactggaggc cacag 25
<210> 58
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 58
ctgccatcta ccacacggat ctgc 24
<210> 59
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 59
cttgccattc caaagcttcc atgc 24
<210> 60
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
CA 02350776 2001-11-09
115
<400> 60
gtacaagtca ggacagtggg tgcg 24
<210> 61
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 61
tggatgatgt cagccagcca ctca 24