Note: Descriptions are shown in the official language in which they were submitted.
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
CLONING OF CDNA OF MAGE'S 5, 8, 9 AND 11 AND THEIR USES IN DIAGNOSIS OF
CANCER
RELATED APPLICATION
This is a continuation in part application of Serial No. 09/260,978, filed
March 2,
1999, incorporated by reference.
FIELD OF THE INVENTION
This invention relates to nucleic acid molecules which are members of the MAGE
family, uses thereof, and a method for determining and quantifying their
expression. Also
a part of the invention are fragments of these nucleic acid molecules,
proteins encoded by
both the whole nucleic acid molecules and the fragments, peptides based
thereon which
form complexes with MHC or HLA molecules, fusion proteins, polytopes, and so
forth.
BACKGROUND AND PRIOR ART
It was shown by Van der Bruggen, et al., Science 254: 1643-1647 (1991), that
there is a family of tumor rejection antigens which complex to human leukocyte
antigens
("HLAs"), and provoke response by autologous, cytolytic T cells. In addition
to Van der
Bruggen et al., supra, see U.S. Patent No. 5,342,774 to Boon, et al.,
incorporated by
reference. These references also describe the isolation of genes which encode
proteins
that are then processed to these tumor lrejection antigens. Further
investigations led to
the discovery of twelve closely related genes. These genes have been found to
be located
in region q28 of the X chromosome. While first named genes MAGE-1 through MAGE-
12, these genes are now referred to as MAGE-A1 through MAGE-A12, in view of
subsequent discoveries. To elaborate, four additional related genes have been
located in
region p21 of the X chromosome, and are referred to as the MAGE-B cluster of
genes.
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
Additional MAGE family members have been located at region q26, and have been
named
MAGE-C 1 and MAGE-C2.
For obvious reasons, it was and is desirable to analyze expression ofMAGE
genes.
There has been extensive work in this area, with patterns of MAGE-A expression
having
been analyzed by reverse transcription-polymerise chain reaction ("RT-PCR"),
in various
tumor samples, tumor cell lines, and normal tissues. Essentially, the level of
transcription
and expression is established, semi-quantitatively, via RT-PCR. This entails
evaluating
intensity of bands on agarose gels, and comparing these to standard dilutions
of material
from a reference or control. This research has established that the genes MAGE-
Al, A2,
A3, A4, A6 and A12 are transcribed, at high levels, in many tumors. Gene MAGE-
A8
was expressed at a high level in one tumor, while MAGE-A5, A9, A10 and A11
appeared
to be transcribed weakly in positive tumors. Collectively, MAGE-A genes were
not found
to be expressed by any normal tissues except in testis and, in a few cases,
placenta. See
Brasseur, et al., Int. J. Cancer 52:839-841 (1992); Brasseur, et al., Int. J.
Canc 63: 375-380
(1995); De Plaen, et al., Immunogenetics 40: 360-369 (1994); van der Bruggen,
et al.,
supra, and Weynants, et al., Int. J. Canc 56: 826-829 ( 1994). The expression
of MAGE-A
genes in testis was restricted to germ line cells in the early phases of
spermatogenesis.
See Takahashi et al., Canc. Res. 55: 3478-3482 (1995). Testis expresses all
MAGE-A
genes, except MAGE-A7. MAGE-A4, A8, A9, A10 and Al l have been found to be
transcribed in placenta.
For CTLs to recognize complexes of TRAs and HLAs, a certain level of
expression of the relevant MAGE-A gene appears to be required. DePlaen, et al.
-2-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
Methods 12:125-142 (1997); Lethe, et al., Melanoma Res 7: Suppl 2: S83-8
(1997) have
shown that in melanoma, the level of expression of MAGE-Al must exceed 10% of
the
level found in reference cell line MZ2-MEL in order to observe recognition of
a MAGE-
A1 peptide presented by HLA-Al. The level of expression of MAGE-A2, A3, A4, A6
and A12 genes has been shown, via semi-quantitative RT-PCR, to be similar to
MAGE-
A1, suggesting that these genes can be processed into TRAs which are presented
for
recognition by CTLs. Several peptides from MAGE-Al and A3 have, in fact, been
found
to be presented by HLAs, and then recognized by autologous CTLs derived from
mixed
lymphocyte-tumor cell culture.
Recently, it was reported that a monoclonal antibody which was reactive with
MAGE-A1 cross reacted with another protein expressed in melanoma. See allowed
U.S.
Patent Application, Serial No. 08/724,774 filed on October 3, 1996 and Carrel,
et al.. Int.
J. Canc 67:417-422 (1996), both of which are incorporated by reference.
Subsequently,
it was found that this cross-reactive protein was encoded by MAGE-A10. In MZ2-
MEL,
the abundance of this protein was similar to that of the MAGE-Al protein.
These results
were surprising, since very low levels of expression of MAGE-A10 had been
found in
MZ2-MEL via RT-PCR. This suggested that the primers used to amplify MAGE-A10
in
the RT-PCR were not very efficient. As a result, investigations were
undertaken to
develop a method for evaluating frequency of expression of a gene which is
independent
of aberrations due to primers. Application of the method, described herein,
led to the
isolation of nucleic acid molecules which are described herein, and are a
feature of the
invention.
-3-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
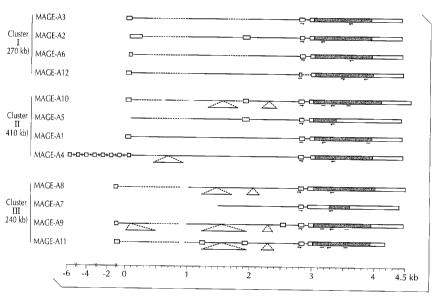
BRIEF DESCRIPTION OF THE FIGURE
Figure 1 presents exon/intron structures of MAGE genes, including for 1VIAGE-
A5, A8, A9 and Al 1 (SEQ ID NOS: 17, 18, 19 and 20).
DETAILED DESCRIPTION
OF PREFERRED EMBODIMENTS
EXAMPLE 1
Experiments were carried out to determine whether the choice of primer
influenced values obtained when quantifying frequency of expression via RT-
PCR. The
frequency of expression of MAGE-A10 was determined using cell line MZ2-MEL,
and
one of two pairs of primers. The first pair is described by De Plaen, et al.,
Immunogenetics 40: 360-369 (1994); i.e.:
CACAGAGCAG CACTGAAGGA G (SEQ ID NO: 1)
and
CTGGGTAAAG ACTCACTGTC TGG (SEQ ID NO: 2),
or
AGCAGCCAAA AGGAGGAGAG TC (SEQ ID NO: 3)
TGACCTCCTC AGGGGTGCAG TA (SEQ ID NO: 4).
SEQ ID NOS: 3&4 correspond to sequences complementary to the last exon of
MAGE-A 10.
The frequency of expression of MAGE-Al was determined using cell line LB 11-
OC1, and two pairs of primers , i.e.:
CGGCCGAAGG AACCTGACCC AG (SEQ ID NO: 5)
-4-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
and
GCTGGAACCC TCACTGGGTT GCC (SEQ ID NO: 6)
or
TCAGGGGACA GGCCAACCC (SEQ ID NO: 7)
and
CTTGCACTGA CCTTGATCAC ATA (SEQ ID NO: 8)
In the experiments, total RNA was extracted from cells. Then, 2 ~.g samples
were
used for reverse transcription, following Weynants, et al., su ra. The PCR was
then
carned out using 1/10 of total cDNA, supplemented with 2.5 q 1 of 10 x PCR
buffer, 2.5
~ 1 of l OmM of dNTP, 10 pmoles of the primers, and 0.5 units of polymerise,
plus water
to a volume of 25 ~ 1. Each mixture was heated to 94°C for 5 minutes,
followed by
amplification for 30 cycles. In the case of SEQ ID NOS: 1-4, 7 & 8, a cycle
was 1 minute
at 94°C, two minutes at 65°C, and three minutes at 72°C.
For SEQ ID NOS: 5 & 6, a
cycle was 1 minute at 94°C, and 3 minutes at 72°C. Cycling was
concluded with a final
extension at 72°C for 5 minutes. Analysis was carned out using 5ql
samples, each of
which was run on a 1% agarose gel, and visualized via standard ethidium
bromide
staining.
When SEQ ID NOS: 1 & 2 were used, a low level of expression of MAGE-A10
was found, whereas use of SEQ ID NOS: 3 & 4 showed a level of expression
equivalent
to that of MAGE-Al. This result is in agreement with the Western blotting work
of
Carrel, et al., supra, which showed equivalent levels of expression.
-5-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
Given that SEQ ID NOS: 3 & 4 corresponded to regions of the last exon of
MAGE-A10, it was possible that contaminating genomic DNA had also been
amplified.
To test this possibility, amplification was carned out with omission of the
reverse
transcription step. No amplification was observed, however, indicating that
there
probably was no contamination. A number of PCR products were sequenced, where
SEQ
ID NOS: 3 & 4 had been used as primers. All corresponded to MAGE-A10.
When results obtained using the primers for MAGE-Al were compared, different
levels of expression were observed, with SEQ ID NOS: 7 & 8 showing higher
levels than
SEQ ID NOS: 5 & 6.
Very low expression levels had been observed previously for MAGE-A5, A9 and
A11. Hence, it was suspected that changing primers might resolve this.
EXAMPLE 2
The results obtained supra suggested that a better method for determining
frequency of expression of genes, MAGE genes for example, was needed. The
method
developed is described in this example.
A cDNA library was prepared from a MAGE-A positive sample, following
standard procedures; see De Plaen, et al., Methods 12: 125-142 (1997),
incorporated by
reference, and was maintained as recombinant plasmids in E. coli bacteria.
Specifically, samples were homogenized in guanidine thiocyanate to form a
lysate,
which was then loaded on top of a CsCI cushion. Then, poly(A)+ RNA was
isolated by
processing total RNA through two successive oligo(dT) cellulose columns. The
isolated
poly(A)+ RNA was converted to cDNA with an oligo(dT) primer which contained a
NotI
-6-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
restriction site. The resulting cDNA was ligated to BstXI adaptors, digested
with NotI,
and then inserted into the BstXI and NotI sites of expression vector
pcDNAI/Amp. The
resulting recombinant plasmids were introduced into E. coli DHS«, using
standard
electroporation techniques, followed by selection with ampicillin (25 ~.g/ml).
All libraries were then diluted in LB medium, supplemented with ampicillin, to
obtain 3-6 clones/~1. Following this, 9.6 mls of each dilution were seeded in
a 96
microwell plate (100 ~ls per microwell). Two or three plates were seeded from
every
library in order to obtain about 100,000 independent clones spread over the
plates. Plates
were then incubated overnight at 37°C, after which 10 ql from every
microwell were
pooled, to obtain 20 different pools from every plate (i.e., 8 pools from
lines, and 12 pools
from columns). Plates and pools were duplicated, the masters frozen (-
70°C, LB medium
containing 20% glycerol), and duplicates were maintained at 4°C for PCR
assays.
The PCR assays were carried out on both the living and frozen bacteria, with
the
first assays being carried out on pools from lines and columns. Positive
microwells were
found at the intersection of a positive line, and a positive column. To carry
out the
amplification assay, 3 gl of living bacteria were supplemented with 2.5 ql of
10 x PCR
buffer, 2.5 ~,l of lOmM dNTP, 10 pmoles of each primer, 0.5 units of
polymerase, and
water to a volume of 25 ~l .
In most cases, the number of positive wells in a plate was less than 20%. In
accordance with Poisson distribution if the percent of positive clones was
less than 20%,
the likelihood of having a single clone in a well should be 90% or greater.
Limiting
_7_
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
dilution could be carned out to the point where less than 10% of the wells are
positive,
with a presumed accuracy of 95%.
In these experiments, as indicated, the number of positives was less than 20%.
It
was then possible to calculate the abundance of the different MAGE-A cDNAs in
the
library, taking the number of independent clones in each well into account.
The experiments were repeated for seven cDNA libraries (five tumor cell lines,
one testis library, one placenta library) for all twelve MAGE-A genes. The
results are
presented in the Table, which follows.
_g_
CA 02366059 2001-08-23
WO PCT/US00/05346
00/52163
N 7, O a~
' +
d o ~. v ~ oo cO w 'n
~
~
o + o of
W + O
+ O
o ~ ~ o ~ o r f~ +
+ + + + ~ w
M N O
+ + ~
O O ~
'+' L
~
'f'
.-. .-. _ _ .
w
N
+ X
O
tn +' N
d O ~ O '~ p O L
O O ~
O ~ O O O p O .~ U
i
~
O + N N p O N
+ + + +
O
U
o N d _ o ~ a~ a~ L
_
~ s > a.
'
L
~ sØ 0.
' aW n a~
b
0
.
4 G
.
gin O
o ~p N
+ 0 0 0 % ~ C by ~
'~
0 0 ~v .~ o o + O O N O
,-. v
'U ,p N~ ~ O O ~
+
~ O_ ~ _~ Cd Vi ~ N
+ N
+ ~ + N C O O
+
N
_
'~
U O
.
~ U N
y Cv
d ~ ~' o ~ ,
o O o '1 O O O U
W .-. v .-. O ~ o ~ T p T3 .,
~ v
~
_
G N CC C ~ CC
~
O O O O .- .-. .D
+
~ w U y
,Y ay o-
00 ~ o ~ o.
0 0 0 ' v
d a~
v~ 0 0 0 o ,~ oo ~
N fz1 +~ . d o L D
o ro+ a+ z.~ ~N
..
d o a o a
U M
_
d-o~.o
.
U W
c y
s~ ~ v
C7 : ~ a~
o d o g o 0 0 0 o d c '~a ~
W O fl U
~ OU
O ~ V V ~ o 1~ .
~ n n ~ O ~ n ~
0 N O N
p,. N d _ y
d N
O O O O O O
0 3 ~ >,
V ~ .9 ~
!/J w U U 0
0
N ~ O O ~ N w N
_ yr O O ~ O L
W O+ o+ o+ + + ~+ ~ ~ O.
o
+ O + O O te
+ + G
+ 00 N ..
d ; , + VJ a
~ ~ ~ ~ o ~ c ~ ~ 3
E
O O C O C U C .D L
O ~ O O C C C
W
w
U O ~ N N O O M
Q i i i + O
d , . o ~ o ~ fV O
~ ~ ~ (C
c
3
- ~ E ca
c '~
o
a
L
~ E
o
Y, v, M O ~ ~ .'0... N ~ O
O O
.... O _. .... ~ t
t7
+ .
+ O n
(
U.. O O ~+ N O+ Q- + N N
n + ~ h
O N
~ O M p \V' 1\ H ~ U L' L w
~ .J N~Q,
~
... _ _. ~, O X
"" T
_
a) N
a7 y
N
N 7, O E"'
w
U
U d ~ - ~i ~- -- o
W ~ + t ~ O O O
i
c7 O O + O ~r~ o+ N ~'~, ~.
g+ + + ~ 3
o+
d o ~ ~~ ~.; ~i D
+ + _ p
~ o '-' a ~- '~
._ .-. ~ _ ._ .-. ~ ~ . U
C
_
_
U 1.U. d 'D n!
oz's o
Q ~ ~ ~ ~ ~ Nn.~
.i 1 O + .~ O ~ V fc$
-~- T
C7 + + ~
a o + N N Y 3 ~ 'o o
+ +
- c o ~ o O ._
_ ~ U fl. "CJ
.
N
E
O
U
~ O c
~
C ~ C U 01
c
d ~ ~ O ~ ~~ O C L O U
.-..+ G
W o=- , o.+.. ov o~ o+ or. s.o
v,+
~ .. O N ~ O N ~ ~ ..O W
+ +
O + + .- _ +
+ + O
+
+ V M ... ~ C ~ 4O
+ +
. 4
-. ~ _I
+
~. x ~.
,..a U
W W
n: O
M y
N M ~ .-. F."
U
a
9
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
The results were compared to the results obtained in RT-PCR assays, as set
forth in
the table supra. MAGE-A1, A9 and A10 were evaluated twice, with different
pairs of
primers, and a level of expression was estimated based upon intensity of
banding on an
agarose gel.
The limiting dilution assay revealed a level of expression of MAGE-A10 which
was
comparable to that obtained with primers SEQ ID NOS: 3 and 4, and higher than
that
obtained with SEQ ID NOS: 1 and 2. These higher values are in agreement with
Western
blotting work reported by Carrel, et al., supra, and in allowed U.S. patent
application Serial
No. 0/724,774 filed October 3, 1996, incorporated by reference. The frequency
was
comparable to that of MAGE-A1 in several of the lines. In other lines, while
the level
decreased, it was still comparable to levels for A1, A2 or A12.
-10-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
The calculated frequencies for A1, A2, A3, A4, A6 and A12 were essentially
consistent with results obtained by RT-PCR. In one library, the results (one
clone in 124,000
analyzed), was consistent with the results obtained with SEQ ID NOS: 7 and 8,
but not SEQ
ID NOS: 5 and 6, suggesting that SEQ ID NOS: S and 6 are more efficient at
determining
transcript present, but SEQ ID NOS: 7 and 8 are better at determining
expression levels.
EXAMPLE 3
One aspect of the results of the experiments described su ra, which provoked
further
investigation was expression of MAGE-A8. Weak expression was always observed,
with the
exception of the cell line TT-THYR, which showed high levels of expression in
a semi-
quantitative PCR assay.
The average size of an insert in the TT-THYR library was only 0.9kb, so
primers were
designed which would amplify a segment of the last 450 nucleotides of MAGE-A8
mRNA.
Similar primers were designed for MAGE-Al, A2, A4 and A8 as well, i.e.:
SEQ ID NO.: 9 GAAGAGAGCGGTCAGTGTTC-3 (sense)
SEQ ID NO.: 10 AATCCAGGTATGCATATATCTTTA (anti-sense)
SEQ ID NO.: 11 GCCTCTTTGAAGAGAGCAGTC (sense)
SEQ ID NO.: 12 CAAAGAAGCAAAAACATACACATA (anti-sense)
SEQ ID NO.: 13 CACTCTGTTTGAAGAAAATAGTC (sense)
SEQ ID NO.: 14 AGTATCTTTTAATTTATCTCACCTA (anti-sense)
SEQ ID NO.: 15 AGCATGTTGGGTGTGAGGGA (sense)
SEQ ID NO.: 16 AGGGTACACTAAGAGGTACAG (anti-sense)
-11-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
(SEQ ID NOS: 9 and 10 amplify Al, NOS: 11 and 12 amplify A2, NOS: 13 and 14
amplify A4, and NOS: 15 and 16 amplify A8)
RT-PCR was carned out as described, supra, using these primers. When
completed, the
frequency of MAGE-A8 expression was found to be much higher; i.e., on a par
with MAGE-
A2.
When the results for testis were analyzed, the level of expression of MAGE-A4
was
found to be higher than MAGE-A1, levels of A2, A3, A8, A9 and A11 were low,
and no
cDNA for A6, A10 or A12 was found. These results are in accordance with those
provided
by Carrel et al. supra, for MAGE-Al AND MAGE-A10.
With respect to placenta, MAGE-A10 showed the highest level of expression,
while
A1-A7 A9 and A12 were not found at all among 230,000 clones analyzed.
EXAMPLE 4
While the literature on the MAGE-A genes is substantial, cDNA for several
members
of the family has never been isolated, notwithstanding inferences regarding
their structure,
based upon the structure of MAGE-A1.
The approach described in example 2, supra, led to isolation of cDNA for MAGE-
A5,
A8, A9, A10 and A11. The cDNA for MAGE-A10 was described in e.g., U.S. patent
application Serial No. 08/724,774, filed October 3, 1996 and incorporated by
reference, but
the others were not known. The cDNA for A5, A8, A9 and Al l is presented as
SEQ ID
NOS: 17 through 20, respectively. Further, knowledge of the sequences of cDNA
led to an
ability to complete exon/intron structures of the genes, as shown in figure 1.
-12-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
The sequencing of MAGE-AS cDNA led to some interesting observations, as it
consisted of the first two exons of MAGE-A10, followed by a sequence
corresponding to a
previously unknown exon, and two exons of MAGE-A5.
The foregoing examples describe the invention, which in addition to nucleic
acid
molecules as described herein includes a method for determining the frequency
of expression
of a particular gene or gene of interest. The method involves preparing cDNA
from a sample,
and then transforming or transfecting the cDNA into cells, to create a library
of
transformants/transfectants. These cells are then divided into a plurality of
samples of
approximately equal size, after which each sample is assayed for presence (as
compared to
quantity), of cDNA. The number of positive samples should be less than or
equal to 20% of
the total number of and, more preferably, equal or less than 10% of the number
of samples
being tested. If the number of positives is greater than the chosen value,
then the library is
diluted, divided into samples and the assay is repeated. When the positive
value is below the
chosen value, the frequency of each MAGE cDNA is determined.
Preferably, the method is carried out by distributing the samples in a
predetermined
array, so that different portions of samples can be pooled. When the samples
are arrayed in
this way, one can determine which samples do contain the cDNA of interest, by
determining
where the two sample pools intersect. For example, consider a rectangular
array of samples,
arranged in vertical and horizontal lines. If the horizontal lines are
represented by letters, i.e.,
"A", "B", "C", etc., and vertical lines by numbers, i.e., "1", "2", "3", etc.,
then one can create
pools "A", "B", "2", "3", etc. Each vertical and horizontal line will
intersect at one point,
-1J-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
these points being represented by codes such as "A1", "B2", "C3", "D4", etc.
Ifboth pool "B"
and pool "7" are positive, then one can conclude that the sample at point "B7"
is positive. By
doing this, one can identify a well containing the sample of interest.
In addition to quantifying expression, the method permits the artisan to
identify cDNA
molecules of interest, especially those which are present at low frequency. As
was described
herein, practice of the invention led to isolation of cDNA for various MAGE
genes. Such
cDNA had not been isolated previously, and is a further feature of the
invention, i.e., isolated
cDNA molecules which encode MAGE-A8, MAGE-A9, and MAGE-11 proteins, such as
cDNA molecules which encode proteins whose amino acid sequence is that of the
protein
encoded by any of SEQ ID NOS: 18, 19 or 20.
Also a part of the invention are newly isolated nucleic acid molecules, such
as the one
set forth in SEQ ID NO: 17 or other similar molecules i.e., those comprising
two exons for
MAGE-AS and two exons for MAGE-A10, separated by a nucleotide sequence between
the
MAGE-AS and MAGE-A10 sequences as well as nucleic acid molecules which
comprise
portions that hybridize to both the MAGE-AS portion, and the MAGE-A10 portion.
These
nucleic acid molecules, i.e., all of the nucleic acid molecules described
herein, can be used
to make expression vectors which comprise the nucleic acid molecule operably
linked to a
promoter. These vectors, as well as the isolated nucleic acid molecules
themselves, can be
used to transform or to transfect cells, to produce recombinant cells thereby.
These nucleic acid molecules can also be used both diagnostically and
therapeutically.
In the diagnostic area, one can examine a sample, such as a cell containing
sample, a cell
-14-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
lysate, etc., for expression of the nucleic acid molecules described herein,
using oligomer
probes in connection with standard methods, such as polymerase chain
reactions, and so forth.
One can also assay such samples by determining presence of the proteins
encoded thereby.
Also a part of the invention are methods based upon these newly identified and
isolated molecules. For example, one can determine expression of a MACE gene
by
contacting a sample with one or more oligonucleotides which hybridize
specifically to a
MAGE nucleic acid molecule of interest. For example SEQ ID NO: 9 and/or SEQ ID
NO:
can be used to determine MAGE-A1, SEQ ID NO: 11 and/or 12 can be used to
determine
MAGE-A2, and so forth. Any form of hybridization based assay can be used, as
exemplified,
10 but not limited to PCR. One can also assay for the MAGE proteins, using
standard
methodologies such as antibody assays, or other systems for determining
proteins.
Also a part of the invention are peptides consisting of from about 8 to about
25 amino
acids concatenated to each other along the amino acid sequence of the MAGE
proteins which
are a part of the invention. Such peptides are specific binders for MHC
molecules, such as
MHC Class I or Class II molecules, including HLA molecules such as HLA-Al, A2,
A3,
A24, B7, B8, B35, B44, B52, and CW6. Determination of relevant sequences can
be carned
out using, e.g., Parker, et al, J. Immunol 152:163 (1994), D'Amaro, et al Hum.
Immunol
43:13-18 (1995), Drijfhout, et al, Hum. Immunol 43:1-12 or Sturniolo, et al,
Nat. Biotechnol
17(6):555-61 (1999) all of which are incorporated by reference, or websites
such the NIH
worldwide web site, found at URLhttp:\\bimas.dcrt.nih.gov., and http://www.uni-
tuebingen.de/uni/kxc, which are incorporated by reference. The tables which
follow list some
-15-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
of these peptides, with reference to the relevant MAGE amino acid sequence.
The complete
amino acid sequences are set out at SEQ ID NOS:21-24, where SEQ ID N0:21 is
that for
MAGE A5, SEQ ID N0:22 is that for MAGE A8, SEQ ID N0:23 is that for MAGE A9,
and
SEQ ID N0:24 is that for MAGE Al l:
MAGE A5: HLA-A1 Binders
98-107 SPDPESVFR
69-78 SAIPTAIDF
32-41 TTEEQEAVS
116-125 LIHFLLLKY
113-122 VADLIHFLL
115-124 DLIHFLLLK
2-11 SLEQKSQHC
77-86 FTLWRQSIK
73-82 TAIDFTLWR
74-83 AIDFTLWRQ
15-24 GLDTQEEAL
MAGE A5: HLA-A2 Binders
112-123 KVADLIHFL
108-117 ALSKKVADL
24-33 GLVGVQAAT
70-79 AIPTAIDFT
-16-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
38-47 AVSSSSPLV
22-31 ALGLVGVQA
15-24 GLDTQEEAL
45-54 LVPGTLGEV
31-40 ATTEEQEAV
71-80 IPTAIDFTL
113-122 VADLIHFLL
25-34 LVGVQAATT
78-87 TLWRQSIKG
48-57 GTLGEVPAA
18-27 TQEEALGLV
MAGE A5: HLA-A3 Binders
115-124 DLIHFLLLK
103-112 SVFRAALSK
108-116 ALSKKVADL
15-23 GLDTQEEAL
77-85 FTLWRQSIK
116-124 LIHFLLLKY
24-32 GLVGVQAAT
73-81 TAIDFTLWR
22-30 ALGLVGVQA
MAGE A5: HLA-A24 Binders
-17-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
112-120 KVADLIHFL
42-50 SSPLVPGTL
17-25 DTQEEALGL
37-45 EAVSSSSPL
76-84 DFTLWRQSI
113-121 VADLIHFLL
71-79 IPTAIDFTL
15-23 GLDTQEEAL
108-116 ALSKKVADL
69-77 SAIPTAIDF
97-105 TSPDPESVF
63-71 KSPQGASAI
109-117 LSKKVADLI
67-75 GASAIPTAI
114-122 ADLIHFLLL
111-119 KKVADLIHF
MAGE A5: HLA-B7 Binders
71-79 IPTAIDFTL
112-123 KVADLIHFL
108-116 ALSKKVADL
37-45 EAVSSSSPL
17-25 DTQEEALGL
42-50 SSPLVPGTL
113-121 VADLIHFLL
-18-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
38-47 AVSSSSPLV
60-68 GPLKSPQGA
54-62 PAAGSPGPL
114-122 ADLIHFLLL
67-75 GASAIPTAI
15-23 GLDTQEEAL
45-53 LVPGTLGEV
30-38 AATTEEQEA
31-39 ATTEEQEAV
100-108 DPESVFRAA
8-16 QHCKPEEGL
25-33 LVGVQAATT
109-117 LSKKVADLI
MAGE A5: HLA-B8 Binders
108-117 ALSKKVADL
37-46 EAVSSSSPL
109-118 LSKKVADLI
71-80 IPTAIDFTL
67-75 GASAIPTAI
42-50 SSPLVPGTL
102-110 ESVFRAALS
MAGE A5: HLA-B35 Binders
-19-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
71-79 IPTAIDFTL
97-105 TSPDPESVF
109-118 LSKKVADLI
42-50 SSPLVPGTL
63-71 KSPQGASAI
112-120 KVADLIHFL
37-46 EAVSSSSPL
69-77 SAIPTAIDF
17-25 DTQEEALGL
60-68 GPLKSPQGA
116-124 LIHFLLLKY
67-75 GASAIPTAI
108-116 ALSKKVADL
113-121 VADLIHFLL
100-107 DPESVFRAA
31-39 ATTEEQEAV
106-114 RAALSKKVA
41-49 SSSPLVPGT
102-110 ESVFRAALS
30-38 AATTEEQEA
MAGE A5: HLA-B44 Binders
69-77 SAIPTAIDF
34-42 EEQEAVSSS
20-28 EEALGLVGV
-20-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
33-41 TEEQEAVSS
90-98 QEEEGPSTS
- 51-59 GEVPAAGSP
114-122 ADLIHFLLL
41-49 SSSPLVPGT
92-100 EEGPSTSPD
97-105 TSPDPESVF
48-56 GTLGEVPAA
91-99 EEEGPSTSP
36-44 QEAVSSSSP
116-124 LIHFLLLKY
56-64 AGSPGPLKS
23-31 LGLVGVQAA
13-23 EEGLDTQEE
75-83 IDFTLWRQS
100-108 DPESVFRAA
17-25 DTQEEALGL
MAGE A5: HLA-B52 Binders
18-26 TQEEALGLV
97-105 TSPDPESVF
45-53 LVPGTLGEV
109-117 LSKKVADLI
71-79 IPTAIDFTL
63-71 KSPQGASAI
-21-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
23-31 LGLVGVQAA
67-75 GASAIPTAI
20-28 EEALGLVGV
69-77 SAIPTAIDF
46-54 VPGTLGEVP
14-22 EGLDTQEEA
106-114 RAALSKKVA
113-121 VADLIHFLL
31-39 ATTEEQEAV
60-68 GPLKSPQGA
76-84 DFTLWRQSI
96-104 STSPDPESV
89-107 NQEEEGPST
112-120 KVADLIHFL
MAGE A5: HLA-CW6 Binders
112-120 KVADLIHFL
108-116 ALSKKVADL
71-79 IPTAIDFTL
113-121 VADLIHFLL
116-124 LIHFLLLKY
105-113 FRAALSKKV
67-75 GASAIPTAI
18-26 TQEEALGLV
37-45 EAVSSSSPL
-22-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
114-122 ADLIHFLLL
45-53 LVPGTLGEV
42-50 SSPLVPGTL
15-23 GLDTQEEAL
8-16 QHCKPEEGL
20-28 EEALGLVGV
17-25 DTQEEALGL
41-49 SSSPLVPGT
63-71 KSPQGASAI
76-84 DFTLWRQSI
109-117 LSKKVADLI
MAGE Al l: HLA-A1 Binders
376-384 GTDPACYEF
281-290 EVDPTSHSY
211-220 LIDPESFSQ
71-80 NLEDRSPRR
142-150 QAEEQEAAF
352-360 FGEPKRLLT
MAGE A11: HLA-A2 Binders
313-321 GLLIIVLGV
350-358 FLFGEPKRL
-23-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
221-229 ILHDKIIDL
89-97 VLWGPITQI
333-341 VMWEVLSIM
384-392 FLWGPRAHA
271-279 MQLLFGIDV
225-233 KIIDLVHLL
398-406 KVLEYIANA
337-345 VLSIMGVYA
289-297 YVLVTSLNL
316-324 IIVLGVIFM
335-353 WEVLSIMGV
MAGE Al 1: HLA-A3 Binders
272-280 QLLFGIDVK
228-236 DLVHLLLRK
89-97 VLWGPITQI
359-367 LTQNWVQEK
313-321 GLLIIVLGV
MAGE Al l: HLA-A24 Binders
343-351 VYAGREHFL
255-263 NYEDYFPEI
-24-
CA 02366059 2001-08-23
WO 00/52163 PCT/CTS00/05346
351-359 LFGEPKRLL
225-234 KIIDLVHLL
288-296 SYVLVTSLN
236-244 KYRVKGLIT
82-90 RITGGEQVL
311-319 KSGLLIIVL
413-421 SYPSLYEDA
MAGE Al l: HLA-B7 Binders
98-106 FPTVRPADL
414-422 YPSLYEDAL
283-291 DPTSHSYVL
64-73 QVFREQANL
76-84 SPRRTQRIT
289-297 YVLVTSLNL
127-135 QAQEEDLGL
307-315 QSMPKSGLL
266-279 EASVCMQLL
147-155 EAAFFSSTL
MAGE Al 1: HLA-B8 Binders
98-106 FPTVRPADL
221-229 ILHDKIIDL
-25-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
241-250 GLITKAEML
234-242 LRKYRVKGL
2-10 ETQFRRGGL
309-317 MPKSGLLII
307-315 QSMPKSGLL
MAGE A 11: HLA-B 5 Binders
374-382 VPGTDPACY
410-418 DPTSYPSLY
102-110 RPADLTRVI
394-402 TSKMKVLEY
309-317 MPKSGLLII
283-291 DPTSHSYVL
181-190 SPTAMDAIF
414-422 YPSLYEDAL
98-106 FPTVRPADL
389-497 RAHAETSKM
48-56 APYGPQLQW
311-319 KSGLLIIVL
378-386 DPACYEFLW
MAGE A11: HLA-B44 Binders
-26-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
143-151 AEEQEAAFF
58-66 QDLPRVQVF
256-269 YEDYFPEIF
392-400 AETSKMKVL
166-174 AESPSPPQS
144-152 EEQEAAFFS
394-402 TSKMKVLEY
280-288 KEVDPTSHS
265-273 REASVCMQL
406-414 ANGRDPTSY
410-418 DPTSYPSLY
335-343 WEVLSIMGV
146-154 QEAAFFSST
331-339 EEVMWEVLS
MAGE Al l: HLA-B52 Binders
309-318 MPKSGLLII
102-110 RPADLTRVI
314-322 LLIIVLGVI
333-341 VMWEVLSIM
271-279 MQLLFGIDV
218-226 SQDILHDKI
181-189 SPTAMDAIF
315-323 LIIVLGVIF
29-37 FGLQVSTMF
-27-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
219-227 QDILHDKII
57-65 SQDLPRVQV
90-98 LWGPITQIF
245-254 KAEMLGSVI
329-338 IPEEVMWEV
256-264 YEDYFPEIF
58-66 QDLPRVQVF
128-136 AQEEDLGLV
283-291 DPTSHSYVL
87-95 EQVLWGPIT
325-334 EGNCIPEEV
MAGE A1 l: HLA-CW6 Binders
225-233 KIIDLVHLL
311-319 KSGLLIIVL
287-295 HSYVLVTSL
221-229 ILHDKIIDL
147-155 EAAFFSSTL
229-237 LVHLLLRKY
269-277 VCMQLLFGI
244-252 TKAEMLGSV
313-321 GLLIIVLGV
222-230 LHDKIIDLV
184-192 AMDAIFGSL
-28-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
MAGE A9: HLA-Al Binders
94-103 SVDPAQLEF
167-176 EVDPAGHSY
262-271 GSDPAHYEF
1354-143MLESVIKNY
153-162 ASEFMQVIF
189-198 LGDGHSMPK
238-247 YGEPRKLLT
112-121 VAELVHFLL
2(4~-255TQDWVQENY
280-289 TSYEKVINY
249-258 WVQENYLEY
MAGE A9: HLA-A2 Binders
199-208 ALLIIVLGV
2123-232ALSVMGVYV
102-111 FMFQEALKL
307-316 VLGEEQEGV
270-279 FLWGSKAHA
175-184 YILVTALGL
~l-166 MQVIFGTDV
140-149 KNYKRYFPV
219-228 VIWEALSVM
290-299 VMLNAREPI
284-293 KVINYLVML
-29-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
221-230 WEALSVMGV
24-33 GLMGAQEPT
187-196 SMLGDGHSM
MAGE A9: HLA-A3 Binders
235-244HMFYGEPRK
114-123ELVHFLLHK
203-212IVLGVILTK
225-234SVMGVYVGK
102-111FMFQEALKL
1L~#-143MLESVIKNY
158-167QVIFGTDVK
118-127FLLHKYRVK
199-208ALLIIVLGV
107-116ALKLKVAEL
?1'~-279FLWGSKAHA
148-157VIFGKASEF
MAGE A9: HLA-A24 Binders
281-290SYEKVINYL
237-246FYGEPRKLL
~-238 VYVGKEHMF
71-80 VYYTLWSQF
-30-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
141-150NYKRYFPVI
236-245MFYGEPRKT.
284-293KVINYLVML
MAGE A9: HLA-B7 Binders
l~-178 DPAGHSYIL
300-309YPSLYEEVL
127-136EPVTKAEML
284-293KVINYLVML
111-120KVAELVHFL
ll9T7-206KAALLIIVL
17-26 EAQGEDLGL
107-116ALKLKVAEL
193-202HSMPKAALL
195-204MPKAALLII
212-237GVYVGKEHM
181-190LGLSCDSML
201-210LIIVLGVIL
173-182HSYILVTAL
175-184YILVTALGL
X101 SSSVDPAQL
67-76 SSISVYYTL
102-111FMFQEALKL
112-121VAELVHFLL
180-189ALGLSCDSM
-31-
CA 02366059 2001-08-23
WO 00/52163 PCT/iJS00/05346 -
MAGE A9: HLA-B8 Binders
107-116ALKLKVAEL
127-136EPVTKAEML
195-204MPKAALLII
1 ~i-202HSMPKAALL
169-178DPAGHSYIL
300-309YPSLYEEVL
MAGE A9: HLA-B52 Binders
195-204MPKAALLII
21~-209LLIIVLGVI
219-228VIWEALSVM
157-166MQVIFGTDV
104-113FQEALKLKV
131-140KAEMLESVI
1Z-161 KASEFMQVI
96-105 DPAQLEFMF
201-210LIIVLGVIL
300-309YPSLYEEVL
212-221DNCAPEEVI
~9-178 DPAGHSYIL
278-287AETSYEKVI
-32-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
141-150 NYKRYFPVI
MAGE A9: HLA-CW6 Binders
197-206 KAALLIIVL
111-120 KVAELVHFL
173-182 HSYILVTAL
107-116 ALKLKVAEL
199-208 ALLIIVLGV
100-109 LEFMFQEAL
130-139 TKAEMLESV
115-124 LVHFLLHKY
201-210 LIIVLGVIL
MAGE A9: HLA-B44 Binders
105-113QEALKLKVA
221-229WEALSVMGV
~'b-SS EEVSAAGSS
296-304EPICYPSLY
280-288TSYEKVINY
217-225EEVIWEALS
255-263LEYRQVPGS
~-286 AETSYEKVI
166-174KEVDPAGHS
-33-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
33-41 GEEEETTSS
96-104 DPAQLEFMF
222-230EALSVMGVY
MAGE A9: HLA-B35 Binders
260-269 VPGSDPAHY
296-305 EPICYPSLY
195-204 MPKAALLII
300-309 YPSLYEEVL
127-136 EPVTKAEML
96-105 DPAQLEFMF
169-178 DPAGHSYIL
280-289 TSYEKVINY
65-74 ASSSISVYY
264-273 DPAHYEFLW
92-101 SSSVDPAQL
18-27 AQGEDLGLM
222-231 EALSVMGVY
64-73 GASSSISVY
197-206 KAALLIIVL
MAGE A8: HLA-Al Binders
-34-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
171-180 EVDPAGHSY
266-275 GSDPVRYEF
138-147 MLESVIKNY
157-166 ASECMQVIF
250-259 TQEWVQENY
98-107 SPDPAHLES
116-125 VAELVRFLL
253-262 WVQENYLEY
193-202 LGDDQSTPK
181-190 LVTCLGLSY
MAGE A8: HLA-B52 Binders
199-208 TPKTGLLII
223-232 AIWEALSVM
161-170 MQVIFGIDV
286-295 YVKVLEHVV
204-213 LLIIVLGMI
135-144 KAEMLESVI
262-271 RQAPGSDPV
205-214 LIIVLGMIL
156-165 KASECMQVI
173-182 DPAGHSYIL
MAGE A8: HLA-A3 Binders
-35-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
280-289 ALAETSYVK
118-127 ELVRFLLRK
122-131 FLLRKYQIK
1-10 MLLGQKS QR
203-212 GLLIIVLGM
210-219 GMILMEGSR
162-171 QVIFGIDVK
138-147 MLESVIKNY
2-11 LLGQKSQRY
111-120 ALDEKVAEL
274-283 FLWGPRALA
29-38 QIPTAEEQK
24-33 GLMDVQIPT
253-262 WVQENYLEY
MAGE A8: HLA-B7 Binders
299-308 RVRISYPSL
304-313 YPSLHEEAL
173-182 DPAGHSYIL
22-31 APGLMDVQI
64-73 SPEGASSSL
240-249 SVYWKLRKL
115-124 KVAELVRFL
37-46 KAASSSSTL
17-26 QAQGEAPGL
-36-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
199-208TPKTGLLII
216-225GSRAPEEAI
196-205DQSTPKTGL
116-125VAELVRFLL
MAGE A8: HLA-B8 Binders
197-206 QSTPKTGLL
199-208 TPKTGLLII
240-249 SVYWKLRKL
297-306 NARVRISYP
111-120 ALDEKVAEL
299-308 RVRISYPSL
MAGE A8: HLA-A2 Binders
288-297 KVLEHVVRV
274-283 FLWGPRALA
24-33 GLMDVQIPT
111-120 ALDEKVAEL
115-124 KVAELVRFL
45-54 LIMGTLEEV
179-188 YILVTCLGL
161-170 MQVIFGIDV
203-212 GLLIIVLGM
-37-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
191-200 GLLGDDQST
223-232 AIWEALSVM
71-80 SLTVTDSTL
279-288 RALAETSYV
251-260 QEWVQENYL
184-193 CLGLSYDGL
MAGE A8: HLA-A24 Binders
241-250 VYWKLRKLL
145-154 NYKNHFPDI
273-282 EFLWGPRAL
121-13 0 RFLLRKYQI
126-135 KYQIKEPVT
115-124 KVAELVRFL
285-294 SYVKVLEHV
303-312 SYPSLHEEA
201-210 KTGLLIIVL
116-125 VAELVRFLL
299-308 RVRISYPSL
37-46 KAASSSSTL
205-214 LIIVLGMIL
17-26 QAQGEAPGL
64-73 SPEGASSSL
131-140 EPVTKAEML
179-188 YILVTCLGL
-3 8-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
185-194LGLSYDGLL
42-51 SSTLIMGTL
111-120ALDEKVAEL
MAGE A8: HLA-CW6 Binders
S 115-1241 KVAELVRFL
201-210 KTGLLIIVL
177-186 HSYILVTCL
240-249 SVYWKLR,KT_.
111-120 ALDEKVAEL
241-250 VYWKLRKLL
119-128 LVRFLLRKY
205-214 LIIVLGMIL
104-113 LESLFREAL
42-51 SSTLIMGTL
134-143 TKAEMLESV
MAGE A8: HLA-B35 Binders
264-273 APGSDPVRY
199-208 TPKTGLLII
304-313 YPSLHEEAL
131-140 EPVTKAEML
173-182 DPAGHSYIL
-39-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
100-109DPAHLESLF
39-48 ASSSSTLIM
268-277DPVRYEFLW
MAGE A8: HLA-B44 Binders
282-291 AETSYVKVL
20-29 GEAPGLMDV
225-234 WEALSVMGL
33-42 AEEQKAASS
234-243 YDGREHSVY
109-118 REALDEKVA
221-230 EEAIWEALS
170-179 KEVDPAGHS
34-43 EEQKAASSS
296-305 VNARVRISY
226-235 EALSVMGLY
237-246 REHSVYWKL
Compositions based upon these molecules are also a part of the invention, such
as
compositions containing a MAGE protein in accordance with the invention, and a
pharmaceutically acceptable adjuvant such as a cytokine, an interleukin (e.g.,
IL-2,IL-4, IL-
12, etc.), or GM-CSF. Similarly, compositions containing one or more of the
peptides
-40-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
discussed supra and an adjuvant, complexes of HLA or MHC molecules and the
peptides plus
adjuvant are also a part of the invention.
These complexes can be combined per se, or on antigen presenting cells, such
as
dendritic cells, which may be treated to be rendered non-proliferative, etc.
The skilled artisan will also recognize that nucleic acid molecules encoding
the
peptides or proteins may be used in the form of appropriate compositions, such
as in liposome
based compositions. Also a part of the invention are isolated cytolytic T cell
lines which are
specific for complexes of these peptides and their MHC binding partner, i.e.,
an HLA
molecule.
The ability of these peptides to bind to HLA molecules makes them useful as
agents
for determining presence of cells positive forparticular HLA molecules such as
HLA-A*0201
positive cells, by determining whether or not the peptides bind to cells in a
sample. This
"ligand/receptor" type of reaction is well known in the art, and various
methodologies are
available for determining it.
A further aspect of the invention are so-called "mini genes" which carry
information
necessary to direct synthesis of modified decapeptides via cells into which
the mini genes are
transfected. Mini genes can be designed which encode one or more antigenic
peptides, and
are then transferred to host cell genomes via transfection with plasmids, or
via cloning into
vaccinia or adenoviruses. See, e.g., Zajac, et al., Int. J. Cancer 71: 496
(1997), incorporated
by reference These recombinant vectors, such as recombinant vaccinia virus
vectors, can be
constructed so as to produce fusion proteins. For example, fusion proteins can
be constructed
-41-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
where one portion of the fusion protein is the desired tumor rejection antigen
precursor, or
tumor rejection antigen, and additional protein or peptide segments can be
included.
Exemplary, but by no means the only types of additional protein or peptide
segments which
can be added to the fusion proteins, are reporter proteins or peptides, i. e.,
proteins or peptides
which give an observable signal so as to indicate that expression has
occurred, such as green
fluoresence protein. Additional reporter proteins include, but are by no means
limited to,
proteins such as [3galactosidase, luciferase, dhfr, and "eGFP", or enhanced
green fluorescent
protein, as described by Cheng, et al., Nature Biotechnology 14:606 ( 1996),
incorporated by
reference, and so forth. The various reporter proteins available to the
skilled artisan can be,
and are used, in different ways. For example, "GFP" and "eGFP" can be used to
visualize
infected cells, thereby facilitating tracking when flow cytometry is used, and
the isolation of
the cells so infected. Other reporter proteins are useful when methods such as
western
blotting, immunoprecipitation, and so forth are used. These techniques are
standard in the
art and need not be reiterated here. Protein or peptide segments which
facilitate the cleavage
of the tumor rejection antigen precursor or tumor rejection antigen from the
fusion peptide
may also be included. The fusion protein can include more than one tumor rej
ection antigen,
as described, supra , and can also include proteins or peptides which
facilitate the delivery of
the tumor rejection antigen or antigens to a relevant MHC molecule. Such
proteins and
peptides are well known to the art, and need not be elaborated herein.
Also a part of the invention are recombinant cells which have been transfected
with
the recombinant vectors described herein. Such cells may be, e.g., any type of
eukaryotic
-42-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
cell, with human cells being especially preferred. Such cells can then be
used, e.g., to
produce tumor rejection antigen precursors or tumor rejection antigens. They
can also be
used, in an ex vivo context, to generate cytolytic T cells specific for
particular complexes of
MHC molecules and tumor rejection antigens. This can be done simply by
contacting the
transfected cells to a source of T cells, such as a blood sample, so as to
provoke the
proliferation of any cells in the sample specific to the complexes of MHC
molecules and
TRAs (i.e., tumorrejection antigens) produced following expression ofthe
fusion protein, and
processing of the TRA. Such cells, when rendered non-proliferative, can also
be used as
vaccine materials, as they will present the relevant complexes on their
surface, and provoke
the same type of T cell response in vivo, as is shown herein. Similarly, the
vectors can be
used as vaccine materials ep r se, and can be administered to a patient in
need of a T cell
response against complexes of MHC molecules and peptide on cell surfaces. Of
course, T
cells generated ex vivo can also be used to treat patients.
The peptides may be combined with peptides from other tumor rejection antigens
to
form'polytopes'. Exemplary peptides include those listed in U.S. Patent
Application Serial
Numbers 08/672,351, 08/718,964, now U.S. Patent No. , 08/487,135 now U.S.
Patent No. 08/530,569 and 08/880,963 all of which are incorporated by
reference.
Additional peptides which can be used are those described in the following
references,
all of which are incorporated by reference: U.S. Patent Nos. 5,405,940;
5,487,974;
5,519,117; 5,530,096; 5,554,506; 5,554,724; 5,558,995; 5,585,461; 5,589,334;
5,648,226; and
-43-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
5,683,886; PCT International Publication Nos. 92/20356; 94/14459; 96/10577;
96/21673;
97/10837; 97/26535; and 97/31017 as well as pending U.S. Application Serial
No.
08/713,354.
Polytopes are groups of two or more potentially immunogenic or immune
stimulating
peptides, which can be joined together in various ways, to determine if this
type of molecule
will stimulate and/or provoke an immune response.
These peptides can be joined together directly, or via the use of flanking
sequences.
See Thompson et al., Proc. Natl. Acad. Sci. USA 92(13): 5845-5849 (1995),
teaching the
direct linkage of relevant epitopic sequences. The use of polytopes as
vaccines is well
known. See, e.g., Gilbert et al., Nat. Biotechnol. 15(12): 1280-1284 (1997);
Thomson et al.,
supra; Thomson et al., J. Immunol. 157(2): 822-826 (1996); Tam et al., J. E~.
Med. 171(1):
299-306 (1990), all of which are incorporated by reference. The Tam reference
in particular
shows that polytopes, when used in a mouse model, are useful in generating
both antibody
and protective immunity. Further, the reference shows that the polytopes, when
digested,
yield peptides which can be and are presented by MHCs. Tam shows this by
showing
recognition of individual epitopes processed from polytope 'strings' via CTLs.
This approach
can be used, e.g., in determining how many epitopes can be joined in a
polytope and still
provoke recognition and also to determine the efficacy of different
combinations of epitopes.
Different combinations may be 'tailor-made' for the patients expressing
particular subsets of
tumor rej ection antigens. These polytopes can be introduced as polypeptide
structures, or via
the use of nucleic acid delivery systems. To elaborate, the art has many
different ways
-44-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
available to introduce DNA encoding an individual epitope, or a polytope such
as is discussed
supra. See, e.g., Allsopp et al., Eur. J. Immunol. 26(8); 1951-1959 (1996),
incorporated by
reference. Adenovirus, pox-virus, Ty-virus like particles, plasmids, bacteria,
etc., can be
used. One can test these systems in mouse models to determine which system
seems most
appropriate for a given, parallel situation in humans. They can also be tested
in human
clinical trials.
Also, a feature of the invention is the use of these peptides to determine the
presence
of cytolytic T cells in a sample. It was shown, supra, that CTLs in a sample
will react with
peptide/MHC complexes. Hence, if one knows that CTLs are in a sample, cells
positive for
particular HLA molecules can be "lysed" by adding the peptides of the
invention to positive
cells, such as HLA-A2 positive cells, and then determining, e.g., radioactive
chromium
release, TNF production, etc. or any other of the methods by which T cell
activity is
determined. Similarly, one can determine whether or not specific tumor
infiltrating
lymphocytes ("TILs") are present in a sample, by adding one of the claimed
peptides with
HLA positive cells to a sample, and determining lysis of the HLA positive
cells via, e.g., 5' Cr
release, TNF presence and so forth. In addition, CTL may be detected by
ELISPOT analysis.
See for example Schmittel et al., (1997). J. Immunol. Methods 210: 167-174 and
Lalvani et
al., (1997). J. Exp. Med. 126: $59 or by FACS analysis of fluorogenic tetramer
complexes
of MHC Class I/peptide (Dunbar et al., (1998), Current Biolo~y 8: 413-416,
Romero, et al.,
J. Exp. Med. 188: 1641-1650 (1998). All are incorporated by reference.
-45-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
Of course, the peptides may also be used to provoke production of CTLs. As was
shown, supra, CTL precursors develop into CTLs when confronted with
appropriate
complexes. By causing such a "confrontation" as it were, one may generate
CTLs. This is
useful in an in vivo context, as well as ex vivo, for generating such CTLs.
Other aspects of the inventions will be clear to the skilled artisan and will
not be
restricted herein.
The terms and expressions which have been employed are used as terms of
description
and not of limitation, and there is no intention in the use of such terms and
expressions of
excluding any equivalents of the features shown and described or portions
thereof, it being
recognized that various modifications are possible within the scope of the
invention.
-46-
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
<110> Serrano, Alfonso
Lethe, Bernard
Lurquin, Christophe
DePlaen, Etienne
Rimoldi, Donati
Boon-Falleur, Thierry
<120> Isolated Nucleic Acid Molecules Encoding MACE Genes,
Proteins
Encoded, Peptides Derived Therefrom, And Uses The
reof
<130> LUD 5566.1
<140>
<141>
<150> US 09/260,975
<151> 1999-03-02
<160> 24
<210> 1
<211> 21
<212> DNA
<213> Homo sapiens
<220>
<400> 1
cacagagcag cactgaagga g 21
<210> 2
<211> 23
<212> DNA
<213> Homo Sapiens
<220>
<400> 2
ctgggtaaag actcactgtc tgg 23
<210> 3
<211> 22
<212> DNA
<213> Homo Sapiens
<220>
<400> 3
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
agcagccaaa aggaggagag tc 22
<210> 4
<211> 22
<212> DNA
<213> Homo Sapiens
<220>
<400> 4
tgacctcctc aggggtgcag to 22
<210> 5
<211> 22
<212> DNA
<213> Homo Sapiens
<220>
<400> 5
cggccgaagg aacctgaccc ag 22
<210> 6
<211> 23
<212> DNA
<213> Homo Sapiens
<220>
<400> 6
gctggaaccc tcactgggtt gcc 23
<210> 7
<211> 19
<212> DNA
<213> Homo Sapiens
<220>
<400> 7
tcaggggaca ggccaaccc 19
<210> 8
<211> 23
<212> DNA
<213> Homo Sapiens
<220>
<400> 8
cttgcactga ccttgatcac ata 23
2
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
<210> 9
<211> 20
<212> DNA
<213> Homo Sapiens
<220>
<400> 9
gaagagagcg gtcagtgttc 20
<210> 10
<211> 24
<212> DNA
<213> Homo Sapiens
<220>
<400> 10
aatccaggta tgcatatatc ttta 24
<210> 11
<211> 21
<212> DNA
<213> Homo Sapiens
<220>
<400> 11
gcctctttga agagagcagt c 21
<210> 12
<211> 24
<212> DNA
<213> Homo sapiens
<220>
<400> 12
caaagaagca aaaacataca cata 24
<210> 13
<211> 23
<212> DNA
<213> Homo sapiens
<220>
<400> 13
cactctgttt gaagaaaata gtc 23
<210> 14
<211> 25
3
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
<212> DNA
<213> Homo Sapiens
<220>
<400> 14
agtatctttt aatttatctc accta 25
<210> 15
<211> 20
<212> DNA
<213> Homo Sapiens
<220>
<400> 15
agcatgttgg gtgtgaggga 20
<210> 16
<211> 21
<212> DNA
<213> Homo Sapiens
<220>
<400> 16
agggtacact aagaggtaca g 21
<210> 17
<211> 1916
<212> DNA
<213> Homo Sapiens
<220>
<400> 17
gcttgagatc ggctgaagag agcgggccca ggctctgtga ggaggcaagg gag
gtgagaa 60
ccttgctctc agagggtgac tcaagtcaac acagggaacc cctcttttct aca
gacacag 120
tgggtcgcag gatctgacaa gagtccagca tcatgcacta tcctgttggg agc
atcctca 180
cctccaagac actgtttggg cctgaggaga aggagtctgc agtgaccctg tcg
tggtatt 240
ttccacaaga attctgaaat gaagcaagca caggttctca ggggacaggc tga
ccaggat 300
caccaggaag ctccagagga tccccaggag gccctagagg agcaccaaag gag
aagatct 360
gccagtgggt ctccattgcc cagctcctgc ccacactcct gcctgttgcg gtg
accagag 420
tcgtcatgtc tcttgagcag aagagtcagc actgcaagcc tgaggaaggc ctt
gacaccc 480
aagaagaggc cctgggcctg gtgggtgtgc aggctgccac tactgaggag cag
4
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
gaggctg 540
tgtcctcctc ctctcctctg gtcccaggca ccctggggga ggtgcctgct acs
gggtcac 600
caggtcctct caagagtcct caaggagcct ccgccatccc cactaccatc aat
tcactc 66r r _
tatggaggca atccattaag ggctccagca accaagaaga ggaggggcca agc
acctccc 720
ctgacccaga gtctgtgttc cgagcagcac tcagtaagaa ggtggctgac ttg
attcatt 780
ttctgctcct caagtattaa gtcaaggagc tggtcacaaa ggcagaaatg ctg
gagagcg 840
tcatcaaaaa ttacaagcgc tgctttcctg agatcttcgg caaagcctcc gag
tccttgc 900
agctggtctt tggcattgac gtgaaggaag cggaccccac cagcaacacc tac
acccttg 960
tcacctgcct gggactccta tgatggcctg ctggttgata ataatcagat cat
gcccaag 1020
acgggcctcc tgataatcgt cttgggcatg attgcaatgg agggcaaatg cgt
ccctgag 1080
gagaaaatct gggaggagct gagtgtgatg aaggtgtatg ttgggaggga gca
cagtgtc 1140
tgtggggagc ccaggaagct gctcacccaa gatttggtgc aggaaaacta cct
ggagtac 1200
cggcaggtgc ccagcagtga tcccatatgc tatgagttac tgtggggtcc aag
ggcactc 1260
gctgcttgaa agtactggag cacgtggtca gggtcaatgc aagagttctc att
tcctacc 1320
catccctgcg tgaagcagct ttgagagagg aggaagaggg agtctgagca tga
gctgcag 1380
ccagggccac tgcgaggggg gctgggccag tgcaccttcc agggctccgt cca
gtagttt 1440
cccctgcctt aatgtgacat gaggcccatt cttctctctt tgaagagagc agt
caacatt 1500
cttagtagtg ggtttctgtt ctattggatg actttgagat ttgtctttgt ttc
cttttgg 1560
aattgttcaa atgtttcttt taatgggtgg ttgaatgaac ttcagcattc aaa
tttatga 1620
atgacagtag tcacacatag tgctgtttat atagtttagg agtaagagtc ttg
tttttta 1680
ttcagattgg gaaatccatt ccattttgtg aattgggaca tagttacagc agt
ggaataa 1740 '
gtattcattt agaaatgtga atgagcagta aaactgatga cataaagaaa tta
aaagata 1800
tttaattctt gcttatactc agtctattcg gtaaaatttt ttttaaaaaa tat
gcatacc 1860
tggatttcct tggcttcttt gagaatgtaa gacaaattaa atctcraataa atc
att 1916
<210> 18
<211> 1765
<212> DNA
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
<213> Homo Sapiens
<220>
<400> 18
c-~ggt~ctga gggg~cgg== -.. gGtcgg~ -~gagggaa4~ gggc~caggg t~.~
gtgagga 60
ggcaaggttc gcagagaaca ggccagccag gaggtcagga ggccccagag aag
cactgaa 120
gaagacctgc ctgtgggtct caattgccca gctccggccc acactctcct get
gccctga 180
cctgagtcat catgcttctt gggcagaaga gtcagcgcta caaggctgag gaa
ggccttc 240
aggcccaagg agaggcacca gggcttatgg atgtgcagat tcccacagct gag
gagcaga 300
aggctgcatc ctcctcctct actctgatca tgggaaccct tgaggaggtg act
gattctg 360
ggtcaccaag tcctccccag agtcctgagg gtgcctcctc ttccctgact gtc
accgaca 420
gcactctgtg gagccaatcc gatgagggtt ccagcagcaa tgaagaggag ggg
ccaagca 480
cctccccgga cccagctcac ctggagtccc tgttccggga agcacttgat gag
aaagtgg 540
ctgagttagt tcgtttcctg ctccgcaaat atcaaattaa ggag.~.cggtc aca
aaggcag 600
aaatgcttga gagtgtcatc aaaaattaca agaaccactt tcctgatatc ttc
agcaaag 660
cctctgagtg catgcaggtg atctttggca ttgatgtgaa ggaagtggac cct
gccggcc 720
actcctacat ccttgtcacc tgcctgggcc tctcctatga tggcctgctg ggt
gatgatc 780
agagtacgcc caagaccggc cLCCtgataa tcgtcctggg catgatctta atg
gagggca 840
gccgcgcccc ggaggaggca atctgggaag cattgagtgt gatggggctg tat
gatggga 900
gggagcacag tgtctattgg aagctcagga agctgctcac ccaagagtgg gtg
caggaga 960
actacctgga gtaccgccag gcgcccggca gtgatcctgt gcgctacgag ttc
ctgtggg 1020
gtccaagggc ccttgctgaa accagctatg tgaaagtcct ggaacatgtg gtc
agggtca 1080
atgcaagagt tcgcatttcc tacccatccc tgcatgaaga ggc-~~tggga gag
gagaaag 1140
gagtttgagc aggagttgca gctagggcca gtggggcagg ttgtgggagg gcc
tgggcca 1200
gtgcacgttc cagggccaca tccaccactt tccctgctct gttacatgag gcc
cattctt 1260
cactctgtgt ttgaagagag cagtcacagt tctcagtagt ggggagcatg ttg
ggtgtga 1320
gggaacacag tgtggacca~ ctctcagttc ctgttctatt gggcgatttg gag
atttatc 1380
tttgtttcct tttggaattg ttccaatgtt ccttctaatg gatggtgtaa tga
acttcaa 1440
6
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
cattcatttt atgtatgaca gtagacagac ttactgcttt ttatatagtt tag
gagtaag 1500
agtcttgctt ttcatttata ctgggaaacc catgttattt cttgaattca gac
actacaa 1560
gagcagagg.~ t taa J g ~ t ' ~ _ ~ 'agaGa ~ J ~~, ~~a'.~. ~.agca.~, ~aau a"
catgaga 1620
taaagacata aagaaattaa acaatagtta attcttgcct tacctgtacc tct
tagtgta 1680
ccctatgtac ctgaatttgc ttggcttctt tgagaatgaa attgaattaa ata
tgaataa 1740
ataagtcaaa aaaaaaaaaa aaaaa
1765
<210> 19
<211> 1831
<212> DNA
<213> Homo Sapiens
<220>
<400> 19
gaggcctcct tctgaggggc ggcttgatac cggtggagga gctccaggaa gca
ggcaggc 60
cttggtctga gacagtgtcc tcaggtcgca gagcagagga gacccaggca gtg
tcagcag 120
tgaaggttct cgggacaggc taaccaggag gacaggagcc ccaagaggcc cca
gagcagc 180
actgacgaag acctgcctgt gggtctccat cgcccagctc ctgcccacgc tcc
tgactgc 240
tgccctgacc agagtcatca tgtctctcga gcagaggagt ccgcactgca agc
ctgatga 300
agaccttgaa gcccaaggag aggacttggg cctgatgggt gcacaggaac cca
caggcga 360
ggaggaggag actacctcct cctctgacag caaggaggag gaggtgtctg ctg
ctgggtc 420
atcaagtcct ccccagagtc ctcagggagg cgcttcctcc tccatttccg tct
actacac 480
tttatggagc caattcgatg agggctccag cagtcaagaa gaggaagagc caa
gctcctc 540
ggtcgaccca gctcagctgg agttcatgtt ccaagaagca ctgaaattga agg
tggctga 600
gttggttcat ttcctgctcc acaaatatcg agtcaaggag ccggtcacaa agg
cagaaat 660
gctggagagc gtcatcaaaa attacaagcg ctactttcct gtgatcttcg gca
aagcctc 720
cgagttcatg caggtgatct ttggcactga tgtgaaggag gtggaccccg ccg
accactc 780
ctacatcctt gtcactgctc ttggcctctcc gtgcgatagc atgctgggtg atg
a~catag 840
catgcccaag gccgccctcc tgatcatta~ ~~tgggtgtg atcctaacca aag
acaacta 900
cgcccctgaa gaggttatct gggaagcgtt gagtgtgatg ggggtgtatg ttg
7
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
ggaagga 960
gcacatgttc tacggggagc ccaggaagct gctcacccaa gattgggtgc agg
aaaacta 1020
cctggagtac cggcaggtgc ccgq_caata_a tcctgcgcac tacgagttcc ta_t
ggggtt~ 1080
caaggcccac gctgaaacca gctatgagaa ggtcataaat tatttggtca tgc
tcaatgc 1140
aagagagccc atctgctacc catcccttta tgaagaggtt ttgggagagg agc
aagaggg 1200
agtctgagca ccagccgcag ccggggccaa agtttgtggg gtcagggccc cat
ccagcag 1260
ctgccctgcc ccatgtgaca tgaggcccat tcttcgctct gtgtttgaag aga
gcaatca 1320
gtgttctcag tggcagtggg tggaagtgag cacactgtat gtcatctctg ggt
tccttgt 1380
ctattgggtg atttggagat ttatccttgc tcccttttgg aattgttcaa atg
ttctttt 1440
aatggtcagt ttaatgaact tcaccatcga agttaatgaa tgacagtagt cac
acatatt 1500
gctgtttatg ttatttagga gtaagattct tgcttttgag tcacatgggg aaa
tccctgt 1560
tattttgtga attgggacaa gataacatag cagaggaatt aataattttt ttg
aaacttg 1620
aacttagcag caaaatagag ctcataaaga aatagtgaaa tgaaaatgta gtt
aattctt 1680
gccttatacc tctttctctc tcctgtaaaa ttaaaacata tacatgtata cct
ggatttg 1740
cttggcttct ttgagcatgt aagagaaata aaaattgaaa gaataaaaaa aaa
aaaaaaa 1800
aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa a
1831
<210>20
<211>1886
<212>DNA
<213>Homo Sapiens
<220>
<400>20
ctctgggatc tgagagaagc gaaagcgtct ttctgagggg tgtcttgaga gtg
gcagagg 60
gcagcgggtc caggctccat gaggaggcaa gccttgggaa tctgagggat gga
gactcag 120
ttccgcagag ggggtctggg gtgcagccct gccagcatca agaggaagaa as
gagggag 180
gactcaggag actttggact ccaggtgagc actatgttct cagaggacga ct~
ccagtca 240
acagaaagag ccccatatgg tccacaac:~ cagtggtccc aggatctgcc as
agtccag 300
gtttttagag aacaggccaa c~tggaaga~ aggagtccca ggagaaccca gao
gatcac~ 360
8
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
ggaggagaac aagtgctgtg gggccccatc acccagatat ttcccacagt tcg
gcctgct 420
gacctaacca gagtcatcat gcctcttgag caaagaagtc agcactgcaa gcc
tgaggaa 480
ggccttcagg cccaagaagG ~. ~ctgggc ctggtgggt~ cacagJ~tc~
c~.a
agctgag 540
gagcaggagg ctgccttctt c~cctctact ctgaatgtgg gcac~ctaga gga
gttgcct 600
gctgctgagt caccaagtcc tccccagagt cctcaggaag agtccttctc tcc
cactgcc 660
atggatgcca tctttgggag cctatctgat gagggctctg gcagccaaga aaa
ggagggg 720
ccaagtacct cgcctgacct gatagaccct gagtcctttt cccaagatat act
acatgac 780
aagataattg atttggttca tttattgctc cgcaagtatc gagtcaaggg get
gatcaca 840
aaggcagaaa tgctggggag tgtcatcaaa aattatgagg actactttcc tga
gatattt 900
agggaagcct ctgtatgcat gcaactgctc tttggcattg atgtgaagga agt
ggacccc 960
actagccact cctatgtcct tgtcacctcc ctcaacctct cttatgatgg cat
acagtgt 1020
aatgagcaga gcatgcccaa gtctggcctc ctgataatag tcctgggtgt aat
cttcatg 1080
gaggggaact gcatccctga agaggttatg tgggaagtcc tgagcattat ggg
ggtgtat 1140
gctggaaggg agcacttcct ctttggggag cccaagaggc tccttaccca aaa
ttgggtg 1200
caggaaaagt acctggtgta ccggcaggtg cccggcactg atcctgcatg cta
tgagttc 1260
ctgtggggtc caagggccca cgctgagacc agcaagatga aagt~cttga gta
catagcc 1320
aatgccaatg ggagggatcc cacttcttac ccatccctgt atgaagatgc ttt
gagagag 1380
gagggagagg gagtctgagc a'gagatgca accagggcca gcgggcaggg aaa
tgggcca 1440
atgcatgctt cagggccaca cccagcagtt tccctgtcct gtgtgaaatc agg
cccattc 1500
ttccctctgt gtttgatgag agaagtcagt gttctcagta gtagaaggca cag
tgaatgg 1560
aagggaacac attgtatact gcctttaggt ttctcttcca tcgga~gact tgg
agatttg 1620
tttttgtttc cctttggtaa ttt~caaata ttgttcctgt aataaaagtt tta
gttagct 1680
tcaacatcta agtgtatgga tgatactgac cacacatgtt gttt~gctta tcc
atttcaa 1740
gtgcaagtgt ttgccatttt gtaaaacatt ttgggaaatc ttccatcttg ctg
tgatttg 1800
caataggtat tttcttggag aa~gtaagaa cttaacaata aagc~gaact ggt
gttgtga 1860
aacagagaaa aaaaaaaaaa aaaaaa
1886
9
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
<210> 21
<211> 124
<212> PRT
<2i3> Ho~r,oSapiens
<220>
<400> 21
Met Ser Glu Gln Lys Ser GlnHis Cys Lys Pro Glu Glu G1
Leu
y Leu
1 5 10 15
Asp Thr Glu Glu Ala Leu GlyLeu Val Gly Val Gln Ala A1
Gln
a Thr
20 25 30
Thr Glu Gln Glu Ala Val SerSer Ser Ser Pro Leu Val Pr
Glu
o Gly
35 40 45
Thr Leu Glu Val Pro Ala AlaGly Ser Pro Gly Pro Leu Ly
Gly
s Ser
50 55 60
Pro Gln Ala Ser Ala Ile ProThr Ala Ile Asp_Phe Thr Le
Gly
a Trp
65 70 75
80
Arg Gln Ser Ile Lys Gly Ser Ser Asn Gln Glu Glu Glu Gly Pr
o Ser
85 90 95
Thr Ser Pro Asp Pro Glu Ser Val Phe Arg Ala Ala Leu Ser Ly
s Lys
100 105 110
Val Ala Asp Leu Ile His Phe Leu Leu Leu Lys Tyr
115 120
<210> 22
<211> 318
<212> PRT
<213> Homo Sapiens
<220>
<400> 22
Met Leu Gly Gln Lys Ser Gln Arg Tyr Lys Ala Glu Glu
Leu G1
y Leu
1 5 10 15
G=in Ala Gly Glu Ala Pro Gly Leu Met Asp Va- Gln Ile
Gln Pr
o Thr
20 25 30
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346 -
Ala Glu Gln Lys Ala Ala SerSer Ser Ser Thr Leu Ile
Glu Me
t Gly
35 40 45
Thr Glu Glu Val Thr Asp SerGiy Ser Pro Ser Pro Pro
Leu G1
n Ser
50 55 60
Pro Gly Ala Ser Ser Ser LeuThr Val Thr Asp Ser Thr
Glu Le
a Trp
65 70 75
80
Ser Gln Ser Asp Glu Gly Ser Ser Ser Asn Glu Glu Glu Gly Pr
o Ser
85 90 95
Thr Ser Pro Asp Pro Ala His Leu Glu Ser Leu Phe Arg Glu Al
a Leu
100 105 110
Asp Glu Lys Val Ala Glu Leu Val Arg Phe Leu Leu Arg Lys Ty
r Gln
115 120 125
Ile Lys Glu Pro Val Thr Lys Ala Glu Met Leu Glu Ser Val I1
a Lys
130 135 140
Asn Tyr Lys Asn His Phe Pro Asp Ile Phe Ser Lys Ala Ser G1
a Cys
145 150 155
160
Met Gln Val Iie Phe Gly Ile Asp Val Lys Glu Val Asp Pro Al
a Gly
165 170 17
His Ser Tyr Ile Leu Val Thr Cys Leu Gly Leu Ser Tyr Asp G1
y Leu
180 185 190
Leu Gly Asp Asp Gln Ser Thr Pro Lys Thr Gly Leu Leu Ile I1
a Val
195 200 205
Leu Gly Met Ile Leu Met Glu Gly Ser Arg Ala Pro Glu Glu Al
a Ile
210 215 220
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
Trp Glu Ala Leu Met Gly
Ser Leu
Val Tyr
Asp
Gly
Arg
Glu
Hi
s Ser
225 230 235
240
Val Tyr Trp Lys LeuArg Lys Leu Leu Thr Gln GluTrp Val G1
n Glu
245 250 25
Asn Tyr Leu Glu TyrArg Gln Ala Pro Gly Ser AspPro Val Ar
g Tyr
260 265 270
Glu Phe Leu Trp GlyPro Arg Ala Leu Ala Glu ThrSer Tyr Va
1 Lys
275 280 285
Val Leu Glu His ValVal Arg Val Asn Ala Arg ValArg Ile Se
r Tyr
290 295 300
Pro Ser Leu His GluGlu Ala Leu Gly Glu Glu LysGly Val
305 310 315
<210> 23
<211> 315
<212> PRT
<213> Homo Sapiens
<220>
<400> 23
Met Ser Leu Glu GlnArg Ser Pro His Cys Lys ProAsp Glu As
p Leu
1 5 10 15
Glu Ala Gln Gly GluAsp Leu Gly Leu Met Gly AlaGln Glu P-r
o Thr
20 25 30
Gly Glu Glu Glu GluThr Thr Ser Ser Ser Asp SerLys Glu G-
a Glu
35 40 45
Val Ser Ala Ala GlySer Ser Ser Pro Pro Gln SerPro Gln G1
y Gly
50 55 60
Ala Ser Ser Ser IleSer Val Tyr Tyr Thr Leu TrpSer Gln P_~
a Asp
65 70 75
80
12
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
Glu Gly Ser SerSer Gln Glu Glu Glu GluPro Ser Ser Ser Va
1 Asp
85 90 95
t'~~ Aia G1?~.i~Cl.._.., ilc~~ v._?'.Gnu' - _
_ _ . P ~ .L ~: L 1
._ _ 1 a y E i
1 a ~, s ~
; .
s Val . .. , ~
100 105 110
Ala Glu Leu ValHis Phe Leu Leu His LysTyr Arg Val Lys G1
a Pro
115 120 125
Val Thr Lys AlaGlu Met Leu Glu Ser ValIle Lys Asn Tyr Ly
s Arg
130 135 140
Tyr Pro Val Ile Phe Gly LysAla Ser Glu Phe Met Gln Va
Phe
1 Ile
145 150 155
160
Phe Thr Asp Val Lys Glu ValAsp Pro Ala Gly His Ser Ty
Gly
r Ile
165 1~0 1~
Leu Thr Ala Leu Gly Leu SerCys Asp Ser Met Leu Gly As
Val
p Gly
180 185 190
His Met Pro Lys A1a Ala LeuLeu Ile Ile Val Leu Gly Va
Ser
1 Ile
195 200 205
Leu Lys Asp Asn Cys Ala ProGlu Glu Val Ile Trp Glu A1
Thr
a Leu
210 215 220
Ser Val Met Gly Val Tyr Val Gly Lys Glu His Met Phe Tyr Gl
y Glu
225 230 235
240
Pro Arg Lys Leu Leu Thr Gln Asp Trp Val Gln Glu Asn Tyr Le
a Glu
245 250 25
5
Tyr Arg Gln Val Pro G-y Ser Asp Pro Ala His Tyr Glu Phe Le
a Trn
13
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
-
260 265 270
Gly Ser Lys AlaHis Ala Glu Thr Ser TyrGlu LysVal Ile As
n Tyr
275 280 285
Leu Val Met LeuAsn Ala Arg Glu Pro IleCys TyrPro Ser Le
a Tyr
290 295 300
Glu Glu Val LeuGly Glu Glu Gln Glu GlyVal
305 310 315
<210> 24
<211> 429
<212> PRT
<213> Homo Sapiens
<220>
<400> 24
Met Glu Thr GlnPhe Arg Arg Gly Gly LeuGly CysSer Pro A1
a Ser
1 5 10 15
Ile Lys Arg LysLys Lys Arg Glu Asp SerGly AspPhe Gly Le
a Gln
20 25 30
Val Ser Thr MetPhe Ser Glu Asp Asp PheGln SerThr Glu Ar
g Ala
35 40 45
Pro Tyr Gly ProGln Leu Gln Trp Ser GlnAsp LeuPro Arg Va
1 Gln
50 55 60
Val Phe Arg GluGln Ala Asn Leu Glu AspArg SerPro Arg Ar
g Thr
65 70 75
80
Gln Arg Ile Thr Gly Gly Glu Gln Val Leu Trp Gly Pro Ile Th
r Gln
85 90 95
Ile Phe Pro Thr Val Arg Pro Ala Asp Leu Thr Arg Val Ile Me
t Pro
100 105 110
Leu Glu Gln Arg Ser Gln His Cys Lys Pro Giu Glu Gly Leu G1
n Ala
115 120 125
14
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
-
Gln Glu Glu Asp Leu GlyLeu Val Gly Ala Gln Ala LeuGln A1
a Glu
130 135 140
Glu G~~:~ Glu Ala Aia Ph Pr:eSer So_ Tn:_Leu Asn
VaiG1~ 1::
r Leu
145 150 155
160
Glu Glu Leu Pro Ala AlaGlu Ser Pro Ser Pro Pro GlnSer Pr
o Gln
165 170 17
Glu Glu Ser Phe Ser ProThr Ala Met Asp Ala Ile PheGly Se
r Leu
180 185 190
Ser Asp Glu Gly Ser GlySer Gln Glu Lys Glu Gly ProSer Th
r Ser
195 200 205
Pro Asp Leu Ile Asp ProGlu Ser Phe Ser Gln Asp IleLeu Hi
s Asp
210 215 220
Lys Ile Ile Asp Leu ValHis Leu Leu Leu Arg Lys TyrArg Va
1 Lys
225 230 235
240
Gly Leu Ile Thr Lys AlaGlu Met Leu Gly Ser Val IleLys As
n Tyr
245 250 25
5
Glu Asp Tyr Phe Pro GluIle Phe Arg Giu Ala Ser ValCys Me
t Gln
260 265 270
Leu Leu Phe Gly Ile AspVal Lys Glu Val Asp Pro ThrSer Hi
s Ser
275 280 285
Tyr Val Leu Val Thr SerLeu Asn Leu Ser Tyr Asp GlyIle Gl
n Cys
290 295 300
Asn Glu Gln Ser Met ProLys Ser Gly Leu Leu Ile IleVal Le
a Gly
305 310 315
320
CA 02366059 2001-08-23
WO 00/52163 PCT/US00/05346
-
Val Ile Phe Met Glu GlyAsn Cys Ile Pro Glu GluVal MetTr
p Glu
325 330 33
J
Val Leu Ser Ile Met GlyVal Tyr Ala Gly Arg GluHis PheLe
a Phe
340 345 350
Gly Glu Pro Lys Arg LeuLeu Thr Gln Asn Trp ValGln GluLy
s Tyr
355 360 365
Leu Val Tyr Arg Gln ValPro Gly Thr Asp Pro AlaCys TyrG1
a Phe
370 375 380
Leu Trp Gly Pro Arg AlaHis Ala Glu Thr Ser LysMet LysVa
1 Leu
385 390 395
400
Glu Tyr Ile Ala Asn AlaAsn Gly Arg Asp Pro ThrSer TyrPr
o Ser
405 410 41
Leu Tyr Glu Asp Ala LeuArg Glu Glu Gly Glu GlyVal
420 425
16