Note: Descriptions are shown in the official language in which they were submitted.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
Description
NOVEL CYTOKINE ZALPHA 1 1 LIGAND
BACKGROUND OF THE INVENTION
Proliferation and differentiation of cells of multicellular organisms are
controlled by hormones and polypeptide growth factors. These diffusable
molecules
io allow
cells to communicate with each other and act in concert to form cells, tissues
and
organs, and to repair damaged tissue. Examples of hormones and growth factors
include the steroid hormones (e.g. estrogen, testosterone), parathyroid
hormone, follicle
stimulating hormone, the interleukins, platelet derived growth factor (PDGF),
epidermal growth factor (EGF), granulocyte-macrophage colony stimulating
factor
(GM-CSF), erythropoietin (EPO) and calcitonin.
Hormones and growth factors influence cellular metabolism by binding
to receptors. Receptors may be integral membrane proteins that are linked to
signaling
pathways within the cell, such as second messenger systems. Other classes of
receptors
are soluble molecules, such as the transcription factors.
Cytokines generally stimulate proliferation or differentiation of cells of
the hematopoietic lineage or participate in the immune and inflammatory
response
mechanisms of the body. Examples of cytokines which affect hematopoiesis are
erythropoietin (EPO), which stimulates the development of red blood cells;
thrombopoietin (TPO), which stimulates development of cells of the
megakaryocyte
lineage; and granulocyte-colony stimulating factor (G-CSF), which stimulates
development of neutrophils. These cytokines are useful in restoring normal
blood cell
levels in patients suffering from anemia, thrombocytopenia, and neutropenia or
receiving chemotherapy for cancer.
The interleukins are a family of cytokines that mediate immunological
responses, including inflammation. The interleukins mediate a variety of
inflammatory
pathologies. Central to an immune response is the T cell. which produce many
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
2
cytokines and adaptive immunity to antigens. Cytokines produced by the T cell
have
been classified as type 1 and type 2 (Kelso, A. Immun. Cell Biol. 76:300-317,
1998).
Type 1 cytokines include IL-2, IFN-y, LT-a, and are involved in inflammatory
responses, viral immunity, intracellular parasite immunity and allograft
rejection. Type
s 2 cytokines include IL-4, IL-5, IL-6, IL-10 and IL-13, and are involved
in humoral
responses, helminth immunity and allergic response. Shared cytokines between
Type 1
and 2 include IL-3, GM-CSF and TNF-a. There is some evidence to suggest that
Type
1 and Type 2 producing T cell populations preferentially migrate into
different types of
inflamed tissue.
Mature T cells may be activated, i.e., by an antigen or other stimulus, to
produce, for example, cytokines, biochemical signaling molecules, or receptors
that
further influence the fate of the T cell population.
B cells can be activated via receptors on their cell surface including B
cell receptor and other accessory molecules to perform accessory cell
functions, such as
production of cytokines.
Natural killer (NK) cells have a common progenitor cell with T cells and
B cells, and play a role in immune surveillance. NK cells, which comprise up
to 15%
of blood lymphocytes, do not express antigen receptors, and therefore do not
use MHC
recognition as requirement for binding to a target cell. NK cells are involved
in the
recognition and killing of certain tumor cells and virally infected cells. In
vivo, NK
cells are believed to require activation, however, in vitro, NK cells have
been shown to
kill some types of tumor cells without activation.
The demonstrated in vivo activities of the cytokine family illustrate the
enormous clinical potential of, and need for, other cytokines, cytokine
agonists, and
cytokine antagonists. The present invention addresses these needs by providing
a new
cytokine that stimulates cells of the hematopoietic cell lineage, as well as
related
compositions and methods.
The present invention provides such polypeptides for these and other
uses that should be apparent to those skilled in the art from the teachings
herein.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
3
BRIEF DESCRIPTION OF THE DRAWINGS
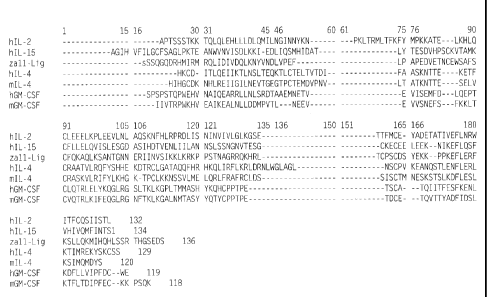
Figure 1 is an illustration of a multiple alignment of human IL-2, human
IL-15, zalphal 1 Ligand (SEQ ID NO: 2), human IL-4, mouse IL-4, human GM-CSF
and mouse GM-CSF.
DETAILED DESCRIPTION OF THE INVENTION
Prior to setting forth the invention in detail, it may be helpful to the
understanding thereof to define the following terms:
o The
term "affinity tag" is used herein to denote a polypeptide segment
that can be attached to a second polypeptide to provide for purification or
detection of
the second polypeptide or provide sites for attachment of the second
polypeptide to a
substrate. In principal, any peptide or protein for which an antibody or other
specific
binding agent is available can be used as an affinity tag. Affinity tags
include a poly-
histidine tract, protein A (Nilsson et al., EMBO J. 4:1075, 1985: Nilsson et
al., Methods
Enzymol. 198:3, 1991), glutathione S transferase (Smith and Johnson, Gene
67:31,
1988), Glu-Glu affinity tag (Grussenmeyer et al., Proc. Natl. Acad. Sci. USA
82:7952-
4, 1985), substance P, FlagTM peptide (Hopp et al.. Biotechnology 6:1204-10,
1988),
streptavidin binding peptide, or other antigenic epitope or binding domain.
See, in
general, Ford et al., Protein Expression and Purification 2: 95-107, 1991.
DNAs
encoding affinity tags are available from commercial suppliers (e.g.,
Pharmacia
Biotech, Piscataway, NJ).
The term "allelic variant" is used herein to denote any of two or more
alternative forms of a gene occupying the same chromosomal locus. Allelic
variation
arises naturally through mutation, and may result in phenotypic polymorphism
within
populations. Gene mutations can be silent (no change in the encoded
polypeptide) or
may encode polypeptides having altered amino acid sequence. The term allelic
variant
is also used herein to denote a protein encoded by an allelic variant of a
gene.
The terms -amino-terminal" and "carboxyl-terminal" are used herein to
denote positions within polypeptides. Where the context allows, these terms
are used
with reference to a particular sequence or portion of a polypeptide to denote
proximity
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
4
or relative position. For example, a certain sequence positioned carboxyl-
terminal to a
reference sequence within a polypeptide is located proximal to the carboxyl
terminus of
the reference sequence, but is not necessarily at the carboxyl terminus of the
complete
polypeptide.
The term "complement/anti-complement pair" denotes non-identical
moieties that form a non-covalently associated, stable pair under appropriate
conditions.
For instance, biotin and avidin (or streptavidin) are prototypical members of
a
complement/anti-complement pair. Other exemplary complement/anti-complement
pairs include receptor/ligand pairs, antibody/antigen (or hapten or epitope)
pairs,
sense/antisense polynucleotide pairs, and the like. Where subsequent
dissociation of
the complement/anti-complement pair is desirable, the complement/anti-
complement
pair preferably has a binding affinity of <109 M-1.
The term "complements of a polynucleotide molecule" denotes a
polynucleotide molecule having a complementary base sequence and reverse
orientation as compared to a reference sequence. For example, the sequence 5'
ATGCACGGG 3' is complementary to 5' CCCGTGCAT 3'.
The term "degenerate nucleotide sequence" denotes a sequence of
nucleotides that includes one or more degenerate codons (as compared to a
reference
polynucleotide molecule that encodes a polypeptide). Degenerate codons contain
different triplets of nucleotides, but encode the same amino acid residue
(i.e., GAU and
GAC triplets each encode Asp).
The term "expression vector" is used to denote a DNA molecule, linear
or circular, that comprises a segment encoding a polypeptide of interest
operably linked
to additional segments that provide for its transcription. Such additional
segments
include promoter and terminator sequences, and may also include one or more
origins
of replication, one or more selectable markers, an enhancer, a polyadenylation
signal.
etc. Expression vectors are generally derived from plasmid or viral DNA, or
may
contain elements of both.
The term "isolated", when applied to a polynucleotide, denotes that the
polynucleotide has been removed from its natural genetic milieu and is thus
free of
other extraneous or unwanted coding sequences. and is in a form suitable for
use within
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
genetically engineered protein production systems. Such isolated molecules are
those
that are separated from their natural environment and include cDNA and genomic
clones. Isolated DNA molecules of the present invention are free of other
genes with
which they are ordinarily associated, but may include naturally occurring 5'
and 3'
5 untranslated regions such as promoters and terminators. The identification
of
associated regions will be evident to one of ordinary skill in the art (see
for example,
Dynan and Tij an, Nature 316:774-78, 1985).
An "isolated" polypeptide or protein is a polypeptide or protein that is
found in a condition other than its native environment, such as apart from
blood and
animal tissue. In a preferred form, the isolated polypeptide is substantially
free of other
polypeptides, particularly other polypeptides of animal origin. It is
preferred to provide
the polypeptides in a highly purified form. i.e. greater than 95% pure, more
preferably
greater than 99% pure. When used in this context, the term "isolated" does not
exclude
the presence of the same polypeptide in alternative physical forms, such as
dimers or
alternatively glycosylated or derivatized forms.
The term "neoplastic", when referring to cells, indicates cells undergoing
new and abnormal proliferation, particularly in a tissue where in the
proliferation is
uncontrolled and progressive, resulting in a neoplasm. The neoplastic cells
can be
either malignant, i.e. invasive and metastatic, or benign.
The term "operably linked", when referring to DNA segments, indicates
that the segments are arranged so that they function in concert for their
intended
purposes, e.g., transcription initiates in the promoter and proceeds through
the coding
segment to the terminator.
The term "ortholog" denotes a polypeptide or protein obtained from one
species that is the functional counterpart of a polypeptide or protein from a
different
species. Sequence differences among orthologs are the result of speciation.
"Paralogs" are distinct but structurally related proteins made by an
organism. Paralogs are believed to arise through gene duplication. For
example, cc-
globin, p-globin, and myoglobin are paralog,s of each other.
A "polynucleotide" is a single- or double-stranded polymer of
deoxyribonucleotide or ribonucleotide bases read from the 5' to the 3' end.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
6
Polynucleotides include RNA and DNA, and may be isolated from natural sources,
synthesized in vitro, or prepared from a combination of natural and synthetic
molecules.
Sizes of polynucleotides are expressed as base pairs (abbreviated "bp"),
nucleotides
("nt"), or kilobases ("kb"). Where the context allows, the latter two terms
may describe
polynucleotides that are single-stranded or double-stranded. When the term is
applied
to double-stranded molecules it is used to denote overall length and will be
understood
to be equivalent to the term "base pairs". It will be recognized by those
skilled in the
art that the two strands of a double-stranded polynucleotide may differ
slightly in length
and that the ends thereof may be staggered as a result of enzymatic cleavage;
thus all
io nucleotides within a double-stranded polynucleotide molecule may not be
paired.
A "polypeptide" is a polymer of amino acid residues joined by peptide
bonds, whether produced naturally or synthetically. Polypeptides of less than
about 10
amino acid residues are commonly referred to as "peptides".
The term "promoter" is used herein for its art-recognized meaning to
is denote a portion of a gene containing DNA sequences that provide for the
binding of
RNA polymerase and initiation of transcription. Promoter sequences are
commonly,
but not always, found in the 5' non-coding regions of genes.
A "protein" is a macromolecule comprising one or more polypeptide
chains. A protein may also comprise non-peptidic components, such as
carbohydrate
20 groups. Carbohydrates and other non-peptidic substituents may be added
to a protein
by the cell in which the protein is produced, and will vary with the type of
cell.
Proteins are defined herein in terms of their amino acid backbone structures;
substituents such as carbohydrate groups are generally not specified, but may
be present
nonetheless.
25 The term "receptor" denotes a cell-associated protein that binds
to a
bioactive molecule (i.e., a ligand) and mediates the effect of the ligand on
the cell.
Membrane-bound receptors are characterized by a multi-peptide structure
comprising
an extracellular ligand-binding domain and an intracellular effector domain
that is
typically involved in signal transduction. Binding of ligand to receptor
results in a
30 conformational change in the receptor that causes an interaction between
the effector
domain and other molecule(s) in the cell. This interaction in turn leads to an
alteration
CA 02366921 2007-12-17
7
in the metabolism of the cell. Metabolic - events -that are linked to receptor-
ligand
interactions include gene transcription, phosphorylation, dephosphorylation,
increases
in cyclic AMP production, mobilization of cellular calcium, mobilization of
membrane
lipids, cell adhesion, hydrolysis of inositol lipidsi and hydrolysis of
phospholipids. In
general, receptors can be membrane bound, cytosolic or nuclear; monomeric
(e.g.,
thyroid stimulating hormone receptor, beta-adrenergic receptor) or multimeric
(e.g.,
PDGF receptor, growth hormone receptor, IL-3 receptor, GM-CSF receptor, G-CSF
receptor, erythropoietin receptor and IL-6 receptor).
The term "secretory signal sequence" denotes a DNA sequence that
o encodes. a polypeptide (a "secretory peptide") that, as a component of a
larger
polypeptide, directs the larger polypeptide thrdugh a secretory pathway of a
cell in
which it is synthesized. The larger polypeptide is commonly cleaved to remove
the
= secretory peptide during transit through the secretory pathway.
The term "splice variant" is used herein to denote alternative forms of
is RNA transcribed from a gene. Splice variation arises naturally through use
of
a_
alternative splicing sites within a transcribed RNA molecule, or less commonly
between separately transcribed RNA molecules, and may result in several mRNAs
transcribed from the same gene. Splice variants may encode polypeptides having
altered amino acid sequence. The term splice variant is also used herein to
denote a
=
20 protein encoded by a splice variant of an mRNA transcribed from a gene.
Moleculpr weights and lengths of 'polymers determined by imprecise
analytical methods (e.g., gel electrophoresis) will be understood to be
approximate
values. When such a value is expressed as "about" X or "approximately" X, the
stated
value of X will be understood to be accurate to 10%.
The present invention is based in part upon the discovery of a novel
DNA sequence that encodes a protein having the structure of a four-helical-
bundle
cytokine. Through processes of cloning, proliferation assays and binding
studies
described in detail herein, a polynucleotide sequence encoding a novel ligand
polypeptide has been identified that is a ligand with high specificity for the
previously
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
8
orphan receptor zalphal 1. This polypeptide ligand, designated zalphal 1
Ligand, was
isolated from a cDNA library generated from activated human peripheral blood
cells
(hPBCs), which were selected for CD3. CD3 is a cell surface marker unique to
cells of
lymphoid origin, particularly T cells.
In the examples which follow, a cell line that is dependent on the
zalphal 1 orphan receptor-linked pathway for survival and growth in the
absence of
other growth factors was used to screen for a source of the cDNA encoding the
zalphal 1 Ligand. The preferred growth factor-dependent cell line that was
used for
transfection and expression of zalphal 1 receptor was BaF3 (Palacios and
Steinmetz,
o Cell
41: 727-734, 1985; Mathey-Prevot et al., Mol. Cell. Biol. 6: 4133-4135, 1986).
However, other growth factor-dependent cell lines, such as FDC-P1 (Hapel et
al., Blood
64: 786-790, 1984), and M07e (Kiss et al., Leukemia 7: 235-240, 1993) are
suitable for
this purpose.
The amino acid sequence for the zalphal 1 receptor indicated that the
encoded receptor belonged to the Class I cytokine receptor subfamily that
includes, but
is not limited to, the receptors for IL-2, IL-4, IL-7, IL-15, EPO, TPO, GM-CSF
and G-
CSF (for a review see, Cosman, "The Hematopoietin Receptor Superfamily" in
Cytokine 5(2): 95-106, 1993). The zalphal 1 receptor is fully described in
commonly-
owned PCT Patent Application No. US99/22149. Analysis of the tissue
distribution of
the mRNA of the zalphal 1 receptor revealed expression in lymph node,
peripheral
blood leukocytes (PBLs), spleen, bone marrow. and thymus. Moreover, the mRNA
was abundant in the Raji cell line (ATCC No. CCL-86) derived from a Burkitt's
lymphoma. The tissue distribution of the receptor suggests that a target for
the
predicted zalphal 1 Ligand is hematopoietic lineage cells, in particular
lymphoid
progenitor cells and lymphoid cells. Other known four-helical-bundle cytokines
that
act on lymphoid cells include IL-2, IL-4, IL-7, and IL-15. For a review of
four-helical-
bundle cytokines, see, Nicola et al., Advances in Protein Chemistry 52:1-65,
1999 and
Kelso, A., Immunol. Cell Biol. 76:300-317, 1998.
Conditioned media (CM) from CD3+ selected, PMA/Ionomycin-
stimulated human peripheral blood cells supported the growth of BaF3 cells
that
expressed the zalphal 1 receptor and were otherwise dependent on IL-3.
Conditioned
CA 02366921 2007-12-17
=
9
medias from cells that were not: 1) PMA/Ionomycin.-stimulated; or were not: 2)
CD3
selected (with or without PMA/Ionomycin stimulation) did not support the
growth of
BaF3/zalphal 1 receptor cells. Control experiments demonstrated that this
proliferative
activity was not attributable to other known growth factors, and that the
ability of such
s conditioned media to stimulate proliferation of zarphal 1 receptor-
expressing cells could
be neutralized by a soluble form of the receptor.
Proliferation of zalphal 1 receptor-expressing BaF3 cells exposed to CM
from CD3+ selected, PMA/Ionomycin-stimulated human peripheral blood cells were
identified by visual inspection of the cultures and/or by proliferation assay.
Many
o suitable proliferation assays are known in the art, and include assays
for reduction of a
dye such as alamarBluend (AccuMed International, Inc. Westlake, Ohio), 3-(4,5-
.
dimethylthiazol-2-y1)-2,5-diphenyl tetrazolium bromide (Mosman, J. Immunol.
Meth.
= 65: 55-63, 1983); 3,(4,5 dimethyl thiazol-2y1)-5-3-carboxymethoxypheny1-
2H-
tetrazolium; 2,3-bis(2-methoxy-4-nitro-5-sulfopheny1)-54(phenylamino)carbonyl]-
2H-
s tetrazolium hydroxide; and cyanoditolyl-tetrazolium _chloride (which are
commercially
available from Polysciences, Inc., Warrington, PA); mitogenesis assays, such
as
measurement of incorporation of 3H-thyznidine; dye exclusion assays using, for
example, naphthalene black or trypan blue; dye uptake using diacetyl
fluorescein; and
chromium release. See, in general, Freshney, Culture of Animal Cells: A Manual
of
20 Basic Technique, 3rd ed., Wiley-Liss, 1994.
A cDNA library was prepared from CD3+ selected, PMA- and
Ionomycin-stimulated primary human peripheral blood cells. The CD3+ selected,
=
PMA- and Ionomycin-stimulated human peripheral blood cells cDNA library was
divided into pools containing multiple cDNA molecules and was transfected into
a host
25 cell line, for example, BHK 570 cells (ATCC accession no. 10314). The
transfected
host cells were cultured in a medium that did not contain exogenous growth
factors and
conditioned medium was collected. The conditioned media were assayed for the
ability
to stimulate proliferation of BaF3 cells transfected with the zalphal 1
receptor. CDNA
pools producing conditioned medium that stimulated BaF3/zalphal 1 receptor
cells were
30 identified. This pooled plasmid cDNA was electroporated into E. colt
CDNA was
isolated from single colonies and transfected individually into BHK 570 cells.
Positive
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
clones were identified by a positive result in the BaF3/zalphal 1 receptor
proliferation
assay, and specificity was tested by neutralization of proliferation using the
soluble
zalphal 1 receptor.
A positive clone was isolated, and sequence analysis revealed that the
5 polynucleotide sequence contained within the plasmid DNA was novel. The
secretory
signal sequence is comprised of amino acid residues 1 (Met) to 31 (Gly), and
the
mature polypeptide is comprised of amino acid residues 32 (Gin) to 162 (Ser)
(as
shown in SEQ ID NO: 2).
In general, cytokines are predicted to have a four-alpha helix structure,
o with helices A, C and D being most important in ligand-receptor
interactions, and are
more highly conserved among members of the family. Referring to the human
zalphal 1 Ligand amino acid sequence shown in SEQ ID NO:2, alignment of human
zalphal 1 Ligand, human IL-15, human IL-4, and human GM-CSF amino acid
sequences it is predicted that zalphall Ligand helix A is defined by amino
acid residues
41-56; helix B by amino acid residues 69-84; helix C by amino acid residues 92-
105;
and helix D by amino acid residues 135-148; as shown in SEQ ID NO: 2.
Structural
analysis suggests that the A/B loop is long, the B/C loop is short and the C/D
loop is
parallel long. This loop structure results in an up-up-down-down helical
organization.
The cysteine residues are absolutely conserved between zalphal 1 Ligand and IL-
15, as
shown in Figure 1. The cysteine residues that are conserved between IL-15 and
zalphal 1 Ligand correspond to amino acid residues 71, 78, 122 and 125 of SEQ
ID
NO: 2. Conservation of some of the cysteine residues is also found in IL-2, IL-
4, GM-
CSF and zalphal 1 Ligand corresponding to amino acid residues 78 and 125 of
SEQ ID
NO: 2, as shown in Figure 1. Consistent cysteine placement is further
confirmation of
the four-helical-bundle structure. Also highly conserved in the family
comprising IL-
15, IL-2, IL-4, GM-CSF and zalphal 1 Ligand is the Glu-Phe-Leu sequence as
shown in
SEQ ID NO: 2 at residues 136-138, as in Figure 1.
Further analysis of zalphal 1 Ligand based on multiple alignments (as
shown in Figure 1) predicts that amino acid residues 44, 47 and 135 (as shown
in SEQ
ID NO: 2) play an important role in zalphal 1 Ligand binding to its cognate
receptor.
Moreover, the predicted amino acid sequence of murine zalphal 1 Ligand shows
57%
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
11
identity to the predicted human protein. Based on comparison between sequences
of
human and murine zalphal 1 Ligand well-conserved residues were found in the
regions
predicted to encode alpha helices A and D. The corresponding polynucleotides
encoding the zalphal 1 Ligand polypeptide regions, domains, motifs, residues
and
sequences described herein are as shown in SEQ ID NO: 1.
Detailed mutational analysis has been performed for IL-4 and IL-2, both
of which are highly related to zalphal 1 ligand. Analysis of murine IL-2
(Zurawski et
al., EMBO J. 12:5113-5119, 1993) shows residues in helices A and C are
important for
binding to IL-2R13; critical residues are Asp34, Asnõ, and Asn103. Multiple
residues
o within murine IL-2 loop A/B and helix B are important for IL-2Ra binding,
while only
a single residue, Gln141 in helix D, is vital for binding with IL-2Ra.
Similarly, helices A
and C are sites of interaction between IL-4 and IL-4Ra (the structurally
similar to IL-
2Ra), and residues within helix D are vital for IL-2Ra interaction (Wang et
al., Proc.
Natl. Acad. Sci. USA 94:1657-1662, 1997; Kruse et al., EMBO J. 11:3237-3244,
1992). In particular, the mutation Tyr124 to Asp in human IL-4 creates an
antagonist,
which binds with IL-4Rcc but not IL-2Ra and therefore cannot signal (Kruse et
al. ibid.
1992).
While helix A is relatively well-conserved between human and murine
zalphal 1 Ligand, helix C is more divergent. While both species have
predominant
acidic amino acids in this region, the differences may account for species
specificity in
interaction between zalphal 1 Ligand and its "beta" type receptor. zalphal 1.
Loop A/B
and helix B of zalphal 1 Ligand are well-conserved between species; although
no
receptor subunit corresponding to IL-2Ra has yet been identified. conservation
through
this region suggests that it is functionally significant. The D helices of
human and
murine zalphal 1 Ligand are also highly conserved. Zalphal 1 receptor
antagonists may
be designed through mutations within zalphal 1 Ligand helix D. These may
include
truncation of the protein from residue Gln145 (SEQ ID NO: 2). or mutations of
Gln145 or
Ile148 (of SEQ ID NO: 2; corresponding to Tyr124 in human IL-4) to residues
such as Ala
or Asp. Any mutation which disrupts the zalphal 1 Ligand helical structure may
abolish
binding with its receptor and thus inhibit signaling.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
12
Four-helical bundle cytokines are also grouped by the length of their
component helices. "Long-helix" form cytokines generally consist of between 24-
30
residue helices, and include IL-6, ciliary neutrotrophic factor (CNTF),
leukemia
inhibitory factor (LIF) and human growth hormone (hGH). "Short-helix" form
cytokines generally consist of between 18-21 residue helices and include IL-2,
IL-4 and
GM-CSF. Zalphal 1 Ligand is believed to be a new member of the short-helix
form
cytokine group. Studies using CNTF and IL-6 demonstrated that a CNTF helix can
be
exchanged for the equivalent helix in IL-6, conferring CTNF-binding properties
to the
chimera. Thus, it appears that functional domains of four-helical cytokines
are
o determined on the basis of structural homology, irrespective of sequence
identity, and
can maintain functional integrity in a chimera (Kallen et al., J. Biol. Chem.
274:11859-
11867, 1999). Therefore, the helical domains of zalphal 1 Ligand will be
useful for
preparing chimeric fusion molecules, particularly with other short-helix form
cytokines
to determine and modulate receptor binding specificity. Of particular interest
are fusion
is proteins engineered with helix A and/or helix D, and fusion proteins
that combine
helical and loop domains from other short-form cytokines such as IL-2, IL-4,
IL-15 and
GM-CSF. The amino acid residues comprising helices A, B, C, and D, and loops
A/B,
B/C and C/D for zalphal 1 Ligand, IL-2, IL-4, IL-15 and GM-CSF are shown in
Table
1.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
13
Table 1
Helix A/B Helix B/C Helix C/D Helix
A Loop B Loop C Loop D
zalphal 1 41-56 57-68 69-84 85-91 92-105 106- 135- SEQ
Ligand 134 148 ID
residues NO:2
IL-2 36-46 47-52 53-75 76-86 87-99 100- 103- SEQ
residues 102 121 ID
NO:
111
IL-4 29-43
44-64 65-83 84-94 95-118 119- 134- SEQ
residues 133 151 ID
NO:
112
IL-15 45-68 69-83 84-101 102- 107- 120- 134- SEQ
residues 106 119 133 160 ID
NO:
113
GM- 30-44
45-71 72-81 82-90 91-102 103- 120- SEQ
CSF 119 131 ID
residues NO:
114
The present invention provides polynucleotide molecules, including
DNA and RNA molecules, that encode the zalphal 1 Ligand polypeptides disclosed
herein. Those skilled in the art will readily recognize that, in view of the
degeneracy of
the genetic code, considerable sequence variation is possible among these
polynucleotide molecules. SEQ ID NO:3 is a degenerate DNA sequence that
o encompasses all DNAs that encode the zalphal 1 Ligand polypeptide of SEQ
ID NO:2.
Those skilled in the art will recognize that the degenerate sequence of SEQ ID
NO:3
also provides all RNA sequences encoding SEQ ID NO:2 by substituting U for T.
Thus, zalphall Ligand polypeptide-encoding polynucleotides comprising
nucleotide 1
or 97 to nucleotide 486 of SEQ ID NO:3 and their RNA equivalents are
contemplated
by the present invention. Table 2 sets forth the one-letter codes used within
SEQ ID
NO:3 to denote degenerate nucleotide positions. "Resolutions" are the
nucleotides
denoted by a code letter. "Complement- indicates the code for the
complementary
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
14
nucleotide(s). For example, the code Y denotes either C or T, and its
complement R
denotes A or G, with A being complementary to T, and G being complementary to
C.
TABLE 2
Nucleotide Resolution Complement Resolution
A A= T T
C C G G
G G C C
T T A A
R AG Y CT
Y CI R AG
M AC K G1T
K GI M AC
S CG S CG
W ALT W AI
H AIM D AIG1T
B CIG1T V WIG
V WIG B CIG1T
D AIG1T H AIM
N AICIG1T N WIG T
The degenerate codons used in SEQ ID NO:3, encompassing all possible
codons for a given amino acid, are set forth in Table 3.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
TABLE 3
One
Amino Letter Codons Degenerate
Acid Code Codon
Cys C TGC TGT TGY
Ser S AGC AGT TCA TCC TCG TCT WSN
Thr T ACA ACC ACG ACT ACN
Pro P CCA CCC CCG.CCT CCN
Ala A GCA GCC GCG GCT GCN
Gly G GGA GGC GGG GGT GGN
Asn N MC MT AAY
Asp D GAC GAT GAY
Glu E GM GAG GAR
Gin Q CAA CAG CAR
His H CAC CAT CAY
Arg R AGA AGG CGA CGC CGG CGT MGN
Lys K AM MG MR
Met M ATG ATG
Ile I ATA ATC ATT ATH
Leu L CIA CTC CTG CTT TTA TTG YTN
Val V GTA GTC GIG GTT GIN
Phe F ITC TIT TTY
Tyr Y TAC TAT TAY
Trp W TGG TGG
Ter . TAA TAG TGA TRR
AsnlAsp B RAY
GlulGln Z SAR
Any X NNN
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
16
One of ordinary skill in the art will appreciate that some ambiguity is
introduced in determining a degenerate codon, representative of all possible
codons
encoding each amino acid. For example, the degenerate codon for serine (WSN)
can, in
some circumstances, encode arginine (AGR), and the degenerate codon for
arginine
s (MGN) can, in some circumstances, encode serine (AGY). A similar
relationship exists
between codons encoding phenylalanine and leucine. Thus, some polynucleotides
encompassed by the degenerate sequence may encode variant amino acid
sequences,
but one of ordinary skill in the art can easily identify such variant
sequences by
reference to the amino acid sequence of SEQ ID NO:2. Variant sequences can be
o readily tested for functionality as described herein.
One of ordinary skill in the art will also appreciate that different species
can exhibit "preferential codon usage." In general, see, Grantham, et al.,
Nuc. Acids
Res. 8:1893-912, 1980; Haas, et al. Curr. Biol. 6:315-24, 1996; Wain-Hobson,
et al.,
Gene 13:355-64, 1981; Grosjean and Fiers, Gene 18:199-209, 1982; Holm, Nuc.
Acids
15 Res. 14:3075-87, 1986; Ikemura, J. Mol. Biol. 158:573-97, 1982. As used
herein, the
term "preferential codon usage" or "preferential codons" is a term of art
referring to
protein translation codons that are most frequently used in cells of a certain
species,
thus favoring one or a few representatives of the possible codons encoding
each amino
acid (See Table 3). For example, the amino acid Threonine (Thr) may be encoded
by
20 ACA, ACC, ACG, or ACT, but in mammalian cells ACC is the most commonly
used
codon; in other species, for example, insect cells, yeast, viruses or
bacteria, different
Thr codons may be preferential. Preferential codons for a particular species
can be
introduced into the polynucleotides of the present invention by a variety of
methods
known in the art. Introduction of preferential codon sequences into
recombinant DNA
25 can, for example, enhance production of the protein by making protein
translation more
efficient within a particular cell type or species. Therefore, the degenerate
codon
sequence disclosed in SEQ ID NO:3 serves as a template for optimizing
expression of
polynucleotides in various cell types and species commonly used in the art and
disclosed herein. Sequences containing preferential codons can be tested and
optimized
30 for expression in various species, and tested for functionality as
disclosed herein.
CA 02366921 2001-09-10
WO 00/53761 PCTTUS00/06067
17
As previously noted, the isolated polynucleotides of the present
invention include DNA and RNA. Methods for preparing DNA and RNA are well
known in the art. In general, RNA is isolated from a tissue or cell that
produces large
amounts of zalphall Ligand RNA. Such tissues and cells are identified by
Northern
s blotting (Thomas, Proc. Natl. Acad. Sci. USA 77:5201, 1980), or by screening
conditioned medium from various cell types for activity on target cells or
tissue. Once
the activity or RNA producing cell or tissue is identified, total RNA can be
prepared
using guanidinium isothiocyanate extraction followed by isolation by
centrifugation in
a CsC1 gradient (Chirgwin et al., Biochemistry 18:52-94, 1979). Poly (A) RNA
is
o
prepared from total RNA using the method of Aviv and Leder (Proc. Natl. Acad.
Sci.
USA 69:1408-12, 1972). Complementary DNA (cDNA) is prepared from poly(A)
RNA using known methods. In the alternative, genomic DNA can be isolated.
Polynucleotides encoding zalphall Ligand polypeptides are then identified and
isolated
by, for example, hybridization or PCR.
15 A full-
length clone encoding zalphal 1 Ligand can be obtained by
conventional cloning procedures. Complementary DNA (cDNA) clones are
preferred,
although for some applications (e.g., expression in transgenic animals) it may
be
preferable to use a genomic clone, or to modify a cDNA clone to include at
least one
genomic intron. Methods for preparing cDNA and genomic clones are well known
and
20 within
the level of ordinary skill in the art, and include the use of the sequence
disclosed herein, or parts thereof, for probing or priming a library.
Expression libraries
can be probed with antibodies to zalphal 1 receptor fragments, or other
specific binding
partners.
Zalphal 1 Ligand polynucleotide sequences disclosed herein can also be
25 used as
probes or primers to clone 5" non-coding regions of a zalphal 1 Ligand gene.
In
view of the tissue-specific expression observed for zalphal 1 Ligand this gene
region is
expected to provide for hematopoietic- and lymphoid-specific expression.
Promoter
elements from a zalphall Ligand gene could thus be used to direct the tissue-
specific
expression of heterologous genes in, for example, transgenic animals or
patients treated
30 with
gene therapy. Cloning of 5' flanking sequences also facilitates production of
zalphal 1 Ligand proteins by "gene activation- as disclosed in U.S. Patent No.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
18
5,641,670. Briefly, expression of an endogenous zalphal 1 Ligand gene in a
cell is
altered by introducing into the zalphal 1 Ligand locus a DNA construct
comprising at
least a targeting sequence, a regulatory sequence, an exon, and an unpaired
splice donor
site. The targeting sequence is a zalphal 1 Ligand 5' non-coding sequence that
permits
homologous recombination of the construct with the endogenous zalphal 1 Ligand
locus, whereby the sequences within the construct become operably linked with
the
endogenous zalphal 1 Ligand coding sequence. In this way, an endogenous
zalphal 1
Ligand promoter can be replaced or supplemented with other regulatory
sequences to
provide enhanced, tissue-specific, or otherwise regulated expression.
The present invention further provides counterpart polypeptides and
polynucleotides from other species (orthologs). These species include, but are
not
limited to mammalian, avian, amphibian, reptile, fish, insect and other
vertebrate and
invertebrate species. Of particular interest are zalphal 1 Ligand polypeptides
from other
mammalian species, including murine, porcine, ovine, bovine, canine, feline,
equine,
and other primate polypeptides. Orthologs of human zalphal 1 Ligand can be
cloned
using information and compositions provided by the present invention in
combination
with conventional cloning techniques. For example, a cDNA can be cloned using
mRNA obtained from a tissue or cell type that expresses zalpha 11 Ligand as
disclosed
herein. Suitable sources of mRNA can be identified by probing Northern blots
with
probes designed from the sequences disclosed herein. A library is then
prepared from
mRNA of a positive tissue or cell line. A zalphal 1 Ligand-encoding cDNA can
then be
isolated by a variety of methods, such as by probing with a complete or
partial human
cDNA or with one or more sets of degenerate probes based on the disclosed
sequences.
A cDNA can also be cloned using the polymerase chain reaction, or PCR (Mullis,
U.S.
Patent No. 4,683,202), using primers designed from the representative human
zalphal 1
Ligand sequence disclosed herein. Within an additional method, the cDNA
library can
be used to transform or transfect host cells, and expression of the cDNA of
interest can
be detected with an antibody to zalphal 1 Ligand polypeptide, binding studies
or
activity assays. Similar techniques can also be applied to the isolation of
genomic
clones.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
19
The polynucleotide sequence for the mouse ortholog of zalphal 1 Ligand
has been identified and is shown in SEQ ID NO: 55 and the corresponding amino
acid
sequence shown in SEQ ID NO: 56. There is 62% identity between the mouse and
human sequences over a 124 amino acid region that corresponds to residues 30
to 153
in SEQ ID NO: 2 and residues 23 to 146 of SEQ ID NO: 56 of zalphal 1 Ligand.
Mature sequence for the mouse zalphal 1 Ligand putatively begins at Hisis (as
shown in
SEQ ID NO: 56), which corresponds to His25 (as shown in SEQ ID NO: 2) in the
human sequence. Because a truncated form of the human polypeptide is active,
it is
likely that an equivalent polypeptide of the mouse zalphal 1 Ligand (i.e.
without
o residues Hisis to Pro22 of SEQ ID NO: 56) is active as well. Tissue
analysis revealed
that expression of mouse zalphal 1 Ligand is found in testis, spleen and
thymus.
Those skilled in the art will recognize that the sequence disclosed in
SEQ ID NO:1 represents a single allele of human zalphal 1 Ligand and that
allelic
variation and alternative splicing are expected to occur. Allelic variants of
this
sequence can be cloned by probing cDNA or genomic libraries from different
individuals according to standard procedures. Allelic variants of the DNA
sequence
shown in SEQ ID NO:1, including those containing silent mutations and those in
which
mutations result in amino acid sequence changes, are within the scope of the
present
invention, as are proteins which are allelic variants of SEQ ID NO:2. cDNAs
generated
from alternatively spliced mRNAs, which retain the properties of the zalphal 1
Ligand
polypeptide, are included within the scope of the present invention, as are
polypeptides
encoded by such cDNAs and mRNAs. Allelic variants and splice variants of these
sequences can be cloned by probing cDNA or genomic libraries from different
individuals or tissues according to standard procedures known in the art.
The zalphal 1 Ligand gene has been mapped to the IL-2 framework
marker SHGC-12342, positioning zalphal 1 Ligand approximately 180 kb from the
IL-2
marker. The use of surrounding markers positions the zalphall Ligand gene in
the 4q27
region on the integrated LDB chromosome 4 map (The Genetic Location Database,
University of Southhampton,). The present invention also provides reagents
which will
find use in diagnostic applications. For example, the zalphal 1 Ligand gene, a
probe
comprising zalphal 1 Ligand DNA or RNA or a subsequence thereof can be used to
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
determine if the zalphall Ligand gene is present on a human chromosome, such
as
chromosome 4, or if a gene mutation has occurred. Based on annotation of a
fragment
of human genomic DNA containing a part of zalphal 1 Ligand genomic DNA
(Genbank
Accession No. AC007458), zalphal 1 Ligand is located at the 4q27 region of
5
chromosome 4. Detectable chromosomal aberrations at the zalphal 1 Ligand gene
locus
include, but are not limited to, aneuploidy, gene copy number changes, loss of
heterogeneity (LOH), translocations, insertions, deletions, restriction site
changes and
rearrangements. Such aberrations can be detected using polynucleotides of the
present
invention by employing molecular genetic techniques, such as restriction
fragment
ia length
polymorphism (RFLP) analysis, short tandem repeat (STR) analysis employing
PCR techniques, and other genetic linkage analysis techniques known in the art
(Sambrook et al., ibid.; Ausubel et. al., ibid.; Marian, Chest 108:255-65,
1995).
The precise knowledge of a gene's position can be useful for a number
of purposes, including: 1) determining if a sequence is part of an existing
contig and
is
obtaining additional surrounding genetic sequences in various forms, such as
YACs,
BACs or cDNA clones; 2) providing a possible candidate gene for an inheritable
disease which shows linkage to the same chromosomal region; and 3) cross-
referencing
model organisms, such as mouse, which may aid in determining what function a
particular gene might have.
20 As
stated previously, human zalphall Ligand gene resides near the IL-2
gene, which is in a region of chromosome 4q that has been shown to have
linkage with
susceptibility to inflammatory bowel disease (IBD) (including Crohn's disease
(CD)
and ulcerative colitis) in some families (Hampe et al. Am. J. Hum. Genet.
64:808-816,
1999; Cho et al. Proc. Natl. Acad. Sei. 95:7502-7507, 1998). In addition, the
zalphal 1
receptor gene maps to 16p11, another genomic region which is associated with
susceptibility to CD (Hugot et al., Nature 379:821-823, 1996; Ohmen et al.,
Hum. Mol.
Genet. 5:1679-1683, 1996). CD is a chronic inflammation of the gut with
frequent
systemic involvement; while the exact etiology is unknown, immunoregulatory
dysfunction involving failure of tolerance to ordinary gut antigens is a major
component (for reviews, see (Braegger et al.. Annals Allergy 72:135-141, 1994;
Sartor,
Am. J. Gastroenterol. 92:55-11S, 1997)). Several studies have found abnormal
NK
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
21
activity in CD patients (see, for example, (Egawa et. al., J. Clin. Lab.
Immunol. 20:187-
192, 1986; Aparicio-Pages et al. J. Clin. Lab. Immunol. 29:119-124, 1989; van
Tol et
al., Scand. J. Gastroenterol. 27:999-1005, 1992)), and defective memory B cell
formation has also been documented (Brogan et al., J. Clin. Lab. Immunol.
24:69-74,
1987). Since zalphal 1 Ligand plays a role in immune regulation, and since the
genes
for both receptor and ligand lie within CD susceptibility regions, both
receptor and
ligand are candidate genes for genetic predisposition to Crohn's disease.
Determination of the involvement of zalphal 1 receptor and/or zalphal 1
Ligand in the pathology of IBD can be accomplished by several methods.
Sequencing
of exons from genomic DNA can reveal coding mutations (including missense,
nonsense, and frameshift mutations), as can sequencing of cDNAs. An additional
advantage of sequencing from genomic DNA is that splice junctions are also
contained
within the sequenced fragments and may reveal splicing abnormalities, which
might not
appear in cDNA samples if, for example, misspliced RNAs were rapidly degraded.
The
genomic structure of zalphal 1 Ligand has been determined. Other methods for
analysis
of zalphal 1 Ligand and receptor in IBD patients include: (1) assessment of
ligand
production from activated T cells from patients vs. normal controls (i.e. by
bioassay);
(2) in situ hybridization of zalphal 1 receptor or zalphal 1 Ligand RNA to
sections of
inflamed intestine from IBD patients, compared to similar sections from normal
controls; (3) immunohistochemistry on sections from IBD patients vs. normal
controls;
and (4) assessment of the responsiveness of patients peripheral B cells to
zalphal 1
Ligand, as measured by mitogenesis assays.
A diagnostic could assist physicians in determining the type of disease
and appropriate associated therapy, or could assist in genetic counseling. As
such, the
inventive anti-zalphal 1 Ligand antibodies, polynucleotides, and polypeptides
can be
used for the detection of zalphal 1 Ligand polypeptide, mRNA or anti-zalphal 1
Ligand
antibodies, thus serving as markers and be directly used for detecting or
genetic
diseases or cancers, as described herein, using methods known in the art and
described
herein. Further, zalphal 1 Ligand polynucleotide probes can be used to detect
abnormalities involving chromosome 4q27 as described herein. These
abnormalities
may be associated with human diseases, or tumorigenesis, spontaneous abortion
or
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
22
other genetic disorders. Thus, zalphal 1 Ligand polynucleotide probes can be
used to
detect abnormalities or genotypes associated with these defects.
As discussed above, defects in the zalphal 1 Ligand gene itself may
result in a heritable human disease state. Molecules of the present invention,
such as
the polypeptides, antagonists, agonists, polynucleotides and antibodies of the
present
invention would aid in the detection, diagnosis prevention, and treatment of
diseases
associated with a zalphal 1 Ligand genetic defect. In addition, zalphal 1
Ligand
polynucleotide probes can be used to detect allelic differences between
diseased or non-
diseased individuals at the zalphal 1 Ligand chromosomal locus. As such, the
zalpha 1 1
Ligand sequences can be used as diagnostics in forensic DNA profiling.
In general, the diagnostic methods used in genetic linkage analysis, to
detect a genetic abnormality or aberration in a patient, are known in the art.
Most
diagnostic methods comprise the steps of (i) obtaining a genetic sample from a
potentially diseased patient, diseased patient or potential non-diseased
carrier of a
recessive disease allele; (ii) producing a first reaction product by
incubating the genetic
sample with a zalphal 1 Ligand polynucleotide probe wherein the polynucleotide
will
hybridize to complementary polynucleotide sequence, such as in RFLP analysis
or by
incubating the genetic sample with sense and antisense primers in a PCR
reaction under
appropriate PCR reaction conditions; (iii) Visualizing the first reaction
product by gel
electrophoresis and/or other known method such as visualizing the first
reaction
product with a zalphal 1 Ligand polynucleotide probe wherein the
polynucleotide will
hybridize to the complementary polynucleotide sequence of the first reaction;
and (iv)
comparing the visualized first reaction product to a second control reaction
product of a
genetic sample from a normal or control individual. A difference between the
first
reaction product and the control reaction product is indicative of a genetic
abnormality
in the diseased or potentially diseased patient, or the presence of a
heterozygous
recessive carrier phenotype for a non-diseased patient, or the presence of a
genetic
defect in a tumor from a diseased patient, or the presence of a genetic
abnormality in a
fetus or pre-implantation embryo. For example, a difference in restriction
fragment
pattern, length of PCR products, length of repetitive sequences at the zalpha
1 1 Ligand
genetic locus, and the like, are indicative of a genetic abnormality, genetic
aberration,
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
23
or allelic difference in comparison to the normal control. Controls can be
from
unaffected family members, or unrelated individuals, depending on the test and
availability of samples. Genetic samples for use within the present invention
include
genomic DNA, mRNA, and cDNA isolated fromm any tissue or other biological
sample from a patient, such as but not limited to, blood, saliva, semen,
embryonic cells,
amniotic fluid, and the like. The polynucleotide probe or primer can be RNA or
DNA,
and will comprise a portion of SEQ ID NO:1, the complement of SEQ ID NO:1, or
an
RNA equivalent thereof. Such methods of showing genetic linkage analysis to
human
disease phenotypes are well known in the art. For reference to PCR based
methods in
o diagnostics see, generally, Mathew (ed.), Protocols in Human Molecular
Genetics
(Humana Press, Inc. 1991), White (ed.), PCR Protocols: Current Methods and
Applications (Humana Press, Inc. 1993), Cotter (ed.), Molecular Diagnosis of
Cancer
(Humana Press, Inc. 1996), Hanausek and Walaszek (eds.), Tumor Marker
Protocols
(Humana Press, Inc. 1998), Lo (ed.), Clinical Applications of PCR (Humana
Press, Inc.
1998), and Meltzer (ed.), PCR in Bioanalysis (Humana Press, Inc. 1998)).
Mutations associated with the zalphal 1 Ligand locus can be detected
using nucleic acid molecules of the present invention by employing standard
methods
for direct mutation analysis, such as restriction fragment length polymorphism
analysis,
short tandem repeat analysis employing PCR techniques. amplification-
refractory
mutation system analysis. single-strand conformation polymorphism detection,
RNase
cleavage methods, denaturing gradient gel electrophoresis, fluorescence-
assisted
mismatch analysis, and other genetic analysis techniques known in the art
(see, for
example, Mathew (ed.), Protocols in Human Molecular Genetics (Humana Press,
Inc.
1991), Marian, Chest 108:255 (1995), Coleman and Tsongalis. Molecular
Diagnostics
(Human Press, Inc. 1996), Elles (ed.) Molecular Diagnosis of Genetic Diseases
(Humana Press, Inc. 1996), Landegren (ed.), Laboratory Protocols for Mutation
Detection (Oxford University Press 1996). Birren et al. (eds.). Genome
Analysis, Vol. 2:
Detecting Genes (Cold Spring Harbor Laboratory Press 1998). Dracopoli et al.
(eds.),
Current Protocols in Human Genetics (John Wiley & Sons 1998), and Richards and
Ward, "Molecular Diagnostic Testing," in Principles of Molecular Medicine,
pages 83-
88 (Humana Press, Inc. 1998). Direct analysis of an zalphal 1 Ligand gene for
a
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
24
mutation can be performed using a subject's genomic DNA. Methods for
amplifying
genomic DNA, obtained for example from peripheral blood lymphocytes, are well-
known to those of skill in the art (see, for example, Dracopoli et al. (eds.),
Current
Protocols in Human Genetics, at pages 7.1.6 to 7.1.7 (John Wiley & Sons
1998)).
Positions of introns in the zalphal 1 Ligand gene were determined by
identification of genomic clones, followed by sequencing the intron/exon
junctions.
The first intron lies between amino acid residue 56 (Leu) and residue 57 (Val)
in Seq.
ID. No. 2, and is 115 base pairs in length. The second intron is the largest
at 4.4
kilobases, and lies between amino acid residue 68 (Glu) and residue 69 (Thr)
in Seq.
o ID. No. 2. The third intron is 2.6 kilobases, and lies between amino acid
residue 120
(Leu) and residue 121 (Thr) in Seq. ID. No. 2. The final intron, 89 base
pairs, lies
between amino acid residue 146 (Lys) and residue 147 (Met) in Seq. ID. No. 2.
The
complete gene spans about 8 kb.
The structure of the zalphal 1 Ligand gene is similar to that of the IL-2
gene (Fujita et al. Proc. Natl. Acad. Sci. 80:7437-7441, 1983), though the
zalphal 1
Ligand gene contains one additional intron (intron 4). The pattern of a short
first intron
and long second and third introns is conserved between the two genes, though
the IL-2
gene is slightly smaller overall (about 6 kb). The IL-15 gene, on the other
hand,
consists of 8 exons and spans at least 34 kb (Anderson et al. Genomics 25:701-
706,
1995). Thus the zalphal 1 Ligand gene is more similar in structure to the IL-2
gene than
to the IL-15 gene.
Within embodiments of the invention, isolated zalphal 1 Ligand-
encoding nucleic acid molecules can hybridize under stringent conditions to
nucleic
acid molecules having the nucleotide sequence of SEQ ID NO:1, to nucleic acid
molecules having the nucleotide sequence of nucleotides 47 to 532 of SEQ ID
NO:1, or
to nucleic acid molecules having a nucleotide sequence complementary to SEQ ID
NO: . In general, stringent conditions are selected to be about 5 C lower than
the
thermal melting point (Tm) for the specific sequence at a defined ionic
strength and pH.
The Tm is the temperature (under defined ionic strength and pH) at which 50%
of the
target sequence hybridizes to a perfectly matched probe.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
A pair of nucleic acid molecules, such as DNA-DNA, RNA-RNA and
DNA-RNA, can hybridize if the nucleotide sequences have some degree of
complementarity. Hybrids can tolerate mismatched base pairs in the double
helix, but
the stability of the hybrid is influenced by the degree of mismatch. The T. of
the
5
mismatched hybrid decreases by 1 C for every 1-1.5% base pair mismatch.
Varying
the stringency of the hybridization conditions allows control over the degree
of
mismatch that will be present in the hybrid. The degree of stringency
increases as the
hybridization temperature increases and the ionic strength of the
hybridization buffer
decreases.
10 It is
well within the abilities of one skilled in the art to adapt these
conditions for use with a particular polynucleotide hybrid. The T. for a
specific target
sequence is the temperature (under defined conditions) at which 50% of the
target
sequence will hybridize to a perfectly matched probe sequence. Those
conditions
which influence the T. include, the size and base pair content of the
polynucleotide
15 probe,
the ionic strength of the hybridization solution, and the presence of
destabilizing
agents in the hybridization solution. Numerous equations for calculating T.
are known
in the art, and are specific for DNA, RNA and DNA-RNA hybrids and
polynucleotide
probe sequences of varying length (see, for example, Sambrook et al.,
Molecular
Cloning: A Laboratory Manual, Second Edition (Cold Spring Harbor Press 1989);
20 Ausubel
et al., (eds.), Current Protocols in Molecular Biology (John Wiley and Sons,
Inc. 1987); Berger and Kimmel (eds.), Guide to Molecular Cloning Techniques,
(Academic Press, Inc. 1987); and Wetmur, Crit. Rev. Biochem. Mol. Biol. 26:227
(1990)). Sequence analysis software such as OLIGO 6.0 (LSR; Long Lake, MN) and
Primer Premier 4.0 (Premier Biosoft International; Palo Alto, CA), as well as
sites on
25 the
Internet, are available tools for analyzing a given sequence and calculating
T. based
on user defined criteria. Such programs can also analyze a given sequence
under
defined conditions and identify suitable probe sequences. Typically,
hybridization of
longer polynucleotide sequences, >50 base pairs, is performed at temperatures
of about
20-25 C below the calculated T.. For smaller probes, <50 base pairs,
hybridization is
typically carried out at the Tõ, or 5-10 C below the calculated T.. This
allows for the
maximum rate of hybridization for DNA-DNA and DNA-RNA hybrids.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
26
Following hybridization, the nucleic acid molecules can be washed to
remove non-hybridized nucleic acid molecules under stringent conditions, or
under
highly stringent conditions. Typical stringent washing conditions include
washing in a
solution of 0.5x - 2x SSC with 0.1% sodium dodecyl sulfate (SDS) at 55 - 65 C.
That
is, nucleic acid molecules encoding a variant zalphal 1 Ligand polypeptide
hybridize
with a nucleic acid molecule having the nucleotide sequence of SEQ ID NO:1 (or
its
complement) under stringent washing conditions, in which the wash stringency
is
equivalent to 0.5x - 2x SSC with 0.1% SDS at 55 - 65 C, including 0.5x SSC
with
0.1% SDS at 55 C, or 2x SSC with 0.1% SDS at 65 C. One of skill in the art can
o readily
devise equivalent conditions, for example, by substituting SSPE for SSC in the
wash solution.
Typical highly stringent washing conditions include washing in a
solution of 0.1x - 0.2x SSC with 0.1% sodium dodecyl sulfate (SDS) at 50 - 65
C. In
other words, nucleic acid molecules encoding a variant zalphal 1 Ligand
polypeptide
hybridize with a nucleic acid molecule having the nucleotide sequence of SEQ
ID NO:1
(or its complement) under highly stringent washing conditions, in which the
wash
stringency is equivalent to 0.1x - 0.2x SSC with 0.1% SDS at 50 - 65 C,
including 0.1x
SSC with 0.1% SDS at 50 C, or 0.2x SSC with 0.1% SDS at 65 C.
The
present invention also provides isolated zalphal 1 Ligand
polypeptides that have a substantially similar sequence identity to the
polypeptides of
SEQ ID NO:2, or their orthologs. The term "substantially similar sequence
identity" is
used herein to denote polypeptides comprising at least 70%, at least 80%, at
least 90%,
at least 95%, or greater than 95% sequence identity to the sequences shown in
SEQ ID
NO:2, or their orthologs. The present invention also includes polypeptides
that
comprise an amino acid sequence having at least 70%, at least 80%, at least
90%, at
least 95% or greater than 95% sequence identity to the sequence of amino acid
residues
1 to 162 or 33 to 162 of SEQ ID NO:2. The present invention further includes
nucleic
acid molecules that encode such polypeptides. Methods for determining percent
identity are described below.
The present invention also contemplates variant zalphal 1 Ligand nucleic
acid molecules that can be identified using two criteria: a determination of
the
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
27
similarity between the encoded polypeptide with the amino acid sequence of SEQ
ID
NO:2, and/or a hybridization assay, as described above. Such zalphal 1 Ligand
variants
include nucleic acid molecules: (1) that hybridize with a nucleic acid
molecule having
the nucleotide sequence of SEQ ID NO:1 (or its complement) under stringent
washing
conditions, in which the wash stringency is equivalent to 0.5x - 2x SSC with
0.1% SDS
at 55 - 65 C; or (2) that encode a polypeptide having at least 70%, at least
80%, at least
90%, at least 95% or greater than 95% sequence identity to the amino acid
sequence of
SEQ ID NO:2. Alternatively, zalphal 1 Ligand variants can be characterized as
nucleic
acid molecules: (1) that hybridize with a nucleic acid molecule having the
nucleotide
sequence of SEQ ID NO:1 (or its complement) under highly stringent washing
conditions, in which the wash stringency is equivalent to 0.1x - 0.2x SSC with
0.1%
SDS at 50 - 65 C; and (2) that encode a polypeptide having at least 70%, at
least 80%,
at least 90%, at least 95% or greater than 95% sequence identity to the amino
acid
sequence of SEQ ID NO:2.
Percent sequence identity is determined by conventional methods. See,
for example, Altschul et al., Bull. Math. Bio. 48:603 (1986), and Henikoff and
Henikoff, Proc. Natl. Acad. Sci. USA 89:10915 (1992). Briefly, two amino acid
sequences are aligned to optimize the alignment scores using a gap opening
penalty of
10, a gap extension penalty of 1, and the "BLOSUM62" scoring matrix of
Henikoff and
Henikoff (ibid.) as shown in Table 4 (amino acids are indicated by the
standard one-
letter codes).
Total number of identical matches
________________________________________________ x 100
[length of the longer sequence plus the
number of gaps introduced into the longer
sequence in order to align the two sequences]
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
28
NH
HN cc)
F LI1NNO
Cf) =t1H T
N N
TI
W zt N N Hr,)H
L.!) ONHHHHH
LC17INHOHO1NN
1 1 1 1 1 1 1
=t1NNOCY1NHNHH
1 1 1 1 1 1
( )
N or) HO cY) N H cr) cn
I I i I I
T I H r N I 7 I I N I N N
0 N N N TN
N T T
I I
I I 1 NO N H T o N N N
II I I I
QI L11NN NON H IOH 71N
I I
0lre)FiCr)M1-1Hre)HNNHHNNH
LONONHHOld-IHNNHOH1M01
I I I I
0'1 0 CD OHNNONNNHO=cP
I I I I
r2
MONNHONONNNHNNHHNNN
1 1 1 1 I I I I I I
I I
g 71-IHNNOHHONHHHHNHHONNO
I I I I
1 1 I 1 1 1 1
g Z U 0 44 u) >
In 0 In 0
-C\1
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
29
Those skilled in the art appreciate that there are many established
algorithms available to align two amino acid sequences. The "FASTA" similarity
search algorithm of Pearson and Lipman is a suitable protein alignment method
for
examining the level of identity shared by an amino acid sequence disclosed
herein and
the amino acid sequence of a putative variant zalphal 1 Ligand. The FASTA
algorithm
is described by Pearson and Lipman, Proc. Nat'l Acad. Sci. USA 85:2444 (1988),
and
by Pearson, Meth. Enzymol. 183:63 (1990).
Briefly, FASTA first =characterizes sequence similarity by identifying
regions shared by the query sequence (e.g., SEQ ID NO:2) and a test sequence
that
have either the highest density of identities (if the ktup variable is 1) or
pairs of
identities (if ktup=2), without considering conservative amino acid
substitutions,
insertions, or deletions. The ten regions with the highest density of
identities are then
rescored by comparing the similarity of all paired amino acids using an amino
acid
substitution matrix, and the ends of the regions are "trimmed" to include only
those
is residues that contribute to the highest score. If there are several
regions with scores
greater than the "cutoff' value (calculated by a predetermined formula based
upon the
length of the sequence and the ktup value), then the trimmed initial regions
are
examined to determine whether the regions can be joined to form an approximate
alignment with gaps. Finally, the highest scoring regions of the two amino
acid
sequences are aligned using a modification of the Needleman-Wunsch-Sellers
algorithm (Needleman and Wunsch, J. Mol. Biol. 48:444 (1970); Sellers, SIAM J.
Appl. Math. 26:787 (1974)), which allows for amino acid insertions and
deletions.
Preferred parameters for FASTA analysis are: ktup=1, gap opening penalty=10,
gap
extension penalty=1, and substitution matrix=BLOSUM62. These parameters can be
introduced into a FASTA program by modifying the scoring matrix file
("SMATRIX"),
as explained in Appendix 2 of Pearson, Meth. Enzymol. 183:63 (1990).
FASTA can also be used to determine the sequence identity of nucleic
acid molecules using a ratio as disclosed above. For nucleotide sequence
comparisons,
the ktup value can range between one to six, preferably from three to six,
most
preferably three, with other parameters set as default.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
Variant zalphal 1 Ligand polypeptides or polypeptides with substantially
similar sequence identity are characterized as having one or more amino acid
substitutions, deletions or additions. These changes are preferably of a minor
nature,
that is conservative amino acid substitutions (see Table 5) and other
substitutions that
5 do not significantly affect the folding or activity of the polypeptide;
small deletions,
typically of one to about 30 amino acids; and amino- or carboxyl-terminal
extensions,
such as an amino-terminal methionine residue, a small linker peptide of up to
about 20-
25 residues, or an affinity tag. The present invention thus includes
polypeptides of
from about 108 to 216 amino acid residues that comprise a sequence that is at
least
o 70%, preferably at least 90%, and more preferably 95% or more identical to
the
corresponding region of SEQ ID NO:2. Polypeptides comprising affinity tags can
further comprise a proteolytic cleavage site between the zalphal 1 Ligand
polypeptide
and the affinity tag. Preferred such sites include thrombin cleavage sites and
factor Xa
cleavage sites.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
31
Table 5
Conservative amino acid substitutions
Basic: arginine
lysine
histidine
Acidic: glutamic acid
aspartic acid
Polar: glutamine
asparagine
Hydrophobic: leucine
isoleucine
valine
Aromatic: phenylalanine
tryptophan
tyrosine
Small: glycine
alanine
serine
threonine
methionine
Determination of amino acid residues that comprise regions or domains
that are critical to maintaining structural integrity can be determined.
Within these
regions one can determine specific residues that will be more or less tolerant
of change
and maintain the overall tertiary structure of the molecule. Methods for
analyzing
sequence structure include, but are not limited to alignment of multiple
sequences with
high amino acid or nucleotide identity, secondary structure propensities,
binary
patterns, complementary packing and buried polar interactions (Barton, Current
Opin.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
32
Struct. Biol. 5:372-376, 1995 and Cordes et al., Current Opin. Struct. Biol.
6:3-10,
1996). In general, when designing modifications to molecules or identifying
specific
fragments determination of structure will be accompanied by evaluating
activity of
modified molecules.
Amino acid sequence changes are made in zalphal 1 Ligand
polypeptides so as to minimize disruption of higher order structure essential
to
biological activity. For example, where the zalphal 1 Ligand polypeptide
comprises
one or more helices, changes in amino acid residues will be made so as not to
disrupt
the helix geometry and other components of the molecule where changes in
conformation abate some critical function, for example, binding of the
molecule to its
binding partners, e.g., A and D helices, residues 44,47 and 135 of SEQ ID NO:
2. The
effects of amino acid sequence changes can be predicted by, for example,
computer
modeling as disclosed above or determined by analysis of crystal structure
(see, e.g.,
Lapthorn et al., Nat. Struct. Biol. 2:266-268, 1995). Other techniques that
are well
known in the art compare folding of a variant protein to a standard molecule
(e.g., the
native protein). For example, comparison of the cysteine pattern in a variant
and
standard molecules can be made. Mass spectrometry and chemical modification
using
reduction and alkylation provide methods for determining cysteine residues
which are
associated with disulfide bonds or are free of such associations (Bean et al.,
Anal.
Biochem. 201:216-226, 1992; Gray, Protein Sci. 2:1732-1748, 1993; and
Patterson et
al., Anal. Chem. 66:3727-3732, 1994). It is generally believed that if a
modified
molecule does not have the same cysteine pattern as the standard molecule
folding
would be affected. Another well known and accepted method for measuring
folding is
circular dichrosism (CD). Measuring and comparing the CD spectra generated by
a
modified molecule and standard molecule is routine (Johnson, Proteins 7:205-
214,
1990). Crystallography is another well known method for analyzing folding and
structure. Nuclear magnetic resonance (NMR), digestive peptide mapping and
epitope
mapping are also known methods for analyzing folding and structurally
similarities
between proteins and polypeptides (Schaanan et al., Science 257:961-964,
1992).
A Hopp/Woods hydrophilicity profile of the zalphal 1 Ligand protein
sequence as shown in SEQ ID NO:2 can be generated (Hopp et al., Proc. Natl.
Acad.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
33
Sci.78:3824-3828, 1981; Hopp, J. Immun. Meth. 88:1-18, 1986 and Triquier et
al.,
Protein Engineering 11:153-169, 1998). The profile is based on a sliding six-
residue
window. Buried G, S, and T residues and exposed II, Y, and W residues were
ignored.
For example, in zalphal 1 Ligand, hydrophilic regions include amino acid
residues 114-
119 of SEQ ID NO: 2, amino acid residues 101-105 of SEQ ID NO: 2, amino acid
residues 126-131 of SEQ ID NO: 2, amino acid residues 113-118 of SEQ ID NO: 2,
and amino acid residues 158-162 of SEQ ID NO: 2.
Those skilled in the art will recognize that hydrophilicity or
hydrophobicity will be taken into account when designing modifications in the
amino
acid sequence of a zalphal 1 Ligand polypeptide, so as not to disrupt the
overall
structural and biological profile. Of particular interest for replacement are
hydrophobic
residues selected from the group consisting of Val, Leu and Ile or the group
consisting
of Met, Gly, Ser, Ala, Tyr and Trp. For example, residues tolerant of
substitution could
include residues 100 and 103 as shown in SEQ ID NO: 2. Cysteine residues at
positions 71, 78, 122 and 125 of SEQ ID NO: 2, will be relatively intolerant
of
substitution.
The identities of essential amino acids can also be inferred from analysis
of sequence similarity between IL-15, IL-2, IL-4 and GM-CSF with zalphal 1
Ligand.
Using methods such as "FASTA" analysis described previously, regions of high
similarity are identified within a family of proteins and used to analyze
amino acid
sequence for conserved regions. An alternative approach to identifying a
variant
zalphal 1 Ligand polynucleotide on the basis of structure is to determine
whether a
nucleic acid molecule encoding a potential variant zalphal 1 Ligand gene can
hybridize
to a nucleic acid molecule having the nucleotide sequence of SEQ ID NO:1, as
discussed above.
Other methods of identifying essential amino acids in the polypeptides
of the present invention are procedures known in the art, such as site-
directed
mutagenesis or alanine-scanning mutagenesis (Cunningham and Wells. Science
244:1081 (1989), Bass et al., Proc. Natl Acad. Sci. USA 88:4498 (1991), Coombs
and
Corey, "Site-Directed Mutagenesis and Protein Engineering," in Proteins:
Analysis and
Design, Angeletti (ed.), pages 259-311 (Academic Press, Inc. 1998)). In the
latter
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
34
technique, single alanine mutations are introduced at every residue in the
molecule, and
the resultant mutant molecules are tested for biological or biochemical
activity as
disclosed below to identify amino acid residues that are critical to the
activity of the
molecule. See also, Hilton et al., J. Biol. Chem. 27/ :4699 (1996).
The present invention also includes functional fragments of zalphal 1
Ligand polypeptides and nucleic acid molecules encoding such functional
fragments. A
"functional" zalphal 1 Ligand or fragment thereof as defined herein is
characterized by
its proliferative or differentiating activity, by its ability to induce or
inhibit specialized
cell functions, or by its ability to bind specifically to an anti- zalphall
Ligand antibody
io or
zalphal 1 receptor (either soluble or immobilized). As previously described
herein,
zalphal 1 Ligand is characterized by a four-helical-bundle structure
comprising helix A
(amino acid residues 41-56), helix B (amino acid residues 69-84), helix C
(amino acid
residues 92-105) and helix D (amino acid residues 135-148), as shown in SEQ ID
NO:
2. Thus, the present invention further provides fusion proteins encompassing:
(a)
polypeptide molecules comprising one or more of the helices described above;
and (b)
functional fragments comprising one or more of these helices. The other
polypeptide
portion of the fusion protein may be contributed by another four-helical-
bundle
cytokine, such as IL-15, IL-2, IL-4 and GM-CSF, or by a non-native and/or an
unrelated secretory signal peptide that facilitates secretion of the fusion
protein.
Thus the present invention provides fusion proteins comprising at least
four polypeptides, wherein the order of polypeptides from N-terminus to C-
terminus
are: a first polypeptide comprises amino acids selected from a group
consisting of: (a)
IL-2 helix A amino acid residues 36-46 of SEQ ID NO: 111; (b) IL-15 helix A
amino
acid residues 29-43 of SEQ ID NO: 112; (c) IL-4 helix A amino acid residues 45-
68 of
SEQ ID NO: 113; (d) GMCSF helix A amino acid residues 30-44 of SEQ ID NO: 114;
and (e) amino acids residues 41 to 56 of SEQ ID NO: 2; a first spacer of 6-27
amino
acids; and a second polypeptide that comprises amino acid residues selected
from the
group consisting of: (a) IL-2 helix B amino acid residues 53-75 of SEQ ID NO:
111;
(b) IL-4 helix B amino acid residues 65-83 of SEQ ID NO: 112; (c) IL-15 helix
B
amino acid residues 84-101 of SEQ ID NO: 113; (d) GMCSF helix B amino acid
residues 72-81 of SEQ ID NO: 114; and (e) amino acid residues 69-84 of SEQ ID
NO:
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
2; a second spacer of 5-11 amino acid residues; a third polypeptide that
comprises a
sequence of amino acid residues selected from the group consisting of: (a) IL-
2 helix C
residues 87-99 of SEQ ID NO: 111; (b) IL-4 helix C residues 95-118 of SEQ ID
NO:
112; (c) IL-15 helix C residues 107-119 of SEQ ID NO: 113; (d) GMCSF helix C
5 residues 91-102 of SEQ ID NO: 114; and (e) amino acid residues 92-105 of
SEQ ID
NO: 2; a third spacer of 3-29 amino acid residues; and a fourth polypeptide
that
comprises amino acid residues selected from the group consisting of: (a) IL-2
helix D
amino acid residues 103-121 of SEQ ID NO: 111 ; (b) IL-15 helix D amino acid
residues 134-157 of SEQ ID NO: 112; (c) IL-4 helix D amino acid residues 134-
160 of
o SEQ ID NO: 113; (d) GMCSF helix D amino acid residues 120-131 of SEQ ID
NO:
114; and (e) amino acid residues 135-148 of SEQ ID NO: 2, wherein at least one
of the
four polypeptides is from zalphall Ligand. In other embodiments that the
spacer
peptides will be selected from the A/B, B/C and C/D loops of zalphal 1 Ligand,
IL-2,
IL-4, IL-15 or GM-CSF, as shown in Table 1.
15 Routine
deletion analyses of nucleic acid molecules can be performed to
obtain functional fragments of a nucleic acid molecule that encodes a zalphal
1 Ligand
polypeptide. As an illustration, DNA molecules having the nucleotide sequence
of
SEQ ID NO:1 or fragments thereof, can be digested with Ba131 nuclease to
obtain a
series of nested deletions. These DNA fragments are then inserted into
expression
20 vectors in proper reading frame, and the expressed polypeptides are
isolated and tested
for zalphal 1 Ligand activity, or for the ability to bind anti-zalphal 1
Ligand antibodies
or zalphal 1 receptor. One
alternative to exonuclease digestion is to use
oligonucleotide-directed mutagenesis to introduce deletions or stop codons to
specify
production of a desired zalphal 1 Ligand fragment. Alternatively, particular
fragments
25 of a zalphal 1 Ligand gene can be synthesized using the polymerase chain
reaction.
Standard methods for identifying functional domains are well-known to
those of skill in the art. For example, studies on the truncation at either or
both termini
of interferons have been summarized by Horisberger and Di Marco, Pharmac.
Ther.
66:507 (1995). Moreover, standard techniques for functional analysis of
proteins are
30 described by, for example, Treuter et al., Molec. Gen. Genet. 240:113
(1993); Content
et al., "Expression and preliminary deletion analysis of the 42 kDa 2-5A
synthetase
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
36
induced by human interferon," in Biological Interferon Systems, Proceedings of
ISIR-TNO Meeting on Interferon Systems, Cantell (ed.), pages 65-72 (Nijhoff
1987);
Herschman, "The EGF Receptor," in Control of Animal Cell Proliferation 1
Boynton
et al., (eds.) pages 169-199 (Academic Press 1985); Coumailleau et al., J.
Biol. Chem.
270:29270 (1995); Fukunaga et al., J. Biol. Chem. 270:25291 (1995); Yamaguchi
et al.,
Biochem. Pharmacol. 50:1295 (1995); and Meisel et al., Plant Molec. Biol. 30:1
(1996).
Multiple amino acid substitutions can be made and tested using known
methods of mutagenesis and screening, such as those disclosed by Reidhaar-
Olson and
Sauer (Science 241:53 (1988)) or Bowie and Sauer (Proc. Nat'l Acad. Sci. USA
86:2152 (1989)). Briefly, these authors disclose methods for simultaneously
randomizing two or more positions in a polypeptide, selecting for functional
polypeptide, and then sequencing the mutagenized polypeptides to determine the
spectrum of allowable substitutions at each position. Other methods that can
be used
include phage display (e.g., Lowman et al., Biochem. 30:10832 (1991), Ladner
et al.,
U.S. Patent No. 5,223,409, Huse, international publication No. WO 92/06204),
and
region-directed mutagenesis (Derbyshire et al., Gene 46:145 (1986), and Ner et
al.,
DNA 7:127, (1988)).
Variants of the disclosed zalphal 1 Ligand nucleotide and polypeptide
sequences can also be generated through DNA shuffling as disclosed by Stemmer,
Nature 370:389 (1994), Stemmer, Proc. Natl Acad. Sci. USA 91:10747 (1994), and
international publication No. WO 97/20078. Briefly, variant DNA molecules are
generated by in vitro homologous recombination by random fragmentation of a
parent
DNA followed by reassembly using PCR, resulting in randomly introduced point
mutations. This technique can be modified by using a family of parent DNA
molecules, such as allelic variants or DNA molecules from different species.
to
introduce additional variability into the process. Selection or screening for
the desired
activity, followed by additional iterations of mutagenesis and assay provides
for rapid
"evolution" of sequences by selecting for desirable mutations while
simultaneously
selecting against detrimental changes.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
37
Mutagenesis methods as disclosed herein can be combined with high-
throughput, automated screening methods to detect activity of cloned,
mutagenized
polypeptides in host cells. Mutagenized DNA molecules that encode biologically
active polypeptides, or polypeptides that bind with anti-zalphal 1 Ligand
antibodies or
s soluble zalphal 1 receptor, can be recovered from the host cells and
rapidly sequenced
using modern equipment. These methods allow the rapid determination of the
importance of individual amino acid residues in a polypeptide of interest, and
can be
applied to polypeptides of unknown structure.
In addition, the proteins of the present invention (or polypeptide
o fragments thereof) can be joined to other bioactive molecules, particularly
other
cytokines, to provide multi-functional molecules. For example, one or more
helices
from zalphal 1 Ligand can be joined to other cytokines to enhance their
biological
properties or efficiency of production.
The present invention thus provides a series of novel, hybrid molecules
15 in
which a segment comprising one or more of the helices of zalphal 1 Ligand is
fused
to another polypeptide. Fusion is preferably done by splicing at the DNA level
to allow
expression of chimeric molecules in recombinant production systems. The
resultant
molecules are then assayed for such properties as improved solubility,
improved
stability, prolonged clearance half-life, improved expression and secretion
levels, and
20 pharmacodynamics. Such hybrid molecules may further comprise additional
amino
acid residues (e.g. a polypeptide linker) between the component proteins or
polypeptides.
Non-naturally occurring amino acids include, without limitation. trans-
3-methylproline, 2,4-methanoproline, cis-4-hydroxyproline, irans-4-
hydroxyproline, N-
25 methylglycine, a//o-threonine, methylthreonine,
hydroxyethylcysteine,
hydroxyethylhomocysteine, nitroglutamine. homoglutamine, pipecolic acid,
thiazolidine carboxylic acid, dehydroproline, 3- and 4-methylproline. 3,3-
dimethylproline, tert-leucine, norvaline, 2-azaphenylalanine, 3-
azaphenylalanine, 4-
azaphenylalanine, and 4-fluorophenylalanine. Several methods are known in the
art for
30 incorporating non-naturally occurring amino acid residues into
proteins. For example,
an in vitro system can be employed wherein nonsense mutations are suppressed
using
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
38
chemically aminoacylated suppressor tRNAs. Methods for synthesizing amino
acids
and aminoacylating tRNA are known in the art. Transcription and translation of
plasmids containing nonsense mutations is typically carried out in a cell-free
system
comprising an E. coli S30 extract and commercially available enzymes and other
reagents. Proteins are purified by chromatography. See, for example, Robertson
et al.,
J. Am. Chem. Soc. 113:2722 (1991), Ellman et al., Methods Enzymol. 202:301
(1991),
Chung et al., Science 259:806 (1993), and Chung et al., Proc. Nat'l Acad. Sci.
USA
90:10145 (1993).
In a second method, translation is carried out in Xenopus oocytes by
microinjection of mutated mRNA and chemically aminoacylated suppressor tRNAs
(Turcatti et al., J. Biol. Chem. 271:19991 (1996)). Within a third method, E.
coli cells
are cultured in the absence of a natural amino acid that is to be replaced
(e.g.,
phenylalanine) and in the presence of the desired non-naturally occurring
amino acid(s)
(e.g., 2-azaphenylalanine, 3-azaphenylalanine, 4-azaphenylalanine, or 4-
fiuorophenylalanine). The non-naturally occurring amino acid is incorporated
into the
protein in place of its natural counterpart. See, Koide et al., Biochem.
33:7470 (1994).
Naturally occurring amino acid residues can be converted to non-naturally
occurring
species by in vitro chemical modification. Chemical modification can be
combined
with site-directed mutagenesis to further expand the range of substitutions
(Wynn and
Richards, Protein Sci. 2:395 (1993). It may be advantageous to stabilize
zalphal 1
Ligand to extend the half-life of the molecule, particularly for extending
metabolic
persistence in an active state. To achieve extended half-life. zalphal 1
Ligand
molecules can be chemically modified using methods described herein.
PEGylation is
one method commonly used that has been demonstrated to increase plasma half-
life,
increased solubility, and decreased antigenicity and immunogenicity (Nucci et
al.,
Advanced Drug Delivery Reviews 6:133-155, 1991 and Lu et al., Int. J. Peptide
Protein
Res. 43:127-138, 1994).
A limited number of non-conservative amino acids, amino acids that are
not encoded by the genetic code, non-naturally occurring amino acids, and
unnatural
amino acids may be substituted for zalphal 1 Ligand amino acid residues.
CA 02366921 2001-09-10
WO 00/53761 PCMJS00/06067
39
The present invention also provides polypeptide fragments or peptides
comprising an epitope-bearing portion of a zalphal 1 Ligand polypeptide
described
herein. Such fragments or peptides may comprise an "immunogenic epitope,"
which is
a part of a protein that elicits an antibody response when the entire protein
is used as an
immunogen. Immunogenic epitope-bearing peptides can be identified using
standard
methods (see, for example, Geysen et al., Proc. Nat'l Acad. Sci. USA 81:3998
(1983)).
In contrast, polypeptide fragments or peptides may comprise an
"antigenic epitope," which is a region of a protein molecule to which an
antibody can
specifically bind. Certain epitopes consist of a linear or contiguous stretch
of amino
1.0 acids, and the antigenicity of such an epitope is not disrupted by
denaturing agents. It is
known in the art that relatively short synthetic peptides that can mimic
epitopes of a
protein can be used to stimulate the production of antibodies against the
protein (see,
for example, Sutcliffe et al., Science 219:660 (1983)). Accordingly, antigenic
epitope-
bearing peptides and polypeptides of the present invention are useful to raise
antibodies
that bind with the polypeptides described herein. Hopp/Woods hydrophilicity
profiles
can be used to determine regions that have the most antigenic potential (Hopp
et al.,
1981, ibid. and Hopp, 1986, ibid.). In zalphal 1 Ligand these regions include:
amino
acid residues 114-119, 101-105, 126-131, 113-118, and 158-162 of SEQ ID NO: 2.
Antigenic epitope-bearing peptides and polypeptides preferably contain
at least four to ten amino acids, at least ten to fourteen amino acids, or
about fourteen to
about thirty amino acids of SEQ ID NO:2 or SEQ ID NO:56. Such epitope-bearing
peptides and polypeptides can be produced by fragmenting a zalphal 1 Ligand
polypeptide, or by chemical peptide synthesis, as described herein. Moreover,
epitopes
can be selected by phage display of random peptide libraries (see, for
example, Lane
and Stephen, Curr. Opin. Immunol. 5:268 (1993); and Cortese el al., Curr.
Opin.
Biotechnol. 7:616 (1996)). Standard methods for identifying epitopes and
producing
antibodies from small peptides that comprise an epitope are described, for
example, by
Mole, "Epitope Mapping,- in Methods in Molecular Biology. Vol. 10, Manson
(ed.),
pages 105-116 (The Humana Press, Inc. 1992); Price, "Production and
Characterization
of Synthetic Peptide-Derived Antibodies,- in Monoclonal Antibodies:
Production,
Engineering, and Clinical Application, Ritter and Ladyman (eds.), pages 60-84
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
(Cambridge University Press 1995), and Coligan et at. (eds.), Current
Protocols in
Immunology, pages 9.3.1 - 9.3.5 and pages 9.4.1 - 9.4.11 (John Wiley & Sons
1997).
Regardless of the particular nucleotide sequence of a variant zalphall
Ligand polynucleotide, the polynucleotide encodes a polypeptide that is
characterized
5 by its
proliferative or differentiating activity, its ability to induce or inhibit
specialized
cell functions, or by the ability to bind specifically to an anti-zalphall
Ligand antibody
or zalphal 1 receptor. More specifically, variant zalphal 1 Ligand
polynucleotides will
encode polypeptides which exhibit at least 50% and preferably, greater than
70%, 80%
or 90%, of the activity of the polypeptide as shown in SEQ ID NO: 2.
10 For any
zalphal 1 Ligand polypeptide, including variants and fusion
proteins, one of ordinary skill in the art can readily generate a fully
degenerate
polynucleotide sequence encoding that variant using the information set forth
in Tables
1 and 2 above.
The present invention further provides a variety of other polypeptide
15 fusions
(and related multimeric proteins comprising one or more polypeptide fusions).
For example, a zalphal 1 Ligand polypeptide can be prepared as a fusion to a
dimerizing protein as disclosed in U.S. Patents Nos. 5,155,027 and 5,567,584.
Preferred dimerizing proteins in this regard include immunoglobulin constant
region
domains. Immunoglobulin- zalphal 1 Ligand polypeptide fusions can be expressed
in
20
genetically engineered cells (to produce a variety of multimeric zalphal 1
Ligand
analogs). Auxiliary domains can be fused to zalphal 1 Ligand polypeptides to
target
them to specific cells, tissues, or macromolecules. For example, a zalphal 1
Ligand
polypeptide or protein could be targeted to a predetermined cell type by
fusing a
zalphal 1 Ligand polypeptide to a ligand that specifically binds to a receptor
on the
25 surface
of that target cell. In this way, polypeptides and proteins can be targeted
for
therapeutic or diagnostic purposes. A zalphal 1 Ligand polypeptide can be
fused to two
or more moieties, such as an affinity tag for purification and a targeting
domain.
Polypeptide fusions can also comprise one or more cleavage sites, particularly
between
domains. See, Tuan et al., Connective Tissue Research 34:1-9, 1996.
30 Using
the methods discussed herein. one of ordinary skill in the art can
identify and/or prepare a variety of polypeptides that have substantially
similar
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
41
sequence identity to residues 1-162 or 33-162 of SEQ ID NO: 2, or functional
fragments and fusions thereof, wherein such polypeptides or fragments or
fusions retain
the properties of the wild-type protein such as the ability to stimulate
proliferation,
differentiation, induce specialized cell function or bind the zalphal 1
receptor or
zalphal 1 Ligand antibodies.
The zalphal 1 Ligand polypeptides of the present invention, including
full-length polypeptides, functional fragments, and fusion polypeptides, can
be
produced in genetically engineered host cells according to conventional
techniques.
Suitable host cells are those cell types that can be transformed or
transfected with
a.o exogenous DNA and grown in culture, and include bacteria, fungal cells,
and cultured
higher eukaryotic cells. Eukaryotic cells, particularly cultured cells of
multicellular
organisms, are preferred. Techniques for manipulating cloned DNA molecules and
introducing exogenous DNA into a variety of host cells are disclosed by
Sambrook et
al., Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor
Laboratory
Press, Cold Spring Harbor, NY, 1989, and Ausubel et al., eds., Current
Protocols in
Molecular Biology, John Wiley and Sons, Inc., NY, 1987.
In general, a DNA sequence encoding a zalphal 1 Ligand polypeptide is
operably linked to other genetic elements required for its expression,
generally
including a transcription promoter and terminator, within an expression
vector. The
vector will also commonly contain one or more selectable markers and one or
more
origins of replication, although those skilled in the art will recognize that
within certain
systems selectable markers may be provided on separate vectors, and
replication of the
exogenous DNA may be provided by integration into the host cell genome.
Selection
of promoters, terminators, selectable markers, vectors and other elements is a
matter of
routine design within the level of ordinary skill in the art. Many such
elements are
described in the literature and are available through commercial suppliers.
To direct a zalpha 1 1 Ligand polypeptide into the secretory pathway of a
host cell, a secretory signal sequence (also known as a leader sequence,
prepro
sequence or pre sequence) is provided in the expression vector. The secretory
signal
sequence may be that of zalpha 1 1 Ligand, or may be derived from another
secreted
protein (e.g., t-PA) or synthesized de novo. The secretory signal sequence is
operably
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
42
linked to the zalphal 1 Ligand DNA sequence, i.e., the two sequences are
joined in the
correct reading frame and positioned to direct the newly synthesized
polypeptide into
the secretory pathway of the host cell. Secretory signal sequences are
commonly
positioned 5' to the DNA sequence encoding the polypeptide of interest,
although
s certain secretory signal sequences may be positioned elsewhere in the DNA
sequence
of interest (see, e.g., Welch et al., U.S. Patent No. 5,037,743; Holland et
al., U.S. Patent
No. 5,143,830).
Alternatively, the secretory signal sequence contained in the
polypeptides of the present invention is used to direct other polypeptides
into the
o secretory pathway. The present invention provides for such fusion
polypeptides. A
signal fusion polypeptide can be made wherein a secretory signal sequence
derived
from amino acid residue 1-31 of SEQ ID NO:2 is be operably linked to a DNA
sequence encoding another polypeptide using methods known in the art and
disclosed
herein. The secretory signal sequence contained in the fusion polypeptides of
the
15 present invention is preferably fused amino-terminally to an additional
peptide to direct
the additional peptide into the secretory pathway. Such constructs have
numerous
applications known in the art. For example, these novel secretory signal
sequence
fusion constructs can direct the secretion of an active component of a
normally non-
secreted protein. Such fusions may be used in vivo or in vitro to direct
peptides through
20 the secretory pathway.
Cultured mammalian cells are suitable hosts within the present
invention. Methods for introducing exogenous DNA into mammalian host cells
include
calcium phosphate-mediated transfection (Wigler et al., Cell 14:725, 1978;
Corsaro and
Pearson, Somatic Cell Genetics 7:603, 1981: Graham and Van der Eb. Virology
25 52:456, 1973), electroporation (Neumann et al., EMBO J. 1:841-5, 1982),
DEAE-
dextran mediated transfection (Ausubel et al., ibid.), and liposome-mediated
transfection (Hawley-Nelson et al.. Focus 15:73, 1993; Ciccarone et al., Focus
15:80,
1993, and viral vectors (Miller and Rosman, BioTechniques 7:980-90. 1989: Wang
and
Finer, Nature Med. 2:714-6, 1996). The production of recombinant polypeptides
in
30 cultured mammalian cells is disclosed, for example, by Levinson et al.,
U.S. Patent No.
4,713,339; Hagen et al., U.S. Patent No. 4,784,950; Palmiter et al., U.S.
Patent No.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
43
4,579,821; and RingoId, U.S. Patent No. 4,656,134. Suitable cultured mammalian
cells
include the COS-1 (ATCC No. CRL 1650), COS-7 (ATCC No. CRL 1651), BHK
(ATCC No. CRL 1632), BHK 570 (ATCC No. CRL 10314), 293 (ATCC No. CRL
1573; Graham et al., J. Gen. Virol. 36:59-72, 1977) and Chinese hamster ovary
(e.g.
CHO-Kl ; ATCC No. CCL 61) cell lines. Additional suitable cell lines are known
in
the art and available from public depositories such as the American Type
Culture
Collection, Manassas, VA. In general, strong transcription promoters are
preferred,
such as promoters from SV-40 or cytomegalovirus. See, e.g., U.S. Patent No.
4,956,288. Other suitable promoters include those from metallothionein genes
(U.S.
o Patent Nos. 4,579,821 and 4,601,978) and the adenovirus major late
promoter.
Drug selection is generally used to select for cultured mammalian cells
into which foreign DNA has been inserted. Such cells are commonly referred to
as
"transfectants". Cells that have been cultured in the presence of the
selective agent and
are able to pass the gene of interest to their progeny are referred to as
"stable
transfectants." A preferred selectable marker is a gene encoding resistance to
the
antibiotic neomycin. Selection is carried out in the presence of a neomycin-
type drug,
such as G-418 or the like. Selection systems can also be used to increase the
expression
level of the gene of interest, a process referred to as "amplification."
Amplification is
carried out by culturing transfectants in the presence of a low level of the
selective
agent and then increasing the amount of selective agent to select for cells
that produce
high levels of the products of the introduced genes. A preferred amplifiable
selectable
marker is dihydrofolate reductase, which confers resistance to methotrexate.
Other
drug resistance genes (e.g. hygromycin resistance, multi-drug resistance,
puromycin
acetyltransferase) can also be used. Alternative markers that introduce an
altered
phenotype, such as green fluorescent protein, or cell surface proteins such as
CD4,
CD8, Class I MHC, placental alkaline phosphatase may be used to sort
transfected cells
from untransfected cells by such means as FACS sorting or magnetic bead
separation
technology.
Other higher eukaryotic cells can also be used as hosts, including plant
cells, insect cells and avian cells. The use of Agrobacterium rhizogenes as a
vector for
expressing genes in plant cells has been reviewed by Sinkar et al., J. Biosci.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
44
(Bangalore) 11:47-58, 1987. Transformation of insect cells and production of
foreign
polypeptides therein is disclosed by Guarino et al., U.S. Patent No. 5,162,222
and
WIPO publication WO 94/06463. Insect cells can be infected with recombinant
baculovirus, commonly derived from Autographa californica nuclear polyhedrosis
virus (AcNPV). See, King, L.A. and Possee, R.D., The Baculovirus Expression
System: A Laboratory Guide, London, Chapman & Hall; O'Reilly, D.R. et al.,
Baculovirus Expression Vectors: A Laboratory Manual, New York, Oxford
University
Press., 1994; and, Richardson, C. D., Ed., Baculovirus Expression Protocols.
Methods
in Molecular Biology, Totowa, NJ, Humana Press, 1995. The second method of
making
recombinant baculovirus utilizes a transposon-based system described by Luckow
(Luckow, V.A, et al., J Virol 67:4566-79, 1993). This system is sold in the
Bac-to-Bac
kit (Life Technologies, Rockville, MD). This system utilizes a transfer
vector,
pFastBac 1 Tm (Life Technologies) containing a Tn7 transposon to move the DNA
encoding the zalphal 1 Ligand polypeptide into a baculovirus genome maintained
in E
is coli as a large plasmid called a "bacmid." The pFastBaclTM transfer
vector utilizes the
AcNPV polyhedrin promoter to drive the expression of the gene of interest, in
this case
zalphal 1 Ligand. However, pFastBacl Tm can be modified to a considerable
degree.
The polyhedrin promoter can be removed and substituted with the baculovirus
basic
protein promoter (also known as Pcor, p6.9 or MP promoter) which is expressed
earlier
in the baculovirus infection, and has been shown to be advantageous for
expressing
secreted proteins. See, Hill-Perkins, M.S. and Possee, R.D., J. Gen. Virol.
71:971-6,
1990; Bonning, B.C. et al., J. Gen. Virol. 75:1551-6, 1994; and, Chazenbalk,
G.D., and
Rapoport, B., J. Biol. Chem. 270:1543-9, 1995. In such transfer vector
constructs, a
short or long version of the basic protein promoter can be used. Moreover,
transfer
vectors can be constructed which replace the native zalphal 1 Ligand secretory
signal
sequences with secretory signal sequences derived from insect proteins. For
example,
a secretory signal sequence from Ecdysteroid Glucosyltransferase (EGT), honey
bee
Melittin (Invitrogen, Carlsbad, CA), or baculovirus gp67 (PharMingen, San
Diego, CA)
can be used in constructs to replace the native zalphal 1 Ligand secretory
signal
sequence. In addition, transfer vectors can include an in-frame fusion with
DNA
encoding an epitope tag at the C- or N-terminus of the expressed zalphal 1
Ligand
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
polypeptide, for example, a Glu-Glu epitope tag (Grussenmeyer, T. et al.,
Proc. Natl.
Acad. Sci. 82:7952-4, 1985). Using techniques known in the art, a transfer
vector
containing zalphal 1 Ligand is transformed into E. Coli, and screened for
bacmids
which contain an interrupted lacZ gene indicative of recombinant baculovirus.
The
5 bacmid DNA containing the recombinant baculovirus genome is isolated, using
common techniques, and used to transfect Spodoptera frugiperda cells, e.g. Sf9
cells.
Recombinant virus that expresses zalphal 1 Ligand is subsequently produced.
Recombinant viral stocks are made by methods commonly used the art.
The recombinant virus is used to infect host cells, typically a cell line
o derived from the fall armyworm, Spodoptera frugiperda. See, in general,
Glick and
Pasternak, Molecular Biotechnology: Principles and Applications of Recombinant
DNA, ASM Press, Washington, D.C., 1994. Another suitable cell line is the High
FiveOTM cell line (Invitrogen) derived from Trichoplusia ni (U.S. Patent
#5.300,435)..
Fungal cells, including yeast cells, can also be used within the present
is invention. Yeast species of particular interest in this regard include
Saccharomyces
cerevisiae, Pichia pastoris, and Pichia methanolica. Methods for transforming
S.
cerevisiae cells with exogenous DNA and producing recombinant polypeptides
therefrom are disclosed by, for example, Kawasaki, U.S. Patent No. 4,599,311;
Kawasaki et al., U.S. Patent No. 4,931,373; Brake, U.S. Patent No. 4,870,008:
Welch et
20 al., U.S. Patent No. 5,037,743; and Murray et al., U.S. Patent No.
4.845,075.
Transformed cells are selected by phenotype determined by the selectable
marker,
commonly drug resistance or the ability to grow in the absence of a particular
nutrient
(e.g., leucine). A preferred vector system for use in Saccharomyces cerevisiae
is the
POT] vector system disclosed by Kawasaki et al. (U.S. Patent No. 4,931.373),
which
25 allows transformed cells to be selected by growth in glucose-containing
media.
Suitable promoters and terminators for use in yeast include those from
dycolytic
enzyme genes (see, e.g., Kawasaki, U.S. Patent No. 4,599,311; Kingsman et al.,
U.S.
Patent No. 4,615,974; and Bitter, U.S. Patent No. 4.977,092) and alcohol
dehydrogenase genes. See also U.S. Patents Nos. 4,990,446; 5.063,154;
5,139,936 and
30 4,661,454. Transformation systems for other yeasts, including Hansenula
polymorpha,
Schizosaccharomyces pombe, Kluyveromyces lactis, Kluyveromyces fragilis,
Ustilago
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
46
maydis, Pichia pastoris, Pichia methanolica, Pichia guillermondii and Candida
maltosa are known in the art. See, for example, Gleeson et al., J. Gen.
Microbiol.
132:3459-65, 1986 and Cregg, U.S. Patent No. 4,882,279. Aspergillus cells may
be
utilized according to the methods of McKnight et al., U.S. Patent No.
4,935,349.
Methods for transforming Acremonium chrysogenum are disclosed by Sumino et
al.,
U.S. Patent No. 5,162,228. Methods for transforming Neurospora are disclosed
by
Lambowitz, U.S. Patent No. 4,486,533.
The use of Pichia methanolica as host for the production of recombinant
proteins is disclosed in WIPO Publications WO 97/17450, WO 97/17451, WO
o 98/02536, and WO 98/02565. DNA molecules for use in transforming P.
methanolica
will commonly be prepared as double-stranded, circular plasmids, which are
preferably
linearized prior to transformation. For polypeptide production in P.
methanolica, it is
preferred that the promoter and terminator in the plasmid be that of a P.
methanolica
gene, such as a P. methanolica alcohol utilization gene (AUGI or AUG2). Other
useful
promoters include those of the dihydroxyacetone synthase (DHAS), formate
dehydrogenase (FMD), and catalase (CAT) genes. To facilitate integration of
the DNA
into the host chromosome, it is preferred to have the entire expression
segment of the
plasmid flanked at both ends by host DNA sequences. A preferred selectable
marker
for use in Pichia methanolica is a P. methanolica ADE2 gene, which encodes
phosphoribosy1-5-aminoimidazole carboxylase (AIRC; EC 4.1.1.21), which allows
ade2 host cells to grow in the absence of adenine. For large-scale, industrial
processes
where it is desirable to minimize the use of methanol, it is preferred to use
host cells in
which both methanol utilization genes (AUG1 and AUG2) are deleted. For
production
of secreted proteins, host cells deficient in vacuolar protease genes (PEP4
and PRB1)
are preferred. Electroporation is used to facilitate the introduction of a
plasmid
containing DNA encodinL, a polypeptide of interest into P. methanolica cells.
It is
preferred to transform P. methanolica cells by electroporation using an
exponentially
decaying, pulsed electric field having a field strength of from 2.5 to 4.5
kV/cm,
preferably about 3.75 kV/cm, and a time constant (Q) of from 1 to 40
milliseconds,
most preferably about 20 milliseconds.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
47
Prokaryotic host cells, including strains of the bacteria Escherichia coli,
Bacillus and other genera are also useful host cells within the present
invention.
Techniques for transforming these hosts and expressing foreign DNA sequences
cloned
therein are well known in the art (see, e.g., Sambrook et al., ibid.). When
expressing a
required. The growth medium will generally select for cells containing the
exogenously added DNA by, for example, drug selection or deficiency in an
essential
nutrient which is complemented by the selectable marker carried on the
expression
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
48
It is preferred to purify the polypeptides of the present invention to
80% purity, more preferably to ?_90% purity, even more preferably 295% purity,
and
particularly preferred is a pharmaceutically pure state, that is greater than
99.9% pure
with respect to contaminating macromolecules, particularly other proteins and
nucleic
s acids, and free of infectious and pyrogenic agents. Preferably, a
purified polypeptide is
substantially free of other polypeptides, particularly other polypeptides of
animal
origin.
Expressed recombinant zalphal 1 Ligand polypeptides (or chimeric
zalphal 1 Ligand polypeptides) can be purified using fractionation and/or
conventional
o purification methods and media. Ammonium sulfate precipitation and acid or
chaotrope extraction may be used for fractionation of samples. Exemplary
purification
steps may include hydroxyapatite, size exclusion, FPLC and reverse-phase high
performance liquid chromatography.
Suitable chromatographic media include
derivatized dextrans, agarose, cellulose, polyacrylamide, specialty silicas,
and the like.
15 PEI, DEAE, QAE and Q derivatives are preferred. Exemplary
chromatographic media
include those media derivatized with phenyl, butyl. or octyl groups, such as
Phenyl-
Sepharose FF (Pharmacia), Toyopearl butyl 650 (Toso Haas, Montgomeryville,
PA),
Octyl-Sepharose (Pharmacia) and the like; or polyacrylic resins, such as
Amberchrom
CG 71 (Toso Haas) and the like. Suitable solid supports include glass beads,
silica-
20 based resins, cellulosic resins, agarose beads, cross-linked agarose
beads, polystyrene
beads, cross-linked polyacrylamide resins and the like that are insoluble
under the
conditions in which they are to be used. These supports may be modified with
reactive
groups that allow attachment of proteins by amino groups, carboxyl groups,
sulfhydryl
groups, hydroxyl groups and/or carbohydrate moieties. Examples of coupling
25 chemistries include cyanogen bromide activation. N-hydroxysuccinimide
activation,
epoxide activation, sulthydryl activation, hydrazide activation, and carboxyl
and amino
derivatives for carbodiimide coupling chemistries. These and other solid media
are
well known and widely used in the art, and are available from commercial
suppliers.
Methods for binding receptor polypeptides to support media are well known in
the art.
30 Selection of a particular method is a matter of routine design and is
determined in part
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
49
by the properties of the chosen support. See, for example, Affinity
Chromatography:
Principles & Methods, Pharmacia LKB Biotechnology, Uppsala, Sweden, 1988.
The polypeptides of the present invention can be isolated by exploitation
of their physical or biochemical properties. For example, immobilized metal
ion
adsorption (IMAC) chromatography can be used to purify histidine-rich
proteins,
including those comprising polyhistidine tags. Briefly, a gel is first charged
with
divalent metal ions to form a chelate (Sulkowski, Trends in Biochem. 3:1-7,
1985).
Histidine-rich proteins will be adsorbed to this matrix with differing
affinities,
depending upon the metal ion used, and will be eluted by competitive elution,
lowering
o the pH, or use of strong chelating agents. Other methods of purification
include
purification of glycosylated proteins by lectin affinity chromatography and
ion
exchange chromatography (Methods in Enzymol., Vol. 182, "Guide to Protein
Purification", M. Deutscher, (ed.), Acad. Press, San Diego, 1990, pp.529-39)
and use of
the soluble zalphal 1 receptor. Within additional embodiments of the
invention, a
fusion of the polypeptide of interest and an affinity tag (e.g., maltose-
binding protein,
an immunoglobulin domain) may be constructed to facilitate purification.
Moreover, using methods described in the art, polypeptide fusions, or
hybrid zalphal 1 Ligand proteins, are constructed using regions or domains of
the
inventive zalphal 1 Ligand in combination with those of other human cytokine
family
proteins (e.g. interleukins or GM-CSF), or heterologous proteins (Sambrook et
al.,
ibid., Altschul et al., ibid., Picard, Cur. Opin. Biology, 5:511-5, 1994, and
references
therein). These methods allow the determination of the biological importance
of larger
domains or regions in a polypeptide of interest. Such hybrids may alter
reaction
kinetics, binding, constrict or expand the substrate specificity, or alter
tissue and
cellular localization of a polypeptide, and can be applied to polypeptides of
unknown
structure.
Fusion proteins can be prepared by methods known to those skilled in
the art by preparing each component of the fusion protein and chemically
conjugating
them. Alternatively, a polynucleotide encoding both components of the fusion
protein
in the proper reading frame can be generated using known techniques and
expressed by
the methods described herein. For example, part or all of a helix conferring a
biological
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
function may be swapped between zalphall Ligand of the present invention with
the
functionally equivalent helices from another family member, such as IL-15, IL-
2, IL-4
or GM-CSF. Such components include, but are not limited to, the secretory
signal
sequence; helices A, B, C, D; loops A/B, B/C, C/D; of four-helical-bundle
cytokines.
5 Such
fusion proteins would be expected to have a biological functional profile that
is
the same or similar to polypeptides of the present invention or other known
four-
helical-bundle cytokine family proteins, depending on the fusion constructed.
Moreover, such fusion proteins may exhibit other properties as disclosed
herein.
Standard molecular biological and cloning techniques can be used to
10 swap
the equivalent domains between the zalphal 1 Ligand polypeptide and those
polypeptides to which they are fused. Generally, a DNA segment that encodes a
domain of interest, e.g., zalphal 1 Ligand helices A through D, or other
domain
described herein, is operably linked in frame to at least one other DNA
segment
encoding an additional polypeptide (for instance a domain or region from
another
15
cytokine, such as the IL-2, or the like), and inserted into an appropriate
expression
vector, as described herein. Generally DNA constructs are made such that the
several
DNA segments that encode the corresponding regions of a polypeptide are
operably
linked in frame to make a single construct that encodes the entire fusion
protein, or a
functional portion thereof. For example, a DNA construct would encode from N-
20
terminus to C-terminus a fusion protein comprising a signal polypeptide
followed by a
mature four helical bundle cytokine fusion protein containing helix A,
followed by
helix B, followed by helix C, followed by helix D. Such fusion proteins can be
expressed, isolated, and assayed for activity as described herein.
Zalpha 1 1 Ligand polypeptides or fragments thereof may also be
25
prepared through chemical synthesis. zalphal 1 Ligand polypeptides may be
monomers
or multimers; glycosylated or non-glycosylated; pegylated or non-pegylated;
and may
or may not include an initial methionine amino acid residue. For example, the
polypeptides can be prepared by solid phase peptide synthesis. for example as
described
by Merrifield, J. Am. Chem. Soc. 85:2149, 1963.
30 The
activity of molecules of the present invention can be measured using
a variety of assays that measure proliferation of and/or binding to cells
expressing the
CA 02366921 2007-12-17
51
zalphall receptor. Of particular interest are changes in zalphal 1 Ligand-
dependent
cells. Suitable cell lines to be engineered to be zalphall Ligand-dependent
include the
IL-3-dependent BaF3 cell line (Palacios and Steinmetz, Cell 41: 727-734, 1985;
Mathey-Prevot et al., Mol. Cell. Biol. 6:4133135, 1986), FDC-P 1 (Hapel et
al.,
Blood 64: 786-790, 1984), and M07e (Kiss et al., Leukemia 7: 235-240, 1993).
Growth factor-dependent cell lines can be established according to published
methods
(e.g. Greenberger et al., Leukemia Res. 8: 363-375, 1984; Dexter et al., in
Baum et al.
Eds., Experimental Hematology Today, 8th Ann. Mtg. Int. Soc. Exp: Hematol.
1979,
145-156, 1980).
=
Proteins of the present invention are useful for stimulating proliferation,
activation, differentiation and/or induction or inhibition of specialized
cell, function of
cells of the involved homeostasis of the hematopoiesis and immune function. In
particular, zalphal 1 Ligand polypeptides are useful for stimulating
proliferation,
activation, differentiation, induction or inhibition of specialized cell
functions of cells
of the hematopoietic lineages, including, but not limited to, T cells, B
cells, NK cells,
dendritic cells, monocytes, and macrophages, as well as epithelial cells.
Proliferation
= = . . . .
and/or differentiation of hematopoietic cells can be measured in vitro using
cultured
cells or in vivo by administering molecules of the claimed invention to the
appropriate
animal model. Assays measuring cell proliferation or differentiation are Well
known in
the art. For example, assays measuring proliferation include such assays as
chemosensitivity to neutral red dye (Cavanaugh et al., Investigational New
Drugs
8:347-354, 1990)
incorporation of radiolabelled
nucleotides (Cook et al., Analytical Biochem. 179:1-7, 1989)
incorporation of 5-bromo-2'-deoxyuridine (BrdU) in the DNA of
proliferating cells (Porstmatm et al., J. Immunol. Methods 82:169-179, 1985)
and use of tetrazolium salts (Mosmann, J. Immunol.
Methods 65:55-63, 1983; Alley et al., Cancer Res. 48:589-601, 1988; Marshall
et al.,
Growth Reg. 5:69-84, 1995; and Scudiero et al., Cancer Res. 48:4827-4833,
1988)
Assays measuring differentiation include, for
example, measuring cell-surface markers associated with stage-specific
expression of a
tissue, enzymatic activity, functional activity or morphological changes
(Watt, FASEB,
CA 02366921 2007-12-17
52
5:281-284, 1991; Francis, Differentiation 5763-75, -1994; Raes, Adv. Anim.
Cell Biol.
Technol. Bioprocesses, 161-171, 1989)
The molecules of the present invention can be assayed in vivo using viral
delivery systems. Exemplary viruses for this purpose include adenovirus,
herpesvirus,
-4
s
retroviruses, vaccinia virus, and adeno-associated:virus (AAV). Adenovirus, a
double-
stranded DNA virus, is currently the best studied gene transfer vector for
delivery of
heterologous nucleic acid (for review, see T.C. Becker et al., Meth. Cell
Biol. 43:161-
89, 1994; and J.T. Douglas and D.T. Curie!, Science & Medicine 4:44-53, 1997).
As a ligand, the activity of zalphal 1 Ligand polypeptide can be
o measured by a silicon-based biosensor microphysiometer which measures the
extracellular acidification rate or proton excretion associated with receptor
binding and
subsequent physiologic cellular responses. An exemplary device is the
CytosensorTM
Microphysiometer manufactured by Molecular Devices, Sunnyvale, CA. A variety
of
cellular responses, such as cell proliferation, ion transport, energy
production,
is inflammatory response, regulatory and receptor activation, and the like,
can be
measured by this method. See, for example, McConnell, H.M. et al., Science
257:1906-1912, 1992; Pitchford, S. et al., Meth. Enzymol. 228:84-108, 1997;
Arimilli,
S. et al., J. Immunol. Meth. 212:49-59, 1998; Van Liefde, I. et al., Eur. J.
Pharmacol.
346-87-95, 1998.
20
Moreover, zalphal 1 Ligand can be used to identify cells, tissues, or cell
lines which respond to a zalphal 1 Ligand-stimulated pathway. The
microphysiometer,
described above, can be used to rapidly identify ligand-responsive cells, such
as cells
responsive to zalphal 1 Ligand of the present invention. Cells can be cultured
in the
presence or absence of zalphal 1 Ligand polypeptide. Those cells which elicit
a
25
measurable change in extracellular acidification in the presence of zalphal 1
Ligand are
responsive to zalphal 1 Ligand. Such cells or cell lines, can be used to
identify
antagonists and agonists of zalphal 1 Ligand polypeptide as described aboVe.
,
In view of the tissue distribution observed for zalphall receptor agonists
(including the natural zalphal 1 Ligand/ substrate/ cofactor/ etc.) and/or
antagonists
30 have enormous potential in both in vitro and in vivo applications.
Compounds
identified as zalphal 1 Ligand agonists are useful for expansion,
proliferation,
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
53
activation, differentiation, and/or induction or inhibition of specialized
cell functions of
cells involved in homeostasis of hematopoiesis and immune function. For
example,
zalphal 1 Ligand and agonist compounds are useful as components of defined
cell
culture media, and may be used alone or in combination with other cytokines
and
hormones to replace serum that is commonly used in cell culture. Agonists are
thus
useful in specifically promoting the growth and/or development of T-cells, B-
cells, NK
cells, cytotoxic lymphocytes, and other cells of the lymphoid and myeloid
lineages in
culture.
Antagonists are also useful as research reagents for characterizing sites
o of ligand-receptor interaction.
Antagonists are useful to inhibit expansion,
proliferation, activation, and/or differentiation of cells involved in
regulating
hematopoiesis. Inhibitors of zalphal 1 Ligand activity (zalphal 1 Ligand
antagonists)
include anti-zalphal 1 Ligand antibodies and soluble zalphal 1 Ligand
receptors, as well
as other peptidic and non-peptidic agents (including ribozymes).
zalphal 1 Ligand can also be used to identify inhibitors (antagonists) of
its activity. Test compounds are added to the assays disclosed herein to
identify
compounds that inhibit the activity of zalphal 1 Ligand. In addition to those
assays
disclosed herein, samples can be tested for inhibition of zalphall Ligand
activity within
a variety of assays designed to measure receptor binding, the
stimulation/inhibition of
zalphal 1 Ligand-dependent cellular responses or proliferation of zalphal 1
receptor-
expressing cells.
A zalphal 1 Ligand polypeptide can be expressed as a fusion with an
immunoglobulin heavy chain constant region, typically an Fc fragment, which
contains
two constant region domains and lacks the variable region. Methods for
preparing such
fusions are disclosed in U.S. Patents Nos. 5,155,027 and 5,567,584. Such
fusions are
typically secreted as multimeric molecules wherein the Fe portions are
disulfide bonded
to each other and two non-Ig polypeptides are arrayed in closed proximity to
each
other. Fusions of this type can be used for example, for dimerization.
increasing
stability and in vivo half-life, to affinity purify ligand, as in vitro assay
tool or
antagonist. For use in assays, the chimeras are bound to a support via the Fe
region
and used in an ELISA format.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
54
A zalphal 1 Ligand-binding polypeptide can also be used for purification
of ligand. The polypeptide is immobilized on a solid support, such as beads of
agarose,
cross-linked agarose, glass, cellulosic resins, silica-based resins,
polystyrene, cross-
linked polyacrylamide, or like materials that are stable under the conditions
of use.
Methods for linking polypeptides to solid supports are known in the art, and
include
amine chemistry, cyanogen bromide activation, N-hydroxysuccinimide activation,
epoxide activation, sulfhydryl activation, and hydrazide activation. The
resulting
medium will generally be configured in the form of a column, and fluids
containing
ligand are passed through the column one or more times to allow ligand to bind
to the
o receptor polypeptide. The ligand is then eluted using changes in salt
concentration,
chaotropic agents (guanidine HCI), or pH to disrupt ligand-receptor binding.
An assay system that uses a ligand-binding receptor (or an antibody, one
member of a complement/ anti-complement pair) or a binding fragment thereof,
and a
commercially available biosensor instrument (BIAcore, Pharmacia Biosensor,
Piscataway, NJ) may be advantageously employed. Such receptor, antibody,
member
of a complement/anti-complement pair or fragment is immobilized onto the
surface of a
receptor chip. Use of this instrument is disclosed by Karlsson, J. Immunol.
Methods
145:229-40, 1991 and Cunningham and Wells, J. Mol. Biol. 234:554-63, 1993. A
receptor, antibody, member or fragment is covalently attached, using amine or
sulfhydryl chemistry, to dextran fibers that are attached to gold film within
the flow
cell. A test sample is passed through the cell. If a ligand, epitope, or
opposite member
of the complement/anti-complement pair is present in the sample, it will bind
to the
immobilized receptor, antibody or member, respectively, causing a change in
the
refractive index of the medium, which is detected as a change in surface
plasmon
resonance of the gold film. This system allows the determination of on- and
off-rates,
from which binding affinity can be calculated, and assessment of stoichiometry
of
binding. Alternatively, ligand/receptor binding can be analyzed using
SELDI(TM)
technology (Ciphergen, Inc., Palo Alto, CA).
Ligand-binding receptor polypeptides can also be used within other
assay systems known in the art. Such systems include Scatchard analysis for
determination of binding affinity (see Scatchard, Ann. NY Acad. Sci. 51: 660-
72, 1949)
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
and calorimetric assays (Cunningham et al., Science 253:545-48, 1991;
Cunningham et
al., Science 245:821-25, 1991).
Zalphal 1 Ligand polypeptides can also be used to prepare antibodies
that bind to zalphal 1 Ligand epitopes, peptides or polypeptides. The zalphal
1 Ligand
5 polypeptide or a fragment thereof serves as an antigen (immunogen) to
inoculate an
animal and elicit an immune response. One of skill in the art would recognize
that
antigenic, epitope-bearing polypeptides contain a sequence of at least 6,
preferably at
least 9, and more preferably at least 15 to about 30 contiguous amino acid
residues of a
zalphal 1 Ligand polypeptide (e.g., SEQ ID NO:2). Polypeptides comprising a
larger
10 portion of a zalphall Ligand polypeptide, i.e., from 30 to 100 residues
up to the entire
length of the amino acid sequence are included. Antigens or immunogenic
epitopes can
also include attached tags, adjuvants and carriers, as described herein.
Suitable
antigens include the zalphal 1 Ligand polypeptide encoded by SEQ ID NO:2 from
amino acid number 32 to amino acid number 162, or a contiguous 9 to 131 amino
acid
15 fragment thereof. Other suitable antigens include, the full length and the
mature
zalphal 1 Ligand, helices A-D, and individual or multiple helices A, B, C, and
D, of the
zalphal 1 Ligand four-helical-bundle structure, as described herein. Preferred
peptides
to use as antigens are hydrophilic peptides such as those predicted by one of
skill in the
art from a hydrophobicity plot, as described herein, for example, amino acid
residues
20 114-119, 101-105, 126-131, 113-118, and 158-162 of SEQ ID NO: 2.
Antibodies from an immune response generated by inoculation of an
animal with these antigens can be isolated and purified as described herein.
Methods
for preparing and isolating polyclonal and monoclonal antibodies are well
known in the
art. See, for example. Current Protocols in Immunology, Cooligan, et al.
(eds.),
25 National Institutes of Health, John Wiley and Sons. Inc., 1995; Sambrook
et al.,
Molecular Cloning: A Laboratory Manual, Second Edition, Cold Spring Harbor,
NY,
1989; and Hurrell, J. G. R., Ed., Monoclonal Hybridoma Antibodies: Techniques
and
Applications, CRC Press, Inc., Boca Raton, FL, 1982.
As would be evident to one of ordinary skill in the art, polyclonal
30 antibodies can be generated from inoculating a variety of warm-blooded
animals such
as horses, cows, goats. sheep, dogs, chickens. rabbits. mice, and rats with a
zalphal 1
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
56
Ligand polypeptide or a fragment thereof The immunogenicity of a zalphal 1
Ligand
polypeptide may be increased through the use of an adjuvant, such as alum
(aluminum
hydroxide) or Freund's complete or incomplete adjuvant. Polypeptides useful
for
immunization also include fusion polypeptides, such as fusions of zalphal 1
Ligand or a
s portion
thereof with an immunoglobulin polypeptide or with maltose binding protein.
The polypeptide immunogen may be a full-length molecule or a portion thereof.
If the
polypeptide portion is "hapten-like", such portion may be advantageously
joined or
linked to a macromolecular carrier (such as keyhole limpet hemocyanin (KLH),
bovine
serum albumin (BSA) or tetanus toxoid) for immunization.
As used herein, the term "antibodies" includes polyclonal antibodies,
affinity-purified polyclonal antibodies, monoclonal antibodies, and antigen-
binding
fragments, such as F(ab')2 and Fab proteolytic fragments. Genetically
engineered intact
antibodies or fragments, such as chimeric antibodies, Fv fragments, single
chain
antibodies and the like, as well as synthetic antigen-binding peptides and
polypeptides,
are also included. Non-human antibodies may be humanized by grafting non-human
CDRs onto human framework and constant regions, or by incorporating the entire
non-
human variable domains (optionally "cloaking" them with a human-like surface
by
replacement of exposed residues, wherein the result is a "veneered" antibody).
In some
instances, humanized antibodies may retain non-human residues within the human
variable region framework domains to enhance proper binding characteristics.
Through
humanizing antibodies, biological half-life may be increased, and the
potential for
adverse immune reactions upon administration to humans is reduced. Moreover,
human antibodies can be produced in transgenic, non-human animals that have
been
engineered to contain human immunoglobulin genes as disclosed in WIPO
Publication
WO 98/24893. It is preferred that the endogenous immunoglobulin genes in these
animals be inactivated or eliminated, such as by homologous recombination.
Antibodies are considered to be specifically binding if: 1) they exhibit a
threshold level of binding activity, and 2) they do not significantly cross-
react with
related polypeptide molecules. A threshold level of binding is determined if
anti-
zalphal 1 Ligand antibodies herein bind to a zalphal 1 Ligand polypeptide,
peptide or
epitope with an affinity at least 10-fold greater than the binding affinity to
control (non-
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
57
zalphal 1 Ligand) polypeptide. It is preferred that the antibodies exhibit a
binding
affinity (Ka) of 106 M-1 or greater, preferably 107 M-1 or greater, more
preferably 108
M-1 or greater, and most preferably 109 M-1 or greater. The binding affinity
of an
antibody can be readily determined by one of ordinary skill in the art, for
example, by
Scatchard analysis (Scatchard, G., Ann. NY Acad. Sci. 51: 660-672, 1949).
Whether anti-zalphall Ligand antibodies do not significantly cross-react
with related polypeptide molecules is shown, for example, by the antibody
detecting
zalphal 1 Ligand polypeptide but not known related polypeptides using a
standard
Western blot analysis (Ausubel et al., ibid.). Examples of known related
polypeptides
o are those disclosed in the prior art, such as known orthologs, and
paralogs, and similar
known members of a protein family. Screening can also be done using non-human
zalphal 1 Ligand, and zalphal 1 Ligand mutant polypeptides. Moreover,
antibodies can
be "screened against" known related polypeptides, to isolate a population that
specifically binds to the zalphal 1 Ligand polypeptides. For example,
antibodies raised
to zalphal 1 Ligand are adsorbed to related polypeptides adhered to insoluble
matrix;
antibodies specific to zalphal 1 Ligand will flow through the matrix under the
proper
buffer conditions. Screening allows isolation of polyclonal and monoclonal
antibodies
non-crossreactive to known closely related polypeptides (Antibodies: A
Laboratory
Manual, Harlow and Lane (eds.), Cold Spring Harbor Laboratory Press, 1988;
Current
Protocols in Immunology, Cooligan, et al. (eds.), National Institutes of
Health. John
Wiley and Sons, Inc., 1995). Screening and isolation of specific antibodies is
well
known in the art. See, Fundamental Immunology, Paul (eds.), Raven Press, 1993;
Getzoff et al., Adv. in Immunol. 43: 1-98, 1988; Monoclonal Antibodies:
Principles and
Practice, Goding, J.W. (eds.), Academic Press Ltd., 1996; Benjamin et al.,
Ann. Rev.
Immunol. 2: 67-101, 1984. Specifically binding anti-zalphal 1 Ligand
antibodies can
be detected by a number of methods in the art, and disclosed below.
A variety of assays known to those skilled in the art can be utilized to
detect antibodies which bind to zalphal 1 Ligand proteins or polypeptides.
Exemplary
assays are described in detail in Antibodies: A Laboratory Manual, Harlow and
Lane
(Eds.), Cold Spring Harbor Laboratory Press. 1988. Representative examples of
such
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
58
assays include: concurrent immunoelectrophoresis, radioimmunoassay,
radioimmuno-
precipitation, enzyme-linked immunosorbent assay (ELISA), dot blot or Western
blot
assay, inhibition or competition assay, and sandwich assay. In addition,
antibodies can
be screened for binding to wild-type versus mutant zalphal 1 Ligand protein or
polypeptide.
Antibodies to zalphal 1 Ligand may be used for tagging cells that
express zalphal 1 Ligand; for isolating zalphall Ligand by affinity
purification; for
diagnostic assays for determining circulating levels of zalphal 1 Ligand
polypeptides;
for detecting or quantitating soluble zalphal 1 Ligand as a marker of
underlying
o pathology or disease; in analytical methods employing FACS; for screening
expression
libraries; for generating anti-idiotypic antibodies; and as neutralizing
antibodies or as
antagonists to block zalphal 1 Ligand activity in vitro and in vivo. Suitable
direct tags
or labels include radionuclides, enzymes, substrates, cofactors, inhibitors,
fluorescent
markers, chemiluminescent markers, magnetic particles and the like; indirect
tags or
labels may feature use of biotin-avidin or other complement/anti-complement
pairs as
intermediates. Antibodies herein may also be directly or indirectly conjugated
to drugs,
toxins, radionuclides and the like, and these conjugates used for in vivo
diagnostic or
therapeutic applications. Moreover, antibodies to zalphall Ligand or fragments
thereof
may be used in vitro to detect denatured zalphal 1 Ligand or fragments thereof
in
assays, for example, Western Blots or other assays known in the art.
Suitable detectable molecules may be directly or indirectly attached to
the polypeptide or antibody, and include radionuclides, enzymes, substrates,
cofactors,
inhibitors, fluorescent markers, chemiluminescent markers, magnetic particles
and the
like. Suitable cytotoxic molecules may be directly or indirectly attached to
the
polypeptide or antibody, and include bacterial or plant toxins (for instance,
diphtheria,
toxin, saporin, Pseudomonas exotoxin, ricin, abrin and the like), as well as
therapeutic
radionuclides, such as iodine-131, rhenium-188 or yttrium-90 (either directly
attached
to the polypeptide or antibody, or indirectly attached through means of a
chelating
moiety, for instance). Polypeptides or antibodies may also be conjugated to
cytotoxic
drugs, such as adriamycin. For indirect attachment of a detectable or
cytotoxic
molecule, the detectable or cytotoxic molecule can be conjugated with a member
of a
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
59
complementary/ anticomplementary pair, where the other member is bound to the
polypeptide or antibody portion. For these purposes, biotin/streptavidin is an
exemplary complementary/ anticomplementary pair.
Binding polypeptides can also act as zalphal 1 Ligand "antagonists" to
block zalphal 1 Ligand binding and signal transduction in vitro and in vivo.
These anti-
zalphal 1 Ligand binding polypeptides would be useful for inhibiting zalphal 1
Ligand
activity or protein-binding.
Polypeptide-toxin fusion proteins or antibody-toxin fusion proteins can
be used for targeted cell or tissue inhibition or ablation (for instance, to
treat cancer
cells or tissues). Alternatively, if the polypeptide has multiple functional
domains (i.e.,
an activation domain or a receptor binding domain, plus a targeting domain), a
fusion
protein including only the targeting domain may be suitable for directing a
detectable
molecule, a cytotoxic molecule or a complementary molecule to a cell or tissue
type of
interest. In instances where the domain only fusion protein includes a
complementary
molecule, the anti-complementary molecule can be conjugated to a detectable or
cytotoxic molecule. Such domain-complementary molecule fusion proteins thus
represent a generic targeting vehicle for cell/tissue-specific delivery of
generic anti-
complementary-detectable/ cytotoxic molecule conjugates.
Zalphal 1 Ligand cytokine fusion proteins or antibody-cytokine fusion
proteins can be used for enhancing in vivo killing of target tissues (for
example, blood
and bone marrow cancers), if the zalphal1 Ligand polypeptide or anti-zalphal 1
Ligand
antibody targets the hyperproliferative blood or bone marrow cell (See,
generally,
Hornick et al., Blood 89:4437-47, 1997). The described fusion proteins enable
targeting of a cytokine to a desired site of action, thereby providing an
elevated local
concentration of cytokine. Suitable zalphal1 Ligand polypeptides or anti-
zalphal 1
Ligand antibodies target an undesirable cell or tissue (i.e.. a tumor or a
leukemia), and
the fused cytokine mediated improved target cell lysis by effector cells.
Suitable
cytokines for this purpose include interleukin 2 and granulocyte-macrophage
colony-
stimulating factor (GM-CSF), for instance.
Differentiation is a progressive and dynamic process, beginning with
pluripotent stem cells and ending with terminally differentiated cells.
Pluripotent stem
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
cells that can regenerate without commitment to a lineage express a set of
differentiation markers that are lost when commitment to a cell lineage is
made.
Progenitor cells express a set of differentiation markers that may or may not
continue to
be expressed as the cells progress down the cell lineage pathway toward
maturation.
s Differentiation markers that are expressed exclusively by mature cells are
usually
functional properties such as cell products, enzymes to produce cell products,
and
receptors. The stage of a cell population's differentiation is monitored by
identification
of markers present in the cell population.
There is evidence to suggest that factors that stimulate specific cell types
io down a pathway towards terminal differentiation or dedifferentiation
affect the entire
cell population originating from a common precursor or stem cell. Thus, the
present
invention includes stimulating or inhibiting the proliferation of lymphoid
cells,
hematopoietic cells and epithelial cells.
Zalphal 1 Ligand was isolated from tissue known to have important
is immunological function and which contain cells that play a role in the
immune system.
Zalphal 1 Ligand is expressed in CD3+ selected, activated peripheral blood
cells, and it
has been shown that zalphal 1 Ligand expression increases after T cell
activation.
Moreover, results of experiments described in the Examples section herein
demonstrate
that polypeptides of the present invention have an effect on the
growth/expansion
20 and/or differentiated state of NK cells or NK progenitors. Additional
evidence
demonstrates that zalphal 1 Ligand affects proliferation and/or
differentiation of T cells
and B cells in vivo. Factors that both stimulate proliferation of
hematopoietic
progenitors and activate mature cells are generally known. NK cells are
responsive to
IL-2 alone, but proliferation and activation generally require additional
growth factors.
25 For example, it has been shown that IL-7 and Steel Factor (c-kit ligand)
were required
for colony formation of NK progenitors. IL-15 + IL-2 in combination with IL-7
and
Steel Factor was more effective (MrOzek et al., Blood 87:2632-2640, 1996).
However,
unidentified cytokines may be necessary for proliferation of specific subsets
of NK
cells and/or NK progenitors (Robertson et. al., Blood 76:2451-2438, 1990). A
30 composition comprising zalphal 1 Ligand and IL-15 stimulates NK
progenitors and NK
CA 02366921 2007-12-17
61
cells, with evidence that this composition is more' Potent than previously
described
factors and combinations of factors.
Assays measuring differentiation include, for example, measuring cell
markers associated with stage-specific expression of a tissue, enzymatic
activity,
4
functional activity or morphological changes (Watt, FASEB, 5:281-284, 1991;
Francis,
Differentiation 57:63-75, 1994; Raes, Adv. Anim. Cell Biol. Technol.
Bioprocesses,
161-171, 1989) Alternatively, zalphal 1
Ligand
polypeptide itself can serve as an additional cell-surface or secreted marker
associated
with stage-specific expression of a tissue. As such, direct measurement of
zalphall
1.0 Ligand polypeptide, or its loss of expression in a tissue as it
differentiates, can serve as
a marker for differentiation of tissues.
Similarly, direct measurement of zalphal 1 Ligand polypeptide, or its
loss of expression in a tissue can be determined in a tissue or in cells as
they undergo
tumor progression. Increases in invasiveness and motility of cells, or the
gain or loss of
expression of zalphal 1 Ligand in a pre-cancerous or cancerous condition, in
comparison to normal tissue, can serve as a diagnostic for transformation,
invasion and
metastasis in tumor progression. As such, knowledge of a tumor's stage of
progression =
or metastasis will aid the physician in choosing the most proper therapy, or
aggressiveness of treatment, for a given individual cancer patient. ' Methods
of
measuring gain and loss of expression (of either mRNA or protein) are Well
known in
the art and described herein and can be applied to zalphall Ligand expression.
For
example, appearance or disappearance of polypeptides that regulate cell
motility can be
used to aid diagnosis and prognosis of prostate cancer (Banyard, J. and
Zetter, B.R.,
Cancer and Metast. Rev. 17:449-458, 1999). As an effector of cell motility,
zalphal 1
Ligand gain or loss of expression may serve as a diagnostic for lymphoid, B-
cell,
epithelial, hematopoietic and other cancers.
Moreover, the activity and effect of zalphal 1 Ligand , on tumor
progression and metastasis can be measured in vivo. Several syngeneic mouse
models
have been developed to study the influence of polypeptides, compounds or other
treatments on tumor progression. In these models, tumor cells passaged in
culture are
implanted into mice of the same strain as the tumor donor. The cells will
develop into
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
62
tumors having similar characteristics in the recipient mice, and metastasis
will also
occur in some of the models. Appropriate tumor models for our studies include
the
Lewis lung carcinoma (ATCC No. CRL-1642) and B16 melanoma (ATCC No. CRL-
6323), amongst others. These are both commonly used tumor lines, syngeneic to
the
C57BL6/J mouse, that are readily cultured and manipulated in vitro. Tumors
resulting
from implantation of either of these cell lines are capable of metastasis to
the lung in
C57BL6/J mice. The Lewis lung carcinoma model has recently been used in mice
to
identify an inhibitor of angiogenesis (O'Reilly MS, et al. Cell 79: 315-
328,1994).
C57BL6/J mice are treated with an experimental agent either through daily
injection of
o recombinant protein, agonist or antagonist or a one time injection of
recombinant
adenovirus. Three days following this treatment, 105 to 106 cells are
implanted under
the dorsal skin. Alternatively, the cells themselves may be infected with
recombinant
adenovirus, such as one expressing zalphal 1 Ligand, before implantation so
that the
protein is synthesized at the tumor site or intracellularly, rather than
systemically. The
mice normally develop visible tumors within 5 days. The tumors are allowed to
grow
for a period of up to 3 weeks, during which time they may reach a size of 1500
- 1800
mm3 in the control treated group. Tumor size and body weight are carefully
monitored
throughout the experiment. At the time of sacrifice, the tumor is removed and
weighed
along with the lungs and the liver. The lung weight has been shown to
correlate well
with metastatic tumor burden. As an additional measure, lung surface
metastases are
counted. The resected tumor, lungs and liver are prepared for
histopathological
examination, immunohistochemistry, and in situ hybridization, using methods
known
in the art and described herein. The influence of the expressed polypeptide in
question,
e.g., zalphal 1 Ligand, on the ability of the tumor to recruit vasculature and
undergo
metastasis can thus be assessed. In addition, aside from using adenovirus, the
implanted cells can be transiently transfected with zalphal 1 Ligand. Use of
stable
zalphal 1 Ligand transfectants as well as use of induceable promoters to
activate
zalphal 1 Ligand expression in vivo are known in the art and can be used in
this system
to assess zalphal 1 Ligand induction of metastasis.
Moreover, purified zalphal 1
Ligand or zalphal 1 Ligand conditioned media can be directly injected in to
this mouse
model, and hence be used in this system. For general reference see, O'Reilly
MS, et al.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
63
Cell 79:315-328, 1994; and Rusciano D, et al. Murine Models of Liver
Metastasis.
Invasion Metastasis 14:349-361, 1995.
Zalphal 1 Ligand will be useful in treating tumorgenesis, and therefore
would be useful in the treatment of cancer. Zalphal 1 Ligand inhibits IL-4
stimulated
proliferation of anti-IgM stimulated normal B-cells and a similar effect is
observed in
B-cell tumor lines suggesting that there may be therapeutic benefit in
treating patients
with the zalphal 1 Ligand in order to induce the B cell tumor cells into a
less
proliferative state. The ligand could be administered in combination with
other agents
already in use including both conventional chemotherapeutic agents as well as
immune
o
modulators such as interferon alpha. Alpha/beta interferons have been shown to
be
effective in treating some leukemias and animal disease models, and the growth
inhibitory effects of interferon-alpha and zalphall Ligand are additive for at
least one
B-cell tumor-derived cell line.
The present invention provides a method of reducing proliferation of a
neoplastic B or T cells comprising administering to a mammal with a B or T
cell
neoplasm an amount of a composition of zalphal 1 Ligand sufficient to reduce
proliferation of the neoplastic B or T cells. In other embodiments, the
composition can
comprise at least one other cytokine selected from the group consisting of IL-
2, IL-15,
IL-4, GM-CSF, F1t3 ligand or stem cell factor.
In another aspect, the present invention provides a method of reducing
proliferation of a neoplastic B or T cells comprising administering to a
mammal with a
B or T cell neoplasm an amount of a composition of zalphal 1 Ligand antagonist
sufficient to reducing proliferation of the neoplastic B or T cells. In
other
embodiments, the composition can comprise at least one other cytokine selected
from
the group consisting of IL-2, IL-15, IL-4, GM-CSF, F1t3 ligand or stem cell
factor.
Furthermore, the zalphal 1 Ligand antagonist can be a ligand/toxin fusion
protein.
A zalphal 1 Ligand-saporin fusion toxin may be employed against a
similar set of leukemias and lymphomas, extending the range of leukemias that
can be
treated with zalphal 1 Ligand. Fusion toxin mediated activation of the zalphal
1
receptor provides two independent means to inhibit the growth of the target
cells, the
first being identical to the effects seen by the ligand alone, and the second
due to
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
64
delivery of the toxin through receptor internalization. The lymphoid
restricted
expression pattern of the zalphal 1 receptor suggests that the ligand-saporin
conjugate
can be tolerated by patients.
When treatment for malignancies includes allogeneic bone marrow or
stem cell transplantion, zalphal 1 Ligand may be valuable in enhancement of
the graft-
vs-tumor effect. zalphal 1 Ligand stimulates the generation of lytic NK cells
from
marrow progenitors and stimulates the proliferation of T-cells following
activation of
the antigen receptors. Therefore, when patients receive allogenic marrow
transplants,
zalphal 1 Ligand will enhance the generation of anti-tumor responses, with or
without
the infusion of donor lymphocytes.
The tissue distribution of a receptor for a given cytokine offers a strong
indication of the potential sites of action of that cytokine. Northern
analysis of zalphal 1
receptor revealed transcripts in human spleen, thymus, lymph node, bone
marrow, and
peripheral blood leukocytes. Specific cell types were identified as expressing
zalphal 1
receptors, and strong signals were seen in a mixed lymphocyte reaction (MLR)
and in
the Burkitt's lymphoma Raji. The two monocytic cell lines, THP-1 (Tsuchiya et
al., Int.
J. Cancer 26:171-176, 1980) and U937 (Sundstrom et al., Int. J. Cancer 17:565-
577,
1976), were negative.
Zalphal 1 receptor is expressed at relatively high levels in the MLR, in
which peripheral blood mononuclear cells (PBMNC) from two individuals are
mixed,
resulting in mutual activation. Detection of high levels of transcript in the
MLR but not
in resting T or B cell populations suggests that zalpha 11 receptor expression
may be
induced in one or more cell types during activation. Activation of isolated
populations
of T and B cells can be artificially achieved by stimulating cells with PMA
and
ionomycin. When sorted cells were subjected to these activation conditions,
levels of
zalphal 1 receptor transcript increased in both cell types, supporting a role
for this
receptor and zalpha 1 1 Ligand in immune responses, especially in autocrine
and
paracrine T and B cell expansions during activation. Zalpha 1 1 Ligand may
also play a
role in the expansion of more primitive progenitors involved in lymphopoiesis.
zalphal 1 receptor was found to be present at low levels in resting T and
B cells, and was upregulated during activation in both cell types.
Interestingly, the B
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
cells also down-regulate the message more quickly than do T cells, suggesting
that
amplitude of signal and timing of quenching of signal are important for the
appropriate
regulation of B cell responses.
In addition, a large proportion of intestinal lamina propria cells show
5 positive hybridization signals with zalphal 1 receptor. This tissue
consists of a mixed
population of lymphoid cells, including activated CD4+ T cells and activated B
cells.
Immune dysfunction, in particular chronic activation of the mucosal immune
response,
plays an important role in the etiology of Crohn's disease; abnormal response
to and/or
production of proinflammatory cytokines is also a suspected factor (Braegger
et al.,
10 Annals Allergy 72:135-141, 1994; Sartor RB Am. J. Gastroenterol. 92:5S-
11S, 1997.
zalphal 1 Ligand in concert with IL-15 expands NK cells from bone marrow
progenitors and augments NK cell effector function. zalphal 1 Ligand also co-
stimulates mature B cells stimulated with anti-CD40 antibodies, but inhibits B
cell
proliferation to signals through IgM. zalphal 1 Ligand enhances T cell
proliferation in
15 concert with a signal through the T cell receptor, and overexpression in
transgenic mice
leads to lymphopenia and an expansion of monocytes and granulocytes. These
pleiotropic effects of zalphal 1 Ligand suggest that it can provide
therapeutic utility for
a wide range of diseases arising from defects in the immune system, including
(but not
limited to) systemic lupus erythematosus (SLE), rheumatoid arthritis (RA),
multiple
20 sclerosis (MS), myasthenia gravis, and diabetes. It is important to note
that these
diseases are the result of a complex network of immune dysfunction (SLE, for
example,
is the manifestation of defects in both T and B cells), and that immune cells
are
dependent upon interaction with one another to elicit a potent immune
response.
Therefore, zalphal 1 Ligand (or an antagonist of the Ligand) that can be used
to
25 manipulate more than one type of immune cell is an attractive
therapeutic candidate for
intervention at multiple stages of disease.
The polypeptides and proteins of the present invention can also be used
ex vivo, such as in autologous marrow culture. Briefly, bone marrow is removed
from a
patient prior to chemotherapy or organ transplant and treated with zalphal 1
Ligand,
30 optionally in combination with one or more other cytokines. The treated
marrow is
then returned to the patient after chemotherapy to speed the recovery of the
marrow or
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
66
after transplant to suppress graft vs. Host disease. In addition, the proteins
of the
present invention can also be used for the ex vivo expansion of marrow or
peripheral
blood progenitor (PBPC) cells. Prior to treatment, marrow can be stimulated
with stem
cell factor (SCF) to release early progenitor cells into peripheral
circulation. These
s
progenitors can be collected and concentrated from peripheral blood and then
treated in
culture with zalphal 1 Ligand, optionally in combination with one or more
other
cytokines, including but not limited to SCF, IL-2, IL-4, IL-7 or IL-15, to
differentiate
and proliferate into high-density lymphoid cultures, which can then be
returned to the
patient following chemotherapy or transplantation.
The present invention provides a method for expansion of hematopoietic
cells and hematopoietic cell progenitors comprising culturing bone marrow or
peripheral blood cells with a composition comprising an amount of zalphal 1
Ligand
sufficient to produce an increase in the number of lymphoid cells in the bone
marrow or
peripheral blood cells as compared to bone marrow or peripheral blood cells
cultured in
is the
absence of zalphal 1 Ligand. In other embodiments., the hematopoietic cells
and
hematopoietic progenitor cells are lymphoid cells. In another embodiment, the
lymphoid cells are NK cells or cytotoxic T cells. Furthermore, the composition
can
also comprise at least one other cytokine selected from the group consisting
of IL-2, IL-
15, IL-4, GM-CSF, Flt3 ligand and stem cell factor.
Alternatively, zalphal 1 Ligand may activate the immune system which
would be important in boosting immunity to infectious diseases, treating
immunocompromised patients, such as HIV+ patients, or in improving vaccines.
In
particular, zalphal 1 Ligand stimulation or expansion of NK cells, or their
progenitors,
would provide therapeutic value in treatment of viral infection, and as an
anti-
neoplastic factor. NK cells are thought to play a major role in elimination of
metastatic
tumor cells and patients with both metastases and solid tumors have decreased
levels of
NK cell activity (Whiteside et. al., Curr. Top, Microbiol. Immunol. 230:221-
244,
1998). Similarly, zalphal 1 Ligand stimulation of the immune response against
viral
and non-viral pathogenic agents (including bacteria, protozoa, and fungi)
would provide
therapeutic value in treatment of such infections by inhibiting the growth of
such
infections agents. Determining directly or indirectly the levels of a pathogen
or
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
67
antigen, such as a tumor cell, present in the body can be achieved by a number
of
methods known in the art and described herein.
The present invention include a method of stimulating an immune
response in a mammal exposed to an antigen or pathogen comprising the steps
of: (1)
determining directly or indirectly the level of antigen or pathogen present in
said
mammal; (2) administering a composition comprising zalphal 1 Ligand
polypeptide in
an acceptable pharmaceutical vehicle; (3) determining directly or indirectly
the level of
antigen or pathogen in said mammal; and (4) comparing the level of the antigen
or
pathogen in step 1 to the antigen or pathogen level in step 3, wherein a
change in the
o level is indicative of stimulating an immune response. In another
embodiment the
zalphal 1 Ligand composition is re-administered. In other embodiments, the
antigen is
a B cell tumor; a virus; a parasite or a bacterium.
In another aspect, the present invention provides a method of stimulating
an immune response in a mammal exposed to an antigen or pathogen comprising:
(1)
determining a level of an antigen- or pathogen-specific antibody; (2)
administering a
composition comprising zalphal 1 Ligand polypeptide in an acceptable
pharmaceutical
vehicle; (3) determining a post administration level of antigen- or pathogen-
specific
antibody; (4) comparing the level of antibody in step (1) to the level of
antibody in step
(3), wherein an increase in antibody level is indicative of stimulating an
immune
response.
Polynucleotides encoding zalphal 1 Ligand polypeptides are useful
within gene therapy applications where it is desired to increase or inhibit
zalphal 1
Ligand activity. If a mammal has a mutated or absent zalphal 1 Ligand gene,
the
zalphal 1 Ligand gene can be introduced into the cells of the mammal. In one
embodiment, a gene encoding a zalphal 1 Ligand polypeptide is introduced in
vivo in a
viral vector. Such vectors include an attenuated or defective DNA virus, such
as, but
not limited to, herpes simplex virus (HSV). papillomavirus, Epstein Barr virus
(EBV),
adenovirus, adeno-associated virus (AAV). and the like. Defective viruses,
which
entirely or almost entirely lack viral genes. are preferred. A defective virus
is not
infective after introduction into a cell. Use of defective viral vectors
allows for
administration to cells in a specific, localized area. without concern that
the vector can
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
68
infect other cells. Examples of particular vectors include, but are not
limited to, a
defective herpes simplex virus 1 (HSV1) vector (Kaplitt et al., Molec. Cell.
Neurosci.
2:320-30, 1991); an attenuated adenovirus vector, such as the vector described
by
Stratford-Perricaudet et al., J. Clin. Invest. 90:626-30, 1992; and a
defective adeno-
s
associated virus vector (Samulski et al., J. Virol. 61:3096-101, 1987;
Samulski et al., J.
Virol. 63:3822-8, 1989).
A zalphal 1 Ligand gene can be introduced in a retroviral vector, e.g., as
described in Anderson et al., U.S. Patent No. 5,399,346; Maim et al. Cell
33:153, 1983;
Temin et al., U.S. Patent No. 4,650,764; Temin et al., U.S. Patent No.
4,980,289;
Markowitz et al., J. Virol. 62:1120, 1988; Temin et al., U.S. Patent No.
5,124,263;
International Patent Publication No. WO 95/07358, published March 16, 1995 by
Dougherty et al.; and Kuo et al., Blood 82:845, 1993. Alternatively, the
vector can be
introduced by lipofection in vivo using liposomes. Synthetic cationic lipids
can be used
to prepare liposomes for in vivo transfection of a gene encoding a marker
(Feigner et
al., Proc. Natl. Acad. Sci. USA 84:7413-7, 1987; Mackey et al., Proc. Natl.
Acad. Sci.
USA 85:8027-31, 1988). The use of lipofection to introduce exogenous genes
into
specific organs in vivo has certain practical advantages. Molecular targeting
of
liposomes to specific cells represents one area of benefit. More particularly,
directing
transfection to particular cells represents one area of benefit. For instance,
directing
transfection to particular cell types would be particularly advantageous in a
tissue with
cellular heterogeneity, such as the immune system, pancreas, liver, kidney,
and brain.
Lipids may be chemically coupled to other molecules for the purpose of
targeting.
Targeted peptides (e.g., hormones or neurotransmitters), proteins such as
antibodies, or
non-peptide molecules can be coupled to liposomes chemically.
It is possible to remove the target cells from the body: to introduce the
vector as a naked DNA plasmid; and then to re-implant the transformed cells
into the
body. Naked DNA vectors for gene therapy can be introduced into the desired
host
cells by methods known in the art, e.g., transfection, electroporation,
microinjection,
transduction, cell fusion, DEAE dextran, calcium phosphate precipitation, use
of a gene
gun or use of a DNA vector transporter. See, e.g., Wu et al., J. Biol. Chem.
267:963-7,
1992; Wu et al., .T. Biol. Chem. 263:14621-4, 1988.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
69
Antisense methodology can be used to inhibit zalphal 1 Ligand gene
transcription, such as to inhibit cell proliferation in vivo. Polynucleotides
that are
complementary to a segment of a zalphal 1 Ligand-encoding polynucleotide
(e.g., a
polynucleotide as set froth in SEQ ID NO:1) are designed to bind to zalphal 1
Ligand-
encoding mRNA and to inhibit translation of such mRNA. Such antisense
polynucleotides are used to inhibit expression of zalphal 1 Ligand polypeptide-
encoding genes in cell culture or in a subject.
Mice engineered to express the zalphall Ligand gene, referred to as
"transgenic mice," and mice that exhibit a complete absence of zalphall Ligand
gene
function, referred to as "knockout mice," may also be generated (Snouwaert et
al.,
Science 257:1083, 1992; Lowell et al., Nature 366:740-42, 1993; Capecchi,
M.R.,
Science 244: 1288-1292, 1989; Palmiter, R.D. et al. Annu Rev Genet. 20: 465-
499,
1986). For example, transgenic mice that over-express zalphal 1 Ligand, either
ubiquitously or under a tissue-specific or tissue-restricted promoter can be
used to ask
whether over-expression causes a phenotype. For example. over-expression of a
wild-
type zalphal1 Ligand polypeptide, polypeptide fragment or a mutant thereof may
alter
normal cellular processes, resulting in a phenotype that identifies a tissue
in which
zalphal 1 Ligand expression is functionally relevant and may indicate a
therapeutic
target for the zalphal 1 Ligand, its agonists or antagonists. For example, a
preferred
transgenic mouse to engineer is one that over-expresses the zalphal 1 Ligand
(amino
acid residues 32-162 of SEQ ID NO:2). Moreover, such over-expression may
result in
a phenotype that shows similarity with human diseases. Similarly, knockout
zalphal 1
Ligand mice can be used to determine where zalphal 1 Ligand is absolutely
required in
vivo. The phenotype of knockout mice is predictive of the in vivo effects of
that a
zalphal 1 Ligand antagonist, such as those described herein. may have. The
human or
mouse zalphal 1 Ligand cDNA can be used to generate knockout mice. These mice
may be employed to study the zalphal1 Ligand gene and the protein encoded
thereby in
an in vivo system, and can be used as in vivo models for corresponding human
diseases.
Moreover, transgenic mice expression of zalphal 1 Ligand antisense
polynucleotides or
ribozymes directed against zalphal 1 Ligand, described herein, can be used
analogously
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
to transgenic mice described above. Studies may be carried out by
administration of
purified zalphal 1 Ligand protein, as well.
For pharmaceutical use, the proteins of the present invention are
formulated for parenteral, particularly intravenous or subcutaneous, delivery
according
5 to
conventional methods. The bioactive polypeptide or antibody conjugates
described
herein can be delivered intravenously, intraarterially or intraductally, or
may be
introduced locally at the intended site of action. Intravenous administration
will be by
bolus injection or infusion over a typical period of one to several hours. In
general,
pharmaceutical formulations will include a zalphal 1 Ligand protein in
combination
io with a
pharmaceutically acceptable vehicle, such as saline, buffered saline, 5%
dextrose
in water or the like. Formulations may further include one or more excipients,
preservatives, solubilizers, buffering agents, albumin to prevent protein loss
on vial
surfaces, etc. Methods of formulation are well known in the art and are
disclosed, for
example, in Remington: The Science and Practice of Pharmacy, Gennaro, ed.,
Mack
15
Publishing Co., Easton, PA, 19th ed., 1995. Therapeutic doses will generally
be in the
range of 0.1 to 100 1..tg/kg of patient weight per day, preferably 0.5-20
jig/kg per day,
with the exact dose determined by the clinician according to accepted
standards, taking
into account the nature and severity of the condition to be treated, patient
traits, etc.
Determination of dose is within the level of ordinary skill in the art. The
proteins may
20 be
administered for acute treatment, over one week or less, often over a period
of one to
three days or may be used in chronic treatment, over several months or years.
In
general, a therapeutically effective amount of zalpha 11 Ligand is an amount
sufficient
to produce a clinically significant change in hematopoietic or immune
function.
25 The
invention is further illustrated by the following non-limiting
examples.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
71
EXAMPLES
Example 1
Construction of MPL-zalphal 1 Polypeptide Chimera: MPL extracellular and TM
domain fused to the zalphal 1 intracellular signaling domain
The extracellular and transmembrane domains of the murine MPL
receptor were isolated from a plasmid containing the murine MPL receptor
(PHZ1/MPL plasmid) using PCR with primers ZC17,212 (SEQ ID NO:5) and
ZC19,914 (SEQ ID NO:6). The reaction conditions were as follows: 95 C for 1
min.;
35 cycles at 95 C for 1 min., 45 C for 1 min., 72 C for 2 min.; followed by 72
C at 10
min.; then a 10 C soak. The PCR product was run on a 1% low melting point
agarose
(Boerhinger Mannheim, Indianapolis, IN) and the approximately 1.5 kb MPL
receptor
fragment isolated using QiaquickTM gel extraction kit (Qiagen) as per
manufacturer's
instructions.
The intracellular domains of human zalphal 1 were isolated from a
plasmid containing zalphal 1 receptor cDNA using PCR with primers ZC19,913
(SEQ
ID NO:8) and ZC20,097 (SEQ ID NO:9). The polynucleotide sequence corresponding
to the zalphal 1 receptor coding sequence is shown in SEQ ID NO:7, and the
corresponding amino acid sequence shown in SEQ ID NO:115. The reaction
conditions were as per above. The PCR product was run on a 1% low melting
point
agarose (Boerhinger Mannheim) and the approximately 900 bp zalphal 1 fragment
isolated using Qiaquick gel extraction kit as per manufacturer's instructions.
Each of the isolated fragments described above were mixed at a 1:1
volumetric ratio and used in a PCR reaction using ZC17,212 (SEQ ID NO:5) and
ZC20,097 (SEQ ID NO:9) to create the MPL-zalphal 1 chimera. The reaction
conditions were as follows: 95 C for 1 min.; 35 cycles at 95 C for 1 min., 55
C for 1
min., 72 C for 2 min.; followed by 72 C at 10 min.; then a 10 C soak. The
entire PCR
product was run on a 1% low melting point agarose (Boehringer Mannheim) and
the
approximately 2.4 kb MPL-zalphal 1 chimera fragment isolated using Qiaquick
gel
extraction kit (Qiagen) as per manufacturer's instructions. The MPL-zalphal 1
chimera
fragment was digested with EcoRI (BRL) and XbaI (Boerhinger Mannheim) as per
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
72
manufacturer's instructions. The entire digest was run on a 1% low melting
point
agarose (Boehringer Mannheim) and the cleaved MPL-zalphal 1 chimera isolated
using
QiaquickTM gel extraction kit (Qiagen) as per manufacturer's instructions. The
resultant cleaved MPL-zalphal 1 chimera was inserted into an expression vector
as
s described below.
Recipient expression vector pZP-5N was digested with EcoRI (BRL)
and HindIII (BRL) as per manufacturer's instructions, and gel purified as
described
above. This vector fragment was combined with the EcoRI and XbaI cleaved MPL-
zalphal 1 chimera isolated above and a XbaI/HindIII linker fragment in a
ligation
reaction. The ligation was run using T4 Ligase (BRL), at 15 C overnight. A
sample of
the ligation was electroporated in to DH1OB ElectroMAXTm electrocompetent E.
coli
cells (25g, 200Q, 2.3V). Transformants were plated on LB+Ampicillin plates and
single colonies screened by PCR to check for the MPL-zalphal 1 chimera using
ZC17,212 (SEQ ID NO:5) and ZC20,097 (SEQ ID NO:9) using the PCR conditions as
described above.
Confirmation of the MPL-zalphal 1 chimera sequence was made by
sequence analyses using the following primers: ZC12,700 (SEQ ID NO:10),
ZC5,020
(SEQ ID NO:11), ZC6,675 (SEQ ID NO:12), ZC7,727 (SEQ ID NO:13), ZC8,290
(SEQ ID NO:14), ZC19,572 (SEQ ID NO:15), ZC6,622 (SEQ ID NO:16), ZC7,736
(SEQ ID NO:17), and ZC9,273 (SEQ ID NO:18). The insert was approximately 2.4
bp,
and was full-length.
Example 2
MPL-zalphal 1 chimera based proliferation in BAF3 assay using Alamar Blue
A. Construction of BaF3 Cells Expressing MPL-zalphal 1 chimera
BaF3, an interleukin-3 (IL-3) dependent pre-lymphoid cell line derived
from murine bone marrow (Palacios and Steinmetz, Cell 41: 727-734. 1985;
Mathey-
Prevot et al., Mol. Cell. Biol. 6: 4133-4135, 1986), was maintained in
complete media
(RPMI medium (JRH Bioscience Inc.. Lenexa, KS) supplemented with 10% heat-
inactivated fetal calf serum. 2ng/m1 murine IL-3 (mIL-3) (R & D, Minneapolis,
MN),
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
73
2mM L-glutaMax-lTm (Gibco BRL), 1 mM Sodium Pyruvate (Gibco BRL), and PSN
antibiotics (GIBCO BRL)). Prior to electroporation, pZP-5N/MPL-zalphal 1
plasmid
DNA (Example 1) was prepared and purified using a Qiagen Maxi Prep kit
(Qiagen) as
per manufacturer's instructions. BaF3 cells for electroporation were washed
once in
RPMI media and then resuspended in RPMI media at a cell density of 107
cells/ml.
One ml of resuspended BaF3 cells was mixed with 30 [tg of the pZP-5N/MPL-
zalphall
plasmid DNA and transferred to separate disposable electroporation chambers
(GIBCO
BRL). Following a 15 minute incubation at room temperature the cells were
given two
serial shocks (800 1Fad/300 V.; 1180 1Fad/300 V.) delivered by an
electroporation
apparatus (CELL-PORATORTm; GIBCO BRL). After a 5 minute recovery time, the
electroporated cells were transferred to 50 ml of complete media and placed in
an
incubator for 15-24 hours (37 C, 5% CO2). The cells were then spun down and
resuspended in 50 ml of complete media containing GeneticinTM (Gibco)
selection (500
[tg/m1 G418) in a T-162 flask to isolate the G418-resistant pool. Pools of the
transfected BaF3 cells, hereinafter called BaF3/MPL-zalphal 1 cells, were
assayed for
signaling capability as described below.
B. Testing the signaling capability of the BaF3/MPL-zalphal 1 cells using an
Alamar
Blue Proliferation Assay
BaF3/MPL-zalphal 1 cells were spun down and washed in the complete
media, described above, but without mIL-3 (hereinafter referred to as "mIL-3
free
media"). The cells were spun and washed 3 times to ensure the removal of the
mIL-3.
Cells were then counted in a hemacytometer. Cells were plated in a 96-well
format at
5000 cells per well in a volume of 1001A1 per well using the mIL-3 free media.
Proliferation of the BaF3/MPL-zalphal 1 cells was assessed using
murine mthrombopoietin (mTP0) diluted with mIL-3 free media to 500 ng/ml,
250ng/ml, 125 ng/ml, 62 ng/ml, 30 ng/ml, 15 ng/ml, 7.5 ng/ml. 3.75 ng/ml, 1.8
ng/ml,
0.9 ng/ml, 0.5 ng/ml and 0.25 ng/ml concentrations. 100 1A1 of the diluted
mTPO was
added to the BaF3/MPL-zalphal 1 cells. The total assay volume is 200 1.
Negative
controls were run in parallel using mIL-3 free media only. without the
addition of
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
74
mTPO. The assay plates were incubated at 37 C, 5% CO2 for 3 days at which time
Alamar Blue (Accumed, Chicago, IL) was added at 20 1/well. Alamar Blue gives a
fluourometric readout based on the metabolic activity of cells, and is thus a
direct
measurement of cell proliferation in comparison to a negative control. Plates
were
s again incubated at 37 C, 5% CO2 for 24 hours. Plates were read on the
FmaxTM plate
reader (Molecular Devices Sunnyvale, CA) using the SoftMaxTm Pro program, at
wavelengths 544 (Excitation) and 590 (Emmission).
Results confirmed the signaling capability of the intracellular portion of
the zalphal 1 receptor, as the thrombopoietin induced proliferation at
approximately 10
fold over back ground at mTPO concentrations of 62 ng/ml and greater.
Example 3
Construction of expression vector expressing full-length zalphall
The entire zalphal 1 receptor was isolated from a plasmid containing
zalphal 1 receptor cDNA (SEQ ID NO:7) using PCR with primers ZC19,905 (SEQ ID
NO:19) and ZC19,906 (SEQ ID NO:20). The reaction conditions were as follows:
95 C for 1 min; 35 cycles at 95 C for 1 min, 55 C for 1 min, 72 C for 2 min;
followed
by 72 C at 10 min; then a 10 C soak. The PCR product was run on a 1% low
melting
point agarose (Boerhinger Mannheim) gel and the approximately 1.5 kb zalphal 1
cDNA isolated using QiaquickTM gel extraction kit (Qiagen) as per
manufacturer's
instructions.
The purified zalphal 1 cDNA was digested with BamHI (Boerhinger
Mannheim) and EcoRI (BRL) as per manufacturer's instructions. The entire
digest was
run on a 1% low melting point agarose (Boerhinger Mannheim) gel and the
cleaved
zalphal 1 fragment was purified the using QiaquickTM gel extraction kit as per
manufacturer's instructions. The resultant cleaved zalphal 1 fragment was
inserted into
an expression vector as described below.
Recipient expression vector pZP-5N was digested with BamHI
(Boerhinger Mannheim) and EcoRI (BRL) as per manufacturer's instructions, and
gel
purified as described above. This vector fragment was combined with the BamHI
and
EcoRI cleaved zalphal 1 fragment isolated above in a ligation reaction using
T4 Ligase
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
(BRL). The ligation was incubated at 15 C overnight. A sample of the ligation
was
electroporated in to DH1OB electroMAXTm electrocompetent E. coli cells (25 F,
200Q,
2.3V). Transformants were plated on LB+Ampicillin plates and single colonies
screened by PCR to check for the zalphal 1 sequence using ZC19,905 (SEQ ID
NO:19)
5 and ZC19,906 (SEQ ID NO:20) using the PCR conditions as described above.
Confirmation of the zalphall sequence was made by sequence analyses
using the following primers: ZC12,700 (SEQ ID NO:10), ZC5,020 (SEQ ID NO:11),
ZC20,114 (SEQ ID NO:21), ZC19,459 (SEQ ID NO:22), ZC19,954 (SEQ ID NO:23),
and ZC20,116 (SEQ ID NO:24). The insert was approximately 1.6 kb, and was full-
o length.
Example 4
Zalphal 1 based proliferation in BAF3 assay using Alamar Blue
A. Construction of BaF3 Cells Expressing zalphal 1 receptor
15 BaF3
cells expressing the full-length zalphal 1 receptor was constructed
as per Example 2A above, using 30[1.g of the zalphal 1 expression vector,
described in
Example 3 above. The BaF3 cells expressing the zalphal 1 receptor mRNA were
designated as BaF3/zalphal 1. These cells were used to screen for zalphall
Ligand as
described below in Examples 5 and 6.
Example 5
Screening for zalphal 1 Ligand using BaF3/Zalphal 1 cells using an Alamar Blue
Proliferation Assay
A. Activation of primary Monkey splenocytes to test for presence of zalphal 1
Ligand
Monkey splenocytes were stimulated in vitro to produce conditioned
media to test for the presence of zalphal 1 Ligand activity as described
below. Monkey
spleens were obtained from 8 year old female A 1. nesestrian monkeys. The
spleens
were teased part to produce a single cell suspension. The mononuclear cells
were
isolated by Ficoll-Paquel) PLUS (Pharmacia Biotech, Uppsala, Sweden) density
gradient. The mononuclear cells were seeded at 2 x 106 cells/ml in RPMI-1640
media
supplemented with 10% FBS and activated with with 5 ng/ml Phorbol-12-myristate-
13-
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
76
acetate (PMA) (Calbiochem, San Diego, CA), and 0.5mg/m1 IonomycinTM
(Calbiochem) for 48hrs. The supernatant from the stimulated monkey spleen
cells was
used to assay proliferation of the BaF3/zalphal 1 cells as described below.
s B. Screening for zalphal 1 Ligand using BaF3/Zalphal 1 cells using an
Alamar Blue
Proliferation Assay
BaF3/Zalphal 1 cells were spun down and washed in mIL-3 free media.
The cells were spun and washed 3 times to ensure the removal of the mIL-3.
Cells
were then counted in a hemacytometer. Cells were plated in a 96-well format at
5000
cells per well in a volume of 100 ill per well using the mIL-3 free media.
Proliferation of the BaF3/Zalphal 1 cells was assessed using conditioned
media from activated monkey spleen (see Example 5A). Conditioned media was
diluted with mIL-3 free media to 50%, 25%, 12.5%, 6.25%, 3.125%, 1.5%, 0.75%
and
0.375% concentrations. 100 I of the diluted conditioned media was added to
the
BaF3/Zalphal 1 cells. The total assay volume is 200 pl. The assay plates were
incubated at 37 C, 5% CO, for 3 days at which time Alamar Blue (Accumed,
Chicago,
IL) was added at 20 I/well. Plates were again incubated at 37 C, 5% CO2 for
24
hours. Plates were read on the FmaxTM plate reader (Molecular devices) as
described
above (Example 2).
Results confirmed the proliferative response of the BaF3/Zalphal 1 cells
to a factor present in the activated monkey spleen conditioned media. The
response, as
measured, was approximately 4-fold over background at the 50% concentration.
The
untransfected BaF3 cells did not proliferate in response to this factor,
showing that this
factor is specific for the Zalphal 1 receptor.
C. Human Primary Source used to isolate zalphal 1 Ligand
100 ml blood draws were taken from each of six donors. The blood was
drawn using 10X 10 ml vacutainer tubes containing heparin. Blood was pooled
from
six donors (600m1), diluted 1:1 in PBS, and separated using a Ficoll-Paqueg
PLUS
(Pharmacia Biotech). The isolated primary human cell yield after separation on
the
ficoll gradient was 1.2X109 cells.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
77
Cells were suspended in 9.6 ml MACS buffer (PBS, 0.5% EDTA, 2mM
EDTA). 1.6 ml of cell suspension was removed and 0.4 ml CD3 microbeads
(Miltenyi
Biotec, Auburn, CA) added. The mixture was incubated for 15 min. at 4 C. These
cells labeled with CD3 beads were washed with 30 ml MACS buffer, and then
resuspended in 2 ml MACS buffer.
A VS+ column (Miltenyi) was prepared according to the manufacturer's
instructions. The VS+ column was then placed in a VarioMACSTm magnetic field
(Miltenyi). The column was equilibrated with 5 ml MACS buffer. The isolated
primary
human cells were then applied to the column. The CD3 negative cells were
allowed to
o pass through. The column was rinsed with 9 ml (3 X 3 ml) MACS buffer. The
column
was then removed from the magnet and placed over a 15 ml falcon tube. CD3+
cells
were eluted by adding 5 ml MACS buffer to the column and bound cells flushed
out
using the plunger provided by the manufacturer. The incubation of the cells
with the
CD3 magnetic beads, washes, and VS+ column steps (incubation through elution)
above were repeated five more times. The resulting CD3+ fractions from the six
column separations were pooled. The yield of CD3+ selected human cells were
3X108
total cells.
A sample of the pooled CD3+ selected human cells was removed for
staining and sorting on a fluorescent antibody cell sorter (FACS) to assess
their purity.
The human CD3+ selected cells were 91% CD3+ cells.
The human CD3+ selected cells were activated by incubating in RPMI +
5% FBS + PMA 10 ng/ml and Ionomycin 0.5 lag/m1 (Calbiochem) for 13 hours 37 C.
The supernatant from these activated CD3+ selected human cells was tested for
zalphall Ligand activity as described below. Moreover, the activated CD3+
selected
human cells were used to prepare a cDNA library, as described in Example 6,
below.
D. Testing supernatant from activated CD3+ selected human cells for zalphal 1
Ligand
using BaF3/Zalphal 1 cells and an Alamar Blue Proliferation Assay
BaF3/Zalphal 1 cells were spun down and washed in mIL-3 free media.
The cells were spun and washed 3 times to ensure the removal of the mIL-3.
Cells
were then counted in a hemacytometer. Cells were plated in a 96-well format at
5000
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
78
cells per well in a volume of 100 ul per well using the mIL-3 free media.
Proliferation of the BaF3/Zalphal1 cells was assessed using conditioned
media from activated CD3+ selected human cells (see Example 5C) diluted with
mIL-3
free media to 50%, 25%, 12.5%, 6.25%, 3.125%, 1.5%, 0.75% and 0.375%
s concentrations. 100
ul of the diluted conditioned media was added to the
BaF3/Zalphal 1 cells. The total assay volume is 200 l. The assay plates were
incubated and assayed as described in Example 5B.
Results confirmed the proliferative response of the BaF3/Zalphal 1 cells
to a factor present in the activated CD3+ selected human Cell conditioned
media. The
response, as measured, was approximately 10-fold over background at the 50%
concentration. The untransfected BaF3 cells did not proliferate in response to
this
factor, showing that this factor is specific for the Zalphal 1 receptor.
Moreover soluble
alphal 1 receptor blocked this proliferative activity in the BaF3/Zalphal 1
cells (see,
Example 11).
Example 6
Cloning of human zalphal 1 Ligand from a human CD3+ selected cell Library
Screening of a primary human activated CD3+ selected cell cDNA
library revealed an isolated cDNA that is a novel member of the four-helix
bundle
cytokine family. This cDNA encoded the zalphal 1 Ligand. The cDNA was
identified
by screening for activity of the zalphal 1 Ligand using the zalphal 1
receptor.
A. The vector for CD3+ selected library construction
The vector for CD3+ selected library construction was pZP7NX. The
pZP7NX vector was constructed as follows: The coding region for the DHFR
selective
marker in vector pZP7 was removed by DNA digestion with Ncol and PstI
restriction
enzymes (Boehringer Mannheim). The digested DNA was run on 1% agarose gel, cut
out and gel purified using the Qiagen Gel Extraction Kit (Qiagen) as per
manufacturer's
instructions. A DNA fragment representing the coding region of Zeocin
selective
marker was amplified by PCR method with primers ZC13,946 (SEQ ID NO:25) and
ZC13,945 (SEQ ID NO:26), and pZeoSV2(+) as a template. There are additional
PstI
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
79
and Bell restriction sites in primer ZC13,946 (SEQ ID NO:25), and additional
NcoI
and SfuI sites in primer ZC13,945 (SEQ ID NO:26). The PCR fragment was cut
with
PstI and NcoI restriction enzymes and cloned into pZP7 vector prepared by
cleaving
with the same two enzymes and subsequent gel purification. This vector was
named
pZP7Z. Then the Zeocin coding region was removed by DNA digestion of vector
pZP7Z with Bell and SfuI restriction enzymes. The digested DNA was run on 1%
agarose gel, cut out and gel purified, and then ligated with a DNA fragment of
Neomycin coding region cut from pZem228 vector (deposited at the American Type
Culture Collection (ATCC), Manassas, VA; ATCC Deposit No. 69446) with the same
restriction enzymes (Bell and SfuI).
This new vector was named pZP7N, in which the coding region for
DHFR selective marker was replaced by the coding region for a Neomycin
selective
marker from vector pZem228. A stuffer fragment including an Xhol site was
added to
pZP7N to create a vector suitable for high efficiency directional cloning of
cDNA; this
new vector was called pZP7NX. To prepare the vector for cDNA, 201.1g of pZP7NX
was digested with 20 units of EcoRl (Life Technologies Gaithersberg,MD) and 20
units of Xho 1 (Boehringer Mannheim Indianapolis,IN) for 5 hours at 37 C. then
68 C
for 15 minutes. The digest was then run on a 0.8% low melt agarose 1XTAE gel
to
separate the stuffer from the vector. The vector band was excised and digested
with
"beta-Agarase" (New England Biolabs, Beverly, MA) following the manufacturer's
recommendations. After ethanol precipitation the digested vector was
resuspended in
water to 45ng/m1 in preparation for ligation of CD3+ selected cDNA library
described
below.
B. Preparation of primary human activated CD3+ selected cell cDNA library
Approximately 1.5X108 primary human CD3+ selected cells stimulated
in ionomycin/PMA were isolated by centrifugation after culturing at 37 C for
13 hours
(Example 5C). Total RNA was isolated from the cell pellet using the "RNeasy
Midi"
kit from Qiagen, Inc. (Valencia, CA). mRNA was isolated from 225 micrograms of
total RNA using the "MPG mRNA purification kit" from CPG Inc. (Lincoln Park,
NJ).
3.4 micrograms of mRNA was isolated and converted to double stranded cDNA
using
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
the following procedure.
First strand cDNA from stimulated human CD3+ selected cells was
synthesized as follows. Nine p.1 Oligo d(T)-selected poly(A) CD3+ RNA at a
concentration of 0.34 jig/ I and 1.0 IA of 1 vig / ,1 first strand primer
ZC18,698 (SEQ
5 ID NO:27) containing an Xhol restriction site were mixed and heated at 65
C for 4
minutes and cooled by chilling on ice. First strand cDNA synthesis was
initiated by the
addition of 9 I of first strand buffer (5x SUPERSCRIPT buffer; (Life
Technologies),
4 1 of 100 mM dithiothreitol and 2 i_t1 of a deoxynucleotide triphosphate
solution
containing 10 mM each of dATP, dGTP, dTTP and 5-methyl-dCTP (Pharmacia Biotech
10 Inc.) to the RNA-primer mixture. The reaction mixture was incubated at
45 C for 4
minutes followed by the addition of 8 IA of 200 U4.11 Superscriptilt, RNase H-
reverse
transcriptase (Life technologies). The reaction was incubated at 45 C for 45
minutes
followed by an incubation ramp of 1 C every 2 minutes to 50 C where the
reaction
was held for 10 minutes. To denature any secondary structure and allow for
additional
15 extension of the cDNA the reaction was then heated to 70 C for 2
minutes then
dropped to 55 C for 4 minutes after which 2 1 of SuperscriptII0 RI was added
and
incubated an additional 15 minutes followed by a ramp up to 70 C 1 minute/1
C.
Unincorporated nucleotides were removed from the cDNA by twice precipitating
in the
presence of 2 g of glycogen carrier, 2.0 M ammonium acetate and 2.5 volume
ethanol,
20 followed by a 100 ,I wash with 70% ethanol. The cDNA was resuspended in
98 I
water for use in second strand synthesis.
Second strand synthesis was performed on the first strand cDNA under
conditions that promoted first strand priming of second strand synthesis
resulting in
DNA hairpin formation. The second strand reaction contained 98 I of the first
strand
25 cDNA, 30 p,1 of 5x polymerase I buffer (100 mM Iris: HC1, pH 7.5, 500 mM
KC1, 25
mM MgC12, 50 mM (NH4)2SO4), 2 ,1 of 100 mM dithiothreitol. 6 I of a solution
containing 10 mM of each deoxynucleotide triphosphate, 5 I of 5 mM b-NAD, 1
1 of
3 U4.1,1 E. coli DNA ligase (New England Biolabs Inc.) and 4 1 of 10 U/ I E.
coli DNA
polymerase I (New England Biolabs Inc.). The reaction was assembled at room
30 temperature and was incubated at room temperature for 2 minutes followed by
the
addition of 4 1 of 3.8 U/ 1 RNase H (Life Technologies). The reaction was
incubated
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
81
at 15 C for two hours followed by a 15 minute incubation at room temperature.
10 1
of 1M TRIS pH7.4 was added to the reaction and extracted twice with
phenol/chloroform and once with chloroform, the organic phases were then back
extracted with 50 1 of TE (10mM TRIS pH 7.4, 1mM EDTA), pooled with the other
aqueous and ethanol precipitated in the presence of 0.3 M sodium acetate. The
pellet
was washed with 100 I 70% ethanol air dried and resuspended in 40 p.1 water.
The single-stranded DNA of the hairpin structure was cleaved using
mung bean nuclease. The reaction mixture contained 40 IA of second strand
cDNA, 5
1 of 10x mung bean nuclease buffer (Life technologies), 5 1 of mung bean
nuclease
o (Pharmacia Biotech Corp.) diluted to 1U/ I in 1X mung bean nuclease
buffer. The
reaction was incubated at 37 C for 45 minutes. The reaction was terminated by
the
addition of 10 11 of 1 M Tris: HC1, pH 7.4 followed by sequential
phenol/chloroform
and chloroform extractions as described above. Following the extractions, the
cDNA
was ethanol precipitated in the presence of 0.3 M sodium acetate. The pellet
was
washed with 100 I 70% ethanol air dried and resuspended in 38 1 water.
The resuspended cDNA was blunt-ended with T4 DNA polymerase.
The cDNA, which was resuspended in 38 1.11 of water, was mixed with 12 1 5x
T4
DNA polymerase buffer (250 mM Tris:HC1, pH 8.0, 250 mM KC1, 25 mM MgC12), 2
1 0.1 M dithiothreitol, 6 pl of a solution containing 10 mM of each
deoxynucleotide
triphosphate and 2 1 of 1 U/ 1 T4 DNA polymerase (Boehrin2er Mannheim Corp.).
After an incubation of 45 minutes at 15 C, the reaction was terminated by the
addition
of 30 1 TE followed by sequential phenol/chloroform and chloroform
extractions and
back extracted with 20 I TE as described above. The DNA was ethanol
precipitated in
the presence of 2 pl Pellet PaintTM (Novagen) carrier and 0.3 M sodium acetate
and
was resuspended 11 1 of water.
Eco RI adapters were ligated onto the 5' ends of the cDNA described
above to enable cloning into an expression vector. 1 1 I of cDNA and 4 pl of
65
pmole/pl of Eco RI hemiphophorylated adaptor (Pharmacia Biotech Corp) were
mixed
with 5 1,11 5x ligase buffer (Life Technologies), 2 1 of 10 mM ATP and 3 p.1
of 1 U/ .1
T4 DNA ligase (Life Technologies), 1 pl 10X ligation buffer (Promega Corp), 9
1
water. The extra dilution with 1X buffer was to prevent the pellet paint from
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
82
precipitating. The reaction was incubated 9 hours in a water bath temperature
ramp
from 10 C to 22 C over 9 hours, followed by 45 minutes at 25 C. The reaction
was
terminated by incubation at 68 C for 15 minutes.
To facilitate the directional cloning of the cDNA into an expression
s vector, the cDNA was digested with Xhol, resulting in a cDNA having a 5'
Eco RI
cohesive end and a 3' Xhol cohesive end. The Xhol restriction site at the 3'
end of the
cDNA had been previously introduced using the ZC18698 (SEQ ID NO:27) primer.
Restriction enzyme digestion was carried out in a reaction mixture containing
35 pl of
the ligation mix described above, 6 1 of 10x H buffer (Boehringer Mannheim
Corp.), 1
p.1 of 2mg/m1 BSA (Biolabs Corp.), 17 p.1 water and 1.0 IA of 40 U/ 1XhoI
(Boehringer
Mannheim). Digestion was carried out at 37 C for 1 hour. The reaction was
terminated by incubation at 68 C for 15 minutes followed by ethanol
precipitation,
washing drying as described above and resuspension in 30 pl water.
The resuspended cDNA was heated to 65 C for 5 minutes and cooled on
15 ice, 4 1 of 5X gel loading dye (Research Genetics Corp.) was added, the
cDNA was
loaded onto a 0.8% low melt agarose 1X TAE gel (SEA PLAQUE GTGTm low melt
agarose; FMC Corp.) and electrophoresed. The contaminating adapters and cDNA
below 0.6 Kb in length were excised from the gel. The electrodes were
reversed,
molten agarose was added to fill in the wells, the buffer was changed and the
cDNA
20 was electrophoresed until concentrated near the lane origin. The area of
the gel
containing the concentrated cDNA was excised and placed in a microfuge tube,
and the
agarose was melted by heating to 65 C for 15 minutes. Following equilibration
of the
sample to 45 C, 2 1 of 1 U/ 1 Beta-agarase I (Biolabs, Inc.) was added, and
the
mixture was incubated for 90 min. at 45 C to digest the agarose. After
incubation, 1
25 tenth volume of 3 M Na acetate was added to the sample, and the mixture
was
incubated on ice for 15 minutes. The sample was centrifuged at 14,000 x g for
15
minutes at room temperature to remove undigested agarose, the cDNA was ethanol
precipitated, washed in 70% ethanol, air-dried and resuspended in 40 I water.
To determine the optimum ratio of cDNA to vector several ligations
30 were assembled and electroporated. Briefly, 2 1.t1 of 5X 14 ligase
buffer(Life
Technologies), 1 1 of 10mM ATP, 1 pl pZP7NX digested with EcoR1-Xho 1 , 111
14
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
83
DNA ligase diluted to 0.25u/ 1 (Life Technologies) water to 10 I and 0.5, 1,2
or 3 I
of cDNA were mixed in 4 separate ligations, incubated at 22 C for 4 hours, 68
C for
20 minutes, sodium acetate-ethanol precipitated, washed, dried and resuspended
in 10
11. A single microliter of each ligation was electroporated into 40 I DH10b
ElectroMaxTm electrocompetent bacteria (Life Technologies) using a 0.1cm
cuvette
(Biorad) and a Genepulser, pulse controllerTM (Biorad) set to 2.5KV, 251F,
200ohms.
These cells were immediately resuspended in 1 ml. SOC broth (Manniatis, et al.
supra.)
followed by 50011 of 50% glycerol-SOC as a preservative. These "glycerol
stocks "
were frozen in several aliquots at -70 C. An aliquot of each was thawed and
plated
o serially on LB-agar plates supplemented with ampicillin at 100 g/ml. Colony
numbers indicated that the optimum ratio of CD3+ cDNA to pZP7NX vector was 1
.1
to 45 ng; such a ligation yielded 4.5 million primary clones.
For the purpose of screening this library using a BaF3-zalphal1 based
proliferation assay (Example 5) glycerol stocks from above were diluted into
liquid
cultures of 100 or 250 clones per pool in deep well microtiter plates, grown
24 hours at
37 C with shaking and plasmid isolated using a Qiagen kit following the
manufacturer's instructions. Such DNA was subsequently transfected into BHK
cells,
media conditioned 72 hours, harvested and placed on 5K BaF3-zalphal1 cells for
72
hours after which proliferation was assessed using an "Alamar blue"
fluorescence assay
(Example 5B and Example 2B)
For the purpose of screening the library by secretion trap cloning, a
complex, amplified form of the library was needed to transfect COS-7 cells.
About 4.8
million clones were plated on 110 15cm LB-agar plates supplemented with 100
g/m1
ampicillin, 10 g/m1 methicillin. After growing the plates overnight at 37 C
the
bacteria were harvested by scraping and pelleted. Plasmid DNA was extracted
from the
pelleted bacteria using a Nucleobond-gigaTM (Clonetech) following the
manufacturer's
instructions. This plasmid was then used to transfect COS-7 cells (ATCC No.
CRL
1651) on slides and screened using the secretion trap technique described
below
(Example 12).
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
84
Example 7
Expression Cloning of human zalphall Ligand
The glycerol stocks from the activated human CD3+ selected cell library
(Example 6) were added to Super Broth JJTM (Becton Dickinson, Cockeysville,
MD) +
0.1 mg/ml ampicillin (amp) at a concentration of 250 cells per 800
microliters. The E.
coli were allowed to equilibrate for 24 hours at room temperature. At the time
of
inoculation, 400 microliters was plated on LB + amp plates to determine the
actual titer
of the inoculation. After 24 hours the plates were counted and then the final
concentration of the SuperBrothllTM + E. coli was adjusted so that the final
o concentration was 250 cells per 1.2 ml. Three times 2 liters were
inoculated for a total
of 6 liters. The media were then plated into 96-well deep well blocks
(Qiagen). Plating
was done on the 8-channel QFill2TM dispenser (Genetix, Christchurch, Dorset,
UK).
The E. coli were grown overnight at 37 C shaking at 250 rotations/min. on a
New
Brunswick Scientific Innova 4900 multi-tier environment shaker. The E. coli
were
spun out of solution at 3000 rpm, using a Beckman GS-6KR centrifuge. These E.
coli
pellets were frozen at -20 C or used fresh before miniprepping the plasmid
DNA. Each
pellet contains approximately 250 cDNA clones from the human CD3+ selected
cell
library.
These pools of 250 cDNA clones were then mini-prepped using
QIAprepTM 96 Turbo Miniprep kit (Qiagen). Plasmid DNA was eluted using 125 [11
of
TE (10 mM Tris pH 8, 1 mM EDTA). This plasmid DNA was then used to transfect
BHK cells.
BHK transfection
BHK cells were plated in 96-well tissue culture plates at a density of
12,000 cells per well in a volume of 100 IA. per well. Culture media was DMEM
(GibcoBRL), 5% heat-inactivated fetal bovine serum, 2 mM L-glutamine
(GibcoBRL),
1X PSN (GibcoBRL), 1 mM NaPyruvate (GibcoBRL).
The following day. BHK cells were washed once with 1001.11 SFA. SFA
is serum-free media which is DMEM/F12 (Gibco/BRL), 2 mM GlutaMaxTm
(Gibco/BRL), 1 mM NaPyruvate, 10 tg/m1 transferrin, 5 pg/m1 insulin, 10 [ig/m1
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
fetuin, 2 g/m1 selenium, 25mM HEPES (Gibco/BRL), 100 M non-essential amino
acids (Gibco/BRL).
A DNA/LipofectamineTM mix is made as follows: 2.2 1
LipofectamineTM reagent (Gibco/BRL) is combined with 102.8 1 SFA at room
5 temperature; approximately 5 I of the plasmid DNA (200 ng/ 1) is then
added to the
LipofectamineTm/SFA to form the DNA/LipofectamineTM mixture, which is
incubated
at room temperature for 30 minutes. The SFA was removed from the BHK cells and
the cells were incubated with 50 1 of the DNA/lipofectamineTM mix for 5 hours
at
37 C with 5% CO2. Fifty I of the DNA/LipofectamineTM mixture was added to
each
io of two wells of the BHK cells, so that transfections were done in
duplicate.
After BHK cells were incubated with DNA/LipofectamineTM mix for 5
hours, the DNA/LipofectamineTM mix was removed and 100 I culture media was
added. Cells were incubated overnight, the media was removed and replaced with
100
,1. culture media. After culturing cells for 72 hours, conditioned media was
removed,
15 frozen at -80 C for a minimum of 20 minutes, thawed, and then 50 1 was
assayed in
the zalphall/BaF3 proliferation assay, described in Examples 2B and Example 5,
to
identify pools of 250 clones with ligand activity.
Thirty-five 96-well plates were screened in a single assay.
This
represented approximately 250 cDNAs/well or 840,000 cDNAs total. Of these,
20 conditioned media from 54 wells (representing 250 cDNAs per well) tested
positive in
the proliferation assay. The conditioned media from these positive pools was
re-tested
in a second assay (secretion trap) with and without the soluble receptor (see,
Example
12). The zalphal 1 CEE soluble receptor (Examlpe 10A) was used at a final
concentration of about 1 g/ml. For all 54 positive pools, essentially all of
the activity
25 was neutralized by addition of the soluble zalphal 1 receptor,
indicating that these pools
contained a cDNA from the zalphall Ligand. Four of these positive pools were
chosen
to break-down and isolate a single cDNA that would encode the zalphal 1
Ligand.
These were 45C5, 46G11, 401412, and 60A1.
For each of these 4 pools, 1 1. of DNA was used to transform
30 ElectroMaxTm DH1OB cells (Gibco/BRL) by electroporation. The
transformants were
plated on LB + amp (100 pg/m1) + methicillin (10 g/m1) plates to give single
colonies.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
86
For each electroporated pool, 960 individual colonies were toothpicked into
ten 96-well
plates containing 1.2 ml of SuperBrothllTM per well. These plates were
numbered #1-
for each of the breakdown pools (45C5, 46G11, 401412, and 60A1). These were
cultured overnight and the plasmid DNA miniprepped as above. For 46G11, 40H12,
5 and
60A1, plasmid DNA from the breakdown plates was transfected into BHK cells as
above.
For 45 C5, a "fast track" protocol was utilized to accelerate the
identification of the zalphal 1 Ligand cDNA. BHK cells were transfected with
plasmid
DNA from the breakdown plates as above, DNA/LipofectamineTM mix was removed
o after a
5 hour incubation, and culture media was added. Since the transfections were
done in duplicate, the culture media was harvested the following day after 24
hours
from one of the transfected BHK plates, and harvested the following day after
48 hours
from the remaining transfected plate. The 24 hour conditioned media was
assayed as
above for zalphal 1 Ligand activity using the proliferation assay as described
herein.
Plasmid DNA was pooled from 45C5 breakdown plates #1-4 and
assayed for binding of zalphal 1 soluble receptor to its ligand by the
"secretion-trap"
protocol (see, Example 12, below). Eight positive clones were identified from
a total of
384 sequences. Results from the proliferation assay confirmed activity of the
zalphal 1
Ligand and correlated with results of the secretion trap assay (see Example
12).
Concurrently, plasmid DNA miniprepped from plates #I-4 of the 45C5 pool
breakdown
was sequenced to determine the DNA sequence of each of the 384 clones.
Several clones that were positively identified in the proliferation and
secretion trap assays were also sequenced using the following primers:
ZC14,063
(SEQ ID NO:28), ZC7,764a (SEQ ID NO:38), ZC7,764b (SEQ ID NO:39), ZC22,034
(SEQ ID NO:40), and ZC22,035 (SEQ ID NO:41). The polynucleotide sequence of
zalphal 1 Ligand was full-length (SEQ ID NO:1) and its corresponding amino
acid
sequence is shown (SEQ ID NO:2).
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
87
Example 8
Construction of mammalian expression vectors that express zalphal 1 soluble
receptors:
zalphal 1 CEE, zalphal 1 CFLG, zalphal 1CHIS and zalphal 1-Fc4
A. Construction of zalphal 1 Mammalian Expression Vector containing
zalphallCEE,
zalphal1CFLG and zalphallCHIS
An expression vector was prepared for the expression of the soluble,
extracellular domain of the zalphal 1 polypeptide, pC4zalphal 1 CEE, wherein
the
construct is designed to express a zalphal 1 polypeptide comprised of the
predicted
initiating methionine and truncated adjacent to the predicted transmembrane
domain,
io and with a C-terminal Glu-Glu tag (SEQ ID NO:29).
A 700 bp PCR generated zalphal 1 DNA fragment was created using
ZC19,931 (SEQ ID NO:30) and ZC19,932 (SEQ ID NO:31) as PCR primers to add
Asp718 and BamHI restriction sites. A plasmid containing the zalphal 1
receptor
cDNA (SEQ ID NO:7) was used as a template. PCR amplification of the zalphal 1
is fragment was performed as follows: Twenty five cycles at 94C for 0.5
minutes; five
cycles at 94 C for 10 seconds, 50 C for 30 seconds, 68 C for 45 seconds,
followed by a
4 C hold. The reaction was purified by chloroform/phenol extraction and
isopropanol
precipitation, and digested with Asp718 and BamHI (Gibco BRL) following
manufacturer's protocol. A band of the predicted size, 700 bp, was visualized
by 1%
20 agarose gel electrophoresis, excised and the DNA was purified using a
QiaexIITM
purification system (Qiagen) according the manufacturer's instructions.
The excised DNA was subcloned into plasmid pC4EE which had been
cut with BamHI and Asp718. The pC4zalphallCEE expression vector uses the
native
zalphal 1 signal peptide and attaches the Glu-Glu tag (SEQ ID NO:29) to the C-
25 terminus of the extracellular portion of the zalphal 1 polypeptide-
encoding
polynucleotide sequence. Plasmid pC4EE, is a mammalian expression vector
containing an expression cassette having the mouse metallothionein-1 promoter,
multiple restriction sites for insertion of coding sequences, a stop codon and
a human
growth hormone terminator. The plasmid also has an E. coil origin of
replication, a
30 mammalian selectable marker expression unit having an SV40 promoter,
enhancer and
origin of replication, a DHFR gene and the SV40 terminator.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
88
About 30 ng of the restriction digested zalphal 1 insert and about 12 ng
of the digested vector were ligated overnight at 16 C. One microliter of each
ligation
reaction was independently electroporated into Dill 0B competent cells (GIBCO
BRL,
Gaithersburg, MD) according to manufacturer's direction and plated onto LB
plates
containing 50 mg/ml ampicillin, and incubated overnight. Colonies were
screened by
restriction analysis of DNA prepared from 2 ml liquid cultures of individual
colonies.
The insert sequence of positive clones was verified by sequence analysis. A
large scale
plasmid preparation was done using a QIAGENO Maxi prep kit (Qiagen) according
to
manufacturer's instructions.
The same process was used to prepare the zalphal 1 soluble receptors
with a C-terminal his tag, composed of 6 His residues in a row; and a C-
terminal flag
(SEQ ID NO:37) tag, zalphal1CFLAG. To
prepare these constructs, the
aforementioned vector has either the HIS or the FLAG tag in place of the glu-
glu tag
(SEQ ID NO:29).
B. Mammalian Expression Construct of Soluble zalphall receptor zalphall-Fc4
An expression plasmid containing all or part of a polynucleotide
encoding zalphall was constructed via homologous recombination. The
extracellular
domain of the zalphal 1 receptor was fused to the Fe region derived from human
IgG,
called "Fc4" (SEQ ID NO:33) which contains a mutation so that it no longer
binds the
Fe receptor. A fragment of zalphal 1 cDNA was isolated using PCR that includes
the
polynucleotide sequence from extracellular domain of the zalphal 1 receptor.
The two
primers used in the production of the zalphall fragment were: (1) The primers
for PCR
each include from 5' to 3' end: 40 bp of the vector flanking sequence (5' of
the insert)
and 17 bp corresponding to the 5' end of the zalphal 1 extracellular domain
(SEQ ID
NO:32); and (2) 40 bp of the 5' end of the Fc4 polynucleotide sequence (SEQ ID
NO:33) and 17 bp corresponding to the 3 end of the zalphal 1 extracellular
domain
(SEQ ID NO:34). The fragment of Fc-4 for fusion with the zalphall was
generated by
PCR in a similar fashion. The two primers used in the production of the Fc4
fragment
were: (1) a 5' primer consisting of 40 bp of sequence from the 3' end of
zalphal 1
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
89
extracellular domain and 17 bp of the 5' end of Fc4 (SEQ ID NO:35); and (2) a
3'
primer consisting of 40 bp of vector sequence (3' of the insert) and 17 bp of
the 3' end
of Fc4 (SEQ ID NO:36).
PCR amplification of the each of the reactions described above was
performed as follows: one cycle at 94 C for 2 minutes; twenty-five cycles at
94 C for
30 seconds, 60 C for 30 seconds, 72 C for 1 minute; one cycle at 72 C for 5
minutes;
followed by a 4 C hold. Ten 1 of the 100 1 PCR reaction was run on a 0.8%
LMP
agarose gel (Seaplaque GTG) with 1 x TBE buffer for analysis. The remaining 90
1 of
the PCR reaction is precipitated with the addition of 5 pl 1 M NaC1 and 250 1
of
o absolute ethanol. The expression vector used was derived from the plasmid
pCZR199
derived from pZP9 (ATCC Deposit No. 98668), and was cut with SmaI (BRL). The
expression vector was derived from the plasmid pCZR199, and is a mammalian
expression vector containing an expression cassette having the CMV immediate
early
promoter, a consensus intron from the variable region of mouse immunoglobulin
heavy
chain locus, multiple restriction sites for insertion of coding sequences, a
stop codon
and a human growth hormone terminator. The expression vector also has an E.
coli
origin of replication, a mammalian selectable marker expression unit having an
SV40
promoter, enhancer and origin of replication, a DHFR gene and the SV40
terminator.
The expression vector used was constructed from pCZR199 by the replacement of
the
metallothionein promoter with the CMV immediate early promoter.
One hundred microliters of competent yeast cells (S. cerevisiae) were
combined with 10 1 containing approximately 1 g each of the zalphal 1 and
Fc4
inserts, and 100 ng of SmaI (BRL) digested expression vector and transferred
to a 0.2
cm electroporation cuvette. The yeast/DNA mixtures were electropulsed at 0.75
kV (5
kV/cm), "infinite" ohms, 25 F. To each cuvette is added 600 p 1 of 1.2 M
sorbitol and
the yeast was plated in two 300 1 aliquots onto two URA-D plates and
incubated at
C.
After about 48 hours, the Ura+ yeast transformants from a single plate
were resuspended in 1 ml H,0 and spun briefly to pellet the yeast cells. The
cell pellet
30 was resuspended in 1 ml of lysis buffer (2% Triton X-100, 1% SDS. 100 mM
NaC1, 10
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
mM Tris, pH 8.0, 1 mM EDTA). Five hundred microliters of the lysis mixture was
added to an Eppendorf tube containing 300 1 acid washed glass beads and 200
1
phenol-chloroform, vortexed for 1 minute intervals two or three times,
followed by a 5
minute spin in a Eppendorf centrifuge at maximum speed. Three hundred
microliters of
5 the aqueous phase was transferred to a fresh tube, and the DNA
precipitated with 600 1
ethanol (Et0H), followed by centrifugation for 10 minutes at 4 C. The DNA
pellet was
resuspended in 100 I H20.
Transformation of electrocompetent E. coli cells (DH10B, GibcoBRL) is
done with 0.5-2 ml yeast DNA prep and 40 ?Al of DH1OB cells. The cells were
10 electropulsed at 2.0 kV, 25 mF and 400 ohms. Following electroporation,
1 ml SOC
(2% BactoTM Tryptone (Difco, Detroit, MI), 0.5% yeast extract (Difco), 10 mM
NaCl,
2.5 mM KC1, 10 mM MgC12, 10 mM MgSO4, 20 mM glucose) was plated in 250 1
aliquots on four LB AMP plates (LB broth (Lennox), 1.8% Bacto Agar (Difco),
100
mg/L Ampicillin).
15 Individual clones harboring the correct expression construct for
zalphal 1 -Fc4 were identified by restriction digest to verify the presence of
the
zalphall-Fc4 insert and to confirm that the various DNA sequences have been
joined
correctly to one another. The insert of positive clones were subjected to
sequence
analysis. Larger scale plasmid DNA is isolated using the Qiagen Maxi kit
(Qiagen)
20 according to manufacturer's instructions.
Example 9
Transfection And Expression Of Zalphal 1 Soluble Receptor Polypeptides
A. Mammalian Expression of soluble zalphal 1 receptor zalphal 10EE,
zalphal1CFLG,
25 and zalphall CHI S
BHK 570 cells (ATCC No. CRL-10314), passage 27, were plated at
1.2X106 cells/well (6-well plate) in 800 I of serum free (SF) DMEM media
(DMEM,
Gibco/BRL High Glucose) (Gibco BRL, Gaithersburg, MD). The cells were
transfected with expression plasmids containing zalphal 1 CEE, zalphal 1 CFLG,
or
30 zalphal 1 CHIS described above (see, Example 8). using LipofectinTM
(Gibco BRL), in
serum free (SF) DMEM. Three micrograms of zalphal 1 CEE, zalphal1CFLG, or
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
91
zalphal 1 CH1S each were separately diluted into 1.5 ml tubes to a total final
volume of
100 tl SF DMEM. In separate tubes, 15 !al of LipofectinTM (Gibco BRL) was
mixed
with 100 1.11 of SF DMEM. The LipofectinTM mix was incubated at room
temperature
for 30-45 minutes then the DNA mix was added and allowed to incubate
approximately
10-15 minutes at room temperature.
The entire DNA: LipofectinTM mixture was added to the plated cells and
distributed evenly over them. The cells were incubated at 37 C for
approximately five
hours, then transferred to separate 150 mm MAXI plates in a final volume of 30
ml
DMEM/5% fetal bovine serum (FBS) (Hyclone, Logan, UT). The plates were
o incubated at 37 C, 5% CO2, overnight and the DNA: LipofectinTM mixture was
replaced with selection media (5% FBS/DMEM with 1 tM methotrexate (MTX))the
next day.
Approximately 10-12 days post-transfection, the plates were washed
with 10 ml SF DMEM. The wash media was aspirated and replaced with 7.25 ml
serum-free DMEM. Sterile Teflon meshes (Spectrum Medical Industries, Los
Angeles,
CA) pre-soaked in SF DMEM were then placed over the clonal cell colonies. A
sterile
nitrocellulose filter pre-soaked in SF DMEM was then placed over the mesh.
Orientation marks on the nitrocellulose were transferred to the culture dish.
The plates
were then incubated for 5-6 hours in a 37 C, 5% CO, incubator.
Following incubation, the filters/meshes were removed, and the media
aspirated and replaced with 5% FBS/DMEM with 1 tM MTX. The filters were then
blocked in 10% nonfat dry milk/Western A buffer (Western A: 50mM Tris pH 7.4.
5
mM EDTA, 0.05% NP-40, 150 mM NaC1 and 0.25% gelatin) for 15 minutes at room
temperature on a rotating shaker. The filters were then incubated with an anti-
Glu-Glu,
anti-FLAG , or anti-HIS antibody-HRP conjugates, respectively, in 2.5% nonfat
dry
milk/Western A buffer for one hour at room temperature on a rotating shaker.
The
filters were then washed three times at room temperature with Western A for 5-
10
minutes per wash. The filters were developed with ultra ECL reagent (Amersham
Corp., Arlington Heights, IL) according the manufacturer's directions and
visualized on
the Lumi-Imager (Roche Corp.)
Positive expressing clonal colonies were mechanically picked to 12-well
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
92
plates in one ml of 5%FCS/DMEM with 5 [tM MTX, then grown to confluence.
Conditioned media samples were then tested for expression levels via SDS-PAGE
and
Western anlaysis. The three highest expressing clones for each construct were
picked;
two out of three were frozen down as back up and one was expanded for
mycoplasma
testing and large-scale factory seeding.
B. Mammalian Expression of soluble zalphal 1 receptor zalphall-Fc4
BHK 570 cells (ATCC NO: CRL-10314) were plated in 10 cm tissue
culture dishes and allowed to grow to approximately 50 to 70% confluency
overnight at
o 37 C , 5% CO2, in DMEM/FBS media (DMEM, Gibco/BRL High Glucose, (Gibco
BRL, Gaithersburg, MD), 5% fetal bovine serum (Hyclone, Logan, UT), 1 mM L-
glutamine (JRH Biosciences, Lenexa, KS), 1 mM sodium pyruvate (Gibco BRL)).
The
cells were then transfected with the plasmid containing zalphall-Fc4 (see,
Example 8),
using LipofectamineTM (Gibco BRL), in serum free (SF) media formulation (DMEM,
10 mg/ml transferrin, 5 mg/ml insulin, 2 mg/ml fetuin, 1% L-glutamine and 1%
sodium
pyruvate). The plasmid containing zalphall-Fc4 was diluted into 15 ml tubes to
a total
final volume of 640 ml with SF media. 35 ml of Lipofectamine' (Gibco BRL) was
mixed with 605 ml of SF medium. The LipofectamineTM mix was added to the DNA
mix and allowed to incubate approximately 30 minutes at room temperature. Five
milliliters of SF media was added to the DNA:LipofectamineTM mixture. The
cells
were rinsed once with 5 ml of SF media, aspirated, and the DNA:Lipofectamine'
mixture is added. The cells were incubated at 37 C for five hours. then 6.4 ml
of
DMEM/10% FBS, 1% PSN media was added to each plate. The plates were incubated
at 37 C overnight and the DNA:LipofectamineTM mixture was replaced with fresh
5%
FBS/DMEM media the next day. On day 2 post-transfection, the cells were split
into
the selection media (DMEM/FBS media from above with the addition of 1 1.1M
methotrexate (Sigma Chemical Co., St. Louis, Mo.)) in 150 mm plates at 1:10.
1:20 and
1:50. The media on the cells was replaced with fresh selection media at day 5
post-
transfection. Approximately 10 days post-transfection, two 150 mm culture
dishes of
methotrexate resistant colonies from each transfection were trypsinized and
the cells are
pooled and plated into a T-162 flask and transferred to large scale culture.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
93
Example 10
Purification of zalphall soluble receptors from BHK 570 cells
A. Purification of zalphallCEE polypeptide from BHK 570
Unless otherwise noted, all operations were carried out at 4 C. The
following procedure was used for purifying zalphal 1 polypeptide containing C-
terminal GluGlu (EE) tags. Thirty liters of cell factory conditioned media was
concentrated to 1.6 liters with an Amicon S1OY3 spiral cartridge on a ProFlux
A30. A
Protease inhibitor solution was added to the concentrated 1.6 liters of cell
factory
conditioned media from transfected BHK 570 cells (see. Example 9) to final
concentrations of 2.5 mM ethylenediaminetetraacetic acid (EDTA, Sigma Chemical
Co.
St. Louis, MO), 0.003 mM leupeptin (Boehringer-Mannheim. Indianapolis, IN),
0.001
mM pepstatin (Boehringer-Mannheim) and 0.4 mM Pefabloc (Boehringer-Mannheim).
Samples were removed for analysis and the bulk volume was frozen at -80 C
until the
purification was started. Total target protein concentrations of the
concentrated cell
factory conditioned media was determined via SDS-PAGE and Western blot
analysis
with the anti-EE HRP conjugated antibody.
A 100 ml column of anti-EE G-Sepharose (prepared as described below)
was poured in a Waters AP-5, 5 cm x 10 cm glass column. The column was flow
packed and equilibrated on a BioCad Sprint (PerSeptive BioSystems, Framingham,
MA) with phosphate buffered saline (PBS) pH 7.4. The concentrated cell factory
conditioned media was thawed, 0.2 micron sterile filtered, pH adjusted to 7.4,
then
loaded on the column overnight with 1 ml/minute flow rate. The column was
washed
with 10 column volumes (CVs) of phosphate buffered saline (PBS, pH 7.4), then
plug
eluted with 200 ml of PBS (pH 6.0) containing 0.5 mg/ml EE peptide (Anaspec,
San
Jose, CA) at 5 ml/minute. The EE peptide used has the sequence EYMPME (SEQ ID
NO:29). The column was washed for 10 CVs with PBS. then eluted with 5 CVs of
0.2M glycine, pH 3Ø The pH of the glycine-eluted column was adjusted to 7.0
with 2
CVs of 5X PBS, then equilibrated in PBS (pH 7.4). Five ml fractions were
collected
over the entire elution chromatography and absorbance at 280 and 215 nM were
monitored; the pass through and wash pools were also saved and analyzed. The
EE-
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
94
polypeptide elution peak fractions were analyzed for the target protein via
SDS-PAGE
Silver staining and Western Blotting with the anti-EE HRP conjugated antibody.
The
polypeptide elution fractions of interest were pooled and concentrated from 60
ml to 5.0
ml using a 10,000 Dalton molecular weight cutoff membrane spin concentrator
(Millipore, Bedford, MA) according to the manufacturer's instructions.
To
separate zalphal 1 CEE from other co-purifying proteins, the
concentrated polypeptide elution pooled fractions were subjected to a POROS HQ-
50
(strong anion exchange resin from PerSeptive BioSystems, Framingham, MA) at pH
8Ø A 1.0 x 6.0 cm column was poured and flow packed on a BioCad Sprint. The
column was counter ion charged then equibrated in 20mM IRIS pH 8.0 (Iris
(Hydroxymethyl Aminomethane)). The sample was diluted 1:13 (to reduce the
ionic
strength of PBS) then loaded on the Poros HQ column at 5 ml/minute. The column
was
washed for 10 CVs with 20mM Iris pH 8.0 then eluted with a 40 CV gradient of
20
mM Iris/ 1 M sodium chloride (NaC1) at 10 ml/minute. 1.5 ml fractions were
collected
over the entire chromatography and absorbance at 280 and 215 nM were
monitored.
The elution peak fractions were analyzed via SDS-PAGE Silver staining.
Fractions of
interest were pooled and concentrated to 1.5-2 ml using a 10,000 Dalton
molecular
weight cutoff membrane spin concentrator (Millipore, Bedford, MA) according to
the
manufacturer's instructions.
To separate zalphal 1 CEE polypeptide from free EE peptide and any
contaminating co-purifying proteins, the pooled concentrated fractions were
subjected
to chromatography on a 1.5 x 90 cm Sephadex S200 (Pharmacia, Piscataway, NJ)
column equilibrated and loaded in PBS at a flow rate of 1.0 ml/min using a
BioCad
Sprint. 1.5 ml fractions were collected across the entire chromatography and
the
absorbance at 280 and 215 nM were monitored. The peak fractions were
characterized
via SDS-PAGE Silver staining, and only the most pure fractions were pooled.
This
material represented purified zalphallCEE polypeptide.
This purified material was finally subjected to a 4 ml ActiClean Etox
(Sterogene) column to remove any remaining endotoxins. The sample was passed
over
the PBS equilibrated gravity column four times then the column was washed with
a
single 3 ml volume of PBS, which was pooled with the "cleaned" sample. The
material
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
was then 0.2 micron sterile filtered and stored at -80 C until it was
aliquoted.
On Western blotted, Coomassie Blue and Silver stained SDS-PAGE
gels, the zalphal 1 CEE polypeptide was one major band of an apparent
molecular
weight of about 50,000 Daltons. The mobility of this band was the same on
reducing
5 and non-reducing gels.
The protein concentration of the purified material was performed by
BCA analysis (Pierce, Rockford, IL) and the protein was aliquoted, and stored
at -80 C
according to our standard procedures. On IEF (isoelectric focusing) gels the
protein
runs with a PI of less than 4.5. The concentration of zalphal 1 CEE
polypeptide was 1.0
10 mg/ml.
To prepare anti-EE Sepharose, a 100 ml bed volume of protein G-
Sepharose (Pharmacia, Piscataway, NJ) was washed 3 times with 100 ml of PBS
containing 0.02% sodium azide using a 500 ml Nalgene 0.45 micron filter unit.
The gel
15 was washed with 6.0 volumes of 200 mM triethanolamine, pH 8.2 (TEA,
Sigma, St.
Louis, MO), and an equal volume of EE antibody solution containing 900 mg of
antibody was added. After an overnight incubation at 4 C, unbound antibody was
removed by washing the resin with 5 volumes of 200 mM TEA as described above.
The resin was resuspended in 2 volumes of TEA, transferred to a suitable
container, and
20 dimethylpimilimidate-2HC1 (Pierce, Rockford, IL) dissolved in TEA, was
added to a
final concentration of 36 mg/ml of protein G-Sepharose gel. The gel was rocked
at
room temperature for 45 min and the liquid was removed using the filter unit
as
described above. Nonspecific sites on the gel were then blocked by incubating
for 10
min. at room temperature with 5 volumes of 20 mM ethanolamine in 200 mM TEA.
25 The gel was then washed with 5 volumes of PBS containing 0.02% sodium
azide and
stored in this solution at
B. Purification of zalphal 1CFLAG polypeptide from BFIK 570
Unless otherwise noted, all operations were carried out at 4 C. The
30 following procedure was used for purifying zalpha 1 1 polypeptide
containing C-
terminal FLAG (FLG) (Sigma-Aldrich Co.) tags. Thirty liters of cell factory
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
96
conditioned media was concentrated to 1.7 liters with an Amicon Si 0Y3 spiral
catridge
on a ProFlux A30. A Protease inhibitor solution was added to the 1.7 liters of
concentrated cell factory conditioned media from transfected BHK 570 cells
(see,
Example 9) to final concentrations of 2.5 mM ethylenediaminetetraacetic acid
(EDTA,
s Sigma Chemical Co. St. Louis, MO), 0.003 mM leupeptin (Boehringer-Mannheim,
Indianapolis, IN), 0.001 mM pepstatin (Boehringer-Mannheim) and 0.4 mM
Pefabloc
(Boehringer-Mannheim). Samples were removed for analysis and the bulk volume
was
frozen at -80 C until the purification was started. Total target protein
concentrations of
the cell factory conditioned media was determined via SDS-PAGE and Western
blot
analysis with the anti-FLAG (Kodak) HRP conjugated antibody. A 125 ml column
of
anti- FLAG M2-Agarose affinity gel (Sigma-Aldrich Co.) was poured in a Waters
AP-5, 5 cm x 10 cm glass column. The column was flow packed and equilibrated
on a
BioCad Sprint (PerSeptive BioSystems, Framingham, MA) with phosphate buffered
saline (PBS) pH 7.4. The concentrated cell factory conditioned media was
thawed, 0.2
micron sterile filtered, pH adjusted to 7.4, then loaded on the column
overnight with 1
ml/minute flow rate. The column was washed with 10 column volumes (CVs) of
phosphate buffered saline (PBS, pH 7.4), then plug eluted with 250 ml of PBS
(p11 6.0)
containing 0.5 mg/ml FLAG (Sigma-Aldrich Co.) peptide at 5 ml/minute. The
FLAG peptide used has the sequence DYKDDDDK (SEQ ID NO:37). The column
was washed for 10 CVs with PBS, then eluted with 5 CVs of 0.2M glycine, pH
3Ø
The pH of the glycine-eluted column was adjusted to 7.0 with 2 CVs of 5X PBS,
then
equilibrated in PBS (pH 7.4). Five ml fractions were collected over the entire
elution
chromatography and absorbence at 280 and 215 nM were monitored; the pass
through
and wash pools were also saved and analyzed. The FLAG -polypeptide elution
peak
fractions were analyzed for the target protein via SDS-PAGE Silver staining
and
Western Blotting with the anti-FLAG HRP conjugated antibody. The polypeptide
elution fractions of interest were pooled and concentrated from 80 ml to 12 ml
using a
10,000 Dalton molecular weight cutoff membrane spin concentrator (Millipore,
Bedford, MA) according to the manufacturer's instructions.
To separate zalphal1CFLG from other co-purifying proteins, the
polypeptide elution pooled fractions were subjected to a POROS HQ-50 (strong
anion
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
97
exchange resin from PerSeptive BioSystems, Framingham, MA) at pH 8Ø A 1.0 x
6.0
cm column was poured and flow packed on a BioCad Sprint. The column was
counter
ion charged then equilibrated in 20mM TRIS pH 8.0 (Tris (Hydroxymethyl
Aminomethane)). The sample was diluted 1:13 (to reduce the ionic strength of
PBS)
then loaded on the Poros HQ-50 column at 5 ml/minute. The column was washed
for
column volumes (CVs) with 20mM Tris pH 8.0 then eluted with a 40 CV gradient
of 20 mM Tris/ 1 M sodium chloride (NaCl) at 10 ml/minute. 1.5 ml fractions
were
collected over the entire chromatography and absorbance at 280 and 215 nM were
monitored. The elution peak fractions were analyzed via SDS-PAGE Silver
staining.
o Fractions of interest were pooled and concentrated to 1.5-2 ml using a
10,000 Dalton
molecular weight cutoff membrane spin concentrator (Millipore, Bedford, MA)
according to the manufacturer's instructions.
To separate zalphal 1CFLG polypeptide from free FLAG peptide and
any contaminating co-purifying proteins, the pooled concentrated fractions
were
subjected to chromatography on a 1.5 x 90 cm Sephacryl S200 (Pharmacia,
Piscataway,
NJ) column equilibrated and loaded in PBS at a flow rate of 1.0 ml/min using a
BioCad
Sprint. 1.5 ml fractions were collected across the entire chromatography and
the
absorbance at 280 and 215 nM were monitored. The peak fractions were
characterized
via SDS-PAGE Silver staining, and only the most pure fractions were pooled.
This
material represented purified zalphal 1CFLG polypeptide.
This purified material was finally sujectd to a 4 ml ActiClean Etox
(Sterogene) column to remove any remaining endotoxins. The sample was passed
over
the PBS equilibrated gravity column four times then the column was washed with
a
single 3 ml volume of PBS, which was pooled with the "cleaned" sample. The
material
was then 0.2 micron sterile filtered and stored at -80 C until it was
aliquoted.
On Western blotted, Coomassie Blue and Silver stained SDS-PAGE
gels, the zalphal 1CFLG polypeptide was one major band of an apparent
molecular
weight of about 50,000 Daltons. The mobility of this band was the same on
reducing
and non-reducing gels.
The protein concentration of the purified material was performed by
BCA analysis (Pierce, Rockford, IL) and the protein was aliquoted. and stored
at -80 C
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
98
according to our standard procedures. On IEF (isoelectric focusing) gels the
protein
runs with a PI of less than 4.5. The concentration of zalphal 1CFLG
polypeptide was
1.2 mg/ml.
C. Purification of zalphal 1 -Fc4 polypeptide from transfected BHK 570 cells
Unless otherwise noted, all operations were carried out at 4 C. The
following procedure was used for purifying zalphal 1 polypeptide containing C-
terminal fusion to human IgG/Fc (zalphall-Fc4; Examples 8 and 9). 12,000 ml of
conditioned media from BHK 570 cells transfected with zalphall-Fc4 (Example 9)
was
filtered through a 0.2 mm sterilizing filter and then supplemented with a
solution of
protease inhibitors, to final concentrations of, 0.001 mM leupeptin
(Boerhinger-
Mannheim, Indianapolis, IN), 0.001 mM pepstatin (Boerhinger-Mannheim) and 0.4
mM Pefabloc (Boerhinger-Mannheim). A protein G sepharose (6 ml bed volume,
Pharmacia Biotech) was packed and washed with 500 ml PBS (Gibco/BRL) The
supplemented conditioned media was passed over the column with a flow rate of
10
ml/minute, follOowed by washing with 1000 ml PBS (BRL/Gibco). zalphal 1-Fc4
was
eluted from the column with 0.1 M Glycine pH 3.5 and 2 ml fractions were
collected
directly into 0.2 ml 2M Tris pH 8.0, to adjust the final pH to 7.0 in the
fractions.
The eluted fractions were characterized by SDS-PAGE and western
blotting with anti-human Fe (Amersham) antibodies. Western blot analysis of
reducing
SDS-PAGE gels reveal an immunoreactive protein of about 80,000 KDa in
fractions 2-
10. Silver stained SDS-PAGE gels also revealed an 80,000 KDa zalpal 1 :Fc
polypeptide in fractions 2-10. Fractions 2-10 were pooled.
The protein concentration of the pooled fractions was performed by
BCA analysis (Pierce, Rockford, IL) and the material was aliquoted, and stored
at -
80 C according to our standard procedures. The concentration of the pooled
fractions
was 0.26 mg/ml.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
99
Example 11
Assay using zalphal 1 soluble receptor zalphallCEE, zalphal1CFLG and zalphall-
Fc4
(mutant) Soluble receptors in competitive inhibition assay
BaF3/Zalphal 1 cells were spun down and washed in mIL-3 free media.
The cells were spun and washed 3 times to ensure the removal of the mIL-3.
Cells
were then counted in a hemacytometer. Cells were plated in a 96-well format at
5000
cells per well in a volume of 100 tl per well using the mIL-3 free media.
Both conditioned media from the monkey spleen cell activation and the
human CD3+ selected cells, described in Example 5, were added in separate
experiments at 50%, 25%, 12.5%, 6.25%, 3.125%, 1.5%, 0.75% and 0.375%
concentrations, with or without zalphal 1 soluble receptors (CEE, C-flag, and
Fc4
constructs; See, Example 9 and 10) at 10 [ig/ml. The total assay volume was
200
The assay plates were incubated 37 C, 5% CO2 for 3 days at which time
Alamar Blue (Accumed) was added at 20 1/well. Plates were again incubated at
37 C,
5% CO2 for 24 hours. Plates were read on the FmaxTM plate reader (Molecular
Devices) as described in Example 2. Results demonstrated complete inhibition
of cell
growth from each of the different zalphal 1 soluble receptor constructs at
10H/ml,
confirming that the factor in each sample was specific for the zalphal 1
receptor.
Titration curves, diluting out the soluble receptors, were also run using
the above stated assay. Both the zalphal 1 CEE and zalphal 1CFLG soluble
zalphal 1
receptors were able to completely inhibit growth at concentrations as low as
20 ng/ml.
The mutant zalphall-Fc4 soluble zalphal 1 receptor was only as effective at
1.5 pg/ml.
Example 12
Secretion trap assay
A secretion trap assay was used to identify the cDNA by for the
zalphal 1 Ligand. The positive DNA pools obtained from the expression cloning
effort
described in Example 7.
The DNA pools of 250 clones were transfected into BHK cells in 96-
well format, and the condition medium were put into the proliferation assay
using
BaF3/zalphal 1 cells described in Examples 4 and Example 5. Several DNA pools
gave
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
100
positive activities which were repeated and neutralized with zalphal 1 soluble
receptors
(see Example 11).
One of the positive DNA pools, 45C5, was transfected into COS cells in
12-well format, using the LipofectamineTM method described below. A secretion
trap
assay was then performed using zalphal 1 soluble receptors (C-terminal Glu-Glu
tagged
either with or without biotinylation; C-terminal Flag tagged; or Fc4 zalphal 1
soluble
receptor fusions) (Example 9) to test the direct binding between potential
ligand of
zalphal 1 receptor in pool 45C5 and zalphal 1 soluble receptor (see below).
The result
was positive. Thus, the DNA of pool 45C5 was electroporated into E. coli, and
single
o colonies were picked into ten 96-well plates. Plates were shaken at 37 C
for 24 hours,
and then DNA minipreps (QiaPrepTM 96 Turbo Miniprep Kit; Qiagen) were prepared
in
96-well format using a TomTech Quadra 9600. The plasmid DNA was then pooled in
the format of rows and columns, transfected into COS cells, and then the
positive pools
were determined by secretion trap as described below.
COS Cell Transfections
The COS cell transfection was performed as follows: Mix 3u1 pooled
DNA and Sul LipofectamineTM in 92u1 serum free DMEM media (5 5mg sodium
pyruvate, 146mg L-glutamine, 5mg transferrin, 2.5mg insulin, 1 ug selenium and
5mg
fetuin in 500m1 DMEM), incubate at room temperature for 30 minutes and then
add
400u1 serum free DMEM media. Add this 500u1 mixture onto 1.5x105 COS
cells/well
plated on 12-well tissue culture plate and incubate for 5 hours at 37 C. Add
500u1 20%
FBS DMEM media (100 ml FBS, 55 mg sodium pyruvate and 146mg L-glutamine in
500m1 DMEM) and incubate overnight.
Secretion Trap Assay
The secretion trap was performed as follows: Media was rinsed off cells
with PBS and then fixied for 15 minutes with 1.8% Formaldehyde in PBS. Cells
were
then washed with TNT (0.1M Tris-HCL, 0.15M NaC1, and 0.05% Tween-20 in H70),
and permeated with 0.1% Triton-X in PBS for 15 minutes, and again washed with
TNT.
CA 02366921 2007-12-17
101 =
Cells were blockd for 1 hour with TNB (0.1M Tris-HCL, 0.15M NaC1 and 0.5%
Blocking Reagent (NEN Renaissance TSA-Direct Kit) in H20), and washed again
with
TNT. If using the biotinylated protein, the cells were blocked for 15 minute
incubations with Avidin and then Biotin (Vectoritabs) washing in-between with
TNT.
s
Depending on which soluble receptor was used;'-the cells were incubated for 1
hour
with: (A) 1-3 g/m1 zalphall soluble receptor zalphall-Fc4 fusion protein
(Example
10); (B) 3 g/m1 zalphall soluble receptor C-terminal FLAG tagged, zalphal
1CFLG
(Example 10); (C) 3 g/m1 zalphal 1 soluble receptor C-terminal. GluGlu
tagged,
= zalphal 1 CEE (Example 10); or (D) 3 g/m1 biotinylated zalphal 1 soluble
receptor
zalphallCEE in TNB. Cells were then washed with TNT. Depending on which
soluble
receptor was used, cells were incubated for another hour with: (A) 1:200
diluted goat-
anti-human Ig-HRP (Fe specific); (B) 1:1000 diluted M2-HRP; (C) 1:1000 diluted
anti-
. GluGlu antibody-HRP; or (D) 1:300 diluted streptavidin-HRP (NEN kit)
in TNB.
Again cells were washed with TNT.
Positive binding wqs detected with fluorescein tyratnide reagent diluted
1:50 in dilution buffer (NEN kit) E_Indincubated for 4-6 minutes, aid washed
with TNT.
Cells were preserved with VectashieldMoiniting Media (Vector Labs Burlingame,
CA).
diluted 1:5 in INT. Cells were visualized using a FITC filter on fluorescent
microscope.
= Example 13
Chromosomal Assignment and Placement of the gene for the zalphal 1 Ligand.
The gene for the zalplial 1 Ligand was mapped to chromosome 4 using
the commercially available version of the "Stanford G3 Radiation Hybrid
Mapping
Panel" (Research Genetics, Inc., Huntsville, AL). The "Stanford G3 RH Panel"
contains
DNAs from each of 83 radiation hybrid clones of the whole human genome, plus
two
control DNAs (the RM donor and the A3 recipient). A publicly available WNW
server
(maintained by the University of Stanford) allows chromosomal localization of
markers.
For the mapping of the zalphal 1 Ligand gene with the "Stanford G3 RH
Panel", 20 I reactions were set up in a 96-well microtiter plate (Stratagene,
La Jolla,
CA) and used in a "RoboCycler Gradient 96" thermal cycler (Stratagene). Each
of the 85
CA 02366921 2007-12-17
, .
=
102
PCR reactions consisted of 2 I 10X KlenTaq PCR reaction buffer (CLONTECH
Laboratories, Inc., Palo Alto, CA), 1.6 I dNTPs mix (2.5 mM each, PERKIN-
ELMER,
Foster City, CA), 1 I sense primer, ZC 22,050 (SEQ ID NO:42), 1 I antisense
primer,
ZC 22,051 (SEQ ID NO:43), 2 I "RediLoad" (Refearch Genetics, Inc.,
Huntsville, AL),
0.4 1 50X Advantage KlenTaq Polymerase Mix1Clontech Laboratories, Inc.), 25
rig of
DNA from an individual hybrid clone or control and ddH20 for a total volume of
20 IA.
The reactions were overlaid with an equal amount of mineral oil and sealed.
The PCR
cycler conditions were as follows: an initial 1 cycle 5 minute denaturation at
94 C, 35
.. =
= cycles of a 45 seconds denaturation at 94 C, 45 seconds annealing at 60 C
and 1 minute
AND 15 seconds extension at 72 C, followed by a final 1 cycle extension of 7
minutes
at 72 C. The reactions were separated by electrophoresis on a 2% agarose gel
(Life
Technologies, Gaithersburg, MD).
The results showed linkage of the zalphal 1 Ligand gene to the IL2
framework marker SHGC-12342 with a LOD score of >12 and at a distance of 6
cR 10000 (approximately 180 kb) from the milker. The use of surrounding
markers
positions the zalphal 1 ligiind - gene in = the 4q27 region on the integrated
LDB
=
chromosome 4 map (The Genetic Location Database, University of Southhampton).
=
Example 14
Identification and cloning of murine zalphal 1 Ligand
Using an EST Sequence to Obtain Full-length murine zalphal 1 Ligand
A. EST sequence of mouse zalphal 1 Ligand
By searching the database with human zalphal 1 Ligand cDNA sequence
(SEQ ID NO:!) as a query, a mouse EST (EST1483966) was identified as potential
partial sequence for mouse zalphal 1 Ligand. The EST1483966 represents a mouse
genomic fragment, in which a peptide sequence derived from two potential exons
shared high sequence identity with a peptide segment of the human zalphal 1
Ligand
(amino acid 13 (Ile) through amino acid 80 (Gin) of the SEQ ID NO:2).
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
103
B. PCR screen of mouse marathon cDNA panel
Eleven mouse Marathon cDNA (Clontech) samples were screened by
PCR described below. The mouse marathon cDNA samples were prepared from brain,
pancreas, kidney, placenta, salivary gland, skin, testis, uterus, embryo, and
spleen
tissues. They were made in-house using a MarathonTM cDNA Amplification Kit
(Clontech) according to manufacturer's instructions. Based on the EST
sequence, two
PCR primers, ZC22,056 (SEQ ID NO:44) and ZC22,057 (SEQ ID NO:45) were used to
identify a source of mouse zalphal 1 Ligand by PCR. The PCR reaction
conditions
were as follows: 94 C for 2 min.; 35 cycles at 94 C for 30 sec., 68 C for 2
min.;
followed by 68 C for 4 min.; then a 10 C soak. The PCR products were run on a
1%
agarose gel. A strong 150 bp band representing an amplified cDNA fragment was
visualized. This indicated mouse spleen marathon cDNA is the source for mouse
zalphal 1 Ligand cDNA cloning. The mouse spleen marathon cDNA contained a
positive cDNA which was subsequently identified by sequence analysis as a
partial
cDNA for mouse zalphal 1 Ligand.
C. A composite sequence for mouse full-length cDNA was generated by 5'- and 3.-
RACE
The 5' and 3' flanking sequences of the mouse zalphal 1 Ligand partial
cDNA sequence were obtained by 5' and 3' RACE amplification. Two rounds of
nested PCR amplification were performed with additional gene-specific oligo
primers
ZC22,205 (SEQ ID NO:46) and ZC22.206 (SEQ ID NO:47), ZC22,056 (SEQ ID
NO:44) and ZC22,057 (SEQ ID NO:45), and two adapter oligo primers ZC9,739 (SEQ
ID NO:48) and ZC9,719 (SEQ ID NO:49). The PCR reactions were run as follows:
94 C for 2 min; 35 cycles at 94 C for 30 sec, 68 C for 2 min; followed by 68 C
for 4
min; then a 10 C soak. The PCR products were run on a 1% agarose gel, and an
approximately 300 bp 5' RACE product and an approximately 800 bp 3' RACE
product were identified. These fragments were isolated using QiaquickTM gel
extraction
kit (Qiagen).
The purified PCR products were sequenced using the following primers:
ZC9,719 (SEQ ID NO:49), ZC22,205 (SEQ ID NO:46) and ZC22,206 (SEQ ID
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
104
NO:47). A preliminary composite full-length mouse zalphal 1 Ligand sequence
was
identified by combining the 5' and 3' RACE fragments. The full length mouse
clone
was isolated as described in Example 15 below.
Example 15
Isolation of mouse zalphall cDNA clone from an activated mouse spleen library
A. Murine Primary Source used to isolate mouse zalphal 1 Ligand
Mouse spleens from Balb/C mice, were collected and mashed between
frosted-end slides to create a cell suspension. The isolated primary mouse
cell yield
io was 6.4X108 cells prior to selection described below.
The spleen cells were suspended in 9.6 ml MACS buffer (PBS, 0.5%
EDTA, 2mM EDTA). 1.6 ml of cell suspension was removed and 0.4 ml CD90
(Thy1.2) microbeads (Miltenyi Biotec) added. The mixture was incubated for 15
min.
at 4 C. These cells labeled with CD90 beads were washed with 30 ml MACS
buffer,
and then resuspended in 2 ml MACS buffer.
A VS+ column (Miltenyi) was prepared according to the manufacturer's
instructions. The VS+ column was then placed in a VarioMACSTm magnetic field
(Miltenyi). The column was equilibrated with 5 ml MACS buffer. The isolated
primary
mouse cells were then applied to the column. The CD3 negative cells were
allowed to
pass through. The column was rinsed with 9 ml (3 X 3 ml) MACS buffer. The
column
was then removed from the magnet and placed over a 15 ml falcon tube. CD90+
cells
were eluted by adding 5 ml MACS buffer to the column and bound cells flushed
out
using the plunger provided by the manufacturer. The incubation of the cells
with the
CD90 magnetic beads, washes, and VS+ column steps (incubation through elution)
above were repeated once more. The resulting CD90+ fractions from the 2 column
separations were pooled. The yield of CD90+ selected mouse spleen cells were
1X108
total cells.
A sample of the pooled CD90+ selected mouse cells was removed for
staining and sorting on a fluorescent antibody cell sorter (FACS) to assess
their purity.
A PE-conjugated hamster anti-mouse CD3E antibody (PharMingen) was used for
staining and sorting the CD90+ selected cells. The mouse CD90+ selected cells
were
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
105
93% CD3+ cells, suggesting the cells were 93% T-cells.
The murine CD90+ selected cells were activated by incubating 3X10'
cells/ml in RPMI + 5% FBS + PMA 10 ng/ml and Ionomycin 0.5 [tg/m1 (Calbiochem)
for overnight at 37 C. The supernatant from these activated CD90+ selected
mouse
cells was tested for zalphal 1 Ligand activity as described below. Moreover,
the
activated CD90+ selected mouse cells were used to prepare a cDNA library, as
described in Example 16, below.
Example 16
Cloning of mouse zalphal 1 Ligand from a mouse CD90+ selected cell library
Screening of a primary mouse activated CD90+ selected cell cDNA
library revealed an isolated cDNA that is a novel member of the four-helix
bundle
cytokine family. This cDNA encoded the mouse ortholog of the human zalphal 1
Ligand. The cDNA was identified by hybridization screening.
A. The vector for CD90+ selected library construction
The vector, pZP7N was used for CD3+ selected library construction
(See Example 6A)
B. Preparation of primary mouse activated CD90+ selected cell cDNA library
Approximately 1.5X108 primary mouse CD90+ selected cells stimulated
in ionomycin/PMA (Example 15) were isolated by centrifugation. Total RNA was
isolated from the cell pellet, and converted to double stranded cDNA as
described in
Example 6B. This DNA was subsequently transfected into BHK cells, as described
in
Example 6B, and proliferation was assessed using an -Alamar blue- fluorescence
assay
(Example 2B).
For the purpose of screening the library by secretion trap cloning, a
complex, amplified form of the library was needed to transfect COS-7 cells.
4.8
million clones were plated on 110 15cm LB-agar plates supplemented with 100
pg/m1
ampicillin, 10 jig/ml methicillin. After growing the plates overnight at 37 C
the
bacteria were harvested by scraping and pelleted. Plasmid DNA was extracted
from the
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
106
pelleted bacteria using a Nucleobond-gigaTM (Clonetech) following the
manufacturer's
instructions. This plasmid was then used to transfect COS-7 cells on slides
and
screened using the secretion trap technique described below (Example 17).
C. Screening the activated mouse cDNA library
Approximately 5X105 clones were plated on 10 LB/Amp Maxi plates.
The colonies were lifted, denatured, neutralized, and cross-linked using the
standard
procedure (Sambrook, J. et al. supra.). Fifty nanograms of the 300 bp 5' RACE
PCR
fragment (Example 14) was labeled with 32P using Prime-Itr RmT random primer
labeling kit (Stratagene). The 10 filters were hybridized with this labeled
probe at 65 C
overnight using ExpressHybTM Hybridization Solution (Clontech). The filters
were then
washed sequentially at 60 C for 1 hour three times with 0.2xSSC (30 mM NaC1, 3
mM
sodium citrate, pH 7.0), 0.1% SDS; and then at 65 C for 1 hour. The filters
were
exposed at -80 C overnight, and the X-ray film were developed. Agar plugs
containing
the positive colonies were pulled, and the clones plated on 10-cm LB/Amp
plates. The
colonies were then filter-lifted and hybridized again following the same
procedure
described above.
One DNA clone, named M1 1 L/pZP7. was isolated and sequenced using
the following primers: ZC14,063 (SEQ ID NO:50), ZC5,020 (SEQ ID NO:51),
ZC22,421 (SEQ ID NO:52), ZC22,604 (SEQ ID NO:53), and ZC22,641 (SEQ ID
NO:54). The polynucleotide sequence of this clone is full-length mouse zalphal
1
Ligand (SEQ ID NO:55) and consistent with the composite sequence obtained from
5'
and 3' RACE products. The corresponding amino acid sequence for the mouse
zalphal 1 Ligand is shown in SEQ ID NO:56.
Example 17
Mouse zalphal 1 Ligand binds to human zalphal 1 soluble receptor in secretion
trap
assay
The DNA for mouse clone M1 1 L/pZP7 was transfected into COS cells,
and the binding of human zalphal 1 soluble receptor zalphal 1-Fc4 (Example
10C) to
the transfected COS cells was tested by a secretion trap assay (Example 12).
The assay
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
107
confirmed that the mouse zalphall Ligand binds to human zalphal 1 soluble
receptor.
The COS cell transfection was performed as per example 12 using 0.7
jig M1 1 L/pZP7 DNA (Example 16) in 3 1.
The secretion trap was performed as as per example 12 using 1 ug/m1
zalphal 1 soluble receptor Fc4 fusion protein (Example 10C) in TNB, and 1:200
diluted
goat-anti-human Ig-HRP (Fe specific) in TNB for the detectable antibody.
Positive
binding of the soluble human zalphal 1 receptor to the prepared fixed cells
was detected
with fluorescein tyramide reagent as per Example 12. Cells were preserved and
visualized according to Example 12.
The positive result indicated the mouse zalphal 1 Ligand binds to human
zalphal 1 soluble receptor.
Example 18
Expression of mouse zalphal 1 Ligand in mammalian cells
A. Construction of expression vector Ml1L/pZP9
An expression vector was prepared for the expression of the mouse
zalphal 1 Ligand in mammalian cells. A 500 bp PCR generated zalphal 1 Ligand
DNA
fragment was created using ZC22,283 (SEQ ID NO:57) and ZC22,284 (SEQ ID
NO:58) as PCR primers to amplify the coding region of mouse zalphal 1 Ligand
and
add XhoI and XbaI restriction sites. The mouse zalphal 1 Ligand clone
Ml1L/pZP7
(Example 16) was used as a template. The PCR reaction conditions were as
follows:
94 C for 2 min.; 25 cycles at 94 C for 30 sec., 68 C for 2 min.; followed by
68 C for 4
min.; then a 10 C soak. A band of the predicted size, about 500 bp, was
visualized by
1% agarose gel electrophoresis, excised and the DNA was purified using a
QiaexIIT'
purification system (Qiagen) according to the manufacturer's instructions. The
purified
DNA was digested with XhoI and XbaI (Boehringer Mannheim) at 37 C for 2 hours.
Then the DNA was gel isolated and purified following the above procedure.
The excised DNA was subcloned into plasmid pZP9 which was cut with
XhoI and XbaI (Boehringer Mannheim). Plasmid pZP9 is a mammalian expression
vector containing an expression cassette having the mouse metallothionein-1
(MT-1)
promoter, multiple restriction sites for insertion of coding sequences, and a
human
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
108
growth hormone terminator. The plasmid also has an E. coli origin of
replication, a
mammalian selective marker expression unit having an SV40 promoter, enhancer
and
origin of replication, a DHFR gene, and the SV40 terminator.
About 30 ng of the restriction digested mouse zalphall Ligand fragment
and about 10 ng of the digested pZP9 vector were ligated at room temperature
for 2
hours. Two u.g of ligation reaction was transformed into INVaF' competent
cells
(Invitrogen) according to manufacturer's protocol and plated onto LB plates
containing
50 ug/m1 ampicillin, and incubated at 37 C overnight. Colonies were screened
by
restriction analysis using XhoI and XbaI (Boerhinger Mannheim) of DNA prepared
o from liquid cultures of individual colonies. The insert sequence of
positive clones was
verified by sequence analysis to be the mouse zalphal 1 Ligand sequence. A
large scale
plasmid preparation was done using a Qiagen Maxi prep kit (Qiagen) according
to
manufacturer's instruction. The expression vector that contains mouse zalphal
1
Ligand was named Ml1L/pZP9.
B. Mammalian expression of mouse zalphal 1 Ligand
BHK 570 cells (ATCC No: CRL-10314) were plated in 10 cm tissue
culture dishes and allowed to grow to approximately 20% confluence overnight
at 37 C,
5% CO2, in DMEM/FBS media (DMEM, Gibco/BRL High Glucose media; Gibco
BRL, Gaithersburg, MD), 5% fetal bovine serum (Hyclone, Logan, UT), 1 mM L-
glutamine (JRH Biosciences, Lenexa, KS), 1 mM sodium pyruvate (Gibco BRL)).
The
cells were then transfected with the plasmid Ml 1L/pZP9 (Example 18A) using a
mammalian stable CaPO4 transfection kit (Stratagene) according to the
manufacturer's
instructions.
One day after transfection, the cells were split 1:10 and 1:20 into the
selection media (DMEM/FBS media with the addition of 1 1tM methotrexate (Sigma
Chemical Co., St. Louis, MO)) in 150 mm plates. The media on the cells was
replaced
with fresh selection media at day 5 post-transfection. Approximately 10 days
post-
transfection, methotrexate resistant colonies were trypsinized and the cells
pooled and
plated into large scale culture flasks. Once the cells were grown to
approximately 90%
confluence, they were rinsed with PBS three times, and cultured with serum-
free
ESTEP2 media (DMEM (Gibco BRL), 0.11 g/1 Na Pyruvate, 3.7g/1 NaHCO,, 2.5 mg/1
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
109
insulin, 5 mg/1 transferrin, pH7.0) conditioned media. The conditioned media
were
collected three days later, and put into a BaF3 proliferation assay using
Alamar Blue,
described in Example 19 below.
Example 19
Mouse zalphal 1 Ligand activates human zalphal 1 receptor in BaF3 assay using
Alamar Blue
Proliferation of BaF3/zalphal 1 cells (Example 4. and 5B) was assessed
using serum-free conditioned media from BHK cells expressing mouse zalphal 1
Ligand (Example 18).
BaF3/Zalphal 1 cells were spun down, washed and plated in mIL-3 free
media as described in Example 5B.
Proliferation of the BaF3/Zalphal 1 cells was assessed using serum-free
conditioned media from BHK cells expressing mouse zalphal 1 Ligand (Example
18).
Conditioned media was diluted with mIL-3 free media to: 50%, 25%, 12.5%,
6.25%,
3.125%, 1.5%, 0.75% and 0.375% concentrations. The proliferation assay was
performed as per Example 5B.
Results confirmed the proliferative response of the BaF3/Zalphal 1 cells
to mouse zalphal 1 Ligand. The response, as measured, was approximately 5-fold
over
background at the 50% concentration.
Example 20
Zalphal 1 Ligand activates human zalphal 1 receptor in luciferase assay
A. Construction of BaF3/KZ134/zalphal1 cell line
The KZ134 plasmid was constructed with complementary
oligonucleotides ZC12,749 (SEQ ID NO:59) and ZC12,748 (SEQ ID NO:60) that
contain STAT transcription factor binding elements from 4 genes. A modified c-
fos Sis
inducible element (m67SIE. or hSIE) (Sadowski, H. et al.. Science 261:1739-
1744,
1993), the p21 SIE1 from the p21 WAF1 gene (Chin, Y. et al.. Science 272:719-
722,
1996), the mammary gland response element of the 13-casein gene (Schmitt-Ney,
M. et
al., Mol. Cell. Biol. 11:3745-3755, 1991), and a STAT inducible element of the
Fcg RI
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
110
gene, (Seidel, H. et al., Proc. Natl. Acad. Sci. 92:3041-3045, 1995).
These
oligonucleotides contain Asp718-XhoI compatible ends and were ligated, using
standard methods, into a recipient firefly luciferase reporter vector with a c-
fos
promoter (Poulsen, L.K. et al., J. Biol. Chem. 273:6229-6232, 1998) digested
with the
s same enzymes and containing a neomycin selectable marker. The KZ134
plasmid was
used to stably transfect BaF3 cells, using standard transfection and selection
methods,
to make the BaF3/KZ134 cell line.
A stable BaF3/KZ134 indicator cell line, expressing the full-length
zalphal 1 receptor was constructed as per Example 2A, using about 30 g of the
zalphal 1 expression vector, described in Example 3. Clones were diluted,
plated and
selected using standard techniques. Clones were screened by luciferase assay
(see
Example 20B, below) using the human zalphal 1 Ligand conditioned media as an
inducer. Clones with the highest luciferase response (via STAT luciferase) and
the
lowest background were selected. A stable transfectant cell line was selected.
The cell
line was called BaF3/KZ134/zalphal 1.
B. Human and mouse Zalphal 1 Ligand activates human zalphal 1 receptor in
BaF3/KZ134/Zalphal 1 luciferase assay
BaF3/KZ134/Zalphal1 cells were spun down and washed in mIL-3 free
media. The cells were spun and washed 3 times to ensure removal of mIL-3.
Cells
were then counted in a hemacytometer. Cells were plated in a 96-well format at
about
30,000 cells per well in a volume of 100 pl per well using the mIL-3 free
media. The
same procedure was used for untransfected BaF3/KZ134 cells for use as a
control in the
subsequent assay.
STAT activation of the BaF3/KZ134/Zalphal 1 cells was assessed using
conditioned media from (1) BHK570 cells transfected with the human zalphal 1
Ligand
(Example 7) or (2) BHK570 cells transfected with the mouse zalphal 1 Ligand
(Example 18), or (4) mIL-3 free media to measure media-only control response.
Conditioned media was diluted with RPMI mIL-3 free media to 50%, 25%, 12.5%,
6.25%, 3.125%, 1.5%, 0.75% and 0.375% concentrations. 100 pl of the diluted
conditioned media was added to the BaF3/KZ134/Zalphal 1 cells. The assay using
the
conditioned media was done in parallel on untransfected BaF3/KZ134 cells as a
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
111
control. The total assay volume was 200 Ill. The assay plates were incubated
at 37 C,
5% CO2 for 24 hours at which time the cells were pelleted by centrifugation at
2000
rpm for 10 min., and the media was aspirated and 25 ?Al of lysis buffer
(Promega) was
added. After 10 minutes at room temperature, the plates were measured for
activation
of the STAT reporter construct by reading them on a luminometer (Labsystems
Luminoskan, model RS) which added 40 pi of luciferase assay substrate
(Promega) at a
five second integration.
Results confirmed the STAT reporter response of the
BaF3/K2134/Zalphal1 cells to the human zalphal 1 Ligand. The response, as
measured, was approximately 50 fold over media-only control at the 50%
concentration. STAT activation in response to human zalphal 1 Ligand was
absent in
the untransfected BaF3/KZ134 control cells, showing that the response is
mediated
through the Zalphal 1 receptor.
Results also confirmed the STAT reporter response of the
BaF3/KZ134/Zalphal 1 cells to the mouse zalphal I Ligand. The response, as
measured, was approximately 40 fold over media-only control at the 50%
concentration. Moreover, STAT activation in response to mouse zalphal 1 Ligand
was
evident (about 5-fold) on the untransfected BaF/KZ134 control cells,
suggesting that
the murine BaF3 cells may have endogenous mouse receptor.
Example 21
Mouse zalphal I Ligand is active in mouse bone marrow assay
A. Isolation of Non-adherent Low Density Marrow Cells:
Fresh mouse femur aspirate (marrow) was obtained from 6-10 week old
male Balb/C or C57BL/6 mice. The marrow was then washed with RPMI+10% FBS
(JRH, Lenexa KS; Hyclone, Logan UT) and suspended in RPMI+10% FBS as a whole
marrow cell suspension. The whole marrow cell suspension was then subjected to
a
density gradient (Nycoprep, 1.077, Animal: Gibco BRL) to enrich for low
density, mostly
mononuclear, cells as follows: The whole marrow cell suspension (About 8 ml)
was
carefully pipeted on top of about 5 ml Nycoprep gradient solution in a 15 ml
conical tube.
and then centrifuged at 600X g for 20 minutes. The interface layer, containing
the low
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
112
density mononuclear cells, was then removed, washed with excess RPMI+10% FBS,
and
pelleted by centrifugation at 400X g for 5-10 minutes. This pellet was
resuspended in
RPM! +10% FBS and plated in a T-75 flask at approximately 106 cells/ml, and
incubated
at 37 C 5% CO2 for approximately 2 hours. The resulting cells in suspension
were Non-
Adherent Low Density (NA LD) Marrow Cells.
B. 96-Well Assay
NA LD Mouse Marrow Cells were plated at 25,000 to 45,000 cells/well
in 96 well tissue culture plates in RPMI +10% FBS + lng/mL mouse Stem Cell
Factor
(mSCF) (R&D Systems, Minneapolis, MN), plus 5% conditioned medium from one of
the following: (1) BHK 570 cells expressing mouse zalphal 1 Ligand (Example
18), (2)
BHK 570 cells expressing human zalphal 1 Ligand (Example 7), or (3) control
BHK
570 cells containing vector and not expressing either Ligand. These cells were
then
subjected to a variety of cytokine treatments to test for expansion or
differentiation of
hematopoietic cells from the marrow. To test, the plated NA LD mouse marrow
cells
were subjected to human Interleukin-15 (hIL-15) (R&D Systems), or one of a
panel of
other cytokines (R&D Systems). Serial dilution of hI1-15, or the other
cytokines, were
tested, with 2-fold serial dilution from about 50 ng/ml down to about 6025
ng/ml
concentration. After 8 to 12 days the 96-well assays were scored for cell
proliferation
by Alamar blue assay as described in Example 5B.
C. Results from the 96-well NA LD Mouse Marrow assay
Conditioned media from the BHK cells expressing both mouse and
human zalphal 1 Ligand acted in synergy with hIL-15 to promote the expansion
of a
population of hematopoietic cells in the NA LD mouse marrow. This expansion of
hematopoietic cells was not shown with control BHK conditioned medium plus IL-
15.
The population hematopoietic cells expanded by the mouse zalphal 1 Ligand with
hIL-
15, and those hematopoietic cells expanded by the human zalphal 1 Ligand with
hIL-
15. were further propagated in cell culture. These hematopoietic cells were
stained with
a Phycoerythrin labeled anti-Pan NK cell antibody (Pharmingen) and subjected
to flow
cytometry analysis, which demonstrated that the expanded cells stained
positively for
this natural killer (NK) cell marker.
The same 96-well assay was run, using fresh human marrow cells
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
113
bought from Poietic Technologies, Gaithersburg, MD. Again, in conjunction with
IL-
15, the mouse and human zalphal 1 Ligand expanded a hematopoietic cell
population
that stained positively for the NK cell marker using the antibody disclosed
above.
Example 22
Constructs for generating human zalphal 1 Ligand Transgenic Mice
A. Construct for expressing human zalphal 1 Ligand from the liver-specific
MT-1
promoter
Oligonucleotides were designed to generate a PCR fragment containing
io a consensus Kozak sequence and the human zalphal 1 Ligand coding region.
These
oligonucleotides were designed with an FseI site at the 5' end and an AscI
site at the 3'
end to facilitate cloning into (a) pMT12-8, our standard transgenic vector, or
(b)
pKF051, a lymphoid-specific transgenic vector (Example 22B).
PCR reactions were carried out with 200 ng human zalphal 1 Ligand
template (Example 7) and oligonucleotides ZC22,143 (SEQ ID NO:61) and ZC22,144
(SEQ ID NO:62). PCR reaction conditions were as follows: 95 C for 5 minutes,
wherein AdvantageTM cDNA polymerase (Clontech) was added; 15 cycles of 95 C
for
60 seconds, 60 C for 60 seconds, and 72 C for 90 seconds; and 72 C for 7
minutes.
PCR products were separated by agarose gel electrophoresis and purified using
a
QiaQuickTM (Qiagen) gel extraction kit. The isolated, 488 bp, DNA fragment was
digested with FseI and AscI (Boerhinger-Mannheim), ethanol precipitated and
ligated
into pMT12-8 previously digested with FseI and AscI. The pMT12-8 plasmid,
designed for expressing a gene of interest in liver and other tissues in
transgenic mice,
contains an expression cassette flanked by 10 kb of MT-1 5 DNA and 7 kb of MT-
1 3'
DNA. The expression cassette comprises the MT-1 promoter. the rat insulin II
intron, a
polylinker for the insertion of the desired clone, and the human growth
hormone (hGH)
poly A sequence.
About one microliter of each ligation reaction was electroporated into
DH1OB ElectroMaxTm competent cells (GIBCO BRL, Gaithersburg, MD) according to
manufacturer's direction and plated onto LB plates containing 100 1.1.g/m1
ampicillin,
and incubated overnight. Colonies were picked and grown in LB media containing
100
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
114
jig/ml ampicillin. Miniprep DNA was prepared from the picked clones and
screened
for the human zalphal 1 Ligand insert by restriction digestion with EcoRI
alone, or FseI
and AscI combined, and subsequent agarose gel electrophoresis. Maxipreps of
the
correct pMT-human zalphal 1 Ligand were performed. A SalI fragment containing
with 5' and 3' flanking sequences, the MT-1 promoter, the rat insulin II
intron, human
zalphal 1 Ligand cDNA and the hGH poly A sequence was prepared to be used for
microinjection into fertilized murine oocytes. Microinjection and production
of
transgenic mice were produced as described in Hogan, B. et al. Manipulating
the Mouse
Embryo, 2"d ed., Cold Spring Harbor Laboratory Press, NY, 1994.
B.
Construct for expressing human zalphal 1 Ligand from the lymphoid-specific
NALCK promoter
Oligonucleotides were designed to generate a PCR fragment containing
a consensus Kozak sequence and the human zalphal 1 Ligand coding region. These
oligonucleotides were designed with an FseI site at the 5' end and an AscI
site at the 3'
end to facilitate cloning into pKF051, a lymphoid-specific transgenic vector.
PCR reactions were carried out with 200 ng human zalphal 1 Ligand
template (Example 7) and oligonucleotides ZC22,143 (SEQ ID NO:61) and ZC22,144
(SEQ ID NO:62). A PCR reaction was performed using AdvantageTM cDNA
polymerase (Clontech) under the following conditions: 95 C for 5 minutes; 15
cycles of
95 C for 60 seconds, 60 C for 60 seconds, and 72 C for 90 seconds; and 72 C
for 7
minutes. PCR products purified as described above. The isolated, 488 bp, DNA
fragment was digested with FseI and AscI (Boerhinger-Mannheim), ethanol
precipitated and ligated into pKF051 previously digested with FseI and AscI.
The
pKF051 transgenic vector is derived from p1026X B.M.,
et al., EMBO J.
16:7019-31, 1997) and contains the T cell-specific lck proximal promoter, the
BIT cell-
specific immunoglobulin f..t heavy chain enhancer. a polylinker for the
insertion of the
desired clone, and a mutated hGH gene that encodes an inactive growth hormone
protein (providing 3' introns and a polyadenylation signal).
About one microliter of each ligation reaction was electroporated,
plated, clones picked and screened for the human zalphall Ligand insert by
restriction
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
115
digestion as described above. A correct clone of pKF051-zalphal1 Ligand was
verified by sequencing, and a maxiprep of this clone was performed. A Noll
fragment,
containing the lck proximal promoter and immunoglobulin enhancer (EIALCK),
zalphal 1 Ligand cDNA, and the mutated hGH gene was prepared to be used for
microinjection into fertilized murine oocytes.
Example 23
Mouse Zalphal 1 Ligand Tissue Distribution
Murine Multiple Tissue Northern Blots (Mouse, Mouse Embryo,
io Clontech; MB1010, MB1012 Origene) were probed to determine the tissue
distribution
of murine zalphal 1 Ligand expression. An approximately 484 bp PCR derived
probe
was amplified using the plasmid M 11L/pZP7 (Example 16) as a template and
oligonucleotide ZC22283 (SEQ ID NO:57) and ZC22284 (SEQ ID NO:58) as primers.
The PCR amplification was carried out as follows: 1 cycle at 94 C for 1.0
minutes; 35
cycles of 94 C for 30 seconds, 50 C for 30 seconds, and 72 C for 30 seconds;
followed
by 1 cycle at 72 C for 10 minutes. The PCR products were visualized by agarose
gel
electrophoresis and the approximately 484 bp PCR product was purified using a
Gel
Extraction Kit (Qiagen) according to manufacturer's instructions. The probe
was
radioactively labeled using the REDIPRIMETm labeling kit (Amersham) according
to
the manufacturer's instructions. The probe was purified using a NUCTRAPTm push
column (Stratagene). EXPRESSHYBTM (Clontech) solution was used for
prehybridization and as a hybridizing solution for the Northern blots.
Hybridization
took place overnight at 65 C using 106 cpm/ml of labeled probe. The blots were
then
washed three times in 2X SSC and 0.1% SDS at room temperature, followed by 2
washes in 0.1X SSC and 0.1% SDS at 55 C. Two transcripts of approximately 1.2
and
3.5 kb were seen in testis. The upper transcript only was seen in thymus.
A murine RNA Master Dot Blot (Clontech) that contained RNAs from
various tissues that were normalized to 8 housekeeping genes was also probed
and
hybridized as described above. Expression was seen in testis.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
116
Example 24
Purification of untagged human and murine zalphall Ligand from BHK 570
Unless other wise stated, all operations were carried out at 4 C. The
following procedure was used for purifying human and murine zalphal 1 Ligand
from
conditioned media from BHK 570 cells transfected with a construct expressing
either
the human zalphal 1 Ligand (Example 25) or the mouse zalphall Ligand
(M11L/pZP9)
(Example 18). The conditioned media was concentrated by standard techniques.
Concentrated conditioned media (CM) was sterile filtered through 0.45 and 0.22
micron
filters. The media was then diluted to low ionic strength (< 2 mS) in 0.01 M
HEPES
(JRH Biosciences, Lenexa, KS) at pH 7Ø The low ionic strength diluted CM was
then
loaded onto a 10 X 66 mm (6 ml) Poros HS 50 column (PerSeptive BioSystems,
Framingham, MA) overnight at 4 ml/min using a BioCAD SPRINT (Perceptive
BioSystems). The column was washed for 10-20 column volumes (CV) with 0.01 M
HEPES pH7Ø The bound proteins were then step eluted with 1 M NaC1
(Mallinckrodt,
Paris, KY) in 0.01 M HEPES pH 7.0 at 5 ml/min; two ml fractions were collected
over
the entire chromatography and absorbence at 280 and 215 nM were monitored.
Peak
absorbence fractions were analyzed by bioassay and by SDS-PAGE Silver (Geno
Technology, St. Louis, MO) and Coomassie (Sigma, St. Louis, MO) staining. Peak
fractions were pooled, sterile filtered and diluted to < 19 mS with Phosphate
buffered
saline (PBS, Gibco BRL)at pH 7.2.
The diluted sample was then loaded at 2 ml/min using a BioCad
SPRINT, onto either a 0.8 ml Poros AL column that had zalphal 1CFLAG soluble
receptor (Example 10B) or zalphal 1 -Fc4 fusion soluble receptor (Example 10C)
immobilized on the resin (see, below). The column was then washed with at
least 20
CV of PBS at 10 ml/min. The column was then rapidly eluted with a 600 .1.1
injection
of 0.1 M glycine (Aminoacetic Acid; Glycocol, Spectrum, Gardena, CA) pH 2.5 at
a
flow rate of 10 ml/min with PBS on a BioCAD 700E. The 1 ml fractions were
collected for 6 seconds each and immediately pH neutralized with 55 [11 of 2 M
TRIS
(Tris (Hydroxymethyl) Aminomethane, EM Science, Gibbstown, NJ) pH 8.8. The
absorbence at 280 and 215 nM were monitored over the entire chromatography.
The peak fractions were analyzed by bioassay and by SDS-PAGE Silver
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
117
(Geno Technology) and Coomassie (Sigma) staining. Two bands, approximately 24
kD
and 18 kD, were seen on both Silver and Coomassie gels for mouse zalphal 1
Ligand.
A single band, at approximately 18 kD, was seen on both Silver and Coomassie
gels for
human zalphal 1 Ligand.
Immobilization of human zalphal 1 soluble receptor polypeptides on POROS AL
media
Poros AL columns having immobilized zalphal 1 CFLAG soluble
receptor (Example 10B) or zalphal 1 -Fc4 fusion soluble receptor (Example 10C)
were
prepared. Approximately 3 mg of zalphal 1CFLAG soluble receptor and
approximately
10 mg of zalphall-Fc4 fusion soluble receptor were used. All operations were
carried
out at room temperature on a BioCAD 700E. A 4.5 X 50 mm column with the POROS
AL media was flow packed in 2 M NaC1 according to manufactures specifications.
The
column was then equilibrated in 1.1 M Na2SO4/50 mM NaPhosphate pH 7.2. The
receptor was concentrated to 4 mg/ml using a Millipore 30 MWKO spin
concentrator
a.s then diluted 1:1 in 1.1 M Na2SO4/50 mM NaPhosphate pH 7.2. The column
was
flowed at 2 ml/min in 1.1 M Na2SO4/50 mM NaPhosphate pH 7.2 and 100 til
injections of the diluted ligand were made ever 9 CVs until a steady state of
saturation,
or break through, was reached. A 62 CV gradient was then run from 1.1 M
Na2SO4/50
mM NaPhosphate pH 7.2., to 550 mM Na2SO4/50 mM NaPhosphate pH 7.2/5 mg/ml
Sodium Cyanoborohydride. The column was then held for about 2 hours to
complete
the immobilization chemistry. The column was then equilibrated in 0.2 M TRIS
pH
7.2/5 mg/ml Sodium Cyanoborohydride and allowed to rest for about 1 hour to
cap the
column. Finally the column was equilibrated in PBS/0.02% Sodium Azide, and
stored
at 4 C until needed. Prior to use, the column was pre-eluted with 0.1 M
glycine to
ensure that non-specific proteins were removed and that the column was not
leaching
the immobilized human zalphal 1 soluble receptor.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
118
Example 25
Expression of human zalphal 1 Ligand in mammalian cells
A. Construction of expression vector PZMP11/zalphal 1Lig
An expression plasmid containing all or part of a polynucleotide
s encoding human zalphal 1 Ligand was constructed via homologous
recombination. A
fragment of human zalphal 1 Ligand cDNA (SEQ ID NO:63) was isolated using PCR.
Two primers were used in the production of the human zalphal 1 Ligand fragment
in a
PCR reaction: (1) Primer ZC22,052 (SEQ ID NO:64), containing 40 bp of the
vector
flanking sequence and 17 bp corresponding to the amino terminus of the human
o zalphal 1 Ligand, and (2) primer ZC22,053 (SEQ ID NO:65), containing 40
bp of the 3'
end corresponding to the flanking vector sequence and 17 bp corresponding to
the
carboxyl terminus of the human zalphal 1 Ligand. The PCR Reaction conditions
were
as follows: 1 cycle at 94 C for 2.0 minutes; 25 cycles of 94 C for 30 seconds,
60 C for
30 seconds, and 72 C for 30 seconds; followed by 1 cycle at 72 C for 5
minutes; 4 C
ls soak. Ten 1 of the 100 1 PCR reaction was run on a 0.8% LMP agarose
gel
(Seaplaque GTG) with 1 x TBE buffer for analysis, and the expected
approximately
560 bp fragment seen. The remaining 90 I of PCR reaction was precipitated
with the
addition of 5 1 1 M NaC1 and 250 I of absolute ethanol to be used for
recombining
onto the recipient vector pZMP11 as described below. The recipient plasmid
pZMP11
20 was previously cut with SmaI.
Plasmid pZMP11 was derived from the plasmid pCZR199 (described
herein, e.g., Example 8). The plasmid pCZR199 is a mammalian expression vector
containing an expression cassette having the CMV immediate early promoter, a
consensus intron from the variable region of mouse immunoglobulin heavy chain
locus,
25 multiple restriction sites for insertion of coding sequences, a stop
codon and a human
growth hormone terminator. The plasmid also has an E. coli origin of
replication, a
mammalian selectable marker expression unit having an SV40 promoter, enhancer
and
origin of replication, a DHFR gene and the SV40 terminator. The vector pZMP11
was
constructed from pCZR199 and includes the replacement of the metallothionein
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
119
promoter with the CMV immediate early promoter, and Kozac sequences at the 5'
end
of the open reading frame.
One hundred microliters of competent yeast cells (S. cerevisiae) were
combined with 10 Ill of a mixture containing approximately 1 p.g of the human
zalphal1 Ligand insert, and 100 ng of SmaI digested pZMP11 vector, and
transferred to
a 0.2 cm electroporation cuvette. The yeast/DNA mixtures were electropulsed at
0.75
kV (5 kV/cm), infinite ohms, 25 p.F. To each cuvette was added 600 11.1 of 1.2
M
sorbitol and the yeast was then plated in two 300 1.t1 aliquots onto two URA-D
plates
and incubated at 30 C.
After about 48 hours, the Ura+ yeast transformants from a single plate
were resuspended in 1 ml H20 and spun briefly to pellet the yeast cells. The
cell pellet
was resuspended in 1 ml of lysis buffer (2% Triton X-100, 1% SDS, 100 mM NaCl,
10
mM Tris, pH 8.0, 1 mM EDTA). Five hundred microliters of the lysis mixture was
added to an Eppendorf tube containing 300 1 acid washed glass beads and 200
[1,1
phenol-chloroform, vortexed for 1 minute intervals two or three times,
followed by a 5
minute spin in a Eppendorf centrifuge at maximum speed. Three hundred
microliters of
the aqueous phase was transferred to a fresh tube, and the DNA precipitated
with 600 I.11
ethanol (Et0H), followed by centrifugation for 10 minutes at 4 C. The DNA
pellet was
resuspended in 100111 H2O.
Transformation of electrocompetent E. coli cells (DH10B, GibcoBRL)
was done with 0.5-2 ml yeast DNA prep and 40 IA of DH1OB cells. The cells were
electropulsed at 2.0 kV, 25 mF and 400 ohms. Following electroporation, 1 ml
SOC
(2% BactoTM Tryptone (Difco, Detroit, MI). 0.5% yeast extract (Difco), 10 mM
NaC1,
2.5 mM KC1, 10 mM MgC12. 10 mM MgSO4, 20 mM glucose) was plated in 250 ?al
aliquots on four LB AMP plates (LB broth (Lennox), 1.8% BactoTM Agar (Difco),
100
mg/L Ampicillin).
Individual clones harboring the correct expression construct for human
zalphal 1 Ligand were identified by restriction digest to verify the presence
of the insert
and to confirm that the various DNA sequences have been joined correctly to
one
another. The insert of positive clones were subjected to sequence analysis.
Larger
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
120
scale plasmid DNA was isolated using the Qiagen Maxi kit (Qiagen) according to
manufacturer's instruction
B. Mammalian Expression of human zalphal 1 Ligand
BHK 570 cells (ATCC NO: CRL-10314) were plated in 10 cm tissue
culture dishes and allowed to grow to approximately 50 to 70% confluence
overnight at
37 C, 5% CO2, in DMEM/FBS media (DMEM, Gibco/BRL High Glucose, (Gibco
BRL, Gaithersburg, MD), 5% fetal bovine serum (Hyclone, Logan, UT), 1 mM L-
glutamine (JRH Biosciences, Lenexa, KS), 1 mM sodium pyruvate (Gibco BRL). The
cells were then transfected with the plasmid PZMP11/zalphal 1 Lig (Example
25A),
using LipofectamineTM (Gibco BRL), in serum free (SF) media formulation (DMEM,
10 mg/ml transferrin, 5 mg/ml insulin, 2 mg/ml fetuin, 1% L-glutamine and 1%
sodium
pyruvate). Zalphal 1 -Fc4/pZMP6 (Example 8B) was diluted into 15 ml tubes to a
total
final volume of 640 1 with SF media. 35 1 of LipofectamineTM (Gibco BRL) was
mixed with 605 p,1 of SF medium. The LipofectamineTM mix was added to the DNA
mix and allowed to incubate approximately 30 minutes at room temperature. Five
milliliters of SF media was added to the DNA:LipofectamineTM mixture. The
cells
were rinsed once with 5 ml of SF media, aspirated, and the DNA:LipofectamineTM
mixture was added. The cells were incubated at 37 C for five hours, then 6.4
ml of
DMEM/10% FBS, 1% PSN media was added to each plate. The plates were incubated
at 37 C overnight and the DNA:Lipofectaminem mixture was replaced with fresh
5%
FBS/DMEM media the next day. On day 5 post-transfection, the cells were split
into
T-162 flask in selection medium (DMEM/ 5% FBS, 1% L-GLU, 1% NaPyr).
Approximately 10 days post-transfection, two 150 mm culture dishes of
methotrexate
resistant colonies from each transfection were trypsinized and the cells are
pooled and
plated into a T-162 flask and transferred to large scale culture. Conditioned
media from
large scale culture was used to purify human zalpha 1 1 Ligand polypeptide as
described
in Example 24.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
121
Example 26
Construct for generating mouse zalphall Ligand Transgenic Mice
A.
Construct for expressing mouse zalphal 1 Ligand from the lymphoid-specific
EuLCK promoter
Oligonucleotides were designed to generate a PCR fragment containing
a consensus Kozak sequence and the mouse zalphal 1 Ligand coding region. These
oligonucleotides were designed with an FseI site at the 5' end and an AscI
site at the 3'
end to facilitate cloning into: (a) pKF051, a lymphoid-specific transgenic
vector, or (b)
pTG12-8, our standard transgenic vector.
PCR reactions were carried out with 200 ng mouse zalphal 1 Ligand
template (SEQ ID NO:55; Example 16) and oligonucleotides ZC23,115 (SEQ ID
NO:66) and ZC23,116 (SEQ ID NO:67). A PCR reaction was performed using
AdvantageTM cDNA polymerase (Clontech) under the PCR conditions described in
Example 22B. PCR product was isolated as described in Example 22B. The
isolated,
440 bp DNA fragment was digested and ligated into pKF051 previously digested
with
FseI and AscI, as described in Example 22B.
About one microliter of each ligation reaction was electroporated,
plated, clones picked and screened for the human zalphal 1 Ligand insert by
restriction
digestion as described in Example 22. A correct pKF051-zalphal 1 Ligand clone
was
verified by sequencing, and a maxiprep of this clone was performed. A NotI
fragment,
containing the lck proximal promoter, immunoglobulin p. enhancer, zalphal 1
Ligand
cDNA, and the mutated hGH gene was prepared to be used for microinjection into
fertilized murine oocytes.
B. Construct for expressing mouse zalphal 1 Ligand from the liver-specific MT-
1
promoter
This same mouse zalphal 1 Ligand insert from Example26A, was
subcloned into the pTG12-8 vector, as described in Example 22A. For this
construct,
about 10 mg of the pKF051-zalphal 1 Ligand maxiprep DNA was digested with FseI
and AscI combined, ethanol precipitated, and the mouse zalphal 1 Ligand
fragment was
purified as described in Example 22. This fragment was then ligated into pTG12-
8
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
122
which had been previously digested with FseI and AscI, as described in Example
22A.
Electroporation, screening of clones and a maxiprep was performed as described
in
Example 22. A Sall fragment containing 5' and 3' flanking sequences, the MT-1
promoter, the rat insulin II intron, mouse zalphal 1 Ligand cDNA and the hGH
poly A
sequence was prepared to be used for microinjection into fertilized murine
oocytes.
Example 27
Mouse zalphall-ligand Polyclonal Antibodies
Polyclonal antibodies were prepared by immunizing 2 female New
a.o Zealand white rabbits with the purified recombinant protein
muzalphallL/MBP-6H
(Example 32). The rabbits were each given an initial intraperitoneal (ip)
injection of
200 mg of purified protein in Complete Freund's Adjuvant followed by booster
ip
injections of 100 mg peptide in Incomplete Freund's Adjuvant every three
weeks.
Seven to ten days after the administration of the second booster injection (3
total
injections), the animals were bled and the serum was collected. The animals
were then
boosted and bled every three weeks.
The muzalphal 1L/MBP-6H specific rabbit serum was pre-adsorbed of
anti-MBP antibodies using a CNBr-SEPHAROSE 4B protein column (Pharmacia
LKB) that was prepared using 10 mg of purified recombinant maltose binding
protein
(MBP) per gram of CNBr-SEPHAROSE. Recombinant MBP was made and purified on
an amylose column in house, using methods well known in the art. The muzalphal
1 -
ligand-specific polyclonal antibodies were affinity purified from the rabbit
serum using
a CNBr-SEPHAROSE 4B protein column (Pharmacia LKB) that was prepared using 10
mg of the specific antigen purified recombinant protein muzalphal 1L/MBP-6H
(Example 32) followed by 20X dialysis in PBS overnight. Muzalphal 1 -ligand-
specific
antibodies were characterized by ELISA using lug/ml of the purified
recombinant
proteins muzalphallUMBP-614 (Example 32) or huzalphal 1L-MBP/61-I (Example 32)
as antibody targets. The lower limit of detection (LLD) of the rabbit anti-
muzalphal 1 L/MBP-6H affinity purified antibody is 100 pg/ml on its specific
purified
recombinant antigen muzalphal 1 L/MBP-6H and 500 pg/ml on purified recombinant
huzalphal1L-MBP/6H.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
123
Example 28
Construction of mammalian expression vector and large-scale Human zalphall
Ligand
expression in CHO DG44 cells
A mammalian expression vector for human zalphal 1 Ligand (SEQ ID
NO:1) designed to add a Sall site at the 5' end and a PmeI site to the 3' end
of the
cDNA, was constructed via amplification by PCR from a plasmid containing human
zalphal 1 Ligand (Example 7) with oligonucleotide primers, ZC22,054 (SEQ ID
NO:70) and ZC22,055 (SEQ ID NO:71). The PCR reaction conditions were as
follows:
94 C for 4 min.; 25 cycles of 94 C for 45 sec., 50 C for 45 seconds, and 72 C
for 3
min.; and 72 C for 10 minutes. The PCR product was isolated as described
herein, and
cut with SalI and PmeI then ligated to plasmid pDC312 previously cut at the
appropriate restriction sites in the polylinker, using standard methods
described herein.
The plasmid pDC312 is described in Morris, A. et al., "Expression Augmenting
Sequence Element (EASE) isolated from Chinese Hamster Ovary Cells," in Animal
Cell Technology, Carrondo, MJT et al (eds.) (1997) Kluwer Academic Publisers,
The
Netherlands, p. 529-534.
The ligated vector was transfected into suspension-adapted CHO DG44
(in house Novo Nordisk, Denmark) cells by lipofection using
LipofectaminePlusTM
reagent (Gibco/BRL) according to manufacturer's instructions. Transfectants
were
selected on PFCHO medium (JRH, Lenexa, Kansas) free of thymidine, and
hypoxanthine, followed by selection on 200 nM methotrexate (Sigma, St. Louis,
MO).
The methotrexate resistant cells were cloned by dilution and assayed for
production of
zalphal 1 Ligand by a BaF3 activity assay (Example 5B).
A productive clone was scaled up and grown in a 7 to 20 liter bioreactor
(Applikon Bioreactors, Schiedam, The Netherlands) in PFCHO medium to produce
material for purification (Example 29).
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
124
Example 29
Large-scale purification of untagged human and murine zalphal 1 Ligand from
BHK
and CHO mammalian expression cell lines.
A. CHO expressed human zalphal 1 Ligand
Unless otherwise stated, all operations were carried out at 4 C. The
following procedure was used for purifying human zalphal 1 Ligand from at
least 30
liters of CHO conditioned media (see, Example 28). Concentrated or non-
concentrated
conditioned media (CM) was sterile filtered through 0.45 and 0.22 micron
filters. The
conditioned media was then buffered with 0.01 M MES (Fluka BioChemika,
io Switzerland)) and the pH adjusted to 6.0, and then loaded onto a 50 X
100 mm (196 ml)
Poros 50 HS column (strong cation exchanger from PerSeptive BioSystems,
Framingham, MA) overnight at 4-10 ml/min. using a BioCAD SPRINT (Perceptive
BioSystems). The column was washed for 10-20 column volumes (CV) with 0.01 M
MES/0.130 M NaC1 (Mallincicrodt, Paris, KY) pH 6Ø The bound proteins were
then
eluted with a 0.130 M to 1 M NaC1 10 CV gradient in 0.01 M MES pH 6.0 at 30
ml/min.; 25 ml fractions were collected over the entire chromatography and
absorbence
at 280 and 215 nM were monitored. Peak absorbence fractions were analyzed by
SDS-
PAGE Silver (Geno Technology, St. Louis , MO), Coomassie (Sigma, St. Louis,
MO)
staining and Western immunological blotting using antibodies against the human
zalphal 1 Ligand (Example 33 and Example 34).
Peak fractions were pooled then concentrated in a stirred cell
concentrator on a YM10 membrane (Millipore/Amicon, Bedford, MA) to a nominal
volume (1-10 m1). The sample was then loaded on an appropriate Sephacryl S-200
(Pharmacia, Uppsala, Sweden) high resolution size exclusion column (52-600 ml)
equilibrated in PBS (Gibco BRL) at flow rates 1-2 ml/ml; 1-2 ml fractions were
collected over the entire chromatography and absorbence at 280 and 215 nM were
monitored. Peak fractions were analyzed by SDS-PAGE Silver (Geno Technology,
St.
Louis, MO), and Coomassie (Sigma, St. Louis, MO) staining.
The fractions of interest were pooled and concentrated with Millipore 5
kD MWKO centrifugal spin concentrators to a minimal volume. The final product
was
then analyzed by SDS PAGE Coomassie staining (Sigma, St. Louis, MO). Western
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
125
Immunological blotting, N-terminal sequencing, Amino Acid Analysis, and BCA
(Pierce, Rockford, Illinois) for protein purity and concentration.
B. BHK 570 expressed murine zalphal 1 Ligand
Unless otherwise stated, all operations were carried out at 4 C. The
following procedure was used for purifying murine zalphal 1 Ligand from BHK
conditioned media (Example 18). Concentrated or non-concentrated conditioned
media
(CM) was sterile filtered through 0.45 and 0.22 micron filters. The media was
then
buffered with 0.01 M MES (Fluka BioChemika, Switzerland)) and the pH adjusted
to
o 6Ø The CM was analyzed, loaded and eluted on an AS column as described in
Example 29A.
Fractions of interest were pooled then concentrated in a stirred cell
concentrator as in Example 29A, to a volume of 20-30 ml. The pH was adjusted
to 7.0
then the sample was loaded onto either a 0.8 ml Poros AL column that had about
3 mg
is of
zalphal 1CFLAG tagged soluble receptor (Example 10B) or one with aboutl 0 mg
of
zalphal 1 -Fc4 fusion receptor (Example 10C) immobilized on the resin (see
method
below) at 1 ml/min on a BioCAD SPRINT. The column was then washed with at
least
20 CV of 0.3 M NaCl/PBS (Gibco BRL) /0.01 M MES at 10 ml/min. The column was
then rapid eluted with a 600 p1 injection of 0.1 M glycine (Aminoacetic Acid;
20
Glycocol, Spectrum, Gardena, CA) pH 2.5 at a flow rate of 10 ml/min with PBS
on a
BioCAD SPRINT. The 1 ml fractions were collected for 6 seconds each and
immediately pH neutralized with 55 p1 of 2 M TRIS pH 8.8 (Tris (Hydroxymethyl)
Aminomethane, EM Science, Gibbstown, NJ). The absorbence at 280 and 215 nM
were monitored over the entire chromatography. The peak fractions were
analyzed
25 SDS-
PAGE Silver (Geno Technology, St. Louis , MO), Coomasise (Sigma, St. Louis,
MO) staining and Western Immunological blotting as above.
Peak fractions were pooled then concentrated in a stirred cell
concentrator as in Example 29A to a minimal volume (1-10 ml). The sample was
then
loaded, equilibrated and analyzed as in Example 29A, on an appropriate
Sephacryl S-
30 200
(Pharmacia) high resolution size exclusion column. Peak fractions were
analyzed
by SDS-PAGE Silver (Geno Technology, St. Louis , MO), and Coomasise (Sigma,
St.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
126
Louis, MO) staining. The fractions of interest were pooled and concentrated
and
analyzed as in Example 29A.
C. Protein immobilization on POROS AL media
All operations were carried out at room temperature on a BioCAD 700E.
Flow packed a 4.5 X 50 mm column with the POROS AL media in 2 M NaC1
according to manufactures specifications. The column was then equilibrated in
1.1 M
Na2SO4 and 50 mM NaPhosphate pH 7.2. The receptor was concentrated to 4 mg/ml
using a Millipore 30 kD MWKO centrifugal spin concentrator then diluted 1:1 in
1.1 M
Na2SO4 and 50 mM NaPhosphate pH 7.2. The column was flowed at 2 ml/min in 1.1
M Na2SO4 and 50 mM NaPhosphate pH 7.2 and 100 p 1 injections of the diluted
ligand
were made ever 9 CVs until a steady state of saturation or break through was
reached.
A 62 CV gradient was then ran from 1.1 M Na2SO4 and 50 mM NaPhosphate pH 7.2
to 550 mM Na2SO4 and 50 mM NaPhosphate pH 7.2 with 5 mg/ml Sodium
is Cyanoborohydride. Column was held for 2 hr. to complete the immobilization
chemistry. The column was then equilibrated in 0.2 M TRIS pH 7.2 with 5 mg/ml
Sodium Cyanoborohydride and allowed to rest for 1 hr. Finally the column was
equilibrated in PBS with 0.02% Sodium Azide, then stored at 4 C until needed.
Prior
to use, the column was pre-eluted with 0.1 M glycine to ensure that non-
specific
proteins were removed and the column is not leaching the immobilized receptor.
Example 30
Expression Vector Construction, Expression and Purification Of Untagged Human
And
Murine Zalphal 1 Ligand From Baculovirus.
A. Construct for Expressing human zalphal 1 Ligand in Baculovirus
An expression vector, pzalphal1L. was prepared to express Human
zalphal 1 Ligand polypeptides in insect cells. A 517 bp fragment containing
sequence
for Human zalphal 1 Ligand and encoded BamH1 and XhoI restriction sites on the
5'
and 3' ends respectively, was generated by PCR amplification from a plasmid
containing human zalphal 1 Ligand cDNA (Example 7) using primers ZC23,444 (SEQ
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
127
ID NO:74) and ZC23,445 (SEQ ID NO:75). The PCR reaction conditions were as
follows: 1 cycle of 94C for 4 minutes, followed by 25 cycles of 94 C for 45
seconds,
50 C for 45 seconds, and 72 C for 2 minutes; 1 cycle at 72 C for 10 min;
followed by a
4 C soak. The fragment was visualized by gel electrophoresis (1% SeaPlaque/1%
NuSieve). The band was excised, diluted to 0.5% agarose with 2 mM MgC12,
melted at
65 C, digested with BamH1 and XhoI (Boerhinger Mannheim), and ligated into an
BamH1/XhoI digested baculovirus expression vector, pZBV3L. The pZBV3L vector
is
a modification of the pFastBac 1 Tm (Life Technologies) expression vector,
where the
polyhedron promoter has been removed and replaced with the late activating
Basic
o Protein Promoter. About 14 nanograms of the restriction digested zalphall
Ligand
insert and about 40 ng of the corresponding vector were ligated overnight at
16 C.
The ligation mix was diluted 3 fold in TE (10 mM Tris-HC1, pH 7.5 and
1 mM EDTA) and about 4 fmol of the diluted ligation mix was transformed into
D115ot
Library Efficiency competent cells (Life Technologies) according to
manufacturer's
direction by heat shock for 45 seconds in a 42 C waterbath. The transformed
DNA and
cells were diluted in 450 I of SOC media (2% BactoTM Tryptone, 0.5% Bacto
Yeast
Extract, 10 ml 1M NaCl, 1.5 mM KC1, 10 mM MgC12, 10 mM MgSO4 and 20 mM
glucose) and plated onto LB plates containing 100 ilg/m1 ampicillin. Clones
were
analyzed by restriction digests and 1 1 of the positive clone was transformed
into 20 1
DH10Bac Max Efficiency competent cells (GIBCO-BRL, Gaithersburg, MD)
according to manufacturer's instruction, by heat shock as described above. The
transformed cells were then diluted in 980 1 SOC media (2% BactoTM Tryptone,
0.5%
BactoTM Yeast Extract, 10 ml 1M NaC1, 1.5 mM KC1, 10 mM MgC12, 10 mM MgSO4
and 20 mM glucose) out grown in shaking incubator at 37 C for four hours and
plated
onto Luria Agar plates containing 50 g/m1 kanamycin, 7 g/m1 gentamicin (Life
Technologies), 10 g/m1 tetracycline, IPTG (Pharmacia Biotech) and Bluo-Gal
(Life
Technologies). The plated cells were incubated for 48 hours at 37 C. A color
selection
was used to identify those cells having Human zalphal 1 Ligand encoding donor
insert
that had incorporated into the plasmid (referred to as a "bacmid"). Those
colonies,
which were white in color, were picked for analysis. Human zalphal 1 Ligand
Bacmid
DNA was isolated from positive colonies using the QiaVac Miniprep8 system
(Qiagen)
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
128
according the manufacturer's directions. Clones were screened for the correct
insert by
amplifying DNA using primers to the transposable element in the bacmid via PCR
using primers ZC447 (SEQ ID NO:76) and ZC976 (SEQ ID NO:77). The PCR
reaction conditions were as follows: 35 cycles of 94 C for 45 seconds, 50 C
for 45
s seconds, and 72 C for 5 minutes; 1 cycle at 72 C for 10 min.; followed by
4C soak.
The PCR product was run on a 1% agarose gel to check the insert size. Those
clones
having the correct insert were used to transfect Spodoptera frugiperda (Sf9)
cells.
B. Expression and generation of material for purification of human zalphal 1
Ligand
from baculovirus
Sf9 cells were seeded at 5 x 106 cells per 35 mm plate and allowed to
attach for 1 hour at 27 C. Five microliters of human zalphal 1 Ligand bacmid
DNA
(above) was diluted with 100 IA Sf-900 IT SFM (Life Technologies). Six IA of
CellFECTIN Reagent (Life Technologies) was diluted with 100 1 Sf-900 II SFM.
The
bacmid DNA and lipid solutions were gently mixed and incubated 30-45 minutes
at
room temperature. The media from one plate of cells were aspirated, the cells
were
washed 1X with 2 ml fresh Sf-900 II SFM media. Eight hundred microliters of Sf-
900
II SFM was added to the lipid-DNA mixture. The wash media was aspirated and
the
DNA-lipid mix added to the cells. The cells were incubated at 27 C for 4-5
hours. The
DNA-lipid mix was aspirated and 2 ml of Sf-900 II media was added to each
plate. The
plates were incubated at 27 C, 90% humidity, for 96 hours after which the
virus was
harvested.
For Primary Amplification 519 cells were grown in 50 ml Sf-900 II SFM
in a 125 ml shake flask to an approximate density of 0.41-0.52 x 105 cells/ml.
They
were then infected with 150 I of the virus stock from above and incubated at
27 C for
3 days after which time the virus was harvested according to standard methods
known
in the art. A 500 I sample submitted for activity in a BaF3 assay (Example 5)
to show
that it was biologically active.
For Secondary Amplification Sf9 cells were grown in 1L of Sf-900 II
SFM in a 2800 ml shake flask to an approximate density of 0.5 x 10 cells/ml.
It was
infected with 500u1 of the Primary viral stock from above and incubated at 27
C for 4
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
129
days after which time the virus was harvested according to standard methods
known in
the art. Virus was titered and grown up large scale for purification of the
baculovirus-
produced human zalphal 1 Ligand (huzalphal1L-Bv), as described in Example 30C
and
Example 30D, below.
C. Large-scaled purification of baculovirus expressed human/murine zalphal 1
Ligand
Unless otherwise stated, all operations were carried out at 4 C. The
following procedure was used for purifying human zalphal 1 Ligand (huzalphal1L-
Bv)
from BV conditioned media (Example 30B). Conditioned media (CM) was sterile
filtered through 0.45 and 0.22 micron filters, then buffered with 0.01 M MES
(Fluka
BioChemika, Switzerland)) and the pH adjusted to 6.0 The CM was then loaded
onto a
POROS 50 HS column and run, fractions collected, analyzed, as described in
Example
29A.
The above peak fractions were pooled, concentrated run on a high
resolution size exclusion column, and analyzed as described in Example 29A.
The fractions of interest from the size exclusion column were pooled and
concentrated with 5 kD MWCO Millipore centrifugal spin concentrators to a
minimal
volume. The final product was then analyzed by SDS-PAGE Coomassie (Sigma, St.
Louis, MO), Western immunological blotting, N-terminal sequencing, Amino Acid
Analysis, and CB (Pierce, Rockford, Illinois) for protein purity and
concentration as
described in Example 29A. Bulk protein was stored at -80 C.
D. Small scale (<2 mg) Purification of Baculovirus-expressed human/murine
zalphal 1
Ligand
Unless other wise stated, all operations were carried out at 4 C. The
following procedure was used for purifying < 2 mg of human or murine zalphal 1
Ligand from BV conditioned media. The CM was filtered, buffered and pH
adjusted as
in Example 30C. The CM was then loaded, eluted and the POROS 50 HS
chromatography was analyzed as in Example 30C.
Fractions were pooled then concentrated via diafiltration in a stirred cell
concentrator on a YM10 membrane (10 kD MWCO) (Millipore/Amicon. Bedford,
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
130
MA) to a nominal volume (20-30 m1). The pH was adjusted to 7.0 then the sample
was
loaded onto either a 0.8 ml Poros AL column that had about 3 mg of zalphal
1CFLAG
soluble receptor (Example 10B) or one with about10 mg of zalphal 1 -Fc4 fusion
soluble receptor (Example 10C) immobilized on the resin (see method in example
29C)
at 1 ml/min on a BioCad SPRINT. The column was then washed with at least 20 CV
of
0.3 M NaCl/PBS(Gibco BRL) /0.01 M MES at 10 ml/min. The column was then rapid
eluted with a 600 .1 injection of 0.1 M glycine (Aminoacetic Acid; Glycocol,
Spectrum, Gardena, CA) pH 2.5 at a flow rate of 10 ml/min with PBS on a BioCAD
SPRINT. The 1 ml fractions were collected for 6 seconds each and immediately
pH
neutralized with 55 111 of 2 M TRIS (Tris (Hydroxymethyl) Aminomethane, EM
Science, Gibbstown, NJ) pH 8.8. The absorbence at 280 and 215 nM were
monitored
over the entire chromatography. Fractions were analyzed as above.
Peak fractions were pooled then concentrated via diafiltration in a stirred
cell concentrator on a YM10 membrane (10 kD MWCO) (Millipore/Amicon, Bedford,
MA) to 1-2 ml. The sample was then loaded on an appropriate Sephacryl S-200
(Pharmacia, Uppsala, Sweden) high resolution size exclusion column
equilibrated in
PBS (Gibco BRL) at an optimal flow rate; fractions were collected over the
entire
chromatography and absorbence at 280 and 215 nM were monitored. Fractions were
analyzed as above.
The fractions of interest were pooled and concentrated with 5 Kd
MWCO Millipore centrifugal spin concentrators to a nominal volume. The final
product was then analyzed by SDS-PAGE Coomassie (Sigma, St. Louis, MO),
Western
immunological blotting, N-terminal sequencing, Amino Acid Analysis, and BCA
(Pierce, Rockford, Illinois) for protein purity and concentration. Bulk
protein stored as
described above.
E. Construct for Expressing mouse zalphal 1 Ligand in Baculovirus: pzalphal 1
lig.M
An expression vector, pzalphallLM. was prepared to express Murine
zalphal 1 Ligand polypeptides in insect cells. A 413 bp fragment containing
sequence
for Murine zalphal 1 Ligand and encoded BspE 1 and Xbal restriction sites on
the 5'
and 3' ends, respectively, was generated by PCR amplification from a plasmid
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
131
containing murine zalphal 1 Ligand cDNA (Example 16) using primers ZC25,970
(SEQ ID NO:109) and ZC25,969 (SEQ ID NO:110) utilizing the Expand High
Fidelity
PCR System (Boerhinger Mannheim) as per manufacturer's instructions. The PCR
conditions were as follows: 1 cycle of 94 C for 2 minutes, followed by 35
cycles of
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
132
primers to the transposable element in the bacmid via PCR using primers ZC447
(SEQ
ID NO:76) and ZC976 (SEQ ID NO:77). The PCR reaction conditions were as
follows: 35 cycles of 94 C for 45 seconds, 50 C for 45 seconds, and 72 C for 5
minutes; 1 cycle at 72 C for 10 min.; followed by 4 C soak. The PCR product
was run
s on a 1%
agarose gel to check the insert size. Those having the correct insert were
used
to transfect Spodoptera frugiperda (Sf9) cells.
F. Expression and generation of material for purification of mouse zalphal 1
from
baculovirus
Sf9 cells were seeded at 1 million cells per 35 mm plate and allowed to
attach for 1 hour at 27 C. The murine zalphal I Ligand bacmid DNA was
transfected
as described in Example 30B and the virus was harvested.
For primary amplification, Sf9 cells were seeded as above and 500 1,11 of
72 hr post transfection supernatant was added and cultures were allowed to
proceed for
96 hr. after which time the virus was harvested according to standard methods.
For Secondary amplification, Sf9 cells were seeded as above and 200 [t1
of the Primary viral stock was added. Cultures were incubated at 27 C for 72
hr., after
which time the virus was harvested according to standard methods.
For Tertiary amplification, 10 l of Secondary Amplified virus stock
was placed on SF9s at 500,000 cells per well in 50 ml of SF900II media in a
250 ml
vol. shake flask for 6 days and virus was harvested as above. Virus was
titered and
grown up large scale for purification of the baculovirus-produced murine
zalphal 1
Ligand (muzalphal1L-Bv), as described in Example 30C and Example 30D.
Presence of predicted molecular weight protein in the supernatant was
determined by western analysis using an anti-muzalphal 1 L/MBP-6H polyclonal
antibody (Example 27). BaF3 based proliferation assay analysis (Example 5)
also
showed that the secreted ligand was active.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
133
Example 31
Expression of human and mouse zalphal I Ligand in E. coli
A. Construction of human zalphall Ligand-MBP fusion expression vector PTAP98/
zalphal 1 Ligand
An expression plasmid containing a polynucleotide encoding part of the
human zalphall Ligand fused N-terminally to maltose binding protein (MBP) was
constructed via homologous recombination. A fragment of human zalphal 1 Ligand
cDNA (SEQ ID NO:1) was isolated using PCR. Two primers were used in the
production of the human zalphall Ligand fragment in a PCR reaction: (1) Primer
ZC22,128 (SEQ ID NO:78) , containing 40 bp of the vector flanking sequence and
26
bp corresponding to the amino terminus of the human zalphal 1 Ligand, and (2)
primer
ZC22,127 (SEQ ID NO:79), containing 40 bp of the 3' end corresponding to the
flanking vector sequence and 28 bp corresponding to the carboxyl terminus of
the
human zalphal 1 Ligand. The PCR reaction conditions were as follows: 25 cycles
of
94 C for 30 seconds, 50 C for 30 seconds, and 72 C for 1 minute; followed by 4
C
soak, run in duplicate. Two I of the 100 I PCR reaction were run on a 1.0%
agarose
gel with 1 x TBE buffer for analysis, showing the expected band of
approximately 472
bp. The remaining 90 1 of PCR reaction was combined with the second PCR tube
precipitated with 400 p1 of absolute ethanol to be used for recombining into
the Smal
cut recipient vector pTAP98 to produce the construct encoding the MBP- zalphal
1
Ligand fusion, as described below.
Plasmid pTAP98 was derived from the plasmids pRS316 and pMAL-c2.
The plasmid pRS316 is a Saccharomyces cerevisiae shuttle vector (Hieter P. and
Sikorski, R., Genetics 122:19-27, 1989). pMAL-C2 (NEB) is an E. colt
expression
plasmid. It carries the tac promoter driving AfalE (gene encoding MBP)
followed by a
His tag, a thrombin cleavage site, a cloning site, and the rrnB terminator.
The vector
pTAP98 was constructed using yeast homologous recombination. 10Ong of EcoR1
cut
pMAL-c2 was recombined with 1 g Pvul cut pRS316, 1 p.g linker. and 11.1g
Scal/EcoR1 cut pRS316. The linker consisted of oligos ZC19,372 (SEQ ID NO:80)
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
134
(100 pmol): ZC19,351 (SEQ ID NO:81) (1 pmol): ZC19,352 (SEQ ID NO:82) (1
pmol), and ZC19,371 (SEQ ID NO:83) (100 pmol) combined in a PCR reaction.
Conditions were as follows: 10 cycles of 94 C for 30 seconds, 50 C for 30
seconds, and
72 C for 30 seconds; followed by 4 C soak. PCR products were concentrated via
100% ethanol precipitation.
One hundred microliters of competent yeast cells (S. cerevisiae) were
combined with 10 1 of a mixture containing approximately 1 g of the human
zalphal 1 Ligand PCR product, and 100 ng of SmaI digested pTAP98 vector, and
transferred to a 0.2 cm electroporation cuvette. The yeast/DNA mixture was
lo electropulsed at 0.75 kV (5 kV/cm), infinite ohms, 25 F. To each cuvette
was added
600 p.1 of 1.2 M sorbitol. The yeast was then plated in two 300 pl aliquots
onto two -
URA D plates and incubated at 30 C.
After about 48 hours, the Ura+ yeast transformants from a single plate
were resuspended in 1 ml H20 and spun briefly to pellet the yeast cells. The
cell pellet
was resuspended in 1 ml of lysis buffer (2% Triton X-100, 1% SDS, 100 mM NaC1,
10
mM Tris, pH 8.0, 1 mM EDTA). Five hundred microliters of the lysis mixture was
added to an Eppendorf tube containing 300 I acid washed glass beads and 200
1.11
phenol-chloroform, vortexed for 1 minute intervals two or three times,
followed by a 5
minute spin in a Eppendorf centrifuge at maximum speed. Three hundred
microliters of
the aqueous phase was transferred to a fresh tube, and the DNA precipitated
with 600 I
ethanol (Et0H), followed by centrifugation for 10 minutes at 4 C. The DNA
pellet
was resuspended in 100 1H2O.
Transformation of electrocompetent E. coli cells (MC1061, Casadaban
et. al. J. Mol. Biol. 138, 179-207) was done with 1 I yeast DNA prep and 40
.1 of
MC1061 cells. The cells were electropulsed at 2.0 kV, 25 1..tF and 400 ohms.
Following electroporation, 0.6 ml SOC (2% BactoTM Tryptone (Difco, Detroit.
MI),
0.5% yeast extract (Difco), 10 mM NaC1, 2.5 mM KC1, 10 mM MgC12, 10 mM MgSO4,
20 mM glucose) was plated in one aliquot on LB AMP plates (LB broth (Lennox),
1.8% BactoTM Agar (Difco), 100 mEIL Ampicillin).
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
135
Individual clones harboring the correct expression construct for human
zalphall Ligand were identified by expression. Cells were grown in Superbroth
II
(Becton Dickinson) with 100 ,g/m1 of ampicillin overnight. 50 vt.1 of the
overnight
culture was used to inoculate 2 ml of fresh Superbroth II +100[tg/m1
ampicillin.
s Cultures were grown at 37 C, shaking for 2 hours. lml of the culture was
induced with
1mM IPTG. 2-4 hours later the 250 vt.1 of each culture was mixed with 250 1,11
acid
washed glass beads and 250 j.il Thorner buffer with 5% PME and dye (8M urea,
100
mM Tris pH7.0, 10% glycerol, 2mM EDTA, 5% SDS). Samples were vortexed for one
minute and heated to 65 C for 5-10 minutes. 20 1,t1 were loaded per lane on a
4%-12%
am PAGE gel (NOVEX). Gels were run in 1XMES buffer. The positive clones were
designated pTAP126 and subjected to sequence analysis. The polynucleotide
sequence
of MBP- human zalphall Ligand fusion within pTAP126 is shown in SEQ ID NO:84,
and the corresponding polypeptide in SEQ ID NO:85.
is B. Bacterial Expression of human zalphall Ligand.
One microliter of sequencing DNA was used to transform strain W3110
(ATCC). The cells were electropulsed at 2.0 kV, 25 !..tF and 400 ohms.
Following
electroporation, 0.6 ml SOC (2% BactoTM Tryptone (Difco, Detroit, MI), 0.5%
yeast
extract (Difco), 10 mM NaCl, 2.5 mM KC1, 10 mM MgC12. 10 mM MgSO4, 20 mM
20 glucose) was plated in one aliquot on LB AMP plates (LB broth (Lennox),
1.8% Bacto
TM Agar (Difco), 100 mg/L Ampicillin).
Individual were expressed. Cells were grown in Superbroth II (Becton
Dickinson) with 100 vtg/ml of ampicillin overnight. 50 jil of the overnight
culture was
used to inoculate 2m1 of fresh Superbroth II +100 ig/m1 ampicillin. Cultures
were
25 grown at 37 C, shaking for 2 hours. lml of the culture was induced with
1mM IPTG.
2-4 hours later the 250 I of each culture was mixed with 250 tti acid washed
glass
beads and 250 [1.1 Thorner buffer with 5% 13ME and dye(8M urea. 100mM Tris
p147.0,
10% glycerol, 2mM EDTA, 5% SDS). Samples were vortexed for one minute and
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
136
heated to 65 C for 10 minutes. 20 [11 were loaded per lane on a 4%-12% PAGE
gel
(NOVEX). Gels were run in 1XMES buffer. The positive clones were used to grow
up
for protein purification of the huzalphal 1 L/MBP-6H fusion protein (Example
32,
below).
C. Construction of mouse zalphal 1 Ligand-MBP fusion expression vector pTAP98/
mouse zalphal 1 Ligand
An expression plasmid containing a polynucleotide encoding part of the
mouse zalphal 1 Ligand fused N-terminally to maltose binding protein (MBP) was
Example 32
Purification of zalphal 1-MBP Ligand or zalphall-MBP Receptor
Unless otherwise stated, all operations were carried out at 4 C. The
following procedure was used for purifying zalphall-MBP Ligand fusions for
human
zalphall-MBP Ligand (huzalphallL/MBP-6H) or murine zalphall-MBP Ligand
(muzalphal 1 L/MBP-6H) from E. coll. Human or mouse zalphal 1 -MBP receptor
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
137
fusions were carried out using the same method. Pre-spun frozen E. coli paste
was
thawed and diluted into 2 liters of Buffer B (0.02 M TRIS (EM Science); 0.2 M
NaC1
(Mallincrodt); 0.01 M 2-mercapto-ethanol (EM Science); pH 8.0; with 5 mg/1
Pepstatin
A (Boerhinger Mannheim); 5 mg/1 Aprotinin (Boerhinger Mannheim); and 1 mg/1
PMSF (Fluka)) plus 1-2 ml of an anti-foaming reagent AF289 antifoam (Sigma).
The
mixture was processed in a pre-chilled French Press cell disrupter (Constant
Systems
LTD) with 20-30 kPSI.
The lysate was then centrifuged at 18,000 x g for 45 minutes at 4 C;
retained the supernatant. A 200 ml slurry of Amylose resin (New England
BioLabs),
pre-equilibrated in Buffer A (0.02 M IRIS (EM Science); 0.2 M NaCl
(Mallincrodt);
0.01 M 2-mercapto-ethanol (EM Science); pH 8.0), was added to the lysate
supernatant
and incubated overnight in 21 roller bottles to allow for maximum batch
absorption of
the MBP fusion protein. The resin was washed in batch column format for > 5
column
volumes with Buffer A, then batch eluted with Buffer C (Buffer A with 0.02 M
Maltose
(Sigma)). Crude fractions were collected and monitored by absorbence 280 nm.
The eluted protein was analyzed by SDS NuPAGE (NOVEX) Coomassie
(Sigma) staining. Sample and bulk protein were stored at -80 C.
Example 33
Human zalphal 1 Ligand Polyclonal Antibodies
Polyclonal antibodies to Human zalphal 1 Ligand were prepared by
immunizing 2 female New Zealand white rabbits with the purified recombinant
protein
huzalphal 1L/MBP-6H (Example 32) or the purified CHO recombinant protein
huzalphal 1 L-CHO (Example 29). The rabbits were each given an initial
intraperitoneal
(ip) injection of 200 mg of purified protein in Complete Freund's Adjuvant
followed by
booster ip injections of 100 mg purified protein in Incomplete Freund's
Adjuvant every
three weeks. Seven to ten days after the administration of the second booster
injection
(3 total injections), the animals were bled and the serum was collected. The
animals
were then boosted and bled every three weeks.
The rabbit serum raised to huzalphallL/MBP-6H was pre-adsorbed of
anti-MBP antibodies using a CNBr-SEPHAROSE 4B protein column (Pharmacia
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
138
LKB) that was prepared using 10 mg of purified recombinant MBP per gram of
CNBr-
SEPHAROSE (Pharmacia). Recombinant MBP was made and purified on an amylose
column in house, using methods well known in the art. The huzalphall-ligand-
specific
polyclonal antibodies were affinity purified from the rabbit serum using a
CNBr-
s SEPHAROSE 4B protein column (Pharmacia LKB) that was prepared using 10 mg
of
the specific antigen purified recombinant protein huzalphal 1 L/MBP-6H or 10
mg of
the purified CHO recombinant protein huzalphal1L-CHO per gram of CNBr-
SEPHAROSE, followed by 20X dialysis in PBS overnight. Huzalphal 1 -ligand-
specific antibodies were characterized by ELISA using 1 i_tg/m1 of the
purified
o recombinant proteins huzalphallL/MBP-6H (Example 32), human zalphal 1
Ligand
(huzalphal 1L-CHO) (Example 29), or muzalphal1L-MBP/6H (Example 32) as
antibody targets.
The lower limit of detection (LLD) of the rabbit anti-huzalphal 1L/MBP-
6H affinity purified antibody was 10 ng/ml on its specific purified
recombinant antigen
15 huzalphal I L/MBP-6H, 500 pg/ml on purified recombinant huzalphal1L-CHO,
and
100 pg/ml on purified recombinant muzalphallL/MBP-6H (Example 32). The LLD of
the rabbit anti-huzalphal 1 L-CHO affinity purified antibody was 20 pg/ml on
its
specific purified recombinant antigen huzalphal1L-CHO, 500 pg/ml on purified
recombinant huzalphal 1L/MBP-6H , and 50 ng/ml on purified recombinant
20 muzalphallL/MBP-6H.
Example 34
Human zalphal 1 Ligand Anti-peptide Antibodies
Polyclonal human zalphal 1 Ligand anti-peptide antibodies were
25 prepared by immunizing 2 female New Zealand white rabbits with the human
zalphal 1
Ligand peptide, huzalphal 1L-1 (SEQ ID NO:72) or huzalphal 1L-3 (SEQ ID
NO:73).
The peptides were synthesized using an Applied Biosystems Model 431A peptide
synthesizer (Applied Biosystems, Inc., Foster City. CA) according to
manufacturer's
instructions. The peptides were then conjugated to the carrier protein keyhole
limpet
30 hemocyanin (KLH) with maleimide-activation. The rabbits were each given
an initial
intraperitoneal (ip) injection of 200 mg of peptide in Complete Freund's
Adjuvant
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
139
followed by booster ip injections of 100 mg peptide in Incomplete Freund's
Adjuvant
every three weeks. Seven to ten days after the administration of the second
booster
injection (3 total injections), the animals were bled and the serum was
collected. The
animals were then boosted and bled every three weeks.
The rabbit sera raised to the human zalphal 1 Ligand peptides were
characterized by an ELISA titer check using 1 p.g/m1 of the respective peptide
used to
make the antibody (SEQ ID NO:72 or SEQ ID NO:73) as an antibody target. The 2
rabbit seras to the huzalphal 1L-1 peptide had titer to their specific peptide
at a dilution
of 1:5,000,000 (1:5E6). The 2 rabbit seras to the huzalphal 11,3 peptide had
titer to
their specific peptide at a dilution of 1:5E6.
Human zalphal 1 Ligand peptide-specific polyclonal antibodies were
affinity purified from the rabbit serum using CNBR-SEPHAROSE 4B protein
columns
(Pharmacia LKB) that were prepared using 10 mg of the respective specific
peptide
(SEQ. ID. NO:72 or SEQ. ID. NO:73) per gram CNBr-SEPHAROSE, followed by 20X
dialysis in PBS overnight. Huzalphal 1 -ligand-specific antibodies were
characterized
by an ELISA titer check using 1 fig/m1 of the appropriate purified peptide
antigen or
purified recombinant full-length proteins as antibody targets.
The lower limit of detection (LLD) of the rabbit anti-huzalphal 1L-1
affinity purified antibody is 500 pg/ml on its specific peptide antigen
(huzalphal IL-1;
SEQ ID NO:72), 500 pg/ml on purified recombinant huzalphal 1 L/MBP-6H (Example
32), and 500 pg/ml on purified CHO recombinant huzalphal1L-CHO (Example 29).
No cross-reactivity was seen to the purified recombinant muzalphal 1 L/MBP-6H
(Example 32). The LLD of the rabbit anti-huzalphal 1L-3 affinity purified
antibody is
50 pg/ml on its specific peptide antigen (huzalphal 1L-l; SEQ ID NO:73), 50
pg/ml on
purified recombinant huzalphal 1L/MBP-6H. 500 pg/ml on purified CHO
recombinant
huzalphal 1L-CHO (Example 29), and 100 pg/ml on purified Baculovirus
recombinant
huzalphal 1L-By (Example 30). Cross-reactivity was seen to the purified
recombinant
muzalphallL/MBP-6H (Example 32) with an LLD of 5 ng/ml.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
140
Example 35
Human zalphal 1 receptor Monoclonal Antibodies
Zalphal 1 receptor Monoclonal antibodies were prepared by immunizing
male BalbC mice (Harlan Sprague Dawley, Indianapolis, IN) with the purified
5 recombinant soluble receptor protein, zalphallCEE (huzalphall-CEE-BHK)
(Example
10A). The mice were each given an initial intraperitoneal (IP) injection of 20
mg of
purified protein in Complete Freund's Adjuvant (Pierce, Rockford, IL) followed
by
booster IP injections of 10 mg purified protein in Incomplete Freund's
Adjuvant every
two weeks. Seven to ten days after the administration of the third booster
injection, the
o animals were bled and the serum was collected.
The mouse sera samples raised to the huzalphal 1 -CEE-BHK were
characterized by an ELISA titer check using purified recombinant CHO huzalphal
1 -Fc
protein (Example 10C) as an antibody target. One mouse serum sample had titer
to the
specific antibody target at a dilution of 1:1,000,000 (1:1E6). Four mouse
serum
samples had titer to the specific antibody target at a dilution of 1:100,000
(1:1E5).
Splenocytes were harvested from the 4 high-titer mice and fused to
murine SP2/0 myeloma cells using PEG 1500 (Boerhinger Mannheim, UK) in two
separate fusion procedures using a 4:1 fusion ratio of splenocytes to myeloma
cells
(Antibodies: A Laboratory Manual, E. Harlow and D. Lane, Cold Spring Harbor
Press).
Following 10 days growth post-fusion, specific antibody-producing hybridomas
were
identified by ELISA using purified recombinant BHK human zalphal 1 -Fc4
protein
(Example 10C) as an antibody target and by FACS using Ba13 cells expressing
the
huzalphal 1 sequence (Example 4, and Example 2) as an antibody target. The
resulting
4 hybridomas positive by both methods were cloned three times by limiting
dilution.
The antibodies were designated: 249.28.2.1.2.2; 247.10.2.15.4.6;
249.19.2.2.3.5; and
249.15.2.4.2.7.
Example 36
Zalphal 1 Ligand Transgenic Mice
A. Generation of transgenic mice expressing human and mouse zalphal 1 Ligand
DNA fragments from transgenic vectors (Example 22 and Example 26 )
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
141
containing 5' and 3' flanking sequences of the respective promoter (MT-1 liver-
specific
promoter (mouse zalphall Ligand (Example 26B) or lymphoid specific LCK
promoter
(mouse and human zalphal 1 Ligand (Examples 26A and 22B), the rat insulin II
intron,
zalphal 1 Ligand cDNA and the human growth hormone poly A sequence were
prepared and used for microinjection into fertilized B6C3f1 (Taconic,
Germantown,
NY) murine oocytes, using a standard microinjection protocol. See, Hogan, B.
et al.,
Manipulating the Mouse Embryo. A Laboratory Manual, Cold Spring Harbor
Laboratory Press, 1994.
Eight transgenic mice expressing human zalphal 1 Ligand from the
lymphoid-specific E[1LCK promoter were identified among 44 pups. Four of these
were pups that died and 4 grew to adulthood. Expression levels were fairly low
in these
animals. Twenty transgenic mice expressing mouse zalphal 1 Ligand from the
lymphoid-specific EIALCK promoter were identified among 77 pups. All 20 grew
to
adulthood. Expression levels were fairly low in these animals. Three
transgenic mice
expressing mouse zalphal 1 Ligand from the liver-specific MT-1 promoter were
identified among 60 pups. Two of these pups died and 1 grew to adulthood.
Expression levels were fairly low in these animals. Tissues were prepared and
histologically examined as describe below.
B. Microscopic evaluation of tissues from transgenic mice
Spleen, thymus, and mesenteric lymph nodes were collected and
prepared for histologic examination from transgenic animals expressing human
and
mouse zalphal 1 Ligand (Example 36A). Other tissues which were routinely
harvested
included the following: Liver, heart, lung, kidney. skin, mammary gland,
pancreas,
stomach, small and large intestine, brain, salivary gland, trachea, espohogus,
adrenal.
pituitary, reproductive tract, accessory male sex glands, skeletal muscle
including
peripheral nerve, and femur with bone marrow. The tissues were harvested from
a
neonatal pup which died unexpectedly, and several adult transgenic mice, as
described
below. Samples were fixed in 10% buffered formalin, routinely processed,
embedded
in paraffin, sectioned at 5 microns, and stained with hematoxylin and eosin.
The slides
were examined and scored as to severity of tissue changes (0=none, 1=mild,
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
142
2=moderate, 3=severe) by a board certified veterinary pathologist blinded to
treatment.
The pup and 2 female adult mice expressing the human zalphal 1
Ligand, and 3 of the 6 male adult mice expressing the mouse zalphal 1 Ligand
showed
inflammatory infiltrates in many of the tissues examined. The organs affected
varied
somewhat from mouse to mouse. The inflammatory infiltrate was composed
primarily
of neutrophils and macrophages in varying numbers and proportions and was
generally
mild to moderate degree in severity. Moreover, these animals showed changes in
lymphoid organs, including moderate to severe lymphopenia in the spleen and
thymus
(human and mouse zalphal 1 Ligand transgenics); and severe lymphopenia (human
zalphal 1 Ligand transgenics), or mild to severe suppurative to
pyogranulomatous
lymphadenitis (mouse zalphal 1 Ligand transgenics) in lymph nodes. In
addition,
increased extramedullary hematopoiesis was evident in the spleens. These
changes
were not observed in age-matched control mice.
C. Flow cytometric analysis of tissues from transgenic mice over expressing
zalphal 1
Ligand
Transgenic animals over expressing either human or mouse
zalphal 1 ligand (Example 36A) were sacrificed for flow cytometric analysis of
peripheral blood, thymus, lymph node, bone marrow, and spleen.
Cell suspensions were made from spleen, thymus and lymph nodes by
teasing the organ apart with forceps in ice cold culture media (500 ml RPMI
1640
Medium (JRH Biosciences. Lenexa, KS); 5 ml 100x L-glutamine (Gibco BRL. Grand
Island, NY); 5 ml 100x Na Pyruvate (Gibco BRL); 5 ml 100X Penicillin,
Streptomycin,
Neomycin (PSN) (Gibco BRL) and then gently pressing the cells through a cell
strainer
(Falcon, VWR Seattle, WA). Peripheral blood (200 ml) was collected in
heparinized
tubes and diluted to 10mls with HBSS containing 10U Heparin/ml. Erythrocytes
were
removed from spleen and peripheral blood preparations by hypotonic lysis. Bone
marrow cell suspensions were made by flushing marrow from femurs with ice cold
culture media. Cells were counted and tested for viability using Trypan Blue
(GIBCO
BRL, Gaithersburg, MD). Cells were resuspended in ice cold staining media
(HBSS,
1% fetal bovine serum, 0.1% sodium azide) at a concentration of ten million
per
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
143
milliliter. Blocking of Fe receptor and non-specific binding of antibodies to
the cells
was achieved by adding 10% normal goat sera and Fe Block (Pharmingen, La
Jolla,
CA) to the cell suspension.
Cell suspensions were mixed with equal volumes of fluorochrome
labeled monoclonal antibodies (PharMingen), incubated on ice for 60 minutes
and then
washed twice with ice cold wash buffer (PBS, 1% fetal bovine serum, 0.1%
sodium
azide) prior to resuspending in 400 ml wash buffer containing lmg/m1 7-AAD
(Molecular Probes, Eugene, OR) as a viability marker in some samples. Flow
data was
acquired on a FACSCalibur flow cytometer (BD Immunocytometry Systems, San
Jose,
CA). Both acquisition and analysis were performed using CellQuest software (BD
Immunocytometry Systems).
The transgenic animals that expressed either the human or mouse
zalphal 1 Ligand at the highest levels had dramatically altered cell
populations in all
lymphoid organs analyzed. Changes seen included complete loss of thymic
cellularity,
complete absence of CD45R positive B cells and increased size and cellularity
of
spleens. Both spleen and bone marrow had increased numbers of myeloid sized
cells,
which was accounted for by increases in both monocytes and neutrophils. The
pan NK
cell marker (DX5) was increased in many populations. Moderate expressing
founders
had less dramatic but still significant changes consistent with the phenotype
seen in the
high expressers. Mice with the lowest level of expression had neither a
significant
increase in myeloid cells nor decrease in B cells numbers. They did show
significant
changes in thymocyte populations with decreases in CD4+CD8+ double positive
cells
and increases in both CD4 and CD8 single positive cells.
Example 37
Zalpha 1 1 Ligand Purified Recombinant Human Protein
Dose-Response Study in Normal Mice
A. Summary
Normal six week old female C57B1/6 (Harlan Sprague Dawley,
Indianapolis, IN). mice were treated by intraperitoneal injection once daily
for either
four or eight days with one of four dose levels of purified recombinant human
zalphal 1
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
144
Ligand (Example 24) at 0.1, 0.5, 5 or 50p.g/mouse/day or with vehicle as a
control.
Body weights and body temperatures were monitored daily. On either day four or
day
nine, four of the eight mice from each protein treatment group and five of the
ten mice
in the vehicle control group were sacrificed. Blood, bone marrow and tissues
were
harvested and analyzed. Potential perturbations in lymphoid tissues were
examined, as
well as general physiologic and toxicological parameters.
There was no evidence of toxicity of human zalphal 1 Ligand protein at
any of the doses tested. Body weights and temperatures were unchanged. There
were
no apparent changes in clinical chemistry parameters. However, there were
consistent
o findings relating to increased percentages of myeloid lineage cells in
bone marrow,
spleen and peripheral blood in mice treated with the highest dose of zalphall
Ligand
compared to the vehicle control. There was a statistically significant
increase in
myeloid lineage sized cells identified by flow cytometric analysis of spleen
homogenate
in the high-dose group. The spleens of the two highest dose groups were
statistically
significantly larger than the other groups. On histopathologic examination,
however,
only a marginal increase in extramedullary hematopoiesis was seen in the
highest dose
group. There was a statistically significant increase in the myeloid to
erythroid ratio of
the bone marrow in the highest dose group compared to the other groups.
Finally, there
were increases seen in peripheral blood both in total white blood cell counts
and in the
percentage of monocytes in the same group.
B. Dosing solution preparation
Purified recombinant human zalphal 1 Ligand (Example 24) was diluted
into sterile phosphate buffered saline (GibcoBRL, Grand Island, NY) at
concentrations
to deliver 50, 5, 0.5 or 0.1 micrograms of protein in 0.1 ml of PBS vehicle.
The doses
for the first four days were made on day 0 and frozen in a frosty -20 C
freezer prior to
use. The doses for days five through eight were made on day five and frozen as
above.
Aliquots of the same PBS were similarly frozen for the vehicle treated control
group.
On the day of administration the appropriate aliquots were thawed and 0.1 ml
of
solution was injected intraperitoneally into the mice each day for either four
or eight
days.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
145
C. Study design
The mice were six weeks old at the start of the study. Each treatment
group consisted of eight mice, except for the vehicle control group that
included ten
mice. One half of the mice in each treatment group were sacrificed after four
days of
treatment and the other half after eight days.
Before treatment each day, each mouse was weighed and her body
temperature recorded using the Portable Programmable Notebook System (BMDS,
Inc,
Maywood, NJ), by scanning the mouse for identification number and body
temperature
from transponders implanted subcutaneously (IPTT-100, BMDS, Maywood, NJ).
At sacrifice, tissues harvested to assess white blood cell populations by
flow cytometric analysis included bone marrow, thymus and spleen. FACS
analysis of
the lymphoid organs and bone marrow was performed with the FACSCalibur,
(Becton
Dickinson, Mansfield, MA). The tissues harvested for histologic examination
for signs
of toxicity of the protein included: spleen, thymus, liver, kidney, adrenal
gland, heart
and lungs. All tissues fixed for histology were kept at 4 C overnight in 10%
Normal
Buffered Saline (NBF) (Surgipath, Richmond, IL). The following day the NBF was
replaced with 70% ethanol and the tissues returned to 4 C until processing for
histology.
The tissues were processed and stained for Hematoxylin and Eosin in
house, then sent to a contract pathologist for histopathologic analysis. Blood
was
collected for complete blood cell counts (CBC) and serum chemistry profiles.
The
CBC's were analyzed in-house with the Cell Dyn 3500 Hematology Analyzer
(Abbott
Diagnostics Division, Abbott Park, IL) and manual differential white blood
cell counts
were analyzed at Phoenix Central Laboratory, (Everett, WA). The serum was kept
frozen at -20 C until submission to Phoenix Central Laboratory for complete
serum
chemistry panels. To assess myeloid:erythroid ratios. the bone marrow from one
femur
was applied to CytoSpin slides (CYTOSPIN 3 CYTOCENTRIFUGE and CYTO
SLIDES, Shandon, Pittsburgh,PA) and sent to Phoenix Central Laboratories for
analysis.
D. Study results
There were no apparent clinical indications of physiologic effects or of
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
146
toxicity of human zalphal 1 Ligand at doses of 50 g/day or lower. Body weights
and
temperatures remained normal for the duration of the treatments. Serum
chemistry
parameters were in normal ranges. Red blood cell and platelet counts appeared
normal.
In the mice receiving 50 g/day for 8 days, manual differential white blood
cell counts
s showed
that the percentage of monocytes was elevated in the peripheral blood, and an
apparent increase in the total white blood cell counts. In bone marrow flushed
from a
femur, myeloid to erythroid ratios were increased in the 50p.g dose group, and
to a
lesser degree the 5 g dose group from the 8-day dose set. In a non-parametric
multiple
column comparison using InStat (InStat MAC; GraphPad Software, Inc., San
Diego,
CA), this difference was statistically significant (p=.0049). The difference
between the
highest dose group and vehicle was also significant, (p=.0286). The increased
white
blood cells in peripheral blood and the significant increase in myeloid
precursors in the
marrow may thus be related.
Histologic evaluation of the following tissues showed no apparent
is
evidence of cytologic or structural changes, mitotic events or necrosis:
thymus, liver,
kidney, adrenal gland, duodenum, pancreas, jejunum, caecum, colon, mesenteric
lymph
nodes, uterus, ovary, salivary gland, heart, trachea, lung, and brain. There
were no
apparent differences between the treatment groups in the weights of the
thymus, kidney,
liver or brain. Of all the tissues examined, only the spleen weights were
significantly
affected.
Each mouse spleen weight was normalized to her brain weight. In the
50 g/day treatment group compared to the vehicle, 0.1 g and 0.5 ug treatment
groups,
the average of the spleen weights was nearly 50% greater after four days of
treatment
and almost 100% greater after eight days than the average spleen weights of
the other
three groups. In the four-day set, the 511g/day group also tended to have
larger spleens
than the control and low dose groups. The difference in the spleen/brain
weights with
data from the four-day and the eight-day sets combined by treatment group was
statistically significant (p = .0072) by Kruskall-Wallace non-parametric
ANOVA,
multiple column comparison test using the InStat program (GraphPad Software).
A marginal increase in extrameduallary hematopoiesis. especially in the
red pulp was seen in spleens of mice from the highest dose group. even in the
mice
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
147
treated for four days. Flow cytometric analysis of the spleens showed a
significant
increase in the proportion of myeloid size cells in the highest dose group
(p=0.01,
Student's t test), representing increases in both monocytes and neutrophils.
This effect
may be related to the increased peripheral blood mononuclear cell percentage,
as well
as the apparent increase in myeloid precursors in the bone marrow, described
above.
Moreover, the transgenic mice derived from insertion of the human zalphal 1
gene had
increased extramedullary hematopoiesis in their spleens compared to non-
transgenic
litter mates.
Several changes were observed in the 50pg per day dose group
1.0 compared to the control group that implicate zalphal 1 Ligand in
production or
development of cells of the myeloid lineage. Taken together. the observed
changes
suggest that zalphal 1 may be useful as a therapeutic protein in such medical
specialties
as cancer and immunologic disorders described herein.
Example 38
Preliminary Elimination and Tissue Distribution Study Of
Purified Recombinant Human Zalphal 1 Ligand Protein
A. Summary
In order to elucidate tissue distribution and elimination patterns of the
purified rhzalphal 1 Ligand, a preliminary pharmacokinetic study was
undertaken.
Nine week old male C57B1/6 mice were given purified recombinant human zalphal
1
Ligand protein labeled with 1"Indium ("'In) (NEN, Boston. MA) by one of three
routes. A single bolus injection was given to each mouse by either the
intravenous (IV),
intraperitoneal (IP), or subcutaneous route (SC). The mice injected by either
the
subcutaneous or intraperitoneal route were sacrificed at either one or three
hours after
injection. The mice injected intravenously were sacrificed after either ten
minutes or
one hour following injection. Blood, plasma and selected tissues were
harvested at
various timepoints and counted by a gamma counter to estimate the approximate
half-
life and tissue distribution of the exogenous labeled protein. The tissues
that were
harvested for counting as well as the intervals of sacrifice were selected
based on
reports of the distribution of other cytokines labeled with radionuclides.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
148
At sacrifice, tissues harvested for counting of radioactivity included
thymus, spleen, kidney, a lobe of liver, a lobe of lung, and urinary bladder.
In the
group receiving the injection intraperitoneally, gut was also counted to
assess incidence
of injection into the gut, and in the subcutaneously dosed mice, skin with
underlying
structures in the area of injection was counted. The cpm for whole liver and
lung were
calculated from a section that was counted and a percentage of the whole organ
weight
represented by the section.
After the end of the study the collected tissues, whole blood and plasma
were counted on the COBRA II AUTO-GAMMA gamma counter (Packard Instrument
Company, Meriden, CT). An aliquot of the original labeled dosing solution was
also
counted at the end of the study with the tissues. This allowed calculation of
percent
total injected radioactivity for each mouse and simultaneous correction of all
counts for
radioactive decay. Approximations of remaining blood volume and organ weights
indicated that the majority of the counts administered were accounted for, and
therefore
the percentage of counts per tissue were a reasonable representation of
distribution of
the counts following labeled zalphal 1 Ligand administration by each route.
B. 111Indium labeling of zalphal 1 Ligand
Purified recombinant human zalpha 1 1 Ligand (Example 29) was
conjugated with a 10 fold molar excess of DTPA (Peirce, Rockford, II) by
incubating
minutes at room temperature in PBS. Unreacted DTPA and hydrolyzates were
removed by buffer exchange on a Biomax-5k NMWL (Ultrafree-15, Millipore.
Bedford, MA). The void volume protein peak was concentrated to 5 mg/ml and an
aliquot taken for testing in a bioassay (anti-CD40 stimulation of murine B-
cells
25 (Example 44)). Upon confirming that the DTPA-conjugate still had full
bioactivity the
conjugate was diluted to 0.5mg/m1 with 1M Na Acetate pH6Ø Two mCi of
"'Indium
was taken up in 0.5ml 1M Na Acetate pH6.0 and mixed with the DTPA-human
zalphal 1 Ligand for 30 min. at room temperature. Unincorporated "'Indium was
removed during buffer exchange to PBS on a PD-10 column (Pharmacia,
Piscataway,
30 NJ). The radio-labeled material was diluted with unlabeled human zalphal
1 Ligand to
give a specific activity of 100 mCi/mg, sterile filtered and stored at 4 C
overnight. One
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
149
hundred percent of the labeled protein was retained on a Biomax-5k NMWL
membrane
(Millipore). The labeled 11 'In-human zalphal 1 Ligand was administered to
mice in the
elimination and pharmacokinetic studies. Fifty g human zalphal 1 Ligand
protein
labeled with 5 uCi of labeled human zalphal 1 Ligand in 0.1 ml of PBS vehicle
was
administered to each animal.
C. Results Of Preliminary Distribution Study
After one and three hours following administration by all three routes,
the highest concentration of " 'In-human zalphal 1 Ligand, was found in kidney
and the
second highest was in urine and urinary bladder, as evinced by these tissues
having the
highest cpm. The average counts recovered from kidneys were from 3 to 8 times
higher
than the whole liver counts, depending on the route of injection and the
sacrifice
timepoint. For example, the average kidney cpm at 60 minutes following IV
injection
was 4.5 times greater than the average counts calculated for whole liver from
the same
group. In the group that was sacrificed ten minutes after intravenous
administration,
the highest cpm was again in kidney, and the second highest accumulation was
equivalent in liver, urinary bladder and urine.
D. Preliminary Pharmacokinetic Study
Blood and plasma collections were done at 10, 30 and 60 minutes
following injection by all three routes. Following injection by the IV route,
a separate
set of mice had blood and plasma samples taken at two, five and ten minutes.
Another
set of mice who received their injections by either the IP or SC route had
blood
sampled at one, two and three hours. For the treatment groups see Table 6. The
short
collection times bracket the reported half-life of IL-2 following intravenous
injection.
The reported T1/2 was in the range of 2.5 to 5.1 minutes. For reference to in
vivo
administration to IL-2, see Donohue JH and Rosenberg SA J Immunol, 130:2203.
1983.
The long timepoints were chosen to outline the anticipated elimination phase.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
150
Table 6
Route of injection Bleed Times(min.) Sacrifice Time
Intravenous Group 1 2, 5, 10 10 min.
Intravenous Group 2 10, 30, 60 60 min.
Intraperitoneal Group 1 10, 30, 60 60 min.
Intraperitoneal Group 2 60, 120, 180 180 min.
Subcutaneous Group 1 10, 30, 60 60 min.
Subcutaneous Group 2 60, 120, 180 180 min.
Un-labeled IL-2 has been shown to be eliminated from the serum with a
half-life of approximately three minutes in mice after IV injection. For
reference see
s Donahue, JH and Rosenburg supra.. Following IP and SC injection of
similar amounts
of IL-2, the duration of persistence of IL-2 activity in serum was prolonged
from 2
units/m1 for less than 30 minutes following IV injection to greater than 2
units/nil for 2
hours following IP and 6 hours following SC injections. The principle route of
clearance of IL-2 appears to be the kidney. Zalphal 1 ligand has been shown to
be
o structurally similar to IL-2, as discussed herein. Preliminary evaluation
of the
elimination of zalphal 1 Ligand appears to be consistent with the apparent
clearance of
IL-2 by the kidneys, based on the accumulation of cpm predominantly in the
kidneys,
followed by the urinary bladder and urine in the present study.
Estimations were made of pharmacokinetic parameters based on non
15 compartmental analysis of the cpm data obtained from the plasma. using
the PK
analysis program WinNonLin, Version 1.1, (Scientific Consulting Inc., Cary,
NC).
Plasma half-lives of zalphal 1 Ligand were estimated using the predicted
terminal
elimination rate constants for intravenous, subcutaneous, and intraperitoneal
administration of a 50 i_tg dose. The pharmacokinetic results were estimations
due to
20 limited data points in the terminal elimination region of the plasma
concentration vs.
time profiles. Moreover, the fit of the terminal elimination phase for SC and
IP dosing
required use of data from timepoints during which absorption of the "'In-human
zalphal 1 Ligand was apparently still occurring. However, estimations of half-
lives
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
151
following intravenous, subcutaneous, and intraperitoneal dosing were 13.6
min., 18.8
min., and 34.3 min., respectively. Since a dosing range was not evaluated it
was not
apparent whether saturable or active elimination (Michaelis Menten kinetics)
was
occurring. Therefore, these half-life calculations are estimations.
Estimates of the bioavailability of the labeled protein were made based
on the area under the curve (AUC) following subcutaneous or intraperitoneal
dosing
compared to that of intravenous dosing. The estimated bioavailability
following
subcutaneous and intraperitoneal injection were 35.8% and 63.9% respectively.
Because only one protein dose was studied, the bioavailability was not
evaluated as a
io
function of dose. The estimated clearance and volume of distribution (based on
the
data from the intravenous injection) were 0.48 ml/min. and 6.1 ml.
respectively.
Although the data are preliminary, the fate of zalphal 1 Ligand
administered IV was similar to that reported for IL-2, another 4-helix bundle
cytokine
(Donahue, JH and Rosenburg, SA supra.). Like 11-2, IV-administered zalphal 1
Ligand
had a plasma half life of only minutes with the main clearance in the kidney.
Three
hours after injection, the majority of the labeled material extracted from
kidney was still
retained in a Biomax 5K NMLW membrane (Millipore). Since it has previously
been
reported that the indium remains associated with protein even during lysosomal
degradation (Staud, F. et al., J. Pharm. Sciences 88:577-585, 1999) zalphal 1
Ligand is
accumulating and may be degraded in the kidney. The current study also showed,
as
observed with many other proteins, including IL-2 (Donahue. JH and Rosenburg,
SA,
supra.), that IP and SC administration significantly prolonged the plasma
levels of
zalphal 1 Ligand.
Example 39
Isolation and Expansion of fresh human bone marrow MNC CD34+ fraction using
zalphal 1 Ligand for assessment of NK Activity
A. Selection and Isolation of CD34+ cells from human Bone Marrow
Fresh human bone marrow mononuclear cells (M-NC) were prepared to
enrich for cells having NK cell activity. Fresh human MNCs were obtained from
Poeitic Technologies (Gaithersburg, MD). 10 ml alpha MEM (JRH, Lenexa, KS)
CA 02366921 2001-09-10
WO 00/53761 PCT/11500/06067
152
containing 10% HIA FBS (Hyclone, Logan, UT) and the antibiotic 1% PSN (Gibco,
BRL, Grand Island, NY) was added to the cell suspension and the cells were
passed
through a 100 Inn sieve. The cells were then counted, pelleted, washed with 10
ml PBS
containing 2%FBS, then pelleted again and resuspended in 1 ml PBS containing
2%FBS. Cells having a CD34 cell surface marker (CD34+ cells) were magnetically
separated using a Detachabead kit with Dynabeads M-450 CD34 ((Dynal, Oslo,
Norway), as per manufacturer's instructions. Both the CD34+ cell and the CD34-
cell
fractions were further analyzed below.
B. Expansion of CD34+ cells using zalphal 1 Ligand
A CD34+ cell fraction was plated into four wells in a 24-well plate.
50,000 positively selected cells suspended in 1 ml Alpha MEM (JRH) containing
10%HIA FBS (Hyclone) and 1% PSN (Gibco/BRL), plus the various cytokines
described below were plated in each of the 4 wells (1-4). Various reagents
were used to
test for zalphal 1 Ligand-induced expansion of the CD34+ selected bone marrow
MNCs: Reagents included human flt3 (R&D, Minneapolis, MN); purified human
zalphal 1 Ligand (Example 30C and Example 30D); human IL-15 (R&D). Reagents
were combined as follows at day 0: In well #1, 2 ng/ml human flt3 was added.
In well
#2, 2ng/m1 human flt3 and 15 ng/ml purified human zalphal 1 Ligand were added.
In
well #3, 2 ng/ml human flt3 and 20 ng/ml human IL15 were added. In well #4, 2
ng/ml
human flt3, 15 ng/ml purified human zalphal 1 Ligand, and 20 ng/ml human IL15
were
added. After incubating for 18 days, the suspension cells from each well were
pelleted,
and then resuspended in 0.5 ml alpha MEM (JRH) containing 10%HIA FBS (Hyclone)
and 1% PSN (Gibco/BRL), and counted to assess proliferation of the CD34+ cell
fraction. A low level of proliferation was seen in the presence of flt3 alone
(control
well #1), but the presence of IL-15 or zalphal 1 in addition to flt3 had not
significant
effect on the expansion (wells, #2 and #3). However, expansion beyond the flt3
control
was evident in well #4 which contained IL-15 and zalphal 1 Ligand in addition
to flt3.
This result suggested that zalphal 1 and IL-15 act in synergy to expand the
human
CD34+ cell population. Moreover, the results of this experiment supported the
results
seen with the mouse zalphall Ligand in the mouse BM assay (Example 21).
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
153
All cell populations were then tested for NK activity and subjected to
flow cytometry analysis, as shown below (Example 41).
C. Expansion of CD34+ or CD34- cells using zalphal 1 Ligand with delayed
addition
s of IL-15
Both CD34 positive and negative (CD34-) fractions were plated
separately into six 12 well plate wells (1-6). Each of six wells contained
100,000
positively or negatively selected cells in 2 ml alpha MEM containing 10%HIA
FBS and
PSN, described above. Reagents used were as described above. In well #1, 2
ng/ml
o human flt3 was added at day 0. In well #2, 2 ng/ml human flt3 was added
at day 0, and
after 5 days incubation 20 ng/ml human IL15 was added. In well #3, 2 ng/ml
human
flt3 and 15 ng/ml human zalphal 1 Ligand were added at day 0. In well #4, 2
ng/ml
human flt3 and 15 ng/ml human zalphall Ligand were added at day 0, and after 5
days
incubation 20 ng/ml human IL15 was added. In well #5, 2 ng/ml human flt3 and
20
15 ng/ml human IL15 were added at day 0. In well #6, 2 ng/ml human flt3, 15
ng/ml
human zalphal 1 Ligand, and 20 ng/ml human IL15 were added at day 0. After
incubating for a total of 15 days from the start of the experiment, the cells
from each
well were harvested and counted.
In the CD34+ population a low level of proliferation was seen in the
20 presence of flt3 alone (control well #1), but the presence of IL-15 or
zalphall added at
day 0 in addition to flt3 had no significant effect on the expansion (wells,
#3 and #5).
Addition of IL-15 after 5 days had some proliferative effect in comparison to
the flt3
control (well #2 compared to well #1) and a proliferative effect in the
presence of
zalphal 1 (well #4 compared to well #3). However, the greatest expansion was
evident
25 in well #6 which contained IL-15 and zalphal 1 Ligand in addition to
flt3 at day 0.
In the CD34- population, no proliferation was seen in the presence of
flt3 alone (control well #1), and in fact a decrease in the cell population
was evident.
The presence of zalphal 1 added at day 0 in addition to flt3 (well #3) was
similar to the
flt3 control. The presence of IL-15 added at day 5 increased proliferation
effect of the
30 cells in the presence (well #4) or absence (well #2) of zalphal 1
Ligand. Again. the
greatest expansion was evident in well #6 which contained IL-15 and zalphal 1
Ligand
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
154
in addition to flt3 at day 0.
All cell populations were then tested for NK activity and subjected to
FACS analysis, as shown below (Example 41).
Example 40
Isolation and Expansion of fresh mouse cells using human and mouse zalphal 1
Ligand
for assessment of NK Activity and NK Cell Markers
A. Isolation and Expansion of fresh mouse low density bone marrow cells using
human
and mouse zalphal 1 Ligand
Fresh mouse marrow cells were isolated by clipping both ends of mouse
femurs, and flushing two to three milliliters of growth medium (see below)
through the
inside of the bone into a collection tube. The growth medium was 500 ml RPMI
1640
Medium (JRH Biosciences. Lenexa, KS); 5 ml 100x L-glutamine (Gibco BRL. Grand
Island, NY); 5 ml 100x Na Pyruvate (Gibco BRL); 5 ml 100X Penicillin,
Streptomycin,
is Neomycin (PSN) (Gibco BRL); and 50 ml heat-inactivated Fetal Bovine
Serum (FBS)
(Hyclone Laboratories. Logan, UT). The marrow cells were then broken-up by
pipeting
the media up and down several times. The cells were then pelleted and washed
once
with growth medium, and passed through a 70-micron sieve. The low-density
mononuclear cells were then isolated by subjecting the marrow cells to a
density
gradient. Marrow cells in five to eight milliliters of growth medium were
carefully
pipetted on top of five to eight milliliters of NycoPrep 1.077 Animal
(Nycomed. Oslo,
Norway) in a centrifuge tube. This gradient was then centrifuged at 600 X g
for 20
minutes. The low density mononuclear cells were harvested from the interface
layer
between the NycoPrep and the medium. These cells were then diluted to
approximately 20 milliliters in growth medium, pelleted and washed. The cells
were
then plated at approximately 0.5-1.5x106 cells per milliliter in growth medium
in a
standard tissue culture flask and incubated at 37 C, 5% CO, for two hours.
The non-adherent, low density (NA LD) marrow cells were then
harvested and plated at 0.5-2.0x105 cells per milliliter in growth medium plus
2.5
nanograms per milliliter mouse 1113 (R and D Systems. Minneapolis, MN) plus 25
to 50
nanograms per milliliter human Interleukin 15 (IL-15) (R and D Systems) with
or
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
155
without 50 to 150 nanograms per milliliter human zalphal 1 Ligand; or with or
without
0.12 to 10 nanograms per milliliter mouse zalphal 1 Ligand.
There was no significant expansion without the addition of the human or
mouse zalphal 1 Ligand. Non-adherent cells were expanded in the cultures
containing
s mouse zalphal 1 Ligand as low as 0.12 ng/ml and in the cultures
containing human
zalphal 1 Ligand as low as 22 ng/ml. In cultures containing both the human and
mouse
zalphal 1 Ligand, non-adherent cell expansion increased with increasing dose
if
zalphal 1 Ligand, with the mouse ligand saturating response at about 5-10
ng/ml and
the human not reaching a saturating response even at the highest dose of 200
ng/ml.
2.o Human zalphal 1 Ligand appeared to be approximately 20 to 100 fold less
potent on
mouse cells as the mouse zalphal 1 Ligand. After approximately five to ten
days the
zalphal 1 Ligand expanded mouse cells were harvested and analyzed by flow
cytometry
(FACSCalibur; Becton Dickinson, Mansfield, MA) to determine what percentage of
them were positive for NK cell antigens, where 46% were positive for the PanNK
cell
ls marker DX5 (Pharmingen).
B. Isolation and Expansion of Fresh lineage Depleted Mouse Marrow Cells
Fresh mouse lineage depleted (lin-) marrow cells were isolated from
fresh mouse marrow cells by first incubating the cells with the following
antibodies:
20 TER119, Gr-1, B220, MAC-1, CD3e and I-Ab (Pharmingen. San Diego. CA).
The lin+
cells were then removed with Dynabeads M-450 sheep anti-rat IgG (Dynal, Lake
Success, NY) as per manufacturer's instructions.
The negatively selected lin- marrow cells were then plated as above in
growth medium plus either 2.5 ng/mL flt3(R&D Systems) and 25ng/mL IL-15 (R&D
25 Systems); or flt3, IL-15 and mouse zalphal 1 Ligand, 2 to 5% BHK mouse
zalphal 1
Ligand conditioned medium. After six days of growth, the cultures were
harvested,
counted and submitted to an NK cell activity assay (Example 41). Cells grown
with
mouse zalphal 1 Ligand were approximately two to three times more effective at
lysing
NK cell target cells (YAC-1 cells) as the cells grown without zalphal 1
Ligand.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
156
C. Isolation and Expansion of CD4- CD8- (Double Negative or DN) Thymocytes
Fresh mouse thymocytes were isolated by chopping and sieving
thymuses from three to eight week old mice. CD4- CD8- (DN) cells were then
negatively selected by incubating the thymocytes with anti-CD4 and anti-CD8
s antibodies (PharMingen), then removing the CD4+ CD8+ cells with Dynabeads
M-450
sheep anti-rat IgG (Dynal) as per manufacturer's instructions.
The DN mouse thymocytes were then grown in growth medium plus 2.5
ng/mL flt3 (R&D Systems), 25 ng/mL IL-15 (R&D Systems) and 10 ng/mL IL-7 (R&D
Systems) with or without mouse zalphal 1 Ligand as above. Six days later the
cells
io were harvested, counted, analyzed by flow cytometry as described above, and
also
submitted to an NK cell activity assay (Example 41).
The culture grown with mouse zalphal 1 Ligand yielded approximately
480,000 cells while the culture without zalphal 1 Ligand yielded only
approximately
160,000 cells. The culture grown with mouse zalphal 1 Ligand was found to be
is approximately 16.2% positive for the NK cell antigen Pan NK, DX5
(PharMingen).
The culture grown without zalphal 1 Ligand was 14.6% positive for DX5. The
cells
grown with zalphal 1 Ligand lysed NK cell target cells. YAC-1, approximately
two
times better than the cells grown without zalphal 1 Ligand. The expanded cells
did not
lyse significantly a negative control target cell line, EL4. These results
suggested that
20 zalphal 1 Ligand selectively expands lytic NK cells.
Example 41
Activity of human and mouse zalphal 1 Ligand expanded cells and mature murine
NK
cells in NK cell cytotoxicity assays
25 A. NK cell assay
NK cell-mediated target cytolysis was examined by a standard 51Cr-
release assay. Target cells (K562 cells (ATCC No. CCL-243) in human assays,
and
YAC-1 cells (ATCC No. TIB-160) in mouse assays) lack expression of major
histocompatability complex (MHC) molecules, rendering them susceptible to NK
cell-
30 mediated lysis. A negative control target cell line in mouse assays is the
MHC
thymoma EL4 (ATCC No. TIB-39). We grew K562. EL4, and YAC-1 cells in RP10
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
157
medium (standard RPMI 1640 (Gibco/BRL, Grand Island, NY) supplemented with
10% FBS (Hyclone, Logan, UT), as well as 4 mM glutamine (Gibco/BRL), 100
I.U./m1
penicillin+100 MCG/ml streptomycin (Gibco/BRL), 50 M 13-mercaptoethanol
(Gibco/BRL) and 10mM HEPES buffer (Gibco/BRL). On the day of assay, 1-2x106
target cells were harvested and resuspended at 2.5-5x106 cells/ml in RP10
medium. We
added 50-100 1.11 of 5 mCi/m1 "Cr-sodium chromate (NEN, Boston, MA) directly
to the
cells and incubated them for 1 hour at 37 C, then washed them twice with 12 ml
of
PBS and resuspended them in 2 ml of RP10 medium. After counting the cells on a
hemacytometer, the target cells were diluted to 0.5-1x105 cells/ml and 100 iii
(0.5-
lx1 04 cells) were mixed with effector cells as described below.
In human assays, effector cells were prepared from selected and
expanded human CD34+ BM cells (Example 39B) which were harvested, washed,
counted, mixed at various concentrations with 'Cr-labeled target cells in 96-
well round
bottomed plates, and incubated for 4 hours at 37 C. After co-incubation of
effector
cells and the labeled target cells, half of the supernatant from each well was
collected
and counted in a gamma counter for 1 min/sample. The percentage of specific
'Cr
release was calculated from the formula 100 x (X-Y)/(Z-Y), where X is "Cr
release in
the presence of effector cells, Y is the spontaneous release in the absence of
effectors,
and Z is the total 5ICr release from target cells incubated with 0.5% Triton X-
100. Data
were plotted as the % specific lysis versus the effector-to-target ratio in
each well.
B. Activity of human zalphal 1 Ligand expanded cells
Isolated CD34+ human HPCs cultured with flt3 +/¨ zalphal 1 Ligand and
flt3 +1L-15 +/¨ zalphal 1 Ligand (Example 39), were harvested the cells on day
15 to
assess their capacity to lyse MHC- K562 cells in a standard "Cr-release assay
as
described above, and to analyze their surface phenotype by flow cytometry. As
expected from previous reports (Mrozek, E et al., Blood 87:2632-2640, 1996:
and Yu.
H et al.. Blood 92:3647-3657, 1998), simultaneous addition of IL-15 and flt3L
did
induce the outgrowth of a small population of CD56- cells. Interestingly,
although BM
cells cultured simultaneously with zalphal 1 Ligand and flt3L did not expand
significantly. there was a significant increase in total cell numbers in
cultures
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
158
containing a combination of flt3L, zalphall Ligand and IL-15 (see, Example
39).
For an assessment of the surface phenotype of these human BM cultures,
we stained small aliquots of the cells for 3-color flow cytometric analysis
with anti-
CD3-FITC, anti-CD56-PE and anti-CD16-CyChrome mAbs (all from PharMingen, San
s Diego, CA) and analyzed them on a FACSCalibur using CellQuest software
(Becton
Dickinson, Mountain View, CA). This flow cytometric analysis confirmed that
the
cells growing out of these cultures were differentiated NK cells, as they were
large and
granular and expressed both CD56 and CD16, and were CD3- (Lanier, LL Annu.
Rev.
Immunol. 16:359-393, 1998). Furthermore, these cells exhibited significantly
higher
io effector function than those cells grown with IL-15 and flt3. More
specifically, cells
grown in all three cytokines lysed more than 40% of the K562 targets at an
effector-to-
target ratio (E:T) of 1.5, whereas cells grown in IL-15+flt3L lysed fewer than
5% of the
targets at an E:T of 2. These data demonstrate that, in combination with IL-
15,
zalphal 1 Ligand stimulates the differentiation of NK cells from CD34 BM
cells.
C. Activity of mouse zalphal 1 Ligand expanded cells
To test the effects of zalphal 1 Ligand on murine hematopoietic
progenitor cells, purified Lineage-negative (Lin¨) bone marrow cells from
C57B1/6
mice were expanded in flt3+IL-15+/¨ zalphal 1 Ligand, as described in Example
40B.
On day 6 of culture, the cells ("effectors") were harvested and counted, then
resuspended in 0.4 ml of RP10 medium (Example 41A). Two aliquots (0.15 ml
each)
of each sample expanded with or without zalphal 1 Ligand (Example 41A) were
diluted
serially 3-fold in duplicate in 96-well round bottomed plates, for a total of
6 wells of
100 [11 each. The remaining 100 I of cells were stained for NK cell surface
markers
with FITC-anti-2B4 and PE-anti-DX5 mAbs (PharMingen) and analyzed by flow
cytometry. Each group of cells exposed to flt3+IL-15 with or without the
presence of
zalphal 1 Ligand had similar fractions of 2B4+DX5+ cells, ranging from 65-75%
positive for both NK markers.
For the NK lysis assay, target cells (YAC-1 and EL4) were labeled with
51Cr as described above. After counting the target cells on a hemacytometer,
the target
cells were diluted to 0.5-1x105 cells/ml and 100 p.1 of YAC-1 or EL4 (0.5-
1x104 cells)
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
159
were mixed with 100 ul effector cells and incubated for 4 hours at 37 C.
Specific lysis
was determined for each well as described above.
We found that cells grown in the presence of flt3+IL-15+zalphal 1
Ligand exhibited enhanced lytic activity (roughly 2-fold) against the YAC-1
targets
(but did not kill the MHC+ control cell line EL4). At an effector-to-target
ratio (E :T) of
5, NK cells generated in the presence of all 3 cytokines (zalphal 1
Ligand+flt3+IL-15)
lysed 12% of the YAC-1 cells, whereas those NK cells expanded with flt3+IL-15
lysed
6% of the YAC-1 targets. Subsequent experiments confirmed this trend.
In a second approach to determine the biological activity of zalphal 1
Ligand on murine NK cells, we isolated immature CD4-CD8- ("double negative",
DN)
mouse thymocytes as described in Example 40C and cultured them with IL-
15+flt3+IL-
7 or IL-15+flt3+IL-2, with or without zalphal 1 Ligand. On day 6 of culture,
the cells
were harvested and assayed for NK lytic activity on YAC-1 and EL4 cells as
described
above. We found that cells cultured in the presence of zalphal 1 Ligand had
the greatest
lytic activity in this assay, with enhanced lytic activity over those cells
cultured in the
presence of the other cytokines. Specifically, DN thymocytes grown with IL-
15+flt3+IL-7 killed 18% of the YAC-1 cells at E:T of 24 while cells grown in
the
presence of IL-15+flt3+IL-7 plus zalphall Ligand killed 48% of the targets at
the same
E:T. DN thymocytes grown in IL-15+flt3+IL-2 killed 15% of the YAC-1 targets at
an
E:T of 6, whereas cells grown with these 3 cytokines and zalphal 1 Ligand
killed 35%
of the YAC-1 cells at an E:T of 9. Flow cytometry was performed on the
cultured cells
one day before the NK lysis assay. As was true for the bone marrow cultures,
despite
the proliferative effect of zalphal 1 Ligand (cell numbers increase
approximately 2-fold
when zalphal 1 Ligand is added), it did not significantly enhance the fraction
of DX5-
cells (17-20% of total cells in the cultures with IL-7, and 35-46% of total in
cultures
with IL-2). These data imply that zalphal 1 Ligand, in combination with IL-15
and flt3,
enhances the lytic activity of NK cells generated from murine bone marrow or
thymus.
D. Activity of mouse zalphal 1 Ligand on Mature murine NK cells
In order to test the effects of mouse zalphal 1 Ligand on mature NK
cells, we isolated spleens from four 5-week old C57B1/6 mice (Jackson
Laboratories,
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
160
Bar Harbor, ME) and mashed them with frosted-end glass slides to create a cell
suspension. Red blood cells were removed by hypotonic lysis as follows: cells
were
pelleted and the supernatant removed by aspiration. We disrupted the pellet
with gentle
vortexing, then added 900 vtl of sterile water while shaking, followed quickly
(less than
s 5 sec
later) by 100 pi of 10X HBSS (Gibco/BRL). The cells were then resuspended in
ml of 1X HBSS and debris was removed by passing the cells over a nylon mesh-
lined cell strainer (Falcon). These RBC-depleted spleen cells were then
pelleted and
resuspended in MACS buffer (PBS+1%I3SA+2mM EDTA) and counted. We stained
300x106 of the cells with anti-DX5-coated magnetic beads (Miltenyi Biotec) and
o
positively selected DX5+ NK cells over a MACS VS+ separation column, according
to
the manufacturer's instructions, leading to the recovery of 8.4x106 DX5+ cells
and
251x106 DX5- cells. Each of these groups of cells were cultured in 24-well
plates
(0.67x106 cells/well, 2 wells per treatment condition) in RP10 medium (Example
41A)
alone or with 1) 30 ng/ml mouse zalphal 1 Ligand, 2) 30 ng/ml recombinant
mouse IL-
is 2 (R&D Systems, Inc., Minneapolis, MN), 3) 30 ng/ml recombinant human IL-15
(R&D), 4) 30 ng/ml each of mouse zalphal 1 Ligand and hIL-15, or 5) 30 ng/ml
each of
mIL-2 and hIL-15. The cells were harvested after 21 hours, washed, and
resuspended
in RP 10 medium and counted. The cells were then assayed for their ability to
lyse
labeled YAC-1 or EL4 targets cells, as described in Example 41A.
In general, there was little NK activity from the DX5- (non-NK cells)
groups, but the DX5- cells cultured with zalphal 1 Ligand and hIL-15 did lyse
25% of
the YAC-1 target cells at an E:T of 82. By comparison, DX5- cells cultured
with hIL-
15 alone lysed 14% of the YAC-1 targets at an E:T of 110. This suggests that
zalphal 1
Ligand and IL-15 are acting together on the residual NK1.1 NK cells in this
cell
preparation. As for the DX5- cell preparation, treatment with mouse zalphal 1
Ligand
alone did not significantly increase their effector function (their lysis of
YAC-1 cells
was similar to the untreated group). As expected, both IL-2 and IL-15
significantly
improved NK activity. The highest level of lysis, however, was detected in the
group
treated with zalphal 1 Ligand and hIL-15 (65% lysis of YAC-1 cells at an E:T
of 3.3,
VS. 45% lysis at an E:T of 4 for the hIL-15 treatment group). Taken together,
these
results suggest that although zalphal 1 Ligand alone may not increase NK cell
lysis
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
161
activity, it does enhance NK lysis activity of mature NK cells, when
administered with
IL-15.
Example 42
Zalphal 1 Ligand Proliferation of Human and Mouse T-cells in a T-cell
Proliferation
assay
A. Murine Zalphal 1 Ligand Proliferation of Mouse T-cells
T cells from C57B1/6 mice (Jackson Laboratories, Bar Harbor, ME)
were isolated from pooled splenocytes and lymphocytes from axillary, brachial,
inguinal, cervical, and mesenteric lymph nodes (LNs). Spleens were mashed with
frosted-end glass slides to create a cell suspension. LNs were teased apart
with forceps
and passed through a cell strainer to remove debris. Pooled splenocytes and LN
cells
were separated into CD8 and CD4+ subsets using two successive MACS magnetic
separation columns, according to the manufacturer's instructions (Miltenyi
Biotec,
Auburn, CA). Whole thymocytes were collected from the same mice.
Cells were cultured at 3x10 cells/well (thymocytes) or 10' cells/well
(mature T cells) with increasing concentrations of purified murine zalphal 1
Ligand (0-
30 ng/ml) (Example 24 and Example 29) in 96-well flat bottomed plates pre-
coated
overnight at 4 C with various concentrations of anti-CD3 mAb 2C11 (PharMingen)
for
3 days at 37 C. The anti-CD3 antibody served to activate the murine T-cells
through
the T-cell receptor. Each well was pulsed with 1 tC'i 31-1-thymidine on day 2
and plates
were harvested and counted 16 hours later to assess proliferation.
When we tested zalphal 1 Ligand in T cell proliferation assays. we found
that it co-stimulated anti-CD3-activated murine thymocytes, leading to an
accelerated
outgrowth of CD8'CD4- cells (the majority of the thymocytes cultured with anti-
CD3+zalphal 1 Ligand were CD8+CD4- by day 3 of culture, while cells cultured
with
anti-CD3 alone did not significantly skew to this phenotype until day 5). We
did not
observe significant levels of proliferation of thymocytes to zalphal 1 Ligand
in the
absence of anti-CD3.
Interestingly, when we assayed mature peripheral murine T cells for
their ability to respond to zalphal 1 Ligand+anti-CD3. we found that only the
CD8-, but
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
162
not the CD4+ subset, responded in a dose-dependent manner to zalphal 1 Ligand.
We
also observed weak but reproducible proliferation of CD8+ cells (but not CD4-
cells) in
response to zalphal 1 Ligand alone. Interestingly, this was not observed for
human T
cells (see Example 42B, below).
B. Human Zalphall Ligand Proliferation of Human T-cells
Human CD4+ and CD8+ T cells were isolated from PBMC as described
in Example 43 (below) Cells were cultured at about 105 cells/well with
increasing
concentrations of purified human zalphal 1 Ligand (0-50 ng/ml) (Example 24) in
96-
o well flat bottomed plates pre-coated overnight at 4 C with various
concentrations of
anti-human CD3 mAb UCHT1 (PharMingen) for 3 days at 37 C. Each well was pulsed
with luCi 31-1-thymidine on day 2 and plates were harvested and counted 16
hours later.
Unlike our results with mouse T cells, our preliminary data suggests that
human
zalphal 1 Ligand co-stimulates CD4+, but not CD8+, human T cells in a dose-
dependent fashion.
Example 43
Real Time PCR shows Zalphal 1 Ligand expression in Human CD4+ cells
A. Purified Human T cells as a Primary Source used to assess human zalphal 1
Ligand
Expression
Whole blood (150 ml) was collected from a healthy human donor and
mixed 1:1 with PBS in 50 ml conical tubes. Thirty ml of diluted blood was then
underlayed with 15 ml of Ficoll Paque Plus (Amersham Pharmacia Biotech,
Uppsala,
Sweden). These gradients were centrifuged 30 min at 500 g and allowed to stop
without braking. The RBC-depleted cells at the interface (PBMC) were collected
and
washed 3 times with PBS. The isolated human PBMC yield was 200x10' prior to
selection described below.
The PBMCs were suspended in 1.5 ml MACS buffer (PBS. 0.5%
EDTA, 2mM EDTA) and 3x106 cells were set aside for control RNA and for flow
cytometric analysis. We next added 0.25 ml anti-human CD8 microbeads (Miltenyi
Biotec) and the mixture was incubated for 15 min at 4 C. These cells labeled
with CD8
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
163
beads were washed with 30 ml MACS buffer, and then resuspended in 2 ml MACS
buffer.
A VS+ column (Miltenyi) was prepared according to the manufacturer's
instructions. The VS+ column was then placed in a VarioMACS magnetic field
s (Miltenyi). The column was equilibrated with 5 ml MACS buffer. The
isolated primary
mouse cells were then applied to the column. The CD8 negative cells were
allowed to
pass through. The column was rinsed with 9 ml (3 X 3 ml) MACS buffer. The
column
was then removed from the magnet and placed over a 15 ml falcon tube. CD8+
cells
were eluted by adding 5 ml MACS buffer to the column and bound cells flushed
out
o using the plunger provided by the manufacturer. The yield of CD8+
selected human
peripheral T cells was about 51x106 total cells. The CD8-negative flow through
cells
were collected, counted, stained with anti-human CD4 coated beads, then
incubated and
passed over a new VS+ column at the same concentrations as described above.
The
yield of CD4+ selected human peripheral T cells was 42x106 total cells.
15 A sample of each of the CD8+ and CD4+ selected human T cells was
removed for staining and sorting on a fluorescence activated cell sorter
(FACS) to
assess their purity. A PE-conjugated anti-human CD4 antibody, an anti-human
CD8-
FITC Ab, and an anti-human CD19-CyChrome Ab (all from PharMingen) were used
for staining the CD8+ and CD4+ selected cells. The CD8-selected cells in this
first
20 experiment were 80% CD8+, and the CD4-selected cells were 85% CD4+. In 2
subsequent experiments (Example 43B), the CD8+ purified cells were 84% and 81%
pure, and the CD4+ cells were 85% and 97% pure, respectively. In one
experiment, we
stained the non-binding (flow-through) cells with anti-human CD19-coated beads
(Miltenyi) and ran them over a third magnetic bead column to isolate CD19+ B
cells
25 (these were 92% pure).
The human CD8+, CD4+ and CD19+ selected cells were activated by
incubating 0.5X10' cells/ml in RPMI + 5% human ultraserum (Gemini Bioproducts,
Calabasas, CA) + PMA 10 ng/ml and Ionomycin 0.5 g/m1 (Calbiochem) for about
4,
16, or 24 hours at 37 C. The T-cells (2.5x1 06/well) were alternately
stimulated in 24-
30 well plates pre-coated overnight with 0.5 g/m1 plate-bound anti-CD3 mAb
UCHT1
(PharMingen) with or without soluble anti-CD28 mAb (PharMingen) at 5 g/ml. At
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
164
each timepoint, the cells were harvested, pelleted, washed once with PBS, and
pelleted
again. The supernatant was removed and the pellets were snap-frozen in a dry
ice/ethanol bath, then stored at ¨80 C for RNA preparation at a later date.
Real Time-PCR was performed on these human CD 8+, CD4+ and
CD19+ selected cells as described in Example 43B and 43C below for assessing
human
zalphall Ligand and receptor expression.
B.
Primers and Probes for Quantitative RT-PCR for human zalphal 1 Ligand
expression
Real-time quantitative RT-PCR using the ABI PRISM 7700 Sequence
Detection System (PE Applied Biosystems, Inc., Foster City, CA) has been
previously
described (see, Heid, CA et al., Genome Research 6:986-994, 1996; Gibson, UEM
et
al., Genome Research 6: 995-1001, 1996; and Sundaresan, S et al.,
Endocrinology
139:4756-4764, 1998). This method incorporates use of a gene specific probe
containing both reporter and quencher dyes. When the probe is intact the
reporter dye
emission is negated due to the proximity of the quencher dye. During PCR
extension
using additional gene-specific forward and reverse primers, the probe is
cleaved by 5'
nuclease activity of Taq polymerase which releases the reporter dye resulting
in an
increase in fluorescent emission.
The primers and probes used for real-time quantitative RT-PCR analyses
were designed using the primer design software Primer ExpressTM (PE Applied
Biosystems). Primers for human zalphal 1 Ligand were designed spanning an
intron-
exon junction to eliminate amplification of genomic DNA. The forward primer,
ZC22,281 (SEQ ID NO:90) and the reverse primer, ZC22,279 (SEQ ID NO:91) were
both used at 300 nM concentration to synthesize an 80 bp product. The
corresponding
zalphal 1 Ligand TaqMan probe, ZG32 (SEQ ID NO:92) was synthesized by PE
Applied Biosystems. The probe was labeled with a reporter fluorescent dye (6-
carboxy-fluorescein) (FAM) (PE Applied Biosystems) at the 5' end and a
quencher
fluorescent dye (6-carboxy-tetramethyl-rhodamine) (TAMRA) (PE Applied
Biosystems) at the 3' end. In order to test the integrity or quality of all
the RNA
samples, they were screened for rRNA using the primer and probe set ordered
from PE
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
165
Applied Biosystems (cat No. 4304483). The reporter fluorescent dye for this
probe is
VIC (PE Applied Biosystems). The rRNA results will allow for the normalization
of
the zalphall Ligand results.
RNA was prepared from pellets provided in Example 43A, using
RNeasy MiniprepTM Kit (Qiagen, Valencia, CA) per the manufacturer's
instructions.
Control RNA was prepared from about 10 million BHK cells expressing human
zalphall Ligand.
C. Primers and Probes for Quantitative RT-PCR for human zalphal 1 Receptor
o expression
Real time PCR was performed to assess the expression of zalphal 1
receptor as per Example 43B and Example 43D, using the cells prepared under
the
conditions detailed in 43A, and probes specific for the zalphal 1 receptor.
The forward
primer, ZC22,277 (SEQ ID NO:93) and the reverse primer, ZC22,276 (SEQ ID
NO:94)
were used in a PCR reaction (above) at about 300 nM concentration to
synthesize a 143
bp product. The corresponding zalphal 1 TaqMan0 probe, designated ZG31 (SEQ ID
NO:95) was synthesized and labeled by PE Applied Biosystems. RNA from BaF3
cells
expressing human zalphal 1 receptor was used to generate appropriate control
for
standard curves for the real-time PCR described in Example 43D below.
D. Real-time quantitative RT-PCR
Relative levels of zalphal 1 Ligand RNA were determined by analysis of
total RNA samples using the One-Step RT-PCR method (PE Applied Biosystems).
RNA from BHK cells expressing human zalphal 1 Ligand was used to generate a
standard curve. The curve consisted of serial dilutions ranging from 2.5-
2.5x10-4ng for
the rRNA screen and 25-0.0025 ng for the zalphal 1 Ligand screen with each
point
analyzed in triplicate. The total RNA samples were also analyzed in triplicate
for
human zalphal 1 Ligand transcript levels and for levels of rRNA as an
endogenous
control. Each One-step RT-PCR reaction consisted of 25 ng of total RNA in
buffer A
(50 mM KCL, 10 mM Tris-HCL, and the internal standard dye, ROX (PE Applied
Biosystems)), appropriate primers (50 nM for rRNA samples, 300 nM for zalphal
1
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
166
Ligand samples) and probe (50 nM for rRNA, 100 nM for zalphal 1 Ligand), 5.5
mM
MgC12, 300 [tIVI each d-CTP, d-ATP, and d-GTP and 600 1.1M of d-UTP, reverse
transcriptase (0.25 U/ 1), AmpliTaq DNA polymerase (0.025 U/111) and RNase
Inhibitor (0.4 U411) in a total volume of 25 1. Thermal cycling conditions
consisted of
an initial RT step at 48 C for 30 minutes, an AmpliTaq Gold activation step of
95 C for
minutes, followed by 40 cycles of amplification for 15 seconds at 95 C and 1
minute
at 60 C. Relative zalphal 1 Ligand RNA levels were determined by the Standard
Curve
Method as described in User Bulletin No. 2 (PE Biosystems; User Bulletin #2:
ABI
Prism 7700 Sequence Detection System, Relative Quantitation of Gene
Expression,
o December 11, 1997) using the rRNA measurements to normalize the zalphall
Ligand
levels. Samples were compared relative to the calibrator within each
experiment. The
calibrator was arbitrarily chosen based on good quality RNA and an expression
level to
which other samples could significantly be compared. Results of the
experiments
analyzing the expression of the zalphall Ligand and zalpha receptor in
stimulated and
unstimulated cells (Example 43A) are as described in Example 43E below.
E. Expression of human zalphal 1 Receptor and Ligand in CD4+, CD8+ and CD19+
cells
The first experiment used RT-PCR, described above, to assess zalphal 1
receptor expression in unstimulated and anti-CD3 stimulated CD4+ and CD8+
samples
at timepoints of Oh (unstimulated ("resting") cells), and at 4h, 15.5h and
24h, after
stimulatoin. The resting CD4+ sample was arbitrarily chosen as the calibrator
and given
a value of 1.00. There was approximately a 4-fold increase in receptor
expression in
unstimulated CD4+ cells from 4h to 24h of culture and about an 8-fold increase
over
the same time period in anti-CD3 stimulated CD4+ cells. The CD8+ cells showed
a 7-
fold increase in zalphal 1 receptor expression that peaked at 4hrs and
decreased over
time. With anti-CD3 stimulation, the CD8+ cells had a constant 8-fold increase
in
receptor expression.
This first experiment also used RT-PCR to assess zalphal 1 Ligand
expression in the same anti-CD3 stimulated and unstimulated CD4+ and CD8+
samples. The 4hr anti-CD3 stimulated CD8+ sample was arbitrarily chosen as the
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
167
calibrator and given a value of 1.00. The results showed that unstimulated
CD4+ and
CD8+ cells do not express zalphal 1 Ligand. We observed a significant
elevation of
expression in the anti-CD3 stimulated CD4+ cells at 4 h, with about a 300-fold
increase
in signal observed at 15.5 h. The CD8+ cells expressed a small amount of
ligand upon
anti-CD3 stimulation, however this is probably due to contamination of the
CD8+
population with a small number of CD4+ cells.
The second experiment used RT-PCR to assess zalphal 1 receptor
expression in anti-CD3-stimulated, PMA + Ionomycin-stimulated and unstimulated
CD4+ and CD8+ samples at timepoints of 0 h, and at 3.5 h. 16 h and 24 h after
activation. The resting CD8+ sample was arbitrarily chosen as the calibrator
and given
a value of 1.00. The resting CD4+ and CD8+ cells did not have significant
amounts of
receptor expression. The expression was about 3 fold higher in the PMA +
Ionomycin-
stimulated CD4+ samples at 3.5 h, 16 h and 24 h after stimulation. The
expression in
anti-CD3 activated CD4+ cells peaked at 10-fold above background levels at 3.5
h after
stimulation, then fell back to levels 4-fold above background at 16 h after
stimulation.
The CD8+ cells showed a 4-fold expression increase at 3.5 h after PMA +
Ionomycin
stimulation, with expression decreasing at subsequent timepoints. As in the
first
experiment, the anti-CD3 stimulated CD8+ cells again exhibited an 8-fold above
background induction of receptor expression.
These samples from the second experiment were also used to assess
zalphall Ligand expression. The 24hr PMA + Ionomycin stimulated CD4+ sample
was arbitrarily chosen as the calibrator and given a value of 1.00. The
results showed
that again none of the unstimulated cells expressed zalphal 1 Ligand. There
was about
a 30-fold induction of ligand expression in the CD4+ cells stimulated with
anti-CD3 at
3.5h, as seen in the previous experiment (at 4h). However, there was only
about a 5-
fold induction with PMA + Ionomycin stimulation at 3.5h that went down at
subsequent timepoints. Again, the CD8+ cells expressed a very small amount of
Ligand that was probably attributed to contaminating CD4+ cells.
The final experiment used RT-PCR to assess zalphal 1 receptor
expression in anti-CD3- and anti-CD3/anti-CD28-stimulated and unstimulated
CD4+
and CD8+ samples at timepoints of 0 h, and at 2 h, 4 h, and 16 h after
stimulation.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
168
CD19+ cells activated with PMA + Ionomycin were also screened for receptor
expression at the same time intervals. The resting CD4+ sample was arbitrarily
chosen
as the calibrator and given a value of 1.00. The 2h anti-CD3 stimulated CD4+
cells
only had a 4-fold induction of receptor, compared to the 10-fold induction
seen at 3.5h
in the previous experiment. The combination of anti-CD3 and anti-CD28
increased
zalphal 1 receptor expression to 8-fold above background. The 16 h anti-
CD3/anti-
CD28 stimulated CD8+ cells had very low zalphall receptor expression levels,
as seen
in the CD8+ cells in previous experiments (above). The CD19+ cells stimulated
with
PMA + Ionomycin had the most significant zalphallreceptor expression with a 19-
fold
increase at 2h, but the expression levels decreased back to those of resting
cells by 16h.
These samples from the final experiment were also used to assess
zalphal 1 Ligand by RT-PCR. The 16h anti-CD3/anti-CD28 stimulated CD8+ sample
was arbitrarily chosen as the calibrator and given a value of 1.00. The
results showed
that at 2 h the CD4+ cells had about a 2-fold induction of zalphal 1 Ligand
expression
with anti-CD3 stimulation and a 5-fold induction with anti-CD3 plus anti-CD28
stimulation. These stimulation conditions induced Ligand expression over time,
with
the 16 h stimulated CD4+ cells exhibiting Ligand expression levels 70-fold
above
background. CD8+ and CD19+ cells showed no zalphal 1 Ligand expression.
A certain amount of variation was expected between blood draws (i.e.
multiple samples at different times from the same patient and between multiple
patients). Therefore, data trends were analyzed within each study or from a
single
blood sample and the three experiments above were compared for an overall
conclusion. The trend from the Real Time PCR experiments described above is
that of
all the cell types tested, CD19+ B cells activated with PMA + ionomycin
expressed the
highest levels of zalphal 1 receptor RNA. CD4+ and CD8+ cells can also be
stimulated
to express receptor, but at lower levels than in B cells. Zalphal 1 Ligand was
expressed
almost exclusively in stimulated CD4+ T cells (and not by CD8+ T cells or
CD19+ B
cells). Although stimulation with PMA + lonomycin induced a good zalphal 1
Ligand
signal in this assay, a significantly higher signal was obtained from CD4+ T
cells
stimulated with anti-CD3 mAb or a combination of anti-CD3 and anti-CD28 mAbs,
conditions that better mimic an antigen encounter in vivo.
=
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
169
Example 44
Zalphal 1 Ligand-dependent Proliferation of B-cell cells stimulated Anti-CD40
or Anti-
IgM
A. Purification of Human B cells
A vial containing 1 x 108 frozen, apheresed human peripheral blood
mononuclear cells (PBMCs) was quickly thawed in a 37 C water bath and
resuspended
in 25 ml B cell medium (RPMI Medium 1640 (JRH Biosciences. Lenexa, KS), 10%
Heat inactivated fetal bovine serum, 5% L-glutamine, 5% Pen/Strep) (Gibco
BRL)) in a
50 ml tube (Falcon VWR, Seattle, WA). Cells were tested for viability using
Trypan
Blue (Gibco BRL). Ten milliliters of Ficoll/Hypaque Plus (Pharmacia LKB
Biotechnology Inc., Piscataway, NJ) was layered under the cell suspension and
spun for
30 minutes at 1800 rpm and allowed to stop with the brake off. The interface
was then
removed and transferred to a fresh 50 ml Falcon tube, brought up to a final
volume of
40 ml with PBS and spun for 10 minutes at 1200 rpm with the brake on. The
viability
of the isolated cells was again tested using Trypan Blue. Alternately fresh
drawn human
blood was diluted 1:1 with PBS (Gibco BRL) and layered over Ficoll/Hypaque
Plus
(Pharmacia), spun and washed as above. Cells isolated from either fresh or
frozen
sources gave equivalent results.
B cells were purified from the Ficoll floated peripheral blood cells of
normal human donors (above) with anti-CD19 magnetic beads (Miltenyi Biotec,
Auburn, CA) following the manufacturer's instructions. The purity of the
resulting
preparations was monitored by flow cytometric analysis with anti-CD22 FITC Ab
(Pharmingen, SanDiego, CA). B cell preparations were typically >90% pure.
B. Purification of Murine B cells
A suspension of murine splenocytes was prepared by teasing adult
C57B1/6 mouse (Charles River Laboratories. Wilmington, MA) spleens apart with
bent
needles in B cell medium. RBCs were removed by hypotonic lysis. CD43 positive
cells
were removed with CD43 magnetic beads (Miltenyi Biotec) following the
manufacturer's instructions. The purity of the resulting preparations was
monitored by
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
170
flow cytometric analysis with anti-CD45R FITC Ab (Pharmingen). B cell
preparations
were typically >90% pure.
C. Proliferation of anti-CD40-stimulated B-Cells in the presence of human or
murine
zalphal 1 Ligand
The B cells from either the human or mouse source were resuspended at
a final concentration of 1 x 106 cells/ml in B cell medium and plated at 100
1/well in a
96 well U bottom plate (Falcon, VWR) containing various stimulation conditions
to
bring the final volume to 200 l/well. For anti-CD40 stimulation human
cultures were
supplemented with 1 iig/ml anti-human CD40 (Genzyme, Cambridge, MA) and mouse
cultures were supplemented with 1 ,g/m1 anti-murine CD40 (Serotec, UK). Human
or
murine zalphal 1 Ligand was added at dilutions ranging from 1 pg/m1-100 ng/ml.
The
specificity of the effect of zalphal 1 Ligand was confirmed by inhibition of
zalphal 1
Ligand with 25mg/m1 soluble human zalphal 1 CEE (Example 10A). All treatments
is were performed in triplicate. The cells were then incubated at 37 C in a
humidified
incubator for 120 hours (human) or 72 hours (mouse). Sixteen hours prior to
harvesting, 1 iCi 11-thymidine (Amersham, Piscataway, NJ) was added to all
wells to
assess whether the B-cells had proliferated. The cells were harvested into a
96 well
filter plate (UniFilter GF/C, Packard, Meriden, CT) using a cell harvester
(Packard) and
collected according to manufacturer's instructions. The plates were dried at
55 C for
20-30 minutes and the bottom of the wells were sealed with an opaque plate
sealer. To
each well was added 0.25 ml of scintillation fluid (Microscint-O, Packard) and
the plate
was read using a TopCount Microplate Scintillation Counter (Packard).
Incubation with Zalphal 1 Ligand at concentrations of 3 ng/ml or more
enhanced the proliferation induced by soluble anti-CD40 in a dose dependent
manner in
both murine and human B cells by as much as 30 fold. The murine and human B
cells
responded equally as well to their respective zalphal 1 Ligand. In both
species, the
stimulation was specific to zalphal 1 Ligand, as it was reversed by the
presence of
soluble zalphal 1 receptor in the culture.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
171
D. Proliferation of anti-IgM-stimulated B-Cells in the presence of human or
murine
zalphal 1 Ligand
The B cells from either human or mouse source as described above
(Example 44A and Example 44B) were plated as described above (Example 44C).
For
anti-IgM stimulation of human cells the plates were pre-coated overnight with
10mg/m1
F(ab')2 anti-human IgM Abs (Southern Biotech Associates, Birmingham, Alabama)
and
washed with sterile media just prior to use. The cultures were supplemented
with 0-10
ng/ml hu rIL-4 (R&D Systems, Minneapolis, MN). For anti-IgM stimulation of
murine
cells soluble anti-IgM (Biosource, Camarillo, CA) was added to the cultures at
10
mg/ml. To each of the preceding anti-IgM/IL-4 conditions, human or murine
Zalphal 1
ligand was added at dilutions ranging from 1 pg/m1-100 ng/ml as described
above. The
specificity of the effect of zalphal 1 Ligand was confirmed by inhibition with
soluble
human zalphal 1 receptor as described above (Example 44C). All treatments were
performed in triplicate. The cells were incubated, labeled with 3H-thymidine,
harvested,
and analyzed as described in Example 44C.
Incubation with Zalphal 1 ligand at concentrations of 0.3 ng/ml or more
inhibited the proliferation induced by insoluble anti-IgM (mouse) or anti-IgM
and IL-4
(human) in a dose-dependent manner. This inhibition was specific to zalphal 1
Ligand,
as it was reversed by the presence of soluble zalphal 1 receptor in the
culture.
Example 45
Expression of Human zalphal 1 soluble receptor in E. coli
A. Construction of expression vector pCZR225 that expresses huzalphall/MBP-6H
fusion polypeptide
An expression plasmid containing a polynucleotide encoding a human
zalphal 1 soluble receptor fused C-terminally to maltose binding protein (MBP)
was
constructed via homologous recombination. A fragment of human zalphal 1 cDNA
(SEQ ID NO:7) was isolated using PCR. The polynucleotide sequence for the MBP-
zalphal 1 soluble receptor fusion polypeptide is shown in SEQ ID NO:96. Two
primers
were used in the production of the human zalphal 1 fragment in a PCR reaction:
(1)
Primer ZC20,187 (SEQ ID NO:98), containing 40 bp of the vector flanking
sequence
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
172
and 25 bp corresponding to the amino terminus of the human zalphal 1, and (2)
primer
ZC20,185 (SEQ ID NO:99), containing 40 bp of the 3' end corresponding to the
flanking vector sequence and 25 bp corresponding to the carboxyl terminus of
the
human zalphal 1. The PCR Reaction conditions were as follows: 25 cycles of 94
C for
s 30 seconds, 50 C for 30 seconds, and 72 C for 1 minute; followed by 4 C
soak, run in
duplicate. Two p.1 of the 100 Ill PCR reaction was run on a 1.0% agarose gel
with 1 x
TBE buffer for analysis, and the expected approximately 660 bp fragment was
seen.
The remaining 90 tl of PCR reaction was combined with the second PCR tube
precipitated with 400 of absolute ethanol. The precipitated DNA used for
recombining into the Smal cut recipient vector pTAP98 (Example 31) to produce
the
construct encoding the MBP-zalphal 1 fusion. Clones were transformed,
identified and
grown up as described in Example 31. The positive clones were designated
pCZR225
and subjected to sequence analysis. The polynucleotide sequence for the MBP-
zalphal 1
soluble receptor fusion polypeptide is shown in SEQ ID NO:96, and
corresponding
polypeptide sequence is shown in SEQ ID NO:97. The positive clones were used
to
grow up in E. coli as described in Example 31 for protein purification of the
huzalphall/MBP-6H fusion protein (Example 46, below).
Example 46
Purification of huzalphal 1/MBP-6H Soluble Receptor From E. coli Fermentation
Unless otherwise noted, all operations were carried out at 4 C. The
following procedure was used for purifying huzalphal 1 /MBP-6H soluble
receptor
polypeptide. E. coli cells containing the pCZR225 construct and expressing
huzalphall/MBP-6H soluble receptor (Example 45) were grown up in SuperBroth II
(12 g/L Casein, 24 g/L Yeast Extract, 11.4 g/L di-potassium phosphate, 1.7 g/L
Mono-
potassium phosphate; Becton Dickenson, Cockeysville. MD), and frozen in 0.5%
glycerol. Twenty grams of the frozen cells in SuperBroth II + Glycerol were
used to
purify the protein. The frozen cells were thawed and diluted 1:10 in a
protease inhibitor
solution (Extraction buffer) prior to lysing the cells and releasing the
huzalphal 1 /MBP-
6H soluble receptor protein. The diluted cells contained final concentrations
of 20 mM
Tris (JT Baker, Philipsburg, NJ) 100 mM Sodium Chloride (NaCl, Mallinkrodt,
Paris,
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
173
KY), 0.5 mM pheynlmethylsulfonyl fluoride (PMSF, Sigma Chemical Co., St.
Louis,
MO), 2 [ig/m1 Leupeptin (Fluka, Switzerland), and 2 lag/m1 Aprotinin (Sigma).
A
French Press cell breaking system (Constant Systems Ltd., Warwick, UK) with
temperature of -7 to -10 C and 30K PSI was used to lyse the cells. The diluted
cells
s were
checked for breakage by A600 readings before and after the French Press. The
lysed cells were centrifuged at 18,000G for 45 minutes to remove the broken
cell
debris, and the supernatant used to purify the protein.
Total target protein
concentrations of the supernatant was determined via BCA Protein Assay
(Pierce,
Rockford, IL), according to manufacturer's instructions.
A 25 ml column of Talon Metal Affinity resin (Clontech, Palo Alto, CA)
(prepared as described below) was poured in a Bio-Rad, 2.5 cm D x 10 cm H
glass
column. The column was packed and equilibrated by gravity with 10 column
volumes
(CVs) of Talon Equilibration buffer (20mM Tris, 100mM NaCl, pH 8.0). The
supernatant was batch loaded to Talon metal affinity resin and was rocked
overnight.
The resin was poured back into the column and was washed with 10 CV's of Talon
Equilibration buffer by gravity, then gravity eluted with 140 ml of Elution
buffer
(Talon Equilibration buffer + 200mM Imidazole-Fluka Chemical). The talon
column
was cleaned with 5 CVs of 20mM 2-(N-Morhpholino) ethanesulfonic acid pH 5.0
(MES, Sigma), 5 CVs of distilled H2O, then stored in 20% Ethanol/0.1% Sodium
Azide. Fourteen ml fractions were collected over the entire elution
chromatography
and the fractions were read with absorbance at 280 and 320 nM and BCA protein
assay:
the pass through and wash pools were also saved and analyzed. The protein
elution
fractions of interest were pooled and loaded straight to Amylose resin (New
England
Biolabs, Beverly, MA).
To obtain more pure huzalphal 1/MBP-6H polypeptide, the talon affinity
elution pooled fractions were subjected to Amylose resin (22m1s) at pH 7.4. A
2.5 cm
D x 10 cm H Bio-Rad column was poured, packed and equilibrated in 10 CVs of
Amylose equilibration buffer-20mM Tris (JT Baker), 100mM NaCl (Mallinkrodt),
1mM PMSF (Sigma), 10mM beta-Mercaptoethanol (BME, ICN Biomedicals Inc.,
Aurora, OH) pH 7.4. The sample was loaded by gravity flow rate of 0.5 ml/min.
The
column was washed for 10 CVs with Amylose equilibration buffer, then eluted
with
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
174
about 2 CV of Amylose equilibration buffer + 10 mM Maltose (Fluka Biochemical,
Switzerland) by gravity. 5 ml fractions were collected over the entire
chromatography
and absorbance at 280 and 320 nM were read. The Amylose column was regenerated
with 1 CV of distilled H20, 5 CVs of 0.1% (w/v) SDS (Sigma), 5 CVs of
distilled H-20,
and then 5 CVs of Amylose equilibration buffer.
Fractions of interest were pooled and dialyzed in a Slide-A-Lyzer
(Pierce) with 4 x 4L PBS pH 7.4 (Sigma) to remove low molecular weight
contaminants, buffer exchange and desalt. After the changes of PBS, the
material
harvested represented the purified huzalphal 1 /MBP-6H polypeptide. The
purified
huzalphal 1/MBP-6H polypeptide was analyzed via SDS-PAGE Coomassie staining
and Western blot analysis with the anti-rabbit HRP conjugated antibody
(Rockland,
Gilbertsville, PA). The concentration of the huzalphall/MBP-6H polypeptide was
1.92
mg/ml as determined by BCA analysis.
Purified huzalphal 1/MBP-6H polypeptide was prepared for injection
into rabbits and sent to R & R Research and Development (Stanwood, WA) for
antibody production. Rabbits were injected to produce anti anti-huzalphal
1/MBP-6H
serum (Example 47, below).
Example 47
Zalphal 1 receptor Polyclonal Antibodies
Polyclonal antibodies were prepared by immunizing two female New
Zealand white rabbits with the purified huzalphal 1 /MBP-6H polypeptide
(Example
46), or the purified recombinant zalphal 1 CEE soluble receptor (Example 10A).
Corresponding polyclonal antibodies were designated rabbit anti-huzalphal
1/MBP-6H
and rabbit anti-huzalphall-CEE-BHK respectively. The rabbits were each given
an
initial intraperitoneal (IP) injection of 200 mg of purified protein in
Complete Freund's
Adjuvant (Pierce, Rockford, IL) followed by booster IP injections of 100 mg
purified
protein in Incomplete Freund's Adjuvant every three weeks. Seven to ten days
after the
administration of the third booster injection, the animals were bled and the
serum was
collected. The rabbits were then boosted and bled every three weeks.
The zalphal 1 -specific polyclonal antibodies were affinity purified from
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
175
the rabbit serum using an CNBr-SEPHAROSE 4B protein column (Pharmacia LKB)
that was prepared using 10 mg of the purified huzalphal 1/MBP-6H polypeptide
(Example 32) per gram CNBr-SEPHAROSE, followed by 20X dialysis in PBS
overnight. Zalphal 1-specific antibodies were characterized by an ELISA titer
check
using 1 mg/ml of the appropriate protein antigen as an antibody target. The
lower limit
of detection (LLD) of the rabbit anti-huzalphall/MBP-6H affinity purified
antibody is
a dilution of 500 pg/ml. The LLD of the rabbit anti-huzalphal 1 -CEE-BHK
affinity
purified antibody is a dilution of 50 pg/ml.
Example 48
Zalphal 1 receptor distribution
To assess zalphal 1 receptor distribution on various cells types, we
generated both rabbit polyclonal and mouse monoclonal antibodies (mAbs)
directed
against the human receptor (Example 35 and Example 47) and conjugated these
antibodies to biotin for use in flow cytometry. We initially used the
polyclonal
antibodies, which were of relatively low affinity, to stain a panel of cell
lines: IL-3
dependent murine pre-B cell line wild-type BaF3 cells (Palacios and Steinmetz,
ibid.;
Mathey-Prevot et al., ibid.); BaF3 cells transfected with human zalphal 1
(Example 4);
human Burkitt's lymphoma cell lines Raji (ATCC No. CCL-86), Ramos (ATCC No.
CRL-1596), RPMI 8226 (ATCC No. CCL-155), and Daudi (ATCC No. CCL-213);
human T cell leukemia cell line Jurkat (ATCC No. TIB-152); human
myelomonocytic
leukemia cell lines Thp-1 (ATCC No. TIB-202) and UT937 (ATCC No.CRL-1593.2);
human pro-myelomonocytic cells HLT60 (ATCC No. CCL-240); murine B cell
lymphoma cell line A20 (ATCC No TIB-208); and murine thymoma cell line EL4
(ATCC No. TIB-39).
The cells were harvested, washed once with FACS wash buffer with
serum (WBS). WBS consisted of Hank's balanced salt solution (Gibco/BRL) + 10mM
HEPES (Gibco/BRL) + 1% BSA (Sigma) + 10% normal goat serum (Gemini
Bioproducts, Woodland, CA) + 10% normal rabbit serum (Sigma). Wash buffer (WB)
was identical to WBS except that it was serum free. After washing, the cells
were
resuspended in 100 1.11 WB containing 10 g/m1 rabbit anti-zalphal 1 polyclonal
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
176
antibodies (Example 47). The cells were kept on ice with Ab for 20 min, then
washed
with WB and resuspended in WB containing goat anti-rabbit-FITC (BioSource,
International), incubated another 20 min on ice, then washed and resuspended
in 400 p.1
WB for analysis on a FACSCalibur flow cytometer (Becton Dickinson). Control
samples were stained with the secondary goat anti-rabbit-FITC Ab only.
Positive
staining was defined as a shift above the staining with secondary alone.
Although the
polyclonal antibodies were of low affinity, we confidently detected zalphal 1
expression
on the BaF3/zalphal 1 transfectant, on all four human Burkitt's lymphomas
(Raji,
Ramos, Daudi, and RPMI 8226), and on Jurkat T cells. Resting
(undifferentiated) HL-
60 cells did not bind the anti-zalphal 1 antibodies, but we did detect a
positive signal on
HL-60 cells activated for 24 hours with PMA (Calbiochem, La Jolla, CA) which
induces HL-60 cell differentiation into a monocyte-like cell. We also saw a
positive
signal on UT937 and Thp-1 cells, although this signal may have been due to non-
specific binding. The polyclonal antibodies weakly cross-reacted on the mouse
B cell
line A20, but we saw no staining of the EL4 murine thymoma.
The four anti-zalphal 1 monoclonal antibodies (Example 35) were
conjugated to biotin, and a subset of the cells described above were screened
for
zalphal 1 receptor expression (BaF3, BaF3/zalphal 1, Raji, Jurkat, and resting
HL-60).
Cells were harvested, washed, then resuspended in 100 pi WB containing 15
p.g/m1 of
one of each of the 4 biotinylated mAbs. The cells were incubated with mAb for
20 min
on ice, then washed with 1.5 ml WB and pelleted in a centrifuge. The
supernatant was
removed by aspiration and the pellets were resuspended in 100 [11 of CyChrome-
conjugated streptavidin (CyC-SA; PharMingen), then incubated on ice for
another 20
min and washed and pelleted as before. Control tubes contained cells stained
only with
CyC-SA. Pellets were resuspended in 400 1 WB and flow cytometry performed as
above. Positive staining was defined as a signal exceeding the background
level of
staining with CyC-SA alone. Using the BaF3/zalphal 1 transfectant as a
control, we
were able to rankthe 4 mAbs in terms of their respective mean fluorescence
intensities
(MFI), which can reflect antibody affinity and/or the extent of biotinylation
of the
mAbs. The mAbs were ranked as follows, from highest to lowest MFI:
249.28.2.1.2.2,
247.10.2.15.4.6. 249.19.2.2.3.5, and 249.15.2.4.2.7. The Raji cells stained
positive
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
177
with the zalphal 1 monoclonal antibodies. The Jurkats cells positively stained
with the
zalphal 1 monoclonal antibodies, but not as strongly as that on B cells
(Raji). Thus the,
zalphal 1 receptor was expressed on these B and T cell lines. The staining
patterns on
non-activated HL60 cells were identical for all the mAbs, and the signal was
very weak.
We believe that this signal does not reflect actual expression of zalphal 1 by
HL-60
cells, but rather may be due to non-specific binding of the mouse mAbs to the
human
cells, probably via Fc-receptors.
Example 49
o Human
zalphal 1 Ligand effect on B-cells and Zalphal 1 Ligand toxic saporin fusion
The effects of human zalphal 1 Ligand were tested on the following
human B-cell lines: and human Burkitt's lymphoma cell lines Raji (ATCC No.CCL-
86), and Ramos (ATCC No. CRL-1596); human EBV B-cell lymphoma cell line RPMI
1788 (ATCC No. CRL-156); human myeloma/plasmacytoma cell line IM-9 (ATCC
No. CRL159); and human EBV transformed B-cell line DAKIKI (ATCC No. TIB-
206), and HS Sultan cells (ATCC No. CRL-1484 ). Following about 2-5 days
treatment
with zalphal 1 Ligand, changes in surface marker expression were found in IM-
9, Raji,
Ramos, and RPMI1788 cell lines, showing that these cells can respond to
zalphal 1
Ligand. Human B-cell lines treated with zalphal 1 Ligand grew much more slowly
than
untreated cells when re-plated in cell culture dishes. These cells also had an
increased
expression of FAS ligand, as assessed by flow cytometry (Example 49D and
Example
49E), and moderately increased sensitivity to an activating FAS antibody
(Example
49A). This results indicate that zalphal 1 Ligand could control some types of
B-cell
neoplasms by inducing them to differentiate to a less proliferative and or
more FAS
ligand sensitive state. Moreover, zalphal 1 receptor is expressed on the
surface of
several of these cell lines (See Example 48). Thus, zalphal 1 Ligand and the
human
zalphal 1 Ligand-saporin immunotoxin conjugate (Example 49B, below), or other
zalphal 1 Ligand-toxin fusion could be therapeutically used in B-cell
leukemias and
lymphomas.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
178
A. The effect of human zalphal 1 Ligand on B-cell lines.
IM-9 cells were seeded at about 50,000 cells per ml +/- 50 ug/m1
purified human zalphal 1 Ligand (Example 29). After 3 days growth the cells
were
harvested, washed and counted then re-plated at about 2500 cells/ml in 96 well
plates in
to wells with 0, 0.033, 0.1 or 0.33 i_tg/m1 anti-FAS antibody (R&D Systems,
Minneapolis). After 2 days an Alamar blue fluorescence assay was performed
(Example 2B) to assess proliferation of the cells.
Zalphal 1 Ligand-treated IM-9 cells grew to only 27% the density of the
untreated cells in the absence of anti-FAS antibody. In the presence of 0.33
ug/m1 anti-
FAS antibody, the zalphal 1 Ligand-treated cells were inhibited an additional
52%
while the untreated cells were inhibited by only 30%. The overall inhibition
of cell
growth with both zalphal 1 Ligand and 0.33 jig/ml anti-FAS antibody treatment
was
86%.
When the IM-9 cells were pretreated for three days with or without
zalphal 1 Ligand and then re-plated at 100 cells per well and grown with or
without
anti-FAS antibody for 6 days, the growth of untreated cells assessed by Alamar
Blue
assay (Example 2B) was inhibited only 25% by anti-FAS antibody while the
growth of
zalphal 1 Ligand-treated cells was inhibited 95% relative to the growth of
untreated
cells in zero anti-FAS antibody.
B. The effect of human zalphal 1 Ligand-saporin immunotoxin on B-cell lines.
The human zalphall Ligand-saporin immunotoxin conjugate
(zalphal 1L-sap) construction and purification is described in Example 50. The
human
zalphal 1L-sap was far more potent than the saporin alone in inhibiting cell
growth.
When the treated cell are re-plated after a three or four day treatment the
human
zalphal 1L-sap treated cells grew very poorly.
IM-9, Ramos and K562 (ATCC No. CCL-243) cells were seeded at
about 2500 cells/well in 96 well plates with zero to 250 ng/ml human zalphal
1L-sap
conjugate or 0-250 ng/ml saporin (Stirpe et al., Biotechnology 10,:405-412.
1992) only
as a control. The plates were incubated 4 days then an Alamar Blue
proliferation assay
was performed (Example 5B). At the maximal concentration of human zalphal 1-
sap
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
179
conjugate, the growth of IM-9 cells and RAMOS cells was inhibited by 79% and
65%
respectively. K562 cells which are low/negative by flow for expression of the
zalphal 1
receptor were not affected by the zalphal 1-sap, thus showing the specificity
of the
conjugate's effect.
IM-9 cells were seeded a 50,000 cells/ml into 6 well plates at zero and
50 ng/ml human zalphal1L-sap conjugate. After 3 days the cells were harvested
and
counted then re-plated from 100 to 0.8 cells per well in 2 fold serial
dilutions, and 12
wells per cell dilution without the human zalphal 1 Ligand-saporin
immunotoxin. After
6 days the number of wells with growth at each cell dilution was scored
according to
io the results of an Alamar blue proliferation assay (Example 2B).
When cell number was assessed, by Alamar blue assay (Example 2B),
after 6 days of growth control cells seeded at about 12.5 and 6.25 cells per
well had
equivalent growth to zalphal 1-sap treated cells seeded at 100 and 50
cells/well
respectively. Thus, the growth of the surviving treated IM-9 cells was
markedly
15 impaired even after the removal, by re-plating, of the zalphal 1-sap
immunotoxin.
The limited tissue distribution of the human zalphal 1 receptor (Example
48), and the specificity of action of the zalphal 1-sap to receptor-expressing
cell lines
suggest that this conjugate may be tolerated in vivo.
20 C. The effect of human zalphal 1 Ligand-saporin immunotoxin on B-cell
line viability.
HS Sultan cells (ATCC No. CRL-1484 ) were seeded at about 40,000
cells per ml into 12 well plates and grown for five days with either no added
cytokines
or 4 0 ng/ml purified human zalphal 1 Ligand (Example 29) or 25 ng/ml human
zalphal 1L-sap conjugate (Example 50, below) or with 20 ng/ml IFN-alpha (RDI)
or
25 zalphal 1 Ligand and IFN-alpha. Zalphal 1 ligand inhibited the outgrowth
of Hs Sultan
cells by 63%. IFN-alpha inhibited the growth by 38%. Zalphal 1 ligand plus IFN-
alpha inhibited growth 78%. indicating that the growth inhibitory effects of
human
zalphal 1 Ligand and IFN-alpha may be additive. The human zalphal IL-sap
inhibited
growth of the HS Sultans by 92%.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
180
The results above support the possible use of zalphall Ligand or human
zalphal 1L-sap in the treatment of malignancies or other diseases that express
the
zalphal1 receptor, particularly those of B-cell origin. The combination of
zalphall
Ligand with IFN-alpha is specifically suggested by their additive effect in
the inhibition
s of HS Sultan cells. Some other types of lymphoid malignancies and
diseases may also
express the zalphal 1 receptor, as activated T-cells also express the receptor
mRNA
(Example 48), and some of these diseases may also be responsive to zalphal 1
Ligand
of zalphall Ligand-toxic fusion therapy.
D. FAS (CD95) Expression on Human B-cell Lines is Increased by human zalphal 1
Ligand Stimulation
Human B-cell lines HS Sultan (ATCC No. CRL-1484), IM-9 (ATCC
No. CRL159), RPMI 8226 (ATCC No. CCL-155), RAMOS (ATCC No. CRL-1596),
DAKIKI (ATCC No. TIB-206), and RPMI 1788 (ATCC No. CRL-156), were all
treated with or without purified 10 to 50 ng/ml human zalphall Ligand (Example
29)
for 2 to 8 days. The cells were then stained with anti-CD95 PE-conjugated
antibody
(PharMingen, San Diego, CA), per manufacturer's protocol, and analyzed on a
FACScalibur (Becton Dickinson, San Jose, CA). In all cell lines, anti-CD95
(FAS or
APO-1) staining was increased, in some cases more than two fold, upon
treatment with
human zalphall Ligand.
E. FAS (CD95) Expression on Primary Mouse Spleen B-cells is Increased by Human
zalphal 1 Ligand Stimulation
Primary mouse splenocytes were obtained by chopping up spleens from
8 to 12 week old C57/BL6 mice. Erythrocytes were lysed by treating the
preparation
for 5 seconds with water then put through a 70 micron sieve. The remaining
splenocytes were washed and plated in RPMI (JRH Bioscience) plus 10% HIA-FBS
(Hyclone, Logan, UT). Interleukin 2 (IL-2) (R and D Systems) with or without
human
zalphall Ligand, as described above. They were then incubated at 37 C, in 5%
CO,
for 5 days. The splenocytes were harvested and stained with anti-CD95 PE
conjugated
antibody (PharMingen) and anti-CD19 FITC conjugated antibody (PharMingen) per
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
181
manufacturer's protocol. The cells were analyzed by flow cytometry on a
FACScalibur
(Becton Dickinson). Upon gating on the CD19+ mouse B-cells, it was found that
anti-
CD95 staining was increased on B-cells treated with IL-2 plus human zalphal 1
Ligand
compared to those in IL-2 alone. The anti-CD95 staining was 37 relative
fluorescent
units (RFU) on the B-cells in IL-2 alone and 55 RFU on the B-cells cultured in
IL-2
and human zalphal 1 Ligand.
Example 50
Construction and purification of Zalphal 1 Ligand toxic fusion
Under a supply contract, 10 mg human zlphal 1 Ligand (Example 29)
was sent to Advanced Targeting Systems (ATS, SanDiego, CA) for conjugation to
the
plant toxin saporin (Stirpe et al., Biotechnology 10,:405-412, 1992).
Zymogenetics
received from ATS 1.3 mg of a protein conjugate comprised of 1.1 molecules
saporin
per molecule of human zlphal 1 Ligand, formulated at a concentration of 1.14
mg/ml in
is 20 nM Sodium phosphate, 300 nM sodium cloride, pH 7.2.
Example 51
Zalphal 1 Ligand toxic fusion in vivo
A. Testing zalphal 1 -saporin conjugate in mice
Zalphall-saporin conjugate (Example 49) was administered to C57BL6
mice (female, 12 weeks of age, purchased from Taconic) at two different
dosages: 0.5
and 0.05 mg/kg. Injections were given i.v. in vehicle consisting of 0.1% BSA
(ICN,
Costa Mesa, CA). Three injections were given over a period of one week (day 0,
2, and
7). Blood samples were taken from the mice on day 0 (pre-injection) and on
days 2 and
8 (post-injection). Blood was collected into heparinized tubes (Bectin
Dickenson,
Franklin Lakes, NJ), and cell counts were determined using an automated
hematology
analyzer (Abbot Cell-Dyn modelNo. CD-3500CS, Abbot Park, IL). Animals were
euthanized and necropsied on day 8 following blood collection. Spleen, thymus,
liver.
kidney and bone marrow were collected for histopathology. Spleen and thymus
were
weighed, and and additional blood sample was collected in serum separator
tubes.
Serum was sent to Pheonix Central Labs. Everett, WA, for testing in a standard
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
182
chemistry panel. Samples were also collected for flow cytometric analysis as
described
herein.
Circulating blood cell counts and serum chemistry measurements did not
differ significantly between zalphall conjugate treated mice and mice treated
with an
equivalent dose of unconjugated toxin (saporin). Histological analysis of
tissues in
zalphall-saporin treated mice showed no significant changes relative to mice
treated
with an equivalent dose of unconjugated toxin. These results indicated that
the saporin
conjugate was not toxic in vivo.
B. Testing Zalphal 1 ligand toxic saporin fusion on B-cell derived tumors in
vivo
The effects of human zalphal 1 Ligand and the human zalphal 1 Ligand
toxic saporin fusion (Example 50) on human tumor cells are tested in vivo
using a
mouse tumor xenograft model described herein. The xenograft models are
initially
tested using cell lines selected on the basis of in vitro experiments, such as
those
described in Example 49. These cell lines include, but are not limited to:
human
Burkitt's lymphoma cell lines Raji (ATCC No.CCL-86), and Ramos (ATCC No. CRL-
1596); human cell line RPMI 1788 (ATCC No. CRL-156); human
myeloma/plasmacytoma cell line IM-9 (ATCC No. CRL159); human cell line DAKIKI
(ATCC No. TIB-206), and HS Sultan cells (ATCC No. CRL-1484). Cells derived
directly from human tumors can also be used in this type of model. In this
way,
screening of patient samples for sensitivity to treatment with zalphall Ligand
or with a
zalphal 1 Ligand toxic saporin fusion can be used to select optimal
indications for use
of zalphal 1 in anti-tumor therapy.
After selection of the appropriate zenograft in vivo model, described
above, zalphal 1 Ligand-induced activity of natural killer cells and/or
zalphal 1 Ligand
effects on B-cell derived tumors are assessed in vivo. Human zalphal 1 Ligand
is tested
for its ability to generate cytotoxic effector cells (e.g. NK cells) with
activity against B-
cell derived tumors using mouse tumor xenograft models described herein.
Moreover,
direct affects of human zalphal 1 Ligand on tumors can be assessed. The
xenograft
models to be carried out are selected as described above. A protocol using
zalphal 1
Ligand stimulated human cells is developed and tested for efficacy in
depleting tumor
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
183
cells and promoting survival in mice innoculated with cell lines or primary
tumors.
Example 52
Identification of P1 Artificial Chromosome clones containing genomic
human zalphall Ligand DNA
The human zalphal 1 Ligand cDNA insert was amplified by PCR using
vector-based primers. The PCR product was 32P-labeled and hybridized to high-
density
filters representing a PAC (P1 Artificial Chromosome) library. Filters and
frozen
library stocks were obtained from Roswell Park Cancer Institute, Buffalo, New
York;
o the library segment was RPCI6, with a four-fold depth of coverage.
Filters were
hybridized overnight at 65 C in ExpressHyb (Clontech) and were washed
according to
manufacturer's suggestions. Decoding the positive signals resulted in
identification of
four PAC clones, designated 23H17, 34A9, 105G9, and 236H14. PCR analysis using
primers specific for the 5' end (ZC22,452 (SEQ ID NO:100) and ZC 22,451 (SEQ
ID
NO:101)) and 3' end (ZC 22,450 (SEQ ID NO:102) and ZC 22A49 (SEQ ID NO:103))
of the coding region showed that PACs 34A9 and 105G9 contained both ends,
while
PACs 23H17 and 236H14 contained the 5' end only. PAC 23H17 was digested with
Eco RI and Not I, and a 9 kb fragment was identified which hybridized with the
zalphal 1 Ligand cDNA probe. This fragment was isolated and subcloned, using
methods described herein, into pBluescript II SK (+) (Stratagene) previously
digested
with Eco RI and Not I. Sequencing revealed that this fragment contained about
380
base pairs of the promoter region, exons 1, 2, and 3, all of introns 1 and 2,
and ended
within intron 3.
The 3' end of the human zalphal 1 Ligand gene was obtained by PCR
using DNA from PAC 34A9 as template, with primers ZC23.771 (SEQ ID NO:104)
and ZC22,449.(SEQ ID NO:103). Taq DNA polymerase was used, with buffer
provided, with the addition of 4% DMSO. Reaction conditions were as follows:
94 C,
5 min.; followed by 35 cycles of 94 C for 30 sec., 52 C for 1 min.. 72 C for 3
min.;
then 72 C for 7 min. This generated a 2.9 kb fragment which contained part of
exon 3,
all of introns 3 and 4, all of exon 4, and the coding portion of exon 5.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
184
The genomic structure of the human zalphall Ligand gene is as follows
from 5' to 3': SEQ ID NO:105, containing about 240 bp of the promoter, exon 1
(nucleotide number 240-455 of SEQ ID NO:105), intron 1 (nucleotide number 456-
562
of SEQ ID NO:105), exon 2 (nucleotide number 563-598 of SEQ ID NO:105), and
part
s of intron 2 containing the 5' 748 base pairs (nucleotide number 599-1347
of SEQ ID
NO:105); a gap of approximately 3 kb; SEQ ID NO:106, containing the 3' 718 bp
of
intron 2, exon 3 (nucleotide number 719-874 of SEQ ID NO:106), and part of the
5'
end of intron 3 (nucleotide number 875-1656 of SEQ ID NO:106); a gap of less
than
about 800 bp; SEQ ID NO:107, containing 644 bp of intron 3; a gap of less than
about
800 bp; and SEQ ID NO:108, containing the 3' 435 bp of intron 3, exon 4
(nucleotide
number 436-513 of SEQ ID NO:108), intron 4 (nucleotide number 514-603 of SEQ
ID
NO:108), and part of the 5' end of exon 5 (nucleotide number 604-645 of SEQ ID
NO:108).
Example 53
125I-labeled human zalphal 1 Ligand binding study in cell lines
micrograms of purified human zalphal 1 Ligand (Example 29) was
labeled with 2 mCI 121 using iodobeads (Pierce, Rockford Illinois), according
to
manufacturer's instructions. This labeled protein was used to asses human
zalphal 1
20 Ligand binding to human Raji cells (ATCC No. CCL-86), using binding to
wild-type
murine BaF3 cells, and BaF3 cells transfected with zalphal 1 receptor
(BaF3/hzalphal 1
cells) as controls. Zalphal 1 Ligand binding to BaF3/hzalphal 1 cells was
expected
(positive control), while no binding to wild-type BaF3 cells was expected
(negative
control), based on proliferation assay results (Example 5). About 5X105 Raji
cells/well,
25 1X106 BaF3/hzalphal 1 and 1X106 BaF3 cells cells/well, were each plated
in 96-well
plates. Ten ng/ml of labeled human zalphall Ligand was added in duplicate to
wells,
with a dilution series of unlabeled human zalphal 1 Ligand competitor added
from 250
fold molar excess in 1:4 dilutions down to .061 fold molar excess. Each point
was run
in duplicate. After the labeled human zalphal 1 Ligand was added to wells, it
was
allowed to incubate at 4 C for 2 h to allow for binding of Ligand to the
cells. The cells
were then washed 3X in binding buffer (RPM-1710 (JRH Bioscience) with 1% BSA
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
185
(Sigma)), and counted on the COBRA II AUTO-GAMMA gamma counter (Packard
Instrument Company, Meriden, CT).
Binding of the labeled zalphal 1 Ligand to cells was evident in the Raji
and the BaF3/hzalphal 1 cells. In addition, for Raji cells, an average 250
fold molar
excess of unlabeled zalphal 1 Ligand decreased binding 3 fold in the presence
of a non-
specific unlabeled competitor (Interferon Gamma from R&D Systems, Minneapolis,
MN), and 3.7 fold relative to no competitor. Competition was observed in a
dose
dependent fashion for the specific unlabeled competitor, human zalphal 1
Ligand.
Thus, the zalphal 1 Ligand binding to Raji cells was specific. Similarly, for
positive
control BaF3/zalphal 1 cells, the 250 fold molar excess of unlabeled zalphal 1
Ligand
decreased binding 2 fold relative to the non-specific competitor and 3.06 fold
relative to
no competitor. Thus, the zalphal 1 Ligand binding to BaF3/zalphal 1 cells also
was
specific. No compeatable binding was observed with the wild-type BaF3 cells.
Thus,
the zalphal 1 Ligand was shown to bind specifically to Raji cells, and to
is Baf3/hzalphal 1 cells, but not to the negative control Baf3 cells.
Example 54
Zalphall Receptor Expression On Human Blood Cells
A. Preparation and Culture of Human Peripheral Blood Cells
Fresh drawn human blood was diluted 1:1 with PBS (GIBCO BRL) and
layered over Ficoll/Hypaque Plus (Pharmacia LKB Biotechnology Inc.,
Piscataway,
NJ) and spun for 30 minutes at 1800 rpm and allowed to stop with the brake off
The
interface layer was removed and transferred to a fresh 50 ml Falcon tube
(Falcon,
VWR, Seattle, WA), brought up to a final volume of 40 ml with PBS and spun for
10
minutes at 1200 rpm with the brake on. The viability of the isolated cells was
tested
using Trypan Blue (GIBCO BRL) and the cells were resuspended at a final
concentration of 1 x 106 cells/ml cell medium (RPMI Medium 1640, 10% Heat
inactivated fetal bovine serum, 5% L-glutamine. 5% Pen/Strep) (GIBCO BRL).
Cells were cultured in 6 well plates (Falcon, VWR) for 0, 4 or 24 hours
with a variety of different stimuli described below. Anti-IgM, anti-CD40 and
anti-CD3
stimulation were done as in Example 44 and Example 42. Phorbol myristate
acetate
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
186
(PMA) and ionomycin (Sigma, St. Louis, MO) (Example 5C) were added to
appropriate wells at 10 ng/ml and 0.5 mg/ml respectively. The cells were
incubated at
37 C in a humidified incubator for various times.
B. Antibody Staining and Analysis
Cells were collected out of the plates, washed and resuspended in ice
cold staining media (HBSS, 1% fetal bovine serum, 0.1% sodium azide) at a
concentration of about ten million cells per milliliter. Blocking of Fc
receptor and non-
specific binding of antibodies to the cells was achieved by adding 10% normal
goat
serum (Gemini Bioproducts, Woodland, CA) and 10% normal human serum
(Ultraserum, Gemini) to the cell suspension. Aliquots of the cell suspensions
were
mixed with a FITC labeled monoclonal antibody against one of the lineage
markers
CD3, CD19 or CD14 (PharMingen, La Jolla, CA) and a biotinylated monoclonal
antibody against the human zalphall receptor (hu-zalphall) (Example 35).
Staining
is specificity was determined by competition using zalphal 1 CEE soluble
receptor
(Example 10A) at a ten fold mass excess. After incubation on ice for 60
minutes the
cells were washed twice with ice cold staining media and resuspended in 50 ml
staining
media containing streptavidin-PE (Caltag, Burlingame, CA). After a 30 minute
incubation on ice, the cells were washed twice with ice cold wash buffer (PBS.
1% fetal
bovine serum, 0.1% sodium azide) and resuspended in wash buffer containing 1
mg/ml
7-AAD (Molecular Probes, Eugene, OR) as a viability marker. Flow data was
acquired
on living cells using a FACSCalibur flow cytometer (BD Immunocytometry
Systems,
San Jose, CA). Both acquisition and analysis were performed using CellQuest
software
(BD Immunocytometry Systems).
Results of staining by anti-zalphal 1 antibody showed that the human
zalphal 1 receptor is expressed on human peripheral blood cells expressing
either CD3,
CD19 or CD14. Staining on CD3 and CD19 cells was specific, as evidenced by
absolute competion with the zalphal 1 soluble receptor. Staining on CD14 cells
showed
some specificity for the Ligand, as evidenced by partial competion with the
soluble
receptor. Activation of either T cells with anti-CD3 or B cells with anti-CD40
resulted
in an increased level of cell surface zalphal 1 at 24 hours. No increase in
the level of
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
187
expression of zalphal 1 was seen at 4 hours with any stimulus on either cell
population.
Treatment of the cells with zalphal 1 ligand resulted in a decrease of zalphal
1 staining
on CD3 positive and CD19 positive cells but not CD14 positive cells at both 4
and 24
hours.
Example 55
Preliminary Evaluation of the Aqueous Stability of human zalphal 1 Ligand
Preliminary studies were conducted to evaluate the aqueous stability
characteristics of human zalphal 1 Ligand in support of bioprocessing,
formulation, and
io in vivo administration. The objectives were to: 1) verify the stability
and recovery from
Alzet Minipumps & general storage and handling, 2) determine the stability-
indicating
nature of several analytical methods including cation-exchange HPLC (CX-HPLC),
reverse-phase HPLC (RP-HPLC), size exclusion HPLC (SEC-HPLC), & bioassay
(BaF3/zalphal 1R proliferation (e.g., Example 2 and Example 4), and 3)
determine the
stability-limiting degradation pathways and their kinetic dependencies.
Aliquots of purified human zalphal 1 Ligand (Example 29) were
prepared by dilution to 2 mg/mL in PBS (pH 7.4) and stored in low density
polyethylene (LDPE) cryovials (Nalgene, 1.8 mL) at -80 C (control), 5 C, 30 C,
and
37 C. Samples were assayed intermittently over 29 days by CX-, RP-, SEC-HPLC,
and bioassay. Aliquots were also stored at -80 C and subjected to freeze-thaw
(f/t)
cycling (-80 C/RT; 5X f/t, 10X f/t). Recovery of human zalphal 1 Ligand was
determined relative to the -80 C control (1 f/t) in all assays.
The remaining human zalphall Ligand solution from the -80 C control
samples were refrozen (-80 C) after analysis. This aliquot (2 f/t) was used to
evaluate
the thermal and conformational stability of human zalphal 1 Ligand as a
function of pH
using circular dichroism (CD). The 2 mg/mL solution was diluted to 100 ug/mL
in
PBS buffers ranging from pH 3.3-8.8. The far-UV CD spectra was monitored over
the
temperature range 5-90 C in 5 C intervals (n=3/pH). The CD spectropolarimeter
used
was a Jasco 715 (Jasco, Easton, MD). The thermal unfolding was monitored by
changes in ellipticity at 222 nm as a function of temperature. Estimates of
the T,, were
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
188
estimated assuming a two-state unfolding model. The data was fit (sigmoidal)
using
Slide Write Plus for Windows v4.1 (Advanced Graphics Software; Encinitas, CA).
Recovery and stability from Alzet Minipumps (Model No. 1007D;
ALZA Corporation, Mountain View, CA) was assessed by filling pumps with 100
!IL
of the 2 mg/mL human zalphal 1 Ligand solution, placing the pumps in 1.8 mL
LDPE
containing 1 mL of PBS (pH 7.4), and storing them at 37 C. The
release/recovery of
human zalphal 1 Ligand from the minipumps was assessed by CX-, RP-, and SEC-
HPLC on days 2, 4, and 7. The activity was assessed by bioassay on day 7. The
study
o was designed to evaluate the release from 3 pumps per sampling time.
The chromatographic data suggested that the CX- & SEC-HPLC
methods were stability-indicating, whereas the RP-HPLC method was not. At
least 3
additional peaks indicating apparent degradation products were observed by CX-
HPLC.
The SEC-HPLC method resolved an apparent human zalphal 1 Ligand aggregate,
eluting prior to human zalphall Ligand. However, no significant additional
peaks were
observed eluting after the human zalphal 1 Ligand peak. This suggests that the
degradation products observed by CX-HPLC most probably result from amino acid
modifications such as deamidation, rather than hydrolysis/proteolysis
processes leading
to clipped variants. A small degree of fronting/tailing was observed by RP-
HPLC
(relative to control) in samples which had been shown to have undergone
significant
degradation by SEC- & CX-HPLC. However, apparent degradation products were not
resolved by RP-HPLC. The degradation observed by CX-HPLC increased as a
function of time-temperature, and followed apparent first-order kinetics. The
% human
zalphal 1 Ligand recovered by CX-HPLC after 29 days at 37 C, 30 C, and 5 C was
39%, 63%, and 98%, respectively. Aggregation also increased in a time-
temperature
dependent fashion. The % aggregate found in preparations stored for 29 days at
37 C,
C, and 5 C was 7.4, 3.4, and below detectable limits (BDL), respectively. No
significant differences were observed by bioassay in any sample, suggesting
the
30 degradation products have equivalent activity to intact human zalphal 1
Ligand. No
degradation was observed by any assay in samples subjected to up to 10 f/t
cycles.
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
189
The release of human zalphal 1 Ligand from Alzet Minipumps was
consistent with the theoretical expected volume release. This suggests that
significant
surface adsorption would not impair the delivery of human zalphal 1 Ligand
using the
Alzet Minipumps with a 2 mg/mL fill concentration. The degradation consistent
with
.5 that previously noted was observed. The % purity determined by CX-HPLC
of human
zalphal1 Ligand released after 2, 4, and 7 days was 96%, 90%, and 79%,
repectively.
It should be recognized that degradation also occurs after human zalphal 1
Ligand is
released into or diluted with release medium. Therefore, the % purity within
the
minipump may be somewhat different than that determined to be in the release
medium. The bioactivity of each sample was consistent with the expected amount
of
human zalphall Ligand released from the minipumps.
The human zalphal 1 Ligand far-UV CD spectra, as expected, was
consistent with interleukins, such as IL-3 (J. Biochem., 23:352-360, 1991), IL-
4
(Biochemistry, 30:1259-1264, 1991), and IL-6 mutants (Biochemistry, 35:11503-
11511, 1996). Gross changes in the far-uv CD spectra as a function of pH were
not
observed. Results showed that the pH of maximum thermal/conformational
stability
was pH 7.4. Analysis of the unfolding curves were based on a two-state
unfolding
mechanism to allow comparison of the thermal/conformational stability as a
function of
pH/composition. However, one or more intermediates may exist during the
unfolding
process since the cooperativity was relatively low, based on the shallowness
of the
unfolding curve. Although studies were not specifically designed to determine
whether
human zalphal 1 Ligand refolds following thermal unfolding to 90 C,
preliminary data
suggests that at least partial refolding occurs after the temperature of the
sample is
cooled back to 20 C.
These studies allow an analytical paradigm to be identified to evaluate
the purity and verify the stability of human zalphal 1 Ligand. For instance,
SEC-HPLC
can be used to characterize the extent and rate of aggregation in aqueous
solution.
Likewise. CX-HPLC can be used to characterize the extent and rate of
degradation of
human zalphal 1 Ligand by mechanisms other than aggregation. The bioassay can
be
used to verify activity of human zalphal 1 Ligand and it's aqueous degradation
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
190
products. For instance, the human zalphal 1 Ligand variants generated in
aqueous
solution & resolved by CX-HPLC may themselves be useful as therapeutic agents,
since they have equivalent bioactivity. Also, the fact that human zalphal 1
Ligand
degrades by several different processes (aggregation, amino acid
modifications)
suggests a preferred or unique formulation which minimizes the rate of each
degradation process may be necessary for long-term stability of a solution
product.
Identification of the nature of the aqueous degradation products and
determination of their kinetic dependencies (pH, concentration, excipients) is
underway. Human zalphal 1 Ligand stability in serum/plasma is determined to
support
the design and interpretation of in vivo studies.
From the foregoing, it will be appreciated that, although specific
embodiments of the invention have been described herein for purposes of
illustration,
various modifications may be made without deviating from the spirit and scope
of the
invention. Accordingly, the invention is not limited except as by the appended
claims.
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
1
SEQUENCE LISTING
<110> ZymoGenetics, Inc.
<120> NOVEL CYTOKINE ZALPHA11 LIGAND
<130> 99-16PC
<150> US 09/264,908
<151> 1999-03-09
<150> US 09/265,992
<151> 1999-03-11
<150> US 60/142,013
<151> 1999-07-01
<160> 115
<170> FastSEQ for Windows Version 3.0
<210> 1
<211> 642
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (47)...(532)
<400> 1
gctgaagtga aaacgagacc aaggtctagc tctactgttg gtactt atg aga tcc 55
Met Arg Ser
1
agt cct ggc aac atg gag agg att gtc atc tgt ctg atg gtc atc ttc 103
Ser Pro Gly Asn Met Gin Arg Ile Val Ile Cys Len Met Val Ile Phe
10 15
ttg ggg aca ctg gtc cac aaa tca agc tcc caa ggt caa gat cgc cac 151
Len Gly Thr Leu Val His Lys Ser Ser Ser Gin Gly Gin Asp Arg His
20 25 30 35
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
2
atg att aga atg cgt caa ctt ata gat att gtt gat cag ctg aaa aat 199
Met Ile Arg Met Arg Gin Leu Ile Asp Ile Val Asp Gin Leu Lys Asn
40 45 50
tat gtg aat gac ttg gtc cct gaa ttt ctg cca gct cca gaa gat gta 247
Tyr Val Asn Asp Leu Val Pro Glu Phe Leu Pro Ala Pro Glu Asp Val
55 60 65
gag aca aac tgt gag tgg tca gct ttt tcc tgt ttt cag aag gcc caa 295
Glu Thr Asn Cys Glu Trp Ser Ala Phe Ser Cys Phe Gin Lys Ala Gin
70 75 80
cta aag tca gca aat aca gga aac aat gaa agg ata atc aat gta tca 343
Leu Lys Ser Ala Asn Thr Gly Asn Asn Glu Arg Ile Ile Asn Val Ser
85 90 95
att aaa aag ctg aag agg aaa cca cct tcc aca aat gca ggg aga aga 391
Ile Lys Lys Leu Lys Arg Lys Pro Pro Ser Thr Asn Ala Gly Arg Arg
100 105 110 115
cag aaa cac aga cta aca tgc cct tca tgt gat tct tat gag aaa aaa 439
Gin Lys His Arg Leu Thr Cys Pro Ser Cys Asp Ser Tyr Glu Lys Lys
120 125 130
cca ccc aaa gaa ttc cta gaa aga ttc aaa tca ctt ctc caa aag atg 487
Pro Pro Lys Glu Phe Leu Glu Arg Phe Lys Ser Leu Leu Gin Lys Met
135 140 145
att cat cag cat ctg tcc tct aga aca cac gga agt gaa gat tcc 532
Ile His Gin His Leu Ser Ser Arg Thr His Gly Ser Glu Asp Ser
150 155 160
tgaggatcta acttgcagtt ggacactatg ttacatactc taatatagta gtgaaagtca 592
tttctttgta ttccaagtgg aggagcccta ttaaattata taaagaaata 642
<210> 2
<211> 162
<212> PRT
<213> Homo sapiens
<400> 2
Met Arg Ser Ser Pro Gly Asn Met Glu Arg Ile Val Ile Cys Leu Met
1 5 10 15
Val Ile Phe Leu Gly Thr Leu Val His Lys Ser Ser Ser Gin Gly Gin
20 25 30
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
3
Asp Arg His Met Ile Arg Met Arg Gin Leu Ile Asp Ile Val Asp Gin
35 40 45
Leu Lys Asn Tyr Val Asn Asp Leu Val Pro Glu Phe Leu Pro Ala Pro
50 55 60
Glu Asp Val Glu Thr Asn Cys Glu Trp Ser Ala Phe Ser Cys Phe Gin
65 70 75 80
Lys Ala Gin Leu Lys Ser Ala Asn Thr Gly Asn Asn Glu Arg Ile Ile
85 90 95
Asn Val Ser Ile Lys Lys Leu Lys Arg Lys Pro Pro Ser Thr Asn Ala
100 105 110
Gly Arg Arg Gin Lys His Arg Leu Thr Cys Pro Ser Cys Asp Ser Tyr
115 120 125
Glu Lys Lys Pro Pro Lys Glu Phe Leu Glu Arg Phe Lys Ser Leu Leu
130 135 140
Gin Lys Met Ile His Gin His Leu Ser Ser Arg Thr His Gly Ser Glu
145 150 155 160
Asp Ser
<210> 3
<211> 486
<212> DNA
<213> Artificial Sequence
<220>
<223> Degenerate polynucleotide sequence for human
zalphall ligand
<221> misc feature
<222> (1)...(486)
<223> n = A,T,C or G
<400> 3
atgmgnwsnw snccnggnaa yatggarmgn athgtnatht gyytnatggt nathttyytn 60
ggnacnytng tncayaarws nwsnwsncar ggncargaym gncayatgat hmgnatgmgn
120
carytnathg ayathgtnga ycarytnaar aaytaygtna aygayytngt nccngartty
180
ytnccngcnc cngargaygt ngaracnaay tgygartggw sngcnttyws ntgyttycar
240
aargcncary tnaarwsngc naayacnggn aayaaygarm gnathathaa ygtnwsnath
300
aaraarytna armgnaarcc nccnwsnacn aaygcnggnm gnmgncaraa rcaymgnytn
360
acntgyccnw sntgygayws ntaygaraar aarccnccna argarttyyt ngarmgntty
420
aarwsnytny tncaraarat gathcaycar cayytnwsnw snmgnacnca yggnwsngar
480
gaywsn
486
<210> 4
<211> 535
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
4
<212> DNA
<213> Mus musculus
<220>
<221> source
<222> (0)...(0)
<223> EST1483966 ; GenBank Acc #AA764063
<400> 4
taaacatgta tcatataagg atatgtcata ataaggatta atattatata attataaata
60
atttataata cttataatat cattgtttgg ttcactaata aatctatgga tacatggtca
120
aaatggaaat gaatattttg ccaattatta atccccaaag tcattgaaaa taagcataac
180
cattctactg acttgttaga ctctaaacta acataaaata cattttcaga aataaattca
240
accgatctta cctttacatc ttgtggagct gatagaagtt caggatccta agaaaattaa
300
ccaaagagta ttagttctga gttggtgata caagtcaaaa ggctcctttt gcattaatta
360
aaaaaatatt atttaaattg cattgtgaca aacatggcct taccaagtca ttttcataga
420
ttttcagctg ttcaacaatg tcaataaggt gacgaagtct aatcaggagg cgatctggcc
480
cttgggggct tgatttatgg gccactgtcc ccaagaagat gactaccaga cagac
535
<210> 5
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC17212
<400> 5
ggggaattcg aagccatgcc ctcttgggcc ctc
33
<210> 6
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer 7C19914
<400> 6
caatggatgg gtctttagca gcagtaggcc
30
<210> 7
<211> 1614
<212> DNA
<213> Homo sapiens
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
<400> 7
atgccgcgtg gctgggccgc ccccttgctc ctgctgctgc tccagggagg ctggggctgc 60
cccgacctcg tctgctacac cgattacctc cagacggtca tctgcatcct ggaaatgtgg
120
aacctccacc ccagcacgct cacccttacc tggcaagacc agtatgaaga gctgaaggac
180
gaggccacct cctgcagcct ccacaggtcg gcccacaatg ccacgcatgc cacctacacc
240
tgccacatgg atgtattcca cttcatggcc gacgacattt tcagtgtcaa catcacagac
300
cagtctggca actactccca ggagtgtggc agctttctcc tggctgagag catcaagccg
360
gctccccctt tcaacgtgac tgtgaccttc tcaggacagt ataatatctc ctggcgctca
420
gattacgaag accctgcctt ctacatgctg aagggcaagc ttcagtatga gctgcagtac
480
aggaaccggg gagacccctg ggctgtgagt ccgaggagaa agctgatctc agtggactca
540
agaagtgtct ccctcctccc cctggagttc cgcaaagact cgagctatga gctgcaggtg
600
cgggcagggc ccatgcctgg ctcctcctac caggggacct ggagtgaatg gagtgacccg
660
gtcatctttc agacccagtc agaggagtta aaggaaggct ggaaccctca cctgctgctt
720
ctcctcctgc ttgtcatagt cttcattcct gccttctgga gcctgaagac ccatccattg
780
tggaggctat ggaagaagat atgggccgtc cccagccctg agcggttctt catgcccctg
840
tacaagggct gcagcggaga cttcaagaaa tgggtgggtg cacccttcac tggctccagc
900
ctggagctgg gaccctggag cccagaggtg ccctccaccc tggaggtgta cagctgccac
960
ccaccacgga gcccggccaa gaggctgcag ctcacggagc tacaagaacc agcagagctg
1020
gtggagtctg acggtgtgcc caagcccagc ttctggccga cagcccagaa ctcggggggc
1080
tcagcttaca gtgaggagag ggatcggcca tacggcctgg tgtccattga cacagtgact
1140
gtgctagatg cagaggggcc atgcacctgg ccctgcagct gtgaggatga cggctaccca
1200
gccctggacc tggatgctgg cctggagccc agcccaggcc tagaggaccc actcttggat
1260
gcagggacca cagtcctgtc ctgtggctgt gtctcagctg gcagccctgg gctaggaggg
1320
cccctgggaa gcctcctgga cagactaaag ccaccccttg cagatgggga ggactgggct
1380
gggggactgc cctggggtgg ccggtcacct ggaggggtct cagagagtga ggcgggctca
1440
cccctggccg gcctggatat ggacacgttt gacagtggct ttgtgggctc tgactgcagc
1500
agccctgtgg agtgtgactt caccagcccc ggggacgaag gacccccccg gagctacctc
1560
cgccagtggg tggtcattcc tccgccactt tcgagccctg gaccccaggc cagc
1614
<210> 8
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19913
<400> 8
ggcctactgc tgctaaagac ccatccattg 30
<210> 9
<211> 33
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
6
<220>
<223> Oligonucleotide primer ZC20097
<400> 9
acatctagat tagctggcct ggggtccagg cgt
33
<210> 10
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC12700
<400> 10
ggaggtctat ataagcagag c
21
<210> 11
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZO5020
<400> 11
cactggagtg gcaacttcca g
21
<210> 12
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC6675
<400> 12
gtggatgccg aacccagtcc
20
<210> 13
<211> 21
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCIYUS00/06067
7
<220>
<223> Oligonucleotide primer ZC7727
<400> 13
tgttcacagc tacctgggct c 21
<210> 14
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC8290
<400> 14
ccaccgagac tgcttggatc accttg 26
<210> 15
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19572
<400> 15
gtcctgtggc tgtgtctcag 20
<210> 16
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC6622
<400> 16
ctgggctgga aactggcaca c 21
<210> 17
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
8
<223> Oligonucleotide primer ZC7736
<400> 17
cactgtcaga aatggagc 18
<210> 18
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC9273
<400> 18
ggtccctccc cgggcaccga gaga 24
<210> 19
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19905
<400> 19
acaggatccg tcagcatgcc gcgtggctgg gccgcc 36
<210> 20
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19906
<400> 20
acagaattct tagctggcct ggggtccagg cgt 33
<210> 21
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC20114
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
9
<400> 21
cctgccttct acatgctgaa gg
22
<210> 22
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19459
<400> 22
ctcctcctgc ttgtcatagt c
21
<210> 23
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19954
<400> 23
actgggctgg gggactgc
18
<210> 24
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC20116
<400> 24
agcacagtca ctgtgtcaat gg
22
<210> 25
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC13946
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
<400> 25
ccctgcagtg atcaacatgg ccaagttgac cagtgccgtt
40
<210> 26
<211> 45
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC13945
<400> 26
gcccatggac tagtttcgaa aggtcgagtg tcagtcctgc tcctc
45
<210> 27
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC18698
<400> 27
tttttttctc gagacttttt tttttttttt tttt
34
<210> 28
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC14063
<400> 28
caccagacat aatagctgac agact
25
<210> 29
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Glu-Glu (CEE) Tag amino acid sequence
<400> 29
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
11
Glu Tyr Met Pro Met Glu
1 5
<210> 30
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19931
<400> 30
ggttggtacc gcaagatgcc gcgtggctgg gccgcc
36
<210> 31
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19932
<400> 31
cggaggatcc gtgagggttc cagccttcc
29
<210> 32
<211> 66
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer spanning vector flanking
region and the 5 end of the zalphall
extracellular domain
<400> 32
tccactttgc ctttctctcc acaggtgtcc agggaattca tcgataatgc cgcgtggctg
60
ggccgc
66
<210> 33
<211> 699
<212> DNA
<213> Homo sapiens
<400> 33
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
12
gagcccagat cttcagacaa aactcacaca tgcccaccgt gcccagcacc tgaagccgag
60
ggggcaccgt cagtcttcct cttcccccca aaacccaagg acaccctcat gatctcccgg
120
acccctgagg tcacatgcgt ggtggtggac gtgagccacg aagaccctga ggtcaagttc
180
aactggtacg tggacggcgt ggaggtgcat aatgccaaga caaagccgcg ggaggagcag
240
tacaacagca cgtaccgtgt ggtcagcgtc ctcaccgtcc tgcaccagga ctggctgaat
300
ggcaaggagt acaagtgcaa ggtctccaac aaagccctcc catcctccat cgagaaaacc
360
atctccaaag ccaaagggca gccccgagaa ccacaggtgt acaccctgcc cccatcccgg
420
gatgagctga ccaagaacca ggtcagcctg acctgcctgg tcaaaggctt ctatcccagc
480
gacatcgccg tggagtggga gagcaatggg cagccggaga acaactacaa gaccacgcct
540
cccgtgctgg actccgacgg ctccttcttc ctctacagca agctcaccgt ggacaagagc
600
aggtggcagc aggggaacgt cttctcatgc tccgtgatgc atgaggctct gcacaaccac
660
tacacgcaga agagcctctc cctgtctccg ggtaaataa
699
<210> 34
<211> 62
<212> DNA
<213> Artificial Sequence
<220>
<223> First Oligonucleotide primer spanning 3' end of
the zalpha11 extracellular domain and the 5' end
of Fc4
<400> 34
gcacggtggg catgtgtgag ttttgtctga agatctgggc tcgtgagggt tccagccttc
60
ct
62
<210> 35
<211> 61
<212> DNA
<213> Artificial Sequence
<220>
<223> Second Oligonucleotide primer spanning 3 end of
the zalpha11 extracellular domain and the 5' end
of Fc4
<400> 35
agacccagtc agaggagtta aaggaaggct ggaaccctca cgagcccaga tcttcagaca
60
a
61
<210> 36
<211> 67
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
13
<220>
<223> Oligonucleotide primer spanning the 3 end of Fc4
and the vector flanking region
<400> 36
gtgggcctct ggggtgggta caaccccaga gctgttttaa tctagattat ttacccggag
60
acaggga
67
<210> 37
<211> 8
<212> PRT
<213> Artificial Sequence
<220>
<223> C-terminal FLAG amino acid sequence
<400> 37
Asp Tyr Lys Asp Asp Asp Asp Lys
1 5
<210> 38
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC7764a
<400> 38
tttttttttt tttttttttt ttttta
26
<210> 39
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC7764b
<400> 39
tttttttttt tttttttttt tttttc
26
<210> 40
<211> 19
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
14
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22034
<400> 40
ttcaaatcac ttctccaaa 19
<210> 41
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22035
<400> 41
ttttggagaa gtgatttgaa 20
<210> 42
<211> 16
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22050
<400> 42
gaatgcgtca acttat 16
<210> 43
<211> 16
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22051
<400> 43
ggaccaagtc attcac 16
<210> 44
<211> 25
<212> DNA
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22056
<400> 44
gtctgtctgg tagtcatctt cttgg 25
<210> 45
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22057
<400> 45
cttgtggagc tgatagaagt tcagg 25
<210> 46
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22205
<400> 46
agctgttcaa caatgtcaat aaggtg 26
<210> 47
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22206
<400> 47
cctcctgatt agacttcgtc acct 24
<210> 48
<211> 27
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
16
<220>
<223> Oligonucleotide primer ZC9739
<400> 48
ccatcctaat acgactcact atagggc
27
<210> 49
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC9719
<400> 49
actcactata gggctcgagc ggc
23
<210> 50
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC14063
<400> 50
caccagacat aatagctgac agact
25
<210> 51
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC5020
<400> 51
cactggagtg gcaacttcca g
21
<210> 52
<211> 20
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
17
<220>
<223> Oligonucleotide primer ZC22421
<400> 52
ctaaaatggc tccttcaaaa 20
<210> 53
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22604
<400> 53
cacacaggcc ggccaccatg ggcttccagc ctccggccgc 40
<210> 54
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22641
<400> 54
atgcgttggt tctgattgtg 20
<210> 55
<211> 3072
<212> DNA
<213> mus musculus
<220>
<221> CDS
<222> (54)...(491)
<400> 55
gagaaccaga ccaaggccct gtcatcagct cctggagact cagttctggt ggc atg 56
Met
1
gag agg acc ctt gtc tgt ctg gta gtc atc ttc ttg ggg aca gtg gcc 104
Glu Arg Thr Leu Val Cys Leu Val Val Ile Phe Leu Gly Thr Val Ala
10 15
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
18
cat aaa tca agc ccc caa ggg cca gat cgc ctc ctg att aga ctt cgt
152
His Lys Ser Ser Pro Gin Gly Pro Asp Arg Leu Leu Ile Arg Leu Arg
20 25 30
cac ctt att gac att gtt gaa cag ctg aaa atc tat gaa aat gac ttg
200
His Leu Ile Asp Ile Val Glu Gin Leu Lys Ile Tyr Glu Asn Asp Leu
35 40 45
gat cct gaa ctt cta tca gct cca caa gat gta aag ggg cac tgt gag
248
Asp Pro Glu Leu Leu Ser Ala Pro Gin Asp Val Lys Gly His Cys Glu
50 55 60 65
cat gca gct ttt gcc tgt ttt cag aag gcc aaa ctc aag cca tca aac
296
His Ala Ala Phe Ala Cys Phe Gin Lys Ala Lys Leu Lys Pro Ser Asn
70 75 80
cct gga aac aat aag aca ttc atc att gac ctc gtg gcc cag ctc agg
344
Pro Gly Asn Asn Lys Thr Phe Ile Ile Asp Leu Val Ala Gin Leu Arg
85 90 95
agg agg ctg cct gcc agg agg gga gga aag aaa cag aag cac ata gct
392
Arg Arg Leu Pro Ala Arg Arg Gly Gly Lys Lys Gin Lys His Ile Ala
100 105 110
aaa tgc cct tcc tgt gat tcg tat gag aaa agg aca ccc aaa gaa ttc
440
Lys Cys Pro Ser Cys Asp Ser Tyr Glu Lys Arg Thr Pro Lys Glu Phe
115 120 125
cta gaa aga cta aaa tgg ctc ctt caa aag atg att cat cag cat ctc
488
Leu Glu Arg Leu Lys Trp Leu Leu Gin Lys Met Ile His Gin His Leu
130 135 140 145
tcc tagaacacat aggacccgaa gattcctgag gatccgagaa gattcccgag
541
Ser
gactgaggag acgccggaca ctatagacgc tcacgaatgc aggagtacat cttgcctctt
601
gggattgcaa gtggagaagt acgatacgtt atgataagaa caactcagaa aagctatagg
661
ttaagatcct ttcgcccatt aactaagcag acattgtggt tccctgcaca gactccatgc
721
tgtcaacatg gaaaatctca actcaacaag agcccagctt cccgtgtcag ggatttctgg
781
tgcttctcaa gctgtggctt catcttattg cccaactgtg acattctttg attggaaggg
841
gaaaactaaa gcttttagca aaaatacagc tagggaattt gtcgatctgc gagagtaaga
901
cctcttatga tcctaacgga atgatgtaag ctggaaataa taagcataag atgaaattga
961
aaattgaagt ctttattctt taagaaaaac tttgtacttg aaagcatgtc tgaagagttt
1021
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
19
actcattacc acaaacatct agcatattga taactaacat ctttatactc tacaagagag
1081
gctttccaga taggtacagt ttttcttctc tattaggtct atcaaaattt aacctattat
1141
gagggtcacc cctggctttc actgtttttc taaagaggca agggtgtagt aagaagcagg
1201
cttaagttgc cttcctccca atgtcaagtt cctttataag ctaatagttt aatcttgtga
1261
agatggcaat gaaagcctgt ggaagtgcaa acctcactat cttctggagc caagtagaat
1321
tttcaagttt gtagctctca cctcaagtgg ttatgggtgt cctgtgatga atctgctagc
1381
tccagcctca gtctcctctc ccacatcctt tcctttcttt cctctttgaa acttctaaga
1441
aaaagcaatc caaacaagtt cagcacttaa gacacattgc atgcacactt ttgataagtt
1501
aaatccaacc atctatttaa aatcaaaatc aggagatgag ccaagagacc agaggttctg
1561
ttccagtttt aaacagactt ttactgaaca tcccaatctt ttaaccacag aggctaaatt
1621
gagcaaatag ttttgccatt tgatataatt tccaacagta tgtttcaatg tcaagttaaa
1681
aagtctacaa agctattttc cctggagtgg tatcatcgct ttgagaattt cttatggtta
1741
aaatggatct gagatccaag catggcctgg gggatggttt tgatctaagg aaaaaggtgt
1801
ctgtacctca cagtgccttt aaaacaagca gagatcccgt gtaccgccct aagatagcac
1861
agactagtgt taactgattc ccagaaaagt gtcacaatca gaaccaacgc attctcttaa
1921
actttaaaaa tatgtattgc aaagaacttg tgtaactgta aatgtgtgac tgttgatgac
1981
attatacaca catagcccac gtaagtgtcc aatggtgcta gcattggttg ctgagtttgc
2041
tgctcgaaag ctgaagcaga gatgcagtcc ttcacaaagc aatgatggac agagagggga
2101
gtctccatgt tttattcttt tgttgtttct ggctgtgtaa ctgttgactt cttgacattg 2161
tgatttttat atttaagaca atgtatttat tttggtgtgt ttattgttct agccttttaa
2221
atcactgaca atttctaatc aagaagtaca aataattcaa tgcagcacag gctaagagct
2281
tgtatcgttt ggaaaagcca gtgaaggctt ctccactagc catgggaaag ctacgcttta
2341
gagtaaacta gacaaaattg cacagcagtc ttgaacctct ctgtgctcaa gactcagcca
2401
gtcctttgac attattgttc actgtgggtg ggaacacatt ggacctgaca cactgttgtg
2461
tgtccatgaa ggttgccact ggtgtaagct ttttttggtt ttcattctct tatctgtaga
2521
acaagaatgt ggggctttcc taagtctatt ctgtatttta ttctgaactt cgtatgtctg
2581
agttttaatg ttttgagtac tcttacagga acacctgacc acacttttga gttaaatttt
2641
atcccaagtg tgatatttag ttgttcaaaa agggaaggga tatacataca tacatacata
2701
catacataca tatatatata tatatataca tatatatata tatatatatg tatatatata
2761
tatatataga gagagagaga gagagagaga gagaaagaga gagaggttgt tgtaggtcat
2821
aggagttcag aggaaatcag ttatggccgt taatactgta gctgaaagtg ttttctttgt
2881
gaataaattc atagcattat tgatctatgt tattgctctg ttttatttac agtcacacct
2941
gagaatttag ttttaatatg aatgatgtac tttataactt aatgattatt tattatgtat
3001
ttggttttga atgtttgtgt tcatggcttc ttatttaaga cctgatcata ttaaatgcta
3061
cccagtccgg a
3072
<210> 56
<211> 146
<212> PRT
<213> mus musculus
<400> 56
Met Glu Arg Thr Leu Val Cys Leu Val Val Ile Phe Leu Gly Thr Val
1 5 10 15
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
Ala His Lys Ser Ser Pro Gln Gly Pro Asp Arg Leu Leu Ile Arg Leu
20 25 30
Arg His Leu Ile Asp Ile Val Glu Gin Leu Lys Ile Tyr Glu Asn Asp
35 40 45
Leu Asp Pro Glu Leu Leu Ser Ala Pro Gin Asp Val Lys Gly His Cys
50 55 60
Glu His Ala Ala Phe Ala Cys Phe Gin Lys Ala Lys Leu Lys Pro Ser
65 70 75 80
Asn Pro Gly Asn Asn Lys Thr Phe Ile Ile Asp Leu Val Ala Gin Leu
85 90 95
Arg Arg Arg Leu Pro Ala Arg Arg Gly Gly Lys Lys Gin Lys His Ile
100 105 110
Ala Lys Cys Pro Ser Cys Asp Ser Tyr Glu Lys Arg Thr Pro Lys Glu
115 120 125
Phe Leu Glu Arg Leu Lys Trp Leu Leu Gin Lys Met Ile His Gin His
130 135 140
Leu Ser
145
<210> 57
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22283
<400> 57
cgctcgagac catggagagg acccttgtct gtct
34
<210> 58
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22284
<400> 58
gctctagaat cttctcggat cctcaggaat c
31
<210> 59
<211> 100
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
21
<220>
<223> Oligonucleotide ZC12749
<400> 59
gtaccttccc gtaaatccct ccccttcccg gaattacaca cgcgtatttc ccagaaaagg
60
aactgtagat ttctaggaat tcaatccttg gccacgcgtc
100
<210> 60
<211> 100
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide ZC12748
<400> 60
tcgagacgcg tggccaagga ttgaattcct agaaatctac agttcctttt ctgggaaata
60
cgcgtgtgta attccgggaa ggggagggat ttacgggaag
100
<210> 61
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer 7C22143
<400> 61
cgtatcggcc ggccaccatg agatccagtc ct
32
<210> 62
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22144
<400> 62
cgtacgggcg cgcctcagga atcttcactt cc
32
<210> 63
<211> 483
<212> DNA
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
22
<213> homo sapiens
<400> 63
tccagtcctg gcaacatgga gaggattgtc atctgtctga tggtcatctt cttggggaca 60
ctggtccaca aatcaagctc ccaaggtcaa gatcgccaca tgattagaat gcgtcaactt 120
atagatattg ttgatcagct gaaaaattat gtgaatgact tggtccctga atttctgcca 180
gctccagaag atgtagagac aaactgtgag tggtcagctt tttcctgttt tcagaaggcc 240
caactaaagt cagcaaatac aggaaacaat gaaaggataa tcaatgtatc aattaaaaag 300
ctgaagagga aaccaccttc cacaaatgca gggagaagac agaaacacag actaacatgc 360
ccttcatgtg attcttatga gaaaaaacca cccaaagaat tcctagaaag attcaaatca 420
cttctccaaa agatgattca tcagcatctg tcctctagaa cacacggaag tgaagattcc 480
tga 483
<210> 64
<211> 57
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22052
<400> 64
tcatataggc cggccatatg cccgggcgcc accatggatt ccagtcctgg caacatg 57
<210> 65
<211> 57
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22053
<400> 65
gtacaacccc agagctgttt taaggcgcgc ctctagatca ggaatcttca cttccgt 57
<210> 66
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC23115
<400> 66
gtatacggcc ggccaccatg gagaggaccc tt 32
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
23
<210> 67
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC23116
<400> 67
cgtatcggcg cgccctagga gagatgctga tg
32
<210> 68
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC20892
<400> 68
gtatacgttt aaacgccacc atgccgcgtg gctgg
35
<210> 69
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC20893
<400> 69
cgtatcggcg cgccttacaa tggatgggtc tt
32
<210> 70
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22054
<400> 70
cccggggtcg acaccatgga ttccagtcct ggcaacatg
39
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
24
<210> 71
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22055
<400> 71
tgcagtttaa actcaggaat cttcacttcc gt
32
<210> 72
<211> 40
<212> PRT
<213> Artificial Sequence
<220>
<223> Huzalphal1L-1 peptide
<400> 72
Gin Asp Arg His Met Ile Arg Met Arg Gin Leu Ile Asp Ile Val Asp
1 5 10 15
Gin Leu Lys Asn Tyr Val Asn Asp Leu Val Pro Glu Phe Leu Pro Ala
20 25 30
Pro Glu Asp Val Glu Thr Asn Cys
35 40
<210> 73
<211> 32
<212> PRT
<213> Artificial Sequence
<220>
<223> Huzalphal1L-3 peptide
<400> 73
Cys Pro Ser Cys Asp Ser Tyr Glu Lys Lys Pro Pro Lys Glu Phe Leu
1 5 10 15
Glu Arg Phe Lys Ser Leu Leu Gin Lys Met Ile His Gin His Leu Ser
20 25 30
<210> 74
<211> 29
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
<220>
<223> Oligonucleotide primer ZC23444
<400> 74
gcccgggcgg atccatggat tccagtcct
29
<210> 75
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC23445
<400> 75
cgcgccctcg agtcaggaat cttcacttcc gt
32
<210> 76
<211> 17
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC447
<400> 76
taacaatttc acacagg
17
<210> 77
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC976
<400> 77
cgttgtaaaa cgacggcc
18
<210> 78
<211> 66
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
26
<220>
<223> Oligonucleotide primer ZC22128
<400> 78
tcaccacgcg aattcggtac cgctggttcc gcgtggatcc caagatcgcc acatgattag 60
aatgcg 66
<210> 79
<211> 68
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22127
<400> 79
tctgtatcag gctgaaaatc ttatctcatc cgccaaaaca tcaggaatct tcacttccgt 60
gtgttcta 68
<210> 80
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19372
<400> 80
tgtcgatgaa gccctgaaag acgcgcagac taattcgagc 40
<210> 81
<211> 60
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19351
<400> 81
acgcgcagac taattcgagc tcccaccatc accatcacca cgcgaattcg gtaccgctgg 60
<210> 82
<211> 60
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
27
<220>
<223> Oligonucleotide primer ZC19352
<400> 82
actcactata gggcgaattg cccgggggat ccacgcggaa ccagcggtac cgaattcgcg 60
<210> 83
<211> 42
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC19371
<400> 83
acggccagtg aattgtaata cgactcacta tagggcgaat tg 42
<210> 84
<211> 1560
<212> DNA
<213> Artificial Sequence
<220>
<221> CDS
<222> (1)...(1560)
<223> MBP-human zalphall Ligand fusion polynucleotide
<400> 84
atg aaa act gaa gaa ggt aaa ctg gta atc tgg att aac ggc gat aaa 48
Met Lys Thr Glu Glu Gly Lys Leu Val Ile Trp Ile Asn Gly Asp Lys
1 5 10 15
ggc tat aac ggt ctc gct gaa gtc ggt aag aaa ttc gag aaa gat acc 96
Gly Tyr Asn Gly Leu Ala Glu Val Gly Lys Lys Phe Glu Lys Asp Thr
20 25 30
gga att aaa gtc acc gtt gag cat ccg gat aaa ctg gaa gag aaa ttc 144
Gly Ile Lys Val Thr Val Glu His Pro Asp Lys Leu Glu Glu Lys Phe
35 40 45
cca cag gtt gcg gca act ggc gat ggc cct gac att atc ttc tgg gca 192
Pro Gin Val Ala Ala Thr Gly Asp Gly Pro Asp Ile Ile Phe Trp Ala
50 55 60
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
28
cac gac cgc ttt ggt ggc tac gct caa tct ggc ctg ttg gct gaa atc 240
His Asp Arg Phe Gly Gly Tyr Ala Gln Ser Gly Leu Leu Ala Glu Ile
65 70 75 80
acc ccg gac aaa gcg ttc cag gac aag ctg tat ccg ttt acc tgg gat 288
Thr Pro Asp Lys Ala Phe Gin Asp Lys Leu Tyr Pro Phe Thr Trp Asp
85 90 95
gcc gta cgt tac aac ggc aag ctg att gct tac ccg atc gct gtt gaa 336
Ala Val Arg Tyr Asn Gly Lys Leu. Ile Ala Tyr Pro Ile Ala Val Glu
100 105 110
gcg tta tcg ctg att tat aac aaa gat ctg ctg ccg aac ccg cca aaa 384
Ala Leu Ser Leu Ile Tyr Asn Lys Asp Leu Leu Pro Asn Pro Pro Lys
115 120 125
acc tgg gaa gag atc ccg gcg ctg gat aaa gaa ctg aaa gcg aaa ggt 432
Thr Trp Glu Glu Ile Pro Ala Leu Asp Lys Glu Leu Lys Ala Lys Gly
130 135 140
aag agc gcg ctg atg ttc aac ctg caa gaa ccg tac ttc acc tgg ccg 480
Lys Ser Ala Leu Met Phe Asn Leu Gin Glu Pro Tyr Phe Thr Trp Pro
145 150 155 160
ctg att gct gct gac ggg ggt tat gcg ttc aag tat gaa aac ggc aag 528
Leu Ile Ala Ala Asp Gly Gly Tyr Ala Phe Lys Tyr Glu Asn Gly Lys
165 170 175
tac gac att aaa gac gtg ggc gtg gat aac gct ggc gcg aaa gcg ggt 576
Tyr Asp Ile Lys Asp Val Gly Val Asp Asn Ala Gly Ala Lys Ala Gly
180 185 190
ctg acc ttc ctg gtt gac ctg att aaa aac aaa cac atg aat gca gac 624
Leu Thr Phe Leu Val Asp Leu Ile Lys Asn Lys His Met Asn Ala Asp
195 200 205
acc gat tac tcc atc gca gaa gct gcc ttt aat aaa ggc gaa aca gcg 672
Thr Asp Tyr Ser Ile Ala Glu Ala Ala Phe Asn Lys Gly Glu Thr Ala
210 215 220
atg acc atc aac ggc ccg tgg gca tgg tcc aac atc gac acc agc aaa 720
Met Thr Ile Asn Gly Pro Trp Ala Trp Ser Asn Ile Asp Thr Ser Lys
225 230 235 240
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
29
gtg aat tat ggt gta acg gta ctg ccg acc ttc aag ggt caa cca tcc 768
Val Asn Tyr Gly Val Thr Val Leu Pro Thr Phe Lys Gly Gin Pro Ser
245 250 255
aaa ccg ttc gtt ggc gtg ctg ago gca ggt att aac gcc gcc agt ccg 816
Lys Pro Phe Val Gly Val Leu Ser Ala Gly Ile Asn Ala Ala Ser Pro
260 265 270
aac aaa gag ctg gca aaa gag ttc ctc gaa aac tat ctg ctg act gat 864
Asn Lys Glu Leu Ala Lys Glu Phe Leu Glu Asn Tyr Leu Leu Thr Asp
275 280 285
gaa ggt ctg gaa gcg gtt aat aaa gac aaa ccg ctg ggt gcc gta gcg 912
Glu Gly Leu Glu Ala Val Asn Lys Asp Lys Pro Leu Gly Ala Val Ala
290 295 300
ctg aag tot tac gag gaa gag ttg gcg aaa gat cca cgt att gcc gcc 960
Leu Lys Ser Tyr Glu Glu Glu Leu Ala Lys Asp Pro Arg Ile Ala Ala
305 310 315 320
acc atg gaa aac gcc cag aaa ggt gaa atc atg ccg aac atc ccg cag
1008
Thr Met Glu Asn Ala Gin Lys Gly Glu Ile Met Pro Asn Ile Pro Gin
325 330 335
atg too got ttc tgg tat gcc gtg cgt act gcg gtg atc aac gcc gcc
1056
Met Ser Ala Phe Trp Tyr Ala Val Arg Thr Ala Val Ile Asn Ala Ala
340 345 350
ago ggt cgt cag act gtc gat gaa gcc ctg aaa gac gcg cag act aat
1104
Ser Gly Arg Gin Thr Val Asp Glu Ala Leu Lys Asp Ala Gin Thr Asn
355 360 365
tog ago too cac cat cac cat cac cac gcg aat tog gta ccg ctg gtt
1152
Ser Ser Ser His His His His His His Ala Asn Ser Val Pro Leu Val
370 375 380
ccg cgt gga too caa gat cgc cac atg att aga atg cgt caa ctt ata
1200
Pro Arg Gly Ser Gin Asp Arg His Met Ile Arg Met Arg Gin Leu Ile
385 390 395 400
gat att gtt gat cag ctg aaa aat tat gtg aat gac ttg gtc cot gaa
1248
Asp Ile Val Asp Gin Leu Lys Asn Tyr Val Asn Asp Leu Val Pro Glu
405 410 415
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
ttt ctg cca gct cca gaa gat gta gag aca aac tgt gag tgg tca gct
1296
Phe Leu Pro Ala Pro Glu Asp Val Glu Thr Asn Cys Glu Trp Ser Ala
420 425 430
ttt tcc tgt ttt cag aag gcc caa cta aag tca gca aat aca gga aac
1344
Phe Ser Cys Phe Gin Lys Ala Gin Leu Lys Ser Ala Asn Thr Gly Asn
435 440 445
aat gaa agg ata atc aat gta tca att aaa aag ctg aag agg aaa cca
1392
Asn Glu Arg Ile Ile Asn Val Ser Ile Lys Lys Leu Lys Arg Lys Pro
450 455 460
cct tcc aca aat gca ggg aga aga cag aaa cac aga cta aca tgc cct
1440
Pro Ser Thr Asn Ala Gly Arg Arg Gin Lys His Arg Leu Thr Cys Pro
465 470 475 480
tca tgt gat tct tat gag aaa aaa cca ccc aaa gaa ttc cta gaa aga
1488
Ser Cys Asp Ser Tyr Glu Lys Lys Pro Pro Lys Glu Phe Leu Glu Arg
485 490 495
ttc aaa tca ctt ctc caa aag atg att cat cag cat ctg tcc tct aga
1536
Phe Lys Ser Leu Leu Gin Lys Met Ile His Gin His Leu Ser Ser Arg
500 505 510
aca cac gga agt gaa gat tcc tga
1560
Thr His Gly Ser Glu Asp Ser *
515
<210> 85
<211> 519
<212> PRT
<213> Artificial Sequence
<220>
<223> MBP-human zalphall Ligand fusion polypeptide
<400> 85
Met Lys Thr Glu Glu Gly Lys Leu Val Ile Trp Ile Asn Gly Asp Lys
1 5 10 15
Gly Tyr Asn Gly Leu Ala Glu Val Gly Lys Lys Phe Glu Lys Asp Thr
20 25 30
Gly Ile Lys Val Thr Val Glu His Pro Asp Lys Leu Glu Glu Lys Phe
40 45
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
31
Pro Gin Val Ala Ala Thr Gly Asp Gly Pro Asp Ile Ile Phe Trp Ala
50 55 60
His Asp Arg Phe Gly Gly Tyr Ala Gin Ser Gly Leu Leu Ala Glu Ile
65 70 75 80
Thr Pro Asp Lys Ala Phe Gin Asp Lys Leu Tyr Pro Phe Thr Trp Asp
85 90 95
Ala Val Arg Tyr Asn Gly Lys Leu Ile Ala Tyr Pro Ile Ala Val Glu
100 105 110
Ala Leu Ser Leu Ile Tyr Asn Lys Asp Leu Leu Pro Asn Pro Pro Lys
115 120 125
Thr Trp Glu Glu Ile Pro Ala Leu Asp Lys Glu Leu Lys Ala Lys Gly
130 135 140
Lys Ser Ala Leu Met Phe Asn Leu Gin Glu Pro Tyr Phe Thr Trp Pro
145 150 155 160
Leu Ile Ala Ala Asp Gly Gly Tyr Ala Phe Lys Tyr Glu Asn Gly Lys
165 170 175
Tyr Asp Ile Lys Asp Val Gly Val Asp Asn Ala Gly Ala Lys Ala Gly
180 185 190
Leu Thr Phe Leu Val Asp Leu Ile Lys Asn Lys His Met Asn Ala Asp
195 200 205
Thr Asp Tyr Ser Ile Ala Glu Ala Ala Phe Asn Lys Gly Glu Thr Ala
210 215 220
Met Thr Ile Asn Gly Pro Trp Ala Trp Ser Asn Ile Asp Thr Ser Lys
225 230 235 240
Val Asn Tyr Gly Val Thr Val Leu Pro Thr Phe Lys Gly Gin Pro Ser
245 250 255
Lys Pro Phe Val Gly Val Leu Ser Ala Gly Ile Asn Ala Ala Ser Pro
260 265 270
Asn Lys Glu Leu Ala Lys Glu Phe Leu Glu Asn Tyr Leu Leu Thr Asp
275 280 285
Glu Gly Leu Glu Ala Val Asn Lys Asp Lys Pro Leu Gly Ala Val Ala
290 295 300
Leu Lys Ser Tyr Glu Glu Glu Leu Ala Lys Asp Pro Arg Ile Ala Ala
305 310 315 320
Thr Met Glu Asn Ala Gin Lys Gly Glu Ile Met Pro Asn Ile Pro Gin
325 330 335
Met Ser Ala Phe Trp Tyr Ala Val Arg Thr Ala Val Ile Asn Ala Ala
340 345 350
Ser Gly Arg Gin Thr Val Asp Glu Ala Leu Lys Asp Ala Gin Thr Asn
355 360 365
Ser Ser Ser His His His His His His Ala Asn Ser Val Pro Leu Val
370 375 380
Pro Arg Gly Ser Gin Asp Arg His Met Ile Arg Met Arg Gin Leu Ile
385 390 395 400
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
32
Asp Ile Val Asp Gin Leu Lys Asn Tyr Val Asn Asp Leu Val Pro Glu
405 410 415
Phe Leu Pro Ala Pro Glu Asp Val Glu Thr Asn Cys Glu Trp Ser Ala
420 425 430
Phe Ser Cys Phe Gin Lys Ala Gin Leu Lys Ser Ala Asn Thr Gly Asn
435 440 445
Asn Glu Arg Ile Ile Asn Val Ser Ile Lys Lys Leu Lys Arg Lys Pro
450 455 460
Pro Ser Thr Asn Ala Gly Arg Arg Gin Lys His Arg Leu Thr Cys Pro
465 470 475 480
Ser Cys Asp Ser Tyr Glu Lys Lys Pro Pro Lys Glu Phe Leu Glu Arg
485 490 495
Phe Lys Ser Leu Leu Gin Lys Met Ile His Gin His Leu Ser Ser Arg
500 505 510
Thr His Gly Ser Glu Asp Ser
515
<210> 86
<211> 64
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22849
<400> 86
tcaccacgcg aattcggtac cgctggttcc gcgtggatcc ccagatcgcc tcctgattag 60
actt 64
<210> 87
<211> 64
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22850
<400> 87
tctgtatcag gctgaaaatc ttatctcatc cgccaaaaca ctaggagaga tgctgatgaa 60
tcat 64
<210> 88
<211> 1533
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
33
<220>
<223> MBP-mouse zalpha11 Ligand fusion polynucleotide
<221> CDS
<222> (1)...(1533)
<400> 88
atg aaa act gaa gaa ggt aaa ctg gta atc tgg att aac ggc gat aaa 48
Met Lys Thr Glu Glu Gly Lys Leu Val Ile Trp Ile Asn Gly Asp Lys
1 5 . 10 15
ggc tat aac ggt ctc gct gaa gtc ggt aag aaa ttc gag aaa gat acc 96
Gly Tyr Asn Gly Leu Ala Glu Val Gly Lys Lys Phe Glu Lys Asp Thr
20 25 30
gga att aaa gtc acc gtt gag cat ccg gat aaa ctg gaa gag aaa ttc 144
Gly Ile Lys Val Thr Val Glu His Pro Asp Lys Leu Glu Glu Lys Phe
35 40 45
cca cag gtt gcg gca act ggc gat ggc cct gac att atc ttc tgg gca 192
Pro Gin Val Ala Ala Thr Gly Asp Gly Pro Asp Ile Ile Phe Trp Ala
50 55 60
cac gac cgc ttt ggt ggc tac gct caa tct ggc ctg ttg gct gaa atc 240
His Asp Arg Phe Gly Gly Tyr Ala Gin Ser Gly Leu Leu Ala Glu Ile
65 70 75 80
acc ccg gac aaa gcg ttc cag gac aag ctg tat ccg ttt acc tgg gat 288
Thr Pro Asp Lys Ala Phe Gin Asp Lys Leu Tyr Pro Phe Thr Trp Asp
85 90 95
gcc gta cgt tac aac ggc aag ctg att gct tac ccg atc gct gtt gaa 336
Ala Val Arg Tyr Asn Gly Lys Leu Ile Ala Tyr Pro Ile Ala Val Glu
100 105 110
gcg tta tcg ctg att tat aac aaa gat ctg ctg ccg aac ccg cca aaa 384
Ala Leu Ser Leu Ile Tyr Asn Lys Asp Leu Leu Pro Asn Pro Pro Lys
115 120 125
acc tgg gaa gag atc ccg gcg ctg gat aaa gaa ctg aaa gcg aaa ggt 432
Thr Trp Glu Glu Ile Pro Ala Leu Asp Lys Glu Leu Lys Ala Lys Gly
130 135 140
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
34
aag agc gcg ctg atg ttc aac ctg caa gaa ccg tac ttc acc tgg ccg
480
Lys Ser Ala Leu Met Phe Asn Leu Gin Glu Pro Tyr Phe Thr Trp Pro
145 150 155 160
ctg att gct gct gac ggg ggt tat gcg ttc aag tat gaa aac ggc aag
528
Leu Ile Ala Ala Asp Gly Gly Tyr Ala Phe Lys Tyr Glu Asn Gly Lys
165 170 175
tac gac att aaa gac gtg ggc gtg gat aac gct ggc gcg aaa gcg ggt
576
Tyr Asp Ile Lys Asp Val Gly Val Asp Asn Ala Gly Ala Lys Ala Gly
180 185 190
ctg acc ttc ctg gtt gac ctg att aaa aac aaa cac atg aat gca gac
624
Leu Thr Phe Leu Val Asp Leu Ile Lys Asn Lys His Met Asn Ala Asp
195 200 205
acc gat tac tcc atc gca gaa gct gcc ttt aat aaa ggc gaa aca gcg
672
Thr Asp Tyr Ser Ile Ala Glu Ala Ala Phe Asn Lys Gly Glu Thr Ala
210 215 220
atg acc atc aac ggc ccg tgg gca tgg tcc aac atc gac acc agc aaa
720
Met Thr Ile Asn Gly Pro Trp Ala Trp Ser Asn Ile Asp Thr Ser Lys
225 230 235 240
gtg aat tat ggt gta acg gta ctg ccg acc ttc aag ggt caa cca tcc
768
Val Asn Tyr Gly Val Thr Val Leu Pro Thr Phe Lys Gly Gin Pro Ser
245 250 255
aaa ccg ttc gtt ggc gtg ctg agc gca ggt att aac gcc gcc agt ccg
816
Lys Pro Phe Val Gly Val Leu Ser Ala Gly Ile Asn Ala Ala Ser Pro
260 265 270
aac aaa gag ctg gca aaa gag ttc ctc gaa aac tat ctg ctg act gat
864
Asn Lys Glu Leu Ala Lys Glu Phe Leu Glu Asn Tyr Leu Leu Thr Asp
275 280 285
gaa ggt ctg gaa gcg gtt aat aaa gac aaa ccg ctg ggt gcc gta gcg
912
Glu Gly Leu Glu Ala Val Asn Lys Asp Lys Pro Leu Gly Ala Val Ala
290 295 300
ctg aag tct tac gag gaa gag ttg gcg aaa gat cca cgt att gcc gcc
960
Leu Lys Ser Tyr Glu Glu Glu Leu Ala Lys Asp Pro Arg Ile Ala Ala
305 310 315 320
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
acc atg gaa aac gcc cag aaa ggt gaa atc atg ccg aac atc ccg cag
1008
Thr Met Glu Asn Ala Gln Lys Gly Glu Ile Met Pro Asn Ile Pro Gln
325 330 335
atg tcc gct ttc tgg tat gcc gtg cgt act gcg gtg atc aac gcc gcc
1056
Met Ser Ala Phe Trp Tyr Ala Val Arg Thr Ala Val Ile Asn Ala Ala
340 345 350
agc ggt cgt cag act gtc gat gaa gcc ctg aaa gac gcg cag act aat
1104
Ser Gly Arg Gln Thr Val Asp Glu Ala Leu Lys Asp Ala Gln Thr Asn
355 360 365
tcg agc tcc cac cat cac cat cac cac gcg aat tcg gta ccg ctg gtt
1152
Ser Ser Ser His His His His His His Ala Asn Ser Val Pro Leu Val
370 375 380
ccg cgt gga tcc cca gat cgc ctc ctg att aga ctt cgt cac ctt att
1200
Pro Arg Gly Ser Pro Asp Arg Leu Leu Ile Arg Leu Arg His Leu Ile
385 390 395 400
gac att gtt gaa cag ctg aaa atc tat gaa aat gac ttg gat cct gaa
1248
Asp Ile Val Glu Gln Leu Lys Ile Tyr Glu Asn Asp Leu Asp Pro Glu
405 410 415
ctt cta tca gct cca caa gat gta aag ggg cac tgt gag cat gca gct
1296
Leu Leu Ser Ala Pro Gln Asp Val Lys Gly His Cys Glu His Ala Ala
420 425 430
ttt gcc tgt ttt cag aag gcc aaa ctc aag cca tca aac cct gga aac
1344
Phe Ala Cys Phe Gln Lys Ala Lys Leu Lys Pro Ser Asn Pro Gly Asn
435 440 445
aat aag aca ttc atc att gac ctc gtg gcc cag ctc agg agg agg ctg
1392
Asn Lys Thr Phe Ile Ile Asp Leu Val Ala Gin Leu Arg Arg Arg Leu
450 455 460
cct gcc agg agg gga gga aag aaa cag aag cac ata gct aaa tgc cct
1440
Pro Ala Arg Arg Gly Gly Lys Lys Gin Lys His Ile Ala Lys Cys Pro
465 470 475 480
tcc tgt gat tcg tat gag aaa agg aca ccc aaa gaa ttc cta gaa aga
1488
Ser Cys Asp Ser Tyr Glu Lys Arg Thr Pro Lys Glu Phe Leu Glu Arg
485 490 495
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
36
cta aaa tgg ctc ctt caa aag atg att cat cag cat ctc tcc tga 1533
Leu Lys Trp Leu Leu Gin Lys Met Ile His Gin His Leu Ser *
500 505 510
<210> 89
<211> 510
<212> PRT
<213> Artificial Sequence
<220>
<223> MBP-mouse zalphall Ligand fusion polypeptide
<400> 89
Met Lys Thr Glu Glu Gly Lys Leu Val Ile Trp Ile Asn Gly Asp Lys
1 5 10 15
Gly Tyr Asn Gly Leu Ala Glu Val Gly Lys Lys Phe Glu Lys Asp Thr
20 25 30
Gly Ile Lys Val Thr Val Glu His Pro Asp Lys Leu Glu Glu Lys Phe
35 40 45
Pro Gin Val Ala Ala Thr Gly Asp Gly Pro Asp Ile Ile Phe Trp Ala
50 55 60
His Asp Arg Phe Gly Gly Tyr Ala Gin Ser Gly Leu Leu Ala Glu Ile
65 70 75 80
Thr Pro Asp Lys Ala Phe Gin Asp Lys Leu Tyr Pro Phe Thr Trp Asp
85 90 95
Ala Val Arg Tyr Asn Gly Lys Leu Ile Ala Tyr Pro Ile Ala Val Glu
100 105 110
Ala Leu Ser Leu Ile Tyr Asn Lys Asp Leu Leu Pro Asn Pro Pro Lys
115 120 125
Thr Trp Glu Glu Ile Pro Ala Leu Asp Lys Glu Leu Lys Ala Lys Gly
130 135 140
Lys Ser Ala Leu Met Phe Asn Leu Gin Glu Pro Tyr Phe Thr Trp Pro
145 150 155 160
Leu Ile Ala Ala Asp Gly Gly Tyr Ala Phe Lys Tyr Glu Asn Gly Lys
165 170 175
Tyr Asp Ile Lys Asp Val Gly Val Asp Asn Ala Gly Ala Lys Ala Gly
180 185 190
Leu Thr Phe Leu Val Asp Leu Ile Lys Asn Lys His Met Asn Ala Asp
195 200 205
Thr Asp Tyr Ser Ile Ala Glu Ala Ala Phe Asn Lys Gly Glu Thr Ala
210 215 220
Met Thr Ile Asn Gly Pro Trp Ala Trp Ser Asn Ile Asp Thr Ser Lys
225 230 235 240
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
37
Val Asn Tyr Gly Val Thr Val Leu Pro Thr Phe Lys Gly Gin Pro Ser
245 250 255
Lys Pro Phe Val Gly Val Leu Ser Ala Gly Ile Asn Ala Ala Ser Pro
260 265 270
Asn Lys Glu Leu Ala Lys Glu Phe Leu Glu Asn Tyr Leu Leu Thr Asp
275 280 285
Glu Gly Leu Glu Ala Val Asn Lys Asp Lys Pro Leu Gly Ala Val Ala
290 295 300
Leu Lys Ser Tyr Glu Glu Glu Leu Ala Lys Asp Pro Arg Ile Ala Ala
305 310 315 320
Thr Met Glu Asn Ala Gin Lys Gly Glu Ile Met Pro Asn Ile Pro Gin
325 330 335
Met Ser Ala Phe Trp Tyr Ala Val Arg Thr Ala Val Ile Asn Ala Ala
340 345 350
Ser Gly Arg Gin Thr Val Asp Glu Ala Leu Lys Asp Ala Gin Thr Asn
355 360 365
Ser Ser Ser His His His His His His Ala Asn Ser Val Pro Leu Val
370 375 380
Pro Arg Gly Ser Pro Asp Arg Leu Leu Ile Arg Leu Arg His Leu Ile
385 390 395 400
Asp Ile Val Glu Gin Leu Lys Ile Tyr Glu Asn Asp Leu Asp Pro Glu
405 410 415
Leu Leu Ser Ala Pro Gin Asp Val Lys Gly His Cys Glu His Ala Ala
420 425 430
Phe Ala Cys Phe Gin Lys Ala Lys Leu Lys Pro Ser Asn Pro Gly Asn
435 440 445
Asn Lys Thr Phe Ile Ile Asp Leu Val Ala Gin Leu Arg Arg Arg Leu
450 455 460
Pro Ala Arg Arg Gly Gly Lys Lys Gin Lys His Ile Ala Lys Cys Pro
465 470 475 480
Ser Cys Asp Ser Tyr Glu Lys Arg Thr Pro Lys Glu Phe Leu Glu Arg
485 490 495
Leu Lys Trp Leu Leu Gin Lys Met Ile His Gin His Leu Ser
500 505 510
<210> 90
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22281
<400> 90
tgtgaatgac ttggtccctg aa
22
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
38
<210> 91
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22279
<400> 91
aacaggaaaa agctgaccac tca 23
<210> 92
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Human zalpha11 Ligand TaqMan probe, ZG32
<400> 92
tctgccagct ccagaagatg tagagacaaa c 31
<210> 93
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22277
<400> 93
ccaggagtgt ggcagctttc 20
<210> 94
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22276
<400> 94
gcttgccctt cagcatgtag a 21
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
39
<210> 95
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Human zalpha11 TaqMan probe, ZG31
<400> 95
cggctccccc tttcaacgtg act 23
<210> 96
<211> 1821
<212> DNA
<213> Artificial Sequence
<220>
<221> CDS
<222> (1)...(1821)
<223> MBP-zalphal1 soluble receptor polynucleotide
sequence
<400> 96
atg aaa atc gaa gaa ggt aaa ctg gta atc tgg att aac ggc gat aaa 48
Met Lys Ile Glu Glu Gly Lys Leu Val Ile Trp Ile Asn Gly Asp Lys
1 5 10 15
ggc tat aac ggt ctc gct gaa gtc ggt aag aaa ttc gag aaa gat acc 96
Gly Tyr Asn Gly Leu Ala Glu Val Gly Lys Lys Phe Glu Lys Asp Thr
20 25 30
gga att aaa gtc acc gtt gag cat ccg gat aaa ctg gaa gag aaa ttc 144
Gly Ile Lys Val Thr Val Glu His Pro Asp Lys Leu Glu Glu Lys Phe
35 40 45
cca cag gtt gcg gca act ggc gat ggc cct gac att atc ttc tgg gca 192
Pro Gln Val Ala Ala Thr Gly Asp Gly Pro Asp Ile Ile Phe Trp Ala
50 55 60
cac gac cgc ttt ggt ggc tac gct caa tct ggc ctg ttg gct gaa atc 240
His Asp Arg Phe Gly Gly Tyr Ala Gln Ser Gly Leu Leu Ala Glu Ile
65 70 75 80
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
acc ccg gac aaa gcg ttc cag gac aag ctg tat ccg ttt acc tgg gat
288
Thr Pro Asp Lys Ala Phe Gin Asp Lys Leu Tyr Pro Phe Thr Trp Asp
85 90 95
gcc gta cgt tac aac ggc aag ctg att gct tac ccg atc gct gtt gaa
336
Ala Val Arg Tyr Asn Gly Lys Leu Ile Ala Tyr Pro Ile Ala Val Glu
100 105 110
gcg tta tcg ctg att tat aac aaa gat ctg ctg ccg aac ccg cca aaa
384
Ala Leu Ser Leu Ile Tyr Asn Lys Asp Leu Leu Pro Asn Pro Pro Lys
115 120 125
acc tgg gaa gag atc ccg gcg ctg gat aaa gaa ctg aaa gcg aaa ggt
432
Thr Trp Glu Glu Ile Pro Ala Leu Asp Lys Glu Leu Lys Ala Lys Gly
130 135 140
aag agc gcg ctg atg ttc aac ctg caa gaa ccg tac ttc acc tgg ccg
480
Lys Ser Ala Leu Met Phe Asn Leu Gin Glu Pro Tyr Phe Thr Trp Pro
145 150 155 160
ctg att gct gct gac ggg ggt tat gcg ttc aag tat gaa aac ggc aag
528
Leu Ile Ala Ala Asp Gly Gly Tyr Ala Phe Lys Tyr Glu Asn Gly Lys
165 170 175
tac gac att aaa gac gtg ggc gtg gat aac gct ggc gcg aaa gcg ggt
576
Tyr Asp Ile Lys Asp Val Gly Val Asp Asn Ala Gly Ala Lys Ala Gly
180 185 190
ctg acc ttc ctg gtt gac ctg att aaa aac aaa cac atg aat gca gac
624
Leu Thr Phe Leu Val Asp Leu Ile Lys Asn Lys His Met Asn Ala Asp
195 200 205
acc gat tac tcc atc gca gaa gct gcc ttt aat aaa ggc gaa aca gcg
672
Thr Asp Tyr Ser Ile Ala Glu Ala Ala Phe Asn Lys Gly Glu Thr Ala
210 215 220
atg acc atc aac ggc ccg tgg gca tgg tcc aac atc gac acc agc aaa
720
Met Thr Ile Asn Gly Pro Trp Ala Trp Ser Asn Ile Asp Thr Ser Lys
225 230 235 240
gtg aat tat ggt gta acg gta ctg ccg acc ttc aag ggt caa cca tcc
768
Val Asn Tyr Gly Val Thr Val Leu Pro Thr Phe Lys Gly Gin Pro Ser
245 250 255
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
41
aaa ccg ttc gtt ggc gtg ctg agc gca ggt att aac gcc gcc agt ccg 816
Lys Pro Phe Val Gly Val Leu Ser Ala Gly Ile Asn Ala Ala Ser Pro
260 265 270
aac aaa gag ctg gca aaa gag ttc ctc gaa aac tat ctg ctg act gat 864
Asn Lys Glu Leu Ala Lys Glu Phe Leu Glu Asn Tyr Leu Leu Thr Asp
275 280 285
gaa ggt ctg gaa gcg gtt aat aaa gac aaa ccg ctg ggt gcc gta gcg 912
Glu Gly Leu Glu Ala Val Asn Lys Asp Lys Pro Leu Gly Ala Val Ala
290 295 300
ctg aag tct tac gag gaa gag ttg gcg aaa gat cca cgt att gcc gcc 960
Leu Lys Ser Tyr Glu Glu Glu Leu Ala Lys Asp Pro Arg Ile Ala Ala
305 310 315 320
acc atg gaa aac gcc cag aaa ggt gaa atc atg ccg aac atc ccg cag
1008
Thr Met Glu Asn Ala Grin Lys Gly Glu Ile Met Pro Asn Ile Pro Gin
325 330 335
atg tcc gct ttc tgg tat gcc gtg cgt act gcg gtg atc aac gcc gcc
1056
Met Ser Ala Phe Trp Tyr Ala Val Arg Thr Ala Val Ile Asn Ala Ala
340 345 350
agc ggt cgt cag act gtc gat gaa gcc ctg aaa gac gcg cag act aat
1104
Ser Gly Arg Gin Thr Val Asp Glu Ala Leu Lys Asp Ala Gin Thr Asn
355 360 365
tcg agc tcc cac cat cac cat cac cac gcg aat tcg gta ccg ctg gtt
1152
Ser Ser Ser His His His His His His Ala Asn Ser Val Pro Leu Val
370 375 380
ccg cgt gga tcc tgc ccc gac ctc gtc tgc tac acc gat tac ctc cag
1200
Pro Arg Gly Ser Cys Pro Asp Leu Val Cys Tyr Thr Asp Tyr Leu Gin
385 390 395 400
acg gtc atc tgc atc ctg gaa atg tgg aac ctc cac ccc agc acg ctc
1248
Thr Val Ile Cys Ile Leu Glu Met Trp Asn Leu His Pro Ser Thr Leu
405 410 415
acc ctt acc tgg caa gac cag tat gaa gag ctg aag gac gag gcc acc
1296
Thr Leu Thr Trp Gln Asp Gin Tyr Glu Glu Leu Lys Asp Glu Ala Thr
420 425 430
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
42
tcc tgc agc ctc cac agg tcg gcc cac aat gcc acg cat gcc acc tac
1344
Ser Cys Ser Leu His Arg Ser Ala His Asn Ala Thr His Ala Thr Tyr
435 440 445
acc tgc cac atg gat gta ttc cac ttc atg gcc gac gac att ttc agt
1392
Thr Cys His Met Asp Val Phe His Phe Met Ala Asp Asp Ile Phe Ser
450 455 460
gtc aac atc aca gac cag tct ggc aac tac tcc cag gag tgt ggc agc
1440
Val Asn Ile Thr Asp Gin Ser Gly Asn Tyr Ser Gin Glu Cys Gly Ser
465 470 475 480
ttt ctc ctg gct gag agc atc aag ccg gct ccc cct ttc aac gtg act
1488
Phe Leu Leu Ala Glu Ser Ile Lys Pro Ala Pro Pro Phe Asn Val Thr
485 490 495
gtg acc ttc tca gga cag tat aat atc tcc tgg cgc tca gat tac gaa
1536
Val Thr Phe Ser Gly Gin Tyr Asn Ile Ser Trp Arg Ser Asp Tyr Glu
500 505 510
gac cct gcc ttc tac atg ctg aag ggc aag ctt cag tat gag ctg cag
1584
Asp Pro Ala Phe Tyr Met Leu Lys Gly Lys Leu Gin Tyr Glu Leu Gin
515 520 525
tac agg aac cgg gga gac ccc tgg gct gtg agt ccg agg aga aag ctg
1632
Tyr Arg Asn Arg Gly Asp Pro Trp Ala Val Ser Pro Arg Arg Lys Leu
530 535 540
atc tca gtg gac tca aga agt gtc tcc ctc ctc ccc ctg gag ttc cgc
1680
Ile Ser Val Asp Ser Arg Ser Val Ser Leu Leu Pro Leu Glu Phe Arg
545 550 555 560
aaa gac tcg agc tat gag ctg cag gtg cgg gca ggg ccc atg cct ggc
1728
Lys Asp Ser Ser Tyr Glu Leu Gin Val Arg Ala Gly Pro Met Pro Gly
565 570 575
tcc tcc tac cag ggg acc tgg agt gaa tgg agt gac ccg gtc atc ttt
1776
Ser Ser Tyr Gin Gly Thr Trp Ser Glu Trp Ser Asp Pro Val Ile Phe
580 585 590
cag acc cag tca gag gag tta aag gaa ggc tgg aac cct cac tag
1821
Gin Thr Gln Ser Glu Glu Leu Lys Glu Gly Trp Asn Pro His *
595 600 605
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
43
<210> 97
<211> 606
<212> PRT
<213> Artificial Sequence
<220>
<223> MBP-zalphall soluble receptor polypeptide sequence
<400> 97
Met Lys Ile Glu Glu Gly Lys Leu Val Ile Trp Ile Asn Gly Asp Lys
1 5 . 10 15
Gly Tyr Asn Gly Leu Ala Glu Val Gly Lys Lys Phe Glu Lys Asp Thr
20 25 30
Gly Ile Lys Val Thr Val Glu His Pro Asp Lys Leu Glu Glu Lys Phe
35 40 45
Pro Gin Val Ala Ala Thr Gly Asp Gly Pro Asp Ile Ile Phe Trp Ala
50 55 60
His Asp Arg Phe Gly Gly Tyr Ala Gin Ser Gly Leu Leu Ala Glu Ile
65 70 75 80
Thr Pro Asp Lys Ala Phe Gin Asp Lys Leu Tyr Pro Phe Thr Trp Asp
85 90 95
Ala Val Arg Tyr Asn Gly Lys Leu Ile Ala Tyr Pro Ile Ala Val Glu
100 105 110
Ala Leu Ser Leu Ile Tyr Asn Lys Asp Leu Leu Pro Asn Pro Pro Lys
115 120 125
Thr Trp Glu Glu Ile Pro Ala Leu Asp Lys Glu Leu Lys Ala Lys Gly
130 135 140
Lys Ser Ala Leu Met Phe Asn Leu Gin Glu Pro Tyr Phe Thr Trp Pro
145 150 155 160
Leu Ile Ala Ala Asp Gly Gly Tyr Ala Phe Lys Tyr Glu Asn Gly Lys
165 170 175
Tyr Asp Ile Lys Asp Val Gly Val Asp Asn Ala Gly Ala Lys Ala Gly
180 185 190
Leu Thr Phe Leu Val Asp Leu Ile Lys Asn Lys His Met Asn Ala Asp
195 200 205
Thr Asp Tyr Ser Ile Ala Glu Ala Ala Phe Asn Lys Gly Glu Thr Ala
210 215 220
Met Thr Ile Asn Gly Pro Trp Ala Trp Ser Asn Ile Asp Thr Ser Lys
225 230 235 240
Val Asn Tyr Gly Val Thr Val Leu Pro Thr Phe Lys Gly Gin Pro Ser
245 250 255
Lys Pro Phe Val Gly Val Leu Ser Ala Gly Ile Asn Ala Ala Ser Pro
260 265 270
Asn Lys Glu Leu Ala Lys Glu Phe Leu Glu Asn Tyr Leu Leu Thr Asp
275 280 285
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
44
Glu Gly Leu Glu Ala Val Asn Lys Asp Lys Pro Leu Gly Ala Val Ala
290 295 300
Leu Lys Ser Tyr Glu Glu Glu Leu Ala Lys Asp Pro Arg Ile Ala Ala
305 310 315 320
Thr Met Glu Asn Ala Gin Lys Gly Glu Ile Met Pro Asn Ile Pro Gln
325 330 335
Met Ser Ala Phe Trp Tyr Ala Val Arg Thr Ala Val Ile Asn Ala Ala
340 345 350
Ser Gly Arg Gln Thr Val Asp Glu Ala Leu Lys Asp Ala Gln Thr Asn
355 360 365
Ser Ser Ser His His His His His. His Ala Asn Ser Val Pro Leu Val
370 375 380
Pro Arg Gly Ser Cys Pro Asp Leu Val Cys Tyr Thr Asp Tyr Leu Gln
385 390 395 400
Thr Val Ile Cys Ile Leu Glu Met Trp Asn Leu His Pro Ser Thr Leu
405 410 415
Thr Leu Thr Trp Gln Asp Gln Tyr Glu Glu Leu Lys Asp Glu Ala Thr
420 425 430
Ser Cys Ser Leu His Arg Ser Ala His Asn Ala Thr His Ala Thr Tyr
435 440 445
Thr Cys His Met Asp Val Phe His Phe Met Ala Asp Asp Ile Phe Ser
450 455 460
Val Asn Ile Thr Asp Gln Ser Gly Asn Tyr Ser Gln Glu Cys Gly Ser
465 470 475 480
Phe Leu Leu Ala Glu Ser Ile Lys Pro Ala Pro Pro Phe Asn Val Thr
485 490 495
Val Thr Phe Ser Gly Gln Tyr Asn Ile Ser Trp Arg Ser Asp Tyr Glu
500 505 510
Asp Pro Ala Phe Tyr Met Leu Lys Gly Lys Leu Gln Tyr Glu Leu Gln
515 520 525
Tyr Arg Asn Arg Gly Asp Pro Trp Ala Val Ser Pro Arg Arg Lys Leu
530 535 540
Ile Ser Val Asp Ser Arg Ser Val Ser Leu Leu Pro Leu Glu Phe Arg
545 550 555 560
Lys Asp Ser Ser Tyr Glu Leu Gln Val Arg Ala Gly Pro Met Pro Gly
565 570 575
Ser Ser Tyr Gln Gly Thr Trp Ser Glu Trp Ser Asp Pro Val Ile Phe
580 585 590
Gln Thr Gln Ser Glu Glu Leu Lys Glu Gly Trp Asn Pro His
595 600 605
<210> 98
<211> 65
<212> DNA
<213> Artificial Sequence
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
<220>
<223> Oligonucleotide primer ZC20187
<400> 98
tcaccacgcg aattcggtac cgctggttcc gcgtggatcc tgccccgacc tcgtctgcta 60
caccg 65
<210> 99
<211> 68
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC20185
<400> 99
tctgtatcag gctgaaaatc ttatctcatc cgccaaaaca ctagtgaggg ttccagcctt 60
cctttaac 68
<210> 100
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22452
<400> 100
tcttcttggg gacactggtc c 21
<210> 101
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer 7C22451
<400> 101
aatcatgtgg cgatcttgac c 21
<210> 102
<211> 21
<212> DNA
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
46
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22450
<400> 102
cagactaaca tgcccttcat g 21
<210> 103
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC22449
<400> 103
ttcacttccg tgtgttctag agg 23
<210> 104
<211> 18
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC23771
<400> 104
accaccttcc acaaatgc 18
<210> 105
<211> 1347
<212> DNA
<213> Homo sapiens
<400> 105
gaattcaccc attctctctt tttcctgtca aagatgcaga tggggcacat ttcgttgact 60
ccatcaatcc ctgcccccac acattagcac atgcacacgt atacctagcc agtggaaaag
120
aaaaaagagt tactcacatt catccatttt acaaagattt ccaggctgca atgggagggc
180
tttacctctc cctgaaggat gaataaatag gtagcttaac tgacaacctg ttctcagtca
240
agctgaagtg aaaacgagac caaggtctag ctctactgtt ggtacttatg agatccagtc
300
ctggcaacat ggagaggatt gtcatctgtc tgatggtcat cttcttgggg acactggtcc
360
acaaatcaag ctcccaaggt caagatcgcc acatgattag aatgcgtcaa cttatagata
420
ttgttgatca gctgaaaaat tatgtgaatg acttggtaag actatatttg tcacaacaaa
480
atctaaatca tacttttcaa ttaatataaa aggagggttt ggcttataaa aataactcag
540
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
47
aacaaatttt cttttgctct aggtccctga atttctgcca gctccagaag atgtagaggt 600
aagaccagtt gaatttattt ctgaaaatac attggacata agtttttaaa tccaataaga 660
aagacattag catgattata taggagtata ctgaatttta atgaacttag cggtctaata 720
attgatgaaa tatttattta tattttggtt aaattcattg atttaccaaa aaccaactaa 780
aaaatgctat attatattcc tcataaacta tgtttatctt caagaatctc taagagtact 840
cctaagtagt attgctgaga cagaataaca aaactagaaa cgaaatctat actctgatca 900
gtttctgaac aatgcacagc tagttactct ttaagagccc ttgggcatga aagcttttga 960
gccttctttg ttatcctacc gaagaaacat agatacatac agtaggaagc agaattaacc
1020
ttttaataac aaacttaaaa aagaaagaaa gaaagaatta gattacaggg acagcatgga
1080
gaaatggtgg tgtggaaatc aaagctgtcc tttagaatat aattcacata taccttggcc
1140
tcagtgagtc ttgtctttgg ccttccgtga ggtcttttga aagaaccatt ttcaacaatt
1200
catcccgtct cttaagccat ttaaatccat tagagttcca ggaagaagag gcctggcatg
1260
agttcagagt gctgtcccgc tgatcttttt ctcagtaact tctacgatct gatcttctgg
1320
tctggtaccc tgaggtataa atgcaat
1347
<210> 106
<211> 1656
<212> DNA
<213> Homo sapiens
<400> 106
cctcaactgc ttgattcagg cagaatccta accctaaact gagctgggag tatgaaaagg 60
gttttagaaa agtcatggtg tgatctatgg caagtatatt gattcttaga tgtaaaatat 120
gctatcagag ggaggtaccc acttcctttc tccaaaggag gggctttaat tcattttctt 180
catctgttaa ctttacaaat atatgttgat cattaactgg caagacacta tgcctggcgc 240
tgtacagaat aaaatgctgc tcaagacatg tcatgataga tacattaaca gaaaccacaa 300
acaaatgaaa aatgttcttc atcagactat aacataattt acccaaagct gccactagtc 360
acagtgtaag ttttagagcc tcataactca gcaaatgtgt cctaaaccga actaactctc 420
ctttataaaa cacaaaggtc ttgtccacca cccagacatc aaaatggtcc tctgtgtagc 480
atcaggaata aagcattgtg aagaagtgag gctcctttct ctcttatctg cgaagcaggg 540
gattgtccct ttttcccatc ccaaagatta agtaggaggt gaaatcatac ctcactcatc 600
tgttgaaacg atgtaatgca cgacattgca gaagagatag aaatagagga ttgggaaagc 660
tatcttttac tttctgaata atgtttgtta acatatatac aaattgttta tctttcagac 720
aaactgtgag tggtcagctt tttcctgttt tcagaaggcc caactaaagt cagcaaatac 780
aggaaacaat gaaaggataa taaatgtatc aattaaaaag ctgaagagga aaccaccttc 840
cacaaatgca gggagaagac agaaacacag actagtaaga ttgtcatttg tcatctctct 900
tatttgtact tataaactat atatcttgca ttacataaac atacacacac acctgtagcc 960
agggctgctg gtgtcttcct tacctatagt tatgccttat tatacatggt gctttttttt
1020
tttaagacag agtctcactc tgtcacccag gctggagtgc agtggcgtga tctctgctca
1080
ccgcaagatc cacctcccgg tttcacgcca ttctcctcct acctcagcct cctgagtacc
1140
tgggactaca ggtgcccgcc accatgcccg gctaattttg tttttgtatt tttagtaaag
1200
acagggtttc accatgttag ccaggatggt ctcgatctcc tgaccccgtg atccgcccgc
1260
cttggcctcc caaagtgtgg ggattacagg catgagccac cgcacccggc ctatacgtgg
1320
tgcattttaa gaagtagggt cactctttta agcccacaga cttgaaagta ttcaaaaacc
1380
caattataat ttcctagtag tccttggcag ctggaatatg ttaatatagc ttctcaaggt
1440
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
48
gaggaagtca ttaggcagag aatccaactg tgattttgga gttaagaact atttcctctc
1500
atatggtcac agataacttg tattcttatt aacaggagct agatcctagc tttctaacaa
1560
gaaaagagcc tacaagaaga ctagggcaaa tcttaaactt tgcctcctct ctaaatcata
1620
ttactatctg tacatcagca gagtcagtat tgaatt
1656
<210> 107
<211> 644
<212> DNA
<213> Homo sapiens
<400> 107
agctaaactt agaactctcc agttaagcat gttcatctta tagatgagga aaagtgagat 60
ctacaaagga gttaagtcac tagccccaag ttccataaat agtgtcagaa tgagaattag 120
aacgtatatc tactatcttt tagtgaaatg ctctcactac aacatcacac tggcattgag 180
atgctaacta ccaagcaatg gcttggtgtt tggatctaaa tagggataaa gacaaagagc 240
ataaactaag aaagcttttt aaaaatctaa gtgagcaatc catatatgaa aaactgttca 300
atctccctag taatcacata aatgcgagtt aaaacaagga aatcctgttt tttccaatta 360
aacattttaa acaataccct ataataataa gaatgctcca agtgaaaaga ggtaaaaccc 420
tttataatgt atatcaaagc cttaaaattt ttatcccttt aatttagtaa ttctacttct 480
aggaatatat caaataagca aagatatata tgaaaaatta tttacagaga tgttctttgg 540
agtaatgtag acaaaaataa aaagttagat acagctgggt gtggtggctc atgcctgtat 600
tcccagcact ttgggaggcc gaggcaggcg gatcacctga gatc 644
<210> 108
<211> 645
<212> DNA
<213> Homo sapiens
<220>
<221> misc feature
<222> (1)...(645)
<223> n = A,T,C or G
<400> 108
aaaaaaaaaa aaaaaaaaaa gttagatgca ccnttgggtc caaaaatagt aagagtgcat 60
cctatgtgga aacagaccaa ccactacatg tcatattttt gaagattatt taacacttag 120
gaaatcctgt gatatgttaa gtgtgaaaaa aaaaaagcaa atcaccaact ggtataaata 180
atgtaaatgc acaataataa ttaaaaatac ccaaaacaca gagagaatat acattaaaac 240
attgcagtgg gattcctatc tctgggaatg ggattacaag gactttttcc attgttactt 300
tccaaacagt tttatgtact tctcgaatgt ttttcagtga acataattta tgtttttaat 360
gaaaaaaaat tttaagaaac attttattac gaaaaaaatt ttaaagaaga ctgttacttt 420
ttcattgatt tctagacatg cccttcatgt gattcttatg agaaaaaacc acccaaagaa 480
ttcctagaaa gattcaaatc acttctccaa aaggtatcta ccttaagttt catttgattt 540
tctgctttat ctttacctat ccagatttgc ttcttagtta ctcacggtat actatttcca 600
cagatgattc atcagcatct gtcctctaga acacacggaa gtgaa 645
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
49
<210> 109
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC25970
<400> 109
atgcattcta gactaggaga gatgctgatg aatcat 36
<210> 110
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Oligonucleotide primer ZC25969
<400> 110
atgcattccg gacataaatc aagcccccaa gggcca 36
<210> 111
<211> 153
<212> PRT
<213> Homo sapiens
<400> 111
Met Tyr Arg Met Gln Leu Leu Ser Cys Ile Ala Leu Ser Leu Ala Leu
1 5 10 15
Val Thr Asn Ser Ala Pro Thr Ser Ser Ser Thr Lys Lys Thr Gln Leu
20 25 30
Gln Leu Glu His Leu Leu Leu Asp Leu Gln Met Ile Leu Asn Gly Ile
35 40 45
Asn Asn Tyr Lys Asn Pro Lys Leu Thr Arg Met Leu Thr Phe Lys Phe
50 55 60
Tyr Met Pro Lys Lys Ala Thr Glu Leu Lys His Leu Gln Cys Leu Glu
65 70 75 80
Glu Glu Leu Lys Pro Leu Glu Glu Val Leu Asn Leu Ala Gln Ser Lys
85 90 95
Asn Phe His Leu Arg Pro Arg Asp Leu Ile Ser Asn Ile Asn Val Ile
100 105 110
Val Leu Glu Leu Lys Gly Ser Glu Thr Thr Phe Met Cys Glu Tyr Ala
115 120 125
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
Asp Glu Thr Ala Thr Ile Val Glu Phe Leu Asn Arg Trp Ile Thr Phe
130 135 140
Cys Gin Ser Ile Ile Ser Thr Leu Thr
145 150
<210> 112
<211> 153
<212> PRT
<213> Homo sapiens
<400> 112
Met Gly Leu Thr Ser Gin Leu Leu Pro Pro Leu Phe Phe Leu Leu Ala
1 5 10 15
Cys Ala Gly Asn Phe Val His Gly His Lys Cys Asp Ile Thr Leu Gin
20 25 30
Glu Ile Ile Lys Thr Leu Asn Ser Leu Thr Glu Gin Lys Thr Leu Cys
35 40 45
Thr Glu Leu Thr Val Thr Asp Ile Phe Ala Ala Ser Lys Asn Thr Thr
50 55 60
Glu Lys Glu Thr Phe Cys Arg Ala Ala Thr Val Leu Arg Gin Phe Tyr
65 70 75 80
Ser His His Glu Lys Asp Thr Arg Cys Leu Gly Ala Thr Ala Gin Gin
85 90 95
Phe His Arg His Lys Gin Leu Ile Arg Phe Leu Lys Arg Leu Asp Arg
100 105 110
Asn Leu Trp Gly Leu Ala Gly Leu Asn Ser Cys Pro Val Lys Glu Ala
115 120 125
Asn Gin Ser Thr Leu Glu Asn Phe Leu Glu Arg Leu Lys Thr Ile Met
130 135 140
Arg Glu Lys Tyr Ser Lys Cys Ser Ser
145 150
<210> 113
<211> 162
<212> PRT
<213> Homo sapiens
<400> 113
Met Arg Ile Ser Lys Pro His Leu Arg Ser Ile Ser Ile Gin Cys Tyr
1 5 10 15
Leu Cys Leu Leu Leu Asn Ser His Phe Leu Thr Glu Ala Gly Ile His
20 25 30
Val Phe Ile Leu Gly Cys Phe Ser Ala Gly Leu Pro Lys Thr Glu Ala
35 40 45
CA 02366921 2001-09-10
WO 00/53761
PCT/US00/06067
51
Asn Trp Val Asn Val Ile Ser Asp Leu Lys Lys Ile Glu Asp Leu Ile
50 55 60
Gin Ser Met His Ile Asp Ala Thr Leu Tyr Thr Glu Ser Asp Val His
65 70 75 80
Pro Ser Cys Lys Val Thr Ala Met Lys Cys Phe Leu Leu Glu Leu Gin
85 90 95
Val Ile Ser Leu Glu Ser Gly Asp Ala Ser Ile His Asp Thr Val Glu
100 105 110
Asn Leu Ile Ile Leu Ala Asn Asn Ser Leu Ser Ser Asn Gly Asn Val
115 120 125
Thr Glu Ser Gly Cys Lys Glu Cys Glu Glu Leu Glu Glu Lys Asn Ile
130 135 140
Lys Glu Phe Leu Gin Ser Phe Val His Ile Val Gin Met Phe Ile Asn
145 150 155 160
Thr Ser
<210> 114
<211> 144
<212> PRT
<213> Homo sapiens
<400> 114
Met Trp Leu Gin Ser Leu Leu Leu Leu Gly Thr Val Ala Cys Ser Ile
1 5 10 15
Ser Ala Pro Ala Arg Ser Pro Ser Pro Ser Thr Gin Pro Trp Glu His
20 25 30
Val Asn Ala Ile Gin Glu Ala Arg Arg Leu Leu Asn Leu Ser Arg Asp
35 40 45
Thr Ala Ala Glu Met Asn Glu Thr Val Glu Val Ile Ser Glu Met Phe
50 55 60
Asp Leu Gin Glu Pro Thr Cys Leu Gin Thr Arg Leu Glu Leu Tyr Lys
65 70 75 80
Gin Gly Leu Arg Gly Ser Leu Thr Lys Leu Lys Gly Pro Leu Thr Met
85 90 95
Met Ala Ser His Tyr Lys Gin His Cys Pro Pro Thr Pro Glu Thr Ser
100 105 110
Cys Ala Thr Gin Ile Ile Thr Phe Glu Ser Phe Lys Glu Asn Leu Lys
115 120 125
Asp Phe Leu Leu Val Ile Pro Phe Asp Cys Trp Glu Pro Val Gin Glu
130 135 140
<210> 115
<211> 538
<212> PRT
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
52
<213> Homo sapiens
<400> 115
Met Pro Arg Gly Trp Ala Ala Pro Leu Leu Leu Leu Leu Leu Gln Gly
1 5 10 15
Gly Trp Gly Cys Pro Asp Leu Val Cys Tyr Thr Asp Tyr Leu Gln Thr
20 25 30
Val Ile Cys Ile Leu Glu Met Trp Asn Leu His Pro Ser Thr Leu Thr
35 40 45
Leu Thr Trp Gln Asp Gln Tyr Glu Glu Leu Lys Asp Glu Ala Thr Ser
50 55 60
Cys Ser Leu His Arg Ser Ala His Asn Ala Thr His Ala Thr Tyr Thr
65 70 75 80
Cys His Met Asp Val Phe His Phe Met Ala Asp Asp Ile Phe Ser Val
85 90 95
Asn Ile Thr Asp Gln Ser Gly Asn Tyr Ser Gln Glu Cys Gly Ser Phe
100 105 110
Leu Leu Ala Glu Ser Ile Lys Pro Ala Pro Pro Phe Asn Val Thr Val
115 120 125
Thr Phe Ser Gly Gin Tyr Asn Ile Ser Trp Arg Ser Asp Tyr Glu Asp
130 135 140
Pro Ala Phe Tyr Met Leu Lys Gly Lys Leu Gln Tyr Glu Leu Gln Tyr
145 150 155 160
Arg Asn Arg Gly Asp Pro Trp Ala Val Ser Pro Arg Arg Lys Leu Ile
165 170 175
Ser Val Asp Ser Arg Ser Val Ser Leu Leu Pro Leu Glu Phe Arg Lys
180 185 190
Asp Ser Ser Tyr Glu Leu Gln Val Arg Ala Gly Pro Met Pro Gly Ser
195 200 205
Ser Tyr Gln Gly Thr Trp Ser Glu Trp Ser Asp Pro Val Ile Phe Gln
210 215 220
Thr Gln Ser Glu Glu Leu Lys Glu Gly Trp Asn Pro His Leu Leu Leu
225 230 235 240
Leu Leu Leu Leu Val Ile Val Phe Ile Pro Ala Phe Trp Ser Leu Lys
245 250 255
Thr His Pro Leu Trp Arg Leu Trp Lys Lys Ile Trp Ala Val Pro Ser
260 265 270
Pro Glu Arg Phe Phe Met Pro Leu Tyr Lys Gly Cys Ser Gly Asp Phe
275 280 285
Lys Lys Trp Val Gly Ala Pro Phe Thr Gly Ser Ser Leu Glu Leu Gly
290 295 300
Pro Trp Ser Pro Glu Val Pro Ser Thr Leu Glu Val Tyr Ser Cys His
305 310 315 320
Pro Pro Arg Ser Pro Ala Lys Arg Leu Gln Leu Thr Glu Leu Gln Glu
325 330 335
CA 02366921 2001-09-10
WO 00/53761 PCT/US00/06067
53
Pro Ala Glu Leu Val Glu Ser Asp Gly Val Pro Lys Pro Ser Phe Trp
340 345 350
Pro Thr Ala Gin Asn Ser Gly Gly Ser Ala Tyr Ser Glu Glu Arg Asp
355 360 365
Arg Pro Tyr Gly Leu Val Ser Ile Asp Thr Val Thr Val Leu Asp Ala
370 375 380
Glu Gly Pro Cys Thr Trp Pro Cys Ser Cys Glu Asp Asp Gly Tyr Pro
385 390 395 400
Ala Leu Asp Leu Asp Ala Gly Leu Glu Pro Ser Pro Gly Leu Glu Asp
405 410 415
Pro Leu Leu Asp Ala Gly Thr Thr Val Leu Ser Cys Gly Cys Val Ser
420 425 430
Ala Gly Ser Pro Gly Leu Gly Gly Pro Leu Gly Ser Leu Leu Asp Arg
435 440 445
Leu Lys Pro Pro Leu Ala Asp Gly Glu Asp Trp Ala Gly Gly Leu Pro
450 455 460
Trp Gly Gly Arg Ser Pro Gly Gly Val Ser Glu Ser Glu Ala Gly Ser
465 470 475 480
Pro Leu Ala Gly Leu Asp Met Asp Thr Phe Asp Ser Gly Phe Val Gly
485 490 495
Ser Asp Cys Ser Ser Pro Val Glu Cys Asp Phe Thr Ser Pro Gly Asp
500 505 510
Glu Gly Pro Pro Arg Ser Tyr Leu Arg Gin Trp Val Val Ile Pro Pro
515 520 525 ,
Pro Leu Ser Ser Pro Gly Pro Gin Ala Ser
530 535