Note: Descriptions are shown in the official language in which they were submitted.
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
NF AT Mediates Cardiac Hypertrophy,
Methods and Reagents Related Thereto
Government Su~nort
This invention was made in the course of work supported by the U.S. Government
and
Howard Hughes Medical Institute, which may have certain rights in this
invention.
Back~!round of the Invention
Heart failure affects approximately three million Americans, developing at a
rate of
approximately 400,000 new cases per year. Current therapy for heart failure is
primarily directed
to using angiotensin-converting enzyme (ACE) inhibitors and diuretics. ACE
inhibitors appear
to slow the progression towards end-stage heart failure in patients; however,
they are unable to
relieve symptoms in more than 60% of heart failure patients and reduce
mortality of heart failure
only by approximately 1 S-20%. Heart transplantation is limited by the
availability of donor hearts.
With the exception of digoxin, the chronic administration of positive
inotropic agents has not
resulted in a useful drug without accompanying adverse side effects, such as
increased arrhythmia,
2 o sudden death, or other deleterious side effects related to survival. These
deficiencies in current
therapy suggest the need for additional therapeutic approaches.
Cardiac muscle hypertrophy is one of the most important adaptive physiological
responses
of the myocardium. In response to increased demands for cardiac work or
following a variety of
pathological stimuli which lead to cardiac injury, the heart adapts through
the activation of a
hypertrophic response in individual cardiac muscle cells, which is
characterized by an increase
in myocyte size, the accumulation of contractile proteins within individual
cardiac cells, the
activation of embryonic gene markers expression, and the lack of a concomitant
effect on muscle
cell proliferation. Although the hypertrophic process can initially be
compensatory, there can be
a pathological transition in which the myocardium becomes dysfunctional
(Braunwald ( 1994) in
3 o Pathoph siolo~v of Heart Failure, (Braunwald, ed.); Saunders,
Philadelphia; Vol.14, pp 393-402).
Studies in an in vitro model system of ventricular muscle cell hypertrophy
have led to the
identification of a number of mechanical, hormonal, growth factor, and
pathological stimuli which
can activate several independent features of hypertrophy (Chien et al. (1991)
A E J. 5:3037-
3046; Knowlton et al. (1991) J. Biol. Chem. 266:7759-7768; Shubeita et al.
{1990) J. Biol. Chem.
265:20555-20562; Thorburn et al. (1993) J. Biol. Chem. 268:2244-2249; LaMorte
et al. (1994)
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
J: Biol. Chem. 269:13490-13496; Knowlton et al. (1993) J. Biol. Chem.
268:15374-15380).
Currently, there are at least two signal transduction pathways, involving both
ras- (Thorburn et
al. (1993) supra), and Gq protein-dependent downstream effectors (LaMorte et
a1. (1994) supra)
implicated in the activation of features of the hypertrophic response in the
in vitro model system.
While a great deal of progress has been made in uncovering the signaling
pathways which activate
the ventricular muscle cell hypertrophic response, relatively little has been
reported as to the
mechanisms which might inhibit or suppress the hypertrophic response in a non-
lethal manner.
There is a clear need for new drugs to treat cardiac disorders, e.g., to
improve heart failure
therapy, such as congestive heart failure and hypertrophic cardiomyopathy.
Methods for
identifying such drugs are thus necessitated.
Summary of the Invention
One aspect of the present invention provides a method for preventing or
reducing cardiac
hypertrophy in a subject, comprising administering to the subject a
pharmaceutically effective
amount of an NF-AT antagonist to decrease the biological activity of NF-AT in
myocardial tissue,
to thereby prevent or reduce cardiac hypertrophy in the subject.
For example, the method of the present invention can use NF-AT antagonists
which
decrease the transcriptional activity of NF-AT, inhibit the nuclear
translocation of NF-AT, and/or
2 0 inhibits dephosphorylation of NF-AT.
In certain embodiments, the antagonists of the present invention inhibit the
formation of
a complex comprising NF-AT, e.g., a protein-protein or protein-DNA
interaction. For example,
the antagonist can inhibit an NF-AT activity by inhibiting interaction of the
protein with a gene
which includes an NF-AT responsive element, or it can inhibit
dephosphorylation of an NF-AT
protein, or inhibit nuclear localization of an NF-AT protein.
The antagonist can be delivered locally or systemically. In the instance of
the former, the
antagonist can be delivered by catheter, and/or perfusion into the myocardial
space.
The present method can be used as part of a treatment for, e.g., congestive
heart disease.
In preferred embodiments, the antagonist selectively inhibits NF-ATc3 and/or
NF-ATc4.
3 o The antagonist can be identified by a method comprising
(i) contacting an isolated NF-AT polypeptide or portion thereof sufficient for
interacting with a molecule, with the molecule and a compound in conditions
under which, but for the presence of the compound, the NF-AT polypeptide or
portion thereof and the molecule interact; and
2
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
(ii) determining the level of interaction between the NF-AT polypeptide or
portion
thereof and the molecule in the presence relative to the absence of the
compound,
such that a weaker interaction between the NF-AT polypeptide or portion
thereof and the
molecule in the presence, relative to the absence, of the compound indicates
that the
compound is an antagonist of the activity of an NF-AT polypeptide.
In still another aspect, the present invention provides a pharmaceutical
composition for
treating or preventing cardiac hypertrophy comprising an NF-AT antagonist in a
pharmaceutically
acceptable delivery vehicle for targeting the NF-AT antagonist to the heart.
The present invention also provides several novel methods and compositions for
modulating the immune response and for screening for modulators of the immune
response.
These methods utilize polynucleotide sequences encoding NF-AT~ recombinant
proteins and
complementary polynucleotides which are substantially identical to NF-AT~
polynucleotide
sequences.
In one aspect of the invention, NF-AT~ polypeptides and compositions thereof
are
provided. NF-AT~ polypeptides comprise polypeptide sequences which are
substantially identical
to a sequence shown in Fig. 1 or a cognate NF-AT~ gene sequence.
Nucleic acid sequences encoding NF-AT~ are provided. The characteristics of
the cloned
sequences are given, including the nucleotide and predicted amino acid
sequence in Fig. 1.
Polynucleotides comprising these sequences can serve as templates for the
recombinant
2 0 expression of quantities of NF-AT~ polypeptides, such as human NF-AT~ and
murine NF-AT~.
Polynucleotides comprising these sequences can also serve as probes for
nucleic acid
hybridization to detect the transcription and mRNA abundance of NF-AT~ mRNA in
individual
lymphocytes (or other cell types) by in ~ hybridization, and in specific
lymphocyte populations
by Northern blot analysis and/or by in ~u hybridization (Alwine et al. (I977)
Proc. Natl. Acad.
~ci. U.S.A. 74: 5350) and/or PCR amplification and/or LCR detection. Such
recombinant
polypeptides and nucleic acid hybridization probes have utility for in vitro
screening methods for
immunomodulatory agents and for diagnosis and treatment of pathological
conditions and genetic
diseases, such as transplant rejection reactions, T cell-mediated immune
responses, lymphocytic
leukemias (e.g., T cell leukemia or lymphoma) wherein NF-AT activity
contributes to disease
3 0 processes, autoimmune disease, arthritis, and the like.
In one embodiment, candidate immunomodulatory agents are identified by their
ability
to block the binding of a NF-AT~ polypeptide to other components of NF-AT
(e.g., AP-1 ) and/or
to block the binding of NF-AT to DNA having an NF-AT recognition site. The DNA
preferably
includes one or more NF-AT binding sites at which a NF-AT protein complex
specifically binds.
3
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
One means for detecting binding of a NF-AT protein comrpising NF-AT~ to DNA is
to
immobilize the DNA, such as by covalent or noncovalent chemical linkage to a
solid support, and
to contact the immobilized DNA with a NF-AT protein complex comprising a NF-
AT~
polypeptide that has been labeled with a detectable marker (e.g., by
incorporation of radiolabeled
amino acid). Such contacting is typically performed in aqueous conditions
which permit binding
of a NF-AT protein to a target DNA containing a NF-AT binding sequence.
Binding of the
labeled NF-AT to the immobilized DNA is measured by determining the extent to
which the
labeled NF-AT~ polypeptide is immobilized as a result of a specific binding
interaction. Such
specific binding may be reversible, or may be optionally irreversible if a
cross-linking agent is
added in appropriate experimental conditions.
In one aspect, candidate immunomodulatory agents are identified as being
agents capable
of inhibiting (or enhancing) intermolecular binding between NF-AT~ and other
polypeptides
which compriss a NF-AT complex (e.g., AP-1, Jung, etc.). The invention
provides methods and
compositions for screening libraries of agents for the capacity to interfere
with binding of NF-AT~
to other NF-AT polypeptide species under aqueous binding conditions.
Typically, at least either
NF-AT~ and/or another NF-AT polypeptide species is labeled with a detectable
label and
intermolecular binding between NF-AT~ and other NF-AT polypeptide species is
detected by the
amount of labeled species captured in NF-AT complexes and the like.
Based at least in part on the observation that NF-AT polypeptides comprise
nuclear
2 0 localization sequences, which allow NF-AT polypeptides to translocate to
the nucleus in the
presence of intracellular calcium, but which are shielded by forming
intramolecular associations
with other domains in the NF-AT polypeptide in the absence of calcium, the
invention also
provides methods for modulating the activity of NF-AT, by modulating
translocation of NF-AT,
such as by modulating intramolecular associations and shielding of a nuclear
localization
2 5 sequence (NLS). In addition, since the NLS of NF-AT form an intramolecular
association with
a phosphorylated domain of NF-AT, the invention provides methods for
modulating NF-AT
activity, comprising modulating NF-AT phosphorylation. Furthermore, since, as
disclosed herein,
NF-AT is phosphorylated by protein kinase A (PKA) and glycogen synthase kinase-
3 (GSK-3),
and dephosphorylated by calcineurin, the state of phosphorylation and thus
activation of NF-AT,
3 0 can be modulated by modulating the activity of these kinases and/or
phosphatase. Also within
the scope of the invention are compounds which modulate nuclear translocation
of NF-AT, as well
as screening assays for identifying additional compounds.
In one aspect, the present invention provides a method for identifying an
agent that
modulates phosphorylation of an NF-AT, comprising contacting test agent with a
mixture
3 5 including GSK-3 kinase activity with an NF-AT polypeptide, or portion
thereof which is a
substrate of the GSK-3 kinase activity, and determining the ability of the
test agent to modulate
4
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27$62
tl~e interaction of the GSK-3 kinase activity with the NF-AT polypeptide
and/or phosphorylation
of the NF-AT polypeptide by the GSK-3 kinase activity.
The subject assay can be carried out in as a cell-based assay, e.g., employing
a
recombinant GSK-3 kinase, NF-AT substrate and/or reporter gene, or in a cell-
free format, e.g.,
using purified or semi-purified preparations of GSK-3 and the NF-AT substrate.
The assay can
be a simple competitive binding assay, or a kinase activity assay which
detects the rate of
phosphorylation of the substrate, or a nuclear translocation assay which
detects the rate of nuclear
localization of the substrate. In preferred embodiments, the subj ect method
includes a further step
of formulating a pharmaceutical preparation including one or more compounds
identified in the
subject assay.
Another aspect of the present invention provides a method for modulating NF-AT
phosphorylation, comprising contacting a cell expressing NF-AT with an agent
that modulates the
phosphorylation of NF-AT by GSK-3, e.g., using a GSK-3 inhibitor, especially
an inhibitor which
inhibits NF-AT nuclear translocation.
Still another aspect of the present invention relates to peptide or
peptidomimetic agents
for modulating nuclear translocation of an NF-AT protein, which agent
corresponds to a portion
of an NF-AT protein involved in intramolecular association of nuclear
localization signals. The
present invention also provides a method for identifying compounds that
modulate nuclear
translocation of NF-AT, comprising contacting a test agent with a an NF-AT
polypeptide, or
2 0 portion thereof which includes a nuclear localization signal, and
determining the ability of the test
agent to bind to the nuclear localization sequence and/or alter the tertiary
structure of a
phosphorylated form of the nuclear localization sequence.As above, such assays
can be carried
out in cell-based and cell-free formats, e.g., as competitive binding assays
or nuclear translocation
assays. In one embodiment, changes in the conformation of the protein which
are dependent on
2 5 phosphorylation of the NLS sequences can be detected photometrically, such
as by CD/ORD or
other means for determining changes in tertiary structure
The invention also provides antisense polynucleotides complementary to NF-AT~
sequences which are employed to inhibit transcription and/or translation of
the cognate mRNA
species and thereby effect a reduction in the amount of the respective NF-AT~
protein in a cell
3 0 (e.g., a T lymphocyte of a patient}. Such antisense polynucleotides can
function as
immunomodulatory drugs by inhibiting the formation of NF-AT protein required
for T cell
activation.
In a variation of the invention, polynucleotides of the invention are employed
for diagnosis
of pathological conditions or genetic disease that involve T cell neoplasms or
T cell hyperfunction
3 5 of hypofunction, and more specifically conditions and diseases that
involve alterations in the
5
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
structure or abundance of NF-AT~ polypeptide, NF-AT~ polynucleotide sequence,
or structure of
the NF-AT~ gene or flanking region(s).
The invention also provides antibodies which bind to NF-AT~ with an affinity
of about at
least 1 x 10' M-~ and which lack specific high affinity binding for other
proteins present in
activated T cells. Such antibodies can be used as diagnostic reagents to
identify T cells (e.g.,
activatable T cells) in a cellular sample from a patient (e.g., a lymphocyte
sample, a solid tissue
biopsy) as being cells which contain an increased amount of NF-AT~ protein
determined by
standardization of the assay to be diagnostic for activated T cells.
Frequently, anti-NF-AT~
antibodies are included as diagnostic reagents for immunohistopathology
staining of cellular
samples in situ. Additionally, anti-NF-AT~ antibodies may be used
therapeutically by targeted
delivery to T cells (e.g., by cationization or by liposome/immunoliposome
delivery).
The invention also provides NF-AT~ polynucleotide probes for diagnosis of
neoplasia or
immune status by detection of NF-AT~ mRNA in cells explanted from a patient,
or detection of
a pathognomonic NF-AT~ allele (e.g., by RFLP or allele-specific PCR analysis).
A
pathognomonic NF-AT~ allele is an allele which is statistically correlated
with the presence of a
predetermined disease or propensity to develop a disease. Typically, the
detection will be by inn
i~tu hybridization using a labeled (e.g., 32P~ ssS~ i4C, 3H, fluorescent,
biotinylated,
digoxigeninylated) NF-AT~ polynucleotide, although Northern blotting, dot
blotting, or solution
hybridization on bulk RNA or poly A+ RNA isolated from a cell sample may be
used, as may PCR
2 0 amplification using NF-ATE specific primers. Cells which contain an
increased amount of NF-
AT~ mRNA as compared to standard control values for cells or cell types other
than activated T
cells or activatable T cells will be thereby identified as activated T cells
or activatable T cells.
Similarly, the detection of pathognomonic rearrangements or amplification of
the NF-AT~ locus
or closely linked loci in a cell sample will identify the presence of a
pathological condition or a
2 5 predisposition to developing a pathological condition (e.g., cancer,
genetic disease).
The present invention also provides a method for diagnosing T cell
hypofunction of
hyperfiznction in a human patient, wherein a diagnostic assay (e.g.,
immunohistochemical
staining of fixed lymphocytic cells by an antibody that specifically binds
human NF-AT~) is used
to determine if a predetermined pathognomonic concentration of NF-AT~ protein
or NF-AT~
3 o mRNA is present in a biological sample from a human patient; if the assay
indicates the presence
of NF-AT~ protein or NF-AT~ mRNA at or above such predetermined pathognomonic
concentration, the patient is diagnosed as having T cell hyperfunction or
hypofimction condition,
or transplant rejection and the like. Alternatively, T cell hypofiznction or
immunosuppression can
be diagnosed by determining the level of nuclear and/or cytoplasmic NF-AT in a
subject and
3 5 comparing the level with that of a normal subj ect. In one embodiment, the
level of nuclear and/or
cytoplasmic NF-AT is determined after incubation of lymphocytes of a subject
with a T cell
6
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
activator. A lower level of nuclear NF-AT relative to the normal subject
indicates that the subject
is immunosuppressed. A similar method can be used to monitor the state of
immunosuppression
in a subject who is being treated with an immunosuppressive drug, e.g.,
cyclosporin A. This
allows more optimal dosages of the immunosuppressive drug to be administered
to the subject.
All publications and patent applications herein are incorporated by reference
to the same
extent as if each individual publication or patent application was
specifically and individually
indicated to be incorporated by reference.
Bri D scri ti f the Drawings
Figure lA-1 shows the nucleotide sequence of the human NF-AT~ cDNA (SEQ ID NO:
45) and the deduced amino acid seqeunce (SEQ ID NO: 46). N inicates that a
seqeunce
ambiguity is present.
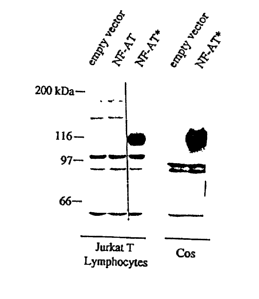
Figure 2 shows the expression of NF-AT~ protein in T cells (Jurkat) and non-T
cells (Cos).
Figure 3A and 3B show that the NF-AT~ cDNA clone encodes a protein that
activates
transcription from an NF-AT site and is capable of activating the IL-2
promoter in non-T cells.
Figure 4 shows homology in the Rel homology domain between NF-AT~, NF-ATP, and
Rel family members. The protein sequences of marine NF-ATp and the Rel
proteins Dorsal (the
Drosophila axis-determining protein) (SEQ ID NO: 47), human c-ReI {SEQ ID NO:
48), NF-rcB
p50 (SEQ ID NO: 49), and NF-KB p65 (SEQ ID NO: 50) are aligned to the sequence
of NF-AT~
2 0 (SEQ ID NO: 51 ) and NF-ATp (SEQ ID NO: 52). Numbering is with respect to
NF-AT~. Identity
to NF-AT~, open boxes; similarity in known residue function or structure,
shaded areas. Stars
indicate regions in which NF-AT~ has: 1) a charge reversal relative to the
majority of other Rel
proteins, or has 2) replaced a potential salt bridge residue with a histidine
or other chelating
residue. Lower portion shows a schematic of NF-AT~ and NF-ATp.
2 5 Figure 5, panel A, shows ribonuclease protection for human NF-AT~ with RNA
from
Jurkat cells (lanes 1-6) or Hela cells (lane 7). The expected specific
ribonuclease-resistant
fragment is 304 nucleotides (arrow). Hela cells were non-stimulated. Jurkat
cells were either
non-stimulated or stimulated with 20ng/ml PMA and 2uM ionomycin for 3 hours,
plus or minus
100ng/ml CsA added at the indicated times after stimulation.
3 0 Figure 5, panel B, shows RNA from the following human cells: KJ (preB cell
ALL), JD-1
(B cell lineage ALL), K562 (erythroleukemia cell line), CML (bone marrow cells
from a patient
with a myeloid leukemia), human muscle tissue, Hep G2 (liver cell line), HPB
ALL (T cell line,
nonstimulated or stimulated with 2ug/mi PHA and SOng/ml PMA for 30 minutes),
and Hela cells
analyzed by ribonuclease protection. A longer exposure of this gel indicates
that the K562 cell
3 5 line contains a small amount of NF-AT~ transcript.
7
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Figure 5, panel C, shows NF-AT~ (upper panel) and NF-AT lower panel) mRNA
expression in mouse tissues and a skin tumor derived from NF-AT-Tag transgenic
mice (Verweij
et al. (1990) J. Biol. Chem ~5: 15788-15795). Cells were either non-stimulated
or stimulated
with 20ng/ml PMA and 2uM ionomycin for 3 hours. RNA was measured by
quantiative
ribonuclease protection using murine cDNA probes. The predicted size of the
fragment
homologous to the probe is indicated by the arrows.
Figure 6, panel A, shows Cos cells and Jurkat cells that were transfected with
reporter
constructs for NF-AT or HNF-1 (,l3Z8). Co-transfected expression vectors for
NF-AT~ (+NF-AT)
or HNF-la (+HNF-1) were included where indicated, otherwise empty pBJS vector
was included.
Cells were stimulated as indicated: PMA, P + I (PMA plus ionomycin).
Figure 6, panel B, shows Cos cells that were transfected with IL-2 luciferase
and with
expression vectors as in panel A. Stimulations were as in panel A. Data in
panel A and panel B
are expressed as fold induction of luciferase activity over nonstimulated
value with empty pBJS
vector. Bars represent mean and range of 2-3 independent transfections.
Figure 6, panel C, shows that expression of NF-AT~ in Cos cells gives rise to
specific
DNA binding activity. Gel mobility shifts using nuclear extracts from Cos
cells transfected with
pBJS (lanes 1 and 3), with NF-AT~ (lanes 2 and 4-7), from non-transfected
Jurkat cells (lanes 8-
11) or using cytosols from pBJS- or NF-ATE transfected Cos cells (lanes 12-13,
15-16) combined
with Hela nuclear extract (lanes 15-16). Lane 14, Hela nuclear extract alone.
Labeled AP-1 (lanes
1-2) or NF-AT (lanes 3-16) probes and cold competitor oligonucleotides are
indicated. Arrows
indicate specific AP-1 and NF-AT complexes.
Figure 6, panel D, shows antisera induced supershift of NF-AT. NF-AT and AP-1
gel
mobility shifts using nuclear extracts from stimulated Jurkat cells or murine
thymocytes. Either
no antisera, preimmune, or one of two different immune antisera was included
as indicated.
Arrows indicate specific NF-AT or AP1 complexes or supershifted NF-AT
complexes (*).
Figure 7 shows dominant-negative NF-AT~. Jurkat Tag cells were transfected
with vector
plasmid (control) or with the dominant negative NF-AT~ plasmid, plus the
indicated secreted
alkaline phosphatase reporter plasmid. Transfected cells were transferred to
fresh culture medium
24 hours after transfection and secreted alkaline phosphatase activity was
measured (Clipstone
and Crabtree (1992) atur 3~7: 695-698) 16 to 24 hours later, after stimulation
with 1 uM
ionomycin plus 20 ng/ml PMA (NF-AT and IL-2 reporters), 20 ng/ml PMA alone
(API reporter)
or no stimulation (RSV reporter). Bars indicate, secreted alkaline phosphatase
activity from cells
transfected with the dominant negative NF-AT~ as a percentage of the activity
from cells
transfected in parallel with control plasmid, and represent data obtained from
(n) independent
3 5 transfections. The dominant negative NF-AT~ consists of a carboxy terminal
truncation of the
epitope tagged NF-AT~ expression plasmid extending to the PvuII site at amino
acid 463.
8
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Figure 8 shows changes in mobility of epitope tagged NF-AT~ expressed in
Jurkat cells.
Cells were transfected with NF-AT~ as in Fig. 2 and stimulated as shown for 2
hrs plus or minus
100ng/ml CsA. Whole cell lysates were analyzed by western blotting as in Fig.
2.
Figure 9, panel A, is a diagram of a FLAG epitoped tagged NF-ATcl protein
indicating
the respective positions in SEQ ID NO: 38 of the Rel Similarity Domain (RSD),
two nuclear
localization sequences (NLS), a conserved domain rich in serines (SRR), and
three repeats rich
is serines and prolines (SP1, SP2, SP3).
Figure 9, panel B, is a diagram showing the percentage of Cos cells
transiently transfected
with NF-ATcI (SH160c) expressing NF-ATcl in the nucleus after various times of
treatment of
the cells with ionomycin and calcium (I + Ca++), or with FK506 plus (I+Ca*' +
FK506) or with
ionomycin plus 2.5 mM EGTA (I + EGTA) for 60 minutes.
Figure 9, panel C, is a diagram of the NF-AT fusion protein C0418-GFP,
containing
amino acids 1-418 of NF-ATcI including the serine rich domain (SRR) and the
three serine-
proline rich domain (SP1, SP2, and SP3) fused to the green fluorescent protein
(GFP).
Figure 9, panel D, shows the percentage of Cos cells transiently transfected
with constructs
encoding NF-ATcI (G) or NF-AT(Cn418)-GFP (O) having cytopiasmic NF-AT after
one hour
of stimulation with ionomycin and calcium and then replacement of the medium
with medium
containing FK506. Cells expressing NF-ATc in the cytoplasm and those
expressing NF-ATc in
both cytoplasm and nucleus were added and divided by the total number of
analyzed expressing
2 0 cells.
Figure 10 depicts a diagram showing the percentage of Cos cells expressing NF-
AT in the
nuclear following transient transfection of the cells with a construct
encoding NF-ATc 1 fused to
zero, one, or two copies of a wild-type NLS from SV40 large T antigen [NLS]
inserted between
the FLAG epitope and the second amino acid of NF-ATcI, or one or two copies of
a mutant form
2 5 of the NLS (NLS-T). Cells were stained with the anti-FLAG antibody.
Figure 11, panel A, shows the amino acid sequence of two conserved putative
NLSs in
NF-ATcI (SEQ ID NO: 53 and SEQ ID NO: 54), and their positions in SEQ ID NO:
38.
Figure 11, panel B, shows a diagram of NF-ATc 1 and the amino acid sequence of
the two
NLSs, above which are shown the amino acid sequence of the mutated NLSs (TRTG
has SEQ ID
3 0 NO: 55 and KRKK has SEQ ID NO: 56). The lower portion of the figure shows
the percentage
of Cos cells expressing wildtype NF-AT or mutated NF-AT (m265, corresponding
to an NF-ATc 1
protein having a mutated N-terminal NLS; and m265 + 682 containing the m265
mutation and
a mutation of the C-terminal NLS) in the nucleus after transient transfection
of the cells and
stimulation with ionomycin and calcium for 60 minutes. The percentage of cells
staining in
3 5 nucleus (lighted shaded areas), cytoplasm (solid areas), or both
compartments (darkly shaded
areas) was determined.
9
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Figure 12, panel A, shows the amino acid sequence of the SRR from NF-ATc 1
(amino
acid 170 to 194 of SEQ ID NO: 38, set forth in SEQ ID NO: 57), and the
cellular localization of
NF-ATcl proteins (nuclear (N) or cytoplasmic (C)} having the indicated serine
to alanine
substitutions (SEQ ID NOs: 58-63).
Figure 12, panel B, is a photograph of a Western blot, showing phosphorylated
GST-NF-
ATcI fusion proteins containing amino acids 196-304 of NF-ATc (WT) or with
serine to alanine
mutations in the three SP repeats (S--~A), after having been phosphorylated by
incubation with
[y 32PJATP and a partially purified preparation of cellular- NF-AT kinase
activity (brain extract)
and then incubated with phosphatases as indicated, prior to separation by
electrophoresis and
autoradiography (top lane). The bottom lane indicates the amount of NF-AT in
each sample, as
visualized by Coomassie staining.
Figure 13, panel A, is a photograph of a Western blot showing the results of
an affinity
purification of extracts of COS, cells that had been transfected with the
empty expression vector
(Vector} or a vector encoding the HA epitope-tagged amino-terminal 418
residues of NF-ATc (2-
418) with glutathione-agarose beads coupled to GST or incubated with beads
coupled to a GST
fusion with the RSD of NF-ATc (GST-RSD). Affinity-selected proteins were
detected by
immonoblotting with the anti-HA 12CA5 antibody. The left part of the panel is
a graphic
representation of the NF-AT polypeptide that was attached to the agarose beads
and the NF-AT
polypeptide which was affinity purified on the beads.
2 0 Figure 13, panel B, is a photograph of a Western blot showing the results
of an affinity
purification of extracts from COS cells transfected with a construct encoding
the HA epitope
tagged amino-terminal 418 residues of NF-ATc (2-418) or vector alone (Vector),
with GST-RSD
or a version with a mutation in the carboxy-terminal NLS (mNLS). Bound
proteins were detected
with the 12CA5 antibody. The left part of the panel is a graphic
representation of the NF-AT
2 5 polypeptides used in the example.
Figure 13, panel C, is a photograph of a Western blot showing the results of
an affinity
purification of extracts from COS cells that had been transfected with the HA
epitone-tagged
amino-terminal 418 residues of NF-ATc ( 1-418 WT) or versions in which S--;A
mutations were
present in the SRR or SP repeats with GST-RSD. The associated proteins were
detected with the
3 0 7A6 antibody. The lower part of the blot shows the amount of NF-AT present
in the cell extracts
prior to affinity purification.
Figure 14 shows a model of the mechanism of NT-ATc nuclear entry. According to
the
model, cytoplasmic NF-ATc is phosphorylated on the SP repeats and SRR masking
the activity
of its two partially redundant NLSs, the sequence KRK at position 265-267, and
the sequence
3 5 KRICK/R at position 682-685 of NF-ATc 1. Dephosphorylation in response to
activation of
calcineurin leads to an alteration in an intramolecular interaction and
perhaps a conformational
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
change, exposing one or more of the NLSs to the nuclear import machinery. Once
in the nucleus,
termination of calcium signaling results in rapid export to the cytosol,
possibly by the exposure
of nuclear export sequences (NES).
Figure 15, panel A, shows the amino acid sequence of amino acids 196-304 of NF-
ATcI
(SEQ ID NO: 38) set forth in SEQ ID NO: 64. Putative overlapping GSK-3
consensus sites
[SPXXS(P)J (Fiol et al., J. Biol. Chem. 269, 32187 (1994)) are overlined. The
nuclear
localization sequence is in bold type, and sites phosphorylated by PKA in
vitro are boxed. The
underlined serines are serines that have been substituted with alanines in
some examples.
Figure 15, panel B, shows a graph depicting the level of GSK-3 activity in
various
fractions eluted from a P-11 column following ammonium sulfate fractionation
of brain extracts.
The lower part of the panel shows autoradiograms of Western blots showing the
ability of the
eluted fractions to phosphorylate GST-NF-AT fusion proteins containing wild-
type (WT) NF
ATcI or NF-ATcI in which the underlined serines of pane A were mutated into
alanines, or NF
ATcl which was in vitro phosphorylated with PKA (WT-PKA prephosph.).
Figure 15, panel C, shows a graph depicting the amount of protein in various
fractions
eluted from a Mono-S column of the P-11 pool of panel B. The lower part of the
panel shows
autoradiograms of Western blots showing the ability of the eluted fractions to
phosphorylate GST-
NF-AT fusion proteins containing wild-type (WT) NF-ATcI or NF-ATcl in which
the underlined
serines of pane A were mutated into alanines, or NF-ATcI which was in vitro
phosphorylated
2 0 with PKA (WT-PKA prephosph.). The bottom two photographs show
autoradiograms of Western
blots containing protein from the eluted fractions incubated with antisera
specific for GSK-3a and
GSK-3~i.
Figure 16, panel A, shows an autoradiogram of a Westen blot of PKA-
prephosphorylated
NF-AT (WT-PKA prephos.) incubated with brain extracts immunodepleted with
antisera to GSK
3a and/or GSK-3(3 or control antibodies in an in vitro kinase reaction with [y-
3zP]ATP, and the
3zP-labeled substrate (upper gel; the lower gel shows Coomassie staining).
Figure 16,
panel B, shows a photograph of an autoradiogram of a Westen blot of PKA-
prephosphorylated
NF-AT (WT-PKA prephos.) or NF-AT (WT) incubated with brain extracts
immunodepleted with
antisera to GSK-3a and GSK-3(i or control antibodies (Ig) in an in vitro
kinase reaction with [y
3 0 32P]ATP.
Figure 17, panel A, shows autoradiograms of a Western blot depicting NF-AT-GST
fusion
proteins phosphorylated in vitro with the indicated purified kinases. In the
rightmost lanes, the
first kinase was permitted to phosphorylate the WT substrate with
nonradioactive ATP to
completion; then, the WT substrate beads were washed to remove the kinase and
the WT beads
3 5 were phosphorylated by the second kinase in the presence of [y 32P]ATP.
11
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Figure 17, panel B, depicts autoradiograms of two-dimensional tryptic
phosphopeptide
maps of the NF-ATcI wild-type fusion protein with the indicated kinases in
vitro. NF-ATc was
overexpressed in COS cells [which support reversible Ca2+-dependent nuclear
localization] and
labeled with [32P]orthophosphate. In the lower right panel, the PKA + GSK-3(3
in vitro
phosphorylated peptides were mixed with the in vivo phosphorylated peptides
before two-
dimensional separation to establish that they are similar. Phosphopeptides
migrating differently
are circled with a dashed Iine.
Figure 18, panel A, shows the activiy of alkaline phosphatase (SEAP) in
extracts from
Jurkat cells transfected with an SEAP reporter gene under the control of an NF-
AT, AP-1 or HIV-
l 0 LTR regulatory element and in which GSK-3~3 is overexpressed or in which
the empty vector was
overexpressed. NF-AT SEAP activity is expressed as a percentage of the
ionomycin-stimulated
and phorbal 12-myristate 13-acetate (PMA)-stimulated control activity; AP-1
and HIV-LTR
SEAP activities are expressed as a percentage of PMA-stimulated activity
(Spencer et al., Science
262, 1019 (1993)).
Figure 18, panel B, shows the percentage of cells expressing NF-ATcI in the
cytoplasm,
nucleus or both after cotransfection of COS cells with FLAG epitope-tagged NF-
ATcI and each
of the indicated serine-threonine kinases. Cells were stimulated with
ionomycin and 10 mM Ca2+,
and the percentages of cells expressing NF-AT localized in the nucleus,
cytoplasm, or both
compartments were scored visually and are presented as a percentage of
expressing cells. The
2 0 transfected ERK kinase was activated by adding PMA (25 ng/ml).
Figure 19 shows the percentage of cells having cytoplasmic NF-ATc, after
overexpression
of GSK-3[3 in COS cells cotransfected with expression constructs encoding FLAG
epitope-tagged
NF-ATcl, calcineurin A and B, and vector (0), GSK-3(3 (0) or GSK-KM (O), a
catalytically
inactive GSK-3~3 (He et al., Nature. 374, 617 (1995)). Cells were also
cotransfected with a version
2 5 of NF-ATcl in which the underlined serines in Fig. lA were changed to
alanines with calcineurn
and GSK-3~i (0). To obtain NF-ATc localization in the nucleus, the transfected
cells were treated
with ionomyin and Ca2+ for 60 min, then the medium was changed to medium with
FK506 (20
ng/ml) to terminate Ca2+ signaling and to block nuclear reentry of NF-ATc.
Transfected NF-ATc
was detected with FLAG mAb M2 by indirect immunofluorescence, and 200
expressing cells were
3 0 scored as expressing NF-ATc in the cytoplasm, nucleus, or both
compartments.
Figure 20A shows a schematic of the endogenous NF-ATc4 gene, the targeting
construct
used to transfect ES cells and the targeted NF-ATc4 locus resulting from
homologus
recombination.
Figure 20B shows a Southern blot of mouse tail DNA digested with EcoRVand
blotted
3 5 with the 3' outside probe
12
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Detailed Description of the Invention
(i) Overview
On a cellular level, the heart functions as a syncytium of myocytes and
surrounding support
cells, called non-myocytes. While non-myocytes are primarily
fibroblast/mesenchymal cells, they
also include endothelial and smooth muscle cells. Indeed, although myocytes
make up most of the
adult myocardial mass, they represent only about 30% of the total cell numbers
present in heart.
Because of their close relationship with cardiac myocytes in vivo, non-
myocytes are capable of
influencing myocyte growth and/or development. This interaction may be
mediated directly
through cell-cell contact or indirectly via production of a paracrine factor.
Such association in vivo
is important since both non-myocyte numbers and the extracellular matrix with
which they interact
are increased in myocardial hypertrophy and in response to injury and
infarction. These changes
are associated with abnormal myocardial function.
Cardiac myocytes are unable to divide shortly after birth. Further growth
occurs through
hypertrophy of the individual cells. Cell culture models of myocyte
hypertrophy have been
developed to understand better the mechanisms for cardiac myocyte hypertrophy.
Simpson et al.,
Circ. Res., 51: 787-801 (1982); Chien et al., FASEB J., 5: 3037-3046 (1991).
Most studies ofheart
myocytes in culture are designed to minimize contamination by non-myocytes.
See, for example,
Simpson and Savion, Cir. Cres., S0: 101-116 (1982); Libby, J. Mol. CeII.
Cardiol., 16: 803-811
(1984); Iwaki et al., J. Biol. Chem., 265:13809-13817 (1990).
2 0 Hypertrophy of adult cardiac ventricular myocytes is a response to a
variety of conditions
which lead to chronic overload. This response is characterized by an increase
in myocyte cell size
and contractile protein content without concomitant cell division, and
activation of embryonic
genes, including the gene for atrial natriuretic peptide (ANP). Chien et al.,
supra. Adult myocyte
hypertrophy is initially beneficial as a short term response to impaired
cardiac function by
2 5 permitting a decrease in the load on individual muscle fibers. With
severe, long-standing
overload, however, the hypertrophied cells begin to deteriorate and die. Katz,
"Heart failure," in
Katz AM, ed., Physiology of the Heart (New York: Raven Press; 1992) pp. 638-
668. Endothelial
cells, smooth muscle cells and fibroblast/mesenchymal cells exist in close
contact with myocytes
in the heart. Nag, Cytobios., 28: 41-61 (1980).
3 0 One aspect of the present invention relates to the use of agents to
inhibitor NF-AT activity,
particularly NF-AT~3 or NF-AT~4, as part of a method for inhibiting or
preventing unwanted
growth of cardiac and other vascular tissues. In considering the potential of
any protein as a target
for drug development, one must consider if the loss-of function of the protein
would be, over the
therapeutic dosage, lethal to the treated subject, e.g., either because of
systemic lethality or
3 5 lethality to the treated tissue. In the art, transgenic animals having
loss-of function mutations to
NF-AT allelles have indicated that inhibition of certain NF-AT functions
includes a lethal
13
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
consequence. Likewise, loss-of function of calcineurin can also result in a
lethal phenotype.
However, as described herein (see, e.g., Example 20), neither loss-of function
of NF-ATc4 or NF-
ATc3 is lethal to the animal, nor do such mutations have any untoward effects
on the pathology
of normal cardiac tissue. Our observations suggest, therefore, that NF-ATc4 or
NF-ATc3 are
appropriate targets for drug development.
Thus, according to the present invention, there is provided a method for the
inhibiting the
growth of cardiac and vascular tissue, e.g., which inhibits growth of myocytic
and/or non-
myocytic cells in such tissue. For example, as described below, the subject
method can be used
to slow the process of cardiac hypertrophy and arteriolar smooth muscle
proliferation, e.g., as part
as of a treatment of vascular smooth muscle hypertrophy or cardiac
hypertrophy. Chronic cardiac
hypertrophy, for example, is a significantly diseased state which is a
precursor to congestive heart
failure and cardiac arrest. Antagonists of NF-AT, and particulary of NF-AT~3
or NF-AT~4, are
useful as part of a treatments for congestive heart failure.
Moreover, based on differences in specificity for DNA recognition elements,
and in
the protein-protein interactions that the various NF-AT paralogs have, the
present invention
specifically contemplates the identification, and use, of NF-AT antagonists
which selectively
inhibit the activity of certain of the NF-ATproteins. In preferred
embodiments, the present
invention contemplates a method for treating cardiac hypertrophy, or other
preventing other
growth of cardiac and vascular tissue, through the use of NF-AT antagonists
which are selective
2 0 for NF-AT~3 and/or NF-ATE, but not NF-ATE 1 or NF-AT~2. For instance, the
NF-AT antagonist
can be selected so as to have an ED50 for inhibition of NF-AT~3 or NF-AT~4 in
vivo of at least
one, and more preferably, two, three, four and even five orders of magnitude
less than its ED50
for inhibition of NF-AT~1 or NF-AT~2 activity.
In other embodiments, antagonists of NF-AT can be used to inhibit growth of
cancers
2 5 and other hyperproliferative disorders involving tissue of muscle origin,
such as in the treatment
of rhabdomyosarcoma and leiomyoma.
(ii) Definitions
Unless defined otherwise, all technical and scientific terms used herein have
the same
3 0 meaning as commonly understood by one of ordinary skill in the art to
which this invention
belongs. Although any methods and materials similar or equivalent to those
described herein can
be used in the practice or testing of the present invention, the preferred
methods and materials are
described. For purposes of the present invention, the following terms are
defined below.
Generally, the nomenclature used hereafter and the laboratory procedures in
cell culture,
3 5 molecular genetics, and nucleic acid chemistry and hybridization described
below are those well
known and commonly employed in the art. Standard techniques are used for
recombinant nucleic
14
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
acid methods, polynucleotide synthesis, and microbial culture and
transformation (e.g.,
electroporation, lipofection). Generally enzymatic reactions and purification
steps are performed
according to the manufacturer's specifications. The techniques and procedures
are generally
performed according to conventional methods in the art and various general
references (~ge_,
general, Sambrook et al. Molecular Cloning: A Laboratory Manual, 2d ed. (
1989) Cold Spring
Harbor Laboratory Press, Cold Spring Harbor, N.Y., which is incorporated
herein by reference)
which are provided throughout this document. The procedures therein are
believed to be well
known in the art and are provided for the convenience of the reader. All the
information contained
therein is incorporated herein by reference.
Oligonucleotides can be synthesized on an Applied Bio Systems oligonucleotide
synthesizer according to specifications provided by the manufacturer.
Methods for PCR amplification are described in the art (PCR Technology:
Principles and
Applications for DNA Amplification ed. HA Erlich, Freeman Press, New York, NY
(1992); P~$
Protocols: A Guide to Methods and A"pnl= ications, eds. Innis, Gelfland,
Snisky, and White,
Academic Press, San Diego, CA (1990); Mattila et al. (1991) Nucleic Acids Res.
~: 4967; Eckert,
K.A. and Kunkel, T.A. (1991) PCR Methods and Applications 1_: 17; _P~I , eds.
McPherson,
Quirkes, and Taylor, IRL Press, Oxford; and U.S. Patent 4,683,202, which are
incorporated herein
by reference).
As used herein, the terms "exemplary NF-AT nucleic acid" and "exemplary NF-AT
2 0 protein" refer to the nucleotide and amino acid sequences, respectively,
of such NF-AT genes as
described in the examples below, as well as in such references as Northrop et
al. (I994) Nature
369:497; Park et al. ( 1996) J. Biol. Chem. 271:20914; Luo et aI. ( 1996) MCB
16:3955; Hoey et al.
(1995) Immunity 2:461; Masuda et al. (1995) Mol. Cell. Biol. 15:2697; and US
Patent Nos.
5,708,158 and 5,612,455. These references, among others, provide the sequences
of human NF-
2 5 AT3 (i.e., human NF-ATc4), and those of three splice variants of human NF-
AT4 (i.e., human NF-
ATc3). The three forms of NF-AT4 have been designated NF-AT4a, NF-AT4b, and NF-
AT4c,
and the positions of the splice junctions in the coding regions are after
proline 699 in NF-AT4a,
and after valine 700 and proline 716 in NF-AT4b and NF-AT4c. An alignment of
the Rel domains
of these NF-AT proteins shows that certain areas are particularly well
conserved. In particular,
3 0 the following amino acid sequences are found in the REL domain of all NF-
AT polypeptides: the
amino acid sequence HHRAHYETEGSRGAVKA (SEQ ID NO: ), the amino acid sequence
PHAFYQVHRITGK (SEQ ID NO: ), the amino acid sequence
DIELRKGETDIGRKNTRVRLVFRVHX1P (SEQ ID NO: ), and the amino acid sequence
PX2ECSQRSAX3ELP (SEQ ID NO: ), wherein each X1 and X2 is hydrophobic residue
such as
3 5 valine or isoleucine, and X3 is any residue, but preferably glutamine or
histidine.
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
The GenBank Accession Numbers of the exemplary human NF-AT nucleic acids and
polypeptides are provided in the following Table:
NF-ATAT GenBank No.
NF-ATc U08015
NF-ATc.b U59736
1~-A~SS'~
NF-ATl I38152
NF-ATpI U43341isoformB
U43342isoform C
NF-ATc3
NF-AT4a I3 815 S
NF-AT4b I38156
NF-AT4c L41067
-A 4
2 0 NF-AT3 L41066
I38154
NF-ATx U 14510
NF-ATx2 U85428
NF-ATx3 U85429
2 5 NF-ATx4 U85430
NF-ATc2 has also been referred to as NFIL2E and NFII-a.
Other examples of NF-AT genes and genes products can be found in GenBank,
particularly
accessions I80836, U36576, U36575, I60722, U02079, AF049606, AF087434, as well
as PRF
3 0 locus 2013343A, PIR locus 545262 and A48753.
"Treatment" refers to both therapeutic treatment and prophylactic or
preventative measures,
wherein the object is to prevent or slow down (lessen) hypertrophy. Those in
need of treatment
include those already with the disorder as well as those prone to have the
disorder or those in
which the disorder is to be prevented. The hypertrophy may be from any cause,
including
3 5 congenital, viral, idiopathic, cardiotrophic, or myotrophic causes, or as
a result of ischemia or
ischemic insults such as myocardial infarction. Typically, the treatment is
performed to stop or
slow the progression of hypertrophy, especially after heart damage, such as
from ischemia, has
occurred. Preferably, for treatment of myocardial infarctions, the agents) is
given immediately
after the myocardial infarction, to prevent or lessen hypertrophy.
4 0 By the term "heart failure" is meant an abnormality of cardiac function
where the heart
does not pump blood at the rate needed for the requirements of metabolizing
tissues. Heart failure
includes a wide range of disease states such as congestive heart failure,
myocardial infarction,
tachyanhythmia, familial hypertrophic cardiomyopathy, ischemic heart disease,
idiopathic dilated
cardiomyopathy, and myocarditis. The heart failure can be caused by any number
of factors,
16
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
including ischemic, congenital, rheumatic, or idiopathic forms. Chronic
cardiac hypertrophy is a
significantly diseased state which is a precursor to congestive heart failure
and cardiac arrest.
Still more specifically, the terms "treating" and "treatment" shall mean
preventing,
alleviating, and/or inhibiting. In this regard, the method of the present
invention can be used as
part of a treatment for, but not limited to, (1) ventricular muscle cell
hypertrophy, e.g., which is
induced by al-adrenergic agonists and/or endothelin, (2) ventricular muscle
cell hypertrophy
induced by drugs which have an adverse side effect of promoting cardiac
hypertrophy, (3) a
medical condition, e.g., heart failure, mediated by ventricular muscle cell
hypertrophy, and (4)
ventricular muscle cell hypertrophy initiated by cardiac injury, such as viral
myocarditis, long-
1 o standing hypertension, cardiomyopathy due to pathological stimuli, and
post-myocardial
infarction.
"Chronic" administration refers to administration of the agents) in a
continuous mode as
opposed to an acute mode, so as to maintain the initial anti-hypertrophic
effect for an extended
period of time.
The term "NF-AT antagonist" as used herein refers to any molecule which blocks
or
prevents the NF-AT dependent transciption. Such antagonists accomplish this
effect in various
ways. For instance, one class of antagonists will bind to an NF-AT protein
with sufficient affinity
and specificity to inhibit NF-AT interaction with its cognate response element
in DNA or protein
factors, such as AP 1. Other classes of antagonist will bind to NF-AT and
inhibit its nuclear
2 0 localization, such as by preventing phosphorylation of NLS sites, or
inhibiting conformation
changes resulting from phosphorylation. Still another class of antagonists can
prevent NF-AT
activity by inhibiting kinases, such as GSK-3 or PKA, from phosphorylating NLS
sites on the NE-
AT protein. Other antagonists are described herein and will be apparent to
those skilled in the art.
"Ventricular muscle cell hypertrophy" is a condition characterized by an
increase in the
2 5 size of individual ventricular muscle cells, the increase in cell size
being sufficient to result in a
clinical diagnosis of the patient or sufficient as to allow the cells to be
determined as larger (e.g.,
2-fold or more larger than non-hypertrophic cells). It may be accompanied by
accumulation of
contractile proteins within the individual cardiac cells and activation of
embryonic gene
expression.
3 0 "Suppression" of ventricular muscle cell hypertrophy means a reduction in
one of the
parameters indicating hypertrophy relative to the hypertrophic condition, or a
prevention of an
increase in one of the parameters indicating hypertrophy relative to the
normal condition. For
example, suppression of ventricular muscle cell hypertrophy can be measured as
a reduction in cell
size relative to the hyertrophic condition. Suppression of ventricular muscle
cell hypertrophy
3 5 means a decrease of cell size of 10% or greater relative to that observed
in the hypertrophic
condition. More preferably, suppression of hypertrophy means a decrease of
cell size of 30% or
17
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
greater; most preferably, suppression of hypertrophy means a decrease of cell
size of SO% or more.
Relative to the hypertrophy score assay when phenylephrine is used as the
inducing agent, these
decreases would correlate with hypertrophy scores of about 6.5 or less, 5.0-
5.5; and 4.0-5.0,
respectively. When a different agent is used as the inducing agent,
suppression is measured
relative to the maximum cell size (or hypertrophic score) measured in the
presence of that inducer.
"Prevention of ventricular muscle cell hypertrophy" is determined by
preventing an
increase in cell size relative to normal cells, in the presence of a
concentration of inducer sufficient
to fully induce hypertrophy. For example, prevention of hypertrophy means a
cell size increase
less than 200% greater than non-induced cells in the presence of a maximally-
stimulating
concentration of inducer. More preferably, prevention of hypertrophy means a
cell size increase
less than 135% greater than non-induced cells; and most preferably, prevention
means a cell size
increase less than 90% greater than non-induced cells. Relative to the
hypertrophy score assay
when phenylephrine is used as the inducing agent, prevention of hypertrophy in
the presence of
a maximally-stimulating concentration of phenylephrine means a hypertrophic
score of about
6.0-6.5, 5.0-5.5, and 4.0-4.5, respectively.
By the term "effective amount" or "therapeutically effective amount"of an NF-
AT
therapeutic, e.g., antagonist is meant an amount of an NF-AT therapeutic
sufficient to obtain the
desired physiological effect, e.g., suppression of ventricular muscle cell
hypertrophy. An
effective amount of an NF-AT therapeutic is determined by the care giver in
each case on the basis
2 0 of factors normally considered by one skilled in the art to determine
appropriate dosages, including
the age, sex, and weight of the subject to be treated, the condition being
treated, and the severity
of the medical condition being treated.
As used herein, the twenty conventional amino acids and their abbreviations
follow
conventional usage (Immunology - A Synthesis, 2nd Edition, E.S. Golub and D.R.
Gren, Eds.,
2 5 Sinauer Associates, Sunderland, Massachusetts (1991), which is
incorporated herein by reference).
Stereoisomers (e.g., D-amino acids) of the twenty conventional amino acids,
unnatural amino acids
such as a,a-disubstituted amino acids, N-alkyl amino acids, lactic acid, and
other unconventional
amino acids may also be suitable components for polypeptides of the present
invention. Examples
of unconventional amino acids include: 4-hydroxyproline, y-carboxyglutamate, E-
N,N,N-
3 o trimethyllysine, E-N-acetyllysine, O-phosphoserine, N-acetylserine, N-
formylmethionine, 3-
methylhistidine, S-hydroxylysine, c.~-N-methylarginine, and other similar
amino acids and imino
acids (e.g., 4-hydroxyproline). In the polypeptide notation used herein, the
lefthand direction is
the amino terminal direction and the righthand direction is the carboxy-
terminal direction, in
accordance with standard usage and convention. Similarly, unless specified
otherwise, the
3 5 lefthand end of single-stranded polynucleotide sequences is the 5' end;
the lefthand direction of
double-stranded polynucleotide sequences is referred to as the 5' direction.
The direction of 5' to
18
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
3' addition of nascent RNA transcripts is referred to as the transcription
direction; sequence regions
on the DNA strand having the same sequence as the RNA and which are 5' to the
5' end of the
RNA transcript are referred to as "upstream sequences"; sequence regions on
the DNA strand
having the same sequence as the RNA and which are 3' to the 3' end of the RNA
transcript are
referred to as "downstream sequences".
The term "naturally-occurnng" as used herein as applied to an object refers to
the fact that
an object can be found in nature. For example, a polypeptide or polynucleotide
sequence that is
present in an organism (including viruses) that can be isolated from a source
in nature and which
has not been intentionally modified by man in the laboratory is naturally-
occurring.
The term "corresponds to" is used herein to mean that a polynucleotide
sequence is
homologous (i.e., is identical, not strictly evolutionarily related) to all or
a portion of a reference
polynucleotide sequence, or that a polypeptide sequence is identical to a
reference polypeptide
sequence. In contradistinction, the term "complementary to" is used herein to
mean that the
complementary sequence is homologous to all or a portion of a reference
polynucleotide sequence.
For illustration, the nucleotide sequence "TATAC" corresponds to a reference
sequence "TATAC"
and is complementary to a reference sequence "GTATA".
The following terms are used to describe the sequence relationships between
two or more
polynucleotides: "reference sequence", "comparison window", "sequence
identity", "percentage
of sequence identity", and "substantial identity". A "reference sequence" is a
defined sequence
2 0 used as a basis for a sequence comparision; a reference sequence may be a
subset of a larger
sequence, for example, as a segment of a full-length cDNA or gene sequence
given in a sequence
listing, such as a polynucleotide sequence of Fig. l, or may comprise a
complete cDNA or gene
sequence. Generally, a reference sequence is at least 20 nucleotides in
length, frequently at least
nucleotides in length, and often at least 50 nucleotides in length. Since two
polynucleotides
2 5 may each (1) comprise a sequence (i.e., a portion of the complete
polynucleotide sequence) that
is similar between the two polynucleotides, and (2) may further comprise a
sequence that is
divergent between the two polynucleotides, sequence comparisons between two
(or more)
polynucleotides are typically performed by comparing sequences of the two
polynucleotides over
a "comparison window" to identify and compare local regions of sequence
similarity. A
3 0 "comparison window", as used herein, refers to a conceptual segment of at
least 20 contiguous
nucleotide positions wherein a polynucleotide sequence may be compared to a
reference sequence
of at least 20 contiguous nucleotides and wherein the portion of the
polynucleotide sequence in
the comparison window may comprise additions or deletions (i.e., gaps) of 20
percent or less as
compared to the reference sequence (which does not comprise additions or
deletions) for optimal
3 5 alignment of the two sequences. Optimal alignment of sequences for
aligning a comparison
window may be conducted by the local homology algorithm of Smith and Waterman
(1981) derv.
19
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Appl~. V~ath. 2: 482, by the homology alignment algorithm of Needleman and
Wunsch (1970) ~
ol. Biol. 48: 443, by the search for similarity method of Pearson and Lipman (
1988) Proc. Natl.
Acad. Sci. l_ .U S.A~ $5: 2444, by computerized implementations of these
algorithms (GAP,
BESTFIT, FASTA, and TFASTA in the Wisconsin Genetics Software Package Release
7.0,
Genetics Computer Group, 575 Science Dr., Madison, WI), or by inspection, and
the best
alignment (i.e., resulting in the highest percentage of homology over the
comparison window)
generated by the various methods is selected. The term "sequence identity"
means that two
polynucleotide sequences are identical (i.e., on a nucleotide-by-nucleotide
basis) over the window
of comparison. The term "percentage of sequence identity" is calculated by
comparing two
Z 0 optimally aligned sequences over the window of comparison, determining the
number of positions
at which the identical nucleic acid base (e.g., A, T, C, G, U, or I) occurs in
both sequences to yield
the number of matched positions, dividing the number of matched positions by
the total number
of positions in the window of comparision (i.e., the window size), and
multiplying the result by
100 to yield the percentage of sequence identity. The terms "substantial
identity" as used herein
denotes a characteristic of a polynucleotide sequence, wherein the
polynucleotide comprises a
sequence that has at least 85 percent sequence identity, preferably at least
90 to 95 percent
sequence identity, more usually at least 99 percent sequence identity as
compared to a reference
sequence over a comparison window of at least 20 nucleotide positions,
frequently over a window
of at least 25-50 nucleotides, wherein the percentage of sequence identity is
calculated by
2 0 comparing the reference sequence to the polynucleotide sequence which may
include deletions or
additions which total 20 percent or less of the reference sequence over the
window of comparison.
The reference sequence may be a subset of a larger sequence, for example, as a
segment of the full-
length human NF-AT~ polynucleotide sequence shown in Fig. 1 or the full-length
murine or bovine
NF-AT~ cDNA sequence.
2 5 As applied to polypeptides, a degree of identity of amino acid sequences
is a function of
the number of identical amino acids at positions shared by the amino acid
sequences. A degree
of homology or similarity of amino acid sequences is a function of the number
of amino acids, i.e.
structurally related, at positions shared by the amino acid sequences. An
"unrelated" or "non-
homologous" sequence shares less than 40 % identity, though preferably less
than 25 % identity,
3 o with one of the NF-ATc sequences of the present invention. The term
"substantial identity" means
that two peptide sequences, when optimally aligned, such as by the programs
GAP or BESTFIT
using default gap weights, share at least 80 percent sequence identity,
preferably at least 90 percent
sequence identity, more preferably at least 95 percent sequence identity or
more (e.g., 99 percent
sequence identity). Preferably, residue positions which are not identical
differ by conservative
3 5 amino acid substitutions. Conservative amino acid substitutions refer to
the interchangeability of
residues having similar side chains. For example, a group of amino acids
having aliphatic side
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
chains is glycine, alanine, valine, leucine, and isoleucine; a group of amino
acids having aliphatic-
hydroxyl side chains is serine and threonine; a group of amino acids having
amide-containing side
chains is asparagine and glutamine; a group of amino acids having aromatic
side chains is
phenylalanine, tyrosine, and tryptophan; a group of amino acids having basic
side chains is lysine,
arginine, and histidine; and a group of amino acids having sulfur-containing
side chains is cysteine
and methionine. Preferred conservative amino acids substitution groups are:
valine-leucine-
isoleucine, phenylalanine-tyrosine, lysine-arginine, alanine-valine, and
asparagine-glutamine.
The term "NF-AT~ native protein" and "full-length NF-AT~ protein" as used
herein refers
to a a naturally-occurring NF-AT~ polypeptide corresponding to the deduced
amino acid sequence
shown in Fig. 1 or corresponding to the deduced amino acid sequence of a
cognate full-length
cDNA. Also for example, a native NF-AT~ protein present in naturally-occurring
lymphocytes
which express the NF-AT~ gene are considered full-length NF-AT~ proteins.
The term "NF-AT~ fragment" as used herein refers to a polypeptide that has an
amino
terminal and/or carboxy-terminal deletion, but where the remaining amino acid
sequence is
identical to the corresponding positions in the NF-AT~ sequence deduced from a
full-length cDNA
sequence (e.g., the cDNA sequence shown in Fig. 1). NF-AT~ fragments typically
are at least 14
amino acids long, preferably at least 20 amino acids long, usually at least SO
amino acids long or
longer.
The term "NF-AT~ analog" as used herein refers to polypeptides which are
comprised of
2 0 a segment of at least 25 amino acids that has substantial identity to a
portion of the deduced amino
acid sequence shown in Fig. 1, and which has at least one of the following
properties: (1) binding
to other polypeptides under suitable binding conditions including (a) other NF-
AT proteins (e.g.,
AP-1 ); (b) a kinase, such as GSK-3 and PKA; (c) a phosphatase, such as
calcineurin; (d) NF-AT
polypeptides, in particular portions thereof, such as an NLS, SRR, SP1, SP2,
and/or SP3; (2)
2 5 binding to a nucleic acid; (3) ability to localize to the nucleus upon T
cell activation; and (4) the
ability to translocate from the nucleus to the cytoplasm after termination of
the stimulatory signal.
Typically, NF-AT~ analog polypeptides comprise a conservative amino acid
substitution (or
addition or deletion) with respect to the naturally-occurring sequence. NF-AT~
analogs typically
are at least 20 amino acids long, preferably at least 50 amino acids long or
longer, most usually
3 0 being as long as full-length naturally-occurnng NF-AT~ (e.g., as shown in
Fig. 1). Some NF-AT~
analogs may lack biological activity but may still be employed for various
uses, such as for raising
antibodies to NF-AT~ epitopes, as an immunological reagent to detect and/or
purify a-NF-AT~
antibodies by affinity chromatography, or as a competitive or noncompetitive
agonist, antagonist,
or partial agonist of native NF-AT~ protein function.
3 5 The term "NF-AT~ polypeptide" which is used herein interchangeably with
the term "NE-
AT" polypeptide, is used herein as a generic term to refer to native protein,
fragments, or analogs
21
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
of NF-AT~. Hence, native NF-AT~, fragments of NF-AT~, and analogs of NF-AT~
are species of
the NF-AT~ polypeptide genus. Preferred NF-AT~ polypeptides include: the human
foil-length
NF-AT~ protein comprising the polypeptide sequence shown in Fig. 1 (which is
also referred to
as "NF-ATc I "), or polypeptides consisting essentially of a sequence shown in
Table II. Thus, the
genus NF-ATc includes all NF-AT polypeptides identified so far as well as
those that have not yet
been identified and which could be identified, e.g., by low stringency
hybridization. In addition
to the NF-ATc having SEQ ID NO: 38, which is also referred to as NF-ATcl, and
homologs of
other species, the NF-ATc genus includes NF-ATc2 (also termed NF-ATp), NF-ATc3
(also termed
NF-AT4 or NF-ATx), NF-ATc4 (also termed NF-AT3), and splice variants thereof.
The term "cognate" as used herein refers to a gene sequence that is
evolutionarily and
functionally related between species. For example but not limitation, in the
human genome, the
human CD4 gene is the cognate gene to the mouse CD4 gene, since the sequences
and structures
of these two genes indicate that they are highly homologous and both genes
encode a protein
which functions in signaling T cell activation through MHC class II-restricted
antigen recognition.
Thus, the cognate marine gene to the human NF-AT~ gene is the marine gene
which encodes an
expressed protein which has the greatest degree of sequence identity to the
human NF-AT~ protein
and which exhibits an expression pattern similar to that of the human NF-AT~
(e.g., expressed in
T lineage cells). Preferred cognate NF-AT~ genes are: rat NF-AT~, rabbit NF-
AT~, canine NF-AT~,
nonhuman primate NF-AT~, porcine NF-ATE, bovine NF-AT~, and hamster NF-AT~.
The term "NF-AT~-dependent gene" is used herein to refer to genes which: (1)
have a NF-
AT binding site (a site which can be specifically footprinted by NF-AT under
suitable binding
conditions) within about 10 kilobases of the first coding sequence of said
gene, and (2) manifest
an altered rate of transcription, either increased or decreased, from a major
or minor transcriptional
start site for said gene, wherein such alteration in transcriptional rate
correlates with the presence
2 5 of NF-AT~ polypeptide in NF-AT complexes, such as in an activated T cell.
The term "altered ability to modulate" is used herein to refer to the capacity
to either
enhance or inhibit a biological activity, e.g., transcription of a gene; such
enhancement or
inhibition may be contingent on the occurrence of a specific event, such as T
cell stimulation. For
example, this alteration may be manifest as an inhibition of the
transcriptional enhancement of the
3 0 IL-2 gene that normally ensues following T cell stimulation. The altered
ability to modulate
transcriptional enhancement or inhibition may affect the inducible
transcription of a gene, such
as in the just-cited IL-2 example, or may effect the basal level transcription
of a gene, or both.
The term "agent" is used herein to denote a chemical compound, a mixture of
chemical
compounds, a biological macromolecule, or an extract made from biological
materials such as
3 5 bacteria, plants, fungi, or animal (particularly mammalian) cells or
tissues. Agents are evaluated
22
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
for potential activity as immunomodulatory agents (e.g., irnmunosuppressants)
by inclusion in
screening assays described hereinbelow.
The term "candidate imunomodulatory agent" is used herein to refer to an agent
which is
identified by one or more screening methods) of the invention as a putative
immuomodulatory
agent. Some candidate immunomodulatory agents may have therapeutic potential
as drugs for
human use.
As used herein, the terms "label" or "labeled" refers to incorporation of a
detectable
marker, g:gs, by incorporation of a radiolabeled amino acid or attachment to a
polypeptide of
biotinyl moieties that can be detected by marked avidin (e.g., streptavidin
containing a fluorescent
l0 marker or enzymatic activity that can be detected by optical or
colorimetric methods). Various
methods of labeling polypeptides and glycoproteins are known in the art and
may be used.
Examples of labels for polypeptides include, but are not limited to, the
following: radioisotopes
(e.g., 3H, 'qC, 35S, 'zsI, '3'I), fluorescent labels (e.g., FITC, rhodamine,
lanthanide phosphors),
enzymatic labels (e.g., horseradish peroxidase, (3-galactosidase, luciferase,
alkaline phosphatase),
biotinyl groups, predetermined polypeptide epitopes recognized by a secondary
reporter (e.g.,
leucine zipper pair sequences, binding sites for secondary antibodies, metal
binding domains,
epitope tags). In some embodiments, labels are attached by spacer arms of
various lengths to
reduce potential steric hindrance.
As used herein, "substantially pure" means an object species is the
predominant species
2 0 present (i.e., on a molar basis it is more abundant than any other
individual species in the
composition), and preferably a substantially purified fraction is a
composition wherein the object
species comprises at least about 50 percent (on a molar basis) of all
macromolecular species
present. Generally, a substantially pure composition will comprise more than
about 80 to 90
percent of all macromolecular species present in the composition. Most
preferably, the object
2 5 species is purified to essential homogeneity (contaminant species cannot
be detected in the
composition by conventional detection methods) wherein the composition
consists essentially of
a single macromolecular species.
As used herein the terms "pathognomonic concentration", "pathognomonic
amount", and
"pathognomonic staining pattern" refer to a concentration, amount, or
localization pattern,
3 0 respectively, of a NF-AT~ protein or mRNA in a sample, that indicates the
presence of a
hypofunctional or hyperfunctional T cell condition or a predisposition to
developing a disease,
such as graft rejection. A pathognomonic amount is an amount of a NF-AT~
protein or NF-AT~
mRNA in a cell or cellular sample that falls outside the range of normal
clinical values that is
established by prospective and/or retrospective statistical clinical studies.
Generally, an individual
3 5 having a neoplastic disease (e.g., lymphocytic leukemia) or T cell-
mediated immune response will
exhibit an amount of NF-AT~ protein or mRNA in a cell or tissue sample that is
higher than the
23
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
range of concentrations that characterize normal, undiseased individuals;
typically the
pathognomonic concentration is at least about one standard deviation above the
mean normal
value, more usually it is at least about two standard deviations or more above
the mean normal
value. However, essentially all clinical diagnostic tests produce some
percentage of false positives
and false negatives. The sensitivity and selectivity of the diagnostic assay
must be sufficient to
satisfy the diagnostic objective and any relevant regulatory requirements. In
general, the
diagnostic methods of the invention are used to identify individuals as
disease candidates,
providing an additional parameter in a differential diagnosis of disease made
by a competent health
professional.
(iii) Exemplary Embodiments
A. NFiAT~ Polvnucleotides
Genomic or cDNA clones encoding NF-AT~ may be isolated from clone libraries
(e.g.,
available from Clontech, Palo Alto, CA) using hybridization probes designed on
the basis of the
15~ nucleotide sequences shown in Fig. 1, or other exemplary NF-AT sequences,
and using
conventional hybridization screening methods (e.g., Benton WD and Davis RW
(1977) Science
1 6: 180; Goodspeed et al. (1989) Gene ~: 1; Dunn et al. (1989) J. Biol. Chem.
~: 13057).
Where a cDNA clone is desired, clone libraries containing cDNA derived from
human mRNA is
preferred. Alternatively, synthetic polynucleotide sequences corresponding to
all or part of the
2 0 sequences shown in Fig. l, or other exemplary NF-AT sequences, may be
constructed by
chemical synthesis of oligonucleotides. Additionally, polymerase chain
reaction (PCR) using
primers based on the sequence of an NF-AT gene may be used to amplify DNA
fragments from
genomic DNA, mRNA pools, or from cDNA clone libraries. U.S. Patents 4,683,195
and 4,683,202
describe the PCR method. Additionally, PCR methods employing one primer that
is based on the
2 5 sequence data disclosed in, e.g., Fig. 1 and a second primer that is not
based on that sequence data
may be used. For example, a second primer that is homologous to or
complementary to a
polyadenylation segment may be used. In an embodiment, a polynucleotide
comprising the 2742
nucleotide long sequence of Fig. 1 can also be readily constructed by those of
skill in the art by
using the degeneracy of the genetic code. Polynucleotides encoding amino acids
418 to 710 of the
3 0 NF-ATc sequence of Fig. 1 can also be constructed by those of skill in the
art.
It is apparent to one of skill in the art that nucleotide substitutions,
deletions, and additions
may be incorporated into the polynucleotides of the invention. Nucleotide
sequence variation may
result from sequence polymorphisms of various NF-AT~ alleles, minor sequencing
errors, and the
like. However, such nucleotide substitutions, deletions, and additions should
not substantially
3 5 disrupt the ability of the polynucleotide to hybridize to one of the
polynucleotide sequences shown
24
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
in Fig. 1 under hybridization conditions that are sufficiently stringent to
result in specific
hybridization.
Specific hybridization is defined herein as the formation of hybrids between a
probe
polynucleotide (e.g., a polynucleotide of the invention which may include
substitutions, deletion,
and/or additions) and a specific target polynucleotide (e.g., a polynucleotide
having the sequence
in Fig. 1, or other exemplary NF-AT sequences), wherein the probe
preferentially hybridizes to
the specific target such that, for example, a single band corresponding to NF-
AT~ mRNA (or bands
corresponding to multiple alternative splicing products of the NF-AT~ gene)
can be identified on
a Northern blot of RNA prepared from a suitable cell source (e.g., a T cell
expressing NF-AT~).
1 o Polynucleotides of the invention and recombinantly produced NF-AT~, and
fragments or analogs
thereof, may be prepared on the basis of the sequence data provided in Fig. 1,
or other exemplary
NF-AT sequences, according to methods known in the art and described in
Maniatis et al.,
Molecular Cloning' A Laboratory Manual, 2nd Ed., (1989), Cold Spring Harbor,
N.Y. and Berger
and Kimmel, Methods in Enz~gy Volume 152, Guide to Molecular Cloning
Techniques
(1987), Academic Press, Inc., San Diego, CA, which are incorporated herein by
reference.
NF-AT~ polynucleotides may be short oligonucleotides (e.g., 25-100 bases
long), such as
for use as hybridization probes and PCR (or LCR) primers. NF-AT~
polynucleotide sequences
may also comprise part of a larger polynucleotide (e.g., a cloning vector
comprising a NF-AT~
clone) and may be fused, by polynucleotide linkage, in frame with another
polynucleotide
2 0 sequence encoding a different protein (e.g., glutathione S-transferase or
~3-galactosidase) for
encoding expression of a fusion protein. Typically, NF-AT~ polynucleotides
comprise at least 25
consecutive nucleotides which are substantially identical to a naturally-
occurring NF-AT~
sequence (e.g., Fig. 1, or other exemplary NF-AT sequences), more usually NF-
AT~
polynucleotides comprise at least 50 to 100 consecutive nucleotides which are
substantially
2 5 identical to a naturally-occurring NF-AT~ sequence. However, it will be
recognized by those of
skill that the minimum length of a NF-AT~ polynucleotide required for specific
hybridization to
a NF-AT~ target sequence will depend on several factors: G/C content,
positioning of mismatched
bases (if any), degree of uniqueness of the sequence as compared to the
population of target
polynucleotides, and chemical nature of the polynucleotide (e.g.,
methylphosphonate backbone,
3 0 phosphorothiolate, etc.), among others.
For example but not limitation, suitable hybridization probes for detecting
and/or
quantifying the presence of NF-AT~ mRNA in a sample generally comprise at
least one, preferably
at least two, and more preferably all of the following human NF-AT~ sequences
shown in Table
I, or their complements:
Table I~ Selected Human NF-AT~~o~ynucleotide Sequences
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
S'-TTC CTC CGG GGC GCG CGG CGT GAG CCC GGG GCG AGG-3' (SEQ ID NO: 1);
S'-CAG CGC GGG GCG GCC ACT TCT CCT GTG CCT CCG CCC GCT GCT-3' (SEQ ID NO: 2);
S'-GCC GCG CGG ATG CCA AGC ACC AGC TTT CCA GTC CCT TCC AAG-3' (SEQ ID NO: 3);
S'-CCA ACG TCA GCC CCG CCC TGC CGC TCC CCA CGG CGC ACT CCA-3' (SEQ ID NO: 4);
S'-TTC AGA CCT CCA CAC CGG GCA TCA TCC CGC CGG CGG-3' (SEQ ID NO: S);
S'-GCC ACA CCA GGC CTG ATG GGG CCC CTG CCC TGG AGA GTC CTC-3' (SEQ ID NO: 6);
S'-AGT CTG CCC AGC CTG GAG GCC TAC AGA GAC CCC TCG TGC CTG-3' (SEQ ID NO: 7);
S'-GTG TCT CCC AAG ACC ACG GAC CCC GAG GAG GGC TTT CCC-3' (SEQ ID NO: 8);
S'-AGC TGG CTG GGT GCC CGC TCC TCC AGA CCC GCG TCC CCT TGC-3' (SEQ ID NO: 9);
S'-TAC AGC CTC AAC GGC CGG CAG CCG CCC TAC TCA CCC CAC CAC-3' (SEQ ID NO: 10);
S'-GAC CAC CGA CAG CAG CCT GGA CCT GGG AGA TGG CGT CCC TGT-3' (SEQ ID NO: 11
);
S'-CCT GGG CAG CCC CCC GCC CCC GGC CGA CTT CGC GCC CGA AGA-3' (SEQ ID NO: 12);
S'-GCT CCC CTA CCA GTG GCG AAG CCC AAG CCC CTG TCC CCT ACG-3' (SEQ ID NO: 13);
S'-CTT CGG ATT GAG GTG CAG CCC AAG TCC CAC CAC CGA GCC CAC-3' (SEQ ID NO: 14);
S'-CAT GGC TAC TTG GAG AAT GAG CCG CTG ATG CTG CAG CTT TTC-3' (SEQ ID NO: 1
S);
S'-AAG ACC GTG TCC ACC ACC AGC CAC GAG GCT ATC CTC TCC AAC-3' (SEQ ID NO: 16);
S'-TCA GCT CAG GAG CTG CCT CTG GTG GAG AAG CAG AGC ACG GAC-3' (SEQ ID NO: 17);
S'-AAC GCC ATC TTT CTA ACC GTA AGC CGT GAA CAT GAG CGC G-3' (SEQ ID NO: 18);
S'-AGA AAC GAC GTC GCC GTA AAG CAG CGT GGC GTG TGG CA-3' (SEQ ID NO: 19); and
2 0 S'-GCA TAC TCA GAT AGT CAC GGT TAT TTT GCT TCT TGC GAA TG-3' (SEQ ID NO:
20).
Also for example but not limitation, the following pair of PCR primers
(amplimers)
may be used to amplify murine or human NF-AT~ sequences (e.g., by reverse
transcriptase
initiated PCR of RNA from NF-AT~ expressing cells):
(forward) S'-AGGGCGCGGGCACCGGGGCGCGGGCAGGGCTCGGAG-3' (SEQ ID NO: 21 )
(reverse) S'-GCAAGAAGCAAAATAACCGTGACTATCTGAGTATGC-3' (SEQ ID NO: 22)
If desired, PCR amplimers for amplifying substantially full-length cDNA copies
may be
3 0 selected at the discretion of the practioner. Similarly, amplimers to
amplify single NF-AT~ exons
or portions of the NF-AT~ gene (murine or human) may be selected.
Each of these sequences may be used as hybridization probes or PCR amplimers
to detect
the presence of NF-AT~ mRNA, for example to diagnose a disease characterized
by the presence
of an elevated NF-AT~ mRNA level in lymphocytes, or to perform tissue typing
(i.e., identify
3 5 tissues characterized by the expression of NF-AT~ mRNA), and the like. The
sequences may also
be used for detecting genomic NF-AT~ gene sequences in a DNA sample, such as
for forensic
DNA analysis (e.g., by RFLP analysis, PCR product lengths) distribution, etc.)
or for diagnosis
of diseases characterized by amplification and/or rearrangements of the NF-AT~
gene.
Disclosure of the full coding sequence for human NF-AT~ shown in Fig. 1 makes
possible
4 o the construction of isolated polynucleotides that can direct the
expression of NF-AT~, fragments
26
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
thereof, or analogs thereof. Further, the sequences in Fig. 1 make possible
the construction of
nucleic acid hybridization probes and PCR primers that can be used to detect
RNA and DNA
sequences encoding NF-AT~.
NF-AT polynucleotides of the invention include full-length NF-AT
polynucleotides or
portions thereof. In one embodiment, the NF-AT polynucleotide is at least
about 60%, 70%, 75%,
80%, 85%, 90%, 95%, more preferably at least about 98% and most preferably at
least about 99%
identical to a nucleotide sequence shown in Figure 1, or set forth in SEQ ID
NO: 45, or other
exemplary NF-AT sequences, or of a portion thereof. Accordingly, the invention
comprises
polynucleotides encoding NF-ATc family members other than NF-ATc having SEQ ID
NO: 38,
which is referred to herein as NF-ATcI. Such family members include NF-ATc2
(GenBank
I38152, U43341, U43342; also referred to as NF-ATp), NF-ATc3 (GenBank L41067,
I38155,
I38156; also referred to as NF-AT4 or NF-ATx), and NF-ATc4 (GenBank L41066,
I38154; also
referred to as NF-AT3) as well as differentially spliced forms thereof, as
described, e.g., in U.S.
Patent No. 5,612,455 issued to Hoey on March 18, 1997 and 5,656,452, issued to
Rao et al., on
August 12, published PCT WO 95/02035 by Arai et al. and WO 94/15964 by Rao et
al.
Preferred portions or fragments of NF-ATc proteins include those comprising
one or more
specific domains. At least the following NF-ATc domains have been identified:
a DNA-binding
domain, corresponding essentially to the Rel Homology Domain (RHD) or Rel
Similary Domain
(RSD); a domain interacting with another protein, e.g., AP-I or a target site
of PKA or GSK-3;
2 0 a nuclear localization sequence (NLS), e.g., comprising amino acids 265-
267 of SEQ ID NO: 38
(N-terminal NLS) or amino acids 681-685 of SEQ ID NO: 38 (C-terminal NLS), or
a domain
interacting with an NLS, e.g., SRR (amino acids 172-194 of SEQ ID NO: 38), SP1
(amino acids
199-2I9 of SEQ ID NO: 38), SP2 (amino acids 233-252 of SEQ ID NO: 38), and SP3
(amino
acids 278-301 of SEQ ID NO: 38). Other potential domains can be identified as
further described
2 5 herein. Accordingly, the invention provides NF-ATc polynucleotides
encoding portions of NF-
ATc polypeptides capable of exercising (agonists) or inhibiting (antagonists)
at least one biological
activity of NF-ATc, e.g., binding to another molecule, such as DNA or another
protein,
translocating across the nuclear membrane of a cell, or inhibiting
translocation across a nuclear
membrane of a cell. Assays for confirming that an NF-AT polypeptide is an
agonist or an
3 0 antagonist of a specific biological activity of NF-AT are further
described herein.
Other preferred nucleic acids of the invention encode an NF-ATc polypeptide or
portion
thereof, e.g., a portion corresponding to a certain domain or having a
specific biological activity
and polypeptides which are at least about 60%, 70%, 75%, 80%, 85%, 90%, 95%,
more preferably
at least about 98% and most preferably at least about 99% identical or similar
to a portion of
3 5 human NF-ATc set forth in SEQ ID NO: 38, or other exemplary NF-AT
sequences. For example,
a preferred nucleic acid of the invention encodes an NF-AT polypeptide which
is capable of
27
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
modulating translocation across the nuclear membrane. The NF-AT polypeptide
can include an
NLS or a region of NF-ATc interacting therewith, such as a domain selected
from the group
consisting of SRR, SP1, SP2, and SP3. Furthermore, the polynucleotides of the
invention can
encode wild-type of mutated forms of NF-ATc polypeptides, such as those
described in the
Examples. For example, preferred polynucleotides encode an NF-ATc polypeptide
having one
or more serine that has been substituted with another amino acid, e.g., an
alanine, to thereby
prevent its phosphorylation. Preferred polypeptides encoded by the nucleic
acids of the invention
are further described herein, e.g., in the section pertaining to NF-AT
polypeptides.
Polynucleotides encoding full-length NF-AT~ or fragments or analogs thereof,
may include
sequences that facilitate transcription (expression sequences) and translation
of the coding
sequences, such that the encoded polypeptide product is produced. Construction
of such
polynucleotides is well known in the art and is described further in Maniatis
et al., of 1
Cloning: A Laboratory Manual, 2nd Ed. (1989), Cold Spring Harbor, N.Y. For
example, but not
for limitation, such polynucleotides can include a promoter, a transcription
termination site
(polyadenylation site in eukaryotic expression hosts), a ribosome binding
site, and, optionally, an
enhancer for use in eukaryotic expression hosts, and, optionally, sequences
necessary for
replication of a vector. A typical eukaryotic expression cassette will include
a polynucleotide
sequence encoding a NF-AT~ polypeptide linked downstream (i.e., in
translational reading frame
orientation; polynucleotide linkage) of a promoter such as the HSV tk promoter
or the pgk
2 0 (phosphoglycerate kinase) promoter, optionally linked to an enhancer and a
downstream
polyadenylation site (e.g., an SV40 large T Ag poly A addition site).
A preferred NF-AT~ polynucleotide encodes a NF-AT~ polypeptide that comprises
at least
one of the following amino acids sequences:
-NAIFLTVSREHERVGC- (SEQ ID NO: 25);
2 5 -LHGYLENEPLMLQLFIGT- (SEQ ID NO: 26);
-PSTSPRASVTEESWLG- (SEQ ID NO: 27);
-GPAPRAGGTMKSAEEEHYG- (SEQ ID NO: 28);
-ASAGGHPIVQ- (SEQ ID NO: 29);
-NTRVRLVFRV- (SEQ ID NO: 30);
30 -AKTDRDLCKPNSLVVEIPPFRN- (SEQ ID NO: 31);
-EVQPKSHHRAHYETEGSR- (SEQ ID NO: 32);
-SPRVSVTDDSWLGNT- (SEQ ID NO: 33);
-SHHRAHYETEGSRGAV- (SEQ ID NO: 34);
-LRNSDIELRKGETDIGR- (SEQ ID NO: 35); and
35 -TLSLQVASNPIEC- (SEQ ID NO: 36}.
28
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
The degeneracy of the genetic code gives a finite set of polynucleotide
sequences encoding these
amino acid sequences; this set of degenerate sequences may be readily
generated by hand or by
computer using commercially available software (Wisconsin Genetics Software
Package Relaes
7.0). Thus, isolated polynucleotides typically less than approximately 10,000
nucleotides in length
and comprising sequences encoding each of the following amino acid sequences:
-NAIFLTVSREHERVGC- (SEQ ID NO: 25);
-LHGYLENEPLMLQLFIGT- (SEQ ID NO: 26);
-PSTSPRASVTEESWLG- (SEQ ID NO: 27);
-GPAPRAGGTMKSAEEEHYG- (SEQ ID NO: 28);
-ASAGGHPIVQ- (SEQ ID NO: 29);
-NTRVRLVFRV- (SEQ ID NO: 30);
-AKTDRDLCKPNSLVVEIPPFRN- (SEQ ID NO: 31);
-EVQPKSHHRAHYETEGSR- (SEQ ID NO: 32);
-SPRVSVTDDSWLGNT- (SEQ ID NO: 33);
-SHHRAHYETEGSRGAV- (SEQ ID NO: 34);
-LRNSDIELRKGETDIGR- (SEQ ID NO: 35); and
-TLSLQVASNPIEC- (SEQ ID NO: 36).
are provided and may be used for, among other uses, the expression of a NF-AT~
polypeptide
which can be used as an immunogen, immunological reagent, and the like. Such
polynucleotides
2 0 typically comprise an operably linked promoter for driving expression in a
suitable prokaryotic
or eukaryotic host cell. One exemplification of such a polynucleotide is the
human NF-AT~ cDNA
sequence of Fig. 1 cloned in operable linkage to the mammalian expression
vector pSRa, many
alternative embodiments will be apparent to those of skill in the art,
including the use of alternative
expression vectors (e.g., pBCI2BI and p91023(B); Hanahan J (1983) J. Mol.
Biol. ~f6 : 577;
Cullen et al. (1985) . Vir ~: 515; Lomedico PT (1982) Proc Natl Acad Sci ( S A
1 ~:
5798; Morinaga et al. (1984) Bio/Technolo~v ~: 636).
Additionally, where expression of a polypeptide is not desired,
polynucleotides of this
invention need not encode a functional protein. Polynucleotides of this
invention may serve as
hybridization probes and/or PCR primers (amplimers) and/or LCR oligomers for
detecting NF-AT~
3 0 RNA or DNA sequences.
Alternatively, polynucleotides of this invention may serve as hybridization
probes or
primers for detecting RNA or DNA sequences of related genes, such genes may
encode
structurally or evolutionarily related proteins. For such hybridization and
PCR applications, the
polynucleotides of the invention need not encode a functional polypeptide.
Thus, polynucleotides
3 5 of the invention may contain substantial deletions, additions, nucleotide
substitutions and/or
29
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
transpositions, so long as specific hybridization or specific amplification to
the NF-AT~ sequence
is retained.
Specific hybridization is defined hereinbefore, and can be roughly summarized
as the
formation of hybrids between a polynucleotide of the invention (which may
include substitutions,
deletions, and/or additions) and a specific target polynucleotide such as
human NF-AT~ mRNA
so that a single band is identified corresponding to each NF-AT~ isoform on a
Northern blot of
RNA prepared from T cells (i.e., hybridization and washing conditions can be
established that
permit detection of discrete NF-AT~ mRNA band(s)). Thus, those of ordinary
skill in the art can
prepare polynucleotides of the invention, which may include substantial
additions, deletions,
substitutions, or transpositions of nucleotide sequence as compared to
sequences shown in Fig. 1,
or other exemplary NF-AT sequences, and determine whether specific
hybridization is a property
of the polynucleotide by performing a Northern blot using RNA prepared from a
T lymphocyte
cell line which expresses NF-AT~ mRNA and/or by hybridization to a NF-AT~ DNA
clone (cDNA
or genomic clone).
Specific amplification is defined as the ability of a set of PCR amplimers,
when used
together in a PCR reaction with a NF-AT~ polynucleotide, to produce
substantially a single major
amplification product which corresponds to a NF-AT~ gene sequence or mRNA
sequence.
Generally, human genomic DNA or mRNA from NF-AT~ expressing human cells (e.g.,
Jurkat cell
line) is used as the template DNA sample for the PCR reaction. PCR amplimers
that exhibit
2 0 specific amplification are suitable for quantitative determination ofNF-
AT~ mRNA by quantitative
PCR amplification. NF-AT~ allele-specific amplification products, although
having sequence
and/or length polymorphisms, are considered to constitute a single
amplification product for
purposes of this definition.
Generally, hybridization probes comprise approximately at least 25 consecutive
nucleotides
2 5 of a sequence shown in Fig. 1 (for human and marine NF-AT~ detection,
respectively), preferably
the hybridization probes contain at least 50 consecutive nucleotides of a
sequence shown in Fig.
l, and more preferably comprise at least 100 consecutive nucleotides of a
sequence shown in Fig.
1. PCR amplimers typically comprise approximately 25 to 50 consecutive
nucleotides of a
sequence shown in Fig. 1, and usually consist essentially of approximately 25
to 50 consecutive
3 0 nucleotides of a sequence shown in Fig. 1 with additional nucleotides, if
present, generally being
at the 5' end so as not to interfere with polymerase-mediated chain extension.
PCR amplimer
design and hybridization probe selection are well within the scope of
discretion of practioners of
ordinary skill in the art.
3 5 B. Antisense Polvnucleotides
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Additional embodiments directed to modulation of NF-AT activity include
methods that
employ specific antisense polynucleotides complementary to all or part of the
sequences shown
in Fig. 1, or other exemplary NF-AT sequences, as well as ribozymes and
molecules forming
triplex structures. Such complementary antisense polynucleotides may include
nucleotide
substitutions, additions, deletions, or transpositions, so long as specific
hybridization to the
relevant target sequence of the NF-AT gene is retained as a functional
property of the
polynucleotide. Complementary antisense polynucleotides include soluble
antisense RNA or
DNA oligonucleotides which can hybridize specifically to NF-AT~ mRNA species
and prevent
transcription of the mRNA species andlor translation of the encoded
polypeptide (Ching et al.
(1989) Proc. Natl. Acad Sci U A $6: 10006; Broder et al. (1990) Ann. Int. Med.
l~: 604;
Loreau et al. (1990) FEBS Letters .~74: 53; Holcenberg et al., W091/11535;
U.S.S.N. 07/530,165;
W091/09865; W091/04753; W090/13641; and EP 386563, each ofwhich is
incorporated herein
by reference}. The antisense polynucleotides therefore inhibit production of
NF-AT~ polypeptides.
Since NF-AT~ protein expression is associated with T lymphocyte activation,
antisense
polynucleotides that prevent transcription and/or translation of mRNA
corresponding to NF-AT~
polypeptides may inhibit T cell activation and/or reverse the the activated
phenotype of T cells.
Compositions containing a therapeutically effective dosage of NF-AT~ antisense
polynucleotides
may be administered for treatment of immune diseases, including lymphocytic
leukemias, and for
inhibition of transplant rejection reactions, if desired. Likewise, NF-AT
Antisense can be used to
2 0 inhibit NF-AT-mediated growth of cardiac and/or vascular tissues, e.g., as
part of a treatment for
cardiac hypertrophy. Antisense polynucleotides of various lengths may be
produced, although
such antisense polynucleotides typically comprise a sequence of about at least
25 consecutive
nucleotides which are substantially identical to a naturally-occurnng NF-AT~
polynucleotide
sequence, and typically which are identical to a sequence shown in Fig. l, or
other exemplary NF
2 5 AT sequences.
Antisense polynucleotides may be produced from a heterologous expression
cassette in a
transfectant cell or transgenic cell, such as a transgenic pluripotent
hematopoietic stem cell used
to reconstitute all or part of the hematopoietic stem cell population of an
individual. Alternatively,
the antisense polynucleotides may comprise soluble oligonucleotides that are
administered to the
3 0 external milieu, either in the culture medium in vitro or in the
circulatory system or interstitial
fluid in vivo. Soluble antisense polynucleotides present in the external
milieu have been shown
to gain access to the cytoplasm and inhibit translation of specific mRNA
species. In some
embodiments the antisense polynucleotides comprise methylphosphonate moieties.
For general
methods relating to antisense polynucleotides, see Antisense RNA and DNA, (
1988), D.A. Melton,
3 5 Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).
31
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
C. Isolation of the Cognate uman NF-AT4 Gene
The human homdlog of the NF-AT~ cDNA is identified and isolated by screening a
human
genomic clone library, such as a human genomic library in yeast artificial
chromosomes, cosmids,
or bacteriophage ~. (e.g., ~, Charon 35), with a polynucleotide probe
comprising a sequence of
about at least 24 contiguous nucleotides (or their complement) of the cDNA
sequence shown in
Fig. 1, or other exemplary NF-AT sequences. Typically, hybridization and
washing conditions
are performed at high stringency according to conventional hybridization
procedures. Positive
clones are isolated and sequenced. For illustration and not for limitation, a
full-length
polynucleotide corresponding to the sequence of Fig. 1 may be labeled and used
as a hybridization
probe to isolate genomic clones from a human or murine genomic clone libary in
~.EMBL4 or
.1GEM11 (Promega Corporation, Madison, Wisconsin); typical hybridization
conditions for
screening plaque lifts (Benton and Davis (1978) c' nc ,~9 : i80) can be: SO%
formamide, 5 x
SSC or SSPE, 1-5 x Denhardt's solution, 0.1-1% SDS, 100-200 pg sheared
heterologous DNA or
tRNA, 0-10% dextran sulfate, 1 x 105 to 1 x 10' cpm/ml of denatured probe with
a specific activity
of about 1 x 10g cpm/pg, and incubation at 42°C for about 6-36 hours.
Prehybridization
conditions are essentially identical except that probe is not included and
incubation time is
typically reduced. Washing conditions are typically 1-3 x SSC, 0.1-1% SDS, 50-
70°C with
change of wash solution at about 5-30 minutes.
Nonhuman NF-AT~ cDNAs and genomic clones (i.e., cognate nonhuman NF-AT~ genes)
2 0 can be analogously isolated fram various nonhuman cDNA and genomic clone
libraries available
in the art (e.g., Clontech, Palo Alto, CA) by using probes based on the
sequences shown in Fig.
1, with hybridization and washing conditions typically being Less stringent
than for isolation of
human NF-AT~ clones.
Polynucleotides comprising sequences of approximately at least 30-50
nucleotides,
2 5 preferably at least 100 nucleotides, corresponding to or complementary to
an NF-AT mRNA can
serve as PCR primers and/or hybridization probes for identifying and isolating
germline NF-AT
genes. These germline genes may be human or may be from a related mammalian
species,
preferably rodents or primates. Such germline genes may be isolated by various
methods
conventional in the art, including, but not limited to, by hybridization
screening of genomic
3 0 libraries in bacteriophage ~, or cosmid libraries, or by PCR amplification
of genomic sequences
using primers derived from the sequences shown in Fig. 1. Human genomic
libraries are publicly
available or may be constructed de novo from human DNA.
Genomic clones of NF-AT~, particularly of the murine cognate NF-ATc gene, may
be used
to construct homologous targeting constructs for generating cells and
transgenic nonhuman
3 5 animals having at least one functionally disrupted NF-AT~ allele,
preferably homozygous for
knocked out NF-AT~ alleles. Guidance for construction of homologous targeting
constructs may
32
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
be found in the art, including: Rahemtulla et al. (1991) Nature X35 : 180;
Jasin et al. (1990) Genes
evel 4: 157; Koh et al. (1992) cie ~5 : 1210; Molina et al. (1992) Nature ~:
161; Grusby
et al. (1991) i nce ?~3: 1417; Bradley et al. (1992) Bio/Technoloev ~Q: 534,
incorporated herein
by reference). Homologous targeting can be used to generate so-called
"knockout" mice, which
are heterozygous or homozygous for an inactivated NF-AT~ allele. Such mice may
be sold
commercially as research animals for investigation of immune system
development, neoplasia, T
cell activation, signal transduction, drug sreening, and other uses.
Chimeric targeted mice are derived according to Hogan, et al., Manipulating
the Mouse
Embrvo~ A LaboratorYManual, Cold Spring Harbor Laboratory (1988) and
Teratocarcinomas a_nd
Embryonic Stem Cells A Practical Approach, E.J. Robertson, ed., IRL Press,
Washington, D.C.,
(1987) which are incorporated herein by reference. Embryonic stem cells are
manipulated
according to published procedures (Teratocarcinomas and Embryonic Stem Cells A
Practical
A r ac , E.J. Robertson, ed., IRL Press, Washington, D.C. (1987); Zjilstra et
al. (1989) Nature
X42:435; and Schwartzberg et al. (1989) ' n a 24~f: 799, each of which is
incorporated herein
by reference).
Additionally, a NF-AT~ cDNA or genomic gene copy may be used to construct
transgenes
for expressing NF-AT~ polypeptides at high levels and/or under the
transcriptional control of
transcription control sequences which do not naturally occur adjacent to the
NF-AT~ gene. For
example but not limitation, a constitutive promoter (e.g., a HSV-tk or pgk
promoter) or a cell-
2 0 lineage specific transcriptional regulatory sequence (e.g., a CD4 or CD8
gene promoter/enhancer)
may be operably linked to a NF-ATE encoding polynucleotide sequence to form a
transgene
(typically in combination with a selectable marker such as a neo gene
expression cassette). Such
transgenes can be introduced into cells (e.g., ES cells, hematopoietic stem
cells) and transgenic
cells and transgenic nonhuman animals may be obtained according to
conventional methods.
2 5 Transgenic cells and/or transgenic nonhuman animals may be used to screen
for antineoplastic
agents and/or to screen for potential immunomodulatory agents, as
overexpression of NF-AT~ or
inappropriate expression of NF-AT~ may result in a hyperimmune state or
enhance graft rejection
reactions.
3 0 D. Expression of NF-ATt Polvmucleotides
The nucleic acid sequences of the present invention capable of ultimately
expressing the
desired NF-AT~ polypeptides can be formed from a variety of different
polynucleotides (genomic
or cDNA, RNA, synthetic oligonucleotides, etc.) as well as by a variety of
different techniques.
As stated previously, the DNA sequences will be expressed in hosts after the
sequences
3 5 have been operably linked to (i.e., positioned to ensure the functioning
of) an expression control
sequence. These expression vectors are typically replicable in the host
organisms either as
33
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
episomes or as an integral part of the host chromosomal DNA. Commonly,
expression vectors
will contain selection markers, g:g_, tetracycline resistance or hygromycin
resistance, to permit
detection and/or selection of those cells transformed with the desired DNA
sequences (she, g.~,
U.S. Patent 4,704,362, which is incorporated herein by reference).
~. coli is one prokaryotic host useful particularly for cloning the DNA
sequences of the
present invention. Other microbial hosts suitable for use include bacilli,
such as cil a i ' ,
and other Enterobacteriaceae, such as Salmonella, ~erratia, and various
Pseudomonas species. In
these prokaryotic hosts, one can also make expression vectors, which will
typically contain
expression control sequences compatible with the host cell (g~g~, an origin of
replication). In
addition, any number of a variety of well-known promoters will be present,
such as the lactose
promoter system, a tryptophan (trp) promoter system, a beta-lactamase promoter
system, or a
promoter system from phage lambda. The promoters will typically control
expression, optionally
with an operator sequence, and have ribosome binding site sequences and the
like, for initiating
and completing transcription and translation.
Other microbes, such as yeast, may also be used for expression. Saccharomyces
is a
preferred host, with suitable vectors having expression control sequences,
such as promoters,
including 3-phosphoglycerate kinase or other glycolytic enzymes, and an origin
of replication,
termination sequences and the like as desired.
In addition to microorganisms, mammalian tissue cell culture may also be used
to express
2 0 and produce the polypeptides of the present invention (sue, Winnacker,
"From Genes to Clones,"
VCH Publishers, N.Y., N.Y. (1987), which is incorporated herein by reference).
Eukaryotic cells
are actually preferred, because a number of suitable host cell lines capable
of secreting intact
human proteins have been developed in the art, and include the CHO cell lines,
various COS cell
lines, HeLa cells, myeloma cell lines, Jurkat cells, etc. Expression vectors
for these cells can
2 5 include expression control sequences, such as an origin of replication, a
promoter, an enhancer
(Queen et al. (1986} Immunol. Rev. $~: 49, which is incorporated herein by
reference), and
necessary processing information sites, such as ribosome binding sites, RNA
splice sites,
polyadenylation sites, and transcriptional terminator sequences. Preferred
expression control
sequences are promoters derived from immunoglobulin genes, SV40, adenovirus,
bovine
3 o papillomavirus, and the like. The vectors containing the DNA segments of
interest (g;~,
polypeptides encoding a NF-AT~ polypeptide) can be transferred into the host
cell by well-known
methods, which vary depending on the type of cellular host. For example, CaCI
transfection is
commonly utilized for prokaryotic cells, whereas CaP04 treatment or
electroporation may be used
for other cellular hosts. (fee, a r I , Maniatis, et al., Molecular Cloning: A
Laboratory
3 5 ual, Cold Spring Harbor Press, (1982), which is incorporated herein by
reference). Usually,
vectors are episomes and are maintained extrachromosomally.
34
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Expression of recombinant NF-ATc protein in cells, particularly cells of the
lymphopoietic
lineage, may be used to identify and isolate genes that are transcriptionally
modulated, either
positively or negatively, by the presence of NF-AT~ protein. Such genes are
typically initially
identified as cDNA clones isolated from subtractive cDNA libraries, wherein
RNA isolated from
cells expressing recombinant NF-ATc and RNA isolated from control cells (i.e.,
not expressing
recombinant NF-ATc) are used to generate the subtractive libraries and
screening probes. In such
a manner, NF-ATE dependent genes may be isolated. NF-AT-dependent genes (or
their regulatory
sequences operably linked to a reporter gene) may be used as a component of an
in vitro
transcription assay employing a NF-AT~ polypeptide as a necessary component
for efficient
transcription; such transcription assays may be used to screen for agents
which inhibit NF-AT~-
dependent gene transcription and are thereby identified as candidate
immunomodulatory agents.
E. ~ Po ypeptides
The nucleotide and amino acid sequences shown in Fig. 1, and those of other
exemplary
NF-AT genes, enable those of skill in the art to produce polypeptides
corresponding to all or part
of the full-length human NF-AT~ polypeptide sequence. Such polypeptides may be
produced in
prokaryotic or eukaryotic host cells by expression of polynucleotides encoding
NF-ATE, or
fragments and analogs thereof. Alternatively, such polypeptides may be
synthesized by chemical
methods or produced by in vitro translation systems using a polynucleotide
template to direct
2 0 translation. Methods for expression of heterologous proteins in
recombinant hosts, chemical
synthesis of polypeptides, and in vitro translation are well known in the art
and are described
further in Maniatis et al., Molecular Cloninw A Laboratory Manual (1989), 2nd
Ed., Cold Spring
Harbor, N.Y. and Berger and Kimmel, Methods in En~ologyLVolume 152 uide to
Molecular
Cloning Techniques (1987), Academic Press, Inc., San Diego, CA.
2 5 Fragments or analogs of NF-AT~ may be prepared by those of skill in the
art. Preferred
amino- and carboxy-termini of fragments or analogs of NF-AT~ occur near
boundaries of
functional domains. For example, but not for limitation, such functional
domains include: (1)
domains conferring the property of binding to other NF-AT components (e.g., AP-
1 ), (2) domains
confernng the property of nuclear localization, and (3) domains conferring the
property of
3 0 enhancing activation of T cells when expressed at sufficient levels in
such cells. Additionally,
such functional domains might include: ( 1 ) domains conferring the property
of binding to RNA
polymerase species, (2} domains having the capacity to directly alter local
chromatin structure,
which may comprise catalytic activities (e.g., topoisomerases, endonucleases)
and/or which may
comprise structural features (e.g., zinc fingers, histone-binding moieties),
and (3) domains which
3 5 may interact with accessory proteins and/or transcription factors.
CA 02352599 2001-05-23
WO 00/30671 PCT/US99127862
One method by which structural and functional domains may be identified is by
comparison of the nucleotide and/or amino acid sequence data shown in Fig. 1,
or other exemplary
NF-AT genes, to public or proprietary sequence databases. Preferably,
computerized comparison
methods are used to identify sequence motifs or predicted protein conformation
domains that occur
in other proteins of known structure and/or function, such as the zinc
fingers. For example, the
NAD-binding domains of dehydrogenases, particularly lactate dehydrogenase and
malate
dehydrogenase, are similar in conformation and have amino acid sequences that
are detectably
homologous (Proteins Structures and Molecular Principles, (1984) Creighton
(ed.), W.H. Freeman
and Company, New York, which is incorporated herein by reference). Further, a
method to
identify protein sequences that fold into a known three-dimensional structure
are known (Bowie
et al. (1991) cience X53: 164). Thus, the foregoing examples demonstrate that
those of skill in
the art can recognize sequence motifs and structural conformations that may be
used to define
structural and functional domains in the NF-AT~ sequences of the invention. ne
example of a
domain is the rel similarity region from amino acid 418 to amino acid 710 of
the NF-AT~
polypeptide sequence of Fig. 1.
Additionally, computerized comparison of sequences shown in Fig. 1 to existing
sequence
databases can identify sequence motifs and structural conformations found in
other proteins or
coding sequences that indicate similar domains of the NF-AT~ protein. For
example but not for
limitation, the programs GAP, BESTFIT, FASTA, and TFASTA in the Wisconsin
Genetics
2 0 Software Package (Genetics Computer Group, 575 Science Dr., Madison, WI)
can be used to
identify sequences in databases, such as GenBank/EMBL, that have regions of
homology with a
NF-AT~ sequences. Such homologous regions are candidate structural or
functional domains.
Alternatively, other algorithms are provided for identifying such domains from
sequence data.
Further, neural network methods, whether implemented in hardware or software,
may be used to:
(1) identify related protein sequences and nucleotide sequences, and (2)
define structural or
functional domains in NF-AT~ polypeptides (Brunak et al. (1991) J. Mol. Biol.
~2( : 49, which is
incorporated herein by reference). For example, the 13-residue repeat motifs -
SPRASVTEESWLG- (SEQ ID NO: 23) and -SPRVSVTDDSWLG- (SEQ ID NO: 24) are
examples of structurally related domains.
3 0 Fragments or analogs comprising substantially one or more functional
domain may be
fused to heterologous polypeptide sequences, wherein the resultant fusion
protein exhibits the
functional property(ies) conferred by the NF-AT~ fragment. Alternatively, NF-
AT~ polypeptides
wherein one or more functional domain have been deleted will exhibit a loss of
the property
normally conferred by the missing fragment.
3 5 By way of example and not limitation, the domain conferring the property
of nuclear
localization and/or interaction with AP-1 may be fused to (3-galactosidase to
produce a fusion
36
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
protein that is localized to the nucleus and which can enzymatically convert a
chromogenic
substrate to a chromophore.
Although one class of preferred embodiments are fragments having amino- and/or
carboxy-
termini corresponding to amino acid positions near functional domains borders,
alternative NF-
AT~ fragments may be prepared. The choice of the amino- and carboxy-termini of
such fragments
rests with the discretion of the practitioner and will be made based on
experimental considerations
such as ease of construction, stability to proteolysis, thermal stability,
immunological reactivity,
amino- or carboxyl-terminal residue modification, or other considerations.
In addition to fragments, analogs of NF-AT~ can be made. Such analogs may
include one
or more deletions or additions of amino acid sequence, either at the amino- or
carboxy-termini, or
internally, or both; analogs may further include sequence transpositions.
Analogs may also
comprise amino acid substitutions, preferably conservative substitutions.
Additionally, analogs
may include heterologous sequences generally linked at the amino- or carboxy-
terminus, wherein
the heterologous sequences) confer a functional property to the resultant
analog which is not
indigenous to the native NF-AT~ protein. However, NF-AT~ analogs must comprise
a segment of
amino acids that has substantial similarity to a portion of the amino acid
sequence shown in
Fig. 1, respectively, and which has at least one of the requisite functional
properties enumerated
in the Definitions (supra). Preferred amino acid substitutions are those
which: (1) reduce
susceptibility to proteolysis, (2) reduce susceptibility to oxidation, (3)
alter post-translational
2 0 modification of the analog, possibly including phosphorylation, and (4)
confer or modify other
physicochemical or functional properties of such analogs, possibly including
interaction with
calcineurin or phophorylation or dephosphorylation thereby. NF-AT~ analogs
include various
muteins of a NF-AT~ sequence other than the naturally-occurring peptide
sequence. For example,
single or multiple amino acid substitutions (preferably conservative amino
acid substitutions) may
2 5 be made in the naturally-occurring NF-AT~ sequence (preferably in the
portion of the polypeptide
outside the functional domains).
Conservative amino acid substitution is a substitution of an amino acid by a
replacement
amino acid which has similar characteristics (e.g., those with acidic
properties: Asp and Glu). A
conservative (or synonymous) amino acid substitution should not substantially
change the
3 0 structural characteristics of the parent sequence (e.g., a replacement
amino acid should not tend
to break a helix that occurs in the parent sequence, or disrupt other types of
secondary structure
that characterizes the parent sequence). Examples of art-recognized
polypeptide secondary and
tertiary structures are described in Proteins Structures and Molecular
Princig~es, (1984) Creighton
(ed.), W.H. Freeman and Company, New York; I ~troduction to Protein tructure,
(1991), C.
3 5 Branden and J. Tooze, Garland Publishing, New York, NY; and Thornton et
al. (1991) Nature 354:
105; which are incorporated herein by reference).
37
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
' The invention further provides phosphorylated NF-AT polypeptides. Preferred
phosphorylated polypeptides comprise at least one phosphoserine, which can be
selected from the
group consisting of serines located in the region from about amino acid 172 to
about amino acid
301. Even more preferred NF-AT polypeptides comprise a phosphorylated serine
in SRR, SP1,
SP2, and/or SP3. Preferred serines in SRR include those at residues 172, 175,
176, 178, 179, 181,
184, 187,188, 192, and 194 of SEQ ID NO: 38. Preferred serines in SP1 include
those at residues
199, 203, 207, and 211 of SEQ ID NO: 38. Preferred serines in SP2 include
those at residues 233,
327, and 245 of SEQ ID NO: 38. Preferred serines in SP3 include those at
residues 278, 282, 286,
290, and 299 of SEQ ID NO: 38. Another preferred NF-AT polypeptide has a
phosphoserine at
1 o position 269.
The invention also provides peptides and peptidomimetics, e.g., for use in
modulating
nuclear translocation of an NF-AT protein. In a preferred embodiment, the
agent comprises a
portion of an NF-AT protein that is involved in translocation of an NF-AT
polypeptide across the
nuclear membrane of a cell, e.g., a portion of an NF-AT polypeptide that forms
an intramolecular
association. In an even more preferred embodiment, the portion of NF-AT
comprises a nuclear
localization signal or sequence (NLS), such as the amino acid sequence KRKK
(SEQ ID NO: 56)
or KRKR (SEQ ID NO: 65) (referred to herein as KRKK/R (SEQ ID NO: 66)),
corresponding to
amino acids 682 to 685 of NF-ATcI (SEQ ID NO: 38), or the amino acid sequence
GKRKK/R
(SEQ ID NO: 67), corresponding to amino acids 681-685 of SEQ ID NO: 38 or
homologous
2 0 sequences in other NF-ATc family members. In fact, this C-terminal NLS, is
also found in the
other NF-ATc family members (see, e.g., Hoey et al. (1995) Immunity 2:461): NF-
ATp (NF-
ATc2) C-terminal NLS has the sequence NGKRKRS (SEQ ID NO: 68) (see figure 4);
NF-ATc3
(NF-AT4) C-terminal NLS has the sequence NGKKKKS (SEQ ID NO: 69); and NF-ATc4
(NF-
AT3) C-terminal NLS has the sequence NGRRKRS (SEQ ID NO: 70) (see Hoey et al.,
supra).
2 5 Accordingly, the invention provides peptides or peptidomimetics comprising
these NLS. In
another embodiment, the invention provides peptides and peptidomimetics
comprising a nuclear
localization signal including the amino acid sequence KRK, corresponding to
amino acids 265-267
of NF-ATcI (SEQ ID NO: 38). Alternatively, the peptide comprises the amino
acid sequence
CNKRKYSLN (SEQ ID NO: 53), corresponding to amino acids 263-271 of SEQ ID NO:
38.
3 0 The presence in a cell expressing NF-AT of a peptide comprising an NF-AT
NLS will
competitively inhibit the interaction of the endogenous NF-AT NLS with the
SR.R, SPl, SP2
and/or SP3 regions of NF-AT, thereby uncovering the NLS and allowing NF-AT to
be translocated
to the nucleus. Thus, the NF-AT NLS peptides of the invention constitute
specific activators of
NF-AT. In view of the sequence similarities between the NF-ATc family members,
the
3 5 intramolecular interaction between NLSs and other domains of the NF-ATcl
molecule set forth
herein, are believed to occur in the other NF-ATc family members and to
regulate their
38
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
translocation across a nuclear membrane. The effect of these activators in a
cell can be reversed
by, e.g., introducing into the cell a peptide that is capable of interacting
with an NF-AT NLS. For
example, if a peptide comprising the NLS KRKIUR (SEQ ID NO: 66) is
administered to a subject,
the administration of a peptide capable of binding to this NLS will inhibit
the activator.
Accordingly, the invention also provides peptides and peptidomimetics that are
capable
of interacting with an NLS in an NF-ATc molecule. Such peptides can be used,
e.g., to stimulate
translocation of NF-AT from the nucleus to the cytoplasm, by interacting with,
and thereby
shielding, the NLS of NF-AT molecules. Preferred peptides include N-terminal
peptides, e.g.,
peptides located in the region from about amino acid 172-301 of SEQ ID NO: 38.
Even more
l0 preferred peptides comprise one or more sequence from the group consisting
of SRR, SP1, SP2,
and SP3. In one embodiment, such a peptide comprises an amino acid sequence of
an SRR
sequence, e.g., corresponding to about amino acids 172-194 of SEQ >D NO: 38.
Shorter peptides
can also be used so long as they are capable of interacting with the NLS.
Other peptides that can
be used for this purpose include peptides comprising one or more sequences
selected from the
group consisting of SP1, SP2 and SP3, e.g., peptides comprising about amino
acids 199-219 of
SEQ ID NO: 38 (corresponding to SP1), about amino acids 233-252 of SEQ ID NO:
38 (SP2),
and/or amino acids 278-301 of SEQ ID NO: 38 (SP3). Other peptides comprising
amino acid
sequence that are homologous (at least about 80%, 85%, 90%, 95%, or preferably
at least about
98% or 99% identical or similar) to the SRR, SP1, SP2, and SP3 sequences set
forth above are also
2 0 within the scope of the invention. In particular, peptides from NF-ATc2,
NF-ATc3 and NF-ATcS
that are capable of forming intramolecular interactions with an NLS are part
of the invention. SP1,
SP2 and SP3 sequences are homologous in all NF-ATc family members (see, e.g.,
Hoey et al.,
supra).
The NF-AT peptides comprising one or more sequences selected from the group of
SRR,
SPl, SP2, and SP3 preferably comprise phosphoserines. Each serine of the
peptide can be
phosphorylated. However, as indicated in the Examples, it is only necessary to
have some serines
phosphorylated (see Figure 12) so that the peptide will interact with an NLS.
Thus, preferred
peptides of the invention are NF-AT peptides having a number of phosphoserines
sufficient to
allow the peptide to interact with an NLS of NF-AT. A peptide of interest can
be phosphorylated
3 0 in vitro according to the method described in the Examples or according to
in vitro
phosphorylation assays known in the art.
The invention also provides NF-AT peptides comprising an NLS or one or more of
the
repeats SRR, SP1, SP2 and SP3 which are homologs, variants, derivatives or
peptidomimtics of
sequences set forth in SEQ ID NO: 38. Preferred homologs, variants,
derivatives, or
peptidomimetics are capable of interacting with a portion of an NF-AT
polypeptide. Peptides or
39
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
peptidomimetics can be screened for those that interact with a portion of NF-
AT using a binding
assay, e.g., the binding assay described in the Examples.
Also within the scope of the invention are NF-AT polypeptides which are
constitutively
active. Such NF-T polypeptides can be useful since they are not dependent on
the presence of
calcium to be activated, but rather they can be activated in a regulated
manner of choice, as further
described below. As shown in the examples, the mutation of certain amino acids
in NF-AT result
in constitutive nuclear localization and thus constitutive activity. Preferred
constitutively active
NF-AT polypeptides have at least one amino acid deletion, addition or
substitution (generally
referred to as "peptide modification") that interferes in the intramolecular
interactions in NF-AT.
Even more preferred NF-AT polypeptides have a peptide modification located in
one or more of
the SRR, SP1, SP2 or SP3 sequences of an NF-AT molecule, such that the ability
of an NF-AT
polypeptide to form an intramolecular association is decreased or inhibited.
The peptide
modification can be a substitution of one or more of the serines in one or
more of the repeats. For
example, a constitutively active NF-AT peptide can have a substitution of all
the serines located
in the SRR region (amino acid 172 to amino acid 194 of SEQ ID NO: 38).
Alternatively, a
constitutively active peptide can have a substitution of the serines at
positions 184, 187 and 188
of SEQ )D NO: 38; a substitution of the serines at positions 172, I75, and 176
of SEQ lD NO: 38;
a substitution of the serines at position 178, I 79, and 181 of SEQ ID NO: 38;
or a substitution of
the serines at position 184, 187, and 188 of SEQ ID NO: 38. Constitutively
active NF-AT
2 0 polypeptides can also be obtained by the substitution of one or more
serine in the SP1, SP2, and/or
SP3 domains. In particular, constitutive NF-AT peptides can comprise a
substitution of the four
serines in SPI (corresponding to amino acids 199-219 of SEQ ID NO: 38);
substitution of the
serines at position 233 and 237 of SP2 (corresponding to amino acids 233-252
of SEQ ID NO: 38);
or substitution of serines at 278, 282, 286, and 299 in SP3 (corresponding to
amino acids 278-301
2 5 of SEQ ID NO: 38). The one or more serines can be substituted with any
amino acid so long as
the substitution reduces intramolecular interactions, and is preferably an
amino acid which cannot
be phosphorylated, e.g., an alanine. Mutations that must be made in an NF-AT
polypeptide to
render it constitutively active can also be identified by screening a library
of peptides. For
example, one can produce a library of degenerate peptides, e.g., peptides
comprising amino acids
3 0 172-I88 of SEQ ID NO: 38, in which one or more serines or other amino acid
is randomly
mutated. This library can then be screened for those peptides which fail to
interact with an NF-AT
NLS, such as by passing the library of degenerate peptides over a column
containing an excess of
NF-AT NLS peptides. The amino acid sequence of the peptides in the flow-
through of the column
can then be determined. Alternatively, individual peptides in a library can be
tagged, and their
3 5 identity determined by detecting and identifying the tag.
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
' Constitutively active NF-AT polypeptides can nevertheless be regulated,
e.g., by having
the gene encoding the constitutively active NF-AT polypeptide under the
control of an inducible
promoter. Alternatively, the NF-AT polypeptide can be fused to another peptide
which can be
regulated. For example, a constitutively active NF-AT polypeptide can be fused
to a ligand
binding domain. Control of the activity of this NF-AT protein can be obtained
by further
expressing in the cell a fusion protein comprising a ligand binding domain and
a cytoplasmic
retention domain, such that in the presence of a dimerizer molecule permitting
cross-hybridization
between the two fusion proteins, the NF-AT fusion protein is retained in the
cytoplasm.
Translocation to the nucleus is then induced by eliminating the dimerizer.
Alternatively, a constitutively active NF-AT polypeptide can be obtained by
fusing the NE-
AT polypeptide to an additional NLS, in particular a heterologous NLS, e.g, a
viral NLS, such as
the SV40 large T antigen NLS. An NF-AT polypeptide can also be fused with two
or more NLS.
The one or more NLS can be fused to the N-terminus of the NF-AT polypeptides.
As shown in
Example 10, an NF-AT polypeptide that is covalently linked to one or two
copies of a
heterologous NLS results in constitutive nuclear localization.
An NLS of NF-AT can also be used to direct a protein, in particular a
heterologous protein
to the nucleus. As shown in Example 11, the addition of a peptide having the
amino acid sequence
from amino acid 263-271 of SEQ ID NO: 38 (N-terminal NLS) or a peptide having
the amino acid
sequence from amino acid 681 to 685 of SEQ ID NO: 38 (C-terminal NLS) to a
heterologous
2 0 polypeptide resulted in constitutive nuclear localization of the
polypeptide. Thus, the NLSs from
NF-AT are sufficient to direct a polypeptide to the nucleus. Accordingly, also
within the scope
of the invention are peptides comprising an NLS from NF-AT, in particular, a
peptide comprising
the amino acid sequence KRK, or preferably CNKRKYSLN (SEQ ID NO: 53) (amino
acids 263-
271 of SEQ ID NO: 38), or even more preferably SPCNKRKYSLNGR (SEQ ID NO: 71)
(amino
acids 261-273 of SEQ ID NO: 38) and/or the amino acid sequence KRKK/R (SEQ ID
NO: 66),
or preferably GKRKK/R (SEQ ID NO: 67) (amino acids 681-685 of SEQ ID NO: 38),
or even
more preferably CNGKRKK/RSQ (SEQ ID NO: 72) (amino acids 679-687 of SEQ ID NO:
38).
A dominant negative NF-AT, or constitutively inactive NF-AT, can be produced
by, e.g.,
3 0 mutating one, or preferably both NLS, such that the NF-AT polypeptide is
incapable of
translocating from the cytoplasm to the nucleus. An NF-AT polypeptide having
one or more
mutated NLS can act as a dominant negative mutant since, the polypeptide is
still capable of
interacting with calcineurin and thus, will compete for calcineurin, thereby
prohibiting calcineurin
to interact with the endogenous NF-AT molecules and activating them. The N-
terminal NLS can
3 5 be mutated, e.g, by substituting residues 265 to 268 (KRK) of SEQ ID NO:
38. For example, these
residues can be changed to QIL. The C-terminal NLS can be mutated, e.g, by
substituting residues
41
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
682-685 (KRKK/R (SEQ ID NO: 66)) of SEQ ID NO: 38. For example, these residues
can be
changed to TRTG (SEQ ID NO: SS). Other mutations are also within the scope of
the invention
an can be identified, e.g, by performing assays, as described in the examples.
Such assays can also
be used to screen libraries of mutated NLS sequences.
Particularly preferred variants are structural mimetics of a dominant negative
NF-AT~
mutants, such as a polypeptide consisting essentially of amino acids 1-418 of
Fig. 1 and
substantially lacking amino acids carboxy-terminal to residue 418. Such
mimetics of dominant-
negative mutant polypeptides can have substantial activity as antagonists or
partial agonists of NE-
AT activation (and hence T cell activation).
Still another aspect of the present invention relates to peptide and
peptidomimetic
inhibitors, derived from the NLS sequence, which inhibit glucan synthase
kinases, e.g., GSK-3.
Native NF-AT~ proteins, fragments thereof, or analogs thereof can be used as
reagents in
DNA binding assays and/or in vitro transcription assays for identifying agents
that interfere with
NF-AT function, said agents are thereby identified as candidate drugs which
may be used, for
example, to block T cell activation or treat T cell lymphocytic leukemias.
Typically, in vitro DNA
binding assays that measure binding of NF-AT to DNA employ double-stranded DNA
that
contains an array of one or more NF-AT recognition sites (as defined by
specific footprinting of
native NF-AT protein). The DNA is typically linked to a solid substrate by any
of various means
2 0 known to those of skill in the art; such linkage may be noncovalent (e.g.,
binding to a highly
charged surface such as Nylon 66) or may be by covalent bonding (e.g.,
typically by chemical
linkage involving a nitrogen position in a nucleotide base, such as
diazotization). NF-AT~
polypeptides are typically labeled by incorporation of a radiolabeled amino
acid. The labeled NF-
AT~ polypeptide, usually reconstituted with an NF-AT nuclear component (e.g.,
AP-1 activity) to
2 5 form an NF-AT complex, is contacted with the immobilized DNA under aqueous
conditions that
permit specific binding in control binding reactions with a binding affinity
of about 1 x 106 M~'
or greater (e.g., 10-250 mM NaCI or KCl and 5-100 m VI Tris HCI pH S-9,
usually pH 6-8),
generally including Zn''- and/or Mn+2 and/or Mg''- in the nanomolar to
micromolar range (1 nM
to 999 ~tM). Specificity of binding is typically established by adding
unlabeled competitor at
3 o various concentrations selected at the discretion of the practitioner.
Examples of unlabeled protein
competitors include, but are not limited to, the following: unlabeled NF-AT~
polypeptide, bovine
serum albumin, and nuclear protein extracts. Binding reactions wherein one or
more agents are
added are performed in parallel with a control binding reaction that does not
include an agent.
Agents which inhibit the specific binding of NF-AT~ polypeptides to DNA, as
compared to a
3 5 control reaction, are identified as candidate immunomodulatory drugs.
Also, agents which prevent
42
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
transcriptional modulation by NF-AT in vitro are thereby identified as
candidate
immunomodulatory drugs.
As set forth above, in addition to NF-AT~ polypeptides consisting only of
naturally
occuring amino acids, NF-ATE peptidomimetics are also provided. Peptide
analogs are commonly
used in the pharmaceutical industry as non-peptide drugs with properties
analogous to those of the
template peptide. These types of non-peptide compound are termed "peptide
mimetics" or
"peptidomimetics" (Fauchere, J. (1986) Adv Dru es ~S: 29; Veber and Freidinger
(1985)
TINS p.392; and Evans et al. (1987) J. Med. Chem ~Q: 1229, which are
incorporated herein by
reference) and are usually developed with the aid of computerized molecular
modeling. Peptide
mimetics that are structurally similar to therapeutically useful peptides may
be used to produce an
equivalent therapeutic or prophylactic effect. Generally, peptidomimetics are
structurally similar
to a paradigm polypeptide (i.e., a polypeptide that has a biological or
pharmacological activity),
such as human NF-AT~, but have one or more peptide linkages optionally
replaced by a linkage
selected from the group consisting of: -CHZNH-, -CHZS-, -CHz-CHZ-, -CH=CH-
(cis and trans),
-COCHZ , -CH(OH)CHz-, and -CHzSO-, by methods known in the art and further
described in the
following references: Spatola, A.F. in "Chemistry and Biochemistry of Amino
Acids, Peptides,
and Proteins," B. Weinstein, eds., Marcel Dekker, New York, p. 267 (1983);
Spatola, A.F., Vega
Data (March 1983), Vol. 1, Issue 3, "Peptide Backbone Modifications" (general
review); Morley,
J.S., Trends Pharm ci (1980) pp. 463-468 (general review); Hudson, D. et al.,
Int J Pept Prot Res
(1979) x:177-185 (-CHzNH-, CHZCHz-); Spatola, A.F. et al., 'f ci (1986) x:1243-
1249 (-
CHZ-S); Hann, M.M., J Chem Soc Perkin Trans I (1982) 307-314 (-CH-CH-, cis and
trans);
Almquist, R.G. et al., J Med Chem (1980) 23:1392-1398 (-COCHZ-); Jennings-
White, C. et al.,
Tetrahedron .ert (1982) 2:2533 (-COCHz ); Szelke, M. et al., European Appln.
EP 45665 (1982)
CA: X7:39405 (1982) (-CH(OH)CHZ-); Holladay, M.W. et al., T~rahedron Lit
(1983) 24:4401-
4404 (-C(OH)CH2-); and Hruby, V.J., Li ' (1982) 31:189-199 (-CHZ-S-); each of
which is
incorporated herein by reference. A particularly preferred non-peptide linkage
is -CHZNH-. Such
peptide mimetics may have significant advantages over polypeptide embodiments,
including, for
example: more economical production, greater chemical stability, enhanced
pharmacological
properties (half life, absorption, potency, efficacy, etc.), altered
specificity (e.g., a broad-spectrum
3 0 of biological activities), reduced antigenicity, and others. Labeling of
peptidomimetics usually
involves covalent attachment of one or more labels, directly or through a
spacer (e.g., an amide
group), to non-interfering positions) on the peptidomimetic that are predicted
by quantitative
structure-activity data and/or molecular modeling. Such non-interfering
positions generally are
positions that do not form direct contacts with the macromolecules(s) (e.g.,
immunoglobulin
3 5 superfamily molecules) to which the peptidomimetic binds to produce the
therapeutic effect.
Derivitization (e.g., labelling) of peptidomimetics should not substantially
interfere with the
43
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
desired biological or pharmacological activity of the peptidomimetic.
Peptidomimetics of NF-AT~
may be used as competitive or noncompetitive agonists or antagonists of NF-AT~
function. For
example, a NF-AT~ peptidomimetic administered to a stimulated T cell
containing NF-AT~ and
may compete with the naturally-occurnng NF-AT~ and reduce NF-AT activity.
Alternatively, an
NF-AT~ peptidomimetic administerd to a T cell lacking NF-AT~ may induce T cell
activation or
the like.
Systematic substitution of one or more amino acids of a consensus sequence
with a D-
amino acid of the same type (e.g., D-lysine in place of L-lysine) may be used
to generate more
stable peptides. In addition, constrained peptides (including cyclized
peptides) comprising a
1 o consensus sequence or a substantially identical consensus sequence
variation may be generated
by methods known in the art (Rizo and Gierasch (1992) Ann. Rev. Biochem ~1:
387, incorporated
herein by reference); for example, by adding internal cysteine residues
capable of forming
intramolecular disulfide bridges which cyclize the peptide.
The amino acid sequences of NF-AT~ polypeptides identified herein will enable
those of
skill in the art to produce polypeptides corresponding to NF-AT~ peptide
sequences and sequence
variants thereof. Such polypeptides may be produced in prokaryotic or
eukaryotic host cells by
expression of polynucleotides encoding a NF-AT~ peptide sequence, frequently
as part of a larger
polypeptide. Alternatively, such peptides may be synthesized by chemical
methods. Methods for
expression of heterologous proteins in recombinant hosts, chemical synthesis
of polypeptides, and
2 0 in vitro translation are well known in the art and are described further
in Maniatis et al., Molecular
Cloninu: A Laboratory Manual (1989), 2nd Ed., Cold Spring Harbor, N.Y.; Berger
and Kimmel,
Methods in Enzvmology Volume 152 ~ ide to Molecular lnnin~TechniaueS (1987),
Academic
Press, Inc., San Diego, CA; Merrifield, J. (1969) J Am Chem oc QI : 501;
Chaiken LM. (1981)
CRC Crit. Rev Biochem 11: 255; Kaiser et al.(1989) Science 43: 187; Mernfield,
B. (1986)
ci nce ~3?: 342; Kent, S.B.H. (1988) Ann. Rev. Biochem ~7,: 957; and Offord,
R.E. (1980)
~nisvnthetic Proteins, Wiley Publishing, which are incorporated herein by
reference).
F. Production and Applications of a NF AT Antibo ie~
Native NF-AT~ proteins, fragments thereof, or analogs thereof, may be used to
immunize
3 0 an animal for the production of specific antibodies. These antibodies may
comprise a polyclonal
antiserum or may comprise a monoclonal antibody produced by hybridoma cells.
For general
methods to prepare antibodies, see Antibodies' A Laboratory Manual, (1988) E.
Harlow and D.
Lane, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, which is
incorporated herein by
reference.
3 5 For example but not for limitation, a recombinantly produced fragment of
human NF-AT~
can be injected into a rat along with an adjuvant following immunization
protocols known to those
44
CA 02352599 2001-05-23
WO 00/306?1 PCT/US99/2?862
of skill in the art so as to generate an immune response. Typically,
approximately at least 1-SO pg
of a NF-AT~ fragment or analog is used for the initial immunization, depending
upon the length
of the polypeptide. Alternatively or in combination with a recombinantly
produced NF-AT~
polypeptide, a chemically synthesized peptide having a NF-AT~ sequence (e.g.,
peptides
exemplified in Table II, infra) may be used as an immunogen to raise
antibodies which bind a NF-
AT~ protein, such as the native human NF-AT~ polypeptide having the sequence
shown essentially
in Fig. 1 or the native human NF-AT~ polypeptide isoform. Immunoglobulins
which bind the
recombinant fragment with a binding affinity of at least 1 x 10' M-' can be
harvested from the
immunized animal as an antiserum, and may be further purified by
immunoaffinity
chromatography or other means. Additionally, spleen cells are harvested from
the immunized
animal (typically rat or mouse) and fused to myeloma cells to produce a bank
of antibody-secreting
hybridoma cells. The bank of hvbridomas can hr crrPanr ~ fn,. r.l.",o~ t~,.,~
,.........~_
immunoglobulins which bind the recombinantly produced NF-AT~ polypeptide (or
chemically
synthesized NF-AT~ polypeptide) with an affinity of at least 1 x 106 M~'.
Animals other than mice
and rats may be used to raise antibodies; for example, goats, rabbits, sheep,
and chickens may also
be employed to raise antibodies reactive with a NF-AT~ protein. Transgenic
mice having the
capacity to produce substantially human antibodies also may be immunized and
used for a source
of a-NF-AT~ antiserum and/or for making monoclonal-secreting hybridomas.
Bacteriophage antibody display libraries may also be screened for binding to a
NF-AT~
2 0 polypeptide, such as a full-length human NF-AT~ protein, a NF-AT~ fragment
(e.g., a peptide
having a sequence shown in Table II, infra), or a fusion protein comprising a
NF-AT~ polypeptide
sequence of at least 14 contiguous amino acids as shown in Fig. 1 or a
polypeptide sequence of
Table II (infra). Combinatorial libraries of antibodies have been generated in
bacteriophage
lambda expression systems which may be screened as bacteriophage plaques or as
colonies of
lysogens (Huse et al. (I989) i c 24~: 1275; Caton and Koprowski (1990) Proc.
Natl. Acad
Sci. U S A 1 ~7: 6450; Mullinax et al (1990) Proc. Natl. Acad Sci (U S A ) 87:
8095; Persson
et al. (1991) Proc. Natl A~ad ci (U S A ) 88: 2432). Various embodiments of
bacteriophage
antibody display libraries and lambda phage expression libraries have been
described (Kang et al.
(1991) Proc. Natl Acad Sci (U S A.) 88: 4363; Clackson et al. (1991) Nature
~?5 : 624;
3 o McCafferty et aI. (1990) ature 48: 552; Burton et al. (1991) Proc Natl
Acad Sci fU S A ) i38:
10134; Hoogenboom et al. (1991) Nucleic cids Red 1_~: 4133; Chang et al.
(199I) J. Immunol.
,~47: 3610; Breitling et al. (1991) en 04: 147; Marks et al. (1991) J. Mol.
Biol. ~: 581;
Barbas et al. (1992) Proc. Natl. Acad ci (U S A ) $9: 4457; Hawkins and Winter
(1992) ~
Immunol. ~2: 867; Marks et al. (1992) Biotechnology ~: 779; Marks et al.
(1992) J. Biol. Chem
3 5 267: 16007; Lowman et al ( 1991 ) Biochemis~y ~0: 10832; Lerner et al. (
1992) ' nce 58: 1313,
incorporated herein by reference). Typically, a bacteriophage antibody display
library is screened
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
with a NF-AT~ polypeptide that is immobilized (e.g., by covalent linkage to a
chromatography
resin to enrich for reactive phage by affinity chromatography) and/or labeled
(e.g., to screenplaque
or colony lifts).
NF-AT~ polypeptides which are useful as immunogens, for diagnostic detection
of a-NF
AT~ antibodies in a sample, for diagnosic detection and quantitation of NF-AT~
protein in a sample
(e.g., by standardized competitive ELISA), or for screening a bacteriophage
antibody display
library, are suitably obtained in substantially pure form, that is, typically
about SO percent (w/w)
or more purity, substantially free of interfering proteins and contaminants.
Preferably, these
polypeptides are isolated or synthesized in a purity of at least 80 percent
(w/w) and, more
preferably, in at least about 95 percent (w/w) purity, being substantially
free of other proteins of
humans, mice, or other contaminants. Preferred immunogens comprise at least
one NF-AT~
polypeptide sequence shown in Table II, either as a discrete peptide or as
part of a fusion
polypeptide (e.g., with a ~i-galactosidase or glutathione S-transferase
sequence). NF-AT~
immunogens comprise at least one, typically several of such immunogenic
epitopes.
For some applications of these antibodies, such as identifying
immunocrossreactive
proteins, the desired antiserum or monoclonal antibody(ies) is/are not
monospecific. In these
instances, it may be preferable to use a synthetic or recombinant fragment of
NF-AT~ as an antigen
rather than using the entire native protein. More specifically, where the
object is to identify
immunocrossreactive polypeptides that comprise a particular structural moiety,
such as a DNA-
2 0 binding domain, it is preferable to use as an antigen a fragment
corresponding to part or all of a
commensurate structural domain in the NF-AT~ protein. Production of
recombinant or synthetic
fragments having such defined amino- and carboxy-termini is provided by the NF-
AT~ sequences
shown in Fig. 1.
If an antiserum is raised to a NF-AT~ fusion polypeptide, such as a fusion
protein
2 5 comprising a NF-AT~ inununogenic epitope fused to (3-galactosidase or
glutathione S-transferase,
the antiserum is preferably preadsorbed with the non-NF-AT~ fusion partner
(e.g, ~i-galactosidase
or glutathione S-transferase) to deplete the antiserum of antibodies that
react (i.e., specifically bind
to) the non-NF-AT~ portion of the fusion protein that serves as the immunogen.
Monoclonal or
polyclonal antibodies which bind to the human and/or murine NF-AT~ protein can
be used to
3 0 detect the presence of human or murine NF-AT~ polypeptides in a sample,
such as a Western blot
of denatured protein (e.g., a nitrocellulose blot of an SDS-PAGE) obtained
from a lymphocyte
sample of a patient. Preferably quantitative detection is performed, such as
by denistometric
scanning and signal integration of a Western blot. The monoclonal or
polyclonal antibodies will
bind to the denatured NF-AT~ epitopes and may be identified visually or by
other optical means
3 5 with a labeled second antibody or labeled Stapyylococcus reus protein A by
methods known in
46
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
the art. Frequently, denatured NF-AT~ will be used as the target antigen so
that more epitopes may
be available for binding.
T I
Selected Human NF-ATE Anti en Pep s~Ps
-NAIFLTVSREHERVGC- (SEQ ID NO: 25);
-LHGYLENEPLMLQLFIGT- (SEQ ID NO: 26);
-PSTSPRASVTEESWLG- (SEQ ID NO: 27);
-GPAPRAGGTMKSAEEEHYG- (SEQ ID NO: 28);
l o -ASAGGHPIVQ- (SEQ ID NO: 29);
-NTRVRLVFRV- (SEQ ID NO: 30);
-AKTDRDLCKPNSLVVEIPPFRN- (SEQ ID NO: 31);
-EVQPKSHHRAHYETEGSR- (SEQ ID NO: 32);
-SPRVSVTDDSWLGNT- (SEQ ID NO: 33);
-SHHRAHYETEGSRGAV- (SEQ ID NO: 34);
-LRNSDIELRKGETDIGR- (SEQ ID NO: 35); and
-TLSLQVASNPIEC- (SEQ ID NO: 36).
Such NF-AT~ sequences as shown in Tables II may be used as an immunogenic
peptide directly (e.g., to screen bacteriophage antibody display libraries or
to immunize a rabbit),
or may be conjugated to a carrier macromolecule (e.g., BSA) or may compose
part of a fusion
protein to be used as an immunogen. A preferred NF-AT~ polypeptide comprises
the following
amino acids sequences:
-NAIFLTVSREHERVGC- (SEQ ID NO: 25);
-PSTSPRASVTEESWLG- (SEQ ID NO: 27);
-SPRVSVTDDSWLGNT- (SEQ ID NO: 33); and
-SHHRAHYETEGSRGAV- (SEQ ID NO: 34);
and may comprise other intervening and/or terminal sequences; generally such
polypeptides are
less than 1000 amino acids in length, more usually less than about 500 amino
acids in length; often
spacer peptide sequences or terminal peptide sequences, if present, correspond
to naturally
3 0 occurring polypeptide sequences, generally mammalian polypeptide
sequences. One application
ofthe preferred NF-AT~ polypeptide just recited is as a commercial immunogen
to raise a-NF-AT~
antibodies in a suitable animal and/or as a commercial immunodiagnostic
reagent for quantitative
ELISA (e.g., competitive ELISA) or competitive RIA in conjunction with the
anti-NF-AT~
antibodies provided by the invention, such as for calibration of
standardization of such
3 5 immunoassays for staging or diagnosis of NF-AT~-expressing lymphocytic
leukemias in humans
or cell typing or identification of T cells (such as activated T cells and/or
activatable T cells). The
47
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
preferred NF-AT~ polypeptide just recited will find many other uses in
addition to serving as an
immunogen or immunological reagent. One or more of the above-listed sequences
may be
incorporated into a fusion protein with a fusion partner such as human serum
albumin, GST, etc.
For such fusion proteins in excess of 1000 amino acids, deletions in the
fusion partner (albumin)
moiety may be made to bring the size to about 1000 amino acids or less, if
desired.
In some embodiments, it will be desirable to employ a polyvalent NF-AT~
antigen,
comprising at least two NF-AT~ immunogenic epitopes in covalent linkage,
usually in peptide
linkage. Such polyvalent NF-AT~ antigens typically comprise multiple NF-AT~
antigenic peptides
from the same species (e.g., human or mouse), but may comprise a mix of
antigenic peptides from
NF-AT~ proteins of different species (i.e., an interspecies NF-AT~ polyvalent
antigen). Frequently,
the spatial order of the antigenic peptide sequences in the primary amino acid
sequence of a
polyvalent antigen occurs in the same orientation as in the naturally
occurring NF-AT~ protein (i.e.,
a first antigenic peptide sequence that is amino-terminal to a second
antigenic peptide sequence
in a naturally occurring NF-AT~r protein will be amino-terminal to said second
antigenic peptide
sequence in a polyvalent antigen. Frequently, spacer peptide sequences will be
used to link
antigenic peptide sequences in a polyvalent antigen, such spacer peptide
sequences may be
predetermined, random, or psuedorandom sequences. Spacer peptide sequences may
correspond
to sequences known to be non-immunogenic to the animal which is to be
immunized with the
polyvalent antigen, such as a sequence to which the animal has been tolerized.
Although many
2 0 examples of such polyvalent antigens may be given, the following
embodiment is provided for
illustration and not limitation:
-NAIFLTVSREHERVGC-(aal) (SEQ ID NO: 25) -AKTDRDLCKPNSLVVEIPPFRN-(aa2)
(SEQ ID NO: 31)-GILKLRNSDIELRKGETD- (SEQ ID NO: 37)
where (aal) and (aa2) are peptide spacers of at least one amino acid and less
than 1000
2 5 amino acids; aal is a peptide sequence selected independently from the aa2
peptide sequence; the
length of aal (which may be composed of multiple different amino acids) is
independent of the
length of aa2 (which may be composed of multiple different amino acids).
Immunogenic NF-AT~ peptides may be used to immunize an animal to raise anti-NF-
AT~
antibodies and/or as a source of spleen cells for making a hybridoma library
from which to select
3 o hybridoma clones which secrete a monoclonal antibody which binds to a NF-
AT~ protein with an
affinity of 1 x 10' M-' or greater, preferably at least 1 x 10g M-' to 1 x 109
1VI' . Such immunogenic
NF-AT~ peptides can also be used to screen bacteriophage antibody display
libraries directly.
One use of such antibodies is to screen cDNA expression libraries, preferably
containing
cDNA derived from human or murine mRNA from various tissues, for identifying
clones
3 5 containing cDNA inserts which encode structurally-related,
immunocrossreactive proteins, that
are candidate novel transcription factors or chromatin proteins. Such
screening of cDNA
48
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
expression libraries is well known in the art, and is further described in
Young et al., Proc. N~1
Acad. Sci. U.S.A. $Q:1194-1198 (1983), which is incorporated herein by
reference] as well as
other published sources. Another use of such antibodies is to identify and/or
purify
immunocrossreactive proteins that are structurally or evolutionarily related
to the native NF-AT~
protein or to the corresponding NF-AT~ fragment (e.g., functional domain; DNA-
binding domain)
used to generate the antibody. It is believed that such antibodies will find
commercial use as such
reagents for research applications, just as other antibodies (and biological
reagents - such as
restriction enzymes and polymerases) are sold commercially.
Various other uses of such antibodies are to diagnose and/or stage leukemias
or other
1 o immunological disease states, and for therapeutic application (e.g., as
cationized antibodies or by
targeted liposomal delivery) to treat neoplasia, hyperimmune function, graft
rejection, and the like.
An example of an NF-ATc polypeptide is a polypeptide having the sequence:
MPSTSFPVPSKFPLGPAAAVFGRGETLGPAPRAGGTMKSAEEEHYGYASSNVSPALPLPTAHS
TLPAPCHNLQTSTPGIIPPADHPSGYGAALDGCPAGYFLSSGHTRPDGAPALESPRIEITSCL
GLYHNNNQFFHDVEVEDVLPSSKRSPSTATLSLPSLEAYRDPSCLSPASSLSSRSCNSEASSY
ESNYSYPYASPQTSPWQSPCVSPKTTDPEEGFPRGLGACTLLGSPQHSPSTSPRASVTEESWL
GARSSRPASPCNKRKYSLNGRQPPYSPHHSPTPSPHGSPRVSVTDDSWLGNTTQYTSSAIVAA
INALTTDSSLDLGDGVPVKSRKTTLEQPPSVALKVEPVGEDLGSPPPPADFAPEDYSSFQHIR
KGGFCDQYLAVPQHPYQWAKPKPLSPTSYMSPTLPALDWQLPSHSGPYELRIEVQPKSHHRAH
2 0 YETEGSRGAVKASAGGHPIVQLHGYLENEPLMLQLFIGTADDRLLRPHAFYQVHRITGKTVST
TSHEAILSNTKVLEIPLLPENSMRAVIDCACILKLRNSDIELRKGETDIGRKNTRVRLVFRVH
VPQPSGRTLSLQVASNPIECSQRSAQELPLVEKQSTDSYPVVGGKKMVLSGHNFLQDSKVIFV
EKAPDGHHVWEMEAKTDRDLCKPNSLWEIPPFRNQRITSPVHVSFYVCNGKRKRSQYQRFTY
LPANGNAIFLTVSREHERVGCFF (SEQ ID NO: 38).
2 5 Other preferred antigens for preparing antibodies include phosphorylated
NF-AT
polypeptides or domains thereof. For example, the invention provides
antibodies binding
specifically to phosphorylated NF-ATc polypeptides, and not to those that are
not phosphorylated.
The NF-ATc polypeptide can be phosphorylated on a serine, such as a serine
located in SRR, SPl,
SP2, SP3 or which is located between these repetitive sequences. Such
antibodies can be used to
3 0 hide or shield these phosphorylated domains from NLSs thereby favoring NF-
ATc localization in
the nucleus of a cell. Alternatively, such antibodies can be used to
specifically detect the
phosphorylated form, e.g., in diagnostics.
G. Identification and Isolation of Proteins That Bind NF-AT~
3 5 Proteins that bind to NF-AT~ and/or a NF-AT-DNA complex are potentially
important
transcriptional regulatory proteins. Such proteins may be targets for novel
immunomodulatory
49
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
agents. These proteins are referred to herein as accessory proteins. Accessory
proteins may be
isolated by various methods known in the art.
One preferred method of isolating accessory proteins is by contacting a NF-AT~
polypeptide to an antibody that binds the NF-AT~ polypeptide, and isolating
resultant immune
complexes. These immune complexes may contain accessory proteins bound to the
NF-AT~
polypeptide. The accessory proteins may be identified and isolated by
denaturing the immune
complexes with a denaturing agent and, preferably, a reducing agent. The
denatured, and
preferably reduced, proteins can be electrophoresed on a polyacrylamide gel.
Putative accessory
proteins can be identified on the polyacrylamide gel by one or more of various
well known
methods (e.g., Coomassie staining, Western blotting, silver staining, etc.),
and isolated by
resection of a portion of the polyacrylamide gel containing the relevant
identified polypeptide and
elution of the polypeptide from the gel portion.
A putative accessory protein may be identified as an accessory protein by
demonstration
that the protein binds to NF-AT~ and/or a NF-AT-DNA complex. Such binding may
be shown in
vitro by various means, including, but not limited to, binding assays
employing a putative
accessory protein that has been renatured subsequent to isolation by a
polyacrylamide gel
electrophoresis method. Alternatively, binding assays employing recombinant or
chemically
synthesized putative accessory protein may be used. For example, a putative
accessory protein
may be isolated and all or part of its amino acid sequence determined by
chemical sequencing,
2 0 such as Edman degradation. The amino acid sequence information may be used
to chemically
synthesize the putative accessory protein. The amino acid sequence may also be
used to produce
a recombinant putative accessory protein by: (1) isolating a cDNA clone
encoding the putative
accessory protein by screening a cDNA library with degenerate oligonucleotide
probes according
to the amino acid sequence data, (2) expressing the cDNA in a host cell, and
(3) isolating the
2 5 putative accessory protein. Alternatively, a polynucleotide encoding a NF-
AT~ polypeptide may
be constructed by oligonucleotide synthesis, placed in an expression vector,
and expressed in a
host cell.
Putative accessory proteins that bind NF-AT~ and/or NF-AT-DNA complex in vitro
are
identified as accessory proteins. Accessory proteins may also be identified by
crosslinking in vivo
3 0 with bifunctional crosslinking reagents (e.g., dimethylsuberimidate,
glutaraldehyde, etc.) and
subsequent isolation of crosslinked products that include a NF-ATc
polypeptide. For a general
discussion of cross-linking, see Kunkel et al. (1981) Mol. Cell. Biochem ~4:
3, which is
incorporated herein by reference. Preferably, the bifunctional crosslinking
reagent will produce
crosslinks which may be reversed under specific conditions after isolation of
the crosslinked
3 5 complex so as to facilitate isolation of the accessory protein from the NF-
AT~ polypeptide.
Isolation of crosslinked complexes that include a NF-AT~ polypeptide is
preferably accomplished
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/2786Z
bybinding an antibody that binds a NF-ATc polypeptide with an affinity of at
least 1 x 10' M'~ to
a population of crosslinked complexes and recovering only those complexes that
bind to the
antibody with an affinity of at least 1 x 10' M''. Polypeptides that are
crosslinked to a NF-AT~
polypeptide are identified as accessory proteins.
Screening assays can be developed for identifying candidate immunomodulatory
agents
as being agents which inhibit binding of NF-AT~ to an accessory protein (e.g.
AP-1) under suitable
binding conditions (see infra).
Yeast two-hybrid systems may be used to screen a mammalian (typically human)
cDNA
expression library, wherein cDNA is fused to a GAL4 DNA binding domain or
activator domain,
and a NF-AT~ polypeptide sequence is fused to a GAL4 activator domain or DNA
binding domain,
respectively. Such a yeast two-hybrid system can screen for cDNAs encoding
proteins which bind
to NF-AT~ sequences. For example, a cDNA library can be produced from mRNA
from a human
mature T cell line or other suitable cell type. Such a cDNA library cloned in
a yeast two-hybrid
expression system (Chien et al. (1991) Proc. Natl. Acad Sci j 1~ A 1 ~; 9578
or ~ ~: 233)
can be used to identify cDNAs which encode proteins that interact with NF-AT~
and thereby
produce expression of the GAL4-dependent reporter gene. Polypeptides which
interact with NF-
AT~can alos be identified by immunoprecipitation of NF-AT~ with antibody and
identification of
co-precipitating species. Further, polypeptides that bind NF-AT~ can be
identified by screening
a peptide library (e.g., a bacteriophage peptide display library, a spatially
defined VLSIPS peptide
2 0 array, and the like) with a NF-AT~ polypeptide.
H Exempla diagnostic and ~ro~nostic methods of the invention
The invention provides diagnostic and prognostic methods, including methods
for
determining the state of immunosuppression in a subject and for determining an
appropriate dose
2 5 of immunosuppressant.
In one preferred embodiment of the invention, hybridization probes that
specifically
identify the NF-AT~ gene may be used in methods for diagnosing genetic
disease. For example,
but not for limitation, the genetic disease thus diagnosed may involve a
lesion in the relevant NF-
AT~ structural or regulatory sequences, or may involve a lesion in a genetic
locus closely linked
3 o to the NF-AT~ locus and which can be identified by restriction fragment
length polymorphism or
DNA sequence polymorphism at the linked NF-AT~ locus. In a fi~rther preferred
embodiment, NF-
AT~ gene probes are used to diagnose or identify genetic disease involving
predisposition to
immunological disease, wherein the amount or functionality of endogenous NF-
AT~ is sufficient
for the individual to exhibit an increased probability of developing an immune
disease, particularly
3 5 an immune deficiency, arthritis, or autoimmune disease.
51
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
The invention also provides a method for determining the state of
invnunosuppression of
a subject. In one embodiment, the method comprises determining the level of
cytoplasmic and/or
nuclear NF-AT polypeptide in a cell, preferably a lymphocyte, of a subject.
The method can
comprise obtaining a blood sample from a subject, and determining the amount
of cytoplasmic
and/or nuclear NF-AT; such as by immunohistochemistry using an antibody that
binds specifically
to NF-AT, as further described herein. In a preferred embodiment, the method
comprises
incubating the blood cells of the subject with a T cell activating compound
and/or calcium
ionophore prior to determining the cellular localization of NF-AT. In fact, if
a patient is
immunosuppressed, stimulation of lymphocytes of the patient will not result in
significant
translocation of NF-AT into the nucleus. A preferred T cell activator is a
polyclonal activator,
including lectins, concanavalin-A (Con-A) and phytohemagglutinin (PHA). Other
activators
include antibodies binding to invariable framework epitopes on the T cell
receptor or CD3.
A preferred method for detecting NF-AT is by immunoflurorescence, using an
antibody
that binds specifically to one or more NF-ATc proteins. A preferred monoclonal
antibody is the
7A6 antibody, described in Northrop et al.(1994) Nature 369:497.
In a preferred embodiment, the test is carried out on an enriched population
of cells from
the subject, such as a blood sample enriched in mononuclear cells. Peripheral
blood mononuclear
cells can be obtained, e.g., by separating the cells from a blood sample on a
huffy coat, according
to methods known in the art.
2 0 The level of nuclear andlor cytoplasmic NF-AT in lymphocytes of a subject
can then be
compared to the Level of nuclear and/or cytoplasmic NF-AT in a control
individual. A "control"
or "normal" subject refers to a subject which has no known disease or
condition involving NF-AT
activation, e.g., inflammation, or autoimmune disease and which are not
subjected to any treatment
at the time the cell sample was obtained. Normal standards can be established
from analysis of
several cell samples from normal subjects. Samples from patients can then be
analyzed and
compared to this set of standards.
In a preferred embodiment, this diagnostic method can be used to monitor the
state of
immunosuppression in a subject who is receiving an immunosuppressive
treatment, e.g.,
cyclosporin A. In one embodiment, cells are obtained from a subject, and the
cellular localization
3 0 of NF-AT is determined prior to and/or after incubation of the cells with
a T cell activator, e.g.,
PHA. If the analysis reveals that the patient contains a high number of
lymphocytes having
nuclear NF-AT, i.e., activated lymphocytes, the patient should be given
additional
immunosuppressive drugs. Thus, a patient can be followed and an adequate
amount of
immunosuppressive drug can be administered to the patient, so that the patient
does not receive
3 5 excessive amounts of immunosuppressants, but received enough
immunosuppressant to maintain
him in an immunosuppressed state.
52
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
In another embodiment, the invention provides a method for determining the
sensitivity
of a subject to a particular immunosuppressive agent, e.g, cyclosporin A. In
one embodiment,
lymphocytes are obtained from a subject, the lymphocytes are incubated in
vitro in the presence
of various amounts of the immunosuppressive drug and for various times, and
the cellular location
of NF-AT is determined. The comparison of the results of this analysis with
those obtained from
the same analysis of lymphocytes from one or more normal subjects will
indicate whether the
subject is more, or alternatively, less sensitive, to the immunosuppressive
drug than an average
person. This analysis can also be performed in vivo. For example, a certain
dose of
immunosuppressive agent is administered to a subject and the cellular
localization of NF-AT in
lymphocytes of the subject is then determined at various time points and
compared to that of a
normal subject. Standards of normal subjects can also be obtained before hand
and the results of
the analysis can then be compared to these standards. Thus, based on the
results of such tests, a
physician can more appropriately predict the effective dose of
immunosuppressant to administer
to a subject, thereby avoiding administering an excessive of amount of
immunosuppressant that
could have toxic effects in the subject.
In other embodiments, the invention provides a method for determining the risk
of a patient
developing a disorder involving unwanted cardiovascular growth, or, if so
diagnosed, identifying
the etiology of the disorder. In the instance of the latter, understanding the
molecular basis of the
disease, including the role of an NF-AT protein in the disorder, can be useful
to determine the
2 0 proper course of therapy (i.e., would a particularly therapy be effective
or not), as well as
proscribing a course of treatment based on NF-AT being a prognostic marker for
reoccurence, etc.
Such embodiments of the subject method include detecting changes to an NF-AT
gene,
e.g., point mutations, deletions, additions, chromosomal rearrangements,
changes in methylation
patterns, etc., as well as changes in the level of expression of the protein,
the rate of turnover of
2 5 the protein (ubiquitin-dependent or independent), the rate of
phosphorylation/dephosphorylation,
and/or the cellular localization of the protein.
I. ~Ietho or rub des~n and screen~~ a savs
The invention further provides screening assays for identifying compounds
which modulate
3 0 the activity of NF-AT proteins. The screening assays can be in vivo or in
vitro and can be cell
based or based on a cell free fomat. These agents include, but are not limited
to, compounds that
either potentiate or inhibit an intrinsic activity of an NF-AT protein or a
complex including an NE
AT protein, compounds that interfere with the interaction of the NF-AT protein
with other
proteins) or nucleic acid, compounds which modify the rate of a certain post-
translational
3 5 modification of NF-AT proteins (e.g., by enzymes such as phosphatases or
kinases), antisense
constructs for inhibiting exrepssion of NF-AT proteins, nucleic acid decoys
for competitively
53
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
inhibiting binding of an NF-AT protein to a response element in genomic DNA,
and compounds
comprising forms of the NF-AT proteins that are altered (mutated) to provide
dominant loss-of
function or gain-of function activity. In one embodiment, the screening
methods of the present
invention are directed to the identif cation of agents capable of modulating,
and in particular of
inhibiting, the development of hypertrophy of a cell, such as a muscle cell or
which are capable
of reducing hypertrophy of a cell.
In preferred embodiments, the subject drug screening assays are carned out in
a manner
designed to detect selectivity for specific NF-AT paralogs. For instance, in
certain preferred
embodiments, the assays are set up to detect NF-AT antagonists which are
selective for NF-AT~3
and/or NF-ATE, but not NF-AT~1 or NF-AT~2. For instance, the NF-AT antagonist
can be
selected so as to have an ED50 for inhibition of NF-AT~3 or NF-AT~4 in vivo of
at least one, and
more preferably, two, three, four and even five orders of magnitude less than
its ED50 for
inhibition of NF-ATE 1 or NF-AT~2 activity.
Such selectivity can take advantage of differences amongst two or more NF-AT
paralogs
in specificity for DNA recognition elements, in the protein-protein
interactions, and/or post
translational modification of NF-AT proteins. Thus, any of the assays
described herein can be run,
e.g.,side-by-side, using two or more different NF-AT paralogs, and compounds
identified not only
on the basis of their ability to affect the biological activity of an NF-AT
protein, but also on their
ability to do so in a manner which is selective as between the NF-AT paralogs
used in the assay.
2 0 In this regard, the present invention provides assays for identifying
agents which are either
agonists or antagonists of the normal cellular function of the subject NF-AT
proteins, or of the role
of those proteins in the pathogenesis of normal or abnormal cell function,
such as muscle cell
hypertrophy and disorders related thereto. In one embodiment, the assay
evaluates the ability of
a compound to modulate binding of an NF-AT protein with other proteins, DNA or
RNA. In
2 5 other embodiments, the assay detects compounds which inhibit or potentiate
the post-translation
modification of an NF-AT protein, such as phosphorylation and/or changes in
folding of the
protein. Still other embodiments detect changes in the cellular localization
of an NF-AT protein.
Compounds identified by the present assay can be used, for example, in the
treatment of diseases
or conditions caused or contributed to by cell hypertrophy, in particular
muscle cell hypertrophy.
3 o For example, compounds of the invention can be use for treating congestive
heart disease.
Agents to be tested for their ability to act as agonists or antagonists of an
NF-AT protein
can be produced, for example, by bacteria, yeast or other organisms (e.g.
natural products),
produced chemically (e.g. small molecules, including peptidomimetics), or
produced
recombinantly. In a preferred embodiment, the test agent is a small organic
molecule having a
35 molecular weight of less than about 2,000 daltons. A high speed screen for
agents that bind
directly to the molecular regulator may employ immobilized or "tagged"
combinatorial libraries
54
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
(or libraries which otherwise readily deconvoluted).
Agents that are identified as active in the drug screening assay are
candidates to be tested
for their capacity to block development of hypertrophy of cells and/or their
capacity to reduce
hypertrophy in a cell. As described below, these agents would be useful for
treating or preventing
cardiac conditions including cardiac infarction and congestive heart disease.
A variety of assay formats will suffice and, in light of the present
disclosure, those not
expressly described herein will nevertheless be comprehended by one of
ordinary skill in the art.
For instance, the assay can be generated in many different formats, and
include assays based on
cell-free systems, e.g. purified proteins or cell lysates, as well as cell-
based assays which utilize
intact cells. Simple binding assays can also be used to detect agents which,
such as those which
detect compounds able to potentiate or disrupt protein-protein or protein-DNA
interaction
involving an NF-AT protein.
In many drug screening programs which test libraries of compounds and natural
extracts,
high throughput assays are desirable in order to maximize the number of
compounds surveyed in
a given period of time. Assays of the present invention which are performed in
cell-free systems,
such as may be derived with purified or semi-purified proteins or with
lysates, are often preferred
as "primary" screens in that they can be generated to permit rapid development
and relatively easy
detection of an alteration in a molecular target which is mediated by a test
compound. Moreover,
the effects of cellular toxicity and/or bioavailability of the test compound
can be generally ignored
2 0 in the in vitro system, the assay instead being focused primarily on the
effect of the drug on the
molecular target as may be manifest in an alteration of binding affinity with
other proteins or
changes in enzymatic properties of the molecular target.
Accordingly, in an exemplary screening assay of the present invention, a
reaction mixture
is generated to include an NF-AT protein, test compound(s), and a "target
molecule", e.g., a
2 5 protein or nucleic acid which interacts with the NF-AT protein. Detection
and quantification of
interaction of the NF-AT protein with the target molecule provides a means for
determining a
compound's efficacy at inhibiting or potentiating interaction between the NF-
AT protein and the
target molecule. The efficacy of the compound can be assessed by generating
dose response curves
from data obtained using various concentrations of the test compound.
Moreover, a control assay
3 0 can also be performed to provide a baseline for comparison. In the control
assay, interaction of the
NF-AT protein and target molecule is quantitated in the absence of the test
compound.
Interaction between an NF-AT protein and an NF-AT binding partner may be
detected by
a variety of techniques. Modulation of the formation of complexes can be
quantitated using, for
example, detectably labeled proteins such as radiolabeled, fluorescently
labeled, or enzymatically
3 5 labeled NF-AT proteins or NF-AT binding partners, by immunoassay, or by
chromatographic
detection.
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Typically, it will be desirable to immobilize either NF-AT or its binding
partner to
facilitate separation of complexes from uncomplexed forms of one or both of
the proteins; as well
as to accommodate automation of the assay. Binding of NF-AT to an NF-AT
binding partner, can
be accomplished in any vessel suitable for containing the reactants. Examples
include microtitre
plates, test tubes, and micro-centrifuge tubes. In one embodiment, a fusion
protein can be
provided which adds a domain that allows the protein to be bound to a matrix.
For example,
glutathione-S-transferase/NF-AT (GST/NF-AT) fusion proteins can be adsorbed
onto glutathione
sepharose beads (Sigma Chemical, St. Louis, MO) or glutathione derivatized
microtitre plates,
which are then combined with the NF-AT binding partner, e.g. an 35S-labeled NF-
AT binding
partner, and the test compound, and the mixture incubated under conditions
conducive to complex
formation, e.g. at physiological conditions for salt and pH, though slightly
more stringent
conditions may be desired. Following incubation, the beads are washed to
remove any unbound
label, and the matrix immobilized and radiolabel determined directly (e.g.
beads placed in
_ scintilant), or in the supernatant after the complexes are subsequently
dissociated. Alternatively,
the complexes can be dissociated from the matrix, separated by SDS-PAGE, and
the level of NE
AT protein or NF-AT binding partner found in the bead fraction quantitated
from the gel using
standard electrophoretic techniques such as described in the appended
examples.
Other techniques for immobilizing proteins on matrices are also available for
use in the
subject assay. For instance, either NF-AT or its cognate binding partner can
be immobilized
2 0 utilizing conjugation of biotin and streptavidin. For instance,
biotinylated NF-AT molecules can
be prepared from biotin-NHS (N-hydroxy-succinimide) using techniques well
known in the art
(e.g., biotinylation kit, Pierce Chemicals, Rockford, IL), and immobilized in
the wells of
streptavidin-coated 96 well plates (Pierce Chemical). Alternatively,
antibodies reactive with NE-
AT can be derivatized to the wells of the plate, and NF-AT trapped in the
wells by antibody
2 5 conjugation. As above, preparations of an NF-AT binding protein and a test
compound are
incubated in the NF-AT presenting wells of the plate, and the amount of
complex trapped in the
well can be quantitated. Exemplary methods for detecting such complexes, in
addition to those
described above for the GST-immobilized complexes, include immunodetection of
complexes
using antibodies reactive with the NF-AT binding partner, or which are
reactive with NF-AT
3 0 protein and compete with the binding partner; as well as enzyme-linked
assays which rely on
detecting an enzymatic activity associated with the binding partner, either
intrinsic or extrinsic
activity. In the instance of the latter, the enzyme can be chemically
conjugated or provided as a
fusion protein with the NF-AT binding partner. To illustrate, the NF-AT
binding partner can be
chemically cross-linked or genetically fused with horseradish peroxidase, and
the amount of
3 5 polypeptide trapped in the complex can be assessed with a chromogenic
substrate of the enzyme,
56
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
e.g. 3,3'-diamino-benzadine terahydrochloride or 4-chloro-1-napthol. Likewise,
a fusion protein
comprising the polypeptide and glutathione-S-transferase can be provided, and
complex formation
quantitated by detecting the GST activity using 1-chloro-2,4-dinitrobenzene
(Habig et al (1974)
J Biol Chem 249:7130).
For processes which rely on immunodetection for quantitating one of the
proteins trapped
in the complex, antibodies against the protein, such as anti-NF-AT antibodies,
can be used.
Alternatively, the protein to be detected in the complex can be "epitope
tagged" in the form of a
fusion protein which includes, in addition to the NF-AT sequence, a second
polypeptide for which
antibodies are readily available (e.g. from commercial sources). For instance,
the GST fusion
1 o proteins described above can also be used for quantification of binding
using antibodies against
the GST moiety. Other useful epitope tags include myc-epitopes (e.g., see
Ellison et al. (1991)
J Biol Chem 266:21150-21157) which includes a 10-residue sequence from c-myc,
as well as the
pFLAG system (International Biotechnologies, Inc.) or the pEZZ-protein A
system (Pharmacia,
NJ).
An interaction between molecules, in particular between NF-AT and an NF-AT
binding
partner, can also be identified by using real-time BIA (Biomolecular
Interaction Analysis,
Pharmacia Biosensor AB) which detects surface plasmon resonance (SPR), an
optical
phenomenon. Detection depends on changes in the mass concentration of
macromolecules at the
biospecific interface, and does not require any labeling of interactants. In
one embodiment, a
2 0 library of test compounds can be immobilized on a sensor surface, e.g.,
which forms one wall of
a micro-flow cell. A solution containing the NF-AT protein, functional
fragment thereof, NF-AT
analog or NF-AT binding partner is then flown continuously over the sensor
surface. A change
in the resonance angle as shown on a signal recording, indicates that an
interaction has occurred.
This technique is further described, e.g., in BIAtechnology Handbook by
Pharmacia.
2 5 The above-described screening assays can generally be performed as
follows. In this
description, one component is NF-AT or a portion thereof and the other
component is generally
termed NF-AT binding partner, which can be, e.g., an NF-AT molecule or portion
thereof, a
kinase, or a phosphatase such as calcineurin. Thus, an exemplary screening
assay of the present
invention includes the steps of (a) forming a reaction mixture including: (i)
an NF-AT polypeptide,
3 0 (ii) an NF-AT binding partner, and (iii) a test compound; and (b)
detecting interaction of the NE-
AT and the NF-AT binding protein. The reaction mixture can be a cell-free
protein preparation,
e.g., a reconsistuted protein mixture or a cell lysate, or it can be a
recombinant cell including a
heterologous nucleic acid recombinantly expressing the NF-AT protein. For
instance, the NF-AT
polypeptide and NF-AT binding partner can be produced recombinantly, purified
from a source,
3 5 e.g., a cell extract, or chemically synthesized, as described herein. A
statistically significant
change (increase or inhibition) in the interaction of the NF-AT and NF-AT
binding protein in the
57
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
presence of the test compound, relative to the interaction in the absence of
the test compound,
indicates a potential agonist (mimetic or potentiator) or antagonist
(inhibitor) of NF-AT bioactivity
for the test compound. The compounds of this assay can be contacted
simultaneously.
Alternatively, an NF-AT protein can first be contacted with a test compound
for an appropriate
amount of time, following which the NF-AT binding partner is added to the
reaction mixture. The
efficacy of the compound can be assessed by generating dose response curves
from data obtained
using various concentrations of the test compound. Moreover, a control assay
can also be
performed to provide a baseline for comparison. In the control assay, isolated
and purified NF-AT
polypeptide or binding partner is added to a composition containing the NF-AT
binding partner
or NF-AT polypeptide, and the formation of a complex is quantitated in the
absence of the test
compound.
The invention also provides screening assays for identifying compounds which
modulate
phosphorylation or depohosphorylation of an NF-AT molecule. In this regard, an
NF-AT
antagonist,atleast with respect to treatment of cardiac hypertrophy, is an
agent which either inhibits
dephosphorylation of an NF-AT protein, or potentiates phosphorylation. In
certain embodiments
of the assay, it may be desirable to directly detect changes in
phosphorylation of an NF-AT
polypeptides.
In one embodiment, the assay is an in vitro assay. In one embodiment, the
assay comprises
contacting a non phosphorylated, or partially phosphorylated NF-AT polypeptide
with a cell
2 o extract, or with one or more purified kinases, such as GSK-3 and PKA, and
other necessary
components of an in vitro kinase assay, including a source of phosphate and
with or without a test
compound and under conditions under which phosphorylation of NF-AT occurs. The
comparison
of the state of phosphorylation of NF-AT in the presence and in the absence of
a test compound
will indicate whether the test compound decreases or inhibits, or
alternatively increases or
2 5 stimulates, the phosphorylation of NF-AT. The kinase assay and preparation
of a cellular extract
can be performed as described in the Examples. When the NF-At polypeptide is
partially
phosphorylated prior to use in the kinase assay, this can be achieved, e.g.,
by prior incubation of
a non-phosphorylated NF-AT with PKA. An NF-AT phosphorylated with PKA can be
use as a
component of an assay for identifying compounds which inhibit phosphorylation
by GSK-3, since
3 0 GSK-3 phosphorylates peptides which have been phosphorylated by PKA. Non
phosphorylated
or partially phosphorylated NF-AT can also be obtained from cells containing
active, e.g., nuclear
NF-AT. Thus, NF-AT substrate for use in this assay can be obtained from or
consist in a nuclear
extract of activated T cell.
In another embodiment, the kinase assay is an in vivo kinase assay. The assay
can
3 5 comprise incubating a cell expressing non phophorylated or partially
phosphorylated NF-AT, e.g,
an activated T cell, with a test compound and comparing the state of
phosphorylation of NF-AT
58
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
in' the presence and in the absence of the test compound. A variation in the
state of
phosphorylation will indicate that the test compound is capable of modulation
phosphorylation of
NF-AT. The state of phosphorylation of NF-AT can be determined by, e.g., by
performing the
incubation of the cells in the presence of labeled, e.g., radioactive,
phosphate (e.g., ATP), and
determining the amount of label present in an immunoprecipitate with an NF-AT
specific
antibody. Alternatively, the state of phosphorylation can be performed by
Western blot analysis,
optionally coupled with immunoprecipitations.
In another embodiment, the invention provides screening assays for identifying
compounds
which modulate dephosphorylation of NF-AT, such as inhibitors of calcineurin-
mediated
dephosphorylation of an NF-AT proteins. In one embodiment, the assay comprises
incubating a
phosphorylated NF-AT polypeptide with a cell extract or with one or more
phosphatases, e.g,
calcineurin, in conditions under which the NF-AT polypeptide is phosphatased,
and a test
compound. NF-AT can be phosphorylated in vitro with PKA and optionally GSK-3,
or NF-AT
can be phosphorylated with a cell extract. NF-AT can also be isolated from or
present in a cell
extract. The comparison of the state of phosphorylation of NF-AT after a
phospatase reaction in
the presence and in the absence of a test compound will indicate whether the
test compound is
capable of modulating dephosphorylation of NF-AT. A higher level of
phosphorylated NF-AT
in the presence of the test compound relative to the level of phosphorylation
in the absence of the
test compound indicates that the compound is an inhibitor of NF-AT
dephosphorylation. A lower
2 0 level indicates that the test compound is a stimulator of
dephosphorylation. The state of
phosphorylation of NF-AT can be determined as described above.
In yet another embodiment, the drug screening assay is derived to include a
whole cell
expressing an NF-AT protein. The ability of a test agent to alter the activity
of the NF-AT protein
can be detected by analysis of the recombinant cell. For example, agonists and
antagonists of the
2 5 NF-AT protein biological activity can by detected by scoring for
alterations in hypertrophy of the
cell. General techniques for detecting such changes are well known, and are
further described
herein. For the cell-based assays, the recombinant cell is preferably a
mammalian cell, e.g., a
human cell. In a preferred embodiment, the cell is a muscle cell, most
preferably a myocyte.
In addition to morphological studies, changes) in the level of an
intracellular second
3 0 messenger responsive to activities dependent on the NF-AT protein can be
detected. For example,
in various embodiments the assay may assess the ability of test agent to cause
changes in
phophorylation patterns of NF-AT or expression of genes whose transcription is
dependent on NE
AT. By detecting changes in intracellular signals, such as alterations in
second messengers or gene
expression, candidate agonists and antagonists to NF-AT protein-dependent
signaling can be
3 5 identified.
By selecting transcriptional regulatory sequences from target genes, e.g.,
that are
59
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
responsible for the up- or down-regulation of these genes having NF-AT
dependent transcriptional
control elements, and operatively linking such promoters to a reporter gene,
the present invention
provides a transcription based assay which is sensitive to the ability of a
specific test compound
to influence signaling pathways dependent on the NF-AT protein.
In an exemplary embodiment, the subject assay comprises detecting, in a cell-
based assay,
changes) in the level of expression of a gene controlled by a transcriptional
regulatory sequence
responsive to signaling by an NF-AT protein. Reporter gene based assays of
this invention
measure the end stage of the above described cascade of events, e.g.,
transcriptional modulation.
Accordingly, in practicing one embodiment of the assay, a reporter gene
construct is inserted into
the reagent cell in order to generate a detection signal dependent on
signaling by the NF-AT
protein. Expression of the reporter gene, thus, provides a valuable screening
tool for the
development of compounds that act as agonists or antagonists of NF-AT protein-
dependent
signalling.
In practicing one embodiment of the assay, a reporter gene construct is
inserted into the
reagent cell in order to generate a detection signal dependent on second
messengers generated by
the NF-AT protein. Typically, the reporter gene construct will include a
reporter gene in operative
linkage with one or more transcriptional regulatory elements responsive to
signal transduction
from the NF-AT protein, with the level of expression of the reporter gene
providing the detection
signal. The amount of transcription from the reporter gene may be measured
using any method
2 0 known to those of skill in the art to be suitable. For example, mRNA
expression from the reporter
gene may be detected using RNAse protection or RNA-based PCR, or the protein
product of the
reporter gene may be identified by a characteristic stain or an intrinsic
activity. The amount of
expression from the reporter gene is then compared to the amount of expression
in either the same
cell in the absence of the test compound or it may be compared with the amount
of transcription
2 5 in a substantially identical cell that lacks the target receptor protein.
Any statistically or otherwise
significant difference in the amount of transcription indicates that the test
compound has in some
manner altered the inductive activity of the NF-AT protein.
As described in further detail below, in preferred embodiments the gene
product of the
reporter is detected by an intrinsic activity associated with that product.
For instance, the reporter
3 o gene may encode a gene product that, by enzymatic activity, gives rise to
a detection signal based
on color, fluorescence, or luminescence. In other preferred embodiments, the
reporter or marker
gene provides a selective growth advantage, e.g., the reporter gene may
enhance cell viability,
relieve a cell nutritional requirement, and/or provide resistance to a drug.
Many reporter genes are
known to those of skill in the art and others may be identified or synthesized
by methods known
3 5 to those of skill in the art. A reporter gene includes any gene that
expresses a detectable gene
product, which may be RNA or protein.
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Preferred reporter genes are those that are readily detectable. The reporter
gene may also
be included in the construct in the form of a fusion gene with a gene that
includes desired
transcriptional regulatory sequences or exhibits other desirable properties.
Examples of reporter
genes include, but are not limited to CAT (chloramphenicoI acetyl transferase)
(Alton and Vapnek
(1979), Nature 282: 864-869) luciferase, and other enzyme detection systems,
such as beta-
galactosidase; firefly luciferase (deWet et al. (1987), Mol. Cell. Biol. 7:725-
737); bacterial
luciferase (Engebrecht and Silverman (1984), PNAS 1: 4154-4158; Baldwin et al.
(1984),
Biochemistry 23: 3663-3667); alkaline phosphatase (Toh et al. (1989) Eur. J.
Biochem. 182: 231-
238, Hall et al. (1983) J. Mol. Appl. Gen. 2: 101), human placental secreted
alkaline phosphatase
l0 (Cullen and Malim (1992) Methods in Enzymol. 216:362-368).
In still another embodiment of a drug screening, a two hybrid assay can be
generated with
an NF-AT protein and target molecule. Drug dependent inhibition or
potentiation of the interaction
can be scored. The two hybrid assay formats described in the art can be
readily adaoted for such
drug screening embodiments. See, for example, U.S. Pat. Nos. 5,283,317,
5,580,736 and
I5 5,695,941; Zervos et al. (1993) Cell 72:223-232; Madura et al. (1993) J
Biol Chem 268:12046-
12054; Bartel et al. (1993) Biotechniques 14:920-924; and Iwabuchi et al.
(1993) Oncogene
8:1693-1696).
The identification of compounds which modulate dephosphorylation of NF-AT can
also
be identified in cell-based assays. For example, a cell containing
phosphorylated NF-AT is
2 o incubated in the presence or absence of a test compound and the state of
phosphorylation of NE
AT is determined as described above. For instance, the ability of compounds to
modulate NF-AT
phosphorylation/de phosphorylation could be screened using colony
immunoblotting (Lyons and
Nelson (1984) PNAS 81:7426-7430) using antibodies against phosphorylated
residues. Reagents
for performing such assays are further described herein.
2 5 In yet another embodiment, the invention provides a cell-based screening
assay for
identifying compounds which modulate nuclear translocation of NF-AT comprising
incubating
or treating a cell with or without a test compound and determining the
localization of NF-AT in
the cell, i.e., whether NF-AT is present in the cytoplasm and/or in the
nucleus of the cell. In one
embodiment, a cell containing NF-AT in the cytoplasm, e.g., a resting T cell
or a Jurkat cell, is
3 0 incubated with a test compound and the cellular localization of NF-AT is
determined. The
presence of NF-AT in the nucleus indicates that the test compound stimulates
the translocation of
NF-AT from the cytoplasm to the nucleus. In another embodiment, a cell
containing NF-AT in
the cytoplasm is incubated with a test compound and an agent which activates
NF-AT, i.e., an
agent which stimulates its translocation to the nucleus and the cellular
localization of NF-AT is
3 5 determined. If more NF-AT is localized in the cytoplasm of the cell
incubated with the test
compound relative to a cell that was not treated with the test compound, the
test compound inhibits
61
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
the translocation of NF-AT from the cytoplasm to the nucleus and is thus an NF-
AT inhibitor.
Alternatively, is more NF-AT is present in the nucleus of the cell incubated
with' the test
compound, relative to the cell that was not incubated with the test compound,
the test compound
is a stimulator or activator of NF-AT.
The invention further provides screening assays for identifying compounds
which modulate
the activity of NF-AT. The screening assays can be in vivo or in vitro and can
be cell based or
based on a cell free format. In a preferred embodiment, the assays allow the
identification of
compounds which modulate NF-AT translocation across the nuclear membrane. In
an even more
preferred embodiment, the screening assay comprises contacting an NF-AT NLS
with an NF-AT
molecule or portion thereof which is sufficient for binding to the NLS and
with a test compound
or a library of test compounds. In one embodiment, the NLS comprises the amino
acid sequence
CNKRKYSLN (SEQ ID NO: 53) (N-terminal NLS). In another embodiment, the NLS
comprises
the amino acid sequence GKRKIUR (SEQ ID NO: 67) (C-terminal NLS). The other
component
of the screening assay can be a peptide comprising the SRR, SP 1, SP2, and/or
SP3 of an NF-AT
molecule. In a preferred embodiment, the screening assay comprises an N-
terminal NLS and a
peptide comprising the SRR region, which is phopshorylated.
In another embodiment, the screening assay comprises a.C-terminal NLS and a
peptide
comprising SPl, SP2 and/or SP3 regions which are phosphorylated.
In another embodiment of the screening assay, one component of the assay is an
NF-AT
2 0 polypeptide of portion thereof sufficient for binding to calcineurin and
the other component is
calcineurin or a portion thereof sufficient for binding to NF-AT. A portion of
NF-AT can be an
N-terminal portion, e.g., amino acids 1-418 of SEQ ID NO: 38. Thus, in one
embodiment, the
screening assay comprises contacting an NF-AT polypeptide with calcineurin and
a test compound
in conditions under which, but for the presence of the test compound, the NF-
AT polypeptide are
2 5 capable of interacting. The comparison of the'binding of the two
components of the assay in the
presence and in the absence of a test compound will indicate whether the test
compound inhibits,
or alternatively stimulates, the interaction between NF-AT and calcineurin.
3 0 In another embodiment, the screening assay comprises incubating a cell
containing nuclear
NF-AT, e.g., an activated T cell such as a T cell treated with ionomycin, is
contacted with a test
compound and the cellular localization of NF-AT is determined. If more NF-AT
is localized in
the nucleus in the cell treated with the test compound versus the cell that
was not treated with the
test compound, the test compound is a compound which is capable of maintaining
NF-AT in an
3 5 activated state, i.e., in the nucleus. If more NF-AT is localized in the
cytoplasm in the cell treated
with the test compound relative to the cell that was not treated with the test
compound, the test
62
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
compound is capable of deactivating NF-AT, i.e., to stimulate its
translocation to the cytoplasm.
The cellular localization of an NF-AT molecule can be determined, by, e.g.,
detecting its
localization within a cell using an antibody or other agent capable of
specifically interacting with
NF-AT. For example, NF-AT can be detected by imrnunfluorescence, such as
described in the
Examples. In another embodiment, the cell can be transfected with a nucleic
acid encoding an NF
AT polypeptide that is fused to a tag or marker, that can be detected. For
example, the cell can be
made to express an NF-AT polypeptide that is fused to a myc tag and the NF-AT
fusion
polypeptide can then be detected with an antibody binding specifically with a
myc tag.
Other agents which are capable of modulating the hypertrophic state of cells
include
nucleic acids which compete away the binding of NF-AT proteins to their DNA
binding site, e.g.,
"decoy" nucleic acids. Preferred nucleic acids comprise a DNA binding site of
NF-AT. These
reagents can be inhibit binding of only a single NF-AT polypeptide , or
alternatively, reagents can
be designed for inhibiting binding of most or all of the different NF-AT
family members to their
DNA binding sites.
In addition to small molecules which may be identified, e.g., by the drug
screening assays
described above, other agents capable of modulating cell hypertrophy may
include peptide
domains (fragments) of the NF-AT protein, as well as mutants of the molecular
regulators. A
"mutant" as used herein refers to a peptide having an amino acid sequence
which differs from that
2 0 of the naturally occurnng peptide or protein by at least one amino acid.
Mutants may have the
same biological and immunological activity as the naturally occurring protein.
However, the
biological or immunological activity of mutants may differ or be lacking. For
example, a protein
mutant may act as an agonist, antagonist (competitive or non-competitive), or
partial agonist of
the function of the naturally occurnng protein.
2 5 For example, homologs of the NF-AT proteins (both agonist and antagonist
forms) can be
generated using, for example, alanine scanning mutagenesis and the like (Ruf
et al. (1994)
Biochemistry 33:1565-1572; Wang et al. (1994) J. Biol. Chem. 269:3095-3099;
Balint et al.
(1993) Gene 137:109-118; Grodberg et al. (1993) Eur. J. Biochem. 218:597-601;
Nagashima et
al. (1993) J. Biol. Chem. 268:2888-2892; Lowman et al. (1991) Biochemistry
30:10832-10838;
30 and Cunningham et al. (1989) Science 244:1081-1085), by linker scanning
mutagenesis (Gustin
et al. (1993) Virology 193:653-660; Brown et al. (1992) Mol. Cell Biol.
12:2644-2652; McKnight
et al. (1982) Science 232:316); by saturation mutagenesis (Meyers et al.
(1986) Science 232:613);
by PCR mutagenesis (Leung et al. (1989) Method Cell Mol Biol 1:11-19); or by
random
mutagenesis (Miller et al. (1992) A Short Course in Bacterial Genetics, CSHL
Press, Cold Spring
3 5 Harbor, N.Y.; and Greener et al. ( 1994) Strategies in Mol Biol 7:32-34).
Linker scanning
matagenesis, particularly in a combinatorial setting, is on attractive method
for identifying
63
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
truncated (such as constitutively active or dominant negative) forms of an NF-
AT protein.
The invention also contemplates the reduction of the subject NF-AT protein to
generate
mimetics, e.g. peptide or non-peptide agents, which are able interfere with,
or mimic, the effect
of the authentic NF-AT protein on the hypertrophic state of cells, such as
muscle cells. Such
peptidomimetics can act as drugs for the modulation of cell hypertrophy.
Peptidomimetics are commonly understood in the pharmaceutical industry to
include non-
peptide drugs having properties analogous to those of the mimicked peptide.
The principles and
practices of peptidomimetic design are known in the art and are described, for
example, in
Fauchere, Adv. Drug es. 15:29 (1986); and Evans et al., J. Med. Chem. 30:1229
(1987).
Peptidomimetics which bear structural similarity to therapeutically useful
peptides may be used
to produce an equivalent therapeutic or prophylactic effect. Typically, such
peptidomimetics have
one or more peptide linkages optionally replaced by a linkage which may
convert desirable
properties such as resistance to chemical breakdown in vivo. These linkages
may include -
CHzNH-, -CHZS-, -CHZ-CHz-, -CH=CH-, -COCHz-, -CH(OH)CHZ-, and -CHZSO-.
Peptidomimetics may exhibit enhanced pharmacological properties (biological
half life, absorption
rates, etc.), different specificity, increased stability, production
economies, lessened antigenicity
and the like which makes their use as therapeutics particularly desirable.
Such mutagenic techniques as described above are also particularly useful for
mapping the
determinants of an NF-AT proteins which participate in protein-protein
interactions involved in,
2 0 for example, binding to a leucine-zipper containing protein. To
illustrate, the critical residues of
an NF-AT protein which are involved in molecular recognition of other cellular
proteins (or
nucleic acid) can be determined and used to generate peptidomimetics which
maintain at least a
portion of that binding activity. By employing, for example, scanning
mutagenesis to map the
amino acid residues involved in binding, peptidomimetic compounds (e.g.
diazepine or
2 5 isoquinoline derivatives) can be generated which mimic those residues in
binding to the kinase.
For instance, non-hydrolyzable peptide analogs of such residues can be
generated using
benzodiazepine (e.g., see Freidinger et al. in Peptides: hemistry a_nd
Biology, G. R. Marshall ed.,
ESCOM Publisher: Leiden, Netherlands, 1988), azepine (e.g., see Huffman et al.
in Pe ide
Chemistry and Biolo~v, G. R. Marshall ed., ESCOM Publisher: Leiden,
Netherlands, 1988),
3 0 substituted gama lactam rings (Garvey et al. in Peptides' Chemistry and
Bioloev, G. R. Marshall
ed., ESCOM Publisher: Leiden, Netherlands, 1988), keto-methylene
pseudopeptides (Ewenson
et al. ( 1986) J. Med. Chem. 29:295; and Ewenson et al. in Peptides: Structure
and Function
(Proceedings of the 9th American Peptide Symposium) Pierce Chemical Co.
Rockland, Ill., 1985),
(3-turn dipeptide cores (Nagai et al. (1985) Tetrahedron Lett 26:647; and Sato
et al. (1986) J Chem
35 Soc Perkin Trans 1:1231), and (3-aminoalcohols (Gordon et al. (1985)
Biochem Biophys Res
o mun 126:419; and Dann et al. (1986) Biochem Biophvs Res Commun 134:71).
64
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
NF-AT~ polypeptides, especially those portions which form direct contacts in
NF-AT
complexes, can be used for rational drug design of candidate NF-AT-modulating
agents (e.g.,
antineoplastics and immunomodulators). The substantially purified NF-AT~ and
the identification
of NF-AT~ as capable of forming intermolecular associations, e.g., with AP-1
and with DNA, as
well as forming intermolecular associations, as provided herein, permits
production of
substantially pure NF-AT polypeptide complexes and computational models which
can be used
for protein X-ray crystallography or other structure analysis methods, such as
the DOCK program
(Kuntz et al. (1982) J. Mol. Biol. 1~: 269; Kuntz ID (1992) ci a 2L,S5 : 1078)
and variants
thereof. Potential therapeutic drugs may be designed rationally on the basis
of structural
information thus provided. In one embodiment, such drugs are designed to
prevent formation of
a NF-AT~ polypeptide: AP-1 polypeptide complex. In another embodiment, such
drugs are
designed to prevent the formation of intramolecular interactions in NF-AT.
Thus, the present
invention may be used to design drugs, including drugs with a capacity to
inhibit binding of NF-
AT~ to form an NF-AT complex.
Cadiac hypertrophy models
Compounds identified by the above-described methods can be further evaluated
as agents
for treating cardiac hypertropy and the like using any of a number of cardiac
hypertrophy models.
Methods for inducing cardiac hypertrophy: induction by a variety of humoral
factors, including
angiotensin II, phenylephrine (PE), and endothelin-1 (ET-1) (Karliner et
aL(1990) ~perientia
46:81, Sadoshima et al. (1993) 'rc. R . 73:424, Leite et al. (1994) Am. J.
Plttysiol. 267: H2193).
Compounds or polypeptides or genes encoding polypeptides which are capable of
inducing
cardiac hypertrophy are referred to herein as "hypertrophy inducing"
compounds, polypeptides or
genes, respectively. Several investigators have shown that endothelin-1, which
is known to be
2 5 produced in endothelial cells, induces hypertrophy of cardiac myocytes in
vitro. Shubeita et al.
(1990) J. Biol. Chem., 265:20555-20562; Ito et al., {1991) it es 69: 209-215;
Suzuki et al.,
(1991) J. Cardiovasc Pharmacol , 17 Suppl 7: 5182-5186. See also U.S. Pat. No.
5,344,644. Yet
another hypertrophy inducing factor is LIF (U.S. Pat. No. 5837241).
In vitro and in vivo methods for determining the presence of muscle cell
hypertrophy, e.g.,
3 0 ventricular muscle cell hypertrophy, are known. In vitro assays for muscle
cell hypertrophy
include those methods described herein, e.g., increased cell size and
increased expression on atrial
natriuretic factor (ANF). Changes in cell size are used in a scoring system to
determine the extent
of hypertrophy. These changes can be viewed with an inverted phase microscope,
and the degree
of hypertrophy scored with an arbitrary scale of 7 to 0, with 7 being fully
hypertrophied cells, and
3 5 3 being non-stimulated cells. The 3 and 7 states may be seen in Simpson et
al., (1982) Circulation
Res. 51:787-801, FIG. 2, A and B, respectively. The correlation between
hypertrophy score and
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
cell surface area (pmt) has been determined to be linear (correlation
coefficient = 0.99). In
phenylephrine-induced hypertrophy, non-exposed (normal) cells have a
hypertrophy score of 3 and
a surface area/cell of 581 mu m2 and fully hypertrophied cells have a
hypertrophy score of 7 and
a surface area/cell of 1811 mu m2, or approximately 200% of normal. Cells with
a hypertrophy
score of 4 have a surface area/cell of 771 p,m2, or approximately 30% greater
size than
non-exposed cells; cells with a hypertrophy score of 5 have a surface
area/cell of 1109 p,m2, or
approximately 90% greater size than non-exposed cells; and cells with a
hypertrophy score of 6
have a surface area/cell of 1366 p,m2, or approximately 135% greater size than
non-exposed cells.
The presence of ventricular muscle cell hypertrophy preferably includes cells
exhibiting an
increased size of about 15% (hypertrophy score 3.5) or more. Inducers of
hypertrophy vary in
their ability to induce a maximal hypertrophic response as scored by the above-
described assay.
For example, the maximal increase in cell size induced by endothelin is
approximately a
hypertrophy score of 5.
A hypertrophic assay can be performed as follows. First, a myocyte cell
suspension can
be prepared as described in Chien et al., (1985) J. Clin. Invest., 75: 1770-
1780 and Iwaki et al.,
supra. Ventricles from the hearts of 1-2 day Sprague-Dawley rat pups are
removed and trisected.
The minced ventricles are digested with a series of sequential collagenase
treatments. Purification
of the resulting single-cell suspension on a discontinuous Percoll gradient
results in a suspension
of 95% pure myocytes.
2 0 The culture of myocytes can then be plated as described in Long et al.,
supra. This method
includes preplating the cell suspension for 30 min. in MEM/5% calf serum. The
unattached
myocytes can then plated in serum-free MEM supplemented with insulin,
transferrin, BrdU, and
bovine serum albumin in 35-mm tissue-culture dishes at a density of 7.5 x 104
cells per mL. The
culture of myocytes can also be plated in D-MEM/199/S% horse serum/5% fetal
calf serum in
2 5 10-cm tissue-culture dishes at 3 x 105 cells per mL. After 24 hr in
culture the cells are washed and
incubated in serum-free D-MEM/199.
Yet another method for culturing myocytes to increase testing capacity with a
miniaturized
assay, is as follows. The wells of 96-well tissue-culture plates are precoated
with D-MEM/F12/4%
fetal calf serum for 8 hr at 37 °C. This medium is removed and the cell
suspension is plated in the
3 0 inner 60 wells at 7.5 x 10a cells per mL in D-MEM/F-12 supplemented with
insulin, transferrin,
and aprotinin. The medium typically also contains an antibiotic such as
penicillin/streptomycin
andglutamine. This medium allows these cells to survive at this low plating
density without
serum. Test substances, e.g., NF-AT antagonists are added directly into the
wells after the cells
have been in culture for 24 hours.
3 5 After stimulation with a-adrenergic agonists or endothelin, neonatal rat
myocardial cells
in culture display several features of the in vivo cardiac muscle cell
hypertrophy seen in congestive
66
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
heart failure, including an increase in cell size and an increase in the
assembly of an individual
contractile protein into organized contractile units. Chien et al., FASEB J.,
. These changes
can be viewed with an inverted phase microscope and the degree of hypertrophy
scored with an
arbitrary scale of 7 to 0, with 7 being fully hypertrophied cells and 3 being
non-stimulated cells.
The 3 and 7 states may be seen in Simpson et al.,(1982) Circulation Research,
51: 787-801. To
facilitate the microscopic readout of the 96-well cultures and to generate a
permanent record, the
myocytes are fixed and stained after the appropriate testing period with
crystal violet stain in
methanol. Crystal violet is a commonly used protein stain for cultured cells.
Additionally, an
aliquot can be taken from the 96-well plates and monitored for the expression
of protein markers
of the response such as release of ANF or ANP (as described, e.g., in U.S.
Pat. No. 5534615).
At a high cell density, myocytes may also begin to self induce hypertrophy.
Thus, in one embodiment, a method for screening for NF-AT antagonists which
inhibit
hypertrophy comprises the following steps:
(a) plating 96-well plates with a suspension of myocytes at a cell density of
about 7.5 x 104
cells per mL in D-MEM/F-12 medium supplemented with at least insulin,
transferrin and
aprotinin;
(b) culturing the cells in the presence of a hypertrophy inducing factor,
e.g., LIF or
endothelin;
(c) adding a test substance to be assayed (an NF-AT antagonist);
2 0 (d) culturing the cells with the test substance; and
(e) measuring for hypertrophy.
The medium can be supplemented with additional elements such as EGF that
ensure a
longer viability of the cells, but such supplements are not essential. D-MEM/F-
12 medium is
available from Gibco BRL, Gaithersburg, Md.
2 5 The preferred hypertrophy assay comprises:
(a) pre-coating the wells of 96-well tissue culture plates with a medium
containing calf
serum, preferably D-MEM/F-12 medium containing 4% fetal calf serum, wherein
preferably the
wells are incubated with the medium for about eight hours at about 37
°C;
(b) removing the medium;
3 0 (c) plating a suspension of myocytes in the inner 60 wells at 7.5 x 104
cells per mL in
D-MEM/F-12 medium supplemented with insulin, transfernn and aprotinin;
(d) culturing the myocytes for at least 24 hours in the presence of a
hypertrophy inducing
factor, e.g, LIF or endothelin;
(e) adding the test substance;
3 5 (f) culturing the cells with the test substance (preferably for about 24-
72 hours, more
preferably for about 48 hours); and
67
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
(g) evaluating hypertrophy, preferably after crystal violet stain, e.g., by
microscopic
examination.
Preferably the medium used in step (c) is a serum-free medium also containing
penicillin/streptomycin (pen/strep) and glutamine. Most preferably, the medium
contains 100 mL
D-MEM/F-12, 100 mu L transferring (10 mg/mL), 20 mu L insulin (5 mg/mL), 50 mu
L aprotinin
(2 mg/mL), 1 mL pen/strep (JRH Biosciences No. 59602-77P), and 1 mL L-
glutamine (200 mM).
The assay capacity of 1000 single samples a week coupled with the small sample
size
requirement of 100 mu L or less has enabled an expression cloning and protein
purification that
would have been impossible to accomplish using the current methods available.
Another method for assaying hypertrophy involves measuring for atrial
natriuretic peptide
(ANP), a marker for myocyte hypertrophy, release by means of an assay that
determines the
competition for binding of <125> I-rat ANP for a rat ANP receptor A-IgG fusion
protein. The
method suitable for use is similar to that used for detenmining GPI30 using a
CD4-IgG fusion
protein described by Chamow et al., Biochemistry, 29: 9885-9891 (1990).
Cultures of myocytes
and hypertrophy induction and assaying are further described in U.S. Pat. No.
5837241.
Alternatively, a compound of the invention for inhibiting cardiac hypertrophy
can be
identified by screening for compounds which inhibit NF-ATc activity.
Preferably, the compound
inhibits NF-ATc4 activity, but does not inhibit the activity of one or more
protein members of the
NF-AT family, i.e., NF-ATcI, NF-ATc2, or NF-ATc3 and splice variants thereof.
Alternatively,
2 0 the antagonists for use in the invention could inhibit the activity of
more than one NF-AT
polyeptide, but not of all of them. For example, an antagonist for use in the
invention could be an
NF-ATc4 and NF-ATc3 antagonist, but not an NF-ATc 1 or NF-ATc2 antagonist.
Compounds that
are antagonists of only certain NF-AT polypeptides, but not of others can be
developed based on
the differences between the NF-AT family members. For example, antagonists
specific for certain
2 5 NF-AT polypeptides can be compounds which inhibit interaction of an NF-AT
polypeptide with
a specific DNA binding sequence. Since the NF-AT polypeptides do not all bind
the same NF-AT
binding sequence, antagonists that are specific for certain NF-AT polypeptides
can be developed.
Alternatively, antagonists that are specific for certain NF-AT polypeptides
can be compounds
which are based on differences in the NF-AT sequences. For example, antisense
compounds can
3 0 be used for specifically inhibiting the production of only some NF-AT
polypeptide and not that
of others. Similarly, ribozymes can be designed to selectively destroy certain
members of the NE-
AT family, without affecting others.
Alternatively, compounds which are capable of inhibiting all of the NF-AT
family
members can also be designed, based on the significant homologies between
these proteins and
3 5 also between the genes encoding them.
Agents which can be used to prevent or treat cardiac hypertrophy can also be
identified in
68
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
animal models. Animal models can be animals which develop cardiac hypertrophy
and animal
models in which these animals are crossed with an NF-AT knock-out mouse of the
invention.
In an illustrative embodiment, the animal model is a transgenic mouse having a
constitutively active NF-AT pathway. Such mice include those having a
constitutive calcineuring
or a constitutive NF-AT, e.g., NF-ATc4, such as those described in Olsen et
al., Cell 93: 215
(1998). These mice can be bred with the NF-ATc4 knock-out mice of the
invention. Thus, as
opposed to transgenic mice having a constitutively active NF-ATc4 signaling
pathway, and which
express wild-type NF-ATc4, mice which have a constitutively active NF-ATc4
pathway, but
which do not express wild-type NF-ATc4 will not develop cardiac hypertrophy.
Other animal models which can be used alone, e.g., in screening assays, or
which can be
crossed with the NF-ATc4 knock-out mice to obtain animal models, include mice
which are
transgenic for a hypertrophy inducing gene, such as those described herein.
Another animal model
of cardiac hypertrophy includes the pressure-overload mouse model wherein the
pulmonary artery
is constricted, resulting in right ventricular failure. A retroviral murine
model of ventricular
dysfunction can also be used. Other animal models include MLC-ras mice and
aortic banding of
heterozygous IGF-I-deficient mice. Additionally, transgenic mice that harbor a
muscle actin
promoter-IGF-I fusion gene display cardiac and skeletal muscle hypertrophy,
without evidence
of myopathy or heart failure. IGF-I-gene-targeted mice display defects in
cardiac myogenesis (as
well as skeletal), including markedly decreased expression of ventricular
muscle contractile
2 0 protein genes. Another useful animal models include the RXR alpha mutant
mouse model (Sucov
et al. (1994) Genes Dev. 8:1997-1018) and RXR alpha -/-embryo model (Dyson et
al. (1995) Proc.
Natl. Acad. Sci. (In Press)). These genetically-based animal models display
important features
of ventricular chamber dysmorphogenesis.
An NF-AT antagonist can also tested in a post-myocardial infarction rat model,
which is
2 5 predictive of human congestive heart failure in producing ANF. A detailed
description of the
procedure for obtaining such rats is described in U.S. Pat. No. 5767155.
Briefly, myocardial
infarction is produced in male Sprague-Dawley (Charles River Breeding
Laboratories, Inc., eight
weeks of age) by left coronary arterial ligation as described by Greenen et
al. (1987) J. Appl.
Physiol. 93:92-96 and Buttrick et al. (1991) Am. J. Physiol. 260:11473-11479.
Four to six weeks
3 0 after ligation, myocardial infarction can develop into heart failure in
rats. The development of
infarcts can be monitored by performing electrocardiograms. The congestive
heart failure in this
model reasonably mimics congestive heart failure in most human patients.
A person of skill in the art will recognize that any of the above described
screening assays
can easily be adapted for use in screening libraries of compounds. The
compounds identified
3 5 using any of the screening assays of the invention are also within the
scope of the invention, as
well as pharmaceutical compositions and kits comprising such.
69
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
' In another aspect, the present invention provides pharmaceutically
acceptable compositions
which comprise a therapeutically-effective amount of one or more of the
compounds described
above, formulated together with one or more pharmaceutically acceptable
carriers (additives)
andJor diluents. As described in detail below, the pharmaceutical compositions
of the present
invention may be specially formulated for administration in solid or liquid
form, including those
adapted for the following: (1) oral administration, for example, drenches
(aqueous or non-aqueous
solutions or suspensions), tablets, boluses, powders, granules, pastes for
application to the tongue;
(2) parenteral administration, for example, by subcutaneous, intramuscular or
intravenous injection
as, for example, a sterile solution or suspension; (3) topical application,
for example, as a cream,
ointment or spray applied to the skin; or (4) intravaginally or intravectally,
for example, as a
pessary, cream or foam.
The phrase "therapeutically-effective amount" as used herein means that amount
of a
compound, material, or composition comprising a peptide or peptidomimetic of
the present
invention which is effective for producing some desired therapeutic effect by
inhibiting an NF-AT
dependent signaling pathway in at least a sub-population of cells in an animal
and thereby
blocking the biological consequences of that pathway in the treated cells, at
a reasonable
benefit/risk ratio applicable to any medical treatment.
The phrase "pharmaceutically acceptable" is employed herein to refer to those
compounds,
materials, compositions, and/or dosage forms which are, within the scope of
sound medical
2 0 judgment, suitable for use in contact with the tissues of human beings and
animals without
excessive toxicity, irritation, allergic response, or other problem or
complication, commensurate
with a reasonable benefit/risk ratio.
The phrase "pharmaceutically-acceptable carrier" as used herein means a
pharmaceutically-
acceptable material, composition or vehicle, such as a liquid or solid filler,
diluent, excipient,
2 5 solvent or encapsulating material, involved in carrying or transporting
the subject peptidomimetic
agent from one organ, or portion of the body, to another organ, or portion of
the body. Each
carrier must be "acceptable" in the sense of being compatible with the other
ingredients of the
formulation and not injurious to the patient. Some examples of materials which
can serve as
pharmaceutically-acceptable carriers include: (1) sugars, such as lactose,
glucose and sucrose; (2)
30 starches, such as corn starch and potato starch; (3) cellulose, and its
derivatives, such as sodium
carboxymethyl cellulose, ethyl cellulose and cellulose acetate; (4) powdered
tragacanth; (S) malt;
(6) gelatin; (7) talc; (8) excipients, such as cocoa butter and suppository
waxes; (9) oils, such as
peanut oil, cottonseed oil, safflower oil, sesame oil, olive oil, com oil and
soybean oil; (10)
glycols, such as propylene glycol; (11) polyols, such as glycerin, sorbitol,
mannitol and
35 polyethylene glycol; (12) esters, such as ethyl oleate and ethyl laurate;
(13) agar; (14) buffering
agents, such as magnesium hydroxide and aluminum hydroxide; (15) alginic acid;
(16) pyrogen-
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
free water; (17) isotonic saline; (18) Ringer's solution; (19) ethyl alcohol;
(20) phosphate buffer
solutions; and (21 ) other non-toxic compatible substances employed in
pharmaceutical
formulations.
J. Exem~ar3r Uses of the invention
The invention provides methods for modulating the activity of NF-AT in a cell.
In one
embodiment, the activity of NF-AT is modulated by modulating its interaction
with another
molecule, such as another protein or a nucleic acid. In particular, the
activity of NF-AT can be
modulated by modulating the interaction between NF-ATc and AP-1 or other basic
domain/leucine
zipper proteins. In another embodiment, the interaction of NF-ATc with DNA,
i.e., the NF-ATc
binding site, can be modulated.
In a preferred embodiment, the method modulates the translocation of NF-AT
through the
nuclear membrane. For example, certain methods of the invention stimulate or
inhibit
translocation of NF-AT proteins from the cytoplasm to the nucleus. Other
methods of the
invention stimulate or inhibit translocation of NF-AT molecules from the
nucleus to the
cytoplasm. For example, the translocation of NF-AT from the cytoplasm into the
nucleus of a cell
can be stimulated or induced by introducing into the cell a compound which
inhibits the interaction
of at least one NLS with another part of the NF-AT molecule, thereby
unshielding at least one NE-
AT NLS, allowing the NF-AT molecule to translocate into the nucleus. A
compound which
2 0 inhibit the interaction between an NLS and another portion of the NF-AT
protein can be a small
molecule, a peptide, peptidomimetic, a nucleic acid or derivatives thereof. A
preferred compound
is peptide or peptidomimetic comprising an NF-AT NLS, or homolog thereof,
which is capable
of forming an intramolecular association with another portion of the NF-AT
molecule, such as
with one or more of the repeats, e.g, SRR, SP1, SP2, or SP3. Such compounds
are further
2 5 described herein. Another preferred compound is a nucleic acid which
encodes such a peptide.
Accordingly, the nucleic acid is introduced into a cell expressing NF-AT and
expressed in the cell.
In another embodiment, the compound is a small molecule, which can be isolated
as described
herein by screening. libraries of small molecules. The compound is preferably
small and able to
cross the cytoplasmic membrane.
3 0 In another embodiment, the invention provides a method for inhibiting the
translocation
of NF-AT from the cytoplasmic to the nucleus, thereby inhibiting NF-AT
dependent biologic
activities. This can be achieved, e.g., by stabilizing the intramolecular
association of at least one
NLS with another portion of an NF-AT molecule. Methods may include introducing
into a cell
a compound which stabilizes the interaction of an NLS with at least one
portion of NF-AT selected
35 from the group consisting of SRR, SP1, SP2 and SP3. The compound is
preferably a small
molecule, which can be obtained as described herein.
71
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
In yet another embodiment, the invention provides a method for stimulating the
translocation of NF-AT from the nucleus to the cytoplasm of a cell, thereby
blocking activation
of NF-AT dependent biological activities. This can be achieved, e.g., by
shielding one or more
of the NLSs in NF-AT molecules. In an illustrative embodiment, an NLS sequence
in an NF-AT
molecule in a cell is shielded by introducing into the cell a compound which
interacts with the
NLS sequence. A preferred compound is a peptide or peptidomimetic, e.g., a
peptide comprising
an amino acid sequence selected from the group consisting of the SRR, SP 1,
SP2, or SP3 sequence
of NF-AT. In a preferred embodiment, two compounds are introduced into a cell,
i.e., a first
compound which interacts with the NLS KRK (amino acids 265-267 of SEQ ID NO:
38) and a
1 o second compound which interacts with the NLS KRKK/R (SEQ B7 NO: 66) (amino
acids 682-685
of SEQ ID NO: 38). For example, a peptide comprising the amino acid sequence
of SRR and a
peptide comprising the amino acid sequence of at least one of SP1, SP2 and SP3
can be
administered to a cell.
Furthermore, also within the scope of the invention are methods for inhibiting
the
translocation of NF-AT molecules from the nucleus to the cytoplasm of a cell,
thereby maintaining
or prolonging NF-AT dependent biological activities. Such methods can comprise
introducing
into the nucleus of a cell comprising NF-AT molecules in the nucleus a
compound which prevents
one or more of the NLSs to form an intramolecular association. This can be
achieved, e.g., by
introducing into the cell a compound which interacts with a portion of NF-AT
which is capable
2 o of interacting with an NLS. For example, the compound can be an NLS
peptide or derivative
thereof, which is capable of binding to a portion of an NF-AT molecule, e.g.,
an SRR, SP1, SP2
and/or SP3 repeat. Based at least on the sequence homologies between the NF-
ATc family
members, a single peptide could interact with at least two NF-ATc family
members. Other
compounds that can be used for this purpose include nucleic acids encoding
such peptides and
2 5 small molecules.
Other methods for modulating translocation of an NF-ATc polypeptide comprise
modulating phosphorylation of NF-ATc, such as the phosphorylation of serines
located in the
region from about amino acid 1 to about amino acid 418 of SEQ ID NO: 38, or
even more
preferably from about amino acid 170 to about amino acid 301 of SEQ ID NO: 38.
Even more
3 o preferably, the method comprises phosphorylation of serines located in
SRR, SP1, SP2, and/or
SP3, and/or in regions located between these repeats.
Phosphorylation of NF-ATc polypeptides can be modulated by a variety of
methods. In
one embodiment, phosphorylation is modulated by modulating the activity of a
kinase which
phosphorylates NF-ATc, such as PKA, GSK-3a and GSK-3~3. Another kinase whose
activity can
3 5 be phosphorylated include JNK (jun kinase), e.g., JNK-1 or JNK-2. The
activity of a kinase can
be modulated by modulating the protein level of the kinase. For example,
increasing the activity
72
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
of a kinase can be accomplished by increasing the endogenous protein level of
the kinase, such as
by increasing transcription of the kinase. Alternatively, the activity of a
kinase can be increased
by introducing a kinase into a cell, such as by introducing a nucleic acid
encoding the kinase. In
fact, as shown herein, overexpression of GSK-3 in T cells inhibited
translocation of NF-AT to the
nucleus, and increased nuclear export of NT-ATc. Stimulation of translocation
of NF-AT from
the cytoplasm to the nucleus can be achieved by inhibiting the activity of a
kinase which
phosphorylates NF-ATc, such as by inhibiting transcription or translation of
the kinase, e.g., by
using antisense technology. The activity of the kinase can also be inhibited
by introducing into
the cell an agent which inhibits its activity, such as an NF-AT peptide,
capable of binding to the
kinase.
In another embodiment, phosphorylation is modulated by modulating the activity
of a
phosphatase, such as calcineurin. This can be achieved by modulating its
phosphorylation
capacity, by, e.g.,contacting it with an NF-AT peptide with which calcineurin
is capable of
interacting. Alternatively, the level of calcineurin in a cell can be
modulated, such as by
modulating its expression or by introducing exogenous calcineurin in the cell
or by introducing
antisense nucleic acids inhibiting calcineurin mRNA translation.
In still another embodiment, the present invention provides compounds which
inhibit either
the phosphorylation of an NF-AT protein by a glucan synthase kinase like GSK-
3, or
dephosphorylation of NF-AT by a phosphatase such as calcineurin. In this
regard, the application
2 0 provides drug screening assays based on monitoring the rate of
phosphorylation of NF-AT, e.g.,
a particularly significant residues, by GSK-3 in the presence of absence of a
test compound.
Likewise, the application describes drug screening assays based on monitoring
the rate of
dephosphorylation of NF-AT, e.g., a particularly significant residues, by
calcineurin in the
presence of absence of a test compound.
2 5 On salient feature to our discovery that GSK-3 is a specific phosphatase
for NF-AT
proteins, and the elucidation of particular residues (e.g., the NLS sites) as
substrates for GSK-3,
the present invention also provides peptide and peptidimimetic inhibitors of
GSK-3. Such
inhibitors can correspond to 4 or more residues of NF-AT, and can be in the
range of 4-25, more
preferably 4-15 and even more preferably 4-10. The inhibitors preferably have
Ki's for inhibition
3 0 of GSK-3 phosphorylation of an NF-AT of 1 pM or less, more preferably of
100mM or less, and
even more preferably of 1nM or less.
Preferably, the peptide or peptidimimetic inhibitor of GSK-3 includes a
phosphoserine
residue, or, even more preferably, an analog
thereof. The phosphoserine moiety can be
3 5 represented by the general formula
-N
H
O
73
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
where R is selected from a group consisting of
D1 O
- (CHz) i-X-P-ORls ~ - (CHz) i-X-P-ORls ~ - (CHz) i-B~ORls
I
ORls Dz ORis
O
- (CH ) i-~-OR I
z I is / - (CHz) i-As'ORls ~ - (CHz) i-BeF3 , and - (CHz) i-A1 F3
OR16 ORis
wherein i is zero or an integer in the range of 1 to 6; X is absent or
represents O, S, or N;
D1 represents O or S; D2 represents N3, SH2, NH2, or N02; and Ri 5 and R16
each independently
represent hydrogen, a lower alkyl, or a pharmaceutically acceptable salt, or
Rls and R16 taken
together with the O-P-O, O-B-O, O-V-O or O-As-O atoms to which they are
attached complete
a heterocyclic ring having from 5 to 8 atoms in the ring structure. In a
preferred embodiment, the
phosphoserine is a non-hydrolyzable phosphoserine analog.
For illustrative purposes, peptide analogs of the present invention can be
generated using
benzodiazepines, substituted gama lactam rings (Garvey et al. in Peptides:
Chemistry and Biology,
G.R. Marshall ed., ESCOM Publisher: Leiden, Netherlands, 1988, p123), C-7
mimics (Huffman
et al. in Peptides: Chemistry and Biologyy, G.R. Marshall ed., ESCOM
Publisher: Leiden,
Netherlands, 1988, p. 105), keto-methylene pseudopeptides (Ewenson et al.
(1986) JMed Chem
29:295; and Ewenson et al. in Peptides: Structure and Function (Proceedings of
the 9th American
Peptide Symposium) Pierce Chemical Co. Rockland, IL, 1985), ~i-turn dipeptide
cores (Nagai et
al. (1985) Tetrahedron Lett 26:647; and Sato et al. (1986) JChem Soc Perkin
Trans 1:1231), ~i-
aminoalcohols (Gordon et al. (1985) Biochem Biophys Res Commun126:419; and
Dann et al.
(1986) Biochem Biophys Res Commun 134:71), diaminoketones (Natarajan et al.
(1984) Biochem
Biophys Res Common 124:141), and methyleneamino-modifed (Roark et al. in
Peptides:
Chemistry and Biology, G.R. Marshall ed., ESCOM Publisher: Leiden,
Netherlands, 1988, p134).
Also, see generally, Session III: Analytic and synthetic methods, in in
Peptides: Chemistry and
Biology, G.R. Marshall ed., ESCOM Publisher: Leiden, Netherlands, 1988)
In other exemplary embodiments, the peptidomimetic can be derived as a retro-
inverso
analog of a peptide sequence, such as that described by the Sisto et al. U.S.
Patent 4,522,752, as
74
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
a'retro-enatio analog of the peptide, as a trans-olefin derivatives such as
can be synthesized
according to the method of Y.K. Shue et al. ( 1987) Tetrahedron Letters
28:3225, or as a
phosphonate derivative, such as can be adapted from such synthesis schemes as,
Loots et al. in
Peptides: Chemistry and Biology, (Escom Science Publishers, Leiden, 1988, p.
118); Petrillo et
al. in Peptides: Structure and Function (Proceedings of the 9th American
Peptide Symposium,
Pierce Chemical Co. Rockland, IL, 1985).
The invention also provides methods for providing activation of NF-AT
dependent
biological activities by introducing into a cell a constitutively active NF-
AT, e.g., an NF-AT
polypeptide in which the NLS cannot form intramolecular interactions with
other parts of the
protein, thereby resulting in an NF-AT protein which constitutively
translocates to the nucleus.
Such NF-AT proteins can, e.g., have substitutions of serines in the SRR, SP1,
SP2, and/or SP3
regions of the NF-AT protein, such that at least one NLS of the NF-AT is
"unshielded". Thus, the
activity of a constitutively active NF-AT protein can be modulated by
modulating the expression
of a gene encoding the protein. For example, the gene encoding the
constitutively active NF-AT
can be placed under the control, e.g., transcriptional control, of an
inducible regulatory element.
Another constitutively active NF-AT polypeptide that can be used according to
the
methods of the invention comprises one or more additional NLS. These can be NF-
AT NLS or
heterologous NLSs, such as the SV40 large T antigen NLS.
Alternatively, NF-AT dependent biological activities can be modulated as
follows. An NF
2 o AT polypeptide that is mutated so that it is constitutively active, is
fused to a ligand binding
domain and expressed in a cell which further expresses a cytoplasmic retention
domain fused to
a ligand binding domain, such that in the presence of a dimerizer, the two
fusion proteins are cross
linked and the NF-AT protein is retained in the cytoplasm. Translocation of
the NF-AT protein
into the nucleus can be induced by the absence of, or removal of, the
dimerizer.
2 5 In yet another embodiment, the invention provides a method for regulating
the expression
of NF-AT by regulating the expression of a dominant negative mutant of NF-AT.
In one
embodiment, a dominant negative mutant is an NF-AT protein that is incapable
of translocating
to the nucleus, e.g., an NF-AT polypeptide in which at least one, but
preferably two NLSs are
inactivated, e.g, by mutation. Such mutant NF-AT polypeptides are still
capable of interacting
3 0 with calcineurin, and will thereby compete away calcineurin from
endogenous NF-AT molecules.
In yet another embodiment, the invention provides a method for regulating
translocation
of a heterologous polypeptide across the nuclear membrane that is not
dependent on intracellular
calcium. Accordingly, a heterologous polypeptide is fused an NLS of NF-AT,
e.g., the C-terminal
NLS, and to an NF-AT portion selected from the group consisting of SRR, SP1,
SP2, and SP3.
3 5 Preferably, the NF-AT portion is an SRR. The cellular localization of a
heterologous polypeptide
fused to these portions of NF-AT in a cell will depend on the presence of
calcium. Thus, in
CA 02352599 2001-05-23
WO 00/30671 PCT/l1S99/27862
normal conditions, it is expected that the protein will be located in the
cytoplasm of the cell and
that, in the presence of a calcium ionophore, the polypeptide will translocate
to the nucleus.
Alternatively, a heterologous polypeptide can be fused to an NLS only and its
intracellular location
modulated by addition to the cell of a compound, e.g, a peptide that interacts
with the NLS.
Moreover, GSK-3 has been shown to be involved in dorsal-ventral pattern
formation in
Xenopus (He et al., Nature. 374, 617 (1995)) and in segment polarity
determination in Drosophila,
where it was discovered as zest white 3 or shaggy (Bourouis et al., EMBO, J.
9, 2877 (1990);
Siegfried et al., Nature 345, 825 (1990)}. GSK-3 is a negative regulator of
the wnt signaling
pathway and it has been shown that loss of function and dominant negative
mutations in GSK-3
1 o beta lead to activation of the wnt pathway in Drosophila and Xenopus.
Furthermore, the Wingless
signaling pathway to GSK-3 is conserved in mammals (Cook et al., EMBO, J. 15,
4526 (1996);
Stambolic et al., Curr. Biol. 6, 1664 (1996)) and the wnt signaling pathway
plays a central role in
the development of invertebrates and vertebrates. Thus, it is likely that the
Wingless signaling
pathway controls the nuclear export of NF-AT family members in the tissues
where these genes
are coexpressed. Accordingly, the invention also provides methods for
regulating NF-AT
translocation in a cell, comprising contacting the cell with a compound which
modulates the
Wingless signaling pathway. For example, translocation of NF-AT from the
cytoplasm to the
nucleus can be stimulated with activators of the wnt signaling pathway. In yet
another
embodiment, the invention provides a method for modulating the Wingless
signaling pathway,
2 0 comprising, e.g., modulating the activity of GSK-3. In one embodiment, the
activity of GSK-3
is inhibited with an NF-AT peptide capable of binding to GSK-3, thereby
activating the Wnt
signaling pathway..
The methods of the invention can be used for treating or preventing in a
subject a disease
2 5 or condition that is associated with abnormal or aberrant T cell
activation. Diseases or conditions
involving, i.e., caused by or contributed to by, an excessive T cell
activation include cancers, such
as leukemias, inflammation, or autoimmune diseases. Alternatively, the methods
of the invention
can also be used to immunosuppress a subject, e.g., a recipient of a graft
such as an organ or bone
marrow transplant patient. Diseases or conditions involving an abnormally low
T cell activation
30 include immunosuppressed states, e.g., AIDS or conditions in which a
subject has an infection.
Thus, for example, a subject having a viral or bacterial infection can be
treated by administering
to the subject a compound which activates NF-ATc thereby activating the T
cells of the subject
and stimulating the immune system of the subject for fighting against the
infection. For example,
one can administer, either locally or systemically to the subject a
pharmaceutically effective
3 5 amount of a compound which increases NF-ATc localization in the nucleus,
such as a compound
which inhibits the intramolecular interaction between an NLS in NF-ATc and
another portion of
76
CA 02352599 2001-05-23
WO 00/30671 PCTlUS99/27862
the NF-AT molecule. On the contrary, where a subject has an autoimmune
disease, it is desirable
to inhibit or reduce T cell activation. Thus, in this situation, one can
administer to the subject a
pharmaceutically effective amount of a compound which increases NF-ATc
localization in the
cytoplasm, e.g., a compound which shields the NLS of NF-ATc. Alternatively, a
compound that
activates GSK-3 and/or PKA activity and NF-AT phosphorylation can be
administered to the
subject.
Furthermore, since the Wingless signaling pathway involves GSK-3, the
invention
provides methods for treating diseases or disorders associated with the
Wingless signaling
pathway, such as cancer, e.g., breast cancer. For example, a disease can be
treated or prevented
by administering to a subject having such a disease a compound which is
capable of inhibiting
GKS-3, e.g, an NF-AT peptide capable of binding to GSK-3.
In one embodiment, the NF-AT antagonists described herein can be used to
inhibit
unwanted vascular tissue proliferation, e.g., either in tissue culture or in
vivo. For example, the
subject antagonists can be used to inhibit growth of cardiac myocytes and/or
nonmyocytes
(fibroblast). In other embodiments, the subject method can be used to inhibit
arteriolar smooth
muscle proliferation.
For example, NF-AT antagonists, particularly antagonists of NF-ATc3 and.or NF-
ATc4,
can be used to inhibit cardiac and/or vascular hypertrophy, e.g., as part of
treatment or prophylaxis
for cardiac injury. For example, the subject antagonists can be used to treat
or prevent
hypertrophic conditions following angioplasty, e.g., coronary angioplasty,
treatment of restenosis
and aortic stenosis, as well as part of a post-myocardial infarction regimen.
The subject antagonists can also be used as part of treatment for
hypertension, especially
hypertensive heart disease.
2 5 In other embodiments, the NEAT antagonists of the present invention can be
used as part
of treatment for cardiomyopathy due to pathological stimuli, such as viral
myocarditis.
In still other embodiments, the antagonists can be used as part of treatment
to counter effect
drugs which have an adverse side effect of promoting cardiac hypertrophy, such
as thyroid
hormone treatment
3 0 Thus, in a preferred embodiment, the invention provides methods for
preventing and
treating diseases and conditions in a mammal, such as a human, relating to NF-
ATc mediated
cellular hypertrophy, in particular, myocyte hypertrophy. In particular, the
invention provides a
method for treating a subject experiencing heart failure to prevent or lessen
hypertrophy. In an
illustrative embodiment, the method comprises administering chronically to a
mammal in need of
3 5 such treatment a therapeutically effective amount of an NF-AT antagonist,
preferably an NF-ATc4
antagonist.
77
CA 02352599 2001-05-23
WO 00/30671 PCT/US99127$62
Optionally, the NF-ATc4 antagonist is chronically administered in combination
with an
effective amount of an antagonist to endothelin or LIF (see U.S. Pat. No.
5837241). Additional
optional components include a cardiotrophin inhibitor such as a CT-1
antagonist, an ACE
inhibitor, such as captopril, and/or human growth hormone and/or IGF-I in the
case of congestive
heart failure, or with another myocardiotrophic, anti-arrhythmic, or inotropic
factor in the case of
other types of heart failure or cardiac disorder, another anti-hypertrophic or
myocardiotrophic
factor in the case of other types of heart failure or cardiac disorder.
Treatment of cardiac
hypertrophy with agents is further described in U.S. Patent No. 5,573,762 by
Ferarra et al.
ACE inhibitors which may be used as part of a conjoint therapeutic regimen are
angiotensin-converting enzyme inhibiting drugs which prevent the conversion of
angiotensin I to
angiotensin II. The ACE inhibitors may be beneficial in congestive heart
failure by reducing
systemic vascular resistance and relieving circulatory congestion. ACE
inhibitors include drugs
designated by the trademarks Accupril Registered TM (quinapril), Altace
Registered TM
(ramipril), Capoten Registered TM (captopril), Lotensin Registered TM
(benazepril), Monopril
Registered TM (fosinopril), Prinivil Registered TM (lisinopril), Vasotec
Registered TM
(enalapril), and Zestril Registered TM (lisinopril).
The present invention can also be combined with the administration of drug
therapies for
the treatment of heart diseases such as hypertension. For example, an NF-AT
antagonist can be
administered with endothelin receptor antagonists, for example, an antibody to
the endothelin
2 0 receptor, and peptide or other small molecule antagonists; beta -
adrenoceptor antagonists such as
carvedilol; alpha 1-adrenoceptor antagonists; anti-oxidants; compounds having
multiple activities
(e.g., beta -blocker/ alpha -blocker/anti-oxidant); carvedilol-like compounds
or combinations of
compounds providing multiple functions found in carvedilol; growth hormone,
etc.
2 5 K. 1t
The compounds of the invention can be provided in the fonm of kits, for use in
treating,
preventing, or diagnosing diseases or conditions in which one desires to
modulate the activity of
T cells. For example, the invention provides kits for activating NF-ATc in a
subject, comprising
a compound which is capable of inhibiting intramolecular interaction of an NLS
in NF-AT, or
3 0 which is capable of inhibiting GSK-3 and/or PKA.
In a preferred embodiment of the invention, the kit of the invention provides
reagents for
determining the level of immunosuppression of a subject, such as a subject who
is undergoing a
treatment with an immunosuppressive drug. In one embodiment, the kit comprises
a reagent for
determining the cellular localization of NF-AT, such as an antibody that
specifically binds to NF-
3 5 AT. Other reagents that can be included in the kit are control reagents or
standards, against which
the results of the test can be compared. In some embodiments, the standard is
a table or curve
78
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
indicating values in normal, e.g., non immunosuppressed, individuals. The kit
may also contain
other reagents, such as a secondary reagent, e.g., fluorescein labeled
antibody and any buffer. In
addition to monitoring the extent of immunosuppression of a subject, the kit
of the invention can
also be used to predict the sensitivity of a subject to a certain drug, e.g.,
an immunosuppressive
drug.
L. Methods for Forensic Identification
The NF-AT~ polynucleotide sequences of the present invention can be used for
forensic
identification of individual humans, such as for identification of decedents,
determination of
paternity, criminal identification, and the like. For example but not
limitation, a DNA sample can
be obtained from a person or from a cellular sample (e.g. , crime scene
evidence such as blood,
saliva, semen, and the like) and subjected to RFLP analysis, allele-specific
PCR, or PCR cloning
and sequencing of the amplification product to determine the structure of the
NF-AT~ gene region.
On the basis of the NF-AT~ gene structure, the individual from which the
sample originated will
be identified with respect to his/her NF-AT~ genotype. The NF-AT~ genotype may
be used alone
or in conjuction with other genetic markers to conclusively identify an
individual or to rule out the
individual as a possible perpetrator.
In one embodiment, human genomic DNA samples from a population of individuals
(typically at least 50 persons from various racial origins) are individually
aliquoted into reaction
2 o vessels (e.g., a well on a microtitre plate). Each aliquot is digested
(incubated) with one or more
restriction enzymes (e.g., EcoRI, HindIII, SmaI, BamHI, SaII, NotI, AccI,
ApaI, BgIII, XbaI, PstI)
under suitable reaction conditions (e.g., Vie, New England Biolabs 1992
catalog). Corresponding
digestion products from each individual are loaded separately on an
electrophoretic gel (typically
agarose), electrophoresed, blotted to a membrane by Southern blotting, and
hybridized with a
2 5 labeled NF-AT~ probe (e.g., a full-length human NF-ATc cDNA sequence of
Fig. 1 ). Restriction
fragments (bands) which are polymorphic among members of the population are
used as a basis
to discriminate NF-AT~ genotypes and thereby classify individuals on the basis
of their NF-AT~
genotype.
Similar categorization of NF-AT~ genotypes may be performed by sequencing PCR
3 0 amplification products from a population of individuals and using sequence
polymorphisms to
identify alleles (genotypes), and thereby identify or classify individuals.
The present invention is further illustrated by the following examples which
should not be
construed as limiting in any way. The contents of all cited references
(including literature
3 5 references, issued patents, published patent applications as cited
throughout this application are
hereby expressly incorporated by reference. The practice of the present
invention will employ,
79
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
sinless otherwise indicated, conventional techniques of cell biology, cell
culture, molecular
biology, transgenic biology, microbiology, recombinant DNA, and immunology,
which are within
the skill of the art. Such techniques are explained fully in the literature.
See, for example,
Molecular Cloning A Laboratory Manual, 2"d Ed., ed. by Sambrook, Fritsch and
Maniatis (Cold
Spring Harbor Laboratory Press: 1989); DNA Cloning, Volumes I and II (D. N.
Glover ed., 1985);
Oligonucleotide Synthesis (M. J. Gait ed., 1984); Mullis et al. U.S. Patent
No: 4,683,195; Nucleic
Acid Hybridization(B. D. Hames & S. J. Higgins eds. 1984); Transcription And
Translation (B.
D. Hames & S. J. Higgins eds. 1984); Culture Of Animal Cells (R. I. Freshney,
Alan R. Liss, Inc.,
1987); Immobilized Cells And Enzymes (IRL Press, 1986); B. Perbal, A Practical
Guide To
Molecular Cloning (1984); the treatise, Methods In Enzymology (Academic Press,
Inc., N.Y.);
Gene Transfer Vectors For Mammalian Cells (J. H. Miller and M. P. Calos eds.,
1987, Cold Spring
Harbor Laboratory); Methods In Enzymology, Vols. 154 and 155 (Wu et al. eds.),
Immunochemical Methods In Cell And Molecular Biology (Mayer and Walker, eds.,
Academic
Press, London, 1987); Handbook Of Experimental Immunology, Volumes I-IV (D. M.
Weir and
C. C. Blackwell, eds., 1986); Manipulating the Mouse Embryo, (Cold Spring
Harbor Laboratory
Press, Cold Spring Harbor, N.Y., 1986).
2 0 Experimental Examples
vervi w
We have purified two related proteins encoded by separate genes that
represent the preexisting or cytosolic components of NF-AT. Expression of a
full length cDNA
for one of these proteins, NF-AT~, activates the IL-2 promoter in non-T
lymphocytes, while a
2 5 dominant negative of NF-AT~ specifically blocks activation of the IL-2
promoter in T
lymphocytes, indicating that NF-AT~ is required for IL-2 gene expression and
is responsible for
the restricted expression of IL-2. NF-AT~ RNA expression is largely restricted
to lymphoid tissues
and is induced upon cell activation. The second protein, NF-ATP, is highly
homologous to NF-
AT~ over a limited domain, but exhibits wider tissue distribution and is
highly expressed in tissues
3 0 characterized by Ca++-dependent regulation. Together these proteins are
members of a new
family of DNA binding proteins, which are distantly related to the Dorsal/Rel
family (Nolan and
Baltimore (1992} Current Biologv Ltd. 2: 211-220). Agents that increase
intracellular Ca++ or
that activate protein kinase C independently produce alterations in the
mobility of NF-AT~,
indicating that distinct signaling pathways converge on NP-AT~ to regulate its
function.
3 5 Since our previous work indicated that the cytosolic component of NF-AT
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
was present at relatively low concentrations in human lymphoid cell lines
(Northrop et al. (1993)
J. Biol. Chem. X6_8: 2917-2923), we chose to purify NF-AT~ from bovine thymus.
Amino acid
sequence, obtained from 6 peptides, was used to isolate two overlapping human
cDNA clones
spanning 2742 nucleotides (Fig. 1 ). The cDNA encodes a protein of 716 amino
acids with a
predicted molecular weight of 77,870. An in-frame stop codon upstream from the
initiator
methionine indicates that the entire NF-AT~ protein is encoded by this cDNA. A
unique repeated
sequence of 13 residues was also identified. The carboxy-terminal half of NF-
AT~ shows limited
similarity to the DNA binding and dimerization regions of the Dorsal/Rel
family of transcription
factors (Fig. 4, for review, Nolan and Baltimore (1992) Current Biolo~v. Ltd.
2_: 211-220)
however, NF-AT~ appears to be the most distantly related member of the group.
There are a
significant number of amino acid changes resulting in charge reversals between
the Rel family
members and NF-AT~, suggesting that charge might be conserved at these
positions to maintain
salt bridges. Six additional peptides obtained from the purified bovine
protein are derived from
the bovine homolog of NF-ATp, a cDNA fragment of which was reported by
McCaffrey et al.
(1993) ci a 262: 750-754). Comparison of NF-AT~ and NF-ATp reveals that they
are products
of distinct genes with 73% amino acid identity in the Rel similarity region
(Fig. 4), however, there
is very little similarity outside this region. A marine cDNA for NF-AT~ was
isolated and the
predicted protein was found to be 87% identical to human NF-AT~, and
distinctly different from
marine NF-ATP.
F~eample 1: Determination of the nucleotide and amino acid seauence of human
NF-
1~T cDNA
This example represents the isolation and purification of this novel human NF-
AT
protein, NF-AT~, the determination of the amino acid sequence of its fragments
and the isolation
2 5 and sequencing of the cDNA clone encoding this protein.
The protein was purified from bovine thymus glands obtained from newborn
calves.
Approximately 20 bovine thymuses were homogenized to make a cytosolic extract
which was then
subjected sequentially to 1 ) ammonium sulfate precipitation, 2) sulphopropyl
Sepharose
chromatography, 3) heparin agarose chromatography, 4) affinity chromatography
using a
3 0 multimerized binding site for NF-AT, with the sequence 5'-
ACGCCCAAAGAGGA.AAATTTGTTTCATACA-3' (SEQ ID NO: 73) coupled to sepharose
CL4B, and 5) HPLC on a reverse phase C4 column. The resulting purified protein
was subjected
to cleavage with LysC/ArgC and fragments isolated by HPLC. The sequences of
these individual
81
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
fragments were then determined by automated Edman degradation. Sequences
obtained included:
LRNSDIELRKGETDIGR (SEQ ID NO: 74) and LRNADIELR.(SEQ ID NO: 75). Degenerate
oligos con-esponding to GETDIG (SEQ ID NO: 76) (reverse primer) and RNADIE
(SEQ ID NO:
77)(forward primer) were made. The degenerate oligo PCR primers had the
following sequences:
A forward: (A/C)GIAA(C/T)GCIGA(C/T)AT(A/C/T)GA(A/G) (SEQ ID NO: 78)
A reverse: ICC(A/G/T)AT(A/G)TCIGT(C/T)TCICC (SEQ ID NO: 79)
To isolate the cDNA, oligonucleotide probes were made corresponding to
the determined amino acid sequence and used as PCR primers to isolate a 45
base fragment from
bovine cDNA prepared from the bovine thymus. The bovine PCR product comprised
the
1 o nucleotide sequence CTG CGG AAA which encodes -L-R-K-. The same 45 by
fragment can be
amplified from human and mouse sources.
This bovine PCR product was then used to screen a cDNA library of the
human Jurkat T cell line. Clones were isolated at frequencies of about 1 in
100,000 to 1 in
200,000. A total of five human cDNA clones of various lengths were isolated.
Two overlapping
clones, one containing the 5' end and one containing the 3' end were ligated
together using a
unique EcoRI restriction site present in each clone, to produce a full-length
cDNA which
corresponded in length to the messenger RNA determined by Northern blotting.
The sequence of the NF-AT~ cDNA was determined by the Sanger method
and the complete nucleotide and predicted amino acid sequence is shown in Fig.
1. The initiator
2 0 methionine indicated in Fig. 1 (boldface, indicated) was determined by
fusing this reading frame
to a glutathione transferase gene and transfecting the resultant clone into
bacteria. The resultant
clone produced a fusion protein of the proper molecular weight, indicating
that the reading frame
designated with the initiator methionine is indeed the correct reading frame.
The position of the
stop codon was determined by a similar procedure. In addition, the stop codon
con-esponds to the
2 5 reading frame for nine of the determined amino acid sequences.
The total NF-AT~ protein structure was aligned against individual Rel
proteins using a Macintosh shareware program called DOTALIGN utilizing the
alignment
parameters of the FASTA programs. Significant homology was observed that
corresponded to the
Rel domains of these proteins. Enhanced amino acid residue alignment was done
using ALIGN
3 0 from the same suite of programs. Alignment of the Rel similarity regions
of NF-ATE and NF-ATp
was done by hand with no insertions necessary, The Miyata alphabet (Miyata et
al. (1979) .J Mol.
vol. ~2: 214-236) was used to determine similar residues. Fig. 4 shows results
of such sequence
alignments.
82
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Example 2: Expression of NF-AT~ in T and non-T cells
The cDNA shown in Fig.- was fused to the Hemophilus influenza hemaglutinin
(HA) 12 amino acid epitope tag in the determined reading frame and operably
linked to the SRa
promoter in the vector pBJS (Lin et al, 1990, Science 249:677-679). The
resultant construct was
transiently transfected by electroporation into Jurkat human T lymphocytes,
and into Cos
fibroblast cells. Expression of the epitope-tagged NF-AT~ protein was
determined by Western
blotting of whole cell extracts prepared from the transfected cells, using an
antibody (12CA5,
Berkeley Antibody Co., CA) that detects the HA epitope. Fig. 2 shows that NF-
AT~ cDNA
construct is able to express a protein of appoximately 120 kDA corresponding
precisely in size to
that of the purified protein, in both Jurkat T cells and Cos cells (see lanes
3 and 6 labeled NF-AT'.
Lane 2 shows as control, NF-AT without the epitope tag which cannot be
detected in the Western
blot).
Example 3: Transfection of NF-ATc activates transcription in both Cos and
Jurkat
The NF-ATc cDNA was operably linked to a portion of the SV40 early gene
promoter and the HIV transcription regulatory regions in the pBJ vector. This
expression vector
was co-tranfected into Jurkat and Cos cells with either a) three copies of NF-
AT binding site
linked to and directing transcription of luciferase (results shown in Fig. 3A
and 3B) the entire IL-2
2 0 enhancer/promoter directing transcription of luciferase (results shown in
Fig. 3B}. Cytosolic
extracts were prepared and luciferase assays carned out by standard procedures
(de Wet et al,
1987, Mol. Cell. Biol. 7:724-837).
The results demonstrate that in both Cos cells and Jurkat cells,
overexpression of
the NF-AT~ protein dramatically enhances NF-AT-dependent transcription by 50-
1000 fold (see
2 5 Fig. 3A). In addition, overexpression of the NF-ATc protein in Cos cells
activates the IL-2
promoter, which in the absence of NF-AT~ cannot otherwise be activated (see
Fig. 3B).
These results indicate that the cDNA clone encodes a functional NF-AT~ protein
and that NF-AT~ is the protein which restricts expression of interleukin-2 to
T cells.
3 0 Example 4: NFAT~ mRNA and Protein Expression
NF-AT~ mRNA is absent in Hela cells (Fig. S, panel _a, lane 7), a cell line
incapable of IL-2 or NF-AT-dependent transcription, but is inducible in Jurkat
cells (Fig. 5, panel
_a) . This induction is sensitive to cyclosporin A, (CsA), indicating that NF-
AT~ may participate
83
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/Z7862
iri an auto-stimulatory loop as CsA has been shown to block its nuclear
association (Flanagan et
al. ( 1991 ) Na- tore ~?5 : 803-807). Two B cell lines, muscle tissue, Hep G2
cells and myeloid
leukemia cells do not express NF-AT~ mRNA (Fig. 5, panel ~). These
observations are consistent
with the observed T cell-restricted pattern of IL-2 transcription and NF-AT
activity. Previous
studies (Verweij et al. (1990) J. Biol. Chem 265: 15788-15795) revealed NF-AT-
dependent
transcription predominantly in spleen, thymus and skin of transgenic mice
expressing an NF-AT-
dependent reporter gene. Consistent with these observations, murine NF-AT~
mRNA shows the
same pattern of expression (Fig. 5 panel ~). Small amounts of NF-AT~
expression are seen in lung
and heart, however, this may be due to contamination with circulating T cells.
Murine NF-ATP
mRNA, also assayed by quantitative ribonuclease protection, was found to be
expressed at
approximately equal levels in brain, heart, thymus and spleen (Fig 5, panel
c_) . In contrast to NF-
AT~, NF-ATP was not inducible by PMA and ionomycin (Fig 5, panel ~).
METHODS. Specific human or mouse NF-AT~ or mouse NF-ATp cDNA
fragments were used as templates for the synthesis of RNA transcripts.
Ribonuclease protection
was done according to Melton et al. (1984) Nucl. Acids. Ress ~: 7035-7056)
using 10 ,ug of total
RNA. Splenocytes and thyrnocytes were isolated and treated as described
(Verweij et al. (1990)
J. Biol. Chem 2,~: 15788-15795) before isolating RNA, otherwise whole tissue
was used.
Example 5: Functional Expression of NF-AT
2 0 NF-AT luciferase and IL-2 luciferase have been described (Northrop et al.
(1993) J. Biol. Chems ~: 2917-2923). (328 luciferase was constructed by
inserting a trimerized
HNF-I recognition site ((328) in place of the NF-AT recognition sites in NF-AT
luciferase. The
plasmid pSV2CAT (Gorman et al. (1982) Mol. Cell. Biol. 2_: 1044-1050) was used
as an internal
control for transfection efficiency. Cells were transfected with 1.5 ug of
luciferase reporter and
3 ug of expression construct as described. After 20 hours of growth, cells
were stimulated for 8
hrs. with 20 ng/ml PMA plus or minus 2 uM ionomycin, and harvested for
luciferase (de Wet et
al. (1987) Mol. Cell. Biol. 7: 725-737) and CAT assays (Gorman et al. (1982)
Mol. Cell. Biol. 2_:
1044-1050).
Cos cells were transfected with epitope tagged NF-AT~ as described. Cos
3 0 cells, Jurkat cells, and murine thymocytes were stimulated for 3hr. with
PMA and ionomycin.
Hela cells were stimulated for 3hr with PMA alone and nuclear extracts
prepared as described
(Fiefing et al. (1990) Genes & Dev. 4: 1823-1834). Cytosols were prepared from
non-stimulated
Cos cells. Gel mobility shifts were performed as previously described
(Flanagan et al. (1991)
84
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
a r 52: 803-807; Northrop et al. (1993) J. Biol. Chem. X68: 2917-2923).
Antisera were raised
in mice immunized with bacterially expressed glutathione S-transferase fusion
proteins using the
vector pGEX-3X (Pharmacia) and purified on glutathione agarose. Fusion
proteins contained NF-
AT~ residues 12 to 143 (immune-1) and 12 to 699 (immune-2).
NF-AT~, expressed in non T cell lines specifically activated transcription
from the NF-AT site and the IL-2 promoter, (Fig. 6 panel ~ (left), and Fig. 6
panel b_) . In
transiently transfected Jurkat cells, overexpression of NF-AT~ activated an NF-
AT-dependent
promoter but not an HNF-1 dependent promoter (Fig. 6 panel ~ (right)) or an AP-
1-dependent
promoter. Transfection of the NF-AT~ cDNA gives rise to DNA binding activity
that is
indistinguishable from endogenous NF-AT (Fig. 6 panel ~, lanes 1-4). Antibody
directed against
the HA epitope encoded by the transfected cDNA induces a supershift of the NF-
AT complex
indicating that NF-AT~ participates in this activity. The nuclear NF-AT
activity in transfected Cos
cells comigrates with, and has the same binding specificity as, the native
nuclear complex in T-
cells (Fig. 6 panel c_, lanes 4-11). cytosolic extracts from NF-ATc,
transfected Cos cells can
reconstitute NF-AT DNA binding activity when mixed with Hela nuclear extract
(Fig. 6 panel ~,
lanes 12-16) as do cytosolic extracts from T-cells (Flanagan et al. (1991) Na
ure 352: 803-807;
Northrop et al. (1993) J. Biol. Chem. 6~8: 2917-2923). Antisera raised against
bacterially
expressed fragments of NF-ATE that have no similarity to NF-ATp are able to
induce a supershift
of the endogenous NF-AT complex, but not the AP-1 complex, from Jurkat cells
or thymocytes
2 0 (immune-I and immune-2 respectively, Fig. 6 panel ~). Immune-2 antisera
reduced the DNA-
protein complex produced using murine thymic nuclear extracts significantly,
consistent with the
relatively equal representation of NF-AT~, and NF-ATP peptides in the purified
protein from
bovine thymus.
2 5 Example 6: NF-AT dominant negative mutant assayed in transient
transfection
ass~rs
A dominant negative NF-AT~, prepared after extensive deletion analysis of
the cDNA, indicated that the amino terminal domain would block NF-AT-dependent
function
without affecting binding. This region of the cDNA is not found in NF-ATp and
hence can be used
3 0 to assess the contribution of NF-AT~ to the activation of the IL-2 gene.
The dominant negative
NF-AT~ used consists of a carboxy terminal truncation of the epitope tagged NF-
AT~ expression
plasmid (supra) extending to the PvuII site at amino acid 463. Transfection of
this dominant
negative resulted in more than 90% inhibition of IL-2 promoter function as
well as transcription
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
directed by the NF-AT site (Fig 7). This effect was highly specific since
transcription directed by
the AP-1 site or the RSV promoter and enhancer were relatively unaffected (Fig
7). These results
strongly indicate that NF-AT~ contributes substantially to IL-2 gene
expression in T cells.
Dominant-negative NF-AT~ polypeptides or peptidomimetics thereof can be used
as pharmaceutical antagonists of NF-AT-mediated activation of T cells. In one
variation, such
drugs can be used as commercial research reagents for laboratory testing and
analysis of T cell
activation and the like, among many other uses (e.g., immunosuppressant).
Example 7: Post-Translational Modification of NF ATi
1 o Post-translational modification of NF-AT~ was investigated in cells
treated
with agents that activate PKC or increase intracellular Ca++. Cells were
transfected with NF-AT~
as described in Fig. 2 and stimulated as shown for 2 hrs plus or minus 1
OOng/ml CsA. Whole cell
lysates were analyzed by western blotting as in Fig. 2. The bulk of NF-AT~ in
cells treated with
ionomycin migrates faster than that in non-treated cells and this mobility
shift is inhibited by CsA
(Fig. 8, lanes 1, 3-4). This is consistent with a dephosphorylation event,
possibly by direct action
of calcineurin (Clipstone and Crabtree (1992) ur ~5 : 695-697), however, any
of a large
number of processes could produce the observed mobility changes. There is
evidence that NF-ATP
is a substrate for calcineurin, however, the mobility shifts produced by
phosphatase treatment of
NF-ATp or NF-AT~ are far greater than those observed in Figure 8. These
observations indicate
2 0 that NF-AT~ is not a direct substrate of calcineurin. PMA treatment
produces a slower migrating
NF-AT~ (Fig. 8, lane 2); therefore, PKC-activated pathways likely contribute
to NF-AT activity
by modification of NF-AT~ in addition to activation of the nuclear component.
Exam In a 8: Cal~ineurin is the rate-limited for NF-ATc nuclear ent
2 5 Most tissues express one of the NF-AT family members. A variety of cell
types,
including lymphocytes and fibroblasts, support the Ca2+-dependant nuclear
localization of
transfected as well as endogenous NF-ATc family members (Shibasaki et al.,
Nature, 382: 370
373). To develop an accurate assay for NF-AT translocation, NF-ATc was
expressed in COS cells
which, unlike lymphocytes, have abundant cytoplasm and hence allow easier
assessment of
3 0 cytoplasmic and nuclear localization.
COS-7 cells were maintained in Dulbecco's modified Eagle medium (DMEM;
Sigma) with 10% fetal calf serum (FCS), 100 pg/ml of penicillin G. 100 pg/ml
of streptomycin,
and 10 mM HEPS (pH 7.4) at 37°C in 5% COz. Cells were transfected by
electroporation with
86
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
1'pg of SH160c Ho et al. (1995) J. Biol. Chem. 270:19898, which encodes the
FLAG epitope
tag inserted at an XbaI site immediately 5' to the second codon of the human
NF-ATcI cDNA in
the pBJS vector (Nature, 369:497-502)), plated on glass coverslips, and
stimulated 18-14 hr post-
transfection in fresh media or fresh media supplemented with ionomycin (2~M
final) plus 10
mM (final) CaCl2 for various amounts of time at 37°C. Ionomycin was
obtained from
Calbiochem and dissolved in DMSO. Cells were also treated with FK506 at 2
ng/ml plus
ionomycin and CaCl2 or with ionomycin plus 2.5 mM EGTA for 60 min. FK506 was
added at
2 ng/ml I S min prior to addition of calcium and ionomycin. FK506 was obtained
from Fukisawa
(Chicago, IL) and dissolved in ethanol. Efficient nuclear translocation of NF-
ATc in COS cells
l0 requires both ionomycin and the elevation of extracelluar calcium. The
reason for this
requirements for Ca2+ may be that ionomycin stimulation of COS cells does not
result in a
intracellular Ca2+ level as high as stimulated lymphocytes. Cells were then
stained with the anti-
FLAG antibody as follows. Cells adhering to coverslips were fixed in 4%
paraformaldehyde and
permeabilized in 0.1% Triton X-100. The FLAG epitope was detected by
incubating with 1
pg/ml of anti-FLAG M2 antibody (Eastman Kodak Co.). The monoclonal antibody
was detected
by incubation with biotin-conjugated anti-mouse IgG (Caltag), followed by
streptavidin-FITC
and DAPI (Molecular Probes). Fluorescence was visualized with a Zeiss Axiophot
fluorescence
microscope. Fluorescent cells in which, the nucleus and plasma membrane could
be identified
were scored as containing predominantly cytoplasmic staining, predominantly
nuclear staining,
2 0 or both cytoplasmic and nuclear staining. At least 100 cells were scored
on each coverslip. Cells
undergoing mitosis or with multiple nuclei were excluded. For all deletion
constructs, the
subcellular localization was confirmed using a confocal imaging fluorescence
microscope.
As shown in the diagram in Figure 9B, as with NF-ATc3(4) (Shibasaki et al.,
Nature, 382: 370-373), the amino terminus ofNF-ATcI was sufficient for Ca2+-
regulated nuclear
2 5 import that was blocked by FK506. Furthermore, transfected NF-ATcl moved
into the nucleus
within S-15 min after ionomycin treatment.
Exit of NF-ATc from the nucleus was determined by stimulating transfected
cells
with I+Ca'~ for 1 hr and then replacing the medium with medium containing
FK506. Slides were
fixed at various time points, and the percentage of cells with cytoplasmic NF-
ATc was
3 0 determined. Cells expressing NF-ATc in the cytoplasm and those expressing
NF-ATc in both
cytoplasm and nucleus were added and divided by the total number of analyzed
expressing cells.
The results, which are shown in Figure 9C indicate that NF-AT moved out of the
nucleus and
into the cytoplasm within 30 min of FK506 addition. These translocations
occurred even if
87
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/27862
protein synthesis was inhibited. The full-length protein behaved similar to a
fusion of the amino-
terminal 418 amino acid with green fluorescent protein (NF-AT(Cn418)-GFP) (Fig
9A).
pSH 160c~418-GFP construct was made by fusing the BamHI-Notl fragment encoding
GFP from
pEGFP-1 (Clonetech) following the PvuII site at codon 418 of SH160c and was
detected using
its autofluorescence.
This time course of nuclear localization is consistent with that observed in
marine
lymphocytes activated by antigen presentation (Timmerman et al., Nature, 383:
837-840) and
indicates that COS cells can support physiologic translocation of NF-ATc. As
with NF-ATc3[4]
(Shibasaki et al., Nature, 382: 370-373), overexpression of calcineurin
enhanced the movement
of NF-AT into the nucleus of COS cells (Fig. 9A), indicating that calcineurin
is rate limiting for
the movement of NF-ATc proteins into the nucleus.
exam lr~ a 9: Addition of heterologous nuclear localization sequences to NF
AT,~
_r~ults in Ca2+-independent FK506-resistant nuclear import
The observation that overexpressed NF-ATc is cytoplasmic in Jurkat T
lymphocytes and COS cells suggests that there is not an easily saturated
cytoplasmic anchoring
protein necessary to retain NF-ATc in the cytoplasm. Transfection of the NF-
ATc 1 expression
construct over a 200-fold ranges of DNA concentration did not result in higher
levels of
constitutive nuclear localization. If NF-ATc was localized by a cytoplasmic
anchoring partner,
2 0 the addition of a fully active nuclear localization sequence (NLS) to NF-
ATc should not
overcome the cytoplasmic retention NF-ATc. Accordingly, NF-ATc 1 expression
constructs with
zero, one, or two copies of either the SV40 large T-antigen NLS encoded
between the FLAG
epitope and the second amino acid of NF-ATcI, or one or two copies of a mutant
form of the
NLS (NLS-T). The constructs bearing the SV40 NLS and mutant (NLS-T) were
created by
insertion of synthetic oligo-nucleotides at the XbaI site of pSH160c (Ho et
al. (1995) J. Biol.
Chem. 270:19898, which encodes the FLAG epitope tag inserted at an XbaI site
immediately 5'
to the second codon of the human NFOATcI cDNA in the pBJS vector (Northrop et
al. ( 1994)
Nature, 369:497-502)). The inserted NLS is CTAGTCCTAAGAAGAAGAGAAAGGTAT
(SEQ ID NO: 80); the sequence of NLS-T is CTAGTCCTAAGACGAAGAGAAAGGTAT
3 0 (SEQ ID NO: 81 ) and substitutes a threonine for a lysine (Cell, 39:499-
509). All point
substitutions were created by sequential overlap extension PCR (J. Biol.
Chem., 270:19898-
19900). Cells were then stained with an anti-FLAG antibody and the percentage
of cells with
nuclear fluorescence determined.
88
CA 02352599 2001-05-23
WO 00/30671 PCT/US99J27862
As shown in Figure 10, expression of NF-ATc 1 with zero, one, or two copies of
the SV40 large T-antigen NLS in COS cells results in a progressive increase in
constitutive
nuclear localization which was insensitive to FK506. In contrast, addition of
the mutant NLS
sequence, NLS-T (Kalderon et al. (1984) Cell, 39: 499-509), to NF-ATcI
resulted in
substantially less nuclear entry. The low level of nuclear localizations
resulting from inclusion
of NLS-T may be attributable to slight activity of this mutant, which, like
the wild-type
sequence, is enhanced when present in multiple copies (Roberts et al. (1987)
Cell, 50: 465-475).
These results argue against a mechanism of cytoplasmic localization dependent
on a dominantly
acting cytoplasmic binding protein.
Example 10: Two NLSs are each sufficient for NF-ATc nuclear tran locatiog~
NF-ATc proteins contain four groups of clustered basic residues that are
conserved among NF-ATc proteins which could possibly be NLSs (see Figure 11A).
To
determine whether these sequences have NLS activity, each of them was linked
individually to
the cytoplasmic exchange factor SOS.
The SOS expression constructs were based on the human SOS cDNA tagged at
the carboxyl terminus by a HA epitope, pSOS-E (Proc. Natl. Acad. Sci. 92:9810-
9814). PSOS-
265 was created by insertion of an oligonucleotides encoding the sequence
LECNKRKYSLNVD
(SEQ ID NO: 82) at the unique SaII site between SOS and the HA epitope. An
expression
2 0 construct encoding SOS (SOS-E) was expressed and visualized with 12CA5
antibody.
Constructs encoding SOS-E attached to residues 263-271 of NF-ATc (SOS-265) or
attached to
residues 681-685 of NF-ATc (SOS-682) were also detected with the 12CA5
antibody.
Immunoflucorescence was as described above, except that the HA epitope was
detected by
incubating with 1:2000 dilution of 12CA5 ascites.
2 5 The results, show that SOS-682, incorporating residues 682-685 of NF-ATc,
and
SOS-265, incorporating residues 265-267, are localized in the nucleus. Thus,
these two
conserved regions within NF-ATC are thus NLSs.
To determine whether these NLSs are required for nuclear import of NF-ATc,
each sequence was mutated separately and in combination within the context of
full-length NF
3 0 ATcl . A diagram of the mutations made in the NLS in the wild-type NF-ATc
sequence is shown
in Figure 11B. The NLS at residues 265-268 was changed to QIL (construct
m265). The NLS
at residues 682-685 was changed to TRTG and the construct containing this
mutation and m265
is referred to as m265 + 682. The mutant expression constructs were
transfected in COS cells,
89
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27$62
arid the cells were stimulated with I+Ca~ for 60 minutes as described above.
The percentage of
cells staining in the nucleus, cytoplasm, or both compartments was determined.
The results, which are shown in Figure 11 B, indicate that mutation of the
sequence at 265-267 from KRK to QIL reduced the extent of nuclear localization
of NF-ATC
in response to ionomycin, but up to 60% of cells show some Caz+-dependent
nuclear
accumulation of NF-ATC. Mutation of the sequence KRKIC (SEQ ID NO: 56) at
position 682-
685 to TRTG (SEQ ID NO: 55), or precise removal of these 4 residues, had no
effect on nuclear
localization of NF-ATC in response to Ca2+ elevation. However, NF-ATc
containing mutations
in both regions remains cytoplasmic after ionomycin treatment. Thus, like
other nuclear proteins
l0 with multiple NLSs (Richardson et al. (1986) Cell, 44: 77-85), the two NLSs
are partially
redundant, as suggested by the observation that either can direct cytoplasmic
SOS to the nucleus
but both must be mutated to prevent nuclear entry. These data also indicated
that both NLSs
must be inactive in the absence of Caz+stimulations.
example 11: Mutation of serines in the amino terminus leads to constitutive
nuclear localization NF-ATc
Since the amino terminus of NF-ATc is sufficient for calcineurin-dependent
nuclear entry, it was determined whether phosphorylation of the amino terminus
directed
subcellular compartmentalization of the transcription factor. The amino
terminus of each NF-
2 0 ATc protein contains three copies of a sequence referred to as the SP-
repeat motif (Ho et al.
(1995) J. Biol. Chem., 270: 19898-19900; Hoey et al. (1995) Immunity, 2: 461-
472.; Masuda et
al. (1995) Mol. Cell Biol., 15:2697-2706). An additional SRR 23 amino acids in
length lies just
amino-terminal to the first SP repeat (Ho et al. (1995) J. Biol. Chem., 270:
19898-19900) Fig.
9A). Phospho-amino acid analysis revealed that all phosphorylation is located
on serines. Two-
2 5 dimensional trypic peptide maps show many phosphopeptides derived from the
amino-tenminal
418 amino acids (see below). To determine whether phosphorylation of these
particular serines
could regulate NF-ATc localization, selected groups of conserved serines in
the SRR and SP
repeats were mutated to alanines and the subcellular localization of these
mutants was
determined in COS cells.
3 0 The SRR was mutagenized by changing the 11 serines to alanines in residues
172-
194 to forni mSRR. The first SP repeat was mutagenized by changing four
serines to alanines
in residues 199-211, to form mSPI. The second SP repeat was mutagenized by
changing serines
at 233 and 237 to alanines, mSP2. The third SP repeat was mutagenized at five
serines at 278,
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
282, 286, 290, and 299, mSP3. Other mutated constructs are shown in Figure 12.
These
substitutions were made in the construct pSH102c0418 (Nature, 369:497-502),
which encodes
the NF-AT carboxy terminal deletion construct (containing amino acid 1- 418)
and a hem-
Agglutinin (HA) epitope tag at the amino terminus.
Immumoblot of cytoplasmic (C) or nuclear (1~ extracts of cells transfected
with
wild-type (WT) or mutant forms of the NF-ATc cDNA were also done. Accordingly,
transfected
cells were treated for 60 min with media without additions (NS) or with
ionomycin and calcium
(I + CA+'), as described above, and then separated into cytoplasmic and
nuclear fractions
according to J. Biol.Chem. 270: 19898-19900, subjected to SDS-PAGE,
transferred to a
membrane and a Western blotting was performed using the M2 or 12CA5 antibody
that was
detected with anti-mouse peroxidase and chemiluminescence (Amersham).
The results indicate that mutation of all the serines within the SRR (mSRR)
leads
to constitutive nuclear localization in 100% of expressing cells that is
unaffected by FK506.
mSRR has an increased mobility on SDS electrophoresis, is present it the
nucleus by Western
blotting, and shows reduced incorporation of 32P after in vivo labeling with
orthophosphate,
consistent with the hypothesis that these serines affect the phosphorylation
state of the protein
in vivo. An NF-ATc mutant in which serines in the first SP repeat were
substituted with alanines
(mSP 1 ) is also constitutively localized to the nucleus in 100% of expressing
cells and shows a
reduction in molecular weight. Similar results were obtained in NF-ATc mutants
with S-~A
2 0 mutations in the first and third SP repeat (mSP 13) and in versions in
which mutations were
engineered in all three SP repeats (mSP123) or combined with the mutations in
the SRR (mSRR
+ SP123). The subcellular localization of each of these mutant forms of NF-ATc
in
constitutively nuclear if they are expressed in Jurkat cells, indicating that
these phosphorserines
control subcellular localization in a variety of cell types. The unregulated
nuclear entry of the
2 5 S-yA mutations is not likely to be caused by denaturation of the protein,
because each of these
mutated forms of the mutated forms of NF-ATc participate in NF-ATc-dependent
transcription.
Since S->A mutation of the SRR resulted in the smallest alteration in apparent
molecular weight, it was likely that this region might contain the smallest
numbers of critical
phosphorserines necessary for cytoplasmic localization. These mutants can be
dephosphorylated
3 0 further after transfection into cells and ionomycin treatment, indicating
that the SRR mutant is
still a substrate for a phosphatase, possibly calcineurin. Thus further
analysis by mutation of
smaller blocks of serines in the SRR were performed. Alanine substitution at
resides 172-176,
178-181, and 184-188, but not residues 191-194, resulted in nuclear
accumulations of NF-ATc
91
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
iri 100% of expressing cells in the absence of Caz+/calcineurin signaling
(see, Figure 12A).
Interestingly, the mutants with constitutive nuclear localization remain in
the nucleus after
adding FK506, a treatment that leads to rapid cytoplasmic accumulation of wild-
type NF-ATc,
NF-ATp, or NF-ATc3 that has been transported to the nucleus by stimulation
(Flanagan et al.
(I991) Nature, 352: 803-807; Shibazaki et al. (1996) Nature, 382: 370-372;
Timmerman et
al.(1996) Nature, 383: 837-840. Thus, these results indicates that
phosphorylation of these
residues is necessary for export of NF-ATc from the nucleus.
~eample 12: Calcineurin dgphosphorylates serines in NF AT
To determine whether calcineurin could be the phosphatase directing nuclear
entry, the ability of calcineurin to specifically dephosphorylate the residues
associated with
nuclear entry was investigated.
NF-AT GST fusion proteins were prepared as follows. Residues 196-304 of NF-
ATcI (Northrop et al. (1994) Nature, 369:497-502) were cloned into the SmaI
site of pGEX-3X
to generate pGSP. A GST fusion protein in which the S-~A substitutions in all
three SP repeats
described above was similarly constructed, pGAP, with 9 S and 10 T residues
remaining. The
GST fusion proteins were phosphorylated by incubating 1 pg of fusion protein
immobilized on
glutathione-Sepharose with whole brain extract (55 p.g protein} (prepared as
described below)
and with 100 um ATP and [y-3zP]ATP (400 uCi/pmole) in 50 ~l of kinase buffer
(20 mtvt Tris
2 0 at pH 7.5, 10 mM MgClz, 1 mNt DTT) for 30 min. at 30°C. Kinase
reactions were teminated by
washing the agarose beads three times in 1 ml of calcineurin buffer. The
fusion proteins were
then incubated with calcineurin as above, or treated with 2 units of shrimp
alkaline phosphatase
(U.S. Biochemical) or 5 units of protein phosphatase I (Boehringer Mannheim)
in the buffer
described by the manufacturer for 30 min at 30°C. Samples were then
electrophoresed and
2 5 exposed for autoradiograpy.
The results, which are presented in Figure 12B, indicate, that once
phosphorylated, the 196-304 WT substrate is readily dephosphorylated by in
vitro treatment with
calcineurin and phosphatase I, which is activated by calcineurin (Cohen (1989)
Annual Rev.
Biochem. 58:453). These results indicate that the conserved serines in the SP
repeats that control
3 0 nuclear localization of NF-ATc are substrates for cellular kinases and
calcineurin. Glycogen
synthase kinase-3 (GSK-3) is a highly conserved proline-directed serine-
threonine kinase that
phosphorylates NF-AT in vivo and opposes Caz+/calcineurin-induced nuclear
entry, see below.
GSK-3 phosphorylates the conserved serines in the SP repeats in vitro. The
serines in the SRR
92
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
and SP repeat motifs conform to a GSK-3 consensus substrate sequence (Fiol et
al. (1994) J.
Biol. Chem., 269: 32187-32197). Taken together, these results indicate that
the conserved
serines in these two motifs are phosphorylated in vivo by cellular kinases and
dephosphorylated
by calcineurin.
Example 13: Phosnhoserines in the SItR control an intramolecular interaction
within NF-ATc
The results described above raise the possibility that both basic NLSs
interact with
phosphoserines on the SP repeats and the serine-rich region to prevent nuclear
entry in the
unstimulated state. Such intramolecular interactions are difficult to discern
because of the
difficulty of expressing separate parts of the same protein at concentrations
that would be
equivalent to the high effective concentration of residues on the same peptide
chain. The
overcome this barner to detecting intramolecular interactions, one part of NF-
ATc was
immobilized and interactions with other regions expressed in extracts of COS
cells, which
phosphorylate and translocate NF-ATc under Caz+/calcineurin control were
analyzed.
Extracts of COS cells that had been transfected with the empty expression
vector
or a vector encoding the HA epitope-tagged amino-terminal 418 residues of NF-
ATc (2-418)
(Northrop (1994) Nature, 369:497-502) were incubated with glutathione-agarose
beads coupled
to GST or incubated v~~ith beads coupled to a GST fusion with the RSD of NF-
ATc (GST-RSD).
2 0 A GST fusion protein consisting of the Rel domain of NF-ATcl, GST-RSD
(residues 41 S-716),
was expressed in bacteria and affinity purified on glutathione-agarose (Gene,
67:31-40).
Residues 1-418 of I''F-ATcI tagged at the amino terminus with the HA epitope
(Nature,
369:497-502) were expressed in COS cells and an extract made by lysis in
buffer A (J.
Biol.Chem. 270: 19898-19900) with protease and phosphatase inhibitors. One
hundred
2 5 micrograms of this extract was incubated with 30 ul of glutathione-agarose
coupled to GST,
GST-RSD, or GST-mIvTLS (-2 pg of fusion protein) in 300 ~1 of incubation
buffer (50 mlvt
HEPES at pH 7.8, 150 mM NaCL, 1 mm EDTA, 50 mM NaP04, 0.5% NP-40) with
protease and
phosphatase inhibitors as in (J. Biol. Chem.270:19898-19900) for 2 hr. at
4°C and washed three
times in incubation buffer. Affinity-selected proteins were eluted from the
washed beads with
3 0 SDS sample buffer and detected by immunoblotting using either the 7A6
(Nature, 369:497-502)
or 12CA5 monoclonal antibodies.
As showm in Figure 13A, when the carboxyl terminus of the protein containing
the Rel similarity domain and one of the two partially redundant NLSs was
immobilized (GST
93
CA 02352599 2001-05-23
WO 00/30671 PCTNS99/2?862
4i5-716), it interacted readily and specifically with the amino-terminal half
of NT-ATcI (1-418)
when expressed in COS cell extracts, as well as interacting with the
endogenous protein in
extracts from lymphocytes.
Because the amino terminus, which contains multiple phosphoserines, might
simply interact with basic residues in the Rel similarity region, the NLS in
the rel similarity
domain was mutated and the binding of this mutated protein to amino-terminal
residues 1-418
was analyzed. As shown in Figure 13B, mutation of the carboxy-terminal NLS
from KRKK
(SEQ ID NO: 56) to TRTG (SEQ ID NO: 55) abolished binding to the amino-
terminal 418
residues.
This mutation is unlikely to result in denaturation because alteration of this
NLS
still permits cooperation with NF-ATn and NF-AT-dependent transcription in
vivo. To
demonstrate that this interaction is sensitive to the presence of
phosphoserines in the amino
terminus, extracts of COS cells that had been transfected with the HA epitope-
tagged amino-
terminal 418 residues of NF-ATc (1-418 WT) or versions in which S-'A mutations
were present
in the SRR or SP repeats were incubated with GST-RSD and then washed. As shown
in Figure
13C, the amino-terminal 418 residues with S-'A mutation in the SRR shows
reduced association
with the carboxyl terminus of the protein, whereas S-~A changes in the three
SP repeats affect
this association less strongly. Each set of mutations results in more rapid
migration on SDS
electrophoresis, indicating that these S-~A mutations prevent phosphorylation.
Nonoverlapping
2 0 S->A mutations within the SRR (Fig. 13C) were also tested in this
intramolecular association
assay. Alanine substitutions in residues 172-176 reduce the association with
the rel similarity
domain (RSD), whereas alanine substitutions in serines between 191 and I94 do
not alter the
association with the RSD. Interestingly, there is a correlation between
binding to the RSD in the
intramolecular association assay and Ca2+/calcineurin-independent nuclear
entry--m172-176 is
2 5 constitutively nuclear and m191-194 undergoes regulated nuclear entry. The
differences in the
in vitro intramolecular association assay between each S->A mutations of NF-
ATc are unlikely
to be attributable to denaturation, as all mutants are immunoprecipitated by a
monoclonal
antibody to the region of NF-ATc in the SP repeats (Northrop et al. ( 1994)
Nature, 369:497-502),
are stable when expressed in cells, and direct NF-AT-dependent transcription.
The binding
3 0 activity is unlikely to indicate a head-to-tail dimer forming between full-
length NF-ATc
molecules, as the protein is a monomer in solution and when bound to DNA (Hoey
et al. (1995)
Immunity, 2:461-472).
Thus, the interaction within NF-ATc is dependent on residues in the SRR as
well
94
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
a's an intact carboxy-terminal NLS. The correlation between the subcellular
localization of
mutants in the SRR and their behavior in in vitro binding assays suggests that
this intramolecular
association controls exposure and function of the carboxy-terminal NLS. The S-
~A changes in
the three SP repeats disturbs binding to the RSD only weakly and suggests that
these
phosphoserines may not participate as strongly in the interaction with the
carboxyl terminus of
the protein. The dephosphorylation of the SP repeat motifs may result in
nuclear localization by
another mechanism, perhaps by exposure of the other NLS that lies between the
second and third
SP repeats. A model representing the interaction between the NLS and
phosphoryated residues
in NF-AT is presented in Figure 14. This model is likely to be extended to the
other members
l0 of the NF-ATc gene family based on the conservation of the NLSs, the SRR,
and the SP repeat
regions. The amino terminus of NF-ATc3 and NF-ATp (Proc. Natl. Acad. Sci.,
93:8907-8912;
Nature, 382:370-373 ) undergo Ca2+-sensitive nuclear entry.
Example 14: NF-A~' kinase activit~r copurifies with GSK~
This example demonstrates that the kinase activity that phosphorylates the N-
terminus of NF-AT copurifies with GSK-3.
Protein extracts from rat brains were tested for NF-AT kinase activity as
follows.
Extracts were prepared from rat brains homogenized in 2 volumes of 20 mM tris
(pH 7.5), 1 mM
EDTA, 5 mM EGTA, 2 mM dithiothreitol (DTT), and SO mM [3-glycerol-phosphate
with
protease and phospatase inhibitors [0.1 mM Na3V04, 1 mM phenylmethylsulfonyl
fluoride,
pepstatin (1 ~cg/ml), aprotinin (1 ,ug/ml), leupeptin (5 ,ug/ml), and 1 mM
benzamidine]. A
portion of the 80,OOOg supernatant was passed over a G-50 sizing column to
remove endogenous
adenosine triphosphate (ATP}, made 10% in glycerol, and used a whole brain
extract (S.Smg of
protein per ml). The NF-AT kinase activity was followed through NH4S04
fractionation and
separation on phosphocellulose (P-11 resin), and elution with 200 mM NaCI. The
active
fractions were pooled and further purified on a Mono-S column (Hughes et al.,
Eur. J. Biochem.
203, 305 ( 1991 )).
Column fractions were assayed for NF-AT kinase activity on wild type and
mutated NF-AT peptides with [y 3zP]ATP and then autoradiographed. Furthermore,
since
3 0 analysis of NF-AT N-terminal portion, in particular amino acids 196-304,
indicated the presence
of putative overlapping GSK-3 consensus sites (SSXXS(P)) (see Figure 15),
column fractions
were also assayed for the phosphorylation of GS-2, the GSK-3-specific peptide
substrate (Welsh
et al., J. Biol. Chent. 271, 11410 (1996)). In addition, since GSK-3 (Hughes
et al., Eur. J.
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Biochem. 203, 305 (1991}) often phosphorylates serines adjacent to serines
previously
phosphorylated by protein kinase A (PKA) or another kinase (Fiol et al., J.
Biol. Chem. 269,
32187 ( 1994)), phosphorylation of PKA-prephosphorylated wild type NF-AT
peptide was also
used as a substrate. Phosphorylation of several sites in NF-ATc by PKA could
produce a series
of phosphorylation-dependent, overlapping GSK-3 consensus sites (Fiol et al.,
J. Biol. Chem.
269, 32187 (1994)) (Fig.lS). The total amount of protein in each fraction was
also determined
(as measured by absorbance at 280 nm).
The substrate NF-AT peptides were prepared by cloning a DNA fragment
encoding residues 196 to 304 of NF-ATcI (Durand et al., Mol. Cell. Biol. 8,
1715 (1988);
Cocerill et al., ibid. 15, 2071 (1995); Chuvpilo et al., NucleicAcids Res. 21,
5694 (1993);Rooney
et al., Immunity 2, 473 (1995); Goldfield et al., J. Exp. Med. 178, 1365
(I993)) into pGEX-3X
to generate pGSP. A GST fusion protein with S--~A substitutions (Fig. 15),
pGAP, was similarly
constructed with 9 serine and 10 threonine residues remaining. Bacterially
expressed proteins
were purified on gluthathione agarose and used at 1 ,ug of fusion protein per
10 ,ul of bead slurry
(D. B. Smith and K. S. Johnson, Gene 67, 31 (1988)). The fusion proteins were
used directly or
were prephosphorylated on agarose by addition of 5 units of PKA (Sigma) per
microgram of
fusion protein at 30°C in kinase buffer [20 mM tris (pH 7.5), 10 mM
MgClz, and 1 mM DTT]
with 1 mM ATP for 2 hours and then washed to remove PKA and ATP. One unit of
PKA is
defined as 1 pmol of 3zP transferred per minute. Kinase assays incubated
fusion protein (1 fig)
2 0 on glutathione Sepharose, 100 ~M ATP with [y 32P]ATP (400 ~cCi/~cmol) in
50 ~cl of kinase
buffer for 30 min at 30°C. Beads were incubated with 10 ~cI of column
fractions or of whole
brain extract (55 ~cg of protein), 2.5 units of purified PKA or GSK-3(3 (New
England Biolabs),
or both. Experiments with crude or partially purified brain extracts included
aprotinin, leupeptin,
and pepstatin (all at 1 ~cg/ml), 0.1 mM (3-glycerol-phosphate, and 1 mM
Na3V04. Kinase
2 5 reactions were terminated by washing the agarose beads twice with 1 ml of
TEN [50 mM tris (pH
7.5), 1 mM EDTA, 150 mM NaCI, and 0.5% NP-40] to remove phosphorylated
cellular proteins,
fractionated on SDS-PAGE, autoradiographed, and stained with Coomassie to
ensure that the
substrate was not degraded.
The result of the kinase assays using fractions from the column, which are
3 0 represented in Figure 15 (panels A and B), show that the chromatographic
behavior of the NF-
AT kinase was similar to that of GSK-3. In particular, NF-AT kinase activity
was shown to be
strongest in about fractions 35-40 of column P-11 (see Figure 15B) and about
fractions 15-25
of Mono-S column (see figure 15C), which are also the fractions which had
strongest GSK-3
96
CA 02352599 2001-05-23
WO 00/30b71 PCT/US99/27862
activity. In fact, the peak of NF-AT kinase activity and GSK-3
immunoreactivity is at fraction
21. Furthermore, the PKA prephosphorylated wild-type NF-AT peptide was also
phosphorylated
by the same column fractions. On the contrary, the active column fractions did
not significantly
phosphorylate the mutated NF-AT substrate peptide.
Protein immunoblotting with antibodies to GSK-3a and GSK-3~i confirmed that
they copurified with the NF-AT kinase (Fig. 15C), and PKA eluted in a
partially overlapping
peak from the Mono-S column. In fact, the peak of PKA immunoreactivity is at
fraction 24.
Thus, these results indicate that NF-ATc is likely be a substrate of GSK-3 and
PKA.
~,xample 15: GSK-3 and another kinase syn~e gize to hosphorvlate NF-AT
The role of GSK-3 in the phosphorylation of NF-ATc was assessed by
immunodepleting GSK-3 from whole brain extracts. Antisera to GSK-3a and GSK-
3~3 or control
antibodies were used to remove these proteins from whole brain extract.
Immunodepletion of
GSK-3 activity in 110 ~cg of whole grain extract was done in 200 ~1 of TEN, 1
mM DTT, and
protease and phosphatase inhibitors (same as used above) with 3 ,ug of anti-
GSK-3a (sheep
polyclonal, Upstate Biotechnolgy), anti-GSK-3~3 (immunoglobulin G1 (lgG1)
monoclonal,
Transduction Labs), or both, and 20 ~cl of protein G-Sepharose at 4°C
for 4 hours. The IgGI
mouse monoclonal antibody (mAb) M2 (Kodak), sheep polyclonal anti-HIVp 17
(NIH), or both
were used as control antibodies. The NF-AT kinase assay used 2.5 ,ul of the
supermatant (1.2
,ug of protein) (Cook et al., EMBO, J. 15, 4526 (1996); Stambolic et al. Curr.
Biol. 6, 1664
( 1996)).
Immunodepleted extracts were incubated with PKA-prephosphorylated NF-AT
in an in vitro kinase reaction with ['y 32P]ATP, and the 3zP-labeled substrate
was detected by
autoradiography. Two substrates were used: NF-AT (WT) to detect the priming
kinase activity,
2 5 and WT-PKA prephoshorylated to detect GSK-3 activity. In one reaction,
five units of purified
GSK-3~i were added to the reaction.
As shown in Figure 16, depletion of GSK-3a and GSK-3(3 from the extracts with
specific antibodies completely and specifically removed the NF-AT kinase
activity toward NF-
ATc prephosphorylated by PKA. However, this immunodepleted extract maintained
the ability
3 0 to phosphorylate NF-ATc (Fig. 16B), which indicated that there are at
least two NF-AT kinase
activities: an activity that can act directly on NF-ATc, and a second activity
that requires prior
phosphorylation of NF-ATc. The second kinase activity is that of GSK-3 {as
shown by
immunodepletion experiments) and the priming kinase activity can be provided
in vitro by PKA.
97
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
However, specific inhibition of PKA in extracts indicates that PKA does not
provide all of the
priming kinase activity in either brain or lymphocyte extracts. It should be
noted, that GSK-3
immunodepletion does not effect the phosphorylation of the unprimed NF-AT
substrate because
under conditions of substrate excess, only a small percentage of substrate
become primed and
hence available for subsequent phosphorylation by GSK-3. It is likely that
enzyme is limiting
in these assays because there is no detectable alteration of the mobility of
the substrate upon
Coomassie staining, which would reflect phosphorylation.
Example 16: PKA and GSK-3 stoichiometrically phosphorvlate NF-ATc
The wild-type NF-AT fusion protein (referred to as "WT substrate") was
phosphorylated in vitro with purified PKA and/or GSK-3 kinases. In the
reactions in which the
WT substrate was incubated with the two kinases, the first kinase was
permitted to phosphorylate
the WT substrate with nonradioactive ATP to completion; then, the WT substrate
beads were
washed to remove the kinase and the WT beads were phosphorylated by the second
kinase in the
presence of [~y_'zP)ATP.
The results, which are shown in Figure 17A, indicate that phosphorylation of
GSK-3~i alone incorporated <0.01 mol of 3zP per mole of NF-AT, whereas PKA
alone gave 1 to
2 mol of 3zP per mole of NF-AT and the combination of GSK-3~i and PKA gave 3
to 7 mol of
'zP per mole of NF-AT. Casein kinase II (CKII) and Caz+-calmodulin-dependent
protein kinase
2 0 II (CaMkII) did not stoichiometrically phosphorylate the glutathione-S-
transferase fusion protein
NF-AT-GST. Furthermore, GSK-3[3 phosphorylated NF-ATc only if it was first
phosphorylated
by PKA. Similar results were obtained using dephosphorylated NF-ATc purified
from
lymphocytes as a substrate.
It was then tested whether PKA and GSK-3(3 contribute to the celullar
2 5 phosphorylation of NF-ATc by comparing the tryptic phosphopeptides from NF-
ATc
phosphorylated in vivo with those derived from in vitro phosphorylation of the
NF-AT fusion
protein. NF-ATc was overexpressed in COS cells (which support reversible Ca2+-
dependent
nuclear localization) and labeled with ['zP)orthophosphate. COS cells
transfected with 3 ~g of
PSH102 (Northrop et al., Nature 369, 497 (1994)) were labeled with
[3zP)orthophosphate (1
3 0 mCi/ml) for 6 hours and immunoprecipitated with the hemagglutinin (HA) mAb
12CA5,
transferred to polyvinylidene difluoride membrane, and digested with trypsin.
Oxidized peptides
(1000 cpm) were separated by electrophoresis on cellulose at pH 1.9 for 30 min
at 1000 V and
then chromatographed in the second dimension using butanol-acetic acid-
pyridine solvent (Boyle
98
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
et al., Methods Enzymol. 201, 110 (1991)). In one reaction, the PKA + GSK-3(3
in vitro
phosphorylated peptides were mixed with the in vivo phosphorylated peptides
before two-
dimensional separation to establish that they are similar.
The results, shown in Figure 17B, indicate that the tryptic phosphopeptides
from
NF-ATc phosphorylated in vivo with thosed derived from in vitro
phosphorylation of the NF-AT
fusion protein were identical, with the exception of one phosphopeptide. These
results suggest
that GSK-3[i and another kinase synergize to phosphorylate NF-AT on the sites
involved in Ca2+-
dependent nuclear localization in vivo.
Example 17: Sites of phosphorylation of NF-T by PK.~ and GSK-3
The sites of phosphorylation by GSK-3 and PKA were defined by Edman
degradation of in vitro '2P-labeled tryptic fragments. Wild-type NF-ATc-GST
fusion protein was
phosphorylated in vitro with PKA with [y-32P]ATP(SOpCi/pmol) and cleaved with
Factor Xa
to release the fusion protein. This was isolated on SDS-polyacrylamide gel
electrophoresis
(PAGE) and cleaved by trypsin, and radioactive fragments were purified by high-
performance
liquid chromatography (HPLC). One radioactive fraction release 32P in the
second Edman
degradation cycle and had the sequence ASVTEESWLGAR (SEQ ID NO: 83) of the
tryptic
peptide with Serz45 in the second position. A second radioactive fraction
released 32P in the third
Edman degradation cycle and had a molecular size indicating the tryptic
peptide KYSLNGR
2 0 encompassing Serz69 in the NF-ATc sequence. A second GST fusion protein
encoding residues
223 to 277 of NF-AT was purified, phosphorylated in vitro with nonradioactive
ATP and PKA,
washed, then phosphorylated with [y 32P]ATP (SOpCi/umol) and GSK-3(3. The
fusion protein
was isolated on SDS-PAGE and cleaved by trypsin, and two radioactive fragments
were purified
by HPLC. One radioactive fragment contained the tryptic peptide
GLGACTLLGSPQHSPSTSPR (SEQ ID NO: 84).
Thus, the results indicate that PKA phosphorylates the NF-ATc fusion protein
at
two serines (Fig. 15A). The PKA site at Serz45 creates a series of overlapping
GSK-3 substrate
sites. Phosphorylation of the PKA-prephosphorylated NF-ATc fusion protein by
GSK-3(3
labeled the peptide that contains this array of GSK-3 sites (Fig. 15A).
Example 18: GSK-3 overexpression blocked Ca++ NF-AT induced nuclear
translocation
The biological importance of NF-ATc phosphorylation by GSK-3(3 was assessed
99
CA 02352599 2001-05-23
PCT/US99/27862
WO 00/30671
by manipulating its activity in cells and determining the effect on the
subcellular localization of
NF-ATc. Cos cells, which like many cells, express GSK-3 (Woodgett, in Methods
in
Enzymology, T. Hunter and B. M. Sefton, Eds. (Academic Press, San Diego, CA,
1991), vol. 200,
p. 564), were cotransfected with 1 ~g of a construct encoding FLAG epitope-
tagged NF-ATcl
and 3 ug of GSK-3 expression vector or with the empty vector and the cells
were left
unstimulated or were treated with 2 pM ionomycin and 10 mM CaCl2 (I + Ca2+) to
induce
nuclear localization of NF-ATc. Human GSK-3~3 cDNA (He et al., Nature 374, 617
(1995))was
cloned into pBJ-5. NF-ATc was visualized with FLAG mAb M2 and indirect
immunofluorescence. COS cell NF-AT translocation assay were done as described
in Northrop
et al., Nature 369, 497 (1994).
The results show that transfected NF-ATc family members (Shibasaki et al.,
Nature 382,
370 (1996); Luo et al., Proc. Natl. Acad. Sci. U.S.A. 93, 8907 (1996)), like
endogenous NF-ATc,
were cytoplasmic and translocated to the nucleus when cells were stimulated by
agents that
increase intracellular Ca2+. Furthermore, overexpression of GSK-3p blocked the
Cap+-
calcineurin-induced nuclear translocation of coexpressed NF-ATc in COS cells.
In another example, endogenous NF-AT-dependent transcription was shown to be
inhibited by overexpression of GSK-3(3. Jurkat-T antigen cells were
transfected with 2 ~g of a
transcription reporter plasmid (NF-AT dependent reporter, AP-1 dependent
reporter, or HIV-
LTR containing reporter linked to a gene encoding the SEAP) and either 3 ~g of
the GSK-3(3
2 0 expression construct or empty vector. NF-AT SEAP activity was measured and
expressed as a
percentage of the ionomycin-stimulated and phorbol 12-myristate 13-acetate
{PMA)-stimulated
control activity; AP-1 and HIV-LTR SEAP activities are expressed as a
percentage of PMA-
stimulated activity (Spencer et al., Science 262, 1019 (1993)). The results,
which are depicted
in Figure 18, panel A, show that GSK-3 overexpression inhibits NF-AT dependent
reporter gene
expression. AP-1 dependent reporter gene expression was also downregulated by
GSK-3
overexpression, probably due to the ability of GSK-3 to produce an inhibitory
phosphorylation
on c-Jun (Mikolakaki et al., Oncogene 8, 833 (1993)). GSK-3 did not have an
inhibitory effect
on the HIV-LTR dependent reporter gene expression.
In yet another example, the ability of various serine-threonine kinases to
inhibit the
3 0 nuclear entry of cotransfected NF-ATc in COS cells was compared.
Accordingly, COS cells
were contransfected with 1 ~g of FLAG epitope-tagged NF-ATcI and 1 pg of the
serine
threonine kinases CKII, CaMkBA, CaMkBB, PKA or PKC or 3 ~.g of GSK-3 ~i or 0.5
pg of ERK.
ERK cDNA was cloned into pBJ-5. Drosophilia CKII cDNA was polymerase chain
reaction
100
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
amplified and cloned into pBJ-5. Murine PKA cDNA and an activated form of PKC-
~i were
cloned into pSRa. The calcineurin A and B expression constructs (Clipstone and
Crabtree,
Nature 357, 695 (1992)), CaMkII constructs (Srinivasan et al. J. Cell Biol.
126, 839 (1994)),
COS cell NF-AT translocation assay, and Jurkat -T antigen cell transcription
reporter assays
(Northrop et al. (1994) Nature 369:497) were as described. Transfected cells
were stimulated
with ionomycin and 10 mM Ca2+, and the percentages of cells expressing NF-AT
localized in the
nucleus, cytoplasm, or both compartments were scored visually and are
presented as a percentage
of expressing cells. The transfected ERK kinase was activated by adding PMA
(25 ng/ml).
Comparison of the relative expression of the HA epitope-tagged kinases was
performed by
immunoblotting 15 ug of whole cell extracts with HA mAb 12CA5.
The results, which are presented in Figure 18 B, indicate that although GSK-3
was
expressed in smaller amounts, it was the most active in inhibiting nuclear
entry of NF-ATc.
Overexpression of PKA had little effect on NF-ATc localization. This may
indicate that
endogenous PKA activity or another kinase is adequate to phosphorylate NF-ATc
in COS cells
or that such phosphorylation is necessary, but not sufficient, for nuclear
export. Thus, these
results indicate that the Caz+-calcineurin signaling pathway is opposed by GSK-
3.
Exam In a 19: OverexpressiQn of GSK-3 enhances the nuclear exhort of NF-AT
This example describes the effects of GSK-3 on the nuclear export of NF-AT by
2 0 first causing its translocation to the nucleus by stimulating cells with
ionomycin, then removing
the CaZ+-calcineurin signal and blocking further nuclear import with the
calcineurin inhibitor
FK506 (Clipstone and G. R. Crabtree, Nature 357, 695 (1992)).
COS cells were cotransfected with expression constructs encoding FLAG epitope-
tagged NF-ATcl (1 pg), calcineurin A and B (0.5 pg each) (N. A. Clipstone and
G. R. Crabtree,
2 5 Nature 357, 695 (1992)), and 2 ~g of vector, GSK-3(3 or GSK-KM, a
catalytically inactive GSK-
3~3 (He et al., Nature. 374, 617 (1995)). Cells were also cotransfected with a
version ofNF-ATcI
in which the underlined serines in Fig. I SA were changed to alanines with
calcineurin and
GSK-3~i. The inclusion of Ca-calcineurin promotes NF-ATc nuclear entry
(Shibasaki et al.,
Nature 382, 370 (1996); Luo et al., Proc. Natl. Acad. Sci. U.S.A. 93, 8907
(1996)) and
3 0 overcomes the cytoplasmic localization of NF-ATc induced by GSK-3 ~3
overexpression. Wild-
type NF-ATc was localized in the cytosol in 98% of unstimulated expressing
cells, whereas 90%
of cells translocated NF-ATc to the nucleus with I + Caz+ treatment; this
translocation was
completely blocked by FKSOG. NF-ATc was localized in the nucleus by treatment
with I + Caz+
101
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
for 60 min, then the medium was changed to medium with FK506 (20 ng/ml) to
terminate Ca2+
signaling and to block nuclear reentry of NF-ATc. Transfected NF-ATc was
detected with
FLAG mAb M2 by indirect immunofluorescence, and 200 expressing cells were
scored as
expressing NF-ATc in the cytoplasm, nucleus, or both compartments.
The results, shown in Figure 19, show that overexpression of GSK-3(3 in
amounts
approximately one-tenth those of NF-ATc enhanced the movement of NF-ATc into
the
cytoplasm relative to that in cells transfected with the vector or with a
catalytically inactive form
of GSK-3~i (He et al., Nature. 374, 617 (1995)). GSK-3~3 overexpression did
not influence the
constitutive nuclear localization of NF-ATc with S--A mutations in the serine-
proline repeats.
These data indicate that GSK-3(3 acts catalytically to direct the nuclear
export of NF-ATc and
that the regulation of nuclear export involves the phosphorylation of NF-ATc
at conserved
serines. Because NF-ATc family members are expressed in many tissues and have
sequence
similarity at the NHS-terminal residues involved in nuclear import and export,
GSK-3 is likely
to control the compartmentalization of each of the four different NF-ATc
family members.
Example 20: NF-AT is a therapeutic tar~~et for cardia hypertronhv
The Ca++ /Calcineurin/NF-AT signaling pathway.
The NF-AT transcription factor was initially described in T cells as a rapidly
inducible
2 0 protein complex binding to the distal antigen receptor response element,
ARRE-2, of the
human IL2 promotor (1,2). The active transcription factor is made up of
cytosolic (NF-ATc)
and nuclear components (NF-ATn) (3). The NF-ATc family of transcription
factors is encoded
by at least four distinct genes, NF-ATcI, c2, c3, c4 (Genome Data Base (GBD)
Nomenclature
Committee) (4-9). NF-ATc family proteins show specific patterns of tissue
distribution (Table
2 5 1).
Table 1
GBD Name Tissue Distribution K O.Phenotype
NF-ATcI Lymphocytes, Heart valves,Failure of heart valve
Muscle, development
Prostate, Colon, ParathyroidIL4, IL2 reduced
3 0 NF-ATc2 Lymphocytes, Brain, Pancreas,Proliferation of lymphocytes
Testis, enhanced
Placenta IL4 enhanced
NF-ATc3 Lymphocytes, Muscle, Heart,Proliferation of lymphocytes
Kidney, enhanced
Brain, Skin IL4 enhanced
NF-ATc4 Heart, Lung, Lymphocytes,N.A.
Kidney,
Brain (hi ocam us, cerebellum),
102
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
Activation of NF-AT-dependent transcription integrates at least three
different signaling
pathways. NF-ATc is sequestered in the cytoplasm and rapidly translocates to
the nucleus in
response to a rise in [Ca++]; (3,10-12). This cytoplasmic to nuclear
translocation requires
dephosphorylation of NF-ATc by calcineurin and is blocked by the
immunosupressive drugs
cyclosporin A (CsA) and FK506, which selectively inhibit the activity of
calcineurin (13-21).
Induction and/or activation of NF-ATn requires signals from the Ras/MAPK
pathway
(7,10,22) and Rac/CDC42-dependent reorganization of the cytoskeleton ((23)).
Each NF-ATc
family member contains two main functional domains, an N-terminal regulatory
region or NF-
AT Homology Region (NHR) and a C-terminal DNA Binding Domain (DBD) or Rel
1 o Homology Domain (RHD). The N-terminal regulatory domain is necessary and
sufficient for
regulated nuclear import and export of NF-ATc proteins. Within the N-terminal
region there
are several conserved motifs, including a serine rich region (SRR) and three
serine-proline (SP)
repeats, which are proline-directed kinase sites (8,19,24). These motifs are
the target of
bidirectional NF-ATc regulation by opposing kinases and phosphatases. The
conserved
phospho-serines in these motifs serve as substrates for calcineurin, while the
serine-threonine
kinase glycogen synthase kinase-3 (GSK-3) phopshorylates NF-ATc at these
conserved serine
residues and opposes the Ca++/calcineurin induced nuclear entry (24). The
highly conserved
C-terminal DNA binding domain, which shows moderate sequence homology to and
shares the
topology with the DNA-binding domain of Rel-family proteins, does not bind to
DNA by itself
2 0 at physiologic concentrations due to substitutions at critical residues
(25-30). Thus, all NF-ATc
proteins require a nuclear partner (NF-ATn) for binding to DNA (10).
Heterodimeric
combinations of the AP-1 family members, Fos and Jun, have been shown to
function as NF-
ATn (25,30,31). NF-ATc and AP-1 cooperatively bind to juxtaposed recognition
sites and
synergistically activate gene expression (25,28,32). Additionally the zinc
finger transcription
2 5 factor GATA4 has been shown to cooperate with NF-ATc4 in the activation of
transcription
in cardiomyocytes (36).
Cardiac Hyperthrophy
In the Unites States half a million new cases of heart failure are diagnosed
each year,
3 0 with a mortality rate of about 50% . Cardiac hypertrophy is a generalized
enlargement of the
myocardium and is initially a compensatory response of the heart muscle that
augments cardiac
output. However, sustained hypertrophy can lead to dilated cardiomyopathy,
functional
103
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
insufficiency, heart failure and sudden death. A variety of underlying disease
states such as
hypertension, myocardial infarction, cardiac arrhythmias, endocrine disorders
and genetic
mutations of cardiac proteins (33-35) genes can lead to hypertrophic
cardiomyopathy (HCM).
Overexpression of a constitutively active /Ca++ independent form of
calcineurin or a
constitutively nuclear form of NF-ATc4 in the heart of transgenic mice evoke
molecular and
pathophysiologic changes typical of cardiac hypertrophy and mimic all
pathological aspects of
human heart failure (36). The development of HCM in the calcineurin transgenic
animals is
blocked by treatment with Cyclosporin A (CsA), demonstrating that the
phosphatase activity
of calcineurin is essential for the induction of HCM. Stimulation of
cardiomyocytes with
to angiotensin II and phenylephrine,which induce hypertrophic responses in
cardiomyocytes,
activate NF-AT-dependent transcription in cardiomyocytes. Furthermore, CsA and
FK506
block the angiotensin and phenylephrine induced hypertrophic response in
vitro. CsA has also
been shown to block the development of HCM in three mouse models based on
mutations in
contractile heart proteins such as tropomodulin, myosin light chain-2, fetal
(3-tropomyosin (33).
Additionally, CsA and FK506 treatment prevented pressure overload induced
hypertrophy
caused by aortic banding of rats(33). These data indicate that the Ca++
/calcineurin/NF-ATc
signaling pathway may play a crucial role in the initiation of HCM. All
hypertrophic stimuli
increase intracellular Ca++ levels (37-39). This increase in intracellular
Ca++ could directly
activate calcineurin. Activation of calcineurin could initiate HCM by
dephosphorylation and
2 o nuclear translocation of NF-ATc proteins. NF-ATc dependent transcription
could induce
hypertrophic response genes.
Voltage gated Ca++ channels (VSCCs} play a central role in the development and
the
contractility of the heart muscle. VSCCs open by membrane depolarization
during the fast
upstroke of the action potential. Prolongation of action potential duration
with delayed
repolarization and alterations in Ca++ homeostasis are common findings in
human heart failure
and animal models of cardiac hypertrophy. Cardiac myocytes express the high
voltage-
activated L-type VSCCs and the low voltage-activated T-type VSCCs. L-type Ca++
channels
play a central role in the excitation-contraction coupling in cardiac muscle
and thus are key
regulators of inotropy. Ca++ influx via L-type VSCCs induces Ca++ release from
the
3 o sarcoplasmic reticulum (SR) during the systole and initiates contraction.
In cardiac muscle,
Ca++ influx through VSCCs is closely associated with the action of
catecholamines. (3-
adrenergic stimulation of cardiomyocytes increases the opening probability of
L-type VSCCs
104
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
via increased levels of intracellular cAMP. Additionally, (3-adrenergic
stimulation of the
cAMP-dependent protein kinase A pathway increases the affinity of the SR Ca*+-
ATPase for
Ca++ via phosphorylation of the regulatory protein phospholamban, thus
increasing the Ca++
filling status of the SR. The filling status of the SR determines the force of
contraction during
the next systole. Additionally, Ca++ influx via L-type VSCCs can trigger gene
transcription
in neurons (40) and skeletal muscle (41). T-type-channels in the heart are
present at lower
density than L-type channels. The role of T-type channels is not as well
defined. T-type
channels are thought to play a role in the rhythmic activity of the heart,
during embryonic heart
development and potentially during pathological conditions such as hypertrophy
(42).
Research Results
We have used homologus recombination to delete exon l, 2 and part of exon 3 of
the
NF-ATc4 gene, coding for amino acid 1-438 of the NF-ATc4 protein which has
been
implicated by Dr Eric Olson's group in cardiac hypertrophy. A 15 kilobase
genomic clone
obtained from a mouse 129 library was used to construct a targeting vector for
homologus
recombination by positive/negative selection. A PGK-neo casette was inserted
in inverse
orientation into the first, second and third exon of the NF-ATc4 gene,
deleting the regulatory
region and part of the DNA binding domain of NF-ATc4. (Figure 1). Targeted ES
cell clones
2 o were used for microinjection into C57B1/6 blastocyts, followed by uterine
transfer into
pseudopregnant CD-1 females. Male chimeric mice were mated to CD-1, C57BI/6
and 129Sv
female mice and the resulting heterozygous mice were bred to homozygosity. The
homozygous NF-ATc4 knock-out mice are viable and hence can be tested for
cardiac function.
105
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
II. Literature Cited
1. J.-P. Shaw, P.J. Utz, D.B. Durand, J.J. Toole, E.A. Emmel, G.R. Crabtree,
Science 241,
202 (1988).
2. D.B. Durand, J.P. Shaw, M.R. Bush, R.E. Replogle, R. Belageje, G.R.
Crabtree, Mol.
Cell Biol. 8, 1715 (1988).
3. W.M. Flanagan, B. Corthesy, R.J. Bram, G.R. Crabtree, Nature 352, 803
(1991).
4. J.P. Northrop, S.N. Ho, L. Chen, et al, Nature 369, 497 (1994).
5. P.G. McCaffrey, C. Luo, T.K. Kerppola, et al, Science 262, 750 (1993).
6. T. Hoey, Y.-L. Sun, K. Williamson, X. Xu, Immun 2, 461 (1995).
7. A. Rao, C. Luo, P.G. Hogan, Annu Rev Immunol 15, 707 (1997).
8. S.N. Ho, D.J. Thomas, L.A. Timmerman, X. Li, U. Francke, G.R. Crabtree, J.
Biol
Chem 270, 19898 (1995).
9. C. Luo, K.T. Shaw, A. Raghavan, et al, Proc. Natl. Acad. Sci U. S. A. 93,
8907 (1996).
10. G.R. Crabtree, Science 243, 355 (1989).
11. L.A. Timmerman, N.A. Clipstone, S.N. Ho, J.P. Northrop, G.R. Crabtree,
Nature 383,
837 (1996).
12. F. Shibasaki, E.R. Price, D. Milan, F. McKeon, Nature 382, 370 (1996).
13. J. Liu, J.D. Farmer, W.S. Lane, J. Friedman, I. Weissman, S.L. Schreiber,
Cell 66, 807
2 0 (1991).
14. N.A. Clipstone, G.R. Crabtree, Nature 357, 695 (1992).
15. E.A. Emmel, C.L. Verweij, D.B. Durand, K.M. Higgins, E. Lacy, G.R.
Crabtree,
Science 246, 1617 (1989).
16. G.R Crabtree, N.A. Clipstone, Annual Review of Biochemistry 63, 1045
(I994).
17. S.L. Schreiber, G.R. Crabtree, Immunology Today 13, I36 (1992).
18. N.A. Clipstone, D.F. Fiorentino, G.R. Crabtree, J. Biol Chem 269, 26431
(1994).
19. C.R. Beals, N.A. Clipstone, S.N. Ho, G.R. Crabtree, Genes Dev. 11, 824
(1997).
20. J. Liu, M.W. Albers, T.J. Wandless, et al, Biochemistry 31, 3896 (1992).
21. D.A. Fruman, C.B. Klee, B.E. Bierer, S.J. Burakoff, Proc. Natl. Acad. Sci
U. S. A. 89,
3686 (1992).
22. V.E. Valge, J.G.P. along, B.M. Datlof, A.J. Sinskey, A. Rao, Cell 55, 101
(1988).
23. L.J Holsinger, LA. Graef, W. Swat, et al, Current Biology 8, 563 (1998).
24. C.R. Beals, C.M. Sheridan, C.W. Turck, P. Gardner, G.R. Crabtree, Science
275, 1930
( 1997).
25. S.A. Wolfe, P. Zhou, V. Dotsch, et al, Nature 385, 172 (1997).
26. L.J. Sun, B.R. Peterson, G.L. Verdine, Proc. Natl. Acad. Sci U. S. A. 94,
4919 (1997).
27. D.A. Erlanson, M. Chytil, G.L. Verdine, Chem Biol 3, 981 (1996).
28. B.R. Peterson, L.J. Sun, G.L. Verdine, Proc. Natl. Acad. Sci U. S. A. 93,
13671 (1996).
29. P. Zhou, L.J. Sun, V. Dotsch, G. Wagner, G.L. Verdine, Cell 92, 687
(1998).
30. L. Chen, J.N. Glover, P.G. Hogan, A. Rao, S.C. Harrison, Nature 392, 42
(1998).
106
CA 02352599 2001-05-23
WO 00/30671 PCT/US99/27862
31. J. Jain, P.G. McCaffrey, V.E. Valge-Archer, A. Rao, Nature 356, 801
(1992).
32. J. Jain, P.G. McCaffrey, Z. Miner, et al, Nature 365, 352 (1993).
33. M.A. Sussman, H.W. Lim, N. Gude, et al, Science 281, 1690 (1998).
34. K.L. Vikstrom, L.A. Leinwand, Curr. Opin. Cell Biol 8, 97 (1996).
35. H. Watkins, J.G. Seidman, C.E. Seidman, Hum. Mol. Genet. 4 Spec No, 1721
(1995).
36. J.D. Molkentin, J.R. Lu, C.L. Antos, et al, Cell 93, 215 (1998).
37. J.O. Bustamante, A. Ruknudin, F. Sachs, J Cardiovasc. Pharmacal. 17 Suppl
2, 5110
(1991).
38. A.D. Wickenden, R. Kaprielian, Z. Kassiri, et al, Cardiovasc. Res. 37, 312
(1998).
39. S.P. Lubic, K.M. Giacomini, J.C. Giacomini, J Mol. Cell Cardiol. 27, 917
(1995).
40. H. Bito, K. Deisseroth, R.W. Tsien, Curr. Opin. Neurobiol. 7, 419 (1997).
41. C.F. Huang, B.E. Flucher, M.M. Schmidt, S.K. Stroud, J. Schmidt, Neuron.
13, 167
( 1994).
42. S. Richard, F. Leclercq, S. Lemaire, C. Piot, J. Nargeot, Cardiovasc. Res.
37, 300
(1998).
43. M.R. Hodge, A.M. Ranger, F. Charles de la Brousse, T. Hoey, M.J. Grusby,
L.H.
Glimcher, Immunity. 4, 397 (1996).
44. J.M. Gardin, F.M. Siri, R.N. Kitsis, J.G. Edwards, L.A. Leinwand, Circ.
Res. 76, 907
(1995).
45. H. Hamada, N. Hakamata, F. Ohsuzu, H. Nakamura, Jpn. Heart J 38, 433
(1997).
46. H.G. Zimmer, J Mol. Med 75, 849 (1997).
47. H.A. Rockman, R.S. Ross, A.N. Harris, et al, Proc. Natl. Acad. Sci U. S.
A. 88, 8277
(1991).
48. R.H. Kehlenbach, A. Dickmanns, L. Gerace, J Cell Biol 141, 863 (1998).
49. P.S. Klein, D.A. Melton, Proc. Natl. Acad. Sci U. S. A. 93, 8455 (1996).
50. V. Stambolic, J.R. Ruel, J.R. Woodgett, Current Biology 6, 1664 (1997).
51. J. Zhu, F. Shibasaki, R. Price, et al, Cell 93, 851 (1998).
52. C.W. Chow, M. Rincon, J. Cavanagh, M. Dickens, R.J. Davis, Science 278,
1638 (1997).
53. J.D. Klemm, C.R. Beals, G.R. Crabtree, Curr. Biol7, 638 (1997).
Equivalents
Those skilled in the art will recognize, or be able to ascertain using no more
than routine
3 5 experimentation, many equivalents of the specific embodiments of the
invention described
herein. Such equivalents are intended to be encompassed by the following
claims.
107