Note: Descriptions are shown in the official language in which they were submitted.
CA 02375320 2012-01-25
-1-
DNA vaccines for pets or for animals used in sports
The present invention relates to improved DNA vaccines
for pets or for animals used in sports, in particular
dogs, cats and horses.
The use of deoxyribonucleic acid (DNA) molecules for
vaccination has been known since the beginning of the
1990s (Wolf et al. Science 1990. 247. 1465-1468) . This
vaccination technique induces cellular and humoral
immunity after in vivo transfection of cells of the
subject to be vaccinated with DNA or RNA molecules
encoding and expressing immunologically active
proteins.
A DNA vaccine is composed of at least one plasmid which
may be expressed by the cellular machinery of the
subject to be vaccinated and of a pharmaceutically
acceptable vehicle or excipient. The nucleotide
sequence of this plasmid encodes and expresses, inter
alia, one or more immunogens, such as proteins or
glycoproteins capable of inducing, in the subject to be
vaccinated, a cellular immune response (mobilization of
the T lymphocytes) and a humoral immune response
(stimulation of the production of antibodies specifi-
cally directed against the imr(iunogen) (Davis H.L.
Current Opinion Biotech. 1997. 8. 635-640).
All the immunogens derived from a pathogen are not
antigens which are naturally sufficiently effective for
inducing an optimum protective immune response in the
animal to be vaccinated. It is therefore necessary to
improve the immune response.
Each route of administration has its own constraints
and difficulties; thus, a DNA vaccine which is effec-
tive via one route of administration may be ineffective
via another.
CA 02375320 2012-01-25
-2-
The choice of the route of administration should take
into account the needs of practitioners and breeders,
the difficulties linked to restraining the animals or
to the nature of the product.
Although the intramuscular route can be used, the
subcutaneous route is of great interest for the
vaccination of pets, especially for animals which are
small in size and which are difficult to handle.
DNA vaccines must therefore be improved in order to
allow their effective administration via various
routes.
DNA vaccines have already been used experimentally, in
particular a DNA vaccine encoding haemagglutinin (HA)
of the measles virus (Etchart et al. J. Gen. Virol.
1997. 78. 1577-1580) whose intranasal administration to
mice proved to be more effective than an oral adminis-
tration. Another example is a DNA vaccine encoding the
envelope (Env) protein of the human immunodeficiency
virus (HIV) whose subcutaneous administration has not
been effective- compared to the administration by the
intramuscular route (Ishii et al. AIDS Res. Hum. Retro.
1997. 13. 1421-1428).
DNA vaccines have also been used experimentally against
animal viruses, in particular against the canine
distemper viruses (CDV). Some CDV immunogens are known,
in particular the nucleocapsid protein (N), the matrix
protein (M), the fusion protein (F) and haemagglutinin
(HA) (WO-A-9741236). However, the subcutaneous adminis-
tration of a DNA vaccine encoding haemagglutinin and
the fusion protein of CDV did not make it possible to
detect the production of antibodies in mice, and
allowed only a small production of antibodies after the
intramuscular administration of this DNA vaccine (Sixt
et al. J. Virol. 1998. 72. 8472-8476).
CA 02375320 2012-01-25
- 3 -
The induction of an immune response and the relation-
ships between the various components of the immune
system coming into play during this response may be
different from one animal species to another. The many
teachings taken from experiments carried out on the
mouse model have made it possible to better understand
the functioning of the immune system in mice, but these
teachings are not directly transposable to other
species, in particular because it is easier to induce
an immune response in mice than in the other species
(van Drunen Little-van den Hurk et al. J. Gen. Virol.
1998. 79. 831-839; Bohm et al. Vaccine 1998. 16.
949-954).
Various routes of administration of the DNA vaccine
have been proposed (intraperitoneal, intravenous,
intramuscular, subcutaneous, intradermal, mucosal, and
the like). Various means of administration have also
been proposed, in particular gold particles coated with
DNA and projected so as to penetrate into the cells of
the skin of the subject to be vaccinated (Tang et al.
Nature 1992. 356. 152-154) and the liquid jet injectors
which make it possible to transfect both skin cells and
cells of the underlying tissues (Furth et al.
Analytical Bioch. 1992. 205. 365-368).
Chemical compounds have been used for the in vitro
transfection of DNA:
A/ - cationic lipids.
The cationic lipids are themselves divided into
four subgroups.
1) The cationic lipids containing quaternary ammonium
salts, such as for example DOTMA (dioleoyloxypropyl-
trimethylammonium, produced by Gibco under the name
Lipofectine), DOTAP (trimethyl-2,3-(octadec-9-eneoyl-
oxy)-l-propaneammonium; Gregoriadis et al. FEBS Letters
CA 02375320 2012-01-25
- 4 -
1997. 402. 107-110), DMRIE (N-(2-hydroxyethyl)-
N,N-dimethyl-2,3-bis(tetradecyloxy)-1-propaneammonium;
WO-A-9634109), DLRIE (N-(2-hydroxyethyl)-N,N-dimethyl-
2, 3-bis(dodecyloxy)-1-propaneammonium; Feigner et al.
Ann. N Y Acad. Sci. 1995. 772. 126-139).
These cationic lipids containing quaternary ammonium
salts may be combined or otherwise with an additional
neutral lipid, such as DOPC (dioleoylphosphatidyl-
choline) or DOPE (dioleoylphosphatidylethanolamine)
(J.P. Behr, Bioconjugate Chemistry 1994. 5. 382-389).
2) The lipoamines, such as for example DOGS (diocta-
decylamidoglycylspermine, produced by Promega under the
name Transfectam; Abdallah et al. Biol. Cell. 1995. 85.
1-7), DC-Chol (dimethylaminoethane-carbamoyl-
cholesterol; Gao and Huang, Biochem. Biophys. Res.
Commun. 1991. 179. 280-285), BGSC (bis-guanidine-
spermidine-cholesterol), BGTC (bis-guanidine-tren-
cholesterol) (Vigneron et al. Proc. Natl. Acad. Sci.
USA 1996. 93. 9682-9686).
3) The cationic lipids containing quaternary ammonium
salts and lipoamines, such as for example DOSPA
(N,N-dimethyl-N-(2-(sperminecarboxamido)ethyl)-2,3-bis-
(dioleoyloxy)-1-propaneimidium, pentahydrochloride,
marketed by Gibco under the name LipofectAmine(D;
Hawley-Nelson et al. Focus 1993. 15. 73-79), GAP-DLRIE
(N-(3-aminopropyl)-N,N-dimethyl-2,3-bis(dodecyloxy)-
1-propaneaminium; Wheeler et al. Proc. Natl. Acad. Sci.
USA 1996. 93. 11454-11459; Norman et al. Vaccine 1997.
15. 801-803).
4) The lipids containing amidine salts, such as for
example ADPDE, ADODE (Ruysschaert et al. Biochem.
Biophys. Res. Commun. 1994. 203. 1622-1628).
B/ - the polymers, such as for example
SuperFectT"' (molecules of activated dendrimers, produced
CA 02375320 2012-01-25
- 5 -
by Qiagen; Xu et al. Mol. Genet. Metab. 1998. 64.
193-197), and
C/ - the biochemical agents, such as for
example toxins, in particular cholera toxins.
Some of these compounds have also been used in the
formulation of DNA vaccines with more than mitigated
results. Knowledge in the field of in vitro trans-
fection is not transposable to DNA vaccination where
the final objective is to ensure a protective immune
reaction. Negative effects on the induction of an
effective immune protection have even been observed
with compounds known to promote transfection in vitro.
Some formulation chemical compounds are toxic at high
doses for the transfected cells.
In the work by Etchart already cited (Etchart et al. J.
Gen. Virol. 1997. 78. 1577-1580), the use of DOTAP did
not have an adjuvant effect during the administration
of the DNA vaccine by the intranasal route, whereas it
had an adjuvant effect by the oral route. DOTAP has
also been used in DNA vaccines encoding the influenza
virus haemagglutinin (HA) on the mouse model which were
administered by the intranasal route (Ban et al.
Vaccine 1997. 15. 811-813), but the addition of DOTAP
inhibited the immune response. The use of DC-Chol or of
DOTAP/DOPE in DNA vaccines encoding the hepatitis B
virus surface protein (S) on the mouse model which were
administered by the intramuscular route made it
possible to increase the antibody response, whereas the
use of Lipofectine (or DOTMA) did not increase this
response (Gregoriadis et al. FEES Letters 1997. 402.
107-110). DC-Chol/DOPE has also been used in DNA
vaccines against the human immunodeficiency virus (HIV,
Env protein) on the mouse model, whose administration
by the intramuscular route induced a more effective
immune response, whereas the administration by the
subcutaneous or intradermal route did not increase it
CA 02375320 2012-01-25
- 6 -
(Ishii et al. AIDS Res. Hum. Retro. 1997. 13.
1421-1428).
Likewise, W0-A-98 40499 proposes preparing nucleic acid
+ cationic lipid complexes for transfecting the mucosal
epithelium in mammals, for gene therapy or the expres-
sion of an antigen intended to induce an immune
response. This document targets the mucosal route, by
inhalation, for example the pulmonary epithelium. It
specifies that its results differ from previous
studies. It also adds that for the intramuscular
(parenteral) route, naked DNA is more effective than a
DNA + lipid mixture.
The addition of certain cytokines, in particular inter-
leukins or interferons, can make it possible to enhance
the immune response induced in particular by DNA
vaccines. Each cytokine triggers a reaction which is
specific to it and orients the immune response to a
greater or lesser degree towards a cellular response or
towards a humoral response (Pasquini et al. Immunol.
Cell. Biol. 1997. 75. 397-401; Kim et al. J. Interferon
Cytokine Res. 1999. 19. 77-84). The adjuvant effects of
a cytokine obtained from a given species are not
necessarily the same if the immune context varies, in
particular if this cytokine is administered to another
species, therefore in a heterologous immune system. The
addition of cytokine may also have no adjuvant effect,
or may even result in a reversal of the effect sought,
that is to 'say a reduction or an inhibition of the
immune response. Thus, a DNA vaccine encoding a single
chain of an immunoglobulin fused with GM-CSF does not
increase the immune response, whereas direct adminis-
tration of this fusion protein to mice is effective, in
the same way as is the administration of a fusion
protein consisting of Fv and of the cytokine IL-lbeta
or the administration of a DNA vaccine encoding the
latter fusion protein (Hakim et al_ J. Immunol. 1996.
157. 5503-5511). The use of plasmids co-expressing the
CA 02375320 2012-01-25
7 -
cytokine IL-2 and the hepatitis B virus envelope
protein in a fused or nonfused conformation results in
an increase in the humoral and cellular immune
responses (Chow et al. J. Virol. 1997. 71. 169-78).
However, the use of a bicistronic plasmid encoding the
human acquired immunodeficiency virus (HIV-1) glyco-
protein gpl20 and the cytokine IL-2 induced a lower
specific anti-gpl20 immune response than that obtained
by the use of a monocistronic plasmid encoding only
gpl20 (Barouch et al. J. Immunol 1998. 161. 1875-1882).
The co-injection, into mice, of two expression vectors,
one coding for the rabies virus G glycoprotein, the
other for murine GM-CSF stimulates the activity of the
B and T lymphocytes, whereas the co-injection with a
plasmid encoding gamma-interferon (in place of murine
GM-CSF) results in a decrease in the immune response
(Xiang et al. Immunity 1995. 2. 129-135).
Certain modifications in the antigens, such as
deletions of part of the nucleotide sequence encoding
the antigen, insertions of a DNA fragment into the
nuclectide sequence encoding the antigen or into non-
translated regions upstream or downstream, can also
enhance the efficacy of DNA vaccines, in particular by
enhancing the level of expression of the antigen or its
presentation.
However in practice, manipulations on the nucleotide
sequence encoding the antigen may bring about a
reduction or loss of the initial immunological
activity. Thus, the deletion of the transmembrane
domain from the gene encoding the rabies virus G
antigen reduced the level of protection induced in the
mouse model after administration by the intramuscular
route of a DNA vaccine encoding this modified antigen
(Xiang et al. Virol. 1995. 209. 569). The deletion of
the transmembrane domain from the gene encoding the
bovine herpesvirus (BHV) gD glycoprotein need not make
it possible to increase the antibody response and
CA 02375320 2012-01-25
-8-
induced only a partial protection in bovines vaccinated by the intramuscular
route (van
Drunen Little-van dcn Hurk et aL J. Gen. Virol. 1998. 79. 831-839). The
humoral and
cellular immune responses and the protection conferred are identical in guinea
pigs
challenged after having been immunized with the aid of either a DNA vaccine
encoding
the Ebola virus GP glycoprotein, or of a DNA vaccine encoding this GP
glycoprotein
but in a secreted form (Xu et al. Nature Medicine 1998.4.37-42).
The insertion of the signal sequence of the human tissue plasminogen activator
(tPA)
into the gene encoding the malaria PB32 antigen did not make it possible to
increase
the antibody response in mice vaccinated by the intramuscular route (Haddad et
al.
FEMS 1997. 18. 193-202). The addition, in phase, of a tPA sequence to the gene
encoding the murine rotavirus VP7 antigen also did not make it possible to
increase the
antibody response in mice vaccinated by the intradermal route, whereas the
fusion
protein consisting of the VP4 antigen and tPA allowed this increase, but
without
16 inducing an effective protection (Choi et al. Virology 1998, 250. 230-240).
The modifications carried out on the nucleotide sequence of one antigen cannot
in
general be directly transposed to another antigen, because antigens do not
always have
the same structural arrangements.
The invention desirably provides the enhancement of the efficacy of DNA
vaccination.
This is in particular to obtain a better immune response and in particular an
effective
protection in pets and animals used in sports, in particular in dogs, cats and
horses, by
DNA vaccination, for various routes of administration, and in particular for
the
subcutaneous route,
The invention desirably provides the production of improved DNA vaccines which
induce an effective and protective immune response against the canine
distemper virus
(CDV), the virus for the respiratory complex or kennel cough (parainfluenza-2
or CPI-2
virus), canine hcrpesvirus (CHV-1) in dogs.
The invention desirably provides the production of improved DNA vaccines which
induce an effective and protective immune response against the feline herpes-
virus
(MV-1) in cats.
CA 02375320 2012-01-25
-9-
The invention desirably provides the production of improved DNA vaccines which
induce an effective and protective immune response against the equine
herpesviuus type
1 (EHV-1), the equine herpesvirus type 4 (EHV-4) in horses.
The invention desirably provides the production of improved DNA vaccines which
make it possible to obtain an effective protection in dogs, comprising at
least one
valency selected from the group consisting of the CDV virus, the CPI-2 virus,
the
CHV-1 virus, the rabies virus (rhabdovirus), the canine parvovirus (CPV), the
canine
coronavirus (CCV) and Borrelta burgdorferi.
The invention desirably provides the production of improved DNA vaccines which
make it possible to obtain an effective protection in cats comprising at least
one
valency selected from the group consisting of the feline herpes-virus (FHV-1),
the feline
calicivirus (FCV), the rabies virus (rhabdovirus), the feline parvovirus
(FPV), the feline
infectious peritonitis virus (FIPV), the feline leukemia virus (FeLV), and the
feline
acquired immunodeficiency syndrome virus (FIV).
The invention desirably provides the production of improved DNA vaccines which
make it possible to obtain an effective protection in horses, comprising at
least
CA 02375320 2012-01-25
- 10 -
one valency selected from the group consisting of the
equine herpesvirus type 1 (EHV-1), the equine herpes-
virus type 4 (EHV-4), the equine influenza virus, the
Eastern equine encephalitis virus, the Western equine
encephalitis virus, the Venezuelan equine encephalitis
virus, the rabies virus, Clostridium tetani, and
Borrelia burgdorferi.
The subject of the invention is improved DNA vaccines
which make it possible to obtain an effective
protection against at least one pathogen which infects
pets or animals used in sports, in particular dogs,
cats and horses. The DNA vaccine is improved: either by
its formulation, or by the addition of GM-CSF, or by
the optimization of the antigen(s), or by combinations
of these propositions.
Preferably, the DNA vaccine is improved by its formula-
tion, and optionally either by the addition of GM-CSF,
or by the optimization of the antigen(s), or finally by
the addition of GM-CSF and by the optimization of the
antigen(s).
By definition,- the DNA vaccine comprises, as active
ingredient, a plasmid encoding and expressing a gene or
gene fragment. The term plasmid covers a DNA trans-
cription unit comprising a polynucleotide sequence
comprising the sequence of the gene to be expressed and
the elements necessary for its expression in vivo. The
circular plasmid form, supercoiled or otherwise, is
preferred. The linear form also falls within the scope
of this invention.
Each plasmid comprises a promoter capable of ensuring,
in the host cells, the expression of the gene inserted
under its control. It is in general a strong eukaryotic
promoter and in particular a cytomegalovirus early
promoter CMV-IE, of human or murine origin, or option-
ally of other origin such as rat or guinea pig. More
CA 02375320 2012-01-25
- 11 -
generally, the promoter is either of viral origin or of
cellular origin. As a viral promoter other than CMV-IE,
there may be mentioned the SV40 virus early or late
promoter or the Rous Sarcoma virus LTR promoter. It may
also be a promoter from the virus from which the gene
is derived, for example the promoter specific to the
gene. As cellular promoter, there may be mentioned the
promoter of a cytoskeleton gene, such as for example
the desmin promoter, or alternatively the actin
promoter. When several genes are present in the same
plasmid, they may be provided in the same transcription
unit or in two different units.
According to a first mode, the DNA vaccines according
to the invention are formulated by adding, as adjuvant,
cationic lipids containing a quaternary ammonium salt,
in particular DMRIE, preferably combined with a neutral
lipid, in particular DOPE, to preferably form
DMRIE-DOPE.
The subject of the present invention is therefore a DNA
vaccine against at least one pathogen affecting pets
and animals used in sports, in particular dogs, cats or
horses, comprising at least one plasmid containing at
least one nucleotide sequence encoding an immunogen of
a pathogen of the animal species considered, under
conditions allowing the in vivo expression of this
sequence, and a cationic lipid containing a quaternary
ammonium salt, of formula:
CH3
R,-O-CHZ-CH-CHZ-N(9 R2-X
I
OR, CH3
in which R1 is a saturated or unsaturated, linear
aliphatic radical having 12 to 18 carbon atoms, R2 is
another aliphatic radical containing 2 or 3 carbon
CA 02375320 2012-01-25
- 12 -
atoms, and X is a hydroxyl or amine group.
Preferably, it is DMRIE (N-(2-hydroxyethyl)-
N,N-dimethyl-2,3-bis(tetradecyloxy)-1-propaneammonium;
WO-A-9634109), preferably combined with a neutral
lipid, in particular DOPE (dioleoylphosphatidylethanol-
amine), to form DMRIE-DOPE.
Preferably, the recombinant vector is mixed with this
adjuvant immediately before use and it is preferable,
before its administration to the animal, to allow the
mixture thus prepared to form a complex, for example
for a period ranging from 10 to 60 minutes, in particu-
lar of the order of 30 minutes.
When DOPE is present, the DMRIE:DOPE molar ratio
preferably ranges from 95:5 to 5:95, and is more
particularly 1:1.
The plasmid:DMRIE or DMRIE-DOPE adjuvant weight ratio
may range in particular from 50:1 to 1:10, in
particular from 10:1 to 1:5, preferably from 1:1 to
1:2.
According to a second mode, GM-CSF (granulocyte
macrophage-colony stimulating factor; Clark S.C. et al.
Science 1987. 230. 1229; Grant S.M. et al. Drugs 1992.
53. 516) is added to the vaccines according to the
invention; this may be carried out by incorporating
GM-CSF protein directly into the vaccinal composition
or preferably by inserting the sequence encoding GM-CSF
into an expression vector under conditions allowing its
expression in vivo. As expression vector, the use of a
plasmid, e.g. the plasmid containing the nucleotide
sequence encoding the antigen(s) of interest or another
plasmid, is preferred. The choice of GM-CSF is made
according to the animal species to be vaccinated; thus,
for dogs, canine GM-CSF is used; for cats, it is feline
GM-CSF, and equine GM-CSF for horses.
CA 02375320 2012-01-25
- 13 -
According to a third mode, the nucleotide sequence(s)
encoding the immunogen are in an optimized form.
Optimization is understood to mean any modification of
the nucleotide sequence, in particular which manifests
itself at least by a higher level of expression of this
nucleotide sequence, by an increase in the stability of
the messenger RNA encoding this antigen, by the
triggered secretion of this antigen into the extra-
cellular medium, and having as direct or indirect
consequence an increase in the immune response induced.
In the present invention, the optimization of the
antigen of interest preferably consists in the deletion
of the fragment of the nucleotide sequence encoding the
transmembrane domain of the antigen of interest
(deletion is understood to mean the complete deletion
or a partial deletion sufficient for the transmembrane
domain to no longer, or no longer substantially, be
functional), and/or in the addition, in phase, of a
nucleotide sequence encoding the tPA (tissue plasmino-
gen activator; Montgomery et al. Cell. Mol. Biol. 1997.
43. 285-292; Harris et al. Mol. Biol. Med 1986. 3.
279-292) signal, and/or in the insertion of a stabiliz-
ing intron upstream of the gene to be expressed. The
deletion of the DNA fragment encoding the transmembrane
domain of the antigen of interest promotes the secre-
tion, into the extracellular medium, of the antigens
thus truncated and thus increases the possibilities of
their coming into contact with the cells of the immune
system. The insertion of the nucleotide sequence
encoding the tPA signal facilitates the translatability
of the messenger RNA to which the tPA signal is joined,
and thus increases the level of expression of this
messenger RNA and therefore the production of antigens.
The tPA signal also plays a role in the secretion of
the antigen synthesized. The insertion of a stabilizing
intron into the gene encoding the antigen of interest
avoids the aberrant splicings of its messenger RNA and
CA 02375320 2012-01-25
- 14 -
maintains the physical integrity of the latter.
Preferably, the tPA signal is of human origin. The
nucleotide sequence of the human tPA signal is
accessible from the GenBank database under the
accession number NM_000930. Preferably, the intron is
intron II of the rabbit beta-globin gene (van Ooyen et
al. Science 1979. 206. 337-344), whose nucleotide
sequence is accessible from the GenBank database under
the accession number V00882 and designated by a
reference under intron No. 2.
The subject of the present invention is an improved DNA
vaccine capable of inducing an effective and protective
immune response in dogs against canine distemper
(Canine Distemper Virus or CDV).
The canine. distemper virus is a Morbillivirus, a member
of the Paramyxoviridae family. This virus infects the
canine species, but also wild-type felines (Harder et
al. J. Gen. Virol. 1996. 77. 397-405).
The present invention makes it possible to obtain an
effective and 'protective DNA vaccine against canine
distemper in dogs, in particular by the subcutaneous
route which had remained up until now a route inducing
a marginal level of protection (Sixt et al. J. Virol.
1998. 72. 8472-8476).
According to the invention, the DNA vaccine against CDV
is preferably improved by its formulation with an
adjuvant according to the invention, in particular
DMRIE, preferably DMRIE-DOPE. Optionally, this may be
combined either with the addition of canine GM-CSF
(Nash et al. Blood 1991. 78. 50-56), or the optimiza-
tion of at least one CDV antigen, or finally the
addition of canine GM-CSF and the optimization of at
least one CDV antigen.
CA 02375320 2012-01-25
- 15 -
A nucleotide sequence encoding canine GM-CSF is acces-
sible from the GenBank database under the accession
number S49738.
The addition of canine GM-CSF may be carried out by the
incorporation of the canine GM-CSF polypeptide into the
vaccinal composition or preferably by the insertion of
the nucleotide sequence encoding the canine GM-CSF into
an in vivo expression vector, preferably a plasmid.
Preferably, the nucleotide sequence encoding canine
GM-CSF is inserted into a second expression plasmid,
different from that (or those) into which the gene(s)
encoding the CDV antigen(s) is(are) inserted.
The optimization of the antigens derived from CDV is
carried out by substitution, by a "signal" sequence, in
particular that of the tPA signal of human origin
(GenBank accession number NM000930), of the sequence
of the signal peptide of haemagglutinin (HA) and/or of
the fusion protein (F), and/or by the deletion of the
DNA fragment encoding the transmembrane domain of HA
and/or of F, and/or by the insertion of an intron, in
particular of intron II of the rabbit beta-globin gene
(whose nucleotide sequence, noted intron No. 2, is
accessible from the GenBank database under the acces-
sion number V00882) upstream of the nucleotide sequence
encoding HA and/or F. The DNA vaccine against CDV
according to the invention can therefore encode and
express a single optimized CDV antigen (HA or F) or
both, that is to say optimized HA and optimized F.
Optionally, the sequence encoding the CDV matrix
protein (M) in its native form (without modification)
and/or the nucleotide sequence encoding the CDV nucleo-
protein (N) in its native form (without modification)
may also be inserted and expressed in a plasmid and
combined with the plasmids containing optimized HA
and/or optimized F.
CA 02375320 2012-01-25
- 16 -
Nucleotide sequences encoding the CDV antigens which
can be used in the present invention and various
constructs of expression vectors are illustrated in the
accompanying examples and in WO-A-9803199, in
particular in Examples 8 and 9 and Figures 2 and 3.
Preferably, according to the invention, the DNA vaccine
against CDV for administration by the intramuscular
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid (e.g. pNS024, Figure 4) encoding
the CDV HA antigen optimized by replacement of the HA
signal sequence with the human tPA signal peptide
sequence, by deletion of the fragment of the nucleotide
sequence encoding the transmembrane domain of HA and by
insertion of intron II of the rabbit beta-globin gene
upstream of the HA gene, and of a second expression
plasmid (e.g. pNS021, Figure 3) encoding the CDV F
antigen optimized by the deletion of the fragment of
the nucleotide sequence encoding the transmembrane
domain and by the insertion of intron II of the rabbit
beta-globin gene upstream of the F gene.
Preferably, according to the invention, the DNA vaccine
against CDV for administration by the subcutaneous
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid encoding canine GM-CSF and the
two plasmids previously defined (e.g. pNS024 and
pNS021).
The subject of the present invention is also an
improved DNA vaccine capable of inducing an effective
and protective immune response in dogs against the
respiratory complex or kennel cough (canine para-
influenza-2 or CPI-2 virus).
The CPI-2 virus is a Paramyxovirus, also a member of
the Paramyxoviridae family (Bittle et al. J. Am. Vet.
Med. Assoc. 1970. 156. 1771-1773; Moloney et al. Aust
Vet J. 1985. 62. 285-286).
CA 02375320 2012-01-25
- 17 -
The DNA vaccine against CPI-2 is preferably formulated
with an adjuvant according to the invention, in
particular DMRIE, preferably DMRIE-DOPE. This may be
optionally combined with either the addition of canine
GM-CSF, or the optimization of at least one CPI-2
antigen, or finally the addition of canine GM-CSF and
the optimization of at least one CPI-2 antigen.
The addition of canine GM-CSF may be carried out as is
described for CDV.
The optimization of the antigens derived from CPI-2 is
carried out by substitution, by a "signal" sequence, in
particular that of the tPA of human origin, of the
signal sequence of the haemagglutinin-neuraminidase
(HN) of CPI-2 and/or of the fusion protein (F) of
CPI-2, and by the deletion of the DNA fragment encoding
the transmembrane domain of HN and/or of F, and by the
insertion of an intron, in particular intron II of the
rabbit beta-globin gene upstream of the nucleotide
sequence encoding FIN and/or F. The DNA vaccine against
CPI-2 according to the invention may therefore encode
and express a single optimized CPI-2 antigen (HN or F)
or both (HN and F).
Nucleotide sequences encoding the CPI-2 antigens which
can be used in the present invention and various
expression vector constructs are given in the accom-
panying examples.
Preferably, according to the invention, the DNA vaccine
against CPI-2 for administration by the intramuscular
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid (e.g. pSB034, Figure 6) encoding
the HN antigen of CPI-2 optimized by the insertion of
the signal sequence of the human tPA in place of the
signal sequence of HN, by the deletion of the fragment
of the nucleotide sequence of HN encoding the trans-
CA 02375320 2012-01-25
- 18 -
membrane domain and by the insertion of intron II of
ream~++~~ o vf F and of a
1,110 Lci1JL1L beta-globin gene up$.,a. c
second expression plasmid (e.g. pSB032, Figure 5)
encoding the F antigen of CPI-2 optimized by the
deletion of the fragment of the nucleotide sequence
encoding the transmembrane domain of F and by the
insertion of intron II of the rabbit beta-globin gene
upstream of F.
Preferably, according to the invention, the DNA vaccine
against CPI-2 for administration by the subcutaneous
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid encoding canine GM-CSF, and of
the two plasmids previously defined (e.g. pSB034 and
pSB032).
The subject of the present invention is also an
improved DNA vaccine capable of inducing an effective
and protective immune response in dogs against the
canine herpesvirus type 1 (CHV-1).
The CHV-1 virus is a member of the Alphaherpesvirinae
family. This virus is responsible for canine viral
rhino trachei ti s'. Nucleotide sequences encoding the gB,
gC and gD glycoproteins are known (Limbach et al. J.
Gen. Virol. 1994. 75. 2029-2039).
The DNA vaccine against CHV-1 is preferably formulated
with an adjuvant according to the invention, in
particular DMRIE, preferably DMRIE-DOPE. This may be
optionally combined with either the addition of canine
GM-CSF, or the optimization of at least one CHV-1
antigen, or finally the addition of canine GM-CSF and
the optimization of at least one CHV-1 antigen.
The addition of canine GM-CSF may be carried out as is
described for CDV.
The optimization of the antigens derived from CHV-1 is
CA 02375320 2012-01-25
- 19 -
carried out by the deletion of the DNA fragment encod-
:a.iay uac a..a uaa~aucuwa uaa~ uvau. ,. ii: vi ~. y., y^a,'cvj..a
and/or of the gC glycoprotein and/or of the gD glyco-
protein of CHV-1. The improved DNA vaccine against
CHV-1 according to the invention may therefore encode
and express a single optimized CHV-1 antigen (gB, gC or
gD) or two of them or the three.
Nucleotide sequences encoding the CHV-1 antigens which
can be used in the present invention and various
expression vector constructs are given in the accom-
panying examples and in WO-A-98/03199, in particular in
Examples 7 and 8, and in Figures 13 and 14.
Preferably, according to the invention, the DNA vaccine
against CHV-1 for administration by the intramuscular
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid (e.g. pSBO16, Figure 7) encoding
the CHV-1 gB antigen optimized by the deletion of the
fragment of the nucleotide sequence encoding the
transmembrane domain, of a second expression plasmid
(e.g. pSBO19, Figure 8) encoding the CHV-1 gC antigen
optimized by the deletion of the fragment of the
nucleotide sequence encoding the transmembrane domain
and of a third expression plasmid (e.g. pSBO17,
Figure 9) encoding the CHV-1 gD antigen optimized by
the deletion of the fragment of the nucleotide sequence
encoding the transmembrane domain.
Preferably, according to the invention, the DNA vaccine
against CHV-1 for administration by the subcutaneous
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid encoding canine GM-CSF, and the
three plasmids previously defined (e.g. pSBO16, pSBO19
and pSBO17).
The subject of the present invention is an improved DNA
vaccine capable of inducing an effective and protective
immune response in cats against the feline herpesvirus
CA 02375320 2012-01-25
- 20 -
type 1 (FHV-1).
The FHV-1 virus is a member of the Alphaherpesvirinae
family, this virus is responsible for feline viral
rhinotracheitis (Fargeaud et al. Arch. Virol. 1984. 80.
69-82).
The DNA vaccine against FHV-l is preferably formulated
with an adjuvant according to the invention, in
particular DMRIE, preferably DMRIE-DOPE. This may be
optionally combined either with the addition of feline
GM-CSF, or the optimization of at least one FHV-1
antigen, or finally the addition of feline GM-CSF and
the optimization of at least one FHV-1 antigen.
The addition of feline GM-CSF may be carried out by the
incorporation of the feline GM-CSF polypeptide into the
vaccinal composition or by the insertion of a
nucleotide sequence encoding the feline GM-CSF (e.g.
accessible from the GenBank database under the
accession number AF053007) into an in vivo expression
vector, preferably a plasmid. Preferably, the nucleo-
tide sequence encoding feline GM-CSF is inserted into a
second expression plasmid, different from that (or
those) into which the gene(s) encoding the FHV-1
antigen(s) is(are) inserted.
The optimization of the antigens derived from FHV-l is
carried out by the deletion of the DNA fragment
encoding the transmembrane domains of the gB glyco-
protein and/or of the gC glycoprotein and/or of the gD
glycoprotein of FHV-1. The improved DNA vaccine against
FHV-1 according to the invention may therefore encode
and express a single optimized FHV-1 antigen (gB, gC or
gD) or two of them or the three.
Nucleotide sequences encoding the FHV-1 antigens which
can be used in the present invention and various
expression vector constructs are given in the accom-
CA 02375320 2012-01-25
- 21 -
panying examples and in WO-A-98/03660, in particular in
Examples 14 and i5 and in Figures 11 and 12.
Preferably, according to the invention, the DNA vaccine
against FHV-1 for administration by the intramuscular
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid (e.g. pSB021, Figure 10) encoding
the FHV-1 gB antigen optimized by the deletion of the
fragment of the nucleotide sequence encoding the
transmembrane domain, of a second expression plasmid
(e.g. pSB023, Figure 11) encoding the FHV-1 gC antigen
optimized by the deletion of the fragment of the
nucleotide sequence encoding the transmembrane domain
and of a third expression plasmid (e.g. pSB024,
Figure 12) encoding the FHV-1 gD antigen optimized by
the deletion of the fragment of the nucleotide sequence
encoding the transmembrane domain.
Preferably, according to the invention, the DNA vaccine
against FHV-1 for administration by the subcutaneous
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid encoding feline GM-CSF, and the
three plasmids previously defined (e.g. pSB021, pSB023
and pSB024).
The subject of the present invention is an improved DNA
vaccine capable of inducing an effective and protective
immune response in horses against the equine herpes-
virus type 1 (EHV-1).
The EHV-1 virus is a member of the Alphaherpesvirinae
family. The EHV-l virus is responsible for equine viral
abortion (Crabb et al. Adv. Virus Res. 1995. 45. 153-
190). The complete genome of this virus has been
determined (Telford et al. Virology 1992. 189.
304-316).
The DNA vaccine against EHV-1 is preferably formulated
with an adjuvant according to the invention, in
CA 02375320 2012-01-25
- 22 -
particular DMRIE, preferably DMRIE-DOPE. This may be
b iacu a -i...... L , aa ,
optionally LLIUJ with c.~.aaci L.aac a"Ul1...'JL vl. r cdui .:
GM-CSF, or the optimization of at least one EHV-1
antigen, or finally the addition of equine GM-CSF and
the optimization of at least one EHV-1 antigen.
The addition of equine GM-CSF may be carried out by the
incorporation of the equine GM-CSF polypeptide into the
vaccinal composition or by the insertion of the nucleo-
tide sequence (e.g. SEQ ID NO 69, Figure 26) encoding
equine GM-CSF into an in vivo expression vector,
preferably a plasmid. Preferably, the nucleotide
sequence encoding equine GM-CSF is inserted into a
second expression plasmid, different from that (or
those) into which the gene(s) encoding the EHV-1
antigen(s) is(are) inserted.
The optimization of the antigens derived from EHV-1 is
carried out by the deletion of the DNA fragment
encoding the transmembrane domains of the gB glyco-
protein and/or of the gC glycoprotein and/or of the gD
glycoprotein of EHV-1. The improved DNA vaccine against
EHV-1 according to the invention may therefore encode
and express a single optimized EHV-1 antigen (gB, gC or
gD) or both of them or the three.
Nucleotide sequences encoding the EHV-1 antigens which
can be used in the present invention and various
expression vector constructs are given in the accom-
panying examples and in WO-A-98/03198, in particular in
Examples 8 and 10, and in Figures 2 and 4.
The intramuscular route is preferred for horses.
Preferably, according to the invention, the DNA vaccine
against EHV-1 for administration by the intramuscular
route is formulated with DMRIE-DOPE, and is composed of
an expressionplasmid (e.g. pSB028, Figure 13) encoding
the EHV-1 gB antigen optimized by the deletion of the
fragment of the nucleotide sequence encoding the
CA 02375320 2012-01-25
- 23 -
transmembrane domain, of a second expression plasmid
(e.g. PSB029, Fiaure 14) encoding t-he EHV-1 gC antigen
optimized by the deletion of the fragment of the
nucleotide sequence encoding the transmembrane domain
and of a third expression plasmid (e.g. pSB030,
Figure 15) encoding the EHV-1 gD antigen optimized by
the deletion of the fragment of the nucleotide sequence
encoding the transmembrane domain.
It is possible, however, to use the subcutaneous route.
In this case, the DNA vaccine against EHV-1 is
preferably formulated with DMRIE-DOPE, and is composed
of an expression plasmid encoding equine GM-CSF, and of
the three plasmids previously defined (e.g. pSB028,
pSB029 and pSB030).
The subject of the present invention is an improved DNA
vaccine capable of inducing an effective and protective
immune response in horses against the equine herpes-
virus type 4 (EHV-4).
The EHV-4 virus is a member of the Alphaherpesvirinae
family. The EHV-4 virus is responsible for equine viral
rhinopneumonia (Crabb et al. Adv. Virus Res. 1995. 45.
153-190). The complete genome of this virus has been
determined (Telford et al. J. Gen. Virol. 1998. 79.
1197-1203).
The DNA vaccine against EHV-4 is preferably formulated
with an adjuvant according to the invention, in
particular DMRIE, preferably DMRIE-DOPE. This may be
optionally combined with either the addition of equine
GM-CSF, or the optimization of at least one EHV-4
antigen, or finally the addition of equine GM-CSF and
the optimization of at least one EHV-4 antigen.
The addition of equine GM-CSF may be made as is
described for EHV-1.
CA 02375320 2012-01-25
- 24 -
The optimization of the antigens derived from EHV-4 is
nM~ ricrl Hitt hts tltca A,=1 IF +-hc rim fragment
w ,J.
encoding the transmembrane domain of the glycoprotein
gB and/or the glycoprotein gC and/or the glycoprotein
gD of EHV-4. The improved DNA vaccine against EHV-4
according to the invention may therefore encode and
express a single optimized EHV-4 antigen (gB, gC or gD)
or two of them or the three.
Nucleotide sequences encoding the EHV-4 antigens which
can be used in the present invention and various
expression vector constructs are given in the accom-
panying examples and in WO-A-98/03198, in particular in
Examples 9 and 11, and in Figures 3 and 5.
Preferably, according to the invention, the DNA vaccine
against EHV-4 for administration by the intramuscular
route is formulated with DMRIE-DOPE, and is composed of
an expression plasmid (e.g. pSB025, Figure 16) encoding
the EHV-4 gB antigen optimized by the deletion of the
fragment of the nucleotide sequence encoding the
transmembrane dornain, of a second expression plasmid
(e.g. pSB026, Figure 17) encoding the EHV-4 gC antigen
optimized by the deletion of the fragment of the
nucleotide sequence encoding the transmembrane domain
and of a third expression plasmid (e.g. pSB027,
Figure 18) encoding the EHV-4 gD antigen optimized by
the deletion of the fragment of the nucleotide sequence
encoding the transmembrane domain.
For the subcutaneous route, the DNA vaccine against
EHV-4 is preferably formulated with DMRIE-DOPE, and is
composed of an expression plasmid encoding equine
GM-CSF, and the three plasmids previously defined (e.g.
pSB025, pSB026 and pSB027).
Although the, invention is described in relation to
specific DNA vaccines, the invention also applies to
DNA vaccines directed against other pathogens of these
CA 02375320 2012-01-25
- 25 -
animal species. Thus, the various valencies described
i n th; c or...l i r-nf i nn mqv ha the ciih~ort of an 4 my-rnvorl
vaccine, in particular by the addition of the adjuvant
of the invention, or of GM-CSF, or optionally gene
optimization, or combinations of these propositions, as
is described here in detail for some valencies.
In the same line of thought, the vaccines according to
the invention may be, for an animal species, combined
with one another and/or with DNA vaccines directed
against other pathogens of the same species.
Thus, the subject of the present invention is also
improved multivalent DNA vaccines which make it
possible to obtain an effective protection in dogs
against at least two canine pathogens selected from the
group consisting of CDV, CPI-2, CHV-1, rabies virus
(rhabdovirus), canine parvovirus (CPV), canine corona-
virus (CCV), Borrelia burgdorferi.
The subject of the present invention is also improved
multivalent DNA vaccines which make it possible to
obtain an effective protection in cats against at least
two feline pathogens selected from the group consisting
of FHV-1, feline calicivirus (FCV), rabies virus
(rhabdovirus), feline parvovirus (FPV), feline infec-
tious peritonitis virus (FIPV), feline leukaemia virus
(FeLV), feline acquired immunodeficiency syndrome virus
(FIV).
The subject of the present invention is also improved
multivalent DNA vaccines which make it possible to
obtain an effective protection in horses against at
least two equine pathogens selected from the group
consisting of EHV-1, EHV-4, equine influenza virus,
Eastern equine encephalitis virus, Western equine
encephalitis virus, Venezuelan equine encephalitis
virus, rabies virus, Clostridium tetani, Borrelia
burgdorferi.
CA 02375320 2012-01-25
- 26 -
m1., .. ..l 4-:iV...=.. a...... . -....i.-,..1- TITTT V h ...i.aa..J ...,:y
.... l.- i ~mtrs.-rnv,rcr~ 1-.t1, 6-k_ i
SASS .iui a......u ,.. ......--
formulation with an adjuvant according to the
invention, in particular with DMRIE, preferably with
DMRIE-DOPE. This may be optionally combined either with
the addition of GM-CSF as previously described, or with
the optimization of at least one antigen of interest as
previously described, or finally by the addition of
GM-CSF and the optimization of at least one antigen of
interest.
The improved multivalent DNA vaccines according to the
invention are composed of one or more expression
plasmids, such that these vaccines lead to the in vivo
expression of at least one immunogen of a first
pathogen and of at least one immunogen of at least one
other pathogen, infecting the same animal species. At
least one of these immunogens is preferably selected
from the members of the following group:
- F of CDV, HA of CDV, F of CPI-2, HN of CPI-2,
gB of CHV-1, gC of CHV-1 and gD of CHV-i for dogs,
- gB of FHV-1, gC of FHV-1 and gD of FHV-1 for
cats, and
- gB of EHV-1, gC of EHV-1, gD of EHV-1, gB of
EHV-4, gC of EHV-4 and gD of EHV-4 for horses.
The improved monovalent or multivalent DNA vaccines
according to the invention may also be combined with at
least one conventional vaccine (inactivated, attenuated
live, subunits) or recombinant vaccine using an in vivo
expression vector (e.g. poxvirus, adenovirus, herpes-
virus) directed against at least one, in particular
different, pathogen infecting the same species.
Persons skilled in the art may refer to WO-A-9803198
for the methods for constructing the plasmids contain-
ing these equine valencies, to WO-A-9803660 for the
feline valencies and to WO-A-9803199 for the canine
CA 02375320 2012-01-25
-27-
valencies.
The subject of the present invention is also a method of vaccinating pets and
animals
used in sports, in particular dogs, cats or horses. This vaccination method
comprises the
administration of one of the monovalent or multivalent improved DNA vaccines
as
described above. This vaccination method comprises the administration of one
or more
doses of the improved DNA vaccine.
The quantity of DNA used in the vaccines according to the present invention is
between about 10 g and about 1000 jig, and preferably between about 50 gg and
about
500 g, for a given plasmid. Persons skilled in the art possess the competence
necessary to precisely define the effective dose of DNA to be used for each
vaccination
protocol.
The dose volumes may be preferably between 0.5 and 5 ml, preferably between I
and 3
ml.
The improved DNA vaccines according to the invention may be administered, in
the
context of this vaccination method, by the various routes of administration
proposed in
the prior art for polynucleotide vaccination and by means of known techniques
of
administration.
According to the two preferred modes of the invention, the methods of
vaccination
comprise the administration of the improved DNA vaccines according to the
invention
by the intramuscular route or by the subcutaneous route.
The present invention provides a DNA vaccine against a pathogen of a host,
wherein
the host is a dog and the pathogen is canine distemper virus (CDV), comprising
at least
one DNA plasmid that contains and expresses, in a host cell, a nucleotide
sequence
encoding an immunogen of the pathogen. and as an adjuvant a cationic lipid
containing
a quaternary ammonium salt, of formula:
CA 02375320 2012-01-25
-27a-
CH3
I+
RI-O-CH2-CH-CH2- i --R2-X
OR1 CH3
in which R1 is a saturated or unsaturated linear aliphatic radical having 12
to 18 carbon
atoms, R2 is another aliphatic radical containing 2 or 3 carbon atoms, and X a
hydroxyl
6 or amine group.
The present invention also provides an immunogenic composition against a
pathogen of
a host, wherein the host is a dog and the pathogen is canine distemper virus
(CDV),
comprising at least one plasmid that contains and expresses, in a host cell, a
nucleotide
sequence encoding an immunogen of the pathogen, and as an adjuvant a cationic
lipid
containing a quaternary ammonium salt, of formula:
CH3
I+
R,-O-CH2- CH-CH2-N _R2-X
OR, CH3
in which Rt is a saturated or unsaturated linear aliphatic radical having 12
to 18 carbon
atoms, R2 is another aliphatic radical containing 2 or 3 carbon atones, and X
a hydroxyl
or amine group.
The present invention finther provides a DNA vaccine against a pathogen of a
host,
wherein the host is a dog and the pathogen is canine distemper virus (CDV),
comprising at least one DNA plasmid that contains and expresses, in a host
cell, a
nucleotide sequence encoding an immunogen of the pathogen, and as an adjuvant
a
cationic lipid comprising DMRIE, wherein the vaccine further comprises
dioleoylphosphatidylethanolamine (DOPE).
26
Throughout this specification the word "comprise", or variations such as
"comprises" or
"comprising", will be understood to Imply the inclusion of a stated element,
integer or
CA 02375320 2012-01-25
- 27b -
step, or group of elements, integers or steps, but not the exclusion of any
other element,
integer or step, or group of elements, integers or steps.
Any discussion of documents, acts, materials, devices, articles or the like
which has
been included in the present specification is solely for the purpose of
providing a
context for the present invention. It is not to be taken as an admission that
any or all of
these matters form part of the prior art base or were common general knowledge
in the
field relevant to the present invention as it existed in Australia before the
priority date
of each claim of this application.
The invention will now be described in greater detail with the aid of
embodiments
taken as nonlimiting examples and referring to the drawing in which:
Figure No. t: Plasmid pAB1l0
CA 02375320 2012-01-25
- 28 -
Figure No. 2: Plasmid pVR1012
Figure ivo. 3: Picisiitiu pNSO21
Figure No. 4: Plasmid pNS024
Figure No. 5: Plasmid pSB032
Figure No. 6: Plasmid pSB034
Figure No. 7: Plasmid pSBO16
Figure No. 8: Plasmid pSBO19
Figure No. 9: Plasmid pSBO17
Figure No. 10: Plasmid pSB021
Figure No. 11: Plasmid pSB023
Figure No. 12: Plasmid pSB024
Figure No. 13: Plasmid pSB028
Figure No. 14: Plasmid pSB029
Figure No. 15: Plasmid pSB030
Figure No. 16: Plasmid pSB025
Figure No. 17: Plasmid pSB026
Figure No. 18: Plasmid pSB027
Figure No..19: Plasmid pJP084
Figure No. 20: sequence of the canine GM-CSF gene
Figure No. 21: Plasmid pJP089
Figure No. 22: sequence of the feline GM-CSF gene 3R3
Figure No. 23: Plasmid pJP090
Figure No. 24: sequence of the feline GM-CSF gene 3R4
Figure No. 25: Plasmid pJP097
Figure No. 26: sequence of the equine GM-CSF gene
Fig-are No. 27: sequence of the CPI-2 F gene
Figure No. 28: sequence of the CPI-2 HN gene
Sequence listing:
SEQ ID NO 1: oligonucleotide NS030
SEQ ID NO 2: oligonucleotide NS031
SEQ ID NO 3: oligonucleotide NS034
SEQ ID NO 4: oligonucleotide NS035
SEQ ID NO -5: olgonucleotide -NS 0-3-6
SEQ ID NO 6: oligonucleotide NS037
SEQ ID NO 7: oligonucleotide SB090
SEQ ID NO 8: oligonucleotide SB091
SEQ ID NO 9: oligonucleotide PB326
SEQ ID NO 10: oligonucleotide PB329
CA 02375320 2012-01-25
- 29 -
SEQ ID NO 11: oligonucleotide PB381
SEQ ID NO 12: oiigonucleotidu PB362
SEQ ID NO 13: oligonucleotide PB383
SEQ ID NO 14: oligonucleotide PB384
SEQ ID NO 15: oligonucleotide SB101
SEQ ID NO 16: oligonucleotide SB102
SEQ ID NO 17: oligonucleotide SB103
SEQ ID NO 18: oligonucleotide SB104
SEQ ID NO 19: oligonucleotide SB105
SEQ ID NO 20: oligonucleotide SB106
SEQ ID NO 21: oligonucleotide SB107
SEQ ID NO 22: oligonucleotide SB108
SEQ ID NO 23: oligonucleotide SB109
SEQ ID NO 24: oligonucleotide SB110
SEQ ID NO 25: oligonucleotide SB111
SEQ ID NO 26: oligonucleotide SB112
SEQ ID NO 27: oligonucleotide SB113
SEQ ID NO 28: oligonucleotide SB114
SEQ ID NO 29: oligonucleotide SB115
SEQ ID NO 30: oligonucleotide SB116
SEQ ID NO 31: oligonucleotide SB117
SEQ ID NO 32: oligonucleotide SB118
SEQ ID NO 33: oligonucleotide AB325
SEQ ID NO 34: oligonucleotide AB326
SEQ ID NO 35: oligonucleotide AB327
SEQ ID NO 36: oligonucleotide AB328
SEQ ID NO 37: oligonucleotide AB329
SEQ ID NO 38: oligonucleotide AB330
SEQ ID NO 39: oligonucleotide NS003
SEQ ID NO 40: oligonucleotide NS004
SEQ ID NO 41: oligonucleotide NS005
SEQ ID NO 42: oligonucleotide NS006
SEQ ID NO 43: oligonucleotide NS007
SEQ ID NO 44: oligonucleotide NSO-08
SEQ ID NO 45: oligonucleotide SB119
SEQ ID NO 46: oligonucleotide SB120
SEQ ID NO 47: oligonucleotide SB121
SEQ ID NO 48: oligonucleotide SB122
SEQ ID NO 49: oligonucleotide SB123
CA 02375320 2012-01-25
- 30 -
SEQ ID NO 50: oligonucleotide SB124
SEQ ID NO 51: oligonucleotide SB125
SEQ ID NO 52: oligonucleotide SB126
SEQ ID NO 53: oligonucleotide SB127
SEQ ID NO 54: oligonucleotide SB128
SEQ ID NO 55: oligonucleotide SB129
SEQ ID NO 56: oligonucleotide SB130
SEQ ID NO.57: oligonucleotide SB131
SEQ ID NO 58: oligonucleotide SB132
SEQ ID NO 59: oligonucleotide SB133
SEQ ID NO 60: oligonucleotide SB134
SEQ ID NO 61: oligonucleotide SB135
SEQ ID NO 62: oligonucleotide SB136
SEQ ID NO 63: oligonucleotide SB137
SEQ ID NO 64: oligonucleotide JP578
SEQ ID NO 65: oligonucleotide JP579
SEQ ID NO 66: sequence of the canine GM-CSF gene (see
Figure 20)=
SEQ ID NO 67: sequence of the feline GM-CSF 3R3 gene
(see Figure 22)
SEQ ID NO 68: sequence of the feline GM-CSF 3R4 gene
(see Figure 24)
SEQ ID NO 69: sequence of the equine GM-CSF gene (see
Figure 26)
SEQ ID NO 70: oligonucleotide JP734
SEQ ID NO 71: oligonucleotide JP735
SEQ ID NO 72: sequence of the CPI-2 F gene (see
Figure 27)
SEQ ID NO 73: sequence of the CPI-2 HN gene (see
Figure 28)
EXAMPLES:
For each of the patnogens considered; each- gene-
encoding the principal surface antigens (native form
and modified form) was the subject of a particular
construction in a eukaryotic expression plasmid. The
secreted forms of the surface antigens were obtained by
deletion of the fragments of genes encoding the
CA 02375320 2012-01-25
- 31 -
transmembrane and cytoplasmic domains. In all cases,
the transmembrane doma i nc of t-h (Ti ~~rnnrntei nc uiern
identified on the basis of the hydropathy profiles of
the corresponding protein sequences. The table given in
Example 11 summarizes the sizes of the wild-type
proteins (in amino acids), the positions identified in
the transmembrane domains, the sizes of the truncated
proteins, as well as the names of the corresponding
expression plasmids.
Example 1: Basic plasmid constructs
The eukaryotic expression plasmid pVR1020 (C.J. Luke et
al. J. of Infectious Diseases 1997, 175: 95-97),
derived from the plasmid pVR1012 (Figure 1, Example 7
of WO-A-9803199, repeated in Figure 2 of the present
application), contains the coding phase of the signal
sequence of the human tissue plasminogen activator
(tPA). The plasmid pVR1020 was modified by BamHI-BglII
digestion and insertion of a sequence containing
several cloning sites (BamHI, NotI, EcoRl, XbaI, PmlI,
PstI, BglII) and resulting from the pairing of the
following oligonucleotides:
PB326 (40 mer) (SEQ ID NO 9)
5' GATCTGCAGCACGTGTCTAGAGGATATCGAATTCGCGGCC 3' and
PB329 (40 mer) (SEQ ID NO 10)
5' GATCCGCGGCCGCGAATTCGATATCCTCTAGACACGTGCT 3'.
The resulting vector, pAB110 (Figure No. 1), was used
for the construction of the plasmids containing the
truncated forms of the genes encoding the canine
distemper virus (CDV) haemagglutinin (HA) and the
parainfluenza virus type 2 (CPI-2) haemagglutinin-
neuraminidase (HN).
Intron II of the rabbit (3-globin gene was cloned into
the vector pCRII (Invitrogen, Carlsbad, CA, USA) after
production of the corresponding DNA fragment by PCR
CA 02375320 2012-01-25
- 32 -
with the aid of the following oligonucleotides:
SB090 (20 mer) (SEQ ID NO 7)
5' TTGGGGACCCTTGATTGTTC 3' and
SB091 (21 mer) (SEQ ID NO 8)
5' CTGTAGGAAAAAGAAGAAGGC 3'
using as template the genomic DNA of rabbit peripheral
blood cells. The resulting plasmid was designated
pNS050-
Example 2: Plasmids encoding the various forms of the
CDV antigens
The genes encoding the fusion protein (F) and the
haemagglutinin (HA) of CDV strain Snyder Hill (SH) were
obtained by RT-PCR from the viral RNA of the SH strain
(accessible from the strain depository American Tissue
Culture Collection under the number ATCC VR-526).
2.1. Plasmids encoding the various forms of CDV-F
2.1.1. pPB229: F gene (native form) cloned into the
vector pVR1012
The cDNA of the CDV F gene was synthesized with the aid
of the primer PB383 and amplified by a PCR reaction
with the aid of the following pair of oligonucleotides:
PB383 (26 mer) (SEQ ID NO 13)
5' TTTCTAGACAGCCGAGCCCCATGCAC 3' and
PB384 (30 mer) (SEQ ID NO 14)
5' TTGGATCCGATATATGACCAGAATACTTCA 3'.
The- PCR product was digested- -with-- BamHI and- Xba-I and
cloned into the expression vector pVR1012 (Example 1)
previously digested with BamHI and XbaI, generating the
plasmid pPB229 (6925 base pairs or bp). The CDV wild-
type F gene encodes a protein of 662 residues.
CA 02375320 2012-01-25
- 33 -
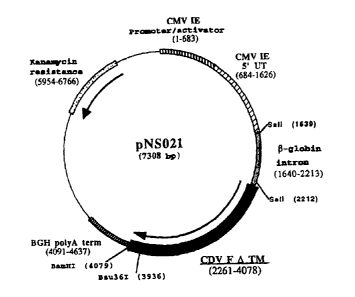
2.1.2. pNS021: F gene (form f3-globin F ATM) cloned into
-,., 4 ..t^.r -IT010-12
The plasmid pNS013 (6735 bp) containing the F gene
truncated of the transmembrane and C-terminal domain
was obtained by ligation of a Bsu361-BamHI fragment
(6593 bp) derived from the plasmid pPB229
(Example 2.1.1), and of a 142-bp fragment obtained by
PCR from the template pPB229 with the aid of the
following oligonucleotides:
NS030 (21 mer) (SEQ ID NO 1)
5' ATGAGCCCACTCTTACAACAA 3' and
NS031 (35 mer) (SEQ ID NO 2)
5' TTTCGCGGATCCATTAAAGGAAGAGCGCCTAACCG 3'
and Bsu36I-BamHI digested. The CDV truncated F gene
encodes a protein of 605 residues.
In a second instance, a sequence corresponding to
intron II of the rabbit P-globin gene was inserted
upstream of the coding sequence of the truncated F
gene, into the Sall site of the plasmid pNS013. The DNA
fragment corresponding to the intron (573 bp) was
obtained by PCR with the aid of the following
oligonucleotides:
NS036 (34 mer) (SEQ ID NO 5)
5' TTTACGCGTCGACTTGGGGACCCTTGATTGTTC 3' and
NS037 (36 mer) (SEQ ID NO 6)
5' TTTACGCGTCGACCTGTAGGAAAAAGAAGAAGGCAT 3'
on the template pNS050 (Example 1), followed by SalI
digestion to release Sall compatible ends. The
derivative of the plasmid pNS013 containing intron II
of the -P=globin -gene -is -designated -pNS-021 (-73-08 bp-)-
(Figure No. 3).
CA 02375320 2012-01-25
- 34 -
2.2. Plasmids encoding the various forms of CDV-HA
2.2.1. pNS018: HA gene (native form) cloned into the
vector pVR1012
The cDNA of the CDV HA gene was synthesized with the
aid of the primer PB381 and amplified by PCR reaction
with the aid of the following pair of oligonucleotides:
PB381 (30 mer) (SEQ ID NO 11)
5' TTCTGCAGATGCTCTCCTACCAAGAYAAGG 3' and
PB382 (28 mer) (SEQ ID NO 12)
5' TTGTCGACATGTGTATCATCATMCTGTC 3'.
The PCR product was cloned into the vector pCRII
(Invitrogen, Carlsbad, CA, USA), generating the plasmid
pPB235. The PstI-SalI fragment of 1846 bp of the
plasmid pPB235 containing the HA gene was then cloned
into the Pstl-Sall digested expression vector pVR1012
(Example 1), generating the plasmid pNS018 (6748 bp).
The CDV wild-type HA gene encodes a protein of 607
residues.
2.2.2. pNS024:' HA gene (form P-globin tPA ATM HA)
cloned into the vector pVR1012
The truncated form of the CDV HA gene was obtained by
deletion of the DNA fragment encoding the first 60
residues of the HA protein. The signal sequence and the
transmembrane sequence of this protein being indistin-
guishable, the secretion of the truncated product is
ensured by the production of a fusion in phase between
the signal sequence of the human tissue plasminogen
activator (tPA) and the truncated HA gene. The plasm-
-35 pNS019, the derivative of pNS018 encoding a fusion
product with the tPA signal, was obtained by the
ligation of the following 3 DNA fragments:
- Fragment A was obtained by BamHI-EcoRV digestion of
CA 02375320 2012-01-25
- 35 -
pAB110 (Example 1).
- Fragment B was obtained by PCR reaction on the
template pNS018 (Example 2.2.1) with the aid of the
following oligonucleotides:
NS034 (30 mer) (SEQ ID NO 3)
5' TTTCGCGGATCCCACAAAGTATCAACTAGC 3' and
NS035 (23 mer) (SEQ ID NO 4)
5' GGGATTTGCTGCCGATGCAATAG 3'
followed by a BamHI-SapI digestion of the PCR product.
- Fragment C is a fragment of SapI-EcoRV digestion of
pNS018.
The hybrid gene tPA ATM HA encodes a protein of 574
residues (1725 bp).
Intron II of the rabbit P-globin gene (Example 2.1.2)
was inserted upstream of the coding frame of the HA
gene into the SalI site of pNS019 to generate the
plasmid pNS024 (Figure No. 4).
Example 3: Plasmids encoding the various forms of the
antigens of the canine parainfluenza virus type 2
(CPI-2)
The F and HN genes of CPI-2 strain D008 (MERIAL) were
obtained by RT-PCR from the viral RNA.
3.1. Plasmids encoding the various forms of CPI-2 F
3.1.1. pAB115: F gene (native form) cloned into the
vector pVR1012
The cDNA of the CPI-2 F gene was synthesized and
amplified by RT-PCR with the aid of the following pair
of oligonucleotides:
CA 02375320 2012-01-25
- 36 -
SB131 (38 mer) (SEQ ID NO 57)
5' AAAAACGCGTCGACATGC;C;TACTATAATTrAATTTCTr 3 '
SB132 (38 mer) (SEQ ID NO 58)
5' TTTTCTAGTCTAGATTATTTATGATAAACAAAATTCTC 3'.
The PCR product was digested with Sall and XbaI,
generating a fragment of 1594 bp, and cloned into the
expression vector pVR1012 (Example 1) previously
digested with the same enzymes, generating the plasmid
pAB115 (6479 bp). The CPI-2 wild-type F gene
(SEQ ID NO 72) (Figure 27) cloned into this plasmid
encodes a protein of 529 residues.
3.1.2. pSB032: F gene (form P-globin F ATM) cloned into
the vector pVR1012
The plasmid pSB031 containing the F gene truncated of
the transmembrane and C-terminal cytoplasmic domains
was obtained in the following manner. A PCR reaction
was carried out with the template pAB115 (Example
3.1.1.) with the aid of the following oligonucleotides:
SB131 (SEQ ID NO 57) and
SB133 (41 mer) =(SEQ ID NO 59)
5' TTTTCTAGTCTAGATTAGTATGTGTCACTTTGTGCTAAGTG 3'
to generate a PCR fragment of about 1450 bp. This
fragment was digested with Sall and XbaI in order to
isolate the Sall-Xbal restriction fragment of 1436 bp.
This fragment was ligated into the vector pVR1012
(Example 1) previously digested with Sall and XbaI to
give the plasmid pSB031. The CPI-2 truncated F gene
encodes a protein of 473 residues.
In- -a second n=stance-, a- sequence -co-r-r-esponding to-
intron II of the rabbit 0-globin gene was inserted
upstream of the coding sequence of the CPI-2 truncated
F gene, into the Sall site of the plasmid pSB031. The
DNA fragment corresponding to the intron (573 bp) was
obtained by PCR with the aid of the oligonucleotides
CA 02375320 2012-01-25
- 37 -
NS036 (SEQ ID NO 5) and NS037 (SEQ ID NO 6) on the
template pNS050 (Example 1), followed by SalI digestion
to release the Sall compatible ends. This Sall-Sall
restriction fragment was ligated into the plasmid
pSB031 previously digested with Sall, and then
dephosphorylated to give the plasmid pSB032 (6884 bp)
(Figure No. 5).
3.2. Plasmids encoding the various forms of CPI-2 IU
3.2.1. pAB114: HN gene (native form) cloned into the
vector pVR1012
The cDNA of the CPI-2 HN gene was synthesized and
amplified with the aid of the following
oligonucleotides:
SB134 (41 mer) (SEQ ID NO 60)
5' AAAAACGCGTCGACATGGTTGCAGAAGATGCCCCTGTTAGG 3'
SB135 (35 mer) (SEQ ID NO 61)
5' TTTTGGAAGATCTTTAGGATAGTGTCACCTGACGG 3'
in order to generate a PCR fragment of about 1720 bp.
This fragment was digested with Sall and Bg1II in order
to isolate the SalI-BglI fragment of 1704 bp. This
fragment was then ligated into the vector pVR1012
(Example 1), previously digested with Sall and BglII,
to give the plasmid pAB114 (6566 bp). The CPI-2 wild-
type HN gene (SEQ ID NO 73) (Figure 28) cloned into
this plasmid encodes a protein of 565 residues.
3.2.2. pSB034: HN gene (form P-globin HN tPA ATM)
cloned into the vector pVR1012
The truncated- form of the. CPI=2 HN- gene-was -obtained by-
deletion of the DNA fragment encoding the first 40
residues of the HN protein. The signal and trans-
membrane sequences of this protein being indistin-
guishable, the secretion of the truncated protein is
ensured by the production of a fusion in phase between
CA 02375320 2012-01-25
- 38 -
the signal sequence of the human tissue plasminogen
activator (tPA) and the truncated HN gene. The nlasmid
pSB033, derived from pAB114 (Example 3.2.1), encoding a
fusion product with tPA, was obtained by ligation of
the EcoRI-PmlI fragment of pAB110 (Example 1), a
derivative of pVR1012 containing an open reading frame
encoding the tPA signal sequence and of a fragment of
(1599 bp) obtained by PCR with the aid of the following
oligonucleotides:
SB136 (37 mer) (SEQ ID NO 62)
5' TTAAAAGAATTCGACCCAAAAGCAAATCATGAGCCAC 3'
SB137 (33 mer) (SEQ ID NO 63)
5' TTAAAAGGCCTTTAGGATAGTGTCACCTGACGG 3-
on the template pAB114 and digested with EcoRI and
EcoRV.
Intron II. of the rabbit 3-globin gene was inserted
upstream of the coding frame of the HN gene into the
Sall site of pSB033 to generate the plasmid pSB034
(Figure No. 6). The Sall fragment containing the intron
was obtained by PCR with the aid of the oligo-
nucleotides NS036 (SEQ ID NO 5) and NS037 (SEQ ID NO 6)
on the template pNS050 (Example 1).
Example 4: Plasmids encoding the various forms of the
CHV-1 virus glycoproteins
The genes encoding the glycoproteins gB, gC and gD of
the Carmichael strain of the canine herpesvirus type I
(CHV-1) were isolated by PCR from the viral genome. The
cloning of the genes encoding gB and gD into the vector
pVR1012 was previously described in patent application
WO=A=-980.3-199 (-plasmids pAB037 and- -pAB03-8-r-espec-t- vely-
in Figures 7 and 8 and in Examples 13 and 14) . On the
other hand, the cloning of the gene encoding gC is
described in this document.
CA 02375320 2012-01-25
- 39 -
4.1. Plasmid encoding the truncated form of CHV-gB
4.1.1. pSBO16: gB gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the CHV-1 gB protein (878 amino acids) is
positioned between residues 702 and 769. The plasmid
containing the truncated form of the gene encoding gB
was obtained by ligation of the following three DNA
fragments: (a) the vector pVR1012 (Example 1) lineari-
zed by a double PstI-XbaI digestion, (b) a fragment of
1997 bp obtained by PstI-NsiI digestion of pAB037
(Example 4) and (c) a fragment of 225 bp obtained by
PCR with the aid of the following oligonucleotides:
SB101 (22 mer) (SEQ ID NO 15)
5' TATATTGAAGGACAACTTGGGG 3' and
SB102 (36 mer) (SEQ ID NO 16)
5' CTAGTCTAGATTAATTATTATCAACTTTTACAACAC 3'
using the plasmid pAB037 as template and digested with
NsiI and XbaI. The resulting plasmid, pSBO16 (6983 bp)
(Figure No. 7) contains a truncated gB gene encoding a
protein of 701 residues.
4.2. Plasmids encoding the various forms of CHV-gC
4.2.1. pSB018: gC gene (native form) cloned into the
vector pVR1012
The DNA fragment containing the open reading frame of
the CHV-1 gC gene was obtained by PCR with the aid of
the following oligonucleotides:
SB105 (32 mer) (SEQ ID NO 19)
5' AAAACTGCAGATGAGTTTTAAAAATTTTTATC 3' and
SB106 (30 mer) (SEQ ID NO 20)
5' CTAGTCTAGATTAGATCTTATTATTTTTTG 3'
using the viral DNA as template. This PCR product was
CA 02375320 2012-01-25
- 40 -
digested with PstI and XbaI generating a fragment of
1400 b^ which wa-S then l i eerl into the .renter Y - % ' % 1 (ll )
r -~ n
(Example 1) linearized by the same double digestion.
The resulting plasmid, pSBO18 (6253 bp), contains the
gene encoding the gC glycoprotein of 459 residues.
4.2.2. pSBO19: gC gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the gC protein is between residues 422 and
452. The plasmid containing the truncated form of the
gene encoding gD was obtained by ligation of the
following three DNA fragments: (a) the vector pVR1012
(Example 1) linearized by a double PstI-Xbal digestion,
(b) a fragment of 934 bp obtained by PstI-StuI
digestion of pSBO18 (preceding example) and (c) a
fragment of 335 bp obtained by PCR with the aid of the
following oligonucleotides:
SB107 (24 mer) (SEQ ID NO 21)
51 TGGATTGACGGTCTTATAACAGGC 3' and
SB108 (37 mer) (SEQ ID NO 22)
5' CTAGTCTAGATTAATTTTCATCCGATGCATCAAACAC 3'
using the plasmid pSBO18 as template and digested with
Stul and XbaI. The resulting plasmid, PSB019 (6139 bp)
(Figure No. 8), contains a truncated gC gene encoding a
protein of 421 residues.
4.3. Plasmid'encoding the truncated form of CHV-gD
4.3.1. pSB017: gD gene (ATM form) cloned into the
vector pVR1012
The transmembrane domain of the CHV-1 gD protein (345
amino acids) is between residues 310 and 328. The
plasmid containing the truncated form of the gene
encoding gD was obtained by ligation of the following
three DNA fragments: (a) the vector pVR1012 (Example 1)
CA 02375320 2012-01-25
- 41 -
linearized by a double PstI-NotI digestion, (b) a
fragment of 6,6,3 hn nhtai nett by Pct T-AvvaTT digestion of
pAB038 (Example 4) and (c) a fragment of 415 bp
obtained by PCR with the aid of the following
oligonucleotides:
SB103 (25 mer) (SEQ ID NO 17)
5' CGAGAAACTTGTTATTTTTCTAAAG 3' and
SB104 (51 mer) (SEQ ID NO 18)
5' ATAAGAATGCGGCCGCAAAGGCTATATATTTTTTGGGGTATTATTTATTGG 3'
using the plasmid pAB038 as template and digested with
Avail and NotI. The resulting plasmid, pSBO17 (5819 bp)
(Figure No. 9), contains a truncated gD gene encoding a
protein of 309 residues.
Example 5: Plasmids encoding the various forms of the
FHV-1 glycoproteins
The genes encoding the glycoproteins gB, gC and gD of
the CO strain of the feline herpesvirus type 1 (FHV-1)
were isolated by PCR from the viral genome. In the
specific case of the gene encoding gD, whose nucleotide
sequence is identical to that of the C-27 strain, we
used the plasmid pAB029, derived from pVR1012
containing the corresponding gene cloned from strain C-
27 and described in patent application WO-A-9803660
(plasmid pAB029, Figure 12 and Example 15).
5.1. Plasmids encoding the various forms of FHV-gB
5.1.1. pSB020: gB gene (native form) cloned into the
vector pVR1012
The DNA fragment containing the open reading frame of
the FHV-l gB gene was obtained by PCR with the aid of
the following oligonucleotides:
SB113 (34 mer) (SEQ ID NO 27)
5' TTTTCTGCAGATGTCCACTCGTGGCGATCTTGGG 3' and
CA 02375320 2012-01-25
- 42 -
SB114 (40 mer) (SEQ ID NO 28)
5' ATAGTTTAGCGGCCGCTTAGACAAGATTTGTTTC'ACTATC 3-
using the viral DNA as template. The PCR product was
digested with PstI and NotI, generating a fragment of
2849 bp, which was then ligated into the vector pVR1012
(Example 1) linearized by the same double digestion.
The resulting plasmid, pSB020 (7728 bp), contains the
gene encoding the gB glycoprotein of 949 residues.
5.1.2. pSB021: gB gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the FHV-1 gB protein is situated between
residues 761 and 834. The plasmid containing the
truncated form of the gene encoding gB was obtained by
ligation of the following three DNA fragments: (a) the
vector pVR1012 (Example 1) linearized by a double PstI-
NotI digestion, (b) a fragment of 1839 bp obtained by
PstI-HindII digestion of pSB020 (preceding example) and
(c) a fragment of 447 bp obtained by PCR with the aid
of the following oligonucleotides:
SB109 (24 mer) (SEQ ID NO 23)
5' CTGTGGACAGAGACCCTAAAACTC 3' and
SB110 (50 mer) (SEQ ID NO 24)
5' TTTCCTTTTGCGGCCGCTTATATGCTGTCTATATCATAAAATTTTAAGGC 3'
using the plasmid pSB020 as template and digested with
Hindll and NotI. The resulting plasmid, pSB021
(7164 bp) (Figure No. 10), contains a truncated gB gene
encoding a protein of 760 residues.
5.2. Plasmids encoding the various forms of FHV-gC
5.2.1. pSB022: gC gene (native form) cloned into the
vector pVR1012
The DNA fragment containing the open reading frame of
the FHV-1 gC gene was obtained by PCR with the aid of
CA 02375320 2012-01-25
- 43 -
the following oligonucleotides:
SB115 (34 mer) (SEQ ID NO 29)
5' TTTTCTGCAGATGAGACGATATAGGATGGGACGC 3' and
SB116 (34 mer) (SEQ ID NO 30)
5'AGTTTAGCGGCCGCTTATAATCGCCGGGGATGAG 3'
using the viral DNA as template. This PCR product was
digested with Pstl and NotI, generating a fragment of
1605 bp, which was then ligated into the vector pVR1012
(Example 1) linearized by the same double digestion.
The resulting plasmid, pSB022 (6483 bp), contains the
gene encoding the gC glycoprotein of 534 residues.
5.2.2. pSB023: gC gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the FHV-1 gC protein is between residues 495
and 526. The plasmid containing the truncated form of
the gene encoding gC was obtained by ligation of the
following two DNA fragments: (a) a fragment of 6198 bp
obtained by BclI and NotI digestion of pSB022
(preceding example) and (b) a fragment of 168 bp
obtained by PCR with the aid of the following
oligonucleotides:
SB117 (24 mer) (SEQ ID NO 31)
5' GTTAAATGTGTACCACGGGACGGG 3' and
SB118 (41 mer) (SEQ ID NO 32)
5' AGTTTAGCGGCCGCTTATTCAGGGGACGCGTCGTAGACTTG 3'
using the plasmid pSB022 as template and digested with
BclI and NotI. The resulting plasmid, pSB023 (6366 bp)
(Figure No. 11), contains a truncated gC gene encoding
a protein of 494 residues.
CA 02375320 2012-01-25
- 44 -
5.3. Plasmid encoding the truncated form of FHV-9D
5.3.1. pSB024: gD gene (ATM form) cloned into the
vector pVR1012
The transmembrane domain of the FHV-1 gD protein (374
amino acids) is between residues 328 and 353. The
plasmid containing the truncated form of the gene
encoding gD was obtained by ligation of the following
two DNA fragments: (a) a fragment of 5712 bp obtained
by XbaI-BglII digestion of pAB029 (Example 5) and (b) a
fragment of 129 bp obtained by PCR with the aid of the
following oligonucleotides:
SB111 (24 mer) (SEQ ID NO 25)
5'GATCGTCCCGCCATACCGTCTGGG 3' and
SB112 (39 mer) (SEQ ID NO 26)
5'TTTGGAAGATCTTTACTGATTATTCATGCCCTTGGGAGG 3'
using the plasmid pAB029 as template and digested with
XbaI and BglII. The resulting plasmid, pSB024 (5841 bp)
(Figure No. 12), contains a truncated gD gene encoding
a protein of 327 residues.
Example 6: Plasmids encoding the various forms of the
EHV-1 glycoproteins
The genes encoding the glycoproteins gB, gC and gD of
the 2234/88-2 strain of EHV-1 were isolated by PCR from
the purified viral DNA.
6.1. Plasmids encoding the various forms of EHV-1 gB
6.1.1. pAB127: gB gene (native form) cloned into the
vector pVR1012
The coding frame of the EHV-1 gB gene was amplified by
PCR with the aid of the following oligonucleotides:
NS003 (30 mer) (SEQ ID NO 39)
CA 02375320 2012-01-25
- 45 -
5' TTCTGCAGATGTCCTCTGGTTGCCGTTCGT 3' and
NS004 (30 mer) (SEQ ID NO 40)
5' TTTCTAGATTAAACCATTTTTTCATTTTCC 3',
the DNA fragment obtained was digested with PstI and
XbaI and ligated into the vector pVR1012 (Example 1)
linearized with PstI and XbaI, generating the plasmid
pAB127 (7818 bp) . The gB gene encodes a protein of 980
amino acids.
6.1.2. pSB028: gB gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the EHV-1 gB protein is positioned between
residues 801 and 875. The plasmid containing the
truncated form of the gene encoding gB was obtained by
ligation of the following two DNA fragments: (a) the
plasmid pAB127 (Example 6.1.1) digested with Afel and
XbaI, and (b) a fragment of 276 bp obtained by PCR with
the aid of the following oligonucleotides:
SB125 (24 mer) (SEQ ID NO 51)
5' AACAACAGAGGGTCGATAGAAGGC 3' and
SB126 (39 mer) (SEQ ID NO 52)
5' AATTTTTCTAGATTACACGTTGACCACGCTGTCGATGTC 3'
using the plasmid pABl27 as template and digested with
Afel and XbaI. The resulting plasmid, pSB028 (7279 bp)
contains a truncated EHV-1 gB gene encoding a protein
of 800 residues.
6.2. Plasmids encoding the various forms of EHV-1 gC
6.2.1. pAB129: gC gene (native form) cloned into the
vector pVR1012
The DNA fragment containing the open reading frame of
the EHV-l gC gene was obtained by PCR with the aid of
the following oligonucleotides:
CA 02375320 2012-01-25
- 46 -
NS005 (31 mer) (SEQ ID NO 41)
5' TTGTCGACATGTGGTTGCCTAATCTCGTGAG 3' and
NS006 (33 mer) (SEQ ID NO 42)
5' TTGGATCCCTAAAAGTCAGACTTCTTGTACGGC 3'.
This PCR product was digested with Sall and BamHI
generating a fragment of 1412 bp, which was then
ligated into the vector pVR1012 (Example 1) linearized
by the same double digestion. The resulting plasmid,
pAB129 (6281 bp), contains the gene encoding the EHV-l
gC glycoprotein and having a size of 468 residues.
6.2.2. pSB029: gC gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the EHV-1 gC protein is between residues 429
and 455. The plasmid containing the truncated form of
the gene encoding gC was obtained by ligation of the
following two DNA fragments: (a) the plasmid pAB129
(Example 6.2.1.) linearized by a double AspI-BamHI
digestion and (b) a fragment of 287 bp obtained by PCR
with the aid of the following oligonucleotides:
SB127 (24 mer) (SEQ ID NO 53)
5' GATCCGGAGGAGGAATACACACCC 3' and
SB128 (39 mer) (SEQ ID NO 54)
5' AATTTTGGATCCCTAAACCGGCCTGTCCTCAACAATCGG 3'
using the plasmid pAB129 as template and digested with
AspI and 'BamHI. The resulting plasmid, pSB029
(6161 bp), contains a truncated gC gene encoding a
protein of 428 residues.
6.3. Plasmids encoding the truncated form of EHV-1 gD
6.3.1. pAB131: gD gene (native form) cloned into the
vector pVR1012
The DNA fragment containing the open reading frame of
CA 02375320 2012-01-25
- 47 -
the EHV-1 gD gene was obtained by PCR with the aid of
the following oligonucleotides:
NS007 (33 mer) (SEQ ID NO 43)
5' TTGTCGACATGTCTACCTTCAAGCTTATGATGG 3' and
NS008 (32 mer) (SEQ ID NO 44)
5' TTGGATCCTTACGGAAGCTGGGTATATTTAAC 3'.
This PCR product was digested with Sall and BamHI
generating a fragment of 1214 bp, which was then
ligated into the vector pVR1012 (Example 1) linearized
by the same double digestion. The resulting plasmid,
pAB138 (6083 bp) contains the gene encoding the gD
glycoprotein of 402 residues.
6.3.2. pSB030: gD gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the EHV-1 gD protein is between residues 348
and 371. The plasmid containing the truncated form of
the gene encoding gD was obtained by ligation of the
following three DNA fragments: (a) the plasmid pVR1012
(Example 1) linearized by a double SalI-BamHI diges-
tion, (b) the fragment of 825 bp derived from the
digestion of pAB131 (Example 6.3.1.) with Sall and BsmI
and (c) a fragment of 239 bp obtained by PCR with the
aid of the following oligonucleotides:
SB129 (24 mer) (SEQ ID NO 55)
5' CGGTTTCTTGGTGAATTCAACTTC 3' and
SB130 (42 mer) (SEQ ID NO 56)
5' AATTTTGGATCCTTACGTAGAGTTGCTCTTAGACGTTTTTGG 3'
using the plasmid pAB131 as template and digested with
BsmI and BamHI. The resulting plasmid pSB030 (5921 bp),
contains a truncated gD gene encoding a protein of 347
residues.
CA 02375320 2012-01-25
- 48 -
Example 7: Plasmids encoding the various forms of the
EHV-4 glycoproteins
The genes encoding the gB, gC and gD glycoproteins of
the KYT445/2 strain of EHV-4 were isolated by PCR from
the purified viral DNA.
7.1. Plasmids encoding the various forms of EHV-4 gB
7.1.1. pAB136: gB gene (native form) cloned into the
vector pVR1012
The coding frame of the EHV-4 gB gene was amplified by
PCR with the. aid of the following oligonucleotides:
AB325 (35 mer) (SEQ ID NO 33)
5' TTTCTGCAGATGTCCACTTGTTGCCGTGCTATTTG 3' and
AB326 (31 mer) (SEQ ID NO 34)
5' TTTTCTAGATTAAACCATTTTTTCGCTTTCC 3'
the DNA fragment obtained was digested with PstI and
XbaI and ligated into the vector pVR1012 (Example 1)
linearized with PstI and XbaI, generating the plasmid
pAB136 (7801 bp). The EHV-4 gB gene encodes a protein
of 975 amino acids.
7.1.2. pSB025: gB gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the EHV-4 gB protein is positioned between
residues 797 and 867. The plasmid containing the
truncated form of the gene encoding gB was obtained by
the ligation of the following two DNA fragments: (a)
the plasmid pAB136 (Example 7.1.1.) digested with SplI
and XbaI, and (b) a fragment of 231 bp obtained by PCR
with the aid of the following oligonucleotides:
SB119 (36 mer) (SEQ ID NO 45)
5' TTTTGGTCTAGATTAGTCCACGTTGACAACGCTGTC 3' and
CA 02375320 2012-01-25
- 49 -
SB120 (23 mer) (SEQ ID NO 46)
5' CGCAAGCTTATCGAGCCGTGCGC 3'
using the plasmid pABl36 as template and digested with
SplI and XbaI. The resulting plasmid, pSB025 (7264 bp),
contains a truncated gB gene encoding a protein of 796
residues.
7.2. Plasmids encoding the various forms of EHV-4 gC
7.2.1. pAB137: gC gene (native form) cloned into the
vector pVR1012
The DNA fragment containing the open reading frame of
the EHV-4 gC gene was obtained by PCR with the aid of
the following oligonucleotides:
AB327 (32 mer) (SEQ ID NO 35)
5' TTTGTCGACATGGGTTTGGTAAATATAATGCG 3' and
AB328 (33 mer) (SEQ ID NO 36)
5' TTTGGATCCTTAGAAGTCTGCTTTCTTGTAGGG 3'.
This PCR product was digested with Sall and BamHI
generating a fragment of 1463 bp, which was then
ligated into the vector pVR1012 (Example 1) linearized
by the same double digestion. The resulting plasmid,
pAB137 (6330 bp), contains the gene encoding the gC
glycoprotein of 485 residues.
7.2.2. pSB026: gC gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the EHV-4 gC protein is between residues 425
and 472. The plasmid containing the truncated form of
the gene encoding gC was obtained by ligation of the
following two DNA fragments (a) the plasmid pAB137
(Example 7.2.1.) linearized by a double AclI-BamHI
digestion and (b) a fragment of 237 bp obtained by PCR
with the aid of the following oligonucleotides:
CA 02375320 2012-01-25
- 50 -
SB121 (24 mer) (SEQ ID NO 47)
5' GTATCAATCCCAGCTGACCCCGAC 3' and
SB122 (41 mer) (SEQ ID NO 48)
5' AATTTTGGATCCTTAGCCGTCCGGGTAACCCTCTATGATGC 3'
using the plasmid pAB137 as template and digested with
AclI and BamHI. The resulting plasmid, pSB026
(6147 bp),. contains a truncated gC gene encoding a
protein of 424 residues.
7.3. Plasmids encoding the truncated form of EHV-4 gD
7.3.1. pAB138: gD gene (native form) cloned into the
vector pVR1012
The DNA fragment containing the open reading frame of
the EHV-4 gD gene was obtained by PCR with the aid of
the following oligonucleotides:
AB329 (33 mer) (SEQ ID NO 37)
5' TTTGTCGACATGTCTACCTTCAAGCCTATGATG 3' and
AB330 (33 mer) (SEQ ID NO 38)
5' TTTGGATCCTTACGGAAGCTGAGTATATTTGAC 3'.
This PCR product was digested with Sall and BamHI
generating a fragment of 1214 bp, which was then
ligated into the vector pVR1012 (Example 1) linearized
by the same double digestion. The resulting plasmid,
pAB138 (6081 bp) contains the gene encoding the gD
glycoprotein of 402 residues.
7.3.2. pSB027: gD gene (ATM form) cloned into the
vector pVR1012
Depending on the hydropathy profile, the transmembrane
domain of the EHV-4 gD protein is between residues 348
and 371. The plasmid containing the truncated form of
the gene encoding gD was obtained by ligation of the
following two DNA fragments: (a) the plasmid pAB138
CA 02375320 2012-01-25
- 51 -
(Example 7.3.1.) linearized by a double EcoRI-BamHI
digestion and (b) a fragment of 310 bp obtained by PCR
with the aid of the following oligonucleotides:
SB123 (24 mer) (SEQ ID NO 49)
5' TTTTCCGTAACAATTCCGAGCAGC 3' and
SB124 (39 mer) (SEQ ID NO 50)
5' AATTTTGGATCCTTACGTAGAGTTGCTATTAGACGCTGG 3'
using the plasmid pAB138 as template and digested with
EcoRI and BamHI. The resulting plasmid, pSB027
(5919 bp), contains a truncated gD gene encoding a
protein of 347 residues.
Example 8: Plasmid encoding the canine GM-CSF
8.1. Preparation of the total RNA of dog lymphocytes
stimulated in vitro by mitogens
Dog blood was collected over a tube containing EDTA by
a blood collection made on a Beagle dog. The
mononucleated cells were harvested by centrifugation on
a Ficoll gradient, and then cultured on a Petri dish
60 mm in diameter. The dog mononucleated cells were
then stimulated with concanavalin A (ConA) (final
concentration of about 4 gg/ml) and with phyto-
haemagglutinin (PHA) (final concentration of about
10 ug/ml). After stimulation, the "ConA" and "PHA"
lymphoblasts were harvested by scraping the culture
dishes, and the total RNA of these cells was extracted
using the kit "mRNA isolation kit for White Blood
Cells" (Boehringer Mannheim/Roche Cat # 1 934 325).
8.2. Isolation of the gene encoding canine GM-CSF and
construction of the plasmid pJP074
The total RNA extracted from the dog lymphoblasts
stimulated by ConA or by PHA (Example 8.1.) served as
template for the synthesis of the complementary DNA
first strand. This complementary DNA first strand was
CA 02375320 2012-01-25
- 52 -
produced by extension of the oligonucleotide p(dT) 15
(Boehringer Mannheim/Roche Cat # 814 270). The single-
stranded complementary DNA obtained was then used as
template for a PCR reaction with the following
oligonucleotides:
JP578 (SEQ ID NO 64) (33 mer)
5' TATGCGGCCGCCACCATGTGGCTGCAGAACCTG 3'
and JP579 (SEQ ID NO 65) (36 mer)
5' TATGCGGCCGCTACGTATCACTTCTTGACTGGTTTC 3'
in order to amplify a PCR fragment of about 450 base
pairs (bp). This fragment was purified by agarose gel
electrophoresis, and then ligated into the vector
pCR2.1 (InVitrogen, Carlsbad, CA, USA) to give the
plasmid pJP074. The sequence of the canine GM-CSF gene
cloned in the plasmid pJP074 was found to be equivalent
to that of the canine GM-CSF sequence available on
GenBank (accession number # 549738).
8.3. Construction of the plasmid pJP084 and sequence of
the canine GM-CSF gene
The plasmid pJP074 (Example 8.2) was digested with NotI
in order to isolate, after agarose gel electrophoresis,
the NotI-NotI fragment of about 450 bp containing the
canine GM-CSF gene. This fragment was ligated into the
plasmid pVR1012 (Example 1). The clone containing the
canine GM-CSF sequence (SEQ ID NO 66, Figure No. 20) in
the correct orientation relative to the hCMV/IE
promoter was identified pJP084. This plasmid has a size
of 5364 bp (Figure No. 19).
Example 9: Plasmid encoding feline GM-CSF
9.1. Preparation of the total RNA of cat lymphocytes
stimulated in vitro by mitogens
Cat blood was collected over a tube containing EDTA by
a blood collection. The mononucleated cells were
CA 02375320 2012-01-25
- 53 -
harvested by centrifugation on a Ficoll gradient, and
then cultured on a Petri dish 60 mm in diameter. The
cat mononucleated cells were then stimulated with
concanavalin A (ConA) (final concentration of about
4 gg/ml) and with phytohaemagglutinin (PHA) (final
concentration of about 10 gg/ml). After stimulation,
the "ConA" and "PHA" lymphoblasts were harvested by
scraping the culture dishes, and the total RNA of these
cells was extracted using the kit "mRNA isolation kit
for White Blood Cells" (Boehringer Mannheim/Roche Cat #
1 934 325).
9.2. Isolation of the gene encoding feline GM-CSF and
construction of the plasmids pJP089 and pJP090
The total RNA extracted from the cat lymphoblasts
stimulated by ConA or by PHA (Example 9.1.) served as
template for the synthesis of the complementary DNA
first strand. This complementary DNA first strand was
produced by extension of the oligonucleotide p(dT) 15
(Boehringer Mannheim/Roche Cat #814 270). The single-
stranded complementary DNA obtained was then used as
template for a PCR reaction with the following
oligonucleotides:
JP578 (SEQ ID NO 64) (33 mer)
5' TATGCGGCCGCCACCATGTGGCTGCAGAACCTG 3'
and JP579 (SEQ ID NO 65) (36 mer)
5' TATGCGGCCGCTACGTATCACTTCTTGACTGGTTTC 3'
in order to amplify a PCR fragment of about 450 base
pairs (bp). This fragment was digested with NotI in
order to isolate, after agarose gel electrophoresis,
the NotI-NotI fragment of 450 bp. This fragment was
then ligated into the plasmid pVR1012 (Example 1). Two
clones containing the feline GM-CSF sequence (SEQ ID NO
67 and SEQ ID NO 68), in the correct orientation
relative to the hCMV/IE promoter were identified pJP089
and pJP090 respectively. These two plasmids have a size
of 5364 bp (Figures No. 21 and 23).
CA 02375320 2012-01-25
- 54 -
The sequence of the feline GM-CSF gene cloned into the
plasmid pJP089 contains 13 differences at the
nucleotide level with the feline GM-CSF sequence
available on GenBank (accession number AF053007). The
most important change is a C -* T change which causes a
Leucine -4 Phenylalanine change for the amino acid
(first base of the amino acid codon # 107; Figure
No. 22). The sequence of the feline GM-CSF gene cloned
into the plasmid pJP090 is equivalent to that contained
in the plasmid pJP089, except that the Leucine -*
Phenylalanine change does not exist for the amino acid
# 107 (Figure No. 24). Verification of the 3' sequence
of the feline GM-CSF gene by means of the 3' RACE kit
showed that, at this position 107, it is possible to
have, in the same cat, the amino acid Leucine or the
amino acid Phenylalanine.
Example 10: Plasmid encoding equine GM-CSF
10.1 Preparation of the total RNA of horse lymphocytes
stimulated in vitro by mitogens
Horse blood was collected over a tube containing EDTA
by a blood collection from the jugular vein. The
mononucleated cells were harvested by centrifugation on
a Ficoll gradient, and then cultured on a Petri dish
60 mm in diameter. The horse mononucleated cells were
then stimulated either with concanavalin A (ConA)
(final concentration of about 5 gg/ml) or with
phytohaemagglutinin (PHA) (final concentration of about
10 g/ml). After stimulation, the "ConA" and "PHA"
lymphoblasts were harvested by scraping the culture
dishes, and the total RNA of these cells was extracted
using the kit "mRNA isolation kit for White Blood
Cells" (Boehringer Mannheim/Roche Cat # 1 934 325).
CA 02375320 2012-01-25
- 55 -
10.2. Isolation of the gene encoding equine GM-CSF
The total RNA extracted from the horse lymphoblasts
stimulated by ConA or by PHA (Example 10.1.) served as
template for the synthesis of the complementary DNA
first strand. This complementary DNA first strand was
produced by extension of the oligonucleotide p(dT) 15
(Boehringer Mannheim/Roche Cat # 814 270). The single-
stranded complementary DNA obtained was then used as
template for a PCR reaction with the following
oligonucleotides:
JP734 (SEQ ID NO 70) (44 mer)
5' CATCATCATGTCGACGCCACCATGTGGCTGCAGAACCTGCTTCT 3'
and JP735 (SEQ ID NO 71) (41 mer)
5' CATCATCATGCGGCCGCTACTTCTGGGCTGCTGGCTTCCAG 3'
in order to amplify a PCR fragment of about 500 base
pairs (bp). This fragment was purified by agarose gel
electrophoresis.
10.3. Construction of the plasmid pJP097 and sequence
of the equine GM-CSF gene
The purified PCR fragment obtained in Example 10.2. was
digested with NotI in order to isolate, after agarose
gel electrophoresis, the NotI-NotI fragment of about
450 bp containing the equine GM-CSF gene. This fragment
was ligated into the plasmid pVR1012 (Example 1). The
clone containing the equine GM-CSF sequence (SEQ ID NO
69, Figure No. 26) in the correct orientation relative
to the hCMV/IE promoter was identified pJP097. This
plasmid has a size of 5334 bp (Figure No. 25).
CA 02375320 2012-01-25
-56-
a)
U
C
()
a)
In
* * * +
q N LD 0) N r=1 M 00 0) O LI) L.D N ro
N N M M H H H N N N N N M N (N N
C) 0 CD C) C) 0 0 0 0 Cl 0 0 0 0 CD 0
U) En U) CQ m m m m W M OQ CJ Oq Oq 00 W W
ro 2 ;4 Ul Ul U) U) En U) Cl) U) U) U) U) C)) U) Cl)
a a a a a aaaaaaaaaaaa 10
'UV'L7~Tl'OV'~broro'd'~bb'D
A 07 t~ G ro
ro ro ro ro ro ro ro ro ro ro ro ro ro ro ro ro C
N 0) CO Lf) d' N a CO (D N 0) N 0) H '..O C` CO
En N H H- M H M N N N N N M M M M ro
N O H H Cl 0 0 0 0 C) H r=1 r-1 p
w A'aaaaa'aaaaaaaaaa a)
ro
ate)
o ~"
0 E U") d' M [, rl r-1 O) O CM ['- C) CO [- l0 ~ [-
U O (D N N U) (D N C) lD O) N O N V' 0) N d' a) ro
r-I O U) 1 44 '-0 L.() ~' u, N V' M N d' M CO -l M N -qr M O) ~-1
z () ri 11
ro Cn
a)
41 0)
44
.,.{ l0 * [~ * O) N co d' l0 M lf) U) .-1 [- N .--I O
N* r=1 * l0 to N M N lf) N lf) N l0 N
lD Lil N V M CO U, M CO 10 M co mot' M
Q O O I 1 I I I I I I I I 1 1 U) N
N N O H L) CO .-1 O) CO N Ln CO .
O 1 N 1 O N H '.0 0) N CD N 0) N V' S-1 H
= ~D d' N 1' M ~' M CO .0 M C- d' M W 0 P4
U
H
x
~ ro
43 U)
O
44 0)
0
~w N N 0) U) CO a) U) ()crOCO N LnLnCN
O a) l0 CD N '0 1- Ln 'I' V' M N 00 %0 O N co CD
' kD lD Lr) Lf) 00 qi m 0) Ul r,) rn V' d+ a) ' zr
4-1 41
4) r1 O 44
O (L)
d) L) En a)
aa OH 2 Pq V A 00 U A m U A CQ V A C U
x x b) b) tr b) b) b) b) tr)
U) CO) U
'o =r1 Cn
=r1 4-1 -r1
.r1
al u b)
N r1 .-1 r-1 d' ri a rH
r ty) > I I 1 I I a U) -0
0 El)
41 U
ro ,A
04 41 .,A
L)
CA 02375320 2012-01-25
- 57 -
Example 12: Molecular biology methods
Culture and purification of the viruses
The viruses were cultured on appropriate cellular
systems until a cytopathic effect was obtained. The
cellular systems to be used for each virus are well
known to persons skilled in the art. Briefly, the
appropriate cells were infected with the viral strain
studied at a multiplicity of infection of one and were
incubated at 37 C for the time necessary to obtain a
cytopathic effect (on average 36 hours).
In the case of the DNA viruses, after the culture, the
supernatant and the lysed cells were harvested and the
cellular debris was removed by centrifugation at 1000 g
and at 4 C for 10 minutes. The viral particles were
harvested by ultracentrifugation at 400,000 g and 4 C
for 1 hour. The pellets were taken up in a minimum
volume of buffer (10 mM Tris, 1 mM EDTA).
The RNA viruses were purified according to standard
purification techniques well known to persons skilled
in the art.
Extraction of the viral genomic DNA
The concentrated viral suspensions were treated with
proteinase K (100 mg/ml final) in the presence of
sodium dodecyl sulphate (SDS) (0.5% final) for 2 hours
at 37 C. The viral DNA was then extracted with the aid
of a phenol /chloroform mixture, and then precipitated
with two volumes of absolute ethanol at -20 C for
16 hours and then centrifuged at 10,000 g for 15
minutes at 4 C. The DNA pellets were dried, and then
taken up in a minimum volume of sterile ultrapure
water.
CA 02375320 2012-01-25
- 58 -
Isolation of viral genomic RNA
The genomic RNA of each virus was extracted using the
"guanidinium thiocyanate/phenol-chloroform" technique
described by P. Chomczynski and N. Sacchi (Anal.
Biochem. 1987. 162. 156-159).
Molecular biology techniques
All the constructions of plasmids were carried out
using the.. standard molecular biology techniques
described by Sambrook et al. (Molecular Cloning: A
Laboratory Manual. 2nd Edition. Cold Spring Harbor
Laboratory, Cold Spring Harbor, New York, 1989). All
the restriction fragments used for the present inven-
tion were isolated with the aid of the "Geneclean" kit
(BIO101 Inc., La Jolla, CA). For all the constructs,
the cloned.DNA fragments, as well as the junctions with
the expression vector, were sequenced by the Sanger
method (Sambrook et al., 1989).
PCR and RT-PCR
The oligonuclebtides specific to the genes or gene
fragments cloned were synthesized, some of them
containing, in some cases, at their 5' end, restriction
sites facilitating the cloning of the amplified
fragments. The reverse transcription (RT) reactions and
the polymerase chain reaction (PCR) were carried out
according to standard techniques (Sambrook et al.,
1989).
Large-scale purification of plasmids
The production, on the scale of about ten mg, of puri-
fied plasmids entering into the vaccinal compositions
was carried out by the caesium chloride-ethidium
bromide gradient method (Sambrook et al., 1989).
CA 02375320 2012-01-25
- 59 -
Example 13: Formulation of the vaccinal plasmids
The DNA solution containing one or more plasmids
according to Examples 2 to 10 is concentrated by
ethanolic precipitation as described in Sambrook et al.
(1989) . The DNA pellet is taken up in a 0.9% NaCl
solution so as to obtain a concentration of 1 mg/ml. A
0.75 mM DMRIE-DOPE solution is prepared by taking up a
lyophilisate of DMRIE-DOPE with an appropriate volume
of sterile H20-
The formation of the plasmid DNA-lipid complexes is
achieved by diluting, in equal parts, the 0.75 mm
DMRIE-DOPE solution with the DNA solution at 1 mg/ml in
0.9% NaCl. The DNA solution is gradually introduced,
with the aid of a seamed 26G needle, along the wall of
the vial containing the cationic lipid solution so as
to avoid the formation of foam. Gentle shaking is
carried out as soon as the two solutions have been
mixed. A composition comprising 0.375 mM of DMRIE-DOPE
and 500 pg/ml of plasmid is finally obtained.
It is desirable for all the solutions used to be at
room temperature for all the operations described
above. The DNA/DMRIE-DOPE complex formation is allowed
to take place at room temperature for 30 minutes before
immunizing the animals.
Example 14: Immunization of dogs against CDV
An injection of 2 ml by the subcutaneous or
intramuscular route repeated 28 days later. The total
mass of plasmid used during each immunization is 100,
500, 1000 or 2000 gg according to the vaccines. Persons
skilled in the art possess the knowledge necessary to
adjust the volume or the concentration as a function of
the plasmid dose required.
CA 02375320 2012-01-25
- 60 -
14.1. Virulent challenge
The challenge strain used corresponds to a ground
product of the spleen removed from a dog infected with
the canine distemper virus, strain Snyder Hill, and
diluted one hundred fold in PBS buffer. The dilution is
kept on crushed ice until used.
After general anaesthetic, the challenge strain,
diluted one hundred fold, is administered by the
intracranial route in a volume of 0.5 ml, 49 days after
the first injection.
14.2. Post-challenge clinical monitoring
This daily clinical monitoring was carried out for
21 days following the challenge and included, for each
dog, a clinical examination to detect possible clinical
signs of canine distemper and taking of rectal
temperature.
The clinical examination comprises:
- An observation of the general state of the
animal on a 4-point scale:
"good" with a score of 0, "apathy" with a score of 1,
"depression" with a score of 2 and "prostration" with a
score of 3. The death of the animal is equivalent to a
clinical score of 10.
- An evaluation of the oculonasal symptoms (test for
serous or purulent discharge, rhinitis and/or
conjunctivitis) . Conjunctivitis/rhinitis with a serous
nasal discharge is equivalent to a score of 1, that
with a purulent nasal discharge is equivalent to a
score of 2.
- An evaluation of the digestive symptoms (test for
signs of gastroenteritis). A mild gastroenteritis is
CA 02375320 2012-01-25
- 61 -
equivalent to a score of 1, a severe gastroenteritis to
a score of 2.
- An evaluation of the nervous symptoms (test for
myoclonias, convulsions and/or paralysis). Myoclonia is
equivalent to a score of 1, convulsion to a score of 2
and paralysis to a score of 3.
- The monitoring of the body temperature of the animal.
A temperature of less than or equal to 37.5 C is
equivalent to a score of 3, a temperature of between
37.5 C and 39.5 C is equivalent to a score of 0, a
temperature equal to 39.5 C or between 39.5 C and 40 C
is equivalent to a score of 1, a temperature equal to
40 C or between 40 C and 40.5 C is equivalent to a
score of 2, and finally a temperature greater than
40.5 C is equivalent to a score of 3.
Of the animals that succumbed to the challenge, the
spleen was removed in order to identify the canine
distemper virus by immunofluorescence.
An overall clinical score was calculated in the
following manner: for each clinical sign, a mean score
was established per group of animals over the
observation period and the total score is the sum of
the mean scores for the five clinical signs considered.
CA 02375320 2012-01-25
-62-
M
m N M co
4J >1 a)
00 bO U) 0 0 O O O
$4 A 0 +I +1 +1 -H -H
14 U1 M m 00 rn U1
r-I ~1 O O O r-1 r1
U
N
(U
41
~', U N '~ l0 co N k D
N N r N O
ri
1J U
A >,
U) 41
) r i ,r Ln Ln 111 Ln
0 410 1- 11
tJ H 0
~I
4-I 0
O
O
iJ - I I I I
I I I I I
N 0
I I I I
.H 41
>1
~ m I ~ ~ b1 tS1 ~ C31 ~ CJl
I CD CD
u A I O o O o 0 0 0 0
4,,,1 in to m L[1 1!1 in 111 in
iJ w x w
U)
w x ~' .~ 'd .O 'O Q
a1 a) a) 0) N a) N 0
'H "'i 4J
J 1 iJ
41
rts 0 rl .~
v z z z
z o 0 0 0 ro
v
rn b
r1 0 a a rp
-r-+
0 0 0
4
Id I Q Q I
U I W W W U)
H H H
1-I
rc: 0 a)
44 41
O -.-1
N co Ol 00 c -c' H r! -W
iJ E
H co 41 c h 0 = m c0/) m E l ) c01) U) col]
N a a a a a a a E
m v
cY.
CA 02375320 2012-01-25
-63-
0 O N N O, 0 M
0 U2 O (D' 0 O O r1 H
A
04
U) N N lD N d~
N .
ro a,
O O N I;v co
0 Ln
ri U N N H N O
r1 N
U
41
=,i
H -~v Ln Ln Ln LO VI Ln
ro
41 Ln (~l -1 (D C) CD
0
[I.
I 1 I I U I 1
0 I I I I I I I
I I I I 1 I
>1
U
U
U)
O Q C C D 0 o 0 0 C C C D
A 0 0 0 0 0 0 0 0 0 0 Co 0 0 0
LU In Ln Ln Ln 141 N Ln Ln Lo Ln Ln
Q1 N 0
(71 '7 N r1 N =r1 N N =N '~ 4J
.14 'rA -H
4J .,I 1J 4-) 11 ro
41 4J 4J
_rj 2 z z z ~, a 4J -W 0, z z aa)
0 0 o 0 o
ro
(a
0
P4 a a a a w
=~ 0 0 0 0 Q A
ro I I I I I I 2
~ W W W W W W ul
H H H H H H
0 A A A A (~ (~ N
4j
H 1 r1
00 O\ OD Ol 'V H -K:V cr d' -4 lp (~ -Li
0
N H N N N N N 00 N N N r-i f" I
O N O N O O 0 O O O 0 O O O L":
o a4 cn m ~n ~n cn cn a cn ~n m v~ v~
aaaaaaaaaaaaaa E
a v
CA 02375320 2012-01-25
- 64 -
The plasmids pNS016 and pNS017 inserting the native
gene M and the native gene N, respectively, were
constructed in the same manner as the plasmid pNS018.
It is surprising to observe that the protection result
obtained with optimized HA alone is greater than or
equal to the results obtained with optimized HA and F
or with optimized HA and F + native M and N.
It should be clearly understood that the invention
defined by, the appended claims is not limited to the
specific embodiments indicated in the description
above, but encompasses the variants which depart
neither from the scope nor the spirit of the present
invention.
CA 02375320 2002-04-09
1
SEQUENCE LISTING
<110> FISCHER, Laurent Jean-Charles
BARZU-LE ROUX, Simona
AUDONNET, Jean-Christophe Francis
<120> Improved pet DNA vaccine
<130> Improved pet DNA vaccine
<140> PCT/FROO/01592
<141> 08/06/2000
<160> 73
<170> Patentln Ver. 2.1
<210> 1
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 1
atgagcccac tcttacaaca a 21
<210> 2
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 2
tttcgcggat ccattaaagg aagagcgcct aaccg 35
<210> 3
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 3
tttcgcggat cccacaaagt atcaactagc 30
<210> 4
<211> 23
CA 02375320 2002-04-09
2
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 4
gggatttgct gccgatgcaa tag 23
<210> 5
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 5
tttacgcgtc gacttgggga cccttgattg ttc 33
<210> 6
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 6
tttacgcgtc gacctgtagg aaaaagaaga aggcat 36
<210> 7
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 7
ttggggaccc ttgattgttc 20
<210> 8
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
CA 02375320 2002-04-09
3
<400> 8
ctgtaggaaa aagaagaagg c 21
<210> 9
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 9
gatctgcagc acgtgtctag aggatatcga attcgcggcc 40
<210> 10
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 10
gatccgcggc cgcgaattcg atatcctcta gacacgtgct 40
<210> 11
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 11
ttctgcagat gctctcctac caagayaagg 30
<210> 12
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 12
ttgtcgacat gtgtatcatc atmctgtc 28
<210> 13
<211> 26
<212> DNA
<213> Artificial Sequence
CA 02375320 2002-04-09
4
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 13
tttctagaca gccgagcccc atgcac 26
<210> 14
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 14
ttggatccga tatatgacca gaatacttca 30
<210> 15
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 15
tatattgaag gacaacttgg gg 22
<210> 16
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 16
ctagtctaga ttaattatta tcaactttta caacac 36
<210> 17
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 17
cgagaaactt gttatttttc taaag 25
CA 02375320 2002-04-09
<210> 18
<211> 51
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 18
ataagaatgc ggccgcaaag gctatatatt ttttggggta ttatttattg g 51
<210> 19
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 19
aaaactgcag atgagtttta aaaattttta tc 32
<210> 20
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 20
ctagtctaga ttagatctta ttattttttg 30
<210> 21
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 21
tggattgacg gtcttataac acgc 24
<210> 22
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
CA 02375320 2002-04-09
6
<400> 22
ctagtctaga ttaattttca tccgatgcat caaacac 37
<210> 23
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 23
ctgtggacag agaccctaaa actc 24
<210> 24
<211> 50
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 24
tttccttttg cggccgctta tatgctgtct atatcataaa attttaaggc 50
<210> 25
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 25
gatcgtcccg ccataccgtc tggg 24
<210> 26
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 26
tttggaagat ctttactgat tattcatgcc cttgggagg 39
<210> 27
<211> 34
<212> DNA
<213> Artificial Sequence
CA 02375320 2002-04-09
7
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 27
ttttctgcag atgtccactc gtggcgatct tggg 34
<210> 28
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 28
atagtttagc ggccgcttag acaagatttg tttcagtatc 40
<210> 29
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 29
ttttctgcag atgagacgat ataggatggg acgc 34
<210> 30
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 30
agtttagcgg ccgcttataa tcgccgggga tgag 34
<210> 31
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 31
gttaaatgtg taccacggga cggg 24
CA 02375320 2002-04-09
8
<210> 32
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 32
agtttagcgg ccgcttattc aggggacgcg tcgtagactt g 41
<210> 33
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 33
tttctgcaga tgtccacttg ttgccgtgct atttg 35
<210> 34
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 34
ttttctagat taaaccattt tttcgctttc c 31
<210> 35
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 35
tttgtcgaca tgggtttggt aaatataatg cg 32
<210> 36
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
CA 02375320 2002-04-09
9
<400> 36
tttggatcct tagaagtctg ctttcttgta ggg 33
<210> 37
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 37
tttgtcgaca tgtctacctt caagcctatg atg 33
<210> 38
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 38
tttggatcct tacggaagct gagtatattt gac 33
<210> 39
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 39
ttctgcagat gtcctctggt tgccgttcgt 30
<210> 40
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 40
tttctagatt aaaccatttt ttcattttcc 30
<210> 41
<211> 31
<212> DNA
<213> Artificial Sequence
CA 02375320 2002-04-09
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 41
ttgtcgacat gtggttgcct aatctcgtga g 31
<210> 42
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 42
ttggatccct aaaagtcaga cttcttgtac ggc 33
<210> 43
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 43
ttgtcgacat gtctaccttc aagcttatga tgg 33
<210> 44
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 44
ttggatcctt acggaagctg ggtatattta ac 32
<210> 45
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 45
ttttggtcta gattagtcca cgttgacaac gctgtc 36
CA 02375320 2002-04-09
11
<210> 46
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 46
cgcaagctta tcgagccgtg cgc 23
<210> 47
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 47
gtatcaatcc cagctgaccc cgac 24
<210> 48
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 48
aattttggat ccttagccgt ccgggtaacc ctctatgatg c 41
<210> 49
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 49
ttttccgtaa caattccgag cagc 24
<210> 50
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
CA 02375320 2002-04-09
12
<400> 50
aattttggat ccttacgtag agttgctatt agacgctgg 39
<210> 51
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 51
aacaacagag ggtcgataga aggc 24
<210> 52
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 52
aatttttcta gattacacgt tgaccacgct gtcgatgtc 39
<210> 53
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 53
gatccggagg aggaatacac accc 24
<210> 54
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 54
aattttggat ccctaaaccg gcctgtcctc aacaatcgg 39
<210> 55
<211> 24
<212> DNA
<213> Artificial Sequence
CA 02375320 2002-04-09
13
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 55
cggtttcttg gtgaattcaa cttc 24
<210> 56
<211> 42
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 56
aattttggat ccttacgtag agttgctctt agacgttttt gg 42
<210> 57
<211> 38
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 57
aaaaacgcgt cgacatgggt actataattc aatttctg 38
<210> 58
<211> 38
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial sequence:
oligonucleotide
<400> 58
ttttctagtc tagattattt atgataaaca aaattctc 38
<210> 59
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 59
ttttctagtc tagattagta tgtgtcactt tgtgctaagt g 41
CA 02375320 2002-04-09
14
<210> 60
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 60
aaaaacgcgt cgacatggtt gcagaagatg cccctgttag g 41
<210> 61
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 61
ttttggaaga tctttaggat agtgtcacct gacgg 35
<210> 62
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 62
ttaaaagaat tcgacccaaa agcaaatcat gagccac 37
<210> 63
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 63
ttaaaaggcc tttaggatag tgtcacctga cgg 33
<210> 64
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
CA 02375320 2002-04-09
<400> 64
tatgcggccg ccaccatgtg gctgcagaac ctg 33
<210> 65
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 65
tatgcggccg ctacgtatca cttcttgact ggtttc 36
<210> 66
<211> 435
<212> DNA
<213> Canis sp.
<400> 66
atgtggctgc agaacctgct tttcttgggc actgtggtct gcagcatctc tgcacccacc 60
cgctcaccca cccttgtcac tcggccctct cagcacgtgg atgccatcca ggaagccctg 120
agccttttga acaacagtaa tgacgtgact gctgtgatga ataaagcagt aaaagtggtc 180
tctgaagtgt ttgaccctga ggggccaaca tgcctggaga cccgcctaca gctgtacaag 240
gagggcctgc agggcagcct caccagcctc aagaatccct taaccatgat ggccaatcac 300
tataagcagc actgtccccc taccccggaa tctccctgtg caacccagaa tattaacttc 360
aaaagtttca aagagaacct gaaggatttt ctgtttaaca tcccctttga ctgctggaaa 420
ccagtcaaga agtga 435
<210> 67
<211> 435
<212> DNA
<213> Felis catus
<400> 67
atgtggctgc agaacctgct tttcctgggc actgtggtct gcagcatctc tgcacccacc 60
agttcaccca gctctgtcac tcggccctgg caacacgtgg atgccatcaa ggaggctctg 120
agccttctga acaacagtag tgaaataact gctgtgatga atgaagcagt agaagtcgtc 180
tctgaaatgt ttgaccctga ggagccgaaa tgcctgcaga ctcacctaaa gctgtacgag 240
cagggcctac ggggcagcct catcagcctc aaggagcctc tgagaatgat ggccaaccat 300
tacaagcagc actgcccctt tactccggaa acgccctgtg aaacccagac tatcaccttc 360
aaaaatttca aagagaatct gaaggatttt ctgtttaaca tcccctttga ctgctggaaa 420
ccagtcaaga agtga 435
<210> 68
<211> 435
<212> DNA
<213> Felis catus
<400> 68
atgtggctgc agaacctgct tttcctgggc actgtggtct gcagcatctc tgcacccacc 60
agttcaccca gctctgtcac tcggccctgg caacacgtgg atgccatcaa ggaggctctg 120
agccttctga acaacagtag tgaaataact gctgtgatga atgaagcagt agaagtcgtc 180
tctgaaatgt ttgaccctga ggagccgaaa tgcctgcaga ctcacctaaa gctgtacgag 240
CA 02375320 2002-04-09
16
cagggcctac ggggcagcct catcagcctc aaggagcctc tgaggatgat ggccaaccat 300
tacaagcagc actgccccct tactccggaa acgccctgtg aaacccagac tatcaccttc 360
aaaaatttca aagagaatct gaaggatttt ctgtttaaca tcccctttga ctgctggaaa 420
ccagtcaaga agtga 435
<210> 69
<211> 435
<212> DNA
<213> Equus sp.
<400> 69
atgtggctgc agaacctgct tcttctgggc actgtggttt acagcatgcc cgcacccacc 60
cgccaaccca gccctgtcac tcggccctgg cagcatgtgg atgccatcaa ggaggccctg 120
agccttctga acaacagtag tgacactgct gctatcatga atgaaacagt agaagtcgtc 180
tctgaaacgt ttgacgccga ggagctgaca tgcctgcaga ctcgcctgaa gctgtacaaa 240
cagggcttgc ggggcagcct catcaagctc gaaggcccct tgaccatgat ggccagccac 300
tacaagcagc actgcccccc caccctggaa acttcctgtg caacccagat gatcaccttc 360
aaaagtttca aaaagaacct gaaggatttt ctgtttgaga tcccgtttga ctgctggaag 420
ccagcccaga agtaa 435
<210> 70
<211> 44
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 70
catcatcatg tcgacgccac catgtggctg cagaacctgc ttct 44
<210> 71
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
oligonucleotide
<400> 71
catcatcatg cggccgctac ttctgggctg ctggcttcca g 41
<210> 72
<211> 1590
<212> DNA
<213> canine parainfluenza virus
<400> 72
atgggtacta taattcaatt tctggtggtc tcctgtctat tggcaggagc aggcagcctt 60
gatctagcag ccctcatgca aatcggtgtc attccaacaa atgtccggca acttatgtat 120
tatactgagg cctcatcagc attcattgtt gtgaagttaa tgcctacaat tgactcgccg 180
attagtggat gtaatataac atcaatttca agctataatg caacagtgac aaaactccta 240
cagccgatcg gtgagaattt ggaaacgatt aggaaccagt tgattccaac tcggaggaga 300
cgccggtttg caggggtggt gattggatta gctgcattag gagtagctac tgccgcacag 360
CA 02375320 2002-04-09
17
gtcactgccg cagtagcact agtaaaggca aataaaaatg ctgcggctat actcaatctc 420
aaaaatgcaa tccaaaaaac aaatacagca gttgcagatg tggtccaggc cacacaatca 480
ctaggaacgg cagttcaagc agttcaagat cacataaaca gtgtggtaag tccagcaatt 540
acagcagcca attgtaaggc ccaagatgct atcattggct caatcctcaa tctctatttg 600
accgagttga caactatctt ccacaatcaa attacaaacc ctgcattgag tcctattaca 660
attcaagctt taaggatcct actggggagt accttgccga ctgtggtcga aaaatctttc 720
aatacccaga taagtgcagc tgagcttctc tcatcagggt tattgacagg ccagattgtg 780
ggattagatt tgacctatat gcagatggtc ataaaaattg agctgccaac tttaactgta 840
caacctgcaa cccagatcat agatctggcc accatttctg cattcattaa caatcaagaa 900
gtcatggccc aattaccaac acgtgttatt gtgactggca gcttgatcca agcctatccc 960
gcatcgcaat gcactattac acccaacact gtgtactgta ggtataatga tgcccaagta 1020
ctctcagatg atacgatggc ttgcctccaa ggtaacttga caagatgcac cttctctcca 1080
gtggttggga gctttctcac tcgattcgtg ctgttcgatg gaatagttta tgcaaattgc 1140
aggtcgatgt tatgcaagtg catgcagcct gctgctgtga tcctacagcc gagttcatcc 1200
cctgtaactg tcattgacat gtacaaatgt gtgagtctgc agcttgacaa tctcagattc 1260
accatcactc aattggccaa tgtaacctac aatagcacca tcaagcttga aacatcccag 1320
atcttgccta ttgatccgtt ggatatatcc cagaatctag ctgcggtgaa taagagtcta 1380
agtgatgcac tacaacactt agcacaaagt gacacatacc tttctgcaat cacatcagct 1440
acgactacaa gtgtattatc cataatggca atctgtcttg gatcgttagg tttaatatta 1500
ataatcttgc tcagtgtagt tgtgtggaag ttattgacca ttgtcactgc taatcgaaat 1560
agaatggaga attttgttta tcataaataa 1590
<210> 73
<211> 1698
<212> DNA
<213> canine parainfluenza virus
<400> 73
atggttgcag aagatgcccc tgttaggggc acttgccgag tattatttcg aacaacaact 60
ttaatttttc tatgcacact actagcatta agcatctcta tcctttatga gagtttaata 120
acccaaaagc aaatcatgag ccacgcaggc tcaactggat ctaattctag attaggaagt 180
atcactgatc ttcttaataa tattctctct gtcgcaaatc agattatata taactctgca 240
gtcgctctac ctctacaatt ggacactctt gaatcaacac tccttacagc cattaagtct 300
cttcaaacca gtgacaagct agaacagaac tgctcgtggg gtgctgcact gattaataat 360
aatagataca ttaatggcat caatcagttc tatttctcaa ttgctgaggg tcgcaatctg 420
acacttggcc cacttcttaa tatacctagt ttcattccaa ctgccacgac accagagggc 480
tgcaccagga tcccatcatt ctcgctcacc aagacacact ggtgttatac acacaatgtt 540
atcctgaatg gatgccagga tcatgtatcc tcaaatcaat ttgtttccat gggaatcatt 600
gaacccactt ctgccgggtt tccatccttt cgaaccctaa agactctata tctcagcgat 660
ggggtcaatc gtaagagctg ctctatcagt acagttccgg ggggttgtat gatgtactgt 720
tttgtctcta ctcaaccaga gagggatgac tacttttcta ccgctcctcc agaacaacga 780
attattataa tgtactataa tgatacaatc gtggagcgca taattaatcc acccggggta 840
ctagatgtat gggcaacatt gaccccagga acaggaagcg gggtatatta tttaggttgg 900
gtgctctttc caatatatgg cggcgtgatt aaagatacga gtttatggaa taatcaagca 960
aataaatact ttatccccca gatggttgct gctctctgct cacaaaacca ggcaactcaa 1020
gtccaaaatg ctaagtcatc atactatagc agctggtttg gcaatcgaat gattcagtct 1080
gggatcctgg catgtcctct tcaacaggat ctaaccaatg agtgtttagt tctgcccttt 1140
tctaatgatc aggtgcttat gggtgctgaa gggagattat acatgtatgg tgactcggtg 1200
tattactacc aaagaagcac tagttggtgg cctatgacca tgctgtataa ggtaaccata 1260
acattcacta atggtcagcc atctgctata tcagctcaga atgtgcccac acagcaggtc 1320
cctagacctg ggacaggagg ctgctctgca acaaatagat gtcccggttt ttgcttgaaa 1380
ggagtgtatg ctgatgcctg gttactgacc aacccttcgt ctaccagtac atttggatca 1440
gaagcaacct tcactggttc ttatctcaac gcagcaactc agcgtatcaa tccgacgatg 1500
tatatcgcga acaacacaca gatcataagc tcacagcaat ttggatcaag cggtcaagaa 1560
gcagcatata gccacacaac ttgttttagg gacacaggct ctgttatggt atactgtatc 1620
tatattattg aattgtcctc atctctctta ggacaatttc agattgtccc atttatccgt 1680
caggtgacac tatcctaa 1698