Note: Descriptions are shown in the official language in which they were submitted.
CA 02365278 2001-10-26
- 1 -
Case 12/203 DI Fa/dc
10 BOEHRINGER INGELHEIM INTERNATIONAL GMBH
55216 Ingelheim am Rhein (D)
Title: Tumour-associated antigen
4
CA 02365278 2001-10-26
- 2 -
The invention relates to the immunotherapy of tumour
diseases.
The immune system has the task of protecting the body from
a number of different microorganisms and actively fighting
these microorganisms. The importance of an :intact immune
system is apparent particularly in the case of inherited
or acquired immunodeficiencies. The use of prophylactic
vaccine programmes proved in many cases to be an extremely
effective and successful immunological intervention in the
fight against viral or bacterial infectious diseases. It
has also been found that the immune system is also
involved to a large extent in eliminating tumour cells.
Recognition of the tumour associated antigens (TAAs) by
components of the immune system plays a crucial role. In
the broadest sense, any (peptidic or non-peptidic)
component of a tumour cell which is recognised by an
element of the immune system and leads to stimulation of
an immune response, can act as an immunogenic tumour
antigen. Those tumour antigens which not only evoke an
immunological reaction but also cause rejection of the
tumour are of particular importance. The identification
of specific antigens which are able to provoke an
immunological reaction of this kind constitutes a major
step in developing a molecularly defined tumour vaccine.
Although it is not yet clear which elements of the immune
system are responsible for rejection of the tumour, there
is nevertheless consensus that CD8-expressing cytotoxic ,
T-lymphocytes (CTLs) play a major part (Coulie, 1997).
Particularly in those types of tumour (such as melanoma
r
and kidney carcinoma) which have a~relativel.y high
spontaneous remission rate, a correlation has been found
between the clinical progress and the increased appearance
of CD8+- and CD4+-T-cells (Schendel et al., 1993;
Mackensen et al., 1993; Halliday et al., 1995; Kawakami et
al., 1995; Kawakami et al., 1996; Wang, 1997; Celluzzi and
CA 02365278 2001-10-26
- 3 -
Falo, 1998). Specific CTL clones were obtained either
from tumour-infiltrating lymphocytes (TIL) or peripheral
mononuclear blood cells (PBMC) after co-cultivation with
generally autologous tumour cells and cytokine stimulation
in vitro. Both in animal models and in human cell culture
systems cultivated in vitro, the T-cell response against
tumour cells was increased by transfection of tumour cells
with cytokines (van Elsas et al., 1997; Gansbacher et al.,
1990; Tepper et al., 1989; Fearon et al., 1990; Dranoff et
al., 1993).
In the light of the correlation between remission and the
involvement of CD8+-T cells, the identification of tumour
associated antigens (TAA) which are recognised by
CD8-positive CTLs is a specific prime objective towards
developing a tumour vaccine (Pardoll, 1998; Bobbins and
Kawakami, 1996). Whether other cell types of the immune
system such as for example CD4+-T-helper cel:Ls play an
important part is not yet clear; a number of studies with
MAGE-3/HLA-A1 peptides in melanoma patients indicated this
(Marchand et al., 1995; Boon et al., 1998). In recent
years a number of TAAs which are recognised by CTLs have
been identified (Boon et al., 1994; van den Eynde and van
der Bruggen, 1997).
T-cells recognise antigens as peptide fragments which are
presented on the cell surfaces of MHC molecules ("major
histocompatibility complex", in man "HLA" - "human
leukocyte antigen"). There are two types of MHC
molecules: MHC-I molecules occur in most cells with a
r
nucleus and present peptides (usually 8-10-mers) which are
produced by proteolytic degradation of endogenous proteins
(so-called "antigen processing"). Peptide: MHC-I
complexes are recognised by CD8-positive CTLs. MHC-II
molecules occur only on so-called "professional antigen-
presenting cells" (APC) and present peptides of exogenous
CA 02365278 2001-10-26
- 4 -
proteins which are absorbed and processed in the course of
endocytosis by APC. Peptide: MHC-II complexes are
recognised by CD4-helper-T cells. By interaction between
the T-cell receptor and peptide:MHC complex, various
effector mechanisms may be triggered which lead to
apoptosis of the target cell in the case of CTLs. This
occurs if either the MHC (e. g. in the case of transplant
rejection) or the peptide (e.g. in the case of
intracellular pathogens) is recognised as foreign. In any
case, not all the presented peptides meet the structural
and functional requirements for effective interaction with
T-cells (as described by Rammensee et al., 1995 and
hereinafter).
In principle, a number of methods of administration are
possible for using TAAs in a tumour vaccine: the antigen
can either be administered as a recombinant protein with
suitable adjuvants or carrier systems or it may be given
as cDNA coding for the antigen in plasmid (DNA vaccine;
Tighe et al., 1998) or viral vectors (Resti:fo, 1997).
Another possibility is to use recombinant bacteria (e. g.
listeria, salmonella) which recombinantly express the
human antigen and have an adjuvant effect as a result of
their additional components (Paterson, 1996; Pardoll,
1998). In all these cases, the antigen has to be
processed and presented by so-called "professional antigen
presenting cells" (APC). Another possibility is to use
synthetic peptides (Melief et al., 1996) which correspond
to the equivalent T-cell epitopes of the antigen and are
either loaded onto the APC from outside (Buschle et al.,
,r
1997; Schmidt et al., 1997) or absorbed by the APC and
transferred intracellularly to the MHC I molecules. The
most therapeutically efficient method of administration of
a specified antigen is generally determined by clinical
trials.
w
CA 02365278 2001-10-26
- 5 -
The antigens or epitopes thereof recognised by the tumour-
specific CTLs include molecules which can come from any
protein classes (e. g. transcription factors, receptors,
enzymes; for a survey see Rammensee et al., 1995; Bobbins
and Kawakami, 1996). These proteins do not necessarily
have to be located on the cell surface, as is necessary
for recognition by antibodies. In order to act as a
tumour specific antigen for recognition by CTLs or in
order to be used for therapy, the proteins must meet
certain conditions: first of all, the antigen should be
expressed exclusively by tumour cells or should occur in
so-called "critical" normal tissues only in smaller
concentrations than in tumours. Critical normal tissues
are essential tissues; an immune reaction directed against
them would have severe, in some cases lethal consequences.
Secondly, the antigen should be present not only in the
primary tumour but also in the metastases. Furthermore,
with a view to broad clinical use of the antigen, it is
desirable for it to be present in high concentrations in
several types of tumour. One further precondition for the
suitability of a TAA as an effective ingredient of a
vaccine is the presence of T-cell epitopes in the amino
acid sequence of the antigen; peptides derived from the
TAA should lead to an in vitro/in vivo T-ce:l1 response
("immunogenic" peptide). Another criterion for selecting
a clinically broadly applicable immunogenic peptide is the
frequency with which the antigen is encountered in a given
population, of patients.
The immunogenic tumour-associated antigens (TAAs), which
,r
have already largely been shown to have T-cell epitopes,
can be divided into a number of categories, including
viral proteins, mutated proteins, overexpressed proteins,
fusion proteins formed by chromosomal translocation,
differentiation antigens, oncofoetal antigens (Van den
CA 02365278 2001-10-26
- 6 -
Eynde and Brichard, 1995; van den Eynde and van der
Bruggen, 1997).
The methods of identifying and characterising TAAs which
form the starting point for the development of a tumour
vaccine are based on the one hand on the use of CTLs which
have already been induced in patients (cellular immune
response) or antibodies (humoral immune response), or are
based on drawing up differential transcription profiles
between tumours and normal tissues. In the former case,
the immunological approach, patient CTLs are used for
screening eukaryotic tumour-cDNA expression libraries
which present the CTL-epitopes via MHC-I molecules (Boon
et al., 1994), whereas by using high affinity patient
antisera prokaryotic cDNA expression libraries, the
presence of TAAs can be searched directly via immunoblot
analysis of the individual plaques (Sahin et. al., 1995).
A combination of CTL reactivity and protein-chemical
processes produces the isolation of peptides isolated from
MHC-I from tumour cells, which are preselected by
reactivity with patient CTLs. The peptides are washed out
of the MHC-I complex and identified by mass spectrometry
(Falk et al., 1991; Woelfel et al., 1994; Cox et al.,
1994). The approaches which use CTLs to characterise
antigens involve substantial costs or are not always
successful, owing to the need to cultivate and activate
CTLs.
Methods of identifying TAAs which are based on comparing
the transcription profile of normal and tumour tissue are
many and varied; these include differential hybridisation,
the establishing of subtraction cDNA banks
("representational difference analysis"; Hubank and
Schatz, 1994; Diatchenko et al., 1996) and the use of DNA
chip technology or the SAGE method (Velculescu et al.,
1995). In contrast to the above-mentioned immunological
CA 02365278 2001-10-26
7 -
method using patient CTLs, when using molecular biological
methods it is necessary to show that the potential antigen
candidates discovered by this method are tumour-specific
(tumour-associated) and do indeed have T-cell epitopes
capable of triggering a cytotoxic T-cell response. In at
least one case (NY-ESO/LAGE-1) an antigen was identified
both by the use of patient sera and by RDA (Chen et al.,
1997; Lethe et al., 1998), and moreover CTL-~epitopes of
this antigen and a simultaneous spontaneous humoral and
T-cell response were described in one patient (lager et
al., 1998).
The aim of the present invention was to provide a new
tumour-associated antigen (TAA).
This objective was achieved by first establishing a cDNA
subtraction library by RDA (representational difference
analysis) between a lung adenocarcinoma cell line (A549)
and normal lung tissue. In order to select the antigens
which were overexpressed in the tumour, the cDNA clones
obtained were then sequenced and compared with sequences
available in databanks. Among the genes identified there
were 321 unknown genes for which there were mostly EST
entries (expressed sequence tags) in the databank. After
further qualitative PCR analysis in cDNA libraries of
critical normal tissues and immunoprivileged tissues as
well as detailed databank searches, the number of
candidate clones was restricted to 56 whose ESTs did not
derive from critical normal tissue. It was established by
RT-PCR that three of the 56 clones investigated showed
expression mainly indifferent tumour tissues and little
or no expression in normal tissue. The quantitative
comparison (using PCR) of the expression of one of the
clones (B99) between tumour tissue and normal tissue
showed overexpression of the B99 cDNA in various tumours.
The expression profile analysed by Northern blot also
CA 02365278 2001-10-26
-
showed that B99 had no or only poor transcription in the
normal tissues investigated.
The human B99 cDNA was cloned; the sequence obtained is
shown in SEQ ID N0:1. The sequence analysis of the cloned
human B99 cDNA showed that from position 427 to position
1743 there is a continuous open reading frame which, at
the nucleotide and protein level, is very similar to the
open reading frame of beta-1,3-galactosyl-o-glycosyl-
glycoprotein beta-1,6-n-acetylglucosaminyltransferase. The
data obtained from Northern Blot experiments lead one to
conclude that the B99 transcript has a length of about
3.0 kb. The cloned region of B99 cDNA is 2216 bp, whilst
the presence of a PolyA tail at the 3'-end of the sequence
is evidence of the completeness of the cDNA in this
region. The difference in the size of the cloned B99-cDNA
compared with the size which can be deduced from the
Northern Blot analysis can be explained by t:he presence of
a PolyA tail of unknown length and an additional sequence
in the 5'-untranslated region of B99. In view of the fact
that there is no continuous reading frame in the 5' region
of the cloned cDNA from position 0 to 427, it can be
concluded that the ATG at position 427 is the start codon
of B99.
Additional information as to the sequence of B99 located
further upstream can be obtained by standard methods of
molecular biology, e.g. by 5'-RACE (rapid amplification of
cDNA ends). In this method, RNA, preferably mRNA, is
reverse transcribed from cells or tissues in which B99 is
r
transcribed (e. g. colon carcinoma tissue or cell lines
derived from lung adenocarcinoma such as A549) and then
ligated with an adaptor of known sequence. A PCR with an
adaptor primer (binds specifically to the adaptor at the
5'-end of the cDNA) and a B99-specific primer (e.g. SEQ ID
N0:8, 10, 11) allows amplification of corresponding B99
CA 02365278 2001-10-26
- 9 -
fragments. These PCR products can be cloned by standard
methods, as described in Example 1, and characterised
particularly by DNA sequencing.
An alternative method of characterising the 5'-end is by
screening cDNA libraries by hybridisation with DNA probes
or antisera which are specific for B99.
If the screening of cDNA libraries does not achieve the
desired outcome, on account of limitations of procedure,
e.g. inefficient reverse transcription caused by marked
secondary structures of the RNA, genomic libraries can be
searched by, for example, isolating clones, as in the
screening of cDNA libraries, by hybridising with DNA
probes specific for B99, said clones containing the
sequence information located upstream of the 5'-end of the
cDNA obtained, e.g. the promoter region of B99.
The isolated cDNA codes for the tumour associated antigen
(TAA) designated B99 with the amino acid sequence given in
SEQ ID N0:2 (B99-1). This sequence is defined by the
start codon at position 427 of the isolated B99-cDNA.
In another attempt at cloning the coding region of B99 in
which cDNA from the lung adenocarcinoma cell line A549 was
used, a sequence was determined which has an insertion of
a nucleotide at position 923, compared with the sequence
shown in SEQ ID N0:1 (SEQ ID N0:3 or SEQ ID N0:5). This
insertion leads to a change in the open reading frame of
B99 with a resulting amended amino acid sequence in the
C-terminal region of 'the B99 protein; the sequence of this
B99 antigen (B99-2) derived from this reading frame is
shown in SEQ ID N0:4. Apart from the insertion, the cDNA
isolated from A549 cells has a nucleotide exchange at
position 622 compared with sequence SEQ ID N0:1. This
nucleotide exchange causes arginine (SEQ ID N0:2, B99-1)
CA 02365278 2001-10-26
- 10 -
to be replaced by tryptophan (SEQ ID N0:4, B99-2) at
position no. 66. Apart from this amino acid exchange the
amino acid sequence of B99-2 up to and including position
166 is identical to B99-1.
The insertion of a nucleotide at position 923 produces a
second potential reading frame from position 845 to 1744
of the sequence shown in SEQ ID N0:3 (or SEQ ID N0:5). A
protein expressed by a cDNA with this reading frame has
the amino acid sequence shown in SEQ ID NO: 6 (B99-3). The
sequence of B99-3 is different from B99-1 from position 1
to 27 and identical to B99-1 from position 28 onwards.
Thus, according to a first aspect, the present invention
relates to a tumour associated antigen designated B99
selected from the group of polypeptides with the amino
acid sequence given in SEQ ID N0:2, SEQ ID rd0:4 or SEQ ID
N0:6.
The amino acid sequences shown in SEQ ID N0:2 (B99-1),
SEQ ID N0:4 (B99-2) and SEQ ID N0:6 (B99-3) may have some
differences, e.g. those caused by the exchamge of amino
acids, if the B99 derivative has the immunogenic
properties desired for use in a tumour vaccine. (An
example of a B99 polymorphism of this kind is the
difference between B99-1 and B99-2 at position 66 caused
by point mutation).
Unless otherwise stated, the term "B99" is used~-
hereinafter to denote B99-1, B99-2 and B99-3.
r
The natural amino acid sequence of B99 (or r_orrespondingly
the sequences of the B99-cDNA) can optionally be modified
by replacing individual amino acids in a B99 CTL-epitope
in order to achieve an increase in the affinity of B99
peptides to MHC-I molecules compared with the natural B99
CA 02365278 2001-10-26
- 11 -
CTL-epitope, and thus bring about increased immunogenicity
and finally greater reactivity to tumours. Modifications
in the region of the B99 epitopes may be carried out on
the whole B99 protein (this is processed by the APCs to
form the corresponding peptides) or on larger B99 protein
fragments or on B99 peptides (cf, below).
According to another aspect the present invention relates
to immunogenic fragments and peptides derived from B99.
The latter are hereinafter referred to as B99 peptides. A
first group are the B99 peptides which trigger a humoral
immune response (induction of antibodies). Such peptides
are selected portions of B99 (at least 12 to 15 amino
acids) which can be determined by so-called prediction
algorithms such as for example the surface probability
plot (Emini et al., 1985), the hydrophobicity blot (Kyte
and Doolittle, 1982) and the antigenic index. (Jameson and
wolf, 1988).
It is known that tumour-associated antigens may have
tumour-specific mutations, which contribute to an
immunological distinction between tumour and normal tissue
(Mandruzzato et al., 1997, Hogan et al., 1998, Gaudi et
al., 1999, Wolfel et al., 1995). In order to determine the
presence of tumour-specific B99 mutations, the B99 cDNA
may be cloned from one or more different tumours,
preferably with probes from the isolated B99 cDNA of the
invention, and the obtained sequences are compared with
B99 cDNAs from normal tissue. It may be expected that
tumour B99 peptides from a sequence section mutated vis a
vis normal tissue wild have an increased immunogenicity in
comparison with B99 peptides from the corresponding
sequence section from normal tissue.
Therefore, the invention relates, in a further aspect, to
B99 peptides derived from tumour-expressed B99, which have
tumor-specific mutations.
CA 02365278 2001-10-26
- 12 -
When selecting B99 peptide candidates the regions of B99-2
and B99-3 which differ from B99-1 deserve special
interest. On the understanding that the insertion of the
B99 DNA which leads to these differences in the amino acid
sequence is a tumour-specific mutation, peptides from this
region can be expected to exhibit greater immunogenicity
than peptides of B99-1. In order to confirm that the
insertion is tumour-specific, antibodies against this
region may be generated and tumour cells investigated for
the expression of B99-2 and B99-3.
B99 Peptides are administered directly or in modified form
(e.g. coupled to~~LH = keyhole limpet hemocyanin) and the
formation of antibodies is determined by normal
immunological assays, e.g. by ELISA.
Other B99 peptides which are preferred within the scope of
the present invention are those which are presented by
MHC-molecules and produce a cellular immune response.
There are two types of MHC-molecules, namely MHC-I
molecules which are recognised by CD8-positive CTLs and
MHC-II molecules which are recognised by CD4-positive
T-helper cells.
In order for a peptide to trigger a cellular immune
response, it must bind to an I~-IC-molecule, whilst the
patient to be,treated must have the MHC molecule in his or
her repertoire. Determining the MHC-subtype of the
patient thus constitutes one of the essential
prerequisites for effective use of 'a peptide in this
patient, with a view to triggering a cellular immune
response.
The sequence of a B99 peptide to be used therapeutically
is determined by the MHC-molecule in questian in terms of
CA 02365278 2001-10-26
- 13 -
the anchor amino acids and length. Defined anchor
positions and length guarantee that a peptide fits the
peptide binding groove of the MHC-molecule of the patient
in question. The result of this is that the immune system
is stimulated and a cellular immune reaction is produced
which is directed against the tumour cells of the patient,
if a peptide derived from a tumour antigen is used.
Immunogenic B99 peptides may be identified by known
methods; one of the basic conditions is the correlation
between MHC-binding and CTL-induction.
Thus, since the sequence of immunogenic peptides can be
predicted on the basis of its peptide binding motif, B99
peptides which constitute CTL-epitopes can be identified
and synthesised on the basis of the B99 protein sequence.
Various methods are available for doing this, which are
used to identify CTL-epitopes of known protein antigens;
e.g. the method described by Stauss et al., 1992 for
identifying T-cell epitopes in human papillama virus.
The allele-specific requirements of each MHC-I allele
product with regard to a peptide which binds to the MHC-
molecule and is presented thereby have been assembled as a
motif (e. g. Falk et al., 1991). Up till now, a large
number of both MHC-peptide motifs and MHC-ligands have
become known. A suitable method, within the scope of the
, present invention, for searching for epitopes of a known
protein which fits a specific MHC-I molecule is described
in a survey by Rammensee et al., 1995. It comprises the
following steps: first, the protein sequence is searched
for fragments which correspond to the anchor motif, whilst
certain variations are possible with regard to peptide
length and anchor occupation. If for example a motif
prescribes a 9-mer with Ile or Leu at the end, 10-mers
with a corresponding C-terminus can also be considered, as
CA 02365278 2001-10-26
- 14 -
can peptides with other aliphatic groups such as Val or
Met at the C-terminus. In this way a number of peptide
candidates is obtained. These are searched for the
presence of as many anchor groups as possib:Le which they
have in common with known ligands and/or to see whether
they have groups which are "preferred" for various
MHC-molecules (according to the Table by Rammensee et al.,
1995). In order to exclude weakly binding peptides,
binding assays are preferably carried out. If the
requirements for the peptide binding for specific
MHC-molecules are known, the peptide candidates can also
be searched for non-anchor groups which have a negative or
positive effect on the binding or which indeed make it
possible at all (Ruppert et al., 1993). However, with
this method, it should be borne in mind that the peptide
binding motif is not the sole deciding factor when
searching for natural ligands; other aspects, e.g. enzyme
specificity during antigen processing, also contribute to
the identity of the ligand, in addition to 'the specificity
of the MHC-binding. One method which takes account of
these aspects and which is suitable for identifying
immunogenic B99 peptides within the scope of the present
invention was used inter alia by Kawakami et al., 1995 for
identifying gp100 epitopes on the basis of known
HLA-A*0201 motifs.
The peptides may also be selected for their ability to
bind to MHC-II molecules. The MHC-II binding motif which
extends over nine amino acids has a higher degree of
degeneration in the anchor positions than t:he MHC-I
binding motif. Methdds have recently been developed,
based on X-ray structural analysis of MHC-II molecules,
which allow accurate analysis of the MHC-II binding motifs
and, based on that, variations in the peptide sequence
(Rammensee et al., 1995, and the original literature cited
therein). Peptides which bind to MHC-II molecules are
CA 02365278 2001-10-26
- 15 -
typically presented to the CD4-T cells by dendritic cells,
macrophages or B-cells. The CD4-T-cells in turn then
activate CTLs directly in sequence by the release of
cytokine, for example, and increase the efficiency of
antigen presentation by APC (dendritic cells, macrophages
and B-cells).
Recently, databanks and prediction algorithms have become
available which allow more reliable prediction of peptide
epitopes which bind to a specific MHC molecule.
Within the scope of the present invention, using the
algorithm described by Parker et al., 1994 and Rammensee
et al., 1995, candidate peptides have been identified for
the most important HLA-types, especially for HLA-A1,
-A*0201, -A3, -B7, -B14 and -B*4403, which c:an be expected
to bind to the corresponding HLA molecules and thus
constitute immunogenic CTL-epitopes; the peptides
discovered are listed in Table 2. Similarly, possibly
using other algorithms which take account of the different
characteristics of the peptides (hydrophobicity, charge,
size) or requirements made of the peptides, such as the
3D structure of the HLA-molecule, it is possible to find
potential peptide epitopes; this also applies to peptide
epitopes of other HLA types.
After selecting B99-peptide candidates using the methods
described, their MHC-binding is tested by peptide binding
assays. First, the immunogenicity of the peptides with
good binding properties is determined (stability of the
peptide-MHC interaction correlates'in most cases with
immunogenicity; van der Burg et al., 1996). In order to
determine the immunogenicity of the selected peptide or
peptide equivalent, methods may be used as described, for
example, by Sette et al., 1994, combined with quantitative
MHC-binding assays. Alternatively, the immunogenicity of
CA 02365278 2001-10-26
- 16 -
the selected peptide may be tested by in vitro CTL-
induction using known methods (as described hereinafter
for ex vivo CTL-induction). The principle of the method,
carried out in several steps, for selecting peptides which
are capable of triggering a cellular immune response is
described in WO 97/30721, the contents of which are hereby
expressly referred to. A general strategy for obtaining
efficient immunogenic peptides which is suitable within
the scope of the present invention has also been described
by Schweighoffer, 1997.
Instead of using the original peptides which. fit the
binding groove of MHC-I or MHC-II molecules, i.e. peptides
which are derived unaltered from B99, variations may be
carried out, adhering to the minimum requirements
regarding anchor positions and length specified on the
basis of the original peptide sequence, provided that
these variations not only do not impair the effective
immunogenicity of the peptide which is made up of its
binding affinity to the MHC-molecule and its ability to
stimulate T-cell receptors, but preferably enhance it. In
this case, artificial peptides or peptide equivalents are
thus used which are designed to correspond t.o the
requirements regarding binding ability to an MHC-molecule.
Peptides modified in this way are referred to as
"heteroclitic peptides". They may be obtained by the
following methods:
First of all, the epitopes of MHC-I or MHC-II ligands or
variations thereof are undertaken,.e.g. using the
principle described by Rammensee et al., 1995. The length
of the peptide preferably corresponds to a minimum
sequence of 8 to 10 amino acids with the necessary anchor
amino acids, if the peptide is being matched to MHC-I
molecules.
CA 02365278 2001-10-26
- 17 -
If desired, the peptide may also be extended at the
C- and/or N-terminus provided that this extension does not
affect the ability to bind to the MFiC-molecule and the
extended peptide can be cellularly processed down to the
minimum sequence.
The modified peptides are then investigated for their
recognition by TILs (tumour infiltrating lymphocytes), for
CTL-induction and for increased MHC-binding and
immunogenicity, as described by Parkhurst et. al., 1996 and
Becker et al., 1997. ,
Another method of finding peptides with greater
immunogenicity than that of the natural B99 peptides,
which is suitable for the purposes of the present
invention, consists in screening peptide lix>raries with
CTLs which recognise the B99 peptides naturally occurring
on tumours, as described by Blake et al., 1996; in
connection with this it is proposed to use combinatorial
peptide libraries in order to design molecules which
imitate tumour epitopes recognised by MHC-I-restricted
CTLs.
The B99 polypeptides according to the present invention or
immunogenic fragments or peptides derived therefrom may be
produced recombinantly or by peptide synthesis, as
described in WO 96/10413,,the disclosure of which is
hereby referred to. Fox recombinant production, the
corresponding DNA molecule is inserted by standard methods
in an expression vector, transfecte'd into a suitable host
cell, the host is cultivated under suitable expression
conditions and the protein is purified. Conventional
methods may be used for the chemical synthesis of B99
peptides, e.g. automatic peptide synthesisers which are
commercially available.
CA 02365278 2001-10-26
- 18 -
Alternatively to natural B99 peptides or heteroclitic
peptides it is also possible to use substances which
imitate such peptides, e.g. "peptidomimetics" or "retro-
inverse peptides". In order to test these molecules with
regard to their therapeutic use in a tumour vaccine the
same methods are used as described above for the natural
B99 peptides or B99 peptide equivalents.
The TAA designated B99 according to the present invention
and the protein fragments, peptides or peptide equivalents
or peptidomimetics derived therefrom may be used in cancer
therapy, e.g. in order to induce an immune response to
tumour cells which express the corresponding antigen
determinants. They are preferably used for the treatment
of B99-positive tumours, particularly in kidney cell-,
lung-, colon-, pancreas-, breast- and stomach-carcinoma.
The immune response in the form of induction of CTLs can
be achieved in vivo or ex vivo.
In order to induce CTLs in vivo, a pharmaceutical
composition containing as active component the TAA B99 or
fragments or a peptide or peptides derived therefrom, is
administered to a patient suffering from a t,umoral disease
associated with the TAA, whilst the quantity of TAA
(peptide) must be sufficient to obtain an effective CTL
response. to the antigen-bearing tumour.
Thus, according to another aspect, the invention relates
to a pharmaceutical composition for parenteral, topical,
oral or local administration. Preferably, the composition
is used parenterally, e.g. for subcutaneous, intradermal
or intramuscular application. The B99-TAAs/peptides are
dissolved or suspended in a pharmaceutically acceptable,
preferably aqueous, carrier. The composition may also
CA 02365278 2001-10-26
- 19 -
contain conventional adjuvants such as buffers etc. The
TAAs/peptides may be used on their own or in conjunction
with adjuvants, e.g. incomplete Freund's adjuvant,
saponines, aluminium salts or, in a preferred embodiment,
polycations such as polyarginine or polylysine. The
peptides may also be bound to components which aid CTL
induction or CTL activation, e.g. T-helper peptides,
lipids or liposomes, or they are administered together
with these substances and/or together with immunostimulant
substances, e.g. cytokines (IL-2, IFN-y). Methods and
formulations which are suitable for the preparation and
administration of the pharmaceutical composition according
to the invention are described in WO 95/04542 and
WO 97/30721, the disclosure of which is hereby referred
to.
B99 (fragments) or B99 peptides may also be used to
trigger a CTL response ex vivo. An ex vivo CTL response
to a tumour which expresses B99 is induced by incubating
the CTL-precursor cells together with APCs and B99
peptides or B99 protein. The activated CTLs are then
allowed to expand, whereupon they are re-administered to
the patient. Alternatively, APCs may be loaded with B99
peptides, which may lead to efficient activation of
cellular immune reactions against B99 positive tumours
(Mayordomo et al., 1995; Zitvogel et al., 1996). One
suitable method of loading peptides onto cells, e.g.
dendritic cells, is disclosed in WO 97/19169.
In one embodiment of the invention a combination of
r
several different B99 peptides or B99 peptide equivalents
is used. In another embodiment, B99 peptides are combined
with peptides derived from other TAAs. The choice of
peptides for such combinations is made in the light of
detecting different MHC-types in order to cover the
broadest possible patient population, and/or it is aimed
CA 02365278 2001-10-26
- 20 -
at the broadest possible spectrum of indications, by
combining peptides from several different tumour antigens.
The number of peptides in a pharmaceutical composition can
fluctuate over a wide range, but typically a clinically
usable vaccine contains 1 to 15, preferably 3 to 10
different peptides.
The peptides according to the invention may also be used
as diagnostic reagents. For example, the peptides may be
used to test the response of a patient to the humoral or
cellular immune response evoked by the immunogenic
peptide. This provides a possibility of improving a
treatment procedure. For example, depending on the form
of administration'(peptide, total protein or DNA vaccine)
of the TAA, the increase of precursor T-cells in the PBLs
which show reactivity against the defined peptide epitope
can be investigated (Bobbins and Kawakami, 1.996 and the
references cited therein). Moreover, the peptides or the
total protein or antibodies directed against: the TAA may
be used to characterise the progression of a B99-positive
tumour (e. g. by immunohistochemical analyses of primary
tumour and metastases). A strategy of this kind has
already proved successful in many cases, e.g. detecting
the oestrogen receptor as the basis for deciding on
endocrine therapy in breast cancer; c-erbB-2 as the
relevant marker in the prognosis and course of therapy in
breast cancer (Ravaioli et al., 1998; Revillion et al.,
1998); PSMA (prostate specific membrane antigen) as a
marker for epithelial cells of prostate carcinoma in the
serum or by using a 111In-labelled monoclonal antibody
.r
against PSMA in immunoscintigraphy~on prostate carcinoma
(Murphy et al., 1998 and references included therein); CEA
(carcinoembryonic antigen) as a serological marker for the
prognosis and progression in patients suffering from
colorectal carcinoma (Jessup and Loda, 1998).
CA 02365278 2001-10-26
- 21 -
According to another aspect the present invention relates
to isolated DNA molecules coding for a protein with the
immunogenic properties of B99 or fragments thereof.
In one aspect, the present invention relates to an
isolated DNA molecule which contains a polynucleotide with
the sequence shown in SEQ ID N0:1 or which contains a
polynucleotide which hybridises with a polynucleotide of
the sequence shown in SEQ ID N0:1, under stringent
conditions.
By "stringent hybridization conditions" as used herein is
meant overnight incubation at 42°C in a solution
comprising: 50~ formamide, 5x SSC (1X SSC = 150 mM NaCl,
l5mM trisodium citrate), 50 mM sodium phosphate (pH 7.6),
5x Denhardt's solution, 10~ dextran sulfate, and 20 ug/ml
denatured, sheared salmon sperm DNA, followed by washing
the filters in 0.1x SSC at about 65°C, or equivalent
conditions.
In another aspect, the present invention relates to an
isolated DNA molecule which contains a polynucleotide with
the sequence shown in SEQ ID N0:3 (or SEQ II) N0:5) or
which contains a polynucleotide which hybridises with a
polynucleotide of the sequence shown in SEQ ID N0:3 (or
SEQ ID N0:5), under stringent conditions.
The DNA molecules according to the invention or fragments
thereof code for (poly)peptides designated B99 (B99-1,
B99-2 or B99-3) with the amino acid sequence shown in
.r
SEQ ID N0:2, SEQ ID N0:4 or SEQ ID N0:6 or for protein
fragments or peptides derived therefrom; this includes DNA
molecules which show deviations from the sequence shown in
SEQ ID N0:1 or SEQ ID N0:3 or SEQ ID N0:5 as a result of
the degeneration of the genetic code.
CA 02365278 2001-10-26
- 22 -
The invention also relates to DNA molecules which have
deviations from the sequence shown in SEQ ID N0:1 or
SEQ ID N0:3 (or SEQ ID N0:5) caused by the conservative
exchange of amino acids, if they code for a B99 derivative
or fragments or peptides with the immunogenic properties
which are desirable for their use as tumour vaccines.
The optionally modified DNA molecules defined above which
code for B99-1, B99-2 or B99-3 or for fragments thereof
are hereinafter referred to as "B99-DNA molecules", unless
stated otherwise.
The B99 DNA molecules of the present invention or the
corresponding RNAs which are also a subject of the present
invention are used, like the (poly)peptides coded by them,
for immunotherapy of cancer diseases.
In one embodiment of the invention, DNA molecules are used
which code for natural B99 polypeptides. A:Lternatively to
the natural B99 cDNA or fragments thereof it is possible
to use modified derivatives. These comprise sequences
with modifications which code for a protein (fragment) or
peptides with greater immunogenicity, whilst the same
considerations apply to modifications at the DNA level as
apply to the peptides described above. Another type of
modification is the lining up of numerous sequences coding
for immunologically relevant peptides like a string of
beads (Toes et al., 1997). The sequences may also be
modified by the addition of auxiliary elements, e.g.
functions, which ensure more efficient release and
processing of the immunogen (Wu et.al., 1995). For
example, the processing and hence the presentation and
finally the immunogenicity of the antigen can be increased
by the addition of a locating sequence in the
endoplasmatic reticulum ("ER targeting sequence").
CA 02365278 2001-10-26
- 23 -
In another aspect, the present invention relates to a
recombinant DNA molecule which contains B99-DNA.
The B99 DNA molecules of the present invention may be
administered, preferably in recombinant form as plasmids,
directly or as part of a recombinant virus or bacterium.
In theory, any method of gene therapy may be used for
immunotherapy of cancer based on DNA ("DNA vaccine") on
B99-DNA, both in vivo and ex vivo.
Examples of in vivo administration are the direct
injection of "naked" DNA, either by intramuscular route or
using a gene gun, which has been shown to lead to the
formation of CTLs against tumour antigens. Examples of
recombinant organisms are vaccinia virus, adenovirus or
listeria monocytogenes (a summary was provided by Coulie,
1997). Moreover, synthetic carriers for nucleic acids
such as cationic lipids, microspheres, micropellets or
liposomes may be used for in vivo administration of
nucleic acid molecules coding for B99 peptide. As with
peptides, different adjuvants which enhance the immune
response may also be administered, e.g. cytokines, either
in the form of proteins or plasmids coding :for them. The
application may optionally be combined with physical
methods, e.g. electroporation.
An example of ex vivo administration is the transfection
of dendritic cells as described by Tuting, 1997, or other
APCs which are used as cellular cancer vaccine.
r
Thus, according to another aspect,~the present invention
relates to the use of cells which express B99, either per
se or, in optionally modified form, after t:ransfection
with the corresponding coding sequence, in order to
produce a cancer vaccine.
CA 02365278 2001-10-26
- 24 -
In another aspect, the invention relates to antibodies
against B99 or fragments thereof. Polyclonal antibodies
are conventionally obtained by immunising animals,
particularly rabbits, by injecting the antigen or
fragments thereof and subsequently purifying' the
immunogl obul in .
Monoclonal anti-B99-antibodies may be obtained by standard
procedures following the principle described by Kohler and
Milstein, 1975, by immunising animals, particularly mice,
then immortalising antibody-producing cells from the
immunised animals, e.g. by fusion with myeloma cells, and
screening the supernatant of the hybridomas obtained by
immunological standard assays for monoclonal. anti-B99-
antibodies. For therapeutic or diagnostic use in humans,
these animal antibodies may optionally be chimerised in
the conventional way (Neuberger et al., 1984., Boulianne et
al., 1984) or humanised (Riechmann et al., 1.988, Graziano
et al., 1995).
Human monoclonal anti-B99-antibodies (or fragments
thereof) may also be obtained from so-called phage display
libraries (Winter et al., 1994, Griffiths et al., 1994,
Kruif et al., 1995, Mc Guiness et al., 1996) and by means
of transgenic animals (Bruggemann et al., 1996, Jakobovits
et al., 1995).
The anti-B99-antibodies according to the invention may be
,used in immunohistochemical analyses for diagnostic
purposes.
r
In another aspect, the invention relates to the use of
B99-specific antibodies for selectively bringing any
desired substances to or into a tumour which expresses
B99. Examples of such substances are cytotoxic agents or
radioactive nuclides the activity of which consists in
CA 02365278 2001-10-26
- 25 -
damaging the tumour in situ. Because of the tumour-
specific expression of B99, no or very few side effects
can be expected. According to another aspect, substances
for showing up tumours which express B99 may be used, with
the aid of B99 antibodies. This is useful f:or the
diagnosis and evaluation of the treatment. Therapeutic
and diagnostic uses of antibodies which apply to anti-B99-
antibodies are described in VETO 95/33771.
The TAA designated B99 according to the present invention
and the protein fragments, peptides or peptide equivalents
or peptidomimetics derived therefrom may be used in cancer
therapy, e.g. to induce an immune response to tumour cells
which express the corresponding antigen determinants.
They are preferably used for the treatment of B99-positive
tumours, particularly in carcinoma of the kidney cells,
lung, colon, pancreas, breast and stomach.
Due to the preferred expression of B99 in tumour cells, it
may be assumed that this protein has an important function
for the tumour, e.g. for tumour formation, infiltration
and growth. B99(DNA) may therefore be employed in
screening assays for identifying substances which
modulate, in particular inhibit, the activity of this
protein. In an embodiment, such an assay may comprise
introducing the B99 protein, or an active fragement
thereof, into cells or expressing B99 DNA in cells, and
determining the proliferation of the cells :in the presence
or absence of a test substance. Substances with a
proliferation-inhibiting effect can be used for the
r
treatment of tumors with strong B99 expression, in
particular carcinoma of the lung, colon, breast, renal
cell carcinoma and for the treatment of Hodgkin lymphoma.
Summary of the drawings
CA 02365278 2001-10-26
- 26 -
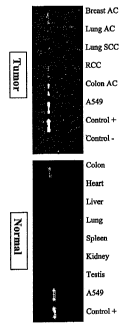
Fig. 1: RT-PCR analysis of cDNA pools of different human
tumour and normal tissues using B99-specific
primers
Fig. 2: RT-PCR analysis of individual cDNAs of different
human tumour and normal tissues using B99-specific
primers
Fig. 3: Transcription of B99 in normal tissues: Northern
Blot analysis of mRNA from 16 normal tissues
Fig. 4: Immunohistochemical analysis of four' different
cases of adenocarcinoma with B99 serum
Fig. 5: MHC stabilization on T2 cells with different
concentrations of B99 peptides
Example 1
RDA (Representational Difference Analysis) of the human
adenocarcinoma cell line of the lung (A549) and normal
lung tissue
The human lung adenocarcinoma cell line A549 (CCL 185)
obtained from the ATCC was cultured in T150 cell culture
bottles. The nutrient medium used was MEM with 10~ heat-
inactivated foetal calf serum and 2 mM L-glutamine. Every
3 to 4 days the cells were cleaved by trypsinisation 1:5
to 1:10 in order to propagate them. After about 80~
confluence was achieved, 4 ml of a trypsin solution
r
(containing per litre: 8 g of NaCl; 0.2 g of KC1, 1.13 g
o f anhydrous Na2HP04 , 0 . 2 g o f KHZ P04 , 10 0 rnl o f 2 . 5 ~
trypsin solution and 1 g of EDTA-Na salt; pH 7.2 to 7.4)
were added to each T150 cell culture flask in order to
harvest the cells. The 4 ml were transferred into a 15 ml
Falcon test tube, 8 ml of PBS were added, the mixture was
CA 02365278 2001-10-26
- 27 -
centrifuged at 1200 rpm in a Haereus bench centrifuge
(Megafuge 2.OR) for 5 minutes at 4°C, the cell pellet was
mixed with 1 ml of lysis buffer (10 mM TrisHCl pH8, 140 mM
NaCl, 1.5 mM MgCl2, 0.5~ NP40), shaken vigorously and
centrifuged in a 2 ml Eppendorf vessel at 1:?000 rpm and at
4°C for 5 minutes in a Sigma bench centrifuge (Sigma
202 MK). The supernatant was transferred into another
Eppendorf vessel and after the addition of 55 ~1 of 20~
SDS solution it was extracted twice with double the volume
of a CHC13/phenol (1:1 v/v) mixture and once with the same
volume of CHC13. The aqueous phase containing RNA was
mixed with 1/10 volume of 3M NaAc (pH5) and twice the
volume of 96~ EtOH and the RNA is precipitated overnight
at -20°C. Starting from 1 mg of total RNA, the
manufacturer's instructions were followed in order to
isolate poly-A(+)RNA using the PolyATtract Kit (Promega).
The storing of the A549 poly-A(+)RNA with a concentration
of 1 mg/ml in DEPC-treated H20 was carried out in aliquots
at -80°C.
In order to carry out representative differential analysis
(RDA; Hubank and Schatz, 1994; Diatchenko et al., 1996)
the poly-A(+)RNA of the lung adenocarcinoma cell line A549
was used as the tester while that of normal lung tissue
(1 mg/ml; Clontech, Palo Alto; #6524-1) was used as
driver. The RDA was carried out using the PCR-selectT'"'
kit (Clontech, Palo Alto) in accordance with the
manufacturer's instructions, except that a rnodifi,ed
primer/adaptor-2-oligonucleotide system having,,the
following sequence was used:
r
5'-TGTAGCGTGAAGACGACAGA.AAGGGCGTGGTACCGAGCTCGAG-3'
(Adaptor-2-alt-1; SEQ ID N0:19),
5'-AGGGCGTGGTACCGAGCTCGAG-3' (nested-PCR-primer-2-alt;
SEQ ID N0:20) and 5'-GGCTCGAGCTC-3' (Adaptor-2-alt-2;
SEQ ID N0:21). The newly generated primer/adaptor
sequences make it possible to excise the relevant cDNA
CA 02365278 2001-10-26
- 28 -
fragments subsequently by means of the presence of three
new restriction enzyme cutting sites (Kpn I, Sac I and
Xho I) in the sequence of the nested-PCR-primer-2-alt
after cloning of the subtracted cDNA fragments into the
pPCRII vector. It was therefore necessary to design a
primer/adaptor sequence with a number of available
restriction enzyme cutting sites because point mutations
were often observed particularly in the primer sequences,
caused by the PCR-amplification steps.
After the synthesis of double-stranded cDNA using
oligo-dT, the cDNA obtained from the tester and driver was
digested with Rsal (RsaI is a 4-base recognising
restriction enzyme and yields 256 by long c:DNA fragments
as a statistical average). Equal parts of tester cDNA
were ligated with either adaptor 1 or 2 and then
separately hybridised with an excess of driver cDNA at
65°C. The two mixtures were then combined and subjected
to a second hybridisation with fresh denatured driver
cDNA. The concentrated tester-specific cDNAs were then
exponentially amplified by PCR with primers specific for
the adaptors 1 or 2. For further concentration an aliquot
from this reaction was subjected to a second PCR with
specific inwardly shifted (nested) primers. The
exponentially amplified cDNA fragments resulting from this
reaction were ligated directly into the pCRII vector
(Invitrogen; "TA cloning vector") and then one third of
the ligation mixture was transfected into competent E.coli
(OneShot~'", Invitrogen) .
r
712 positive transformants (blue-white selection) were
obtained and cultivated in 96-well blocks in LB-Amp medium
(1.3 ml per well) at 37°C for 48 hours. 750 ~.1 of the
E. coli suspensions were used per well for the preparation
of the plasmid DNA (96-well mini preparation method of
QIAgen in accordance with the manufacturer's
CA 02365278 2001-10-26
- 29 -
instructions). The bacterial cultures remaining were
stored as glycerine stock cultures at -80°C.
A cDNA subtraction library consisting of 712 individual
clones was obtained, which was present both in the form of
E. coli glycerine stock cultures and also in. the form of
purified plasmids.
Example 2
DNA sequencing and annotation of TAA candidates
The isolated plasmid-DNA of all 712 clones (see Example 1)
was sequenced by the Sanger method using an ABI Prism
device. The sequences obtained were annotated using the
BioScout Software (LION, Heidelberg) and subjected to
databank comparisons (Genbank). Of 712 clones, 678 were
able to be sequenced and annotated. The rest (34) had
only poly(A) sequences as inserts or corresponded to a
relegated vector or could not be sequenced. Of the 678
annotable sequences, 357 proved to be genes with a known
function. The remaining 321 represented clones coding for
genes of unknown function; 59 of these did not even have
entries in the human EST databank. Known genes were not
treated further. For those unknown genes for which an EST
entry was available, the expression profile was evaluated:
all those ES'~s with >95~ identity (BLAST), which belonged
to the correspondingly experimentally determined sequence
of the subtraction libraries, were examined. In the
r
annotation, a subdivision was made into i) c:ritical normal
tissue, ii) foetal, "disposable" and immunoprivileged
tissue and iii) tumours and tumour cell lines. On the
basis of this "virtual mRNA profile", 200 clones for which
no ESTs were found in group i) were selected for further
experimental analyses (including the 59 clones for which
CA 02365278 2001-10-26
- 30 -
there was no EST entry). In order to narrow down the
candidate clones still further, oligonucleotide primer
pairs were designed and synthesised from the sequences
determined from the 200 selected clones. First of all,
8 different cDNA libraries derived from human tissue
(GibcoBRL SUPERSCRIPTTT') which are directionally cloned in
pCMV SPORT, were tested by qualitative PCR for the
presence of the candidates in question. The cDNA
libraries used originated from heart tissue (#10419-018),
liver (#10422-012), leukocytes (#10421-022), kidney
(#10420-016), lung (#10424-018), testis (#10426-013),
brain (#10418-010) and foetal brain (#10662-013). The PCR
conditions were as follows: 20 ~l of total volume per PCR
mixture contained 1x TaqPol buffer (50 mM KC1, 10 mM
TrisHCl pH9, 0.1~ Triton X-100), 1.5 mM MgCl2, 0.2 mM
dNTPs (Promega), 0.025 U/~1 Taq-DNA-polymerase (Promega),
5 pM of specific oligonucleotide primer SEQ ID N0:7 and
SEQ ID N0:8 and 100 ng of the plasmid DNA to be
investigated. As a control, specific primers for GAPDH
(SEQ ID N0:14 and 15) were used. In order t:o check the
selective evidence the primer pairs were also tested in
parallel on the isolated plasmid. The deter_tability of
fragments of the expected length in one of the critical
normal tissues (heart, liver, lung, kidney and leukocyte)
but not in the cDNA libraries of immunoprivileged tissues
(brain, foetal brain and testis) under these PCR
conditions (1 cycle: 3' 94°C; 35 cycles: 1' 94°C - 1'
55°C
- 1' 72°C; 1 cycle: 7' 72°C) was defined as the selection
criterion. Using this qualitative PCR analysis the number
of candidates could be narrowed down to 56.
.r
Example 3
Transcriptional analysis of the candidate clones in
various tumour and normal tissues
CA 02365278 2001-10-26
- 31 -
For RT-PCR analysis, cDNA pools were used which had been
prepared from 3 ~g of total RNA from 3 different tissues
of the same type. The 9 ~g total RNA per tissue pool of
tumour or normal tissues was reverse transcribed by means
of AMV-RT (Promega) in accordance with the manufacturer's
recommendations. To avoid contamination with genomic DNA,
the RNA was previously incubated with DNAse I (Boehringer
Mannheim). The quality and quantity of the cDNAs were
tested by PCR with GAPDH specific primers SEQ ID N0:14 and
15) after 20 cycles (30", 95°C, 90" 60°C). B99-cDNA was
amplified by 25, 30 and 35 cycles of the program 1' 95°C,
1' 55°C, 1' 72°C with the B99-specific primers according
to SEQ ID N0:7 and 8. The other 55 candidate clones were
investigated analogously with specific primers. The PCR
products were detected by agarose gel electrophoresis and
ethidium bromide staining. For 3 of the 56 clones
investigated, RT-PCR showed increased expression in
different tumour tissues compared with normal tissues. An
example of candidate B99 is shown in Fig. 1: the RT-PCR
analysis of cDNA pools of different human tumour and
normal tissues by means of B99-specific primers produced a
strong signal in colon carcinoma and in the lung
adenocarcinoma line A549 and a weak signal in breast
carcinoma and in kidney cell carcinoma. Of all the normal
tissues investigated, a weak signal could be found only in
colon tissue. The candidate B99 was evaluated more
precisely thereafter.
Example 4
Expression profile of B99 in tumour and normal tissue
One of the 56 candidates (designated: B99) showed a signal
by means of RT-PCR transcription in 3 out of 5 tumour
CA 02365278 2001-10-26
- 32 -
tissue pools tested. In the majority of normal tissues
tested, on the other hand, no transcription could be
detected. For a more detailed investigation of the B99
expression, cDNAs of individual tumour and normal tissue
samples were analysed by PCR as follows. It was found
that the majority of all the colon carcinomas investigated
(17/ 23), all the pancreas carcinomas (3/ 3;) and all the
stomach carcinomas (4/ 4) express B99. In corresponding
normal tissues, expression was detected in :L/ 4, 0/ 3 and
2/ 4 cases, respectively. All the results are shown in
Table 1, whilst Fig. 2 shows exemplary resu:Lts for
pancreas and colon tissue: by RT-PCR analysis of
individual cDNAs of different human tumour <~nd normal
tissue by means of B99-specific primers it was possible to
detect B99-cDNA in 6 out of 7 tumour samples, whereas only
one of the normal tissues investigated (1/ 5) showed a
weak expression of B99.
TABLE 1
Tumour tissue Normal tissue
Type POSITIVE Type POSITIVE
CASES/CASES CASES/CASES
TESTED TESTED
Colon carcinoma 17/ 23 Colon
Liver carcinoma 0/3 Liver 0/4
Lung carcinoma 2/9 Lung 0/4
Breast carcinoma0/10 n.d. ,
Pancreas- 3/3 Pancreas 0/4
carcinoma
r
Kidney cell- 1/10 Kidney 0/4
carcinoma
Stomach 4/4 Stomach 2/4
carcinoma
Lymphocyte 0/1
Small 1/1
CA 02365278 2001-10-26
- 33 -
intestine
Ovary 0/1
Testis 0/4
Prostate 0/1
Thymus 0/1
Spleen 0/4
Muscle 0/1
Brain 0/1
Heart 0/4
For the Northern blot analysis, human multiple tissue
Northern blots (Clontech, Palo Alto) were hybridised at
65°C for 16 hours with the [a-32P]dCTP (NEN, Boston)
labelled 271 by B99 PCR product. Visualisation was
carried out by standard autoradiography (Xomat AR film,
Kodak) and exposure on a phosphoirnager (Molecular
Dynamics). Fig. 3 shows the results of thi:~ analysis of
16 normal tissues. A transcript of ~3.0 kb in size was
found only in the colon and, much weaker, in the duodenum.
The low intensity of the signal leads one to conclude that
immunologically relevant expression is improbable.
The results of all the tests regarding the mRNA-profile of
B99 (compiled from RT-PCR and Northern blot analysis) are
collected in Table 1. Example 4 shows that B99 is clearly
transcribed in a high percentage of tumours of various
indications, whereas no or only isolated cases of
transcription were found in all the normal tissues
investigated. ,
Example 5
Detection of B99 protein expression in human tumours
CA 02365278 2001-10-26
- 34 -
To detect B99 protein expression, B99-specific antibodies
were generated in rabbits. For immunisation, the bacterial
fusion protein pGEX-ORF-1/1, in which Glutat:hione-S
transferase is fused to a portion of the B99 ORF (position
1278 to 1740 in SEQ ID N0: 1) was used. ThE: obtained
serum was affinity-purified with the peptide B99-KML
(SEQ ID NO: 61). To test the specific reactivity of the
serum, the complete B99 open reading frame was transiently
expressed in COS cells as a GFP fusion protein, and the
transfected cells were tested with the serum in a Western
Blot. It was shown that the serum clearly reacts with the
expressed B99 fusion protein. Subsequently, 56 samples
from different tumour types were analysed for B99
expression by immunohistochemistry with the B99 serum
(Table 2). In 53 cases, an expression of B99 could be
shown in the tumour cells. Examples can be seen in Fig. 4,
which shows the immunohistochemical analysis of four
different cases of adenocarcinoma (a: colon, b: breast,
c: pancreas, d: stomach). In all cases a clear staining of
the tumour cells can be seen, while the tumour stroma and
the vessels did not show staining. It can also be seen
from Fig. 4a that the residual normal colon_~c mucosa does
not show reactivity with the antibody. The sections were
counterstained with hematoxylin. In a selection of cases,
the positive reactivity in the immunohistochemistry was
confirmed at RNA level by RT-PCR. In this experiment, also
breast tumours yielded a positive PCR signal. The
difference of this result from Example 4 can be explained
by the use of different PCR primers than in Example 4.
Table 2
Tumour type positive cases/ tested cases
breast adenocarcinoma 8/ 8
colon adenocarcinoma 10% 11
lung adenocarcinoma 7/ 8
lung squamous cell 8/ 8
CA 02365278 2001-10-26
- 35 -
carcinoma
_ __
pancreas adenocarcinoma ~ 11/ 12
stomach adenocarcinoma 9/ 9
Example 6
Cloning of B99
Clone B99 has a 271 by long insert of an unknown human
gene between the adaptors introduced by the RDA. In order
to clone the human sequence fully the following procedure
was used: a UniGene Analysis (National Centre for
Biotechnology Information) produced the following ESTs
homologous to B99: AA315469, AA345780, AA295520. Using
these ESTs the B99 sequence could be extended to
439 nucleotides: New primers within this sequence were
synthesised (SEQ ID N0:9 to 12). By PCR with various
combinations of these primers the theoretical fragment
lengths could be amplified from A549 cDNA.
Cloning of the 3' end: 10 ~g of total RNA from the kidney
cell carcinoma cell line 786-0 were reverse transcribed
using oligo-dT primer and one aliquot of this cDNA was
subjected to a PCR the program of which begins with high
annealing temperatures so that only the gene-specific
primer binds to the cDNA (primer SEQ ID N0:'7 or 9),
whereas the second primer (Tm~53°C, SEQ ID N0:13) only
binds to the newly synthesised DNA substrate at lower
temperatures. This so-called "touch-down PCR"
(Mastercycler Gradient, Eppendorf) was carried out under
the following conditions: 20 cycles of {15" 95°C, 30" 75°C
(reduced by 0.7°C per cycle)}, 1 cycle of 7' 72°C and
20 cycles of (15" 95°C, 30" 50°C) , 1 cycle 7' 72°C} .
One aliquot of the above mixture was PCR amplified once
more using a second primer combination (SEQ ID N0:7 and 13
CA 02365278 2001-10-26
- 36 -
or SEQ ID N0:12 and 13) under the same conditions as
before. Aliquots of the PCR mixtures were ligated
directly into the pGEM-T easy-vector (Promega) and
subsequently transformed into competent E. roll J109
(Promega). Positive clones were sequenced after PCR
selection. The sequencing yielded an agreement with the
former sequence from the primer used for PCR amplification
and in addition 1777 additional nucleotides up to the
start of a PolyA sequence in the 3'-region of the
sequence. PCR amplification of cDNA from tumour cell
lines (786-0, A549) and tissue samples from colon
carcinomas with the primers according to SEQ ID N0:16 to
18 yielded, with the original primers, the fragments
expected after cloning. In the~cDNA fragment now
consisting of 2216 bp, a continuous reading frame could be
identified from position 427 to 1743. No additional
reading frames could be identified further on in the
5'-region of the sequence, leading one to conclude that
the region from 0 to 427 already belongs to the
5'-untranslated region of the B99 mRNA.
Using the primers SEQ ID N0:12 and SEQ ID N0:17 the entire
coding region of B99 from A549-cDNA was amplified, cloned
and sequenced. In the sequencing, a number of clones were
obtained with an insertion of a nucleotide at position 923
compared with SEQ ID N0:1 (codes for B99-1) (SEQ ID N0:3
or SEQ ID N0:5).
This insertion leads to a change in the open reading frame
of B99 with a resulting, altered amino acid sequence in
r
the C-terminal region of the B99 protein; the sequence of
the B99 antigen derived from this reading frame (B99-2) is
shown in SEQ ID N0:4.
One of the clones isolated from A549 cells exhibited, in
addition to the above-mentioned insertion, a nucleotide
CA 02365278 2001-10-26
- 37 -
exchange at position 622 compared with sequence SEQ ID
N0:1. This nucleotide exchange causes, at position No. 66
of the amino acid sequence, arginine (SEQ ID N0:2, B99-1)
to be replaced by tryptophan (SEQ ID N0:4, B99-2). Apart
from this amino acid exchange, the amino acid sequence of
B99-2 is identical to B99-1 as far as position 166.
Moreover, the above-mentioned insertion results in a
second potential reading frame from position 845 to 1744
of the sequence shown in SEQ ID N0:3 (or SEQ ID N0:5). A
protein expressed by a cDNA with this reading frame has
the amino acid sequence shown in SEQ ID N0:6 (B99-3). The
sequence of B99-3 is different from B99-1 from positions 1
to 27 and identical to B99-1 from position 28 onwards.
Example 7:
Potential MHC-binding peptides in the coding region of B99
Potential peptide epitopes within the coding region of
B99-1 according to SEQ ID N0:2 (amino acid position:
1-438; Table 3A), of B99-2 according to SEQ ID N0:4 (amino
acid position 150-190; Table 3B) and of B99-3 according to
SEQ ID N0:6 (amino acid position 1-40; Table 3C) was
carried out using the algorithms described by Parker et
al., 1994, on the basis of known motifs (Rammensee et al.,
1995). For the most important HLA types, particularly for
HLA-A1, -A*0201, -A3, -B7, -B14 and -B*4403, 9-mer
candidate peptides were identified, which can be expected
to bind to the corresponding HLA molecules and therefore
constitute immunogenic CTL-epitopes; the peptides found
are listed in Table 3. Other potential peptide epitopes
for other HLA types or 8- and 10-mer peptides can be
determined by proceeding analogously.
CA 02365278 2001-10-26
- 38 -
Table 3A
Immunogenic B99-peptide candidates (B99-1)
Starting HLA
position Sequence
in SEQ
ID N0:2
5 Lys Arg Leu Cys Gln Leu B14
His Tyr Leu
12 Tyr Leu Trp Ala Leu Gly A*0201
Cys Tyr Met
lg Tyr Met Leu Leu Ala Thr A*0201
Val Ala Leu
21 Leu Leu Ala Thr Val Ala A*0201
Leu Lys Leu
85 Ile Leu Asn Asn Leu Glu A3
Val Lys Lys
90 Glu Val Lys Lys Lys Arg A3
Glu Pro Phe
95 Arg Glu Pro Phe Thr Asp B*4403
Thr His Tyr
96 Glu Pro Phe Thr Asp Thr 87
His Tyr Leu
123 Pro Leu Ser Lys Glu Glu A3
Val Glu Phe
127 Glu Glu Val Glu Phe Pro B*4403 '
Ile Ala Tyr
147 Glu Arg Leu f~euArg Ala B14
Val Tyr Ala
187 Phe Ile Ala Ser Lys Leu A*0201
Val Arg Val
CA 02365278 2001-10-26
- 39 -
206 Asp Leu AsnCys Met Glu A*0201
Asp Leu Leu
209 Cys Met GluAsp Leu Leu A*0201
Gln Ser SerVal
251 Ser Met GluSer Glu Val A1, A3
Pro Pro Lys
275 Thr Leu HisLeu Thr Asn A3
Lys Lys Lys
305 Phe Val GlnHis Val Leu A3
Lys Asn ProLys
318 Leu Ile GluTrp Val Lys A1
Asp Thr Tyr
400 Trp Met LeuGln Asn His A*0201
His Leu Leu
407 Leu Leu AlaAsn Lys Phe A3
Asp Pro Lys
413 Asp Pro LysVal Asp Asp B7
Asn Ala Leu
422 Gln Cys LeuGlu Glu Tyr A1
Leu Arg Tyr
423 Cys Leu GluGlu Tyr Leu A1, A3
Arg Tyr Lys
CA 02365278 2001-10-26
- 40 -
Table 3B
Immunogenic B99-peptide-candidates (B99-2)
Starting - _
position
in SEQ
ID N0:4
152 Ala Val Tyr Ala Pro Gln Al, A3
Asn Ile Tyr
154 Tyr Ala Pro Gln Asn Ile A*0201
Tyr Cys Val
160 Tyr Cys Val His Val Asp A*0201
Glu Glu Val
163 His Val Asp Glu Glu Val A1
Pro Arg Asn
164 Val Asp Glu Glu Val Pro A1, B*4403
Arg Asn Phe
172 Phe Gln Arg Gly Gly Gln Al
Ser Asn Tyr
177 Gln Ser Asn Tyr Phe Leu A1
Leu Pro Lys
182 Leu Leu Pro Lys Cys Leu A*0201
His Ser Gln
r
CA 02365278 2001-10-26
- 41 -
Table 3C
Immunogenic B99-peptide-candidates (B99-3)
Starting
position
in SEQ
ID N0:6
2 Arg Arg Leu Lys Thr Leu A1, B14
Lys Gly Tyr
5 Lys Thr Leu Lys Gly Tyr A*0201
Cys Glu Leu
Tyr Cys Glu Leu Cys Met A1
Pro Leu Arg
12 Glu Leu Cys Met Pro Leu A1, A3
Arg Thr Tyr
14 Cys Met Pro Leu Arg Thr A*0201
Tyr Thr Val
16 Pro Leu Arg Thr Tyr Thr A*0201
Val Ser Met
10 Example 8
MHC stabilisation assay with potential B99-specific
MHC binding peptides
r
In a T2 peptide loading assay, potential B99 MHC binding
peptides were tested for their ability to stabilise HLA-A2
molecules, which is a hint to their MHC binding capacity.
The assay was conducted as described by Bohm et al., 1998.
The stabilisation was measured by means of FACS analysis
CA 02365278 2001-10-26
- 42 -
with an HLA-A2 specific antibody (BB7.2). Five peptides
exhibited a stabilisation effect, which is presented by an
increase of the mean fluorescence intensity in comparison
with a control containing either no peptide or a MAGE-3 A1
control peptide which does not bind.
Table 4
peptide sequence mean
fluorescence
intensity
no peptide 2,27
MAGE-3 A1 Glu Val Asp Pro Ile Gly His Leu 2,25
negative Tyr
control
B99-12 Tyr Leu Trp Ala Leu Gly Cys Tyr 7,46
Leu
B99-19 Tyr Met Leu Leu Ala Thr Val Ala 7,43
Leu
B99-21 Leu Leu Ala Thr Val Ala Leu Lys 3,32
Leu
B99-187 Phe Ile Ala Ser Lys Leu Val Arg 3,57
Val
B99-209 Cys Met Glu Asp Leu Leu Gln Ser 4,72
Ser Val
For further analysis, the peptides were serially diluted
and tested in the same test system in order to show
whether their binding is concentration-dependent. Fig. 5
shows the MHC stabilisation on T2 cells with various
concentrations of B99 peptides. Tyrosinase was used as
positive control, a HLA-A1-specific MAGE-3 peptide as
negative control. In particular, the peptide's B99-19,
B99-187 and B99-209 exhibited a clear dependency of MHC
stabilisation on their concentration, which makes them
preferred candidates for immunisation stratE~gies.
CA 02365278 2001-10-26
- 43 -
Bibliography
Becker, D., Kuhn, U., Lempertz, U., Enk, A., Saloga, J.,
and Knop, J. (1997), J. Immunol. Methods 243, 171-180.
Blake, J., Johnston, J.V., Hellstrom, K.E., Marquardt, H.,
and, Chen, L. (1996), J. Exp. Med. 1f34, 121-130.
Boon, T, J. C. Cerottini, B. Van den Eynde, P. van der
Bruggen, and A. Van Pel (1994), Annu.Rev.Immunol. 12.:
337-365
Boon T ,(1998). Tumor antigens recognized by cytolytic
T cells. Cancer Vaccine Week - Interantional
Symposium, New York, Oct 1998; abstract S01
C. M. Bohm, M. L. Hanski, S. Stefanovic, H. G. Rammensee,
H. Stein, J. Taylor-Papadimitriou, E. 0. Riecken, and
C. Hanski. Int.J.Cancer 1998.Mar.2. 75:688-693
Boulianne, G. L., et al., (1984), Nature 312: 643-646
Bruggemann, M. and Neuberger, M.S., (1996), Immunol. Today
17: 391-397
Buschle M, Schmidt W, Zauner W, Mechtler K, Trska B,
Kirlappos H, Birnstiel M (1997), Proc. :lVatl. Acad. v
Sci. U.S.A. ,9:3256-3261
r
Celluzzi, CM and Falo, LD, Jr. (1998), J. Immunol. 1~Q.:
3081-3085
Chen, Y.T., Scanlan, M.J., Sahin, U., Tureci, 0., Gure,
A.O., Tsang, S., Williamson, B., Stockert, E.,
CA 02365278 2001-10-26
- 44 -
Pfreundschuh, M., and Old, L.J. (1997), Proc. Natl.
Acad. Sci. U. S. A. 24, 1914-1918.
Coulie, P.G. (1997) , Mol. Med. Today ~-: 261--268
Cox, AL, Skipper, J, Chen, Y, Henderson, RA, barrow, TL,
Shabanowitz, J, Engelhard, VH, Hunt, DF, and
Slingluff, CL, Jr. (1994), Science X64: 716-719
Diatchenko, L., Lau, Y.F., Campbell, A.P., Chenchik, A.,
Moqadam, F., Huang, B., Lukyanov, S., Lukyanov, K.,
Gurskaya, N., Sverdlov, E.D., and Siebert, P.D.
(1996), Proc. Natl. Acad. Sci. U. S. A. 9~., 6025-6030.
Dranoff, G., Jaffee, E., Lazenby, A., Golumbek, P.,
Levitsky, H., Brose, K., Jackson, V., Hamada, H.,
Pardoll, D., and Mulligan, R. (1993), Proc. Natl.
Acad. Sci. U. S. A 2Q: 3539-3543.
Emini, EA, Hughes, J, Perlow, D, and Boger, J (1985), J.
Virol. ~5.: 836-839
Falk, K., Rotzschke, O., Stevanovi'c, S., Jung, G., and
Rammensee, H-G (1991), Nature ~: 290-296.
Fearon, E.R., Pardoll, D.M., Itaya, T., Golumbek, P.,
Levitsky, H.I., Simons, J.4V., Karasuyama, H.,
Vogelstein, B., and Frost, P. (1990), Cell f Q:
397-403.
Gansbacher, B., Zier,'K., Daniels, B., Cronin, K.,
Bannerji, R., and Gilboa, E. (1990), J. Exp. Med. 172:
1217-1224.
Gaudi C, Krerner F, Angevin E, Scott V, Triebel F (1999), J
Immunol 162:1730-1738
CA 02365278 2001-10-26
- 45 -
Graziano, R.F., et al., (1995), J. Immunol. 155: 4996-5002
Griffiths, A.D., et al., (1994), EMBO J. 13: 3245-3260
Halliday, GM, Patel, A, Hunt, MJ, Tefany, FJ, and
Barnetson RS (1995), World J. Surg. 1~: 352-358
Hogan KT, Eisinger DP, Cupp SBC, Lekstrom KJ, Deacon DD,
Shabanowitz J, Hunt DF, Engelhard VH, S:Lingluff CL,
Ross MM (1998), Cancer Res 58:5144-5150
Hubank, M. and Schatz, D.G., (1994), Nucleic. Acids. Res.
2,,2., 5640-5648.
Japer, E., Chen, Y.T., Drijfhout, J.W., Karbach, J.,
Ringhoffer, M., Japer, D., Arand, M., W<~da, H.,
Noguchi, Y., Stockert, E., Old, L.J., and Knuth, A.
(1998), J. Exp. Med. 1$Z, 265-270.
Jakobovits, A., (1995), Curr. Opin. Biotechnol. 6: 561-566
Jameson, BA and Wolf, H (1988) The antigenic index: a
novel algorithm for predicting antigenic determinants.
Comput. Appl. Biosci. 4: 181-186
Jessup, JM and Loda, M (1998) Prognostic markers in rectal
carcinoma. Semin. Surg: Oncol. 15.: 131-:L40.
Kawakami, Y., Eliyahur, S., Jennings, C., Sakaguchi, K.,
King, X., Southwood, S., Bobbins, P.F., Sette, A.,
Appella, E., and Rosenberg, S.A. (1995), J. Immunol.
154: 3961-3968.
Kawakami, Y, Bobbins, PF, and Rosenberg, SA (1996), Keio
J. Med. 4~.: 100-108
CA 02365278 2001-10-26
- 46 -
Kohler, G. and Milstein, C. (1975), Nature 265, 495-497
Kruif, J., et al., (1995), Proc. Natl. Acad. Sci. USA 92:
3938-3942
Kyte, J and Doolittle, RF (1982), J. Mol. Biol. 157:
105-132
Lethe, B, Lucas, S, Michaux, L, De Smet, C, Godelaine, D,
Serrano, A, De Plaen, E, and Boon, T (1998), Int. J.
Cancer ~: 903-908
Mackensen A, Ferradini L, Carcelain G, Triebel F, Faure F,
Viel S, and Hercend T (1993), Cancer Res.
3569-3573
Mandruzzato S, Brasseur F, Andry G, Boon T, van der
Bruggen P (1997) , J Exp Med 186:785-79:3
Marchand M, Weynants P, Rankin E et al (1995), Int. J.
Cancer x:883-885
Mayordomo, J.I., Zorina, T., Storkus, W.J., Zitvogel, L.,
Celluzzi, C., Falo, L.D., Melief, C.J., Ildstad, S.T.,
Kast, W.M., DeLeo, A.B., and Lotze, M.T. (1995),
Nature Medicine 1. 1297-1302.
McGuinnes, B.T., et al., (1996), Nature Biotechnol. 14,
1149
r
Melief CJ, Offringa R, Toes RE, Kast WM (1996), Curr.
Opin. Immunol. $:651-657
Murphy, G.P., Elgamal, A.A., Su, S.L., Bostwick, D.G., and
Holmes, E.H. (1998), Cancer $3., 2259-2269
CA 02365278 2001-10-26
- 47 -
Neuberger, M.S., et al., (1984), Nature 312: 604-608
Pardoll, D. M.(1998) Nature Medcine 4: 525-531
Parker, K. C., M. A. Bednarek, and J. E. Coligan. (1994),
J. Immunol. 15:163.
Parkhurst, M.R., Salgaller, M.L., Southwood, S., Bobbins,
P.F., Sette, A., Rosenberg, S.A., and Kawakami, Y.
(1996), J. Immunol. 157, 2539-2548.
Paterson Y, Ikonomidis G (1996), Curr. Opin. Immunol. $:
664-9
Rammensee HG, Friede T, Stevanovic S (1995),
Immunogenetics 41:178-228
Rapellino, M, Pecchio, F, Baldi, S, Scappaticci, E, and
Cavallo, A (1995), Anticancer Res. 1.5.: 1065-1070
Ravaioli, A., Bagli, L., Zucchini, A., and Monti, F.
(1998), Cell. Prolif. 31, 113-126
Revillion, F, Bonneterre, J, and Peyrat, JP (1998), Eur.
J. Cancer ,3~: 791-808
Restifo NP (1996), Curr. Opin. Immunol. 5.: 658-63
Riechmann, L., et al., (1988), Nature 332: 323-327
.r
Bobbins, PF and Kawakami, Y (1996), Curr. Opin. Immunol.
$: 628-636
Ruppert, J., Sidney, J., Celis, E., Kubo, R.T., Grey,
H.M., and Sette, A. (1993), Cell ,~, 929-937.
CA 02365278 2001-10-26
- 48 -
Sahin, U., Tiireci, O., Schmitt, H., Cochlovius, B.,
Johannes, T., Schmits, R., Stenner, F., Luo, G.,
Schobert, I., and Pfreundschuh, M. (1995), Proc. Natl.
Acad. Sci. U.S.A. ~, 11810-11813
Sambrook, J., Fritsch, E.F., and Maniatis, T. Molecular
Cloning: A Laboratory Manual. ColdSpring Harbor
Laboratory Press, 1989
Schendel DJ, Gansbacher B, Oberneder R, Kriegmair M,
Hofstetter A, Riethmuller G, and Segurado OG (1993),
J. Immunol. 151: 4209-4220
Schmidt W, Buschle M, Zauner W, Kirlappos H, Mechtler K,
Trska B, Birnstiel M (1997), Proc. Natl. Acad. Sci.
U.S.A 24:3262-3267
Schweighoffer, T. (1997), Onc. Res. ~., 164-:176
Sette, A., Vitiello, A., Reherman, B., Fowler, P.,
Nayersina, R., Kast, W.M., Melief, C.J.M., Oseroff,
C., Yuan, L., Ruppert, J., Sidney, J., del Guercio,
M.-F., Southwood, S., Kubo, R.T., Chesmut, R.W., Grey,
H.M., and Chisari, F.V. (1994), J. Immunol. 15
5586-5592.
Stauss, H.J., Davies, H., Sadovnikova, E., Chain, B.,
Horowitz, N., and Sinclair, C. (1992), Proc. Natl.
Acad. Sci. U.S.A $.3, 7871-7875.
r
Tepper, R.I., Pattengale, P.K., and Leder, P. (1989), Cell
~Z: 503-512.
Tighe H, Corr, M, Roman M, and Raz E (1998), Immunol.
Today 12:89-97
CA 02365278 2001-10-26
- 49 -
Toes, R.E., Hoeben, R.C., Van der Voort, E., Ressing,
M.E., Van-der-Eb, A.J. Melief, C.J.M., and Offringa,
R. (1997), Proc. Natl. Acad. Sci. U.S.A. 2f-(26):
14660-14665
Tuting, T., DeLeo, A.B., Lotze, M.T., and Storkus, W.J.,
(1997), Eur. J. Immunol. 2~Z, 2702-2707,..
Van den Eynde, B. and Brichard, V.G. (1995),, Curr. Opin.
Immunol. Z, 674-681.
Van den Eynde, BJ, and van der Bruggen, P (1997), Curr.
Opin. Immunol. .x:684-693
van der Bruggen, P., C. Traversari, P. Chomez, C. Lurquin,
E. De Plaen, B. Van den Eynde, A. Knuth, and T. Boon.
(1991, Science ,54:1643-1647
van der Burg, S.H., et al., (1996), J. Immunol. 156,
3308-3314
van Elsas, A., van der Minne, C.E., Borghi, M., van der
Spek, C.W., Braakman, E., Osanto, S., arid Schrier,
P.I. (1996), CTL Recognition of an IL-2 Producing
Melanoma Vaccine. In: Immunology of human melanoma.
Tumor-host interaction and immunotherapy, edited by M.
Maio, Amsterdam: IOS, 1996, p. 165-173
van Elsas, A., Aarnoudse, C., van der Minne, C.E., van der
Spek, C.W., Brouv~enstijn, N., O.santo, S., and Schrier,
P.I. (1997), J. Immunother. 2~Q: 343-353.
Vaughan, T.J., et al., (1998), Nature Biotechnol. 16,
535-539
CA 02365278 2001-10-26
Case 12/203
SEQUENCE LISTING
<110> Boehringer Ingelheim International GmbH et al.
<120> Tumor associated antigene
<130> seqlist12203
<140> ~ .
<141>
<160> 61
<170> PatentIn Ver. 2.1
.
<210> 1
<211> 2216
<212> DNA
<213> Homo sapiens
<220>
<221> 5'UTR
<222> (1)..(426)
<220>
<221> CDS
<222> (427)..(1743)
<220>
<221> 3'UTR
<222> (1744)..(2216)
<400> 1 .
G'lCACC~OC~A CZGCCCTIUC '1~~ CTGCCCZTIA C'I~''~IT TI~C~'IT~.'IL3G 60
GA~GCCCTGG GATTCIGCTA ATACC~ C~AGGTGC ~AAOOGAAA C~GATGAACA 120
p~p,~CCIC A~pGAGCTTC CTGmAA7CA GAAGACCAAG C'IGALGCCPG GCAAACATAT 180
4 5 TAA~GGAG CCZGAAACIG TI;CCIT~AC ATCITA' "~' A TG'1'C~rAAAA TACCI'I~f'IGG
240
AGGGTT~AF~ Gp.TCAGGGGA CA'ICdGTIGIT ~C3CI' GCCAC13GAAC ACCG~C 300
T'I~AC.TIC3GG AF~CAGAAT~' CGCC~1.G~ AGAGATCATC CCTAAGCAOG AC~1C~CI~.1 360
C'IT~AAC~ATT GIGTACTCCT CCACCITCOC T~CI~GT CIC.'C1~C(..'i~T CTCCCATTCT 420
G~ ATG GTT CAA TC~G AAG ~ GTC Tip CAG CTG CAT TAC TTU TGG 468
5 0 Met Val Gln Trp Lys An3 Leu Cys Gln Leu His Tyr Leu Trp
1 5 10
GGT CIG GGC ZC,'C TAT ATG CIG CIG GCC ACT GTG GCT CIG AAA CTT TCr 516
Ala Leu Gly Cys Tyr Met Leu Leu Ala 'Thr Val Ala Leu Lys Leu Ser
5 5 15 20 25 30
T'IC AGG TIG AAG T~' CSC TCT GAC CAC TIG GGT C'IG GAG TCC FAG GAP. 564
1
Case 12/203
CA 02365278 2001-10-26
Phe Arg Leu Lys Cys Asp Ser Asp His Leu Gly Leu
Glu Ser An3 Glu
35 40 45
TCT CAA PGC CPG TAC Z~'T AOG AAT ATC TIG TAT AAT 612
TTC CTG AAA CIT .
Ser Gln Ser Gln Tyr Cys Arg Asn Ile Leu Tyr Asn
Phe Leu Lys Leu
50 55 60
CCA GCA AAG AGG TCT ATC AAC zGT TCA GOG GTC ACC 660
CGA GOG GAC QUA
Ptro Ala Lys And Ser Ile Asn Cys Ser Gly Val Thr
Arg Gly Asp Gln
65 70 75
~ ~ ~ ~T (~G GCT ATT CIG AAT AzIC CIG GAG GTC 708
AAG AAG AAG
Glu Ala Val Leu Gln Ala Ile Leu Asn Asn Leu Glu
Val Lys Lys Lys
80 85 ~ 90
CU~A GAG CCT!' TI'C ACF1 CAC ACC C~.C TAC ChC 756
TCC ChC ACC PGA G'AC 'IGT
An3 Glu Pro Phe Zhr Asp Thr His Tyr Leu Ser Leu
Thr Arg Asp Gds
95 100 105 11.0
2 G~ cAC Trc A~ ccr c~ ~ AAG Trc ATA c~ Trc ccA ao4
0 cIG ~C A~
Glu His Phe Lys Ala Glu A~ Lys Phe Ile Gln Phe
Pro Leu Ser Lys
115 120 125
GAA GAG GIG GAG TrC CCT ATT GCA TAC TCT AZG GIG 852
ATT CAT GAG AAG
2 Glu Glu Val Glu Phe Piro Ile Ala Tyr Ser lit Val
5 Ile His Glu Lys
130 135 140
ATT GAA AAC TIT G~1A AC~G CTA CIG 03A GCT GIs 900
TAT GCC CCT CAG AAC
Ile Glu Asn Phz Glu Arg Leu Leu Azg Ala Val Tyr
Ala Piro Gln Asn
3 145 150 155
0
ATA TAC TuT GTC CAT GIG GAT GAG AAG TCC CC~1 GAA 948
ACT TIC AAA G~1G
Ile Tyr Cys Val His Val Asp Glu Lys Ser Pno Glu
'Thr Phe Lys Glu
160 165 170
35
GCG GTC AAA GCA ATT ATT TC.'r TC~C TI'C CC'A AAT 996
GTC TTC ATA GCC AGT
Ala Val Lys Ala Ile Ile Ser Cys Phe Pro Asn Val
Phe Ile Ala Ser
175 180 185 190
4 A~ CIG GIT Ct3G GIG GTT TAT GCC TG'C TC~G TCC 1044
0 AGG GIG CAA GCT GAC
Lys Leu Val Azg Val Val Tyr Ala Ser Trp Ser Azg
Val Gln Ala Asp .
195 200 205
CTC AAC TGC A'I~ GAA GAC TIG GTC CI~G pCC TCA 1092
GIG CCG 'Ir3G AAA TAC
4 Leu Asn Cys I~t Glu Asp Leu Leu Gln Ser Ser Val
5 Piro Trp Lys Tyr
210 215 220
TIC CIG AAT AC3~ TGT GG1G ACS GAC TIT CCT ATA 1140
AAG PGC AAT GCA GAG
Phe Leu Asn Thr Cys Gly Thr Asp Phe Pro Ile Lys
Ser Asn Ala Glu
5 225 X130 235
0
AZG GTC CpG GGT CTC A'9G ATG TIG AAT GGG PGG AAT 1188
1~GC AZG GAG TCA
Met Val Gln Ala Leu Lys Met Leu Asn Gly Arg Asn
Ser Met Glu Ser
240 245 250
55
GAG GTA CCT CCT AAG CAC AAA GAA ACC CGC 'IC~G 1236
AAA TAT C~ TTT GAG
Glu Val Pro Pro Lys His Lys Glu Thr AnJ Trp Lys
Tyr His Phe Glu
2
' . CA 02365278 2001-10-26
Case 12/203
255 260 265 270
GTP. GZG ACS GAC ACS TTA CAC CIA ACC AAC APB 1284
AAG AAG GAT CCT CCC
Val Val Arg Asp Thr Leu His Leu Thr Asn Lys Lys
Lys Asp Pro Faro
275 280 285
CCT TAT AAT TIA ACT A'IG TIT A~'A GGG AAT GCG 1332
TAC ATT GIG C,~C,T TCC
pro Tyr Asn Leu Thr Met Phe Thr Gly Asn Ala Tyr
Ile Val Ala Ser
290 295 300
CuA GAT TTC GhC CAA CAT GTT TIG APG AP.C CCT 1380
AAA TCC CAA CAA CIG
An3 Asp Phe Val Gln H:is Val Leu Lys Asn Pro
Lys Ser Gln Gln Leu
305 310 315
1 ATT GAA TGG GTA AAA G~ ACr TAT AGC CC~ GAT GAA 1428
5 CAC CTC ZC3G GCC
Ile Glu Tzp Val Lys Asp Thr Tyr Ser Pro Asp Glu
His Leu Txp A7:a
320 325 330
ACC CIT CF1G CGT GCA ~G 'IGG AZG CCT GGC TCT 1476
GTr CCC AAC C~ CCC
2 Thr Leu Gln Arg Ala Arg Trp Met Pm Gly Ser Val
0 Pro Asn His Pro
335 340 ' 345 350
AAG TAC GAC ATC Tt~ CSC ATG ACT TCT ATT GCC ~ 1524
CIG GTC A~G 'IC3G
Lys Tyr Asp Ile Ser Asp Met Thr Ser Ile Ala Arg
Leu Val Lys Trp
2 355- 360 365
5
CAG GGT CAT GaG GGA GAC ATC GAT ApG GGT GGT C~.T1572
TAT GCT CCC 2~'.C .
Gln Gly His Glu Gly Asp Ile Asp Lys Gly Ala Pro
Tyr Ala Pro Cys
370 375 380
30
TCT GGA ATC CAC CAG COG GCT ATC Tv~C GIT TAT 1620
GGG GG'I' GGG GAC T1G
Ser Gly Ile His Gln And Ala Ile Cys Val Tyr Gly
Ala Gly Asp Leu
385 390 395
3 AAT T3G ATG CIT CAA AAC CAT CAC CTG TTG GCC AAC 1668
5 AAG TIT CAC CC~
Asn Trp Met Leu Gln Asn His His Leu Leu Ala Asn
Lys Phe Asp Pro
400 405 . 410
AAG G'IP. GAT GAT AAT C'CT CTT CAG TGC TTA GAA 1716
GAA TAC C'IP. CST TAT
4 Lys Val Asp Asp Asn Ala Leu Gln Cys Leu Glu Glu
0 Tyr Leu Arg Tyr
415 420 425 430
AAG GCC ATC TAT OGG ACT CAA CIT TC~. GACp~AC1'AT1763
C~1~~IZG
Lys Ala Ile Tyr Gly Thr Glu Leu
4 435
5
CTACCIGIC3G GGC~l~GAGC'A TGT~ TGCI~GAAC TIGCICY3GAC1823
AGIGIGGGTG
GG~?GACCI~GG GCITIGCAAT ~GCATC CrTTAGGGATA AG~~(30GCIGC1883
TATTAG~TIG
50
ZGC3GTAP~GIT3 GATG'1TTIGC CTIGCAAATT C~GCCIC~OG 1943
~AA~ TTGI''CCTCCC
ACCCC1~1A~CC CTAC~TAGrPC CTCC~C1T~AC TIZC'T'CAC'IP~2003
AGIG<~1AZG ~1GAACIGCIG
5 ~,G ~~; ~,TGIGG T1~C~CIT GATTTCAGIT GAATC~C1GC 2063
5
2123
3
CA 02365278 2001-10-26
Case 12/203
AA~tT ZGAZC3GAAAG AC~A~CTTGC CTTC'!G'~' GI~1A~A AAA'I~AATAG 2183
ChGC'IGATTC AAAGTAAAAA AAAAAAAAAA AAA 2216
<210> 2
<211> 439
<212> PRT
<213> Homo sapiens
<400> 2
1 5 Met Val Gln Trp Lys Azg Leu Cys Gln.Leu His Tyr Leu Trp Ala Leu
1 5 10 15
G1Y Cps Tyr Met L~eu Leu Ala Tt~r Val Ala Leu Lys I~eu Ser Phe Arg
25 30
Izu Lys Cys Asp Ser Asp His~Leu Gly I~eu Glu Ser Arg Glu 5er Gln
35 40 45
Ser Gln Tyr Cps Azg Asn Ile Leu Tyr Asn Phe Leu Lys Leu Pzro Al.a
2 5 50 55 60
Lys Arg Ser Ile Asn Cys Ser Gly Val Thr Azg Gly Asp Gln Glu Al.a .
65 70 75 80
3 0 Val Leu Gln Ala Ile Leu Asn Asn Leu Glu Val Lys Lys Lys Azg Glu
85 90 95
Pzro Phe Thr Asp Thr His Tyr Leu Ser 1xu Thr Arg Asp Cys Glu His
100 105 110
Phe Lys Ala Glu Arg Lys Phe Ile Gln Phe Piro Leu Ser Lys Glu Glu
115 120 125
Val Glu Phe Pro Ile Ala Tyr Ser Met Val Ile His Glu Lys Ile Glu
4 0 130 135 140
Asn Phe Glu Arg Leu Leu Azg Ala Val Tyr Ala Pro Gln Asn Ile Tyr
145 150 155 160
4 5 Cps Val His Val Asp Glu Lys Ser Pn~ Glu Thr Phe Lys Glu Ala Val
165 170 175
Lys Ala Ile Ile Ser Cys Phe Pro Asn Val Phe Ile Ala Ser Lys Leu
180 185 190
Val Arg Val Val Tyr Ala Ser Tzp Ser Azg Val G1n Ala Asp Leu Asn
195 200 205
5 5 Cys Met Glu Asp Leu Leu Gln Ser Ser Val Piro Trp Lys Tyr Phe Leu
210 215 220
4
' , ~ CA 02365278 2001-10-26
Case 12/203
Asn Thr Cys Gly 'Il~r Asp Phe Pro Ile Lys Ser Asn Ala Glu Met Val
225 230 235 240
Gln Ala Leu Lys Met Leu,Asn Gly Arg Asn Ser Met Glu Ser Glu Val
245 250 255
Pro Pro Lys His Lys Glu Thr Axg Trp Lys Tyr His Phe Glu Val Val
260 265 270
1 0 Azg Asp Thr Leu His Leu T'hr Asn Lys Lys Lys Asp Pro Pro Pro 'I~'
275 280 285
Asn Leu Thr Met Phe Tnr Gly Asn Ala Tyr Ile Val Ala Ser Axg Asp
290 295 300
Phe Val Gln His Val Leu Lys~Asn Pro Lys Ser Gln Gln Leu Ile Glu '
305 310 315 320
Trp Val Lys Asp Zhr 'Tyr Ser Pro Asp Glu His Leu Trp Ala Thr Leu ,
2 0 325 330 335
Gln An3 Ala Axg Trp Met Pro Gly Ser Val Pro Asn His Piro Lys 'Iyr
340 345 350
2 5 Asp Ile Ser Asp Met 'rhr Ser Ile Ala Azg Leu Val Lys Tip Gln Gly
355 360 ' 365
His Glu Gly Asp Ile Asp Lys Gly Ala Ptro Tyr Ala Pro Cys Ser Gly
370 375 380
Ile His Gln Azg Ala Ile Cys Val Tyr Gly Ala Gly Asp Leu Asn Tip
385 390 395 400
Met Lsu Gln Asn His His Leu Leu Ala Asn Lys Phe Asp Pro Lys Val
3 5 ~ 405 410 415
Asp Asp Asn Ala Leu Gln Cys Leu Glu Glu Tyr Leu Axg Tyr Lys Ala
420 425 430
4 0 Ile Zyr Gly I'hr Glu Leu
435
45 <210> 3
<211> 2217
<212> DNA
<213> Homo sapiens
50 <220>
<221> 5'UTR
<222> (1)..(426)
<220>
55 <221> CDS
<222> (427)..(999)
CA 02365278 2001-10-26
Case 12/203
<220> .
<221> 3'UTR
<222> (1000)..(2217)
<220>
<221> 5'UTR
<222> (1)..(844)
<220>
<221> CDS
<222> (845)..(1744)
<220>
<221> 3'UTR
<222> (1745)..(2217)
<400> 3
GTCA~JOCGAA CIGCCC.t~CyC TAC.'ITGIGAC CIGOC~TTT'A C't'C~CCAGIT TTTGThCIGG 60
C~1GCCC~G GATIATACCTATCA CIGI~G'IGC ~AAOGCd~AA CAGA~ 120
2 5 AO'A~Ci'C AAGGAGCITC CMG~C~A'n'aP. C~AG~CAAG CIGA~CCIG GCAAF~GATAT 180
TAAAC~F~C~~G CCI~AAACIG TTCC.TInC'~C AAA ~GAAA1~ TAOCITT'Ir3G 240
AODGTTAGAA GA'ICPGGC3C~GA CA'IC~GTI~IT CACATTIGCT GCCALC'~AAC AGTC 300
TIC~C,'ILC3CG AAC~P.TrA CCCCTIGZ'Gp. AGAGA~C~~'C CGTAAGCAGG AGAGAAC~TA 360
~CIP~AAC3GATT GTGT~~CCT CCACCTI'CCC TGIGCT03Gr G'TCCACCICGT ~'IT'CT 420
3 5 G~ A'I~ GTT CAA TC~G AFG Ate. CIC 'IGC CAG CIG CAT TAC TIG 'IC~',~ 468
Met Val Gln Trp Lys Arg Leu Cys Gln Leu His Tyr Leu Trp
1 5 10
GCr CIG GGC TGC TAT A'I~ CIA CIG GCC ACT GIG GCT CIG AAA CIT TCT 516
4 0 Ala Leu Gly Cys Tyr Met Leu Leu Ala Thr Val Ala Leu Lys Leu Sex
15 20 25 30
TIC AC~G TIG AAG ~T GAC TCT GAC CAC TIG GGT CIG GAG TCC ~1C~G GF1A 564
Phe Arg Leu Lys Cys Asp Ser Asp His Leu Gly Leu Glu Ser Azg Glu
4 5 35 40 45
'1'CI' CAA AGC C~1G TAC SGT hC~G AAT A'I~ dT'IG T'AT AAT TIC CIG AAA CIT 612
Ser Gln Ser Gln Tyr Cys Azg Asn Ile Leu Tyr Asn Phe Leu Lys Leu
50 55 60
50 v
CCA GCA AAG TGG TCI' ATC .'4AC 'IC~T TCA GOG GTC ACC f~A GGG GAC CAA 660
Pro Ala Lys Trp Ser Ile Asn Cys Ser Gly Val Thr Azg Gly Asp G:In
65 70 75
5 5 GF1G GCA G'IG CIT CAG GCT ATT CIG AAT AAC CIG GAG GTC AAG AAG AAG 708
Glu Ala Val Leu Gln Ala Ile Leu Asn Asn Leu Glu Val Lys Lys Lys
80 85 90
6
CA 02365278 2001-10-26
Case 12/203
CGA GAG OGT T1L~ ACA GAC ACC C3~C TAC CTC TCC CTC ACC AGA GAC 'IGT 756
Azg Glu Pro Phe Thr Asp Thr His Tyr Leu Ser Leu Thr Axg Asp Cys
95 100 105 11.0
GAG CAC TTC AAG GCT GAA AGG AAG TTC ATA C~G TTC CC~. CIG AGC AAA 804
Glu His Phe Lys Ala Glu Azg Lys Phe Ile Gln Phe Pro Leu Ser Lys
115 120 125
1 0 GAA GAG GTG GPG TIC CC.T ATr GCA TAC TCP A'IG GIG ATT CAT GAG AAG 852
Glu Glu Val Glu Phe Pzn Ile Ala Tyr Ser Met Val Ile His Glu Lys
130 135 140
ATT GAA AAC TTT GAA A(,3G CTA CIG CGA.GGT GIG TAT GCC CCT CAG AAC 900
1 5 Ile Glu Asn Phe Glu Arg Leu Leu Arg Ala Val Tyr Ala Pro Gln Asn
145 150 155
ATA TAC ~T GTC CAT GIG GAT GAA G~AA GhC CCC ~1GA AAC TIT CAA AC3P. 948
Ile Tyr Cys Val His Val Asp Glu Glu Val Pro Axg Asn Phe Gln Axg
2 0 160 165 170
GGC GGT CAA PGC AAT 'IP.T TIC TIG CIT CCC AAA TGT CTT CAT AGC C~G 996
Gly Gly Gln Ser Asn Tyr Phe I~eu Leu Pro Lys C'ys Leu His Ser Gln
175 180 185 190
TAA GCIGGITJOG GIC3C~TTI'ATG CCrCC~'C CAOOGTGCAA GCIGACCTC~. 1049
* ,
3 0 ACIGCATOGA AG~'A~.'~IGChC CAGAGCIC'AG ZGCLGInC~FIA ATACI'PCCIG
AATAC'A'IGIG 1109
GCCTT TCCTATAAAG AGC~AZGC~ AGA'I~Y'A GGChCIC~AG A'IV'!'IGAA'IG 1169
pCApG~,ATAG CAIr3G~ GA~~'ACC:TC CTA~CAC~A AGAAACCGGC TGC~AAT~.TC 1229
~~~~ pCC Tpp,~,ACAF. G~GAA~3GAT CCICCCCCTT 1289
A~,ATITAAC Tp,~CA C~AATGC~T ACATIGIGGC ThCCCGAGAT ZTCGTC'CAAC 1349
4 0 CAF. G~1ACCCTAAA T'~P~CAAC ZGATIC~ATG GGTAAAAGAC P~~CC 1409
C~,TG~1ACA CC~'IGGGCC ACCCrTCAGC GTGCACX3GZG GFiIGCCIC3GC TCIGI'hCCCA 1469
ACCACCCCAA GTACGACATC TC~1GACAIGA CIT4'rATIGC CACGCIC3GI'C AAG'rGGCAG3G 1529
GTC~1TGAC~~ l~CACp.TCGAT a~ACIC~CzIGCI'C CITACIGCTC'lGGA ATCCACX,'AGC 1589
G~CTKTCIG Q~GG GCIC3GOGACT ~AA2'IC~GAT GCTICA~AAC CA~~ 1649
5 0 TG~C~ACAP. GIfiiG'9CCCA P~AC~GI~A'IG ATAAZGCTCr TQ1G:IGCITA GAAC~1ATAGC
1709
TACGTTAATAA GGCCATC'IAT ~~AC~AC TI'IC~GA~CAC ACS .'TACC 1769
~G~, ~p~ ~GCI'C ~CITGCT GOGACAGIGT GGGZGGGAGP. 1829
C~~GGGCLZT GC~ATIGGIG GCA'IGCITIA GGATAAGAC~G GCIGGTATTA GATIG'.~' 1889
7
CA 02365278 2001-10-26
Case 12/203
p,~T TPIGCCIT~ AAATIGCICC CIC3GGG'IC~~AT GCIGCITGrT CTCTC'~!CC.C 1949
Th,ACaTAGT ~GIT~~CA CI~1A~.TTTCr CF1,G'TAAGIGA GAATG~AC 1GCIGIC'~ATA 2009
GC~GAC'PDGAG3GATA T<'~3GIp1'~G CAC1IGATIT CAG'I7GAAZG CGIG2069
GCITTIGCAT TCIGIGC~AGC TGC'~I'tL''CT AA'I~1A'~I~C~ C3GI'IT3GTAG ~'IC~G~AG 2129
AACIrl'IGATG GAAAG~C~ CI'I'CCCrI'C'T GAP. CTTA,AAAP.TA AATAGCI'C<.'r 2189
~ p~ p~,~A 2217
<210> 4
<211> 190
<212> PRT
<213> Homo sapiens
<400> 4 '
Met Val Gln Trp Lys Azg Leu Cys Gln Leu His Tyr Leu Trp Ala Leu
1 5 10 15
Gly Cys Tyr Met I~eu Leu Ala Thr Val Ala Leu Lys Lzu Ser Phe A~eg
20 25 30
Leu Lys Cys Asp Ser Asp His Leu Gly Leu Glu Ser Axg Glu Ser Gln
3 0 35 40 45
Ser Gln Tyr Cys Azg Asn Ile Leu Tyr Asn Phe Leu Lys Leu Pro Ala
50 55 60
3 5 Lys Tzp Ser Ile Asn Cys Ser Gly Val Thr Arg Gly Asp G1n Glu Ala
65 70 75 80
Val Leu Gln Ala Ile Leu Asn Asn Leu Glu Val Lys Lys Lys Azg Glu
85 90 95
Pro Phe Thr Asp Thr His Tyr Leu Ser Leu T'hr Axg Asp Cps Glu His
100 105 110
Phe Lys Ala Glu Arg Lys Phe Ile Gln Phe Pro Leu Ser Lys Glu Glu
4 5 115 120 125
Val Glu Phe Pro Ile Ala Tyr Ser Met Val Ile His Glu Lys Ile Glu
130 135 140
5 0 Asn Phe Glu Azg Leu Leu Arg l~la Val Tyr Ala Pro Gln Asn Ile Tyr
145 150 155 160
Cys Val His Val Asp Glu Glu Val Pro Azg Asn Phe Gln Arg Gly Gly
165 170 175
Gln Ser Asn Tyr Phe Leu Leu Piro Lys Cys Leu His Ser Gln
180 185 190
g
CA 02365278 2001-10-26
Case 12/203
<210> 5
<211> 2217
<212> DNA
<213> Homo sapiens
<220>
1 <221> 5'UTR
0
<222> (1) . . (844)
<220>
<221> CDS
<222> (845)..(1744)
<220>
<221> 3'UTR '
<222> (1745)..(.2217) w
<400> 5
GI'CA030GAA CIGCCCTIGC Zi,GAC CIGCCCITlA CTCAGCAGIT60
'I~G
GAE~GC~G GATI~'IGC'm ATACCI~.TCA CIGI~OGIGC TGF~C~OGAAA120
CAGA'IGAAGA
A~'ATG'P~CTC AA~GCTrC CIGrCAA~ C-AAGACC~AG G'IGA~'7GCCrG180
C'CAAAGAT~1T ~
3 TAAAGAGGAG CCl~~AAAC'.I~ TTCCITaGAC AZ'CITATGAA 240
0 ~G~AAA TACCTTT1GG
AGGG'T17!GAA C~TC~GGGp. CA'IC3GI~Tr CAC~T'IT~.T 300
GCC~GOGAAC ACC~CC~1GI'C
TTC~CIZ~~OG 19AC~P.1CA CGCCr'~IG~ AG~TCATC CCTAAGCAGG360
AGF~GAAC~C'.IT~
CTAA~GP.TI' GTC'zTAG!'CGT CCACCTI~CC ~Cr00CT CTC'CACCIGT420
CTCCCAITGT
GI~~.~G TmAAT3GAA G~PL~I~'~C CF~GCIC~~.TT AGTIGIC30CC480
TC'IC~GC.'ICdC
4 TATA'IGCZGC 'IC3GCCACIGT GGCI~AAA CTI'I'C'ITTCPr 540
0 OG~'l~AAGIG ~.'rCl.~AC
C~ICrIC3GGTC 'I~C'.~IC.''C~1G GGAAT~a AGCCAGTACr 600
GIAC~AATP.T CTIG'lATAAT
TI~CCIGAAAC T'ICCA~~AA GIGGrCTA'rC AACIC~'~TCpG 660
GGGII~CCCG Af3GGGP.C:C~A
C~AGIGC TTCAGG<.'rAT TCIGAA'rAAC C~CaGAf3G'TC'A 720
AGAAG~1AGCG PG~1GC~TrC
ACI~C~CCC ACIP,~JCrCTC CCrCA~ G~P~C AGC ~AAALaC'~780
5 cr Tcec~~crG~ C~~ GrGc crATyc~TA cz~~~Gr~ a4o
0
ATIC ATG PC,~ AGA TIG AAA ACT TIG AAA GGC TAC 889
'IGC C~ CIG ~T A~
Met Arg Arg Leu Lys Thr Leu Lys Gly Tyr Cys Glu
Leu Cys Met
1 5 10 15
CCC CTC AGA ACA TAT ACT GIG TCC A'I~ ZC3G A~ AAG 937
AAG Ti C GC'A GAA
Ptro Leu Azg Thr Tyr Thr Val Ser Met Trp Met Lys
Lys Ser Ftro G1u
CA 02365278 2001-10-26
Case 12/203
20 . 25 30
AL'r TTC AAA GAG GCS GIC AAA GCA ATT ATT TCT IGC 985
TTC CCA AAT GTC
Thr Phe Lys Glu Ala Val Lys Ala Ile Ile Ser Cys
Phe Pro Asn Val
35 40 45
TIC ATA GCC AGT .'9AG CIG GTT 0f3G GIG GTT TAT 1033
GCC TCC Tu~G TCC AGG
Phe Ile Ala Ser Lys Leu Val Azg Val Val Tyr Ala
Sex Trp Ser Azg
50 55 . 60
Grc cAA ccT c-~c cTC AAC TcC A~ c~ c~.c TIC c-rc foal
c~c Arc TCA cIC
Val G1n Ala Asp Leu Asn Cys Met Glu Asp Leu Leu
Gln Ser Ser Val
65 70 75
C~ 'IC3G AAA TAC TTC C1G AAT ACA TC~T GGG ACJ 1129
S GAC TIT CCT ATA AFG
Pn~ Trp Lys Tyr Phe Leu Asn Thr Cys Gly Thr Asp
Phe Pro Ile Lys
80 85 90 95
P~GC AAT GCA GAG A'IG GTC CAG GCT CTC AAG ATG 1177
TIG AAT GGG PDG AAT
2 Ser Asn Ala Glu Met Val Gln Ala Leu Lys Met Leu
0 Asn Gly Arg Asn
~
~ 105 110
100
AGC ATG GAG Tv~ GPG GTP. GCT CGT AAG C~.C AAA 1225
GAA ACC CSC 'n.3G AAA
Ser Met Glu Ser Glu Val Pro Pro Lys His Lys Glu
Zhr Arg Trp Lys
2 115 120 125
5
TAT CAC TIT GAG GTA GIG AGA CAC AO'A Tg1 CAC CI'A1273
ACC AAC AAG AFG .
Tyr His Phe Glu Val Val Ar3 Asp 'I~r Leu His Leu
Thr Asn Lys Lys
130 135 140
30
AAG GAT CCT C'CC C'C'T TAT AAT TTA ACT ATG TTT 1321
ACA GOG AAT GCG TAC
Lys Asp Pro Pro Pro Tyr Asn Leu Thr Met Phe Thr
Gly Asn Ala Tyr
145 150 155
3 ATT GIG GGT TCC GGA GAT TIC GIT CAA CAT GIT TIG 1369
5 AFG AAC CGT AAA
Ile Val Ala Ser Arg Asp Phe Val Gln His Val Leu
Lys Asn Pro Lys
160 165 170 175
TCC CAA CAA CIG ATT GAA ~ G'IA AAA GAC ACT TAT 1417
AGC CC~1 GAT GAF.
4 Ser Gln Gln Leu Ile Glu Trp Val Lys Asp Thr Tyr
0 Ser Piro Asp Glu
180 185 190
CAC CIC 'IC~G GCC ACC CTT CFG CGT GCA 03G ~G AZG 1465
CCT GGC TCT GIZ'
His Leu.Trp Ala Thr Leu Gln Axg Ala An3 Txp Met
Pzro Gly Ser Va1
4 195 200 205
5
CCC AAC C~ OCC AAG T~1C GAC ATC TCA GAC AZG ACT 1513
TCT ATT GCC PLUG
Pro Asn His Pm Lys Tyr Asp Ile Ser Asp Met Thr
Ser I1~ Ala Arg
210 215 220
50 v
CIG G1'C AFG n3G CAG GGT CAT GAG GGA GAC ATC GAT 1561
AAG C3GT GCT C'G'I'
Letz Val Lys Trp Gln Gly His Glu Gly Asp Ile Asp
Lys Gly Ala Pro
225 230 235
5 TAT GCT . CCC IGC TCT GGA ATC CAC C~SG CMG GCT 1609
5 ATC 'IC'C GTr TAT GOG
Tyr Ala Pn~ Cys Ser Gly Ile His Gln Azg Ala Ile
Cys Val Tyr Gly
240 245 250 255
CA 02365278 2001-10-26
Case 12/203
Gcr c~oG GG~cc Tlc AAT TGG ~ crr c~A AAC cAT c~c cIC TIC c~ AAC 1657
Ala Gly Asp Leu Asn Trp Met Leu Gln Asn His His Leu Leu Ala Asn
260 265 270
AAG TIT GAC CC~1 AAG GTA GAT GAT AP.T GCT CTT CAG Tu"C TTA GAA GAA 1705
Lys Phe Asp Pro Lys Val Asp Asp Asn Ala Leu Gln Cys Leu Glu Glu
275 280 285
TAC CTA QiT TAT AAG GCC ATC TAT GGG ACT GAA CTT ~ C~ACACAO.'~AT 1754
Tyr Leu Arg Tyr Lys Ala Ile Tyr Gly Thr Glu Txu
290 295 300
G~AG~G CTAGCIGIC~G GC'~CAAG~GGA ZG. ~CZG~AC TIGCI~~C 1814
.~C~IGZ~IG GGAGACCAGG GCITIGCAAT T~1~~GC'P.'I~ C'1'ITI~CACCIL;C 1874
Tp~T~,~ TuG~ ~C C?'IGG~AATT GCIGG'C'tC~3G TuAAZGC'1~"'.~C 1934
2 0 'I~TCPCPC ACCCGI'AACC CT~GI~'ITC CTCCACTAAC TTTC'11CACTA PA'IG 1994
~~ ~CGPG FLIT C'~ C~TGIC3G TPGA(sCAC.TT C3ATTIC~IT 2054
GAATGCCIG~C C.'CIT ~CAT'tGTG'I' GGAGCIGCCG TICCTAA'I~A TTCCAGGTIT 2114
GGTAC'~DG AGGAGAAC'TT WC~AAG P~ACCTI'CC C'II!' GTT~ACZTAA 2174
AAATAAP.TAG CTCCIGATl'C AAAGI'AAAAA AAAAAAAAAA AAA ~ ~ 2217
<210> 6
<211> 300
<212> PRT
3 5 <213> Homo sapiena ,
<400> 6
4 4 Met Arg Azg Leu Lys Thr Leu Lys Gly Tyr Cps Glu Leu Cys Met Pro
1 5 10 15
Leu Azg Thr Tyr Thr Val Ser Met Tzp Met Lys Lys Ser Pzro Glu Thzv
20 25 30
Phe Lys Glu Ala Val Lys Ala Ile Ile Ser Cys Phe Pro Asn Val Phe
35 40 45
Ile Ala Ser Lys Leu Val An3 Val Val Tyr Ala Ser Trp Ser An3 Val
5 0 50 55 v 60
Gln Ala Asp Leu Asn Cys Met Glu Asp Leu Leu Gln Ser Ser Val Pra
65 70 75 80
5 5 Trp Lys Tyr Phe Leu Asn Thr Cys Gly Thr Asp Phe Pro Ile Lys Ser
85 90 95
11
CA 02365278 2001-10-26
Case 12/203
Asn Ala Glu Met Val Gln Ala Leu Lys Met Leu Asn Gly Azg Asn Ser
100 105 110
Met Glu Ser Glu Val Pro Pro Lys His Lys Glu Thr An3 Trp Lys Tyr
115 120 ~5
His Phe Glu Val Val Arg Asp Thr Leu His Leu 'Il~r Asn Lys Lys Lys
130 135 140
Asp Pro Pro Pro Tyr Asn Leu 'iii' Met Phe Thr Gly Asn Ala Tyr Ile
145 150 155 160
Val Ala Ser Axg Asp Phe Val G7n His Val Leu Lys Asn Pro Lys Ser
165 170 175
Gln Gln Leu Ile Glu Txp Val Lys Asp Thr Tyr Ser Pro Asp Glu His
180 185 190
Leu Tzp Ala Thr Leu Gln Axg Ala Azg Trp Met Pro Gly Ser Val Pro
2 0 195 200 205
Asn His Pro Lys Tyr Asp Ile,Ser Asp Met~I'hr Ser Ile Ala An3 Leu
210 215 220
2 5 Val Lys Trp Gln Gly His Glu Gly Asp Ile Asp Lys Gly Ala Pro Tyr
225 230 235 240
Ala Pro Cys Ser Gly Ile His Gln Azg Ala Ile Cys Val Tyr Gly Ala
245 250 255
Gly Asp Leu Asn Trp Met Isu Gln Asn His His 7xu Ixu Ala Asn Lys
260 265 270
Phe Asp Pro Lys. Val Asp Asp Asn Ala Leu Gln Cys Leu Glu Glu Tyr
3 5 275 280 285
Lieu Azg Tyr Lys Ala Ile Tyr Gly Thr Glu Leu
290 295 300
<210> 7
<211> 25
<212> PRT
4 5 <213> Homo Sapiens
<400> 7 °
TCAATGAGAA GACCAAGCTG ACGCC 25
<210> 8
<211> 22
<212> DNA
<213> Artificial sequence
1L
CA 02365278 2001-10-26
Case 12/203
<220>
<223> Primer
<400> 8
GGGAGACAGG TGGAGACCGA GC 22
<210> 9
<211> 26
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 9 _
TCACGGGAAC TGCCCTTGCT ACTTGT 26
<210> 10
<211> 27
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 10
3 5 GCTCCTTGAG GTCATGTTCT TCATCTG 27
<210> 11
<211> 21
<212> DNA
<213> Artificial sequence
<220>
4 5 <223> Primer
<400> 11
GGAGACCGAG CACAGGGAAG G 21
<210> 12
<211> 21
5 5 <212> DNA
<213> Artificial sequence
13
CA 02365278 2001-10-26
Case 12/203
<220> .
<223> Primer
<400> 12
CCTTCCCTGT GCTCGGTCTC C 21
<210> 13
<211> 20
<212> DNA
<213> Artificial sequence
<a2o>
<223> Primer
<400> 13
TCGAGTCGAC TATATGTACC 20
<210> 14
<211> 22
<212> DNA
<213> Artificial sequence .
<220>
<223> Primer
<400> 14
AAGGTGAAGG TCGGAGTCAA CG 22
<210> 15
<211> 24
4 0 <212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 15
GGCAGAGATG ATGACCCTTT TGGC 24
~
<210> 16
<211> 27
<212> DNA
5 5 <213> Artificial sequence
<220>
1~
CA 02365278 2001-10-26
Case 12/203
<223> Primer
<400> 16
GACCAGGAGG CATAAACCAC CCGAACC 27
<210> 17
<211> 28
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 17
TTCCATCAAA GTTCTCCTCC ACGCTACC ~~ ~ 28
<210> 18
<211> 28
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 18
CATCCAATTC AAGTCCCCAG CCCCATAA 28
<210> 19
<211> 43
<212> DNA
4 <213> Artificial sequence
0
<220>
<223> Primer
<400> 19
TGTAGCGTGA
AGACGACAGA
AAGGGCGTGG
TACCGAGCTC
GAG
43
v
<210> 20
<211> 22
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
Case 12/203
<400> 20
AGGGCGTGGT ACCGAGCTCG AG 22
<210> 21
<211> 11
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 21
GGCTCGAGCT C 11
<210> 22
<211> 9
<212> PRT
<213> Homo Sapiens
<400> 22
Lys Arg Leu Cys Gln Leu His Tyr Leu
1 5
<210> 23 '
<211> 9
<212> PRT
<213>.Homo Sapiens
<400> 23
Tyr Leu Trp Ala Leu Gly Cys Tyr Met
1 5
<210> 24 .
<211> 9
5 0 <212> PRT t
<213> Homo Sapiens
<400> 24
Ile Leu Asn Asn Leu Glu Val Lys Lys
1 S
't6
CA 02365278 2001-10-26
CA 02365278 2001-10-26
Case 12/203
<210> 25
<211> 9
<212> PRT
<213> Homo Sapiens
<400> 25
Glu Val Lys Lys Lys Arg Glu Pro Phe
1 5
<210> 26
<211> 9
<212> PRT
2 0 <213> Homo Sapiens
<400> 26
2 5 Arg Glu Pro Phe Thr Asp.Thr His Tyr
1 5
30 <210> 27
<211> 9
<212> PRT
<213> Homo sapiens
40
<400> 27
Glu Pro Phe Thr Asp Thr His Tyr Leu
1 5
<210> 28
<211> 9
4 5 <212> PRT
<213> Homo sapiens
<400> 28
Pro Leu Ser Lys Glu Glu Val Glu Phe
1 5
<210> 29
<211> 9
17
CA 02365278 2001-10-26
Case 12/203
<212> PRT
<213> Homo sapiens
<400> 29
Glu Glu Val Glu Phe Pro Ile Ala Tyr
1 5
<210> 30
<211> 9
<212> PRT
<213> Homo sapiens
<400> 30
Glu Arg Leu Leu Arg Alal'Val Tyr Ala
1 5
30
<210> 31
<211> 9 .
<212> PRT
<213> Homo Sapiens
<400> 31
Phe Ile Ala Ser Lys Leu Val Arg Val
1 5
<210> 32
<211> 9
<212> PRT
<213> Homo sapiens
<400> 32
Ash Leu Asn Cys Met Glu Asp Leu Leu
1 5
<210> 33
<211> 9
<212> PRT
<213> Homo sapiens
18
CA 02365278 2001-10-26
Case 12/203
<400> 33
Ser Met Glu Ser Glu Val Pro Pro Lys
1 5
<210> 34
<211> 9
<212> PRT
<213> Homo sapiens
<400> 34
Thr Leu His Leu Thr Asn Lys Lys Lys
1 5
<210> 35
<211> 10 '
<212> PRT
<213> Homo sapiens
<400> 35 '
Phe Val Gln His~Val Leu Lys Asn Pro Lys
3 0 1 5 10
<210> 36
<211> 9
<212> PRT
<213> Homo sapiens
<400> 36
Leu Ile Glu Trp Val Lys Asp Thr Tyr
1 5
<210> 37
<211> 9
<212> PRT
5 0 <213> Homo sapiens r
<400> 37
5 5 Trp Met Leu Gln Asn His His Leu Leu
1 5
CA 02365278 2001-10-26
Case 12/203
<210> 3a
<211> 9
<212> PRT
<213> Homo Sapiens
<400> 38
Leu Leu Ala Asn Lys Phe Asp Pro Lys
1 5
20
<210> 39'
<211> 9
<212> PRT
<213> Homo Sapiens
<400> 39
Asp Pro Lys Val Asp Asp Asn Ala Leu
1 5
<210> 40
<211> 9
<212 > PRT
<213> Homo sapiens
<400> 40
Gln Cys Leu Glu Glu Tyr Leu Arg Tyr
1 5
<210> 41
<211> 9
<212> PRT
4 5 <213> Homo sapiens
<400> 41
Cys Leu Glu Glu Tyr Leu Arg Tyr Lys
1 5
<210> 42
<211> 9
<212> PRT
CA 02365278 2001-10-26
Case 12/203
<213> Homo sapiens~
<400> 42
Ala Val Tyr Ala Pro Gln Asn Ile Tyr
1 5
<210> 43
<211> 9
<212> PRT
<213> Homo sapiens
<400> 43
Tyr Ala Pro Gln Asn Ile Tyr Cys Val
1 5
<210> 44
<211> 9
<212> PRT
<213> Homo sapiens
<400> 44
Tyr Cys Val His Val Asp Glu Glu Val
1 5
<210> 45
<211> 9
<212> PRT
4 <213> Homo sapiens
0
<400> 45
4 His
5 Val
Asp
Glu
Glu
Val
Pro
Arg
Asn
1 5
50 <210> 46
<211> 9
<212> PRT
<213> Homo sapiens
55
<400> 46
z~
CA 02365278 2001-10-26
Case 12/203
Val Asp Glu Glu Val Pro Arg Asn Phe
1 5
<210> 47
<211> 9
<212> PRT
<213> Homo sapiens
<400> 47
Phe Gln Arg Gly Gly Gln Ser Asn Tyr
1 5
<210> 4a
<211> 9
<212> PRT
<213> Homo sapiens
<400> 48
Gln Ser Asn Tyr Phe Leu Leu Pro Lys
1 5
<210> 49
<211> 9
<212> PRT
<213> Homo sapiens
<400> 49
4 0 Leu Leu Pro Lys Cys Leu His Ser Gln
1 5
<210> 50
<211> 9
<212> PRT
<213> Homo sapiens
<400> 50
Arg Arg Leu Lys Thr Leu Lys Gly Tyr
1 5
ZZ
CA 02365278 2001-10-26
Case 12/203
<210> 51
<211> 9
<212> PRT
<213> Homo sapiens
<400> 51
Lys Thr Leu Lys Gly Tyr Cys Glu Leu
1 5
<210> 52
<211> 9
<212> PRT
<213> Homo sapiens
<400> 52
Tyr Cys Glu Leu Cys Met Pro Leu Arg
1 5
<210> 53
<211> 9
<212> PRT
3 0 <213> Homo Sapiens
<400> 53
Glu Leu Cys Met Pro Leu Arg Thr Tyr
1 5
<210> 54
<211> 9
<212> PRT
<213> Homo Sapiens
50
<400> 54 "
Cys Met Pro Leu Arg Thr Tyr Thr Val
1 5
_ <210 > 55
<211> 9
5 5 <212> PRT
<213> Homo sapiens
Z3
' , CA 02365278 2001-10-26
Case 12/203
<400> 55
Pro Leu Arg Thr Tyr Thr Val Ser Met
1 5
<210> 56
<211> 10
<212> PRT
<213> Homo sapiens
'
<400> 56
Tyr Leu Trp Ala 2eu Gly Cys Tyr Met Leu
1 5 - 10
<210> 57
<211> 9
<212> PRT
<213> Homo sapiens
<400> 57
Tyr Met Leu Leu Ala Thr Val Ala Leu
1 5
<210> 58
<211> 9
<212> PRT
4 0 <213> Homo sapiens
<400> 58
4 5 Leu Leu Ala Thr Val Ala Leu Lys Leu
° 1 5
r
50 <210> 59
<211> 10
<212> PRT
<213> Homo sapiens
Z~
CA 02365278 2001-10-26
Case 12/203
<400> 59
Cys Met Glu Asp Leu Leu Gln Ser Ser Val
1 5 10
<210> 60
<211> 9
1 0 <212> PRT
<213> Homo sapiens
<400> 60
Glu Val Asp Pro Ile Gly His Leu Tyr
1 5
<210> 61
<211> 218
<212> PRT
<213> Homo sapiens
<400> 61
3 0 Asn Lys Phe Asp Pro Lys Val Asp Asp Asn Ala Leu Gln. Cys Leu
1 5 10 15
Glu Glu Tyr
.r
zs
CA 02365278 2001-10-26
- 50 -
Velculescu, VE, Zhang, L, Vogelstein, B, and Kinzler, KW
(1995), Science 270: 484-487
Wang, L., et al., (1997), Mol. Immunol. 34: 609-618
Wang, RF (1997), Mol. Med. 3.: 716-731
Wax, S.D., Rosenfield, C.L., and Taubman, M.B., (1994), J.
Biol. Chem. 269, 13041-13047.
Wax, S.D., Tsao, L., Lieb, M.E., Fallon, J.'f., and
Taubman, M.B. (1996), Lab. Invest. 24, ',797-808.
Winter, G., et al., (1994), Annu. Rev. Immunol. 12,
433-455
Woelfel, T, Schneider, J, Zum Buschenfelde, Meyer, KH,
Rammensee, HG, Rotzschke, 0, and Falk, K (1994), Int.
J. Cancer 52: 413-418
Woelfel T, Hauer M, Schneider J, Serrano M, Wolfel C,
Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum
Biischenfelde KH, Beach D (1995), Science 269:
1281-1284
Wu, T.C., Guarnieri, F.G., Staveley-O'Carrol.l, K.F.,
Viscidi, R.P., Levitsky, H.I., Hedrick, L., Cho, K.R.,
August, J.T., and Pardoll, D.M. (1995), Proc. Natl.
Acad. Sci. U.S.A. x(25): 11671-11675.
Zitvogel, L., Mayordomo, J.I., Tjandrawan, T., DeLeo,
A.B., Clarke, M.R., Lotze, M.T., and Storkus, W.J.
(1996), J. Exp. Med. 1~3, 87-97.