Note: Descriptions are shown in the official language in which they were submitted.
CA 02362004 2001-11-13
TITLE: FUSION PROTEINS
FIELD OF THE INVENTION
The present invention relates to conjugate or fusion type proteins
(polypeptides) comprising,
for example, C3 (see below) (i.e., C3-like protein, C3 chimeric proteins).
Although, in the
following, fusion-type proteins of the present invention, will be particularly
discussed in
relation to the use to facilitate regeneration of axons and neuroprotection,
it is to be
understood that the fusion proteins may be exploited in other contexts.
The present invention in particular pertains to the field of mammalian nervous
system repair
(e.g. repair of a central nervous system (CNS) lesion site or a peripheral
nervous system
(PNS) lesion site), axon regeneration and axon sprouting, neurite growth and
protection from
neurodegeneration and ischemic damage.
The Rho family GTPases regulates axon growth and regeneration. Inactivation of
Rho with
Clostridium botulinum C3 exotransferase (hereinafter simply referred to as C3)
can stimulate
regeneration and sprouting of injured axons; C3 is a toxin purified from
Clostridium
2o botulinum (see Saito et al., 1995, FEBS Lett 371:105-109; Wilde et al 2000.
J. Biol. Chem.
275:16478). Compounds of the C3 family from Clostridium botulinum inactivate
Rho by
ADP-ribosylation and thus act as antagonists of Rho effect or function (Rho
antagonists).
The present invention in particular relates to a means of intracellular
delivery of C3 protein
(e.g. C3 itself or other active analogues such as C3-like transferases - see
below) or other Rho
antagonists to repair damage in the nervous system, to prevent ischemic cell
death, and to
treat various disease where the inactivation of Rho is required. The means of
delivery may
take the form of chimeric (i.e. conjugate) C3-like Rho antagonists. These
conjugate
antagonists provide a significant improvement over C3 compounds (alone)
because they are 3
to 4 orders of magnitude more potent with respect to the stimulation of axon
growth on
inhibitory substrates than recombinant C3 alone. Examples of these Rho
antagonists have
been made as recombinant proteins created to facilitate penetration of the
cell membrane (i.e.
to enhance cell uptake of the antagonists), improve dose-response when applied
to neurons to
2
CA 02362004 2001-11-13
stimulate growth on growth inhibitory substrates, and to inactivate Rho.
Examples of these
conjugate Rho antagonists are described below in relation to the designations
C3APL,
C3APLT, C3APS, C3-TL, C3-TS, C3Basicl, C3Basic2 and C3Basic3.
BACKGROUND OF THE INVENTION
Traumatic injury of the spinal cord results in permanent functional
impairment. Most of the
deficits associated with spinal cord injury result from the loss of axons that
are damaged in
the central nervous system (CNS). Similarly, other diseases of the CNS are
associated with
axonal loss and retraction, such as stroke, human immunodeficiency virus (HIV)
dementia,
to prion diseases, Parkinson's disease, Alzheimer's disease, multiple
sclerosis and glaucoma.
Common to all of these diseases is the loss of axonal connections with their
targets, and cell
death. The ability to stimulate growth of axons from the affected or diseased
neuronal
population would improve recovery of lost neurological functions, and
protection from cell
death can limit the extent of damage. For example, following a white matter
stroke, axons
are damaged and lost, even though the neuronal cell bodies are alive, and
stroke in grey
matter kills many neurons and non-neuronal (glial) cells. Treatments that are
effective in
eliciting sprouting from injured axons are equally effective in treating some
types of stroke
(Boston life sciences, Sept. 6, 2000 Press release ). Neuroprotective agents
often tested as
potential compounds that can limit damage after stroke. Compounds which show
both
2o growth-promotion and neuroprotection are especially good candidates for
treatment of stroke
and neurodegenerative diseases. Similarly, although the following discussion
will generally
relate to delivery of Rho antagonists, etc. to a traumatically damaged nervous
system, this
invention may also be applied to damage from unknown causes, such as during
stroke,
multiple sclerosis, HIV dementia, Parkinson's disease, Alzheimer's disease,
prion diseases or
other diseases of the CNS were axons are damaged in the CNS environment. Also,
Rho is an
important target for treatment of cancer and metastasis (Clark et al (2000)
Nature 406:532
535), and hypertension (Uehata et al. (1997) Nature 389:990) and RhoA is
reported to have a
cardioprotective role (Lee et al. FASEB J. 15:1886-1884). Therefore, the new
C3-like
proteins are expected to be useful for a variety of diseases were inhibition
of Rho activity is
required.
3
CA 02362004 2001-11-13
It has been proposed to use various Rho antagonists as agents to stimulate
regeneration of
(cut) axons, i.e. nerve lesions; please see, for example, Canadian Patent
application nos.
2,304,981 (McKerracher et al) and 2,300,878 (Strittmatter). These patent
application
documents propose the use of known Rho antagonists such as for example C3,
chimeric C3
proteins, etc. (see blow) as well as substances selected from among known
trans-4
amino(alkyl)-1-pyridylcarbamoylcyclohexane compounds (also see below) or Rho
kinase
inhibitors for use in the regeneration of axons. C3 inactivates Rho by ADP-
ribosylation and
is fairly non-toxic to cells (Dillon and Feig (1995) Methods in Enzymology:
Small GTPases
l0 and their regulators Part. B.256:174-184).
While the following discussion will generally relate or be directed at repair
in the CNS, the
techniques described herein may be extended to use in many other diseases
including, but not
restricted to, cancer, metastasis, hypertentension, cardiac disease, stroke,
diabetic neuropathy,
and neurodegenerative disorders such as stroke, Alzheimer's disease,
Parkinson's disease,
amyotrophic lateral sclerosis (ALS). Treatment with Rho antagonists would be
used to
enhance the rate of axon growth of peripheral nerves and thereby be effective
for repair of
peripheral nerves after surgery, for example after reattaching severed limbs.
Also, treatment
with our fusion compounds (proteins) is expected to be effective for the
treatment of various
2o peripheral neuropathies because of their axon growth promoting effects.
As mentioned above, traumatic injury of the spinal cord results in permanent
functional
impairment. Axon regeneration does not occur in the adult mammalian CNS
because
substrate-bound growth inhibitory proteins block axon growth. Many compounds,
such as
trophic factors, enhance neuronal differentiation and stimulate axon growth in
tissue culture.
However, most factors that enhance growth and differentiation are not able to
promote axon
regenerative growth on inhibitory substrates. To demonstrate that a compound
known to
stimulate axon growth in tissue culture most accurately reflects the potential
for therapeutic
use in axon regeneration in the CNS, it is important for the cell culture
studies to include the
demonstration that a compound can permit axon growth on growth inhibitory
substrates. An
4
CA 02362004 2001-11-13
example of trophic and differentiation factors that stimulate growth on
permissive substrates
in tissue culture, are neurotrophins such as nerve growth factor (NGF) and
brain-derived
growth factor. NGF, however, does not promote growth on inhibitory substrates
(Lehmann,
et al. (1999) 19: 7537-7547) and it has not been effective in promoting axon
regeneration in
vivo. Brain derived neurotrophic factor (BDNF) is not effective to promote
regeneration in
vivo either (Mansour-Robaey, et al. J. Neurosci. (1994) 91: 1632-1636). BDNF
does not
promote neurite growth on growth inhibitory substrates (Lehmann et al supra).
Targeting intracellular signalling mechanisms involving Rho and the Rho kinase
for
1o promoting axon regeneration has been proposed (see, for example, the above
mentioned
Canadian Patent application nos. 2,304,981 (McKerracher et al)). For
demonstration that
inactivation of Rho promotes axon regeneration on growth inhibitory
substrates, recombinant
C3, a protein that inactivates Rho by ADP ribosylation of the effector domain
was used.
While such a C3 protein can effectively promote regeneration, it has been
noted that such a
C3 protein does not easily penetrate into cells, and high doses must therefore
be applied for it
to be effective.
The high dose of recombinant C3 needed to promote functional recovery presents
a practical
constraint or limitation on the use of C3 in vivo to promote regeneration
(Lehmann, et al.
(1999) J. Neurosci. 19: 7537-7547; Morii, N and Narumiya, S. (1995) Methods in
Enzymology, Vol 256 part B, pg.196-206. In tissue culture studies, it has, for
example, been
determined that the minimum amount of C3 that can be used to induce growth on
inhibitory
substrates is 25 ug/ml (Lehmann, et al. (1999) J. Neurosci. 19: 7537-7547;
Morii, N and
Narumiya, S. (1995) Methods in Enzymology, Vol 256 part B, pg.196-206. If the
cells are
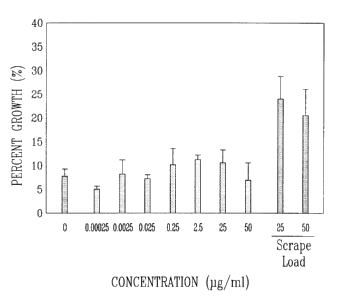
not triturated, even this dose is ineffective (Figure 1). In the context of
the present invention
it has been determined, for example, that at least 40 ug/20 g mouse needs to
be applied to
injured mouse spinal cord or rat optic nerve (McKerracher, Canadian patent
application No.:
2,325,842). Calculating doses that would be required to treat an adult human
on an
equivalent dose per weight scale up used for our rat and mice experiments, it
would be
3o necessary to apply 120 mg/kg of C3 (i.e. alone) to the injured human spinal
cord. This large
amount of recombinant protein creates significant problems for manufacturing,
due to the
5
CA 02362004 2001-11-13
large scale protein purification and cost. It also limits the dose ranging
that can be tested
because of the large amount of protein needed for minimal effective doses.
Another related limitation with respect to the use of C3 to promote repair in
the injured CNS
is that it does not easily penetrate the plasma membrane of living cells. In
tissue culture
studies when C3 is applied to test biological effects it has been
microinjected directly into the
cell (Ridley and Hall (1992) Cell 70: 389-399), or applied by trituration of
the cells to break
the plasma membrane (Lehmann, et al. (1999) J. Neurosci.l9: 7537-7547, Jin and
Strittmatter
(1997) J. Neurosci. 17: 6256-6263). In the case of axon injury in vivo, the C3
protein is
likely able to enter the cell because injured axons readily take up substances
from their
environment. However, C3-like proteins of the present invention are likely to
act also on
surrounding undammaged neurons and help them make new connections as well,
thus
facilitating recovery. After incomplete SCI, there is plasticity of motor
systems attributed to
cortical and subcortical levels, including spinal cord circuitry (Raineteau,
O., and Schwab, M.
E. (2001) Nat Rev Neurosci 2: 263-73). This plasticity may be attributed to
axonal or
dendritic sprouting of collaterals and synaptic strengthening or weakening.
Additionally, it
has been shown that sparing of a few ventrolateral fibers may translate into
significant
differences in locomotor performance since these fibers are important in the
initiation and
control of locomotor pattern through spinal central pattern generators
(Brustein, E., and
2o Rossignol, S. (1998) JNeurophysiol 80: 1245-67). It is well documented that
reorganization
of spared collateral cortical spinal fibres occurs after spinal cord injury
and this contributes to
functional recovery (Weidner et al, 2001 Proc. Natl. Acad. Sci. 98: 3513-
3518). The process
of reorganization and sprouting of spared fibers would be enhanced by
treatment with C3-like
proteins able to enter non-injured neurons. This would enhance spontaneous
plasticity of
axons and dendritic remodeling known to help functional recovery.
Other methods of delivery of C3 in vitro have been to make a recombinant
protein that
can be taken up by a receptor-mediated mechanism (Boquet, P. et al. (1995)
Meth. Enzymol.
256: 297-306). The disadvantage of this method is that the cells needing
treatment must
express the necessary receptor. Lastly, addition of a C2II binding protein to
the tissue culture
medium, along with a C21N-C3 fusion toxin allows uptake of C3 by receptor-
mediated
6
CA 02362004 2001-11-13
endocytosis (Barthe et al. (1998) Infection and Immunity 66:1364). The
disadvantage of this
system is that much of the C3 in the cell will be restrained within a membrane
compartment.
More importantly, two different proteins must be added separately for
transport to occur
(Wahl et al. 2000. J. Cell Biol. 149:263), which make this system difficult to
apply to for
treatment of disease in vivo.
SUMMARY OF THE INVENTION
The term "Rho antagonists" as used herein includes, but is not restricted to,
(known ) C3,
including C3 chimeric proteins, and like Rho antagonists.
to
The term "C3 protein" refers to ADP-ribosyl transferase C3 isolated from
Clostridium
botulinum or a recombinant ADP-ribosyl transferase.
The term "C3-like protein", "ADP-ribosyl transferase C3-like protein", "ADP-
ribosyl
transferase C3 analogue", "C3-like transferase" or "C3 chimeric proteins" as
used herein
refers to any protein (polypeptide) having a biological activity similar
(e.g., the same,
substantially similar), to ADP-ribosyl transferase C3. Examples of such C3-
like protein
include, for example, but are not restricteed to C3APL, C3APLT, C3APS, C3-TL,
C3-TS,
C3Basicl, C3Basic2 and C3Basic3 and the protein defined in SEQ ID NO.: 20.
The term "nerve injury site" refers to a site of traumatic nerve injury or
nerve injury caused
by disease. The nerve injury site may be a single nerve (eg sciatic nerve) or
a nerve tract
comprised of many nerves (eg. damaged region of the spinal cord). The nerve
injury site may
be in the central nervous system or peripheral nervous system or in any region
needing repair.
The nerve injury site may form as a result of damage caused by stroke. The
nerve injury site
may be in the brain as a result of surgery, brain tumour removal or therapy
following a
cancerous lesion. The nerve injury site may result from stroke, Parkinson's
disease,
7
CA 02362004 2001-11-13
Alzheimer's disease, amyotrophic lateral sclerosis (ALS), diabetes or any
other type of
neurodegenerative disease.
"Polynucleotide" generally refers to any polyribonucleotide or
polydeoxyribonucleotide,
which may be unmodified RNA or DNA, or modified RNA or DNA. "Polynucleotides"
include, without limitation single- and double-stranded DNA, DNA that is a
mixture of
single- and double-stranded regions, single- and double-stranded RNA, and RNA
that is a
mixture of single- and double-stranded regions, hybrid molecules comprising
DNA and RNA
that may be single-stranded or, more typically, double-stranded or a mixture
of single- and
to double-stranded regions. In addition, "polynucleotide" refers to triple-
stranded regions
comprising RNA or DNA or both RNA and DNA. The term polynucleotide also
includes
DNAs or RNAs containing one or more modified bases and DNAs or RNAs with
backbones
modified for stability or for other reasons. "Modified" bases include, for
example, tritylated
bases and unusual bases such as inosine. A variety of modifications has been
made to DNA
and RNA; thus "polynucleotide" embraces chemically, enzymatically or
metabolically
modified forms of polynucleotides as typically found in nature, as well as the
chemical forms
of DNA and RNA characteristic of viruses and cells. "Polynucleotide" includes
but is not
limited to linear and end-closed molecules. "Polynucleotide" also embraces
relatively short
polynucleotides, often referred to as oligonucleotides.
"Polypeptides" refers to any peptide or protein comprising two or more amino
acids joined to
each other by peptide bonds or modified peptide bonds (i.e., peptide
isosteres).
"Polypeptide" refers to both short chains, commonly referred as peptides,
oligopeptides or
oligomers, and to longer chains generally referred to as proteins. As
described above,
polypeptides may contain amino acids other than the 20 gene-encoded amino
acids.
As used herein the term "analogues" relates to mutants, variants, chimeras,
fusions, deletions,
additions and any other type of modifications made relative to a given
polypeptide. The term
"analogue" is synonym of homologue, derivative and chemical equivalent or
biological
3o equivalent.
As used herein, the term "homologous" sequence relates to nucleotide or amino
acid
8
CA 02362004 2001-11-13
sequence derived from the DNA sequence or polypeptide sequence of C3APL,
C3APLT,
C3APS, C3-TL, C3-TS, C3Basicl, C3Basic2 and C3Basic3.
As used herein, the term "heterologous" sequence relates to DNA sequence or
amino acid
sequence of a heterologous polypeptide and includes sequence other than that
of C3APL,
C3APLT, C3APS, C3-TL, C3-TS, C3Basicl, C3Basic2 and C3Basic3.
As used herein the term "basic amino acid rich region" relates to a region of
a protein with a
high content of the basic amino acids such as Arginine, Histidine, Asparagine,
Glutamine,
1o Lysine (Lys). "Basic amino acid rich region" may have, for example 15% or
more (up to
100%) of basic amino acids. In some instance, a "basic amino acid rich region"
may have
less than 15% of basic amino acids and still function as a transport agent
region.
As used herein the term "proline rich region" refers to a region of a protein
with 5 % or more
15 (up to 100%) of proline in its sequence. In some instance a "proline rich
region" may have
between 5% and 10% of proline and still function as a transport agent region.
As used herein the term "to help neuron make new connections with other cells"
or "helping
neurones to make new cell connection" means that upon treatment of cells
(e.g., neuron(s))
20 or tissue with a drug delivery construct, a conjugate, a fusion-protein, a
polypeptide or a
pharmaceutical compositions of the present invention, neurons may grow
(develop) for
example new dendrite, new axon or new neurite (i.e., cell bud), or already
existing
dendrite(s), axon or neurite (i.e., cell bud) are induce to grow to a greater
extent.
25 As used herein, the term "vector" refers to an autonomously replicating DNA
or RNA
molecule into which foreign DNA or RNA fragments are inserted and then
propagated in a
host cell for either expression or amplification of the foreign DNA or RNA
molecule. The
term "vector" comprises and is not limited to a plasmid (e.g., linearized or
not) that can be
used to transfer DNA sequences from one organism to another.
The term "pharmaceutically acceptable carrier" or "adjuvant" and
"physiologically
acceptable vehicle" and the like are to be understood as refernng to an
acceptable carrier or
adjuvant that may be administered to a patient, together with a compound of
this invention,
9
CA 02362004 2001-11-13
and which does not destroy the pharmacological activity thereof. Further, as
used herein
"pharmaceutically acceptable carrier" or "pharmaceutical carrier" are known in
the art and
include, but are not limited to, 0.01-0.1 M and preferably 0.05 M phosphate
buffer or 0.8
saline. Additionally, such pharmaceutically acceptable carriers may be aqueous
or non-
aqueous solutions, suspensions, and emulsions. Examples of non-aqueous
solvents are
propylene glycol, polyethylene glycol, vegetable oils such as olive oil, and
injectable organic
esters such as ethyl oleate. Aqueous carriers include water, alcoholic/aqueous
solutions,
emulsions or suspensions, including saline and buffered media. Parenteral
vehicles include
sodium chloride solution, Ringer's dextrose, dextrose and sodium chloride,
lactated Ringer's
to orfixed oils. Intravenous vehicles include fluid and nutrient replenishers,
electrolyte
replenishers such as those based on Ringer's dextrose, and the like.
Preservatives and other
additives may also be present, such as, for example, antimicrobials,
antioxidants, collating
agents, inert gases and the like.
As used herein, "pharmaceutical composition" means therapeutically effective
amounts of the
agent together with pharmaceutically acceptable diluents, preservatives,
solubilizers,
emulsifiers, adjuvant and/or Garners. A "therapeutically effective amount" as
used herein
refers to that amount which provides a therapeutic effect for a given
condition and
administration regimen. Such compositions are liquids or lyophilized or
otherwise dried
2o formulations and include diluents of various buffer content (e.g., Tris-
HCI., acetate,
phosphate), pH and ionic strength, additives such as albumin or gelatin to
prevent absorption
to surfaces, detergents (e.g., Tween 20, Tween 80, Pluronic F68, bile acid
salts). Solubilizing
agents (e.g., glycerol, polyethylene glycerol), anti-oxidants (e.g., ascorbic
acid, sodium
metabisulfite), preservatives (e.g., thimerosal, benzyl alcohol, parabens),
bulking substances
or tonicity modifiers (e.g., lactose, mannitol), covalent attachment of
polymers such as
polyethylene glycol to the protein, complexation with metal ions, or
incorporation of the
material into or onto particulate preparations of polymeric compounds such as
polylactic acid,
polyglycolic acid, hydrogels, etc, or onto liposomes, microemulsions,
micelles, unilamellar or
multilamellar vesicles, erythrocyte ghosts, or spheroplasts. Such compositions
will influence
3o the physical state, solubility, stability, rate of in vivo release, and
rate of in vivo clearance.
Controlled or sustained release compositions include formulation in lipophilic
depots (e.g.,
fatty acids, waxes, oils). Also comprehended by the invention are particulate
compositions
CA 02362004 2001-11-13
coated with polymers (e.g., poloxamers or poloxamines). Other embodiments of
the
compositions of the invention incorporate particulate forms protective
coatings, protease
inhibitors or permeation enhancers for various routes of administration,
including parenteral,
pulmonary, nasal and oral routes. In one embodiment the pharmaceutical
composition is
administered parenterally, paracancerally, transmucosally, transdermally,
intramuscularly,
intravenously, intradermally, subcutaneously, intraperitonealy,
intraventricularly,
intracranially intratumorally or more preferably, directly at a central
nervous system (CNS)
lesion site or a peripheral nervous system (PNS) lesion site.
1o As may be appreciated, a number of modifications may be made to the
polypeptides of the
present invention, such as for example the active agent region (e.g., ADP-
ribosyl transferase
C3 or ADP-ribosyl transferase C3 analogue) or the transport agent region
(e.g., a subdomain
of HIV Tat protein, or a homeodomain of antennapedia) and fragments thereof
without
deleteriously affecting the biological activity of the polypeptides or
fragments. Polypeptides
15 of the present invention comprises for example, those containing amino acid
sequences
modified either by natural processes, such as posttranslational processing, or
by chemical
modification techniques which are known in the art. Modifications may occur
anywhere in a
polypeptide including the polypeptide backbone, the amino acid side-chains and
the amino or
carboxy termini. It will be appreciated that the same type of modification may
be present in
20 the same or varying degrees at several sites in a given polypeptide. Also,
a given polypeptide
may contain many types of modifications. Polypeptides may be branched as a
result of
ubiquitination, and they may be cyclic, with or without branching. Cyclic,
branched and
branched cyclic polypeptides may result from posttranslational natural
processes or may be
made by synthetic methods. Modifications comprise for example, without
limitation,
25 acetylation, acylation, addition of acetomidomethyl (Acm) group, ADP-
ribosylation,
amidation, covalent attachment to fiavin, covalent attachment to a heme
moiety, covalent
attachment of a nucleotide or nucleotide derivative, covalent attachment of a
lipid or lipid
derivative, covalent attachment of phosphatidylinositol, cross-linking,
cyclization, disulfide
bond formation, demethylation, formation of covalent cross-links, formation of
cystine,
30 formation of pyroglutamate, formylation, gamma-carboxylation,
glycosylation, GPI anchor
formation, hydroxylation, iodination, methylation, myristoylation, oxidation,
proteolytic
processing, phosphorylation, prenylation, racemization, selenoylation,
sulfation, transfer-
RNA mediated addition of amino acids to proteins such as arginylation and
ubiquitination
11
CA 02362004 2001-11-13
(for reference see, Protein-structure and molecular proterties, 2°d
Ed., T.E. Creighton, W.H.
Freeman and Company, New-York, 1993).
Other type of polypeptide modification may comprises for example, amino acid
insertion
(i.e., addition), deletion and substitution (i.e., replacement), either
conservative or non-
conservative (e.g., D-amino acids, desamino acids) in the polypeptide sequence
where such
changes do not substantially alter the overall biological activity of the
polypeptide.
Polypeptides of the present invention comprise for example, biologically
active mutants,
variants, fragments, chimeras, and analogues; fragments encompass amino acid
sequences
1o having truncations of one or more amino acids, wherein the truncation may
originate from the
amino terminus (N-terminus), carboxy terminus (C-terminus), or from the
interior of the
protein. Analogues of the invention involve an insertion or a substitution of
one or more
amino acids. Variants, mutants, fragments, chimeras and analogues may have the
biological
properties of polypeptides of the present invention which comprise for example
(without
being restricted to the present examples) to facilitate neuronal axon growth,
to suppress the
inhibition of neuronal axon growth, to facilitate neurite growth, to inhibit
apoptosis, to treat
nerve injury, to regenerate injured axon and/or to act as a Rho antagonist.
As it may be examplified (Example 13: reverse Tat sequence), in some instance,
the order of
2o the amino acids in a particular polypeptide is not critical. As for the
transport agent region
described herein, the transport function of this region may be preserved even
if the amino
acids are not in their original (as it is found in nature) order (sequence).
Example of substitutions may be those, which are conservative (i.e., wherein a
residue is
replaced by another of the same general type). As is understood, naturally
occurring amino
acids may be sub-classified as acidic, basic, neutral and polar, or neutral
and non-polar.
Furthermore, three of the encoded amino acids are aromatic. It may be of use
that encoded
polypeptides differing from the determined polypeptide of the present
invention contain
substituted codons for amino acids, which are from the same group as that of
the amino acid
3o be replaced. Thus, in some cases, the basic amino acids Lys, Arg and His
may be
interchangeable; the acidic amino acids Asp and Glu may be interchangeable;
the neutral
polar amino acids Ser, Thr, Cys, Gln, and Asn may be interchangeable; the non-
polar
aliphatic amino acids Gly, Ala, Val, Ile, and Leu are interchangeable but
because of size Gly
12
CA 02362004 2001-11-13
and Ala are more closely related and Val, Ile and Leu are more closely related
to each other,
and the aromatic amino acids Phe, Trp and Tyr may be interchangeable.
It should be further noted that if the polypeptides are made synthetically,
substitutions by
amino acids, which are not naturally encoded by DNA may also be made. For
example,
alternative residues include the omega amino acids of the formula
NH2(CH2)nCOOH
wherein n is 2-6. These are neutral nonpolar amino acids, as are sarcosine, t-
butyl alanine, t-
butyl glycine, N-methyl isoleucine, and norleucine. Phenylglycine may
substitute for Trp,
Tyr or Phe; citrulline and methionine sulfoxide are neutral nonpolar, cysteic
acid is acidic,
to and ornithine is basic. Proline may be substituted with hydroxyproline and
retain the
conformation confernng properties.
It is known in the art that mutants or variants may be generated by
substitutional mutagenesis
and retain the biological activity of the polypeptides of the present
invention. These variants
have at least one amino acid residue in the protein molecule removed and a
different residue
inserted in its place (one or more nucleotide in the DNA sequence is changed
for a diferent
one using known molecular biology techniques, giving a different amino acid
upon
translation of the corresponding messenger RNA to a polypeptide). For example,
one site of
interest for substitutional mutagenesis may include but are not restricted to
sites identified as
2o the active site(s), or immunological site(s). Other sites of interest may
be those, for example,
in which particular residues obtained from various species are identical.
These positions may
be important for biological activity. Examples of substitutions identified as
"conservative
substitutions" are shown in Table 1. If such substitutions result in a change
not desired, then
other type of substitutions, denominated "exemplary substitutions" in Table 1,
or as further
described herein in reference to amino acid classes, are introduced and the
products screened.
In some cases it may be of interest to modify the biological activity of a
polypeptide by
amino acid substitution, insertion, or deletion. For example, modification of
a polypeptide
may result in an increase in the polypeptide's biological activity, may
modulate its toxicity,
may result in changes in bioavailability or in stability, or may modulate its
immunological
activity or immunological identity. Substantial modifications in function or
immunological
identity are accomplished by selecting substitutions that differ significantly
in their effect on
maintaining (a) the structure of the polypeptide backbone in the area of the
substitution, for
13
CA 02362004 2001-11-13
example, as a. sheet or helical conformation. (b) the charge or hydrophobicity
of the molecule
at the target site, or (c) the bulk of the side chain. Naturally occurring
residues are divided
into groups based on common side chain properties:
(1) hydrophobic: norleucine, methionine (Met), Alanine (Ala), Valine (Val),
Leucine
(Leu), Isoleucine (Ile)
(2) neutral hydrophilic: Cysteine (Cys), Serine (Ser), Threonine (Thr)
(3) acidic: Aspartic acid (Asp), Glutamic acid (Glu)
(4) basic: Asparagine (Asn), Glutamine (Gln), Histidine (His), Lysine (Lys),
Arginine
i o (fig)
(5) residues that influence chain orientation: Glycine (Gly), Proline (Pro);
and
(6) aromatic: Tryptophan (Trp), Tyrosine (Tyr), Phenylalanine (Phe)
Non-conservative substitutions will entail exchanging a member of one of these
classes for another.
14
CA 02362004 2001-11-13
TABLE 1. Preferred amino acid substitution
Original residueExemplary substitutionConservative substitution
Ala (A) Val, Leu, Ile Val
Arg (R) Lys, Gln, Asn Lys
Asn (N) Gln, His, Lys, Arg Gln
Asp (D) Glu Glu
Cys (C) Ser Ser
Gln (Q) Asn Asn
Glu (E) Asp Asp
Gly (G) Pro Pro
His (H) Asn, Gln, Lys, Arg Arg
Ile (I) Leu, Val, Met, Ala,Leu
Phe,
norleucine
Leu (L) Norleucine, Ile, Ile
Val, Met,
Ala, Phe
Lys (K) Arg, Gln, Asn Arg
Met (M) Leu, Phe, Ile Leu
Phe (F) Leu, Val, Ile, Ala Leu
Pro (P) Gly Gly
Ser (S) Thr Thr
Thr (T) Ser Ser
Trp (W) Tyr Tyr
Tyr (Y) Trp, Phe, Thr, Ser Phe
Val (V) Ile, Leu, Met, Phe,Leu
Ala,
norleucine
Amino acids sequence insertions (e.g., additions) include amino and/or
carboxyl-terminal
fusions ranging in length from one residues to polypeptides containing a
hundred or more
residues, as well as intrasequence insertions of single or multiple amino acid
residues. Other
insertional variants include the fusion of the N- or C-terminus of the protein
to a homologous
CA 02362004 2001-11-13
or heterologous polypeptide forming a chimera. Chimeric polypeptides (i.e.,
chimeras,
polypeptide analogue) comprise sequence of the polypeptides of the present
invention fused
to homologous or heterologous sequence. Said homologous or heterologous
sequence
encompass those which, when formed into a chimera with the polypeptides of the
present
invention retain one or more biological or immunological properties.
Other type of chimera generated by homologous fusion includes new polypeptides
formed by
the repetition of two or more polypeptides of the present invention. The
number of repeat
may be, for example, between 2 and 50 units (i.e., repeats). In some instance,
it may be
to useful to have a new polypeptide with a number of repeat greater than 50.
For example, it
may be useful to fuse (using cross-linking techniques or recombianant DNA
technology
techniques) polypeptides such as C3APL, C3APLT, C3APS, C3-TL, C3-TS, C3Basicl,
C3Basic2 and C3Basic3 either to themselves (e.g., C3APLT fused to C3APLT) or
to another
polypeptide of the present invention (e.g., C3APLT fused to C3APL).
In addition, a transport agent such as for example, a subdomain of HIV Tat
protein, and a
homeodomain of antennapedia may be repeated more than one time in a
polypeptide
comprising the ADP-ribosyl transferase C3 or ADP-ribosyl transferase C3
analogues. The
transport agent region may be either at the amino-terminal region of an ADP-
ribosyl
2o transferase C3 or ADP-ribosyl transferase C3 analogues or at its carboxy-
terminal region or
at both regions. The repetition of a transport agent region may affect
(increase) the uptake of
the ADP-ribosyl transferase C3 or ADP-ribosyl transferase C3 analogues by a
desired cell.
Heterologous fusion includes new polypeptides made by the fusion of
polypeptides of the
present invention with heterologous polypeptides. Such polypeptides may
include but are not
limited to bacterial polypeptides (e.g., betalactamase, glutathione-S-
transferase, or an enzyme
encoded by the E.coli trp locus), yeast protein, viral proteins, phage
proteins, bovine serum
albumin, chemotactic polypeptides, immunoglobulin constant region (or other
immunoglobulin regions), albumin, or ferntin.
Other type of polypeptide modification includes amino acids sequence deletions
(e.g.,
truncations). Those generally range from about 1 to 30 residues, more
preferably about 1 to
10 residues and typically about 1 to 5 residues.
16
CA 02362004 2001-11-13
Mutants, variants and analogues proteins
Mutant polypeptides will possess one or more mutations, which are deletions
(e.g.,
truncations), insertions (e.g., additions), or substitutions of amino acid
residues. Mutants can
be either naturally occurring (that is to say, purified or isolated from a
natural source) or
synthetic (for example, by performing site-directed mutagenesis on the
encoding DNA or
made by other synthetic methods such as chemical synthesis). It is thus
apparent that the
polypeptides of the invention can be either naturally occurnng or recombinant
(that is to say
prepared from the recombinant DNA techniques).
A protein at least 50 % identical, as determined by methods known to those
skilled in the art
(for example, the methods described by Smith, T.F. and Waterman M.S. (1981)
Ad.
Appl.Math., 2:482-489, or Needleman, S.B. and Wunsch, C.D. (1970) J.Mol.Biol.,
48: 443-
453), to those polypeptides of the present invention, for example C3APL,
C3APLT, C3APS,
C3-TL, C3-TS, C3Basicl, C3Basic2 and C3Basic3 are included in the invention,
as are
proteins at least 70 % or 80 % and more preferably at least 90 % identical to
the protein of the
present invention. This will generally be over a region of at least 5,
preferably at least 20
contiguous amino acids.
"Variant" as the term used herein, is a polynucleotide or polypeptide that
differs from
reference polynucleotide or polypeptide respectively, but retains essential
properties. A
typical variant of a polynucleotide differs in nucleotide sequence from
another, reference
polynucleotide. Changes in the nucleotide sequence of the variant may or may
not alter the
amino acid sequence of a polypeptide encoded by the reference polynucleotide.
Nucleotide
changes may result in amino acid substitutions, additions, deletions, fusion
and truncations in
the polypeptide encoded by the reference sequence, as discussed herein. A
typical variant of
a polypeptide differs in amino acid sequence from another, reference
polypeptide. Generally,
differences are limited so that the sequence of the reference polypeptide and
the variant are
closely similar overall and, in many regions, identical. A variant and
reference polypeptide
may differ in amino acid by one or more substitutions, additions, deletions,
or any
combination therefore. A substituted or inserted amino acid residue may or may
not be one
encoded by the genetic code. A variant polynuclotide or polypeptide may be a
naturally
occurring such as an allelic variant, or it may be a variant that is not known
to occur
17
CA 02362004 2001-11-13
naturally. Non-naturally occurring variants of polynucleotides and
polypeptides may be
made by mutagenesis techniques or by direct synthesis.
Amino acid sequence variants may be prepared by introducing appropriate
nucleotide
changes into DNA, or by in vitro synthesis of the desired polypeptide. Such
variant include,
for example, deletions, insertions, or substitutions of residues within the
amino acid
sequence. A combination of deletion, insertion and substitution can be made to
arrive at the
final construct, provided that the final protein product possesses the desired
biological
activity, or characteristics. The amino acid changes also may alter
posttranslational processes
to such as changing the number or position of the glycocylation sites,
altering the membrane
anchoring characteristics, altering the intra-cellular location by inserting,
deleting or
otherwise affecting the transmembrane sequence of the native protein, or
modifying its
susceptibility to proteolytic cleavage.
Unless otherwise indicated, the recombinant DNA techniques utilized in the
present invention
are standard procedures, known to those skilled in the art. Example of such
techniques are
explained in the literature in sources such as J. Perbal, A Practical Guide to
Molecular
Cloning, John Wiley and Sons (1984), J. Sambrook et al ., Molecular Cloning: A
Laboratory
2o Manual, Cold Spring Harbor Laboratory Press (1989), T.A. Brown (editor),
Essential
Molecular Biology: A Practical Approach, Volumes 1 and 2, IRL Press (1991),
D.M. Glover
and B.D. Hames (editors), DNA Cloning: A Practical Approach, Volumes 1-4, IRL
Press
(1995 and 1996), and F.M. Ausubel et al. (editors), Current Protocols in
Molecular Biology,
Greene Pub. Associates and Wiley-Interscience (1988, including all updates
until present)
and are incorporated herein by reference.
It is to be understood herein, that if a "range" or "group of substances" is
mentioned with
respect to a particular characteristic (e.g. amino acid groups, temperature,
pressure, time and
the like) of the present invention, the present invention relates to and
explicitly incorporates
3o herein each and every specific member and combination of sub-ranges or sub-
groups therein
whatsoever. Thus, any specified range or group is to be understood as a
shorthand way of
refernng to each and every member of a range or group individually as well as
each and
18
CA 02362004 2001-11-13
every possible sub-ranges or sub-groups encompassed therein; and similarly
with respect to
any sub-ranges or sub-groups therein. Thus, for example,
- with respect to a sequence comprising up to 50 base units it is to be
understood as
specifically incorporating herein each and every individual unit, as well as
sub-
range of units;
- with respect to reaction time, a time of 1 minute or more is to be
understood as
specifically incorporating herein each and every individual time, as well as
sub-
range, above 1 minute, such as for example 1 minute, 3 to 15 minutes, 1 minute
to
20 hours, 1 to 3 hours, 16 hours, 3 hours to 20 hours etc.;
- with respect to polypeptides, a polypeptide analogue comprising a particular
sequence and having an addition of at least one amino acid to its amino-
terminus
or to its carboxy terminus is to be understood as specifically incorporating
each
and every individual possibility, such as for example one, two, three, ten ,
eighteen, forty, etc.;
- with respect to polypeptides, a polypeptide analogue having at least 90 % of
its
amino acid sequence identical to a particular amino acid sequence is to be
understood as specifically incorporating each and every individual possibility
(excluding 100 %), such as for example, a polypeptide analogue having 90 %,
2o 90.5%, 91%, 93.7%, 97%, 99%, etc., of its amino acid sequence identical to
a
particular amino acid sequence.
- with respect to polypeptides, a polypeptide analogue having at least 70 % of
its
amino acid sequence identical to a particular amino acid sequence is to be
understood as specifically incorporating each and every individual possibility
(excluding 100 %), such as for example, a polypeptide analogue having 70 %,
72.3%, 73%, 88.6%, 98% etc., of its amino acid sequence identical to a
particular
amino acid sequence.
- with respect to polypeptides, a polypeptide analogue having at least 50 % of
its
amino acid sequence identical to a particular amino acid sequence is to be
19
CA 02362004 2001-11-13
understood as specifically incorporating each and every individual possibility
(excluding 100 %), such as for example, a polypeptide analogue having 50 %,
54%, 66.7%, 70.2%, 84%, 93% etc., of its amino acid sequence identical to that
particular amino acid sequence.
- with respect to polypeptide, a polypeptide comprising at least one transport
agent
region is to be understood as specifically incorporating each and every
individual
possibility, such as for example a polypeptide having one, two, five, ten,
etc.,
transport agent region.
- and similarly with respect to other parameters such as low pressures,
concentrations, elements, etc...
It is also to be understood herein that "g" or "gm" is a reference to the gram
weight unit; that
"C", or " °C " is a reference to the Celsius temperature unit; and
"psig" is a reference to
"pounds per square inch guage".
CA 02362004 2001-11-13
Table 2: Abbreviations
Abbreviation Full name
C3 ADP-ribosyl transferase C3
NGF Nerve growth factor
BDNF Brain-derived neurotrophic factor
C or C Degree Celcius
ml milliliter
~l or u1 microliter
~M or uM micromolar
mM millimolar
M molar
N normal
CNS Central nervous system
PNS Peripheral nervous system
HIV Human immunodeficiency virus
kDa kilodalton
GST Glutathione S-transferase
MTS Membrane transport sequence
SDS-PAGE Sodium dodecyl sulfte polyacrylamide gel
electrophoresis
PBS Phosphate buffered saline
U unit
BBB Basso, Beattie Breshnahan behavior recovery
scale
IPTG Isopropyl /3-D-thiogalactopyranoside
rpm Rotation per minutes
DTT dithiothreitol
PMSF Phenylmethylsulfonyl fluoride
NaCI Sodium chloride
21
CA 02362004 2001-11-13
Table 2: Abbreviations; continued
Abbreviation Full name
MgCl2 Magnesium chloride
HBSS Hank's balanced salt solution
NaOH Sodium hydroxide
CSPG chondroitin sulfate proteoglycan
PKN Protein kinase N
RSV Rous sarcoma virus
MMTV Mouse mammary tumor virus
LTR Long terminal repeat
HL Hind limb
FL Fore limb
neo neomycin
hygro hygromycin
IN-1 monoclonal antibody called IN-1
ADP Adenosine di-phosphate
ATP Adenosine tri-phosphate
"P Isotope 32 of phosphorus
DHFR Dihydrofolate reductase
PCR Polymerase chain reaction
In accordance with the present invention a conjugate or fusion protein
comprising a
therapeutically active agent is provided whereby the active agent may be
delivered across a
cell wall membrane, the conjugate or fusion protein comprising at least a
transport
subdomain(s) or moiety(ies) in addition to an active agent moiety(ies). More
particularly, as
discussed herein, in acccordance with the present invention a conjugate or
fusion protein is
provided wherein the therapeutically active agent is one able to faciliate
(for facilitating) axon
22
CA 02362004 2001-11-13
(or dendrite, or neurite) growth (e.g. regeneration) i.e. a conjugate or
fusion protein in the
form of a conjugate Rho antagonist.
The present invention in acccordance with an aspect thereof provides a drug
delivery
construct or conjugate [e.g. able to (for) suppressing) the inhibition of
neuronal axon growth
at a central nervous system (CNS) lesion site or a peripheral nervous system
(PNS) lesion
site] comprising at least one transport agent region and an active agent
region not naturally
associated with the active agent region, wherein the transport agent region is
able to facilitate
(i.e. facilitates) the uptake of the active agent region into a mammalian
(i.e. human or animal)
1o tissue or cell, and wherein the active agent region is an active
therapeutic agent region able
(i.e. has the capacity or capability) to facilitate axon growth for example on
growth inhibitory
substrates (e.g. regeneration), either in vivo (in a mammal (e.g., human or
animal)) or in vitro
(in cell culture), including a derivative or homologue thereof (i.e.
pharmaceutically
acceptable chemical equivalents thereof - pharmaceutically acceptable
derivative or
homologue).
In accordance with the present invention the active agent region may be an ADP-
ribosyl
transferase C3 region. In accordance with the present invention the ADP-
ribosyl transferase
C3 may be selected from the group consisting of ADP-ribosyl transferase (e.g.,
ADP-ribosyl
transferase C3) derived from Clostridium botulinum and a recombinant ADP-
ribosyl
transferase (e.g., recombinant ADP-ribosyl transferase C3) that includes the
entire C3 coding
region, or only a part (fragment) of the C3 coding region that retains the ADP-
ribosyl
transferase activity, or analogues (derivatives) of C3 that retains the ADP-
ribosyl transferase
activity, or enough of the C3 coding region to be able to effectively
inactivate Rho. The
active agent could also be selected from other known ADP-ribosyl transferases
that act on
Rho (Wilde et al. 2000 J. Biol. Chem. 275-16478-16483; Wilde et al 2001. J.
Biol. Chem.
276:9537-9542).
In accordance with another aspect the present invention provides a drug
conjugate consisting
of a transport polypeptide moiety (e.g. rich in basic amino acids e.g.
arginine, lysine,
23
CA 02362004 2001-11-13
histidine, asparagine, glutamine) covalently linked to an active cargo moiety
(e.g. by a
peptide bond or a labile bond (i.e. a bond readily cleavable or subject to
chemical change in
the interior target cell environment)) wherein the transport polypeptide
moiety is able to or
has the capability to facilitates) the uptake of the active cargo moiety into
a mammalian (e.g.
human or animal) tissue or cell (for example, a transport subdomain of HIV Tat
protein, a
homeoprotein transport sequence (refered also as a transport homeoprotein)
(e.g. the
homeodomain of antennapedia), a Histidine tag (ranging in length from 4 to 30
histidine
repeat) or a variation derivative or homolog thereof, (i.e. pharmaceutically
acceptable
chemical equivalents thereof)) [by a receptor independent process] and wherein
the active
cargo moiety is an active therapeutic moiety able (i.e. has the capacity or
capability) to
facilitate (i.e. for facilitating) axon growth (e.g. regeneration, budding) or
neuroprotection
(prevention of cell death) either in vivo (in a mammal (e.g., human or
animal)) or in vitro (in
cell culture).
In accordance with the present invention the transport polypeptide moiety may
be selected
from the group consisting of a transport subdomain of HIV Tat protein such as
for example
SEQ ID NO.: 46, SEQ ID N0.:47, a homeodomain of antennapedia, such as for
example
SEQ ID NO.: 44, SEQ ID NO.: 45, SEQ ID NO.: 48, a Histidine tag and a
functional
derivative and analogue thereof (e.g., SEQ ID NO.: 21, SEQ ID NO.: 26, SEQ ID
NO.: 31)
[i.e. by the addition of polyamine, or any random sequence enriched in basic
amino acids] -
[i.e. pharmaceutically acceptable chemical equivalents thereof) and wherein
the active cargo
moiety is selected from the group consisting of C3 protein able (i.e. has the
capacity or
capability) to facilitate (i.e. for facilitating) axon growth (e.g.
regeneration, budding) or
neuroprotection (prevention of cell death) either in vivo (in a mammal (e.g.,
human or
2s animal)) or in vitro (in cell culture).
In accordance with the present invention the C3 protein may be selected from
the group
consisting of ADP-ribosyl transferase C3 and ADP-ribosyl transferase C3
analogue. In
accordance with the present invention the ADP-ribosyl transferase C3 may be
selected from
3o the group consisting of ADP-ribosyl transferase (e.g., ADP-ribosyl
transferase C3) derived
from Clostridium botulinum and a recombinant ADP-ribosyl transferase (e.g.,
recombinant
24
CA 02362004 2001-11-13
ADP-ribosyl transferase C3). The ADP-ribosyl transferase may be a protein with
a C3-like
activity, such as that derived from Staphylococcus aureus (Wilde et al 2001.
J. Biol. Chem.
276:9537-9542). The ADP-ribosyl transferase may be any other transferase that
acts to
inactivate RhoA, RhoB and/or RhoC such as those derived from Clostridium
limosum, and
Bacillus cereus (Wilde et al 2000. J. Biol. Chem. 275:16478-16483). In
accordance with the
present invention the transport polypeptide moiety may include an active
contiguous amino
acid sequence as described herein.
In accordance with an additional aspect the present invention provides a
fusion protein [e.g.
able to (for) suppressing) the inhibition of neuronal axon growth at a central
nervous system
(CNS) lesion site or a peripheral nervous system (PNS) lesion site] consisting
of a carboxy
terminal active cargo moiety and an amino terminal transport moiety, wherein
the amino
terminal transport moiety is selected from the group consisting of a transport
subdomain of
HIV Tat protein, homeoprotein transport sequence (refered also as a transport
homeoprotein)
(e.g. the homeodomain of antennapedia), a Histidine tag and a functional
derivatives and
analogues thereof (i.e. pharmaceutically acceptable chemical equivalents
thereof) and
wherein the active cargo moiety consists of a C3 protein.
The present invention in particular provides a fusion protein (e.g. able to
(for) suppressing the
2o inhibition of neuronal axon growth at a central nervous system (CNS) lesion
site or a
peripheral nervous system (PNS) lesion site) consisting of a carboxy terminal
active cargo
moiety and an amino terminal transport moiety, wherein the amino terminal
transport moiety
consists of the homeodomain of antennapedia and the active cargo moiety
consists of a C3
protein (i.e. as described herein).
The present invention also in particular provides a fusion protein (e.g. able
to (for)
suppressing the inhibition of neuronal axon growth at a central nervous system
(CNS) lesion
site or a peripheral nervous system (PNS) lesion site) consisting of a carboxy
terminal active
cargo moiety and an amino terminal transport moiety, wherein the amino
terminal transport
CA 02362004 2001-11-13
moiety consists of a transport subdomain of HIV Tat protein and the active
cargo moiety
consists of a C3 protein (i.e. as described herein).
In accordance with the present invention the C3 protein may be selected from
the group
consisting of ADP-ribosyl transferase C3 and ADP-ribosyl transferase C3
analogues. In
accordance with the present invention the ADP-ribosyl transferase C3 is
selected from the
group consisting of ADP-ribosyl transferase (e.g., ADP-ribosyl transferase C3)
derived from
Clostridium botulinum and a recombinant ADP-ribosyl transferase (e.g.,
recombinant ADP-
ribosyl transferase C3).
In accordance with an additional aspect the present invention provides a
fusion protein [e.g.
able to (for) suppressing) the inhibition of neuronal axon growth at a central
nervous system
(CNS) lesion site or a peripheral nervous system (PNS) lesion site] consisting
of an amino
terminal active cargo moiety and a carboxy terminal transport moiety, wherein
the carboxy
terminal transport moiety is selected from the group consisting of a transport
subdomain of
HIV Tat protein, a homeoprotein transport sequence (refered also as a
transport
homeoprotein) (e.g. the homeodomain of antennapedia), a Histidine tag and a
functional
derivatives and analogues thereof (i.e. pharmaceutically acceptable chemical
equivalents
thereof) and wherein the active cargo moiety consists of a C3 protein.
The present invention in particular provides a fusion protein (e.g. able to
(for) suppressing the
inhibition of neuronal axon growth at a central nervous system (CNS) lesion
site or a
peripheral nervous system (PNS) lesion site) consisting of an amino terminal
active cargo
moiety and a carboxy terminal transport moiety, wherein the carboxy terminal
transport
moiety consists of the homeodomain of antennapedia and the active cargo moiety
consists of
a C3 protein (i.e. as described herein).
The present invention also in particular provides a fusion protein (e.g. able
to (for)
suppressing the inhibition of neuronal axon growth at a central nervous system
(CNS) lesion
26
CA 02362004 2001-11-13
site or a peripheral nervous system (PNS) lesion site) consisting of an amino
terminal active
cargo moiety and a carboxy terminal transport moiety, wherein the carboxy
terminal transport
moiety consists of a transport subdomain of HIV Tat protein and the active
cargo moiety
consists of a C3 protein (i.e. as described herein).
In accordance with the present invention the C3 protein may be selected from
the group
consisting of ADP-ribosyl transferase C3 and ADP-ribosyl transferase C3
analogues. In
accordance with the present invention the ADP-ribosyl transferase C3 is
selected from the
group consisting of ADP-ribosyl transferase C3 derived from Clostridium
botulinum and a
l0 recombinant ADP-ribosyl transferase C3.
The present invention in a further aspect provides for the use of a member
selected from the
group consisting of a drug delivery construct as described herein, a drug
conjugate as
described herein and a fusion protein as described herein (e.g. including
pharmaceutically
acceptable chemical equivalents thereof) for suppressing the inhibition of
neuronal axon
growth.
The present invention in a further aspect relates to a pharmaceutical
composition (e.g. for
suppressing the inhibition of neuronal axon growth), the pharmaceutical
composition
2o comprising a pharmaceutically acceptable diluent or Garner and an effective
amount of an
active member selected from the group consisting of a drug delivery construct
as described
herein, a drug conjugate as described herein, and a fusion protein as
described herein (e.g.
including pharmaceutically acceptable chemical equivalents thereof).
The present invention further provides for the use of a member selected from
the group
consisting of a drug delivery construct as described herein, a drug conjugate
as described
herein, and a fusion protein as described herein (e.g. including
pharmaceutically acceptable
chemical equivalents thereof) for the manufacture of a pharmaceutical
composition (e.g. for
suppressing the inhibition of neuronal axon growth).
27
CA 02362004 2001-11-13
The present invention also relates to a method for preparing a conjugate or
fusion protein as
defined above comprising
- cultivating a host cell (bacterial or eukaryotic) under conditions which
provide for the expression of the conjugate or fusion protein within the
cell; the protein could also be expressed to be produced in an animals,
such as, for example, the production of recombinant proteins in the milk
of farm animals
- recovering the conjugate or fusion protein by purification
to
The purification of the fusion protein may be by affinity methods, ion
exchange
chromatography, size exclusion chromatography, hydrophobicity or any other
purification
technique typically used for protein purification.
The present invention also relates to the expression of the fusion protein in
a mammalian cell,
which when used with a signal sequence, will allow expression and secretion of
the fusion
protein into the extracellular milieu.
The present invention in particular provides a fusion protein selected from
the group
2o consisting of C3APL (SEQ ID NO.: 4), C3APLT (SEQ ID NO.: 37), C3APS (SEQ ID
N0.:6), C3-TL (SEQ ID N0.:14), C3-TS (SEQ ID NO.: 18), C3Basicl (SEQ ID
N0.:25),
C3Basic2 (SEQ ID NO.: 30), C3Basic3 (SEQ ID N0.:35), SEQ ID NO.: 20, and SEQ
ID
NO.: 43 and pharmaceutically acceptable chemical equivalents thereof.
The present invention in another aspect provides a method of suppressing the
inhibition of
neuronal axon growth comprising administering (e.g.delivering) a member
selected from the
group consisting of a drug delivery construct as described herein, a drug
conjugate as
described herein and a fusion protein as described herein (e.g. including
pharmaceutically
28
CA 02362004 2001-11-13
acceptable chemical equivalents thereof) (e.g. directly) to (at) a central
nervous system (CNS)
lesion site or a peripheral nervous system (PNS) lesion site (of a patient),
in an amount
effective to counteract said inhibition. Such application could be useful for
treatment of a
wide variety of peripheral neuropathies, such as diabetic neuropathy.
The present invention, for example, provides recombinant Rho antagonists
comprising C3
enzymes with basic stretches of amino acids added to the C3 coding sequence to
facilitate the
uptake thereof into tissue or cells for the repair and/or promotion of repair
or promotion of
growth in the CNS, even in the lack of traumatic axon damage.
to
In accordance with an additional aspect, the present invention provides a
pharmaceutical
composition comprising a polypeptide selected from the group consisting of
C3APL (SEQ ID
N0.:4), C3APLT (SEQ ID N0.:37), C3APS (SEQ ID N0.:6), C3-TL (SEQ ID N0.:14),
C3-
TS (SEQ ID N0.:18), C3Basicl(SEQ ID N0.:25), C3Basic2 SEQ ID N0.:30), C3Basic3
(SEQ ID N0.:35), SEQ ID NO.: 20 and SEQ ID NO.: 43, and a pharmaceutically
acceptable
Garner.
In accordance with the present invention, the pharmaceutical composition may
further
comprise a biological adhesive, such as, for example, fibrin (fibrin glue).
In accordance with the present the transport agent region may be selected from
the group
consisting of a basic amino acid rich region (region comprising some basic
amino acid (e.g.,
arginine, lysine, histidine, glutamine, and/or asparagine)) and a proline rich
region (e.g.
region comprising some proline).
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein (e.g., SEQ ID NO.:
46, SEQ ID
NO.: 47, or any other subdomain of Tat, that could act as a transport
sequence), a
homeodomain of antennapedia (e.g., SEQ ID NO.: 44, SEQ ID NO.: 45, SEQ ID NO.:
48, or
3o any other domain of antennapedia, that could act as a transport sequence),
a homeoprotein
transport sequence, a Histidine tag, and analogues thereof (e.g., SEQ ID NO.:
21, SEQ ID
29
CA 02362004 2001-11-13
NO.: 26, SEQ ID N0.:31 ).
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
to
In a further aspect the present invention provides a pharmaceutical
composition comprising a
polypeptide comprising at least one (one or more) transport agent region and
an active agent
region, said active agent region being selected from the group consisting of
ADP-ribosyl
transferase C3 and ADP-ribosyl transferase C3 analogues, and a
pharmaceutically acceptable
Garner.
In accordance with the present invention, the pharmaceutical composition may
further
comprise a biological adhesive, such as, for example, fibrin (fibrin glue).
2o In accordance with the present the transport agent region may be selected
from the group
consisting of a basic amino acid rich region (region comprising some basic
amino acid (e.g.,
arginine, lysine, histidine, glutamine, and/or asparagine)) and a proline rich
region (e.g.
region comprising some proline).
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein (e.g., SEQ ID NO.:
46, SEQ ID
NO.: 47, or any other subdomain of Tat, that could act as a transport
sequence), a
homeodomain of antennapedia (e.g., SEQ ID NO.: 44, SEQ ID NO.: 45, SEQ ID NO.:
48, or
any other domain of antennapedia, that could act as a transport sequence), a
homeoprotein
3o transport sequence, a Histidine tag, and analogues thereof (e.g., SEQ ID
NO.: 21, SEQ ID
NO.: 26, SEQ ID N0.:31 ).
CA 02362004 2001-11-13
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In an additional aspect, the present invention provides a polypeptide
comprising at least one
(one or more) transport agent region and an active agent region, said active
agent region
being selected from the group consisting of ADP-ribosyl transferase C3 and ADP-
ribosyl
transferase C3 analogues.
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein (e.g., SEQ ID NO.:
46, SEQ ID
NO.: 47, or any other subdomain of Tat, that could act as a transport
sequence), a
2o homeodomain of antennapedia and analogues thereof (e.g., SEQ ID NO.: 21,
SEQ ID NO.:
26, SEQ ID N0.:31 ).
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof. (e.g., SEQ ID NO.: 44, SEQ ID NO.: 45,
SEQ ID NO.:
48, or any other domain of antennapedia, that could act as a transport
sequence), a
homeoprotein transport sequence, a Histidine tag,
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
31
CA 02362004 2001-11-13
In an additional aspect, the present invention provides a polypeptide
consisting of a carboxy-
terminal active agent moiety and an amino-terminal transport moiety region and
wherein said
carboxy-terminal active agent moiety may be selected from the group consisting
of ADP-
ribosyl transferase C3 and ADP-ribosyl transferase C3 analogues thereof.
In accordance with the present invention, the carboxy-terminal transport
moiety region may
be selected from the group consisting of a basic amino acid rich region and a
proline rich
region.
to In accordance with the present invention, the basic amino acid rich region
may be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21 (Basic 1 ), SEQ ID NO.: 26 (Basic2),
SEQ ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
2o group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In a further aspect, the present invention relates to a polypeptide consisting
of an amino-
terminal active agent moiety and a carboxy-terminal transport moiety region,
wherein said
amino-terminal active agent moiety may be selected from the group consisting
of ADP-
ribosyl transferase C3 and ADP-ribosyl transferase C3 analogues thereof.
In accordance with the present invention, the carboxy-terminal transport
moiety region may
be selected from the group consisting of a basic amino acid rich region and a
proline rich
region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag and analogues thereof.
32
CA 02362004 2001-11-13
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21 (Basic 1 ), SEQ ID NO.: 26 (Basic2),
SEQ ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
1o In yet a further aspect, the present invention relates to a conjugate
comprising at least one
transport agent region (including one, two, three or more transport agent
region) and an active
agent region, said active agent region being selected from the group
consisting of ADP-
ribosyl transferase C3 and ADP-ribosyl transferase C3 analogues, wherein said
transport
agent region is covalently linked to said active agent region.
In accordance with the present invention, the transport agent region may be
cross-linked (e.g.,
chemically cross-linked, L1V cross -linked) to the active agent region (C3-
like proteins of the
present invention and analogues thereof).
2o In accordance with the present invention, the transport agent region may be
fused to ADP-
ribosyl transferase C3 and ADP-ribosyl transferase C3 analogues according to
recombinant
DNA technology (e.g., cloning the DNA sequence of the transport agent region
in frame with
the DNA sequence of the ADP-ribosyl transferase C3 or an ADP-ribosyl
transferase C3
analogue comprising or not a spacer DNA sequence (multiple cloning site,
linker) or any
other DNA sequence that would not interfere with the activity of the C3-like
protein once
expressed).
In an additional aspect, the present invention relates to the use of a
polypeptide selected from
the group consisting of C3APL (SEQ ID NO.: 4), C3APLT (SEQ ID N0.:37), C3APS
(SEQ
3o ID N0.:6), C3-TL (SEQ ID N0.:14), C3-TS (SEQ ID N0.:18), C3Basicl (SEQ ID
N0.:25),
C3Basic2 (SEQ ID N0.:30), C3Basic3 (SEQ ID N0.:35), SEQ ID NO.: 20 and SEQ ID
NO.:
43, for the manufacture of a pharmaceutical composition.
33
CA 02362004 2001-11-13
In other aspects, the present invention relates to the use of a polypeptide
comprising at least
one (one or more) transport agent region and an active agent region, for the
manufacture of a
pharmaceutical composition, or to facilitate (for facilitating) axon growth or
for treating (in
the treatment of) nerve injury (e.g., nerve injury arising from traumatic
nerve injury or nerve
injury caused by disease), or for preventing (diminishing, inhibiting
(partially or totally)) cell
apoptosis (cell death, such as following ischemia in the CNS), or for
suppressing the
inhibition of neuronal axon growth, or for the treatment of ischemic damage
related to stroke,
or for suppressing Rho activity, or to regenerate (for regenerating) injured
axon (helping
injured axon to recover, partially or totally, their function), or to help
(for helping) neurons to
make new connections (developing axon, dendrite, neurite) with other
(surrounding) cells, in
a mammal, (e.g., human, animal), wherein said active agent region being
selected from the
group consisting of ADP-ribosyl transferase C3 and ADP-ribosyl transferase C3
analogues.
In accordance with the present invention, the transport agent region may be at
the amino-
terminal end of the protein and the ADP-ribosyl transferase C3 or ADP-ribosyl
transferase
C3 analogue may be at the carboxy-terminal end of the protein.
In accordance with the present invention, the transport agent region may be at
the carboxy
terminal end of the protein and the ADP-ribosyl transferase C3 or ADP-ribosyl
transferase
2o C3 analogue may be at the amino-terminal end of the protein.
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invnetion, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
3o the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2),
SEQ ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
34
CA 02362004 2001-11-13
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In a further aspect, the present invention provides a method of (for)
suppressing the inhibition
of neuronal axon growth (e.g., in a mammal, (e.g., human, animal)) comprising
delivering a
polypeptide or conjugate comprising at least one (one or more) transport agent
region and an
active agent region selected from the group consisting of ADP-ribosyl
transferase C3 and
ADP-ribosyl transferase C3 analogues directly at a central nervous system
(CNS) lesion site
l0 or a peripheral nervous system (PNS) lesion site, in an amount effective to
counteract said
inhibition.
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
2o In accordance with the present invention, the basic amino acid region may
be selected from
the group consisting of SEQ ID NO.: 21 (Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.:
31 (Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL),
SEQ ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In yet a further aspect, the present invention provides a method of (for)
facilitating axon
growth (e.g., in a mammal, (e.g., human, animal)) comprising delivering a
polypeptide or
3o conjugate comprising at least one transport agent region and an active
agent region selected
from the group consisting of ADP-ribosyl transferase C3 and ADP-ribosyl
transferase C3
analogues directly at a central nervous system (CNS) lesion site or a
peripheral nervous
system (PNS) lesion site, in an amount effective to facilitate said growth.
CA 02362004 2001-11-13
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
1o the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2),
SEQ ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
15 group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In an additional aspect, the present invention provides a method of (for)
treating nerve injury
(e.g., in a mammal, (e.g., human, animal)) comprising delivering a polypeptide
or conjugate
comprising at least one transport agent region and an active agent region
selected from the
2o group consisting of ADP-ribosyl transferase C3 and ADP-ribosyl transferase
C3 analogues
directly at (to) a central nervous system (CNS) lesion site or a peripheral
nervous system
(PNS) lesion site.
In accordance with the present invention, the transport agent region may be
selected from the
25 group consisting of a basic amino acid rich region and a proline rich
region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21 (basic 1 ), SEQ ID NO.: 26 (basic 2),
SEQ ID NO.:
36
CA 02362004 2001-11-13
31 (basic 3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (Tat
long),
SEQ ID NO.: 47 (Tat short), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In yet an additional aspect, the present invention provides a method of (for)
preventing cell
apoptosis (e.g., in a mammal, (e.g., human, animal)) comprising delivering a
polypeptide or
conjugate comprising at least one transport agent region and an active agent
region selected
to from the group consisting of ADP-ribosyl transferase C3 and ADP-ribosyl
transferase C3
analogues directly at a central nervous system (CNS) lesion site or a
peripheral nervous
system (PNS) lesion site.
In accordance with the present invention, the transport agent region may be
selected from the
15 group consisting of a basic amino acid rich region and a proline rich
region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In another aspect, the present invention provides a method of (for) treating
ischemic damage
3o related to stroke (e.g., in a mammal, (e.g., human, animal)) comprising
delivering a
polypeptide or conjugate comprising at least one transport agent region and an
active agent
region selected from the group consisting of ADP-ribosyl transferase C3 and
ADP-ribosyl
37
CA 02362004 2001-11-13
transferase C3 analogues directly at a central nervous system (CNS) lesion
site (to said
mammal).
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In yet another aspect, the present invention provides a method of (for)
suppressing Rho
2o activity comprising delivering a polypeptide or conjugate comprising at
least one transport
agent region and an active agent region selected from the group consisting of
ADP-ribosyl
transferase C3 and ADP-ribosyl transferase C3 analogues directly at a central
nervous system
(CNS) lesion site or a peripheral nervous system (PNS) lesion site, in an
amount effective to
suppress said activity.
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
3o from the group consisting of a subdomain of HIV Tat protein, a homeodomain
of
antennapedia, a Histidine tag, and analogues thereof.
38
CA 02362004 2001-11-13
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In accordance with an additional aspect, the present invention provides a
method of (for)
1o regenerating injured axon (e.g., in a mammal, (e.g., human, animal))
comprising delivering a
polypeptide or conjugate comprising at least one transport agent region and an
active agent
region selected from the group consisting of ADP-ribosyl transferase C3 and
ADP-ribosyl
transferase C3 analogues directly at a central nervous system (CNS) lesion
site or a peripheral
nervous system (PNS) lesion site (e.g., in a mammal), in an amount effective
to regenerate
said injured axon.
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
2o In accordance with the present invention, the basic amino acid rich region
may be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21 (Basicl ), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
3o group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In accordance with a further aspect, the present invention provides a method
of (for) helping
neurones to make new cell connection (developing axon, dendrite, neurite with
other
39
CA 02362004 2001-11-13
(surrounding) cells) comprising delivering a polypeptide or conjugate
comprising at least one
transport agent region and an active agent region selected from the group
consisting of ADP-
ribosyl transferase C3 and ADP-ribosyl transferase C3 analogues directly at a
central nervous
system (CNS) lesion site or a peripheral nervous system (PNS) lesion site.
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
1o from the group consisting of a subdomain of HIV Tat protein, a homeodomain
of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
15 (Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL),
SEQ ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In another aspect, the present invention, provides a polypeptide consisting of
a basic amino
acid rich region and an active agent region, wherein, amino acids from said
basic rich region
comprises amino acids selected from the group consisting of Histidine,
Asparagine,
Glutamine, Lysine and Arginine and wherein the active agent region is ADP-
ribosyl
transferase C3.
In yet another aspect, the present invention relates to the use of a
polypeptide comprising at
least one transport agent region and an active agent region, said active agent
region being
selected from the group consisting of ADP-ribosyl transferase C3 and ADP-
ribosyl
3o transferase C3 analogues for the manufacture of a medicament for
suppressing the inhibition
of neuronal axon growth.
CA 02362004 2001-11-13
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
1o (Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL),
SEQ ID
NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may selected
from the group
consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In a further aspect, the present invention relates to the use of a polypeptide
comprising at
least one transport agent region and an active agent region, said active agent
region being
selected from the group consisting of ADP-ribosyl transferase C3 and ADP-
ribosyl
transferase C3 analogues for the manufacture of a medicament for facilitating
axon growth.
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
41
CA 02362004 2001-11-13
In accordance with the present invention, the proline rich region may selected
from the group
consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In yet a further aspect the present invention relates to the use of of a
polypeptide comprising
at least one (one or more) transport agent region and an active agent region,
said active agent
region being selected from the group consisting of ADP-ribosyl transferase C3
and ADP-
ribosyl transferase C3 analogues for the manufacture of a medicament for
treating nerve
injury (e.g., in a mammal, (e.g., human, animal)).
1o In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
15 antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
2o NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may selected
from the group
consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
25 In accordance with another aspect, the present invention relates to the use
of a polypeptide
selected from the group consisting of C3APL (SEQ ID NO.: 4), C3APLT (SEQ ID
N0.:37),
C3APS (SEQ ID N0.:6), C3-TL (SEQ ID N0.:14), C3-TS (SEQ ID N0.:18), C3Basicl
(SEQ ID N0.:25), C3Basic2 (SEQ ID N0.:30), C3Basic3 (SEQ ID N0.:35), SEQ ID
NO.:
20 and SEQ ID NO.: 43 for the manufacture of a pharmaceutical composition for
suppressing
3o the inhibition of neuronal axon growth (e.g., in a mammal, (e.g., human,
animal)).
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
42
CA 02362004 2001-11-13
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
1o
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In yet another aspect, the present invention relates to the use of a
polypeptide selected from
the group consisting of C3APL (SEQ ID N0.:4), C3APLT (SEQ ID N0.:37), C3APS
(SEQ
ID N0.:6), C3-TL (SEQ ID N0.:14), C3-TS (SEQ ID N0.:18), C3Basicl (SEQ ID
N0.:25),
C3Basic2 (SEQ ID N0.:30), C3Basic3 (SEQ ID N0.:35), SEQ ID NO.: 20 and SEQ ID
NO.:
43 for the manufacture of a pharmaceutical composition for facilitating axon
growth (e.g., in
a mammal, (e.g., human, animal)).
In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
NO.: 47 (TS), and analogues thereof.
43
CA 02362004 2001-11-13
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In a further aspect, the present invention relates to the use of a polypeptide
selected from the
group consisting of C3APL (SEQ ID NO.: 4), C3APLT (SEQ ID N0.:37), C3APS (SEQ
ID
N0.:6), C3-TL (SEQ ID N0.:14), C3-TS (SEQ ID N0.:18), C3Basicl (SEQ ID
N0.:25),
C3Basic2 (SEQ ID N0.:30), C3Basic3 (SEQ ID N0.:35), SEQ ID NO.: 20 and SEQ ID
NO.:
43 for the manufacture of a pharmaceutical composition for treating nerve
injury.
1o In accordance with the present invention, the transport agent region may be
selected from the
group consisting of a basic amino acid rich region and a proline rich region.
In accordance with the present invention, the basic amino acid rich region may
be selected
from the group consisting of a subdomain of HIV Tat protein, a homeodomain of
antennapedia, a Histidine tag, and analogues thereof.
In accordance with the present invention, the basic amino acid region may be
selected from
the group consisting of SEQ ID NO.: 21(Basicl), SEQ ID NO.: 26 (Basic2), SEQ
ID NO.: 31
(Basic3), SEQ ID NO.: 44 (APL), SEQ ID NO.: 45 (APS) SEQ ID NO.: 46 (TL), SEQ
ID
2o NO.: 47 (TS), and analogues thereof.
In accordance with the present invention, the proline rich region may be
selected from the
group consisting of SEQ ID NO.: 48 (APLT) and analogues thereof.
In another aspect, the present invention provides an isolated polynucleotide
comprising at
least the polynucleotide sequence (for example the polynucleotide sequence
disclosed herein
in addition with (or in some cases without) a suitable backbone (e.g.,
plasmid, viral vector))
selected from the group consisting of SEQ ID NO.: 3, SEQ ID NO.: 5, SEQ ID
NO.: 13, SEQ
ID NO.: 17, SEQ ID NO.: 19, SEQ ID NO.: 24, SEQ ID NO.: 29, SEQ ID NO.: 34,
SEQ ID
NO.: 36, and SEQ ID NO.: 42.
In yet another aspect, the present invention provides a cell transformed
(transfected,
transduced, infected, electroporated, micro-injected, etc.) with an isolated
polynucleotide
44
CA 02362004 2001-11-13
comprising at least the polynucleotide sequence (for example the
polynucleotide sequence
disclosed herein in addition with (or in some cases without) a suitable
backbone (e.g.,
plasmid, viral vector)) selected from the group consisting of SEQ ID NO.: 3,
SEQ ID NO.: 5,
SEQ ID NO.: 13, SEQ ID NO.: 17, SEQ ID NO.: 19, SEQ ID NO.: 24, SEQ ID NO.:
29,
SEQ ID NO.: 34, SEQ ID NO.: 36, and SEQ ID NO.: 42.
The invention in particular provides C3-like proteins, which may have
additional amino acids
added to the carboxy terminal end of the C3 proteins. Examples of such
proteins includes:
1o C3APL: (C3 antennapedia -long) created by annealing sequences from the
antennapedia
transcription factor to the 5' end of the sequence encoding C3 cDNA. The long
antennapedia
sequence of 60 amino acids containing the homeodomain of antennapedia, was
used;
C3APLT: (C3 antennapedia -truncated) created by annealing sequences from the
antennapedia transcription factor to the 5' end of the sequence encoding C3
cDNA. This
clone with a frameshift mutation gives a proline-rich transport peptide with
good transport
activity. This sequence is tuncated ie shorter than C3APL.
C3APS: A short 11 amino acid sequence of antennapedia that has transmembrane
transport
2o properties was fused to the carboxy terminal of C3 to create C3APS;
C3-TL: C3 Tat-long created by fusing amino acids 27 to 72 of Tat to the
carboxy terminal of
C3 protein;
30
C3-TS: C3 Tat-short created by fusing the amino acids YGRKRRQRRR to the C3
protein;
C3Basicl : a random basic charge sequence added to the C-terminal of C3;
C3Basic2 : a random basic charge sequence added to the C-terminal of C3;
CA 02362004 2001-11-13
C3Basic3: C3 Tat-short created by fusing the reverse sequence of Tat amino
acids
RRQRRKKR to the C3 protein.
It has been found that conjugates or fusion proteins (C3-like proteins) Rho
antagonists of the
present invention are effective to stimulate repair in the CNS after spinal
cord injury. The
increased cell permeability of new Rho antagonist (new chimeric C3) would now
allow
treatment of victims of stroke and neurodegenerative disease because Rho
signalling pathway
is important in repair after stroke (Hitomi, et al. (2000) 67: 1929-39. Trapp
et al 2001.
Mol.Cell. Neurosci. 17: 883-84). Treatment with Rho antagonists in the
adhesive delivery
to system could be used to enhance the rate of axon growth in the PNS. Also,
evidence in the
literature now links Rho signalling with formation of Alzheimer's disease
tangles through its
ability to activate PKN which then phosphorylates tau and neurofilaments
(Morissette, et al.
(2000) 278: H1769-74., Kawamata, et al. (1998) 18: 7402-10., Amano, et al.
(1996) 271:
648-50., Watanabe, et al. (1996) 271: 645-8.). Therefore, Rho antagonists are
expected to be
useful in the treatment of Alzheimer's disease. The new chimeric C3 drugs
should be able to
diffuse readily and therefore can promote repair for diseases that are
neurodegenerative.
Examples include, but are not limited to stroke, traumatic brain injury,
Parkinson's disease,
Alzheimer's disease and ALS. Moreover, it is now well established that Rho
signalling
antagonists are effective in the treatment of other diseases. These include,
but are not limited
2o to eye diseases such as glaucoma (Honjo, et al. (2001) 42: 137-44., Rao, et
al. (2001) 42:
1029-1037.), cancer cell migration and metastasis (Sahai, et al. (1999) 9: 136-
45., Takamura,
et al. (2001) 33: 577-81., Imamura, et al. (2000) 91: 811-6.). The effect of
the Rho signalling
pathway on smooth muscle relaxation are well established. This has led to the
identification
of Rho signalling antagonists as effective in treatment of hypertension
(Chitaley, et al. (2001)
3: 139-144., Somlyo (1997) 389: 908-911, Uehata, et al. (1997) 389: 990-994),
asthma
(Nakahara, et al. (2000) 389: 103-6., Ishizaki, et al. (2000) 57: 976-83), and
vascular disease
(Miyata, et al. (2000) 20: 2351-8., Robertson, et al. (2000) 131: 5-9.) as
well as penile erectile
dysfunction (Chitaley, et al. (2001) 7: 119-22.). Rho is also important as a
cardioprotective
protein (Lee et al. 2001. FASEB J. 15:1886-1894).
46
CA 02362004 2001-11-13
Rho GTPases include members of the Rho, Rac and Cdc42 family of proteins. Our
invention
concerns Rho family members of the Rho class. Rho proteins consist of
different variants
encoded by different genes. For example, PC-12 cells express RhoA, RhoB and
RhoC
(Lehmann et al 1999 supra); PC-12 cells: Pheochromocytom cell line (Greene A
and
Tischler, A S PNAS 73:2424 (1976). To inactivate Rho proteins inside cells,
Rho antagonists
of the C3 family type are effective because they inactivate all forms of Rho
(eg. RhoA, Rho B
etc). In contrast, gene therapy techniques, such as introduction of a dominant
negative RhoA
family member into a diseased cell; will only inactivate that specific RhoA
family member.
1o Recombinant C3 proteins, or C3 proteins that retain the ribosylation
activity are also effective
in our delivery system and are covered by this invention. In addition, Rho
kinase is a well-
known target for active Rho, and inactivating Rho kinase has the same effect
as inactivating
Rho, at least in terms of neurite or axon growth (Kimura and Schubert (1992)
Journal of Cell
Biology.116:777-783, Keino-Masu, et al. (1996)Ce11.87:175-185, Matsui, et al.
(1996)EMBO
J.15:2208-2216, Matsui, et al. (1998)J. Cell Bio1.140:647-657, Ishizaki
(1997)FEBS
Lett.404:118-124), the biological activity that concerns this invention.
The C3 polypeptides of the present invention include biologically active
fragments and
analogues of C3; fragments encompass amino acid sequences having truncations
of one or
2o more amino acids , wherein the truncation may originate from the amino
terminus, carboxy
terminus, or from the interior of the protein. Fragments containing Glu(173)
of C3 are
included in this invention (Saito et al. 1995. FEBS Lett. 371-105). Analogues
of the
invention involve an insertion or a substitution of one or more amino acids.
Fragments and
analogues will have the biological property of C3 that is capable of
inactivating Rho GTPase
on Asn(41 ) on Rho. Also encompassed by the invention are chimeric
polypeptides
comprising C3 amino acid sequences fused to heterologous amino acid sequences.
Said
heterologous sequences encompass those which, when formed into a chimera with
C3 retain
one or more biological or immunological properties of C3. A host cell
transformed or
transfected with nucleic acids encoding C3 protein or C3 chimeric protein are
also
3o encompassed by the invention. Any host cell which produces a polypeptide
having at least
one of the biological properties of C3 may be used. Specific examples include
bacterial,
47
CA 02362004 2001-11-13
yeast, plant, insect or mammalian cells. In addition, C3 protein may be
produced in
transgenic animals. Transformed or transfected host cells and transgenic
animals are
obtained using materials and methods that are routinely available to one
skilled in the art.
Host cells may contain nucleic acid sequences having the full-length gene for
C3 protein
including a leader sequence and a C-terminal membrane anchor sequence (see
below) or,
alternatively, may contain nucleic acid sequences lacking one or both of the
leader sequence
and the C-terminal membrane anchor sequence. In addition, nucleic acid
fragments, variants
and analogues which encode a polypeptide capable of retaining the biological
activity of C3
may also be resident in host expression systems.
C3 is produced as a 26 kDa protein. The full length C3 protein inactivates Rho
by ADP-
ribosylating asparagine 41 of Rho A (Han et al. (2001) J. Mol. Biol. 305: 95).
Truncated,
elongated or altered C3 proteins or C3-derived peptides that retain the
ability to ribosylate
Rho are included in this invention and can be used to make fusion proteins.
The crystal
structure of C3 has been determined giving insight to elements of the C3
protein that could be
changed without affecting ribosylating activity (Han et al. (2001 ) J. Mol.
Biol. 305: 95).
The Rho antagonist that is a recombinant proteins can be made according to
methods present
in the art. The proteins of the present invention may be prepared from
bacterial cell extracts,
or through the use of recombinant techniques. In general, C3 proteins
according to the
invention can be produced by transformation (transfection, transduction, or
infection) of a
host cell with all or part of a C3-encoding DNA fragment in a suitable
expression vehicle.
Suitable expression vehicles include: plasmids, viral particles, and phages.
For insect cells,
baculovirus expression vectors are suitable. The entire expression vehicle, or
a part thereof,
can be integrated into the host cell genome. In some circumstances, it is
desirable to employ
an inducible expression vector.
Those skilled in the field of molecular biology will understand that any of a
wide variety of
expression systems can be used to provide the recombinant protein. The precise
host cell used
3o is not critical to the invention. The C3 and C3-like proteins may be
produced in a prokaryotic
48
CA 02362004 2001-11-13
host (e.g., E. coli or B. subtilis) or in a eukaryotic host (e.g.,
Saccharomyces or Pichia;
mammalian cells, e.g., COS, NIH 3T3, CHO, BHK, 293, or HeLa cells; or insect
cells).
Proteins and polypeptides may also be produced by plant cells. For plant cells
viral
expression vectors (e.g., cauliflower mosaic virus and tobacco mosaic virus)
and plasmid
expression vectors (e.g., Ti plasmid) are suitable. Such cells are available
from a wide range
of sources (e.g., the American Type Culture Collection, Rockland, Md.). The
methods of
transformation or transfection and the choice of expression vehicle will
depend on the host
system selected.
The host cells harbouring the expression vehicle can be cultured in
conventional nutrient
media adapted as need for activation of a chosen gene, repression of a chosen
gene, selection
of transformants, or amplification of a chosen gene. One expression system is
the mouse 3T3
fibroblast host cell transfected with a pMAMneo expression vector (Clontech,
Palo Alto,
Cali~). pMAMneo provides an RSV-LTR enhancer linked to a dexamethasone-
inducible
MMTV-LTR promotor, an SV40 origin of replication which allows replication in
mammalian
systems, a selectable neomycin gene, and SV40 splicing and polyadenylation
sites. DNA
encoding a C3 or C3-like protein would be inserted into the pMAMneo vector in
an
orientation designed to allow expression. The recombinant C3 or C3-like
protein would be
2o isolated as described below. Other preferable host cells that can be used
in conjunction with
the pMAMneo expression vehicle include COS cells and CHO cells (ATCC Accession
Nos.
CRL 1650 and CCL 61, respectively).
C3 polypeptides can be produced as fusion proteins. For example, expression
vectors may be
used to create lacZ fusion proteins. The pGEX vectors can be used to express
foreign
polypeptides as fusion proteins with glutathione S-transferase (GST). In
general, such fusion
proteins are soluble and can be easily purified from lysed cells by adsorption
to glutathione-
agarose beads followed by elution in the presence of free glutathione. The
pGEX vectors are
designed to include thrombin or factor Xa protease cleavage sites so that the
cloned target
49
CA 02362004 2001-11-13
gene product can be released from the GST moiety. Another stategy to make
fusion proteins
is to use the His tag system.
In an insect cell expression system, Autographa californica nuclear
polyhedrosis virus
AcNPV), which grows in Spodoptera frugiperda cells, is used as a vector to
express foreign
genes. A C3 coding sequence can be cloned individually into non-essential
regions (for
example the polyhedrin gene) of the virus and placed under control of an AcNPV
promoter,
e.g., the polyhedrin promoter. Successful insertion of a gene encoding a C3 or
C3-like
protein (polypeptide) will result in inactivation of the polyhedrin gene and
production of non-
occluded recombinant virus (i.e., virus lacking the proteinaceous coat encoded
by the
polyhedrin gene). These recombinant viruses are then used to infect Spodoptera
frugiperda
cells in which the inserted gene is expressed (see, Lehmann et al for an
example of making
recombinant MAG protein).
In mammalian host cells, a number of viral-based expression systems can be
utilised. In
cases where an adenovirus is used as an expression vector, the C3 nucleic acid
sequence can
be ligated to an adenovirus transcription/translation control complex, e.g.,
the late promoter
and tripartite leader sequence. This chimeric gene can then be inserted into
the adenovirus
genome by in vitro or in vivo recombination. Insertion into a non-essential
region of the viral
2o genome (e.g., region El or E3) will result in a recombinant virus that is
viable and capable of
expressing a C3 gene product in infected hosts.
Specific initiation signals may also be required for efficient translation of
inserted nucleic
acid sequences. These signals include the ATG initiation codon and adjacent
sequences. In
cases where an entire native C3 gene or cDNA, including its own initiation
codon and
adjacent sequences, is inserted into the appropriate expression vector, no
additional
translational control signals may be needed. In other cases, exogenous
translational control
signals, including, perhaps, the ATG initiation codon, must be provided.
Furthermore, the
initiation codon must be in phase with the reading frame of the desired coding
sequence to
3o ensure translation of the entire insert. These exogenous translational
control signals and
CA 02362004 2001-11-13
initiation codons can be of a variety of origins, both natural and synthetic.
The efficiency of
expression may be enhanced by the inclusion of appropriate transcription
enhancer elements,
transcription terminators.
In addition, a host cell may be chosen which modulates the expression of the
inserted
sequences, or modifies and processes the gene product in a specific, desired
fashion. Such
modifications (e.g., glycosylation) and processing (e.g., cleavage) of protein
products may be
important for the function of the protein. Different host cells have
characteristic and specific
mechanisms for the post-translational processing and modification of proteins
and gene
1o products. Appropriate cell lines or host systems can be chosen to ensure
the correct
modification and processing of the foreign protein expressed. To this end,
eukaryotic host
cells that possess the cellular machinery for proper processing of the primary
transcript,
glycosylation, and phosphorylation of the gene product can be used. Such
mammalian host
cells include, but are not limited to, CHO, VERO, BHK, HeLa, COS, MDCK, 293,
3T3,
15 WI38, and in particular, choroid plexus cell lines.
Alternatively, a C3 protein can be produced by a stably-transfected mammalian
cell line. A
number of vectors suitable for stable transfection of mammalian cells are
available to the
public; methods for constructing such cell lines are also publicly available.
In one example,
2o cDNA encoding the C3 protein may be cloned into an expression vector that
includes the
dihydrofolate reductase (DHFR) gene. Integration of the plasmid and,
therefore, the C3 or
C3-like protein-encoding gene into the host cell chromosome is selected for by
including
0.01-300 ~,M methotrexate in the cell culture medium (as described in Ausubel
et al., supra).
This dominant selection can be accomplished in most cell types. Recombinant
protein
25 expression may be increased by DHFR-mediated amplification of the
transfected gene.
Methods for selecting cell lines bearing gene amplifications are known in the
art; such
methods generally involve extended culture in medium containing gradually
increasing levels
of methotrexate. DHFR-containing expression vectors commonly used for this
purpose
include pCVSEII-DHFR and pAdD26SV(A). Any of the host cells described above
or,
3o preferably, a DHFR-deficient CHO cell line (e.g., CHO DHFR cells, ATCC
Accession No.
51
CA 02362004 2001-11-13
CRL 9096) are among the host cells preferred for DHFR selection of a stably-
transfected cell
line or DHFR-mediated gene amplification.
A number of other selection systems may be used, including but not limited to
the herpes
simplex virus thymidine kinase, hypoxanthine-guanine
phosphoribosyltransferase, and
adenine phosphoribosyltransferase genes can be employed in tk, hgprt, or aprt
cells,
respectively. In addition, gpt, which confers resistance to mycophenolic acid;
neo, which
confers resistance to the aminoglycoside G-418; and hygro, which confers
resistance to
hygromycin may be used.
Alternatively, any fusion protein can be readily purified by utilising an
antibody specific for
the fusion protein being expressed. For example, a system described in
Janknecht et al.
(1981) Proc. Natl. Acad. Sci. USA 88, 8972, allows for the ready purification
of non-
denatured fusion proteins expressed in human cell lines. In this system, the
gene of interest is
1 s subcloned into a vaccinia recombination plasmid such that the gene's open
reading frame is
translationally fused to an amino-terminal tag consisting of six histidine
residues. Extracts
from cells infected with recombinant vaccinia virus are loaded onto Ni2+
nitriloacetic acid-
agarose columns, and histidine-tagged proteins are selectively eluted with
imidazole-
containing buffers.
Alternatively, C3, C3-like protein or a portion (fragment) thereof, can be
fused to an
immunoglobulin Fc domain. Such a fusion protein can be readily purified using
a protein A
column.
To test Rho antagonists for activity, a tissue culture bioassay system was
used. This bioassay
is used to define activity of Rho antagonists that will be effective in
promoting axon
regeneration in spinal cord injury, stroke or neurodegenerative disease.
s2
CA 02362004 2001-11-13
Neurons do not grow neurites on inhibitory myelin substrates. When neurons are
placed on
inhibitory substrates in tissue culture, they remain rounded. When an
effective Rho
antagonist is added, the neurons are able to grow neurites on myelin
substrates. The time that
it takes for neurons to growth neurites upon the addition of a Rho antagonist
is the same as if
neurons had been plated on growth permissive substrate such as laminin or
polylysine,
typically 1 to 2 days in cell culture. The results can be scored visually. If
needed, a
quantitative assessment of neurite growth can be performed. This involved
measuring the
neurite length in a) control cultures where neurons are plated on myelin
substrates and left
untreated b) in positive control cultures, such as neurons plated on
polylysine c) or treating
1o cultures with different concentrations of the test antagonist.
To test C3 in tissue culture, it has been found that the best concentration is
25-50 ug/ml.
(Lehmann et al, 1999. J.Neurosci. 19: 7537-7547; Jin & Strittmatter, 1997. J.
Neurosci. 17:
6256-6263). Thus, high concentrations of this Rho antagonist are needed as
compared to the
growth factors used to stimulate neurite outgrowth. Growth factors, such as
nerve growth
factor (NGF) are used at concentrations of 1- 100 ng/ml in tissue culture.
However, growth
factors are not able to overcome growth inhibition by myelin. Our tissue
culture experiments
are all performed in the presence of the growth factor BDNF for retinal
ganglion cells, or
NGF for PC-12 cells. When growth factors have been tested in vivo, typically
the highest
2o concentrations possible are used, in the ug/ml range. Also they are often
added to the CNS
with the use of pumps for prolonged delivery (eg. Ramer et al, supra). For in
vivo
experiments the highest concentrations possible was used when working with C3
stored as a
frozen 1 mg/ml solution.
The Rho antagonist C3 is stable at 37 °C for at least 24 hours. The
stability of C3 was tested
in tissue culture with the following experiment. The C3 was diluted in tissue
culture medium,
left in the incubator at 37 °C for 24 hours, then added to the bioassay
system described above,
using retinal ganglion cells as the test cell type. These cells were able to
extend neurites on
inhibitory substrates when treated with C3 stored for 24 hours at 37
°C. Therefore , the
53
CA 02362004 2001-11-13
minimum stability is 24 hours. This is in keeping with the stability
projection based on
amino acid composition (see sequence data, below).
A compound can be confirmed as a Rho antagonist in one of the following ways:
a) Cells are cultured on a growth inhibitory substrate as above, and exposed
to the
candidate Rho antagonist;
b) Cells of step a) are homogenized and a pull-down assay is performed. This
assay is based on the capability of GST-Rhotektin to bind to GTP-bound Rho.
Recombinant GST-Rhotektin or GST rhotektin binding domain (GST-RBD) is
1o added to the cell homogenate made from cells cultured as in a). It has been
found that inhibitory substrates activate Rho, and that this activated Rho is
pulled down by GST-RBD. Rho antagonists will block activation of Rho, and
therefore, an effective Rho antagonist will block the detection of Rho when
cell
are cultured as described by a) above;
c) An alternate method for this pull-down assay would be to use the GTPase
activating protein, Rho-GAP as bait in the assay to pull down activated Rho,
as
described (Diekmann and Hall, 1995. In Methods in Enzymology Vol. 256 part
B 207-215).
2o Another method to confirm that a compound is a Rho antagonist is as
follows: When added
to living cells antagonists that inactivate Rho by ADP-ribosylation of the
effector domain can
be identified by detecting a molecular weight shift in Rho (Lehmann et al,
1999 supra). The
molecular weight shift can be detected after treatment of cells with Rho
antagonist by
homogenizing the cells, separating the proteins in the cellular homogenate by
SDS
polyacrylamide gel electrophoresis. The proteins are transferred to
nitrocellulose paper, then
Rho is detected with Rho-specific antibodies by a Western blotting technique.
Another method to confirm that compound is a Rho-kinase antagonist is as
follows:
54
CA 02362004 2001-11-13
a) Recombinant Rho kinase tagged with myc epitope tag, or a GST tag or any
suitable tag is expressed in Hela cells or another suitable cell type by
transfection;
b) The kinase is purified from cell homogenates by immunoprecipation using
antibodies directed against the specific tag (e.g., myc tag or the GST tag);
c) The recovered immunoprecipitates from b) are incubated with [32P] ATP and
histone type 2 as a substrate in the presence or absence of the Rho kinase
inhibitor. In the absence of Rho kinase inhibitor activity , the Rho kinase
1o phosphorylated histone. In the presence of Rho kinase inhibitor the
phosphorylation activity of Rho kinase (i.e. phosphorylation of histone) is
blocked, and as such identified the compound as a Rho kinase antagonist.
Turning now to the transport side of the conjugates of the present invention ,
known methods
are available to add transport sequences that allow proteins to penetrate into
the cell;
examples include membrane translocating sequence (Rojas (1998) 16: 370-375),
Tat-
mediated protein delivery (Vives (1997) 272: 16010-16017), polyargine
sequences (Wender
et al. 2000, PNAS 24: 13003-13008) and antennapedia (Derossi (1996) 271: 18188-
18193).
Examples of known tranport agents, moities, subdomains and the like are also
shown for
2o example in Canadian patent document no. 2,301,157 (conjugates containing
homeodomain of
antennapedia) as well as in U.S. patent 5,652,122, 5,670,617, 5,674,980,
5,747,641, and
5,804604 (conjugates containing amino acids of Tat HIV protein (hereinafter
Tat HIV protein
is sometimes simply referred to as Tat); the entire contents of each of these
patent documents
is incorporated herein by reference.
Several receptor-mediated transport strategies have been used to try and
improve function of
ADP ribosylases: these methods include fusing C2 and C3 sequences (Wilde, et
al. (2001)
276: 9537-9542.) and use of receptor-mediated transport with the diptheria
toxin receptor
(Aullo, et al. (1993) 12: 921-31 ; Boquet, P. et al. (1995) Meth. Enzymol.
256: 297-306).).
CA 02362004 2001-11-13
These methods have not been demonstrated to dramatically increase the potency
of C3.
Moreover, these proteins require receptor-mediated transport. This means that
the cells must
express the receptor, and must express sufficient quantities of the receptor
to significantly
improve transport. Moreover, when C3 enters the cell by endocytosis, it will
be locked
within a membrane compartment, and therefore most of it will not be available
to inactivate
Rho. In the case of dipthera toxin, not all cells express the appropriate
receptor, limiting its
potential use. The clinical importance for any of these has not been tested or
shown. A
C2/C3 fusion protein has also been made to try and improve the effectiveness
of C3. In this
case, the addition of a C2II binding protein to the tissue culture medium is
needed, along with
to the C2-C3 fusion toxin to allow uptake of C3 by receptor-mediated
endocytosis (Barthe et al.
(1998) Infection and Immunity 66:1364). The disadvantage of this system is
that much of the
C3 in the cell will be restrained within a membrane compartment. More
importantly, two
different proteins must be added separately for transport to occur (Wahl et
al. 2000. J. Cell
Biol. 149:263), which make this system difficult to apply to in vivo for
treatment of disease.
Moreover, none of the methods to inactivate Rho with C3 or C3 analogues (C3-
like protein)
have been demonstrated to be sufficient to overcome growth inhibition in
tissue culture, or to
promote recovery after CNS damage in vivo.
One strategy which may be used in accordance with the present invention is to
exploit the
2o antennapedia homeodomain that is able to transport proteins across the
plasma membrane by
a receptor-independent mechanism (Derossi (1996) 271: 18188-18193); an
alternate strategy
is to exploit Tat-mediated delivery (Vives (1997) 272: 16010-16017, Fawell
(1994) 91: 664-
668, Frankel (1988) 55: 1189-1193).
The Antennapedia strategy has been used for protein translocation into neurons
(Derossi
(1996) 271: 18188-18193). Antennapedia has, for example, been used to
transport biotin-
labelled peptides in order to demonstrate the efficacy of the technique; see
U.S. Patent No.
6,080,724 (the entire contents of this patent are incorporated herein by
reference).
Antennapedia enhances growth and branching of neurons in vitro (Bloch-Gallego
(1993) 120:
485-492). Homeoproteins are transcription factors that regulate development of
body
organization, and antennapedia is a Drosophila homeoprotein. Tat on the other
hand is a
56
CA 02362004 2001-11-13
regulatory protein from human immunodeficiency virus (HIV). It is a highly
basic protein
that is found in the nucleus and can transport reporter genes into cell.
Moreover, Tat-linked
proteins can penetrate cells after intraperotoneal injection, and it can even
cross the blood
brain barrier to enter cells within the brain (Schwarze, et al. (1999) 285:
1569-72).
Other transport sequences that have been tested in other contexts, (i.e., to
show that they work
through the use of reporter sequences), are known. One transport peptide, a 12
mer,
AAVLLPVLLAAP, is rich in proline. It was made as a GST-MTS fusion protein and
is
derived from the h region of the Kaposi FGF signal sequence (Royas et al. 1998
Nature
to Biotech. 16: 370-375. Another example is the sperm fertiline alpha peptide,
HPIQIAAFLARIPPISSIGTCILK (This is reviewed in Pecheur, J. Sainte-Marie, A.
Bienveniie, D. Hoekstra. 1999. J. Membrane Biol. 167: 1-17). It must be noted
however that
the alpha helix-breaking propensity of proline (Pro) residues is not a general
rule, since the
putative fusion peptide of sperm fertilin alpha displays a high alpha helical
content in the
presence of liposomes. However, the Pro-Pro sequence is required for efficient
fusion
properties of fertilin. The C3APLT fusion protein that we tested fits the
requirement of
having a two prolines for making an effective transport peptide. Therefore,
proline-rich
sequences and random sequences that have helix-breaking propensity that act as
effective
transporters would also be effective if fused to C3.
In the context of axon growth on inhibitory substrates, axon regeneration
after injury, or axon
regeneration in the brain or spinal cord, no method using these transport
sequences has been
devised. In particular, it should be noted that the ability of antennapedia to
enhance growth
was tested with neurons placed on laminin-coated coverslips. Laminin supports
axon growth
and overndes growth inhibition (David, et al. (1995) 42: 594-602) thus, it is
not a suitable
substrate to test the potential for regeneration. There is an enormous wealth
of literature over
the last 20 years on substances that promote axon growth under such favourable
tissue culture
conditions, but none of these has lead to clinical advances in the treatment
of spinal cord
injury. The effect of antennapedia was shown to act as similar to a growth
factors. Growth
3o factors do not overcome growth inhibition by CNS growth inhibitory
substrates (Lehmann, et
al. (1999) 19: 7537-7547, Cai, et al. (1999) 22: 89-101). Growth factors
applied in vivo do
not support regeneration, only sprouting (Schnell, et al. (1994) 367: 170-
173).
57
CA 02362004 2001-11-13
The transport sequence may be added to the N-terminal (amino-terminal)
sequence of the C3
protein. Alternatively, the transport sequence may be added on the C-terminal
(carboxy-
terminal) end of the C3 protein; because the C-terminal is already quite
basic, this should
enhance further the transport properties. This is likely one of the reasons
that C3APLT
shows activity in addition to its basic charge and the proline-rich sequences.
The new chimeric C3 may be used to treat spinal cord injury to promote
functional repair. We
have demonstrated that both C3APLT and C3APS can overcome growth inhibition on
complex inhibitory substrates that include myelin and mixed chondroitin
sulfate
proteoglycans. Further, we demonstrate that C3APLT can promote functional
recovery after
application to injured spinal cord in adult mice. The new chimeric protein may
be used to
promote axon regeneration and reduce scarnng after CNS injury. Scarring is a
barrier to
nerve regeneration.
The advantage of the new chimeric C3 is the ability to treat the injured axons
after a
significant delay between the injury and the treatment. Also, the new
recombinant protein
may be useful in the treatment of chronic injury. The chimeric C3 can also be
used to treat
neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease
where
2o penetration of the Rho antagonist to the affected neuronal population is
required for effective
treatment. The chimeric C3 (fusion proteins) will also be of benefit for the
treatment of
stroke and traumatic brain injury. Moreover, much evidence suggests efficacy
in the
treatment of cancer cell migration. Rho antagonists are also useful in the
treatment of disease
involving smooth muscle, such as vascular disease, hypertension, asthma, and
penile
dysfunction.
For treatment of spinal cord injury, the conjugate Rho antagonists of the
present invention
may be used in conjunction with cell transplantation. Many different cell
transplants have
been extensively tested for their potential to promote regeneration and
repair, including , but
58
CA 02362004 2001-11-13
not restricted to, Schwann cells, fibroblasts modified to express growth
factors, fetal spinal
cord transplants, macrophages, embryonic or adult stem cells, and olfactory
ensheathing glia.
C3 fusion proteins may be used in conjunction with neurotrophins, apoptosis
inhibitors, or
other agents that prevent cell death. They may be used in conjunction with
cell adhesion
molecules such as L1, laminin, and artifical growth matrices that promote axon
growth. The
chimeric C3 constructs of the present invention may also be used in
conjunction with the use
of antibodies that block growth inhibitory protein substrates to promote axon
growth.
Examples of such antibody methods are the use of IN-1 or related antibodies
(Schnell and
Schwab (1990) 343: 269-272) or through the use of therapeutic vaccine
approaches (Huang
(1999) 24: 639-647).
BRIEF DESCRIPTION OF THE FIGURES
In drawings which illustrate example embodiments of the present invention:
Figure 1 illustrates the dose response of normal C3 with and without
trituration;
Figure 2 illustrates ADP ribosylation by C3APLT and C3APS, but not C3 after
passively
adding the compounds to PC-12 cells;
Figure 3A illustrates that C3APLT penetrates cells;
Figure 3B illustrates a lower level of cell penetration by C3 as compared to
Figure 3A;
2o Figure 4 illustrates the effectiveness of C3APLT and C3APS at low doses;
Figure 5 illustrates the effectiveness of C3APLT and C3APS at low doses;
Figure 6 illustrates the effectiveness of C3APLT to stimulate axon
regeneration of primary
neurons;
Figure 7 illustrates the effectiveness of C3APLT to promote functional
recovery after spinal
cord injury;
Figure 8 illustrates effectiveness of Tat transport sequences to enhance
growth as C3-Tat (C3-
TL and C3-TS) chimeras;
59
CA 02362004 2001-11-13
Figures 9A and 9B illustrate axon regeneration after spinal cord injury and
treatment with
C3APLT;
Figure 10 illustrates effectiveness of C3APLT to prevent cell death after
spinal cord injury,
thereby showing that it is neuroprotective; and,
Figure 11 illustrates a comparison of C3APLT and C3Basic3 to promote neurite
outgrowth.
Referring to Figure 1, PC-12 cells were plated on inhibitory myelin substrates
(0).
Unmodified C3 added to the tissue culture medium at concentration from 0.00025
- 50 ug/ml
did not significantly improve neurite outgrowth over the untreated control
(grey bars). C3
1o was only effective in stimulating neurite outgrowth for cells plated on
myelin substrates after
scrape -loading (black bars). This Figure demonstrates the limited or no
penetration in cells
when passively added to the tissue culture medium. Please see Example 4 below
for
techniques.
Refernng to Figure 2, this Figure provides a demonstration that C3APLT and
C3APS, ADP
ribosylate Rho. Western blot showing RhoA in untreated cells (lane 1 ), and
cells treated with
C3APLT (lane 2) or C3APS (lane 3). When Rho is ADP ribosylated by C3 it
undergoes a
molecular weight shift (Lehmann et al supra), as observed for lanes 2 and 3.
Please see
Example 4 below for techniques.
Referring to Figure 3, this Figure shows intracellular activity after
treatment with C3APLT.
Detection that the new fusion C3 penetrates into the cells.
Immunocytochemistry with anti-
C3 antibody of PC-12 cells plated on myelin and treated with C3 (A) or C3APLT
(B). Cells
in A (Figure 3A) are not immunoreactive because C3 has not penetrated into the
cells. Cells
in B (Figure 3B) are immunoreactive and they are able to extend neurites on
myelin
substrates. Please see Example 4 below for techniques.
CA 02362004 2001-11-13
Turning to Figure 4, this Figure shows that C3-antennapedia fusion proteins
promote growth
on inhibitory substrates. The percent of neurons that grow neurites was
counted for each
treatment. The dose response experiment shows that C3APLT and C3APS promote
more
neurite growth per cell than control PC-12 cells plated on myelin (0). PC-12
cells were
plated on myelin and either scrape loaded with unmodified C3 (C3 50) left
untreated (0) or
treated with various concentrations of C3APLT. Compared to C3 used at 25
ug/ml, C3APS
is effective at stimulating more cells to grow neurites at 0.0025 ug/ml, a
dose 10,000 X less.
Please see Example 4 below for techniques.
1o Figure 5 shows a dose-response experiment showing that C3APLT and C3APS
elicit long
neurites to grow when cells are plated on inhibitory substrates. The length of
neurites was
measured for each treatment. PC-12 cells were plated on myelin and either
scrape loaded
with unmodified C3 (C3 50) left untreated (0) or treated with various
concentrations of
C3APLT. Compared to C3 used at 25 ug/ml, C3APS is effective at stimulating
more cells to
longer neurite growth at 0.0025 ug/ml, a dose 10,000 X less. Please see
Example 4 below for
techniques.
As may be seen Figure 6 shows primary neurons growing on inhibitory substrates
after
treatment with C3APLT. Rat retinal ganglion cells were plated on myelin
substrates and
2o treated with different concentrations of C3APLT. Concentrations of 0.025
and above
promoted significantly longer neurites. This dose is 1000X lower than that of
C3 needed to
promote growth on myelin.
Refernng to Figure 7, this Figure shows behavioral recovery after treatment of
adult mice
with C3APLT in a dose-response experiment. Mice received a dorsal hemisection
of the
spinal cord and were left untreated (transection), were treated with fibrin
alone (fibrin) or
were treated with fibrin plus C3APLT at the indicated concentrations given in
ug/mouse.
Each point represents one animal. The BBB score (see Example 6 for details)
was assessed
24 hours after treatment. Animals treated with C3APLT exhibited a significant
improvement
in behavioural recovery compared to untreated animals. The effective dose of
0.5 ug is 100X
61
CA 02362004 2001-11-13
less than unmodified C3 used (see previous experiment shown in Canadian patent
application
2,325,842). Please see Example 6.
Refernng to Figure 8, this Figure shows promotion of axon growth by C3-Tat
chimeric
proteins. The dose-response experiment shows that C3-TS and C3-TL promote more
neurite
growth per cell than control PC-12 cells plated on myelin. PC-12 cells were
plated on myelin
and either scrape loaded with unmodified C3 (scrape load) left untreated
(myelin) or treated
with various concentrations of C3-TS (grey bars) or C3-TL (black bars).
Compared to C3
used at 25 ug/ml, C3-TL is effective at stimulating more cells to grow
neurites at 0.0025
1o ug/ml, a dose 10,000 X less than C3.
Refering to Figure 9A and 9B, these Figures show axon regeneration in injured
spinal cord,
i.e. anatomical regeneration after treatment with C3APLT. Section of the
spinal cord after
anterograde labeling with horseradish peroxidase conjugated to wheat germ
agglutinin
(WGA-HRP). A) Sprouting of cut axons into the dorsal white matter. Arrows show
regenerating axons distal to the lesion. B) Same section 3 mm from the lesion
site. Arrows
show regenerating axons.
Referring to Figure 10, this Figure shows that C3-APLT protected neurons from
cell death
2o following spinal cord injury. Apoptotic (dying) cells were counted
following TLTNEL
labelling (see Example 16) 2mm rostral to the lesion (Rostral) at the lesion
site (lesion) and 2
mm caudal to the lesion site (caudal). Bars show average counts of Tunel
positive cells from
4 animals treated with fibrin only after spinal cord injury as control (white
bars), or with
C3APLT in fibrin at lug (black bars). Treatment with C3APLT show significantly
reduced
numbers of Tunel-labeled cells (dying cells). Non-inj ured spinal cord samples
were also
processed and these spinal cords did not show Tunel labelling, as expected.
62
CA 02362004 2001-11-13
Referring to Figure 11, this Figure shows that C3APLT and C3Basic3 promote
rapid neurite
outgrowth compared to untreated cells when cells are plated on plastic as part
of a rapid
bioassay (see Example 4).
DETAILED DESCRIPTION
Method for making the C3APL, C3APLT, and C3APS
C3APL is the name given to the protein made by ligating a cDNA encoding C3
(Dillon and Feig (1995) 256: 174-184) with cDNA encoding the antennapedia
homeodomain
to (Block-Gallego (1993) 120: 485-492). The stop codon at the 3' end of the
DNA was replaced
with an EcoR I site by polymerase chain reaction (PCR) using the primers 5'GAA
TTC TTT
AGG ATT GAT AGC TGT GCC 3' (SEQ ID NO: 1) and 5'GGT GGC GAC CAT CCT
CCA AAA 3' (SEQ ID NO: 2). The PCR product was sub-cloned into a pSTBlue-1
vector
(Novagen, city), then cloned into a pGEX-4T vector using BamH I and Not I
restriction site.
This vector was called pGEX-4T/C3. The antennapedia sequence used to add to
the 3' end of
C3 in pGEX-4T/C3 was created by PCR from the pET-3a vector (Block-Gallego
(1993) 120:
485-492, Derossi (1994) 269: 10444-10450), subcloned into a pSTBlue-1 blunt
vector, then
cloned into the pGEX-4T/C3, using the restriction sites EcoR I and Sal I,
creating pGEX-
4T/C3APL. Another clone (C3APLT) with a frameshift mutation was selected, and
the
2o protein made and tested. When the cultures tested positive despite the
mutation, the clone
was resequenced by another company to confirm the mutation, and this clone was
called
C3APLT. To confirm the sequence of C3APLT, the coding sequence from both
strands was
sequenced. The sequence for this clone is given in Examples 16 and 17
(nucleotide sequence
of C3APLT; SEQ ID NO: 42, amino acid sequence of C3APLT; SEQ ID N0:43).
A shorter version of the Antennapedia (pGEX-4T/C3APS) was also made. This
chimeric
sequence was made by ligating oligonucleotides encoding the short antennapedia
peptide
(Maizel (1999) 126: 3183-3190) into the pGEX-4T/C3 vector cut with EcoR I and
Sal I. The
63
CA 02362004 2001-11-13
recombinant C3APLT and C3APS cDNAs were separately transformed into bacteria ,
and
after the recombinant proteins were produced, a bacterial homogenate was
obtained by
sonication, and the homogenate cleared by centrifugation. Glutathione-agrose
beads (Sigma)
were added to the cleared lysate and placed on a rotating plate for 2-3 hours,
then washed
extensively. To remove the glutathione S transferase sequence from the
recombinant protein,
20U (unit) of Thrombin was added, the beads were left on a rotator overnight
at 4°C. After
cleavage with thrombin, the beads were loaded into an empty 20m1 column, and
the proteins
eluted with PBS (phosphate buffered saline). Aliquots containing recombinant
protein were
pooled and 100,1 p-aminobenzamidine agrose beads (Sigma) were added and left
mixing for
45 minutes at 4°C to remove thrombin, then recombinant protein was
isolated from the beads
by centrifugation. Purity of the sample was determined by sodium dodecyl
sulfate
polyacrylamide gel electrophoresis (SDS-PAGE), and bioactivity bioassay with
PC-12 cells
was performed (See Lehmann et al supra).
Other possible methods for making bioactive chimeric proteins include anion
exhange
chromatography. For this, the GST tag is not required and can be removed. The
cDNA can
then be cloned into a high expression bacterial vector, such as pET, as given
in Example 16.
The Rho antagonist is a recombinant protein and can be made according to
methods present
2o in the art. The proteins of the present invention may be prepared from
bacterial cell extracts,
or through the use of recombinant techniques by transformation, transfection,
or infection of a
host cell with all or part of a C3-encoding DNA fragment with an antennapedia-
derived
transport sequence in a suitable expression vehicle. Those skilled in the
field of molecular
biology will understand that any of a wide variety of expression systems can
be used to
provide the recombinant protein. The precise host cell used is not critical to
the invention.
Any fusion protein can be readily purified by utilising either affinity
purification techniques
or more traditional column chromatography. Affinity techniques include, but
are not
restricted to GST (gluathionie -S-transferase), or the use of an antibody
specific for the
64
CA 02362004 2001-11-13
fusion protein being expressed, or the use of a histidinetag. Alternatively,
recombinant
protein can be fused to an immunoglobulin Fc domain. Such a fusion protein can
be readily
purified using a protein A column. It is envisioned that small molecule
mimetics of the
above described antagonists are also encompassed by the invention.
Testing the bioactivity of C3APLT, C3APS, C3-TL and C3-TS
To test the efficacy of C3APLT, C3APS, C3-TL and C3-TS a number of experiments
were
performed with PC-12 cells, a neural cell line, grown on growth inhibitory
substrates (see
Lehmann et al supra). PC-12 cells were plated on myelin substrates as
described (Lehmann
1o et al , supra). C3, C3APLT, C3APS, C3-TL or C3-TS were added at different
concentrations
without trituration (please refer to Figures 4, 5 and 8 for concentrations
used). C3 added
passively to the culture medium in this way was not able to promote neurite
growth in the
growth inhibitory substrates because cells must be triturated for C3 to enter
the cells and be
active (Figure 1). Both C3APLT and C3APS were able to ADP ribosylate Rho to
cause a
shift in the molecular weight of RhoA (Figure 2). Both C3APLT and C3APS were
able to
promote neurite growth and enter neurons after being added passively to the
culture medium
(Figure 3, Figures 4 and 5). Dose-response experiment where concentrations of
0.25ng/ml,
2.5 ng/ml, 25 ng/ml, 250 ng/ml and 2.5 ~,g/ml and 25 ~,g/ml were tested and
showed that
C3APLT and C3APS helped more neurons differentiate neurites at doses 10,000
fold less
2o than C3 (Figure 4). Dose response experiments where concentrations of
0.25ng/ml, 2.5
ng/ml, 25 ng/ml, 250 ng/ml and 2.5 ~,g/ml and 25 ~.g/ml were tested and showed
that
C3APLT was able to promote long neurite growth when added at a minimum
concentration
of 0.0025 ug/ml (Figure 5). These concentrations of 2.5 ng/ml and 25 ng/ml for
C3APLT
and C3APS, represent 10,000 and 1,000 times less than the dose needed with C3,
respectively. Moreover, at the highest concentration tested, 50 ug/ml, these
two new Rho
antagonists did not exhibit toxic effects on PC-12 cells, and were able to
stimulate neurite
outgrowth on growth inhibitory substrates.
CA 02362004 2001-11-13
C3-TL and C3-TS also were tested at concentrations of 0.25ng/ml, 2.5 ng/ml, 25
ng/ml, 250
ng/ml and 2.5 ~,g/ml and 25 ~.g/ml and were found to be able to promote
neurite growth on
myelin substrates at doses significantly less than C3 (Figure 8). C3Basic3 was
tested at 50
ug/ml in a fast growth assay (Figure 11).
To verify the ability of C3APLT and C3APS to promote growth from primary
neurons,
primary retinal cultures were prepared, and the neurons were plated on myelin
substrates as
described with respect to Example 5. In the absence of treatment with C3APLT
or C3APS,
the cells remained round and were not able to grow neurites. When treated with
C3APLT or
to C3APS, retinal neurons were able to extend long neurites on inhibitory
myelin substrates
(Figure 6).
Next, was tested the ability of C3APLT and C3APS to promote growth on a
different type of
growth inhibitory substrate relevant to the type of growth inhibitory proteins
found at glial
scars. Chamber slides were coated with a mixture of chondroitin sulfate
proteoglycans
(Chemicon), and then plated with retinal neurons. The neurons were not able to
extend
neurites on the proteoglycan substrates, but when treated with C3APLT or
C3APS, they
extended long neurites (not shown). These studies demonstrate that C3APLT and
C3APS
can be used to promote neurite growth on myelin and on proteoglycans, the
major classes of
inhibitory substrates that prevent repair after injury in the CNS.
Testing ability of C3APLT to promote regeneration and functional recovery
after spinal
cordinjury
To test if C3APLT could promote repair after spinal cord injury, fully adult
mice were used
(as described with respect to Example 6). A dorsal hemisection was made at T8
(thoracic
spinal level 8), and mice were treated with different amounts (Figure 7) of
C3APL in a fibrin
glue as described (McKerracher, US patent pending (delivery patent)). In
previous known
experiments with C3, it was found that 40-50 ug was needed to promote
anatomical
66
CA 02362004 2001-11-13
regeneration in optic nerve (Lehmann et all supra). We tested different doses
(see Figure 7)
of C3APLT ranging from 1 ug to 50 ug and assessed animals for behavioural
recovery
according the BBB scale (Basso ( 1995) 12: 1-21 )
The day following surgery and application of C3APLT, behavioural testing
began. The
animals were placed in an open field environment that consisted of a rubber
mat
approximately 4' X 3' in size. The animals were left to move randomly, the
movement of the
animals were videotaped. For each test two observers scored the animals for
ability to move
ankle, knee and hip joints in the early phase of recovery. Previously C3
treatment of mice
1o was seen to lead to functional recovery observable 24 hours after
treatment. In mice treated
with C3APLT, functional recovery could be observed as early as 24 hours after
spinal cord
injury (Figure 7). Untreated mice exhibit a function recovery score according
to the BBB
scale averaging 0, whereas mice treated with C3 are able to walk and have a
BBB score
averaging 8 (Figure 7). At higher concentrations of 50 ug, about 50 % of the
mice treated
with C3APLT died within 24 hours. However, of the mice that survived, they
exhibited good
long-term functional recovery. These results demonstrate that C3APLT
effectively promotes
functional recovery early after spinal cord injury, and that it is effective
at much lower doses
than C3. However, at high concentrations, C3APLT appears to exhibit toxicity,
and therefore
careful doing will be required for clinical use.
Qualitative observations of the video tapes showed that only animals that
received C3APLT
reached the late phase of recovery after 30 days of treatment. Untreated
control animals did
not typically pass beyond the early phase of recovery. These results indicate
that the
application of C3APLT improved long-term functional recovery after spinal cord
injury
compared to no treatment, injury alone, or fibrin adhesive alone.
To test if the early recovery was due to neuroprotection, spinal cord sections
were examined
for apoptosis by Tunel labelling (see Example 16 for details). C3APLT was able
to reduce
67
CA 02362004 2001-11-13
the number of dying cells observed at the lesion site. Therefore, C3APLT
should be an
effective neuroprotective agent for treatment of ischemia, such as follows
stroke.
EXAMPLE 1. DNA AND PROTEIN SEQUENCE DETAILS OF C3APL
Nucleotide sequence of C3APL.
It has been reported that the long version of antennapedia transport sequence
can enhance
neurite growth ( Bloch-Gallego,E.,LeRoux,L, Joliot,A.H., Volovitch,M.,
Henderson,C.E.,
Prochiantz,A. 1993. J. Cell Biol. 120:485). Therefore, this sequence is
expected to enhance
neurite growth. For the sequence given below, the start site, is in the GST
sequence of the
1o plasmid (not shown). The vector with the GST sequence is commercially
available and thus
the entire GST sequence including the start was not sequenced. It was desired
to determine
only the sequence located 3' to the thrombin cleavage site which releases C3
conjugate from
the GST sequence. The GST sequence is cleaved with thrombin.
The APL transport sequence (SEQ ID NO.: 44) is as follows:
VMESRKRARQTYTRYQTLELEKEFHFNRYLTRRRRIEIAHALCLTERQIKIWFQNRR
MKWKKEN
Nucleotide sequence of C3APL (SEQ ID NO: 3)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
2o CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
68
CA 02362004 2001-11-13
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC GTG ATG GAA TCC CGC AAA CGC GCA AGG
CAG ACA TAC ACC CGG TAC CAG ACT CTA GAG CTA GAG AAG GAG TTT CAC
TTC AAT CGC TAC TTG ACC CGT CGG CGA AGG ATC GAG ATC GCC CAC GCC
CTG TGC CTC ACG GAG CGC CAG ATA AAG ATT TGG TTC CAG AAT CGG CGC
1o ATG AAG TGG AAG AAG GAG AAC TGA 3'
Amino acid sequence of C3APL (SEQ ID NO: 4)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIV S
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYL
GTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFK
VAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPK
EFVMESRKRARQTYTRYQTLELEKEFHFNRYLTRRRRIEIAHALCLTERQIKIWFQNR
RMKWKKEN
2o Physical characteristics of C3APL
Molecular Weight 34098.03 Daltons
295 Amino Acids
48 Strongly Basic(+) Amino Acids (K,R)
28 Strongly Acidic(-) Amino Acids (D,E)
89 Hydrophobic Amino Acids (A,I,L,F,W,V)
94 Polar Amino Acids (N,C,Q,S,T,Y)
69
CA 02362004 2001-11-13
9.847 Isolectric Point
20.524 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 79.48
EXAMPLE 2. DNA AND PROTEIN SEQUENCE DETAILS OF C3APS
Nucleotide sequence of C3APS (SEQ ID NO: S). The start site, is in the GST
sequence of the
plasmid, not shown here.
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
1o GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
2o ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC CGC CAG ATC AAG ATT TGG TTC CAG AAT
CGT CGC ATG AAG TGG AAG AAG GTC GAC TCG AGC GGC CGC ATC GTG ACT
GAC TGA 3'
The APS transport sequence (SEQ ID NO.: 45)is as follows:
RQIKIWFQNRRMKWKKVDS
CA 02362004 2001-11-13
Amino acid sequence for C3APS (SEQ ID NO: 6)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYL
GTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFK
VAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPK
EFRQIKI WFQNRRMKWKKV D S S GRI V TD
Physical characteristics of C3APS
1o Molecular Weight 29088.22 Daltons
257 Amino Acids
38 Strongly Basic(+) Amino Acids (K,R)
23 Strongly Acidic(-) Amino Acids (D,E)
79 Hydrophobic Amino Acids (A,I,L,F,W,V)
83 Polar Amino Acids (N,C,Q,S,T,Y)
9.745 Isolectric Point
15.211 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 78.34
EXAMPLE 3. METHOD FOR MAKING THE C3APLT AND C3APS PROTEINS
C3APL (amino acid sequence: SEQ ID NO.: 4) and C3APLT (amino acid sequence;
SEQ ID
NO: 37) are the names given to the proteins encoded by cDNAs made by ligating
the
functional domain of C3 transferase and the homeobox region of the
transcription factor
71
CA 02362004 2001-11-13
called antennapedia (Block-Gallego (1993) 120: 485-492) in the following way.
A cDNA
encoding C3 (Dillon and Feig (1995) 256: 174-184) cloned in the plasmid vector
pGEX-2T
was used for the C3 portion of the chimeric protein. The stop codon at the 3'
end of the DNA
was replaced with an EcoR I site by polymerase chain reaction using the
primers 5'GAA
TTC TTT AGG ATT GAT AGC TGT GCC 3' (SEQ ID NO: 1) and 5'GGT GGC GAC CAT
CCT CCA AAA 3' (SEQ ID NO: 2). The PCR product was sub-cloned into a pSTBlue-1
vector (Novagen, city), then cloned into a pGEX-4T vector using BamH I and Not
I
restriction site. This vector was called pGEX-4T/C3. The pGEX-4T vector has a
5'
glutathione S transferase (GST) sequence for use in affinity purification. The
antennapedia
to sequence used to add to the 3' end of C3 in pGEX-4T/C3 was created by PCR
from the pET-
3a vector (Block-Gallego (1993) 120: 485-492, Derossi (1994) 269: 10444-
10450). The
primers used were 5'GAA TCC CGC AAA CGC GCA AGG CAG 3' (SEQ ID NO: 7) and
S'TCA GTT CTC CTT CTT CCA CTT CAT GCG 3' (SEQ ID NO: 8). The PCR product
obtained from the reaction was subcloned into a pSTBlue-1 blunt vector, then
cloned into the
pGEX-4T/C3, using the restriction sites EcoR I and Sal I, creating pGEX-
4T/C3APL and
C3APLT. C3APLT was selected for the presence of a frameshift mutation giving a
transport
region moiety rich in prolines.
A shorter version of the antennapedia (pGEX-4T/C3AP-short) (amino acid
sequence of
2o C3APS; SEQ ID NO.: 6) was also made. This chimeric sequence was made by
ligating
oligonucleotides encoding the short antennapedia peptide (Maizel (1999) 126:
3183-3190)
into the pGEX-4T/C3 vector cut with EcoR I and Sal I. For pGEX-4T/C3AP-short
the
sequences of the oligos made were 5'AAT TCC GCC AGA TCA AGA TTT GGT TCC
AGA ATC GTC GCA TGA AGT GGA AGA AGG 3' (SEQ ID NO: 9) and 5'GGC GGT
CTA GTT CTA AAC CAA GCT CTT AGC AGC GTA GTT CAC CTT CTT CCA GCT 3'
(SEQ ID NO: 10). The two strands were annealed together by mixing equal
amounts of the
oligonucleotides, heating at 72 °C for 5 minutes and then leaving them
at room temperature
for 15 minutes. The oligonucleotides were ligated into the pGEX4T/C3 vector
and clones
were picked and analyzed.
72
CA 02362004 2001-11-13
To prepare recombinant C3APLT (SEQ ID NO.: 37) and C3APS (SEQ ID NO.: 6)
proteins,
the plasmids containing the corresponding cDNAs (pGEX-4T/C3APLT and pGEX-
4T/C3AP-short) were transformed into bacteria, strain XL-1 blue competent E.
coli. The
bacteria were grown in L-broth (lOg/L Bacto-Tryptone, 5g/L Yeast Extract,
lOg/L NaCI)
with ampicillin at 50 ug/ml (BMC-Roche), in a shaking incubator for 1 hr at 37
°C and 300
rpm. Isopropyl ~3-D-thiogalactopyranoside (IPTG), (Gibco) was added to a final
concentration of 0.5 mM to induce the production of recombinant protein and
the culture was
grown for a further 6 hours at 37 °C and 250 rpm. Bacteria pellets were
obtained by
centrifugation in 250 ml centrifuge bottles at 7000 rpm for 6 minutes at 4
°C. Each pellet
1o was re-suspended in 10 ml of Buffer A (50mM Tris, pH 7.5, 50 mM NaCI, 5mM
MgCl2_,
1mM DTT) plus 1mM PMSF. All re-suspended pellets were pooled and transferred
to a 100
ml plastic beaker on ice. The remaining Buffer A with PMSF was added to the
pooled
sample. The bacteria sample was sonicated 6 x 20 seconds using a Branson
Sonifier 450
probe sonicator. Both the bacteria and probe were cooled on ice 1 minute
between
sonications. The sonicate was centrifuged in a Sorvall SS-34 rotor at 16,000
rpm for 12
minutes at 4 °C to clarify the supernatant. The supernatant was
transferred into fresh SS-34
tubes and re-spun at 12,000 rpm for 12 minutes at 4 °C. Up to 20 ml of
Glutathione-agarose
beads (Sigma) were added to the cleared lysate and placed on a rotating plate
for 2-3 hours.
The beads were washed 4 times with buffer B, (Buffer A, NaCI is 150 mM, no
PSMF) then 2
2o times with Buffer C (Buffer B + 2.5 mM CaCl2). The final wash was poured
out till the
beads created a thick slurry. To remove the glutathione S transferase sequence
from the
recombinant protein, 20U of Thrombin (Bovine, Plasminogen-free, Calbiochem)
was added,
the beads were left on a rotator overnight at 4 °C. After cleavage with
thrombin the beads
were loaded into an empty 20 ml column. Approximately 20 aliquots of 1 ml were
collected
by elution with PBS. Samples of each aliquot of 0.5 u1 were spotted on
nitrocellulose and
stained with Amido Black to determine the protein peak. Aliquots containing
fusion proteins
were pooled and 100 ~,1 p-aminobenzamidine agarose beads (Sigma) were added
and left
mixing for 45 minutes at 4 °C. This last step removed the thrombin from
the recombinant
protein sample. The recombinant protein was centrifuged to remove the beads
and then
3o concentrated using a centriprep-10 concentrator (Amicon). The concentrated
recombinant
protein was desalted with a PD-10 column (Pharmacia, containing Sephadex G-
25M) and ten
0.5 ml aliquots were collected. A dot-blot was done on these samples to
determine the
73
CA 02362004 2001-11-13
protein peak, and the appropriate aliquots pooled, filter-sterilized, and
stored at - 80°C. A
protein assay (DC assay, Biorad) was used to determine the concentration of
recombinant
protein. Purity of the sample was determined by SDS-PAGE, and bioactivity
bioassay with
PC-12 cells.
EXAMPLE 4: TESTING OF EFFICACY OF C3APLT AND C3APS IN TISSUE CULTURE
To test the ability of C3APLT and C3APS to overcome growth inhibition, PC-12
cells were
plated on myelin, a growth inhibitory substrate. The myelin was purified from
bovine brain
(Norton and Poduslo (1973) 21: 749-757). In some other experiments chondroitin
sulfate
to proteoglycan (CSPG) substrates were made from a purchased protein
composition
(Chemicon). Before coating coverslips or wells of a 96 well plate, they were
coated with
poly-L-lysine (0.025~,g/ml) (Sigma, St. Louis, MO), washed with water and
allowed to dry.
Myelin stored as a 1 mg/ml solution at -80 °C was thawed at 37
°C, and vortexed. The myelin
was plated at 8 ug/well in a 8 well chamber Lab-Tek slides (Nuc, Naperville,
IL). The myelin
15 solution was left to dry overnight in a sterile tissue culture hood. The
next morning the
substrate was washed gently with phosphate buffered saline, and then cells in
media were
added to the substrate. PC-12 cells (Lehmann et al., 1999) were grown in DMEM
with 10%
horse serum (HS) and 5% fetal bovine serum (FBS). Two days prior to use the PC-
12 cells
were differentiated by SO ng/ml of nerve growth factor (NGF). After the cells
were primed,
2o Sml of trypsin was added to the culture dish to detach the cells, the cells
were pelleted and re-
suspended in 2 ml of DMEM with 1% HS and SO ng/ml of nerve growth factor.
Approximately, 5000 to 7000 cells were then plated on 8 well chamber Lab-Tek
slides (Nuc,
Naperville, IL) coated myelin. The cells were placed on the test substrates at
37 °C for 3-4
hours to allow the cells to settle. The original media was carefully removed
by aspiration,
25 taking care not to disrupt the cells and replaced with DMEM with 1% HS,
SOng/ml of NGF
and varying amounts of the C3, C3APLT, or C3APS, depending on the dose
desired. After
two days, the cells were fixed (4% paraformaldehyde and 0.5% glutaraldehyde).
For control
experiments with unmodified C3, NGF primed PC-12 cells were trypsinized to
detach them
from the culture dish, the cells were washed once with scrape loading buffer
(114 mM KCL,
30 15 mM NaCI, 5.5 mM MgClz, and 10 mM Tris-HCL) and then the cells were
scraped with a
rubber policeman into 0.5 ml of scraping buffer in the presence of 25 or 50
~,g/ml of C3. The
74
CA 02362004 2001-11-13
cells were pelleted and resuspended in 2m1 of DMEM, 1% HS and SO ng/ml nerve
growth
factor before plating. At least four experiments were analyzed for each
treatment. For each
well, twelve images were collected with a 20X objective using a Zeiss Axiovert
microscope.
For each image, the numbers of cells with and without neurites were counted
and the lengths
of the neurites were determined. Since myelin is phase dense, cells plated on
myelin
substrates were immuno-stained with anti-(3III tubulin antibody before
analysis. Quantitative
analysis of neurite outgrowth was with the aid of Northern Eclipse software
(Empix Imaging,
Mississauga, Ontario, Canada). Data analysis and statistics were with
Microsoft Excel.
1o For a fast bioassay, the compounds were tested in tissue culture as
described above, except
that the cells were plated on the tissue culture plastic rather than on
inhibitory substrates. For
these experiments the plates were fixed and the neurites counted five hours
after plating the
cells. The test compounds (C3APLT and C3Basic3) were able to promote faster
growth on
tissue culture plastic than cells plated without treatment (Figure 11 ).
To examine ADP ribosylation by C3, C3APLT, and C3APS, the compounds were added
to
PC-12 cell cultures, as described above. The cells were harvested by
centrifugation, cell
homogenates prepared and the proteins separated by SDS polyacrylamide gel
electrophoresis.
The proteins were then transfered to nitrocellulose and the Western blots
probed with anti-
2o RhoA antibody (Santa Cruz).
EXAMPLE 5: TESTING ABILITY OF C3APLT AND C3APS TO OVERRIDE INHIBITION OF
MULTIPLE GROWTH INHIBITORY PROTEINS.
Myelin substrates were made as described in Example 4 and plated on tissue
culture chamber
slides. P1 to P3 rat pups were decapitated, the heads washed in ethanol and
the eye removed
and placed in a petri dish with Hanks buffered saline solution (HBSS, from
Gibco). A hole
was cut in the cornea, the lens removed, and the retina squeezed out.
Typically, four retinas
per preparation were used. The retinas were removed to a 15 ml tube and the
volume brought
CA 02362004 2001-11-13
to 7 ml. A further 7 ml of dissociation enzymes and papain were added. The
dissociation
enzyme solution was made as follows: 30 mg DL cysteine was added to a 15 ml
tube (Sigma
DL cystein hydrochloride), and 70 ml HBSS, 280 u1 of lOmg.ml bovine serum
albumin were
added and the solution mixed and pH adjusted to 7 with 0.3 N NaOH. The
dissociate solution
was filter-sterilized and kept frozen in 7 ml aliquots, and before use 12.5
units papain per ml
(Worthington) was added. After adding the dissociation solution to the retina,
the tube was
incubated for 30 minutes on a rocking tray at 37 °C. The retinas were
then gently triturated,
centrifuged and washed with HBSS. The HBSS was replaced with growth medium
(DMEM
(Gibco), 10 % fetal bovine serum, and 50 ng/ml brain derived neurotrophic
factor (BDNF)
1o vitamins, penicillin-streptomycin, in the presence or absence of C3APLT or
C3APS. Cells
were plated on test substrates of myelin or CSPG in chamber slides prepared as
described in
Example 4, above. A quantitative analysis was completed as described for
Example 4 above.
Neurons were visualized by fluorescent microscopy with anti-(3III tubulin
antibody, which
detects growing retinal ganglion cells (RGCs).
EXAMPLE 6. TREATMENT OF INJURED MOUSE SPINAL CORD WITH C3APLT AND
MEASUREMENT OF RECOVERY OF MOTOR FUNCTION IN TREATED MICE.
Adult Balb-c mice were anaesthetized with 0.6 ml/kg hypnorm, 2.5 mg/kg
diazepam and 35
mg/kg ketamine. This does gives about 30 minutes of anaesthetic, which is
sufficient for the
2o entire operation. A segment of the thoracic spinal column was exposed by
removing the
vertebrae and spinus process with microrongeurs (Fine Science Tools). A spinal
cord lesion
was then made dorsally, extending past the central canal with fine scissors,
and the lesion was
recut with a fine knife. This lesion renders all of the control animals
paraplegic. The
paravertebral muscle were closed with reabsorbable sutures, and the skin was
closed with 2.0
silk sutures. After surgery, the bladder was manually voided every 8-10 hours
until the
animals regained control, typically 2-3 days. Food was placed in the cage for
easy access,
and sponge-water used for easy accessibility of water after surgery. Also,
animals received
subcutaneous injection Buprenorphine (0.05 a 0.1 mg/kg) every 8-12 hours for
the first 3
days. Any animals that lost 1 S-20% of body weight were killed.
76
CA 02362004 2001-11-13
Rho antagonists (C3 or C3-like proteins) were delivered locally to the site of
the lesion by a
fibrin-based tissue adhesive delivery system (McKerracher, Canadian patent
application No.
2,325,842). Recombinant C3APLT was mixed with fibrinogen and thrombin in the
presence
of CaCl2. Fibrinogen is cleaved by thrombin, and the resulting fibrin monomers
polymerize
into a three-dimensional matrix. We added C3APLT as part of a fibrin adhesive,
which
polymerized within about 10 seconds after being placed in the injured spinal
cord. We tested
C3APLT applied to the spinal cord lesion site after the lesion was made. For
control we
injected fibrin adhesive alone, or transected the cord without further
treatment. For
behavioural testing, the BBB scoring method was used to examine locomotion in
an open
field environment (Basso (1995) 12: 1-21). The envirorunent was a rubber mat
approximately 4' X 3' in size, and animals were placed on the mat and
videotaped for about 4
minutes. Care was taken not to stimulate the peroneal region or touch the
animals
excessively during the taping session. The video tapes were digitized and
observed by two
observers to assign BBB scores. The BBB score, modified for mice, was as
follows:
Score Description
1 No observable hindlimb (HL) movement.
2 Slight movement of one or two joints.
3 Extensive movement of one joint and /or slight movement of one other joint..
4 Extensive movement of two joints.
5 Slight movement of all three joints of the HL.
6 Slight movement of two joints and extensive movement of the third.
7 Extensive movement of two joints and slight movement of the third.
8 Extensive movement of all three joints of the HL walking with no weight
support.
9 Extensive movement of all three joints, walking with weight support.
10 Frequent to consistent dorsal stepping with weight support.
11 Frequent plantar stepping with weight support.
12 Consistent plantar stepping with weight support, no coordination.
13 Consistent plantar stepping with consistent weight support, occasional FL-
HL
coordination.
14 Consistent plantar stepping with consistent weight support, frequent FL-HL
coordination.
77
CA 02362004 2001-11-13
15 Consistent plantar stepping with consistent weight support, consistent FL-
HL
coordination; predominant paw position during locomotion is rotated internally
or
externally, or consistent FL-HL coordination with occasional dorsal stepping.
16 Consistent plantar stepping with consistent weight support, consistent FL-
HL
coordination; predominant paw position is parallel to the body; frequent to
consistent
toe drag, or curled toes, trunk instability.
17 Consistent plantar stepping with consistent weight support, consistent FL-
HL
coordination; predominant paw position is parallel to the body, no toe drag,
some
trunk instability.
18 Consistent plantar stepping with consistent weight support, consistent FL-
HL
coordination; predominant paw position is parallel to the body, no toe drag
and
consistent stability in the locomotion.
EXAMPLE . 7 TREATMENT OF INJURED MOUSE SPINAL CORD WITH C3APLT AND
ASSESSMENT OF ANATOMICAL RECOVERY.
Mice that received a spinal cord injury and treated as controls or with
C3APLT, as described
for Example 6 were assessed for morphological changes to the scar and for axon
regeneration. To study axon regeneration, the corticospinal axons were
identified by
anterograde labeling. For anterograde labeling studies, the animals were
anaesthetized as
above, and the cranium over the motor cortex was removed. With the fine glass
micropipetter (about 100 um in diameter) the cerebral cortex was injected with
2-4 u1 of horse
radish peroxidase conjugated to wheat germ agglutinin (2%), a marker that is
taken up by
nerve cells and transported anterogradely into the axon that extends into the
spinal cord.
After injection of the anterograde tracer, the cranium was replaced, and the
skin closed with
S-0 silk sutures. The animals were sacrificed with chloral hydrate (4.9 mg/10
g) after 48
hours, and perfused with 4% paraformaldehyde in phosphate buffer as a
fixative. The spinal
3o cord was removed, cryoprotected with sucrose and cryostat sections placed
on slides for
histological examination.
CA 02362004 2001-11-13
EXAMPLE 8. DNA AND PROTEIN SEQUENCE DETAILS OF C3-TL.
The Tat coding sequence was obtained by polymerase chain reaction of the
plasmid SVCMV-
TAT (obtained form Dr. Eric Cohen, Universite de Montreal) that contains the
entire Tat
coding sequence. To isolate the transport sequence of the Tat protein, PCR was
used. The
first primer (5'GAATCCAAGCACCAGGAAGTCAGCC 3' (SEQ ID NO.: 11)) and the
second primer (5' ACC AGCCACCACCTTCTGATA 3' (SEQ ID NO.: 12)) used
corresponded to amino acids 27 to 72 of the HIV Tat protein. Upon verification
and
purification, the PCR product was sub cloned into a pSTBlue-1 blunt vector.
This transport
segment of the Tat protein was then cloned into pGEX-4T/C3 at the 3' end of
C3, using the
restriction sites EcoR I and Sac I. The new C3-Tat fusion protein was called
C3-TL.
Recombinant protein was made as described in Example 3.
DNA sequence of C3-TL (SEQ ID NO.: 13)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
2o GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
3o ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AAG CAT CCA GGA AGT CAG CCT AAA ACT
GCT TGT ACC AAT TGC TAT TGT AAA AAG TGT TGC TTT CAT TGC CAA GTT
7
CA 02362004 2001-11-13
TGT TTC ATA ACA AAA GCC TTA GGC ATC TCC TAT GGC AGG AAG CGG AGA
CAG CGA CGA AGA GCT CAT CAG AAC AGT CAG ACT CAT CAA GCT TCT CTA
TCA AAG CAG TAA 3'
The TL transport peptide sequence by itself is as follows:(SEQ ID NO.: 46)
KHPGSQPKTACTNCYCKKCCFHCQVCFITKALGISYGRKRRQRRRAHQNSQTHQAS
LSKQ.
The protein sequence of C3-TL (SEQ ID NO.: 14)
l0
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYL
GTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITQFK
VAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAlNPK
EFKHPGSQPKTACTNCYCKKCCFHCQVCFITKALGISYGRKRRQRRRAHQNSQTHQ
ASLSKQ.
Molecular Weight 32721.40 Daltons
291 Amino Acids
43 Strongly Basic(+) Amino Acids (K,R)
21 Strongly Acidic(-) Amino Acids (D,E)
82 Hydrophobic Amino Acids (A,I,L,F,W,V)
104 Polar Amino Acids (N,C,Q,S,T,Y)
9.688 Isolectric Point
22.655 Charge at PH 7.0
Total number of bases translated is 876
A = 37.44 [328]
G = 17.58[154]
% T = 28.31 [248]
C = 16.67 [ 146]
8
CA 02362004 2001-11-13
EXAMPLE 9. DNA AND PROTEIN SEQUENCE DETAILS OF C3-TS.
A shorter Tat construct was also made (C3-TS). To make the shorter C3 Tat
fusion protein
the following oligonucleotrides were 5'AAT TCT ATG GTC GTA AAA AAC GTC GTC
AAC GTC GTC GTG 3' (SEQ ID NO.: 15) and 5' GAT ACC AGC ATT TTT TGC AGC
AGT TGC AGC AGC ACA GCT 3' (SEQ ID NO.: 16). The two oligonucleotide strands
were annealed together by combining equal amounts of the oligonucleotides,
heating at 72 °C
for 5 minutes and then letting the oligonucleotide solution cool at room
temperature for 15
1o minutes. The oligonucleotides were ligated into the pGEX4T/C3 vector at the
3' end of C3.
The construct was sequenced. All plasmids were transformed into XL-1 blue
competent
cells. Recombinant protein was made as described in Example 3.
Nucleotide sequence of C3-TS (SEQ ID NO.: 17)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
2o TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTC TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA CAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
3o TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC TAT GGT GCT AAA AAA CGT CGT CAA CGT
CGT CGT GTC GAC TCG AGC GGC CCG CAT CGT GAC TGA 3'
s
CA 02362004 2001-11-13
The TS transport peptide sequence by itself is as follows: (SEQ ID NO.: 47)
YGAKKRRQRRRVDSSGPHRD
The protein sequence of C3-TS (SEQ ID NO.: 18)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYL
GTEFQNTLLNSNGTINKTAFEKAKAKF'LNKDRLEYGYISTSLMNVSQFAGRPIITQFK
1o VAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPK
EFYGAKKRRQRRRVDSSGPHRD
Molecular Weight 26866.62 Daltons
238 Amino Acids
36 Strongly Basic(+) Amino Acids (K,R)
21 Strongly Acidic(-) Amino Acids (D,E)
71 Hydrophobic Amino Acids (A,I,L,F,W,V)
78 Polar Amino Acids (N,C,Q,S,T,Y)
9.802 Isolectric Point
15.212 Charge at PH 7.0
Total number of bases translated is 717
A = 38.91 [279]
% G = 17.43 [125]
T = 28.45 [204]
C = 15.20 [ 109]
8
CA 02362004 2001-11-13
EXAMPLE 10.
The following example illustrates how a coding sequence can be modified
without affecting
the efficacy of the translated protein. The example shows modifications to
C3Basic3 that
would not affect the activity. Sequences may include the entire GST sequence,
as shown here
that includes the start site, which would not be removed enzymatically. Also,
the transport
sequence shown in this example has changes in amino acid composition
surrounding the
active sequence due to a difference in the cloning strategy, and the His tag
has been omitted.
However, the active region is : R R K Q R R K R R. This sequence is contained
in the
1o C3Basic3, and is the active transport sequence in the sequence below. Also
note that the C-
terminal region of the protein after this active region differs from C3Basic3.
That is because
the cloning strategy was changed, the restriction sites differ, and therefore
non-essential
amino acids 3' terminal to the transport sequence are transplanted and
included in the protein.
Nucleic acid sequence: (SEQ ID NO.: 19)
1413 base pairs
single strand
linear sequence
5' ATG TCC CCT ATA CTA GGT TAT TGG AAA ATT AAG GGC CTT GTG CAA CCC
ACT CGA CTT CTT TTG GAA TAT CTT GAA GAA AAA TAT GAA GAG CAT TTG
TAT GAG CGC GAT GAA GGT GAT AAA TGG CGA AAC AAA AAG TTT GAA TTG
GGT TTG GAG TTT CCC AAT CTT CCT TAT TAT ATT GAT GGT GAT GTT AAA
TTA ACA CAG TCT ATG GCC ATC ATA CGT TAT ATA GCT GAC AAG CAC AAC
ATG TTG GGT GGT TGT CCA AAA GAG CGT GCA GAG ATT TCA ATG CTT GAA
GGA GCG GTT TTG GAT ATT AGA TAC GGT GTT TCG AGA ATT GCA TAT AGT
AAA GAC TTT GAA ACT CTC AAA GTT GAT TTT CTT AGC AAG CTA CCT GAA
3o ATG CTG AAA ATG TTC GAA GAT CGT TTA TGT CAT AAA ACA TAT TTA AAT
GGT GAT CAT GTA ACC CAT CCT GAC TTC ATG TTG TAT GAC GCT CTT GAT
GTT GTT TTA TAC ATG GAC CCA ATG TGC CTG GAT GCG TTC CCA AAA TTA
GTT TGT TTT AAA AAA CGT ATT GAA GCT ATC CCA CAA ATT GAT AAG TAC
8
CA 02362004 2001-11-13
TTG AAA TCC AGC AAG TAT ATA GCA TGG CCT TTG CAG GGC TGG CAA GCC
ACG TTT GGT GGT GGC GAC CAT CCT CCA AAA TCG GAT CTG GTT CCG CGT
GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
1o TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AGA AGG AAA CAA AGA AGA A AA AGA
AGA CTG CAG GCG GCC GCA TCG TGA 3'
Amino acid sequence (SEQ ID NO: 20)
479 amino acids
linear, single strand
MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRIVI~KF'ELGLEFPNLPY
YIDGDVKLTQSMAIIRYIADKHNMLGGCPKERAEISMLEGAVLDIRYGVSRIAYSKDF
ETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALDVVLYMDPMCL
DAFPKLVCFKKRIEAIPQIDKYLKSSKYIAWPLQGWQATFGGGDHPPKSDLVPRGSSR
3o VDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVSYTKS
ASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYLGTEFQ
NTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITKFKVAKGS
8
CA 02362004 2001-11-13
KAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPKEFRRK
QRRKRRLQAAAS.
Molecular Weight 53813.02 Daltons
470 Amino Acids
68 Strongly Basic(+) Amino Acids (K,R)
55 Strongly Acidic(-) Amino Acids (D,E)
149 Hydrophobic Amino Acids (A,I,L,F,W,V)
121 Polar Amino Acids (N,C,Q,S,T,Y)
9.137 Isolectric Point
14.106 Charge at PH 7.0
Total number of bases translated is 1413
% A = 34.61[489]
G = 19.75[279]
T = 29.51 [417]
C = 15.99[226]
Ambiguous 0.14 [2]
=
A+T = 64.12 [906]
C+G = 35.74 [505]
Davis,Botstein,Roth Melting Temp C. 79.20
EXAMPLE 11. ADDITIONAL CHIMERIC C3 PROTEINS THAT WOULD BE EFFECTIVE TO
STIMULATE REPAIR IN THE CNS.
3o The following sequences could be added to the amino terminal or carboxy
terminal of C3 or a
truncated C3 that retains its enzymatic activity.
8
CA 02362004 2001-11-13
1) Sequences of polyarginine as decribed (Wender, et al. (2000) 97: 13003-8.).
These
could be from 6 to 9 or more arginines.
2) Sequences of poly-lysine
3) Sequences of poly-histidine
4) Sequences of arginine and lysine mixed.
5) Basic stretchs of amino acids containing non-basic amino acids stretch
where the
sequence added retains transport characteristics.
6) Sequences of 5- 15 amino acids containing at least 50 % basic amino acids
7) Sequences longer than 15 -30 amino acids containing at least 30 % basic
amino acids.
l0 8) Sequences longer than 50 amino acids containing at least 18 % basic
amino acids.
9) Any of the above where the amino acids are chemically modified, such as by
addition
of cyclohexyl side chains, other side chains, different alkyl spacers.
10) Sequences that have proline residues with helix-breaking propensity to act
as
effective transporters.
EXAMPLE 12. ADDITIONAL CHIMERIC C3 PROTEINS THAT WOULD BE EFFECTIVE TO
STIMULATE REPAIR IN THE CNS.
C3Basicl: C3 fused to a randomly designed basic tail
2o C3Basic2: C3 fused to a randomly designed basic tail
C3 Basic3: C3 fused to the reverse Tat sequence
We have designed the following DNA encoding a chimeric C3 with membrane
transport
properties. The protein is designated C3Basicl. This sequence was designed
with C3 fused
to a random basic sequence. The construct was made to encode the peptide given
below.
K R R R R R P K K R R R A K R R (SEQ ID NO.: 21)
3o The construct was made by synthesizing the two oligonucleotides given
below, annealing
them together, and ligating them into the pGEX-4T/C3 vector with an added
histidine tag.
86
CA 02362004 2001-11-13
s' AAG AGA AGG CGA AGA AGA CCT AAG AAG AGA CGA AGG GCG AAG
AGG AGA 3' (SEQ ID NO.: 22)
s' TTC TCT TCC GCT TCT TCT GGA TTC TTC TCT GCT TCC CGC TTC
TCC TCT 3' (SEQ ID NO.: 23)
DNA sequence of C3Basicl (SEQ ID NO: 24)
1o s' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
is AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
2o TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AAG AGA AGG CGA AGA AGA CCT AAG
2s AAG AGA CGA AGG GCG AAG AGG AGA CAC CAC CAC CAC CAC CAC GTC
GAC TCG AGC GGC CGC ATC GTG ACT GAC TGA 3'
Protein sequence of C3Basicl (SEQ ID NO: 25)
3o GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASE1NGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYL
GTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITKFK
87
CA 02362004 2001-11-13
VAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPK
EFI~WRRRRPKKRRRAKRRHHHHHHVDSSGRIVTD.
Molecular Weight 29897.03 Daltons
263 Amino Acids
44 Strongly Basic(+) Amino Acids (K,R)
23 Strongly Acidic(-) Amino Acids (D,E)
75 Hydrophobic Amino Acids (A,I,L,F,W,V)
l0 79 Polar Amino Acids (N,C,Q,S,T,Y)
10.024 Isolectric Point
22.209 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 78.56
EXAMPLE 13. ADDITIONAL CHIMERIC C3 PROTEIN THAT WOULD BE EFFECTIVE TO
STIMULATE REPAIR IN THE CNS.
We have designed the following DNA encoding a chimeric C3 with membrane
transport
2o properties. The protein is designated C3Basic2. This sequence was designed
with C3 fused
to a random basic sequence. The construct was made to encode the peptide given
below.
K R R R R K K R R Q R R R (SEQ ID NO.: 26)
The construct was made by synthesizing the two oligonucleotides given below,
annealing
them together, and ligating them into the pGEX4T/C3 vector with an added
histidine tag.
5' AAG CGT CGA CGT AGA AAG AAA CGT AGA CAG CGT AGA CGT 3' (SEQ
ID NO.: 27)
5' TTC GCA GCT GCA TCT TTC TTT GCA TCT GTC GCA TCT GCA 3'
(SEQ ID NO.: 28)
88
CA 02362004 2001-11-13
DNA sequence of C3Basic2 (SEQ ID NO.: 29)
5' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
to AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
2o GCT ATC AAT CCT AAA GAA TTC AAG CGT CGA CGT AGA AAG AAA CGT
AGA CAG CGT AGA CGT CAC CAC CAC CAC CAC CAC GTC GAC TCG AGC
GGC CGC ATC GTG ACT GAC TGA 3'
Protein sequence of C3Basic2 (SEQ ID NO.: 30)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYL
GTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITKFK
3o VAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPK
EFKRRRRKKRRQRRRHHHHHHVDSSGRIVTD.
Molecular Weight 29572.61 Daltons
89
CA 02362004 2001-11-13
260 Amino Acids
42 Strongly Basic(+) Amino Acids (K,R)
23 Strongly Acidic(-) Amino Acids (D,E)
74 Hydrophobic Amino Acids (A,I,L,F,W,V)
80 Polar Amino Acids (N,C,Q,S,T,Y)
9.956 Isolectric Point
20.210 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 78.45
EXAMPLE 14. ADDITIONAL CHIMERIC C3 PROTEIN THAT WOULD BE EFFECTIVE TO
STIMULATE REPAIR IN THE CNS.
We have designed the following DNA encoding a chimeric C3 with membrane
transport
properties. The protein is designated C3Basic3. This sequence was designed
with C3 fused
to a reverse Tat sequence. The construct was made to encode the peptide given
below
2o R R K Q R R K R R (SEQ ID NO.: 31 )
The construct was made by synthesizing the two oligonucleotides given below,
annealing
them together, and ligating them into the pGEX4T/C3 vector with an added
histidine tag,
then subcloning root pGEX-4T/C3.
5' AGA AGG AAA CAA AGA AGA AAA AGA AGA 3' (SEQ ID NO.: 32)
5' TCT TCC TTT GTT TCT TCT TTT TCT TCT 3' (SEQ ID NO.: 33)
CA 02362004 2001-11-13
DNA sequence of C3Basic3 (SEQ ID NO.: 34)
S' GGA TCC TCT AGA GTC GAC CTG CAG GCA TGC AAT GCT TAT TCC ATT AAT
CAA AAG GCT TAT TCA AAT ACT TAC CAG GAG TTT ACT AAT ATT GAT CAA
GCA AAA GCT TGG GGT AAT GCT CAG TAT AAA AAG TAT GGA CTA AGC AAA
TCA GAA AAA GAA GCT ATA GTA TCA TAT ACT AAA AGC GCT AGT GAA ATA
AAT GGA AAG CTA AGA CAA AAT AAG GGA GTT ATC AAT GGA TTT CCT TCA
AAT TTA ATA AAA CAA GTT GAA CTT TTA GAT AAA TCT TTT AAT AAA ATG
AAG ACC CCT GAA AAT ATT ATG TTA TTT AGA GGC GAC GAC CCT GCT TAT
to TTA GGA ACA GAA TTT CAA AAC ACT CTT CTT AAT TCA AAT GGT ACA ATT
AAT AAA ACG GCT TTT GAA AAG GCT AAA GCT AAG TTT TTA AAT AAA GAT
AGA CTT GAA TAT GGA TAT ATT AGT ACT TCA TTA ATG AAT GTT TCT CAA
TTT GCA GGA AGA CCA ATT ATT ACA AAA TTT AAA GTA GCA AAA GGC TCA
AAG GCA GGA TAT ATT GAC CCT ATT AGT GCT TTT CAG GGA CAA CTT GAA
ATG TTG CTT CCT AGA CAT AGT ACT TAT CAT ATA GAC GAT ATG AGA TTG
TCT TCT GAT GGT AAA CAA ATA ATA ATT ACA GCA ACA ATG ATG GGC ACA
GCT ATC AAT CCT AAA GAA TTC AGA AGG AAA CAA AGA AGA AAA AGA
AGA CAC CAC CAC CAC CAC CAC GTC GAC TCG AGC GGC CGC ATC GTG ACT
GAC TGA 3'
2o
Protein sequence of C3Basic3 (SEQ ID NO.: 35)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYL
GTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITKFK
VAKGSKAGYIDPISAFQGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPK
EFRRKQRRKRRHHHHHHVDSSGRIVTD.
3o Molecular Weight 29441.47 Daltons
260 Amino Acids
39 Strongly Basic(+) Amino Acids (K,R)
23 Strongly Acidic(-) Amino Acids (D,E)
91
CA 02362004 2001-11-13
76 Hydrophobic Amino Acids (A,I,L,F,W,V)
80 Polar Amino Acids (N,C,Q,S,T,Y)
9.833 Isolectric Point
17.211 Charge at PH 7.0
Davis,Botstein,Roth Melting Temp C. 78.29
EXAMPLE 15. SEQUENCES FOR C3APLT
One of the clones that was selected from the subcloning of C3APL into pGEX
encoded a
protein that was not the expected size but had good biological activity. This
clone that had a
frameshift mutation leading to a tuncation, and this clone was called C3APLT.
The clone
was resquenced and the chromotograms analyzed to confirm the sequence. To
confirm the
sequences of C3APLT, the coding sequence from both strands of pGEX-4T/C3APLT
were
sequenced by double strand sequencing of the full length of the clone (BioS&T,
Montreal,
Quebec).
The DNA sequence for C3APLT is as follows: (SEQ ID NO.: 36)
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTTTCTCAA 480
TTTGCAGGAA GACCAATTAT TACAAAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TGCAGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCGTGATGA ATCCCGCAAA CGCGCAAGGC 720
AGACATACAC CCGGTACCAG ACTCTAGAGC TAGAGAAGGA GTTTCACTTC AATCGCTACT 780
TGACCCGTCG GCGAAGGATC GAGATCGCCC ACGCCCTGTG CCTCACGGAG CGCCAGATAA 840
AGATTTGGTT CCAGAATCGG CGCATGAAGT GGAAGAAGGA GAACTGA 887
92
CA 02362004 2001-11-13
The APLT transport peptide sequence by itself is as follows (SEQ ID NO.: 48):
VMNPANAQGRHTPGTRL
The protein sequence for C3APLT is as follows: (SEQ ID NO.: 37)
GSSRVDLQACNAYSINQKAYSNTYQEFTNIDQAKAWGNAQYKKYGLSKSEKEAIVS
YTKSASEINGKLRQNKGVINGFPSNLIKQVELLDKSFNKMKTPENIMLFRGDDPAYL
GTEFQNTLLNSNGTINKTAFEKAKAKFLNKDRLEYGYISTSLMNVSQFAGRPIITKFK
VAKGSKAGYIDPISAFAGQLEMLLPRHSTYHIDDMRLSSDGKQIIITATMMGTAINPK
to EFVMNPANAQGRHTPGTRL
Molecular Weight 27574.42 Daltons
248 Amino Acids
33 Strongly Basic(+) Amino Acids (K,R)
21 Strongly Acidic(-) Amino Acids (D,E)
76 Hydrophobic Amino Acids (A,I,L,F,W,V)
80 Polar Amino Acids (N,C,Q,S,T,Y)
9.636 Isolectric Point
12.379 Charge at PH 7.0
EXAMPLE 16. SUBCLONING AND SEQUENCES FOR C3APLT IN PET
C3 has been reported to be stably expressed in E. coli by both pGEX- series
and pET-series
vectors (e.g., Dillon and Feig, 1995 Meth. Enzymol. 256: 174-184. Small
GTPases and Their
Regulators. Part B. Rho Family. W.E. Balch, C.J. Der, and A. Hall, eds.
Lehmann et al., 1999 supra; Han et al., 2001. J. Mol. Biol. 395: 95-107). The
fusion
proteins were expressed well in the pGEX vector, for synthesis and testing.
However, for
large-scale production it is more efficient to synthesize recombinant proteins
without an
affinity tag that increases the size of the protein produced. Also, it is more
economical to
93
CA 02362004 2001-11-13
sythesize proteins in large scale by affinity chromatography using automated
FPLC systems.
The polymerise chain reaction was used to transfer recombinant construct
C3APLT into the
pET T7 polymerise based system E. coli expression system (reviewed by Studier
et al., 1990.
Meth. Enzyrnol. 185: 60-89. Gene Expression Technology. D.V. Goeddel, ed.). A
similar
PCR approach is suitable for others in the fusion protein series of C3-based
constructs with
transport sequences. The pET3a vector DNA was obtained from Dr. Jerry
Pelletier, McGill
University. PCR primers were obtained from Invitrogen. The upper (5') primer
was 5'-GGA
TCT GGT TCC GCG TCA TAT GTC TAG AGT CGA CCT G-3' (37 b) (SEQ ID N0.:38).
Underlined is the Nde I site that was introduced into the primer to replace
the BamHI site in
1o pGEX4T-C3APLT. The lower primer was 5'-CGC GGA TCC ATT AGT TCT CCT TCT
TCC ACT TC-3' (32 b) (SEQ ID N0.:39). This primer introduced two changes in
the coding
strand DNA of pGEX4T-C3APLT, replacing the EcoRI site from pGEX4T- C3APLT with
a
BamH I site (underlined) and replacing a TGA stop codon with the strong stop
sequence
TART (the italicized ATTA sequence in the complementary primer). Compared to
pGEX4T-
C3APLT, the predicted N-terminal sequence of pET3a- C3APLT is Met-Ser rather
than Gly-
Ser-Ser, a loss of one serine and a substitution of Met for Gly. There were no
changes in
amino acid sequence at the C-terminus of C3APLT.
The target C3APLT gene was amplified using Pfu polymerise (Invitrogen/Canadian
Life
2o Technologies) with buffer, DNA and deoxyribonucleotide concentrations
recommended by
the manufacturer. The PCR was carned out as follows : 95 °C for 5
minutes, 10 cycles of 94
°C for 2 minutes followed by 56 °C for 2 minutes then extension
at 70 °C for 2 minutes, then
30 cycles of 94 °C for 2 minutes followed by 70 °C. Completed
reactions were stored at 4 °C.
The QIAEXII kit (Qiagen) was used to purify the agarose gel slice containing
DNA band.
The purified PCR product DNA and the vector were digested with BamH I and Nde
I (both
obtained from New England BioLabs) following the instructions of the
manufacturer. The
digestion products were separated from extraneous DNA by agarose gel
electrophoresis and
purified with the QIAEXII kit. . The insert and vector DNA were incubated
together
overnight at 16 °C with T4 DNA ligase according to directions provided
by the manufacturer
3o (New England BioLabs). Competent E. coli (DHSa, obtained from
Invitrogen/Canadian Life
Technologies) were transformed with the ligation mixture.
DNA was prepared from purified colonies using the Qiagen plasmid midi kit, and
the entire
94
CA 02362004 2001-11-13
insert and junction sequences were verified by double strand sequencing of the
full length of
the clone (BioS&T, Montreal, Quebec) with forward primer 5' AAA TTA ATA CGA
CTC
ACT ATA GGG 3' (24 bases) (SEQ ID NO.: 40)and reverse T7 terminator sequencing
primer 5' GCT AGT TAT TGC TCA GCG G 3' (19 bases) (SEQ ID NO.: 41). The
sequence
of the C3APLT cDNA in pET is given in SEQ ID NO.: 42. The amino acid sequence
is
given in SEQ. ID NO.: 43.
EXAMPLE 17: MODIFICATIONS OF SEQUENCES.
to Any of sequences given in Examples 1, 2, 8, 9, 10, 11, 12 and 13, 15 and 16
could be
modified to retain C3 enzymatic activity and effective transport sequences.
For example
amino acids encoded from DNA at the 3' end of the sequence that represents the
translation of
the restriction sites used in cloning may be removed without affecting
activity. Some of the
amino terminal amino acids may also be removed without affecting activity. The
minimal
amount of sequence needed for biological activity of the C3 portion of the
fusion protein is
not known but could be easily determined by known techniques. For example,
increasingly
more of the 5' end of the cDNA encoding C3 could be removed, and the resulting
proteins
made and tested for biological activity. Similarly, increasing amounts of the
3' end could be
removed and the fragments tested for biological activity. Next, fragments
testing the central
2o region could be tested for retention of C3 activity. Therefore, the C3
portion of the protein
could be truncated to include just the amino acids needed for activity.
Alternatively
mutations could be made in the coding regions of C3, and the resulting
proteins tested for
activity. The transport sequences could be modified to add or remove one or
more amino
acids or to completely change the transport peptide, but retain the transport
characteristics in
terms of effective dose compared to C3 in our tissue culture bioassay (Example
4). New
transport sequences could be tested for biological activity to improve the
efficiency of C3
activity by plating neurons and testing them on inhibitory substrates, as
described in Example
4.
CA 02362004 2001-11-13
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i)APPLICANT:
LISA
MCKERRACHER
(ii) TITLE
OF INVENTION:
FUSION
PROTEINS
(iii) NUMBER OF SEQUENCES: 48
(iv) CORRESPONDENCE
ADDRESS:
(A) ADRESSEE: BROULLETTE KOSIE
(B) STREET: 1100 RENE-LEVESQUE BLVD
WEST
(C) PROV/STATE: QUEBEC
(D) COUNTRY: CANADA
(E) POSTAL/ZIP CODE: H3B 5C9
(v) COMPUTER
READABLE
FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: ASCII (TEXT)
(vi) CURRENT
APPLICATION
DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE:
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA
(A) APPLICATION NUMBER: 2,342,970
(B) FILING DATE: 2001-04-12
(C) CLASSIFICATION:
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: BROULLETTE KOSIE
(B) REGISTRATION NO.: 3939
(C) REFERENCE/DOCKET NO.: 06447-004-CA-02
(D) TEL. NO.: (514) 397 8500
(E) FAX NO.: (514) 397 8515
(2) INFORMATION
FOR SEQ
ID N0:
1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
96
CA 02362004 2001-11-13
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
(G) DATE:
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID N0:1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 1:
GAATTCTTTA GGATTGATAG CTGTGCC 27
(2) INFORMATION FOR SEQ ID N0: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 2:
GGTGGCGACC ATCCTCCAAA A 21
97
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 888
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTCTCTCAA 480
TTTGCAGGAA GACCAATTAT TACACAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCGTGATGG AATCCCGCAA ACGCGCAAGG 720
CAGACATACA CCCGGTACCA GACTCTAGAG CTAGAGAAGG AGTTTCACTT CAATCGCTAC 780
TTGACCCGTC GGCGAAGGAT CGAGATCGCC CACGCCCTGT GCCTCACGGA GCGCCAGATA 840
AAGATTTGGT TCCAGAATCG GCGCATGAAG TGGAAGAAGG AGAACTGA 888
(2) INFORMATION FOR SEQ ID N0: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 295
(B) TYPE: AMINO ACIDS
(C) STRANDEDNESS SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
98
CA 02362004 2001-11-13
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 4:
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Val Met Glu Ser Arg Lys Arg Ala Arg
230 235 240
Gln Thr Tyr Thr Arg Tyr Gln Thr Leu Glu Leu Glu Lys Glu Phe
245 250 255
His Phe Asn Arg Tyr Leu Thr Arg Arg Arg Arg Ile Glu Ile Ala
260 265 270
His Ala Leu Cys Leu Thr Glu Arg Gln Ile Lys Ile Trp Phe Gln
275 280 285
99
CA 02362004 2001-11-13
Asn Arg Arg Met Lys Trp Lys Lys Glu Asn
290 295
(2) INFORMATION FOR SEQ ID N0: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 774
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 5:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTCTCTCAA 480
TTTGCAGGAA GACCAATTAT TACACAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCCGCCAGA TCAAGATTTG GTTCCAGAAT 720
CGTCGCATGA AGTGGAAGAA GGTCGACTCG AGCGGCCGCA TCGTGACTGA CTGA 774
(2) INFORMATION FOR SEQ ID N0: 6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 257
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
100
CA 02362004 2001-11-13
(vii) IMMEDIATE SOURCE:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 6
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
S 1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Arg Gln Ile Lys Ile Trp Phe Gln Asn
230 235 240
Arg Arg Met Lys Trp Lys Lys Val Asp Ser Ser Gly Arg Ile Val
245 250 255
Thr Asp
101
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID N0: 7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 7:
GAATCCCGCA AACGCGCAAG GCAG 24
(2) INFORMATION FOR SEQ ID N0: 8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 8:
TCAGTTCTCC TTCTTCCACT TCATGCG 27
(2) INFORMATION FOR SEQ ID N0: 9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 9:
AATTCCGCCA GATCAAGATT TGGTTCCAGA ATCGTCGCAT GAAGTGGAAG AAGG 54
1~2
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID N0: 10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 10:
GGCGGTCTAG TTCTAAACCA AGCTCTTAGC AGCGTAGTTC ACCTTCTTCC AGCT 54
(2) INFORMATION FOR SEQ ID N0: 11:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 11:
GAATCCAAGC ATCCAGGAAG TCAGCC 26
(2) INFORMATION FOR SEQ ID N0: 12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
103
CA 02362004 2001-11-13
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 12:
ACCAGCCACC ACCTTCTGAT A
(2) INFORMATION FOR SEQ ID N0: 13:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 876
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 13:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTCTCTCAA 480
TTTGCAGGAA GACCAATTAT TACACAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCAAGCATC CAGGAAGTCA GCCTAAAACT 720
GCTTGTACCA ATTGCTATTG TAAAAAGTGT TGCTTTCATT GCCAAGTTTG TTTCATAACA 780
AAAGCCTTAG GCATCTCCTA TGGCAGGAAG CGGAGACAGC GACGAAGAGC TCATCAGAAC 840
AGTCAGACTC ATCAAGCTTC TCTATCAAAG CAGTAA 876
(2) INFORMATION FOR SEQ ID N0: 14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 291
104
CA 02362004 2001-11-13
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 14:
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
15 Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
20 80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Lys His Pro Gly Ser Gln Pro Lys Thr
230 235 240
105
CA 02362004 2001-11-13
Ala Cys Thr Asn Cys Tyr Cys Lys Lys Cys Cys Phe His Cys Gln
245 250 255
Val Cys Phe Ile Thr Lys Ala Leu Gly Ile Ser Tyr Gly Arg Lys
260 265 270
Arg Arg Gln Arg Arg Arg Ala His Gln Asn Ser Gln Thr His Gln
275 280 285
Ala Ser Leu Ser Lys Gln
290
(2) INFORMATION FOR SEQ ID NO: 15:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 15:
AATTCTATGG TCGTAAAAAA CGTCGTCAAC GTCGTCGTG 39
(2) INFORMATION FOR SEQ ID N0: 16:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 16:
GATACCAGCA TTTTTTGCAG CAGTTGCAGC AGCACAGCT 39
106
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID N0: 17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 756
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 17:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTCTCTCAA 980
TTTGCAGGAA GACCAATTAT TACACAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCTATGGTG CTAAAAAACG TCGTCAACGT 720
CGTCGTGTCG ACTCGAGCGG CCCGCATCGT GACTGA 756
(2) INFORMATION FOR SEQ ID N0: 18:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 251
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 18:
107
CA 02362004 2001-11-13
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu SerLysSer GluLysGlu AlaIleVal SerTyrThr LysSer
50 55 60
Ala SerGluIle AsnGlyLys LeuArgGln AsnLysGly ValIle
65 70 75
Asn GlyPhePro SerAsnLev IleLysGln ValGluLeu LeuAsp
80 85 90
Lys SerPheAsn LysMetLys ThrProGlu AsnIleMet LeuPhe
95 100 105
Arg GlyAspAsp ProAlaTyr LeuGlyThr GluPheGln AsnThr
110 115 120
Leu LeuAsnSer AsnGlyThr IleAsnLys ThrAlaPhe GluLys
125 130 135
Ala LysAlaLys PheLeuAsn LysAspArg LeuGluTyr GlyTyr
140 145 150
Ile SerThrSer LeuMetAsn ValSerGln PheAlaGly ArgPro
155 160 165
Ile IleThrGln PheLysVal AlaLysGly SerLysAla GlyTyr
170 175 180
Ile AspProIle SerAlaPhe GlnGlyGln LeuGluMet LeuLeu
185 190 195
Pro ArgHisSer ThrTyrHis IleAspAsp MetArgLeu SerSer
200 205 210
Asp GlyLysGln IleIleIle ThrAlaThr MetMetGly ThrAla
215 220 225
Ile AsnProLys GluPheTyr GlyAlaLys LysArgArg GlnArg
230 235 240
Arg ArgValAsp SerSerGly ProHisArg Asp
245 250
(2) INFORMATION FOR SEQ ID N0: 19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1413
(B) TYPE: NUCLEIC ACID
108
CA 02362004 2001-11-13
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 19:
ATGTCCCCTA TACTAGGTTATTGGAAAATTAAGGGCCTTGTGCAACCCACTCGACTTCTT60
TTGGAATATC TTGAAGAAAAATATGAAGAGCATTTGTATGAGCGCGATGAAGGTGATAAA120
TGGCGAAACA AAAAGTTTGAATTGGGTTTGGAGTTTCCCAATCTTCCTTATTATATTGAT180
GGTGATGTTA AATTAACACAGTCTATGGCCATCATACGTTATATAGCTGACAAGCACAAC240
ATGTTGGGTG GTTGTCCAAAAGAGCGTGCAGAGATTTCAATGCTTGAAGGAGCGGTTTTG300
GATATTAGAT ACGGTGTTTCGAGAATTGCATATAGTAAAGACTTTGAAACTCTCAAAGTT360
GATTTTCTTA GCAAGCTACCTGAAATGCTGAAAATGTTCGAAGATCGTTTATGTCATAAA420
ACATATTTAA ATGGTGATCATGTAACCCATCCTGACTTCATGTTGTATGACGCTCTTGAT480
GTTGTTTTAT ACATGGACCCAATGTGCCTGGATGCGTTCCCAAAATTAGTTTGTTTTAAA540
AAACGTATTG AAGCTATCCCACAAATTGATAAGTACTTGAAATCCAGCAAGTATATAGCA600
TGGCCTTTGC AGGGCTGGCAAGCCACGTTTGGTGGTGGCGACCATCCTCCAAAATCGGAT660
CTGGTTCCGC GTGGATCCTCTAGAGTCGACCTGCAGGCATGCAATGCTTATTCCATTAAT720
CAAAAGGCTT ATTCAAATACTTACCAGGAGTTTACTAATATTGATCAAGCAAAAGCTTGG780
GGTAATGCTC AGTATAAAAAGTATGGACTAAGCAAATCAGAAAAAGAAGCTATAGTATCA840
TATACTAAAA GCGCTAGTGAAATAAATGGAAAGCTAAGACAAAATAAGGGAGTTATCAAT900
GGATTTCCTT CAAATTTAATAAAACAAGTTGAACTTTTAGATAAATCTTTTAATAAAATG960
AAGACCCCTG AAAATATTATGTTATTTAGAGGCGACGACCCTGCTTATTTAGGAACAGAA1020
TTTCAAAACA CTCTTCTTAATTCAAATGGTACAATTAATAAAACGGCTTTTGAAAAGGCT1080
AAAGCTAAGT TTTTAAATAAAGATAGACTTGAATATGGATATATTAGTACTTCATTAATG1140
AATGTTTCTC AATTTGCAGGAAGACCAATTATTACAAAATTTAAAGTAGCAAAAGGCTCA1200
AAGGCAGGAT ATATTGACCCTATTAGTGCTTTTCAGGGACAACTTGAAATGTTGCTTCCT1260
AGACATAGTA CTTATCATATAGACGATATGAGATTGTCTTCTGATGGTAAACAAATAATA1320
ATTACAGCAA CAATGATGGGCACAGCTATCAATCCTAAAGAATTCAGAAGGAAACAAAGA1380
AGAAAAAGAA GACTGCAGGCGGCCGCATCGTGA 1413
(2) INFORMATION FOR SEQ ID N0: 20:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 470
(B) TYPE: AMINO ACIDS
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
109
CA 02362004 2001-11-13
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 20:
Met Ser Pro Ile Leu Gly Tyr Trp Lys Ile Lys Gly Leu Val Gln
1 5 10 15
Pro Thr Arg Leu Leu Leu Glu Tyr Leu Glu Glu Lys Tyr Glu Glu
20 25 30
His Leu Tyr Glu Arg Asp Glu Gly Asp Lys Trp Arg Asn Lys Lys
35 40 45
Phe Glu Leu Gly Leu Glu Phe Pro Asn Leu Pro Tyr Tyr Ile Asp
50 55 60
Gly Asp Val Lys Leu Thr Gln Ser Met Ala Ile Ile Arg Tyr Ile
65 70 75
Ala Asp LysHisAsn MetLeuGly GlyCysPro LysGluArg Ala
80 85 90
Glu Ile SerMetLeu GluGlyAla ValLeuAsp IleArgTyr Gly
95 100 105
Val Ser ArgIleAla TyrSerLys AspPheGlu ThrLeuLys Val
110 115 120
Asp Phe LeuSerLys LeuProGlu MetLeuLys MetPheGlu Asp
125 130 135
Arg Leu CysHisLys ThrTyrLeu AsnGlyAsp HisValThr His
140 145 150
Pro Asp PheMetLeu TyrAspAla LeuAspVal ValLeuTyr Met
155 160 165
Asp Pro MetCysLeu AspAlaPhe ProLysLeu ValCysPhe Lys
170 175 180
Lys Arg IleGluAla IleProGln IleAspLys TyrLeuLys Ser
185 190 195
Ser Lys TyrIleAla TrpProLeu GlnGlyTrp GlnAlaThr Phe
200 205 210
Gly Gly GlyAspHis ProProLys SerAspLeu ValProArg Gly
215 220 225
Ser Ser ArgValAsp LeuGlnAla CysAsnAla TyrSerIle Asn
230 235 240
Gln Lys AlaTyrSer AsnThrTyr GlnGluPhe ThrAsnIle Asp
245 250 255
Gln Ala LysAlaTrp GlyAsnAla GlnTyrLys LysTyrGly Leu
260 265 270
110
CA 02362004 2001-11-13
Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser Ala
275 280 285
Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile Asn
290 295 300
Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp Lys
305 310 315
Ser PheAsnLysMet LysThrPro GluAsnIle MetLeuPhe Arg
320 325 330
Gly AspAspProAla TyrLeuGly ThrGluPhe GlnAsnThr Leu
335 340 345
Leu AsnSerAsnGly ThrIleAsn LysThrAla PheGluLys Ala
350 355 360
Lys AlaLysPheLeu AsnLysAsp ArgLeuGlu TyrGlyTyr Ile
365 370 375
Ser ThrSerLeuMet AsnValSer GlnPheAla GlyArgPro Ile
380 385 390
Ile ThrLysPheLys ValAlaLys GlySerLys AlaGlyTyr Ile
395 400 405
Asp ProIleSerAla PheGlnGly GlnLeuGlu MetLeuLeu Pro
410 915 420
Arg HisSerThrTyr HisIleAsp AspMetArg LeuSerSer Asp
425 430 435
Gly LysGlnIleIle IleThrAla ThrMetMet GlyThrAla Ile
440 445 950
Asn ProLysGluPhe ArgArgLys GlnArgArg LysArgArg Leu
455 460 465
Gln Ala Ala Ala Ser
470
(2) INFORMATION FOR SEQ ID N0: 21
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:16
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PEPTIDE
111
CA 02362004 2001-11-13
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 21:
Lys Arg Arg Arg Arg Arg Pro Lys Lys Arg Arg Arg
1 5 10
Ala Lys Arg Arg
10
(2) INFORMATION FOR SEQ ID NO: 22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:48
15 (B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 22:
AAGAGAAGGC GAAGAAGACC TAAGAAGAGA CGAAGGGCGA AGAGGAGA 48
(2) INFORMATION FOR SEQ ID N0: 23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 48
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 23:
TTCTCTTCCG CTTCTTCTGG ATTCTTCTCT GCTTCCCGCT TCTCCTCT 48
112
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID N0: 24:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 792
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 24:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTTTCTCAA 480
TTTGCAGGAA GACCAATTAT TACAAAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCAAGAGAA GGCGAAGAAG ACCTAAGAAG 720
AGACGAAGGG CGAAGAGGAG ACACCACCAC CACCACCACG TCGACTCGAG CGGCCGCATC 780
GTGACTGACT GA 792
(2) INFORMATION FOR SEQ ID N0: 25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 263
. (B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 25:
113
CA 02362004 2001-11-13
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn GlyPheProSer AsnLeu IleLysGlnVal GluLeuLeu Asp
80 85 90
Lys SerPheAsnLys MetLys ThrProGluAsn IleMetLeu Phe
95 100 105
Arg GlyAspAspPro AlaTyr LeuGlyThrGlu PheGlnAsn Thr
110 115 120
Leu LeuAsnSerAsn GlyThr IleAsnLysThr AlaPheGlu Lys
125 130 135
Ala LysAlaLysPhe LeuAsn LysAspArgLeu GluTyrGly Tyr
140 145 150
Ile SerThrSerLeu MetAsn ValSerGlnPhe AlaGlyArg Pro
155 160 165
Ile IleThrLysPhe LysVal AlaLysGlySer LysAlaGly Tyr
170 175 180
Ile AspProIleSer AlaPhe GlnGlyGlnLeu GluMetLeu Leu
185 190 195
Pro ArgHisSerThr TyrHis IleAspAspMet ArgLeuSer Ser
200 205 210
Asp GlyLysGlnIle IleIle ThrAlaThrMet MetGlyThr Ala
215 220 225
Ile AsnProLysGlu PheLys ArgArgArgArg ArgProLys Lys
230 235 290
Arg ArgArgAlaLys ArgArg HisHisHisHis HisHisVal Asp
245 250 255
Ser SerGlyArgIle ValThr Asp
260
(2) INFORMATION FOR SEQ ID N0: 26:
(i) SEQUENCE CHARACTERISTICS:
114
CA 02362004 2001-11-13
(A) LENGTH: 13
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PEPTIDE
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 26:
Lys Arg Arg Arg Arg Lys Lys Arg Arg Gln Arg Arg Arg
1 5 10
(2) INFORMATION FOR SEQ ID N0: 27:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 27:
AAGCGTCGAC GTAGAAAGAA ACGTAGACAG CGTAGACGT 39
(2) INFORMATION FOR SEQ ID NO: 28:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 28:
115
CA 02362004 2001-11-13
TTCGCAGCTG CATCTTTCTT TGCATCTGTC GCATCTGCA 39
(2) INFORMATION FOR SEQ ID 29:
N0:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 783
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBL E
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID 29:
N0:
GGATCCTCTA GAGTCGACCT GCAGGCATGCAATGCTTATTCCATTAATCAAAAGGCTTAT60
TCAAATACTT ACCAGGAGTT TACTAATATTGATCAAGCAAAAGCTTGGGGTAATGCTCAG120
TATAAAAAGT ATGGACTAAG CAAATCAGAAAAAGAAGCTATAGTATCATATACTAAAAGC180
GCTAGTGAAA TAAATGGAAA GCTAAGACAAAATAAGGGAGTTATCAATGGATTTCCTTCA290
AATTTAATAA AACAAGTTGA ACTTTTAGATAAATCTTTTAATAAAATGAAGACCCCTGAA300
AATATTATGT TATTTAGAGG CGACGACCCTGCTTATTTAGGAACAGAATTTCAAAACACT360
CTTCTTAATT CAAATGGTAC AATTAATAAAACGGCTTTTGAAAAGGCTAAAGCTAAGTTT420
TTAAATAAAG ATAGACTTGA ATATGGATATATTAGTACTTCATTAATGAATGTTTCTCAA480
TTTGCAGGAA GACCAATTAT TACAAAATTTAAAGTAGCAAAAGGCTCAAAGGCAGGATAT540
ATTGACCCTA TTAGTGCTTT TCAGGGACAACTTGAAATGTTGCTTCCTAGACATAGTACT600
TATCATATAG ACGATATGAG ATTGTCTTCTGATGGTAAACAAATAATAATTACAGCAACA660
ATGATGGGCA CAGCTATCAA TCCTAAAGAATTCAAGCGTCGACGTAGAAAGAAACGTAGA720
CAGCGTAGAC GTCACCACCA CCACCACCACGTCGACTCGAGCGGCCGCATCGTGACTGAC780
TGA
(2) INFORMATION FOR SEQ ID N0: 30:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 260
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
116
CA 02362004 2001-11-13
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 30:
Gly SerSerArg ValAspLeu GlnAlaCys AsnAlaTyrSer Ile
1 5 10 15
Asn GlnLysAla TyrSerAsn ThrTyrGln GluPheThrAsn Ile
20 25 30
Asp GlnAlaLys AlaTrpGly AsnAlaGln TyrLysLysTyr Gly
35 90 45
Leu SerLysSer GluLysGlu AlaIleVal SerTyrThrLys Ser
50 55 60
Ala SerGluIle AsnGlyLys LeuArgGln AsnLysGlyVal Ile
65 70 75
Asn GlyPhePro SerAsnLeu IleLysGln ValGluLeuLeu Asp
80 85 90
Lys SerPheAsn LysMetLys ThrProGlu AsnIleMetLeu Phe
95 100 105
Arg GlyAspAsp ProAlaTyr LeuGlyThr GluPheGlnAsn Thr
110 115 120
Leu LeuAsnSer AsnGlyThr IleAsnLys ThrAlaPheGlu Lys
125 130 135
Ala LysAlaLys PheLeuAsn LysAspArg LeuGluTyrGly Tyr
140 145 150
Ile SerThrSer LeuMetAsn ValSerGln PheAlaGlyArg Pro
155 160 165
Ile IleThrLys PheLysVal AlaLysGly SerLysAlaGly Tyr
170 175 180
Ile AspProIle SerAlaPhe GlnGlyGln LeuGluMetLeu Leu
185 190 195
Pro ArgHisSer ThrTyrHis IleAspAsp MetArgLeuSer Ser
200 205 210
Asp GlyLysGln IleIleIle ThrAlaThr MetMetGlyThr Ala
215 220 225
Ile AsnProLys GluPheLys ArgArgArg ArgLysLysArg Arg
230 235 240
Gln ArgArgArg HisHisHis HisHisHis ValAspSerSer Gly
245 250 255
Arg IleValThr Asp
260
117
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID N0: 31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 9
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PEPTIDE
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 31:
Arg Arg Lys Gln Arg Arg Lys Arg Arg
1 5
(2) INFORMATION FOR SEQ ID N0: 32:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 32:
AGAAGGAAAC AAAGAAGAAA AAGAAGA 27
(2) INFORMATION FOR SEQ ID N0: 33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
118
CA 02362004 2001-11-13
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 33:
TCTTCCTTTG TTTCTTCTTT TTCTTCT 27
(2) INFORMATION FOR SEQ ID N0: 34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 771
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 34:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTTTCTCAA 480
TTTGCAGGAA GACCAATTAT TACAAAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCAGAAGGA AACAAAGAAG AAAAAGAAGA 720
CACCACCACC ACCACCACGT CGACTCGAGC GGCCGCATCG TGACTGACTG A 771
(2) INFORMATION FOR SEQ ID N0: 35:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 256
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
119
CA 02362004 2001-11-13
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 35:
Gly Ser SerArgVal AspLeuGln AlaCysAsn AlaTyrSer Ile
1 5 10 15
Asn Gln LysAlaTyr SerAsnThr TyrGlnGlu PheThrAsn Ile
20 25 30
Asp Gln AlaLysAla TrpGlyAsn AlaGlnTyr LysLysTyr Gly
35 40 45
Leu Ser LysSerGlu LysGluAla IleValSer TyrThrLys Ser
50 55 60
Ala Ser GluIleAsn GlyLysLeu ArgGlnAsn LysGlyVal Ile
65 70 75
Asn Gly PheProSer AsnLeuIle LysGlnVal GluLeuLeu Asp
80 85 90
Lys Ser PheAsnLys MetLysThr ProGluAsn IleMetLeu Phe
95 100 105
Arg Gly AspAspPro AlaTyrLeu GlyThrGlu PheGlnAsn Thr
110 115 120
Leu Leu AsnSerAsn GlyThrIle AsnLysThr AlaPheGlu Lys
125 130 135
Ala Lys AlaLysPhe LeuAsnLys AspArgLeu GluTyrGly Tyr
140 145 150
Ile Ser ThrSerLeu MetAsnVal SerGlnPhe AlaGlyArg Pro
155 160 165
Ile Ile ThrLysPhe LysValAla LysGlySer LysAlaGly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Arg Arg Lys Gln Arg Arg Lys Arg Arg
230 235 240
His His His His His His Val Asp Ser Ser Gly Arg Ile Val Thr
245 250 255
Asp
120
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID N0: 36:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 887
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 36:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTTTCTCAA 480
TTTGCAGGAA GACCAATTAT TACAAAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TGCAGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCGTGATGA ATCCCGCAAA CGCGCAAGGC 720
AGACATACAC CCGGTACCAG ACTCTAGAGC TAGAGAAGGA GTTTCACTTC AATCGCTACT 780
TGACCCGTCG GCGAAGGATC GAGATCGCCC ACGCCCTGTG CCTCACGGAG CGCCAGATAA 840
AGATTTGGTT CCAGAATCGG CGCATGAAGT GGAAGAAGGA GAACTGA 887
(2) INFORMATION FOR SEQ ID NO: 37:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 248
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
121
CA 02362004 2001-11-13
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 37:
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Val Met Asn Pro Ala Asn Ala Gln Gly
230 235 240
Arg His Thr Pro Gly Thr Arg Leu
245
122
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 38:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 37
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 38:
GGATCTGGTT CCGCGTCATA TGTCTAGAGT CGACCTG 37
(2) INFORMATION FOR SEQ ID N0: 39:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 39:
CGCGGATCCA TTAGTTCTCC TTCTTCCACT TC 32
(2) INFORMATION FOR SEQ ID N0: 40:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 40:
123
CA 02362004 2001-11-13
AAATTAATAC GACTCACTAT AGGG 24
(2) INFORMATION FOR SEQ ID N0: 41:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 41:
GCTAGTTATT GCTCAGCGG 19
(2) INFORMATION FOR SEQ ID N0: 42:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 888
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 42:
ATGTCTAGAG TCGCACTGCA GGCATGCAAT GCTTATTCCA TTAATCAAAA GGCTTATTCA 60
AATACTTACC AGGAGTTTAC TAATATTGAT CAAGCAAAAG CTTGGGGTAA TGCTCAGTAT 120
AAAAAGTATG GACTAAGCAA ATCAGAAAAA GAAGCTATAG TATCATATAC TAAAAGCGCT 180
AGTGAAATAA ATGGAAAGCT AAGACAAAAT AAGGGAGTTA TCAATGGATT TCCTTCAAAT 240
TTAATAAAAC AAGTTGAACT TTTAGATAAA TCTTTTAATA AAATGAAGAC CCCTGAAAAT 300
ATTATGTTAT TTAGAGGCGA CGACCCTGCT TATTTAGGAA CAGAATTTCA AAACACTCTT 360
CTTAATTCAA ATGGTACAAT TAATAAAACG GCTTTTGAAA AGGCTAAAGC TAAGTTTTTA 420
AATAAAGATA GACTTGAATA TGGATATATT AGTACTTCAT TAATGAATGT TTCTCAATTT 480
124
CA 02362004 2001-11-13
GCAGGAAGAC CAATTATTAC AAAATTTAAA GTAGCAAAAG GCTCAAAGGC AGGATATATT 540
GACCCTATTA GTGCTTTTGC AGGACAACTT GAAATGTTGC TTCCTAGACA TAGTACTTAT 600
CATATAGACG ATATGAGATT GTCTTCTGAT GGTAAACAAA TAATAATTAC AGCAACAATG 660
ATGGGCACAG CTATCAATCC TAAAGAATTC GTGATGAATC CCGCAAACGC GCAAGGCAGA 720
CATACACCCG GTACCAGACT CTAGAGCTAG AGAAGGAGTT TCACTTCAAT CGCTACTTGA 780
CCCGTCGGCG AAGGATCGAG ATCGCCCACG CCCTGTGCCT CACGGAGCGC CAGATAAAGA 840
TTTGGTTCCA GAATCGGCGC ATGAAGTGGA AGAAGGAGGA CTAACTGA 888
(2) INFORMATION FOR SEQ ID N0: 43:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 247
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 43:
Met Ser Arg Val Asp Leu Gln Ala Cys Asn A1a Tyr Ser Ile Asn
1 5 10 15
Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile Asp
20 25 30
Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly Leu
35 40 45
Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser Ala
50 55 60
Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile Asn
65 70 75
Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp Lys
80 85 90
Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe Arg
95 100 105
Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr Leu
110 115 120
Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys Ala
125 130 135
Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr Ile
140 145 150
125
CA 02362004 2001-11-13
Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro Ile
155 160 165
Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr Ile
170 175 180
Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu Pro
185 190 195
Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser Asp
200 205 210
Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala Ile
215 220 225
Asn Pro Lys Glu Phe Val Met Asn Pro Ala Asn Ala Gln Gly Arg
230 235 240
His Thr Pro Gly Thr Arg Leu
245
(2) INFORMATION FOR SEQ ID N0: 44:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 64
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
xi) SEQUENCE DESCRIPTION: SEQ ID N0: 44:
Val Met Glu Ser Arg Lys Arg Ala Arg Gln Thr Tyr Thr Arg Tyr
1 5 10 15
Gln Thr Leu Glu Leu Glu Lys Glu Phe His Phe Asn Arg Tyr Leu
20 25 30
Thr Arg Arg Arg Arg Ile Glu Ile Ala His Ala Leu Cys Leu Thr
40 45
Glu Arg Gln Ile Lys Ile Trp Phe Gln Asn Arg Arg Met Lys Trp
35 50 55 60
Lys Lys Glu Asn
126
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 45:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19
(B) TYPE: AMINO ACID
$ (C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 45:
Arg Gln Ile Lys Ile Trp Phe Gln Asn Arg Arg Met Lys Trp Lys
1 5 10 15
Lys Val Asp Ser
(2) INFORMATION FOR SEQ ID N0: 46:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 60
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 46:
Lys His Pro Gly Ser Gln Pro Lys Thr Ala Cys Thr Asn Cys Tyr
1 5 10 15
Cys Lys Lys Cys Cys Phe His Cys Gln Val Cys Phe Ile Thr Lys
20 25 30
Ala Leu Gly Ile Ser Tyr Gly Arg Lys Arg Arg Gln Arg Arg Arg
40 45
Ala His Gln Asn Ser Gln Thr His Gln Ala Ser Leu Ser Lys Gln
50 55 60
(2) INFORMATION FOR SEQ ID N0: 47:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20
127
CA 02362004 2001-11-13
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 47:
Tyr Gly Ala Lys Lys Arg Arg Gln Arg Arg Arg Val Asp Ser Ser
1 5 10 15
Gly Pro His Arg Asp
(2) INFORMATION FOR SEQ ID NO: 48:
15 (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
20 (ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 48:
Val Met Asn Pro Ala Asn Ala Gln Gly Arg His Thr Pro Gly Thr
1 5 10 15
Arg Leu
128
CA 02362004 2001-11-13
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i)APPLICANT: LISA MCKERRACHER
(ii) TITLE OF INVENTION: FUSION PROTEINS
(iii) NUMBER OF SEQUENCES: 48
(iv) CORRESPONDENCE ADDRESS:
(A) ADRESSEE: BROULLETTE KOSIE
(B) STREET: 1100 RENE-LEVESQUE BLVD WEST
(C) PROV/STATE: QUEBEC
(D) COUNTRY: CANADA
(E) POSTAL/ZIP CODE: H3B 5C9
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: ASCII (TEXT)
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE:
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA
(A) APPLICATION NUMBER: 2,342,970
(B) FILING DATE: 2001-04-12
(C) CLASSIFICATION:
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: BROULLETTE KOSIE
(B) REGISTRATION NO.: 3939
(C) REFERENCE/DOCKET NO.: 06447-004-CA-02
(D) TEL. NO.: (514) 397 8500
(E) FAX NO.: (514) 397 8515
(2) INFORMATION FOR SEQ ID NO: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE:
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
1
CA 02362004 2001-11-13
(vii) IMMEDIATE SOURCE:
(ix) FEATURE:
(A) NAME/KEY:
(B) LOCATION:
(D) OTHER INFORMATION:
(x) PUBLICATION INFORMATION:
(A) AUTHORS:
(B) TITLE:
(C) JOURNAL:
(D) VOLUME:
(E) ISSUE:
(F) PAGES:
( G ) DAT E
(H) DOCUMENT NO.:
(I) FILING DATE:
(J) PUBLICATION DATE:
(K) RELEVANT RESIDUES IN SEQ ID N0:1
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: l:
GAATTCTTTA GGATTGATAG CTGTGCC 2'7
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2:
GGTGGCGACC ATCCTCCAAA A 21
(2) INFORMATION FOR SEQ ID NO: 3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 888
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
2
CA 02362004 2001-11-13
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTCTCTCRA 480
TTTGCAGGAA GACCAATTAT TACACAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 590
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCGTGATGG AATCCCGCAA ACGCGCAAGG 720
CAGACATACA CCCGGTACCA GACTCTAGAG CTAGAGAAGG AGTTTCACTT CAATCGCTAC 780
TTGACCCGTC GGCGAAGGAT CGAGATCGCC CACGCCCTGT GCCTCACGGA GCGCCAGATA 840
AAGATTTGGT TCCAGAATCG GCGCATGAAG TGGAAGAAGG AGAACTGA 888
(2) INFORMATION FOR SEQ ID NO: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 295
(B) TYPE: AMINO ACIDS
(C) STRANDEDNESS SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(v) FRAGMENT TYPE:
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 4:
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
3
CA 02362004 2001-11-13
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Val Met Glu Ser Arg Lys Arg Ala Arg
230 235 240
Gln Thr Tyr Thr Arg Tyr Gln Thr Leu Glu Leu Glu Lys Glu Phe
245 250 255
His Phe Asn Arg Tyr Leu Thr Arg Arg Arg Arg Ile Glu Ile Ala
260 265 270
His Ala Leu Cys Leu Thr Glu Arg Gln Ile Lys Ile Trp Phe Gln
275 280 285
Asn Arg Arg Met Lys Trp Lys Lys Glu Asn
290 295
(2) INFORMATION FOR SEQ ID NO: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 774
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
4
CA 02362004 2001-11-13
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATRGACTTGA ATATGGATAT ATTAGTACTT CATTAATGRA TGTCTCTCAA 480
TTTGCAGGAA GACCAATTAT TACACAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCCGCCAGA TCAAGATTTG GTTCCAGAAT 720
CGTCGCATGA AGTGGAAGAA GGTCGACTCG AGCGGCCGCA TCGTGACTGA CTGA 774
(2) INFORMATION FOR SEQ ID NO: 6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 257
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 6
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
CA 02362004 2001-11-13
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Va1 Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Arg Gln Ile Lys Ile Trp Phe Gln Asn
230 235 240
Arg Arg Met Lys Trp Lys Lys Val Asp Ser Ser Gly Arg Ile Val
245 250 255
Thr Asp
(2) INFORMATION FOR SEQ ID NO: 7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 7:
GAATCCCGCA AACGCGCAAG GCAG 24
6
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 8:
(1) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 8:
TCAGTTCTCC TTCTTCCACT TCATGCG 27
(2) INFORMATION FOR SEQ ID N0: 9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 9:
AATTCCGCCA GATCAAGATT TGGTTCCAGA ATCGTCGCAT GAAGTGGAAG AAGG 54
(2) INFORMATION FOR SEQ ID NO: 10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 10:
GGCGGTCTAG TTCTAAACCA AGCTCTTAGC AGCGTAGTTC ACCTTCTTCC ACCT 54
7
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 11:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 11:
GAATCCAAGC ATCCAGGAAG TCAGCC 26
(2) INFORMATION FOR SEQ ID NO: 12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 12:
ACCAGCCACC ACCTTCTGAT A 21
(2) INFORMATION FOR SEQ ID NO: 13:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 876
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(Vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 13:
g
CA 02362004 2001-11-13
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAA.AGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTCTCTCAA 480
TTTGCAGGAA GACCAATTAT TACACAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCAAGCATC CAGGAAGTCA GCCTAAAACT 720
GCTTGTACCA ATTGCTATTG TAAAAAGTGT TGCTTTCATT GCCAAGTTTG TTTCATAACA 780
AAAGCCTTAG GCATCTCCTA TGGCAGGAAG CGGAGACAGC GACGAAGAGC TCATCAGAAC 840
AGTCAGACTC ATCAAGCTTC TCTATCAAAG CAGTAA 876
(2) INFORMATION FOR SEQ ID NO: 14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 291
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(Vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 14:
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
9
CA 02362004 2001-11-13
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala G1y Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Lys His Pro Gly Ser Gln Pro Lys Thr
230 235 240
Ala Cys Thr Asn Cys Tyr Cys Lys Lys Cys Cys Phe His Cys Gln
245 250 255
Val Cys Phe Ile Thr Lys Ala Leu Gly Ile Ser Tyr Gly Arg Lys
260 265 270
Arg Arg Gln Arg Arg Arg Ala His Gln Asn Ser Gln Thr His Gln
275 280 285
Ala Ser Leu Ser Lys Gln
290
(2) INFORMATION FOR SEQ ID NO: 15:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 15:
AATTCTATGG TCGTAAAAAA CGTCGTCAAC GTCGTCGTG 39
1~
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 16:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(vii) IMMEDIATE SOURCE:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 16:
GATACCAGCA TTTTTTGCAG CAGTTGCAGC AGCACAGCT 39
(2) INFORMATION FOR SEQ ID NO: 17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 756
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 17:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTCTCTCAA 480
TTTGCAGGAA GACCAATTAT TACACAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCTATGGTG CTAAAAAACG TCGTCAACGT 720
CGTCGTGTCG ACTCGAGCGG CCCGCATCGT GACTGA 756
11
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 18:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 251
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 18:
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
12
CA 02362004 2001-11-13
Ile Asn Pro Lys Glu Phe Tyr Gly Ala Lys Lys Arg Arg Gln Arg
230 235 240
Arg Arg Val Asp Ser Ser Gly Pro His Arg Asp
245 250
(2) INFORMATION FOR SEQ ID NO: 19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1913
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 19:
ATGTCCCCTA TACTAGGTTA TTGGAAAATT AAGGGCCTTG TGCAACCCAC TCGACTTCTT 60
TTGGAATATC TTGAAGAAAA ATATGAAGAG CATTTGTATG AGCGCGATGA AGGTGATAAA 120
TGGCGAAACA AAAAGTTTGA ATTGGGTTTG GAGTTTCCCA ATCTTCCTTA TTATATTGAT 180
GGTGATGTTA AATTAACACA GTCTATGGCC ATCATACGTT ATATAGCTGA CAAGCACAAC 240
ATGTTGGGTG GTTGTCCAAA AGAGCGTGCA GAGATTTCAA TGCTTGAAGG AGCGGTTTTG 300
GATATTAGAT ACGGTGTTTC GAGAATTGCA TATAGTAAAG ACTTTGAAAC TCTCAAAGTT 360
GATTTTCTTA GCAAGCTACC TGAAATGCTG AAAATGTTCG AAGATCGTTT ATGTCATAAA 420
ACATATTTAA ATGGTGATCA TGTAACCCAT CCTGACTTCA TGTTGTATGA CGCTCTTGAT 480
GTTGTTTTAT ACATGGACCC AATGTGCCTG GATGCGTTCC CAAAATTAGT TTGTTTTAAA 540
AAACGTATTG AAGCTATCCC ACAAATTGAT AAGTACTTGA AATCCAGCAA GTATATAGCA 600
TGGCCTTTGC AGGGCTGGCA AGCCACGTTT GGTGGTGGCG ACCATCCTCC AAAATCGGAT 660
CTGGTTCCGC GTGGATCCTC TAGAGTCGAC CTGCAGGCAT GCAATGCTTA TTCCATTAAT 720
CAAAAGGCTT ATTCAAATAC TTACCAGGAG TTTACTAATA TTGATCAAGC AAAAGCTTGG 780
GGTAATGCTC AGTATAAAAA GTATGGACTA AGCAAATCAG AAAAAGAAGC TATAGTATCA 840
TATACTAAAA GCGCTAGTGA AATAAATGGA AAGCTAAGAC AAAATAAGGG AGTTATCAAT 900
GGATTTCCTT CAAATTTAAT AAAACAAGTT GAACTTTTAG ATAAATCTTT TAATAAAATG 960
AAGACCCCTG AAAATATTAT GTTATTTAGA GGCGACGACC CTGCTTATTT AGGAACAGAA 1020
TTTCAAAACA CTCTTCTTAA TTCAAATGGT ACAATTAATA AAACGGCTTT TGAAAAGGCT 1080
AAAGCTAAGT TTTTAAATAA AGATAGACTT GAATATGGAT ATATTAGTAC TTCATTAATG 1140
AATGTTTCTC AATTTGCAGG AAGACCAATT ATTACAAAAT TTAAAGTAGC AAAAGGCTCA 1200
AAGGCAGGAT ATATTGACCC TATTAGTGCT TTTCAGGGAC AACTTGAAAT GTTGCTTCCT 1260
AGACATAGTA CTTATCATAT AGACGATATG AGATTGTCTT CTGATGGTAA ACAAATAATA 1320
ATTACAGCAA CAATGATGGG CACAGCTATC AATCCTAAAG AATTCAGAAG GAAACAAAGA 1380
AGAAAAAGAA GACTGCAGGC GGCCGCATCG TGA 1413
13
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 20:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 470
(B) TYPE: AMINO ACIDS
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 20:
Met Ser Pro Ile Leu Gly Tyr Trp Lys Ile Lys Gly Leu Val Gln
1 5 10 15
Pro Thr Arg Leu Leu Leu Glu Tyr Leu Glu Glu Lys Tyr Glu Glu
20 25 30
His Leu Tyr Glu Arg Asp Glu Gly Asp Lys Trp Arg Asn Lys Lys
35 40 45
Phe Glu Leu Gly Leu Glu Phe Pro Asn Leu Pro Tyr Tyr Ile Asp
50 55 60
Gly Asp Val Lys Leu Thr Gln Ser Met Ala Ile Ile Arg Tyr Ile
65 70 75
Ala Asp Lys His Asn Met Leu Gly Gly Cys Pro Lys Glu Arg Ala
80 85 90
Glu Ile Ser Met Leu Glu Gly A1a Val Leu Asp Ile Arg Tyr Gly
95 100 105
Val Ser Arg Ile Ala Tyr Ser Lys Asp Phe Glu Thr Leu Lys Val
110 115 120
Asp Phe Leu Ser Lys Leu Pro Glu Met Leu Lys Met Phe Glu Asp
125 130 135
Arg Leu Cys His Lys Thr Tyr Leu Asn Gly Asp His Val Thr His
140 145 150
Pro Asp Phe Met Leu Tyr Asp Ala Leu Asp Val Val Leu Tyr Met
155 160 165
Asp Pro Met Cys Leu Asp Ala Phe Pro Lys Leu Val Cys Phe Lys
170 175 180
Lys Arg Ile Glu Ala Ile Pro Gln Ile Asp Lys Tyr Leu Lys Ser
185 190 195
Ser Lys Tyr Ile Ala Trp Pro Leu Gln Gly Trp Gln Ala Thr Phe
200 205 210
Gly Gly Gly Asp His Pro Pro Lys Ser Asp Leu Val Pro Arg Gly
215 220 225
14
CA 02362004 2001-11-13
Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile Asn
230 235 240
Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile Asp
245 250 255
Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly Leu
260 265 270
Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser Ala
275 280 285
Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile Asn
290 295 300
Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp Lys
305 310 315
Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe Arg
320 325 330
Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr Leu
335 340 345
Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys Ala
350 355 360
Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr Ile
365 370 375
Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro Ile
380 385 390
Ile Thr Lys Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr Ile
395 400 405
Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu Pro
410 415 420
Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser Asp
425 430 435
Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala Ile
440 445 450
Asn Pro Lys Glu Phe Arg Arg Lys Gln Arg Arg Lys Arg Arg Leu
455 460 465
Gln Ala Ala Ala Ser
470
(2) INFORMATION FOR SEQ ID NO: 21
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:16
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
1$
CA 02362004 2001-11-13
(ii) MOLECULE TYPE: PEPTIDE
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 21:
Lys Arg Arg Arg Arg Arg Pro Lys Lys Arg Arg Arg
1 5 10
Ala Lys Arg Arg
(2) INFORMATION FOR SEQ ID NO: 22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH:48
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 22:
AAGAGAAGGC GAAGAAGACC TAAGAAGAGA CGAAGGGCGA AGAGGAGA 48
(2) INFORMATION FOR SEQ ID NO: 23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 48
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 23:
TTCTCTTCCG CTTCTTCTGG ATTCTTCTCT GCTTCCCGCT TCTCCTCT 48
(2) INFORMATION FOR 5EQ ID NO: 24:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 792
16
CA 02362004 2001-11-13
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 24:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTTTCTCAA 480
TTTGCAGGAA GACCAATTAT TACAAAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCAAGAGAA GGCGAAGAAG ACCTAAGAAG 720
AGACGAAGGG CGAAGAGGAG ACACCACCAC CACCACCACG TCGACTCGAG CGGCCGCATC 780
GTGACTGACT GA 792
(2) INFORMATION FOR SEQ ID NO: 25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 263
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 25:
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
17
CA 02362004 2001-11-13
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr A1a Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Lys Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu G1u Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Lys Arg Arg Arg Arg Arg Pro Lys Lys
230 235 240
Arg Arg Arg Ala Lys Arg Arg His His His His His His Val Asp
245 250 255
Ser Ser Gly Arg Ile Val Thr Asp
260
(2) INFORMATION FOR SEQ ID NO: 26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PEPTIDE
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 26:
18
CA 02362004 2001-11-13
Lys Arg Arg Arg Arg Lys Lys Arg Arg Gln Arg Arg Arg
1 5 10
(2) INFORMATION FOR SEQ ID NO: 27:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 27:
AAGCGTCGAC GTAGAAAGAA ACGTAGACAG CGTAGACGT 39
(2) INFORMATION FOR SEQ ID NO: 28:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 28:
TTCGCAGCTG CATCTTTCTT TGCATCTGTC GCATCTGCA 39
(2) INFORMATION FOR SEQ ID NO: 29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 783
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
19
CA 02362004 2001-11-13
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 29:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTTTCTCAA 480
TTTGCAGGAA GACCAATTAT TACAAAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCAAGCGTC GACGTAGAAA GAAACGTAGA 720
CAGCGTAGAC GTCACCACCR CCACCACCAC GTCGACTCGA GCGGCCGCAT CGTGACTGAC 780
TGA 783
(2) INFORMATION FOR SEQ ID NO: 30:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 260
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 30:
Gly Ser Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile
1 5 10 15
Asn Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile
20 25 30
Asp Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly
35 40 45
Leu Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser
50 55 60
Ala Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile
65 70 75
Asn Gly Phe Pro 5er Asn Leu Ile Lys Gln Val Glu Leu Leu Asp
80 85 90
CA 02362004 2001-11-13
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr A1a Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Lys Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Lys Arg Arg Arg Arg Lys Lys Arg Arg
230 235 240
Gln Arg Arg Arg His His His His His His Val Asp Ser Ser Gly
245 250 255
Arg Ile Val Thr Asp
260
(2) INFORMATION FOR SEQ ID NO: 31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 9
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PEPTIDE
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 31:
Arg Arg Lys Gln Arg Arg Lys Arg Arg
1 5
21
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 32:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 32:
AGAAGGAAAC ARAGAAGAAA AAGAAGA 27
(2) INFORMATION FOR SEQ ID NO: 33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 33:
TCTTCCTTTG TTTCTTCTTT TTCTTCT 27
(2) INFORMATION FOR SEQ ID NO: 34:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 771
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 34:
22
CA 02362004 2001-11-13
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTTTCTCAA 480
TTTGCAGGAA GACCAATTAT TACAAAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TCAGGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCRACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCAGAAGGA AACAAAGAAG AAAAAGAAGA 720
CACCACCACC ACCACCACGT CGACTCGAGC GGCCGCATCG TGACTGACTG A 771
(2) INFORMATION FOR SEQ ID NO: 35:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 256
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 35:
GlySer SerArgVal AspLeuGln AlaCysAsn AlaTyrSer Ile
1 5 10 15
AsnGln LysAlaTyr SerAsnThr TyrGlnGlu PheThrAsn Ile
20 25 30
AspGln AlaLysAla TrpGlyAsn AlaGlnTyr LysLysTyr Gly
35 40 45
LeuSer LysSerGlu LysGluAla IleValSer TyrThrLys Ser
50 55 60
AlaSer GluIleAsn GlyLysLeu ArgGlnAsn LysGlyVal Ile
65 70 75
AsnGly PheProSer AsnLeuIle LysGlnVal GluLeuLeu Asp
80 85 90
23
CA 02362004 2001-11-13
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
190 195 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Lys Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Arg Arg Lys Gln Arg Arg Lys Arg Arg
230 235 240
His His His His His His Val Asp Ser Ser Gly Arg Ile Val Thr
245 250 255
Asp
(2) INFORMATION FOR SEQ ID NO: 36:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 887
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: DOUBLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 36:
GGATCCTCTA GAGTCGACCT GCAGGCATGC AATGCTTATT CCATTAATCA AAAGGCTTAT 60
24
CA 02362004 2001-11-13
TCAAATACTT ACCAGGAGTT TACTAATATT GATCAAGCAA AAGCTTGGGG TAATGCTCAG 120
TATAAAAAGT ATGGACTAAG CAAATCAGAA AAAGAAGCTA TAGTATCATA TACTAAAAGC 180
GCTAGTGAAA TAAATGGAAA GCTAAGACAA AATAAGGGAG TTATCAATGG ATTTCCTTCA 240
AATTTAATAA AACAAGTTGA ACTTTTAGAT AAATCTTTTA ATAAAATGAA GACCCCTGAA 300
AATATTATGT TATTTAGAGG CGACGACCCT GCTTATTTAG GAACAGAATT TCAAAACACT 360
CTTCTTAATT CAAATGGTAC AATTAATAAA ACGGCTTTTG AAAAGGCTAA AGCTAAGTTT 420
TTAAATAAAG ATAGACTTGA ATATGGATAT ATTAGTACTT CATTAATGAA TGTTTCTCAA 480
TTTGCAGGAA GACCAATTAT TACAAAATTT AAAGTAGCAA AAGGCTCAAA GGCAGGATAT 540
ATTGACCCTA TTAGTGCTTT TGCAGGACAA CTTGAAATGT TGCTTCCTAG ACATAGTACT 600
TATCATATAG ACGATATGAG ATTGTCTTCT GATGGTAAAC AAATAATAAT TACAGCAACA 660
ATGATGGGCA CAGCTATCAA TCCTAAAGAA TTCGTGATGA ATCCCGCAAA CGCGCAAGGC 720
AGACATACAC CCGGTACCAG ACTCTAGAGC TAGAGAAGGA GTTTCACTTC AATCGCTACT 780
TGACCCGTCG GCGAAGGATC GAGATCGCCC ACGCCCTGTG CCTCACGGAG CGCCAGATAA 840
AGATTTGGTT CCAGAATCGG CGCATGAAGT GGAAGAAGGA GAACTGA 887
(2) INFORMATION FOR SEQ ID NO: 37:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 248
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 37:
GlySerSerArgVal AspLeuGln AlaCysAsn AlaTyrSer Ile
1 5 10 15
AsnGlnLysAlaTyr SerAsnThr TyrGlnGlu PheThrAsn Ile
20 25 30
AspGlnAlaLysAla TrpGlyAsn AlaGlnTyr LysLysTyr Gly
35 40 45
LeuSerLysSerG1u LysGluAla IleValSer TyrThrLys Ser
50 55 60
AlaSerGluIleAsn GlyLysLeu ArgGlnAsn LysGlyVal Ile
65 70 75
AsnGlyPheProSer AsnLeuIle LysGlnVal GluLeuLeu Asp
80 85 90
CA 02362004 2001-11-13
Lys Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe
95 100 105
Arg Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr
110 115 120
Leu Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys
125 130 135
Ala Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr
140 145 150
Ile Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro
155 160 165
Ile Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr
170 175 180
Ile Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu
185 190 195
Pro Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser
200 205 210
Asp Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala
215 220 225
Ile Asn Pro Lys Glu Phe Val Met Asn Pro Ala Asn Ala Gln Gly
230 235 240
Arg His Thr Pro Gly Thr Arg Leu
245
(2) INFORMATION FOR SEQ ID NO: 38:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 37
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 38:
GGATCTGGTT CCGCGTCATA TGTCTAGAGT CGACCTG 37
(2) INFORMATION FOR SEQ ID NO: 39:
26
CA 02362004 2001-11-13
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 39:
CGCGGATCCA TTAGTTCTCC TTCTTCCACT TC 32
(2) INFORMATION FOR SEQ ID NO: 90:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 40:
AAATTAATAC GACTCACTAT AGGG 24
(2) INFORMATION FOR SEQ ID NO: 41:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
27
CA 02362004 2001-11-13
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 41:
GCTAGTTATT GCTCAGCGG 19
(2) INFORMATION FOR SEQ ID NO: 42:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 888
(B) TYPE: NUCLEIC ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 42:
ATGTCTAGAG TCGCACTGCA GGCATGCAAT GCTTATTCCA TTAATCAAAA GGCTTATTCA 60
AATACTTACC AGGAGTTTAC TAATATTGAT CAAGCAAAAG CTTGGGGTAA TGCTCAGTAT 120
AAAAAGTATG GACTAAGCAA ATCAGAAAAA GAAGCTATAG TATCATATAC TAAAAGCGCT 180
AGTGAAATAA ATGGAAAGCT AAGACAAAAT AAGGGAGTTA TCAATGGATT TCCTTCAAAT 240
TTAATAAAAC AAGTTGAACT TTTAGATAAA TCTTTTAATA AAATGAAGAC CCCTGAAAAT 300
ATTATGTTAT TTAGAGGCGA CGACCCTGCT TATTTAGGAA CAGAATTTCA AAACACTCTT 360
CTTAATTCAA ATGGTACAAT TAATAAAACG GCTTTTGAAA AGGCTAAAGC TAAGTTTTTA 420
AATAAAGATA GACTTGAATA TGGATATATT AGTACTTCAT TAATGAATGT TTCTCAATTT 480
GCAGGAAGAC CAATTATTAC AAAATTTAAA GTAGCAAAAG GCTCAAAGGC AGGATATATT 540
GACCCTATTA GTGCTTTTGC AGGACAACTT GAAATGTTGC TTCCTAGACA TAGTACTTAT 600
CATATAGACG ATATGAGATT GTCTTCTGAT GGTAAACAAA TAATAATTAC AGCAACAATG 660
ATGGGCACAG CTATCAATCC TAAAGAATTC GTGATGAATC CCGCAAACGC GCAAGGCAGA 720
CATACACCCG GTACCAGACT CTAGAGCTAG AGAAGGAGTT TCACTTCAAT CGCTACTTGA 780
CCCGTCGGCG AAGGATCGAG ATCGCCCACG CCCTGTGCCT CACGGAGCGC CAGATAAAGA 840
TTTGGTTCCA GAATCGGCGC ATGAAGTGGA AGAAGGAGGA CTAACTGA 888
28
CA 02362004 2001-11-13
(2) INFORMATION FOR SEQ ID NO: 43:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 247
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: PROTEIN
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 43:
Met Ser Arg Val Asp Leu Gln Ala Cys Asn Ala Tyr Ser Ile Asn
1 5 10 15
Gln Lys Ala Tyr Ser Asn Thr Tyr Gln Glu Phe Thr Asn Ile Asp
20 25 30
Gln Ala Lys Ala Trp Gly Asn Ala Gln Tyr Lys Lys Tyr Gly Leu
35 40 45
Ser Lys Ser Glu Lys Glu Ala Ile Val Ser Tyr Thr Lys Ser Ala
50 55 60
Ser Glu Ile Asn Gly Lys Leu Arg Gln Asn Lys Gly Val Ile Asn
65 70 75
Gly Phe Pro Ser Asn Leu Ile Lys Gln Val Glu Leu Leu Asp Lys
80 85 90
Ser Phe Asn Lys Met Lys Thr Pro Glu Asn Ile Met Leu Phe Arg
95 100 105
Gly Asp Asp Pro Ala Tyr Leu Gly Thr Glu Phe Gln Asn Thr Leu
110 115 120
Leu Asn Ser Asn Gly Thr Ile Asn Lys Thr Ala Phe Glu Lys Ala
125 130 135
Lys Ala Lys Phe Leu Asn Lys Asp Arg Leu Glu Tyr Gly Tyr Ile
140 145 150
Ser Thr Ser Leu Met Asn Val Ser Gln Phe Ala Gly Arg Pro Ile
155 160 165
Ile Thr Gln Phe Lys Val Ala Lys Gly Ser Lys Ala Gly Tyr Ile
170 175 180
Asp Pro Ile Ser Ala Phe Gln Gly Gln Leu Glu Met Leu Leu Pro
185 190 195
29
CA 02362004 2001-11-13
Arg His Ser Thr Tyr His Ile Asp Asp Met Arg Leu Ser Ser Asp
200 205 210
Gly Lys Gln Ile Ile Ile Thr Ala Thr Met Met Gly Thr Ala Ile
215 220 225
Asn Pro Lys Glu Phe Val Met Asn Pro Ala Asn Ala Gln Gly Arg
230 235 240
His Thr Pro Gly Thr Arg Leu
245
(2) INFORMATION FOR SEQ ID NO: 44:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 64
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM
xi) SEQUENCE DESCRIPTION: SEQ ID NO: 44:
Val Met Glu ArgLys ArgAlaArg Gln Tyr ThrArgTyr
Ser Thr
1 5 10 15
Gln Thr Leu LeuGlu LysGluPhe His Asn ArgTyrLeu
Glu Phe
20 25 30
Thr Arg Arg ArgIle GluIleAla His Leu CysLeuThr
Arg Ala
35 40 45
Glu Arg Gln LysIle TrpPheGln Asn Arg MetLysTrp
Ile Arg
50 55 60
Lys Lys Glu Asn
(2) INFORMATION FOR SEQ ID NO: 45:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19
CA 02362004 2001-11-13
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 45:
Arg Gln Ile Lys Ile Trp Phe Gln Asn Arg Arg Met Lys Trp Lys
1 5 10 15
Lys Val Asp Ser
(2) INFORMATION FOR SEQ ID NO: 46:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 60
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(Vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID N0: 46:
Lys His Pro Gly Ser Gln Pro Lys Thr Ala Cys Thr Asn Cys Tyr
1 5 10 15
Cys Lys Lys Cys Cys Phe His Cys Gln Val Cys Phe Ile Thr Lys
20 25 30
Ala Leu Gly Ile Ser Tyr Gly Arg Lys Arg Arg Gln Arg Arg Arg
35 40 45
Ala His Gln Asn Ser Gln Thr His Gln Ala Ser Leu Ser Lys Gln
50 55 60
(2) INFORMATION FOR SEQ ID NO: 47:
(i) SEQUENCE CHARACTERISTICS:
31
CA 02362004 2001-11-13
(A) LENGTH: 20
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 47:
Tyr Gly Ala Lys Lys Arg Arg Gln Arg Arg Arg Val Asp Ser Ser
1 5 10 15
Gly Pro His Arg Asp
(2) INFORMATION FOR SEQ ID NO: 48:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 17
(B) TYPE: AMINO ACID
(C) STRANDEDNESS: SINGLE
(D) TOPOLOGY: LINEAR
(ii) MOLECULE TYPE: Peptide
(vi) ORIGINAL SOURCE:
(A) ORGANISM:
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 48:
Val Met Asn Pro Ala Asn Ala G1n Gly Arg His Thr Pro Gly Thr
1 5 10 15
Arg Leu
32