Note: Descriptions are shown in the official language in which they were submitted.
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
HUMAN CANCER VIRTUAL SIMULATION SYSTEM
BACKGROUND OF THE INVENTION
Field of the Invention
This invention generally relates to computer-implemented simulation systems
using mathematical and
descriptive algorithms and data, and more specifically relates to a computer-
implemented system that simulates
events of cancer in the human body. The invention uses computer-generated
simulations of biochemical and
morphological human cellular transformation from normal cells to metastatic
tumors and provides where, when and
how the distant metastases of cancer can take place. The invention uses
information from molecular biology and
medical science to model and predict cell to cancerous tumor to metastatic
occurrence using parameters related to
living organisms.
Background and Prior Art
The simulation of human biological and medical processes, and in particular
human cancer processes from
cell to tumor to distant metastatic sites would provide many advantages to
students, researchers, physicians and
patients. Currently, no mathematical and descriptive approach exists that can
provide practical results to students,
researchers, physicians and their patients about the origin and future
behavior of cancer in the human body. At
present, cancer is diagnosed and treated in real-time and in the real world
based on the best clinical information
available to health care providing teams. The present invention improves this
situation by utilizing a mathematical
and descriptive process to develop vital information for a human cancer
virtual simulation, thus providing a
computer based and interactive environment in which to examine origins,
current characteristics and future
outcomes of the human cancer process.
One or two models currently are under development to simulate normal cellular
processes and describe
biologic properties of normal cells, not human cancer cells as proposed in
this invention. Some models are under
construction based on scanned images or construction of tumors in a 3-
dimensional computer simulation. These are
representational models and differ from the present invention in that they do
not contain predictive, interactive or
retroactive analytical subroutines capable of modification of the program
output in real time. No descriptive or
mathematical engines have been developed to depict a cell undergoing
transformation from a normal cell and distant
cancer sites within the human body using specific molecular biological
cellular changes for human cancer. The
simulation engine that is provided by the present invention has the specific
ability to simulate human cancer
behavior and in an interactive environment that has practical diagnostic,
research and treatment approaches
regarding the origin of human cancer and possible future behavior of cancer in
the human body.
SUMMARY OF THE INVENTION
The present invention provides a model that simulates human cancer cell
behavior for medical and
physiological functions has unique educational value. It provides medical
students the ability to repeat a lesson. The
invention reduces the use of animals and the concomitant uncertainty with
animal information extrapolated to
human beings. The invention can improve medical education by offering a
dynamic flexible learning environment
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
never before possible due to the small number and availability of real live
patients with cancer cases at all stages of
disease. It allows students to make some therapeutic choices and examine the
efficacy and results in a virtual
environment. It links probability to the medical learning process in a hands
on way. No model can replace the live
patient experience; however the invention augments it and ethically provides a
realistic, yet forgiving environment
for learning. It provides students opportunities to make choices and to learn
from them.
For researchers, the interactive human cancer model according to the present
invention provides
descriptive simulations useful for the study of new and current pharmaceutical
applications in cancer treatment, and
can reduce costs and improve therapeutic interventions. A computer based
mathematical and descriptive engine of
the molecular to metastases process can provide simulated outcomes in a non-
clinical environment to help make
decisions when experimental clinical applications are being considered. Some
of the interfaces envisioned in the
invention will allow drug response to be modeled at the molecular, cellular,
tissue, and metastatic levels, with
simulated results. The results will be generated around biochemical parameters
of importance for a drug's
effectiveness or to help identify the patient's individual characteristics
that could benefit from a drug's treatment.
The invention has the ability examine timing cycles for drug delivery to
improve the effectiveness of anti-cancer
action, enable the modeling of a drug's effects on a tumor at multiple times
using the invention under a variety of
conditions enhancing the statistical and probable confidence of outcome.
The human cancer virtual simulation engine is capable of continual refinement
and improvement in
predictive accuracy through medical research by comparing and contrasting
results from the engine with results
from the actual clinical settings. This allows testing, modification and
improvement of the engine's mathematical
algorithms as new medical and scientific discoveries are uncovered. The
invention, because it is a computer based
mathematical and descriptive engine model of the human cancer process,
provides a researcher with a powerful tool
because it can be experimented with and modified using information from past
research and clinical studies. This
allows a researcher to test new and current hypotheses and algorithmic
expressions of the human cancer process.
The invention thus provides a means to test ideas and improve scientific
knowledge in a clinical or non-clinical
environment using retrospective information before clinical trials on real
patients begin. If new and improved
assumptions add new algorithms, based on molecular biology and medical
science, the invention can be modified
and improved in its ability to provide useful information to students,
researchers and physicians in understanding
and predicting the path of human cancer in the human body.
For treatment of the patient by a physician, the simulation system according
to the invention will allow
input of clinical findings specific to an individual cancer patient to be
entered into the engine. The function of
allowing input will allow the physician flexibility to make a range of
judgments of medical importance to be entered
into the engine and then the engine will develop a probable and statistical
prediction of several possible clinical
outcomes. The possible clinical outcomes will include where or when high
concentrations of cancerous emboli will
cause metastases to appear in the human body. Once a tumor and its location is
discovered, the engine will allow a
simulation backwards in time to determine the possible range of times for
cancer or tumor origin, cellular
biochemical changes that possibly gave rise to carcinogenesis, vascular
formation within the tumor and micro
metastatic behavior.
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
The engine's results and reports will be of importance in understanding the
etiology of cancer in an
individual patient and examining treatment options and evaluating prognosis.
The reports from the invention will
potentially allow diagnostic testing to be performed in localized areas,
looking for smaller cancer presence,
increasing the likelihood that earlier, less expensive, less invasive
treatment can be performed, thus improving the
quality of treatment outcomes. The invention will provide useful information
to assist the physician with short term
and long term patient follow-up and to compare and contrast the patient's
response to treatment in a real clinical
setting with simulations developed by the human cancer virtual simulation
engine. The engine will allow interface
and insertion of new information at any time along a time line permitting the
module's in the engine to be modified
to conform to new assumptions on a daily, weekly, monthly or yearly basis. The
engine would become another tool
in the arsenal of the oncologist for examining possible outcomes of treatment
as they develop treatment regimes.
The simulation system is progressive and is capable of being modified and
improved in conjunction with
new advances in molecular biology and medical science. The continual updating
of databases to improve the
calculation abilities of the engine and its subroutines, described later in
this application, is one way to accomplish
this. Another dynamic aspect of the invention is that the descriptions
generated from some parts, specifically the
molecular driven portions of the metastatic, tissue, and tumor modules will
provide estimations of tumor size, and
location in the human body and other cancer tumor morphology characteristics
that can be coupled with
visualization, display and imaging technology. As described hereinafter, the
invention describes processes involved
in cellular transformation from a nom~al cell then to a cancerous tumor and
describe probable metastases elsewhere
in the human body. The invention also has the capability to provide
projections about the origin and future of
human cancer manifestations.
The invention synthesizes fundamental molecular biology and medical knowledge
into a simulation
system. The system allows questions to be asked and provides answers of
practical value to the human cancer
process. It provides a virtual description beyond the present tense, such as
possible future metastatic sites and past
origin of the human cancer process. As new discoveries are made certain
algorithms can and will be modified and
improved, but the fundamental workings of the engine will remain the same.
The principal object of the present invention is to provide a descriptive and
mathematical engine for a
human cancer virtual simulation system that applies information from molecular
biology and medical science to
simulate the occurrence and metastases of cancer in the human body. The
invention uses a computer as an
information-input apparatus and a visual monitor for output. A series of
software program modules employing a
system of specially written programs and databases are employed which allow
the user to enter into the programs
individual patient information thereby producing information, results and
reports about simulated human cancer.
The invention has two major configurations, medical and educational
applications. The medical application
configuration generates human cancer simulation information, reports and
staristical and predictive results along
applicable time lines of the future or the past. This configuration allows
selection of modules, subroutines,
parameters, and patient input to be entered into the engine selectively.
Depending on the selection, the user can
move forward or backward in time to generate simulated human cancer results
and reports. The system provides the
capability to predict future courses of human cancer to describe the possible
origin of human cancer in a patient.
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
The results from the engine is virtual in that it produces simulated
descriptions of human cancer in the body in the
present, the past and the future.
The educational configuration will use pre-programmed information, but allows
limited interaction for
medical student educational purposes. The medical applications configuration
will allow diagnostic, treatment and
research human cancer simulations to be performed.
Brief Description of The Drawings
Fig. 1 is a block diagram illustrating the operator's relationship to the
engine including data flow;
Fig. 2 is a block diagram illustrating the operator's relationship as
described above with the engine configurations;
Fig. 3 is a block diagram of the overall engine module structure according to
one preferred embodiment of the
invention;
Figs. 4-10 are flow diagrams illustrating the various subroutines for each of
the modules of the simulation engine
according to the present invention;
Fig. 11 is a block diagram illustrating data flow interactions of manual user
input with the various modules of Fig. 3
and a patient information database;
Fig. 12 is a block diagram illustrating data flow interactions between the
various modules of Fig. 3;
Fig. 13 is a block diagram of a system for receiving data from external data
sources, and analyzing and distributing
the received data into various data types for incorporation into model
algorithms according to one preferred
embodiment of the invention;
Fig. 14 is a flow diagram detail of the cell cycle routine shown in Fig. 3
according to one preferred embodiment of
the invention; and
Fig. 15 is a flow diagram of an algorithm for simulating cell protein
production within a cell life cycle, according to
one preferred embodiment of the invention.
Detailed Description of the Preferred Embodiments
Description of Databases
The Human Cancer Virtual Simulation (HCVS) Tumor Databases hereafter will be
referred to as the
databases. The HCVS system of databases is defined as the organized
accumulation of information needed for the
numerical subroutines to perform their functions. The databases all operate in
the following manner. Based on input
or instructions from the user module interfaces the connection module in the
HCVS system will access databases,
provide information to numerical solution modules and their subroutines to
generate results and reports relevant to
the human cancer virtual simulation under study. The databases described in
this version are the DNA (genetic)
database, biomarker (molecular) database, statistical database of information
from actual patients, and a metastatic
database.
Genetic Database
The DNA database includes information concerning genes of human cells as part
of the carcinogenesis.
4
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
The information will include cellular genes that are mutated and/or deleted.
This information will include why,
when, how, and other parameters of need for the simulation system's
subroutines.
Molecular Database
The molecular database includes information that provides biochemical evidence
of human carcinogenesis.
This database will include human normal cell and cancer cellular molecular
information, for example the phenotype
of the mutated genes from the genetic database. This information will include
biochemical mechanisms, protein
functions and possible biological significant compounds.
Statistical Database
The statistical database includes information from studies performed for the
specific forms of human
cancer the engine is asked to examine. The statistical database will also
include lifestyle issues related to specific
cancers. This information is useful for all of the applications of the
invention. Examples include information on the
average age of a cancer patient at death, physiological information concerning
the average size of an adenoma stage
1 tumor, etc.
Metastatic Database
The metastatic database includes all types of previously mentioned information
specific to metastasis. This
metastatic information will support the metastatic module of the HCVS.
System Overview and Operation
The invention is implemented on a computer, including an information input
device, such as a keyboard,
and a visual monitor or printer for output. The simulation system includes a
series of software program modules
each employing a system of specially written programs in conjunction with the
above-described databases that
utilizes a computer to perform its operations and generate results. These
programs allow the user to enter individual
patient information, if desired, or to use pre-programmed information
producing information results in the form of
reports about simulated human cancer. As shown in Fig. 1, the operator
interface (including keyboard, mouse and
display device in operative connection with a central processing unit) sends
data and instructions to the engine,
where they are processed, and processed data and instructions are sent by the
engine back to the operator interface
for output to the operator in the form of graphical displays, textual
displays, or printed reports.
As illustrated by Fig. 2, the invention has two major configurations,
educational and medical applications.
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
The medical applications configuration allows diagnostic, treatment and
research human cancer simulations to be
performed. The educational configuration uses pre-programmed information, but
allows limited interaction for the
medical student's or training professional's educational proposes. The medical
application configuration generates
human cancer simulation information. This information can be in the form of
reports of present information, and/or
statistical and predictive information correlated to applicable time lines in
the future or from the past. This
configuration allows selection of modules and patient input to be entered into
the engine selectively. Depending on
the module selection, the user can move into the future or backward in time to
generate simulated human cancer
results. The system provides the capability to predict future courses of human
cancer, and to describe the possible
origin sight and initial biological traits of human cancer in a patient. The
result from the engine is virtual in that it
produces simulated descriptions of human cancer in the body in the present,
the past, and the future.
The biological process for a cell's transformation from a normal cell to a
cancerous cell and then to metastatic
activity is described within six modules in this invention. As shown in Fig.
3, each section (biological stage and
view) is organized into a module. The six modules are the tumor origin,
cellular, colony, tissue, tumor and
metastatic modules. Each module contains one or more subroutines. These
subroutines carry out smaller descriptive
and mathematical processes needed to simulate human cancer biology. Each of
the subroutines will produce results
in forms needed by the user to describe the biological process the subroutine
simulates. There are fourteen
different subroutines within the system of the invention. The subroutines are
as follows:
The genetic mutation subroutine and diagnostic subroutine of the tumor origin
module,
The cell cycle subroutine and the physical properties subroutine of the
cellular module,
The interaction between cells subroutine and the structure subroutine of the
colony module,
The interaction between cells subroutine and the tissue structure subroutine
of the tissue module,
The interaction between cells subroutine, the tissue structure subroutine, and
the physical properties of the
tumor subroutine of the tumor module, and
The statistical and clinical outcome subroutine, the molecular biological
subroutine, and the cancer originlrun
forward subroutine of the metastatic module.
The results are in the form of reports, generated by the subroutines of the
modules, and placed into data sets.
The data sets can then be viewed in the interface corresponding to the module
that the reports came from.
As illustrated in Fig. 3, the invention has six module user interfaces, the
molecular, cellular, cellular expansion,
pre-neoplastic, neoplastic and metastatic interfaces. These interfaces each
correspond to a respective module and act
as information input and output points. This is where the human user receives
instructions or requests, where
choices about subroutines or their information output reports are chosen and
where input information about a patient
is entered to allow the programs to operate. It is at the user interface where
the reports, that the subroutine
generated and places into data sets, will be viewed. Thus the input
information goes in through the user interfaces
and the output information sets come back out of the user interface (see Fig 1
).
A general example of the entire process that the simulation system uses is
described in Fig 4. In operation, the
user selects a particular module for operation. Each module and its
subroutines then follows a process of operating
the associated interface using the computer monitor and keyboard. The module
then initiates activation of the
6
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
central processing unit through the software programs with the connection
module connecting the module to the
databases of information needed for the subroutines. The modules then engage
the numerical solutions module and
the subroutines associated with the numerical solutions module and finally
provide the user with reported solutions
to user requests through the monitor and various other displays. All of the
operations take place in the basic
computer hardware and software previously described.
The connection module is activated whenever the input from the user is
completed and the user initiates the
subroutines. The connection module processes the input from the user, gathers
information from the various
databases necessary for the execution of the mathematical and descriptive
algorithms within the various numerical
solution modules and transmit it to its proper location. If the program is
stopped at the user interface along any
aspect of the time line, and input is modified, the connection module will
automatically engage to transmit to the
numerical solutions module the new information. The numerical solutions module
will then generate the sequential
processing. If the program is paused, the connector module will disengage. The
numerical solutions module
subroutines carry out the task of generating the results for the reports. Once
started they will generate the previously
mentioned results and arrange them into reports. These reports will be sent to
the user interface for viewing on the
monitor or for printing. If continual output is chosen and the user stops the
program, the numerical module (s) will
store results generated up to that point in time. The user may at any time
request reports. If the program is restarted
it will begin where it left off, include the changed parameters in the next
sequential report, continue calculations
with modified parameters and produce a new report. If paused, the numerical
module will resume its calculations
when the pause is ended, generate its results, report back to the user
interface and the process is completed.
Detailed descriptions of the various subroutines of each module will now be
described with reference to
Figs. 5-10.
TUMOR ORIGIN MODULE
Referring to Fig. 5, the HCVS Tumor Origin Module has two purposes. The first
purpose is to use
mathematical calculations to go back in time to the original site of
carcinogenesis. This will include the original
genetic mutations found to be the initiatory step of the disease. The
initiatory step is the first mutation of a normal
cell that leads to the phenotype of the specified cancer.
The second purpose of the Tumor Origin Module is to diagnose cancer at the
genetic level. This will be the
cutting edge of the early detection technology. For example, in colon cancer
there are set genetic steps to
carcinogenicity. If the patient had a biopsy of a benign polyp and the
molecular (i.e. genetic) information was
extracted, it could be entered into the HCVS and the Tumor Origin Module would
diagnose the probability of a
future malignancy.
The operator enters information about the patient and their current tumor into
the system through the
operator interface. This will include the age, weight, and other relevant
physiological information about the patient.
This information is then run through the tumor origin subroutines in the Tumor
Origin Module. This information
will then result in a reports) of information about the location and molecular
properties of the tumor's origin.
Prostate cancer is a good example of the function of this module. In prostate
cancer the disease's initiatory
step is not fully understood. The main reason for the complexity of prostate
cancer is the heterogenicity of the
7
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
tumors. In the prostate there are several primary tumors at detection. Within
the detected prostate tumors there are
several different genotypes. This complexity makes early detection of prostate
cancer for a pathologist difficult. The
tumor origin module of the HCVS will be able to trace the tumors' progress
from the state of the detected tumor
back in time using the databases and calculations of the subroutines. The
product of the subroutine executions will
be molecular information showing the probability of which mutation or series
of mutations could be the initiatory
step.
The display of the resultant information is described in the molecular
interface.
Genetic Mutation Subroutine of the Tumor Origin Module
The purpose of the genetic subroutine of the tumor origin module is to
calculate the different mutations
that can occur in cellular genetic code involving cell transformation steps
leading to, and resulting in, a cancerous
state. The genetic mutations can occur by three different methods (deletion,
insertion, and substitution) these three
methods lead to codon mutations (chain termination mutation, frameshift
mutation, missence mutation, and
nonsense mutations). Other somatic mutations also exist, these mutations only
occur in somatic cells. By the
pathways of mutation being limited we can categorize a mutation to have a
specific phenotype. The genetic
mutation will then effect the cell by the expression or non-expression of the
gene's protein.
For example, in order to inactivate the APC (Adenmatous Polyposis Coli) gene
both alleles must be lost. In
the situation of Familial Adenomatous Polyposis Syndrome a recessive APC
allele is inherited. At this point only
one mutation (i.e. a somatic mutation) needs to occur in order to lose APC
function. (See Human Cancer Virtual
Simulation Cellular Module to see the results of an APC deletion) By
simulating these relationships, the tumor
origin module will be able to examine the existing situation (disease) and
estimate back to the tumors origin. These
genetic mutations and functional relationships will be mathematically operated
in the genetic subroutine of the
origin module.
Diagnosis from Molecular Information Subroutine of the Tumor Origin Module
The purpose of the diagnosis from the molecular information subroutine in the
tumor origin module is to
improve early detection of cancer. For example, in colon cancer, the accepted
initiatory step is the mutation of APC.
If, as described earlier, a user had the molecular information for a benign
polyp of a patient and this information
was entered into the HCVS the diagnosis from molecular information subroutine
would calculate the probable
timing and initial intensity of a future malignancy.
Tumor Origin Module Information Sets
Both of the tumor origin module subroutines, genetic mutation and diagnosis
from molecular information
will have information outputs that will be inputted into the molecular
interface.
Genetic Mutation Subroutine
~The type and location of the genetic mutation.
Using the information from the genetic database the genetic mutation
subroutine will show the type and
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
location of the genetic mutation that has occurred.
~The phenotypic result of the genetic mutation.
Using the genetic database as well as the molecular database the genetic
mutation subroutine will express
the phenotypic results of the mutated genes. This will include protein
expression, protein reactions, and etc.
~The gene products lost in the mutation.
Using the molecular database the genetic mutation subroutine will determine
the gene products that are lost
due to the genetic mutation. The operator will be able to activate and
deactivate genes therefore effecting the gene
products. This information will then be available to the operator to view and
manipulate.
Diagnnosis from Molecular Information Subroutine
~The possible initiatory step and the probability it has occurred
Using the genetic and the molecular databases, the diagnosis from molecular
information subroutine will
calculate a list of possible initiatory steps and their probability. This
information will be displayed on the molecular
interface, and the operator will be able to manipulate the scenarios using
this interface.
~The mutations that have occurred and follow the carcinogenic path and the
mutations along that same path
that have not occurred.
Using the genetic and the molecular databases, the diagnosis from molecular
information subroutine will
determine the most likely genetic pathway. From the most probable carcinogenic
pathway, the diagnosis from
molecular information subroutine will determine the genetic variations that
have occurred, and the genetic
variations that will occur, if the selected pathway is followed.
~The probability and proposed carcinogenic pathway.
Using the genetic and molecular databases, the diagnosis from molecular
information subroutine will
determine all the possible carcinogenic pathways and their corresponding
probability. This information will be
displayed and the operator will be able to select a pathway and the engine
will analyze it.
Molecular Interface
The molecular interface will display and allow user interaction with the
module for the information on the
actual interaction of the genes and molecules. The reports will include
chemical mechanisms, bond strengths, timing
of mutations, possible alternative mutations, etc.
The cell bonding strength is an example of the information included in the
molecular level. In the cell
bonding the APC product must undergo homo-oligomerization to produce the beta-
catenin that is essential for the
production of the cell bonding cadherin. In the situation where an APC
deletion would occur, the cell bond strength
would be reduced. The information will be available concerning the reaction
mechanism, time of reaction,
9
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
conditions of reaction, and etc.
CELLULAR MODULE
As shown in Fig. 6, the Cellular Module is the portion of the HCVS engine that
controls the cellular
information. The purpose of the Cellular Module is to use mathematical
calculations to describe the behavior of an
individual cell and its surrounding environment. The Cellular Module will
accomplish its tasks by mathematically
calculating the properties of the cells. This is described in the subroutines
of this module.
The control of an individual cell and it's environment is shown by an example
of p53 gene loss. The p53
gene is a tumor suppressor gene. The p53 gene activates the p21 protein. The
p21 protein has the function of
arresting cell cycle progression so the cell can either undergo repair or
apoptosis. This is accomplished by inhibiting
cyclin cdk complexes. When p53 is mutated the p21 protein doesn't activate at
the levels necessary and the cell
continues through a complete cycle to produce two mutated daughter cells. This
process, and many others not
described, are part of carcinogenesis, and will be controlled through said
mathematical calculations in the cellular
module.
The display of the resultant information is described in the cellular level
interface.
Cell Cycle Subroutine of the Cellular Module
The purpose of the cell cycle subroutine of the cellular module is to simulate
the aspects of a cell life cycle.
These aspects will include the timing of the cell cycle, control of the stage
of the cell cycle, etc. The required
information will be in the molecular database. The cell cycle subroutine will
use statistical and chemical information
to run its calculations.
The control of the cells cycle properties is shown in the p53 example (see
Cellular Module). The effects of
the mutation of the p53 gene will affect the timing of mitosis. This effect is
an increase in speed because the cell
doesn't stop the process of cell cycle progression to repair the mutation.
Physical Properties of the Cell Subroutine of the Cellular Module
The purpose of the physical properties of the cell subroutine of the cellular
module is to describe
mathematically the size, shape and structure of the cell changes throughout
carcinogenesis. For example, in the
shape of the cells in large cell undifferentiated carcinoma there is a
classification of cells named clear cell. This
nomenclature came about due to the cytoplasm of a cell turning clear during
carcinogenesis. This state occurs
because of large deposits of glycogen in the cytoplasm.
Cellular Module Information Sets
Both of the cellular module subroutines, cell cycle and physical properties of
the cell will generate
information from its calculations and return this information to the cellular
level interface.
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
Cell Cycle Subroutine
~Rate of mitosis
Using the molecular and the statistical databases the cell cycle database will
produce information on the
rate of mitosis in the form of time/cell cycle. This information will be
useful in determining the speed of the
diseases' progression. The operator will be able to adjust the cycle in order
to view hypothetical situations, but the
system will always default back to the calculated "real life" rate.
~Survival rate of the cells
Using the molecular and statistical database the cell cycle subroutine will
determine the mean and standard
deviation/time of the survival rates of cells. This subroutine will also
determine the number of cells that induce
apoptosis as a normal function. As described in the cellular module many
mutated cells will induce apoptosis to
deter a genotype or phenotypic mutation from progressing. In carcinogenesis
one of the factors is the situation that
the gene products that are responsible for apoptosis are themselves mutated.
This information will be displayed in
the cellular level interface.
Physical Properties of the Cell Subroutine
~Physical properties of the cell.
Using the molecular and statistical databases the physical properties of the
cell subroutine will determine
the size, shape, physical makeup, and etc. This information will be displayed
on the cellular level interface and will
be used as information by other modules.
Cellular Interface
The cellular level of information will display the rate of mitosis, the rate
and level of mutation, the size and
shape of the cell, etc. generated by the Cellular Module of the HCVS engine.
The rate of mitosis is an example of
important information at the cellular level. The HCVS will be able to give the
information concerning the shape size
and rate of replication (mitosis). The time and stage of the disease will
reference this information from the cell. The
operator will be able to select the information set according to time to see
the size and shape of the cells at various
times in the past, present or future. The other variables will be available
for adjustment as well. The operator will
have tailored the HCVS for the specific case and will then enter the size and
shape of the cells they are looking at.
The operator will then enter that information and the HCVS will generate the
information for the probable time and
stage of the disease.
COLONY MODULE
Referring to Fig. 7, the purpose of the Colony Module is to mathematically
describe small cellular
populations. The importance of this can be shown by nutrition consumption.
When a pre-neoplastic growth occurs,
the nutrition consumption of the growth increases from its standard rate of
consumption. This happens for many
reasons, but an example is cellular growth rate. The pre-neoplastic growth
occurred partly because of an increased
11
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
growth rate. This increased growth rate needs more nutrition from the body.
This imbalance will lead to the cells
around the growth to give up some of their needed nutrition to the growth; the
colony module through mathematical
calculations will determine this dynamic relationship.
The display of the resultant information is described in the cellular
expansion interface.
Interaction between Cells Subroutine of the Colony Module
The purpose of the interaction between cells subroutine of the colony module
is to control the interaction
of simulated cells through mathematical calculations. The nutritional
consumption described in the colony module is
good example of this relationship. Other modules in the invention will use the
information from this subroutine.
Structure Subroutine of the Colony Module
The purpose of the structure subroutine of the colony module is to show the
structure of the cells together
in this small population. The structure subroutine will include the physical
characteristics of the individual cells but
will also include the structural relationship of the cells together. In large
cell undifferentiated carcinoma of the lung,
the physical properties of the cell along with this intracellular structure
are the keys to diagnosis and classification.
Colony Module Information Sets
Both of the subroutines of the colony module, interaction between cells and
structure, will produce
information and return it to the cellular expansion interface.
Interaction between Cells Subroutine
~Nutrition Consumption
Using the molecular and statistical databases, the interaction between cells
subroutine will produce
information in the measurement of moles/unit. This information will include
all standard compounds that are
biochemically necessary, but also will include a section to input a new
compound along with its chemical properties
and the interaction between cells subroutine will calculate the involvement.
~Cell Bonding
Using the molecular database the interaction between cells subroutine will
calculate the bond strength
between cells. Some of the contemplated modes of measurement are kcal/mol
necessary to break the bond and etc.
This information will be displayed on the cellular expansion interface, but
will also go into the information input
area to fuel other modules.
Structure Subroutine
~Physical Properties of the Individual Cells
Using the molecular and statistical databases the physical properties of the
cell subroutine will determine
the size, shape, physical makeup, and etc. This information will be displayed
on the cellular level interface and will
12
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
be used as information by other modules.
~Intracellular Structure
Using the molecular and statistical database the structure subroutine will
produce information for the
structure of the tissue. This structure will take on many forms and functions.
This information will be displayed on
the cellular expansion interface.
Cellular Expansion Interface
The cellular expansion interface will be where the information form the
subroutines of the Colony Module
(concerning small populations of cells and their interactions) will be shown
and described to the user. The operator
will be able to adjust the variables such as oxygen distribution in order to
see the interaction between the simulated
cells described by the subroutine programs. The value of the information
displayed here to the user is that these
relationships would be difficult to see in the later interfaces.
TISSUE MODULE
Referring to Fig. 8, the purpose of the Tissue Module is to describe with
mathematical calculations a large
population of different cells interacting in a normal and a malignant
environment. Within the tissue of every organ
there are several different types of cells. The different types of cells have
different functions and therefore different
locations. The Tissue Module will describe small abnormal growths. The
interactions between the small growth and
the outer levels of the tissue, are important to describing and understanding
the path of carcinogenesis in practical
terms. These interactions are described mathematically by the tissue module.
The display of the resultant information is described in the pre-neoplastic
interface.
Interaction between Cells Subroutine of the Tissue Module
The purpose of the interaction between cells subroutine of the tissue module
is to mathematically describe
the intracellular interaction among large populations of simulated cells
within the Tissue Module. The nutritional
consumption described in the Colony Module is a good example of this
relationship. Other modules in the
invention will use the information from this subroutine.
Tissue Structure Subroutine of the Tissue Module
The purpose of the tissue structure subroutine of the Tissue Module is to
mathematically control the
intracellular structure among large populations of simulated cells within the
Tissue Module. As described in the
Structure Subroutine of the Colony Module, the structure of the intracellular
relationship is important not only in
diagnosis but in cellular classification. In the Tissue Module, this
subroutine will also begin to generate the different
layers of the tissue cross section. In all tissues there are several levels of
tissue within the tissue sample. This
subroutine will generate the information needed to represent these levels and
their interaction with each other.
13
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
Tissue Module Information Set
Both of the subroutines of the tissue module, interaction between cells and
tissue structure, will produce
information and return it to the pre-neoplastic interface.
Interaction between Cells Subroutine
~Nutrition Consumption
Using the molecular and statistical databases, this subroutine will produce
information in the measurement
of tissue level nutritional consumption. The simulation will generate
information in chemical terms (i.e. moles)
relevant to the user. The unit may vary dependent on the organ simulation.
This information will include all
standard compounds that are biochemically necessary, but also will include a
section to input a new compound
along with its chemical properties and the interaction between cells
subroutine. The subroutine will then calculate
the chemical interacrion of the new compounds at the tissue level and display
the information at the pre-neoplastic
interface.
~Cell Bonding
Using the molecular database the interaction between cells subroutine will
calculate the bond strength
between simulated cells in the tissue module. One of the contemplated modes of
measurement are kcal/mol
necessary to break the bond among others not described. This information will
be displayed on the pre-neoplastic
interface, but will also go into the information input area to fuel other
modules.
Tissue Structure Subroutine
~Physical Properties of the Individual Cells
Using the molecular and statistical databases the physical properties of the
cell subroutine will determine
the size, shape, physical makeup, and etc. of the simulated cells within the
tissue module. This information will be
displayed on the pre-neoplastic interface and will be used as information by
other modules.
~Intracellular Structure
Using the molecular and statistical database the structure subroutine will
produce information for the
structure of the simulated tissue. This structure will take on many forms and
functions. This information will be
displayed on the pre-neoplastic interface
~Tissue Levels
Using the molecular and statistical database the tissue structure subroutine
will generate information
concerning the development, size, shape, and etc. of the tissue. This
information will be presented on the pre-
neoplastic interface.
14
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
Pre-neoplastic Interface
The pre-neoplastic interface will display information from the Tissue Module
information set, such as
nutritional consumption and the other information sets described which are
generated by the Tissue Module
subroutines. This level will show information from the earliest stages of a
simulated primary tumor or tumors. This
information will also be available for the operator to manipulate as in the
previous levels.
TUMOR MODULE
Referring to Fig. 9, the purpose of the Tumor Module is to control complex
relationships of simulated
tumor growth through mathematical calculations for the majority of
tumorigenesis. This process needs its own
module because of the complexity of the interactions between the disease and
the surrounding cells.
The development of blood vessels is a good example of the complexity of the
systems of the Tumor
Module. The primary tumor will continue to grow at an accelerated rate as long
as the nutrients are available for the
growth. (When the threshold is reached where the nutrients are limited, this
corresponds to the weak cell bonds
discussed earlier.) This weakness allows for the nutrient carrying blood to
reach the tumor and form small blood
vessels. This relationship, along with many others, will be controlled
mathematically by the Tumor Module.
The display of the resultant information is described in the neoplastic
interface.
Interaction between Cells Subroutine of the Tumor Module
The purpose of the interaction between cells subroutine of the Tissue Module
is to mathematically run the
intracellular interaction. The nutritional consumption described in the Colony
Module is a good example of this
relationship. Other modules in the invention will use the information from
this subroutine.
Tissue Structure Subroutine of the Tumor Module
The purpose of the tissue structure subroutine of the Tumor Module is to
mathematically control the
intracellular structure. As described in the Structure Subroutine of the
Colony Module, the structure of the
intracellular relationship important not only in diagnosis but in
classification. In the Tissue Module this subroutine
will also begin to generate the different layers of the tissue cross section.
As in all tissues there are several levels of
tissue with in the tissue sample. This subroutine will generate the
information needed to represent these levels and
their interaction with each other.
Physical Properties of the Tumor Subroutine of the Tumor Module
The purpose of the physical properties of the tumor subroutine of the Tumor
Module is to calculate the
behavior of the virtual disease. This subroutine will handle information such
as tumor mass, growth rate, cell
structure, etc. The information describing that characteristic will be
generated in the physical properties of the tumor
subroutine.
An example of the physical properties of a tumor is prostate cancer, as
mentioned previously (see Tumor
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
Origin Module) where there are multiple primary tumors.
Tumor Module Information Sets
Each of the three subroutines; interaction between cells, tissue structure,
and physical properties of the
tumor subroutine, will generate information for the neoplastic interface. The
first two subsections have already been
described and are referenced. The last of the three subroutines' results is
described below.
Physical Properties of the Tumor
~Tumor Mass
The physical properties of the tumor will describe the size, shape, and
overall mass of the primary tumor.
This information will be shown in a metric scale and percentage of total area.
This information will also be sent to
the input information area to help run other modules.
~Tumor Growth Rate
The physical properties of the tumor, generate tumor growth information
concerning the growth of a tumor
over time. This information will also include the major factors driving the
growth. This will be sent to the input
information area, as was the tumor mass information.
~Genetic Mutations Present
Using all the databases except the metastatic database, the physical
properties of the tumor subroutine will
determine the genetic changes that have occurred and will occur during the
simulation.
~Vascular Construction
Using the molecular and statistical database the physical properties of the
tumor database will generate
information for concerning vascular construction. This information will also
come from the tissue module. This
information will be sent to the information-input area to help run the
metastatic module.
Neoplastic Interface
The neoplastic interface will display the information from the Tumor Module.
The complex relationships
will be shown and available for adjustment. This interface will show the most
information concerning the primary
growth.
After entering all the information concerning the patient and the disease, the
operator will then be able to
adjust the size of the tumor to determine the stage that the patient is in.
The operator will also be able to adjust the
information to see the corresponding information, the order of mutations that
have occurred and see the information
for the mutations that have yet to occur.
16
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
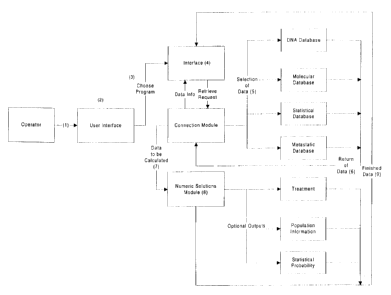
METASTATIC MODULE
Referring to Fig. 10, the Metastatic Module is a human cancer predictive and
prognostic engine used to
simulate and generate simulations and reports about cancer behavior in the
human body. It uses information
supplied to its various subroutine computer software programs or using pre-
programmed instructions to accomplish
this. The purpose of the Metastatic Module is to provide useful information
from mathematical and descriptive
algorithms to help forecast and predict the course of cancer progression in
the human body.
A second use of the module is to examine the possible effects of treatment
intervention steps using input
supplied to its programs. The input supplied to the programs within the
Metastatic Module may be specific to an
individual or the program can examine possible futures based on assumptions
within the program. This module has
significant diagnostic and therapeutic applications for human cancer patients
and the physicians that treat them. Due
to the importance of this module and the predictive and prognostic
applications, its operation will be described in
more detail and serve as a fuller example of what the invention is intended to
do.
The Metastatic Module Subroutines, Their Scientific Assumptions and Mode of
Operation
There are three metastatic cancer predictive and prognostic subroutines that
appear on the user interface
1 S once the Metastatic Model is selected. The first would be a clinical and
statistical outcome, the second would be
molecular biological, and the third would be cancer origin/run forward.
The Statistical and Clinical Outcome Subroutine of the Metastatic Module
The Statistical and Clinical Outcome subroutine will use epidemiological
information (morbidity,
mortality, height, weight, treatment steps such as chemotherapy, surgery and
radiation; when treatments were
administered, patient response to treatment or estimates thereof, initial
tumor size and volume, tumor geometric
shape, tumor location, patient age, health conditions of relevance to patient
such as HIV status, immune system
strength through secondary biomarkers such as white blood cell and T-cell
count, family history of relevance to
cancer epidemiology, other complicating factors or diseases, local and distant
metastases location, biomarkers of
human cancer such as estrogen receptivity in breast cancer and others, mitosis
rate, cell diploidy, etc.) assembled
into databases previously described within the invention and gathered from
credible scientific sources from real
human cancer patients relating to the course of human cancer. The purpose and
resulting information from this
subroutine is to predict the future course of a patient's human cancer by
relating user inputted information related to
the database described and using linear mathematics within the subroutine
coupled with inputted information and
assumptions to predict possible clinical disease outcomes for the
parient/physician. The underlying assumption is
that future cancer behavior will be similar to that documented from the past
and the engine information will prove
useful for predictive and prognostic purpose.
Molecular Biological Subroutine of the Metastatic Module
The molecular biological subroutine will use information known about a cancer
or tumor (pathology report
information such as tumor size and volume, tumor geometric shape, type of
cancer, tumor location, level of
17
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
vascularity within the tumor, location to veins and artery in target organ,
mitosis rate, biomarkers of relevance, cell
differentiation, cell diploidy etc.) in addition to patient characteristics of
importance to the engine (height, weight,
treatment steps such as chemotherapy, surgery and radiation; when treatments
were administered, patient response
to treatment or estimates thereof, patient age, health conditions of relevance
to patient such as HIV status, immune
system strength through secondary biomarkers such as white blood cell and T-
cell count, family history of relevance
to cancer epidemiology, other complicating factors or diseases, local and
distant metastases location, etc.) and
generate information related to micro metastasis rate and behavior of cancer
cells or emboli (clusters of cancer cells)
and their ability to move to new sites developing possible and probable
scenarios of human cancer reoccurrence and
growth within the body. The underlying assumption in this subroutine is to
provide populations of individual cells
with instructions, based on what is known about the patient and their cancer's
molecular biological behavior, that
will influence cancer cell growth, death, and other disease characteristics
within the body. The molecular biological
subroutine will then begin functioning, using programs and assumptions
inputted by the physician about a patient to
provide information useful for prognosis and prediction of human cancer.
Cancer Origin/Run Forward Subroutine of the Metastatic Module
The cancer origin/run forward subroutine is a sequential operation of the
Cancer Origin module which will
be a reverse operation of the subroutines within the tumor, tissue, colony,
cellular and finally tumor origin modules
and then followed by the molecular biological subroutine. The underlying
assumption of the cancer origin module is
beginning at a point in time when the cancerous tumor is discovered a
simulation with the HCVS engine can be
developed running backward in time using the various modules to reach the
origin of cancer development. The
HCVS engine can then be run forward through the present and into the future.
It is envisioned this will provide
useful information for prognosis and prediction of human cancer behavior. One
advantage of the cancer origin/mn
forward subroutine is that it can test engine input for efficacy. When
information from this subroutine generates
information that closely resembles the patient's disease condition in the
present, that result may be used to decide
parameters to drive simulations in the future. Additional information that
could be provided by using this technique
would be additional micro metastasis not seen at the primary site of the
tumor.
Metastatic Module Information Set
Each of the three Metastatic Module subroutines, statistical and clinical
outcome , molecular biological and
cancer origin/run forward will contain numerical solution modules to perform
the following estimations and
descriptions and generate reports back to the user interface and can be
selected by the user: a) metastatic occurrence,
target organ and percentage possibility, b) percentage disease free survival,
c) micro metastatic and metastases
volume, d) projected cancer cellular mitosis phase table, e) projected blood
biomarker concentration over time.
Considering now that the system herein described will have 15 different
numerical solution modules
described in the diagram for this part of the invention in greater detail,
each numerical solution module within the 3
Metastatic Module subroutines will be described as to what it will do beyond
the general function of a numerical
solution module above and how it will perform its said function:
18
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
The Statistical and Clinical Outcome Numerical Solution Module Output
Subroutines
~Metastatic occurrence, target organ and percentage possibility.
Using information from the epidemiological database and from other databases
within the invention an
algorithm will be constructed that will make use of medical and personal input
about an individual patient's present
condition to determine the probable organ sites of distant metastatic human
cancer projected in a future time frame.
The algorithms will use linear and curvilinear analysis and other mathematical
means to achieve this endpoint.
Where possible the algorithm will use statistical analysis to determine the
confidence of its predictions.
~Percentage disease free survival
Using information from the epidemiological database and from other databases
within the invention, an
algorithm will be constructed that will make use of medical and personal input
about an individual patient's present
condition to determine the morbidity and mortality of distant metastatic
human, cancer projected in a future time
frame to generate predictions of disease free survival. The algorithms will
use linear and curvilinear analysis and
other mathematical means to achieve this endpoint. Where possible, the
algorithm will use statistical analysis to
determine the confidence of its predictions.
~Micro metastatic and metastases volume
Using information from the epidemiological database and from other databases
within the invention an
algorithm will be constructed that will make use of medical and personal input
about an individual patient's present
condition to determine the micro metastatic and metastases volume of distant
metastatic human cancer projected in a
future time frame. The algorithms will use linear and curvilinear analysis and
other mathematical means to achieve
this endpoint. It is envisioned that this description will encompass
assumptions about micro metastatic behavior not
visibly seen but that can be inferred from the scientific literature. Where
possible, the algorithm will use statistical
analysis to determine the confidence of its predictions.
~Projected cancer cellular mitosis phase table
Using information from the epidemiological database and from other databases
within the invention, an
algorithm will be constructed that will make use of medical and personal input
about an individual patient's present
condition to determine the projected cancer cellular mitosis phase table of
metastatic human cancer, projected in a
future time frame to generate predictions about cancer cell populations and
their behavior. It is expected that this
program will generate information about cellular functions of interest, such
as S-phase, mitosis or m-phase doubling
rate and many others to be specified. The algorithms will use linear and
curvilinear analysis and other
mathematical means to achieve this endpoint. Where possible the algorithm will
use statistical analysis to determine
the confidence of its predictions.
~Projected blood biomarker concentration over time.
19
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
Using information from the epidemiological database and from other databases
within the invention an
algorithm will be constructed that will make use of medical and personal input
about an individual patient's present
condition to determine the projected blood biomarker concentration over time
of metastatic human cancer projected
in a future time frame. It is expected that this program will generate
information about chemical behavior of cancer
cells, including biomarkers but not limited to them, chemical metabolic
factors associated with cancer may be
included. It is envisioned that numerical solution program will be
mathematically linked to predictions generated by
the micro metastatic and metastases volume, projected cancer cellular mitosis
phase table and the metastatic
occurrence, target organ and percentage numerical module subroutines, or their
predictions, within this module.
These numerical solution modules will probably include mathematical expression
to determine uptake by various
organs in the body and other mathematical modeling expressions to provide a
close to reality prediction of the
concentration of chemicals of interest in the bloodstream. The algorithms will
use linear and curvilinear analysis
and other mathematical means to achieve this endpoint. Where possible, the
algorithm will use statistical analysis to
determine the confidence of its predictions.
Molecular Bioloeical Numerical Solution Module Output Subroutines
~Metastatic occurrence, target organ and percentage possibility
Using information from the GENETIC database and from other databases within
the invention an
algorithm will be constructed that will make use of medical and personal input
about an individual patient's present
condition to determine the probable organ sites of distant metastatic human
cancer projected in a future time frame.
The algorithms will use linear, curvilinear, geometric, algebraic functions
and other mathematical means to achieve
this endpoint. The algorithm (s) within this numerical solution module will
extend and improve upon the
mathematical expressions of the HCVS engine modules that will simulate genetic
changes of a cell to cancerous
tumor and cellular overgrowth to include descriptions of breakout or micro
metastatic shed rate, new blood vessel
formation; blood, tissue and lymph vessel invasion, survival in the
bloodstream, tissue transport and new colony
formation at secondary or tertiary sites. It is envisioned that information
useful for motion, shape, path and particle
behavior of cancer in the body will be generated. Where possible, the
algorithm will use statistical analysis to
determine the confidence of its predictions.
~Percentage disease free survival.
Using information from the GENETIC database and from other databases within
the invention an
algorithm will be constructed that will make use of medical and personal input
about an individual patient's present
condition to determine the percentage of disease free survival from distant
metastatic human cancer projected in a
future time frame. The algorithms will use linear, curvilinear, geometric,
algebraic functions and other
mathematical means to achieve this endpoint. The algorithm (s) within this
numerical solution module will extend
and improve upon the mathematical expressions of the HCVS engine modules that
will simulate genetic changes of
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
a cell to cancerous tumor and cellular overgrowth to include descriptions of
breakout or micro metastatic shed rate,
new blood vessel formation; blood, tissue and lymph vessel invasion, survival
in the bloodstream, tissue transport
and new colony formation at secondary or tertiary sites. It is envisioned that
information useful for motion, shape,
path and particle behavior of cancer in the body will be generated. Where
possible, the algorithm will use statistical
analysis to determine the confidence of its predictions.
~Micro metastatic and metastases volume
Using information from the GENETIC database and from other databases within
the invention an
algorithm will be constructed that will make use of medical and personal input
about an individual patient's present
condition to determine the micro metastatic and metastases volume of distant
metastatic human cancer projected in a
future time frame. The algorithms will use linear, curvilinear, geometric,
algebraic functions and other
mathematical means to achieve this endpoint. The algorithm (s) within this
numerical solution module will extend
and improve upon the mathematical expressions of the HCVS engine modules that
will simulate genetic changes of
a cell to cancerous tumor and cellular overgrowth to include descriptions of
breakout or micro metastatic shed rate,
new blood vessel formation; blood, tissue and lymph vessel invasion, survival
in the bloodstream, tissue transport
and new colony formation at secondary or tertiary sites. It is envisioned that
information useful for motion, shape,
path and particle behavior of cancer in the body will be generated. Where
possible, the algorithm will use statistical
analysis to determine the confidence of its predictions.
~Projected cancer cellular mitosis phase table.
Using information from the genetic database and from other databases within
the invention an algorithm
will be constructed that will make use of medical and personal input about an
individual patient's present condition
to determine the projected cancer cellular mitosis phase table of distant
metastatic human cancer projected in a
future time frame. The algorithms will use linear, curvilinear, geometric,
algebraic functions and other
mathematical means to achieve this endpoint. The algorithm (s) within this
numerical solution module will extend
and improve upon the mathematical expressions of the HCVS engine modules that
will simulate genetic changes of
a cell to cancerous tumor and cellular overgrowth to include descriptions of
breakout or micro metastatic shed rate,
new blood vessel formation; blood, tissue and lymph vessel invasion, survival
in the bloodstream, tissue transport
and new colony formation at secondary or tertiary sites. It is envisioned that
information useful for motion, shape,
path and particle behavior of cancer in the body will be generated. Where
possible, the algorithm will use statistical
analysis to determine the confidence of its predictions.
~ Projected blood biomarker concentration over time.
Using information from the genetic database and from other databases within
the invention, an algorithm
will be constructed that will make use of medical and personal input about an
individual patient's present condition
to determine the projected blood biomarker concentration, over time, of
distant metastatic human cancer projected
in a future time frame. The algorithms will use linear, curvilinear,
geometric, algebraic functions and other
21
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
mathematical means to achieve this endpoint. The algorithm (s) within this
numerical solution module will extend
and improve upon the mathematical expressions of the HCVS engine modules that
will simulate genetic changes of
a cell to cancerous tumor and cellular overgrowth to include descriptions of
breakout or micro metastatic shed rate,
new blood vessel formation; blood, tissue and lymph vessel invasion, survival
in the bloodstream, tissue transport
and new colony formation at secondary or tertiary sites. It is expected that
this program will generate information
about chemical behavior of cancer cells, including biomarkers, but not limited
to them. Chemical metabolic factors
associated with cancer may be included. It is envisioned that numerical
solution program will be mathematically
linked to predictions generate by the micro metastatic and metastases volume,
projected cancer cellular mitosis
phase table and the metastatic occurrence, target organ and percentage
numerical module subroutines, or their
predictions, within this module. This numerical solution module will probably
include mathematical expressions to
determine uptake by various organs in the body and other mathematical modeling
expressions to provide a close to
reality prediction of the concentration of chemicals of interest in the
bloodstream. Where possible, the algorithm
will use statistical analysis to determine the confidence of its predictions.
Cancer Origin/Run Forward Numerical Solution Module Output Subroutines
~Metastatic occurrence, target organ and percentage possibility
Using information from the genetic database, the molecular database and from
other databases within the
invention, an algorithm will be constructed that will make use of medical and
personal input about an individual
patient's present condition to determine the probable organ sites of distant
metastatic human cancer projected in a
future time frame. The algorithm will reverse the process to the origin of
cancer and then move forward through the
present to future time frames. The algorithms will use linear, curvilinear,
geometric, algebraic functions and other
mathematical means to achieve this endpoint. The algorithm (s) within this
numerical solution module will extend
and improve upon the mathematical expressions of the Cancer Origin and HCVS
engine modules that will simulate
genetic changes of a cell to cancerous tumor and cellular overgrowth to
include descriptions of breakout or micro
metastatic shed rate, new blood vessel formation; blood, tissue and lymph
vessel invasion, survival in the
bloodstream, tissue transport and new colony formation at secondary or
tertiary sites. It is envisioned that
information useful for motion, shape, path and particle behavior of cancer in
the body will be generated. Where
possible, the algorithm will use statistical analysis to determine the
confidence of its predictions.
~Percentage disease free survival
Using information from the cancer origin database, the molecular database and
from other databases within
the invention, an algorithm will be constructed that will make use of medical
and personal input about an individual
patient's present condition to determine the percentage of disease free
survival from distant metastatic human cancer
projected in a future time frame. The algorithm will reverse the process to
the origin of cancer and then move
forward through the present to future time frames. The algorithms will use
linear, curvilinear, geometric, algebraic
22
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
functions and other mathematical means to achieve this endpoint. The algorithm
(s) within this numerical solution
module will extend and improve upon the mathematical expressions of the
cellular module to the tumor module that
will simulate genetic changes of a cell to cancerous tumor and cellular
overgrowth to include descriptions of
breakout or micro metastatic shed rate, new blood vessel formation; blood,
tissue and lymph vessel invasion,
survival in the bloodstream, tissue transport and new colony formation at
secondary or tertiary sites. It is envisioned
that information useful for morion, shape, path and particle behavior of
cancer in the body will be generated. Where
possible, the algorithm will use statistical analysis to determine the
confidence of its predictions.
~Micro metastatic and metastases volume
Using information from the cancer origin, the genetic database and from other
databases within the
invention, an algorithm will be constructed that will make use of medical and
personal input about an individual
patient's present condition to determine the micro metastatic and metastases
volume of distant metastatic human
cancer projected in a future time frame. The algorithm will reverse the
process to the origin of cancer and then move
forward through the present to future time frames. The algorithms will use
linear, curvilinear, geometric, algebraic
funcrions and other mathematical means to achieve this endpoint. The algorithm
(s) within this numerical solution
module will extend and improve upon the mathematical expressions of the Cancer
Origin and HCVS engine
modules that will simulate genetic changes of a cell to cancerous tumor and
cellular overgrowth to include
descriptions of breakout or micro metastatic shed rate, new blood vessel
formation; blood, tissue and lymph vessel
invasion, survival in the bloodstream, tissue transport and new colony
formation at secondary or tertiary sites. It is
envisioned that information useful for motion, shape, path and particle
behavior of cancer in the body will be
generated. Where possible, the algorithm will use statistical analysis to
determine the confidence of its predictions.
~Projected cancer cellular mitosis phase table
Using information from the cancer origin, the statistical and metastatic
database and from other databases
within the invention, an algorithm will be constructed that will make use of
medical and personal input about an
individual patient's present condition to determine the probable organ sites
of distant metastatic human cancer
projected in a future time frame. The algorithm will reverse the process to
the origin of cancer and then move
forward through the present to future time frames. The algorithms will use
linear, curvilinear, geometric, algebraic
functions and other mathematical means to achieve this endpoint. The algorithm
(s) within this numerical solution
module will extend and improve upon the mathematical expressions of the Cancer
Origin and HCVS engine
modules that will simulate genetic changes of a cell to cancerous tumor and
cellular overgrowth to include
descriptions of breakout or micro metastatic shed rate, new blood vessel
formation; blood, tissue and lymph vessel
invasion, survival in the bloodstream, tissue transport and new colony
formation at secondary or tertiary sites. It is
envisioned that information useful for motion, shape, path and particle
behavior of cancer in the body will be
generated. Where possible, the algorithm will use statistical analysis to
determine the confidence of its predictions.
~Projected blood biomarker concentration over time.
23
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/1'7810
Using information from the cancer origin, the statistical and metastatic
database and from other databases
within the invention, an algorithm will be constructed that will make use of
medical and personal input about an
individual patient's present condition to determine the projected blood
biomarker concentration over time of distant
metastatic human cancer projected in a future time frame. The algorithm will
reverse the process to the origin of
cancer and then move forward through the present to future time frames. The
algorithms will use linear, curvilinear,
geometric, algebraic functions and other mathematical means to achieve this
endpoint. The algorithm (s) within this
numerical solution module will extend and improve upon the mathematical
expressions of the HCVS engine
modules that will simulate genetic changes of a cell to cancerous tumor and
cellular overgrowth to include
descriptions of breakout or micro metastatic shed rate, new blood vessel
formation; blood, tissue and lymph vessel
invasion, survival in the bloodstream, tissue transport and new colony
formation at secondary or tertiary sites. It is
expected that this program will generate information about chemical behavior
of cancer cells, including biomarkers
but not limited to them. Chemical metabolic factors associated with cancer may
be included. It is envisioned that
numerical solution program will be mathematically linked to predictions
generate by the micro metastatic and
metastases volume, projected cancer cellular mitosis phase table and the
metastatic occurrence, target organ and
percentage numerical module subroutines, or their predictions, within this
module. This numerical solution module
will probably include mathematical expressions to determine uptake by various
organs in the body and other
mathematical modeling expressions to provide a close to reality prediction of
the concentration of chemicals of
interest in the bloodstream. Where possible the algorithm will use statistical
analysis to determine the confidence of
its predictions.
Metastatic Interface
In operation, the Metastatic Module and its subprograms follow the system
described in this invention of
engaging a user interface using a standard computer monitor and keyboard. The
Metastatic Module then activates
the central processing unit through the software programs with the connector
module connecting to databases of
information needed for the numerical and descriptive processing. The
Metastatic Module then engages the
numerical solution module and subroutines and finally provides the user with
reported solutions to their requests
through the monitor and various displays. All of the operations take place in
the basic software described in figure
3-10.
Similar to other modules described in this invention, the first phase of the
Metastatic Module involves a
fact gathering process for information input and the selection of programs and
patient information to be entered to
enable operation of the subprograms selected through a keyboard. Assuming the
software is loaded and operational
through conventional means, the user will be prompted to select modules of
operation. When the Metastatic Module
is selected, a menu with instructions is provided for all of the subroutines.
Patient information is entered through
the keyboard and when completed a prompt is given indicating information entry
is finished and to run the engine.
At this point the software engines) selected engages the central processing
unit and begins operations.
24
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
Metastatic Interface Communication to Connector Module
The user interface activates the connection module program and uses input
received from the user to make
decisions and gather information from various databases to execute programs
selected by the user in the numerical
solution module. Reports and information from other modules in the invention,
tumor origin, cellular module,
colony module, tissue module, tumor module will be retrieved from their
interfaces if needed automatically as part
of the program. It is envisioned that information and report links will exist
between the molecular, cellular, cellular
expansion, pre-neoplastic , neoplastic interfaces and the numerical solution
module subroutines in the metastatic
module to facilitate exchange from other parts of the invention so
calculations and descriptions can be performed.
Modification and Customization of Variables in the Metastatic Module
Subroutines
All of the Metastatic Module's subroutines may be stopped and input parameters
modified at points along
the time line of operation according to the instructions at the user
interface, and then allowed to continue provide
human cancer simulation reports with a record of when and what modifications
were made to input assumptions.
Default Parameters/Pre-Programmed Inputs in the Metastatic Module Subroutines
As with other modules described in the invention, the metastatic module
subroutines and the algorithms
within them will have pre-programmed default parameters in the event that
information supplied by the user is
incomplete. This default parameter will allow the subroutines to run and/or
instruct the user saying that the program
subroutine cannot operate.
Connection Module Communication to Numerical Solution Module of the Metastatic
Module
The connection module retrieves information from the databases, other module
output reports as necessary
and synthesizes it with the patient input from the user so subroutines within
the numerical solution module can
begin generating information relating to the future course of human cancer
within the human body.
Connection Module/Database Operation in the Metastatic Module Subroutines
The connection module will be engaged whenever the input from the user is
completed and the subroutines
are designated to start. The connection module will process the input from the
user and gather information from the
various databases necessary for the execution of the mathematical and
descriptive algorithms within the various
numerical solution modules and transmit it to its proper location. If the
program is stopped at the user interface
along any aspect of the time line, and input is modified, the connection
module will again automatically be engaged
and then the new information will be gathered and transmitted to the numerical
solution module for sequential
processing. If the program is paused, the connector module will not be
engaged.
Numerical Solution Modules) in the Metastatic Module Subroutines
The numerical solutions modules) carries out the generation of results and
information for the reports.
Once started they will complete their calculations, generate results and their
reports and return them to the user
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
interface for viewing on the monitor or for printing as per the computer
hardware description. If continual output is
chosen and the user stops the program the numerical solutions module (s) will
store results generated up to that
point in time or report if requested. If the program is restarted it will
begin where it left off, include the changed
parameters in the next sequential report, continue calculations with modified
parameters and produce the report. If
paused the numerical module will resume its calculations when the pause is
ended and generate its results and
report.
Results and Reports/LJser Interface in the Metastatic Module Output
Subroutines
The final result of Metastatic Module and its subroutines will be a series of
informational reports from the
numerical solution modules and the information gathered from the databases to
assist in running the mathematical
and descriptive algorithms, continuously reported or sequentially reported,
summarizing results or providing
information at continuous or pre-selected discrete time intervals by request
of the user along the time line of the
human cancer virtual simulation engine's operation domain through the user
interface. Similarly the reports can be
requested by the user at various points in the future, along the time line of
the operation of the module so that
information generated by these subroutines can be reported. If the Metastatic
Modules subroutines are stopped and
restarted the report will contain the sequence of information generated by the
numerical solution modules in the
order of the instructions received and containing the modifications for ease
of understanding by the user.
Specific Operation Example of the Metastatic Module and its Subroutines
As an example of use, imagine a physician who wishes to examine the possible
metastatic behavior of a
patient's breast cancer through the human cancer virtual simulation
information engine. Information will be
generated by the engine to help answer two questions, is there an optimal time
to administer the adjuvant therapy?
What kind of reoccurrence scenarios on the microscopic (micro metastatic) and
macroscopic (distant metastases)
level may occur in the future?
1. After the program is started the user would be prompted by the start-up
screen from a menu at the user
interface to select the module to be used. The Metastatic Module is selected.
2. Next the three selections of metastatic module subroutines would appear,
statistical and clinical outcome
extrapolation, molecular biological and cancer origin/run forward. For this
example the user would select all three.
3. Next the user interface would ask how many models were desired to be run by
the engine in each
subroutine. The physician is interested in looking at the potential outcomes
for a prediction of an excellent to a
moderate response to a single course of treatment so she selects two models
for each.
4. Next the user interface would ask which model is to be run first and if
continuous reporting was desired.
If continual reporting was desired, the interface would ask which subroutine
was to be run first. The user interface
will allow one subroutine to operate continuously on display with a selection
of desired parameters of information
continually reporting the selected information. The user could select all the
information that the subroutine is
capable of producing or be limited to the selections the user makes. The
subroutine would allow for this. For
simplicity we will say that the user wishes to run the clinical and
statistical outcome, molecular biological and
26
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
cancer origin/run forward in that order and two models for each to allow for
variation in the treatment regimens and
assumptions for the models. We will assume that the user in this example
requests that final reports be produced, not
continually displayed, although at the end of this example we will demonstrate
the continual display option as a
replay of one of the stored programs.
5. At this point, the user interface will display a menu of inputs needed for
the statistical and clinical
outcome extrapolation. The user will be requesting information about the
patient's name, height, weight, age,
treatment steps such as chemotherapy, surgery and radiation; when treatments
are planned, when treatment's took
place, patient response to treatment or estimates thereof, initial tumor size
and volume, tumor geometric shape,
tumor location, health conditions of relevance to patient such as HIV status,
immune system strength through
secondary biomarkers such as white blood cell and T-cell count, family history
of relevance to cancer epidemiology,
other complicating factors or diseases, local and distant metastases location,
biomarkers of human cancer such as
estrogen receptivity in breast cancer and others, mitosis rate, cell diploidy,
etc. Additionally it is envisioned that
some of the input information may change and improve as the engine is
developed, or may be tailored for certain
kinds of cancer to provide better predictive and prognostic information to the
user. For instance, for the breast
cancer patient example, in addition to the information above a prompt may
appear to request staging of patient 1-4,
specific questions related to the staging such as numbers of lymph nodes
involved if radical or modified radical
mastectomy has occurred, the presence of the BRCA1 gene, p53 tumor suppressor
gene activity in the tumor, HER
2 new expression, breast micro calcifications from X-ray, date of first
estrus, menstruation cycle and other factors
that could be useful for subroutine algorithms. The user enters this
information and if unavailable a prompt for
default will be provided with any additional instructions. The user is then
prompted to go to the next step, the
selection and customization of the report.
For this example we will say that the patient is a pre-menopausal 40 year old
African American woman
with a discovered 2.5 cm pear shaped invasive ductal carcinoma tumor with
micro calcifications in her right breast.
The tumor is removed and biopsied. The tumor was showed mild vascularity and
the margins around the biopsy
sample were not clean indicating spread beyond the tumor. The pathology report
indicated HER 2 new expression,
p53 loss, estrogen receptivity, a mitosis rate of 1 due to a low S phase
fraction, tumor cell DNA intact (high
diploidy) and good cellular differentiation, by some factors a slow to
moderate growing tumor but invasive. Due to
micro calcifications and of the other tumor factors that indicate the tumor
may be active, breast conservation surgery
is abandoned and the breast is removed and lymph nodes sampled, no lymph node
activity is seen and the patient is
classified as a stage 2 patient. The physician has entered all the above
information into prompted questions from the
program. She is considering adjuvant chemotherapy and the type and time to
administer it. The adjuvant therapy she
is considering will be the standard Cytoxan, Methotrexate and 5-Fluorouracil
(CMF). The woman is healthy in all
other respects. The physician will enter two scenarios into the metastatic
program, one in which the patient response
to chemotherapy will be considered superior, the other where it is moderate, a
reflection of the aggressive genetic
tumor factors versus its similarity to normal cells by good cell
differentiation and only a low mitosis grade, making
it more difficult to eradicate with chemotherapy. As stated earlier,
information will be generated by the engine to
help answer two questions, is there an optimal time to administer the adjuvant
therapy? What kind of reoccurrence
27
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
scenarios on the microscopic (micro metastatic) and macroscopic (distant
metastases) level may occur in the future?
The Metastatic Module will assist answering these two questions.
6. The physician now sees a menu of other report selections from the
metastatic clinical and statistical
outcome subroutine on the screen before her. The possible reports available to
her include a) metastatic occurrence,
target organ and percentage possibility, b) percentage disease free survival,
c) micro metastatic and metastases
volume, d) projected cancer cellular mitosis phase table, e) projected blood
biomarker concentration over time. By
keyboard selection the physician chooses all five.
7. After the physician enters the basic patient information and selects the
reports to be issued, she will see a
menu screen with a variety of options to provide the information back in the
form of reports. The first selection is
the final time domain of the engine's simulation. This is the point in the
future from the present that the subroutine
programs selected should end their calculations and generate results. This
could range from short periods of time of
days to approximately 20 years. The upper limit may change and will be bounded
by the scientific information
available to provide information useful to the subroutine calculation. The
lower range will be determined by the
information the engine could produce that would be of value beyond
observation. Each simulation would be
bounded by time but for example, the user could chose five metastatic
simulation each with longer and longer time
frames, or stop the simulation at various points and request a report or begin
new simulations with a desired new
time frame sequentially on the previous one. For this example we will say the
physician user requests a 5 year
projection for the two simulations and display the information for the first
six months by day for subroutine d) the
projected cellular mitosis phase table. This enables information to be
provided about follow up in reasonable time
increments to be estimated from the engine's programs, given the young woman's
age, medical aspects of the case
and the fact that follow-up adjuvant chemotherapy is being considered.
8. Now the interface prompts the physician to begin and she hits start. The
invention does the rest of the
work until the reports appear at the user interface. That sequence of events
is described next.
9. The computer now engages the connection module, a computer software program
which takes the
information supplied by the physician about the patient and extracts
information from the epidemiological database
and other databases as needed and automatically transfers retrieved
information to the five numerical sub-module
selected.
10. The numerical sub-modules automatically conduct their calculations and
descriptions using their
algorithms and other aspects of their programs, produce their reports and send
them to the user interface.
11. After the engine has completed its work as described in steps 9 and 10,
after starting the program in
step 8, what the physician would receive, in this example first, is five
reports for two simulations from the clinical
and statistical outcome subroutines. Each is described here in example of what
the inventions subroutines could
produce with a visualization of what could appear in the content and its
possible decision making value:
Metastatic occurrence, target organ and percentage possibility. This report
would contain a table of the target organs
of reoccurrence, as an estimate this would be the left breast, chest wall,
lungs, skeletal system and brain among
others, with a percentage range in the 5 year time frame selected that
reoccurrence would appear in various organs
for the two scenarios selected. The optimal response would likely contain a
lower percentage but given other factors
28
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
in this case and the evaluation of information from the epidemiological
database the percentage differences may be
great or small and provide useful information for follow up. The report would
be structured in a table of predictions
at 6-month intervals in this example.
Percentage disease free survival. The report would provide an estimate of no
reoccurrence for the optimal
and moderate breast cancer patient response scenarios. It is envisioned that
confidence intervals could be applied to
this value based on the information in the database to allow better
statistical value. Mortality and morbidity
statistics would be reported in 6-month intervals over the 5-year time frame,
in this example the optimal and
moderate response scenarios could be compared and contrasted. Again the
examination of the database information
may show large or small differences, and possible improvement in the lessening
of reoccurrence in certain organs
over time. This could be reported positively to the patient and helps with
decision making about follow-up activity.
Micro metastatic and metastases volume. This report would estimate the size,
shape and potential volume of a
recurrent tumor in the various target organs in six month increments over five
years in a tabular format. The report
would also indicate an estimate of the total micro metastatic volume, or an
estimate of the total cancer localized in
tumors and non-localized within the whole body and in organs at given times in
a tabular format. The value of this
report would be an indication of when cancer mass may be large enough to be
detected by various imaging
techniques or other means in distant organs. In this example the physician
would have an indication, say in the left
breast, when micro metastasis would reach a point where micro calcifications
may appear in the breast before small
tumors in a mammogram appear in a moderately responsive patient. This could
assist the physician in optimal
timing patient follow-up for maximum probability of detection of any cancer
spreading. If follow up mammograms
are performed and micro calcification are not seen at the times predicted,
this could be used as an indication of more
optimal response to therapy, assisting the physician by focusing parameters
for the subroutine assumptions in this
invention if future simulations are conducted in the future.
I. Projected cancer cellular mitosis phase table. This report would contain an
estimate of populations of
cancer cellular growth, cancer cell death, and micro metastatic growth in
terms of numbers of cells in
various phases of cell division in the whole body and various target organs.
What the physician would see,
in this example since a daily report over the next 6 month time frame was
selected would be a daily table
indicating the number of potential cancer cells and what phase of growth or
division they were in for the
two optimal and moderate response to chemotherapy.
In this example chemotherapy has not been administered yet, but the physician
is estimating potential
responses. Also we know that the cell grade, mitosis rate and general DNA
structure are fairly intact. Therefore, for
example, we would expect to see a table representing shallowly rising curves
indicative of a slow doubling rate.
Given the more regular, rather than erratic cell reproduction rate, an
estimation may appear in the daily tables
indicative of high points when the remaining micro metastatic cancer left
after surgery would be in certain phases of
reproduction or cell cycles. The physician could use this information in the
near term to plan to administer
chemotherapeutic drugs so that the maximum concentration of the standard CMF
regimen would be available in the
target organs or whole body to interfere or destroy the cancer the best. In
short this information may help physicians
estimate how to get the medicine where and when it is needed most. She may
even plan smaller dosages of
29
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
chemotherapeutic agents in the regimen at later times and specifically to
target organs to correspond to estimated
cancer cellular peaks to destroy potential remaining cells, not caught on the
first go around with a particular agent or
to improve the effectiveness of response in a sub-optimally responding
patient. This could be especially useful in
the case of breast cancer patients in the use of Adriamycin, which has
cardiotoxic side effects at high dosages but is
one of the most effective anti-cancer agents. Adriamycin and possibly other
chemotherapy agents could be timed
and effectively used at large, as well as smaller dosages, for maximum
positive effect while diminishing negative
side effects.
Projected blood biomarker concentration over time. The report would estimate
the concentration in
nanograms per milliliter of a variety of cancer biomarkers in the bloodstream
at six-month interval over the 5-year
time period selected in this example. This area is new and ripe for discovery
and will be a more valuable piece of
report information in the future than today. For this example possibly the
presence of HCG (human chorionic
gonadotrophin) would be listed since it has been implicated with many cancers,
CEA (carcinoembryionic antigen)
has been shown to rise in severely metastatic breast cancer patients but
current test methods may not be sensitive or
the CEA may not expressed sufficiently to be useful for a stage 2 breast
cancer patient at this time. Others may
provide useful indications of microscopic molecular activity indicative of
micro metastasis. What the physician
would see in the reports would be two estimates based on an optimal and
moderate response of concentrations of
chemicals that could be present in the bloodstream pertinent to the cancer
selected and a report of other metabolic
factors deemed useful for predictive and prognostic applications.
At the conclusion of reading the reports, the physician decides she would like
to view the projected cancer
cellular mitosis phase table in a continual fashion. She would only need to
request that of the user interface when the
program is brought back to start in step 4.
The process for the physician in this example steps 1 through 11 and the
process of the operation of the
invention in carrying out its functions is the same for the other two
subroutine modules, the molecular biological
and the cancer orzgin/run forward with the same title reports issued back to
the user interface.
Because the underlying assumptions and the mathematical and descriptive
calculation are different in these
numerical modules, as described earlier in the numerical module section for
the metastatic module in total, the
reports may reach similar or very different conclusions. For example, it is
fully expected that because DNA and
molecular mathematical and descriptive algorithms are used to generate the
information in the molecular biology
and the cancer run forward metastatic subroutines that predictions in the
cellular mitosis, micro metastatic volume
and biomarker reports will be more precise and capable of generating
meaningful predictions down to single cellular
cycles and will vastly improve over time. Initially the statistical and
clinical outcome extrapolation will provide the
most useful predictions for disease free survival and distant metastases,
target organ and probability predictions
because it is based on real world information and subject to less uncertainty.
All three approaches provide useful
information for comparison against real world patient behavior, decision
making and for research and educational
purposes.
For ease of usage, the input to run all three metastatic sub-modules and their
five numerical solution
subroutines is envisioned to be the same. So the user need only enter the
information one time, or can modify
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
parameters and input selectively and generate 15 reports for human cancer
metastatic behavior, or 5 reports each to
provide predictive and prognostic information from 3 modeling approaches.
The Metastatic Module and all its subroutines and the mathematical and
descriptive algorithms include a
plurality of components to provide information that simulates the functions of
living systems, in this case the
behavior of cancer in the human body. The preparation of these modules is not
a static occurrence and the
numerical solution modules and the invention's databases, will be subject to
modification and improvement with
advancing scientific knowledge. Information supplied by these modules can be
used to drive animations or other
types of visual display beyond tables and reports to provide informative
visualization and expression of the
predictive and prognostic information that is derived from them.
DATA FLOW TO MODULES
Fig. 11 is a block diagram illustrating the types of data and infonmation sent
to each of the modules from a
user interface (GUI) and from a patient information database. For each
patient, the user inputs into the tumor origin
module data and results of genetic tests, family history information, and
information pertaining to life style
(smoking, drug use, etc.). The tumor origin module also receives from the
database information on genetic
relationships and possible mutations, including data on protein reactions
relating to the synthesis of genetic material,
and genetic information related to the interaction between cells in a normal
environment, such as cell adhesion,
intracellular structure, etc. The tumor origin module further receives
statistical data from the statistical database
regarding which genetic markers should exist and which genes, if any, are
mutated.
The cellular module receives from the databases data pertaining to cell life
cycle control, such as which
genes are responsible for cell cycle control, for example Cyclin Dependent
Kinesis (CDK), statistical data on cell
cycle control and physical properties of cells, and data pertaining to the
compounds and associated concentrations
required for proper cell function.
The colony module receives from the databases data related to the interaction
between cells, such as the
identification of genes responsible for cell interaction (e.g. for production
of proteins used in cell adhesion, etc.), the
identification of genes responsible for physical properties of cells (e.g.
cell shape and size), and statistical data
concerning cell interaction (e.g. cell bond strength, nutrition distribution,
etc.), and genetic information responsible
for cell structure (e.g. cell cycle, bond strength, membrane strength).
The tissue module receives from the databases data related to the proteins and
other biochemical
compounds and elements involved in the interaction between cells and in tissue
structure, both in a normal
environment and in a malignant environment, and other genetic and
protein/biochemical information not specific to
metastatic spread.
The tumor module receives from the databases data related to genetic,
biochemical and statistical
information concerning cell cycle, bond strength, membrane strength, tissue
structure, interaction between cells, etc.
in a malignant environment.
The metastatic module receives from the databases data relating to the
mechanisms of metastatic spread,
31
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
and statistical data relating to cellular activity, both specific to
metastatic spread and generally.
DATA FLOW BETWEEN MODULES
Fig. 12 is a block diagram illustrating the types of data and information
passed between the various
modules and between the user interface and database. The subroutines of the
tumor origin module develop from the
inputted data, data relating to cell cycle rate, genetic changes in cellular
DNA, and protein expression. This
information is inputted to the cellular module for use in the cellular module
subroutines. The tumor origin module
also develops diagnosis data from the inputted patient information and
relevant data from the databases, concerning
possible genetic changes and expression of unusual proteins as indicating
possible staging of disease.
The cellular module subroutines utilize the information developed by the tumor
origin module to calculate
cell cycle rates, genetic changes and protein expression on a cellular level,
and to determine cell size, shape, growth
rate, and protein expression related to cell structure. This information is
passed to the colony module, where the
subroutines of the colony module use this information in conjunction with the
data received from the databases to
calculate cell structure, size, shape, etc. of a tissue matrix, which
information is inputted to the tissue module.
The tissue module in turn utilizes this data to calculate the size, shape,
structure, bond strength, etc. of
tissue, which information is passed on to the tumor module. The tumor module
in turn utilizes this information to
predict tumor growth, shape and size, etc. into the future. The metastatic
module takes the tumor-related
information and utilizes it to predict metastatic spread of carcinogenic cells
to other systems of the body into the
future.
DATABASE FORMATION, UPDATING, AND MODEL DEVELOPMENT
Fig. 13 is a block diagram illustrating the building and updating of the
various databases used in the HCVS
system. As shown, data from various external sources, such as research and
development institutions, medical and
scientific journals, textbooks, university databases, public and private
databases, public research institutions,
research laboratories and insurance companies, is inputted to an error
screening module for filtering of the data to
eliminate erroneous, irrelevant or incomplete data. The filtered data is then
inputted to data type distribution
module which separates the data and groups it by type, such as input data,
relationship data, or output data, and
formats the data into a matrix. The data matrix is then inputted into a
modeling data spreadsheet module for
preparation of data sets. The data sets are then inputted to a learning system
module, for development and updating
of the various models used in the subroutines. The models are stored in a
model staging storage memory, from
which they are inputted to the application environment of the system, for use
with the specific patient information to
calculate diagnosis and predictive results which are then displayed to the
user on a graphical user interface.
CELL CYCLE ALGORITHMS
Fig. 15 illustrates a general algorithm for determining particular protein
expressions in cell life cycle
process. At step 150, the maximum possible amount of protein capable of being
produced by the cell under study.
At step 151, the percentage of this maximum possible amount being produced is
calculated. At step 152, it is
32
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
determined whether any other processes are involved upon which protein
expression is dependent. If not (step 153),
then the determined protein concentration is outputted at step 154. If it is
determined that protein production is
dependent on a following process (step 155), at step 156 the calculation is
paused to await the information from the
following process, and at step 157 the needed information is imported from the
following process algorithm, and the
process advances to step 160.
If it is determined (step 158) that the following process itself is dependent
on the protein at issue, then at
step 164 the calculated protein concentration is outputted. If it is
determined (step 159) that the protein at issue is
dependent on a previous process, then at step 160 the dependence ratio or
relationship is determined. The value of
the dependence is then determined at step 161 and the operation needed to
simulate the dependence is selected at
step 162. At step 163 the percentage of protein production controlled by the
dependence is calculated, and the
determined protein concentration is then outputted at step 164.
Fig. 14 provides an example of a cell cycle algorithm for calculating cell
life cycle from the G1 (start)
cycle phase through to the M phase. In each phase, the production of various
free proteins such as cyclin A, cyclin
B, cyclin D, cyclin E, Cdk 1, Cdk 2, Cdk4, and 1tB are calculated, according
to the algorithm of Fig. 15. The results
of each calculation are then used to calculate the next protein/ protein
complex in the cell cycle phase, leading to the
production of growth factors. If complex failure is detected at any point
during the calculation run, a cell cycle halt
is triggered. The calculated growth factors are then transported as expressed
growth factors to the next phase of the
cell cycle.
Specific Operation Ezample of the Tissue Level Module and the Metastatic
Module
In this example we will assume that a physician has a patient with a large
colon tumor, 7 cm in diameter
and the patient is 75 year old male in frail health. Given the health of the
patient possibly radiation treatment may be
a better option than surgery. The physician would like to use the tissue level
and then the metastatic level simulation
engine to examine the effects of radiation on the tumor and decide if this is
a viable option for treatment in lieu of
surgery.
l.First the program would be loaded as in the previous example and the
physician would choose the tissue
module and would see the pre-neoplastic interface, additionally the tumor
module and the neoplastic interface and
the metastatic module and the metastatic interface would be activated by the
physician in this example.
2. The physician would then see the menu of options that would allow patient
information to be entered.
3. The physician, similar to the response information in the previous breast
cancer example, would have the
option to estimate the response that radiation would have on the tumor and the
surrounding tissues. For this
example we will assume two scenarios, a simulation where no radiation
treatment is given and a simulation where a
high dose of radiation is given within a week of the simulation. In the
operational version of the invention it is
envisioned that a wide variety of parameters would be available to the
physician based upon the latest indices of
cancer radiation treatment to easily enable entry of the strength, type and
dose of radiation and times of treatment to
assist estimation of the number of tissue and tumor cells destroyed in the
process.
4. The physician would then need to follow instructions and enter the types of
reports he would like to see
and the frequency of information from the simulation. For the tissue module
example we will look at daily reports
33
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
rate of mitosis, survival rates of cells and a daily readout of the projected
cell mitosis phase report from the
metastatic module for the next sixty days or two months.
5. The order of the reports that the physician requests are the no radiation
treatment first, and then the high
dose radiation treatment reports.
6. Once the patient tumor information is entered into the tissue module
interface, the physician would hit
the start button.
7. The engine now goes through the same process of taking the input to the
connection module and then
retrieving information from the inventions database necessary to run the
algorithms in the tissue and metastatic level
numerical solution modules, when the tissue level and metastatic numerical
solution modules have completed their
work, they produce the reports and send them back to the user interface.
8. What the physician sees at the user interface is six reports waiting to be
reviewed. Four related to the
increase in tissue growth (rate of mitosis and survival rate of cells) from
the tissue module in a no intervention
versus a high intervention scenario. The last two would be an analysis of cell
population, death and cell growth
phases, with numbers of cells and times from the metastatic module's projected
cell phase mitosis report for a no
intervention versus high dose radiation intervention scenario.
The physician could, if desired, have activated other subroutines through the
pre neo-plastic interface
accessing the tissue module to generate reports such as physical properties of
cells, nutritional consumption, cell
bonding and intracellular structure. The physician could, if desired, have
activated other subroutines through the
neo-plastic interface for the tumor module to generate reports such as tumor
mass, tumor growth rate, genetic
mutations present and vascular construction. In this example the metastatic
reports requested include some of these
areas but not all.
The physician now has some comparisons of what the effectiveness of a high
dose radiation treatment may
be at the tissue level for a tumor of certain size and molecular biological
properties characteristic of the individual
patient. Several pieces of information can be gleaned from reports of this
kind. For the sake of example we will
explore them.
1. First in the no treatment example, the tissue level reports of projected
cell mitosis phases and metastases
volume can be very useful in estimating the behavior of the tumor in the next
two months for treatment options. Is it
slow growing or fast? Based on patient input what kind of mitosis patter is
expected in the next sixty days? Is the
mitosis patter in the tissue of the tumor periodic or erratic? The report we
expect will have statistical information
associated with the engines predictions based partly on the quality and
quantity of the information inputted and also
on the information available from the database to assist in its predictions
and will help answer these questions at the
tissue level. How does this compare with predictions in the metastatic module
from its report?
2. The physician will have a readout of exactly one week from the present day
on a kill off of the cancer
cells in the tumor and then a projection of growth for the next 53 days after
that. How much would the tissue
metastases shrink?
3. Does the projected cell mitosis report indicate a patter that would help in
the optimal timing of a high
dose radiation treatment? Radiation is most effective in disrupting and
destroying mammalian cells when applied
34
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
during the mitotic phase during actual cell division. The mitosis phase can be
very brief and the shortest phase in the
cell life cycle. Possibly the physician would see from the HCVS reports a
period of statistically high mitotic activity
within the tumor in 2 weeks or sixty day period and would delay treatment
until that period. The invention could
and most likely would be rerun to estimate the effects of radiation treatment
on a colon cancer under these
potentially better conditions to see what the effect would be.
4. Following the pattern of inquiry further, could lower doses at different
times in the next sixty days be as
effective as one high dose? Would there be a patient benefit? Again comparing
the result of the two scenarios and
the reports associated with each may indicate directly that this could be the
case or point to the need for further
simulations with the invention to fully explore radiation treatment effect on
colon cancer tumor tissue in the near
term.
In the beginning of this example we mentioned the metastatic module in
conjunction with the tissue
module.
As Fig 3 shows the linear progression of interfaces and modules, the next
logical step to take would be to do a more
distant metastatic projection on the patient. The physician would have two new
pieces of information to achieve
this, a scenario in the next sixty days that could be plugged into the
metastatic module indicating no treatment and
projections could be made from the present line, second information on tumor
shrinkage based on a high dose
radiation treatment. The process would be the same as in the previous example
for the metastatic module and once
the metastatic module would be activated the interface would ask various
question of value in making a longer-term
prediction. At this point, the morbidity and mortality statistics from the
clinical outcome report would be of interest
In considering this example, it is worth thinking that a logical process of
usage of the information from this
simulation would be for the physician to:
1. Examine the options and efficacy for radiation treatment using the tissue
module simulation
2. Decide the best treatment at the tissue level and administer it
3. Compare the tissue level modules results against the actual patient
response
4. Enter the patient actual response information into the metastatic module at
a later time and validate or
change assumption parameter for the metastatic module interface based on real
world results to improve predictions
and maximize the possibilities for a favorable outcome.
System Configuration
There are two general configurations for the HCVS system according to one
preferred embodiment of the
invention. The first of the two is medical. This is by far the most complex
and interactive. The purpose and
usefulness of the diagnostic, treatment, and research configuration is to deal
with real life patients. This
configuration will use the information entered into the HCVS to artificially
generate in the computer a replication of
the actual situation at hand. This configuration will run with preprogrammed
cases of cancer. The purpose and
usefulness of this configuration is to train and prepare present and future
healthcare professionals. The second of the
CA 02376831 2001-12-28
WO 01/00083 PCT/US00/17810
configurations is the educational configuration.
While particular specifics of the present invention have been disclosed, it is
to be understood that various
different modifications are possible and are contemplated within the true
spirit and scope of the claims. All such
modifications and variations of the invention herein described as would be
apparent to those skilled in the art are
intended to be encompassed within the following claims.
36