Note: Descriptions are shown in the official language in which they were submitted.
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
RECEPTOR SELECTIVITY MAPPING
BACKGROUND OF THE INVENTION
The present invention relates generally to a combination of chemoinformatics
and
bioinformatics and data on chemical-molecular target interactions to create
multi-
dimensional databases. More particularly, this invention relates to databases
comprising
chemical compound, molecular target, and biological or clinical information in
which
patterns or relationships of interactions between chemical compounds and
molecular
targets are determined and compared with other information in the database in
order to
draw conclusions that are useful for drug discovery and development and for
related areas.
The worldwide pharmaceutical industry spends more than $30 billion a year on
research and development, of which nearly one-third is spent on the discovery
and early
development phase, which is the period leading up to the selection of a drug
candidate for
preclinical and clinical development. Some critical steps in drug discovery
include (1)
sequencing DNA comprising segments of the human genome; (2) identification of
genes
within the genome that are associated with specific diseases or biological
functions; (3)
production of a protein such as a receptor or enzyme that corresponds to, or
is encoded by,
the functional gene and which then becomes a biological or molecular target
for drug
discovery; (4) screening a library of chemical compounds for activity against
the
molecular target (high throughput screening); (5) screening the most potent
active
compounds against other biological targets (particularly other receptors or
enzymes) to
assess the compounds' selectivity or specificity for the intended
biological/molecular target
and potential to cause undesirable side effects through activity at other
targets; (6)
evaluating the most potent and selective compounds for their activity in a
range of other
assays designed to measure such properties as toxicity, absorption,
distribution,
metabolism, excretion, etc.; (7) assessing the most promising compounds based
on
empirical judgments using the above information, and then sending that
information to a
chemical synthesis group to produce analogs (or modified but related chemical
structures)
of the initial active compounds; (8) retesting the chemical analogs through
Steps (4), (5)
and (6), then repeating Step (7) until an optimized lead compound or series of
compounds
is identified; and (9) forwarding the optimized lead compounds to further
preclinical and
clinical testing.
-1-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
Throughout this process of discovery and development, compounds go through
successively narrower filters, and compounds are eventually selected for the
more
expensive phases of preclinical and clinical development. Unfortunately, the
selection
process often leads to preclinical testing and clinical testing of compounds
that will fail at
these stages and never reach commercialization. These failures lead to
extremely high
average costs, estimated to exceed $300 million, to develop and launch a new
drug. If,
however, the optimal drug candidate is correctly identified early in the
discovery and
development process and successfully passes preclinical and clinical testing,
the actual
cost to develop that drug may be reduced by as much as 75%. Clearly, a major
goal of
pharmaceutical R&D should be to enhance the predictability of early drug
development
tests such as outlined above.
With the revolution of new techniques in biotechnology and the evolution of
tools
to automate many laboratory processes, two dominant trends have emerged in
recent years
that are having an important impact on pharmaceutical R&D. First, the number
of
molecular targets (such as new receptors and enzymes) available for discovery
screening
programs continues to increase dramatically due to progress in sequencing the
human
genome. About 400 molecular taxgets have been explored for drug discovery;
estimates of
the number of potential molecular targets that may be elucidated from the
human genome
project range in the thousands to more than 10,000. Second, the size of
chemical
compound libraries available for discovery screening programs has expanded
nearly ten-
fold (to more than a million compounds in many drug companies) due to
automation and
new technologies such as combinatorial chemistry. These two factors hold
tremendous
promise for new drug discovery, but they also create significant potential
problems having
adverse consequences on the cost of drug development. More targets and more
compounds will result in many more bioactive compounds being discovered,
leading to
greater difficulty in selecting the optimal drug candidates to advance to
preclinical testing,
as well as increased development costs due to more compounds entering
preclinical and
clinical testing and potentially more failures at these stages.
These factors point to an increased need for rapid, inexpensive, in vitro
("test-tube"
or microplate-based) assays for lead compound selection, optimization, and
validation.
Such rapid assays may help identify the most promising of these active
compounds before
-2-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
they enter the later, more expensive, stages of drug development. These
factors further
point to a need for more effective methods to manage and interpret the vast
amount of data
on genes and gene products (molecular targets), chemical structures, and
screening results.
One application of in vitro assays that is gaining increased importance in
pharmaceutical R&D is "profiling." The assignee of this patent application
pioneered the
concept of profiling in the late 1980's. Drug companies are provided with an
extraordinarily broad array of in vitro assays for characterizing the
pharmaceutical activity
and the potential side effects of compounds under development as new drugs.
Currently
there are more than 200 different assays that may be performed on a routine
basis based on
molecular targets, called receptors and enzymes, that play a key role in a
wide range of
human diseases, including those associated with central nervous system
disorders, immune
diseases, pain and inflammation, infectious diseases, cancer, metabolism or
growth factors,
cardiovascular function, and the endocrine system. Pharmaceuticals accounting
for more
than one-half of the worldwide market function by interacting with cellular
receptors. In
addition, many side effects of pharmaceuticals are also mediated through their
interactions
with receptors or enzymes.
Through profiling, a drug company's lead compounds, generally those entering
preclinical development, are tested in a battery of receptor and enzyme
assays.
Information from the profiling process about interactions between the drug
company's
compound and certain receptors is important for the process of lead compound
optimization and selection and can suggest possible side effects or secondary
therapeutic
activities of the compound. This knowledge can potentially save the drug
company
millions of dollars in wasted time and expense during preclinical and/or
clinical
development of the compound.
While profiling services have been practiced for many years, the data
generated
from these tests are generally used empirically by drug companies. Most drugs,
even
highly selective drugs, interact with numerous receptors or other molecular
targets.
Interpreting data produced by profiling, therefore, depends on the experience
and
knowledge of the scientist from the drug company who reviews the data on both
the
chemical structure of the compounds and the binding interactions of the
compounds with
specific receptors. Unfortunately, even the most experienced pharmacologist
has an
-3-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
incomplete knowledge of the interaction of different drug compounds with the
broad range
of receptors relevant to drug development.
The need for more effective methods to manage, collate, interpret, and utilize
the
vast amount of data on genes and gene products (molecular targets), chemical
structures,
and screening results has led to the creation of new opportunities in
bioinformatics and
chemoinformatics, or managing biological and chemical data. The stages of
generating
large pools of information for drug discovery can be broken down into (1) DNA
sequences
(code of genetic material or genes that are blueprints for the cell to make
gene products or
proteins); (2) functional genomics (process of conversion of DNA sequences to
expression
of corresponding gene products or proteins via mRNA production, especially in
response
to drugs or changes in biological function);
(3) proteomics (identification of the amino acid sequence and/or three-
dimensional
structure of gene products or proteins, such as receptors, for which the genes
code);
(4) small molecule pharmacology/toxicology (molecular binding or interactions
between
gene products, like receptors, and small organic chemicals that are potential
drugs); and
(S) chemical structure (of small molecule, drug-like compounds).
Databases for DNA sequences (Group 1) are well established and include
GenBank, The Genome Center, and others. Similarly, databases of chemical
structures
(Group 5) are well known and provided by vendors such as MDL (Isis) and Oxford
Molecular. Databases for proteomics (Group 3), such as SWISS-I'~~a~T, ProLink,
and
PDB, are also being established. Each of these databases can be considered as
one-
component, in that they contain structural information and can be used to
determine
patterns in that one dimension or single component of structural or sequence
information.
Databases for Groups 2 and 4 are not well established, but should be valuable
additions to
the information pool for drug discovery and development. These latter two
forms of
datasets would be two-component or two-dimensional in that they would contain
data
relating to the interaction between two structures, such as genes to proteins
(Group 2) and
proteins to chemicals (Group 4). Such relationship databases add a significant
level of
complexity compared with the one-component databases.
Partial databases or datasets for Group 4 relationships have been or are being
established. For example, profiles of the binding of single compounds against
a broad set
-4-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
of receptor targets by the assignee for its clients is a partial dataset for
Group 4-type
databases. Similarly, data generated through high throughput screening
projects in which
thousands to hundreds of thousands of chemicals, such as might be contained in
a
chemical structure database (Group 5), are screened for activity against a
specific receptor
target (a single point in a Group 3 database), would represent a partial Group
4 database.
Although such partial Group 4 datasets will be helpful aids for drug discovery
and
development, they suffer from two major drawbacks. First, they are directed
toward
specific two-component analyses, such as the binding selectivity of a single
compound or
limited set of compounds across a range of receptors (profile) or of many
compounds at
one receptor target (high throughput screening). In both cases, the breadth of
the dataset is
insufficient to allow statistical correlations to be drawn among a
multiplicity of receptor
targets and a multiplicity of chemical structures. Second, and importantly,
these partial
datasets are being generated on chemical compounds selected for their
structural novelty
and therefore proprietary potential as new drugs. Since these are novel
compounds, there
does not exist any biological information about the activity of these
compounds in animals
or humans. Such approaches therefore suffer the same limitations as the
pharmacologist
trying to empirically interpret the data of a profile, as described above.
SUMMARY OF THE INVENTION
Accordingly, it is an object of the present invention to meet the foregoing
needs by
providing systems and methods for analyzing data relevant to drug discovery
and
development. A full-rank screening database including positive and negative
data
resulting from a large number of chemical compounds being tested against a
large number
of molecular targets is provided. The number of combinations of chemical
compounds
and molecular targets must be large enough such that a person of ordinary
skill in the art of
statistical or other data mining methods can use the screening database
together with the
corresponding chemical compound database and molecular target database to
produce a
reliable prediction of which chemical compounds are suitable for clinical
testing and have
an enhanced probability to be safe and effective drugs.
Specifically, systems and methods for meeting the foregoing needs are
disclosed.
The system includes a computer system comprising a first database containing
records
corresponding to a plurality of chemical compounds and records corresponding
to
-5-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
biological information related to effects of the plurality of chemical
compounds on
biological systems of humans or animals, and a second database containing
records
corresponding to a plurality of molecular targets. The computer system further
comprises
a third database containing records corresponding to tests of binding,
reactivity, or other
interactions between compounds in the first database and molecular targets in
the second
database, the tests including information on the effect that a compound from
the plurality
of compounds in the first database has on the interaction between a selected
compound
(e.g., a reference agent or standard) known to interact with a specific
molecular target from
among the plurality of molecular targets, said tests being performed for a
plurality of the
molecular targets in the second database. Means for setting an interaction
test threshold
corresponding to said effect and means for selecting the compound, sets of
compounds,
and/or information associated with such compounds) when the results of the
testing of the
effect meet the interaction test threshold are also included in the computer
system. A user
interface is provided to allow a user to view and manipulate or analyze
information from
the first database, the second database, and the third database as it relates
to one or more
compound records in the first database and/or as it relates to one or more
molecular target
records in the second database, especially with respect to compounds,
molecular targets, or
other database records associated with results that meet the interaction test
threshold(s).
Furthermore, the invention relates to using methods of statistical analysis
and other
data mining methods as applied to these multidimensional databases to
determine
correlations or patterns that are relevant to drug discovery and development.
Both the foregoing general description and the following detailed description
provide examples and explanations only. They do not restrict the claimed
invention.
DESCRIPTION OF THE DRAWINGS
The accompanying drawings, which are incorporated in and constitute a part of
this
specification, illustrate embodiments of the invention and, together with the
description,
explain the advantages and principles of the invention.
Fig. 1A illustrates a chemical compound table in the receptor selectivity
mapping
database according to one embodiment of the present invention;
-6-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
Fig. 1B illustrates a snap-shot of a chemical compound record containing
spatial
coordinates of a compound in the receptor selectivity mapping database
according to one
embodiment of the present invention;
Fig. 2 illustrates several logical tables that may be used to access the
molecular
target information in the receptor selectivity mapping database according to
one
embodiment of the present invention;
Fig. 3 illustrates a biological information table in the receptor selectivity
mapping
database according to one embodiment of the present invention;
Fig. 4 illustrates the use of a receptor selectivity mapping database as part
of a
screening process according to one embodiment of the present invention;
Fig. 5A illustrates the use of a receptor selectivity mapping database as part
of a
screening process to discover and select new compounds as potential new drug
candidates
for further development;
Fig. 5B illustrates the use of a receptor selectivity database as part of a
screening
process to identify new targets as potential validated targets to use to
discover new drug
candidates for specific disease indications;
Fig. 6A illustrates the use of a database for predicting the drug potential of
a new
compound; and
Fig. 6B illustrates the use of a database to validate the disease relevance
and/or the
biological function of a new molecular target.
DETAILED DESCRIPTION
Reference will now be made to preferred embodiments of this invention,
examples
of which are shown in the accompanying drawings and will be obvious from the
description of the invention. In the drawings, the same reference numbers
represent the
same or similar elements in the different drawings whenever possible.
Systems and methods consistent with the present invention allow the analysis
of
data relevant to drug discovery and development, for example, for predicting
the potential
of a new compound's suitability for progression to preclinical and clinical
tests with an
enhanced probability of becoming a safe or effective new drug. For purposes of
the
following description, the systems and methods consistent with the present
invention are
described with respect to a relational database containing multiple main
tables and with the
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
use of the binding between chemical compounds and molecular targets as a
measurement
of the interactions between the two. The description should also be understood
to apply in
general for any database structure having multiple main components and to the
measurement of any interactions between chemical compounds and molecular
targets.
The present invention relates to the novel design, construction, and
application of a
database relating information-rich chemicals, molecular targets, especially
proteins or
other macromolecules, and biological activity of the chemicals. Furthermore,
the present
invention relates to the primary use of known drugs and drug candidates that
have failed in
clinical or preclinical trials as a source of the chemical library for the
database, together
with preclinical or clinical data generated for such chemicals describing
their side effects,
mechanism of action and other medically relevant data. The present invention
further
relates to determining the binding or other interactions between the chemicals
and the
molecular targets in the database, then using methods of relationship analysis
and data
mining to correlate patterns of these interactions with specific biological
activities that are
relevant to drug discovery and development, or with specific chemical
structures,
substructures, or other features of compounds exhibiting such interactions, or
with
biochemical, structural, or other features of molecular targets exhibiting
such interactions.
Examples of such data mining techniques can be found in the following
references, which
are incorporated by reference in their entirety:
a) Chen et al., Recursive Partitioning Analysis of a Large ~s.r~xcture-
Activity Data
Set Using Three-Dimensional Descriptors, Journal of Chemical Information and
Computer
Sciences, October 1998;
b) Hawkins et al., Analysis of a Large Structure-Activity Data Set Using
Recursive
Partitioning, Quant. Struct.-Act. Relat., 16:296-302 (1997);
c) DePriest et al., 3D-QSAR of angiotensin-converting enzyme and thermolysin
inhibitors; a comparison of CoMFA models based on deduced and experimentally
determined active site geometrics, J. Am. Chem. Soc., 115:5372-84 (1993);
d) Good et al., in Reviews in Computational Chemistry; Lipkowitz, K. B., Boyd,
D.
B. (eds.), VCH, New York, Vol. 7, pp 67-117 (1996);
e) Marshal et al., in Computer Assessed Drug Design; ACS Symposium Scrica
112; American Chemical Society: Washington, DC, 1979; pp 205--226;
_g_
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
f) Moloc et al., A three-dimensional structure activity relationships and
biological
receptor mapping, in Mathematics and Computational Concepts in Chemistry;
Ellis
Horwood; Chichester, 1985; pp 225-251;
g) Mayer et al., A unique geometry of the active site of angiotensin-
converting-
enzyme consistent with structure activity studies, J. Comput. Aided Mol. Des.,
1:3-16.
( 1987);
h) Sheridan et al., The ensemble approach to distance geometry: application to
the
nicotinic pharmacophone, J. Med Chem. 29:899-906 (1986);
i) Martin et al., A fast new approach to pharmacophone mapping and its
application to dopaminergic and benzodiazepine agonists, J. Comput. Aided Mol.
Des.,
7;83-102 (1993);
j) Catalyst/Hypo Tutorial, version 2.0, BioCAD Corp. Mountain View, CA, 1993
k) Sprague, P. W., Automaied chemical hypothesis generation and database
searching with Catalyst, Perspect. Drug Discov. Des., 3:1-20 (1995);
1) Barnum et al. Identification of common functional configurations among
molecules, J. Chem. Inf. Comput. Sci., 1996, 36:563-71 (1996).
m) HipHop Tutorial, version 2.3; Molecular Simulation Inc.; Sunnyvale, CA,
1995;
n) Davies, K. and Upinn, R., 3D pharmacophore searching, Net. Sci.,
(http://www.netsci.org/Science/Cheminform/feature02.htm1);
0) Golender, V. and Vesterman, B., APEX 3D expert system for drug design, Net.
Sci. (http://www.awod.com/netsci/Science/Compchem/feature09.html);
p) Van Drie, J., Strategies for the determination of pharmacophoric 3D
database
queries, J. Comput. Aided Mol. Des., 11:39-52 (1997);
q) Van Drie, J. and Nugent, R., Addressing the challenges posed by combination
chemistry: 3D databases, pharmacophon; recognition and beyond, SAR QSAR
Environ.
Res., 9:1-21 (1998);
r) Finn et al., Pharmacophore discovery using the inductive logic programming
progol, in Machine Learning, Special Issue on Applications and Knowledge
Discovery,
Kluwer Academic Publishers: Boston, 1998, pp 1-33; and
s) Jain et al., Compass: a shape-based machine learning tool for drug design.
J.
Comput. Aided Mol. Des., 8:635-52 (1994).
-9-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
The background section suggests that, contrary to standard operating
procedures in
the pharmaceutical industry, a Group 4 database should be established having
more
components than a two-component database, and that it should cover a
substantial breadth
of both receptor or enzyme targets and chemical compounds. By way of example,
a three-
s component database would be created by first selecting a broad set of
chemical compounds
that are rich in information of direct relevance to drug discovery and
development. The
most relevant information is often obtained by actual experience of testing
such chemical
compounds in humans through clinical trials and/or post-marketing surveillance
or in
animals through preclinical testing. Other relevant biological information may
come from
natural products that demonstrate one or more observed bioactivities, as well
as chemical
reference standards that have been used in the industry to characterize the
biology of
receptors. Accordingly, one embodiment of information-rich chemical compounds
selected for such a Group 4 database includes marketed pharmaceuticals, drugs
that have
failed in clinical or preclinical trials, bioactive natural products or
natural extracts, and
reference agents used for receptor binding assays.
One may construct such a database using screening data obtained from the
scientific literature. While this approach could yield partial datasets, it
may have
limitations. First, literature references generally provide only positive
information (e.g.,
reports of inhibition of binding of a specific compound to a specific
receptor) and not
negative data (e.g., a lack of inhibition of binding and therefore lack of
activity). In
determining useful comparisons of information, negative data can be as
valuable as
positive data. Furthermore, certain statistical analyses may not be applicable
to datasets
that lack completeness of both positive and negative data. Second, separate
quantitative
reports of binding data for one compound against a receptor in one article vs.
reports of
binding data for a second compound at the same receptor may not be comparable
because
of variations in the way the assays were performed. Therefore, one embodiment
for
creation of a Group 4 three-component database would be to screen a broad
array of
compounds through a broad array of receptor or enzyme targets in order to
obtain
consistent comparative results and ensure the collection of both positive and
negative data.
The Chemical Compound Component: Selection of Chemical
-10-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
Libraries and Inclusion of Chemical Data
The present invention relates to databases that contain, as one component,
chemical
compounds about which information is known concerning biological activity
relevant to
pharmaceutical research and development. The biological activity information
may be
included in the chemical compound database or table.
For example, these information-rich chemicals include:
(a) Compounds that are pharmacological reference agents or reference standards
for measuring the interaction or molecular binding between unknown chemical
compounds
and a specific molecular target, such as a receptor or enzyme. Examples of
such reference
compounds include those compounds that are used for characterizing binding
interactions
between test compounds and molecular targets including receptors or enzymes.
Other
reference agents could include chemicals selected from the catalog of Research
Biochemicals Inc. (RBI), a unit of Sigma Aldrich Corp., and from other sources
that are
well known in the industry. These pharmacological reference compounds often
have been
tested previously and/or marketed as pharmaceuticals or are natural products
with
characterized biological activity and therefore may overlap with compounds in
the
following three categories;
(b) Compounds that are known pharmaceuticals that are currently or have
previously been marketed for clinical use, and for which there is a
substantial amount of
biological information available. These compounds are well-known and are
listed in
publications available from U.S. government agencies such as the Food and Drug
Administration (FDA), as well as publications by private or non-profit
organizations. One
such publication by a non-profit organization is the United States
Pharmacopeial
Convention Inc.'s USP DI Series, including Volume I. Drug Information for the
Health
Care Professional, which is updated monthly by USP DI Update. As new drugs are
approved for marketing, they would be included in this category. Marketed
pharmaceuticals or drugs approved by the FDA or equivalent foreign regulatory
bodies are
a matter of public record so that one normally skilled in the art can easily
identify chemical
compounds that would be included in this category;
(c) Compounds that have been approved for testing in humans, such as compounds
that had been granted IND (Investigational New Drug) status, as potential
drugs but that
-11-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
failed to achieve sufficient efficacy or safety in clinical trials to gain
approval from the
FDA or otherwise did not reach the status of marketed pharmaceuticals.
Compounds in
this category may also include those compounds that have been approved by the
FDA for
commercialization but that have later been withdrawn from the market. These
compounds
also would have a significant amount of biological information available and
would be
especially useful for purposes of this invention. The identity of failed drugs
can be
obtained from numerous sources, including public announcements by drug and
biotechnology companies, publications such as the "Pink Sheets," and lists
maintained by
the FDA; and
(d) Compounds that are obtained from natural sources such as plants,
microorganisms, animals, etc., that exhibit biological activity. These natural
products may
include toxins, antimicrobial agents, behavioral modifiers, defensive agents,
and other
categories of compounds that provide information relevant to drug discovery
and
development. The identity of natural products can be found in numerous
publications,
including but not limited to, the RBI catalog and Sigma Aldrich catalog of
chemical
compounds.
For each compound included in the database, chemical structure, chemical
formulae, physical-chemical characteristics, chemical space coordinates or
other chemical
structure descriptors (e.g., Smiles codes), solubility, and other relevant
data, to the extent
such information is available, are entered into fields in the databas~:~.
Those skilled in the
art would recognize other parameters that might be included. Chemicals can be
organized
by chemical structure relatedness in the database or by other relationships.
Fig. 1A illustrates a chemical compound table 300 in a relational database
system.
The table 300 lists a number of chemical compounds and includes records (rows
1-N) of a
number of compounds N. For each compound there may be a number of
corresponding
columns 301-307 containing information related to the compound. For example,
in Fig.
1A column 301 contains the name of the compound; column 302 includes the
compound
type (e.g., compounds that have been approved for testing in humans, etc.);
column 303
includes information related to the chemical structure, for example, a
hyperlink that brings
up a screen containing a drawing of the structure (see snap-shot 310 in Fig. 1
B); column
304 includes the chemical formula for the compound; column 305 includes
information
-12-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
about the physical-chemical characteristics of the compound; column 306
includes
chemical space coordinates of the compound; and column 307 includes solubility
information of the compound.
Additional columns may be added in order to include other relevant data
related to
each chemical compound 301 listed in the table 300. These additional columns
may
include biological activity of the compound, rendering the chemical compound
database a
two component database (see also database 500).
Fig. 1 B illustrates a snapshot 310 that may include information corresponding
to a
record in the table 300. For example, the chemical formula 304 of a compound
may be
included in the snapshot of the record as well as the compound's structure
303.
The Molecular Target Component: Selection of Receptors, Enzymes, and Other
Molecular Targets and Inclusion of Molecular Target Data
Molecular targets such as receptors, enzymes, other proteins, nucleic acids,
carbohydrates, and other macromolecules relevant to drug discovery and
development, are
representative of the second component of the databases comprising this
invention. In one
embodiment of this invention, receptors and enzymes are the principal
molecular targets.
Receptors mediate much of the molecular communication among cells and organs
in the
body. Enzymes often amplify such communications through, for example,
secondary
messenger systems and cell signaling pathways.
Receptors include classical families of receptors such as dopamine receptors,
serotonin receptors, opiate receptors, muscarinic receptors, adrenergic
receptors, adenosine
receptors, etc. These receptor groups include subtypes of the receptor type
(such as
dopamine-1, dopamine-2, dopamine-3, dopamine-4, and dopamine-5 receptors).
Certain
subtypes have further variations (such as dopamine 4.2, dopamine 4.4, and
dopamine 4.7)
or can have different forms (such as dopamine 2 short and dopamine 2 long).
Splice
variants of receptors can also occur, as can mutations in the genes encoding
specific
receptors which might lead to a subset of a population that has a receptor
with slightly
different binding affinity for drugs or other compounds compared with the
normal receptor
type. Receptors can be grouped by family, superfamily, or subfamily. Some
groupings
include G-Protein Coupled Receptors, 7 transmembrane receptors, nuclear
receptors, etc.
-13-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
Receptors can be grouped by the degree of homology of the DNA sequence of
their
corresponding genes. Receptors can also be grouped by their amino acid
sequence and
related three-dimensional conformations. Receptors can be classified by their
location of
expression in tissues or across different cell types.
Enzymes can include proteases, carbohydrases, kinases, phosphatases, DNA-
modifying enzymes, transferases, P450's, and others known to those skilled in
the art.
Other receptors, receptor sources, and corresponding assays are constantly
being
developed by the assignee to be added to the content of the database.
Additional receptors
and receptor assays are well known to those skilled in the art. Lists and
descriptions of
certain receptors relevant to drug discovery and development can be found in
numerous
publications known to those skilled in the art. These publications include the
RBI
Handbook of Receptor Classification and the IUPHAR receptor classification
book.
Furthermore, as new receptors and receptor subtypes are discovered, they can
be added to
the content of the database.
Enzymes and enzyme assays are well known to those skilled in the art. Lists
and
descriptions of certain receptors relevant to drug discovery and development
can be found
in numerous publications known to those skilled in the art.
Fig. 2 illustrates tables 400, 410, and 420 forming part of a relational
database
system which may be used to access molecular target information. Table 400
lists the
targets and includes records (rows 1-M) of a number of targets M. Column 401
lists the
names of the target, while column 402 specifies the target type corresponding
to each
target name.
Table structures may vary according to the target type specified in column
402.
Table 410 includes information about those targets listed in table 400 which
are classified
as receptors. Records from table 410 may be accessed by querying the database
for a
particular receptor name. The receptor names found in table 410 may be
accessed, in turn,
by querying table 400 for those target names for which column 402 reads
"Receptor."
In table 410, column 411 contains the name of the receptor, which is also the
name
of the target in column 401 in table 400; column 412 includes receptor family
information;
column 413 includes receptor superfamily information; column 414 includes
receptor
subfamily information; column 415 includes the information about the degree of
-14-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
homology of the DNA sequence of corresponding genes; and column 416 includes
information on amino acid sequence. The amino acid sequence is one of a number
of
molecular descriptors that may be included in the database. Other molecular
descriptors
417, for example, could include hydropathy plots corresponding to the amino
acid
sequence. Because the molecular target database represented by tables 400,
410, and 420
includes target information and associated biological information related to
the targets is
included in the database (see table 600), this database may be considered a
two-component
database. The columns shown are illustrative of the types of information that
may be
included in the database and should not be construed as limiting the
invention.
Table 420 includes information about those targets in table 400 that are
classified
as enzymes. Records from table 420 may be accessed by querying the database
for a
particular enzyme name. The enzyme names found in table 420 may be accessed,
in turn,
by querying table 400 for those target names for which the target type column
402 reads
"Enzyme."
In table 420, column 421 contains the name of the enzyme, which is also the
name
of the target in column 401 of table 400 and column 422 includes enzyme type
information. Column 423 is labeled as "Other relevant information" and is
included in the
table for purposes of illustrating that additional columns may be added to
table 420
depending on other enzyme information that a user of the database might want
to access,
including amino acid sequence and molecular descriptors.
Although only tables 410 and 420 are shown to describe the access of molecular
target information by using the target type, additional tables may be added to
the relational
database system corresponding to the number of molecular target types
available in the
database.
The Biological Information Component: Selection of
Biolo~ical/Clinical Information Parameters
Biological information forming part of the database includes material that
would
relate to side effects, mechanism of drug action, metabolism of a drug,
toxicity, adsorption,
distribution, and excretion, for example. This information is available on FDA-
approved
labels of marketed drugs, or from literature sources and publications for
drugs that have
-15-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
failed in clinical trials. Examples of some specific parameters are toxicity,
LDso,
LDSO/EDso, teratogenicity, mechanism of toxicity, target organ for toxicity,
in vitro toxicity
battery, induction of apoptosis, bioavailability, absorption, blood-brain
barrier, oral
absorption, mucosal absorption, % absorbed, distribution, blood protein bound,
half life,
onset of action, duration of action, peak concentration in blood, metabolism,
major
pathway, minor pathway, active metabolites, excretion, primary excretion mode,
secondary excretion modes, in vivo effects, therapeutic indication, animal
behavioral
effects, side effects, primary known target, other organ/system targets, and
known receptor
interactions.
Fig. 3 shows table 500 which includes some of the biological information
parameters mentioned above. Table 500 comprises N rows (1 through N) which
correspond to all the possible chemical compounds in the first database.
Column 501
includes the compound name; column 502 includes the therapeutic indication
(for
marketed or failed drugs); column 503 includes toxicity information; column
504 includes
side effects information; and column 505 includes information on the mechanism
of drug
action. Table 500 would be associated with table 300, for example, to form a
two
component chemical compound and biological activity table.
Fig. 3 also shows table 600, which includes biological information parameters
associated with the molecular targets in the database. Table 600 includes P
rows (1
through P) which correspond to all the possible targets in the seco~u
database. Column
601 includes the target name; column 602 includes the therapeutic indication
(for marketed
or failed drugs); column 603 includes toxicity information; and column 604
includes side
effects information. Similarly, table 600 would be associated with table 400,
for example,
to form a two-component molecular target and biological activity table. Tables
500 and
600 together may be a full-rank database (e.g., including all possible
combinations
between compounds and molecular targets in a relational database system)
including
molecular target information, chemical compound information, and biological
activity
information associated with each of the molecular targets and with each of the
chemical
compounds, and may be considered a multidimensional database. Additional
columns
may be included in tables 500 and 600 without departing from the invention.
-16-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
Determining Binding Information
A key feature of this invention is the establishment of several components of
information which, by way of illustration, comprise chemicals, molecular
targets, and
biological information, and measuring the binding, reactivity or other
interactions between
the chemicals and molecular targets. This binding or reactivity information
can then be
related back to the known biological information in order to distinguish
patterns and
relationships that can be used for drug discovery and development. An
important aspect of
this invention is to generate broad and consistent binding or reactivity data
between the
chemicals and molecular targets in order to provide as complete a dataset as
possible in
order to be able to identify relevant patterns or relationships and to provide
both positive
and negative binding or reactivity information for the datasets. In one
embodiment, the
binding data is established as a numerical descriptor that either satisfies or
does not satisfy
a threshold set, for example, for a specific molecular target or set of
molecular targets.
The numerical descriptor may relate to the activity or lack of activity for
each compound
and each receptor or other molecular target measured at a concentration deemed
near the
appropriate threshold for relevance to the biological system or biological
information set.
For example, chemicals can be tested at 10-5 M (10 micromolar) for their
ability to inhibit
binding at a threshold of 30% between a receptor and its specific reference
compound.
Other initial concentrations or percentage inhibition thresholds can be
selected. Also, in
one embodiment, those chemicals that demonstrate inhibition of binding above
the
threshold in the initial yes/no testing are further tested for the potency of
the binding
inhibition. These active chemicals are tested at a series of concentrations
that might, for
example, include tests at 7-14 different concentrations within the range of 10-
5 to 10-9 M,
such that an ICSO and/or Ki value can be determined for the active compound at
the specific
receptor. Fewer or more concentrations may be used for such determinations and
concentrations above or below 10-5 to 10-9 M may be required. These data then
yield a
matrix of relative degree of activity or relative potency for each active
compound at each
molecular target.
In order to generate these screening data, chemicals are first solubilized in
a
suitable solvent system, such as 4% DMSO, although other concentrations of
DMSO and
other solvents are also acceptable. These chemical stock solutions are then
diluted to the
-17-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
appropriate concentration and made available as repositories. For each assay
measuring
the interactions between the chemical and molecular target, the reagents and
protocols for
the assay will vary. Each such assay needs to be characterized and routinely
established
for consistency. Appropriate controls need to be run each time the assay is
performed.
Any assay format that can generate the desired type and accuracy of
information can be
used. Numerous assay detection systems, such as radioactive labels,
fluorescence,
fluorescence polarization, time-resolved fluorescence, fluorescence
correlation
spectroscopy, chemiluminescence, UV absorption, colorimetric, etc., can be
used.
In one embodiment, a receptor-binding assay or enzyme activity assay is used
to
generate data on molecular interactions. As an example, for a receptor binding
assay,
chemicals from a repository are tested for their ability to inhibit the
binding interaction
between the receptor and a reference agent selected for that receptor. The
receptor may be
derived from a tissue source, such as animal or human tissue, or from a cell
line expressing
the receptor, or from a transfected cell line containing the gene for the
receptor. The
receptor source is prepared for the assays, for example, by preparing a
membrane fraction
containing the receptor. Alternatively, the receptor may be partially
purified. The
reference compound, or ligand, is preferably selected for its potent and/or
specific binding
to the specific receptor and may have a radioactive tracer such as Iodine-125
or tritium or
carbon-14 or other marker to enable a bound ligand to be distinguished from an
unbound
ligand. Coincident with testing the chemicals for binding data to include in
the database,
positive and negative controls are run, as is a reference curve with varying
concentrations
of the reference (radio)ligand to ensure the quality of the assay run.
A plurality of methods and systems may measure the interactions between
targets
and compounds as would be recognized by a person of ordinary skill. The
radioligand,
receptor preparation, and test compounds are incubated together for an
appropriate time, in
an appropriate buffer, and at an appropriate temperature, often with the
objective of
reaching equilibrium of the binding reactions. The amount of bound versus
unbound
radioligand is determined by a separation step, such as filtration, or by use
of a method,
such as SPA (scintillation proximity assay), and measured by liquid
scintillation or gamma
counting. The amount of specific binding of the test compound is then
determined by
-18-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
comparing assay results for the test chemicals) vs. the positive and negative
controls. The
per cent inhibition of the test chemicals) is calculated from these data.
Fig. 4 shows table 200 as an illustration of a screening results and assay
database in
which, for example, chemical compounds included in database 300 (comprising 1
to N
chemical compounds) are tested for their effect against molecular targets
included in
database 400. Numerous forms of table 200 are possible. For example, in table
210
screening results are entered in a "yes" or "no" entry with respect to whether
the screening
result for each of a plurality of chemical compounds tested against each of a
plurality of
molecular targets was above or below the selected threshold test result for
each set of
determinations.
As another example, in table 220 screening results are entered as a numerical
descriptor identifying the potency or magnitude of the binding or other effect
(e.g., the Ki
for chemical:receptor interactions) for each of a plurality of chemical
compounds tested
against each of a plurality of molecular targets. In a preferred embodiment,
all such matrix
points for chemicals x targets in tables 210 and 220 are determined and
entered into the
database such that a full-rank dataset is derived. The screening results and
assay database
200 may also include other measurements of chemicalaarget interactions,
including raw
data of screening results and measurements derived from the raw data, assay
protocols and
performance characteristics, and other relevant information.
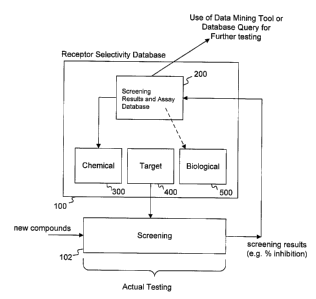
Figs. 5A and SB illustrate the use of a database 100, here shown as a receptor
selectivity database, by way of example, as part of a screening process to
discover and
select new compounds as potential new drug candidates for further development
(Fig. 5A)
or new targets as potential validated targets to use to discover new drug
candidates for
specific disease indications (Fig. 5B). The database 100 may include a
chemical
compound component 300; a molecular target component 400; biological
information
components 500 and 600; and a screening results and assay database 200.
A new compound or set of compounds is introduced to a screening process 102
for
determining whether it is effective in inhibiting the binding of a specific
chemical
compound (e.g., a reference agent) and a molecular target (see Fig. 5A). The
screening
process may use target information from the molecular target component 400.
-19-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
The results of the screening process 102 may be stored in an intermediate
database
or entered into the screening results and assay database 200 of the receptor
selectivity
database 100. The results may also be stored in the biological information
database 500 as
particular parameters (e.g., cytotoxicity, etc.) as well as in the chemical
compound
database 300 (e.g., name of the compound, etc.).
The complete set of results from the screening process 102 may be stored in
the
screening results and assay database 200. The database 200 may be queried for
those new
compounds that exhibit an inhibitory effect on the binding of molecular
targets and
chemical compounds (e.g., reference agent) so that those new compounds can
further be
tested.
Alternatively, a new molecular target, such as, for example, an "orphan"
receptor
about which the structure is known but the function or disease relevance is
not known, is
introduced to a screening process to be to be tested against the chemical
compounds in the
chemical compound database 300 (see Fig. 5B). Results of the screening
process,
including identification of chemicals that interacted with the new molecular
target, are
incorporated into the screening results database 200. Queries are made within
database
100 to determine further steps to identify the function of the new molecular
target and/or
validate the disease relevance of the new target.
Fig. 6A illustrates the use of the database 100 for predicting the drug
potential of a
new compound. A table 710 relies on information from the chemi~;al compound
(300),
molecular target (400), biological information (500 and 600), and screening
results (200)
databases. The table 710 is filled in with information from one or more of
these databases
(or tables) by executing an automatic query script to retrieve the information
once a user
provides the database 100 with information about a new chemical compound.
The query script used for the creation of table 710 may select chemical
compounds
from the chemical compound database 300 upon receiving the new compound
information.
The selection may be based on similar characteristics, such as chemical
structure or other
properties, between the new compound and the compounds already included in the
database 300.
After the selection of chemical compounds, the query script selects targets
from the
target database 400 that are known to react (e.g., bind) with the selected
compounds.
-20-
CA 02371093 2001-10-26
WO 00/65421 PCT/US00/11073
Finally, the combination of selected chemical compounds and selected molecular
targets
may be used for querying the biological information databases 500 and 600 and
inserting
biological information corresponding to chemical compound-molecular target
pairings into
table 710. Alternatively, the user may enter a specific biological information
category of
interest (e.g., toxicity) so that the biological information included in table
710 is limited to
that category.
The table 710 may be queried by the user to produce information relevant to
the
predictability of the potential use of the new compound as a drug. An example
of this
would be a query of the molecular targets known to react with chemical
compounds
associated with the new compound, and the known side effects produced by the
chemical
compounds when combined with the retrieved targets.
Fig. 6B illustrates the use of the database 100 to validate the disease
relevance
and/or the biological function of a new molecular target using an approach
similar to that
used to predict the drug potential of a new compound, but with the data inputs
and queries
shown in Fig. 6B.
All patent, patent applications, and publications mentioned are incorporated
by
reference in their entirety into this application.
The foregoing description of embodiments of the present invention provides an
exemplary illustration and description, but is not intended to be exhaustive
or to limit the
invention to the precise form disclosed. Modifications and variations are
possible in light
of the above teachings or may be acquired from practice of the invention.
-21-