Note: Descriptions are shown in the official language in which they were submitted.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
1
NEW INTERFERON BETA-LIKE MOLECULES
FIELD OF THE INVENTION
s
The present invention relates to new interferon (3 conjugates, methods of
preparing such
conjugates and the use of such conjugates in therapy, in particular for the
treatment of multiple
sclerosis.
~o BACKGROUND OF THE INVENTION
Interferons are important cytokines characterized by antiviral,
antiproliferative, and
immunomodulatory activities. These activities form a basis for the clinical
benefits that have
been observed in a number of diseases, including hepatitis, various cancers
and multiple
t s sclerosis. The interferons are divided into the type I and type II
classes. Interferon (3 belongs to
the class of type I interferons, which also includes interferons a, i and c~,
whereas interferon y
is the only known member of the distinct type II class.
Human interferon ~i is a regulatory polypeptide with a molecular weight of 22
kDa consisting of 166 amino acid residues. It can be produced by most cells in
the body, in
2o particular fibroblasts, in response to viral infection or exposure to other
biologics. It binds to a
multimeric cell surface receptor, and productive receptor binding results in a
cascade of
intracellular events leading to the expression of interferon [3 inducible
genes which in turn
produces effects which can be classified as antiviral, antiproliferative and
immunomodulatory.
The amino acid sequence of human interferon ~3 was reported by Taniguchi, Gene
2s 10:11-15, 1980, and in EP 83069, EP 41313 and US 4686191.
Crystal structures have been reported for human and marine interferon (3,
respectively (Proc. Natl. Acad. Sci. USA 94:11813-11818, 1997. J. Mol. Biol.
253:187-207,
1995). They have been reviewed in Cell Mol. Life Sci. 54:1203-1206, 1998.
Relatively few protein-engineered variants of interferon (3 have been reported
30 (WO 9525170, WO 9848018, US 5545723, US 4914033, EP 260350, US 4588585, US
4769233, Stewart et al, DNA Vol 6 not 1987 pp. 119-128, Runkel et al, 1998,
Jour. Biol.
Chem. 273, No. 14, pp. 8003-8008).
SUBSTITUTE SHEET (RULE 26)
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
2
Expression of interferon (3 in CHO cells has been reported (US 4966843, US
5376567 and US 5795779).
Redlich et al, Proc. Natl. Acad. Sci., USA, Vol. 88, pp. 4040-4044, 1991
disclose
immunoreactivity of antibodies against synthetic peptides corresponding to
peptide stretches of
s recombinant human interferon ~i with the mutation C17S.
Interferon ~3 molecules with a particular glycosylation pattern and methods
for
their preparation have been reported (EP 287075 and EP 539300).
Various references disclose modification of polypeptides by polymer
conjugation or glycosylation. Polymer modification of native interferon (3 or
a C17S variant
t o thereof has been reported (EP 229108, US 5382657, EP 593868, US 4917888
and WO
99/55377). US 4,904,584 discloses PEGylated lysine depleted polypeptides,
wherein at least
one lysine residue has been deleted or replaced with any other amino acid
residue. WO
99/67291 discloses a process for conjugating a protein with PEG, wherein at
least one amino
acid residue on the protein is deleted and the protein is contacted with PEG
under conditions
i s sufficient to achieve conjugation to the protein. WO 99/03887 discloses
PEGylated variants
of polypeptides belonging to the growth hormone superfamily, wherein a
cysteine residue has
been susbstituted with a non-essential amino acid residue located in a
specified region of the
polypeptide. Interferon (3 is mentioned as one example of a polypeptide
belonging to the
growth hormone superfamily. WO 00/23114 discloses glycosylated and pegylated
interferon
20 (3. WO 00/23472 discloses interferon (3 fusion proteins. WO 00/26354
discloses a method of
producing a glycosylated polypeptide variant with reduced allergenicity, which
as compared
to a corresponding parent polypeptide comprises at least one additional
glycosylation site.
US 5,218,092 discloses modification of granulocyte colony stimulating factor
(G-CSF) and
other polypeptides so as to introduce at least one additional carbohydrate
chain as compared
2s to the native polypeptide. Interferon (3 is mentioned as one example among
many
polypeptides that allegedly can be modified according to the technology
described in US
5,218,092.
Commercial preparations of interferon (3 are sold under the names Betaseron~
(also termed interferon ~ilb, which is non-glycosylated, produced using
recombinant bacterial
30 - cells, has a deletion of the N-terminal methionine residue and the C 175
mutation), and
AvonexTM and Rebi~ (also termed interferon (31 a, which is glycosylated,
produced using
recombinant mammalian cells) for treatment of patients with multiple
sclerosis, and have
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
3
shown to be effective in reducing the exacerbation rate, and more patients
remain exacerbation-
free for prolonged periods of time as compared with placebo-treated patients.
Furthermore, the
accumulation rate of disability is reduced (Neurol. S 1:682-689, 1998).
Comparison of interferon X31 a and (31 b with respect to structure and
function has
s been presented in Pharmaceut. Res. 15:641-649, 1998.
Interferon (3 is the first therapeutic intervention shown to delay the
progression of
multiple sclerosis, a relapsing then progressive inflammatory degenerative
disease of the
central nervous system. Its mechanism of action, however, remains largely
unclear. It appears
that interferon (3 has inhibitory effects on the proliferation of leukocytes
and antigen
Io presentation. Furthermore, interferon (3 may modulate the profile of
cytokine production
towards an anti-inflammatory phenotype. Finally, interferon (3 can reduce T-
cell migration by
inhibiting the activity of T-cell matrix metalloproteases. These activities
are likely to act in
concert to account for the mechanism of interferon (3 in MS (Neurol. S 1:682-
689, 1998).
In addition, interferon (3 may be used for the treatment of osteosarcoma,
basal cell
is carcinoma, cervical dysplasia, glioma, acute myeloid leukemia, multiple
myeloma, Hodgkin's
disease, breast carcinoma, melanoma, and viral infections such as papilloma
virus, viral
hepatitis, herpes genitalia, herpes zoster, herpetic keratitis, herpes
simplex, viral encephalitis,
cytomegalovirus pneumonia, and rhinovirus.
Various side effects are associated with the use of current preparations of
2o interferon (3, including injection site reactions, fever, chills, myalgias,
arthralgias, and other flu-
like symptoms (Clin. Therapeutics, 19:883-893, 1997).
In addition, 6-40% of patients develop neutralizing antibodies to interferon
(3 (Int.
Arch. Allergy Immunol. 118:368-371, 1999). It has been shown that development
of interferon
~3-neutralizing antibodies decreases the biological response to interferon (3,
and causes a trend
2s towards decreased treatment effect (Neurol. 50:1266-1272, 1998).
Neutralizing antibodies will
likely also impede the therapeutic utility of interferon (3 in connection with
treatment of other
diseases (Immunol. Immuther. 39:263-268, 1994).
Given the magnitude of side effects with current interferon (3 products, their
association with frequent injection, the risk of developing neutralizing
antibodies impeding the
desired therapeutic effect of interferon ~3, and the potential for obtaining
more optimal
therapeutic interferon (3 levels with concomitant enhanced therapeutic effect,
there is clearly a
need for improved interferon [3-like molecules.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
4
BRIEF DISCLOSURE OF THE INVENTION
This application discloses improved interferon (3 molecules providing one or
s more of the aforementioned desired benefits. In particular conjugates are
disclosed that exhibit
interferon (3 activity and comprise at least one non-polypeptide moiety
covalently attached to
an interferon ~i polypeptide that comprises an amino acid sequence that
differs from that of
wildtype human interferon (3 with the amino acid sequence shown in SEQ ID NO 2
in at least
one amino acid residue selected from an introduced or removed amino acid
residue comprising
to an attachment group for the non-polypeptide moiety. Conjugates of the
present invention have
a number of improved properties as compared to human interferon (3, including
reduced
immunogenicity, increased functional in vivo half life, increased serum half
life, and/or
increased bioavailability. Consequently, the conjugate of the invention offers
a number of
advantages over the currently available interferon (3 compounds, including
longer duration
i s between injections, fewer side effects, and/or increased efficiency due to
reduction in
antibodies. Moreover, higher doses of active protein and thus a more effective
therapeutic
response may be obtained by use of a conjugate of the invention. Furthermore,
conjugates of
the invention have demonstrated significantly reduced cross-reactivity with
sera from patients
treated with currently available interferon ~3 products as defined
hereinbelow.
2o In one aspect the invention relates to a conjugate exhibiting interferon (3
activity
and comprising at least one first non-polypeptide moiety covalently attached
to an interferon (3
polypeptide, the amino acid sequence of which differs from that of wild-type
human interferon
~i in at least one introduced and at least one removed amino acid residue
comprising an
attachment group for said first non-polypeptide moiety.
2s In another aspect the invention relates to a conjugate exhibiting
interferon (3
activity and comprising at least one first non-polypeptide moiety conjugated
to at least one
lysine residue of an interferon ~i polypeptide, the amino acid sequence of
which differs from
that of wild-type human interferon ~i in at least one introduced and/or at
least one removed
lysine residue.
3o In yet another aspect the invention relates to a conjugate exhibiting
interferon (3
activity and comprising at least one first non-polypeptide moiety conjugated
to at least one
cysteine residue of an interferon (3 polypeptide, the amino acid sequence of
which differs from
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
at least one introduce cysteine residue into a position that in wild-type
human interferon (3 is
occupied by a surface exposed amino acid residue.
In yet another aspect the invention relates to a conjugate exhibiting
interferon (3
activity and comprising at least one first non-polypeptide moiety having an
acid group as an
s attachment group, which moiety is conjugated to at least one aspartic acid
or glutamic acid
residue of an interferon ~3 polypeptide, the amino acid sequence of which
differs from that of
wild-type human interferon (3 in at least one introduced and/or at least one
removed aspartic
acid or glutamic acid residue.
In yet another aspect the invention relates to a conjugate exhibiting
interferon (3
to activity and comprising at least one polymer molecule and at least one
sugar moiety covalently
attached to an interferon (3 polypeptide, the amino acid sequence of which
differs from that of
wild-type human interferon ~i in
a) at least one introduced and/or at least one removed amino acid residue
comprising an
attachment group for the polymer molecule, and
is b) at least one introduced and/or at least one removed amino acid residue
comprising an
attachment group for the sugar moiety,
provided that when the attachment group for the polymer molecule is a cysteine
residue, and
the sugar moiety is an N-linked sugar moiety, a cysteine residue is not
inserted in such a
manner that an N-glycosylation site is destroyed.
2o In yet another aspect the invention relates to a conjugate exhibiting
interferon (3
activity and comprising an interferon (3 polypeptide, the amino acid sequence
of which differs
from that of wild-type human interferon (3 in at least one introduced
glycosylation site, the
conjugate further comprising at least one un-PEGylated sugar moiety attached
to an introduced
glycosylation site.
2s In yet another aspect the invention relates to a conjugate exhibiting
interferon (3
activity and comprising an interferon (3 polypeptide, the amino acid sequence
of which differs
from that of wild-type human interferon ~i in that a glycosylation site has
been introduced or
removed by way of introduction or removal of amino acid residues) constituting
a part of a
glycosylation site in a position that in wildtype human interferon ~3 is
occupied by a surface
3o exposed amino acid residue.
In a still further aspect the invention relates to a conjugate exhibiting
interferon (3
activity and comprising a sugar moiety covalently attached to an interferon (3
polypeptide, the
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
6
amino acid sequence of which differs from that of wild-type human interferon
(3 in at least one
removed glycosylation site.
In still further aspects the invention relates to means and methods for
preparing a
conjugate or interferon (3 polypeptide for use in the invention, including
nucleotide sequences
s and expression vectors encoding the polypeptide as well as methods for
preparing the
polypeptide or the conjugate.
In final aspects the invention relates to a therapeutic composition comprising
a
conjugate of the invention, to a conjugate or composition of the invention for
use in therapy, to
the use of a conjugate or composition in therapy or for the manufacture of a
medicament for
~ o treatment of diseases.
BRIEF DESCRIPTION OF THE DRAWINGS
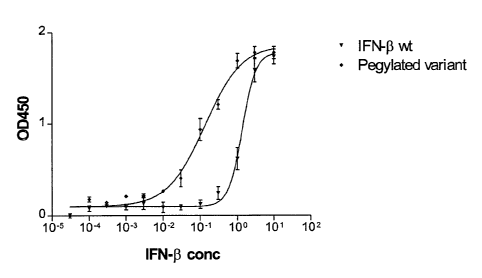
Fig. 1 illustrates the antiviral activity of a conjugate of the invention,
t s Fig 2 the yield of interferon (3 production obtained according to Example
8.
DETAILED DISCLOSURE OF THE INVENTION
In the present application a number of references are referred to. They are
all intended to be
2o incorporated herein by reference.
Definitions
In the context of the present application and invention the following
definitions apply:
The term "conjugate" (or interchangeably "conjugated polypeptide") is intended
2s to indicate a heterogeneous (in the sense of composite or chimeric)
molecule formed by the
covalent attachment of one or more polypeptide(s) to one or more non-
polypeptide moieties.
The term covalent attachment means that the polypeptide and the non-
polypeptide moiety are
either directly covalently joined to one another, or else are indirectly
covalently joined to one
another through an intervening moiety or moieties, such as a bridge, spacer,
or linkage moiety
30 or moieties using an attachment group present in the polypeptide.
Preferably, the conjugate is
soluble at relevant concentrations and conditions, i.e. soluble in
physiological fluids such as
blood. Examples of conjugated polypeptides of the invention include
glycosylated and/or
PEGylated polypeptides. The term "non-conjugated polypeptide" may be used
about the
polypeptide part of the conjugate.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
7
The term "non-polypeptide moiety" is intended to indicate a molecule that
is capable of conjugating to an attachment group of the polypeptide of the
invention. Preferred
examples of such molecule include polymer molecules, sugar moieties,
lipophilic compounds,
or organic derivatizing agents. When used in the context of a conjugate of the
invention it will
s be understood that the non-polypeptide moiety is linked to the polypeptide
part of the
conjugate through an attachment group of the polypeptide.
The term "polymer molecule" is defined as a molecule formed by covalent
linkage of two or more monomers, wherein none of the monomers is an amino acid
residue,
except where the polymer is human albumin or another abundant plasma protein.
The term
"polymer" may be used interchangeably with the term "polymer molecule". The
term is
intended to cover carbohydrate molecules attached by in vitro glycosylation,
i.e. a synthetic
glycosylation performed in vitro normally involving covalently linking a
carbohydrate
molecule to an attachment group of the polypeptide, optionally using a cross-
linking agent.
Carbohydrate molecules attached by in vivo glycosylation, such as N- or O-
glycosylation (as
t s further described below)) are referred to herein as "a sugar moiety" .
Except where the
number of non-polypeptide moieties, such as polymer molecules) or sugar
moieties in the
conjugate is expressly indicated every reference to "a non-polypeptide moiety"
contained in a
conjugate or otherwise used in the present invention shall be a reference to
one or more non-
polypeptide moieties, such as polymer molecules) or sugar moieties, in the
conjugate.
2o The term "attachment group" is intended to indicate an amino acid residue
group
of the polypeptide capable of coupling to the relevant non-polypeptide moiety.
For instance,
for polymer, in particular PEG, conjugation a frequently used attachment group
is the s-amino
group of lysine or the N-terminal amino group. Other polymer attachment groups
include a free
carboxylic acid group (e.g. that of the C-terminal amino acid residue of of an
aspartic acid or
2s glutamic acid residue), suitably activated carbonyl groups, oxidized
carbohydrate moieties and
mercapto groups.
For in vivo N-glycosylation, the term "attachment group" is used in an
unconventional way to indicate the amino acid residues constituting an N-
glycosylation site
(with the sequence N-X'-S/T/C-X", wherein X' is any amino acid residue except
proline,
30 X" any amino acid residue that may or may not be identical to X' and
preferably is different
from proline, N is asparagine and S/T/C is either serine, threonine or
cysteine, preferably
serine or threonine, and most preferably threonine). Although the asparagine
residue of the
N-glycosylation site is the one to which the sugar moiety is attached during
glycosylation,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
8
such attachment cannot be achieved unless the other amino acid residues of the
N-
glycosylation site is present. Accordingly, when the non-polypeptide moiety is
an N-linked
sugar moiety, the term "amino acid residue comprising an attachment group for
the non-
polypeptide moiety" as used in connection with alterations of the amino acid
sequence of the
s parent polypeptide is to be understood as amino acid residues constituting
an N-glycosylation
site is/are to be altered in such a manner that either a functional N-
glycosylation site is
introduced into the amino acid sequence or removed from said sequence.For an
"O-
glycosylation site" the attachment group is the OH-group of a serine or
threonine residue.
The term "one difference" or "differs from" as used in connection with
specific
1 o mutations is intended to allow for additional differences being present
apart from the specified
amino acid difference. For instance, in addition to the removal and/or
introduction of amino
acid residues comprising an attachment group for the non-polypeptide moiety
the interferon (3
polypeptide may comprise other substitutions that are not related to
introduction and/or
removal of such amino acid residues. The term "at least one" as used about a
non-polypeptide
i s moiety, an amino acid residue, a substitution, etc is intended to mean one
or more. The terms
"mutation" and "substitution" are used interchangeably herein.
In the present application, amino acid names and atom names (e.g. CA, CB,
CD, CG, SG, NZ, N, O, C, etc) are used as defined by the Protein DataBank
(PDB)
(www.pdb.org) which are based on the IUPAC nomenclature (IUPAC Nomenclature
and
2o Symbolism for Amino Acids and Peptides (residue names, atom names e.t.c.),
Eur. J.
Biochem. , 138, 9-37 ( 1984) together with their corrections in Eur. J.
Biochem. , 152, 1
(1985). CA is sometimes referred to as Ca, CB as Chi. The term "amino acid
residue" is
intended to indicate an amino acid residue contained in the group consisting
of alanine (Ala or
A), cysteine (Cys or C), aspartic acid (Asp or D), glutamic acid (Glu or E),
phenylalanine (Phe
2s or F), glycine (Gly or G), histidine (His or H), isoleucine (Ile or I),
lysine (Lys or K), leucine
(Leu or L), methionine (Met or M), asparagine (Asn or N), proline (Pro or P),
glutamine (Gln
or Q), arginine (Arg or R), serine (Ser or S), threonine (Thr or T), valine
(Val or V), tryptophan
(Trp or V~, and tyrosine (Tyr or Y) residues. The terminology used for
identifying amino acid
positions/substitutions is illustrated as follows: C17 (indicates positiom#17
occupied by a
3o cysteine residue in the amino acid sequence shown in SEQ ID NO 2). C17S
(indicates that the
cysteine residue of position 17 has been replaced with a serine). The
numbering of amino acid
residues made herein is made relative to the amino acid sequence shown in SEQ
ID NO 2.
"M 1 del" is used about a deletion of the methionine residue occupying
position 1. Multiple
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
9
substitutions are indicated with a "+", e.g. R71N+D73T/S means an amino acid
sequence
which comprises a substitution of the arginine residue in position 71 with an
asparagine and a
substitution of the aspartic acid residue in position 73 with a threonine or
serine residue,
preferably a threonine residue. T/S as used about a given substitution herein
means either a T
s or a S residue, preferably a T residue.
The term "nucleotide sequence" is intended to indicate a consecutive stretch
of
two or more nucleotide molecules. The nucleotide sequence may be of genomic,
cDNA, RNA,
semisynthetic, synthetic origin, or any combinations thereof.
The term "interferon (3 protein sequence family" is used in its conventional
meaning, i.e. to indicate a group of polypeptides with sufficiently homologous
amino acid
sequences to allow alignment of the sequences, e.g. using the CLUSTALW
program. An
interferon (3 sequence family is available, e.g. from the PFAM families,
version 4.0, or may be
prepared by use of a suitable computer program such as CLUSTALW version 1.74
using
default parameters (Thompson et al., 1994, CLUSTAL W: improving the
sensitivity of
i s progressive multiple sequence alignment through sequence weighting,
position-specific gap
penalties and weight matrix choice, Nucleic Acids Research, 22:4673-4680).
The term "polymerase chain reaction" or "PCR" generally refers to a method for
amplification of a desired nucleotide sequence in vitro, as described, for
example, in US
4,683,195. In general, the PCR method involves repeated cycles of primer
extension synthesis,
2o using oligonucleotide primers capable of hybridising preferentially to a
template nucleic acid.
"Cell", "host cell", "cell line" and "cell culture" are used interchangeably
herein
and all such terms should be understood to include progeny resulting from
growth or culturing
of a cell. "Transformation" and "transfection" are used interchangeably to
refer to the process
of introducing DNA into a cell.
2s "Operably linked" refers to the covalent joining of two or more nucleotide
sequences, by means of enzymatic ligation or otherwise, in a configuration
relative to one
another such that the normal function of the sequences can be performed. For
example, the
nucleotide sequence encoding a presequence or secretory leader is operably
linked to a
nucleotide sequence for a polypeptide if it is expressed as a preprotein that
participates in the
3o secretion of the polypeptide: a promoter or enhancer is operably linked to
a coding sequence if
it affects the transcription of the sequence; a ribosome binding site is
operably linked to a
coding sequence if it is positioned so as to facilitate translation.
Generally, "operably linked"
means that the nucleotide sequences being linked are contiguous and, in the
case of a secretory
leader, contiguous and in reading phase. Linking is accomplished by ligation
at convenient
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
restriction sites. If such sites do not exist, then synthetic oligonucleotide
adaptors or linkers are
used, in conjunction with standard recombinant DNA methods.
The term "introduce" is primarily intended to mean substitution of an existing
amino acid residue, but may also mean insertion of an additional amino acid
residue. The term
s "remove" is primarily intended to mean substitution of the amino acid
residue to be removed
by another amino acid residue, but may also mean deletion (without
substitution) of the amino
acid residue to be removed.
The term "immunogenicity" as used in connection with a given substance is
intended to indicate the ability of the substance to induce a response from
the immune system.
to The immune response may be a cell or antibody mediated response (see, e.g.,
Roitt: Essential
Immunology (8'h Edition, Blackwell) for further definition of immunogenicity).
Immunogenicity may be determined by use of any suitable method known in the
art, e.g. in
vivo or in vitro, e.g. using the in vitro immunogenicity test outlined in the
Materials and
Methods section below.The term "reduced immunogenicity" is intended to
indicate that the
is conjugate or polypeptide of the present invention gives rise to a
measurably lower immune
response than a reference molecule, such as wildtype human interferon (3, e.g.
Rebif or
Avonex, or a variant of wild-type human interferon (3 such as Betaseron, as
determined under
comparable conditions. When reference is made herein to commercially available
interferon (3
products (i.e. Betaseron, Avonex and Rebif), it should be understood to mean
either the
2o formulated product or the interferon (3 polypeptide part of the product (as
appropriate).
Normally, reduced antibody reactivity (e.g. reactivity towards antibodies
present in serum from
patients treated with commercial interferon ~i products) is an indication of
reduced
immunogenicity.
The term "functional in vivo half life" is used in its normal meaning, i.e.
the time
2s at which 50% of a given functionality of the polypeptide or conjugate is
retained (such as the
time at which 50% of the biological activity of the polypeptide or conjugate
is still present in
the body/target organ, or the time at which the activity of the polypeptide or
conjugate is 50%
of the initial value). As an alternative to determining functional in vivo
half life, "serum half
life" may be determined, i.e. the time in which 50% of the polypeptide or
conjugate molecules
3o circulate in the plasma or bloodstream prior to being cleared.
Determination of serum half life
is often more simple than determining functional in vivo half life and the
magnitude of serum
half life is usually a good indication of the magnitude of functional in vivo
half life.
Alternative terms to serum half life include "plasma half life", "circulating
half life",
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
11
"serum clearance", "plasma clearance" and "clearance half life". The
functionality to be
retained is normally selected from antiviral, antiproliferative,
immunomodulatory or receptor
binding activity. Functional in vivo half life and serum half life may be
determined by any
suitable method known in the art as further discussed in the Materials and
Methods section
s hereinafter.
The polypeptide or conjugate is normally cleared by the action of one or more
of
the reticuloendothelial systems (RES), kidney, spleen or liver, or by specific
or unspecific
proteolysis. Clearance taking place by the kidneys may also be referred to as
"renal clearance"
and is e.g. accomplished by glomerular filtration, tubular excretion or
tubular elimination.
io Normally, clearance depends on physical characteristics of the conjugate,
including molecular
weight, size (diameter) (relative to the cut-off for glomerular filtration),
charge, symmetry,
shape/rigidity, attached carbohydrate chains, and the presence of cellular
receptors for the
protein. A molecular weight of about 67 kDa is considered to be an important
cut-off value for
renal clearance.
~ s Reduced renal clearance may be established by any suitable assay, e.g. an
established in vivo assay. Typically, the renal clearance is determined by
administering a
labelled (e.g. radiolabelled or fluorescence labelled) polypeptide conjugate
to a patient and
measuring the label activity in urine collected from the patient. Reduced
renal clearance is
determined relative to the corresponding non-conjugated polypeptide or the non-
conjugated
2o corresponding wild-type polypeptide or a commercial interferon (3 product
under comparable
conditions.
The term "increased" as used about the functional in vivo half life or serum
half
life is used to indicate that the relevant half life of the conjugate or
polypeptide is statistically
significantly increased relative to that of a reference molecule, such as an
un-conjugated
2s wildtype human interferon (3 (e.g. Avonex or RebifJ or an unconjugated
variant human
interferon (3 (e.g. Betaseron) as determined under comparable conditions.
The term "reduced immunogenicity and/or increased functional in vivo half life
and/or increased serum half life" is to be understood as covering any one, two
or all of these
properties. Preferably, a conjugate or polypeptide of the invention has at
least two or these
3o properties, i.e. reduced immunogenicity and increased functional in vivo
half life, reduced
immunogenicity and increased serum half life or increased functional in vivo
half life and
increased serum half life. Most preferably, the conjugate or polypeptide of
the invention has all
properties.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
12
The term "exhibiting interferon (3 activity" is intended to indicate that the
polypeptide or conjugate has one or more of the functions of native interferon
(3, in particular
human wildtype interferon ~i with the amino acid sequence shown in SEQ ID NO 2
(which is
the mature sequence) optionally expressed in a glycosylating host cell or any
of the
s commercially available interferon (3 products. Such functions include
capability to bind to an
interferon receptor that is capable of binding interferon ~i and initiating
intracellular signaling
from the receptor, in particular a type I interferon receptor constituted by
the receptor subunits
IFNAR-2 and IFNAR-1 (Domanski et al., The Journal of Biological Chemistry,
Vol. 273, No.
6, pp3144-3147, 1998, Mogensen et al., Journal of Interferon and Cytokine
Research, 19:
1069-1098, 1999), and antiviral, antiproliferative or immunomodulatory
activity (which can be
determined using assays known in the art (e.g. those cited in the following
disclosure)).
Interferon [3 activity may be assayed by methods known in the art as
exemplified in the
Materials and Methods section hereinafter.
The polypeptide or conjugate "exhibiting" or "having" interferon (3 activity
is
is considered to have such activity, when it displays a measurable function,
e.g. a measurable
receptor binding and stimulating activity (e.g. as determined by the primary
or secondary assay
described in the Materials and Methods section). The polypeptide exhibiting
interferon (3
activity may also be termed "interferon (3 molecule" or "interferon (3
polypeptide" herein. The
term "interferon ~3 polypeptide" is primarily used herein about modified
polypeptides of the
20 invention (having introduced or removed attachment groups for the relevant
non-polypeptide
moiety).
The term "parent interferon (3" is intended to indicate the starting molecule
to
be improved in accordance with the present invention. While the parent
interferon (3 may be
of any origin, such as vertebrate or mammalian origin (e.g. any of the origins
defined in WO
2s 00/23472), the parent interferon (3 is preferably wild-type human
interferon (3 with SEQ ID
NO 2 or a variant thereof. A "variant" is a polypeptide, which differs in one
or more amino
acid residues from a parent polypeptide, normally in 1, 2,3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13,
14 or 15 amino acid residues. Examples of wild-type human interferon ~i
include the
polypeptide part of Avonex or Rebif. An example of a parent interferon (3
variant is
3o Betaseron. Alternatively, the parent interferon (3 polypeptide may comprise
an amino acid
sequence, which is a hybrid molecule between interferon ~i and another
homologous
polypeptide, such as interferon a, optionally containing one or more
additional substitutions
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
13
introduced into the hybrid molecule. Such a hybrid molecule may contain an
amino acid
sequence, which differs in more than 10 amino acid residues from the amino
acid sequence
shown in SEQ ID NO 2. In order to be useful in the present invention the
hybrid molecule
exhibits interferon (3 activity (e.g. as determined in the secondary assay
described in the
s Materials and Methods section herein).
The term "functional site" as used about a polypeptide or conjugate of the
invention is intended to indicate one or more amino acid residues which is/are
essential for or
otherwise involved in the function or performance of interferon (3, and thus
"located at" the
functional site. The functional site is e.g. a receptor binding site and may
be determined by
to methods known in the art, preferably by analysis of a structure of the
polypeptide complexed to
a relevant receptor, such as the type I interferon receptor constituted by
IFNAR-1 and IFNAR-
2.
Conjugate of the invention .
t s As stated above, in a first aspect the invention relates to a conjugate
exhibiting interferon (3
activity and comprising at least one first non-polypeptide moiety covalently
attached to an
interferon [3 polypeptide, the amino acid sequence of which differs from that
of wildtype
human interferon (3 in at least one introduced and at least one removed amino
acid residue
comprising an attachment group for said first non-polypeptide moiety.
20 . By removing and/or introducing amino acid residues comprising an
attachment
group for the non-polypeptide moiety it is possible to specifically adapt the
polypeptide so as
to make the molecule more susceptible to conjugation to the non-polypeptide
moiety of choice,
to optimize the conjugation pattern (e.g. to ensure an optimal distribution of
non-polypeptide
moieties on the surface of the interferon [3 molecule and thereby, e.g.,
effectively shield
2s epitopes and other surface parts of the polypeptide without significantly
impairing the function
thereof). For instance, by introduction of attachment groups, the interferon
~3 polypeptide is
boosted or otherwise altered in the content of the specific amino acid
residues to which the
relevant non-polypeptide moiety binds, whereby a more efficient, specific
and/or extensive
conjugation is achieved. By removal of one or more attachment groups it is
possible to avoid
3o conjugation to the non-polypeptide moiety in parts of the polypeptide in
which such
conjugation is disadvantageous, e.g. to an amino acid residue located at or
near a functional site
of the polypeptide (since conjugation at such a site may result in
inactivation or reduced
interferon ~i activity of the resulting conjugate due to impaired receptor
recognition). Further, it
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
14
may be advantageous to remove an attachment group located closely to another
attachment
group in order to avoid heterogeneous conjugation to such groups.
It will be understood that the amino acid residue comprising an attachment
group for a non-polypeptide moiety, either it be removed or introduced, is
selected on the
s basis of the nature of the non-polypeptide moiety and, in most instances, on
the basis of the
conjugation method to be used. For instance, when the non-polypeptide moiety
is a polymer
molecule, such as a polyethylene glycol or polyalkylene oxide derived
molecule, amino acid
residues capable of functioning as an attachment group may be selected from
the group
consisting of lysine, cysteine, aspartic acid, glutamic acid and arginine.
When the non-
to polypeptide moiety is a sugar moiety the attachment group is an in vivo
glycosylation site,
preferably an N-glycosylation site.
Whenever an attachment group for a non-polypeptide moiety is to be introduced
into or removed from the interferon ~i polypeptide in accordance with the
present invention, the
position of the interferon ~i polypeptide to be modified is conveniently
selected as follows:
t s . The position is preferably located at the surface of the interferon (3
polypeptide,
and more preferably occupied by an amino acid residue which has more than 25%
of its side
chain exposed to the solvent, preferably more than 50% of its side chain
exposed to the solvent.
Such positions have been identified on the basis of an analysis of a 3D
structure of the human
interferon [3 molecule as described in the Methods section herein.
2o Alternatively or additionally, the position to be modified is identified on
the basis
of an analysis of an interferon (3 protein sequence family. More specifically,
the position to be
modified can be one, which in one or more members of the family othei than the
parent
interferon (3, is occupied by an amino acid residue comprising the relevant
attachment group
(when such amino acid residue is to be introduced) or which in the parent
interferon (3, but not
2s in one or more other members of the family, is occupied by an amino acid
residue comprising
the relevant attachment group (when such amino acid residue is to be removed).
In order to determine an optimal distribution of attachment groups, the
distance
between amino acid residues located at the surface of the interferon ~i
molecule is calculated on
the basis of a 3D structure of the interferon ~3 polypeptide. More
specifically, the distance
3o between the CB's of the amino acid residues comprising such attachment
groups, or the
distance between the functional group (NZ for lysine, CG for aspartic acid, CD
for glutamic
acid, SG for cysteine) of one and the CB of another amino acid residue
comprising an
attachment group are determined. In case of glycine, CA is used instead of CB.
In the
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
interferon (3 polypeptide part of a conjugate of the invention, any of said
distances is preferably
more than 8 t~, in particular more than 10~r in order to avoid or reduce
heterogeneous
conjugation.
Furthermore, in the interferon (3 polypeptide part of a conjugate of the
invention
s attachment groups located at the receptor-binding site of interferon (3 has
preferably been
removed, preferably by substitution of the amino acid residue comprising such
group.
A still further generally applicable approach for modifying an interferon ~3
polypeptide is to shield, and thereby destroy or otherwise inactivate an
epitope present in the
parent interferon Vii, by conjugation to a non-polypeptide moiety. Epitopes of
human interferon
t o (3 may be identified by use of methods known in the art, also known as
epitope mapping, see,
e.g. Romagnoli et al., J. Biol Chem, 1999, 380(5):553-9, DeLisser HM, Methods
Mol Biol,
1999, 96:11-20, Van de Water et al., Clin Immunol Immunopathol, 1997,
85(3):229-35, Saint-
Remy JM, Toxicology, 1997, 119(1):77-81, and Lane DP and Stephen CW, Curr Opin
Immunol, 1993, 5(2):268-71. One method is to establish a phage display library
expressing
~s random oligopeptides of e.g. 9 amino acid residues. IgGI antibodies from
specific antisera
towards human interferon (3 are purified by immunoprecipitation and the
reactive phages are
identified by immunoblotting. By sequencing the DNA of the purified reactive
phages, the
sequence of the oligopeptide can be determined followed by localization of the
sequence on the
3D-structure of the interferon ~3. Alternatively, epitopes can be identified
according to the
2o method described in US 5,041,376. The thereby identified region on the
structure constitutes an
epitope that then can be selected as a target region for introduction of an
attachment group for
the non-polypeptide moiety. Preferably, at least one epitope, such as two,
three or four epitopes
of human recombinant interferon (3 (optionally comprising the C17S mutation)
are shielded by
a non-polypeptide moiety according to the present invention. Accordingly, in
one embodiment,
2s the conjugate of the invention has at least one shielded epitope as
compared to wild type
human interferon (3, optionally comprising the C17S mutation, including any
commercially
available interferon (3. Preferably, the conjugate of the invention comprises
a polypeptide that
is modified so as to shield the epitope located in the vicinity of amino acid
residue Q49 and/or
F 111. This may be done by introduction of an attachment group for a non-
polypeptide moiety
3o into a position located in the vicinity of (i.e. within 4 amino acid
residues in the primary
sequence or within about 10~ in the tertiary sequence) of Q49 and/or F 111.
The 10~ distance
is measured between CB's (CA's in case of glycine). Such specific
introductions are described
in the following sections.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
16
In case of removal of an attachment group, the relevant amino acid residue
comprising such group and occupying a position as defined above is preferably
substituted
with a different amino acid residue that does not comprise an attachment group
for the non-
polypeptide moiety in question.
s In case of introduction of an attachment group, an amino acid residue
comprising
such group is introduced into the position, preferably by substitution of the
amino acid residue
occupying such position.
The exact number of attachment groups available for conjugation and present in
the interferon (3 polypeptide is dependent on the effect desired to be
achieved by conjugation.
to The effect to be obtained is, e.g., dependent on the nature and degree of
conjugation (e.g. the
identity of the non-polypeptide moiety, the number of non-polypeptide moieties
desirable or
possible to conjugate to the polypeptide, where they should be conjugated or
where
conjugation should be avoided, etc.). For instance, if reduced immunogenicity
is desired, the
number (and location of) attachment groups should be sufficient to shield most
or all epitopes.
t s This is normally obtained when a greater proportion of the interferon ~i
polypeptide is shielded.
Effective shielding of epitopes is normally achieved when the total number of
attachment
groups available for conjugation is in the range of 1-10 attachment groups, in
particular in the
range of 2-8, such as 3-7.
Functional in vivo half life is i.a. dependent on the molecular weight of the
2o conjugate and the number of attachment groups needed for providing
increased half life thus
depends on the molecular weight of the non-polypeptide moiety in question. In
one
embodiment, the conjugate of the invention has a molecular weight of at least
67 kDa, in
particular at least 70 kDa as measured by SDS-PAGE according to Laemmli, U.K.,
Nature
Vol 227 (1970), p680-85. Interferon (3 has a molecular weight of about 20 kDa,
and therefore
2s - additional about SOkDa is required to obtain the desired effect. This may
be, e.g., be provided
by 5 l OkDa PEG molecules or as otherwise described herein.
In order to avoid too much disruption of the structure and function of the
parent
human interferon (3 molecule the total number of amino acid residues to be
altered in
accordance with the present invention (as compared to the amino acid sequence
shown in SEQ
3o ID NO 2) typically does not exceed 15. Preferably, the interferon (3
polypeptide comprises an
amino acid sequence, which differs in 1-15 amino acid residues from the amino
acid sequence
shown in SEQ ID NO 2, such as in 1-8 or in 2-8 amino acid residues, e.g. in 1-
5 or in 2-5
amino acid residues from the amino acid sequence shown in SEQ ID NO 2. Thus,
normally the
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
17
interferon (3 polypeptide comprises an amino acid sequence that differs from
the amino acid
sequence shown in SEQ ID NO 2 in 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14
or 15 amino acid
residues. Preferably, the above numbers represent either the total number of
introduced or the
total number of removed amino acid residues comprising an attachment group for
the relevant
s non-polypeptide moiety, or the total number of introduced and removed amino
acid residues
comprising such group.
In the conjugate of the invention it is preferred that at least about SO% of
all
conjugatable attachment groups, such as at least about 80% and preferably all
of such groups
are occupied by the relevant non-polypeptide moiety. Accordingly, in a
preferred embodiment
~o the conjugate of the invention comprises, e.g., 1-10 non-polypeptide
moieties, such as 2-8 or 3-
6.
The conjugate of the invention has one or more of the following improved
properties:
Reduced immunogenicity as compared to wild-type human interferon (3 (e.g.
Is Avonex or Rebifj or to Betaseron, e.g. a reduction of at least 25%, such as
at least 50%, and
more preferably at least 75%;
Increased functional in vivo half life and/or increased serum half life as
compared
to wild-type human interferon (3 (e.g. Avonex or Rebif) or to Betaseron;
Reduced or no reaction with neutralizing antibodies from patients treated with
2o wildtype human interferon ~3 (e.g. Rebif or Avonex) or with Betaseron, e.g.
a reduction of
neutralisation of at least 25%, such as of at least 50%, and preferably of at
least 75%.
The magnitude of the antiviral activity of a conjugate of the invention may
not be
critical, and thus be reduced (e.g. by up to 75%) or increased (e.g. by at
least 5%) or equal to
that of wild-type human interferon (3 ((e.g. Avonex or Rebif) or to Betaseron;
2s Furthermore, the degree of antiviral activity as compared to
antiproliferative
activity of a conjugate of the invention may vary, and thus be higher, lower
or equal to that of
wildtype human interferon (3.
Conjugate of the invention, wherein the non polypeptide moiety is a molecule
that has lysine as
3o an attachment group
In a preferred embodiment the first non-polypeptide moiety has lysine as an
attachment group, and thus the interferon (3 polypeptide is one that comprises
an amino acid
sequence that differs from that of wildtype human interferon ~i in at least
one introduced and/or
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
1~
at least one removed lysine residue. While the non-polypeptide moiety may be
any of those
binding to a lysine residue, e.g. the s-amino group thereof, such as a polymer
molecule, a
lipophilic group, an organic derivatizing agent or a carbohydrate moiety, it
is preferably any of
the polymer molecule mentioned in the section entitled "Conjugation to a
polymer molecule",
s in particular a branched or linear PEG or polyalkylene oxide. Most
preferably, the polymer
molecule is PEG and the activated molecule to be used for conjugation is SS-
PEG, NPC-PEG,
aldehyd-PEG, mPEG-SPA, mPEG-SCM, mPEG-BTC from Shearwater Polymers, Inc, SC-
PEG from Enzon, Inc., tresylated mPEG as described in US 5,880,255, or
oxycarbonyl-oxy-N-
dicarboxyimide-PEG (US 5,122,614). Normally, for conjugation to a lysine
residue the non-
t o polypeptide moiety has a molecular weight of about S or 10 kDa.
In one embodiment the amino acid sequence of the interferon ~3 polypeptide
differs from that of human wildtype interferon ~i in at least one removed
lysine residue, such as
1-5 removed lysine residues, in particular 1-4 or 1-3 removed lysine residues.
The lysine
residues) to be removed, preferably by replacement, is selected from the group
consisting of
~s K19, K33, K45, K52, K99, K105, K108, K115, K123, K134, and K136. The lysine
residues)
may be replaced with any other amino acid residue, but is preferably replaced
by an arginine or
a glutamine residue in order to give rise to the least structural difference.
In particular, the
polypeptide part may be one, wherein K19, K45, K52 and/or K123, preferably
K19, K45
and/or K123 has/have been replaced with another any other amino acid residue,
preferably
2o arginine or glutamine. For instance, the interferon ~i polypeptide part of
a conjugate of the
invention comprises a combination of amino acid substitutions selected from
the following list:
K 19R+K45R+K 1238;
K19Q+K45R+K123R;
K19R+K45Q+K123R;
2s K19R+K45R+K123Q;
K 19Q+K45Q+K 1238;
K19R+K45Q+K123Q;
K19Q+K45R+K123Q;
K19Q+K45Q+K123Q;
30- K45R+K123R;
K45Q+K123R;
K45 Q+K 123 Q;
K45R+K123Q;
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
19
K19R+K123R;
K19Q+K123R;
K19R+K123Q;
K19Q+K123Q;
s K19R+K45R;
K19Q+K45R;
K19R+K45Q; or
K19Q+K45Q.
In addition or alternatively to the amino acid substitutions mentioned in the
above
~ o list the polypeptide part may comprise at least one substitution selected
from the group
consisting of K33R, K33Q, K52R, K52Q, K99R, K99Q, K105R, K105Q, K108R, K108Q,
K115R, K115Q, K134R, K134Q, K136R, and K136Q, e.g. at least one of the
following
substitutions:
K52R+K 1348;
~s K99R+K136R;
K33R+K105R+K136R;
K52R+K108R+K134R;
K99R+K11 SR+K136R;
K19R+K33R+K45R+K123R;
2o K19R+K45R+K52R+K123R;
K19R+K33R+K45R+K52R+K123R; or
K19R+K45R+K52R+K99R+K123R.
In a further embodiment the amino acid sequence of the interferon (3
polypeptide
differs from that shown in SEQ ID NO 2 in that a lysine residue has been
introduced by
2s substitution of at least one amino acid residue occupying a position that
in the parent interferon
~i molecule is occupied by a surface exposed amino acid residue, preferably an
amino acid
residue having at least 25%, such as at least 50% of its side chain exposed to
the surface.
Preferably, the amino acid residue to be substituted is selected from the
group consisting of N4,
. F8, L9, R11, 512, F15, Q16, Q18, L20, W22, Q23, G26, R27, L28, E29, Y30,
L32, R35, M36,
3o N37, D39, P41, E42, E43, L47, Q48, Q49, T58, Q64, N65, F67, A68, R71, Q72,
D73, 575,
S76, G78, N80, E81, I83, E85, N86, A89, N90, Y92, H93, H97, T100, L102, E103,
L106,
E107, E109, D110, F111, 8113, 6114, L116, M117, L120, H121, 8124, 6127, 8128,
L130,
H131, E137, Y138, H140, I145, 8147, V148, E149, 8152, Y155, F156, N158, 8159,
6162,
Y 163, 8165 and N 166 of SEQ ID NO 2.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
2U
More preferably, the amino acid sequence of the interferon ~i polypeptide
differs
from the amino acid sequence shown in SEQ ID NO 2 in that a lysine residue has
been
introduced, by substitution, of at least one amino acid residue occupying a
position selected
from the group consisting of N4, F8, L9, R 11, S 12, G26, R27, E29, R35, N37,
D39, E42, L47,
s Q48, Q49, A68, R71, Q72, D73, S75, G78, N80, E85, N86, A89, Y92, H93, D110,
F111,
8113, L116, H121, 8124, 6127, 8128, 8147, V148, Y155, N158, 8159, 6162 and
8165, even
more preferably selected from the group consisting of N4, R11, G26, R27, Q48,
Q49, R71,
D73, 575, N80, E85, A89, Y92, H93, F111, 8113, L116, 8124, 6127, 8128, Y155,
N158 and
6162, and most preferably selected from the group consisting of R11, Q49, R71,
575, N80,
t o E85, A89, H93, F 111, R 113, L 116 and Y 155, and most preferably Q49 and
F 111.
In accordance with this embodiment, the interferon [3 polypeptide comprises a
substitution to lysine in one or more of the above positions, in particular in
1-15, such as 1-8 or
1-5, and preferably in at least two positions, such as 2-8 or 2-5 positions.
In a further embodiment the amino acid sequence of the interferon ~i
polypeptide
t s . part of a conjugate differs in at least one removed and at least one
introduced lysine residue,
such as 1-5 or 2-5 removed lysine residues and 1-5 or 2-5 introduced lysine
residues. It will be
understood that the lysine residues to be removed and introduced preferably
are selected from
those described in the present section.
In accordance with this embodiment of the invention, the total number of
2o conjugatable lysine residues is preferably in the range of 1-10, such as 2-
8 or 3-7.
For instance, the interferon ~i polypeptide part of the conjugate according to
this
embodiment may comprise at least one of the following substitutions: R11K,
Q48K, Q49K,
R71K, S75K, N80K, E85K, A89K, H93K, Fl 11K, R113K, L116K and Y155K; more
preferably R11K, Q49K, R71K, S75K, N80K, E85K, A89K, H93K, F111K, R113K, L116K
2s and Y155K, in combination with at least one of the substitutions: K19R/Q
K33R/Q K45R/Q,
K52R/Q, K99R/Q, K105R/Q, K108R/Q, K115R/Q, K123R/Q, K134R/Q, and K136R/Q,
wherein R/Q indicates substitution to an R or a Q residue, preferably an R
residue. More
preferably, the interferon (3 polypeptide comprises at least one of the
following substitutions
R11K, Q49K, R71K, S75K, N80K, E85K, A89K, H93K, F111K, R113K, L116K and Y155K,
3o in particular Q49K, F111K and/or N80K, in combination with substitution of
at least one of
K19, K45, K52 and/or K123, preferably to an R or a Q residue. In particular,
the interferon (3
polypeptide comprises at least one of the substitutions Q49K, F 111 K and N80K
in combination
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
21
with at least one of the substitutions mentioned above for removal of a lysine
residue. For
instance, the interferon ~3 polypeptide may comprise the following
substitutions:
Y+Z+K19R+K45R+K123R;
Y+Z+K 19Q+K45R+K123R;
s Y+Z+K19R+K45Q+K123R;
Y+Z+K19R+K45R+K123Q;
y+Z+K19Q+K45Q+K123R;
Y+Z+K19R+K45Q+K123Q;
y+Z+K19Q+K45R+K123Q;
~o Y+Z+K19Q+K45Q+K123Q;
Y+Z+K45R+K123R;
Y+Z+K45Q+K123R;
Y+Z+K45Q+K123Q;
y+Z+K45R+K123Q;
is Y+Z+K19R+K123R;
y+Z+K19Q+K123R;
y+Z+K 19R+K 123 Q;
y+Z+K19Q+K123Q;
y+Z+K19R+K45R;
2o Y+Z+K19Q+K45R;
y+Z+K19R+K45Q; or
Y+Z+K19Q+K45Q, wherein Y is selected from the group of Q49K, F111K, N80K,
Q49K+F 111 K, Q49K+N80K, F 111 K+N80K and Q49K+F 111 K+N80K and Z is absent or
comprises at least one substitution selected from the group consisting of
K33R, K33Q, K52R,
2s K52Q, K99R, K99Q, K105R, K105Q, K108R, K108Q, K115R, K115Q, K134R, K134Q,
K136R, and K136Q. Preferably, the interferon (3 polypeptide comprises the
following
substitution Y+Z+K19R+K45Q+K123R, wherein Y and Z have the above meaning.
More specifically, according to this embodiment the interferon (3 polypeptide
may comprise one of the following substitutions:
3o K19R+K45R+F111K+K123R;
K 19R+K45 R+Q49K+F 111 K+K 123 R;
K19R+K45R+Q49K+K123R;
K19R+K45R+ F111K;
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
22
K 19R+K45R+Q49K+F 111 K;
K19R+Q49K+K123R;
K19R+Q49K+F 111 K+K123R;
K45Q+F 111 K+K123Q;
s K45R+Q49K+K123R; or
K45 R+Q49K+F 111 K+K 123 R.
Especially for expression in a non-glycosylating host such as E. coli the
interferon [3 polypeptide may contain the substitution N80K or C17S+N80K,
optionally in
combination with one or more of K19R/Q; K45R/Q; K52R/Q or K123R/Q. The
substitution
to N80K is of particular interest, when the interferon ~i polypeptide is
expressed in a non-
glycosylating host cell, since N80 constitutes part of an inherent
glycosylation site of human
interferon (3 and conjugation at such site may mimick natural glycosylation.
Furthermore, it is preferred that the conjugate according to this aspect
comprises
at least two first non-polypeptide moieties, such as 2-8 moieties.
~s
Conjugate of the invention wherein the non polypeptide moiety binds to a
cysteine residue
In a still further aspect, the invention relates a conjugate exhibiting
interferon (3
activity and comprising at least one first non-polypeptide conjugated to at
least one cysteine
residue of an interferon (3 polypeptide, the amino acid sequence of which
differs from that of
2o wildtype human interferon (3 in that at least one cysteine residue has been
introduced,
prefererably by substitution, into a position that in the parent interferon (3
molecule is occupied
by an amino acid residue that is exposed to the surface of the molecule,
preferably one that has
at least 25%, such as at least 50% of its side chain exposed to the surface.
For instance, the
amino acid residue is selected from the group consisting of F8, L9, R11, 512,
F15, Q16, Q18,
2s L20, W22, L28, L32, M36, P41, T58, Q64, N65, F67, I83, E85, N86, A89, N90,
Y92, H93,
H97, T100, L102, E103, L106, M117, L120, H121, 8124, 6127, 8128, L130, H131,
H140,
I145, R 147, V 148, E 149, R 152, Y 155, and F 156 of SEQ ID NO 2.
Additionally or alternatively, the substitution is preferably performed at a
position occupied by a threonine or serine residue. For instance, such
position is selected from
so the group consisting of S2, 512, 513, T58, S74, 575, S76, T77, T82, T100,
T112, 5118, 5119,
5139, T144, and T161, more preferably S2, 512, 513, 574, 575, 576, T77, T82,
T100, T112,
S 118, S 119, S 139, and T 144 (side chain surface exposed), still more
preferably S2, S 12, 575,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
23
576, T82, T100, 5119 and 5139 (at least 25% of its side chain exposed), and
even more
preferably 512, 575, T82 and T100 (at least 50% of its side chain exposed).
Of the above threonine or serine substitutions, serine substitutions are
preferred.
Accordingly, in even more preferred embodiments, the position is selected from
the group
s consisting of S2, S 12, S 13, S74, 575, S76, S 118, S 119 and S 139, more
preferably S2, S 12,
S 13, 574, 575, 576, S 118, S 119 and S 139, even more preferably S2, S 12,
S75, 576, S 119 and
S 139, and still more preferably S 12 and S75.
In one embodiment, only one cysteine residue is introduced into the interferon
~i
polypeptide in order to avoid formation of disulphide bridges between two or
more introduced
cysteine residues. In this connection C 17 present in wildtype human
interferon (3 may be
removed, preferably by substitution, in particular by substitution with S or
A. In another
embodiment, two or more cysteine residues are introduced, such as 2-6 or 2-4
cysteine
residues. Preferably, the interferon (3 polypeptide part of the conjugate
according to this
embodiment of the invention comprises the mutation L47C, Q48C, Q49C, D 11 OC,
F 111 C or
is R113C, in particular only one of these mutations, optionally in combination
with the mutation
C17S. Also, the interferon (3 polypeptide may comprise the substitution
C17S+N80C.
While the first non-polypeptide moiety according to this aspect of the
invention
may be any molecule which, when using the given conjugation method has
cysteine as an
attachment group (such as a carbohydrate moiety, a lipophilic group organ
organic derivatizing
2o agent), it is preferred that the non-polypeptide moiety is a polymer
molecule. The polymer
molecule may be any of the molecules mentioned in the section entitled
"Conjugation to a
polymer molecule", but is preferably selected from the group consisting of
linear or branched
polyethylene glycol or polyalkylene oxide. Most preferably, the polymer
molecule is VS-PEG.
The conjugation between the polypeptide and the polymer may be achieved in any
suitable
2s . manner, e.g. as described in the section entitled "Conjugation to a
polymer molecule", e.g. in
using a one step method or in the stepwise manner referred to in said section.
When the
interferon ~3 polypeptide comprises only one conjugatable cysteine residue,
this is preferably
conjugated to a first non-polypeptide moiety with a molecular weight of at
least 20kDa, either
directly conjugated or indirectly through a low molecular weight polymer (as
disclosed in WO
30 99/55377). When the conjugate comprises two or more first non-polypeptide
moieties,
normally each of these has a molecular weight of S or l OkDa.
Conjugate of the invention wherein the non polypeptide moiety binds to an acid
group
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
24
In a still further aspect the invention relates to a conjugate exhibiting
interferon ~i
activity and comprising at least one first non-polypeptide moiety having an
acid group as the
attachment group, which moiety is conjugated to at least one aspartic acid
residue or one
glutamic acid residue of an interferon (3 polypeptide, the amino acid sequence
of which differs
s from that of wildtype human interferon (3 in at least one introduced and/or
at least one removed
aspartic acid or glutamic acid residue, respectively. The relevant amino acid
residue may be
introduced in any position occupied by a surface exposed amino acid residue,
preferably by an
amino acid residue having more than 25% of its side chain surface exposed.
Preferably, at least
one amino acid residue occupying a position selected from the group consisting
of N4, L5, L6,
~ o F8, L9, Q 10, R 11, S 12, S 13, F 15, Q 16, Q 18, K19, L20, W22, Q23, L24,
N25, G26, R27, Y30,
M36, Q46, Q48, Q49, I66, F67, A68, I69, F70, R71, 575, T82, I83, L87, A89,
N90, V91, Y92,
H93, Q94, I95, N96, H97, K108, F111, L116, L120, K123, 8124, Y126, 6127, 8128,
L130,
H131, Y132, K134, A135, H140, T144, 8147, Y155, F156, N158, 8159, 6162, Y163
and
8165 has been substituted with an aspartic acid residue or a glutamic acid
residue.
~ s More preferably, the position is selected from the group consisting of N4,
L5, F8,
L9, R 11, S 12, F 15, Q 16, Q 18, K 19, W22, Q23, G26, R27, Y30, M36, Q46,
Q48, Q49, A68,
R71, 575, T82, A89, N90, Y92, H93, N96, H97, K108, F111, L116, L120, K123,
8124, 6127,
8128, L130, H131, K134, A135, H140, Y155, N158, 8159, 6162, Y163 and 8165,
such as
from the group consisting ofN4, L5, F8, 512, F15, Q16, K19, W22, Q23, R27,
Y30, M36,
2o Q46, Q48, Q49, R71, 575, T82, A89, Y92, H93, K108, F111, L116, K123, 8124,
6127, H131,
K134, A135, Y155 and 8165, still more preferably from the group consisting
ofN4, L5, F8,
512, F15, Q16, K19, W22, Q23, R27, Y30, Q46, Q48, Q49, 575, T82, A89, Y92,
H93, K108,
F 111, L 116, R 124, G 127, H 131, K 134, Y 1 S S and R 165, such as from the
group consisting of
L5, F8, S 12, F 1 S, Q 16, K 19, W22, Q23, Q48, Q49, Y92, H93, 8124, G 127, H
131 and Y 155,
2s even more preferably from the group consisting of 512, Q16, K19, Q23, Q48,
Q49, Y92, H93,
R 124, G 127, H 131 and Y 1 S5, such as from the group consisting of S 12, Q
16, K19, Q23, Q48,
Y92, H93, 8124, 6127, H 131 and Y 155, in particular from the group consisting
of S 12, Q 16,
K19, Q23, Q48, H93 and H 131, even more preferably from the group consisting
of S 12, Q 16,
K19, Q48, H93 and H131, and most preferably from the group consisting of Q16
and Q48.
3o Furthermore, in order to obtain a sufficient number of non-polypeptide
moieties it
is preferred that that least two aspartic acid residues or at least two
glutamic acid residues be
introduced, preferably in two positions selected from any of the above lists.
Also, it is preferred
that the conjugate according to this aspect comprises at least two first non-
polypeptide
moieties.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
In case of removal of an amino acid residue, the amino acid sequence of the
interferon (3 polypeptide differs from that of human wildtype interferon (3 in
at least one
removed aspartic acid or glutamic acid residue, such as 1-5 removed residues,
in particular
1-4 or 1-3 removed aspartic acid or glutamic acid residues. The residues) to
be removed,
s preferably by replacement, is selected from the group consisting of D34,
D39, D54, D73,
D110, E29, E42, E43, E53, E61, E81, E85, E103, E104, E107, E109, E137 and
E149. The
aspartic acid or glutamic acid residues) may be replaced with any other amino
acid residue,
but is preferably replaced by an arginine or a glutamine residue.first non-
polypeptide moiety
can be any non-polypeptide moiety with such property, it is presently
preferred that the non-
to polypeptide moiety is a polymer molecule or an organic derivatizing agent
having an acid
group as an attachment group, in particular a polymer molecule such as PEG,
and the conjugate
is prepared, e.g., as described by Sakane and Pardridge, Pharmceutical
Research, Vol. 14, No.
8, 1997, pp 1085-1091. Normally, for conjugation to an acid group the non-
polypeptide moiety
has a molecular weight of about 5 or 10 kDa.
Conjugate of the invention comprising a second non polypeptide moiety
In addition to a first non-polypeptide moiety (as described in the preceding
sections), the conjugate of the invention may comprise a second non-
polypeptide moiety of a
different type as compared to the first non-polypeptide moiety. Preferably, in
any of the above
2o described conjugates wherein the first non-polypeptide moiety is, e.g., a
polymer molecule
such as PEG, a second non- polypeptide moiety is a sugar moiety, in particular
an N-linked
sugar moiety. While the second non-polypeptide moiety may be attached to a
natural
glycosylation site of human interferon (3, e.g. the N-linked glycosylation
site defined by N80, it
is normally advantageous to introduce at least one additional glycosylation
site in the interferon
2s (3 polypeptide. Such site is e.g. any of those described in the immediately
preceding section
entitled "Conjugate of the invention wherein the non-polypeptide moiety is a
sugar moiety".
Furthermore, in case at least one additional glycosylation site is introduced
this may be
accompanied by removal of an existing glycosylation site as described below.
It will be understood that in order to obtain an optimal distribution of
attached
3o first and second non-polypeptide moieties, the interferon (3 polypeptide
may be modified in the
number and distribution of attachment groups for the first as well as the
second non-
polypeptide moiety so as to have e.g. at least one removed attachment group
for the first non-
polypeptide moiety and at least one introduced attachment group for the second
non-
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
26
polypeptide moiety or vice versa. For instance, the interferon (3 polypeptide
comprises at least
two (e.g. 2-5) removed attachment groups for the first non-polypeptide moiety
and at least one
(e.g. 1-5) introduced attachment groups for the second non-polypeptide moiety
or vice versa.
Of particular interest is a conjugate wherein the first non-polypeptide moiety
is a polymer
s molecule such as PEG having lysine as an attachment group, and the second
non-polypeptide
moiety is an N-linked sugar moiety.
More specifically, the conjugate of the invention may be one exhibiting
interferon
(3 activity and comprising at least one polymer molecule, preferably PEG, and
at least one
sugar moiety covalently attached to an interferon (3 polypeptide, the amino
acid sequence of
to which differs from that of wild-type human interferon ~i in -
a) at least one introduced and/or at least one removed amino acid residue
comprising an
attachment group for the polymer molecule; and
b) at least one introduced and/or at least one removed in vivo glycosylation
site, in particular an
N-glycosylation site,
t s provided that when the attachment group for the polymer molecule is a
cysteine residue, and
the sugar moiety is an N-linked sugar moiety, a cysteine residue is not
inserted in such a
manner that an N-glycosylation site is destroyed. WO 99/03887 suggests that a
cysteine residue
can be introduced into the natural N-glycosylation site of interferon ~3.
In a specific embodiment, the interferon (3 polypeptide comprises one of the
2o following sets of mutations:
K 19R+K45R+Q49N+Q$1 T+F 111 N+R 113T+K 1238;
K19R+K45R+Q49N+QS1T+F111N+R113T; or
K 19R+K45R+Q49N+QS 1 T+ K 1238.
25 Conjugate of the invention wherein the non polypeptide moiety is a sugar
moiety
When the conjugate of the invention comprises at least one sugar moiety
attached
to an in vivo glycosylation site, in particular an N-glycosylation site, this
is either the natural N-
glycosylation site of wild-type human interferon ~3 at position N80, i.e.
defined by amino acid
residues N80, E81, T82 and I83, or a new in vivo glycosylation site introduced
into the
3o interferon (3 polypeptide. The in vivo glycosylation site may be an O-
glycosylation site, but is
preferably an N-glycosylation site.
More specifically, in one aspect the invention relates to a conjugate
exhibiting
interferon (3 activity and comprising an interferon (3 polypeptide, the amino
acid sequence of
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
27
which differs from that of wild-type human interferon (3 in at least one
introduced
glycosylation site, the conjugate further comprising at least one un-PEGylated
sugar moiety
attached to an introduced glycosylation site.
In another aspect the invention relates to a conjugate exhibiting interferon
(3
s activity and comprising an interferon (3 polypeptide, the amino acid
sequence of which differs
from that of wild-type human interferon (3 in that a glycosylation site has
been introduced or
removed, provided that if only a glycosylation site is removed (and thus that
no glycosylation
site is introduced) the interferon (3 polypeptide does not comprise one or
more of the following
substutions: N80C, E81C or T82C. The latter substitution is suggested in WO
99/03887.
to For instance, an in vivo glycosylation site is introduced into a position
of the
parent interferon ~i molecule occupied by an amino acid residue exposed to the
surface of the
molecule, preferably with more than 25% of the side chain exposed to the
solvent, in particular
more than SO% exposed to the solvent (these positions are identified in the
Methods section
herein). The N-glycosylation site is introduced in such a way that the N-
residue of said site is
is located in said position. Analogously, an O-glycosylation site is
introduced so that the S or T
residue making up such site is located in said position. Furthermore, in order
to ensure efficient
glycosylation it is preferred that the in vivo glycosylation site, in
particular the N residue of the
N-glycosylation site or the S or T residue of the O-glycosylation site, is
located within the first
141 amino acid residues of the interferon (3 polypeptide, more preferably
within the first 116
20 - amino acid residues. Still more preferably, the in vivo glycosylation
site is introduced into a
position wherein only one mutation is required to create the site (i.e. where
any other amino
acid residues required for creating a functional glycosylation site is already
present in the
molecule).
Substitutions that lead to introduction of an additional N-glycosylation site
at
2s positions exposed at the surface of the interferon (3 molecule and occupied
by amino acid
residues having more than 25% of the side chain exposed to the surface
include:
S2N+N4S/T, L6S/T, LSN+G7S/T, F8N+Q10S/T, L9N+R11S/T, R11N, R11N+S13T,
S 12N+N 14S/T, F 1 SN+C 17S/T, Q 16N+Q 18S/T, Q 18N+L20S/T, K19N+L21 S/T,
W22N+L24S/T, Q23N+H25S/T, G26N+L28S/T, R27N+E29S/T, L28S+Y34S/T,
3o Y30N+L32S/T, L32N+D34S/T, K33N+R35S/T, R35N+N37S/T, M36N+F38S/T, D39S/T,
D39N+P41S/T, E42N+I44S/T, Q43N+K45S/T, K45N+L47S/T, Q46N+Q48S/T,
L47N+Q49T/S, Q48N+FSOS/T, Q49N+QS 1 S/T, QS 1N+E53S/T, K52N+D54S/T,
L57N+I59S/T, Q64N+I66S/T, A68N+F70S/T, R71N+D73S/T, Q72N, Q72N+S74T, D73N,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
28
D73N+S75T, S75N+T77S, S75N, S76N+G78S/T, E81N+I83S/T, T82N+V84S/T,
E85N+L87S/T, L88S/T, A89N+V91 S/T, Y92S/T, Y92N+Q94S/T, H93N+I95S/T, L98S/T,
H97N+K99S/T, K99N+V 1 O 1 S/T, T 1 OON+L 102S/T, E 103N+K1 OS S/T, E 104N+L
106S/T,
K 1 OSN+E 107 S/T, E 107N+E 109S/T, K 108N+D 11 OS/T, E 109N+F 111 S/T, D 11
ON+T 1125,
s D110N, F111N+R113S/T, R113N+K115S/T, G114N+L116S/T, K115N+M117S/T, L116N,
L116N+S118T, S119N+H212S/T, L120N+L122S/T, H121N+K123S/T, K123N+Y125S/T,
R124N+Y126S/T, G127N+I129S/T, R128N+L130S/T, L130N+Y132S/T, H131N+L133S/T,
K134N+K136S/T, A135N+E137S/T, K136N+Y138S/T, E137N, Y138N+H140S/T,
H140N+A142S/T, V148N+I150S/T, R152N+F154S/T, Y155N+I157S/T, L160S/T,
~o R159N+T161S, R159N, G162N+L164S/T, and Y163N+R165S/T.
Substitutions that lead to introduction of an additional N-glycosylation site
at
positions exposed at the surface of the interferon ~i molecule having more
than 50% of the side
chain exposed to the surface include:
L6S/T, LSN+G7S/T, F8N+Q10S/T, L9N+R11S/T, S12N+N14S/T, F15N+C17S/T,
is Q16N+Q18S/T, K19N+L21S/T, W22N+L24S/T, Q23N+H25S/T, G26N+L28S/T,
R27N+E29S/T, Y30N+L32S/T, K33N+R35S/T, R35N+N37S/T, M36N+F38S/T, D39S/T,
D39N+P41 S/T, E42N+I44S/T, Q46N+Q48S/T, Q48N+FSOS/T, Q49N+Q51 S/T,
QS 1N+E53S/T, K52N+D54S/T, L57N+I59S/T, R71N+D73S/T, D73N, D73N+S75T,
S75N+T77S, S75N, S76N+G78S/T, E81N+I83S/T, T82N+V84S/T, E85N+L87S/T,
2o A89N+V91S/T, Y92S/T, Y92N+Q94S/T, H93N+I95S/T, T100N+L102S/T,
E103N+K105S/T,
E 104N+L 106 S/T, E 107N+E 109S/T, K 108N+D 11 OS/T, D 11 ON+T 112 S, D 11 ON,
F111N+R113S/T, R113N+K115S/T, L116N, L116N+S118T, K123N+Y125S/T,
R124N+Y126S/T, G127N+I129S/T, H131N+L133S/T, K134N+K136S/T, A135N+E137S/T,
E 137N, V 148N+I1 SOS/T, and Y 1 SSN+I157S/T.
2s Among the substitutions mentioned in the above lists, those are preferred
that
have the N residue introduced among the 141 N-terminal amino acid residues, in
particular
among the 116 N-terminal amino acid residues.
Substitutions that lead to introduction of an N-glycosylation site by only one
amino acid substitution include: L6S/T, R11N, D39S/T, Q72N, D73N, S75N,
L88S/T,
3o Y92S/T, L98S/T, D110N, L116N, E137N, R159N and L160S/T. Among these, a
substitution
is preferred that is selected from the group consisting of L6S/T, R11N,
D39S/T, Q72N, D73N,
S75N, L88S/T, Y92S/T, L98S/T, D110N and L116N, more preferably from the group
consisting of L6S/T, D39S/T, D73N, S75N, L88S/T, D110N, L116N and E137N; and
most
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
29
preferably selected from the group consisting of L6S/T, D39S/T, D73N, S75N,
L88S/T,
D 11 ON and L 116N.
The presently most preferred interferon ~i polypeptide according to this
aspect
include at least one of the following substitutions: ..
s S2N+N4T/S, L9N+R 11 T/S, R 11 N, S 12N+N 14T/S, F 1 SN+C 17 S/T, Q 16N+Q
18T/S,
K19N+L21T/S, Q23N+H25T/S, G26N+L28T/S, R27N+E29T/S, L28N+Y30T/S, D39T/S,
K45N+L47T/S, Q46N+Q48T/S, Q48N+FSOT/S, Q49N+QS 1 T/S, QS 1N+E53T/S,
R71N+D73T/S, Q72N, D73N, S75N, S76N+G78T/S, L88T/S, Y92T/S, N93N+I95T/S,
L98T/S, E 103N+K 1 OST/S, E 104N+L 106T/S, E 107N+E 109T/S, K108N+D 1 l OT/S,
D 11 ON,
~o F111N+R113T/S, or L116N, more preferably at least one of the following
substitutions:
S2N+N4T, L9N+R11T, 49N+QS1T or F111N+R113T or R71N+D73T, in particular
49N+Q$1T or F111N+R113T or R71N+D73T. For instance, the interferon (3
polypeptide
comprises one of the following sets of substitutions
Q49N+QS 1 T+F 111 N+R 113T ;
~s Q49N+QS1T+R71N+D73T+F111N+R113T;
S2N+N4T+ Fl 11N+R113T ;
S2N+N4T+Q49N+QS 1 T ;
S2N+N4T+Q49N+QS 1 T+F 111 N+R 113T ;
S2N+N4T+L9N+R 11 T+Q49N+QS 1 T ;
2o S2N+N4T+L9N+R11T+F111N+R113T ;
S2N+N4T+L9N+R 11 T+Q49N+QS 1 T+F 111 N+R 113T ;
L9N+R 11 T+Q49N+Q$1 T;
L9N+R 11 T+Q49N+QS 1 T+F 111 N+R 113 T ; or
L9N+R 11 T+F 111 N+R 113T
2s It will be understood that in order to introduce a functional in vivo
glycosylation
site the amino acid residue in between the N-residue and the S/T residue is
different from
proline. Normally, the amino acid residue in between will be that occupying
the relevant
position in the amino acid sequence shown in SEQ ID NO 2. For instance, in the
polypeptide
comprising the substitutions Q49N+Q$1 S, position 50 is the position in
between.
3o The interferon ~i polypeptide part of a conjugate of the invention may
contain a
single in vivo glycosylation site. However, in order to obtain efficient
shielding of epitopes
present on the surface of the parent polypeptide it is often desirable that
the polypeptide
comprises more than one in vivo glycosylation site, in particular 2-7 in vivo
glycosylation sites,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
such as 2, 3, 4, S, 6 or 7 in vivo glycosylation sites. Thus, the interferon
(3 polypeptide may
comprise one additional glycosylation site, or may comprise two, three, four,
five, six, seven or
more introduced in vivo glycosylation sites, preferably introduced by one or
more substitutions
described in any of the above lists.
As indicated above, in addition to one or more introduced glycosylation sites,
existing glycosylation sites may have been removed from the interferon (3
polypeptide. For
instance, any of the above listed substitutions to introduce a glycosylation
site may be
combined with a substitution to remove the natural N-glycosylation site of
human wild-type
interferon (3. For instance, the interferon ~3 polypeptide may comprise a
substitution of N80,
to e.g. one of the substitutions N80K/C/D/E, when a first non-polypeptide
polypeptide is one
having one of K, C, D, E as an attachment group. For instance, the interferon
(3 polypeptide
may comprise at least one of the following substitutions: S2N+N4T/S,
L9N+R11T/S, R11N,
S 12N+N 14T/S, F 1 SN+C 17 S/T, Q 16N+Q 18T/S, K 19N+L21 T/S, Q23N+H25T/S,
G26N+L28T/S, R27N+E29T/S, L28N+Y30T/S, D39T/S, K45N+L47T/S, Q46N+Q48T/S,
~s Q48N+FSOT/S, Q49N+QS1T/S, QS1N+E53T/S, R71N+D73T/S, Q72N, D73N, S75N,
S76N+G78T/S, L88T/S, Y92T/S, N93N+I95T/S, L98T/S, E103N+K105T/S,
E 104N+L 106T/S, E 107N+E 109T/S, K 108N+D 11 OT/S, D 11 ON, F 111 N+R 113T/S,
or L 116N
in combination with N80K/C/D/E. More specifically, the interferon (3
polypeptide may
comprise the substitution: Q49N+QS1T or Fl 11N+R113T or R71N+D73T, in
particular
20. Q49N+QS1T+F111N+R113T or Q49N+QS1T+R71N+D73T+ F111N+ R113T, in combination
with N80K/C/D/E.
Any of the glycosylated variants disclosed in the present section having
introduced and/or removed at least one glycosylation site, such as the variant
comprising the
substitutions Q48N+FSOT/S, Q48N+FSOT/S+F111N+R113T/S, Q49N+QS1T/S,
zs F111N+R113T/S, or Q49N+QS1T/S+F111N+R113T/S, may further be conjugated to a
polymer molecule, such as PEG, or any other non-polypeptide moiety. For this
purpose the
conjugation may be achieved by use of attachment groups already present in the
interferon (3
polypeptide or attachment groups may have been introduced and/or removed, in
particular such
that a total of 1-6, in particular 3-4 or 1, 2, 3, 4, 5, or 6 attachment
groups are available for
3o conjugation. Preferably, in a conjugate of the invention wherein the
interferon (3 polypeptide
comprises two glycosylation sites, the number and molecular weight of the non-
polypeptide
moiety is chosen so as that the total molecular weight added by the non-
polypeptide moiety is
in the range of 20-40 kDa, in particular about 20 kDa or 30 kDa.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
31
In particular, the glycosylated variant may be conjugated to a non-polypeptide
moiety via a lysine attachment group, and one or more lysine residues of the
parent polypeptide
may have been removed, e.g. by any of the substitutions mentioned in the
section entitled
"Conjugate of the invention, wherein the non-polypeptide moiety is a molecule
which has
s lysine as an attachment group", in particular the substitutions
K19R+K45R+K123R.
Alternatively or additionally, a lysine residue may have been introduced, e.g.
by any of the
substitutions mentioned in said section, in particular the substitution R71K.
Accordingly, one
specific conjugate of the invention is one, which comprises a glycosylated
interferon ~3
polypeptide comprising the mutations Q49N + QS 1T + F 111N + R113T + K19R +
K45R +
~o K123R or Q49N + QS 1T + F 111N + R113T + K19R + K45R + K123R + R71K further
conjugated to PEG. The glycosylated polypeptide part of said conjugate is
favourably produced
in CHO cells and PEGylated subsequent to purification using e.g. SS-PEG, NPC-
PEG,
aldehyd-PEG, mPEG-SPA, mPEG-SCM, mPEG-BTC from Shearwater Polymers, Inc, SC-
PEG from Enzon, Inc., tresylated mPEG as described in US 5,880,255, or
oxycarbonyl-oxy-N-
ts dicarboxyimide-PEG (US 5,122,614).
Alternatively, to PEGylation via a lysine group, the glycosylated conjugate
according to this embodiment may be PEGylated via a cysteine group as
described in the
section entitled "Conjugate of the invention, wherein the non-polypeptide
moiety is a molecule
that has cysteine as an attachment group" (for this purpose the interferon ~3
polypeptide may,
2o e.g. comprising at least one of the mutations N80C, R71C and C17S), via an
acid group as
described in the section entitled "Conjugation of the invention wherein the
non-polypeptide
moiety binds to an acid group", or via any other suitable group.
Other conjugates of the invention
2s In addition to the introduction and/or removal of amino acid residues
comprising
an attachment group for the non-polypeptide moiety of choice (as described in
any of the
sections above entitled "Conjugate of the invention ....") the interferon. ~i
polypeptide part of
the conjugate may contain further substitutions. A preferred example is a
substitution of any of
the residues, M1, C17, N80 or V101, e.g. one or more of the following
substitutions: C17S;
3o N80K/C/D/E; V 1 O 1 Y/W/F/,H; a deletion of M 1; or M 1 K. The substitution
M 1 K is of
particular interest when the interferon (3 polypeptide is expressed with a
tag, e.g. a His-l4tag,
where such tag is to be removed by DAP (diaminopeptidase) subsequent to
purification and/or
conjugation.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
32
Non-polypeptide moiety of the conjugate of the invention
As indicated further above the non-polypeptide moiety of the conjugate of the
invention is preferably selected from the group consisting of a polymer
molecule, a lipophilic
s compound, a sugar moiety (by way of in vivo glycosylation) and an organic
derivatizing agent.
All of these agents may confer desirable properties to the polypeptide part of
the conjugate, in
particular reduced immunogenicity and/or increased functional in vivo half
life and/or
increased serum half life. The polypeptide part of the conjugate may be
conjugated to only one
type of non-polypeptide moiety, but may also be conjugated to two or more
different types of
to non-polypeptide moieties, e.g. to a polymer molecule and a sugar moiety, to
a lipophilic group
and a sugar moiety, to an organic derivating agent and a sugar moiety, to a
lipophilic group and
a polymer molecule, etc. The conjugation to two or more different non-
polypeptide moieties
may be done simultaneous or sequentially. The choice of non-polypeptide
moiety/ies, e.g.
depends on the effect desired to be achieved by the conjugation. For instance,
sugar moieties
~ s have been found particularly useful for reducing immunogenicity, whereas
polymer molecules
such as PEG are of particular use for increasing functional in vivo half life
and/or serum half
life. Using a polymer molecule as a first non-polypeptide moiety and a sugar
moiety as a
second non-polypeptide moiey may result in reduced immunogenicity and
increased functional
in vivo or serum half life.
Methods of preparing a conjugate of the invention
In the following sections "Conjugation to a lipophilic compound", "Conjugation
to a polymer molecule", "Conjugation to a sugar moiety" and "Conjugation to an
organic
derivatizing agent" conjugation to specific types of non-polypeptide moieties
is described.
2s
Conjugation to a lipophilic compound
For conjugation to a lipophilic compound the following polypeptide groups may
function as attachment groups: the N-terminal or C-terminal of the
polypeptide, the hydroxy
groups of the amino acid residues Ser, Thr or Tyr, the s-amino group of Lys,
the SH group of
3o Cys or the carboxyl group of Asp and Glu. The polypeptide and the
lipophilic compound may
be conjugated to each other, either directly or by use of a linker. The
lipophilic compound may
be a natural compound such as a saturated or unsaturated fatty acid, a fatty
acid diketone, a
terpene, a prostaglandin, a vitamine, a carotenoide or steroide, or a
synthetic compound such as
a carbon acid, an alcohol, an amine and sulphonic acid with one or more alkyl-
, aryl-, alkenyl-
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
33
or other multiple unsaturated compounds. The conjugation between the
polypeptide and the
lipophilic compound, optionally through a linker may be done according to
methods known in
the art, e.g. as described by Bodanszky in Peptide Synthesis, John Wiley, New
York, 1976 and
in WO 96/12505.
Conjugation to a polymer molecule
The polymer molecule to be coupled to the polypeptide may be any suitable
polymer molecule, such as a natural or synthetic homo-polymer or
heteropolymer, typically with a
molecular weight in the range of 300-100,000 Da, such as 300-20,000 Da, more
preferably in the
to range of 500-10,000 Da, even more preferably in the range of 500-5000 Da.
Examples of homo-polymers include a polyol (i.e. poly-OH), a polyamine (i.e.
poly-
NH2) and a polycarboxylic acid (i.e. poly-COOH). A hetero-polymer is a
polymer, which
comprises one or more different coupling groups, such as, e.g., a hydroxyl
group and an amine
group.
t s Examples of suitable polymer molecules include polymer molecules selected
from
the group consisting of polyalkylene oxide (PAO), including polyalkylene
glycol (PAG), such as
polyethylene glycol (PEG) and polypropylene glycol (PPG), branched PEGs, poly-
vinyl alcohol
(PVA), poly-carboxylate, poly-(vinylpyrolidone), polyethylene-co-malefic acid
anhydride,
polystyrene-co-malic acid anhydride, dextran including carboxymethyl-dextran,
or any other
2o biopolymer suitable for reducing immunogenicity and/or increasing
functional in vivo half life
and/or serum half life. Another example of a polymer molecule is human albumin
or another
abundant plasma protein. Generally, polyalkylene glycol-derived polymers are
biocompatible,
non-toxic, non-antigenic, non-immunogenic, have various water solubility
properties, and are
easily excreted from living organisms.
2s PEG is the preferred polymer molecule to be used, since it has only few
reactive
groups capable of cross-linking compared, e.g., to polysaccharides such as
dextran, and the like.
In particular, monofunctional PEG, e.g monomethoxypolyethylene glycol (mPEG),
is of interest
since its coupling chemistry is relatively simple (only one reactive group~is
available for conju-
gating with attachment groups on the polypeptide). Consequently, the risk of
cross-linking is
3o eliminated, the resulting polypeptide conjugates are more homogeneous and
the reaction of the
polymer molecules with the polypeptide is easier to control.
To effect covalent attachment of the polymer molecules) to the polypeptide,
the
hydroxyl end groups of the polymer molecule must be provided in activated
form, i.e. with
reactive functional groups (examples of which include primary amino groups,
hydrazide (HZ),
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
34
thiol, succinate (SUC), succinimidyl succinate (SS), succinimidyl succinamide
(SSA),
succinimidyl proprionate (SPA), succinimidy carboxymethylate (SCM),
benzotriazole
carbonate (BTC), N-hydroxysuccinimide (NHS), aldehyde, nitrophenylcarbonate
(NPC), and
tresylate (TRES)). Suitably activated polymer molecules are commercially
available, e.g. from
s Shearwater Polymers, Inc., Huntsville, AL, USA. Alternatively, the polymer
molecules can be
activated by conventional methods known in the art, e.g. as disclosed in WO
90/13540.
Specific examples of activated linear or branched polymer molecules for use in
the present
invention are described in the Shearwater Polymers, Inc. 1997 and 2000
Catalogs
(Functionalized Biocompatible Polymers for Research and pharmaceuticals,
Polyethylene
Glycol and Derivatives, incorporated herein by reference). Specific examples
of activated PEG
polymers include the following linear PEGs: NHS-PEG (e.g. SPA-PEG, SSPA-PEG,
SBA-
PEG, SS-PEG, SSA-PEG, SC-PEG, SG-PEG, and SCM-PEG), and NOR-PEG), BTC-PEG,
EPOX-PEG, NCO-PEG, NPC-PEG, CDI-PEG, ALD-PEG, TRES-PEG, VS-PEG, IODO-PEG,
and MAL-PEG, and branched PEGs such as PEG2-NHS and those disclosed in US
5,932,462 and
is US 5,643,575, both of which references are incorporated herein by
reference. Furthermore, the
following publications, incorporated herein by reference, disclose useful
polymer molecules
and/or PEGylation chemistries: US 5,824,778, US 5,476,653, WO 97/32607, EP
229,108, EP
402,378, US 4,902,502, US 5,281,698, US 5,122,614, US 5,219,564, WO 92/16555,
WO
94/04193, WO 94/14758, WO 94/17039, WO 94/18247, WO 94/28024, WO 95/00162, WO
zo 95/11924, W095/13090, WO 95/33490, WO 96/00080, WO 97/18832, WO 98/41562,
WO
98/48837, WO 99/32134, WO 99/32139, WO 99/32140, WO 96/40791, WO 98/32466, WO
95/06058, EP 439 508, WO 97/03106, WO 96/21469, WO 95/13312, EP 921 131, US
5,736,625, WO 98/05363, EP 809 996, US 5,629,384, WO 96/41813, WO 96/07670, US
5,473,034, US 5,516,673, EP 605 963, US 5,382,657, EP 510 356, EP 400 472, EP
183 503
2s and EP 154 316.
The conjugation of the polypeptide and the activated polymer molecules is
conducted by use of any conventional method, e.g. as described in the
following references
(which also describe suitable methods for activation of polymer molecules):
Harris and Zalipsky,
eds., Polyethylene glycol) Chemistry and Biological Applications, AZC,
Washington; R.F.
so Taylor, ( 1991 ), "Protein immobilisation. Fundamental and applications",
Marcel Dekker, N.Y.;
S.S. Wong, (1992), "Chemistry of Protein Conjugation and Crosslinking'.', CRC
Press, Boca
Raton; G.T. Hermanson et al., (1993), "Immobilized Affinity Ligand
Techniques", Academic
Press, N.Y.). The skilled person will be aware that the activation method
and/or conjugation
chemistry to be used depends on the attachment groups) of the interferon (3
polypeptide as well as
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
the functional groups of the polymer (e.g. being amino, hydroxyl, carboxyl,
aldehyde or sulfy-
dryl). The PEGylation may be directed towards conjugation to all available
attachment groups
on the polypeptide (i.e. such attachment groups that are exposed at the
surface of the
polypeptide) or may be directed towards specific attachment groups, e.g. the N-
terminal amino
s group (US 5,985,265). Furthermore, the conjugation may be achieved in one
step or in a
stepwise manner (e.g. as described in WO 99/55377).
It will be understood that the PEGylation is designed so as to produce the
optimal
molecule with respect to the number of PEG molecules attached, the size and
form (e.g.
whether they are linear or branched) of such molecules, and where in the
polypeptide such
i o molecules are attached. For instance, the molecular weight of the polymer
to be used may be
chosen on the basis of the desired effect to be achieved. For instance, if the
primary purpose of the
conjugation is to achieve a conjugate having a high molecular weight (e.g. to
reduce renal
clearance) it is usually desirable to conjugate as few high Mw polymer
molecules as possible to
obtain the desired molecular weight. When a high degree of epitope shielding
is desirable this may
t s be obtained by use of a sufficiently high number of low molecular weight
polymer (e.g. with a
molecular weight of about 5,000 Da) to effectively shield all or most epitopes
of the polypeptide.
For instance, 2-8, such as 3-6 such polymers may be used.
In connection with conjugation to only a single attachment group on the
protein
(as described in US 5,985,265), it may be advantageous that the polymer
molecule, which may
2o be linear or branched, has a high molecular weight, e.g. about 20 kDa.
Normally, the polymer conjugation is performed under conditions aiming at
reacting
all available polymer attachment groups with polymer molecules. Typically, the
molar ratio of
activated polymer molecules to polypeptide is 1000-1, in particular 200-1,
preferably 100-1, such
as 10-1 or 5-1 in order to obtain optimal reaction. However, also equimolar
ratios may be used.
2s It is also contemplated according to the invention to couple the polymer
molecules
to the polypeptide through a linker. Suitable linkers are well known to the
skilled person. A
preferred example is cyanuric chloride (Abuchowski et al., (1977), J. Biol.
Chem., 252,
3578-3581; US 4,179,337; Shafer et al., (1986), J. Polym. Sci. Polym. Chem.
Ed., 24, 375-378.
Subsequent to the conjugation residual activated polymer molecules are blocked
so according to methods known in the art, e.g. by addition of primary amine to
the reaction
mixture, and the resulting inactivated polymer molecules removed by a suitable
method.
Covalent in vitro coupling of a carbohydrate moiety to amino acid residues of
interferon (3 may be used to modify or increase the number or profile of
carbohydrate
substituents. Depending on the coupling mode used, the carbohydrates) may be
attached to a)
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
36
arginine and histidine (Lundblad and Noyes, Chemical Reagents for Protein
Modification,
CRC Press Inc. Boca Raton, FI), b) free carboxyl groups (e.g. of the C-
terminal amino acid
residue, asparagine or glutamine), c) free sulfhydryl groups such as that of
cysteine, d) free
hydroxyl groups such as those of serine, threonine, tyrosine or
hydroxyproline, e) aromatic
s residues such as those of phenylalanine or tryptophan or fj the amide group
of glutamine.
These amino acid residues constitute examples of attachment groups for a
carbohydrate
moiety, which may be introduced and/or removed in the interferon ~3
polypeptide. Suitable
methods of in vitro coupling are described in WO 87/05330 and in Aplin etl
al., CRC Crit
Rev. Biochem., pp. 259-306, 1981. The in vitro coupling of carbohydrate
moieties or PEG
t o to protein- and peptide-bound Gln-residues can also be carried out by~
transglutaminases
(TGases), e.g. as described by Sato et al., 1996 Biochemistry 35, 13072-13080
or in EP
725145
Coupling to a sugar moiety
is In order to achieve in vivo glycosylation of an interferon (3
polypeptidethat has
been modified by introduction of one or more glycosylation sites (see the
section "Conjugates
of the invention wherein the non-polypeptide moiety is a sugar moiety"), the
nucleotide
sequence encoding the polypeptide part of the conjugate must be inserted in a
glycosylating,
eucaryotic expression host. The expression host cell may be selected from
fungal (filamentous
2o fungal or yeast), insect, mammalian animal cells, from transgenic plant
cells or from
transgenic animals. Furthermore, the glycosylation may be achieved in the
human body when
using a nucleotide sequence encoding the polypeptide part of a conjugate of
the invention or a
polypeptide of the invention in gene therapy. In one embodiment the host cell
is a mammalian
cell, such as an CHO cell, BHK or HEK cell, e.g. HEK293, or an insect cell,
such as an SF9
2s cell, or a yeast cell, e.g. Saccharomyces cerevisiae, Pichia pastoris or
any other suitable
glycosylating host, e.g. as described further below. Optionally, sugar
moieties attached to the
interferon ~i polypeptide by in vivo glycosylation are further modified by use
of
glycosyltransferases, e.g. using the glycoAdvanceTM technology marketed by
Neose, Horsham,
PA, USA. Thereby, it is possible to, e.g., increase the sialyation of the
glycosylated interferon
30 (3 polypeptide following expression and in vivo glycosylation by CHO cells.
Coupling to an organic derivatizing agent
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
37
Covalent modification of the interferon (3 polypeptide may be performed by
reacting (an) attachment groups) of the polypeptide with an organic
derivatizing agent.
Suitable derivatizing agents and methods are well known in the art. For
example, cysteinyl
residues most commonly are reacted with a-haloacetates (and corresponding
amines), such as
s chloroacetic acid or chloroacetamide, to give carboxymethyl or
carboxyamidomethyl
derivatives. Cysteinyl residues also are derivatized by reaction with
bromotrifluoroacetone, a-
bromo-(3-(4-imidozoyl)propionic acid, chloroacetyl phosphate, N-
alkylmaleimides, 3-vitro-2-
pyridyl disulfide, methyl 2-pyridyl disulfide, p-chloromercuribenzoate, 2-
chloromercuri-4-
nitrophenol, or chloro-7-nitrobenzo-2-oxa-1,3-diazole. Histidyl residues are
derivatized by
reaction with diethylpyrocarbonateat pH 5.5-7.0 because this agent is
relatively specific for the
histidyl side chain. Para-bromophenacyl bromide also is usefixl; the reaction
is preferably
performed in 0.1 M sodium cacodylate at pH 6.O.Lysinyl and amino terminal
residues are
reacted with succinic or other carboxylic acid anhydrides. Derivatization with
these agents has
the effect of reversing the charge of the lysinyl residues. Other suitable
reagents for
t s - derivatizing a-amino-containing residues include imidoesters such as
methyl picolinimidate;
pyridoxal phosphate; pyridoxal; chloroborohydride; trinitrobenzenesulfonic
acid; O-
methylisourea; 2,4-pentanedione; and transaminase-catalyzed reaction with
glyoxylate. Arginyl
residues are modified by reaction with one or several conventional reagents,
among them
phenylglyoxal, 2,3-butanedione, 1,2-cyclohexanedione, and ninhydrin.
Derivatization of
2o arginine residues requires that the reaction be performed in alkaline
conditions because of the
high pKa of the guanidine functional group. Furthermore, these reagents may
react with the
groups of lysine as well as the arginine guanidino group. Carboxyl side groups
(aspartyl or
glutamyl or C-terminal amino acid residue) are selectively modified by
reaction with
carbodiimides (R-N=C=N-R'), where R and R' are different alkyl groups, such as
1-
2s cyclohexyl-3-(2-morpholinyl-4-ethyl) carbodiimide or 1-ethyl-3-(4-azonia-
4,4-dimethylpentyl)
carbodiimide. Furthermore, aspartyl and glutamyl residues are converted to
asparaginyl and
glutaminyl residues by reaction with ammonium ions.
Blocking of functional site
so It has been reported that excessive polymer conjugation can lead to a loss
of
activity of the interferon (3 polypeptide to which the polymer is conjugated.
This problem can
be eliminated, e.g., by removal of attachment groups located at the functional
site or by
blocking the fiznctional site prior to conjugation. These latter strategies
constitute further
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
3~
embodiments of the invention (the first strategy being exemplified further
above, e.g. by
removal of lysine residues which may be located close to a functional site).
More specifically,
according to the second strategy the conjugation between the interferon (3
polypeptide and the
non-polypeptide moiety is conducted under conditions where the functional site
of the
s polypeptide is blocked by a helper molecule capable of binding to the
functional site of the
polypeptide. Preferably, the helper molecule is one, which specifically
recognizes a functional
site of the polypeptide, such as a receptor, in particular the type I
interferon receptor.
Alternatively, the helper molecule may be an antibody, in particular a
monoclonal antibody
recognizing the interferon (3 polypeptide. In particular, the helper molecule
may be a
t o neutralizing monoclonal antibody.
The polypeptide is allowed to interact with the helper molecule before
effecting
conjugation. This ensures that the fiznctional site of the polypeptide is
shielded or protected and
consequently unavailable for derivatization by the non-polypeptide moiety
such, as a polymer.
Following its elution from the helper molecule, the conjugate between the non-
polypeptide
t s moiety and the polypeptide can be recovered with at least a partially
preserved functional site.
The subsequent conjugation of the polypeptide having a blocked functional site
to
a polymer, a lipophilic compound, an organic derivatizing agent or any other
compound is
conducted in the normal way, e.g. as described in the sections above entitled
"Conjugation to
2o Irrespectively of the nature of the helper molecule to be used to shield
the
functional site of the polypeptide from conjugation, it is desirable that the
helper molecule is
free from or comprises only a few attachment groups for the non-polypeptide
moiety of choice
in parts) of the molecule, where the conjugation to such groups will hamper
the desorption of
the conjugated polypeptide from the helper molecule. Hereby, selective
conjugation to
2s attachment groups present in non-shielded parts of the polypeptide can be
obtained and it is
possible to reuse the helper molecule for repeated cycles of conjugation. For
instance, if the
non-polypeptide moiety is a polymer molecule such as PEG, which has the
epsilon amino
group of a lysine or N-terminal amino acid residue as an attachment group, it
is desirable that
the helper molecule is substantially free from conjugatable epsilon amino
groups, preferably
3o free from any epsilon amino groups. Accordingly, in a preferred embodiment
the helper
molecule is a protein or peptide capable of binding to the functional site of
the polypeptide,
which protein or peptide is free from any conjugatable attachment groups for
the non-
polypeptide moiety of choice.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
39
In a further embodiment the helper molecule is first covalently linked to a
solid
phase such as column packing materials, for instance Sephadex or agarose
beads, or a surface,
e.g. reaction vessel. Subsequently, the polypeptide is loaded onto the column
material carrying
the helper molecule and conjugation carried out according to methods known in
the art, e.g. as
s described in the sections above entitled "Conjugation to ....". This
procedure allows the
polypeptide conjugate to be separated from the helper molecule by elution. The
polypeptide
conjugate is eluated by conventional techniques under physico-chemical
conditions that do not
lead to a substantive degradation of the polypeptide conjugate. The fluid
phase containing the
polypeptide conjugate is separated from the solid phase to which the helper
molecule remains
t o covalently linked. The separation can be achieved in other ways: For
instance, the helper
molecule may be derivatised with a second molecule (e.g. biotin) that can be
recognized by a
specific binder (e.g. streptavidin). The specific binder may be linked to a
solid phase thereby
allowing the separation of the polypeptide conjugate from the helper molecule-
second
molecule complex through passage over a second helper-solid phase column which
will retain,
~ s upon subsequent elution, the helper molecule-second molecule complex, but
not the
polypeptide conjugate. The polypeptide conjugate may be released from the
helper molecule in
any appropriate fashion. De-protection may be achieved by providing conditions
in which the
helper molecule dissociates from the functional site of the interferon ~i to
which it is bound. For
instance, a complex between an antibody to which a polymer is conjugated and
an anti-
2o idiotypic antibody can be dissociated by adjusting the pH to an acid or
alkaline pH.
Conjugation of a tagged interferon ~i polypeptide
In an alternative embodiment the interferon ~i polypeptide is expressed, as a
fusion protein, with a tag, i.e. an amino acid sequence or peptide stretch
made up of typically
2s 1-30, such as 1-20 or I-15 or 1-10 amino acid residues. Besides allowing
for fast and easy
purification, the tag is a convenient tool for achieving conjugation between
the tagged
polypeptideand the non-polypeptide moiety. In particular, the tag may be used
for achieving
conjugation in microtiter plates or other carriers, such as paramagnetic
beads, to which the
tagged polypeptide can be immobilised via the tag. The conjugation to the
tagged polypeptide
in, e.g., microtiter plates has the advantage that the tagged polypeptide can
be immobilised in
the microtiter plates directly from the culture broth (in principle without
any purification) and
subjected to conjugation. Thereby, the total number of process steps (from
expression to
conjugation) can be reduced. Furthermore, the tag may function as a spacer
molecule ensuring
an improved accessibility to the immobilised polypeptide to be conjugated. The
conjugation
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
using a tagged polypeptide may be to any of the non-polypeptide moieties
disclosed herein,
e.g. to a polymer molecule such as PEG.
The identity of the specific tag to be used is not critical as long as the tag
is
capable of being expressed with the polypeptide and is capable of being
immobilised on a
s suitable surface or Garner material. A number of suitable tags are
commercially available, e.g.
from Unizyme Laboratories, Denmark. For instance, the tag may be any of the
following
sequences:
His-His-His-His-His-His
Met-Lys-His-His-His-His-His-His
Met-Lys-His-His-Ala-His-His-Gln-His-His
Met-Lys-His-Gln-His-Gln-His-Gln-His-Gln-His-Gln-His-Gln .
(vectors useful for providing such tags are available from Unizyme
Laboratories, Denmark)
or any of the following:
EQKLI SEEDL (a C-terminal tag described in Mol. Cell. Biol. 5:3610-16, 1985)
i s DYKDDDDK (a C- or N-terminal tag)
YPYDVPDYA
Antibodies against the above tags are commercially available, e.g. from ADI,
Aves Lab and
Research Diagnostics.
A convenient method for using a tagged polypeptide for PEGylation is given in
2o the Materials and Methods section below.
The subsequent cleavage of the tag from the polypeptide may be achieved by use
of commercially available enzymes.
Polypeptides of the Invention .
2s In further aspects the invention relates to generally novel interferon (3
polypeptides described herein that, as compared to human wildtype interferon
~3 has at least one
introduced and/or at least one removed attachment group for a non-polypeptide
moiety . The
novel polypeptides are important intermediate compounds for the preparation of
a conjugate of
the invention. In addition, the polypeptides themselves may have interesting
properties.
30 . Examples of such polypeptides include those that comprises an amino acid
sequence which differs from that of wild-type human interferon [3 in that at
least one amino
acid residue selected from the group consisting of N4, F8, L9, Q10, R11, 513,
L24, N25, G26,
L28, E29, N37, F38, Q48, Q49, Q64, N65, I66, F67, A68, I69, F70, R71, Q72,
D73, 574, 575,
S76, T77, G78, W79, N80, E81, T82, I83, V84, L87, L88, A89, N90, V91, Y92,
H93, Q94,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
41
D110, F111, T112, 8113, 8128, H140, T144, I145, 8147, V148, L151, 8152, F154,
Y155,
N 158 and N 166 is replaced with a different amino acid residue selected from
the group
consisting of K, R, D, E, C and N. The amino acid residues specified above are
located in
positions, which are exposed at the surface of human interferon (3 molecule as
demonstrated by
s . the solved 3D structure of human interferon (3. By replacing one or more
of these residue with
either of K, R, D, E, C and N attachment groups) for a non-polypeptide moiety,
in particular a
polymer attachment group or an amino acid residue susceptible to modification
by a
carbohydrate moiety, is/are introduced into human interferon (3. The resulting
modified human
interferon (3 molecule is a suitable starting compound for the preparation of
an interferon (3
to conjugate having improved properties as compared to the unmodified human
interferon ~i
molecule.
In a further aspect the invention relates to an interferon (3 polypeptide
comprising
an amino acid sequence which differs from that of wild-type human interferon
~i in that at least
one amino acid residue selected from the group consisting of N4, F8, L9, Q 10,
R 11, S 12, S 13,
is L24, N25, G26, L28, E29, N37, F38, D39, Q48, Q49, Q64, N65, I66, F67, A68,
I69, F70, R71,
Q72, D73, S74, 575, 576, T77, G78, W79, N80, E81, T82, I83, V84, E85, L87,
L88, A89,
N90, V91, Y92, H93, Q94, D 110, F 111, T 112, 8113, 8128, H 140, T 144, I 145,
8147, V 148,
L 151, R 152, F 154, Y 155, N 1 S 8, G 162, and N 166 is replaced with a
lysine residue, provided
that the polypeptide is different from the one having the amino acid sequence
of wild-type
2o human interferon (3 with the following substitutions: D54N+E85K+V91I+V101M
and different
from one which is a hybrid molecule between interferon (3 and interferon a
which as a
consequence of being a hybrid has a lysine in position 39. The first of the
disclaimed
polypeptides is disclosed by Stewart et al, DNA Vol 6 not 1987 p119-128 and
was found to be
inactive, the second is disclosed in US 4,769,233 and was constructed with the
purpose of
2s improving the biological activity of interferon Vii. None of the disclaimed
polypeptides were
made for or described as being suitable intermediates for the preparation of
interferon (3
conjugates with reduced immunogenicity and/or prolonged functional in vivo
half life and/or
serum half life.
A still further example includes an interferon (3 polypeptide comprising an
amino acid
3o sequence which differs from that of SEQ ID NO 2 in one or more
substitutions selected from
the group consisting ofN4K, F15K, Q16K, R27K, R35K, D39K, Q49K, E85K, A89K,
E103K,
E109K, R124K, E137K and R159K, provided that when the substitution is R27K the
polypeptide is different from the one having the amino acid sequence of wild-
type human
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
42
interferon (3 with the following substitutions: R27K+E43K. The disclaimed
polypeptide is
disclosed by Stewart et al, DNA Vol 6 not 1987 p1 19-128 and was found to have
a low
activity. The polypeptide was made in the course of a study of function-
structure relationship
and was not mentioned as a possible intermediate product for the preparation
of improved
s - interferon ~3 conjugate molecules. For instance, the interferon ~3
polypeptide comprises an
amino acid sequence, which differs from that of SEQ ID NO 2 in that it
comprises the
substitution R27K in combination with at least one additional substitution
that is different from
E43K, or the substitution R35K in combination with at least one additional
substitution
provided that the polypeptide has an amino acid sequence which is different
from the amino
~ o acid sequence of wild-type human interferon (3 modified with the following
substitutions:
G7E+S12N+C17Y+R35K. The disclaimed polypeptide is disclosed by Stewart et al,
DNA Vol
6 not 1987 p1 19-128 as having a retained antiproliferative activity on Daudi
cells relative to
their antiviral activity, but reduced overall activity as compared to wild
type interferon (3. The
disclaimed polypeptide was not prepared with the purpose of reducing the
immunogenicity
~ s and/or increasing the functional in vivo half life and/or serum half life,
but was made in the
course of a study of the structural functional relationship of interferon Vii.
The polypeptide of the invention may, in addition to any of the above
specified
substitutions, additionally comprise the substitution C 17S and/or a deletion
of M 1 or the
substitution M 1 K. Furthermore, the polypeptide of the invention may comprise
an amino acid
2o sequence, which further differs from that of SEQ ID NO 2 in the removal,
preferably by
substitution, of at least one lysine residue selected from the group
consisting of K19, K33,
K45, K52, K99, K105, K108, K115, K123, K134, and K136. The lysine residues)
may be
replaced with any other amino acid residue, but is preferably replaced by an
arginine or a
glutamine. In particular, the polypeptide of the invention may be one, wherein
K45, K52 and/or
2s K123 has/have been replaced with another amino acid residue, but preferably
an arginine or a
glutamine residue. Also, the polypeptide may be expressed with a tag, e.g. as
described in the
section further above entitled "Conjugation of a tagged interferon (3
polypeptide".
A still further example of an interferon ~3 polypeptide of the invention
includes
one, that comprises an amino acid sequence which differs from that of wild-
type human
3o interferon ~3 in that at least one lysine residue selected from the group
consisting of K19, K33,
K45, K52, K99, K105, K108, K115, K123, K134, and K136 has been replaced with
any other
amino acid residue, provided that the interferon (3 polypeptide is different
from a hybrid
between interferon (3 and interferon a, which as a consequence of being a
hybrid has a
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
43
phenylalanine in position 45. Preferably, at least K19, K45, K52 and/or K123
is/are are
replaced. While the lysine residue may be deleted in accordance with this
aspect of the
invention, it is preferred that it be replaced with any other amino acid
residue, preferably an
arginine or a glutamine. Normally, the polypeptide of the invention comprises
an amino acid
s sequence which differs in 1-15 amino acid residues from the amino acid
sequence shown in
SEQ ID NO 2 as further discussed above. Examples of polypeptides of the
invention are
selected from the group consisting of those that comprises an amino acid
sequence, which
differs from that of SEQ ID NO 2 in at least the following substitutions:
R27K+R159K;
to R27K+K45R+R159K;
R27K+Q49K+E85K+A89K;
R27K+K45R+Q49K+E85K+A89K;
R27K+D39K+Q49K+E85K+A89K;
R27K+D39K+K45R+Q49K+E85K+A89K;
t s N4K+R27K+D39K+Q49K+E85K+A89K;
N4K+R27K+D39K+K45R+Q49K+E85K+A89K;
R27K+K123R+R159K;
R27K+K45R+K123R+R159K;
R27K+Q49K+E85K+A89K+K123R;
2o R27K+K45R+Q49K+E85K+A89K+K123R;
R27K+D39K+Q49K+E85K+A89K+K123R;
R27K+D39K+K45R+Q49K+E85K+A89K+K123R;
N4K+R27K+D39K+Q49K+E85K+A89K+K123R; and
N4K+R27K+D39K+K45R+Q49K+E85K+A89K+K123R.
2s It will be understood that any of the polypeptides of the invention
disclosed
herein may be used to prepare a conjugate of the invention, i.e. be covalently
coupled to any of
the non-polypeptide moieties disclosed herein. In particular, when a
polypeptide of the
invention is expressed in a glycosylating microorganism the polypeptide may be
provided in
glycosylated form.
Methods of preparing an interferon (3 polypeptide for use in the invention
The polypeptide of the present invention or the polypeptide part of a
conjugate of
the invention, optionally in glycosylated form, may be produced by any
suitable method known
in the art. Such methods include constructing a nucleotide sequence encoding
the polypeptide
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
44
and expressing the sequence in a suitable transformed or transfected host.
However,
polypeptides of the invention may be produced, albeit less efficiently, by
chemical synthesis or
a combination of chemical synthesis or a combination of chemical synthesis and
recombinant
DNA technology.
The nucleotide sequence of the invention encoding an interferon (3 polypeptide
may be constructed by isolating or synthesizing a nucleotide sequence encoding
the parent
interferon (3, e.g. with the amino acid sequence shown in SEQ ID NO 2, and
then changing the
nucleotide sequence so as to effect introduction (i.e. insertion or
substitution) or deletion (i.e.
removal or substitution) of the relevant amino acid residue(s).
io The nucleotide sequence is conveniently modified by site-directed
mutagenesis in
accordance with well-known methods, see, e.g., Mark et al., "Site-specific
Mutagenesis of the
Human Fibroblast Interferon Gene", Proc. Natl. Acad. Sci. USA, 81, pp. 5662-66
(1984); and
US 4,588,585.
Alternatively, the nucleotide sequence is prepared by chemical synthesis, e.g.
by
t s . using an oligonucleotide synthesizer, wherein oligonucleotides are
designed based on the
amino acid sequence of the desired polypeptide, and preferably selecting those
codons that are
favored in the host cell in which the recombinant polypeptide will be
produced. For example,
several small oligonucleotides coding for portions of the desired polypeptide
may be
synthesized and assembled by PCR, ligation or ligation chain reaction (LCR).
The individual
20 oligonucleotides typically contain 5' or 3' overhangs for complementary
assembly.
Once assembled (by synthesis, site-directed mutagenesis or another method),
the
nucleotide sequence encoding the interferon (3 polypeptide is inserted into a
recombinant vector
and operably linked to control sequences necessary for expression of the
interferon (3 in the
desired transformed host cell.
2s It should of course be understood that not all vectors and expression
control
sequences function equally well to express the nucleotide sequence encoding a
polypeptide
variant described herein. Neither will all hosts function equally well with
the same expression
system. However, one of skill in the art may make a selection among these
vectors, expression
control sequences and hosts without undue experimentation. For example, in
selecting a
3o vector, the host must be considered because the vector must replicate in it
or be able to
integrate into the chromosome. The vector's copy number, the ability to
control that copy
number, and the expression of any other proteins encoded by the vector, such
as antibiotic
markers, should also be considered. In selecting an expression control
sequence, a variety of
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
factors should also be considered. These include, for example, the relative
strength of the
sequence, its controllability, and its compatibility with the nucleotide
sequence encoding the
polypeptide, particularly as regards potential secondary structures. Hosts
should be selected by
consideration of their compatibility with the chosen vector, the toxicity of
the product coded
s for by the nucleotide sequence, their secretion characteristics, their
ability to fold the
polypeptide correctly, their fermentation or culture requirements, and the
ease of purification of
the products coded for by the nucleotide sequence.
The recombinant vector may be an autonomously replicating vector, i.e. a
vector
which exists as an extrachromosomal entity, the replication of which is
independent of
Io chromosomal replication, e.g. a plasmid. Alternatively, the vector is one
which, when
introduced into a host cell, is integrated into the host cell genome and
replicated together with
the chromosomes) into which it has been integrated.
The vector is preferably an expression vector, in which the nucleotide
sequence
encoding the polypeptide of the invention is operably linked to additional
segments required
~ s for transcription of the nucleotide sequence. The vector is typically
derived from plasmid or
viral DNA. A number of suitable expression vectors for expression in the host
cells mentioned
herein are commercially available or described in the literature. Useful
expression vectors for
eukaryotic hosts, include, for example, vectors comprising expression control
sequences from
SV40, bovine papilloma virus, adenovirus and cytomegalovirus. Specific vectors
are, e.g.,
20 pCDNA3.1 (+)\Hyg (Invitrogen, Carlsbad, CA, USA) and pCI-neo (Stratagene,
La Jola, CA,
USA). Useful expression vectors for bacterial hosts include known bacterial
plasmids, such as
plasmids from E. coli, including pBR322, pET3a and pETl2a (both from Novagen
Inc., WI,
USA), wider host range plasmids, such as RP4, phage DNAs, e.g., the numerous
derivatives of
phage lambda, e.g. , NM989, and other DNA phages, such as M13 and filamentous
single
2s stranded DNA phages. Useful expression vectors for yeast cells include the
2p plasmid and
derivatives thereof, the POT1 vector (US 4,931,373), the pJS037 vector
described in (Okkels,
Ann. New York Acad. Sci. 782, 202-207, 1996) and pPICZ A, B or C (Invitrogen).
Useful
vectors for insect cells include pVL941, pBG311 (Cate et al., "Isolation of
the Bovine and
Human Genes for Mullerian Inhibiting Substance And Expression of the Human
Gene In
so Animal Cells", Cell, 45, pp. 685-98 (1986), pBluebac 4.5 and pMelbac (both
available from
Invitrogen).
Other vectors for use in this invention include those that allow the
nucleotide
sequence encoding the polypeptide variant to be amplified in copy number. Such
amplifiable
vectors are well known in the art. They include, for example, vectors able to
be amplified by
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
46
DHFR amplification (see, e.g., Kaufman, U.S. Pat. No. 4,470,461, Kaufman and
Sharp,
"Construction Of A Modular Dihydrofolate Reductase cDNA Gene: Analysis Of
Signals
Utilized For Efficient Expression", Mol. Cell. Biol., 2, pp. 1304-19 (1982))
and glutamine
synthetase ("GS") amplification (see, e.g., US 5,122,464 and EP 338,841).
s The recombinant vector may further comprise a DNA sequence enabling the
vector to replicate in the host cell in question. An example of such a
sequence (when the host
cell is a mammalian cell) is the SV40 origin of replication. When the host
cell is a yeast cell,
suitable sequences enabling the vector to replicate are the yeast plasmid 2p
replication genes
REP 1-3 and origin of replication.
~o The vector may also comprise a selectable marker, e.g. a gene the product
of
which complements a defect in the host cell, such as the gene coding for
dihydrofolate
reductase (DHFR) or the Schizosaccharomyces pombe TPI gene (described by P.R.
Russell,
Gene 40, 1985, pp. 125-130), or one which confers resistance to a drug, e.g.
ampicillin,
kanamycin, tetracyclin, chloramphenicol, neomycin, hygromycin or methotrexate.
For
~ s filamentous fungi, selectable markers include amdS, pyre, arcB, niaD, sC.
The term "control sequences" is defined herein to include all components,
which are necessary or advantageous for the expression of the polypeptide of
the invention.
Each control sequence may be native or foreign to the nucleic acid sequence
encoding the
polypeptide. Such control sequences include, but are not limited to, a leader,
polyadenylation
2o sequence, propeptide sequence, promoter, enhancer or upstream activating
sequence, signal
peptide sequence, and transcription terminator. At a minimum, the control
sequences include
a promoter.
A wide variety of expression control sequences may be used in the present
invention. Such useful expression control sequences include the expression
control sequences
2s associated with structural genes of the foregoing expression vectors as
well as any sequence
known to control the expression of genes of prokaryotic or eukaryotic cells or
their viruses, and
various combinations thereof.
Examples of suitable control sequences for directing transcription in
mammalian
cells include the early and late promoters of SV40 and adenovirus, e.g. the
adenovirus 2 major
30 late promoter, the MT-1 (metallothionein gene) promoter, the human
cytomegalovirus
immediate-early gene promoter (CMV), the human elongation factor 1 a (EF-1 a)
promoter, the
Drosophila minimal heat shock protein 70 promoter, the Rous Sarcoma Virus
(RSV) promoter,
the human ubiquitin C (UbC) promoter, the human growth hormone terminator,
SV40 or
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
47
adenovirus Elb region polyadenylation signals and the Kozak consensus sequence
(Kozak, M.
JMoI Biol 1987 Aug 20;196(4):947-50).
In order to improve expression in mammalian cells a synthetic intron may be
inserted in the 5' untranslated region of the nucleotide sequence encoding the
polypeptide of
s interest. An example of a synthetic intron is the synthetic intron from the
plasmid pCI-Neo
(available from Promega Corporation, WI, USA).
Examples of suitable control sequences for directing transcription in insect
cells
include the polyhedrin promoter, the P 10 promoter, the Autographa californica
polyhedrosis
virus basic protein promoter, the baculovirus immediate early gene 1 promoter
and the
baculovirus 39K delayed-early gene promoter, and the SV40 polyadenylation
sequence.
Examples of suitable control sequences for use in yeast host cells include the
promoters of the yeast a-mating system, the yeast triose phosphate isomerase
(TPI) promoter,
promoters from yeast glycolytic genes or alcohol dehydogenase genes, the ADH2-
4c promoter
and the inducible GAL promoter.
~ s Examples of suitable control sequences for use in filamentous fungal host
cells
include the ADH3 promoter and terminator, a promoter derived from the genes
encoding
Aspergillus oryzae TAKA amylase triose phosphate isomerase or alkaline
protease, an A. niger
a-amylase, A. niger or A. nidulans glucoamylase, A. nidulans acetamidase,
Rhizomucor miehei
aspartic proteinase or lipase, the TPI1 terminator and the ADH3 terminator.
zo Examples of suitable control sequences for use in bacterial host cells
include
promoters of the lac system, the trp system, the TAC or TRC system and the
major promoter
regions of phage lambda.
The nucleotide sequence of the invention encoding an interferon (3
polypeptide,
whether prepared by site-directed mutagenesis, synthesis or other methods, may
or may not
2s also include a nucleotide sequence that encode a signal peptide. The signal
peptide is present
when the polypeptide is to be secreted from the cells in which it is
expressed. Such signal
peptide, if present, should be one recognized by the cell chosen for
expression of the
polypeptide. The signal peptide may be homologous (e.g. be that normally
associated with
human interferon (3) or heterologous (i.e. originating from another source
than human
30 interferon (3) to the polypeptide or may be homologous or heterologous. to
the host cell, i.e. be a
signal peptide normally expressed from the host cell or one which is not
normally expressed
from the host cell. Accordingly, the signal peptide may be prokaryotic, e.g.
derived from a
bacterium such as E. coli, or eukaryotic, e.g. derived from a mammalian, or
insect or yeast cell.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
48
The presence or absence of a signal peptide will, e.g., depend on the
expression host cell
used for the production of the polypeptide, the protein to be expressed
(whether it is an
intracellular or extracellular protein) and whether it is desirable to obtain
secretion. For use in
filamentous fungi, the signal peptide may conveniently be derived from a gene
encoding an
s Aspergillus sp. amylase or glucoamylase, a gene encoding a Rhizomucor miehei
lipase or
protease or a Humicola lanuginosa lipase. The signal peptide is preferably
derived from a gene
encoding A. oryzae TAKA amylase, A. niger neutral a-amylase, A. niger acid-
stable amylase,
or A. niger glucoamylase. For use in insect cells, the signal peptide may
conveniently be
derived from an insect gene (cf. WO 90/05783), such as the lepidopteran
Manduca sexta
adipokinetic hormone precursor, (cf. US 5,023,328), the honeybee melittin
(Invitrogen),
ecdysteroid UDPglucosyltransferase (egt) (Murphy et al., Protein Expression
and Purification
4, 349-357 (1993) or human pancreatic lipase (hpl) (Methods in Enzyrnology
284, pp. 262-272,
1997).
A preferred signal peptide for use in mammalian cells is that of human
interferon (3 apparent
1 s from the examples hereinafter or the murine Ig kappa light chain signal
peptide (Coloma, M
(1992) J. Imm. Methods 152:89-104). For use in yeast cells suitable signal
peptides have been
found to be the a-factor signal peptide from S. cereviciae. (cf. US
4,870,008), the signal
peptide of mouse salivary amylase (cf. O. Hagenbuchle et al.,Nature 289, 1981,
pp. 643-646), a
modified carboxypeptidase signal peptide (cf. L.A. Valls et al., Cell 48,
1987, pp. 887-897), the
2o yeast BAR1 signal peptide (cf. WO 87/02670), and the yeast aspartic
protease 3 (YAP3) signal
peptide (cf. M. Egel-Mitani et al., Yeast 6, 1990, pp. 127-137).
Any suitable host may be used to produce the interferon ~3 polypeptide,
including
bacteria, fungi (including yeasts), plant, insect, mammal, or other
appropriate animal cells or
cell lines, as well as transgenic animals or plants. Examples of bacterial
host cells include
2s grampositive bacteria such as strains of Bacillus, e.g. B. brevis or B.
subtilis, Pseudomonas or
Streptomyces, or gramnegative bacteria, such as strains of E. coli. The
introduction of a vector
into a bacterial host cell may, for instance, be effected by protoplast
transformation (see, e.g.,
Chang and Cohen, 1979, Molecular General Genetics 168: 111-11 S), using
competent cells
(see, e.g., Young and Spizizin, 1961, Journal of Bacteriology 81: 823-829, or
Dubnau and
3o Davidoff Abelson, 1971, Journal ofMolecular Biology 56: 209-221),
electroporation (see, e.g.,
Shigekawa and Dower, 1988, Biotechniques 6: 742-751), or conjugation (see,
e.g., Koehler and
Thorne, 1987, Journal ofBacteriology 169: 5771-5278).
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
4y
Examples of suitable filamentous fungal host cells include strains of
Aspergillus,
e.g. A. oryzae, A. niger, or A. nidulans, Fusarium or Trichoderma. Fungal
cells may be
transformed by a process involving protoplast formation, transformation of the
protoplasts, and
regeneration of the cell wall in a manner known per se. Suitable procedures
for transformation
s of Aspergillus host cells are described in EP 238 023 and US 5,679,543.
Suitable methods for
transforming Fusarium species are described by Malardier et al., 1989, Gene
78: 147-156 and
WO 96/00787. Yeast may be transformed using the procedures described by Becker
and
Guarente, In Abelson, J.N. and Simon, M.L, editors, Guide to Yeast Genetics
and Molecular
Biology, Methods in Enzymology, Volume 194, pp 182-187, Academic Press, Inc.,
New York;
Ito et al., 1983, Journal ofBacteriology 153: 163; and Hinnen et al., 1978,
Proceedings of the
National Academy of Sciences USA 75: 1920.
Examples of suitable yeast host cells include strains of Saccharomyces, e.g.
S.
cerevisiae, Schizosaccharomyces, Klyveromyces, Pichia, such as P. pastoris or
P. methanolica,
Hansenula, such as H. Polymorpha or Yarrowia. Methods for transforming yeast
cells with
t s heterologous DNA and producing heterologous polypeptides therefrom are
disclosed by
Clontech Laboratories, Inc, Palo Alto, CA, USA (in the product protocol for
the YeastmakerTM
Yeast Tranformation System Kit), and by Reeves et al., FEMS Microbiology
Letters 99 ( 1992)
193-198, Manivasakam and Schiestl, Nucleic Acids Research, 1993, Vol. 21, No.
18, pp. 4414-
4415 and Ganeva et al., FEMS Microbiology Letters 121 (1994) 159-164.
20 Examples of suitable insect host cells include a Lepidoptora cell line,
such as
Spodoptera frugiperda (S~ or SfZI) or Trichoplusioa ni cells (High Five) (US
5,077,214).
Transformation of insect cells and production of heterologous polypeptides
therein may be
performed as described by Invitrogen.
Examples of suitable mammalian host cells include Chinese hamster ovary
2s (CHO) cell lines, (e.g. CHO-K1; ATCC CCL-61), Green Monkey cell lines (C05)
(e.g. COS 1
(ATCC CRL-1650), COS 7 (ATCC CRL-1651)); mouse cells (e.g. NS/O), Baby Hamster
Kidney (BHK) cell lines (e.g. ATCC CRL-1632 or ATCC CCL-10), and human cells
(e.g.
HEK 293 (ATCC CRL-1573)), as well as plant cells in tissue culture. Additional
suitable cell
lines are known in the art and available from public depositories such as the
American Type
3o Culture Collection, Rockville, Maryland. Also, the mammalian cell, such as
a CHO cell, may
be modified to express sialyltransferase, e.g. 1,6-sialyltransferase, e.g. as
described in US
5,047,335, in order to provide improved glycosylation of the interferon ~i
polypeptide.
Methods for introducing exogeneous DNA into mammalian host cells include
calcium phosphate-mediated transfection, electroporation, DEAE-dextran
mediated
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
SO
transfection, liposome-mediated transfection, viral vectors and the
transfection methods
described by Life Technologies Ltd, Paisley, UK using Lipofectamin 2000 and
Roche
Diagnostics Corporation, Indianapolis, USA using FuGENE 6. These methods are
well known
in the art and e.g. described by Ausbel et al. (eds.), 1996, Current Protocols
in Molecular
s Biology, John Wiley & Sons, New York, USA. The cultivation of mammalian
cells are
conducted according to established methods, e.g. as disclosed in (Animal Cell
Biotechnology,
Methods and Protocols, Edited by Nigel Jenkins, 1999, Human Press Inc, Totowa,
New Jersey,
USA and Harrison MA and Rae IF, General Techniques of Cell Culture, Cambridge
University
Press 1997).
io In the production methods of the present invention, the cells are
cultivated in a
nutrient medium suitable for production of the polypeptide using methods known
in the art.
For example, the cell may be cultivated by shake flask cultivation, small-
scale or large-scale
fermentation (including continuous, batch, fed-batch, or solid state
fermentations) in laboratory
or industrial fermenters performed in a suitable medium and under conditions
allowing the
t s polypeptide to be expressed and/or isolated. The cultivation takes place
in a suitable nutrient
medium comprising carbon and nitrogen sources and inorganic salts, using
procedures known
in the art. Suitable media are available from commercial suppliers or may be
prepared
according to published compositions (e.g., in catalogues of the American Type
Culture
Collection). If the polypeptide is secreted into the nutrient medium, the
polypeptide can be
2o recovered directly from the medium. If the polypeptide is not secreted, it
can be recovered
from cell lysates.
The resulting polypeptide may be recovered by methods known in the art. For
example, the polypeptide may be recovered from the nutrient medium by
conventional
procedures including, but not limited to, centrifugation, filtration,
extraction, spray drying,
2s evaporation, or precipitation.
The polypeptides may be purified by a variety of procedures known in the art
including, but not limited to, chromatography (e.g., ion exchange, affinity,
hydrophobic,
chromatofocusing, and size exclusion), electrophoretic procedures (e.g.,
preparative
isoelectric focusing), differential solubility (e.g., ammonium sulfate
precipitation), SDS-
3o PAGE, or extraction (see, e.g., Protein Purification, J.-C. Janson and Lars
Ryden, editors,
VCH Publishers, New York, 1989). Specific methods for purifying polypeptides
exhibiting
interferon (3 activity are disclosed in US 4,289,689, US 4,359,389, US
4,172,071, US
4,551,271, US 5,244,655, US 4,485,017, US 4,257,938 and US 4,541,952. A
specific
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
$1
purification method is based on immunoaffinity purification (see, e.g.,
Okamura et al.,
"Human Fibroblastoid Interferon: Immunosorbent Column Chromatography And N-
Terminal
Amino Acid Sequence", Biochem., 19, pp. 3831-35 (1980)). Furthermore,
purification may
be based on the use of IFNAR 1 and/or IFNAR 2, in particular IFNAR 2.
The biological activity of the interferon (3 polypeptides can be assayed by
any
suitable method known in the art. Such assays include antibody neutralization
of antiviral
activity, induction of protein kinase, oligoadenylate 2,5-A synthetase or
phosphodiesterase
activities, as described in EP 41313 B 1. Such assays also include
immunomodulatory assays
(see, e.g., US 4,753,795), growth inhibition assays, and measurement of
binding to cells that
express interferon receptors. Specific assays for determining the biological
activity of
polypeptides or conjugates of the invention are disclosed in the Materials and
Methods section
hereinafter.
Cell culture of the invention
In a further aspect the invention relates to a cell culture comprising a) a
host cell transformed
~ s with a nucleotide sequence encoding a polypeptide exhibiting interferon (3
activity, and b) a
culture medium comprising said polypeptide produced by expression of said
nucleotide
sequence in a concentration of at least 800,000 IU/ml of medium, preferably in
a concentration
in the range of 800,000-3,500,000 IU/ml medium. While the polypeptide
exhibiting interferon
a activity may be a wild-type interferon ~3, e.g. human interferon (3 or a
variant thereof (e.g.
2o interferon (3 la or 1b) the polypeptide is preferably an interferon ~i
polypeptide as described
herein.
In a still further aspect the invention relates to a method of producing an
interferon (3
polypeptide as described herein, the method comprising:
(a) culturing a cell expressing an interferon ~i polypeptide variant in a
culture
2s , medium, such that the concentration of the interferon ~3 polypeptide
variant in the medium is at
least 800,000 IU/ml medium, in particular in the range of between 800,000 and
3,500,000
IU/ml medium; and
(b) recovering the interferon (3 polypeptide.
3o Other methods of the invention
In a still further aspect the invention relates to a method reducing
immunogenicity and/or of increasing functional in vivo half life and/or~serum
half life of an
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
52
interferon (3 polypeptide, which method comprises introducing an amino acid
residue
constituting an attachment group for a first non-polypeptide moiety into a
position exposed at
the surface of the protein that does not contain such group and/or removing an
amino acid
residue constituting an attachment group for a first non-polypeptide moiety
and subjecting the
s resulting modified polypeptide to conjugation with the first non-polypeptide
moiety.
Preferably, the amino acid residue to be introduced and/or removed is as
defined
in the present application. The non-polypeptide moiety is normally selected
from the group
consisting of a polymer molecule, a sugar moiety, a lipophilic group and an
organic
derivatizing agent.
In a still further aspect the invention relates to a method for preparing a
conjugate
of the invention, wherein the interferon (3 polypeptide is reacted with the
non-polypeptide
moiety to which it is to be conjugated under conditions conducive for the
conjugation to take
place, and the conjugate is recovered.
~ s Pharmaceutical compositition and uses of a conjugate of the invention
The interferon ~i polypeptide or the conjugate of the invention is
administered at a
dose approximately paralleling that employed in therapy with human interferon
(3 such as
Avonex, Rebif and Betaseron, or a higher dosis. The exact dose to be
administered depends on
the circumstances. Normally, the dose should be capable of preventing or
lessening the severity
20 or spread of the condition or indication being treated. It will be apparent
to those of skill in the
art that an effective amount of a polypeptide, conjugate or composition of the
invention
depends, inter alia, upon the disease, the dose, the administration schedule,
whether the
polypeptide or conjugate or composition is administered alone or in
conjunction with other
therapeutic agents, the serum half life of the compositions, and the general
health of the
2s patient.
The polypeptide or conjugate of the invention can be used "as is" and/or in a
salt
form thereof. Suitable salts include, but are not limited to, salts with
alkali metals or alkaline
earth metals, such as sodium, potassium, lithium, calcium and magnesium, as
well as e.g. zinc
salts. These salts or complexes may by present as a crystalline and/or
amorphous structure.
The polypeptide or conjugate of the invention is preferably administered in a
composition including a pharmaceutically acceptable Garner or excipient.
"Pharmaceutically
acceptable" means a carrier or excipient that does not cause any untoward
effects in patients to
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
53
whom it is administered. Such pharmaceutically acceptable carriers and
excipients are well
known in the art.
The polypeptide or conjugate of the invention can be formulated into
pharmaceutical compositions by well-known methods. Suitable formulations are
described in
s US 5,183,746, Remington's Pharmaceutical Sciences by E.W.Martin, 18th
edition, A. R.
Gennaro, Ed., Mack Publishing Company [1990]; Pharmaceutical Formulation
Development of
Peptides and Proteins, S. Frokjaer and L. Hovgaard, Eds., Taylor & Francis
[2000]; and
Handbook of Pharmaceutical Excipients, 3rd edition, A. Kibbe, Ed.,
Pharmaceutical Press
[2000]).
The pharmaceutical composition of the polypeptide or conjugate of the
invention
may be formulated in a variety of forms, including liquid, gel, lyophilized,
pulmonary
dispersion, or any other suitable form, e.g. as a compressed solid. The
preferred form will
depend upon the particular indication being treated and will be apparent to
one of skill in the
art.
~ s The pharmaceutical composition containing the polypeptide or conjugate of
the
invention may be administered orally, intravenously, intracerebrally,
intramuscularly,
intraperitoneally, intradermally, subcutaneously, intranasally,
intrapulmonary, by inhalation, or
in any other acceptable manner, e.g. using PowderJect or Protease technology.
The preferred
mode of administration will depend upon the particular indication being
treated and will be
2o apparent to one of skill in the art.
Parentals
An example of a pharmaceutical composition is a solution designed for
parenteral
administration. Although in many cases pharmaceutical solution formulations
are provided in
2s liquid form, appropriate for immediate use, such parenteral formulations
may also be provided
in frozen or in lyophilized form. In the former case, the composition must be
thawed prior to
use. The latter form is often used to enhance the stability of the active
compound contained in
the composition under a wider variety of storage conditions, as it is
recognized by those skilled
in the art that lyophilized preparations are generally more stable than their
liquid counterparts.
3o Such lyophilized preparations are reconstituted prior to use by the
addition of one or more
suitable pharmaceutically acceptable diluents such as sterile water for
injection or sterile
physiological saline solution.
In case of parenterals, they are prepared for storage as lyophilized
formulations or
aqueous solutions by mixing, as appropriate, the polypeptide having the
desired degree of
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
54
purity with one or more pharmaceutically acceptable carriers, excipients or
stabilizers typically
employed in the art (all of which are termed "excipients"), for example
buffering agents,
stabilizing agents, preservatives, isotonifiers, non-ionic detergents,
antioxidants and/or other
miscellaneous additives.
s Buffering agents help to maintain the pH in the range which approximates
physiological
conditions. They are typically present at a concentration ranging from about 2
mM to about 50
mM Suitable buffering agents for use with the present invention include both
organic and
inorganic acids and salts thereof such as citrate buffers (e.g., monosodium
citrate-disodium
citrate mixture, citric acid-trisodium citrate mixture, citric acid-monosodium
citrate mixture,
io etc.), succinate buffers (e.g., succinic acid-monosodium succinate mixture,
succinic acid-
sodium hydroxide mixture, succinic acid-disodium succinate mixture, etc.),
tartrate buffers
(e.g., tartaric acid-sodium tartrate mixture, tartaric acid-potassium tartrate
mixture, tartaric
acid-sodium hydroxide mixture, etc.), fumarate buffers (e.g., fumaric acid-
monosodium
fumarate mixture, fumaric acid-disodium fizmarate mixture, monosodium
fiunarate-disodium
~s fumarate mixture, etc.), gluconate buffers (e.g., gluconic acid-sodium
glyconate mixture,
gluconic acid-sodium hydroxide mixture, gluconic acid-potassium glyuconate
mixture, etc.),
oxalate buffer (e.g., oxalic acid-sodium oxalate mixture, oxalic acid-sodium
hydroxide
mixture, oxalic acid-potassium oxalate mixture, etc.), lactate buffers (e.g.,
lactic acid-sodium
lactate mixture, lactic acid-sodium hydroxide mixture, lactic acid-potassium
lactate mixture,
2o etc.) and acetate buffers (e.g., acetic acid-sodium acetate mixture, acetic
acid-sodium
hydroxide mixture, etc.). Additional possibilities are phosphate buffers,
histidine buffers and
trimethylamine salts such as Tris.
Preservatives are added to retard microbial growth, and are typically added in
amounts of about
0.2%-1% (w/v). Suitable preservatives for use with the present invention
include phenol,
2s benzyl alcohol, meta-cresol, methyl paraben, propyl paraben,
octadecyldimethylbenzyl
ammonium chloride, benzalkonium halides (e.g. benzalkonium chloride, bromide
or iodide),
hexamethonium chloride, alkyl parabens such as methyl or propyl paraben,
catechol,
resorcinol, cyclohexanol and 3-pentanol.
Isotonicifiers are added to ensure isotonicity of liquid compositions and
include polyhydric
so sugar alcohols, preferably trihydric or higher sugar alcohols, such as
glycerin, erythritol,
arabitol, xylitol, sorbitol and mannitol. Polyhydric alcohols can be present
in an amount
between 0.1 % and 25% by weight, typically 1 % to 5%, taking into account the
relative
amounts of the other ingredients.
Stabilizers refer to a broad category of excipients which can range in
function from a bulking
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
agent to an additive which solubilizes the therapeutic agent or helps to
prevent denaturation or
adherence to the container wall. Typical stabilizers can be polyhydric sugar
alcohols
(enumerated above); amino acids such as arginine, lysine, glycine, glutamine,
asparagine,
histidine, alanine, omithine, L-leucine, 2-phenylalanine, glutamic acid,
threonine, etc., organic
s sugars or sugar alcohols, such as lactose, trehalose, stachyose, mannitol,
sorbitol, xylitol,
ribitol, myoinisitol, galactitol, glycerol and the like, including cyclitols
such as inositol;
polyethylene glycol; amino acid polymers; sulfur-containing reducing agents,
such as urea,
glutathione, thioctic acid, sodium thioglycolate, thioglycerol, a-
monothioglycerol and sodium
thiosulfate; low molecular weight polypeptides (i.e. <10 residues); proteins
such as human
serum albumin, bovine serum albumin, gelatin or immunoglobulins; hydrophilic
polymers such
as polyvinylpyrrolidone; monosaccharides such as xylose, mannose, fructose and
glucose;
disaccharides such as lactose, maltose and sucrose; trisaccharides such as
raffinose, and
polysaccharides such as dextran. Stabilizers are typically present in the
range of from 0.1 to
10,000 parts by weight based on the active protein weight.
i s Non-ionic surfactants or detergents (also known as "wetting agents") may
be present to help
solubilize the therapeutic agent as well as to protect the therapeutic
polypeptide against
agitation-induced aggregation, which also permits the formulation to be
exposed to shear
surface stress without causing denaturation of the polypeptide. Suitable non-
ionic surfactants
include polysorbates (20, 80, etc.), polyoxamers (184, 188 etc.), Pluronic~
polyols,
2o polyoxyethylene sorbitan monoethers (Tween~-20, Tween~-80, etc.).
Additional miscellaneous excipients include bulking agents or fillers (e.g.
starch),
chelating agents (e.g. EDTA), antioxidants (e.g., ascorbic acid, methionine,
vitamin E) and
cosolvents. The active ingredient may also be entrapped in microcapsules
prepared, for
example, by coascervation techniques or by interfacial polymerization, for
example
zs hydroxymethylcellulose, gelatin or poly-(methylmethacylate) microcapsules,
in colloidal drug
delivery systems (for example liposomes, albumin microspheres, microemulsions,
nano-
particles and nanocapsules) or in macroemulsions. Such techniques are
disclosed in
Remington's Pharmaceutical Sciences, supra.
Parenteral formulations to be used for in vivo administration must be sterile.
This
3o is readily accomplished, for example, by filtration through sterile
filtration membranes.
Sustained release preparations
Suitable examples of sustained-release preparations include semi-permeable
matrices of solid hydrophobic polymers containing the polypeptide or
conjugate, the matrices
having a suitable form such as a film or microcapsules. Examples of sustained-
release matrices
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
56
include polyesters, hydrogels (for example, poly(2-hydroxyethyl-methacrylate)
or
poly(vinylalcohol)), polylactides, copolymers of L-glutamic acid and ethyl-L-
glutamate, non-
degradable ethylene-vinyl acetate, degradable lactic acid-glycolic acid
copolymers such as the
ProLease~ technology or Lupron Depot~ (injectable microspheres composed of
lactic acid-
s glycolic acid copolymer and leuprolide acetate), and poly-D-(-)-3-
hydroxybutyric acid. While
polymers such as ethylene-vinyl acetate and lactic acid-glycolic acid enable
release of
molecules for long periods such as up to or over 100 days, certain hydrogels
release proteins
for shorter time periods. When encapsulated polypeptides remain in the body
for a long time,
they may denature or aggregate as a result of exposure to moisture at
37°C, resulting in a loss
i o of biological activity and possible changes in immunogenicity. Rational
strategies can be
devised for stabilization depending on the mechanism involved. For example, if
the
aggregation mechanism is discovered to be intermolecular S-S bond formation
through thio-
disulfide interchange, stabilization may be achieved by modifying sulthydryl
residues,
lyophilizing from acidic solutions, controlling moisture content, using
appropriate additives,
~ s and developing specific polymer matrix compositions.
Pulmonary delivery
Conjugate formulations suitable for use with a nebulizer, either jet or
ultrasonic, will typically
comprise the conjugate dissolved in water at a concentration of, e.g., about
0.01 to 25 mg of
2o conjugate per mL of solution, preferably about 0.1 to 10 mg/mL. The
formulation may also
include a buffer and a simple sugar (e.g., for protein stabilization and
regulation of osmotic
pressure), and/or human serum albumin ranging in concentration from 0.1 to 10
mg/ml.
Examples of buffers that may be used are sodium acetate, citrate and glycine.
Preferably, the
buffer will have a composition and molarity suitable to adjust the solution to
a pH in the range
2s of 3 to 9. Generally, buffer molarities of from 1 mM to 50 mM are suitable
for this purpose.
Examples of sugars which can be utilized are lactose, maltose, mannitol,
sorbitol, trehalose,
and xylose, usually in amounts ranging from 1 % to 10% by weight of the
formulation.
The nebulizer formulation may also contain a surfactant to reduce or prevent
surface induced aggregation of the protein caused by atomization of the
solution in forming the
3o aerosol. Various conventional surfactants can be employed, such as
polyoxyethylene fatty acid
esters and alcohols, and polyoxyethylene sorbitan fatty acid esters. Amounts
will generally
range between 0.001 % and 4% by weight of the formulation. An especially
preferred surfactant
for purposes of this invention is polyoxyethylene sorbitan monooleate.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
57
Specific formulations and methods of generating suitable dispersions of liquid
particles of the invention are described in WO 9420069, US 5915378, US
5960792, US
5957124, US 5934272, US 5915378, US 5855564, US 5826570 and US 5522385 which
are
hereby incorporated by reference.
Three specific examples of commercially available nebulizers suitable for the
practice of this invention are the Ultravent nebulizer, manufactured by
Mallinckrodt, Inc., St.
Louis, Mo., the Acorn II nebulizer, manufactured by Marquest Medical Products,
Englewood,
Colorado, and the AERx pulmonary drug delivery system manufactured by Aradigm
Corporation, Hayward, California.
to Conjugate formulations for use with a metered dose inhaler device will
generally
comprise a finely divided powder. This powder may be produced by lyophilizing
and then
milling a liquid conjugate formulation and may also contain a stabilizer such
as human serum
albumin (HSA). Typically, more than 0.5% (w/w) HSA is added. Additionally, one
or more
sugars or sugar alcohols may be added to the preparation if necessary.
Examples include
~ s lactose maltose, mannitol, sorbitol, sorbitose, trehalose, xylitol, and
xylose. The amount added
to the formulation can range from about 0.01 to 200% (w/w), preferably from
approximately 1
to 50%, of the conjugate present. Such formulations are then lyophilized and
milled to the
desired particle size.
The properly sized particles are then suspended in a propellant with the aid
of a
2o surfactant. The propellant may be any conventional material employed for
this purpose, such as
a chlorofluorocarbon, a hydrochlorofluorocarbon, a hydrofluorocarbon, or a
hydrocarbon,
including trichlorofluoromethane, dichlorodifluoromethane,
dichlorotetrafluoroethanol, and
1,1,1,2-tetrafluoroethane, or combinations thereof. Suitable
surfactants~include sorbitan
trioleate and soya lecithin. Oleic acid may also be useful as a surfactant.
This mixture is then
2s loaded into the delivery device. An example of a commercially available
metered dose inhaler
suitable for use in the present invention is the Ventolin metered dose
inhaler, manufactured by
Glaxo Inc., Research Triangle Park, N.C.
Such conjugate formulations for powder inhalers will comprise a finely divided
dry powder containing conjugate and may also include a bulking agent, such as
lactose,
so sorbitol, sucrose, or mannitol in amounts which facilitate dispersal of the
powder from the
device, e.g., 50% to 90% by weight of the formulation. The particles of the
powder shall have
aerodynamic properties in the lung corresponding to particles with a density
of about 1 g/cmz
having a median diameter less than 10 micrometers, preferably between 0.5 and
5 micrometers,
most preferably of between 1.5 and 3.5 micrometers.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
58
An example of a powder inhaler suitable for use in accordance with the
teachings
herein is the Spinhaler powder inhaler, manufactured by Fisons Corp., Bedford,
Mass.
The powders for these devices may be generated and/or delivered by methods
disclosed in US 5997848, US 5993783, US 5985248, US 5976574, US 5922354, US
5785049
s and US 55654007.
The pharmaceutical composition containing the conjugate of the invention may
be administered by a wide range of mechanical devices designed for pulmonary
delivery of
therapeutic products, including but limited to nebulizers, metered dose
inhalers, and powder
inhalers, all of which are familiar to those of skill in the art.
io Some specific examples of commercially available devices suitable for the
practice of this invention are the Ultravent nebulizer, manufactured by
Mallinckrodt, Inc., St.
Louis, Missouri; the Acorn II nebulizer, manufactured by Marquest Medical
Products,
Englewood, Colorado; the Ventolin metered dose inhaler, manufactured by Glaxo
Inc.,
Research Triangle Park, North Carolina; the Spinhaler powder inhaler,
manufactured by Fisons
1 s Corp., Bedford, Massachusetts; the "standing cloud" device of Inhale
Therapeutic Systems,
Inc., San Carlos, California; the AIR inhaler manufactured by Alkermes,
Cambridge,
Massachusetts; and the AERx pulmonary drug delivery system manufactured by
Aradigm
Corporation, Hayward, California.
The pharmaceutical composition of the invention may be administered in
2o conjunction with other therapeutic agents. These agents may be incorporated
as part of the
same pharmaceutical composition or may be administered separately from the
polypeptide or
conjugate of the invention, either concurrently or in accordance with any
other acceptable
treatment schedule. In addition, the polypeptide, conjugate or pharmaceutical
composition of
the invention may be used as an adjunct to other therapies.
2s Accordingly, this invention provides compositions and methods for treating
most
types of viral infections, cancers or tumors (e.g. breast carcinoma, non-small
cell lung cancer)
or tumour angiogenesis, Chrohn's disease, ulcerative colitis, Guillain-Barre
syndrome, glioma,
idiopathic pulmonary fibrosis, abnormal cell growth, or for immunomodulation
in any suitable
animal, preferably mammal, and in particular human. In particular the
polypeptide, conjugate
30 or composition of the invention may be used for the treatment of multiple
sclerosis (MS), such
as any of the generally recognized four types of MS (benign, relapsing
remitting MS (RRMS),
primary progressive MS (PPMS) and secondary progressive MS (SPMS)) and for
monosymptomatic MS), hepatitis, or a herpes infection (the latter treatment
optionally being
combined with a treatment with IL-10).
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
59
In a further aspect the invention relates to a method of treating a mammal
having
circulating antibodies against interferon (3 la, such as AvonexTM or Rebif~,
or 1b, such as
Betaseron~, which method comprises administering a compound which has the
bioactivity of
interferon (3 and which has a reduced or no reaction with said antibodies. The
compound is
s administered in an effective amount. The compound is preferably a conjugate
as described
herein and the mammal is preferably a human being. The mammals to be treated
may suffer
from any of the diseases listed above for which interferon (3 is a useful
treatment. In particular,
this aspect of the invention is of interest for the treatment of multiple
sclerosis (any of the types
listed above) or cancer. Furthermore, the invention relates to a method of
making a
io pharmaceutical product for use in treatment of mammals having circulating
antibodies against
interferon ~i la, such as AvonexTM or RebiR~, or 1b, such as Betaseron~,
wherein a compound
which has the bioactivity of interferon (3 and which does not react with such
is formulated into
an injectable or otherwise suitable formulation. The term "circulating
antibodies" is intended to
indicate antibodies, in particular neutralizing antibodies, formed in a mammal
in response to
~ s having been treated with any of the commercially available interferon (3
preparations (Rebif,
Betaseron, Avonex).
In a further aspect the invention relates to a method of treating a patient in
need
of treatment with a pharmaceutical composition with at least some of the
therapeutically
beneficial properties of interferon (3 comprising administering a composition
comprising a
2o compound with at least part of the therapeutically beneficial activity of
interferon (3, said
treatment having reduced or removed adverse psychological effects as compared
to treatment
with interferon ~3, wherein said compound is a non-naturally occurring
conjugate of a
polypeptide with interferon ~i activity and a non-polypeptide moiety, in
particular a conjugate
according to the present invention.
2s In a still further aspect the invention relates to a pharmaceutical
composition for
the treatment of a patient in need of treatment with a compound having at
least part of the
therapeutically beneficial properties of interferon (3, said composition
comprising a compound
which is a non-naturally occurring conjugate of interferon ~i and a non-
polypeptide moiety,
said treatment further giving rise to fewer adverse psychological effects than
treatment with
interferon ~3. The conjugate is preferably a conjugate of the invention.
Also contemplated is use of a nucleotide sequence encoding a polypeptide of
the
invention in gene therapy applications. In particular, it may be of interest
to use a nucleotide
sequence encoding a polypeptide as described in the section above entitled
"Glycosylated
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
Polypeptides of the Invention modified to incorporate additional glycosylation
sites". The
glycosylation of the polypeptides is thus achieved during the course of the
gene therapy, i.e.
after expression of the nucleotide sequence in the human body.
Gene therapy applications contemplated include treatment of those diseases in
s which the polypeptide is expected to provide an effective therapy due to its
antiviral activity,
e.g., viral diseases, including hepatitis such as hepatitis C, and
particularly HPV, or other
infectious diseases that are responsive to interferon (3 or infectious agents
sensitive to interferon
~3. Furthermore, the conjugate or polypeptide of the invention may be used in
the treatment of
chronic inflammatory demyelinating polyradiculoneuropathy, and of severe
necrotising
~o cutaneous lesions. Also, gene therapy in connection with the treatment of
any MS type is
contemplated. Similarly, this invention contemplates gene therapy applications
for
immunomodulation, as well as in the treatment of those diseases in which
interferon (3 is
expected to provide an effective therapy due to its antiproliferative
activity, e.g., tumors and
cancers, or other conditions characterized by undesired cell proliferation,
such as restenosis. A
1 s further description of such gene therapy is provided in WO 95/25170.
Local delivery of interferon ~i using gene therapy may provide the therapeutic
agent to the target area while avoiding potential toxicity problems associated
with non-specific
administration.
Both in vitro and in vivo gene therapy methodologies are contemplated.
2o Several methods for transfernng potentially therapeutic genes to defined
cell
populations are known. For further reference see, e.g., Mulligan, "The Basic
Science Of Gene
Therapy", Science, 260, pp. 926-31 (1993). These methods include:
Direct gene transfer, e.g., as disclosed by Wolff et al., "Direct Gene
transfer Into
Mouse Muscle In vivo", Science 247, pp. 1465-68 (1990);
2s Liposome-mediated DNA transfer, e.g., as disclosed by Caplen et al.,
"Liposome-
mediated CFTR Gene Transfer to the Nasal Epithelium Of Patients With Cystic
Fibrosis"
Nature Med., 3, pp. 39-46 (1995); Crystal, "The Gene As A Drug", Nature Med.,
1, pp.- 15-17
(1995); Gao and Huang, "A Novel Cationic Liposome Reagent For Efficient
Transfection of
Mammalian Cells", Biochem.Biophys Res. Comm., 179, pp. 280-85 (1991);
3o Retrovirus-mediated DNA transfer, e.g., as disclosed by Kay et al., "In
vivo Gene
Therapy of Hemophilia B: Sustained Partial Correction In Factor IX-Deficient
Dogs", Science,
262, pp. 117-19 (1993); Anderson, "Human Gene Therapy", Science, 256, pp.808-
13(1992);
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
61
DNA Virus-mediated DNA transfer. Such DNA viruses include adenoviruses
(preferably Ad-2 or Ad-5 based vectors), herpes viruses (preferably herpes
simplex virus based
vectors), and parvoviruses (preferably "defective" or non-autonomous
parvovirus based
vectors, more preferably adeno-associated virus based vectors, most preferably
AAV-2 based
vectors). See, e.g., Ali et al., "The Use Of DNA Viruses as Vectors for Gene
Therapy", Gene
Therapy, 1, pp. 367-84 (1994); US 4,797,368, and US 5,139,941.
The invention is further described in the following examples. The examples
should not, in any manner, be understood as limiting the generality of the
present specification
and claims.
MATERIALS AND METHODS
Materials
HeLa cells - (available from American Type Culture Collection (ATCC)
is ISRE-Luc (Stratagene, La Jolla USA)
pCDNA 3.1/hygro (Invitrogen, Carlsbad USA)
pGL3 basic vector (Promega)
Human genomic DNA (CloneTech, USA)
DMEM medium: Dulbecco's Modified Eagle Media (DMEM), 10% fetal bovine serum
20 (available from Life Technologies A/S, Copenhagen, Denmark) -
Assays
Interferon Assay Outline
It has previously been published that interferon (3 interacts with and
activates Interferon type I
2s receptors on HeLa cells. Consequently, transcription is activated at
promoters containing an
Interferon Stimulated Response Element (ISRE). It is thus possible to screen
for agonists of
interferon receptors by use of an ISRE coupled luciferase reporter gene (ISRE-
luc) placed in
HeLa cells.
so Primary Assay
HeLa cells are co-transfected with ISRE-Luc and pCDNA 3.1/hygro and foci (cell
clones) are
created by selection in DMEM media containing Hygromycin B. Cell clones are
screened for
luciferase activity in the presence or absence of interferon (3. Those clones
showing the highest
ratio of stimulated to unstimulated luciferase activity are used in further
assays.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
62
To screen muteins, 15,000 cells/well are seeded in 96 well culture plates and
incubated overnight in DMEM media. The next day muteins as well as a known
standard are
added to the cells in various concentrations. The plates are incubated for 6
hours at 37 C in a
5% C02 air atmosphere LucLite substrate (Packard Bioscience, Groningen The
Netherlands )
s is subsequently added to each well. Plates are sealed and luminescence
measured on a
TopCount luminometer (Packard) in SPC (single photon counting) mode. Each
individual plate
contains wells incubated with interferon ~i as a stimulated control and other
wells containing
normal media as an unstimulated control. The ratio between stimulated and
unstimulated
luciferase activity serves as an internal standard for both mutein activity
and experiment-to-
experiment variation.
Secondary Assay
Currently, there are 18 non-allelic interferon a genes and one interferon (3
gene.
These proteins exhibit overlapping activities and thus it is critical to
ensure that muteins retain
1 s the selectivity and specificity of interferon (3.
The (3-R1 gene is activated by interferon ~3 but not by other interferons. The
transciption of (3-R1 thus serves as a second marker of interferon ~i
activation and is used to
ensure that muteins retain interferon ~i activity. A 300 by promoter fragment
of (3-R1 shown to
drive interferon sensitive transcription (Rare. M.R. et al (1996) JBC 27122878-
22884 ) was
20 isolated by PCR from human genomic DNA and inserted into the pGL3 basic
vector
(Promega). The resulting (3-Rl :luciferase gene is used in assays similar to
the primary assay
described above. In astrocytoma cells, the resulting (3-Rl:luciferase gene has
been described to
show 250 fold higher sensitivity to interferon ~i than to interferon a (Rani
et al. op city.
25 ELISA assay
The concentration of IFN-(3 is quantitated by use of a commercial sandwich
immunoassay (PBL Biomedical Laboratories, New Brunswick, NJ, USA). The kit is
based on
an ELISA with monoclonal mouse anti-IFN-(3 antibodies for catching and
detection of IFN-(3
in test samples. The detecting antibody is conjugated to biotin.
3o Tests samples and recombinant human IFN-[3 standard are added in 0.1 mL in
concentrations from 10-0.25 ng/mL to microtiter plates, precoated with
catching antibody. The
plates are incubated at RT for 1 hr. Samples and standard are diluted in kit
dilution buffer.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
63
The plates are washed in the kit buffer and incubated with the biotinylated
detecting antibody
in 0.1 mL for 1 hr at RT. After another wash the streptavidin-
horseradishperoxidase conjugate
is added in 0.1 mL and incubated for 1 hr at RT.
The reaction is visualised by addition of 0.1 mL Tetramethylbenzidine (TMB)
substrate chromogen. The plates are incubated for 15 minutes in the dark at RT
and the
reaction is stopped by addition of stop solution. The absorbanse is read at
450nm using an
ELISA reader.
Receptor binding assay
The receptor binding capability of a polypeptide or conjugate of the invention
can
be determined using the assay described in WO 95/25170 entitled "Analysis Of
IFN-~i(Phe,ol)
For Receptor Binding"(which is based on Daudi or A549 cells). Soluble domains
of IFNAR1
and IFNAR2 can be obtained essentially as described by Arduini et al, Protein
Science, 1999,
vol. 8, 1867-1877 or as described in Example 9 herein.
Alternatively, the receptor binding capability is determined using a
crosslinking
agent such as disuccinimidyl suberate (DSS) available from Pierce, Rockford,
IL, USA as
follows:
The polypeptide or conjugate is incubated with soluble IFNAR-2 receptor in the
presence or absence of DSS in accordance with the manufacturer's instructions.
Samples are
2o separated by SDS-PAGE, and a western blot using anti-interferon (3 or anti-
IFNAR2 antibodies
is performed. The presence of a functional interferon ~i
polypeptide/conjugate: receptor
interaction is apparent by an increase in the molecular size of receptor and
interferon (3 in the
presence of DSS.
Furthermore, a crosslinking assay using a polypeptide or conjugate of the
2s invention and both receptor subunits (IFNAR-1 and IFNAR-2) can establish
Interferon
receptor 1 binding ability. In this connection it has been published that
IFNAR-1 binds only
after an interferon (3: IFNAR-2 complex is formed (Mogensen et al., Journal of
Interferon and
Cytokine Research, 19:1069-1098, 1999).
In vitro immunogenicity tests of interferon (3 conjugates
so Reduced immunogenicity of a conjugate or polypeptide of the invention is
determined by use of an ELISA method measuring the immunoreactivity of the
conjugate or
polypeptide relative to a reference molecule or preparation. The reference
molecule or
preparation is normally a recombinant human interferon (3 preparation such as
Avonex, Rebif
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
64
or Betaseron, or another recombinant human interferon (3 preparation produced
by a method
equivalent to the way these products are made. The ELISA method is based on
antibodies from
patients treated with one of these recombinant interferon ~3 preparations. The
immunogenicity
is considered to be reduced when the conjugate or polypeptide of the invention
has a
s statistically significant lower response in the assay than the reference
molecule or preparation.
Another method of determining immunogenicity is by use of sera from patients
treated with interferon beta (i.e. any commercial interferon ~i product) in an
analogous manner
to that described by Ross et al. J. Clin Invest. 95, 1974-78, 1995. In the
antiviral neutralisation
bioassay reduced immunogenicity results in reduced inhibition of a conjugate
of the invention
~ o . by patient sera compared to a wt IFN-beta reference molecule.
Furthermore, in the biochemical
IFN binding assay a less immunogenic conjugate is expected to bind to patient
IgG to a lesser
extent than reference IFN-beta molecules.
For the neutralisation assay, the reference and conjugate molecules are added
in a
concentration that produces approximately 80% virus protection in the
antiviral neutralisation
~ s bioassay. The IFN-(3 proteins are mixed with patient sera in various
dilutions (starting at 1:20).
Antiviral activity
The antiviral bioassay is performed using A549 cells (CCL 185, American tissue
culture collection) and Encephalomyocarditis (EMC) virus (VR-129B, American
tissue culture
2o collection).
The cells are seeded in 96 well tissue culture plates at a concentration of
10,000
cells/well and incubated at 37°C in a 5% C02 air atmosphere. A
polypeptide or conjugate of
the invention is added in concentrations from 100-0.0001 IU/mL in a total of
100,1 DMEM
medium containing fetal calf serum and antibiotics.
2s After 24 hours the medium is removed and 0.1 mL fresh medium containing
EMC virus is added to each well. The EMC virus is added in a concentration
that causes 100%
cell death in IFN-(3 free cell cultures after 24 hours.
After another 24 hrs, the antiviral effect of the polypeptide or conjugate is
measured using the WST-1 assay. 0.01 mL WST-1 (WST-1 cell proliferation agent,
Roche
3o Diagnostics GmbH, Mannheim, Germany) is added to 0.1 mL culture and
incubated for '/Z-2
hours at 37°C in a 5% C02 air atmosphere The cleavage of the
tetrazolium salt WST-1 by
mitochondrial dehydrogenases in viable cells results in the formation of
forrnazan that is
quantified by measuring the absorbance at 450 nm.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
Neutralisation of activity in Interferon Stimulated Response Element (ISRE)
assay
The interferon (3 neutralising effect of anti-interferon ~3 sera are analysed
using
the ISRE-Luciferase activity assay.
Sera from interferon (3 treated patients or from immunised animals are used.
Sera
are added either in a fixed concentration (dilution 1:20-1:500 (pt sera) or 20-
600 ng/mL
(animal sera)) or in five-fold serial dilutions of sera starting at 1 /20 (pt
sera) or 600 ng/mL
(animal sera). Interferon (3 is added either in five fold-dilutions starting
at 25.000 IU/mL or in a
fixed concentration (0.1-10 IU/mL) in a total volume of 80p,1 DMEM medium +
10% FCS. The
io sera are incubated for 1 hr. at 37°C with IFN-~3.
The samples are then transferred to 96 well tissue culture plates containing
HeLa
cells transfected with ISRE-Luc grown from 24 hrs before (15,000 cells/well)
in DMEM
media. The cultures are incubated for 6 hours at 37°C in a 5% COz air
atmosphere. LucLite
substrate (Packard Bioscience, Groningen, The Netherlands) is subsequently
added to each
t s well. Plates are sealed and luminescence measured on a TopCount
luminometer (Packard) in
SPC (single photon counting) mode.
When interferon (3 samples are titrated in the presence of a fixed amount of
serum, the neutralising effect was defined as fold inhibition (FI) quantified
as EC50(w.
serum)/EC50 (w/o serum). The reduction of antibody neutralisation of
interferon (3 variant
zo proteins is defined as
FI variant
( 1 - ) x 100%
FI wt
zs Biological half life measurement of a PEG - interferon (3 conjugate
Measurement of biological half life can be carned out in a number of ways
described in the literature. One method is described by Munafo et al (European
Journal of
Neurology 1998, vol 5 No2 p 187-193), who used an ELISA method to detect serum
levels of
interferon (3 after subcutaneous and intramuscular administration of
interferon [3.
3o The rapid decrease of interferon (3 serum concentrations after i.v.
administration
has made it important to evaluate biological responses to interferon ~i
treatment. However it is
contemplated that the conjugates of the present invention will have prolonged
serum half lifes
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
66
also after i.v. administration making it possible to measure by e.g. an ELISA
method or by the
primary screening assay.
Different pharmacodynamic markers (e.g. serum neupterin and beta2
microglobulin) have also been studied (Clin Drug Invest (1999) 18(1):27-34).
These can
equally well be used to evaluate prolonged biological effect. These
experiments may also be
carried out in suitable animal species, e.g. rats.
Assays to assess the biological effects of interferon ~3 such as antiviral,
antiproliferative and immunomodulatory effects (as described in e.g. Annals of
Neurology
1995 vol 37 No 1 p 7-15) can be used together with the primary and secondary
screening
assays described herein to evaluate the biological efficacy of the conjugate
in comparison to
wild type interferon (3.
Finally an animal model such as the commonly used experimental autoimmune
encephalomyelitis (EAE) model can be used to establish efficacy of a conjugate
or polypeptide
of the invention. In the EAE model immunization with myelin or myelin derived
proteins
~ s elicits a disease mimicking the majority of the inflammatory and
neurologic features of
multiple sclerosis in humans. EAE has been used in mice, rats, rabbits, and
marmosets
(Cannella et al. PNAS, 95, 10100-5, 1998, Zaprianova et al. Morfologiia, 112,
25-8, 1997,
Hassouna et al. J.Urology, 130, 806-10, 1983, Genain & Hauser J. Mol. Med. 75,
187-
97,1997). Other models include Theiler's marine encephalomyelitis virus (TMEV)
model
20 (Murray et al. J.Neurosci. 18, 7306-14, 1998). will be used to establish
efficacy of the
interferon (3 conjugate.
PEGylation in microtiter plates of a tagged polypeptide with interferon (3
activity
The method comprises
2s Expressing the interferon (3 polypeptide with a suitable tag, e.g. any of
the tags
exemplified in the general description above.
Transferring culture broth to one or more wells in a microtiter plate capable
of
immobilising the tagged polypeptide. When the tag is His-His-His-His-His-His
(Casey et al, J.
Immunol. Meth., 179, 105 (1995)), a Ni-NTA HisSorb microtiter plate
commercially available
3o from QiaGen can be used.
After allowing for immobilising the tagged polypeptide to the microtiter
plate, the
wells are washed in a buffer suitable for binding and subsequent PEGylation.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
67
Incubating the wells with the activated PEG of choice. As an example, M-SPA-
5000 from
Shearwater Polymers is used. The molar ratio of activated PEG to polypeptide
has to be
optimised, but will typically be greater than 10:1 more typically greater than
100:1.
After a suitable reaction time at ambient temperature, typically around 1
hour, the reaction is
s stopped by removal of the activated PEG solution. The conjugated protein is
eluted from the
plate by incubation with a suitable buffer. Suitable elution buffers may
contain Imidazole,
excess NTA or another chelating compound.
The conjugated protein is assayed for biological activity and immunogenicity
as
appropriate.
to This tag may optionally be cleaved off using a method known in the art,
e.g.
using diaminopeptidase and the Gln in pos -1 will be converted to pyroglutamyl
with GCT
(glutamylcyclotransferase) and finally cleaved off with PGAP (gyro-glutamyl-
aminopeptidase)
giving the native protein. The process involves several steps of metal chelate
affinity
chromatography. Alternatively, the tagged polypeptide may be conjugated.
is
PEGylation of a receptor-bound interferon a polypeptide
In order to optimize PEGylation of an interferon (3 polypeptide in a manner
excluding
PEGylation of lysines involved in receptor recognition, the following method
has been
developed:
2o The soluble domains of IFNAR1 and IFNAR2 are obtained essentially as
described in Arduini et al, Protein Science (1999), vol 8: 1867-1877 or as
described in
Example 9.
A ternary complex consisting of an interferon (3 polypeptide, a soluble domain
of
IFNAR1 and a soluble domain of IFNAR2 in a 1:1:1 stoichiometry is formed in a
PBS buffer
2s at pH 7-9. The concentration of Interferon (3 polypeptide is approximately
20 ug/ml or 1 uM
and the receptors are present at equimolar concentration.
M-SPA-5000 from Shearwater Polymers, Inc is added at 3 different concentration
levels corresponding to 5, 20 or 100 molar excess of interferon (3
polypeptide. The reaction
time is 30 min at RT. After the 30 min reaction period, the pH of the reaction
mixture is
so adjusted to pH 2.0 and the reaction mixture is applied to a Vydac C18
column and eluted with
an acetonitrile gradient essentially as described (Utsumi etal, J. Biochem.,
vol 101, 1199-1208,
(1987). Alternatively and more elegantly, an isopropanol gradient can be used.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
68
Fractions are analyzed using the primary screening assay described herein and
active PEGylated interferon-(3 polypeptide obtained by this method stored at -
80 C in PBS, pH
7 containing 1 mg/ml HSA.
Alternatively, to the procedure described above a soluble domain of IFNAR2 is
s used as the only receptor component to form a binary complex. Furthermore,
IFNAR2 may be
immobilized on a suitable resin (e.g. Epoxy activated Sepharose 6B) according
to the
manufactures instructions prior to forming the binary complex. After
PEGylation, the
PEGylated Interferon-(3 is eluted with a 0.1 M Glycin, pH 2 buffer and
activity measured as
described after pH adjustment to neutral.
io
Accessible Surface Area (ASA)
The computer program Access (B. Lee and F.M.Richards, J. Mol.Biol. 55: 379-
400 ( 1971 )) version 2 (Copyright (c) 1983 Yale University) are used to.
compute the accessible
surface area (ASA) of the individual atoms in the structure. This method
typically uses a probe-
is size of 1.4~ and defines the Accessible Surface Area (ASA) as the area
formed by the centre of
the probe. Prior to this calculation all water molecules and all hydrogen
atoms are removed
from the coordinate set, as are other atoms not directly related to the
protein. Alternative
programs are available for computing ASA, e.g. the program WhatIf G.Vriend, J.
Mol. Graph.
(1990) 8, 52-56, electronically available at the WWW interface on
http://swift.embl-
2o heidelberg.de/servers2/ (R.Rodriguez et.al. CABIOS (1998) 14, 523-528.)
using the option
Accessibility to calculate the accessible molecular surface.
Fractional ASA of side chain
The fractional ASA of the side chain atoms is computed by division of the sum
of
2s the ASA of the atoms in the side chain with a value representing the ASA of
the side chain
atoms of that residue type in an extended ALA-x-ALA tripeptide. See Hubbard,
Campbell &
Thornton (1991) J.Mol.Bio1.220,507-530. For this example the CA atom is
regarded as a part
of the side chain of Glycine residues but not for the remaining residues. The
following table
indicates the 100% ASA standard for the side chain:
Ala 69.23
t~2
Arg 200.35
~2
Asn 106.25
~2
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
69
Asp 102.06
Az
Cys 96.69
~2
Gln 140.58
~2
Glu 134.61
A2
Gly 32.28
~2
His 147.00
~2
Ile 137.91
~r2
Leu 140.76
~2
Lys 162.50
~2
Met 156.08
t~2
Phe 163.90
AZ
Pro 119.65
~2
Ser 78.16
~2
Thr 101.67
t~2
Trp 210.89
~2
Tyr 176.61
~2
Val 114.14
~ ~2
Determining surface exposed amino acid residues
The three-dimensional crystal structure of human interferon'beta at 2.2 ~
resolution (Karpusas et al. Proc. Nat. Acad. Sci. USA (1997) 94:11813-11818 is
available from
s the Protein Data Bank (PDB) (Bernstein et.al. J. Mol. Biol. (1977) 112 pp.
535) and
electronically available via The Research Collaboratory for Structural
Bioinformatics PDB at
http://www.pdb.org/ under accession code IAUI. This crystal structure contain
two
independent molecules of human interferon beta in this example the A molecule
is used.
t 0 Surface exposure:
Using the WhatIf program as described above the following residues were found
to have zero surface accessibility for their side chain atoms (for Gly the
accessibility of the CA
atom is used): G7, N14, C17, L21, I44, A55, A56, T58, I59, M62, L63, L98,
L122, Y125,
I129, L133, A142, W143, V146, I150, N153, I157, L160, T161, and L164.
t s Fractional surface exposure
For further analysis it was necessary to remodel the side chains of residues
R71,
R 113, K 115, L 116, M 117 due to steric clashes. The remodelling was done
using Modeler 98,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
MSI INC. Performing fractional ASA calculations using the Access computer
program on the
remodelled interferon beta molecule (only including the amino acid residues
and excluding the
N-linked sugar moiety) resulted in the following residues having more than 25%
of their side
chain exposed to the surface: S2, N4, L5, F8, L9, R11, 512, F15, Q16 Q18, K19,
W22, Q23,
s G26, R27, L28, E29, Y30, L32, K33, R35, M36, N37, D39, E42, K45, Q46, L47,
Q48, Q49,
Q51, K52, Q64, A68, R71, Q72, D73, 575, S76, G78, N80, E81, T82, E85, N86,
A89, Y92,
H93, N96, H97, K99, T 100, E 103, E 104, K 1 O5, E 107, K 1 O8, E 109, D 110,
F 111, R 113, G 114,
K115, L116, 5119, L120, H121, K123, 8124, 6127, 8128, L130, H131, K134, A135,
K136,
E137, Y138, 5139, H140, V148, 8152, Y155, N158, 6162, Y163, 8165, and N166.
and the
1 o following residues have more than 50% of their side chain exposed to the
surface: N4, L5, F8,
S12, F15, Q16, K19, W22, G26, R27, E29, Y30, K33, R35, N37, D39, E42, Q46,
Q48, Q49,
QS1, K52, R71, D73, S75, G78, N80, E81, T82, E85, N86, A89, Y92, H93, K99,
T100, E103,
E104, E107, K108, D110, F111, L116, K123, 8124, 6127, H131, K134, E137, V148,
Y155,
R 165, and N 166.
is
EXAMPLE 1
Design of an expression cassette for expression of interferon ~i in mammalian
and insect cells
The DNA sequence, GenBank accession number M28622 (shown in SEQ ID NO
20 1 ), encompassing a full length cDNA encoding human interferon (3 with its
native signal
peptide, was modified in order to facilitate high expression in mammalian
cells. First the ATG
start codon context was modified according to the Kozak consensus sequence
(Kozak, M. J
Mol Biol 1987 Aug 20;196(4):947-SO), such that there is a perfect match to the
consensus
sequence upstream of the ATG start codon. Secondly the codons of the native
human
zs interferon (3 was modified by making a bias in the codon usage towards the
codons frequently
used in highly expressed human genes. Subsequently, certain nucleotides in the
sequence were
substituted with others in order to introduce recognition sites for DNA
restriction
endonucleases (this allows for easier modification of the DNA sequence later).
Primers were
designed such that the gene could be synthesised:
3o CBProFprl:
S'GGCTAGCGTTTAAACTTAAGCTTCGCCACCATGACCAACAAGTGCCTGCTCCAGA
TCGCCCTGCTCCTGT-3', SEQ ID 3,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
71
CBProFpr2:
5'ACAACCTGCTCGGCTTCCTGCAGAGGAGTTCGAACTTCCAGTGCCAGAAGCTCCT
GTGGCAGCTGAACGG-3', SEQ ID 4,
CBProFpr3:
s 5'GAACTTCGACATCCCCGAGGAAATCAAGCAGCTGCAGCAGTTCCAGAAGGAGGA
CGCCGCTCTGACCATC-3', SEQ ID S,
CBProFpr4
5'TTCCGCCAGGACTCCAGCTCCACCGGTTGGAACGAGACCATCGTGGAGAACCTG
CTGGCCAACGTGTACC-3', SEQ ID 6,
t o . CBProFprS
5'AGGAGAAGCTGGAGAAGGAGGACTTCACCCGCGGCAAGCTGATGAGCTCCCTGC
ACCTGAAGCGCTACTA-3', SEQ ID 7,
CBProFpr6
5'GGAGTACAGCCACTGCGCCTGGACCATCGTACGCGTGGAGATCCTGCGCAACTT
t s CTACTTCATCAACCGC-3', SEQ ID 8,
CBProFpr9
5'CACCACACTGGACTAGTGGATCCTTATCAGTTGCGCAGGTAGCCGGTCAGGCGG
TTGATGAAGTAGAAGT-3', SEQ ID 9,
CBProFprlO
20 5'AGGCGCAGTGGCTGTACTCCTTGGCCTTCAGGTAGTGCAGGATGCGGCCATAGT
AGCGCTTCAGGTGCAG-3', SEQ ID 10,
CBProFprl l
5'CTCCTTCTCCAGCTTCTCCTCCAGCACGGTCTTCAGGTGGTTGATCTGGTGGTACA
CGTTGGCCAGCAGG-3', SEQ ID 11,
2s CBProFprl2
5'GAGCTGGAGTCCTGGCGGAAGATGGCGAAGATGTTCTGCAGCATCTCGTAGATG
GTCAGAGCGGCGTCCT-3', SEQ ID 12,
CBProFprl3
5'CCTCGGGGATGTCGAAGTTCATCCTGTCCTTCAGGCAGTACTCCAGGCGCCCGTT
30 CAGCTGCCACAGGAG-3', SEQ ID 13,
CBProFprl4
5'CAGGAAGCCGAGCAGGTTGTAGCTCATCGATAGGGCCGTGGTGCTGAAGCACAG
GAGCAGGGCGATCTGG-3', SEQ ID 14,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
72
The primers were assembled to the synthetic gene by one step PCR using
Platinum Pfx- polymerase kit (Life Technologies) and standard three step PCR
cycling
parameters. The assembled gene was amplified by PCR using the same conditions.
A cDNA encoding a N-terminal extended form of human interferon (3 was
s synthesised using the same PCR conditions as described above but with the
primers
CBProFprl and -14 substituted with the primers:
CBProFpr7
5'CTGCTCCAGATCGCCCTGCTCCTGTGCTTCAGCACCACGGCCCTATCGATGAAGC
ACCAGCACCAGCATC-3', SEQ ID 15,
CBProFpr8
5'CACTGCTTACTGGCTTATCGAAATTAATACGACTCACTATAGGGAGACCCAAGCT
GGCTAGCGTTTAAAC-3', SEQ ID 16,
CBProFprl S
5'CAGGAAGCCGAGCAGGTTGTAGCTCATCTGTTGGTGTTGATGTTGGTGCTGATGC
i s TGGTGCTGGTGCTTC-3', SEQ ID 17,
CBProFprl6
5'AGCAGGGCGATCTGGAGCAGGCACTTGTTGGTCATGGTGGCGAAGCTTAAGTTT
AAACGCTAGCCAGCTT-3', SEQ ID 18,
in order to incorporate a purification TAG in the interferon (3 molecule.
2o The synthesised genes were cloned into pcDNA3.IlHygro (Invitrogen) between
the HindIII site at the 5' end and the BamHI at the 3', resulting in pCBProF 1
and pCBProF2.
The synthetic intron from pCI-Neo (Promega) was amplified using standard PCR
conditions as described above and the primers:
CBProFpr37 S'-CCGTCAGATCCTAGGCTAGCTTATTGCGGTAGTTTATCAC-3', SEQ
2s ID 19,
CBProFpr38 5'-GAGCTCGGTACCAAGCTTTTAAGAGCTGTAAT-3', SEQ ID 20,
resulting in a 332 by PCR fragment which was cut with NheI and HindIII and
inserted in the
S'UTR of the plasmids pCBProF 1 and pCBProF2 resulting in pCBProF4 and
pCBProFS.
Codons for individual amino acids were changed by amplifying relevant regions
30 of the coding region by PCR in such a way that the PCR introduced changes
in the sequence
can be introduced in the expression plasmids by classical cloning techniques.
E.g. the primers:
Lys45arg-5 primer (NarI/KasI):
5'GCTGAACGGGCGCCTGGAGTACTGCCTGAAGGACAGGATGAACTTCGACATCCC
CGAGGAAATCCGCCAGCTGCAGC-3', SEQ ID 21,
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
73
Lys45mut-3 primer (BsiWI): 5'TCTCCACGCGTACGATGGTCCAGGCGCAGTGGCTG-3',
SEQ ID 22,
were used to introduce a K45R substitution in the PCR-fragment spanning the
region from
position 1055 to 1243 in pCBProF 1. Both the PCR fragment and pCBProF 1 was
cut with NarI
s and BsiWI which are both unique. The PCR fragment and the vector backbone of
pCBProF 1
are purified and ligated resulting in substitution of the Lys45 codon AAG with
the Arg codon
CGC in pCBProF 1.
Furthermore, SOE (sequence overhang extension) PCR was used for introduction
of amino acid substitutions. In the SOE-PCR both the N-terminal part and the C-
terminal part
Io of the INFB molecule were first amplified in individual primary PCRs.
For these primary PCRs the central complementary primers were synthesised
such that the codon(s) for the amino acids) to be substituted is/are changed
to the desired
codon(s). The terminal primers were standard primers defining the N- and C-
terminal of the
INF(3 molecule respectively. Further the terminal primers provided a
restriction enzyne site
1 s enabling subsequent cloning of the full-length PCR product. Thus, the
central (nonsense)
primer and the N-terminal (sense) primer were used to amplify the N-terminal
part of the INF(3
coding region in one of the primary PCRs and equivalently for the C-terminal
part. Once
amplified the N- and C-terminal parts are assembled into the full-length
product in a secondary
PCR and cloned into a modified version of pCDNA3.1 /Hygro as described above.
For instance,
20 the following primers were used to introduce the mutations for the
substitutions F111N and
R113T:
CBProFprimer9(Sense):
CACCACACTGGACTAGTGGATCCTTATCAGTTGCGCAGGTAGCCGGTCAGGCGGTTG ATG
AAGTAGAAGT (SEQ ID NO 23),
2s CBProFprimer231 (Antisense):
CATCAGCTTGCCGGTGGTGTTGTCCTCCTTC (SEQ ID NO 24),
CBProFprimer230 (Sense):
GAAGGAGGACAACACCACCGGCAAGCTGATG (SEQ ID NO 25),
CBProFprimer42 (Antisense):
30 CACACTGGACTAGTAAGCTTTTATCAGTTGCGCAGGTAGC (SEQ ID NO 26),
Furthermore, in cases where the introduced mutations) were sufficiently close
to
a unique restriction endo-nuclease site in the expression plasmid variant
genes were
constructed using construction procedure encompassing a single PCR step and a
subsequent
cloning. For instance, the substitution K19R was introduced by use of the PCR
primer:
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
74
CBProFpr58:
GAGGAGTTCGAACTTCCAGTGCCAGCGCCTCCTGTGGCAGCTGAACG (SEQ ID NO 27),
and CBProFprimer9:
The PCR product was subsequently cloned using the restriction endo-nuclease
s sites BsiWI and BstBI.
EXAMPLE 2
Expression of human interferon (3 in a baculoviruslinsect cell system
In order to express the synthetic gene, encoding human interferon (3 harboured
in
pCBProF 1 (described in example 1 ) in the baculovirus/insect cell system the
gene was excised
with NheI and XhoI and ligated into the transfer vector pBlueBac 4.5, which is
included in the
MaxBac 2.0 Transfection kit obtained from Invitrogen (San Diego, USA). All
methods used
i s for generation of recombinant baculovirus and expression in insect cells
are described in the
"MaxBac 2.0 Transfection and Expression Manual" included in the kit.
In brief, together with liniarized AcMNPV DNA (Bac-N-Blue DNA) pBlueBac
4.5-interferon ~i CBProFI) was transfected into SF9 cells. 3 days post-
transfection the
transfection supernatant was harvested and a plaque assay with appropriate
viral dilutions was
2o prepared. Blue distinct plaques were visible after 7 days and 6 individual
plaques were
collected for propagation in a 6-well plate. After 5 days 2 ml virus
supernatant (P-1 stock) was
harvested from each well. 0.75 ml was taken out from the P 1 stocks and viral
genomic DNA
was isolated. The viral genomic DNA's were analysed in PCR reactions with
forward/reverse
primers in order to be able to select the recombinant baculoviruses among the
six P-1 stocks. A
2s small aliquot from the recombinant P-1 stock was tested in a human
interferon (3 specific
ELISA (available from PBL Biomedical Laboratories) in order to ensure that
recombinant
human interferon (3 was present in the supernatant.
For further propagation of chosen recombinant baculovirus 6 x 106 SF9 cells
were seeded in a T-80 culture flask and infected with 200 ~l of the P-1 stock.
After S days the
3o supernatant (P-2 stock) was harvested and 2 ml of the P-2 stock was used to
infect a 100 ml
suspension culture (1 x 106 SF9 cells/ml) in a 500 ml Erlenmeyer flask
(Corning). After 5 days
the supernatant (P-3 stock) was harvested and the virus titer was determined
by plaque assay.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
In order to produce human interferon (3 for purification 1 x 109 SF9 cells
were
harvested from a backup suspension culture. In a 50 ml screw-cap tube the SF9
cells were
infected with recombinant baculovirus from the P-3 stock (MOI = 2) in a period
of 1 S minutes.
Hereafter the cells were spun down and washed one time in serum-free medium
(Sf 900 II
s SFM, Gibco BRL) and transferred to a 2800 ml Triple Baffle Fernbach Flask
(Bellco)
containing 1 1 serum-free medium. 3 days post-infection the medium supernatant
was harvested
and the recombinant human interferon (3 was purified.
Purification of interferon (3 molecules
~ o The fermentation broth is concentrated and/or pH adjusted to approximately
4.5
after dilution to suitable ionic strength. Suitable is intended to mean that
the ionic strength is so
low that interferon (3 will bind to a Mono S cation exchange column
(Pharmacia) equilibrated
in 4 mM acetic acid pH 4.5 (buffer A). After application, the column is washed
with 3 column
volumes buffer A and interferon (3 is eluted with a linear gradient from
buffer A to buffer A
~ s including 1 M NaCI. Alternatively purification can be obtained as
described for Interferon a
(Analytical Biochemistry 247, 434-440 (1997) using a TSK-gel SP-SPW column
(Toso Haas)).
Alternatively His tagged interferon ~i can be purified using IMAC (Immobilized
Metal Affinity Chromatography) in accordance with well known methods, e.g., as
described by
UniZyme Laboratories, Denmark.
2o Another purification method makes use of monoclonal or polyclonal
antibodies.
Interferon (3 fermentation broth is adjusted to pH 7 and 0.5 M NaCI and
applied to a column
with immobilized monoclonal antibody to recombinant human interferon (3. The
column is
equilibrated with e.g. 10 mM Tris, 0.5 M NaCI, pH 7 (Buffer B) prior to
application. After
application the column is washed with 3 column volumes Buffer B and eluted
with a suitable
2s buffer at low pH (e.g. pH 2-3).
Alternatively, if the interferon ~i is tagged with e.g. the c-Myc peptide
(EQKLI
SEEDL), monoclonal antibodies raised against the c-Myc peptide, can be used in
a similar
fashion. Immobilization of antibody to the column is achieved using e.g. CNBr-
Sepharose
(Pharmacia) according to the manufacturers instructions.
3o A combination of Cation exchange chromatography, IMAC and/or antibody
chromatography may be applied if necessary to obtain relevant purity for
further experiments.
Purity, identity, quantity and activity of eluted fractions from the
abovementioned
columns can be determined using a combination of methods known by the person
skilled in the
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
76
art. These may include one or more of the following assays and methods or
other relevant
methods known by the person skilled in the art: the primary and secondary
assays described
above, ELISA methods, SDS-PAGE, western blotting, IEF, HPLC, amino acid
sequencing,
mass spectrometry and amino acid analysis.
Following purification, the modified interferon ~i polypeptide may be
subjected to
conjugation to a polymer molecule such as M-SPA-5000 from Shearwater Polymers
according
to the manufacturer's instructions. Preferably, the receptor recognition site
of the purified
modified interferon ~i polypeptide is blocked prior to conjugation as
described in the Materials
and Methods section herein.
io
EXAMPLE 3
Expression of human interferon (3 in HEK293 cells
In order to express the synthetic gene, encoding human interferon (3,
harboured by
Is pCBProFI (described in example 1), in HEK293 cells (ATCC Cat. No: CRL-1573)
the gene
was PCR-amplified with the two primers PBR 7 (5'-
CGCGGATCCATATGACCAACAAGTGCCTG-3') (SEQ ID NO 28) and PBR 2 (5'-
CGCGGATCCTTATCAGTTGCGCAG-3') (SEQ ID NO 29) and cloned into the BamHI site
of pcDNA3.1 (-) (Invitrogen, USA) in correct orientation, giving the plasmid
pPR9.
2o For transfection of the HEK293 cell line a T-25 culture flask was seeded to
50%
confluency in DMEM medium (Life Technologies, USA) containing 10% FBS and
incubated
over night. By usage of FuGENE 6 Transfection Reagent (Roche, USA) pPR9 was
transfected
into the cells: To 95 ~1 serum-free DMEM medium was added 5 ~1 FuGENE 6 and
1.7 ~l (2
fig) pPR9 and incubated at room temperature for 20 minutes. The transfection
complex was
2s then added drop-wise to the cells and the culture flask was returned to the
incubator. Next day
the cells were trypsinized and seeded into a T-80 culture flask in DMEM medium
containing
10% FBS and S00 ~g Geneticin (Life Technologies) per ml.
At confluency it was confirmed, by usage of a human interferon (3 specific
ELISA, that the primary transfection-pool was expressing the wished protein
and the cells were
3o sub-cloned by limited dilution. In this way a high-producing HEK293 clone
was identified
expressing human interferon (3.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
77
EXAMPLE 4
High level expression of Interferon (3 in CHO cells
The cell line CHO K1 [p22]-E4 (ATCC # CCL-61) stably expressing human
s interferon (3 was passed 1:10 from a confluent culture and propagated as
adherent cells in T-25
flasks in serum containing medium (MEMa w/ ribonucleotides and
deoxyribonucleotides
(GibcoBRL Cat # 32571), 10% FCS (GibcoBRL Cat # 10091), penicillin and
streptomycin
(Gibco/BRL Cat # 15140-114) until confluence. The media was then changed to
serum free
media (RenCyte CHO; MediCult Cat.# 22600140) for 24 hours before including 5
mM Sodium
t o Butyrate (Merck Cat # 8.17500) during a medium change. The cells were then
allowed to
express interferon ~3 for 48 hours prior to harvest of the medium. The
interferon (3
concentration in the duplicate cultures were determined to be 854,797 IU/ml
(with lower and
upper 95% confidence interval at 711,134 IU/ml and 1,032,012 IU/ml)
respectively).
In a separate set of experiments, the cell line CHO K1 [p22]-E4 stably
expressing
~ s human interferon ~3 was passed 1:10 from a confluent culture and
propagated as adherent cells
in serum containing medium (MEMa w/ ribonucleotides and deoxyribonucleotides
(Gibco/BRL Cat # 32571), 10% FCS (GibcoBRL Cat # 10091), penicillin and
streptomycin
(GibcoBRL Cat # 15140-114) until confluence in a 10 layer cell factory (NUNC #
165250).
'The media was then changed to serum free media; DMEM/F 12 (Gibco/BRL # 11039-
021 ) with
2o the addition of 1:100 ITS-A (GibcoBRL # S 1300-044) and 1:500 EX-CYTE VLE
(Serological
Proteins Inc. # 81-129-1) and 1:100 penicillin and streptomycin (GibcoBRL Cat
# 15140-114)
for 48 hours before changing the medium with the further addition of S~mM
butyrate (Merck
Cat # 8.17500). The cells were then allowed to express interferon (3 for 48
hours prior to
harvest of the medium. The interferon [i concentration was determined to be
824,791 IU/ml
2s (with lower and upper 95% confidence interval at 610,956 IU/ml and
1,099,722 IU/ml)
respectively).
It is contemplated that interferon (3 polypeptides of the invention may be
produced in
equally high yields in the same manner as any of those described above.
3o EXAMPLE 5
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
78
Construction and expression of interferon (3 variant with one introduced
glycosylation site
In order to insert an extra N-linked glycosylation site at position 111 in
hINF-[3,
the synthetic gene (hinf [3) encoding hINF-(3 (described in example 1 ) was
altered by site
directed PCR mutagenesis. Using BIO-X-ACT (Bioline, UK) and the plasmid PFO50
[hinf
s ~i)/pcDNA3.1 (-)Hygro/Intron (a derivative of pcDNA3.1 (-)Hygro (Invitrogen,
USA) in which
a chimeric intron obtained from pCI-neo (Promega, USA) had been inserted
between the
BamHI and NheI sites in the MCS of the vector] as template, two PCR reactions
were
performed with two overlapping primer-sets [CB41 (5'-
TTTAAACTGGATCCAGCCACCATGACCAACAAG-3') (SEQ ID NO 30) lCBSS (5'-
to CGGCCATAGT
AGCGCTTCAGGTGCAGGGAGCTCATCAGCTTGCCGGTGGTGTTGTCCTCCTTC-3')
(SEQ ID NO 31 ) and CB42 (SEQ ID 26) / CB86 (5'-
GAAGGAGGACAACACCACCGGCAAGCTGATGAGCTCCCTGCACCTGAAGCGCTAC
TATGGCC G-3') (SEQ ID NO 32) resulting in two fragments of 446 and 184 base
pairs,
is respectively. These two fragments were assembled in a third PCR with the
flanking primers
CB41 and CB42. The resulting gene was inserted into the mammalian expression
vector
pcDNA3.1 (-)Hygro/Intron and confirmed by DNA sequencing to have the correct
base changes
leading to the substitutions F111N and R113T in hINF-(3 (plasmid designated
PF085).
To test the activity of the [F111N+ R113T]hINF-[i variant, PF085 was
2o transfected into the CHO K 1 cell line (ATCC #CCL-61 ) by use of
Lipofectamine 2000 (Life
Technologies, USA) as transfection agent. 24 hours later the culture medium
was harvested
and assayed for INF-(3 activity/concentration:
Activity: 56046 IU/ml [primary assay]
ELISA: 80 ng/ml
2s Specific activity: 7x108IU/mg
As seen, the [F111N+R113T]hINF-(3 variant has a very high specific activity,
about twice the specific activity of wt hINF-(3.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
79
EXAMPLE 6
Construction and expression of interferon (3 with another introduced
glycosylation site
~Q49N+Q51 TJ
s Analogously to what is described in Example 5 an extra N-linked
glycosylation
site was introduced in position 49 by means of the substitutions Q49N and
QS1T. Using PF043
(hinf [i /pcDNA3.1 (Invitrogen, USA)) as template, two PCR reactions were
performed with
two overlapping primer-sets [PBR7 (SEQ ID NO 28) lPBR78 (5'-
GGCGTCCTCCTTGGTGAAGTTCTGCAGCTG-3') (SEQ ID NO 33) and PBR8 (5'-
~o ATATATCCCAAGCTTTTATCAGTTGCGCAGGTAGCCGGT-3') (SEQ ID NO 34)lPBR77
(5'- CAGCTGCAGAACTTCACCAAGGAGGACGCC-3') (SEQ ID NO 35) resulting in two
fragments of 228 and 369 base pairs, respectively. These two fragments were
assembled in a
third PCR with the flanking primers PBR7 and PBRB. The resulting gene was
inserted into the
mammalian expression vector pcDNA3.1 (-)Hygro/Intron and confirmed by DNA
sequencing
~s to have the correct base changes leading to [Q49N,QS1T]hINF-(3 (plasmid
designated PF104).
To test the activity of the [Q49N+QS 1 T]hINF-(3 variant, PF 104 was
transfected
into the CHO K1 cell line by use of Lipofectamine 2000 (Life Technologies,
USA) as
transfection agent. 24 hours later the culture medium was harvested and
assayed for INF-[i
activity/concentration:
2o Activity: 17639 IU/ml [primary assay]
ELISA: 10 ng/ml
Specific activity: 1.7x109IU/mg
As observed here the [Q49N+QS 1 T]hINF-(3 variant has a high specific
activity.
This may be due to poor recognition by one of the monoclonal antibodies used
in the ELISA.
2s
EXAMPLE 7
Construction and expression of interferon [i with two introduced glycosylation
sites
The additional glycosylation sites described in Examples 5 and 6 were
introduced
3o into human interferon [3 by means of the substitutions Q49N, QS1T, F111N,
and R113T.
Using PF085 (described in example 5) as template, two PCR reactions were
performed with two overlapping primer-sets [PBR89
(5'CGCGGATCCAGCCACCATGACCAACAAGTGCCTG) (SEQ ID NO 36)/
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
PBR78 (SEQ ID NO 33) and PBR8 (SEQ ID NO 34) )lPBR77 (SEQ ID NO 35)] resulting
in
two fragments of 228 and 369 base pairs, respectively.
These two fragments were assembled in a third PCR with the flanking primers
PBR89 and PBRB. The resulting gene was inserted into the mammalian expression
vector
s pcDNA3.1 (-)Hygro/Intron and confirmed by sequencing to have the correct
base changes
leading to [Q49N, QS1T, F111N, R113T] hINF-[i (plasmid designated PF123).
PF123 was transfected into CHO K1 cells by use of Fugene 6 (Roche) as
transfection agent. 24 hours later the culture medium was harvested and
assayed for INF-(3
activity/concentration:
to Activity: 29401 IU/ml [primary assay]
ELISA: 14 ng/ml
Specific activity: 2.1x109IU/ml
As observed here the [Q49N+QS1T+ F111N+ R113T]hINF-(3 variant also has a
high specific activity.
~ s T'he variant was found to have receptor binding activity in the receptor
binding
assay described in the Materials and Methods section, which is based on the
use of the
crosslinking agent DSS.
EXAMPLE 8
Production of ~Q49N+ QSI T+ FlIlN+ R113TJInterferon-(3 glycosylation variant
in Roller
Bottles
A CHOK1 sub-clone (5/G-10) producing the [Q49N+QS1T+F111N+R113T]
glycosylation variant was seeded into 2 roller bottles, each with an expanded
surface of 1700
2s cm2 (Corning, USA), in 200 ml DMEM/F-12 medium (LifeTechnologies; Cat. #
31330)
supplemented with 10% FBS and penicillin/streptomycin (P/5). After 2 days the
medium was
exchanged. After another 2 days the two roller bottles were nearly 100%
confluent and the
medium was shifted to 300 ml serum-free UltraCHO medium (BioWhittaker; Cat. #
12-724)
supplemented with 1/500 EX-CYTE (Serologicals Proteins; Cat. # 81129N) and
P/S. Growing
so the cells in this medium promotes a higher cell mass, higher than can be
achieved in the serum
containing medium. After 2 days the medium was renewed. After another 2 days
the medium
was shifted to the production medium: DMEM/F-12 medium (Life Technologies;
Cat. #
21041) supplemented with 1/100 ITSA (Life Technologies; Cat. # 51300-044)
[ITSA stands
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
81
for Insulin (1.0 g/L) - Transferrin (0.55 g/L) - Selenium (0.67 mg/L)
supplement for Adherent
cultures], 1 /500 EC-CYTE and P/S. In Fig. 2 the production run is shown,
where 300 ml
medium was harvested from each roller bottle every day. The harvested media
from the two
roller bottles were pooled before a medium sample was taken out for interferon
(3 activity
s determination.
As seen in fig. 2 the production run was terminated after 26 days. After a lag-
period of 5 days the activity mediated by the [49N+ QS 1 T+F 111 N+R
113T]Interferon-~i variant
increased dramatically and for the rest of the production run the harvested
interferon-~i activity
per day, in average, was 2.4 million IU/ml x 600 ml = 1.440 billion IU. In
total 3.2 x 101° IU
1 o was produced corresponding to 160 mg protein (with a hypothetical specific
activity of 2 x 1 O8
IU/mg).
EXAMPLE 9
t s Production, purification, and PEGylation of the Interferon-(3 variant
K19R+ K45R+ KI23R
To end up with 100 ml serum-free medium containing the Interferon-[3 variant
K19R+K45R+K123R, 3 T-175 flasks were seeded with COS-7 cells in DMEM medium
(Life
technologies; Cat. # 21969-035) supplemented with 10% FBS plus Glutamine and
penicillin/streptomycin. On the day of transfection (at nearly 100%
confluency) the medium
2o was renewed with 30 ml fresh medium 4 - 5 hours before the transfection. To
prepare the
transfection, 1890 ~1 DMEM medium without supplements was aliquoted into a 14
ml
polypropylene tube (Corning). 210 p1 Fugene 6 (Roche) was added directly into
the medium
and incubated for 5 min at RT. In the meantime 168 ~g plasmid DNA ([K19R,
K45R,
K123R]INF-(3/pcDNA3.1(-)Hygro; PF #161) was aliquoted into another 14 ml
polypropylene
2s tube. After 5 min incubation the Fugene 6 mix was added directly to the DNA
solution and
incubated for 15 min at RT. After incubation about 700 p1 was added drop wise
to each of the
three cell media.
Next day the transfection medium was substituted with 35 ml serum-free
production medium. The serum-free medium is based on DMEM medium (Life
Technologies;
30 . Cat. # 31053-028) supplemented with Glutamine, Sodium Pyruvate,
penicillin/streptomycin,
1 % ITSA (Life Technologies; Cat. # 51300-044), and 0.2% Ex-Cyte (Serologicals
Proteins;
Cat. # 81-129). Before the production medium was added the cell layers were
washed two
times in the DMEM medium without additives.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
82
Three days post-transfection the 100 ml serum-free medium was harvested for
purification and PEGylation of the Interferon-~3 variant.
pH was adjusted to 6.8 and conductivity adjusted to < 10 mS/cm with Milli Q
water. Then the broth was batch adsorbed to 1 ml SP SSO cation exchange resin
(TosoHaas)
s preequilibrated with buffer A (20 mM phosphate, 100 mM NaCI, pH 7). After 2
h rotation end
over end, the resin was allowed to sediment and transferred to a column. The
resin was washed
with 5 column volumes buffer A and eluted with 2 ml buffer B (20 mM phosphate,
800 mM
NaCI, pH 7). The eluate was concentrated to 500 u1 on VivaSpin (cutoff 10 kDa)
after addition
of 5 % ethyleneglycol. The concentrate was adjusted to 50 mM phosphate, 0.3 M
NaCI, 20
to ethyleneglycol, pH 8 in a final volume of 2 ml and further concentrated to
0.5 ml.
The final concentrate was PEGylated as follows: to 100 u1 of the final
concentrate, 25 u1 of activated mPEG-SPA (5000 kDa, Shearwater, Alabama)
freshly prepared
in phosphate buffer, pH 8 were added to make final concentrations of activated
PEG of 0, 5, 10
25 or 50 mg/ml. The reaction was allowed to proceed for 30 min at room
temperature and
is then quenched by addition of 50 mM glycine buffer. Samples were frozen
immediately at -
80 C and bioactivity was measured as described (Primary Assay). Western blots
of each
sample were performed in order to evaluate the amount of unreacted Interferon-
~i variant
present in the PEGylated sample.
Results demonstrate that at 25 mg activated PEG/ml, nonPEGylated Interferon-[3
2o variant was absent as judged by western blot and the variant retained 50 %
of its bioactivity
compared to the control sample (treated identically, but with 0 mg/ml
activated PEG).
EXAMPLE 10
2s Expression and purification of soluble IFNAR2
The cDNA's encoding the extracellular domain of IFNAR-1 and IFNAR-2
(termed IFNARlec and IFNAR2ec, respectively) were amplified from HeLa cell
cDNA using
PCR with primers corresponding to the first 10 amino acid residues and the
final 10 amino acid
residues of the extracellular domain of IFNAR-2 (the nucleotide sequence of
which is apparent
3o from Novick et al., Cell, Vol. 77, pp 391-400, 1994) and the first 10 amino
acid residues and
the final 10 amino acid residues of the extracellular domain of IFNAR-1 (the
nucleotide
sequence of which is apparent from Uze et al., Cell Vol. 60, 225-234, 1990).
The cDNA's were
subcloned into the pBlueBac 4.5/VS-His-TOPO vector (Invitrogen) and a
recombinant
Baculovirus obtained by homologous recombination, plaque purification, and
propagation in
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
83
S~ cells. Sf9 cells were infected with the recombinant Baculovirus and
expression from the
resulting cells was obtained essentially as described in Example 2.
IFNAR 1 ec and IFNAR2ec protein was observed in culture supernatants two to
three days after infection of S~ cells with recombinant baculovirus. The
activity of soluble
s receptors was observed in an Interferon antagonist assay. Briefly, Hela
cells containing the
ISRE element (as described in the Primary Assay above) are stimulated with a
sub-maximal
dose of human wild-type Interferon ~3 in the presence of varying
concentrations of IFNARec
supernatant. The antagonist effect of the supernatant is directly proportional
to the amount of
soluble receptor present.
io IFNAR2ec was purified from filtered culture supernatants using ion
exchange,
and affinity chromatography. Culture supernatants positive for IFNAR2ec were
pH adjusted to
7.5 and loaded onto an anion exchange column, and the bound recombinant
protein was eluted
using SOOmM NaCI. The partially pure IFNAR2ec was then diluted and pH adjusted
to 8.0,
before further purification using binding to a TALONTM Metal Affinity Resin
and elution with
~ s . imidazol. The final preparation was frozen in aliquots. IFNAR1 ec can be
purified as described
for IFNAR2ec with the exception that cation exchange chromatography at pH6.0
will be used
as the ion exchange step.
EXAMPLE 11
Use of soluble IFNAR2 for purification and PEGylation of Interferon-(3 and
variants thereof
Purified IFNAR2 obtained as described in Example 9 is immobilized either
through amino or carboxyl groups using e.g. CNBr-activated Sepharose 4B or EAH
Sepharose
4B according to the manufacturer's instructions (Amersham Pharmacia Biotech,
Affinity
2s Chromatography, Principles and Methods, 18-1022-29, edition AB). It is
critically important
that the coupling method allows fixnctional IFNAR2 to be immobilized and this
is tested
through optimization of the coupling conditions (pH, coupling buffer, ratio of
IFNAR 2 to
activate matrix etc). Another critical parameter is the blocking of excess
active groups.
Subsequently, testing of binding capacity by addition of interferon-~3 and
measurement of
3o breakthrough is carried out.
Optimally immobilized IFNAR2 is used for purification of Interferon-(3 as
follows. A 5 ml column with 1 mg IFNAR 2 immobilized per ml gel is
equilibrated with buffer
A (20 mM phosphate, 300 mM NaCI, pH 7). Then the column is loaded with a 2 mg
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
~4
Interferon-(3 sample in buffer A and subsequently washed with S column volumes
buffer A.
Elution is obtained by pumping 2 column volumes of buffer B onto the column.
Fractions of 1
ml are collected and assayed for bioactivity. Optimal elution conditions are
dependent on the
immobilization method, but examples of elution conditions include pH 1.5 - 3
(e.g. 0.1 M
s glycine pH 2.3 in 0.5 M NaCI), pH 11.5 - 12, 3.5 M MgCl2, 6M urea or the
like.
EXAMPLE 12
Use of immobilized IFNAR2 for PEGylation of interferon ~3 (variants)
io In addition to the use described in Example 10, immobilized IFNAR 2 may be
used for optimal PEGylation, wherein PEGylation of the part of Interferon-~i
or variants thereof
interacting with the receptor is avoided.
A S ml column with 1 mg IFNAR 2 immobilized per ml gel is equilibrated with
buffer A (20 mM phosphate, 300 mM NaCI, pH 7). Then the column is loaded with
a 2 mg
is Interferon-~3 sample in buffer A and subsequently washed with 5 column
volumes buffer A. A
solution of activated mPEG-SPA (1-50 mg/ml in buffer A) is pumped on the
column and
allowed to react for 15 min-12 h depending on temperature. One preferred range
of
combination of residence time and temperature is 15-60 min, 10-20 C; another
is 30 min to 5
h, 2-8 C. After the indicated time period, elution is obtained by pumping 2
column vdumes of
2o buffer B onto the column. Fractions of 1 ml are collected and assayed for
bioactivity using the
primary screening assay. Optimal elution conditions are dependent on the
immobilization
method, but examples of elution conditions include pH 1.5-3 (e.g. 0.1 M
glycine pH 2.3 in 0.5
M NaCI), pH 11.5-12, 3.5 M MgCl2, 6M Urea or the like.
EXAMPLE 13
Antiviral activity of PEGylated variant
The pegylated IFN-~i variant protein, K19R+K45R+K123R, was assayed using
the antiviral bioassay. Wild-type and variant proteins were added to A549
cells in
concentrations from 10-0.0001 IU/mL in triplicate cultures.
3o The pegylated IFN-~i variant showed total inhibition of EMC virus induced
cell
death at a concentration of 3 IU/mL, with an EC50 of 0.13 IU/mL (Figure 1 ).
The wild-type
standard shows virus inhibition with an EC50 of 1.4 IU/mL.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
These results demonstrate that the pegylation of the modified interferon ~i
polypeptide resulted in a conjugate with full anti-viral activity.
s EXAMPLE 14
Antibody neutralisation of glycosylated variant
The antibody neutralisation of wild-type and glycosylated IFN-(3 variant
protein,
Q49N+QS1T+F111N+R113T, was assayed using the ISRE neutralisation assay.
Interferon (3
~ o wild-type and variant proteins (in five fold dilutions starting at 12500
IU/mL) were incubated
with polyclonal rabbit anti-interferon (3 antibody (PBL Biomedical
Laboratories) in
concentrations 0, 40 and 200 ng/mL.
In the presence of 200 ng/mL polyclonal rabbit anti-serum the activity of the
wild
type interferon (3 protein was reduced 11.8 times whereas the activity of the
glycosylated
~ s interferon (3 variant only was reduced 3.0 times. Thus the degree of
antibody recognition of the
interferon [3 variant was reduced by 75% of the wt level, see Table 1 below.
These results
demonstrate that the recognition of the glycosylated mutant interferon ~i by
polyclonal
antibodies raised in animals immunised with wild-type human interferon (3 is
highly reduced.
Thus, a large portion of the immunogenic epitopes in wild-type human
interferon ~i have been
2o removed/shielded by the modifications made in the variant molecule.
Table 1
Antibody conc.Protein ECSO Fold inhibitionReduction of
(ng/mL) antibody
neutralisation
0 wt 0.00039 - -
variant 0.00020 - -
40 wt 0.00190 4.8 -
variant 0.00020 1.0 79%
200 wt 0.00461 11.8 -
variant 0.00059 3.0 75%
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
86
EXAMPLE 1 S
Construction and expression of interferon ~3 molecules with modified N
terminal
s N-terminally modified variants of interferon ~3 were constructed as
described in
the preceding examples.
For the construction of an expression plasmid for the interferon [3 variant,
INFB
S(-1 )A + M 1 Q the following primers were used:
CBProFpr110:
AAC TGG ATC CAG CCA CCA TGA CCA ACA AGT GCC TGC TCC AGA TCG CCC TGC TCC TGT
GCT
TCA GCA CCA CGG CCC TAG CCC AGA GCT AC (SEQ ID NO 37) and CBProFpr42 (SEQ ID
NO
26).
For the construction of an expression plasmid for the interferon variant,
INF(3 S(-
1)AQ (indicating substitution of the S residue located in position (-1) with
an A and a Q
i s residue) the following primers were used:
CBProFpr109:
AAC TGG ATC CAG CCA CCA TGA CCA ACA AGT GCC TGC TCC AGA TCG CCC
TGC TCC TGT GCT TCA GCA CCA CGG CCC TAG CCC AGA TGA GCT AC (SEQ ID
NO 38) and CBProFpr42 (SEQ ID NO 26).
2o To test the activity of these variants the respective plasmids; pF 154 and
pF 163
were transfected into CHO K1 cells using Lipofectamine 2000 (Life
Technologies, USA) as
transfection reagent. The supernatants were harvested 24 hours post
transfection and assayed in
the primary activity assay and in the ELISA as described in the Materials and
Methods section.
The following results were obtained:
2s INFB S-lA + MI Q (pF154):
Activity: 106410 IU/ml
ELISA: 333 ng/ml
Specific activity: 3.2 x 108 IU/mg
INFB S IAQ (pF163):
3o Activity: 90634 IU/ml
ELISA: 193 ng/ml
Specific activity: 4.7 x 108 IU/mg
These molecules are as active as wild type human interferon ~3.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
87
EXAMPLE 16
s Preparation of pegylated IFN ~3 variants
50 microliters of a 0.3 mg/ml solution of recombinant human IFN-(3 polypeptide
comprising the mutations Q49N+QS 1T+ K19R+ K45R+ K123R in SO mM Na-acetate,
35%
ethylene glycol, pH S.5 were mixed with 10 X10.5 M Na-phosphate, pH 8.0 and 20
~l 50 mM
Na-phosphate, 0.1 M NaCI, 30% ethylene glycol, pH 8.0 containing 0.02 mg/ml
SPA-mPEG
(N-succinimidyl Propionate methoxy polyethylene glycol). This corresponds to a
10 molar
excess of SPA-mPEG to IFN-(3.
After %Z hour with gentle rotation at room temperature, the reaction was
quenched
by addition of 5 X120 mM Glycine, pH 8Ø At this stage, the reaction mixture
contained a
mixture of unmodified as well as pegylated forms of recombinant human IFN-(3.
~ s In vitro testing using the primary screening assay demonstrated that the
pegylated
material retained 40 % activity, as compared to the unmodified recombinant
human IFN-(3.
In another experiment, 50 p1 of a 0.14 mg/ml solution of recombinant human IFN
~3 polypeptide comprising the mutations Q49N+QS1T in 50 mM Na-acetate, 35%
ethylene
glycol, pH 5.5 was mixed with 10 X10.5 M Na-phosphate, pH 8.0 and 20 p1 50 mM
Na-
2o phosphate, 0.1 M NaCI, 30% ethylene glycol, pH 8.0 containing 0.03 mg/ml
SPA-mPEG. This
gave a 10 molar excess of SPA-mPEG to IFN-(3.
After %z hour with gentle rotation at room temperature, the reaction was
quenched
by addition of 5 X120 mM Glycine, pH 8Ø At this stage, the reaction mixture
contained a
mixture of unmodified as well as pegylated forms of recombinant human IFN-(3.
2s In vitro testing using the primary screening assay demonstrated that the
pegylated
material retained 20 % activity, as compared to the unmodified recombinant
human IFN-(3.
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
SEQUENCE LISTING
<110> Maxygen ApS
<120> New Interferon Beta-like Molecules
<130> 2wo2
<140>
<141>
<160> 38
<170> PatentIn Ver. 2.1
<210> 1
<211> 840
<212> DNA
<213> Homo sapiens
<400> 1
acattctaac tgcaaccttt cgaagccttt gctctggcac aacaggtagt aggcgacact 60
gttcgtgttg tcaacatgac caacaagtgt ctcctccaaa ttgctctcct gttgtgcttc 120
tccactacag ctctttccat gagctacaac ttgcttggat tcctacaaag aagcagcaat 180
tttcagtgtc agaagctcct gtggcaattg aatgggaggc ttgaatactg cctcaaggac 240
aggatgaact ttgacatccc tgaggagatt aagcagctgc agcagttcca gaaggaggac 300
gccgcattga ccatctatga gatgctccag aacatctttg ctattttcag acaagattca 360
tctagcactg gctggaatga gactattgtt gagaacctcc tggctaatgt ctatcatcag 420
ataaaccatc tgaagacagt cctggaagaa aaactggaga aagaagattt caccagggga 480
aaactcatga gcagtctgca cctgaaaaga tattatggga ggattctgca ttacctgaag 540
gccaaggagt acagtcactg tgcctggacc atagtcagag tggaaatcct aaggaacttt 600
tacttcatta acagacttac aggttacctc cgaaactgaa gatctcctag cctgtgcctc 660
tgggactgga caattgcttc aagcattctt caaccagcag atgctgttta agtgactgat 720
ggctaatgta ctgcatatga aaggacacta gaagattttg aaatttttat taaattatga 780
gttattttta tttatttaaa ttttattttg gaaaataaat tatttttggt gcaaaagtca 840
<210> 2
<211> 166
<212> PRT
<213> Homo sapiens
<400> 2
Met Ser Tyr Asn Leu Leu Gly Phe Leu Gln Arg Ser Ser Asn Phe Gln
1 5 10 15
Cys Gln Lys Leu Leu Trp Gln Leu Asn Gly Arg Leu Glu Tyr Cys Leu
20 25 30
Lys Asp Arg Met Asn Phe Asp Ile Pro Glu Glu Ile Lys Gln Leu Gln
35 40 45
Gln Phe Gln Lys Glu Asp Ala Ala Leu Thr Ile Tyr Glu Met Leu Gln
50 55 60
1
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
Asn Ile Phe Ala Ile Phe Arg Gln Asp Ser Ser Ser Thr Gly Trp Asn
65 70 75 80
Glu Thr Ile Val Glu Asn Leu Leu Ala Asn Val Tyr His Gln Ile Asn
85 90 95
His Leu Lys Thr Val Leu Glu Glu Lys Leu Glu Lys Glu Asp Phe Thr
100 105 110
Arg Gly Lys Leu Met Ser Ser Leu His Leu Lys Arg Tyr Tyr Gly Arg
115 120 125
Ile Leu His Tyr Leu Lys Ala Lys Glu Tyr Ser His Cys Ala Trp Thr
130 135 140
Ile Val Arg Val Glu Ile Leu Arg Asn Phe Tyr Phe Ile Asn Arg Leu
145 150 155 160
Thr Gly Tyr Leu Arg Asn
165
<210> 3
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 3
ggctagcgtt taaacttaag cttcgccacc atgaccaaca agtgcctgct ccagatcgcc 60
ctgctcctgt 70
<210> 4
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 4
acaacctgct cggcttcctg cagaggagtt cgaacttcca gtgccagaag ctcctgtggc 60
agctgaacgg 70
<210> 5
<211> 70
<212> DNA
<213> Artificial Sequence
2
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
<220>
<223> Description of Artificial Sequence: primer
<400> 5
gaacttcgac atccccgagg aaatcaagca gctgcagcag ttccagaagg aggacgccgc 60
tctgaccatc 70
<210> 6
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 6
ttccgccagg actccagctc caccggttgg aacgagacca tcgtggagaa cctgctggcc 60
aacgtgtacc . 70
<210> 7
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 7
aggagaagct ggagaaggag gacttcaccc gcggcaagct gatgagctcc ctgcacctga 60
agcgctacta 70
<210> 8
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 8
ggagtacagc cactgcgcct ggaccatcgt acgcgtggag atcctgcgca acttctactt 60
catcaaccgc 70
<210> 9
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
3
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
<223> Description of Artificial Sequence: primer
<400> 9
caccacactg gactagtgga tccttatcag ttgcgcaggt agccggtcag gcggttgatg 60
aagtagaagt 70
<210> 10
c211> 70
<212> DNA
<213> Artificial Sequence
c220>
<223> Description of Artificial Sequence: primer
<400> 10
aggcgcagtg gctgtactcc ttggccttca ggtagtgcag gatgcggcca tagtagcgct 60
tcaggtgcag 70
<210> 11
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 11
ctccttctcc agcttctcct ccagcacggt cttcaggtgg ttgatctggt ggtacacgtt 60
ggccagcagg 70
<210> 12
<211> 70
<212> DNA
<213> Artificial Sequence
c220>
<223> Description of Artificial Sequence: primer
<400> 12
gagctggagt cctggcggaa gatggcgaag atgttctgca gcatctcgta gatggtcaga 60
gcggcgtcct 70
<210> 13
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
4
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
<400> 13
cctcggggat gtcgaagttc atcctgtcct tcaggcagta ctccaggcgc ccgttcagct 60
gccacaggag 70
<210> 14
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 14
caggaagccg agcaggttgt agctcatcga tagggccgtg gtgctgaagc acaggagcag 60
ggcgatctgg
<210> 15
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 15
ctgctccaga tcgccctgct cctgtgcttc agcaccacgg ccctatcgat gaagcaccag 60
caccagcatc ' 70
<210> 16
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 16
cactgcttac tggcttatcg aaattaatac gactcactat agggagaccc aagctggcta 60
gcgtttaaac 70
<210> 17
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 17
caggaagccg agcaggttgt agctcatctg ttggtgttga tgttggtgct gatgctggtg 60
5
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
ctggtgcttc 70
<210> 18
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 18
agcagggcga tctggagcag gcacttgttg gtcatggtgg cgaagcttaa gtttaaacgc 60
tagccagctt 70
<210> 19
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 19
ccgtcagatc ctaggctagc ttattgcggt agtttatcac 40
<210> 20
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 20
gagctcggta ccaagctttt aagagctgta at _ 32
<210> 21
<211> 77
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 21
gctgaacggg cgcctggagt actgcctgaa ggacaggatg aacttcgaca tccccgagga 60
aatccgccag ctgcagc ~ 77
<210> 22
6
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 22
tctccacgcg tacgatggtc caggcgcagt ggctg 35
<210> 23
<211> 70
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 23
caccacactg gactagtgga tccttatcag ttgcgcaggt agccggtcag gcggttgatg 60
aagtagaagt 70
<210> 24
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 24
catcagcttg ccggtggtgt tgtcctcctt c 31
<210> 25
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 25
gaaggaggac aacaccaccg gcaagctgat g 31
<210> 26
<211> 40
<212> DNA
<213> Artificial Sequence
<220>
7
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
<223> Description of Artificial Sequence: primer
<400> 26
cacactggac tagtaagctt ttatcagttg cgcaggtagc 40
<210> 27
<211> 47
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 27
gaggagttcg aacttccagt gccagcgcct cctgtggcag ctgaacg 47
<210> 28
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 28
cgcggatcca tatgaccaac aagtgcctg ' 29
<210> 29
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 29
cgcggatcct tatcagttgc gcag 24
<210> 30
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 30
tttaaactgg atccagccac catgaccaac aag . 33
8
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
<210> 31
<211> 63
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 31
cggccatagt agcgcttcag gtgcagggag ctcatcagct tgccggtggt gttgtcctcc 60
ttc 63
<210> 32
<211> 63
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 32
gaaggaggac aacaccaccg gcaagctgat gagctccctg cacctgaagc gctactatgg 60
ccg 63
<210> 33
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 33
ggcgtcctcc ttggtgaagt tctgcagctg 30
<210> 34
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 34
atatatccca agcttttatc agttgcgcag gtagccggt 39
<210> 35
<211> 30
<212> DNA
<213> Artificial Sequence
9
CA 02380760 2002-O1-30
WO 01/15736 PCT/DK00/00471
<220>
<223> Description of Artificial Sequence: primer
<400> 35
cagctgcaga acttcaccaa ggaggacgcc 30
<210> 36
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 36
cgcggatcca gccaccatga ccaacaagtg cctg 34
<210> 37
<211> 89
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 37
aactggatcc agccaccatg accaacaagt gcctgctcca gatcgccctg ctcctgtgct 60
tcagcaccac ggccctagcc cagagctac 89
<210> 38
<211> 92
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: primer
<400> 38
aactggatcc agccaccatg accaacaagt gcctgctcca gatcgccctg ctcctgtgct 60
tcagcaccac ggccctagcc cagatgagct ac 92