Note: Descriptions are shown in the official language in which they were submitted.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
1
TITLE OF THE INVENTION
DIAGNOSIS, PROGNOSIS AND TREATMENT OF
TRINUCLEOTIDE REPEAT-ASSOCIATED DISEASES AND INTRANUCLEAR
INCLUSIONS-ASSOCIATED DISEASES
FIELD OF THE INVENTION
The present invention relates to neurodegenerative disorders.
More particularly, the invention relates to Machado-Joseph disease (MJD). The
present invention also relates to trinucleotide repeat expansions and more
particularly to CAG repeats, also termed expansions of a coding CAG repeat
(exp-CAG), and GCG repeats. More particularly, the invention relates to: (1 )
exp-CAG associated diseases; (2) intranuclear inclusions (1N1) in patients and
cellular models of exp-CAG associated diseases; and (3) the elucidation of the
mechanism responsible for the toxic effects of such repeats. The present
invention therefore relates to the diagnosis, prognosis and treatment of
repeat-
associated diseases and INI-associated diseases as well as to assays for the
identification of agents which could be used for the treatment of such
diseases or
disorders.
BACKGROUND OF THE INVENTION
Coding CAG triplet repeat expansions cause several
neurodegenerative disorders, including Machado-Joseph disease (MJD)~5. The
presence of intranuclear filamentous inclusions (1N1) containing expanded
protein
in MJD, as well as in other expanded CAG repeat disorders (exp-CAG), have lead
to a nuclear toxicity model '~ ~9. Similar INI are found in oculopharyngeal
muscular dystrophy, which is caused by a short expansion of analanine encoding
GCG repeat. According to the present invention, it is proposed that
transcriptional or translational frameshifts occurring within expanded CAG
tracts
result in the production and accumulation of polyalanine-containing mutant
proteins. These alanine polymers might deposit in cells forming INI and lead
to
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
2
nuclear toxicity. Support for this disease model is provided using lymphoblast
cells from MJD patients, as well as in pontine neurons of MJD brain and in in
vitro
cell culture models of the disease. Evidence that alanine polymers alone are
toxic to cells is also provided and strongly suggests that a similar
pathogenic
mechanism underlies the other CAG repeat disorders.
Indeed, recent reviews describe a significant number of
neurodegenerative diseases, including Huntington disease as well as spinal
cerebellar ataxias which are caused by CAG repeat expansions. Of note, CAG
repeats code for polyglutamine in the protein containing same. It is commonly
believed that these polyglutamine stretches in proteins are toxic to cells,
and
these repeats are also termed CAG/polyglutamine repeats (Iver et al. 1999,
Nature Medicine 5:383-384). These diseases, also termed polyglutamine
diseases, are thought to occur by "a gain function mechanism". Unfortunately,
the mechanism explaining toxicity of the polyglutamine diseases, apparently
through an aggregation in nuclear inclusions, has yet to be provided, although
transgenic mice bearing a polyglutamine repeat in a recombinant protein were
shown to display intranuclear polyglutamine inclusions (Hardy et al. 1998,
Science 282:1076-1079). Of relevance, although the pathogenic effect of these
inclusion bodies is not clearly understood, it is recognized that numerous
types
of genes can contain these so-called CAG repeats, and that while these repeats
are linked to the disease, the genes containing these repeats are "largely
irrelevant to the disease process" (Hardy et al. ipid., supra).
INI are also found in oculopharyngeal muscular dystrophy
(OPMD)'°, which is caused by short expansions of a polyalanine
(polyAla)
encoding GCG tract in the PA8P2 gene" (also see PCT/CA98/01133). In
contrast to the CAG repeat disorders, where expansions frequently involve the
addition of 20 or more codons, very small GCG expansions (exp-GCG) of 2 to 7
additional codons are seen in dominant OPMD, suggesting thatpolyalanine tracts
are prone to aggregation and may be very toxic". This contention is supported
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
3
by the observation that polyAla peptides containing more than 9 alanines (Ala)
in
a row form [3-pleated sheet fibrillar macromolecules spontaneously in vitro'z,
which in turn are extremely resistant to chemical and enzymatic degradation'3.
Short trinucleotide repeat expansions causing a human
disease have been first described in PCT application number PCT/CA98/0113,
of Rouleau et al., which teaches that the addition of only two GCG repeats
(which
encode the amino acid alanine [ALA]) is sufficient to cause dominant OPMD.
OPMD expansions do not share the cardinal features of "dynamic mutations". The
GCG expansions are not only short they are also meiotically quite stable.
Furthermore, there is a clear cut-off between the normal and abnormal alleles:
a
single GCG expansion causing a recessive phenotype. The PAB II (GCC~7 allele
was thus the first example of a relatively frequent allele which can act as
either
a modifier of a dominant phenotype or as a recessive mutation. A dosage effect
of these repeats is also disclosed in PCT/CA98/01133, since a patient having
an
expansion in the polyalanine tract of the HOXD13 protein (Akarsu,. et al.,
1996,
Hum. Mol. Genet. 5: 945-952) has more severe deformities. A duplication
causing
a similar polyalanine expansion in the subunit 1 gene of the core-binding
transcription factor CBF(1 ) has also been found to cause dominant cleido-
cranial
dysplasia (Mundlos, S. et a1.,1997, Cell 89:773-779). Of note, however, the
mutations in these two rare diseases are not triplet-repeats. They are
duplications
of "cryptic repeats" composed of mixed synonymous codons and are thought to
result from unequal crossing over (Warren, 1997, Science275: 408-409). In the
case of OPMD, slippage during replication causing a reiteration of the GCG
codon is a more likely mechanism (Wells, 1996, J. Biol. Chem. 271: 2875-2878).
Different observations converge to suggest that a gain of
function of PAB II may cause the accumulation of nuclear filaments observed in
OPMD (Tome et al., 1980, Acta Neuropath. 49: 85-87). PAB II is found mostly in
dimeric and oligomeric form (Nemeth, et a1.,1995, Nucleic Acids Res.23: 4034-
4041 ). It is possible that the polyalanine tract plays a role in
polymerization.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
4
Polyalanine stretches have been found in many other nuclear proteins such as
the HOX proteins, but their function is still unknown (Davies, et al., 1997,
Ce1190:
537-548). Alanine is a highly hydrophobic amino acid present in the cores of
proteins. In dragline spider silk, polyalanine stretches are thought to form B-
sheet
structures important in ensuring the fibers' strength (Simmons, A.H. et al.,
Science 271:4-87 (1996)). Polyalanine oligomers have also been shown to be
extremely resistant to chemical denaturation and enzymatic degradation (Forood
et al. 1995, Bioch. and Biophy. Res. Com. 211:7-13). Their role in the disease
process, however, has still not been clearly identified. The more severe
phenotypes observed in homozygotes for the (GCG)9 mutations and compound
heterozygotes for the (GCG)9 mutation and (GCG)7 allele may correspond to the
fact that in these cases PAB II oligomers are composed only of mutated
proteins.
The ensuing faster filament accumulation could cause accelerated cell
death.The
recent description of nuclear filament inclusions in Huntington's disease,
raises
the possibility that "nuclear toxicity" caused by the accumulation of mutated
homopolymeric domains is involved in the molecular pathophysiology of other
triplet repeat diseases (Davies, S.W. et al., Ce1190:537-548 (1997);
Scherzinger,
E. et al., Cell 90:549-558 (1997); DiFiglia, M. et al., Science 277:1990-1993
(1997)). Additional data, including immunocytochemical and expression studies
will have to be provided to test this pathophysiological hypothesis and
provide
some insight into why certain muscle groups are more affected, while all
tissues
express PAB II.
There thus remains a need to elucidate the mechanism by
which GCG and CAG expansions are toxic to cells. There also remains a need
to provide diagnosis and/or prognosis and/or treatment tools for diseases
associated with GCG or CAG repeats.
The present invention seeks to meet these and other needs.
The present description refers to a number of documents, the
content of which is herein incorporated by reference in their entirety.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
SUMMARY OF THE INVENTION
The invention therefore concerns the identification of the
mechanistic action of expanded CAG tracts in cell pathogenesis, cell death or
disease. More specifically, the invention relates to the identification that
5 mutational events within these CAG enable them to encodepolyalanine
stretches
which accumulate in intranuclear filamentous inclusions and somehow trigger
toxicity in cells. In particular, the present invention relates
totranslational and/or
transcriptional frameshift events occurring within the CAG tracts, thereby
resulting
in the production and accumulation of polyalanine-containing mutant proteins.
The present invention, in addition, relates to a formal
identification of the toxic effects ofpolyalanine stretches on nuclear
toxicity. The
Applicant is thus the first to have demonstrated that CAG expansions can give
rise to production of mutant proteins containing polyalanine stretches. The
Applicant is also the first to have demonstrated that polyalanine stretches in
proteins are indeed toxic to cells.
The instant invention also relates to gene replacement
technologies aimed at deleting a repeat-containing protein giving rise to a
mutant
protein by a normal corresponding protein (i.e. lacking the repeat or having a
smaller repeat).
The present invention also provides the means to determine
a predisposition to developing a disease or condition associated with the
expression of a polyamino acid-containing protein such as polyalanine-
containing
proteins. This determination could thus enable a better prognosis of the
disease
and condition and enable a determination of the best treatment or prevention
of
the disease or condition.
Another aim of the present invention is thus to provide means
to screen humans (and more broadly animals) to identify those that might have
a predisposition to developing a disease associated with the expression of
genes
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
6
that lead to polyalanine-containing proteins such as CAG, GCG-repeat
containing
genes.
It is thus an aim of the present invention to provide the means
to better manage such disease prevention and intervention programs.
Before the present invention, CAG repeat expansions (also
known as CAG repeats or CAG/polyglutamine stretches) were recognized as a
common denominator in numerous neurological diseases and were thought to
code for polyglutamine stretches in the mutant protein. These polyglutamine
stretches were thought to confer "a gain of toxic property to these proteins"
(Iver et al. supra) by a mechanism that was not understood (Hardy et al.
supra.
Indeed, CAG tract toxicity was also referred to aspolyglutamine diseases. The
present invention demonstrates that these CAG repeats actually encode
polyalanine stretches.
Prior to the present invention, the identification of CAG
repeats in a gene correlating with a neurological disease led to the
classification
of such disease in the polyglutamine diseases (Hardy et al. supra). The
present
invention now demonstrates that polyalanine stretches, as opposed to
polyglutamine stretches, are responsible for the diseases.
In view of the above, the present invention opens the way to
numerous methods of diagnosing, prognosing or treating CAG repeat-dependent
diseases or conditions. In addition, it provides means to diagnose,prognose
and
treat diseases or conditions associated with polyalanine-containing proteins
(i.e.
GCG repeats). Non-limiting examples thereof comprise methods usingligands
(i.e. polyclonal and monoclonal antibodies), nucleic acid sequences,
restriction
length polymorphisms (RFLPs) and the like.
While the instant invention is more particularly directed to
neurological diseases, as is demonstrated with Machado-Joseph disease, it
should be understood that the present invention should not be so limited.
Indeed,
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
7
polyalanine stretches have been shown to be responsible for non-neurological
diseases such as, for example, a muscle disease (OPMD; PCT/CA98/01133).
In order to better understand the disease process associated
with the presence of INI in exp-CAG and exp-GCG associated disorders, a direct
assessment of whether polyAla stretches accumulated as intranuclear protein
aggregates was carried out. Experiments were thus performed to analyze
whether rare transcriptional or translational frameshifts in large CAG
stretches
resulting in new reading frames with the formation of a hybrid protein
containing
a mixed polyglutamine/poly-alanine tract occurred. Additionally, it was of
importance to assess whether the resultant polyAla peptides accumulate in
nuclei
where they form INI.
Surprisingly, it was discovered that an antiserum raised
against the hypothetical COOH terminus of the predicted polyAla containing
frameshifted ataxin-3 protein (MJD-Ala) detects the frameshifted species in
lymphoblastoid cells from MJD patients with large CAG tracts. This antiserum
detects these polyAla tracts as insoluble macromolecules on Western blots, and
as intranuclear inclusions by immunocytochemistry. Frameshifted species were
also present in INI in pontine neurons of MJD brain. Transfection of COS-7
cells
with full-length MJD-1 fused to the enhanced green fluorescence protein (EGFP~
gene in the alternative polyAla reading frame also leads to EGFP accumulation
preferentially when the CAG tract is expanded.
Of interest, it is also demonstrated that long CAG repeats are
prone to frameshifts, which result in accumulation of the predicted polyAla-
containing inclusions. Transfection of COS-7 cells with mutated MJD-1
constructs containing alanine-coding GCA stretches results in a more severe
phenotype when compared to their CAG counterparts. Furthermore,transfected
polyAla-encoding GCA stretches alone are toxic and form aggregates.
How these accumulations lead to cell death still needs to be
elucidated.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
8
A frameshift error occurring within a CAG tract thus results in
the alternate alanine-encoding GCA frame. Many authors have reported
frameshifts at the level of transcription or translation. The observation of
transcriptional errors of the ~3-amyloid precursor protein and ubiquitin-B in
Alzheimer's disease'4, and apoB86'S, supports the existence of such errors,
and
their role in disease pathogenesis. Translational errors have also been shown
to
occur and may be the basis for the formation of frameshifted proteins'6.
In accordance with one embodiment of the present invention,
there is therefore provided a method for the diagnosis of a disease associated
with protein accumulation in intranuclear inclusions, which comprises
obtaining
a sample of a patient and determining a presence of the protein accumulation
in
the intranuclear inclusions, wherein this protein accumulation is indicative
of a
disease related thereto.
In accordance with another embodiment of the present
invention, there is also provided a method for the screening of agents which
can
modulate at least one of (1 ) a polyamino acid-containing protein expression;
(2)
accumulation of polyamino acid-containing proteins in intranuclear inclusions;
and
(3) toxicity to cells, which comprises: a) incubating a cell harboring an
expression
vector of the present invention, comprising a repeat domain which can give
rise
to a polyamino acid-containing protein associated with a disease or condition
in
an animal, with a compound; and b) assessing one of (1) an expression of the
polyamino acid-containing protein; (2) accumulation of the polyamino acid-
containing protein; and (3) toxicity to cells; whereby a modulator is selected
when
the agent significantly modulates one of the expression, accumulation and
toxicity, as compared to a control agent.
The instant invention also relates to GCG repeats encoding
polyalanine stretches and their association with protein accumulation in a
cell
nucleus, swallowing difficulty and/or ptosis in a patient. In accordance with
another embodiment of the present invention, there is provided a method for
the
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
9
diagnosis or prognosis of a disease associated with protein accumulation in a
cell
nucleus, and/or swallowing difficulty and/or ptosis in a human patient, which
comprises:
a) obtaining a sample of a patient; and
b) determining the extract of the polyalanine stretch in an alanine
stretch-containing protein having the amino acid sequence:
Met (Ala)6+~ Ala,
wherein n is selected from 0 to 7, and
whereby an n equal to 1 to 7 is indicative of a disease related
with the protein accumulation in the nucleus, and/or a swallowing difficulty
and/or
ptosis in the patient. In a related aspect of the present invention, there is
provided
a human PAB II protein comprising a polymorphic GCG repeat encoding a
polyalanine stretch having the sequence
Met (Ala)6,~ Ala,
wherein n is 0, and wherein the sequence is indicative of a non-disease
phenotype associated with protein accumulation in a cell nucleus, swallowing
difficulty, and/or ptosis in a human patient.
In order to provide a clear and consistent understanding of
terms used in the present description, a number of definitions are provided
hereinbelow.
Nucleotide sequences are presented herein by single strand,
in the 5' to 3' direction, from left to right, using the one letter nucleotide
symbols
as commonly used in the art and in accordance with the recommendations of the
IUPAC-IUB Biochemical Nomenclature Commission.
Unless defined otherwise, the scientific and technological
terms and nomenclature used herein have the same meaning as commonly
understood by a person of ordinary skill to which this invention pertains.
Generally, the procedures for cell cultures, infection, molecular biology
methods
and the like are common methods used in the art. Such standard techniques can
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
be found in reference manuals such as for example Sambrook et al. (1989,
Molecular Cloning - A Laboratory Manual, Cold Spring Harbor Laboratories) and
Ausubel et al. (1994, Current Protocols in Molecular Biology, Wiley, New
York).
The present description refers to a number of routinely used
5 recombinant DNA (rDNA) technology terms. Nevertheless, definitions of
selected
examples of such rDNA terms are provided for clarity and consistency.
As used herein, "nucleic acid molecule", refers to a polymer of
nucleotides. Non-limiting examples thereof include DNA (i.e. genomic DNA,
cDNA) and RNA molecules (i.e. mRNA). The nucleic acid molecule can be
10 obtained by cloning techniques or synthesized. DNA can be double-stranded
or
single-stranded (coding strand or non-coding strand [antisense]).
The term "recombinant DNA" as known in the art refers to a
DNA molecule resulting from the joining of DNA segments. This is often
referred
to as genetic engineering.
The term "DNA segment", is used herein, to refer to a DNA
molecule comprising a linear stretch or sequence of nucleotides. This sequence
when read in accordance with the genetic code, can encode a linear stretch or
sequence of amino acids which can be referred to as a polypeptide, protein,
protein fragment and the like.
The terminology "amplification pair" refers herein to a pair of
oligonucleotides (oligos) of the present invention, which are selected to be
used
together in amplifying a selected nucleic acid sequence by one of a number of
types of amplification processes, preferably a polymerase chain reaction.
Other
types of amplification processes include ligase chain reaction, strand
displacement amplification, or nucleic acid sequence-based amplification, as
explained in greater detail below. As commonly known in the art, theoligos are
designed to bind to a complementary sequence under selected conditions.
The nucleic acid (i.e. DNA or RNA) for practicing the present
invention may be obtained according to well known methods.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
11
As used herein, the term "physiologically relevant" is meant to
describe a frameshifting event which can result in the production of a toxic
protein
in vivo.
Oligonucleotide probes or primers of the present invention
may be of any suitable length, depending on the particular assay format and
the
particular needs and targeted sequences employed. In general, the
oligonucleotide probes or primers are at least 12 nucleotides in length,
preferably
between 15 and 24 molecules, and they may be adapted to be especially suited
to a chosen nucleic acid amplification system. As commonly known in the art,
the
oligonucleotide probes and primers can be designed by taking into
consideration
the melting point of hydrizidation thereof with its targeted sequence (see
below
and in Sambrook et al., 1989, Molecular Cloning - A Laboratory Manual, 2nd
Edition, CSH Laboratories; Ausubel et al., 1989, in Current Protocols in
Molecular
Biology, John Wiley & Sons Inc., N.Y.).
The terms "oligonucleotide" or "DNA" molecule or sequence
refers to a molecule comprised of thedeoxyribonucleotides adenine (A), guanine
(G), thymine (T) and/or cytosine (C), in a double-stranded or single-stranded
form. The term "oligonucleotide" or "DNA" can be found in linear DNA molecules
or fragments, viruses, plasmids, vectors, chromosomes or synthetically derived
DNA. As used herein, particular DNA sequences may be described according to
the normal convention of giving only the sequence in the 5' to 3' direction.
"Nucleic acid hybridization" refers generally to the
hybridization of two single-stranded nucleic acid molecules having
complementary base sequences, which under appropriate conditions will form a
thermodynamically favored double-stranded structure. Examples of hybridization
conditions can be found in the two laboratory manuals referred above ~Sambrook
et al., 1989, supra and Ausubel et al., 1989, supra) and are commonly known in
the art. In the case of a hybridization to a nitrocellulose filter, as for
example in the
well known Southern blotting procedure, a nitrocellulose filter can be
incubated
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
12
overnight at 65°C with a labeled probe in a solution containing
50%formamide,
high salt (5 x SSC or 5 x SSPE), 5 x Denhardt's solution, 1 % SDS, and 100
Ng/ml
denatured carrier DNA (i.e. salmon sperm DNA). The non-specifically binding
probe can then be washed off the filter by several washes in 0.2 x SSC/0.1 %
SDS
at a temperature which is selected in view of the desired stringency: room
temperature (low stringency), 42°C (moderate stringency) or 65°C
(high
stringency). The selected temperature is based on the melting temperature (Tm)
of the DNA hybrid (Sambrook et al. 1989, supra). Of course, RNA-DNA hybrids
can also be formed and detected. In such cases, the conditions of
hybridization
and washing can be adapted according to well known methods by the person of
ordinary skill. Stringent conditions will be preferably used (Sambrook et
a1.,1989,
supra).
Probes or primers of the invention can be utilized with
naturally occurring sugar-phosphate backbones as well as modified backbones
including phosphorothioates, dithionates, alkyl phosphonates and a-nucleotides
and the like. Modified sugar phosphate backbones are generally taught by
Miller,
1988, Ann. Reports Med. Chem. 23:295 and Moran et al., 1987, Nucleic acid
molecule. Acids Res., 14:5019. Probes or primers of the invention can be
constructed of either ribonucleic acid (RNA) or deoxyribonucleic acid (DNA),
and
preferably of DNA.
The types of detection methods in which probes can be used
include Southern blots (DNA detection), dot or slot blots (DNA, RNA), and
Northern blots (RNA detection). Although less preferred, labeled proteins
could
also be used to detect a particular nucleic acid sequence to which it binds.
Other
detection methods include kits containing probes on a dipstick setup and the
like.
Although the present invention is not specifically dependent
on the use of a label for the detection of a particular nucleic acid sequence,
such
a label might be beneficial, by increasing the sensitivity of the detection.
Furthermore, it enables automation (the same can also be said of detection of
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
13
proteins using ligands such as antibodies). Probes can be labeled according to
numerous well known methods (Sambrook et al., 1989, supra). Non-limiting
examples of detectable markers include ligands, fluorophores, chemiluminescent
agents, enzymes, and antibodies. Other detectable markers for use with probes,
which can enable an increase in sensitivity of the method of the invention,
include
biotin and radionucleotides. It will become evident to the person of ordinary
skill
that the choice of a particular label dictates the manner in which it is bound
to the
probe.
As commonly known, radioactive nucleotides can be
incorporated into probes of the invention by several methods. Non-limiting
examples thereof include kinasing the 5' ends of the probes using gamma 3ZP
ATP and polynucleotide kinase, using the Klenow fragment of Pol I of E. coli
in
the presence of radioactive dNTP (i.e. uniformly labeled DNA probe using
random
oligonucleotide primers in low-melt gels), using the SP6/T7 system to
transcribe
a DNA segment in the presence of one or more radioactive NTP, and the like.
As used herein, "oligonucleotides" or "oligos" define a
molecule having two or more nucleotides (ribo or deoxyribonucleotides). The
size
of the oligo will be dictated by the particular situation and ultimately on
the
particular use thereof and adapted accordingly by the person of ordinary
skill. An
oligonucleotide can be synthetised chemically or derived by cloning according
to
well known methods.
As used herein, a "primer" defines an oligonucleotide which is
capable of annealing to a target sequence, thereby creating a double stranded
region which can serve as an initiation point for DNA synthesis under suitable
conditions.
Amplification of a selected, or target, nucleic acid sequence
may be carried out by a number of suitable methods. See generally Kwoh et al.,
1990, Am. Biotechnol. Lab. 8:14-25. Numerous amplification techniques have
been described and can be readily adapted to suit particular needs of a person
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
14
of ordinary skill. Non-limiting examples of amplification techniques include
polymerase chain reaction (PCR), ligase chain reaction (LCR), strand
displacement amplification (SDA), transcription-based amplification, the Q(3
replicase system and NASBA (Kwoh et al., 1989, Proc. Natl. Acad. Sci. USA 86,
1173-1177; Lizardi et al., 1988, BioTechnology 6:1197-1202; Malek et al.,
1994,
Methods Mol. Biol., 28:253-260; and Sambrook et al., 1989, supra). Preferably,
amplification will be carried out using PCR.
Polymerase chain reaction (PCR) is carried out in accordance
with known techniques. See, e.g., U.S. Pat. Nos. 4,683,195; 4,683,202;
4,800,159; and 4,965,188 (the disclosures of all three U.S. Patent are
incorporated herein by reference). In general, PCR involves, a treatment of a
nucleic acid sample (e.g., in the presence of a heat stable DNA polymerase)
under hybridizing conditions, with one oligonucleotide primer for each strand
of
the specific sequence to be detected. An extension product of eachprimer which
is synthesized is complementary to each of the two nucleic acid strands, with
the
primers sufficiently complementary to each strand of the specific sequence to
hybridize therewith. The extension product synthesized from each primer can
also
serve as a template for further synthesis of extension products using the same
primers. Following a sufficient number of rounds of synthesis of extension
products, the sample is analysed to assess whether the sequence or sequences
to be detected are present. Detection of the amplified sequence may be carried
out by visualization following EtBr staining of the DNA following gel
electrophores,
or using a detectable label in accordance with known techniques, and the like.
For
a review on PCR techniques (see PCR Protocols, A Guide to Methods and
Amplifications, Michael et al. Eds, Acad. Press, 1990).
Ligase chain reaction (LCR) is carried out in accordance with
known techniques (Weiss, 1991, Science 254:1292). Adaptation of the protocol
to meet the desired needs can be carried out by a person of ordinary skill.
Strand
displacement amplification (SDA) is also carried out in accordance with known
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
techniques or adaptations thereof to meet the particular needs (Walker et al.,
1992, Proc. Natl. Acad. Sci. USA 89:392-396; and ibid., 1992, Nucleic Acids
Res.
20:1691-1696).
As used herein, the term "gene" is well known in the art and
5 relates to a nucleic acid sequence defining a single protein or polypeptide.
A
"structural gene" defines a DNA sequence which is transcribed into RNA and
translated into a protein having a specific amino acid sequence thereby giving
rise
the a specific polypeptide or protein. It will be readily recognized by the
person of
ordinary skill, that the nucleic acid sequence of the present invention can be
10 incorporated into anyone of numerous established kit formats which are well
known in the art.
A "heterologous" (i.e. a heterologous gene) region of a DNA
molecule is a subsegment segment of DNA within a larger segment that is not
found in association therewith in nature. The term 'heterologous" can be
similarly
15 used to define two polypeptidic segments not joined together in nature. Non-
limiting examples of heterologous genes include reporter genes such as green
fluorescence protein, luciferase, chloramphenicol acetyl transferase, (3-
galactosidase, and the like which can be juxtaposed or joined to heterologous
control regions or to heterologous polypeptides.
The term "vector" is commonly known in the art and defines a
plasmid DNA, phage DNA, viral DNA and the like, which can serve as a DNA
vehicle into which DNA of the present invention can be cloned. Numerous types
of vectors exist and are well known in the art.
The term "expression" defines the process by which a gene is
transcribed into mRNA (transcription), the mRNA is then being translated
(translation) into one polypeptide (or protein) or more.
The terminology "expression vector" defines a vector or
vehicle as described above but designed to enable the expression of an
inserted
sequence following transformation into a host. The cloned gene (inserted
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
16
sequence) is usually placed under the control of control element sequences
such
as promoter sequences. The placing of a cloned gene under such control
sequences is often refered to as being operably linked to control elements or
sequences.
Operably linked sequences may also include two segments
that are transcribed onto the same RNA transcript. Thus, two sequences, such
as a promoter and a "reporter sequence" are operably linked if transcription
commencing in the promoter will produce an RNA transcript of the reporter
sequence. In order to be "operably linked" it is not necessary that two
sequences
be immediately adjacent to one another.
Expression control sequences will vary depending on whether
the vector is designed to express the operably linked gene in a prokaryotic or
eukaryotic host or both (shuttle vectors) and can additionally contain
transcriptional elements such as enhancer elements, termination sequences,
tissue-specificity elements, and/or translational initiation and termination
sites.
Prokaryotic expressions are useful for the preparation of large
quantities of the protein encoded by the DNA sequence of interest. This
protein
can be purified according to standard protocols that take advantage of the
intrinsic properties thereof, such as size and charge (i.e. SDS gel
electrophoresis,
gel filtration, centrifugation, ion exchange chromatography...). In addition,
the
protein of interest can be purified via affinity chromatography using
polyclonal or
monoclonal antibodies. The purified protein can be used for diagnostic or
therapeutic applications.
The DNA construct can be a vector comprising a promoter
that is operably linked to an oligonucleotide sequence of the present
invention,
which is in turn, operably linked to a heterologous gene, such as the gene for
the
luciferase reporter molecule. "Promoter" refers to a DNA regulatory region
capable of binding directly or indirectly to RNA polymerase in a cell and
initiating
transcription of a downstream (3' direction) coding sequence. For purposes of
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
17
the present invention, the promoter is bound at its 3' terminus by the
transcription
initiation site and extends upstream (5' direction) to include the minimum
number
of bases or elements necessary to initiate transcription at levels detectable
above
background. Within the promoter will be found a transcription initiation site
(conveniently defined by mapping with S1 nuclease), as well as protein binding
domains (consensus sequences) responsible for the binding of RNA polymerase.
Eukaryotic promoters will often, but not always, contain "TATA" hoses and
"CCAT" boxes. Prokaryotic promoters contain Shine-Dalgarno sequences in
addition to the -10 and -35 consensus sequences.
As used herein, the designation "functional derivative"
denotes, in the context of a functional derivative of a sequence whether an
nucleic acid or amino acid sequence, a molecule that retains a biological
activity
(either function or structural) that is substantially similar to that of the
original
sequence. This functional derivative or equivalent may be a natural derivative
or
may be prepared synthetically. Such derivatives include amino acid sequences
having substitutions, deletions, or additions of one or more amino acids,
provided
that the biological activity of the protein is conserved. The same applies to
derivatives of nucleic acid sequences which can have substitutions, deletions,
or
additions of one or more nucleotides, provided that the biological activity of
the
sequence is generally maintained. When relating to a protein sequence, the
substituting amino acid as chemico-physical properties which are similar to
that
of the substituted amino acid. The similarchemico-physical properties include,
similarities in charge, bulkiness, hydrophobicity, hydrophylicity and the
like. The
term "functional derivatives" is intended to include "fragments", "segments",
"variants", "analogs" or "chemical derivatives" of the subject matter of the
present
invention.
Thus, the term "variant" refers herein to a protein or nucleic
acid molecule which is substantially similar in structure and biological
activity to
the protein or nucleic acid of the present invention.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
18
The functional derivatives of the present invention can be
synthesized chemically or produced through recombinant DNA technology. All
these methods are well known in the art.
As used herein, "chemical derivatives" is meant to cover
additional chemical moieties not normally part of the subject matter of the
invention. Such moieties could affect the physico-chemical characteristic of
the
derivative (i.e. solubility, absorption, half life and the like, decrease of
toxicity).
Such moieties are examplified in Remington's Pharmaceutical Sciences (1980).
Methods of coupling these chemical-physical moieties to a polypeptide are well
known in the art.
The term "allele" defines an alternative form of a gene which
occupies a given locus on a chromosome.
As commonly known, a "mutation" is a detectable change in
the genetic material which can be transmitted to a daughter cell. As well
known,
a mutation can be, for example, a detectable change in one or more
deoxyribonucleotide. For example, nucleotides can be added, deleted,
substituted
for, inverted, or transposed to a new position. Spontaneous mutations and
experimentally induced mutations exist. A mutant polypeptide can be encoded
from a mutant nucleic acid molecule. In addition, mutant proteins can be
produced through aberrant events during replication, transcription and/or
translation. Frameshifting (the switching from a particular reading frame to
another) is such a mechanism that can modify the sequence of the translated
protein.
As used herein, the term "purified" refers to a molecule having
been separated from a cellular component. Thus, for example, a "purified
protein"
has been purified to a level not found in nature. A "substantially pure"
molecule
is a molecule that is lacking in all other cellular components.
As used herein, the terms "molecule", "compound", "agent", or
"ligand" are used interchangeably and broadly to refer to natural, synthetic
or
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
19
semi-synthetic molecules or compounds. The term "molecule" therefore denotes
for example chemicals, macromolecules, cell or tissue extracts (from plants or
animals) and the like. Non limiting examples of molecules include nucleic acid
molecules, peptides, antibodies, carbohydrates and pharmaceuticalkherapeutical
agents. The agents can be selected and screened by a variety of means
including random screening, rational selection and by rational design using
for
example protein or ligand modelling methods such as computer modelling. The
terms "rationally selected" or "rationally designed" are meant to define, for
example, compounds which have been chosen based on the configuration of the
polyalanine domains of the present invention. As will be understood by the
person of ordinary skill, macromolecules having non-naturally occurring
modifications are also within the scope of the term "molecule". For example,
peptidomimetics, well known in the pharmaceutical industry and generally
referred to as peptide analogs can be generated by modelling as mentioned
above. Similarly, in one embodiment, the polypeptides of the present invention
can be modified to enhance or decrease their stability. It should be
understood
that in most cases this modification should not alter the biological activity
of the
polyalanine domain (its toxic effect or INI localization property). The
molecules
identified in accordance with the teachings of the present invention have a
therapeutic value in diseases or conditions in which the physiology or
homeastasis of the cell and/or tissue is compromised by a production of
polyalanine-containing proteins or polypeptides. Alternatively, the molecules
identified in accordance with the teachings of the present invention find
utility in
the development of more efficient molecule to lower and/or abrogate the
toxicity
of such proteins and/or to reduce or eliminate the production of such mutant
proteins. It will be understood that agents can be screened, in accordance
with
the present invention, with libraries of compounds, using for example
automated
screening methods (e.g. array technologies).
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
The level of gene expression of the reporter gene (e.g. the
level of luciferase, or (3-gal, produced) within the treated cells can be
compared
to that of the reporter gene in the absence of the molecules(s). The
difference
between the levels of gene expression indicates whether the molecules) of
5 interest agonizes the aforementioned interaction. The magnitude of the level
of
reporter gene product expressed (treated vs. untreated cells) provides a
relative
indication of the strength of that molecules) as an agonist. The same type of
approach can also be used in the presence of an antagonist(s). Thus,
modulators of the production of such proteins can be identified and selected.
10 Non-limiting examples of modulators in accordance with the present
invention
include frameshift mutants or suppressors, relievers of codon rareness (i.e.
relieving the limitation of rare codons which favor frameshifting events),
agents
which degrade polyalanine stretches, tRNAs, tRNA suppressors and the like.
One skilled in the art will realize that the assays to identify compounds that
15 modulate frameshifting of CAG repeats and the like, could be carried out
using
other repeats as well as other genes known to promote frameshifting. Such
genes are known in the art.
The present invention also provides antisense nucleic acid
molecules which can be used for example to modulate the expression of the
20 mutant proteins of the present invention. An antisense nucleic acid
molecule
according to the present invention refers to a molecule capable of forming a
stable duplex or triplex with a portion of its targeted nucleic acid sequence
(DNA
or RNA). The use of antisense nucleic acid molecules and the design and
modification of such molecules is well known in the art as described for
example
in WO 96/32966, WO 96/11266, WO 94/15646, WO 93/08845 and
USP 5,593,974. Antisense nucleic acid molecules according to the present
invention can be derived from the nucleic acid sequences and modified in
accordance to well known methods. For example, some antisense molecules can
be designed to be more resistant to degradation to increase their affinity to
their
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
21
targeted sequence, to affect their transport to chosen cell types or cell
compartments, and/or to enhance their lipid solubilitybu using nucleotide
analogs
andlor substituting chosen chemical fragments thereof, as commonly known in
the art.
Alternatively, an indicator cell in accordance with the present
invention can be used to identify antagonists. For example, the test molecule
or
molecules are incubated with the host cell in conjunction with one or more
agonists held at a fixed concentration. An indication and relative strength of
the
antagonistic properties of the molecules) can be provided by comparing the
level
of gene expression in the indicator cell in the presence of the agonist, in
the
absence of test molecules vs in the presence thereof. Of course, the
antagonistic
effect of a molecule can also be determined in the absence ofagonist, simply
by
comparing the level of expression of the reporter gene product in the presence
and absence of the test molecule(s).
It shall be understood that the "in vivo" experimental model
can also be used to carry out an "in vitro" assay. For example, cellular
extracts
from the indicator cells can be prepared and used in one of the aforementioned
"in vitro" tests.
As used herein the recitation "indicator cells" refers to, for
example, cells that express a fusion protein comprising a polyalanine segment
(e.g. a "CAG" repeat) and an identifiable or selectable phenotype or
characteristic
which enables an assessment of the level of fusion protein expression (e.g. a
reporter protein). Such indicator cells can be used in the screening assays of
the
present invention. In certain embodiments, the indicator cells have been
engineered so as to express a chosen derivative, fragment, homolog, or mutant
of a repeat. It should be understood that the repeats should not be limited to
CAG
repeats. Indeed, GCG repeats can also be used. In addition, the invention
should
not be limited to polyalanine repeats, since the present invention provides
for the
testing of polyserine fragments and other polyamino acids which could be
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
22
expressed from a frameshifting event. The cells can be yeast cells or higher
eukaryotic cells such as mammalian cells (WO 96/41169). Preferably, the
indicator cells are higher eukaryotic cells. Non-limiting examples of such
cells
and vectors are exemplified herein below (i.e. Examples 2-4). In one
particular
embodiment, the indicator cell could be used to test a compound or a library
thereof.
As exemplified herein below in one embodiment, a
polyalanine polypeptide segment of the present invention is provided as a
fusion
protein. The design of constructs therefor and the expression and production
of
fusion proteins are exemplified herein and are well known in the art (Sambrook
et al., 1989, supra; and Ausubel et al., 1994, supra).
Non limiting examples of such fusion proteins include a
hemaglutinin fusions (HA) and Gluthione-S-transferase (GST) fusions. In
certain
embodiments, it might be beneficial to introduce a protease cleavage site
between the two polypeptide sequences which have been fused. Such protease
cleavage sites between two heterologously fused polypeptides are well known in
the art.
In certain embodiments, it might also be beneficial to
introduce a linker (commonly known) between the repeat segment of the protein
and the heterologous polypeptide portion (e.g. reporter protein portion). Such
fusion protein find utility in the assays of the present invention as well as
for
purification purposes, detection purposes and the like.
For certainty, the sequences and polypeptides useful to
practice the invention include without being limited thereto mutants,
homologs,
subtypes, alleles and the like. It shall be understood that generally, the
sequences of the present invention should encode a functional (albeit
defective)
repeat domain. It will be clear to the person of ordinary skill that whether a
repeat
domain of the present invention, variant, derivative, or fragment thereof
retains
its function in enabling a concentration of the protein containing same in INI
or
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
23
triggering toxicity in cells or animals, can be readily determined by using
the
teachings and assays of the present invention and the general teachings of the
art.
As exemplified herein below, the repeat domains of the
present invention can be modified, for example by in vitro mutagenesis, to
dissect
the structure-function relationship thereof and permit a better design and
identification of modulating compounds. However, some derivative or analogs
having lost their biological function may still find utility, for example for
raising
antibodies. These antibodies could be used for detection or purification
purposes.
In addition, these antibodies could also act as competitive or non-competitive
inhibitor and be found to be modulators of the biological activity of the
repeat
domain.
A host cell or indicator cell has been "transfected" by
exogenous or heterologous DNA (e.g. a DNA construct) when such DNA has
been introduced inside the cell. The transfecting DNA may or may not be
integrated (covalently linked) into chromosomal DNA making up the genome of
the cell. In prokaryotes, yeast, and mammalian cells for example,
thetransfecting
DNA may be maintained on a episomal element such as a plasmid. With respect
to eukaryotic cells, a stably transfected cell is one in which the
transfecting DNA
has become integrated into a chromosome so that it is inherited by daughter
cells
through chromosome replication. This stability is demonstrated by the ability
of
the eukaryotic cell to establish cell lines or clones comprised of a
population of
daughter cells containing the transfecting DNA. Transfection methods are well
known in the art (Sambrook et al., 1989, supra; Ausubel et al., 1994 supra).
In general, techniques for preparing antibodies (including
monoclonal antibodies and hybridomas) and for detecting antigens using
antibodies are well known in the art (Campbell, 1984, In "Monoclonal Antibody
Technology: Laboratory Techniques in Biochemistry and Molecular Biology",
Elsevier Science Publisher, Amsterdam, The Netherlands) and in Harlow et al.,
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
24
1988 (in: Antibody- A Laboratory Manual, CSH Laboratories). The present
invention also provides polyclonal, monoclonal antibodies, or humanized
versions
thereof, chimeric antibodies and the like which are specific to the repeat
domains
of the present invention.
From the specification and appended claims, the term
therapeutic agent should be taken in a broad sense so as to also include a
combination of at least two such therapeutic agents. Further, the therapeutic
agent according to the present invention can be introduced into individuals in
a
number of ways. The therapeutic agent can also be delivered through a vehicle
such as a liposome, which can be designed to be targeted to a specific cell
type,
and engineered to be administered through different routes. Having shown that
a polyalanine segment is toxic to cells, the present invention provides the
means
to trigger toxicity in cells by expressing thereinto or delivering thereto a
polyalanine-containing protein (or polyalanine-encoding nucleic acid). In
accordance with known methods, a chosen cell population could be targeted.
For administration to humans, the prescribing medical
professional will ultimately determine the appropriate form and dosage for a
given
patient, and this can be expected to vary according to the chosen therapeutic
regimen (i.e. DNA construct, protein, cells), the response and condition of
the
patient as well as the severity of the disease.
Composition within the scope of the present invention should
contain the active agent (i.e. fusion protein, nucleic acid, and molecule) in
an
amount effective to achieve the desired therapeutic effect while avoiding
adverse
side effects. Typically, the nucleic acids, fusion proteins and molecules in
accordance with the present invention can be administered to mammals (i.e.
humans) in doses ranging from 0.005 to 1 mg per kg of body weight per day of
the mammal which is treated. Pharmaceutically acceptable preparations and
salts of the active agent are within the scope of the present invention and
are well
known in the art (Remington's Pharmaceutical Science, 16th Ed., Mack Ed.). For
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
the administration of polypeptides, antagonists, agonists and the like, the
amount
administered should be chosen so as to avoid adverse side effects. The dosage
will be adapted by the clinician in accordance with conventional factors such
as
the extent of the disease and different parameters from the patient.
Typically,
5 0.001 to 50 mg/kglday will be administered to the mammal.
The present invention also relates to a kit for diagnosing a
disease or condition associated with the expression of a repeat domain,
encoding
for example a polyalanine stretch, or a predisposition to contracting same
comprising a nucleic acid, a protein or a ligand in accordance with the
present
10 invention. For example, a compartmentalized kit in accordance with the
present
invention includes any kit in which reagents are contained in separate
containers.
Such containers include small glass containers, plastic containers or strips
of
plastic or paper. Such containers allow the efficient transfer of reagents
from one
compartment to another compartment such that the samples and reagents are not
15 cross-contaminated and the agents or solutions of each container can be
added
in a quantitative fashion from one compartment to another. Such containers
will
include a container which will accept the test sample (DNA protein or cells),
a
container which contains the primers used in the assay, containers which
contain
enzymes, containers which contain wash reagents, and containers which contain
20 the reagents used to detect the extension products.
BRIEF DESCRIPTION OF THE DRAWINGS
Having thus generally described the invention, reference will
now be made to the accompanying drawings, showing by way of illustration a
preferred embodiment thereof, and in which:
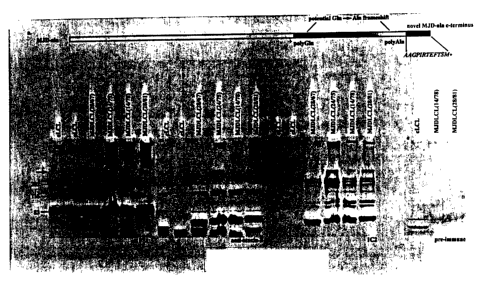
25 Figure 1 shows the Western blot analysis of lymphoblastoid
cells from controls and MJD patients. a, Schematic representation of the MJD-
Ala protein that results from a frameshift in the CAG tract showing the new C-
terminus (italicized; used to raise the FS1 and FS2 antibodies). Western blots
of
two control lymphoblastoid cell lines (cLCL) and four MJD lymphoblastoid cell
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
26
lines (MJDLCL) immunoprobed with FS1 (b), anti-ataxin-3 (c), 1C2 (d) and FS1
pre-immune serum (e). Arrow indicates the threshold between stacking and
resolving portions of the gel. Panels b-d represent serial probing of a single
membrane.
Figure 2 shows the immunocytochemical detection of
intranuclear deposits in lymphoblastoid cells. Immunocytochemistry of control
LCL versus MJD LCL: absence of INI in control LCL probed with FS1 (a), and
anti-ubiquitin (c), and detection of INI in MJD LCL with FS1 (b) and
anti~biquitin
(d). e, Immuno detection of MJD LCL with FS1 pre-immune serum. For all
panels, the magnification before publication is 400x (left) and 1000x (right).
These results have been replicated in three separate experiments.
Figure 3 shows the immunohistochemical detection of INI in
MJD pontine neurons. Immunoprobing with FS1 antiserum in MJD pons (a) and
control pons (b); immunoprobing with anti-ubiquitin in MJD pons (c) and
control
pons (d). INI in pontine neurons are indicated by arrowheads. Double labeling
immunofluorescence analysis of MJD pons showing ubiquitin-labeled INI (e, h)
and FS1-labeled INI (f, i), and the composite image of bothlabelings (g, j).
For all
panels, the magnification before publication is 1000x, before reproduction.
Figure 4 shows the constructs used in the transfection
experiments. All constructs, with the exception of pMJD11, represent full-
length
MJD-1. Solid black boxes indicate the repeat portion of the constructs.
Staggered ends indicate that EGFP will only be expressed if aframeshift
occurs.
Encircled blown up detail of pMJD1 is also present in pMJD2 and pMJD3; details
of pMJDS are present in pMJD6; details of pMJD7 are present in pMJD8 and
details of pMJD9 are the same as pMJD10.
Figure 5 shows the transfection experiments with different
MJDIEGFP constructs. a, DNA sequence of the clones with EGFP out of frame
(pMJD1, pMJD2 and pMJD3) and b, with EGFP inglutamine frame (pMJD4), both
(a, b) showing the predicted amino acid sequence. c-e, Fluorescence at 72
hours
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
27
of COS-7 cells transfected with pMJD1 (c), pMJD2 (d) and pMJD3 (e) where the
MJD(CAG)~IEGFP fusion protein is translated only when frameshifts to GCA-
polyAla occur. f, COS-7 cells transfected with pMJD4 where EGFP is in frame
with CAG-Gln. g, COS-7 cells transfected with the vector pEGFP-N1. h,
perinuclear fluorescent aggregates observed in cells transfected with pMJD3.
Pictures of sections c, d, e, f and g were taken at 25 x magnification for a
fixed
exposure time of 60 seconds before reproduction; picture shown in h, is at 400
x before reproduction. i-k, Western blots of protein isolated from cells shown
in
c, d, e, f, g and from mock-transfected cells immunoprobed with 1 C2 (i), anti-
ataxin-3 Q) and anti-HA (k). Arrows on the right of panel j indicate the
proteins
detected. Arrowhead on panel k indicates threshold between stacking and
resolving portions of the gel. Rpts stands for repeats.
Figure 6 shows the time-course immunocytochemical analysis
of COS-7 cells transfected with constructs encoding ataxin-3 with either
apolyAla
or a polyGln tract. Immunoprobing with anti-HA antibody at time-points 8
hours:
(a) pMJD7, (b) pMJD9, (c) pMJDB, (d) pMJD10 and (e) vector pEGFP-N1 alone;
hours: (f) pMJD7, (g) pMJD9, (h) pMJDB, ~) pMJD10 and Q) vector pEGFP-N1
alone; 24 hours: (k) pMJD7, (I) pMJD9, (m) pMJDB, (n) pMJD10 and (o) vector
pEGFP-N1 alone; 48 hours: (p) pMJD7, (q) pMJD9, (r) pMJDB, (s) pMJD10 and
20 (t) vector pEGFP-N1 alone. For all panels pictures were taken at 1000x
magnification, before reproduction.
Figure 7 shows the western analysis of transfected COS-7
cells (in figure 5). Blots were probed with (a) anti-HA, (b) 1 C2. Arrow
indicates
threshold between stacking and resolving portions of the gel. Cells were
harvested at 72 hours after transfection.
Figure 8 shows the immunocytochemical analysis of COS-7
cells transfected with a truncated polyAla-encoding construct. Immunoprobing
with anti-HA antibody in: (a) cells transfected with 42A and (b)
mock~transfected
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
28
cells. For all panels pictures were taken at 1000x magnification, before
reproduction.
Other objects, advantages and features of the present
invention will become more apparent upon reading of the following
nonrestrictive
description of preferred embodiments with reference to the accompanying
drawing which is exemplary and should not be interpreted as limiting the scope
of the present invention.
DESCRIPTION OF THE PREFERRED EMBODIMENT
The data herein presented strongly suggest the fact that (1 )
frameshifts occur within CAG repeats and are responsible for the production of
alanine-containing proteins, and (2) these proteins accumulate as toxic
aggregates. Detection of the hypothetical peptide in lymphoblastoid and
neuronal
cells of MJD patients and not in controls, and the production of green
fluorescence preferentially in cells transfected with MJD-1 bearing large CAG
tracts with an out of frame EGFP, supports the occurrence of rareframeshifts
during transcription and/or translation of long CAG tracts resulting from the
use
of the alternative GCA/Ala reading frame. The relatively small proportion of
frameshifted product may explain the absence of loss-of-function of the
protein
and the relatively late-onset of these diseases. This model of slow
accumulation,
due to relatively rare frameshift events, better explains the late-onset
nature of
these diseases given that, in light of the high expression of these proteins
in
affected brain regions, a glutamine toxicity model would be expected to result
in
much earlier cell death and disease onsetz'-Z8.
Slippage into the third possible frame, AGC/Ser, may also be
occurring. However, the absence of diseases associated with tracts
ofpolyserine
and the physical nature of alanine polymers, resulted in an exclusive focus on
the
GCA/Ala frame. While the relative frequencies of differentframeshifts remain
unknown, the fact that mosttranslational frameshift errors cause a +2 shift in
the
framez9 suggests that GCA/Ala may be the more frequent of the mutant species.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
29
The present invention also assessed whether polyAla tracts
are toxic and may be the initiating event in the formation of the INI seen in
expanded CAG tract diseases. The evidence presented herein shows that in
cells transfected with the CAG~GIn constructs, frameshifting into the alanine
frame
is progressive and frameshifted products are slowly accumulating in the
nucleus
as INI (see Fig. 5). Given the expected low frequency of frameshifts, the
finding
of frameshifted protein in INI as early as 12 hours aftertransfection argues
that
polyAla accumulation is a very early event in the formation of these
structures. In
fact, in cells transfected with as few as 14CAGs, frameshifts are occurring
and
polyalanine-containing protein is accumulating in the nucleus as INI. This
finding
is consistent with a recent report by Perez et al. showing that transfection
of
normal sized CAG repeats leads to IN13°. In contrast with the slow,
progressive
accumulation detected for the CAG~GIn constructs, transfection with the
GCA/Ala
constructs results in early and rapid accumulation ofalanine-containing
product.
In this case the cells seem unable to cope with the presence of the toxic
product
resulting in an earlier and much more severe phenotype. The pattern of
expression of polyAla products, consisting of one major juxtanuclear aggregate
per cell and several smaller inclusions, is similar to that found in
aggresomes,
structures that form when the capacity of the proteasome degradation pathway
is exceeded3'. This seems to indicate that the direct expression ofpolyalanine
in cells is likely to be extremely toxic. The presence of more "classical" INI
in cells
expressing CAG/Gln constructs is thought to result from rareframeshift events
that produce alanine-containing protein, which slowly accumulates in the
nucleus
as aggregates. This model therefore better depicts what may be happening in
diseased tissue cells.
The proposed model of toxic aggregates resulting from
frameshifts into the alanine frame is consistent with previous experiments
performed with MJD, as well as other exp-CAG diseases. For example, in
Huntington disease (HD) and MJD, INI and protein accumulation are most clearly
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
detected using antibodies against epitopes to the N-terminus side of the CAG
repeat'~4, likely because frameshifting will truncate the protein shortly
after the
repeat. This model could also explain the correlation observed in all CAG
triplet
diseases between the size of the repeat and the severity of the symptoms32. It
is
5 suggested that if the frameshift errors occur randomly, the longer the
repeat the
more frequently such errors would arise. This is supported by the observations
that: (1) no accumulation was yet detected in the lymphoblasts of a MJD
patient
with the shortest (CAG)s, mutation; (2) fluorescence increased with the size
of the
CAG repeat in the transfection experiments; and (3) at any time-point, more
than
10 twice as many cells have inclusions containing frameshifted species when
transfected with a (CAG)s2 construct than with a (CAG),5 construct.
Another puzzling finding previously reported by other groups,
and which can be explained by the model offrameshifts into the alternate
alanine
frame and the production of prematurely truncated proteins herein proposed, is
15 the presence of short cleavage products in inclusions in HD and spinal and
bulbar
muscular atrophy (SBMA)33-ss, These short products seem to be too small to be
the result of caspase-3 cleavage, but are of a size consistent with premature
termination of transcription or translation due to the generation of an early
STOP
codon in the alanine shifted frame.
20 The model presented herein might also explain the
observation that while both CAA and CAG codons encode glutamine, only
uninterrupted CAG tracts cause disease. In spinocerebellar ataxia type 1 (SCA1
),
for polyglutamine tracts of similar length the presence of a CAAcodon
interrupting
a CAG tract is used to differentiate a normal from a disease causing allelezs.
as, 3'.
25 These observations, while difficult to explain if polyglutamine is toxic,
are
predicted by the proposed polyAla nuclear toxicity model of the present
invention,
where interruption of a repeat sequence may lead to more stable transcription
or
translation.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
31
Finally, the present invention may predict a similar pathogenic
mechanism in the other diseases caused by expanded CAG tracts. It has been
shown here that constructs containing almost exclusively a GCA tract are toxic
to cells and lead to the formation of aggregates. This seems to indicate that
the
full MJD protein is not necessary to obtain a disease phenotype and that the
presence of an expanded CAG repeat within any protein may be sufficient to
cause disease. This would support the contention that the protein or gene
causing the disease is indeed irrelevant to the disease process (Hardy et al.
1998). In addition, it is possible that polyAla accumulation may play a role
in
aging of certain cell types. For example, if frameshifts causing polyAla
tracts
occur with all CAG repeat-containing genes even in the "normal range" as is
seen
in the 14 CAG- and 37 CAG-containing constructs of the present invention, very
slow accumulation of polyAla polymers may occur, leading to cell death. This
could explain the observation that fewer large repeats are found in healthy
elders
compared to younger controls 38. The discovery of expanded polyalanine
domains in three diseases"~ 3~4' and the fact that CAG repeat frameshift
mutant
proteins can cause disease if they code forpolyAla, pinpoints this homopolymer
as a potential target for drug design. Polyalanine nuclear toxicity may well
be a
frequent cause of premature cell death in different tissues.
The present invention is illustrated in further detail by the
following non-limiting examples.
EXAMPLE 1
Frameshifts occur in CAG tracts resulting in polyalanine-
containing proteins
Two immunopurified polyclonal antisera were raised against
a synthetic peptide corresponding to the 12 last amino acids predicted to
result
from polyAla tract producing frameshifts within the CAG repeat of theMJD-1
gene
(FS1 and FS2)(Fig. 1a). This new amino acid sequence has no homology to any
known protein. Both antisera detected high molecular weight aggregates in the
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
32
stacking gels in Western blots of total lymphoblast protein from three MJD
cases
(Fig. 1 b). In the two controls and in the MJD patient with the shortest
(CAG)6,
repeat no aggregates were observed (Fig. 1 b). Both FS antibodies also
recognize, in all samples, an 83kD protein of unknown origin. In order to test
if the
signal detected in the stacking gels was the putative MJD-Ala protein, the
same
blots were probed with antisera raised against MJD protein epitopes (anti-
ataxin-
3; Fig. 1 c)" and against polyGln domains (1 C2; Fig. 1 d)'8. These results
are
compatible with the presence of the predicted hybrid protein in the stacking
gel.
An anti-ubiquitin polyclonal antibody (data not shown) also detected the
accumulations, which is consistent with previous reports' 5~ 8~ ~9. zo_ These
accumulations are compatible with those reported by Paulson et al. who studied
cells expressing mutant CAG tract expanded MJD-1 using an antibody raised
against the expressed ataxin-3 fusion proteins'.
To test if the MJD-Ala protein accumulates in nuclei,
immunocytochemistry on lymphoblasts from four controls and three MJD patients
was performed. As Fig. 2 depicts, FS1 positive INI are observed in MJD cell
lines
and not in controls (Fig. 2a, b). Similar to the Western blot results, the
anti-
ubiquitin antibody (Fig. 2c, d) and the anti-ataxin-3 antibody (data not
shown) also
detect INI in MJD cell lines.
In an attempt to show the presence of the frameshifted
species in affected MJD brain regions, immunohistochemical FS1 and anti-
ubiquitin staining in diseased and control pons, a region known to be affected
in
MJD was performed (Fig. 3). Both antibodies stain intranuclear structures in
neurons of this MJD brain area (Fig. 3a, c), whereas pontine neurons in
control
brain have no INI (Fig. 3b, d). In immunofluorescence studies, it was shown
that
the frameshifted product colocalizes with ubiquitin in INI of pontine neurons
(Fig.
3e, f, g, h, i and j), suggesting that the intranuclear structures detected by
both
antibodies are the same.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
33
~Yennpi ~ ~
Frameshifts into alanine frame occur preferentially
with expanded CAG tracts
To test if frameshifts producing GCA/polyAla occur
preferentially within expanded CAG tracts, an in vitro system to examine the
effect
of CAG repeat length on the frequency offrameshifts was designed. COS-7 cells
were transfected with constructs bearing the full length MJD-1 sequence with
either a (CAG),4, (CAG)3, or (CAG)ez repeat fused out of frame to theEGFP gene
and driven by the CMV promoter (Fig. 4 and Fig. 5a). Whileframeshifts would
occur with all CAG tracts, it was hypothesized that they would occur more
frequently within larger repeats. The pMJD1, pMJD2 and pMJD3 constructs
would yield EGFP-containing fusion proteins only if a frameshift occurs and
produces the GCA/polyAla reading frame and the MJD-Ala protein. At 72 hours,
cells transfected with pMJD2 and, especially pMJD3, showed green fluorescence
(Fig. 5c, d, e). Green fluorescence was observed within 24 hours in the
positive
control cells transfected with the pEGFP-N1 vector alone or pMJD4 construct,
where the EGFP coding sequence was in the glutamine frame in MJD-1 (Fig. 5f,
g). At 96 hours, and at higher magnification, frequent EGFPpositive
perinuclear
inclusions were observed in cells transfected with the pMJD3 construct (82
CAGs) but not in the construct with 14 CAGs, and rarely in cellstransfected
with
the 37 CAG construct (Fig. 5h). These perinuclear inclusions are similar to
those
found in cell culture models of MJD, as well as other CAG tract disorders,
such
as Huntington disease'~4.
Western blots of protein extracted from thetransfected cells
were probed to confirm this interpretation (Fig. 5i, j). While 1C2 detects
only the
expanded MJD gene products bearing either 37 or 82 polyglutamine repeats in
cells transfected with pMJD2 and pMJD3 respectively (Fig. 5i), anti-ataxin-3
detects all three different size polyglutamine-tract containing gene products
(Fig.
5j), as expected. With both antibodies, protein accumulation in the wells for
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
34
pMJD3 and pMJD4 was detected. In order to further determine the nature of the
accumulated protein seen for pMJD3, the pMJD1 and pMJD3 constructs were
modified by adding epitopes for FS1 and HA in the alanine frame (pMJD5 and
pMJD6, Fig. 4). Western blots of cells transfected with these constructs were
probed with anti-HA. No signal for the 14 CAG-bearing pMJD5 construct was
detected, but a band corresponding to aggregated protein was detected at the
top
of the gel for pMJD6, showing that with 82 repeats,frameshifts are occurring.
The
absence of a band corresponding to the predicted size of the ataxin-3/HA
protein
(Fig. 5K) indicates that all frameshifted protein is accumulating as insoluble
aggregates. These experiments further demonstrate thatframeshifts do occur
and that their frequency increases with the size of the CAG repeat.
It shall be recognized by the skilled artisan that the in vitro
system described above can be modified at will and still enable a
determination
of the frameshifting frequencies. Non-limiting examples of such modifications
include the use of longer or shorter CAG-tracts, different reporter gene (i.e.
luciferase) and different epitopes. In addition, such systems could be used to
screen for drugs or compounds which modulate frameshifting and/or affect the
level of INI formation and/orpolyalanine-protein aggregation andlor cell
toxicity.
It should also be recognized that the in vivo methods shown
above (and others) could also be used to screen drugs or compounds which can
modulate the level of polyalanine formation, INI formation and the like. In
addition, these in vivo methods (and others) could be used to validate the
effect
of compounds or drugs identified in an in vitro assay.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
GYennpi G ~
Polyalanine-containing proteins are present in
aggregates and are toxic to transfected COS-7 cells
5 in a time-dependant manner
It has been shown that frameshifting into the alternate alanine
frame occurs both in vitro and in vivo. To determine if these products are
toxic
or are simply harmless byproducts with no real consequences to the cell, a new
set of MJD-1 constructs where the reading frame immediately before the CAG
10 repeat was mutated to code for a polyalanine stretch was designed. These
constructs, pMJD9 and pMJD10, contained stretches of 14 GCA repeats and 82
GCA repeats respectively, and were fused in frame to the EGFP gene (Fig. 4). A
HA tag was also added at the COOH-terminus in frame with the GCA stretch.
COS-7 cells were transfected with these constructs, and with pMJD7 and pMJDB,
15 which contained 14 CAG repeats and 82 CAG repeats respectively, as well as
EGFP fused in frame with the CAG tracts. In addition, a HAepitope was also
added at the COOH-terminus in frame with the GCA tracts, and so only
detectable if frameshifts occurred (Fig. 4).
In a time-course experiment using anti-HA antibody as a
20 probe to detect only protein frameshifted to the alanine frame, cells were
collected
and immunostained cells at 8, 12, 16, 20, 24, 48 and 72 hours.
Cellstransfected
with the CAG/Gln constructs pMJD7 and pMJD8 showed faint background
staining at 8 hours (Fig. 6a, c). Positive signal was detected for these two
constructs at 12 hours (not shown) in the form ofintranuclear inclusions
(typically
25 one or two per nucleus). At 20 hours, nuclei of cellstransfected with pMJD7
and
pMJD8 contained inclusions, but were morphologically normal (Fig. 6f, h). At
24
hours, cells transfected with the shorter construct remained morphologically
normal (Fig. 6k), but cells transfected with the construct bearing 82 CAG
repeats
started showing some perinuclear and cytoplasmic inclusions in addition to the
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
36
intranuclear aggregates (Fig. 6m). At this time-point about 85% oftransfected
cells have inclusions when transfected with pMJD8, whereas only 40% of
transfected cells show inclusions when transfected with pMJD7, suggesting that
frameshifts occur with both constructs, but more frequently for the longer
repeat.
Nuclei of cells stained at 48 hours showed some membrane disintegration with
both constructs, but cells containing the longer CAG repeat also
hadperinuclear
and cytoplasmic inclusions (Fig. 6p, r), indicating a more severe phenotype.
It is
useful to reiterate that probing cells transfected with the CAG~GIn constructs
with
anti-HA will only detect frameshifted protein. These species are detected
early
in the transfection and only as INI, suggesting that, despite the probable
rarity of
frameshifts, they are producing proteins that accumulate as highly insoluble
INI.
Inclusions for both GCA/Ala constructs (14A and 82A) as
early as 8 hours after transfection were detected (Fig. 6b, d). Typically,
cells have
one major perinuclear or cytoplasmic inclusion and abnormal nuclear
morphology. The cellular phenotype progresses rapidly with time in cells
transfected with either pMJD9 (Fig. 6g, I, and q) or pMJD10 (Fig. 6i, n, s),
and
was extremely severe when compared with the CAGIGIn counterparts of the
same repeat size (for example: compare panels m and n in Fig. 6). Cells
transfected with the GCA/Ala constructs showed abnormal nuclear structure and
aggregate formation mainly in the cytoplasm, usually with one
majorjuxtanuclear
inclusion and what appears to be cytoskeletal reorganization. Similar results
were obtained in cells transfected in parallel with the same constructs but
probed
with FS1 antiserum (results not shown). At all time-points cellstransfected
with
the pEGFP-N1 vector alone showed only background staining and were devoid
of inclusions (Fig. 6e, j, o, t).
Western blots of protein extracted from thetransfected cells
were performed to investigate the nature of the inclusions found. Probing the
blots with anti-HA detected a signal for both GCA/Ala constructs (pMJD9 and
pMJD10), where the HA tag is in the main reading frame (figure 7a). No signal
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
37
was detected with equivalent exposure for the CAG/Gln constructs, but
overexposure of the blots revealed the presence of smears and protein in the
wells for these constructs as well. In addition, while discrete bands are
resolved
with pMJDB, signal is only seen in the stacking gel with pMJD10, suggesting
that
the larger polyAla-containing proteins are very insoluble and do not migrate
into
the gel at all. Probing the same blots with 1 C2 confirms the presence
ofpolyGln-
containing proteins in the cells transfected with pMJDB, and the absence of
such
proteins in cells transfected with frameshifted constructs (figure 7b). The
presence of signal in the stacking gel for pMJD8 may result from the presence
of
enough Gln residues to allow detection by the 1 C2 antibody. It could also be
due
to the recruitment of the intact non-frameshifted protein into the insoluble
aggregates, recruitment of hybrid polyAla/polyGln protein, or polyGln protein
accumulation independent of polyAla polymers.
FYAMPI F d
PoIyGCAlpolyAla stretches atone are toxic and sufficient
for formation of aggregates
In order to determine whether a construct with only a GCA
tract encoding an alanine peptide, outside the context of the ataxin-3
protein,
would be sufficient to produce a cellular phenotype, a truncated (GCP)4z
construct
with FS1 and HA epitopes in frame with GCA was transfected into COS-7 cells
(pMJD11, Fig. 4). In this construct, theMJD-1 sequence was truncated so that
the resulting protein will only have 25 amino acid residues left upstream from
the
repeat, and only the FS1 and HA epitopes after the GCA tract. At 24 hours,
cells
showed perinuclear and cytoplasmic aggregates and an abnormal nuclear
morphology (Fig. 8a), a phenotype that is very similar to the one obtained by
transfection of the full-length GCA/Ala constructs described above. Mock-
transfected cells showed background staining and absence of inclusions (Fig.
8b). These findings indicate that the presence of a frameshifted CAG repeat
product is sufficient for toxicity, independent of the protein context, and
suggest
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
38
a similar pathogenic mechanism may be operating in the other diseases caused
by expanded CAG tracts. This finding is thus of relevance to expand the
teachings of the present invention to all diseases or conditions caused by
expanded CAG tracts and to diseases and conditions caused by the accumulation
of polyalanine-containing proteins.
FYAMPI F
Antisera production
A 12-mer peptide corresponding to the new predicted COOH-
terminus of the ataxin-3 protein afterframeshift occurs (AAGPIRTEFTSM) was
used to raise antisera from two rabbits, FS1 and FS2. Injections were
performed
using 0.5 mg of peptide conjugated to KLH and sera were collected using
standard protocols°2.
~YeMPi ~ a
Western Blots
For protein extraction, MJD and control lymphoblastoid cells
were lysed in buffer containing NP-40. Equal amounts of protein were
electrophoresed in 8% SDS-polyacrylamide gels and transblotted to
nitrocellulose
membranes (as commonly known). Immunodetection was performed using FS1
(1:300), FS2 (1:300), anti-ataxin-3 (1:1000), 1C2 (1:2000), anti-ubiquitin
(1:400;
DAKO) and FS1 and FS2 pre-immune serum (1:300). Results for FS1 and FS2
were always consistent; all experiments were repeated three times on different
blots. HRP conjugated secondary antibodies were used at a 1:10,000 dilution.
COS-7 cells transfected with various constructs were collected, washed and
lysed
in Sample Loading Buffer. 100 Ng of each sample was used to run on a 10%
SDS-PAGE and transblotted onto nitrocellulose membranes. Immunodetection
was performed using antisera at following dilutions: 1C2 (1:5000), anti-ataxin-
3
(1:2000) and anti-HA (1:1000). Results were visualized by chemiluminescence
(RENAISSANCE).
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
39
~YeMVI F ~
Immunocytochemistry
MJD and control lymphoblastoid cells were harvested and a
total of 50x10' cells from each cell line were plated onto poly-D-lysine
coated
slides and fixed with acetone/methanol (1:1). Immunodetection was performed
using FS1 (1:300), anti-ubiquitin (1:300), anti-ataxin-3 (1:500) and FS1 pre-
immune serum (1:300). Biotinylated secondary antibodies were used at a 1:500
dilution and an amplification step was performed using the ABC kit (VECTOR).
Reaction product was visualized using the VIP kit (VECTOR). Immunocyto-
chemistry on COS-7 cells was performed on cover slips. At each time-point
cells
were fixed with 4% paraformaldehyde and immunodetection was performed using
anti-HA probe (1:500) and FS1 (1:300). Secondary antibodies and subsequent
amplification and detection procedures were carried out as described above.
GYeMPI F R
Immunohistochemistry
For immunohistochemistry of brain sections, 5 Nm sections of
paraffin-embedded tissue from the pons of an MJD patient and a control subject
were used. Sections were deparaffinized, permeabilized and immunostained with
FS1 (1:50) and anti-ubiquitin (DAKO) (1:300). Biotinylated secondary
antibodies
were used at a 1:500 dilution and an amplification step was performed using
the
ABC kit (VECTOR). Reaction product was visualized using the VIP kit
(VECTOR). For coimmunofluorescence of FS1 and anti-ubiquitin antibodies in
brain sections, the same procedure for preparation of samples was followed.
Immunodetection was performed using FS1 (1:50) and monoclonal anti~biquitin
(ZYMED) (1:300). A mixture of Cy3-conjugated anti-mouse antibody (1:100) and
fluorescein-conjugated anti-rabbit antibody (1:50) was used as secondary
probe.
Sections were mounted in SIowFade (Molecular Probes).
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
wnnno~ c o
Plasmid construction, transfection and cell culture
DNA amplification was performed using Pfu DNA polymerase
(STRATAGENE). Primer MJD-5': TTTTAAGCTTAGACAAA-TAAACATGGAG
5 (SEQ ID N0:1) was used in conjunction with MJD-3':
CCGGTGGATCCCTCATCCTGATAGGTCCCGCTGCTG (SEQ ID N0:2) for
pMJD1, pMJD2 and pMJD3, or MJD-3'c: CCGGTGGATCCCTCA-
TTGATAGGTCCCGCTGCTG (SEQ ID N0:3) for pMJD4 (STRATAGENE).
Primer MJD-5'(HIII): TTTAAGCTTCCCACCATGGAGT-CATCTTCCA (SEQ ID
10 N0:4) was used in conjunction with MJD-3'(BI):
CCGGTGGATCCCTCAGGGCGTAGTCGGGGACGTCGTAGGGGTACATGGAT
GTGAACTCTGTCCTGATAGGTCCCGCTG (SEQ ID N0:5). for pMJDS and
pMJD6, or MJD-3'c(BI): CCGGTGGATCCCAGGGCGTAGTCGGGGACGTCG-
TAGGGGTACATGGATGTGAACTCTGTCCTGATAGGTCCCGCTG (SEQ ID
15 N0:6) for pMJD7 and pMJDB, or MJD-Ala:
CGGAAGAGACGAGAAGCCTACTCCGGAAAAACAGCAGCAAAA-GCAGC
(SEQ ID N0:7) for pMJD9, pMJD10 pMJD11. Amplified products were digested
with BamHl and Hindlll and cloned into plasmid pEGFP-N1 (CLONTECH), except
for pMJD11, for which the amplified product was cloned into a modified version
20 of the pEGFP-N1 plasmid lacking the EGFP gene. All constructs were
confirmed
by sequencing (as commonly known). COS-7 cells were seeded in Dulbecco's
modified Eagle's medium (DMEM) containing 10% fetal calf serum the day before
transfection at 2 x 105 per well in 6-well plates containing sterile
coverslips. COS-
7 cells were transfected with plasmid DNA (2.0 Ng) using lipofectamine reagent
25 (GIBCO BRL) according to the manufacturer's instructions. For the
experiment
depicted in figure 4, after 72-96 hours, the cells were fixed with PBS/4%
paraformaldehyde and observed under a fluorescent microscope with FITC filter
in four independent experiments.
CONCLUSION
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
41
The present invention thus shows that transcriptional or
translational frameshifts occurring within expanded CAG tracts result in the
production and accumulation of polyalanine-containing mutant proteins. These
alanine polymers might deposit in cells forming INI and lead to nuclear
toxicity.
Support for this disease model is provided using lymphoblast cells from MJD
patients, as well as in pontine neurons of MJD brain and in in vitro cell
culture
models of the disease. Evidence that alanine polymers alone are toxic to cells
is
also provided and strongly suggests that a similar pathogenic mechanism
underlies the other CAG repeat disorders. How these accumulations lead to cell
death still needs to be elucidated.
Although the present invention has been described
hereinabove by way of preferred embodiments thereof, it can be modified,
without
departing from the spirit and nature of the subject invention as defined in
the
appended claims.
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
42
REFERENCES:
1. Paulson et al., 1997, Neuron 19:333-344.
2. Roizin et al., 1979, Adv Neurol 23:95-122.
3. Jackson et al., 1995, Neuropathology and Applied
Neurobiology
21:18-26.
4. DiFiglia et al., 1997, Science 277:1990-1993.
5. Holmberg et al., 1998, Human Molecular Genetics,
913-918.
6. Davies et al., 1997, Cell 90:537-548.
7. Ordway et al., 1997, Cell 91:753-763.
8. Skinner et al., 1997, Nature 389:971-974.
9. Davies et al., 1998, Lancet 351:131-133.
10. Tome et al., 1980, Acta Neuropathologica 49:85-87.
11. Brais et al., 1998, Nature Genetics 18:164-167.
12. Blondelle et al., 1997, Biochemistry 36:8393-8400.
13. Forood et al., 1995, Biochemical and Biophysical
Research
Communications 211:7-13.
14. van Leeuwen et al., 1998, Science 279:242-247.
15. Linton et al., 1997, Journal of Biological Chemistry272:14127-14132.
16. Lurland et al., 1996, Current Opinion in Biotechnology
7:489-493.
17. Paulson et al., 1997, Annals of Neurology 41:453-462.
18. Trottier et al., 1995, Nature 378:403-406.
19. Igarashi et al., 1998, Nature Genetics 18:111-117.
20. Cummings et al., 1998, Nature Genetics 19:148-154.
SUBSTITUTE SHEET (RULE 26)
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
43
21. Llinas, 1992, Trends in Neurosciences 15:351-355.
22. Ikeda et al., 1996, Nature Genetics 13:196-202.
23. Orr et al., 1993, Nature Genetics 4:221-226.
24. Sharp et al., 1995, Neuron 14:1065-1074.
25. David et al., 1997, Nature Genetics 17:65-70.
26. Imbert et al., 1996, Nature Genetics 14:285-291.
27. Sanpei et al., 1996, Nature Genetics 14:227-284.
28. Servadio et al., 1995, Nature Genetics 10:84-98.
29. Farabaugh, P.J., 1996, Annual Review in Genetics
30:507-28.
30. Perez et al., 1998, Journal of Cell Biology 143:1457-1470.
31. Johnston et al., 1998, Journal of Cell Biology
143:1883-1898.
32. Paulson et al., 1996, Annual Review in Neurosciences
19:79-107.
33. Cooper et al., 1998, Human Molecular Genetics
7:783-790.
34. Lunkes et al., 1998, Human Molecular Genetics
7:1355-1361.
35. Merry et al., 1998, Human Molecular Genetics 7:693-701.
36. Rubinsztein et al., 1995, Human Molecular Genetics
4:1585-1590.
37. Chung et al., 1993, Nature Genetics 5:254-258.
38. O'Donovan et al., 1996, Lancet 348:1739-1740.
39. Akarsu et al., 1996, Human Molecular Genetics
5:945-952.
40. Muragaki et al., 1996, Science 272:548-551.
41. Mundlos et al., 1997, Cell 89:773-779 (1997).
42. Harlow et al., 1988, in "Antibodies" CSH Laboratory
Press, Cold Spring
Harbor.
SUBSTITUTE SHEET (RULE 26)
CA 02384146 2002-03-06
WO 01/18544 PCT/CA00/01052
1/1
SEQUENCE LISTING
SEQ ID N0:1 TTTTAAGCTTAGACAAATAAACATGGAG
SEQ ID N0:2 CCGGTGGATCCCTCATCCTGATAGGTCCCGCTGCTG
SEQ ID N0:3 CCGGTGGATCCCTCATTGATAGGTCCCGCTGCTG
SEQ ID N0:4 TTTAAGCTTCCCACCATGGAGTCATCTTCCA
SEQ ID N0:5 CCGGTGGATCCCTCAGGGCGTAGTCGGGGACGTCG-
TAGGGGTACATGGATGTGAACTCTGTCCTGATAGGTCCCG
CTG
SEQ ID N0:6: CCGGTGGATCCCAGGGCGTAGTCGGGGACGTCGTAGGG-
GTACATGGATGTGAACTCTGTCCTGATAGGTCCCGCTG
SEQ ID N0:7 CGGAAGAGACGAGAAGCCTACTCCGGAAAAACAGCAGCA-
AAAGCAGC
SUBSTITUTE SHEET (RULE 26)