Note: Descriptions are shown in the official language in which they were submitted.
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-1-
p193 PROTEINS AND NUCLEIC ACIDS,
AND USES THEREOF
REFERENCE TO RELATED APPLICATION
This application claims the benefit of U.S. Patent Application Serial No.
60/150,266 filed August 23, 1999, which is hereby incorporated by reference in
its
entirety.
BACKGROUND
The present invention relates generally to cell physiology, and more
particularly to cell cycle regulatory proteins. Specifically, the present
invention
relates to a novel apoptosis associated protein designated p 193 and modified
forms
thereof; to nucleotide sequences encoding p 193 proteins; and to products and
processes involved in the cloning, preparation and expression of nucleotide
sequences encoding p 193.
Normal development is dependent upon an intricate balance between cell
proliferation and programmed cell death (apoptosis). Alteration of this
balance can
have significant pathophysiological consequences; tumorigenesis results when
cell
proliferation is favored whereas autoimmune and/or degenerative disorders
result
when apoptosis is favored.
In mammalian cells, apoptosis can be induced by at least two independent
regulatory pathways. The first pathway relies on direct activation of the
death
receptors (members of the tumor necrosis factor receptor superfamily, reviewed
in
Ashkenazi, A. et al. (1998) Science 281, 1305-1308). For example, activation
of
the TNFR1 or CD95 receptors initiates a signal transduction cascade primarily
through FADD (Fas-associated death domain) which rapidly activates caspase 8,
thereby initiating apoptosis. Apoptosis can also be regulated through the
activities
of Bcl-2 family members (reviewed in Adams, J.M. et al. ( 1998) Science 281,
1322-1326). The prototypical family member, Bcl-2, was originally identified
as a
gene activated by chromosomal translocation in some human lymphomas
(Tsujimoto, Y. et al. (1984) Science 226, 1097-1099; Bakhshi, A. et al.
(1985),
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-2-
Cell 41, 899-906; Cleary, M.L. et al. (1986) Cell 47, 19-28). Subsequent
analyses
have identified a family of approximately 20 proteins which share homology to
Bcl-2 at one or more domains (known as Bcl-2 Homology domains BH1 through
BH4). Functional analyses have shown that family members with the greatest
homology to Bcl-2 tend to promote cell survival while those more distantly
related
tend to promote apoptosis. The pro-apoptosis group is further subdivided into
the
Bax sub-family (which contain BH1, 2 and 3 domains, see Oltvai, Z.N. et al.
(1993) Cell 74, 609-619; Chittenden, T. et al. (1995) Nature 374, 733-736;
Kiefer,
M.C., et al. ( 1995) Nature 374, 736-739; Furrow, S.N. et al. ( 1995) Nature
374,
731-733; Hsu, Y.T. et al. ( 1997) Proc. Nat). Acad. Sci. USA 94, 3668-3672)
and
the BH3 only sub-family (which as the name implies contain only BH3 domains,
see Boyd, J.M. et al. (1994) Cell 79, 341-351; Boyd, J.M., et al., (1995)
Oncogene
11, 1921-1928; Yang, E. et al., (1995) Cell 80, 285-291; Wang, K. et al.
(1996)
Genes Dev. 10, 2859-2869; Inohara, N. et al. (1997) EMBO J. 16, 1686-1694;
Conradt, B. et al. (1998) Cell 93, 519-529; O'Connor, L. et al. (1998) EMBO J.
17,
384-395; Hegde, R. et al. ( 1998) Journal of Biological. Chemistry 273, 7783-
7786.
11-18).
Commitment to apoptosis is governed, at least in part, by the relative levels
of pro-survival and pro-apoptosis Bcl-2 family members which, in turn,
regulate
the activity of Apaf-1 (an activator of caspase 8). Thus the caspase family of
cysteine proteases are the downstream effectors of apoptosis, regardless of
the
initial regulatory pathway. Once activated, the caspases effect cell death by
initiating a proteolytic cascade which destroys cellular organelles thereby
giving
rise to distinct morphologic changes which are diagnostic for apoptosis
(reviewed
in Thornberry, N.A. et al. (1998) Science 281, 1312-1316). These include
nuclear
condensation, fragmentation of DNA at nucleosomal junctions, mitochondria)
disintegration and ultimately autolysis of the cell.
The DNA tumor virus oncoproteins have provided a more useful model
system with which to dissect the molecular regulation of cell growth and
death.
The transforming activities of these proteins (as exemplified by S V40 Large T
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-3-
Antigen and Adenovirus E 1 A) reside largely in their ability to bind to, and
thereby
alter the activity of, endogenous cell cycle and cell death regulatory
proteins
(reviewed in Ludlow, J.W. et al. (1995) Virus Research 35, 113-121; and Moran,
E. ( 1993) FASEB Journal 7, 880-885). In the case of T Antigen (T-Ag), amino
acid residues 105 through 115 are required for binding to members of the
Retinoblastoma family (RB and the related proteins p 107 and p 130, see
DeCaprio,
J.A. et al. ( 1988) Cell 54, 275-283; Ewen, M.E. et al. ( 1991 ) Cell 66, 1155-
1164;
Li, Y. et al. (1993) Genes Dev. 7, 2366-2377; and Hannon, G.J. et al. (1993)
Genes
Dev. Dec. 7, 2378-2391 ). T-Ag/RB binding blocks sequestration of E2F family
members (which are maintained in an inactive state by binding to RB). Once
released, these transcription factors activate expression of a large number of
genes
needed for S phase entry (reviewed in Nevins, J.R. ( 1992) Science 258, 424-
429;
Hatakeyama, M. et al. (1995) Prog. Cell cycle Res. 1, 9-19; and La Thangue,
N.B.
( 1996) Bioch. Soc. Trans. 24, 54-59). The discontinuous region localized
between
T-Ag amino acid residues 350 through 450 and 532 through 625 is required for
binding to p53 (Kierstead, T.D. et al. (1993) J. Virol. 67, 1817-1829). Among
other activities, p53 functions as a transcriptional co-activator of both pro-
apoptosis and growth inhibitory genes. T-Ag/p53 binding prevents
transcriptional
activation of these genes, and concomitantly inhibits their activities (Bates,
S. et al.
( 1999) Cell. c~ Mol. Life Sci. 55, 28-37; and Ko, L.J. et al. ( 1996) Genes
Dev. 10,
1054-1072).
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-4-
DESCRIPTION OF THE FIGURES
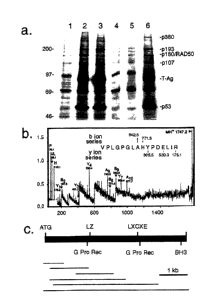
Figure 1. (a) Immune complex from metabolically labeled AT-2
cardiomyocytes generated with anti-T-AG or anti-p53 monoclonal antibodies.
p193 is present in anti-T-AG (lane 3) and anti-p53 (lanes 2 and 6) immune
complex from 35S-methionine labeled AT-2 cardiomyocytes, but not in immune
complex prepared with IgG subtype-matched nonspecific control antibodies
(lanes
1 and 5), nor in controls lacking primary antibody (lane 4). Molecular weight
standards are indicated on the left. (b) PSD MALDI mass spectrum and sequence
of a p 193 tryptic peptide. The b and y ions and immonium ions that were
detected
are shown. (c) Schematic diagram of p 193 protein and cDNAs. The positions of
several structural motifs are shown. Horizontal black lines indicate the
relative
position of the cDNA clones.
Figure 2. (a) Deduced amino acid sequence of p 193. Underlined
sequences correspond to the peptides identified by PSD mass spectrometry. Bold
sequence corresponds to the BH3 domain homology. (b) Comparison of the BH3
domain in p 193 and several other apoptosis regulatory proteins.
Figure 3. (a) p193 binds to T-Ag in NIH-3T3 cells. Protein prepared from
cells co-transfected with CMV-p 193myc (which encodes a p 193 protein
harboring
a c-terminal myc epitope tag) and CMV-T-Ag (which encodes SV40 T-Ag) was
reacted with the indicated antibodies, and the resulting immune complex was
analyzed by Western blotting using anti-myc and anti-T-Ag antibodies. Tfx,
transfection; Tot. Pro., total protein; IP, immune precipitation. (b) In vitro
translated p 193 binds to recombinant T-Ag. Radiolabeled in vitro translated p
193
was mixed with recombinant T-Ag, and then reacted with the indicated
antibodies.
The resulting immune complexes were displayed on a polyacrylamide gel and
transferred to nylon membranes. p 193 was visualized by autoradiography, and T-
Ag was visualized by Western blot. (c) Northern blot analysis of p193
expression
in adult mice. Total RNA ( 10 micrograms) prepared from the indicated tissues
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-5-
was probed with a full-length p193 cDNA. The integrity of the RNA samples was
confirmed by staining the Northern blots with methylene blue (lower panel).
Figure 4. p 193 binds to the N-terminus of T-Ag. The schematic diagram
depicts the T-Ag constructs used in the mapping experiments. These products
were translated in vitro and mixed with in vitro translated full length p 193.
Immune complex generated with anti-T-Ag antibody PAb419 was resolved on a
polyacrylamide gel and visualized by autoradiography. Construct 1-92myc
encoded a myc epitope-tag at the C-terminus.
Figure 5. p 193 promotes apoptosis. (a) DNA content distribution for NIH-
3T3 cells expression CMV-f3GALmyc at 40 hrs post-transfection. (b) DNA
content distribution for NIH-3T3 cells expressing CMV-p193myc at 40 hrs post-
transfection. (c) Time course of cell death and DNA synthesis in synchronized
cultures of NIH-3T3 cells transfected with CMV-p 193myc. The % survival of
CMV-p193myc expressing cells (squares), the thymidine labeling index for CVM-
pl93myc transfected cells (circles), and the thymidine labeling index for non-
transfected NIH-3T3 cells on the same chamber slides (diamonds) are shown. (d
and e) p193myc immune localization (signal corresponds to anti-myc epitope tag
immune reactivity) in NIH-3T3 cells at 8 and 14 hrs, respectively, post serum
replenishment. (f) Co-expression of Bcl-XL or T-Ag antagonizes p193-induced
apoptosis. NIH-3T3 cells were transfected as indicated; the total number of p
193
positive cells at 68 hours post-transfection is shown. Also note that cells
transfected with the p193deltaBH (which harbors a deletion spaning the p193
BH3
domain) are viable.
Figure 6. (a and b) pl93myc (panel a, signal corresponds to anti-p193myc
immune reactivity) and T-Ag (panel b, signal corresponds to anti-T-Ag immune
reactivity) are sequestered in the cytoplasm in cells co-expressing CMV-
p193myc
and CMV-T-Ag. (c) The percentage thymidine positive cells with cytoplasmic T-
Ag immune reactivity is plotted against the number of hours post S-phase (as
determined by pulse-chase experiments). (d and e) Autoradiographic and anti-T-
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-6-
Ag immune cytologic analysis, respectively, of two 3H-thymidine positive
daughter cells after cytokinesis (from the 10 hrs chase time point in panel
c).
Figure 7. (A). NIH-3T3 colony growth assay with expression constructs
encoding p193 in the sense (CMV-p193s) and anti-sense (CMV-p193as)
orientation. Expression vector lacking insert (CMV-null) was used as a
control.
(B). RT-PCR analysis from cells expressing the CMV-null vector, from cells
expressing the CMV-p193as vector, or from non-transfected NIH-3T3 cells.
Figure 8. (A). Structure of CMV expression vectors with nested p193 C-
terminal truncations, as described in Example 4. (B). Colony growth assay
using
expression constructs of Figure 8A, as described in Example 4. (C) DNA
fragmentation studies confirming that p 193dn encodes dominant negative
activity
which blocks MMS-induced apoptosis, as described in Example 4.
Figure 9. Schematic diagram of MHC-p 193dn transgene used to generate
transgenic mice, as further described in Example 5.
Figure 10. Northern blot of transgene expression in MHC-p 193dn
transgenic mouse lines designated 4, 5, 6, 7, 9, 10 and 13, as further
described in
Example 5.
Figure 11. Heart sections showing myocardial damage in response to
isoproterenol infusion in control and MCH-p 193dn transgenic mice, obtained as
described in Example 6. Sections were stained with sirius red (which reacts
with
collagen to produce a dark signal) and counterstained with fast green (which
reacts
with cardiomyocytes to produce a light signal).
Figure 12. ES cell-derived cardiomyocyte colony growth assay showing
the effects of p53dn, pl93dn, and ElA gene expression, alone or in
combination,
as further described in Example 7.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCTNS00/23161
Figure 13. (A). Western blot analysis of protein prepared from the ES cell-
derived cardiomyocyte colony growth assay shown in Figure 12 with anti-ElA or
anti-T-Ag antibodies; (B). DNA fragmentation studies showing that ElA
expression in the absence of co-expression of both p l3dn and p 193dn induced
apoptosis (see Example 7).
Figure 14. p 193 is expressed in G1/S of the cell cycle (see Example 8).
(A) Plot of % tritiated thymidine postive cells over time showing that the NIH-
3T3
culture studies were well synchronized (B). Western analysis of p193
expression
over the same time period, as described in Example 8. The Western analyses
indicate that p193 is expressed during G1/S.
Figure 15. Colony growth assay demonstrating that isoproternol induces
growth in cardiomyocytes which co-express 193dn and p53dn, as described in
Example 9.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
_g_
SUMMARY OF THE INVENTION
A feature of the present invention is the identification and characterization
of an apoptosis associated protein, designated p 193. p 193 is a S V40 T-Ag
binding
protein and appears to be a new member of the BH3 only pro-apoptosis family.
This is supported by the observation that p193 expression promoted a prompt
apoptotic response in NIH-3T3 cells. Immune cytologic analysis indicated that
p 193 is a cytoplasmic protein, and that co-expression of T-Ag resulted in the
cytoplasmic localization of both proteins. p 193-induced apoptosis occurs in
G,,
and pulse chase experiments revealed that T-Ag is also localized in the
cytoplasm
(albeit transiently) at the same point of the cell cycle. The data are
consistent with
the conclusion that T-Ag possesses an anti-apoptosis activity, independent of
p53
sequestration, which is actuated by T-Ag/p193 binding in the cytoplasm.
Accordingly, one aspect of the present invention concerns a method for
modifying the cell cycle of a cell which involves modulating the level of p193
protein within the cell and/or interfering with the p 193 protein signal
transduction
pathway in the cell. Particularly, increasing the wild-type pro-apototic p 193
activity can be used to induce apoptosis, and decreasing the level of pro-
apoptotic
p 193 activity in the cell (including interfering with the p 193 signal
transduction
pathway) can be used to suppress apoptosis and/or promote cellular
proliferation.
Increases in pro-apoptotic p193 activity can be achieved, for example, by
expression of introduced DNA encoding a pro-apoptotic p 193 protein. Decreases
in pro-apoptotic p193 activity can be achieved, illustratively, by decreasing
the
level of expression of the native p193 of the cell (e.g. by antisense
technology),
and/or by interference with the pathway through which the native p 193 acts,
for
example by the introduction of a dominant negative p 193 protein which
antagonizes at least a portion of the biological function of the native p 193
protein.
In certain aspects of the invention, methods for modifying the cell cycle of a
cell
include decreasing the level of expression of the native p 193 protein of the
cell
and/or interfering with the p193 pathway, in conjunction with decreasing the
level
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-9-
of expression of p53 protein in the cell or interfering with the p53 pathway,
and/or
in conjunction with increasing the level of expression of ElA protein in the
cell.
In another aspect, the present invention provides an expression vector
including nucleic acid encoding a p 193 polypeptide. Such vectors can be used
in
inventive methods to genetically transduce host cells, and in the case of pro-
apoptotic p193 polypeptides to induce apoptosis in the cells. In the case of
p193
polypeptides with a dominant negative character, such transduction may be used
to
effectively suppress apoptosis or induce proliferation.
Another preferred embodiment of the invention provides an isolated p 193
protein, preferably an isolated, recombinant p193 protein. Such proteins can
be
combined with an appropriate pharmaceutically acceptable carrier to produce
pharmaceutical compositions, also constituting a part of the present
invention.
Such proteins can also be used in the preparation of inventive antibodies to p
193.
The present invention also concerns a method for producing a p 193 protein,
comprising culturing a host cell having introduced DNA encoding a p 193
protein
under conditions suitable from expression of said introduced DNA.
The present invention provides a newly characterized apoptosis associated
protein designated p 193, and novel modified p 193 proteins, including those
exhibiting a dominant negative character; nucleotide sequences encoding such
p 193 proteins; products and processes involved in the cloning, preparation
and
expression of nucleotide sequences encoding p 193 proteins; methods and
materials
for modifying the cell cycle in cells, for example regulating apoptosis and/or
proliferation of cells; and methods for screening for pharmacological or other
chemical agents for effect on cell cycle which involve assessing their impact
on
p 193 or its signal transduction pathway in cells. Additional embodiments as
well
as features and advantages of the invention will be apparent from the
descriptions
herein.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
- 10-
DESCRIPTION OF THE PREFERRED EMBODIMENTS
For the purpose of promoting an understanding of the principles of the
invention, reference will now be made to certain preferred embodiments thereof
and specific language will be used to describe the same. It will nevertheless
be
understood that no limitation of the scope of the invention is thereby
intended,
such alterations, further modifications and applications of the principles of
the
invention as described herein being contemplated as would normally occur to
one
skilled in the art to which the invention relates.
As disclosed above, the present invention provides a novel apoptosis
associated protein designated p193 and modified forms thereof; nucleotide
sequences encoding p193 proteins; and products and processes involved in the
cloning, preparation and expression of nucleotide sequences encoding p 193
proteins.
SEQ. LD. NO. 1 shows the nucleotide sequence and deduced amino acid
sequence (see also SEQ. LD. NO. 2) for mouse p 193 as utilized in the Examples
herein. SEQ. LD. NO. 3 shows the nucleotide sequence and deduced amino acid
sequence (see also SEQ. LD. NO. 4) for human p193. In this regard, the term
"nucleotide sequence," as used herein, is intended to refer to a natural or
synthetic
sequential array of nucleotides and/or nucleosides, and derivatives thereof.
The
term amino acid sequence is intended to refer to a natural or synthetic
sequential
array of amino acids and/or derivatives thereof. The terms "encoding" and
"coding" refer to the process by which a nucleotide sequence, through the
mechanisms of transcription and translation, provides the information to a
cell
from which a series of amino acids can be assembled into a specific amino acid
sequence to produce a polypeptide.
It will be understood that the present invention also encompasses the use of
nucleotide sequences and amino acid sequences which differ from the specific
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-11-
p193 sequences disclosed herein, but which have substantial identity thereto
and
exhibit pro-apoptotic or proliferative activities as identified herein. Such
sequences will be considered to provide p 193 nucleic acid and p 193 proteins
for
use in the various aspects of the present invention. For example, nucleic acid
sequences encoding variant amino acid sequences are within the scope of the
invention. Modifications to a sequence, such as deletions, insertions, or
substitutions in the sequence, which produce "silent" changes that do not
substantially affect the functional properties of the resulting polypeptide
molecule
are expressly contemplated by the present invention. For example, it is
understood
that alterations in a nucleotide sequence which reflect the degeneracy of the
genetic code, or which result in the production of a chemically equivalent
amino
acid at a given site, are contemplated. Thus, a codon for the amino acid
alanine, a
hydrophobic amino acid, may be substituted by a codon encoding another less
hydrophobic residue, such as glycine, or a more hydrophobic residue, such as
valine, leucine, or isoleucine. Similarly, changes which result in
substitution of
one negatively charged residue for another, such as aspartic acid for glutamic
acid,
or one positively charged residue for another, such as lysine for arginine,
can also
generally be expected to produce a biologically equivalent product.
It has also been discovered that modifications to the p193 sequence which
substantially affect the functional properties of the resulting polypeptide
can be
made, and such changes are also expressly contemplated by the present
invention.
For example, modifications of the p193 amino acid sequence can be used to
produce dominant-negative p 193 proteins which antagonize at least a portion
of the
wild-type p 193 activity, and which lead to suppression of apoptotic activity
in the
cells and/or an enhanced proliferative capacity of the cells.
In one manner of defining the invention, nucleic acid (e.g. DNA) may be
used that has a coding sequence that differs from that set forth in SEQ. LD.
NO. 1
(nucleotides 62-5128) or SEQ. LD. NO. 3 (nucleotides 87-5183), wherein the
nucleic acid, or at least the coding portion thereof, will bind to nucleic
acid having
nucleotides 62-5128 of SEQ. LD. NO. 1 or nucleotides 87-5183 of SEQ. LD. NO.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-12-
3, or at least about nucleotides 62-3517 of SEQ. LD. NO. 1 or about
nucleotides
87-3615 of SEQ. LD. NO. 3, under stringent conditions. Such nucleic acid will
desirably encode a polypeptide having pro-apoptotic p 193 activity, or a
dominant-
negative p193 polypeptide. "Stringent conditions" are sequence dependent and
will be different in different circumstances. Generally, stringent conditions
are
selected to be about 5°C lower than the thermal melting point (Tin) for
the specific
sequence at a defined ionic strength and pH. The Tm is the temperature (under
defined ionic strength and pH) at which 50% of the target sequence hybridizes
to a
perfectly matched probe. Typically, stringent conditions will be those in
which the
salt concentration is at least about 0.02 molar at pH 7 and the temperature is
at
least about 60°C.
In another manner of defining the invention, nucleic acid may be used that
encodes a polypeptide that has an amino acid sequence which has at least about
70% identity, more preferably at least about 80% identity, most preferably a
least
about 90% identity, with the amino acid sequence set forth in SEQ. LD. NO. 2
or
in SEQ. LD. NO. 4, or with at least one significant length (i.e. at least 40
amino
acid residues) segment thereof, and which polypeptide possesses a pro-
apoptotic
p 193 activity or a dominant-negative p 193 character. The polypeptide may,
for
example, have an amino acid sequence which has at least about 70% , 80%, or
90% identity with at least about amino acid residues 1-1152 of SEQ. LD. NO. 2
or
about amino acid residues 1- 1173 of SEQ. LD. NO. 4, or with amino acid
residues
1-1689 of SEQ LD. NO. 2 or amino acid residues 1-1698 of SEQ. LD. NO. 4. Such
polypeptides, especially when a functional pro-apoptotic protein is desired,
will
preferably include the characteristic p193 BH3 domain occurring at residues
1566
to 1572 of SEQ. LD. NO. 2 or at residues 1575 to 1581 of SEQ. LD. NO. 4:
Leu Lys Ala His Gly Asp Glu
Percent identity, as used herein, is intended to mean percent identity as
determined by comparing sequence information using the advanced BLAST
computer program, version 2Ø8, available from the National Institutes of
Health,
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-13-
USA. The BLAST program is based on the alignment method of Karlin and
Altschul, Proc. Natl. Acad. Sci. USA 87:2264-68 (1990) and as discussed in
Altschul, et al., J. Mol. Biol. 215:403-10 (1990); Karlin and Altschul, Proc.
Natl.
Acad. Sci. USA 90:5873-7 ( 1993); and Altschul et al. ( 1997) Nucleic Acids
Res.
25:3389-3402. Briefly, the BLAST program defines identity as the number of
identical aligned symbols (i.e., nucleotides or amino acids), divided by the
total
number of symbols in the shorter of the two sequences. The program may be used
to determine percent identity over the entire length of the proteins being
compared.
Preferred default parameters for the BLAST program, blastp, include: ( 1 )
description of 500; (2) Expect value of 10; (3) Karlin-Altschul parameter ~, _
0.270; (4) Karlin-Altschul parameter K = 0.0470; (5) gap penalties: Existence
1 l,
Extension 1; (6) H value = 4.94e-3za; (6) scores for matched and mismatched
amino
acids found in the BLOSUM62 matrix as described in Henikoff, S. and Henikoff,
J.G., Proc. Natl. Acad. Sci. USA 89:10915-10919 (1992); Pearson, W.R., Prot.
Sci.
4:1145-1160 (1995); and Henikoff, S. and Henikoff, J.G., Proteins 17:49-61
(1993). The program also uses an SEG filter to mask-off segments of the query
sequence as determined by the SEG program of Wootton and Federhen Computers
and Chemistry 17:149-163, (1993).
In another form, nucleic acid may be used that includes a coding sequence
that has at least about 70% identity with the coding portion of the nucleotide
sequence set forth in SEQ. LD. NO. 1 (nucleotides 62 to 5128) or in SEQ. LD.
NO.
3 (nucleotides 87 to 5183), or with at least one significant length (i.e. at
least 100
nucleotides) segment thereof, and which nucleic acid encodes a polypeptide
possessing pro-apoptotic p 193 activity or dominant-negative 193 activity as
identified herein. The nucleic acid may, for example, have a coding sequence
which has at least about 70% at least about 80%, or at least about 90%,
identity
with nucleotides 62 to 5128 of SEQ. LD. NO. 1 or with nucleotides 87 to 5183
of
SEQ. LD. NO. 3, or at least with about nucleotides 62 to 3517 of SEQ. LD. NO.
1
or about nucleotides 87 to 3615 of SEQ. LD. NO. 3.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCTNS00/23161
- 14-
The p193 nucleotide sequence may be operably linked to a promoter
sequence as known in the art to provide recombinant nucleic acid useful in a
variety of applications including, for example, in the provision of vehicles
such as
vectors for functionally introducing the nucleic acid in to mammalian or other
eukaryotic cells, such as cardiomyocytes, hepatocytes, smooth muscle cells,
hemotpoietic stem cells, tumorogenic cells, and the like. As defined herein, a
nucleotide sequence is "operably linked" to another nucleotide sequence (e.g.
a
regulatory element such as a promoter) when it is placed into a functional
relationship with the other nucleotide sequence. For example, if a nucleotide
sequence is operably linked to a promoter sequence, this generally means that
the
nucleotide sequence is contiguous with the promoter and the promoter exhibits
the
capacity to promote transcription of the gene. A wide variety of promoters are
known in the art, including cell-specific promoters, inducible promoters and
constitutive promoters. The promoters may be selected so that the desired
product
produced from the nucleotide sequence template is produced constitutively in
the
target cells. Alternatively, promoters, such as inducible promoters, may be
selected that require activation by activating elements known in the art, so
that
production of the desired product may be regulated as desired. Still further,
promoters may be chosen that promote transcription of the gene in one or more
selected cell types, e.g. the so-called cell-specific promoters.
Expression vectors in accordance with the present invention can be
designed to effectively increase wild-type p 193 activity in a cell thus
inducing
apoptosis, or to interfere with wild-type p193 activity in a cell thus
suppressing
apoptosis and/or inducing proliferation. For example, expression vectors
incorporating nucleic acid encoding a pro-apoptotic p193 polypeptide can be
employed to increase apoptotic activity in a cell. On the other hand, vectors
incorporating nucleic acid encoding a modified p 193 polypeptide, for example
truncation mutants of p193 exhibiting activity consistent with dominant
negative
(p 193dn), can be used to interfere with wild-type p 193 activity and thereby
suppress apoptosis in the cell and/or induce proliferation of the cell.
Genetic
transduction of cells with vectors incorporating antisense (as) p 193
nucleotide
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-15-
sequences can also be used to effectively suppress apoptotic activity and/or
induce
proliferation in the cells. Similarly, p 193 antisense RNA may be administered
to
cells so as to decrease p 193 and apoptotic activity and/or induce
proliferation in the
cells.
In a preferred aspect of the invention, the p 193 nucleotide sequence is
operably linked to a cell-specific promoter, for example, providing for
constitutive
expression of the nucleotide sequence in a selected cell type. Illustrative
candidates for such promoters include cardiomyocyte-specific promoters such as
the a-myosin heavy chain (a.-MHC) promoter, the (3-myosin heavy chain ((3-
MHC) promoter, the myosin light chain-2V (MLC-2V) promoter, the atrial
natriuretic factor (ANF) promoter, and the like. Additional cell-specific
promoters
include liver-specific cells such as PePCK, albumin, transthyretin, and major
urinary protein (MUP). Any cell type expressing endogenous gene, and its
exressed ubiquitous, lung, heart, liver, eyes. Such constructs enable the
expression
of the p 193 nucleic acid selectively in selected tissues.
Another aspect of the invention provides recombinant nucleic acid that
includes a p193 nucleotide sequence encoding a p193 polypeptide operably
linked
to an inducible promoter. The p 193 nucleotide sequence may, for instance,
encode
a pro-apoptosis polypeptide, such that expression and induces of apoptosis in
cells,
or an apoptosis-suppressing and/or proliferation-inducing polypeptide, such
that
expression suppresses apoptosis and/or promotes cellular proliferation. Using
an
inducible promoter, expression of the polypeptide encoded by the cells
incorporating the nucleic acid can be upregulated in response to an inducing
agent.
Illustrative candidate inducible promoter systems include, for example, the
metallothionein (MT) promoter system, wherein the MT promoter is induced by
heavy metals such as copper sulfate; the tetracycline regulatable system,
which is a
binary system wherein expression is dependent upon the presence or absence of
tetracycline; a glucocorticoid responsive promoter, which uses a synthetic
sequence derived from the glucocorticoid response element and is inducible in
vivo
by administering dexamethasome (cells having the appropriate receptor); a
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
- 16-
muristerone-responsive promoter, which uses the ganodotropin-releasing hormone
promoter and is inducible with muristerone (cells having the appropriate
receptor);
and TNF responsive promoters. Additional inducible promoters which may be
used, and which are more preferred, include the ecdysone promoter system,
which
is inducible using an insect hormone (ecdysone) and provides complete ligand-
dependent expression in mammals; the ~i-GAL system, which is a binary system
utilizing an E. coli lac operon operator and the I gene product in trans, and
a
gratuitous inducer (IPTG) is used to regulate expression; and, the RU486
inducible
system, which uses the CYP3A5 promoter and is inducible by RU486, a well
defined pharmaceutical. These and other similar inducible promoter systems are
known, and their use in the present invention is within the purview of those
skilled
in the area.
One aspect of the present invention concerns the discovery that blocking
p 193 and p53 activity (by expression of dominant negative cDNA variants)
protects against proliferation-induced apoptotic signals. This in turns
renders
cardiac myocytes responsive to the pro-proliferation signals, such signals
encoded
for example by E 1 A. Therapeutic approaches may be adopted which promote
controlled regeneration of cardiac tissue, or alternatively controlled
proliferation of
engrafted cardiomyocytes, which rely upon the use of regulatable promoters to
drive expression of the dominant negative cDNAs in addition to the growth
promoting gene. An alternative approach may rely on pharmaceutical blockade of
the p53 and/or the p 193 pathways, in conjunction with expression of growth-
promoting genes in combination with a regulatable promoter. For example,
Gudkov and colleagues (Science (1999) 285; 1733-1737) developed an agent
which inhibits p53-dependent transcriptional activation and apoptosis. Similar
reagents to block p 193 activity are readily generated by one skilled in the
art. This
approach has the advantage of an intrinsic cell cycle check-point in the event
of
illegitimate promoter activity (e.g. induction of promoter activity in the
absence of
inducing agent). Specifically, if the activity of one or both of the pro-
apoptotic
genes are blocked pharmaceutically, and that pharmaceuticals) is withdrawn
after
regenerative growth is completed, illegitimate activation of the growth
promoting
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
- 17-
gene would result in the apoptotic death of the cell because the anti-
apoptotic
activity would not be present. This approach may be used to provide a
clinically
safer modality to effect controlled cardiomyocyte proliferation in vivo.
An additional or alternative safeguard approach would encompass inclusion
of a conditionally lethal gene in the expression cassette, as for example the
well-
known Herpes simplex virus thymidine kinase (HS V-TK) gene. The HS V-TK
gene can incorporate normal nucleotides as well as the nucleotide analog
gancyclovir at a high efficiency whereas mammalian thymidine kinase does not
incorporate gancyclovir into cells at high efficiency. Incorporation of
gancyclovir
is cytotoxic. Thus, in this mode of operation, illegitimate activation of the
regulatable promoter would result in expression of the anti-apoptosis (e.g.
p53dn
and p193dn) and pro-growth (e.g. ElA) genes, as well as the HSV-TK gene.
Inappropriately growing cells (e.g. those where illegitimate promoter activity
has
occurred) can be eliminated by simple treatment with gancyclovir.
The present invention also concerns vectors which incorporate a p 193
nucleotide sequence and which are useful in the genetic transduction of cells
in
vitro or in vivo. A variety of vector systems are suitable for these purposes.
These
include, for example, viral vectors such as adenovirus vectors as disclosed
for
example in Franz et al., Cardiovasc. Res. 35(3):560-566 (1997); Inesi et al.,
Am. J.
Physiol. 274 (3 Pt. 1):C645-653 (1998); Kohout et al., Circ. Res. 78(6):971-
977
(1996); Leor et al., J. Mol. Cell Cardiol. 28(10):2057-2067 (1996); March et
al.,
Clin. Cardiol. 22( 1 Suppl. 1 ):I23-29 ( 1999); and Rothman et al., Gene Ther.
3( 10):919-926 ( 1996). Adeno-Associated Virus (AAV) vectors are also
suitable,
and are illustratively disclosed in Kaptlitt et al., Ann. Thora. Surg.
62(6):1669-
1676 (1996); and Svensson et al., Circulation 99(2):201-205 (1999). Additional
viral vectors which may be used include retroviral vectors (see e.g. Prentice
et al.,
J. Mol. Cell Cardiol. 28(1):133-140 (1996); and Petropoulos et al., J. Virol.
66(6):3391-3397 (1992)), and Lenti (HN-1) viral vectors as disclosed in
Rebolledo et al., Circ. Res. 83(7):738-742 (1998). A preferred class of
expression
vectors will incorporate the p 193 nucleic acid operably linked to a
cardiomyocyte-
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
- 18-
specific promoter, such as one of those identified above. Still further, AAV
vectors are highly compatible for use in transfection of myocardial and other
cells
and tissue, and are preferred from among those identified above.
In accordance with the invention, cells can also be genetically transduced
with p 193 nucleic acid in vitro or in vivo using liposome-based transduction
systems. A variety of liposomal transduction systems are known, and have been
reported to successfully deliver recombinant expression vectors to a variety
of
cells. Illustrative teachings may be found for example in R.W. Zajdel, et al.,
Developmental Dynamics. 213(4):412-20 (1998); Y. Sawa, et al., Gene
Therapy.5( 11 ):1472-80 ( 1998); Y. Kawahira, et al., Circulation 98( 19
Suppl):II262-7; discussion II267-8 ( 1998); G. Yamada, et al., Cellular &
Molecular Biology 43(8):1165-9 ( 1997); M. Aoki, et al., Journal of Molecular
&
Cellular Cardiology 29(3):949-59 ( 1997); Y. Sawa, et al., Journal of Thoracic
&
Cardiovascular Surgery 113(3):512-8; discussion 518-9 (1997); and I. Aleksic,
et
al., Thoracic & Cardiovascular Surgeon 44(2):81-5 (1996). Thus, liposomal
recombinant expression vectors including p 193 DNA can also be utilized to
tranduce cells in vitro and in vivo for the purposes described herein.
Nucleic acid constructs can be used for example to introduce nucleotide
sequences encoding a p193 protein into cells in vivo or in vitro, to achieve a
level
of intracellular p 193 activity that is increased relative to the native level
of the
cells. Such increased activity can induce apoptosis in the cells. Induction of
apoptotic activity can be evidenced, for example, by cell death and other
characteristic morphological changes such as cell shrinkage and nuclear
condensation and fragmentation. Alternatively or in addition, purified (e.g.
purified recombinant) p 193 protein may be introduced into cells to increase p
193
activity (e.g. by fusogenic liposomes or other macromolecular delivery
systems),
or the cells can be treated with pharmacologic agents which increase p 193
activity,
to provide increased apoptotic activity to the cells.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-19-
Nucleic acid constructs can also be used to introduce modified p 193
nucleotide sequences into cells in vivo or in vitro, wherein the sequences
provide
characteristics of a dominant negative gene and effectively antagonize wild-
type
p 193 activity, resulting in as for example a suppression of apoptosis and/or
an
increase in the proliferative capacity of the cells. In a similar approach, a
dominant
negative p 193 protein or another molecule can be introduced into the cells
which
interferes with or antagonizes wild-type p 193 activity, and thereby
suppresses
apoptosis and/or induces proliferation in the cells. Illustratively, vectors
incorporating antisense (as) p 193 nucleotide sequences can be used, and/or
small
synthetic organic molecules serving as pharmacologic agents can be used, to
effectively interfere with the expression of or the activity of wild-type p
193
protein.
The present invention makes available methods which can be applied in vitro
or in vivo for research, therapeutic, screening or other purposes. Methods for
the in
vitro culture of cells expressing introduced p 193 DNA (in sense or antisense
orientation) can be used, for example, in the study and understanding of the
cell
cycle, in screening for chemical or physical agents which modulate p 193
activity
or other aspects of the cell cycle, or in the culture of cells having
suppressed
apoptotic activity and/or increased proliferative potential for subsequent
engraftment into mammals, including humans.
Cells to be cultured in accordance with the invention can be derived from a
variety of sources. For example, they may be harvested from a mammal for
culture and subsequent engraftment into that mammal (autografts) or another
mammal of the same species (allografts) or a different species (xenografts).
Cardiomyocyte or other cells may also be derived from the differentiation of
stem
cells such as embryonic stem cells, somatic stem cells or other similar
pluripotent
cells. General methodology for such derivations is disclosed in U.S. Patent
Nos.
5,602,301 and 5,733,727 to Field et al. In this regard, when so derived, the
genetic
modification to incorporate the p 193 nucleic acid may take place at the stem
cell
level, for instance utilizing one or more vectors to introduce the p 193
nucleic acid
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-20-
operably linked to a tissue-specific promoter, and nucleic acid enabling the
selection of a target cell type from other cells differentiating from the stem
cell
and/or at a differentiated level e.g., including a selectable marker gene
operably
linked to a tissue-specific promoter. Nucleic acid enabling selection of
transduced
from non-transduced stem cells may also be used in such strategies. Such
selection
of the stem and/or differentiated cell types may be achieved, illustratively,
utilizing
a gene conferring resistance to an antibiotic (e.g. neomycin or hygromycin) or
other chemical agent operably linked to an appropriate promoter.
Using stem-cell derived cells, the genetic modification to incorporate the p
193
and potentially other nucleic acid may also occur after differentiation of the
stem
cells. For example, a differentiated cell population enriched in
cardiomyocytes or
another target cell type, for instance containing 90% or more of the target
cell type,
may be transformed with a vector having p 193 nucleic acid (especially
antisense or
including a dominant negative mutation) operably linked to a promoter
(optionally
tissue specific), as described above. The same or a different vector may also
be
used to introduce other functional nucleic acid to the cells, for example
providing a
reporter gene and/or selectable marker, or providing for the expression of a
growth
factor andlor another cell cycle regulatory protein.
Illustratively, in certain embodiments of the invention, decreasing the level
of
p 193 protein or interfering with the p 193 signal transduction pathway can be
used
in conjunction with other means of effecting the cell cycle. For example, such
modifications of p193 and/or its pathway (effected e.g. by an introduced
antisense
p 193 nucleic acid or a nucleic acid having a dominant negative mutation) can
be
used in combination with a p53 nucleic acid (especially antisense or a
dominant
negative mutation), an E 1 A nucleic acid, or a combination of the two. Still
further,
such modifications of p 193 and/or its pathway may be used in conjunction with
other methods of relaxing or facilitating the G,/S transit, for example by
manipulating key regulators at the restriction point of the cell cycle such as
inhibiting RB family members, overexpressing D-type cyclin or cyclin-dependent
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-21
kinase activities, inhibiting cyclin-dependent kinase inhibitors,
overexpressing
downstream targets, and the like.
In one mode of carrying out the invention, left ventricular, right
ventricular,
left atrial, or right atrial cardiomyocytes, or a mixture of some or all of
these, may
be genetically modified in vitro to incorporate anti-apoptotic andlor
proliferative
p193 nucleic acid using a suitable vector as disclosed above. Cells to be
genetically transduced in such protocols may be obtained for instance from
animals
at different developmental stages, for example fetal, neonatal and adult
stages.
Suitable animal sources include mammals such as bovine, porcine, equine, ovine
and murine animals. Human cells may be obtained from human donors or from a
patient to be treated. The modified cardiomyocytes may thereafter be implanted
into a mammal, for example into the left or right atrium or left or right
ventricle, to
establish a cellular graft in the mammal. Implantation of the cells may be
achieved
by any suitable means, including for instance by injection or catheterization.
In
addition to the p 193 nucleic acid, the cells may also be modified in vitro to
contain
other functional nucleic acid sequences which can be expressed to provide
other
proteins, for example or one or more additional cell cycle regulatory
proteins. In
one preferred embodiment, cells are modified with nucleic acid encoding p 193
and
with nucleic acid encoding at least one other cell cycle regulatory protein,
for
example combining forced expression of p193 and p53 dominant (Mowat, M.,
Nature Vol. 314, p. 633-636 ( 1985); Munroe, D.G. Mol. Cell. Biol., Vol. 10,
3307-3313 ( 1990) so as to suppress apoptosis in the cells.
Cells for culture, and potential implantation, may also be obtained from a
transgenic animal (especially mammal) expressing introduced p 193 nucleic
acid.
Using known techniques, transgenic animals which harbor introduced p 193
nucleic
acid in essentially all of their cells can be raised, and used as sources for
harvesting
culturable cells (e.g. cardiomyocytes), tissues or organs, or may be used as
animal
models for research or screening purposes. For instance, transgenic bovine,
porcine, equine, ovine or murine animals may be used as sources for cells,
tissues
or organs, or as animal models for study. Illustratively, transgenic animals
having
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-22-
reduced levels of wild-type p 193 protein and/or expressing an introduced
dominant
negative p193 protein, can be used as a source for apoptotically-suppressed
and/or
proliferatively enhanced cells, tissue or organs, which will be protected
against
fibrosis or other similar damage. Such materials will thus possess significant
advantages for use in transplantation into other animals, such as humans.
The present invention also provides for the genetic modification of cells in
vivo to increase p 193 activity (using pro-apoptotic protein) or decrease p
193
activity (using p 193dn) in the cells (impacting transduction pathway). An
expression vector containing the p 193 nucleic acid, for instance one as
described
above, may be delivered to tissue of a recipient mammal, to achieve
transduction
of cells in the tissue. In preferred modes, the p 193 nucleic acid in such
vectors will
be operably linked to a tissue-specific promoter, for instance a cardiomyocyte-
specific promoter. The delivery of the vector can be suitably achieved, for
instance, by injection, catheterization, or infusion into the blood stream, or
by other
known means. It will be understood that any mode of delivery which enables the
establishment of transduced cells within the recipient mammal is contemplated
as
being within the present invention. A single delivery of the vector may be
used, or
multiple deliveries nearly simultaneous or over time may be used, in order to
establish a substantial population of transduced cells within the recipient.
The
transduced cells will then express the encoded p 193 polypeptide, for instance
under the control of a constitutive, inducible or tissue-specific promoter,
and
thereby exhibit a suppressed or induced level of apoptosis.
The implantation of cells cultured in vitro or the delivery of the vector for
in
vivo genetic transduction may be directed to a selected site or sites within
the
recipient. For example, in the case of apoptosis-suppressed and/or
proliferatively
enhanced cardiomyocyte engraftment or corresponding in vivo transduction, such
site or sites may be in the left or right atrium or left or right ventricle of
the
recipient, or any combination of these. Commonly, the implantation or delivery
site or sites will occur in the left or right ventricle of the recipient. The
sites) may,
for instance, be ones) in which there is a need for additional viable cells,
for
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-23-
example in a damaged or diseased area of the heart such as in cases of
myocardial
infarcts and cardiomyopathies. The sites) may also be targets for the delivery
of
other proteins such as growth factors, e.g. nerve growth or angiogenic
factors, via
expression in the grafted or in viva transduced cells.
Cellular engraftment and/or in viva genetic modification in accordance with
the invention can be used, for example, to deliver therapy to mammals,
including
humans. A variety of ex viva cellular transplantation and implantation
techniques
and gene therapy techniques are thus contemplated as forming a part of the
invention. For example, these techniques may be used to provide cells in the
mammal having a reduced level of wild-type p 193 protein and/or having a
disrupted or partially disrupted p 193 signal transduction pathway, the cells
thereby
exhibiting decreased apoptotic activity and/or an enhanced proliferative
capacity.
Illustratively, such an approach may be used to target an improvement or
protection of the contractile function of the heart of the patient, for
example in the
treatment of contractile losses due to infarcts or cardiomyopathies. They may
also
be used to target an improvement and/or protection of the function of other
tissue
or organs in the patient, for example the liver or lungs of the patient. The
use of a
p193 protein having a dominant negative mutation will be especially
advantageous
for such purposes. In addition, the delivery of pro-apoptotic p 193 protein to
cells,
for example by in viva genetic transduction with an appropriate p 193 nucleic
acid
and consequent expression of the pro-apoptotic protein, can be used to promote
apoptosis in cells in which apoptosis is desired, for example in the case of
inappropriately proliferative cells.
The present invention also provides access to antibodies having specificity
to one or more epitopes present on the p 193 peptide, or an idiotype on the p
193
(see e.g. Figure 14 and accompanying discussion in Examples). Such antibodies
can be polyclonal or monoclonal, and can be made with the p 193 polypeptide or
fragment thereof as the immunogen. In this regard, the term "antibody" (Ab) or
"monoclonal antibody" (Mab) as used herein is meant to include intact
molecules
as well as fragments thereof capable of binding an antigen. Antibodies to p
193 can
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-24-
be used, for example, to detect the presence of the p 193 protein in a human
or
other mammalian tissue sample. This may involve contacting the sample with a
detectably labeled antibody and detecting the label, thereby establishing the
presence of the p193 protein in the sample. Detection can be carried out by
imaging in vivo. The p193 protein can also be detected by known immunoassay
techniques, including, for example, RIA, ELISA, etc., using appropriate
antibodies
according to the invention.
Antibodies of the invention can be prepared by any of a variety of known
methods. For example, cells expressing the p 193 protein can be administered
to an
animal in order to induce the production of serum containing polyclonal
antibodies
that are capable of binding the p 193 protein. For example, the p 193 protein
or
fragment thereof is chemically synthesized and purified by HPLC to render it
substantially free of contaminants. Such a preparation is then introduced into
ari
animal in order to produce polyclonal antisera of high specific activity.
Polyclonal antibodies can be generated in any suitable animal including, for
example, mice, rabbits or goats. The p 193 immunogenic peptide or fragment
thereof can be injected by itself or linked to an appropriate immunoactivating
carrier.
Monoclonal antibodies can be prepared in various ways using techniques
well understood by those having ordinary skill in the art. For example,
monoclonal
antibodies can be prepared using hybridoma technology (Kohler, et al., Nature
256:495 ( 1975); Kohler, et al., Eur. J. imninol. 6:511 ( 1976); Kohler, et
al., Eur. J.
Immunol. 6:292 ( 1976); Hammerling, et al., In: Monoclonal Antibodies and T-
Cell
Hybridomas, Elsevier, N.Y., pp. 563-681 (1981)); Roger H. Kennett, et al.,
Eds.,
Monoclonal Antibodies - Hybridomas: A New Dimension in Biological Analysis,
Plenum Press ( 1980). In general, such procedures involve immunizing an animal
with the present p 193 protein, or a fragment thereof. Splenocytes from such
animals are extracted and fused with a suitable mycloma cell line. Any
suitable
mycloma cell line may be employed in accordance with the present invention.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-25-
After fusion, the resulting hybridoma cells are selectively maintained in HAT
medium, and then cloned by limiting dilution as described by Wands, et al.,
Gastroenterol. 80:225-232 ( 1981 ). The hybridoma cells obtained through such
a
selection are then assayed to identify clones which secrete antibodies capable
of
binding the p 193 protein.
These and other general techniques in relation to the production and use of
antibodies will be apparent to and readily utilizable by those of ordinary
skill in the
art to produce and use p 193 antibodies of the present invention.
For the purpose of promoting a further understanding of the present
invention and its advantages, the following specific Examples are provided. It
will
be understood that these Examples are illustrative, and not limiting, of the
invention.
EXAMPLES
EXAMPLES I -3: METHODS
EXAMPLE 1
Isolation and Sequence Analysis of p193 Proteins.
AT-2 cardiomyocytes were homogenized in 20 ml of NET, pre-cleared
with protein A sepharose beads, and mixed with anti-T-Ag monoclonal antibody
PAb419 (90 min., 4°C). Immune complexes were collected with
Protein A-
sepharose, displayed on polyacrylamide gels and visualized by staining with
Coomassie Brilliant Blue. The region of the gel containing p193 was excised,
alkylated with isopropylacetamide, and digested with F-trypsin (0.2 pg
trypsin,
37°C, 17 hrs) as described ( Shevchenko, A., Wilm, M., Vorm, O., and
Mann, M.
(1996) Anal. Chem. 68, 850-858). The peptides were then extracted with 5%
formic acid/50% acetonitrile and separated on a C 18 0.32 x 100 mm capillary
column (LC Packing, Inc.). An aliquot of each of the isolated HPLC fractions
was
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-26-
applied to a pre-made spot of matrix (0.5 ml of 20 mg/ml a-cyano-4-
hydroxycinammic acid + 5 mg/ml nitrocellulose in 50% acetone/50% 2-propanol)
on the target plate. Ions were formed by matrix-assisted laser
desorption/ionization with a nitrogen laser, 337 nm. Spectra were acquired
with a
PerSeptive Biosystems Voyager Elite time-of-flight mass spectrometer, operated
in
linear delayed extraction mode. Subsequently, fragment ions for selected
precursor masses were obtained from post-source decay (PSD) experiments
(Kaufman, R., Kirsch, D. and Spengler, B. ( 1994) International J. Mass Spec.
and
lon Proc. 131, 355-385). Automated protein sequencing was performed on a
model 470A Applied Biosystems sequencer equipped with an on-line PTH
analyzer using modified cycles as described (Henzel, W.J., Grimley, C.,
Bourell,
J.H., Billeci, T.M., Wong, S.C. and Stults, J.T. (1994) Methods: A companion
to
Methods in Enzymology 6, 239-247). Peaks were integrated with Justice
Innovation software using Nelson Analytic 760 interfaces. Sequence
interpretation
was performed on a DEC 5900 (Henzel, W.J., Rodriguez, H., and Watanabe, C.
(1987) J. Chromatogr. 404, 41-52).
EXAMPLE 2
Isolation and Molecular Analysis of p 193 cDNAs.
p 193 cDNAs were isolated from an adult heart cDNA library generated
from C3HeB/FeJ inbred mice (Kim, K.K., Daud, A.L, Wong, S.C., Pajak, L., Tsai,
S.C., Wang, H., Henzel, W.J., and Field, L.J. (1996) J. Biol. Chem. 271, 29255-
29264). Plaque hybridizations, phage DNA isolation and subcloning were
performed using standard methodologies (Sambrook, J., Fritsch, E.F., and
Maniatis, T. ( 1989) Molecular cloning, Cold Spring Harbor Laboratory Press,
Cold
Spring Harbor, N.Y.). Sequence was determined for both strands of the cDNA
clone examined using the dideoxy chain terminating approach (Sequenase, United
States Biochemicals, Cleveland OH).
To demonstrate p 193/T-Ag binding, a full length p 193 cDNA was
subcloned into the pcDNA3.l/Myc-His expression vector (Invitrogen, Carlsbad
CA) such that the epitope tag was incorporated into the C-terminus of the
molecule (construct designated CMV-p 193myc). A T-Ag cDNA was subcloned
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-27-
into pcDNA3.1 expression vector (which lacks the epitope tag; construct
designated CMV-T-Ag). For IP/Western analyses, NIH-3T3 cells (ATCC,
Rockville MD) were co-transfected with CVM-5-Ag and CMV-p193myc using the
calcium phosphate approach (Sambrook, J., Fritsch, E.F., and Maniatis, T.
(1989)
Molecular cloning, Cold Spring Harbor Laboratory Press, Cold Spring Harbor,
N.Y.). Protein (1 mg) prepared from the transfected cells was reacted with an
anti-
T-Ag (PAb419), an anti-myc (9E10, Santa Cruz Biotech.), or an IgG subtype-
matched non-specific antibody (anti-GST, Pharmacia), and the resulting immune
complexes were subjected to Western blot analysis. 100 p.g of total protein
from
non-transfected and transfected cells were included as controls. The blots
were
probed with an anti-myc (9E10) or anti-T-Ag (PAb416) antibody, and signal was
developed using the ECL method. To demonstrate p 193/T-Ag binding in vitro,
3sS_methionine labeled in vitro transcription/translation (TNT kit, Promega)
product obtained from a full length p193 cDNA subcloned into pBluescript IISK
(Stratagene, LaJolla CA) was mixed with 1.2 pg of recombinant SV40 T-Ag
(Molecular Biology Resource), and reacted with anti-T-Ag (PAb419) or an IgG
subtype-matched nonspecific control antibody (anti-MAP kinase #D2, Santa Cruz
Biotech.). Immune complex was then visualized via autoradiography (p193) or
Western blotting (T-Ag) as described above.
For Northern blots, 10 pg of total RNA was denatured with glyoxal,
displayed on agarose gels, transferred to Genescreen (NEN) and reacted with a
nick-translated full-length p193 cDNA as described (Sambrook, J., Fritsch,
E.F.,
and Maniatis, T. ( 1989) Molecular cloning, Cold Spring Harbor Laboratory
Press,
Cold Spring Harbor, N.Y.). For mapping the p193 binding site on T-Ag, in vitro
translation products from a full length p 193 cDNA clone and various t-Ag
deletion
constructs were mixed, and immune complex was generated with an N-terminal
specific anti-T-Ag monoclonal antibody (PAb419). Immune complex was then
visualized by autoradiography. The various T-Ag deletion constructs were
generated via PCR amplification using oligonucleotide primers incorporating
stop
codons or base pair substitutions as indicated in the text. The fidelity of
each
construct was confirmed by sequence analysis.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-28-
EXAMPLE 3
Functional Analyses of p 193
For FACS analyses, NIH-3T3 cells transfected with either CMV-(3-
GALmyc or CMV-p193myc were labeled with Hoechst, trypsinized and fixed in
5% acetic acid in ethanol, rehydrated in PBS and reacted with FITC-conjugated
anti-myc antibody (9E 10, Oncogene Sciences) as described (Esser, C.,
Gottlinger,
C., Kremer, J., Hundeiker, C., and Radbruch, A. ( 1995) Cytometry 21, 382-386;
Brown, D.R., Thomas, C.A., and Deb, S.P. (1998) EMBO J., 17, 2513-2525). The
DNA content of FTTC-positive cells was then analyzed on a Becton Dickinson
FACS-PLUS instrument. Immune cytologic analyses were as described (44), and
images were captured using a BioRad laser scanning confocal microscope or
photographed directly using conventional light or fluorescent microscopy.
For the cell death time course experiment, NIH-3T3 cells synchronized by
two rounds of serum depletion (starvation media contained 0.1 % FBS in DMEM)
were transfected with either CMV-p193myc or CMV-(3GALmyc using Lipofectin
(Gibco-Life Sciences, Grand Island NY) for 24 hrs. The cultures were then
rinsed
with PBS, and cultured for an additional 6 hrs in starvation media. Media
containing 3H-thymidine (26 Ci/mmol, Amersham, Buckinghamshire, England)
and 10% FBS in DMEM was then added, and cells were processed for
autoradiography and immune cytology at various points thereafter as described
(Klug, M.G., Soonpaa, M.H., Koh, G.Y., and Field, L.J. (1996) J. Clin. Invest.
98,
216-224). To localize T-Ag during the cell cycle, subconfluent culture of AT-2
cells in DMEM containing 10% FBS received a 40 minute pulse of 3H-thymidine.
The cells were then rinsed, and then cultured with DMEM containing 10% FBS.
The cells were then processed for autoradiography and immune cytology at
various
points thereafter as described (Klug, M.G., Soonpaa, M.H., Koh, G.Y., and
Field,
L.J. ( 1996) J. Clin. Invest. 98, 216-224). For the colony growth assay, NIH-
3T3
cells transfected with CMV-null, CMV-p193s or CMV-p193as were selected in
6418 for 15 days (the expression vectors also encoded a CMV-neo' cassette).
The
dishes were then fixed and stained with gentian violet.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-29-
EXAMPLES 1-3: RESULTS
Cloning of p193
To identify the T-Ag binding proteins in cardiomyocytes, immune
complexes were generated using protein prepared form 35S-methionine labeled AT
2 cells, a cell line derived form the transgenic heart tumors (Daud, A.L,
Lanson,
N.A., Jr., Claycomb, W.C., and Field, L.J. (1993) Am. J. Physiol. 264, H1693-
700). Proteins with apparent molecular weights of 380, 193 and 120 kd (see
Figure 1 a) were detected in immune complex generated with either anti-T-Ag
(PAb419, lane 3) or anti-p53 (PAb421 and PAb246, lanes 2 and 6, respectively)
monoclonal antibodies. These proteins were not present in immune complexes
generated with IgG subtype-matched nonspecific control antibodies (DYS1, lane
1;
PAb240, lane 5), nor in controls lacking primary antibody (lane 4). Previous
studies have shown that the 120 kd protein is p 107 (45, 46), and that the 180
kd
protein (present only in PAb421 anti-p53 immune complex) is the murine
homologue of RAD50, a protein involved in the repair of dsDNA breaks in yeast
(Kim, K.K., Daud, A.L, Wong, S.C., Pajak, L., Tsai, S.C., Wang, H., Henzel,
W.J.,
and Field, L.J. ( 1996) J. Biol. Chem. 271, 29255-29264). The 380 kd protein
has
not yet been characterized.
To clone p 193, large scale anti-T-Ag immune complex preparations were
resolved on polyacrylamide gels and visualized by Coomassie blue staining. The
region containing p 193 was excised, digested with trypsin in situ,
fractionated by
HPLC and analyzed by mass spectroscopy using post source decay (PSD,
Figure 1b). Information obtained from the PSD experiment was used to search a
protein sequence database using a modified version of Frag-Fit. The search
indicated that p 193 was homologous to a previously identified open reading
frame
of unknown function isolated from a human immature myeloid cell line (Nomura,
N., Nagase, T., Miyajima, N., Sazuka, T., Tanaka, A., Sato, S., Seki, N.,
Kawarabayasi, Y., Ishikawa, K., and Tabata, S. (1994) DNA Res. 1, 251-262).
Reverse transcriptase-polymerase chain reaction was used to generate a short
cDNA clone spanning the region homologous to the largest p 193 peptide. This
clone was then used to screen an adult mouse heart cDNA library.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-30-
Six overlapping cDNA clones were ultimately obtained (Figure lc).
Sequence analysis revealed an open reading frame 5067 nucleotides in length
which encoded a protein of 1689 amino acid residues and with a deduced
molecular weight of 192,346 d (Figure 2a). All of the p 193 proteolytic
peptides
identified in the PSD experiment were present in the deduced amino acid
sequence
of the cDNA clones. Analysis of the sequence revealed the presence of a
leucine
zipper at amino acid residues 593-619, an LXCXE motif (the consensus sequence
for binding to retinoblastoma family proteins, see Gibson, T.J., Thompson,
J.D.,
Blocker, A., and Kouzarides, T. ( 1994) Nucleic. Acids. Res. 22, 946-952) at
amino
acid residues 1052-1056, and two G protein receptor motifs at amino acid
residues
1566-1572: No other BH domains were present in p193. Sequence alignment of
the BH3 domain of p193 and those of the BH3 only family members is shown in
Figure 2b.
Cloned p 193 binds to T-Ag
To confirm that these clones encoded the 193 kd T-Ag binding protein, a
full length p193 cDNA was subcloned into a CMV-promoted expression vector
which incorporated a short myc-epitope tag at the C-terminus. The resulting
clone,
designated CMV-p 193myc, was co-transfected alone with CMV-T-Ag (an
expression construct encoding T-Ag) into NIH-3T3 cells. Protein prepared from
cells 24 hrs. post transfection was subjected to Immune Precipitation/Western
blot
analyses using anti-T-Ag and anti-myc antibodies (Figure 3a). p193myc was
detected in anti-T-Ag immune complex, and T-Ag was detected in anti-myc
immune complex. Neither protein was present in immune complex generated with
an AgG subtype-matched nonspecific control antibody. Immune Precipitation
analyses of mixtures of in vitro translated p 193 and recombinant T-Ag were
also
performed (Figure 3b). Radiolabeled p 193 was present in immune complex
generated with anti-T-Ag antibody, but not in immune complex generated with an
IgG subtype-matched nonspecific control antibody, confirming that the 193 kd T-
Ag binding protein was successfully cloned. Northern blots revealed a somewhat
restricted pattern of p193 expression in adult mouse tissues (Figure 3c).
Relatively
high levels of p 193 mRNA were detected in the heart, as might be anticipated
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-31 -
given that the protein was originally identified in cell lines derived from
cardiac
tumors.
p 193 binds to the N-terminus of T-Ag
To identify the region of T-Ag which binds to p 193, in vitro translation
products from a series of T-Ag deletion constructs were mixed with full length
in
vitro translated p 193, and immune complexes generated with anti-T-Ag antibody
were resolved on polyacrylamide gels and visualized by autoradiography
(Figure 4). p 193 was present in immune complex generated with T-Ag mutants
with deletions encompassing as much as amino acid residues 147 through 708,
indicating that the p 193 binding site resides within T-Ag amino acid residues
1
through 147. In contrast, p 193 was not present in immune complex generated
with
a T-Ag mutant in which amino acid residues 92 through 708 where deleted,
indicating that the C-terminal boundary of the binding site lies within T-Ag
amino
acid residues 107 and 108 which disrupt binding of RB family members did not
effect p 193 binding (Figure 4, construct 1-147 ~ RB). Thus, p 193 binds to
the N-
terminal region of T-Ag distinct from the RB family member binding site.
Expression of p193 promotes apoptosis
To determine the effects of p 193expression on cell growth, NIH-3T3 cells
were transfected with either CMV-(3GALmyc (an expression construct encoding (3-
galactosidase with a myc-epitope tag) or CMV-p 193myc. At 48 hrs. post-
transfection, FACS analyses using a FITC-conjugated anti-myc antibody revealed
that most cells expressing CMV-~iGALmyc had a normal 2C DNA content
(Figure Sa; the inset shows a CMV-(3GALmyc transfected cell: image on the left
shows anti-myc immune fluorescence (see arrow), image on right shows nuclear
morphology via Hoechst fluorescence (see arrow)). In contrast the
preponderance
of cells expressing CMV-p193myc exhibited hypodiploid DNA content, indicative
of apoptotic cell death (Figure Sb). Visual inspection of the cultures
confirmed
that the bulk of the transfected cells were dying and had markedly condensed
chromatin (Figure SB inset: image on the left shows anti-myc immune
SUBSTITUTE SHEET (RULE Z6)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-32-
fluorescence (see arrow), image on right shows nuclear morphology via Hoechst
fluorescence (see arrow)). Thus expression of p 193 can induce apoptosis.
To determine at what point in the cell cycle p 193 induced cell death,
serum-starved NIH-3T3 cells were transfected with CMV-p193myc. Media
containing serum and 3H-thymidine was then added, and the cultures were
processed for anti-myc immune cytology and autoradiography at various time
points thereafter. Most cells expressing CMV-p193myc were dead by 20 hrs. post-
serum replenishment (Figure Sc, trace with square symbols), and DNA synthesis
never reinitiated in these cells (Figure Sc, trace with diamond symbols. This
suggests that p193-induced apoptosis occurs during G,. In contrast, the
preponderance of non-transfected cells on the same chamber slide reinitiated
DNA
synthesis by 14 hrs. post-serum replenishment (Figure Sc trace with circle
symbols), thus establishing the fidelity of the synchronization protocol. In
control
experiments cells expressing CMV-~3GALmyc reinitiated DNA synthesis at a rate
comparable to the non-transfected cells (not shown), indicating that transgene
expression per se did not impact on cell cycle progression or viability.
p193myc
immune reactivity in the synchronized cultures was initially localized
uniformly
throughout the cytoplasm, but became restricted to the perinuclear region
prior to
the onset of cell death (Figures Sd and e, respectively). Bax, a well
characterized
pro-apoptosis protein, undergoes a similar cytoplasmic to perinuclear
redistribution
during apoptosis (Hsu, Y.T., Wolter, K.G., and Youle, R.J. (1997) Proc. Natl.
Acad. Sci. USA 94, 3668-3672). Finally, it is of interest to note that p193myc
expressing cells are viable if cell cycle progression is blocked; >90% of p193
expressing cells were viable at 40 hrs. post transfection if maintained under
low
serum conditions. This suggests that some degree of cell cycle progression is
needed to actuate apoptosis.
Cell death induction by BH3 only family members can be antagonized by
co-expression of pro-survival members of the Bcl-2 family. To determine if
p193
shares this trait, NIH-3T3 cells were transfected with CMV-p193myc alone or co-
transfected with CMV-p193myc and CMV-Blc-XL (a construct encoding human
Bcl-XL). The preponderance of cells transfected with p193 alone were dead at
68 hrs. post transfection, whereas co-transfection with Bcl-XL markedly
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-33-
antagonized p193-induced apoptosis (Figure 5f). In control experiments,
virtually
no viable cells were seen following co-transfection with CMV-pl93myc and
CMV-GFP (an expression construct encoding green fluorescent protein),
indicating
that co-expression of two CMV-driven constructs does not abate p 193-induced
cell
death (Figure 5f).
To establish the importance of the BH3 domain in p193-induced apoptosis,
the CMV-pl93myc expression construct was modified such that amino acid
residues 1563 through 1576 were deleted (VRILKAHGDEGLHV). This
modification resulted in the deletion of the BH3 motif (amino acid residues
1566-
1572,), and the resulting construct was designated CMV-p193[delta symbol]BH.
NIH-3T3 cells transfected with CMV-p193[delta symbol]BH were viable (Figure
5f). Indeed, the survival was similar to that obtained for cells co-
transfected with
CMV-p 193 plus CMV-Bcl-XL. Thus the BH3 domain is required for p 193-
mediated apoptosis.
T-AQ is transiently localized in the c~plasm during M and G~
Given that p193 was originally identified as a T-Ag binding protein, and
that T-Ag is a nuclear oncoprotein, the cytoplasmic/perinuclear localization
of
p 193myc was somewhat surprising. To further address this paradox, NIH-3T3
cells co-transfected with CMV-193myc and CMV-T-Ag were examined. Survival
was greatly enhanced in cells co-expressing p 193 and T-Ag (Figure 5f),
indicating
that, like Bcl-XL, T-Ag can antagonize p 193-induced apoptosis. Moreover
immune cytologic analyses indicated that p 193myc and T-Ag co-localized to the
cytoplasm in the majority (ca. 63%) of the co-transfected cells (Figures 6a
and
brespectively). In contrast, co-transfection with CMV-(3GALmyc and CMV-T-Ag
did not result in prominent cytoplasmic T-Ag immune reactivity (not shown).
These results raised the possibility that p 193-T-Ag binding might normally
occur
in the cytoplasm.
Cytoplasmic T-Ag immune reactivity has previously been noted in mitotic
cells (Stenman, S., Zeuthen, J., and Ringertz, N.R. (1975) International.
Journal.
Of. Cancer 15, 547-554; Davis, D. and Wynford-Thomas, D. (1986) Experimental.
Cell Research. 166, 94-102). A pulse chase experiment was used to determine
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-34-
how long T-Ag persists in the cytoplasm of AT-2 cardiomyocytes. Sub-confluent
cultures received a 40 min. pulse of 3H-thymidine (to mark cells in S-phase)
followed by a chase with radioisotope free media. The cultures were processed
for
anti-T-Ag immune cytology and autoradiography at various time points
thereafter.
Significant percentages of the thymidine positive cells exhibited cytoplasmic
T-Ag
immune reactivity from 4 hrs. through 10 hrs. post S-phase (Figure 6c). In
contrast, 3H-thymidine positive cells with mitotic figures were only observed
at 4
and 6 hrs. post S-phase. Thus, cytoplasmic T-Ag localization persisted through
cytokinesis and well into G,. This point is further illustrated by the
presence of
~H-thymidine positive daughter cells with cytoplasmic T-Ag immune reactivity
at
8-12 hrs. post S-phase (Figures 6d and e). Thus T-Ag is transiently located in
the
cytoplasm at the same point of the cell cycle (G,) when p193-induced cell
death
occurs.
Loss of p 193 activity_promotes proliferation
The data presented above indicate that forced expression of p 193 promotes
apoptosis prior to the onset of S-phase. A colony growth assay employing a p
193
antisense construct (CMV-p193as) was used to determine the consequences of
diminished p193 expression. Transfection of NIH-3T3 cells with CMV-p193as
resulted in markedly increased colony size as compared to transfection with
CMV-
null (a control expression vector lacking insert, see Figure 7A). Northern
blots of
parallel cultures indicated that p193 transcripts were markedly diminished in
cultures transfected with the anti-sense construct (data not shown). As
expected,
transfection with CMV-193s (an expression vector encoding p193 in the sense
orientation) yielded no visible colonies (Figure 7A), consistent with the pro-
apoptotic activity of p 193 noted above.
To confirm that expression of the antisense construct effected levels of the
endogenous p193 transcript, RT-PCR analyses were performed (see Figure 7B).
The amplification primers were selected to permit co-amplification of a
fragment
of the beta-actin transcript and a fragment of the p 193 transcript present in
the
endogenous gene but not in the expression vector. The relative ratio of the
beta-
actin amplification products and the p 193 amplification products provides a
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-35-
quantitative assessment steady-state levels of endogenous p 193 transcripts.
RNA
was prepared from cells expressing the CMV-null vector, from cells expressing
the
CMV-p193as vector, or from non-transfected NIH-3T3 cells. The relative level
of
the p 193 amplification products are reduced in RNA from cells expressing the
CMV-p193as construct vs. the control cells, indicating that the antisense
intervention was successful at decreasing steady state levels of the
endogenous
transcripts (Figure 7B). Note however that p193 expression was not completely
blocked.
EXAMPLES 1-3: DISCUSSION
We have shown that p 193, a T-Ag binding protein present in the AT-2
cardiomyocyte tumor cell line, is a new member of the BH3 only pro-apoptosis
gene family. Like other BH3 only proteins, p193-induced apoptosis can be
antagonized by co-expression of pro-survival Cl-2 family members (in our case,
Bcl-X, was tested). p193 differs markedly in size as compared to other BH3
only
family members; the next largest family member, BID, is only 21.95 kd (Wang,
K.,
Yin, X.M., Chao, D.T., Milliman, C.L., and Korsmeyer, S.J. (1996) Genes Dev.
10,
2859-2869). Co-expression of T-Ag antagonizes p 193-induced apoptosis in
transiently transfected cells, and results in the cytoplasmic sequestration of
both
proteins. Moreover, T-Ag is localized in the cytoplasm of AT-2 cardiomyocytes
during G 1, the same point of the cell cycle where p 193 induces apoptosis.
These
data are consistent with the notion that T-Ag/p193 binding in the cytoplasm
may
modify or abrogate p193 activity in SV40 transformed cells.
If p193 binding is important for T-Ag mediated transformation, we would
anticipate that mutations at and/or near the p193 binding site would diminish
T-Ag
transforming activity. Indeed, previous mutational analyses have identified
transformation activity at the N-terminus of T-Ag. For example, Kohrman and
Imperiale (Kohrman, D.C. and Imperiale, M.J. (1992) J. Virol. 66, 1752-1760)
demonstrated that amino acid residues 1-108 were required to effectively
transform
B2-1 cells, and that a ca. 185 kd protein bound to this region of T-Ag.
Moreover,
binding between T-Ag and the 185 kd protein was not disrupted by point
mutations
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-36-
abrogating the binding of RB family members. Given the similarity in molecular
weight and binding specificity, p 193 may be the same protein as p 185.
Other studies have demonstrated that mutations at T-Ag amino acid
residues 1-82 (Marsilio, E., Cheng, S.H., Schaffhausen, B., Paucha, E., and
Livingston, D.M. (199) J. Virol. 65, 5647-5652), 3-35 (Zhu, J., Rice, P.W.,
Gorsch,
L., Abate, M., and Cole, C.N. (1992) J. Virol. 66, 2780-2791), and 17-27
(Srinivasan, A., McClellan, A.J., Vartikar, J., Marks, L, Cantalupo, P., Li,
Y.,
Whyte, P., Rundell, K., Brodsky, J.L., and Pipas, J.M. ( 1997) Mol. Cell.
Biol. 17,
4761-4773) all impact upon transforming activity in selected cell types. Some
of
these mutants are thought to disrupt the N-terminal J domain, a sequence motif
which functions as a DnaJ molecular chaperone (Srinivasan, A., McClellan,
A.J.,
Vartikar, J., Marks, L, Cantalupo, P., Li, Y., Whyte, P., Rundell, K.,
Brodsky, J.L.,
and Pipas, J.M. (1997) Mol. Cell. Biol. 17, 4761-4773; Campbell, K.S.,
Mullane,
K.P., Aksoy, LA., Stubdal, H., Zalvide, J., Pipas, J.M., Silver, P.A.,
Roberts, T.M.,
Schaffliausen, B.S., and DeCaprio, J.A. (1997) Genes Dev. 11, 1098-1110). DnaJ
binds to members of the 70 kd heat shock protein family, and this complex
facilitates correct protein folding, formation of mufti-protein complexes, and
protein transport across intracellular membranes (Gething, M.J. and Sambrook,
J.
( 1992) Nature 355, 33-45). Although our data indicate that the C-terminal
boundary of the p193 binding site resides between T-Ag amino acids 92 through
147, the N-terminal boundary of the binding site is not yet mapped. Mutations
encompassing residues upstream of amino acid 92 could alter p 193/T-Ag
binding,
by direct disruption to the binding domain or by altering the tertiary
structure of T-
Ag. Confirmation of the important of p 193 binding for T-Ag transforming
activity
requires precise mapping of the binding site followed by assessment of
transforming activity with appropriately mutated T-Ag expression constructs.
Finally, given the relative proximity of the p 193 and RB family member
binding
sites, it will be of interest to determine if RB, p 107 and/or p 193
sterically compete
for T-Ag binding. Such a mechanism could account for the absence of RB in anti-
T-Ag immune precipitates from the myocardial cell lines, despite the presence
of
hypophosphorylated RB in total protein prepared from these cells (Figure 1,
see
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-37-
also Kim, K.K., Soonpa, M.H., Daud, A.L, Koh, G.Y., Kim, J.S., and Field, L.J.
( 1994) J. Biol. Chem. 269, 22607-22613).
p193 also appears to be unique among the BH3 only family members with
respect to its ability to bind to T-Ag. However it is of interest to note that
the BH3
only proteins Bik and BNIP-3 (as well as Bax and Bak, pro-apoptosis proteins
containing BH1, BH2 and BH3 domains) are able to bind to adenoviral E1B 19K
protein (Furrow, S.N., White, J.H., Martinou, L, Raven, T., Pun, K.T.,
Grinham,
C.J., Martinou, J.C., and Brown, R. (1995) Nature 374, 731-733; Boyd, J.M.,
Malstrom, S., Subramanian, T., Venkatesh, L.K., Schaeper, U., Elangovan, B.,
D'Sa-Eipper, C., and Chinnadurai, G. (1994) Cell79, 341-351; Boyd, J.M.,
Gallo,
G.J., Elangovan, B., Houghton, A.B., Malstrom, S., Avery, B.J., Ebb, R.G.,
Subramanian, T., Chittenden, T., Lutz, R.J., and et al. (1995) Oncogene 11,
1921-
1928; Han, J., Sabbatini, P., Perez, D. Rao, L., Modha, D., and White, E.
(1996)
Genes Dev. 10, 461-477). It is thought that the anti-apoptotic activity of the
E1B
19K protein is due at least in part to binding with pro-apoptosis Bcl-2 family
members (White, E. ( 1995) Current Topics in Micro. & Immuno. 34-58). Previous
studies have identified a T-Ag anti-apoptotic activity at amino acid residues
525
through 541 which appeared to act independently of p53 sequestration (Conzen,
S.D., Snay, C.A., and Cole, C.N. ( 1997) J. Virol. 71, 4536-4543). These
authors
noted a significant degree of sequence homology between this region of T-Ag
and
amino acid residues 77 through 93 in E1B 19K as well as Bcl-2 amino acid
residues 133 through 151. Although these observations suggest that the binding
activity at T-Ag amino acid residues 525-542 might be functionally similar to
E1B
19K protein sequestration of pro-apoptosis proteins, experiments aimed at
establishing direct binding of T-Ag to Bak were unsuccessful (Conzen, S.D.,
Snay,
C.A., and Cole, C.N. (1997) J. Virol. 71, 4536-4543).
The results from the antisense transfection experiment indicated that loss of
p 193 activity is associated with marked growth enhancement in NIH-3T3 cells.
The increase in growth rate is in excess to that which we would anticipate
from
simple inhibition of apoptosis in the NIH-3T3 cells, which occurs somewhat
infrequently under the growth conditions employed. In support of this,
preliminary
experiments have shown that cells expressing the CMV-p193as construct exhibit
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-38-
higher DNA synthesis labeling indices as compared to cells expressing control
constructs (Tsai, unpublished results). This observation is consistent with
the
notion that p 193 may function at a cell cycle checkpoint, and that transit
through
the checkpoint is accelerated in the absence of p 193 activity. This
hypothesis is
supported in part by the serum starvation experiment described above, which
indicated that at least a limited degree of cell cycle progression is required
for
actuation of the p 193-mediated cell death program. Thus p 193 is only able to
trigger cell death after transit through a specific point in G~ (i.e. the
presumed cell
cycle check point), and accumulation of the protein in itself is not harmful
to the
cell. The observation that cytoplasmic T-Ag localization occurs during the
same
point of the cell cycle lends additional credence to this notion. Further
insight into
the molecular pathway of p 193 must await the generation of additional loss of
function models.
Our efforts to characterize p 193 were motivated in part by the hope of
identifying potential therapeutic targets with which to engender regenerative
growth in diseased hearts. In this regard it is of interest to note that
transfection of
primary cardiomyocyte cultures with ElA or E2F-1 results in a prompt apoptotic
response which is only partially abated by co-expression of E 1 B, Bcl-2 or
abrogation of p53 activity (Kirshenbaum, L.A. and Schneider, M.D. ( 1995) J.
Biol.
Chem. 270, 7791-7794; Kirshenbaum, L.A., Abdellatif, M., Chakraborty, S., and
Schneider, M.D. ( 1996) Dev. Biol. 179, 402-411; Liu, Y. and Kitsis, R.N. (
1996) J.
Cell Biol. 133, 325-334; Agah, R., Kirshenbaum, L.A., Abdellatif, M.,
Truong, L.D., Chakraborty, S., Michael, L.H., and Schneider M.D. ( 1997) J.
Clin.
Invest. 100, 2722-2728; Bishopric, N.H., Zeng, G.Q., Sato, B., and Webster,
K.A.
( 1997) J. Biol. Chem. 272, 20584-20594). In contrast, transformation with T-
Ag
results in sustained cardiomyocyte proliferation. This observation suggests
that, in
cardiomyocytes, T-Ag possesses an anti-apoptotic activity which is lacking in
ElA
and E2F-1. Given that p193 is a pro-apoptotic T-Ag binding protein, and that T-
Ag expression does not elicit an apoptotic response in cardiomyocytes, it will
be of
interest to determine if abrogation of p 193 activity can antagonize E 1 A
and/or
E2F-1 induced cardiomyocyte apoptosis.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-39-
Abrogation of p 193 activity may also have a cardioprotective effect under
pathophysiological conditions which promote cardiomyocyte apoptosis.
Numerous descriptive studies have established the presence of apoptotic
cardiomyocytes in a variety of cardiovascular diseases including dilated
cardiomyopathy, ischemic cardiomyopathy, arrhythmogenic right ventricular
dysplasia, acute myocardial infarction, myocarditis, allograft rejection, and
preexcitation syndromes (reviewed in Haunstetter, A. and Izumo, S. (1998)
Circulation Research 82, 1111-1129). In particular apoptosis and resulting
cardiac
remodeling may contribute to the onset of dilated cardiomyopathy and heart
failure
(reviewed in Anversa, P., Leri, A., Beltrami, C.A., Guerra, S., and Kajstura,
J.
(1998) Lccb. Invest. 78, 767-786). Studies in transgenic mice have implicated
a
number of signal transduction pathways, including the IL-6 cytokine
family/gp130/LIF receptor (Hirota, H., Chen, J., Betz, U.A., Rajewsky, K., Gu,
Y.,
Ross, J., Jr., Muller, W., and Chien, K.R. ( 1999) Cell 97, 189-98), the TNF-
a/TNFRI (Kubota, T., McTiernan, C.F., Frye, C.S., Slawson, S.E., Lemster,
B.H.,
Koretsky, A.P., Demetris, A.J., and Feldman, A.M. ( 1997) Circulation Research
81, 627-635; Bryant, D., Becker, L., Richardson, J., Shelton, J., Franco, F.,
Peshock, R., Thompson, M., and Giroir, B. ( 1998) Circulation 97, 1375-1381 ),
catecholamine/Gsalpha (Geng, Y.J., Ishikawa, Y., Vatner, D.E., Wagner, T.E.,
Bishop, S.P., Vatner, S.F., and Homcy, C.J. ( 1999) Circulation Research 84,
34-
42), and cAMP/CREB (Fentzke, R.C., Korcarz, C.E., Lang, R.M., Lin, H., and
Leiden, J.M. ( 1998) J. Clin. Invest. 101, 2415-2426) cascades. The role to
which
p 193 may participate in these processes remains to be established.
In summary, the data presented here indicate that p 193 is a new member of
the BH3 only pro-apoptosis gene family. p193 promotes cell death during G,,
prior to the onset of DNA synthesis. T-Ag is localized in the cytoplasm during
the
same phase of the cell cycle, and co-expression of T-Ag antagonizes p193-
induced
cell death and results in the cytoplasmic localization of both proteins. p 193
binds
to the N-terminus of T-Ag in a region which contributes to transforming
activity in
some cell types. Collectively, these results suggest that T-Ag possesses an
anti-
apoptosis activity, independent of p53 sequestration, which is actuated by T-
Ag/p 193 binding in the cytoplasm.
SUBSTITUTE SKEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-40-
EXAMPLE 4
Characterization of a Dominant-Negative p193 Mutation
Colony growth assay in NIH-3T3 cells indicate that decreased p 193 activity
as a consequence of anti-sense expression results in increased rates of cell
growth
(Figure 7). A priori, expression of dominant negative variants of p193 should
also
result in increased rates of cell growth. We generated a nested series of p
193
cDNAs harboring progressively greater C-terminal truncations. The cDNAs were
subcloned into a CMV expression vector. The structure of the p 193 variants
are
depicted in Figure 8A. The expression vectors also carried a neomycin-
resistance
cassette. NIH-3T3 cells were transfected with the various expression vectors,
and
the cells were cultured in the presence of 6418. After 15 days of selection
the
cultures were fixed and stained with gentian violet. Representative cultures
of
cells transfected with the various constructs are shown in Figure 8B. Cells
transfected with the CMV-null vector represent the negative control (this
reflects
the rate of growth in the absence of any positive or negative cell cycle
regulators,
see culture plate A). Consistent with the pro-apoptotic activity of p193, no
colonies were observed in cultures transfected with full-length p 193 (amino
acid
residues 1-1689; culture plate B). A slight enhancement in cell growth was
detected in cells transfected with a vector expressing p 193 amino acid
residues 1
through 1342 (culture plate C). Marked growth enhancement was observed in
cells
transfected with a vector expressing p 193 amino acid residues 1 through 1152
(culture plate D). Little or no growth enhancement was observed in cells
transfected vectors expressing p193 molecules 1-912, 1-309, and 1-243 (culture
plates E-G, respectively), or with a vector expressing a p193 molecule where
only
the BH3 domain was deleted (culture plate F). These data indicate that
expression
of p193 amino acid residues 1-1152 promotes growth in NIH-3T3 cells, a trend
which is also observed with expression of p 193 anti-sense constructs (see
Figure
7). Based on this, sequences encoded by p193 amino acid residues 1-1152 have
been designated "p 193dn", for p 193 dominant negative. The greater effect of
the
p 193dn on cells growth as compared to the p 193 antisense constructs likely
reflects
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-41 -
the fact that expression of the antisense construct does not completely
eliminate
endogenous p 193 transcripts (Figure 7).
The above-described experiments provide a preliminary characterization of
sequence modifications which bestow a dominant negative phenotype on p 193 (as
evidenced by the property of bestowing enhanced growth and anti-apoptotic
activity in NIH-3T3 cells, and blocking apoptosis in cardiomyocytes). Further
delineations of the p 193 amino acid residues responsible for these
characteristics is
easily accomplished by one skilled in the art. For example, fine scale
deletions
encompassing the regions defined in the experiments described above would
further delineate the amino acids required to bestow the dominant negative
phenotype. The use of amino acid substitutions which retain gross protein
structure but inhibit specific amino acid interactions are readily performed
with
generic molecular biology techniques. The NIH-3T3, ES-derived cardiomyocyte
growth assays and targeted cardiac expression in transgenic mice, as described
in
other sections of this patent application, provide the requisite experimental
endpoints with which to characterize the modified p 193 constructs.
To further confirm that p 193dn encodes dominant negative activity, we
tested its ability to block apoptosis in response to treatment with methyl
methanesulfonate (MMS). NIH-3T3 cells were transfected with a CMV-null
expression construct, or a CMV-p193dn expression construct, and stable cell
lines
were generated. The cells were then incubated in growth medium supplemented
with MMS (0 mM, 0.1 mM, 0.5 mM or 1 mM) for 3 hrs. at 37°C. Cells were
then
harvested and apoptosis was measured by determining the degree of DNA
fragmentation (nucleosomal cleavage of DNA is diagnostic for apoptosis).
Extensive fragmentation is apparent in DNA prepared from the CMV-null control
cells cultured in the presence of 1 mM MMS; in contrast no DNA fragmentation
is
apparent DNA prepared from CMV-p 193dn cells following 1 mM MMS treatment
(see Figure 8C). These data indicate that the p193dn construct blocks MMS
induced apoptosis in NIH-3T3 cells, and supports the notion that this variant
encodes dominant negative activity.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-42-
EXAMPLE 5
Generation of TransQenic Mice Expressin~~p193dn in the Heart
Transgenic mice were generated to examine the potential cardioprotective
effects of p193dn on the heart. Cardiac expression was targeted using the a-
cardiac myosin heavy chain (MHC) promoter. The MHC promoter consisted of
4.5 kb of 5' flanking sequence and 1 kb of the gene encompassing exons 1
through
3 up to but not including the initiation codon (Gulick, J., A. Subramaniam, J.
Neumann, and J. Robbins ( 1991 ) Isolation and characterization of the mouse
cardiac myosin heavy chain genes. Journal of Biological Chemistr~~ 266:9180-
9185). A cDNA encoding p193dn was inserted downstream of the promoter,
followed by the SV40 early region transcription terminator (SV40 nucleotide
residues #2586-2452, see Reddy, V. B., B. Thimmappaya, R. Dhar, K. N.
Subramanian, B. S. Zain, J. Pan, P. K. Ghosh, M. L. Celma, and S. M. Weissman
( 1978) The genome of simian virus 40. Science 200:494-502.). The resulting
transgene was designated MHC-p 193dn. A schematic diagram of the transgene is
presented in Figure 9. To generate transgenic mice, the transgene DNA was
digested with restriction enzymes to separate the MHC-p 193 sequences from the
vector, and the insert purified from an agarose gel using Geneclean glass
beads
(Bio 101, Vista CA). Purified insert DNA was microinjected into inbred
C3HeB/FeJ (Jackson Laboratories,Bar Harbor MA) zygotes using standard
methodologies [3]. The microinjected embryos were cultured in vitro to the two
cell stage, and then implanted into pseudopregnant SW/Taconic (Taconic Farms,
Germantown NY) female mice. For all surgeries, mice were anesthetized with
2.5% Avertin (0.015 ml/g bodyweight IP, Fluka Biochemicals, Ronkomkoma NY).
All manipulations were performed according to NIH and Institutional Animal
Care
and Use Guidelines.
Pups derived from the microinjected embryos were screened for the
presence of the transgene using diagnostic PCR amplification as described
(Steinhelper, M. E., K. L. Cochrane, and L. J. Field ( 1990) Hypotension in
transgenic mice expressing atrial natriuretic factor fusion genes.
Hypertension
16:301-307). 13 transgenic founder animals were obtained from the embryos
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCTNSOOI23161
- 43 -
microinjected with the MHC-pl93dn construct. Seven of the founders were
randomly selected to establish transgenic lineages. Adult heart RNA prepared
from Fl transgenic animals was used to stratify the levels of p193dn
expression
between the different lines. High levels of pl93dn transcripts were observed
in all
of the lines (designated MHC-p 193dn line 4, 5, 6, 7, 9, 10 and 13, see Figure
10):
based on these analyses line 13 was selected for additional experiments.
EXAMPLE 6
Demonstration that Expression of p 193dn is Cardioprotective in vivo
Myocardial damage in response to chronic isoproterenol infusion was
monitored in control and MHC-p 193dn transgenic mice to determine if transgene
expression was cardioprotective. Non-transgenic control and MHC-p 193dn
transgenic mice were identified and sequestered until they reached 11 weeks of
age. Continuous isoproterenol infusion was administered using implanted
osmotic
mini-pumps (model 2001, Alzet, Palo Alto CA, flow rate of 1 pl/hr) filled with
0.028 g/ml isoproterenol (dissolved in saline). After seven days of treatment
the
mice were sacrificed, the hearts harvested, cryoprotected and sectioned using
standard histologic techniques (Bullock, G. R. and P. Petrusz (1982)
Techniques in
immunocytochemistry, Academic Press, London; New York.). Heart sections
were then stained with Sirius red (which reacts with collagen to produce a
dark
signal in the images presented) and counter-stained with fast green (which
reacts
with muscle cells to produce a light signal in the images presented). The
results
are shown in Figure 11. Panels A and B depict sections of a nontransgenic
heart
after seven days of isoproterenol infusion. Abundant Sirius red staining is
apparent
throughout the ventricular myocardium (panel A shows the left ventricular
myocardium near the apex of the heart, panel B shows the ventricle myocardium
near the base of the). The dark staining is indicative of extensive fibrosis
which
resulted from isoproterenol-induced cardiomyocyte death. Panel C and D depict
similar analysis of a MHC-193dn transgenic heart after seven days of
isoproterenol
infusion. Essentially no dark staining is detected, indicating the absence of
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-44-
fibrosis in the isoproterenol-treated transgenic hearts. This result indicates
that
expression of the p193dn transgene protects the myocardium from isoproterenol-
induced fibrosis. Other studies (Communal C; Singh K; Pimentel DR; Colucci WS
( 1998) Norepinephrine stimulates apoptosis in adult rat ventricular myocytes
by
activation of the beta-adrenergic pathway, Circulation 29:98(13):1329-34) have
shown that isoproterenol treatment induces cardiomyocyte apoptosis: thus p
193dn
expression blocks cardiomyocyte apoptosis and the ensuing fibrosis.
EXAMPLE 7
Demonstration that co-expression of p 193dn and p53dn blocks
ElA induced apoptosis and promotes proliferation in ES derived cardiomyocytes
Previous studies have shown that expression of the Adenoviral ElA
oncoprotein can reactivate cell cycle ion cardiomyocytes, but this
reactivation is
immediately followed by apoptotic cardiomyocyte death (Kirshenbaum, L. A. and
M. D. Schneider. Adenovirus ElA represses cardiac gene transcription and
reactivates DNA synthesis in ventricular myocytes, via alternative pocket
protein-
and p300-binding domains, J. Biol. Chem. (1995) 270: 7791-7794). Moreover,
blocking the p53-regulated apoptotic pathway only partially rescues the
cardiomyocytes. A study was therefore employed to determine if co-expression
of
p 193dn and p53dn can block E 1 A-induced cardiomyocyte apoptosis. The
experiment utilized a previously described technique to generate enriched
cardiomyocyte cultures from differentiating ES cells (U.S. Patent Nos.
5,602,301
and 5,733,727 to Field et al.; and Klug, M. G., M. H. Soonpaa, G. Y. Koh, and
L.
J. Field ( 1996) Genetically selected cardiomyocytes from differentiating
embronic
stem cells form stable intracardiac grafts, J. Clin. Invest, 98: 216-224).
Undifferentiated ES cells were transfected with an MHC-neor/pGK-hygror
transgene alone or in combination with a MHC-E 1 A, MHC-p 193dn and/or MHC
p53dn transgenes. Transfected undifferentiated ES cells were then selected on
the
basis of hygromycin resistance.
When the hygromycin-resistant clones were sufficiently amplified, the
cultures were induced to differentiate. Once cardiomyocytes were apparent in
the
culture (as evidenced by the presence of beating cells, which usually occurs
at 8
SUBSTITUTE SKEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
- 45 -
days post-induction), the cultures were subjected to 6418 selection. Since the
neor
cassette is under the regulation of the cardiac MHC promoter, only
cardiomyocytes
survive this selection procedure. After 60 days of 6418 selection, the
cultures
were fixed and stained with PAS to permit visualization of the cardiomyocytes.
Control plates (transfected with the MHC-neor/pGK-hygror transgene
alone) gave rise to numerous colonies of beating myocytes (see the control
plate,
Figure 12). This is indicative of the normal rate of ES-derived cardiomyocyte
growth in the absence of positive or negative factors. Control dishes
transfected
with p 193dn and p53dn alone are also shown: expression of these genes
resulted in
a slight increase in cardiomyocyte yield, consistent with their anti-apoptotic
activities. In contrast, very few cardiomyocytes were observed in the plate
transfected with the MHC-ElA transgene, consistent with previous studies
(Kirshenbaum, L. A. and M. D. Schneider. Adenovirus ElA represses cardiac
gene transcription and reactivates DNA synthesis in ventricular myocytes, via
alternative pocket protein- and p300-binding domains. J. Biol. Chem. 270: 7791-
7794, 1995). Co-transfection of E 1 A and p53dn or E 1 A and p 193dn did not
result
in a marked increase in cardiomyocyte viability.
In marked contrast to these results, transfection with MHC-E 1 A; MHC
p53dn and MHC-p 193dn gave rise to numerous and substantively larger colonies
of cardiomyocytes. Cardiomyocyte colony size was much greater than that
observed for the control plates. This result indicates that the combinatorial
effect
of p53dn and pl93dn effectively and completely blocks ElA-induced apoptosis.
Moreover, the increase in colony size indicates enhanced proliferation in the
ES-
derived cardiomyocytes expressing all three transgenes. Thus, co-expression of
p193dn and p53dn blocks ElA induced apoptosis and in so doing permits ElA-
induced cell cycle activation in ES derived cardiomyocytes.
To further characterize the cardiomyocytes, protein was prepared from
representative dishes from each of the transfections depicted in Figure 12.
The
protein was then subjected to Western blot analysis with anti-ElA or anti-T-Ag
antibodies using standard protocols (Figure 13A). Importantly, no ElA protein
was detected in cells expressing E 1 A alone, or E 1 A + p53dn, or E 1 A + p
193. In
contrast, abundant levels of ElA were detected in cells expressing ElA + P53dn
+
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-46-
p193dn. This suggests that ElA is lethal in cardiomyocytes unless both the p53
and p 193 pathways are blocked. Moreover, these data indicate that the
cardiomyocytes on the E 1 A alone, or E 1 A + p53dn, or E 1 A + p 193 culture
dishes
probably arose from progenitors which did not take up (or alternatively did
not
express) the E 1 A construct. The experiment was performed on 60 day old
cultures.
To confirm that ElA expression in the absence of co-expression of both
p l3dn and p 193dn induced apoptosis, DNA prepared from 13 day old cultures
was
analyzed for the degree of DNA fragmentation (nucleosomal cleavage of DNA is
diagnostic for apoptosis) (Figure 13B). Extensive fragmentation is apparent in
DNA prepared from cells expressing E 1 A alone, or E 1 A + p53dn, or E 1 A + p
193.
In contrast, no fragtmentation was observed in cells expressing E 1 A + P53dn
+
p 193dn. These data confirm that co-expression of p53dn and p 193dn blocks E 1
A-
induced apoptosis in cardiomyocytes.
EXAMPLE 8
Demonstration that p 193 is expressed in a cell-cycle dependent fashion
Many cell cycle regulatory proteins are expressed and/or active during
discrete phases of the cell cycle. To determine if this is the case for p 193,
anti-
p 193 monoclonal antibodies were produced. A recombinant protein encoding
p193 amino acid residues 1153-1689 was used as the immunogen, and monoclonal
antibodies were raised, screened and validated using standard approaches. To
monitor p 193 expression during the cell cycle, NIH-3T3 cells were
synchronized
by two rounds of serum depletion (starvation media contained 0.1 % FBS in
DMEM). To monitor cell synchronization, some of the cells were incubated in
media containing 3H-thymidine (26 Ci/mmol, Amersham, Buckinghamshire,
England) and 10% FBS in DMEM; the cells were processed for autoradiography at
various points thereafter to monitor DNA synthesis. The preponderance of non-
transfected cells on the same chamber slide reinitiated DNA synthesis by 14
hrs.
post-serum replenishment (Figure 14A), thus establishing the fidelity of the
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
-47-
synchronization protocol. Protein prepared at similar time points from
parallel
dishes was used for Western blot analysis (Figure 14B). No p193 expression was
detected at 2, 4, or 6 hours following the addition of serum. Prominent p 193
expression was detected at 8, 10 and 12 hours post serum addition, roughly
concomitant with the onset of DNA synthesis. The levels of p 193 were markedly
reduced in subsequent time points. These data indicate that p 193 expression
is
tightly regulated during cell cycle progression, and that peak levels occur at
the
Gl/S boundary. Interestingly, this is the precise point of the cell cycle
where
forced expression of p193 induces apoptosis, and is also a point in the cell
cycle
where T-Ag is localized in the cytoplasm.
EXAMPLE 9
Data sup estin~ that blockage of p193 and p53 activity can result in a
proliferative
response to l~pertrophic stimuli.
Many forms of cardiac injury result in an initial phase of hypertrophic
growth which compensates for the loss of functional myocytes. Over time, the
hypertrophic heart can decompensate, a process which leads to cardiac dilation
and
ultimately heart failure. Cardiomyocyte apoptosis is frequently observed
during
this process. It is also well established a large number of gene products
normally
associated with cell proliferation are induced during cardiac hypertrophy (see
for
example Izumo, S. et al Proc. Natl. Acad. Sci. USA 85, 339-343; Mulvagh, S.L.
et
al., Biochem. Biophys. Res. Commun. 147, 627-636; Simpson, P.C. Annual
Review of Physiology, 51, 189-202). It is possible that hypertrophic stimuli
are in
fact mitogenic stimuli, and that in the mature cardiac myocyte the response to
such
stimuli is to first increase cell size, and then transit G1/S. Our data
clearly indicate
that two pro-apoptotic pathways (the p53 and p 193 pathways) are activated in
cardiomyocytes which are experimentally induced to proliferate. The apoptotic
response observed during the process of decompensation might result from the
initiation of cell cycle activity in the presence of active p53 and p 193
pathways.
It follows then that if the p 193 and p53 pathways are blocked, hypertrophic
stimuli might result in direct cell cycle activity. To test this,
undifferentiated ES
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
- 48 -
cells were transfected with the MHC-neor/pGK-hygror transgene in combination
with both the MHC-p193dn and MHC-p53dn transgenes. Transfected cells were
enriched by virtue of their resistance to hygromycin, and then induced to
differentiate. The ES-derived cardiomyocytes were then enriched by virtue of
their
resistance to 6418. Cardiomyocyte growth was then compared in these cultures
in
the presence vs. absence of exogenous isoproterenol (1 pM mg/ml) for 58 days.
Markedly enhanced cardiomyocyte colony size was apparent in the isoproterenol
treated cultures, consistent with the presence of increase cell numbers (see
Figure
16). This suggests that relaxation of cell cycle apoptotic check-points
renders
cardiomyocytes proliferative to what would otherwise be a hypertrophic
stimuli.
While the invention has been illustrated and described in detail in the
drawings and foregoing description, the same is to be considered as
illustrative and
not restrictive in character, it being understood that only the preferred
embodiment
has been shown and described and that all changes and modifications that come
within the spirit of the invention are desired to be protected.
All publications cited herein are indicative of the level of skill in the art
and
are hereby incorporated by reference as if each had been individually
incorporated
by reference and fully set forth.
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
1
SEQUENCE LISTING
<110> Field, Loren J.
Tsai, Shih-Chong
<120> P193 PROTEINS AND NUCLEIC ACIDS, AND USES THEREOF
<130> IU99-PCT
<140> Not Yet Known
<141> 2000-08-23
<150> 60/150,266
<151> 1999-08-23
<160> 4
<170> PatentIn Ver. 2.1
<210> 1
<211> 5217
<212> DNA
<213> Mus muscul;a
<220>
<221> CDS
<222> (62)..(5123)
<400> 1
ttcctagctc tgcaaaggac aggcctcgcg caggatcccg gcggacttct gaggtgccac 60
g atg gta ggg gag cta cga tac agg gaa ttc agg gtg ccc ctg ggg cct 109
Met Val Gly G1~.: Leu Arg Tyr Arg Glu Phe Arg Val Pro Leu Gly Pro
1 5 10 15
ggc ttg cac gcg tat ccg gat gaa ttg atc cgc caa cgg gtt ggc cat 157
Gly Leu His Ala '~-r Pro Asp Glu Leu Ile Arg Gln Arg Val Gly His
20 25 30
aat ggg cac ccc gag tat cag atc cgc tgg ctc atc ctc agg cgc ggg 205
Asn Gly His Pro Glu Tyr Gln Ile Arg Trp Leu Ile Leu Arg Arg Gly
35 40 45
gat gat ggg gac cgg gac tct aca gtg gac tgc aag get gag cat atc 253
Asp Asp Gly Asp Arg Asp Ser Thr Val Asp Cys Lys Ala Glu His Ile
50 55 60
ctg tta tgg atg tct gac gat gag atc tat gcc aac tgc cac aag atg 301
Leu Leu Trp Met Ser Asp Asp Glu Ile Tyr Ala Asn Cys His Lys Met
65 70 75 80
ctg ggc gag aat ggc caa gtc atc gca cct tcc cgg gag tcc act gag 349
Leu Gly Glu Asn Gly Gln Val Ile Ala Pro Ser Arg Glu Ser Thr Glu
g5 90 95
gca ggg gcc ctc gac aag tct gtg ctg ggg gag atg gaa aca gat gtg 397
Ala Gly Ala Leu ?sp Lys Ser Val Leu G1y Glu Met Glu Thr Asp Val
100 105 110
aag tcc ttg att cag agg gcc ctt cgg cag ctg gag gag tgc gtg ggc 445
Lys Ser Leu Ile G'_n Arg Ala Leu Arg Gln Leu Glu Glu Cys Val Gly
115 120 125
acc gtg cct cct gcc cct ctc ctt cac acg gtc cat gta ctc agt gcc 493
Thr Val Pro Pro Ala Pro Leu Leu His Thr Val His Val Leu Ser Ala
. 130 135 140
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
2
tatgccagcatcgag cccctcactggc atcttcaaagaccgcagggtt 541
TyrAlaSerIleGlu ProLeuThrGly IlePheLysAspArgArgVal
145 i50 155 160
gtgaacttgctcatg cacatgttgagc agtcctgattatcagatccgc 589
ValAsnLeuLeuMet HisMetLeuSer SerProAspTyrGlnIleArg
165 170 175
tggagcgcaggccgg atgatccaaget ctgtcctcccacgatgetggg 637
TrpSerAlaGlyArg MetIleGlnAla LeuSerSerHisAspAlaGly
180 185 190
acccggacccagatc cttctgtcattg agccaacaagaggccattgaa 685
ThrArgThrGlnIle LeuLeuSerLeu SerGlnGlnGluAlaIleGlu
195 200 205
aagcacctggatttt gatagccgctgc getctgcttgcactgttcgcc 733
LysHisLeuAspPhe AspSerArgCys AlaLeuLeuAlaLeuPheAla
210 215 220
caggetactctcacg gaacacccgatg tctttcgagggcgttcagctg 781
GlnAlaThrLeuThr GluHisProMet SerPheGluGlyValGlnLeu
225 230 235 240
ccacaggtcccagga cggctgctcttc tccctggtaaaacgctacctg 829
ProGlnValProGly ArgLeuLeuPhe SerLeuValLysArgTyrLeu
245 250 255
cacgtcaccttcctc ctggatcggctg aacggcgatgcaggggatcaa 877
HisValThrPheLeu LeuAspArgLeu AsnGlyAspA1aGlyAspGln
260 265 270
ggagcccagaacaac tttagtcctgag gagttgaatgtagggaggggc 925
GlyAlaGlnAsnAsn PheSerProGlu 'GluLeuAsnValGlyArgGly
275 280 285
cggctggaactggaa ttcagtatggcc atgggcactctgatctctgag 973
ArgLeuGluLeuGlu PheSerMetAla MetGlyThrLeuIleSerGlu
290 295 300
ctggtgcaggccatg cgctgggacggg gcctcaagcagaccagagagt
1021
LeuValGlnAlaMet ArgTrpAspGly AlaSerSerArgProGluSer
305 310 315 320
tcttcctcctccacc ttccagcctcgg ccagcacagttccgcccctac
1069
SerSerSerSerThr PheGlnProArg ProAlaGlnPheArgProTyr
325 330 335
acccagcgtttcagg aggtcgaggcgg tttcgcccccgtgcctcgttt
1117
ThrGlnArgPheArg ArgSerArgArg PheArgProArgAlaSerPhe
340 345 350
gccagttttaatacc tatgccttgtat gtgcgggacacgctgcggccc
1165
AlaSe_PheAsnThr TyrAlaLeuTyr ValArgAspThrLeuArgPro
355 360 365
gggatgcgggtacgg atgctggagaat tacgaggagatcgetgetggg
1213
GlyMetArgValArg MetLeuGluAsn TyrGluGluIleAlaAlaGly
370 375 380
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
3
gat gag ggc cag ttc cga cag agc aat gat ggc gtt ccc cca gcg cag
1261
Asp Glu Gly Gln Phe Arg Gln Ser Asn Asp Gly Val Pro Pro Ala Gln
385 390 395 400
gtg ttg tgg gat tca aca ggc cat acc tac tgg gtg cac tgg cac atg
1309
Val Leu Trp Asp Ser Thr Gly His Thr Tyr Trp Val His Trp His Met
405 410 415
ctg gag atc ttg ggc ttt gag gaa gac atc gag gat gtg att gat att
1357
Leu Glu Ile Leu Gly Phe Glu Glu Asp Ile Glu Asp Val Ile Asp Ile
420 425 430
gaa gag tta cag gag cta ggg gcc aat gga gca ctg agc atc gtc ccg
1405
Glu Glu Leu Gln Glu Leu Gly Ala Asn G1y Ala Leu Ser Ile Val Pro
435 440 445
ccg tcc cag cgc tgg aag ccc ata act cag ctc ttt gcc gag cct tac
1453
Pro Ser Gln Arg Trp Lys Pro Ile Thr Gln Leu Phe Ala Glu Pro Tyr
450 455 460
gtg gta ccc gag gag gaa gac agg gaa gag agc gag aac ttg acc cag
1501
Val Val Pro Glu Glu Glu Asp Arg GIu Glu Ser Glu Asn Leu Thr Gln.
470 475 480
465
get gag tgg tgg gag ctc ctc ttc ttc atc cgg cag ttg agt gag gca
1549
Ala Glu Trp Trp Glu Leu Leu Phe Phe Ile Arg Gln Leu Ser Glu Ala
485 490 495
gag cgc ctt cac a=c gtg gat ctc ctg caa gac cac ctg gaa gag gag
1597
Glu A_g Leu His Ile Val Asp Leu Leu Gln Asp His Leu Glu Glu Glu
500 505 510
cgc gtt ctg gac tac gat atg ctg cct gag ctg acc gtg ccc gtt gac
1545
Arg Val Leu Asp Tyr Asp Met Leu Pro Glu Leu Thr Val Pro Vai Asp
515 520 525
ttg gcc cag gat ctg ctg ttg tct ctg cct cag caa ctt gag gac agt
1693
Leu Ala Gln Asp Leu Leu Leu Ser Leu Pro Gln Gln Leu Glu Asp Ser
530 535 540
get ctg agg gac ctg ttc agc tgc agt gtc tac agg aag tat ggg ccc
1741
Ala Leu Arg Asp Leu Phe Ser Cys Ser Val Tyr Arg Lys Tyr Gly Pro
550 555 560
545
gaa gtc ctg gta ggg cat cta agc tac cca ttt gtg cca ggt gcc cag
1789
Glu Val Leu Val Gly His Leu Ser Tyr Pro Phe Va1 Pro Gly Ala Gln
565 570 575
cca aat tta ttc gga gcc aat gaa gag tct gaa gcc aaa gat ccc cca
1837
pro Asn Leu Phe Gly Ala Asn Glu Glu Ser Glu Ala Lys Asp Pro Pro
580 585 590
c,tt cag agt gcc agc cct gcc ctg cag cgc ctg gtg gag agc ttg ggc
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
4
1885
Leu Gln Ser A1a Ser Pro Ala Leu Gln Arg Leu Val Glu Ser Leu Gly
595 600 605
ccc gaa ggg gag gtc ctt gtg gaa ctg gaa caa gcc cte ggc tcc gag
1933
Pro Glu Gly Glu Val Leu Val Glu Leu Glu Gln Ala Leu Gly Ser Glu
610 615 620
get ccc cag gaa act gag gtc aag tcc tgc ttg ttg cag ctc cag gag
1981
Ala Pro Gln Glu Thr Glu Va1 Lys Ser Cys Leu Leu Gln Leu Gln Glu
625 630 635 640
cag ccc cag ccc ttc ctc get ctg atg cgg agc ctg gac act tcc gcc
2029
Gln Pro Gln Pro Phe Leu Ala Leu Met Arg Ser Leu asp Thr Ser Ala
645 650 655
agc aac aag acc ctg cac ctc act gtg ctc aga atc tta atg cag ctg
2077
Ser Asn Lys Thr Leu His Leu Thr Val Leu Arg Ile Leu Met Gln Leu
660 665 670
gtg aac ttc cca gag gcg ctg ttg cta ccc tgg cac gag gcc atg gat
2125
Val Asn Phe Pro G'_u Ala Leu Leu Leu Pro Trp His G1;: Ala Met Asp
675 680 685
gcc tgc gtg acc tgc ctt cgg tcc ccc aat act gac cga gag gtg ctc
2173
Ala Cys Val Thr Cys Leu Arg Ser Pro Asn Thr Asp Arg Glu Val Leu
690 695 700
cag gaa cta atc t~t ttc ctg cac cgc ctg acc acc aca agc cgg gac
2221
Gln G'_u Leu Ile P'e Phe Leu His Arg Leu Thr Thr T..r Ser Arg Asp
705 710 715 720
tat gcg gtg ata cta aac cag cat gga gcc cgg gac gcc atc tcc aaa
2269
Tyr -Ala 'Jal I1a Leu Asn Gln His Gly Ala Arg Asp Ala Ile Ser Lys
725 730 735
gtc ctg gaa aag cac cga ggg aaa ctg gag ttg get cag gag ctg cgg
2317
Val Leu Glu Lys His Arg Gly Lys Leu Glu Leu Ala Gln G1u Leu Arg
740 745 750
gat atg gtg tcc aag tgt gag aag cat gcc cac ctc tac cgg aaa ctc
2365
Asp Met Val Ser Lys Cys Glu Lys His Ala His Leu Tyr Arg Lys Leu
755 760 765
acc acc aac atc ctg ggc ggt tgc atc cag atg gtc ctg ggt cag att
2413
Thr Thr Asn Ile Leu Gly Gly Cys Ile Gln Met Val Leu G1y Gln Ile
770 775 780
gaa gac cac aga cga acc cac cgg ccc atc caa atc cca ttc ttt gat
2461
Glu Asp His Arg A=g Thr His Arg Pro Ile Gln Ile Pro Phe Phe Asp
785 790 795 800
gtg ttt ctc aga tat ctg tgc cag ggc tcc agt gag gaa atg aag aaa
2509
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
Val Phe Leu Arg Tyr Leu Cys Gln Gly Ser Ser Glu Glu Met Lys Lys
805 810 815
aac agg tac tgg gag aag gtg gag gtg tcc tcc aac cca cag cgg gcc
2557
Asn Arg Tyr Trp Glu Lys Val Glu Val Ser Ser Asn Pro Gln Arg Ala
820 825 830
agc agg ctg acg gac cgc aac ccc aag acc tac tgg gag tcc agt ggc
2605
Ser Arg Leu Thr Asp Arg Asn Pro Lys Thr Tyr Trp Glu Ser Ser Gly
835 840 845
agg gcc ggc tcc cac ttc atc acc tta cac atg cgc cca ggt gtc atc
2653
Arg Ala Gly Ser His Phe Ile Thr Leu His Met Arg Pro Gly Val Ile
850 855 860
atc agg cag ctg act cta ctg gtg get ggc gag gac tca agc tac atg
2701
Ile Arg Gln Leu Thr Leu Leu Val Ala Gly Glu Asp Ser Ser Tyr Met
865 870 875 880
cca gcc tgg gtg gtg gta tgc ggg ggc aac agc atc aag tcc gtt aat
2749
Pro Ala Trp Val Val Val Cys Gly Gly Asn Ser Ile Lys Ser Val Asn
885 890 895
aaa gaa ctc aac acg gta aac gtg atg ccc tct gcc agc cgg gtg acc
2797
Lys G'_u Leu Asn Thr Val Asn Val Met Pro Ser Ala Ser Arg Val Thr
900 905 910
ctc ctg gag aac ctg acc cgc ttc tgg ccc atc atc caa atc aga ata
2845
Leu Leu Glu Asn Leu Thr Arg Phe Trp Pro Ile Ile Gln Ile Arg Ile
915 920 925
aag cgc tgc cag cag ggt ggc att aac acg cgc atc cgg ggc cta gag
2893
Lys Arg Cys Gln Gln Gly Gly Ile Asn Thr Arg Ile Arg G1y Leu Glu
930 935 940
gtg ctg ggc ccc aag ccc acc ttc tgg cca gtg ttc cga gag caa ctg
2941
Val Leu Gly Pro Lys Pro Thr Phe Trp Pro Val Phe Arg Glu Gln Leu
995 950 955 960
tgc cgg cac acg cgc ctc ttc tac atg gtt cgg gcc cag gca tgg agt
2989
Cys Arg His Thr Arg Leu Phe Tyr Met Val Arg Ala Gln Ala Trp Ser
965 970 975
cag gac ata gca gag gac cgc cgg agc ctt ctg cac ctg agt tct agg
3037
Gln Asp Ile Ala Glu Asp Arg Arg Ser Leu Leu His Leu Ser Ser Arg
980 985 990
cta aat ggg get ctg cgc cat gaa cag aat ttt gca gag cgc ttc ctt
3085
Leu Asn Gly Ala Leu Arg His Glu Gln Asn Phe Ala Glu Arg Phe Leu
995 1000 1005
cct gat atg gag gcc gcc caa gca ctg agc aag acc tgc tgg gag gcg
3133
Pro Asp Met Glu Ala Ala Gln Ala Leu Ser Lys Thr Cys Trp Glu Ala
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
6
1010 1015 1020
ctg gtc agc ccc ctg gtg cag aac att aca tct ccc gat gag gac agc
3181
Leu Val Ser Pro Leu Val Gln Asn Ile Thr Ser Pro Asp Glu Asp Ser
1025 1030 1035 1040
acc agc tcc ttg ggc tgg ctg ctg gat cag tac ttg gga tgc agg gag
3229
Thr Ser Ser Leu Gly Trp Leu Leu Asp Gln Tyr Leu Gly Cys Arg Glu
1045 1050 1055
get gcc tac aat ccc cag agc agg get get get ttc tcc tcc cgg gtt
3277
Ala Ala Tyr Asn Pro Gln Ser Arg Ala Ala Ala Phe Ser Ser Arg Val
1060 1065 1070
cgc cgc ctt acc cac ctc ctg gtc cat gtg gag ccc cgt gag gca gca
3325
Arg Arg Leu Thr His Leu Leu Val His Val Glu Pro Arg Glu Ala Ala
1075 1080 1085
cct ccg gtg gtg gcc atc cct cga tcc aag ggc agg aac aga atc cat
3373
Pro Pro Va1 Val 3_a T_le Pro Arg Ser Lys Giy Arg Asn Arg Ile His
1090 1095 1100
gac tgg agc tac ttg atc acc cgg ggc ctt cca agt agc atc atg aag
3421
Asp Trp Ser Tyr Leu Ile Thr Arg Gly Leu Pro Ser Ser Ile Met Lys
1105 1110 1115 1120
aac ctg acc cgc tgt tgg cgg tct gtg gtg gag gag cag atg aac aag
3469
Asn Leu Thr Arg Cys Trp Arg Ser Val Val G_:: Glu Gln Met Asn Lys
1125 1130 1135
ttt ctc agt gcg tcc tgg aaa gac gat gat ttc gta ccc cgc tat tgc
3517
Phe Leu Ser Ala Ser Trp Lys Asp Asp Asp P::e Val Pro Arg Tyr Cys
1140 1145 1150
gag cgc tat tac gtc ctg cag aag tcc agc tca gag ctg ttt ggg cca
3565
Glu Arg Tyr Tyr Val Leu Gln Lys Ser Ser Ser Glu Leu Phe Gly Pro
1155 1160 1165
cga get gcc ttc ttg ctg gca atg cgg aat ggc tgt get gat get gtg
3613
Arg Ala Ala Phe Leu Leu Ala Met Arg Asn Gly Cys Ala Asp Ala Val
1170 1175 1180
cgg agg ctc cct ttc ctc agg gcc gcc cat gtg aag cag cag ttt get
3661
Arg Arg Leu Pro Phe Leu Arg Ala Ala His Val Lys Gln Gln Phe Ala
1185 1190 1195 1200
cgg cac att gac cag agg atc caa ggc agt agg atg ggt gga gcc cgg
3709
Arg His I1e Asp Gln Arg Ile Gln Gly Ser Arg Met Gly Gly Ala Arg
1205 1210 1215
gga atg gag atg ctg gca cag ttg cag cga tgc ctg gag tct gtc ctg
3757
Gly Met Glu Met Leu Ala G1:: Leu Gln Arg Cys Leu Glu Ser Val Leu
. 1220 1225 1230
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
7
att ttc tct ccc ctg gag ata gcc acc acc ttt gag cat tac tac cag
3805
Ile Phe Ser Pro Leu Glu Ile Ala Thr Thr Phe G1u His Tyr Tyr Gln
1235 1240 1245
cac tac atg get gac cgt ctc ctg agc gta ggc tcc agc tgg ctg gag
3853
His Tyr Met Ala Asp Arg Leu Leu Ser Val Gly Ser Ser Trp Leu Glu
1250 1255 1260
ggg gcc gta ctg gag cag atc ggt ccc tgc ttc ccc agc cgt ctt ccc
3901
Gly Ala Val Leu Glu Gln Ile Gly Pro Cys Phe Pro Ser Arg Leu Pro
1265 1270 1275 1280
cag cag atg cta cag agc ctg aac gtc tca gag gag ttg cag cgt cag
3949
Gln Gln Met Leu Gln Ser Leu Asn Val Ser Glu Glu Leu Gln Arg Gln
1285 1290 1295
ttc cac gtt tac cag ctg cag cag ctt gat cag gag ctc ctg aag ctg
3997
Phe His Val Tyr Gln Leu Gln Gln Leu Asp Gln Glu Leu Leu Lys Leu
1300 1305 1310
gaa gac acg gaa aag aag ata cag gtg gcc cat gag gac agt ggc aga
4045
Glu Asp Thr Glu Lys Lys I1e Gln Val Ala His Glu Asp Ser Gly Arg
1315 1320 1325
gag gac aag agc aag aag gaa gaa gcc att gga gaa gcc gcg get gtg
4093
Glu Asp Lys Ser Lys Lys Glu Glu A1a Ile Gly Glu Ala Ala A'_a Val
1330 1335 1340
get atg gca gag gag gag gat caa ggg aag aag gag gag gga gag gag
4141
Ala Met Ala Glu G'_u Glu Asp Gln Gly Lys Lys Glu Glu Gly Glu Giu
1345 1350 1355 1360
gaa gag gag gga gag gat gag gag gaa gag cgc tat tat aaa gga aca
4189
Glu Gly Glu Gly Glu Asp Glu Glu Glu Glu Arg Tyr Tyr Lys G1y Thr
1365 1370 1375
atg cca gaa gtg tgt gta ctt gtc gtg acg cca cgc ttc tgg cct gtc
4237
Met Pro Glu Val Cys Va1 Leu Val Val. Thr Pro Arg Phe Trp Pro Val
1380 1385 1390
gcc tcc gtc tgc caa atg ctc aac ccg gca acg tgc ctg ccc gca tac
4285
Ala Ser Val Cys Gln Met Leu Asn Pro Ala Thr Cys Leu Pro Ala Tyr
1395 1400 1405
ctg cgg ggg acc ata aac cac tac acc aac ttt tac agc aag agt cag
4333
Leu Arg Gly Thr Ile Asn His Tyr Thr Asn Phe Tyr Ser Lys Ser Gln
1410 1415 1420
agc cgc tcc agc tta gag aaa gag cca cag agg cgg ctg cag tgg acc
4381
Se_~ Arg Ser Ser Leu Glu Lys Glu Pro Gln Arg Arg Leu G1n Trp Thr
1425 1430 1435 1440
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
s
tgg cag ggc cgg gca gaa gtg cag ttc ggg ggt cag att ctg cat gtg
4429
Trp Gln Gly Arg Ala Glu Val Gln Phe Gly Gly Gln Ile Leu His Val
1445 1450 1455
tcc aca gta cag atg tgg ctg ctg ctg cat ctc aac aac caa aag gag
4477
Ser Thr Val Gln Met Trp Leu Leu Leu His Leu Asn Asn Gln Lys Glu
1460 1465 1470
gtg tct gtc gag agc ctg cag get atc tcg gag ctc cct cca gat gtg
4525
Val Ser Val Glu Ser Leu Gln Ala Ile Ser Glu Leu Pro Pro Asp Val
1475 1480 1485
ctt cac agg gcc atc ggg cct ctc acc tca tca aga ggt ccc ttg gac
4573
Leu :?is Arg Ala Ile Gly Pro Leu Thr Ser Ser Arg Gly Pro Leu Asp
1490 1495 1500
ctg cag gag cag aag aac gta cca gga ggg gtg ctc aag att cga gat
4621
Leu G_z Glu Gln Lys Asn Val Pro Gly Gly Val Leu Lys Ile Arg Asp
150 1510 1515 '_520
gac agt gag gaa ccc agg ccg agg agg ggc aac gtg tgg ctg atc cca
4669
Asp Ser Glu Glu Pro Arg Pro Arg Arg Gly Asn Val Trp Leu Ile Pro
1525 1530 1535
cct cag aca tac cta caa get gag gcc gaa gag ggc cgg aac atg gag
471.
Pro ~~n Thr Tyr Leu Gln Ala Glu A1a Glu Glu Gly Arg Asn Met Glu
1540 1545 1550
aag a~a agg aat ctt ctg aat tgc ctt gtt gtc cga atc ctc aag get
476
Lys :~=g Arg Asn Leu Leu Asn Cys Leu Val Val Arg Ile Leu Lys Ala
1555 1560 1565
cac ggg gat gaa ggc ttg cat gtt gac cgg ctc gtc tat ctg gtg cta
48'_3
His G'_y Asp Glu Gly Leu His Val Asp Arg Leu Val Tyr Leu Val Leu
1570 1575 1580
gaa gcg tgg gag aaa ggc ccg tgt cct get agg ggt ctg gtc agc agc
486
Glu =.'_a Trp Glu Lys Gly Pro Cys Pro Ala Arg Gly Leu Val Ser Ser
1585 1590 1595 1600
ctc ggc agg gga gca acc tgc agg agc tct gat gtc ctc tcc tgc atc
4909
Leu Gly Arg Gly Ala Thr Cys Arg Ser Ser Asp Val Leu Ser Cys Ile
1605 1610 1615
ctg cac ctc ctg gtc aag ggc acg ctg aga cgc cat gac gac cgg ccg
4957
Leu ~vs Leu Leu Val Lys Gly Thr Leu Arg Arg His Asp Asp Arg Pro
1620 1625 1630
cag g~g ctg tac tat gca gtt cct gta act gtg atg gag ccc cac atg
5005
Gln '!al Leu Tyr Tyr Ala Val Pro Val Thr Val Met Glu Pro His Met
1635 1640 1645
gag tcc ctg aac cct ggc tcg gca ggc ccc aat cca ccc ctc acc ttc
SUBSTITUTE SHEET (RULE Z6)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
9
5053
Glu Ser Leu Asn Pro Gly Ser Ala Gly Pro Asn Pro Pro Leu Thr Phe
1650 1655 1660
cac acc ctg cag att cga tcc cgg ggt gtg cct tac gcc tcc tgc act
5101
His Thr Leu Gln Ile Arg Ser Arg Gly Val Pro Tyr Ala Ser Cys Thr
1665 1670 1675 1680
gat aac cac acc ttc tcc act ttc cgg tagccctgga tatggggttg
5148
Asp Asn His Thr Phe Ser Thr Phe Arg
1685
ggggtaggtg aggctggggc tttcatcaga aaataaaatg ctggaatttg aaaaaaaaaa
5208
aaaaaaaaa
5217
<210> 2
<211> 1689
<212> PTtT
<213> Mus muscuius
<400> 2
Met Val Gly G1u Leu Arg Tyr Arg Glu Phe Arg Val Pro Leu Gly Pro
1 5 10 15
Gly Leu His Ala Tyr Pro Asp Glu Leu Ile Arg Gln Arg Val Gly His
20 25 30
Asn Gly His Pro Glu Tyr Gln Ile Arg Trp Leu I1e Leu Arg Arg Gly
35 40 45
Asp Asp Gly Asp A=g Asp Ser Thr Val Asp Cys Lys Ala Glu His I'_e
50 55 60
Leu Leu Trp Met Ser Asp Asp Glu T_le Tyr Ala Asn Cys ::is Lys Met
65 70 75 80
Leu Gly Glu Asn Gly Gln Val Ile Ala Pro Ser Arg Glu Ser Thr Glu
85 90 95
Ala Gly Ala Leu Asp Lys Ser Va1 Leu Gly Glu Met Glu Thr Asp Val
100 105 110
Lys Ser Leu Ile Gln Arg Ala Leu Arg Gln Leu Glu Glu Cys Val Gly
115 120 125
Thr Val Pro Pro Ala Pro Leu Leu His Thr Val His Val Leu Ser Ala
130 135 140
Tyr Ala Ser Ile Glu Pro Leu Thr Gly Ile Phe Lys Asp Arg Arg Val
145 150 155 160
Val Asn Leu Leu Met His Met Leu Ser Ser Pro Asp Tyr Gln Ile Arg
165 170 175
Trp Ser Ala Gly Arg Met Ile Gln Ala Leu Ser Ser His Asp Ala Gly
180 185 190
Thr Arg Thr Gln Ile Leu Leu Ser Leu Ser Gln Gln Glu A1a Ile Glu
195 200 205
Lys His Leu Asp Phe Asp Ser Arg Cys Ala Leu Leu Ala Leu Phe Ala
210 215 220
Gln Ala Thr Leu Thr Glu His Pro Met Ser Phe Glu Gly Val Gln Leu
225 230 235 240
Pro Gln Val Pro Gly Arg Leu Leu Phe Ser Leu Val Lys Arg Tyr Leu
245 250 255
His Val Thr Phe Leu Leu Asp Arg Leu Asn Gly Asp Ala Gly Asp G1n
260 265 270
Gly Ala Gln Asn Asn Phe Ser Pro Glu Glu Leu Asn Val Gly Arg Gly
275 280 285
Arg Leu Glu Leu Glu Phe Ser Met Ala Met Gly Thr Leu Ile Ser Glu
. 290 295 300
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
Leu Val Gln Ala Met Arg Trp Asp Gly Ala Ser Ser Arg Pro Glu Ser
305 310 315 320
Ser Ser Ser Ser Thr Phe Gln Pro Arg Pro Ala Gln Phe Arg Pro Tyr
325 330 335
Thr Gln Arg Phe Arg Arg Ser Arg Arg Phe Arg Pro Arg Ala Ser Phe
340 345 350
Ala Ser Phe Asn Thr Tyr Ala Leu Tyr Val Arg Asp Thr Leu Arg Pro
355 360 365
Gly Met Arg Val Arg Met Leu Glu Asn Tyr Glu Glu Ile Ala Ala Gly
370 375 380
Asp Glu Gly Gln Phe Arg Gln Ser Asn Asp Gly Val Pro Pro Ala Gln
385 390 395 400
Val Leu Trp Asp Ser Thr Gly His Thr Tyr Trp Val His Trp His Met
405 410 415
Leu Glu Ile Leu Gly Phe Glu Glu Asp Ile Glu Asp Val Ile Asp Ile
420 425 430
Glu Glu Leu Gln Glu Leu Gly Ala Asn Gly Ala Leu Ser Ile Val Pro
435 440 445
Pro Ser Gln Arg Trp Lys Pro Ile Thr Gln Leu Phe Ala Glu Pro Tyr
450 455 460
Val Va1 Pro Glu Glu Glu Asp Arg Glu Glu Ser Glu Asn Leu Thr G1n
465 470 475 480
Ala Glu Trp Trp Glu Leu Leu Phe Phe Ile Arg Gln Leu Ser Glu Ala
485 490 495
Glu Arg Leu His I'_e Val Asp Leu Leu Gln Asp His Leu Glu Glu Glu
500 505 510
Arg Va1 Leu Asp Tyr Asp, Met Leu Pro Glu Leu Thr Val Pro Val Asp
515 520 525
Leu Ala Gin Asp Leu Leu Leu Ser Leu Pro Gln Gln Leu Glu Asp Ser
530 535 540
Ala Leu Arg Asp Leu Phe Ser Cys Ser Val Tyr Arg Lys Tyr Gly Pro
545 550 555 560
Glu Val Leu Val Gly His Leu Ser Tyr Pro Phe Val Pro Gly Ala Gln
565 570 575
Pro As.~. Leu Phe Gly Ala Asn Glu Glu Ser Glu Ala Lys Asp Pro Pro
580 585 590
Leu Gln Ser Ala Se. Pro Ala Leu Gln Arg Leu Val Glu Ser Leu Gly
595 600 605
Pro Glu Gly Glu Va'_ Leu Val Glu Leu Glu Gln Ala Leu G1y Ser Glu
610 615 620
Ala Pro Gln Glu Thr Glu Val Lys Ser Cys Leu Leu Gln Leu Gln Glu
625 630 635 640
Gln Pro Gln Pro Phe Leu Ala Leu Met Arg Ser Leu Asp Thr Ser Ala
645 650 655
Ser Asn Lys Thr Leu His Leu Thr Val Leu Arg Ile Leu Met Gln Leu
660 665 670
Val Asn Phe Pro G-a Ala Leu Leu Leu Pro Trp His Glu Ala Met Asp
675 680 685
Ala Cys Val Thr Cys Leu Arg Ser Pro Asn Thr Asp Arg Glu Val Leu
690 695 700
Gln Glu Leu Ile P'.~.e Phe Leu His Arg Leu Thr Thr Thr Ser Arg Asp
705 710 715 720
Tyr Ala Val Ile Leu Asn Gln His Gly Ala Arg Asp Ala Ile Ser Lys
725 730 735
Va1 Leu G1u Lys His Arg Gly Lys Leu Glu Leu Ala Gln Glu Leu Arg
740 745 750
Asp Met Val Ser Lys Cys Glu Lys His Ala His Leu Tyr Arg Lys Leu
755 760 765
Thr Thr Asn Ile Leu Gly Gly Cys Ile Gln Met Val Leu Gly Gln Ile
770 775 780
Glu Asp His Arg A=g Thr His Arg Pro Ile Gln Ile Pro Phe Phe Asp
785 790 795 800
Val Phe Leu Arg Tyr Leu Cys Gln Gly Ser Ser Glu Glu Met Lys Lys
805 810 815
Asn Arg Tyr Trp Glu Lys Val Glu Val Ser Ser Asn Pro Gln Arg Ala
820 825 830
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
11
Ser Arg Leu Thr Asp Arg Asn Pro Lys Thr Tyr Trp Glu Ser Ser Gly
835 840 845
Arg Ala Gly Ser His Phe Ile Thr Leu His Met Arg Pro Gly Val Ile
850 855 860
Ile Arg Gln Leu Thr Leu Leu Val Ala Gly Glu Asp Ser Sez Tyr Met
865 870 875 880
Pro Ala Trp Val Val Val Cys Gly Gly Asn Ser Ile Lys Ser Val Asn
885 890 895
Lys Glu Leu Asn Thr Val Asn Val Met Pro Ser Ala Ser Arg Val Thr
900 905 910
Leu Leu Glu Asn Leu Thr Arg Phe Trp Pro Ile Ile Gln Ile Arg Ile
915 920 925
Lys Arg Cys Gln Gln G1y Gly Ile Asn Thr Arg Ile Arg Gly Leu Glu
930 935 940
Val Leu Gly Pro Lys Pro Thr Phe Trp Pro Va1 Phe Arg Glu Gln Leu
945 950 955 960
Cys Arg His Thr Arg Leu Phe Tyr Met Val Arg Ala Gln Ala Trp Ser
965 970 975
Gln Asp Ile Ala Glu Asp Arg Arg Ser Leu Leu His Leu Ser Ser Arg
980 985 990
Leu Asn Gly Ala Leu Arg His Glu Gln Asn Phe Ala Glu Arg Phe Leu
995 1000 1005
Pro Asp Met Glu Ala A1a Gln Ala Leu Ser Lys Thr Cys Trp Glu Ala
1010 1015 1020
Leu Val Ser Pro Leu Val Gln Asn Ile Thr Ser P=o Asp Glu Asp Ser
1025 1030 1035 1040
Thr Ser Ser Leu Gly Trp Leu Leu As_p Gln Tyr Leu G1y Cys Arg Glu
1045 1050 1055
Ala Ala Tyr Asn Pro Gln Ser Arg Ala Ala Ala Phe Se_- Ser Arg Val
1060 1065 1070
Arg Arg Leu Thr His Leu Leu Val His Val Glu Pro Arg Glu Ala Ala
1075 1080 1085
Pro Pro Val Val Ala Ile Pro Arg Ser Lys Gly Arg Asn Arg Ile His
1090 1095 1100
Asp Trp Ser Tyr Leu Ile Thr Arg Gly-Leu Pro Ser Ser Ile Met Lys
1105 1110 1115 1120
Asn Leu Thr Arg Cys Trp Arg Ser Val Val Glu Glu Gln Met Asn Lys
1125 1130 1135
Phe Leu Ser Ala Ser Trp Lys Asp Asp Asp Phe Val Pro Arg Tyr Cys
1140 1145 1150
Glu Arg Tyr Tyr Val Leu Gln Lys Ser Ser Ser Glu Leu Phe Gly Pro
1155 1160 1165
Arg Ala Ala Phe Leu Leu Ala Met Arg Asn G1y Cys Ala Asp Ala Val
1170 1175 1180
Arg Arg Leu Pro Phe Leu Arg Ala Ala His Val Lys G1n Gln Phe Ala
1185 1190 1195 1200
Arg His Ile Asp Gln Arg Ile Gln Gly Ser Arg Met Gly Gly Ala Arg
1205 1210 1215
Gly Met G1u Met Leu Ala Gln Leu Gln Arg Cps Leu Glu Ser Val Leu
1220 1225 1230
Ile Phe Ser Pro Leu Glu Ile Ala Thr Thr Phe Glu His Tyr Tyr Gln
1235 1240 1245
His Tyr Met Ala Asp Arg Leu Leu Ser Val Gly Ser Ser Trp Leu Glu
1250 1255 1260
Gly Ala Val Leu Glu Gln Ile G1y Pro Cys Phe Pro Ser Arg Leu Pro
1265 1270 1275 1280
Gln Gln Met Leu Gln Ser Leu Asn Val Ser Glu Glu Leu Gln Arg Gln
1285 1290 1295
Phe His Val Tyr Gln Leu Gln Gln Leu Asp Gln Glu Leu Leu Lys Leu
1300 1305 1310
Glu Asp Thr Glu Lys Lys Ile Gln Val Ala His Glu Asp Ser Gly Arg
1315 1320 1325
Glu Asp Lys Ser Lys Lys Glu Glu Ala Ile Gly G1u Ala A1a Ala Val
1330 1335 1340
Ala Met Ala Glu G1u Glu Asp Gln Gly Lys Lys Glu Glu Gly Glu Glu
1345 1350 1355 1360
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
12
Glu Gly Glu Gly Glu Asp Glu Glu Glu Glu Arg Tyr Tyr Lys Gly Thr
1365 1370 1375
Met Pro Glu Val Cys Val Leu Val Val Thr Pro Arg Phe Trp Pro Val
1380 1385 1390
Ala Ser Val Cys Gln Met Leu Asn Pro Ala Thr Cys Leu Pro Ala Tyr
1395 1400 1405
Leu Arg Gly Thr Ile Asn His Tyr Thr Asn Phe Tyr Ser Lys Ser Gln
1410 1415 1420
Ser Arg Ser Ser Leu Glu Lys Glu Pro Gln Arg Arg Leu Gln Trp Thr
1425 1430 1435 1440
Trp Gln Gly Arg Ala Glu Val Gln Phe Gly Gly Gln Ile Leu His Val
1445 1450 1455
Ser Thr Val Gln Met Trp Leu Leu Leu His Leu Asn Asn Gln Lys Glu
1460 1465 1470
Val Ser Val Glu Ser Leu Gln Ala Ile Ser Glu Leu Pro Pro Asp Val
1475 1480 1485
Leu His Arg Ala I1e G1y Pro Leu Thr Ser Ser Arg Gly Pro Leu Asp
1490 1495 1500
Leu Gln Glu Gln Lys Asn Val Pro Gly Gly Val Leu Lys Ile Arg Asp
1505 1510 1515 1520
Asp Ser Glu Glu Pro Arg Pro Arg Arg Gly Asn Val Trp Leu Ile Pro
1525 1530 1535
Pro Gln Thr Tyr Leu G1~ Ala Glu Ala Glu Glu Gly Arg Asn Met Glu
1540 1545 1550
Lys Arg Arg Asn Leu Leu Asn Cys Leu Val Val Arg Ile Leu Lys Ala
1555 1560 1565
His Gly Asp Glu Gly Leu His Val AsF Arg Leu Val Tyr Leu Val Leu
1570 1575 1580
Glu A1a Trp Glu Lys Gly Pro Cys Pro Ala Arg Gly Leu Va1 Ser Ser
1585 1590 1595 1600
Leu Gly Arg Gly Ala Thr Cys Arg Ser Ser Asp Val Leu Ser Cys Ile
1605 1610 1615
Leu His Leu Leu Val Lys Gly Thr Leu Arg Arg His Asp Asp Arg Pro
1620 1625 1630
Gln Va'_ Leu Tyr Tyr Ala Val Pro Val Thr Val Met Glu Pro His Met
1635 1640 1645
Glu Ser Leu Asn Pro Gly Ser Ala Gly Pro Asn Pro Pro Leu Thr Phe
1650 1655 1660
His Thr Leu Gln Ile A_rg Ser Arg Gly Val Pro Tyr Ala Ser Cys Thr
1665 1670 1675 1680
Asp Asn His Thr Phe Ser Thr Phe Arg
1685
<210> 3
<211> 5253
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (87)..(5183)
<400> 3
gtcgccgcca gcgtctgtgc cgcgtccctt gctctgtgaa ggacaggcct cgcgccagga 60
ccccggtgga cttctgaggt gccagg atg gtg gga gaa ctc cgc tac agg gaa 113
Met Val Gly Glu Leu Arg Tyr Arg Glu
1 5
ttc agg gtg ccc ctg ggg ccc ggc tta cat gcc tat cct gat gag ctg 161
Phe Arg Val Pro Leu Gly Pro Gly Leu His Ala Tyr Pro Asp Glu Leu
15 20 25
atc cgc cag cgc g~g ggc cat gat ggg cat cct gag tac cag atc cgt 209
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
13
Ile Arg Gln Arg Val Gly His Asp Gly His Pro Glu Tyr Gln Ile Arg
30 35 40
tgg ctc atc ctg cgg cgt ggc gat gag ggg gac ggg ggc tct ggc caa 257
Trp Leu Ile Leu Arg Arg Gly Asp Glu Gly Asp Gly Gly Ser Gly Gln
45 50 55
gtg gac tgc aag get gag cac atc ctg ctg tgg atg tcc aag gat gag 305
Val Asp Cys Lys Ala Glu His Ile Leu Leu Trp Met Ser Lys Asp Glu
60 65 70
atc tat gcc aac tgc cac aag atg ctg ggc gag gat ggc cag gtc atc 353
Ile Tyr Ala Asn Cys His Lys Met Leu Gly Glu Asp Gly Gln Val Ile
75 80 85
ggg ccc tcc cag gag tct gca ggg gag gtt ggg gcc ctg gac aaa tct 401
Gly Pro Ser Gln Glu Ser Ala Gly Glu Val G'_y A1a Leu Asp Lys Ser
90 95 100 105
gtg ctg gag gag atg gaa acc gat gtg aag tcc ctc att cag aga gcc 449
Val Leu Glu Glu Met Glu Thr Asp Val Lys Ser Leu Ile Gln Arg Ala
110 115 120
ctt cgg cag ctg gag gag tgt gtg ggc act atc cct cct get ect eta 497
Leu Arg Gln Leu Glu Glu Cys Va1 Gly Thr :'_e Pro Pro Ala Pro Leu
125 130 135
ctt cac act gtc cac gtg ctc agc gcc tat gcc agc att gag ccc ctc 545
Leu F-fis Thr Val ::is Val Leu Ser Ala Ty= Ala Ser Ile G1u Pro Leu
140 145 150
act gga gta ttc aag gac cca agg gtc ctg gac ttg ctc atg cac atg 593
Thr Gly Val Phe Lys Asp Pro Arg Val Leu Asp Leu Leu Met His Met
155 160 165
ttg agt agt cct gat tat cag att cgc tgg agt gca ggc cgg atg ata 641
Leu Ser Ser Pro ~sp Tyr Gln Ile Arg Trp Ss_- Ala Gly Arg Met Ile
170 175 130 185
caa gcc ctg tcc tec cat gac get ggg ace egg act cag atc ett ctg 689
Gln Ala Leu Ser Ser His Asp Ala Gly Thr Arg Thr Gln Ile Leu Leu
190 195 200
tca ctg agc caa caa gaa gcc att gag aaa cac ctg gat ttt gac agc 737
Ser Leu Ser G1n Gln Glu Ala Ile Glu Lys His Leu Asp Phe Asp Ser
205 210 215
cgc tgt get ctg eta gca ctg ttt gca cag gec acg ctc tct gaa cac 785
Arg Cys Ala Leu Leu Ala Leu Phe Ala Gln Ala Thr Leu Ser Glu His
220 225 230
ccc atg tct ttc gag ggc att cag cta cca cag gtc cca gga agg gtg 833
Pro Met Ser Phe Glu Gly Ile Gln Leu Pro G'_.~. Val Pro Gly Arg Val
235 240 245
ctc ttc tcc ctg gtg aag cgg tat ttg cat gtc acc tcg ctc ctg gat 881
Leu Phe Ser Leu Val Lys Arg Tyz Leu His Val Thr Ser Leu Leu Asp
250 255 250 265
cag ctg aac gac agt get gcg gag cca gga gcc cag aac acc tct get 929
Gln Leu Asn Asp Ser Ala Ala Glu Pro Gly Ala Gln Asn Thr Ser Ala
270 275 280
cct gag gag ttg agt ggg gag agg ggt caa c:g gag ctg gag ttc agt 977
Pro Glu Glu Le:: Ser Gly G1u Arg Gly Gln Leu Glu Leu Glu Phe Ser
285 290 295
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
14
atg gcc atg ggc acc ctg atc tcg gag ctg gtg caa gcc atg cgc tgg
1025
Met Ala Met Gly Thr Leu Ile Ser G'_u Leu Va1 Gln Ala Met Arg Trp
300 305 310
gac cag gcc tca gac aga cca agg agc tca gca cgg tcc ccc ggt tcc
1073
Asp Gln Ala Ser Asp Arg Pro Arg Ser Ser Ala Arg Ser Pro Gly Ser
315 320 325
atc ttc cag cct cag ctg gca gat gtg agc cca ggg ctc ccc get gcc
1121
Ile Phe Gln Pro Gln Leu Ala Asp Val Ser Pro Gly Leu Pro Ala Ala
330 335 340 345
cag get cag ccc tcc ttc agg agg tca aga cgt ttt cgc cct cgt tct
1169
Gln Ala Gln Pro Ser Phe Arg Arg Ser Arg Arg Phe Arg Pro Arg Ser
350 355 360
gag ttc gca agt ggc aat acc tat get ttg tat gtg cgg gac aca ctg
1217
Glu Phe Ala Ser Gly Asn Thr Tyr Ala Leu Tyr Val Arg Asp Thr Leu
365 370 375
cag ccg ggg atg cga gtg cgg atg ctg gat gat tat gag gag atc agt
1265
Gln P=o Gly Met Arg Val Arg Met Leu Asp Asp Tyr Glu Glu Ile Ser
380 385 390
gcc ggg gat gag ggc gag ttt cgg cag agc aac aac ggt gtg cct cct
1313
Ala Gly Asp Glu Gly G1u P'.~.e Arg Gln Ser Asn Asn Gly VaI Pro Pro
395 400 405
gtg cag gta ttt tgg gag tca aca ggc cgc acc tat tgg gtg cac tgg
1361
Val Gln Val Phe Trp Glu Ser Thr Gly Arg Thr Tyr Trp Val His Trp
410 415 420 425
cac atg ctg gag atc ttg ggc ttt gag gaa gac att gag gac atg gtt
1409
His Met Leu Glu Ile Leu Gly Phe Glu Glu Asp Ile Glu Asp Met Val
430 435 440
gag get gat gag tac caa ggg gca gtg gcc agt aga gtc ctg ggt aga
1457
Glu Ala Asp Glu Tyr Gln Gly Ala Val Ala Ser Arg Val Leu Gly Arg
445 450 455
gcc ctg cct gcc tgg cgc tgg agg ccc atg aca gaa ctc tat get gtg
1505
Ala Leu Pro Ala Trp Arg Trp Arg Pro Met Thr Glu Leu Tyr Ala Val
460 465 470
cct tat gtg ctg cct gag gat gag gac act gag gag tgt gaa cac ctg
1553
Pro Tyr Val Leu Pro Glu Asp Glu Asp Thr G1u Glu Cys Glu His Leu
475 480 985
acc ctg get gag tgg tgg gaa ctc cte ttc ttc atc aag aag ctg gat
1601
Thr Leu Ala Glu Trp Trp Glu Leu Leu Phe Phe I1e Lys Lys Leu Asp
490 495 500 505
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
gga cct gac cat cag gag gtt ctc cag atc ctc cag gag aac cta gat
1649
Gly Pro Asp His Gln Glu Val Leu Gln Ile Leu Gln Glu Asn Leu Asp
510 515 520
ggg gag att ctg gat gat gag atc cta get gaa ctg gcc gtg ccc ata
1697
Gly G1u Ile Leu Asp Asp Glu Ile Leu Ala Glu Leu Ala Va1 Pro Ile
525 530 535
gaa ttg gcc cag gac ttg ctg ctg act ctg cca cag cga ctc aat gac
1745
Glu Leu Ala Gln Asp Leu Leu Leu Thr Leu Pro Gln Arg Leu Asn Asp
540 545 550
agt gcc ctc agg gac ctg atc aac tgc cat gtc tac aag aag tat ggg
1793
Ser Ala Leu Arg Asp Leu Ile Asn Cys His Val Tyr Lys Lys Tyr Gly
555 560 565
cct gaa gcc cta gca ggg aac caa gcc tac cca tcc ctt cta gaa gcc
1841
Pro Glu Ala Leu Aia Gly Asn Gln Ala Tyr Pro Ser Leu Leu Glu Ala
570 575 580 585
caa gaa gat gtc c=c ctg cta gac gcg cag gcc cag get aag gac tca
1889
Gln Glu Asp Val Le'3 Leu Leu Asp Ala Gln Ala Gln Ala Lys Asp Ser
590 595 600
gaa gat gca gcc aaa gtg gaa gca aaa gaa ccc cca tct cag agt ccc
1937
Glu Asp Ala Ala Lys Val Glu Ala Lys Glu Pro Pro Ser Gln Ser Pro
605 610 615
aac act ccc ctg cag cgt ctg gtg gag ggt tat ggt cca get ggg aaa
1985
Asn Thr Pro Leu G:z Arg Leu Val Glu Gly Tyr Gly Pro Ala Gly Lys
620 625 630
atc ctc ctg gat c_a gag caa gcc ctc agc tca gag ggg acc cag gag
2033
Ile Leu Leu Asp Leu Glu Gln Ala Leu Ser Ser Glu Gly Thr Gln Glu
635 640 645
aac aag gtc aag cca ctc ctg ctg cag ctg cag cgg cag ccg cag ccc
2081
Asn Lys Val Lys Pro Leu Leu Leu Gln Leu Gln Arg Gln Pro Gln Pro
650 655 660 665
ttc ctg gca ctg atg cag agc ctg gac act ccg gag act aac agg acc
2129
Phe Leu Ala Leu Met Gln Ser Leu Asp Thr Pro Glu Thr Asn Arg Thr
670 675 680
ctg cac ctg act gtg ctg aga atc ctg aag cag ctg gtg gac ttc ccc
2177
Leu His Leu Thr Val Leu Arg Ile Leu Lys Gln Leu Val Asp Phe Pro
685 690 695
gag gca ctg ctg ctc ccc tgg cac gag gcc gtg gat gcc tgc atg gcc
2225
Glu Ala Leu Leu Leu Pro Trp His Glu Ala Val Asp Ala Cys Met Ala
700 705 710
tg~ ctg cgg tcc cca aac act gat cga gag gtg ctc cag gaa ctg att
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
16
2273
Cys Leu Arg Ser Pro Asn Thr Asp Arg Glu Val Leu Gln Glu Leu Ile
715 720 725
ttc ttc ctg cac cgc ctg acc tca gtg agc agg gac tat gcc gtg gtg
2321
Phe Phe Leu His Arg Leu Thr Ser Va1 Ser Arg Asp Tyr Ala Val Val
730 735 740 745
ctg aat cag ctg gga gca aga gac get atc tcc aag gcc ctg gaa aag
2369
Leu Asn Gln Leu Gly Ala Arg Asp Ala Ile Ser Lys Ala Leu Glu Lys
750 755 760
cac ctg gga aag ctg gag ctg get cag gag ctg cgg gac atg gtg ttc
2417
His Leu Gly Lys Leu Glu Leu Ala Gln Glu Leu Arg Asp Met Val Phe
765 770 775
aag tgt gag aag cat gcc cac ctc tac cgc aaa ctc atc acc aac atc
2465
Lys Cys Glu Lys His Ala His Leu Tyr Arg Lys Leu Ile Thr Asn Ile
780 785 790
ctg gga ggc tgc atc cag atg gtg ctg ggc cag atc gaa gac cac aga
2513
Leu Gly Gly Cys T_le G'_n Met Val Leu Gly Gln Ile Glu Asp His Arg
795 800 805
cga acc cac cgg ccc atc aac atc cct ttc ttt gat gtg ttc ctc aga
2561
Arg Thr His Arg P~o Ile Asn Ile Pro Phe Phe Asp Val Phe Leu Arg
810 815 820 825
tac ctg tgc cag ggc tcc agt gtg gaa gtg aag gag gac aag tgc tgg
2609
Tyr Leu Cys Gln G'_y Ser Ser Val Glu Val Lys Glu Asp Lys Cys Trp
830 835 840
gag aag gtg gag gtg tcc tcc aac ccg cac cgg gcc agc aag ctg acg
2657
Glu Lys Va1 Glu Val Ser Ser Asn Pro His Arg Ala Ser Lys Leu Thr
845 850 855
gac cac aac ccc aag acc tat tgg gag tcc aac ggc agc gcc ggc tcc
2705
Asp His Asn Pro Lys Thr Tyr Trp Glu Ser Asn Gly Ser Ala Gly Ser
860 865 870
cac tac atc acc ctg cac atg cgc cgg ggc atc ctc atc agg caa ctg
2753
His Tyr Ile Thr Leu His Met Arg Arg Gly Ile Leu Ile Arg Gln Leu
875 880 885
act ctg ctt gtg get agt gag gac tcg agt tac atg ccg gcc cga gtg
2801
Thr Leu Leu Val Ala Ser G1u Asp Ser Ser :'yr Met Pro Ala Arg Val
890 895 900 905
gtg gtg tgc ggg ggt gat agc act agc tct ctt cac acg gaa ctc aac
2849
Val Val Cys Gly Gly Asp Ser Thr Ser Ser Leu His Thr Glu Leu Asn
910 915 920
tcg gtg aat gtg atg ccc tct gcc agc cgg gtg atc ctc ctg gag aac
2897
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
17
Ser Val Asn Val Met Pro Ser Ala Ser Arg Val Ile Leu Leu Glu Asn
925 930 935
ctg acc cgc ttc tgg ccc atc atc cag atc cgc ata aag cgc tgc cag
2945
Leu Thr Arg Phe Trp Pro Ile Ile Gln Ile Arg Ile Lys Arg Cys Gln
940 945 950
cag ggt ggc att gat acg cgc att cgg ggg tta gag atc cta ggc ccc
2993
Gln Gly Gly Ile Asp Thr Arg Ile Arg Gly Leu Glu Ile Leu Gly Pro
955 960 965
aag ccc acg ttc tgg cca gtg ttc cgg gag cag ctc tgt cgt cac aca
3041
Lys Pro Thr Phe Trp Pro Val Phe Arg Glu Gln Leu Cys Arg His Thr
970 975 980 985
cgc ctc ttc tac atg gtt cgg gca cag gcc tgg agc cag gac atg gca
3089
Arg Leu Phe Tyr Met Val Arg Ala Gln Ala Trp Ser Gln Asp Met Ala
990 995 1000
gag gac cgc agg agc ctc ctg cac ctg agt tct aga ctc aac ggt get
3137
Glu Asp Arg Arg S2_- Leu Leu His Leu Ser Ser Arg Leu Asn Gly Ala
1005 1010 1015
ctg cgc cag gag cag aat ttt get gac cgc ttc ctc cct gat gac gag
3185
Leu Arg Gln G1u Gln Asn Phe Ala Asp Arg Phe Leu Pro Asp Asp Glu
1020 1025 1030
get gcc caa get ctg ggc aag acc tgc tgg gag gcc ctg gtc agc ccc
3233
Ala Ala Gln A1a Leu Gly Lys Thr Cys Trp Glu Ala Leu Val Ser Pro
1035 1040 1045
gtg gtg cag aac atc acc tcc cct gat gag gat ggc att agc ccc ctg
3281
Val Val Gln Asn Ile Thr Ser Pro Asp Glu Asp Gly Ile Ser Pro Leu
1050 1055 1060 1065
ggt tgg ctg ctg gac cag tac ctg gag tgt cag gaa get gtc ttc aac
3329
Gly Trp Leu Leu Asp Gln Tyr Leu Glu Cys Gln Glu Ala Val Phe Asn
1070 1075 1080
ccc cag agc cgc ggc cca get ttc ttc tcg cgg gtg cgc cgt ctc act
3377
Pro Gln Ser Arg Gly Pro Ala Phe Phe Ser Arg Val Arg Arg Leu Thr
1085 1090 1095
cac ctg ctg gtg cat gtc gag ccc tgt gag gca ccc cct cct gtg gtg
3425
His Leu Leu Val His Val Glu Pro Cys Glu Ala Pro Pro Pro Val Val
1100 1105 1110
gcc act cct cgg ccc aaa ggc aga aac aga agc cac gac tgg agc tcc
3473
Ala Thr Pro Arg Pro Lys G1y Arg Asn Arg Ser His Asp Trp Ser Ser
1115 1120 1125
ttg get acc cgg ggc ctt cca agc agc atc atg aga aac ctg acg cgc
3521
Leu Ala Thr Arg Gly Leu Pro Ser Ser Ile Met Arg Asn Leu Thr Arg
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
18
1130 1135 1140 1145
tgt tgg cgg gcc gtg gtg gag aag cag gtg aac aat ttt ctg acc tca
3569
Cys Trp Arg Ala Val Val Glu Lys Gln Val Asn Asn Phe Leu Thr Ser
1150 1155 1160
tcc tgg cgg gat gat gac ttt gtg cca cgc tac tgt gag cac ttt aat
3617
Ser Trp Arg Asp Asp Asp Phe Val Pro Arg Tyr Cys Glu His Phe Asn
1165 1170 1175
att ctg cag aac tca agc tct gaa ctg ttt ggg cct cgg gca gcc ttc
3665
Ile Leu Gln Asn Ser Ser Ser Glu Leu Phe Gly Pro Arg Ala Ala Phe
1180 1185 1190
ttg ctg gcg ctg caa aat ggc tgt gcg gga gcc ttg ctg aag ctc cct
3713
Leu Leu Ala Leu Gln Asn Gly Cys Ala Gly Ala Leu Leu Lys Leu Pro
1195 1200 1205
ttt ctc aaa get gcc cac gtg agt gag cag ttc gcc cgg cac att gac
3761
Phe Leu Lys Ala Ala His Val Ser Glu Gln Phe Ala Arg His Ile Asp
1210 1215 1220 1225
cag cag atc cag ggc agc cgg atc ggt gga gcc cag gaa atg gag agg
3809
Gln Gln Ile Gln G1y Ser Arg Ile Gly G1y A1a Gln Glu Met Glu Arg
1230 1235 1240
ctg gca cag ctg cag caa tgc ctg caa get gtc ctg att ttc tcc ggc
3857
Leu A'_a Gln Leu Gln Gln Cys Leu Gln Ala Vai Leu Ile Phe Ser Gly
1245 1250 1255
ttg gag ata gcc acc act ttt gag cat tat tac cag cac tac atg gcg
3905
Leu Glu Ile Ala Thr Thr Phe Glu His Tyr Tyr G1n His Tyr Met Ala
1260 1265 1270
gac cgt ctc ctg ggc gtg gtc tcg agc tgg ctg gag ggg gcc gtg ctg
3953
Asp Arg Leu Leu Gly Val Val Ser Ser Trp Leu Glu Gly Ala Val Leu
1275 1280 1285
gag cag atc ggt ccc tgc ttc ccc aac cgc ctc ccc cag cag atg ttg
4001
Glu Gln Ile Gly Pro Cys Phe Pro Asn Arg Leu Pro Gln Gln Met Leu
1290 1295 1300 1305
cag agc ctg agc acc tct aag gag ctg cag cgc cag ttc cac gtc tac
4049
Gln Ser Leu Ser Thr Ser Lys Glu Leu Gln Arg Gln Phe His Val Tyr
1310 1315 1320
cag c=c cag cag ctg gat cag gaa ctc cta aag ctg gag gat aca gag
4097
Gln Leu Gln Gln Leu Asp Gln Glu Leu Leu Lys Leu Glu Asp Thr Glu
1325 1330 1335
aag aaa ata cag gtg ggc ctt ggg gcc agt ggc aag gag cac aag agc
4145
Lys Lys Ile G1.~. Val Gly Leu Gly Ala Ser Gly Lys Glu His Lys Ser
1340 1345 1350
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
19
gag aag gaa gag gaa get ggg gca gca gca gtg gtg gat gtg gcg gag
4193
Glu Lys Glu Glu Glu Ala Gly Ala Ala Ala Val Val Asp Val Ala Glu
1355 1360 1365
gga gag gag gaa gag gag gag aat gag gac ctc tac tat gaa ggg gca
4241
Gly Glu Glu Glu Glu Glu Glu Asn Glu Asp Leu Tyr Tyr Glu Gly Ala
1370 1375 1380 1385
atg cca gaa gtg tct gtg ctt gtc ctg tcc cga cac tcc tgg cct gtt
4289
Met Pro Glu Val Ser Val Leu Val Leu Ser Arg His Ser Trp Pro Val
1390 1395 1400
gcc tca atc tgc cac aca ctg aac ccc aga acc tgc ctg ccc tcc tac
4337
Ala Ser Ile Cys His Thr Leu Asn Pro Arg Thr Cys Leu Pro Ser Tyr
1405 1410 1415
ctg agg ggc act ttg aac aga tac tcc aac ttc tac aac aag agt cag
4385
Leu Arg Gly Thr Leu Asn Arg Tyr Ser Asn Phe Tyr Asn Lys Ser Gln
1420 1425 1430
agc cac cct gcc ctt gag cga ggc tca cag agg cga ctg cag tgg acg
4433
Ser His Pro Ala Leu Glu Arg Gly Ser Gln Arg Arg Leu Gln Trp Thr
1435 1440 1445
tgg ctg ggc tgg get gag ctg cag ttt ggg aac cag acc ctg cat gtg
4481
Trp Leu Gly Trp Ala G1u Leu Gln Phe Gly Asn Gln Thr Leu His Val
1450 1455 1460 1465
tcc acc gtg cag atg tgg cta ctg ctg tat ctc aac gac ctg aag gcg
4529
Ser Thr Val Gln Met Trp Leu Leu Leu Tyr Leu Asn Asp Leu Lys Ala
1470 1475 1480
gtc tct gtg gag agt ctg ctg gcg ttc tca ggg ctc tcc gca gac atg
4577
Val Ser Val Glu Ser Leu Leu Ala Phe Ser Gly Leu Ser Ala Asp Met
1485 1490 1495
ctc aat cag gcg att ggg ccc ctc acc tct tca aga ggc ccc ctg gac
4625
Leu Asn Gln Ala Ile Gly Pro Leu Thr Ser Ser Arg Gly Pro Leu Asp
1500 1505 1510
ctt cac gag caa aag gat ata cca gga ggg gtc ctc aag att cga gat
4673
Leu His Glu Gln Lys Asp Ile Pro Gly Gly Val Leu Lys Ile Arg Asp
1515 1520 1525
ggc agc aag gaa ccc agg tcg aga tgg gac att gtg cgg ctc atc cca
4721
Gly Ser Lys G1u Pro Arg Ser Arg Trp Asp Ile Val Arg Leu Ile Pro
1530 1535 1540 1545
cct cag acg tac ctg caa get gag ggt gaa gac ggc cag aac ttg gag
4769
Pro Gln Thr Tyr Leu Gln Ala Glu G1y Glu Asp Gly Gln Asn Leu Glu
1550 1555 1560
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
aag aga cgg aat ctt ctg aac tgc ctc atc gtc cga atc ctc aag gcc
4817
Lys Arg Arg Asn Leu Leu Asn Cys Leu Ile Val Arg Ile Leu Lys Ala
1565 1570 1575
cat gga gat gag ggg ctg cac att gac cag ctt gtc tgt ctg gtg ctg
4865
His Gly Asp Glu Gly Leu His Ile Asp Gln Leu Val Cys Leu Val Leu
1580 1585 1590
gag get tgg cag aag ggc ccg tgt cct ccc agg ggt ttg gtc agc agc
4913
Glu Ala Trp Gln Lys Gly Pro Cys Pro Pro Arg Gly Leu Val Ser Ser
1595 1600 1605
ctt ggt aag ggg tct gca tgc agc agc act gac gtc ctc tcc tgc atc
4961
Leu G1y Lys Gly Ser Ala Cys Ser Ser Thr Asp Val Leu Ser Cys Ile
1610 1615 1620 1625
cta cac ctc ctg ggc aag ggc acg ctg aga cgc cat gac gac cgg ccc
5009
Leu His Leu Leu Gly Lys Gly Thr Leu Arg Arg His Asp Asp Arg Pro
1630 1635 1640
cag gtg ctg tcc tat gca gtc cct gtg act gtc atg gag cct cac act
5057
Gln Val Leu Ser Tyr Ala Val Pro V-al Thr Val Met Glu Pro His Thr
1645 1650 1655
gag tcc ctg aac cca ggc tcc tca ggc ccc aac cca ccc ctc acc ttc
5105
Glu Ser Leu Asn Pro Gly Ser Ser Gly Pro Asn Pro Pro Leu Thr Phe
1660 1665 1670
cat acc cta cag att cgc tcc cgg ggt gtg ccc tat gcc tcc tgc act
5153
His Thr Leu Gln Ile Arg Ser Arg G1y Val P=o Tyr Ala Ser Cys Thr
1675 1680 1685
gcc acc cag agc ttc tct acc ttc cgg tag ccctagactt ggggtcaggg
5203
Ala Thr Gln Ser Phe Ser Thr Phe Arg
1690 1695
gaaggtagag ctggagcttt tacagaaatt aaacccaaga gtttgattat
5253
<210> 4
<211> 1698
<212> PRT
<213> Homo sapiens
<400> 4
Met Val Gly Glu Leu Arg Tyr Arg Glu Phe Arg Va1 Pro Leu Gly Pro
1 5 10 15
Gly Leu His Ala Tyr Pro Asp Glu Leu Ile A_~g Gln Arg Val G1y His
20 25 30
Asp G1y His Pro G1u Tyr Gln Ile Arg Trp Leu Ile Leu Arg Arg Gly
35 40 45
Asp Glu Gly Asp Gly Gly Ser Gly Gln Val Asp Cys Lys Ala Glu His
50 55 60
Ile Leu Leu Trp Met Ser Lys Asp Glu Ile Tyr Ala Asn Cys His Lys
65 70 75 80
Met Leu Gly Glu Asp Gly Gln Val Ile Gly Pro Ser Gln Glu Ser Ala
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
2l
85 90 95
Gly Glu Val Gly Ala Leu Asp Lys Ser Val Leu Glu Glu Met Glu Thr
100 105 110
Asp Val Lys Ser Leu I1e Gln Arg Ala Leu Arg Gln Leu Glu Glu Cys
115 120 125
Val Gly Thr Ile Pro Pro Ala Pro Leu Leu His Thr Val His Val Leu
130 135 140
Ser Ala Tyr Ala Ser Ile Glu Pro Leu Thr Gly Val Phe Lys Asp Pro
145 150 155 160
Arg Val Leu Asp Leu Leu Met His Met Leu Ser Ser Pro Asp Tyr Gln
165 170 175
Ile Arg Trp Ser Ala Gly Arg Met Ile Gln Ala Leu Ser Ser His Asp
180 185 190
A-la Gly Thr Arg Thr Gln Ile Leu Leu Ser Leu Ser Gln Gln Glu Ala
195 200 205
Ile Glu Lys His Leu Asp Phe Asp Ser Arg Cys Ala Leu Leu Ala Leu
210 215 220
Phe Ala Gln Ala Thr Leu Ser Glu His Pro Met Ser Phe Glu Gly Ile
225 230 235 240
Gln Leu Pro Gln Val Pro Gly Arg Val Leu Phe Ser Leu Val Lys Arg
245 250 255
Tyr Leu His Val Thr Ser Leu Leu Asp Gln Leu Asn Asp Ser Ala Ala
260 265 270
Glu Pro Gly Ala Gln Asn Thr Ser Ala Pro Glu Glu Leu Ser Gly Glu
275 280 285
Arg Gly Gln Leu Glu Leu Glu Phe Ser Met Ala :~!et Gly Thr Leu Ile
290 295 300
Ser G1u Leu Val Gln Ala Met Arg Trp Asp Gln Ala Ser Asp Arg Pro
305 ~ 310 315 320
Arg Ser Ser Ala Arg Ser Pro Gly Ser Ile Phe Gln Pro Gln Leu Ala
325 330 335
Asp Val Ser Pro Gly Leu Pro Ala Ala Gln Ala Gln Pro Ser Phe Arg
340 345 350
Arg Ser Arg Arg Phe Arg Pro Arg Ser Glu Phe Ala Ser Gly Asn Thr
355 360 365
Tyr Ala Leu Tyr Val r..rg Asp Thr Leu Gln Pro Gly M_et Arg Val Arg
370 375 ' 380
Met Leu Asp Asp Tyr Glu Glu I1e Ser Ala Gly Asp Glu Gly Glu Phe
385 390 395 400
Arg Gln Ser Asn Asn Gly Val Pro Pro Val G1n Val Phe Trp Glu Ser
405 410 415
Thr Gly Arg Thr Tyr Trp Val His Trp His Met Leu Glu Ile Leu Gly
420 425 430
Phe Glu Glu Asp Ile Glu Asp Met Val Glu Ala Asp Glu Tyr Gln Gly
435 940 445
Ala Val Ala Ser Arg Val Leu Gly Arg Ala Leu Pro Ala Trp Arg Trp
450 455 460
Arg Pro Met Thr Glu Leu Tyr Ala Val Pro Tyr Val Leu Pro Glu Asp
465 470 475 480
Glu Asp Thr Glu Glu Cys Glu His Leu Thr Leu Ala Glu Trp Trp Glu
485 490 495
Leu Leu Phe Phe Ile Lys Lys Leu Asp Gly Pro Asp His Gln Glu Val
500 505 510
Leu Gln Ile Leu Gln Glu Asn Leu Asp Gly Glu Ile Leu Asp Asp Glu
515 520 525
Ile Leu Ala Glu Leu Ala Val Pro Ile Glu Leu Ala Gln Asp Leu Leu
530 535 540
Leu Thr Leu Pro Gln Arg Leu Asn Asp Ser Ala Leu Arg Asp Leu Ile
545 550 555 560
Asn Cys His Val Tyr Lys Lys Tyr Gly Pro G'_u ala Leu Ala Gly Asn
565 570 575
Gln Ala Tyr Pro Ser Leu Leu Glu Ala Gln Glu Asp Val Leu Leu Leu
580 585 590
Asp Ala Gln Ala Gln Ala Lys Asp Ser Glu Asp Ala Ala Lys Val Glu
595 600 605
Al.a Lys Glu Pro Pro Ser Gln Ser Pro Asn Thr Pro Leu Gln Arg Leu
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
22
610 615 620
Val Glu Gly Tyr Gly Pro Ala Gly Lys Ile Leu Leu Asp Leu Glu Gln
625 630 635 640
Ala Leu Ser Ser Glu Gly Thr Gln Glu Asn Lys Val Lys Pro Leu Leu
645 650 655
Leu Gln Leu Gln Arg Gln Pro Gln Pro Phe Leu Ala Leu Met Gln Ser
660 665 670
Leu Asp Thr Pro G1u Thr Asn Arg Thr Leu His Leu Thr Val Leu Arg
675 680 685
Ile Leu Lys Gln Leu Val Asp Phe Pro Glu Ala Leu Leu Leu Pro Trp
690 695 700
His Glu Ala Val Asp Ala Cys Met Ala Cys Leu Arg Ser Pro Asn Thr
705 710 715 720
Asp Arg Glu Val Leu Gln Glu Leu Ile Phe Phe Leu His Arg Leu Thr
725 730 735
Ser Val Ser Arg Asp Tyr Ala Val Val Leu Asn Gln Leu Gly Ala Arg
740 745 750
Asp Ala Ile Ser Lys Ala Leu Glu Lys His Leu Gly Lys Leu Glu Leu
755 760 765
Ala Gln Glu Leu Azg Asp Met Val Phe Lys Cys Glu Lys His Ala His
770 775 780
Leu Tyr Arg Lys Leu Ile Thr Asn Ile Leu Gly Gly Cys Ile Gln Met
785 790 795 800
Val Leu Gly G1n Ile G1u Asp His Arg Arg Thr His Arg Pro Ile Asn
805 810 815
Ile Pro Phe Phe Asp Val Phe Leu Arg Tyr Leu Cys Gln Gly Ser Ser
820 825 830
Val Glu Val Lys G'_u Asp Lys Cys Trp Glu Lys Val Glu Val Ser Ser
835 840 845
Asn Pro His Arg A'_a Ser Lys Leu Thr Asp His Asn Pro Lys Thr Tyr
850 855 860
Trp Glu Ser Asn G'_y Ser A1a Gly Ser His Tyz Ile Thr Leu His Met
865 870 875 880
Arg Arg Gly I1e Le;: Ile Arg Gln Leu Thr Leu Leu Val Ala Ser Glu
885 890 895
Asp Se_- Ser Tyr Me~ Pro Ala Arg Val Val Va'_ Cys Gly Gly Asp Ser
900 905 910
Thr Se_- Ser Leu His Thr Glu Leu Asn Ser Va'_ Asn Val Met Pro Ser
915 920 925
Ala Ser Arg Val I'_e Leu Leu Glu Asn Leu Thr Arg Phe Trp Pro Ile
930 935 940
Ile Gln Ile Arg Ile Lys Arg Cys Gln Gln Gly Gly Ile Asp Thr Arg
945 950 955 960
Ile Arg Gly Leu Glu Ile Leu Gly Pro Lys Pro Thr Phe Trp Pro Val
965 970 975
Phe Arg Glu Gln Leu Cys Arg His Thr Arg Leu Phe Tyr Met Val Arg
980 985 990
Ala Gln Ala Trp Se_- Gln Asp Met Ala Glu Asp Arg Arg Ser Leu Leu
995 1000 1005
His Leu Ser Ser Arg Leu Asn Gly Ala Leu Arg Gln Glu Gln Asn Phe
1010 1015 1020
Ala Asp Arg Phe Leu Pro Asp Asp Glu Ala Ala Gln Ala Leu Gly Lys
1025 1030 1035 1040
Thr Cys Trp Glu Ala Leu Val Ser Pro Val Val Gln Asn Ile Thr Ser
1045 1050 1055
Pro Asp Glu Asp G'_y Ile Ser Pro Leu Gly Trp Leu Leu Asp Gln Tyr
1060 1065 1070
Leu Glu Cys Gln Glu Ala Val Phe Asn Pro Gln Ser Arg Gly Pro Ala
1075 1080 1085
Phe Phe Ser Arg Val Arg Arg Leu Thr His Leu Leu Val His Val Glu
1090 1095 1100
Pro Cys Glu Ala P=o Pro Pro Val Val Ala Thr Pro Arg Pro Lys Gly
1105 1110 1115 1120
Arg Asn Arg Ser His Asp Trp Ser Ser Leu Ala Thr Arg Gly Leu Pro
1125 1130 1135
Ser Sez Ile Met Arg Asn Leu Thr Arg Cys Trp Arg Ala Val Val Glu
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
23
1140 1145 1150
Lys Gln Val Asn Asn Phe Leu Thr Ser Ser Trp Arg Asp Asp Asp Phe
1155 1160 1165
Val Pro Arg Tyr Cys Glu His Phe Asn Ile Leu Gin Asn Ser Ser Ser
1170 1175 1180
Glu Leu Phe Gly Pro Arg Ala Ala Phe Leu Leu Ala Leu Gln Asn Gly
1185 1190 1195 1200
Cys Ala Gly Ala Leu Leu Lys Leu Pro Phe Leu Lys Ala Ala His Val
1205 1210 1215
Ser Glu Gln Phe Ala Arg His Ile Asp Gln Gln Ile Gln Gly Ser Arg
1220 1225 1230
Ile Gly Gly Ala Gln Glu Met Glu Arg Leu Ala Gln Leu Gln Gln Cys
1235 1240 1245
Leu Gln Ala Val Leu Ile Phe Ser Gly Leu Glu Ile Ala Thr Thr Phe
1250 1255 1260
Glu His Tyr Tyr Gln His Tyr Met Ala Asp Arg Leu Leu Gly Val Val
1265 1270 1275 1280
Ser Ser Trp Leu Glu Gly Ala Val Leu Glu Gln Ile Gly Pro Cys Phe
1285 1290 1295
Pro Asn Arg Leu Pro Gln Gln Met Leu Gln Ser Leu Ser Thr Ser Lys
1300 1305 1310
Glu Leu Gln Arg Gln Phe His Val Tyr Gln Leu Gln Gln Leu Asp Gln
1315 1320 1325
Glu Leu Leu Lys Leu Glu Asp Thr Glu Lys Lys I=a Gln Val Gly Leu
1330 1335 1340
Gly Ala Ser Gly Lys Glu His Lys Ser Glu Lys G'_u Glu Glu Ala Gly
1345 1350 1355 1360
Ala A,la Ala Val Val Asp Val Ala Glu Gly Glu G'_u Glu Glu Glu Glu
1365 1370 1375
Asn Glu Asp Leu Tyr Tyr Glu Gly Ala Met Pro Gnu Val Ser Val Leu
1380 1385 1390
Val Leu Ser Arg vis Ser Trp Pro Val Ala Ser I'_e Cys His Thr Leu
1395 1400 1405
Asn Pro Arg Thr Cys Leu Pro Ser Tyr_Leu Arg G'_y Thr Leu Asn Arg
1410 1415 1420
Tyr Ser Asn Phe Tyr Asn Lys Ser Gln Ser His ?_-o Ala Leu Glu Arg
1425 1430 1435 1440
Gly Ser Gln Arg Arg Leu Gln Trp Thr Trp Leu G'_y Trp Ala Glu Leu
1445 1450 1455
Gln Phe Gly Asn Gln Thr Leu His Val Ser Thr Val Gln Met Trp Leu
1460 1465 1470
Leu Leu Tyr Leu Asn Asp Leu Lys Ala Val Ser Val Glu Ser Leu Leu
1475 1980 1485
Ala Phe Ser Gly Leu Ser Ala Asp Met Leu Asn Gln Ala Ile Gly Pro
1490 1495 1500
Leu Thr Ser Ser Arg Gly Pro Leu Asp Leu His Glu Gln Lys Asp Ile
1505 1510 1515 1520
Pro Gly Gly Val Leu Lys Ile Arg Asp Gly Ser Lys Glu Pro Arg Ser
1525 1530 1535
Arg Trp Asp Ile Val Arg Leu Ile Pro Pro Gln Thr Tyr Leu Gln Ala
1540 1545 1550
Glu Gly Glu Asp Gly Gln Asn Leu G1u Lys Arg Arg Asn Leu Leu Asn
1555 1560 1565
Cys Leu Ile Val Arg Ile Leu Lys Ala His Gly Asp Glu Gly Leu His
1570 1575 1580
Ile Asp Gln Leu Val Cys Leu Val Leu G1u Ala Trp Gln Lys Gly Pro
1585 1590 1595 1600
Cys Pro Pro Arg Gly Leu Val Ser Ser Leu Gly :,yrs Gly Ser Ala Cys
1605 1610 1615
Ser Ser Thr Asp Val Leu Ser Cys Ile Leu His Leu Leu Gly Lys G1y
1620 1625 1630
Thr Leu Arg Arg His Asp Asp Arg Pro Gln Val Leu Ser Tyr Ala Va1
1635 1640 1645
Pro Val Thr Val Met Glu Pro His Thr Glu Ser Leu Asn Pro Gly Ser
1650 1655 1660
Ser Gly Pro Asn P~o Pro Leu Thr Phe His Thr Leu Gln Ile Arg Ser
SUBSTITUTE SHEET (RULE 26)
CA 02383371 2002-02-21
WO 01/14418 PCT/US00/23161
24
1665 1670 1675 1680
Arg Gly Val Pro Tyr Ala Ser Cys Thr Ala Thr Gln Ser Phe Ser Thr
1685 1690 1695
Phe Arg
SUBSTITUTE SHEET (RULE 26)