Note: Descriptions are shown in the official language in which they were submitted.
r
CA 02370387 2001-10-19
1
GENETIC SEQUENCE WHICH CODES FOR THE FLAVONE SYNTHASE II
ENZYME AND USE OF THE SAME
The present invention relates to genetic sequences encoding enzymes of the
flavonoid metabolism, in particular flavone synthase II (FNS II) or
derivatives
thereof as well as to the use thereof for the targeted modification of the
color of
flowers, for altering the flavone content or the flavone pattern,
respectively, in
leaves, flowers and other tissues of plants and other organisms, moreover, it
relates to the use in expression systems for the synthesis of natural,
functional
flavones for medical or similar applications, for example for the treatment of
cancer or for improving the human immune defense.
Flavonoids and their function in plants
Flavonoids are the most important and most widespread plant pigments which
have been detected in various tissues, such as flowers, leaves or roots.
Furthermore, they are among the best characterized secondary metabolites in
plants. Up to now, more than 3000 different flavonoids have been
characterized.
They have been divided into different subclasses (e.g. flavones, flavonols, or
anthocyans) based on the degree of oxidation of the central C ring.
Additionally,
each type may be further modified by hydroxylation, acylation or glycosylation
(Heller and Forkmann, 1994). Due to the different physico-chemical properties
of
the molecules, the subclasses may in part exhibit very different biological
functions.
The accumulation of certain flavonoids in a plant cell depends on the
availability
of the corresponding enzymes wherein the availability of the enzymes
ultimately is
dependent on the expression of the respective gene. Regulation of the
expression
of the genes of flavonoid biosynthesis is substantially determined by the
plant
species, the developmental stage and environmental conditions.
Flavonoids play an important role both within and outside of the plant. Thus,
for
example, certain flavonols have been shown to be required for growth of the
pollen tube. If the accumulation of flavonols is suppressed by blocking their
biosynthetic pathway, sterile pollens are obtained (Taylor and Jorgensen,
1992).
Both biotic and abiotic signals may result in an accumulation of flavonoids
during
CA 02370387 2001-10-19
2
interaction of a plant with its environment. Thus, for example, UV irradiation
leads to an accmnulation of flavonols and flavones. This is achieved by
induction
of the transcription of the respective flavonoid biosynthesis genes in
different
species (Kubasek et al., 1992). But also other stress factors such as
wounding,
extreme temperature variations and water stress may induce flavonoid
accumulation and/or gene expression in different species (Hradzina, 1982).
Flavonoids have been assigned a double role in interactions of plants with
other
organisms. On the one hand, as phenolic compounds flavonoids have a
phytoalexin effect against various pathogens and as a deterrent against
predators
(Harborne and Grayer, 1994), on the other hand they are responsible for the
communication between plants of the family of Leguminosae and particular
microorganisms. In this respect, flavonoids serve as signaling agents for
nitrogen
fixing bacteria which then express genes required for establishing a symbiosis
with the plant (Redmond et al., 1986). In flowers, leaves, and fruits, the
flavonoids
and particularly the colored anthocyans but also chalcones, aurones, flavones,
and
flavonols, are responsible for the coloring and the patterns of various
secondary
metabolites. Together with other characteristics such as e.g. t:he scent the
latter is
important for recognition by various animals but also for humans using the
plant
as decoration or food stuff (Harborne and Grayer, 1994). Furthermore,
particular
flavonoids such as the flavone apigenin and the flavonol quercetin, have an
effect
on the auxin transport within a plant (Jacobs and Rubery, 1988)
The flavonoid biosynthesis pathway (Fib. 1_A~
The structure of flavonoids includes two aromatic rings (A and B) and a
central
heterocycle (C) (Fig. 1B). Within the plant, they are synthesized starting
from L-
phenylalanine via the phenyl propanoid pathway by the enzymatic reaction of
phenylalanine ammonia lyase (PAL) and cinnamonic acid 4-h;ydroxylase (4CL). 4-
Cumaroyl CoA resulting from this reaction together with 3 molecules of malonyl
CoA yields tetrahydroxychalcone. This reaction is catalyzed by chalcone
synthase
(CHS), the key enzyme of flavonoid biosynthesis (Fig. lA). Generally,
tetrahydroxychalcone (THC) is quickly isomerized to yield naringenin (NAR) by
the enzyme chalcone isomerase (CHI). Different subsequent reactions generate
the
anthocyans. Flavones are formed via a side pathway by the action of FNS I or
FNS
II, a cytochrome P450 enzyme. This class of enzymes is widespread in nature,
and
CA 02370387 2001-10-19
3
various genes for cytochrome P450 enzymes have been isolated and sequenced
from vertebrates, insects, yeasts, fungi, bacteria, and plants.
Flavone synthase uses different flavanones such as NAR or eriodyctol (ERI) as
substrates to synthesize the corresponding flavones apigenin (Ap) and luteolin
(Lu). For this purpose, a double bond is introduced between positions C2 and
C3,
as shown in Figure 1B. Flavones may be present in plants in glycosylated as
well
as in methylated forms.
The formation of flavones from flavanones in vitro has been observed first in
enzyme preparations of UV irradiated Petroselium crispum cell suspension
cultures. The corresponding enzyme is a soluble 2-oxoglutarate-dependent
dioxygenase which has been referred to as flavone synthase I (FNS I). In the
flowers of various flavone-producing plants, e.g. Si;nningia cardinalis,
Antirrhinum majus, Verbena hybrida, Columnea hybrida, Chrysanthemum
morifolium, Gerbera hybrids and osmotically induced cell suspension cultures
of
Glycine max, this reaction, however, is catalyzed by the above-mentioned FNS
II,
an NADPH-dependent microsomal enzyme belonging to the class of P450
cytochromes (Heller and Forkmann, 1994). The formation of flavones in flowers
of Gerbera hybrids is regulated by the Fns locus. Flavone synthesis is only
detected in lines which carry the dominant allele while no FNS II activity is
detectable in homozygous recessive lines (Fig. 2; Martens and Forkmann, 1998).
Three types of pigments are responsible for the color in flowers: betalains,
carotenoids, and flavonoids. Betalains are present only in a few families of
the
Centrospermae in which they are responsible for yellow, orange, red, and
purple
colors. Carotenoids result in orange and yellow shades and ane the main
pigments
of most of the orange and yellow flowers. Flavonoids are the most important
and
most widespread pigments in flowers and plants, respectively. This group
includes
the coloring anthocyans which are present in the vacuole in glycosylated and
often
acylated forms. Different anthocyans are able to produce different shades. The
color of flowers is further affected by the pH of the vacuole, the
complexation
with metals and the pattern of glycosylation and acylation, respectively
(Forkmann, 1991 ). Another important factor for the generation of the various
flower colors is copigmentation of anthocyans with colorless flavonoids, such
as
CA 02370387 2001-10-19
4
flavones or flavonols, or also with tannins (Scott-Moncrieff, 1936).
Anthocyans
which are copigmented with flavones may attain different colors depending on
the
basic structure of the anthocyan which may vary between purple and blue (Asen
and Horowitz, 1974; Goto and Kondo, 1991). Moreover, several flavones, such as
isoetin, have been identified as yellow flower pigments (Harborne, 1978).
It is an important aim in horticultural plant breeding to develop novel
varieties of
flowering ornamental plants. In the past, classical methods of breeding have
been
partly successful in establishing a number of different colors in many
economically important ornamental plants. The gene pool of the individual
species, however, limits possibilities of such approaches carried out in a
natural
manner. This is the reason why today there are only a few species showing the
whole spectrum of colors. Furthermore, the alteration generated by means of
classical methods of breeding cannot be targeted. Since the aesthetic value of
a
flower is determined by various factors, such as form, scent and color, and an
alteration of one of this factors by crossing generally can only be achieved
to the
expense of similar other, visible characteristics and is extremely lengthy and
laborious, an effective way to achieve novel varieties must be utilized. The
possibility of altering the color of plant flowers in a targeted manner
provides
clear advantages as compared to other methods. This is particularly important
in
an area with a high product turnover in which novelty is an important market
factor. For example, the development of blue flowering varieties of the main
cut
flower species, such as roses, chrysanthemums, carnations, lilies, tulips, and
gerbera would lead to a substantial market advantage on the cut flower market,
but
also on the potted plant market. The possibility of controlling the synthesis
of
copigments, e.g. flavones, in plants is a beneficial application of the
targeted
alteration of flower colors. In addition, besides flowers this application may
also
be applied to fruits and other agricultural plants, e.g. fruit and vegetable
plants,
and to leaves, e.g. of ornamental plants.
Besides their contribution to the color of flowers, the flavonoids and
particularly
the flavones also have other biological properties and effects. For example,
in
some plants they have been found to be a feed stimulant for monophagous and
oligophagous insects (Harborne and Grayer, 1994). In most cases, the
glycosides
exhibit a higher effect than the corresponding aglycones presumably due to an
CA 02370387 2001-10-19
5
improved solubility of the glycosides. Furthermore, insects are able to
distinguish
between different sugar residues whereby the active components are further
differentiated. In addition, also the basic structure of the aglycones may
result in
different effects. Compared to many other secondary plant metabolites, the
flavonoids and flavones, respectively, obviously have no excessive toxic
effect on
insects. Nevertheless there are some flavones which already in very low
concentrations are able to act as a deterrent for feed insects or to severely
inhibit
the growth of the animals. In this respect, no effect of the type of
glycosylation
could be demonstrated. However, an effect of the type of hydroxylation or
methoxylation, respectively, of the flavone could be demonstrated (Harborne
and
Grayer, 1994).
Moreover, several flavones stimulate the egg laying of butterflies on specific
plants. It has been shown that egg laying does not occur until the animals
have
recognized the stimulus. Such stimulating substances include for example
flavones the vicenin-2 and various luteolin derivatives. If the synthesis of
these
substances is blocked in the respective host plants it is possible to inhibit
egg
laying by the butterflies and, thus, feed damages by their caterpillars.
Utilizing the natural chemical defense mechanisms of the plants may avoid the
problems encountered with the use of synthetic insecticides such as e.g.
environmental pollution caused by residues of the substances used in fruits
and
soil. In addition resistance generation observed with most of the synthetic
pesticides is avoided or at least delayed, and the sometimes extensive costs
for
agents and applications are saved.
Another important biological property of the flavonoids relates to the
activation of
nodulation genes in various rhizobium species. These bacteria infect
leguminous
plants and form nitrogen-fixing root nodules. In this process, the flavonoids
produced by the host plant act as a "signaling agent" whereby the bacteria
induce
the process of infection. These plant-specific active compounds also include
various flavones such as apigenin, luteolin, and 7,4'-dihydroxyflavone (Firmin
et
al., 1986; Redmond et al., 1986). Altering the production and delivery of
flavones
by the root or the flavone pattern, respectively, in this tissue provides a
possibility
CA 02370387 2001-10-19
6
to improve the nitrogen fixation and probably to establish this mechanism also
in
non-leguminous plants.
Utilizing this natural symbiotic mechanism may reduce nitrogen fertilization
and
thereby the environmental pollution by washing away of the nutrients.
Moreover,
the costs for fertilizers and for their spreading are saved.
Within a plant, several naturally occurring flavonoids such as the flavone
apigenin
or the flavonols kaempferol and quercitin affect the transport of auxins in
different
plant tissues and transport systems. In this respect, they act similar to
chemical
transport inhibitors. As growth regulators of plants the auxins themselves
affect
cell extension, cell division, apical dominance, reformation of roots and
shoot as
well as parthenocarpy. An induced, altered flavonoid concentration (endogenous
change and/or exogenous application), thus, may have a significant effect on
plant
growth via their interaction with auxins. This may allow for a substitution of
synthetic growth inhibitors.
Moreover, as bioactive substances flavonoids have a non-negligible role in the
diet
of men and animals. They are found in fruits, vegetables, nuts, seeds, shoots
but
also in tea and wine. Since some time, anti-allergic, anti-inflammatory, anti-
viral,
proliferation-reducing and anti-cancer properties are attributed to flavonoids
and
also to several flavones. But also an effect on the metabolism and the highly
complex immune system of humans and animals has been described. In this
respect, flavonoids and flavones, respectively, have an effect on a large
number of
different enzymes (e.g. on hyaluronidase or aldose reductase), they have
important
enzyme inducing and anti-oxidative properties, and are capable of scavenging
free
radicals, chelating several metal canons, and have an effect on cellular
protein
phosphorylation (Middleton and Kandaswami, 1994).
If agricultural plants important for the human and animal nutrition because of
their
content in health-promoting flavonoids can be improved by a targeted
alteration of
the content or pattern of the respective compounds this would greatly
contribute to
a healthy diet of men and animals.
CA 02370387 2001-10-19
7
Therefore, it has been an object of the present invention to provide means and
methods to alter and control the flavonoid biosynthesis and the formation of
flavones, respectively, in plants in a targeted manner in order to e.g. alter
the color
of a plant flower or to improve the resistance properties and capability to
establish
symbioses of a plant.
It has been another object of the present invention to provide means and
methods
useful for a targeted synthesis of defined flavones. Flavones obtained in such
manner may find use i.a. in cancer therapy or may contribute to the health of
men
and animals in the form of medicaments.
According to the present invention, this object has been achieved by the
claims.
Therefore, the present invention relates to a nucleic acid sequence encoding a
flavone synthase II (FNS II). In one embodiment, the present invention relates
to a
nucleic acid sequence as shown in SEQ ID NO:1, or to a portion thereof. In
another embodiment, the present invention relates to a nucleic acid sequence
hybridizing to a nucleic acid sequence as shown in SEQ II) NO:1 or a portion
thereof and/or having a homology of at least 40%, more preferably at least
45%,
further preferred at least 55% or most preferably at least 65-70% or most
preferably a homology of more than 85% on the level of the nucleic acid
sequence
or amino acid sequence to at least one or more regions (preferably to the
whole
region) of the sequence as shown in SEQ ID NO:1. Preferably, the nucleic acid
sequence encodes a protein or a polypeptide having the biological activity of
a
flavone synthase. In another embodiment, the present invention relates to a
nucleic
acid sequence which is degenerated with respect to a nucleic acid sequence
according to the above-mentioned embodiments. In a prefen-ed embodiment; the
nucleic acid sequence according to the present invention is DNA or RNA and is
derived from a flavone-containing plant such as gerbera (Gerbera hybrids),
aster
(Callistephus chinensis), snapdragon (Antirrhinium majus), chrysanthemum
(Chrysanthemum indicum), dahlia (Dahlia hybrids), gloxinia (Sinningia
hybrids),
verbena (Verbena hybrids), and Streptocarpus (S. hybrids). In another
preferred
embodiment the nucleic acid sequence according to the present invention is a
recombinant nucleic acid sequence. Furthermore, the present invention relates
to a
CA 02370387 2001-10-19
8
nucleic acid sequence complementary to the sequence encoding flavone synthase
II (FNS In.
Accordingly, the present invention provides an isolated nucleic acid sequence
comprising a nucleic acid sequence encoding flavone synthase II (FNS I~ or a
functional derivative of said enzyme or a nucleic acid sequence complementary
thereto. The term "FNS II enzyme" means enzymes of the flavonoid biosynthetic
pathway using flavanones such as naringenin and eriodictyol or also other
compounds of this class as a substrate for the synthesis of the corresponding
flavones.
Preferred is a nucleic acid according to the present invention which has been
isolated from its natural environment or chemically synthesized. Particularly
preferred are nucleic acid molecules which are formed or obtained in vitro
including genomic DNA fragments, recombinant and synthetic molecules and
nucleic acids in combination with heterologous nucleic acids. This also
comprises
genomic DNA or cDNA or portions thereof encoding FNS II or portions thereof in
a reverse orientation to its own or another promoter. Further comprised are
naturally occurring, closely related nucleic acid sequences.
The term "nucleic acid sequence encoding a flavone synthase II" is used herein
in
its most general form and comprises any sequential order of nucleotide bases
defining, directly or via a complementary array of bases, an ~unino acid
sequence
of an FNS II.
A polypeptide having a portion or the complete amino acid sequence of flavone
synthase II means a full length FNS II or an active incomplete form thereof.
In another embodiment, the present invention relates to oligonucleotides which
may be used as genetic probes or "antisense" molecules for controlling the
expression of the corresponding gene in plants or other organisms. An
"antisense"
molecule as described herein also comprises a gene construct consisting of a
structural, genomic or cDNA gene or a portion thereof in reverse orientation
with
respect to its own or any other promoter.
CA 02370387 2001-10-19
9
In another embodiment, the nucleic acid sequence encoding FNS II or various
functional derivatives thereof is used to reduce the activity of endogenous
FNS II,
or alternatively a nucleic acid sequence encoding said enzyme or various
derivatives or portions thereof is used in antisense orientation to reduce the
activity of FNS II. Moreover, it is also possible that an antisense transcript
of FNS
II or a fragment or a portion of FNS II (e.g. an oligonucleotide molecule)
forms a
duplex with the whole or portions of the naturally occurring mRNA which
specifies the enzyme and thus inhibits an accumulation of or the translation
of the
mRNA into the active enzyme. Another possibility is the use of ribozymes to
inactivate specific nucleic acid sequences.
Alterations of the FNS II activity mentioned herein relate to an increase or
decrease of the activity of up to 30% or more preferably 30 to 50% or still
more
preferably 50 to 75% and most preferably of 75% or even higher or lower,
respectively, as compared to the normal, endogenous or existing activity
value.
The amount of the activity can be tested easily using the method described in
Martens and Forkmann (1998) (see Example 3).
The nucleic acid described in the present invention may also be a ribonucleic
acid
or a deoxyribonucleic acid existing in the form of a single stranded or double
stranded and linear or covalently closed, circular molecule. Generally, the
nucleic
acid molecule is present in the form of cDNA. The present invention also
comprises other nucleic acid molecules hybridizing to the nucleic acid
molecules
of the invention or specifically to the sequence shown in SEQ m NO:1 or a
portion or region thereof under conditions of low~, preferably medium and most
preferably high stringency. A particularly preferred embodiment relates to a
nucleic acid molecule comprising the nucleic acid sequence shown in SEQ >D
NO:1 or a molecule having a similarity of at least 40%, more preferably at
least
45%, still more preferably at least SS% or most preferably at least 65-70% or
most
preferably a similarity of more than 85% on the level of the mucleic acid
sequence
or amino acid sequence to at least one or more regions (preferably over the
whole
region) of the sequence shown in SEQ >Z7 NO:1 and wherein the nucleic acid
encodes or is complementary to a sequence encoding an enzyme having FNS II
activity.
CA 02370387 2001-10-19
10
Furthermore, the present invention comprises nucleic acid molecules in the
form
of oligonucleotide primers or competent probes for hybridization with a
portion of
the nucleic acid molecules described above and specifically to that shown in
SEQ
ID NO:1. The hybridization may be earned out under conditions of low,
preferably
medium and most preferably high stringency. Preferably, said portion
corresponds
to the 5' or the 3' end of the gene. For the purposes herein, the 5' end is
defined as
the region extending mainly between the start codon of the structural gene
sequence to the medium region of the gene. The 3' end is considered herein as
being the region defining the structural genetic sequence between the medium
region of the gene and the stop codon. Therefore, it is obvious that
oligonucleotides or probes are able to hybridize to the 5' end or the 3' end
or to a
region which is common to both the S' or the 3' end. The present invention
comprises all such probes. Preferred oligonucleotides are presented in Example
4.
The nucleic acid or the complementary form thereof may encode the full length
enzyme or a portion or derivative thereof. "Derivative" means single or
multiple
amino acid substitutions, deletions and/or additions with respect to the
naturally
occurnng enzyme while, preferably, the flavone synthase II activity is
maintained.
In this respect, the nucleic acid according to the present invention comprises
the
naturally occurnng nucleotide sequence encoding FNS II and single or multiple
nucleotide substitutions, deletions, and/or additions. The nucleic acid
according to
the present invention or the complementary form thereof may also encode a
portion of FNS II which is either active or inactive. Such a nucleic acid
molecule
may be used as an oligonucleotide probe, as a primer for polymerase chain
reaction, in different mutagenesis techniques, or for the preparation of
antisense
molecules.
Furthermore, the present invention relates to a recombinant DNA molecule
containing a nucleic acid sequence according to the present invention. In a
preferred embodiment, the recombinant DNA molecule is a vector or a vector
having a promoter. In a particularly preferred embodiment the promoter is
capable
of expressing the nucleic acid sequence according to the present invention.
The
nucleic acid molecules of the invention may be present in combination with a
vector molecule, e.g. an expression vector, in both orientations. In this
respect, the
term "vector molecule" is used in its most general meaning to comprise any
CA 02370387 2001-10-19
11
intermediary vehicles of the nucleic acid molecule which enable the transfer
of the
nucleic acid into cells, particularly in plant cells and/or its integration
into a
genome. Preferably, these vector molecules or portions thereof are used for
integration into a plant genome. Such vector molecules may be replicated
and/or
expressed in prokaryotic cells and/or in eukaryotic cells. An intermediary
vehicle
may be for example adapted to the use in electroporation, microprojectile
bombardement, in the transfer using agrobacteria or in the insertion via DNA
or
RNA viruses. The intermediary vehicle and/or the nucleic acid molecule
contained
therein may be stably integrated into the plant genome. Additionally, the
nucleic
acid molecule may contain also a promoter sequence useful for the initiation
of
expression of the nucleic acid molecule in a cell, particularly in a plant
cell. The
nucleic acid molecule and the promoter may also be introduced into the cell
using
different methods (see above).
The present invention also relates to host cells containing the DNA molecules
according to the present invention. The host cells may be prokaryotic or
eukaryotic
cells, particularly yeast cells, insect cells, plant cells, and mammalian
cells.
The present invention further relates to a polypeptide encoded by a nucleic
acid
according to the invention. In a preferred embodiment the present invention
relates
to a polypeptide having the amino acid sequence as shown in SEQ >D N0:2 or a
portion or derivatives thereof. In another preferred embodiment the
polypeptide is
derived from a flavone-containing plant, such as e.g. from gerbera (Gerbera
hybrids), aster (Callistephus chinensis), snapdragon (Antirrhinium majus),
chrysanthemum (Chrysanthemum indicum), dahlia (Dahlia hybrids), gloxinia
(Sinningia hybrids), verbena (Verbena hybrids), and Streptocarpus (S.
hybrids). In
a particularly preferred embodiment the polypeptide of the present invention
has
flavone synthase II activity.
Derivatives in the sense of the present invention are amino acid insertion
derivatives, deletion derivatives and/or substitution amino acid variants of
the
amino acid sequence of SEQ ID N0:2.
Amino acid insertion derivatives of FNS II according to the present invention
comprise both amino and carboxyl fusions as well as insertions of single or
CA 02370387 2001-10-19
12
multiple amino acids within the sequence. Insertion amino acid sequence
variants
are those in which one or more amino acid residues have been introduced into
the
protein at a predetermined site, although a random insertion together with
appropriate screening of the product is also possible. Deletion variants are
characterized by the removal of one or more amino acids from the sequence.
Substitution amino acid variants are those wherein at least one residue of the
sequence has been removed and another residue has been introduced at the same
site. Typical substitutions are presented in Table 1.
TABLE 1
Suitable residues for amino acid substitution
Original amino acid Exemplary substitution by
Aln Ser
Arg Lys
Asn Gln; Fps
Asp Glu
Cys Ser
Gln Asn
Glu Asp
Gly ~'o _ _ .. _
~s Asn; Gln
Ile Leu; Val
Leu IIe; Val
Lys Arg; G(n; Glu
Met Leu; Ile
Phe Met; Leu; Tyr
S er Thr
Thr Ser
Trp Tyr .
Tyr Trp; Phe
Val IIe; Leu
In general, amino acids are substituted by amino acids having similar
properties
such as hydrophobicity, hydrophilicity, electronegativity, very bulky side
chains
and the like. Amino acid substitutions generally relate to only a single
residue
whereas insertions typically are directed to a region of 1 to 10 amino acid
residues
and deletions to a region of 1 to 20 residues. Preferably, deletions and
insertions
are carried out on adjacent pairs, e.g. a deletion of two residues or an
insertion of
two residues.
CA 02370387 2001-10-19
13
The amino acid variants described above of derivatives according to the
present
invention may be prepared easily using known techniques for peptide synthesis
such as e.g. by solid phase synthesis and similar methods or by recombinant
DNA
manipulations. Techniques for introducing substitution mutations at
predetermined sites in DNA having a known or partially known sequence are well-
known and include for example M13 mutagenesis. The manipulation of DNA
sequences for the preparation of proteins carrying substitutions, insertions
or
deletions is detailed for example in Sambrook et al. (1989).
Other examples of recombinant or synthetic mutants and derivatives of FNS II
according to the present invention comprise single or multiple substitutions,
deletions and/or additions of any molecule associated with the enzyme such as
carbohydrates, lipids and/or proteins or polypeptides.
The terms "analogue" and "derivative" also comprise any functional chemical
equivalent of FNS II and also any amino acid derivative as already described
above.
Another aspect of the present invention relates to recombinant forms of FNS
II.
Recombinant forms of the enzyme provide the possibility to develop for example
more active enzymes or systems for production of various flavones in vitro for
the
use in various fields such as for example in human medicine. The latter system
may be for example of use in cancer research.
In another embodiment, the present invention relates to transgenic plants
containing a nucleic acid sequence according to the present invention. In a
preferred embodiment, the nucleic acid sequence is suitable for expression and
optionally can be controlled or is controlled within the plant in conjunction
with
its development. In another preferred embodiment the tra.nsgenic plant is
selected
from the group of flavone-containing plants such as gerbera. (Gerbera
hybrids),
aster (Callistephus chinensis), snapdragon (Antirrhinium majus), chrysanthemum
(Chrysanthemum indicum), dahlia (Dahlia hybrids), gloxinia (Sinningia
hybrids),
verbena (Verbena hybrids), and Streptocarpus (S. hybrids), contains an
CA 02370387 2001-10-19
14
endogenous FNS II and further contains a non-endogenous FNS II nucleic acid
sequence according to the present invention.
According to the present invention, a nucleic acid sequence encoding an FNS II
or
a derivative or portion thereof may be introduced into a plant in one of two
possible orientations and may be expressed therein thereby providing the
possibility to convert either naringenin (NAR) and/or other suitable
substrates, if
synthesized within the plant cell, which eventually results in the formation
of
different flavones. Moreover, the formation of said metabolites may be
inhibited
by a reduction or elimination of endogenous or existing FNS II activity. In
gerbera,
the synthesis of flavones results in an alteration of the flower color. Using
the
flavone-containing, orange colored variety "Th 58" as an example which is
heterozygous for the Fns locus (fns+ fns) a color variation from dark red
(flavone-
free; genotype fns fns) via different shades of orange-red (flavone-
containing;
genotype fns+ fns) to a yellow-orange color (very high flavone content;
genotype
fns+ fns+) could be demonstrated in autogamy progeny. This experiment may also
be applied to other gerbera varieties heterozygous for the Fns locus (see also
Example 2). Expression of the nucleic acid sequence in one of two possible
orientations within the plant may be constitutively, inducible or dependent on
the
development and also tissue-specific. The term "expression" is used in its
most
general meaning to include the production of RNA or both :RNA and protein. It
also comprises the partial expression of nucleic acid molecules.
The present invention relates to a method for the preparation of transgenic
plants
capable of synthesizing FNS II or active mutants or derivatives. Said method
comprises the stable transformation of a cell of a suitable plant with a
nucleic acid
molecule comprising a nucleotide sequence encoding said FNS II under
conditions
achieving the possible expression of said nucleic acid molecule, regeneration
of a
transgenic plant from the cell and growth of said transgenic plant for a
particular
time and under conditions suitable to achieve expression of the nucleic acid.
The
transgenic plant may exhibit higher values of FNS II activity compared to the
value measured in comparable non-transgenic plants, or the values may be lower
compared to those of comparable non-transgenic plants.
CA 02370387 2001-10-19
15
One aspect of the present invention relates to a method for the preparation of
a
transgenic plant having a reduced, endogenous or existing hNS II activity.
This
method comprises the stable transformation of a cell of a suitable plant with
a
nucleic acid molecule comprising a nucleotide sequence encoding a sequence or
a
complementary sequence of FNS II, the regeneration of a transgenic plant from
the
cell, and, if necessary, the raising of this transgenic plant under conditions
suitable
to achieve expression of nucleic acids.
Another aspect of the present invention relates to a method fox the
preparation of a
genetically engineered plant having a reduced, endogenous or existing FNS II
activity. This method comprises the alteration of the FNS II gene by a
modification of the endogenous sequence via homologous recombination starting
from an appropriately modified gene of an FNS If or a derivative or portion
thereof. The gene is introduced into the plant and a genetically engineered
plant is
regenerated from the cell.
Another aspect of the present invention relates to a method for the
preparation of a
transgenic flowering plant having altered flower characteristics. This method
comprises the introduction of the nucleic acid sequence according to the
present
invention into a cell of a suitable flavone-free plant, the regeneration of a
transgenic plant from the cell, and raising a transgenic plant for a time and
under
conditions to achieve the expression of the introduced nucleic acid sequence
according to the present invention. The transgenic plant may be for example
selected from the group of flavone-containing plants consisting of euphorbia
(E.
pulcherrima), cyclamen (Cyclamen persicum), rose (Rosa hybrida), pelargonium
(P. spec.), begonia (B. spec.), carnation (Dianthus caryophyllus), and tulip
(Tulipa
hybrids). In another preferred embodiment, the transgenic plant is capable of
expressing an endogenous flavone synthase II. Such a transgenic plant may be
for
example selected of the group of plants consisting of gerbera (Gerbera
hybrids),
aster (Callistephus chinensis), snapdragon (Antirrhinium majus), chrysanthemum
(Chrysanthemum indicum), dahlia (Dahlia hybrids), gloxinia (Sinningia
hybrids),
verbena (Verbena hybrids), and Streptocarpus (S. hybrids). In another
preferred
embodiment this endogenous flavone synthase II is coexpressed during
expression
of the nucleic acid introduced according to the present invention.
CA 02370387 2001-10-19
16
In one embodiment, the endogenously existing flavone synthase II activity is
reduced by the introduction of the nucleic acid sequence. This method
comprises
the stable transformation of a cell of a suitable plant with a nucleic acid
sequence
according to the invention or a sequence complementary thereto, the
regeneration
of a transgenic plant from the cell and raising this transgenic plant for a
time and
under conditions suitable to alter the amount of activity of the endogenous or
existing FNS II. Preferably, the altered level is lower than the endogenous or
existing level of FNS II activity in a comparable non-transgenic plant.
Optionally,
for the reduction of the endogenous FNS II activity it is necessary to express
the
nucleic acid sequence introduced or a complementary sequence thereof. Thus, an
expression of the genetic sequence introduced or of its complementary analogue
may be required to achieve the desired effect. This substantially means a
flowering
plant with altered flower characteristics.
In this respect the present invention relates to a method for the preparation
of a
flowering plant showing different flower characteristics. This method
comprises
alterations of the FNS II gene by modification of the endogenous sequences via
homologous recombination of an appropriately altered gene of FNS II or a
derivative or portion thereof, the introduction into the plant cell and the
regeneration of the genetically engineered plant from the cell.
Moreover, the nucleic acid molecule according to the present invention may be
regulated in a development-depending manner. Generally, an altered
inflorescence
rules out the possibility of producing a flower having a weaker color or other
shades depending on the physiological conditions of the recipient plant.
"Recipient
plant" refers to a plant which produces a measurable amount of substrate of
FNS II
enzyme or FNS II itself and has the corresponding physiological properties and
the
genotype necessary for the development of the desired colors. This includes
but is
not restricted to the following plants: gloxinia (Sinningia Izybrids),
snapdragon
(Antirrhinium majus), columnea (Columnea hybrids), dahlia (Dahlia variabilis),
gloxinia (Sinningia cardinalis), Streptocarpus (Streptocarpus hybridus),
verbena
(Verbena hybrida), chrysanthemum (Chrysanthemum indicum); peace lily
(Spathiphyllum wallisii), petunia (Petunia hybrida), cyclamen (Cyclamen
persicum), rose (Rosa hybrids), and pelargonium (P. spec.).
CA 02370387 2001-10-19
17
Accordingly, the present invention relates to a method for the preparation of
a
transgenic plant expressing, to a measurable extent, a recombinant gene
encoding
FNS II or a portion thereof or carrying a nucleic acid sequence which is
substantially complementary to the full length or a portion of the mRNA
molecule
which may be easily transcribed, if necessary, to achieve the regulation of
FNS II.
This method comprises the stable transformation of a cell of a suitable plant
with
the isolated nucleic acid comprising a nucleotide sequence encoding FNS II or
a
derivative or a portion thereof or a sequence complementary to the coding
nucleotide sequence, if necessary under conditions allowing an expression of
said
isolated nucleic acid, and the regeneration of a transgenic plant from the
cell.
The skilled artisan immediately appreciates the possibilities for use of the
present
invention such as e.g. for an increase or decrease in the expression of
enzymes
which occur naturally in a target plant. This will result in different novel
flower
color shades, for example various orange and dark red shades.
Therefore, the present invention relates to any transgenic plant containing
the full
length or a portion of the nucleic acid sequence according to the present
invention
and/or containing a homologous or related form thereof or an antisense form of
any of those described, and in particular those transgenic plants exhibiting
varying
flower characteristics. The transgenic plants may contain nucleic acid
molecules
introduced therein comprising a nucleotide sequence encoding FNS II or a
complementary sequence thereof. Generally, the nucleic acid is stably
introduced
into the plant genome although the present invention also comprises the
introduction of an FNS II nucleic acid sequence within an autonomously
replicating nucleic acid sequence such as for example DNA or RNA viruses
capable of replicating in a plant cell. Moreover, the present invention also
comprises seeds of the transgenic plant, particularly those containing
flavones.
Using the FNS II gene according to the present invention, the biosynthesis of
flavonoids and the formation of flavones, respectively, in plants may be
altered in
a targeted manner in different ways.
To begin with, two different approaches must be considered in the field of
plants.
On the one hand, plants are available which naturally form flavones and the
CA 02370387 2001-10-19
18
glycosides thereof, such as Antirrhinium and Verbena in flowers, Clerodendron
and Citrus in leaves, Althaea and Sophora (leguminosae) in roots, and Prunus
and
Pinus spec. in hardwood (Wollenweber, 1994; Williams and Harborne, 1994).
Besides this very large group there are also some plants which lack flavone
synthesis in certain tissues. This is the case e.g. in several flowers of
important
ornamental plants such as Pelargonium, Cyclamen, and Petunia as well as in
apple leaves.
Accordingly, the present invention relates to an altered FNS II activity in
plants
and other organisms which may be achieved both by increasing and by decreasing
the naturally occurnng FNS II activity by an introduction of the sequence of
the
present invention. A reduction of the amount of FNS II activity may also be
referred to as down regulation.
In flavone-containing plants, the flavone synthesis may be up or down
regulated or
switched off completely in a targeted manner by means of suitable methods.
This
has several consequences for the plant. Basically, an alteration of the
biosynthetic
pathway of this type has an effect on the whole flavone biosynthesis since
there is
a permanent competition of several enzymes (FNS Ii, FHT, F3'H, F3',5'H) for
the
substrate (flavonones, e.g. naringenin). This means in detail that e.g. an up-
regulation of FNS II reduces the synthesis of flavonoids in a downstream
biosynthetic pathway (Fig. lA) whereby for example a paler flower color may be
achieved. In addition, by this targeted increase in flavone content which may
be
restricted to specific tissues, the resistance properties and the capability
to form
symbioses of the plant may be markedly improved. In contrast, a down-
regulation
results in an increase in the synthesis of other flavonoids which may be more
beneficial for the plant. This may affect both the flavonols important for W
protection and the coloring anthocyans or the resistance-inducing
proanthocyanidines and phytoalexins. A complete suppression of flavone
formation results in an enhancement of the effects of a down-regulation and
may
remove possible stimulants for insects and thus contribute to the resistance
generation.
An entirely new synthesis of flavones may be achieved by the targeted
introduction of FNS II into plants lacking a natural activity. Depending on
the
CA 02370387 2001-10-19
19
substrates present, the flavone content and the flavone pattern may thus be
controlled. By establishing this step, flower colors, resistance properties,
and the
capability to form symbioses with nitrogen-fixing bacteria may be altered in a
targeted manner. Moreover, opening a novel biosynthetic pathway may reduce the
formation of flavonoids which are less beneficial for the plant (e.g. feed
stimulants), and the synthesis of biflavones generated from flavones may be
established.
Using these approaches, the flavonoid pattern and content of important flavone-
containing or t7avone-free agricultural plants may be altered to optimize
their
positive properties with respect to the biology of men and animals.
In addition, the present invention relates to methods for the targeted
alteration of
the flavone content and flavone pattern, respectively, in various plant
tissues
(flowers, roots, leaves etc.) and other organisms, and thus in general it
relates to an
alteration of the flavonoid composition, particularly the alteration of flower
colors
by means of copigmentation, of resistance properties, and of the capability of
nodulation in the case of leguminous plants.
In another embodiment, the present invention relates to the use of a
polypeptide
according to the present invention for the synthesis of flavones. In suitable
expression systems, the enzyme itself and thereby eventually the natural
flavones
may be synthesized starting from appropriate substrates. A possibility is the
use of
suitable expression systems for obtaining health-promoting, natural flavones.
These expression systems may be of plant origin or consist of cell cultures or
yeast
cultures, respectively. The expression of FNS II in plants or in cell cultures
may be
used to directly obtain the respective flavones wherein the yeast expression
system
may be preferred for obtaining the intact enzyme. Using this enzyme, chemical
or
natural precursors (flavanones) may then be reacted to form the corresponding
flavone. The flavones synthesized in this manner may for example find use in
cancer therapy or contribute to human and animal health in the form of
medicaments.
The present invention is explained in detail with respect to the following
Figures
and Examples. The present invention is illustrated with respect to a nucleic
acid
CA 02370387 2001-10-19
20
sequence derived form Gerbera hybrids. It will now be obvious that similar
sequences may be isolated easily from various other sources such as e.g. other
plants or specific microorganisms. Examples of other flavone-producing plants
include but are not restricted to gloxinia (Sinningia hybrids), snapdragon
(Antirrhinium majus), columnea (Columnea hybrids), dahlia (Dahlia variabilis),
gloxinia (Sinningia cardinalis), Streptocarpus (Streptocarpu.r hybridus),
verbena
(Verbena hybrida), chrysanthemum (Chrysanthemum indicum), peace lily
(Spathiphyllum wallisii). All these nucleic acid sequences encoding, directly
or
indirectly, an enzyme of the flavonoid biosynthesis pathway and particularly
FNS
II are encompassed by the present invention disregarding the source thereof.
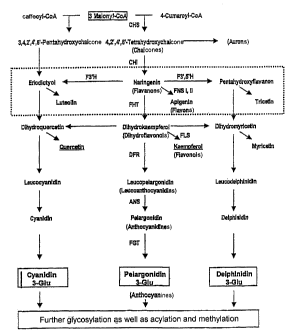
FIGURE lA-C shows a schematic representation of the general flavonoid
biosynthesis pathway and the chemical structures of several flavonoids.
The enzymes involved are abbreviated in Figs. lA and 1B as follows: CHS =
chalcone synthase; CHI = chalcone isomerase; FHT = flavanone 3-hydroxylase;
DFR = dihydroflavonol 4-reductase; ANS = anthocyanidine synthase; FGT =
flavonoid 3-glycosyl transferase; FNS II = flavone synthase II; FLS = flavonol
synthase; F3'H = flavonoid 3'-hydroxylase; F3',5'-H = flavonoid 3',5'-
hydroxylase.
The level of flavone formation is particularly indicated (++-+-+) in Figure
lA. In
the upper portion of Figure 1B the FNS II reaction is shown in the presence of
NADPH and several common flavanones. In the lower portion, other important
flavonoids are shown. Figure 1C describes the flavonoid biosynthesis pathway
as
present in Gerbera hybrids. The following abbreviations have been used for the
different flavonoids: THC = tetrahydroxychalcone; PHC = pentahydroxychalcone;
NAR = naringenin; ERI = eriodictyol; Ap = apigenin; Lu = luteolin; DHK =
dihydrokaempferol; DHQ = dihydroquercetin; Km = kaempferol; Qu = quercetin;
LPg = leucopelargonidin; LCy = leucocyanidin; Pg = pelargonidin; Cy =
cyanidin.
FIGURE 2 shows the activity or the lack of activity, respectively, of FNS II
in
enzyme extract from petals of different gerbera lines: "Th 58" (genotype fns+
fns),
"147-150" (fns+ .), and "147-146" (genotype fns fns). The lines "147-150" and
"147-146" are autogamy progeny of "Th 58". The FNS II activity was measured by
means of the turnover of '4C-labeled naringenin to the corresponding flavone
apigemn.
CA 02370387 2001-10-19
21
FIGURE 3 shows the FNS II activity (~) and the accumulation of flavones (p) in
line "Th 58" (fns+ fns) over different developmental stages of the flower. The
different flower stages are defined in Example 2. The flavone content was
determined by extraction with ethylacetate and HPLC detecaion as described in
Martens and Forkmann (1998).
FIGURE 4A+B (according to Schopfer and Ebel, 1998) shows the known
structure of cytochrome P450 sequences containing regions of high sequence
conservation. The prolin-rich, the oxygen binding and the heme binding regions
are indicated. In Fig. 4B the heme binding region is shown in detail in
addition to
the primers derived therefrom for DD-RT PCR. A set of eight non-degenerated 5'
primers was prepared according to the putative nucleotide sequence.
FIGURE 5 shows a schematic representation of the different cytochrome P450
DNA fragments generated. All clones contain the heme binding site indicated.
pDDd7a: a 358 by fragment could be generated via PCR using the
oligonucleotides "Decamer d7" and "Oligo A" with a DNA template recovered
from a differentially expressed band.
pTABATA: a 1519 by fragment was isolated starting from gerbera "Th 58" cDNA
via a PCR-supported RACE method using oligonucleotides "GSP7", "GSP8",
GSP9", and "AAP" (GIBCO-BRL) or by "backrace", respectively.
pCYPFNS 1: a 1589 by fragment containing an open reading frame was isolated
via PCR using oligonucleotides "CypFNS 1 H" and "CypFNS 1 R". cDNA of
gerbera "Th 58" was used as the template.
FIGURES 6 and 7 are representations of the nucleic acid and the amino acid
sequence derived therefrom, respectively, of the full length clone. The start
codon
and the different stop codons are indicated separately.
Figure 8 shows a diagram of the restriction sites present for standard
restriction
enzymes.
CA 02370387 2001-10-19
22
FIGURE 9 shows a FNS II assay using yeast microsomes. [14C]-naringenin was
used as the substrate. Microsomes were prepared from transformed yeasts
(INV Sc 1 - CypFNS 1 ) and untransformed yeasts (INV Sc 1 ). The
autoradiograph
shows the conversion of [14C]-naringenin to the corresponding flavone, [14C]-
apigenin, using an extract of transformed yeast (INVScl -- CypFNSI). In the
control experiment (INVScl) no activity was measured. The product was
identified by co-chromatography with authentic apigenin in four different
eluents.
FIGURE 10 shows an autoradiography of an RNA gel blot hybridized with a 32P-
labeled cDNA of insert CypFNS 1. Each lane contains 20 ~g of total RNA which
was applied as follows: (1) Simm (genotype fns+ .), (2) Delphi (fns+ .), (3)
T3 (fns
fns), (4) 147-150 (fns+ .), (5) clivia (fns fns), (6) 147-146 (fns fns), (7)
Regina
(fns+ .), (8) gerbera leaves (fns fns), (9) pool of 10 (fns+ .), (10) pool of
10 (fns
fns).
EXAMPLE 1
l~~Tora,-;.~ta
Chemicals, enzymes and radiochemicals
Naringenin, eriodictyol, apigenin, and luteolin were obtained from Carl Roth
(Karlsruhe, Germany). [14C]-naringenin was prepared from ['4C]-malonyl CoA
(ARC, St. Louis, USA) and p-cumaroyl CoA (Dr. Werner Heller, GSF,
Neuherberg, Germany) according to the method described in Britsch et al.
(1981)
using partially purified chalcone synthase (CHS) and chalcone isomerase from
parsley suspension culture. All other enzymes were obtained from commercial
suppliers and used according to their specifications.
Bacterial and yeast strains
The following Escherichia coli strains were used: TOP10F' and TOP10, both from
Invitrogen (Groningen, Netherlands). In addition, the following yeast strain
was
used: INVScl (Invitrogen).
Cloning vectors pCR2.1 and pYES2 were obtained from lnvitrogen.
The ligation of insert and vector pCR2.1 and the transformation of bacteria,
respectively, was carried out according to the instructions of the
manufacturer.
Plant materials
CA 02370387 2001-10-19
23
Chemogenetically defined Gerbera clonal varieties (Tyrach and Horn, 1997) and
autogamy progeny of line "Th 58" were available. In addition, the current cut
gerbera varieties "Regina" of Terra Nigra company (DeKwakel, Netherlands) and
"Delphi" of Florist company (DeKwakel, Netherlands) were included. A detailed
description of the plant material may be found in Table 2.
TABLE 2
Plant material used
Gerbera line Genotype Source
~5g fns ns Tyrach and Horn, 1997
Delphi fns+ fns Florist, Tyrach and Horn,
1997
Sign fnsi fns Tyrach and Horn, 1997
T3 fns &~s T;yrach and Horn, 1997
147-150 fns+ fns Th 58 x S
Clivia fns fns T;yrach and Horn, 1997
147-146 fns fits Th 58 x S
Regina fnsi . Terra Nigra
Regina leaves fns fns Terra Nigra
pool of 10 (147-...)fns' . Th 58 x S
pool of 10 (147_,._?fns fns Th 58 x S
EXAMPLE 2
Plant cultivation, crossing methods and flower stages
Plants of Gerbera hybrids were cultivated in a green house under conditions
common in practice. The day length was at least 14 h at a light intensity of
10,000
lux and at 22°C.
Autogamy of the variety "Th 58" (genotype fns+ fns) was carried out within the
same inflorescence or between inflorescences of the same plant. This autogamy
experiment may be carried out with any variety or line heterozygous for the
Fns
locus. The corresponding chemogenetic and biochemical methods are detailed in
Tyrach and Horn (1997) and Martens and Forkmann (1998). Controlled flowering
was achieved by glassine bags put over the flowers. Ideally, the pollinations
were
repeated up to four times in daily intervals. Further descriptions with
respect to the
methods of pollination and floral morphology in Gerbera may be found in Maurer
( 1967).
CA 02370387 2001-10-19
24
Gerbera flowers were harvested in different developmental stages defined as
follows (according to Martens and Forkmann, 1998):
Stage 1: bud closed, petals smaller than 5 mm;
Stage 2: ray flowers visible, 5-10 mm long;
Stage 3: ray flowers 10-15 mm long;
Stage 4: beginning pigmentation, length 15-23 mm;
Stage 5: ligula of ray flowers pigmented, 23-26 mm long;
Stage 6: ray flowers 26-3 S mm long;
Stage 7: inflorescence half open; 35-40 mm long;
Stage 8: inflorescence completely opened, 40-50 mm long;
Stage 9: ray flowers 50-SS mm long;
Stage 10: ray flowers 55-60 mm long;
Stage 11: senescent inflorescence, SS-60 mm long.
FX A MP1.F ~
Biochemical and enzymolo~ical characterization of the autogamy progeny
Known standard methods (Marbry et al., 1970; Harborne, 19fi7) were used for
the
extraction and identification of flavonoids. Flavones were additionally
detected
under LTV light (243 nm) prior and after vaporization with ammonia. Flavanones
were identified by reduction with sodium borohydride and subsequent treatment
with hydrochloric acid vapors (Eigen et al., 1957).
Thin layer chromatography was carried out on precoated cellulose plates 61440
of
Schleicher & Schull company (Dassel, Germany). For this purpose, the following
eluents were used: (1) chloroform-acetic acid-water (10:9:1); (2) 30% acetic
acid;
(3) acetic acid-hydrochloric acid-water (30:3:10); and (4) tert-butanol-acetic
acid-
water (3:1:1).
The flavone content of buds and flowers during development was determined by
extraction of the pigments from petals of the different stages using
ethylacetate.
The ratio of tissue to extracting agent was 1:40 (g/ml) and the; period of
extraction
was 48 h at 4°C in the dark. Characterization and quantification were
performed
using HPLC. 10 ~1 of 75% methanolic extract were applied to and separated on a
Spherisorb ODS II column (particle size 5 Vim, 250 x 4.6 mm, Bischoff,
Leonberg,
Germany) (Lange et al., 1994). The detection was done using a diode array
detector Model 168 (Beckman, Munich, Germany).
CA 02370387 2001-10-19
25
Enzyme preparations and FNS II assays were performed as described in Martens
and Forkmann (1998). The preparation was carried out in 6.0 ml Tris-HCl buffer
(pH 7.5) containing 28 mmoles/1 2-mercaptoethanol and l0 mmoles/1 sodium
ascorbate at 4°C with 1.0 g of petals, 0.5 g Dowex (equilibrated in
Tris buffer, pH
7.5), and 0.5 g sea sand. After homogenizing in a cooled mortar the homogenate
was transferred into Eppendorf tubes and centrifuged twice for 5 min at 10,000
x
g. The cleared supernatant was used as raw extract or was used for the
precipitation of microsomes with MgCl2 according to Diesperger et al., (1974).
The protein content of the preparations was determined according to the method
of
Bradford (1976).
The standard assay for FNS II contained in a total volume of 200 pl: 175 pl
Tris-
HCl buffer (pH 7.5), 0.3 nmol radiolabeled substrate (83 Bq; naringenin), 2.0
nmol of unlabeled substrate, 10 ul of 20 mmoles/1 NADPH and 15 ~1 raw extract
or microsomal preparation. After an incubation of 20 min at 25°C the
reaction was
stopped by addition of 20 pl methanol containing a mixture of the
corresponding
flavonoids. The extraction of the reaction mixture was carried out twice with
100
and 50 ~l of ethylacetate. The upper phase was chromatographed on cellulose
thin
layer plates in eluent 1 (see above). The radioactivity was localized and
quantified
using a Fuji BAS 1000 Bio-Imaging Analyzer (Fuji Photo Filrn Co., Tokio,
Japan)
and the TINA software package (Raytest, Straubenhardt, Gerniany).
Fig. 2 exemplarily shows the result of the enzyme assays carried out with the
Gerbera . variety "Th 58" and the lines "147-150" and "147-146". The
corresponding genotypes are shown in Table 2.
CA 02370387 2001-10-19
26
EXAMPLE 4
Oli~onucleotide synthesis
The oligonucleotides were synthesized by Metabion company (Martinsried,
Germany). The following oligonucleotides were used (5'-3'):
Oligo A 5'- T(A,C,G)A-3' (SEQ ID NO:3)
:
Oligo C 5'- T(A,C,G)G3'
: (SEQ lD N0:4)
Oligo G 5'-TTfITTTTTT T(A,C,G)G-3' {SEQ ID N0:5)
:
Decamer 5'-CGCCATTTGG-3' (SEQ ID N0:6)
1 :
Decamer 5'-CGCCATTCGG-3'
2 : {SEQ lD N0:7)
Decamer 5'-CGCCCTTTGG-3' (SEQ ID N0:8)
3 :
Decamer 5'-CGCCCTTCGG-3' (SEG1 ID N0:9)
4 :
Decamer 5'-CGCCGTTTGG-3' (SEQ lD N0:10)
5 :
Decainer6 5'-CGCCGTTCGG-3' (SEQ ID N0:11)
:
Decamet 5'-CGCCTTTTGG-3'
7 : (SEQ ID N0:12)
Decamer 5'-CGCCTTTCGG-3' (SEQ ID N0:13)
8 :
GSP7 : 5'-ATCTTCAAAGTGTTTCCTCGTTCC-3' (SEQ ID NO:i4)
GSPB : 5'-AATGGAACACACACAAAATCTACG3' {SEQ ID N0:15)
GSP9 : 5'-TCACCACTGAGAGTTCTCTCATGG-3'
(SEQ ID N0:16)
AAP : 5'-GGCCACGCGTCGACTAGTACGGGIlGGGIIGGGI1G-3'
(SEQ ID No:l7)
Backrace 5'-GCCACGCGTCGACTAGTACG-3' (SEQ ID N0:18)
:
CypFNSIH 5'-CAAAGGATCCCAACACCATGAATACACTGG3' (SEQ lD N0:19)
:
CypFNS1 5'-AGATAGACCGACTGCCATCAAGAAAGG3' (SEQ ID N0:20)
R :
The three
oligomers
and eight
decamers
were synthesized
according
to Schopfer
and Ebel
(1998).
EXAMPLE 5
Cloning of a P450 fragment from Gerbera hybrids
Isolation of total RNA form Gerbera petals
Total RNA from Gerbera petals was isolated according to a method described in
Guiliano et al. (1993) from various defined genotypes (fns+ . or fns frcs;
Tab. 2) of
stages 2-4 in which the FNS II activity increases (Fig. 3). 1.2 g of plant
material
frozen in liquid nitrogen was ground to a fine powder in a cooled mortar and
transferred into a cooled Corex tube. The tissue was homogenized by thorough
vortexing in 3 ml of charged extraction buffer consisting of 4 M guanidinium
thiocyanate, 0.15 M sodium acetate {pH 5.3), 0.2% sodium s~ucosinate and 0.7%
CA 02370387 2001-10-19
27
13-mercaptoethanol and 2.4 ml water-equilibrated phenol (saturated with 0.1 M
citrate buffer, pH 4.3, Sigma, Deisenhofen, Germany). After addition of 0.6 ml
chloroform the thoroughly mixed homogenate was kept on ice for 20 min and then
centrifuged at 15,000 x g (Sorvall RC-SB plus; SS34). The removed upper phase
was added with 1 vol. of isopropanol and incubated for another 60 min on ice.
After centrifugation for 30 min at 15,000 x g (Sorvall, see above) the upper
phase
was discarded and the pellet resuspended in sterile HzO. To remove the
polysaccharides, the solution was added with 100% ethanol (20% (v/v) final
concentration), incubated for 20 min on ice, and centrifuged 10 min at 10,000
x g
and 4°C. The supernatant containing the nucleic acids was added with
1/3 vol. of
8 M lithium chloride. After incubation for 30 min on ice centrifugation for 20
min
at 15,000 x g was earned out and the supernatant discarded. The precipitated
RNA
was washed twice each with 1 ml 80% ethanol and then resuspended in 50 p.l H20
and stored at -70°C. Determination of the RNA concentration was carried
out
spectrophotometrically at a wave length of 260 nm (Pharmacia Biochrome 4060).
If necessary, poly (A)+ RNA was additionally isolated from the total RNA by
two
cycles of oligo (dT) cellulose chromatography (Sambrook et a:L., 1989).
Reverse transcription of RNA
5 ~g of total RNA or 500 ng of poly (A)+ RNA isolated from the different
genotypes or flower stages, respectively, as described above were transcribed
into
cDNA in a 25 pl sample using one of the three oligo (dT) primers each by
reverse
transcriptase (SuperScriptTM II, GIBCO BRL, Paisley, Great Britain). The RNA
was added with 1 ~l of the respective oligo (dT) primer (1 pM final) and water
up
to a volume of 17 pl. The sample was then denatured at 70°C for 10 min
and
afterwards placed on ice. After addition of 4 pl 5 x Superscript first strand
buffer,
2 ~1 100 mM dithiothreitol, 1 pl 10 mM dNTP mix, the sample was preincubated
for 2 min at 42°C. Afterwards, 1 ~l (200 U) of SuperScriptTM reverse
transcriptase
was pipetted directly into the sample. The reaction was incubated for 60 min
at
42°C and then for 15 min at 70°C. This provided the following
cDNAs: fns+ A, C
or G and corespondingly for the recessive line fns- A, C or G. These cDNAs
were
used directly as templates for the PCR sample.
PCR amplification using p450-specific primers
CA 02370387 2001-10-19
28
The amplification of different cDNAs was performed by means of PCR wherein
besides the oligo (dT) anchor primer a second non-degenerated P450-specific
primer (Decamer 1-8) was used. The PCR sample contained the following
components in a total volume of 20 pl: 6.2 ~ l water, 1 pl 20 x polymerase
buffer,
2.5 mM MgCl2, 0.2 pM dNTP mix, 1 pl 35S-ATP [1000 Ci/mmol] (ICN
Pharmaceuticals, Irvine, USA), 0.5 ~M of one of eight decarner primers, 1 pM
of
oligo (dT) primer specific for the cDNA, 4 ~l of the cDNA sample described
above, and 0.2 ~l 5 U/~1 Replitherm polymerase (Epicentre, Madison, USA). The
following PCR parameters were used: denaturation step at 94°C for 10
min, then
40 cycles at 94°C for 30 seconds, 40°C (annealing) for 2 min,
and 72°C
(extension) for 30 sequence, followed by a final extension at 72°C for
7 min. 4 ~l
of the PCR product were then added with 2 pl formamide loading buffer (80%
formamide; 10 mM EDTA (pH 8.0), 1 mg/ml xylencyanol FF and 1 mg/ml
bromophenol Blue, mixed, denatured at 95°C for 2 min, and then placed
directly
on ice. The sample prepared in this manner were loaded to a 5% denaturing
polyacrylamide gel and separated for 3 hours at a maximum of 40 W.
Subsequently, the gel was transferred to Whatman paper and fixed in a gel
dryer.
The radioactivity or differential bands, respectively, were localized using a
Fuji
BAS 1000 Bio-Imaging Analyzer (Fuji) and the TINA software package (Raytest).
Differentially expressed bands at a size of 300 to 500 by were cut together
with
the Whatman paper from the fixed gel using a sharp blade, transferred to an
Eppendorf tube and rehydrated in 100 pl water for 10 min at room temperature.
Subsequently, the tube was incubated at 100°C for 10 min and
centrifuged for 2
min at full speed in a table centrifuge to remove residual gel and paper. The
supernatant was transferred into a new Eppendorf tube. After addition of 10 ~l
3
M sodium acetate, 5 ~l 10 mg/ml glycogen as a earner and 400 pl 100% ethanol
the DNA was precipitated for 60 min at -70°C. The precipitated DNA was
then
pelleted by centrifugation at 4°C and 14,000 rpm, washed twice with 85%
ice-cold
ethanol, dried in air and resuspended in lOpl water. The reamplification of
the
cDNA obtained in this manner was carried out in PCR samples of 50 pl
containing the following components: 27.8 pl water, 2 pl 20 x polymerase
buffer,
4 ~l 25 mM MgCl2, 3.2 ul 500 pM dNTP mix, 4 ~l of S~M of the corresponding
decamer primer, 4 ~1 25 ~M of the corresponding oligo (dT;l primer, 4 ~I of
the
eluted DNA described above and 0.5 ~1 SU/~1 Replitherm pol;ymerase
(Epicentre).
The PCR parameters were the same as in the first amplification. The PCR
CA 02370387 2001-10-19
29
products were separated using a 1.5% agarose gel and the corresponding
amplificates were cloned into vector TOPO pCR2.l and then transformed into
TOPO lOF' one-shot competent cells (Invitrogen) according to the instructions
of
the manufacturer. Plasmid isolation from bacteria identified by means of blue-
white screening was performed using the Plasmid Miniprep Quantum Prep kit
(Bio-Rad, Munich, Germany) according to the instructions of the manufacturer.
The inserts identified after digestion with the appropriate restriction
enzymes (e.g.
Eco RI; Boehringer, Mannheim, Germany) and separation on a 1.5% agarose gel
having a length between 300 and 500 by were eluted from the gel using the
QUIAEX II gel elution kit (QIJIAGEN, Hilden, Germany). These DNA fragments
were labeled with 32P using the Rediprime Labelling kit (Amersham,
Braunschweig, Germany) and used for Northern blot analyses. DNA sequencing of
these and other clone was carried out substantially according to the method
described by Sanger et al. (1977) using Sequenase enzyme Version 2.1
(Amersham, Braunschweig, Germany).
EXAMPLE 6
Northern blot analysis
Total RNA was isolated as described in Example 5. 10 ~g of different total RNA
samples were electrophoresed on a 2.2 M formaldehyde/l.2°,% (w/v)
agarose gel.
The running buffer contained 20 mM MOPS (pH 7.0), S mM sodium acetate and 1
mM EDTA (pH 7.0). The RNA was transferred to Hybond-NX membrane
(Amersham) according to the instructions of the manufacturer and hybridized
with
a 32P-labeled Eco RI-Eco RI pDDd7a cDNA fragment. Prehybridization ( 1 to 3
hours at 42°C) and hybridization (16 to 24 hours at 42°C) were
performed in SO%
deionized formamide, 5 x SSPE, 5 X Denhardt's, 0.5% SDS. Denatured herring
sperm DNA (100 ~g/ml) was added in the hybridization step together with the
32P-
labeled probe. The filters were washed twice for 15 min at 42°C in 2 x
SSPE, 1
SDS (w/v) and then once or twice at 65°C with 1 x SSPE, 1% SDS
(w/v). The
radioactivity was localized and quantified using a Fuji BAS 1000 Bio-Imaging
Analyzer (Fuji) and the TINA software package (Raytest).
Northern blot analyses revealed that the gene corresponding to cDNA clone
pDDd7a is only expressed in lines carrying the fns+ . genotype. In lines
having the
recessive genotype fns fns no hybridization signal could be detected (Fig.
10).
Moreover, the expression pattern over the different flower stages is parallel
to the
CA 02370387 2001-10-19
30
enzyme activity and the flavone accumulation, respectively, measured in the
flowers (Martens and Forkmann, 1998).
EXAMPLE 7
Isolation of a full-length clone of pDDd7a
cDNA clone pDDd7a is no full length clone but covers only the region from the
heme binding site to the poly (A+) end (3' end) of the sequence including
several
stop codons. To obtain the full length clone of pDDd7a a PC'.R-supported RACE
method according to Frohmann et al. (1988) using the 5' RACE system version
2.0
(GibcoBRL) was used.
First, several gene-specific 5'-RACE primers (GSP7-9) on the basis of pDDd7a
and in addition the nested amplification primer "backrace" were constructed.
Using GSP7, the total RNA was transcribed into cDNA according to the method
described in Example 5. Subsequently, the first strand product was purified
from
the excess of nucleotides and GSP7 using the High Pure PCR Product
Purification
kit (Boehringer Mannheim) according to the instructions of the manufacturer.
To
the purified cDNA an oligo-dC tail was added using terminal transferase (TdT).
The tailing sample was as follows: 6.5 ~I water, 5.0 ul 5 x tailing buffer,
2.5 ~l 2
mM dCTP and 10 pl cDNA. This mix was denatured for 3 min at 94°C and
then
placed on ice for 1 min. The reaction was started by addition of 1 pl TdT and
then
incubated for 10 min at 37°C. Inactivation of the enzyme was carried
out by
incubation for 10 min at 65°C. PCR amplification of the dC tailed cDNA
was
performed in 0.5 ml thin-walled PCR tubes according to the following protocol:
single denaturation for 2 min at 94°C, 35 subsequent cycles consisting
of 1 min at
94°C, 57°C for 1 min, and 72°C for 2 min. A final
extension step of 7 min was
also performed. The PCR sample contained the following components: 31.5 ~1
water, 5.0 pl 1 CI x PCR buffer, 3.0 ul 25 mM MgCl2, 1.0 ~l 10 mM dNTPs, 2.0
~.1
10 ~M GSPB, 2.0 pl 10 ~M AAP, 5.0 ~1 dC tailed cDNA and 0.5 ~1 5 U/~l Taq
DNA polymerase (Promega, Madison, USA). 15 pl of 5'-RACE product were
analyzed on a 1.5% agarose gel using an appropriate lenl,~th marker. Specific
single bands in a region of 1.5 kb were cloned and verified by Northern blot
and
sequence analysis, respectively.
Using the 5'-RACE a specific 1.5 kb fragment (pTABATA) was amplified which
hybridizes only with total RNA of gerbera lines having the fns+ . genotype and
not
with RNA from recessive genotypes. Within the region of the gene-specific
primer
CA 02370387 2001-10-19
31
(GSPB) up to the heme binding site, this novel fragment is homologous to
fragment pDDd7a. A full length clone (1698 bp) contains an open reading frame
and shows a homology of 58% on the amino acid level to cytochrome P450 clone
CYP93B 1 (Akashi et al., 1998).
EXAMPLE 8
Expression of pCYPFNS 1 in yeast
Construction of pYeCYPFNS 1
A 1.5 kb Bam HI-Eco RI fragment corresponding to pCYPFNS 1 was ligated into
Bam HI-Eco RI opened yeast expression vector pYES2. The resulting plasmid
referred to as pYeCYPFNS 1 contained the pCYPFNS 1 cDNA fragment in the
sense orientation as the insert.
Yeast transformation
The yeast strain INVScI was transformed with plasmid pYeCYPFNSl following
the protocol according to Gietz et al. (1992). The selection of transformed
yeast
cells was earned out via a complementation marker.
Preparation of yeast microsomes for flavone synthase II assays
Individual colonies of INVScI/pYeCYPFNS1 and INVScl grown on selection
medium SGI (20 g glucose (w/v), 1 g peptone (Fluka), 6.7 g yeast nitrogen base
without amino acids (Difco) and 20 mg L-tryptophane (Fluka) per liter) were
then
inoculated in 5 x 5 ml SGI broth and incubated at 200 rpm and 30°C for
24 hours.
At an ODboo of 0.2-0.4 of a 1:10 dilution of the preculture it was completely
inoculated into 250 ml YPGE (5 g glucose, 10 g peptone., 10 g yeast extract
(Fluka) and 3 vol.% ethanol per liter). The main culture was incubated at
30°C and
120 rpm. At an OD6oo of 0.8 to 1.2 the induction was earned out by addition of
27
ml 200 g/1 sterile galactose solution.
After 12 to 15 hours at an OD6oo of 0.6 to 1.2 of a 1:10 dilution of the main
culture
the yeast cells were harvested by centrifugation, washed once with TEK (50 mM
Tris-HCI, pH 7.4, 1 mM EDTA, and 0.1 M KCI) and resuspended in TES-B* (50
mM Tris-HCI pH 7.4, 1 mM EDTA, 0.6 mM sorbitol, and 2 mM DTT).
Disruption of the yeast cells was performed at 4°C using 15 g glass
beads (Sigma)
per sample. The glass beads were then washed three times with 5 ml TES-B* and
the combined supernatant was adjusted with 4 M NaCI to a final concentration
of
0.15 M. The microsomes were precipitated by addition of 2.5 g PEG-4000 (Fluka)
CA 02370387 2001-10-19
32
and following a washing step with 2 ml TES-B* were homogenized in 2.5 ml
TEG* (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 2 mM DT'T) in a Potter. The
resulting homogenate served as microsomal enzyme source for FNS II assays.
The FNS II activity was measured according to the method of Martens and
Forkmann (1998). The standard assay for flavone synthase II contained in a
total
volume of 200 ~l: 140 ~l Tris-HCl buffer (pH 7.5), 0.3 nmoles of radiolabeled
substrate (83 Bq; [14C]-naringenin), 10 ~l 20 mmoles/1 NADPH and 50 ~1 of the
yeast microsomal preparation. After an incubation for 20 min at 25°C
the reaction
was stopped by addition of 20 ul methanol containing a mixture of naringenin
and apigenin (product). Extraction of the reaction mixture was carned out
twice
with 100 or 50 ~1 ethylacetate, respectively. The upper phase was
chromatographed on cellulose thin layer plates in eluents 1 to 4 (see above).
The
radioactivity was localized and quantified using a Fuji BAS 1000 Bio-Imaging
Analyzer (Fuji) and the TINA software package (Raytest).
The enzyme extract prepared from INVScI/pYeCYPFNSl showed a clear FNS II
activity whereas the corresponding fraction from untransformed yeasts showed
no
activity (Fig. 9). The results of the yeast expression confirm that the cDNA
insert
pCYPFNS 1 encodes an FNS II enzyme. Moreover, the result reveals that
expression of the enzyme encoded by the Gerbera cDNA clone is sufficient in
yeast to achieve a direct formation of flavones. This indicates that only a
single
enzyme is required for introducing the double bond between C2 and C3 (Fig. 1B)
and that the cDNA clone described by Akashi et al., (1998) does not represent
FNS II but rather a flavanone 2-hydroxylase.
CA 02370387 2001-10-19
33
References
AKASHI, T., AOKI, T. and AYABE, S.L, 1998. Identification of a cytochrome
P450 cDNA encoding (25)-flavanone 2-hydroxylase of licorice (Glycyrrhiza
echinata L; Fabaceae) which represents licodione synthase and flavone synthase
II. FEBS Letters 431, 287-290.
ASEN, S. and HOROWITZ, R.M., 1974. Apigenin 4'-O-(3-D-glucoside 7-O-13-D-
glucuronide: the copigment in the blue pigment of Centaurea cyanus.
Phytochemistry 13, 1219-1223.
BRADFORD, M.M., 1976. A rapid and sensitive method for the quantitation of
microgram quantities of protein utilizing the principle of proteindye binding.
Analytical Biochemistry 72, 248-254.
BRITSCH, L., HELLER, W. and GRISEBACH, H., 1981. Conversion of
flavanone to flavone, dihydroflavonol and flavonol with an enzyme system from
cell cultures of parsley. Zeitschrift fur Naturforschung 36c, 742-750.
DIESPERGER, H., MULLER, C.R. and SANDERMANN, H. jr., 1974. Rapid
isolation of a plant microsomal fraction by Mg2+ precipitation. FEBS Letter
43,
15-158.
EIGEN, E., BLITZ, M. and GINSBERG, E., 1957. The detection of some
naturally occuring flavanone compounds on paper chromatograms. Archives of
Biochemistry and Biophysics 68, 1501.
FIRMIN, J.L., WILSON, K.E., ROSSEN, L. and JOHNSTON, W.B., 1986.
Flavonoid activation of nodulation genes in Rhizobium reversed by other
compounds present in plants. Nature 324, 90-92.
FORKMANN, G., 1991. Flavonoids as Flower Pigments: The Formation of the
Natural Spectrum and its Extension by Genetic Engineering. .Plant Breeding
106,
1-26.
FROHMAN, M.A., DUSH, M.K. and MARTIN, G.R., 1988. Rapid production of
full-length cDNAs from rare transcipts: Amplification using a single gene-
specific
oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998-9002.
GIETZ, D., ST. JEAN, A., WOODS, R.A. and ScHIESTL, R,.H., 1992. Improved
method for high efficiency transformation of intact yeast cells. Nucleic Acids
Res.
20, 1425.
GIULIANO, G., BARTLEY, G.E. and SCOLNIK, P.A., 1993. Regulation of
carotenoid biosynthesis during tomato development Plant Cell 5, 379-387.
CA 02370387 2001-10-19
34
GOTO, T. and KONDO, T., 1991. Struktur and molekulare Stapelung von
Anthocyanen -Variation der Bliitenfarben. Angew. Chem. 103, 17-33.
HARBORNE, J.B., 1967. Comparative Biochemistry of the Flavonoids. Academic
Press, New York.
HARBORNE, J.B., 1978. The rare Flavone Isoetin as a Yellow Flower pigment in
Heywoodiella ollgocephala and in other Cichorieae. Phytochemistry 17, 915-917.
HARBORNE, J.B. and GRAYER, R.J., 1994. Flavonoids and Insects. In : The
Flavonoids - Advances in Research since 1986 (Ed.: J.B. HARBORNE).
Chapman & Hall, London, 589-618.
HELLER, W. and FORKMANN, G., 1994. Biosynthesis of Flavonoids. In : The
Flavonoids - Advances in Research since 1986 (Ed.: J.B. HARBORNE).
Chapman & Hall, London, 499-536.
HRAZDINA, G., 1982. Anthocyans. In: The Flavonoids - Advances in Research
(Ed.: J.B. HARBORNE and MABRY, T.J.). Chapman & Hall, London, 135-188.
JACOBS, M. and RUBERY, P.H., 1988. Naturally occuring auxin transport
regulators. Science 241, 346-349.
KUBASEK, W.L., SHIRLEY, B.W., Mc KILLOP A., GOODMAN, H.M.,
BRIGGS, W. and AUSUBEL, F.M., 1992. Regulation of flavonoid biosynthethic
genes in germinating Arabidopsis seedlings. Plant Cell 4, 1229-1236.
LANGE, B.M., TROST, M., HELLER, W., LANGEBARTELS, C. and
SANDERMANN jr, H., 1994. Elicitor-induced formation of free and cell-wall-
bound stilbenes in cell-suspension cultures of Scots pine (Pinus sylvestris
L.).
Planta 194, 143-148.
MARBRY, T.L., MARKHAM, K.R. and THOMAS, M.B., 1970. The Systematic
Identification of Flavonoids. Springer Verlag, Berlin, Heidelberg, New York.
MARTENS, S. and FORKMANN, G., 1998. Genetic Control of Flavone Synthase
II Activity in Flowers of Gerbera Hybrids. Phytochemistry 49,1953-1958.
MAURER, J., 1967. Untersuchung zur Bliitenbiologie an Gerbera jamesonii J.
Bolus. Z. Pflanzenziichtg. 59, 288-303.
MIDDLETON JR, E. and KANDASWAMI, C., 1994. The impact of plant
flavonoids on mammalian biology: implications for immunity, inflammation and
cancer. In : The Flavonoids - Advances in Research since 1986 (Ed.: J.B.
HARBORNE). Chapman & Hall, London, 619-652.
REDMOND, J.W., BATLEY, M., DJORDJEVIC, M.A., INNES, R.W.,
KUEMPEL, P.L and ROLFE, B.G., 1986. Flavones induce expression of
CA 02370387 2001-10-19
35
nodulation genes in Rhizobium. Nature 323, 632-635.
SAMBROOK, J., FRITSCH, E.F. and MANIAT1S, T., 1989. :Molecular cloning: a
laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
SANGER, F., NICKLEN, S. and COULSON, A., 1977. DNA sequencing with
chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463-5467.
SCOTT-MONCRIEFF, R., 1936. A biochemical survey of some mendelian
factors for flower colour. J. Genet. 32, 117-170.
SCHOPFER, C.R. and EBEL, J., 1998. Identification of elicitor-induced
cytocnrome P450s of soybean (Glycine max L.) using differential display of
mRNA. Mol. Gen. Genet. 258, 315-322.
TAYLOR, L.P. and JORGENSEN, R., 1992. Conditional male fertility in
chalcone synthase deficient petunia. J. Heredity 83, 11-17.
TYRACH, A. and HORN, W., 1997. Inheritance of flower colour and flavonoid
pigments in Gerbera. Plant Breeding 116, 377-381.
WILLIAMS, C.A. and HARBORNE, J.B., 1994. Flavone and flavonol glycosides.
In: The Flavonoids - Advances in Research since 1986 (Ed.: J.B. HARBORNE).
Chapman & Hall, London, 337-386.
WOLLENWEBER, E., 1994. Flavone and flavonols. In : The Flavonoids -
Advances in Research since 1986 (Ed.: J.B. HARBORNE). Chapman & Hall,
London, 259-336.
CA 02370387 2001-10-19
36
SEQUENCE LISTING
GENERAL INFORMATION:
APPLICANT:
NAME: Martens, Stefan
STREET: Waldweg 14
TOWN: Eching
COUNTRY: Germany
POSTAL CODE: 83386
APPLICANT:
NAME: Forkmann, Gert
STREET: In der Point 17
TOWN: Tiefenbach
COUNTRY: Germany
POSTAL CODE: 84184
NAME OF THE INVENTION:
Genetic sequence which codes for the flavone synthase
II enzyme and use of the same
NUMBER OF SEQUENCES: 20
COMPUTER READABLE FORM:
DATA MEDIUM: Diskette
COMPUTER: IBM PC-compatible
OPERATING SYSTEM: Windows 3.11
SOFTWARE: Microsoft Word 6.0
INFORMATION FOR SEQ ID-NO:1:
SEQUENCE CHARACTERISTICS:
LENGTH: 1697 base pairs
TYPE: nucleic acid
STRANDEDNESS: double
TOPOLOGY: linear
MOLECULE TYPE: DNA
SEQUENCE DESCRIPTION: SEQ ID-N0:1:
ATGTCCTAAC ACAACCCAAC ACCATGAATA CACTCCAACT CATCTTCCTC
CTCTTCTTCT 60
TCCCAACCTT ACTCTTCCTC TACTGTCTCC CCTACAAAAG AAACCAAAAC
CACCGCCGTC 120
TTCCGCCGTC CCCGCCATCT TTTCCGATCA TCGGCCACCT CCF.CCATCTC
GGCCCACTCA 180
TCCACCAATC CTTCCACGCT CTCTCCACTC GCTACGGCTC TCTAATCCAC
CTCCGTCTCG 240
GCTCAGTCCC ATGCGTCGTC GTTTCAACCC CAGACCTCGC CAAAGACTTC
CTCAAAACAA 300
ACGAACTCGC GTTCTCATCA AGAAAACACT CCTTAGCCAT .CGA.CCACATC
ACCTATGGCG 360
CA 02370387 2001-10-19
37
TAGCATTTGC ATTCGCACCA TATGGAACTT ACTGGAAGTT CATCAAGAAA
CTCTTCACAG 420
TGGAGCTTTT GGGCACCCAG AATCTCAGCC ATTTCCTACC CATTCGAACC
CATGAAATTC 480
GCGAGCTTCT TCGAACGTTA ATGGTGAAAT CTAGGGCAAA GGAGAGAGTA
AACTTGACGG 540
AAGAGTTGTT GAAGTTGACC AACAATGTGA TAAGTCAAAT GATGATGAGC
ATTAGGTGTT 600
CGGGGACGAA TAGTGAGGCT GATGAAGCAA AGAATCTTGT TCGGGAAGTG
ACCAAAATTT 660
TTGGACAGTT TAATGTTTCA GATTTCATAT GGTTTTGTAA GAACATAGAT
TTGCAAGGGT 720
TTAAGAAGAG GTACGAGGGT ACACATAGAA GATATGATGC TT7.'GCTTGAA
AGGATTATAA 780
TGGGGAGGGA AGAAAATAGA AGAAGAGGGA AGATAAAAGA TGGTGAAGGG
AAAGATTTTC B40
TTGATATGTT ACTTGATGTT TTGGAGGATG GTAAGGCAGA GATTAAAATT
ACTAGAGACC 900
ACATCAAAGC CTTGATTTTG GACTTTCTTA CAGCTGGGAC GGATACCACC
GCGATTGCAA 960
TTGAATGGGC ACTAGTCGAA TTGATAAACA ACCCGAACGC TC7.'CGAGAAA
GCAAGACAAG 1020
AGATTGATCA GGTCATCGGT GATGAGAGGC TAGTTCAAGA ATCAGACACG
CCTAACCTCC 1080
CTTATATCCA AGCTATCATA AAGGAAGCCC TACGACTTCA CCC_'ACCAATC
CCAATGTTGA 1140
TTCGCAAGTC AACAGAAAAT GTAATTGTTC AGGGGTATGA CATCCCAGCC
GGCACCTTGT 1200
TGTTTGTCAA TATTTGGTCC ATTGGAAGAA ACCCTCAATG TTGGGAAACC
CCTTTAGAGT 1260
TCAAGCCTCA TCGGTTTTTG GATGGTGGTG ACCTTAAAAG CTCTTTAGAT
ATTAAAGGCC 1320
ACAATTTTCA ACTATTGCCT TTTGGGACGG GGAGGAGAGG GTGTCCTGGT
GTTAATTTGG 1380
CCATGAGAGA ACTCTCAGTG GTGATTGCAA ACCTCATACA ATGCTTTGAT
TGGGATGTTG 1440
TAGGTGAACG ACTATTGAAT ACAGATGAAC GTGCTGGATT GACGGCTCCA
AGGGCGGTAG 1500
ATTTTGTGTG TGTTCCATTG GAACGAGGAA ACACTTTGAA GA'.CTCTTGGT
TCAAACTAAA 1560
CA 02370387 2001-10-19
38
TTTATTTGTT GTTGCTTTCT TGATGGCAGT CGGTCTATCT ATAGGTCATA
ATACCTTGGG 1620
ACTCACGTGT TTGAATCTTA ATACGCTTTT AGTACATTGC TTATCGTATA
TCTTGGGTAT 1680
GCATGAAAAA AAAAAAA
INFORMATION FOR SEQ ID-N0:2:
SEQUENCE CHARACTERISTICS:
LENGTH: 511 amino acids
TYPE: amino acid
TOPOLOGY: linear
MOLECULE TYPE: protein
SEQUENCE DESCRIPTION: SEQ ID-N0:2:
Met Asn Thr Leu Gln Leu Ile Phe Leu Leu Ph<= Phe Phe Pro
Thr Leu
5 10
15
Leu Phe Leu Tyr Cys Leu Pro Tyr Lys Arg Asn Gln Asn His
Arg Arg
20 25 30
Leu Pro Pro Ser Pro Pro Ser Phe Pro Ile Ile: Gly His Leu His
His
35 40 45
Leu Gly Pro Leu Ile His Gln Ser Phe His Ala Leu Ser Thr Arg
Tyr
50 55 60
Gly Ser Leu Ile His Leu Arg Leu Gly Ser Val Pro Cys Val Val
Val
65 70 75
80
CA 2001-10-19
02370387
39
Ser Thr Pro Asp Leu Ala Lys Asp Phe Leu Lys Thr Asn Glu Leu
Ala
85 90 95
Phe Ser Ser Arg Lys His Ser Leu Ala Ile Asp His Ile Thr Tyr
Gly
100 105 110
Val Ala Phe Ala Phe Ala Pro Tyr Gly Thr Tyr Trp Lys Phe Ile
Lys
115 120 125
Lys Leu Phe Thr Val Glu Leu Leu Gly Thr Gln Asn Leu Ser His
Phe
130 135 140
Leu Pro Ile Arg Thr His Glu Ile Arg Glu Leu Leu Arg Thr Leu
Met
145 150 155
160
Val Lys Ser Arg Ala Lys Glu Arg Val Asn Leu Thr Glu Glu Leu
Leu
165 170 175
Lys Leu Thr Asn Asn Val Ile Ser Gln Met Met:Met Ser Ile Arg
Cys
180 185 190
Ser Gly Thr Asn Ser Glu Ala Asp Glu Ala Ly:;Asn Leu Val Arg
Glu
195 200 205
Val Thr Lys Ile Phe Gly Gln Phe Asn Val Ser Asp Phe Ile Trp
Phe
210 215 220
Cys Lys Asn Ile Asp Leu Gln Gly Phe Lys Lys Arg Tyr Glu Gly
Thr
CA 02370387 2001-10-19
40
225 230 235
240
His Arg Arg Tyr Asp Ala Leu Leu Glu Arg Ile Ile Met Gly Arg
Glu
245 250 255
Glu Asn Arg Arg Arg Gly Lys Ile Lys Asp Gly Glu Gly Lys Asp
Phe
260 265 270
Leu Asp Met Leu Leu Asp Val Leu Glu Asp Gly Lys Ala Glu Ile
Lys
275 280 285
Ile Thr Arg Asp His Ile Lys Ala Leu Ile Leu Asp Phe Leu Thr
Ala
290 295 300
Gly Thr Asp Thr Thr Ala Ile Ala Ile Glu Trp Ala Leu Val Glu
Leu
305 310 315
320
Ile Asn Asn Pro Asn Ala Leu Glu Lys Ala Arc3 Gln Glu Ile Asp
Gln
325 330 335
Val Ile Gly Asp Glu Arg Leu Val Gln Glu Ser Asp Thr Pro Asn
Leu
340 345 350
Pro Tyr Ile Gln Ala Ile Ile Lys Glu Ala Leu Arg Leu His Pro
Pro
355 360 365
Ile Pro Met Leu Ile Arg Lys Ser Thr Glu Asn Val Ile Val Gln
Gly
370 375 380
CA 02370387 2001-10-19
41
Tyr Asp IlePro Ala Gly Thr Leu Leu Phe Val Asn Ile Trp Ser
Ile
385 390 395
400
Gly Arg AsnPro Gln Cys Trp Glu Thr Pro Leu Glu Phe Lys Pro
His
405 410 415
Arg Phe LeuAsp Gly Gly Asp Leu Lys Ser Ser Leu Asp Ile Lys
Gly
420 425 430
His Asn PheGln Leu Leu Pro Phe Gly Thr Gly Arg Arg Gly Cys
Pro
435 440 445
Gly Val AsnLeu Ala Met Arg Glu Leu Ser Va1 Val Ile Ala Asn
Leu
450 455 460
Ile Gln CysPhe Asp Trp Asp Val Val Gly Glu Arg Leu Leu Asn
Thr
465 470 475
480
Asp Glu Arg Ala Gly Leu Thr Ala Pro Arg Ala Val Asp Phe Val
Cys
485 490 495
Val Pro Leu Glu Arg Gly Asn Thr Leu Lys Ile Leu Gly Ser Asn
500 505 510
INFORMATION FOR SEQ ID-N0:3:
SEQUENCE CHARACTERISTICS:
LENGTH: 12 base pairs
CA 02370387 2001-10-19
42
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:3:
TTTTTTTTTT NA
12
INFORMATION FOR SEQ ID-N0:4:
SEQUENCE CHARACTERISTICS:
LENGTH: 12 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:4:
TTTTTTTTTT NC
12
INFORMATION FOR SEQ ID-N0:5:
SEQUENCE CHARACTERISTICS:
LENGTH: 12 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:5:
TTTTTTTTTT NG
12
INFORMATION FOR SEQ ID-N0:6:
SEQUENCE CHARACTERISTICS:
LENGTH: 10 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:6:
CGCCATTTGG
10
CA 02370387 2001-10-19
43
INFORMATION FOR SEQ ID-N0:7:
SEQUENCE CHARACTERISTICS:
LENGTH: 10 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:7:
CGCCATTCGG
INFORMATION FOR SEQ ID-NO: B:
SEQUENCE CHARACTERISTICS:
LENGTH: 10 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:8:
CGCCCTTTGG
INFORMATION FOR SEQ ID-N0:9:
SEQUENCE CHARACTERISTICS:
LENGTH: 10 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:9:
CGCCCTTCGG
INFORMATION FOR SEQ ID-NO:10:
SEQUENCE CHARACTERISTICS:
LENGTH: 10 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
i'OPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-NO:10:
_._~.- ~-._ ~__~,
CA 02370387 2001-10-19
44
CGCCGTTTGG
10
INFORMATION FOR SEQ ID-N0:11:
SEQUENCE CHARACTERISTICS:
LENGTH: 10 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-NO:11:
CGCCGTTCGG
10
INFORMATION F'OR SEQ ID-N0:12:
SEQUENCE CHARACTERISTICS:
LENGTH: 10 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:12:
CGCCTTTTGG
10
INFORMATION FOR SEQ ID-N0:13:
SEQUENCE CHARACTERISTICS:
LENGTH: 10 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:13:
CGCCTTTCGG
10
INFORMATION FOR SEQ ID-N0:14:
SEQUENCE CHARACTERISTICS:
LENGTH: 24 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
CA 02370387 2001-10-19
45
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:14:
ATCTTCAAAG TGTTTCCTCG TTCC
24
INFORMATION FOR SEQ ID-N0:15:
SEQUENCE CHARACTERISTICS:
LENGTH: 24 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:15:
AATGGAACAC ACACAAAATC TACO
24
INFORMATION FOR SEQ ID-N0:16:
SEQUENCE CHARACTERISTICS:
LENGTH: 24 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:16:
TCACCACTGA GAGTTCTCTC ATGG
24
INFORMATION FOR SEQ ID-N0:17:
SEQUENCE CHARACTERISTICS:
LENGTH: 36 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:17:
GGCCACGCGT CGACTAGTAC GGGNNGGGNN GGGNNG
36
CHARACTERISTIC:
NAME / KEY: other characteristics
LOCATION:24
OTHER INFORMATION: N is i
CA 02370387 2001-10-19
46
CHARACTERISTIC:
NAME / KEY: other characteristics
LOCATION:25
OTHER INFORMATION: N is i
CHARACTERISTIC:
NAME / KEY: other characteristics
LOCATION:29
OTHER INFORMATION: N is i
MERKMAL:
CHARACTERISTIC:
NAME / KEY: other characteristics
LOCATION:30
OTHER INFORMATION: N is i
CHARACTERISTIC:
NAME / KEY: other characteristics
LOCATION:34
OTHER INFORMATION: N is i
CHARACTERISTIC:
NAME / KEY: other characteristics
LOCATION:35
OTHER INFORMATION: N is i
INFORMATION FOR SEQ ID-N0:18:
SEQUENCE CHARACTERISTICS:
LENGTH: 20 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:18:
GCCACGCGTC GACTAGTACG
20
INFORMATION FOR SEQ ID-N0:19:
SEQUENCE CHARACTERISTICS:
LENGTH: 30 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:19:
CAAAGGATCC CAACACCATG AATACACTCC
30
INFORMATION FOR SEQ ID-N0:20:
CA 02370387 2001-10-19
47
SEQUENCE CHARACTERISTICS:
LENGTH: 27 base pairs
TYPE: nucleic acid
STRANDEDNESS: single
TOPOLOGY: linear
MOLECULE TYPE: synthetic oligonucleotide
SEQUENCE DESCRIPTION: SEQ ID-N0:20:
AGATAGACCG ACTGCCATCA AGAAAGC
27