Note: Descriptions are shown in the official language in which they were submitted.
CA 02363020 2001-08-15
is
25
WO 00/49540 PCT/US00/04331
TITLE: METHOD AND SYSTEI~~I FOR DYNAMIC STORAGE
RETRIEVAL AND ANALYSIS OF EXPERIMENTAL D_ ATA WITH
DETERMINED RELATIONSHIPS
FIELD OF THE INVENTION
This invention relates to storing, retrieving and analyzing experimental
information. More specifically, it relates to a method and system for dynamic
3o storing, retrieving and analyzing experimental information with determined
relationships.
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
BACKGROUND OF THE INVENTION
Traditionally, cell biology research has largely been a manual, labor
intensive activity. With the advent of tools that can automate cell biology
experimentation (see for example U.S. Patent Application SN 08/810,983 filed
February 2.7, 1997, assigned to the same Assignee as the present application.)
the rate at which complex information is generated about the functioning of
cells has increased dramatically. As a result, cell biology is not only an
academic discipline, but also the new frontier for large-scale drug discovery.
1o Cells are the basic units of life and integrate information from
Deoxyribonucleic Acid ("DNA"), Ribonucleic Acid ("RNA"), proteins,
metabolites, ions and other cellular components. New drug compounds that
may look promising at a nucleotide level may be toxic at a cellular level.
Thus, cell biology is becoming increasingly important to test now drug
compounds. Florescence-based reagents can be applied to cells to determine
ion concentrations, membrane potentials, enzyme activities, gene expression,
as well as the presence of metabolites, proteins, lipids, carbohydrates, and
other cellular components.
Innovations in automated screening systems for biological and other
2o research are capable of generating enormous amounts of data. The massive
volumes of feature-rich data being generated by these systems and the
effective management and use of information from the data has created a
number of very challenging problems. As is known in the art, "feature-rich"
data includes data wherein one or more individual features of an obj ect of
interest (e.g., a cell) can be collected.
2
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
For more information on feature-rich cell screening see "High content
fluorescence-based screening," by Kenneth A. Guiliano, et al., Journal of
Biomolecular Screening, Vol. 2, No. 4, pp. 249-259, Winter 1997. ISSN 1087-
0571, "PTH receptor internalization," Bruce R. Conway, et al., Journal of
Biomolecular Screening, Vol. 4, No. 2, pp. 75-68, April 1999, ISSN 1087-
0571, "Fluorescent-protein biosensors: new tools for drug discovery," Kenneth
A. Giuliano and D. Lansing Taylor, Trends in Biotechnology, ("TIBTECH"),
Vol. 16, No. 3, pp. 99-146, March 1998, ISSN 0167-7799.
To fully exploit the potential of data from high-volume data generating
to screening instrumentation, there is a need for new informatic and
bioinformatic tools. As is known in the art, "bioinformatic" techniques are
used to address problems related to the collection, processing, storage,
retrieval and analysis of biological information including cellular
information.
Bioinformatics is defined as the systematic development and application of
information technologies and data processing techniques for collecting,
analyzing and displaying data obtained by experiments, modeling, database
searching, and instrumentation to make observations about biological
processes. How to present, organize and analyze the complex information
about cell functioning so that new knowledge can be generated is critical for
2o both pharmaceutical research and basic cell biology research.
There are several problems associated with using bioinformatic
systems and techniques known in the art to capture and display biological
information, such as cellular information. One problem is that biological
information is typically collected and displayed as textual information in a
uni-dimensional formation. This format prevents a user from visually
3
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
interacting with identified dimensions of biological information at the same
time and dynamically navigating through those dimensions to find out the
relationship of one piece of information with other pieces of information.
This
prevents the abstraction of knowledge from information.
Another problem is that biological pathways can not be adequately
displayed with uni-dimensional textual information. Graphical representation
of biological pathways is typically required to capture biological knowledge
such as cellular knowledge. Biological pathway knowledge obtained from
graphical representations is then typically used as a portal to unite other
to biological information, thus enabling the synthesis of new knowledge by
investigating the inner relationship of this information.
Another problem is that bioinformatic systems known in the art only allow
input and display of a small amount of uni-dimensional biological
information. Such systems may use present only a subset of a total amount of
known information associated with a biological entity or transformations.
Another problem is that bioinformatic systems known in the art typically
present a static graphical representation of a biological pathway cannot be
input, edited or otherwise altered by a user. Another problem is that a user
typically cannot navigate, expand or contract a portion of a presented
biological pathway. Another problem is that collected biological information
cannot be easily linked to other private or public databases to provide access
to additional known or related information.
There have been attempts to solve some of these problems associated
with inputting and displaying biological information associated with
biological
pathways. Such attempts include for example, "Ecocyc" from Pangea (see,
4
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
e.g., Nucleic Acids Research 26:50-53 (1998), Ismb 2:203-211 (1994));
"KEGG" pathway database from Institute for Chemical Research, Kyoto
University (see, e.g., Nucleic Acids Research 27:377-379 (1999), Nucleic
Acids Research 27:29-34 (1999)); "CSNDB" links to from Japanese National
Institute of Health Sciences (see, e.g., Pac Symp. Biocomput 187-197 (1997));
"SPAD" from Graduate School of Genetic Resources Technology, Kyushu
University, Japan; "PUMA" now called "WIT" from Computational Biology
in the Mathematics and Computer Science Division at Argonne National
Laboratory; and others. However, these solutions still suffer from one or more
of the problems described above.
Thus, it is desirable to provide a bioinformatic system that enables the easy
storage, retrieval and analysis of biological information associated with
biological pathways. The bioiniformatic system should include the ability to
dynamically input, edit and generate biological pathways and to provide the
ability to access hierarchical information associated with the biological
pathways from plural private and public databases.
5
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
SUMMARY OF THE INVENTION -
In accordance with preferred embodiments of the present invention,
some of the problems associated with inputting and displaying biological
information associated with biological pathways are overcome. A method and
system for dynamic storage, retrieval and display of experimental information
with determined relationships is presented.
One aspect of the invention includes a method for storing
experimental information with determined relationships. The method includes
providing a graphical user interface from which shapes and arrows
representing biological entities and transformations respectively, can be
input
and edited. Multi-dimensional information based on a pre-determined, but
expandable hierarchy is input to link the entities and transformations to
additional information about the entities and transformations. Related
information, if any, is input to link the entities and transformations to
other
information in plural external databases on a public network such as the
Internet. Information associated with plural shapes connected with plural
arrows is saved as a biological pathway with determined relationships in a
database. The biological pathway defines a hierarchical representation of a
2o biological function with determined relationships between entities and
transformations.
Another aspect of the invention includes a method for dynamically
displaying experimental information with determined relationships. A
biological pathway is selected from a list of biological pathways with
determined relationships. A display mode is selected that is used to display
6
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
the biological pathway. A graphical representation including shapes and
arrows representing entities and transformations respectively is dynamically
generated using a first set of colors. The first set of colors is used to
indicate a
level of generalization in a hierarchy or a directed graph used to display the
biological pathway with determined relationships.
Another aspect of the invention includes a system for dynamically
storing, retrieving and displaying of experimental information with determined
relationships. The system includes a graphical user interface and a database.
The graphical user interface is used for dynamically inputting or editing
to information associated with biological pathway with determined
relationships
using shapes and arrows to represent entities and transformations and to
capture information associated with biological pathway as it is drawn, for
saving information associated with a biological pathway in a database, for
retrieving information associated with selected biological entities or
transformations from a database, for dynamically generating graphical
representation of a biological pathway with multiple colors from information
retrieved from a database, and for navigating through a hierarchy or a
directed
graph of information associated with a generated biological pathway.
The database is used for saving information associated with a
2o plurality of shapes connected with a plurality of arrows as a biological
pathway with determined relationships. The biological pathway defines a
hierarchical representation of a biological function with determined
relationships between the entities and transformations.
The present invention may provide the following advantages.
Biological pathway diagrams with determined relationships may be
7
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
dynamically input, edited and dynamically generated to represent biological
functions, such as cellular functions, to enable a user to visually interact
with
identified dimensions of biological information. A user may dynamically
navigate through identified dimensions of biological infornlation with
different display colors to find out a relationship of a specific piece of
biological information with other pieces of biological information. The
biological pathways are linked to plural databases on local private and remote
public networks (e.g. the Internet), including information related to the
biological pathway. This may help facilitate the abstraction of knowledge
l0 from information.
The present invention may also be used to further facilitate a user's
understanding of biological functions, such as cell functions, to design
experiments more intelligently and to analyze experimental results more
thoroughly. Specifically, the present invention may help drug discovery
scientists select better targets for pharmaceutical intervention in the hope
of
curing diseases.
The foregoing and other features and advantages of preferred
embodiments of the present invention will be more readily apparent from the
following detailed description. The detailed description proceeds with
2o references to the accompanying drawings.
8
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
BRIEF DESCRIPTION OF THE DRAWINGS
Preferred embodiments of the present invention are described with
reference to the following drawings, wherein:
FIG. 1 illustrates an exemplary experimental data storage system for
storing experimental data with determined relationships;
FIGS. 2A and 2B are a flow diagram illustrating a method for storing
experimental information with determined relationships;
to FIG. 3 is a block diagram illustrating a screen display of a graphical
user interface used to create, store and analyze biological pathways with
determined relationships;
FIG. 4 is a block diagram illustrating an exemplary multi-dimensional
hierarchy;
FIG. 5 is a block diagram illustrating an exemplary multi-dimensional
hierarchy for a biological entity;
FIG. 6 is a block diagram illustrating an exemplary multi-dimensional
hierarchy for a transformation;
FIG. 7 is a flow diagram illustrating a method for dynamically
2o displaying experimental information including determined relationships;
FIG. 8 is a block diagram illustrating an exemplary mufti-dimensional
information page dynamically and created for a user in a summary display
mode;
FIG. 9 is a block diagram illustrating an exemplary entity multi-
dimensional information page dynamically created and displayed for a user in
a dimension display mode;
9
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
FIG. I O is a block diagram illustrating an exemplary related
information page that dynamically created and displayed for a user in a link
display mode; and
FIG. 11 is a flow diagram illustrating a method for dynamically
displaying experimental information including determined relationships
displaying from a remote computer.
to
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
DETAILED DESCRIPTION OF PREFERRED EMBODIMENTS
Exemplary data storage system
FIG. 1 illustrates an exemplary experimental data storage system 10 for
one embodiment of the present invention. The data storage system 10 includes
one or more internal user computers 12, 14, (only two of which are
illustrated)
for inputting, retrieving and analyzing experimental data on a private local
area
network ("LAN") 16 (e.g., an intranet). The LAN 16 is connected to one or
more internal proprietary databases 18, 20 (only two of which are illustrated)
used to store private proprietary experimental information that is not
available to
to the public.
The LAN 16 is connected to an internal database server 22 that is
connected to one or more internal experimental information databases 24, 26
(only two of which are illustrated) comprising a private part and publicly
part of
a data store for experimental data. The internal database server 22 is
connected
to a public network 28 (e.g., the Internet). One or more external user
computers,
30, 32, 34, 36 (only four of which are illustrated) are connected to the
public
network 28, to plural public domain databases 38, 40, 42 (only three of which
are
illustrated) and internal databases 24, 26 including experimental data and
other
related experimental information available to the public. However, more, fewer
or other equivalent data store components can also be used and the present
invention is not limited to the data storage system 10 components illustrated
in
FIG. 1.
In one specific exemplary embodiment of the present invention, data
storage system 10 includes the following specific components. However, the
present invention is not limited to these specific components and other
similar
11
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
or equivalent components may also be used. The one or more internal user
computers, 12, 14, and the one or more external user computers, 30, 32, 34,
36,
are conventional personal computers that include a display application that
provide a Graphical User Interface ("GUI") application (See FIG. 3). The GUI
application is used to lead a scientist or lab technician through input,
retrieval,
analysis of experimental data with determined relationships and supports
custom viewing capabilities. The GUI application also supports data exported
into standard desktop tools such as spreadsheets, graphics packages, and word
processors.
to The internal user computers 12, 14, connect to the one or more private
proprietary databases 18, 20, the database server 22 and the one or more or
more
internal databases 24, 26 over the LAN 16. In one embodiment of the present
invention, the LAN 16 is a 100 Mega-bit ("Mbit") per second or faster
Ethernet, LAN. However, other types of LANs could also be used (e.g.,
optical or coaxial cable networks). In addition, the present invention is not
limited to these specific components and other similar components may also
be used.
In one specific embodiment of the present invention, one or more
protocols from the Internet Suite of protocols are used on the LAN 16 so LAN
16 comprises a private intranet. Such a private intranet can communicate with
other public or private networks using protocols from the Internet Suite. As
is
known in the art, the Internet Suite of protocols includes such protocols as
the
Internet Protocol ("IP"), Transmission Control Protocol ("TCP"), User
Datagram Protocol ("UDP"), Hypertext Transfer Protocol ("HTTP"),
12
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
Hypertext Markup Language ("HTML"), eXtensible Markup Language
("XML") and others.
The one or more private proprietary databases 18, 20, and the one or
more internal databases 24, 26 are mufti-user, mufti-view databases that store
experimental data. The databases 18, 20, 24, 26 use relational database tools
and structures. The data stored within the one or more internal proprietary
databases 18, 20 is not available to the public. Selected portions of the
internal
experimental information databases 24, 26, may be available to the public
through database server 22 using selected security features (e.g., login,
password,
1 o firewall, etc.
The one or more external user computers, 30, 32, 34, 36, are connected to
the public network 28 and to plural public domain databases 38, 40, 42. The
plural public domain databases 38, 40, 42 include experimental data and
information in the public domain and are also mufti-user, mufti-view
databases.
The plural public domain databases 38, 40, 42, include such well known
databases such as provided by Medline, Gen Bank, SwissProt, PDB, etc.
An operating environment for components of the data storage system
10 for preferred embodiments of the present invention include a processing
system with one or more speed Central Processing Units) ("CPU") and a
2o memory. In accordance with the practices of persons skilled in the art of
computer programming, the present invention is described below with
reference to acts and symbolic representations of operations or instructions
that are performed by the processing system, unless indicated otherwise. Such
acts and operations or instructions are referred to as being
"computer-executed" or "CPU executed."
13
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
It will be appreciated that acts and symbolically represented operations
or instructions include the manipulation of electrical signals by the CPU. An
electrical system represents data bits which cause a resulting transformation
or
reduction of the electrical signals, and the maintenance of data bits at
memory
locations in a memory system to thereby reconfigure or otherwise alter the
CPU's operation, as well as other processing of signals. The memory locations
where data bits are maintained are physical locations that have particular
electrical, magnetic, optical, or organic properties corresponding to the data
bits.
to The data bits may also be maintained on a computer readable medium
including magnetic disks, optical disks, organic memory, and any other
volatile (e.g., Random Access Memory ("RAM")) or non-volatile (e.g., Read-
Only Memory ("ROM")) mass storage system readable by the CPU. The
computer readable medium includes cooperating or interconnected computer
readable medium, which exist exclusively on the processing system or be
distributed among multiple interconnected processing systems that may be
local or remote to the processing system.
Storing experimental information with determined relationships
FIGS. 2A and 2B are a flow diagram illustrating a Method 46 for
2o storing experimental information with determined relationships. In FIG. 2A
at
Step 48, a shape is selected from a menu on graphical user interface on a
computer. The shape represents an entity that participates in a biological
pathway. At Step 50, the shape is placed at a desired location in an
electronic
window on the graphical user interface. At Step 52, an arrow is selected from
the graphical user interface. The arrow represents a transformation between
14
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
entities that participate in a biological pathway. At Step 54, the arrow and
the
shape are connected. This provides a graphical representation of a
transformation of an entity with a determined relationship. At Step 56, multi-
dimensional information is input to link the shape and arrow to multi-
dimensional information specifying entity and transformation. The multi-
dimensional information is stored in a database with a pre-determined format.
In FIG. 2B at Step 58, related information, if any, is input to link the
shape and arrow to other information related to the entity and transformation
from plural external databases. At Step 60, a test is conducted to determine
if
to a desired number of iterations of Steps 50, 52, 54, 56 and 58 have been
completed. If so, at Step 62, information associated with the plural shapes
connected with plural arrows is saved in a database as a biological pathway
with determined relationships between entities and transformations. If a
desired number of iterations have not been completed at Step 60, a loop
continues at Step 48 of FIG. 2A until the desired number of iterations has
been
completed. The biological pathway defines a hierarchical representation of a
biological function with determined relationships between entities and
transformations.
In another embodiment of the present invention, Method 46 allows all
shapes for all entities selected and placed at one time. In such an
embodiment,
a loop would be entered to repeat steps 48 and 50 a desired number of times,
and then Step 62 would be executed. (not illustrated in FIG. 2).
In another embodiment of the present invention, Method 46 allows
arrows for all transformations to be connected to entities at one time. In
such
an embodiment, a loop would be entered to repeat steps 52 and 54 a desired
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
number of times, and then Step 62 would be executed (not illustrated in FIG.
2).
Either of these embodiments, all shapes and/or all arrows would be
input at one before any multi-dimensional information, or any related
information is input. This allows a user to spatially layout one or more
desired
biological and then go back and input the mufti-dimensional and/or related
information at a later time.
In one embodiment of the present invention only an indication of the
types of shapes and arrows and their absolute or relative locations on the
graphical user interface is saved in the database. In such an embodiment,
when the saved biological pathway is displayed, the shapes are arrows
representing entities and transformations with determined relationships are
dynamically re-generated from the saved information. Such an embodiment
requires less storage space to store biological pathway and also allows for a
quicker re-generation and display of a saved biological pathway with
determined relationship. In another embodiment of the present invention, the
graphical shapes and arrows are saved in the database along with the
associated information.
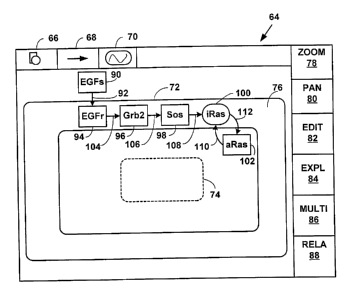
FIG. 3 is a block diagram illustrating a screen display of a Graphical
2o User Interface ("GUI") 64 used to create, display and analyze biological
pathways with Method 64 (FIGS. 2A and 2B). The GUI 64 includes a
graphical button for selecting a shape 66, selecting an arrow 68, and
selecting
a cell organelle or compartment 70. The GUI 64 also illustrates an outline of
a
cell 72, an outline of a nucleus 74 within the cell 72, and an outline of a
cell
membrane 76. The cell membrane 76 is exaggerated in FIG. 3 to present a
16
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
specific example of a cell signaling pathway in the cell membrane 76. FIG. 3
illustrates only one cell 72. However, the present invention is not limited to
one cell and multiple cells, multiple organelles and multiple compartments,
inside and outside of a cell can also be illustrated with GUI 64.
GUI 64 further comprises a graphical button for zooming in and out
78, panning 80, editing a new or a previously saved biological pathway 82,
exploring a saved biological pathway 84, specifying and/or examining multi-
dimensional information associated with a pathway 86 and its components,
and examining related information associated with a biological pathway 88
1o and its componets. However, the present invention is not limited to a GUI
64
with the graphical buttons and associated functionality illustrated in FIG. 3
and more, fewer or equivalent graphical buttons and functionality can also be
used.
In one embodiment of the present invention, shape graphical button 66
on the GUI 64 provides a menu for shapes including rectangles, ovals, circles,
hexagons, pie-shapes or other shapes to be selected. The different shapes
represent different types of biological entities. For example, the rectangles
represent active entities. The ovals represent inactive entities. The circles
represent entity inhibitors. The hexagons represent factors exchanged between
2o entities. The pie-shapes represent intermediate entity transformation
products.
However, the present invention is not limited to the shapes or entities
listed,
and more fewer or equivalent entities can also be represented by more fewer
or equivalent shapes.
The arrows represent biological transformations. The biological
transformations include, for example, transcription factor activation,
cellular
17
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
hypertrophy, protein kinase activation, protease activation, gene expression,
receptor activation, apoptosis, material translocation such as internalization
of
cell surface receptor proteins, mitochondria) potential, neurite outgrowth,
cell
viability or a miotic index for a cell. However, the present invention is not
limited to this list of biological transformations and more, fewer or
equivalent
biological transformations can also be represented by the arrows.
The cell organelle and compartment graphical button 70 allows
graphical representations of cell organelles and compartments including,
chromosomes, nucleolus, mitochondria, golgi bodies, ribosomes, micro-
to tubules, smooth endoplasmic reticulum, rough endoplasmic reticulum, and
other cell organelles to be created. Compartments, such a region surrounding
a stress fiber, can be defined as needed by specific biological pathways.
However, more, fewer or equivalent cell organelles and compartments can
also be used and the present invention is not limited to the cell organelles
15 listed. The cell organelles and compartments may participate in selected
biological pathways or be the location of compartments of selected pathways.
Method 64 (FIG. 2) is illustrated with GUI 64 (FIG. 3) with a portion
of a extracellular Epidermal Growth Factor ("EGF") signaling pathway
known in the biological arts. However, the present invention is not limited to
2o this specific example associated with this specific signaling pathway and
virtually any biological pathway can be used with Method 46 and GUI 64. As
is known in the biological arts, a biological pathway is a pathway for any
biological entity and any transformation upon or between biological entities.
In such a specific embodiment in FIG. 2A at Step 48, the edit graphical
25 button 82 is selected from GUI 64 to input a new biological pathway. A
shape
is
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
90 is selected using the shape graphical button 66 on GUI 64 (FIG. 3),
wherein the shape 90 represents an entity that participates-in the EGF cell
signaling pathway. At Step 50, the shape 90 is placed at a desired location in
an electronic window on the GUI. For example, a rectangle 90 (FIG. 3) is
selected using shape graphical button 66 menu and placed outside the cell
outline 72. In this specific example, the rectangle 90 represents an active
extracellular EGF signaling molecule ("EGFs") that initially effects the cell
72
from outside the cell 72.
At Step 52, an arrow 92 is selected from the arrow graphical button 68
on the GUI 64. The arrow 92 represents a transformation between entities that
participate in a pathway. At Step 54, the arrow and the shape are connected.
This provides a graphical representation of a transformation of an entity with
a
determined relationship to the cell 72 (i.e., extracellular signal) as is
illustrated
in FIG. 3.
At Step 56, mufti-dimensional information is input to link the shape
and arrow to mufti-dimensional information specifying the entity and
transformation. In one embodiment of the present invention, general multi-
dimensional information is input at Step 56 and is organized in a hierarchical
fashion that allows electronic links to other associated information. In one
2o embodiment of the present invention, the general mufti-dimensional
information includes, general information for a species, an experimental
system, functional types to classify an entity, transformation types to
classify a
transformation, and a compartment where an entity or transformation occurs
(See FIG. 4). However, more, fewer or equivalent dimensions and other
mufti-dimensional information can also be input.
19
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
In one embodiment of the present invention, when a user is creating a
biological pathway and selects a shape or arrow, (e.g., by '-'clicking" on
it), an
electronic input form is presented to the user so the user can input any known
general mufti-dimensional information about an entity or transformation.
In such an embodiment, an electronic input form created in the Hyper
Text Mark-up Language ("HTML"), or the eXtensible Mark-up Language
("XML") or other hardware independent mark-up languages known in the art
is displayed for a user. However, virtually any programming language can be
used to create and display the electronic input form (e.g., C, C++, Visual
to Basic, Visual C++, Java, etc.) and the present invention is not limited to
hardware independent mark-up languages. The user then inputs any known
general mufti-dimensional information for the entity or transformation.
FIG. 4 is a block diagram illustrating an exemplary general multi-
dimensional hierarchy 114 for used to input mufti-dimensional information at
Step 56. However, the present invention is not limited to this exemplary
general hierarchy, and other types or equivalent mufti-dimensional
information storage schemes can also be used to input mufti-dimensional
information at Step 56.
In addition, the general mufti-dimensional information can be
2o represented with a directed graph. As is known in the computer science
arts, a
"directed graph" is a graph whose edges have a direction. An edge in a
directed graph not only relates two nodes in a graph, but it also specifies a
predecessor-successor relationship. A "directed path" through a directed
graph is a sequence of nodes, n~, n2 ,... nk , such that there is a directed
edge
from n; to n;+~ for all appropriate i. The general mufti-dimensional
information
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
can be represented exclusively by a hierarchy, exclusively by a directed
graph,
by both a hierarchy and a directed graph, or any combination thereof.
The hierarchy 114 includes, a species 116 (e.g., human), an
experimental system 118 (e.g., skeletal system), functional types 120
including classifications for biological entities (e.g., organ, tissue, cell,
sub-
cell component, molecule) and transformation types 122 including
classifications of transformations and a level for a compartment 124 where an
entity or transformation occurs. This multi-dimensional information is stored
in an internal database (e.g., 18,20,24,26). Each component in the hierarchy
l0 114 represents a hierarchy, so hierarchy 114 actually includes five
parallel
hierarchies.
FIG. 5 is a block diagram illustrating an exemplary mufti-dimensional
hierarchy 126 for a functional type including a biological entity (e.g., a
cell)
from hierarchy 114. In addition, the mufti-dimensional information for a
biological entity can also be represented with a directed graph or a
combination of a hierarchy and/or a directed graph as was described above.
However, the present invention is not limited to this exemplary hierarchy, and
other types or equivalent mufti-dimensional information storage schemes can
also be used to input mufti-dimensional information for an entity.
2o In one embodiment of the present invention, a separate hierarchy for
providing specific mufti-dimensional information a biological entity or a
transformation is not used. Only the general hierarchy 114 is used. In another
embodiment of the present invention, separate specific hierarchies are used
for
both biological entities and transformations to specific further provide multi-
dimensional information about an entity or a transformation.
21
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
The entity hierarchy 126 includes a first level for a biological entity
128. A second level includes a component view 130, a morphology 132 of the
biological entity 128, an optional electron microscope ("EM") photograph 134
and an optional fluorescent view 136 of the biological entity 128. The
component view 130 includes a third level. The third level includes basic
information 138, site information 140, function information 142, enzyme
information, if any, 144, reaction information 146, transport system
information 148 and a pathway view 150. Multi-dimensional information that
is input for a biological entity 128 is stored in a local database using the
1 o hierarchy 126.
In one embodiment of the present invention, the biological entity 128
is assumed to be a sub-component of a cell, or a cell. In another embodiment
of the present invention, the hierarchy 126 also includes additional levels
above the first level for the biological entity 128 from lowest to highest for
tissues, organs, systems, or organisms. These additional levels are not
illustrated in FIG. 4, but may also be used to input and display specific
multi-
dimensional information for an entity.
In such an embodiment, an aggregation of plural cells comprise a
tissue. An aggregation of plural tissues comprise an organ. An aggregation of
plural organs comprise a system. An aggregation of plural systems comprise
an organism. An aggregation of plural organism comprise a species.
In one embodiment of the present invention, when a user is creating a
general biological pathway and selects a shape, (e.g., by "clicking" on it),
an
input electronic form for hierarchy 114 and/or a transformation hierarchy is
22
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
presented to a user, so the user can input any known mufti-dimensional
information.
In such an embodiment, an electronic input fornl is created in the
HTML or XML or other hardware independent mark-up languages known in
the art is displayed for a user. However, virtually any programming language
can be used to create and display the electronic input form (e.g., C, C++,
Visual Basic, Visual C++, Java, etc.) and the present invention is not limited
to hardware independent mark-up languages. The user then inputs any known
general mufti-dimensional information for the entity or transformation. The
1o user then inputs any known general or specific mufti-dimensional
information
for an entity or transformation.
Not all of the categories of mufti-dimensional information can be input
for every biological entity 128. For some biological entities 128, all of the
categories of mufti-dimensional information may be known. For other
biological entities 128, only some of the categories mufti-dimensional
information may be known, so only the known information is input.
Table 1 illustrates exemplary general mufti-dimensional information
input that maybe by a user at Step 56 for general mufti-dimensional hierarchy
114 for the exemplary EGF pathway. However, the present invention is not
limited to the general mufti-dimensional information illustrated in Table 1 or
the hierarchy 114 for inputting general mufti-dimension information. More,
less or equivalent general mufti-dimensional information can be used.
Cate o Descri tion
S ecies 116 Human
Ex erimental S stem 118 Skeletal Muscle
Functional T a of Entit 120 EGF, EGF rece for
Transformation 122 EGF bindin to EGF rece for
Compartment 124 Cell membrane
Table 1.
23
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
Table 2 illustrates exemplary specific mufti-dimensional information
that may be input by a user for EGF signaling molecule 90_ (i.e., a functional
type for an entity) at Step 56 based on the entity hierarchy 126 (FIG. 5).
However, the present invention is not limited to the mufti-dimensional
information illustrated in Table 2 or the entity hierarchy 126.
In addition, a morphology 132 of the biological entity 128, an optional
electron microscope ("EM") photograph 134 and an optional fluorescent view
136 of the biological entity 128 may also be input by a user (e.g., by
inputting
a link to a file or location including such information).
Cate o Descri tion
Basic Information 138 EGF 78 is a globular protein
of 6.4 kDa
comprising 53 amino acids. It
includes
three intra-molecular disulfide
bonds
essential for biological activity.
Site 140 Extracellular si nalin molecule.
Function 142 Activates encoding of an intrinsic
tryosine-specific protein kinase
activity.
This kinase activity catalyses
the
transfer of the gamma-phosphate
of
ATP to a tryosine resiude of
the
receptor and also of some other
intra-
cellular roteins.
Enz me 144 T osine-s ecific rotein kinase
Reactions 146 The EGF precursor is N-glycosylated
and contains a hydrophobic domain
allowing it to be anchored in
the cell
membrane. In cells that do not
cleave
this precursor (e.g., Kidney
cells), the
membrane-bound form of the precursor
may itself serve as a receptor
for yet
unknown ligands. EGF 78 may
be
involved in Juxtacrine growth
control
mechanisms.
Trans ort S stem 148 NA
Pathwa s 150 NA
Table 2.
FIG. 6 is a block diagram illustrating an exemplary mufti-dimensional
hierarchy for a transformation 152. The transformation hierarchy 152 includes
a first level for a transformation identifier 154, type 156, name 158, role
160,
24
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
and group type 162. The transformation hierarchy includes a second level for a
transformation input 164, output 166, 1<ey 168, and effectors 170. Multi-
dimensional information input for a transformation is stored as a local
database. In addition, the mufti-dimensional information for a transformation
can also be represented with a directed graph or a combination of a hierarchy
and/or a directed graph as was described above. However the present
invention is not limited to this exemplary transformation hierarchy and have
fewer or equivalent transformation levels can also be used.
Table 3 illustrates exemplary specific mufti-dimensional information
1o input by a user for the transformation 92 from EGF signaling molecule 90 at
Step 56 based on the transformation hierarchy 152 (FIG. 6). However, the
present invention is not limited to the specific mufti-dimensional information
illustrated in Table 3 or the transformation hierarchy 152.
Cate o Descri tion
Transformation Identifier 156 EGFs
T a 158 Rece tor/li and interaction
Name 160 EGF
Role 162 Extracellular si nalin
Group Type 164 Currently used for a group of
transformations. A group type
can be
simultaneous, cou led, etc.
In ut 166 EGF molecule 90, EGF rece for
Out ut 168 EGF, EGR rece for com lex
Ke 170 EGF1
Effectors 172 EGF rece for 94
Table 3.
In one specific embodiment of the present invention, when the shape or
arrow is placed at a location on the GUI 64, it is placed with a first color
(e.g.,
red). When mufti-dimensional information is input, the shape or arrow is
changed from a first color to second color (e.g., green). The colors allow a
user to visually determine if mufti-dimension information has been input for
2o the entity or transformation. The second color allows a user to visually
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
determine an aggregated view of the multi-dimensional information for the
shape or arrow.
In FIG. 2B at Step 58, related information, if any, is input to link the
shape and arrow to other information related to the entity and transformation
from plural external databases. In one exemplary embodiment of the present
invention, the related information is input and stored in a hierarchy. In
another exemplary embodiment of the present invention, the related
information is input and stored in a non-hierarchical manner.
In one exemplary embodiment of the present includes specifying
to related information including information about entities, including
detailed
information such as assays including an experimental protocol used to test the
entity or transformation; compounds, including compounds that are effective
on selected entities or transformations; diseases, including known diseases
that
are related to the selected entities or transformations; authors, including
other
15 authors who have expertise in the selected entities or transformations;
expression, including gene expression related to the selected entity or
transformation; validation, including a level of credibility of the existence
and
role of the selected entity or transformation; or other pathways, including
other
pathways that the selected entities or transformations participate in.
However,
2o more or fewer related information can also be specified and the present is
not
limited to this list of related information.
In one embodiment of the present invention, a validation level is
assigned in one of two ways: (1) manual assignment by an editorial board; or
(2) using an automated method. If manual assignment is completed, an
25 editorial board made up of scientists will confer to manually assess the
26
CA 02363020 2001-08-15
WO 00/49540 PCT/US00104331
credibility of the information associated with an entity or transformation. A
validation weight (e.g., from zero to ten) is assigned. A validation weight of
zero indicates a lowest level of validity for the information (e.g., results
from a
single experiment). A validation weight of ten indicates a very high level of
validity for the information (e.g., similar results obtained from many
different
experiments).
If automatic assignment is completed, an automated method is used to
take into account multiple pre-determined factors that contribute to the
validity
of a piece of biological information. The predetermined factors are evaluated
1 o to calculate a validation weight. The pre-determined factors may include,
but
are not limited to, such factors as a number of experiments or references used
to create the information, a quality of a source of an experiment or
reference,
what type of experiment was used to acquire the information, a reputation, if
any, of the researcher that supplied the information, etc.
In one embodiment of the present invention, when a user is creating a
biological pathway and has selected a shape or arrow, (e.g., by "clicking" on
it), an input form is presented to the user so the user can input any known
related information for an entity or transformation.
In such an embodiment, an electronic input form created in HTML,
XML or other hardware independent mark-up languages known in the art is
displayed for a user. However, virtually any programming language can be
used to create and display the electronic input form (e.g., C, C++, Visual
Basic, Visual C++, Java, etc.) and the present invention is not limited to
hardware independent mark-up languages. The user then inputs any known
related information.
27
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
As was discussed above for mufti-dimensional information, not all
categories of related information can be input for every entjty. For some
entity all of the categories of related information may be known. For other
entities, only some of related information may be known, so only known
information is input. For still other entities, none of the categories of
related
information may be known, so no related information will be input.
Table 4 illustrates exemplary related information input by a user for
EGF signaling molecule 90 at Step 58. However, the present invention limited
to the related information illustrated in Table 4.
Cate o Descri tion
Assa s 220 NA
Com ounds 222 NA
Diseases 224 Human Cancers
Authors 226 Shigeo Tsuchiya, et al.,
Solution Structure of SH2 Domain
of
Grb2/Ash Complexed with EGF
Receptor-Derived Phosphotyrosine-
Containing Peptide, J. Biochem.
125,
1151-1159 1999 .
Ex ression 228 NA
Validation 230 10
Other Pathwa s 232 PDGF
to 1 able 4.
In one specific embodiment of the present invention, when related
information is input, if any, the shape is changed from a second color (e.g.,
green) to a third color (e.g., blue). The third color allows a user to
visually
determine if both mufti-dimensional and related information has been input for
the shape.
Returning to FIG. 2B at Step 60, a test is conducted to determine if a
desired number of iterations of Steps 50, 52, 54, 56 and 58 have been
completed. If a desired number of iterations have not been reached at Step 60,
a loop continues at Step 48 of FIG. 2A until the desired number of iterations
has been completed.
28
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
In this specific example Steps 50, 52, 54, 56 and 58 are repeated five
times adding shapes 94, 96, 98, 100 and 102 with connecting arrows 104, 106,
108, 110 and 112, respectively, via the GUI 64 of FIG. 3. In this specific
illustrative example, shape 94 represents an active entity for an EGF receptor
("EGFr"). Shape 96 represents an active entity for a Growth factor receptor
bound protein 2 ("Grb2"). Shape 98 represents an active entity for Son of
sevenless ("Sos"). Shape 100 represents an inactive entity for Ras ("iRAS")
Shape 102 represents an active entity for Ras ("aRAS"). The function of these
shapes as used in the exemplary EGF pathway is explained below.
1o As is known in the biological arts, the EGF receptor 94 is a 170 kDa
transmembrane glycoprotein. An extra cellular receptor domain contains an
EGF binding site and also binds mammalian TGF-alpha. An intracellular
receptor domain encodes an intrinsic tyrosine-specific protein kinase. This
kinase catalyses the transfer of the gamma-phosphate of ATP to a tyrosine
residue of the receptor and also of some other intracellular proteins. The
intracellular kinase domain of the EGF receptor 94 is activated by binding of
EGF or TGF-alpha to the extracellular receptor domain. The EGF receptor 94
is also phosphorylated by protein kinase-C at serine and threonine residues.
Grb2 96 is an adaptor protein with a domain structure (SH3-SH2-
2o SH3). The two SH3 domains bind to protein sequences in a carboxyl terminal
region of a guanine nucleotide to exchange Sos 98. Upon EGF stimulation 90,
Grb2 96 binds to the EGR receptor 92 directly or indirectly through proteins
such as Shc, FAK, Syp and IRS-l, by recognizing phosphotyrosine-containing
sequences to allow interaction with inactive Ras 100.
29
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
Sos 98 is a guanine nucleotide exchange factor for inactive Ras 100
that binds to Grb2 96. Sos 98 mediates the coupling of receptor tyrosine
kinases for inactive Ras 100 activation. Sos 98 is also associated with ligand-
activated tyrosine kinase receptors which bind Grb2 96. At the cell membrane
76, Sos 98 can catalyze the exchange of GDP for GTP bound to inactive Ras
100, thereby activating active Ras 102 from inactive Ras 100.
Ras 100,102 is a super-family of small GTPases including a single
GTPase domain. Ras is active 102 in its GTP bound state. Ras is inactive 100
in its GDP state. Ras 100,102 activity is positively regulated by EGF's 90.
1 o Inactive Ras 100 proteins are generally associated with cell membranes 76
via
prenylation near their C-terminus. Active Ras 102 proteins are generally
associated with cell cytoplasm.
A desired number of iterations have been completed at Step 60 when a
portion of a biological pathway or a complete biological pathway has been
input. In the specific example, after five iterations at Step 62, information
associated with the plural shapes connected with plural arrows is saved as a
biological pathway with pre-determined relationships in database in a pre-
determined format.
In one exemplary preferred embodiment of the present invention,
2o information associated with a biological pathway, whose structure is
defined
by a hardware independent XML Document Type Definition ("DTD") that is
stored in a local file. A specific exemplary XML document used to store a
biological pathway is illustrated in Table 5. However, the present invention
is
not limited to the XML DTD in Table 5 or to storing a biological pathway in
an XML format and other similar or equivalent formats can also be used.
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
COPYRIGHT ~ 1999, by Cellomics, Inc. All rights reserved.
<!--A DTD for cellular pathway information: PML.dtd
<!--Author(s): Jian Wang -->
<!__ Copyright ~ 1999, Cellomics, Inc. All rights reserved.-->
<!ELEMENT Pathways (Pathway*)>
<!-- ref defines the references used in an xml doc. Reflink at this level
links to the references that is generic to the whole pathway. Reflink at other
levels are
references specific to that level -->
<!ELEMENT Pathway
((BioSysIComponent~Cell_Compartment~Cellular-Process~Functional-
unit~Transformat
ionslFeatureinfolRef)*,RefLink?, Notes*)>
<!ATTLIST Pathway
Pathway-ID ID #REQUIRED
PathwayName CDATA #IMPLIED>
<!ELEMENT BioSys (Organism?,System?,Organ?,Tissue?,Cell?,Notes*)>
<!ATTLIST BioSys
BioSys - ID ID #REQUIRED>
<!ELEMENT Organism EMPTY>
<!ATTLIST Organism
Organism CDATA #REQUIRED
DevStage CDATA #IMPLIED>
<!ELEMENT System EMPTY>
<!ATTLIST System
System CDATA #REQUIRED
DevStage CDATA #IMPLIED>
<!ELEMENT Organ EMPTY>
<IATTLIST Organ
Organ CDATA #REQUIRED
DevStage CDATA #IMPLIED>
<!ELEMENT Tissue EMPTY>
<!ATTLIST Tissue
Tissue CDATA #REQUIRED
DevStage CDATA #IMPLIED>
<!ELEMENT Cell EMPTY>
<!ATTLIST Cell
Cell CDATA #REQUIRED
CeIICycIeStage CDATA #IMPLIED
DevStage CDATA #IMPLIED>
<!ELEMENT Cell - Compartment (#PCDATAINotes)*>
<!ATTLIST Cell Compartment
Compartment - ID ID #REQUIRED
Compartment-Name CDATA #REQUIRED>
<!ELEMENT Cellular Process (#PCDATAINotes)*>
<IATTLIST Cellular Process
Process ID ID #REQUIRED
Process-Name CDATA #REQUIRED>
<!ELEMENT Component ((Abbreviation~Modification~Synonym)*, Notes*)>
31
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
<!Al-fLIST Component
Component ID ID #REQUIRED _
Component Name CDATA #REQUIRED BioSys IDREF #REQUIRED>
<!ELEMENT Modification (#PCDATAINotes)*>
<!ATTLIST Modification
Modification Site CDATA #IMPLIED Modification Type CDATA #REQUIRED>
EMENT Functional - Unit (ComponentLink*, Synonym*, RefLink?, Notes*)>
<!ATTLIST Entity
Functional Unit
Unit ID ID #REQUIRED
Unit Name CDATA #REQUIRED
Unit-Abbr CDATA #IMPLIED
BioSys IDREF #REQUIRED
X Coord CDATA
Y-Coord CDATA #IMPLIED
Shape (CIRCLE~POLYGON~SQUARE~OVAL~RECTANGLE) "CIRCLEII>
<!__ the following "SimpIeLink" points to the ID of a defined component or
functional - unit
or cell_compartment or cellular_process. The above can be accomplished by
using
IDREF instead of Simple Links However, it may be more extensible using links
since we
know that the component definitions will be on the server somewhere (outside
of any
specific xml doc) in the future. -->
:LEMENT ComponentLink (SimpIeLink, Notes*)> <!ATTLIST ComponentLink
NumberOfComponent CDATA #IMPLIED
InCompartment IDREF #REQUIRED
UniformlnCompartment (TRUEIFALSE) "TRUE">
<!ELEMENT Synonym (Abbreviat-i-on*, Notes*)>
<IATTLIST Synonym
Synonym DDATA #REQUIRED>
<!ELEMENT Abbreviation (#PCDATAINotes)*>
<!ATTLIST Abbreviation
Abbreviation CDATA #REQUIRED>
<!-- having a RefLink element is for the sole purpose of making the xml doc
more
readable; otherwise one would not know what the extended link is all about
since the
"ExtendedLink" element is reused extensively. In this case, the href attribute
should point
to some defined reference in the same xml doc using XPointers: "#IDo"-->
<!ELEMENT RefLink (ExtendedLink)>
LEMENT Ref ((PublicationlPerson~Organization)*, Notes*)> <!ATTLIST Ref
Ref ID ID #REQUIRED
Date-Month CDATA #IMPLIED
Date-Day CDATA #IMPLIED
Date-Year CDATA #IMPLIED>
<!-- the following simplelink links to a medline record --> <!ELEMENT
Publication
(Person*, SimpIeLink, Note?)>
<!ATTLIST Publication
Title CDATA #IMPLIED
Journal CDATA #IMPLIED
32
CA 02363020 2001-08-15
WO 00/49540 - PCT/US00/04331
Publisher CDATA #IMPLIED
PageStart CDATA #IMPLIED
PageEnd CDATA #IMPLIED
Volume CDATA #IMPLIED
Issue CDATA #IMPLIED
Type CDATA #IMPLIED
Date - month CDATA #IMPLIED
Date_Day CDATA #IMPLIED
Date - Year CDATA #IMPLIED >
<!ELEMENT Person (Organization*, Notes*)> <!ATTLIST Person .
FirstName CDATA #IMPLIED
Middlelnit CDATA #IMPLIED
LastName CDATA #IMPLIED
StreetAddress CDATA #IMPLIED
City CDATA #IMPLIED
State CDATA #IMPLIED
ZipCode CDATA #IMPLIED
AreaCode CDATA #IMPLIED
PhoneNum CDATA #IMPLIED
Ext CDATA #IMPLIED
Email CDATA #IMPLIED
Web CDATA #IMPLIED
Role CDATA #IMPLIED>
<!ELEMENT Organization (#PCDATAINotes)*>
<!ATTLIST Organization
Name CDATA #REQUIRED
Type (commerciallAcademiciGovernment) #REQUIRED>
<!-- "Role" describes the function of some item in a collection, such as "rate
limiting" -->
<!ELEMENT Transformations ((TransformationlTransformationslEffectors)*,
RefLink?, Notes*)>
<!ATTLIST Transformations
Transformations - ID ID #REQUIRED
Transformations-Type CDATA #IMPLIED
Transformations Name CDATA #IMPLIED
Role CDATA #IMPLIED
Group Type CDATA #IMPLIED>
<!ELEMENT Transformation (Input+,Output+,Effectors*,RefLink?, Notes*)>
<!ATTLIST Transformation
33
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
Transformation ID ID #REQUIRED
TransformationType CDATA #IMPLIED
Transformation Name CDATA #IMPLIED
Role CDATA #IMPLIED>
<!-- Input, Output and Effector reference Unit -->
<!ELEMENT Input (#PCDATAINotes)*>
<!ATTLIST Input
Input-ID IDREP #REQUIRED>
<!ELEMENT Output (#PCDATAINotes)*>
<!ATTLIST Output
Output - ID IDREF #REQUIRED>
<!ELEMENT Effectors (Effector+, Notes*)>
<!ATTLIST Effectors
Group Type (synergismlxyz) "synergism">
<!ELEMENT Effector (#PCDATAINotes)*>
<!ATTLIST Effector
Effector ID IDREF #REQUIRED
Effect-Type CDATA #IMPLIED
Role CDATA #IMPLIED
Is-Positive (TRUEIFALSE) "TRUE">
<!-- Feature-ID references an object of the type specified by Feature Type -->
<!ELEMENT Featurelnfo (ExtendedLink, Notes*)>
<!ATTLIST Featurelnfo
Feature ID IDREF #REQUIRED
Feature Type (ComponentlunitiTransformations)
Info Type
(EntitylAssayICompoundlReferencelPathwaylDiseaselCredibility) "Entity">
<!ELEMENT ExtendedLink (LinkLocator*, Notes*)>
<!ATTLIST ExtendedLink
XML-LINK CDATA #FIXED "EXTENDED"
ROLE CDATA #IMPLIED
TITLE CDATA #IMPLIED
INLINE (TRUEIFASLE) "TRUE"
SHOW (EMBEDIREPLACEINEW) "REPLACE"
ACTUATE (AUTOJUSER) "USER">
<!ELEMENT LinkLocator (#PCDATAINotes)*>
<!ATTLIST LinkLocator -
XML-LINK CDATA #FIXED "LOCATOR-'
ROLE CDATA #IMPLIED
HREF CDATA #REQUIRED
TITLE CDATA #IMPLIED
SHOW (EMBEDIREPLACEINEW) "REPLACE"
34
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
ACTUATE (AUTOJUSER) "USER">
<!ELEMENT SimpIeLink (#PCDATAINotes)*>
<!ATTLIST SimpIeLink
XML-LINK CDATA #FIXED "SIMPLE"
HREF CDATA #REQUIRED
TITLE CDATA #IMPLIED>
<!ELEMENT Notes #PCDATA >
Table 5.
FIG. 3 illustrates a portion of the exemplary EGF pathway including
spatial information and determined relationships between entities and
transformations from the extracellular EGF signal 90, through the cell
membrane 76 via EGF receptor 94, Grb2 96, Sos 98 and inactive Ras 100, and
into the cell cytoplasm via active Ras 102.
Method 46 allows a user to dynamically build (or edit) and save
information associated with a biological pathway that represents a biological
function with determined relationships. Method 46 allows information about a
1o biological entity to be organized into a hierarchy including multiple
dimensions of information. Spatial information about each entity or
transformation is captured by associating an entity or transformation with a
specific cellular component (e.g., cell membrane 76). Varying shapes are used
to represent different entities and transformations in a biological pathway.
Displaying experimental information with determined relationships from
a local computer
FIG. 7 is a flow diagram illustrating a Method 174 for dynamically
displaying experimental information including determined relationships. At
Step 176, a biological pathway with determined relationships is selected from
2t1 a list of biological pathways displayed on a graphical user interface on a
computer. At Step 178, a display mode used to display the biological pathway
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
is selected from the graphical user interface. The display mode allows
hierarchical information associated with the selected biolo-gical pathway with
determined relationships to be displayed on the graphical user interface. At
Step 180, a graphical representation of the selected biological pathway with
determined relationships is dynamically generated on the graphical user
interface using associated information from a database for the selected
biological pathway and the selected mode of operation. The graphical
representation of the selected biological pathway is not stored in a database,
but dynamically generated from information in a database. The selected
1o biological pathway is dynamically generated using a first set of colors to
indicate a level of generalization in a multi-dimensional hierarchy used to
display individual components of the biological pathway.
Method 174 (FIG. 7) is illustrated with GUI 64 (FIG. 3) with the
portion of the cellular Epidermal Growth Factor ("EGF") signaling pathway
15 input and stored using Method 46 (FIG. 2). However, the present invention
is
not limited to such an embodiment and Method 174 can be used with
biological pathways that were input and stored with other methods.
In such an embodiment at Step 176, the EGF biological pathway is
selected from a list of biological pathways displayed on a graphical user
20 interface on an internal or local computer 12, 14. The information
associated
with the biological pathways was stored in a local database using Method 46.
In this embodiment, the information associated with the biological pathways
includes the mufti-dimensional and related information described above for
Method 46. In such an embodiment, the list of biological pathways can be
25 displayed by selecting the graphical explore button 84 from the GUI 64.
36
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
When the graphical explore button 84 is selected a list of saved biological
pathways is displayed for a user. In one embodiment of the present invention,
the list of biological pathways is created dynamically from a database. In
another embodiment of the present invention, the list of biological pathways
is
displayed from a static list saved in a database.
At Step 178, a display mode to display the biological pathway is
selected from the graphical user interface. The display mode allows
hierarchical information associated with the selected biological pathway with
determined relationships to be displayed on the graphical user interface. In
to this specific embodiment, the display mode of operation includes a summary,
dimension and a link display mode. However, the present invention is not
limited to these display modes and more, fewer or equivalent display modes
can also be used. The display modes allow a user to view information
associated with an entity or transformation in a hierarchical fashion from
general to specific.
The "summary" display mode allows a user to view general multi-
dimensional information about a selected entity or transformation in a
selected
biological pathway (e.g., from hierarchy 114). The summary display mode
includes displaying graphical shapes of varying colors and arrows representing
2o a general level for entities and transformations in a biological pathway.
Visiting a pre-determined level in the summary mode may automatically
switch the user into the dimension mode and/or the link mode.
The "dimension" display mode allows a user to view general or
specific mufti-dimensional information associated with entities or
transformations in a biological pathway (e.g. from hierarchy 126 and 152).
37
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
The dimension display mode allows a user to electronically link to other multi-
dimensional information stored in the internal databases. In such an
embodiment of the present invention, the mufti-dimensional information is
obtained exclusively from local databases. In another embodiment of the
present invention, the mufti-dimensional information is obtained from the
local databases as well as from the public domain databases.
The "link" display mode allows a user to view related information
stored in external databases associated with entities and/or transformations
in a
biological pathway. In one embodiment of the present invention, related
to information for the link mode is obtained exclusively from external
databases.
As a result, the link mode includes use of additional network security
features
(e.g., logins, passwords, firewalls, encryption, other secure transfer, etc.)
to
protect the integrity of the private network 16. In other embodiment of the
present invention, related information for the link mode is obtained from the
15 external databases and the internal databases. In such an embodiment, all
or
selected portions of related information from the external databases may be
cached in one or more of the internal databases or in random access memory
for quicker access and display after any of the related information is
accessed
once from the external databases.
2o At Step 178, a graphical representation of the selected EGF biological
pathway with determined shapes and arrows is dynamically generated with a
first set of colors on the graphical user interface using associated
information
from the internal database for the selected biological pathway and the
selected
mode of operation. The first set of colors is used to indicate a level of
25 generalization in a mufti-dimensional hierarchy used to display the
biological
38
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
pathway. The first set of colors may include, for example, red, orange,
yellow, green, blue, indigo and violet to indicate a highest level or general
level, to a lowest level, or most specific level, in the mufti-dimensional
hierarchy.
For example, FIG. 3 illustrates a portion of the EGF pathway as
displayed in the summary mode. The graphical representation of the EGF
pathway as illustrated in FIG. 3 is not stored in a database. That is, shapes
92,
94, 96, 98, 100, 102 and arrows 104, 106, 108, 110, 112 are not stored in a
database. Instead an identifier for the shapes and arrows are stored in a
to database as pathway information (e.g., XCoord, YCoord and Shape indication
of a Functional-Unit in the XML DTD from Table 5). When database records
are read for a selected biological pathway information in the database records
are used to dynamically generate a graphical shape in a desired location that
is
displayed on the GUI 64 as is illustrated in FIG. 3.
FIG. 8 is a block diagram illustrating an exemplary general multi-
dimensional information page 182 that is dynamically created and displayed
for a user in the summary display mode. A similar page may be dynamically
created and displayed to input general mufti-dimensional information. A
general mufti-dimensional information electronic display page is dynamically
2o created from information in the local databases in a hardware independent
mark-up language and displayed for the user.
For example, an electronic display page is created HTML, XML or
other hardware independent mark-up languages known in the art. However,
virtually any programming language can be used to create and display the
electronic display page (e.g., C, C++, Visual Basic, Visual C++, Java, etc.)
39
CA 02363020 2001-08-15
WO 00/49540 PCTlUS00/04331
and the present invention is not limited to hardware independent mark-up
languages.
The general multi-dimensional information page 182 includes a display
field for a species 184, an experimental system 186, a functional unit for an
entity 188, a transformation 190 and a compartment 192. The contents of
these fields were discussed above for the multi-dimensional hierarchy 114
(FIG. 4). The multi-dimensional information page 182 illustrated in FIG. 8 is
dynamically created from the exemplary multi-dimensional information input
at Step 56 (FIG. 2A) and illustrated in Table 1 above. Such multi-dimension
to information is created from a hierarchy and/or a directed graph as was
discussed above.
The mufti-dimensional information page 182 also includes electronic
links to other mufti-dimensional information. For example, in the functional
unit display field 188, the letters "EGF" is underlined indicating an
electronic
link to additional specific mufti-dimensional information for a cell (e.g.,
from
entity hierarchy 126 of FIG. 5).
The summary mode also allows a user to "zoom in" and "zoom out" to
view more detailed information associated with an entity or transformation in
a selected biological pathway. The zooming is completed by selecting the
2o graphical zoom button 78 on the GUI 64. Zooming to a pre-determined level
in the summary mode may automatically switch the user into the dimension
mode and/or the link mode. The summary mode also allows a user to pan
back and forth on the GUI 64 to view multiple cells displayed on the GUI 64
for a biological pathway that may be inter-cellular. The panning is completed
by selecting the graphical pan button 80 on the GUI 64.
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
FIG. 9 is a block diagram illustrating an exemplary specific multi-
dimensional information page 194 for a biological entity such as a cell. This
display page can be dynamically displayed by selecting the "MULTI Button"
86 from the GUI 64 (FIG. 3) or by access from another display (e.g., clicking
on the word CELL in the functional unit display field 188 (FIG. 8) mode. The
mufti-dimensional information page 194 includes a display field for a
morphology 196, an optional EM photograph 198 and an optional fluorescent
view 200. These display fields correspond to the mufti-dimensional
information from the entity hierarchy 126 (FIG. 5) that was input at Step 56
(FIG. ZA). Such mufti-dimension information is created from a hierarchy
and/or a directed graph as was discussed above. The mufti-dimensional
information page 194 also includes display fields for basic information 202,
site information 204, functions 206, enzymes 208, if any, reactions 210, a
transport system 212, and a pathway view 214.
FIG. 9 illustrates an exemplary specific mufti-dimensional information
page 194 at an entity level in a mufti-dimensional hierarchy that might be
displayed in a dimension display mode for extracellular EGF signal 90 on the
GUI 64 (FIG. 3). Such mufti-dimension information is created from a
hierarchy and/or a directed graph as was discussed above. The multi-
2o dimensional information page 194 illustrated in FIG. 9 is dynamically
created
from the exemplary mufti-dimensional information input at Step 56 (FIG. 2A)
and illustrated in Table 2 above. Other entities in the EGF pathway would
have similar mufti-dimensional information pages. Transformations in the
EGF pathway would also have similar mufti-dimensional information pages
dynamically generated and displayed (e.g., based on hierarchy 152).
41
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
The specific mufti-dimensional information page 194 also includes
electronic links to other information. For example, in the basic information
display field 202, the letters "EGF" are underlined indicating an electronic
link
to additional related information in a local database. In this example,
selecting
the electronic link for EGF would link the user to a three-dimensional
graphical display of the EGF signal molecule. The remaining underlined text
on the mufti-dimensional information page 194 also indicates electronic links
to additional information in local databases.
FIG. 10 is a block diagram illustrating an exemplary related
1o information page 216 that is dynamically created and displayed for a user
in a
link display mode. A related information electronic display page is
dynamically created from information in external databases and/or cached in
local databases in a hardware independent mark-up language and displayed for
the user.
For example, an electronic display page is created in XML, HTML or
other hardware independent mark-up languages known in the art. However,
virtually any programming language can be used to create and display the
electronic display page (e.g., C, C++, Visual Basic, Visual C++, Java, etc.)
and the present invention is not limited to hardware independent mark-up
languages.
The related information page 216 includes, but is not limited to, a
display field for entities 218, assays 220, compounds 222, diseases 224,
authors 226, expression 228, validations 230 and other known pathways 232
this entity or transformation participates in.
42
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
FIG. 10 illustrates an exemplary related information page 216 that
might be displayed in the link mode for extracellular EGF signal 90 on the
GUI 64 (FIG. 3). The related information page 216 illustrated in FIG. 10 is
dynamically created from the exemplary related information input at Step 58
(FIG. 2A), illustrated in Table 4 above and stored in external databases.
The related information page 216 may also include electronic links to
other remote information. Such electronic links are also illustrated with
underlined text in FIG. 10. For example, in the authors field 226 the author,
SHIGEO TSUCHIYA, is underlined indicating an electronic link to other
1 o related works by the same author stored in external databases on a public
network like the Internet.
As was discussed above, the link display mode includes use of
additional network security features (e.g., logins, passwords, firewalls,
encryption, other secure transfer, etc.) to protect the integrity of the
private
network 16. Selected portions of related information from the external
databases may be cached in one or more of the internal databases for quicker
access and display after any of the related information is accessed once from
the external databases.
Displaying experimental information with determined relationships from
2o a remote computer
The present invention has been described with respect to use from
internal or local computers 12,14 on private LAN 16. In such an
embodiments, information associated with a biological pathway with
determined relationships may be stored in a local proprietary database 18, 20
43
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
without public access. Such information may be used for private research and
may never be made available to the public.
In another embodiment, information associated with a biological
pathway with determined relationships may be stored in a local database with
a public access portion 24, 26. Such information may be made available to the
public when the research used to generate the information is at a stage
appropriate for public review or public disclosure. Such information can be
used to quickly make the new research information available to a large number
of people via the public network 28 for critical review.
1o However, the present invention can also be used from external
computers 30, 32, 34, 36 via public network 28 to input and/or access and
display information from a private organization. For example, Method 46
(FIG. 2) may be used from external computers 30, 32, 34, 36, to input and/or
edit a biological pathway with determined relationships that can immediately
15 be shared by a large number of people via the public network 28.
In such an embodiment, any information associated with a biological
pathway with determined relationships may be temporarily stored in a local
database associated with the external computers (not illustrated in FIG. 1 )
and
then transferred to the internal databases with public access 24, 26 on the
2o private LAN 16. The information may also be transferred directly to the
internal databases with public access 24, 26 on the private LAN 16 as the
information is input. Related information may also be transferred to one or
more of the plural public domain databases 38, 40, 42, indirectly or directly.
An organization that owns the private intranet LAN 16 may designate its
25 internal databases with public access 24, 26 as an information repository
and
44
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
allow members of the public to input, access, display and share such
information to aid and further advance biological research on a world-wide
basis.
FIG. 11 is a flow diagram illustrating a Method 234 for dynamically
displaying experimental information including determined relationships
displaying from a remote computer. At Step 236, a request is made on a
graphical user interface on remote computer connected to a public network, to
select a biological pathway with determined relationships from a private
database server connected a private network. The private network includes
to plural private databases with public access including information
associated
with plural of biological pathways with determined relationships. At Step
238, a display mode is selected to display the biological pathway with
determined relationship from the graphical user interface on the remote
computer. The display modes allows hierarchical information associated with
15 the biological pathway with determined relationships to be displayed on the
graphical user interface. At Step 240, a first portion of information
associated
with the selected biological pathway with determined relationships is received
from the plural private databases via the private database server on the
private
network in a hardware independent mark-up language on the remote
2o computer. At Step 242, a second portion of information associated with the
selected biological pathway with determined relationships from plural public
databases via one or more public database servers on the public network. At
Step 244, a graphical representation of the selected biological pathway with
determined relationships is dynamically generated on the graphical user
25 interface on the remote computer using the selected display mode, the first
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
portion of information from the private network and the second portion of
information from the public network, thereby creating a graphical
representation of the selected biological pathway with determined
relationships with information from a plurality of private databases and with
information from a plurality of public databases.
Method 232 (FIG. 11) is illustrated with a specific example from
remote computer 30 including GUI 64 (FIG. 3). However, the present
invention is not limited to this specific example virtually any biological
pathway can be input, displayed and manipulated from a remote computer
1o using Method 232 and GUI 64.
In such an embodiment, at Step 234 a request is made on the GUI 64
on the remote computer 30 connected to the Internet 28, to select a biological
pathway (e.g., the EGF signaling pathway) with determined relationships from
a private database server 22 connected a private intranet LAN 16. The
15 selection includes inputting a new biological or requesting a previously
saved
biological pathway with determined relationships. At Step 236, a display
mode is selected to display the biological pathway from the GUI 64 on the
remote computer 30. The display mode includes the summary, dimension and
link display modes described above. However, other display modes can also
2o be used on the present invention is not limited to these display modes.
Step 238, a first portion of information associated with the selected
biological pathway is received from the plural private databases 24, 26 via
the
private database server 22 on the private intranet LAN 16 in a hardware
independent mark-up language on the remote computer. The first portion of
25 information includes information in XML, HTML or other hardware
46
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
independent mark-up languages. At Step 240, a second portion of infornlation
associated with the selected biological pathway with determined relationships
from plural public databases 38, 40, 42 via one or more public database
servers on the Internet 28. The second portion of information also includes
information in XML, HTML or other hardware independent mark-up
languages.
In one embodiment of the present invention, the first portion of
information includes the XML conforming to the DTD illustrated in Table 5.
The second portion of the information includes XML data (e.g., electronic
to links or actual information) that are used with the XML DTD to dynamically
generate the biological pathway and related information.
In another embodiment of the present invention, the first portion of
information and the second portion of information each include discrete XML
data that is combined and used to dynamically generate a graphical
15 representation of the selected biological pathway with determined
relationships. However, other types of data can also be used for the first
portion and the second portion of information, and the present invention is
not
limited to the XML data described.
At Step 242, a graphical representation of the selected biological
2o pathway with determined relationships is dynamically generated on the GUI
64 on the remote computer 30 using the selected display mode, the first
portion of information from the private intranet LAN 16 and the second
portion of information from the Internet 28. This creates a graphical
representation of the selected biological pathway with determined
47
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/0433t
relationships with information from a plural private databases 24, 26 and with
information from a plural public databases 38, 40, 42
In one embodiment of the present invention, the first portion of
information includes general and/or mufti-dimensional information (e.g.,
FIGS. 8 and 9) for a biological entity or a transformation is stored in the
plurality of private databases 24, 26 on the private network 16. The second
portion of information includes related information (e.g., FIG. 10) for a
biological entity or transformation is stored in the plural public databases
38,
40, 42, on the public network 28. The second portion of the information may
to include electronic links to related information or actual electronic
information.
In one embodiment of the present invention, Step 242 includes
dynamically generating the graphical representation of the selected biological
pathway with a first set of colors on the GUI 64 on the remote computer 30.
As was described above, the first set of colors is used to indicate a level of
generalization in a hierarchy or directed graph used to display the biological
pathway on the GUI 64.
The graphical representation of the selected biological pathway is
generated "seamlessly" so a user is not able to visually determine by
observing
the selected biological pathway that information used to create it came from
2o plural databases on private and public networks.
A user on a remote computer can also input and/or modify information
and/or dynamically generate a selected biological pathway with Method 46
(FIG. 2) or Method 232 (FIG. 11 ). In such an embodiment, a request is
received to change the selected biological pathway with determined
relationships. Any changes relating to the first portion of information used
to
48
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
display the selected biological pathway is sent to the private database server
22 on the private network 1 G to update appropriate private databases in the
plural local databases 24,26 on the private network 16. Any changes relating
to the second portion of information used to display the selected biological
pathway is sent to an appropriate public database server on the public network
28 to update appropriate public databases in the plural databases 38, 40, 42,
on
the public network 28. In one embodiment of the present invention, only
changes to the first portion of the information is allowed. In another
embodiment, only changes to the second portion of the information is allowed.
In another embodiment of the present invention, changes to both the first
portion and the second portion of information are allowed.
The methods and system described herein may provide at least the
following advantages. An input/edit tool (e.g., GUI 64) is provided to
input/edit a biological pathway with determined relationships using predefined
entities and transformation templates (e.g., shapes and arrows) to capture
information about that pathway as it is drawn. Spatial information about
entities and transformations is captured by associating an entities and
transformations with specific biological compartments. Graphical biological
pathway diagrams are dynamically generated to represent biological functions.
2o A navigation tool (e.g., GUI 64) is provided to retrieve information
associated with selected biological entities or transformations from local and
remote databases. Information is presented hierarchically, from more general
to more specific. Color-coding is used to reflect levels of generalization.
Entity and transformation information is organized into hierarchical
dimensions. Users can selectively expand and/or collapse parts of the
49
CA 02363020 2001-08-15
WO 00/49540 PCT/US00/04331
graphical pathway, or rearrange the layout of the pathway. The methods and
system may also be used to provide new bioinformatic techniques used to
make observations about biological pathways, such as cell pathways, with
determined relationships.
It should be understood that the programs, processes, methods and systems
described herein are not related or limited to any particular type of computer
or network system (hardware or software), unless indicated otherwise.
Various types of general purpose or specialized computer systems may be
used with or perform operations in accordance with the teachings described
1 o herein.
In view of the wide variety of embodiments to which the principles of
the present invention can be applied, it should be understood that the
illustrated embodiments are exemplary only, and should not be taken as
limiting the scope of the present invention.
For example, the steps of the flow diagrams may be taken in
sequences other than those described, and more or fewer elements may be
used in the block diagrams. While various elements of the preferred
embodiments have been described as being implemented in software, in other
embodiments in hardware or firmware implementations may alternatively be
2o used, and vice-versa.
The claims should not be read as limited to the described order or
elements unless stated to that effect. Therefore, all embodiments that come
within the scope and spirit of the following claims and equivalents thereto
are
claimed as the invention.