Note: Descriptions are shown in the official language in which they were submitted.
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
DIFFERENTIAL EXPRESSION OF ORGANELLAR GENE PRODUCTS
TECHNICAL FIELD
The invention relates to factors encoded by genes that are differentially
expressed in cellular models of particular disease states associated with
organelles in
cells as compared to control cells, or in cells response to various compounds
or
conditions thought to influence organellar function. Differentially expressed
genes and
factors in organelle-associated diseases include organellar factors, i. e.,
macromolecules
found within or associated with organelles, and cellular factors that
negatively or
positively influence, either directly or indirectly, the amount and/or
activity of such
l0 macromolecules. Organellar factors include nucleic acids and proteins that
are
expressed from genes that are derived from a cell's or organism's nuclear
genome, as
well as those expressed from the genomes of organelles such as mitochondria or
chloroplasts. Cells and cellular models useful in the invention include
cybrids and rho-
zero (po) cells. Cybrids are cellular hybrids having a nucleus derived from a
first cell
line and a cytoplasmic component (which may include organelles) derived from a
second cell line or from an organism suffering from, or suspected of being
prone to
develop, a disease or disorder. Rho~ cells are cells derived from an organism
or from
cell lines that have been treated so as to eliminate the genomes of their
mitochondria
and/or chloroplasts. Differential expression can reflect a comparison between
p~ and
2o control cells; between cybrids and control cells; between cells, including
cybrids and p~
cells, that have been exposed to one or more stressors.
BACKGROUND OF THE INVENTION
The cell is the basic unit of life and comprises a variety of subcellular
compartments including, e.g., organelles. An organelle is a structural
component of a
cell that is physically separated, typically by one or more membranes, from
other
cellular components, and which carries out specialized cellular functions.
1
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Mitochondria and chloroplasts are two organelles of particular interest
with regard to the present invention as each contains its own DNA genome.
These
organellar genomes encode a fraction of the gene products required for
organellar
function, the remainder of such gene products being encoded by the nuclear
genome.
Relatively little is known about the mechanisms by which mitochondrial and
chloroplast gene products, which may be encoded by nuclear sequences or
sequences
found in the respective organellar genomes, are coordinately regulated (Surpin
and
Chory, Essays Biochem. 32:113-125, 1997).
Because of the role of mitochondria in various diseases and disorders,
1 o there is a need to identify genetic sequences, present in either the
nuclear or
mitochondria) genomes (or both), that encode mitochondria) gene products and
that are
differentially expressed in such diseases and disorders. There is also a need
for nucleic
acids comprising such genetic sequences that can be used as probes in
diagnostic,
prognostic and pharmacogenomic assays, useful in the therapeutic management of
such
diseases and disorders. Such nucleic acids can also be used to produce gene
products
that can be used as novel targets in methods for identifying therapeutic
compounds,
including high through-put screening, useful to treat such diseases and
disorders.
Additionally, in view of the economic desirability of enhanced crop
production, and the role of chloroplasts in processes such as photosynthesis
that are
essential for producing biomass, there is a need to identify genetic sequences
present in
the nuclear or chloroplast genomes (or both), that encode chloroplast gene
products that
are differentially expressed under different environmental conditions or in
response to
extraneously added agents. Such nucleic acids can be used to identify and
produce gene
products that may be used as novel targets in methods for identifying
compounds and
conditions that promote or optimize photosynthesis and other biomass producing
processes.
A number of difficulties are also associated with killing eukaryotic
pathogens and parasites without harming their eukaryotic hosts, such that
species-to-
species variation in organellar functions may be exploited to develop novel
antibiotics.
3o There is thus a need to identify genetic sequences encoding organellar
functions that are
2
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
differentially expressed in a species-specific fashion in response to
compounds,
particularly compounds that are known or candidate antibiotics that kill or
slow the
growth of eukaryotic pathogens and parasites without harming their eukaryotic
hosts.
Such nucleic acids can be used to identify and produce gene products that may
be used
as novels targets in methods for identifying antibiotics, including high
throughout
screening, useful to treat diseases and disorders resulting from such
eukaryotic
pathogens and parasites.
The present invention fulfills these and other needs. These and other
advantages of the present invention will become more apparent by the detailed
I o description of the invention provided herein.
Mitochondria
The organelle known as the mitochondrion (plural, mitochondria) is the
main energy source in cells of higher organisms. Mitochondria provide direct
and
indirect biochemical regulation of a wide array of cellular respiratory,
oxidative and
metabolic processes. These include electron transport chain (ETC) activity,
which
drives oxidative phosphorylation to produce metabolic energy in the form of
adenosine
triphosphate (ATP), and which also underlies a central mitochondria) role in
intracellular calcium homeostasis. In addition to their role in energy
production in
growing cells, mitochondria (or, at least, mitochondria) components)
participate in
2o programmed cell death (PCD), also known as apoptosis (Newmeyer et al., Cell
79:3~3-
364, 1994; Liu et al., Cell 86:147-157, 1996; for general reviews of
apoptosis, and the
role of mitochondria therein, see Green and Reed (Science 281:1309-1312,
1998),
Green (Cell 94:695-698, 1998) and Kromer (Nature Medicine 3:614-620, 1997).
Mitochondria) ultrastructural characterization reveals the presence of an
outer mitochondria) membrane that serves as an interface between the organelle
and the
cytosol, a highly folded inner mitochondria) membrane that appears to form
attachments
to the outer membrane at multiple sites, and an intermembrane space between
the two
mitochondria) membranes. The subcompartment within the inner mitochondria)
membrane is commonly referred to as the mitochondria) matrix. (For a review,
see,
3
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
e.g., Ernster and Schatz, J. Cell Biol. 91:227s-255s, 1981.) The cristae,
originally
postulated to occur as infoldings of the inner mitochondria) membrane, have
recently
been characterized using three-dimensional electron tomography as also
including tube-
like conduits that may form networks, and that can be connected to the inner
membrane
by open, circular junctions (Perkins et al., Journal of Structural Biology
119:260-272,
1997). While the outer membrane is freely permeable to ionic and non-ionic
solutes
having molecular weights less than about ten kilodaltons, the inner
mitochondria)
membrane exhibits selective and regulated permeability for many small
molecules,
including certain cations, and is impermeable to large (> ~10 kDa) molecules.
1 o Chloroplasts
The chloroplast is an organelle found in plant cells wherein
photosynthesis takes place. Photosynthesis, in addition to being an integral
part of a
plant cell's metabolism, is an important process that impacts many other
living
organisms as well. The reason for this is twofold: photosynthesis "fixes"
atmospheric
1 s COZ into biologically usable carbohydrate (CHO)~ molecules and also
produces Oz
which is required by all aerobic organisms.
Like mitochondria, chloroplasts have a double (outer and inner)
membrane, contain their own DNA and have translation factors (ribosomes,
tRNAs,
etc.) that are distinct from those found in the cytoplasm (Sugiura, Essays
Biochem.
20 30:49-57, 1995). Electron microscopy demonstrates that, like mitochondria.
chloroplasts have a highly organized internal ultrastructure which includes
flattened
membranous bodies known as lamellae or thykaloid discs. Chloroplasts are,
however,
typically much larger than mitochondria; in higher plants they are generally
cylindrical
in shape and range from about 5 to 10 micrometers in length and from 0.5 to 2
2s micrometers in diameter. Like mitochondria, which are present in greater
numbers in
certain tissues (e.g., liver) than others, chloroplasts have greater copy
numbers in some
tissues than others. For example, mature leaves contain many chloroplasts and
the total
amount of chloroplast DNA in such leaves is about twice that of nuclear DNA
(Dope et
al., J. Cell. Biol. 79:631-636, 1978).
4
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Mitochondria) Electron Transport Chain, O~I', and Pore Transition
The electron transport chain (ETC) is a mitochondria) activity that drives
oxidative phosphorylation to produce metabolic energy in the form of adenosine
triphosphate (ATP). Four of the five multisubunit protein complexes (Complexes
I, III,
IV and V) that mediate ETC activity are localized to the inner mitochondria)
membrane;
the remaining ETC complex (Complex II) is situated in the mitochondria)
matrix. In at
least three distinct chemical reactions known to take place within the ETC,
protons are
moved from the mitochondria) matrix, across the inner membrane, to the
intermembrane space. This disequilibrium of charged species creates an
1 o electrochemical potential of approximately 220 mV referred to as the
"protonmotive
force" (PMF). PMF, which is often represented by the notation Op, corresponds
to the
sum of the electric potential (OLl'm) and the pH differential (OpH) across the
inner
mitochondria) membrane according to the equation
Op = O~I'm - ZOpH,
wherein Z stands for -2.303 RT/F. The value of Z is -59 at 25°C when Op
and ~~m are
expressed in mV and OpH is expressed in pH units (see, e.g., Ernster et al.,
1981 J. Cell
Biol. 91:227s-255s and references cited therein).
Many mitochondria) functions depend in part or entirely on 4~m. For
example, OLYm provides the energy for phosphorylation of adenosine diphosphate
(ADP) to yield ATP by ETC Complex V, a process that is coupled
stoichiometrically
with transport of a proton into the matrix. Furthermore, 4~1'm is also the
driving force
for the influx of cytosolic Ca2+ into the mitochondrion. Even fundamental
biological
processes, such as translation of mRNA molecules to produce polypeptides,
appear to
be dependent on O~I'm (Cote et al., J. Biol. Chem. 265:7532-7538, 1990).
Under normal metabolic conditions, the inner membrane is impermeable
to proton movement from the intermembrane space into the matrix, leaving ETC
Complex V as the sole means whereby protons can return to the matrix. When,
however, the integrity of the inner mitochondria) membrane is compromised, as
occurs
during mitochondria) permeability transition (MPT) that accompanies certain
diseases
associated with altered mitochondria) function, protons are able to bypass the
conduit of
5
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Complex V without generating ATP, thereby uncoupling respiration. During MPT,
0~'m collapses and mitochondria) membranes lose the ability to selectively
regulate
permeability to solutes both small (e.g., ionic Ca2l, Na+, K+, H+) and large
(e.g.,
proteins).
Mitochondria) Defects, Diseases and Disorders
Mitochondria (or, at least, mitochondria) components) participate in
programmed cell death (PCD), also known as apoptosis (Newmeyer et al., Cell
79:353-
364, 1994; Liu et al., Cell 86:147-157, 1996), which is apparently required
for normal
development of the nervous system and functioning of the immune system.
Moreover,
1 o some disease states are thought to be associated with either insufficient
or excessive
levels of apoptosis (e.g., cancer and autoimmune diseases in the first
instance, and
stroke damage and neurodegeneration in Alzheimer's disease in the latter
case). Thus,
agents that affect apoptotic events, including those associated with
mitochondria)
components, might have a variety of palliative, prophylactic and therapeutic
uses.
Altered or defective mitochondria) activity, including but not limited to
failure at any step of the ETC, may result in the generation of highly
reactive free
radicals that have the potential of damaging cells and tissues. These free
radicals may
include reactive oxygen species (ROS) such as superoxide, peroxynitrite and
hydroxyl
radicals, and potentially other reactive species that may be toxic to cells.
For example,
2o oxygen free radical induced lipid peroxidation is a well established
pathogenetic
mechanism in central nervous system (CNS) injury such as that found in a
number of
degenerative diseases, and in ischemia (i.e., stroke).
In addition to free radical mediated tissue damage, there are at least two
deleterious consequences of exposure to reactive free radicals arising from
mitochondria) dysfunction that adversely impact the mitochondria themselves.
First,
free radical mediated damage may inactivate one or more of the myriad proteins
of the
ETC. Second, free radical mediated damage may result in catastrophic
mitochondria)
collapse that has been termed "permeability transition" (PT) or "mitochondria)
permeability transition" (MPT). According to generally accepted theories of
6
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
mitochondria) function, proper ETC respiratory activity requires maintenance
of an
electrochemical potential (O~fm) in the inner mitochondria) membrane by a
coupled
chemiosmotic mechanism, as described herein. Free radical oxidative activity
may
dissipate this membrane potential, thereby preventing ATP biosynthesis and
halting the
production of a vital biochemical energy source. In addition, mitochondria)
proteins
such as cytochrome c and "apoptosis inducing factor" may leak out of the
mitochondria
after permeability transition and may induce the genetically programmed cell
suicide
sequence known as apoptosis or programmed cell death (PCD). Therefore, mere
determination of free radical induced damage, such as lipid peroxidation, is
not an
1 o accurate or early indicator of mitochondria) dysfunction.
Altered mitochondria) function characteristic of the mitochondria
associated diseases may also be related to loss of mitochondria) membrane
electrochemical potential by mechanisms other than free radical oxidation, and
permeability transition may result from direct or indirect effects of
mitochondria) genes,
gene products or related downstream mediator molecules and/or
extramitochondrial
genes, gene products or related downstream mediators, or from other known or
unknown causes. Loss of mitochondria) potential therefore may be a critical
event in
the progression of diseases associated with altered mitochondria) function,
including
degenerative diseases.
2o Mitochondria) defects, which may include defects related to the discrete
mitochondria) genome that resides in mitochondria) DNA and/or to the
extramitochondrial genome, which includes nuclear chromosomal DNA and other
extramitochondrial DNA, may contribute significantly to the pathogenesis of
diseases
associated with altered mitochondria) function. For example, alterations in
the
structural and/or functional properties of mitochondria) components comprising
subunits encoded directly or indirectly by mitochondria) and/or
extramitochondrial
DNA, including alterations deriving from genetic and/or environmental factors
or
alterations derived from cellular compensatory mechanisms, may play a role in
the
pathogenesis of any disease associated with altered mitochondria) function. A
number
of degenerative, hyperproliferative and other types of diseases are thought to
be caused
7
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
by, or to be associated with, alterations in mitochondria) function. These
include, for
example, Alzheimer's Disease, Parkinson's Disease, Huntington's disease,
diabetes
mellitus, and hyperproliferative disorders, such as cancer, tumors and
psoriasis. The
extensive list of mitochondria associated diseases, i.e., diseases associated
with altered
mitochondria) function and/or mitochondria) mutations, continues to expand as
aberrant
mitochondria) or mitonuclear activities are implicated in particular disease
processes.
SUMMARY OF THE INVENTION
The invention relates to factors encoded by genes that are differentially
expressed in cellular models of particular disease states associated with
organelles in
to cells as compared to control cells, or in cells in response to various
compounds or
conditions thought to influence organellar function, or in a species-specific
manner. In
brief, the present invention provides methods for identifying factors that
directly or
indirectly influence organellar function, or which are over- or under-
expressed in
organelle-associated diseases and disorders, including but not limited to
diseases and
disorders associated with mitochondria. Differentially expressed genes and
factors in
organelle-associated diseases include organellar factors, i. e.,
macromolecules found
within or associated with organelles, and cellular factors that negatively or
positively
influence, either directly or indirectly, the amount and/or activity of such
macromolecules. Organellar factors may be macromolecules found within or
associated
2o with organelles, or cellular factors that negatively or positively
influence, either directly
or indirectly, the amount and/or activity of such macromolecules. Such factors
(e.g.,
gene products) include nucleic acids and proteins that are expressed from
genes that are
derived from a cell's or an organism's nuclear genome, as well as those
expressed from
the genomes of organelles such as mitochondria or chloroplasts (e.g.,
extranuclear
genomes). Of particular interest are nucleic acids that are differentially
expressed in
particular disease states, in response to various compounds or conditions, or
in a
species-specific fashion.
Thus in one aspect the present invention provides a method for
identifying organellar factors encoded by genes that are differentially
expressed,
8
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
comprising providing at least one cell in a first state; providing at least
one cell in a
second state, determining the expression of genes in such cells, and
identifying genes
that are differentially expressed in cells in the first state relative to
cells in the second
state. The cells) in either state may be treated with one or more stressors
known or
thought to influence organellar function, and the cells) in the other state
may be control
(e.g., untreated) cells.
In another aspect, the invention provides a method for identifying
differentially expressed organellar genes in manipulated cells, comprising
providing at
least one first cell that is not a manipulated cell, providing at least one
second cell that
is a manipulated cell, determining the expression of genes in the first cells)
and the
second cell(s), and identifying genes that are differentially expressed in the
first cell
relative to the second cell. Manipulated cells include but are not limited to
(a) po and
cybrid cells, (b) cells that have been genetically engineered to over- or
under-express
factors known or thought to directly or indirectly influence organellar
function, and (c)
cells that have been treated with an agent (e.g., an antisense
oligonucleotide) that
influences organellar function and/or expression of factors associated with
organellar
function and diseases or disorders. Manipulated cells also includes cells that
fall into
two or more of the categories (a), (b) and (c); these categories are not
mutually
exclusive. It is also possible to compare gene expression in a cybrid cell
line to po cells
2o from which the cybrids were prepared.
In an aspect of the invention related to category (c) of the preceding
paragraph (i.e., cells that have been treated with an agent (e.g., an
antisense
oligonucleotide) that influences organellar function and/or expression of
factors
associated with organellar function and diseases or disorders), a method is
provided for
identifying nucleic acids that are differentially expressed during apoptosis,
comprising
providing at least one first cell that is not in an apoptotic state, providing
at least one
second cell that is in an apoptotic state, determining the expression of genes
in the first
cells) and the second cell(s), and identifying genes that are differentially
expressed in
first cells) relative to said second cell(s). Apoptosis can be induced by a
variety of
3o treatments, as detailed below. In a related aspect of the invention, other
agents may
9
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
effect, alter (e.g., increase or decrease), influence or otherwise regulate
organellar
function, including apoptogens at concentrations where apoptosis is not
induced.
Examples of such compounds include but are not limited to Ruthenium Red, which
blocks the action of the mitochondria) calcium uniporter; ionophores such as
ionomycin, which increase the intracellular concentration of ions such as
Ca++; and
uncouplers and/or blockers of the electron transport chain.
It is another aspect of the present invention to provide a method for
identifying nucleic acids that are differentially expressed in a species-
specific manner,
comprising providing at least one cell from a first species, providing at
least one cell
to that is from a second species, determining the expression of genes in the
cells) from the
first species and the cells) from the second species, and identifying genes
that are
differentially expressed in the cells) from the first species as compared to
the cells)
from the second species. This aspect of the invention includes methods in
which a
candidate species-specific agent is tested for its ability to impact the
expression of
related (homologous) genes in one species and not the other. The cells can
additionally
or alternatively be treated with an agent that influences organellar function
and/or
expression of factors associated with organellar function and diseases or
disorders, and
can be manipulated cells, including but not limited to p~ and cybrid cells.
Accordingly, and as provided herein, in certain aspects the present
2o invention provides a method for identifying a factor encoded by a gene that
is
differentially expressed, comprising comparing (i) expression of a plurality
of genes in
at least one first cell that is in a first state to (ii) expression of a
plurality of genes in at
least one second cell that is in a second state, thereby identifying a gene
that is
differentially expressed in said first state relative to said second state,
and therefrom
identifying a factor encoded by a gene that is differentially expressed. In
one
embodiment the first cell is a manipulated cell and in certain further
embodiments the
second cell is a manipulated cell. In certain further embodiments the
manipulated cell
is a cybrid cell, while in certain other embodiments the manipulated cell is a
p° cell. In
one embodiment the first cell is a manipulated cell and the second cell is a
manipulated
3o cell, and in certain further embodiments at least one of said first and
second cells is a
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
cybrid cell. In certain other further embodiments both of said first and
second cells are
cybrid cells. In another embodiment at least one of said first and second
cells is a p°
cell, and in another embodiment both of said first and second cells are
p° cells.
In certain embodiments the factor is an organellar factor, which in
certain other embodiments is protein and in certain other embodiments is a
nucleic acid.
In certain other embodiments the factor is differentially expressed in an
organelle
associated disease. In certain other embodiments the factor is differentially
expressed in
response to treatment with an agent that alters at least one organellar
function, which in
certain further embodiments is a mitochondria) function and in certain still
further
to embodiments is electron transport chain activity, oxidative
phosphorylation, ATP
production, intracellular calcium homeostasis, apoptosis, mitochondria)
permeability
transition or free radical production. In certain other embodiments the factor
is
differentially expressed in response to treatment with an agent that is a
stressor or an
apoptogen. In certain other embodiments the factor is differentially expressed
in a
species specific fashion.
In yet another embodiment, the first state and the second state are
different and at least one of the first and second states is a disease state.
In one such
embodiment, the disease is an organelle associated disease. In another
embodiment, the
first state and the second state are different and at least one of the first
and second states
is a response to a stressor, which in certain further embodiments is a
molecule and in
certain other further embodiments is an environmental factor. In certain
embodiments
of the present invention, the step of comparing comprises determining mRNA in
each of
the first and second cells, while in certain other embodiments the step of
comparing
comprises determining protein in each of the first and second cells. According
to
certain embodiments, the first and second cells are derived from the same
clone, while
in certain other embodiments the first and second cells are derived from
different
species. In another embodiment, the first state and the second state are
different and at
least one of the first and second states is a metabolic state, a respiratory
state, a cell
cycle state, a pathologic state, a differentiative state, a maturational
state, a genetic state,
3o an apoptotic state, an excitotoxic state or a pharmacological state.
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
In another embodiment, the invention provides a method of diagnosing a
disease comprising contacting a biological sample from an individual suspected
of
having the disease with at least one factor identified according to the above
described
method for identifying a factor encoded by a gene that is differentially
expressed,
comprising comparing (i) expression of a plurality of genes in at least one
first cell that
is in a first state to (ii) expression of a plurality of genes in at least one
second cell that
is in a second state, thereby identifying a gene that is differentially
expressed in said
first state relative to said second state, and therefrom identifying a factor
encoded by a
gene that is differentially expressed. In one embodiment the factor is a
nucleic acid,
1o which in certain further embodiments may have the sequence of SEQ ID NOS:B,
9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 or 22; the reverse complements of
SEQ ID
NOS:B, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 or 22; or an
equivalent thereof.
It is another aspect of the present invention to provide a method of
diagnosing a disease comprising contacting a biological sample from an
individual
suspected of having the disease with an antibody that specifically binds a
factor
identified according to the above described method for identifying a factor
encoded by a
gene that is differentially expressed, comprising comparing (i) expression of
a plurality
of genes in at least one first cell that is in a first state to (ii)
expression of a plurality of
genes in at least one second cell that is in a second state, thereby
identifying a gene that
2o is differentially expressed in the first state relative to the second
state, and therefrom
identifying a factor encoded by a gene that is differentially expressed. In a
further
embodiment, the factor is a protein.
In another aspect, the invention provides the cybrid cell lines 1685,
ATCC 207149 and ATCC 207150.
These and other aspects of the present invention will become apparent
upon reference to the following detailed description and attached drawings.
BRIEF DESCRIPTION OF THE DRAWINGS
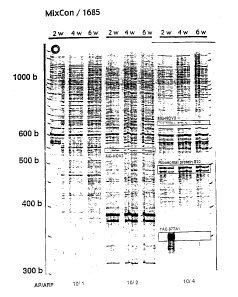
Figure 1 is an electrophoretic gel showing the results (fluorescently
labeled PCR products) from a typical differential display (DD) experiment with
control
12
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
(MixCon) and Alzheimer's (1685) cybrids. The positions of molecular weight
markers
(b, number of bases) are indicated on the left. Primer pairs (AP, anchored
primer; ARP,
arbitrary primer) are indicated on the bottom (as an example, "10/1" indicates
that the
primers AP10 and Ml3r-ARP1 were used). The numbers on the top indicate the
times
at which samples were taken ("2w" = 2 weeks; "4w" = 4 weeks; "6w" = 6 weeks).
Duplicate reactions were prepared and run in parallel in adjacent lanes. In
the figure,
certain nucleic acids of interest are boxed and labeled, including MG-NOV2
(a.k.a.
1685 DD-Sequence #4, SEQ ID NO:10), MG-NOV3 (a.k.a. 1685 DD-Sequence #5,
SEQ ID NO:11) and YAC 377A1 (a.k.a. 1685 DD-Sequence #2, SEQ ID N0:8).
l0 Figure 2 shows an alignment between 1685 DD-Sequence #1 (SEQ ID
N0:7) and human nucleotide sequences derived from the gene encoding 3-
hydroxyisobutyryl-coenzyme A hydrolase (GenBank accession No. U66669; SEQ ID
N0:64).
Figure 3 shows an alignment between 1685 DD-Sequence #2 (SEQ ID
N0:8) and human nucleotide sequences derived from YAC clone 377A1 (GenBank
accession No. AF009203; SEQ ID N0:65) and a cDNA encoding an uncharacterized
protein designated KIAA0711 (GenBank accession No. AB018254; SEQ ID N0:66).
Figure 4 shows an alignment between 1685 DD-Sequence #3 (SEQ ID
N0:9) and human nucleotide sequences derived from BAC clone CIT987-SKA-237H1
(GenBank accession No. AC002287; SEQ ID N0:67).
Figures 5-32 show, respectively, sequences UNKl-UNK28 (SEQ ID
NOS: 23-58).
Figure 33 shows an alignment of UNKS (SEQ ID N0:27), UNK10-5'
(SEQ ID N0:32) and UNK10-3' (SEQ ID N0:33) nucleotide sequences.
Figure 34 shows an alignment of UNK19 (SEQ ID N0:45) and UNK18
(SEQ ID N0:44) nucleotide sequences.
Figure 35 shows an alignment of KIAA0138 (encoded by a cDNA that
overlaps SEQ ID N0:8) with two human proteins having related amino acid
sequences,
and a consensus sequence (SEQ ID N0:63) derived therefrom. KIAA0138,
3o uncharacterized protein KIAA0138 (Accession No. ; SEQ ID N0:62); AK000867,
13
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
uncharacterized protein AK000867 (Accession No. ; SEQ ID N0:61 ); Factor B
(SEQ
ID N0:60), scaffold attachment factor. Upper case residues in the consensus
sequence
are conserved in all three proteins; lower case residues indicate variable
positions.
Figure 36 shows a sequence (SEQ ID N0:59) that aligns with and
overlaps a cDNA (Accession No. X01662) that encodes SOD-1 (superoxide
dismutase).
Figure 37 shows the results of various homology searches as explained
in the Examples.
Figure 38 shows the results of an EST database sequence alignment
search using SEQ ID N0:8.
to Figure 39 shows the results of homology searching with an UNKS-
derived consensus sequence (SEQ ID N0:8).
FREC~UENTLY USED SYMBOLS AND ABBREVIATIONS
O~r, Dym mitochondria) membrane potential
OpH pH differential across the inner mitochondria) membrane
AD Alzheimer's disease
ETC electron transport chain
MixCon mixed control
MPT Mitochondria) Permeability Transition
mtDNA mitochondria) DNA
2o NAO nonyl acridine orange
PD Parkinson's disease
PMF, Op protonmotive force
rho°, p° lacking mtDNA
DETAILED DESCRIPTION OF THE INVENTION
In certain embodiments, the present invention is directed to a method of
identifying organellar factors encoded by genes that directly or indirectly
alter or
influence organellar function; and/or that are differentially expressed in
particular
disease states including organelle associated diseases and disorders including
those
14
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
described herein; and/or which are differentially expressed in response to
treatment with
one or more agents thought or known to impact, either directly or indirectly,
one or
more organellar functions; and/or which are differentially expressed in cells,
including
manipulated cells, derived from one species relative to cells derived from a
second
species; and/or that are differentially expressed in response to various
stressors or in a
species-specific fashion. By "differentially expressed," it is meant that the
gene is over-
or under-expressed in one cell type, or under one set of conditions, relative
to another;
accordingly, in certain embodiments the corresponding gene product is present
in
greater amounts in one cell type, or under one set of conditions, than in
another.
1 o Thus, the present invention provides methods for identifying factors,
including organellar factors as provided herein, that directly or indirectly
influence
organellar function, or which are over- or under-expressed in organelle-
associated
diseases and disorders, including but not limited to diseases and disorders
associated
with mitochondria. As noted above, organellar factors may be macromolecules
found
within or associated with organelles, or cellular factors that negatively or
positively
influence, either directly or indirectly, the amount and/or activity of such
macromolecules. Such factors (e.g., gene products) include nucleic acids and
proteins
that are expressed from genes that are derived from a cell's or an organism's
nuclear
genome, as well as those expressed from the genomes of organelles such as
mitochondria or chloroplasts. Of particular interest are nucleic acids that
are
differentially expressed in particular disease states, in response to various
compounds or
conditions, or in a species-specific fashion. Therefore, differentially
expressed genes
and factors in organelle associated diseases as provided herein include
organellar
factors.
In one aspect of the present invention there is provided a method for
identifying factors, which in certain embodiments are organellar factors,
encoded by
genes that are differentially expressed, comprising providing at least one
cell in a first
state, providing at least one cell in a second state, determining the
expression of genes
in such cells, and identifying genes that are differentially expressed in
cells in the first
3o state relative to cells in the second state. The cells) in either state may
be treated with
CA 02363496 2001-09-14
WO 00/55323 PCT/US00107311
one or more stressors known or thought to influence organellar function, and
the cells)
in the other state may be control (untreated) cells. The state of a cell as
provided herein
includes the biological or physiological status or condition of the cell, for
example, the
metabolic, respiratory, cell cycle (e.g., mitotic), pathologic,
differentiative,
maturational, genetic (e.g., ploidy, homoplasmic, heteroplasmic, nuclear
genetic,
extranuclear genetic, etc.), apoptotic, electrochemical, adhesive,
activational,
excitotoxic or pharmacological status or the like. Preferably, the first state
and the
second state are different regarding a particular disease state, which may in
certain
embodiments be an organelle associated disease state. In certain other
embodiments the
to first state and the second state may differ with respect to the presence
and/or effects of a
stressor. The stressor can be any stressor, but is preferably a molecule or an
environmental factor. The determining step preferably includes determining the
mRNA
or protein in the cells) in the first state or the cells) in the second state,
preferably
both. Preferably, the cells) in the first state and the cells) in the second
state are
clonally derived and/or are derived from the same organism. The identifying
step
preferably includes comparing the mRNA or protein in the cells) in the first
state and
the cells) in the second state. Accordingly, in certain preferred embodiments
of the
invention there is provided a method of identifying a differentially expressed
factor that
is an organellar factor as provided herein.
2o In another aspect the invention provides a method for identifying
differentially expressed genes, for example organellar genes, in manipulated
cells,
comprising providing at least one first cell that is not a manipulated cell,
providing at
least one second cell that is a manipulated cell, determining the expression
of genes in
the first cells) and the second cell(s), and identifying genes that are
differentially
expressed in the first cell relative to the second cell. Manipulated cells
include but are
not limited to (a) po and cybrid cells, (b) cells that have been genetically
engineered to
over- or under-express factors known or thought to directly or indirectly
influence
organellar function, and (c) cells that have been treated with an agent (e.g.,
an antisense
oligonucleotide) that influences organellar function and/or expression of
factors
3o associated with organellar function and diseases or disorders. Manipulated
cells also
16
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
includes cells that fall into two or more of these categories (a), (b) and
(c); these
categories are not mutually exclusive.
In an aspect of the invention related to category (c) of the preceding
paragraph (cells that have been treated with an agent (e.g., an antisense
oligonucleotide)
that influences organellar function and/or expression of factors associated
with
organellar function and diseases or disorders), a method is provided for
identifying
nucleic acids that are differentially expressed during apoptosis, comprising
providing at
least one first cell that is not in an apoptotic state, providing at least one
second cell that
is in an apoptotic state, determining the expression of genes in the first
cells) and the
to second cell(s), and identifying genes that are differentially expressed in
first cells)
relative to said second cell(s). Apoptosis can be induced by a variety of
treatments, as
detailed below. In a related aspect of the invention, other agents that impact
organellar
function, including apoptogens at concentrations where apoptosis is not
induced.
Examples of such compounds include but are not limited to Ruthenium Red, which
blocks the action of the mitochondria) calcium uniporter; ionophores such as
ionomycin, which increase the intracellular concentration of ions such as
Ca++; and
uncouplers and blockers of the electron transport chain.
The invention also provides, in another aspect, a method for identifying
nucleic acids that are differentially expressed in a species-specific manner,
comprising
2o providing at least one cell from a first species, providing at least one
cell that is from a
second species, determining the expression of genes in the cells) from the
first species
and the cells) from the second species, and identifying genes that are
differentially
expressed in the cells) from the first species as compared to the cells) from
the second
species. This aspect of the invention includes methods in which a candidate
species-
specific agent is tested for its ability to impact the expression of related
(homologous)
genes in one species and not the other. The cells can additionally or
alternatively be
treated with an agent that influences organellar function and/or expression of
factors
associated with organellar function and diseases or disorders, and can be
manipulated
cells, including but not limited to p~ and cybrid cells.
17
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Definitions and General Methods
The following definitions and general methods are applicable to the
present invention. Unless defined otherwise, all technical and scientific
terms used
herein have the same meaning as commonly understood by one of ordinary skill
in the
art to which this invention belongs. Generally, the nomenclature used herein
and the
laboratory procedures in cell culture, chemistry, microbiology, molecular
biology, cell
science and cell culture described below are well known and commonly employed
in
the art. Conventional methods are used for these procedures, such as those
provided in
the art and various general references (Sambrook et al., Molecular Cloning: A
1o Laboratory Manual, 2nd edition, Cold Spring Harbor Press, Cold Spring
Harbor, N.Y.
( 1989)). Where a term is provided in the singular, the inventors also
contemplate the
plural of that term. The nomenclature used herein and the laboratory
procedures
described below are those well known and commonly employed in the art.
Detecting Differentially Expressed Nucleic Acids
A variety of methods and means for detecting differentially expressed
nucleic acids may be used in the methods of the invention. Differential
Display (DD)
and Quantitative Real-Time Polymerase Chain Reaction (Q-RTPCR) are described
in
detail in the Examples of the disclosure; some other methods and means
include,
without limitation, the following methodologies. It should be noted that,
regardless of
2o which method is used to initially identify candidate differentially
expressed genes, a
second independent method is preferably used to verify the results obtained
from the
first method.
Subtractive Hybridization: In a typical procedure for applying the
technique of subtraction hybridization (Hedrick et al., Nature 308:149-153,
1984) to
investigate differences in the active genes of a certain sample of test or
target cells, e.g.,
from tumor tissues, as compared with the active genes of a sample of reference
cells,
e.g., cells from corresponding normal tissue, total cell mRNA is extracted
(using any
preferred method) from both samples of cells. The mRNA in the extract from the
test
or target cells is then used in a conventional manner to synthesize
corresponding single
18
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
stranded cDNA using an appropriate primer and a reverse transcriptase in the
presence
of the necessary deoxynucleoside triphosphates, and the template mRNA is
subsequently degraded by alkaline hydrolysis or RNase H to leave only the
single
stranded cDNA. The single stranded cDNA thus derived from the mRNA expressed
by
the test or target cells is then mixed under hybridizing conditions with an
excess
quantity of the mRNA extract from the reference (normal) cells; this mRNA is
generally termed the subtraction hybridization "driver" since it is this mRNA
or other
single stranded nucleic acid present in excess which "drives" the subtraction
process.
As a result; cDNA strands having common complementary sequences anneal with
the
to mRNA strands to form mRNA/cDNA duplexes and are thus subtracted from the
single
stranded species present. The only single stranded DNA remaining is then the
unique
cDNA that is derived specifically from the mRNA produced by genes which are
expressed solely by the test or target cells.
From this point onwards, to complete the subtraction process and use the
single stranded unique cDNA, for example for producing labeled probes that may
perhaps then be used for detecting or identifying corresponding cloned copies
in a
cDNA clone colony (labeling of such probes is frequently introduced by using
labeled
deoxynucleoside triphosphates in synthesis of the cDNA), it is generally
necessary to
physically to separate out the common mRNA/cDNA duplexes, using for example
2o hydroxyapatite (HAP) or (strept)avidin-biotin in a chromatographic
separation method.
Finally, one or more repeat rounds of the subtraction hybridization may be
carried out
to improve the extent of recovery of the desired product, although other means
may be
employed (see, e.g., U.S. Patent No. 5,589,339).
High Density Array: Multiple sample nucleic acid hybridization
analysis can be carried out on micro-formatted multiplex or matrix devices
(e.g., DNA
or RNA chips, filters and microarrays) (see, e.g., Bains, BiolTechnology
10:757-758,
1992). These hybridization formats are micro-scale versions of the
conventional "dot
blot" and "sandwich" hybridization systems. In these methods, specific DNA
sequences are typically attached to, or synthesized on, very small specific
areas of a
3o solid support, allowing large numbers of different DNA sequences to be
placed in a
19
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
small area. The high density arrays comprise target elements, i.e., target
nucleic acid
molecules bound to a solid support. The nucleic acids for both the target
elements and
the probes may be, for example, RNA, DNA, or cDNA. In one type of array,
target
elements comprising nucleic acid elements that are short synthetic
oligonucleotides
derived from mRNA, cDNA or EST sequences are used to carry out serial analysis
of
gene expression (SAGE; U.S. Patent No. 5,866,330).
In methods for comparing two nucleic acid collections, nucleic acid
molecules in the test and control collections (which may be, e.g., mRNA
preparations
from a diseased and undiseased human) are detectably labeled. The first and
second
labeled probes thus formed are each contacted to an identical high density
array
comprising a plurality of target elements under conditions such that nucleic
acid
hybridization to the target elements can occur.
After contacting the probes to the target elements the amount of binding
to each target element in each of the two arrays is measured, and the binding
ratio (i.e.,
amount bound in the disease sample / amount bound in the control sample) is
determined for each target element. A binding ratio > 1 indicates that nucleic
acids
hybridizing to the particular target element are ''up-regulated" in the
nucleic acid
collection prepared from the diseased patient relative to the nucleic acid
prepared from
the control individual, whereas a binding ratio <1 indicates that nucleic
acids
2o hybridizing to the particular target element are "down-regulated" in the
diseased patient.
High density cDNA arrays that may be used in the invention include but
are not limited to GeneChipTM arrays comprising synthetic oligonucleotides
(Affymetrix, Inc., Santa Clara, CA); GeneFiltersTM yeast or human cDNA arrays
(Research Genetics, Huntsville, AL); ATLASTM cDNA arrays (Clontech); and GEMTM
and Gene Display Arrays (GDA) cDNA arrays (Genome Systems, Inc., St. Louis,
MO).
Furthermore, one method for building a microarrayer (a machine that produces
microarrays) is available on-line at http://cmgm.stanford.edu/pbrown/mguide/
index.html.
One type of high density arrays uses electronic hybridization, i.e., a
3o method that directs sample DNA molecules to, and concentrates them at, test
sites on a
CA 02363496 2001-09-14
WO 00/55323 PCT/IJS00/07311
microchip that can be electronically activated by a positive charge. Because
DNA
molecules in solution have strong negative charges, they are attracted to
activated sites.
The electronic hybridization of sample DNA molecules at each test site
promotes rapid
hybridization of the sample DNAs with the nucleic acids of the target
elements.
Materials for electronic hybridization are available from Nanogen (San Diego,
CA) and
the method is described in U.S. Patent No. 5,849,486.
Manipulated Cells
In the present disclosure, the term "manipulated cells" refers to cells that
have been altered by human manipulation, such manipulation often (but not
necessarily)
1 o occurring in vitro. Manipulated cells include, but are not limited to,
cybrids, rho° cells,
and cells that have been genetically manipulated in one fashion or another.
It is known in the art to prepare cellular hybrids (cybrids) having a
cytoplasmic component, which typically includes organelles such as
mitochondria or
chloroplasts, from one cell line and a nuclear component from another cell
line.
Experiments with such cybrids have demonstrated that cellular defects
associated with
diseased cells are transferred with cytoplasmic elements (mitochondria) from
diseased
cells to cybrids. Human diseases that have been demonstrated to have a
cytoplasmic
component in this manner include Alzheimer's disease and Parkinson's disease
(Swerdlow et al., Neurology 49:918-925, 1997; Davis et al., Proc. Natl. Acad.
Sci.
(USA) 94:4526-4531, 1997; Swerdlow et al., Annals of'Neurology 40:663-671,
1996).
In some embodiments of the invention, differentially expressed factors
are defined as factors that have a pattern of expression in "disease cybrids"
(i.e., cybrids
having a cytoplasmic component derived from one or more individuals known to
have
or suspected of having a disease of interest) that is different from the
pattern of
expression observed in "control cybrids" (i.e., cybrids having a cytoplasmic
component
derived from one or more individuals not having the disease of interest). One
advantage of using cybrid cells for experiments designed to identify the
differential
expression of factors involved in organellar functions is that disease and
control cybrids
share commonly-derived nuclear components. Differences in expression patterns
21
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
between various cybrids are thus more likely to be due solely to differences
in
cytoplasmic components and not to differences in the nuclear genome.
With regard to animal cells, methods for preparing cellular hybrids
(cybrids) comprising the nucleus of one cell type and organelles
(mitochondria) from
another cell type have been described (see published PCT application No.
PCT/L1S95/04063, U.S. patent application Serial No. 09/069,489, and U.S.
Patent No.
5,840,493, all of which are hereby incorporated by reference). In a particular
embodiment of the invention, differentiable cybrid cell lines are used to
carry out
differential expression experiments (see U.S. patent application Serial No.
08/397,808,
to now U.S. Patent No. 5,888,498, hereby incorporated by reference).
Cybrid plant cells have also been described (see, for example, U.S.
Patents 4,751,347 and 5,360,725, hereby incorporated by reference). In one
embodiment of the invention, plant cybrids are used in differential expression
experiments to identify factors related to functions of organelles
(mitochondria and/or
chloroplasts) in plants. In another embodiment of the invention, factors that
are
differentially expressed in plant cells comprising genetically engineered
chloroplasts
(U.S. Patent No. 5,693,507, hereby incorporated by reference) relative to
plant cells
having wildtype chloroplasts are identified. Factors identified by these
embodiments of
the invention are useful for agricultural applications such as, e.g.,
increasing the
lifespan, productive capacity, and/or insecticide or herbicide resistance of
crops.
In general, cybrids are prepared by first preparing cells that lack
mitochondria; such cells are known as rho° cells. In a further
embodiment of the
invention, a differentially expressed factor is defined as a factor that has a
pattern of
expression in rho° cells that is different from the pattern of
expression observed in the
parent rho+ (mitochondria-containing) cells. Methods for preparing rho°
cells for a
variety of cell types (animal, fungal, etc.) are known in the art. By way of
example and
not limitation, yeast rho° cells can be prepared by ethanol treatment
(Ibeas and Jimenez,
Appl. Environ. Microbiol. 63:7-12, 1997), and a variety of mammalian
rho° cells can be
prepared by treatment with ditercalinium (moue et al., Biochem. Biophys. Res.
Commun. 239:257-260, 1997), ethidium bromide (King and Attardi, Science
246:500-
22
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
503, 1989; Cavalli et al., Cell Growth Differ. 8:1189-1198, 1997; Miller et
al., J.
Neurochem. 67:1897-1907, 1996) and various antiviral agents (U.S. patent
application
Serial No. 09/069,489).
Methods and compositions for the genetic manipulation of the
mitochondria) genome of the yeast species Saccharomyces cerevisiae have been
described in the art (Steele et al., Proc. Natl. Acad. Sci. U.S.A. 93:5253-
5257, 1996).
Another embodiment of the invention is drawn to the identification and
isolation of
factors that are differentially expressed in yeast cells having genetically
engineered
mitochondria) genomes relative to yeast cells having wildtype mitochondria)
genomes.
to Manipulated cells includes the preceding cell types in which an
organellar genome has been altered by human manipulation; additionally or
alternatively, such cells may comprise alterations in their nuclear genomes
(such as,
e.g., point mutations or "knock-outs" in chromosomal nucleic acid sequences)
or in
non-organellar, extrachromosomal elements (such as, e.g., plasmids, viruses,
and the
like). In the latter instance, genetic elements from a species different from
that to which
the host cell belongs may be introduced into the manipulated cell on the
extrachromasomal element, in which case differentially expressed factors are
those
factors having an altered pattern of expression in response to the exogenic
element(s).
Nucleic Acids and Nucleotide Sequences
A "nucleic acid of interest" is defined herein as a nucleic acid that is
differentially expressed in a particular disease state, under particular
conditions, in
manipulated cells, or in a species-specific manner, as described above. Once a
nucleic
of interest has been identified, it can be used to generate other useful
nucleic acids
having related sequences, including without limitation deoxyribonucleic acids
(DNA).
In a preferred embodiment, an RNA of interest is used to generate a cDNA
molecule
that can be used to detect nucleic acids having the sequence of interest, or
to produce a
polypeptide encoded by the sequence of the RNA of interest.
For example, it is known in the art to isolate mRNAs of interest and have
them reverse-transcribed. Reverse transcription is a process by which a
reverse
23
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
complementary DNA (cDNA) is produced from an RNA molecule which acts as a
template. The RNA portion of the resultant (RNA:DNA) hybrid may then be
displaced
or enzymatically degraded, after which the single-stranded DNA (ssDNA) is used
as a
template for one or more rounds of DNA polymerization, the product of which is
a
double-stranded DNA (dsDNA) molecule. The dsDNA molecule includes the sequence
of the RNA of interest (except that uridine residues in the RNA are replaced
by thymine
residues in the DNA). The nucleotide sequence of the dsDNA is then determined
and
analyzed; additionally or alternatively, the dsDNA is cloned, i. e. ,
incorporated into a
vector DNA that is capable of replication in an appropriate host cell. If the
dsDNA
to molecule includes a sequence that encodes a polypeptide, a preferred vector
is an
expression vector.
A DNA molecule prepared according to the methods of the invention can
be a full-length cDNA, i.e., one comprising a nucleotide sequence that encodes
an entire
protein. At a minimum, a full-length cDNA will encompass a "start"
(translation
initiating) codon, a "stop" (translation terminating) codon, and all the
polypeptide-
encoding sequences in-between. Such an assemblage of elements is known in the
art as
an open reading frame (ORF).
Alternatively, a DNA molecule prepared according to the methods of the
invention can be an Expressed Sequence Tag (EST), i.e., one which does not
comprise a
2o complete ORF but which does comprise a nucleotide sequence that is a
portion of an
ORF or of an mRNA comprising an ORF. An EST is useful in of itself as, e.g., a
probe
in methods for detecting a mRNA of interest. Because a full-length cDNA is
required
for, e.g., recombinant DNA expression of a protein encoded by a mRNA interest,
it may
also be desirable to use an EST as a tool to isolate a full-length cDNA
according to a
variety of methods. For example, a nucleic acid comprising an EST sequence of
interest can be labeled and used to probe preparations of cellular DNA or RNA
for
hybridizing sequences, and such hybridizing sequences can be isolated,
amplified and
cloned according to known methods. As another example, the sequence of an EST
can
be used to prepare primers for inverse PCR, a process by which sequences
flanking an
3o EST of interest can be determined (see, e.g., Benkel and Fong, Genet. Anal.
13:123-
24
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
127, 1996; Silverman, Methods Mol. Biol. 54:145-155, 1996; Pang and Knecht,
BioTechniques 22:1046-1048, 1997; Huang, Methods Mol. Biol. 69:89-96, 1997;
Huang, Methods Mol. Biol. 67:287-294, 1997; and Offringa and van der Lee,
Methods
Mol. Biol. 49:181-195, 1996; all of which are hereby incorporated by
reference).
In methods of cloning full-length cDNAs from ESTs, and as a useful
method in its own right, it is desirable to screen mRNA or cDNA libraries
prepared
from various cells and tissues in order to identify cells and tissues that
express relatively
high levels of a nucleic acid of interest. For example, a nucleic acid of
interest initially
identified in a first disease state (e.g., Alzheimer's disease) can be used to
probe cells
1o from patients suffering from a second disease state (e.g., Parkinson's
disease, MELAS,
MERFF, diabetes, cancer, arthritis, etc.) in order to determine if the nucleic
acid of
interest is differentially expressed in such second disease states. If a
nucleic acid of
interest is differentially expressed in a concordant manner in one or more
second
disease states, then applications developed from a first disease state (e.g.,
diagnostic,
I S prognostic, pharmacogenomic, compound screening methods and therapeutic
compounds and compositions) may be applied to such second disease states.
As another example, a nucleic acid of interest can be used to examine
tissue- or temporal-specific patterns of expression of a nucleic acid of
interest in a
variety of methods known in the art. The nucleic acid of interest can be
detestably
20 labeled and used to probe (i) an immobilized collection of mRNA molecules
(e.g., RNA
Master BlotsTM or Multiple Tissue Northern, MTNTM, Blots from Clontech) or
(ii) a
cDNA library (prepared according to methods known in the art or available
from, e.g.,
Clontech or from depositories such as the American Type Culture Collection,
ATCC,
Manassas, VA). Alternatively or additionally, a sequence of interest can be
used to
25 design specific PCR primers that can be used in amplification reactions in
96-well
plates wherein each well comprises first strand cDNAs from a particular tissue
(such as,
e.g., the Rapid-ScanTM gene expression panel from OriGene Technologies, Ins.,
Rockville, MD); in this embodiment, automated, semi-automated or robotic means
may
be used to carry out such assays.
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Regardless of the method used, the RNA or cDNA that is examined may
be from a variety of species, including without limitation mammals such as
porcine
species, rabbits, bovine species, rodent species (rats and mice) and primates
including
humans; avian species such as chicken or turkey; fish such as Fugu species;
and simple
or complex plants such as Arabidopsis species, Zea mays, potatoes, soybeans,
rice,
wheat and the like. Mammalian tissues that may be examined include but are not
limited to brain (including, by way of example but not limitation, whole brain
and
subsections thereof, e.g., amygdala, caudate nucleus, cerebellum, cerebral
cortex,
frontal lobe, hippocampus, medulla oblongata, occipital lobe, putamen,
substantia nigra,
l0 temporal lobe, thalamus, acumens, subthalamic nucleus), heart, kidney,
spleen, liver,
colon, lung, small intestine, stomach, skeletal muscle, smooth muscle, testis,
uterus,
bladder, lymph nodes, spinal cord, trachea, bone marrow, placenta, salivary
glands,
thyroid glands, thymus, adrenal glands, pancreas, ovary, uterus, prostate,
skin, bone
marrow, fetal brain and fetal liver.
Cell types that can be probed in this manner include, without limitation,
plant and animal cybrids and rho° cells; cells from organisms such as,
for example, any
unicellular organism, multicellular organism, yeast, fungi, protozoa,
parasites,
helminths, invertebrates or vertebrates or other organisms as they are known
in the art
or later identified having mitochondria, chloroplasts or other organelles,
such as, for
example, Caenorhabditis, Neurospora, Spodoptera, Trichopolusia, Phycomycetes,
Ascomycetes, Basidiomycetes, Deuteromycetes, Mycosporum, Trichophyton,
Nannizia,
Arthroderma, Crytptococcus, Coccidioides, Histoplasma, Blastomyces, Candidia,
Cryptococcus, Histoplasma, Saccharomyces, Trichosporon, Coccidioides,
Aspergillus,
Phycomycetes, Sporothrix, Microsporum, Penicillium, Cladosporium, Alternaria,
Geotrichum, Fusarium, Acremonium. Scopulariopsis, Beauveria, Trichophyton,
Eidermophyton, Fusarium, Trichosporon, Phialophora, Trichophyton,
Epidermophyton, Paracoccidioides, Sporothrix, Pityriasis, Entamoeba,
Balantidium,
Naegleria, Acanthamoeba, Giardia, Isospora, Cryptosporidium, Enterocytozoon,
Trichomonas, Plasmodium, Babesia, Trypanosoma, Leishmania, Toxoplasma,
Caenorhabditis elegans, Neurospora crassa, Saccharomyces cerevisae, Spodoptera
26
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
frugiperda, Trichopolusia ni, Xenopus laevis any species or related species
thereof
(Davis et al., Microbiology, Harper and Row, Philadelphia (1980); O'Learly,
Practical
Handbook of Microbiology, CRC Press, Boca Raton, (1989); Baron et al.,
Diagnostic
Microbiology, The C.V. Mosby Company, St. Louis (1990) and Robbins, Pathologic
Basis of Disease, W.B. Saunders Co, Philadelphia (1994); culturable insect
cell lines
such as Sf~ and Sf2l; cells isolated from mammals such as peripheral blood
leukocytes
(PBLs), chondriocytes, and the like; culturable mammalian cell lines such as
differentiable and differentiated cell lines, cultured neuronal cell lines
such as SH-
SYSY or NT2 cells, cultured tumor or cancer cell lines such as Hela cells,
cells isolated
1o from or primary cell cultures derived from human patient suffering from
diseases and
disorders known or suspected of having a mitochondrial component (as defined
herein)
and manipulated cells (as defined herein) derived from any of the preceding.
Such cells
are obtained with informed consent from patients suffering from such diseases
or
disorders, or, in the case of culturable cell lines, are available from a
variety of
commercial sources or from depositories such as the ATCC.
In order to identify tissues or cells from which a cDNA corresponding to
an EST of interest can optimally be prepared, mRNA or cDNA libraries or arrays
derived from the organism from which the EST of interest was isolated are
probed.
Tissues or cells having a high level of expression of the nucleic acid of
interest are
preferably used as sources for full-length nucleic acids, i. e. , nucleic
acids containing all
the genetic information required to express a complete gene product of
interest. The
full-length nucleic acids are used, e.g., to express the gene product (i.e.,
RNA or
protein) of interest or to prepare manipulated cells or transgenic animals in
which the
level of expression or activity, or tissue- or temporal-specific patterns of
expression, of
the gene product of interest is altered relative to the wildtype condition.
Another utility of ESTs and full-length cDNAs is to search in silico for
corresponding protein sequences, in order to identify proteins of interest
encoded
thereby and to prepare antibodies thereto. For example, the nucleotide
sequence of an
EST or cDNA of interest is translated in silico in all six potential reading
frames (three
3o reading frames on each strand of a dsDNA), and the resulting amino acid
sequences are
27
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
used as probes to search protein databases for a match to a portion of a
protein having a
known amino acid sequence. In the case of mitochondria) proteins, it is
desirable to
perform such in silico translations using both the "universal" genetic code
and the
somewhat different genetic code utilized in mitochondria (Table 1 ), as
different amino
acid sequences will result in each case.
TABLE 1: Differences Between the "Universal" and Mitochondria) Genetic Codes
"Universal" Yeast Mitochondria)Mammalian Mitochondria)
Codon Genetic Code Genetic Code Genetic Code
AGA Arg Arg (stop)
AGG Arg Arg (stop)
AUA Ile Met Met
CUA Leu Thr Leu
UGA (stop) Trp Trp
Nucleic acids having or comprising a sequence of interest can be
prepared by a variety of methods known in the art. For example, such nucleic
acids can
be made using molecular biology or synthetic techniques (Sambrook et al.,
Molecular
Cloning, A Laboratory Manual, Cold Spring Harbor Press (1989)). Many
equivalent
bases in nucleotide sequences are known in the art. For example, thymine (T)
residues
in DNA are transcribed into uracil (U) residues in RNA molecules but, because
both T
and U specifically pair with adenine (A) residues, these changes do not impact
hybridization specificity. Nucleic acids comprising such equivalent
substitutions are
within the scope of the disclosure.
As another example, such nucleic acids can be oligonucleotides,
including oligodeoxyribonucleotides and oligodeoxynucleotides synthesized in
vitro by,
for example, the phosphotriester, phosphoramidite or H-phosphanate
methodologies
(see, respectively, Christodoulou, "Oligonucleotide Synthesis: Phosphotriester
Approach," Chapter 2 In: Protocols for Oligonucleotides and Analogs: Synthesis
and
Properties, Agrawal, ed., Methods in Molecular Biology Vol. 20, Humana Press,
Totowa, NJ (1993); Beaucage, "Oligodeoxyribonucleotide Synthesis:
Phosphoramidite
28
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Approach," Chapter 3, Id.; and Froehler, "Oligodeoxynucleotide Synthesis: H-
phosphonate Approach," Chapter 4, Id., all of which are hereby incorporated by
reference).
The length of a nucleic acid according to the present invention can be
chosen by one skilled in the art depending on the particular purpose for which
the
nucleic acid is intended. For PCR primers and antisense oligonucleotides, the
length of
the nucleic acid is preferably from about 10 to about 50 base nucleotides
(nt), more
preferably from about 12 to about 30 nt, and most preferably from about 15 to
about 25
nt. For probes, the length of the nucleic acid is preferable from about 10 to
about 5,000
1o nt, more preferably from about 15 to about 500 nt, and most preferably from
about 20 to
about 100 nt.
Appropriate chemical modifications to nucleic acids of the invention are
also readily chosen by one skilled in the art. Such modifications may include,
for
example, means by which the nucleic acid is detectably labeled for use as a
probe.
Typical detectable labels include radioactive moieties and reporter groups
such as, e.g.,
enzymes and fluorescent or luminescent moieties. Other chemical modifications
appropriate for particular uses, such as antisense applications, as explained
herein.
Detectably labeled nucleic acids are preferred for diagnostic, prognostic
and pharmacogenomic methods of the invention. Whether labeled or unlabeled,
nucleic
2o acids of the invention can be provided in kit form, e.g., in a single or
separate container,
along with other reagents, buffers, enzymes or materials to be used in
practicing at least
one method of the invention. The kit can be provided in a container that can
optionally
include instructions or software for performing a method of the invention.
Such
instructions or software can be provided in any language or human- or machine
readable format.
Machine Readable Formats and Data Processing Systems
The invention is drawn not only to nucleic acids having or comprising a
nucleotide sequence of interest or proteins or polypeptides having or
comprising an
amino acid sequence of interest, but also to such sequences per se when
provided in a
29
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
format, such as data, such as data in a patentable format. Thus, for example,
the present
invention encompasses a format such as a machine-readable format comprising
data
such as one or more nucleotide sequences or amino acid sequences of interest
as
determined or isolated according to the present invention. The format can also
include
one or more nucleotide sequences or amino acid sequences obtained from other
sources,
such as databases of such sequences.
For example, the invention includes data in any format, preferably
provided in a medium of expression such as printed medium, perforated medium,
magnetic medium, holographs, plastics, polymers or copolymers such as
cycoolifin
1 o polymers. Such data can be provided on or in the medium of expression as
an
independent article of manufacture, such as a disk, tape or memory chip, or be
provided
as part of a machine, such as a computer, that is either processing or not
processing the
data, such as part of memory or part of a program. The data can also be
provided as at
least a part of a database. Such database can be provided in any format,
leaving the
choice or selection of the particular format, language, code, selection of
data, form of
data or arrangement of data to the skilled artisan. Such data is useful, for
example, for
comparing sequences obtained by the present invention with known sequences to
identify novel sequences.
One aspect of the invention is a data processing system for storing and
2o comparing at least a portion of data provided by the present invention. The
data
processing system is useful for a variety of purposes, for example, for
storing, sorting or
arranging such data in, for example, database format, and for comparing such
data to
other data, including data of the present invention or from other sources (for
example,
GENBANK or SWISPROT). Such a data processing system can include two or more
of the following elements in any combination:
I. A computer processing system, such as a central processing unit
(CPU). A storage medium or means for storing data, including at least a
portion of the
data of the present invention or at least a portion of compared data, such as
a medium of
expression, such as a magnetic medium or polymeric medium;
CA 02363496 2001-09-14
WO 00/55323 PCT/~1500/07311
II. A processing program or means for sorting or arranging data,
including at least a portion of the data of the present invention, preferably
in a database
format, such as a database program or an appropriate portion thereof such as
they are
known in the art (for example EXCEL or QUATROPRO);
III. A processing program or means for comparing data, including at
least a portion of the data of the present invention, which can result in
compared data,
such as nucleic acid or amino acid comparing programs or an appropriate
portion
thereof, such as they are known in the art [for example BLAST
(http://ncbi.nlm.nih.gov/BLAST (March 7, 1999) and Altschul et al., Nucleic
Acids
to Res. 25:3389-3402 (1997)), ALLIGN, GAP, BESTFIT, FASTA and TFASTA
(Wisconsin Genetics Software Page Release 7.0, Genetics Computer Groups,
Madison,
WI )];
IV. A processing program or means for analyzing at least a portion
of the data of the present invention, compared data, or a portion thereof,
particularly
statistical analysis, such as programs for analyzing nucleic acid or amino
acid sequences
or statistical analysis programs or an appropriate portion thereof as they are
known in
the art [for example SAS, BLAST (http://ncbi.nlm.nih.gov/BLAST (March 7, 1999)
and
Altschul et al., Nucleic Acids Res. 25:3389-3402 (1997)), ALLIGN, GAP,
BESTFIT,
FASTA and TFASTA (Wisconsin Genetics Software Page Release 7.0; Genetics
2o Computer Groups, Madison, WI )];
V. A formatting processing program or means that can format an
output from the data processing system, such as data of the present invention
or a
portion thereof or compared data or a portion thereof, such as database
management
programs or word-processing programs, or appropriate portions thereof as they
are
known in the art; or
VI. An output program or means to output data, such as data of the
present invention or a portion thereof or compared data or a portion thereof
in a format
useful to an end user, such as a human or another data processing system, such
as
database management programs or word-processing programs or appropriate
portions
3o thereof as they are known in the art. Such formats useful to an end user
can be any
31
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
appropriate format in any appropriate form, such as in an appropriate language
or code
in an appropriate medium of expression.
See, generally, United States Patent No. 5,138,695 to Means et al.,
issued August 1 l, 1992; United States Patent No. 5,325,298 to Gallant, issued
June 28,
1994; United States Patent No. 5,398,300 to Levey, issued March 14, 1995;
United
States Patent No. 5,471,627 to Means et al., issued November 28, 1995; United
States
Patent No. 5,619,709 to Caid et al., issued April 8, 1997; United States
Patent No.
5,745,654 to Titan, issued April 28, 1998; United States Patent No. 5,687,306
to Blank,
issued November 11, 1997; United States Patent No. 5,577,179 to Blank, issued
1o November 19, 1996; United States Patent No. 5,469,536 to Blank, issued
November 21,
1995 and United States Patent No. 5,345,313 to Blank, issued September 6,
1994.
When the nucleotide sequence of interest encodes a protein, the
invention is further drawn to the corresponding polypeptide sequences provided
in such
formats. Such formats are useful in, e.g., diagnostic, prognostic or
pharmacogenomic
assays useful in the methods of the invention, or in methods for searching in
silico for
homologs of the sequences of interest.
Expression Systems
In order to produce a gene product of interest in sufficient quantities for
further embodiments of the invention, the nucleotide sequence of interest or
its
functional equivalent, is inserted into an appropriate "expression vector,"
i.e., a genetic
element, often capable of autonomous replication, which contains the necessary
elements for the transcription and, in instances where the gene product is a
protein,
translation of the inserted nucleotide sequence. A genetic element that
comprises an
expression vector and a nucleic acid of interest in an arrangement appropriate
for
expression of a gene product of interest is referred to herein as an
"expression
construct."
Methods which are well known to those skilled in the art can be used to
prepare expression constructs containing a nucleotide sequence of interest and
appropriate transcriptional and translational controls. These methods include
in vitro
32
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
recombinant DNA techniques, synthetic techniques and in vivo recombination or
genetic recombination. Such techniques are known in the art (see, e.g.,
Sambrook et al.,
Molecular Cloning: A Laboratory Manual, Second Edition, Cold Spring Harbor
Press,
Plainview N.Y., 1989; Ausubel et al., eds., Short Protocols in Molecular
Biology,
Second Edition, John Wiley & Sons, New York N.Y., 1992).
A variety of expression vector/host systems may be utilized to contain
and express a nucleotide sequence of interest. These include but are not
limited to
microorganisms such as bacteria transformed with recombinant bacteriophage,
plasmid
or cosmid DNA expression vectors; yeast transformed with yeast expression
vectors;
to insect cell systems infected with virus expression vectors (e.g.,
baculovirus); plant cell
systems transfected with virus expression vectors (e.g., cauliflower mosaic
virus,
CaMV; tobacco mosaic virus, TMV) or transformed with bacterial expression
vectors
(e.g., Ti or pBR322 plasmid); or animal cell systems.
The "control elements" or "regulatory sequences" of these systems,
which may vary in their strength and specificities, are those non-translated
regions of
the vector, enhancers, promoters, and 5' and 3' untranslated regions, which
interact with
host cellular proteins to carry out transcription and, where the gene product
of interest is
a protein, translation. Depending on the vector system and host utilized, any
number of
suitable transcription and translation elements, including constitutive and
inducible
2o promoters, may be used. For example, when cloning in bacterial systems,
inducible
promoters such as the hybrid lacZ promoter of the BluescriptTM phagemid
(Stratagene,
La Jolla, CA.) or pSportl (Life Technologies, Inc., Rockville, MD) and ptrp-
lac hybrids
and the like may be used. In insect cells, the baculovirus polyhedrin promoter
may be
used in insect cells. Promoters and/or enhancers derived from the genomes of
plant
cells (e.g., heat shock, RUBISCO; and storage protein genes) or from plant
viruses (e.g.,
viral promoters or leader sequences) may be cloned into the vector. In
mammalian cell
systems, promoters from mammalian genes or from mammalian viruses are
appropriate.
If it is necessary to generate a cell line that contains multiple copies of
the nucleotide
sequence of interest, vectors based on SV40 or EBV may be used with an
appropriate
3o selectable marker.
33
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
In bacterial systems, a number of expression vectors may be selected
depending upon the use intended for expressed gene product of interest. For
example,
when large quantities of a protein of interest are needed for the induction of
antibodies,
vectors which direct high level expression of the protein of interest, or
fusion proteins
derived therefrom that are more readily assayed and/or purified, may be
desirable.
Such vectors include, but are not limited to, Escherichia coli cloning and
expression vectors such as pET (Stratagene, La Jolla, CA), pRSET (Invitrogen,
Carlsbad, CA) or pGEMEXTM (Promega, Madison, WI) vectors, in which the
sequence
encoding a protein of interest is ligated downstream from a bacteriophage T7
promoter
l0 and ribosome binding site so that, when the expression construct is
transformed into E.
coli expressing the T7 RNA polymerise, large levels of the polypeptide of
interest are
produced; pGEMTM vectors (Promega), in which inserts into sequences encoding
the
lacZ a-peptide may be detected using colorimetric screening; and the like. For
polypeptides that are relatively insoluble, it may be desirable to produce
thioredoxin
fusion proteins using, for example, pBAD/Thio-TOPO vectors (Invitrogen).
Plasmids such as pGEX vectors (Amersham Pharmacia Biotech,
Piscataway, NJ) may be used to express polypeptides of interest as fusion
proteins.
Such vectors comprise a promoter operably linked to a glutathione S-
transferase (GST)
gene from Schistosoma japonicum. (Smith et al., 1988, Gene 67:31-40), the
coding
sequence of which has been modified to comprise a thrombin cleavage site-
encoding
nucleotide sequence immediately 5' from a multiple cloning site. GST fusion
proteins
can be detected by Western blots with anti-GST or by using a colorimetric
assay; the
latter assay utilizes glutathione and 1-chloro-2-4-dinitrobenzene (CDNB) as
substrates
for GST and yields a yellow product detectable at 340 nm (Habig et al., 1974,
J. Biol.
Chem. 249:7130-7139). GST fusion proteins produced from expression constructs
derived from this expression vector can be purified by, e.g., adsorption to
glutathione-
agarose beads followed by elution in the presence of free glutathione. Another
series of
expression vectors of this type are the pBAD/His vectors (Guzman et al., J.
Bact.
177:4121-4130, 1997; Invitrogen, Carlsbad, CA), which contains the following
3o elements operably linked in a S' to 3' orientation: the inducible, but
tightly regulatable,
34
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
araBAD promoter; optimized E. coli translation initiation signals; an amino
terminal
polyhistidine(6xHis)-encoding sequence (also referred to as a "His-tag"); an
XPRESS~
epitope-encoding sequence; an enterokinase cleavage site which can be used to
remove
the preceding N-terminal amino acids following protein purification, if so
desired; a
multiple cloning site; and an in-frame termination codon. Fusion proteins made
from
pBAD/His expression constructs can be purified using substrates or antibodies
that
specifically bind to the His-tag, and assayed by Western analysis using the
Anti-
XpressTM antibody. Proteins made in such systems are designed to include
heparin,
thrombin, enterokinase, factor XA or other protease cleavage sites so that the
cloned
1o polypeptide of interest can be released from the GST moiety by treatment
with the
appropriate protease.
Expression vectors derived from bacteriophage, including cosmids and
phagemids, may also be used to express nucleic acids of interest in bacterial
cells. Such
vectors include, but are not limited to, Lambda FIXTM, Lambda DASHTM, Lambda
ZAPTM, Lambda EMBL3 and EMBL4 bacteriophage vectors, pBluescriptTM phagemids,
SuperCos and pWElS vectors (all available from Stratagene) and the pSL1180
Superlinker Phagemid (Amersham Pharmacia Biotech).
In yeast such as Saccharomyces cerevisiae or Pichia pastoris, a number
of vectors containing constitutive or inducible promoters such as those for
mating factor
alpha, GALI , TEFI , AOXI or GAP may be used. Appropriate expression vectors
include various pYES, pYD and pTEF derivatives (Invitrogen) (see, for example,
Grant
et al., Methods in Enzymology 153:516-544, 1987; Lundblad et al., Units 13.4
to 13.7 of
Chapter 13 in: Short Protocols in Molecular Biology, 2nd Ed., Ausubel et al.,
eds., John
Wiley & Sons, New York, New York, 1992, pages 13-19 to 13-33).
In cases where plant expression vectors are used, the expression of a
nucleotide sequence of interest may be driven by any of a number of promoters.
For
example, viral promoters such as the 35S and 19S promoters of CaMV (Brisson et
al.,
Nature 310:511-514, 1984) may be used alone or in combination with the omega
leader
sequence from TMV (Takamatsu et al., EMBO J. 6:307-311, 1987). Alternatively,
3o plant promoters such as the small subunit of RUBISCO (Coruzzi et al., EMBO
J.
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
3:1671-1680, 1984; Brogue et al., Science 224:838-843, 1984); or heat shock
promoters
(Winter and Sinibaldi, Results Probl. Cell. Differ. 17:85-105, 1991) may be
used.
These constructs can be introduced into plant cells by direct DNA
transformation or
pathogen-mediated transfection. For reviews of such techniques, see Gossen et
al.
(Curr. Opin. Biotechnol. 5:516-520, 1994), Porta and Lomonossoff (Mol.
Biotechnol.
3:209-221, 1996) and Turner and Foster (Mol. Biotechnol. 3:225-36, 1995).
Another expression system which may be used to express a gene product
of interest is an insect system. In one such system, Autographa californica
nuclear
polyhedrosis virus (AcNPV) is used as a vector to express foreign genes in
Spodoptera
to frugiperda cells or in Trichoplusia larvae. The nucleotide sequence of
interest may be
cloned into a nonessential region of the virus, such as the polyhedrin gene,
and placed
under control of the polyhedrin promoter. Successful insertion of the sequence
of
interest will render the polyhedrin gene inactive and produce recombinant
virus lacking
coat protein. The recombinant viruses are then used to infect S. frugiperda
cells or
Trichoplusia larvae in which the gene product of interest is expressed (see
"Piwnica-
Worms, Expression of Proteins in Insect Cells Using Baculovirus Vectors,"
Section II
of Chapter 16 in: Short Protocols in Molecular Biology, 2nd Ed., Ausubel et
al., eds.,
John Wiley & Sons, New York, New York, 1992, pages 16-32 to 16-48; Lopez-
Ferber
et al., Chapter 2 in: Baculovirus Expression Protocols, Methods in Molecular
Biology,
2o Vol. 39, C.R. Richardson, Ed., Humana Press, Totawa, NJ, 1995, pages 25-
63). S.
frugiperda cells (Sf~, Sf21 or High FiveTM cells) and appropriate baculovirus
transfer
vectors are commercially available from, e.g., Invitrogen. Expression systems
utilizing
Drosophila S2 cells (also available from Invitrogen) may also be utilized.
Expression constructs for expressing nucleic acids of interest in
mammalian cells are prepared in a stepwise process. First, expression
cassettes that
comprise a promoter (and associated regulatory sequences) operably linked to a
nucleic
acid of interest are constructed in bacterial plasmid-based systems; these
expression
cassette-comprising constructs are evaluated and optimized for their ability
to produce
the gene product of interest in mammalian cells that are transiently
transfected
3o therewith. Second, these expression cassettes are transferred to viral
systems that
36
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
produce recombinant proteins during lytic growth of the virus (e.g., SV40,
BPV, EBV,
adenovirus; see below) or from a virus that can stably integrate into and
transduce a
mammalian cellular genome (e.g., a retroviral expression construct).
With regard to the first step, commercially available "shuttle" (i.e.,
capable of replication in both E coli and mammalian cells) vectors that
comprise
promoters that function in mammalian cells and can be operably linked to a
nucleic acid
of interest include, but are not limited to, SV40 late promoter expression
vectors (e.g.,
pSVL, Amersham Pharmacia Biotech), glucocorticoid-inducible promoter
expression
vectors (e.g., pMSG, Amersham Pharmacia Biotech), Rous sarcoma enhancer-
promoter
1o expression vectors (e.g., pRc/RSV, Invitrogen) and CMV early promoter
expression
vectors, including derivatives thereof having selectable markers to agents
such as
Neomycin, Hygromycin or ZEOCINTM (e.g., pRc/CMV2, pCDMB, pcDNAl.l,
pcDNA 1.1 /Amp, pcDNA3. l , pcDNA3 .1 /Zeo and pcDNA3. l /Hygro, Invitrogen).
In
general, preferred shuttle vectors for nucleic acids of interest are those
having selectable
markers (for ease of isolation and maintenance of transformed cells) and
inducible, and
thus regulatable, promoters as overexpression of a gene product of interest
may have
toxic effects.
Methods for transfecting mammalian cells are known in the art (see,
Kingston et al., "Transfection of DNA into Eukaryotic Cells," Section I of
Chapter 9 in:
Short Protocols in Molecular Biology, 2nd Ed., Ausubel et al., eds., John
Wiley &
Sons, New York, New York, 1992, pages 9-3 to 9-16). A control plasmid, such as
pCH110 (Pharmacia), may be cotransfected with the expression construct being
examined so that levels of the gene product of interest can be normalized to a
gene
product expressed from the control plasmid. Preferred expression cassettes,
consisting
essentially of a promoter and associated regulatory sequences operably linked
to a
nucleic acid of interest, are identified by the ability of cells transiently
transformed with
a vector comprising a given expression cassette to express high levels of the
gene
product of interest, or a fusion protein derived therefrom, when induced to do
so.
Expression may be monitored by Northern or Western analysis or, in the case of
fusion
37
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
proteins, by a reporter moiety such as an enzyme or epitope. Effective
expression
cassettes are then incorporated into viral expression vectors.
Nucleic acids, preferably DNA, comprising preferred expression
cassettes are isolated from the transient expression constructs in which they
were
prepared, characterized and optimized. A preferred method of isolating such
expression
cassettes is by amplification by PCR, although other methods (e.g., digestion
with
appropriate restriction enzymes) can be used. Preferred expression cassettes
are
introduced into viral expression vectors, preferably retroviral expression
vectors, in the
following manner.
to A DNA molecule comprising a preferred expression cassette is
introduced into a retroviral transfer vector by ligation. Two types of
retroviral transfer
vectors are known in the art: replication-incompetent and replication-
competent.
Replication-incompetent vectors lack viral genes necessary to produce
infectious
particles but retain cis-acting viral sequences necessary for viral
transmission. Such cis-
acting sequences include the ~I' packaging sequence, signals for reverse
transcription
and integration, and viral promoter, enhancer, polyadenylation and other
regulatory
sequences. Replication-competent vectors retain all these elements as well as
genes
encoding virion structural proteins (typically, those encoded by genes
designated gag,
pol and env) and can thus form infectious particles in a variety of cell
lines. In contrast,
these functions are supplied in traps to replication-incompetent vectors in a
packaging
cell line, i.e, a cell line that produces mRNAs encoding gag, pol and env
genes but
lacking the 'f packaging sequence. See, generally, Cepko, Unit 9.10 of Chapter
9 in:
Short Protocols in Molecular Biology, 2nd Ed., Ausubel et al., eds., John
Wiley &
Sons, New York, New York, 1992, pages 9-30 to 9-35.
A retroviral construct comprising an expression cassette comprising a
nucleic acid of interest produces RNA molecules comprising the cassette
sequences and
the ~I' packaging sequence. These RNA molecules correspond to viral genomes
that are
encapsidated by viral structural proteins in an appropriate cell line (by
''appropriate" it
is meant that, for example, a packaging cell line must be used for constructs
based on
3o replication-incompetent retroviral vectors). Infectious viral particles are
then produced,
38
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
and released into the culture supernatant, by budding from the cellular
membrane. The
infectious particles, which comprise a viral RNA genome that includes the
expression
cassette for the gene product of interest, are prepared and concentrated
according to
known methods. It may be desirable to monitor undesirable helper virus, i. e.
, viral
particles which do not comprise the expression cassette for the gene product
of interest.
See, generally, Cepko, Units 9.11, 9.12 and 9.13 of Chapter 9 in: Short
Protocols in
Molecular Biology, 2nd Ed., Ausubel et al., eds., John Wiley & Sons, New York,
New
York, 1992, pages 9-36 to 9-45.
Viral particles comprising an expression cassette for the gene product of
1o interest are used to infect in vitro (e.g., cultured cells) or in vivo
(e.g., cells of a rodent,
or of an avian species, which are part of a whole animal). Tissue explants or
cultured
embryos may also be infected according to methods known in the art. See,
generally,
Cepko, Unit 9.14 of Chapter 9 in: Short Protocols in Molecular Biology, 2nd
Ed.,
Ausubel et al., eds., John Wiley & Sons, New York, New York, 1992, pages 9-45
to 9-
48. Regardless of the type of cell used, production of the gene product of
interest is
directed by the recombinant viral genome.
In eukaryotic expression systems, host cells may be chosen for its ability
to modulate the expression of the inserted sequences or, when the gene product
of
interest is a protein, to process the protein of interest in the desired
fashion. Such
2o modifications of proteins include, but are not limited to, acetylation,
carboxylation,
glycosylation, phosphorylation, lipidation and acylation. Post-translational
processing
which cleaves a "prepro" form of the protein of interest may also be important
for its
correct intracellular localization, folding and/or function. Different host
cells such as
CHO, HeLa, MDCK, HEK293, WI38, etc. have specific cellular machinery and
characteristic mechanisms for such post-translational activities and may be
chosen to
ensure the correct modification and processing of a protein of interest.
It may be desirable to use expression systems that can be tightly
regulated, particularly in mammalian cells. By ''tightly regulated" it is
meant that the
expression system is normally repressed (i.e., kept from expressing the gene
of interest)
3o but can be induced to high levels of expression upon the addition of an
inducing agent
39
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
to the cells harboring the expression construct. Such tightly regulated
expression
systems include, but are not limited to, ecdysone-inducible mammalian
expression
systems, tetracycline-regulated expression systems (such as the T-RExTM
system,
Invitrogen), and the GeneSwitchTM system (Invitrogen).
Expression systems of the invention also include the few systems in
which a nucleic acid of interest is expressed from an organellar genome. Means
for the
genetic manipulation of the mitochondria) genome of Saccharomyces cerevisiae
(Steele
et al., Proc. Natl. Acad. Sci. U.S.A. 93:5253-5257, 1996) and systems for the
genetic
manipulation of plant chlorplasts (U.S. Patent No. 5,693,507; Daniel) et al.,
Nature
1o Biotechnology 16:345-348, 1998) have been described. Naturally, nucleic
acids that
encode polypeptide sequences have to be altered in organellar expression
systems in
order to reflect the differences in the genetic codes of organelles (see Table
1 ).
Genetic Modulation of Nucleic Acids and Gene Products
Various antisense-based methodologies may be used to modulate (reduce
or eliminate) the expression of a nucleic acid of interest, and the
corresponding gene
product, in organelles, cells, tissues, organs and organisms. Such antisense
modulation
may be used to validate the role of a gene of interest in a disease or
disorder or, when
the causes or symptoms of a disease or disorder result from the over-
expression of a
nucleic acid of interest, as therapeutic agents.
2o The term "antisense" refers to nucleic acids that comprise one or more
sequences that are the reverse complement of the "sense" strand of a gene,
i.e., the
strand that is transcribed and, in the case of protein-encoding sequences,
translated.
Because antisense nucleic acids bind with high specificity to their targeted
nucleic
acids, selectivity is high and toxic side effects resulting from misdirection
of the
compounds can be minimal.
In general, antisense compositions are of two types: (i) synthetic
antisense oligonucleotides, including enzymatic ones such as, e.g., ribozymes;
and (ii)
antisense expression constructs. One skilled in the art will be able to
utilize either
modality as is appropriate to the given situation.
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Synthetic antisense oligonucleotides are prepared from the reverse
complement of a nucleic acid of interest. An antisense oligonucleotide
consists of
nucleic acid sequences corresponding to the reverse complement of a
differentially
expressed RNA. When introduced into cells expressing the RNA of interest, the
antisense oligonucleotides specifically bind to the RNA molecules and
interfere with
their function by preventing secondary structures from forming or blocking the
binding
of regulatory or RNA-stabilizing factors. In addition, in the case of protein-
encoding
RNA species, oligonucleotides can inhibit RNA splicing, polyadenylation or
protein
translation, thus limiting or preventing the amount of protein made from such
mRNAs.
1 o Additionally or alternatively, such oligoncuelotides can bind to double-
stranded DNA
molecules and form triplexes therewith, and thus interfere with the
transcription of such
sequences.
In instances where it is desired to target antisense oligonucleotides to
RNAs produced from organellar genomes, peptide nucleic acids (PNAs) are
preferred
synthetic oligonucleotides. In PNAs, the sugar-phosphate backbone of
biological
nucleic acids has been replaced with a polypeptide-like chain. Targeting
sequences that
direct proteins to organelles can be conjugated to the backbone of antisense
PNAs, with
the result being that such conjugates are preferentially delivered to the
targeted
organelle (see, for example, published PCT applications WO 97/41150 and WO
99/05302, and Taylor et al., Nature Genetics 15:212-215, 1997).
Antisense oligonucleotides may be inherently enzymatic in nature, that
is, capable of degrading the RNA molecule towards which they are targeted;
such
molecules are generally referred to as "ribozymes." A variety of increasingly
short
synthetic ribozyme frameworks that can be modified to comprise a nucleic acid
sequence of interest have been described (Couture and Stinchcomb, Trends
Genet.
12:510-515, 1996), including but not limited to hairpin ribozymes (Hampel,
Prog.
Nucleic Acid Res. Mol. Biol. X8:1-39, 1998), hammerhead ribozymes (Birikh et
al., Eur.
J. Biochem. 245:1-16, 1997) and minizymes (Kuwabara et al., Nature
Biotechnology
16:961-965, 1998).
41
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
In the case of non-catalytic antisense nucleic acids in general, and
ribozymes in particular, antisense modulation in a cell can also be achieved
by
expression constructs that direct the transcription of the reverse complement
of a
nucleotide sequence of interest in vivo. For example, in order to express non-
catalytic
antisense transcripts in mammalian or plant cells, all that may be required is
the
"flipping" (i.e., reversing the orientation) of a nucleic acid of interest
that has been
cloned into a mammalian or plant expression vector, respectively. It is not
necessary to
maintain the proper relationship of elements such as translation signals and
the like as
the minimum requirement for an antisense expression construct of this type is
a
1o promoter operably linked to the reverse complement of a nucleic acid of
interest. It is
also possible to design expression constructs that express ribozymes in cells.
Antisense
and ribozyme expression constructs are also used to produce transgenic animals
in
which the level of expression of a gene of interest can be modulated in a
temporal- or
tissue-specific manner (see Sokol and Murray, Transgenic Res. x:363-371, 1996,
for a
review).
Nucleic acid sequences derived according to the present invention may
also be used to design "RNA decoys," i.e., short RNA molecules corresponding
to cis-
acting regulatory sequences that bind traps-acting regulatory factors. When
overexpressed in a cell or administered in excess thereto, such RNA decoys
2o competitively inhibit the binding and thus action of the traps-acting
regulatory factors,
and thus limit or prevent the ability of such factors to carry out processes
that stabilize
(or destabilize) the RNA of interest, or enhance (or decrease) the
polyadenylation,
splicing nuclear transport, or translation of the RNA (Sullenger et al., J.
Yirol. 65:6811-
6816, 1991 ). Expression of the RNA of interest may thus be either enhanced or
decreased for therapeutic purposes.
Transgenic and Transmitochondrial Animals
Transgenic animals, modified with regard to a nucleic acid of interest,
may be prepared. Such animals are useful for developing animal models of human
disease and for evaluating the safety and effectiveness of therapeutic agents
of the
42
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
invention. In general, such transgenic animals are of three types: (i)
"transgenic knock-
outs," in which the animal's homolog of a gene of interest is disrupted or
removed, with
a resulting more-or-less total loss of function of the corresponding gene
product; (ii)
"regulatable transgenics," in which the gene of interest is operably linked to
an
inducible promoter; and (iii) "replacement transgenics," in which the animal's
homolog
of the gene of interest has been replaced with the human gene of interest,
which may be
expressed from an endogenous or inducible promoter.
The non-human transgenic animals of the invention comprise any animal
that can be genetically manipulated to produce one or more of the above-
described
1o classes of transgenic animals. Such non-human animals include vertebrates
such as
rodents, non-human primates, sheep, dog, cow, amphibians, reptiles, etc.
Preferred
non-human animals are selected from non-human mammalian species of animals,
including without limitation animals from the rodent family including but not
limited to
rats and mice, most preferably mice (see, e.g., U.S. Patents 5,675,060 and
5,850,001).
Other non-human transgenic animals that may be prepared include without
limitation
rabbits (U.S. Patent No. 5,792,902), pigs (U.S. Patent No. 5,573,933), bovine
species
(U.S. Patents 5,633,076 and 5,741,957) and ovine species such as goats and
sheep (U.S.
Patents 5,827690; 5,831,141; and 5,849,992).
The transgenic animals of the invention are animals into which has been
2o introduced by non-natural means (i.e., by human manipulation), one or more
genes that
do not occur naturally in the animal, e.g., foreign genes, genetically
engineered
endogenous genes, etc. The non-naturally introduced genes, known as
transgenes, may
be from the same or a different species as the animal but not naturally found
in the
animal in the configuration and/or at the chromosomal locus conferred by the
transgene.
Transgenes may comprise foreign DNA sequences, i.e., sequences not normally
found
in the genome of the host animal. Alternatively or additionally, transgenes
may
comprise endogenous DNA sequences that are abnormal in that they have been
rearranged or mutated in vitYO in order to alter the normal in vivo pattern of
expression
of the gene, or to alter or eliminate the biological activity of an endogenous
gene
3o product encoded by the gene. (Watson et al., in Recombinant DNA, 2d Ed.,
W.H.
43
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Freeman & Co., New York, 1992), pages 255-272; Gordon, Intl. Rev. Cytol.
115:171-
229, 1989; Jaenisch, Science 240:1468-1474, 1989; Rossant, Neuron 2:323-334,
1990).
The transgenic non-human animals of the invention are produced by
introducing transgenic constructs comprising sequences of interest, or the
host animal's
homologs thereof, into the germline of the non-human animal. Embryonic target
cells
at various developmental stages are used to introduce the transgenes of the
invention.
Different methods are used depending on the stage of development of the
embryonic
target cell(s).
Microinjection of zygotes is the preferred method for incorporating
l0 transgenes into animal genomes in the course of practicing the invention. A
zygote, a
fertilized ovum that has not undergone pronuclei fusion or subsequent cell
division, is
the preferred target cell for microinjection of transgenic DNA sequences. The
murine
male pronucleus reaches a size of approximately 20 micrometers in diameter, a
feature
which allows for the reproducible injection of 1-2 picoliters of a solution
containing
transgenic DNA sequences. The use of a zygote for introduction of transgenes
has the
advantage that, in most cases, the injected transgenic DNA sequences will be
incorporated into the host animal's genome before the first cell division
(Brinster et al.,
Proc. Natl. Acad. Sci. U.S.A. 82:4438-4442, 1985). As a consequence, all cells
of the
resultant transgenic animals (founder animals) stably carry an incorporated
transgene at
2o a particular genetic locus, referred to as a transgenic allele. The
transgenic allele
demonstrates Mendelian inheritance: half of the offspring resulting from the
cross of a
transgenic animal with a non-transgenic animal will inherit the transgenic
allele, in
accordance with Mendel's rules of random assortment.
Viral integration can also be used to introduce the transgenes of the
invention into an animal. The developing embryos are cultured in vitro to the
developmental stage known as a blastocyte. At this time, the blastomeres may
be
infected with appropriate retroviruses (Jaenisch, Proc. Natl. Sci. U.S.A.
73:1260-1264,
1976; Soriano and Jaenisch, Cell 46:19-29, 1986). Infection of the blastomeres
is
enhanced by enzymatic removal of the zona pellucida (Hogan, et al., in
Manipulating
the Mouse Embryo, Cold Spring Harbor Press, Cold Spring Harbor, N.Y., 1986).
44
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Transgenes are introduced via viral vectors which are typically replication-
defective but
which remain competent for integration of viral-associated DNA sequences,
including
transgenic DNA sequences linked to such viral sequences, into the host
animal's
genome (Jahneret al., Proc. Natl. Acad Sci. U.S.A. 82:6927-6931, 1985; Van der
Putten
et al., Proc. Natl. Acad. Sci. U.S.A. 82:6148-6152, 1985). Transfection is
easily and
efficiently obtained by culture of blastomeres on a mono-layer of cells
producing the
transgene-containing viral vector (Van der Putten et al., Proc. Natl. Acad.
Sci. U.S.A.
82:6148-6152, 1985; Stewart, et al., EMBO J. 6:383-388, 1987). Alternatively,
infection may be performed at a later stage, such as a blastocoele (Jahneret
al., Nature
298:623-628, 1982). In any event, most transgenic founder animals produced by
viral
integration will be mosaics for the transgenic allele; that is, the transgene
is
incorporated into only a subset of all the cells that form the transgenic
founder animal.
Moreover, multiple viral integration events may occur in a single founder
animal,
generating multiple transgenic alleles which will segregate in future
generations of
offspring. Introduction of transgenes into germline cells by this method is
possible but
probably occurs at a low frequency (Jahner et al., Nature 298:623-628, 1982).
However, once a transgene has been introduced into germline cells by this
method,
offspring may be produced in which the transgenic allele is present in all of
the animal's
cells, i.e., in both somatic and germline cells.
2o Embryonic stem (ES) cells can also serve as target cells for introduction
of the transgenes of the invention into animals. ES cells are obtained from
pre-
implantation embryos that are cultured in vitro (Evans et al., Nature 292:154-
156.
1981; Bradley et al., Nature 309:255-258, 1984; Gossler et al., Proc. Natl.
Acad. Sci.
U.S.A. 83:9065-9069, 1986; Robertson et al., Nature 322:445-448, 1986;
Robertson,
E.J., in Teratocarcinomas and Embryonic Stem Cells: A Practical Approach,
Robertson, E.J., ed., IRL Press, Oxford, 1987, pp. 71-112). ES cells, which
are
commercially available (from, e.g., Genome Systems, Inc., St. Louis, MO), can
be
transformed with one or more transgenes by established methods (Lovell-Badge,
R.H.,
in Teratocarcinomas and Embryonic Stem Cells: A Practical Approach, Robertson,
3o E.J., ed., IRL Press, Oxford, 1987, pp. 153-182). Transformed ES cells can
be
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
combined with an animal blastocyst, whereafter the ES cells colonize the
embryo and
contribute to the germline of the resulting animal, which is a chimera
(composed of
cells derived from two or more animals) (Jaenisch, Science 240:1468-1474,
1988;
Bradley in: Teratocarcinomas and Embryonic Stem Cells: A Practical Approach,
Robertson, E.J., ed., IRL Press, Oxford 1987, pp. 113-151). Again, once a
transgene
has been introduced into germline cells by this method, offspring may be
produced in
which the transgenic allele is present in all of the animal's cells, i.e., in
both somatic
and germline cells.
However it occurs, the initial introduction of a transgene is a Lamarckian
(non-Mendelian) event. However, the transgenes of the invention may be stably
integrated into germ line cells and transmitted to offspring of the transgenic
animal as
Mendelian loci. Other transgenic techniques result in mosaic transgenic
animals, in
which some cells carry the transgenes and other cells do not. In mosaic
transgenic
animals in which germ line cells do not carry the transgenes, transmission of
the
transgenes to offspring does not occur. Nevertheless, mosaic transgenic
animals are
capable of demonstrating phenotypes associated with the transgenes.
Offspring that have inherited the transgenes of the invention are
distinguished from littermates that have not inherited transgenes by analysis
of genetic
material from the offspring for the presence of biomolecules that comprise
unique
2o sequences corresponding to sequences of, or encoded by, the transgenes of
the
invention. For example, biological fluids that contain polypeptides uniquely
encoded
by the transgenes of the invention may be immunoassayed for the presence of
the
polypeptides. A more simple and reliable means of identifying transgenic
offspring
comprises obtaining a tissue sample from an extremity of an animal, e.g., a
tail, and
analyzing the sample for the presence of nucleic acid sequences corresponding
to the
DNA sequence of a unique portion or portions of the transgenes of the
invention. The
presence of such nucleic acid sequences may be determined by, e.g.,
hybridization
("Southern") analysis with DNA sequences corresponding to unique portions of
the
transgene, analysis of the products of PCR reactions using DNA sequences in a
sample
3o as substrates and oligonucleotides derived from the transgene's DNA
sequence, etc.
46
CA 02363496 2001-09-14
WO 00/55323 PCT/IJS00/07311
Cloned animals, transgenic and otherwise, of the invention may also be
prepared (for a review of mammalian cloning techniques, see Wolf et al., J.
Assist.
Reprod. Genet. 15:235-239, 1998). Such cloned animals include, without
limitation,
ovine species such as sheep (Campbell et al., Nature 380:64-66, 1996; Wells et
al., Biol.
Reprod. 57:385-393, 1997) rodents such as mice (Wakayama et al., Nature
394:369-
374, 1998) and non-human primates such as rhesus monkeys (Meng et al., Biol.
Reprod.
57:454-459, 1997).
The transgenic and cloned animals of the invention may be used as
animal models of human disease states and to evaluate potential therapies for
such
l0 disease states. For example, in such methods, a first transgenic animal
having a disease
state (or one or more symptomatic components thereof) is given a known dose of
a
candidate therapeutic composition or exposed to a candidate therapeutic
treatment, and
a second (control) transgenic animal is given a placebo or not exposed to the
candidate
therapeutic treatment. Symptoms and/or clinical end-points relevant to the
disease state
are measured in both animals over appropriate intervals of time, and the
results are
compared. Therapeutic (desirable) compositions and treatments are identified
as those
which ameriolate, delay the onset of or eliminate such symptoms and end-points
in the
treated animal relative to the control animal. In like fashion, undesirable
compositions
and treatments that aggravate or accelerate the disease state are identified
as those
2o which enhance the degree of such symptoms and end-points and/or hasten
their onset.
Because of their high degree of genetic identity, cloned transgenic animals
are preferred
in such methods.
With regard to transmitochondrial animals, two types of such animals
presently exist. First, because of the way they are generated ("nuclear
transfer"),
"Dolly-like" cloned animals are cybrid-like transmitochondrial animals. In
nuclear
transfer, a donor somatic cell is electrofused with a recipient enucleated
oocyte; this
method was used to produce Dolly, the first mammal reported to have been
cloned
(Wilmut et al., Nature 385:810-813, 1997). When the mitochondria) DNA (mtDNA)
in
Dolly and in nine other nuclear transfer-derived sheep generated from fetal
cells was
3o examined, it was found that the mtDNA of each of the ten nuclear-transfer
sheep was
47
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
derived exclusively from recipient enucleated oocytes. There was no detectable
contribution of mtDNA from the respective somatic donor cells. Thus, although
these
ten sheep are authentic nuclear clones, they are in fact "cybrid animals",
containing
mtDNA that is (apparently) derived from the oocyte, and nuclear DNA derived
from the
somatic cells used in the cloning process (Evans et al., Nature Genetics 23:90-
93,
1999).
A second type of transmitochondrial animal is a heteroplasmic animal,
i.e., one that has been manipulated so that the animal contains mitochondria)
genomes
from two or more animals. Such animals may (or may not) contain heteroplasmic
cells
1 o in which two different mitochondria) genomes are contained, and/or may be
chimeric
with regard to their heteroplasmy (i.e., some cells contain only a first
mitochondria)
genome, whereas other cells only contain a second mitochondria) genome.
In any event, heteroplasmic transmitochondrial animals can be generated
in at least two ways. In one method of generating heteroplasmic
transmitochondrial
animals, purified mitochondria from a first animal having one mitchondrial
genome are
micro-injected into ova derived from a second animal having a different
mitochondria)
genome, and the manipulated ova are then implanted into pseudopregnant mice
(see
Pinkert et al., Transgenic Research 6:379-383, 1997; Irwin et al., Transgenic
Research
8:119-123, 1999; and WO 99/05259). In a second method of generating
heteroplasmic
2o transmitochondrial animals, one-cell embryos of one strain of animal are
electrofused to
cytoplasts recovered from zygotes of another strain of animal (Jenuth et al.,
Nature
Genetics 14:146-151, 1996).
Polypeptides and Proteins
The nucleic acids of interest identified according to the methods of the
invention may encode amino acid sequences. Such amino acid sequences may
correspond to a full-length protein or to a polypeptide portion thereof.
In instances wherein a full-length protein is encoded by a nucleic acid of
interest, the protein may be a known protein that is commercially available or
one to
which antibodies are known and can be used to isolate the protein from
appropriate
48
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
biological samples. If a full-length protein of the invention has not
previously been
described, it may be produced via recombinant DNA methodologies or prepared
from
biological samples using known biochemical techniques. Short (i. e. , having
less than
about 30 amino acids) polypeptides that are encoded by short (i.e., having
less than
about 100 nucleotides) nucleic acids of the invention or derived from the
amino acid
sequences encoded by longer nucleic acids or from full-length proteins can be
synthesized in vitro by methods known in the art. Fusion proteins comprising
amino
acid sequences of interest may also be prepared and are included within the
scope of the
polypeptides and proteins of the invention.
1o Regardless of the means by which they are prepared, the polypeptides
and proteins of the invention have a variety of applications. They may be used
to
generate antibodies or to screen for ligands that may serve as therapeutic
agents, or may
themselves be used as therapeutic agents. Full-length proteins of the
invention may
have the activity of the wildtype protein and may thus be used to treat
conditions
resulting from a loss of such activity. Polypeptides of the invention may also
have such
activities, or may competitively inhibit a protein of interest in vivo by
binding a ligand
of the protein. If the ligand is an activator of the protein, such
polypeptides may be
used to treat conditions resulting from the over-expression or over-activation
of the
protein in vivo. If the ligand is a toxin or activator of cell death
(apoptosis or necrosis),
2o administration of a protein or polypeptide that binds such a ligand to a
patient in need
thereof will have the beneficial effect of competitively inhibiting the action
of the toxin
or cell death activator.
Antibodies
Antibodies to a protein or polypeptide of interest are prepared according
to a variety of methods known in the art. In general, such antibodies may be
polyclonal,
monoclonal or monospecific antibodies. Primary antibodies of the invention
bind
specifically to a particular protein or polypeptide of interest and are thus
used in assays
to detect and quantitate such proteins and polypeptides. In such assays,
generally
referred to in the art as immunoassays, a primary antibody of the invention is
detectably
49
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
labeled or is specifically recognized and monitored by a detectably labeled
secondary
antibody or a combination of a secondary antibody and a tertiary molecule
(which may
also be an antibody) that is detectably labeled. Regardless of the specific
format, the
primary antibody of the invention provides a means by which a protein or
polypeptide
s of interest is specifically bound and subsequently detected. One preferred
assay format
is the Enzyme-Linked Immunosorbent Assay (ELISA) format.
A nucleic acid of interest may encode a known protein or a portion
thereof, or a polypeptide sequence that is homologous to a known protein. In
such
instances, antisera to the known protein, or the known protein itself, may be
1 o commercially available. In the latter instance, or when the nucleic acid
of interest can
be used to produce a protein of interest (or a polypeptide portion thereof
greater than
about 30 amino acids in length) via recombinant DNA expression techniques, the
known or recombinantly-produced protein can be used to immunize a mammal of
choice (e.g., a rabbit, mouse or rat) in order to produce antisera from which
polyclonal
15 antibodies can be prepared (see, e.g., Cooper and Paterson, Units 11.12 and
11.13 in
Chapter 11 in: Short Protocols in Molecular Biology, 2nd Ed., Ausubel et al.,
eds., John
Wiley & Sons, New York, New York, 1992, pages 11-37 to 11-41).
In the event that a nucleic acid sequence of interest encodes a
polypeptide sequence for which no complete protein (or homolog thereof) is
known, is
2o too short to encode more than about 30 amino acids (i.e., the nucleic acid
of interest is
less than about 100 nucleotides in length), or encodes more than one
polypeptide
sequence of potential interest, such candidate amino acid sequences can be
used to
synthesize one or more polypeptide molecules, each of which has a defined
amino acid
sequence. Such synthetic polypeptides can then be used to immunize animals
(e.g.,
25 rabbits) according to methods known in the art (Collawn and Paterson, Units
11.14 and
11.15 in Chapter 11 in: Short Protocols in Molecular Biology, 2nd Ed., Ausubel
et al.,
eds., John Wiley & Sons, New York, New York, 1992, pages l l-42 to 11-46;
Cooper
and Paterson, Units 11.12 and 11.13 in Chapter 11 in: Short Protocols in
Molecular
Biology, 2nd Ed., Ausubel et al., eds., John Wiley & Sons, New York, New York,
1992,
3o pages 11-37 to 11-41). The resulting antisera, which is specific for a
particular peptide
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
and is sometimes referred to as "monospecific," may then be used to probe
cells from
which the nucleic acid of interest was isolated. A positive response to a
given
antiserum indicates that the candidate reading frame from which the synthetic
polypeptide used to raise the antiserum was derived is a reading frame used to
encode at
least one protein in the cells) so examined. Moreover, such an antiserum can
be used
to identify proteins of interest in the cells from which the nucleic acid of
interest was
isolated.
Because of their high degree of specificity and homogeneity, monoclonal
antibodies are often the preferred type of antibody for a variety of
applications.
to Methods for producing and preparing monoclonal antibodies are known in the
art (see,
e.g., Fuller et al., Units 11.4 to 11.11 in Chapter 11 in: Short Protocols in
Molecular
Biology, 2nd Ed., Ausubel et al., eds., John Wiley & Sons, New York, New York,
1992,
pages 11-22 to 11-36). Murine monoclonal antibodies may be "humanized" and
used as
therapeutic agents (see, e.g., Gussow and Seemann, Methods in Enzymology
203:99-
121, 1991; Vaughan et al., Nature Biotechnology 16:535-539, 1998).
Antibodies to proteins and polypeptides of interest are used to detect
such proteins and polypeptides in a variety of assay formats. Such
immunoassays may
useful in diagnostic, prognostic or pharmacogenomic methods of the invention,
or in
methods in which various cell types, tissues or organs are probed for the
presence of a
protein of interest. Monoclonal antibodies are generally preferred for such
methods due
to their high degree of specificity and homogeneity.
Diagnostic Prognostic and Pharmaco~enomic Methods
Assays for or utilizing one or more of the antibodies, polypeptides and
proteins, ligands therefor and nucleic acid probes and primers of the
invention are used
in diagnostic, prognostic and pharmacogenomic methods of the invention. The
term
"diagnostic" refers to assays that provide results which can be used by one
skilled in the
art, typically in combination with results from other assays, to determine if
an
individual is suffering from a disease or disorder of interest, whereas the
term
"prognostic" refers to the use of such assays to evaluate the response of an
individual
51
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
having such a disease or disorder to therapeutic or prophylactic treatment.
The term
"pharmacogenomic" refers to the use of assays to predict which individual
patients in a
group will best respond to a particular therapeutic or prophylactic
composition or
treatment.
The terms "disease" and "disorder" refer without limitation to illnesses
and abnormal conditions resulting from infection by one or more pathogens or
parasites,
exposure to toxic compounds or harmful physical conditions, genetic
deficiencies such
as inborn errors of metabolism, hyperproliferative diseases such as tumors and
cancers,
auto-immune disorders, psychological and metal disorders, undesirable results
of the
to aging process, inabilities to perform sexual activities, damage resulting
from physical
trauma or environmental conditions and the like. Neither disease nor disorder
encompasses pregnancy per se but certain diseases and disorders may
particularly
impact pregnant individuals or fetuses and embryos.
In diagnostic applications of the invention, samples from individuals are
assayed with regard to the relative or absolute amounts of a "marker," i.e., a
nucleic
acid or protein of interest, or an endogenous ligand of or antibody to a
nucleic acid or
protein of interest. An increased or decreased level of a marker relative to
control levels
indicates that the individual from which the sample was taken has, has had, or
is likely
to develop the disease or disorder of interest. The term "control level"
refers to the
level of marker present in samples taken from one or more individuals known to
not
have the disease or disorder of interest, or to the level of marker present in
a sample
taken from the individual in question before of after the diagnostic sample.
Additionally or alternatively, a number of individuals known to not have the
disease or
disorder of interest are tested for levels of the marker, and an absolute
amount or
concentration corresponding to a normal level of the marker is established; in
this
embodiment, effected individuals are identified as those having a level of
marker that is
significantly lower or higher than the normal value.
In prognostic applications of the invention, samples from individuals are
assayed as in the preceding paragraph, but (i) the individuals in question are
known to
3o be suffering from the disease or disorder of interest and (ii) the results
of the assays are
52
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
put to a related but different use. Specifically, such assays are used to
evaluate the
response of an individual having a disease or disorder to therapeutic or
prophylactic
treatment, and to predict the course of recovery therefrom or to determine the
need for
additional or alternative treatments.
In pharmacogenomic applications of the invention, patients suffering
from a disease or disorder of interest are stratified with regard to desirable
or
undesirable responses using one or more assays of the invention. A therapeutic
composition and/or treatment known to be more effective, or which produces
more
side-effects, in some patients as compared to others is administered a group
of patients
suffering from a disease or disorder of interest. A method of identifying
which patients
having the disease are more likely to respond to a therapeutic composition
and/or
treatment comprises providing samples from a group of patients having the
disease;
measuring the amount of a protein or polypeptide of interest, or of a nucleic
acid of
interest, or a ligand therefor or antibody thereto, present in the samples;
providing the
therapeutic composition and/or treatment to the patients; measuring the
degree,
frequency, rate or extent of responses of the patients to the therapeutic
composition
and/or treatment; and determining if a correlation exists between the amount
of amount
of the protein or polypeptide of interest, or of a nucleic acid of interest,
or a ligand
therefor or antibody thereto present in the samples and the degree, frequency,
rate or
2o extent of such responses.
The resulting correlations are used to stratify patients in the following
manner. If such a correlation is a positive correlation, the presence of such
correlation
indicates that patients yielding samples having an increased amount of the
protein or
polypeptide of interest, or the ligand therefor, or of the nucleic acid of
interest are more
likely to respond to the treatment. In contrast, if the correlation is a
negative correlation,
the presence of the correlation indicates that patients yielding samples
having an
increased amount of the protein or polypeptide of interest, or the ligand
therefor, or of
the nucleic acid of interest are less likely to respond to the treatment.
The responses) that are measured in these methods can be desirable
3o response(s), in which case it is preferred to provide the therapeutic
composition and/or
53
CA 02363496 2001-09-14
WO 00/55323 PCT/iJS00/07311
treatment to patients having a relatively high level of the protein or
polypeptide of
interest, or the ligand therefor, or of the nucleic acid of interest present.
Alternatively,
the responses) that are measured in these methods can be undesirable
response(s), in
which case it is preferred to avoid providing the therapeutic composition
and/or
treatment to patients having a relatively high level of the protein or
polypeptide of
interest, or the ligand therefor, or of the nucleic acid of interest.
The assays for the preceding methods may be performed at a laboratory
to which patient-derived samples or delivered, or at the site of patient
treatment. In the
latter instance, kits for performing one or more assays of the invention are
preferred.
1o Antibodies, polypeptides and proteins, ligands therefor and nucleic acid
probes and
primers of the invention can be provided in kit form, e.g., in a single or
separate
container, along with other reagents, buffers, enzymes or materials to be used
in
practicing at least one method of the invention. Such kits can be provided in
a container
that can optionally include instructions or software for performing a method
of the
invention. Such instructions or software can be provided in any language or
human- or
machine-readable format.
Compound Screening, including High-Throughput Assays
The nucleic acids, proteins, polypeptides, antibodies and transgenic
animals of the invention may be used to validate the role of a gene product of
interest in
2o a particular disease, disorder or undesirable response, and to screen for
conditions or
compounds that can be used to treat such diseases, disorders and undesirable
responses,
preferably using high-throughput screening methods such as they are known in
the art
or later developed. Such treatment can be remedial, therapeutic, palliative,
rehabilitative, preventative, impeditive or prophylactic in nature. Diseases
and
disorders to which the invention may be applied, including organellar
associated
diseases as provided herein, include without limitation, mitochondria
associated
diseases, including but not limited to neurodegenerative disorders such as
Alzheimer's
disease (AD) and Parkinson's disease (PD); auto-immune diseases; diabetes
mellitus,
including Type I and Type II; MELAS, MERFF, arthritis, NARP (Neuropathy;
Ataxia;
54
CA 02363496 2001-09-14
WO 00/55323 PCT/iJS00/07311
Retinitis Pigmentosa); MNGIE (Myopathy and external ophthalmoplegia;
Neuropathy;
Gastro-Intestinal; Encephalopathy), LHON (Leber's; Hereditary; Optic;
Neuropathy),
Kearns-Sayre disease; Pearson's Syndrome; PEO (Progressive External
Ophthalmoplegia); congenital muscular dystrophy with mitochondria) structural
abnormalities; Wolfram syndrome (DIDMOAD; Diabetes Insipidus, Diabetes
Mellitus,
Optic Atrophy, Deafness), Leigh's Syndrome, fatal infantile myopathy with
severe
mtDNA depletion, benign "later-onset" myopathy with moderate reduction in
mtDNA;
dystonia; schizophrenia; mitochondria) encephalopathy, lactic acidosis, and
stroke
(MELAS); mitochondria) diabetes and deafness (MIDD); myoclonic epilepsy ragged
1o red fiber syndrome (MERFF); and hyperproliferative disorders, such as
cancer, tumors
and psoriasis.
The term "undesirable response" refers to a biological or biochemical
response by one or more cells of an organism to one or more physical
conditions,
chemical agents, or combinations thereof that leads to an undesirable
consequence. An
undesirable response can occur at the organellar level (e.g., loss of ~y in
mitochondria,
inhibition of photosynthesis in chloroplasts), the cellular level (e.g., cell
death such as
apoptosis or necrosis), in tissues (e.g., ischemia), in organs (e.g., ischemic
heart disease)
or to the organism as a whole (e.g., death; loss of reproductive capacity or
cognitive
processes).
2o Physical conditions that may produce an undesirable response include,
without limitation, hypothermia, hyperthermia, dehydration, exposure to
ultraviolet and
other types of radiation, micro-gravity, physical trauma, tensile stress, and
exposure to
electrical or magnetic fields. Chemical agents that may produce an undesirable
response include without limitation reactive oxygen species (ROS), apoptogens,
and the
like.
Nucleic acids of the invention are used to screen for conditions or
compounds that can be used to treat disease states and undesirable responses
in the
following manner. Treatment of cells with antisense molecules, including
ribozymes,
or introduction therein of antisense constructs, specific for a given gene
product of
3o interest should result in such cells demonstrating at least one of the
biochemical or
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
biological defects associated with the disease or disorder for which the gene
product is
being validated. In like fashion, transgenic animals comprising constructs
directing the
over-expression of a gene of interest, or an antisense or ribozyme expression
construct,
or animals to which antisense, ribozyme or molecular decoy oligonucleotides
are
administered, will demonstrate at least one of the biochemical or biological
defects
associated with the disease or disorder of interest if the nucleic acid
encodes a gene
product that is a valid target for the disease or disorder.
Similarly, for proteins of interest that may be targets for therapeutic
intervention, cells may be contacted with one or more antibodies specific for
the
1o protein, and the presentation of responses associated with the disease or
disorder will be
seen with valid targets. Polypeptides and proteins of the invention are also
used to
screen for conditions or compounds that can be used to treat disease states
and
undesirable responses in the following manner. The protein of interest, or a
polypeptide
derived therefrom having at least one activity of the protein of interest, is
produced by
recombinant DNA methods or in vitro synthetic techniques. The protein or
polypepeptide, which may be attached to a solid support, is contacted with a
detestably
labeled ligand (including, for example, an antibody). A compound is then
introduced to
the reaction vessel, and active compounds are identified as those that cause
the release
of the detestably labeled ligand.
2o Therapeutic Applications
Therapeutic agents derived therefrom according to the above
embodiments can be employed in combination with conventional excipients, i. e.
,
pharmaceutically acceptable organic or inorganic carrier substances suitable
for
parenteral application which do not deleteriously react with the active
compound.
Suitable pharmaceutically acceptable carriers include, but are not limited to,
water, salt
solutions, alcohol, vegetable oils, polyethylene glycols, gelatin, lactose,
amylose,
magnesium stearate, talc, silicic acid, viscous paraffin, perfume oil, fatty
acid
monoglycerides and diglycerides, petroethral fatty acid esters,
hydroxymethylcellulose,
polyvinylpyrrolidone, etc. The pharmaceutical preparations can be sterilized
and if
56
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
desired, mixed with auxiliary agents, e.g., lubricants, preservatives,
stabilizers, wetting
agents, emulsifiers, salts for influencing osmotic pressure, buffers,
colorings, flavoring
and/or aromatic substances and the like which do not deleteriously react with
the active
compounds. For parenteral application, particularly suitable vehicles consist
of
solutions preferably oily or aqueous solutions, as well as suspensions,
emulsions, or
implants. Aqueous suspensions may contain substances which increase the
viscosity of
the suspension and include, for example, sodium carboxymethyl cellulose,
sorbitol,
and/or dextran. Optionally, the suspension may also contain stabilizers (see
generally
WO 98/13353 to Whitney, published April 2, 1998).
to The term "therapeutically effective amount," for the purposes of the
invention, refers to the amount of a therapeutic agent which is effective to
achieve its
intended purpose. While individual needs vary, determination of optimal ranges
for
effective amounts of a therapeutic agent is within the skill of the art. Human
doses can
be extrapolated from animal studies (Fingle and Woodbury, Chapter 1 in Goodman
and
Gilman's The Pharmacological Basis of Therapeutics, 5th Ed., MacMillan
Publishing
Co., New York (1975), pages 1-46). Generally, the dosage required to provide
an
effective amount of the composition, and which can be adjusted by one of
ordinary skill
in the art will vary, depending on the age, health physical condition, weight,
extent of
disease of the recipient, frequency of treatment and the nature and scope of
the desired
effect.
Therapeutic agents of the invention can be delivered to mammals via
intermittent or continuous intravenous injection of one or more these
compositions or of
a liposome (Rahman and Schein, in Liposomes as Drug Carriers, Gregoriadis,
ed., John
Wiley, New York (1988), pages 381-400; Gabizon, A., in Drug Carrier Systems,
Vol.
9, Roerdink et al., eds., John Wiley, New York, 1989, pp. 185-212) or
microparticle
(Tice et al., U.S. Patent 4,542,025) formulation comprising one or more of
these
compositions; via subdermal implantation of drug-polymer conjugates (Duncan,
Anti-
cancer Drugs 3:175-210, 1992; via microparticle bombardment (Sanford et al.,
U.S.
Patent 4,945,050); via infusion pumps (Blackshear and Rohde, in: Drug Carrier
3o Systems, Vol. 9, Roerdink et al., eds., John Wiley, New York, 1989, pp. 293-
310) or by
57
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
other appropriate methods known in the art (see, generally, Remington 's
Pharmaceutical Sciences, 18th Ed., Gennaro, ed., Mack Publishing Co., Easton,
PA,
1990). Anti-cancer therapeutic compositions of the invention may be used in
combination with other anti-cancer compositions known in the art.
ASPECTS OF THE INVENTION
I. Identification of Differentially Expressed Or~anellar Factors
It is an object of the invention to identify organellar factors encoded by
genes that are differentially expressed in particular disease states,
apoptosis, in response
to various stressors or in a species-specific fashion. By "differentially
expressed," it is
1o meant that the gene product is present in greater amounts in one cell type,
or under one
set of conditions, than in another.
Organellar factors may be macromolecules found within or associated
with organelles, or cellular factors that negatively or positively influence,
either directly
or indirectly, the amount and/or activity of such macromolecules. Such factors
include
gene products that are expressed from genes that are derived from a cell's or
organism's
nuclear genome, as well as those expressed from the genomes of organelles such
as
mitochondria or chloroplasts. Nuclear genomes and genes may include organellar
"pseudogene" sequences, i.e., sequences originally present in organellar
genomes that
have been translocated from the organellar genome to the nuclear genome.
Pseudogene
2o sequences are generally not normally expressed but may become active in
certain
disease states or in response to certain conditions such as, e.g., cellular
stress.
A gene product may be a RNA molecule or a protein. Of particular
interest are those genes and gene products that are differentially expressed
in a disease
state (i.e., differentially expressed in cells from a diseased organism
relative to cells
from an undiseased, control organism of the same species), in manipulated
cells versus
wildtype cells, or in a species-specific manner (i.e., differentially
expressed in cells
from one species relative to cells from a second species). Thus, for example,
an "RNA
of interest," a "gene of interest" and a ''protein of interest" refer to,
respectively, a RNA,
gene and protein that are differentially expressed with regard to a disease
state, in
58
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
manipulated cells or in a species-specific manner. As one example of a gene of
interest
that does not directly encode a mitochondria) gene product, a nucleic acid of
interest
may be an antisense regulator of a mitochondria) gene product (Shayiq, J.
Biol. Chem.
272:4050-4057 (1997)). "RNAs of interest" include RNA molecules that are not
mRNA molecules but are themselves gene products such as, for example,
ribosomal
RNA (rRNA) molecules, transfer RNA (tRNA) molecules, ribozymes, RNA molecules
that form part of a nucleoprotein complex, and antisense transcripts.
As regards genes and gene products that are differentially expressed in a
disease or disorder, "mitochondria associated disorders," i.e., diseases
associated or
to thought to be associated with altered mitochondria) function and/or
mitochondria)
mutations, are of particular interest. Mitochondria associated disorders may
include
without limitation AD, PD, auto-immune diseases, diabetes mellitus, MELAS,
MERFF,
arthritis, NARP (Neuropathy; Ataxia; Retinitis Pigmentosa); MNGIE (Myopathy
and
external ophthalmoplegia; Neuropathy; Gastro-Intestinal; Encephalopathy), LHON
(Leber's; Hereditary; Optic; Neuropathy), Kearns-Sayre disease; Pearson's
Syndrome;
PEO (Progressive External Ophthalmoplegia); congenital muscular dystrophy with
mitochondria) structural abnormalities; Wolfram syndrome (DIDMOAD; Diabetes
Insipidus, Diabetes Mellitus, Optic Atrophy, Deafness), Leigh's Syndrome,
fatal
infantile myopathy with severe mtDNA depletion, benign "later-onset" myopathy
with
moderate reduction in mtDNA; dystonia; schizophrenia; mitochondria)
encephalopathy,
lactic acidosis, and stroke (MELAS); mitochondria) diabetes and deafness
(MIDD);
myoclonic epilepsy ragged red fiber syndrome (MERFF); and hyperproliferative
disorders, such as cancer, tumors and psoriasis.
One aspect of the present invention is a method for identifying organellar
factors encoded by genes that are differentially expressed, comprising:
providing one or
more cells in a first state, providing one or more cells in a second state,
determining the
expression of genes in the first state and the second state, and identifying
genes or
proteins that are differentially expressed in the first state and the second
state.
The cells) in the first state and the cells) in the second state can be the
3o same or different and can be any cell or population of cells, such as a
primary cell line,
59
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
a continuous cell line, a population of clones, a population of cells, a
manipulated cell
line, a population of manipulated cells, or a cell or population of cells
derived from the
same or different organism or species of organism, such as a sample, fluid,
tissue or
organ, or any combination of the foregoing. "Derived from," as used in this
context,
refers to cells whose lineage can be traced to a taxonomical kingdom, phylum,
class or
order; preferably a family of genus; and more preferably a species, and most
preferably
an identified organism. An organism can be a transmitochondrial organism, a
transgenic organism or a non-transgenic organism. Reference to an organism
refers to a
particular organism or a group of organisms. When a group of organisms is used
in a
to method of the present invention, the organisms can be from the same
species, but that
need not be the case.
The first state and the second state can be different regarding a particular
disease state. For example, the cells) in the first state can be derived from
a first
organism having a diseased state and the cells) in the second state can be
derived from
a second organism not having the diseased state or from a normal organism. For
example, the cells) in the first state can be from a patient diagnosed as
having
Alzheimer's disease and the cells) in the second state can be from a patient
not being
diagnosed as having Alzheimer's disease.
In addition, the first and second states can be different based on the
2o different source of the sample, fluid, tissue or organ. In this aspect of
the invention, the
cells) in the first state can be derived from a different sample, fluid,
tissue or organ as
the cells) in the second state. For example, the cells) in the first state can
be one or
more muscle cells and the cells) in the second state can be one or more
central nervous
system cells.
Furthermore, the first state and the second state can be different based on
the different treatments or the course of treatments of at least one organism.
In this
aspect of the present invention, the cells) in the first state can be derived
from the same
or different organism provided a treatment of a course of treatment, such as
environment, diet, or administration of compounds, such as proteins, peptides,
nucleic
3o acids (such as in a vector, such as a viral vector), drugs, chemicals or
toxins, as the
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
cells) in the second state is (are) derived from. A sample, fluid, tissue or
organ can be
taken at different times over the course of such treatment from one or more
organisms
that receive a treatment, do not receive a treatment or receive a different
treatment.
These samples, fluids, tissues or organs can be the source of the cells) in
the first state
or the cells) in the second state. For example, the cells) in the first state
can be
derived from an organism before being provided a treatment and the cells) in
the
second state can be derived from the same or different organism at different
times
during such treatment. By way of further example, the cells) in the first
state can be
derived from an organism receiving a first treatment and the cells) in the
second state
1o can be derived from a different organism receiving a second treatment.
In addition, the first state and the second state can be different based on
treatment of at least one of the cells) in the first state or the cells) in
the second state
with at least one compound. For example, the cells) in the first state can be
treated
with a compound, such as a protein, peptide, nucleic acid (such as in a
vector, such as a
viral vector), drug, chemical or toxin and the cells) in the second state not
be treated
with the compound used to treat the at least one first cell, be treated with a
compound
different from the compound used to treat the cells) in the first state, or be
treated with
the compound used to treat the cells) in the first state but at a different
concentration.
Furthermore, the first state and the second state can be different based on
2o the presence of one or more cellular stressors. The cellular stressor(s)
can be any
cellular stressor, but is preferably an environmental factor such as
temperature, ionic
strength or partial pressure of gasses such as, for example, oxygen, carbon
dioxide or
carbon monoxide. For example, the cells) in the first state can be treated
with a
cellular stressor and the cells) in the second state not be treated with a
cellular stressor,
be treated with a cellular stressor different from the cellular stressor used
to treat the
cells) in the first state, or be treated with the cellular stressor used to
treat the cells) in
the first state but at a different concentration.
The determining step preferably includes determining the mRNA or
protein in the cells) in the first state or the cells) in the second state,
preferably both,
3o using methods known in the art or later developed, such as nucleic acid
hybridization
61
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
methods, nucleic acid arrays, immunoassays or peptidometrics. The identifying
step
preferably includes comparing the mRNA or protein in the cells) in the first
state and
the cells) in the second state. Such comparing can utilize automation and be
computer
assisted using, for example, pattern recognition or data mining (United States
Patent
No. 5,13 8,695 to Means et al., issued August 11, 1992; United States Patent
No.
5,325,298 to Gallant, issued June 28, 1994; United States Patent No. 5,398,300
to
Levey, issued March 14, 1995; United States Patent No. 5,471,627 to Means et
al.,
issued November 28, 1995; United States Patent No. 5,619,709 to Caid et al.,
issued
April 8, 1997; United States Patent No. 5,745,654 to Titan, issued April 28,
1998;
1o United States Patent No. 5,687,306 to Blank, issued November 11, 1997;
United States
Patent No. 5,577,179 to Blank, issued November 19, 1996; United States Patent
No.
5,469,536 to Blank, issued November 21, 1995 and United States Patent No.
5,345,313
to Blank, issued September 6, 1994).
II. Identification of Differentially Expressed Genes in Manpulated Cells
In another embodiment of the invention, differentially expressed
organellar genes are identified in manipulated cells. Such cells include, but
are not
limited to (i) cybrid cells, i.e., cell lines having a commonly derived
nuclear component
that has, in the case of a particular cybrid, been combined with a distinct
cytoplasmic
(mitochondria and/or chloroplast containing) component; (ii) rho°
cells, i.e., cells in
2o which the amount of DNA in an organellar genome has been reduced or
eliminated; and
(iii) cells in which the wildtype genomic DNA (nuclear and/or organellar) has
been
mutated, added to or otherwise altered.
This aspect of the invention includes a method for identifying
differentially expressed organellar genes in manipulated cells, including:
providing one
that is not a manipulated cell, providing at least one second cell that is a
manipulated
cell, determining the ~ expression of genes in the first cell and the second
cell, and
identifying genes that are differentially expressed in the first cells) and
the second
cell(s). Preferably, the manipulated cell is a cybrid cell and the cell that
is not a
62
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
manipulated cell is a parent cell of the manipulated cell, but this need not
be the case.
The first cells) and the second cells) can be provided in the same or
different states.
Preferably, methods of the present invention use normal cells and cybrid
cells (such as 1685) for a particular disease state, such as diabetes or
Alzheimer's
disease, to identify genes or proteins that are differentially expressed in
the particular
disease state. Optionally, the nucleic acid molecules and proteins identified
by the
methods of the present invention can be used to investigate cells, samples or
tissues
from normal and diseased states. In this aspect of the present invention,
nucleic acid
molecules identified by the present invention are used to interrogate cDNA
libraries
1o made from cells, samples or tissues that are appropriate for a particular
disease state
using, for example, nucleic hybridization methods. For example, for diabetes,
tissue
samples from skeletal muscle would be preferable, and for Alzheimer's disease,
samples from the central nervous system, such as the brain, spinal column or
fluids
(preferably as soon after death as possible is the samples are taken post-
mortem). The
presence, absence, increased amount or decreased amount of a nucleic acid
molecule
identified by the present invention in cDNA libraries make from cells, samples
or
tissues of a diseased state as compared to cDNA libraries made using similar
cells,
samples or tissues of a non-diseased state indicates an association of that
nucleic acid
molecule, or the protein encoded by that nucleic acid molecule, with the
disease state
2o investigated. Optionally, a protein identified by the methods of the
present invention
can be measured in such samples using established methods, such as
immunoassays or
two-dimensional gel electrophoresis. The presence, absence, increased amount
or
decreased amount of a protein identified by the present invention in cells,
samples or
tissues of a diseased state as compared to cells, samples or tissues of a non-
diseased
state indicates an association of that protein, with the disease state
investigated.
III. Identification of Differentiall~pressed Genes during Cell Death
Another aspect of the invention involves the identification of nucleic
acids that are differentially expressed during apoptosis (a.k.a. PCD,
programmed cell
death) and necrosis. Mutations and other alterations that limit a cell's
response to
63
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
apoptosis may be events that occur during oncogenesis; that is, some cancer
cells may
represent the progeny of cells that have escaped apoptosis (Evan and
Littlewood,
Science 281:1317-1322, 1998). Nucleic acids that are differentially expressed
during
apoptosis, or biochemical events associated with apoptosis, can be used as
probes in
diagnostic, prognostic and pharmacogenomic assays useful in the therapeutic
management of such diseases and disorders. Such nucleic acids can also be used
to
produce gene products that can be used as novel targets in methods for
identifying pro-
apoptotic agents useful to treat hyperproliferative diseases and disorders, as
well as anti-
apoptotic agents that can be used to treat, e.g., degenerative diseases and
disorders that
1 o are known to have or suspected of having an apoptotic component, including
by way of
non-limiting example, neurodegenerative diseases and disorders such as
Alzheimer's
disease and stroke (Barinaga, Science 281:1302-1304, 1998).
This aspect of the invention preferably includes a method for identifying
nucleic acids that are differentially expressed during apoptosis, including:
providing at
least one first cell that is not apoptotic providing at least one second cell
that is
apoptotic state, determining the expression of genes in the first cell and the
second cell,
and identifying genes that are differentially expressed in the first cell and
the second
cell. An apoptotic cell is a cell that is expressing at least one gene, gene
product or
protein that can lead to apoptosis or have cellular conditions, such as redox
potential or
2o concentrations of ions or proteins in the cytosol or within or on an
organelle, that can
lead to apoptosis. The at least one first cell and the at least one second
cell can also be
provided in the same or different states.
In this embodiment of the invention, differentially expressed nucleic
acids are identified in cells that have been induced to undergo apoptosis, or
apoptotis-
related processes, relative to cells that have not been so treated. Compounds
generally
known as apoptogens may induce apoptosis. Some apoptogens act only on cells
having
specific receptors; these include, as non-limiting examples, Tumor Necrosis
Factor
(TNF), Fast, NMDA, corticosterone and the like. However, many apoptogens do
not
require specific receptors, including by way of example and not limitation,
herbimycin
3o A, paraquat, ethylene glycols, protein kinase inhibitors (such as, e.g.,
staurosporine,
64
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
calphostin C and caffeic acid phenethyl), chelerythrine chloride, Genistein, 1-
(5-
isoquinolinesulfonyl)-2-methylpiperazine, Quercitin, N-[2-((p-
bromocinnamyl)amino)ethyl]-5-5-isoquinolinesulfonamide, KN-93, d-erythro-
sphingosine derivatives, MAP kinase inducers (such as, e.g., anisomycin and
anandamine), cell cycle blockers (such as, e.g., aphidicolin, colcemid, 5-
fluorouracil
and homoharringtonine), acetylcholineesterase inhibitors (such as, e.g.,
berberine),
anti-estrogens (such as, e.g., Tamoxifen), pro-oxidants (such as, e.g., tert-
butyl
peroxide and hydrogen peroxide), free radicals (such as, e.g., nitrous oxide),
inorganic
metal ions, such as, e.g., Cadmium), DNA synthesis inhibitors (such as, e.g.,
1o Actinomycin D, Bleomycin sulfate, Mitomycin C, camptothecin, daunorubicin,
hydroxyurea, methotrexate and intercalators such as, e.g., doxorubicin),
protein
synthesis inhibitors (such as, e.g., cyclohexamide, puromycin and rapamycin),
agents
that affect microtubulin formation or stability (such as, e.g., vinblastine,
vincristine,
colchicine, 4-hydroxyphenylretinamide and paclitaxel), and ionophores (such
as, e.g.,
ionomycin and valinomycin). Apoptosis may also be induced in some cell types
by the
withdrawal of growth factors such as, e.g., interleukin-3 (IL-3). Furthermore,
physical
treatments, such as ultraviolet radiation, can induce apoptosis, as can
intracellular
bacteria such as Staphylococcus aureus (Bayles et al., Infection and Immunity
66:336-
342, 1998).
2o IV. Identification of Genes that are Differentially Expressed in a
Species-Specific Manner
Another aspect of the invention involves the identification of nucleic
acids that are differentially expressed in a species-specific manner. By
"species-specific
manner" it is meant that nucleic acids encoding homologous gene products are
up-
regulated or down-regulated in a first organism belonging to one species but
not in a
second organism belonging to another species when cells from such species are
exposed
to a particular chemical compound or set of physical conditions. This
embodiment of
the invention is used in a variety of methods.
This aspect of the present invention includes a method for identifying
3o nucleic acids that are differentially expressed in a species-specific
manner, including:
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
providing one or more cells from a first species, providing one or more cells
from a
second species, determining the expression of genes in the cells) from the
first species
and the cells) from the second species and identifying genes that are
differentially
expressed in the cells) from the first species and the cells) from the second
species.
Preferably, the cells) from the first species and the cells) from the second
species are
cultured under the same or similar conditions, but that need not be the case.
The cells)
from the first species and the cells) from the second species can be provided
in the
same or different states.
For example, this embodiment of the invention can be used to identify
to homologous nucleic acids that are differentially expressed in a species-
specific manner
during apoptosis, and used to develop novel antibiotics. For example, species-
specific
nucleic acids of interest include without limitation homologs that are
differentially
expressed in apoptotic human cells relative to apoptotic cells from a
eukaryotic
pathogen or parasite, such as e.g., trypanasomes (Ashkenazi and Dixit, 1998
Science
281:1305-1308) or insects. Such nucleic acids can be used to identify and
produce gene
products that can be used as novel targets in methods for identifying
antibiotics that
induce apoptosis in such pathogens and parasites but which do not induce
apoptosis in
the cells of their mammalian hosts. Alternatively, such nucleic acids can be
used to
identify and produce gene products that can be used as novel targets in
methods for
identifying compounds which protect mammalian cells from pro-apoptotic agents
but
which do not prevent or limit apoptosis in the cells of the eukaryotic
pathogen or
parasite. Such agents are expected to be useful for the prophylactic or
therapeutic
management of such pathogens and parasites.
In a related embodiment of the invention, nucleic acids that are
differentially expressed in a species-specific manner include those that are
up- or down-
regulated during apoptosis in cells from undesirable plants (e.g., weeds) but
not in cells
from desirable plants (e.g., crops); or in cells from undesirable insects (in
particular,
members of the family Lepidoptera and other crop-damaging insects) but not in
cells
from desirable insects (e.g., bees) or desirable plants. Such nucleic acids
can be used to
3o identify and produce gene products that can be used as novel targets in
methods for
66
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
identifying herbicides and pesticides, respectively, that act by inducing
apoptosis in
such undesirable plants and insects but which do not induce apoptosis in the
cells of
desirable plants and insects. Alternatively, such nucleic acids can be used to
identify
and produce gene products that can be used as novel targets in methods for
identifying
compounds which protect cells from desirable plant and insect species from pro-
apoptotic agents but which do not prevent or limit apoptosis in cells from
undesirable
plant and insect species exposed to such pro-apoptotic agents. Such agents are
expected
to be useful for the prophylactic or therapeutic management of such pathogens
and
parasites.
1 o In a related aspect of this embodiment of the invention, the genomes of
organelles of a desirable plant species are engineered to express a nucleic
acid of
interest that directs the production of a gene product which protects the
cells of the
desirable plant from herbicides (e.g., paraquat) and insecticides that act by
inducing
apoptosis or by interfering with organellar functions (see, e.g., Daniell et
al., Nature
Biotechnology 16:345-348, 1998). The nucleic acid that is introduced into the
organellar genome may be one that is endogenous (i.e., derived from the
desirable
plant) or one that is exogenous (derived from some other plant) in origin.
EXAMPLES
The following examples illustrate the invention and are not intended to
limit the same. Those skilled in the art will recognize, or be able to
ascertain through
routine experimentation, numerous equivalents to the specific substances and
procedures described herein. Such equivalents are considered to be within the
scope of
the present invention.
EXAMPLE 1
PREPARATION OF A CYBRID CELL LINE FOR DIFFERENTIAL GENE
EXPRESSION EXPERIMENTS OF ALZHEIMER'S DISEASE
Gene expression in cybrid cells derived from a patient having
Alzheimer's disease were compared to appropriate control cybrid cells. In
particular,
3o RNA species (or cDNA molecules derived therefrom) from the cybrid cell line
67
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
designated "1685 AD" were analyzed and compared to "MixCon" control cells.
"MixCon" designates a Mixed Control composed of cybrids prepared using
platelets
from n normal patients (n = 2-3, depending on the particular experiment).
Procedures for preparing cybrid cells comprising mitochondria derived
from patients having Alzheimer's disease have been previously described
(Miller et al.,
J. Neurochem. 67:1897-1907, 1996; Swerdlow et al., Neurology 49:918-925, 1997;
and
U.S. patent application Serial No. 08/397,808, hereby incorporated by
reference). The
1685 cybrid cell line is one example of a cybrid cell line of this type. The
1685 cybrid
cell line was created by fusing platelets from an AD donor with SH-SYSY
1 o neuroblastoma cells that had been made rho° by extended treatment
with ethidium
bromide.
To rule out the possibility of inadvertent transfection of donor nuclear
DNA during cybrid formation (due to, e.g., the presence of white blood cells
in the
platelet preparation), ApoE genotyping was performed with DNA isolated from
the AD
donor, parental SH-SYSY cells and AD cybrids by a primer extension assay that
uses
primers having the sequences 5'-GGCACGGCTGTCCAAGG (sense strand, SEQ ID
NO:I) and 5'-CCCGGCCTGGTACACTG (antisense strand, SEQ ID N0:2). Various
changes in the nucleotide sequence present in the ApoE gene between these two
primers
correspond to the ApoEl, ApoE2, ApoE3 and ApoE4 alleles (Mahley, Science
240:622-
630, 1988). Primer extension using this primer pair thus interrogates a
particular DNA
sample for the presence or absence of these alleles (Livak and Haimer, Hum.
Mutat.
3:379-385, 1994). Lymphocytes from the AD donor exhibited a heterozygous
(ApoE3/ApoE4) allelic pattern. In contrast, the SH-SYSY cells and 1685 cybrid
cells
displayed a homozygous (ApoE3/ApoE3) allelic pattern, thus indicating that the
1685
cybrid cells have the same nuclear complement as the parental SH-SYSY cell
line.
Mitochondrial DNAs from cell lines were also examined in order to
confirm the transfer of the mitochondrial genome from the Alzheimer's patient.
Total
cellular DNA was prepared from a blood sample from the AD patient, rho°
SH-SYSY
cells, parental SH-SYSY cells, the 1685 AD cybrids and the MixCon cybrids. A
multiplex primer extension assay was used to simultaneously interrogate mtDNA
68
CA 02363496 2001-09-14
WO 00/55323 PCT/LJS00/07311
positions 6366 and 6483 in PCR-generated fragments that encompass both loci
(see
pending U.S. patent application Serial No. 08/810,599, hereby incorporated by
reference). In contrast to the parental SH-SYSY and MixCon cybrids, total
cellular
DNA prepared from the 1685 cybrids and from a blood sample from the AD patient
demonstrated a homoplasmic mutation at mtDNA position 6366 and the wildtype
base
at mtDNA position 6483.
In a typical differential gene expression experiment using cybrid cells,
the following protocol was followed. MixCon and 1685 cybrid cells were thawed
and
cultured for approximately 2, 4 or 6 weeks. At the end of the culture period,
the
1o activities of two different components of the ETC (Complex I and Complex
IV) in the
cybrids was measured using the methods of Miller et al. (J. Neurochem. 67:1897-
1907,
1996). These mitochondria) enzymes have been previously shown to be
differentially
active in AD platelets and in AD brains post mortem, and in cybrids in which
the
cytoplasmic component is derived from AD cells, in the following manner.
Relative to
control cybrids (i.e., those in which the cytoplasmic component is derived
from normal,
undiseased cells) Complex IV (cytochrome c oxidase, COX) activity is
significantly
decreased in AD cybrids, whereas Complex I (NADH:ubiquinone oxidoreductase)
activity is not significantly different between the two (Davis et al., Proc.
Natl. Acad.
Sci. USA 94:4526-4531, 1997; Ghosh et al., "Mitochondria) Dysfunction and
Alzheimer's Disease," Chapter 10 in: Progress in Alzheimer's and Parkinson's
Diseases, Fisher et al., eds., Plenum Press, New York, 1998, pages 59-66; see
also PCT
application No. PCT/L1S95/04063, published as WO 95/269?3, the entire contents
of
which are hereby incorporated by reference).
The activities of Complexes I and IV are monitored to ensure that the
AD cybrids retain a phenotype associated with Alzheimer's disease. The results
of a
typical experiment are shown in Table 2. At the same time that samples were
taken
from the cybrids for the Complex I and IV assays, samples were also taken for
preparation of total cellular RNA.
69
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
TABLE 2: Complex I and IV Activities in 1685 AD Cybrids
MixCon 1685
AD
Cybrids
Days Complex Complex Complex Complex
Out Passage I ActivityIV ActivityPassageI ActivityIV Activity
23 107 23.0 2.00 106 35.5 1.41
37 108 33.5 1.84 107 23.6 1.47
58 112 28.8 2.23 112 33.3 1.18
EXAMPLE 2
PREPARATION OF RNA
In the present Example, RNA was prepared from MixCon cybrids and
1685 (AD) cybrids after 2, 4 and 6 weeks of culture. RNA was prepared from the
cybrids using the TRIZOL~ reagent (Life Technologies, Gaithersburg, MD; see
U.S.
Patent No. 5,346,994, hereby incorporated by reference) essentially according
to the
manufacturer's instructions. To remove DNA from the RNA preparations, samples
1 o were treated with RNase-free DNase I (Promega or Ambion) at a
concentration of 1 to S
u/uL for 20 to 30 minutes at 37°C.
EXAMPLE 3
REVERSE TRANSCRIPTION FOR DIFFERENTIAL DISPLAY
A. Design of Primers for Reverse Transcription
In order to generate DNA templates for amplification and analysis, it is
necessary to reverse transcribe the RNA molecules in a sample. Of particular
interest
are those RNA molecules that encode polypeptides, known as messenger RNA
(mRNA)
molecules. In eukaryotic systems, nuclear mRNA molecules have a 5' poly(A+)
"tail"
consisting of about 200 to 600 adenylic (A) residues that are added to the RNA
2o molecule after transcription whereas, in the case of mitochondrial mRNAs,
the 5'
poly(A+) "tail" is often somewhat shorter, i.e., about 50 to 60 adenylic
residues. Either
type of transcript is amenable to the procedure described below.
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Reverse transcription and PCR amplification of subsets of the RNA
molecules present in the samples was performed using the HIEROGLYPHS mRNA
Profile System (Genomyx Corp., Foster City, CA). The system is composed of
five
mRNA Profile Kits, each of which comprises 12 anchored oligonucleotide primers
(AP-
l, AP-2, etc.) in combination with 4 of 20 arbitrary 5' oligonucleotide
primers (ARP-1,
ARP-2, etc.).
Each anchored primer (AP) oligonucleotide has the sequence 5'-(dT),o_
,ZNM, where "NM" is, in each of the 12 AP oligonucleotides, GA, GC, GG, GT,
CA,
CC, CG, AA, AC, AG, AT or CT. Thus, each AP oligonucleotide is complementary
to
to the 3' ends of some mRNA molecules, which have a poly(A+) "tail." However,
the
identity of the "NM" nucleotides limits exact complementarity of a given AP
oligonucleotide to a subset of the poly(A) RNA molecules in a sample. For
example, an
AP oligonucleotide having the sequence 5'-TTTTTTTTTTTTCG (SEQ ID N0:3) will
have exact complementarity to only those mRNA molecules having the sequence 5'-
CGAAAAAAAAAAAA (SEQ ID N0:4) at the beginning of their poly(A+) "tail."
Assuming that the identity of the two nucleotides immediately 5' from the
first base of
the poly(A+) "tail" is random, each AP oligonucleotide will have exact
complementarity
to, and thus hybridize specifically to, 1 out of 12 (about 8%) of all of the
mRNA species
present in a sample.
2o B. Reverse Transcription
Regardless of which set of anchored primer (AP) oligonucleotides is or
was employed, the RNA samples were combined with individual AP primer and
heated
(by incubation at 70°C for 5 minutes) and then chilled quickly on ice.
Moloney murine
leukemia virus (Mo-MLV or M-MLV) reverse transcriptase is used, in the
presence of
appropriate buffers and a combination of the 4 dNTPs necessary for DNA
synthesis
(i.e., dATP, dCTP, dGTP and dTTP), to carry out reverse transcription of the
mRNA
molecules according to protocols known in the art (see, e.g., Dorit, "cDNA
Amplification Using One-Sided (Anchored) PCR," Unit 15.6 in: Short Protocols
in
Molecular Biology, 2nd Ed., Ausubel et al., eds., John Wiley & Sons, New York,
New
71
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
York, 1992, pages 15-21 to 15-27). More specifically, the reactions were
carried out
essentially according to the manufacturer's (Genomyx Corp.) instructions for
first-
strand cDNA synthesis reactions. Each reaction mix consisted of 20 uL (7.8 uL
sterile
nuclease-free H20; 4.0 uL Sx Superscript II RT buffer; dNTP mix, 1:1:1:1,
dATP:dTTP:dCTP:dGTP, 250 uM each; 100 mM DTT, 2.0 uL; and 0.2 uL of 200
Units/uL of Superscript II RT enzyme). In the control -RT (no Reverse
Transcriptase)
reaction, 8.0 uL of sterile nuclease-free H20 was added. Reactions were
carried out in a
thermal cycler with a heated lid and the following cycles were used: (I)
42°C for 5
minutes, (II) 50°C for 50 minutes, (III) 70°C for 15 minutes and
(IV) hold at 4°C.
The products of the reverse transcription reactions are a group of
DNA:RNA hybrid molecules, the DNA strand of each of which has a sequence that
is
the reverse complement of an mRNA molecule capable of specifically hybridizing
to
the specific AP oligonucleotide used in the particular instance. These
reaction mixtures,
referred to as "RT mixes," were stored at -20°C in a nonfrost-free
freezer.
EXAMPLE 4
DIFFERENTIAL DISPLAY (DD) IN AD CYBRIDS
Following reverse transcription using the anchored primer, which
produces a collection of RNA:DNA hybrid molecules, it was desirable to (a)
prepare,
2o amplify and label a set of the corresponding double-stranded cDNA molecules
and (b)
separate and evaluate the labeled double-stranded cDNA molecules. In the
present
instance, fluorescently labeled versions of the anchored and arbitrary primers
were used
in order to prepare labeled cDNA molecules, but it is also possible to labeled
cDNA
molecules by other means such as, e.g., labeling via radioactive isotopes.
These
reactions were carried out in duplicate in order to verify reproducibility.
Second-strand cDNA synthesis was primed using, in separate reactions,
one of 20 arbitrary primers (e.g., Ml3r-ARP1, Ml3r-ARP2, etc. to Ml3r-ARP20;
Genomyx Corp.). In each case, the arbitrary primer (ARP), corresponding to
sense
strand sequences located 5' from the poly-A tail of specific mRNA molecules,
was
3o hybridized to heat-denatured single-stranded (ss) DNA molecules. The
reaction mixes
72
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
also contained labeled and unlabeled versions of the same anchored primer (AP)
used in
the reverse transcription reactions of the preceding Example. The fluorescent
label used
in the present Example was tetramethylrhodamine (TMR).
More specifically, each reaction mix contained 1.95 uL of sterile,
nuclease-free H20; 1.0 uL of PCR Buffer II (without MgCl2); 1.5 uL of 25 mM
MgCl2;
2.0 uL of dNTP mix, 1:1:1:1, dATP:dTTP:dCTP:dGTP, 250 uM each; 1.75 uL of 2 uM
appropriate ARP primer (non-fluorescent version); 0.7 uL of fluorescent (TMR-
labeled)
version of 5 uM appropriate 3' AP primer (preceding reagents from Geonomyx
Corp.);
1.0 uL of a specific "RT mix" (see preceding Example); and 0.1 uL of AmpliTaq~
to thermostable DNA polymerase (Perkin Elmer). The reaction mixes were
incubated in a
thermal cycler with a heated lid according to the following set of cycles: (I)
95°C for 2
minutes; (II) 4 cycles of 92°C for 15 seconds, 50°C for 30
seconds, and 72°C for 2
minutes; (III) 30 cycles 92°C for 15 seconds, 60°C for 30
seconds, and 72°C for 2
minutes; (IV) 72°C for 7 minutes; and (V) hold at 4°C. In
general, caution was taken to
avoid introducing nucleases into the reagents and the areas where the
reactions were
prepared and carried out, and aerosol-barrier, sterile, nuclease-free pipet
tips were used.
Each of the resultant "cDNA reactions" contains a set of fluorescently labeled
PCR
products corresponding to a particular subset of RNAs.
Four uL of each cDNA reaction was combined with 1.5 uL of fluoroDD
loading dye in uncapped tubes. The DNAs were denatured and concentrated by
heating
the uncapped tubes at 95°C for 2 minutes in a thermal cycler with the
lid open. The
entire volume of the concentrated samples (about 2.5 to 3 uL) was loaded and
electrophoresed on 5.6% polyacrylamide HR-1000TM clear denaturing gels
(Genomyx).
Gels containing the electrophoresed labeled PCR products were imaged using the
genomyxSC scanner. Some representative results are shown in Figure 1.
Labeled PCR products from pairs of control and AD cybrid experiments
were compared for bands of interest. Such bands include both (i) "up-
regulated" genes,
i.e., bands that show an increased signal in the experimental (AD cybrid)
lanes relative
to the corresponding control (MixCon cybrid) lanes and (ii) "down-regulated"
genes,
73
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
i.e., bands that show a decreased signal in the AD cybrid lanes relative to
the
corresponding control lanes.
Bands of interest were cloned in order to determine their nucleotide
sequences (see following Example). Sequences were given "UNK" designations
(i.e., UNK1, UNK2, etc.; see Figures 5 through 32) until further
characterized. In some
instances, UNK sequences found to encode proteins of uncharacterized function
were
given "MG-UC" designations, and apparently novel UNK sequences were given "MG-
NOV" designations.
As can be seen in Figure l, both up-regulated and down-regulated
1 o nucleic acid species were identified in the AD cybrids in the present
example. In
particular, nucleic acids having the nucleotide sequences designated 1685 DD-
Sequences #3 (UNK4, a.k.a. MG-UC2; SEQ ID N0:9), #5 (MG-NOV3; SEQ ID
NO:11 ), and #6 (SEQ ID NO:~ showed decreased expression in the 1685 AD
cybrids, as did LINKS, UNK10, UNK18 and UNK19 (SEQ ID NOS: 27, 32, 33, 44, and
45, respectively).
In contrast, nucleic acids having the nucleotide sequences designated
1685 DD-Sequences #1 (3-HICAH; SEQ ID N0:7), #2 (UNK3, a.k.a. MG-UC1; SEQ
ID N0:8), and #4 (UNK2, a.k.a. MG-NOV2; SEQ ID NO:10), showed increased
expression in the 1685 AD cybrids, ), as did nucleic acids encoding SOD-1
(CuZnSOD;
see below).
EXAMPLE 5
DETERMINATION OF NUCLEOTIDE SEQUENCES OF DIFFERENTIALLY
DISPLAYED NUCLEIC ACIDS FROM AD CYBRIDS
The differentially expressed sequences of the preceding example were
further characterized by determination of their nucleotide sequences. These
sequences
were determined as follows:
Labeled bands of interest (i. e., either up- or down-regulated) were
excised from gels by generating a digital image from the scanned gel and a
virtual grid
3o was used as an overlay to define the location of a band of interest. This
location was
74
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
then transferred to a physical grid that was transferred to the actual gel.
Gel fragments
derived from the location of the band of interest were physically removed from
the gel
using a scalpel or similar instrument. DNA was eluted from the gel matrix by
adding
50 uL of lOmM Tris to the excised gel fragments and incubation at 37°C
for 30 to 60
minutes. One to 4 uL of the gel band eluent was subjected to further
amplification in
reaction mixes that further contained 19.4 to 16.4 uL, respectively, of
sterile, nuclease-
free HZO (i. e., the total volume of the gel band eluent and H20 was 20.4 uL;
8.0 uL of
Genomyx Sx Re-Amp Buffer; 3.2 uL of dNTP mix, 1:1:1:1, dATP:dTTP:dCTP:dGTP,
250 uM each; 4.0 uL of each primer (non-labeled versions of the pair of
anchored and
arbitrary primers used in the DD reactions were used); and 0.4 uL of 5
Units/uL
AmpliTaq~ thermostable DNA polymerase (Perkin Elmer). The reaction mixes were
incubated in a thermal cycler with a heated lid according to the following set
of cycles:
(I) 95°C for 2 minutes; (II) 4 cycles of 92°C for 15 seconds,
60°C for 30 seconds, and
72°C for 2 minutes; (III) 25 cycles 92°C for 15 seconds,
60°C for 30 seconds, and 72°C
for 2 minutes; (IV) 72°C for 7 minutes; and (V) hold at 4°C.
The resulting PCR products were cloned directly into linearized pCR2.1
vector DNA essentially according to the manufacturer's (Invitrogen, Carlsbad,
CA)
instructions using the "Original TA Cloning~ Kit" (see
http://www.invitrogen.com/manuals.html and U.S. Patent No. 5,487,993 for
details).
2o This linearized vector DNA is provided with single 3' deoxythymidine (dT)
overhangs
on each strand. Amplified DNA molecules produced by Taq polymerase have single
3'
deoxyadenine (dA) residues and are thus complementary to, and can be ligated
without
further manipulation into, the linearized pCR2. l DNA. (As will be appreciated
by those
skilled in the art, amplification products resulting from polymerases
containing
extensive 3' to 5' exonuclease activity, e.g., Vent and Pfu polymerases, lack
such dA
overhangs and would thus have to be further treated prior to ligation.)
Taq-amplified DNAs were combined with linearized pCR2.1 DNA and
ligated using T4 DNA ligase and manufacturer (Invitrogen) supplied Iigation
buffer.
The ligated DNAs were used to transform Escherichia coli cells. The E. coli
strain used
3o was XL1-BIueTM cells (Stratagene) having the phenotype recAl endAl gyrA96
thi-1
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
hsdRl7 supE44 relAl lac [F' proAB lacIqZOMIS TnlO (TetR)]. Transformants were
isolated as ampicillin-resistant colonies.
Strains MKN2 and MKN3, comprising pMKN2 (containing 1685 DD-
Sequence #4, SEQ ID NO:10) and pMKN3 (containing 1685 DD-Sequence #5, SEQ
ID NO:11 ), respectively, were deposited at the American Type Culture
Collection
(Manassas, VA) on March 4, 1999. Strain MKN2 was given the Accession No.
207149, and strain MKN3 was given the Accession No. 207150.
Plasmid DNA was isolated from transformants using the Wizard~ Plus
Series 9600 Miniprep Reagent System (Promega). The nucleotide sequences of the
1 o inserts in the isolated plasmid DNAs were determined in sequencing
reactions that used
primers that hybridize to regions present in the vector adjacent to the
inserted DNAs
[i.e., a universal M13 reverse primer (5'-CAGGAAACAGCTATGAC, SEQ ID N0:5)
and a T7 promoter primer (5'-TAATACGACTCACTATAGGG, SEQ ID N0:6), both
from Invitrogen], and Prism~ sequencing reagents (Perkin Elmer). Sequencing
reaction products were purified by ethanol precipitation and then
electrophoresed and
analyzed using an ABI Prism 373A DNA Sequencer (Perkin Elmer) essentially
according to the manufacturer's instructions. In some instances, the sequences
of both
the 5' and 3' ends of the insert were determined, resulting in sequences
designated, for
example, UNK10-5' and UNK10-3'.
2o The Sequence NavigatorTM software (Perkin Elmer) was used for
analysis of sequence data. Nucleotide sequences, and corresponding polypeptide
sequences derived via in silico translation, were used to search the GenBank
and
Swissprot databases, respectively.
76
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
EXAMPLE 6
ANALYSIS OF NUCLEOTIDE SEQUENCES OF DIFFERENTIALLY DISPLAYED NUCLEIC ACIDS
FROM AD CYBRIDS
A. Overlapp~ DD Sequences
As an initial matter, the LTNK sequences were compared with each other
in order to determine if any transcripts had been identified as differentially
expressed in
the cybrids more than once. This result is possible, as different pairs of
primers used in
differential display can result in PCR products that are of different length
even though
they are derived from the same transcript.
to Several differentially displayed sequences were indeed found to overlap
one another. In particular, LINKS overlaps LTNK10-S' and UNK10-3' (see Figure
33).
In addition, I1NK18 and UNK19 overlap one another (see Figure 34). These
sequences
are of particular interest as they indicate that the same transcript has been
identified as
differentially expressed in AD cybrids in two independent experiments, each of
which
uses a different set of PCR primers.
B. apes of Sequences and Homologies
In general, nucleotide sequences identified as being differentially
displayed in the AD cybrids have been found to have nucleotide sequences that
( 1 ) are
identical (or nearly so, reflecting sequence errors in the databases) to human
nucleotide
2o sequences present in the databases examined, (2) encode putative
polypeptide sequences
having some homology to the amino acid sequence of a known protein in humans
and/or other species, and (3) have no apparent homology to any previously
described
nucleotide or polypeptide sequences (novel sequences). Sequences in classes (
1 ) and
(2) may be further characterized as being either (a) sequences encoding a gene
product
having characterized functions) or (b) previously described sequences that
encode a
gene product whose function is unknown. In the present example, sequences of
each
type were identified by the preceding differential display (DD) methodology
(Table 3).
77
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
TABLE 3: Differentially Expressed Genes in AD Cybrids as Determined by
Differential Display (DD)
SEQ Gene Product Identity Change in Expression
ID NO: (if known) in AD Cybrids
1685 DD-Sequence #1, a.k.a. 3-HICAH
7 (3-hydroxyisobutyryl coenzyme A hydrolase)Increased expression
1685 DD-Sequence #2, a.k.a. MG-UC 1
8 (uncharacterized; corresponds to YAC377A1)Increased expression
1685 DD-Sequence #3, a.k.a. MG-UC2
(UNK4)
9 (corresponds to uncharacterized proteinDecreased expression
KIAA0711 )
1685 DD-Sequence #4, a.k.a. MG-NOV2
(UNK2)
(unknown; novel sequence) Increased expression
1685 DD-Sequence #5, a.k.a. MG-NOV3
(UNK3)
11 (unknown; novel sequence) Decreased expression
C. Previously Described Genetic Seguences
5 The sequences of interest in AD cybrids included nucleic acids encoding
known gene products. Examples of such gene products included, but were not
limited
to, the following sequences:
1. UNKI (1685 DD-Sequence #l; SEQ ID N0:7) was used to probe
DNA databases and demonstrated a significant overlap with the cDNA for 3-
to hydroxyisobutyryl coenzyme A hydrolase (a.k.a. 3-HICAH; SEQ ID N0:7; see
also
Figure 2 and GenBank accession No. U66669).
2. SOD-1 (superoxide dismutase is an enzyme encoded by a cDNA
(Accession No. X01662) having a sequence that overlaps an UNK sequence (SEQ ID
NO:; Figure 36). The DD results indicate that SOD-1 expression is decreased in
AD cybrids.
3. UNK19 and UNK18 (SEQ ID NOS: 44 and 45, respectively; see
also Figures 22, 23 and 34), which overlap and have increased expression in AD
cybrids, were translated in silico in all six reading frames, and the
resultant amino acid
sequences were used to probe polypeptide and putative protein sequences. The
search
2o results yielded a number of matches to a reverse transcriptase homolog
(designated
78
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
"ORF2" or "p150") found in long interspersed nuclear elements (LINEs). Many
copies
of LINES are present in mammalian genomes; it is estimated that there are
100,000
LINES in the human genome, of which 3,000 to 4,000 are full-length. It has
been
reported that many LINEs are capable of retrotransposition (Sassaman et al.,
Nature
Genetics 16:37-43, 1997), so these results may signify that, for whatever
reason, LINES
are more likely to express p150, and thus retrotranspose, in AD cybrids.
However,
because many LINEs of nearly identical sequence are present in the genome, the
present
results do not allow one to distinguish between increased expression
associated with
one, as opposed to many LINES. Accordingly, one possibility by way of non-
limiting
theory is that the increased expression of UNK19 and UNK18 may reflect the up-
regulation of a single LINE, which may in turn result in the overexpression
(e.g.,
through trans-activation), or inappropriate expression, of genes located near
that
particular LINE.
D. Uncharacterized Genetic Seguences
Several previously described sequences of uncharacterized function were
identified by the DD methodology.
1. MG-UCl (a.k.a. UNKS, 1685 DD-Sequence #2, SEQ ID N0:8),
which exhibited increased expression in AD cybrids, was used to probe
databases for
homologous and/or overlapping nucleotide sequences. A good match (E value = e-
148)
corresponds to sequences present on a cDNA encoding an uncharacterized protein
designated "KIAA0711" (see Nagase et al., DNA Res. 5:277-286, 1998, and
GenBank
accession No. AB018254). When used to probe an EST database, SEQ ID N0:8
yielded many identical matches to several ESTs (Figure 38); this result
indicates that
MG-UC 1 is expressed in a variety of tissues, including but not limited to,
brain, testis,
pineal gland, kidney, pancreas, liver, lung, etc., in adult, as well as in
fetal and infant
tissues, in many instances.
The KIIA0711 putative protein has homology (E value = e-11 to e-10) to
members of the family of proteins related to the Kelch protein of Drosophila
melanogaster, which is a component of ring canals that regulates the flow of
cytoplasm
79
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
between cells during oogenesis and other processes. However, another match of
note (E
value = 2e-10) occurs between KIIA0711 and the marine Keapl protein. Keapl
represses the nuclear activation of antioxidant responsive elements by Nrf2
(Itoh et al.,
Genes. Dev. 13:76-86, 1999). Accordingly, by way of non-limiting theory, if
the
expression of Keapl is increased in AD, the expected consequence would be that
activation of antioxidant responsive elements would be decreased. This effect
would
work to increase the damage wrought by reactive oxygen species (ROS), where
increased ROS production has been reported in AD cybrids and has been proposed
as a
possible contributing factor to neuronal death in AD (Swerdlow et al.,
Neurology
l0 49:918-925, 1997).
2. MG-UC2 (a.k.a. LINK , 1685 DD-Sequence #3, SEQ ID
N0:9), the expression of which was decreased in AD cybrids, contains sequences
corresponding to a bacterial artificial chromosome (BAC) clone known as BAC
CIT987-SKA-237H1 that contains sequences from the p12 region of human
chromosome 16 (see Figure 4 and GenBank accession No. AC002287). Like UNK19
and UNK18 (see above), the sequences in SEQ ID N0:9 are part of a set of
repeated
elements known as Alu elements, and, as a result, until further sequence
information is
obtained, one cannot be certain if the expression of a particular Alu element,
or a gene
associated with a particular Alu sequence, is increased in AD cybrids versus
overexpression of two or more Alu elements and/or genes.
3. LINKS, UNK10-5' and UNK10-3' (SEQ ID NOS: 27 , 32 and 33,
respectively) sequences overlap each other (Figure 33) and showed decreased
expression in the AD cybrids. Although candidate homologs for LINKS and UNK10
have been identified using other search strategies (see below), the following
search
strategy also yielded results. The nucleotide sequence "LINKS" (SEQ ID N0:27)
was
analyzed using the BLASTx program (Gish et al., Nature Genetics 3:266-272,
1993).
This program translated, in silico, the LINKS sequence in all six potential
reading
frames, and the resultant amino acid sequences were used to search for
homologous
amino acid sequences. The most homologous (E value = 4e-89) protein to UNKS-
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
encoded peptides is a putative polypeptide given the designation "AK000867"
encoded
by Accession No. dbj~BAA91401.1.
The AK000867 amino acid sequence was then used to probe polypeptide
and putative amino acid sequences resulting from the in silico translation of
nucleotide
databases. The best-matching results were the uncharacterized putative protein
"KIIA0138" (Accession No. gb~AAC14666.1) and scaffold attachment factor B
("Factor B"; Accession Nos. ref ~ NP 002958.1 and gb~AAC18697.1). Amino acid
sequences from a conserved portion of the three polypeptide sequences were
aligned (as
shown in Figure 35) in order to generate the consensus sequence:
N1WVSGLSStTrAtDLKNLFsKYGKVvgAKVVTNARSPGArCYGfVTMStseE
atkCIaHLHrTELHGkmISVEKaKnEPagKKmSDkndeKSSkekssdvdr
(SEQ ID N0:63),
wherein upper case amino acid residues are absolutely conserved in all
three amino acid sequences, and lower case amino acids represent the amino
acid in two
of the three sequences in most cases and the most neutral amino acid in those
few
positions where the three sequences each differed with respect to one another.
The amino acid consensus sequence was in turn used as a probe of
peptide sequences in various databases. The search results (Figure 39) include
a
plethora of RNA-binding proteins, some of which are found in organelles
(mitochondria
or chloroplasts), one of which is a ribosomal protein. Thus, by way of non-
limiting
theory, the transcript from which L1NK5, which is down-regulated in AD
cybrids,
ultimately derives from a gene encoding a protein that is likely to be a RNA-
binding
protein. This RNA-binding protein may be localized to an organelle, and may
further
be part of one or more ribonucleoprotein complexes, where such complexes
include but
are not limited to ribosomal subunits and ribosomes.
81
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
E. Novel Genetic Sequences
Several apparently novel sequences were identified in the DD screening
described in this example. These are designated MG-NOV2 (a.k.a. UNK2; SEQ ID
NO:10) and MG-NOV3 (SEQ ID NO:11). According to the DD results, MG-NOV2
expression is increased, whereas MG-NOV3 expression is decreased, in AD
cybrids.
Some of the sequences in MG-NOV2 (SEQ ID NO:10) are derived from Alu
sequences,
repetitive elements present in multiple copies in the human nuclear genome.
SEQ ID
N0:12 defines a non-repetitive portion of MG-NOV2 that can be used to
specifically
probe for nucleic acids or nucleotide sequences corresponding to MG-NOV2.
Other
to apparently novel sequences include LTNK4, LJNK6, LTNK7, UNK11, L1NK12,
LTNK13,
UNK16, UNK17, UNK20, UNK21-5', LJNK21-3', UNK23, UNK24, UNK25-5',
UNK25-3', UNK26-5', and UNK26-3'.
F. Further Analyses
In addition to the database searches for homology of differentially
expressed sequences disclosed herein (e.g., the various UNK sequences) to
other
nucleotide sequences, additional homology searches using different search
strategies
were carried out to help identify the function of the differentially displayed
sequences.
The results of these searches are shown in Figure 37. The figure indicates the
results
from the following search strategies:
"Genbank nt'' indicates the results from searches using each LJNK
nucleotide sequence as a probe of the Genbank DNA database.
"Genbank nr'' indicates the results from a search wherein each LTNK
nucleotide sequence was translated in silico in all 6 potential reading frames
to yield
peptide sequences that were compared to peptide sequences in various
databases.
"Human EST" indicates the results from searches using each I1NK
nucleotide sequence as a probe of the Expressed Sequence Tag (EST) DNA
database.
Because the EST database is generally considered to have relatively poor
quality sequences, the Unigene database was also searched. This database
assembles
various EST sequences into virtual transcripts, a process that is believed to
eliminate
82
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
many sequencing errors in the EST sequences. The results of these searches are
given
under the heading "Unigene".
In Figure 37, the degree of homology was calculated according to E
values, which are presented therein. An "E value" (expectation value) is a
result of a
FASTA analysis that indicates the probability that a match between two
sequences is
due to random chance (Pearson et al., Proc. Natl. Acad. Sci. U.S.A. 85:2444-
2448,
1988). E values are typically presented in exponential form (i.e., "E-43" is
an
abbreviation for 1-43). The closer the E value is to zero, the greater the
likelihood that
the homology between the sequences being compared is not due to random chance.
For
Io example, "E-50" is a smaller number than "E-10" and thus represents a
better potential
"match" between the sequences.
Some candidate homologies of note included, but were not limited to,
those of UNK9 and UNK11 to neuronal thread protein (NTP), a protein that has
been
implicated in AD; UNK15 (both 3' and 5') to related tyrosine kinases; UNK16
(3' and
5') to DNA repair enzymes; UNK22-3' to mitochondria) uncoupling protein 2; and
UNK11 and UNK12 to ribosomal proteins.
EXAMPLE 7
CONFIRMATION OF DIFFERENTIAL EXPRESSION IN AD HYBRIDS BY Q-RTPCR
In order to confirm the differential expression of a particular gene
product, it is necessary to validate the results from a first method of
monitoring
differential expression (in this instance, the above-described differential
display) via a
second, independent method. In the present example, quantitative real-time
polymerase
chain reaction (Q-RTPCR) was used to validate the six sequences of interest
identified
in the preceding Example.
A. Reverse Transcription for Q-RTPCR
The RNA prepared from normal and AD cybrids according to Example 2
was used in reverse transcription reactions. First strand cDNA was synthesized
with the
SuperScriptTM pre-amplification system (Life Technologies) using an oligo(dT)
primer.
83
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
B. Design of Primers for Q-RTPCR
In the remainder of the Example, the RNA:DNA hybrid molecules
produced by these reactions were used as templates in PCR amplification
reactions
using primers derived from the nucleotide sequences determined as in the
preceding
Example. The sequences of these oligonucleotide primers, designed to
correspond to
(reverse primers) or be complementary to (forward primers) sense strand
sequences in
the 3' region of the nucleotide sequences of interest, are described in Table
4.
TABLE 4 Sequences of Primers for Quantitative Real Time PCR (Q-RTPCR)
Template
SEQ Oligonucleotide Sequence (5' ~ Nucleic AcidTemplate
ID NO: 3') of Interest Coordinates'
13 GGATTCAGACTAAAAGGAAGAGATGTG 3-HICAHZ 40 ~ 66
(f)
14 AAATCTTCCTCTAACATGGCCAACT 3-HICAHz 131 ~ 107
(r)
CGCCAAGTGGATGGATTTG MG-UC13 12 ~ 30
(f)
16 GGAGGAGCTTTGATCTCACATGA MG-UC 13 82 ~ 63
(r)
17 GATTCAGAGCTTGCCCTAGCA MG-UC24 96 ~ 116
(f)
18 CCAGTGTGAACCTTTTTCACTGTT MG-UC2~ 178 ~ 155
(r)
19 AGAAAATTTGTGAGACATCTTTGTGTAAA MG-NOV25 352 -~ 360
(f)
CTGGTTATAAGTTATATCCTCGCAGCTA MG-NOV25 432 ~ 405
(r)
21 GAGCTGATACTATTCCCACTGAAACTATT MG-NOV36 448 ~ 476
(f)
22 TGTCTCTACCAGGTTTTGGTATTAGGA MG-NOV36 550 ~ 524
(r)
l0 Notes for Table 6:
' "f', forward; "r", reverse.
z SEQ ID N0:7, 1685 DD-Sequence #1, 3-hydroxyisobutyryl coenzyme A hydrolase.
3 SEQ ID N0:8, 1685 DD-Sequence #2, Uncharacterized sequence MG-UC1, 3' region
similar to YAC clone 377A1 and cDNA for uncharacterized protein KIAA0711.
15 4 SEQ ID N0:9, 1685 DD-Sequence #3, Uncharacterized sequence MG-UC2, 3'
region
similar to BAC clone CIT987-SKA-237H1.
5 SEQ ID NO:10, 1685 DD-Sequence #4. Novel sequence MK-NOV2.
6 SEQ ID NO:1 l, 1685 DD-Sequence #5, Novel sequence MK-NOV3.
84
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
C. Confirmation of Primer Specificity
The Q-RTPCR analyses described in the present Example involve the
quantification of amplified DNA based on the fluorescence of an intercalating
dye,
SYBR~ Green (Perkin Elmer Applied Biosystems, Foster City, CA; see
http://www2.perkinelmer.com/ab/techsupp/doclib/ pcr/protocols/pdf/SYBR
Green.pdf
and U.S. Patent No. 4,304,886, hereby incorporated by reference). Because the
SYBR~ Green dye fluoresces to a greater degree when bound to any double-
stranded
(ds) DNA, it is necessary to perform an initial set of PCR reactions to
confirm that the
to PCR primers of choice amplify a single DNA species.
PCR reactions were carned out using the primers described in Table 4
and the DNA templates produced by the reverse transcription reactions
described in
section A of this Example. The RNA:DNA molecules produced by reverse
transcription were used as templates and the appropriate primers were added to
reaction
mixtures. Amplification was carried out using Taq DNA polymerase (Perkin
Elmer)
and the following cycles: (I) 95°C, 10 minutes; (II) 30 cycles of
95°C, 1 minute, 60°C,
1 minute, 72°C, 1 minute; (II) 72°C for 4 minutes; then (III)
hold at 4°C.
The PCR products, and appropriate molecular size markers, were
electrophoresed, stained with ethidium bromide and visualized via
fluorescence. In
2o each instance, a single band of the predicted molecular weight was
detected, confirming
that the primer pair amplifies a sequence corresponding to the specific
nucleic acid of
interest.
D. Quantitation of Nucleic Acids of Interest via Q-RTPCR
The use of real time PCR to quantitate levels of specific nucleic acids
has been described in the art (Heid et al., Genome Research 6:986-994, 1996;
Gibson et
al., Genome Research 6:995-1001, 1996; see Freeman et al., BioTechniques
26:112-
125, 1999, for a recent review; all references being hereby incorporated by
reference).
For ease of understanding, a brief explanation of quantitative real time PCR
(Q-
RTPCR) follows.
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
Until recently, the traditional means of measuring the products of a
specific PCR reaction was the "end-point" method of analysis, in which the
reaction
products are measured and quantitated after the amplification reactions are
completed.
In contrast, "real-time" PCR monitors amplification reactions in the thermal
cycler as
they progress. Q-RTPCR provides for improved quantification, because
quantification
is achieved most accurately during the linear range of amplification, and more
information about the amplification reactions is obtained for each cycle. For
example,
the normalized (i. e., to a passive reference dye that does not bind DNA)
fluorescence
intensity ("0R""), which indicates the magnitude of the signal generated by a
given set
to of PCR conditions, can be measured during each cycle.
From such data, the cycle at which a statistically significant increase in 4
R" is first detected can be determined. The "threshold cycle" or "C-i. value"
is
determined at one log above the signal first detected and provides a
quantitative
measure of the amount of the input nucleic acid template of interest present
in the
original sample.
In order to correct for sample-to-sample variation, an internal RNA
normalizer is used in Q-RTPCR. The RNA normalizer may be an endogenous RNA
species, for example, an mRNA encoding a constitutively-expressed protein like
actin
or glyceraldehyde-3-phosphate dehydrogenase (GAPDH), or a ribosomal RNA such
as
18S or 28S rRNA; RNA molecules produced in vitro may also be used as
normalizers.
Results of Q-RTPCR analyses are thus often expressed as relative amounts.
For instance, when the normalizer is actin and the nucleic acid that is
being quantitated is 3-hydroxyisobutyryl coenzyme A hydrolase (3-HICAH; SEQ ID
N0:7), the relative amount of 3-HICAH RNA in a sample is determined as
compared to
the normalizer actin according to standard curves created for both gene
sequences for
each RNA sample (i. e., AD and control). Standard curves were typically
prepared using
4 to 5 different amounts of input RNA in triplicate reactions. For example,
the
following amounts of input RNA might be evaluated in triplicate: (I) 0.1 ng,
0.5 ng, 1
ng and 5 ng or (II) 0.3 ng, 1 ng, 3 ng and 10 ng). Standard curves were
plotted as log
86
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
input ng (x axis) versus Ct (y axis, also log scale). For each standard curve,
the slope
(m) and the y-intercept (b) were calculated using standard analysis software.
The log input amount for the normalizer (nN) is calculated for a given Ct
(Ct°). For example, when Ct° = 20,
(20-bN)
nN -
mrr
l0 For a specific target (T) sequence of interest, CtT (the Ct required to
reach a log input amount equal to nN) is determined by the formula:
CtT - ( m.r. x nN) + bT
The normalized target Ct (normalized CtT) is calculated according to the
formula:
normalized CtT - CtT - Ct°
2o The Change in Expression, i.e., the comparative ratio of the target
sequence of interest in AD (1685) versus control (MixCon) cybrids is
calculated
according to the formula:
Change in Expression - 2 (Control normalized CtT) - AD normalized
CtT)
In the present Example, PCR reactions were performed using Taq DNA
polymerase and the primers described in Table 6 with the following cycles: (I)
50°C for
2 minutes, 95°C for 10 minutes; (II) 40 cycles of 95°C for 15
minutes, 60°C for 1
3o minute; and then (III) cooling to room temperature. PCR products were
detected with
SYBR~ Green detection reagents (Perkin Elmer) using the ABI Prism 7700
Sequence
Detection System (Perkin Elmer).
87
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
The relative (normalized) amounts of each candidate. gene of interest
(a.k.a. DD-Sequences #1 to #5) compared to the normalizer gene (actin) were
calculated
according to the preceding formulae: Comparative ratios of [the normalized
amount of
DD-Sequence in the 1685 AD cybrids] to [the normalized amount of DD-Sequence
in
MixCon control cybrids] were calculated for each DD-Sequence. The results are
shown
in Table 5.
TABLE 5: Differentially Expressed Genes in AD Cybrids as Determined by
Differential Display (DD) and Quantitative Real Time PCR (Q-RTPCR)
SEQ Change in Expression Change in Expression
ID NO: Gene Product (AD vs. control): DD (AD vs. control): Q-RTPCR
7 3-HICAH T T 2.2x
8 MG-UC I T T 1.9x
9 MG-UC2 sL J2.5x
MG-NOV2 T T 3.3x
11 MG-NOV3 J
These results confirmed the differential expression of RNAs having
sequences corresponding to 3-HICAH (SEQ ID N0:7). MG-UCI (SEQ ID N0:8). MG-
UC2 (SEQ ID N0:9) and MG-NOV2 (SEQ ID NO:10). and these sequences are thus
derived from bona ode differentially expressed genes in AD cvbrids. The gene
products corresponding to these sequences are therefore implicated in
Alzheimer~s
disease and may be used to develop diagnostic, prognostic and therapeutic
compositions
and methods.
For the accompanying SEQUENCE LISTING, the indicated summary
comments for the indicated SEQ ID NOs: are provided:
SEQ ID NO Summary Comments
1 Forward PCR primer for ApoE genotyping
2 Reverse PCR primer for ApoE genotyping
88
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
SEQ ID NO Summary Comments
3 None
4 None
M13 reverse primer
6 T7 Promoter primer
7 1685 DD-Sequence #1
3-hydroxyisobutyryl coenzyme A hydrolase
8 1685 DD-Sequence #2
Uncharacterized sequence MG-UC 1
3' region similar to YAC clone 3 77A l and
to cDNA for
uncharacterized protein KIAA0711
9 1685 DD-Sequence #3
Uncharacterized sequence MG-UC2
3' region similar to BAC clone CIT987-SKA-237H1
1685 DD-Sequence #4
Novel sequence MG-NOV2
11 1685 DD-Sequence #5
Novel sequence MG-NOV3
12 Non-repetitive portion of 1685 DD-sequence
#~
Novel sequence MG-NOV2
13 Forward primer for Q-RTPCR I
For 1685 DD-Sequence # 1
3-hydroxyisobutyryl coenzyme A hydrolase
14 3-HICAH reverse primer for Q-RTPCR
For 1685 DD-Sequence # 1
3-hydroxyisobutyryl coenzyme A hydrolase
Forward primer for Q-RTPCR
For 1685 DD-Sequence #2
3' region similar to YAC clone 377A1
89
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
SEQ ID NO Summary Comments
16 Reverse primer for Q-RTPCR
For 1685 DD-Sequence #2
Uncharacterized sequence MG-UC 1
3' region similar to YAC clone 377A1
17 Forward primer for Q-RTPCR
For 1685 DD-Sequence #3
Uncharacterized sequence MG-UC2
3' regions similar to BAC clone 987-SKA-237H1
18 Reverse primer for Q-RTPCR
For 1685 DD-Sequence #3, Uncharacterized sequence
MG-UC2
3' region similar to BAC clone CIT987-SKA-237H1
19 Forward primer for Q-RTPCR
For 1685 DD-Sequence #4
Novel sequence MG-NOV2
20 Reverse primer for Q-RTPCR
For 1685 DD-Sequence #4
Novel sequence MG-NOV?
21 Forward primer for Q-RTPCR
For 1685 DD-Sequence #~
Novel sequence MG-NOV3
22 Reverse primer for Q-RTPCR
For 1685 DD-Sequence #s
Novel sequence MG-NOV3
From the foregoing, it will be appreciated that although specific
embodiments of the invention have been described herein for purposes of
illustration.
various modifications may be made without deviating from the spirit and scope
of the
invention. All publications, including patent documents and scientific
articles, referred
to in this application are incorporated by reference in their entirety for all
purposes to
the same extent as if each individual publication were individually
incorporated by
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
reference. All headings are for the convenience of the reader and should not
be used to
limit the meaning of the text that follows the heading, unless so specified.
91
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
1 °'
SEQUENCE LISTING
<110> Mitokor
Hernnstadt, Corinna
Miller, Scott W.
Davis, Robert E.
<120> DIFFERENTIAL EXPRESSION OF ORGANELLAR
GENE PRODUCTS
<130> 660088.419PC
<140> PCT
<141> 2000-03-16
<160> 67
<170> FastSEQ for Windows Version 3.0
<210> 1
<211> 17
<212> DNA
<213> Homo sapien
<400> 1
ggcacggctg tccaagg 17
<210> 2
<211> 17
<212> DNA
<213> Homo sapien
<400> 2
cccggcctgg tacactg 17
<210> 3
<211> 14
<212> DNA
<213> Homo sapien
<400> 3
tttttttttt ttCg 14
<210> 4
<211> 14
<212> DNA
<213> Homo sapien
<400> 4
cgaaaaaaaa aaaa 14
<210> 5
<211> 17
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
2 -"
<212> DNA
<213> Homo sapien
<400> 5
caggaaacag ctatgac 17
<210> 6
<211> 19
<212> DNA
<213> Homo sapien
<400> 6
taatacgact cactatagg 19
<210> 7
<211> 390
<212> DNA
<213> Homo sapien
<400>
7
cgactccaaggaaaacttggttacttccttgcattaacaggattcagactaaaaggaaga 60
gatgtgtacagagcaggaattgctacacactttgtatattctgaaaagttggccatgtta 120
gaggaagatttgttagccttgaaatctccttcaaaagaaaatattgcatctgtcttagaa 180
aattaccatacagagtctaagattgatcgagacaagtcttttatacttgaggaacacatg 240
gacaaaataaacagttgtttttcagccaatactgtggaaagaaattattgaaaacttaca 300
gcaagatggttcatctttttgcccctagaacaattgaaggtaattaataaatgttctccc 360
aacatctcttaaagatccaccctaaggccc 390
<210> 8
<211> 314
<212> DNA
<213> Homo sapien
<400> 8
gctagcagacacgccaagtggatggatttggattgaacgcatatgaaacaggagacgggt 60
tctcatgtgagatcaaagctcctccaaagcctgttcaagctctaagcgattctcaaatgt 120
taccatttattaaaggtaaactacacctgttgaaggccaagttcagggcagctgttgtga 180
tctgtgtagttaatgtatttattaatgcttgacttttaaaatcctgggcataaatagtgc 240
agagcctcgtatgtttgtcagctcatgccgagatgaaataaatcacgcagaaagtgccag 300
tcctaaaaaaaaaa 314
<210> 9
<211> 395
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(395)
<223> n = A,T,C or G
<400> 9
gaccattgca tcctactaca tctgcattcc actcagcagg aagagggtgt agaaataaat 60
gaagactatc caagagagag caagcagagg tcattgattc agagcttgcc ctagcaaaga 120
gtcttgcatt tggcagaaac tcacaggctg gcagaacagt gaaaaaggtt cacactggaa 180
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
3 °'
aagagagaag gcttcagggg tgcctgattg gaggtagttg gcgtangaaa gctggaagtg 240
ggctcattan aagtggggca tccggctggg tgcagcagct cacacctata atcccagcac 300
tttgggaggc taaggctggc agatcccttg agcctaggag tgcgagacca gcctgggcaa 360
catggcaaaa ccctgtctct atgaaaaaaa aaaaa 395
<210> 10
<211> 510
<212> DNA
<213> Homo sapien
<400> 10
gctagcatggccaacatggtgaaaccccgtctctacaaaagaaaaaaatacaaaaattag 60
ctgggtgttgtggtgtatgcctgtaatcccaactatttgggtggctgaggcacgagagtc 120
gcttggacttggggggcggaggttgcagtgagctgagatcgtgccactgcactccagcct 180
gggtgacagactgagacagtctcaaaaaaaaaaaaaagaaaataatggatttgcagagac 240
ttgctatttagatttcagacatctgttaactaaaacacatgtgtaggcttttgttactta 300
tttcagtaatctgtaaatatctttatatttgagaaaatttgtgagacatctttgtgtaaa 360
ttataacttgaagaacctctcttacaagcaggcatattggtaagtagctgcgaggatata 420
acttataaccagattgaagtgtataattataatatgttattattctggggttctataaaa 480
aataaaatctttgaatctaaaaaaaaaaaa 510
<210> 11
<211> 622
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1) . . (622)
<223> n = A,T,C or G
<400>
11
gctagcagacgacaagaaataaccaagatcagagctgaactgaaggagattgagacacaa 60
aaaaatttaaaagatcaatgaatccagaaactcattctttgaaaaaactcagtaaaatag 120
actgctagctagactaataaaaaagaaaagagagaagattcaaataaacacnatcagaag 180
taataagggggataataccactgaccccacagaactacaaacaaccattagaggagtcta 240
tatntataaactggaaaatgtagaagaactggatacattnctggacacgtacacctccca 300
agactgaccaggaagaattgatccctgatagactaattcatggaattctggaaattgagt 360
cagtaataaatagcttaccaaccagaaacaagcccaggatcagacagattcacagctaaa 420
ttctaccagatgtacaaagaagagctgatactattcccactgaaactattccaaaaattg 480
aggaggagggactcttctctaacatgctatgaggccagcatcatcctaataccaaaacct 540
ggtagagacacaacaaaaaaaaataaaacttcaggccaatatccttgatgaacattgacg 600
caaaaatcctaaaaaaaaaaas 622
<210> 12
<211> 214
<212> DNA
<213> Homo sapien
<400> 12
atctgttaac taaaacacat gtgtaggctt ttgttactta tttcagtaat ctgtaaatat 60
ctttatattt gagaaaattt gtgagacatc tttgtgtaaa ttataacttg aagaacctct 120
cttacaagca ggcatattgg taagtagctg cgaggatata acttataacc agattgaagt 180
gtataattat aatatgttat tattctgggg ttct 214
CA 02363496 2001-09-14
WO 00!55323 PCT/US00/07311
4 '-
<210> 13
<211> 27
<212> DNA
<213> Homo sapien
<400> 13
ggattcagac taaaaggaag agatgtg 27
<210> 14
<211> 25
<212> DNA
<213> Homo sapien
<400> 14
aaatcttcct ctaacatggc caact 25
<210> 15
<211> 19
<212> DNA
<213> Homo sapien
<400> 15
cgccaagtgg atggatttg 19
<210> 16
<211> 23
<212> DNA
<213> Homo sapien
<400> 16
ggaggagctt tgatctcaca tga 23
<210> 17
<211> 21
<212> DNA
<213> Homo sapien
<400> 17
gattcagagc ttgccctagc a 21
<210> 18
<211> 24
<212> DNA
<213> Homo sapien
<400> 18
ccagtgtgaa cctttttcac tgtt 24
<210> 19
<211> 29
<212> DNA
<213> Homo sapien
<400> 19
agaaaatttg tgagacatct ttgtgtaaa 29
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
<210> 20
<211> 28
<212> DNA
<213> Homo sapien
<400> 20
ctggttataa gttatatcct cgcagcta 28
<210> 21
<211> 29
<212> DNA
<213> Homo sapien
<400> 21
gagctgatac tattcccact gaaactatt 29
<210> 22
<211> 27
<212> DNA
<213> Homo sapien
<400> 22
tgtctctacc aggttttggt attagga 27
<210> 23
<211> 394
<212> DNA
<213> Homo sapien
<400> 23
cgactccaagataggcagattgtggagaaataaatatttccctagtcattgtgattacat 60
tcctaatggaccttctctggtgctgatactgaaatagtacaaaaagttgtcagtaccttt 120
caattctgttggtcaaaaatatgtttttcctttttttgtgtgtgtgttttttttttcctt 180
taaaatgaacatatacttccaacatagaagttgtaacctttatatttaaccaagtttcca 240
gttgaagccagtttggggtgtgcatgtgtgtgcatgtgtctatatgcgtgtgtgtgtata 300
tacacacacaccaattatatatatagtatgcatgtgtgtatgtacatacagagaattttt 360
gagctggggcctttttagcagtaaaaaaaaaaaa 394
<210> 24
<211> 510
<212> DNA
<213> Homo sapien
<400> 24
gctagcatggccaacatggtgaaaccccgtctctacaaaagaaaaaaatacaaaaattag 60
ctgggtgttgtggtgtatgcctgtaatcccaactatttgggtggctgaggcacgagagtc 120
gcttggacttggggggcggaggttgcagtgagctgagatcgtgccactgcactccagcct 180
gggtgacagactgagacagtctcaaaaaaaaaaaaaagaaaataatggatttgcagagac 240
ttgctatttagatttcagacatctgttaactaaaacacatgtgtaggcttttgttactta 300
tttcagtaatctgtaaatatctttatatttgagaaaatttgtgagacatctttgtgtaaa 360
ttataacttgaagaacctctcttacaagcaggcatattggtaagtagctgcgaggatata 420
acttataaccagattgaagtgtataattataatatgttattattctggggttctataaaa 480
aataaaatctttgaatctaaaaaaaaaaaa 510
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
6 °'
<210> 25
<211> 622
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(622)
<223> n = A,T,C or G
<400> 25
gctagcagacgacaagaaataaccaagatcagagctgaactgaaggagattgagacacaa 60
aaaaatttaaaagatcaatgaatccagaaactcattctttgaaaaaactcagtaaaatag 120
actgctagctagactaataaaaaagaaaagagagaagattcaaataaacacnatcagaag 180
taataagggggataataccactgaccccacagaactacaaacaaccattagaggagtcta 240
tatntataaactggaaaatgtagaagaactggatacattnctggacacgtacacctccca 300
agactgaccaggaagaattgatccctgatagactaattcatggaattctggaaattgagt 360
cagtaataaatagcttaccaaccagaaacaagcccaggatcagacagattcacagctaaa 420
ttctaccagatgtacaaagaagagctgatactattcccactgaaactattccaaaaattg 480
aggaggagggactcttctctaacatgctatgaggccagcatcatcctaataccaaaacct 540
ggtagagacacaacaaaaaaaaataaaacttcaggccaatatccttgatgaacattgacg 600
caaaaatcctaaaaaaaaaaas 622
<210> 26
<211> 537
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(537)
<223> n = A,T,C or G
<400> 26
gaccattgcattataatggaagagggaccatataaagagccagaattactgggtgctaat 60
tctaaccatgcctgggctgccctcatctctatgactgttcttcctgagttctttgcaggt 120
taatttcctttgtgtagtcataaaatgataaattcgccctgaataacagtctaggctcta 180
ctctaaccccacactatcttctgagtaggcttacaaagcctaannnttacaaagcngagn 240
ngnatcnagctgtcaaaagtgattagaaattttaaatgatcantccagcctttaatttgg 300
tatatgcaccatattaagtcatttaagtgagtcagtaaatgtggcttgtaatataagaat 360
gacagtaatatctatatgtgtatattctttgattgtcagtgatgcatcaatttaccaaaa 420
acagcagataacaacttaaaatatactttactattttcaaattgcagtttgattaagtgc 480
aattgcaattgtacttaattgaaaaaaatcaagttttatatgaacaaaaaaaaaaaa 537
<210> 27
<211> 691
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(691)
<223> n = A,T,C or G
<400> 27
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
7
gaccattgcacagagcccggagaaggaaagcaaggattatgagatgaatgcgaaccataa 60
agatggtaagaaggaagactgcgtgaagggtgaccctgtcgagaaggaagccagagaaag 120
ttctaagagagcagaatctggagacaaagaaaaggatactttgaagaaagggccctcgtc 180
tactggggcctctggtcaagcaaagagctcttcaaaggaatctaaagacagcaagacatc 240
atctaaagatgacaaaggaagtacaagtngtactagtggtagcagtggaagctcaactaa 300
aaatatctgggttagtgaactttcatctaataccaaagctgctgatttgaagaactcttt 360
ggcaaatatggaaaggttctgagtgcaaaagtagttacaaatgctcgaagtcctggggca 420
aaatgctatggcattgtaactatgtcttcaagcacagaggtgtccaggtgtattgcacat 480
cttcatcgcactgagctgcatggacagctgatttctgttgaaaaagtaaaaggtgatccc 540
tctaagaaagaaacgaagaaagaaaatgatgaaaagagtagttcaagaagtcctggagat 600
aaaaaaaatacgagtgatagaagtagcaagacacaagcctctgtcaaaaaagaagagaaa 660
agatcgtctgagaaatctcaaaaaaaaaaaa 691
<210> 28
<211> 392
<212> DNA
<213> Homo sapien
<400>
28
cgactccaagccctgactctttgctgcgcctgagacaaaataaactttccataaaagact60
gagaatagaatacaaagtagtatacatagctataaccaataaacaaattatgtctttaaa120
aatatcccaaatgtgtgcagaaaaaaacattaacagtgaccgtctttgagtagtagatat180
gaccaatattattctctttgctataaatagtattccaaattttaataatacactttttaa240
atatttgtatacatacttttatatttcactatactgtgttgaaaagtatatattgtaata300
agctattttatacatgaaagaaaaaaatttttgcatcataagttgtatatattataataa360
actatttttaagttaccttgaaaaaaaaaaas 392
<210> 29
<211> 567
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(567)
<223> n = A,T,C or G
<400> 29
gctagcatggcaagcgcccataaatgccagtaactgtggatgctgccagaagtcagctgt 60
ttccagggacacagtgtagctgggttgcattttacagtttaatgtaactcgggtcgtctt 120
ctgtgggagtaaactcatgtttttgtgactgttttatgggtttgtccctcatattggagc 180
ttagtctaagctgcgcctcagactcctgtgtctgtcatgctgggagcctttggagaacgg 240
tccgtttgtccaacgtccagtttgctgagcatttttaaatccaactctgcacttacacct 300
ggccaggcaggaatgctcccagaatgggtcggcagtgtagaaagagatcctgagaagtgg 360
gtttctntcttttggtcaaaacttacctgttttgcatgaacatttaaaagtctgtcttga 420
tcccaatttggaacaatatgcctcaaaaccataaaggttgtatttaccagcctgatgttg 480
atttgactaatgttaatttgcgagagatgaatattagtatcttttaaataaaaaatgcct 540
gcctatttcactatcaaaaaaaaaaaa 567
<210> 30
<211> 567
<212> DNA
<213> Homo sapien
<400> 30
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
8 °'
gctagcatgggtgatagagtgagatcgtctccaaactctcctttctgaaattttacttag 60
ctaaattttttcctaattcctcctcagtatttccacttgattcccccacagaatgtaatt 120
gtaatgtatttattagtatctttgaacatcttttattatttgcctatcatacttctctac 180
aacaaaatatatgtaagttaataaaaatatttcctgtgtacatgatattgtcttaaattt 240
cttctatatttagttattacattacatttattattaggacatggcaattgaaggagcata 300
aatatactttgttttgccaaactagtatgaaacatttaaaaatgaaattttactgaatat 360
atgcattagtgagaaggatggtccttattaatatagttgtaggtgaatattaagctagaa 420
tggtagtgttcattaattctctcttcctattttctatttttatatatgtgaattctaaaa 480
aaccttatttacataatgtttttagtgcacatggaagtttttgataactttttaaattga 540
atttcttctgaattataagtcaaaaaa 567
<210> 31
<211> 460
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1) . . (460)
<223> n = A,T,C or G
<400> 31
gntagcagacacattttcaaagggtcatattcttggcttgttggtaatcagaatcgggca60
ggagaagtggggtggatgcagaccagctgaccacactggcaccaccagcagtttcagttt120
cgtcttgattgtaaagaggaaatatctaatcttaaaactcattaggggcctggcgcagtg180
gctcatacctgtattcccaacactttgggaggccgaggcaggcagatcacccgaggtcag240
gattttgagaccagcctggccaacatggtgaaaccccatctctactaaaaatacaaaact300
tagctaggcgtgatggcaggcacctctaatcccagttacttgggaggctgaggcaggaga350
atcacttgaacccggaaggcagacgttgcagtgagccaagatcgtgccactgcactccag420
cctgggcaactagagcaagactccatctaaaaaaaaaaaa 460
<210> 32
<211> 258
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(258)
<223> n = A,T,C or G
<400> 32
gaccattgcacagagcccggagaaggaaagcaaggattatgagatgaatgcgaaccataa60
agatggtaagaaggangactgcgtgaagggtgaccctgtcgagaaggaagccagagaaag120
ttctaagaaagcagaatctggagacaaagaaaaggatactttgaagaaagggccctcgtc180
tactggggcctctggtcaagcaaagagctcttcaaaggaatctaaagacagcaagacatc240
atctaaagatgacaaagg 258
<210> 33
<211> 259
<212> DNA
<213> Homo sapien
<220>
<221> misc feature
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
9 "'
<222> (1)...(259)
<223> n = A,T,C or G
<400> 33
ggcattgtaactatgtcttcaagcacagaggtgtccaggtgtattgcacatcttcatcgc 60
actgagctgcatggacagctgatttctgttgaaaaagtaaaaggtgatccctctaagaaa 120
gaaatgaagaaagaaaangangaaaagagtagttcaagaagttctggagataaaaaaata 180
cgaatgatagaagtagcaagacacaagcctctgtcaaaaaagaagagaaaagatcgtntg 240
agaaatcaaaaaaaaaaaa 259
<210> 34
<211> 696
<212> DNA
<213> Homo sapien
<400> 34
ggaccattgcattaaaatgttttggatacctgtttgaataacattgccttaatgttaata 60
aatccataatggtcacacaggcaggggtggtgtgtgaatcaccctggaagggatgttcat 120
taatcagttacttggggcttttttctttat.tcattccctcctagggtttgtacctgtgag 180
gaagcagcctaccttctttgcagccatctagcatctatattctagaatcatttttcccta 240
tgatggtcaaatccagattatctacacagaagaataaaataacctgagtaatccaaagtg 300
agtcataagtttttaaaagtctgggccaggcacagtgtctcatgcctgtaatcccagcat 360
tttaggaggcccaggagggaggatcacttgagcttaggagctcgagaccagcctgagcaa 420
catagtgagaccccatttctaccaaaaatagttttaaaaatagccagacatggtggtgca 480
tccctgtggtcccaggcagttggtggctgaggtgggaggatcctttgaacccaggaggtt 540
gaggttggagtgagctatgatggatcacaccactgcactccagcctgggcaaocgagtga 600
aaccctttctcaaaaatatgcattgtcctttggaatatgttctgtattcgaacatggatg 660
tagctaatgtttgattttaattacaaaaaaaaaaaa 696
<210> 35
<211> 393
<212> DNA
<213> Homo sapien
<400>
35
gaccattgcaaaatactgtagaagaactgtttagcttgcttcatttcttggaaccgtcac 60
aatttccctcagaatcagagtttctcaaggactttggggatctcaagacagaggaacagg 120
ttcaaaagctacaggccattcttaagccaatgatgctgagaagactcaaagaggatgttg 180
aaaaaaacttggcacccaaacaggaaacaattattgaagtagagctgactaatatccaga 240
agaaatactatcgggctattttggagaagaatttctccttcctttccaaaggggcaggtc 300
ataccaacatgcctaatctacttaacacaatgatggagttgcgcaagtgctgcaaccacc 360
catatctcatcaatggtgctgaaaaaaaaaaaa 393
<210> 36
<211> 253
<212> DNA
<213> Homo sapien
<400> 36
gaccattgcacagtaaatccattgtaggctttctttatgggtggcgggggaatctctaaa60
ggtcaggagtccagattgcttcaaataaacatccagaatctcagatgcttttttgaaaca120
agcccaagtttatctgaacctctttctctggtttggaaatcaggctgaaaatgtcacaga180
aacagattttcttgtgagatctcagaatgttgtggtttaagtaaagtaataaacaaagtc240
gaaaaaaaaaaaa 253
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
°'-
<210> 37
<211> 307
<212> DNA
<213> Homo sapien
<400> 37
ggccattgcataaaagtaactttacaagaatattagcactaaataattcctaacatcttg 60
gagacaattttcaaaataatgcgtttgtttactcattcaagatgtatttactgagctacc 120
actgttatatgccaagtgatgttctaggtcctagacatgtagcaaaaaccaaactgaaaa 180
aaaaattaactcttgtagattttcaaagcaactatagcagcaagaggaagagacgaacac 240
cgaaaaaaaaaaagccctatagtgagtcgtattaagccgaattctgcagatatccatcac 300
actggcg 307
<210> 38
<211> 481
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(481)
<223> n = A,T,C or G
<400>
38
gaccattgcaatgaatccccaataattgcagaactaaactcatttataaagctaaaataa 60
ccggatatatacatagcatgacatttctttgtgct-.ttggcttacttgtttaaaaaaaaaa 120
aaaactaatccaacctgttagatttngcaggtgaagtcagcagcttaaaaatgtctttcc 180
cagatttcaatgatttttttccccctacctcccaaaatctgagactgttaaaacattttt 240
ctcctatgaacactgctcagacctgcttcgacatgccataggagtggcgtgcacatctct 300
ctctcttccagcaggaggagcccgtgagcacgcacagctgccctgtctgctcacccgaag 360
gcaccgggctcacctggacctcccaggaaagggagaaagagcctccagaaactgctctgt 420
gtttagaaaggaatatctttaagaatccaagtttttcatttccacaaatttcctatatcc 480
a 481
<210> 39
<211> 450
<212> DNA
<213> Homo sapien
<400>
39
gatttgtttggacaatgtagttgggaagaactaagattctaatctgtgaagaaccttata 60
gggccttctaaaacataagagtttcctttgttgcttcaaatatttgaacattatgttaaa 120
gatcaagtattaattttagttgtactctagaaagctaaagtgccacattcggggctattt 180
ttatgattcagcaatcttttctaaattgtgtagcatgtgtatgagactatttatacccaa 240
ggatatgaaggaatataagtgactacaaggctctaataagccacggtggcaggaggttca 300
agcggttctgttcactaaatttttctcctgtaagctttgaatggaaacttctgtatcaca 360
tgatgtgtttcacttatgctg-ttgtgtatatacctaatatttctatttttgattttattt 420
taatacacctcgtccaataaaaaaaaaaaa 450
<210> 40
<211> 420
<212> DNA
<213> Homo sapien
<220>
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
11
<221> misc_feature
<222> (1). .(420)
<223> n = A,T,C or G
<400> 40
gctagcagacccacttaaggatgaattaaaccttgctgattctgaagtggataaccaaaa 60
acgagggaaacgacnttatgaagaaaaacaaaaagaacacttggataccttaaataaaaa 120
gaaacgagaactggatatgaaagagaaagaactagaggagaaaatgtcacaagcaagaca 180
aatctgcccagagcgtatagaagtagaaaaatctgcatcaattctggacaaagaaattaa 240
tcgattaaggcagaagatacaggcagaacatgctagtcatggagatcgagaggaaataat 300
gaggcagtaccaagaagcaagagagacctatcttgatctggatagtaaagtgaggacttt 360
aaaaaagtttattaaattactgggagaaatcatggagcccagattccagacatatcaacc 420
<210> 41
<211> 507
<212> DNA
<213> Homo sapien
<400> 41
gaccacaagatgaaattctaagtatatcagttcagcctggagaaggaaataaagctgctt 60
tcaatgacatgagagccttgtctggaggtgaacgttctttctccacagtgtgttttattc 120
tttccctgtggtccatcgcagaatctcctttcagatgcctggatgaatttgatgtctaca 180
tggatatggttaataggagaattgccatggacttgatactgaagatggcagattcccagc 240
gttttagacagtttatcttgctcacacctcaaagcatgagttcacttccatccagtaaac 300
tgataagaattctccgaatgtctgatcctgaaagaggacaaactacattgcctttcagac 360
ctgtgactcaagaagaagatgatgaccaaaggtgatttgtaacttaacatgccttgtcct 420
gatgttgaaggatttgtgaagggaaaaaaaattctgaactctttgatataataaaatgag 480
actggaggcattctcaaaaaaaaaaaa 507
<21G> 42
<211> 513
<212> DNA
<213> Homo sapien
<400> 42
gctagcagactatcattaaccaaataaattatgggattttgtcttaattatatacatata 60
catatacacacacatacacatacacatacatgtgtatatattccctaaaacttaataaag 120
ctcaaataataaaatcagatttcttaagtattccaattccctttaaaatgtaaatcagat 180
tttataattcttttgttcaaaactgtccattggctcccatttcacttaaatcaaaagcta 240
gtttttacaataagctaaggtagcaaacattattatctatttacttatgagttacttatg 300
taactcagcatccaataacactgtaggtgctcaataaaatagttgctgaatggataactt 360
tcactatttggatgagatccaacagaaaagaatactcttagcttgacaaacaatggtaaa 420
cagaagttaacattagaacactagatccttgctcactaaaatcagacataattatatgtt 480
tgtgtgtgtgtgtaaatataaacgtatatatgt 513
<210> 43
<211> 489
<212> DNA
<213> Homo sapien
<400> 43
tacatatttg aattaaatga aatatatcag aatttgtggt aacaacggat taaagcttag 60
ttcagaaaag aagaaagttt tcaaatcagc gatataataa tttccaaact taagaaacta 120
gaagagcaaa ttgaaccaaa gcaggcagaa tggaagaaag aataagataa gaaaatcaat 180
gaaattaaaa gcaacagaaa ctaaggccag gtgcagtggc tcatgcctgt aatcccaaca 240
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
12
cttcgggaggccgaggtgggcaggtcacgtgaggtcaggagtttgagaccagcctaacca 300
tcatggcaaaaccatctctactaaaaatacaaaaataagctgggcatggtggcaggcacc 360
agtaatcccagctactcgggagactgaggcagaagaatcactctgggaggcagaggctgt 420
agtgagctgagattgccactgcactctagcctgggctacagagtgagactccatctcaaa 480
aaaaaaaaa 489
<210> 44
<211> 505
<212> DNA
<213> Homo sapien
<400>
44
ggttttttgaaaggatcaacaaaattgatagatctctagcaagactaataagaaaagaga 60
gaagaatcaagtggatgcaataaaaaatgataaaggggatatcaccactgattccataga 120
agtacaaactaccatcagagaatactacaaacacctctacacaaataaactagaaaatct 180
agaagaaatggataaattcctggacacatacacccacccaagactaaaccaggaagaagt 240
tgaatctctgaatagaccagtaacaggctctgaaatggaggcaataattaatagcttacc 300
aaccaaaaaaagtccaggaccagatggaatcacagctgaattctgtcagaggtacaaaaa 360
ggagctggtaccattccttctgaaactattccaatcaataggaaaagagggaatcctccc 420
taactcattttatgaggccagcaccatcctgataccaaagcctggcagagacgcgacaaa 480
aagaattttacaccaaaaaaaaaaa 505
<210> 45
<211> 506
<21?.> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(506)
<223> n = A,T,C or G
<400> 45
gctagcagacggcgagaaataactaaaatcagagcacaactgaaggaaatagagacacaa60
aaaacccttcaaaaaattaaggaatccaggagctggttttttgaaaggatcaacaaaatt120
gatagaccactagcaagactaataaagaagaaaagagagaagantcaaatagacgcaata180
aaaaatgataaaggggaaatcaccaccaatcccacagaaatacaaactaccatcagagaa240
tactacaaacacctctatgcaaataaactagaaaatctagaagaaatggataaattcctc300
gacacatacaccctcccaagactaaaccaggaagaagttgaatctctgaatagtccaata360
acaggctctgaaattgtggcaataatcaatagcttaccaaccaaaaagagtccaggacca420
gatggattcacagccaaattctaccagangtttaaggaagaactggtaccattccttctg480
aaactactctaatcnatagaaaaaga 506
<210> 46
<211> 488
<212> DNA
<213> Homo sapien
<400> 46
gctagcagaccacaaaggacgttgatccctgagggaggtgaatcctatgaaggctctggc 60
ttaagcccgtgaaggtttcttagcagtggcacaggaagggcaactaactcaagggaggca 120
aactccatgaattgaggaaacagagctaaagatatgggataataaagtagctagagctta 180
caggacagagttcaggagagagcagatgcacagacaacaatctcttgaaatctgcagaga 240
gtctcagagatctggatatgtgcatgaggaagctacccaaggctgaggaaagaagcatcg 300
gaaataattatacggggaacagtacctggcaccttcctagggctggaaataggccttttc 360
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
13 -'
tcaccagtca gaatggaaaa tctcttaatt catatgcaat taggtagaac ctcagtagtg 420
agaaatgagg tagactatgc actgctctgg tctctcctag ctaacatttt aatcccaaaa 480
aaaaaaaa 488
<210> 47
<211> 267
<212> DNA
<213> Homo sapien
<400>
47
atggtagtctcatcacacactacaattaccttcccttacattactaatttgaagcataat 60
taacacaagcctcacatacttggtaaagtttgctatgttatagttaaagtctgtcttcac 120
agatcactacgtttgtgactcattgcgagttcaataatcaaagttcatgaactcgaggtg 180
attgatacacagtgtcctcatcagtgaacctggtgttaatgtagtatttgtccagaaagt 240
tattgtgaggactgtataaacccttgc 267
<210> 48
<211> 309
<212> DNA
<213> Homo sapien
<400> 48
gtaaaatgggtgataacagtagcaaattccaggtattgctgtgagataatagggtactta 60
gaacagggcttaacacttagtattgcatagtcattatttgctgttattaaagaataatgt 120
tttggaaagggcctggcacataaaaaagctattaatattaaatactattattagtatcaa 180
gaataaaagattagatatcactactggttctacattcagtaaagaataacatgataattt 240
acaaataatgttatgacaataagcctgacaacttaaataaaaatgacaaatccctcgaaa 300
aaaaaaaaa 309
<210> 49
<211> 217
<212> DNA
<213> Homo sapien
<400> 49
atggtagtct aagtaaaaaa aaaaaagccc tatagtgagt cgtattacaa gccgaattcc 60
agcacactgg cggccgttac tagtggatcc gagctcggta ccaagcttgg cgtaatcatg 120
gtcatagctg tttcctgtgt gaaattgtta tccgctcaca attccacaca acatacgagc 180
cggaagcata aagtgtaaac cctggggtcc ctaatga 217
<210> 50
<211> 349
<212> DNA
<213> Homo sapien
<400> 50
gtaacccaccacacccgcggcggttaatgggccgctacagggcgcgtccattcgccattc 60
aggctggcgcaactgttgggaagggcgatcggtgcgggcctcttcgctattacgccagct 120
ggcgaaagggggatgtgctgcaaggcgattaagttgggtaacgccagggttttcccagtc 180
acgacgttgtaaaacgacggccagtgaattgtaatacgactcactatagggcgaattggg 240
ccctctagatgcatgctcgagcggccgccagtgtgatggatatctgcagaattcggctta 300
gcggataacaatttcacacaggaatggtagtctaagtaaaaaaaaaaaa 349
<210> 51
<211> 433
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
14 -"
<212> DNA
<213> Homo sapien
<400>
51
atgtagtctacatttgacatacactggtgacattcaaaggtatagttctggtaaaataaa 60
attgaacatatggtggcaccagcactgagagcttggttcttttcctgatcagcagtttgg 120
ctcttcatcagttaactgcctgggcctcagtttctcagctgttaaattgaaggaggtgga 180
tgagttataacgttcctttctagttcttacacagaatgagtttcttgagttccaatatgc 240
tggagaagaaaaatagaagagtttggccactaatttataacagaagtagtatataccagg 300
acacgtgataaattatagacattttctgttagggagacttgtctgaagactagttttatt 360
actttcatttcttcctcaaagatcctttcataaaaaacaaacaaacaaaaaacaaaaaac 420
gaaaaaaaaaaaa 433
<210> 52
<211> 222
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(222)
<223> n = A,T,C or G
<400> 52
atggtagtct aatcatcaga gaaattacag ctgtagtgaa attgtgatga agataatgtt 60
ggattgacta cctaccagca tacctgagac atagtcgatg ctcaatgata ttaggtcctt 120
tctgtaatga aaaaatctcg tatattccaa tccccttttt caccaattta tgaacatgtg 180
ngtatgtgtt tataaacaca catgtgcttg tgtgatttgg gg 222
<210> 53
<211> 337
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(337)
<223> n = A,T,C or G
<400> 53
atggtagtctaggagaagagagggctcacanccagacacacctgggtggggctngggtca 60
agtgtcttcatctctctgagtctatctccccaacttttaaaaacagacagtgtatgtacn 120
acataggaggggctctcataactgccatcccttccactttcctaactttgcccccataca 180
ccctcacccccatcaagcccttgcccaggacagatttggacagctctcctctactcagat 240
acgaaaacaaaaaaaagccctataagccgaattctgcagatatccatcacactggncggc 300
cgctcgagcancncatctagagggcccaattcgccct 337
<210> 54
<211> 89
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(89)
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
15 °'-
<223> n = A,T,C or G
<400> 54
tttgccccca tacaccctca cccccatcaa gcccttgccc aggacagatt tggacagctc 60
tcctctactc agatanaaaa accaaaaaa 89
<210> 55
<211> 298
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(298)
<223> n = A,T,C or G
<400>
55
cagccncagnacacacctgggtggggctngggncaagngtctncatctctcngagnctat 60
ctccccaacnnnnaaaaacagacagcgnatgnactacataggaggggctctcataactgc 120
catcccntccactnnnctaactnngcccccatacaccctcacccccatcaagcccttgcc 180
caggacagacnnggacagctctcccctactcagacacgaaaaaaaaaaaagccctatagn 240
gagncgcanaacaagccgaacncngcaganatccatcacacnggcggccgcccgagca 298
<210> 56
<211> 85
<2i2> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1) . . (85)
<223> n = A,T,C or G
<400> 56
cccccataca ccctcacccc catcaagccc ttgcccagga cagatttgga cagctctcct 60
ctactcagat ncgaaaaaaa aaaaa g5
<210> 57
<211> 684
<212> DNA
<213> Homo sapien
<400>
57
atggtagtcttaagtcaactttgacaggaaataaagtgtttaattgtatttcctattctc 60
cactttgtaaacgtttagcatctggaagcacagataggcatatcagactgtgggatcccc 120
gaactaaagatggttctttggtgtcgctgtccctaacgtcacatactggttgggtgacat 180
cagtaaaatggtctcctacccatgaacagcagctgatttcaggatctttagataacattg 240
ttaagctgtgggatacaagaagttgtaaggctcctctctatgatctggctgctcatgaag 300
acaaagttctgagtgtagactggacagacacagggctacttctgagtggaggagcagaca 360
ataaattgtattcctacagatattcacctaccgcttcccatgttggggcatgaaagtgaa 420
caataatttgactatagagattatttctgtaaatgaaattggtagagaaccatgaaatta 480
catagatgcagatgcagaaagcagccttttgaagtttatataatgttttcacccttcata 540
acagctaacgtatcactttttcttattttgtatttataataagataggttgtgtttataa 600
aatacaaactgtggcatacattctctatacaaacttgaaattaaactgagttttacattt 660
ctctttaaaggtaaaaaaaaaaaa 684
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
16
<210> 58
<211> 694
<212> DNA
<213> Homo sapien
<400>
58
atggtagtctgtccagtggataaggtgtttctctcactttttatgtaacaactgagtaat 60
gacaacaaagtttacctaccactccttaggatataaggcccagtaaggcagagtttttgt 120
ttttcttttttcctactttattcactgctatgtcccagcccctagaacaactagttacaa 180
ctaggcagttgtaactgcctagtacataatagggactcaaaaatatttgtaaatgaatga 240
ataaatccactttcccagaattaccaaggcacatatttctgttgtcagaagtagagactc 300
ttaaactttgttgtacatcagaaccacagatgcagcatcttaatgtacatgtcccttccc 360
cagccctagtttactgtatgtgtatttggaaaggaatccacagatgattctgacatgtga 420
aaggctaagaagcagggagcttgccaggaaggttgaaattaaaatctgaaagttgtgggg 480
agtcttagaatattaagtgttactttttgttggaaatggcttcttttgtctttattaaag 540
ttaggaatgtgttttctgaaaagcttactttttgatattaatttccatttttaaagaaat 600
aacttgagattacaggcgtgaaccaccgcgcccggccgacttcaggagatctttaggcat 660
cattggtttgtgttcttcaggtaaaaaaaaaaaa 694
<210> 59
<211> 499
<212> DNA
<213> Homo sapien
<400> 59
gaccattgcatcattggccgcacactggtggtccatgaaaaagcagatgacttgggcaaa 60
ggtggaaatggagaaagtacaaagacaggaaacgctggaagtcgtttggcttgtggtgta 120
attgggatcgcccaataaacattcccttggatgtagtctgaggccccttaactcatctgt 180
tatcctgctagctgtagaaatgtatcctgataaacattaaacactgtaatcttaaaagtg 240
taattgtgtgactttttcagagttgctttaaagtacctgtagtgagaaactgatttatga 300
tcacttggaagatttgtatagttttataaaactcagttaaaatgtctgtttcaatgacct 360
gtattttgccagacttaaatcacagatgggtattaaacttgtcagaatttctttgtcatt 420
caagcctgtgaataaaaaccctgtatggcacttattatgaggctattaaaagaatccaaa 480
ttcaaaataaaaaaaaaaa 499
<210> 60
<211> 112
<212> PRT
<213> Homo sapien
<400> 60
Asn Phe Trp Val Ser Gly Leu Ser Ser Thr Thr Arg Ala Thr Asp Leu
1 5 10 15
Lys Asn Leu Phe Ser Lys Tyr Gly Lys Val Val Gly Ala Lys Val Val
20 25 30
Thr Asn Ala Arg Ser Pro Gly Ala Arg Cys Tyr Gly Phe Val Thr Met
35 40 45
Ser Thr Ala Glu Glu Ala Thr Lys Cys Ile Asn His Leu His Lys Thr
50 55 60
Glu Leu His Gly Lys Met Ile Ser Val Glu Lys Ala Lys Asn Glu Pro
65 70 75 80
Val Gly Lys Lys Thr Ser Asp Lys Arg Asp Ser Asp Gly Lys Lys Glu
85 90 95
Lys Ser Ser Asn Ser Asp Arg Ser Thr Asn Leu Lys Arg Asp Asp Lys
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
17 -"
100 105 110
<210> 61
<211> 116
<212> PRT
<213> Homo sapien
<400> 61
Asn Ile Trp Val Ser Gly Leu Ser Ser Asn Thr Lys Ala Ala Asp Leu
1 5 10 15
Lys Asn Leu Phe Gly Lys Tyr Gly Lys Val Leu Ser Ala Lys Val Val
20 25 30
Thr Asn Ala Arg Ser Pro Gly Ala Lys Cys Tyr Gly Ile Val Thr Met
35 40 45
Ser Ser Ser Thr Glu Val Ser Arg Cys Ile Ala His Leu His Arg Thr
50 55 60
Glu Leu His Gly Gln Leu Ile Ser Val Glu Lys Val Lys Gly Asp Pro
65 70 75 80
Ser Lys Lys Glu Met Lys Lys Glu Asn Asp Glu Lys Ser Ser Ser Arg
85 90 95
Ser Ser Gly Asp Lys Lys Asn Thr Ser Asp Arg Ser Ser Lys Thr Gln
100 105 110
Ala Ser Val Lys
115
<210> 62
<211> 106
<212> PRT
<213> Homo sapien
<400> 62
Asn Leu Trp Val Ser Gly Leu Ser Ser Thr Thr Arg Ala Thr Asp Leu
1 5 10 15
Lys Asn Leu Phe Ser Lys Tyr Gly Lys Val Val Gly Ala Lys Val Val
20 25 30
Thr Asn Ala Arg Ser Pro Gly Ala Arg Cys 'I~r Gly Phe Val Thr Met
35 40 45
Ser Thr Ser Asp Glu Ala Thr Lys Cys Ile Ser His Leu His Arg Thr
50 55 60
Glu Leu His Gly Arg Met Ile Ser Val Glu Lys Ala Lys Asn Glu Pro
65 70 75 80
Ala Gly Lys Lys Leu Ser Asp Arg Lys Glu Cys Glu Val Lys Lys Glu
85 90 95
Lys Leu Ser Ser Val Asp Arg His His Ser
100 105
<210> 63
<211> 103
<212> PRT
<213> Unknown
<220>
<223> Consensus sequence
<400> 63
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
18 °'-
Asn Leu Trp Val Ser Gly Leu Ser Ser Thr Thr Arg Ala Thr Asp Leu
1 5 10 15
Lys Asn Leu Phe Ser Lys Tyr Gly Lys Val Val Gly Ala Lys Val Val
20 25 30
Thr Asn Ala Arg Ser Pro Gly Ala Arg Cys Tyr Gly Phe Val Thr Met
35 40 45
Ser Thr Ser Glu Glu Ala Thr Lys Cys Ile Ala His Leu His Arg Thr
50 55 60
Glu Leu His Gly Lys Met Ile Ser Val Glu Lys Ala Lys Asn Glu Pro
65 70 75 80
Ala Gly Lys Lys Met Ser Asp Lys Asn Asp Glu Lys Ser Ser Lys Glu
85 90 95
Lys Ser Ser Asp Val Asp Arg
100
<210> 64
<211> 383
<212> DNA
<213> Homo sapien
<400> 64
cgactccaaggaaaacttggttacttccttgcattaacggattcagactaaaaggaagag 60
atgtgtacagagcaggaattgctacacactttgtagattctgaaaagttggccatgttag 120
aggaagatttgttagccttgaaatctccttcaaaagaaaatattgcatctgtcttagaaa 180
attaccatacagagtctaagattgatcgagacaagtcttttatacttgaggaacacatgg 240
acaaaataaacagttgtttttcagccaatactgtggaagaaattattgaaaacttacagc 300
aagatggttcatcttttgccctagagcaattgaaggtaattaataaaatgtctccaacat 360
ctctaaagatcacactaaggcaa 383
<210> 65
<211> 364
<212~ DNA
<213> Homo sapien
<400> 65
ggaagcctggactgtgcagccttcgggcacccggcacagacactgtgctggcaggagctt 60
cagacacgccaagtggatggatttggattgaacgcatatgaaacaggagacgggttctca 120
tgtgagatcaaagctcctccaaagcctgttcaagctctaagcgattctcaaatgttacca 180
tttattaaaggtaaactacacctgttgaaggccaagttcagggcagctgttgtgatctgt 240
gtagttaatgtatttattaatgcttgacttttaaaaycctgggcataaatagtgcagagc 300
ctcgtatgtttgtcagttcatgccgagatgaaataaatcacgcagaaagtgccagtcaaa 360
aaaa 364
<210> 66
<211> 357
<212> DNA
<213> Homo sapien
<400>
66
ggaagcctggactgtgcagccttcgggcacccggcacagacactgtgctggcaggagctt 60
cagacacgccaagtggatggatttggattgaacgcatatgaaacaggagacgggttctca 120
tgtgagatcaaagctcctccaaagcctgttcaagctctaagcgattctcaaatgttacca 180
tttattaaaggtaaactacacctgttgaaggccaagttcagggcagctgttgtgatctgt 240
gtagttaatgtatttattaatgcttgacttttaaaatcctgggcataaatagtgcagagc 300
ctcgtatgtttgtcagttcatgccgagatgaaataaatcacgcagaaagtgccagtc 357
CA 02363496 2001-09-14
WO 00/55323 PCT/US00/07311
19 -=-
<210> 67
<211> 420
<212> DNA
<213> Homo sapien
<400> 67
gacccttgcatcctactacatctgcattccactcagcaggaagagggtgtagaaataaat 60
gaagactatccaaaagagagcaagcagaggtcattgattcagagcttgccctagcaaaga 120
gtcttgcatttggcagaaactcacaggctggcagaacagtgaaaaaggttcacactggaa 180
aagagagaaggcttcaggggtgcctgattggaggtagttggcgtaggaaagctggaagtg 240
ggctcattagaagtggggcatccggctgggtgcagcagctcacacctataatcccagcac 300
tttgggaggctaaggctggcagatcccttgagcctaggagtgcgagaccagcctgggcaa 360
catggcaaaaccctgtctctatgaaaaaaaaacaaaagaaaagaaaaaatagctgggcat 420