Note: Descriptions are shown in the official language in which they were submitted.
CA 02363803 2001-07-05
WO 00/40730 PCT/LTS00/00364
POLYHYDROXYALKANOATE BIOSYNTHESIS ASSOCIATED PROTEINS AND
CODING REGION IN BACILLUS MEGATERIUM
FIELD OF THE INVENTION
The invention relates to nucleic acid and amino acid sequences involved in
polyhydroxyalkanoate biosynthesis, and more specifically, to
polyhydroxyalkanoate biosynthesis
sequences isolated from Bacillus megaterium. In particular, nucleic acid
sequences phaP, phaQ,
phaR, phaB, phaC, and their encoded amino acid sequences are disclosed.
BACKGROUND OF THE INVENTION
This patent application is related to U.S. Provisional Application Serial
Number
~0 60/115,092, filed on January 7, 1999. The government may own partial rights
to the present
invention pursuant to grant number MCB 9604450 from the National Science
Foundation.
Polyhydroxyalkanoic acids (PHA) are a class of aliphatic polyesters that
accumulate in
inclusion-bodies in many bacteria and archaea (2, 41 ). Their physiological
role in the cell is that
of carbon and energy reserves, and as a sink for reducing power. The most
studied PHA have
~s repeating subunits of: -[O-CH(R)(CHZ)XCO]-, where the most common form is
polyhydroxybutyrate (PHB), with R = CH3 and x = 1 (45). The PHA biosynthetic
pathway has
been determined for Alcaligenes eutrophus (17, 18. 44). In this organism two
molecules of
acetyl-Coenzyme A (CoA) are condensed by (3-ketothiolase (PhaA), followed by a
stereo-
specific reduction catalyzed by an NADPH dependent acetoacetyl-CoA reductase
(PhaB) to
Zo produce the monomer D-(-)-(3-hydroxybutyryl-CoA, which is polymerized by
PHA synthase
(PhaC). These 3 pha genes are coded on the phaCAB operon. which is speculated
to be
constitutively expressed, but PHA is not constitutively synthesized.
Alternative pathways for
synthesis of the monomer in other organisms have been suggested, most notably
in the
Pseudomonas species where the side chain, R, is longer than CH3 and its
composition is
z~ influenced by carbon substrates in the growth medium (7, 45). In addition
to A. eutrophus, phaC
has been cloned from more than twenty different bacteria (26, 43). Other genes
associated with
PHA synthesis, phaA, phaB, phaZ (PHA depolymerase) and genes for inclusion-
body associated
proteins and other low molecular weight proteins of unknown function, have
also been cloned
from some of these bacteria, in many cases by virtue of the fact that they are
clustered with
~o phaC.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-2-
PHA inclusion-bodies are 0.2 to O.Sq.m in diameter, but their structural
details are largely
unknown. They were described originally for some species of Bacillus (6, 8,
15, 30, 47) and
later for many more bacteria including Pseudomonas, Alcaligenes and
Rhodococcus (5, 11, 12,
25, 42). Those from Bacillus megaterium were shown to contain 97.7% PHA, 1.87%
protein
s and 0.46% lipid with protein and lipid forming an outer layer (15). More
recent reports show the
presence of a 14 kDa protein (GA14) on PHA inclusion-bodies of R. Tuber (36,
37), and a 24
kDa protein (GA24) with similarities to GA14 on the inclusion-bodies of A.
eutrophus (48).
These proteins are not essential for PHA accumulation but have been shown to
influence the size
of PHA inclusion-bodies and the rate of PHA accumulation (37, 48). GA14 and
GA24 have
to been named "phasins" due to some similarities with oleosins, which are
proteins on the surface
of oil bodies in plant seeds (21 ). Granule associated proteins are wide-
spread in PHA
accumulating bacteria (49).
The pattern of PHA inclusion-body growth and proliferation throughout the
growth cycle
of Bacillus megaterium has been described (32).
is There exists a need for additional nucleic acid and amino acid sequences
useful for the
production of polymers in biological systems.
SUMMARY OF THE INVENTION
This invention is the result of a study of PHA inclusion-body associated
proteins from
Bacillus megaterium and the cloning and analysis of their coding region. The
transcription starts
Zo were identified, the functional expression of several of the sequences was
confirmed in
Escherichia coli and in PHA negative mutants of Bacillus megaterium and
Pseudomonas putida,
and PhaP and PhaC were localized to PHA inclusion-bodies throughout growth.
A nucleic acid fragment encoding proteins involved in polyhydroxyalkanoate
biosynthesis was isolated from Bacillus megaterium. Nine nucleic acid
sequences and their
Zs encoded amino acid sequences are disclosed. Sequences encoding PhaB and
PhaC display not
insignificant percent identity and similarity to known acetoacetyl-CoA
reductase and
polyhydroxyalkanoate synthase proteins, while sequences encoding PhaP, PhaQ,
and PhaR do
not display significant similarity to known sequences. YkoY is similar to
known toxic anion
resistance proteins; YkoZ is similar to known RNA polymerase sigma factors;
YkrM is similar
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-3-
to known Na+-transporting ATP synthase proteins; and SspD matches the known B.
megaterium
spore specific DNA binding protein.
While several PHA related sequences were expressed in two organisms, it is
envisioned
that the sequences may be expressed in a wide array of organisms, and that the
nucleic acid
s sequences themselves may be modified to change the sequence and properties
of the encoded
proteins.
DESCRIPTION OF THE FIGURES
The following figures form part of the present specification and are included
to further
demonstrate certain aspects of the present invention. The invention may be
better understood by
io reference to one or more of these drawings in combination with the detailed
description of
specific embodiments presented herein.
Figure 1. PHA inclusion-body associated proteins. SDS-polyacrylamide gel
electrophoresis of proteins released from purified PHA inclusion-bodies. Lane
1, molecular
weight markers in kDa, 14, 18, 29, 43, 68 and 97. Lane 2, proteins from
inclusion-bodies of
~s cells harvested at late exponential growth phase. Lane 3, same as lane 2
except this part of the
gel was stained following 45 minutes transfer of proteins (seen in lane 2) to
PVDF membrane.
The bands were visualized by staining with Coomassie Blue.
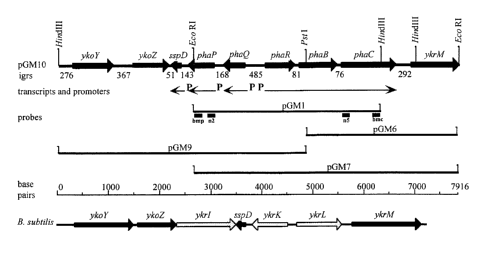
Figure 2 (A): The pha sequence cluster and flanking sequences. Map of cloned
fragment
in pGMlO carrying the pha genes (stripped arrows), intergenic regions (igrs)
and flanking genes
ao (thick black arrows) from Bacillus megaterium. The thin arrows indicate the
locations and
directions of transcripts; P, indicates promoter positions. pGMI, pGM6, pGM9
and pGM7
indicate the cloned DNA fragments in these plasmids (Table 1 ). Probes used to
identify and
clone the pha cluster are indicated by thick short lines under pGMI; n2 and n5
are degenerate
probes; bmp and bmc are homologous probes to the ends of the pGMl fragment.
Ruler of
Zs sequence in base pairs is for Bacillus megaterium and B. subtilis. Map of
yko, sspD and ykr
region in the B. subtilis genome; genes with homology to those of Bacillus
megaterium in this
region are indicted by thick black arrows; non-homologous genes are indicated
by thick gray
arrows. Gene annotations are horizontal over each gene symbol. Relevant
restriction enzyme
sites are vertical.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-4-
Figure 2 (B): Putative promoter regions for phaRBC, -Q, -P and sspD. Curved
arrows
indicate transcription start (+1), -10 and -35 nucleotides. The closest
resemblance to known -10
and -35 promoter sequences are in lower case letters below putative pha
promoter sequences.
Immediately downstream from the PhaP stop codon, the previously described (9)
sspD putative
s promoter is boxed, and putative hairpin structure is underlined.
Figure 2 (C): Mapping of the 5' ends of the phaRBC, -Q and -P transcripts (see
Example
11 ). Lanes G, A, T and C show the dideoxy sequencing ladders obtained with
the same primers
used in primer extension analysis; nucleotide sequences are complementary to
the transcripts.
Lane P is the primer extension product. Lane M is a DNA molecular size marker
measured in
~o nucleotides. The primer extension product is indicated by an arrowhead and
the 5' end of the
transcript within the sequence is indicated by a star. Only regions of the gel
containing extension
product bands are shown.
Figure 3: Pairwise alignment of PhaC from Bacillus megaterium (this study) and
P.
oleovorans (SWISS-PROT accession no. P26494); amino acid identities are shown
in black.
Is The Clustal method with PAM250 residue weight table was used.
Figure 4. pha: : gfp fusion plasmids and precursors. Only relevant restriction
sites are
shown. Annotations are as Figure 2. In all fusions the c-terminus excluding
the stop codon, of
either phaC or phaP, is fused to the gfp gene by the pGFPuv polylinker. For
more details, see
Table 1.
Zo Figure 5 (A): Time-course analysis of Bacillus megaterium (pGMl6.2) by
phase contrast,
green fluorescence, light image, and PHA fluorescence. Time (hours) are hours
post-inoculation
as indicated.
Figure 5 (B): Growth curve for Figure 5 (A); arrowheads indicate a decrease in
PhaP::GFP fluorescence.
as Figure 5 (C): Bacillus megaterium (pGM16.2) sampled at 2 days post-
inoculation. Top
image is phase contrast, bottom image is GFP fluorescence.
Figure 5 (D): Bacillus megaterium (pGMl3) sampled at 2 days post-inoculation,
left -
whole cells, right -lysed cells. Top image is phase contrast, bottom image is
GFP fluorescence.
Figure 5 (E): Bacillus megaterium (pGMl3C) sampled at 9 hours post-
inoculation. Top
so image is phase contrast, bottom image is GFP fluorescence.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-5-
Figure 5 (F): Bacillus megaterium (pHPS9) showed no fluorescence at any time
point.
Top image is phase contrast, bottom image is GFP fluorescence.
Figure 6: Hydrophilicity plot of PhaP protein.
Figure 7: Hydrophilicity plot of PhaQ protein.
s Figure 8: Hydrophilicity plot of PhaR protein.
Figure 9: Pairwise alignment of PhaC from Bacillus megaterium (this study) and
T.
violacea (SWISS-PROT accession no. P45366); amino acid identities are
indicated by a star (*),
and amino acid similarities are indicated by a period (.) below the sequences.
The ClustalW
method with PAM350 residue weight table was used.
~o Figure 10: Proposed biosynthetic pathway for the preparation of C8
copolymers.
DESCRIPTION OF THE SEQUENCE LISTINGS
The following sequence listings form part of the present specification and are
included to
further demonstrate certain aspects of the present invention. The invention
may be better
understood by reference to one or more of these sequences in combination with
the detailed
i s description of specific embodiments presented herein.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-6-
SEQ ID NO Description
1 Bacillus megaterium 7,916 by fragment
2 phaP nucleic acid sequence, 2566-3075 reverse
complement
3 PhaP amino acid sequence, 170 amino acids
4 phaQ nucleic acid sequence, 3247-3684 reverse
complement
PhaQ amino acid sequence, 146 amino acids
6 phaR nucleic acid sequence, 4170-4673
7 PhaR amino acid sequence, 168 amino acids
8 phaB nucleic acid sequence, 4758-5498
9 PhaB amino acid sequence, 247 amino acids
phaC nucleic acid sequence, 5578-6663
11 PhaC amino acid sequence, 362 amino acids
12 oligonucleotide probe n2, 39 bases
13 oligonucleotide probe n5, 30 bases
14 oligonucleotide probe bmp, 19 bases
oligonucleotide probe bmc, 22 bases
16 oligonucleotide primer for phaP transcription
start, 20 bases
17 oligonucleotide primer for phaQ transcription
start, 19 bases
18 oligonucleotide primer for phaRBC transcription
start, 19 bases
19 N-terminal amino acid sequence of 14 kDa protein
N-terminal amino acid sequence of 20 kDa protein
21 N-terminal amino acid sequence of 41 kDa protein
22 ykoYnucleic acid sequence, 277-1089
23 YkoY amino acid sequence, 271 amino acids
24 ykoZ nucleic acid sequence, 1460-2167
YkoZ amino acid sequence, 236 amino acids
26 ykrM nucleic acid sequence, 6959-7916 (partial)
27 YkrM amino acid sequence, 319 amino acids (partial)
28 sspD nucleic acid sequence, 2419-2225 reverse
complement
29 SspD amino acid sequence, 65 amino acids
DEFINITIONS
The following definitions are provided in order to aid those skilled in the
art in
understanding the detailed description of the present invention.
"C-terminal region" refers to the region of a peptide, polypeptide, or protein
chain from
s the middle thereof to the end that carries the amino acid having a free a
carboxyl group (the C-
terminus).
"CoA" refers to coenzyme A.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-7_
The phrases "coding sequence", "open reading frame", and "structural sequence"
refer to
the region of continuous sequential nucleic acid triplets encoding a protein,
polypeptide, or
peptide sequence.
The term "encoding DNA" or "encoding nucleic acid" refers to chromosomal
nucleic
s acid, plasmid nucleic acid, cDNA, or synthetic nucleic acid which codes on
expression for any of
the proteins or fusion proteins discussed herein.
The term "genome" as it applies to bacteria encompasses both the chromosome
and
plasmids within a bacterial host cell. Encoding nucleic acids of the present
invention introduced
into bacterial host cells can therefore be either chromosomally-integrated or
plasmid-localized.
~o The term "genome" as it applies to plant cells encompasses not only
chromosomal DNA found
within the nucleus, but organelle DNA found within subcellular components of
the cell. Nucleic
acids of the present invention introduced into plant cells can therefore be
either chromosomally-
integrated or organelle-localized.
"Identity" refers to the degree of similarity between two nucleic acid or
protein
is sequences. An alignment of the two sequences is performed by a suitable
computer program. A
widely used and accepted computer program for performing sequence alignments
is
CLUSTALW v1.6 (Thompson, et al. Nucl. Acids Res., 22: 4673-4680, 1994). The
number of
matching bases or amino acids is divided by the total number of bases or amino
acids, and
multiplied by 100 to obtain a percent identity. For example, if two 580 base
pair sequences had
Zo 145 matched bases, they would be 25 percent identical. If the two compared
sequences are of
different lengths, the number of matches is divided by the shorter of the two
lengths. For
example, if there were 100 matched amino acids between 200 and a 400 amino
acid proteins,
they are 50 percent identical with respect to the shorter sequence. If the
shorter sequence is less
than 150 bases or 50 amino acids in length, the number of matches are divided
by 150 (for
as nucleic acids) or 50 (for proteins); and multiplied by 100 to obtain a
percent identity.
The terms "microbe" or "microorganism" refer to algae, bacteria, fungi, and
protozoa.
"N-terminal region" refers to the region of a peptide, polypeptide, or protein
chain from
the amino acid having a free amino group to the middle of the chain.
"Nucleic acid" refers to ribonucleic acid (RNA) and deoxyribonucleic acid
(DNA).
so A "nucleic acid segment" is a nucleic acid molecule that has been isolated
free of total
genomic DNA of a particular species, or that has been synthesized. Included
with the term
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
_g_
"nucleic acid segment" are DNA segments, recombinant vectors, plasmids,
cosmids, phagemids,
phage, viruses, etcetera.
"Overexpression" refers to the expression of a polypeptide or protein encoded
by a DNA
introduced into a host cell, wherein said polypeptide or protein is either not
normally present in
s the host cell, or wherein said polypeptide or protein is present in said
host cell at a higher level
than that normally expressed from the endogenous gene encoding said
polypeptide or protein.
The term "plastid" refers to the class of plant cell organelles that includes
amyloplasts,
chloroplasts, chromoplasts, elaioplasts, eoplasts, etioplasts, leucoplasts,
and proplastids. These
organelles are self replicating, and contain what is commonly referred to as
the "chloroplast
to genome," a circular DNA molecule that ranges in size from about 120 to
about 217 kb,
depending upon the plant species, and which usually contains an inverted
repeat region (Fosket,
Plant growth and Development, Academic Press, Inc., San Diego, CA, p. 132,
1994).
"Polyadenylation signal" or "polyA signal" refers to a nucleic acid sequence
located 3' to
a coding region that directs the addition of adenylate nucleotides to the 3'
end of the mRNA
i s transcribed from the coding region.
The term "polyhydroxyalkanoate (or PHA) synthase" refers to enzymes that
convert
hydroxyacyl-CoAs to polyhydroxyalkanoates and free CoA.
The term "promoter" or "promoter region" refers to a nucleic acid sequence,
usually
found upstream (5') to a coding sequence, that controls expression of the
coding sequence by
Zo controlling production of messenger RNA (mRNA) by providing the recognition
site for RNA
polymerise and/or other factors necessary for start of transcription at the
correct site. As
contemplated herein, a promoter or promoter region includes variations of
promoters derived by
means of ligation to various regulatory sequences, random or controlled
mutagenesis, and
addition or duplication of enhancer sequences. The promoter region disclosed
herein, and
as biologically functional equivalents thereof, are responsible for driving
the transcription of coding
sequences under their control when introduced into a host as part of a
suitable recombinant
vector, as demonstrated by its ability to produce mRNA.
"Regeneration" refers to the process of growing a plant from a plant cell
(e.g., plant
protoplast or explant).
30 "Transformation" refers to a process of introducing an exogenous nucleic
acid sequence
(e.g., a vector, recombinant nucleic acid molecule) into a cell or protoplast
in which that
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-9-
exogenous nucleic acid is incorporated into a chromosome or is capable of
autonomous
replication.
A "transformed cell" is a cell whose nucleic acid has been altered by the
introduction of
an exogenous nucleic acid molecule into that cell.
s A "transformed plant" or "transgenic plant" is a plant whose nucleic acid
has been altered
by the introduction of an exogenous nucleic acid molecule into that plant, or
by the introduction
of an exogenous nucleic acid molecule into a plant cell from which the plant
was regenerated or
derived.
DETAILED DESCRIPTION OF THE INVENTION
io This invention was developed in the pursuit of proteins which are
associated with
polyhydroxyalkanoate inclusion bodies, and in the pursuit of novel nucleic
acid and amino acid
sequences from the bacteria Bacillus megaterium. A 7,916 base pair nucleic
acid fragment was
isolated and sequenced (SEQ ID NO:1). This fragment was found to contain nine
open reading
frames, five of which encode proteins suspected of being involved in
polyhydroxyalkanoate
1 > biosynthesis.
Genomic fragment
An embodiment of the invention is a nucleic acid segment at least about 80%
identical to
SEQ ID NO:1. More preferably, the nucleic acid segment is at least about 82%,
84%, 86%,
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:1.
Zo Alternatively, the nucleic acid segment may be a nucleic acid segment that
hybridizes under
stringent conditions to SEQ ID NO:1, or to the complement thereof. The nucleic
acid segment
may be obtained from a natural source, may be mutagenized, may be genetically
engineered by
mutagenesis or other methods, or may be synthetic.
The invention is further directed to nucleic acid segments, proteins,
recombinant vectors,
Zs recombinant host cells, genetically transformed plant cells, genetically
transformed plants,
methods of preparing host cells, methods of preparing plants, fusion proteins,
and nucleic acid
segments encoding fusion proteins.
phaP and PhaP
A nucleic acid segment may comprise a nucleic acid sequence encoding a
3o polyhydroxyalkanoate inclusion body associated protein, wherein the nucleic
acid sequence is
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-10-
selected from the group consisting of: a nucleic acid sequence at least about
80% identical to
SEQ ID N0:2; a nucleic acid sequence that hybridizes under stringent
conditions to SEQ ID
N0:2 or the complement thereof; a nucleic acid sequence encoding a protein at
least about 80%
identical to SEQ ID N0:3; and a nucleic acid sequence encoding a protein that
is
s immunoreactive with an antibody prepared using SEQ ID N0:3 as an antigen,
the antibody being
immunoreactive with SEQ ID N0:3. More preferably, the nucleic acid sequence is
at least about
82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to
SEQ ID
N0:2. The nucleic acid segment may be obtained from a natural source, may be
mutagenized,
may be genetically engineered by mutagenesis or other methods, or may be
synthetic. The
to nucleic acid sequence preferably encodes a protein at least about 82%, 84%,
86%, 88%, 90%,
92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:3.
An isolated polyhydroxyalkanoate inclusion body associated protein may
comprise an
amino acid sequence selected from the group consisting of: an amino acid
sequence at least
about 80% identical to SEQ ID N0:3; and an amino acid sequence that is
immunoreactive with
~s an antibody prepared using SEQ ID N0:3 as an antigen, the antibody being
immunoreactive with
SEQ ID N0:3. The protein is preferably at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:3
A recombinant vector may comprise in the 5' to 3' direction: a) a promoter
that directs
transcription of a structural nucleic acid sequence encoding a
polyhydroxyalkanoate inclusion
Zo body associated protein; b) a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
inclusion body associated protein; wherein the structural nucleic acid
sequence is selected from
the group consisting of: a nucleic acid sequence at least about 80% identical
to SEQ ID N0:2; a
nucleic acid sequence that hybridizes under stringent conditions to SEQ ID
N0:2 or the
complement thereof; a nucleic acid sequence encoding a protein at least about
80% identical to
Zs SEQ ID N0:3; and a nucleic acid sequence encoding a protein that is
immunoreactive with an
antibody prepared using SEQ ID N0:3 as an antigen, the antibody being
immunoreactive with
SEQ ID N0:3; and c) a 3' transcription terminator. More preferably, the
nucleic acid sequence
is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or
100%
identical to SEQ ID N0:2. The nucleic acid segment may be obtained from a
natural source,
3o may be mutagenized, may be genetically engineered by mutagenesis or other
methods, or may be
synthetic. The nucleic acid sequence preferably encodes a protein at least
about 82%, 84%,
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-11-
86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
N0:3. The
promoter may generally be any promoter, and more preferably is a tissue
selective or tissue
specific promoter. The promoter may be constitutive or inducible. The promoter
may be a viral
promoter. The promoter may be a CMV35S, enhanced CMV35S, an FMV35S, a
Lesquerella
s hydroxylase, or a 7S conglycinin promoter.
A recombinant host cell may comprise a nucleic acid segment encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the nucleic
acid segment is
selected from the group consisting of: a nucleic acid sequence at least about
80% identical to
SEQ ID N0:2; a nucleic acid sequence that hybridizes under stringent
conditions to SEQ ID
io N0:2 or the complement thereof; a nucleic acid sequence encoding a protein
at least about 80%
identical to SEQ ID N0:3; and a nucleic acid sequence encoding a protein that
is
immunoreactive with an antibody prepared using SEQ ID N0:3 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:3. More preferably, the nucleic acid sequence is
at least about
82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to
SEQ ID
is N0:2. The nucleic acid segment may be obtained from a natural source, may
be mutagenized,
may be genetically engineered by mutagenesis or other methods, or may be
synthetic. The
nucleic acid sequence preferably encodes a protein at least about 82%, 84%,
86%, 88%, 90%,
92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:3. The host
cell may
generally be any host cell, and preferably is a bacterial, fungal, mammalian,
or plant cell. The
ao bacterial cell is preferably an Escherichia coli, Bacillus, Pseudomonas, or
Ralstonia eutropha
cell. The fungal cell is preferably a Saccharomyces cerevisiae or
Schizosaccharomyces pombe
cell. The plant cell is preferably a tobacco, wheat, potato, Arabidopsis, and
high oil seed plants
such as corn, soybean, canola, oil seed rape, sugarbeet, sunflower, flax,
peanut, sugarcane,
switchgrass, or alfalfa cell.
Zs A genetically transformed plant cell may comprise in the 5' to 3'
direction: a) a promoter
that directs transcription of a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
inclusion body associated protein; b) a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein; wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
3o identical to SEQ ID N0:2; a nucleic acid sequence that hybridizes under
stringent conditions to
SEQ ID N0:2 or the complement thereof; a nucleic acid sequence encoding a
protein at least
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-12-
about 80% identical to SEQ ID N0:3; and a nucleic acid sequence encoding a
protein that is
immunoreactive with an antibody prepared using SEQ ID N0:3 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:3; c) a 3' transcription terminator; and d) a 3'
polyadenylation
signal sequence that directs the addition of polyadenylate nucleotides to the
3' end of RNA
s transcribed from the structural nucleic acid sequence. More preferably, the
nucleic acid
sequence is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%,
99.5%, or
100% identical to SEQ ID N0:2. The nucleic acid segment may be obtained from a
natural
source, may be mutagenized, may be genetically engineered by mutagenesis or
other methods, or
may be synthetic. The nucleic acid sequence preferably encodes a protein at
least about 82%,
84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ
ID N0:3.
The plant may generally be any plant, and more preferably a monocot, dicot, or
conifer. The
plant is preferably a tobacco, wheat, potato, Arabidopsis, and high oil seed
plants such as corn,
soybean, canola, oil seed rape, sugarbeet, sunflower, flax, peanut, sugarcane,
switchgrass, or
alfalfa plant.
Is A method of preparing host cells useful to produce a polyhydroxyalkanoate
inclusion
body associated protein may comprise a) selecting a host cell; b) transforming
the selected host
cell with a recombinant vector having a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
ao identical to SEQ ID N0:2; a nucleic acid sequence that hybridizes under
stringent conditions to
SEQ ID N0:2 or the complement thereof; a nucleic acid sequence encoding a
protein at least
about 80% identical to SEQ ID N0:3; and a nucleic acid sequence encoding a
protein that is
immunoreactive with an antibody prepared using SEQ ID N0:3 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:3; and c) obtaining transformed host cells. More
preferably,
Zs the nucleic acid sequence is at least about 82%, 84%, 86%, 88%, 90%, 92%,
94%, 96%, 98%,
99%, 99.5%, or 100% identical to SEQ ID N0:2. The nucleic acid segment may be
obtained
from a natural source, may be mutagenized, may be genetically engineered by
mutagenesis or
other methods, or may be synthetic. The nucleic acid sequence preferably
encodes a protein at
least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical
3o to SEQ ID N0:3. The host cell may generally be any host cell, and
preferably is a bacterial,
fungal, mammalian, or plant cell. The bacterial cell is preferably an
Escherichia coli, Bacillus,
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-13-
Pseudomonas, or Ralstonia eutropha cell. The fungal cell is preferably a
Saccharomyces
cerevisiae or Schizosaccharomyces pombe cell. The plant cell is preferably a
tobacco, wheat,
potato, Arabidopsis, and high oil seed plants such as corn, soybean, canola,
oil seed rape,
sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa cell.
s A method of preparing plants useful to produce a polyhydroxyalkanoate
inclusion body
associated protein may comprise a) selecting a host plant cell; b)
transforming the selected host
plant cell with a recombinant vector having a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
~o identical to SEQ ID N0:2; a nucleic acid sequence that hybridizes under
stringent conditions to
SEQ ID N0:2 or the complement thereof; a nucleic acid sequence encoding a
protein at least
about 80% identical to SEQ ID N0:3; and a nucleic acid sequence encoding a
protein that is
immunoreactive with an antibody prepared using SEQ ID N0:3 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:3; c) obtaining transformed host plant cells;
and d)
~s regenerating the transformed host plant cells. More preferably, the nucleic
acid sequence is at
least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical
to SEQ ID N0:2. The nucleic acid segment may be obtained from a natural
source, may be
mutagenized, may be genetically engineered by mutagenesis or other methods, or
may be
synthetic. The nucleic acid sequence preferably encodes a protein at least
about 82%, 84%,
20 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
N0:3. The
plant (and plant cell) may generally be any plant, and more preferably a
monocot, dicot, or
conifer. The plant is preferably a tobacco, wheat, potato, Arabidopsis, and
high oil seed plants
such as corn, soybean, canola, oil seed rape, sugarbeet, sunflower, flax,
peanut, sugarcane,
switchgrass, or alfalfa plant.
as The invention also relates to fusion proteins. A fusion protein may
comprise a green
fluorescent protein subunit; and a polyhydroxyalkanoate inclusion body
associated protein
subunit; wherein the polyhydroxyalkanoate inclusion body associated protein
subunit comprises
an amino acid sequence selected from the group consisting of: an amino acid
sequence at least
about 80% identical to SEQ ID N0:3; and an amino acid sequence that is
immunoreactive with
3o an antibody prepared using SEQ ID N0:3 as an antigen, the antibody being
immunoreactive with
SEQ ID N0:3. The polyhydroxyalkanoate inclusion body associated protein
subunit is
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-14-
preferably at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%,
99.5%, or
100% identical to SEQ ID N0:3
A nucleic acid segment encoding a fusion protein may comprise a nucleic acid
sequence
encoding a green fluorescent protein subunit; and a nucleic acid sequence
encoding a
s polyhydroxyalkanoate inclusion body associated protein subunit; wherein the
nucleic acid
sequence encoding a polyhydroxyalkanoate inclusion body associated protein
subunit is selected
from the group consisting of: a nucleic acid sequence at least about 80%
identical to SEQ ID
N0:2; a nucleic acid sequence that hybridizes under stringent conditions to
SEQ ID N0:2 or the
complement thereof; a nucleic acid sequence encoding a protein at least about
80% identical to
io SEQ ID N0:3; and a nucleic acid sequence encoding a protein that is
immunoreactive with an
antibody prepared using SEQ ID N0:3 as an antigen, the antibody being
immunoreactive with
SEQ ID N0:3. More preferably, the nucleic acid sequence is at least about 82%,
84%, 86%,
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:2.
The nucleic
acid sequence may be obtained from a natural source, may be mutagenized, may
be genetically
is engineered by mutagenesis or other methods, or may be synthetic. The
nucleic acid sequence
preferably encodes a protein subunit at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%, 96%,
98%, 99%, 99.5%, or 100% identical to SEQ ID N0:3.
phaQ and PhaO
A nucleic acid segment may comprise a nucleic acid sequence encoding a
zo polyhydroxyalkanoate inclusion body associated protein, wherein the nucleic
acid sequence is
selected from the group consisting of: a nucleic acid sequence at least about
80% identical to
SEQ ID N0:4; a nucleic acid sequence that hybridizes under stringent
conditions to SEQ ID
N0:4 or the complement thereof; a nucleic acid sequence encoding a protein at
least about 80%
identical to SEQ ID NO:S; and a nucleic acid sequence encoding a protein that
is
zs immunoreactive with an antibody prepared using SEQ ID NO:S as an antigen,
the antibody being
immunoreactive with SEQ ID NO:S. More preferably, the nucleic acid sequence is
at least about
82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to
SEQ ID
N0:4. The nucleic acid segment may be obtained from a natural source, may be
mutagenized,
may be genetically engineered by mutagenesis or other methods, or may be
synthetic. The
3o nucleic acid sequence preferably encodes a protein at least about 82%, 84%,
86%, 88%, 90%,
92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:S.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-15-
An isolated polyhydroxyalkanoate inclusion body associated protein may
comprise an
amino acid sequence selected from the group consisting of: an amino acid
sequence at least
about 80% identical to SEQ ID N0:5; and an amino acid sequence that is
immunoreactive with
an antibody prepared using SEQ ID N0:5 as an antigen, the antibody being
immunoreactive with
s SEQ ID N0:5. The protein is preferably at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:5
A recombinant vector may comprise in the 5' to 3' direction: a) a promoter
that directs
transcription of a structural nucleic acid sequence encoding a
polyhydroxyalkanoate inclusion
body associated protein; b) a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
io inclusion body associated protein; wherein the structural nucleic acid
sequence is selected from
the group consisting of: a nucleic acid sequence at least about 80% identical
to SEQ ID N0:4; a
nucleic acid sequence that hybridizes under stringent conditions to SEQ ID
N0:4 or the
complement thereof; a nucleic acid sequence encoding a protein at least about
80% identical to
SEQ ID N0:5; and a nucleic acid sequence encoding a protein that is
immunoreactive with an
is antibody prepared using SEQ ID N0:5 as an antigen, the antibody being
immunoreactive with
SEQ ID N0:5; and c) a 3' transcription terminator. More preferably, the
nucleic acid sequence
is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or
100%
identical to SEQ ID N0:4. The nucleic acid segment may be obtained from a
natural source,
may be mutagenized, may be genetically engineered by mutagenesis or other
methods, or may be
zo synthetic. The nucleic acid sequence preferably encodes a protein at least
about 82%, 84%,
86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
N0:5. The
promoter may generally be any promoter, and more preferably is a tissue
selective or tissue
specific promoter. The promoter may be constitutive or inducible. The promoter
may be a viral
promoter. The promoter may be a CMV35S, enhanced CMV35S, an FMV35S, a
Lesquerella
Zs hydroxylase, or a 7S conglycinin promoter.
A recombinant host cell may comprise a nucleic acid segment encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the nucleic
acid segment is
selected from the group consisting of: a nucleic acid sequence at least about
80% identical to
SEQ ID N0:4; a nucleic acid sequence that hybridizes under stringent
conditions to SEQ ID
so N0:4 or the complement thereof; a nucleic acid sequence encoding a protein
at least about 80%
identical to SEQ ID N0:5; and a nucleic acid sequence encoding a protein that
is
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-16-
immunoreactive with an antibody prepared using SEQ ID NO:S as an antigen, the
antibody being
immunoreactive with SEQ ID NO:S. More preferably, the nucleic acid sequence is
at least about
82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to
SEQ ID
N0:4. The nucleic acid segment may be obtained from a natural source, may be
mutagenized,
s may be genetically engineered by mutagenesis or other methods, or may be
synthetic. The
nucleic acid sequence preferably encodes a protein at least about 82%, 84%,
86%, 88%, 90%,
92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:S. The host
cell may
generally be any host cell, and preferably is a bacterial, fungal, mammalian,
or plant cell. The
bacterial cell is preferably an Escherichia coli, Bacillus, Pseudomonas, or
Ralstonia eutropha
~o cell. The fungal cell is preferably a Saccharomyces cerevisiae or
Schizosaccharomyces pombe
cell. The plant cell is preferably a tobacco, wheat, potato, Arabidopsis, and
high oil seed plants
such as corn, soybean, canola, oil seed rape, sugarbeet, sunflower, flax,
peanut, sugarcane,
switchgrass, or alfalfa cell.
A genetically transformed plant cell may comprise in the 5' to 3' direction:
a) a promoter
is that directs transcription of a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
inclusion body associated protein; b) a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein; wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
identical to SEQ ID N0:4; a nucleic acid sequence that hybridizes under
stringent conditions to
Zo SEQ ID N0:4 or the complement thereof; a nucleic acid sequence encoding a
protein at least
about 80% identical to SEQ ID NO:S; and a nucleic acid sequence encoding a
protein that is
immunoreactive with an antibody prepared using SEQ ID NO:S as an antigen, the
antibody being
immunoreactive with SEQ ID NO:S; c) a 3' transcription terminator; and d) a 3'
polyadenylation
signal sequence that directs the addition of polyadenylate nucleotides to the
3' end of RNA
Zs transcribed from the structural nucleic acid sequence. More preferably, the
nucleic acid
sequence is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%,
99.5%, or
100% identical to SEQ ID N0:4. The nucleic acid segment may be obtained from a
natural
source, may be mutagenized, may be genetically engineered by mutagenesis or
other methods, or
may be synthetic. The nucleic acid sequence preferably encodes a protein at
least about 82%,
30 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to
SEQ ID NO:S.
The plant may generally be any plant, and more preferably a monocot, dicot, or
conifer. The
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-17-
plant is preferably a tobacco, wheat, potato, Arabidopsis, and high oil seed
plants such as corn,
soybean, canola, oil seed rape, sugarbeet, sunflower, flax, peanut, sugarcane,
switchgrass, or
alfalfa plant.
A method of preparing host cells useful to produce a polyhydroxyalkanoate
inclusion
s body associated protein may comprise a) selecting a host cell; b)
transforming the selected host
cell with a recombinant vector having a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
identical to SEQ ID N0:4; a nucleic acid sequence that hybridizes under
stringent conditions to
io SEQ ID N0:4 or the complement thereof; a nucleic acid sequence encoding a
protein at least
about 80% identical to SEQ ID NO:S; and a nucleic acid sequence encoding a
protein that is
immunoreactive with an antibody prepared using SEQ ID NO:S as an antigen, the
antibody being
immunoreactive with SEQ ID NO:S; and c) obtaining transformed host cells. More
preferably,
the nucleic acid sequence is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%,
96%, 98%,
is 99%, 99.5%, or 100% identical to SEQ ID N0:4. The nucleic acid segment may
be obtained
from a natural source, may be mutagenized, may be genetically engineered by
mutagenesis or
other methods, or may be synthetic. The nucleic acid sequence preferably
encodes a protein at
least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical
to SEQ ID NO:S. The host cell may generally be any host cell, and preferably
is a bacterial,
zo fungal, mammalian, or plant cell. The bacterial cell is preferably an
Escherichia coli, Bacillus,
Pseudomonas, or Ralstonia eutropha cell. The fungal cell is preferably a
Saccharomyces
cerevisiae or Schizosaccharomyces pombe cell. The plant cell is preferably a
tobacco, wheat,
potato, Arabidopsis, and high oil seed plants such as corn, soybean, canola,
oil seed rape,
sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa cell.
Zs A method of preparing plants useful to produce a polyhydroxyalkanoate
inclusion body
associated protein may comprise a) selecting a host plant cell; b)
transforming the selected host
plant cell with a recombinant vector having a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
3o identical to SEQ ID N0:4; a nucleic acid sequence that hybridizes under
stringent conditions to
SEQ ID N0:4 or the complement thereof; a nucleic acid sequence encoding a
protein at least
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-18-
about 80% identical to SEQ ID N0:5; and a nucleic acid sequence encoding a
protein that is
immunoreactive with an antibody prepared using SEQ ID N0:5 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:5; c) obtaining transformed host plant cells;
and d)
regenerating the transformed host plant cells. More preferably, the nucleic
acid sequence is at
s least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical
to SEQ ID N0:4. The nucleic acid segment may be obtained from a natural
source, may be
mutagenized, may be genetically engineered by mutagenesis or other methods, or
may be
synthetic. The nucleic acid sequence preferably encodes a protein at least
about 82%, 84%,
86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
N0:5. The
~o plant (and plant cell) may generally be any plant, and more preferably a
monocot, dicot, or
conifer. The plant is preferably a tobacco, wheat, potato, Arabidopsis, and
high oil seed plants
such as corn, soybean, canola, oil seed rape, sugarbeet, sunflower, flax,
peanut, sugarcane,
switchgrass, or alfalfa plant.
The invention also relates to fusion proteins. A fusion protein may comprise a
green
~s fluorescent protein subunit; and a polyhydroxyalkanoate inclusion body
associated protein
subunit; wherein the polyhydroxyalkanoate inclusion body associated protein
subunit comprises
an amino acid sequence selected from the group consisting of: an amino acid
sequence at least
about 80% identical to SEQ ID N0:5; and an amino acid sequence that is
immunoreactive with
an antibody prepared using SEQ ID N0:5 as an antigen, the antibody being
immunoreactive with
Zo SEQ ID N0:5. The polyhydroxyalkanoate inclusion body associated protein
subunit is
preferably at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%,
99.5%, or
100% identical to SEQ ID N0:5
A nucleic acid segment encoding a fusion protein may comprise a nucleic acid
sequence
encoding a green fluorescent protein subunit; and a nucleic acid sequence
encoding a
Zs polyhydroxyalkanoate inclusion body associated protein subunit; wherein the
nucleic acid
sequence encoding a polyhydroxyalkanoate inclusion body associated protein
subunit is selected
from the group consisting of: a nucleic acid sequence at least about 80%
identical to SEQ ID
N0:4; a nucleic acid sequence that hybridizes under stringent conditions to
SEQ ID N0:4 or the
complement thereof; a nucleic acid sequence encoding a protein at least about
80% identical to
3o SEQ ID N0:5; and a nucleic acid sequence encoding a protein that is
immunoreactive with an
antibody prepared using SEQ ID N0:5 as an antigen, the antibody being
immunoreactive with
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-19-
SEQ ID NO:S. More preferably, the nucleic acid sequence is at least about 82%,
84%, 86%,
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:4.
The nucleic
acid sequence may be obtained from a natural source, may be mutagenized, may
be genetically
engineered by mutagenesis or other methods, or may be synthetic. The nucleic
acid sequence
s preferably encodes a protein subunit at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%, 96%,
98%, 99%, 99.5%, or 100% identical to SEQ ID NO:S.
phaR and PhaR
A nucleic acid segment may comprise a nucleic acid sequence encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the nucleic
acid sequence is
~o selected from the group consisting of: a nucleic acid sequence at least
about 80% identical to
SEQ ID N0:6; a nucleic acid sequence that hybridizes under stringent
conditions to SEQ ID
N0:6 or the complement thereof; a nucleic acid sequence encoding a protein at
least about 80%
identical to SEQ ID N0:7; and a nucleic acid sequence encoding a protein that
is
immunoreactive with an antibody prepared using SEQ ID N0:7 as an antigen, the
antibody being
is immunoreactive with SEQ ID N0:7. More preferably, the nucleic acid sequence
is at least about
82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to
SEQ ID
N0:6. The nucleic acid segment may be obtained from a natural source, may be
mutagenized,
may be genetically engineered by mutagenesis or other methods, or may be
synthetic. The
nucleic acid sequence preferably encodes a protein at least about 82%, 84%,
86%, 88%, 90%,
zo 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:7.
An isolated polyhydroxyalkanoate inclusion body associated protein may
comprise an
amino acid sequence selected from the group consisting of: an amino acid
sequence at least
about 80% identical to SEQ ID N0:7; and an amino acid sequence that is
immunoreactive with
an antibody prepared using SEQ ID N0:7 as an antigen, the antibody being
immunoreactive with
zs SEQ ID N0:7. The protein is preferably at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:7
A recombinant vector may comprise in the 5' to 3' direction: a) a promoter
that directs
transcription of a structural nucleic acid sequence encoding a
polyhydroxyalkanoate inclusion
body associated protein; b) a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
30 inclusion body associated protein; wherein the structural nucleic acid
sequence is selected from
the group consisting of: a nucleic acid sequence at least about 80% identical
to SEQ ID N0:6; a
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-20-
nucleic acid sequence that hybridizes under stringent conditions to SEQ ID
N0:6 or the
complement thereof; a nucleic acid sequence encoding a protein at least about
80% identical to
SEQ ID N0:7; and a nucleic acid sequence encoding a protein that is
immunoreactive with an
antibody prepared using SEQ ID N0:7 as an antigen, the antibody being
immunoreactive with
s SEQ ID N0:7; and c) a 3' transcription terminator. More preferably, the
nucleic acid sequence
is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or
100%
identical to SEQ ID N0:6. The nucleic acid segment may be obtained from a
natural source,
may be mutagenized, may be genetically engineered by mutagenesis or other
methods, or may be
synthetic. The nucleic acid sequence preferably encodes a protein at least
about 82%, 84%,
io 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
N0:7. The
promoter may generally be any promoter, and more preferably is a tissue
selective or tissue
specific promoter. The promoter may be constitutive or inducible. The promoter
may be a viral
promoter. The promoter may be a CMV35S, enhanced CMV35S, an FMV35S, a
Lesquerella
hydroxylase, or a 7S conglycinin promoter.
Is A recombinant host cell may comprise a nucleic acid segment encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the nucleic
acid segment is
selected from the group consisting o~ a nucleic acid sequence at least about
80% identical to
SEQ ID N0:6; a nucleic acid sequence that hybridizes under stringent
conditions to SEQ ID
N0:6 or the complement thereof; a nucleic acid sequence encoding a protein at
least about 80%
Zo identical to SEQ ID N0:7; and a nucleic acid sequence encoding a protein
that is
immunoreactive with an antibody prepared using SEQ ID N0:7 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:7. More preferably, the nucleic acid sequence is
at least about
82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to
SEQ ID
N0:6. The nucleic acid segment may be obtained from a natural source, may be
mutagenized,
Zs may be genetically engineered by mutagenesis or other methods, or may be
synthetic. The
nucleic acid sequence preferably encodes a protein at least about 82%, 84%,
86%, 88%, 90%,
92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:7. The host
cell may
generally be any host cell, and preferably is a bacterial, fungal, mammalian,
or plant cell. The
bacterial cell is preferably an Escherichia coli, Bacillus, Pseudomonas, or
Ralstonia eutropha
3o cell. The fungal cell is preferably a Saccharomyces cerevisiae or
Schizosaccharomyces pombe
cell. The plant cell is preferably a tobacco, wheat, potato, Arabidopsis, and
high oil seed plants
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-21 -
such as corn, soybean, canola, oil seed rape, sugarbeet, sunflower, flax,
peanut, sugarcane,
switchgrass, or alfalfa cell.
A genetically transformed plant cell may comprise in the 5' to 3' direction:
a) a promoter
that directs transcription of a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
s inclusion body associated protein; b) a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein; wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
identical to SEQ ID N0:6; a nucleic acid sequence that hybridizes under
stringent conditions to
SEQ ID N0:6 or the complement thereof; a nucleic acid sequence encoding a
protein at least
io about 80% identical to SEQ ID N0:7; and a nucleic acid sequence encoding a
protein that is
immunoreactive with an antibody prepared using SEQ ID N0:7 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:7; c) a 3' transcription terminator; and d) a 3'
polyadenylation
signal sequence that directs the addition of polyadenylate nucleotides to the
3' end of RNA
transcribed from the structural nucleic acid sequence. More preferably, the
nucleic acid
is sequence is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%,
99%, 99.5%, or
100% identical to SEQ ID N0:6. The nucleic acid segment may be obtained from a
natural
source, may be mutagenized, may be genetically engineered by mutagenesis or
other methods, or
may be synthetic. The nucleic acid sequence preferably encodes a protein at
least about 82%,
84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ
ID N0:7.
zo The plant may generally be any plant, and more preferably a monocot, dicot,
or conifer. The
plant is preferably a tobacco, wheat, potato, Arabidopsis, and high oil seed
plants such as corn,
soybean, canola, oil seed rape, sugarbeet, sunflower, flax, peanut, sugarcane,
switchgrass, or
alfalfa plant.
A method of preparing host cells useful to produce a polyhydroxyalkanoate
inclusion
z> body associated protein may comprise a) selecting a host cell; b)
transforming the selected host
cell with a recombinant vector having a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
identical to SEQ ID N0:6; a nucleic acid sequence that hybridizes under
stringent conditions to
3o SEQ ID N0:6 or the complement thereof; a nucleic acid sequence encoding a
protein at least
about 80% identical to SEQ ID N0:7; and a nucleic acid sequence encoding a
protein that is
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-22-
immunoreactive with an antibody prepared using SEQ ID N0:7 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:7; and c) obtaining transformed host cells. More
preferably,
the nucleic acid sequence is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%,
96%, 98%,
99%, 99.5%, or 100% identical to SEQ ID N0:6. The nucleic acid segment may be
obtained
s from a natural source, may be mutagenized, may be genetically engineered by
mutagenesis or
other methods, or may be synthetic. The nucleic acid sequence preferably
encodes a protein at
least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical
to SEQ ID N0:7. The host cell may generally be any host cell, and preferably
is a bacterial,
fungal, mammalian, or plant cell. The bacterial cell is preferably an
Escherichia coli, Bacillus,
~o Pseudomonas, or Ralstonia eutropha cell. The fungal cell is preferably a
Saccharomyces
cerevisiae or Schizosaccharomyces pombe cell. The plant cell is preferably a
tobacco, wheat,
potato, Arabidopsis, and high oil seed plants such as corn, soybean, canola,
oil seed rape,
sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa cell.
A method of preparing plants useful to produce a polyhydroxyalkanoate
inclusion body
is associated protein may comprise a) selecting a host plant cell; b)
transforming the selected host
plant cell with a recombinant vector having a structural nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein, wherein the structural
nucleic acid
sequence is selected from the group consisting of: a nucleic acid sequence at
least about 80%
identical to SEQ ID N0:6; a nucleic acid sequence that hybridizes under
stringent conditions to
ao SEQ ID N0:6 or the complement thereof; a nucleic acid sequence encoding a
protein at least
about 80% identical to SEQ ID N0:7; and a nucleic acid sequence encoding a
protein that is
immunoreactive with an antibody prepared using SEQ ID N0:7 as an antigen, the
antibody being
immunoreactive with SEQ ID N0:7; c) obtaining transformed host plant cells;
and d)
regenerating the transformed host plant cells. More preferably, the nucleic
acid sequence is at
zs least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or
100% identical
to SEQ ID N0:6. The nucleic acid segment may be obtained from a natural
source, may be
mutagenized, may be genetically engineered by mutagenesis or other methods, or
may be
synthetic. The nucleic acid sequence preferably encodes a protein at least
about 82%, 84%,
86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical .to SEQ ID
N0:7. The
3o plant (and plant cell) may generally be any plant, and more preferably a
monocot, dicot, or
conifer. The plant is preferably a tobacco, wheat, potato, Arabidopsis, and
high oil seed plants
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
- 23 -
such as corn, soybean, canola, oil seed rape, sugarbeet, sunflower, flax,
peanut, sugarcane,
switchgrass, or alfalfa plant.
The invention also relates to fusion proteins. A fusion protein may comprise a
green
fluorescent protein subunit; and a polyhydroxyalkanoate inclusion body
associated protein
s subunit; wherein the polyhydroxyalkanoate inclusion body associated protein
subunit comprises
an amino acid sequence selected from the group consisting of: an amino acid
sequence at least
about 80% identical to SEQ ID N0:7; and an amino acid sequence that is
immunoreactive .with
an antibody prepared using SEQ ID N0:7 as an antigen, the antibody being
immunoreactive with
SEQ ID N0:7. The polyhydroxyalkanoate inclusion body associated protein
subunit is
~o preferably at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%,
99.5%, or
100% identical to SEQ ID N0:7
A nucleic acid segment encoding a fusion protein may comprise a nucleic acid
sequence
encoding a green fluorescent protein subunit; and a nucleic acid sequence
encoding a
polyhydroxyalkanoate inclusion body associated protein subunit; wherein the
nucleic acid
is sequence encoding a polyhydroxyalkanoate inclusion body associated protein
subunit is selected
from the group consisting of: a nucleic acid sequence at least about 80%
identical to SEQ ID
N0:6; a nucleic acid sequence that hybridizes under stringent conditions to
SEQ ID N0:6 or the
complement thereof; a nucleic acid sequence encoding a protein at least about
80% identical to
SEQ ID N0:7; and a nucleic acid sequence encoding a protein that is
immunoreactive with an
Zo antibody prepared using SEQ ID N0:7 as an antigen, the antibody being
immunoreactive with
SEQ ID N0:7. More preferably, the nucleic acid sequence is at least about 82%,
84%, 86%,
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:6.
The nucleic
acid sequence may be obtained from a natural source, may be mutagenized, may
be genetically
engineered by mutagenesis or other methods, or may be synthetic. The nucleic
acid sequence
is preferably encodes a protein subunit at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%, 96%,
98%, 99%, 99.5%, or 100% identical to SEQ ID N0:7.
phaB and PhaB
A nucleic acid segment may comprise a nucleic acid sequence encoding a 3-keto-
acyl
CoA reductase protein, wherein the nucleic acid sequence is selected from the
group consisting
30 of: a nucleic acid sequence at least about 80% identical to SEQ ID N0:8; a
nucleic acid sequence
that hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-24-
acid sequence encoding a protein at least about 80% identical to SEQ ID N0:9;
and a nucleic
acid sequence encoding a protein that is immunoreactive with an antibody
prepared using SEQ
ID N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9.
More
preferably, the nucleic acid sequence is at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
s 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:8. The nucleic acid
segment may be
obtained from a natural source, may be mutagenized, may be genetically
engineered by
mutagenesis or other methods, or may be synthetic. The nucleic acid sequence
preferably
encodes a protein at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%,
99%, 99.5%,
or 100% identical to SEQ ID N0:9.
~o An isolated 3-keto-acyl-CoA reductase protein may comprise an amino acid
sequence
selected from the group consisting of: an amino acid sequence at least about
80% identical to
SEQ ID N0:9; and an amino acid sequence that is immunoreactive with an
antibody prepared
using SEQ ID N0:9 as an antigen, the antibody being immunoreactive with SEQ ID
N0:9. The
protein is preferably at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%,
98%, 99%,
is 99.5%, or 100% identical to SEQ ID N0:9
A recombinant vector may comprise in the 5' to 3' direction: a) a promoter
that directs
transcription of a structural nucleic acid sequence encoding a 3-keto-acyl-CoA
reductase protein;
b) a structural nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein; wherein the
structural nucleic acid sequence is selected from the group consisting of: a
nucleic acid sequence
ao at least about 80% identical to SEQ ID N0:8; a nucleic acid sequence that
hybridizes under
stringent conditions to SEQ ID N0:8 or the complement thereof; a nucleic acid
sequence
encoding a protein at least about 80% identical to SEQ ID N0:9; and a nucleic
acid sequence
encoding a protein that is immunoreactive with an antibody prepared using SEQ
ID N0:9 as an
antigen, the antibody being immunoreactive with SEQ ID N0:9; and c) a 3'
transcription
Zs terminator. More preferably, the nucleic acid sequence is at least about
82%, 84%, 86%, 88%,
90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:8. The
nucleic acid
segment may be obtained from a natural source, may be mutagenized, may be
genetically
engineered by mutagenesis or other methods, or may be synthetic. The nucleic
acid sequence
preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%,
96%, 98%,
30 99%, 99.5%, or 100% identical to SEQ ID N0:9. The promoter may generally be
any promoter,
and more preferably is a tissue selective or tissue specific promoter. The
promoter may be
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-25-
constitutive or inducible. The promoter may be a viral promoter. The promoter
may be a
CMV35S, enhanced CMV35S, an FMV35S, a Lesquerella hydroxylase, or a 7S
conglycinin
promoter.
A recombinant host cell may comprise a nucleic acid segment encoding a 3-keto-
acyl-
s CoA reductase protein, wherein the nucleic acid segment is selected from the
group consisting
of: a nucleic acid sequence at least about 80% identical to SEQ ID N0:8; a
nucleic acid sequence
that hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic
acid sequence encoding a protein at least about 80% identical to SEQ ID N0:9;
and a nucleic
acid sequence encoding a protein that is immunoreactive with an antibody
prepared using SEQ
to ID N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9.
More
preferably, the nucleic acid sequence is at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
96%, 98%, 99%, 99.5%, or I00% identical to SEQ ID N0:8. The nucleic acid
segment may be
obtained from a natural source, may be mutagenized, may be genetically
engineered by
mutagenesis or other methods, or may be synthetic. The nucleic acid sequence
preferably
is encodes a protein at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%,
98%, 99%, 99.5%,
or 100% identical to SEQ ID N0:9. The host cell may generally be any host
cell, and preferably
is a bacterial, fungal, mammalian, or plant cell. The bacterial cell is
preferably an Escherichia
coli, Bacillus, Pseudomonas, or Ralstonia eutropha cell. The fungal cell is
preferably a
Saccharomyces cerevisiae or Schizosaccharomyces pombe cell. The plant cell is
preferably a
Zo tobacco, wheat, potato, Arabidopsis, and high oil seed plants such as corn,
soybean, canola, oil
seed rape, sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or
alfalfa cell.
A genetically transformed plant cell may comprise in the 5' to 3' direction:
a) a promoter
that directs transcription of a structural nucleic acid sequence encoding a 3-
keto-acyl-CoA
reductase protein; b) a structural nucleic acid sequence encoding a 3-keto-
acyl-CoA reductase
zs protein; wherein the structural nucleic acid sequence is selected from the
group consisting o~ a
nucleic acid sequence at least about 80% identical to SEQ ID N0:8; a nucleic
acid sequence that
hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic acid
sequence encoding a protein at least about 80% identical to SEQ ID N0:9; and a
nucleic acid
sequence encoding a protein that is immunoreactive with an antibody prepared
using SEQ ID
so N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9; c)
a 3' transcription
terminator; and d) a 3' polyadenylation signal sequence that directs the
addition of polyadenylate
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-26-
nucleotides to the 3' end of RNA transcribed from the structural nucleic acid
sequence. More
preferably, the nucleic acid sequence is at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:8. The nucleic acid
segment may be
obtained from a natural source, may be mutagenized, may be genetically
engineered by
s mutagenesis or other methods, or may be synthetic. The nucleic acid sequence
preferably
encodes a protein at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%,
99%, 99.5%,
or 100% identical to SEQ ID N0:9. The plant may generally be any plant, and
more preferably a
monocot, dicot, or conifer. The plant is preferably a tobacco, wheat, potato,
Arabidopsis, and
high oil seed plants such as corn, soybean, canola, oil seed rape, sugarbeet,
sunflower, flax,
io peanut, sugarcane, switchgrass, or alfalfa plant.
A method of preparing host cells useful to produce a 3-keto-acyl-CoA reductase
protein
may comprise a) selecting a host cell; b) transforming the selected host cell
with a recombinant
vector having a structural nucleic acid sequence encoding a 3-keto-acyl-CoA
reductase protein,
wherein the structural nucleic acid sequence is selected from the group
consisting of: a nucleic
is acid sequence at least about 80% identical to SEQ ID N0:8; a nucleic acid
sequence that
hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic acid
sequence encoding a protein at least about 80% identical to SEQ ID N0:9; and a
nucleic acid
sequence encoding a protein that is immunoreactive with an antibody prepared
using SEQ ID
N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9; and c)
obtaining
Zo transformed host cells. More preferably, the nucleic acid sequence is at
least about 82%, 84%,
86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
N0:8. The
nucleic acid segment may be obtained from a natural source, may be
mutagenized, may be
genetically engineered by mutagenesis or other methods, or may be synthetic.
The nucleic acid
sequence preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%,
Zs 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:9. The host cell may
generally be any
host cell, and preferably is a bacterial, fungal, mammalian, or plant cell.
The bacterial cell is
preferably an Escherichia coli, Bacillus, Pseudomonas, or Ralstonia eutropha
cell. The fungal
cell is preferably a Saccharomyces cerevisiae or Schizosaccharomyces pombe
cell. The plant
cell is preferably a tobacco, wheat, potato, Arabidopsis, and high oil seed
plants such as corn,
3o soybean, canola, oil seed rape, sugarbeet, sunflower, flax, peanut,
sugarcane, switchgrass, or
alfalfa cell.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-27-
A method of preparing plants useful to produce a 3-keto-acyl-CoA reductase
protein may
comprise a) selecting a host plant cell; b) transforming the selected host
plant cell with a
recombinant vector having a structural nucleic acid sequence encoding a 3-keto-
acyl-CoA
reductase protein, wherein the structural nucleic acid sequence is selected
from the group
s consisting of: a nucleic acid sequence at least about 80% identical to SEQ
ID N0:8; a nucleic
acid sequence that hybridizes under stringent conditions to SEQ ID N0:8 or the
complement
thereof; a nucleic acid sequence encoding a protein at least about 80%
identical to SEQ ID N0:9;
and a nucleic acid sequence encoding a protein that is immunoreactive with an
antibody prepared
using SEQ ID N0:9 as an antigen, the antibody being immunoreactive with SEQ ID
N0:9; c)
io obtaining transformed host plant cells; and d) regenerating the transformed
host plant cells.
More preferably, the nucleic acid sequence is at least about 82%, 84%, 86%,
88%, 90%, 92%,
94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:8. The nucleic acid
segment
may be obtained from a natural source, may be mutagenized, may be genetically
engineered by
mutagenesis or other methods, or may be synthetic. The nucleic acid sequence
preferably
is encodes a protein at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%,
98%, 99%, 99.5%,
or 100% identical to SEQ ID N0:9. The plant (and plant cell) may generally be
any plant, and
more preferably a monocot, dicot, or conifer. The plant is preferably a
tobacco, wheat, potato,
Arabidopsis, and high oil seed plants such as corn, soybean, canola, oil seed
rape, sugarbeet,
sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa plant.
zo The invention also relates to fusion proteins. A fusion protein may
comprise a green
fluorescent protein subunit; and a 3-keto-acyl-CoA reductase protein subunit;
wherein the 3-
keto-acyl-CoA reductase protein subunit comprises an amino acid sequence
selected from the
group consisting of: an amino acid sequence at least about 80% identical to
SEQ ID N0:9; and
an amino acid sequence that is immunoreactive with an antibody prepared using
SEQ ID N0:9
zs as an antigen, the antibody being immunoreactive with SEQ ID N0:9. The 3-
keto-acyl-CoA
reductase protein subunit is preferably at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:9
A nucleic acid segment encoding a fusion protein may comprise a nucleic acid
sequence
encoding a green fluorescent protein subunit; and a nucleic acid sequence
encoding a 3-keto
3o acyl-CoA reductase protein subunit; wherein the nucleic acid sequence
encoding a 3-keto-acyl
CoA reductase protein subunit is selected from the group consisting of: a
nucleic acid sequence
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-28-
at least about 80% identical to SEQ ID N0:8; a nucleic acid sequence that
hybridizes under
stringent conditions to SEQ ID N0:8 or the complement thereof; a nucleic acid
sequence
encoding a protein at least about 80% identical to SEQ ID N0:9; and a nucleic
acid sequence
encoding a protein that is immunoreactive with an antibody prepared using SEQ
ID N0:9 as an
s antigen, the antibody being immunoreactive with SEQ ID N0:9. More
preferably, the nucleic
acid sequence is at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%,
99%, 99.5%,
or 100% identical to SEQ ID N0:8. The nucleic acid sequence may be obtained
from a natural
source, may be mutagenized, may be genetically engineered by mutagenesis or
other methods, or
may be synthetic. The nucleic acid sequence preferably encodes a protein
subunit at least about
l0 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical
to SEQ ID
N0:9.
phaC and PhaC
A nucleic acid segment may comprise a nucleic acid sequence encoding a
polyhydroxyalkanoate synthase protein, wherein the nucleic acid sequence is
selected from the
is group consisting of: a nucleic acid sequence at least about 80% identical
to SEQ ID NO:10; a
nucleic acid sequence that hybridizes under stringent conditions to SEQ ID
NO:10 or the
complement thereof; a nucleic acid sequence encoding a protein at least about
80% identical to
SEQ ID NO:11; and a nucleic acid sequence encoding a protein that is
immunoreactive with an
antibody prepared using SEQ ID NO:11 as an antigen, the antibody being
immunoreactive with
zo SEQ ID NO:11. More preferably, the nucleic acid sequence is at least about
82%, 84%, 86%,
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:10.
The
nucleic acid segment may be obtained from a natural source, may be
mutagenized, may be
genetically engineered by mutagenesis or other methods, or may be synthetic.
The nucleic acid
sequence preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%,
zs 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:11.
An isolated polyhydroxyalkanoate synthase protein may comprise an amino acid
sequence selected from the group consisting of: an amino acid sequence at
least about 80%
identical to SEQ ID NO:11; and an amino acid sequence that is immunoreactive
with an
antibody prepared using SEQ ID NO:11 as an antigen, the antibody being
immunoreactive with
3o SEQ ID NO:11. The protein is preferably at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:11
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-29-
A recombinant vector may comprise in the 5' to 3' direction: a) a promoter
that directs
transcription of a structural nucleic acid sequence encoding a
polyhydroxyalkanoate synthase
protein; b) a structural nucleic acid sequence encoding a polyhydroxyalkanoate
synthase protein;
wherein the structural nucleic acid sequence is selected from the group
consisting of: a nucleic
s acid sequence at least about 80% identical to SEQ ID NO:10; a nucleic acid
sequence that
hybridizes under stringent conditions to SEQ ID NO:10 or the complement
thereof; a nucleic
acid sequence encoding a protein at least about 80% identical to SEQ ID NO:11;
and a nucleic
acid sequence encoding a protein that is immunoreactive with an antibody
prepared using SEQ
ID NO:11 as an antigen, the antibody being immunoreactive with SEQ ID NO:11;
and c) a 3'
io transcription terminator. More preferably, the nucleic acid sequence is at
least about 82%, 84%,
86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
NO:10. The
nucleic acid segment may be obtained from a natural source, may be
mutagenized, may be
genetically engineered by mutagenesis or other methods, or may be synthetic.
The nucleic acid
sequence preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%,
is 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:11. The promoter may
generally be
any promoter, and more preferably is a tissue selective or tissue specific
promoter. The promoter
may be constitutive or inducible. The promoter may be a viral promoter. The
promoter may be
a CMV35S, enhanced CMV35S, an FMV35S, a Lesquerella hydroxylase, or a 7S
conglycinin
promoter.
ao A recombinant host cell may comprise a nucleic acid segment encoding a
polyhydroxyalkanoate synthase protein, wherein the nucleic acid segment is
selected from the
group consisting of: a nucleic acid sequence at least about 80% identical to
SEQ ID NO:10; a
nucleic acid sequence that hybridizes under stringent conditions to SEQ ID
NO:10 or the
complement thereof; a nucleic acid sequence encoding a protein at least about
80% identical to
Zs SEQ ID NO:11; and a nucleic acid sequence encoding a protein that is
immunoreactive with an
antibody prepared using SEQ ID NO:11 as an antigen, the antibody being
immunoreactive with
SEQ ID NO:11. More preferably, the nucleic acid sequence is at least about
82%, 84%, 86%,
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:10.
The
nucleic acid segment may be obtained from a natural source, may be
mutagenized, may be
3o genetically engineered by mutagenesis or other methods, or may be
synthetic. The nucleic acid
sequence preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%,
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-30-
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:11. The host cell may
generally be
any host cell, and preferably is a bacterial, fungal, mammalian, or plant
cell. The bacterial cell is
preferably an Escherichia coli, Bacillus, Pseudomonas, or Ralstonia eutropha
cell. The fungal
cell is preferably a Saccharomyces cerevisiae or Schizosaccharomyces pombe
cell. The plant
s cell is preferably a tobacco, wheat, potato, Arabidopsis, and high oil seed
plants such as corn,
soybean, canola, oil seed rape, sugarbeet, sunflower, flax, peanut, sugarcane,
switchgrass, or
alfalfa cell.
A genetically transformed plant cell may comprise in the 5' to 3' direction:
a) a promoter
that directs transcription of a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
io synthase protein; b) a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
synthase protein; wherein the structural nucleic acid sequence is selected
from the group
consisting of: a nucleic acid sequence at least about 80% identical to SEQ ID
NO:10; a nucleic
acid sequence that hybridizes under stringent conditions to SEQ ID NO:10 or
the complement
thereof; a nucleic acid sequence encoding a protein at least about 80%
identical to SEQ ID
i s NO:11; and a nucleic acid sequence encoding a protein that is
immunoreactive with an antibody
prepared using SEQ ID NO:11 as an antigen, the antibody being immunoreactive
with SEQ ID
NO:11; c) a 3' transcription terminator; and d) a 3' polyadenylation signal
sequence that directs
the addition of polyadenylate nucleotides to the 3' end of RNA transcribed
from the structural
nucleic acid sequence. More preferably, the nucleic acid sequence is at least
about 82%, 84%,
zo 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
NO:10. The
nucleic acid segment may be obtained from a natural source, may be
mutagenized, may be
genetically engineered by mutagenesis or other methods, or may be synthetic.
The nucleic acid
sequence preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:I 1. The plant may
generally be any
zs plant, and more preferably a monocot, dicot, or conifer. The plant is
preferably a tobacco, wheat,
potato, Arabidopsis, and high oil seed plants such as corn, soybean, canola,
oil seed rape,
sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa plant.
A method of preparing host cells useful to produce a polyhydroxyalkanoate
synthase
protein may comprise a) selecting a host cell; b) transforming the selected
host cell with a
3o recombinant vector having a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
synthase protein, wherein the structural nucleic acid sequence is selected
from the group
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-31-
consisting of: a nucleic acid sequence at least about 80% identical to SEQ ID
NO:10; a nucleic
acid sequence that hybridizes under stringent conditions to SEQ ID NO:10 or
the complement
thereof; a nucleic acid sequence encoding a protein at least about 80%
identical to SEQ ID
NO:11; and a nucleic acid sequence encoding a protein that is immunoreactive
with an antibody
s prepared using SEQ ID NO:11 as an antigen, the antibody being immunoreactive
with SEQ ID
NO:11; and c) obtaining transformed host cells. More preferably, the nucleic
acid sequence is at
least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical
to SEQ ID NO:10. The nucleic acid segment may be obtained from a natural
source, may be
mutagenized, may be genetically engineered by mutagenesis or other methods, or
may be
io synthetic. The nucleic acid sequence preferably encodes a protein at least
about 82%, 84%,
86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
NO:11. The
host cell may generally be any host cell, and preferably is a bacterial,
fungal, mammalian, or
plant cell. The bacterial cell is preferably an Escherichia coli, Bacillus,
Pseudomonas, or
Ralstonia eutropha cell. The fungal cell is preferably a Saccharomyces
cerevisiae or
is Schizosaccharomyces pombe cell. The plant cell is preferably a tobacco,
wheat, potato,
Arabidopsis, and high oil seed plants such as corn, soybean, canola, oil seed
rape, sugarbeet,
sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa cell.
A method of preparing plants useful to produce a polyhydroxyalkanoate synthase
protein
may comprise a) selecting a host plant cell; b) transforming the selected host
plant cell with a
2o recombinant vector having a structural nucleic acid sequence encoding a
polyhydroxyalkanoate
synthase protein, wherein the structural nucleic acid sequence is selected
from the group
consisting of: a nucleic acid sequence at least about 80% identical to SEQ ID
NO:10; a nucleic
acid sequence that hybridizes under stringent conditions to SEQ ID NO:10 or
the complement
thereof; a nucleic acid sequence encoding a protein at least about 80%
identical to SEQ ID
as NO:11; and a nucleic acid sequence encoding a protein that is
immunoreactive with an antibody
prepared using SEQ ID NO:11 as an antigen, the antibody being immunoreactive
with SEQ ID
NO:11; c) obtaining transformed host plant cells; and d) regenerating the
transformed host plant
cells. More preferably, the nucleic acid sequence is at least about 82%, 84%,
86%, 88%, 90%,
92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:10. The nucleic
acid
3o segment may be obtained from a natural source, may be mutagenized, may be
genetically
engineered by mutagenesis or other methods, or may be synthetic. The nucleic
acid sequence
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-32-
preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%,
96%, 98%,
99%, 99.5%, or 100% identical to SEQ ID NO:11. The plant (and plant cell) may
generally be
any plant, and more preferably a monocot, dicot, or conifer. The plant is
preferably a tobacco,
wheat, potato, Arabidopsis, and high oil seed plants such as corn, soybean,
canola, oil seed rape,
s sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa
plant. ,
The invention also relates to fusion proteins. A fusion protein may comprise a
green
fluorescent protein subunit; and a polyhydroxyalkanoate synthase protein
subunit; wherein the
polyhydroxyalkanoate synthase protein subunit comprises an amino acid sequence
selected from
the group consisting o~ an amino acid sequence at least about 80% identical to
SEQ ID NO:11;
io and an amino acid sequence that is immunoreactive with an antibody prepared
using SEQ ID
NO:11 as an antigen, the antibody being immunoreactive with SEQ ID NO:11. The
polyhydroxyalkanoate synthase protein subunit is preferably at least about
82%, 84%, 86%,
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:11
A nucleic acid segment encoding a fusion protein may comprise a nucleic acid
sequence
is encoding a green fluorescent protein subunit; and a nucleic acid sequence
encoding a
polyhydroxyalkanoate synthase protein subunit; wherein the nucleic acid
sequence encoding a
polyhydroxyalkanoate synthase protein subunit is selected from the group
consisting of: a
nucleic acid sequence at least about 80% identical to SEQ ID NO:10; a nucleic
acid sequence
that hybridizes under stringent conditions to SEQ ID NO:10 or the complement
thereof; a nucleic
ao acid sequence encoding a protein at least about 80% identical to SEQ ID
NO:11; and a nucleic
acid sequence encoding a protein that is immunoreactive with an antibody
prepared using SEQ
ID NO:11 as an antigen, the antibody being immunoreactive with SEQ ID NO:11.
More
preferably, the nucleic acid sequence is at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:10. The nucleic acid
sequence may
zs be obtained from a natural source, may be mutagenized, may be genetically
engineered by
mutagenesis or other methods, or may be synthetic. The nucleic acid sequence
preferably
encodes a protein subunit at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%,
96%, 98%,
99%, 99.5%, or 100% identical to SEQ ID NO:11.
PHA biosynthesis methods: phaB and phaC
3o A method for the preparation of polyhydroxyalkanoate may comprise: a)
obtaining a cell
comprising: a nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein; and a nucleic
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-33-
acid sequence encoding a PHA synthase protein; wherein: the nucleic acid
sequence encoding a
3-keto-acyl-CoA reductase protein is not naturally found in the cell; the
nucleic acid sequence
encoding a PHA synthase protein is not naturally found in the cell; the
nucleic acid sequence
encoding a 3-keto-acyl-CoA reductase protein is selected from the group
consisting of: a nucleic
s acid sequence at least about 80% identical to SEQ ID N0:8; a nucleic acid
sequence that
hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic acid
sequence encoding a protein at least about 80% identical to SEQ ID N0:9; and a
nucleic acid
sequence encoding a protein that is immunoreactive with an antibody prepared
using SEQ ID
N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9; and
the nucleic acid
io sequence encoding a PHA synthase protein is selected from the group
consisting of: a nucleic
acid sequence at least about 80% identical to SEQ ID NO:10; a nucleic acid
sequence that
hybridizes under stringent conditions to SEQ ID NO:10 or the complement
thereof; a nucleic
acid sequence encoding a protein at least about 80% identical to SEQ ID NO:11;
and a nucleic
acid sequence encoding a protein that is immunoreactive with an antibody
prepared using SEQ
i s ID NO:11 as an antigen, the antibody being immunoreactive with SEQ ID
NO:11; and b)
culturing the cell under conditions suitable for the preparation of
polyhydroxyalkanoate. The
nucleic acid sequence encoding a 3-keto-acyl-CoA reductase protein more
preferably is at least
about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical to
SEQ ID N0:8. The nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein may be
Zo obtained from a natural source, may be mutagenized, may be genetically
engineered by
mutagenesis or other methods, or may be synthetic. The nucleic acid sequence
encoding a 3-
keto-acyl-CoA reductase protein preferably encodes a protein at least about
82%, 84%, 86%,
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:9.
The nucleic
acid sequence encoding a PHA synthase protein more preferably is at least
about 82%, 84%,
is 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
NO:10. The
nucleic acid sequence encoding a PHA synthase protein may be obtained from a
natural source,
may be mutagenized, may be genetically engineered by mutagenesis or other
methods, or may be
synthetic. The nucleic acid sequence encoding a PHA synthase protein
preferably encodes a
protein at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%,
99.5%, or 100%
3o identical to SEQ ID NO:11. The cell may generally be any cell, and
preferably is a bacterial,
fungal, mammalian, or plant cell. The bacterial cell is preferably an
Escherichia coli, Bacillus,
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-34-
Pseudomonas, or Ralstonia eutropha cell. The fungal cell is preferably a
Saccharomyces
cerevisiae or Schizosaccharomyces pombe cell. The plant cell is preferably a
tobacco, wheat,
potato, Arabidopsis, and high oil seed plants such as corn, soybean, canola,
oil seed rape,
sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa cell.
The
s polyhydroxyalkanoate may be a homopolymer or copolymer. The
polyhydroxyalkanoate may be
a polyhydroxybutyrate, polyhydroxyvalerate, polyhydroxyhexanoate,
polyhydroxyoctanoate,
polyhydroxydecanoate, or copolymers thereof.
A method for the preparation of polyhydroxyalkanoate may comprise: a)
obtaining a
plant comprising: a nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein; and a
to nucleic acid sequence encoding a PHA synthase protein; wherein: the nucleic
acid sequence
encoding a 3-keto-acyl-CoA reductase protein is not naturally found in the
plant; the nucleic acid
sequence encoding a PHA synthase protein is not naturally found in the plant;
the nucleic acid
sequence encoding a 3-keto-acyl-CoA reductase protein is selected from the
group consisting of:
a nucleic acid sequence at least about 80% identical to SEQ ID N0:8; a nucleic
acid sequence
is that hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic
acid sequence encoding a protein at least about 80% identical to SEQ ID N0:9;
and a nucleic
acid sequence encoding a protein that is immunoreactive with an antibody
prepared using SEQ
ID N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9; and
the nucleic
acid sequence encoding a PHA synthase protein is selected from the group
consisting of: a
2o nucleic acid sequence at least about 80% identical to SEQ ID NO:10; a
nucleic acid sequence
that hybridizes under stringent conditions to SEQ ID NO:10 or the complement
thereof; a nucleic
acid sequence encoding a protein at least about 80% identical to SEQ ID NO:1
l; and a nucleic
acid sequence encoding a protein that is immunoreactive with an antibody
prepared using SEQ
ID NO:11 as an antigen, the antibody being immunoreactive with SEQ ID NO:11;
and b)
zs growing the plant under conditions suitable for the preparation of
polyhydroxyalkanoate. The
nucleic acid sequence encoding a 3-keto-acyl-CoA reductase protein more
preferably is at least
about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical to
SEQ ID N0:8. The nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein may be
obtained from a natural source, may be mutagenized, may be genetically
engineered by
3o mutagenesis or other methods, or may be synthetic. The nucleic acid
sequence encoding a 3-
keto-acyl-CoA reductase protein preferably encodes a protein at least about
82%, 84%, 86%,
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-35-
88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID N0:9.
The nucleic
acid sequence encoding a PHA synthase protein more preferably is at least
about 82%, 84%,
86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID
NO:10. The
nucleic acid sequence encoding a PHA synthase protein may be obtained from a
natural source,
s may be mutagenized, may be genetically engineered by mutagenesis or other
methods, or may be
synthetic. The nucleic acid sequence encoding a PHA synthase protein
preferably encodes a
protein at least about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%,
99.5%, or 100%
identical to SEQ ID NO:11. The plant is preferably a tobacco, wheat, potato,
Arabidopsis, and
high oil seed plants such as corn, soybean, canola, oil seed rape, sugarbeet,
sunflower, flax,
io peanut, sugarcane, switchgrass, or alfalfa plant. The polyhydroxyalkanoate
may be a
homopolymer or copolymer. The polyhydroxyalkanoate may be a
polyhydroxybutyrate,
polyhydroxyvalerate, polyhydroxyhexanoate; polyhydroxyoctanoate,
polyhydroxydecanoate, or
copolymers thereof.
PHA biosynthesis methods: ~haB
is A method for the preparation of polyhydroxyalkanoate may comprise: a)
obtaining a cell
comprising: a nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein; and a nucleic
acid sequence encoding a PHA synthase protein; wherein: the nucleic acid
sequence encoding a
3-keto-acyl-CoA reductase protein is not naturally found in the cell; the
nucleic acid sequence
encoding a 3-keto-acyl-CoA reductase protein is selected from the group
consisting o~ a nucleic
zo acid sequence at least about 80% identical to SEQ ID N0:8; a nucleic acid
sequence that
hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic acid
sequence encoding a protein at least about 80% identical to SEQ ID N0:9; and a
nucleic acid
sequence encoding a protein that is immunoreactive with an antibody prepared
using SEQ ID
N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9; and b)
culturing the
Zs cell under conditions suitable for the preparation of polyhydroxyalkanoate.
The nucleic acid
sequence encoding a 3-keto-acyl-CoA reductase protein more preferably is at
least about 82%,
84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ
ID N0:8.
The nucleic acid sequence encoding a 3-keto-acyl-CoA reductase protein may be
obtained from
a natural source, may be mutagenized, may be genetically engineered by
mutagenesis or other
3o methods, or may be synthetic. The nucleic acid sequence encoding a 3-keto-
acyl-CoA reductase
protein preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%, 96%,
CA 02363803 2001-07-05
WO 00/40730 PCT/IJS00/00364
-36-
98%, 99%, 99.5%, or 100% identical to SEQ ID N0:9. The cell may generally be
any cell, and
preferably is a bacterial, fungal, mammalian, or plant cell. The bacterial
cell is preferably an
Escherichia coli, Bacillus, Pseudomonas, or Ralstonia eutropha cell. The
fungal cell is
preferably a Saccharomyces cerevisiae or Schizosaccharomyces pombe cell. The
plant cell is
s preferably a tobacco, wheat, potato, Arabidopsis, and high oil seed plants
such as corn, soybean,
canola, oil seed rape, sugarbeet, sunflower, flax, peanut, sugarcane,
switchgrass, or alfalfa cell.
The polyhydroxyalkanoate may be a homopolymer or copolymer. The
polyhydroxyalkanoate
may be a polyhydroxybutyrate, polyhydroxyvalerate, polyhydroxyhexanoate,
polyhydroxyoctanoate, polyhydroxydecanoate, or copolymers thereof.
io A method for the preparation of polyhydroxyalkanoate may comprise: a)
obtaining a
plant comprising: a nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein; and a
nucleic acid sequence encoding a PHA synthase protein; wherein: the nucleic
acid sequence
encoding a 3-keto-acyl-CoA reductase protein is not naturally found in the
plant; the nucleic acid
sequence encoding a 3-keto-acyl-CoA reductase protein is selected from the
group consisting of:
is a nucleic acid sequence at least about 80% identical to SEQ ID N0:8; a
nucleic acid sequence
that hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic
acid sequence encoding a protein at least about 80% identical to SEQ ID N0:9;
and a nucleic
acid sequence encoding a protein that is immunoreactive with an antibody
prepared using SEQ
ID N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9; and
b) growing
Zo the plant under conditions suitable for the preparation of
polyhydroxyalkanoate. The nucleic acid
sequence encoding a 3-keto-acyl-CoA reductase protein more preferably is at
least about 82%,
84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ
ID N0:8.
The nucleic acid sequence encoding a 3-keto-acyl-CoA reductase protein may be
obtained from
a natural source, may be mutagenized, may be genetically engineered by
mutagenesis or other
Zs methods, or may be synthetic. The nucleic acid sequence encoding a 3-keto-
acyl-CoA reductase
protein preferably encodes a protein at least about 82%, 84%, 86%, 88%, 90%,
92%, 94%, 96%,
98%, 99%, 99.5%, or 100% identical to SEQ ID N0:9. The plant may generally be
any plant,
and preferably is a tobacco, wheat, potato, Arabidopsis, high oil seed plants
such as corn,
soybean, canola, oil seed rape, sugarbeet, sunflower, flax, peanut, sugarcane,
switchgrass, or
3o alfalfa plant. The polyhydroxyalkanoate may be a homopolymer or copolymer.
The
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-37-
polyhydroxyalkanoate may be a polyhydroxybutyrate, polyhydroxyvalerate,
polyhydroxyhexanoate, polyhydroxyoctanoate, polyhydroxydecanoate, or
copolymers thereof.
PHA biosynthesis methods: phaC
A method for the preparation of polyhydroxyalkanoate may comprise: a)
obtaining a cell
s comprising: a nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein; and a nucleic
acid sequence encoding a PHA synthase protein; wherein: the nucleic acid
sequence encoding a
PHA synthase protein is not naturally found in the cell; the nucleic acid
sequence encoding a
PHA synthase protein is selected from the group consisting of: a nucleic acid
sequence at least
about 80% identical to SEQ ID NO:10; a nucleic acid sequence that hybridizes
under stringent
io conditions to SEQ ID NO:10 or the complement thereof; a nucleic acid
sequence encoding a
protein at least about 80% identical to SEQ ID NO:11; and a nucleic acid
sequence encoding a
protein that is immunoreactive with an antibody prepared using SEQ ID NO:11 as
an antigen, the
antibody being immunoreactive with SEQ ID NO:11; and b) culturing the cell
under conditions
suitable for the preparation of polyhydroxyalkanoate. The nucleic acid
sequence encoding a
is PHA synthase protein more preferably is at least about 82%, 84%, 86%, 88%,
90%, 92%, 94%,
96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:10. The nucleic acid
sequence
encoding a PHA synthase protein may be obtained from a natural source, may be
mutagenized,
may be genetically engineered by mutagenesis or other methods, or may be
synthetic. The
nucleic acid sequence encoding a PHA synthase protein preferably encodes a
protein at least
Zo about 82%, 84%, 86%, 88%, 90%, 92%, 94%, 96%, 98%, 99%, 99.5%, or 100%
identical to
SEQ ID NO:11. The cell may generally be any cell, and preferably is a
bacterial, fungal,
mammalian, or plant cell. The bacterial cell is preferably an Escherichia
coli, Bacillus,
Pseudomonas, or Ralstonia eutropha cell. The fungal cell is preferably a
Saccharomyces
cerevisiae or Schizosaccharomyces pombe cell. The plant cell is preferably a
tobacco, wheat,
Zs potato, Arabidopsis, and high oil seed plants such as corn, soybean,
canola, oil seed rape,
sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa cell.
The
polyhydroxyalkanoate may be a homopolymer or copolymer. The
polyhydroxyalkanoate may be
a polyhydroxybutyrate, polyhydroxyvalerate, polyhydroxyhexanoate,
polyhydroxyoctanoate,
polyhydroxydecanoate, or copolymers thereof.
3o A method for the preparation of polyhydroxyalkanoate may comprise: a)
obtaining a
plant comprising: a nucleic acid sequence encoding a 3-keto-acyl-CoA reductase
protein; and a
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-38-
nucleic acid sequence encoding a PHA synthase protein; wherein: the nucleic
acid sequence
encoding a PHA synthase protein is not naturally found in the plant; the
nucleic acid sequence
encoding a PHA synthase protein is selected from the group consisting of: a
nucleic acid
sequence at least about 80% identical to SEQ ID NO:10; a nucleic acid sequence
that hybridizes
s under stringent conditions to SEQ ID NO:10 or the complement thereof; a
nucleic acid sequence
encoding a protein at least about 80% identical to SEQ ID NO:11; and a nucleic
acid sequence
encoding a protein that is immunoreactive with an antibody prepared using SEQ
ID NO:11 as an
antigen, the antibody being immunoreactive with SEQ ID NO:11; and b) growing
the plant under
conditions suitable for the preparation of polyhydroxyalkanoate. The nucleic
acid sequence
~o encoding a PHA synthase protein more preferably is at least about 82%, 84%,
86%, 88%, 90%,
92%, 94%, 96%, 98%, 99%, 99.5%, or 100% identical to SEQ ID NO:10. The nucleic
acid
sequence encoding a PHA synthase protein may be obtained from a natural
source, may be
mutagenized, may be genetically engineered by mutagenesis or other methods, or
may be
synthetic. The nucleic acid sequence encoding a PHA synthase protein
preferably encodes a
is protein at least about 82% 84% 86% 88% 90% 92% 94% 96% 98% 99% 99.5% or
100%
> > > > > > > > > > >
identical to SEQ ID NO:11. The plant may generally be any plant, and
preferably is a tobacco,
wheat, potato, Arabidopsis, high oil seed plants such as corn, soybean,
canola, oil seed rape,
sugarbeet, sunflower, flax, peanut, sugarcane, switchgrass, or alfalfa plant.
The
polyhydroxyalkanoate may be a homopolymer or copolymer. The
polyhydroxyalkanoate may be
zo a polyhydroxybutyrate, polyhydroxyvalerate, polyhydroxyhexanoate,
polyhydroxyoctanoate,
polyhydroxydecanoate, or copolymers thereof.
Methods for preparing higher polyhydroxyalkanoates
Polyhydroxyalkanoate may be prepared by a method comprising: a) obtaining a
recombinant host cell comprising: a nucleic acid sequence encoding a (3-
ketothiolase protein; a
zs nucleic acid sequence encoding a 3-ketoacyl-CoA reductase protein; a
nucleic acid sequence
encoding a polyhydroxyalkanoate synthase protein; a nucleic acid sequence
encoding a (3-
hydroxyacyl-CoA dehydrase; and a nucleic acid sequence encoding an acyl-CoA
dehydrogenase
protein or an enoyl-CoA reductase protein; and b) culturing the recombinant
host cell under
conditions suitable for the preparation of polyhydroxyalkanoate; wherein: the
3o polyhydroxyalkanoate comprises C6, C8, or C10 monomer subunits; the nucleic
acid sequence
encoding a 3-keto-acyl-CoA reductase protein is selected from the group
consisting of: a nucleic
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-39-
acid sequence at least about 80% identical to SEQ ID N0:8; a nucleic acid
sequence that
hybridizes under stringent conditions to SEQ ID N0:8 or the complement
thereof; a nucleic acid
sequence encoding a protein at least about 80% identical to SEQ ID N0:9; and a
nucleic acid
sequence encoding a protein that is immunoreactive with an antibody prepared
using SEQ ID
s N0:9 as an antigen, the antibody being immunoreactive with SEQ ID N0:9.
Primers, probes, and antibodies
The sequences disclosed in the sequence listing may also be used to prepare
primers,
probes, and monoclonal or polyclonal antibodies.
SEQ ID NOS:1, 2, 4, 6, 8, 10, 22, 24, 26, and 28, and the their complementary
strands
io may be used to design oligonucleotide primers and probes. Primers and
probes are typically at
least 15 nucleotides in length, and more preferably are at least 20, 22, 24,
26, 28, 30, 40, or 50
nucleotides in length. Contiguous nucleotide sequences from a given sequence
are chosen based
upon favorable hybridization conditions, including minimization of hairpin or
other detrimental
sequences. The identification of suitable primer or probe sequences is well
known to those of
is skill in the art, and is facilitated by commercially available software
such as MacVector (Oxford
Molecular Group) and Xprimer (http://alces.med.umn.edu/rawprimer.html).
Primers and probes
may be used for the screening of libraries, for PCR amplification, and other
routine molecular
biological applications. Primers and probes may also be used for antisense
applications.
SEQ ID NOS:3, 5, 7, 9, 11, 23, 25, 27, and 29 may be used for the generation
of
Zo monoclonal or polyclonal antibodies. The entire sequences may be used, or
antigenic fragments
thereof. Alternatively, portions of the full length sequences may be
synthesized and covalently
attached to antigenic proteins such as keyhole limpet hemocyanin (KLH).
Portions of the full
length sequences may be used for the preparation of multi-antigenic peptides
(52). The
generation of monoclonal and polyclonal antibodies is well known to those of
skill in the art.
Zs The following Examples are included to demonstrate preferred embodiments of
the
invention. It should be appreciated by those of skill in the art that the
techniques disclosed in the
examples which follow represent techniques discovered by the inventors to
function well in the
practice of the invention, and thus can be considered to constitute preferred
modes for its
practice. However, those of skill in the art should, in light of the present
disclosure, appreciate
so that many changes can be made in the specific embodiments which are
disclosed and still obtain
a like or similar result without departing from the spirit and scope of the
invention.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-40-
EXAMPLES
Example 1: Bacterial strains and plasmids
Table 1. Strains
Strains Relevant characteristicsa Source
or
Reference
E. coli deoR endAl gyrA96 hsdRl7 (r,; mk*) recAl relAlClontech
DHSa supE44 thi-1
A~'(lacZYA-argFY169) ~80lacZ~EMIS F-~,-. Cloning
host and
for expression of pha genes
B. Wild type, used to clone pha genes ATCC
megaterium
11561
B. phaP, -Q, -R, -B and -C deletion derivative This
of B. megaterium
megaterium 11561 Applicatio
PHA05 n
P. PHA positive control ATCC
oleovorans
29347
P. putida PHA negative mutant obtained by chemical mutagenesis(22)
GPp 104
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-41 -
Table 2. Plasmids
Plasmids Relevant characteristicsa Source
or
Referenc
a
pBluescriptIISCloning vector, ColEl oriV, Amp' Stratagen
K a
pGFPuv Source of gfp gene, ColEl oriV, Amp' Clontech
pHPS9 Bacillus-Escherichia coli shuttle vector, ColEl(16)
and pTA1060
oriV, Em', Cm'
pSUP104 Pseudomonas-Escherichia coli shuttle vector, (40)
Q-type and minil5
oriV, Em', Tc'
pGM 1 EcoRI in phaP to HindIII in phaC, cloned into This
the EcoRI-HindIII
sites of pBluescriptIISK, Amp' applicatio
n
pGM6 PstI in phaB to EcoRI in ykrM, cloned into This
the PstI-EcoRI sites of
pBluescriptIISK, Amp' applicatio
n
pGM7 EcoRI in phaP to EcoRI in ykrM, cloned into This
the EcoRI site of
pBluescriptIISK, Amp' applicatio
n
pGM9 HindIII upstream of ykoY to PstI in phaB, clonedThis
into the HindIII -
PstI sites of pBluescriptIISK, Amp' applicatio
n
pGMlO HindIII upstream of ykoYto EcoRI in ykrM, clonedThis
into the
HindIII -EcoRI sites of pBluescriptIISK, Amp' applicatio
n
pGM7H EcoRI in phaP to EcoRI in ykrM, cloned into This
the EcoRI site of
pHPS9, Cm' applicatio
n
pC/GFP2 PhaC::GFP out-of frame fusion plasmid. This
Fragment shown in Figure 4A cloned in pBluescriptIISK,applicatio
Amp'
n
pC/GFP3 PhaC::GFP in-frame fusion plasmid. This
Fragment shown in Figure 4B cloned in pBluescriptIISK,applicatio
Amp'
n
pGMl3 PhaC::GFP in-frame fusion plasmid. This
Fragment shown in Figure 4C cloned in pHPS9, applicatio
Em'Lm'
n
pGMl3C GFP localization control plasmid. Part of phaBThis
and phaC deleted.
Fragment shown in Figure 4D cloned in pHPS9, applicatio
Em'Lm' '
n
pP/GFP3 PhaP::GFP in-frame fusion plasmid. This
Fragment shown in Figure 4E cloned in pBluescriptIISK,applicatio
Amp'
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-42-
n
pGMl6.2 PhaP::GFP in-frame fusion plasmid. This
Fragment shown in Figure 4F cloned in pHPS9, applicatio
EmrLmr
n
pGM107 EcoRI in phaP to EcoRI in ykrM, cloned as a This
BamHI-SaII
fragment from pGM7, into the BamHI and SaII applicatio
sites of
pSUP104, Cm~ n
pDRl PstI in phaB to EcoRI in ykrM, cloned as a This
SmaI-EcoRV fragment
from pGM6 into the two DraI sites of pSUP 104 applicatio
in same
orientation as the Cm gene, with phaC expressedn
from the Cm
promoter, Tcr
pGM61 Derived from pGMl3. It carries an in-frame This
594 by deletion in
phaR, extending from 96 by upstream of the Applicati
phaR initiation
codon through codon 144. on
pGM73 Derived from pGM61. Carries a transcriptional This
fusion between
the promoter of phaP and the coding region Applicati
plus translation
signals of phaR. A 663 by DNA fragment harboringon
phaR was
cloned into the SnaBI site in phaP in the sense
orientation.
"P;m', erythromycin resistant; Lm', lincomycin resistant; Cm', chlorampheW col
resistant; Amp',
ampicillin resistant; Tc', Tetracycline resistant. bATCC, American Type
Culture Collection.
Origin of replication.
Example 2: Media and growth conditions
s Cultures were grown at 37°C (unless otherwise stated) in liquid
media, aerated by
rotation at 250 rpm in either Luria-Bertani (LB) broth (33) or M9 Minimal
Salts (Life
Technologies, Bethesda, MD) with 1 % (w/v) glucose. For growth on plates, the
above media
with 1.5% agar (Sigma, A4550) was used. For plasmid selections, the
appropriate antibiotics
were included in the media: ampicillin (200 ~g/mL [AMPZOO]), chloramphenicol
(25 ~g/mL
io [CM25]), erythromycin (200 ~,g/mL [EM2°°]), or tetracycline
(12.~ ~g/mL [TC12~']) for plasmid
selection in Escherichia coli; chloramphenicol (12 ~g/mL [CM12]), or
erythromycin (1 ~g/mL
[EM1]) plus lincomycin (25 ~.g/mL [LM25]) for plasmid selection in Bacillus
megaterium;
chloramphenicol (160 ~g/mL [CMlbo]), or tetracycline (30 ~g/mL [TC3°])
for selection in
Pseudomonas.
1 s Example 3 : Transformations
Escherichia coli and Pseudomonas putida were transformed by electroporation of
competent cells using an electroporator (Eppendorfj and following the
manufacturers
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
- 43 -
instructions. Bacillus megaterium was transformed using a biolistic
transformation procedure
(39).
Example 4: Microsco~y-
For phase contrast microscopy, wet mounts of cultures were visualized at
x1,000
s magnification in a light microscope with phase contrast attachments
(Labophot-2 Microscope,
Nikon, Inc.). To view PHA inclusion-bodies, samples were heat fixed, stained
with 1 % (w/v)
Nile Blue A (Sigma) for 15 minutes at 55°C, destained for 30 seconds in
8% (v/v) acetic acid,
water washed, air dried, and viewed at x1000 magnification under fluorescence
using filters;
excitation, 446/10 nm; barrier filter, 590 nm; dichroic mirror, 580 nm. To
view GFP, wet
Io mounts of cultures with or without 1% (w/v) agarose were viewed at x1000
magnification under
fluorescence using filters; excitation, 390-450 nm; barrier filter, 480-520
nm; dichroic mirror,
470 nm.
Example 5: Codon usage in Bacillus megaterium
Bacillus megaterium uses three codons as start codons in protein coding
sequences.
~ s ATG, TTG, and GTG all encode methionine when present at the start of a
coding region. TTG
and GTG encode leucine and valine when present within a coding region,
respectively. Bacillus
megaterium uses TGA, TAA, and TAG as stop codons.
Bacillus megaterium sequences starting with TTG or GTG may require mutagenesis
to
ATG if the sequences are to be expressed in organisms that use ATG exclusively
as a start
Zo codon.
Example 6: Set~aration of nolvneptides associated with PHA inclusion-bodies.
In an attempt to determine their relevance, proteins that co-purify with PHA
inclusion-
bodies were separated by electrophoreses on an SDS-polyacrylamide gel (Figure
1 ).
Inclusion-bodies were purified (32) followed by suspension in TE buffer (10 mM
Tris-
Zs HCl pH 8, 1 mM EDTA) with 2% (w/v) SDS. An equal volume of 2x sample buffer
(100 mM
Tris-HCl (pH 6.8), 4% SDS, 4 mM EDTA, 20% glycerol, 2% 2-mercaptoethanol, 0.1%
bromophenol blue) was added prior to boiling for 5 minutes and samples were
centrifuged for 3
minutes to pellet PHA; the supernatant was loaded on a 12% SDS-polyacrylamide
gel and run at
8 mA overnight at 4°C to separate proteins. The gel was stained with
Coomassie Blue for 5
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-44-
minutes prior to transfer of proteins to a polyvinylidene difluoride membrane
using a semi-dry
electroblotter at 400 mA for 45 minutes.
There were at least thirteen such proteins present in various quantities. Some
or all of
these proteins could be intrinsic structural components of PHA inclusion-
bodies, enzymes
s involved with PHA metabolism or possibly scaffolding components involved in
inclusion-body
assembly. Alternatively, they could have been acquired by the inclusion-bodies
during the
purification procedure. The three most abundant proteins had molecular weights
of
approximately 14, 20 and 41 kDa.
The N-terminal amino acid sequence for the three most prevalent proteins were
io determined. Membrane carrying the proteins of interest was cut for use in N-
terminal amino acid
sequence determination by Edman Degradation using a minimum quantity of 200
pmols of each
protein. The N-terminal amino acid sequence of the 14 kDa protein was
KVFGRXELAAAMKRXGL (SEQ ID N0:19), the 20 kDa protein was
NTVKYXTVIXAMXXQ (SEQ ID N0:20), and the 41 kDa proteins was AIPYVQEXEKL
i s (SEQ ID N0:21 ). A BLASTp search (( 1 ), performed with NCBI Entrez
database;
http://www.ncbi.nlm.nih.gov/Entrezn revealed that the 14 kDa protein was
lysozyme and the
other two N-terminal sequences were novel. It was concluded that the lysozyme
used in the cell
lysis procedure had co-purified with the PHA inclusion bodies. This result
confirms that not
necessarily all of the proteins that co-purify with PHA inclusion-bodies are
associated with them
Zo in vivo, as was also shown for Chromatium vinosum (27).
Example 7: Cloning the pha re ion
Purification of genomic and plasmid DNA, Southern blot, hybridization and
cloning were
by standard procedures (38). To clone the DNA sequences that coded for the two
most abundant
proteins on purified PHA inclusion-bodies, degenerate oligonucleotide probes
based on their N-
Zs terminal amino acid sequences were used. The probes were:
AAYACRGTNAAATAYNNNACRGTNATYNNNGCDATGATG (n2, SEQ ID N0:12) and
GCDATYCCDTAYGTNCARGAAGGHTTYAAA (n5, SEQ ID N0:13) for the 20 kDa and 41
kDa proteins, respectively (Figure 1 ).
Both probes, used in separate 38°C Southern blotting hybridization
experiments,
3o identified a 6.4 kb HindIII, a 5.2 kb EcoRI, and a 3.7 kb HindIII to EcoRI
DNA fragment of
CA 02363803 2001-07-05
WO 00/40730 PCT/LTS00/00364
- 45 -
DNA, indicating that the 5' ends of the coding regions for both of these
proteins were located
less than 3.7 kb apart in the genome. The three fragments were purified from
agarose following
electrophoresis, and cloned into plasmid pBluescriptIISK.
Positive clones were identified by hybridization to the same degenerate
probes, thus
s yielding plasmid pGMl containing the 3.7 kb fragment. Sequences contiguous
with and
overlapping this primary cloned fragment were cloned in a similar manner
except that probes
based on the ends of the sequenced DNA fragment were used, and hybridization
was performed
at 55°C. The probes used were GCTTCATGCGTGCGGTTTG (bmp, SEQ ID N0:14)
and
GGACCGTTCGGAAAATCAGCGG (bmc, SEQ ID NO:15), yielding respectively, pGM9 and
io pGM6 (Figure 2).
DNA fragments of pGMl, pGM6 and pGM9 were subcloned into pBluescriptIISK, and
sequenced, from both ends using universal primers and internally by primer
walking on both
strands, using dye terminator chemistry, cycle sequencing and an ABI Prism 377
sequencer
(Applied Biosystems). Sequence assembly and analysis was performed using
Lasergene
Is (DNAStar, Inc.), and Gapped BLAST and PSI BLAST (1).
The 3.7 kb fragment contained 5 ORFs (Figure 2), whose predicted amino acid
sequences
encode PhaP (20 kDa protein), PhaQ, PhaR, PhaB and PhaC (41 kDa protein). The
20 and 41
kDa proteins were identified by their N-terminal amino acid sequences. Since
the C-terminus for
each of these two proteins extended beyond the boundaries of pGMI, the
remaining sequence
Zo were obtained from plasmids pGM6 and pGM9.
Example 8: The pha locus.
The 7,916 by region (SEQ ID NO:1) containing pha genes from Bacillus
megaterium
was cloned, sequenced and characterized. It was shown to carry 8 complete and
1 incomplete
open reading frame (Figure 2, Tables 3 and 4). Coding sequences in this region
were assigned
Zs on the basis of homology to known sequences, N-terminal amino acid
sequences, putative
ribosome binding sites and operon location. The complement and arrangement of
genes flanking
the pha genes in Bacillus megaterium are very similar to a region of Bacillus
subtilis 168 (Figure
2). This strain is negative for PHA and no known pha genes or sequences occur
in its genome,
for which the complete sequence is available (24). In place of pha genes in
this region of B.
CA 02363803 2001-07-05
WO 00/40730 PCT/LTS00/00364
-46-
subtilis are ykrl, ykrK and ykrL, which, respectively, code for putative
proteins similar to two
unknown proteins, and a probable heat shock protein.
Table 3: Sequence analysis results
Sequence Number of amino Mol mass DaltonsIsoelectric
acids point
ykoY 271 29,996 6.89
ykoZ 236 27,662 9.36
sspD 65 7,027 8.58
phaP 170 19,906 5.29
phaQ 146 16,686 5.09
phaR 168 19,150 5.10
phaB 247 26,098 7.39
phaC 362 41,463 8.31
-
--
ykrM 318a ND ND
aPartial protein.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-47-
Table 4: Sequence homologies
Sequen Homologies to known IdentitSimilaritFunction or putative
and function
ce putative genes (accessiony y
n0.) a
ykoY YkoY, B. subtilis 64% 73% Toxic anion resistance
(Z99110) protein (24)
ykoZ YkoZ, B. subtilis 57% 74% RNA polymerase sigma
(Z99111 ) factor (24)
sspD SspD, Bacillus 100% Spore specific, DNA
binding
megaterium (P10572) protein (4, 10)
SspD, B. subtilis 73% 87%
(P04833)
phaP None PHA inclusion-body
structure, shape and
size (49)
phaQ None Unknown
phaR None Unknown
phaB FabG, Synechocystis 50% 66% Fatty acid biosynthesis
(23)
(D90907) 48% 64% 3-ketoacyl-CoA reductase
PhaB, C. vinosum 47% 67% (28)
D
(P45375) Fatty acid biosynthesis
(35)
FabG, B. subtilis
(P51831)
phaC PhaC, T violacea 38% 59% PHA synthase (29,
23, 28)
(P45366) 37% 56%
PhaC, Synechocystis 35% 55%
(D90906)
PhaC, C. vinosum
(P45370)
ykrM YkrM, B. subtilis 55% 71% Na'~-transporting
ATP
(Z99111 ) synthase (24)
°Accession numbers are SWISS-PROT, EMBL or DDBJ; None, No discernible
similarity to
known sequences.
Example 9: The pha nucleic acid and encoded protein sequences
s The deduced amino acid sequence of PhaP shows a 20 kDa extremely hydrophilic
product with no obvious similarity to known sequences (Figure 6). Inclusion-
body associated
low molecular weight proteins (phasins) have been described in many bacteria
(49), but where
sequences were available no similarities of identifiable significance with
PhaP of Bacillus
megaterium were found.
io Low molecular weight, PHA inclusion-body abundant proteins play an
important role in
PHA producing cells, since they are involved in determining inclusion-body
size and shape, and
are present in quantities up to 5% of total protein in the case of PHA
producing A. eutrophus
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-48-
(48). It is an interesting observation that the amino acid sequences of phasin
proteins are so
dissimilar, even in closely related bacteria. Some similarity between such
proteins would be
expected in closely related bacteria, were they to have a role in inclusion-
body biogenesis,
however, conservation of sequence would be entirely unnecessary should they
have a role as
s storage proteins.
The deduced amino acid sequences of PhaQ and PhaR also revealed small
hydrophilic
proteins with no significant identifiable similarity to known proteins
(Figures 7 and 8). Figure 1
(lane 2) shows that purified inclusion-bodies have proteins represented by
bands of the
approximate sizes of PhaQ (17 kDa) and PhaR (19 kDa), but the roles of these
proteins are
to unknown. They may be non-orthologous replacements for the small putative
gene products,
whose roles are also unknown, coded in known pha gene clusters. The deduced
amino acid
sequence of PhaB, is similar in size and amino acid sequence to known phaB and
fabG gene
products (Table 2). The deduced amino acid sequence of PhaC shows that while
it has low
homology overall to known PhaC proteins, it is most similar to that of T.
violacea, Synechocystis
is and C. vinosum. PhaC proteins from these three bacterial strains,
respectively, have 355, 378,
and 355 amino acids while PhaC from Bacillus megaterium has 362 amino acids.
All other
PhaC proteins studied are larger in size, and range from 559 amino acids for
that of P.
oleovorans (22) to 636 amino acids for that of Rhizobium etli (3). Alignment
studies of
sequences of all previously known PhaC proteins show that the synthases are
either large single
zo subunit enzymes (PhaC) or smaller two subunit enzymes (PhaC and PhaE). The
Bacillus
megaterium PhaC protein aligns poorly with large, single subunit enzymes such
as the P.
oleovorans PhaC (Figure 3).
Example 10: Functionality of the pha gene cluster
It has been demonstrated that the phaP, -Q, -R, -B and -C gene cluster can
complement a
zs deletion mutant of B. megaterium. This mutant PHA05 was constructed by a
gene substitution
technique. A plasmid (based on pGMlO) in which the pha genes were substituted
by the
erythromycin gene, was propagated in B. megaterium 11561. Selection on
erythromycin allowed
isolation of the PHA05 mutant that was negative for PHA synthesis.
Complementation with the
phaP, -Q, -R, -B and -C gene cluster was obtained when pGM7H or pGMl3 was
introduced into
30 the PHA05 strain.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-49-
Experiments introducing a phaR deletion of pGM 13 (pGM61 ) into PHA05 suggests
that
the presence of phaR may be preferred for PHA synthesis. This result was
confirmed by the
recloning of phaR into pGM61 (pGM73) as it was isolated from PHA05(pGM61)
strain,
followed by the introduction of pGM73 into PHA05. Accumulation of PHA in
PHA05(pGM73)
s confirmed the preference for phaR. It has been previously demonstrated that
the small type
PhaCs (see Example 17) is not sufficient for PHA synthesis; another peptide,
PhaE of
approximate size 30 kDa, is also required (51). These complementation studies
suggest that it is
preferable to combine PhaC of B. megaterium (also a small type PhaC) and phaR
(19 kDa),
however there is no sequence similarity between phaR of B. megaterium and phaE
of other
io organisms.
Example 11: Mapp~ transcription starts
The transcription start points were mapped in the region from the EcoRl
restriction site in
phaP to the HindIII site in ykrM by primer extension analysis, using the
Promega system for
primer extension on RNA templates. DNA oligonucleotide primers, 17 to 20
nucleotides in
is length, were synthesized to match target sequences, initially at
approximately 500 base pair
intervals and subsequently at about 50 to 250 nucleotides down-stream from the
predicted
transcription start points. The 32P 5' end-labeled primers were extended with
reverse
transcriptase using total RNA (10 ~g per reaction) purified from Bacillus
megaterium (31). The
fragment length initially, and transcription start nucleotides subsequently,
were determined by
zo running the cDNA on a 8% denaturing polyacrylamide gel along-side the
products of sequencing
reactions, which were generated using the same 5'-end labeled primers. The
primers used to
identify the transcription start nucleotides for the phaP, phaQ, and phaRBC
promoters were,
respectively, CCCCTTTGTCCATTGTTCCC (SEQ ID N0:16); CCATGTAGATTCCACCCTC
(SEQ ID N0:17); and CTCCATCTCCTTTCTTGTG (SEQ ID N0:18).
zs Primer extension products showed a single band from each reaction,
indicating one
transcript, while control reactions in which RNA was omitted showed no bands.
The extension
products run alongside sequencing reaction products obtained with the same
primer (Figure 2C),
identified the 5' ends of the transcripts thus allowing the putative promoter
sequences at
approximately -10 and -35 -by for phaP, -Q and -R to be identified. The
arrangement of genes in
3o the pha cluster of Bacillus megaterium is unique among those already
published and phaA is
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-50-
notably absent. The phaP, -Q, -R, -B and -C genes were shown to be in a 4,104 -
by region, with
phaP and -Q transcribed in one orientation, each from a separate promoter,
while phaR, -B and -
C were divergently transcribed from a promoter in front of phaR. The putative
promoters
responsible for transcription of phaQ and phaR, phaB and phaC show strong
similarity to both
s Bacillus subtilis Sigma A type (34) and Escherichia coli, Sigma 70 type
promoters (14), which
can express constitutively. This is in keeping with previous data for
Alcaligenes eutrophus
showing that phaC is constitutively synthesized, but PHA is not constitutively
accumulated (19).
The third putative promoter in this region, the phaP promoter, resembles a
Sigma D (SigD) type
promoter known to control the expression of a regulon of genes associated with
flagellar
io assembly, chemotaxis and motility (13, 20, 46). In Bacillus subtilis Sigma
D is expressed in the
exponential phase and peaks in late exponential phase of growth. This
parallels the pattern of
PHA accumulation previously described for Bacillus megaterium 11561 (32).
However, further
experiments are required to test the hypothesis that PHA accumulation in
regulated by sigma D
or products of its resulting transcripts. The phaP gene has 18 -by duplicate
sequences that could
~s base-pair to form a rho-independent terminator close to its translational
stop codon (Figure 2B).
The fact that the -35 promoter region of sspD is within this putative hairpin
structure, suggests
that transcription of phaP and sspD could be mutually exclusive, thus allowing
the expression of
phaP to play a regulatory role in the expression of sspD (spore specific
storage protein).
Example 12~ Expression of Bacillus megaterium pha genes in Escherichia coli
and Pseudomonas
2o up tidy
Functionality of the Bacillus megaterium putative pha gene cluster was tested
in
Escherichia coli, which is naturally PHA negative, and Pseudomonas putida
GPp104, a phaC-
mutant. Plasmids carrying one or more of these genes were introduced and the
resulting
transformants were tested for PHA accumulation following growth on LB or M9
medium with
is various carbon sources and the appropriate antibiotic for plasmid
selection.
Triplicate 500 mL cultures, were grown in 2 liter flasks at 30°C,
rotating at 250, using
1 % inocula of 16 hour cultures, which had been grown in LB, centrifuged and
resuspended in
equal volumes of 0.9% saline. At 48 hours samples were removed for microscopy
and cells were
harvested, washed once in dH20 and lyophilized. For PHA extraction,
lyophilized cells were
so suspended in 10 volumes of 5% (w/v) bleach, shaken at 65°C for 1
hour and centrifuged. The
pellet was resuspended in 10 volumes of 5% bleach and centrifuged followed by
sequentially
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-51 -
washing in water and 95% ethanol. The amount of PHA is expressed as percent
PHA per mass
of vacuum dried cells (w/w).
Escherichia coli carrying pGM7 or pGMlO accumulated low levels of PHA while
Escherichia coli carrying pGMI or pGM6 accumulated no PHA. Fluorescence
microscopy of
s Nile Blue A stained cells showed approximately 1 cell in 20 had one or
several inclusion-bodies
and the quantity of PHA produced was approximately 5% of cell dry weight.
Since Escherichia
coli does not have PhaA, a low level or no PHA is the expected result.
However, in
Pseudomonas where PhaA is not known to be required, Pseudomonas putida GPp104
(pGM107)
accumulated PHA on rich as well as minimal medium with various carbon sources
to >50% of
io cell dry weight, and 90 to 100% of cells appeared full of PHA (Table 5).
The positive control P.
oleovorans, (equivalent to wild-type Pseudomonas putida) accumulated PHA only
when grown
on longer chain carbon sources, and not on LB. No PHA was accumulated by the
negative
control or by Pseudomonas putida carrying phaC alone (pDRI ). These results
showed that this
Bacillus megaterium gene cluster is functional in both Escherichia coli and
Pseudomonas putida.
is It is not known if the negative results obtained with pDRI was due to PhaC
alone being
insufficient to complement PhaC- Pseudomonas putida or to synthesize PHA in
Escherichia coli,
or if the expression of phaC on pDRI was not successful in producing protein.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-52-
Table 5: Cells with PHA as a percents of total cells following growth on
different carbon
sources
Substrates Source Positive Negative phaP"QRBC phaC:
of
(no. C atoms)genes: control: control,
vector
only:
Bacillus P. PseudomonaPseudomona Pseudomona
megateriumoleovoranss putida s putida s putida
GPp 104 GPp 104 GPp 104
(pSUP 104)(pGM 107) (pDRI )
LB 100 0 0 90 0
LB/Glucose,100 0 0 92 0
1%
M9/Caproate,no growth 88 0 100* 0
12 mM (C6)
M9/Octanoatno growth 90 0 92 0
e, 12 mM
(C8)
100%, PHA in all cells; 0%, no PHA in any cell; data averaged from >5 fields
of each of 3
different cultures, error less than 5%. °N-terminus only present. *
Cell shape distorted by large
s quantity of PHA.
These results suggest that the B. megaterium gene cluster, phaP, -Q, -R, -B,
and -C, is
functional in both E. coli and P. putida in so far as accumulation of PHA
polymer. It is not
known if the negative results obtained with pDRl were due to PhaC alone being
insufficient to
complement the PhaC mutant of P. putida or to synthesize PHA in E. coli.
~o Example 13: Localization of PhaP and PhaC proteins
Proteins associated with purified PHA inclusion-bodies may not accurately
reflect the
localization of the these proteins within the growing cell. Visualization of
pha:: gfp gene product
fusion proteins in living cells throughout culture growth is a useful method
for determining both
the localization of the pha gene products and their comparative levels in
growing cells. PhaP
is and PhaC, as fusion proteins (Figure 4), localized to PHA inclusion-bodies
at all time points
tested throughout growth of Bacillus megaterium 11561. The negative control
(pHPS9) showed
no fluorescence at any time point. The localization control (pGMl3C) showed
non-localized
green fluorescence at all time points. The profiles of PHA accumulation in
these two control
strains were similar to that of the wild-type, where the quantity of PHA
decreased during the lag
ao phase, increased during exponential phase, and continued to increase at a
lower steady state rate
in stationary phase growth (32).
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-53-
At time 0, cultures of Bacillus megaterium carrying, pGM16.2, pGMl3, pGMl3C or
pHPS9, grown in LB with LM25 EMl for 24 hours at 35°C, were inoculated
(5% v/v) into 75 mL
of fresh media of the same composition, in 300 mL Naphelco flasks, and growth
was continued
at 27°C, 250 rpm. Optical densities of cultures were monitored and
samples were removed for
s microscopy at time points starting at time 0, for up to 24 hours. One part
of each sample was
immediately observed for green fluorescence by embedding in 1 % low melting
point agarose for
viewing in phase contrast and under fluorescence for GFP, magnification x1000.
Another part of
each sample was stained for PHA and viewed under light microscopy and by
fluorescence for
PHA inclusion bodies, magnification x1000. Images were recorded using
identical parameters
~o for all samples to allow comparison of fluorescence and light intensities
(f stop, 1/15; brightness,
0.6; sharpness, 1.0; contrast, 0.8; color, 0.3; see also methods and
materials). Results are shown
in Figure 5 (A-F).
PhaP, monitored as a PhaP::GFP fusion protein in pGM16.2 (Figures SA and SB),
decreased significantly during the first half (2 hours) of lag phase growth,
increased during late
is lag phase and early to mid-exponential phase, decreased in mid to late
exponential phase and
increased during stationary phase growth. A possible explanation for the rapid
decrease of PhaP
in lag phase is that PhaP may be a storage protein that is degraded as a
source of amino acids.
The profile of PHA accumulation in these cells (carrying pGM16.2) followed a
similar pattern to
that of PhaP except that PHA decreased only in the lag phase and continued to
accumulate
Zo throughout other phases of culture growth. This data is consistent with PHA
inclusion-bodies
being a source of carbon, reducing equivalents and amino acids when the
organism is first
provided with fresh medium. Possible explanations as to why the level of PhaP
and not PHA
decreased at mid to late exponential phase are that either PhaP was
synthesized at a slower rate
than that of PHA, or PhaP was used as a source of amino acids at this phase of
growth or both
Zs scenarios may apply.
PhaC, monitored as a PhaC::GFP fusion protein in pGMl3 showed a similar
profile of
expression to that of PhaP with one exception: PhaC did not reduce in level
during lag phase
growth. It did, however, reduce in level in mid to late exponential phase
growth, as did PhaP.
The profile of PHA accumulation in these cells carrying PhaC::GFP was similar
to that of cells
3o carrying PhaP::GFP, except that the PHA level did not reduce during lag
phase growth. The
increased quantity of PhaC in the cell is a likely explanation since PhaC
remained functional in
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-54-
the fusion protein PhaC::GFP. This was indicated by the fact that Escherichia
coli DHSa
(pC/GFP3) and Escherichia coli DHSa (pGM7) accumulated PHA to equivalent low
levels,
while the host strain alone, or carrying pGFPuv accumulated no PHA, as
visualized by
fluorescence microscopy of Nile Blue A stained cells. The reduction in level
of PhaC in mid to
s late exponential phase, as was also seen with PhaP, is consistent with both
PhaC and PhaP being
synthesized at a slower rate than that of PHA.
In cells of all growth phases, inclusion-bodies were rarely visible under
light in stained
heat fixed cells while larger inclusion-bodies were visible in phase contrast
of living cells (Figure
SC-F). In older cultures (2 days and older) some cells were lysed, and showed
PhaP::GFP and
Io PhaC::GFP localized to free PHA inclusion-bodies (Figure SD). Both free and
intracellular
inclusion-bodies had doughnut shaped localization of GFP at some focal planes
while at other
focal planes the same inclusion-bodies appeared completely covered in GFP. We
interpret this
data as a difference in quantity of GFP that is visible when viewed through
the edge or the center
of the inclusion-bodies.
is Example 14~ Analysis of Bacillus megaterium 3-ketoacyl-CoA reductase PhaB
Stereospecificity assays were conducted on the Bacillus megaterium reductase
using
various chain length enoyl-CoA esters (C4-C8, Table 6). The assay was done
using crotonase
from Sigma (L-hydroxy acids) or hydratase from Rhodosprillum rubrum (D-hydroxy
acids) to
form the 3-hydroxyacyl-CoA compounds from the enoyl-CoA esters. Acetoacetyl-
CoA reductase
Zo activity was monitored spectrophotometrically as the reduction of NADP+
while 3-hydroxyacyl-
CoAs were oxidized. Based on the assay results (Table 6) the Bacillus
megaterium reductase is a
D-specific enzyme with a preference for C6 carbon chains. Enzyme reactions
using NADH as
electron donor for 3-ketoacyl-CoA reduction did not indicate significant
enzyme activity with
this cofactor.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-55-
Table 6: Analysis for stereo-specificity of the Bacillus megaterium 3-ketoacyl-
CoA reductase.
Clone D-stereoisomerSpec. Clone L-stereoisomerSpec. act.
#a (hydratase)act. # (crotonase) U/mg
U/mg
B1-30 Crotonyl 0.155 B1-30 Crotonyl CoA 0.014
CoA
B1-30 C5 0.15 Bl-30 C5 0.009
B1-30 C6 0.39 B1-30 C6 0.017
B1-30 C8 0.014 B1-30 C8 0.039
B5-20 Crotonyl 0.077 B5-20 Crotonyl CoA 0.004
CoA
B5-20 C5 0.074 B5-20 C5 0.01
B5-20 C6 0.219 B5-20 C6 0.012
B5-20 C8 0.003 B5-20 C8 0.001
NegativeCrotonyl 0.02 NegativeCrotonyl CoA 0.001
CoA
NegativeCS 0.011 NegativeC5 0.003
NegativeC6 0.006 NegativeC6 0.008
NegativeC8 0.033 NegativeC8 0.003
Clone B1-30 contains pMUN48213; clone >35-~u contains pmmv4a~m.
Example 15~ Verification of the Bacillus megaterium 3-ketoacvl-CoA reductase
for PHA
an~mmWatW n
s The functionality of the Bacillus megaterium sequence for PHA accumulation
in a
recombinant system was assayed. Escherichia coli DHSa harboring either
pMON48222
(phaARe, phaBBm, phaCRe) only, or two of the following plasmids: pJM9238 DAB
(phaA and
phaB deleted by FseI digest and religation) or pJM9117 DAB (phaA and phaB
deleted by FseI
digest and religation) and pMON48220 (phaARe, phaBBm,) was grown in LB +
mannitol in
~o concentrations of 1 or 2 % (w/v), respectively. Cultures were induced for
PHA accumulation at
OD6oo = 0.6. Percentage PHA (Table 7) and enzyme activity (Table 8) were
determined.
Plasmid pMON48213 contains the same pha sequences as pMON48220, but was
constructed
with pSE380 (Invitrogen, Carlsbad, CA), a high level expression vector.
Plasmid pMON48221
contains the same pha sequences as pMON48220, but lacks a small fragment of
the multicloning
i s site between phaARe and phaBBm.
3-Ketoacyl-CoA reductase was monitored in a total volume of 1 mL containing
100 mM
potassium phosphate buffer pH 7.0, 50 ~.M acetoacetyl-CoA and 150 ~M NADPH.
The reaction
mixture contained between 5 and 50 pL cell extract. Assays were monitored
spectrophotometrically at 340 nm.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-56-
Table 7: Application of the Bacillus megaterium 3-ketoacyl-CoA reductase for
PHA formation in
Escherichia coli
Vectors % PHA Standard deviation
pMON48222-4 12.9
pMON48222-8 19.2
Average16.1 ~ 4.5
pJM9238 pMON 48220 23.7
4AB
pJM9238 pMON48220 18.9
DAB
Average21.3 ~ 3.4
pJM9238 Average1.5 ~ 1.5
DAB
pJM9117 pMON 48220 12.5
DAB
pJM9117 pMON48220 3.9
DAB
Average8.2 ~ 6.1
pJM9117 Average0.7 ~ 0.1
DAB
Table 8: Enzyme activity of the Bacillus megaterium 3-ketoacyl-CoA reductase
using
pMON48220 and pMON48213
Vector acetoacetyl-CoA reductase[U/mg]
Negative control 0.08
pMON48220-2 0.24
0.15
pMON48220-9 0.22
0.23
Average 0.21 0.04
pMON48213 4.0
Table 9: Verification of the Bacillus megaterium 3-ketoacyl-CoA reductase
functionality
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-57-
E. coli DHSa containing Relevant genotype PHB content %
plasmids CDW
pJM92380AB, pMON34610 phaCRe nd
pJM9238~AB, pMON34575 phaCRe, phaARe 1.2 ~ 0.4
pJM92380AB, pMON48221 phaCRe, phaARe, phaBB~"22.2 ~ 4.7
nd = not detectable
Example 16: Additional sequences in ~enomic fragment
The 7,916 base pair genomic fragment (SEQ ID NO: l ) additionally contained
three
complete open reading frames and one incomplete open reading frame encoding
proteins in
s addition to PhaP, PhaQ, PhaR, PhaB, and PhaC. As indicated in Tables 3 and
4, sequence
comparisons suggest that ykoY (SEQ ID N0:22) encodes toxic anion resistance
protein YkoY
(SEQ ID N0:23), ykoZ (SEQ ID N0:24) encodes RNA polymerase sigma factor
protein YkoZ
(SEQ ID N0:25), and ykrM (SEQ ID N0:26) encodes a portion of the Na+ -
transporting ATP
synthase protein YkrM (SEQ ID N0:27). Sequence sspD (SEQ ID N0:28) matches the
known
io Bacillus megaterium sequence (4, 10) encoding SspD (SEQ ID N0:29). While
the activity of
the proteins is identified by their similarity to other known proteins, it is
possible that the
proteins may have additional functionality involved in polyhydroxyalkanoate
biosynthesis.
These nucleic acid and amino acid sequences may be used in nucleic acid
segments,
recombinant vectors, transgenic host cells, and transgenic plants.
~ a Example 17: One and two subunit PHA synthase proteins
PHA synthases have been identified to be either one or two subunit enzymes
(51). Single
subunit enzymes have only the PhaC protein, while two subunit enzymes have
PhaC and PhaE
protein subunits. Nucleic acid sequences encoding PhaE subunits have been
found to be located
adjacent to the nucleic acid sequences encoding PhaC.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-58-
Table 10: One and two subunit PHA synthases
Source organism (Reference) Subunits PhaC Amino acids
T. violacea (P45366, D48376)2 355
C. vinosum (P45370, 529274) 2 355
T. pfennigii (WO 96/08566) 2 357
Synechocystis sp. PCC6803 2 378
(50,
D90906, 577327)
P. oleovorans (22, A38604) 1 559
P. aeruginosa (S29305) 1 559
R. Tuber (S25725) 1 562
R. eutropha (A34371 ) 1 589
A. caviae (D88825) 1 594
P. denitrificans (JC6023) 1 624
R. etli (3, U30612) 1 636
B. megaterium (SEQ ID NO:11) 362
Based on the number of amino acids in the deduced sequence and homology to
known
PhaC proteins, the B. megaterium would be expected to be part of a two subunit
synthase.
However, the nucleic acid sequences adjacent to phaC in the 7,916 base pair
genomic fragment
s show no significant similarity to a phaE sequence. Upstream of phaC is phaB,
and downstream
is ykrM, a suspected Na+ transporting ATP synthase (Table 4). In combination
with the
observation that the B. megaterium sequences were able to complement P. putida
GPp104 to
accumulate PHA, this suggests that the B. megaterium phaC may encode a novel
class of PHA
synthase, i.e. a single subunit synthase with a molecular weight in the range
of two subunit PhaC
to proteins.
Example 18: Pathway for the~roduction of C4/C6/C8/C 10 PHA copolymers
Figure 10 outlines a proposed biosynthetic pathway for the production of PHA
copolymers incorporating C4 and/or C6 monomer units. Produced polymers may
include C4-co-
C6, C4-co-C8, C4-co-C6-co-C8, C6-co-C8, C6, and C8. A recombinant host cell or
plant may
is be constructed to contain the nucleic acid sequences encoding the required
enzymes.
The ~3-ketothiolase is preferably BktB (53, WO 98/00557). The ~3-ketothiolase
can
condense two molecules of acetyl-CoA to form acetoacetyl-CoA. This product may
be reduced
to 3HB-CoA by the Bacillus megaterium 3-keto-acyl-CoA reductase protein. 3HB-
CoA may be
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-59-
converted to crotonyl-CoA by a hydratase such as that from Aeromonas caviae
(54). Subsequent
reduction to butyryl-CoA is performed by a butyryl-CoA dehydrogenase such as
that cloned
from Clostridium acetobutylicum (55). This product may be condensed with
acetyl-CoA by the
(3-ketothiolase to afford 3-ketohexanoyl-CoA. This is the preferred substrate
of the Bacillus
s megaterium reductase, leading to the production of 3-hydroxyhexanoyl-CoA.
This product may
be incorporated into C6 polymers or copolymers (e.g. C4-co-C6) by a PHA
synthase having a
broad substrate specificity (e.g. (56)). An additional round of condensation
may lead to
production of the C8 monomer, allowing the introduction of C8 into PHA
polymers or
copolymers. A further additional round of condensation may lead to production
of the C 10
~o monomer, allowing the introduction of C10 into PHA polymers or copolymers.
Example 19: Nucleic acid mutation and hybridization
Variations in the nucleic acid sequence encoding a protein may lead to mutant
protein
sequences that display equivalent or superior enzymatic characteristics when
compared to the
sequences disclosed herein. This invention accordingly encompasses nucleic
acid sequences
is which are similar to the sequences disclosed herein, protein sequences
which are similar to the
sequences disclosed herein, and the nucleic acid sequences that encode them.
Mutations may
include deletions, insertions, truncations, substitutions, fusions, shuffling
of subunit sequences,
and the like.
Mutations to a nucleic acid sequence may be introduced in either a specific or
random
Zo manner, both of which are well known to those of skill in the art of
molecular biology. A myriad
of site-directed mutagenesis techniques exist, typically using
oligonucleotides to introduce
mutations at specific locations in a nucleic acid sequence. Examples include
single strand rescue
(Kunkel, T. Proc. Natl. Acad. Sci. U.S.A., 82: 488-492, 1985), unique site
elimination (Deng and
Nickloff, Anal. Biochem. 200: 81, 1992), nick protection (Vandeyar, et al.
Gene 65: 129-133,
zs 1988), and PCR (Costa, et al. Methods Mol. Biol. 57: 31-44, 1996). Random
or non-specific
mutations may be generated by chemical agents (for a general review, see
Singer and Kusmierek,
Ann. Rev. Biochem. 52: 655-693, 1982) such as nitrosoguanidine (Cerda-Olmedo
et al., J. Mol.
Biol. 33: 705-719, 1968; Guerola, et al. Nature New Biol. 230: 122-125, 1971 )
and 2-
aminopurine (Rogan and Bessman, J. Bacteriol. 103: 622-633, 1970), or by
biological methods
3o such as passage through mutator strains (Greener et al. Mol. Biotechnol. 7:
189-195, 1997).
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-60-
Nucleic acid hybridization is a technique well known to those of skill in the
art of DNA
manipulation. The hybridization properties of a given pair of nucleic acids is
an indication of
their similarity or identity. Mutated nucleic acid sequences may be selected
for their similarity to
the disclosed nucleic acid sequences on the basis of their hybridization to
the disclosed
s sequences. Low stringency conditions may be used to select sequences with
multiple mutations.
One may wish to employ conditions such as about 0.15 M to about 0.9 M sodium
chloride, at
temperatures ranging from about 20°C to about 55°C. High
stringency conditions may be used
to select for nucleic acid sequences with higher degrees of identity to the
disclosed sequences.
Conditions employed may include about 0.02 M to about 0.15 M sodium chloride,
about 0.5% to
~o about 5% casein, about 0.02% SDS and/or about 0.1% N-laurylsarcosine, about
0.001 M to
about 0.03 M sodium citrate, at temperatures between about 50°C and
about 70°C. More
preferably, high stringency conditions are 0.02 M sodium chloride, 0.5%
casein, 0.02% SDS,
0.001 M sodium citrate, at a temperature of 50°C.
Example 20: Determination of homologous and degenerate nucleic acid seguences
~s Modification and changes may be made in the sequence of the proteins of the
present
invention and the nucleic acid segments which encode them and still obtain a
functional
molecule that encodes a protein with desirable properties. The following is a
discussion based
upon changing the amino acid sequence of a protein to create an equivalent, or
possibly an
improved, second-generation molecule. The amino acid changes may be achieved
by changing
zo the codons of the nucleic acid sequence, according to the codons given in
Table 11.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-61 -
Table 11: Codon degeneracies of amino acids
I Ammo acid One letterThree letter Codons
Alanine A Ala GCA GCC GCG GCT
Cysteine C Cys TGC TGT
Aspartic acidD Asp GAC GAT
Glutamic acidE Glu GAA GAG
PhenylalanineF Phe TTC TTT
Glycine G Gly GGA GGC GGG GGT
Histidine H His CAC CAT
Isoleucine I Ile ATA ATC ATT
Lysine K Lys AAA AAG
Leucine L Leu TTA TTG CTA
CTC
CTG
CTT
Methionine M Met ATG
Asparagine N Asn AAC AAT
Proline P Pro CCA CCC CCG CCT
Glutamine Q Gln CAA CAG
Arginine R Arg AGA AGG CGA CGC CGG CGT
Serine S Ser AGC AGT TCA TCC TCG TCT
Threonine T Thr ACA ACC ACG ACT
Valine V Val GTA GTC GTG
GTT
Tryptophan W Trp TGG
Tyrosine Y Tyr TAC TAT I
Certain amino acids may be substituted for other amino acids in a protein
sequence
without appreciable loss of enzymatic activity. It is thus contemplated that
various changes may
be made in the peptide sequences of the disclosed protein sequences, or their
corresponding
s nucleic acid sequences without appreciable loss of the biological activity.
In making such changes, the hydropathic index of amino acids may be
considered. The
importance of the hydropathic amino acid index in conferring interactive
biological function on a
protein is generally understood in the art (Kyte and Doolittle, J. Mol. Biol.,
157: 105-132, 1982).
It is accepted that the relative hydropathic character of the amino acid
contributes to the
io secondary structure of the resultant protein, which in turn defines the
interaction of the protein
with other molecules, for example, enzymes, substrates, receptors, DNA,
antibodies, antigens,
and the like.
Each amino acid has been assigned a hydropathic index on the basis of their
hydrophobicity and charge characteristics. These are: isoleucine (+4.5);
valine (+4.2); leucine
(+3.8); phenylalanine (+2.8); cysteine/cystine (+2.5); methionine (+1.9);
alanine (+1.8); glycine
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-62-
(-0.4); threonine (-0.7); serine (-0.8); tryptophan (-0.9); tyrosine (-1.3);
proline (-1.6); histidine (-
3.2); glutamate/glutamine/aspartate/asparagine (-3.5); lysine (-3.9); and
arginine (-4.5).
It is known in the art that certain amino acids may be substituted by other
amino acids
having a similar hydropathic index or score and still result in a protein with
similar biological
s activity, i.e., still obtain a biologically functional protein. In making
such changes, the
substitution of amino acids whose hydropathic indices are within ~2 is
preferred, those within ~1
are more preferred, and those within ~0.5 are most preferred.
It is also understood in the art that the substitution of like amino acids may
be made
effectively on the basis of hydrophilicity. U.S. Patent No. 4,554,101 (Hopp,
T.P., issued
io November 19, 1985) states that the greatest local average hydrophilicity of
a protein, as governed
by the hydrophilicity of its adjacent amino acids, correlates with a
biological property of the
protein. The following hydrophilicity values have been assigned to amino
acids: arginine/lysine
(+3.0); aspartate/glutamate (+3.0 ~1); serine (+0.3); asparagine/glutamine
(+0.2); glycine (0);
threonine (-0.4); proline (-0.5 ~1); alanine/histidine (-0.5); cysteine (-
1.0); methionine (-1.3);
is valine (-1.5); leucine/isoleucine (-1.8); tyrosine (-2.3); phenylalanine (-
2.5); and tryptophan (-
3.4).
It is understood that an amino acid may be substituted by another amino acid
having a
similar hydrophilicity score and still result in a protein with similar
biological activity, i.e., still
obtain a biologically functional protein. In making such changes, the
substitution of amino acids
zo whose hydropathic indices are within ~2 is preferred, those within ~1 are
more preferred, and
those within ~0.5 are most preferred.
As outlined above, amino acid substitutions are therefore based on the
relative similarity
of the amino acid side-chain substituents, for example, their hydrophobicity,
hydrophilicity,
charge, size, and the like. Exemplary substitutions which take various of the
foregoing
zs characteristics into consideration are well known to those of skill in the
art and include: arginine
and lysine; glutamate and aspartate; serine and threonine; glutamine and
asparagine; and valine,
leucine, and isoleucine. Changes which are not expected to be advantageous may
also be used if
these resulted in functional fusion proteins.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
- 63 -
Plant Vectors
In plants, transformation vectors capable of introducing nucleic acid
sequences encoding
polyhydroxyalkanoate biosynthesis enzymes are easily designed, and generally
contain one or
more nucleic acid coding sequences of interest under the transcriptional
control of 5' and 3'
s regulatory sequences. Such vectors generally comprise, operatively linked in
sequence in the 5'
to 3' direction, a promoter sequence that directs the transcription of a
downstream heterologous
structural nucleic acid sequence in a plant; optionally, a 5' non-translated
leader sequence; a
nucleic acid sequence that encodes a protein of interest; and a 3' non-
translated region that
encodes a polyadenylation signal which functions in plant cells to cause the
termination of
~o transcription and the addition of polyadenylate nucleotides to the 3' end
of the mRNA encoding
the protein. Plant transformation vectors also generally contain a selectable
marker. Typical 5'-
3' regulatory sequences include a transcription initiation start site, a
ribosome binding site, an
RNA processing signal, a transcription termination site, and/or a
polyadenylation signal. Vectors
for plant transformation have been reviewed in Rodriguez et al. (Vectors: A
Survey of Molecular
~s Cloning Vectors and Their Uses, Butterworths, Boston., 1988), Glick et al.
(Methods in Plant
Molecular Biology and Biotechnology, CRC Press, Boca Raton, Fla., 1993), and
Croy (Plant
Molecular Biology Labfax, Hames and Rickwood (Eds.), BIOS Scientific
Publishers Limited,
Oxford, UK., 1993).
Plant Promoters
2o Plant promoter sequences can be constitutive or inducible, environmentally-
or
developmentally-regulated, or cell- or tissue-specific. Often-used
constitutive promoters include
the CaMV 35S promoter (Odell, J.T. et al., Nature 313: 810-812. 1985), the
enhanced CaMV
35S promoter, the Figwort Mosaic Virus (FMV) promoter (Richins et al., Nucleic
Acids Res. 20:
8451-8466, 1987), the mannopine synthase (mas) promoter, the nopaline synthase
(nos)
2, promoter, and the octopine synthase (ocs) promoter. Useful inducible
promoters include
promoters induced by salicylic acid or polyacrylic acids (PR-I, Williams , S.
W. et al,
Biotechnology 10: 540-543, 1992), induced by application of safeners
(substituted
benzenesulfonamide herbicides, Hershey, H.P. and Stoner, T.D., Plant Mol.
Biol. 17: 679-690,
1991), heat-shock promoters (Ou-Lee et al., Proc. Natl. Acad Sci U.S.A. 83:
6815-6819, 1986;
3o Ainley et al., Plant Mol. Biol. 14: 949-967, 1990), a nitrate-inducible
promoter derived from the
spinach nitrite reductase gene (Back et al., Plant Mol. Biol. 17: 9-18. 1991),
hormone-inducible
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-64-
promoters (Yamaguchi-Shinozaki, K. et al., Plant Mol. Biol. 15: 905-912, 1990;
Kares et al.,
Plant Mol. Biol. 15: 225-236, 1990), and light-inducible promoters associated
with the small
subunit of RuBP carboxylase and LHCP gene families (Kuhlemeier et al., Plant
Cell 1: 471,
1989; Feinbaum, R.L. et al., Mol. Gen. Genet. 226: 449-456, 1991; Weisshaar.
B. et al., EMBO
s J. 10: 1777-1786, 1991; Lam, E. and Chua, N.H., J. Biol. Chem. 266: 17131-
17135, 1990;
Castresana, C. et al., EMBO J. 7: 1929-1936, 1988; Schulze-Lefert et al., EMBO
J. 8: 651,
1989). Examples of useful tissue-specific, developmentally-regulated promoters
include the (3-
conglycinin 7S promoter (Doyle, J.J. et al., J. Biol. Chem. 261: 9228-9238,
1986; Slighton and
Beachy, Planta 172: 356-363, 1987), and seed-specific promoters (Knutzon, D.S.
et al., Proc.
~o Natl. Acad. Sci U.S.A. 89: 2624-2628, 1992; Bustos, M.M. et al., EMBO J.
10: 1469-1479, 1991;
Lam and Chua, Science 248: 471, 1991; Stayton et al., Aust. J. Plant. Physiol.
18: 507, 1991).
Plant functional promoters useful for preferential expression in seed plastids
include those from
plant storage protein genes and from genes involved in fatty acid biosynthesis
in oilseeds.
Examples of such promoters include the 5' regulatory regions from such genes
as napin (Kridl et
~ s al., Seed Sci. Res. 1: 209-219, 1991 ), phaseolin, zero, soybean trypsin
inhibitor, ACP, stearoyl-
ACP desaturase, and oleosin. Seed-specific gene regulation is discussed in EP
0 255 378.
Promoter hybrids can also be constructed to enhance transcriptional activity
(Comai, L. and
Moran, P.M., U.S. Patent No. 5,106,739, issued April 21, 1992), or to combine
desired
transcriptional activity and tissue specificity. A developing seed selective
promoter may be
zo obtained from the fatty acid hydroxylase gene of Lesquerella (P-lh) (Broun,
P. and C.
Somerville. Plant Physiol. 113: 933-942, 1997).
Plant transformation and regeneration
A variety of different methods can be employed to introduce such vectors into
plant
protoplasts, cells, callus tissue, leaf discs, meristems, etcetera, to
generate transgenic plants,
zs including Agrobacterium-mediated transformation, particle gun delivery,
microinjection,
electroporation, polyethylene glycolmediated protoplast transformation,
liposome-mediated
transformation, etcetera (reviewed in Potrykus, I. Ann. Rev. Plant Physiol.
Plant Mol. Biol. 42:
205-225, 1991). In general, transgenic plants comprising cells containing and
expressing DNAs
encoding polyhydroxyalkanoate biosynthesis proteins can be produced by
transforming plant
30 cells with a DNA construct as described above via any of the foregoing
methods; selecting plant
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-65-
cells that have been transformed on a selective medium; regenerating plant
cells that have been
transformed to produce differentiated plants; and selecting a transformed
plant which expresses
the protein-encoding nucleotide sequence.
Specific methods for transforming a wide variety of dicots and obtaining
transgenic
s plants are well documented in the literature (Gasser and Fraley, Science
244: 1293-1299, 1989;
Fisk and Dandekar, Scientia Horticulturae 55: 5-36, 1993; Christou, Agro Food
Industry Hi
Tech, p.17, 1994; and the references cited therein).
Successful transformation and plant regeneration have been reported in the
monocots as
follows: asparagus (Asparagus officinalis; Bytebier et al., Proc. Natl. Acad.
Sci. U.S.A. 84: 5345-
~0 5349, 1987); barley (Hordeum vulgarae; Wan and Lemaux, Plant Physiol. 104:
37-48, 1994);
maize (Zea mays; Rhodes, C.A. et al., Science 240: 204-207, 1988; Gordon-Kamm
et al., Plant
Cell 2: 603-618, 1990; Fromm, M.E. et al., BiolTechnology 8: 833-839, 1990;
Koziel et al.,
BiolTechnology l l: 194-200, 1993); oats (Avena saliva; Somers et al.,
BiolTechnology 10: 1589-
1594, 1992); orchardgrass (Dactylis glomerata; Horn et al., Plant Cell Rep. 7:
469-472, 1988);
is rice (Oryza saliva, including indica and japonica varieties; Toriyama et
al., BiolTechnology 6:
10, 1988; Zhang et al., Plant Cell Rep. 7: 379-384, 1988; Luo and Wu, Plant
Mol. Biol. Rep. 6:
165-174, 1988; Zhang and Wu, Theor. Appl. Genet. 76: 835-840, 1988; Christou
et al.,
BiolTechnology 9: 957-962, 1991); rye (Secale cereale; De la Pena et al.,
Nature 325: 274-276,
1987); sorghum (Sorghum bicolor; Casas, A.M. et al., Proc. Natl. Acad. Sci.
U.S.A. 90: 11212-
Zo 11216, 1993); sugar cane (Saccharum spp.; Bower and Birch, Plant J. 2: 409-
416, 1992); tall
fescue (Festuca arundinacea; Wang, Z.Y. et al., BiolTechnology 10: 691-696,
1992); turfgrass
(Agrostis palustris; Zhong et al., Plant Cell Rep. 13: 1-6, 1993); wheat
(Triticum aestivum; Vasil
et al., BiolTechnology 10: 667-674, 1992; Weeks, T. et al., Plant Physiol.
102: 1077-1084, 1993;
Becker et al., Plant J. 5: 299-307, 1994), and alfalfa (Masoud, S.A. et al.,
Transgen. Res. 5: 313,
2s 1996).
Host lp ants
Particularly useful plants for polyhydroxyalkanoate production include those
that produce
carbon substrates, including tobacco, wheat, potato, Arabidopsis, and high oil
seed plants such as
corn, soybean, canola, oil seed rape, sugarbeet, sunflower, flax, peanut,
sugarcane, switchgrass,
3o and alfalfa.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-66-
Example 21: Plastid transformation
Alternatively, polyhydroxyalkanoate biosynthesis enzymes facilitating the
increase in oil
content of plants and/or herbicide resistance discussed herein can be
expressed in situ in plastids
by direct transformation of these organelles with appropriate recombinant
expression constructs.
s Constructs and methods for stably transforming plastids of higher plants are
well known in the
art (Svab, Z. et al., Plant Mol. Biol. 14(2): 197-205, 1990; Svab et al.,
Proc. Natl. Acad. Sci. U S
A. 90(3): 913-917, 1993; Staub et al., EMBO J. 12(2): 601-606, 1993; Maliga et
al., U.S. Patent
No. 5,451,513; PCT International Publications WO 95/16783, WO 95/24492, and WO
95/24493). These methods generally rely on particle gun delivery of DNA
containing a
~ o selectable or scorable marker in addition to introduced DNA sequences for
expression, and
targeting of the DNA to the plastid genome through homologous recombination.
Transformation
of a wide variety of different monocots and dicots by particle gun bombardment
is routine in the
art (Hinchee et al., 1994; Walden and Wingender, 1995). The plastid may be
transformed by
using protoplast and PEG (polyethylene glycol) (Koop, et al., Physiol. Plant.
85: 339, 1992;
Golds et al., BiolTechnol. 11: 95-97, 1993), cocultivation of protoplasts and
Agrobacteria
carrying transformation vectors (De Block et al., EMBO J. 4: 1367-1372, 1985),
and by
electroporation (Kin-Ying et al., Plant J. 4: 737, 1996).
Nucleic acid constructs for plastid transformation generally comprise a
targeting
segement comprising flanking nucleic acid sequences substantially homologous
to a
zo predetermined sequence of a plastid genome, which targeting segment enables
insertion of
nucleic acid coding sequences of interest into the plastid genome by
homologous recombination
with the predetermined sequence; a selectable marker sequence, such as a
sequence encoding a
form of plastid 16S ribosomal RNA that is resistant to spectinomycin or
streptomycin, or that
encodes a protein which inactivates spectinomycin or streptomycin (such as the
aadA gene),
z, disposed within the targeting segment, wherein the selectable marker
sequence confers a
selectable phenotype upon plant cells, substantially all the plastids of which
have been
transformed with the nucleic acid construct; and one or more nucleic acid
coding sequences of
interest disposed within the targeting segment relative to the selectable
marker sequence so as
not to interfere with conferring of the selectable phenotype. In addition,
plastid expression
3o constructs also generally include a plastid promoter region and a
transcription termination region
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-67-
capable of terminating transcription in a plant plastid, wherein the regions
are operatively linked
to the nucleic acid coding sequences of interest.
A further refinement in chloroplast transformation/expression technology that
facilitates
control over the timing and tissue pattern of expression of introduced nucleic
acid coding
s sequences in plant plastid genomes has been described in PCT International
Publication WO
95/16783. This method involves the introduction into plant cells of constructs
for nuclear
transformation that provide for the expression of a viral single subunit RNA
polymerise and
targeting of this polymerise into the plastids via fusion to a plastid transit
peptide.
Transformation of plastids with nucleic acid constructs comprising a viral
single subunit RNA
~o polymerise-specific promoter specific to the RNA polymerise expressed from
the nuclear
expression constructs operably linked to nucleic acid coding sequences of
interest permits
control of the plastid expression constructs in a tissue and/or developmental
specific manner in
plants comprising both the nuclear polymerise construct and the plastid
expression constructs.
Expression of the nuclear RNA polymerise coding sequence can be placed under
the control of
~ s either a constitutive promoter, or a tissue- or developmental stage-
specific promoter, thereby
extending this control to the plastid expression construct responsive to the
plastid-targeted,
nuclear-encoded viral RNA polymerise. The introduced nucleic acid coding
sequence can be a
single encoding region, or may contain a number of consecutive encoding
sequences to be
expressed as an engineered or synthetic operon. The latter is especially
attractive where, as in
zo the present invention, it is desired to introduce multigene biochemical
pathways into plastids.
This approach is more complex using standard nuclear transformation techniques
since each
gene introduced therein must be engineered as a monocistron, including an
encoded transit
peptide and appropriate promoter and terminator signals. Individual gene
expression levels may
vary widely among different cistrons, thereby possibly adversely affecting the
overall
2> biosynthetic process. This can be avoided by the chloroplast transformation
approach.
All of the compositions and/or methods disclosed and claimed herein can be
made and
executed without undue experimentation in light of the present disclosure.
While the
compositions and methods of this invention have been described in terms of
preferred
embodiments, it will be apparent to those of skill in the art that variations
may be applied to the
3o compositions and/or methods and in the steps or in the sequence of steps of
the methods
described herein without departing from the concept, spirit and scope of the
invention. More
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-68-
specifically, it will be apparent that certain agents which are both
chemically and physiologically
related may be substituted for the agents described herein while the same or
similar results would
be achieved. All such similar substitutes and modifications apparent to those
skilled in the art
are deemed to be within the spirit, scope and concept of the invention.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-69-
REFERENCES
The following references, to the extent that they provide exemplary procedural
or other
details supplementary to those set forth herein, are specifically incorporated
herein by reference.
s 1. Altschul, S.F., T.L. Madden, A.A. Schaffer, J. Zhang, Z. Zhang, W.
Miller, and D.J.
Lipman. 1997. Gapped BLAST and PSI BLAST: a new generation of protein database
search programs. Nucleic Acids Res., 25: 3389-3402.
2. Anderson, A. and E.A. Dawes. 1990. Occurrence, metabolism, metabolic role,
and
industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev., 54: 450-
472.
~0 3. Cevallos, M.A., S. Encarnacion, A. Leija, Y. Mora, and J. Mora. 1996.
Genetic and
physiological characterization of a Rhizobium etli mutant strain unable to
synthesize
poly-beta-hydroxybutyrate. J. Bacteriol., 178: 1646-1654.
4. Connors, M.J., J.M. Mason, and P. Setlow. 1986. Cloning and nucleotide
sequencing of
genes for three small, acid soluble proteins Bacillus subtilis spores. J.
Bacteriol., 166:
Is 417-425.
5. deSmet, M.J., G. Eggink, B. Witholt, J. Kingma, and H. Wynberg. 1983.
Characterization
of intracellular inclusions formed by Pseudomonas oleovorans during growth on
octane.
J. Bacteriol., 154: 870-878.
6. Dunlop, W. and A.W. Robards. 1973. Ultrastructural study of poly-(3-
hydroxybutyrate
ao granules from Bacillus cereus. J. Bacteriol., 114: 1271-1280.
7. Eggink, G., P. de Waard, and G.N.M. Huijberts. 1992. The role of fatty acid
biosynthesis
and degradation in the supply of substrates for poly(3-hydroxyalkanoate)
formation in
Pseudomonas putida. FEMS Microbiol. Rev., 103: 159-164.
8. Ellar, D., D.G. Lundgren, K. Okamura, and R.H. Marchessault. 1968.
Morphology of
is poly-~3-hydroxybutyrate granules. J. Mol. Biol., 35: 489-502.
9. Fliss, E.R., A.C. Loshon, and P. Setlow. 1986. Genes for Bacillus
megaterium small,
acid-soluble spore proteins: Cloning and nucleotide sequence of three
additional genes
from this multigene family. J. Bacteriol., 165: 467-473.
10. Fliss, E.R. and P. Setlow. 1984. Bacillus megaterium spore protein C-3:
nucleotide
so sequence of its gene and the amino acid sequence at its spore cleavage
site. Gene, 30:
167-172.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-70-
11. Fuller, R.C., J.P. O'Donnell, J Saulnier, T.E. Redlinger, J. Foster, and
R.W. Lenz. 1992.
The supramolecular architecture of the polyhydroxyalkanoate inclusions in
Pseudomonas
oleovorans. FEMS Microbiol. Rev., 103: 279-288.
12. Gerngross, T.U., P. Reilly, J. Stubbe, A.J. Sinskey, and O.P. Peoples.
1993.
s Immunocytochemical analysis of poly-(3-hydroxybutyrate (PHB) synthase enzyme
at the
surface of PHB granules. J. Bacteriol., 175: 5289-5293.
13. Gilman, M.Z., J.L. Wings, and M.J. Chamberlin. 1981. Nucleotide sequence
of two
Bacillus subtilis promoters used by Bacillus subtilis sigma-28 RNA polymerase.
Nucleic
Acids Res., 9: 5991-6000.
~0 14. Gitt, M.A., L.F. Wang, and R.H. Doi. 1985. A strong sequence homology
exists between
RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J.
Biol. Chem.,
260: 7178-7185.
15. Griebel, R., Z. Smith, and M. Merrick. 1968. Metabolism of poly-(3-
hydroxybutyrate. 1.
Purification, composition, and properties of native poly-(3-hydroxyburyrate
granules from
is Bacillus megaterium. Biochem., 7: 3676-3681.
16. Haima, P., D. van Sinderen, H. Scholting, S. Bron, and G. Venema. 1990.
Development
of (3-galactosidase a,-complementation system for molecular cloning in
Bacillus subtilis.
Gene, 86: 63-69.
17. Haywood, G.W., A.J. Anderson, L. Chu, and E.A. Dawes. 1988. The role of
NADH- and
ao NADPH-linked acetoacetyl-CoA reductases in the poly-3-hydroxybutyrate
synthesizing
organism Alcaligenes eutrophus. FEMS Microbiol. Lett., 52: 259-264.
18. Haywood, G.W., A.J. Anderson, L. Chu, and E.A. Dawes. 1988.
Characterization of two
3-ketothiolases in the polyhydroxyalkanoate synthesizing organism Alcaligenes
eutrophus. FEMSMicrobiol. Lett., 52: 91-96.
Zs 19. Haywood, G.W., A.J. Anderson, and E.A. Dawes. 1989. The importance of
PHB-
synthase substrate specificity in polyhydroxyalkanoate synthesis by
Alcaligenes
eutrophus. FEMSMicrobiol. Lett., 57: 1-6.
20. Helmann, J.D. 1991. Alternative sigma factors and the regulation of
flagellar gene
expression. Mol. Microbiol., 5: 2875-2882.
30 21. Huang, A.H.C. 1992. Oil bodies and oleosins in seeds. Annu. Rev. Plant
Physiol. Plant
Mol. Biol., 43: 177-200.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-71 -
22. Huisman, G.W., E. Wonink, R. Meima, B. Kazemier, P. Terpstra, and B.
Witholt. 1991.
Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. J.
Biol.
Chem., 266: 2191-2198.
23. Kaneko, T. et al. 1996. Sequence analysis of the genome of the unicellular
s cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination
of the
entire genome and assignment of potential protein-coding regions. DNA Res., 3:
109-
136.
24. Kunst, N. et al. 1997. The complete genome sequence of the Gram-positive
bacterium
Bacillus subtilis. Nature, 390: 249-256.
~0 25. Lauzier, C., R.H. Marchessault, P. Smith, and H. Chanzy. 1992.
Structural study of
isolated poly((3-hydroxybutyrate) granules. Polymer, 33: 823-827.
26. Lee, S.Y. 1995. Bacterial polyhydroxyalkanoates. Biotechnology &
Engineering, 49: 1-14.
27. Liebergesell, M., B. Schmidt, and A. Steinbiichel. 1992. Isolation and
identification of
granule-associated proteins relevant for poly(hydroxyalkanoic acid)
biosynthesis in
is Chromatium vinosum D. FEMS Microbiol. Lett., 99: 227-232.
28. Liebergesell, M. and A. Steinbiichel. 1992. Cloning and nucleotide
sequences of genes
relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum
strain
D. Eur. J. Biochem., 209: 135-150.
29. Liebergesell, M. and A. Steinbiichel. 1993. Cloning and molecular analysis
of the poly
Zo (3-hydroxybutyric acid) biosynthetic genes of Thiocystis violacea. Appl.
Microbiol.
Biotechnol., 38: 493-501.
30. Lundgren, D.G., R.M. Pfister, and J.M. Merrick. 1964. Structure of poly-(3-
hydroxybutyric acid granules. J. Gen. Microbiol., 34: 441-446.
31. Magni, C., P. Marini, and D. de Mendoza. 1995. Extraction of RNA from gram-
positive
is bacteria. Biotechniques, 19: 882-884.
32. McCool, G.J., T. Fernandez, N. Li, and M.C. Cannon. 1996.
Polyhydroxyalkanoate
inclusion-body growth and proliferation in Bacillus megaterium. FEMS
Microbiol. Lett.,
137: 41-48.
33. Miller, J.H. 1972. Experiments in molecular genetics. Cold Spring Harbor
Laboratory,
3o Cold Spring Harbor, NY:
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-72-
34. Moran, C.P. Jr., N. Lang, S.F.J. LeGrice, G. Lee, M. Stephens, A.L.
Sonnenshein, J.
Pero, and R. Losick. 1982. Nucleotide sequences that signal the initiation of
transcription
and translation in Bacillus subtilis. Mol. Gen. Genet., 186: 339-346.
35. Morbidoni, H.R., D. de Mendoza, and J.E. Cronan. 1996. Bacillus subtilis
acyl carrier
s protein is encoded in a cluster of lipid biosynthesis genes. J. Bacteriol.,
178: 4794-4800.
36. Pieper-Furst, U., M.H. Madkour, F. Mayer, and A. Steinbiichel. 1994.
Purification and
characterization of a 14-kilodalton protein that is bound to the surface of
polyhydroxyalkanoic acid granules in Rhodococcus ruber. J. Bacteriol., 176:
4328-4337.
37. Pieper-Furst, U., M.H. Madkour, F. Mayer, and A. Steinbiichel. 1995.
Identification of
to the region of a 14-kilodalton protein of Rhodococcus ruber that is
responsible for the
binding of this Phasin to polyhydroxyalkanoic acid granules. J. Bacteriol.,
177: 2513-
2523.
38. Sambrook, J., E.F. Fritsch & T. Maniatis. 1989. Molecular Cloning: A
Laboratory
Manual., 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
is 39. Shark, K.B., F.D. Smith, P.R. Harpending, and J.L. Rasmussen. 1991.
Biolistic
transformation of a procaryote, Bacillus megaterium. Appl. Environ.
Microbiol., 57: 480-
485.
40. Simon, R., Priefer, U. & Piihler, A. 1983. In A. Puhler (Ed.), Molecular
genetics of the
bacteria-plant interaction. Springer, Berlin. p. 98-106.
ao 41. Steinbiichel, A. 1991. Polyhydroxyalkanoic acids. p. 123-213. In D.
Byrom (Ed.),
Biomaterials, novel materials from biological sources. Macmillan Publishers
Ltd.,
Basingstoke, England.
42. Steinbiichel, A., K. Aerts, W. Babel, C. Follner, M. Liebergesell, M.H.
Madkour, F.
Mayer, U. Pieper-Furst, A. Pries, H.E. Valentin, and R. Wieczorek. 1995.
Considerations
Zs on the structure and biochemistry of bacterial polyhydroxyalkanoic acid
inclusions. Can.
J. Microbiol., 41: 94-105.
43. Steinbiichel, A., E. Hustede, M. Liebergesell, U. Pieper, A. Timm, and H.
Valentin. 1992.
Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids
in
bacteria. FEMS Microbiol. Rev., 103: 217-230.
30 44. Steinbiichel, A. and H.G. Schlegel. 1991. Physiology and molecular
genetics of poly((3-
hydroxyalkanoic acid) synthesis in Alcaligenes eutrophus. Mol. Microbiol., 5:
535-542.
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-73-
45. Steinbiichel, A. and H.E. Valentin. 1995. Diversity of bacterial
polyhydroxyalkanoic
acids. FEMS Microbiol. Lett., 128: 219-228.
46. Vary, P. 1993. The genetic map of Bacillus megaterium, p. 475-481. In A.L.
Sonenshein, J.A. Hoch & R. Losich (Eds.), Bacillus subtilis and other gram
positive
s bacteria. American Society for Microbiology, Washington, D.C.
47. Wang, W.S. and D.G. Lundgren. 1969. Poly-(3-hydroxybutyrate in the
chemolithotrophic
bacterium Ferrobacillus ferrooxidans. J. Bacteriol., 97: 947-950.
48. Wieczorek, R., A. Pries, A. Steinbiichel, and F. Mayer. 1995. Analysis of
a 24-kilodalton
protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes
eutrophus.
io J. Bacteriol., 177: 2425-2435.
49. Wieczorek, R., A. Steinbiichel, and B. Schmidt. 1996. Occurrence of
polyhydroxyalkanoic acid granule-associated proteins related to the
Alcaligenes
eutrophus H16 GA24 protein in other bacteria. FEMS Microbiol. Lett., 135: 23-
30.
50. Hein, S., Tran, H., and A. Steinbiichel. 1998. Synechocystis sp. PCC6803
possesses a
is two-component polyhydroxyalkanoic acid synthase similar to that of
anoxygenic purple
sulfur bacteria. Arch. Microbiol., 170(3): 162-70.
51. Steinbiichel, A., Hustede, E., Liebergesell, M., Pieper, U., Timm, A., and
H. Valentin.
1992. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic
acids in
bacteria. FEMS Microbiol. Rev. 103: 217-230.
zo 52. Basak, A., Boudreault, A., Chen, A., Chretien, M., Seidah, N.G., and C.
Lazure. 1995.
Application of the multiple antigenic peptides (MAP) strategy to the
production of
prohormone convertases antibodies: synthesis, characterization and use of 8-
branched
immunogenic peptides. J. Pept. Sci. 1(6): 385-95.
53. Slater, S., K.L. Houmiel, M. Tran, T.A. Mitsky, T.B. Taylor, S.R.
Padgette, and K.J.
Zs Gruys. 1998. Multiple (3-ketothiolases mediate poly(~3-hydroxyalkanoate)
copolymer
synthesis in Ralstonia eutropha. J. Bacteriol. 180: 1979-1987.
54. Fukui, T., N. Shiomi, and Y. Doi. 1998. Expression and characterization of
(R)-specific
enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis in
Aeromonas caviae. J. Bacteriol. 180: 667-6673.
30 5~. Boynton, Z.L., G.N. Bennett, and F.B. Rudolph. 1996. Cloning,
sequencing, and
expression of clustered genes encoding b-hydroxybutyryl-coenzyme A (CoA)
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
-74-
dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium
acetobutylicum ATCC 824. J. Bacteriol. 178: 3015-3024.
56. Liebergesell, M., F. Mayer, and A. Steinbiichel. 1993. Analysis of
polyhydroxyalkanoic
acid-biosynthesis genes of anoxygenic phototrophic bacteria reveals synthesis
of a
polyester exhibiting an unusual composition. Appl. Microbiol. Biotechnol.
40:292-300.
CA 02363803 2001-07-05
WO 00/40730 PCT/CTS00/00364
1
SEQUENCE LISTING
<110> McCool, Gabriel J.
Cannon, Maura C.
Cannon, Francis C.
Valentin, Henry E.
Gruys, Kenneth J.
<120> POLYHYDROXYALKANOATE BIOSYNTHESIS ASSOCIATED PROTEINS
AND CODING REGION IN BACILLUS MEGATERIUM
<130> MOBT212
<140> 60/115,092
<141> 1999-01-07
<160> 29
<170> PatentIn Ver. 2.1
<210> 1
<211> 7916
<212> DNA
<213> Bacillus megaterium
<400> 1
aagcttaaca aaaggttaga gggcttttaa ttgacttatt aatgaacctt ttataaaata 60
aaaacaactt aatacattta cttcttacgg agtaaagggg agtagcgtca gggaaacctg 120
aaacaaagtc gtcattacat ggagtattcc atcggctttg ttggcataat gattatgctt 180
agcaagacct ttgccatatt tggcaaaggt ctttttgtgt tttattccgg taatgaggat 240
aaataaactt atacaaagaa acgagagggg atttttatgg atgcatcact tttgttagag 300
tatggatggg tattgctagt gctggttgca ttagaaggaa ttttggcggc ggataatgct 360
cttgtgatgg ctattatggt caaacattta ccggaagaaa aacgcaagaa ggcattattt 420
tacggattag ccggtgcctt tatttttaga tttggttcgt tgttcttgat ttcattttta 480
gtcgacgtat ggcagcttca agctatagga gccatttact tattgttcat ttccattaat 540
catattgtga agcgatatgt gaaaaaagac gatcatgaaa aagtgaaaga agcagacgag 600
aaaaagggct caggtttctg gatgacggtt ttaaaagtag aaatagcaga cattgctttt 660
gccgttgatt caattttggc cgctgtggct ctcgccgtta cgttgccaac aacaaatctt 720
cctcaaattg gcggactcga cggcggacaa ttcttggtga tcttcgccgg aggaattatg 780
ggattaatta ttatgcgttt tgctgcaact tggttcgtca agctattaaa tacgcgccca 840
ggcctagaaa cggcggcttt tgctattgta ggctgggtag gagttaagtt agcggtctat 900
acccttgctc atccagagtt aggtattatt aatgaacatt tccctgaatc aaaagtgtgg 960
aaaattacgt tttggattgt gttacttggc atagctgctt caggctggtt tctatctaaa 1020
aataaagaac aaactgatct tgaaggctca gagaaagaaa aagaatcgtt aaaaaaaatt 1080
gaaaatcaat aataaaaaaa acgcgccttc aatgttaatt gaaggcgcgt tttttatagg 1140
gggataaaat acctagttta gattgtttaa ataaagggta aaagaattaa ttactgttta 1200
attactgtat atccgaatgt tttgcttttt cgttcatatt ctgtagagaa cagagccgtg 1260
agtaattcag ttcaatgaag tttatttcag tttataaaca tatatttcaa atgtatgtta 1320
attggtaaaa ttgagctatt atgggttata ataaaggaaa attaaaagga gcagagcgct 1380
tctcaaccct gttcgtcatc aaaagaatgg atattaacat ggtataaagc aactgagtat 1440
agtaaaagag gtgaagccaa tgctcacaaa agttcaaacg cctccatcgc ttgaaacgct 1500
tgtactgacg attcagcaag gggataaaca attacataat gaaatgattc aacaatataa 1560
accgtttatt gctaaagttg tttcagctgt atgtaaacgt tatataagtg aagctgacga 1620
tgaatttagc attggtctga ttgcatttaa tgaagccatt gaaaattaca caatccaaaa 1680
aggacgatct cttcttgcat ttgcggaact tattattaaa agaagagtaa tcgactatat 1740
tcgaaaagaa aagcgaaatc aaacgctgct ctataaccga attgaaaatg aaggttttat 1800
tcaaggtaag gtagaaaggg atatatcgct ttctaactat aaaaggcaaa gtgaaacttc 1860
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
2 _.
atatattcaa gaggaaatga cttatttttg tcaggcgcta aaattgttta aattaactct 1920
tgaagacatt attaacacgt ctcctaaaca taaggatgca aggggaaatg cagtggaagt 1980
tgcatctttt atcgtcaatg aaaaagaatt aaaagataag ctgtttttaa agcggcagct 2040
tcctattcgc ttgattgaaa aacatgtcaa agtaagccgg aaaacaattg aaagaaaccg 2100
taaatatatt atcgcgatgg ttattatatt agcgggggac tacgtgtatt taaaagacta 2160
tattatgtaa gaaaaaggca cacgcaggtg ccttttttta gccatgttat gaaattatgt 2220
tttatttttg tgtgctgccg cttaagtgag cttgagcttg ttgcaccaag cgttttgtga 2280
tttctccgcc tactgaaccg ttgctgcgtg cagcagtgtc agaacctaga gttaccccaa 2340
attcttgagc gatttcatat ttatattgat ctaaaaattg ttctactcct ggtgttaata 2400
gtttatttgt tctagccatg attagtcatc tcctttttta gtggtacagc agttcactgc 2460
tgtacctgta ttatttgttg tttcctattt tgcatactgg taatggtttg caagaaagta 2520
taaaaaagcc cagctttcaa agaagctggg cttttttata tattatttta caactgcata 2580
ttgctcaaga gactgagcaa atgacttttg aagagatttt acttgttcta cgtatgcatc 2640
agatacgtgt tggaattctt ggcgcgtttt cttttgctct tctactaatt gtgttactac 2700
ttgatgatat tgttcttgcg cttgctttac taaagaatag cttgatttgc tttggttaaa 2760
aaaaagctct tgtaatttat ttaatgcttc atgcgtgcgg tttgtccatt cttcataaga 2820
atcggcaaca gcgtttccag ctgttttacg taagttttca actgtttttt gttgaaggtc 2880
ttccaattca gctttccatt gtttgtctgt tgcttgaagt tgttcaactg cttttgttac 2940
aaattcttgc tgttgctcaa gtgcttttaa cgtccattgc tcaatttgtt tgtttccgtc 3000
tgcaatgttt tgtaacccct ttgtccattg ttcccacatt gcatcaatta ctgtatcata 3060
ctttactgtt gacataatca aattcctcct tgaataattt ggtctggcga acagttatcg 3120
taatgaacaa aacgttaaag aagaaattcg tagttgtgtg ataaacctaa cattccgcgc 3180
tcgctggttg accgtgaaat ttaacattat attaggaaac gcagatgtac tgcgtgaatc 3240
cctttaatcg ttttcttcct cttttgattc ttttgacttt ttagaagaag aagagctaaa 3300
agggaacaac atgtcggtat acatgtgaaa aaatgaatct aacatgtttt gatactgttc 3360
aagtgaatta gcccacatac ttaaatattg ttcgccggca tctgttaatg aataaatccg 3420
gcgtgcaggt ccttcagctg acgtatccca ttgcgatgta atcaagttgt ctttttcaag 3480
ctgtctcagc gtgcggtaga catttccctg atcaactgat gtgaatccaa agctcattag 3540
ttgctgaatg agcttgtaac catgtagatt ccaccctctt aaacttaaaa gaagaaaagg 3600
aaccatcaag ttttttggtg caccgctaat cgatttttct aagttgtttg aattagaggt 3660
gttatctttt tcacttgatc ccaaggtaat caccccttcg aaaaaagaga atttgttttc 3720
catatgtgta atttacacct agatgaaaaa attgtcaaca ctttaaaaca ggtctttcaa 3780
atacaggaaa aaagaacgat tttttggatt tcgacaaaaa gcgtcaaaaa aattcaaaaa 3840
taaacaaaga tttagaattg tttattttgg aaaacgtatt tataatagta catgtaggta 3900
atttttataa catttataat ttgttggtaa gtgaattgtg aaaagattta cattatccaa 3960
taaataaacg caaaattggt tgcgtttact tagagcttct aatatgtcgc ttgtcacatt 4020
aagtgtccat attgaaagcc atcactttta acgtacacaa gaaaggagat ggagttttgg 4080
aacagcaaaa agtatttgat ccgtttcaag catggaaaga cgtatatgac aaaaccgaat 4140
cttactgggg taaagttatt ggggacaata tgaatcgtga agaattttcc cagctcatgg 4200
gaaatgtgct aaatatgaac cttcaatatc aacaagcagt aaatgaagta acggggcgct 4260
atctgcacca agtaaatgta ccaacaaaag aagatgtagc aaacgttgcg tcattagtca 4320
tcaatgtgga agaaaaagta gaattattag aagagcaatt tgacgatcgt tttgatgaat 4380
tagaagcaca gcaagaaagt gcatctgctt tgaaaaaaga tgtcactaag ctgaaatctg 4440
atgtcaaatc gttagacaaa aaactcgata aagttttatc tcttcttgaa gggcagcaaa 4500
aaacacaaga cgagttaaaa gaaacgattc aaaaacaaat taaaactcaa ggtgagcagc 4560
ttcaggctca gctgttagaa aaacaagaaa aattagctga aaagccaaag gcagaagcta 4620
aatctgaagc aaaaccatca aacgctcaaa aaactgagca gccggctcgc aagtaaggta 4680
tcggagattt tataacaaca ttaactgctg tatttacagt aaaaatcatc gctgaagaag 4740
caagggggaa atttttcatg acaacattac aaggtaaagt agcaatcgta acaggcggat 4800
ctaaaggtat cggggcagca attacacgtg agcttgcttc taatggagta aaagtagcag 4860
taaactataa cagcagtaaa gaatctgcag aagcaattgt aaaagaaatt aaagacaacg 4920
gcggagaagc tattgcggtt caagctgacg tgtcttatgt agatcaagca aaacacctaa 4980
tcgaagaaac aaaagctgcg tttggtcaat tagacattct agtaaacaat gctggaatta 5040
cgcgcgaccg ttcattcaag aagttaggtg aagaagattg gaaaaaagta attgatgtaa 5100
acttacatag cgtatacaac acaacatcag ctgcgctaac gcacctttta gaatctgaag 5160
gtggtcgtgt tatcaatatt tcatcaatta ttggtcaagc gggcggattt ggtcaaacaa 5220
actactcagc tgctaaagca ggtatgctag gattcactaa atcattagct cttgaactag 5280
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
3
ctaagacagg cgtaacggtt aatgcaattt gcccaggatt tattgaaacg gaaatggtga 5340
tggcaattcc tgaagatgtt cgtgcaaaaa ttgttgcgaa aattccaact cgtcgcttag 5400
gtcacgctga agaaattgca cgtggagttg tttacttagc aaaagacggc gcgtacatta 5460
caggacaaca gttaaacatt aacggcggct tatacatgta ataaatgctg gccctgactt 5520
ttgtcggggc tgtgcttgtt aactaactac taatggaatg aaagggtgtg tatattcgtg 5580
gcaattcctt acgtgcaaga gtgggaaaaa ttaatcaaat caatgccaag tgaatataaa 5640
agttctgcaa gacgttttaa gcgtgcatat gaaattatga caacagaagc ggaaccggaa 5700
gttggattaa caccaaaaga ggttatttgg aaaaagaaca aagcgaaatt atatcgctat 5760
acgccagtaa aagataacct gcataaaaca ccaatcttac tcgtatatgc attgatcaat 5820
aaaccgtata ttttggattt aacacctgga aacagccttg ttgaatactt attaaaccgc 5880
ggttttgacg tgtatttgct tgactgggga actcctgggc ttgaagacag caatatgaag 5940
ctagatgatt atattgtaga ttatattcca aaagcggcga aaaaggtgct gcgcacttct 6000
aaatctcctg atttgtctgt tcttggttac tgcatgggcg gaactatgac atctattttt 6060
gctgcattaa atgaagactt gccgattaaa aacttaattt ttatgacaag tccatttgat 6120
ttttcggata caggtttata cggagcattc ctagatgatc gctactttaa tttagataaa 6180
gcagtagata cattcggaaa catccctcca gagatgattg actttggaaa caagatgtta 6240
aagccaatca cgaatttcta cggcecgtat gtaacgttgg tggaccgttc ggaaaatcag 6300
cggtttgttg aaagctggaa gctaatgcaa aagtgggttg ctgacggaat cccatttgct 6360
ggcgaagctt atcgtcagtg gattcgtgac ttctatcaac aaaacaaact aatcaatggt 6420
2o gaacttgaag ttcgcggacg caaagtagat ttaaaaaata ttaaagctaa tattttaaac 6480
attgctgcta gccgtgatca tattgcgatg ccgcatcaag tggcagcttt aatggacgct 6540
gtttcaagtg aagataaaga gtataaattg ttgcaaacag gtcacgtatc tgttgtattt 6600
ggtccaaaag cagtgaagga aacatatcct tcaatcggcg attggctaga aaaacgctct 6660
aaataaaata aagacgaggc tgagacaaaa gtattttagc cgaagtgaaa aacgaaccac 6720
tgatatcagt ggttcgtttt tttgtataaa cagacaatag cgagtgatga ttctttatct 6780
ggactgatgg gatttatgat atctaatgac aagtgagatg acttctttta tactaatgta 6840
gtcaccttct taaacatggg attttttaca tggatatagc tattcatgta acaatgagta 6900
tgctttgaga agaaagaaga aaactattag tattataatg aaaaaggaat gtcagattat 6960
gccacaacca tggaaacgac gagtaaggca gatgtcttca gctcaaatta ttgttacatt 7020
3o ttacatagtg gctgttacgc ttgggtttct attacttagt attccagaag ctttaaggcc 7080
aggagcaaag ttagcattta ttgatcgctt atttattgcc gttagtgcgg taagtgtaac 7140
agggctgaca cctgtctcga ctccagatac atttagtaca acgggctatt ttttactcgt 7200
ttttattttt caaatcggtg gtattggtgt aatgacactc agtacattta tttggatgat 7260
tttaggtaaa aaaatcggtc tgaaggaacg tcagctcatt atgacggacc ataatcaatc 7320
ccgtttatca ggattagttg atttgatgag aaatatttta tttattattt ttgccattga 7380
actagttggc gccattattt tagggttaca ttttctccgt tattattcga gctggacaga 7440
tgcgtttttg catggtttct ttgcttctgt cagtgctaca acaaatgctg gcttcgatat 7500
tacaggatct tcatttattc cgtatgccca tgattatttc gtacaagtgg taaccgttat 7560
tttaattacg cttggagcga ttggattccc tgtattaatt gaaatcaagc actatttttt 7620
aacatttaaa gataagcgta aatttcaatt ttcgctattt acgaagctaa cgactattat 7680
gttttttctg ctgttaggag ggggaacaat cttgattctt gtgctagagc attcaggatt 7740
tctagcagat aagtcttggg atgaatcgtt tttttatgcg tttttccaat ccgctgccac 7800
aaggagcgga ggagtggcga ccatgaatat taatgagttt tcacttccta cgttaattat 7860
gatgagcgcg atgatgttta tcggtgcttc accgagttca gtagggggag gaattc 7916
<210> 2
<211> 510
<212> DNA
<213> Bacillus megaterium
<400> 2
atgtcaacag taaagtatga tacagtaatt gatgcaatgt gggaacaatg gacaaagggg 60
ttacaaaaca ttgcagacgg aaacaaacaa attgagcaat ggacgttaaa agcacttgag 120
caacagcaag aatttgtaac aaaagcagtt gaacaacttc aagcaacaga caaacaatgg 180
aaagctgaat tggaagacct tcaacaaaaa acagttgaaa acttacgtaa aacagctgga 240
aacgctgttg ccgattctta tgaagaatgg acaaaccgca cgcatgaagc attaaataaa 300
CA 02363803 2001-07-05
WO 00/40730 PCT/LTS00/00364
4
ttacaagagc ttttttttaa ccaaagcaaa tcaagctatt ctttagtaaa gcaagcgcaa 360
gaacaatatc atcaagtagt aacacaatta gtagaagagc aaaagaaaac gcgccaagaa 420
ttccaacacg tatctgatgc atacgtagaa caagtaaaat ctcttcaaaa gtcatttgct 480
cagtctcttg agcaatatgc agttgtaaaa 510
<210> 3
<211> 170
<212> PRT
<213> Bacillus megaterium
<400> 3
Met Ser Thr Val Lys Tyr Asp Thr Val Ile Asp Ala Met Trp Glu Gln
1 5 10 15
Trp Thr Lys Gly Leu Gln Asn Ile Ala Asp Gly Asn Lys Gln Ile Glu
25 30
Gln Trp Thr Leu Lys Ala Leu Glu Gln Gln Gln Glu Phe Val Thr Lys
20 35 40 45
Ala Val Glu Gln Leu Gln Ala Thr Asp Lys Gln Trp Lys Ala Glu Leu
50 55 60
Glu Asp Leu Gln Gln Lys Thr Val Glu Asn Leu Arg Lys Thr Ala Gly
65 70 75 80
Asn Ala Val Ala Asp Ser Tyr Glu Glu Trp Thr Asn Arg Thr His Glu
85 90 95
Ala Leu Asn Lys Leu Gln Glu Leu Phe Phe Asn Gln Ser Lys Ser Ser
100 105 110
Tyr Ser Leu Val Lys Gln Ala Gln Glu Gln Tyr His Gln Val Val Thr
115 120 125
Gln Leu Val Glu Glu Gln Lys Lys Thr Arg Gln Glu Phe Gln His Val
130 135 140
Ser Asp Ala Tyr Val Glu Gln Val Lys Ser Leu Gln Lys Ser Phe Ala
145 150 155 160
Gln Ser Leu Glu Gln Tyr Ala Val Val Lys
165 170
<210> 4
<211> 438
<212> DNA
<213> Bacillus megaterium
<400> 4
ttgggatcaa gtgaaaaaga taacacctct aattcaaaca acttagaaaa atcgattagc 60
ggtgcaccaa aaaacttgat ggttcctttt cttcttttaa gtttaagagg gtggaatcta 120
catggttaca agctcattca gcaactaatg agctttggat tcacatcagt tgatcaggga 180
aatgtctacc gcacgctgag acagcttgaa aaagacaact tgattacatc gcaatgggat 240
acgtcagctg aaggacctgc acgccggatt tattcattaa cagatgccgg cgaacaatat 300
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
ttaagtatgt gggctaattc acttgaacag tatcaaaaca tgttagattc attttttcac 360
atgtataccg acatgttgtt cccttttagc tcttcttctt ctaaaaagtc aaaagaatca 420
aaagaggaag aaaacgat 438
5
<210> 5
<211> 146
<212> PRT
<213> Bacillus megaterium
<400> 5
Met Gly Ser Ser Glu Lys Asp Asn Thr Ser Asn Ser Asn Asn Leu Glu
1 5 10 15
Lys Ser Ile Ser Gly Ala Pro Lys Asn Leu Met Val Pro Phe Leu Leu
25 30
Leu Ser Leu Arg Gly Trp Asn Leu His Gly Tyr Lys Leu Ile Gln Gln
35 40 45
Leu Met Ser Phe Gly Phe Thr Ser Val Asp Gln Gly Asn Val Tyr Arg
50 55 60
Thr Leu Arg Gln Leu Glu Lys Asp Asn Leu Ile Thr Ser Gln Trp Asp
65 70 75 80
Thr Ser Ala Glu Gly Pro Ala Arg Arg Ile Tyr Ser Leu Thr Asp Ala
85 90 95
Gly Glu Gln Tyr Leu Ser Met Trp Ala Asn Ser Leu Glu Gln Tyr Gln
100 105 110
Asn Met Leu Asp Ser Phe Phe His Met Tyr Thr Asp Met Leu Phe Pro
115 120 125
Phe Ser Ser Ser Ser Ser Lys Lys Ser Lys Glu Ser Lys Glu Glu Glu
130 135 140
Asn Asp
145
<210> 6
<211> 504
<212> DNA
<213> Bacillus megaterium
<400> 6
atgaatcgtg aagaattttc ccagctcatg ggaaatgtgc taaatatgaa ccttcaatat 60
caacaagcag taaatgaagt aacggggcgc tatctgcacc aagtaaatgt accaacaaaa 120
gaagatgtag caaacgttgc gtcattagtc atcaatgtgg aagaaaaagt agaattatta 180
gaagagcaat ttgacgatcg ttttgatgaa ttagaagcac agcaagaaag tgcatctgct 240
ttgaaaaaag atgtcactaa gctgaaatct gatgtcaaat cgttagacaa aaaactcgat 300
aaagttttat ctcttcttga agggcagcaa aaaacacaag acgagttaaa agaaacgatt 360
caaaaacaaa ttaaaactca aggtgagcag cttcaggctc agctgttaga aaaacaagaa 420
aaattagctg aaaagccaaa ggcagaagct aaatctgaag caaaaccatc aaacgctcaa 480
aaaactgagc agccggctcg caag 504
CA 02363803 2001-07-05
WO 00/40730 PCT/C1S00/00364
6
<210> 7
<211> 168
<212> PRT
<213> Bacillus megaterium
<400> 7
Met Asn Arg Glu Glu Phe Ser Gln Leu Met Gly Asn Val Leu Asn Met
1 5 10 15
Asn Leu Gln Tyr Gln Gln Ala Val Asn Glu Val Thr Gly Arg Tyr Leu
25 30
IS His Gln Val Asn Val Pro Thr Lys Glu Asp Val Ala Asn Val Ala Ser
35 40 45
Leu Val Ile Asn Val Glu Glu Lys Val Glu Leu Leu Glu Glu Gln Phe
50 55 60
Asp Asp Arg Phe Asp Glu Leu Glu Ala Gln Gln Glu Ser Ala Ser Ala
65 70 75 80
Leu Lys Lys Asp Val Thr Lys Leu Lys Ser Asp Val Lys Ser Leu Asp
85 90 95
Lys Lys Leu Asp Lys Val Leu Ser Leu Leu Glu Gly Gln Gln Lys Thr
100 105 110
Gln Asp Glu Leu Lys Glu Thr Ile Gln Lys Gln Ile Lys Thr Gln Gly
115 120 125
Glu Gln Leu Gln Ala Gln Leu Leu Glu Lys Gln Glu Lys Leu Ala Glu
130 135 140
Lys Pro Lys Ala Glu Ala Lys Ser Glu Ala Lys Pro Ser Asn Ala Gln
145 150 155 160
Lys Thr Glu Gln Pro Ala Arg Lys
16 5
<210> 8
<211> 741
<212> DNA
<213> Bacillus megaterium
<400> 8
atgacaacat tacaaggtaa agtagcaatc gtaacaggcg gatctaaagg tatcggggca 60
gcaattacac gtgagcttgc ttctaatgga gtaaaagtag cagtaaacta taacagcagt 120
aaagaatctg cagaagcaat tgtaaaagaa attaaagaca acggcggaga agctattgcg 180
gttcaagctg acgtgtctta tgtagatcaa gcaaaacacc taatcgaaga aacaaaagct 240
gcgtttggtc aattagacat tctagtaaac aatgctggaa ttacgcgcga ccgttcattc 300
aagaagttag gtgaagaaga ttggaaaaaa gtaattgatg taaacttaca tagcgtatac 360
aacacaacat cagctgcgct aacgcacctt ttagaatctg aaggtggtcg tgttatcaat 420
atttcatcaa ttattggtca agcgggcgga tttggtcaaa caaactactc agctgctaaa 480
gcaggtatgc taggattcac taaatcatta gctcttgaac tagctaagac aggcgtaacg 540
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
7
gttaatgcaa tttgcccagg atttattgaa acggaaatgg tgatggcaat tcctgaagat 600
gttcgtgcaa aaattgttgc gaaaattcca actcgtcgct taggtcacgc tgaagaaatt 660
gcacgtggag ttgtttactt agcaaaagac ggcgcgtaca ttacaggaca acagttaaac 720
attaacggcg gcttatacat g 741
<210> 9
<211> 247
<212> PRT
<213> Bacillus megaterium
<400> 9
Met Thr Thr Leu Gln Gly Lys Val Ala Ile Val Thr Gly Gly Ser Lys
1 5 10 15
Gly Ile Gly Ala Ala Ile Thr Arg Glu Leu Ala Ser Asn Gly Val Lys
25 30
Val Ala Val Asn Tyr Asn Ser Ser Lys Glu Ser Ala Glu Ala Ile Val
20 35 40 45
Lys Glu Ile Lys Asp Asn Gly Gly Glu Ala Ile Ala Val Gln Ala Asp
50 55 60
Val Ser Tyr Val Asp Gln Ala Lys His Leu Ile Glu Glu Thr Lys Ala
65 70 75 80
Ala Phe Gly Gln Leu Asp Ile Leu Val Asn Asn Ala Gly Ile Thr Arg
85 90 95
Asp Arg Ser Phe Lys Lys Leu Gly Glu Glu Asp Trp Lys Lys Val Ile
100 105 110
Asp Val Asn Leu His Ser Val Tyr Asn Thr Thr Ser Ala Ala Leu Thr
115 120 125
His Leu Leu Glu Ser Glu Gly Gly Arg Val Ile Asn Ile Ser Ser Ile
130 135 140
Ile Gly Gln Ala Gly Gly Phe Gly Gln Thr Asn Tyr Ser Ala Ala Lys
145 150 155 160
Ala Gly Met Leu Gly Phe Thr Lys Ser Leu Ala Leu Glu Leu Ala Lys
165 170 175
Thr Gly Val Thr Val Asn Ala Ile Cys Pro Gly Phe Ile Glu Thr Glu
180 185 190
Met Val Met Ala Ile Pro Glu Asp Val Arg Ala Lys Ile Val Ala Lys
195 200 205
Ile Pro Thr Arg Arg Leu Gly His Ala Glu Glu Ile Ala Arg Gly Val
210 215 220
Val Tyr Leu Ala Lys Asp Gly Ala Tyr Ile Thr Gly Gln Gln Leu Asn
225 230 235 240
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
8
Ile Asn Gly Gly Leu Tyr Met
245
<210> 10
<211> 1086
<212> DNA
<213> Bacillus megaterium
<400> 10
gtggcaattc cttacgtgca agagtgggaa aaattaatca aatcaatgcc aagtgaatat 60
aaaagttctg caagacgttt taagcgtgca tatgaaatta tgacaacaga agcggaaccg 120
gaagttggat taacaccaaa agaggttatt tggaaaaaga acaaagcgaa attatatcgc 180
tatacgccag taaaagataa cctgcataaa acaccaatct tactcgtata tgcattgatc 240
aataaaccgt atattttgga tttaacacct ggaaacagcc ttgttgaata cttattaaac 300
cgcggttttg acgtgtattt gcttgactgg ggaactcctg ggcttgaaga cagcaatatg 360
aagctagatg attatattgt agattatatt ccaaaagcgg cgaaaaaggt gctgcgcact 420
tctaaatctc ctgatttgtc tgttcttggt tactgcatgg gcggaactat gacatctatt 480
tttgctgcat taaatgaaga cttgccgatt aaaaacttaa tttttatgac aagtccattt 540
2o gatttttcgg atacaggttt atacggagca ttcctagatg atcgctactt taatttagat 600
aaagcagtag atacattcgg aaacatccct ccagagatga ttgactttgg aaacaagatg 660
ttaaagccaa tcacgaattt ctacggcccg tatgtaacgt tggtggaccg ttcggaaaat 720
cagcggtttg ttgaaagctg gaagctaatg caaaagtggg ttgctgacgg aatcccattt 780
gctggcgaag cttatcgtca gtggattcgt gacttctatc aacaaaacaa actaatcaat 840
ggtgaacttg aagttcgcgg acgcaaagta gatttaaaaa atattaaagc taatatttta 900
aacattgctg ctagccgtga tcatattgcg atgccgcatc aagtggcagc tttaatggac 960
gctgtttcaa gtgaagataa agagtataaa ttgttgcaaa caggtcacgt atctgttgta 1020
tttggtccaa aagcagtgaa ggaaacatat ccttcaatcg gcgattggct agaaaaacgc 1080
tctaaa 1086
<210> 11
<211> 362
<212> PRT
<213> Bacillus megaterium
<400> 11
Met Ala Ile Pro Tyr Val Gln Glu Trp Glu Lys Leu Ile Lys Ser Met
1 5 10 15
Pro Ser Glu Tyr Lys Ser Ser Ala Arg Arg Phe Lys Arg Ala Tyr Glu
20 25 30
Ile Met Thr Thr Glu Ala Glu Pro Glu Val Gly Leu Thr Pro Lys Glu
35 40 45
Val Ile Trp Lys Lys Asn Lys Ala Lys Leu Tyr Arg Tyr Thr Pro Val
55 60
50 Lys Asp Asn Leu His Lys Thr Pro Ile Leu Leu Val Tyr Ala Leu Ile
65 70 75 80
Asn Lys Pro Tyr Ile Leu Asp Leu Thr Pro Gly Asn Ser Leu Val Glu
85 90 95
Tyr Leu Leu Asn Arg Gly Phe Asp Val Tyr Leu Leu Asp Trp Gly Thr
100 105 110
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
9
Pro Gly Leu Glu Asp Ser Asn Met Lys Leu Asp Asp Tyr Ile Val Asp
115 120 125
Tyr Ile Pro Lys Ala Ala Lys Lys Val Leu Arg Thr Ser Lys Ser Pro
130 135 140
Asp Leu Ser Val Leu Gly Tyr Cys Met Gly Gly Thr Met Thr Ser Ile
145 150 155 160
Phe Ala Ala Leu Asn Glu Asp Leu Pro Ile Lys Asn Leu Ile Phe Met
165 170 175
Thr Ser Pro Phe Asp Phe Ser Asp Thr Gly Leu Tyr Gly Ala Phe Leu
180 185 190
Asp Asp Arg Tyr Phe Asn Leu Asp Lys Ala Val Asp Thr Phe Gly Asn
195 200 205
Ile Pro Pro Glu Met Ile Asp Phe Gly Asn Lys Met Leu Lys Pro Ile
210 215 220
Thr Asn Phe Tyr Gly Pro Tyr Val Thr Leu Val Asp Arg Ser Glu Asn
225 230 235 240
Gln Arg Phe Val Glu Ser Trp Lys Leu Met Gln Lys Trp Val Ala Asp
245 250 255
Gly Ile Pro Phe Ala Gly Glu Ala Tyr Arg Gln Trp Ile Arg Asp Phe
260 265 270
Tyr Gln Gln Asn Lys Leu Ile Asn Gly Glu Leu Glu Val Arg Gly Arg
275 280 285
Lys Val Asp Leu Lys Asn Ile Lys Ala Asn Ile Leu Asn Ile Ala Ala
290 295 300
Ser Arg Asp His Ile Ala Met Pro His Gln Val Ala Ala Leu Met Asp
305 310 315 320
Ala Val Ser Ser Glu Asp Lys Glu Tyr Lys Leu Leu Gln Thr Gly His
325 330 335
Val Ser Val Val Phe Gly Pro Lys Ala Val Lys Glu Thr Tyr Pro Ser
340 345 350
Ile Gly Asp Trp Leu Glu Lys Arg Ser Lys
355 360
<210> 12
<211> 39
<212> DNA
<213> Artificial Sequence
SS
<220>
<223> Description of Artificial Sequence: Synthetic
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
<400> 12
aayacrgtna aataynnnac rgtnatynnn gcdatgatg 39
5
<210> 13
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
<400> 13
gcdatyccdt aygtncarga agghttyaaa 30
<210> 14
<211> 19
<212> DNA
<213> SYNTHETIC
<400> 14
gcttcatgcg tgcggtttg 19
<210> 15
<211> 22
<212> DNA
<213> SYNTHETIC
<400> 15
ggaccgttcg gaaaatcagc gg 22
<210> 16
<211> 20
<212> DNA
<213> SYNTHETIC
<400> 16
cccctttgtc cattgttccc 20
<210> 17
<211> 19
<212> DNA
<213> SYNTHETIC
<400> 17
ccatgtagat tccaccctc 19
<210> 18
~5 <211> 19
<212> DNA
<213> SYNTHETIC
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
11
<400> 18
ctccatctcc tttcttgtg 19
10
<210> 19
<211> 17
<212> PRT
<213> Bacillus megaterium
<400> 19
Lys Val Phe Gly Arg Xaa Glu Leu Ala Ala Ala Met Lys Arg Xaa Gly
1 5 10 15
Leu
<210> 20
<211> 15
<212> PRT
<213> Bacillus megaterium
<400> 20
Asn Thr Val Lys Tyr Xaa Thr Val Ile Xaa Ala Met Xaa Xaa Gln
1 5 10 15
<210> 21
<211> 11
<212> PRT
<213> Bacillus megaterium
<400> 21
Ala Ile Pro Tyr Val Gln Glu Xaa Glu Lys Leu
1 5 10
<210> 22
<211> 813
<212> DNA
<213> Bacillus megaterium
<400> 22
atggatgcat cacttttgtt agagtatgga tgggtattgc tagtgctggt tgcattagaa 60
ggaattttgg cggcggataa tgctcttgtg atggctatta tggtcaaaca tttaccggaa 120
gaaaaacgca agaaggcatt attttacgga ttagccggtg cctttatttt tagatttggt 180
tcgttgttct tgatttcatt tttagtcgac gtatggcagc ttcaagctat aggagccatt 240
tacttattgt tcatttccat taatcatatt gtgaagcgat atgtgaaaaa agacgatcat 300
gaaaaagtga aagaagcaga cgagaaaaag ggctcaggtt tctggatgac ggttttaaaa 360
gtagaaatag cagacattgc ttttgccgtt gattcaattt tggccgctgt ggctctcgcc 420
gttacgttgc caacaacaaa tcttcctcaa attggcggac tcgacggcgg acaattcttg 480
gtgatcttcg ccggaggaat tatgggatta attattatgc gttttgctgc aacttggttc 540
gtcaagctat taaatacgcg cccaggccta gaaacggcgg cttttgctat tgtaggctgg 600
gtaggagtta agttagcggt ctataccctt gctcatccag agttaggtat tattaatgaa 660
catttccctg aatcaaaagt gtggaaaatt acgttttgga ttgtgttact tggcatagct 720
gcttcaggct ggtttctatc taaaaataaa gaacaaactg atcttgaagg ctcagagaaa 780
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
12
gaaaaagaat cgttaaaaaa aattgaaaat caa 813
<210> 23
<211> 271
<212> PRT
<213> Bacillus megaterium
<400> 23
Met Asp Ala Ser Leu Leu Leu Glu Tyr Gly Trp Val Leu Leu Val Leu
1 5 10 15
Val Ala Leu Glu Gly Ile Leu Ala Ala Asp Asn Ala Leu Val Met Ala
25 30
Ile Met Val Lys His Leu Pro Glu Glu Lys Arg Lys Lys Ala Leu Phe
35 40 45
Tyr Gly Leu Ala Gly Ala Phe Ile Phe Arg Phe Gly Ser Leu Phe Leu
50 55 60
Ile Ser Phe Leu Val Asp Val Trp Gln Leu Gln Ala Ile Gly Ala Ile
65 70 75 80
Tyr Leu Leu Phe Ile Ser Ile Asn His Ile Val Lys Arg Tyr Val Lys
85 90 95
Lys Asp Asp His Glu Lys Val Lys Glu Ala Asp Glu Lys Lys Gly Ser
100 105 110
Gly Phe Trp Met Thr Val Leu Lys Val Glu Ile Ala Asp Ile Ala Phe
115 120 125
Ala Val Asp Ser Ile Leu Ala Ala Val Ala Leu Ala Val Thr Leu Pro
130 135 140
Thr Thr Asn Leu Pro Gln Ile Gly Gly Leu Asp Gly Gly Gln Phe Leu
145 150 155 160
Val Ile Phe Ala Gly Gly Ile Met Gly Leu Ile Ile Met Arg Phe Ala
165 170 175
Ala Thr Trp Phe Val Lys Leu Leu Asn Thr Arg Pro Gly Leu Glu Thr
180 185 190
Ala Ala Phe Ala Ile Val Gly Trp Val Gly Val Lys Leu Ala Val Tyr
195 200 205
Thr Leu Ala His Pro Glu Leu Gly Ile Ile Asn Glu His Phe Pro Glu
210 215 220
Ser Lys Val Trp Lys Ile Thr Phe Trp Ile Val Leu Leu Gly Ile Ala
225 230 235 240
Ala Ser Gly Trp Phe Leu Ser Lys Asn Lys Glu Gln Thr Asp Leu Glu
245 250 255
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
13
Gly Ser Glu Lys Glu Lys Glu Ser Leu Lys Lys Ile Glu Asn Gln
260 265 270
S <210> 24
<211> 708
<212> DNA
<213> Bacillus megaterium
<400> 24
atgctcacaa aagttcaaac gcctccatcg cttgaaacgc ttgtactgac gattcagcaa 60
ggggataaac aattacataa tgaaatgatt caacaatata aaccgtttat tgctaaagtt 120
gtttcagctg tatgtaaacg ttatataagt gaagctgacg atgaatttag cattggtctg 180
attgcattta atgaagccat tgaaaattac acaatccaaa aaggacgatc tcttcttgca 240
tttgcggaac ttattattaa aagaagagta atcgactata ttcgaaaaga aaagcgaaat 300
caaacgctgc tctataaccg aattgaaaat gaaggtttta ttcaaggtaa ggtagaaagg 360
gatatatcgc tttctaacta taaaaggcaa agtgaaactt catatattca agaggaaatg 420
acttattttt gtcaggcgct aaaattgttt aaattaactc ttgaagacat tattaacacg 480
tctcctaaac ataaggatgc aaggggaaat gcagtggaag ttgcatcttt tatcgtcaat 540
gaaaaagaat taaaagataa gctgttttta aagcggcagc ttcctattcg cttgattgaa 600
aaacatgtca aagtaagccg gaaaacaatt gaaagaaacc gtaaatatat tatcgcgatg 660
gttattatat tagcggggga ctacgtgtat ttaaaagact atattatg 708
<210> 25
<211> 236
<212> PRT
<213> Bacillus megaterium
<400> 25
Met Leu Thr Lys Val Gln Thr Pro Pro Ser Leu Glu Thr Leu Val Leu
1 5 10 15
Thr Ile Gln Gln Gly Asp Lys Gln Leu His Asn Glu Met Ile Gln Gln
3~ 20 25 30
Tyr Lys Pro Phe Ile Ala Lys Val Val Ser Ala Val Cys Lys Arg Tyr
40 45
Ile Ser Glu Ala Asp Asp Glu Phe Ser Ile Gly Leu Ile Ala Phe Asn
55 60
Glu Ala Ile Glu Asn Tyr Thr Ile Gln Lys Gly Arg Ser Leu Leu Ala
65 70 75 80
Phe Ala Glu Leu Ile Ile Lys Arg Arg Val Ile Asp Tyr Ile Arg Lys
85 90 95
Glu Lys Arg Asn Gln Thr Leu Leu Tyr Asn Arg Ile Glu Asn Glu Gly
~0 100 105 110
Phe Ile Gln Gly Lys Val Glu Arg Asp Ile Ser Leu Ser Asn Tyr Lys
115 120 125
Arg Gln Ser Glu Thr Ser Tyr Ile Gln Glu Glu Met Thr Tyr Phe Cys
130 135 140
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
14
Gln Ala Leu Lys Leu Phe Lys Leu Thr Leu Glu Asp Ile Ile Asn Thr
145 150 155 160
Ser Pro Lys His Lys Asp Ala Arg Gly Asn Ala Val Glu Val Ala Ser
165 170 175
Phe Ile Val Asn Glu Lys Glu Leu Lys Asp Lys Leu Phe Leu Lys Arg
180 185 190
Gln Leu Pro Ile Arg Leu Ile Glu Lys His Val Lys Val Ser Arg Lys
195 200 205
Thr Ile Glu Arg Asn Arg Lys Tyr Ile Ile Ala Met Val Ile Ile Leu
210 215 220
Ala Gly Asp Tyr Val Tyr Leu Lys Asp Tyr Ile Met
225 230 235
<210> 26
<211> 957
<212> DNA
<213> Bacillus megaterium
<400> 26
atgccacaac catggaaacg acgagtaagg cagatgtctt cagctcaaat tattgttaca 60
ttttacatag tggctgttac gcttgggttt ctattactta gtattccaga agctttaagg 120
ccaggagcaa agttagcatt tattgatcgc ttatttattg ccgttagtgc ggtaagtgta 180
acagggctga cacctgtctc gactccagat acatttagta caacgggcta ttttttactc 240
3o gtttttattt ttcaaatcgg tggtattggt gtaatgacac tcagtacatt tatttggatg 300
attttaggta aaaaaatcgg tctgaaggaa cgtcagctca ttatgacgga ccataatcaa 360
tcccgtttat caggattagt tgatttgatg agaaatattt tatttattat ttttgccatt 420
gaactagttg gcgccattat tttagggtta cattttctcc gttattattc gagctggaca 480
gatgcgtttt tgcatggttt ctttgcttct gtcagtgcta caacaaatgc tggcttcgat 540
attacaggat cttcatttat tccgtatgcc catgattatt tcgtacaagt ggtaaccgtt 600
attttaatta cgcttggagc gattggattc cctgtattaa ttgaaatcaa gcactatttt 660
ttaacattta aagataagcg taaatttcaa ttttcgctat ttacgaagct aacgactatt 720
atgttttttc tgctgttagg agggggaaca atcttgattc ttgtgctaga gcattcagga 780
tttctagcag ataagtcttg ggatgaatcg tttttttatg cgtttttcca atccgctgcc 840
acaaggagcg gaggagtggc gaccatgaat attaatgagt tttcacttcc tacgttaatt 900
atgatgagcg cgatgatgtt tatcggtgct tcaccgagtt cagtaggggg aggaatt 957
<210> 27
<211> 319
<212> PRT
<213> Bacillus megaterium
<400> 27
Met Pro Gln Pro Trp Lys Arg Arg Val Arg Gln Met Ser Ser Ala Gln
1 5 10 15
Ile Ile Val Thr Phe Tyr Ile Val Ala Val Thr Leu Gly Phe Leu Leu
20 25 30
Leu Ser Ile Pro Glu Ala Leu Arg Pro Gly Ala Lys Leu Ala Phe Ile
35 40 45
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
Asp Arg Leu Phe Ile Ala Val Ser Ala Val Ser Val Thr Gly Leu Thr
50 55 60
5 Pro Val Ser Thr Pro Asp Thr Phe Ser Thr Thr Gly Tyr Phe Leu Leu
65 70 75 80
Val Phe Ile Phe Gln Ile Gly Gly Ile Gly Val Met Thr Leu Ser Thr
85 90 95
Phe Ile Trp Met Ile Leu Gly Lys Lys Ile Gly Leu Lys Glu Arg Gln
100 105 110
Leu Ile Met Thr Asp His Asn Gln Ser Arg Leu Ser Gly Leu Val Asp
115 12 0 12 5
Leu Met Arg Asn Ile Leu Phe Ile Ile Phe Ala Ile Glu Leu Val Gly
130 135 140
Ala Ile Ile Leu Gly Leu His Phe Leu Arg Tyr Tyr Ser Ser Trp Thr
145 150 155 160
Asp Ala Phe Leu His Gly Phe Phe Ala Ser Val Ser Ala Thr Thr Asn
165 170 175
Ala Gly Phe Asp Ile Thr Gly Ser Ser Phe Ile Pro Tyr Ala His Asp
180 185 190
Tyr Phe Val Gln Val Val Thr Val Ile Leu Ile Thr Leu Gly Ala Ile
195 200 205
Gly Phe Pro Val Leu Ile Glu Ile Lys His Tyr Phe Leu Thr Phe Lys
210 215 220
Asp Lys Arg Lys Phe Gln Phe Ser Leu Phe Thr Lys Leu Thr Thr Ile
225 230 235 240
Met Phe Phe Leu Leu Leu Gly Gly Gly Thr Ile Leu Ile Leu Val Leu
245 250 255
Glu His Ser Gly Phe Leu Ala Asp Lys Ser Trp Asp Glu Ser Phe Phe
260 265 270
Tyr Ala Phe Phe Gln Ser Ala Ala Thr Arg Ser Gly Gly Val Ala Thr
275 280 285
Met Asn Ile Asn Glu Phe Ser Leu Pro Thr Leu Ile Met Met Ser Ala
290 295 300
Met Met Phe Ile Gly Ala Ser Pro Ser Ser Val Gly Gly Gly Ile
305 310 315
<210> 28
<211> 195
<212> DNA
<213> Bacillus megaterium
CA 02363803 2001-07-05
WO 00/40730 PCT/US00/00364
16
<400> 28
atggctagaa caaataaact attaacacca ggagtagaac aatttttaga tcaatataaa 60
tatgaaatcg ctcaagaatt tggggtaact ctaggttctg acactgctgc acgcagcaac 120
ggttcagtag gcggagaaat cacaaaacgc ttggtgcaac aagctcaagc tcacttaagc 180
ggcagcacac aaaaa 195
<210> 29
<211> 65
<212> PRT
<213> Bacillus megaterium
<400> 29
Met Ala Arg Thr Asn Lys Leu Leu Thr Pro Gly Val Glu Gln Phe Leu
1 5 10 15
Asp Gln Tyr Lys Tyr Glu Ile Ala Gln Glu Phe Gly Val Thr Leu Gly
25 30
Ser Asp Thr Ala Ala Arg Ser Asn Gly Ser Val Gly Gly Glu Ile Thr
35 40 45
Lys Arg Leu Val Gln Gln Ala Gln Ala His Leu Ser Gly Ser Thr Gln
50 55 60
Lys
30