Note: Descriptions are shown in the official language in which they were submitted.
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
SUNFLOWER RhoGAP, LOX, ADH, AND SCIP-1
POLYNUCLEOT)Z7ES AND METHODS OF USE
FIELD OF THE INVENTION
The invention relates to the field of the genetic manipulation of plants,
particularly the modulation of gene activity and development in plants and
increased
disease resistance.
BACKGROUND OF THE INVENTION
Disease in plants is caused by biotic and abiotic causes. Biotic causes
include
fungi, viruses, bacteria, and nematodes. An example of the importance of plant
disease is illustrated by phytopathogenic fungi, which cause significant
annual crop
yield losses as well as devastating epidemics. Plant disease outbreaks have
resulted in
catastrophic crop failures that have triggered famines and caused major social
change.
Pathogenic fungi attack all of the approximately 300,000 species of flowering
plants,
however, a single plant species can be host to only a few fungal species, and
similarly,
most fungi usually have a limited host range. Generally, the best strategy for
plant
disease control is to use resistant cultivars selected or developed by plant
breeders for
1 S this purpose. However, the potential for serious crop disease epidemics
persists
today, as evidenced by outbreaks of the Victoria blight of oats and southern
corn leaf
blight. Molecular methods of crop protection have the potential to implement
novel
mechanisms for disease resistance and can also be implemented more quickly
than
traditional breeding methods. Accordingly, molecular methods are needed to
supplement traditional breeding methods to protect plants from pathogen
attack.
A host of cellular processes enable plants to defend themselves against
disease
caused by pathogenic agents. These defense mechanisms are activated by initial
pathogen infection in a process known as elicitation. In elicitation, the host
plant
recognizes a pathogen-derived compound known as an elicitor; the plant then
activates disease gene expression to limit further spread of the invading
microorganism. It is generally believed that to overcome these plant defense
mechanisms, plant pathogens must find a way to suppress elicitation as well as
to
-1-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
overcome more physically-based barriers to infection, such as reinforcement
and/or
rearrangement of the actin filament networks near the cell's plasma membrane.
Thus, the present invention solves needs for enhancement of the plant's
defensive elicitation response via a molecularly based mechanism that can be
quickly
incorporated into commercial crops.
SUMMARY OF THE INVENTION
The present invention provides nucleotide sequences that may find use in
modulating development, developmental pathways, and the plant pathogen defense
system. Particularly, the nucleotide and amino acid sequences for a sunflower
rhoGTPase-Activating Protein (rhoGAP), Lipoxygenase (LOX), Alcohol
Dehydrogenase (ADH), and Sclerotinia-Inducible Protein-1 (SCIP-1) are
provided.
In particular, the methods and compositions can be used to modulate plant
development. More specifically, methods and compositions of the invention may
be
1 S used for enhancing resistance to plant pathogens including fungal
pathogens, plant
viruses, and the like. The method involves stably transforming a plant with a
nucleotide sequence capable of modulating the plant pathogen defense system
operably linked with a promoter capable of driving expression of a gene in a
plant
cell. The disease resistance genes of the present invention additionally find
use in
manipulating these processes in transformed plants and plant cells.
Transformed plants, plant cells, and seeds, as well as methods for making such
plants, plant cells, and seeds are additionally provided. It is recognized
that a variety
of promoters will be useful in the invention, the choice of which will depend
in part
upon the desired level of expression of the disclosed nucleotide sequences. It
is
recognized that the levels of expression can be controlled to modulate the
levels of
expression in the plant cell.
Methods and compositions for regulating gene expression in a plant are also
provided. Novel nucleotide sequences for inducible plant promoters derived
from the
LOX and SCIP-1 genes are provided. The methods comprise transforming a plant
with a nucleotide sequence of interest operably linked to the LOX or SCIP-1
promoters. Exposure of the transformed plant to a stimulus activates, within
the
exposed tissue of the plant, transcription of the nucleotide sequence of
interest.
-2-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
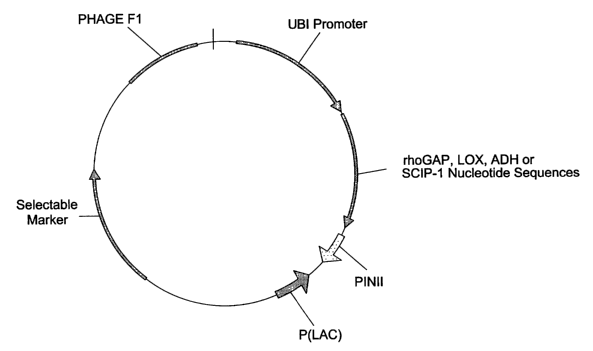
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 schematically illustrates an expression vector containing the
ubiquitin
promoter operably linked the rhoGAP, LOX, ADH, and SCIP-1 nucleotide
sequences.
Figure 2 schematically illustrates an expression vector used for plant
transformation containing the LOX or SCIP-1 promoter nucleotide sequences
operably linked to the nucleotide sequences encoding the GUS reporter protein.
DETAILED DESCRIPTION OF THE INVENTION
Overview
The present invention provides, inter alia, compositions and methods for
modulating the total level of proteins of the present invention and/or
altering their
ratios in a plant. By "modulation" is intended an increase or a decrease in a
particular
character, quality, substance, or response.
The compositions comprise sunflower nucleic acid and amino acid sequences.
Particularly, the nucleotide and amino acid sequence for a sunflower rhoGAP
(SEQ
ID NOS: 1 and 2), LOX (SEQ ID NOS: 3 and 4), ADH (SEQ ID NOS: 6 and 7) and
SCIP-1 (SEQ ID NOS: 8 and 9) are provided. As discussed in more detail below,
the
sequences of the invention are involved in many basic biochemical pathways
that
regulate plant growth, development, and pathogen resistance. Methods are
provided
for the expression of these sequences in a host plant to modulate plant
development,
developmental pathways, and defense responses. The method involves stably
transforming a plant with a nucleotide sequence capable of modulating the
plant
pathogen defense system operably linked with a promoter capable of driving
expression of the nucleotide sequence in a plant cell.
Also provided are LOX and SCIP-1 promoter sequences set forth in SEQ ID
NO: 5 and SEQ ID NO: 10, respectively. Methods are provided for the regulated
expression of a nucleotide sequence of interest that is operably linked to the
LOX or
SCIP-1 promoter sequences disclosed herein. Nucleotide sequences operably
linked
to the LOX or SCIP-1 promoters are transformed into a plant cell. Exposure of
the
transformed plant to a stimulus, induces transcriptional activation of the
nucleotide
sequences operably linked to the LOX or SCIP-1 promoters.
-3-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
rhoGTPase-activating protein (rhoGAP)
A nucleic acid sequence encoding a rhoGAP polypeptide from sunflower is
provided. The rhoGAP sequence shares homology to the conserved rhoGAP genes
from humans. The sunflower rhoGAP amino acid sequence shares about 40%
homology with the human p50rhoGAP sequence and about 80% homology with an
Arabidopsis hypothetical 23.6 kDa protein.
RhoGAPs are a central part of an evolutionarily conserved regulatory system
that are involved in cell growth and differentiation. Thus the sequence of the
invention finds use in controlling or modulating cell division,
differentiation,
development, as well as the defense response. Transformed plants can be
obtained
having altered metabolic states with respect to cell division and cellular
processes as
well as development and defense response; hence, the methods and compositions
may
find uses in affecting or studying differentiation.
RhoGAP proteins have been shown to interact with rho members of the ras
superfamily. Ras oncogenes were initially found to play an important role in
human
cancers and have since been shown to play important roles in regulation of
cell
growth and differentiation. Further, the rhoGAP proteins affect the activity
of
rhoGTPases (also called rho proteins), which act as molecular switches to
regulate
affected processes. The rho family of "G proteins" have a GTP-bound form and a
GDP-bound form; the relative amount of the GDP-bound form is increased by
GTPase activating proteins, or GAPS, which stimulate the intrinsic GTPase
activity of
the rho proteins.
Processes affected by GAPS include the transduction of hormone signals
across cell plasma membranes and the regulation of intracellular transport
pathways.
For example, rhoGTP-binding proteins have been shown to control signal
transduction pathways connecting the activation of actin polymerization to
activation
of cellular growth factor receptors. Hence, the compositions and methods of
the
invention find use in the activation or modulation of the cellular actin
cytoskeleton.
Although there is a great deal of conservation among members of the rhoGAP
family,
there are a large number of different proteins that contain the rhoGAP domain,
and
many of these proteins are large and multifunctional. Thus, the rhoGAP genes
and/or
proteins may contain different elements or motifs or sequence patterns that
modulate
or affect the activity, subcellular localization, and/or target of the rhoGAP
protein.
-4-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Such elements, motifs, or sequence patterns may be useful in engineering novel
enzymes for reducing or enhancing gene expression in particular tissues.
RhoGAP proteins activate rho genes and the related rac genes, which both
stimulate actin polymerization. The rho proteins in mammalian systems have
been
shown to regulate the formation of mufti-molecular complexes that are
associated
with polymerized actin located at the plasma membrane of the cell. Such
complexes
include actin stress fibers and focal adhesions in fibroblasts as well as the
actin-driven
phenomenon called membrane ruffling, which is exhibited by many cell types in
response to extracellular stimuli. Rho proteins have also been shown to play
roles in
epithelial cell migration in response to wounding. Expression of the sequences
of the
invention can be used to modulate or regulate the expression of corresponding
GTP-
binding proteins, i.e., rho, rac, etc. Hence, the compositions and methods of
the
invention find use in the activation or modulation of the cellular actin
cytoskeleton
and other actin-based structures and actin-related processes.
The RhoGAP gene of the present invention additionally finds use in enhancing
the plant pathogen defense system. Early plant-cell defense responses include
the
rearrangement of the cellular actin cytoskeleton to protect the cell from
attack.
RhoGAP genes are involved in cellular signaling cascades such as the oxidative
burst
that comprises part of the early defense response in plants. Hence, the
compositions
and methods of the invention can be used for enhancing resistance to plant
pathogens
including fungal pathogens, plant viruses, and the like.
Lipoxygenase
A nucleic acid sequence encoding a LOX polypeptide from sunflower is also
provided. The sunflower LOX polypeptide shares homology with other known LOX
proteins from potato, tomato, cowpea, Arabidopsis, and rice.
The LOX protein has been implicated in a number of important plant
developmental processes. LOX catalyzes the hydroperoxidation of
polyunsaturated
fatty acids containing cis, cis-1,4-pentadiene-conjugated double-bonds. The
primary
products of LOX-catalyzed reactions are fatty acid hydroperoxides, which are
typically metabolized into molecules with known or suspected regulatory
activity.
For example, LOX derived fatty acid hydroperoxides are precursors to traumatin
and
jasmonic acid. Traumatin induces cell division and may be involved in the
plant
-5-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
wounding response (Zimmerman et al. (1979) Plant Physiol. 63:536-541).
Jasmonates have been implicated as signal transduction molecules in the
response of
plants to stress, particularly wounding and pathogen attack (Farmer et al.
(1992) Cell
4:129-134). Therefore, the sunflower LOX gene may play an important role in
cell
division and defense signal transduction pathways that are regulated by the
biosynthesis of traumatin and jasmonic acid.
The LOX gene has also been implicated in the regulation of coordinated gene
activation in response to wounding. It is speculated that resistance to
pathogen attack
is the result of the coordinated accumulation of secondary metabolites and
protein
products. Some of these products, such as proteinase inhibitors, may directly
interfere
with digestibility of the injected tissue whereas others products may affect
food
intake. A potato LOX gene, 13-LOX, has been shown to control the expression
levels
of proteinase inhibitors in a wounding response to insect feeding (Royo et al.
(1999)
Proc. Natl. Acad. Sci. USA 96:1146-11 S 1 ). Therefore, the LOX sequences of
the
present invention may find use in an antifeedant stragedy by regulating
proteinase
inhibitor levels in plants, and thereby controlling insect and nematode
pathogens.
Additionally, LOX-derived fatty acid hydroperoxides and free radical species
are cytotoxic and are capable of damaging membranes, proteins, and DNA
(Hildebrand et al. (1998) Curr. Top. Plant Biochem. Physiol. 7:201-219).
Therefore,
LOXs may play a role in membrane degradation observed during senescence,
wounding, and the hypersensitive response to pathogen attack.
LOX proteins may also play an important role in plant growth and
development. There is a positive correlation between LOX activity levels
within an
organ and its rate of elongation. The concomitant increase in LOXs and the
enzymes
involved in the metabolism of LOX-derived fatty hydroperoxides is consistent
with a
role for LOX in generating lipid-derived growth regulators.
Furthermore, in plants, the LOX proteins may be involved in lipid turnover
and fat mobilization. Hence, the compositions and methods of the invention
find use
the turnover of lipids in the developing seedling.
Hence, the LOX sequences of the present invention may be used to modulate
many important developmental processes, such as, cell division, seed
germination,
plant growth and senescence, and/or to enhanced plant resistance to
environmental
stresses, such as, wounding and pathogen attacks.
-6-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
The present invention also provides the nucleotide sequences of the LOX
promoter. The promoter sequence contains cis-elements that resemble W-boxes,
TTGACC (nucleotides 322-327 of SEQ ID NO: 5), and G-boxes, CACGTG
(nucleotides 722-727 of SEQ ID NO: 5).
G-box and G-box like sequences are involved in the regulation of a variety of
unrelated genes and activate transcription in response to various stimuli
including:
exposure to visible and UV light (Chattopadhyay et al. (1998) Plant Cell
10:673-683),
dehydration-stress (Lam et al. (1991) JBiol Chem 266: 17131-17135 and Dolferus
et
al. (1994) Plant Physiol 105: 1075-1087), cold stress (Dolferus et al. (1994)
Plant
Physiol 105: 1075-1087), abscisic acid (Marcotte et al. (1989) Plant Cell I:
969-976),
sucrose (Urwin et al. (1997) Plant Mol Biol 35:929-942), and plant pathogen
defense
response (Wolfgang et al. (1997) EMBO Journal 16:726-738). In addition, G-box-
like sequences were also found to determine tissue-preferred expression
patterns
(Saunas et al. (1992) Plant Cell 4: 1485-1493 and Thomas (1993) Plant Cell 5:
1401-
1410). Functional analysis of G-box containing promoters has shown that the
nucleotide sequences immediately flanking the G-box and/or additional cis-
acting
promoter elements are often required for the G-box to influence transcription
activation.
The W-box promoter elements are involved in elicitor-induced gene
expression. W-boxes have been identified in a several plant promoters
including, for
example, members of the WRKY family (Eulgem et al. (1999) EMBO J. 18:4689-
4699) and members of the pathogenesis-related protein family (Rushton et al.
(1996)
EMBO J. 15:5690-5700). The fungal elicitor responsiveness of these genes is
mediated mainly by the presence of the W-boxes in the promoter elements.
Hence, the LOX promoter sequences find use in the regulated expression of an
operably linked heterologous gene of interest. More specifically, the
nucleotide
sequence may find use as an inducible promoter, more specifically, a pathogen-
inducible promoter.
Alcohol Dehydrogenase
A nucleic acid sequence encoding an ADH protein from sunflower is also
provided. Sequences of the sunflower alcohol dehydrogenase protein (ADH) share
about 85-95% sequence homology with plant alcohol dehydrogenases from garden
_7_
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
lettuce, potato, tomato, apple, and maize. ADH is an important enzyme in
anaerobic
metabolism, and it is usually encoded by a small multigene family in flowering
plants.
In both maize and Arabidopsis, the gene is expressed in seeds, roots, and
pollen
grains, whereas green aerial plant parts are devoid of detectable ADH
activity.
ADH has been implicated in responses to a number of environmental stresses,
including low oxygen, drought, salinity, cold acclimation, freezing tolerance,
flooding, and wounding. See, for example, Zeevaart et al. (1988) Annu. Rev.
Plant
Physiol. Plant Mol. Biol. 39:439-473; Sanchez et al. (1991 ) Abscisic Acid,
Physiology
and Biochemistry, Bio Scientific Publishers, Oxford, UK, pp. 210-216; and
Bruxelles
et al. (1996) Plant Physiol. 111:381-391. The ADH sequence of the present
invention
may find use in modulating a plant's response to adverse environmental
stresses.
The sunflower ADH sequences may also find use in modulating the
developmental process of fruit ripening. ADH reduces aldehydes to alcohols.
Modulation of ADH levels in ripening fruit has been shown to influence the
balance
between some of the aldehydes and the corresponding alcohols associated with
flavor
production. Hence, the compositions and methods of the present invention may
find
use in the modulation of ADH protein levels leading to a more intense "ripe
fruit"
flavor. See, for example, Speirs et al. (1998) Plant Physiol. 117:1047-1058.
The ADH sequences of the invention may additionally find use in enhancing
plants defense response. Under low oxygen conditions, i.e., a hypotic state,
ADH
plays a crucial role in cell survival. ADH serves as the major terminal
dehydrogenase
in regenerating oxidizing power in mature roots, thereby allowing glycolysis
to
continue in the absence of oxygen. Treatment of a plant with an elicitor
increases the
levels of active oxygen species in the plant cells and leads to a transient
state of
oxidative stress. See, for example, Robertson et al. (1995) Plant Molecular
Biology
27:59-67. Since aerobic respiration is compromised as a result of elicitor
action, the
ADH sequences of the present invention may find use in modulating a plant's
defense
against pathogens.
Sclerotinia-inducible protein-1 (SCIP-1)
The nucleic acid sequence encoding a novel sunflower protein, designated
Sclerotinia-Inducible Protein-l, SLIP-1, is also provided. SCIP-1 has limited
homology with hypothetical proteins from several bacteria.
_g_
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Transcript levels of SCIP-1 increase in both lesion mimic transgenic plants
and Sclerotinia-infected plants. The accumulation of SCIP-1 in lesion mimic
and
infected sunflower plants implicates that the protein is involved in the plant
defense
response to Sclerotinia and other pathogens. Hence, the compositions and
methods of
S the invention may find uses for enhancing resistance to plant pathogens,
including
fungal pathogens, plant viruses, and the like.
Furthermore, a PSI-Blast search revealed that the SCIP-1 sequence of the
invention fall into a class of flowering-related plant proteins (CEN and
others), as
well as some phosphatidylethanolamine-binding proteins (PEBP). The CEN-related
proteins are known to be related to a class of phosphatidylethanolamine-
binding
proteins (PEBP). Banfield et al. ((2000) JMoI Biol 297:1159-70) determined the
crystal structure of the centroradialis protein from Anthirrhinum. The
structure
confirmed what had been suspected by sequence homology studies: that the CEN
plant proteins are a subset of the family of PEBPs. Mammalian forms of PEBP
are
involved in inhibition of MAP kinase signaling, which is a central signaling
cascade
regulating cell differentiation (Banfield et al. (2000) JMoI Biol 297:1159-
70). The
structure of these proteins suggests that they may play a role in membrane
signal
transduction (Banfield et al. (1998) Structure 6.' 1245-54). In addition,
another recent
study (Kuramitsu et al. (2000) Electrophoresis 21:660-4) showed that a line of
mammalian cells resistant to tumor necrosis factor-alpha contained elevated
levels of
a protein identified as a PEBP. The report suggested that this PEBP could be
responsible for the resistance of certain cell lines to tumor necrosis factor
induced cell
death. Hence, the SCIP-1 polypeptide of the invention may play a role in
signaling,
membrane transduction, or in the regulation of cell death.
Furthermore, flowering plants exhibit two types of inflorescence architecture:
determinate and indeterminate. The centroradialis mutation causes the normally
indeterminate inflorescence of Antirrhinum to terminate in a flower. CEN-
related
protein have therefore been shown to influence maintenance of the
indeterminate state
of inflorescence meristems (Pnueli et al. (1998) Development 125:1979-1989;
Bradley et al. (1996) Nature 379:791-7; Bradley-Desmond et al. (1997) Science
275:80-83; and Amaya et al. (1999) Plant Cell 11:1405-1417). In addition, the
SCIP-
1 shares homology to Terminal Flower 1 (TFL 1 ) from both Arabidopsis thaliana
and
Brassica. TFL1 has also been shown to influence inflorescence meristem
identity.
-9-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
See, for example, Mimida-Naozumi et al. (1999) Plant-Science 142: 155-162 and
Ohshima et al. (1997) Mol Gen Genet 254:186-94. Hence, the SCIP-1 sequences of
the invention find use in influencing the state of inflorescence of meristem
development.
The present invention also provides the nucleotide sequences of the SCIP-1
promoter. The promoter sequence contains cis-elements that resemble W-boxes,
GTCAA (nucleotides 364-368 and 371-375 of SEQ ID NOS: 8 and 10), and G-boxes,
CACGTG (nucleotides 415-420 of SEQ ID NOS: 8 and 10). As with the LOX
promoter sequences, the SCIP-1 promoter sequences may find use in the
regulated
expression of an operably linked heterologous gene of interest. More
specifically, the
nucleotide sequence may fmd use as an inducible promoter, more specifically, a
pathogen-inducible promoter.
Compositions
Compositions of the invention include the polynucleotide sequences of the
sunflower rhoGAP, LOX, ADH, and SCIP-1 genes. In addition, the LOX and SCIP-1
promoter nucleotide sequences are also provided. The polypeptides encoded by
those
sequences may be involved in various plant developmental processes, including
the
plant pathogen defense response.
In particular, the present invention provides for isolated nucleic acid
molecules comprising nucleotide sequences encoding the amino acid sequences
shown in SEQ ID NOS: 2, 4, 7, and 9 or the nucleotide sequences encoding the
DNA
sequences deposited in a bacterial host as Patent Deposit Nos. PTA-284 and PTA-
285
(DNA sequences corresponding to rhoGAP), PTA-286 (DNA sequences
corresponding to ADH), PTA-287 (DNA sequences corresponding to LOX), PTA-
288 (DNA sequences corresponding to SCIP-1), or the DNA sequences obtained
from
the overlapping clones deposited in a bacterial host as Patent Deposit Nos.
PTA-284
and PTA-285. Further provided are polypeptides having an amino acid sequence
encoded by a nucleic acid molecule described herein, for example those set
forth in
SEQ ID NOS: l, 3, 6, and 8 or those deposited in a bacterial host as Patent
Deposit
Nos. PTA-284, PTA-285, PTA-286, PTA-287, PTA-288 and fragments and variants
thereof.
-10-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
The present invention further provides for isolated nucleic acid molecules
comprising nucleotide sequences shown in SEQ ID NO: 5 and SEQ ID NO: 10, or
nucleotide sequences encoding the DNA sequences deposited in a bacterial host
as
Patent Deposit Nos. PTA-559 and PTA-1721, and fragments and variants thereof.
By "DNA sequences obtained from the overlapping clones" is intended that
the complete DNA sequence of the rhoGAP sequence of the invention (SEQ ID NO:
1 ) can be obtained by sequencing the two individual clones that together
comprise the
entire rhoGAP sequence.
Plasmids containing the nucleotide sequences of the invention were deposited
with the Patent Depository of the American Type Culture Collection (ATCC),
Manassas, Virginia, and assigned Accession Nos. PTA-284, PTA-285, PTA-286,
PTA-287, PTA-288, PTA-559, and PTA-1721. Plasmids having the Accession Nos.
PTA-284, PTA-285, PTA-286, PTA-287, and PTA-288 were deposited on June 30,
1999. The plasmid having Accession No. PTA-559 was deposited on August 20,
1999, and the plasmid deposited as Accession No. PTA-1721 was deposited on
April
26, 2000. Two of these plasmids, designated as Accession No. PTA-284 and
Accession No. PTA-285, contained overlapping clones. The plasmids deposited as
PTA-284 and PTA-285 comprise the S' and the 3' end of the rhoGAP sequence,
respectively. It is noted, however, that clones deposited as PTA-284 and PTA-
285
contain common sequences at the regions where they overlap. One of skill in
the art
by sequencing the clones and aligning the overlap may obtain the entire
sequence of
the sunflower rhoGAP. These deposits will be maintained under the terms of the
Budapest Treaty on the International Recognition of the Deposit of
Microorganisms
for the Purposes of Patent Procedure. These deposits were made merely as a
convenience for those of skill in the art and are not an admission that a
deposit is
required under 35 U.S.C. ~112.
The invention encompasses isolated or substantially purified nucleic acid or
protein compositions. An "isolated" or "purified" nucleic acid molecule or
protein, or
biologically active portion thereof, is substantially free of other cellular
material, or
culture medium when produced by recombinant techniques, or substantially free
of
chemical precursors or other chemicals when chemically synthesized.
Preferably, an
"isolated" nucleic acid is free of sequences (preferably protein encoding
sequences)
that naturally flank the nucleic acid (i.e., sequences located at the 5' and
3' ends of the
-11-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
nucleic acid) in the genomic DNA of the organism from which the nucleic acid
is
derived. For example, in various embodiments, the isolated nucleic acid
molecule can
contain less than about 5 kb, 4 kb, 3 kb, 2 kb, 1 kb, 0.5 kb, or 0.1 kb of
nucleotide
sequences that naturally flank the nucleic acid molecule in genomic DNA of the
cell
from which the nucleic acid is derived. A protein that is substantially free
of cellular
material includes preparations of protein having less than about 30%, 20%,
10%, 5%,
(by dry weight) of contaminating protein. When the protein of the invention or
biologically active portion thereof is recombinantly produced, preferably
culture
medium represents less than about 30%, 20%, 10%, or 5% (by dry weight) of
chemical precursors or non-protein-of interest chemicals.
Fragments and variants of the disclosed nucleotide sequences and proteins
encoded thereby are also encompassed by the present invention. By "fragment"
is
intended a portion of the nucleotide sequence or a portion of the amino acid
sequence
and hence protein encoded thereby. Fragments of a nucleotide sequence may
encode
protein fragments that retain the biological activity of the native protein
and hence
affect development, developmental pathways, and defense response by retaining
rhoGAP-, LOX-, ADH-, or SCIP-1-like activity. Alternatively, fragments of a
nucleotide sequence that are useful as hybridization probes generally do not
encode
fragment proteins retaining biological activity. Thus, fragments of a
nucleotide
sequence may range from at least about 20 nucleotides, about 50 nucleotides,
about
100 nucleotides, and up to the full-length nucleotide sequence encoding the
proteins
of the invention.
A fragment of a rhoGAP nucleotide sequence that encodes a biologically
active portion of a rhoGAP protein of the invention will encode at least 12,
25, 30, 50,
100, 150, or 200 contiguous amino acids, or up to the total number of amino
acids
present in a full-length rhoGAP protein of the invention (for example, 201
amino
acids for SEQ ID NO: 2).
A fragment of a LOX nucleotide sequence that encodes a biologically active
portion of a LOX protein of the invention will encode at least 22, 30, 50,
100, 150,
200, 300, 400, 500, 600, 700, 800, 900 contiguous amino acids, or up to the
total
number of amino acids present in a full-length LOX protein of the invention
(for
example, 901 amino acids for SEQ ID NO: 3).
-12-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
A fragment of an ADH nucleotide sequence that encodes a biologically active
portion of an ADH protein of the invention will encode at least 92, 100, 150,
200,
250, 300, 350, 400 contiguous amino acids, or up to the total number of amino
acids
present in a full-length ADH protein of the invention (for example, 381 amino
acids
for SEQ ID NO: 7).
A fragment of a SCIP-1 nucleotide sequence that encodes a biologically active
portion of a SCIP-1 protein of the invention will encode at least 8, 15, 25,
30, 50, 100,
or 150 contiguous amino acids, or up to the total number of amino acids
present in a
full-length SCIP-1 protein of the invention (for example, 168 amino acids for
SEQ ID
NO: 9).
Fragments of a rhoGAP, LOX, ADH, and SCIP-1 nucleotide sequence that are
useful as hybridization probes or PCR primers generally need not encode a
biologically active portion of a rhoGAP, LOX, ADH, or SCIP-1 protein. Thus, a
fragment of a rhoGAP, LOX, ADH, or SCIP-1 nucleotide sequence may encode a
biologically active portion of a rhoGAP, LOX, ADH, or SCIP-1 protein, or it
may be
a fragment that can be used as a hybridization probe or PCR primer using
methods
disclosed below. A biologically active portion of a rhoGAP, LOX, ADH, or SCIP-
1
protein can be prepared by isolating a portion of one of the rhoGAP, LOX, ADH,
or
SCIP-1 nucleotide sequences of the invention, expressing the encoded portion
of the
rhoGAP, LOX, ADH, or SCIP-1 protein (e.g., by recombinant expression in
vitro),
and assessing the activity of the encoded portion of the rhoGAP, LOX, ADH, or
SCIP-1 protein. Nucleic acid molecules that are fragments of a rhoGAP, LOX,
ADH,
SCIP-1 nucleotide sequence comprise at least 16, 20, 50, 75, 100, 150, 200,
250, 300,
350, 400, 450, 500, 550, 600, 650, 700, or 800 nucleotides, or up to the
number of
nucleotides present in a full-length rhoGAP, LOX, ADH, or SCIP-1 nucleotide
sequence disclosed herein (for example, 824 nucleotides for SEQ ID NO: 1, 3806
nucleotides for SEQ ID NO: 3, 1403 nucleotides for SEQ ID NO: 6, and 746
nucleotide sequences for SEQ ID NO: 8).
By "variants" is intended substantially similar sequences. For nucleotide
sequences, conservative variants include those sequences that, because of the
degeneracy of the genetic code, encode the amino acid sequence of one of the
polypeptides of the invention. Naturally occurring allelic variants such as
these can
be identified with the use of well-known molecular biology techniques, as, for
-13-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
example, with polymerase chain reaction (PCR) and hybridization techniques as
outlined below. Variant nucleotide sequences also include synthetically
derived
nucleotide sequences, such as those generated, for example, by using site-
directed
mutagenesis but which still encode a rhoGAP, LOX, ADH, or SCIP-1 protein of
the
S invention. Generally, variants of a particular nucleotide sequence of the
invention will
have at least 40%, 50%, 60%, 70%, generally at least 75%, 80%, 85%, preferably
about 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, and more preferably about 98%,
99%, or more sequence identity to that particular nucleotide sequence as
determined
by sequence alignment programs described elsewhere herein using default
parameters.
By "variant" protein is intended a protein derived from the native protein by
deletion (so-called truncation) or addition of one or more amino acids to the
N-
terminal and/or C-terminal end of the native protein; deletion or addition of
one or
more amino acids at one or more sites in the native protein; or substitution
of one or
more amino acids at one or more sites in the native protein. Variant proteins
encompassed by the present invention are biologically active, that is they
continue to
possess the desired biological activity of the native protein, hence they will
continue
to possess rhoGAP, LOX, ADH, or SCIP-1 activity. Such variants may result
from,
for example, genetic polymorphism or from human manipulation. Biologically
active
variants of a native rhoGAP, LOX, ADH, or SCIP-1 protein of the invention will
have
at least 40%, 50%, 60%, 70%, generally at least 75%, 80%, 85%, preferably
about
90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more sequence identity to
the amino acid sequence for the native protein as determined by sequence
alignment
programs described elsewhere herein using default parameters. A biologically
active
variant of a protein of the invention may differ from that protein by as few
as 1-15
amino acid residues, as few as 1-10, such as 6-10, as few as 5, as few as 4,
3, 2, or
even 1 amino acid residue.
Biological activity of the rhoGAP, LOX, ADH and SCIP-1 polypeptides can
be assayed by any method known in the art. Assays to measure the developmental
pathways and defense responses that are influenced by the rhoGAP, LOX, ADH and
SCIP-1 polypeptides having rhoGAP-, LOX-, ADH-, and SCIP-1-like activity are
well known in the art. Furthermore, assays to detect rhoGAP-like activity
include
GTP binding assays (Borg et al. (1994) Plant Mol. Biol. 27:175-187);
interactions
with Rac or Ras (Diekman et al. (1995) EMBO J. 14:5297-5305 and Van Aelst et
al.
-14-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
(1996) EMBO J. 15:3778-3786); and GTPase and GTPase activating activity assays
(Borg et al. (1999) FEBSLetters 453:341-345). Assays to detect LOX-like
activity
include, for example, assays to measure LOX enzymatic activity (Maach et al.
(1997)
Plant Physiol. 114:1561-1566, Royo et al. (1996) J. Biol. Chem. 35:21012-21019
and
Voros et al. (1998) FEBSLetters 251:36-44). Assays to detect ADH-like activity
include, for example, ADH enzymatic activity assays (Tomes et al. ( 1976)
Biochem.
Genetics 14:87-97).
The proteins of the invention may be altered in various ways including amino
acid substitutions, deletions, truncations, and insertions. Methods for such
manipulations are generally known in the art. For example, amino acid sequence
variants of the rhoGAP, LOX, ADH, or SCIP-1 proteins can be prepared by
mutations
in the DNA. Methods for mutagenesis and nucleotide sequence alterations are
well
known in the art. See, for example, Kunkel (1985) Proc. Natl. Acad. Sci. USA
82:488-492; Kunkel et al. (1987) Methods in Enzymol. 154:367-382; US Patent
No.
4,873,192; Walker and Gaastra, eds. (1983) Techniques in Molecular Biology
(MacMillan Publishing Company, New York) and the references cited therein.
Guidance as to appropriate amino acid substitutions that do not affect
biological
activity of the protein of interest may be found in the model of Dayhoff et
al. (1978)
Atlas of Protein Sequence and Structure (Natl. Biomed. Res. Found.,
Washington,
D.C.), herein incorporated by reference. Conservative substitutions, such as
exchanging one amino acid with another having similar properties, may be
preferred.
Thus, the genes and nucleotide sequences of the invention include both the
naturally occurring sequences as well as mutant forms. Likewise, the proteins
of the
invention encompass both naturally occurring proteins as well as variations
and
modified forms thereof. Such variants will continue to possess the desired
rhoGAP-,
LOX-,
ADH-, or SCIP-1-like activity. Obviously, the mutations that will be made in
the
DNA encoding the variant must not place the sequence out of reading frame and
preferably will not create complementary regions that could produce secondary
mRNA structure. See, EP Patent Application Publication No. 75,444.
The deletions, insertions, and substitutions of the protein sequences
encompassed herein are not expected to produce radical changes in the
characteristics
of the protein. However, when it is difficult to predict the exact effect of
the
-15-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
substitution, deletion, or insertion in advance of doing so, one skilled in
the art will
appreciate that the effect will be evaluated by routine screening assays. That
is, the
activity can be evaluated by either an enhanced resistance to pathogens or a
modulation in a plant developmental process when expression of the protein
sequence
is altered.
Variant nucleotide sequences and proteins also encompass sequences and
proteins derived from a mutagenic and recombinogenic procedure such as DNA
shuffling. With such a procedure, one or more different rhoGAP, LOX, ADH, or
SCIP-1 coding sequences can be manipulated to create a new rhoGAP, LOX, ADH,
or SCIP-1 possessing the desired properties. In this manner, libraries of
recombinant
polynucleotides are generated from a population of related sequence
polynucleotides
comprising sequence regions that have substantial sequence identity and can be
homologously recombined in vitro or in vivo. For example, using this approach,
sequence motifs encoding a domain of interest may be shuffled between the
rhoGAP,
LOX, ADH, or SCIP-1 gene of the invention and other known rhoGAP, LOX, ADH,
or SCIP-1 genes to obtain a new gene coding for a protein with an improved
property
of interest, such as an increased Km in the case of an enzyme. Strategies for
such
DNA shuffling are known in the art. See, for example, Stemmer (1994) Proc.
Natl.
Acad. Sci. USA 91:10747-10751; Stemmer (1994) Nature 370:389-391; Crameri et
al.
(1997) Nature Biotech. 15:436-438; Moore et al. (1997) J. Mol. Biol. 272:336-
347;
Zhang et al. (1997) Proc. Natl. Acad. Sci. USA 94:4504-4509; Crameri et al.
(1998)
Nature 391:288-291; and U.S. Patent Nos. 5,605,793 and 5,837,458.
The compositions of the invention also include isolated nucleic acid molecules
comprising the promoter nucleotide sequences set forth in SEQ ID NO: 5 and SEQ
ID
NO: 10. By "promoter" is intended a regulatory region of DNA usually
comprising a
TATA box (nucleotides 808-901 of SEQ ID NO: 5) capable of directing RNA
polymerase II to initiate RNA synthesis at the appropriate transcription
initiation site
for a particular coding sequence. A promoter may additionally comprise other
recognition sequences generally positioned upstream or 5' to the TATA box,
referred
to as upstream promoter elements, which influence the transcription initiation
rate.
Such elements include a W-box (nucleotide sequence 322-327 of SEQ ID NO: 5;
nucleotide sequence 364-368 and 371-375 of SEQ ID NO: 10) and a G-box
(nucleotide sequence 722-727 of SEQ ID NO: 5; nucleotide sequence 415-420 of
-16-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
SEQ ID NO:10). The promoter sequences of the present invention "regulate"
(i.e.,
repress or activate) transcription from the promoter region. The regulation of
transcription by the promoter sequences of the present invention is defined
herein as
"inducible." By "inducible" is intended the ability of the promoter sequences
to
regulate expression of an operably linked nucleotide sequence in response to a
stimulus.
It is recognized that having identified the nucleotide sequences for the
promoter regions disclosed herein, it is within the state of the art to
isolate and
identify additional regulatory element in the 5' untranslated region upstream
from the
particular promoter regions defined herein. Thus for example, the promoter
regions
disclosed herein may further comprise upstream regulatory elements that confer
tissue-preferred expression of heterologous nucleotide sequences operably
linked to
the disclosed promoter sequence. See particularly, Australian Paten No. AU-A-
77751/94 and U.S. Patent Nos. 5,466,785 and 5,635,618.
Fragments and variants of the disclosed LOX and SCIP-1 promoter nucleotide
sequences are also encompassed by the present invention. By "fragment" is
intended
a portion of the nucleotide sequence. Fragments of a nucleotide sequence may
retain
biological activity and hence retain their transcriptional regulatory
activity. Thus, for
example, less than the entire promoter sequence disclosed herein may be
utilized to
drive expression of an operably linked nucleotide sequence of interest, such
as a
nucleotide sequence encoding a heterologous protein. Alternatively, a fragment
of
promoter sequence may retain the ability to regulate transcription in the
presence of a
stimulus when operably linked to a heterologous transcriptional initiation
region.
Alternatively, fragments of a nucleotide sequence that are useful as
hybridization
probes generally do not retain biological activity. Thus, fragments of a
nucleotide
sequence may range from at least about 20 nucleotides, about 50 nucleotides,
about
100 nucleotides, and up to the full-length nucleotide sequence of the
invention.
Thus, a fragment of a LOX or SCIP-1 promoter nucleotide sequence may
encode a biologically active portion of the LOX or SCIP-1 promoter, or it may
be a
fragment that can be used as a hybridization probe or PCR primer using methods
disclosed below. A biologically active portion of a LOX or SCIP-1 promoter can
be
prepared by isolating a portion of one of the LOX or SCIP-1 promoter
nucleotide
sequences of the invention, and assessing the activity of the portion of the
LOX or
-17-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
SCIP-1 promoter. Nucleic acid molecules that are fragments of a LOX or SCIP-1
promoter nucleotide sequence comprise at least 16, 20, 50, 75, 100, 150, 200,
250,
300, 350, 400, 450, 500, 550, 600, 650, 700, or 800 nucleotides, or up to the
number
of nucleotides present in a full-length LOX or SCIP-1 promoter nucleotide
sequence
disclosed herein (for example, 880 nucleotide for SEQ ID NO: 5; 510 nucleotide
for
SEQ ID NO: 10). Assays to determine the activity of a promoter sequence are
well
known in the art. For example, a LOX or SCIP-1 promoter fragment or variant
may
be operably linked to the nucleotide sequence encoding any reporter protein,
such as
the (3-glucuronidase protein (GUS reporter) or the luciferase protein. The DNA
construct is inserted into the genome of a plant or plant cell and the mRNA or
protein
level of the reporter sequence is determined. See, for example, Eulgem et al.
( 1999)
EMBO. 18: 4689-4699.
Thus, isolated sequences that have promoter activity and which hybridize
under stringent conditions to the LOX and SCIP-1 sequences disclosed herein,
or to
fragments thereof, are encompassed by the present invention. Such sequences
will be
at least about 40% to 50% homologous, about 60%, 65%, or 70% homologous, and
even at least about 75% 80% 85% 90% 91% 92% 93% 94% 95% 96% 97%
> > > > > > > > > > >
98%, 99% or more homologous with the disclosed sequences. That is, the
sequence
identity of sequences may range, sharing at least about 40% to 50%, about 60%,
65%,
or 70%, and even at least about 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%,
96%, 97%, 98%, 99% or more sequence identity.
By "variants" of the promoter sequences is intended substantially similar
sequences. For nucleotide sequences naturally occurring variants such as these
can be
identified with the use of well-known molecular biology techniques, as, for
example,
with polymerise chain reaction (PCR) and hybridization techniques as outlined
below. Variant nucleotide sequences also include synthetically derived
nucleotide
sequences, such as those generated, for example, by using site-directed
mutagenesis.
Generally, variants of a particular nucleotide sequence of the invention will
have at
least about 40%, 50%, 60%, 65%, 70%, generally at least about 75%, 80%, 85%,
preferably at least about 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, and more
preferably at least about 98%, 99% or more sequence identity to that
particular
nucleotide sequence as determined by sequence alignment programs described
elsewhere herein using default parameters.
-18-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
The nucleotide sequences of the invention can be used to isolate
corresponding sequences from other organisms, particularly other plants, more
particularly other monocots. In this manner, methods such as PCR,
hybridization, and
the like can be used to identify such sequences based on their sequence
homology to
the sequences set forth herein. Sequences isolated based on their sequence
identity to
the entire disease resistant sequences set forth herein or to fragments
thereof are
encompassed by the present invention. Such sequences include sequences that
are
orthologs of the disclosed sequences. By "orthologs" is intended genes derived
from
a common ancestral gene and which are found in different species as a result
of
speciation. Genes found in different species are considered orthologs when
their
nucleotide sequences and/or their encoded protein sequences share substantial
identity
as defined elsewhere herein. Functions of orthologs are often highly conserved
among species.
In a PCR approach, oligonucleotide primers can be designed for use in PCR
reactions to amplify corresponding DNA sequences from cDNA or genomic DNA
extracted from any plant of interest. Methods for designing PCR primers and
PCR
cloning are generally known in the art and are disclosed in Sambrook et al.
(1989)
Molecular Cloning: A Laboratory Manual (2d ed., Cold Spring Harbor Laboratory
Press, Plainview, New York). See also Innis et al., eds. (1990) PCR Protocols:
A
Guide to Methods and Applications (Academic Press, New York); Innis and
Gelfand,
eds. (1995) PCR Strategies (Academic Press, New York); and Innis and Gelfand,
eds.
(1999) PCR Methods Manual (Academic Press, New York). Known methods of PCR
include, but are not limited to, methods using paired primers, nested primers,
single
specific primers, degenerate primers, gene-specific primers, vector-specific
primers,
partially-mismatched primers, and the like.
In hybridization techniques, all or part of a known nucleotide sequence is
used
as a probe that selectively hybridizes to other corresponding nucleotide
sequences
present in a population of cloned genomic DNA fragments or cDNA fragments (i.
e.,
genomic or cDNA libraries) from a chosen organism. The hybridization probes
may
be genomic DNA fragments, cDNA fragments, RNA fragments, or other
oligonucleotides, and may be labeled with a detectable group such as 32P, or
any other
detectable marker. Thus, for example, probes for hybridization can be made by
labeling synthetic oligonucleotides based on the disease resistant sequences
of the
-19-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
invention. Methods for preparation of probes for hybridization and for
construction of
cDNA and genomic libraries are generally known in the art and are disclosed in
Sambrook et al. (1989) Molecular Cloning: A Laboratory Manual (2d ed., Cold
Spring Harbor Laboratory Press, Plainview, New York).
For example, an entire sequence disclosed herein, or one or more portions
thereof, may be used as a probe capable of specifically hybridizing to the
corresponding LOX, SCIP-1, rhoGAP, or ADH sequences and messenger RNAs. To
achieve specific hybridization under a variety of conditions, such probes
include
sequences that are unique among disease resistant sequences and are preferably
at
least about 10 nucleotides in length, and most preferably at least about 20
nucleotides
in length. Such probes may be used to amplify corresponding sequences from a
chosen organism by PCR. This technique may be used to isolate additional
coding
sequences from a desired organism or as a diagnostic assay to determine the
presence
of coding sequences in an organism. Hybridization techniques include
hybridization
screening of plated DNA libraries (either plaques or colonies; see, for
example,
Sambrook et al. ( 1989) Molecular Cloning: A Laboratory Manual (2d ed., Cold
Spring Harbor Laboratory Press, Plainview, New York).
Hybridization of such sequences may be carried out under stringent
conditions. By "stringent conditions" or "stringent hybridization conditions"
is
intended conditions under which a probe will hybridize to its target sequence
to a
detectably greater degree than to other sequences (e.g., at least 2-fold over
background). Stringent conditions are sequence-dependent and will be different
in
different circumstances. By controlling the stringency of the hybridization
and/or
washing conditions, target sequences that are 100% complementary to the probe
can
be identified (homologous probing). Alternatively, stringency conditions can
be
adjusted to allow some mismatching in sequences so that lower degrees of
similarity
are detected (heterologous probing). Generally, a probe is less than about
1000
nucleotides in length, preferably less than 500 nucleotides in length.
Typically, stringent conditions will be those in which the salt concentration
is
less than about 1.5 M Na ion, typically about 0.01 to 1.0 M Na ion
concentration (or
other salts) at pH 7.0 to 8.3 and the temperature is at least about
30°C for short probes
(e.g., 10 to 50 nucleotides) and at least about 60°C for long probes
(e.g., greater than
50 nucleotides). Stringent conditions may also be achieved with the addition
of
-20-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
destabilizing agents such as formamide. Exemplary low stringency conditions
include hybridization with a buffer solution of 30 to 35% formamide, 1 M NaCI,
1%
SDS (sodium dodecyl sulphate) at 37°C, and a wash in 1X to 2X SSC (20X
SSC = 3.0
M NaCI/0.3 M trisodium citrate) at 50 to 55°C. Exemplary moderate
stringency
conditions include hybridization in 40 to 45% formamide, 1.0 M NaCI, 1% SDS at
37°C, and a wash in 0.5X to 1X SSC at 55 to 60°C. Exemplary high
stringency
conditions include hybridization in 50% formamide, 1 M NaCI, 1% SDS at
37°C, and
a wash in O.1X SSC at 60 to 65°C. Duration of hybridization is
generally less than
about 24 hours, usually about 4 to about 12 hours.
Specificity is typically the function of post-hybridization washes, the
critical
factors being the ionic strength and temperature of the final wash solution.
For DNA-
DNA hybrids, the Tm can be approximated from the equation of Meinkoth and Wahl
(1984) Anal. l3iochem. 138:267-284: Tm = 81.5°C + 16.6 (log M) + 0.41
(%GC) -
0.61 (% form) - 500/L; where M is the molarity of monovalent canons, %GC is
the
percentage of guanosine and cytosine nucleotides in the DNA, % form is the
percentage of formamide in the hybridization solution, and L is the length of
the
hybrid in base pairs. The Tm is the temperature (under defined ionic strength
and pH)
at which 50% of a complementary target sequence hybridizes to a perfectly
matched
probe. Tm is reduced by about 1 °C for each 1 % of mismatching; thus,
Tm,
hybridization, and/or wash conditions can be adjusted to hybridize to
sequences of the
desired identity. For example, if sequences with >90% identity are sought, the
Tm can
be decreased 10°C. Generally, stringent conditions are selected to be
about 5°C lower
than the thermal melting point (Tm) for the specific sequence and its
complement at a
defined ionic strength and pH. However, severely stringent conditions can
utilize a
hybridization and/or wash at 1, 2, 3, or 4°C lower than the thermal
melting point (Tm);
moderately stringent conditions can utilize a hybridization and/or wash at 6,
7, 8, 9, or
10°C lower than the thermal melting point (Tm); low stringency
conditions can utilize
a hybridization and/or wash at 11, 12, 13, 14, 15, or 20°C lower than
the thermal
melting point (Tm). Using the equation, hybridization and wash compositions,
and
desired Tm, those of ordinary skill will understand that variations in the
stringency of
hybridization and/or wash solutions are inherently described. If the desired
degree of
mismatching results in a Tm of less than 45°C (aqueous solution) or
32°C (formamide
solution), it is preferred to increase the SSC concentration so that a higher
temperature
-21-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
can be used. An extensive guide to the hybridization of nucleic acids is found
in
Tijssen (1993) Laboratory Techniques in Biochemistry and Molecular Biolo~y-
Hybridization with Nucleic Acid Probes, Part I, Chapter 2 (Elsevier, New
York); and
Ausubel et al., eds. (1995) Current Protocols in Molecular Biology, Chapter 2
(Greene Publishing and Wiley-Interscience, New York). See Sambrook et al.
(1989)
Molecular Cloning.' A Laboratory Manual (2d ed., Cold Spring Harbor Laboratory
Press, Plainview, New York).
In general, sequences that have promotor activity or encode a rhoGAP, LOX,
ADH or SCIP-1 polypeptide and which hybridize under stringent conditions to
the
rhoGAP, LOX, ADH or SCIP-1 sequences disclosed herein, or to fragments
thereof,
are encompassed by the present invention. Such sequences will be at least 40%
to
50% homologous, about 60%, 65%, or 70% homologous, and even at least about 75%
homologous, and even about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%,
97%, 98%, 99% homologous or more with the disclosed sequences. That is, the
sequence identity of the sequences may range, sharing at least 40% to 50%,
about
60%, 65%, or 70%, and even about 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%,
95%, 96%, 97%, 98%, 99% or more sequence identity.
The following terms are used to describe the sequence relationships between
two or more nucleic acids or polynucleotides: (a) "reference sequence", (b)
"comparison window", (c) "sequence identity", (d) "percentage of sequence
identity",
and (e) "substantial identity".
(a) As used herein, "reference sequence" is a defined sequence used as a
basis for sequence comparison. A reference sequence may be a subset or the
entirety
of a specified sequence; for example, as a segment of a full-length cDNA or
gene
sequence, or the complete cDNA or gene sequence.
(b) As used herein, "comparison window" makes reference to a contiguous
and specified segment of a polynucleotide sequence, wherein the polynucleotide
sequence in the comparison window may comprise additions or deletions (i.e.,
gaps)
compared to the reference sequence (which does not comprise additions or
deletions)
for optimal alignment of the two sequences. Generally, the comparison window
is at
least 20 contiguous nucleotides in length, and optionally can be 30, 40, 50,
100, or
longer. Those of skill in the art understand that to avoid a high similarity
to a
-22-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
reference sequence due to inclusion of gaps in the polynucleotide sequence a
gap
penalty is typically introduced and is subtracted from the number of matches.
Methods of alignment of sequences for comparison are well known in the art.
Thus, the determination of percent identity between any two sequences can be
accomplished using a mathematical algorithm. Non-limiting examples of such
mathematical algorithms are the algorithm of Myers and Miller (1988) CABIOS
4:11-
17; the local homology algorithm of Smith et al. (1981) Adv. Appl. Math.
2:482; the
homology alignment algorithm of Needleman and Wunsch (1970) J. Mol. Biol.
48:443-453; the search-for-similarity-method of Pearson and Lipman (1988)
Proc.
Natl. Acad Sci. 85:2444-2448; the algorithm of Karlin and Altschul (1990)
Proc.
Natl. Acad Sci. USA 872264, modified as in Karlin and Altschul (1993) Proc.
Natl.
Acad Sci. USA 90:5873-5877.
Computer implementations of these mathematical algorithms can be utilized
for comparison of sequences to determine sequence identity. Such
implementations
include, but are not limited to: CLUSTAL in the PC/Gene program (available
from
Intelligenetics, Mountain View, California); the ALIGN program (Version 2.0)
and
GAP, BESTFIT, BLAST, FASTA, and TFASTA in the Wisconsin Genetics Software
Package, Version 8 (available from Genetics Computer Group (GCG), 575 Science
Drive, Madison, Wisconsin, USA). Alignments using these programs can be
performed using the default parameters. The CLUSTAL program is well described
by
Higgins et al. (1988) Gene 73:237-244 (1988); Higgins et al. (1989) CABIOS
5:151-
153; Corpet et al. (1988) Nucleic Acids Res. 16:10881-90; Huang et al. (1992)
CABIOS 8:155-65; and Pearson et al. (1994) Meth. Mol. Biol. 24:307-331. The
ALIGN program is based on the algorithm of Myers and Miller (1988) supra. A
PAM120 weight residue table, a gap length penalty of 12, and a gap penalty of
4 can
be used with the ALIGN program when comparing amino acid sequences. The
BLAST programs of Altschul et al (1990) J. Mol. Biol. 215:403 are based on the
algorithm of Karlin and Altschul (1990) supra. BLAST nucleotide searches can
be
performed with the BLASTN program, score = 100, wordlength = 12, to obtain
nucleotide sequences homologous to a nucleotide sequence encoding a protein of
the
invention. BLAST protein searches can be performed with the BLASTX program,
score = 50, wordlength = 3, to obtain amino acid sequences homologous to a
protein
or polypeptide of the invention. To obtain gapped alignments for comparison
-23-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
purposes, Gapped BLAST (in BLAST 2.0) can be utilized as described in Altschul
et
al. (1997) Nucleic Acids Res. 25:3389. Alternatively, PSI-BLAST (in BLAST 2.0)
can be used to perform an iterated search that detects distant relationships
between
molecules. See Altschul et al. (1997) supra. When utilizing BLAST, Gapped
BLAST,
PSI-BLAST, the default parameters of the respective programs (e.g., BLASTN for
nucleotide sequences, BLASTX for proteins) can be used. See
http://www.ncbi.hlm.nih.gov. Alignment may also be performed manually by
inspection.
Unless otherwise stated, sequence identity/similarity values provided herein
refer to the value obtained using GAP Version 10 using the following
parameters:
identity using GAP Weight of 50 and Length Weight of 3; % similarity using Gap
Weight of 12 and Length Weight of 4, or any equivalent program. By "equivalent
program" is intended any sequence comparison program that, for any two
sequences
in question, generates an alignment having identical nucleotide or amino acid
residue
matches and an identical percent sequence identity when compared to the
corresponding alignment generated by the preferred program.
GAP uses the algorithm of Needleman and Wunsch (1970) J. Mol. Biol. 48:
443-453, to fmd the alignment of two complete sequences that maximizes the
number
of matches and minimizes the number of gaps. GAP considers all possible
alignments
and gap positions and creates the alignment with the largest number of matched
bases
and the fewest gaps. It allows for the provision of a gap creation penalty and
a gap
extension penalty in units of matched bases. GAP must make a profit of gap
creation
penalty number of matches for each gap it inserts. If a gap extension penalty
greater
than zero is chosen, GAP must, in addition, make a profit for each gap
inserted of the
length of the gap times the gap extension penalty. Default gap creation
penalty values
and gap extension penalty values in Version 10 of the Wisconsin Genetics
Software
Package for protein sequences are 8 and 2, respectively. For nucleotide
sequences
the default gap creation penalty is 50 while the default gap extension penalty
is 3.
The gap creation and gap extension penalties can be expressed as an integer
selected
from the group of integers consisting of from 0 to 200. Thus, for example, the
gap
creation and gap extension penalties can be 0, l, 2, 3, 4, 5, 6, 7, 8, 9, 10,
15, 20, 25,
30, 35, 40, 45, 50, 55, 60, 65 or greater.
-24-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
GAP presents one member of the family of best alignments. There may be
many members of this family, but no other member has a better quality. GAP
displays four figures of merit for alignments: Quality, Ratio, Identity, and
Similarity.
The Quality is the metric maximized in order to align the sequences. Ratio is
the
quality divided by the number of bases in the shorter segment. Percent
Identity is the
percent of the symbols that actually match. Percent Similarity is the percent
of the
symbols that are similar. Symbols that are across from gaps are ignored. A
similarity
is scored when the scoring matrix value for a pair of symbols is greater than
or equal
to 0.50, the similarity threshold. The scoring matrix used in Version 10 of
the
Wisconsin Genetics Software Package is BLOSUM62 (see Henikoff and Henikoff
(1989) Proc. Natl. Acad. Sci. USA 89:10915).
For purposes of the present invention, comparison of nucleotide or protein
sequences for determination of percent sequence identity to the rhoGAP, LOX,
ADH,
and SCIP-1 sequences disclosed herein is preferably made using the ClustalW
program (Version 1.7 or later) with its default parameters or any equivalent
program.
By "equivalent program" is intended any sequence comparison program that, for
any
two sequences in question, generates an alignment having identical nucleotide
or
amino acid residue matches and an identical percent sequence identity when
compared to the corresponding alignment generated by the preferred program.
(c) As used herein, "sequence identity" or "identity" in the context of two
nucleic acid or polypeptide sequences makes reference to the residues in the
two
sequences that are the same when aligned for maximum correspondence over a
specified comparison window. When percentage of sequence identity is used in
reference to proteins it is recognized that residue positions which are not
identical
often differ by conservative amino acid substitutions, where amino acid
residues are
substituted for other amino acid residues with similar chemical properties
(e.g., charge
or hydrophobicity) and therefore do not change the functional properties of
the
molecule. When sequences differ in conservative substitutions, the percent
sequence
identity may be adjusted upwards to correct for the conservative nature of the
substitution. Sequences that differ by such conservative substitutions are
said to have
"sequence similarity" or "similarity". Means for making this adjustment are
well
known to those of skill in the art. Typically this involves scoring a
conservative
substitution as a partial rather than a full mismatch, thereby increasing the
percentage
-25-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
sequence identity. Thus, for example, where an identical amino acid is given a
score
of 1 and a non-conservative substitution is given a score of zero, a
conservative
substitution is given a score between zero and 1. The scoring of conservative
substitutions is calculated, e.g., as implemented in the program PC/GENE
(Intelligenetics, Mountain View, California).
(d) As used herein, "percentage of sequence identity" means the value
determined by comparing two optimally aligned sequences over a comparison
window, wherein the portion of the polynucleotide sequence in the comparison
window may comprise additions or deletions (i.e., gaps) as compared to the
reference
sequence (which does not comprise additions or deletions) for optimal
alignment of
the two sequences. The percentage is calculated by determining the number of
positions at which the identical nucleic acid base or amino acid residue
occurs in both
sequences to yield the number of matched positions, dividing the number of
matched
positions by the total number of positions in the window of comparison, and
multiplying the result by 100 to yield the percentage of sequence identity.
(e)(i) The term "substantial identity" of polynucleotide sequences means that
a polynucleotide comprises a sequence that has at least 70% sequence identity,
preferably at least 80%, more preferably at least 90%, and most preferably at
least
95%, compared to a reference sequence using one of the alignment programs
described using standard parameters. One of skill in the art will recognize
that these
values can be appropriately adjusted to determine corresponding identity of
proteins
encoded by two nucleotide sequences by taking into account codon degeneracy,
amino acid similarity, reading frame positioning, and the like. Substantial
identity of
amino acid sequences for these purposes normally means sequence identity of at
least
60%, more preferably at least 70%, 80%, 90%, and most preferably at least 95%.
Another indication that nucleotide sequences are substantially identical is if
two molecules hybridize to each other under stringent conditions. Generally,
stringent conditions are selected to be about 5°C lower than the
thermal melting point
(Tm) for the specific sequence at a defined ionic strength and pH. However,
stringent
conditions encompass temperatures in the range of about 1 °C to about
20°C,
depending upon the desired degree of stringency as otherwise qualified herein.
Nucleic acids that do not hybridize to each other under stringent conditions
are still
substantially identical if the polypeptides they encode are substantially
identical. This
-26-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
may occur, e.g., when a copy of a nucleic acid is created using the maximum
codon
degeneracy permitted by the genetic code. One indication that two nucleic acid
sequences are substantially identical is when the polypeptide encoded by the
first
nucleic acid is immunologically cross reactive with the polypeptide encoded by
the
second nucleic acid.
(e)(ii) The term "substantial identity" in the context of a peptide indicates
that
a peptide comprises a sequence with at least 70% sequence identity to a
reference
sequence, preferably 80%, more preferably 85%, most preferably at least 90% or
95%
sequence identity to the reference sequence over a specified comparison
window.
Preferably, optimal alignment is conducted using the homology alignment
algorithm
of Needleman and Wunsch (1970) J. Mol. Biol. 48:443-453. An indication that
two
peptide sequences are substantially identical is that one peptide is
immunologically
reactive with antibodies raised against the second peptide. Thus, a peptide is
substantially identical to a second peptide, for example, where the two
peptides differ
only by a conservative substitution. Peptides that are "substantially similar"
share
sequences as noted above except that residue positions that are not identical
may
differ by conservative amino acid changes.
Disease and Pests
Compositions and methods for controlling pathogenic agents are provided.
The anti-pathogenic compositions comprise sunflower nucleotide and polypeptide
sequences. Particularly, the sunflower nucleic acid and amino acid sequences
are
selected from rhoGAP-1, LOX, ADH, and SCIP-1. Accordingly, the compositions
and methods are useful in protecting plants against fungal pathogens, viruses,
nematodes, insects and the like.
By "disease resistance" or "pathogen resistance" is intended that the plants
avoid the disease symptoms which are the outcome of plant-pathogen
interactions.
That is, pathogens are prevented from causing plant diseases and the
associated
disease symptoms, or alternatively, the disease symptoms caused by the
pathogen is
minimized or lessened. The methods of the invention can be utilized to protect
plants
from disease, particularly those diseases that are caused by plant pathogens.
By "antipathogenic compositions" is intended that the compositions of the
invention have antipathogenic activity and thus are capable of suppressing,
-27-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
controlling, and/or killing the invading pathogenic organism. An
antipathogenic
composition of the invention will reduce the disease symptoms resulting from
pathogen challenge by at least about S% to about SO%, at least about 10% to
about
60%, at least about 30% to about 70%, at least about 40% to about 80%, or at
least
about 50% to about 90% or greater. Hence, the methods of the invention can be
utilized to protect plants from disease, particularly those diseases that are
caused by
plant pathogens.
Assays that measure antipathogenic activity are commonly known in the art,
as are methods to quantitate disease resistance in plants following pathogen
infection.
See, for example, U.S. Patent No. 5,614,395, herein incorporated by reference.
Such
techniques include, measuring over time, the average lesion diameter, the
pathogen
biomass, and the overall percentage of decayed plant tissues. For example, a
plant
either expressing an antipathogenic polypeptide or having an antipathogenic
composition applied to its surface shows a decrease in tissue necrosis (i.e.,
lesion
diameter) or a decrease in plant death following pathogen challenge when
compared
to a control plant that was not exposed to the antipathogenic composition.
Alternatively, antipathogenic activity can be measured by a decrease in
pathogen
biomass. For example, a plant expressing an antipathogenic polypeptide or
exposed
to an antipathogenic composition is challenged with a pathogen of interest.
Over
time, tissue samples from the pathogen-inoculated tissues are obtained and RNA
is
extracted. The percent of a specific pathogen RNA transcript relative to the
level of a
plant specific transcript allows the level of pathogen biomass to be
determined. See,
for example, Thomma et al. (1998) Plant Biology 95:15107-15111, herein
incorporated by reference.
Furthermore, in vitro antipathogenic assays include, for example, the addition
of varying concentrations of the antipathogenic composition to paper disks and
placing the disks on agar containing a suspension of the pathogen of interest.
Following incubation, clear inhibition zones develop around the discs that
contain an
effective concentration of the antipathogenic polypeptide (Liu et al. (1994)
Plant
Biology 91:1888-1892, herein incorporated by reference). Additionally,
microspectrophotometrical analysis can be used to measure the in vitro
antipathogenic
properties of a composition (Hu et al. (1997) Plant Mol. Biol. 34:949-959 and
-28-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Cammue et al. (1992) J. Biol. Chem. 267: 2228-2233, both of which are herein
incorporated by reference).
Methods for increasing pathogen resistance in a plant are provided. The
methods involve stably transforming a plant with a DNA construct comprising an
anti-pathogenic nucleotide sequence of the invention operably linked to
promoter that
drives expression in a plant. Such methods may find use in agriculture
particularly in
limiting the impact of plant pathogens on crop plants. The anti-pathogenic
nucleotide
sequences comprise the sunflower rhoGAP, LOX, ADH, or SCIP-1 nucleic acid
molecules. While the choice of promoter will depend on the desired timing and
location of expression of the anti-pathogenic nucleotide sequences, preferred
promoters include constitutive and pathogen-inducible promoters.
Additionally, the compositions can be used in formulation use for their
disease
resistance activities. The proteins of the invention can be formulated with an
acceptable carrier into a pesticidal compositions) that is for example, a
suspension, a
solution, an emulsion, a dusting powder, a dispersible granule, a wettable
powder, and
an emulsifiable concentrate, an aerosol, an impregnated granule, an adjuvant,
a
coatable paste, and also encapsulations in, for example, polymer substances.
Additionally, transformed plants, plant cells, plant tissues and seeds thereof
are provided.
It is understood in the art that plant DNA viruses and fungal pathogens
remodel the control of the host replication and gene expression machinery to
accomplish their own replication and effective infection. The present
invention may
be useful in preventing such corruption of the cell.
As discussed above, the sequences encoding the sunflower rhoGAP, LOX,
ADH, and SCIP-1 are involved in many basic biochemical pathways and cellular
functions that influence the plant defense response. Hence, the sequences of
the
invention may find use in disrupting cellular function of plant pathogens or
insect
pests as well as altering the defense mechanisms of a host plant to enhance
resistance
to disease or insect pests. While the invention is not bound by any particular
mechanism of action, the gene products, probably proteins or polypeptides,
function
to inhibit or prevent plant diseases in a plant. Such gene products may be
anti-
pathogenic. It is recognized that the present invention is not dependent upon
a
particular mechanism of defense. Rather, the genes and methods of the
invention
-29-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
work to increase resistance of the plant to pathogens independent of how that
resistance is increased or achieved.
The methods of the invention can be used with other methods available in the
art for enhancing disease resistance in plants. Similarly, the plant defense
mechanisms described herein may be used alone or in combination with other
proteins
or agents to protect against plant diseases and pathogens. Although any one of
a
variety of second nucleotide sequences may be utilized, specific embodiments
of the
invention encompass those second nucleotide sequences that, when expressed in
a
plant, help to increase the resistance of a plant to pathogens. It is
recognized that such
second nucleotide sequences may be used in either the sense or antisense
orientation
depending on the desired outcome. Other plant defense proteins include those
described in WO 99/43823 and WO 99/43821, all of which are herein incorporated
by
reference.
Additionally, the LOX and SCIP-1 promoter nucleotide sequences disclosed
herein are also useful for genetic engineering of plants to express a
phenotype of
interest. The promoter sequences may be used to regulate expression of any
heterologous nucleotide sequence. Alternatively, the LOX or SCIP-1 promoter
sequence may be used to drive expression of its native, i.e., naturally
occurring, LOX
or SCIP-1 gene sequence disclosed herein. In a specific embodiment, the LOX or
SCIP-1 promoter sequences are operably linked to an anti-pathogenic nucleotide
sequence and drive expression of said sequence in a plant cell. The LOX or
SCIP-1
promoter sequences may therefore be used in creating or enhancing pathogen or
disease resistance in a transformed plant.
Pathogens of the invention include, but are not limited to, viruses or
viroids,
bacteria, insects, nematodes, fungi, and the like. Viruses include any plant
virus, for
example, tobacco or cucumber mosaic virus, ringspot virus, necrosis virus,
maize
dwarf mosaic virus, etc. Specific fungal and viral pathogens for the major
crops
include: Soybeans: Phytophthora megasperma ~sp. glycinea, Macrophomina
phaseolina, Rhizoctonia solani, Sclerotinia sclerotiorum, Fusarium oxysporum,
Diaporthe phaseolorum var. sojae (Phomopsis sojae), Diaporthe phaseolorum var.
caulivora, Sclerotium rolfsii, Cercospora kikuchii, Cercospora sojina,
Peronospora
manshurica, Colletotrichum dematium (Colletotichum truncatum), Corynespora
cassiicola, Septoria glycines, Phyllosticta sojicola, Alternaria alternata,
-30-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Pseudomonas syringae p.v. glycinea, Xanthomonas campestris p.v. phaseoli,
Microsphaera diffuse, Fusarium semitectum, Phialophora gregata, Soybean mosaic
virus, Glomerella glycines, Tobacco Ring spot virus, Tobacco Streak virus,
Phakopsora pachyrhizi, Pythium aphanidermatum, Pythium ultimum, Pythium
debaryanum, Tomato spotted wilt virus, Heterodera glycines Fusarium solani;
Canola: Albugo candida, Alternaria brassicae, Leptosphaeria maculans,
Rhizoctonia
solani, Sclerotinia sclerotiorum, Mycosphaerella brassiccola, Pythium ultimum,
Peronospora parasitica, Fusarium roseum, Alternaria alternate; Alfalfa:
Clavibater
michiganese subsp. insidiosum, Pythium ultimum, Pythium irregulare, Pythium
splendens, Pythium debaryanum, Pythium aphanidermatum, Phytophthora
megasperma, Peronospora trifoliorum, Phoma medicaginis var. medicaginis,
Cercospora medicaginis, Pseudopeziza medicaginis, Leptotrochila medicaginis,
Fusarium, Xanthomonas campestris p.v. alfalfae, Aphanomyces euteiches,
Stemphylium herbarum, Stemphylium alfalfae; Wheat: Pseudomonas syringae p.v.
atrofaciens, Urocystis agropyri, Xanthomonas campestris p.v. translucens,
Pseudomonas syringae p.v. syringae, Alternaria alternate, Cladosporium
herbarum,
Fusarium graminearum, Fusarium avenaceum, Fusarium culmorum, Ustilago tritici,
Ascochyta tritici, Cephalosporium gramineum, Collotetrichum graminicola,
Erysiphe
graminis f.sp. tritici, Puccinia graminis f.sp. tritici, Puccinia recondite
~sp. tritici,
Puccinia striiformis, Pyrenophora tritici-repentis, Septoria nodorum, Septoria
tritici,
Septoria avenge, Pseudocercosporella herpotrichoides, Rhizoctonia solani,
Rhizoctonia cerealis, Gaeumannomyces graminis var. tritici, Pythium
aphanidermatum, Pythium arrhenomanes, Pythium ultimum, Bipolaris sorokiniana,
Barley Yellow Dwarf Virus, Brome Mosaic Virus, Soil Borne Wheat Mosaic Virus,
Wheat Streak Mosaic Virus, Wheat Spindle Streak Virus, American Wheat Striate
Virus, Claviceps purpurea, Tilletia tritici, Tilletia laevis, Ustilago
tritici, Tilletia
indica, Rhizoctonia solani, Pythium arrhenomannes, Pythium gramicola, Pythium
aphanidermatum, High Plains Virus, European wheat striate virus; Sunflower:
Orobanche, Plasmophora halstedii, Sclerotinia sclerotiorum, Aster Yellows,
Septoria
helianthi, Phomopsis helianthi, Alternaria helianthi, Alternaria zinniae,
Botrytis
cinerea, Phoma macdonaldii, Macrophomina phaseolina, Erysiphe cichoracearum,
Rhizopus oryzae, Rhizopus arrhizus, Rhizopus stolonifer, Puccinia helianthi,
Verticillium dahliae, Erwinia carotovorum pv. carotovora, Cephalosporium
-31-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
acremonium, Phytophthora cryptogea, Albugo tragopogonis; Corn: Fusarium
moniliforme var. subglutinans, Erwinia stewartii, Fusarium moniliforme,
Gibberella
zeae (Fusarium graminearum), Stenocarpella maydi (Diplodia maydis), Pythium
irregulare, Pythium debaryanum, Pythium graminicola, Pythium splendens,
Pythium
ultimum, Pythium aphanidermatum, Aspergillus flavus, Bipolaris maydis O, T
(Cochliobolus heterostrophus), Helminthosporium carbonum I, II & III
(Cochliobolus
carbonum), Exserohilum turcicum I, II & III, Helminthosporium pedicellatum,
Physoderma maydis, Phyllosticta maydis, Kabatiella-maydis, Cercospora sorghi,
Ustilago maydis, Puccinia sorghi, Puccinia polysora, Macrophomina phaseolina,
Penicillium oxalicum, Nigrospora oryzae, Cladosporium herbarum, Curvularia
lunata, Curvularia inaequalis, Curvularia pallescens, Clavibacter michiganense
subsp. nebraskense, Trichoderma viride, Maize Dwarf Mosaic Virus A & B, Wheat
Streak Mosaic Virus, Maize Chlorotic Dwarf Virus, Claviceps sorghi,
Pseudonomas
avenae, Erwinia chrysanthemi pv. zea, Erwinia carotovora, Corn stunt
spiroplasma,
Diplodia macrospora, Sclerophthora macrospora, Peronosclerospora sorghi,
Peronosclerospora philippinensis, Peronosclerospora maydis, Peronosclerospora
sacchari, Sphacelotheca reiliana, Physopella zeae, Cephalosporium maydis,
Cephalosporium acremonium, Maize Chlorotic Mottle Virus, High Plains Virus,
Maize Mosaic Virus, Maize Rayado Fino Virus, Maize Streak Virus, Maize Stripe
Virus, Maize Rough Dwarf Virus; Sor hum: Exserohilum turcicum, Colletotrichum
graminicola (Glomerella graminicola), Cercospora sorghi, Gloeocercospora
sorghi,
Ascochyta sorghina, Pseudomonas syringae p.v. syringae, Xanthomonas campestris
p.v. holcicola, Pseudomonas andropogonis, Puccinia purpurea, Macrophomina
phaseolina, Perconia circinata, Fusarium moniliforme, Alternaria alternata,
Bipolaris sorghicola, Helminthosporium sorghicola, Curvularia lunata, Phoma
insidiosa, Pseudomonas avenae (Pseudomonas alboprecipitans), Ramulispora
sorghi,
Ramulispora sorghicola, Phyllachara sacchari, Sporisorium reilianum
(Sphacelotheca reiliana), Sphacelotheca cruenta, Sporisorium sorghi, Sugarcane
mosaic H, Maize Dwarf Mosaic Virus A & B, Claviceps sorghi, Rhizoctonia
solani,
Acremonium strictum, Sclerophthona macrospora, Peronosclerospora sorghi,
Peronosclerospora philippinensis, Sclerospora graminicola, Fusarium
graminearum,
Fusarium oxysporum, Pythium arrhenomanes, Pythium graminicola, etc.
-32-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Nematodes include parasitic nematodes such as root-knot, cyst, lesion, and
renniform nematodes, etc.
Insect pests include insects selected from the orders Coleoptera, Diptera,
Hymenoptera, Lepidoptera, Mallophaga, Homoptera, Hemiptera, Orthoptera,
Thysanoptera, Dermaptera, Isoptera, Anoplura, Siphonaptera, Trichoptera, etc.,
particularly Coleoptera and Lepidoptera. Insect pests of the invention for the
major
crops include: Maize: Ostrinia nubilalis, European corn borer; Agrotis
ipsilon, black
cutworm; Helicoverpa zea, corn earworm; Spodoptera frugiperda, fall armyworm;
Diatraea grandiosella, southwestern corn borer; Elasmopalpus lignosellus,
lesser
cornstalk borer; Diatraea saccharalis, surgarcane borer; Diabrotica virgifera,
western
corn rootworm; Diabrotica longicornis barberi, northern corn rootworm;
Diabrotica
undecimpunctata howardi, southern corn rootworm; Melanotus spp., wireworms;
Cyclocephala borealis, northern masked chafer (white grub); Cyclocephala
immaculata, southern masked chafer (white grub); Popillia japonica, Japanese
beetle;
Chaetocnema pulicaria, corn flea beetle; Sphenophorus maidis, maize billbug;
Rhopalosiphum maidis, corn leaf aphid; Anuraphis maidiradicis, corn root
aphid;
Blissus leucopterus leucopterus, chinch bug; Melanoplus femurrubrum, redlegged
grasshopper; Melanoplus sanguinipes, migratory grasshopper; Hylemya platura,
seedcorn maggot; Agromyza parvicorni.s, corn blot leafminer; Anaphothrips
obscrurus, grass thrips; Solenopsis milesta, thief ant; Tetranychus urticae,
twospotted
spider mite; Sor hum: Chilo partellus, sorghum borer; Spodoptera frugiperda,
fall
armyworm; Helicoverpa zea, corn earworm; Elasmopalpus lignosellus, lesser
cornstalk borer; Feltia subterranea, granulate cutworm; Phyllophaga crinita,
white
grub; Eleodes, Conoderus, and Aeolus spp., wireworms; Oulema melav~opus,
cereal
leaf beetle; Chaetocnema pulicaria, corn flea beetle; Sphenophorus maidis,
maize
billbug; Rhopalosiphum maidis; corn leaf aphid; Sipha flava, yellow sugarcane
aphid;
Blissus leucopterus leucopterus, chinch bug; Contarinia sorghicola, sorghum
midge;
Tetranychus cinnabarinus, carmine spider mite; Tetranychus urticae, twospotted
spider mite; Wheat: Pseudaletia unipunctata, army worm; Spodoptera frugiperda,
fall armyworm; Elasmopalpus lignosellus, lesser cornstalk borer; Agrotis
orthogonia,
western cutworm; Elasmopalpus lignosellus, lesser cornstalk borer; Oulema
melanopus, cereal leaf beetle; Hypera punctata, clover leaf weevil; Diabrotica
undecimpunctata howardi, southern corn rootworm; Russian wheat aphid;
Schizaphis
-33-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
graminum, greenbug; Macrosiphum avenae, English grain aphid; Melanoplus
femurrubrum, redlegged grasshopper; Melanoplus differentialis, differential
grasshopper; Melanoplus sanguinipes, migratory grasshopper; Mayetiola
destructor,
Hessian fly; Sitodiplosis mosellana, wheat midge; Meromyza americana, wheat
stem
maggot; Hylemya coarctata, wheat bulb fly; Frankliniella fusca, tobacco
thrips;
Cephus cinctus, wheat stem sawfly; Aceria tulipae, wheat curl mite; Sunflower:
Suleima helianthana, sunflower bud moth; Homoeosoma electellum, sunflower
moth;
zygogramma exclamationis, sunflower beetle; Bothyrus gibbosus, carrot beetle;
Neolasioptera murtfeldtiana, sunflower seed midge; Cotton: Heliothis
virescens,
cotton budworm; Helicoverpa zea, cotton bollworm; Spodoptera exigua, beet
armyworm; Pectinophora gossypiella, pink bollworm; Anthonomus grandis grandis,
boll weevil; Aphis gossypii, cotton aphid; Pseudatomoscelis seriatus, cotton
fleahopper; Trialeurodes abutilonea, bandedwinged whitefly; Lygus lineolaris,
tarnished plant bug; Melanoplus femurrubrum, redlegged grasshopper; Melanoplus
differentialis, differential grasshopper; Thrips tabaci, onion thrips;
Franklinkiella
fusca, tobacco thrips; Tetranychus cinnabarinus, carmine spider mite;
Tetranychus
urticae, twospotted spider mite; Rice: Diatraea saccharalis, sugarcane borer;
Spodoptera frugiperda, fall armyworm; Helicoverpa zea, corn earworm; Colaspis
brunnea, grape colaspis; Lissorhoptrus oryzophilus, rice water weevil;
Sitophilus
oryzae, rice weevil; Nephotettix nigropictus, rice leafliopper; Blissus
leucopterus
leucopterus, chinch bug; Acrosternum hilare, green stink bug; Soybean:
Pseudoplusia includens, soybean looper; Anticarsia gemmatalis, velvetbean
caterpillar; Plathypena scabra, green cloverworm; Ostrinia nubilalis, European
corn
borer; Agrotis ipsilon, black cutworm; Spodoptera exigua, beet armyworm;
Heliothis
virescens, cotton budworm; Helicoverpa zea, cotton bollworm; Epilachna
varivestis,
Mexican bean beetle; Myzus persicae, green peach aphid; Empoasca fabae, potato
leafhopper; Acrosternum hilare, green stink bug; Melanoplus femurrubrum,
redlegged
grasshopper; Melanoplus differentialis, differential grasshopper; Hylemya
platura,
seedcorn maggot; Sericothrips variabilis, soybean thrips; Thrips tabaci, onion
thrips;
Tetranychus turkestani, strawberry spider mite; Tetranychus urticae,
twospotted
spider mite; Barley: Ostrinia nubilalis, European corn borer; Agrotis ipsilon,
black
cutworm; Schizaphis graminum, greenbug; Blissus leucopterus leucopterus,
chinch
bug; Acrosternum hilare, green stink bug; Euschistus servus, brown stink bug;
Delia
-34-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
platura, seedcorn maggot; Mayetiola destructor, Hessian fly; Petrobia latens,
brown
wheat mite; Oil Seed Rape: Brevicoryne brassicae, cabbage aphid; Phyllotreta
cruciferae, Flea beetle; Mamestra configurata, Bertha armyworm; Plutella
xylostella,
Diamond-back moth; Delia ssp., Root maggots.
Expression of Sequences
The nucleic acid sequences of the present invention can be expressed in a host
cell such as bacteria, yeast, insect, mammalian, or preferably plant cells. It
is
expected that those of skill in the art are knowledgeable in the numerous
expression
systems available for expression of a nucleic acid encoding a protein of the
present
invention. No attempt to describe in detail the various methods known for the
expression of proteins in prokaryotes or eukaryotes will be made.
As used herein, "heterologous" in reference to a nucleic acid is a nucleic
acid
that originates from a foreign species, or, if from the same species, is
substantially
modified from its native form in composition and/or genomic locus by
deliberate
human intervention. For example, a promoter operably linked to a heterologous
structural gene is from a species different from that from which the
structural gene
was derived, or, if from the same species, one or both are substantially
modified from
their original form. A heterologous protein may originate from a foreign
species, or,
if from the same species, is substantially modified from its original form by
deliberate
human intervention.
By "host cell" is meant a cell, which comprises a heterologous nucleic acid
sequence of the invention. Host cells may be prokaryotic cells such as E coli,
or
eukaryotic cells such as yeast, insect, amphibian, or mammalian cells.
Preferably,
host cells are monocotyledonous or dicotyledonous plant cells. A particularly
preferred monocotyledonous host cell is a maize host cell.
The disease resistant sequences of the invention are provided in expression
cassettes or DNA constructs for expression in the plant of interest. The
cassette will
include 5' and 3' regulatory sequences operably linked to a rhoGAP, LOX, ADH,
or
SCIP-1 sequence of the invention. By "operably linked" is intended a
functional
linkage between a promoter and a second sequence, wherein the promoter
sequence
initiates and mediates transcription of the DNA sequence corresponding to the
second
sequence. Generally, operably linked means that the nucleic acid sequences
being
-3 5-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
linked are contiguous and, where necessary to join two protein coding regions,
contiguous and in the same reading frame. The cassette may additionally
contain at
least one additional gene to be cotransformed into the organism.
Alternatively, the
additional genes) can be provided on multiple expression cassettes.
Such an expression cassette is provided with a plurality of restriction sites
for
insertion of the disease resistant sequence to be under the transcriptional
regulation of
the regulatory regions. The expression cassette may additionally contain
selectable
marker genes.
The expression cassette will include in the 5'-3' direction of transcription,
a
transcriptional and translational initiation region, a disease resistant DNA
sequence of
the invention, and a transcriptional and translational termination region
functional in
plants. The transcriptional initiation region, the promoter, may be native or
analogous
or foreign or heterologous to the plant host. Additionally, the promoter may
be the
natural sequence or alternatively a synthetic sequence. By "foreign" is
intended that
the transcriptional initiation region is not found in the native plant into
which the
transcriptional initiation region is introduced. As used herein, a chimeric
gene
comprises a coding sequence operably linked to a transcription initiation
region that is
heterologous to the coding sequence.
While it may be preferable to express the sequences using heterologous
promoters, the native promoter sequences may be used. Such constructs would
change expression levels of the disease resistant RNA/protein in the plant or
plant
cell. Thus, the phenotype of the plant or plant cell is altered.
The termination region may be native with the transcriptional initiation
region,
may be native with the operably linked DNA sequence of interest, or may be
derived
from another source. Convenient termination regions are available from the Ti-
plasmid of A. tumefaciens, such as the octopine synthase and nopaline synthase
termination regions. See also Guerineau et al. (1991) Mol. Gen. Genet. 262:141-
144;
Proudfoot ( 1991 ) Cell 64:671-674; Sanfacon et al. ( 1991 ) Genes Dev. 5:141-
149;
Mogen et al. (1990) Plant Cell 2:1261-1272; Munroe et al. (1990) Gene 91:151-
158;
Ballas et al. (1989) Nucleic Acids Res. 17:7891-7903; and Joshi et al. (1987)
Nucleic
Acid Res. 15:9627-9639.
Where appropriate, the genes) may be optimized for increased expression in
the transformed plant. That is, the genes can be synthesized using plant-
preferred
-36-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
codons for improved expression. Methods are available in the art for
synthesizing
plant-preferred genes. See, for example, U.S. Patent Nos. 5,380,831, and
5,436,391,
and Murray et al. (1989) Nucleic Acids Res. 17:477-498, herein incorporated by
reference.
Additional sequence modifications are known to enhance gene expression in a
cellular host. These include elimination of sequences encoding spurious
polyadenylation signals, exon-intron splice site signals, transposon-like
repeats, and
other such well-characterized sequences that may be deleterious to gene
expression.
The G-C content of the sequence may be adjusted to levels average for a given
cellular host, as calculated by reference to known genes expressed in the host
cell.
When possible, the sequence is modified to avoid predicted hairpin secondary
mRNA
structures.
The expression cassettes may additionally contain 5' leader sequences in the
expression cassette construct. Such leader sequences can act to enhance
translation.
Translation leaders are known in the art and include: picornavirus leaders,
for
example, EMCV leader (Encephalomyocarditis 5' noncoding region) (Elroy-Stein
et
al. (1989) PNAS USA 86:6126-6130); potyvirus leaders, for example, TEV leader
(Tobacco Etch Virus) (Allison et al. (1986); MDMV leader (Maize Dwarf Mosaic
Virus); hirology 154:9-20), and human immunoglobulin heavy-chain binding
protein
(BiP), (Macejak et al. (1991) Nature 353:90-94); untranslated leader from the
coat
protein mRNA of alfalfa mosaic virus (AMV RNA 4) (Jobling et al. (1987) Nature
325:622-625); tobacco mosaic virus leader (TMV) (Gallie et al. (1989) in
Molecular
Biology of RNA, ed. Cech (Liss, New York), pp. 237-256); and maize chlorotic
mottle
virus leader (MCMV) (Lommel et al. (1991) Virology 81:382-385). See also,
Della-Cioppa et al. (1987) Plant Physiol. 84:965-968. Other methods known to
enhance translation can also be utilized, for example, introns, and the like.
In preparing the expression cassette, the various DNA fragments may be
manipulated, so as to provide for the DNA sequences in the proper orientation
and, as
appropriate, in the proper reading frame. Toward this end, adapters or linkers
may be
employed to join the DNA fragments or other manipulations may be involved to
provide for convenient restriction sites, removal of superfluous DNA, removal
of
restriction sites, or the like. For this purpose, in vitro mutagenesis, primer
repair,
-3 7-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
restriction, annealing, resubstitutions, e.g., transitions and transversions,
may be
involved.
Generally, the expression cassette will comprise a selectable marker gene for
the
selection of transformed cells. Selectable marker genes are utilized for the
selection of
transformed cells or tissues. Marker genes include genes encoding antibiotic
resistance,
such as those encoding neomycin phosphotransferase II (NEO) and hygromycin
phosphotransferase (HPT), as well as genes conferring resistance to herbicidal
compounds, such as glufosinate ammonium, bromoxynil, imidazolinones, and 2,4-
dichlorophenoxyacetate (2,4-D). See generally, Yarranton (1992) Curr. Opin.
Biotech.
3:506-511; Christopherson et al. ( 1992) Proc. Natl. Acad Sci. USA 89:6314-
6318; Yao
et al. (1992) Cell 71:63-72; Reznikoff (1992) Mol. Microbiol. 6:2419-2422;
Barkley et
al. (1980) in The Operon, pp. 177-220; Hu et al. (1987) Cell 48:555-566; Brown
et al.
(1987) Cell 49:603-612; Figge et al. (1988) Cell 52:713-722; Deuschle et al.
(1989)
Proc. Natl. Acad. Aci. USA 86:5400-5404; Fuerst et al. (1989) Proc. Natl. Acad
Sci.
USA 86:2549-2553; Deuschle et al. (1990) Science 248:480-483; Gossen (1993)
Ph.D.
Thesis, University of Heidelberg; Refines et al. (1993) Proc. Natl. Acad. Sci.
USA
90:1917-1921; Labow et al. (1990) Mol. Cell. Biol. 10:3343-3356; Zambretti et
al.
(1992) Proc. Natl. Acad Sci. USA 89:3952-3956; Baim et al. (1991) Proc. Natl.
Acad.
Sci. USA 88:5072-5076; Wyborski et al. (1991) Nucleic Acids Res. 19:4647-4653;
Hillenand-Wissman ( 1989) Topics Mol. Struc. Biol. 10:143-162; Degenkolb et
al. ( 1991 )
Antimicrob. Agents Chemother. 35:1591-1595; Kleinschnidt et al. (1988)
Biochemistry
27:1094-1104; Bonin (1993) Ph.D. Thesis, University of Heidelberg; Gossen et
al.
(1992) Proc. Natl. Acad Sci. USA 89:5547-5551; Oliva et al. (1992) Antimicrob.
Agents
Chemother. 36:913-919; Hlavka et al. (1985) Handbook of Experimental
Pharmacology,
Vol. 78 ( Springer-Verlag, Berlin); Gill et al. (1988) Nature 334:721-724.
Such
disclosures are herein incorporated by reference.
The above list of selectable marker genes is not meant to be limiting. Any
selectable marker gene can be used in the present invention.
A number of promoters can be used in the practice of the invention. The
promoters can be selected based on the desired outcome. That is, the nucleic
acids
can be combined with constitutive, tissue-preferred, or other promoters for
expression
in plants. Such constitutive promoters include, for example, the core promoter
of the
Rsyn7 (PCT Application Serial No. U599/03863); Scpl promoter (U.5. Patent
-3 8-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Application Serial No. 09/028,819), rice actin (McElroy et al. (1990) Plant
Cell
2:163-171); ubiquitin (Christensen et al. (1989) Plant Mol. Biol. 12:619-632
and
Christensen et al. (1992) Plant Mol. Biol. 18:675-689); pEMU (Last et al.
(1991)
Theor. Appl. Genet. 81:581-588); MAS (Velten et al. (1984) EMBO J. 3:2723-
2730);
ALS promoter (U.S. Application Serial No. 08/409,297), and the like. Other
constitutive promoters include, for example, U.S. Patent Nos. 5,608,149;
5,608,144;
5,604,121; 5,569,597; 5,466,785; 5,399,680; 5,268,463; and 5,608,142.
Generally, it will be beneficial to express the gene from an inducible
promoter,
particularly from a pathogen-inducible promoter. Such promoters include those
from
pathogenesis-related proteins (PR proteins), which are induced following
infection by
a pathogen; e.g., PR proteins, SAR proteins, beta-1,3-glucanase, chitinase,
etc. See,
for example, Redolfi et al. (1983) Neth. J. Plant Pathol. 89:245-254; Uknes et
al.
(1992) Plant Cell 4:645-656; and Van Loon (1985) Plant Mol. Irirol. 4:111-116.
See
also the copending application entitled "Inducible Maize Promoters", U.S.
Patent
Application Serial No. 09/257,583, filed February 25, 1999.
Of interest are promoters that are expressed locally at or near the site of
pathogen infection. See, for example, Marineau et al. (1987) Plant Mol. Biol.
9:335-
342; Matton et al. (1989) Molecular Plant-Microbe Interactions 2:325-331;
Somsisch
et al. (1986) Proc. Natl. Acad. Sci. USA 83:2427-2430; Somsisch et al. (1988)
Mol.
Gen. Genet. 2:93-98; and Yang (1996) Proc. Natl. Acad. Sci. USA 93:14972-
14977.
See also, Chen et al. (1996) Plant J. 10:955-966; Zhang et al. (1994) Proc.
Natl.
Acad. Sci. USA 91:2507-2511; Warner et al. (1993) Plant J. 3:191-201; Siebertz
et al.
(1989) Plant Cell 1:961-968; U.S. Patent No. 5,750,386 (nematode-inducible);
and
the references cited therein. Of particular interest is the inducible promoter
for the
maize PRms gene, whose expression is induced by the pathogen Fusarium
moniliforme (see, for example, Cordero et al. (1992) Physiol. Mol. Plant Path.
41:189-200).
Additionally, as pathogens find entry into plants through wounds or insect
damage, a wound-inducible promoter may be used in the constructions of the
invention. Such wound-inducible promoters include potato proteinase inhibitor
(pin
II) gene (Ryan (1990) Ann. Rev. Phytopath. 28:425-449; Duan et al. (1996)
Nature
Biotechnology 14:494-498); wunl and wun2, US Patent No. 5,428,148; winl and
win2 (Stanford et al. (1989) Mol. Gen. Genet. 215:200-208); systemin (McGurl
et al.
-3 9-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
(1992) Science 225:1570-1573); WIP1 (Rohmeier et al. (1993) Plant Mol. Biol
22:783-792; Eckelkamp et al. (1993) FEBS Letters 323:73-76); MPI gene
(Corderok
et al. (1994) Plant J. 6(2):141-150); and the like, herein incorporated by
reference.
Chemical-regulated promoters can be used to modulate the expression of a
gene in a plant through the application of an exogenous chemical regulator.
Depending upon the objective, the promoter may be a chemical-inducible
promoter,
where application of the chemical induces gene expression, or a chemical-
repressible
promoter, where application of the chemical represses gene expression.
Chemical-
inducible promoters are known in the art and include, but are not limited to,
the maize
In2-2 promoter, which is activated by benzenesulfonamide herbicide safeners,
the
maize GST promoter, which is activated by hydrophobic electrophilic compounds
that
are used as pre-emergent herbicides, and the tobacco PR-la promoter, which is
activated by salicylic acid. Other chemical-regulated promoters of interest
include
steroid-responsive promoters (see, for example, the glucocorticoid-inducible
promoter
in Schena et al. (1991) Proc. Natl. Acad. Sci. USA 88:10421-10425 and McNellis
et
al. (1998) Plant J. 14(2):247-257) and tetracycline-inducible and tetracycline-
repressible promoters (see, for example, Gatz et al. (1991) Mol. Gen. Genet.
227:229-
237, and U.S. Patent Nos. 5,814,618 and 5,789,156), herein incorporated by
reference.
The method of transformation/transfection is not critical to the instant
invention; various methods of transformation or transfection are currently
available.
Thus, any method, which provides for effective transformation/transfection may
be
employed. Transformation protocols as well as protocols for introducing
nucleotide
sequences into plants may vary depending on the type of plant or plant cell,
i.e.,
monocot or dicot, targeted for transformation. Suitable methods of introducing
nucleotide sequences into plant cells and subsequent insertion into the plant
genome
include microinjection (Crossway et al. (1986) Biotechniques 4:320-334),
electroporation (Riggs et al. (1986) Proc. Natl. Acad. Sci. USA 83:5602-5606,
Agrobacterium-mediated transformation (Townsend et al., U.S. Patent No.
5,563,055;
Zhao et al., U.S. Patent No. 5,981,840), direct gene transfer (Paszkowski et
al. (1984)
EMBO J. 3:2717-2722), and ballistic particle acceleration (see, for example,
Sanford
et al., U.S. Patent No. 4,945,050; Tomes et al. (1995) "Direct DNA Transfer
into
Intact Plant Cells via Microprojectile Bombardment," in Plant Cell, Tissue,
and
-40-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Organ Culture: Fundamental Methods, ed. Gamborg and Phillips (Springer-Verlag,
Berlin); and McCabe et al. (1988) Biotechnology 6:923-926). Also see
Weissinger et
al. (1988) Ann. Rev. Genet. 22:421-477; Sanford et al. (1987) Particulate
Science and
Technology 5:27-37 (onion); Christou et al. (1988) Plant Physiol. 87:671-674
(soybean); McCabe et al. (1988) BiolTechnology 6:923-926 (soybean); Finer and
McMullen (1991) In Vitro Cell Dev. Biol. 27P:175-182 (soybean); Singh et al.
(1998)
Theor. Appl. Genet. 96:319-324 (soybean); Datta et al. (1990) Biotechnology
8:736-740 (rice); Klein et al. (1988) Proc. Natl. Acad. Sci. USA 85:4305-4309
(maize); Klein et al. (1988) Biotechnology 6:559-563 (maize); Tomes, U.S.
Patent
No. 5,240,855; Buising et al., U.S. Patent Nos. 5,322,783 and 5,324,646; Tomes
et al.
(1995) "Direct DNA Transfer into Intact Plant Cells via Microprojectile
Bombardment," in Plant Cell, Tissue, and Organ Culture: Fundamental Methods,
ed.
Gamborg (Springer-Verlag, Berlin) (maize); Klein et al. (1988) Plant Physiol.
91:440-444 (maize); Fromm et al. (1990) Biotechnology 8:833-839 (maize);
Hooykaas-Van Slogteren et al. (1984) Nature (London) 311:763-764; Bytebier et
al.
(1987) Proc. Natl. Acad Sci. USA 84:5345-5349 (Liliaceae); De Wet et al.
(1985) in
The Experimental Manipulation of Ovule Tissues, ed. Chapman et al. (Longman,
New
York), pp. 197-209 (pollen); Kaeppler et al. (1990) Plant Cell Reports 9:415-
418 and
Kaeppler et al. ( 1992) Theor. Appl. Genet. 84:560-566 (whisker-mediated
transformation); D'Halluin et al. (1992) Plant Cell 4:1495-1505
(electroporation); Li
et al. (1993) Plant Cell Reports 12:250-255 and Christou and Ford (1995)
Annals of
Botany 75:407-413 (rice); Osjoda et al. (1996) Nature Biotechnology 14:745-750
(maize via Agrobacterium tumefaciens); all of which are herein incorporated by
reference.
The cells that have been transformed may be grown into plants in accordance
with conventional ways. See, for example, McCormick et al. (1986) Plant Cell
Reports 5:81-84. These plants may then be grown, and either pollinated with
the
same transformed strain or different strains, and the resulting hybrid having
constitutive expression of the desired phenotypic characteristic identified.
Two or
more generations may be grown to ensure that constitutive expression of the
desired
phenotypic characteristic is stably maintained and inherited and then seeds
harvested
to ensure constitutive expression of the desired phenotypic characteristic has
been
achieved.
-41-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
The present invention may be used for transformation of any plant species,
including, but not limited to, monocots and dicots. Examples of plants of
interest
include, but are not limited to, corn (Zea mays), Brassica sp. (e.g., B.
napus, B. rapa,
B. juncea), particularly those Brassica species useful as sources of seed oil,
alfalfa
(Medicago sativa), rice (Oryza sativa), rye (Secale cereale), sorghum (Sorghum
bicolor, Sorghum vulgare), millet (e.g., pearl millet (Pennisetum glaucum),
proso
millet (Panicum miliaceum), foxtail millet (Setaria italica), finger millet
(Eleusine
coracana)), sunflower (Helianthus annuus), safflower (Carthamus tinctorius),
wheat
(Triticum aestivum), soybean (Glycine max), tobacco (Nicotiana tabacum),
potato
(Solanum tuberosum), peanuts (Arachis hypogaea), cotton (Gossypium barbadense,
Gossypium hirsutum), sweet potato (Ipomoea batatus), cassava (Manihot
esculenta),
coffee (Coffea spp.), coconut (Cocos nucifera), pineapple (Ananas comosus),
citrus
trees (Citrus spp.), cocoa (Theobroma cacao), tea (Camellia sinensis), banana
(Musa
spp.), avocado (Persea americana), fig (Ficus casica), guava (Psidium
guajava),
mango (Mangifera indica), olive (Olea europaea), papaya (Carica papaya),
cashew
(Anacardium occidentale), macadamia (Macadamia integrifolia), almond (Prunus
amygdalus), sugar beets (Beta vulgaris), sugarcane (Saccharum spp.), oats,
barley,
vegetables, ornamentals, and conifers.
Vegetables include tomatoes (Lycopersicon esculentum), lettuce (e.g., Lactuca
sativa), green beans (Phaseolus vulgaris), lima beans (Phaseolus limensis),
peas
(Lathyrus spp.), and members of the genus Cucumis such as cucumber (C.
sativus),
cantaloupe (C. cantalupensis), and musk melon (C. melo). Ornamentals include
azalea
(Rhododendron spp.), hydrangea (Macrophylla hydrangea), hibiscus (Hibiscus
rosasanensis), roses (Rosa spp.), tulips (Tulipa spp.), daffodils (Narcissus
spp.), petunias
(Petunia hybrida), carnation (Dianthus caryophyllus), poinsettia (Euphorbia
pulcherrima), and chrysanthemum. Conifers that may be employed in practicing
the
present invention include, for example, pines such as loblolly pine (Pinus
taeda), slash
pine (Pinus elliotii), ponderosa pine (Pinus ponderosa), lodgepole pine (Pinus
contorta),
and Monterey pine (Pinus radiata); Douglas-fir (Pseudotsuga menziesii);
Western
hemlock (Tsuga canadensis); Sitka spruce (Picea glauca); redwood (Sequoia
sempervirens); true firs such as silver fir (Abies amabilis) and balsam fir
(Abies
balsamea); and cedars such as Western red cedar (Thuja plicata) and Alaska
yellow-cedar (Chamaecyparis nootkatensis). Preferably, plants of the present
invention
-42-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
are crop plants (for example, corn, alfalfa, sunflower, Brassica, soybean,
cotton,
safflower, peanut, sorghum, wheat, millet, tobacco, etc. ), more preferably
corn and
soybean plants, yet more preferably corn plants.
Prokaryotic cells may be used as hosts for expression. Prokaryotes most
frequently are represented by various strains of E. coli; however, other
microbial
strains may also be used. Commonly used prokaryotic control sequences which
are
defined herein to include promoters for transcription initiation, optionally
with an
operator, along with ribosome binding sequences, include such commonly used
promoters as the beta lactamase (penicillinase) and lactose (lac) promoter
systems
(Chang et al. (1977) Nature 198:1056), the tryptophan (trp) promoter system
(Goeddel et al. (1980) Nucleic Acids Res. 8:4057) and the lambda derived P L
promoter and N-gene ribosome binding site (Shimatake et al. (1981) Nature
292:128).
The inclusion of selection markers in DNA vectors transfected in E coli. is
also
useful. Examples of such markers include genes specifying resistance to
ampicillin,
tetracycline, or chloramphenicol.
The vector is selected to allow introduction into the appropriate host cell.
Bacterial vectors are typically of plasmid or phage origin. Appropriate
bacterial cells
are infected with phage vector particles or transfected with naked phage
vector DNA.
If a plasmid vector is used, the bacterial cells are transfected with the
plasmid vector
DNA. Expression systems for expressing a protein of the present invention are
available using Bacillus sp. and Salmonella (Palva et al. (1983) Gene 22:229-
235);
Mosbach et al. (1983) Nature 302:543-545).
A variety of eukaryotic expression systems such as yeast, insect cell lines,
plant and mammalian cells, are known to those of skill in the art. As
explained briefly
below, a polynucleotide of the present invention can be expressed in these
eukaryotic
systems. In some embodiments, transformed/transfected plant cells, as
discussed
infra, are employed as expression systems for production of the proteins of
the instant
invention.
Synthesis of heterologous nucleotide sequences in yeast is well known
(Sherman et al. (1982) Methods in Yeast Genetics, Cold Spring Harbor
Laboratory).
Two widely utilized yeasts for production of eukaryotic proteins are
Saccharomyces
cerevisiae and Pichia pastoris. Vectors, strains, and protocols for expression
in
Saccharomyces and Pichia are known in the art and available from commercial
-43-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
suppliers (e.g., Invitrogen). Suitable vectors usually have expression control
sequences, such as promoters, including 3-phosphoglycerate kinase or alcohol
oxidase, and an origin of replication, termination sequences and the like as
desired.
A protein of the present invention, once expressed, can be isolated from yeast
by lysing the cells and applying standard protein isolation techniques to the
lists. The
monitoring of the purification process can be accomplished by using Western
blot
techniques or radioimmunoassay of other standard immunoassay techniques.
The sequences of the present invention can also be ligated to various
expression vectors for use in transfecting cell cultures of, for instance,
mammalian,
insect, or plant origin. Illustrative cell cultures useful for the production
of the
peptides are mammalian cells. A number of suitable host cell lines capable of
expressing intact proteins have been developed in the art, and include the
HEK293,
BHK21, and CHO cell lines. Expression vectors for these cells can include
expression control sequences, such as an origin of replication, a promoter
(e.g. the
CMV promoter, a HSV tk promoter or pgk (phosphoglycerate kinase) promoter), an
enhancer (Queen et al. (1986) Immunol. Rev. 89:49), and necessary processing
information sites, such as ribosome binding sites, RNA splice sites,
polyadenylation
sites (e.g., an SV40 large T Ag poly A addition site), and transcriptional
terminator
sequences. Other animal cells useful for production of proteins of the present
invention are available, for instance, from the American Type Culture
Collection.
Appropriate vectors for expressing proteins of the present invention in insect
cells are usually derived from the SF9 baculovirus. Suitable insect cell lines
include
mosquito larvae, silkworm, armyworm, moth and Drosophila cell lines such as a
Schneider cell line (See, Schneider (1987) J. Embryol. Exp. Morphol. 27:353-
365).
As with yeast, when higher animal or plant host cells are employed,
polyadenylation or transcription terminator sequences are typically
incorporated into
the vector. An example of a terminator sequence is the polyadenylation
sequence
from the bovine growth hormone gene. Sequences for accurate splicing of the
transcript may also be included. An example of a splicing sequence is the VP1
intron
from SV40 (Sprague et a1.(1983) J. Virol. 45:773-781). Additionally, gene
sequences
to control replication in the host cell may be incorporated into the vector
such as those
found in bovine papilloma virus type-vectors (Saveria-Campo (1985) DNA Cloning
-44-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Vol. II a Practical Approach, D.M. Glover, Ed., IRL Press, Arlington,
Virginia, pp.
213-238).
Animal and lower eukaryotic (e.g., yeast) host cells are competent or rendered
competent for transfection by various means. There are several well-known
methods
of introducing DNA into animal cells. These include: calcium phosphate
precipitation, fusion of the recipient cells with bacterial protoplasts
containing the
DNA, treatment of the recipient cells with liposomes containing the DNA, DEAE
dextrin, electroporation, biolistics, and micro-injection of the DNA directly
into the
cells. The transfected cells are cultured by means well known in the art
(Kuchler
(1997) Biochemical Methods in Cell Culture and Virology, Dowden, Hutchinson
and
Ross, Inc.).
It is recognized that with these nucleotide sequences, antisense
constructions,
complementary to at least a portion of the messenger RNA (mRNA) for the
disease
resistant sequences can be constructed. Antisense nucleotides are constructed
to
hybridize with the corresponding mRNA. Modifications of the antisense
sequences
may be made as long as the sequences hybridize to and interfere with
expression of
the corresponding mRNA. In this manner, antisense constructions having 70%,
preferably 80%, more preferably 85% sequence identity to the corresponding
antisensed sequences may be used. Furthermore, portions of the antisense
nucleotides
may be used to disrupt the expression of the target gene. Generally, sequences
of at
least 50 nucleotides, 100 nucleotides, 200 nucleotides, or greater may be
used.
The nucleotide sequences of the present invention may also be used in the
sense orientation to suppress the expression of endogenous genes in plants.
Methods
for suppressing gene expression in plants using nucleotide sequences in the
sense
orientation are known in the art. The methods generally involve transforming
plants
with a DNA construct comprising a promoter that drives expression in a plant
operably linked to at least a portion of a nucleotide sequence that
corresponds to the
transcript of the endogenous gene. Typically, such a nucleotide sequence has
substantial sequence identity to the sequence of the transcript of the
endogenous gene,
preferably greater than about 65% sequence identity, more preferably greater
than
about 85% sequence identity, most preferably greater than about 95% sequence
identity. See, U.S. Patent Nos. 5,283,184 and 5,034,323; herein incorporated
by
reference.
-45-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
The nucleotide sequences of the present invention may also be used in the
sense orientation to suppress the expression of endogenous genes in plants.
Methods
for suppressing gene expression in plants using nucleotide sequences in the
sense
orientation are known in the art. The methods generally involve transforming
plants
with a DNA construct comprising a promoter that drives expression in a plant
operably linked to at least a portion of a nucleotide sequence that
corresponds to the
transcript of the endogenous gene. Typically, such a nucleotide sequence has
substantial sequence identity to the sequence of the transcript of the
endogenous gene,
preferably greater than about 65% sequence identity, more preferably greater
than
about 85% sequence identity, most preferably greater than about 95% sequence
identity. See, U.S. Patent Nos. 5,283,184 and 5,034,323; herein incorporated
by
reference.
In some embodiments, the content and/or composition of polypeptides of the
present invention in a plant may be modulated by altering, in vivo or in
vitro, the
promoter of a gene to up- or down- regulate gene expression. In some
embodiments,
the coding regions of native genes of the present invention can be altered via
substitution, addition, insertion, or deletion to decrease activity of the
encoded
enzyme. See, e.g., Kmiec, U.S. Patent 5,565,350; Zarling et al.,
PCT/US93/03868.
And in some embodiments, an isolated nucleic acid (e.g., a vector) comprising
a
promoter sequence is transfected into a plant cell. Subsequently, a plant cell
comprising the promoter operably linked to a polynucleotide of the present
invention
is selected for by means known to those of skill in the art such as, but not
limited to,
Southern blot, DNA sequencing, or PCR analysis using primers specific to the
promoter and to the gene and detecting amplicons produced therefrom. A plant
or
plant part altered or modified by the foregoing embodiments is grown under
plant
forming conditions for a time sufficient to modulate the concentration and/or
composition of polypeptides of the present invention in the plant. Plant
forming
conditions are well known in the art and discussed briefly, supra.
In general, concentration or composition is increased or decreased by at least
5%, 10%, 20%, 30%, 40%, SO%, 60%, 70%, 80%, or 90% relative to a native
control
plant, plant part, or cell lacking the aforementioned recombinant expression
cassette.
Modulation in the present invention may occur during and/or subsequent to
growth of
the plant to the desired stage of development. Modulating nucleic acid
expression
-46-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
temporally and/or in particular tissues can be controlled by employing the
appropriate
promoter operably linked to a polynucleotide of the present invention in, for
example,
sense or antisense orientation as discussed in greater detail, supra.
Induction of
expression of a polynucleotide of the present invention can also be controlled
by
exogenous administration of an effective amount of inducing compound.
Inducible
promoters and inducing compounds, which activate expression from these
promoters,
are well known in the art. In preferred embodiments, the polypeptides of the
present
invention are modulated in monocots, particularly maize.
Methods of Use for LOX and SCIP-1 Promoter Sequences
The nucleotide sequences for the LOX and SCIP-1 promoters disclosed in the
present invention, as well as variants and fragments thereof, are useful in
the genetic
manipulation of any host cell, preferably plant cell, when assembled with a
DNA
construct such that the promoter sequence is operably linked to a nucleotide
sequence
encoding a heterologous protein of interest. In this manner, the nucleotide
sequences
of the LOX and SCIP-1 promoter of the invention are provided in expression
cassettes
along with heterologous nucleotide sequences for expression in the host cell
of
interest.
The promoters for the LOX and SCIP-1 genes may regulate expression of
operably linked nucleotide sequences in an inducible manner. That is,
expression of
the operably linked nucleotide sequences in a host cell (i.e., plant cell) is
induced in
response to a stimulus. By "stimulus" is intended a chemical, which may be
applied
externally or may accumulate in response to another external stimulus; a
pathogen,
which may, for example, induce expression as a result of invading a plant
cell; or
other factor such as environmental stresses, including but not limited to,
drought,
temperature, and salinity.
As such, the stimulus either directly or indirectly regulates the activity
(i.e., an
increase in initiation or expression) of an inducible promoter. By "direct
action" is
intended that the stimulus regulates transcription via a direct interaction
between the
stimulus and the DNA sequence. By "indirect action" is meant that the
regulation
occurs via an interaction between the stimulus and some other endogenous or
exogenous component in the system, the ultimate result of the indirect action
being
regulation of the inducible promoter. The stimulus can result from a biotic or
abiotic
-47-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
stress, including for example, tissue wounding (i.e., insect herbivory, wind,
intentional abiotic infliction of tissue injury or wounding for the purpose of
experimentation and/or expression analysis); wound-responsive chemicals (i.e.,
chemicals that result in the activation of wound-response signal transduction
pathways, including, various hormones, jasmonic acid, abscissic acid,
linolenic acid,
ethylene, their chemical analogues, derivatives, precursors, and the like);
pathogens
(i.e, fungi, bacteria, nematodes, mycoplasmas, viruses, and insects and the
like); and
various environmental stresses (i.e., heat, drought, cold, reactive oxygen
species
and/or radiation). Hence, the promoter of the present invention can be used in
combination with a nucleotide sequence that enhances disease resistance, and
the
compositions therefor find use in the defense of a plant against disease,
pathogens,
and the like.
Synthetic hybrid promoter regions are known in the art. Such regions
comprise upstream promoter elements of one nucleotide sequence operably linked
to
1 S the promoter element of another nucleotide sequence. In an embodiment of
the
invention, heterologous gene expression is controlled by a synthetic hybrid
promoter
comprising the LOX or SCIP-1 promoter sequences of the invention, or a variant
or
fragment thereof, operably linked to upstream promoter elements) from a
heterologous promoter. Upstream promoter elements that are involved in the
plant
defense system have been identified and may be used to generate a synthetic
promoter. See, for example, Rushton et al. (1998) Curr. Opin. Plant Biol.
1:311-315.
Alternatively, a synthetic LOX or SCIP-1 promoter sequence may comprise
duplications of the upstream promoter elements found within the LOX or SCIP-1
promoter sequence. Such elements include, for example the G-box (nucleotides
722-
727 of SEQ ID NO: 5; nucleotides 415-420 of SEQ ID NO: 10) or W-box
(nucleotides 322-327 of SEQ ID NO: S; nucleotides 364-368 and 371-375 of SEQ
ID
NO: 10).
It is recognized that the promoter sequence of the invention may be used with
its native LOX or SCIP-1 coding sequences. A DNA construct comprising the LOX
or SCIP-1 promoter operably linked with its native LOX or SCIP-1 gene may be
used
to transform any plant of interest to bring about a desired phenotypic change,
such as
enhanced disease resistance. Where the promoter and its native gene is
naturally
occurring within the plant, i.e., in sunflower, transformation of the plant
with these
-48-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
operably linked sequences also results in either a change in phenotype, such
as
enhanced disease resistance, the insertion of operably linked sequences within
a
different region of the chromosome thereby altering the plant's genome, or the
modulation in the level of expression of the nucleotide sequence of interest.
In another embodiment of the invention, expression cassettes will comprise a
transcriptional initiation region comprising the LOX or SCIP-1 promoter
nucleotide
sequences disclosed herein, or variants or fragments thereof, operably linked
to the
heterologous nucleotide sequence whose expression is to be controlled by the
inducible promoter of the invention.
The promoter nucleotide sequences and methods disclosed herein are useful in
regulating expression of any heterologous nucleotide sequence in a host plant
in order
to vary the phenotype of a plant. Various changes in phenotype are of interest
including modifying the fatty acid composition in a plant, altering the amino
acid
content of a plant, altering a plant's pathogen defense mechanism, and the
like. These
results can be achieved by providing expression of heterologous products or
increased
expression of endogenous products in plants. Alternatively, the results can be
achieved by providing for a reduction of expression of one or more endogenous
products, particularly enzymes or cofactors in the plant. These changes result
in a
change in phenotype of the transformed plant.
Genes of interest are reflective of the commercial markets and interests of
those involved in the development of the crop. Crops and markets of interest
change,
and as developing nations open up world markets, new crops and technologies
will
emerge also. In addition, as our understanding of agronomic traits and
characteristics
such as yield and heterosis increase, the choice of genes for transformation
will
change accordingly. General categories of genes of interest include, for
example,
those genes involved in information, such as zinc fingers, those involved in
communication, such as kinases, and those involved in housekeeping, such as
heat
shock proteins. More specific categories of transgenes, for example, include
genes
encoding important traits for agronomics, insect resistance, disease
resistance,
herbicide resistance, sterility, grain characteristics, and commercial
products. Genes
of interest include, generally, those involved in oil, starch, carbohydrate,
or nutrient
metabolism as well as those affecting kernel size, sucrose loading, and the
like.
-49-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Agronomically important traits such as oil, starch, and protein content can be
genetically altered in addition to using traditional breeding methods.
Modifications
include increasing content of oleic acid, saturated and unsaturated oils,
increasing
levels of lysine and sulfur, providing essential amino acids, and also
modification of
starch. Hordothionin protein modifications are described in U.S. Application
Serial
Nos. 08/838,763, filed April 10, 1997; U.S. Patent Nos. 5,703,049, 5,885,801,
and
5,885,802; herein incorporated by reference. Another example is lysine and/or
sulfur
rich seed protein encoded by the soybean 2S albumin described in U.S. Patent
No.
5,850,016, and the chymotrypsin inhibitor from barley, described in Williamson
et al.
(1987) Eur. J. Biochem. 165:99-106, the disclosures of which are herein
incorporated
by reference.
Derivatives of the coding sequences can be made by site-directed mutagenesis
to increase the level of preselected amino acids in the encoded polypeptide.
For
example, the gene encoding the barley high lysine polypeptide (BHL) is derived
from
barley chymotrypsin inhibitor, U.S. Application Serial No. 08/740,682, filed
November l, 1996, and PCT/LTS97/20441, filed October 31, 1997, the disclosures
of
which are herein incorporated by reference. Other proteins include methionine-
rich
plant proteins such as from sunflower seed (Lilley et al. (1989) Proceedings
of the
World Congress on hegetable Protein Utilization in Human Foods and Animal
Feedstuffs, ed. Applewhite (American Oil Chemists Society, Champaign,
Illinois), pp.
497-502; herein incorporated by reference); corn (Pedersen et al. (1986) J.
Biol.
Chem. 261:6279; Kirihara et al. (1988) Gene 71:359; both of which are herein
incorporated by reference); and rice (Musumura et al. ( 1989) Plant Mol. Biol.
12:123,
herein incorporated by reference). Other agronomically important genes encode
latex,
Floury 2, growth factors, seed storage factors, and transcription factors.
Insect resistance genes may encode resistance to pests that have great yield
drag such as rootworm, cutworm, European Corn Borer, and the like. Such genes
include, for example, Bacillus thuringiensis toxic protein genes (U.5. Patent
Nos.
5,366,892; 5,747,450; 5,736,514; 5,723,756; 5,593,881; and Geiser et al.
(1986) Gene
48:109); lectins (Van Damme et al. (1994) Plant Mol. Biol. 24:825); and the
like.
Genes encoding disease resistance traits include detoxification genes, such as
against fumonosin (U.5. Patent No. 5,792,931); avirulence (avr) and disease
-50-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
resistance (R) genes (Jones et al. (1994) Science 266:789; Martin et al.
(1993) Science
262:1432; and Mindrinos et al. (1994) Cell 78:1089); and the like.
Herbicide resistance traits may include genes coding for resistance to
herbicides that act to inhibit the action of acetolactate synthase (ALS), in
particular
the sulfonylurea-type herbicides (e.g., the acetolactate synthase (ALS) gene
containing mutations leading to such resistance, in particular the S4 and/or
Hra
mutations), genes coding for resistance to herbicides that act to inhibit
action of
glutamine synthase, such as phosphinothricin or basta (e.g., the bar gene), or
other
such genes known in the art. The bar gene encodes resistance to the herbicide
basta,
the nptll gene encodes resistance to the antibiotics kanamycin and geneticin,
and the
ALS-gene mutants encode resistance to the herbicide chlorsulfuron.
Sterility genes can also be encoded in an expression cassette and provide an
alternative to physical detasseling. Examples of genes used in such ways
include
male tissue-preferred genes and genes with male sterility phenotypes such as
QM,
described in U.S. Patent No. 5,583,210. Other genes include kinases and those
encoding compounds toxic to either male or female gametophytic development.
The quality of grain is reflected in traits such as levels and types of oils,
saturated and unsaturated, quality and quantity of essential amino acids, and
levels of
cellulose. In corn, modified hordothionin proteins, described in U.S.
Application
Serial No. 08/838,763 (filed April 10, 1997) and U.S. Patent Nos. 5,703,049,
5,885,801, and 5,885,802, provide descriptions of modifications of proteins
for
desired purposes.
Commercial traits can also be encoded on a gene or genes that could increase
for example, starch for ethanol production, or provide expression of proteins.
Another important commercial use of transformed plants is the production of
polymers and bioplastics such as described in U.S. Patent No. 5,602,321. Genes
such
as (3-Ketothiolase, PHBase (polyhydroxyburyrate synthase), and acetoacetyl-CoA
reductase (see Schubert et al. (1988) J. Bacteriol. 170:5837-5847) facilitate
expression of polyhyroxyalkanoates (PHAs). Exogenous products include plant
enzymes and products as well as those from other sources including procaryotes
and
other eukaryotes. Such products include enzymes, cofactors, hormones, and the
like.
The level of proteins, particularly modified proteins having improved amino
acid
-51-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
distribution to improve the nutrient value of the plant, can be increased.
This is
achieved by the expression of such proteins having enhanced amino acid
content.
The following examples are offered by way of illustration and not by way of
limitation.
EXPERIMENTAL
1e 1. The Isolation of Sunflower Disease Resistant Genes and the LOX and
SCIP-1 Promoter Sequences.
Materials and Methods
Plant Material
Sunflower plants were grown in the greenhouse and growth chamber. The
sunflower line SMF 3 and oxox-transgenic sunflower were used for an RNA
profiling
study by CuraGen. Sunflower pathogen, Sclerotinia sclerotiorum was maintained
on
PDA plate at 20 °C in dark.
Preparation of Total RNA for RNA Proftling Study and Northern Analysis
Plant materials were ground in liquid nitrogen and total RNA was extracted by
the Tri-agent Method. For each RNA profiling study, RNA samples from 6-week-
old
sunflower leaves and stems of transgenic sunflower plants expressing a wheat
oxalate
oxidase gene (oxox) were compared with RNA samples from the non-transformed
parent sunflower line SMF3. Total RNA (20 ug) was separated in a 1 % agarose
gel
containing formaldehyde. Ethidium bromide was included to verify equal loading
of
RNA. After transfer onto Hybond N+ membrane, the blots were hybridized with
32P-
labelled rhoGAP, LOX, ADH, or SCIP-1 cDNA probes. A duplicate blot was
hybridized with a ribosomal 18S RNA probe as a control. Hybridization and
washing
conditions were performed according to Church and Gilbert.
RNA Profiling
Differences in the expression of specific genes between sunflower plants
expressing a wheat oxalate oxidase gene and the sunflower line SMF3 were
determined using gene expression profiling. Total RNA was analyzed using the
gene
-52-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
expression profiling process (GeneCalling~) as described in U.S. Patent No.
5,871,697, herein incorporated by reference.
Isolation of Full-Length or Flanking Sequences by PCR Amplification of cDNA
Ends
The four cDNAs of the present invention were isolated by using RNA
profiling and PCR-based technologies. RNA profiling studies were conducted
through the collaboration with CuraGen Corp. The sequence information
generated
by the CuraGen study was used to design gene specific primers to amplifying
both 3'
and/or 5' end regions of the target genes using PCR-based RACE technology.
Sclerotinia-infected and oxox-induced cDNA libraries or cDNAs made by the
Marathon cDNA Amplification Kit (Clontech) were used as template for PCR
amplification. To facilitate cloning full-length cDNAs from initial cloned
regions, a
pair of 28 by vector primers were designed that flanked the cDNAs on both ends
(3'
and 5') of the pBS vector. Amplification of either the 5' or 3' end of the
cDNA was
accomplished using a vector primer (pBS-upper or pBS-lower) paired with a gene
specific primer. The 5' end of a specific gene with the intact ATG start codon
was
cloned and sequenced. The full-length cDNA was amplified by using a new gene
specific 5' primer containing sequences upstream of the ATG start codon and a
3'
primer containing vector sequences.
PCR reactions were performed in a total volume of 25 ~.l in 10 mM Tris-HCL,
pH 8.3; 1.5 mM MgCl2; 50 mM KCI; 0.1 mM dNTPs; 0.25 ~M of each primer with
0.5 units of advantage cDNA polymerase mix (Clontech) or Pwo DNA polymerase
(Boehringer). Genomic DNA and/or cDNA library mixtures were used as template
for PCR amplification.
Isolation of Disease Inducible Promoters
The promoter regions of the LOX and SCIP-1 genes were isolated from
sunflower genomic DNA using Universal Genomic Walker Kit (clontech) according
to the manufactures instructions. Restriction digested genomic DNAs were
ligated
with an adapter to construct pools of genomic DNA fragments for walking by
PCR.
-53-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Analysis ofAmplifred PCR Products
Amplified PCR fragments with the expected sizes were individually sliced out
of the gel for second round PCR re-amplification. The second round of PCR was
carried out using the same conditions as initial PCR. Each second round PCR
product
that showed a single band of the expected size was cloned into the TA vector
(Clontech) according to the manufacturer's instructions. Positive clones were
selected for DNA sequencing using an Applied Biosystems 373A (ABI) automated
sequencer at the Nucleic Acid Analysis Facility of Pioneer Hi-Bred Int'1 Inc.
DNA
sequence analysis was carried out with the Sequencer (3.0). Multiple-sequence
alignments (Clustal W) of the DNA sequence were carried out with the Curatool
(CuraGen).
Construction of the Sclerotinia-Infected and Resistance-Enhanced (Oxox-
Induced)
Sunflower cDNA Libraries
Six-week old SMF3 sunflower plants were infected with Sclerotinia
sclerotrium by petiole inoculation with Sclerotinia-infected carrot plugs. Six
days
after infection, leaf and stem tissues were collected from infected plants for
total RNA
isolation. Total RNA was also isolated from 6-week-old sunflower oxox-
transgenic
plants (line 610255) expressing a wheat oxalate oxidase gene. Our previous
studies
have shown that elevated levels of H202, SA, and PR1 protein were detected in
oxox-
transgenic plants at the 6-week stage and that the plants showed more
resistance to
Sclerotinia infection. The mRNAs were isolated using a mRNA purification kit
(BRL) according to the manufacture's instructions. The cDNA libraries were
constructed using the ZAP-cDNA synthesis kit and the pBluescript phagemid
(Stratagene). A cDNA library mixture for PCR cloning was made of oxox-
transgenic
stem and Sclerotinia-infected leaf libraries (1:2 mix).
Fungal Infection and Chemical Treatments
Sunflower plants SMF3 were planted in 4-inch pots and grown in greenhouse
for first four weeks. After transfer to growth chamber, plants were maintained
under
12-hour photoperoid at 22°C with an 80% relative humidity. Six-week old
plants
were inoculated with Sclerotinia-infected carrot plugs or sprayed with four
different
-54-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
chemicals at the given concentration. For each plant, three petioles were
inoculated
and wrapped with 1x2 inch parafilm. Plant tissue samples were harvested at
different
time points and immediately frozen in liquid nitrogen and then stored at -
80°C.
CuraGen Analysis and Database Search
An RNA profiling study identified 7 bands that were induced in oxox-
transgenic and Sclerotinia-infected sunflower plants (Table 1 ).
Table 1. Summary of RNA profiling results from four sets of experiments.
Stem&Leaf Leaf Leaf Stem
Oxox-48d Oxox-48d Infection Infection
Gene name Fold Band Id
Diff
GTPase-activating +2.6mOvO-120.8+2.58 +3.15 0 0
protein (rhoGAP)
Lipoxygenase +11.8mln0-257.60 0 0 +11.82
Lipoxygenase +6.4wOhO-279.10 0 0 +6.42
Lipoxygenase +3.4mOrO-276.70 0 0 +3.37
Alcohol dehydrogenase+7.7i0a0-289.5+7.69 0 0 +2.21
SCIP-1 -+3.210m0-273.90 0 +23.24 0
SCIP-1 +12.310m0-94.20 +.74 +12.28 0
Note: three lipoxygenase bands belong to same isolated sunflower LOX cDNA and
the two SCIP-1
bands are the same cDNA.
The CuraGen band mOvO-120.8 was induced 2 to 3 fold in oxox-transgenic
plants and showed lesion mimic phenotype (Table 1 ). The translated product of
CuraGen band mOvO-120.8 shares (amino acids 23-118 of SEQ ID NO: 2) 78% amino
acid identity with an Arabidopsis hypothetical 23.6 kDa protein (Accession No.
081806). Band mOvO-120.8 represents amino acids 23-118 of SEQ ID NO: 2. The
nucleotide sequence encoding the rhoGAP polypeptide is shown in SEQ ID NO: 1.
The sunflower rhoGAP clone is 824 by long with an ORF from nucleotides 35 to
nucleotide 637 from the 5' end. It encodes a 201 amino acid protein (SEQ ID
NO: 2)
with a molecular weight of 23.4 kDa and pI of 8.1. A BLASTX (version 2.0)
search
indicated that the sunflower rhoGAP-1 protein has sequence homology (about
40%)
with human p50rhoGAP (Accession No. 223024) and also shares sequence homology
with the Lotus japonicus rhoGAP (Accession No. AF064787).
Three of the CuraGen bands (mln0-257.6, wOhO-279.1 and mOrO-276.7) were
identified as lipoxygenase homologues by blast search of Genbank. The bands
were
-55-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
induced 3.37- to 11.82- fold higher in Sclerotinia-infected samples compared
with
uninfected control samples. The sunflower LOX nucleotide sequence is shown in
SEQ ID NO: 3. The LOX ORF extends from nucleotide 881 to nucleotide 3583. The
LOX ORF encodes a 901 amino acid protein (SEQ ID NO: 4) with a predicted
molecular weight of 101.37 kDa and a pI of 5.61. The sunflower LOX protein
shares
homology to other LOX genes from potato (Accession No. 034370), tomato
(Accession No. Q96573), cowpea (Accession No. P93698), Arabidopsis (Accession
No. LOXC_ARATH) and rice (Accession No. LOXC ORYSA).
The CuraGen band i0a0-289.5 was induced 7.7-fold higher in Sclerotinia-
infected samples compared with uninfected control samples. PCR-based cloning
of
this band allowed the isolation of a cDNA encoding a 381 amino acid
polypeptide,
which has 85-95% amino acid identity with known plant ADH sequences from
garden
lettuce (Accession No. Q40249), potato (Accession No. P 14674), tomato
(Accession
No. P28032), apple (Accession No. P48977), and maize (Accession No. P00333).
The CuraGen bands 10m0-273.9 and 10m0-94.2 were induced 23.2- and 12.3-
fold higher in Sclerotinia-infected samples compared with uninfected control
samples,
respectfully. PCR-based cloning of this band allowed two partial cDNA clones
to be
isolated. The full length cDNA of SCIP-1 encodes a 168 amino acid polypeptide.
A
BLASTP (version 2.0) sequence alignment showed that SCIP-1 has about 41%
identity from amino acid 10-167 with a conserved protein from Methanobacterium
thermoautotrophicum (Accession No. 026373), about 34% identity from amino acid
residuesl-163 with a hypothetical 20.7 kDa protein from Pyrococcus abyssi
(Accession No. CAB50064), about 35% identity from amino acid residues 13-159
with a conserved hypothetical protein from Archaeoglobus fulgidus (Accession
No.
028575), about 36% identity with a hypothetical 16.5 kDa protein from
Chlamydia
trachomatis (Accession No. 084741), about 36% identity from amino acid
residues
21-163, about 36% identity from amino acid 21 to 163 with a long hypothetical
protein from Pyrococcus horikoshii (Accession No.058984), about 35% identity
with
a hypothetical 18.9 kDa protein from Aquifex aeolicus (Accession No. 067293),
about 35% identity from amino acid residues 21-167 with a hypothetical 17.1
kDa
protein from E. coli (Accession No. P12994), and about 35% identity from amino
acid
residues 8-135 with a hypothetical 19.5 kDa protein from E coli (Accession No.
P77368).
-56-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
As discussed further in example 2, the transcript encoding SCIP-1 was up-
regulated in infected tissue as compared to control tissue. The accumulation
of SCIP-
1 in lesion mimic and infected sunflower plants suggests that the polypeptide
may be
involved in the plant defense response to Sclerotinia and other pathogens.
Position specific iterative BLAST (PSI-BLAST)
(http://www.ncbi.nlm.nih.gov/Education/BLASTinfo/psil.html) was performed on
the SCIP-1 sequence of the invention. PSI-BLAST refers to a feature of BLAST
2.0
in which a rp ofile (or position specific scoring matrix, PSSM) is constructed
(automatically) from a multiple alignment of the highest scoring hits in an
initial
BLAST search. The PSSM is generated by calculating position-specific scores
for
each position in the alignment. Highly conserved positions receive high scores
and
weakly conserved positions receive scores near zero. The profile is used to
perform a
second (BLAST search and the results of each "iteration" used to refine the
profile.
The second profile is used to perform another BLAST search and so on, until
the
profile no longer brings back new information and is said to have "converged".
This
iterative searching strategy results in increased sensitivity. PSI-BLAST
searches thus
can identify subtle homologies to annotated entries in the database. PSI-BLAST
is an
important tool for predicting both biochemical activities and function from
sequence
relationships.
A search was set up using the Lion BioSCOUT implementation of the PSI-
Blast algorithm. The query was the SCIP-1 polypeptide sequence, and the NR
database provided with Lion BioScout was searched. Parameters were set to
match as
closely as possible the defaults recommended by NCBI.
The best PSI-Blast hits found in the database for SLIP-1 were the group of
archeal and eubacterial hypothetical proteins found in earlier BLASTP
searches. As
the PSI-Blast search progressed, however, another group of proteins entered
the
alignments. Table 2 shows the 14 best hits from the PSI-Blast search other
than the
hypothetical proteins. It is notable that the areas of alignment are found
across the
entire length of these proteins (data not shown).
The proteins found in the PSI-Blast search fall into a class of flowering-
related
plant protein (CEN and others), as well as some phosphatidylethanolamine-
binding
proteins (PEBP). The CEN-related proteins are known to be related to a class
of
phosphatidylethanolamine-binding proteins (PEBP). The alignment of the SCIP-1
-57-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
protein with both the CEN proteins and PEBP suggests that SCIP-1 may be
related to
this class of PEBP proteins. By analogy to other reported PEBP-type proteins,
SCIP-
1 may play a role in signaling, in membrane transduction, or in the regulation
of cell
death.
Table 2. Selected hits from PSI-Blast search with SCIP-1 against the NR
protein
database. From the entire set of PSI-Blast results, the I4 best hits were
selected that
were not from bacteria, and that had functional annotations associated with
the
molecule.
Hit (AccessionPSI-BlastDescription Reference
Numbers) expectation
trembl~AB017525~AB1e-29 gene: "BNTFLI-1";BrassicaMimidaetal.
(1999)
017525 1 napus BNTFL1-1 gene,Plant Science
complete
cds 142:155-162
swiss~P54186~D11e-29 D1 PROTEIN (FRAGMENT)Erttmann et
ON al. (1996)
CVO from Onchocerca volvulusGene 174:203-7
(nematode)
trembl~AB017528~AB3e-29 gene: "BRTFL1-I "; Mimida et al.
Brassica (1999)
017528 I rapa BRTFL1-1 gene, Plant Science
complete
cds 142:155-162
trembl~AF145261~AF11e-28 gene: "CET4"; product:Amaya et al.
"CEN- (1999)
45261 1 like protein 4"; Plant Cell
Nicotiana 11:1405-
tabacum CEN-like 1417
protein 4
(CET4) mRNA, complete
cds.
tremb1~D87130~ATD12e-28 gene: "terminal flowerlOhshima et
"; al. (1997)
30 1 product: "terminal Mol Gen Genet
flowed ";
Arabidopsis thaliana254:186-94
DNA for
terminal flowed,
complete cds.
trembl~U84140~U84144e-28 gene: "sp"; product:Pnueli et al.
"self (1989)
_0 1 pruning protein"; Development
Lycopersicon
esculentum self pruning125:1979-1989
protein
(sp) mRNA, complete
cds
trembl~AB027456~AB1e-27 gene: "CiFT"; CitrusKobayashi et
unshiu al.
027456 1 CiFT mRNA, complete (1999) Science
cds.
286:1960-2
swiss~016264~PBPH3e-27 PHOSPHATIDYLETHANOLAUnpublished
CAEEL MINE-BINDING PROTEIN
HOMOLOG F40A3.3.
trembl~AB027506~AB6e-27 gene: "TSF"; product:Kobayashi et
"TSF"; al.
027506 1 Arabidopsis thaliana(1999) Science
TSF (TWIN
SISTER OF FT) mRNA, 286:1960-2
complete cds
swiss~Q41261~CEN3e-26 CENTRORADIALISPROTEIN.Bradley et
A al. (1996)
NTMA . [Antirrhinum=snapdragons,Nature 379:791-7
and
mRNA, 929 nt]. Bradley et
al. (1997)
Science 275:80-83.
trembI~AB027504~AB5e-26 gene: "FT"; product:Kobayashi et
"FT"; al.
027504 1 Arabidopsis thaliana(1999) Science
FT
(FLOWERING LOCUS 286:1960-2
T)
mRNA, complete cds
-5 8-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
trembl~AF159882~AF18e-26 gene: "Fdr2"; product:Unpublished
"Cen-like
59882 1 protein FDR2"; Oryza
saliva
Cen-like protein FDR2
(Fdr2)
mRNA, complete cds
trembl~AB024712~AB1 e-24 gene: "ATC"; ArabidopsisUnpublished
024712 1 thaliana ATC (centroradialis)
gene, complete cds,
strain: Landsberg
trembl~D1611 1e-23 product: "human homologueHori et al.
1~HSHR of (1994)
PBP 1 rat phosphatidylethanolamineGene 140:293-4
binding protein";
Human mRNA
for human homologue
of rat
phosphatidylethanolamine
binding protein, complete
cds
Example 2: Northern Analysis of mRNA Levels of Disease Resistance Genes
Following Biotic and Abiotic Stresses
The expression of many plant defense genes are induced by biotic and abiotic
stresses. Salicylic acid (SA), Jasmonic acid (JA), and H202 have been
implicated in
playing a central role of plant disease resistance and systemic acquired
resistance.
Oxalic acid (OA), a compound produced by Sclerotinia and many other fungal
pathogens in planta, plays an important role in the disease infection process.
The expression of LOX, rhoGAP, SCIP-1, and ADH mRNA in the presence of
compounds known to induce either systemic acquired resistance or disease
response
was determined. Six-week-old sunflower leaves were sprayed until runoff with 5
mM
SA, 45 1tM JA, 5 mM of oxalic acid, and 5 mM H202. Leaf samples from chemical
treated plants were collected at 0, 6, 12, and 24 hours after foliar
application.
Northern analysis indicated that there was a significant increase in the
steady-
state levels of ADH mRNA in SA and H202 treated leaves (data not shown). The
highest level of ADH mRNA expression was detected at 6 hours after application
of
H202 and 12 hours following the administration of SA. There was about a 2-to 3-
fold increase in rhoGAP transcripts in response to both the H202 and OA
application
(data not shown). The mRNA levels of SCIP-1 increased upon treatment with OA.
JA, a product of the LOX pathway, significantly induced the steady-state
levels of LOX mRNA. Therefore, LOX mRNA seems to be controlled by a positive
feedback loop. In contrast, foliar application of SA and H20z repressed the
expression of LOX mRNA in sunflower. Northern and RNA profiling results
revealed that sunflower LOX mRNA was elevated by Sclerotinia infection and
oxalic
acid, a pathogenic factor produced by the fungus (data not shown).
-59-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
The effects of Sclerotinia infection and oxox on SCIP-1, rhoGAP, and ADH
mRNA levels was also determined. RNA levels in Sclerotinia-infected sunflower
and
oxox-transgenic sunflower plants were determined using Northern analysis. RNA
was
isolated from leaves from 6-week-old non-transformed SMF3 plants and from 6-
S week-old oxox-transgenic plants. Steady state levels of SCIP-1 mRNA
significantly
increased in 6-week-old leaf tissue from the oxox-transgenic plants as
compared to
the control SMF3 leaf samples. RNA was also isolated from stem tissue from 6-
week-old non-transformed SMF3 plants and from 6-week-old oxox-transgenic
plants.
An increase in steady state levels of SCIP-1 mRNA was not detected in the stem
tissue (data not shown).
RNA was also isolated from 6-week-old SMF3 plants that were infected 5-
days prior to sample collection with Sclerotinia. The steady state levels of
SCIP-1
RNA following Sclerotinia infection increased significantly when compared to
RNA
levels from uninfected leaf tissue. No change in SCIP-1 RNA levels was seen in
the
stem tissue following Sclerotinia infection (data not shown).
Induction of SCIP-1, ADH, and rhoGAP expression in oxox-transgenic
sunflower leaf and stem tissue during development was analyzed. RNA was
isolated
from leaf and stem tissue from SMF3 plants and oxox-transgenic sunflower
plants at
4-week-old, 6-week-old, and 8-week-old stages. Northern blot analysis using
RNA
samples from SMF3 sunflower plants and samples from oxox-transgenic plants
demonstrated SCIP-1 RNA levels increased in leaf tissues in the oxox-
transgenic
sunflowers. The increase in SCIP-1 RNA levels was most significant at the 8-
week-
old stage. No detectable induction of SCIP-1 was seen in the stem tissue.
The steady-state-level of ADH mRNA is much higher in stem than in leaf
tissue. Adh expression was induced in 8-week-old oxox-transgenic leaf tissue.
However, its expression was repressed in oxox-stem tissue. CuraGen QEA assay
results indicate that Adh expression was induced by Sclerotinia infection in
stem and
leaf tissues (data not shown).
In the leaf tissue, rhoGAP mRNA level was induced by oxox expression at the
4-week-old stage, and then was repressed with development. In stem tissue,
rhoGAP
expression was slightly induced by oxox (data not shown).
LOX expression in response to wounding and the ABA signal was determined.
Six-week-old sunflower plants (SMF3) were sprayed with 100 ~M ABA until the
-60-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
chemical solution started to run off the leaves. For the wounding experiment,
each
wounded leaf was crushed by a hemostat 20 times and three leaves were treated
from
each plant. At each time point, six leaves were collected from two treated
plants and
immediately frozen in liquid N2. Total RNA was isolated and Northern analysis
was
performed. Northern blot analysis indicated that wounding significantly
induced the
steady state levels of LOX mRNA. The peak of the LOX mRNA accumulation was
detected at 6 hours after wounding and high levels of LOX expression was
maintained
through 72 hours after initial treatment. ABA treatment showed only a slight
induction of LOX expression at 12 hours after treatment (data not shown).
Example 3: Transformation and Regeneration of Maize Transgenic Plants
The nucleotide sequences of the present invention can be used to transform
sunflower, maize, or other plants using Agrobacterium or particle-gun methods.
Examples 3, 4, and 5 provide methods for sunflower, maize, and soybean
transformations.
Immature maize embryos from greenhouse donor plants are bombarded with a
plasmid containing a rhoGAP, LOX, ADH or SCIP-1 nucleotide sequence operably
linked to a ubiquitin promoter (Figure 1 ). The plasmids also contain the
selectable
marker gene PAT (Wohlleben et al. (1988) Gene 70:25-37) that confers
resistance to
the herbicide Bialaphos. Transformation is performed as follows. All media
recipes
are given in the Appendix. Alternatively, the plant can be transformed with a
plasmid
comprising the LOX or SCIP-1 promoter sequences of the invention operably
linked
to the nucleotide sequence encoding the GUS reporter protein (Figure 2).
Preparation of Target Tissue
The ears are surface sterilized in 30% Chlorox bleach plus 0.5% Micro
detergent for 20 minutes, and rinsed two times with sterile water. The
immature
embryos are excised and placed embryo axis side down (scutellum side up), 25
embryos per plate, on 560Y medium for 4 hours and then aligned within the 2.5-
cm
target zone in preparation for bombardment.
-61-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Preparation of DNA
A plasmid vector shown in Figures 1 or 2 is precipitated onto 1.1 ~m (average
diameter) tungsten pellets using a CaCl2 precipitation procedure as follows:
100 ~1 prepared tungsten particles in water
p1 ( 1 fig) DNA in TrisEDTA buffer ( 1 ~g total)
100 X12.5 M CaC 1 Z
10 p1 0.1 M spermidine
Each reagent is added sequentially to the tungsten particle suspension, while
10 maintained on the multitube vortexer. The final mixture is sonicated
briefly and
allowed to incubate under constant vortexing for 10 minutes. After the
precipitation
period, the tubes are centrifuged briefly, liquid removed, washed with 500 ml
100%
ethanol, and centrifuged for 30 seconds. Again the liquid is removed, and 105
p1
100% ethanol is added to the final tungsten particle pellet. For particle gun
bombardment, the tungsten/DNA particles are briefly sonicated and 10 ~I
spotted
onto the center of each macrocarrier and allowed to dry about 2 minutes before
bombardment.
Particle Gun Treatment
The sample plates are bombarded at level #4 in particle gun #HE34-1 or
#HE34-2. All samples receive a single shot at 650 PSI, with a total of ten
aliquots
taken from each tube of prepared particles/DNA.
Subsequent Treatment
Following bombardment, the embryos are kept on 560Y medium for 2 days,
then transferred to 5608 selection medium containing 3 mg/liter Bialaphos, and
subcultured every 2 weeks. After approximately 10 weeks of selection,
selection-
resistant callus clones are transferred to 288J medium to initiate plant
regeneration.
Following somatic embryo maturation (2-4 weeks), well-developed somatic
embryos
are transferred to medium for germination and transferred to the lighted
culture room.
Approximately 7-10 days later, developing plantlets are transferred to 272V
hormone-
free medium in tubes for 7-10 days until plantlets are well established.
Plants are then
-62-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
transferred to inserts in flats (equivalent to 2.5" pot) containing potting
soil and grown
for I week in a growth chamber, subsequently grown an additional 1-2 weeks in
the
greenhouse, then transferred to classic 600 pots (1.6 gallon) and grown to
maturity.
Plants transformed with the plasmid shown in Figure I comprising the
rhoGAP, LOX, ADH, or SCIP-1 nucleotide sequences are monitored and scored for
altered defense response, or altered rhoGAP, LOX, ADH, or SCIP-1 activity.
Plants transformed with the plasmid shown in Figure 2 comprising the LOX or
SCIP-1 promoter sequences operably linked to the GUS reporter sequences are
monitored for LOX or SCIP-1 promoter activity. Following exposure to various
stimuli, such as for example, Sclerotinia and oxalic acid, LOX promoter
activity is
measured using the reporter gene GUS. GUS activity in various tissues is
measured
by a fluorogenic assay. The fluorogenic assay determines the specific activity
of (3-
glucuronidase (GUS) in various maize tissue extracts. The specific activity of
the
enzyme is expressed as moles of 4-methyl umbelliferone (MU) released/pg
protein/hour. MU is produced when the enzyme (GUS) in plant cell extracts
cleaves
the glucuronide moiety from the 4-methyl umbelliferyl-(3-D-glucuronide (MUG)
substrate.
Bombardment and Culture Media
Bombardment medium (560Y) comprises 4.0 g/1 N6 basal salts (SIGMA C-
1416), 1.0 m1/1 Eriksson's Vitamin Mix (1000X SIGMA-1511), 0.5 mg/1 thiamine
HCI, 120.0 g/1 sucrose, 1.0 mg/12,4-D, and 2.88 g/1 L-proline (brought to
volume
with D-I HZO following adjustment to pH 5.8 with KOH); 2.0 g/1 Gelrite (added
after
bringing to volume with D-I H20); and 8.5 mg/1 silver nitrate (added after
sterilizing
the medium and cooling to room temperature). Selection medium (560R) comprises
4.0 g/1 N6 basal salts (SIGMA C-1416), 1.0 m1/1 Eriksson's Vitamin Mix (1000X
SIGMA-151 I), 0.5 mg/1 thiamine HC1, 30.0 g/1 sucrose, and 2.0 mg/12,4-D
(brought
to volume with D-I H20 following adjustment to pH 5.8 with KOH); 3.0 g/1
Gelrite
(added after bringing to volume with D-I H20); and 0.85 mg/1 silver nitrate
and 3.0
mg/1 bialaphos(both added after sterilizing the medium and cooling to room
temperature).
Plant regeneration medium (288J) comprises 4.3 g/1 MS salts (GIBCO 11117-
074), 5.0 m1/1 MS vitamins stock solution (0.100 g nicotinic acid, 0.02 g/1
thiamine
-63-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
HCL, 0.10 g/1 pyridoxine HCL, and 0.40 g/1 glycine brought to volume with
polished
D-I HZO) (Murashige and Skoog (1962) Physiol. Plant. 15:473), 100 mg/1 myo-
inositol, 0.5 mg/1 zeatin, 60 g/1 sucrose, and 1.0 m1/1 of 0.1 mM abscisic
acid (brought
to volume with polished D-I HZO after adjusting to pH 5.6); 3.0 g/1 Gelrite
(added
after bringing to volume with D-I HZO); and 1.0 mg/1 indoleacetic acid and 3.0
mg/1
bialaphos (added after sterilizing the medium and cooling to 60°C).
Hormone-free
medium (272V) comprises 4.3 g/1 MS salts (GIBCO 11117-074), 5.0 m1/1 MS
vitamins stock solution (0.100 g/1 nicotinic acid, 0.02 g/1 thiamine HCL, 0.10
g/1
pyridoxine HCL, and 0.40 g/1 glycine brought to volume with polished D-I H20),
0.1
g/1 myo-inositol, and 40.0 g/1 sucrose (brought to volume with polished D-I
H20 after
adjusting pH to 5.6); and 6 g/1 bacto-agar (added after bringing to volume
with
polished D-I H20), sterilized and cooled to 60° C.
Example 4: Agrobacterium-mediated Transformation
For Agrobacterium-mediated transformation of maize with rhoGAP, LOX,
ADH, or SCIP-1 nucleotide sequences of the invention or a nucleotide sequence
operably linked to the LOX or SCIP-1 promoter sequence of the invention,
preferably
the method of Zhao is employed (U.5. Patent No. 5,981,840, and PCT patent
publication
W098/32326; the contents of which are hereby incorporated by reference).
Briefly,
immature embryos are isolated from maize and the embryos contacted with a
suspension of Agrobacterium, where the bacteria are capable of transferring
the DNA
constructs of interest to at least one cell of at least one of the immature
embryos (step
1: the infection step). In this step the immature embryos are preferably
immersed in
an Agrobacterium suspension for the initiation of inoculation. The embryos are
co-
cultured for a time with the Agrobacterium (step 2: the co-cultivation step).
Preferably the immature embryos are cultured on solid medium following the
infection step. Following this co-cultivation period an optional "resting"
step is
contemplated. In this resting step, the embryos are incubated in the presence
of at
least one antibiotic known to inhibit the growth of Agrobacterium without the
addition of a selective agent for plant transformants (step 3: resting step).
Preferably
the immature embryos are cultured on solid medium with antibiotic, but without
a
selecting agent, for elimination of Agrobacterium and for a resting phase for
the
infected cells. Next, inoculated embryos are cultured on medium containing a
-64-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
selective agent and growing transformed callus is recovered (step 4: the
selection
step). Preferably, the immature embryos are cultured on solid medium with a
selective agent resulting in the selective growth of transformed cells. The
callus is
then regenerated into plants (step 5: the regeneration step), and preferably
calli grown
on selective medium are cultured on solid medium to regenerate the plants.
Example 5: Transformation and Regeneration of Sunflower Plants
The intact meristem method is used for transformation of sunflower plants and
expression of the LOX, ADH, rhoGAP, or SCIP-1 nucleotide sequences as follows.
Alternatively, the same method could be used to express a nucleotide sequence
of
interest under the control of the LOX or SCIP-1 promoter sequence of the
invention.
Explant Preparation
Seeds are dehulled and surface-sterilized for 20 minutes in a 20% ChloroxTM
bleach solution with the addition of two to three drops of Tween 20 per 100 ml
of
solution, and then rinsed three times with distilled water. Sterilized seeds
are imbibed
in the dark at 26°C for 20 hours on filter paper moistened with water.
Meristem
explants are created by removing cotyledons and root radicle from imbibed
seeds, and
then culturing overnight at 26°C in the dark on 374E medium (1X MS
salts, Shepards
vitamins, 40 mg/1 adenine sulfate, 30 g/1 sucrose, 0.5 mg/1 BAP, 0.25 mg/1
IAA, 0.1
mg/1 IAA, pH 5.6, 8g/1 phytagar). Primary leaves are then removed and explants
are
transferred to 374M medium (374E except 12 g/1 phytagar), arranged in a manner
suitable for particle gun bombardment, and cultured overnight at 26°C
in the dark.
Preparation of DNA
A plasmid vector comprising the rhoGAP, LOX, ADH, or SCIP-1 nucletoide
sequences operably linked to a ubiquitin promoter is constructed (Figure 1 ).
Alternatively, a plasmid vector comprising the LOX promoter or the SCIP-1
nucleotide sequence operably linked to the nucleotide sequence encoding the
GUS
reporter protein is constructed (Figure 1). Both of these plasmids contain a
kanamycin selectable marker gene. The transformation is performed as follows.
-65-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Transformation
Approximately 18.8 mg of 1.8 ~m tungsten particles are suspended in 150 ~1
absolute ethanol, and sonicated for 2-4 seconds. After sonication, 10 ~l of
the
suspension is dropped on the center of the surface of a macrocarrier. Each
plate of
meristem explants is bombarded twice with 650 psi rupture discs in the top
shelf at 26
mm of Hg helium gun vacuum, using a BioRad helium gun.
The plasmid vector shown in Figure 1 or in Figure 2 is introduced into
Agrobacterium strain EHA 105 (see above) via freeze-thawing as described by
Holsters et al. (1978) Mol. Gen. Genet. 163:181-187. Actively growing,
transformed
Agrobacteria were maintained in shaking liquid cultures using 60A medium with
kanamycin (YEP, 50 mg/1 kanamycin: 10 g/1 yeast extract, 10 g/1 bactopeptone,
5 g/1
NaCI, pH 7.0, 50 mg/1 kanamycin). On the day before the Agrobacterium strain
is to
be used, new liquid cultures are initiated in 60A with kanamycin from the
active
maintenance culture. They are cultured with shaking at 26°C until they
reach an
optical density (OD vis = 600 nm) of about 1Ø When the cultures have
established
this density, they are centrifuged (6000 rpm, 5 min), the supernatant is
discarded, and
the pellet of bacteria is resuspended in inoculation medium (l2.SmM 2-(N-
morpholino) ethanesulfonic acid, 1 g/1 NH4C1, and 0.3 g/1 MgS04, at pH 5.7),
to a
final calculated concentration of Agrobacteria of 4.0 at OD 600. The particle
bombarded explants are inoculated with Agrobacterium by first spreading the
explants
apart on the 374M medium, then placing a droplet of the above suspension
directly
onto the top of each meristem. The explants are co-cultivated on the medium
for 4
days, after which the explants are transferred to 374 C medium (GBA with 1%
sucrose and with no BAP, IAA, or GA3, and supplemented with 250 ~g/ml
cefotaxime). The explants are cultured on this medium for about 2 weeks under
16
hours of daylight, at 26°C.
Recovering Nodes and Plants
Following the 4 days of co-cultivation time on 374M medium, the explants are
transferred to 374D (374C medium with 50 mg/1 kanamycin) selection medium
containing kanamycin. After 2 weeks of selection, explants with associated
shoots are
transferred to 374C medium and selection resistant shoots are screened using
NPTII
-66-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
ELISA. Positive shoots are removed for recovery by in vitro grafting and
transformation verified by further NPTII ELISA analysis. Negative shoots are
discarded. Explants with smaller shoots which could not be assayed following
the 2
weeks on 374D are transferred to 3746 (374E with 250 mg/1 cefotaxime) for 3-4
days
then back to 374C for 2 additional weeks. Assays are then done to identify
positive
shoots which are too small to sample in the first round and recovery
initiated.
Recovered positive shoots are grafted to Pioneer sunflower hybrid in vitro-
grown sunflower seedling rootstock. The seeds are dehulled and surface-
sterilized for
20 minutes in a 20% ChloroxTM bleach solution with two to three drops of
Tween20
per 100 ml total volume, and rinsed three times with distilled water. The
sterilized
seeds are germinated for three days on filter paper moistened with water, then
transferred into "48 Medium" (one-half strength MS salts, 0.5% sucrose, 0.3%
gelrite,
at pH 5.0) and grown at 26°C at 26 in the dark for 3 days, then
incubated at 16 hour
day culture conditions. The upper portions of selected seedlings are removed,
a
vertical slice is made in each hypocotyl, and a transformed shoot is inserted
into the
vertical slice. The cut area is wrapped with parafilm, and after one week
culture on
the medium, the grafted plants are transferred to soil. In the first two weeks
they are
maintained under high humidity conditions to acclimatize to the greenhouse
environment.
Transformed sectors of TO plants are identified by additional NPTII assays of
the greenhouse established positive grafted shoots. After assay, non-
transformed
sectors are trimmed off to promote auxillary bud development and auxiliary
buds
from transgenic sectors are recovered so as to establish the best probability
to
encompass the sector of transformation in germ line cells so that the
transformation
event is recovered in the next generation. Seed from TO plants are collected,
de-
hulled, surface sterilized, and germinated on filter paper wetted with water.
T1
seedlings are then sampled for NPTII ELISA by removing green cotyledon pieces
followed by transfer to seedling growth medium 48P (0.1 X MS salts, 0.5%
sucrose,
pH 5.6, 0.3% gelrite). NPTII positive, actively growing T1 seedlings are
transferred
at the two leaf stage to soil for growth in the greenhouse. Seed from the
confirmed T1
transgenics is used to grow T2 plants.
-67-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
T2 seeds are planted in a greenhouse. Positive plants are screened by NPTII
assay. Various plant tissues are harvested at 80-day-old stage after planting.
The
harvested material is put in mini-tubes, frozen and stored at -80°C.
Plants transformed with the plasmid shown in Figure 1 comprising the
rhoGAP, LOX, ADH, or SCIP-1 nucleotide sequences are monitored and scored for
an altered defense response, or a modulation in rhoGAP, LOX, ADH or SCIP-1
activity.
Plants transformed with the plasmid shown in Figure 2 comprising the LOX
promoter sequences operably linked to the GUS reporter sequences are monitored
for
LOX promoter activity. Following exposure to various stimuli that induce the
LOX
promoter, LOX promoter activity is measured by assaying for GUS activity. GUS
activity in various tissues is measured by a fluorogenic assay. The
fluorogenic assay
determines the specific activity of ~i-glucuronidase (GUS) in various
sunflower tissue
extracts. The specific activity of the enzyme is expressed as moles of 4-
methyl
umbelliferone (MU) released/ug protein/hour. MU is produced when the enzyme
(GUS) in plant cell extracts cleaves the glucuronide moiety from the 4-methyl
umbelliferyl-(3-D-glucuronide (MUG) substrate.
Harvested T2 tissue samples stored at -80°C are homogenized in 400
p1 lysis
buffer (40 mM Phosphate, pH 7.0, 10 mM EDTA, 10 mM (3-mercaptoethanol), and
then centrifuged in the Jouan GR422 centrifuge for 10 minutes at 4000 rpm. The
total
protein concentration of the supernatant is measured using the Bio-Rad
Bradford
Method (Bio-RAD) with BSA as the standard protein according to manufacture's
protocol. Ten p1 of diluted supernatant (about 4 ~g of total protein) is used
for the
GUS activity assay. GUS activity is assayed according to Jefferson et al.
(1987)
EMBOJ. 6: 3901-3907 using MUG as substrate.
As an alternative to the intact meristem method, the split embryonic axis
method may be used as described in Malone-Schoneberg et al. ( 1994) Plant
Science
X03:193-207, in transforming sunflower plants with either the plasmid shown in
Figure 3 or Figure 4 and generating T2 plants. T2 seeds are planted in a
greenhouse
and positive plants are screened by NPTII assay. Plant tissues are harvested
at 80-
day-old stage after planting. The harvested material is put in mini-tubes,
frozen, and
stored at -80°C.
-68-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Example 6: Soybean Embryo Transformation
Soybean embryos are bombarded with a plasmid containing the SCIP-l,
rhoGAP, LOX, or ADH sequences operably linked to a ubiquitin promoter (Figure
1 )
as follows. Alternatively, the soybean embryos can be bombarded with a DNA
construct containing the SCIP-1 or LOX promoter operably linked to a
nucleotide
sequence of interest (Figure 2). To induce somatic embryos, cotyledons, 3-5 mm
in
length dissected from surface-sterilized, immature seeds of the soybean
cultivar
A2872, are cultured in the light or dark at 26°C on an appropriate agar
medium for six
to ten weeks. Somatic embryos producing secondary embryos are then excised and
placed into a suitable liquid medium. After repeated selection for clusters of
somatic
embryos that multiplied as early, globular-staged embryos, the suspensions are
maintained as described below.
Soybean embryogenic suspension cultures can maintained in 35 ml liquid media
on a rotary shaker, 150 rpm, at 26°C with florescent lights on a 16:8
hour day/night
schedule. Cultures are subcultured every two weeks by inoculating
approximately
35 mg of tissue into 35 ml of liquid medium.
Soybean embryogenic suspension cultures may then be transformed by the
method of particle gun bombardment (Klein et al. (1987) Nature (London) 327:70-
73,
U.S. Patent No. 4,945,050). A Du Pont Biolistic PDS1000/HE instrument (helium
retrofit) can be used for these transformations.
A selectable marker gene that can be used to facilitate soybean transformation
is
a transgene composed of the 35S promoter from Cauliflower Mosaic Virus (Odell
et
al. (1985) Nature 313:810-812), the hygromycin phosphotransferase gene from
plasmid pJR225 (from E. coli; Gritz et al. (1983) Gene 25:179-188), and the 3'
region
of the nopaline synthase gene from the T-DNA of the Ti plasmid of
Agrobacterium
tumefaciens. The expression cassette comprising the DNA construct can be
isolated
as a restriction fragment. This fragment can then be inserted into a unique
restriction
site of the vector carrying the marker gene.
To 50 ~l of a 60 mg/ml 1 ~m gold particle suspension is added (in order): 5 ~l
DNA (1 ~g/pl), 20 ~l spermidine (0.1 M), and 50 p.1 CaCl2 (2.5 M). The
particle
preparation is then agitated for three minutes, spun in a microfuge for 10
seconds and
the supernatant removed. The DNA-coated particles are then washed once in 400
~l
-69-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
70% ethanol and resuspended in 40 ~1 of anhydrous ethanol. The DNA/particle
suspension can be sonicated three times for one second each. Five microliters
of the
DNA-coated gold particles are then loaded on each macro carrier disk.
Approximately 300-400 mg of a two-week-old suspension culture is placed in
S an empty 60x15 mm petri dish and the residual liquid removed from the tissue
with a
pipette. For each transformation experiment, approximately 5-10 plates of
tissue are
normally bombarded. Membrane rupture pressure is set at 1100 psi, and the
chamber
is evacuated to a vacuum of 28 inches mercury. The tissue is placed
approximately
3.5 inches away from the retaining screen and bombarded three times. Following
bombardment, the tissue can be divided in half and placed back into liquid and
cultured as described above.
Five to seven days post bombardment, the liquid media may be exchanged with
fresh media, and eleven to twelve days post-bombardment with fresh media
containing 50 mg/ml hygromycin. This selective media can be refreshed weekly.
Seven to eight weeks post-bombardment, green, transformed tissue may be
observed
growing from untransformed, necrotic embryogenic clusters. Isolated green
tissue is
removed and inoculated into individual flasks to generate new, clonally
propagated,
transformed embryogenic suspension cultures. Each new line may be treated as
an
independent transformation event. These suspensions can then be subcultured
and
maintained as clusters of immature embryos or regenerated into whole plants by
maturation and germination of individual somatic embryos.
All publications and patent applications mentioned in the specification are
indicative of the level of those skilled in the art to which this invention
pertains. All
publications and patent applications are herein incorporated by reference to
the same
extent as if each individual publication or patent application was
specifically and
individually indicated to be incorporated by reference.
Although the foregoing invention has been described in some detail by way of
illustration and example for purposes of clarity of understanding, it will be
obvious
that certain changes and modifications may be practiced within the scope of
the
appended claims.
-70-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
INDICATIONS RELATING TO DEPOSITED MICROORGANISM
OR OTHER BIOLOGICAL MATERIAL
A. The indications made below relate
to the deposited microorganism
or other biological material referred
to in the description on page 10,
lines
23, 27 and 31
B. IDENTIFICATION OF DEPOSIT Further
deposits are identified on an additional
sheet 0
Name of depository institution
American Type Culture Collection
Address of depositary institution
(including postal code and country)
10801 University Blvd.
Manassas, VA 20110-2209 US
Date of deposit Accession Number
30 June 1999 (30.06.99) PTA-284
C. ADDITIONAL INDICATIONS (leave
blank if not applicable) This information
is continued on an additional sheet
Page 11, lines 11, 13, 16, 18 and
19; Page 78, lines 9 and 13; Page
79, lines 4 and 6; Page 80, lines
8 and
10; Page 81, lines 11 and 13; Page
83, lines 11 and 13
D. DESIGNATED STATES FOR WHICH INDICATIONS
ARE MADE (if the indicators are
not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS
(leave blank if not applicable)
The indications listed below will
be submitted to the International
Bureau later (specify the general
nature of the indications e.g.,
Accession
Number of Deposit's
For receiving Office use only For International Bureau use only
This sheet was received with the international application ~ ~ ~ This sheet
was received with the International Bureau on:
Authorized officer
-71-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
INDICATIONS RELATING TO DEPOSITED MICROORGANISM
OR OTHER BIOLOGICAL MATERIAL
A. The indications made below relate
to the deposited microorganism
or other biological material referred
to in the description on page
10, lines
23, 28 and 31
B. IDENTIFICATION OF DEPOSIT Further
deposits are identified on an
additional sheet
Name of depository institution
American Type Culture Collection
Address of depositary institution
(including postal code and country)
10801 University Blvd.
Manassas, VA 20110-2209 US
Date of deposit Accession Number
30 June 1999 (30.06.99) PTA-285
C. ADDITIONAL INDICATIONS (leave
blank if not applicable) This
information is continued on an
additional sheet 0
Page 11, lines 11, 13, 17-19; Page
78, lines 9 and 13; Page 79, lines
4 and 6; Page 80, lines 8 and
10; Page
81, lines 11 and 13; Page 83, lines
11 and 13
D. DESIGNATED STATES FOR WHICH
INDICATIONS ARE MADE (if the indicators
are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS
(leave blank if not applicable)
The indications listed below will
be submitted to the International
Bureau later (specify the general
nature of the indications e.g.,
Accession
Number of Deposit
For receiving Office use only For International Bureau use only
.This sheet was received with the international application I I 0 This sheet
was received with the International Bureau on:
Authorized officer
-72-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
INDICATIONS RELATING TO DEPOSITED MICROORGANISM
OR OTHER BIOLOGICAL MATERIAL
A. The indications made below relate
to the deposited microorganism
or other biological material referred
to in the description on page 10,
lines
24 and 31
B. IDENTIFICATION OF DEPOSIT Further
deposits are identified on an additional
sheet
Name of depository institution
American Type Culture Collection
Address of depositary institution
(including postal code and country)
10801 University Blvd.
Manassas, VA 20110-2209 US
Date of deposit Accession Number
30 June 1999 (30.06.99) PTA-286
C. ADDITIONAL INDICATIONS (leave
blank ifnot applicable) This information
is continued on an additional sheet
Page 11, lines 11 and 13; Page 78,
line 9; Page 79, line 4; Page 80,
line 8; Page 81, line 11; Page
83, line 11
D. DESIGNATED STATES FOR WHICH INDICATIONS
ARE MADE (if the indicators are
not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS
(leave blank ifnot applicable)
The indications listed below will
be submitted to the International
Bureau later (specify the general
nature of the indications e.g.,
Accession
Number of Deposit')
For receiving Office use only For International Bureau use only
This sheet was received with the international application I I ~ This sheet
was received with the International Bureau on: .
Authorized officer
-73-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
INDICATIONS RELATING TO DEPOSITED MICROORGANISM
OR OTHER BIOLOGICAL MATERIAL
A. The indications made below relate
to the deposited microorganism
or other biological material referred
to in the description on page 10,
lines
25 and 31
B. IDENTIFICATION OF DEPOSIT Further
deposits are identified on an additional
sheet 0
Name of depository institution
American Type Culture Collection
Address of depositary institution
(including postal code and country)
10801 University Blvd.
Manassas, VA 20110-2209 US
Date of deposit Accession Number
30 June 1999 (30.06.99) PTA-287
C. ADDITIONAL INDICATIONS (leave
blank if not applicable) This information
is continued on an additional sheet
0
Page 11, lines 12 and 13; Page 78,
line 9; Page 79, line 4; Page 80,
line 8; Page 81, line 11; Page
83, line 11
D. DESIGNATED STATES FOR WHICH INDICATIONS
ARE MADE (if the indicators are
not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS
(leave blank if not applicable)
The indications listed below will
be submitted to the International
Bureau later (specify the general
nature of the indications e.g.,
Accession
Number of Deposit
For receiving Office use only For International Bureau use only
sheet was received with the international application I I 0 This sheet was
received with the International Bureau on:
Authorized officer
-74-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
INDICATIONS RELATING TO DEPOSITED MICROORGANISM
OR OTHER BIOLOGICAL MATERIAL
A. The indications made below relate
to the deposited microorganism
or other biological material referred
to in the description on page
10, lines
25 and 31
B. IDENTIFICATION OF DEPOSIT Further
deposits are identified on an
additional sheet
Name of depository institution
American Type Culture Collection
Address of depositary institution
(including postal code and country)
10801 University Blvd.
Manassas, VA 20110-2209 US
Date of deposit Accession Number
30 June 1999 (30.06.99) ' PTA-288
C. ADDITIONAL INDICATIONS (leave
blank if not applicable) This
information is continued on an
additional sheet
Page 11, lines 12 and 13; Page
78, line 10; Page 79, line 4;
Page 80, line 8; Page 81, line
11; Page 83, line
11
D. DESIGNATED STATES FOR WHICH
INDICATIONS ARE MADE (if the indicators
are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS
(leave blank ifnot applicable)
The indications listed below will
be submitted to the International
Bureau later (specify the general
nature of the indications e.g.,
'Accession
Number of Deposit
For receiving Office use only -.-~ ~- For International Bureau use only
This sheet was received with the international application ~ ~ ~ This sheet
was received with the International Bureau on:
Authorized officer
-75-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
INDICATIONS RELATING TO DEPOSITED MICROORGANISM
OR OTHER BIOLOGICAL MATERIAL
A. The indications made below relate
to the deposited microorganism
or other biological material referred
to in the description on page
11, lines
4, 12 and 14
B. IDENTIFICATION OF DEPOSIT Further
deposits are identified on an
additional sheet 0
Name of depository institution
American Type Culture Collection
Address of depositary institution
(including postal code and country)
10801 University Blvd.
Manassas, VA 20110-2209 US
Date of deposit Accession Number
20 August 1999 (20.08.99) PTA-559
C. ADDITIONAL INDICATIONS (leave
blankifnot applicable) This information
is continued on an additional
sheet
Page 82, line 5; Page 84, lines
4 and 17; Page 85, lines 1 and
15
D. DESIGNATED STATES FOR WHICH
INDICATIONS ARE MADE (if the indicators
are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS
(leave blankifnot applicable)
The indications listed below will
be submitted to the International
Bureau later (specify the general
nature of the indications e.g.,
Accession
Number of Deposit')
For receiving Office use only For International Bureau use only
This sheet was received with the international application I I ~ This sheet
was received with the International Bureau on:
Authorized officer
-76-
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
INDICATIONS RELATING TO DEPOSITED MICROORGANISM
OR OTHER BIOLOGICAL MATERIAL
A. The indications made below relate to the deposited microorganism or other
biological material referred to in the description on page 11, lines
4, 12 and 15
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional
sheet 0
Name of depository institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
10801 University Blvd.
Manassas, VA 20110-2209 US
Date of deposit Accession Number
26 April 2000 (26.04.00) PTA-1721
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is
continued on an additional sheet
Page 82, line 6; Page 84, lines 5 and 18; Page 85, lines 2 and 16
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indicators are not
for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank ifnot applicable)
The indications listed below will be submitted to the International Bureau
later (specify the general nature of the indications e.g., Accession
Number of Deposit')
For receiving Office use only For International Bureau use only
sheet was received with the international application I I ~ This sheet was
received with the International Bureau on:
officer / I n I I Authorized officer
_77_
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
SEQUENCE LISTING
<110> Bidney, Dennis
Duvick, Jon
Hendrick, Carol
Hu, Xu
Lu, Guihua
Crasta, Oswald
<120> Sunflower RhoGAP, LOX, ADH and SCIP -
Polynucleotides and Methods of Use
<130> 35718/202438
<150> US 60/166,128
<151> 1999-11-18
<150> US 60/201,837
<151> 2000-05-03
<160> 10
<170> FastSEQ for Windows Version 4.0
<210> 1
<211> 824
<212> DNA
<213> Helianthus annus
<220>
<221> misc_feature
<222> (0). .(0)
<223> rhoGAP
<221> CDS
<222> (35)...(637)
<400> 1
ttcggcacga gtccaaatcc aatcttcaat cacc atg get gaa gaa caa ctg ccg
Met Ala Glu Glu Gln Leu Pro
1 5
cct gat caa att aaa ctc att cac aag ctt aat ttg ttc aaa atc aaa
103
Pro Asp Gln Ile Lys Leu Ile His Lys Leu Asn Leu Phe Lys Ile Lys
10 15 20
ggc aga gat aaa cac aat cgc aaa atc tta cga att gtc gga aaa aac
151
Gly Arg Asp Lys His Asn Arg Lys Ile Leu Arg Ile Val Gly Lys Asn
25 30 35
ttt cca get aag agt ttg acc gtt gac ctg ttg aaa aaa tat cta gaa
199
Phe Pro Ala Lys Ser Leu Thr Val Asp Leu Leu Lys Lys Tyr Leu Glu
40 45 50 55
gtg aaa att ttc ccc aaa ctt gaa cga ccg ttt gtg gtg gtt tac gtt
247
Val Lys Ile Phe Pro Lys Leu Glu Arg Pro Phe Val Val Val Tyr Val
65 70
1
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
cac act gat gtt cag aag agc gag aat ttc cct gga ata tcc gtt ctc
295
His Thr Asp Val Gln Lys Ser Glu Asn Phe Pro Gly Ile Ser Val Leu
75 80 85
cgg tca gtt tac gac gcg att ccg atg acc gtg aaa caa tat ctt gag
343
Arg Ser Val Tyr Asp Ala Ile Pro Met Thr Val Lys Gln Tyr Leu Glu
90 95 100
gcg gtt tac ttt gtt cat ccg gat ctg cag tcc aga att ttt ctg get
391
Ala Val Tyr Phe Val His Pro Asp Leu Gln Ser Arg Ile Phe Leu Ala
105 110 115
aca ttt ggc cgg ctt atc ttc acc gga ggg tta tat gca aag ctg aga
439
Thr Phe Gly Arg Leu Ile Phe Thr Gly Gly Leu Tyr Ala Lys Leu Arg
120 125 130 135
ttt gtg agt cga ttg gcg tat ctg tgg gaa cat gtg aaa agg aac gag
487
Phe Val Ser Arg Leu Ala Tyr Leu Trp Glu His Val Lys Arg Asn Glu
190 145 150
atc gag atc cca gag ttt gtc tac gat cat gat gag gat ctg gag tac
535
Ile Glu Ile Pro Glu Phe Val Tyr Asp His Asp Glu Asp Leu Glu Tyr
155 160 165
cgt ccg atg atg gat tac ggg ata gag agt gac cac get aga gtt tat
583
Arg Pro Met Met Asp Tyr Gly Ile Glu Ser Asp His Ala Arg Val Tyr
170 175 180
gga gcg ccc gcg gtt gat tcc tct gtg gcg get tat tcc atg agg tgt
631
Gly Ala Pro Ala Val Asp Ser Ser Val Ala Ala Tyr Ser Met Arg Cys
185 190 195
atc tca taggggaaat agttgttttt tcttttgttt ttgaaaatag gtgctaaaag
687
Ile Ser
200
aagtgcaata tatagtattt agcaatattt cgggtgttgt agtatgttga taacgggctt
747
ttcttataac attcattgtt ctagttttct tttgtaaaaa ttatttgata aattctttgt
807
aaaaaaaaaa aaaaaaa
824
<210> 2
<211> 201
<212> PRT
<213> Helianthus annus
<400> 2
Met Ala Glu Glu Gln Leu Pro Pro Asp Gln Ile Lys Leu Ile His Lys
1 5 10 15
Leu Asn Leu Phe Lys Ile Lys Gly Arg Asp Lys His Asn Arg Lys Ile
20 25 30
Leu Arg Ile Val Gly Lys Asn Phe Pro Ala Lys Ser Leu Thr Val Asp
35 40 45
Leu Leu Lys Lys Tyr Leu Glu Val Lys Ile Phe Pro Lys Leu Glu Arg
50 55 60
Pro Phe Val Val Val Tyr Val His Thr Asp Val Gln Lys Ser Glu Asn
2
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
65 70 75 80
Phe Pro Gly Ile Ser Val Leu Arg Ser Val Tyr Asp Ala Ile Pro Met
85 90 95
Thr Val Lys Gln Tyr Leu Glu Ala Val Tyr Phe Val His Pro Asp Leu
100 105 110
Gln Ser Arg Ile Phe Leu Ala Thr Phe Gly Arg Leu Ile Phe Thr Gly
115 120 125
Gly Leu Tyr Ala Lys Leu Arg Phe Val Ser Arg Leu Ala Tyr Leu Trp
130 135 190
Glu His Val Lys Arg Asn Glu Ile Glu Ile Pro Glu Phe Val Tyr Asp
145 150 155 160
His Asp Glu Asp Leu Glu Tyr Arg Pro Met Met Asp Tyr Gly Ile Glu
165 170 175
Ser Asp His Ala Arg Val Tyr Gly Ala Pro Ala Val Asp Ser Ser Val
180 185 190
Ala Ala Tyr Ser Met Arg Cys Ile Ser
195 200
<210> 3
<211> 2993
<212> DNA
<213> Helianthus annus
<220>
<221> misc_feature
<222> (0). .(0)
<223> lox cDNA
<221> CDS
<222> (18)...(2720)
<400> 3
ggcacgagaa gaaaacc atg ttg aat tct caa atc aac cat tct cac cct
Met Leu Asn Ser Gln Ile Asn His Ser His Pro
1 5 10
ctt aac aac cta cta cca atc cgc aaa gcc ttt gtc cat ggt gac acc
98
Leu Asn Asn Leu Leu Pro Ile Arg Lys Ala Phe Val His Gly Asp Thr
15 20 25
act aac cat tcc tcc tcc aac gcc tac tcc ccc gcc aac ctt cgc caa
146
Thr Asn His Ser Ser Ser Asn Ala Tyr Ser Pro Ala Asn Leu Arg Gln
30 35 40
cac gcg tcc acc aag aaa tcc aat get acc cgt gca cga tcc acc tca
194
His Ala Ser Thr Lys Lys Ser Asn Ala Thr Arg Ala Arg Ser Thr Ser
45 50 55
act gcg ggt aac att aaa gcc ata tca atc ccc ttt ctt acc aag gag
242
Thr Ala Gly Asn Ile Lys Ala Ile Ser Ile Pro Phe Leu Thr Lys Glu
65 70 75
acc acc gtc aag tgt gtc atc acc gtc caa cca acc att agt tcc gcc
290
Thr Thr Val Lys Cys Val Ile Thr Val Gln Pro Thr Ile Ser Ser Ala
80 85 90
att get ggt gta ggc gtt ggt ggt att gtt gat ggt gtt tct aat ctt
338
Ile Ala Gly Val Gly Val Gly Gly Ile Val Asp Gly Val Ser Asn Leu
95 100 105
3
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
cta ggg ttg tca ttt ttg ttg gag ctc gtt tca aat gac ctc gat tca
386
Leu Gly Leu Ser Phe Leu Leu Glu Leu Val Ser Asn Asp Leu Asp Ser
110 115 120
aaa gga aac caa aag aca gtg aag get tat gca aga tac aac gca ctg
934
Lys Gly Asn Gln Lys Thr Val Lys Ala Tyr Ala Arg Tyr Asn Ala Leu
125 130 135
gat ttg gac att agc gtg tac aca tac aaa tgc gac ttc gac gtc cct
482
Asp Leu Asp Ile Ser Val Tyr Thr Tyr Lys Cys Asp Phe Asp Val Pro
190 145 150 155
gaa gat ttt ggg gag ata gga get gtg ttg gta gaa aat gag tat agc
530
Glu Asp Phe Gly Glu Ile Gly Ala Val Leu Val Glu Asn Glu Tyr Ser
160 165 170
aag aag atg ttt ttc aag aac att gtt ctt aac aac ggt gtt acc ttc
578
Lys Lys Met Phe Phe Lys Asn Ile Val Leu Asn Asn Gly Val Thr Phe
175 180 185
aca tgc gag tca tgg gtt cac tcc aaa tac gat aac cct gag aaa aga
626
Thr Cys Glu Ser Trp Val His Ser Lys Tyr Asp Asn Pro Glu Lys Arg
190 195 200
ata ttt ttc acc gac aag tcg tat cta ccg ttg gaa acg ccg acg gca
679
Ile Phe Phe Thr Asp Lys Ser Tyr Leu Pro Leu Glu Thr Pro Thr Ala
205 210 215
ctg aag ccg tta cga gag aaa gat atg gaa tcg ctt cga gga aac ggc
722
Leu Lys Pro Leu Arg Glu Lys Asp Met Glu Ser Leu Arg Gly Asn Gly
220 225 230 235
gaa gga gaa cgt aaa tca ttc gag cgg ata tat gat tat gat gtg tac
770
Glu Gly Glu Arg Lys Ser Phe Glu Arg Ile Tyr Asp Tyr Asp Val Tyr
240 295 250
aac gat ctc gga gat ccg gat gga agc tta gat cta gca cgg ccg gtg
818
Asn Asp Leu Gly Asp Pro Asp Gly Ser Leu Asp Leu Ala Arg Pro Val
255 260 265
ctc ggt ggc gag aca cat ccg tac cct agg cgg tgc cgt act ggt cgc
866
Leu Gly Gly Glu Thr His Pro Tyr Pro Arg Arg Cys Arg Thr Gly Arg
270 275 280
aaa atg tcc tct aaa gat ccg tta aca gaa agc aga act acg ctc cct
919
Lys Met Ser Ser Lys Asp Pro Leu Thr Glu Ser Arg Thr Thr Leu Pro
285 290 295
ttt tat gta cct gcg gat gaa gat ttt tca gag ata aag agt gtg aac
962
Phe Tyr Val Pro Ala Asp Glu Asp Phe Ser Glu Ile Lys Ser Val Asn
300 305 310 315
ttt gga gca aaa act tta tac tct gtg ctt cat gga gtt gta cca atg
1010
4
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Phe Gly Ala Lys Thr Leu Tyr Ser Val Leu His Gly Val Val Pro Met
320 325 330
cta gac tca att gta aca gac aaa gac aag ggg ttt cca tta ttc aca
1058
Leu Asp Ser Ile Val Thr Asp Lys Asp Lys Gly Phe Pro Leu Phe Thr
335 340 395
tcc ata gat ttg ctt tat aat gaa ggt gtt aat gtt cct tct cct gac
1106
Ser Ile Asp Leu Leu Tyr Asn Glu Gly Val Asn Val Pro Ser Pro Asp
350 355 360
aat gga att cta agt get tta cct aga ctt gtc aaa ggg get act gat
1154
Asn Gly Ile Leu Ser Ala Leu Pro Arg Leu Val Lys Gly Ala Thr Asp
365 370 375
gcc gca aat acc gtt atc aag ttc gag acc ccc gaa acc att gat aga
1202
Ala Ala Asn Thr Val Ile Lys Phe Glu Thr Pro Glu Thr Ile Asp Arg
380 385 390 395
gac gca ttc tca tgg ttc cgt gat gaa gag ttc tgc cgg caa atg ctt
1250
Asp Ala Phe Ser Trp Phe Arg Asp Glu Glu Phe Cys Arg Gln Met Leu
400 405 410
gcc ggt att aat cct tgt cgc ata caa ttg gtt acg gaa tgg cca ttg
1298
Ala Gly Ile Asn Pro Cys Arg Ile Gln Leu Val Thr Glu Trp Pro Leu
415 920 425
atg agt aaa ctg gac cct gaa atc tat gga cca get gag tca gca att
1346
Met Ser Lys Leu Asp Pro Glu Ile Tyr Gly Pro Ala Glu Ser Ala Ile
930 435 940
aca aag gag att gta gag gaa gag att aaa ggt ttc atg act ctt gag
1394
Thr Lys Glu Ile Val Glu Glu Glu Ile Lys Gly Phe Met Thr Leu Glu
445 450 955
gag get tta gca caa aag aag ctg ttt atg ctg gat tat cat gat ctg
1492
Glu Ala Leu Ala Gln Lys Lys Leu Phe Met Leu Asp Tyr His Asp Leu
460 465 470 475
ctc ttg cct tat gtt aac aaa acg gag get gaa ggg aga act ttg tat
1490
Leu Leu Pro Tyr Val Asn Lys Thr Glu Ala Glu Gly Arg Thr Leu Tyr
480 485 490
ggt tca aga act tta atg ttc ctt act cct get gga aca tta agg cca
1538
Gly Ser Arg Thr Leu Met Phe Leu Thr Pro Ala Gly Thr Leu Arg Pro
495 500 505
cta gcc att gag ctg act cgc cca cca att gat ggg aaa cca cag tgg
1586
Leu Ala Ile Glu Leu Thr Arg Pro Pro Ile Asp Gly Lys Pro Gln Trp
510 515 520
aaa cat gtt tac aca ccc get tgg gat get aca ggt gca tgg ctt tgg
1634
Lys His Val Tyr Thr Pro Ala Trp Asp Ala Thr Gly Ala Trp Leu Trp
525 530 535
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
aag cta gcc aag get cat gtc ctt gcc cat gat tct agc tat cac caa
1682
Lys Leu Ala Lys Ala His Val Leu Ala His Asp Ser Ser Tyr His Gln
540 545 550 555
ctt gtt agc cat tgg cta aga aca cat tgt get acc gaa cct tac att
1730
Leu Val Ser His Trp Leu Arg Thr His Cys Ala Thr Glu Pro Tyr Ile
560 565 570
att get acc aat cgc caa ctc agt caa atg cat cca att cga cga ttt
1778
Ile Ala Thr Asn Arg Gln Leu Ser Gln Met His Pro Ile Arg Arg Phe
575 580 585
cta ctc cct cac ttt cgt tac act atg caa att aat tct cta get aga
1826
Leu Leu Pro His Phe Arg Tyr Thr Met Gln Ile Asn Ser Leu Ala Arg
590 595 600
ctt tta ctc gtc aat gcc atg ggt atc ata gag tca aca ttt tct cct
1874
Leu Leu Leu Val Asn Ala Met Gly Ile Ile Glu Ser Thr Phe Ser Pro
605 610 615
gga aga tat tgt atg caa att tcc tct gat gca tat gat cag caa tgg
1922
Gly Arg Tyr Cys Met Gln Ile Ser Ser Asp Ala Tyr Asp Gln Gln Trp
620 625 630 635
cgt ttt gat cat gaa gcg ctt ccg gcc gac cta att agc agg ggt atg
1970
Arg Phe Asp His Glu Ala Leu Pro Ala Asp Leu Ile Ser Arg Gly Met
690 645 650
gcg gtt gaa gat cca acc gca cca tat ggt gta aaa cta aca atc gag
2018
Ala Val Glu Asp Pro Thr Ala Pro Tyr Gly Val Lys Leu Thr Ile Glu
655 660 665
gat tac cca tat gca aat gat ggt tta ctc att tat gat acc att aaa
2066
Asp Tyr Pro Tyr Ala Asn Asp Gly Leu Leu Ile Tyr Asp Thr Ile Lys
670 675 680
caa tgg gca act tct tat gtc aac cac tat tac cca cca gcg aat cta
2114
Gln Trp Ala Thr Ser Tyr Val Asn His Tyr Tyr Pro Pro Ala Asn Leu
685 690 695
gtg gaa tct gat gaa gag ctt caa gca tgg tgg aat gaa atc cgt aca
2162
Val Glu Ser Asp Glu Glu Leu Gln Ala Trp Trp Asn Glu Ile Arg Thr
700 705 710 715
gtt ggt cat gga gat aag aaa gat gaa cca tgg tgg cca caa ctc aaa
2210
Val Gly His Gly Asp Lys Lys Asp Glu Pro Trp Trp Pro Gln Leu Lys
720 725 730
acc caa gat gat ttg att gga att gtt tca acc atc ttg tgg gtg acc
2258
Thr Gln Asp Asp Leu Ile Gly Ile Val Ser Thr Ile Leu Trp Val Thr
735 790 745
6
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
tct ggt caa cat tca gca gtc aac ttc ggt caa tat gat ttc gcg ggt
2306
Ser Gly Gln His Ser Ala Val Asn Phe Gly Gln Tyr Asp Phe Ala Gly
750 755 760
tat ttc cct aac agg ccg aca att tcc aga acc aaa atg ccc aac gaa
2354
Tyr Phe Pro Asn Arg Pro Thr Ile Ser Arg Thr Lys Met Pro Asn Glu
765 770 775
gac ccc aca gac gaa gaa tgg cag tcg ttt ata aag cga ccc gag gat
2402
Asp Pro Thr Asp Glu Glu Trp Gln Ser Phe Ile Lys Arg Pro Glu Asp
780 785 790 795
get tta ttg aaa tgc ttc cca tcc caa atc caa get aca aaa gtg atg
2450
Ala Leu Leu Lys Cys Phe Pro Ser Gln Ile Gln Ala Thr Lys Val Met
800 805 810
gcg att ttg gat gtt tta tca agt cat tca cca gat gaa gaa tat atc
2498
Ala Ile Leu Asp Val Leu Ser Ser His Ser Pro Asp Glu Glu Tyr Ile
815 820 825
ggt gga aat att gag gcg gca tgg gag gcg gag cct get ata aaa gca
2546
Gly Gly Asn Ile Glu Ala Ala Trp Glu Ala Glu Pro Ala Ile Lys Ala
830 835 840
gcc ttt gag gag ttc cgt gga agg ctc aat gag ctg gaa gca atc ata
2594
Ala Phe Glu Glu Phe Arg Gly Arg Leu Asn Glu Leu Glu Ala Ile Ile
845 850 855
gac tca agg aac acg gat ccc aat ttg aag aat cgt agt ggt gcg ggg
2642
Asp Ser Arg Asn Thr Asp Pro Asn Leu Lys Asn Arg Ser Gly Ala Gly
860 865 870 875
ttg gtt ccg tat caa ctt ctc aaa ccg tat tct gaa aaa ggt gtg acc
2690 '
Leu Val Pro Tyr Gln Leu Leu Lys Pro Tyr Ser Glu Lys Gly Val Thr
880 885 890
ggg aga ggt gtt cca aac agc ata tcc att tagttggatt ggtttggttc
2740
Gly Arg Gly Val Pro Asn Ser Ile Ser Ile
895 900
ctaatgctcg aggaatagtc tatgtggtgt aataaggcca tgatccatgg tttagttgtg
2800
ttttattgtt atttggaata agttcactta tgtgccttct tgtattataa gccaacatta
2860
tcgaacttta tattgtatgt gtattattgt tatttggaat aacatggcat agcaccattc
2920
ttgttaaaaa aaaaaaaaaa aaa
2993
<210> 9
<211> 901
<212> PRT
<213> Helianthus annus
<400> 4
Met Leu Asn Ser Gln Ile Asn His Ser His Pro Leu Asn Asn Leu Leu
1 5 10 15
7
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Pro Ile Arg Lys Ala Phe Val His Gly Asp Thr Thr Asn His Ser Ser
20 25 30
Ser Asn Ala Tyr Ser Pro Ala Asn Leu Arg Gln His Ala Ser Thr Lys
35 40 45
Lys Ser Asn Ala Thr Arg Ala Arg Ser Thr Ser Thr Ala Gly Asn Ile
50 55 60
Lys Ala Ile Ser Ile Pro Phe Leu Thr Lys Glu Thr Thr Val Lys Cys
65 70 75 80
Val Ile Thr Val Gln Pro Thr Ile Ser Ser Ala Ile Ala Gly Val Gly
85 90 95
Val Gly Gly Ile Val Asp Gly Val Ser Asn Leu Leu Gly Leu Ser Phe
100 105 110
Leu Leu Glu Leu Val Ser Asn Asp Leu Asp Ser Lys Gly Asn Gln Lys
115 120 125
Thr Val Lys Ala Tyr Ala Arg Tyr Asn Ala Leu Asp Leu Asp Ile Ser
130 135 ' 140
Val Tyr Thr Tyr Lys Cys Asp Phe Asp Val Pro Glu Asp Phe Gly Glu
145 150 155 160
Ile Gly Ala Val Leu Val Glu Asn Glu Tyr Ser Lys Lys Met Phe Phe
165 170 175
Lys Asn Ile Val Leu Asn Asn Gly Val Thr Phe Thr Cys Glu Ser Trp
180 185 190
Val His Ser Lys Tyr Asp Asn Pro Glu Lys Arg Ile Phe Phe Thr Asp
195 200 205
Lys Ser Tyr Leu Pro Leu Glu Thr Pro Thr Ala Leu Lys Pro Leu Arg
210 215 220
Glu Lys Asp Met Glu Ser Leu Arg Gly Asn Gly Glu Gly Glu Arg Lys
225 230 235 240
Ser Phe Glu Arg Ile Tyr Asp Tyr Asp Val Tyr Asn Asp Leu Gly Asp
245 250 255
Pro Asp Gly Ser Leu Asp Leu Ala Arg Pro Val Leu Gly Gly Glu Thr
260 265 270
His Pro Tyr Pro Arg Arg Cys Arg Thr Gly Arg Lys Met Ser Ser Lys
275 280 285
Asp Pro Leu Thr Glu Ser Arg Thr Thr Leu Pro Phe Tyr Val Pro Ala
290 295 300
Asp Glu Asp Phe Ser Glu Ile Lys Ser Val Asn Phe Gly Ala Lys Thr
305 310 315 320
Leu Tyr Ser Val Leu His Gly Val Val Pro Met Leu Asp Ser Ile Val
325 330 335
Thr Asp Lys Asp Lys Gly Phe Pro Leu Phe Thr Ser Ile Asp Leu Leu
340 395 350
Tyr Asn Glu Gly Val Asn Val Pro Ser Pro Asp Asn Gly Ile Leu Ser
355 360 365
Ala Leu Pro Arg Leu Val Lys Gly Ala Thr Asp Ala Ala Asn Thr Val
370 375 380
Ile Lys Phe Glu Thr Pro Glu Thr Ile Asp Arg Asp Ala Phe Ser Trp
385 390 395 400
Phe Arg Asp Glu Glu Phe Cys Arg Gln Met Leu Ala Gly Ile Asn Pro
405 410 415
Cys Arg Ile Gln Leu Val Thr Glu Trp Pro Leu Met Ser Lys Leu Asp
420 425 430
Pro Glu Ile Tyr Gly Pro Ala Glu Ser Ala Ile Thr Lys Glu Ile Val
935 440 445
Glu Glu Glu Ile Lys Gly Phe Met Thr Leu Glu Glu Ala Leu Ala Gln
450 455 460
Lys Lys Leu Phe Met Leu Asp Tyr His Asp Leu Leu Leu Pro Tyr Val
465 470 975 480
Asn Lys Thr Glu Ala Glu Gly Arg Thr Leu Tyr Gly Ser Arg Thr Leu
485 990 495
Met Phe Leu Thr Pro Ala Gly Thr Leu Arg Pro Leu Ala Ile Glu Leu
500 505 510
Thr Arg Pro Pro Ile Asp Gly Lys Pro Gln Trp Lys His Val Tyr Thr
515 520 525
Pro Ala Trp Asp Ala Thr Gly Ala Trp Leu Trp Lys Leu Ala Lys Ala
530 535 540
His Val Leu Ala His Asp Ser Ser Tyr His Gln Leu Val Ser His Trp
g
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
545 550 555 560
Leu Arg Thr His Cys Ala Thr Glu Pro Tyr Ile Ile Ala Thr Asn Arg
565 570 575
Gln Leu Ser Gln Met His Pro Ile Arg Arg Phe Leu Leu Pro His Phe
580 585 590
Arg Tyr Thr Met Gln Ile Asn Ser Leu Ala Arg Leu Leu Leu Val Asn
595 600 605
Ala Met Gly Ile Ile Glu Ser Thr Phe Ser Pro Gly Arg Tyr Cys Met
610 615 620
Gln Ile Ser Ser Asp Ala Tyr Asp Gln Gln Trp Arg Phe Asp His Glu
625 630 635 640
Ala Leu Pro Ala Asp Leu Ile Ser Arg Gly Met Ala Val Glu Asp Pro
645 650 655
Thr Ala Pro Tyr Gly Val Lys Leu Thr Ile Glu Asp Tyr Pro Tyr Ala
660 665 670
Asn Asp Gly Leu Leu Ile Tyr Asp Thr Ile Lys Gln Trp Ala Thr Ser
675 680 685
Tyr Val Asn His Tyr Tyr Pro Pro Ala Asn Leu Val Glu Ser Asp Glu
690 695 700
Glu Leu Gln Ala Trp Trp Asn Glu Ile Arg Thr Val Gly His Gly Asp
705 710 715 720
Lys Lys Asp Glu Pro Trp Trp Pro Gln Leu Lys Thr Gln Asp Asp Leu
725 730 735
Ile Gly Ile Val Ser Thr Ile Leu Trp Val Thr Ser Gly Gln His Ser
740 795 750
Ala Val Asn Phe Gly Gln Tyr Asp Phe Ala Gly Tyr Phe Pro Asn Arg
755 760 765
Pro Thr Ile Ser Arg Thr Lys Met Pro Asn Glu Asp Pro Thr Asp Glu
770 775 780
Glu Trp Gln Ser Phe Ile Lys Arg Pro Glu Asp Ala Leu Leu Lys Cys
785 790 795 800
Phe Pro Ser Gln Ile Gln Ala Thr Lys Val Met Ala Ile Leu Asp Val
805 810 815
Leu Ser Ser His Ser Pro Asp Glu Glu Tyr Ile Gly Gly Asn Ile Glu
820 825 830
Ala Ala Trp Glu Ala Glu Pro Ala Ile Lys Ala Ala Phe Glu Glu Phe
835 890 845
Arg Gly Arg Leu Asn Glu Leu Glu Ala Ile Ile Asp Ser Arg Asn Thr
850 855 860
Asp Pro Asn Leu Lys Asn Arg Ser Gly Ala Gly Leu Val Pro Tyr Gln
865 870 875 880
Leu Leu Lys Pro Tyr Ser Glu Lys Gly Val Thr Gly Arg Gly Val Pro
885 890 895
Asn Ser Ile Ser Ile
900
<210> 5
<211> 883
<212> DNA
<213> Helianthus annus
<220>
<221> promoter
<222> (1)...(880)
<223> lox promoter
<221> misc_feature
<222> (322)...(327)
<223> w-box
<221> misc_feature
<222> (722)...(727)
<223> G-box
<221> misc_feature
<222> (808)...(811)
<223> TATA box
9
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
<400> 5
agggcacgcg tggtcgacgg accgggctgg gtatctcatt catcttagct cggttttgga
cgtggtttag ttcgttgcgt acctcttcca acatagaaca caaacccaca aataagtaca
120
taaaacccgc tcattttatt agatttattt ctagtccaaa atacgaaaaa atcattgtcc
180
tatatttgtg aaaaatgcta ttttcactta tttttccaac aacacataca cagagagggc
240
aacaatcggt taacaaactc accagagttg tgaaaattat gaggacttct atctgtcatg
300
caatttttta tatatttcct tttgaccaaa acatgtatac atgactaact aaaaatatag
360
ttgcgagttg gaaaagggtt atacactata actcatattt acacaatatt gccttgaaca
420
ttattaacta attacacggt gttgaataat tttgataaaa acttttctat gtgttgaggt
480
atatctgaac tattaaaata aataccatta atacttcaag attataacat gagaacatta
540
catatattgt gattttatat tataatttaa taattatttt tttttgaaag gcataattta
600
ataattataa gcgacaatac ttctacgttt atagtactag gtactttttc caacccacaa
660
tcaaatgcat tctagccgta gattgtaaat tattaatgca accctgaaca ataatgcata
720
acacgtgaaa tcaatgcaga aatgtatcat tcttatccga tgttttccca ttaaataaaa
780
accttaaaat atagcacatt tcctctctat aaatagagct attttttcaa cttccagatc
840
acacaaaaca agagtgagag tagagtgact aaagaaaacc atg
883
<210> 6
<211> 1403
<212> DNA
<213> Helianthus annus
<220>
<221> misc_feature
<222> (0). .(0)
<223> ADH cDNA
<221> CDS
<222> (74)...(1216)
<400> 6
ttcggcacga gccaaaactc acaatttaat ctcatttcaa gaatattctc tctttcaccg
atcaaacaaa agt atg tcg tcg acc act aca ggc caa gtt att cga tgc
109
Met Ser Ser Thr Thr Thr Gly Gln Val Ile Arg Cys
1 5 10
aaa gcc gcg gtg acg tgg gaa gcc gga aaa ccg ctg gtg atc gaa gaa
157
Lys Ala Ala Val Thr Trp Glu Ala Gly Lys Pro Leu Val Ile Glu Glu
15 20 25
gtg gag gtg gcg cca ccg cag aaa atg gaa gtc cgg att aag atc ctc
205
Val Glu Val Ala Pro Pro Gln Lys Met Glu Val Arg Ile Lys Ile Leu
30 35 40
ttc act tcc ctc tgc cac act gat gtt tac ttc tgg gaa gcc aaa gga
253
Phe Thr Ser Leu Cys His Thr Asp Val Tyr Phe Trp Glu Ala Lys Gly
1~
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
95 50 55 60
caa aat cct gta ttc cca aga att tta gga cat gaa get gga ggg gtt
301
Gln Asn Pro Val Phe Pro Arg Ile Leu Gly His Glu Ala Gly Gly Val
65 70 75
gtg gag agt gtt ggg gaa gga gtg act gat ctt cag cca ggg gat cat
399
Val Glu Ser Val Gly Glu Gly Val Thr Asp Leu Gln Pro Gly Asp His
80 85 90
gtt ctt ccc gtt ttc acc gga gaa tgc aaa gag tgt get cac tgt aag
397
Val Leu Pro Val Phe Thr Gly Glu Cys Lys Glu Cys Ala His Cys Lys
95 100 105
tcc gaa gag agc aac atg tgt gac ctt ctc agg atc aac acc gac agg
495
Ser Glu Glu Ser Asn Met Cys Asp Leu Leu Arg Ile Asn Thr Asp Arg
110 115 120
gga gtc atg ctt cac gat cag aaa tct cga ttc tcg atc aac ggc aaa
993
Gly Val Met Leu His Asp Gln Lys Ser Arg Phe Ser Ile Asn Gly Lys
125 130 135 140
ccc atc ttc cat ttt gtg ggg act tct act ttc agc gag tac acg gtt
541
Pro Ile Phe His Phe Val Gly Thr Ser Thr Phe Ser Glu Tyr Thr Val
195 150 155
gtt cat gtt gga tgt ctt gca aag atc aac cct ctt gcc cct ctt gat
589
Val His Val Gly Cys Leu Ala Lys Ile Asn Pro Leu Ala Pro Leu Asp
160 165 170
aaa gtt tgt gtt ctc agc tgt ggg atc tcc aca ggg ctg ggt get act
637
Lys Val Cys Val Leu Ser Cys Gly Ile Ser Thr Gly Leu Gly Ala Thr
175 180 185
ttg aat gtt gca aaa ccg aaa aaa ggc tct tcg gtg gcg gtt ttc ggt
685
Leu Asn Val Ala Lys Pro Lys Lys Gly Ser Ser Val Ala Val Phe Gly
190 195 200
ctg ggg gca gtg gga ctt get get get gaa ggt gca aga att tct ggg
733
Leu Gly Ala Val Gly Leu Ala Ala Ala Glu Gly Ala Arg Ile Ser Gly
205 210 215 220
get tca aga atc att ggt gtt gat ctc aat gcc aat aga ttc gag ctt
781
Ala Ser Arg Ile Ile Gly Val Asp Leu Asn Ala Asn Arg Phe Glu Leu
225 230 235
gca aag aaa ttt ggg gtt aca gag ttt gtg aac cca aaa gat tat aag
829
Ala Lys Lys Phe Gly Val Thr Glu Phe Val Asn Pro Lys Asp Tyr Lys
240 245 250
aag ccg gtg caa gaa gtg att gca gag atg aca aat gga gga gtt gac
877
Lys Pro Val Gln Glu Val Ile Ala Glu Met Thr Asn Gly Gly Val Asp
255 260 265
11
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
agg agt gtt gaa tgc act ggt cat att gat get atg atc tct get ttt
925
Arg Ser Val Glu Cys Thr Gly His Ile Asp Ala Met Ile Ser Ala Phe
270 275 280
gaa tgt gtt cat gat ggg tgg ggt gtt get gtt cta gta ggt gtt ccg
973
Glu Cys Val His Asp Gly Trp Gly Val Ala Val Leu Val Gly Val Pro
285 290 , 295 300
cat aaa gat gcc gtg ttc aag acc agt ccc atg aat ctg ttg aac gaa
1021
His Lys Asp Ala Val Phe Lys Thr Ser Pro Met Asn Leu Leu Asn Glu
305 310 315
agg act ctg aag ggt acc ttc ttt gga aac tat aaa ccg cga tct gat
1069
Arg Thr Leu Lys Gly Thr Phe Phe Gly Asn Tyr Lys Pro Arg Ser Asp
320 325 330
att cct tcg gtt gtc gaa aag tat atg aac aag gaa ctt gag gtg gag
1117
Ile Pro Ser Val Val Glu Lys Tyr Met Asn Lys Glu Leu Glu Val Glu
335 340 345
aag ttc ata aca cat gaa gtg cca ttt tca gag atc aat aag ccc ttt
1165
Lys Phe Ile Thr His Glu Val Pro Phe Ser Glu Ile Asn Lys Pro Phe
350 355 360
gac ttg atg ctt aaa ggt gaa ggt ctt cgt tgc att att cga atg gat
1213
Asp Leu Met Leu Lys Gly Glu Gly Leu Arg Cys Ile Ile Arg Met Asp
365 370 375 380
gcc taaataattt caaactgtgc aagagagagc agtaggagtc gtctattcgt
1266
Ala
aaagatatat gtgtgtgttc tcgtctctca tcgtcgtaaa tgtgtcctta agatcttggt
1326
ttgttaattg ttacccataa aagattttga atttgaataa caatagaaat tgatgtctaa
1386
aaaaaaaaaa aaaaaaa
1403
<210> 7
<211> 381
<212> PRT
<213> Helianthus annus ADH
<400> 7
Met Ser Ser Thr Thr Thr Gly Gln Val Ile Arg Cys Lys Ala Ala Val
1 5 10 15
Thr Trp Glu Ala Gly Lys Pro Leu Val Ile Glu Glu Val Glu Val Ala
20 25 30
Pro Pro Gln Lys Met Glu Val Arg Ile Lys Ile Leu Phe Thr Ser Leu
35 40 45
Cys His Thr Asp Val Tyr Phe Trp Glu Ala Lys Gly Gln Asn Pro Val
50 55 60
Phe Pro Arg Ile Leu Gly His Glu Ala Gly Gly Val Val Glu Ser Val
65 70 75 80
Gly Glu Gly Val Thr Asp Leu Gln Pro Gly Asp His Val Leu Pro Val
85 90 95
Phe Thr Gly Glu Cys Lys Glu Cys Ala His Cys Lys Ser Glu Glu Ser
100 105 110
12
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
Asn Met Cys Asp Leu Leu Arg Ile Asn Thr Asp Arg Gly Val Met Leu
115 120 125
His Asp Gln Lys Ser Arg Phe Ser Ile Asn Gly Lys Pro Ile Phe His
130 135 190
Phe Val Gly Thr Ser Thr Phe Ser Glu Tyr Thr Val Val His Val Gly
145 150 155 160
Cys~Leu Ala Lys Ile Asn Pro Leu Ala Pro Leu Asp Lys Val Cys Val
165 170 175
Leu Ser Cys Gly Ile Ser Thr Gly Leu Gly Ala Thr Leu Asn Val Ala
180 185 190
Lys Pro Lys Lys Gly Ser Ser Val Ala Val Phe Gly Leu Gly Ala Val
195 200 205
Gly Leu Ala Ala Ala Glu Gly Ala Arg Ile Ser Gly Ala Ser Arg Ile
210 215 220
Ile Gly Val Asp Leu Asn Ala Asn Arg Phe Glu Leu Ala Lys Lys Phe
225 230 235 290
Gly Val Thr Glu Phe Val Asn Pro Lys Asp Tyr Lys Lys Pro Val Gln
245 250 255
Glu Val Ile Ala Glu Met Thr Asn Gly Gly Val Asp Arg Ser Val Glu
260 265 270
Cys Thr Gly His Ile Asp Ala Met Ile Ser Ala Phe Glu Cys Val His
275 280 285
Asp Gly Trp Gly Val Ala Val Leu Val Gly Val Pro His Lys Asp Ala
290 295 300
Val Phe Lys Thr Ser Pro Met Asn Leu Leu Asn Glu Arg Thr Leu Lys
305 310 315 320
Gly Thr Phe Phe Gly Asn Tyr Lys Pro Arg Ser Asp Ile Pro Ser Val
325 330 335
Val Glu Lys Tyr Met Asn Lys Glu Leu Glu Val Glu Lys Phe Ile Thr
340 395 350
His Glu Val Pro Phe Ser Glu Ile Asn Lys Pro Phe Asp Leu Met Leu
355 360 365
Lys Gly Glu Gly Leu Arg Cys Ile Ile Arg Met Asp Ala
370 375 380
<210> 8
<211> 747
<212> DNA
<213> Helianthus annus
<220>
<221> misc_feature
<222> (0)...(0)
<223> SCIP-1 cDNA
<221> CDS
<222> (15)...(518)
<400> 8
ttcggcacga gcaa atg gcg aac gca agc gat gag ttc aga cta gcg tct
Met Ala Asn Ala Ser Asp Glu Phe Arg Leu Ala Ser
1 5 10
tcc ggc atc gat cat gaa ggc cga cta cca cga aaa tac acc ggt gac
98
Ser Gly Ile Asp His Glu Gly Arg Leu Pro Arg Lys Tyr Thr Gly Asp
15 20 25
ggt caa ggt aca aaa aaa gac ata tca cca ccg tta gaa tgg tac aac
146
Gly Gln Gly Thr Lys Lys Asp Ile Ser Pro Pro Leu Glu Trp Tyr Asn
30 35 40
gtt ccg gag ggg aca aaa aca cta gca cta gtg gtg gag gac atc gat
194
Val Pro Glu Gly Thr Lys Thr Leu Ala Leu Val Val Glu Asp Ile Asp
13
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
45 50 55 60
gca ccg gac cca gaa gcg ccg ctg gtt ccg tgg act gtg tgg gtg gtg
242
Ala Pro Asp Pro Glu Ala Pro Leu Val Pro Trp Thr Val Trp Val Val
65 70 75
gtc aat ata cca cct act ttg aag ggg ctc cca gag gga ttt tcc ggg
290
Val Asn Ile Pro Pro Thr Leu Lys Gly Leu Pro Glu Gly Phe Ser Gly
80 85 90
aaa gag ggg gac atg ggt ggc gat tat get aat gtt aaa gaa gga cat
338
Lys Glu Gly Asp Met Gly Gly Asp Tyr Ala Asn Val Lys Glu Gly His
95 100 105
aat gac ttt aag gtg cct gga tgg cgc gca ccg aag atg ccc tca tcc
386
Asn Asp Phe Lys Val Pro Gly Trp Arg Ala Pro Lys Met Pro Ser Ser
110 115 120
gga cac cgg ttc gag ttt aag ctg tat gcg ttg gat gaa caa gtt gag
934
Gly His Arg Phe Glu Phe Lys Leu Tyr Ala Leu Asp Glu Gln Val Glu
125 130 135 140
ttg ggg aat aag gtg act aag gag aag ttg ctg gag gcg att gat ggc
482
Leu Gly Asn Lys Val Thr Lys Glu Lys Leu Leu Glu Ala Ile Asp Gly
195 150 155
cat gtg gtt ggg gag get gtt ctg atg gcc gta aat taaattgaga
528
His Val Val Gly Glu Ala Val Leu Met Ala Val Asn
160 165
atggtttata tatatgttag ttgtgtgact tgtgtcatgt gtgatgttct tgttttaacg
588
tattttgaaa cagaagtgac gagagagaga gagtgtttgt tgtgtgtttt tcttgagaga
648
tcgtgaatta attatgctgt tttgcttcaa ggaatcaagc tttataaagt aaaatacaaa
708
tgtaatgctt caaccgagct aaaaaaaaaa aaaaaaaaa
747
<210> 9
<211> 168
<212> PRT
<213> Helianthus annus SCIP-1
<400> 9
Met Ala Asn Ala Ser Asp Glu Phe Arg Leu Ala Ser Ser Gly Ile Asp
1 5 ' 10 15
His Glu Gly Arg Leu Pro Arg Lys Tyr Thr Gly Asp Gly Gln Gly Thr
20 25 30
Lys Lys Asp Ile Ser Pro Pro Leu Glu Trp Tyr Asn Val Pro Glu Gly
35 90 45
Thr Lys Thr Leu Ala Leu Val Val Glu Asp Ile Asp Ala Pro Asp Pro
50 55 60
Glu Ala Pro Leu Val Pro Trp Thr Val Trp Val Val Val Asn Ile Pro
65 70 75 80
Pro Thr Leu Lys Gly Leu Pro Glu Gly Phe Ser Gly Lys Glu Gly Asp
85 90 95
Met Gly Gly Asp Tyr Ala Asn Val Lys Glu Gly His Asn Asp Phe Lys
100 105 110
Val Pro Gly Trp Arg Ala Pro Lys Met Pro Ser Ser Gly His Arg Phe
14
CA 02392065 2002-05-17
WO 01/36464 PCT/US00/31187
115 120 125
Glu Phe Lys Leu Tyr Ala Leu Asp Glu Gln Val Glu Leu Gly Asn Lys
130 135 140
Val Thr Lys Glu Lys Leu Leu Glu Ala Ile Asp Gly His Val Val Gly
195 150 155 160
Glu Ala Val Leu Met Ala Val Asn
165
<210> 10
<211> 513
<212> DNA
<213> Helianthus annus
<220>
<221> promoter
<222> (1)...(510)
<223> SCIP promoter
<221> misc_feature
<222> (369)...(368)
<223> W-box
<221> misc_feature
<222> (371)...(375)
<223> w-box
<221> misc_feature
<222> (415)...(420)
<223> G-box
<400> 10
cttccctatt ttcggtaaca cttgtgcggc aaaggggttg gcagtggtta ccgctcggtg
ccgaaccact ttgccgctgc cactccgggc agcctaaata atgctatata tgtgacattt
120
ttgcactgaa ttctactttt tatttaccat acgcgatgaa aaggcattgg ttttttatta
180
tattatattt cagtttctat ttttggacgg caaaaatgaa ttttattaaa agtaaacgaa
290
tttaaaaata ttcggataat tactttttct tttgaatctt gattcggata agttgttacg
300
aattttaaaa cgacaattga ttgaaaatga gtgatgtagc tctttctagc gtaccacgta
360
tctgtcaagt gtcaacatgc tacagcttct caaaactgct agaactctta actacacgtg
920
tccacaaacc cacaaaatcc taaccatcca taacactata agaacttgat caacagatct
480
gtttagtaac aagttattga aggtacaaca atg
513