Note: Descriptions are shown in the official language in which they were submitted.
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
NUCLEIC ACID PROBE ARRAYS
Field of the Invention
The invention relates generally to nucleic acids and more particularly to
arrays of
nucleic acids for identifying nucleic acids in a population of nucleic acids.
Backeround of the Invention
Many diseases are associated with particular DNA sequences. The DNA sequences
are
often referred to as DNA sequence polymorphisms to indicate that the DNA
sequence
associated with a diseased state differs from the corresponding DNA sequence
in non-afflicted
individuals. DNA sequence polymorphisms can include, e.g., insertions,
deletions, or
substitutions of nucleotides in one sequence relative to a second sequence. An
example of a
particular DNA sequence polymorphism is 5'-ATCG-3', relative to the sequence
5'-ATGG-3'.
The first nucleotide 'G' in the latter sequence has been replaced by the
nucleotide 'C' in the
former sequence. The former sequence is associated with a particular disease
state, whereas
the latter sequence is found in individuals not suffering from the disease.
Thus, the presence
of the nucleotide sequence '5-ATCG-3' indicates the individual has the
particular disease.
This particular type of sequence polymorphism is known as a single-nucleotide
polymorphism,
or SNP, because the sequence difference is due to a change in one nucleotide.
Techniques that allow for the rapid detection of as little as a single DNA
base change
are therefore important methods for use in genetic analysis. Because the size
of the human
genome is large, on the order of 3 billion base pairs, techniques for
identifying polymorphisms
must be sensitive enough to specifically identify the sequence containing the
polymorphism in
a potentially large population of nucleic acids.
Typically a DNA sequence polymorphism analysis is performed by isolating DNA
from an individual, manipulating the isolated DNA, e.g., by digesting the DNA
with restriction
enzymes and/or amplifying a subset of sequences in the isolated DNA. The
manipulated DNA
is then examined further to determine if a particular sequence is present.
Techniques for analyzing DNA sequences include gel-based electrophoretic
analysis,
scanning tunnel electron microscopy, sequencing by hybridization, and solid
substrate-based
nucleic acid analyses. These solid supports can include, e.g., glass surfaces,
plastic microtiter
plates, and plastic sheets. The substrates typically contain a plurality of
linked
oligonucleotides or polynucleotides that can be, e.g., adsorbed or covalently
attached to the
support.
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
Substrate-based nucleic acid analyses can include applying a sample nucleic
acid
known or suspected of containing a particular sequence polymorphism to an
array of probes
attached to the solid substrate. The nucleic acids in the population are
allowed to hybridize to
complementary sequences attached to the substrate, if present. Hybridizing
nucleic acid
sequences are then detected in a detection step.
Solid support matrix-based hybridization and sequencing methods can require a
high
sample-DNA concentration and can be hampered by the relatively slow
hybridization kinetics
of nucleic acid samples with immobilized oligonucleotide probes. Often, only a
small amount
of template DNA is available.
Summary of the Invention
The invention is based in part on the discovery of a sensitive method for
generating a
clonally amplified nucleic acid at a discrete location in an array of nucleic
acids. The method
is preferably performed by annealing a nucleic acid template to an anchor
primer attached to a
surface of the array. At least one anchor primer in the array has a sequence
complementary to
sequences at the ~' and 3' termini of a target nucleic acid. Annealing of a
desired target
nucleic acid to the anchor primer results in juxtaposition, or near
juxtaposition, of the 5' and 3'
ends of the linear target nucleic acid.
The annealed linear target nucleic acid is circularized using one or two
ligation
reactions. In one embodiment, one ligation is used. Annealing of the linear
nucleic acid
results in juxtaposition of the 5' and 3' termini of the target nucleic acid
on the anchor primer.
Addition of a ligase results in circularization of the target nucleic acid.
This circularized
nucleic acid is a template for extension of the anchor primer in a rolling
circle amplification
reaction. An extended anchor primer containing multiple copies of a sequence
complementary
to the circular nucleic acid is formed. A nucleic acid containing an anchor
primer covalently
linked to two or more copies of a sequence complementary to the target nucleic
acid is also
referred to herein as a anchor primer nucleic acid -nucleic acid concatamer.
The presence of multiple copies of the complementary sequence facilitates
detection of
the nucleic acid. Thus, the method provides for a highly sensitive method of
detecting a
desired nucleic acid attached at a discrete location on the array. Because
only a few anchor
primers in the array will typically be extended in a given reaction, arrays
containing high
densities of anchor primers can be prepared. Thus, the methods and
compositions of the
2
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
invention provide high density arrays in which desired nucleic acids can be
detected with high
sensitivity.
In an alternative embodiment, two ligation events occur. The 5' and 3' termini
of the
annealed target nucleic acids are separated by a gap. An oligonucleotide
complementary to the
gap is added. Ligation of the termini of the annealed target nucleic acid to
the gap
oligonucleotide results in formation of a covalently closed circular molecule
that can act as a
template for rolling circle amplification.
When either one or two ligation events are used, extension of the anchor
primer using
as a template the circular nucleic acid molecule results in clonally amplified
product at a
discrete location in the substrate.
The requirement of one or two ligation reactions increases allows for
increased
sensitivity in identifying a desired nucleic acid molecule because it requires
a perfect pairing
between the terminal nucleotides of the target nucleic acid molecule and the
corresponding
nucleotides in the anchor primer. Lipase molecules preferentially ligate
substrates in which
the 3' and 5' terminal nucleotides are base-paired to complementary
nucleotides. Accordingly,
duplexes of a target nucleic acid and an anchor primer containing a mismatched
oligonucleotide at or near the termini of the target nucleic acid are not
substrates for the lipase
and therefore do not produce a covalently closed circle.
Thus, the invention provides a highly sensitive method for using an array to
identify
and clonally amplify a target nucleic acid in a population of nucleic acid
molecules.
A preferred polynucleotide anchor primer includes a first region, a second
region, and
a third region. The first region includes a sequence complementary to a 5'
region of a target
nucleic acid sequence, and the second region includes a sequence complementary
to a 3'
region of the target nucleic acid sequence. The first and second regions can
each be 4-25, 4-
20, 8-20, 10-18, or 12-16 nucleotides in length. In particular embodiments,
the first region or
second region, or both, is, e.g., 4, 6, 8, 10, 12, 16, 18, 20, or 25
nucleotides in length. The
third region can be, e.g., 4-20, 6-18, 8-16, or 10-14 nucleotides in length.
In particular
embodiments, the third region is, e.g., 4, 6, 8, 10, 12, 14, 16, 18, or 20
nucleotides in length.
The target nucleic acid sequence can include, e.g., a restriction fragment
produced by
digesting a starting population of nucleic acids with one or more restriction
enzymes.
In some embodiments, the first region includes a sequence complementary to
some or
all of the recognition sequence of a first restriction endonuclease, and a
region complementary
to a sequence of a target nucleic acid sequence. The complementary region is
preferably 3' to
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
the recognition sequence for the first restriction endonuclease in the target
nucleic acid
sequence.
The first region may in addition include a query nucleotide complementary to a
nucleotide defining a sequence polymorphism in a target nucleic acid.
Preferably, the query
nucleotide is at the 3' terminus of the first region, i.e., at the 3' terminus
of the polynucleotide.
In some embodiments, the third region includes a sequence complementary to
some or
all of a recognition sequence of a second restriction endonuclease, and a
region
complementary to a sequence of the target nucleic acid sequence. The
complementary region
is preferably located 5' to the recognition sequence for the second
restriction endonuclease in
the target nucleic acid sequence.
The first and second restriction enzymes can be identical or different. In
addition, the
restriction enzymes can be, e.g., type II or type IIS restriction enzymes.
In some embodiments, the oligonucleotide can be, e.g., 4-25, 8-20 or 6-12
nucleotides
in length. For example, in particular embodiments, the oligonucleotide can be,
e.g., 4, 6, 8, 10,
12, 16, 18, 20, or 25 nucleotides in length.
The target nucleic acid may contain, or be suspected of containing, a sequence
polymorphism. The polymorphism can in some embodiments be located within,
e.g., 200,
100, 75, 50, 25, or 15 nucleotides of the 5' or 3' terminus of the target
nucleic acid sequence,
or at the 5' or 3' terminus of the target nucleic acid.
In a further aspect, the invention provides an array of oligonucleotide anchor
primers
(either of identical or differing sequences). The array includes a support
having a first surface
linked thereto a plurality of oligonucleotide anchor primers attached to the
first surface of the
solid support. The anchor primers attached to the surface include a first
region complementary
to a 5' region of a target nucleic acid sequence, a second region
complementary to a 3' region
of the target nucleic acid sequence, and an optional third region located
between the first
region and the second region. Each of the different oligonucleotides is
preferably attached to
the surface of the solid support in a different predetermined region and has a
different
predetermined sequence.
Attachment of the anchor primer to the surface can occur before, during, or
subsequent
to extension of the annealed anchor primer. Thus, in one embodiment, one or
more anchor
primers are linked to the solid substrate, after which the anchor primer is
annealed to a target
nucleic acid and extended in the presence of a polymerase. Alternatively, in a
second
embodiment, an anchor primers is first annealed to a target nucleic acid, and
a 3'0H terminus
4
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
of the annealed anchor primer is extended with a polymerase. The extended
anchor primer is
then linked to the solid substrate. By varying the sequence of anchor primers,
it is possible to
specifically amplify distinct target nucleic acids present in a population of
nucleic acids.
In some embodiments, the anchor primer is 12-100, 18-SO or 24-40 nucleotides
in
length. In particular embodiments the anchor primer is, e.g., 12, 18, 24, 40,
or 50 nucleotides
in length. The array can include at least 100, or even 1,000, 10,000, 100,000,
or 1,000,000, or
10,000,000 or more different oligonucleotides attached to a solid support.
Preferably, each of the predefined regions is physically separated from each
of the
other predetermined regions, e.g., the defined regions can be separated by 1-
400 Vim, 40-150
Vim, or 100-150 qm.
In another aspect, the invention provides a method of determining the sequence
of a
nucleic acid. The method includes providing a substrate having a plurality of
different anchor
primers, e.g., the anchor primers described above. In some embodiments, the
plurality
includes anchor primers of known sequence at known locations on the array.
The anchor primers include a first region complementary to a 5' region of a
target
nucleic acid sequence, a second region complementary to a 3' region of the
target nucleic acid
sequence, and, optionally, a third region lying between the first and second
region.
One or more of the anchor primers on the array are contacted with a population
of
circular target nucleic acid molecules. The target circular nucleic acids can
be open circles or
closed circles.
In some embodiments, the anchor primers on the substrate are contacted with a
circular
nucleic acid having 5' and 3' regions complementary to the first and third
regions of an anchor
primer having the first and third regions.
In some embodiments, the circular target nucleic acid is formed by annealing
an anchor
primer in the array with a linear target nucleic acid having 5' and 3' termini
under conditions
which allow the 5' and 3' termini of the target nucleic acid to anneal to
complementary
sequences in an anchor primer in the array. The annealed termini of the target
nucleic acid are
then contacted with a gap oligonucleotide complementary to the third region,
(which can also
be called a gap region) of the anchor primer and a ligase under conditions
sufficient allow for
ligation of the 5' and 3' termini of the target nucleic acid to the gap
oligonucleotide. This
results in formation of a closed circular target nucleic acid.
In some embodiments, sequence homologous to the closed circular molecule are
amplified prior to determining their nucleotide sequence. Amplification take
place, e.g., by
5
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
extending the 3' OH terminus of an anchor primer annealed to a closed circular
molecule.
Extension occurs with a polymerase under conditions that allow for formation
of a
polynucleotide product complementary to one or more copies of the covalently
closed circular
target nucleic acid. The polynucleotide product is identified, thereby
determining the sequence
of the circularized target nucleic acid.
The target nucleic acid can contain a sequence polymorphism, e.g., a single
nucleotide
sequence polymorphism. Preferably, the single nucleotide polymorphism is
complementary to
a query nucleotide in said first region of said anchor primer, e.g., a query
nucleotide at the 3'
terminus of the anchor primer.
In some embodiments, the covalently closed circular nucleic acid is formed by
contacting one or more of the anchor primers on the array with a linear target
nucleic acid
under conditions that allow the 5' and 3' termini of the target nucleic acid
to anneal to the
anchor primers. The 5' and 3' termini may in some embodiments by separated by
a third, "gap
region" of at least 4 nucleotides in the anchor primer. If the gap region is
present, the 3'
terminus of the target nucleic acid is then extended with a polymerase under
condition
sufficient to form an extended product whose 3' terminal nucleotide abuts the
5' terminus of
the annealed target nucleic acid.
Next, the 3' terminal nucleotide of the extended product is ligated to the 5'
terminus of
the annealed target nucleic acid, thereby forming a closed circular molecule.
The target molecule can come from any population of nucleic acid molecules,
e.g., a
population of DNA or RNA molecules.
In some embodiments, the anchor primers on the substrate are contacted with a
linear
target nucleic acid under conditions to allow the 5' and 3' termini of the
target nucleic acid to
anneal to the anchor primers. The 5' and 3' termini, when annealed to the
anchor primers, are
separated by a gap region of at least 6 nucleotides on the anchor primer.
Next, the annealed termini of the target nucleic acid are contacted with a gap
oligonucleotide and a ligase. The gap oligonucleotide is complementary to the
gap region of
the anchor primer. The ligase is added under conditions sufficient to allow
for ligation of the
5' and 3' termini of the target nucleic acid to the gap oligonucleotide,
thereby forming a
covalently closed circular molecule that includes the nucleotide sequence of
the target nucleic
acid and the gap oligonucleotide. The nucleotide sequence of at least a
portion of the
covalently closed circular molecule is determined, thereby determining the
sequence of the
target nucleic acid.
6
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
In some embodiments, the circular (closed or open) target nucleic acid is
sequenced
directly, e.g., by extending the anchor primer using the annealed target
nucleic acid sequence
as a template. The target nucleic acid to be sequenced may contain, or be
suspected of
containing, a sequence polymorphism. The polymorphism can in some embodiments
be
located within, e.g., 200, 100, 75, 50, 25, or 15 nucleotides of the 5' or 3'
terminus (prior to of
the target nucleic acid sequence, or at the 5' or 3' terminus of the target
nucleic acid. In some
embodiments, the polymorphism is at a nucleotide complementary to a query
nucleotide
present in the 3' terminal region of the anchor primer, e.g., at the 3'
terminus of the anchor
primer.
In other embodiment, the circularized target nucleic acid is amplified prior
to
determining its nucleotide sequence. Amplification can occur, e.g., by
extending an annealed
anchor primer annealed to a circular target molecule to generate a concatemer
of sequences
complementary to the circular target molecule. At least a portion of the
amplified
polynucleotide product is then identified, thereby determining the sequence of
the circularized
target nucleic acid.
The linear target nucleic acid molecule can contain, e.g., a sequence
polymorphism
such as a single nucleotide polymorphism.
In a further aspect, the invention provides a method of determining the
sequence of a
nucleic acid. The method includes providing a substrate having a plurality of
different anchor
primers of known sequence at known locations. The anchor primers include a
first region
complementary to a 5' region of a target nucleic acid sequence, a second
region
complementary to a 3' region of the target nucleic acid sequence, and,
optionally, a third
region located between the first region and second region. The substrate is
then contacted with
a linear target nucleic acid under conditions to allow the 5' and 3' termini
of the target nucleic
acid to anneal to the anchor primers. In preferred embodiments, the 5' and 3'
termini are
separated by a region corresponding to the second region (which is also known
as a gap
region) of at least 6 nucleotides on the anchor primer. Next, the 3' terminus
of the target
nucleic acid is extended with a polymerase under condition sufficient to form
an extended
product whose 3' terminal nucleotide abuts the 5' terminus of the annealed
target nucleic acid.
The 3' terminal nucleotide of the extended product is ligated to the 5'
terminus of the annealed
target nucleic acid, thereby forming a covalently closed circular molecule
comprising the
nucleotide sequence of the target nucleic acid and the gap oligonucleotide.
The nucleotide
7
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
sequence of at least a portion of the covalently closed circular molecule is
determined as
described above.
Also provided is a method for comparing the relative amount of a target
nucleic acid in
a first and second population of nucleic acid molecules. The method includes
providing one or
more or more nucleic acid anchor primers linked to a solid support, as well as
a f rst plurality
of circular single-stranded nucleic acid templates derived from the first
population of nucleic
acid molecules and a second plurality of open or closed circular single-
stranded nucleic acid
molecules derived from the second population of nucleic acid molecules. An
effective amount
of a nucleic acid anchor primer is then annealed to at least one of the single-
stranded circular
templates derived from the first plurality of circular single-stranded nucleic
acid molecules, to
yield a first primed single-stranded circular template. An effective amount of
the nucleic acid
anchor primers is also annealed to at least one of the single-stranded
circular templates from
the second plurality of circular single-stranded nucleic acid molecule to
yield a second primed
single-stranded circular template. The first and second primed anchor primer-
circular template
complexes are used to determine the sequence of the first and second nucleic
acid molecules.
In some embodiments, the sequences are determined directly. In other
embodiments, the
sequences are determined after amplifying the target nucleic acids, e.g.,
after amplification
with a polymerase to generate multiple copies of the circular nucleic acid
template derived
from the first and second populations of nucleic acid molecules.
The sequence of the target nucleic acids can be determined by annealing an
effective
amount of a sequencing primer to the first and second circular nucleic acid
templates to yield
first and second primed sequencing primer-circular nucleic acid template
complexes. The
complexes are then extended to yield a first and second sequencing product
associated with the
first and second primed sequencing primer-circular nucleic acid template
complexes. Levels
of the first and second sequencing products are then compared. The relative
level of the first
and second sequencing products indicates the relative level of the target
nucleic acid in the
first and second population of nucleic acid sequences.
The first and second population of nucleic acid molecules can be, e.g.,
genomic DNA
sequences, or derived from RNA sequences. The population of nucleic acid
sequences can
include, e.g., a first population of nucleic acids is from a cell population
that has been exposed
to a stimulus and the second population of nucleic acids is from the same cell
population that
has not been exposed to the stimulus. Stimuli can include, e.g., a physical
stimulus (e.g.,
heat), a mitogen, and a ligand for a receptor expressed by cells in the cell
population. In
8
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
addition, the first and second population of nucleic acids can be obtained
from cells of two
different individuals.
If desired, levels of nucleic acid sequences from multiple populations can be
compared,
e.g., nucleic acid molecules in 3, 5, 10, 25, S0, 100, 1,000, or 10,000 or
more populations.
In a still further aspect, the invention provides a method for detecting an
RNA
molecule in a population of RNA molecules. The method includes providing a
population of
cDNA molecules and a substrate having a plurality of different anchor primers
of known
sequence at known locations. The anchor primers include a first region
complementary to a 5'
region of a target nucleic acid sequence, a second complementary to a 3'
region of the target
nucleic acid sequence, and, optionally, a third region (also known as a gap
region) located
between the first and third regions. The substrate is then contacted with a
linear target nucleic
acid in the population of cDNA molecules under conditions sufficient to allow
the 5' and 3'
termini of the target nucleic acid to anneal to the anchor primers. The 5' and
3' termini are
preferably separated by a gap region of at least 6 nucleotides on the anchor
primer. The
armealed termini are then contacted with a gap oligonucleotide complementary
to the gap
region of the anchor primer and a ligase under conditions sufficient allow for
ligation of the 5'
and 3' termini of the target nucleic acid to the gap oligonucleotide. As a
result, a covalently
closed circular molecule including the nucleotide sequence of the target
nucleic acid and the
gap oligonucleotide is formed. The nucleotide sequence of at least a portion
of the covalently
closed circular molecule is then determined, thereby detecting the RNA
molecule.
The details of one or more embodiments of the invention are set forth in the
accompanying description below. Although any methods and materials similar or
equivalent
to those described herein can be used in the practice or testing of the
present invention, the
preferred methods and materials are now described. Other features, objects,
and advantages of
the invention will be apparent from the description and from the claims. In
the specification
and the appended claims, the singular forms also include the plural unless the
context clearly
dictates otherwise. Unless defined otherwise, all technical and scientific
terms used herein
have the same meaning as commonly understood by one of ordinary skill in the
art to which
this invention belongs. All patents and publications cited in this
specification are incorporated
by reference.
Brief Description of the Drawings
Figs. 1A and 1B are schematic drawings of an anchor primer according to the
invention.
9
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
FIG. 2 is a schematic drawing of a second anchor primer according to the
invention.
FIG. 3 is a schematic drawing of a third anchor primer according to the
invention.
FIG. 4 is a schematic drawing of rolling circle based amplification product
linked to an
anchor primer.
Detailed Description of the Invention
The invention provides arrays of polynucleotide and oligonucleotide anchor
probes that
can be used, e.g., in identifying and sequencing nucleic acids.
Structure of Anchor Probes
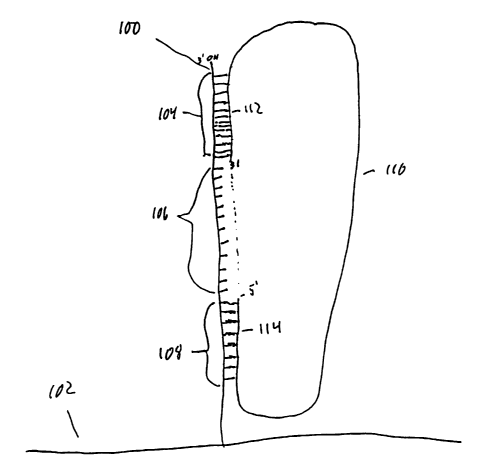
An anchored primer according to the invention is shown in FIG. 1. An anchor
primer
100 is attached at its S' end to a substrate 102. The anchor primer includes
regions 104, 106,
and 108, which are contiguous with one another.
Also shown in FIG. 1 is a linear target nucleic acid 110 having region 114 at
its 5'
terminus and region 112 at its 3' terminus. Regions 104 and 108 of the anchor
primer 100
contain nucleotides complementary to regions 112 and 114, respectively. Thus,
sequences in
regions 104 and 108 allow for capture of the termini of a linear target
nucleic acid. For some
applications, the terminal regions 104 and 108 can include recognition
sequences for
restriction enzymes, along with cDNA or genomic sequences adjacent to these
recognition
sequences, e.g. sequences at the termini of fragments of cDNA or genomic DNA
digested with
restriction enzymes. Thus, regions 104 and 108 can be designed to capture
single stranded
nucleic acids corresponding to restriction enzyme-generated fragments.
Annealing of the target nucleic acid 110 to the primer 100 as shown in FIG. 1
leaves
the 3'-OH terminus of the target nucleic acid 110 available for either primer
extension or
ligation to a polynucleotide. In primer extension, the 3' hydroxyl group at
the terminus of the
target nucleic acid 112 is extended in the presence of a DNA polymerase,
nucleotide
triphosphates and any desired associated factors using the gap region 106 of
anchor primer 100
as a template. Complete extension of the primer results in juxtaposition of
the free 3'-OH
group of the extended primer and the 5'-P04 of the linear target nucleic acid
110. In the
presence of a ligase, the 5'P04 and 3'-OH can be covalently attached. This
results in
circularization of the target nucleic acid 110. While regions 104 and 108 of
the anchor primer
100 preferably contain nucleotides perfectly complementary to regions 112 and
114,
respectively, in the primer extension this requirement, some mismatch can be
tolerated
between the anchor primer and target nucleic acid molecules.
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
In an alternative embodiment, the annealed target nucleic acid 110 can be
circularized
using a gap oligonucleotide 116 using annealing and ligation. In this
embodiment, the
terminal regions 112 and 114 of the target nucleic acid are preferably
perfectly complementary
to regions 104 and 108 of the anchor primer 100. The gap oligonucleotide 116
is
complementary to nucleotides in the gap region 106 of the anchor primer and
mixed with the
anchor primer 106 under conditions that allow for it to anneal to the anchor
primer. In the
presence of ligase, the gap oligonucleotide 116 will become covalently linked
by its 3'0H
group to the 5'-PO~ of the target nucleic acid. The S'POa of the gap
oligonucleotide will be
similarly linked to the 3'-OH of the target nucleic acid sequences.
Preferably, the gap
oligonucleotide 116 is perfectly complementary to the gap region 106. A highly
sensitive
system for detecting a nucleic acid system is therefore provided by requiring
that a circular
nucleic acid be produced based on (a) homology at the 5' and 3' ends of a
linear target nucleic
acid sequence, and (b) two separate ligation events.
If desired, an annealed circularized nucleic acid can be sequenced directly.
In preferred
1 S embodiments, the 3' terminal region of the anchor primer includes a query
nucleotide, e.g., the
3' terminus can be within 15, 10, 5, 4, 3, 2, 1, nucleotides of, or can be at,
the 3' terminus of
the anchor primer. The query nucleotide is a nucleotide able to detect a
nucleotide associated
with a predetermined SNP. A target nucleic acid under is annealed with the
anchor primer
under conditions that result in hybridization of the target nucleic acid
molecule if it contains a
nucleotide homologous to the query nucleotide. A successful annealing event
allows for
primed extension of the anchor primer using the target nucleic acid molecule
as a template.
Conversely, attempted annealing of a target nucleic acid sequence lacking a
nucleotide
complementary to the query nucleotide results in a mismatch between the query
nucleotide in
the anchor primer and the target nucleic acid. The anchor primer cannot be
extended in this
case.
For a given SNP, anchor primer probes vary in their query nucleotides
according to the
nucleotides associated with the SNP. Thus, for a SNP defined by the presence
of an 'A'
nucleotide in some members of a population, and 'C' nucleotide at other
positions, the
corresponding query nucleotides in the anchor primer can be 'C' and 'T',
respectively.
Polymerases lacking exonucleolytic activity e.g., exo- or mutS polymerase,
particularly suited
using this approach.
11
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
Alternatively, the circularized nucleic acid can be amplified and the sequence
of the
amplified products determined. The 3'-OH terminus of the anchor primer 100 is
available to
prime a sequencing reaction, e.g., a dideoxy sequencing reaction.
The 3'-OH terminus of the anchor primer 100 can alternatively prime
amplification,
e.g., rolling-circle amplification (RCA), with the circularized nucleic acid
acting as the
template nucleic acid. RCA using the 3'-OH of the anchor primer results in
molecule
covalently linked to the substrate 102 that contains multiple copies of a
sequence
complementary to the circularized nucleic acid template.
Another anchor primer according to the invention is shown in FIG. 2. An anchor
primer 200 is attached at its 5' end to a substrate 202. The anchor primer
includes two
adjoining regions, 204 and 206, which are complementary to regions.
Annealed to the anchor primer 200 in FIG. 2 is a linear target nucleic acid
210 having
regions 212 at its 3' end and region 214 at its 5' end. Regions 204 and 206 of
the anchor
primer 200 anneal to regions 212 and 214, respectively. Thus, by including
sequences
complementary to the termini of a restriction enzyme generated fragment in
regions 204 and
206, the anchor primer 200 can capture desired restriction fragments. Suitable
fragments
include those containing, or suspected of containing, polymorphisms, such as
single nucleotide
polymorphisms.
A third type of anchor primer is shown in FIG. 3. The figure shows an anchor
primer
300 attached at its 5' end to a substrate 302. The anchor primer 300 includes
near its 5' end a
nucleotide region 304.
Also shown in FIG.3 is a circular target nucleic acid 310. The target nucleic
acid 310
includes a region 312 complementary to the nucleotide region 304.
Also present in the target nucleic acid 310 is a nucleic region 350. In
preferred
embodiments, the nucleotide region 304 is complementary to a nucleotide region
312 present
in a plurality of target nucleic acids. In still more preferred embodiments,
the nucleotide
region 312 is present in a sequence, e.g., a vector, which has been ligated to
members of a
starting population of nucleic acids.
An anchor primer may optionally contain additional elements, e.g., spacer
sequences at
its 5' terminus, one or more restriction enzyme recognition sites, RNA
polymerase binding
sites (e.g., a T7 promoter site).
One or more of the adapter regions may include, e.g., a restriction enzyme
recognition
site or sequences present in identified DNA sequences, e.g., sequences present
in known
12
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
genes. One or more adapter regions may also include sequences that identify
sequence known
to flank and/or encompass sequence polymorphisms. Examples of such sequences
include the
query nucleotides. Sequence polymorphisms include nucleotide substitutions,
insertions,
deletions, or other rearrangements that result in a sequence difference
between two otherwise
identical nucleic acid sequences. An example of a sequence polymorphism is a
single
nucleotide polymorphism (SNP).
In other embodiments, the anchor primer arrays described herein can be used to
identify or otherwise characterize target nucleic acid sequences that are
circularized prior to
being added to the arrays.
Linkine of Anchor Primers to a Solid Support
In general, any nucleic acid capable of base pairing can be used as an anchor
primer.
In some embodiments, the anchor primer is an oligonucleotide. As utilized
herein the term
oligonucleotide includes linear oligomers of natural or modified monomers or
linkages, e.g.,
deoxyribonucleosides, ribonucleosides, anomeric forms thereof, peptide nucleic
acids (PNAs),
and the like, that are capable of specifically binding to a target
polynucleotide by way of a
regular pattern of monomer-to-monomer interactions. These types of
interactions can include,
e.g., Watson-Crick type of base-pairing, base stacking, Hoogsteen or reverse-
Hoogsteen types
of base-pairing, or the like. Generally, the monomers are linked by
phosphodiester bonds, or
analogs thereof, to form oligonucleotides ranging in size from, e.g., 3-200, 8-
150, 10-100, 20
80, or 25-50 monomeric units.
The oligonucleotides of the present invention can include non-natural
nucleotide
analogs. However, where, for example, processing by enzymes is required, or
the like,
oligonucleotides comprising naturally occurring nucleotides are generally
required for
maintenance of biological function.
Any material can be used as the solid support material, as long as the surface
allows for
stable attachment of the primers and detection of nucleic acid sequences. The
solid support
material can be planar. In some embodiments, the solid support is optically
transparent, e.g.,
glass.
The anchor primer can be linked to the solid support to reside on or within
the solid
support. The distance between anchor primers on the array will be determined
in part by the
method and apparatus used to detect and further analyze extended anchor primer
templates.
Methods for detecting extended anchor primers are discussed below. Apparatuses
for
detecting extended anchor primers are also discussed below and in addition
include those
13
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
known in the art for detecting macromolecules. PCT publication WO 00/06770,
for example,
discusses apparatuses such as confocal scanning microscopy, scanning near-
field optical
microscopy (SNOM), scanning tunneling microscopy, and atomic force microscopy.
These
apparatuses allow for resolution on the order of tens of manometers.
Thus, in some embodiments, the plurality of anchor primers is linked to the
solid
support so they are spaced regular intervals within an array. The distance
between primers on
a solid substrate can be, e.g., 1 nm to 150~m, 10-400 gym, 50-150 gym, 100-150
gym, or 150 gym.
In preferred embodiments, arrays are spaced at manometer resolution, e.g., 10
run X 10 nm.
Construction of arrays with manometer resolution is described in PCT
application
WO 00/06770.
An array of attachment sites on the optically transparent solid support is
constructed
using lithographic techniques commonly used in the construction of electronic
integrated
circuits. These techniques are described in, e.g., U.S. Patent Nos.
5,5143,854, 5,445,934,
5,744,305, and 5, 800,992; Chee et al., Science 274: 610-614 (1996); Fodor et
al., Nature 364:
555-556 (1993); Fodor et al., Science 251: 767-773 (1991); Gushin, et al.,
Anal. Biochem. 250:
203-211 (1997); Kinosita et al., Cell 93: 21-24 (1998); Kato-Yamada et al., J.
Biol. Chem.
273: 19375-19377 (1998); and Yasuda et al., Cell 93: 1117-1124 (1998).
Photolithography
and electron beam lithography sensitize the solid support or substrate with a
linking group that
allows attachment of a modified biomolecule (e.g., proteins or nucleic acids).
See e.g.,
Service, Science 283: 27-28 (1999); Rai-Choudhury, HANDBOOK OF
MICROLITHOGRAPHY,
MICROMACHINING, AND MICROFABRICATION, VOLUME I: MICROLITHOGRAPHY, Volume PM39,
SPIE Press (1997). Alternatively, an array of sensitized sites can be
generated using thin-film
technology as described in Zasadzinski et al., Science 263: 1726-1733 (1994).
Additional methods for attaching nucleic acids to solid substrates use
sialinized
surfaces and are disclosed in US Patent No. 6,136,962. Attachment using
disulfide bonds is
described in US. Patent No.6,030,782. The contents of all of these patents and
publications are
incorporated by reference in their entirety.
Anchor primers are linked to the solid substrate at the sensitized sites. A
region of a
solid substrate containing a linked primer is an anchor pad. Thus, by
specifying the sensitized
sites on the solid support, it is possible to form an array or matrix of
anchored pads. The
anchor pads can, e.g., small diameter spots etched at evenly spaced intervals
on the solid
support.
14
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
Each sensitized site on a solid support is potentially capable of attaching
multiple
anchor primers. Thus, each anchor pad may include one or more anchor primers.
It is
preferable to maximize the number of pads that have only a single productive
reaction center
(e.g., the number of pads that, after the extension reaction, have only a
single sequence
extended from the anchor primer). This can be accomplished by techniques which
include, but
are not limited to: (i) varying the dilution of biotinylated anchor primers
that are washed over
the surface; (ii) varying the incubation time that the biotinylated primers
are in contact with the
avidin surface; or (iii) varying the concentration of open- or closed-circular
template so that,
on average, only one primer on each pad is extended to generate the sequencing
template.
In some embodiments, each individual pad contains just one linked anchor
primer.
Pads having only one anchor primer can be made by performing limiting
dilutions of a
selected anchor primer on to the solid support such that, on average, only one
anchor primer is
deposited on each pad. The concentration of anchor primer to be applied to a
pad can be
calculated utilizing, for example, a Poisson distribution model.
In order to maximize the number of reaction pads that contain a single anchor
primer, a
series of dilution experiments are performed in which a range of anchor primer
concentrations
or circular template concentrations are varied. For highly dilute
concentrations of primers,
primers and circular templates binding to the same pad will be independent of
each other, and
a Poisson distribution will characterize the number of anchor primers extended
on any one
pad. Although there will be variability in the number of primers that are
actually extended, a
maximum of 37% of the pads will have a single extended anchor primer (the
number of pads
with a single anchor oligonucleotide). This number can be obtained as follows.
Let NP be the average number of anchor primers on a pad and f be the
probability that
an anchor primer is extended with a circular template. Then the average number
of extended
anchor primers per pad is NPf, which is defined as the quantity a. There will
be variability in
the number of primers that are actually extended. In the low-concentration
limit, primers and
circular templates binding to the same pad will be independent of each other,
and a Poisson
distribution P(n) will characterize the number of anchor primers n extended on
any pad. This
distribution may be mathematically defined by: P(n) _ ( a"/ n!)exp(-a), with
P(1) = a exp(-a).
The probability P(1) assumes it maximum value exp(-1) for a = 1-, with 37% of
pads having a
single extended anchor primer.
A range of anchor primer concentrations and circular template concentrations
may be
subsequently scanned to find a value of Npf closest to 1. A preferable method
to optimize this
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
distribution is to allow multiple anchor primers on each reaction pad, but use
a limiting
dilution of circular template so that, on average, only one primer on each pad
is extended to
generate the sequencing template.
Alternatively, at low concentrations of anchor primers,, at most one anchor
primer will
likely be bound on each reaction pad. A high concentration of circular
template may be used
so that each primer is likely to be extended.
Where the reaction pads are arrayed on a planar surface or a fiber optic
array. (FORA),
the individual pads are approximately 10 ~m on a side, with a 100 ~m spacing
between
adjacent pads. Hence, on a 1 cm surface a total of approximately 10,000
microreaetors could
be deposited, and, according to the Poisson distribution, approximately 3700
of these will
contain a single anchor primer. In certain embodiments, after the primer
oligonucleotide has
been attached to the solid support, modified, e.g., biotinylated, enzymes are
deposited to bind
to the remaining, unused avidin binding sites on the surface.
In other embodiments multiple anchor primers are attached to any one
individual pad
in an array. Limiting dilutions of a plurality of circular nucleic acid
templates (described in
more detail below) may be hybridized to the anchor primers so immobilized such
that, on
average, only one primer on each pad is hybridized to a nucleic acid template.
Library
concentrations to be used may be calculated utilizing, for example, limiting
dilutions and a
Poisson distribution model.
The anchor primer can be attached to the solid support via a covalent or non-
covalent
interaction. Examples of such linkages common in the art include
Ni'+/hexahistidine,
streptavidin/biotin, avidinlbiotin, glutathione S-transferase
(GST)/glutathione, monoclonal
antibody/antigen, and maltose binding protein/maltose. Samples containing the
appropriate
tag are incubated with the sensitized substrate so that a single molecule
attaches at each
sensitized site.
The biotin-(strept-)avidin methodology provides several different ways to
immobilize
the anchor on the solid support. One biotin-(strept-)avidin-based anchoring
method uses a
thin layer of a photoactivatable biotin analog dried onto a solid surface.
(Hengsakul and Cass,
1996. Biocongjugate Chem. 7: 249-254) . The biotin analog is then exposed to
white light
through a mask, so as to create defined areas of activated biotin. Avidin (or
streptavidin) is
then added and allowed to bind to the activated biotin. The avidin possesses
free biotin
binding sites that can be utilized to "anchor" the biotinylated
oligonucleotides through a
biotin-(strept-)avidin linkage.
16
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
Alternatively, the anchor primer can be attached to the solid support with a
biotin
derivative possessing a photo-removable protecting group. This moiety is
covalently bound to
bovine serum albumin (BSA), which is attached to the solid support, e.g., a
glass surface. See
Pirrung and Huang, 1996. Bioconjugate Chem. 7: 317-321. A mask is then used to
create
S activated biotin within the defined irradiated areas. Avidin may then be
localized to the
irradiated area, with biotinylated DNA subsequently attached through a BSA-
biotin-avidin-
biotin link. If desired, an intermediate layer of silane is deposited in a
self assembled
monolayer on a silicon dioxide silane surface that can be patterned to
localize BSA binding in
defined regions. See e.g., Mooney, ET al., 1996. Proc. Natl. Acad. Sci. USA
93: 12287-12291.
Each sensitized site on a solid support is potentially capable of attaching
multiple
anchor primers. Thus, each anchor pad may include one or more anchor primers.
It is
preferable to maximize the number of pads that have only a single productive
reaction center
(e.g., the number of pads that, after the extension reaction, have only a
single sequence
extended from the anchor primer). This can be accomplished by, e.g.: (i)
varying the dilution
of biotinylated anchor primers that are washed over the surface; (ii) varying
the incubation
time that the biotinylated primers are in contact with the avidin surface; or
(iii) varying the
concentration of open- or closed-circular template to maximize the number of
pads on which
only primer is extended to generate the sequencing template.
Preparing template nucleic acids
The template nucleic acids can in general be derived from any source, as long
as at
least a portion of the template nucleic acids are linear and single-stranded
when annealed to the
anchor primer arrays. Preferably a template nucleic acid containing a target
nucleic acid is in
the form an open circle. An "open circle" is a linear single-stranded nucleic
acid molecule
having a 5' phosphate group and a 3' hydroxyl group, i.e., the ends of a given
open circle
nucleic acid molecule can be ligated by DNA ligase. Open circles are described
in detail in
Lizardi, U.S. Pat. No. 5, 854,033. An open circle can be converted to a closed
circle in the
presence of a DNA ligase (for DNA) or RNA ligase following, e.g., annealing of
the open
circle to an anchor primer.
In preferred embodiments, at least one linear target nucleic acid in a
population of
nucleic acids will have sequences at its 5' and 3' ends, i.e., regions 112 and
114 as shown in
FIG. l, that are complementary to regions 104 and 108 of an anchor primer.
The 5'- and 3'-terminal regions 112 and 114 of the oligonucleotides are
designed to
basepair adjacent to one another on a specific target sequence strand, thus
the termini of the
17
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
linear oligonucleotide are brought into juxtaposition by hybridization to the
target sequence.
This juxtaposition allows the two probe segments (if properly hybridized) to
be covalently-
bound by enzymatic ligation (e.g., with T; DNA ligase), thus converting the
probes to
circularly-closed molecules which are catenated to the specific target
sequences (see e.g.,
Nilsson, et al., 1994. Science 265: 2085-2088). The resulting probes are
suitable for the
simultaneous analysis of many gene sequences (see e.g., Lizardi, et al., 1998.
Nat. Genet. 19:
225-232; Nilsson, et al., 1997. Nat. Genet. 16: 252-255).
The starting population of nucleic acid can be either single-stranded or
double-
stranded, as long as it includes a region that, if present in the library, is
available for annealing,
or can be made available for annealing, to an anchor primer sequence.
Library templates can include multiple elements, including, but not limited
to, one or
more regions that are complementary to the anchor primer. For example, the
template libraries
may include a region complementary to a sequencing primer, a control
nucleotide region, and
an insert sequence containing a sequence of interest, i.e., the sequencing
template to be
subsequently characterized. As utilized herein the term "complement" refers to
nucleotide
sequences that are able to hybridize to a specific nucleotide sequence to form
a matched
duplex.
In one embodiment, a library template includes: (i) two distinct regions,
e.g., regions
114 and 112, that are complementary to the anchor primer, (ii) one region
complementary to a
sequencing primer, (iii) one control nucleotide region, (iv) an insert
sequence of 30 - 100
nucleotides that is to be sequenced. The template can, of course, include two,
three, or all four
of these features.
The template nucleic acid can be constructed from any source of nucleic acid,
e.g., any
cell, tissue, or organism, and can be generated by any art-recognized method.
Suitable
methods include, e.g., sonication of genomic DNA and digestion with one or
more restriction
endonucleases (RE) to fragment a population of nuclei acid molecules, e.g.,
genomic DNA.
The restriction enzymes can recognize eight or six base recognition sequences,
or can
recognize degenerate sequences. In some embodiments, the restriction enzymes
are Type II or
Type IIS restriction enzymes. Preferably, one or more of the restriction
enzymes have distinct
four-base recognition sequences. Examples of such enzymes include, e.g.,
Sau3Al, MspI, and
TaqI. Preferably, the enzymes are used in conjunction with anchor primers
having regions
containing recognition sequences for the corresponding restriction enzymes. In
some
embodiments, the one or both adapter regions anchor primers contain additional
sequences
18
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
adjoining known restriction enzyme recognition sequences, thereby allowing for
capture or
annealing of specific restriction fragments of interest to the anchor primer.
The target nucleic acid can also be generated by amplification, e.g., primed
amplification of a desired target sequence.
Alternatively, template libraries can be made by generating a complementary
DNA
(cDNA) library from RNA, e.g., messenger RNA (mRNA). The cDNA library can, if
desired,
be further processed with restriction endonucleases and/or primed
amplification to obtain
either 3' signature sequences, internal fragments, or 5' fragments. The
libraries can be
alternatively be constructed in vectors having sequences complementary to
adapter regions in
an anchor primer. In some embodiments, the libraries contain a sequence of
interest, e.g., a
known or suspected sequence polymorphism on a restriction fragment.
Annealins and Amplification of Primer-Template
Nucleic Acid Complexes
Libraries of nucleic acids are annealed to anchor primer sequences using
recognized
techniques (see, e.g., Hatch, et al., 1999. Genet. Anal. Biomol. Engineer. 15:
35-40; Kool, U.S.
Patent No. 5,714, 320 and Lizardi, U.S. Patent No. 5,854,033). In general, any
procedure for
annealing the anchor primers to the template nucleic acid sequences is
suitable as long as it
results in formation of specific, i.e., perfect or nearly perfect,
complementarity between the
adapter region or regions in the anchor primer sequence and a sequence present
in the template
library.
In some embodiments, the circularized template is amplified. A number of in
vitro
nucleic acid amplification techniques may be utilized to extend the anchor
primer sequence.
Preferably, the size of the amplified DNA should be smaller than the size of
the anchor pad
and also smaller than the distance between anchoring pads.
The amplification is typically performed in the presence of a polymerise,
e.g., a DNA
or RNA-directed DNA polymerise, and one, two, three, or four types of
nucleotide
triphosphates, and, optionally, auxiliary binding proteins. In general, any
polymerise capable
of extending a primed 3'-OH group can be used a long as it lacks a 3' to 5'
exonuclease
activity. Suitable polymerises include, e.g., the DNA polymerises from
Bacillus
stearothermophilus, Thermus acquaticus, Pyrococcus furiosis, Thermococcus
litoralis, and
Thermus thermophilus, bacteriophage T4 and T~, and the E. coli DNA polymerise
I Klenow
fragment. Suitable RNA-directed DNA polymerises include, e.g., the reverse
transcriptase
19
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
from the Avian Myeloblastosis Virus, the reverse transcriptase from the
Moloney Murine
Leukemia Virus, and the reverse transcriptase from the Human Immunodeficiency
Virus-I.
A number of in vitro nucleic acid amplification techniques have been
described. These
amplification methodologies may be differentiated into those methods: (i)
which require
temperature cycling - polymerase chain reaction (PCR) (see e.g., Saiki, et
al., 1995. Science
230: 1350-1354), ligase chain reaction (see e.g., Barany, 1991. Proc. Natl.
Acad. Sci. USA 88:
189-193; Barnnger, et al., 1990. Gene 89: 117-122) and transcription-based
amplification (see
e.g., Kwoh, et al., 1989. Proc. Natl. Acad. Sci. USA 86: 1173-1177) and (ii)
isothermal
amplification systems - self sustaining, sequence replication (see e.g.,
Guatelli, et al., 1990.
Proc. Natl. Acad. Sci. USA 87: 1874-1878); the Q(3 replicase system (see e.g.;
Lizardi, et al.,
1988. BioTechnology 6: 1197-1202); strand displacement amplification Nucleic
Acids Res.
1992 Apr 11;20(7):1691-6.; and the methods described in PNAS 1992 Jan
1;89(1):392-6; and
NASBA J Virol Methods. 1991 Dec;35(3):273-86.
Isothermal amplification also includes rolling circle-based amplification
(RCA). RCA
is discussed in, e.g., Kool, U.S. Patent No. 5,714,320 and Lizardi, U.S.
Patent No. 5,854,033;
Hatch, et al., 1999. Genet. Anal. Biomol. Engineer. 15: 35-40. The result of
the RCA is a
single DNA strand extended from the 3' terminus of the anchor primer (and thus
is linked to
the solid support matrix) and including a concatamer containing multiple
copies of the circular
template annealed to a primer sequence. Typically, 10,000 or more copies of
circular
templates, each having a size of approximately 100 nucleotides size range, can
be obtained
with RCA.
The product of RCA amplification following annealing of a circular nucleic
acid
molecule to an anchor primer is shown schematically in FIG. 4. A circular
template nucleic
acid 402 is annealed to an anchor primer 404, which has been linked to a
surface 106 at its 5'
end and has a free 3' OH available for extension. The circular template
nucleic acid 402
includes two adapter regions 408 and 410 that are complementary to regions of
sequence in the
anchor primer 404. Also included in the circular template nucleic acid 402 is
an insert 412 and
a region 414 complementary to a sequencing primer, which is used in the
sequencing reactions
described below.
Upon annealing, the free 3'-OH on the anchor primer 404 can be extended using
sequences within the template nucleic acid 402. The anchor primer 402 can be
extended along
the template multiple times , with each iteration adding to the sequence
extended from the
anchor primer a sequence complementary to the circular template nucleic acid.
Four iterations,
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
or four rounds of rolling circle amplification, are shown in FIG.4 as the
extended anchor
primer amplification product 414. Extension of the anchor primer results in an
amplification
product covalently attached to the substrate 406.
RCA-based amplification has recently been utilized to obtain an isothermal
cascade
amplification reaction of circularized padlock probes in vitro in order to
detect single-copy
genes in human genomic DNA samples (see Lizardi, et al., 1998. Nat. Genet. 19:
225-232). In
addition, RCA has also been utilized to detect single DNA molecules in a solid
phase-based
assay (see Lizardi, et al., 1998. Nat. Genet. 19: 225-232).
Rolling-circle amplification (RCA) can replicate circularized oligonucleotide
probes
with either linear or geometric kinetics under isothermal conditions. In the
presence of two
primers (one hybridizing to the + strand, and the other, to the - strand of
DNA), concatamers of
both strands can be simultaneously synthesized. This allows for 1 x 109 or
more copies of each
circle in a short period of time (i.e., less-than 90 minutes), enabling the
detection of single-
point mutations within the human genome.
Using a single primer, RCA generates hundreds of randomly linked copies of a
covalently closed circle in several minutes. If solid support matrix-
associated, the DNA
product remains bound at the site of synthesis, where it may be labeled,
condensed, and
imaged as a point light source. For example, linear oligonucleotide probes,
which can
generate RCA signals, have been bound covalently onto a glass surface. The
color of the
signal generated by these probes indicates the allele status of the target,
depending upon the
outcome of specific, target-directed ligation events. As RCA permits millions
of individual
probe molecules to be counted and sorted, it is particularly amenable for the
analysis of rare
somatic mutations. RCA also shows promise for the detection of padlock probes
bound to
single-copy genes in cytological preparations.
In addition, solid-phase RCA methods have also been developed to provide an
effective method of detecting constituents within a solution. Initially, a
recognition step is
used to generate a complex consisting of a DNA primer duplexed with a circular
template is
bound to a surface. A polymerase enzyme is then used to amplify the bound
complex. RCA
uses small DNA probes that are amplified to provide an intense signal using
detection
methods, including the methods described in more detail below.
Other examples of isothermal amplification systems include, e.g., (i) self
sustaining,
sequence replication (see e.g., Guatelli, et al., 1990. Proc. Natl. Acad. Sci.
USA 87: 1874-
1878), (ii) the Q~3 replicase system (see e.g., Lizardi, et al., 1988.
BioTechnology 6: 1197-
21
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
1202), and (iii) nucleic acid sequence-based amplification (NASBAN; see
Kievits, et al., 1991.
J. Virol. Methods 35: 273-286).
Methods of identifying extended anchor primers
In general, the oligonucleotide arrays described herein can be used in
conjunction with
any substrate-based sequencing method known in the art. The methods can also
be used in
association with conventional optic, as well as fiber-optic based detection
signal detection
methods. The extended anchor primers produced using the arrays and methods
disclosed
herein can be further analyzed using methods known in the art. For example,
the sequence of
an extended anchor primer can be determined, or sequences homologous to a
probe molecule
can be identified. Some of these methods are described below.
One or more sequences in the extended anchor primer can be determined by
subjecting
the extended anchor primer product to dideoxy terminator sequencing. Analysis
of the
sequencing products allows the identification of one or more nucleotides in
the extended
anchor primer.
Alternatively, pyrophosphate-based sequencing methods can be used to identify
nucleotides in an extended anchor primer. For example, Nyren et al.
(Analytical Biochemistry
208:171-175, 1993) have described a method relying on the detection of DNA
polymerase
activity by an enzymatic luminometric inorganic pyrophosphate detection assay
(ELIDA)
These methods can be used in as part of microsequencing assays. These assays
are
based on extension of a single nucleotide from a sequencing primer that
hybridizes just
upstream of a polymorphic base of interest in the extended anchor primer. A
polymerase is
used to specifically extend the 3' end of the primer with one single ddNTP
(chain terminator)
complementary to the selected nucleotide at the polymorphic site. Next, the
identity of the
incorporated nucleotide is determined.
In one embodiment, microsequencing reactions are carried out using fluorescent
ddNTPs, and the extended microsequencing primers are analyzed to determine the
identity of
the incorporated nucleotide. Microsequencing reactions are described in EP 412
883.
Incorporated ddNTPs can alternatively be radiolabeled (Syvanen, Clinica
Chimica Acta
226:225-236, 1994) or linked to fluorescein (Livak and Hainer, Human Mutation
3:379-
385,1994). The detection of radiolabeled ddNTPs can be achieved through
scintillation-based
techniques. The detection of fluorescein-linked ddNTPs can be based on the
binding of
antifluorescein antibody conjugated with alkaline phosphatase, followed by
incubation with a
chromogenic substrate (such as p-nitrophenyl phosphate). Other possible
reporter-detection
22
CA 02392474 2002-05-24
WO 01/38580 PCT/LTS00/32131
pairs include, e.g., ddNTP linked to dinitrophenyl (DNP) and anti-DNP alkaline
phosphatase
conjugate (Harju et al., Clin. Chem. 39/11 2282-2287, 1993) or biotinylated
ddNTP and
horseradish peroxidase-conjugated streptavidin with o-phenylenediamine as a
substrate (WO
92/ 15712).
Other approaches recognized in the art can be used to detect the nucleotide
added to the
microsequencing primer. For example, homogeneous phase detection method based
on
fluorescence resonance energy transfer has been described by Chen et al.,
Nucleic Acids
Research 25:347-353 1997) and Chen et al., Proc. Natl. Acid. Sci USA 94/20
10756-
10761,1997). Amplified genomic DNA fragments containing polymorphic sites are
incubated
with a 5'-fluorescein-labeled primer in the presence of allelic dye-labeled
dideoxyribonucleoside triphosphates and a modified Taq polymerise. The dye-
labeled primer
is extended one base by the dye-terminator specific for the allele present on
the template. At
the end of the genotyping reaction, the fluorescence intensities of the two
dyes in the reaction
mixture are analyzed directly without separation or purification. All these
steps can be
performed in the same tube and the fluorescence changes can be monitored in
real time.
Alternatively, the extended primer may be analyzed by MALDI-TOF Mass
Spectrometry. The base at the polyrnorphic site is identified by the mass
added onto the
microsequencing primer (see Haff L.A. and Smirnov LP., Genome Research, 7:378-
388,
1997).
The extended anchor primers can also be detected using mismatch detection
assays
based on polymerises and/or ligases in enzyme based mismatch detection assays.
The terms
"enzyme based mismatch detection assay" are used herein to refer to any method
of
determining the allele of a biallelic marker based on the specificity of
ligases and polymerises.
An example of these methods is the oligonucleotide ligation assay ("OLA"). OLA
uses two oligonucleotides which are designed to be capable of hybridizing to
abutting
sequences of a single strand of a target molecules. One of the
oligonucleotides is biotinylated,
and the other is detectably labeled. If the precise complementary sequence is
found in a target
molecule, the oligonucleotides will hybridize such that their termini abut,
and create a ligation
substrate that can be captured and detected. OLA is described in Nickerson
D.A. et al. (Proc.
Natl. Acid Sci. US.A. 87:8923-8927, 1990). In preferred embodiments, an extra
amplification
step, such as PCR, is used to achieve the exponential amplification of target
DNA, which is
then detected using OLA.
23
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
A second ligation-based method is ligase chain reaction ("LCR"). The sequences
of
each pair of oligonucleotides in LCR is selected to permit the pair to
hybridize to abutting
sequences of the same strand of the target. Such hybridization forms a
substrate for a
template-dependent ligase. LCR can be performed on extended anchor primers
with
oligonucleotides having the proximal and distal sequences of the same strand
of an extended
anchor primer. In one embodiment, either oligonucleotide will be designed to
include a site of
interest on the extended anchor primer. In such an embodiment, the reaction
conditions are
selected such that the oligonucleotides can be ligated together only if the
target molecule either
contains or lacks the specific nucleotides) that is complementary to the site
of interest on the
oligonucleotide. In an alternative embodiment, the oligonucleotides will not
include the
biallelic marker, such that when they hybridize to the target molecule, a
"gap" is created as
described in WO 90/0 1069. This gap is then "filled" with complementary dNTPs
(as
mediated by DNA polymerase), or by an additional pair of oligonucleotides.
Thus at the end
of each cycle, each single strand has a complement capable of serving as a
target during the
next cycle and exponential allele-specific amplification of the desired
sequence is obtained.
Ligase/Polymerase-mediated Genetic Bit AnalysisTM is another method for
determining
the identity of a nucleotide at a preselected site in a nucleic acid molecule
(WO 95/21271).
This method includes incorporation of a nucleoside triphosphate that is
complementary to a
nucleotide present at the preselected site onto the terminus of a primer
molecule, and
subsequent ligation of the primer to a second oligonucleotide. The reaction is
monitored by
detecting a specific label attached to the reaction's solid phase or by
detection in solution.
A further method of determining the identity of an extended anchor primer
involves
nucleic acid hybridization. Specific probes can be designed that hybridize
preferentially or
specifically to one form of an extended anchor primer. Hybridization
conditions should be
sufficiently stringent that there is a significant difference in hybridization
intensity between
alternative sequences of an extended anchor primer. For example, conditions
are preferably
chosen to discriminate between hybridization of a probe sequence to a
perfectly matched
homologous extended anchor primer sequence and an extended anchor primer
sequence
containing one or more mismatches with the probe sequence. Stringent, sequence
specific
hybridization conditions, under which a probe will hybridize only to the
exactly
complementary target sequence are well known in the art (see, e.g., Sambrook
et al., Molecular
Cloning - A Laboratory Manual, Second Edition, Cold Spring Harbor Press, N.Y.,
1989).
Stringent conditions are sequence dependent and will be different in different
circumstances.
24
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
Generally, stringent conditions are selected to be about 5°C lower than
the thermal melting
point (Tm) for the specific sequence at a defined ionic strength and pH. By
way of example
and not limitation, procedures using conditions of high stringency are as
follows:
Prehybridization of filters containing DNA is carried out for 8 h to overnight
at 65° C in buffer
composed of 6X SSC, 50 mM Tris-HC 1 (pH 7.5), 1 mM EDTA, 0.02% PVP, 0.02%
Ficoll,
0.02% BSA, and S00 pg/ml denatured salmon sperm DNA. Filters are hybridized
for 48 h at
65°C, the preferred hybridization temperature, in prehybridization
mixture containing 100
pg/ml denatured salmon sperm DNA and 5-20 X 106 cpm of 3'P-labeled probe.
Alternatively,
the hybridization step can be performed at 65°C in the presence of SSC
buffer, 1 x SSC
corresponding to O.15M NaCI and 0.05 M Na citrate. Subsequently, filter washes
can be done
at 37°C for 1 h in a solution containing 2X SSC, 0.01% PVP, 0.01%
Ficoll, and 0.01% BSA,
followed by a wash in O.1X SSC at SO°C for 45 min. Alternatively,
filter washes can be
performed in a solution containing 2 x SSC and 0.1% SDS, or 0.5 x SSC and 0.1%
SDS, or 0.1
x SSC and 0.1 % SDS at 68°C for 15 minute intervals. Following the wash
steps, the
1 S hybridized probes are detectable by autoradiography.
Alternatively, in some embodiments, extended anchor primers containing a
limited
number of mismatches to a target sequence can be detected with a probe
sequence. Such
extended anchor primers can be detected using conditions of intermediate
stringency. By way
of example and not limitation, procedures using conditions of intermediate
stringency are as
follows: Filters containing DNA are prehybridized, and then hybridized at a
temperature of
60°C in the presence of a 5 x SSC buffer and labeled probe.
Subsequently, filters washes are
performed in a solution containing 2x SSC at 50°C and the hybridized
probes are detectable by
autoradiography.
Highly sensitive hybridization assays with no need for separations or washes
have
been described (see Landegren et at., Genome Research, 8:769-776,1998). For
example, the
TaqManTM assay takes advantage of the S' nuclease activity of Taq DNA
polymerase to digest
a DNA probe annealed specifically to the accumulating amplification product.
TaqManThi
probes are labeled with a donor-acceptor dye pair that interacts via
fluorescence energy
transfer. Cleavage of the TaqManTM probe by the advancing polymerase during
amplification
dissociates the donor dye from the quenching acceptor dye, greatly increasing
the donor
fluorescence. All reagents necessary to detect two allelic variants can be
assembled at the
beginning of the reaction and the results are monitored in real time (see
Livak et al., Nature
Genetics, 9:341-342, 1995).
CA 02392474 2002-05-24
WO 01/38580 PCT/US00/32131
In an alternative homogeneous hybridization-based procedure, molecular beacons
are
used for allele discriminations. Molecular beacons are hairpin-shaped
oligonucleotide probes
that report the presence of specific nucleic acids in homogeneous solutions.
When they bind to
their targets they undergo a conformational reorganization that restores the
fluorescence of an
internally quenched fluorophore (Tyagi et al., Nature Biotechnology, 16:49-53,
1998).
OTHER EMBODIMENTS
It is to be understood that while the invention has been described in
conjunction with
the detailed description thereof, the foregoing description is intended to
illustrate and not limit
the scope of the invention, which is defined by the scope of the appended
claims. Other
aspects, advantages, and modifications are within the scope of the following
claims.
26