Note: Descriptions are shown in the official language in which they were submitted.
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
CRYSTALLIZATION AND STRUCTURE DETERMINATION OF
STAPHYLOCOCCUSAUREUS ELONGATION FACTOR P
This application claims the benefit of U.S. Provisional
Application Serial~No. 60/147,851 filed 6 August 1999, which is incorporated
herein by reference in its entirety.
FIELD OF THE INVENTION
1 S This invention relates to the crystallization and structure
determination of Staphylococcus aureus elongation factor P (S aureus EF-P).
BACKGROUND OF THE INVENTION
Translation is fundamental to the biochemical and cellular
process of all cells; therefore, it is not surprising that many antibacterial
agents
target this process. Preparation for translation begins with the binding of
the
mRNA to the ribosome placing the first codon, AUG, in position for interaction
with the fMet-tRNA. Translation is initiated with the binding of the fMet-tRNA
to the 30S subunit in the P site. Subsequently, the second tRNA is transported
to the ribosome via the GTP dependent elongation factor-TU which situates the
tRNA in the A site enabling the first peptide bond to be synthesized. After
synthesis, the newly free tRNA localizes to the E site while the tRNA
containing the growing amino acid chain moves to the P site vacating the A
site
for the next aminoacyl-tRNA. This translocation step is catalyzed by the GTP
dependent elongation factor G.
Several decades ago it was observed by Ganoza that another
purified factor, EF-P, could increase the rate of formation of the first
peptide
bond as demonstrated in a model system by its stimulation of the synthesis of
N
formylmethionyl-puromycin from fMet-tRNA and puromycin which serves as a
mimic of an aminoacyl tRNA (M.C. Ganoza et al., Eur. J. Biochem; 146:287-94
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
(1985); B.R. Glick & M.C. Ganoza, Proc. Natl. Acad. Sci. U.S.A.; 72:4257-60
(1975)). The precise mechanism for stimulation by elongation factor P has not
yet been determined, although experiments have shown a selectivity of EF-P for
the stimulation of peptide bond synthesis with small to medium amino acids
such as Gly and Leu rather than larger amino acids such as Phe, Met, and Lys
(B.R. Glick et al., Eur. J. Biochem; 97:23-28 (1979)). The gene for EF-P from
Escherichia coli has been cloned (H. Aoki et al., Nucl. Acid Res; 19:6215-20
(1991)) and it has been shown to be essential (H. Aoki et al., J. Biol. Chem.
272:32254-59 (1997)). The quantities of EF-P within E. coli are about one EF-
P molecule per ten ribosomes suggesting that it plays a catalytic role in
translation (G. An, Can. J. Biochem; 58:1312-14 (1980)). This 193 amino acid
protein has homologs throughout bacteria and eukaryotes, although the
sequence identity with higher organisms is quite low.
SUMMARY OF THE INVENTION
In one aspect, the present invention provides a method of
crystallizing an S. aureus EF-P molecule or molecular complex that includes
preparing purified S. aureus EF-P at a concentration of about 1 mg/ml to about
50 mg/ml; and crystallizing S. aureus EF-P from a solution including about 0
wt. % to about 50 wt. % polyethylene glycol and 0 to about 20 wt. % DMSO,
and buffered to a pH of about 3.5 to about 5.5.
In another aspect, the present invention provides crystalline
forms of S. aureus EF-P. In one embodiment, a crystal of S. aureus EF-P is
provided having the orthorhombic space group symmetry P2,2,21.
In another aspect, the present invention provides a scalable three
dimensional configuration of points wherein at least a portion of the points
are
derived from structure coordinates of a least a portion of an S. aureus EF-P
molecule or molecular complex listed in Figure 4, preferably comprising amino
acids Val 29, Lys30, Pro3l, G1y32, Lys 33, Gly 34, Ser 35, and Ala 36. In one
2
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
embodiment, at least a portion of the points are derived from S aureus EF-P
structure coordinates representing the locations of at least the backbone
atoms
of amino acids defining an S. aureus EF-P or EF-P-like binding surface, the
binding surface comprising amino acids selected from the surface residues
listed
in Table 1. In another embodiment, at least a portion of points are derived
from
S. aureus EF-P structure coordinates representing the backbone atoms of amino
acids within 4 ~, preferably within 7 ~, more preferably within 10 ~, and most
preferably within 15 ~ of Lys 33, as shown in Table 2. In another aspect, the
present invention provides a scalable three dimensional configuration of
points
with at least a portion of the points derived from structure coordinates of at
least
a portion of a molecule or a molecular complex that is structurally homologous
to an S. aureus EF-P molecule or molecular complex, On a molecular scale,
with points derived from a molecule or molecular complex preferably have a
root mean square deviation of less than about 1.9 A from the structure
coordinates.
In another aspect, the present invention provides a molecule or
molecular complex that includes at least a portion of an S. aureus EF-P
binding
surface. In one embodiment the binding surface comprises amino acids selected
from the surface residues listed in Table 1. In one embodiment, the binding
surface is further defined by a set of points having a root mean square
deviation
of less than about 1.9 A from points representing the backbone atoms of amino
acids Val 29, Lys30, Pro3l, G1y32, Lys 33, Gly 34, Ser 35, and Ala 36 as
represented by the structure coordinates listed in Figure 4. In another
embodiment, the binding surface is further defined by a set of points having a
root mean square deviation of less than about 1.9 A from points representing
the
backbone atoms of the amino acids that are within 4 ~ of Lys33, preferably
within 7 ~ of Lys33, more preferably within 10 A of Lys33, and most
preferably within 15 A of Lys33, as shown in Table 2 and represented by the
structure coordinates listed in Figure 4.
3
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
TABLE 1
Identified Surface Residues for S. aureus EF-P
GLY 2 GLY 58 TYR 105 TYR 159
ILE 3 GLU 59 LEU 106 THR 160
SER 4 LYS 60 LYS 107 LEU 161
VAL 5 VAL 61 GLU 108 ASN 162
ASN 6 GLU 62 GLY 109 VAL 163
ASP 7 PRO 63 MET 110 PRO 164
PHE 8 ALA 64 GLU 111 LEU 165
LYS 9 MET 65 VAL 112 PHE 166
THR 10 ILE 66 GLN 113 ASN 168
GLY 11 GLU 67 ILE 114 GLU 169
LEU 12 ASN 68 GLN 115 GLY 170
THR 13 ARG 69 THR 116 ASP 171
ILE 14 ARG 70 TYR 117 VAL 172
SER 15 MET 71 GLU 118 ILE 174
ALA 19 GLN 72 GLY 119 ASN 176
ILE 20 TYR 73 GLU 120 THR 177
TRP 21 LEU 74 THR 121 GLY 178
LYS 22 TYR 75 ILE 122 ASP 179
ILE 24 ALA 76 GLY 123 GLY 180
ASP 25 ASP 77 VAL 124 SER 181
PHE 26 GLY 78 GLU 125 TYR 182
GLN 27 ASP 79 LEU 126 ILE 183
HIS 28 ASN 80 PRO 127 SER 184
VAL 29 HIS 81 LYS 128 ARG 185
LYS 30 VAL 82 THR 129 GLY 186
PRO 31 MET 84 VAL 130
GLY 32 ASP 85 GLU 131
LYS 33 ASN 86 LEU 132
GLY 34 GLU 87 THR 133
SER 35 SER 88 VAL 134
ALA 36 PHE 89 THR 135
PHE 37 GLU 90 GLU 136
ARG 39 GLN 91 THR 137
SER 40 THR 92 GLU 138
LYS 41 GLU 93 PRO 139
LEU 41 LEU 94 GLY 140
ARG 43 SER 95 ALA 149
ALA 49 SER 96 THR 150
ILE 50 ASP 97 LYS 151
GLN 51 TYR 98 SER 152
GLU 52 LEU 99 ALA 153
LYS 53 LYS 100 THR 154
THR 54 GLU 101 VAL 155
PHE 55 GLU 102 GLU 156
ARG 56 LEU ~ 103 THR~ 157
I
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
ALA 57 ASN 104 GLY 158
Table 2.
Residues that are near Lys33 in S. aureus EF-P
Atoms with in 4 A of Lys 33
PRO 31
GLY 32
GLY 34
Atoms within 7 t~ of Lys 33
LYS 30
PRO 31
GLY 32
GLY 34
SER 35
PHE 37
ARG 56
Atoms within 10 t~ of Lys 33
VAL 29
LYS 30
3 0 PRO 31
GLY 32
GLY 34
SER 35
ALA 36
PHE 37
ARG 56
Atoms within 15 A of Lys 33
VAL 5
ASN 6
GLN 27
HIS 28
VAL 29
LYS 30
PRO 31
GLY 32
GLY 34
SER 35
ALA 36
PHE 37
VAL 38
ARG 39
THR 54
5
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
PHE 55
ARG 56
ALA 57
GLY 58
GLU 59
In another aspect, the present invention provides molecules or
molecular complexes that are structurally homologous to an S. aureus EF-P
molecule or molecular complex.
In another aspect, the present invention provides a machine
readable storage medium including the structure coordinates of all or a
portion
of an S. aureus EF-P molecule, molecular complex, a structurally homologous
molecule or complex, including structurally equivalent structures, as defined
herein, particularly a binding surface thereof, or a similarly shaped
homologous
binding surface. A storage medium encoded with these data is capable of
displaying on a computer screen, or similax viewing device, a three-
dimensional
graphical representation of a molecule or molecular complex which comprises a
binding surface or a similarly shaped homologous binding surface.
In another aspect, the present invention provides a method for
identifying inhibitors, ligands, and the like for an S. aureus EF-P molecule
by
providing the coordinates of a molecule of S. aureus EF-P to a computerized
modeling system; identifying chemical entities that are expected to bind to or
interfere with the molecule (e.g., screening a small molecule library); and,
optionally, procuring or synthesizing then assaying the compounds or analogues
derived therefrom for bioactivity. In another aspect, the present invention
provides methods for designing inhibitors, ligands, and the like by providing
the
coordinates of a molecule of S. aureus EF-P to a computerized modeling
system; designing a chemical entity that is likely to bind to or interfere
with the
molecule; and optionally, synthesizing the chemical entity and assaying the
chemical entity for bioactivity. In another aspect, the present invention
provides
inhibitors and ligands designed or identified by the above method. In one
embodiment, a composition is provided that includes an inhibitor or ligand
6
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
designed or identified by the above method. In another embodiment, the
composition is a pharmaceutical composition.
In another aspect, the present invention provides a method
involving molecular replacement to obtain structural information about a
molecule or molecular complex of unknown structure. The method includes
crystallizing the molecule or molecular complex, generating an x-ray
diffraction
pattern from the crystallized molecule or molecular complex, and applying at
least a portion of the EF-P structure coordinates set forth in Fig. 4 to the x-
ray
diffraction pattern to generate a three-dimensional electron density map of at
least a portion of the molecule or molecular complex.
1 S In another aspect, the present invention provides a method for
homology modeling an S. aureus EF-P homolog.
DEFINITIONS
Two crystallographic data sets (with structure factors F) are
considered isomorphous if, after scaling,
OF ~~F,-F2~
F ~ F,
is less than about 35% for the reflections between 8 A and 4 A.
ABBREVIATIONS
The following abbreviations are used throughout this disclosure:
Staphylococcus aureus elongation factor P (S. aureus EF-P)
Isopropylthio-(3-n-galactoside (IPTG).
Dithiothreitol (DTT).
Dimethyl sulfoxide (DMSO).
Multiple anomalous dispersion (MAD).
Polyethylene glycol (PEG)
7
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
The following amino acid abbreviations are used throughout this
disclosure:
A = Ala = Alanine I T = Thr = Threonine
V = = Valine C Cys Cysteine
Val = =
L = = Leucine Y Tyr Tyrosine
Leu = =
I Ile Isoleucine N Asn = Asparagine
= = =
P = Pro = Proline Q = Gln = Glutamine
F = Phe = Phenylalanine D = Asp = Aspartic Acid
W = Trp = Tryptophan E = Glu = Glutamic Acid
M = Met = Methionine K = Lys = Lysine
G = Gly = Glycine R = Arg = Arginine
S = Ser = Serine H = His = Histidine
BRIEF DESCRIPTION OF THE FIGURES
Figure 1 shows a crystal of S. aureus elongation factor P grown
in 0.1 M sodium acetate pH 5.4, 1 % PEG 4000. The width of the crystal is
approximately 0.350 mm.
Figure 2 shows anomalous difference Patterson maps for
selenomethionine EF-P at the inflection point of the selenium K edge (7~ _
0.979746 t~, 10-1.9A resolution). a) x Hacker section, b) y Hacker section,
and
c) z Hacker section.
Figure 3 shows electron density maps for EF-P for residues 73
77 for a) multiple anomalous dispersion map after solvent flattening
calculated
to 2.3 t~ and b) the final 2Fo F~ map calculated to 1.9 A resolution.
8
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Figure 4 lists the atomic structure coordinates for S. aureus EF-P
as derived by x-ray diffraction from a crystal of that complex. The following
abbreviations are used in Figure 1:
"Atom" refers to the element whose coordinates are measured.
The second column defines the number of the atom in the structure. The letters
in the third column define the element. The fourth and fifth columns define
the
amino acid and the number of the amino acid in the structure, respectively.
"X, Y, Z" crystallographically define the atomic position of the
element measured.
"Occ" is an occupancy factor that refers to the fraction of the
molecules in which each atom occupies the position specified by the
coordinates. A value of " 1 " indicates that each atom has the same
conformation, i.e., the same position, in all molecules of the crystal.
"B" is a thermal factor that measures movement of the atom
around its atomic center.
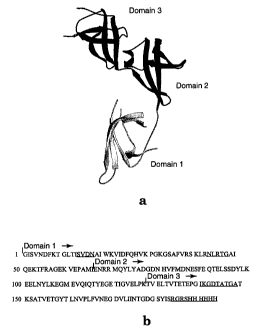
Figure 5 shows structure and sequence of S. aureus EF-P by a)
ribbon representation of the X-ray crystal structure of S. aureus EF-P, and b)
the amino acid sequence of recombinant EF-P (SEQ ID NO:1, including a Hiss
tag) with the beginning of each domain indicated by an arrow. Disordered
residues are underlined.
Figure 6 shows a secondary structure diagram for S. aureus EF-
P.
Figure 7 shows conserved secondary and tertiary structure
between domains 2 and 3 of EF-P. Two views (a and b) of a superposition of
domain 2 and domain 3 from S. aureus elongation factor P are shown.
Figure 8 shows two stereoviews (a and b) of EF-P.
Figure 9 shows a surface representation of S. aureus EF-P.
Alternative views (a and b) of the surface charge density (180°
apart) of
elongation factor P are shown.
Figure 10 shows a hypothetical superposition of EF-P and
9
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
tRNA~'" from E. coli (tRNA from M.A. Rould et al., Science 246:1135-42
(1989), PDB access code lgtr). a) EF-P is oriented with domain 3 at the
anticodon stem and domain 1 at the acceptor stem. b) Another possible
orientation of EF-P with domain 1 at the anticodon stem and domain 3 at the
acceptor stem.
Figure 11 shows a structural comparison and sequence alignment
of EF-P homologs. The three solved structures from a) S. aureus, b)
Methanococcus jannaschii, and c) Pyrobaculum aerophilum are shown. d)
Sequence alignment of recombinant S. aureus EF-P (SEQ ID NO:1, which
includes a His6 tag not present in the compared sequences) with four EF-P
homologs: EF-P from E. coli (SEQ ID N0:2), eIFSA from Methanococcus
jannaschii (SEQ ID N0:3), IFSA from Pyrobaculum aerophilum (SEQ ID
N0:4), and eIFSa from humans (SEQ ID NO:S). Identical residues have been
shaded or boxed.
Figure 12 shows a superposition of S aureus EF-P (dark) and
eIFSA from Methanococcus jannaschii (light). a) Alignment of domain 1
(residues 2-15, 19-33, 34-43, and 49-65 from S. aureus EF-P and residues 10-
23, 27-41, 43-52, 58-74 from M. jannaschii). b) Alignment of domain 2
(residues 66-95 and 99-128 from S. aureus EF-P and residues 75-103 and 104-
132 from M. jannaschii).
Figure 13 shows the superposition of S. aureus EF-P (dark) and
eIFSA from Pyrobaculum aerophilum (light). a) Alignment of domain 1
(residues 2-15, 19-33, 34-43, and 49-65 from S. aureus EF-P and residues 11-
24, 28-42, 44-53, and 59-75 from P. aerophilum). b) Alignment of domain 2
(residues 66-95 and 99-128 from S. aureus EF-P and residues 76-105 and 109-
139 from P. aerophilum).
Figure 14 shows certain residues of interest in S. aureus EF-P.
Lys33 is the proposed site for post-translational modification based on the
hypusine modification found in EF-P homologs in eukaryotic systems.
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Figure 15 lists the structure factors and multiple anomalous
dispersion phases for the crystal structure of S. aureus EF-P (SEQ ID NO:1 ).
"INDE" refers to the indices h, k, and 1 (columns 2, 3, and 4 respectively) of
the
lattice planes. "FOBS" refers to the structure factors of the observed
reflections.
"SIGMA" is the standard deviation for the observations. "PHAS" refers to the
phase used for the observations. "FOM" refers to the figure of merit.
11
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
DETAILED DESCRIPTION OF THE INVENTION
Crystalline Forms) and Method of Making
Applicants have produced crystals comprising S. aureus EF-P
which are suitable for x-ray crystallographic analysis. The three-dimensional
structure of S. aureus EF-P was solved using high resolution x-ray
crystallography. Preferably, the crystal has orthorhombic space group
symmetry P2,2,2,. More preferably, the crystal comprises rectangular shaped
unit cells, each unit cell having the dimensions a, b, and c, wherein a is
about 25
~ to about 50 A, b is about 35 ~ to about 60 A, and c is about 85 ~ to about
110 ~; and a = (3 = y = 90°. The crystallized enzyme is a monomer and
has one
molecule in the asymmetric unit.
Purified S. aureus EF-P at a concentration of about 1 mg/ml to
about 50 mg/ml may be crystallized, for example, using the hanging drop
procedure from a solution including about 0 wt. % to about 50 wt.
polyethylene glycol (PEG, preferably having a number average molecular
weight between about 200 and about 20,000), 0 to about 20 wt. % DMSO, and
buffered to a pH of about 3.5 to about 5.5. Use of a buffer having a pKa of
between 2.5 and 6.5 is preferred. In a particularly preferred embodiment of
the
method, the buffer includes about 10 mM to about 300 mM sodium acetate.
Variation in buffer and buffer pH as well as other additives such as PEG is
apparent to those skilled in the art and may result in similar crystals.
The invention further includes an S. aureus EF-P crystal or S.
aureus EF-P/ligand crystal that is isomorphous with an S. aureus EF-P crystal
characterized by a unit cell having the dimensions a, b, and c, wherein a is
about
25 t~ to about 50 A, b is about 35 A to about 60 A, and c is about 85 ~ to
about
110~;anda=(3=y=90°.
X-ray Crystallographic Analysis
Crystals of recombinant S. aureus EF-P (Figure 1) were obtained
12
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
from a crystallization screening solution that contained 100 mM sodium acetate
at pH 4.6 and 4% PEG 4000. The recombinant S. aureus EF-P used for
crystallization contains a six-residue polyhistidine tag at the C-terminus in
order
to facilitate purification of the recombinant protein. Refinement of the
conditions resulted in ideal crystal growth occurring at pH 5.2-5.4. Since
there
was no homologous structure available for molecular replacement,
selenomethionine incorporated EF-P was prepared by the downregulation of
methionine (T.E. Benson et al., Nat. Struct. Biol 2:644-53 (1995); G.D. Van
Duyne et al., J. Mol. Biol; 229:105-24 (1993)). Selenomethionine EF-P
crystallized in a similar manner to the native protein and these crystals
diffracted to 1.9 ~ resolution at the synchrotron (Advance Photon Source,
Argonne, IL). The crystals belong to the orthorhombic space group P2,2121 with
cell constants a=37.5 !~, b=47.3 A, c=97.5 A, a=~i=y=90°. Anomalous and
dispersive difference Patterson maps (Figure 2) revealed two of the four
potential selenium sites while a third site was found via cross difference
Fourier
methods. The fourth site could not be located by either method due to multiple
conformations of the methionine as observed later in the electron density
maps.
Maximum likelihood phasing with solvent-flattening resulted in a disconnected
but interpretable electron density map (Figure 3). The structure has been
refined to an R-factor of 25.6% with a Free R-factor of 29.0%. Details of the
structure determination and refinement are described in Tables 3 and 4.
13
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Table 3. Data collection and phasing statistics for EF-P.
~, 1.0332 A ~, 0.979746 A ~, 0.979617 ~
(12000 eV) (12654.8 eV) (12656.5 eV)
Resolution 1.9 t~ 1.9 ~ 1.9 ~
No. observations 105,951 106,013 104,323
No. unique refl. 12,373 14,350 14,349
completeness 100% 100% 100%
RsYm 0.063 0.085 0.084
R~~ns acentrics - 0.587 0.605
R~~,";5 anomalous 0.990 0.683 0.694
Phasing power
centrics - 1.533 1.018
acentrics - 2.417 2.260
Mean figure of merit (to 1.9 ~ resolution)
before solvent flattening 0.559
after solvent flattening 0.895
14
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Table 4. Refinement Statistics for EF-P.
R-factor Free R-factor No. of reflections
10-1.9 ~ F> 2a 0.256 0.290 13,812
Bonds (~) Angles(°)
r.m.s deviation from ideal geometry 0.011 1.508
1 S Number of atomsAverage B-factor
Protein 1327 25.5
Waters 109 35.0
Total 1436 26.2
Each of the constituent amino acids of S. aureus EF-P is defined
by a set of structure coordinates as set forth in Figure 4. The term
"structure
coordinates" refers to Cartesian coordinates derived from mathematical
equations related to the patterns obtained on diffraction of a monochromatic
beam of x-rays by the atoms (scattering centers) of an S. aureus EF-P complex
in crystal form. The diffraction data are used to calculate an electron
density
map of the repeating unit of the crystal. The electron density maps are then
used to establish the positions of the individual atoms of the S. aureus EF-P
protein or protein/ligand complex.
Slight variations in structure coordinates can be generated by
mathematically manipulating S. aureus EF-P structure coordinates. For
example, the structure coordinates set forth in Figure 4 could be manipulated
by
crystallographic permutations of the structure coordinates, fractionalization
of
the structure coordinates, integer additions or subtractions to sets of the
structure coordinates, inversion of the structure coordinates or any
combination
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
of the above. Alternatively, modifications in the crystal structure due to
mutations, additions, substitutions, and/or deletions of amino acids, or other
changes in any of the components that make up the crystal, could also yield
variations in structure coordinates. Such slight variations in the individual
coordinates will have little effect on overall shape. If such variations are
within
an acceptable standard error as compared to the original coordinates, the
resulting three-dimensional shape is considered to be structurally equivalent.
Structural equivalence is described in more detail below.
It should be noted that slight variations in individual structure
coordinates of the S. aureus EF-P would not be expected to significantly alter
the nature of chemical entities such as ligands that could associate with the
binding surfaces. In this context, the phrase "associating with" refers to a
condition of proximity between a chemical entity, or portions thereof, and an
S.
aureus EF-P molecule or portions thereof. The association may be non-
covalent, wherein the juxtaposition is energetically favored by hydrogen
bonding, van der Waals forces, or electrostatic interactions, or it may be
covalent.
Thus, for example, a ligand that bound to or interfered with a
binding surface of S. aureus EF-P would also be expected to bind to or
interfere
with another binding surface whose structure coordinates define a shape that
falls within the acceptable error.
It will be readily apparent to those of skill in the art that the
numbering of amino acids in other isoforms of S. aureus EF-P may be different
than that of S. aureus EF-P expressed in E coli.
Overview of the Structure with Implications for Function
Elongation factor P is primarily comprised of (3 strands
organized into three distinct domains (Figures 5 and 6). Domain 1 contains
four
antiparallel (3 strands and a single turn of a helix. Domains two and three
are
16
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
both five stranded antiparallel ~i barrels similar to the putative
oligonucleotide-
oligosaccharide binding fold (A.G. Murzin, EMBO. J.; 12:861-67 (1993)).
Domain 2 also contains a single turn of a 3,o helix between strands X37 and
(38.
Superposition of the Ca from domains 2 and 3 resulted in an r.m.s. deviation
of
1.55 ~ (Figure 7). Although there are several loop regions that are not
conserved (including the absence of a true helical region in domain 3), the
general fold is maintained. This fold has been observed in many other proteins
some of which bind RNA such as IF1 (M. Sette et al., EMBO. J.; 16:1436-43
(1997)), CspA (W. Jiang et al., J. Biol. Chem.; 272:196-202 (1997); H.
Schindelin et al., Proc. Natl. Acad. Sci. U.S.A.; 91:5119-23 (1994)) and EF-Tu
(P. Nissen et al., Science; 270:1464-72 (1995)). This suggests that domains 2
and 3 of EF-P may play a role in interacting with RNA - probably either tRNA
or rRNA. Other structures that utilize this putative oligonucleotide-
oligosaccharide binding fold show specific interactions with their respective
ligands with the loops between strands 1 and 2, strand 3 and the a helix, or
strands 4 and S (A.G. Murzin, EMBO. J.; 12:861-67 (1993)). Identification of
these residues within the two (3 barrels for EF-P reveals potential sites for
interaction with RNA (Figure 8). Based on evidence from related structures,
residues for S. aureus ER-P that could be involved in oligonucleotide binding
include residues 77-80, 99-105, and 117-120 from domain 2 and residues 149-
150, 164-169, and 177-181 from domain 3. These residues correspond to the
loop between strand 1 and strand 2, the loop from strand 3 through helix 1,
and
the loop between strand 4 and strand 5 in a model beta barrel oligonucleotide
binding fold as described in A.G. Murzin, EMBO. J.; 12:861-67 (1993).
An intriguing feature of EF-P is its polarity of surface charges as
shown in a surface representation (Figure 9). Such polarity in a protein which
most likely interacts with RNA suggests that the positively charged face of
the
protein (Figure 9a) would interact with the negatively charged
oligonucleotide.
In addition, this surface representation reveals that EF-P resembles the shape
and dimensionality of tRNA (Figure 10) (M.A. Rould et al., Science 246:1135-
17
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
42 (1989)). This similarity in shape to tRNA may provide a hypothesis for the
mechanism of action of EF-P. In models of translation, three distinct sites on
the large subunit of the ribosome have been proposed - the P site for the tRNA
containing the elongating amino acyl chain, the A site for the incoming tRNA
and the E site for the deacylated tRNA. During the first step of translation
the E
site would be unoccupied since there is no tRNA which has yet been deacylated.
One possibility is that EF-P could be binding in the E site during translation
initiation to act as a mimic of a deacylated tRNA. This function might serve
to
bring the ribosome into a more active initial conformation which would enhance
the synthesis of the first peptide bond.
Comparison of EF-P to Related Structures
Two related structures have been solved from archaebacteria (M.
jannaschii and P. aerophilum) which are more closely related to the human
protein, eIF-SA (K.K. Kim et al., Proc. Natl. Acad. Sci. U.S.A; 95:10419-24
(1998); T.S. Peat et al., Structure; 6:1207-14 (1998)). These archaebacterial
proteins contain homology to domains 1 and 2 of the bacterial EF-P, but domain
3 is surprisingly absent (Figure 11). While it is not apparent why domain 3 is
absent in the archaebacteria and eukaryotic sequences but is present in E coli
and S. aureus EF-P, the similarity of domain 3 to domain 2 and the
oligonucleotide binding fold suggests the interaction with rRNA or tRNA may
be somewhat different for E. coli and S. aureus. Superposition of domain 1
from S. aureus and M. jannaschii gave a r.m.s.d. for Ca atoms of 2.02 ~ and
superposition of domain 2 of these two structures gave a r.m.s.d. for Ca atoms
of 3.02 ~. Superposition of domain 1 from S. aureus and P. aerophilum gave a
r.m.s.d. for Ca atoms of 3.29 ~ and superposition of domain 2 of these two
structures gave a r.m.s.d. for Ca atoms of 4.25 t~. While there is good
agreement for each domain individually, the relative orientation of domains 1
and 2 is quite flexible as illustrated in Figures 12 and 13. A second crystal
form
of eIFSA from M. jannaschii has been solved which reveals some flexibility in
18
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
the relative orientation of domains 1 and 2 (K.K. Kim et al., Proc. Natl.
Acad.
Sci. U.S.A; 95:10419-24 (1998)). Since we have only solved one crystal, it is
unclear how flexible this linker is for S. aureus EF-P.
19
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
The structures of S. aureus, M. jannaschii, and P. aerophilum
possess a highly conserved motif within domain 1 - xKxGKGxA - which has
been identified in yeast and human as a site for post-translational
modification
of the second lysine to NE-(4-aminobutyl)lysine (also called deoxyhypusine).
This modification has been shown to be essential for cell viability in yeast
(J.
Schnier et al., Mol. Cell. Biol; 11:3105-14 (1991)) and occurs only with eIFSA
by the enzyme deoxyhypusine synthase (D.I. Liao et al., Structure; 6:23-32
(1998)). The location of this conserved loop in EF-P is between X32 and (33
with K33 of S. aureus EF-P being the conserved residue which is modified in
eukaryotic EF-P homologs. This exposed lysine is illustrated in Figure 14
projecting out from this loop. In bacteria, hypusination of the homologous EF-
P
K33 amino acid has not been observed, and, in fact, analysis of the electron
density for this residue in S. aureus EF-P does not reveal evidence of a post
translational modification in this recombinant protein. Similar investigation
of
the electron density for the EF-P homologs from archaebacteria did not reveal
any modification (K.K. Kim et al., Proc. Natl. Acad. Sci. U.S.A; 95:10419-24
(1998); T.S. Peat et al., Structure; 6:1207-14 (1998)). Mass spectrometry of
the
S. aureus EF-P sample (both methionine and selenomethionine) did not reveal
any additional mass that would be accounted for by a post-translational
modification. Since S. aureus EF-P was purified from an EF-P overexpression
strain, it is possible that the modification pathway was overwhelmed and
unable
to sufficiently carry out the post-translational modification. There is also
no
direct evidence that hypusination occurs in bacteria, and it may be that
another
type of modification is incorporated for S aureus EF-P.
Binding Surfaces and Other Structural Features
Applicants' invention has provided, for the first time,
information about the shape and structure of a putative oligonucleotide
binding
surface of S. aureus EF-P.
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Binding surfaces are of significant utility in fields such as drug
discovery. The association of natural ligands or substrates with the binding
surfaces of their corresponding receptors or enzymes is the basis of many
biological mechanisms of action. Similarly, many drugs exert their biological
effects through association with the binding surfaces of receptors and
enzymes.
Such associations may occur with all or any parts of the binding surface. An
understanding of such associations helps lead to the design of drugs having
more favorable associations with their target, and thus improved biological
effects. Therefore, this information is valuable in designing potential
inhibitors
of S. aureus EF-P-like binding surfaces, as discussed in more detail below.
A "molecular complex" means a protein in colvalent or non-
covalent association with a chemical entity. The term "binding surface" as
used
herein, refers to a region of a molecule or molecular complex, that, as a
result of
its shape, favorably associates with another chemical entity.
The amino acid constituents of an S. aureus EF-P
oligonucleotide binding surface as defined herein, as well as selected
constituent
atoms thereof, are positioned in three dimensions in accordance with the
structure coordinates listed in Figure 4. In one aspect, the structure
coordinates
defining the binding surface of S. aureus EF-P include structure coordinates
of
substantially all atoms in the constituent amino acids; in another aspect, the
structure coordinates of the binding surface include structure coordinates of
just
the backbone atoms of the constituent atoms.
A specific chemical entity may bind to any of the amino acid
surface residues of S. aureus EF-P as listed in Table 1. Preferably, the
surface
residues that comprise the binding surface include amino acid K33. Even more
preferably, the surface residues that comprise the binding surface include
residues whose backbone atoms are situated within 4 A, preferably within 7 ~,
more preferably within 10 ~, and most preferably within 1 S ~ of K33 as listed
in Table 2.
21
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
The term "S. aureus EF-P-like binding surface" refers to a
portion of a molecule or molecular complex whose shape is sufficiently similar
to at least a portion of a binding surface of S aureus EF-P as to be expected
to
bind common or structurally related ligands. A structurally equivalent binding
surface is defined by a root mean square deviation from the structure
coordinates of the backbone atoms of the amino acids that make up the binding
surfaces in S. aureus EF-P (as set forth in Figure 4) of at most about 1.9
fir.
How this calculation is obtained is described below.
Accordingly, the invention thus provides molecules or molecular
complexes comprising an S. aureus EF-P oligonucleotide binding surface or S.
aureus EF-P-like binding surface, as defined by the sets of structure
coordinates
described above.
Three-Dimensional Configurations
X-ray structure coordinates define a unique configuration of
points in space. Those of skill in the art understand that a set of structure
coordinates for protein or a protein/ligand complex, or a portion thereof,
define
a relative set of points that, in turn, define a configuration in three
dimensions.
A similar or identical configuration can be defined by an entirely different
set of
coordinates, provided the distances and angles between coordinates remain
essentially the same. In addition, a scalable configuration of points can be
defined by increasing or decreasing the distances between coordinates by a
scalar factor while keeping the angles essentially the same.
The present invention thus includes the scalable three-
dimensional configuration of points derived from the structure coordinates of
at
least a portion of an S. aureus EF-P molecule or molecular complex, as listed
in
Figure 4, as well as structurally equivalent configurations, as described
below.
Preferably, the scalable three-dimensional configuration includes points
derived
from structure coordinates representing the locations of a plurality of the
amino
acids defining the S. aureus EF-P binding surface. In one embodiment, the
22
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
scalable three-dimensional configuration includes points derived from
structure
coordinates representing the locations the backbone atoms of a plurality of
amino acids defining the S. aureus EF-P binding surface, preferably those
amino acids listed in Table 1. In another embodiment, the scalable three-
dimensional configuration includes points derived from structure coordinates
representing the locations of the side chain and the backbone atoms (other
than
hydrogens) of a plurality of the amino acids defining the S. aureus EF-P
binding
surface, preferably those amino acids listed in Table 1. Alternatively, the
scalable three-dimensional configuration of points are derived from structure
coordinates representing the locations of backbone and, optionally, side chain
atoms (other than hydrogens) of amino acids within 4 ~, preferably within 7
t~,
more preferably within 10 ~, and most preferably within 15 ~ of Lys33 as
shown in Table 2.
Likewise, the invention also includes the scalable three-
dimensional configuration of points derived from structure coordinates of
molecules or molecular complexes that are structurally homologous to S. aureus
EF-P, as well as structurally equivalent configurations. Structurally
homologous molecules or molecular complexes are defined below.
Advantageously, structurally homologous molecules can be identified using the
structure coordinates of S. aureus EF-P (Figure 4) according to a method of
the
invention.
The configurations of points in space derived from structure
coordinates according to the invention can be visualized as, for example, a
holographic image, a stereodiagram, a model or a computer-displayed image,
and the invention thus includes such images, diagrams or models.
Structurally Equivalent Crystal Structures
Various computational analyses can be used to determine
whether a molecule or the binding surface portion thereof is "structurally
equivalent," defined in terms of its three-dimensional structure, to all or
part of
23
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
S. aureus EF-P or its binding surfaces. Such analyses may be carried out in
current software applications, such as the Molecular Similarity application of
QUANTA (Molecular Simulations Inc., San Diego, CA) version 4.1, and as
described in the accompanying User's Guide.
The Molecular Similarity application permits comparisons
between different structures, different conformations of the same structure,
and
different parts of the same structure. The procedure used in Molecular
Similarity to compare structures is divided into four steps: (1) load the
structures to be compared; (2) define the atom equivalences in these
structures;
(3) perform a fitting operation; and (4) analyze the results.
Each structure is identified by a name. One structure is
identified as the target (i.e., the fixed structure); all remaining structures
are
working structures (i.e., moving structures). Since atom equivalency within
QUANTA is defined by user input, for the purpose of this invention equivalent
atoms are defined as protein backbone atoms (N, Ca, C, and O) for all
conserved residues between the two structures being compared. A conserved
residue is defined as a residue that is structurally or functionally
equivalent.
Only rigid fitting operations are considered.
When a rigid fitting method is used, the working structure is
translated and rotated to obtain an optimum fit with the target structure. The
fitting operation uses an algorithm that computes the optimum translation and
rotation to be applied to the moving structure, such that the root mean square
difference of the fit over the specified pairs of equivalent atom is an
absolute
minimum. This number, given in angstroms, is reported by QUANTA.
For the purpose of this invention, any molecule or molecular
complex or binding surface thereof, or any portion thereof, that has a root
mean
square deviation of conserved residue backbone atoms (N, Ca, C, O) of less
than about 1.9 ~, when superimposed on the relevant backbone atoms described
by the reference structure coordinates listed in Figure 4, is considered
"structurally equivalent" to the reference molecule. That is to say, the
crystal
24
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
structures of those portions of the two molecules are substantially identical,
within acceptable error. Particularly preferred structurally equivalent
molecules
or molecular complexes are those that are defined by the entire set of
structure
coordinates in Figure 4, ~ a root mean square deviation from the conserved
backbone atoms of those amino acids of not more than 1.9 ~. More preferably,
the root mean square deviation is less than about 1.0 ~.
The term "root mean square deviation" means the square root of
the arithmetic mean of the squares of the deviations. It is a way to express
the
deviation or variation from a trend or object. For purposes of this invention,
the
"root mean square deviation" defines the variation in the backbone of a
protein
from the backbone of S. aureus EF-P or a binding surface portion thereof, as
defined by the structure coordinates of S. aureus EF-P described herein.
Machine Readable Storage Media
Transformation of the structure coordinates for all or a portion of
S. aureus EF-P or the S. aureus EF-P/ligand complex, for structurally
homologous molecules as defined below, or for the structural equivalents of
any
of these molecules or molecular complexes as defined above, into three-
dimensional graphical representations of the molecule or complex can be
conveniently achieved through the use of commercially-available software.
The.invention thus further provides a machine-readable storage
medium comprising a data storage material encoded with machine readable data
which, when using a machine programmed with instructions for using said data,
is capable of displaying a graphical three-dimensional representation of any
of
the molecule or molecular complexes of this invention that have been described
above. In a preferred embodiment, the machine-readable data storage medium
comprises a data storage material encoded with machine readable data which,
when using a machine programmed with instructions for using said data, is
capable of displaying a graphical three-dimensional representation of a
molecule or molecular complex comprising all or any parts of an S. aureus EF-P
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
binding surface or an S. aureus EF-P-like binding surface, as defined above.
In
another preferred embodiment, the machine-readable data storage medium is
capable of displaying a graphical three-dimensional representation of a
molecule or molecular complex defined by the structure coordinates of all of
the
amino acids in Figure 4, ~ a root mean square deviation from the backbone
atoms of said amino acids of not more than 1.9 ~.
In an alternative embodiment, the machine-readable data storage
medium comprises a data storage material encoded with a first set of machine
readable data which comprises the Fourier transform of the structure
coordinates
set forth in Figure 4, and which, when using a machine programmed with
instructions for using said data, can be combined with a second set of machine
readable data comprising the x-ray diffraction pattern of a molecule or
molecular complex to determine at least a portion of the structure coordinates
corresponding to the second set of machine readable data.
For example, a system for reading a data storage medium may
include a computer comprising a central processing unit ("CPU"), a working
memory which may be, e.g., RAM (random access memory) or "core" memory,
mass storage memory (such as one or more disk drives or CD-ROM drives), one
or more display devices (e.g., cathode-ray tube ("CRT") displays, light
emitting
diode ("LED") displays, liquid crystal displays ("LCDs"), electroluminescent
displays, vacuum fluorescent displays, field emission displays ("FEDs"),
plasma
displays, projection panels, etc.), one or more user input devices (e.g.,
keyboards, microphones, mice, touch screens, etc.), one or more input lines,
and
one or more output lines, all of which are interconnected by a conventional
bidirectional system bus. The system may be a stand-alone computer, or may
be networked (e.g., through local area networks, wide area networks,
intranets,
extranets, or the Internet) to other systems (e.g., computers, hosts, servers,
etc.).
The system may also include additional computer controlled devices such as
consumer electronics and appliances.
26
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Input hardware may be coupled to the computer by input lines
and may be implemented in a variety of ways. Machine-readable data of this
invention may be inputted via the use of a modem or modems connected by a
telephone line or dedicated data line. Alternatively or additionally, the
input
hardware may comprise CD-ROM drives or disk drives. In conjunction with a
display terminal, a keyboard may also be used as an input device.
Output hardware may be coupled to the computer by output lines
and may similarly be implemented by conventional devices. By way of
example, the output hardware may include a display device for displaying a
graphical representation of a binding surface of this invention using a
program
such as QUANTA as described herein. Output hardware might also include a
printer, so that hard copy output may be produced, or a disk drive, to store
system output for later use.
In operation, a CPU coordinates the use of the various input and
output devices, coordinates data accesses from mass storage devices, accesses
to
and from working memory, and determines the sequence of data processing
steps. A number of programs may be used to process the machine-readable data
of this invention. Such programs are discussed in reference to the
computational methods of drug discovery as described herein. References to
components of the hardware system are included as appropriate throughout the
following description of the data storage medium.
Machine-readable storage devices useful in the present invention
include, but are not limited to, magnetic devices, electrical devices, optical
devices, and combinations thereof. Examples of such data storage devices
include, but are not limited to, hard disk devices, CD devices, digital video
disk
devices, floppy disk devices, removable hard disk devices, magneto-optic disk
devices, magnetic tape devices, flash memory devices, bubble memory devices,
holographic storage devices, and any other mass storage peripheral device. It
should be understood that these storage devices include necessary hardware
27
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
(e.g., drives, controllers, power supplies, etc.) as well as any necessary
media
(e.g., disks, flash cards, etc.) to enable the storage of data.
Structurally Homologous Molecules, Molecular Complexes, and Crystal
Structures
The structure coordinates set forth in Figure 4 can be used to aid
in obtaining structural information about another crystallized molecule or
molecular complex. The method of the invention allows determination of at
least a portion of the three-dimensional structure of molecules or molecular
complexes which contain one or more structural features that are similar to
structural features of S. aureus EF-P. These molecules are referred to herein
as
"structurally homologous" to S. aureus EF-P. Similar structural features can
include, for example, regions of amino acid identity, conserved active site or
binding site motifs, and similarly arranged secondary structural elements
(e.g., a
helices and (3 sheets). Optionally, structural homology is determined by
aligning the residues of the two amino acid sequences to optimize the number
of
identical amino acids along the lengths of their sequences; gaps in either or
both
sequences are permitted in making the alignment in order to optimize the
number of identical amino acids, although the amino acids in each sequence
must nonetheless remain in their proper order. Preferably, two amino acid
sequences are compared using the Blastp program, version 2Ø9, of the BLAST
2 search algorithm, as described by Tatiana et al., FEMS Microbiol Lett 174,
247-50 (1999), and available at http://www.ncbi.nlm.nih.gov/gorf/bl2.html.
Preferably, the default values for all BLAST 2 search parameters are used,
including matrix = BLOSUM62; open gap penalty = 11, extension gap penalty
= 1, gap x dropoff = 50, expect = 10, wordsize = 3, and filter on. In the
comparison of two amino acid sequences using the BLAST search algorithm,
structural similarity is referred to as "identity." Preferably, a structurally
homologous molecule is a protein that has an amino acid sequence sharing at
least 65% identity with a native or recombinant amino acid sequence of S.
28
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
aureus EF-P (for example, SEQ ID NO: 1). More preferably, a protein that is
structurally homologous to S. aureus EF-P includes at least one contiguous
stretch of at least 50 amino acids that shares at least 80% amino acid
sequence
identity with the analogous portion of the native or recombinant S. aureus EF-
P
(for example, SEQ ID NO: 1). Methods for generating structural information
about the structurally homologous molecule or molecular complex are well
known and include, for example, molecular replacement techniques.
Therefore, in another embodiment this invention provides a
method of utilizing molecular replacement to obtain structural information
about a molecule or molecular complex whose structure is unknown comprising
the steps o~
(a) crystallizing the molecule or molecular complex of unknown
structure;
(b) generating an x-ray diffraction pattern from said crystallized
molecule or molecular complex; and
(c) applying at least a portion of the structure coordinates set forth in
Figure 4 to the x-ray diffraction pattern to generate a three-dimensional
electron
density map of the molecule or molecular complex whose structure is unknown.
By using molecular replacement, all or part of the structure
coordinates of S. aureus EF-P or the S. aureus EF-P/ligand complex as provided
by this invention (for example as set forth in Figure 4) can be used to
determine
the structure of a crystallized molecule or molecular complex whose structure
is
unknown more quickly and efficiently than attempting to determine such
information ab initio.
Molecular replacement provides an accurate estimation of the
phases for an unknown structure. Phases are a factor in equations used to
solve
crystal structures that cannot be determined directly. Obtaining accurate
values
for the phases, by methods other than molecular replacement, is a time-
consuming process that involves iterative cycles of approximations and
refinements and greatly hinders the solution of crystal structures. However,
29
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
when the crystal structure of a protein containing at least a structurally
homologous portion has been solved, the phases from the known structure
provide a satisfactory estimate of the phases for the unknown structure.
Thus, this method involves generating a preliminary model of a
molecule or molecular complex whose structure coordinates are unknown, by
orienting and positioning the relevant portion of S. aureus EF-P according to
Figure 4 within the unit cell of the crystal of the unknown molecule or
molecular complex so as best to account for the observed x-ray diffraction
pattern of the crystal of the molecule or molecular complex whose structure is
unknown. Phases can then be calculated from this model and combined with
the observed x-ray diffraction pattern amplitudes to generate an electron
density
map of the structure whose coordinates are unknown. This, in turn, can be
subjected to any well-known model building and structure refinement
techniques to provide a final, accurate structure of the unknown crystallized
molecule or molecular complex (E. Lattman, "Use of the Rotation and
Translation Functions," in Meth. Enzymol., 115, pp. 55-77 (1985); M.G.
Rossman, ed., "The Molecular Replacement Method," Int. Sci. Rev. Ser., No.
13, Gordon & Breach, New York (1972)).
Structural information about a portion of any crystallized
molecule or molecular complex that is sufficiently structurally homologous to
a
portion of S. aureus EF-P can be resolved by this method. In addition to a
molecule that shares one or more structural features with S. aureus EF-P as
described above, a molecule that has similar bioactivity, such as the same
substrate specificity or ligand binding activity as S. aureus EF-P, may also
be
sufficiently structurally homologous to S. aureus EF-P to permit use of the
structure coordinates of S aureus EF-P to solve its crystal structure.
In a preferred embodiment, the method of molecular replacement
is utilized to obtain structural information about a molecule or molecular
complex, wherein the molecule or molecular complex comprises at least one S.
aureus EF-P subunit or homolog. A "subunit" of S. aureus EF-P is an S. aureus
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
EF-P molecule that has been truncated at the N-terminus or the C-terminus, or
both. In the context of the present invention, a "homolog" of S. aureus EF-P
is
a protein that contains one or more amino acid substitutions, deletions,
additions, or rearrangements with respect to the amino acid sequence of S.
aureus EF-P, but that, when folded into its native conformation, exhibits or
is
reasonably expected to exhibit at least a portion of the tertiary (three-
dimensional) structure of S aureus EF-P. For example, structurally
homologous molecules can contain deletions or additions of one or more
contiguous or noncontiguous amino acids, such as a loop or a domain.
Structurally homologous molecules also include "modified" S. aureus EF-P
molecules that have been chemically or enzymatically derivatized at one or
more constituent amino acid, including side chain modifications, backbone
modifications, and N- and C- terminal modifications including acetylation,
hydroxylation, methylation, amidation, and the attachment of carbohydrate or
lipid moieties, cofactors, and the like.
A heavy atom derivative of S. aureus EF-P is also included as an
S. aureus EF-P homolog. The term "heavy atom derivative" refers to
derivatives of S. aureus EF-P produced by chemically modifying a crystal of S.
aureus EF-P. In practice, a crystal is soaked in a solution containing heavy
metal atom salts, or organometallic compounds, e.g., lead chloride, gold
thiomalate, thiomersal or uranyl acetate, which can diffuse through the
crystal
and bind to the surface of the protein. The locations) of the bound heavy
metal
atoms) can be determined by x-ray diffraction analysis of the soaked crystal.
This information, in turn, is used to generate the phase information used to
construct three-dimensional structure of the protein (T.L. Blundell and N.L.
Johnson, Protein Crystallography, Academic Press (1976)).
Because S aureus EF-P can crystallize in more than one crystal
form, the structure coordinates of S. aureus EF-P as provided by this
invention
are particularly useful in solving the structure of other crystal forms of S.
aureus
EF-P or S. aureus EF-P complexes.
31
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
The structure coordinates of S. aureus EF-P as provided by this
invention are particularly useful in solving the structure of S. aureus EF-P
mutants. Mutants may be prepared, for example, by expression of S. aureus EF-
P cDNA previously altered in its coding sequence by oligonucleotide-directed
mutagenesis. Mutants may also be generated by site-specific incorporation of
unnatural amino acids into EF-P proteins using the general biosynthetic method
of C.J. Noren et al., Science, 244:182-188 (1989). In this method, the codon
encoding the amino acid of interest in wild-type S aureus EF-P is replaced by
a
"blank" nonsense codon, TAG, using oligonucleotide-directed mutagenesis. A
suppressor tRNA directed against this codon is then chemically aminoacylated
in vitro with the desired unnatural amino acid. The aminoacylated tRNA is then
added to an in vitro translation system to yield a mutant S. aureus EF-P with
the
site-specific incorporated unnatural amino acid.
Selenocysteine or selenomethionine may be incorporated into
wild-type or mutant S. aureus EF-P by expression of S. aureus EF-P-encoding
cDNAs in auxotrophic E coli strains (W.A. Hendrickson et al., EMBO J.,
9(5):1665-1672 (1990)). In this method, the wild-type or mutagenized S.
aureus EF-P cDNA may be expressed in a host organism on a growth medium
depleted of either natural cysteine or methionine (or both) but enriched in
selenocysteine or selenomethionine (or both). Alternatively, selenomethionine
analogues may be prepared by down regulation methionine biosynthesis. (T.E.
Benson et al., Nat. Struct. Biol., 2:644-53 (1995); G.D. Van Duyne et al., J.
Mol. Biol. 229:105-24 (1993)).
The structure coordinates of S aureus EF-P in Figure 4 are also
particularly useful to solve the structure of crystals of S. aureus EF-P, S.
aureus
EF-P mutants or S. aureus EF-P homologs co-complexed with a variety of
chemical entities. This approach enables the determination of the optimal
sites
for interaction between chemical entities, including candidate S. aureus EF-P
inhibitors. Potential sites for modification within the various binding site
of the
molecule can also be identified. This information provides an additional tool
32
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
for determining the most efficient binding interactions, for example,
increased
hydrophobic interactions, between S. aureus EF-P and a chemical entity. For
example, high resolution x-ray diffraction data collected from crystals
exposed
to different types of solvent allows the determination of where each type of
solvent molecule resides. Small molecules that bind tightly to those sites can
then be designed and synthesized and tested for their S. aureus EF-P
inhibition
activity.
All of the complexes referred to above may be studied using
well-known x-ray diffraction techniques and may be refined versus 1.5-3 A
resolution x-ray data to an R value of about 0.20 or less using computer
software, such as X-PLOR (Yale University, 81992, distributed by Molecular
Simulations, Inc.; see, e.g., Blundell & Johnson, supra; Meth. Enzymol., Vol.
114 & 115, H.W. Wyckoff et al., eds., Academic Press (1985)). This
information may thus be used to optimize known S. aureus EF-P inhibitors, and
more importantly, to design new S. aureus EF-P inhibitors.
The invention also includes the unique three-dimensional
configuration defined by a set of points defined by the structure coordinates
for
a molecule or molecular complex structurally homologous to S. aureus EF-P as
determined using the method of the present invention, structurally equivalent
configurations, and magnetic storage media comprising such set of structure
coordinates.
Further, the invention includes structurally homologous
molecules as identified using the method of the invention.
Homology Modeling
Using homology modeling, a computer model of an S. aureus
EF-P homolog can be built or refined without crystallizing the homolog. First,
a preliminary model of the S. aureus EF-P homolog is created by sequence
alignment with S. aureus EF-P, secondary structure prediction, the screening
of
structural libraries, or any combination of those techniques. Computational
33
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
software may be used to carry out the sequence alignments and the secondary
structure predictions. Structural incoherences, e.g., structural fragments
around
insertions and deletions, can be modeled by screening a structural library for
peptides of the desired length and with a suitable conformation. For
prediction
of the side chain conformation, a side chain rotamer library may be employed.
If the S aureus EF-P homolog has been crystallized, the final homology model
can be used to solve the crystal structure of the homolog by molecular
replacement, as described above. Next, the preliminary model is subjected to
energy minimization to yield an energy minimized model. The energy
minimized model may contain regions where stereochemistry restraints are
violated, in which case such regions are remodeled to obtain a final homology
model. The homology model is positioned according to the results of molecular
replacement, and subjected to further refinement comprising molecular
dynamics calculations.
Rational Drub Design
Computational techniques can be used to screen, identify, select
and/or design chemical entities capable of associating with S. aureus EF-P or
structurally homologous molecules. Knowledge of the structure coordinates for
S. aureus EF-P permits the design and/or identification of synthetic compounds
and/or other molecules which have a shape complementary to the conformation
of the S. aureus EF-P binding site. In particular, computational techniques
can
be used to identify or design chemical entities, such as inhibitors, agonists
and
antagonists, that associate with an S. aureus EF-P binding surface or an S.
aureus EF-P-like binding surface. Inhibitors may bind to or interfere with all
or a portion of the binding surface of S. aureus EF-P, and can be competitive,
non-competitive, or uncompetitive inhibitors; or interfere with dimerization
by
binding at the interface between the two monomers. For example, inhibitors
that are bound to a binding surface of S. aureus EF-P may interfere with
binding
of S. aureus EF-P to a ribosomal protein or ribosomal RNA during translation.
34
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Once identified and screened for biological activity, these
inhibitors/agonists/antagonists may be used therapeutically or
prophylactically
to block S aureus EF-P activity and, thus, inhibit growth of the bacteria or
cause its death. Structure-activity data for analogs of ligands that bind to
or
interfere with S. aureus EF-P or S. aureus EF-P-like binding surfaces can also
be obtained computationally.
The term "chemical entity," as used herein, refers to chemical
compounds, complexes of two or more chemical compounds, and fragments of
such compounds or complexes. Chemical entities that are determined to
associate with S. aureus EF-P are potential drug candidates.
Data stored in a machine-readable storage medium that is
capable of displaying a graphical three-dimensional representation of the
structure of S. aureus EF-P or a structurally homologous molecule, as
identified
herein, or portions thereof may thus be advantageously used for drug
discovery.
The structure coordinates of the chemical entity are used to generate a three-
dimensional image that can be computationally fit to the three-dimensional
image of S. aureus EF-P or a structurally homologous molecule. The three-
dimensional molecular structure encoded by the data in the data storage medium
can then be computationally evaluated for its ability to associate with
chemical
entities. When the molecular structures encoded by the data is displayed in a
graphical three-dimensional representation on a computer screen, the protein
structure can also be visually inspected for potential association with
chemical
entities.
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
One embodiment of the method of drug design involves
evaluating the potential association of a known chemical entity with S. aureus
EF-P or a structurally homologous molecule, particularly with an S. aureus EF-
P binding surface or S. aureus EF-P-like binding surface. The method of drug
design thus includes computationally evaluating the potential of a selected
chemical entity to associate with any of the molecules or molecular complexes
set forth above. This method comprises the steps of: (a) employing
computational means to perform a fitting operation between the selected
chemical entity and a binding surface of the molecule or molecular complex;
and (b) analyzing the results of said fitting operation to quantify the
association
between the chemical entity and the binding surface.
In another embodiment, the method of drug design involves
computer-assisted design of chemical entities that associate with S. aureus EF-
P, its homologs, or portions thereof. Chemical entities can be designed in a
step-wise fashion, one fragment at a time, or may be designed as a whole or
"de
novo."
To be a viable drug candidate, the chemical entity identified or
designed according to the method must be capable of structurally associating
with at least part of an S. aureus EF-P or S. aureus EF-P-like binding
surfaces,
and must be able, sterically and energetically, to assume a conformation that
allows it to associate with the S. aureus EF-P or S. aureus EF-P-like binding
surface. Non-covalent molecular interactions important in this association
include hydrogen bonding, van der Waals interactions, hydrophobic
interactions, and electrostatic interactions. Conformational considerations
include the overall three-dimensional structure and orientation of the
chemical
entity in relation to the binding surface, and the spacing between various
functional groups of an entity that directly interact with the S. aureus EF-P-
like
binding surface or homologs thereof.
Optionally, the potential binding of a chemical entity to an S.
aureus EF-P or S. aureus EF-P-like binding surface is analyzed using computer
36
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
modeling techniques prior to the actual synthesis and testing of the chemical
entity. If these computational experiments suggest insufficient interaction
and
association between it and the S. aureus EF-P or S. aureus EF-P-like binding
surface, testing of the entity is obviated. However, if computer modeling
indicates a strong interaction, the molecule may then be synthesized and
tested
for its ability to bind to or interfere with an S. aureus EF-P or S. aureus EF-
P-
like binding surface. binding assays to determine if a compound actually binds
to S. aureus EF-P can also be performed and are well known in the art. binding
assays may employ kinetic or thermodynamic methodology using a wide variety
of techniques including, but not limited to, microcalorimetry, circular
dichroism, capillary zone electrophoresis, nuclear magnetic resonance
spectroscopy, fluorescence spectroscopy, and combinations thereof.
One skilled in the art may use one of several methods to screen
chemical entities or fragments for their ability to associate with an S.
aureus EF-
P or S. aureus EF-P-like binding surface. This process may begin by visual
inspection of, for example, an S. aureus EF-P or S. aureus EF-P-like binding
surface on the computer screen based on the S. aureus EF-P structure
coordinates in Figure 4 or other coordinates which define a similar shape
generated from the machine-readable storage medium. Selected fragments or
chemical entities may then be positioned in a variety of orientations, or
docked,
within the binding surface. Docking may be accomplished using software such
as QUANTA and SYBYL, followed by energy minimization and molecular
dynamics with standard molecular mechanics forcefields, such as CHARMM
and AMBER.
Specialized computer programs may also assist in the process of
selecting fragments or chemical entities. Examples include GRID (P.J.
Goodford, J. Med. Chem. 28:849-857 (1985); available from Oxford University,
Oxford, UK); MCSS (A. Miranker et al., Proteins: Struct. Funct. Gen.,11:29-34
(1991); available from Molecular Simulations, San Diego, CA); AUTODOCK
(D.S. Goodsell et al., Proteins: Struct. Funct. Genet. 8:195-202 (1990);
available
37
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
from Scripps Research Institute, La Jolla, CA); and DOCK (LD. Kuntz et al., J.
Mol. Biol. 161:269-288 (1982); available from University of California, San
Francisco, CA).
Once suitable chemical entities or fragments have been selected,
they can be assembled into a single compound or complex. Assembly may be
preceded by visual inspection of the relationship of the fragments to each
other
on the three-dimensional image displayed on a computer screen in relation to
the structure coordinates of S. aureus EF-P. This would be followed by manual
model building using software such as QUANTA or SYBYL (Tripos
Associates, St. Louis, MO).
Useful programs to aid one of skill in the art in connecting the
individual chemical entities or fragments include, without limitation, CAVEAT
(P.A. Bartlett et al., in Molecular Recognition in Chemical and Biological
Problems," Special Publ., Royal Chem. Soc., 78:182-196 (1989); G. Lauri et
al.,
J. Comput. Aided Mol. Des. 8:51-66 (1994); available from the University of
California, Berkeley, CA); 3D database systems such as ISIS (available from
MDL Information Systems, San Leandro, CA; reviewed in Y.C. Martin, J. Med.
Chem. 35:2145-2154 (1992)); and HOOK (M.B. Eisen et al., Proteins: Struc.,
Funct., Genet. 19:199-221 (1994); available from Molecular Simulations, San
Diego, CA).
S aureus EF-P binding compounds may be designed "de novo"
using either an empty binding site or optionally including some portions) of a
known inhibitor(s). There are many de novo ligand design methods including,
without limitation, LUDI (H.-J. Bohm, J. Comp. Aid. Molec. Design. 6:61-78
(1992); available from Molecular Simulations Inc., San Diego, CA); LEGEND
(Y. Nishibata et al., Tetrahedron, 47:8985 (1991); available from Molecular
Simulations Inc., San Diego, CA); LeapFrog (available from Tripos Associates,
St. Louis, MO); and SPROUT (V. Gillet et al., J. Comput. Aided Mol. Design
7:127-153 (1993); available from the University of Leeds, UK).
38
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Once a compound has been designed or selected by the above
methods, the efficiency with which that entity may bind to or interfere with
an
S. aureus EF-P or S. aureus EF-P-like binding surface may be tested and
optimized by computational evaluation. For example, an effective S. aureus
EF-P or S. aureus EF-P-like binding surface inhibitor must preferably
demonstrate a relatively small difference in energy between its bound and free
states (i.e., a small deformation energy of binding). Thus, the most efficient
S.
aureus EF-P or S. aureus EF-P-like binding surface inhibitors should
preferably
be designed with a deformation energy of binding of not greater than about 10
kcal/mole; more preferably, not greater than 7 kcal/mole. S aureus EF-P or S.
aureus EF-P-like binding surface inhibitors may interact with the binding
surface in more than one conformation that is similar in overall binding
energy.
In those cases, the deformation energy of binding is taken to be the
difference
between the energy of the free entity and the average energy of the
conformations observed when the inhibitor binds to the protein.
An entity designed or selected as binding to or interfering with
an S. aureus EF-P or S. aureus EF-P-like binding surface may be further
computationally optimized so that in its bound state it would preferably lack
repulsive electrostatic interaction with the target enzyme and with the
surrounding water molecules. Such non-complementary electrostatic
interactions include repulsive charge-charge, dipole-dipole, and charge-dipole
interactions.
Specific computer software is available in the art to evaluate
compound deformation energy and electrostatic interactions. Examples of
programs designed for such uses include: Gaussian 94, revision C (M.J. Frisch,
Gaussian, Inc., Pittsburgh, PA 81995); AMBER, version 4.1 (P.A. Kollman,
University of California at San Francisco, 81995); QUANTA/CHARMM
(Molecular Simulations, Inc., San Diego, CA 81995); Insight II/Discover
(Molecular Simulations, Inc., San Diego, CA 81995); Delphi (Molecular
Simulations, Inc., San Diego, CA 81995); and AMSOL (Quantum Chemistry
39
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Program Exchange, Indiana University). These programs may be implemented,
for instance, using a Silicon Graphics workstation such as an Indigo2 with
"IMPACT" graphics. Other hardware systems and software packages will be
known to those skilled in the art.
Another approach encompassed by this invention is the
computational screening of small molecule databases for chemical entities or
compounds that can bind in whole, or in part, to a S. aureus EF-P or S. aureus
EF-P-like binding surface. In this screening, the quality of fit of such
entities to
the binding site may be judged either by shape complementarity or by estimated
interaction energy (E.C. Meng et al., J. Comp. Chem., 13, pp. 505-524 (1992)).
This invention also enables the development of chemical entities
that can isomerize to short-lived reaction intermediates in the chemical
reaction
of a substrate or other compound that binds to or interferes with or with S.
aureus EF-P. Time-dependent analysis of structural changes in S. aureus EF-P
during its interaction with other molecules is carried out. The reaction
intermediates of S. aureus EF-P can also be deduced from the reaction product
in co-complex with S. aureus EF-P. Such information is useful to design
improved analogs of known S. aureus EF-P inhibitors or to design novel classes
of inhibitors based on the reaction intermediates of the S. aureus EF-P and
inhibitor co-complex. This provides a novel route for designing S. aureus EF-P
inhibitors with both high specificity and stability.
Yet another approach to rational drug design involves probing
the S. aureus EF-P crystal of the invention with molecules comprising a
variety
of different functional groups to determine optimal sites for interaction
between
candidate S. aureus EF-P inhibitors and the protein. For example, high
resolution x-ray diffraction data collected from crystals soaked in or co-
crystallized with other molecules allows the determination of where each type
of solvent molecule sticks. Molecules that bind tightly to those sites can
then be
further modified and synthesized and tested for their EF-P inhibitor activity
(J.
Travis, Science, 262:1374 (1993)).
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
In a related approach, iterative drug design is used to identify
inhibitors of S. aureus EF-P. Iterative drug design is a method for optimizing
associations between a protein and a compound by determining and evaluating
the three-dimensional structures of successive sets of protein/compound
complexes. In iterative drug design, crystals of a series of protein/compound
complexes are obtained and then the three-dimensional structures of each
complex is solved. Such an approach provides insight into the association
between the proteins and compounds of each complex. This is accomplished by
selecting compounds with inhibitory activity, obtaining crystals of this new
protein/compound complex, solving the three dimensional structure of the
complex, and comparing the associations between the new protein/compound
complex and previously solved protein/compound complexes. By observing
how changes in the compound affected the protein/compound associations,
these associations may be optimized.
A compound that is identified or designed as a result of any of
these methods can be obtained (or synthesized) and tested for its biological
activity, e.g., inhibition of S. aureus EF-P activity.
Pharmaceutical Compositions
Pharmaceutical compositions of this invention comprise an
inhibitor of S. aureus EF-P activity identified according to the invention, or
a
pharmaceutically acceptable salt thereof, and a pharmaceutically acceptable
carrier, adjuvant, or vehicle. The term "pharmaceutically acceptable carrier"
refers to a carriers) that is "acceptable" in the sense of being compatible
with
the other ingredients of a composition and not deleterious to the recipient
thereof. Optionally, the pH of the formulation is adjusted with
pharmaceutically
acceptable acids, bases, or buffers to enhance the stability of the formulated
compound or its delivery form.
Methods of making and using such pharmaceutical compositions
are also included in the invention. The pharmaceutical compositions of the
41
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
invention can be administered orally, parenterally, by inhalation spray,
topically, rectally, nasally, buccally, vaginally, or via an implanted
reservoir.
Oral administration or administration by injection is preferred. The term
parenteral as used herein includes subcutaneous, intracutaneous, intravenous,
intramuscular, intra-articular, intrasynovial, intrasternal, intrathecal,
intralesional, and intracranial injection or infusion techniques.
Dosage levels of between about 0.01 and about 100 mg/kg body
weight per day, preferably between about 0.5 and about 75 mg/kg body weight
per day of the S. aureus EF-P inhibitory compounds described herein are useful
for the prevention and treatment of S. aureus EF-P mediated disease.
Typically,
the pharmaceutical compositions of this invention will be administered from
about 1 to about 5 times per day or alternatively, as a continuous infusion.
Such
administration can be used as a chronic or acute therapy. The amount of active
ingredient that may be combined with the carrier materials to produce a single
dosage form will vary depending upon the host treated and the particular mode
of administration. A typical preparation will contain from about 5% to about
95% active compound (w/w). Preferably, such preparations contain from about
20% to about 80% active compound.
In order that this invention be more fully understood, the
following examples are set forth. These examples are for the purpose of
illustration only and are not to be construed as limiting the scope of the
invention in any way.
EXAMPLES
Example 1: Analysis of the Structure of S. aureus EF-P
Expression of EF-P and Incorporation of Selenomethionine
The Escherichia coli construct M15 (pREP4) (pQE-60 EF-P)
which expresses S. aureus EF-P was obtained from Human Genome Sciences.
Various genes and polypeptides derived from S. aureus are published in WO
42
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
0012678. For preparation of the selenomethionine analog of EF-P, the
construct was grown in a minimal salts medium, M9, which contained glucose
and NH4Cl as the sources of carbon and nitrogen. Endogenous methionine
biosynthesis was then inhibited while adding an excess of selenomethionine to
the growth medium just prior to IPTG induction of EF-P synthesis (T.E.
Benson et al., Nat. Struct. Biol 2:644-53 (1995); G.D. Van Duyne et al., J.
Mol.
Biol; 229:105-24 (1993)). The formulation of basal M9 media was Na2HP04, 6
g; KHzP04, 3 g; NH4C1, 1.0 g; and NaCI, 0.5 g per L of deionized water. The
pH was adjusted to 7.4 with concentrated KOH and the medium was sterilized
by autoclaving. Prior to inoculation, the following filter sterilized
solutions
were added per L of basal medium: 1 M MgS04, 1.0 ml; 1 M CaClz, 0.1 ml;
trace metal salts solution, 0.1 ml, 10 mM thiamin, 1.0 ml; and 20% glucose, 20
ml. The trace metal salts solution contained per L of deionized water: MgCl2'
6H20, 39.44 g; MnS04' HZO, 5.58 g; FeS04' 7H20, 1.11 g; Na2Mo04' 2H20,
0.48 g; CaCl2, 0.33 g; NaCI, 0.12 g; and ascorbic acid, 1.0 g. Filter
sterilized
ampicillin and kanamycin were added to the medium at final concentrations of
100 p,g/ml and 30 pg/ml, respectively.
Fermentations were prepared in 100 ml volumes of M9 medium
contained in 500 ml wide mouth flasks. A 0.1 ml aliquot of the stock culture
was inoculated into the medium and allowed to grow at 37°C for 18 - 20
hours
with a shaking rate of 200 rpm. The seed culture was harvested by
centrifugation and then resuspended in an equal volume of M9 medium. The
resuspended seed was used to inoculate expression fermentations at a rate of
3%. For expression, the culture was grown under the same conditions to an Aboo
of 0.6. At this point, methionine biosynthesis was down regulated by the
addition of L-lysine, L-threonine, and L-phenylalanine at a final
concentration
for each of 100 ~,g/ml and L-leucine, L-isoleucine, and L-valine at 50 ~g/ml
each. D,L-selenomethionine was added simultaneously to a final concentration
of 100 ~g/ml. After 15 - 20 minutes, protein expression was induced by
addition of IPTG (isopropyl thio-(3-D-galactosidase, Gibco BRL) to 1 mM.
43
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Growth of the culture was continued for an additional 3 hours until an A6oo of
2.0 - 2.2. Cells were then harvested by centrifugation and frozen at -
80°C.
Under these conditions, the average yield of cell paste was 4.0 g/L and
approximately 20 mg/L of selenomethionine EF-P was produced.
Purification of Selenomethionine EF-P
E. coli cell paste resulting from 1 liter of induced cell culture was
resuspended in 25 mL of buffer A (50 mM Tris pH 7.6 containing 300 mM
NaCI and 10% glycerol). 10 mg of DNAse I and 1 Completes protease
inhibitor tablet was added to the suspension. The cells were then lysed by
passing the suspension three times through a French Press (Spectronic
Instruments, Rochester, NY) at a pressure of 1000 PSI using a chilled 40 mL
high pressure cell. The soluble cytosol was prepared from the lysate by
ultracentrifugation at 100,000 x g for 60 min at 4°C. Benzonase (25~L)
was
added to the cytosol to remove any residual polynucleotides. The clarified
cytosol was injected onto a HR10/10 FPLC column (Pharmacia Biotech,
Piscataway, NJ) packed with Ni-NTA Superflow resin (Qiagen, Chatsworth,
CA) which had been previously equilibrated in buffer A. All of the column
chromatography was performed on an AKTA 100 Explorer biochromatography
system (Pharmacia Biotech, Piscataway, NJ) at a flow rate of 1 mL/minute.
After injection, the column was rinsed with buffer A until the absorbance at
280
nm of the column eluate was less than 0.1 AUFS. The material was then eluted
from the column by a linear gradient of buffer B (500 mM imidazole in buffer
A). The gradient consisted of 0-8% buffer B in 20 minutes, followed by a 10
minute segment of 8% buffer B, and then a linear gradient of 8-100% buffer B
over 20 minutes. The column was washed with 100% buffer B for 30 minutes.
4 mL fractions were collected starting at the beginning of the gradient.
Selected
fractions were analyzed by SDS-PAGE for purity before making the final pool
for crystallographic studies.
In a manner similar to that for selenomethionine EF-P, S. aureus
44
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
EF-P was prepared and purified.
Protein Crystallization
The final purified EF-P protein sample was exchanged into 10
mM Tris pH 7.6 (5 mM DTT was added for selenomethionine EF-P) and
concentrated to 12 mg/mL for crystallization experiments. Initial crystals of
EF-P were obtained from a standard library of crystallization screening
solutions - Hampton Screen 1, condition 37, 0.1 M sodium acetate pH 4.6, 8%
PEG 4000. Optimization of the crystallization condition was conducted using
EF-P at a concentration of 12 mg/ml. Crystals grew best at O.1M sodium acetate
pH 5.2-5.4 and 1-4% PEG 4000. The crystals were grown by vapor diffusion in
hanging drops by the addition of a 1:1 starting ratio of protein to well
solution.
Since the pH of the crystallization buffer was at or very near the pI of the
protein, protein precipitation was always observed after adding well solution
to
the protein solution. Over the course of equilibration, the protein
resolubilized
and eventually crystallized during a period of 1-5 days. Subsequently, it was
determined that EF-P crystals could be grown by vapor diffusion in sitting
drops
where no well solution was added to the protein drop and equilibration
occurred
through the vapor phase overnight (this method avoided the initial
precipitation
event before crystal formation). Because of the lack of access to coordinates
for
either one of the eIFSA structures during the early stages of this project,
selenomethionine EF-P was overexpressed by downregulating methionine
biosynthesis (T.E. Benson et al., Nat. Struct. Biol 2:644-53 (1995); G.D. Van
Duyne et al., J. Mol. Biol; 229:105-24 (1993)) and purified using immobilized
metal affinity chromatography (see above). The purified selenomethionine EF-
P was maintained in 50 mM Tris pH 7.8 and 5 mM DTT. Crystals of the
selenomethionine EF-P grew in a manner similar to the methionine EF-P.
Crystals grown under these conditions diffracted to 1.9 A resolution at the
synchrotron.
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Data Collection and Structure Determination.
Multiwavelength anomalous dispersion data was collection at the
Advanced Photon Source located at Argonne National Labs in Argonne, IL on
beamline 17-ID on the sodium acetate crystal form. Complete data sets were
collected at three different wavelengths - one remote, low energy wavelength
(1.0332 ~), one wavelength at the inflection point of the selenium X-ray
fluorescence spectrum where the dispersive differences would be maximal
(0.979746 ~) and one wavelength at the peak of the X-ray fluorescence
spectrum where the anomalous differences would be maximal (0.979617 ~).
Each of these individual data sets was indexed and integrated separately (see
Table 1 for integration statistics). The data sets were scaled to each other
using
the program SCALEIT in the CCP4 Program Suite (Collaborative
Computational Project N4, Acta Cryst.; D50:760-63 (1994)). Patterson maps
initially revealed only two of the four selenium sites. Solution of the
Patterson
function was complicated by the presence of a cross vector between the two top
selenium sites on a Harker section. Cross difference Fourier maps revealed a
weaker third selenium site that indeed corresponded to a selenomethionine
position in the EF-P structure.
Heavy atom refinement and phase calculations carried out in
MLPHARE (Z. Otwinowski, in Isomorphous Replacement and Anomalous
Scattering 80-86 (W. Wolf et al., eds., SERC Daresbury Laboratory,
Warrington, 1991) and subsequently followed by solvent flattening in DM
(K.D. Cowtan & P. Main, Acta Cryst.; D49:148-57 (1993)) yielded electron
density maps that were severely disconnected. At this point in the structure
solution after investigating several alternative space groups, heavy atom
refinement and phase calculations were conducted using SHARP (E. La Fortelle
et al., A Maximum-Likelihood Heavy-Atom Parameter Refinement and
Phasing Program for the MIR and MAD Methods, P. Bourne & K. Watenpaugh,
eds., Crystallographic Computing 7 (1997)). Phases calculated in SHARP were
solvent flattened using the program SOLOMON (Collaborative Computational
46
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Project N4, Acta Cryst.; D50:760-63 (1994)) and gave a significantly improved
electron density map. At this stage in the structure solution, the coordinates
for
eIFSA from M. jannaschii became available and greatly aided the process of
model building. Model building was done using the program CHAIN ( J.S.
Sack, J. Mol. Graph; 6:224-25 (1988)) and LORE (B.C. Finzel, Meth.
Enzymol.; 277:230-42 (1997)). Refinement was carried out with XPLOR98
(A.T. Brunger, X-PLOR version 3.1: A system for X-ray Crystallography and
NMR, New Haven: Yale Univ. Press (1992)) incorporating bulk solvent
correction during the refinement (J.S. Jiang & A.T. Brunger, J. Mol. Biol;
243:100-15 (1994)). Progress of the refinement was monitored by a decrease in
both the R-factor and Free R-factor. Stereochemistry of the model was checked
using PROCHECK (R.A. Laskowski et al., Journal of Applied Crystallography;
26:283-91 (1993)) revealing no residues in disallowed regions of the
Ramachandran plot. Figures 3 and 10 were made using SETOR (S.V. Evans, J.
Mol. Graphics; 11:134-38 (1993)) and figures 5, 7, 8, 11-14 were produced in
MOLSCRIPT (P. Kraulis, J. Appl. Cryst.; 24:946-50 (1991)).
47
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
SEQUENCE LISTING
<110> Pharmacia & Upjohn
<120> CRYSTALLIZATION AND STRUCTURE DETERMINATION OF
STAPHYLOCCCUS AUREUS ELONGATION FACTOR P
<130> 6240.PCP
<140> Unassigned
<141> 2000-08-04
<150> 60/147,851
<151> 1999-08-06
<160> 5
<170> PatentIn Ver. 2.1
<210> 1
<211> 193
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Recombinant
Staphylococcus Aureus Elongation Factor P
<400> 1 -
Gly Ile Ser Val Asn Asp Phe Lys Thr Gly Leu Thr Ile Ser Val Asp
1 . 5 10 15
Asn Ala Ile Trp Lys Val Ile Asp Phe Gln His Val Lys Pro Gly Lys
20 25 30
Gly Ser Ala Phe Val Arg Ser Lys Leu Arg Asn Leu Arg Thr Gly Ala
35 40 45
Ile Gln Glu Lys Thr Phe Arg Ala Gly Glu Lys Val Glu Pro Ala Met
50 55 60
Ile Glu Asn Arg Arg Met Gln Tyr Leu Tyr Ala Asp Gly Asp Asn His
65 70 75 80
Val Phe Met Asp Asn Glu Ser Phe Glu Gln Thr Glu Leu Ser Ser Asp
85 90 95
Tyr Leu Lys Glu Glu Leu Asn Tyr Leu Lys Glu Gly Met Glu Val Gln
1
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
100 105 110
Ile Gln Thr Tyr Glu Gly Glu Thr Ile Gly Val Glu Leu Pro Lys Thr
115 120 125
Val Glu Leu Thr Val Thr Glu Thr Glu Pro Gly Ile Lys Gly Asp Thr
130 135 140
Ala Thr Gly Ala Thr Lys Ser Ala Thr Val Glu Thr Gly.:fiyr Thr Leu
145 150 155 160
Asn Val Pro Leu Phe Val Asn Glu Gly Asp Val Leu Ile Ile Asn Thr
165 170 175
Gly Asp Gly Ser Tyr Ile Ser Arg Gly Arg Ser His His His His His
180 185 190
His
<210> 2
<211> 188
<212> PRT
<213> Escherichia coli
<400> 2
Met Ala Thr Tyr Tyr Ser Asn Asp Phe Arg Ala Gly Leu Lys Ile Met
1 5 . 10 15
Leu Asp Gly Glu Pro Tyr Ala Val Glu Ala Ser Glu Phe Val Lys Pro
20 25 30
Gly Lys Gly Gln Ala Phe Ala Arg Val Lys Leu Arg Arg Leu Leu Thr
35 40 45
Gly Thr Arg Val Glu Lys Thr Phe Lys Ser Thr Asp Ser Ala Glu Gly
50 55 60
Ala Asp Val Val Asp Met Asn Leu Thr Tyr Leu Tyr Asn Asp Gly Glu
65 70 75 80
Phe Trp His Phe Met Asn Asn Glu Thr Phe Glu Gln Leu Ser Ala Asp
85 90 95
Ala Lys Ala Ile Gly Asp Asn Ala Lys Trp Leu Leu Asp Gln Ala Glu
100 105 110
2
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Cys Ile Val Thr Leu Trp Asn Gly Gln Pro Ile Ser Val Thr Pro Pro
115 120 125
Asn Phe Val Glu Leu Glu Ile Val Asp Thr Asp Pro Gly Leu Lys Gly
130 135 140
Asp Thr Ala Gly Thr Gly Gly Lys Pro Ala Thr Leu Ser Thr Gly Ala
145 150 ~ 155 160
Val Val Lys Val Pro Leu Phe Val Gln Ile Gly Glu Val Ile Lys Val
165 170 175
Asp Thr Arg Ser Gly Glu Tyr Val Ser Arg Val Lys
180 185
<210> 3
<211> 134
<212> PRT
<213> Methanococcus jannaschi
<400> 3
Val Ile Ile Met Pro Gly Thr Lys Gln Val Asn Val Gly Ser Leu Lys
1 ~ 5 10 15
Val Gly Gln Tyr Val Met Ile Asp Gly Val Pro Cys Glu Ile Val Asp
20 25 30
Ile Ser Val Ser Lys Pro Gly Lys His Gly Gly Ala Lys Ala Arg Val
35 40 45
Val Gly Ile Gly Ile Phe Glu Lys Val Lys Lys Glu Phe Val Ala Pro
50 55 60
Thr Ser Ser Lys Val Glu Val Pro Ile Ile Asp Arg Arg Lys Gly Gln
65 70 75 80
Val Leu Ala Ile Met Gly Asp Met Val Gln Ile Met Asp Leu Gln Thr
85 90 95
Tyr Glu Thr Leu Glu Leu Pro Ile Pro Glu Gly Ile Glu Gly Leu Glu
100 105 110
Pro Gly Gly Glu Val Glu Tyr Ile Glu Ala Val Gly Gln Tyr Lys Thr
115 120 125
Arg Val Ile Gly Gly Lys
130
3
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
<210> 4
<211> 136
<212> PRT
<213> Pyrobaculum aerophilum
<400> 4
Lys Trp Val Xaa Ser Thr Lys Tyr Val Glu Ala Gly G7,~.'Leu Lys Glu
1 5 10 15
Gly Ser Tyr Val Val Ile Asp Gly Glu Pro Cys Arg Val Val Glu Ile
20 25 30
Glu Lys Ser Lys Thr Gly Lys His Gly Ser Ala Lys Ala Arg Ile Val
35 40 45
Ala Val Gly Val Phe Asp Gly Gly Lys Arg Thr Leu Ser Leu Pro Val
50 55 60
Asp Ala Gln Val Glu Val Pro Ile Ile Glu Lys Phe Thr Ala Gln Ile
65 70 75 80
Leu Ser Val Ser Gly Asp Val Ile Gln Leu Xaa Asp Xaa Arg Asp Tyr
85 ' 90 95
Lys Thr Ile Glu Val Pro Xaa Lys Tyr Val Glu Glu Glu Ala Lys Gly
100 105 110
Arg Leu Ala Pro Gly Ala Glu Val Glu Val Trp Gln Ile Leu Asp Arg
115 120 125
Tyr Lys Ile Ile Arg Val Lys Gly
130 135
<210> 5
<211> 154
<212> PRT
<213> Homo Sapiens
<400> 5
Met Ala Asp Asp Leu Asp Phe Glu Thr Gly Asp Ala Gly Ala Ser Ala
1 5 10 15
Thr Phe Pro Met Gln Cys Ser Ala Leu Arg Lys Asn Gly Phe Val Val
20 25 30
4
SUBSTITUTE SHEET (RULE 26)
CA 02376065 2001-12-27
WO 01/10906 PCT/US00/21528
Leu Lys Gly Arg Pro Cys Lys Ile Val Glu Met Ser Thr Ser Lys Thr
35 40 45
Gly Lys His Gly His Ala Lys Val His Leu Val Gly Ile Asp Ile Phe
50 55 60
Thr Gly Lys Lys Tyr Glu Asp Ile Cys Pro Ser Thr His Asn Met Asp
65 70 . 75 80
Val Pro Asn Ile Lys Arg Asn Asp Phe Gln Leu Ile Gly Ile Gln Asp
85 90 95
Gly Tyr Leu Ser Leu Leu Gln Asp Ser Gly Glu Val Arg Glu Asp Leu
100 105 110
Arg Leu Pro Glu Gly Asp Leu Gly Lys Glu Ile Glu Gln Lys Tyr Asp
115 120 125
Cys Gly Glu Glu Ile Leu Ile Thr Val Leu Ser Ala Met Thr Glu Glu
130 135 140
Ala Ala Val Ala Ile Lys Ala Met Ala.Lys
145 150
SUBSTITUTE SHEET (RULE 26)