Note: Descriptions are shown in the official language in which they were submitted.
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
~~ TTENANT LES PAGES 1 A 281
NOTE : Pour les tomes additionels, veuillez contacter 1e Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 2
CONTAINING PAGES 1 TO 281
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME
NOTE POUR LE TOME / VOLUME NOTE:
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
COMPOSITIONS AND METHODS FOR THE THERAPY AND DIAGNOSIS OF
PROSTATE CANCER
S TECHNICAL FIELD OF THE INVENTION
The present invention relates generally to therapy and diagnosis of
cancer, such as prostate cancer. The invention is more specifically related to
polypeptides, comprising at least a portion of a prostate-specific protein,
and to
polynucleotides encoding such polypeptides. Such polypeptides and
polynucleotides
are useful in pharmaceutical compositions, e.g., vaccines, and other
compositions for
the diagnosis and treatment of prostate cancer.
BACKGROUND OF THE INVENTION
Cancer is a significant health problem throughout the world. Although
Cancer is a significant health problem throughout the world. Although advances
have
1 S been made in detection and therapy of cancer, no vaccine or other
universally successful
method for prevention or treatment is currently available. Current therapies,
which are
generally based on a combination of chemotherapy or surgery and radiation,
continue to
prove inadequate in many patients.
Prostate cancer is the most common form of cancer among males, with
an estimated incidence of 30% in men over the age of S0. Overwhelming clinical
evidence shows that human prostate cancer has the propensity to metastasize to
bone,
and the disease appears to progress inevitably from androgen dependent to
androgen
refractory status, leading to increased patient mortality. This prevalent
disease is
currently the second leading cause of cancer death among men in the U.S.
2S In spite of considerable research into therapies for the disease, prostate
cancer remains- difficult to treat. Commonly, treatment is based on surgery
and/or
radiation therapy, but these methods are ineffective in a significant
percentage of cases.
Two previously identified prostate specific proteins - prostate specific
antigen (PSA)
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
2
and prostatic acid phosphatase (PAP) - have limited therapeutic and diagnostic
potential. For example, PSA levels do not always correlate well with the
presence of
prostate cancer, being positive in a percentage of non-prostate cancer cases,
including
benign prostatic hyperplasia (BPH). Furthermore, PSA measurements correlate
with
S prostate volume, and do not indicate the level of metastasis.
In spite of considerable research into therapies for these and other
cancers, prostate cancer remains diff cult to diagnose and treat effectively.
Accordingly,
there is a need in the art for improved methods for detecting and treating
such cancers. .
The present invention fulf lls these needs and further provides other related
advantages.
SUMMARY OF THE INVENTION
In one aspect, the present invention provides polynucleotide
compositions comprising a sequence selected from the group consisting of:
(a) sequences provided in SEQ ID NO: 1-111, IlS-171, I73-175,
I77, 179-305, 307-315, 326, 328, 330, 332-335, 340-375, 381, 382 and 384-476,
524,
IS 526, 530, 531, 533, S3S, 536, 552, S69-572, 587, 591, S93-606, 6I8-626,
630, 631, 634,
636, 639-6SS, 674, 680, 681, 711, 713, 716, 720-722, 735, 737-739, 751, 753,
764, 765,
773-776 and 786-788;
(b) complements of the sequences provided in SEQ ID NO: 1-111,
11S-171, 173-175, 177, 179-305, 307-315, 326, 328, 330, 332-335, 340-375, 381,
382
and 384-476, 524, 526, 530, 531, 533, S3S, 536, SS2, S69-572, 587, 591, S93-
606, 618-
626, 630, 631, 634, 636, 639-6SS, 674, 680, 681, 71 l, 713, 716, 720-722, 735
737-739,
751, 753, 764, 765, 773-776 and 786-788;
(c) sequences consisting of at least 20 contiguous residues of a
sequence provided in SEQ ID NO: 1-111, 11S-171, 173-175, 177, 179-305, 307-
315,
2S 326, 328, 330, 332-335, 340-375, 381, 382 and 384-476, 524, 526, 530, 531,
533, 535,
536, SS2, S69-572, 587, 591, S93-606, 618-626, 630, 631, 634, 636, 639-6SS,
674, 680,
681, 71 l, 713, 716, 720-722, 735, 737-739, 751, 753, 764, 765, 773-776 and
786-788;
(d) sequences that hybridize to a sequence provided in SEQ ID NO:
1-111, 11S-17I, 173-175, 177, 179-305, 307-315, 326, 328, 330, 332-335, 340-
375,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
3
381, 382 and 384-476, 524, 526, 530, 531, 533, S3S, 536, SS2, S69-572, 587,
591, 593-
606, 6I8-626, 630, 63I, 634, 636, 639-6SS, 674, 680, 68I, 7I l, 713, 716, 720-
722, 735,
737-739, 751, 753, 764, 765, 773-776 and 786-788, under moderately stringent
conditions;
S (e) sequences having at least 7S% identity to a sequence of SEQ ID
NO: I-111, 115-171, 173-175, 177, 179-305, 307-315, 326, 328, 330, 332-335,
340-
375, 381, 382 and 384-476, 524, 526, 530, 531, 533, S3S, 536, SS2, 569-572,
587, 591,
593-606, 618-626, 630, 631, 634, 636, 639-6SS, 674, 680, 681, 711, 713, 716,
720-722,
735, 737-739, 751, 753, 764, 765, 773-776 and 786-788;
(f) sequences having at least 90% identity to a sequence of SEQ ID
NO: 1-111, 1I5-171, 173-175, 177, 179-305, 307-315, 326, 328, 330, 332-335,
340=
375, 38I, 382 and 384-476, 524, 526, 530, 531, 533, S3S, 536, SS2, 569-572,
587, 591,
593-606, 618-626, 630, 631, 634, 636, 639-6SS, 674, 680, 681, 711; 713, 716,
720-722,
735, 737-739, 7S 1, 753, 764, 765, 773-776 and 786-788; and
1 S (g) degenerate variants of a sequence provided in SEQ ID NO: I-
111, 115-171, 173-175, 177, 179-305, 307-315, 326, 328, 330, 332-335, 340-375,
381,
382 and 384-476, 524, 526, 530, 531, 533, S3S, 536, SS2, 569-572, 587, 591,
593-606,
618-626, 630, 631, 634, 636, 639-6SS, 674, 680, 681, 711, 713, 716, 720-722,
735, 737-
739, 751, 753, 764, 765, 773-776 and 786-788.
In one preferred embodiment, the polynucleotide compositions of the
invention are expressed in at least about 20%, more preferably in at least
about 30%,
and most preferably in at least about SO% of prostate tissue samples tested,
at a level
that is at least about 2-fold, preferably at least about S-fold, and most
preferably at least
about 10-fold higher than that for other normal tissues.
2S The present invention, in another aspect, provides polypeptide
compositions comprising an amino acid sequence that is encoded by a
polynucleotide
sequence described above.
The present invention further provides polypeptide compositions
comprising an amino acid sequence selected from the group consisting of
sequences
recited in SEQ ID NO: 112-114, 172, 176, 178, 327, 329, 331, 336, 339, 376-
380, 383,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
4
477-483, 496, 504, SOS, 519, 520, 522, 525, 527, 532, 534, S37-SS1, 553-568,
S73-586,
S88-590, 592, 627-629, 632, 633, 635, 637, 638, 6S6-671, 675, 683, 684, 710,
712, 714,
715, 717-719, 723-734, 736, 740-750, 752, 754, 755, 766-772, 777-785 and 789-
791.
In certain preferred embodiments, the polypeptides and/or
polynucleotides of the present invention are immunogenic, i.e., they are
capable of
eliciting an immune response, particularly a humoral and/or cellular immune
response,
as further described herein.
The present invention further provides fragments, variants and/or
derivatives of the disclosed polypeptide and/or polynucleotide sequences,
wherein the
fragments, variants and/or derivatives preferably have a level of immunogenic
activity
of at least about SO%, preferably at least about 70% and more preferably at
least about
90% of the Level of irmnunogenic activity of a polypeptide sequence set forth
in SEQ ID
NO: 112-114, 172, 176, 178, 327, 329, 331, 336, 339, 376-380, 383, 477-483,
496, S04,
505, 519, 520, 522, 52S, 527, 532, 534, 537-SSI, 553-568, 573-586, 588-590,
592, 627-
629, 632, 633, 635, 637, 638, 656-671, 675, 683, 684, 710, 712, 714, 715, 717-
719,
723-734, 736, 740-750, 752, 754, 7SS, 766-772, 777-785 or 789-791, or a
polypeptide
sequence encoded by a polynucleotide sequence set forth in SEQ ID NO: 1-111,
115-
171, 173-175, 177, 179-305, 307-315, 326, 328, 330, 332-335, 340-375, 381, 382
and
384-476, 524, 526, 530, 531, 533, 535, 536, 552, 569-572, 587, 591, 593-606,
618-626,
630, 631, 634, 636, 639-655, 674, 680, 681, 711, 713, 716, 720-722, 735, 737-
739, 751,
753, 764, 765, 773-776 and 786-788.
The present invention fiu ther provides polynucleotides that encode a
polypeptide described above, expression vectors comprising such
polynucleotides and
host cells transformed or transfected with such expression vectors.
Within other aspects, the present invention provides pharmaceutical
compositions comprising a polypeptide or polynucleotide as described above and
a
physiologically acceptable carrier.
Within a related aspect of the present invention, pharmaceutical
compositions, e.g., vaccine compositions, are provided for prophylactic or
therapeutic
applications. Such compositions generally comprise an immunogenic polypeptide
or
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
S
polynucleotide of the invention and an immunostimulant, such as an adjuvant,
together
with a physiologically acceptable carrier.
The present invention further provides pharmaceutical compositions that
comprise: (a) an antibody or antigen-binding fragment thereof that
specifically binds to
a polypeptide of the present invention, or a fragment thereof; and (b) a
physiologically
acceptable carrier.
Within further aspects, the present invention provides pharmaceutical
compositions comprising: (a) an antigen presenting cell that expresses a
polypeptide as
described above and (b) a pharmaceutically acceptable carrier or excipient.
Illustrative
antigen presenting cells include dendritic cells, macrophages, monocytes,
fibroblasts
and B cells.
Within related aspects, pharmaceutical compositions are provided that
comprise: (a) an antigen presenting cell that expresses a polypeptide as
described above
and (b) an immunostimulant.
The present invention further provides, in other aspects, fusion proteins
that comprise at least one polypeptide as described above, as well as
polynucleotides
encoding such fusion proteins, typically in the form of pharmaceutical
compositions,
e.g., vaccine compositions, comprising a physiologically acceptable carrier
and/or an
immunostimulant. The fusions proteins may comprise multiple immunogenic
polypeptides or portions/variants thereof, as described herein, and may
further comprise
one or more. polypeptide segments for facilitating and/or enhancing the
expression,
purification and/or immunogenicity of the polypeptide(s).
Within further aspects, the present invention provides methods for
stimulating an immune response in a patient, preferably a T cell response in a
human
patient, comprising administering a pharmaceutical composition described
herein. The
patient may be afflicted with prostate cancer, in which case the methods
provide
treatment for the disease, or a patient considered to be at risk for such a
disease may be
treated prophylactically.
Within further aspects, the present invention provides methods for
inhibiting the development of a cancer in a patient, comprising administering
to a
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
6
patient a pharmaceutical composition as recited above. The patient may be
afflicted
with prostate cancer, in which case the methods provide treatment for the
disease, or a
patient considered to be at risk for such a disease may be treated
prophylactically.
The present invention further provides, within other aspects, methods for
removing tumor cells from a biological sample, comprising contacting a
biological
sample with T cells that specifically react with a polypeptide of the present
invention,
wherein the step of contacting is performed under conditions and for a time
sufficient to
permit the removal of cells expressing the polypeptide from the sample.
Within related aspects, methods are provided for inhibiting the
development of a cancer in a patient, comprising administering to a patient a
biological
sample treated as described above.
Methods are further provided, within other aspects, for stimulating
and/or expanding T cells specific for a polypeptide of the present invention,
comprising
contacting T cells with one or more of: (i) a polypeptide as described above;
(ii) a
polynucleotide encoding such a polypeptide; and (iii) an antigen presenting
cell that
expresses such a polypeptide; under conditions and for a time sufficient to
permit the
stimulation and/or expansion of T cells. Isolated T cell populations
comprising T cells
prepared as described above are also provided.
Within further aspects, the present invention provides methods for
inhibiting the development of a cancer in a patient, comprising administering
to a
patient an effective amount of a T cell population as described above.
The present invention further provides methods for inhibiting the
development of a cancer in a patient, comprising the steps o~ (a) incubating
CD4+
and/or CD8+ T cells isolated from a patient with one or more of-. (i) a
polypeptide
comprising at least an immunogenic portion of polypeptide disclosed herein;
(ii) a
polynucleotide encoding such a polypeptide; and (iii) an antigen-presenting
cell that
expressed such a polypeptide; and (b) administering to the patient an
effective amount
of the proliferated T cells, thereby inhibiting the development of a cancer in
the patient.
Proliferated cells may, but need not, be cloned prior to administration to the
patient.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
7
Within further aspects, the present invention provides methods for
determining the presence or absence of a cancer, preferably a prostate cancer,
in a
patient comprising: (a) contacting a biological sample obtained from a patient
with a
binding agent that binds to a polypeptide as recited above; (b) detecting in
the sample an
amount of polypeptide that binds to the binding agent; and (c) comparing the
amount of
polypeptide with a predetermined cut-off value, and therefrom determining the
presence
or absence of a cancer in the patient. Within preferred embodiments, the
binding agent
is an antibody, more preferably a monoclonal antibody.
The present invention also provides, within other aspects, methods for
monitoring the progression of a cancer in a patient. Such methods comprise the
steps
of (a) contacting a biological sample obtained from a patient at a first point
in time
with a binding agent that binds to a polypeptide as recited above; (b)
detecting in the
sample an amount of polypeptide that binds to the binding agent; (c) repeating
steps (a)
and (b) using a biological sample obtained from the patient at a subsequent
point in
I S time; and (d) comparing the amount of polypeptide detected in step (c)
with the amount
detected in step (b), and therefrom monitoring the progression of the cancer
in the
patient.
The present invention further provides, within other aspects, methods for
determining the presence or absence of a cancer in a patient, comprising the
steps of: (a)
contacting a biological sample obtained from a patient with an oligonucleotide
that
hybridizes to a polynucleotide of the present invention; (b) detecting in the
sample a
level of a polynucleotide, preferably mRNA, that hybridizes to the
oligonucleotide; and
(c) comparing the level of polynucleotide that hybridizes to the
oligonucleotide with a
predetermined cut-off value, and therefrom determining the presence or absence
of a
cancer in the patient. Within certain embodiments, the amount of mRNA is
detected
via polymerase chain reaction using, for example, at least one oligonucleotide
primer
that hybridizes to a polynucleotide of the present invention, or a complement
of such a
polynucleotide. Within other embodiments, the amount of mRNA is detected using
a
hybridization technique, employing an oligonucleotide probe that hybridizes to
an
inventive polynucleotide, or a complement of such a polynucleotide.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
8
In related aspects, methods are provided for monitoring the progression
of a cancer in a patient, comprising the steps of (a) contacting a biological
sample
obtained from a patient with an oligonucleotide that hybridizes to a
polynucleotide of
the present invention; (b) detecting in the sample an amount of a
polynucleotide that
hybridizes to the oligonucleotide; (c) repeating steps (a) and (b) using a
biological
sample obtained from the patient at a subsequent point in time; and (d)
comparing the
amount of polynucleotide detected in step (c) with the amount detected in step
(b), and
therefrom monitoring the progression of the cancer in the patient.
Within further aspects, the present invention provides antibodies, such as
monoclonal antibodies, that bind to a polypeptide as described above, as well
as
diagnostic kits comprising such antibodies. Diagnostic kits comprising one or
more
oligonucleotide probes or primers as described above are also provided.
These and other aspects of the present invention will become apparent
upon reference to the following detailed description and attached drawings.
All
references disclosed herein are hereby incorporated by reference in their
entirety as if
each was incorporated individually.
BRIEF DESCRIPTION OF THE DRAWINGS AND SEQUENCE IDENTIFIERS
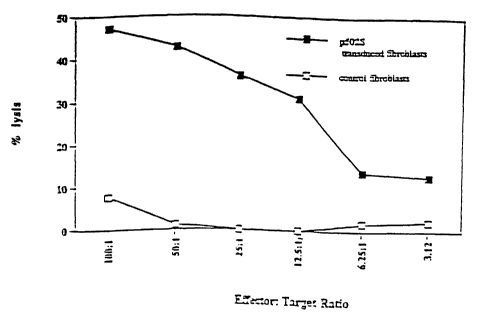
Figure 1 illustrates the ability of T cells to kill fibroblasts expressing the
representative prostate-specific polypeptide P502S, as compared to control
fibroblasts.
The percentage lysis is shown as a series of effectoraarget ratios, as
indicated.
Figures 2A and 2B illustrate the ability of T cells to recognize cells
expressing the representative prostate-specific polypeptide P502S. In each
case, the
number of y-interferon spots is shown for different numbers of responders. In
Figure
2A, data is presented for fibroblasts pulsed with the P2S-12 peptide, as
compared to
fibroblasts pulsed with a control E75 peptide. In Figure 2B, data is presented
for
fibroblasts expressing P502S, as compared to fibroblasts expressing HER-2/neu.
Figure 3 represents a peptide competition binding assay showing that the
P1S#10 peptide, derived from PSO1S, binds HLA-A2. Peptide P1S#10 inhibits HLA-
A2 restricted presentation of fluM58 peptide to CTL clone D150M58 in TNF
release
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
9
bioassay. D150M58 CTL is specific for the HLA-A2 binding influenza matrix
peptide
fluMS 8.
Figure 4 illustrates the ability of T cell lines generated from P1S#10
immunized mice to specifically lyse P1S#10-pulsed Jurkat A2Kb targets and
P501S-
transduced Jurkat A2Kb targets, as compared to EGFP-transduced Jurkat A2Kb.
The
percent lysis is shown as a series of effector to target ratios, as indicated.
Figure 5 illustrates the ability of a T cell clone to recognize and
specifically lyse Jurkat A2Kb cells expressing the representative prostate-
specific
polypeptide PSO1S, thereby demonstrating that the P1S#10 peptide may be a
naturally
processed epitope ofthe PSOlS polypeptide.
Figures 6A and 6B are graphs illustrating the specificity of a CD8+ cell
line (3A-1) for a representative prostate-specific antigen (P501 S). Figure 6A
shows the
results of a SICr release assay. The percent specific lysis is shown as a
series of
effectoraarget xatios, as indicated. Figure 6B shows the production of
interferon-
I 5 gamma by 3A-1 cells stimulated with autologous B-LCL transduced with P501
S, at
varying effectoraarget rations as indicated.
Figure 7 is a Western blot showing the expression of P501 S in
baculovirus.
Figure 8 illustrates the results of epitope mapping studies on P501 S.
Figure 9 is a schematic representation of the P501 S protein showing the
location of transmembrane domains and predicted intracellular and
extracellular
domains.
Figure 10 is a genomic map showing the location of the prostate genes
P775P, P704P, B305D, P712P and P774P within the Cat Eye Syndrome region of
chromosome 22q1 I.2
Figure 11 shows the results of an ELTSA assay to determine the
specificity of rabbit polyclonal antisera raised against PSOI S.
SEQ ID NO: 1 is the determined cDNA sequence for F1-13
SEQ ID NO: 2 is the determined 3' cDNA sequence for FI-12
SEQ ID NO: 3 is the determined 5' cDNA sequence for Fl-12
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
SEQ ID NO: 4 is the determined 3' cDNA sequence fox F1-16
SEQ ID NO: S is the determined 3' cDNA sequence for Hl-1
SEQ ID NO: 6 is the determined 3' cDNA sequence for HI-9
SEQ ID NO: 7 is the determined 3' cDNA sequence for H1-4
S SEQ ID NO: 8 is the determined 3' cDNA sequence for J1-17
SEQ ID NO: 9 is the determined S' cDNA sequence for JI-17
SEQ ID NO: 10 is the determined 3' cDNA sequence for L1-12
SEQ ID NO: 11 is the determined S' cDNA sequence for Ll-12
SEQ ID NO: 12 is the determined 3' cDNA sequence for N1-1862
10 SEQ ID NO: 13 is the determined S' cDNA sequence for Nl-1862
SEQ ID NO: 14 is the determined 3' cDNA sequence for Jl-13
SEQ ID NO: 1 S is the determined S' cDNA sequence for Jl-13
SEQ ID NO: 16 is the determined 3' cDNA sequence for J1-19
SEQ ID NO: 17 is the determined S' cDNA sequence for J1-19
1 S SEQ ID NO: 18 is the determined 3' cDNA sequence for J 1-25
SEQ ID NO: 19 is the determined S' cDNA sequence for Jl-2S
SEQ ID NO: 20 is the determined S' cDNA sequence for J1-24
SEQ ID NO: 21 is the determined 3' cDNA sequence for J1-24
SEQ ID NO: 22 is the determined S' cDNA sequence for KI-S8
SEQ ID NO: 23 is the determined 3' cDNA sequence for K1-S8
SEQ ID NO: 24 is the determined S' cDNA sequence for Kl-63
SEQ ID NO: 2S is the determined 3' cDNA sequence for Kl-63
SEQ ID NO: 26 is the determined S' cDNA sequence for Ll-4
SEQ ID NO: 27 is the determined 3' cDNA sequence for Ll-4
2S SEQ ID NO: 28 is the determined S' cDNA sequence for L1-14
SEQ ID NO: 29 is the determined 3' cDNA sequence for L1-14
SEQ ID NO: 30 is the determined 3' cDNA sequence for Jl-12
SEQ ID NO: 31 is the determined 3' cDNA sequence for Jl-16
SEQ ID NO: 32 is the determined 3' cDNA sequence for Jl-21
SEQ ID NO: 33 is the determined 3' cDNA sequence for Kl-48
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
11
SEQ ID NO: 34 is the cDNA sequence for
determined 3' K1-55
SEQ ID NO: 35 is the cDNA sequence for
determined 3' L1-2
SEQ ID NO: 36 is the cDNA sequence for
determined 3' Ll-6
SEQ ID NO: 37 is the cDNA sequence for
determined 3' N1-1858
SEQ ID NO: 38 is the determined 3' cDNA sequence for N1-1860
SEQ ID NO: 39 is the determined 3' cDNA sequence for N1-1861
SEQ ID NO: 40 is the determined 3' cDNA sequence forNl-1864
SEQ ID NO: 41 is the determined cDNA sequence fox PS
SEQ ID NO: 42 is the determined cDNA sequence fox P8
SEQ ID NO: 43 is the determined cDNA sequence for P9
SEQ ID NO: 44 is the determined cDNA sequence for P18
SEQ ID NO: 45 is the determined cDNA sequence for P20
SEQ ID NO: 46 is the deteixnined cDNA sequence for P29
SEQ ID NO: 47 is the determined cDNA sequence for P30
SEQ ID NO: 48 is the determined cDNA sequence for P34
SEQ ID NO: 49 is the determined cDNA sequence for P36
SEQ ID NO: 50 is the determined cDNA sequence for P38
SEQ ID NO: 51 is the determined cDNA sequence for P39
SEQ ID NO: 52 is the determined cDNA sequence for P42.
SEQ ID NO: 53 is the determined cDNA sequence for P47
SEQ ID NO: 54 is the determined cDNA sequence for P49
SEQ ID NO: 55 is the determined cDNA sequence for P50
SEQ ID NO: 56 is the determined cDNA sequence for P53
SEQ ID NO: 57 is the determined cDNA sequence for P55
SEQ ID NO: 58 is the determined cDNA sequence for P60
SEQ ID NO: 59 is the determined cDNA sequence for P64
SEQ ID NO: 60 is the determined cDNA sequence for P65
SEQ ID NO: 61 is the determined cDNA sequence for P73
SEQ ID NO: 62 is the determined cDNA sequence for P75
SEQ ID NO: 63 is the determined cDNA sequence for P76
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
12
SEQ ID NO: 64 is the determined cDNA sequence for P79
SEQ ID NO: 65 is the determined cDNA sequence for P84
SEQ ID NO: 66 is the determined cDNA sequence for P68
SEQ ID NO: 67 is the determined cDNA sequence for P80 (also referred
to as P704P)
SEQ ID NO: 68 is the determined cDNA sequence for P82
SEQ ID NO: 69 is the determined cDNA sequence for U1-3064
SEQ ID NO: 70 is the determined cDNA sequence for U1-3065
SEQ ID NO: 71 is the determined cDNA sequence for V 1-3692
IO SEQ ID NO: 72 is the determined cDNA sequence for 1A-3905
SEQ ID NO: 73 is the determined cDNA sequence for V1-3686
SEQ ID NO: 74 is the determined cDNA sequence for Rl-2330
SEQ ID NO: 75 is the determined cDNA sequence for 1B-3976
SEQ ID NO: 76 is the determined cDNA sequence for VI-3679
SEQ ID NO: 77 is the determined cDNA sequence for 1G-4736
SEQ ID NO: 78 is the determined cDNA sequence for 1G-4738
SEQ ID NO: 79 is the determined cDNA sequence for I G-4741
SEQ ID NO: 80 is the determined cDNA sequence for 1 G-4744
SEQ ID NO: 81 is the determined cDNA sequence for I G-4734
SEQ ID NO: 82 is the determined cDNA sequence for IH-4774
SEQ ID NO: 83 is the determined cDNA sequence for 1H-4781
SEQ ID NO: 84 is the determined cDNA sequence for 1H-4785
SEQ ID NO: 85 is the determined cDNA sequence for 1H-4787
SEQ ID NO: 86 is the determined cDNA sequence for 1 H-4796
SEQ ID NO: 87 is the determined cDNA sequence for 1I-4807
SEQ ID NO: 88 is the determined cDNA sequence for 1I-4810
SEQ ID NO: 89 is the determined cDNA sequence for 1I-4811
SEQ ID NO: 90 is the determined cDNA sequence for 1 J-4876
SEQ ID NO: 91 is the determined cDNA sequence for 1K-4884
SEQ ID NO: 92 is the determined cDNA sequence for 1K-4896
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
13
SEQ ID NO: 93 is the determined cDNA sequence for 1 G-4761
SEQ ID NO: 94 is the determined cDNA sequence for 1 G-4762
SEQ ID NO: 95 is the determined cDNA sequence for 1H-4766
SEQ. ID NO: 96 is the determined cDNA sequence for 1 H-4770
SEQ ID NO: 97 is the determined cDNA sequence for 1 H-4771
SEQ ID NO: 98 is the determined cDNA sequence for 1 H-4772
SEQ ID NO: 99 is the determined cDNA sequence for 1D-4297
SEQ ID NO: 100 is the determined cDNA sequence for 1D-4309
SEQ ID NO: 101 is the determined cDNA sequence for 1D.1-4278
SEQ ID NO: 102 is the determined cDNA sequence for 1D-4288
SEQ ID NO: 103 is the determined cDNA sequence for 1D-4283
SEQ ID NO: 104 is the determined cDNA sequence for 1D-4304
SEQ ID NO: 105 is the determined cDNA sequence for 1D-4296
SEQ ID NO: 106 is the determined cDNA sequence for ID-4280
SEQ ID NO: 107 is the determined full length cDNA sequence for Fl-12
(also referred to as P504S)
SEQ ID NO: 108 is the predicted amino acid sequence for Fl-12
SEQ ID NO: 109 is the determined full length cDNA sequence for J1-17
SEQ ID NO: 110 is the determined full length cDNA sequence for Ll-12
(also referred to as P501 S)
SEQ ID NO: 111 is the determined full 'length cDNA sequence for N1-
1862 (also referred to as P503S)
SEQ ID NO: 112 is the predicted amino acid sequence for J1-17
SEQ ID NO: 113 is the predicted amino acid sequence for Ll-12 (also
referred to as P501 S)
SEQ ID NO: 114 is the predicted amino acid sequence for N1-1862 (also
referred to as P503S)
SEQ ID NO: 115 is the determined cDNA sequence for P89
SEQ ID NO: 116 is the determined cDNA sequence for P90
SEQ ID NO: 117 is the determined cDNA sequence for P92
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
14
SEQ ID NO: I 18 is the determined cDNA sequence for P95
SEQ ID NO: 119 is the determined cDNA sequence for P98
SEQ ID NO: 120 is the determined cDNA sequence for P102
SEQ ID NO: 121 is the determined cDNA sequence for P 110
SEQ ID NO: 122 is the determined cDNA sequence for P 111
SEQ ID NO: 123 is the determined cDNA sequence for P 114
SEQ ID NO: 124 is the determined cDNA sequence for P115
SEQ ID NO: 125 is the determined cDNA sequence for P116
SEQ ID NO: 126 is the determined cDNA sequence for P 124
SEQ ID NO: 127 is the determined cDNA sequence for P126
SEQ ID NO: 128 is the determined cDNA sequence for P130
SEQ ID NO: 129 is the determined cDNA sequence for P133
SEQ ID NO: 130 is the determined cDNA sequence for P138
SEQ ID NO: 131 is the determined cDNA sequence for P143
SEQ ID NO: 132 is the determined cDNA sequence for P151
SEQ ID NO: 133 is the determined cDNA sequence for P156
SEQ ID NO: 134 is the determined cDNA sequence for P157
SEQ ID NO: 135 is the determined cDNA sequence for P166
SEQ ID NO: 136 is the determined cDNA sequence for P176
SEQ ID NO: 137 is the determined cDNA sequence for P178
SEQ ID NO: 138 is the determined cDNA sequence for P179
SEQ ID NO: 139 is the determined cDNA sequence for P 185
SEQ ID NO: 140 is the determined cDNA sequence for P 192
SEQ ID NO: 141 is the determined cDNA sequence fox P201
SEQ ID NO: 142 is the determined cDNA sequence for P204
SEQ ID NO: 143 is the determined cDNA sequence fox P208
SEQ ID NO: 144 is the determined cDNA sequence for P211
SEQ ID NO: 145 is the determined cDNA sequence for P213
SEQ ID NO: 146 is the determined cDNA sequence for P219
SEQ ID NO: 147 is the determined cDNA sequence for P237
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
SEQ ID NO: 148 is the determined cDNA sequence for P239
SEQ ID NO: 149 is the determined cDNA sequence for P248
SEQ ID NO: 150 is the determined cDNA sequence for P251
SEQ ID NO: 151 is the determined cDNA sequence for P255
5 SEQ ID NO: 152 is the determined cDNA sequence for P256
SEQ ID NO: 153 is the determined cDNA sequence for P259
SEQ ID NO: 154 is the determined cDNA sequence for P260
SEQ ID NO: 155 is the determined cDNA sequence for P263
SEQ ID Nb: 156 is the determined cDNA sequence for P264
10 SEQ ID NO: 157 is the determined cDNA sequence for P266
SEQ ID NO: 158 is the determined cDNA sequence for P270
SEQ ID NO: 159 is the determined cDNA sequence for P272
SEQ ID NO: 160 is the determined cDNA sequence for P278
SEQ ID NO: 161 is the determined cDNA sequence for P105
I 5 SEQ ID NO: I 62 is the determined cDNA sequence for P 107
SEQ ID NO: 163 is the determined cDNA sequence for PI37
SEQ ID NO: 164 is the determined cDNA sequence for PI94
SEQ ID NO: 165 is the determined cDNA sequence for P 195
SEQ ID NO: 166 is the determined cDNA sequence for P 196
SEQ ID NO: 167 is the determined cDNA sequence for P220
SEQ ID NO: 168 is the determined cDNA sequence for P234
SEQ ID NO: 169 is the determined cDNA sequence for P235
SEQ ID NO: I70 is the determined cDNA sequence for P243
SEQ ID NO: 171 is the determined cDNA sequence for P703P-DE1
SEQ ID NO: I72 is the predicted amino acid sequence for P703P-DEI
SEQ ID NO: I73 is the determined cDNA sequence for P703P-DE2
SEQ ID NO: 174 is the determined cDNA sequence for P703P-DE6
SEQ ID NO: 175 is the determined cDNA sequence for P703P-DE13
SEQ ID NO: 176 is the predicted amino acid sequence for P703P-DE13
SEQ ID NO: 177 is the determined cDNA sequence for P703P-DE14
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
16
SEQ ID NO: I78 is the predicted amino acid sequence for P703P-DE14
SEQ ID NO: 179 is the determined extended cDNA sequence for I G-
4736
SEQ ID NO: 180 is the determined extended cDNA sequence for I G-
4738
SEQ ID NO: 181 is the determined extended cDNA sequence for 1 G-
4741
SEQ ID NO: 182 is the determined extended cDNA sequence for 1 G-
4744
IO SEQ ID NO: I83 is the determined extended cDNA sequence for 1H-
4774
SEQ ID NO: 184 is the determined extended cDNA sequence for 1H-
4781
SEQ ID NO: 185 is the determined extended cDNA sequence for 1H-
IS 4785
SEQ ID NO: 186 is the determined extended cDNA sequence for 1H-
4787
SEQ ID NO: 187 is the determined extended cDNA sequence for 1H-
4796
20 SEQ ID NO: 188 is the determined extended cDNA sequence fox 1I-
4807
SEQ ID NO: 189 is the determined 3' cDNA sequence for 1I-4810
SEQ ID NO: 190 is the determined 3' cDNA sequence for 1I-4811
SEQ ID NO: 191 is the determined extended cDNA sequence fox 1 J-
25 4876
SEQ ID NO: 192 is the determined extended cDNA sequence for 1K-
4884
SEQ ID NO: 193 is the determined extended cDNA sequence for 1 K-
4896
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
17
SEQ ID NO: 194 is the determined extended cDNA sequence for 1 G-
4761
SEQ ID NO: 195 is the determined extended cDNA sequence for 1 G-
4762
SEQ ID NO: 196 is the determined extended cDNA sequence for 1 H-
4766
SEQ ID NO: 197 is the determined 3' cDNA sequence for 1H-4770
SEQ ID NO: 198 is the determined 3' cDNA sequence for 1H-4771
SEQ ID NO: 199 is the determined extended cDNA sequence for 1H-
4772
SEQ ID NO: 200 is the determined extended cDNA sequence for 1D-
4309
SEQ ID NO: 201 is the determined extended cDNA sequence for 1D.1-
4278
SEQ ID NO: 202 is the determined extended cDNA sequence for 1 D-
4288
SEQ ID NO: 203 is the determined extended cDNA sequence for 1D-
4283
SEQ ID NO: 204 is the determined extended cDNA sequence for 1 D-
4304
SEQ ID NO: 205 is the determined extended cDNA sequence for 1D-
4296
SEQ ID NO: 206 is the determined extended cDNA sequence fox 1D-
4280
SEQ ID NO: 207 is, the determined cDNA sequence for 10-d8fwd
SEQ ID NO: 208 is the determined cDNA sequence for 10-HlOcon
SEQ ID NO: 209 is the determined cDNA sequence for 11-CBrev
SEQ ID NO: 210 is the determined cDNA sequence for 7.g6fwd
SEQ ID NO: 211 is the determined cDNA sequence for 7.g6rev
SEQ ID NO: 212 is the determined cDNA sequence for 8-b5fwd
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
18
SEQ ID NO: 213 is the determined cDNA sequence for 8-b5rev
SEQ ID NO: 214 is the determined cDNA sequence for 8-b6fwd
SEQ ID NO: 215 is the determined cDNA sequence for 8-b6 rev
SEQ ID NO: 216 is the determined cDNA sequence for 8-d4fwd
SEQ ID NO: 217 is the determined cDNA sequence for 8-d9rev
SEQ ID NO: 218 is the determined cDNA sequence for 8-g3fwd
SEQ ID NO: 219 is the determined cDNA sequence for 8-g3rev
SEQ ID NO: 220 is the determined cDNA sequence for 8-hl lrev
SEQ ID NO: 221 is the determined cDNA sequence for g-fl2fwd
SEQ ID NO: 222 is the determined cDNA sequence for g-f3rev
SEQ ID NO: 223 is the determined cDNA sequence for P509S
SEQ ID NO: 224 is the determined cDNA sequence for PS l OS
SEQ ID NO: 225 is the determined cDNA sequence for P703DE5
SEQ ID NO: 226 is the determined cDNA sequence for 9-Al 1
SEQ ID NO: 227 is the determined cDNA sequence for 8-C6
SEQ ID NO: 228 is the determined cDNA sequence for 8-H7
SEQ ID NO: 229 is the determined cDNA sequence for JPTPN13
SEQ ID NO: 230 is the determined cDNA sequence for JPTPN14
SEQ ID NO: 231 is the determined cDNA sequence for JPTPN23
SEQ ID NO: 232 is the determined cDNA sequence for JPTPN24
SEQ ID NO: 233 is the determined cDNA sequence for JPTPN25
SEQ ID NO: 234 is the determined cDNA sequence for JPTPN30
SEQ ID NO: 235 is the determined cDNA sequence for JPTPN34
SEQ ID NO: 236 is the determined cDNA sequence for PTPN35
SEQ ID NO: 237 is the determined cDNA sequence for JPTPN36
SEQ ID NO: 238 is the determined cDNA sequence for JPTPN38
SEQ ID NO: 239 is the determined cDNA sequence for JPTPN39
SEQ ID NO: 240 is the determined cDNA sequence for JPTPN40
SEQ ID NO: 241 is the determined cDNA sequence for JPTPN41
SEQ ID NO: 242 is the determined cDNA sequence for JPTPN42
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
19
SEQ ID NO: 243 is the determined cDNA sequence for JPTPN45
SEQ ID NO: 244 is the determined cDNA sequence for JPTPN46
SEQ ID NO: 245 is the determined cDNA sequence for JPTPN51
SEQ ID NO: 246 is the determined cDNA sequence fox JPTPN56
SEQ ID NO: 247 is the determined cDNA sequence for PTPN64
SEQ ID NO: 248 is the determined cDNA sequence for JPTPN65
SEQ ID NO: 249 is the determined cDNA sequence for JPTPN67
SEQ ID NO: 250 is the determined cDNA sequence for JPTPN76
SEQ ID NO: 251 is the determined cDNA sequence for JPTPN84
SEQ ID NO: 252 is the determined cDNA sequence for JPTPN85
SEQ ID NO: 253 is the determined cDNA sequence for JPTPN86
SEQ ID NO: 254 is the determined cDNA sequence for JPTPN87
SEQ ID NO: 255 is the determined cDNA sequence for JPTPN88
SEQ ID NO: 256 is the determined cDNA sequence for JP1F1
SEQ ID NO: 257 is the determined cDNA sequence for JP1F2
SEQ ID NO: 258 is the determined cDNA sequence for JP1C2
SEQ ID NO: 259 is the determined cDNA sequence for JP 1 B 1
SEQ ID NO: 260 is the determined cDNA sequence for JP 1 B2
SEQ ID NO: 261 is the determined cDNA sequence for JP1D3
SEQ ID NO: 262 is the determined cDNA sequence for JP 1 A4
SEQ ID NO: 263 is the determined cDNA sequence for JP1F5
SEQ ID NO: 264 is the determined cDNA sequence for JP 1 E6
SEQ ID NO: 265 is the determined cDNA sequence for JP1D6
SEQ ID NO: 266 is the determined cDNA sequence for JP1B5
SEQ ID NO: 267 is the determined cDNA sequence for JP 1 A6
SEQ ID NO: 268 is the determined cDNA sequence for JP1E8
SEQ ID NO: 269 is the determined cDNA sequence for JP 1 D7
SEQ ID NO: 270 is the determined cDNA sequence for JP1D9
SEQ ID NO: 271 is the determined cDNA sequence for JP1C10
SEQ ID NO: 272 is the determined cDNA sequence for JP1A9
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
SEQ ID NO: 273 is the determined cDNA sequence for JP1F12
SEQ ID NO: 274 is the determined cDNA sequence for JPlEl2
SEQ ID NO: 275 is the determined cDNA sequence for JP 1D 11
SEQ ID NO: 276 is the determined cDNA sequence for JP 1 C 11
5 SEQ ID NO: 277 is the determined cDNA sequence for JP 1 C 12
SEQ ID NO: 278 is the determined cDNA sequence for JP1B12
SEQ ID NO: 279 is the determined cDNA sequence for JPlAl2
SEQ ID NO: 280 is the determined cDNA sequence for JP8G2
SEQ ID NO: 281 is the determined cDNA sequence for JP8H1
10 SEQ ID NO: 282 is the determined cDNA sequence for JP8H2
SEQ ID NO: 283 is the determined cDNA sequence for JP8A3
SEQ ID NO: 284 is the determined cDNA sequence for JP8A4
SEQ ID NO: 285 is the determined cDNA sequence for JP8C3
SEQ ID NO: 286 is the determined cDNA sequence for JP8G4
IS SEQ ID NO: 287 is the determined cDNA sequence for JP8B6
SEQ ID NO: 288 is the determined cDNA sequence for JP8D6
SEQ ID NO: 289 is the determined cDNA sequence for JP8F5
SEQ ID NO: 290 is the determined cDNA sequence for JP8A8
SEQ ID NO: 291 is the determined cDNA sequence for JP8C7
20 SEQ ID NO: 292 is the determined cDNA sequence for JP8D7
SEQ ID NO: 293 is the determined cDNA sequence for P8D8
SEQ ID NO: 294 is the determined cDNA sequence for JP8E7
SEQ ID NO: 295 is the determined cDNA sequence for JP8F8
SEQ ID NO: 296 is the determined cDNA sequence for JP8G8
SEQ ID NO: 297 is the determined cDNA sequence for JPBB 10
SEQ ID NO: 298 is the determined cDNA sequence for JP8C10
SEQ ID NO: 299 is the determined cDNA sequence for JP8E9
SEQ ID NO: 300 is the determined cDNA sequence for JP8E10
SEQ ID NO: 301 is the determined cDNA sequence for JP8F9
SEQ ID NO: 302 is the determined cDNA sequence for JP8H9
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
21
SEQ ID NO: 303 is the determined cDNA sequence for JP8C12
SEQ ID NO: 304 is the determined cDNA sequence for JP8E11
SEQ ID NO: 305 is the determined cDNA sequence for JP8E12
SEQ ID NO: 306 is the amino acid sequence for the peptide PS2#12
SEQ ID NO: 307 is the determined cDNA sequence for P711P
SEQ ID NO: 308 is the determined cDNA sequence for P712P
SEQ ID NO: 309 is the determined cDNA sequence for CLONE23
SEQ ID NO: 310 is the determined cDNA sequence for P774P
SEQ ID NO: 311 is the determined cDNA sequence for P775P
SEQ ID NO312 is the determined cDNA sequence for P715P
SEQ ID NO: 313 is the determined cDNA sequence for P710P
' SEQ ID NO: 314 is the determined cDNA sequence for P767P
SEQ ID NO: 315 is the determined cDNA sequence for P768P
SEQ ID NO: 316-325 are the determined cDNA sequences of previously
isolated genes
SEQ ID NO: 326 is the determined cDNA sequence for P703PDE5
SEQ ID NO: 327 is the predicted amino acid sequence fox P703PDE5
SEQ ID NO: 328 is the determined cDNA sequence for P703P6.26
SEQ ID NO: 329 is the predicted amino acid sequence fox P703P6.26
SEQ ID NO: 330 is the determined cDNA sequence for P703PX-23
SEQ ID NO: 331 is the predicted amino acid sequence for,P703Px-23
SEQ ID NO: 332 is the determined full length cDNA sequence for
P509S
SEQ ID NO: 333 is the determined extended cDNA sequence for P707P
(also referred to as 11-C9)
SEQ ID NO: 334 is the determined cDNA sequence for P714P
SEQ ID NO: 335 is the determined cDNA sequence for P705P (also
referred to as 9-F3)
SEQ ID NO: 336 is the predicted amino acid sequence for P705P
SEQ ID NO: 337 is the amino acid sequence of the peptide P1S#10
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
22
SEQ ID NO: 338 is the amino acid sequence of the peptide p5
SEQ ID NO: 339 is the predicted amino acid sequence of P509S
SEQ ID NO: 340 is the determined cDNA sequence for P778P
SEQ ID NO: 341 is the determined cDNA sequence for P786P
SEQ ID NO: 342 is the determined cDNA sequence for P789P
SEQ ID NO: 343 is the determined cDNA sequence for a clone showing
homology to Homo Sapiens MM46 mRNA
SEQ ID NO: 344 is the determined cDNA sequence for a clone showing
homology to Homo Sapiens TNF-alpha stimulated ABC protein (ABC50) mRNA
SEQ ID NO: 345 is the determined cDNA sequence for a clone showing
homology to Homo sapiens mRNA for E-cadherin
SEQ ID NO: 346 is the determined cDNA sequence for a clone showing
homology to Human nuclear-encoded mitochondria) serine
hydroxymethyItransferase
(SHMT)
SEQ ID NO: 347 is the determined cDNA sequence for a clone showing
homology to Homo sapiens natural resistance-associated macrophage protein2
(NRAMP2)
SEQ ID NO: 348 is the determined cDNA sequence for a clone showing
homology to Homo Sapiens phosphoglucomutase-related protein (PGMRP)
SEQ ID NO: 349 is the determined cDNA sequence for a clone showing
homology to Human mRNA for proteosome subunit p40
SEQ ID NO: 350 is the determined cDNA sequence for P777P
SEQ ID NO: 351 is the determined cDNA sequence for P779P
SEQ ID NO: 352 is the determined cDNA sequence for P790P
SEQ ID NO: 353 is the determined cDNA sequence for P784P
SEQ ID NO: 354 is the determined cDNA sequence for P776P
SEQ ID NO: 355 is the determined cDNA sequence for P780P
SEQ ID NO: 356 is the determined cDNA sequence for P544S
SEQ ID NO: 357 is the determined cDNA sequence for P745S
SEQ ID NO: 358 is the determined cDNA sequence for P782P
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
23
SEQ ID NO: 359 is the determined cDNA sequence for P783P
SEQ ID NO: 360 is the determined cDNA sequence for unknown 17984
SEQ ID NO: 361 is the detemnined cDNA sequence for P787P
SEQ ID NO: 362 is the determined cDNA sequence for P78~8P
SEQ ID NO: 363 is the determined cDNA sequence for unknown 17994
SEQ ID NO: 364 is the determined cDNA sequence for P781P
SEQ ID NO: 365 is the determined cDNA sequence for P785P
SEQ ID NO: 366-375 are the determined cDNA sequences for splice
variants of B305D.
SEQ ID NO: 376 is the predicted amino acid sequence encoded by the
sequence of SEQ ID NO: 366.
SEQ ID NO: 377 is the predicted amino acid sequence encoded by the
sequence of SEQ ID NO: 372.
SEQ ID NO: 378 is the predicted amino acid sequence encoded by the
sequence of SEQ ID NO: 373.
SEQ ID NO: 379 is the predicted amino acid sequence encoded by the
sequence of SEQ ID NO: 374.
SEQ ID NO: 380 is the predicted. amino acid sequence encoded by the
sequence of SEQ ID NO: 375.
SEQ ID NO: 381 is the determined cDNA sequence for B716P.
SEQ ID NO: 382 is the determined full-length cDNA sequence for
P711P.
SEQ ID NO: 383 is the predicted amino acid sequence for P711P.
SEQ ID NO: 384 is the cDNA sequence for P1000C.
SEQ ID NO: 385 is the cDNA sequence for CGI-82.
SEQ ID N0:386 is the cDNA sequence for 23320.
SEQ ID N0:387 is the cDNA sequence for CGI-69.
SEQ ID N0:388 is the cDNA sequence for L-iditol-2-dehydrogenase.
SEQ ID N0:389 is the cDNA sequence for 23379.
SEQ ID N0:390 is the cDNA sequence for 23381.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
24
SEQ ID N0:391 is the cDNA sequence for KIAA0122.
SEQ ID N0:392 is the cDNA sequence for 23399.
SEQ ID N0:393 is the cDNA sequence for a previously identified gene.
SEQ ID N0:394 is the cDNA sequence for HCLBP.
SEQ ID N0:3~95 is the cDNA sequence for transglutaminase.
SEQ ID N0:396 is the cDNA sequence for a previously identified gene.
SEQ ID N0:397 is the cDNA sequence for PAP.
SEQ ID N0:398 is the cDNA sequence for Ets transcription factor
PDEF.
SEQ ID N0:399 is the cDNA sequence for hTGR.
SEQ ID N0:400 is the cDNA sequence for KIAA0295.
SEQ ID N0:401 is the cDNA sequence for 22545.
SEQ ID N0:402 is the cDNA sequence for 22547.
SEQ ID N0:403 is the cDNA sequence for 22548.
SEQ ID N0:404 is the cDNA sequence for 22550.
t
SEQ ID N0:405 is the cDNA sequence for 22551.
SEQ ID N0:406 is the cDNA sequence for 22552.
SEQ ID N0:407 is the cDNA sequence for 22553 (also known as
P 1020C).
SEQ ID N0:408 is the cDNA sequence for 22558.
SEQ ID N0:409 is the cDNA sequence for 22562.
SEQ ID N0:410 is the cDNA sequence for 22565.
SEQ ID N0:411 is the cDNA sequence for 22567.
SEQ ID N0:412 is the cDNA sequence for 22568.
SEQ ID N0:413 is the cDNA sequence for 22570.
SEQ ID N0:414 is the cDNA sequence for 22571.
SEQ ID N0:415 is the cDNA sequence for 22572.
SEQ ID N0:416 is the cDNA sequence for 22573.
SEQ ID N0:417 is the cDNA sequence for 22573.
SEQ ID N0:418 is the cDNA sequence for 22575.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
SEQ ID NO:419 is the cDNA sequence for 22580.
SEQ ID N0:420 is the cDNA sequence for 22581.
SEQ ID N0:421 is the cDNA sequence for 22582.
SEQ ID N0:422 is the cDNA sequence for 22583.
5 SEQ ID N0:423 is the cDNA sequence for 22584.
SEQ ID N0:424 is the cDNA sequence for 22585.
SEQ ID N0:425 is the cDNA sequence fox 22586.
SEQ ID N0:426 is the cDNA sequence fox 22587.
SEQ ID N0:427 is the cDNA sequence for 22588:
10 SEQ ID N0:428 is the cDNA sequence for 22589.
SEQ ID N0:429 is the cDNA sequence for 22590.
SEQ ID N0:430 is the cDNA sequence for 22591.
SEQ ID N0:431 is the cDNA sequence for 22592.
SEQ ID N0:432 is the cDNA sequence for 22593.
1 S SEQ ID N0:433 is the cDNA sequence for 22594.
SEQ ID N0:434 is the cDNA sequence for 22595.
SEQ ID NO:435 is the cDNA sequence for 22596.
SEQ ID N0:436 is the cDNA sequence for 22847.
SEQ ID N0:437 is the cDNA sequence for 22848.
20 SEQ ID N0:438 is the cDNA sequence for 22849.
SEQ ID N0:439 is the cDNA sequence for 22851.
SEQ ID N0:440 is the cDNA sequence for 22852.
SEQ ID N0:441 is the cDNA sequence for 22853.
SEQ ID N0:442 is the cDNA sequence for 22854.
25 SEQ ID N0:443 is the cDNA sequence for 22855.
SEQ ID N0:444 is the cDNA sequence for 22856.
SEQ ID N0:445 is the cDNA sequence for 22857.
SEQ ID N0:446 is the cDNA sequence for 23601.
SEQ ID N0:447 is the cDNA sequence for 23602.
SEQ ID N0:448 is the cDNA sequence for 23605.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
26
SEQ ID N0:449 is the cDNA sequence for 23606.
SEQ ID N0:450 is the cDNA sequence for 23612.
SEQ ID N0:451 is the cDNA sequence for 23614.
SEQ ID N0:452 is the cDNA sequence for 23618.
SEQ ID N0:453 is the cDNA sequence for 23622.
SEQ ID N0:454 is the cDNA sequence for folate hydrolase.
SEQ ID N0:455 is the cDNA sequence for LIM protein.
SEQ ID N0:456 is the cDNA sequence for a known gene.
SEQ ID N0:457 is the cDNA sequence for a known gene.
SEQ ID N0:458 is the cDNA sequence for a previously identified gene.
SEQ ID N0:459 is the cDNA sequence for 23045.
SEQ ID N0:460 is the cDNA sequence for 23032.
SEQ ID N0:461 is the cDNA sequence for clone 23054.
SEQ ID N0:462-467 are cDNA sequences for known genes.
SEQ ID N0:468-471 are cDNA sequences for P710P.
SEQ ID N0:472 is a cDNA sequence for P 1001 C.
SEQ ID NO: 473 is the determined cDNA sequence for a first splice
variant of P775P (referred to as 27505).
SEQ ID NO: 474 is the determined cDNA sequence for a second splice
variant of P775P (referred to as 19947).
SEQ ID NO: 475 is the determined cDNA sequence for a third splice
variant of P775P (referred to as 19941 ).
SEQ ID NO: 476 is the determined cDNA sequence for a fourth splice
variant of P775P (referred to as 19937).
SEQ ID NO: 477 is a first predicted amino acid sequence encoded by the
sequence of SEQ ID NO: 474.
SEQ ID NO: 478 is a second predicted amino acid sequence encoded by
the sequence of SEQ ID NO: 474.
SEQ ID NO: 479 is the predicted amino acid sequence encoded by the
sequence of SEQ ID NO: 475.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
27
SEQ ID NO: 480 is a first predicted amino acid sequence encoded by the
sequence of SEQ ID NO: 473.
SEQ ID NO: 481 is a second predicted amino acid sequence encoded by
the sequence of SEQ ID NO: 473.
SEQ ID NO: 482 is a third predicted amino acid sequence encoded by
the sequence of SEQ ID NO: 473.
SEQ ID NO: 483 is a fourth predicted amino acid sequence encoded by
the sequence of SEQ ID NO: 473.
SEQ ID NO: 484 is the first 30 amino acids of the M. tuberculosis
antigen Ral2.
SEQ ID NO: 485 is the PCR primer AW025.
SEQ ID NO: 486 is the PCR primer AW003.
SEQ ID NO: 487 is the PCR primer AW027.
SEQ ID NO: 488 is the PCR primer AW026.
SEQ ID NO: 489-501 are peptides employed in epitope mapping studies.
SEQ ID NO: 502 is the determined cDNA sequence of the
complementarity determining region for the anti-P503S monoclonal antibody
20D4.
SEQ ID NO: 503 is the determined cDNA sequence of the
complementarily determining region for the anti-P503 S monoclonal antibody JA
1.
SEQ ID NO: 504 & 505 are peptides employed in epitope mapping
studies.
SEQ ID NO: 506 is the determined cDNA sequence of the
complementarity determining region for the anti-P703P monoclonal antibody 8H2.
SEQ ID NO: 507 is the determined cDNA sequence of the
complementarily determining region for the anti-P703P monoclonal antibody 7H8.
SEQ ID NO: 508 is the determined cDNA sequence of the
complemenlarity determining region for the anti-P703P monoclonal antibody 2D4.
SEQ ID NO: 509-522 are peptides employed in epitope mapping studies.
SEQ ID NO: 523 is a mature form of P703P used to raise antibodies
against P703P.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
28
SEQ ID NO: 524 is the putative full-length cDNA sequence of P703P.
SEQ ID NO: 525 is the predicted amino acid sequence encoded by SEQ
ID NO: 524.
SEQ ID NO: 526 is the full-length cDNA sequence for P790P.
SEQ ID NO: 527 is the predicted amino acid sequence for P790P.
SEQ ID NO: 528 & 529 are PCR primers.
SEQ ID NO: 530 is the cDNA sequence of a splice variant of SEQ ID
NO: 366.
SEQ ID NO: 531 is the cDNA sequence of the open reading frame of
SEQ ID NO: 530.
SEQ ID NO: 532 is the predicted amino acid encoded by the sequence of
SEQ ID NO: 531.
SEQ ID NO: 533 is the DNA sequence of a putative ORF of P775P.
SEQ ID NO: 534 is the predicted amino acid sequence encoded by SEQ
ID NO: 533.
SEQ ID NO: 535 is a first full-length cDNA sequence for PS 105.
SEQ ID NO: 536 is a second full-length cDNA sequence for PS10S.
SEQ ID NO: 537 is the predicted amino acid sequence encoded by SEQ
ID NO: 535.
SEQ ID NO: 538 is the predicted amino acid sequence encoded by SEQ
ID NO: 536.
SEQ ID NO: 539 is the peptide PSO1S-370.
SEQ ID NO: 540 is the peptide P501 S-376.
SEQ ID NO: 541-551 are epitopes of P501 S.
SEQ ID NO: 552 is an extended cDNA sequence for P712P.
SEQ ID NO: 553-568 are the amino acid sequences encoded by
predicted open reading frames within SEQ ID NO: 552.
SEQ ID NO: 569 is an extended cDNA sequence for P776P.
SEQ ID NO: 570 is the determined cDNA sequence for a splice variant
of P776P referred to as contig 6.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
29
SEQ ID NO: 571 is the determined cDNA sequence for a splice variant
of P776P referred to as contig 7.
SEQ ID NO: 572 is the determined cDNA sequence for a splice variant
of P776P referred to as contig 14.
SEQ ID NO: 573 is the amino acrd sequence encoded by a first predicted
ORF of SEQ ID NO: 570.
SEQ ID NO: 574 is the amino acid sequence encoded by a second
predicted ORF of SEQ ID NO: 570.
SEQ ID NO: 575 is the amino acid sequence encoded by a predicted
ORF of SEQ ID NO: 571.
SEQ ID NO: 576-586 are amino acid sequences encoded by predicted
ORFs of SEQ ID NO: 569.
SEQ ID NO: 587 is a DNA consensus sequence of the sequences of
P767P and P777P.
IS SEQ ID NO: 588-590 are amino acid sequences encoded by predicted
ORFs of SEQ ID NO: 587.
SEQ ID NO: 591 is an extended cDNA sequence for P1020C.
SEQ ID NO: 592 is the predicted amino acid sequence encoded by the
sequence of SEQ ID NO: P 1020C.
SEQ ID NO: 593 is a splice variant of P775P referred to as 50748.
SEQ ID NO: 594 is a splice variant of P775P referred to as 50717.
SEQ ID NO: 595 is a splice variant of P775P referred to as 45985.
SEQ ID NO: 596 is a splice variant of P775P referred to as 38769.
SEQ ID NO: 597 is a splice variant of P775P referred to as 37922.
SEQ ID NO: 598 is a splice variant of PS l OS referred to as 49274.
SEQ ID NO: 599 is a splice variant of PS l OS referred to as 39487.
SEQ ID NO: 600 is a splice variant of P504S referred to as 5167.16.
SEQ ID NO: 601 is a splice variant of P504S referred to as 5167.1.
SEQ ID NO: 602 is a splice variant of P504S referred to as 5163.46.
SEQ ID NO: 603 is a splice variant of P504S referred to as 5163.42.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
SEQ ID NO: 604 is a splice variant of P504S referred to as 5163.34.
SEQ ID NO: 605 is a splice variant of P504S referred to as 5163.17.
SEQ ID NO: 606 is a splice variant of P501 S referred to as 10640.
SEQ ID NO: 607-615 are the sequences of PCR primers.
5 SEQ ID NO: 616 is the determined cDNA sequence of a fusion of P703P
and PSA.
SEQ ID NO: 617 is the amino acid sequence of the fusion of P703P and
PSA.
SEQ ID NO: 618 is the cDNA sequence of the gene DD3.
10 SEQ ID NO: 619 is an extended cDNA sequence for P714P.
SEQ ID NO: 620-622 are the cDNA sequences for splice variants of
P704P.
SEQ ID NO: 623 is the cDNA sequence of a splice variant of P553S
referred to as P553S-14.
15 SEQ ID NO: 624 is the cDNA sequence of a splice variant of P553S
referred to as P553S-12.
SEQ ID NO: 625 is the cDNA sequence of a splice variant of P553S
referred to as P553S-10.
SEQ ID NO: 626 is the cDNA sequence of a splice variant of P553S
20 referred to as P553S-6.
SEQ ID NO: 627 is the amino acid sequence encoded by SEQ ID NO:
626.
SEQ ID NO: 628 is a first amino acid sequence encoded by SEQ ID NO:
623.
25 SEQ ID NO: 629 is a second amino acid sequence encoded by SEQ ID
NO: 623.
SEQ ID NO: 630 is a first full-length cDNA sequence for prostate-
specific transglutaminase gene (also referred to herein as P558S).
SEQ ID NO: 631 is a second full-length cDNA sequence for prostate-
30 specific transglutaminase gene.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
31
SEQ ID NO: 632 is the amino acid sequence encoded by the sequence of
SEQ ID NO: 630.
SEQ ID NO: 633 is the amino acid sequence encoded by the sequence of
SEQ ID NO: 631.
SEQ ID NO: 634 is the full-length cDNA sequence for P788P.
SEQ ID NO: 635 is the amino acid sequence encoded by SEQ ID NO:
634.
SEQ ID NO: 636 is the determined cDNA sequence for a polymorphic
variant of P788P.
SEQ ID NO: 637 is the amino acid sequence encoded by SEQ ID NO:
636.
SEQ ID NO: 638 is the amino acid sequence of peptide 4 from P703P.
SEQ ID NO: 639 is the cDNA sequence that encodes peptide 4 from
P703 P.
SEQ ID NO: 640-655 are cDNA sequences encoding epitopes of P703P.
SEQ ID NO: 656-671 are the amino acid sequences of epitopes of
P703P.
SEQ ID NO: 672 and 673 are PCR primers.
SEQ ID NO: 674 is the cDNA sequence encoding an N-terminal portion
of P788P expressed in E. coli.
SEQ ID NO: 675 is the amino acid sequence of the N-terminal portion of
P788P expressed in E. coli.
SEQ ID NO: 676 is the amino acid sequence of the M. tuberculosis
antigen Ral2.
SEQ ID NO: 677 and 678 are PCR primers.
SEQ ID NO: 679 is the cDNA sequence for the Ral2-PS10S-C
construct.
SEQ ID NO: 680 is the cDNA sequence for the PS l OS-C construct.
SEQ ID NO: 681 is the cDNA sequence for the PS l OS-E3 construct.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
32
SEQ ID NO: 682 is the amino acid sequence for the Ral2-PS l0S-C
construct.
SEQ ID NO: 683 is the amino acid sequence for the PS l OS-C construct.
SEQ ID NO: 684 is the amino acid sequence for the PS l OS-E3 construct.
SEQ ID NO: 685-690 are PCR primers.
SEQ ID NO: 691 is the cDNA sequence of the construct Ral2-P775P-
ORF3.
SEQ ID NO: 692 is the amino acid sequence of the construct Ral2-
P775P-ORF3.
SEQ ID NO: 693 and 694 are PCR primers.
SEQ ID NO: 695 is the determined amino acid sequence for a P703P His
tag fusion protein.
SEQ ID NO: 696 is the determined cDNA sequence fox a P703P His tag
fusion protein.
SEQ ID NO: 697 and 698 are PCR primers.
SEQ ID NO: 699 is the determined amino acid sequence for a P705P His
tag fusion protein.
SEQ ID NO: 700 is the determined cDNA sequence for a P705P His tag
fusion protein.
SEQ ID NO: 701 and 702 are PCR primers.
SEQ ID NO: 703 is the determined amino acid sequence for a P711P His
tag fusion protein.
SEQ ID NO: 704 is the determined cDNA sequence for a P711P His tag
fusion protein.
SEQ ID NO: 705 is the amino acid sequence of the M. tuberculosis
antigen Ral2.
SEQ ID NO: 706 and 707 are PCR primers.
SEQ ID NO: '708 is the determined cDNA sequence for the construct
Ral2-P501 S-E2.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
33
SEQ ID NO: 709 is the determined amino acid sequence for the
construct Ral2-PSOlS-E2.
SEQ ID NO: 710 is the amino acid sequence for an epitope of P501 S.
SEQ ID NO: 711 is the DNA sequence encoding SEQ ID NO: 710.
SEQ ID NO: 712 is the amino acid sequence for an epitope of P501 S.
SEQ ID NO: 713 is the DNA sequence encoding SEQ ID NO: 712.
SEQ ID NO: 714 is a peptide employed in epitope mapping studies.
SEQ ID NO: 715 is the amino acid sequence for an epitope of P501 S.
SEQ ID NO: 716 is the DNA sequence encoding SEQ ID NO: 715.
SEQ ID NO: 717-719 are the amino acid sequences for CD4 epitopes of
P501 S.
SEQ ID NO: 720-722 are the DNA sequences encoding the sequences of
SEQ ID NO: 717-719.
SEQ ID NO: 723-734 are the amino acid sequences for putative CTL
epitopes of P703P.
SEQ ID NO: 735 is the full-length cDNA sequence for P789P.
SEQ ID NO: 736 is the amino acid sequence encoded by SEQ ID NO:
735.
SEQ ID NO: 737 is the determined full-length cDNA sequence for the
splice variant of P776P referred to as contig 6.
SEQ ID NO: 738-739 are determined full-length cDNA sequences for
the splice variant of P776P referred to as contig 7.
SEQ ID NO: 740-744 are amino acid sequences encoded by SEQ ID NO:
737.
SEQ ID NO: 745-750 are amino acid sequences encoded by the splice
variant of P776P referred to as contig 7.
SEQ ID NO: 751 is the full-length cDNA sequence for human
transmembrane protease serine 2.
SEQ ID NO: 752 is the amino acid sequence encoded by SEQ ID NO:
751.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
34
SEQ ID NO: 753 is the cDNA sequence encoding the first 209 amino
acids of human ~transmembrane protease serine 2.
SEQ ID NO: 754 is the first 209 amino acids of human transmembrane
protease serine 2.
SEQ ID NO: 755 is the amino acid sequence of peptide 296-322 of
P501 S.
SEQ ID NO: 756-759 are PCR primers.
SEQ ID NO: 760 is the determined cDNA sequence of the Vb chain of a
T cell receptor for the P501 S-specific T cell clone 4E5.
SEQ ID NO: 761 is the determined cDNA sequence of the Va chain of a
T cell receptor for the P501 S-specific T cell clone 4E5.
SEQ ID NO: 762 is the amino acid sequence encoded by SEQ ID NO
760.
SEQ ID NO: 763 is the amino acid sequence encoded by SEQ ID NO
761.
SEQ ID NO: 764 is the full-length open reading frame for P768P
including stop codon.
SEQ ID NO: 765 is the full-length open reading frame for P768P without
stop codon.
SEQ ID NO: 766 is the amino acid sequence encoded by SEQ ID NO:
765.
SEQ ID NO: 767-772 are the amino acid sequences for predicted
domains of P768P.
SEQ ID NO: 773 is the full-length cDNA sequence of P835P.
SEQ ID NO: 774 is the cDNA sequence of the previously identified
clone FLJ13581.
SEQ ID NO: 775 is the cDNA sequence of the open reading frame for
P835P with stop codon.
SEQ ID NO: 776 is the cDNA sequence of the open reading frame for
P835P without stop codon.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
SEQ ID NO: 777 is the full-length amino acid sequence for P835P.
SEQ ID NO: 778-785 are the amino acid sequences of extracellular and
intracellular domains of P835P.
SEQ ID NO: 786 is the full-length cDNA sequence for P1000C.
5 SEQ ID NO: 787 is the cDNA sequence of the open reading frame for
P I 000C, including stop codon.
SEQ ID NO: 788 is the cDNA sequence of the open reading frame for
P 1 OOOC, without stop codon.
SEQ ID NO: 789 is the full-length amino acid sequence for P1000C.
10 SEQ ID NO: 790 is amino acids 1-100 of SEQ ID NO: 789.
SEQ ID NO: 791 is amino acids 100-492 of SEQ ID NO: 789.
SEQ ID NO: 792 is the amino acid sequence of an a prepro-P501 S
recombinant protein.
I S DETAILED DESCRIPTION OF THE INVENTION
The present invention is directed generally to compositions and their use
in the therapy and diagnosis of cancer, particularly prostate cancer. As
described further
below, illustrative compositions of the present invention include, but are not
restricted
to, polypeptides, particularly immunogenic polypeptides, polynucleotides
encoding such
20 polypeptides, antibodies and other binding agents, antigen presenting
.cells (APCs) and
immune system cells (e.g., T cells).
The practice of the present invention will employ, unless indicated
specifically to the contrary, conventional methods of virology, immunology,
microbiology, molecular biology and recombinant DNA techniques within the
skill of
25 the art, many of which are described below fox the purpose of illustration.
Such
techniques are explained fully in the literature. See, e.g., Sambrook, et al.
Molecular
Cloning: A Laboratory Manual (2nd Edition, 1989); Maniatis et al. Molecular
Cloning:
A Laboratory Manual (1982); DNA Cloning: A Practical Approach, vol. I & II (D.
Glover, ed.); Oligonucleotide Synthesis (N. Gait, ed., 1984); Nucleic Acid
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
36
Hybridization (B. Hames & S. Higgins, eds., 1985); Transcription and
Translation (B.
Hames & S. Higgins, eds., 1984); Animal Cell Culture (R. Freshney, ed:, 1986);
Perbal,
A Practical Guide to Molecular Cloning (I984).
All publications, patents and patent applications cited herein, whether
supra or infra, are hereby incorporated by reference in their entirety.
As used in this specification and the appended claims, the singular forms
"a," "an" and "the" include plural references unless the content clearly
dictates
otherwise.
Polypeptide Compositions
As used herein, the term "polypeptide" " is used in its conventional
meaning, i.e., as a sequence of amino acids. The polypeptides are not limited
to a
specific length of the product; thus, peptides, oligopeptides, and proteins
are included
within the definition of polypeptide, and such terms may be used
interchangeably herein
unless specifically indicated otherwise. This term also does not refer to or
exclude post-
expression modifications of the polypeptide, for example, glycosylations,
acetylations,
phosphorylations and the like, as well as other modifications known in the
art, both
naturally occurring and non-naturally occurring. A polypeptide may be an
entire
protein, or a subsequence thereof. Particular polypeptides of interest in the
context of
this invention are amino acid subsequences comprising epitopes, i.e.,
antigenic
determinants substantially responsible for the immunogenic properties of a
polypeptide
and being capable of evoking an immune response.
Particularly illustrative polypeptides of the present invention comprise
those encoded by a polynucleotide sequence set forth in any one of SEQ ID NOs:
1-11 l,
115-171, 173-175, 177, 179-305, 307-315, 326, 328, 330, 332-335, 340-375, 381,
382
and 384-476, 524, 526, 530, 531, 533, 535, 536, 552, 569-572, 587, 591, 593-
606, 618-
626, 630, 631, 634, 636, 639-655, 674, 680, 681, 711, 713, 716, 720-722, 735,
737-739,
751, 753, 764, 765, 773-776 and 786-788, or a sequence that hybridizes under
moderately stringent conditions, or, alternatively, under highly stringent
conditions, to a
polynucleotide sequence set forth in any one of SEQ ID NOs: 1-111, 115-171,
173-175,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
37
177, 179-305, 307-315, 326, 328, 330, 332-335, 340-375, 381, 382 and 384-476,
524,
526, 530, 531, 533, 535, 536, 552, 569-572, 587, 591, 593-606, 618-626, 630,
631, 634,
636, 639-655, 674, 680, 681, 711, 713, 716, 720-722, 735, 737-739, 751, 753,
764, 765,
773-776 and 786-788. In specific embodiments, the polypeptides of the
invention
comprise amino acid sequences as set forth in any one of SEQ ID NO: 112-114,
172,
176, 178, 327, 329, 331, 336, 339, 376-380, 383, 477-483, 496, 504, 505, 519,
520,
522, 525, 527, 532, 534, 537-551, 553-568, 573-586, 588-590, 592, 627-629,
632, 633,
635, 637, 638, 656-671, 675, 683, 684, 710, 712, 714, 715, 717-719, 723-734,
736, 740-
750, 752, 754, 755, 766-772, 777-785 and 789-791.
The polypeptides of the present invention are sometimes herein referred
to as prostate-specific proteins or prostate-specific polypeptides, as an
indication that
their identification has been based at least in part upon their increased
levels of
expression in prostate tissue samples. Thus, a "prostate-specific polypeptide"
or
"prostate-specif c protein," refers generally to a polypeptide sequence of the
present
invention, or a polynucleotide sequence encoding such a polypeptide, that is
expressed
in a substantial proportion of prostate tissue samples, for example preferably
greater
than about 20%, more preferably greater than about 30%, and most preferably
greater
than about 50% or more of prostate tissue samples tested, at a level that is
at least two
fold, and preferably at least five fold, greater than the level of expression
in other
normal tissues, as determined using a representative assay provided herein. A
prostate-
specific polypeptide sequence of the invention, based upon its increased level
of
expression in tumor cells, has particular utility both as a diagnostic marker
as well as a
therapeutic target, as further described below.
In certain preferred embodiments, the polypeptides of the invention are
immunogenic, i.e., they react detectably within an immunoassay (such as an
ELISA or
T-cell stimulation assay) with antisera and/or T-cells from a patient with
prostate
cancer. Screening for immunogenic activity can be performed using techniques
well
known to the skilled artisan. For example, such screens can be performed using
methods such as those described in Harlow and Lane, Antibodies: A Laboratory
Manual, Cold Spring Harbor Laboratory, 1988. In one illustrative example, a
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
38
polypeptide may be irmnobilized on a solid support and contacted with patient
sera to
allow binding of antibodies within the sera to the immobilized polypeptide.
Unbound
sera may then be removed and bound antibodies detected using, for example,
Izsl-
labeled Protein A.
As would be recognized by the skilled artisan, immunogenic portions of
the polypeptides disclosed herein are also encompassed by the present
invention. An
"immunogenic portion," as used herein, is a fragment of an immunogenic
polypeptide
of the invention that itself is immunologically reactive (i. e., specifically
binds) with the
B-cells and/or T-cell surface antigen receptors that recognize the
polypeptide.
Immunogenis portions may generally be identified using well known techniques,
such
as those summarized in Paul, Fundamental Immunology, 3rd ed., 243-247 (Raven
Press,
1993) and references cited therein. Such techniques include screening
polypeptides for
the ability to react with antigen-specific antibodies, antisera and/or T-cell
lines or
clones. As used herein, antisera and antibodies are "antigen-specific" if they
specifically bind to an antigen (i.e., they react with the protein in an ELISA
or other
immunoassay, and do not react detestably with unrelated proteins). Such
antisera and
antibodies may be prepared as described herein, and using well-known
techniques.
In one preferred embodiment, an immunogenic portion of a polypeptide
of the present invention is a portion that reacts with antisera and/or T-sells
at a level that
is not substantially less than the reactivity of the full-length polypeptide
(e.g., in an
ELISA and/or T-cell reactivity assay). Preferably, the level of immunogenic
activity of
the immunogenic portion is at least about 50%, preferably at least about 70%
and most
preferably greater than about 90% of the immunogenicity for the full-length
polypeptide. In some instances, preferred immunogenic portions will be
identified that
have a level of immunogenic activity greater than that of the corresponding
full-length
polypeptide, e.g., having greater than about 100% or 150% or more immunogenic
activity.
In certain other embodiments, illustrative immunogenic portions may
include peptides in which an N-terminal leader sequence and/or transmembrane
domain
has been deleted. Other illustrative immunogenic portions will contain a small
N-
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
39
and/or C-terminal deletion (e.g., 1-30 amino acids, preferably S-1S amino
acids),
relative to the mature protein.
In another embodiment, a polypeptide composition of the invention may
also comprise one or more polypeptides that are immunologically reactive with
T cells
S and/or antibodies generated against a polypeptide of the invention,
particularly a
polypeptide having an amino acid sequence disclosed herein, or to an
immunogenic
fragment or variant thereof.
In another embodiment of the invention, polypeptides are provided that
comprise one or more polypeptides that are capable of eliciting T cells and/or
antibodies
that are immunologically reactive with one or more polypeptides described
herein, or
one or more polypeptides encoded by contiguous nucleic acid sequences
contained in
the polynucleotide sequences disclosed herein, or immunogenic fragments or
variants
thereof, or to one or more nucleic acid sequences which hybridize to one or
more of
these sequences under conditions of moderate to high stringency.
1 S The present invention, in another aspect, provides polypeptide fragments
comprising at least about S, 10, 1S, 20, 2S, S0, or 100 contiguous amino
acids, or more,
including all intermediate lengths, of a polypeptide composition set forth
herein, such as
those set forth in SEQ ID NO: 112-114, 172, 176, 178, 327, 329, 331, 336, 339,
376-
380, 383, 477-483, 496, 504, SOS, 519, 520, 522, S2S, 527, 532, 534, S37-SS1,
SS3-568,
S73-586, S88-590, 592, 627-629, 632, 633, 635, 637, 638, 6S6-671, 675, 683,
684, 710,
712, 714, 715, 717-719, 723-734, 736, 740-750, 752, 754, 7SS, 766-772, 777-78S
and
789-791, or those encoded by a polynucleotide sequence set forth in a sequence
of SEQ
ID NO: 1-11I, 11S-17I, I73-175, I77, I79-305, 307-315, 326, 328, 330, 332-335,
340-
375, 381, 382 and 384-476, 524, 526, 530, 531, 533, S3S, 536, SS2, S69-572,
587, 591,
2S S93-606, 618-626, 630, 631, 634, 636, 639-6SS, 674, 680, 681, 711, 713,
716, 720-722,
735, 737-739, 751, 753, 764, 765, 773-776 and 786-788.
In another aspect, the present invention provides variants of the
polypeptide compositions described herein. Polypeptide variants generally
encompassed by the present invention will typically exhibit at least about
70%, 7S%,
80%, 8S%, 90%, 91%, 92%, 93%, 94%, 9S%, 96%, 97%, 98%, or 99% or more identity
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
(determined as described below), along its length, to a polypeptide sequence
set forth
herein.
In one preferred embodiment, the polypeptide fragments and variants
provided by the present invention are immunologically reactive with an
antibody and/or
5 T-cell that reacts with a full-length polypeptide specifically set forth
herein.
In another preferred embodiment, the polypeptide fragments and variants
provided by the present invention exhibit a level of immunogenic activity of
at least
about 50%, preferably at least about 70%, and most preferably at least about
90% or
more of that exhibited by a full-length polypeptide sequence specifically set
forth
10 herein.
A polypeptide "variant," as the term is used herein, is a polypeptide that
typically differs from a polypeptide specifically disclosed herein in one or
more
substitutions, deletions, additions and/or insertions. Such variants may be
naturally
occurring or may be synthetically generated, for example, by modifying one or
more of
15 the above polypeptide sequences of the invention and evaluating their
immunogenic
activity as described herein using any of a number of techniques well known in
the art.
For example, certain illustrative vaxiants of the polypeptides of the
invention include those in which one or more portions, such as an N-terminal
leader
sequence or transmembrane domain, have been removed. Other illustrative
variants
20 include variants in which a small portion (e.g., 1-30 amino acids,
preferably 5-15 amino
acids) has been removed from the N- and/or C-terminal of the mature protein.
In many instances, a variant will contain conservative substitutions. A
"conservative substitution" is one in which an amino acid is substituted for
another
amino acid that has similar properties, such that one skilled in the art of
peptide
25 chemistry would expect the secondary structure and hydropathic nature of
the
polypeptide to be substantially unchanged. As described above, modifications
may be
made in the structure of the polynucleotides and polypeptides of the present
invention
and still obtain a functional molecule that encodes a variant or derivative
polypeptide
with desirable characteristics, e.g., with immunogenic characteristics. When
it is
30 desired to alter the amino acid sequence of a polypeptide to create an
equivalent, or
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
41
even an improved, immunogenic variant or portion of a polypeptide of the
invention,
one skilled in the art will typically change one or more of the codons of the
encoding
DNA sequence according to Table 1.
For example, certain amino acids may be substituted for other amino
acids in a protein structure without appreciable loss of interactive binding
capacity with
structures such as, for example, antigen-binding regions of antibodies or
binding sites
on substrate molecules. Since it is the interactive capacity and nature of a
protein that
defines that protein's biological functional activity, certain amino acid
sequence
substitutions can be made in a protein sequence, and, of course, its
underlying DNA
coding sequence, and nevertheless obtain a protein with like properties. It is
thus
contemplated that various changes may be made in the peptide sequences of the
disclosed compositions, or corresponding DNA sequences which encode said
peptides
without appreciable loss of their biological utility or activity.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
42
TABLE 1
Amino Acids Codons
Alanine AIa A GCA GCC GCG GCU
Cysteine Cys C UGC UGU
Aspartic acid Asp D GAC GAU
Glutamic acid Glu E GAA GAG
Phenylalanine Phe F UUC UUU
Glycine Gly G GGA GGC GGG GGU
Histidine His H CAC CAU
Isoleucine Ile I AUA AUC AUU
Lysine Lys K AAA AAG
Leucine Leu L UUA UUG CUA CUC GUG CUU
Methionine Met M AUG
Asparagine Asn N AAC AAU
Proline Pro P CCA CCC CCG CCU
Glutamine Gln Q CAA CAG
Arginine Arg R AGA AGG CGA CGC CGG CGU
Serine Ser S AGC AGU UCA UCC UCG UCU
Threonine Thr T ACA ACC ACG ACU
Valine Val V GUA GUC GUG GUU
Tryptophan Trp W UGG 1
Tyrosine Tyr Y UAC UAU
In making such changes, the hydropathic index of amino acids may be
considered. The importance of the hydropathic amino acid index in conferring
interactive biologic function on a protein is generally understood in the art
(Kyte and
Doolittle, 1982, incorporated herein by reference). It is accepted that the
relative
hydropathic character of the amino acid contributes to the secondary structure
of the
resultant protein, which in turn defines the interaction of the protein with
other
molecules, for example, enzymes, substrates, receptors, DNA, antibodies,
antigens, and
the like. Each amino acid has been assigned a hydropathic index on the basis
of its
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
43
hydrophobicity and charge characteristics (Kyte and Doolittle, 1982). These
values are:
isoleucine (+4.5); saline (+4.2); leucine (+3.8); phenylalanine (+2.8);
cysteine/cystine
(+2.5); methionine (+1.9); alanine (+1.8); glycine (-0.4); threonine (-0.7);
serine (-0.8);
tryptophan (-0.9); tyrosine (-1.3); proline (-1.6); histidine (-3.2);
glutamate (-3.5);
glutamine (-3.5); aspartate (-3.5); asparagine (-3.5); lysine (-3.9); and
arginine (--4.5).
It is known in the art that certain amino acids may be substituted by other
amino acids having a similar hydropathic index or score and still result in a
protein with
similar biological activity, i. e. still obtain a biological functionally
equivalent protein.
In making such changes, the substitution of amino acids whose hydropathic
indices are
within ~2 is preferred, those within ~1 are particularly preferred, and those
within ~0.5
are even more particularly preferred. It is also understood in the art that
the substitution
of like amino acids can be made effectively on the basis of hydrophilicity. U.
S. Patent
4,554,1 O1 (specifically incorporated herein by reference in its entirety),
states that the
greatest local average hydrophilicity of a protein, as governed by the
hydrophilicity of
its adjacent amino acids, correlates with a biological property of the
protein.
As detailed in U. S. Patent 4,554,101, the following hydrophilicity values
have been assigned to amino acid residues: arginine (+3.0); lysine (+3.0);
aspartate
(+3.0 ~ 1); glutamate (+3.0 ~ 1); serine (+0.3); asparagine (+0.2); glutamine
(+0.2);
glycine (0); threonine (-0.4); proline (-0.5 ~ 1); alanine (-0.5); histidine (-
0.5); cysteine
(-1.0); methionine (-1.3); valine (-1.5); leucine (-1.8); isoleucine (-1.8);
tyrosine (-
2.3); phenylalanine (-2.5); tryptophan (-3.4). It is understood that an amino
acid can be
substituted for another having a similar hydrophilicity value and still obtain
a
biologically equivalent, and in particular, an immunologically equivalent
protein. In
such changes, the substitution of amino acids whose hydrophilicity values are
within ~2
is preferred, those within ~1 axe particularly preferred, and those within
~0.5 are even
more particularly preferred.
As outlined above, amino acid substitutions are generally therefore based
on the relative similarity of the amino acid side-chain substituents, for
example, their
hydrophobicity, hydrophilicity, charge, size, and the like. Exemplary
substitutions that
take various of the foregoing characteristics into consideration are well
known to those
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
44
of skill in the art and include: arginine and lysine; glutamate and aspartate;
serine and
threonine; glutamine and asparagine; and valine, leucine and isoleucine.
In addition, any polynucleotide may be further modified to increase
stability i~r vivo. Possible modifications include, but are not limited to,
the addition of
flanking sequences at the 5' and/or 3' ~ ends; the use of phosphorothioate or
2' O-methyl
rather than phosphodiesterase linkages in the backbone; and/or the inclusion
of
nontraditional bases such as inosine, queosine ~ and wybutosine, as well as
acetyl-
methyl-, thio- and other modified forms of adenine, cytidine, guanine,
thyrnine and
uridine.
Amino acid substitutions may further be made on the basis of similarity
in polarity, charge, solubility, hydrophobicity, hydrophilicity and/or the
amphipathic
nature of the residues. For example, negatively charged amino acids include
aspartic
acid and glutamic acid; positively charged amino acids include lysine and
arginine; and
amino acids with uncharged polar head groups having similar hydrophilicity
values
I S include leucine, isoleucine and valine; glycine and alanine; asparagine
and glutamine;
and serine, threonine, phenylalanine and tyrosine. Other groups of amino acids
that may
represent conservative changes include: (1) ala, pro, gly, glu, asp, gln, asn,
ser, thr;
(2) cys, ser, tyr, thr; (3) val, ile, leu, met, ala, phe; (4) lys, arg, his;
and (5) phe, tyr, trp,
his. A variant may also, or alternatively, contain nonconservative changes. In
a
preferred embodiment, variant polypeptides differ from a native sequence by
substitution, deletion or addition of five amino acids or fewer. Variants may
also (or
alternatively) be modified by, for example, the deletion or addition of amino
acids that
have minimal influence on the immunogenicity, secondary structure and
hydropathic
nature of the polypeptide. .
As noted above, polypeptides may comprise a signal (or leader) sequence
at the N-terminal end of the protein, which co-translationally or post-
translationally
directs transfer of the protein. The polypeptide may also be conjugated to a
linker or
other sequence for ease of synthesis, purification or identification of the
polypeptide
(e.g., poly-His), or to enhance binding of the polypeptide to a solid support.
For
example, a polypeptide may be conjugated to an immunoglobulin Fc region.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
When comparing polypeptide sequences, two sequences are said to be
"identical" if the sequence of amino acids in the two sequences is the same
when
aligned for maximum correspondence, as described below. Comparisons between
two
sequences are typically performed by comparing the sequences over a comparison
5 window to identify and compare local regions of sequence similarity. A
"comparison
window" as used herein, refers to a segment of at least about 20 contiguous
positions,
usually 30 to about 75, 40 to about 50, in which a sequence may be compared to
a
reference sequence of the same number of contiguous positions after the two
sequences
are optimally aligned.
10 Optimal alignment of sequences for comparison may be conducted using
the Megalign program in the Lasergene suite of bioinformatics software
(DNASTAR,
Inc., Madison, WI), using default parameters. This program embodies several
alignment schemes described in the following references: Dayhoff, M.O. (1978)
A
model of evolutionary change in proteins - Matrices for detecting distant
relationships.
15 In Dayhoff, M.O. (ed.) Atlas of Protein Sequence and Structure, National
Biomedical
Research Foundation, Washington DC Vol. 5, Suppl. 3, pp. 345-358; Hein J.
(1990)
Unified Approach to Alignment and Phylogenes pp. 626-645 Methods in Enzymology
vol. 183, Academic Press, Inc., San Diego, CA; Higgins, D.G. and Sharp, P.M.
(1989)
CABIOS 5:151-153; Myers, E.W. and Muller W. (1988) CABIOS 4:11-17; Robinson,
20 E.D. (1971) Cornb. Theor 11:105; Santou, N. Nes, M. (1987) Mol. Biol. Evol.
4:406-
425; Sneath, P.H.A. and Sokal, R.R. (1973) Numerical Taxonomy - the Principles
and
Practice of Numerical Taxonomy, Freeman Press, San Francisco, CA; Wilbur, W.J.
and
Lipman, D.J. (1983) Proc. Natl. Acad., Sci. USA 80:726-730.
Alternatively, optimal alignment of sequences for comparison may be
25 conducted by the local identity algorithm of Smith and Waterman (1981) Add.
APL.
Math 2:482, by the identity alignment algorithm of Needleman and Wunsch (1970)
J.
Mol. Biol. 48:443, by the search fox similarity methods of Pearson and Lipman
(1988)
Proc. Natl. Acad. Sci. USA 85: 2444, by computerized implementations of these
algorithms (GAP, BESTFIT, BLAST, FASTA, and TFASTA in the Wisconsin Genetics
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
46
Software Package, Genetics Computer Group (GCG), 575 Science Dr., Madison,
WI),
or by inspection.
One preferred example of algorithms that are suitable for determining
percent sequence identity and sequence similarity are the BLAST and BLAST 2.0
algorithms, which are described in Altschul et al. (1977) Nucl. Acids Res.
25:3389-3402
and Altschul et al. (1990) J. Mol. Biol. 215:403-410, respectively. BLAST and
BLAST
2.0 can be used, for example with the parameters described herein, to
determine percent
sequence identity for the polynucleotides and polypeptides of the invention.
Software
for performing BLAST analyses is publicly available through the National
Center for
Biotechnology Information. For amino acid sequences, a scoring matrix can be
used to
calculate the cumulative score. Extension of the word hits in each direction
are halted
when: the cumulative alignment score falls off by the quantity X from its
maximum
achieved value; the cumulative score goes to zero or below, due to the
accumulation of
one or more negative-scoring residue alignments; or the end of either sequence
is
reached. The BLAST algorithm parameters W, T and X determine the sensitivity
and
speed of the alignment.
In one preferred approach, the "percentage of sequence identity" is
determined by comparing two optimally aligned sequences over a window of
comparison of at least 20 positions, wherein the portion of the polypeptide
sequence in
the comparison window may comprise additions or deletions (i.e., gaps) of 20
percent
or less, usually 5 to 15 percent, or 10 to 12 percent, as compared to the
reference
sequences (which does not comprise additions or deletions) for optimal
alignment of the
two sequences. The percentage is calculated by determining the number of
positions at
which the identical amino acid residue occurs in both sequences to yield the
number of
matched positions, dividing the number of matched positions by the total
number of
positions in the reference sequence (i. e., the window size) and multiplying
the results by
100 to yield the percentage of sequence identity.
Within other illustrative embodiments, a polypeptide may be a fusion
polypeptide that comprises multiple polypeptides as described herein, or that
comprises
at least one polypeptide as described herein and an unrelated sequence, such
as a known
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
47
tumor protein. A fusion partner may, for example, assist in providing T helper
epitopes
(an immunological fusion partner), preferably T helper epitopes recognized by
humans,
or may assist in expressing the protein (an expression enhancer) at higher
yields than the
native recombinant protein. Certain preferred fusion partners are both
immunological
and expression enhancing fusion partners. Other fusion partners may be
selected so as
to increase the solubility of the polypeptide or to enable the polypeptide to
be targeted to
desired intracellular compartments. Still further fusion partners include
affinity tags,
which facilitate purification of the polypeptide.
Fusion polypeptides may generally be prepared using standard
techniques, including chemical conjugation. Preferably, a fusion polypeptide
is
expressed as a recombinant polypeptide, allowing the production of increased
levels,
relative to a non-fused polypeptide, in an expression system. Briefly, DNA
sequences
encoding the polypeptide components may be assembled separately, and ligated
into an
appropriate expression vector. The 3' end of the DNA sequence encoding one
polypeptide component is ligated, with or without a peptide linker, to the 5'
end of a
DNA sequence encoding the second polypeptide component so that the reading
frames
of the sequences are in phase. This permits translation into a single fusion
polypeptide
that retains the biological activity of both component polypeptides.
A peptide linker sequence may be employed to separate the first and
second polypeptide components by a distance sufficient to ensure that each
polypeptide
folds into its secondary and tertiary structures. Such a peptide linker
sequence is
incorporated into the fusion polypeptide using standard techniques well known
in the
art. Suitable peptide linker sequences may be chosen based on the following
factors:
(1) their ability to adapt a flexible extended conformation; (2) their
inability to adopt a
secondary structure that could interact with functional epitopes on the first
and second
polypeptides; and (3) the Lack of hydrophobic or charged residues that might
react with
the polypeptide functional epitopes. Preferred peptide linker sequences
contain Gly,
Asn and Ser residues. Other near neutral amino acids, such as Thr and Ala may
also be
used in the linker sequence. Amino acid sequences which may be usefully
employed as
linkers include those disclosed in Maratea et al., Gene 40:39-46, 1985; Murphy
et al.,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
48
Pr~oc. Natl. Acad Sci. USA 83:8258-8262, 1986; U.S. Patent No. 4,935,233 and
U.S.
Patent No. 4,751,180. The linker sequence may generally be from 1 to about 50
amino
acids in length. Linker sequences are not required when the first and second
polypeptides have non-essential N-terminal amino acid regions that can be used
to
separate the functional domains and prevent steric interference.
The ligated DNA sequences are operably linked to suitable
transcriptional or translational regulatory elements. The regulatory elements
responsible for expression of DNA are located only 5' to the DNA sequence
encoding
the first polypeptides. Similarly, stop codons required to end translation and
transcription termination signals are only present 3' to the DNA sequence
encoding the
second polypeptide.
The fusion polypeptide can comprise a polypeptide as described herein
together with an unrelated immunogenic protein, such as an immunogenic protein
capable of eliciting a recall response. ~ Examples of such proteins include
tetanus,
I 5 tuberculosis and hepatitis proteins (see, for example, Stoute et aI. New
Engl. J. Med.,
336:86-91, 1997).
In one preferred embodiment, the immunological fusion partner is
derived from a Mycobacterium sp., such as a Mycobacter°ium tuberculosis-
derived Ral2
fragment. Ral2 compositions and methods for their use in enhancing the
expression
and/or immunogenicity of heterologous polynucleotide/polypeptide sequences is
described in U.S. Patent Application 60/158,585, the disclosure of which is
incorporated herein by reference in its entirety. Briefly, Ral2 refers to a
polynucleotide
region that is a subsequence of a Mycobacterium tuberculosis MTB32A nucleic
acid.
MTB32A is a serine protease of 32 KD molecular weight encoded by a gene in
virulent
and avirulent strains of M. tuberculosis. The nucleotide sequence and amino
acid
sequence of MTB32A have been described (for example, U.S. Patent Application
60/158,585; see also, Skeiky et al., Infection aid Imrnun. (1999) 67:3998-
4007,
incorporated herein by reference). C-terminal fragments of the MTB32A coding
sequence express at high levels and remain as a soluble polypeptides
throughout the
purification process. Moreover, Ral2 may enhance the immunogenicity of
heterologous
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
49
immunogenic polypeptides with which it is fused. One preferred Ral2 fusion
polypeptide comprises a 14 KD C-terminal fragment corresponding to amino acid
residues 192 to 323 of MTB32A. Other preferred Ral2 polynucleotides generally
comprise at least about 15 consecutive nucleotides, at least about 30
nucleotides, at least
about 60 nucleotides, at least about 100 nucleotides, at least about 200
nucleotides, or at
least about 300 nucleotides that encode a portion of a Ral2 polypeptide. Ral2
polynucleotides may comprise a native sequence (i. e., an endogenous sequence
that
encodes a Ral2 polypeptide or a portion thereof) or may comprise a variant of
such a
sequence. Ral2 polynucleotide variants may contain one or more substitutions,
additions, deletions and/or insertions such that the biological activity of
the encoded
fusion polypeptide is not substantially diminished, relative to a fusion
polypeptide
comprising a native Ral2 polypeptide. Variants preferably exhibit at least
about 70%
identity, more preferably at least about 80% identity and most preferably at
least about
90% identity to a polynucleotide sequence that encodes a native Ral2
polypeptide or a
portion thereof.
Within other preferred embodiments, an immunological fusion partner is
derived from protein D, a surface protein of the gram-negative bacterium
Haemophilus
influenza B (WO 91/18926). Preferably, a protein D derivative comprises
approximately the first third of the protein (e.g., the first N-terminal 100-
110 amino
acids), and a protein D derivative may be lipidated. Within certain preferred
embodiments, the first 109 residues of a Lipoprotein D fusion partner is
included on the
N-terminus to provide the polypeptide with additional exogenous T-cell
epitopes and to
increase the expression level in E. coli (thus functioning as an expression
enhancer).
The lipid tail ensures optimal presentation of the antigen to antigen
presenting cells.
Other fusion partners include the non-structural protein from influenzae
virus, NS 1
(hemaglutinin). Typically, the N-terminal 81 amino acids are used, although
different
fragments that include T-helper epitopes may be used.
In another embodiment, the immunological fusion partner is the protein
known as LYTA, or a portion thereof (preferably a C-terminal portion). LYTA is
derived from Streptococcus pheunaoniae, which synthesizes an N-acetyl-L-
alanine
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
amidase known as amidase LYTA (encoded by the LytA gene; Gene 43:265-292,
1986).
LYTA is an autolysin that specifically degrades certain bonds in the
peptidoglycan
backbone. The C-terminal domain of the LYTA protein is responsible for the
affinity to
the choline or to some choline analogues such as DEAE. This property has been
5 exploited for the development of E. coli C-LYTA expressing plasmids useful
for
expression of fusion proteins. Purification of hybrid proteins containing the
C-LYTA
fragment at the amino terminus has been described (see Biotechnology 10:795-
798,
1992). Within a preferred embodiment, a repeat portion of LYTA may be
incorporated
into a fusion polypeptide. A repeat portion is found in the C-terminal region
starting at
10 residue 178. A particularly preferred repeat portion incorporates residues
188-305.
Yet another illustrative embodiment involves fusion polypeptides, and
the polynucleotides encoding them, wherein the fusion partner comprises a
targeting
signal capable of directing a polypeptide to the endosomal/lysosomal
compartment, as
described in U.S. Patent No. 5,633,234. An immunogenic polypeptide of the
invention,
15 when fused with this targeting signal, will associate more efficiently with
MHC class II
molecules and thereby provide enhanced in vivo stimulation of CD4+ T-cells
specific
for the polypeptide.
Polypeptides of the invention are prepared using any of a vaxiety of well
known synthetic and/or recombinant techniques, the latter of which are further
20 described below. Polypeptides, portions and other variants generally less
than about
150 amino acids can be generated by synthetic means, using techniques well
known to
those of ordinary skill in the art. In one illustrative example, such
polypeptides are
synthesized using any of the commercially available solid-phase techniques,
such as the
Merrifield solid-phase synthesis method, where amino acids are sequentially
added to a
25 growing amino acid chain. See Merrifield, J. Am. Chem. Soc. 85:2149-2146,
1963.
Equipment for automated synthesis of polypeptides is coW mercially available
from
suppliers such as Perkin Elmer/Applied BioSystems Division (Foster City, CA),
and
may be operated according to the manufacturer's instructions.
In general, polypeptide compositions (including fusion polypeptides) of
30 the invention are isolated. An "isolated" polypeptide is one that is
removed from its
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
51
original environment. For example, a naturally-occurring protein or
polypeptide is
isolated if it is separated from some or all of the coexisting materials in
the natural
system. Preferably, such polypeptides are also purified, e.g., are at least
about 90%
pure, more preferably at least about 95% pure and most preferably at least
about 99%
pure.
Polynucleotide Compositions
The present invention, in other aspects, , provides polynucleotide
compositions. The terms "DNA" and "polynucleotide" are used essentially
interchangeably herein to refer to a DNA molecule that has been isolated free
of total
genomic DNA of a particular species. "Isolated," as used herein, means that a
polynucleotide is substantially away from other coding sequences, and that the
DNA
molecule does not contain large portions of unrelated coding DNA, such as
large
chromosomal fragments or other functional genes or polypeptide coding regions.
Of
course, this refers to the DNA molecule as originally isolated, and does not
exclude
genes or coding regions later added to the segment by the hand of man.
As will be understood by those skilled in the art, the polynucleotide
compositions of this invention can include genomic sequences, extra-genomic
and
plasmid-encoded sequences and smaller engineered gene segments that express,
or may
be adapted to .express, proteins, polypeptides, peptides and the like. Such
segments may
be naturally isolated, or modified synthetically by the hand of man.
As will be also recognized by the skilled artisan, polynucleotides of the
invention may be single-stranded (coding or antisense) or double-stranded, and
may be
DNA (genomic, cDNA or synthetic) or RNA molecules. RNA molecules may include
HnRNA molecules, which contain introns and correspond to a DNA molecule in a
one-
to-one manner, and mRNA molecules, which do not contain introns. Additional
coding
or non-coding sequences may, but need not, be present within a polynucleotide
of the
present invention, and a polynucleotide may, but need not, be linked to other
molecules
and/or support materials.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
52
Polynucleotides may comprise a native sequence (i. e., an endogenous
sequence that encodes a polypeptide/protein of the invention or a portion
thereof) or
may comprise a sequence that encodes a variant or derivative, preferably an
immunogenic variant or derivative, of such a sequence.
Therefore, according to another aspect of the present invention,
polynucleotide compositions are provided that comprise some or all of a
polynucleotide
sequence set forth in any one of SEQ ID NOs: 1-111, 115-171, 173-175, 177, 179-
305,
307-315, 326, 328, 330, 332-335, 340-375, 381, 382 and 384-476, 524, 526, 530,
531,
533, 535, 536, 552, 569-572, 587, 591, 593-606, 618-626, 630, 631, 634, 636,
639-655,
674, 680, 681, 711, 713, 716, 720-722, 735, 737-739, 751, 753, 764, 765, 773-
776 and
786-788, complements of a polynucleotide sequence set forth in any one of SEQ
ID
NOs: I-111, 115-171, 173-175, I77, I79-305, 307-315, 326, 328, 330, 332-335,
340-
375, 381, 382 and 384-476, 524, 526, 530, 531, 533, 535, 536, 552, 569-572,
587, 591,
593-606, 618-626, 630, 631, 634, 636, 639-655, 674, 680, 681, 711, 713, 716,
720-722,
735, 737-739, 751, 753, 764, 765, 773-776 and 786-788, and degenerate variants
of a
polynucleotide sequence set forth in any one of SEQ ID NOs: I-11 l, 115-171,
173-175,
177, 179-305, 307-315, 326, 328, 330, 332-335, 340-375, 381, 382 and 384-476,
524,
526, 530, 531, 533, 535, 536, 552, 569-572, 587, 591, 593-606, 618-626, 630,
631, 634,
636, 639-655, 674, 680, 681, 711, 713, 716, 720-722, 735, 737-739, 751, 753,
764, 765,
773-776 and 786-788. In certain preferred embodiments, the polynucleotide
sequences
set forth herein encode immunogenic polypeptides, as described above.
In other related embodiments, the present invention provides
polynucleotide variants having substantial identity to the sequences disclosed
herein in
SEQ ID NOs: 1-111, 115-171, 173-175, 177, 179-305, 307-315, 326, 328, 330, 332-
335, 340-375, 381, 382 and 384-476, 524, 526, 530, 531, 533, 535, 536, 552,
569-572,
587, 591, 593-606, 618-626, 630, 631, 634, 636, 639-655, 674, 680, 681, 71 l,
713, 716,
720-722, 735, 737-739, 751, 753, 764, 765, 773-776 and 786-788, fox example
those
comprising at least 70% sequence identity, preferably at least 75%, 80%, 85%,
90%,
95%, 96%, 97%, 98%, or 99% or higher, sequence identity compared to a
polynucleotide sequence of this invention using the methods described herein,
(e.g.,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
53
BLAST analysis using standard parameters, as described below). One skilled in
this art
will recognize that these values can be appropriately adjusted to determine
corresponding identity of proteins encoded by two nucleotide sequences by
taking into
account codon degeneracy, amino acid similarity, reading frame positioning and
the
like.
Typically, polynucleotide variants will contain one or more substitutions,
additions, deletions and/or insertions, preferably such that the
immunogenicity of the
polypeptide encoded by the variant polynucleotide is not substantially
diminished
relative to a polypeptide encoded by a polynucleotide sequence specifically
set forth
herein). The term "variants" should also be understood to encompasses
homologous
genes of xenogenic origin.
In additional embodiments, the present invention provides
polynucleotide fragments comprising various lengths of contiguous stretches of
sequence identical to, or complementary to, one or more of the sequences
disclosed
I S herein. For example, polynucleotides are provided by this invention that
comprise at
least about I0, 15, 20, 30, 40, 50, 75, 100, I50, 200, 300, 400, 500 or 1000
or more
contiguous nucleotides of one or more of the sequences disclosed herein as
well as all
intermediate lengths there between. It will be readily understood that
"intermediate
lengths", in this context, means any length between the quoted values, such as
16, 17,
18, 19, etc.; 21, 22, 23, etc.; 30, 31, 32, etc.; 50, 51, 52, 53, etc.; 100,
101, 102, 103,
etc.; 150, 151, I52, 153, etc.; including all integers through 200-500; 500-
1,000, and the
like.
In another embodiment of the invention, polynucleotide compositions are
provided that are capable of hybridizing under moderate to high stringency
conditions to
a polynucleotide sequence provided herein, or a fragment thereof, or a
complementary
sequence thereof. Hybridization techniques are well known in the art of
molecular
biology. For purposes of illustration, suitable moderately stringent
conditions for
testing the hybridization of a polynucleotide of this invention with other
polynucleotides
include prewashing in a solution of 5 X SSC, 0.5% SDS, 1.0 mM EDTA (pH 8.0);
hybridizing at 50°C-60°C, 5 X SSC, overnight; followed by
washing twice at 65°C for
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
54
20 minutes with each of 2X, O.SX and 0.2X SSC containing 0.1% SDS. One skilled
in
the art will understand that the stringency of hybridization can be readily
manipulated,
such as by altering the salt content of the hybridization solution and/or the
temperature
at which the hybridization is performed. For example, in another embodiment,
suitable
highly stringent hybridization conditions include those described above, with
the
exception that the temperature of hybridization is increased, e.g., to 60-
65°C or 65-
70°C.
In certain preferred embodiments, the polynucIeotides described above,
e.g., polynucleotide variants, fragments and hybridizing sequences, encode
polypeptides
that are immunologically cross-reactive with a polypeptide sequence
specifically set
forth herein. In other preferred embodiments, such ' polynucleotides encode
polypeptides that have a level of immunogenic activity of at least about 50%,
preferably
at least about 70%, and more preferably at least about 90% of that for a
polypeptide
sequence specifically set forth herein.
The polynucleotides of the present invention, or fragments thereof,
regardless of the length of the coding sequence itself, may be combined with
other DNA
sequences, such as promoters, polyadenylation signals, additional restriction
enzyme
sites, multiple cloning sites, other coding segments, and the like, such that
their overall
length may vary considerably. It is therefore contemplated that a nucleic acid
fragment
of almost any length may be employed, with the total length preferably being
limited by
the ease of preparation and use in the intended recombinant DNA protocol. For
example, illustrative polynucleotide segments with total lengths of about
10,000, about
5000, about 3000, about 2,000, about 1,000, about 500, about 200, about 100,
about 50
base pairs in length, and the like, (including all intermediate lengths) are
contemplated
to be useful in many implementations of this invention.
When comparing polynucleotide sequences, two sequences are said to be
"identical" if the sequence of nucleotides in the two sequences is the same
when aligned
for maximum correspondence, as described below. Comparisons between two
sequences are typically perforn~ed by comparing the sequences over a
comparison
window to identify and compare local regions of sequence similarity. A
"comparison
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
window" as used herein, refers to a segment of at least about 20 contiguous
positions,
usually 30 to about 75, preferably 40 to about 50, in which a sequence may be
compared
to a reference sequence of the same number of contiguous positions after the
two
sequences are optimally aligned.
5 Optimal alignment of sequences for comparison may be conducted using
the Megalign program in the Lasergene suite of bioinformatics software
(DNASTAR,
Inc., Madison, WI), using default parameters. This program embodies several
alignment schemes described in the following references: Dayhoff, M.O. (1978)
A
model of evolutionary change in proteins - Matrices for detecting distant
relationships.
10 In Dayhoff, M.O. (ed.) Atlas of Protein Sequence and Structure, National
Biomedical
Research Foundation, Washington DC Vol. 5, Suppl. 3, pp. 345-358; Hein J.
(1990)
Unified Approach to Alignment and Phylogenes pp. 626-645 Methods in
EnzynZOlogy
vol. 183, Academic Press, Inc., San Diego, CA; Higgins, D.G. and Sharp, P.M.
(1989)
CABIOS 5:151-153; Myers, E.W. and Muller W. (1988) CABIOS 4:11-17; Robinson,
15 E.D. (197I) Comb. Theor 11:105; Santou, N. Nes, M. (1987) Mol. Biol. Evol.
4:406-
425; Sneath, P.H.A. and Sokal, R.R. (1973) Numerical Taxonomy - the Principles
and
Practice of Numerical Taxonomy, Freeman Press, San Francisco, CA; Wilbur, W.J.
and
Lipman, D.J. (1983) Proc. Natl. Acad., Sci. USA 80:726-730.
Alternatively, optimal alignment of sequences for comparison may be
20 conducted by the local identity algorithm of Smith and Waterman (1981) Add.
APL.
Math 2:482, by the identity alignment algorithm of Needleman and Wunsch (1970)
J.
Mol. Biol. 48:443, by the search for similarity methods of Pearson and Lipman
(1988)
Pf-oc. Natl. Acad. Sci. USA 85: 2444, by computerized implementations of these
algorithms (GAP, BESTFIT, BLAST, FASTA, and TFASTA in the Wisconsin Genetics
25 Software Package, Genetics Computer Group (GCG), 575 Science Dr., Madison,
WI),
or by inspection.
One preferred example of algorithms that are suitable for determining
percent sequence identity and sequence similarity are the BLAST and BLAST 2.0
algorithms, which are described in Altschul et al. (1977) Nucl. Acids Res.
25:3389-3402
30 and Altschul et al. (1990) J. Mol: Biol. 215:403-410, respectively. BLAST
and BLAST
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
56
2.0 can be used, for example with the parameters described herein, to
determine percent
sequence identity for the polynucleotides of the invention. Software for
performing
BLAST analyses is publicly available through the National Center for
Biotechnology
Information. In one illustrative example, cumulative scores can be calculated
using, for
nucleotide sequences, the parameters M (reward score for a pair of matching
residues;
always >0) and N (penalty score for mismatching residues; always <0).
Extension of
the word hits in each direction are halted when: the cumulative alignment
score falls off
by the quantity X from its maximum achieved value; the cumulative score goes
to zero
or below, due to the accumulation of one or more negative-scoring residue
alignments;
or the end of either sequence ~is reached. The BLAST algorithm parameters W, T
and X
determine the sensitivity and speed of the alignment. The BLASTN program (for
nucleotide sequences) uses as defaults a wordlength (W) of 11, and expectation
(E) of
10, and the BLOSUM62 scoring matrix (see Henikoff and Henikoff (1989) Proc.
Natl.
Acad. Sci. USA 89:10915) alignments, (B) of 50, expectation (E) of 10, M=5, N=-
4 and
I 5 a comparison of both strands.
Preferably, the "percentage of sequence identity" is determined by
comparing two optimally aligned sequences over a window of comparison of at
least 20
positions, wherein the portion of the polynucleotide sequence in the
comparison
window may comprise additions or deletions (i. e., gaps) of 20 percent or
less, usually 5
to 15 percent, or 10 to I2 percent, as compared to the reference sequences
(which does
not comprise additions or deletions) for optimal alignment of the two
sequences. The
percentage is calculated by determining the number of positions at which the
identical
nucleic acid bases occurs in both sequences to yield the number of matched
positions,
dividing the number of matched positions by the total number of positions in
the
reference sequence (i.e., the window size) and multiplying the results by 100
to yield the
percentage of sequence identity.
It will be appreciated by those of ordinary skill in the art that, as a result
of the degeneracy of the genetic code, there are many nucleotide sequences
that encode
a polypeptide as described herein. Some of these polynucleotides bear minimal
homology to the nucleotide sequence of any native gene. Nonetheless,
polynucleotides
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
57
that vary due to differences in codon usage are specifically contemplated by
the present
invention. Further, alleles of the genes comprising the polynucleotide
sequences
provided herein are within the scope of the present invention. Alleles are
endogenous
genes that are altered as a result of one or more mutations, such as
deletions, additions
andlor substitutions of nucleotides. The resulting mRNA and protein may, but
need not,
have an altered structure or function. ~ Alleles may be identified using
standard
techniques (such as hybridization, amplification and/or database sequence
comparison).
Therefore, in another embodiment of the invention, a mutagenesis
approach, such as site-specific mutagenesis, is employed for the preparation
of
immunogenic variants and/or derivatives of the polypeptides described herein.
By this
approach, specific modifications in a polypeptide sequence can be made through
mutagenesis of the underlying polynucleotides that encode them. These
techniques
provides a straightforward approach to prepare and test sequence variants, for
example,
incorporating one or more of the foregoing considerations, by introducing one
or more
I S nucleotide sequence changes into the polynucleotide.
Site-specific mutagenesis allows the production of mutants through the
use of specific oligonucleotide sequences which encode the DNA sequence of the
desired mutation, as well as a sufficient number of adjacent nucleotides, to
provide a
primer sequence of sufficient size and sequence complexity to form a stable
duplex on
both sides of the deletion junction being traversed. Mutations may be employed
in a
selected polynucleotide sequence to improve, alter, decrease, modify, or
otherwise
change the properties of the polynucleotide itself, and/or alter the
properties, activity,
composition, stability, or primary sequence of the encoded polypeptide.
In certain embodiments of the present invention, the inventors
contemplate the mutagenesis of the disclosed polynucleotide sequences to alter
one or
more properties of the encoded polypeptide, such as the immunogenicity of a
polypeptide vaccine. The techniques of site-specific mutagenesis are well-
known in the
art, and are widely used to create variants of both polypeptides and
polynucleotides. For
example, site-specific mutagenesis is often used to alter a specific portion
of a DNA
molecule. In such embodiments, a primer comprising typically about 14 to about
25
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
58
nucleotides or so in length is employed, with about 5 to about 10 residues on
both sides
of the junction of the sequence being altered.
As will be appreciated by those of skill in the art, site-specific
mutagenesis techniques have often employed a phage vector that exists in both
a single
stranded and double stranded form. Typical vectors useful in site-directed
mutagenesis
include vectors such as the M13 phage. These phage are readily commercially
available
and their use is generally well-known to those skilled in the art. Double-
stranded
plasmids are also routinely employed in site directed mutagenesis that
eliminates the
step of transferring the gene of interest from a plasmid to a phage.
In general, site-directed mutagenesis in accordance herewith ~ is
performed by first obtaining a single-stranded vector or melting apart of two
strands of a
double-stranded vector that includes within its sequence a DNA sequence that
encodes
the desired peptide. An oligonucleotide primer bearing the desired mutated
sequence is
prepared, generally synthetically. This primer is then annealed with the
single-stranded
vector, and subjected to DNA polymerizing enzymes such as E. coli polymerase I
Klenow fragment, in order to complete the synthesis of the mutation-bearing
strand.
Thus, a heteroduplex is formed wherein one strand encodes the original non-
mutated
sequence and the second strand bears the desired mutation. This heteroduplex
vector is
then used to transform appropriate cells, such as E. coli cells, and clones
are selected
which include recombinant vectors bearing the mutated sequence arrangement.
The preparation of sequence variants of the selected peptide-encoding
DNA segments using site-directed mutagenesis provides a means of producing
potentially useful species and is not meant to be limiting as there are other
ways in
which sequence variants of peptides and the DNA sequences encoding them may be
obtained. For example, recombinant vectors encoding the desired peptide
sequence
may be treated with mutagenic agents, such as hydroxylamine, to obtain
sequence
variants. Specific details regarding these methods and protocols are found in
the
teachings of Maloy et al., 1994; Segal, 1976; Prokop and Bajpai, 1991; Kuby,
1994; and
Maniatis et al., 1982, each incorporated herein by reference, for that
purpose.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
59
As used herein, the term "oligonucleotide directed mutagenesis
procedure" refers to template-dependent processes and vector-mediated
propagation
which result in an increase in the concentration of a specific nucleic acid
molecule
relative to its initial concentration, or in an increase in the concentration
of a detectable
signal, such as amplification. As used herein, the teen "oligonucleotide
directed
mutagenesis procedure" is intended to refer to a process that involves the
template-dependent extension of a primer molecule. The term template dependent
process refers to nucleic acid synthesis of an RNA or a DNA molecule wherein
the
sequence of the newly synthesized strand of nucleic acid is dictated by the
well-known
rules of complementary base pairing (see, for example, Watson, 1987).
Typically,
vector mediated methodologies involve the introduction of the nucleic acid
fragment
into a DNA or RNA vector, the clonal amplification of the vector, and the
recovery of
the amplified nucleic acid fragment. Examples of such methodologies are
provided by
U. S. Patent No. 4,237,224, specifically incorporated herein by reference in
its entirety.
In another approach for the production of polypeptide variants of the
present invention, recursive sequence recombination, as described in U.S.
Patent No.
5,837,458, may be employed. In this approach, iterative cycles of
recombination and
screening or selection are performed to "evolve" individual polynucleotide
variants of
the invention having, for example, enhanced immunogenic activity.
In other embodiments of the present invention, the polynucleotide
sequences provided herein can be advantageously used as probes or primers for
nucleic
acid hybridization. As such, it is contemplated that nucleic acid segments
that comprise
a sequence region of at least about I S contiguous nucleotides that has the
same
sequence as, or is complementary to, a 15 nucleotide long contiguous sequence
disclosed herein will find particular utility. Longer contiguous identical or
complementary sequences, e.g., those of about 20, 30, 40, 50, 100, 200, 500,
1000
(including all intermediate lengths) and even up to full length sequences will
also be of
use in certain embodiments.
The ability of such nucleic acid probes to specifically hybridize to a
sequence of interest will enable them to be of use in detecting the presence
of
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
complementary sequences in a given sample. However, other uses are also
envisioned,
such as the use of the sequence information for the preparation of mutant
species
primers, or primers for use in preparing other genetic constructions.
Polynucleotide molecules having sequence regions consisting of
S contiguous nucleotide stretches of 10-14, 1S-20, 30, S0, or even of 100-200
nucleotides
or so (including intermediate lengths as well), identical or complementary to
a
polynucleotide sequence disclosed herein, are particularly contemplated as
hybridization
probes for use in, e.g., Southern and Northern blotting. This would allow a
gene
product, or fragment thereof, to be analyzed, both in diverse cell types and
also in
10 various bacterial cells. The total size of fragment, as well as the size of
the
complementary stretch(es), will ultimately depend on the intended use or
application of
the particular nucleic acid segment. Smaller fragments will generally fmd use
in
hybridization embodiments, wherein the length of the contiguous complementary
region
may be varied, such as between about 1 S and about 100 nucleotides, but larger
1 S contiguous complementarity stretches may be used, according to the length
complementary sequences one wishes to detect.
The use of a hybridization probe of about 1 S-2S nucleotides in length
allows the formation of a duplex molecule that is both stable and selective.
Molecules
having contiguous complementary sequences over stretches greater than 1 S
bases in
20 length are generally preferred, though, in order to increase stability and
selectivity of the
hybrid, and thereby improve the quality and degree of specific hybrid
molecules
obtained. One will generally pxefer to design nucleic acid molecules having
gene-
complementary stretches of 1 S to 2S contiguous nucleotides, or even longer
where
desired.
2S Hybridization probes may be selected from any portion of any of the
sequences disclosed herein. All that is required is to review the sequences
set forth
herein, or to any continuous portion of the sequences, from about 1 S-2S
nucleotides in
length up to and including the full length sequence, that one wishes to
utilize as a probe
or primer. The choice of probe and primer sequences may be governed by various
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
61
factors. For example, one may wish to employ primers from towards the termini
of the
total sequence.
Small polynucleotide segments or fragments may be readily prepared by,
for example, directly synthesizing the fragment by chemical means, as is
commonly
practiced using an automated oligonucleotide synthesizer. Also, fragments may
be
obtained by application of nucleic acid reproduction technology, such as the
PCRTM
technology of U. S. Patent 4,683,202 (incorporated herein by reference), by
introducing
selected sequences into recombinant vectors for recombinant production, and by
other
recombinant DNA techniques generally known to those of skill in the art of
molecular
IO biology.
The nucleotide sequences of the invention may be used for their ability to
selectively form duplex molecules with complementary stretches of the entire
gene or
gene fragments of interest. Depending on the application envisioned, one will
typically
desire to employ varying conditions of hybridization to achieve varying
degrees of
selectivity of probe towards target sequence. For applications requiring high
selectivity,
one will Typically desire to employ relatively stringent conditions to form
the hybrids,
e.g., one will select relatively low salt and/or high temperature conditions,
such as
provided by a salt concentration of from about 0.02 M to about 0.15 M salt at
temperatures of from about 50°C to about 70°C. Such selective
conditions tolerate
little, if any, mismatch between the probe and the template or target strand,
and would
be particularly suitable for isolating related sequences.
Of course, for some applications, for example, where one desires to
prepare mutants employing a mutant primer strand hybridized to an underlying
template, less stringent (reduced stringency) hybridization conditions will
typically be
needed in order to allow formation of the heteroduplex. In these
circumstances, one
may desire to employ salt conditions such as those of from about 0.15 M to
about 0.9 M
salt, at temperatures ranging from about 20°C to about 55°C.
Cross-hybridizing species
can thereby be readily identified as positively hybridizing signals with
respect to control
hybridizations. In any case, it is generally appreciated that conditions can
be rendered
more stringent by the addition of increasing amounts of formamide, which
serves to
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
62
destabilize the hybrid duplex in the same manner as increased temperature.
Thus,
hybridization conditions can be readily manipulated, and thus will generally
be a
method of choice depending on the desired results. ,
According to another embodiment of the present invention,
polynucleotide compositions comprising antisense oligonucleotides are
provided.
Antisense oligonucleotides have been demonstrated to be effective and targeted
inhibitors of protein synthesis, and, consequently, provide a therapeutic
approach by
which a disease can be treated by inhibiting the synthesis of proteins that
contribute to
the disease. The efficacy of antisense oligonucleotides for inhibiting protein
synthesis
is well established. For example, the synthesis of polygalactauronase and the
muscarine
type 2 acetylcholine receptor are inhibited by antisense oligonucleotides
directed to their
respective mRNA sequences (U. S. Patent 5,739,119 and U. S. Patent 5,759,829).
Further, examples of antisense inhibition have been demonstrated with the
nuclear
protein. cyclin, the multiple drug resistance gene (MDG1), ICAM-l, E-selectin,
STK-1,
striatal GABAA receptor and human EGF (Jaskulski et al., Science. 1988 Jun
10;240(4858):1544-6; Vasanthakumar and Ahmed, Cancer Commun. 1989;1(4):225-
32; Peris et al., Brain Res Mol Brain Res. 1998 Jun 15;57(2):310-20; U. S.
Patent
5,801,154; U.S. Patent 5,789,573; U. S. Patent 5,718,709 and U.S. Patent
5,610,288).
Antisense constructs have also been described that inhibit and can be used to
treat a
variety of abnormal cellular proliferations, e.g. cancer (U. S. Patent
5,747,470; U. S.
Patent 5,591,317 and U. S. Patent 5,783,683).
Therefore, in certain embodiments, the present invention provides
oligonucleotide sequences that comprise all, or a portion of, any sequence
that is
capable of specifically binding to polynucleotide sequence described herein,
or a
complement thereof. In one embodiment, the antisense oligonucleotides comprise
DNA
or derivatives thereof. In another embodiment, the oligonucleotides comprise
RNA or
derivatives thereof. In a third embodiment, the oligonucleotides are modified
DNAs
comprising a phosphorothioated modified backbone. In a fourth embodiment, the
oligonucleotide sequences comprise peptide nucleic acids or derivatives
thereof. In
each case, preferred compositions comprise a sequence region that is
complementary,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
63
and more preferably substantially-complementary, and even more preferably,
completely complementary to one or more portions of polynucleotides disclosed
herein.
Selection of antisense compositions specif c for a given gene sequence is
based upon
analysis of the chosen target sequence and determination of secondary
structure, Tm,
binding energy, and relative stability. Antisense compositions may be selected
based
upon their relative inability to form dimers, hairpins, or other secondary
structures that
would reduce or prohibit specific binding to the target mRNA in a host cell.
Highly
preferred target regions of the mRNA, are those which are at or near the AUG
translation initiation codon, and those sequences which are substantially
complementary
to 5' regions of the mRNA. These secondary structure analyses and target site
selection
considerations can be performed, for example, using v.4 of the OLIGO primer
analysis
software and/or the BLASTN 2Ø5 algorithm software (Altschul et al., Nucleic
Acids
Res. 1997 Sep 1;25(17):3389-402).
The use of an antisense delivery method employing a short peptide
I S vector, termed MPG (27 residues), is also contemplated. The MPG peptide
contains a
hydrophobic domain derived from the fusion sequence of HIV gp41 and a
hydrophilic
domain from the nuclear localization sequence of SV40 T-antigen (Morris et
al.,
Nucleic Acids Res. 1997 Jul 15;25(14):2730-6). It has been demonstrated that
several
molecules of the MPG peptide coat the antisense oligonucleotides and can be
delivered
into cultured mammalian cells in less than 1 hour with relatively high
efficiency (90%).
Further, the interaction with MPG strongly increases both the stability of the
oligonucleotide to nuclease and the ability to cross the plasma membrane.
According to another embodiment of the invention, the polynucleotide
compositions described herein are used in the design and preparation of
ribozyme
molecules for inhibiting expression of the tumor polypeptides and proteins of
the
present invention in tumor cells. Ribozymes are RNA-protein complexes that
cleave
nucleic acids in a site-specific fashion. Ribozymes have specific catalytic
domains that
possess endonuclease activity (Kim and Cech, Proc Natl Acad Sci U S A. 1987
Dec;84(24):8788-92; Forster and Symons, Cell. 1987 Apr 24;49(2):211-20). For
example, a large number of ribozymes accelerate phosphoester transfer
reactions with a
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
64
high degree of specificity, often cleaving only one of several phosphoesters
in an
oligonucleotide substrate (Cech et al., Cell. 1981 Dec;27(3 Pt 2):487-96;
Michel and
Westhof, J Mol Biol. 1990 Dec 5;216(3):585-610; Reinhold-Hurek and Shub,
Nature.
1992 May 14;357(6374):173-6). This specificity has been attributed to the
requirement
that the substrate bind via specific base-pairing interactions to the internal
guide
sequence ("IGS") of the ribozyme prior to chemical reaction.
Six basic varieties of naturally-occurring enzymatic RNAs are known
presently. Each can catalyze the hydrolysis of RNA phosphodiester bonds ih
t~a~s (and
thus can cleave other RNA molecules) under physiological conditions. In
general,
enzymatic nucleic acids act by first binding to a target RNA. Such binding
occurs
through the target binding portion of a enzymatic nucleic acid which is held
in close
proximity to an enzymatic portion of the molecule that acts to cleave the
target RNA.
Thus, the enzymatic nucleic acid first recognizes and then binds a target RNA
through
complementary base-pairing, and once bound to the correct site, acts
enzymatically to
cut the target RNA. Strategic cleavage of such a target RNA will destroy its
ability to
direct synthesis of an encoded protein. After an enzymatic nucleic acid has
bound and
cleaved its RNA target, it is released from that RNA to search for another
target and can
repeatedly bind and cleave new targets.
The enzymatic nature of a ribozyme is advantageous over many
technologies, such as antisense technology (where a nucleic acid molecule
simply binds
to a nucleic acid target to block its translation) since the concentration of
ribozyme
necessary to affect a therapeutic treatment is lower than that of an antisense
oligonucleotide. This advantage reflects the ability of the ribozyme to act
enzymatically. Thus, a single ribozyme molecule is able to cleave many
molecules of
target RNA. In addition, the ribozyme is a highly specific inhibitor, with the
specificity
of inhibition depending not only on the base pairing mechanism of binding to
the target
RNA, but also on the mechanism of target RNA cleavage. Single mismatches, or
base-
substitutions, near the site of cleavage can completely eliminate catalytic
activity of a
ribozyme. Similar mismatches in antisense molecules do not prevent their
action
(Woolf et al., Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7305-9). Thus, the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
specificity of action of a ribozyme is greater than that of an antisense
oligonucleotide
binding the same RNA site.
The enzymatic nucleic acid molecule may be formed in a hammerhead,
hairpin, a hepatitis 8 virus, group I intron or RNaseP RNA (in association
with an RNA
5 guide sequence) or Neurospora VS RNA motif. Examples of hammerhead motifs
are
described by Rossi et al. Nucleic Acids Res. 1992 Sep 11;20(17):4559-65.
Examples of
hairpin motifs are described by Hampel et al. (Eur. Pat. Appl. Publ. No. EP
0360257),
Hampel and Tritz, Biochemistry 1989 Jun 13;28(12):4929-33; Hampel et al.,
Nucleic
Acids Res. 1990 Jan 25;18(2):299-304 and U. S. Patent 5,631,359. An example of
the
10 hepatitis ~ virus motif is described by Perrotta and Been, Biochemistry.
1992 Dec
1;31(47):11843-52; an example of the RNaseP motif is described by Guerrier-
Takada
et al., Cell. 1983 Dec;35(3 Pt 2):849-57; Neurospora VS RNA ribozyme motif is
described by Collins (Saville and Collins, Cell. 1990 May 18;61 (4):685-96;
Saville and
Collins, Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8826-30; Collins and
Olive,
15 Biochemistry. 1993 Mar 23;32(11):2795-9); and an example of the Group I
intron is
described in (U. S. Patent 4,987,071). All that is important in an enzymatic
nucleic acid
molecule of this invention is that it has a specific substrate binding site
which is
complementary to one or more of the target gene RNA regions, and that it have
nucleotide sequences within or surrounding that substrate binding site which
impart an
20 RNA cleaving activity to the molecule. Thus the ribozyme constructs need
not be
limited to specific motifs mentioned herein.
Ribozymes may be designed as described in Int. Pat. Appl. Publ. No.
WO 93/23569 and Int. Pat. Appl. Publ. No. WO 94/02595, each specifically
incorporated herein by reference) and synthesized to be tested in
vitf°o and in vivo, as
25 described. Such ribozymes can also be optimized for delivery. While
specific
examples are provided, those in the art will recognize that equivalent RNA
targets in
other species can be utilized when necessary.
Ribozyme activity can be optimized by altering the length of the
ribozyme binding arms, or chemically synthesizing ribozymes with modifications
that
30 prevent their degradation by serum ribonucleases (see e.g., Int. Pat. Appl.
Publ. No. WO
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
66
92/07065; Int. Pat. Appl. Publ. No. WO 93/15187; Int. Pat. Appl. Publ. No. WO
91/03162; Eur. Pat. Appl. Publ. No. 92110298.4; U. S. Patent 5,334,711; and
Int. Pat.
Appl. Publ. No. WO 94/13688, which describe various chemical modifications
that can
be made to the sugar moieties of enzymatic RNA molecules), modifications which
enhance their efficacy in cells, and removal of stem II bases to shorten RNA
synthesis
times and reduce chemical requirements.
Sullivan et al. (Int. Pat. Appl. Publ. No. WO 94/02595) describes the
general methods for delivery of enzymatic RNA molecules. Ribozymes may be
administered to cells by a variety of methods known to those familiar to the
art,
including, but not restricted to, encapsulation in liposomes, by
iontophoresis, or by
incorporation into other vehicles, such as hydrogels, cyclodextrins,
biodegradable
nanocapsules, and bioadhesive microspheres. For some indications, ribozymes
may be
directly delivered ex vivo to cells or tissues with or without the
aforementioned vehicles.
Alternatively, the RNA/vehicle combination may be locally delivered by direct
inhalation, by direct injection or by use of a catheter, infusion pump or
stmt. Other
routes of delivery include, but are not limited to, intravascular,
intramuscular,
subcutaneous or joint injection, aerosol inhalation, oral (tablet or pill
form), topical,
systemic, ocular, intraperitoneal and/or intrathecal delivery. More detailed
descriptions
of ribozyme delivery and administration are provided in Int. Pat. Appl. Publ.
No. WO
94/02595 and Int. Pat. Appl. Publ. No. WO 93/23569, each specifically
incorporated
herein by reference.
Another means of accumulating high concentrations of a ribozyme(s)
within cells is to incorporate the ribozyme-encoding sequences into a DNA
expression
vector. Transcription of the ribozyme sequences are driven from a promoter for
eukaryotic RNA polymerase I (pol I), RNA polymerase II (pol II), or RNA
polymerase
III (pol III). Transcripts from pol II or pol III promoters will be expressed
at high levels
in all cells; the levels of a given pol II promoter in a given cell type will
depend on the
nature of the gene regulatory sequences (enhancers, silencers, etc.) present
nearby.
Prokaryotic RNA polymerase promoters may also be used, providing that the
prokaryotic RNA polymerase enzyme is expressed in the appropriate cells
Ribozymes
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
67
expressed from such promoters have been shown to function in mammalian cells.
Such
transcription units can be incorporated into a variety of vectors for
introduction into
mammalian cells, including but not restricted to, plasmid DNA vectors, viral
DNA
vectors (such as adenovirus or adeno-associated vectors), or viral RNA vectors
(such as
retroviral, semliki forest virus, sindbis virus vectors).
In another embodiment of the invention, peptide nucleic acids (PNAs)
compositions are provided. PNA is a DNA mimic in which the nucleobases are
attached to a pseudopeptide backbone (Good and Nielsen, Antisense Nucleic Acid
Drug
Dev. 1997 7(4) 431-37). PNA is able to be utilized in a number methods that
traditionally have used RNA or DNA. Often PNA sequences perform better in
techniques than the corresponding RNA or DNA sequences and have utilities that
are
not inherent to RNA or DNA. A review of PNA including methods of making,
characteristics of, and methods of using, is provided by Corey (Tends
Biotechnol 1997
Jun;lS(6):224-9). As such, in certain embodiments, one may prepare PNA
sequences
that are complementary to one or more portions of the ACE mRNA sequence, and
such
PNA compositions may be used to regulate, alter, decrease, or reduceahe
translation of
ACE-specific mRNA, and thereby alter the level of ACE activity in a host cell
to which
such PNA compositions have been administered.
PNAs have 2-aminoethyl-glycine linkages replacing the normal
phosphodiester backbone of DNA (Nielsen et al., SciefZCe 1991 Dec
6;254(5037):1497-
500; Hanvey et al., Science. 1992 Nov 27;258(5087):1481-5; Hyrup and Nielsen,
Bioorg Med Chem. 1996 Jan;4(1):5-23). This chemistry has three important
consequences: firstly, in contrast to DNA or phosphorothioate
oligonucleotides, PNAs
are neutral molecules; secondly, PNAs are achiral, which avoids the need to
develop a
stereoselective synthesis; and thirdly, PNA synthesis uses standard Boc or
Fmoc
protocols for solid-phase peptide synthesis, although other methods, including
a
modified Merrifield method, have been used.
PNA monomers or ready-made oligomers are 'commercially available
from PerSeptive Biosystems (Framingham, MA). PNA syntheses by either Boc or
Fmoc protocols are straightforward using manual or automated protocols (Norton
et al.,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
68
Bioorg Med Chem. 1995 Apr;3(4):437-45). The manual protocol lends itself to
the
production of chemically modified PNAs or the simultaneous synthesis of
families of
closely related PNAs.
As with peptide synthesis, the success of a particular PNA synthesis will
depend on the properties of the chosen sequence. For example, while in theory
PNAs
can incorporate any combination of nucleotide bases, the presence of adjacent
purines
can lead to deletions of one or more residues in the product. In expectation
of this
difficulty, it is suggested that, in producing PNAs with adjacent purines, one
should
repeat the coupling of residues likely to be added inefficiently. This should
be followed
IO by the purification of PNAs by reverse-phase high-pressure liquid
chromatography,
providing yields and purity of product similar to those observed during the
synthesis of
peptides.
Modifications of PNAs for a given application may be accomplished by
coupling amino acids during solid-phase synthesis or by attaching compounds
that
contain a carboxylic acid group to the exposed N-terminal amine.
Alternatively, PNAs
can be modified after synthesis by coupling to an introduced lysine or
cysteine. The
ease with which PNAs can be modified facilitates optimization for better
solubility or
for specif c functional requirements. Once synthesized, the identity of PNAs
and their
derivatives can be confirmed by mass spectrometry. Several studies have made
and
utilized modifications of PNAs (for example, Norton et al., Bioorg Med Chem.
1995
Apr;3(4):437-45; Petersen et al., J Pept Sci. 1995 May-Jun;l(3):175-83; Orum
et~ al.,
Biotechniques. 1995 Sep;l9(3):472-80; Footer et al., Biochemistry. 1996 Aug
20;35(33):10673-9; Griffith et al., Nucleic Acids Res. 1995 Aug 11;23(15):3003-
8;
Pardridge et al., Proc Natl Acad Sci U S A. 1995 Jun 6;92(12):5592-6; Boffa et
al.,
Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1901-5; Gambacorti-Passerini et
al.,
Blood. 1996 Aug 15;88(4):1411-7; Armitage et al., Proc Natl Acad Sci U S A.
1997
Nov 11;94(23):12320-5; Seeger et al., Biotechniques. 1997 Sep;23(3):512-7).
U.5.
Patent No. 5,700,922 discusses PNA-DNA-PNA chimeric molecules and their uses
in
diagnostics, modulating protein in organisms, and treatment of conditions
susceptible to
therapeutics.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
69
Methods of characterizing the antisense binding properties of PNAs are
discussed in Rose (Anal Chem. 1993 Dec 15;65(24):3545-9) and Jensen et al.
(Biochemistry. 1997 Apr 22;36(16):5072-7). Rose uses capillary gel
electrophoresis to
determine binding of PNAs to their complementary oligonucleotide, measuring
the
relative binding kinetics and stoichiometry. Similar types of measurements
were made
by Jensen et al. using BIAcoreTM technology.
Other applications of PNAs that have been described and will be
apparent to the skilled artisan include use in DNA strand invasion, antisense
inhibition,
mutational analysis, enlnancers of transcription, nucleic acid purification,
isolation of
transcriptionally active genes, blocking of transcription factor binding,
genome
cleavage, biosensors, in situ hybridization, and the like.
Polynucleotide Identification, Characterization and Expression
Polynucleotide compositions of the present invention may be identified,
prepared and/or manipulated using any of a variety of well established
techniques (see
generally, Sambrook et al., Molecular Clo~ihg: A Laboratory Manual, Cold
Spring
Harbor Laboratories, Cold Spring Harbor, NY, 1989, and other like references).
For
example, a polynucleotide may be identified, as described in more detail
below, by
screening a microarray of cDNAs for tumor-associated expression (i.e.,
expression that
is at least two fold greater in a tumor than in normal tissue, as determined
using a
representative assay provided herein). Such screens may be performed, for
example,
using the microarray technology of Affymetrix, Inc. (Santa Clara, CA)
according to the
manufacturer's instructions (and essentially as described by Schena et al.,
Pr°oc. Natl.
Acad. Sci. USA 93:10614-10619, 1996 and Heller et al., Proc. Natl. Acad. Sci.
USA
94:2150-2155, 1997). Alternatively, polynucleotides may be amplified from cDNA
prepared from cells expressing the proteins described herein, such as tumor
cells.
Many template dependent processes are available to amplify a target
sequences of interest present in a sample. One of the best known amplification
methods
is the polymerase chain reaction (PCRTM) which is described in detail in U.S.
Patent
Nos. 4,683,195, 4,683,202 and 4,800,159, each of which is incorporated herein
by
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
70 .
reference in its entirety. Briefly, in PCRTM, two primer sequences are
prepared which
are complementary to regions on opposite complementary strands of the target
sequence. An excess of deoxynucleoside triphosphates is added to a reaction
mixture
along with a DNA polymerise (e.g., Taq polymerise). If the target sequence is
present
in a sample, the primers will bind to the target and the polymerise will cause
the
primers to be extended along the target sequence by adding on nucleotides. By
raising
and lowering the temperature of the reaction mixture, the extended primers
will
dissociate from the target to form reaction products, excess primers will bind
to the
target and to the reaction product and the process is repeated. Preferably
reverse
transcription and PCRTM amplification procedure may be performed in order to
quantify
the amount of mRNA amplified. Polymerise chain reaction methodologies are well
known in the art.
Any of a number of other template dependent processes, many of which
are variations of the PCR TM amplification technique, are readily known and
available in
the art. Illustratively, some such methods include the ligase chain reaction
(referred to
as LCR), described, for example, in Eur. Pat. Appl. Publ. No. 320,308 and U.S.
Patent
No. 4,883,750; Qbeta Replicase, described in PCT Intl. Pat. Appl. Publ. No.
PCT/US87/00880; Strand Displacement Amplification (SDA) and Repair Chain
Reaction (RCR). Still other amplification methods are described in Great
Britain Pat.
Appl. No. 2 202 328, and in PCT Intl. Pat. Appl. Publ. No. PCT/LJS89/01025.
Other
nucleic acid amplification procedures include transcription-based
amplification systems
(TAS) (PCT Intl. Pat. Appl. Publ. No. WO 88/10315), including nucleic acid
sequence
based amplification (NASBA) and 3SR. Eur. Pat. Appl. Publ. No. 329,822
describes a
nucleic acid amplification process involving cyclically synthesizing single-
stranded
RNA ("ssRNA"), ssDNA, and double-stranded DNA (dsDNA). PCT Intl. Pat. Appl.
Publ. No. WO 89/06700 describes a nucleic acid sequence amplification scheme
based
on the hybridization of a promoter/primer sequence to a target single-stranded
DNA
("ssDNA") followed by transcription of many RNA copies of the sequence. Other
amplification methods such as "RACE" (Frohman, 1990), and "one-sided PCR"
(Ohara,
1989) are also well-known to those of skill in the art.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
71
An amplified portion of a polynucleotide of the present invention may be
used to isolate a full length gene from a suitable library (e.g., a tumor cDNA
library)
using well known techniques. Within such techniques, a library (cDNA or
genomic) is
screened using one or more polynucleotide probes or primers suitable for
amplification.
Preferably, a library is size-selected to include larger molecules. Random
primed
libraries may also be preferred for identifying 5' and upstream regions of
genes.
Genomic libraries are preferred for obtaining introns and extending 5'
sequences.
For hybridization techniques, a partial sequence may be labeled (e.g., by
nick-translation or end-labeling with 32P) using well known techniques. A
bacterial or
bacteriophage library is then generally screened by hybridizing filters
containing
denatured bacterial colonies (or lawns containing phage plaques) with the
labeled probe
(see Sambrook et al., Molecular Cloning: A Labof~ato~y Manual, Cold Spring
Harbor
Laboratories, Cold Spring Harbor, NY, 1989). Hybridizing colonies or plaques
are
selected and expanded, and the DNA is isolated for further analysis. cDNA
clones may
be analyzed to determine the amount of additional sequence by, for example,
PCR using
a primer from the partial sequence and a primer from the vector. Restriction
maps and
partial sequences may be generated to identify one or more overlapping clones.
The
complete sequence may then be determined using standard techniques, which may
involve generating a series of deletion clones. The resulting overlapping
sequences can
then assembled into a single contiguous sequence. A full length cDNA molecule
can be
generated by ligating suitable fragments, using well known techniques.
Alternatively, amplification techniques, such as those described above,
can be useful for obtaining a full length coding sequence from a partial cDNA
sequence.
One such amplification technique is inverse PCR (see Triglia et al., Nucl.
Acids Res.
16:8186, 1988), which uses restriction enzymes to generate a fragment in the
known
regioxi of the gene. The fragment is then circularized by intramolecular
ligation and
used as a template for PCR with divergent primers derived from the known
region.
Within an alternative approach, sequences adjacent to a partial sequence may
be
retrieved by amplification with a primer to a linker sequence and a primer
specific to a
known region. The amplified sequences are typically subjected to a second
round of
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
72
amplification with the same linker primer and a second primer specific to the
known
region. A variation on this procedure, which employs two primers that initiate
extension in opposite directions from the known sequence, is described in WO
96138591. Another such technique is known as "rapid amplification of cDNA
ends" or
RACE. This technique involves the use of an internal primer and an external
primer,
which hybridizes to a polyA region or vector sequence, to identify sequences
that are 5'
and 3' of a known sequence. Additional techniques include capture PCR
(Lagerstrom et
al., PCR Methods Applic. 1:l I 1-19, 1991) and walking PCR (Parker et al.;
Nucl. Acids.
Res. 19:3055-60, 1991). Other methods employing amplification may also be
employed
to obtain a full length cDNA sequence.
In certain instances, it is possible to obtain a full length cDNA sequence
by analysis of sequences provided in an expressed sequence tag (EST) database,
such as
that available from GenBank. Searches for overlapping ESTs may generally be
performed using well known programs (e.g., NCBI BLAST searches), and such ESTs
may be used to generate a contiguous full length sequence. Full length DNA
sequences
may also be obtained by analysis of genomic fragments.
In other embodiments of the invention, polynucleotide sequences or
fragments thereof which encode polypeptides of the invention, or fusion
proteins or
functional equivalents thereof, may be used in recombinant DNA molecules to
direct
expression of a polypeptide in appropriate host cells. Due to the inherent
degeneracy of
the genetic code, other DNA sequences that encode substantially the same or a
functionally equivalent amino acid sequence may be produced and these
sequences may
be used to clone and express a given polypeptide.
As will be understood by those of skill in the art, it may be advantageous
in some instances to produce polypeptide-encoding nucleotide sequences
possessing
non-naturally occurring codons. For example, codons preferred by a particular
prokaryotic or eukaryotic host can be selected to increase the rate of protein
expression
or to produce a recombinant RNA transcript having desirable properties, such
as a half
life which is longer than that of a transcript generated from the naturally
occurring
sequence.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
73
Moreover, the polynucleotide sequences of the present invention can be
engineered using methods generally known in the art in order to alter
polypeptide
encoding sequences for a variety of reasons, including but not limited to,
alterations
which modify the cloning, processing, and/or expression of the gene product.
For
example, DNA shuffling by random fragmentation and PCR reassembly of gene
fragments and synthetic oligonucleotides may be used to engineer the
nucleotide
sequences. In addition, site-directed mutagenesis may be used to insert new
restriction
sites, alter glycosylation patterns, change codon preference, produce splice
variants, or
introduce mutations, and so forth.
In another embodiment of the invention, natural, modified, or
recombinant nucleic acid sequences may be ligated to a heterologous sequence
to
encode a fusion protein. For example, to screen peptide libraries for
inhibitors of
polypeptide activity, it may be useful to encode a chimeric protein that can
be
recognized by a commercially available antibody. A fusion protein may also be
engineered to contain a cleavage site located between the polypeptide-encoding
sequence and the heterologous protein sequence, so that the polypeptide may be
cleaved
and purified away from the heterologous moiety.
Sequences encoding a desired polypeptide may be synthesized, in whole
or in part, using chemical methods well known in the art (see Caruthers, M. H.
et al.
(1980) Nucl. Acids Res. Syynp. Ser. 215-223, Horn, T. et al. (1980) Nucl.
Acids Res.
Syrup. Ser. 225-232). Alternatively, the protein itself may be produced using
chemical
methods to synthesize the amino acid sequence of a polypeptide, or a portion
thereof.
Fox example, peptide synthesis can be performed using various solid-phase
techniques
(Roberge, J. Y. et al. (1995) Science 269:202-204) and automated synthesis may
be
achieved, for example, using the ABI 431 A Peptide Synthesizer (Perkin Elmer,
Palo
Alto, CA).
A newly synthesized peptide may be substantially purified by preparative
high performance liquid chromatography (e.g., Creighton, T. (1983) Proteins,
Structures
and Molecular Principles, WH Freeman and Co., New York, N.Y.) or other
comparable
techniques available in the art. The composition of the synthetic peptides may
be
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
74
confirmed by amino acid analysis or sequencing (e.g., the Edman degradation
procedure). Additionally, the amino acid sequence of a polypeptide, or any
part thereof,
may be altered during direct synthesis and/or combined using chemical methods
with
sequences from other proteins, or any part thereof, to produce a variant
polypeptide.
In order to express a desired polypeptide, the nucleotide sequences
encoding the polypeptide, or functional equivalents, may be inserted into
appropriate
expression vector, i.e., a vector which contains the necessary elements for
the
transcription and translation of the inserted coding sequence. Methods which
are well
known to those skilled in the art may be used to construct expression vectors
containing
sequences encoding a polypeptide of interest and appropriate transcriptional
and
translational control elements. These methods include in vitro recombinant DNA
techniques, synthetic techniques, and in vivo genetic recombination. Such
techniques
are described, for example, in Sambrook, J. et al. (1989) Molecular Cloning, A
Laboratory Manual, Cold Spring Harbor Press, Plainview, N.Y., and Ausubel, F.
M. et
al. (1989) Current Protocols in Molecular Biology, John Wiley & Sons, New
York.
N.Y.
A variety of expression vector/host systems may be utilized to contain
and express polynucleotide sequences. These include, but are not limited to,
microorganisms such as bacteria transformed with recombinant bacteriophage,
plasmid,
or cosmid DNA expression vectors; yeast transformed with yeast expression
vectors;
insect cell systems infected with virus expression vectors (e.g.,
baculovirus); plant cell
systems transformed with virus expression vectors (e.g., cauliflower mosaic
virus,
CaMV; tobacco mosaic virus, TMV) or with bacterial expression vectors (e.g.,
Ti or
pBR322 plasmids); or animal cell systems.
The "control elements" or "regulatory sequences" present in an
expression vector are those non-translated regions of the vector--enhancers,
promoters,
5' and 3' untranslated regions--which interact with host cellular proteins to
carry out
transcription and translation. Such elements may vary in their strength and
specificity.
Depending on the vector system and host utilized, any number of suitable
transcription
and translation elements, including constitutive and inducible promoters, may
be used.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
Fox example, when cloning in bacterial systems, inducible promoters such as
the hybrid
lacZ promoter of the PBLUESCRIPT phagemid (Stratagene, La Jolla, Calif.) or
PSPORT1 plasmid (Gibco BRL, Gaithersburg, MD) and the like may be used. In
mammalian cell systems, promoters from mammalian genes or from mammalian
viruses
5 are generally preferred. If it is necessary to generate a cell line that
contains multiple
copies of the sequence encoding a polypeptide, vectors based on SV40 or EBV
rnay be
advantageously used with an appropriate selectable marker.
In bacterial systems, any of a number of expression vectors may be
selected depending upon the use intended for the expressed polypeptide. For
example,
10 when large quantities are needed, for example for the induction of
antibodies, vectors
which direct high level expression of fusion proteins that are readily
purified may be
used. Such vectors include, but are not limited to, the multifunctional E,
coli cloning
and expression vectors such as BLUESCRIPT (Stratagene), in which the sequence
encoding the polypeptide of interest may be Iigated into the vector in frame
with
15 sequences for the amino-terminal Met and the subsequent 7 residues of
.beta.-
galactosidase so that a hybrid protein is produced; pIN vectors (Van Heeke, G.
and S.
M. Schuster (I989) J. Biol. Chem. 264:5503-5509); and the like. pGEX Vectors
(Promega, Madison, Wis.) may also be used to express foreign polypeptides as
fusion
proteins with glutathione S-transferase (GST). In general, such fusion
proteins are
20 soluble and can easily be purified from lysed cells by adsorption to
glutathione-agarose
beads followed by elution in the presence of free glutathione. Proteins made
in such
systems may be designed to include heparin, thrombin, or factor XA protease
cleavage
sites so that the cloned polypeptide of interest can be released from the GST
moiety at
will.
25 In the yeast, Saccharomyces cerevisiae, a number of vectors containing
constitutive or inducible promoters such as alpha factor, alcohol oxidase, and
PGH may
be used. For reviews, see Ausubel et al. (supra) and Grant et al. (1987)
Methods
Enzymol. 153:516-544.
In cases where plant expression vectors are used, the expression of
30 sequences encoding polypeptides may be driven by any of a number of
promoters. For
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
76
example, viral promoters such as the 35S and 19S promoters of CaMV may be used
alone or in combination with the omega leader sequence from TMV (Takamatsu, N.
(1987) EMBO J. 6:307-311. Alternatively, plant promoters such as the small
subunit of
RUBISCO or heat shock promoters may be used (Coruzzi, G. et al. (1984) EMBO J.
3:1671-1680; Brogue, R. et al. (1984) Science 224:838-843; and Winter, J. et
al. (1991)
Results Probl. Cell Differ. 17:85-105). These constructs can be introduced
into plant
cells by direct DNA transformation or pathogen-mediated transfection. Such
techniques
are described in a number of generally available reviews (see, for example,
Hobbs, S. or
Murry, L. E. in McGraw Hill Yearbook of Science and Technology (1992) McGraw
Hill, New York, N.Y.; pp. 191-196).
An insect system may also be used to express a polypeptide of interest.
For example, in one such system, Autographa californica nuclear polyhedrosis
virus
(AcNPV) is used as a vector to express foreign genes in Spodoptera frugiperda
cells or
in Trichoplusia larvae. The sequences encoding the polypeptide may be cloned
into a
non-essential region of the virus, such as the polyhedrin gene, and placed
under control
of the polyhedrin promoter. Successful insertion of the polypeptide-encoding
sequence
will render the polyhedrin gene inactive and produce recombinant virus lacking
coat
protein. The recombinant viruses may then be used to infect, for example, S.
frugiperda
cells or Trichoplusia larvae in which the polypeptide of interest may be
expressed
(Engelhard, E. K. et al. (1994) Proc. Natl. Acad. Sci. 91 :3224-3227).
In mammalian host cells, a number of viral-based expression systems are
generally available. For example, in cases where an adenovirus is used as an
expression
vector, sequences encoding a polypeptide of interest may be ligated into an
adenovirus
transcriptionltranslation complex consisting of the late promoter and
tripartite leader
sequence. Insertion in a non-essential El or E3 region of the viral genome may
be used
to obtain a viable virus which is capable of expressing the polypeptide in
infected host
cells (Logan, J. and Shenk, T. (1984) P~oc. Natl. Acad. Sci. 81:3655-3659). In
addition,
transcription enhancers, such as the Rous sarcoma virus (RSV) enhancer, may be
used
to increase expression in mammalian host cells.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
77
Specific initiation signals may also be used to achieve more efficient
translation of sequences encoding a polypeptide of interest. Such signals
include the
ATG initiation codon and adjacent sequences. In cases where sequences encoding
the
polypeptide, its initiation codon, and upstream sequences are~~ inserted into
the
appropriate expression vector, no additional transcriptional or translational
control
signals may be needed. Howevex, in cases where only coding sequence, or a
portion
thereof, is inserted, exogenous translational control signals including the
ATG initiation
codon should be provided. Furthermore, the initiation codon should be in the
correct
reading frame to ensure translation of the entire insert. Exogenous
translational
elements and initiation codons may be of various origins, both natural and
synthetic.
The efficiency of expression may be enhanced by the inclusion of enhancers
which are
appropriate for the particular cell system which is used, such as those
described in the
literature (Scharf, D. et al. (1994) Results Probl. Cell Differ. 20:125-162).
In addition, a host cell strain may be chosen for its ability to modulate
IS the expression of the inserted sequences or to process the expressed
protein in the
desired fashion. Such modifications of the polypeptide include, but are not
limited to,
acetylation, carboxylation. glycosylation, phosphorylation, lipidation, and
acylation.
Post-translational processing which cleaves a "prepro" form of the protein may
also be
used to facilitate correct insertion, folding and/or function. Different host
cells such as
CHO, COS, HeLa, MDCK, HEK293, and WI38, which have specific cellular machinery
and characteristic mechanisms for such post-translational activities, may be
chosen to
ensure the correct modification and processing of the foreign protein.
For long-term, high-yield production of recombinant proteins, stable
expression is generally preferred. For example, cell lines which stably
express a
polynucleotide of interest may be transformed using expression vectors which
may
contain viral origins of replication andlor endogenous expression elements and
a
selectable marker gene on the same or on a separate vector. Following the
introduction
of the vector, cells may be allowed to grow for 1-2 days in an enriched media
before
they are switched to selective media. The purpose of the selectable marker is
to confer
resistance to selection, and its presence allows growth and recovery of cells
which
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
78
successfully express the introduced sequences. Resistant clones of stably
transformed
cells may be proliferated using tissue culture techniques appropriate to the
cell type.
Any number of selection systems may be used to recover transformed
cell lines. These include, but are not limited to, the herpes simplex virus
thymidine
kinase (Wigler, M. et al. (1977) Cell 11:223-32) and adenine
phosphoribosyltransferase
(Lowy, I. et al. (1990) Cell 22:817-23) genes which can be employed in tk-
or
aprt- cells, respectively. Also, antimetabolite, antibiotic or herbicide
resistance can
be used as the basis for selection; for example, dhfr which confers resistance
to
methotrexate (Wigler, M. et al. (1980) Proc. Natl. Acad. Sci. 77:3567-70);
npt, which
confers resistance to the aminoglycosides, neomycin and G-418 (Colbere-
Garapin, F. et
al (1981) J. Mol. Biol. 150:1-14); and als or pat, which confer resistance to
chlorsulfuron and phosphinotricin acetyltransferase, respectively (Marry,
supra).
Additional selectable genes have been described, for example, trpB, which
allows cells
to utilize indole in place of tryptophan, or hisD, which allows cells to
utilize histinol in
place of histidine (Hartman, S. C. and R. C. Mulligan (1988) Py~oc. Natl.
Acad. Sci.
85:8047-51 ). The use of visible markers has gained popularity with such
markers as
anthocyanins, beta-glucuronidase and its substrate GUS, and Iuciferase and its
substrate
luciferin, being widely used not only to identify transformants, but also to
quantify the
amount of transient or stable protein expression attributable to a specific
vector system
(Rhodes, C. A. et al. (1995) Methods Mol. Biol. 55:121-131).
Although the presence/absence of marker gene expression suggests that
the gene of interest is also present, its presence and expression may need to
be
confirmed. For example, if the sequence encoding a polypeptide is inserted
within a
marker gene sequence, recombinant cells containing sequences can be identified
by the
absence of marker gene function. Alternatively, a marker gene can be placed in
tandem
with a polypeptide-encoding sequence under the control of a single promoter.
Expression of the marker gene in response to induction or selection usually
indicates
expression of the tandem gene as well.
Alternatively, host cells that contain and express a desired
polynucleotide sequence may be identified by a variety of procedures known to
those of
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
79
skill in the art. These procedures include, but are not limited to, DNA-DNA or
DNA-
RNA hybridizations and protein bioassay or immunoassay techniques which
include,
for example, membrane, solution, or chip based technologies for the detection
and/or
quantification of nucleic acid or protein.
A variety of protocols for detecting and measuring the expression of
polynucleotide-encoded products, using either polyclonal or monoclonal
antibodies
specific for the product are known in the art. Examples include enzyme-linked
immunosorbent assay (ELISA), radioimmunoassay (RIA), and fluorescence
activated
cell sorting (FACS). A two-site, monoclonal-based immunoassay utilizing
monoclonal
antibodies reactive to two non-interfering epitopes on a given polypeptide may
be
preferred for some applications, but a competitive binding assay may also be
employed.
These and other assays are described, among other places, in Hampton, R. et
al. (1990;
Serological Methods, a Laboratory Manual, APS Press, St Paul. Minn.) and
Maddox, D.
E. et al. (1983; J. Exp. Med. 158:1211-1216).
A wide variety of labels and conjugation techniques are known by those
skilled in the art and may be used in various nucleic acid and amino acid
assays. Means
for producing labeled hybridization or PCR probes for detecting sequences
related to
polynucleotides include oligolabeling, nick translation, end-labeling or PCR
amplification using a labeled nucleotide. Alternatively, the sequences, or any
portions
thereof may be cloned into a vector for the production of an mRNA probe. Such
vectors
are known in the art, axe commercially available, and may be used to
synthesize RNA
probes in vitro by addition of an appropriate RNA polymerase such as T7, T3,
or SP6
and labeled nucleotides. These procedures may be ' conducted using a variety
of
commercially available kits. Suitable reporter molecules or labels, which may
be used
include radionuclides, enzymes, fluorescent, chemiluminescent, or chromogenic
agents
as well as substrates, cofactors, inhibitors, magnetic particles, and the
like.
Host cells transformed with a polynucleotide sequence of interest may be
cultured under conditions suitable for the expression and recovery of the
protein from
cell culture. The protein produced by a recombinant cell may be secreted or
contained
intracellularly depending on the sequence and/or the vector used. As will be
understood
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
by those of skill in the art, expression vectors containing polynucleotides of
the
invention may be designed to contain signal sequences which direct secretion
of the
encoded polypeptide through a prokaryotic or eukaryotic cell membrane. Other
recombinant constructions may be used to join sequences encoding a polypeptide
of
5 interest to nucleotide sequence encoding a polypeptide domain which will
facilitate
I
purification of soluble proteins. Such purif cation facilitating domains
include, but are
not limited to, metal chelating peptides such as histidine-tryptophan modules
that allow
purification on immobilized metals, protein A domains that allow purification
on
immobilized immunoglobulin, and the domain utilized in the FLAGS
extension/affinity
10 purification system (Immunex Corp., Seattle, Wash.). The inclusion of
cleavable linker
sequences such as those specific for Factor XA or enterokinase (Invitrogen.
San Diego,
Calif.) between the purification domain and the encoded polypeptide may be
used to
facilitate purification. One such expression vector provides for expression of
a fusion
protein containing a polypeptide of interest and a nucleic acid encoding 6
histidine
15 residues preceding a thioredoxin or an enterokinase cleavage site. The
histidine residues
facilitate purification on IMIAC (immobilized metal ion affinity
chromatography) as
described in Porath, J. et al. (1992, Prot. Exp. Purif. 3:263-281) while the
enterokinase
cleavage site provides a means for purifying the desired polypeptide from the
fusion
protein. A discussion of vectors which contain fusion proteins is provided in
Kroll, D. J.
20 et aI. (1993; DNA Cell Biol. 12:441-453).
In addition to recombinant production methods, polypeptides of the
invention, and fragments thereof, may be produced by direct peptide synthesis
using
solid-phase techniques (Merrifield J. (1963) J. Am. Chem. Soc. 85:2149-2154).
Protein
synthesis may be performed using manual techniques or by automation. Automated
25 synthesis may be achieved, for example, using Applied Biosystems 431A
Peptide
Synthesizer (Perkin Elmer). Alternatively, various fragments may be chemically
synthesized separately and combined using chemical methods to produce the full
length
molecule.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
81
Antibod Compositions, Fragments Thereof and Other Binding Agents
According to another aspect, the present invention further provides
binding agents, such as antibodies and antigen-binding fragments thereof, that
exhibit
immunological binding to a tumor polypeptide disclosed herein, or to a
portion, variant
or derivative thereof. An antibody, or antigen-binding fragment thereof, is
said to
"specifically bind," "immunogically bind," and/or is "immunologically
reactive" to a
polypeptide of the invention if it reacts at a detectable level (within, for
example, an
ELISA assay) with the polypeptide, and does not react detestably with
unrelated
polypeptides under similar conditions.
Immunological binding, as used in this context, generally refers to the
non-covalent interactions of the type which occur between an immunoglobulin
molecule and an antigen for which the immunoglobulin is specific. The
strength, or
affinity of immunological binding interactions can be expressed in terms of
the
dissociation constant (Kd) of the interaction, wherein a smaller Kd represents
a greater
affinity. Immunological binding properties of selected polypeptides can be
quantified
using methods well known in the art. One such method entails measuring the
rates of
antigen-binding site/antigen complex formation and dissociation, wherein those
rates
depend on the concentrations of the complex partners, the affinity of the
interaction, and
on geometric parameters that equally influence the rate in both directions.
Thus, both
the "on rate constant" (Ko") and the "off rate constant" (Ko~) can be
determined by
calculation of the concentrations and the actual rates of association and
dissociation.
The ratio of Koff lKon enables cancellation of all parameters not related to
affinity, and is
thus equal to the dissociation constant Kd. See, generally, Davies et al.
(1990) Annual
Rev. Biochem. 59:439-473.
An "antigen-binding site," or "binding portion" of an antibody refers to
the part of the immunoglobulin molecule that participates in antigen binding.
The
antigen binding site is formed by amino acid residues of the N-terminal
variable ("V")
regions of the heavy ("H") and light ("L") chains. Three highly divergent
stretches
within the V regions of the heavy and light chains are referred to as
"hypervariable
regions" which are interposed between more conserved flanking stretches known
as
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
82
"framework regions," or "FRs". Thus the term "FR" refers to amino acid
sequences
which axe naturally found between and adjacent to hypervariable regions in
immunoglobulins. In an antibody molecule, the three hypervariable regions, of
a light
chain and the three hypervariable regions of a heavy chain axe disposed
relative to each
S other in three dimensional space to form an antigen-binding surface. The
antigen-
binding surface is complementary to the three-dimensional surface of a bound
antigen,
and the three hypervariable regions of each of the heavy and light chains are
referred to
as "complementarity-determining regions," or "CDRs."
Binding agents may be further capable of differentiating between patients
with and without a cancer, such as prostate cancer, using the representative
assays
provided herein. For example, antibodies or other binding agents that bind to
a tumor
protein will preferably generate a signal indicating the presence of a cancer
in at least
about 20% of patients with the disease, more preferably at least about 30% of
patients.
Alternatively, or in addition, the antibody will generate a negative signal
indicating the
absence of the disease in at least about 90% of individuals without the
cancer. To
determine whether a binding agent satisfies this requirement, biological
samples (e.g.,
blood, sera, sputum, urine and/or tumor biopsies) from patients with and
without a
cancer (as determined using standard clinical tests) may be assayed as
described herein
for the presence of polypeptides that bind to the binding agent. Preferably, a
statistically
significant number of samples with and without the disease will be assayed.
Each
binding agent should satisfy the above criteria; however, those of ordinary
skill in the
art will recognize that binding agents may be used in combination to improve
sensitivity.
Any agent that satisfies the above requirements may be a binding agent.
For example, a binding agent may be a ribosome, with or without a peptide
component,
an RNA molecule or a polypeptide. In a preferred embodiment, a binding agent
is an
antibody or an antigen-binding fragment thereof. Antibodies may be prepared by
any of
a variety' of techniques known to those of ordinary skill in the art. See,
e.g., Harlow and
Lane, Antibodies: A Labof~atory Manual, Cold Spring Harbor Laboratory, 1988.
In
general, antibodies can be produced by cell culture techniques, including the
generation
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
83
of monoclonal antibodies as described herein, or via transfection of antibody
genes into
suitable bacterial or mammalian cell hosts, in order to allow for the
production of
recombinant antibodies. In one technique, an immunogen comprising the
polypeptide is
initially injected into any of a wide variety of mammals (e.g., mice, rats,
rabbits, sheep
or goats). In this step, the polypeptides of this invention may serve as the
immunogen
without modification. Alternatively, particularly for relatively short
polypeptides, a
superior immune response may be elicited if the polypeptide is joined to a
carrier
protein, such as bovine serum albumin or keyhole limpet hemocyanin. The
immunogen
is injected into the animal host, preferably according to a predetermined
schedule
incorporating one or more booster immunizations, and the animals are bled
periodically.
Polyclonal antibodies specific for the polypeptide may then be purified from
such
antisera by, for example, affinity chromatography using the polypeptide
coupled to a
suitable solid support.
Monoclonal antibodies specific for an antigenic polypeptide of interest
may be prepared, for example, using the technique of Kohler and Milstein, Eur.
,I.
Immunol. 6:511-519, 1976, and improvements thereto. Briefly, these methods
involve
the preparation of immortal cell lines capable of producing antibodies having
the
desired specificity (i.e., reactivity with the polypeptide of interest). Such
cell lines may
be produced, for example, from spleen cells obtained from an animal immunized
as
described above. The spleen cells are then immortalized by, for example,
fusion with a
myeloma cell fusion partner, preferably one that is syngeneic with the
immunized
animal. A variety of fusion techniques may be employed. For example, the
spleen cells
and myeloma cells may be combined with a nonionic detergent for a few minutes
and
then plated at low density on a selective medium that supports the growth of
hybrid
cells, but not myeloma cells. A preferred selection technique uses HAT
(hypoxanthine,
aminopterin, thymidine) selection. After a sufficient time, usually about 1 to
2 weeks,
colonies of hybrids are observed. Single colonies are selected and their
culture
supernatants tested for binding activity against the polypeptide. Hybridomas
having
high reactivity and specif city axe preferred.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
84
Monoclonal antibodies may be isolated from the supernatants of growing
hybridoma colonies. In addition, various techniques may be employed to enhance
the
yield, such as injection of the hybridoma cell line into the peritoneal cavity
of a suitable
vertebrate host, such as a mouse. Monoclonal antibodies may then be harvested
from
the ascites fluid or the blood. Contaminants may be removed from the
antibodies by
conventional techniques, such as chromatography, gel filtration,
precipitation, and
extraction. The polypeptides of this invention may be used in the purification
process
in, for example, an affinity chromatography step.
A number of therapeutically useful molecules are known in the art which
comprise antigen-binding sites that are capable of exhibiting immunological
binding
properties of an antibody molecule. The proteolytic enzyme papain
preferentially
cleaves IgG molecules to yield several fragments, two of which (the "F(ab)"
fragments)
each comprise a covalent heterodimer that includes an intact antigen-binding
site. The
enzyme pepsin is able to cleave IgG molecules to provide several fragments,
including
I S the "F(ab')a " fragment which comprises both antigen-binding sites. An
"Fv" fragment
can be produced by preferential proteolytic cleavage of an IgM, and on rare
occasions
IgG or IgA immunoglobulin molecule. Fv fragments are, however, more commonly
derived using recombinant techniques known in the art. The Fv fragment
includes a
non-covalent VH::Vz heterodimer including an antigen-binding site which
retains much
of the antigen recognition and binding capabilities of the native antibody
molecule.
mbar et al. (1972) Proc. Nat. Acad. Sci. USA 69:2659-2662; Hochman et al.
(1976)
Biochem 15:2706-2710; and Ehrlich et al. (1980) Biochem 19:4091-4096.
A single chain Fv ("sFv") polypeptide is a covalently linked VH::VL
heterodimer which is expressed from a gene fusion including VH- and VL-
encoding
genes linked by a peptide-encoding linker. Huston et al. (1988) Proc. Nat.
Acad. Sci.
USA 85(16):5879-5883. A number of methods have been described to discern
chemical
structures for converting the naturally aggregated--but chemically separated--
light and
heavy polypeptide chains from an antibody V region into an sFv molecule which
will
fold into a three dimensional structure substantially similar to the structure
bf an
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
antigen-binding site. See, e.g., U.S. Pat. Nos. 5,091,513 and 5,132,405, to
Huston et al.;
and U.S. Pat. No. 4,946,778, to Ladner et al.
Each of the above-described molecules includes a heavy chain and a
light chain CDR set, respectively interposed between a heavy chain and a light
chain FR
5 set which provide support to the CDRS and define the spatial relationship of
the CDRs
relative to each other. As used herein, the term "CDR set" refers to the three
hypervariable regions of a heavy or light chain V region. Proceeding from the
N-
terminus of a heavy or light chain, these regions are denoted as "CDRl,"
"CDR2," and
"CDR3" respectively. An antigen-binding site, therefore, includes six CDRs,
10 comprising the CDR set from each of a heavy and a light chain V region. A
polypeptide
comprising a single CDR, (e.g., a CDRl, CDR2 or CDR3) is referred to herein as
a
"molecular recognition unit." Crystallographic analysis of a number of antigen-
antibody
complexes has demonstrated that the amino acid residues of CDRs form extensive
contact with bound antigen, wherein the most extensive antigen contact is with
the
15 heavy chain CDR3. Thus, the molecular recognition units are primarily
responsible for
the specificity of an antigen-binding site.
As used herein, the term "FR set" refers to the four flanking amino acid
sequences which frame the CDRs of a CDR set of a heavy or light chain V
region.
Some FR residues may contact bound antigen; however, FRs are primarily
responsible
20 for folding the V region into the antigen-binding site, particularly the FR
residues
directly adjacent to the CDRS. Within FRs, certain amino residues and certain
structural
features are very highly conserved. In this regard, all V region sequences
contain an
internal disulfide loop of axound 90 amino acid residues. When the V regions
fold into a
binding-site, the CDRs axe displayed as projecting loop motifs which form an
antigen-
25 binding surface. It is generally recognized that there are conserved
structural regions of
FRs which influence the folded shape of the CDR loops into certain "canonical"
structures--regardless of the precise CDR amino acid sequence. Further,
certain FR
residues are known to participate in non-covalent interdomain contacts which
stabilize
the interaction of the antibody heavy and Light chains.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
86
A number of "humanized" antibody molecules comprising an antigen-
binding site derived from a non-human immunoglobulin have been described,
including
chimeric antibodies having rodent V regions and their associated CDRs fused to
human
constant domains (Winter et al. (1991) Nature 349:293-299; Lobuglio et al.
(1989)
Proc. Nat. Acad. Sci. USA 86:4220-4224; Shaw et al. (1987) J Immunol. 138:4534-
4538; and Brown et al. (1987) Cancer Res. 47:3577-3583), rodent CDRs grafted
into a
human supporting FR prior to fusion with an appropriate human antibody
constant
domain (Riechmann et al. (1988) Nature 332:323-327; Verhoeyen et al. (1988)
Science
239:1534-1536; and Jones et al. (1986) Nature 321:522-525), and rodent CDRs
supported by recombinantly veneered rodent FRs (European Patent Publication
No.
519,596, published Dec. 23, 1992). These "humanized" molecules are designed to
minimize unwanted immunological response toward rodent antihuman antibody
molecules which limits the duration and effectiveness of therapeutic
applications of
those moieties in human recipients.
As used herein, the terms "veneered FRs" and "recombinantly veneered
FRs" refer to the selective replacement of FR residues from, e.g., a rodent
heavy or light
chain V region, with human FR residues in order to provide a xenogeneic
molecule
comprising an antigen-binding site which retains substantially all of the
native FR
polypeptide folding structure. Veneering techniques are based on the
understanding that
the ligand binding characteristics of an antigen-binding site are determined
primarily by
the structure and relative disposition of the heavy and light chain CDR sets
within the
antigen-binding surface. Davies et al. (1990) Ann. Rev. Biochem. 59:439-473.
Thus,
antigen binding specificity can be preserved in a humanized antibody only
wherein the
CDR structures, their interaction with each other, and their interaction with
the rest of
the V region domains are carefully maintained. By using veneering techniques,
exterior
(e.g., solvent-accessible) FR residues which are readily encountered by the
immune
system are selectively replaced with human residues to provide a hybrid
molecule that
comprises either a weakly immunogenic, or substantially non-irnmunogenic
veneered
surface.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
87
The process of veneering makes use of the available sequence data for
human antibody variable domains compiled by Kabat et al., in Sequences of
Proteins of
Immunological Interest, 4th ed., (U.S. Dept. of Health and Human Services,
U.S.
Government Printing Office, 1987), updates to the Kabat database, and other
accessible
U.S. and foreign databases (both nucleic acid and protein). Solvent
accessibilities of V
region amino acids can be deduced from the known three-dimensional structure
for
human and marine antibody fragments. There are two general steps in veneering
a
marine antigen-binding site. Initially, the FRs of the variable domains of an
antibody
molecule of interest are compared with corresponding FR sequences of human
variable
domains obtained from the above-identified sources. The most homologous human
V
regions are then compared residue by residue to corresponding marine amino
acids. The
residues in the marine FR which differ from the human counterpart are replaced
by the
residues present in the human moiety using recombinant techniques well known
in the
art. Residue switching is only carried out with moieties which are at least
partially
exposed (solvent accessible), and care is exercised in the replacement of
amino acid
residues which may have a significant effect on the tertiary structure of V
region
domains, such as proline, glycine and charged amino acids.
In this manner, the resultant "veneered" marine antigen-binding sites are
thus designed to retain the marine CDR residues, the residues substantially
adjacent to
the CDRs, the residues identified as buried or mostly buried (solvent
inaccessible), the
residues believed to participate in non-covalent (e.g., electrostatic and
hydrophobic)
contacts between heavy and light chain domains, and the residues from
conserved
structural regions of the FRs which are believed to influence the "canonical"
tertiary
structures of the CDR loops. These design criteria are then used to prepare
recombinant
nucleotide sequences which combine the CDRs of both the heavy and light chain
of a
marine antigen-binding site into human-appearing FRs that can be used to
transfect
mammalian cells for the expression of recombinant human antibodies which
exhibit the
antigen specificity of the marine antibody molecule.
In another embodiment of the invention, monoclonal antibodies of the
present invention may be coupled to one or more therapeutic agents. Suitable
agents in
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
88
this regard include radionuclides, differentiation inducers, drugs, toxins,
and derivatives
thereof. Preferred radionuclides include 9oY, ~23h Lash i3~I~ ls6Re, 188Re,
2nAt, and
z~2Bi. preferred drugs include methotrexate, and pyrimidine and purine
analogs.
Preferred differentiation inducers include phorbol esters and butyric acid.
Preferred
toxins include ricin, abrin, diptheria toxin, cholera toxin, gelonin,
Pseudomonas
exotoxin, Shigella toxin, and pokeweed a~tiviral protein.
A therapeutic agent may be coupled (e.g., covalently bonded) to a
suitable monoclonal antibody either directly or indirectly (e.g., via a linker
group). A
direct reaction between an agent and an antibody is possible when each
possesses a
substituent capable of reacting with the other. For example, a nucleophilic
group, such
as an amino or sulfhydryl group, on one may be capable of reacting with a
carbonyl-
containing group, such as an anhydride or an acid halide, or with an alkyl
group
containing a good leaving group (e.g., a halide) on the other.
Alternatively, it may be desirable to couple a therapeutic agent and an
antibody via a linker group. A linker group can function as a spacer to
distance an
antibody from an agent in order to avoid interference with binding
capabilities. A linker
group can also serve to increase the chemical reactivity of a substituent on
an agent or
an antibody, and thus increase the coupling efficiency. An increase in
chemical
reactivity may also facilitate the use of agents, or functional groups on
agents, which
otherwise would not be possible.
It will be evident to those skilled in the art that a variety of bifunctional
or polyfunctional reagents, both homo- and hetero-functional (such as those
described in
the catalog of the Pierce Chemical Co., Rockford, IL), may be employed as the
linker
group. Coupling may be effected, for example, through amino groups, carboxyl
groups,
sulfliydryl groups or oxidized carbohydrate residues. There are numerous
references
describing such methodology, e.g., U.S. Patent No. 4,671,958, to Rodwell et
al.
Where a therapeutic agent is more potent when free from the antibody
portion of the immunoconjugates of the present invention, it may be desirable
to use a
linker group which is cleavable during or upon internalization into a cell. A
number of
different cleavable linker groups have been described. The mechanisms for the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
89
intracellular release of an agent from these linker groups include cleavage by
reduction
of a disulfide bond (e.g., U.S. Patent No. 4,489,710, to Spider), by
irradiation of a
photolabile bond (e.g., U.S. Patent No. 4,625,014, to Senter et al.), by
hydrolysis of
derivatized amino acid side chains (e.g., U.S. Patent No. 4,638,045, to Kohn
et al.), by
serum complement-mediated hydrolysis (e.g., U.S. Patent No. 4,67I,9S8, to
Rodwell
et al.), and acid-catalyzed hydrolysis (e.g., U.S. Patent No. 4,569,789, to
Blattler et al.).
It may be desirable to couple more than one agent to an antibody. In one
embodiment, multiple molecules of an agent are coupled to one antibody
molecule. In
another embodiment, more than one type of agent may be coupled to one
antibody.
Regardless of the particular embodiment, immunoconjugates with more than one
agent
may be prepared in a variety of ways. For example, more than one agent may be
coupled directly to an antibody molecule, or linkers that provide multiple
sites for
attachment can be used. Alternatively, a carrier can be used.
A carrier may bear the agents in a variety of ways, including covalent
bonding either directly or via a linker group. Suitable carriers include
proteins such as
albumins (e.g., U.S. Patent No. 4,507,234, to Kato et al.), peptides and
polysaccharides
such as aminodextran (e.g., U.S. Patent No. 4,699,784, to Shih et al.). A
carrier may
also bear an agent by noncovalent bonding or by encapsulation, such as within
a
liposome vesicle (e.g., U.S. Patent Nos. 4,429,008 and 4,873,088). Carriers
specific for
radionuclide agents include radiohalogenated small molecules and chelating
compounds. For example, U.S. Patent No. 4,735,792 discloses representative
radiohalogenated small molecules and their synthesis. A radionuclide chelate
may be
formed from chelating compounds that include those containing nitrogen and
sulfur
atoms as the donor atoms for binding the metal, or metal oxide, radionuclide.
For
example, U.S. Patent No. 4,673,562, to Davison et al. discloses representative
chelating
compounds and their synthesis.
T Cell Compositions
The present invention, in another aspect, provides T cells specific for a
tumor polypeptide disclosed herein, or for a variant or derivative thereof.
Such cells
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
may generally be prepared in vitro or ex vivo, using standard procedures. For
example,
T cells may be isolated from bone marrow, peripheral blood, or a fraction of
bone
marrow or peripheral blood of a patient, using a commercially available cell
separation
system, such as the IsolexTM System, available from Nexell Therapeutics, Inc.
(Irvine,
5 CA; see also U.S. Patent No. 5,240,856; U.S. Patent No. 5,215,926; WO
89/06280; WO
91/16116 and WO 92/07243). Alternatively, T cells may be derived from related
or
unrelated humans, non-human mammals, cell lines or cultures.
T cells may be stimulated with a polypeptide, polynucleotide encoding a
polypeptide and/or an antigen presenting cell (APC) that expresses such a
polypeptide.
10 Such stimulation is performed under conditions and for a time sufficient to
permit the
generation of T cells that are specific for the polypeptide of interest.
Preferably, a tumor
polypeptide or polynucleotide of the invention is present within a delivery
vehicle, such
as a microsphere, to facilitate the generation of specific T cells.
T cells are considered to be specific for a polypeptide of the present
I5 invention if the T cells specifically proliferate, secrete cytokines or
kill target cells
coated with the polypeptide or expressing a gene encoding the polypeptide. T
cell
specificity may be evaluated using any of a variety of standard techniques.
For
example, within a chromium release assay or proliferation assay, a stimulation
index of
more than two fold increase in lysis and/or proliferation, compared to
negative controls,
20 indicates T cell specificity. Such assays may be performed, for example, as
described in
Chen et al., Cahce~ Res. 54:1065-1070, 1994. Alternatively, detection of the
proliferation of T cells may be accomplished by a variety of known techniques.
For
example, T cell proliferation can be detected by measuring an increased rate
of DNA
synthesis (e.g., by pulse-labeling cultures of T cells with tritiated
thymidine and
25 measuring the amount of tritiated thymidine incorporated into DNA). Contact
with a
tumor polypeptide (100 ng/ml - 100 ~glml, preferably 200 ng/ml - 25 ~g/ml) for
3 - 7
days will typically result in at least a two fold increase in proliferation of
the T cells.
Contact as described above for 2-3 hours should result in activation of the T
cells, as
measured using standard cytokine assays in which a two fold increase in the
level of
30 cytokine release (e.g., TNF or IFN y) is indicative of T cell activation
(see Coligan et
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
91
al., Current Protocols in Immunology, vol. 1, Wiley Interscience (Greene
1998)). T
cells that have been activated in response to a tumor polypeptide,
polynucleotide or
polypeptide-expressing APC may be CD4+ and/or CD8+. Tumor polypeptide-specific
T
cells may be expanded using standard techniques. Within preferred embodiments,
the T
cells are derived from a patient, a related donor or an unrelated donor, and
are
administered to the patient following stimulation and expansion.
For therapeutic purposes, CD4+ or CD8+ T cells that proliferate in
response to a tumor polypeptide, polynucleotide or APC can be expanded in
number
either ih vitf°o or i~ vivo. Proliferation of such T cells in vitro may
be accomplished in a
I O variety of ways. For example, the T cells can be re-exposed to a tumor
polypeptide, or a
short peptide corresponding to an immunogenic portion of such a polypeptide,
with or
without the addition of T cell growth factors, such as interleukin-2, and/or
stimulator
cells that synthesize a tumor polypeptide. Alternatively, one or more T cells
that
proliferate in the presence of the tumor polypeptide can be expanded in number
by
cloning. Methods for cloning cells are well known in the art, and include
limiting
dilution.
Pharmaceutical Compositions
In additional embodiments, the present invention concerns formulation
of one or more of the polynucleotide, polypeptide, T-cell and/or antibody
compositions
disclosed herein in pharmaceutically-acceptable carriers for administration to
a cell or
an animal, either alone, or in combination with one or more other modalities
of therapy.
It will be understood that, if desired, a composition as disclosed herein
may be administered in combination with other agents as well, such as, e.g.,
other
proteins or polypeptides or various pharmaceutically-active agents. In fact,
there is
virtually no limit to other components that may also be included, given that
the
additional agents do not cause a significant adverse effect upon contact with
the target
cells or host tissues. The compositions may thus be delivered along with
various other
agents as required in the particular instance. Such compositions may be
purified from
host cells or other biological sources, or alternatively may be chemically
synthesized as
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
92
described herein. Likewise, such compositions may further comprise substituted
or
derivatized RNA or DNA compositions.
Therefore, in another aspect of the present invention, pharmaceutical
compositions are provided comprising one or more of the polynucleotide,
polypeptide,
antibody, and/or T-cell compositions described herein in combination with a
physiologically acceptable carrier. In certain preferred embodiments, the
pharmaceutical compositions of the invention comprise immunogenic
polynucleotide
and/or polypeptide compositions of the invention for use in prophylactic and
theraputic
vaccine applications. Vaccine preparation is generally described in, for
example, M.F.
Powell and M.J. Newman, eds., "Vaccine Design (the subunit and adjuvant
approach),"
Plenum Press (NY, 1995). Generally, such compositions will comprise one or
more
polynucleotide and/or polypeptide compositions of the present invention in
combination
with one or more immunostimulants.
It will be apparent that any of the pharmaceutical compositions described
herein can contain pharmaceutically acceptable salts of the polynucleotides
and
polypeptides of the invention. Such salts can be prepared, for example, from
pharmaceutically acceptable non-toxic bases, including organic bases (e.g.,
salts of
primary, secondary and tertiary amines and basic amino acids) and inorganic
bases (e.g.,
sodium, potassium, lithium, ammonium, calcium and magnesium salts).
In another embodiment, illustrative immunogenic compositions, e.g.,
vaccine compositions, of the present invention comprise DNA encoding one or
more of
the polypeptides as described above, such that the polypeptide is generated in
situ. As
noted above, the polynucleotide may be administered within any of a variety of
delivery
systems known to those of ordinary skill in the art. Indeed, numerous gene
delivery
techniques are well known in the art, such as those described by Rollar~d,
Crit. Rev.
Therap. Drwg Ca~r~ier Systems 15:143-198, 1998, and references cited therein.
Appropriate polynucleotide expression systems will, of course, contain the
necessary
regulatory DNA regulatory sequences for expression in a patient (such as a
suitable
promoter and terminating signal). Alternatively, bacterial delivery systems
may involve
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
93
the administration of a bacterium (such as Bacillus-Calfyaette-Gue~r~iya) that
expresses an
immunogenic portion of the polypeptide on its cell surface or secretes such an
epitope.
Therefore, in certain embodiments, polynucleotides encoding
immunogenic polypeptides described herein are introduced into suitable
mammalian
host cells for expression using any of a number of known viral-based systems.
In one
illustrative embodiment, retroviruses provide a convenient and effective
platform for
gene delivery systems. A selected nucleotide sequence encoding a polypeptide
of the
present invention can be inserted into a vector and packaged in retroviral
particles using
techniques known in the art. The recombinant virus can then be isolated and
delivered
to a subject. A number of illustrative retroviral systems have been described
(e.g., U.S.
Pat. No. 5,219,740; Miller and Rosman (1989) BioTechniques 7:980-990; Miller,
A. D.
(1990) Human Gene Therapy 1:5-14; Scarpa et al. (1991) Virology 180:849-852;
Burns
et al. (1993) Proc. Nat!. Acad. Sci. USA 90:8033-8037; and Boris-Lawrie and
Temin
(1993) Cur. Opin. Genet. Develop. 3:102-109.
In addition, a number of illustrative adenovirus-based systems, have also
been described. Unlike retroviruses which integrate into the host genome,
adenoviruses
persist extrachromosomally thus minimizing the risks associated with
insertional
mutagenesis (Haj-Ahmad and Graham (1986) J. Virol. 57:267-274; Bett et al.
(1993) J.
Virol. 67:5911-5921; Mittereder et al. (1994) Human Gene Therapy 5:717-729;
Seth et
al. (1994) J. Virol. 68:933-940; Barn et al. (1994) Gene Therapy 1:51-58;
Berkner, I~. L.
(1988) BioTechniques 6:616-629; and Rich et al. (1993) Human Gene Therapy
4:461-
476).
Vaxious adeno-associated virus (AAV) vector systems have also been
developed for polynucleotide delivery. AAV vectors can be readily constructed
using
techniques well known in the art. See, e.g., U.S. Pat. Nos. 5,173,414 and
5,139,94.1;
International Publication Nos. WO 92101070 and WO 93/03769; Lebkowski et al.
(1988) Molec. Cell. Biol. 8:3988-3996; Vincent et al. (1990) Vaccines 90 (Cold
Spring
Harbor Laboratory Press); Carter, B. J. (1992) Current Opinion in
Biotechnology 3:533-
539; Muzyczka, N. (1992) Current Topics in Microbiol. and Immunol. 158:97-129;
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
94
Kotin, R. M. (1994) Human Gene Therapy 5:793-801; Shelling and Smith (1994)
Gene
Therapy 1:165-169; and Zhou et al. (1994) J. Exp. Med. 179:1867-1875.
Additional viral vectors useful for delivering the polynucleotides
encoding polypeptides of the present invention by gene transfer include.those
derived
from the pox family of viruses, such as vaccinia virus and avian poxvirus. By
way of
example, vaccinia virus recombinants expressing the novel molecules can be
constructed as follows. The DNA encoding a polypeptide is first inserted into
an
appropriate vector so that it is adjacent to a vaccinia promoter and flanking
vaccinia
DNA sequences, such as the sequence encoding thymidine kinase (TK). This
vector is
then used to transfect cells which are simultaneously infected with vaccinia.
Homologous recombination serves to insert the vaccinia promoter plus the gene
encoding the polypeptide of interest into the viral genome. The resulting
TK(-)
recombinant can be selected by culturing the cells in the presence of 5-
bromodeoxyuridine and picking viral plaques resistant thereto.
A vaccinia-based infection/transfection system can be conveniently used
to provide for inducible, transient expression or coexpression of one or more
polypeptides described herein in host cells of an organism. In this particular
system,
cells are first infected in vitro with a vaccinia virus recombinant that
encodes the
bacteriophage T7 RNA polymerase. This polymerase displays exquisite
specificity in
that it only transcribes templates bearing T7 promoters. Following infection,
cells are
transfected with the polynucleotide or polynucleotides of interest, driven by
a T7
promoter. The polymerase expressed in the cytoplasm from the vaccinia virus
recombinant transcribes the transfected DNA into RNA which is then translated
into
polypeptide by the host translational machinery. The method provides for high
level,
transient, cytoplasmic production of large quantities of RNA and its
translation
products. See, e.g., Elroy-Stein and Moss, Proc. Natl. Acad. Sci. USA (1990)
87:6743-
6747; Fuerst et al. Proc. Natl. Acad. Sci. USA (1986) 83:8122-8126.
Alternatively, avipoxviruses, such as the fowlpox and canarypox viruses,
can also be used to deliver the coding sequences of interest. Recombinant
avipox
viruses, expressing immunogens from mammalian pathogens, are known to confer
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
protective immunity when administered to non-avian species. The use of an
Avipox
vector is particularly desirable in human and other mammalian species since
members
of the Avipox genus can only productively replicate in susceptible avian
species and
therefore are not infective in mammalian cells. Methods for producing
recombinant
5 Avipoxviruses are known in the art and employ genetic recombination, as
described
above with respect to the production of vaccinia viruses. See, e.g., WO
91/12882; WO
89/03429; and WO 92/03545.
Any of a number of alphavirus vectors can also be used for delivery of
polynucleotide compositions of the present invention, such as those vectors
described in
10 U.S. Patent Nos. 5,843,723; 6,015,686; 6,008,035 and 6,015,694. Certain
vectors based
on Venezuelan Equine Encephalitis (VEE) can also be used, illustrative
examples of
which can be found in U.S. Patent Nos. 5,505,947 and 5,643,576.
Moreover, molecular conjugate vectors, such as the adenovirus chimeric
vectors described in Michael et al. J. Biol. Chem. (1993) 268:6866-6869 and
Wagner et
15 al. Proc. Natl. Acad. Sci. USA (1992) 89:6099-6103, can also be used for
gene delivery
under the invention.
Additional illustrative information on these and other known viral-based
delivery systems can be found, for example, in Fisher-Hoch et al., Proc. Natl.
Acad. Sci.
USA 86:317-321, 1989; Flexner et al., Anh. N Y. Acad. Sci. 569:86-103, 1989;
Flexner
20 et al., haccine 8:17-21, 1990; U.S. Patent Nos. 4,603,112, 4,769,330, and
5,017,487;
WO 89/01973; U.S. Patent No. 4,777,127; GB 2,200,651; EP 0,345,242;
WO 91/02805; Berkner, Biotechniques 6:616-627, 1988; Rosenfeld et al., Science
252:431-434, 1991; Kolls et al., Proc. Natl. Acad. Sci. USA 91:215-219, 1994;
Kass-Eisler et al., P~oc. Natl. Acad. Sci. USA 90:11498-11502, 1993; Guzman et
al.,
25 CirAculation 88:2838-2848, 1993; and Guzman et al., Cir. Res. 73:1202-1207,
1993.
In certain embodiments, a polynucleotide may be integrated into the
genome of a target cell. This integration may be in a specific location and
orientation
via homologous recombination (gene replacement) or it may be integrated in a
random,
non-specific location (gene augmentation). In yet further embodiments, the
30 polynucleotide may be stably maintained in the cell as a separate, episomal
segment of
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
96
DNA. Such polynucleotide segments or "episomes" encode sequences sufficient to
permit maintenance and replication independent of or in synchronization with
the host
cell cycle. The manner in which the ,expression construct is delivered to a
cell and
where in the cell the polynucleotide remains is dependent on the type of
expression
construct employed.
In another embodiment of the invention, a polynucleotide is
administered/delivered as "naked" DNA, for example as described in Ulmer et
al.,
Science 259:1745-1749, 1993 and reviewed by Cohen, SciefZCe 259:1691-1692,
1993.
The uptake of naked DNA may be increased by coating the DNA onto
biodegradable'
beads, which are efficiently transported into the cells.
In still another embodiment, a composition of the present invention can
be delivered via a particle bombardment approach, many of which have been
described.
In one illustrative example, gas-driven particle acceleration can be achieved
with
devices such as those manufactured by Powderject Pharmaceuticals PLC (Oxford,
UK)
and Powderject Vaccines Inc. (Madison, WI), some examples of which are
described in
U.S. Patent Nos. 5,846,796; 6,010,478; 5,865,796; 5,584,807; and EP Patent No.
0500
799. This approach offers a needle-free delivery approach wherein a dry powder
formulation of microscopic particles, such as polynucleotide or polypeptide
particles,
are accelerated to high speed within a helium gas jet generated by a hand held
device,
propelling the particles into a target tissue of interest.
In a related embodiment, other devices and methods that may be useful
for gas-driven needle-less injection of compositions of the present invention
include
those provided by Bioject, Inc. (Portland, OR), some examples of which are
described
in U.S. Patent Nos. 4,790,824; 5,064,413; 5,312,335; 5,383,851; 5,399,163;
5,520,639
and 5,993,412.
According to another embodiment, the pharmaceutical compositions
described herein will comprise one or more immunostimulants in addition to the
immunogenic polynucleotide, polypeptide, antibody, T-cell and/or APC
compositions
of this invention. An immunostimulant refers to essentially any substance that
enhances
or potentiates an immune response (antibody and/or cell-mediated) to an
exogenous
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
97
antigen. One preferred type of immunostimulant comprises an adjuvant. Many
adjuvants contain a substance designed to protect the antigen from rapid
catabolism,
such as aluminum hydroxide or mineral oil, and a stimulator of immune
responses, such
as lipid A, Bortadella pe~tussis or Mycobacterium tuberculosis derived
proteins.
Certain adjuvants axe commercially available as, for example, Freund's
Incomplete
Adjuvant and Complete Adjuvant (Difco Laboratories, Detroit, MI); Merck
Adjuvant
65 (Merck and Company, Inc., Rahway, NJ); AS-2 (SmithKline Beecham,
Philadelphia,
PA); aluminum salts such as aluminum hydroxide gel (alum) or aluminum
phosphate;
salts of calcium, iron or zinc; an insoluble suspension of acylated tyrosine;
acylated
sugars; cationically or anionically derivatized polysaccharides;
polyphosphazenes; .
biodegradable microspheres; monophosphoryl lipid A and quil A. Cytokines, such
as
GM-CSF, interleukin-2, -7, -12, and other like growth factors, may also be
used as
adj uvants.
Within certain embodiments of the invention, the adjuvant composition
is preferably one that induces an immune response predominantly of the Thl
type. High
levels of Thl-type cytokines (e.g., IFN-y, TNFa,, IL-2 and IL-12) tend to
favor the
induction of cell mediated immune responses to an administered antigen. In
contrast,
high levels of Th2-type cytokines (e.g., IL-4, IL-5, IL-6 and IL-10) tend to
favor the
induction of humoral immune responses. Following application of a vaccine as
provided herein, a patient will support an immune response that includes Thl-
and Th2-
type responses. Within a preferred embodiment, in which a response is
predominantly
Thl-type, the level of Thl-type cytokines will increase to a greater extent
than the level
of Th2-type cytokines. The levels of these cytokines may be readily assessed
using
standard assays. For a review of the families of cytokines, see Mosmann and
Coffman,
Ann. Rev. Immunol. 7:145-173, 1989.
Certain preferred adjuvants for eliciting a predominantly Thl-type
response include, for example, a combination of monophosphoryl lipid A,
preferably 3-
de-O-acylated monophosphoryl lipid A, together with an aluminum salt.
MPL°
adjuvants are available from Corixa Corporation (Seattle, WA; see, for
example, US
Patent Nos. 4,436,727; 4,877,611; 4,866,034 and 4,912,094). CpG-containing
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
98
oligonucleotides (in which the CpG dinucleotide is unmethylated) also induce a
predominantly Thl response. Such oligonucleotides are well known and are
described,
for example, in WO 96/02555, WO 99/33488 and U.S. Patent Nos. 6,008,200 and
5,856,462. Immunostimulatory DNA sequences are also described, for example, by
Sato et al., Science 273:352, 1996. Another preferred adjuvant comprises a
saponin,
such as Quil A, or derivatives thereof, including QS21 and QS7 (Aquila
Biopharmaceuticals Inc., Framingham, MA); Escin; Digitonin; or GypsoplZila or
Chenopodium quinoa saponins . Other preferred formulations include more than
one
saponin in the adjuvant combinations of the present invention, for example
combinations of at least two of the following group comprising QS21, QS7, Quil
A, (3-
escin, or digitonin.
Alternatively the saponin formulations may be combined with vaccine
vehicles composed of chitosan or other polycationic polymers, polylactide and
polylactide-co-glycolide particles, poly-N-acetyl glucosamine-based polymer
matrix,
particles composed of polysaccharides or chemically modified polysaccharides,
liposomes and lipid-based particles, particles composed of glycerol
monoesters, etc. The
saponins may also be formulated in the presence of cholesterol to form
particulate
structures such as liposomes or ISCOMs. Furthermore, the saponins may be
formulated
together with a polyoxyethylene ether or ester, in either a non-particulate
solution or
suspension, or in a particulate structure such as a paucilamelar liposome or
ISCOM. The
saponins may also be formulated with excipients such as CarbopolR to increase
viscosity, or may be formulated in a dry powder form with a powder excipient
such as
lactose.
In one preferred embodiment, the adjuvant system includes the
combination of a monophosphoryl lipid A and a saponin derivative, such as the
combination of QS21 and 3D-MPL~t adjuvant, as described in WO 94/00153, or a
less
reactogenic composition where the QS21 is quenched with cholesterol, as
described in
WO 96/33739. Other preferred formulations comprise an oil-in-water emulsion
and
tocopherol. Another particularly preferred adjuvant formulation employing
QS21, 3D-
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
99
MPL~ adjuvant and tocopherol in an oil-in-water emulsion is described in WO
95/17210.
Another enhanced adjuvant system involves the combination of a CpG-
containing oligonucleotide and a saponin derivative particularly the
combination of
CpG and QS2I is disclosed in WO 00/09159. Preferably the formulation
additionally
comprises an oiI in water emulsion and tocopherol.
Additional illustrative adjuvants for use in the pharmaceutical
compositions of the invention include Montanide ISA 720 (Seppic, France), SAF
(Chiron, California, United States), ISCOMS (CSL), MF-59 (Chiron), the SBAS
series
of adjuvants (e.g., SBAS-2 or SBAS-4, available from SmithKline Beecham,
Rixensart,
Belgium), Detox (Enhanzyn~; Corixa, Hamilton, MT), RC-529 (Corixa, Hamilton,
MT)
and other aminoalkyl glucosaminide 4-phosphates (AGPs), such as those
described in
pending U.S. Patent Application Serial Nos. 08/853,826 and 09/074,720, the
disclosures
of which are incorporated herein by reference in their entireties, and
polyoxyethylene
IS ether adjuvants such as those described in WO 99/52549A1.
Other preferred adjuvants include adjuvant molecules of the general
formula
(I): HO(CH2CH20)n A-R,
wherein, n is 1-50, A is a bond or -C(O)-, R is C1_so alkyl or Phenyl Cl_so
alkyl.
One embodiment of the present invention consists of a vaccine
formulation comprising a polyoxyethylene ether of general formula (I), wherein
fZ is
between 1 and 50, preferably 4-24, most preferably 9; the R component is C1_so
preferably C4-C2o alkyl and most preferably C~2 alkyl, and A is a bond. The
concentration of the polyoxyethylene ethers should be in the range 0.1-20%,
preferably
from 0.1-IO%, and most preferably in the range 0.1-I%. Preferred
polyoxyethylene
ethers are selected from the following group: polyoxyethylene-9-lauryl ether,
polyoxyethylene-9-steoryl ether, polyoxyethylene-8-steoryl ether,
polyoxyethylene-4-
lauryl ether, polyoxyethylene-35-lauryl ether, and polyoxyethylene-23-lauryl
ether.
Polyoxyethylene ethers such as polyoxyethylene lauryl ether are described in
the Merck
index (12th edition: entry 7717). These adjuvant molecules are described in WO
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
100
99/52549. The polyoxyethylene ether according to the general formula (I) above
may, if
desired, be combined with another adjuvant. For example, a preferred adjuvant
combination is preferably with CpG as described in the pending UK patent
application
GB 9820956.2.
According to another embodiment of this invention, an immunogenic
composition described herein is delivered to a host via antigen presenting
cells (APCs),
such as dendritic cells, macrophages, B cells, monocytes and other cells that
may be
engineered to be efficient APCs. Such cells may, but need not, be genetically
modified
to increase the capacity for presenting the antigen, to improve activation
and/or
maintenance of the T cell response, to have anti-tumor effects per' se and/or
to be
immunologically compatible with the receiver (i. e., matched HLA haplotype).
APCs
may generally be isolated from any of a variety of biological fluids and
organs,
including tumor and peritumoral tissues, and may be autologous, allogeneic,
syngeneic
or xenogeneic cells.
Certain preferred embodiments of the present invention use dendritic
cells or progenitors thereof as antigen-presenting cells. Dendritic cells are
highly potent
APCs (Banchereau and Steinman, Natuf~e 392:245-251, 1998) and have been shown
to
be effective as a physiological adjuvant for eliciting prophylactic or
therapeutic
antitumor immunity (see Timmerman and Levy, Afzn. Rev. Med. 50:507-529, 1999).
In
general, dendritic cells may be identified based on their typical shape
(stellate in situ,
with marked cytoplasmic processes (dendrites) visible i~ vity~o), their
ability to take up,
process and present antigens with high efficiency and their ability to
activate naive T
cell responses. Dendritic cells may, of course, be engineered to express
specific cell-
surface receptors or ligands that are not commonly found on dendritic cells
i~c vivo or ex
vivo, and such modified dendritic cells are contemplated by the present
invention. As
an alternative to dendritic cells, secreted vesicles antigen-loaded dendritic
cells (called
exosomes) may be used within a vaccine (see Zitvogel et al., Nature Med. 4:594-
600,
1998).
Dendritic cells and progenitors may be obtained from peripheral blood,
bone marrow, tumor-infiltrating cells, peritumoral tissues-infiltrating cells,
lymph
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
101
nodes, spleen, skin, umbilical cord blood or any other suitable tissue or
fluid. For
example, dendritic cells may be differentiated ex vivo by adding a combination
of
cytokines such as GM-CSF, IL-4, IL-I3 and/or TNFa to cultures of monocytes
harvested from peripheral blood. Alternatively, CD34 positive cells harvested
from
S peripheral blood, umbilical cord blood or bone marrow may be differentiated
into
dendritic cells by adding to the culture medium combinations of GM-CSF, IL-3,
TNFa,
CD40 ligand, LPS, flt3 Iigand and/or other compounds) that induce
differentiation,
maturation and proliferation of dendritic cells.
Dendritic cells are conveniently categorized as "immature" and "mature"
cells, which allows a simple way to discriminate between two well
characterized
phenotypes. However, this nomenclature should not be construed to exclude all
possible intermediate stages of differentiation. Immature dendritic cells are
characterized as APC with a high capacity for antigen uptake and processing,
which
correlates with the high expression of Fcy receptor and mannose receptor. The
mature
1 S phenotype is typically characterized by a lower expression of these
markers, but a high
expression of cell surface molecules responsible for T cell activation such as
class I and
class II MHC, adhesion molecules (e.g., CDS4 and CD11) and costimulatory
molecules
(e.g., CD40, CD80, CD86 and 4-1BB).
APCs may generally be transfected with a polynucleotide of the
invention (or portion or other variant thereof) such that the encoded
polypeptide, or an
immunogenic portion thereof, is expressed on the cell surface. Such
transfection may
take place ex vivo, and a pharmaceutical composition comprising such
transfected cells
may then be used for therapeutic purposes, as described herein. Alternatively,
a gene
delivery vehicle that targets a dendritic or other antigen presenting cell may
be
ZS administered to a patient, resulting in transfection that occurs in vivo.
Ih vivo and ex
vivo transfection of dendritic cells, for example, may generally be performed
using any
methods known in the art, such as those described in WO 97/24447, or the gene
gun
approach described by Mahvi et al., hn~au~ology ahd cell Biology 75:456-460,
1997.
Antigen loading of dendritic cells may be achieved by incubating dendritic
cells or
progenitor cells with the tumor polypeptide, DNA (naked or within a plasmid
vector) or
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
102
RNA; or with antigen-expressing recombinant bacterium or viruses (e.g.,
vaccinia,
fowlpox, adenovirus or lentivirus vectors). Prior to loading, the polypeptide
may be
covalently conjugated to an immunological partner that provides T cell help
(e.g., a
carrier molecule). Alternatively, a dendritic cell may be pulsed with a non-
conjugated
immunological partner, separately or in the presence of the polypeptide.
While any suitable carrier known to those of ordinary skill in the art may
be employed in the pharmaceutical compositions of this invention, the type of
carrier
will typically vary depending on the mode of administration. Compositions of
the
present invention may be formulated for any appropriate manner of
administration,
including for example, topical, oral, nasal, mucosal, intravenous,
intracranial,
intraperitoneal, subcutaneous and intramuscular administration.
Carriers for use within such pharmaceutical compositions are
biocompatible, and may also be biodegradable. In certain embodiments, the
formulation preferably provides a relatively constant level of active
component release.
In other embodiments, however, a more rapid rate of release immediately upon
administration may be desired. The formulation of such compositions is well
within the
level of ordinary skill in the art using known techniques. Illustrative
carriers useful in
this regard include microparticles of poly(lactide-co-glycolide),
polyacrylate, latex,
starch, cellulose, dextran and the like. Other illustrative delayed-release
carriers
include supramolecular biovectors, which comprise a non-liquid hydrophilic
core (e.g.,
a cross-linked polysaccharide or oligosaccharide) and, optionally, an external
layer
comprising an amphiphilic compound, such as a phospholipid (see e.g., U.S.
Patent No.
5,151,254 and PCT applications WO 94/20078, WO/94/23701 and WO 96/06638). The
amount of active compound contained within a sustained release formulation
depends
upon the site of implantation, the rate and expected duration of release and
the nature of
the condition to be treated or prevented.
In another illustrative embodiment, biodegradable microspheres (e.g.,
polylactate polyglycolate) are employed as carriers for the compositions of
this
invention. Suitable biodegradable microspheres are disclosed, for example, in
U.S.
Patent Nos.4,897,268; 5,075,109; 5,928,647; 5,811,128; 5,820,883; 5,853,763;
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
103
5,814,344, 5,407,609 and 5,942,252. Modified hepatitis B core protein carrier
systems.
such as described in WO/99 40934, and references cited therein, will also be
useful for
many applications. Another illustrative carrier/delivery system employs a
carrier
comprising particulate-protein complexes, such as those described in U.S.
Patent No.
5,928,647, which are capable of inducing a class I-restricted cytotoxic T
lymphocyte
responses in a host.
The pharmaceutical compositions of the invention will often further
comprise one or more buffers (e.g., neutral buffered saline or phosphate
buffered
saline), carbohydrates (e.g., glucose, mannose, sucrose or dextrans),
mannitol, proteins,
polypeptides or amino acids such as glycine, antioxidants, bacteriostats,
chelating
agents such as EDTA or glutathione, adjuvants (e.g., aluminum hydroxide),
solutes that
render the formulation isotonic, hypotonic or weakly hypertonic with the blood
of a
recipient, suspending agents, thickening agents and/or preservatives.
Alternatively,
compositions of the present invention may be formulated as a lyophilizate.
The pharmaceutical compositions described herein may be presented in
unit-dose or mufti-dose containers, such as sealed ampoules or vials. Such
containers
are typically sealed in such a way to preserve the sterility and stability of
the
formulation until use. In general, formulations may be stored as suspensions,
solutions
or emulsions in oily or aqueous vehicles. Alternatively, a pharmaceutical
composition
may be stored in a freeze-dried condition requiring only the addition of a
sterile liquid
carrier immediately prior to use.
The development of suitable dosing and treatment regimens for using the
particular compositions described herein in a variety of treatment regimens,
including
e.g., oral, parenteral, intravenous, intranasal, and intramuscular
administration and
formulation, is well known in the art, some of which are briefly discussed
below for
general purposes of illustration.
In certain applications, the pharmaceutical compositions disclosed herein
may be delivered via oral administration to an animal. As such, these
compositions
may be formulated with an inert diluent or with an assimilable edible carrier,
or they
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
104
may be enclosed in hard- or soft-shell gelatin capsule, or they rnay be
compressed into
tablets, or they may be incorporated directly with the food of the diet.
The active compounds rnay even be incorporated with excipients and
used in the form of ingestible tablets, buccal tables, troches, capsules,
elixirs,
suspensions, syrups, wafers, and the like (see, for example, Mathiowitz et
al., Nature
1997 Mar 27;386(6623):410-4; Hwang et al., Crit Rev Ther Drug Carrier Syst
1998;15(3):243-84; U. S. Patent 5,641,515; U. S. Patent 5,580,579 and U. S.
Patent
5,792,451). Tablets, troches, pills, capsules and the like may also contain
any of a
variety of additional components, for example, a binder, such as gum
tragacanth, acacia,
cornstarch, or gelatin; excipients, such as dicalcium phosphate; a
disintegrating agent,
such as corn starch, potato starch, alginic acid and the like; a lubricant,
such as
magnesium stearate; and a sweetening agent, such as sucrose, lactose or
saccharin may
be added or a flavoring agent, such as peppermint, oil of wintergreen, or
cherry
flavoring. When the dosage unit form is a capsule, it may contain, in addition
to
materials of the above type, a liquid carrier. Various other materials may be
present as
coatings or to otherwise modify the physical form of the dosage unit. For
instance,
tablets, pills, or capsules may be coated with shellac, sugar, or both. Of
course, any
material used in preparing any dosage unit form should be pharmaceutically
puxe and
substantially non-toxic in the amounts employed. In addition, the active
compounds
may be incorporated into sustained-release preparation and formulations.
Typically, these formulations will contain at least about 0.1 % of the
active compound or.more, although the percentage of the active ingredients)
may, of
course, be varied and may conveniently be between about 1 or 2% and about 60%
or
70% or more of the weight or volume of the total formulation. Naturally, the
amount of
active compounds) in each therapeutically useful composition may be prepared
is such
a way that a suitable dosage will be obtained in any given unit dose of the
compound.
Factors such as solubility, bioavailability, biological half life, route of
administration,
product shelf life, as well as other pharmacological considerations will be
contemplated
by one skilled in the art of preparing such pharmaceutical formulations, and
as such, a
variety of dosages and treatment regimens may be desirable.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
105
For oral administration, the compositions of the present invention may
alternatively be incorporated with one or more excipients in the form of a
mouthwash,
dentifrice, buccal tablet, oral spray, or sublingual orally-administered
formulation.
Alternatively, the active ingredient may be incorporated into an oral solution
such as
one containing sodium borate, glycerin and potassium bicarbonate, or dispersed
in a
dentifrice, or added in a therapeutically-effective amount to a composition
that may
include water, binders, abrasives, flavoring agents, foaming agents, and
humectants.
Alternatively the compositions may be fashioned into a tablet or solution form
that may
be placed under the tongue or otherwise dissolved in the mouth.
In certain circumstances it will be desirable to deliver the pharmaceutical
compositions disclosed herein parenterally, intravenously, intramuscularly, or
even
intraperitoneally. Such approaches are well known to the skilled artisan, some
of which
are further described, for example, in U. S. Patent 5,543,158; U. S. Patent
5,641,515
and U. S. Patent 5,399,363. In certain embodiments, solutions of the active
compounds
as free base or pharmacologically acceptable salts may be prepared in wafer
suitably
mixed with a surfactant, such as hydroxypropylcellulose. Dispersions may also
be
prepared in glycerol, liquid polyethylene glycols, and mixtures thereof and in
oils.
Under ordinary conditions of storage and use, these preparations generally
will contain a
preservative to prevent the growth of microorganisms.
Illustrative pharmaceutical forms suitable for injectable use include
sterile aqueous solutions or dispersions and sterile powders for the
extemporaneous
preparation of sterile injectable solutions or dispersions (for example, see
U. S. Patent
5,466,468). In all cases the form must be sterile and must be fluid to the
extent that
easy syringability exists. It must be stable under the conditions of
manufacture and
storage and must be preserved against the contaminating action of
microorganisms,
such as bacteria and fungi. The carrier can be a solvent or dispersion medium
containing, for example, water, ethanol, polyol (e.g., glycerol, propylene
glycol, and
liquid polyethylene glycol, and the like), suitable mixtures thereof, and/or
vegetable
oils. Proper fluidity may be maintained, for example, by the use of a coating,
such as
lecithin, by the maintenance of the required particle size in the case of
dispersion and/or
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
106
by the use of surfactants. The prevention of the action of microorganisms can
be
facilitated by various antibacterial and antifungal agents, for example,
parabens,
chlorobutanol, phenol, sorbic acid, thimerosal, and the like. In many cases,
it will be
preferable to include isotonic agents, for example, sugars or sodium chloride.
Prolonged absorption of the injectable compositions can be brought about by
the use in
the compositions of agents delaying absorption, for example, aluminum
monostearate
and gelatin.
In one embodiment, for parenteral administration in an aqueous solution,
the solution should be suitably buffered if necessary and the liquid diluent
first rendered
isotonic with sufficient saline or glucose. These particular aqueous solutions
are
especially suitable for intravenous, intramuscular, subcutaneous and
intraperitoneal
administration. In this connection, a sterile aqueous medium that can be
employed will
be known to those of skill in the art in light of the present disclosure. For
example, one
dosage may be dissolved in 1 ml of isotonic NaCI solution and either added to
1000 ml
of hypodermoclysis fluid or injected at the proposed site of infusion, (see
for example,
"Remington's Pharmaceutical Sciences" 15th Edition, pages 1035-1038 and 1570
1580). Some variation in dosage will necessarily occur depending on the
condition of
the subject being treated. Moreover, for human administration, preparations
will of
course preferably meet sterility, pyrogenicity, and the general safety and
purity
standards as required by FDA Office of Biologics standards.
In another embodiment of the invention, the compositions disclosed
herein may be formulated in a neutral or salt form. Illustrative
pharmaceutically-acceptable salts include the acid addition salts (formed with
the free
amino groups of the protein) and which are formed with inorganic acids such
as, for
example, hydrochloric or phosphoric acids, or such organic acids as acetic,
oxalic,
tartaric, mandelic, and the like. Salts formed with the free carboxyl groups
can also be
derived from inorganic bases such as, for example, sodium, potassium,
ammonium,
calcium, or ferric hydroxides, and such organic bases as isopropylamine,
trimethylamine, histidine, procaine and the like. Upon formulation, solutions
will be
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
107
administered in a manner compatible with the dosage formulation and in such
amount
as is therapeutically effective.
The carriers can further comprise any and all solvents, dispersion media,
vehicles, coatings, diluents, antibacterial and antifungal agents, isotonic
and absorption
delaying agents, buffers, carrier solutions, suspensions, colloids, and the
like. The use
of such media and agents for pharmaceutical active substances is well known in
the art.
Except insofar as any conventional media or agent is incompatible with the
active
ingredient, its use in the therapeutic compositions is contemplated.
Supplementary
active ingredients can also be incorporated into the compositions. The phrase
"pharmaceutically-acceptable" refers to molecular entities and compositions
that do not
produce an allergic or similar untoward reaction when administered to a human.
In certain embodiments, the pharmaceutical compositions may be
delivered by intranasal sprays, inhalation, and/or other aerosol delivery
vehicles.
Methods for delivering genes, nucleic acids, and peptide compositions directly
to the
lungs via nasal aerosol sprays has been described, e.g., in U. S. Patent
5,756,353 and U.
S. Patent 5,804,212. Likewise, the delivery of drugs using intranasal
microparticle
resins (Takenaga et al., J Controlled Release 1998 Mar 2;52(1-2):81-7) and
lysophosphatidyl-glycerol compounds (U. S. Patent 5,725,871) are also well-
known in
the pharmaceutical arts. Likewise, illustrative transmucosal drug delivery in
the form of
a polytetrafluoroetheylene support matrix is described in U. S. Patent
5,780,045.
In certain embodiments, liposomes, nanocapsules, microparticles, lipid
particles, vesicles, and the like, are used for the introduction of the
compositions of the
present invention into suitable host cells/organisms. In particular, the
compositions of
the present invention may be formulated for delivery either encapsulated in a
lipid
particle, a liposome, a vesicle, a nanosphere, or a nanoparticle or the like.
Alternatively,
compositions of the present invention can be bound, either covalently or non-
covalently,
to the surface of such carrier vehicles.
The formation and use of liposome and liposome-like preparations as
potential drug carriers is generally known to those of skill in the art (see
for example,
Lasic, Trends Biotechnol 1998 Ju1;16(7):307-21; Takakura, Nippon Rinsho 1998
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
108
Mar;56(3):691-5; Chandran et al., Indian J Exp Biol. 1997 Aug;35(8):801-9;
Margalit,
Crit Rev Ther Drug Carrier Syst. 1995;12(2-3):233-61; U.S. Patent 5,567,434;
U.S.
Patent 5,552,157; U.S. Patent 5,565,213; U.S. Patent 5,738,868 and U.S. Patent
5,795,587, each specifically incorporated herein by reference in its
entirety).
Liposomes have been used successfully with a number of cell types that
are normally difficult to transfect by other procedures, including T cell
suspensions,
primary hepatocyte cultures and PC 12 cells (Renneisen et al., J Biol Chem.
1990 Sep
25;265(27):16337-42; Muller et al., DNA Cell Biol. 1990 Apr;9(3):221-9). In
addition,
liposomes are free of the DNA length constraints that are typical of viral-
based delivery
systems. Liposomes have been used effectively to introduce genes, various
drugs,
radiotherapeutic agents, enzymes, viruses, transcription factors, allosteric
effectors and
the like, into a variety of cultured cell lines and animals. Furthermore, he
use of
liposomes does not appear to be associated with autoimmune responses or
unacceptable
toxicity after systemic delivery.
I S In certain embodiments, liposomes are formed from phospholipids that
are dispersed in an aqueous medium and spontaneously form multilamellar
concentric
bilayer vesicles (also termed multilamellar vesicles (MLVs).
Alternatively, in other embodiments, the invention provides for
pharmaceutically-acceptable nanocapsule formulations of the compositions of
the
present invention. Nanocapsules can generally entrap compounds in a stable and
reproducible way (see, for example, Quintanar-Guerrero et al., Drug Dev Ind
Pharm.
1998 Dec;24(12):1113-28). To avoid side effects due to intracellular polymeric
overloading, such ultrafine particles (sized around 0.1 Vim) may be designed
using
polymers able to be degraded is vivo. Such particles can be made as described,
for
example, by Couvreur et al., Crit Rev Ther Drug Carrier Syst. 1988;5(1):1-20;
zur
Muhlen et al., Eur J Pharm Biopharm. 1998 Mar;45(2):149-55; Zambaux et al. J
Controlled Release. 1998 Jan 2;50(1-3):31-40; and U. S. Patent 5,145,684.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
109
Cancer Therapeutic Methods
In further aspects of the present invention, the pharmaceutical
compositions described herein may be used for the treatment of cancer,
particularly for
the immunotherapy of prostate cancer. Within such methods, the pharmaceutical
compositions described herein are administered to a patient, typically a warm-
blooded
animal, preferably a human. A patient may or may not be afflicted with cancer.
Accordingly, the above pharmaceutical compositions may be used to prevent the
development of a cancer or to treat a patient afflicted with a cancer.
Pharmaceutical
compositions and vaccines may be administered either prior to or following
surgical
removal of primary tumors and/or treatment such as administration of
radiotherapy or
conventional chemotherapeutic drugs. As discussed above, administration of the
pharmaceutical compositions may be by any suitable method, including
administration
by intravenous, intraperitoneal, intramuscular, subcutaneous, intranasal,
intradermal,
anal, vaginal, topical and oral routes.
Within certain embodiments, immunotherapy may be active
immunotherapy, in which treatment relies on the i~ vivo stimulation of the
endogenous
host immune system to react against tumors with the administration of immune
response-modifying agents (such as polypeptides and polynucleotides as
provided
herein).
Within other embodiments, immunotherapy may be passive
immunotherapy, in which treatment involves the delivery of agents with
established
tumor-immune reactivity (such as effector cells or antibodies) that can
directly or
indirectly mediate antitumor effects and does not necessarily depend on an
intact host
immune system. Examples of effector cells include T cells as discussed above,
T
lymphocytes (such as CD~+ cytotoxic T lymphocytes and CD4~ T-helper tumor-
infiltrating lymphocytes), killer cells (such as Natural Killer cells and
lymphokine-
activated killer cells), B cells and antigen-presenting cells (such as
dendritic cells and
macrophages) expressing a polypeptide provided herein. T cell receptors and
antibody
receptors specific for the polypeptides recited herein may be cloned,
expressed and
transferred into other vectors or effector cells for adoptive immunotherapy.
The
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
110
polypeptides provided herein may also be used to generate antibodies or anti-
idiotypic
antibodies (as described above and in U.S. Patent No. 4,918,164) for passive
immunotherapy.
Effector cells may generally be obtained in sufficient quantities for
adoptive immunotherapy by growth in vitro, as described herein. Culture
conditions for
expanding single antigen-specific effector cells to several billion in number
with
retention of antigen recognition ih vivo are well knov~m in the art. Such in
vita°o culture
conditions typically use intermittent stimulation with antigen, often in the
presence of
cytokines (such as IL-2) and non-dividing feeder cells. As noted above,
immunoreactive polypeptides as provided herein may be used to rapidly expand
antigen-specific T cell cultures in order to generate a sufficient number of
cells for
immunotherapy. In particular, antigen-presenting cells, such as dendritic,
macrophage,
monocyte, fibroblast and/or B cells, may be pulsed with immunoreactive
polypeptides
or transfected with one or more polynucleotides using standard techniques well
known
in the art. For example, antigen-presenting cells can be transfected with a
polynucleotide having a promoter appropriate for increasing expression in a
recombinant virus or other expression system. Cultured effector cells for use
in therapy
must be able to grow and distribute widely, and to survive long term in vivo.
Studies
have shown that cultuxed effector cells can be induced to grow in vivo and to
survive
long term in substantial numbers by repeated stimulation with antigen
supplemented
with IL-2 (see, for example, Cheever et al., Immuhological Reviews 157:177,
1997).
Alternatively, a vector expressing a polypeptide recited herein may be
introduced into antigen presenting cells taken from a patient and clonally
propagated ex
vivo for transplant back into the same patient. Transfected cells may be
reintroduced
into the patient using any means known in the art, preferably in sterile form
by
intravenous, intracavitary, intraperitoneal or intratumor administration.
Routes and frequency of administration of the therapeutic compositions
described herein, as well as dosage, will vary from individual to individual,
and may be
readily established using standard techniques. In general, the pharmaceutical
compositions and vaccines may be administered by injection (e.g.,
intracutaneous,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
11I
intramuscular, intravenous or subcutaneous), intranasally (e.g., by
aspiration) or orally.
Preferably, between 1 and 10 doses may be administered over a 52 week period.
Preferably, 6 doses are administered, at intervals of 1 month, and booster
vaccinations
may be given periodically thereafter. Alternate protocols may be appropriate
for
individual patients. A suitable dose is an amount of a compound that, when
administered as described above, is capable of promoting an anti-tumor immune
response, and is at least 10-50% above the basal (i. e., untreated) level.
Such response
can be monitored by measuring the anti-tumor antibodies in a patient or by
vaccine-
dependent generation of cytolytic effector cells capable of killing the
patient's tumor
cells in vitro. Such vaccines should also be capable of causing an immune
response that
leads to an improved clinical outcome (e.g., more frequent remissions,
complete or
partial or longer disease-free survival) in vaccinated patients as compared to
non-
vaccinated patients. In general, for pharmaceutical compositions and vaccines
comprising one or more polypeptides, the amount of each polypeptide present in
a dose
ranges from about 25 ~.g to 5 mg per kg of host. Suitable dose sizes will vary
with the
size of the patient, but will typically range from about 0.1 mL to about 5 mL.
In general, an appropriate dosage and treatment regimen provides the
active compounds) in an amount sufficient to provide therapeutic and/or
prophylactic
benefit. Such a response can be monitored by establishing an improved clinical
outcome (e.g., more frequent remissions, complete or partial, or longer
disease-free
survival) in treated patients as compared to non-treated patients. Increases
in
preexisting immune responses to a tumor protein generally correlate with an
improved
clinical outcome. Such immune responses may generally be evaluated using
standard
proliferation, cytotoxicity or cytokine assays, which may be performed using
samples
obtained from a patient before and after treatment.
Cancer Detection and Diagnostic Compositions, Methods and Kits
In general, a cancer may be detected in a patient based on the presence of
one or more prostate tumor proteins and/or polynucleotides encoding such
proteins in a
biological sample (for example, blood, sera, sputum urine and/or tumor
biopsies)
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
112
obtained from the patient. In other words, such proteins may be used as
markers to
indicate the presence or absence of a cancer such as prostate cancer. In
addition, such
proteins may be useful for the detection of other cancers. The binding agents
provided
herein generally permit detection of the level of antigen that binds to the
agent in the
S biological sample. Polynucleotide primers and probes may be used to detect
the level of
mRNA encoding a tumor protein, which is also indicative of the presence or
absence of
a cancer. In general, a prostate tumor sequence should be present at a level
that is at
least three fold higher in tumor tissue than in normal tissue
There are a variety of assay formats known to those of ordinary skill in
the art for using a binding agent to detect polypeptide markers in a sample.
See, e.g.,
Harlow and Lane, Antibodies: A Laborato~ y Manual, Cold Spring Harbor
Laboratory,
1988. In general, the presence or absence of a cancer in a patient may be
determined by
(a) contacting a biological sample obtained from a patient with a binding
agent; (b)
detecting in the sample a level of polypeptide that binds to the binding
agent; and (c)
comparing the level of polypeptide with a predetermined cut-off value.
In a preferred embodiment, the assay involves the use of binding agent
immobilized on a solid support to bind to and remove the polypeptide from the
remainder of the sample. The bound polypeptide may then be detected using a
detection
reagent that contains a reporter group and specifically binds to the binding
agent/polypeptide complex. Such detection reagents may comprise, for example,
a
binding agent that specifically binds to the polypeptide or an antibody or
other agent
that specifically binds to the binding agent, such as an anti-immunoglobulin,
protein G,
protein A or a lectin. Alternatively, a competitive assay may be utilized, in
which a
polypeptide is labeled with a reporter group and allowed to bind to the
immobilized
binding agent after incubation of the binding agent with the sample. The
extent to
which components of the sample inhibit the binding of the labeled polypeptide
to the
binding agent is indicative of the reactivity of the sample with the
immobilized binding
agent. Suitable polypeptides for use within such assays include full length
prostate
tumor proteins and polypeptide portions thereof to which the binding agent
binds, as
described above.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
113
The solid support may be any material known to those of ordinary skill
in the art to which the tumor protein may be attached. For example, the solid
support
may be a test well in a microtiter plate or a nitrocellulose or other suitable
membrane.
Alternatively, the support may be a bead or disc, such as glass, fiberglass,
Iatex or a
plastic material such as polystyrene or polyvinylchloride. The support may
also be a
magnetic particle or a fiber optic sensor, such as those disclosed, for
example, in U.S.
Patent No. 5,359,681. The binding agent may be immobilized on the solid
support
using a variety of techniques known to those of skill in the art, which are
amply
described in the patent and scientific literature. Tn the context of the
present invention,
the term "immobilization" refers to both noncovalent association, such as
adsorption,
and covalent attachment (which may be a direct linkage between the agent and
functional groups on the support or may be a linkage by way of a cross~linking
agent).
Tmmobilization by adsorption to a well in a microtiter plate or to a membrane
is
preferred. In such cases, adsorption may be achieved by contacting the binding
agent, in
a suitable buffer, with the solid support for a suitable amount of time. The
contact time
varies with temperature, but is typically between about 1 hour and about 1
day. In
general, contacting a well of a plastic microtiter plate (such as polystyrene
or
polyvinylchloride) with an amount of binding agent ranging from about 10 ng to
about
10 ~.g, and preferably about 100 ng to about 1 fig, is sufficient to
immobilize an
adequate amount of binding agent.
Covalent attachment of binding agent to a solid support may generally be
achieved by first reacting the support with a bifunctional reagent that will
react with
both the support and a functional group, such as a hydroxyl or amino group, on
the
binding agent. For example, the binding agent may be covalently attached to
supports
having an appropriate polymer coating using benzoquinone or by condensation of
an
aldehyde group on the support with an amine and an active hydrogen on the
binding
partner (see, e.g., Pierce Immunotechnology Catalog and Handbook, 1991, at
A12-A13).
In certain embodiments, the assay is a two-antibody sandwich assay.
This assay may be performed by first contacting an antibody that has been
immobilized
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
114
on a solid support, commonly the well of a microtiter plate, with the sample,
such that
polypeptides within the sample are allowed to bind to the immobilized
antibody.
Unbound sample is then removed from the immobilized polypeptide-antibody
complexes and a detection reagent (preferably a second antibody capable of
binding to a
different site on the polypeptide) containing a reporter group is added. The
amount of
detection reagent that remains bound to the solid support is then determined
using a
method appropriate for the specific reporter group.
More specifically, once the antibody is immobilized on the support as
described above, the remaining protein binding sites on the support are
typically
blocked. Any suitable blocking agent known to those of ordinary skill in the
art, such as
bovine serum albumin or Tween 20TM (Sigma Chemical Co., St. Louis, MO). The
immobilized antibody is then incubated with the sample, and polypeptide is
allowed to
bind to the antibody. The sample may be diluted with a suitable diluent, such
as
phosphate-buffered saline (PBS) prior to incubation. In general, an
appropriate contact
I S time (i. e., incubation time) is a period of time that is sufficient to
detect the presence of
polypeptide within a sample obtained from an individual with prostate cancer.
Preferably, the contact time is sufficient to achieve a level of binding that
is at least
about 95% of that achieved at equilibrium between bound and unbound
polypeptide.
Those of ordinary skill in the art will recognize that the time necessary to
achieve
equilibrium may be readily determined by assaying the level of binding that
occurs over
a period of time. At room temperature, an incubation time of about 30 minutes
is
generally sufficient.
Unbound sample may then be removed by washing the solid support
with an appropriate buffer, such as PBS containing 0.1% Tween 20TM. The second
antibody, which contains a reporter group, may then be added to the solid
support.
Preferred reporter groups include those groups recited above.
The detection reagent is then incubated with the immobilized antibody-
polypeptide complex for an amount of time suff dent to detect the bound
polypeptide.
An appropriate amount of time may generally be determined by assaying the
level of
binding that occurs over a period of time. Unbound detection reagent is then
removed
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
115
and bound detection reagent is detected using the reporter group. The method
employed
for detecting the reporter group depends upon the nature of the reporter
group. For
radioactive groups, scintillation counting or autoradiographic methods are
generally
appropriate. Spectroscopic methods may be used to detect dyes, luminescent
groups
and fluorescent groups. Biotin may be detected using avidin, coupled to a
different
reporter group (commonly a radioactive or fluorescent group or an enzyme).
Enzyme
reporter groups may generally be detected by the addition of substrate
(generally for a
specific period of time), followed by spectroscopic or other analysis of the
reaction
products.
To determine the presence or absence of a cancer, such as prostate
cancer, the signal detected from the reporter group that remains bound to the
solid
support is generally compared to a signal that corresponds to a predetermined
cut-off
value. In one preferred embodiment, the cut-off value for the detection of a
cancer is
the average mean signal obtained when the immobilized antibody is incubated
with
samples from patients without the cancer. In general, a sample generating a
signal that
is three standard deviations above the predetermined cut-off value is
considered positive
for the cancer. In an alternate preferred embodiment, the cut-off value is
determined
using a Receiver Operator Curve, according to the method of Sackett et al.,
Clinical
Epidemiology: A Basic Science for Clinical Medicine, Little Brown and Co.,
1985,
p. 106-7. Briefly, in this embodiment, the cut-off value may be determined
from a plot
of pairs of true positive rates (i.e., sensitivity) and false positive rates
(100%-specificity)
that correspond to each possible cut-off value for the diagnostic test result.
The cut-off
value on the plot that is the closest to the upper left-hand corner (i.e., the
value that
encloses the largest area) is the most accurate cut-off value, and a sample
generating a
2S signal that is higher than the cut-off value determined by this method may
be considered
positive. Alternatively, the cut-off value may be shifted to the left along
the plot, to
minimize the false positive rate, or to the right, to minimize the false
negative rate. In
general, a sample generating a signal that is higher than the cut-off value
determined by
this method is considered positive for a cancer.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
116
In a related embodiment, the assay is performed in a flow-through or
strip test format, wherein the binding agent is immobilized on a membrane,
such as
nitrocellulose. In the flow-through test, polypeptides within the sample bind
to the
immobilized binding agent as the sample passes through the membrane. A second,
labeled binding agent then binds to the binding agent-polypeptide complex as a
solution
containing the second binding agent flows through the membrane. The detection
of
bound second binding agent may then be performed as described above. In the
strip test
format, one end of the membrane to which binding agent is bound is immersed in
a
solution containing the sample. The sample migrates along the membrane through
a
region containing second binding agent and to the area of immobilized binding
agent.
Concentration of second binding agent at the area of immobilized antibody
indicates the
presence of a cancer. Typically, the concentration of second binding agent at
that site
generates a pattern, such as a line, that can be read visually. The absence of
such a
pattern indicates a negative result. In general, the amount of binding agent
immobilized
on the membrane is selected to generate a visually discernible pattern when
the
biological sample contains a level of polypeptide that would be sufficient to
generate a
positive signal in the two-antibody sandwich assay, in the format discussed
above.
Preferred binding agents for use in such assays are antibodies and antigen-
binding
fragments thereof. Preferably, the amount of antibody immobilized on the
membrane
ranges from about 25 ng to about 1 ~,g, and more preferably from about 50 ng
to about
500 ng. Such tests can typically be performed with a very small amount of
biological
sample.
Of course, numerous other assay protocols exist that are suitable for use
with the tumor proteins or binding agents of the present invention. The above
descriptions are intended to be exemplary only. For example, it will be
apparent to
those of ordinary skill in the art that the above protocols may be readily
modified to use
tumor polypeptides to detect antibodies that bind to such polypeptides in a
biological
sample. The detection of such tumor protein specific antibodies may correlate
with the
presence of a cancer.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
117
A cancer may also, or alternatively, be detected based on the presence of
T cells that specifically react with a tumor protein in a biological sample.
Within
certain methods, a biological sample comprising CD4+ and/or CD8+ T cells
isolated
from a patient is incubated with a tumor polypeptide, a polynucleotide
encoding such a
polypeptide and/or an APC that expresses at least an immunogenic portion of
such a
polypeptide, and the presence or absence of specific activation of the T cells
is detected.
Suitable biological samples include, but are not limited to, isolated T cells.
For
example, T cells may be isolated from a patient by routine techniques (such as
by
Ficoll/Hypaque density gradient centrifugation of peripheral blood
lymphocytes). T
cells may be incubated in vitro for 2-9 days (typically 4 days) at 37°C
with polypeptide
(e.g., 5 - 25 pg/ml). It may be desirable to incubate another aliquot of a T
cell sample in
the absence of tumor polypeptide to serve as a control. For CD4+ T cells,
activation is
preferably detected by evaluating proliferation of the T cells. For CD8+ T
cells,
activation is preferably detected by evaluating cytolytic activity. A level of
proliferation
that is at least two fold greater and/or a level of cytolytic activity that is
at least 20%
greater than in disease-free patients indicates the presence of a cancer in
the patient.
As noted above, a cancer may also, or alternatively, be detected based on
the level of mRNA encoding a tumor protein in a biological sample. Fox
example, at
least two oligonucleotide primers may be employed in a polymerase chain
reaction
(PCR) based assay to amplify a portion of a tumor cDNA derived from a
biological
sample, wherein at least one of the oligonucleotide primers is specific for
(i. e.,
hybridizes to) a polynucleotide encoding the tumor protein. The amplified cDNA
is
then separated and detected using techniques well known in the art, such as
gel
electrophoresis. Similarly, oligonucleotide probes that specifically hybridize
to a
polynucleotide encoding a tumor protein may be used in a hybridization assay
to detect
the presence of polynucleotide encoding the tumor protein in a biological
sample.
To permit hybridization under assay conditions, oligonucleotide primers
and probes should comprise an oligonucleotide sequence that has at least about
60%,
preferably at least about 75% and more preferably at least about 90%, identity
to a
portion of a polynucleotide encoding a tumor protein of the invention that is
at least 1f
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
118
nucleotides, and preferably at least 20 nucleotides, in length. Preferably,
oligonucleotide primers and/or probes hybridize to a polynucleotide encoding a
polypeptide described herein under moderately stringent conditions, as defined
above.
Oligonucleotide primers and/or probes which may be usefully employed in the
diagnostic methods described herein preferably are at least 10-40 nucleotides
in length.
In a preferred embodiment, the oligonucleotide primers comprise at least 10
contiguous
nucleotides, more preferably at least 15 contiguous nucleotides, of a DNA
molecule
having a sequence as disclosed herein. Techniques for both PCR based assays
and
hybridization assays are well known in the art (see, for example, Mullis et
al., Cold
Spring Harbor Symp. Quart. Biol., 51:263, 1987; Erlich ed., PCR Technology,
Stockton
Press, NY, 1989).
One preferred assay employs RT-PCR, in which PCR is applied in
conjunction with reverse transcription. Typically, RNA is extracted from a
biological
sample, such as biopsy tissue, and is reverse transcribed to produce cDNA
molecules.
PCR amplification using at least one specific primer generates a cDNA
molecule, which
may be separated and visualized using, for example, gel electrophoresis.
Amplification
may be performed on biological samples taken from a test patient and from an
individual who is not afflicted with a cancer. The amplification reaction may
be
performed on several dilutions of cDNA spanning two orders of magnitude. A two-
fold
or greater increase in expression in several dilutions of the test patient
sample as
compared to the same dilutions of the non-cancerous sample is typically
considered
positive.
In another embodiment, the compositions described herein may be used
as markers for the progression of cancer. In this embodiment, assays as
described above
for the diagnosis of a cancer may be performed over time, and the change in
the level of
reactive polypeptide(s) or polynucleotide(s) evaluated. For example, the
assays may be
performed every 24-72 hours for a period of 6 months to 1 year, and thereafter
performed as needed. In general, a cancer is progressing in those patients in
whom the
level of polypeptide or polynucleotide detected increases over time. In
contrast, the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
119
cancer is not progressing when the level of reactive polypeptide or
polynucleotide either
remains constant or decreases with time.
Certain in vivo diagnostic assays may be performed directly on a tumor.
One such assay involves contacting tumor cells with a binding agent. The bound
binding agent may then be detected directly or indirectly via a reporter
group. Such
binding agents may also be used in histological applications. Alternatively,
polynucleotide probes may be used within such applications.
As noted above, to improve sensitivity, multiple tumor protein markers
may be assayed within a given sample. It will be apparent that binding agents
specific
for different proteins provided herein may be combined within a single assay.
Further,
multiple primers or probes may be used concurrently. The selection of tumor
protein
markers may be based on routine experiments to determine combinations that
results in
optimal sensitivity. In addition, or alternatively, assays for tumor proteins
provided
herein may be combined with assays for other known tumor antigens.
The present invention further provides kits for use within any of the
above diagnostic methods. Such kits typically comprise two or more components
necessary for performing a diagnostic assay. Components may be compounds,
reagents,
containers and/or equipment. For example, one container within a kit may
contain a
monoclonal antibody or fragment thereof that specifically binds to a tumor
protein.
Such antibodies or fragments may be provided attached to a support material,
as
described above. One or more additional containers may enclose elements, such
as
reagents or buffers, to be used in the assay. Such kits may also, or
alternatively, contain
a detection reagent as described above that contains a reporter group suitable
for direct
or indirect detection of antibody binding.
Alternatively, a kit may be designed to detect the level of mRNA
encoding a tumor protein in a biological sample. Such kits generally comprise
at least
one oligonucleotide probe or primer, as described above, that hybridizes to a
polynucleotide encoding a tumor protein. Such an oligonucleotide may be used,
for
example, within a PCR or hybridization assay. Additional components that may
be
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
120
present within such kits include a second oligonucleotide and/or a diagnostic
reagent or
container to facilitate the detection of a polynucleotide encoding a tumor
protein.
The following Examples are offered by way of illustration and not by
way of limitation.
EXAMPLES
EXAMPLE 1
ISOLATION AND CHARACTERIZATION OF PROSTATE-SPECIFIC POLYPEPTIDES
This Example describes the isolation of certain prostate-specific
polypeptides from a prostate tumor cDNA library.
A human prostate tumor cDNA expression library was constructed from
prostate tumor poly A+ RNA using a Superscript Plasmid System for cDNA
Synthesis
and Plasmid Cloning kit (BRL Life Technologies, Gaithersburg, MD 20897)
following
the manufacturer's protocol. Specifically, prostate tumor tissues were
homogenized
with polytron (Kinematics, Switzerland) and total RNA was extracted using
Trizol
reagent (BRL Life Technologies) as directed by the manufacturer. The poly A+
RNA
was then purified using a Qiagen oligotex spin column mRNA purification kit
(Qiagen,
Santa Clarita, CA 91355) according to the manufacturer's protocol. First-
strand cDNA
was synthesized using the NotI/Oligo-dTl8 primer. Double-stranded cDNA was
synthesized, ligated with EcoRIBAXI adaptors (Invitrogen, San Diego, CA) and
digested with Notl. Following size fractionation with Chroma Spin-1000 columns
(Clontech, Palo Alto, CA), the cDNA was ligated into the EcoRI/NotI site of
pCDNA3.1 (Invitrogen) and transformed into ElectroMax E. coli DHlOB cells (BRL
Life Technologies) by electroporation.
Using the same procedure, a normal human pancreas cDNA expression
library was prepared from a pool of six tissue specimens (Clontech). The cDNA
libraries were characterized by determining the number of independent
colonies, the
percentage of clones that carried insert, the average insert size and by
sequence analysis.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
121
The prostate tumor library contained 1.64 x 107 independent colonies, with 70%
of
clones having an insert and the average insert size being 1745 base pairs. The
normal
pancreas cDNA library contained 3.3 x 106 independent colonies, with 69% of
clones
having inserts and the average insert size being 1120 base pairs. For both
libraries,
sequence analysis showed that the majority of clones had a full length cDNA
sequence
and were synthesized from mRNA, with minimal rRNA and mitochondria) DNA
contamination.
cDNA library subtraction was performed using the above prostate tumor
and normal pancreas cDNA libraries, as described by Hara et al. (Blood, 84:189-
199,
1994) with some modifications. Specifically, a prostate tumor-specific
subtracted
cDNA library was generated as follows. Normal pancreas cDNA library (70 ~,g)
was
digested with EcoRI, NotI, and SfuI, followed by a filling-in reaction with
DNA
polymerase Klenow fragment. After phenol-chloroform extraction and ethanol
precipitation, the DNA was dissolved in 100 ~l of H20, heat-denatured and
mixed with
100 ~1 (100 ~.g) of Photoprobe biotin (Vector Laboratories, Burlingame, CA).
As
recommended by the manufacturer, the resulting mixture was irradiated with a
270 W
sunlamp on ice for 20 minutes. Additional Photoprobe biotin (50 ~.1) was added
and
the biotinylation reaction was repeated. After extraction with butanol five
times, the
DNA was ethanol-precipitated and dissolved in 23 p1 H20 to form the driver
DNA.
To form the tracer DNA, 10 ~,g prostate tumor cDNA libxary was
digested with BamHI and XhoI, phenol chloroform extracted and passed through
Chroma spin-400 columns (Clontech). Following ethanol precipitation, the
tracer DNA
was dissolved in 5 ~,1 H20. Tracer DNA was mixed with 15 ~,1 driver DNA and 20
~l of
2 x hybridization buffer (1.5 M NaCI/10 mM EDTA/50 mM HEPES pH 7.5/0.2%
sodium dodecyl sulfate), overlaid with mineral oil, and heat-denatured
completely. The
sample was immediately transferred into a 68 °C water bath and
incubated for 20 hours
(long hybridization [LH]). The reaction mixture was then subjected to a
streptavidin
treatment followed by phenol/chloroform extraction. This process was repeated
three
more times. Subtracted DNA was precipitated, dissolved in 12 ~l H20, mixed
with 8 ~l
driver DNA and 201.1 of 2 x hybridization buffer, and subjected to a
hybridization at 68
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
122
°C for 2 hours (short hybridization [SH]). After removal of
biotinylated double-
stranded DNA, subtracted cDNA was ligated into BamHI/XhoI site of
chloramphenicol
resistant pBCSK+ (Stratagene, La Jolla, CA 92037) and transformed into
ElectroMax E.
coli DHlOB cells by electroporation to generate a prostate tumor specific
subtracted
cDNA library (referred to as "prostate subtraction 1 ").
To analyze the subtracted cDNA library, plasmid DNA was prepared
from 100 independent clones, randomly picked from the subtracted prostate
tumor
specific library and grouped based on insert size. Representative cDNA clones
were
further characterized by DNA sequencing with a Perkin Elmer/Applied Biosystems
Division Automated Sequencer Model 373A (Foster City, CA). Six cDNA clones,
hereinafter referred to as F1-13, F1-12, F1-16, H1-l, H1-9 and Hl-4, were
shown to be
abundant in the subtracted prostate-specific cDNA library. The determined 3'
and 5'
cDNA sequences for F1-12 are provided in SEQ ID NO: 2 and 3, respectively,
with
determined 3' cDNA sequences for F1-13, Fl-16, HI-l, Hl-9 and Hl-4 being
provided
in SEQ ID NO: l and 4-7, respectively.
The cDNA sequences for the isolated clones were compared to known
sequences in the gene bank using the EMBL and GenBank databases (release 96).
Four
of the prostate tumor cDNA clones, Fl-13, Fl-16, H1-1, and H1-4, were
determined to
encode the following previously identified proteins: prostate specific antigen
(PSA),
human glandular kallikrein, human tumor expression enhanced gene, and
mitochondria
cytochrome C oxidase subunit II. Hl-9 was found to be identical to a
previously
identified human autonomously replicating sequence. No significant homologies
to the
cDNA sequence for F 1-12 were found.
Subsequent studies led to the isolation of a full-length cDNA sequence
for F1-12 (also referred to as P504S). This sequence is provided in SEQ ID NO:
107,
with the corresponding predicted amino acid sequence being provided in SEQ ID
NO:
108. cDNA splice variants of P504S are provided in SEQ ID NO: 600-605.
To clone less abundant prostate tumor specific genes, cDNA library
subtraction was performed by subtracting the prostate tumor cDNA library
described
above with the normal pancreas cDNA library and with the three most abundant
genes
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
123
in the previously subtracted prostate tumor specific cDNA library: human
glandular
kallikrein, prostate specific antigen (PSA), and mitochondria cytochrome C
oxidase
subunit II. Specifically, 1 p.g each of human glandular kallikrein, PSA and
mitochondria cytochrome C oxidase subunit II cDNAs in pCDNA3.1 were added to
the
driver DNA and subtraction was performed as described above to provide a
second
subtracted cDNA library hereinafter referred to as the "subtracted prostate
tumor
specific cDNA library with spike".
Twenty-two cDNA clones were isolated from the subtracted prostate
tumor specific cDNA library with spike. The determined 3' and 5' cDNA
sequences for
the clones referred to as J1-17, L1-12, N1-1862, J1-13, J1-19, J1-25, J1-24,
Kl-58, Kl-
63, Ll-4 and Ll-14 are provided in SEQ ID NOS: 8-9, 10-11, 12-13, 14-15, 16-
17, 18-
19, 20-21, 22-23, 24-25, 26-27 and 28-29, respectively. The determined 3' cDNA
sequences for the clones referred to as J1-12, Jl-16, J1-21, Kl-48, Kl-55, L1-
2, Ll-6,
Nl-1858, Nl-1860, Nl-I86I, Nl-I864 are provided in SEQ ID NOS: 30-40,
I S respectively. Comparison of these sequences with those in the gene bank as
described
above, revealed no significant homologies to three of the five most abundant
DNA
species, (J1-17, Ll-12 and Nl-1862; SEQ ID NOS: 8-9, 10-11 and 12-13,
respectively).
Of the remaining two most abundant species, one (J1-12; SEQ ID N0:30) was
found to
be identical to the previously identified human pulmonary surfactant-
associated protein,
and the other (K1-48; SEQ ID N0:33) was determined to have some homology to R.
nor-vegicus mRNA for 2-arylpropionyl-CoA epimerase. Of the 17 less abundant
cDNA
clones isolated from the subtracted prostate tumor specific cDNA library with
spike,
four (J1-16, Kl-55, Ll-6 and Nl-1864; SEQ ID NOS:31, 34, 36 and 40,
respectively)
were found to be identical to previously identified sequences, two (J1-21 and
Nl-1860;
SEQ ID NOS: 32 and 38, respectively) were found to show some homology to non-
human sequences, and two (L1-2 and N1-1861; SEQ ID NOS: 35 and 39,
respectively)
were found to show some homology to known human sequences. No significant
homologies were found to the polypeptides J1-13, JI-19, Jl-24, Jl-25, K1-58,
K1-63,
Ll-4, L1-14 (SEQ ID NOS: 14-15, 16-17, 20-21, 18-19, 22-23, 24-25, 26-27, 28-
29,
respectively).
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
124
Subsequent studies led to the isolation of full length cDNA sequences for
Jl-17, L1-12 and N1-1862 (SEQ ID NOS: 109-111, respectively). The
corresponding
predicted amino acid sequences are provided in SEQ ID NOS: 112-114. L1-12 is
also
referred to as P501 S. A cDNA splice variant of P501 S is provided in SEQ ID
NO: 606.
In a further experiment, four additional clones were identified by
subtracting a prostate tumor cDNA library with normal prostate cDNA prepared
from a
pool of three normal prostate poly A+ RNA (referred to as "prostate
subtraction 2").
The determined cDNA sequences for these clones, hereinafter referred to as U1-
3064,
U1-3065, V1-3692 and 1A-3905, are provided in SEQ ID NO: 69-72, respectively.
Comparison of the determined sequences with those in the gene bank revealed no
significant homologies to Ul-3065.
A second subtraction with spike (referred to as "prostate subtraction
spike 2") was performed by subtracting a prostate tumor specific cDNA library
with
spike with normal pancreas cDNA library and further spiked with PSA, J1-17,
pulmonary surfactant-associated protein, mitochondria) DNA, cytochrome c
oxidase
subunit II, Nl-1862, autonomously replicating sequence, Ll-12 and tumor
expression
enhanced gene. Four additional clones, hereinafter referred to as V1-3686, R1-
2330,
1B-3976 and V1-3679, were isolated. The determined cDNA sequences for these
clones are provided in SEQ ID N0:73-76, respectively. Comparison of these
sequences
with those in the gene bank revealed no significant homologies to V1-3686 and
Rl-
2330.
Further analysis of the three prostate subtractions described above
(prostate subtraction 2, subtracted prostate tumor specific cDNA library with
spike, and
prostate subtraction spike 2) resulted in the identification of sixteen
additional clones,
referred to as 1 G-4736, 1 G-473 8, 1 G-4741, 1 G-4744, 1 G-4734, 1 H-4774, 1
H-4781,
1H-4785, 1H-4787, 1H-4796, 1I-4810, 1I-4811, 1J-4876, 1K-4884 and 1K-4896. The
determined cDNA sequences for these clones are provided in SEQ ID NOS: 77-92,
respectively. Comparison of these sequences with those in the gene bank as
described
above, revealed no significant homologies to 1 G-4741, 1 G-4734, 1I-4807, 1 J-
4876 and
1K-4896 (SEQ ID NOS: 79, 81, 87, 90 and 92, respectively). Further analysis of
the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
125
isolated clones led to the determination of extended cDNA sequences for 1 G-
4736, 1 6
4738, 1G-4741, 1G-4744, 1H-4774, 1H-4781, 1H-4785, 1H-4787, 1H-4796, 1I-4807,
1J-4876, 1K-4884 and 1K-4896, provided in SEQ ID NOS: 179-188 and 191-193,
respectively, and to the determination of additional partial cDNA sequences
for 1I-4810
and 1I-4811, provided in SEQ ID NOS: 189 and 190, respectively.
Additional studies with prostate subtraction spike 2 resulted in the
isolation of three more clones. Their sequences were determined as described
above
and compared to the most recent GenBank. All tlvree clones were found to have
homology to known genes, which'are Cysteine-rich protein, KIAA0242, and
KIAA0280
(SEQ ID NO: 317, 319, and 320, respectively). Further analysis of these clones
by
Synteni microarray (Synteni, Palo Alto, CA) demonstrated that all three clones
were
over-expressed in most prostate tumors and prostate BPH, as well as in the
majority of
normal prostate tissues tested, but low expression in all other normal
tissues.
An additional subtraction was performed by subtracting a normal
I S prostate cDNA Library with normal pancreas cDNA (referred to as "prostate
subtraction
3"). This led to the identification of six additional clones referred to as 1G-
4761, 16-
4762, 1H-4766, 1H-4770, 1H-4771 and 1H-4772 (SEQ ID NOS: 93-98). Comparison
of these sequences with those in the gene bank revealed no significant
homologies to
1G-4761 and 1H-4771 (SEQ ID NOS: 93 and 97, respectively). Further analysis of
the
isolated clones Led to the determination of extended cDNA sequences for I G-
4761, 1 6-
4762, 1H-4766 and 1H-4772 provided in SEQ ID NOS: 194-196 and 199,
respectively,
and to the determination of additional partial cDNA sequences for 1H-4770 and
1H-
4771, provided in SEQ ID NOS: 197 and 198, respectively.
Subtraction of a prostate tumor cDNA library, prepared from a pool of
polyA+ RNA from three prostate cancer patients, with a normal pancreas cDNA
library
(prostate subtraction 4) led to the identification of eight clones, referred
to as 1D-4297,
1D-4309, 1D.1-4278, 1D-4288, 1D-4283, 1D-4304, 1D-4296 and 1D-4280 (SEQ ID
NOS: 99-107). These sequences were compared to those in the gene bank as
described
above. No significant homologies were found to 1D-4283 and 1D-4304 (SEQ ID
NOS:
103 and I04, respectively). Further analysis of the isolated clones led to the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
126
determination of extended cDNA sequences for 1D-4309, 1D.1-4278, 1D-4288, 1D-
4283, 1D-4304, 1D-4296 and 1D-4280, provided in SEQ ID NOS: 200-206,
respectively.
cDNA clones isolated in prostate subtraction 1 and prostate subtraction
2, described above, were colony PCR amplified and their mRNA expression levels
in
prostate tumor, normal prostate and in various other normal tissues were
determined
using microarray technology (Synteni, Palo Alto, CA). Briefly, the PCR
amplification
products were dotted onto slides in an array format, with each product
occupying a
unique location in the array. mRNA was extracted from the tissue sample to be
tested,
reverse transcribed, and fluorescent-labeled cDNA probes were generated. The
microarrays were probed with the labeled cDNA probes, the slides scanned and
fluorescence intensity was measured. This intensity correlates with the
hybridization
intensity. Two clones (referred to as P509S and PS l OS) were found to be over-
expressed in prostate tumor and normal prostate and expressed at low levels in
all other
normal tissues tested (liver, pancreas, skin, bone marrow, brain, breast,
adrenal gland,
bladder, testes, salivary gland, large intestine, kidney, ovary, lung, spinal
cord, skeletal
muscle and colon). The determined cDNA sequences for P509S and PS10S are
provided in SEQ ID NO: 223 and 224, respectively. Comparison of these
sequences
with those in the gene bank as described above, revealed some homology to
previously
identified ESTs.
Additional, studies led to the isolation of the full-length cDNA sequence
for P509S. This sequence is provided in SEQ ID NO: 332, with the corresponding
predicted amino acid sequence being provided in SEQ ID NO: 339. Two variant
full-
length cDNA sequences for P51 OS are provided in SEQ ID NO: 535 and 536, with
the
corresponding predicted amino acid sequences being provided in SEQ ID NO: 537
and
538, respectively. Additional splice variants of PS l OS are provided in SEQ
ID NO: 598
and 599.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
127
EXAMPLE 2
DETERMINATION OF TISSUE SPECIFICITY OF PROSTATE-SPECIFIC POLYPEPTIDES
Using gene specific primers, mRNA expression levels for the
representative prostate-specific polypeptides F1-16, Hl-l, J1-17 (also
referred to as
P502S), L1-12 (also referred to as PSOlS), F1-12 (also referred to as P504S)
and N1-
1 X62 (also referred to as P503S) were examined in a variety of normal and
tumor tissues
using RT-PCR.
Briefly, total RNA was extracted from a variety of normal and tumor
tissues using Trizol reagent as described above. First strand synthesis was
carried out
using 1-2 ~,g of total RNA with Superscript II reverse transcriptase (BRL Life
Technologies) at 42 °C for one hour. The cDNA was then amplified by PCR
with gene-
specific primers. To ensure the semi-quantitative nature of the RT-PCR, (3-
actin was
used as an internal control for each of the tissues examined. First, serial
dilutions of the
first strand cDNAs were prepared and RT-PCR assays were performed using (3-
actin
specific primers. A dilution was then chosen that enabled the linear range
amplification
of the (3-actin template and which was sensitive enough to reflect the
differences in the
initial copy numbexs. Using these conditions, the [3-actin levels were
determined for
each reverse transcription reaction from each tissue. DNA contamination was
minimized by DNase treatment and by assuring a negative PCR result when using
first
strand cDNA that was prepared without adding reverse transcriptase.
mRNA Expression levels were examined in four different types of tumor
tissue (prostate tumor from 2 patients, breast tumor from 3 patients, colon
tumor, lung
tumor), and sixteen different normal tissues, including prostate, colon,
kidney, liver,
lung, ovary, pancreas, skeletal muscle, skin, stomach, testes, bone marrow and
brain.
F1-16 was found to be expressed at high levels in prostate tumor tissue, colon
tumor
and normal prostate, and at lower levels in normal Liver, skin and testes,
with expression
being undetectable in the other tissues examined. H1-1 was found to be
expressed at
high levels in prostate tumor, lung tumor, breast tumor, normal prostate,
normal colon
and normal brain, at much lower levels in normal lung, pancreas, skeletal
muscle, skin,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
128
small intestine, bone marrow, and was not detected in the other tissues
tested. J1-17
(P502S) and L1-12 (PSOlS) appear to be specifically over-expressed in
prostate, with
both genes being expressed at high levels in prostate tumor and normal
prostate but at
low to undetectable levels in all the other tissues examined. N1-1862 (P503S)
was
S found to be over-expressed in 60% of prostate tumors and detectable in
normal colon
and kidney. The RT-PCR results thus indicate that Fl-16, H1-1, Jl-17 (P502S),
N1-
1862 (P503S) and L1-12 (PSO1S) are either prostate specific or are expressed
at
significantly elevated levels in prostate.
Further RT-PCR studies showed that F1-12 (P504S) is over-expressed in
60% of prostate tumors, detectable in normal kidney but not detectable in all
other
tissues tested. Similarly, Rl-2330 was shown to be over-expressed in 40% of
prostate
tumors, detectable in normal kidney and liver, but not detectable in all other
tissues
tested. UI-3064 was found to be over-expressed in 60% of prostate tumors, and
also
expressed in breast and colon tumors, but was not detectable in normal
tissues.
RT-PCR characterization of Rl-2330, U1-3064 and 1D-4279 showed
that these three antigens are over-expressed in prostate and/or prostate
tumors.
Northern analysis with four prostate tumors, two normal prostate
samples, two BPH prostates, and normal colon, kidney, liver, lung, pancrease,
skeletal
muscle, brain, stomach, testes, small intestine and bone marrow, showed that
Ll-12
(P501 S) is over-expressed in prostate tumors and normal prostate, while being
undetectable in other normal tissues tested. J1-17 (P502S) was detected in two
prostate
tumors and not in the other tissues tested. N1-1862 (P503S) was found to be
over
expressed in three prostate tumors and to be expressed in normal prostate,
colon and
kidney, but not in other tissues tested. Fl-12 (P504S) was found to be highly
expressed
in two prostate tumors and to be undetectable in all other tissues tested.
The microarray technology described above was used to determine the
expression levels of representative antigens described herein in prostate
tumor, breast
tumor and the following normal tissues: prostate, liver, pancreas, skin, bone
marrow,
brain, breast, adrenal gland, bladder, testes, salivary gland, large
intestine, kidney,
ovary, lung, spinal cord, skeletal muscle and colon. L1-12 (PSOlS) was found
to be
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
129
over-expressed in normal prostate and prostate tumor, with some expression
being
detected in normal skeletal muscle. Both JI-12 and Fl-12 (P504S) were found to
be
over-expressed in prostate tumor, with expression being lower or undetectable
in all
other tissues tested. N1-1862 (P503S) was found to be expressed at high levels
in
prostate tumor and normal prostate, and at low levels in normal large
intestine and
normal colon, with expression being undetectable in all other tissues tested.
R1-2330
was found to be over-expressed in prostate tumor and normal prostate, and to
be
expressed at lower levels in all other tissues tested. 1 D-4279 was found to
be over
expressed in prostate tumor and normal prostate, expressed at lower levels in
normal
spinal cord, and to be undetectable in all other tissues tested.
Further microarray analysis to specifically address the extent to which
P501 S (SEQ ID NO: 110) was expressed in breast tumor revealed moderate over
expression not only in breast tumor, but also in metastatic breast tumor (2/31
), with
negligible to low expression in normal tissues. This data suggests that P501 S
may be
over-expressed in various breast tumors as well as in prostate tumors.
The expression levels of 32 ESTs (expressed sequence tags) described by
Vasmatzis et al. (Proc. Natl. Acad. Sci. USA 9,5:300-304, 1998) in a variety
of tumor
and normal tissues were examined by microarray technology as described above.
Two
of these clones (referred to as P1000C and P1001C) were found to be over-
expressed in
prostate tumor and normal prostate, and expressed at low to undetectable
levels in all
other tissues tested (normal aorta, thymus, resting and activated PBMC,
epithelial cells,
spinal cord, adrenal gland, fetal tissues, skin, salivary gland, large
intestine, bone
marrow, liver, lung, dendritic cells, stomach, lymph nodes, brain, heart,
small intestine,
skeletal muscle, colon and kidney. The determined cDNA sequences for P1000C
and
P 1001 C are provided in SEQ ID NO: 3 84 and 472, respectively. The sequence
of
PIOOlC was found to show some homology to the previously isolated Human mRNA
for JM27 protein. Subsequent comparison of the sequence of SEQ ID NO: 3 84
with
sequences in the public databases, led to the identification of a full-length
cDNA
sequence of P1000C (SEQ ID NO: 786), which encodes a 492 amino acid sequence.
Analysis of the amino acid sequence using the PSORT II program led to the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
130
identification of a putative transmembrane domain from amino acids 84-100. The
cDNA sequence of the open reading frame of P 1 OOOC, including the stop codon,
is
provided in SEQ ID NO: 787, with the open reading frame without the stop codon
being
provided in SEQ ID NO: 788. The full-length amino acid sequence of P1000C is
provided in SEQ ID NO: 789. SEQ ID NO: 790 and 791 represent amino acids 1-100
and 100-492 of P 1 OOOC, respectively.
The expression of the polypeptide encoded by the full length cDNA
sequence for Fl-12 (also referred to as P504S; SEQ ID NO: 108) was
investigated by
immunohistochemical analysis. Rabbit-anti-P504S polyclonal antibodies were
generated against the full length P504S protein by standard techniques.
Subsequent
isolation and characterization of the polyclonal antibodies were also
performed by
techniques well known in the art. Immunohistochemical analysis showed that the
P504S polypeptide was expressed in 100% of prostate carcinoma samples tested
(n=5).
The rabbit-anti-P504S polyclonal antibody did not appear to label benign
I S prostate cells with the same cytoplasmic granular staining, but rather
with light nuclear
staining. Analysis of normal tissues revealed that the encoded polypeptide was
found to
be expressed in some, but not all normal human tissues. Positive cytoplasmic
staining
with rabbit-anti-P504S polyclonal antibody was found in normal human kidney,
liver,
brain, colon and lung-associated macrophages, whereas heart and bone marrow
were
negative.
This data indicates that the P504S polypeptide is present in prostate
cancer tissues, and that there are qualitative and quantitative differences in
the staining
between benign prostatic hyperplasia tissues and prostate cancer tissues,
suggesting that
this polypeptide may be detected selectively in prostate tumors and therefore
be useful
in the diagnosis of prostate cancer.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
131
EXAMPLE 3
ISOLATION AND CHARACTERIZATION OF PROSTATE-SPECIFIC
POLYPEPTIDES BY PCR-BASED SUBTRACTION
A cDNA subtraction library, containing cDNA from normal prostate
subtracted with ten other normal tissue cDNAs (brain, heart, kidney, liver,
lung, ovary,
placenta, skeletal muscle, spleen and thymus) and then submitted to a first
round of
PCR amplification, was purchased from Clontech. This library was subjected to
a
second round of PCR amplification, following the manufacturer's protocol. The
resulting cDNA fragments were subcloned into the vector pT7 Blue T-vector
(Novagen,
Madison, WI) and transformed into XL-1 Blue MRF' E. coli (Stratagene). DNA was
isolated from independent clones and sequenced using a Perkin Elmer/Applied
Biosystems Division Automated Sequencer Model 373A.
Fifty-nine positive clones were sequenced. Comparison of the DNA
sequences of these clones with those in the gene bank, as described above,
revealed no
significant homologies to 25 of these clones, hereinafter referred to as P5,
P8, P9, P18,
P20, P30, P34, P36, P38, P39, P42, P49, P50, P53, P55, P60, P64, P65, P73,
P75, P76,
P79 and P84. The determined cDNA sequences for these clones are provided in
SEQ
ID NO: 41-45, 47-52 and 54-65, respectively. P29, P47, P68, P80 and P82 (SEQ
ID
NO: 46, 53 and 66-68, respectively) were found to show some degree of homology
to
previously identified DNA sequences. To the best of the inventors' knowledge,
none of
these sequences have been previously shown to be present in prostate.
Further studies employing the sequence of SEQ ID NO: 67 as a probe in
standard full-length cloning methods, resulted in the isolation of three cDNA
sequences
which appear to be splice variants of P80 (also known as P704P). These
sequences are
provided in SEQ ID NO: 620-622.
Further studies using the PCR-based methodology described above
resulted in the isolation of more than 180 additional clones, of which 23
clones were
found to show no significant homologies to known sequences. The determined
cDNA
sequences for these clones are provided in SEQ ID NO: l I5-I23, 127, 131, 137,
I45,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
132
147-151, 153, 156-158 and 160. Twenty-three clones (SEQ ID NO: 124-126, 128-
130,
132-136, 138-144, 146, 152, 154, 155 and 159) were found to show some homology
to
previously identified ESTs. An additional ten clones (SEQ ID NO: 161-170) were
found to have some degree of homology to known genes. Larger cDNA clones
containing the P20 sequence represent splice variants of a gene referred to as
P703P.
The determined DNA sequence for the variants referred to as DE1, DE13 and DE14
are
provided in SEQ ID NOS: 171, 175 and 177, respectively, with the corresponding
predicted amino acid sequences being provided in SEQ ID NO: 172, 176 and 178,
respectively. The determined cDNA sequence for an extended spliced form of
P703 is
provided in SEQ ID NO: 225. The DNA sequences for the splice variants referred
to as
DE2 and DE6 are provided in SEQ ID NOS: 173 and 174, respectively.
mRNA Expression levels for representative clones in tumor tissues
(prostate (n=5), breast (n=2), colon and lung) normal tissues (prostate (n=5),
colon,
kidney, liver, lung (n=2), ovary (n=2), skeletal muscle, skin, stomach, small
intestine
and brain), and activated and non-activated PBMC was determined by RT-PCR as
described above. Expression was examined in one sample of each tissue type
unless
otherwise indicated.
P9 was found to be highly expressed in normal prostate and prostate
tumor compared to all normal tissues tested except for normal colon which
showed
comparable expression. P20, a portion of the P703P gene, was found to be
highly
expressed in normal prostate and prostate tumor, compared to all twelve normal
tissues
tested. A modest increase in expression of P20 in breast tumor (n=2), colon
tumor and
lung tumor was seen compared to all normal tissues except lung ( 1 of 2).
Increased
expression of P 18 was found in normal prostate, prostate tumor and breast
tumor
compared to other normal tissues except lung and stomach. A modest increase in
expression of PS was observed in normal prostate compared to most other normal
tissues. However, some elevated expression was seen in normal lung and PBMC.
Elevated expression of PS was also observed in prostate tumors (2 of 5),
breast tumor
and one lung tumor sample. For P30, similar expression levels were seen in
normal
prostate and prostate tumor, compared to six of twelve other normal tissues
tested.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
133
Increased expression was seen in breast tumors, one lung tumor sample and one
colon
tumor sample, and also in normal PBMC. P29 was found to be over-expressed in
prostate tumor (5 of 5) and normal prostate (5 of 5) compared to the majority
of normal
tissues. However, substantial expression of P29 was observed in normal colon
and
normal lung (2 of 2). P80 was found to be over-expressed in prostate tumor (5
of 5) and
normal prostate (5 of 5) compared to all other normal tissues tested, with
increased
expression also being seen in colon tumor.
Further studies resulted in the isolation of twelve additional clones,
hereinafter referred to as 10-d8, 10-h10, 11-c8, 7-g6, 8-b5, 8-b6, 8-d4, 8-d9,
$-g3, 8-
hl l, 9-f12 and 9-f3. The determined DNA sequences for 10-d8, 10-h10, 11-c8, 8-
d4, 8-
d9, 8-hl l, 9-f12 and 9-f3 are provided in SEQ ID NO: 207, 208, 209, 216, 217,
220,
221 and 222, respectively. The determined forward and reverse DNA sequences
for 7-
g6, 8-b5, 8-b6 and 8-g3 are provided in SEQ ID NO: 210 and 211; 212 and 213;
214
and 215; and 218 and 219, respectively. Comparison of these sequences with
those in
the gene bank revealed no significant homologies to the sequence of 9-f3. The
clones
10-d8, 11-c8 and 8-hll were found to show some homology to previously isolated
ESTs, while 10-h10, 8-b5, 8-b6, 8-d4, 8-d9, 8-g3 and 9-f12 were found to show
some
homology to previously identified genes. Further characterization of 7-G6 and
8-G3
showed identity to the known genes PAP and PSA, respectively.
mRNA expression levels for these clones were determined using the
micro-array technology described above. The clones 7-G6, 8-G3, 8-B5, 8-B6, 8-
D4, 8-
D9, 9-F3, 9-F12, 9-H3, 10-A2, 10-A4, 11-C9 and 11-F2 were found to be over-
expressed in prostate tumor and normal prostate, with expression in other
tissues tested
being low or undetectable. Increased expression of 8-F 11 was seen in prostate
tumor
and normal prostate, bladder, skeletal muscle and colon. Increased expression
of 10-
H10 was seen in prostate tumor and normal prostate, bladder, lung, colon,
brain and
large intestine. Increased expression of 9-B 1 was seen in prostate tumor,
breast tumor,
and normal prostate, salivary gland, large intestine and skin, with increased
expression
of 11-C8 being seen in prostate tumor, and normal prostate and large
intestine.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
134
An additional cDNA fragment derived from the PCR-based normal
prostate subtraction, described above, was found to be prostate specific by
both micro
array technology and RT-PCR. The determined cDNA sequence of this clone
(referred
to as 9-Al 1) is provided in SEQ ID NO: 226. Comparison of this sequence with
those
in the public databases revealed 99% identity to the known gene HOXB 13.
Further studies led to the isolation of the clones 8-C6 and 8-H7. The
determined cDNA sequences for these clones are provided in SEQ ID NO: 227 and
228,
respectively. These sequences were found to show some homology to previously
isolated ESTs.
PCR and hybridization-based methodologies were employed to obtain
longer cDNA sequences for clone P20 (also referred to as P703P), yielding
three
additional cDNA fragments that progressively extend the 5' end of the gene.
These
fragments, referred to as P703PDE5, P703P6.26, and P703PX-23 (SEQ ID NO: 326,
328 and 330, with the predicted corresponding amino acid sequences being
provided in
SEQ ID NO: 327, 329 and 331, respectively) contain additional 5' sequence.
P703PDE5 was recovered by screening of a cDNA library (#141-26) with a portion
of
P703P as a probe. P703P6.26 was recovered from a mixture of three prostate
tumor
cDNAs and P703PX 23 was recovered from cDNA library (#438-48). Together, the
additional sequences include all of the putative mature serine protease along
with part of
the putative signal sequence. The full-length cDNA sequence for P703P is
provided in
SEQ ID NO: 524, with the corresponding amino acid sequence being provided in
SEQ
ID NO: 525.
Using computer algorithms, the following regions of P703P were
predicted to represent potential HLA A2-binding CTL epitopes: amino acids 164-
172
of SEQ ID NO: 525 (SEQ ID NO: 723); amino acids 160-168 of SEQ ID NO: 525
(SEQ ID NO: 724); amino acids 239-247 of SEQ ID NO: 525 (SEQ ID NO: 725);
amino acids 118-126 of SEQ ID NO: 525 (SEQ ID NO: 726); amino acids 112-120 of
SEQ ID NO: 525 (SEQ ID NO: 727); amino acids 155-164 of SEQ ID NO: 525 (SEQ
ID NO: 728); amino acids 117-126 of SEQ ID NO: 525 (SEQ ID NO: 729); amino
acids
164-173 of SEQ ID NO: 525 (SEQ ID NO: 730); amino acids 154-163 of SEQ ID NO:
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
135
525 (SEQ ID NO: 731); amino acids 163-I72 of SEQ ID NO: 525 (SEQ ID NO: 732);
amino acids 58-66 of SEQ ID NO: 525 (SEQ ID NO: 733); and amino acids 59-67 of
SEQ ID NO: 525 (SEQ ID NO: 734).
P703P was found to show some homology to previously identified
proteases, such as thrombin. The thrombin receptor has been shown to be
preferentially
expressed in highly metastatic breast carcinoma cells and breast carcinoma
biopsy
samples. Introduction of thrombin receptor antisense cDNA has been shown to
inhibit
the invasion of metastatic breast carcinoma cells in culture. Antibodies
against
thrombin receptor inhibit thrombin receptor activation and thrombin-induced
platelet
activation. Furthermore, peptides that resemble the receptor's tethered ligand
domain
inhibit platelet aggregation by thrombin. P703P may play a role in prostate
cancer
through a protease-activated receptor on the cancer cell or on stromal cells.
The
potential trypsin-like protease activity of P703P may either activate a
protease-activated
receptor on the cancer cell membrane to promote tumorgenesis or activate a
protease-
activated receptor on the adjacent cells (such as stromal cells) to secrete
growth factors
and/or proteases (such as matrix metalloproteinases) that could promote tumor
angiogenesis, invasion and metastasis. P703P may thus promote tumor
progression
and/or metastasis through the activation of protease-activated receptor.
Polypeptides
and antibodies that block the P703P-receptor interaction may therefore be
usefully
employed in the treatment of prostate cancer.
To determine whether P703P expression increases with increased
severity of Gleason grade, an indicator of tumor stage, quantitative PCR
analysis was
performed on prostate tumor samples with a range of Gleason scores from 5 to >
8. The
mean level of P703P expression increased with increasing Gleason score,
indicating that
P703P expression may correlate with increased disease severity.
Further studies using a PCR-based subtraction library of a prostate tumor
pool subtracted against a pool of normal tissues (referred to as JP: PCR
subtraction)
resulted in the isolation of thirteen additional clones, seven of which did
not share any
significant homology to known GenBank sequences. The determined cDNA sequences
for these seven clones (P711P, P712P, novel 23, P774P, P775P, P710P and P768P)
axe
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
136
provided in SEQ ID NO: 307-31 l, 313 and 315, respectively. The remaining six
clones
(SEQ ID NO: 316 and 321-325) were shown to share some homology to known genes.
By micxoarray analysis, all thirteen clones showed three or more fold over-
expression in
prostate tissues, including prostate tumors, BPH and normal prostate as
compared to
normal non-prostate tissues. Clones P711P, P712P, novel 23 and P768P showed
over-
expression in most prostate tumors and BPH tissues tested (n=29), and in the
majority
of normal prostate tissues (n=4), but background to low expression levels in
all normal
tissues. Clones P774P, P775P and P710P showed comparatively lower expression
and
expression in fewer prostate tumors and BPH samples, with negative to low
expression
in normal prostate.
Further studies led to the isolation of an extended cDNA sequence for
P712P (SEQ ID NO: 552). The amino acid sequences encoded by 16 predicted open
reading frames present within the sequence of SEQ ID NO: 552 are pxovided in
SEQ ID
NO: 553-568.
The Rill-length cDNA for P711P was obtained by employing the partial
sequence of SEQ ID NO: 307 to screen a prostate cDNA library. Specifically, a
directionally cloned prostate cDNA library was prepaxed using standard
techniques.
One million colonies of this library were plated onto LB/Amp plates. Nylon
membrane
filters were used to lift these colonies, and the cDNAs which were picked up
by these
filters were denatured and cross-linked to the filters by UV light. The P711P
cDNA
fragment of SEQ ID NO: 307 was radio-labeled and used to hybridize with these
filters.
Positive clones were selected, and cDNAs were prepared and sequenced using an
automatic Perkin Elmer/Applied Biosystems sequences. The determined full-
length
sequence of P711P is provided in SEQ ID NO: 382, with the corresponding
predicted
amino acid sequence being provided in SEQ ID NO: 3 83.
Using PCR and hybridization-based methodologies, additional cDNA
sequence information was derived for two clones described above, 11-C9 and 9-
F3,
herein after referred to as P707P and P714P, respectively (SEQ ID NO: 333 and
334).
After comparison with the most recent GenBank, P707P was found to be a splice
variant of the known gene HoxBl3. In contrast, no significant homologies to
P714P
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
137
were found. Further studies employing the sequence of SEQ ID NO: 334 as a
probe in
standard full-length cloning methods, resulted in an extended cDNA sequence
for
P714P. This sequence is provided in SEQ ID NO: 619. This sequence was found to
show some homology to the gene that encodes human ribosomal L23A protein.
Clones 8-B3, P89, P98, P130 and P201 (as disclosed in U.S. Patent
Application No. 09/020,956, filed February 9, 1998) were found to be contained
within
one contiguous sequence, referred to as P705P (SEQ ID NO: 335, with the
predicted
amino acid sequence provided in SEQ ID NO: 336), which was determined to be a
splice variant of the known gene NKX 3.1.
Further studies on P775P resulted in the isolation of four additional
sequences (SEQ ID NO: 473-476) which are all splice variants of the P775P
gene. The
sequence of SEQ ID NO: 474 was found to contain two open reading frames
(ORFs).
The predicted amino acid sequences encoded by these ORFs are provided in SEQ
ID
NO: 477 and 478. The cDNA sequence of SEQ ID NO: 475 was found to contain an
ORF which encodes the amino acid sequence of SEQ ID NO: 479. The cDNA
sequence of SEQ ID NO: 473 was found to contain four ORFs. The predicted amino
acid sequences encoded by these ORFs are provided in SEQ ID NO: 480-483.
Additional splice variants of P775P are provided in SEQ ID NO: 593-597.
Subsequent studies led to the identification of a genomic region on
chromosome 22q11.2, known as the Cat Eye Syndrome region, that contains the
five
prostate genes P704P, P712P, P774P, P775P and B305D. The relative location of
each
of these five genes within the genomic region is shown in Fig. 10. This region
may
therefore be associated with malignant tumors, and other potential tumor genes
may be
contained within this region. These studies also led to the identification of
a potential
open reading frame (ORF) for P775P (provided in SEQ ID NO: 533), which encodes
the amino acid sequence of SEQ ID NO: 534.
Comparison of the clone of SEQ ID NO: 325 (referred to as P558S) with
sequences in the GenBank and GeneSeq DNA databases showed that P558S is
identical
to the prostate-specific transglutaminase gene, which is known to have two
forms. The
full-length sequences for the two forms are provided in SEQ ID NO: 630 and
63I, with
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
138
the corresponding amino acid sequences being provided in SEQ ID NO: 632 and
633,
respectively. The cDNA sequence of SEQ ID NO: 631 has a 15 pair base insert,
resulting in a 5 amino acid insert in the corresponding amino acid sequence
(SEQ ID
NO: 633). This insert is not present in the sequence of SEQ ID NO: 630.
Further studies on P768P (SEQ ID NO: 315) led to the identification of
the putative full-length open reading frame (ORF). The cDNA sequence of the
ORF
with stop codon is provided in SEQ ID NO: 764. The cDNA sequence of the ORF
without stop codon is provided in SEQ ID NO: 765, with the corresponding amino
acid
sequence being provided in SEQ ID NO: 766. This sequence was found to show 86%
identity to a rat calcium transporter protein, indicating that P768P may
represent a
human calcium transporter protein. The locations of transmembrane domains
within
P768P were predicted using the PSORT II computer algorithm. Six transmernbrane
domains were predicted at amino acid positions 118-134, 172-188, 211-227, 230-
246,
282-298 and 348-364. The amino acid sequences of SEQ ID NO: 767-772 represent
amino acids 1-I34, 135-I88, 189-227, 228-246, 247-298 and 299-511 of P768P,
respectively.
EXAMPLE 4
SYNTHESIS OF POLYPEPTIDES
Polypeptides may be synthesized on a Perkin Elmer/Applied Biosystems
430A peptide synthesizer using FMOC chemistry with HPTU (O-Benzotriazole-
N,N,N',N'-tetramethyluronium hexafluorophosphate) activation. A Gly-Cys-Gly
sequence may be attached to the amino terminus of the peptide to provide a
method of
. conjugation, binding to an immobilized surface, or labeling of the peptide.
Cleavage of
the peptides from the solid support may be carried out using the following
cleavage
mixture: trifluoroacetic acid:ethanedithiolahioanisole:water:phenol
(40:1:2:2:3). After
cleaving for 2 hours, the peptides may be precipitated in cold methyl-t-butyl-
ether. The
peptide pellets may then be dissolved in water containing 0.1 %
trifluoroacetic acid
(TFA) and lyophilized prior to purification by C18 reverse phase HPLC. A
gradient of
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
139
0%-60% acetonitrile (containing 0.1 % TFA) in water (containing 0.1 % TFA) may
be
used to elute the peptides. Following lyophilization of the pure fractions,
the peptides
may be characterized using electrospray or other types of mass spectrometry
and by
amino acid analysis.
EXAMPLE 5
FURTHER ISOLATION AND CHARACTERIZATION OF
PROSTATE-SPECIFIC POLYPEPTIDES BY PCR-BASED SUBTRACTION
A cDNA library generated from prostate primary tumor mRNA as
described above was subtracted with cDNA from normal prostate. The subtraction
was
performed using a PCR-based protocol (Clontech), which was modified to
generate
larger fragments. Within this protocol, tester and driver double stranded cDNA
were
separately digested with five restriction enzymes that recognize six-
nucleotide
restriction sites (MIuI, MscI, PvuII, SaII and StuI). This digestion resulted
in an average
cDNA size of 600 bp, rather than the average size of 300 by that results from
digestion
with RsaI according to the Clontech protocol. This modification did not affect
the
subtraction efficiency. Two tester populations were then created with
different
adapters, and the driver library remained without adapters.
The tester and driver libraries were then hybridized using excess driver
cDNA. In the first hybridization step, driver was separately hybridized with
each of the
two tester cDNA populations. This resulted in populations of (a) unhybridized
tester
cDNAs, (b) tester cDNAs hybridized to other tester cDNAs, (c) tester cDNAs
hybridized to driver cDNAs and (d) unhybridized driver cDNAs. The two separate
hybridization reactions were then combined, and rehybridized in the presence
of
additional denatured driver cDNA. Following this second hybridization, in
addition to
populations (a) through (d), a fifth population (e) was generated in which
tester cDNA
with one adapter hybridized to tester cDNA with the second adapter.
Accordingly, the
second hybridization step resulted in enrichment of differentially expressed
sequences
which could be used as templates for PCR amplification with adaptor-specific
primers.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
140
The ends were then filled in, and PCR amplification was performed
using adaptor-specific primers. Only population (e), which contained tester
cDNA that
did not hybridize to driver cDNA, was amplified exponentially. A second PCR
amplification step was then performed, to reduce background and further enrich
differentially expressed sequences.
This PCR-based subtraction technique normalizes differentially
expressed cDNAs so that rare transcripts that are overexpressed in prostate
tumor tissue
may be recoverable. Such transcripts would be difficult to recover by
traditional
subtraction methods.
In addition to genes known to be overexpressed in prostate tumor,
seventy-seven further clones were identif ed. Sequences of these partial cDNAs
are
provided in SEQ ID NO: 29 to 305. Most of these clones had no significant
homology
to database sequences. Exceptions were JPTPN23 (SEQ ID NO: 231; similarity to
pig
valosin-containing protein), JPTPN30 (SEQ ID NO: 234; similarity to rat mRNA
for
proteasome subunit), JPTPN45 (SEQ ID NO: 243; similarity to rat
fzo~°vegicus cytosolic
NADP-dependent isocitrate dehydrogenase), JPTPN46 (SEQ ID NO: 244; similarity
to
human subclone H8 4 d4 DNA sequence), JP1D6 (SEQ ID NO: 265; similarity to G.
gallus dynein light chain-A), JP8D6 (SEQ ID NO: 288; similarity to human BAC
clone
RG016J04), JP8F5 (SEQ ID NO: 289; similarity to human subclone H8 3 b5 DNA
sequence), and JP8E9 (SEQ ID NO: 299; similarity to human Alu sequence).
Additional studies using the PCR-based subtraction library consisting of
a prostate tumor pool subtracted against a normal prostate pool (referred to
as PT-PN
PCR subtraction) yielded three additional clones. Comparison of the cDNA
sequences
of these clones with the most recent release of GenBank revealed no
significant
homologies to the two clones referred to as P715P and P767P (SEQ ID NO: 312
and
314). The remaining clone was found to show some homology to the known gene
KIAA0056 (SEQ ID NO: 318). Using microarray analysis to measure mRNA
expression levels in various tissues, alI three clones were found to be over-
expressed in
prostate tumors and BPH tissues. Specifically, clone P715P was over-expressed
in most
prostate tumors and BPH tissues by a factor of three or greater, with elevated
expression
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
141
seen in the majority of normal prostate samples and in fetal tissue, but
negative to low
expression in all other normal tissues. Clone P767P was over-expressed in
several
prostate tumors and BPH tissues, with moderate expression levels in half of
the normal
prostate samples, and background to low expression in all other normal tissues
tested.
Further analysis, by microatTay as described above, of the PT-PN PCR
subtraction library and of a DNA subtraction library containing cDNA from
prostate
tumor subtracted with a pool of normal tissue cDNAs, led to the isolation of
27
additional clones (SEQ ID NO: 340-365 and 381) which were determined to be
over-
expressed in prostate tumor. The clones of SEQ ID NO: 341, 342, 345, 347, 348,
349,
351, 355-359, 361, 362 and 364 were also found to be expressed in normal
prostate.
Expression of all 26 clones in a variety of normal tissues was found to be low
or
undetectable, with the exception of P544S (SEQ ID NO: 356) which was found to
be
expressed in small intestine. Of the 26 clones, 11 (SEQ ID NO: 340-349 and
362) were
found to show some homology to previously identified sequences. No significant
homologies were found to the clones of SEQ ID NO: 350, 351, 353-361, and 363-
365.
Comparison of the sequence of SEQ ID NO: 362 with sequences in the
GenBank and GeneSeq DNA databases showed that this clone (referred to as
P788P) is
identical to GeneSeq Accession No. X27262, which encodes a protein found in
the
GeneSeq protein Accession No. Y00931. The full length cDNA sequence of P788P
is
provided in SEQ ID NO: 634, with the corresponding predicted amino acid being
provided in SEQ ID NO: 635. Subsequently, a full-length cDNA sequence for
P788P
that contains polymorphisms not found in the sequence of SEQ ID NO: 634, was
cloned
multiple times by PCR amplification from cDNA prepared from several RNA
templates
from three individuals. This determined cDNA sequence of this polymorphic
variant of
P788P is provided in SEQ ID NO: 636, with the corresponding amino acid
sequence
being provided in SEQ ID NO: 637. The sequence of SEQ ID NO: 637 differs from
that of SEQ ID NO: 635 by six amino acid residues. The P788P protein has 7
potential
transmembrane domains at the C-terminal portion and is predicted to be a
plasma
membrane protein with an extracellular N-terminal region.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
142
Further studies on the clone of SEQ ID NO: 352 (referred to as P790P)
led to the isolation of the full-length cDNA sequence of SEQ ID NO: 526. The
corresponding predicted amino acid is provided in SEQ ID NO: 527. Data from
two
quantitative PCR experiments indicated that P790P is over-expressed in 11/15
tested
prostate~tumor samples and is expressed at low levels in spinal cord, with no
expression
being seen in all other normal samples tested. Data from fiu ther PCR
experiments and
microarray experiments showed over-expression in normal prostate and prostate
tumor
with little or no 'expression in other tissues tested. P790P was subsequently
found to
show significant homology to a previously identified G-protein coupled
prostate tissue
receptor.
Additional studies on the clone of SEQ ID NO: 354 (referred to as
P776P) led to the isolation of an extended cDNA sequence, provided in SEQ ID
NO:
569. The determined cDNA sequences of three additional splice variants of
P776P are
provided in SEQ ID NO: 570-572. The amino acid sequences encoded by two
predicted
open reading frames (ORFs) contained within SEQ ID NO: 570, one predicted ORF
contained within SEQ ID NO: 571, and 11 predicted ORFs contained within SEQ ID
NO: 569, are provided in SEQ ID NO: 573-586, respectively. Further studies led
to the
isolation of the full-length sequence for the clone of SEQ ID NO: 570
(provided in SEQ
ID NO: 737). Full-length cloning efforts on the clone of SEQ ID NO: 571 led to
the
isolation of two sequences (provided in SEQ ID NO: 738 and 739), representing
a
single clone, that are identical with the exception of a polymorphic
insertion/deletion at
position 1293. Specifically, the clone of SEQ ID NO: 739 (referred to as clone
F1) has
a C at position 1293. The clone of SEQ ID NO: 738 (referred to as clone F2)
has a
single base pair deletion at position 1293. The predicted amino acid sequences
encoded
by 5 open reading frames located within SEQ ID NO: 737 are provided in SEQ ID
NO:
740-744, with the predicted amino acid sequences encoded by the clone of SEQ
ID NO:
738 and 739 being provided in SEQ ID NO: 745-750.
Comparison of the cDNA sequences for the clones P767P (SEQ ID NO:
314) and P777P (SEQ ID NO: 350) with sequences in the GenBank human EST
database showed that the two clones matched many EST sequences in common,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
143
suggesting that P767P and P777P may represent the same gene. A DNA consensus
sequence derived from a DNA sequence alignment of P767P, P777P and multiple
EST
clones is provided in SEQ ID NO: 587. The amino acid sequences encoded by
three
putative ORFs located within SEQ ID NO: 587 are provided in SEQ ID NO: 588-
590.
The clone of SEQ ID NO: 342 (referred to as P789P) was found to show
homology to a previously identified gene. The full length cDNA sequence for
P789P
and the corresponding amino acid sequence are provided in SEQ ID NO: 735 and
736,
respectively.
EXAMPLE 6
PEPTIDE PRIMING OF MICE AND PROPAGATION OF CTL LINES
6.1. This Example illustrates the preparation of a CTL cell line specific
for cells expressing the P502S gene.
Mice expressing the transgene for human HLA A2Kb (provided by Dr L.
Sherman, The Scripps Research Institute, La Jolla, CA) were immunized with
P2S#12
peptide (VLGW'VAEL; SEQ ID NO: 306), which is derived from the P502S gene
(also
referred to herein as Jl-17, SEQ ID NO: 8), as described by Theobald et al.,
P~oc. Natl.
Acad. Sci. ZISA 92:11993-11997, 1995 with the following modifications. Mice
were
immunized with IOO~g of P2S#I2 and I20~,g of an I-Ab binding peptide derived
from
hepatitis B Virus protein emulsified in incomplete Freund's adjuvant. Three
weeks later
these mice were sacrificed and using a nylon mesh single cell suspensions
prepared.
Cells were then resuspended at 6 x 106 cells/ml in complete media (RPMI-1640;
Gibco
BRL, Gaithersburg, MD) containing 10% FCS, 2mM Glutamine (Gibco BRL), sodium
pyruvate (Gibco BRL), non-essential amino acids (Gibco BRL), 2 x IO-5 M 2-
mercaptoethanol, SOU/ml penicillin and streptomycin, and cultured in the
presence of
irradiated (3000 rads) P2S#12-pulsed (Smg/ml P2S#12 and lOmg/ml [32-
microglobulin)
LPS blasts (A2 transgenic spleens cells cultured in the presence of 7pg/ml
dextran
sulfate and 25~.g1m1 LPS for 3 days). Six days later, cells (5 x 105/m1) were
restimulated with 2.5 x 106/mI peptide pulsed irradiated (20,000 rads) EL4A2Kb
cells
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
144
(Sherman et al, Science 258:815-818, 1992) and 3 x 106/m1 A2 transgenic spleen
feeder
cells. Cells were cultured in the presence of 20U/ml IL-2. Cells continued to
be
restimulated on a weekly basis as described, in preparation fox cloning the
line.
P2S#12 line was cloned by limiting dilution analysis with peptide pulsed
EL4 A2Kb tumor cells (1 x 104 cells/ well) as stimulators and A2 transgenic
spleen
cells as feeders ( 5 x 105 cells/ well) grown in the presence of 30U/ml IL-2.
On day 14,
cells were restimulated as before. On day 21, clones that were growing were
isolated
and maintained in culture. Several of these clones demonstrated significantly
higher
reactivity (lysis) against human fibroblasts (HLA A2Kb expressing) transduced
with
I O P502S than against control fibroblasts. An example is presented in Figure
I.
This data indicates that P2S #12 represents a naturally processed epitope
of the P502S protein that is expressed in the context of the human HLA A2Kb
molecule.
I5 6.2. This Example illustrates the preparation of murine CTL lines and
CTL clones specific for cells expressing the P501 S gene.
This series of experiments were performed similarly to that described
above. Mice were immunized with the P1S#10 peptide (SEQ ID NO: 337), which is
20 derived from the P501S gene (also referred to herein as L1-I2, SEQ ID NO:
1I0). The
P 1 S#10 peptide was derived by analysis of the predicted polypeptide sequence
for
P501 S for potential HLA-A2 binding sequences as defined by published HLA-A2
binding motifs (Parker, KC, et al, J. Immunol., 152:163, 1994). P 1 S# 10
peptide was
synthesized as described in Example 4, and empirically tested for HLA-A2
binding
25 using a T cell based competition assay. Predicted A2 binding peptides were
tested for
their ability to compete HLA-A2 specific peptide presentation to an HLA-A2
restricted
CTL clone (D150M58), which is specific for the HLA-A2 binding influenza matrix
peptide fluM58. D150M58 CTL secretes' TNF in response to self presentation of
peptide fluM58. In the competition assay, test peptides at 100-200 ~,g/ml were
added to
30 cultures of DI50M58 CTL in order to bind HLA-A2 on the CTL. After thirty
minutes,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
145
CTL cultured with test peptides, or control peptides, were tested for their
antigen dose
response to the fluM58 peptide in a standard TNF bioassay. As shown in Figure
3,
peptide P1S#10 competes HLA-A2 restricted presentation of f1uM58,
demonstrating
that peptide Pl S#10 binds HLA-A2.
Mice expressing the transgene for human HLA A2Kb were immunized
as described by Theobald et al. (P~oc. Natl. Acad. Sci. USA 92:11993-11997,
1995)
with the following modifications. Mice were immunized with 62.S~,g of P1S #10
and
120~,g of an I-Ab binding peptide derived from Hepatitis B Virus protein
emulsified in
incomplete Freund's adjuvant. Three weeks later these mice were sacrificed and
single
cell suspensions prepared using a nylon mesh. Cells were then resuspended at 6
x 106
cells/ml in complete media (as described above) and cultured in the presence
of
irradiated (3000 rads) P1S#10-pulsed (2~g/ml P1S#10 and lOmg/ml (32-
microglobulin)
LPS blasts (A2 transgenic spleens cells cultured in the presence of 7~g/ml
dextran
sulfate and 25~.g/ml LPS for 3 days). Six days later cells (5 x 10$/m1) were
restimulated
with 2.5 x 106/m1 peptide-pulsed irradiated (20,000 rads) EL4A2Kb cells, as
described
above, and 3 x 106/m1 A2 transgenic spleen feeder cells. Cells were cultured
in the
presence of 20 U/ml IL-2. Cells were restimulated on a weekly basis in
preparation for
cloning. After three rounds of in vitro stimulations, one line was generated
that
recognized P1S#10-pulsed Jurkat A2Kb targets and PSO1S-transduced Jurkat
targets as
shown in Figure 4.
A P1S#10-specific CTL line was cloned by limiting dilution analysis
with peptide pulsed EL4 A2Kb tumor cells (1 x 104 cells/ well) as stimulators
and A2
transgenic spleen cells as feeders (5 x 105 cells/ well) grown in the presence
of 30U/ml
IL-2. On day 14, cells were restimulated as before. On day 21, viable clones
were
isolated and maintained in culture. As shown in Figure 5, five of these clones
demonstrated specific cytolytic reactivity against P501 S-transduced Jurkat
A2Kb
targets. This data indicates that P1S#10 represents a naturally processed
epitope of the
P501 S protein that is expressed in the context of the human HLA-A2.1
molecule.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
146
EXAMPLE 7
PRIMING OF CTL IN VIVO USING NAKED DNA IMMUNIZATION
WITH A PROSTATE ANTIGEN
The prostate-specific antigen Ll-12, as described above, is also referred
to as P501 S. HLA A2Kb Tg mice (provided by Dr L. Sherman, The Scripps
Research
Institute, La Jolla, CA) were immunized with 100 ~g P501 S in the vector
VR1012
either intramuscularly or intradermally. The mice were immunized three times,
with a
two week interval between immunizations. Two weeks after the last
immunization,
immune spleen cells were cultured with Jurkat A2Kb-P501 S transduced
stimulator
cells. CTL lines were stimulated weekly. After two weeks of ih vitro
stimulation, CTL
activity was assessed against P501 S transduced targets. Two out of 8 mice
developed
strong anti-P501 S CTL responses. These results demonstrate that P501 S
contains at
least one naturally processed HLA-A2-restricted CTL epitope.
EXAMPLE 8
ABILITY OF HUMAN T CELLS TO RECOGNIZE PROSTATE-SPECIFIC POLYPEPTIDES
This Example illustrates the ability of T cells specific for a prostate
tumor polypeptide to recognize human tumor.
Human CD8+ T cells were primed in vitro to the P2S-12 peptide (SEQ
ID NO: 306) derived from P502S (also referred to as Jl-17) using dendritic
cells
according to the protocol of Van Tsai et al. (Critical Reviews in Immunology
18:65-75,
1998). The resulting CD8+ T cell microcultures were tested for their ability
to
recognize the P2S-12 peptide presented by autologous fbroblasts or fibroblasts
which
were transduced to express the P502S gene in a y-interferon ELISPOT assay (see
Lalvani et al., J. Exp. Med. 186:859-865, 1997). Briefly, titrating numbers of
T cells
were assayed in duplicate on 104 fibroblasts in the presence of 3 ~,g/ml human
(32-
microglobulin and 1 ~g/ml P2S-12 peptide or control E75 peptide: In addition,
T cells
were simultaneously assayed on autologous fibroblasts transduced with the
P502S gene
or as a control, fibroblasts transduced with HER-2/neu. Prior to the assay,
the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
147
fibroblasts were treated with 10 ng/ml y-interferon for 48 hours to upregulate
class I
MHC expression. One of the microcultures (#5) demonstrated strong recognition
of
both peptide pulsed fibroblasts as well as transduced fibroblasts in a y-
interferon
ELISPOT assay. Figure 2A demonstrates that there was a strong increase in the
number
of y-interferon spots with increasing numbers of T cells on fibroblasts pulsed
with the
P2S-12 peptide (solid bars) but not with the control E75 peptide (open bars).
This
shows the ability of these T cells to specifically recognize the P2S-12
peptide. As
shown in Figure 2B, this microculture also demonstrated an increase in the
number of y-
interferon spots with increasing numbers of T cells on fibroblasts transduced
to express
the P502S gene but not the HER-2/ueu gene. These results provide additional
confirmatory evidence that the P2S-12 peptide is a naturally processed epitope
of the
P502S protein. Furthermore, this also demonstrates that there exists in the
human T cell
repertoire, high affinity T cells which are capable of recognizing this
epitope. These T
cells should also be capable of recognizing human tumors which express the
P502S
gene.
EXAMPLE 9
ELICITATION OF PROSTATE ANTIGEN-SPECIFIC CTL RESPONSES
IN HUMAN BLOOD
This Example illustrates the ability of a prostate-specific antigen to elicit
a CTL response in blood of normal humans.
Autologous dendritic cells (DC) were differentiated from monocyte
cultures derived from PBMC of normal donors by growth for five days in RPMI
medium containing 10% human serum, 50 ng/ml GMCSF and 30 ng/ml IL-4.
Following culture, DC were infected overnight with recombinant P501 S-
expressing
vaccinia virus at an M.O.I. of 5 and matured for 8 hours by the addition of 2
microgramslml CD40 ligand. Virus was inactivated by UV irradiation, CD8+ cells
were
isolated by positive selection using magnetic beads, and priming cultures were
initiated
in 24-well plates. Following five stimulation cycles using autologous
fibroblasts
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
148
retrovirally transduced to express P501 S and CD80, CD8+ lines were identified
that
specifically produced interferon-gamma when stimulated with autologous P501 S-
transduced fibroblasts. The P501 S-specific activity of cell line 3A-1 could
be
maintained following additional stimulation cycles on autologous B-LCL
transduced
with . P501 S. Line 3A-1 was shown to specifically recognize autologous B-LCL
transduced to express P501 S, but not EGFP-transduced autologous B-LCL, as
measured
by cytotoxicity assays (SICr release) and interferon-gamma production
(Interferon-
gamma Elispot; see above and Lalvani et al., J. Exp. Med. I X6:859-865, 1997).
The
results of these assays are presented in Figures 6A and 6B.
EXAMPLE 10
IDENTIFICATION OF A NATURALLY PROCESSED CTL EPITOPE CONTAINED WITHIN THE
PROSTATE-SPECIFIC ANTIGEN P703P
The 9-mer peptide p5 (SEQ ID NO: 338) was derived from the P703P
antigen (also referred to as P20). The p5 peptide is immunogenic in human HLA-
A2
donors and is a naturally processed epitope. Antigen specific human CD8+ T
cells can
be primed following repeated i~ vitro stimulations with monocytes pulsed with
p5
peptide. These CTL specifically recognize p5-pulsed and P703P-transduced
target cells
in both ELISPOT (as described above) and chromium release assays.
Additionally,
immunization of HLA-A2Kb transgenic mice with p5 leads to the generation of
CTL
lines which recognize a variety of HLA-A2Kb or HLA-A2 transduced target cells
expressing P703P.
Initial studies demonstrating that p5 is a naturally processed epitope were
done using HLA-A2Kb transgenic mice. HLA-A2Kb transgenic mice were immunized
subcutaneously in the footpad with 100 pg of p5 peptide together with 140 ~g
of
hepatitis B virus core peptide (a Th peptide) in Freund's incomplete adjuvant.
Three
weeks post immunization, spleen cells from immunized mice were stimulated iyz
vitro
with peptide-pulsed LPS blasts. CTL activity was assessed by chromium release
assay
five days after primary in vitro stimulation. Retrovirally transduced cells
expressing the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
149
control antigen P703P and HLA-A2Kb were used as targets. CTL lines that
specifcally
recognized both p5-pulsed targets as well as P703P-expressing targets were
identified.
Human in vitf°o priming experiments demonstrated that the p5
peptide is
immunogenic in humans. Dendritic cells (DC) were differentiated from monocyte
cultures derived from PBMC of normal human donors by culturing for five days
in
RPMI medium containing 10% human serum, 50 ng/ml human GM-CSF and 30 ng/ml
human IL-4. Following culture, the DC were pulsed with 1 ug/ml p5 peptide and
cultured with CD8+ T cell enriched PBMC. CTL lines were restimulated on a
weekly
basis with p5-pulsed monocytes. Five to six weeks after initiation of the CTL
cultures,
CTL recognition of p5-pulsed target cells was demonstrated. CTL were
additionally
shown to recognize human cells transduced to express P703P, demonstrating that
p5 is
a naturally processed epitope.
Studies identifying a further peptide epitope (referred to as peptide 4)
derived from the prostate tumor-specific antigen P703P that is capable of
being
recognized by CD4 T cells on the surface of cells in the context of HLA class
II
molecules were carried out as follows. The amino acid sequence for peptide 4
is
provided in SEQ ID NO: 638, with the corresponding cDNA sequence being
provided
in SEQ ID NO: 639.
Twenty 15-mer peptides overlapping by 10 amino acids and derived
from the carboxy-terminal fragment of P703P were generated using standard
procedures. Dendritic cells (DC) were derived from PBMC of a normal female
donor
using GM-CSF and IL-4 by standard protocols. CD4 T cells were generated from
the
same donor as the DC using MACS beads and negative selection. DC were pulsed
overnight with pools of the 15-mer peptides, with each peptide at a final
concentration
of 0.25 microgram/ml. Pulsed DC were washed and plated at 1 x 104 cells/well
of 96-
well V-bottom plates and purified CD4 T cells were added at 1 x 105/well.
Cultures
were supplemented with 60 ng/ml IL-6 and 10 ng/ml IL-12 and incubated at 37
°C.
Cultures were restimulated as above on a weekly basis using DC generated and
pulsed
as above as antigen presenting cells, supplemented with 5 ng/ml IL-7 and 10
u/ml IL-2.
Following 4 in vitro stimulation cycles, 96 lines (each line corresponding to
one well)
were tested for specific proliferation and cytokine production in response to
the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
150
stimulating pools with an irrelevant pool of peptides derived from mammaglobin
being
used as a control.
One line (referred to as 1-F9) was identified from pool #1 that
demonstrated specific proliferation (measured by 3H proliferation assays) and
cytokine
production (measured by interferon-gamma ELISA assays) in response to pool #1
of
P703P peptides. This line was further tested for specif c recognition of the
peptide
pool, specific recognition of individual peptides in the pool, and in HLA
mismatch
analyses to identify the relevant restricting allele. Line 1-F9 was found to
specifically
proliferate and produce interferon-gamma in response to peptide pool #1, and
also to
peptide 4 (SEQ ID NO: 638). Peptide 4 corresponds to amino acids 126-140 of
SEQ ID
NO: 327. Peptide titration experiments were conducted to assess the
sensitivity of line
1-F9 fox the specific peptide. The line was found to specifically respond to
peptide 4 at
concentrations as low as 0.25 ng/ml, indicating that the T cells are very
sensitive and
therefore likely to have high affinity for the epitope.
To determine the HLA restriction of the P703P response, a panel of
antigen presenting cells (APC) was generated that was partially matched with
the donor
used to generate the T cells. The APC were pulsed with the peptide and used in
proliferation and cytokine assays together with line 1-F9. APC matched with
the donor
at HLA-DRB0701 and HLA-DQB02 alleles were able to present the peptide to the T
cells, indicating that the P703P-specific response is restricted to one of
these alleles.
Antibody blocking assays were utilized to determine if the restricting
allele was HLA-DR0701 or HLA-DQ02. The anti-HLA-DR blocking antibody L243
or an irrelevant isotype matched IgG2a were added to T cells and APC cultures
pulsed with the peptide RMPTVLQCVNVSVVS (SEQ ID NO: 638) at 250 nglml.
Standard interferon-gamma and proliferation assays were performed. Whereas the
control antibody had no effect on the ability of the T cells to recognize
peptide-pulsed
APC, in both assays the anti-HLA-DR antibody completely blocked the ability of
the
T cells to specifically recognize peptide-pulsed APC.
To determine if the peptide epitope RMPTVLQCVNVSVVS (SEQ ID
NO: 638) was naturally processed, the ability of line 1-F9 to recognize APC
pulsed with
recombinant P703P protein was examined. For these experiments a number of
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
151
recombinant P703P sources were utilized; E. coli-derived P703P, Pichia-derived
P703P
and baculovirus-derived P703P. Irrelevant protein controls used were E. coli-
derived
L3E a lung-specific antigen) and baculovirus-derived mammaglobin. In
interferon-
gamma ELISA assays, line 1-F9 was able to efficiently recognize both E. coli
forms of
P703P as well as Pichia-derived recombinant P703P, while baculovirus-derived
P703P
was recognized less efficiently. Subsequent Western blot analysis revealed
that the E
coli and Pichia P703P protein preparations were intact while the baculovirus
P703P
preparation was approximately 75% degraded. Thus, peptide RMPTVLQCVNVSVVS
(SEQ ID NO: 638) from P703P is a naturally processed peptide epitope derived
from
P703P and presented to T cells in the context of HLA-DRB-0701
In further studies, twenty-four 15-rner peptides overlapping by 10 amino
acids and derived from the N-terminal fragment of P703P (corresponding to
amino
acids 27-154 of SEQ ID NO: 525) were generated by standard procedures and
their
ability to be recognized by CD4 cells was determined essentially as described
above.
DC were pulsed overnight with pools of the peptides with each peptide at a
final
concentration of 10 microgram/ml. A large number of individual CD4 T cell
lines
(65/480) demonstrated significant proliferation and cytokine release (IFN-
gamma) in
response to the P703P peptide pools but not to a control peptide pool. The CD4
T cell
lines which demonstrated specific activity were restimulated on the
appropriate pool of
P703P peptides and reassayed on the individual peptides of each pool as well
as a
peptide dose titration of the pool of peptides in a IFN-gamma release assay
and in a
proliferation assay.
Sixteen immunogenic peptides were recognized by the T cells from the
entire set of peptide antigens tested. The amino acid sequences of these
peptides are
provided in SEQ ID NO: 656-671, with the corresponding cDNA sequences being
provided in SEQ ID NO: 640-655, respectively. In some cases the peptide
reactivity of
the T cell line could be mapped to a single peptide, however some could be
mapped to
more than one peptide in each pool. Those CD4 T cell lines that displayed a
representative pattern of recognition from each peptide pool with a reasonable
affinity
for peptide were chosen for further analysis (I-lA, -6A; II-4C, -SE; III-6E,
IV-4B, -3F, -
9B, -IOF, V-SB, -4D, and -10F). These CD4 T cells lines were restimulated on
the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
152
appropriate individual peptide and reassayed on autologous DC pulsed with a
truncated
form of recombinant P703P protein made in E coli (a.a. 96 - 254 of SEQ ID NO:
525),
full-length P703P made in the baculovirus expression system, and a fusion
between
influenza virus NS1 and P703P made in E. coli. Of the T cell lines tested,
line I-lA
recognized specifically the truncated form of P703P (E. coli) but no other
recombinant
form of P703P. This line also recognized the peptide used to elicit the T
cells. Line 2-
4C recognized the truncated form of P703P (E. coli) and the full length form
of P703P
made in baculovirus, as well as peptide. The remaining T cell lines tested
were either
peptide-specific only (II-5E, II-6F, IV-4B, IV-3F, IV-9B, IV-lOF, V-5B and V-
4D) or
were non-responsive to any antigen tested (V-lOF). These results demonstrate
that the
peptide sequence RPLLANDLMLIKLDE (SEQ ID NO: 671; corresponding to a.a. 110-
124 of SEQ ID NO: 525) recognized by the T cell line I-lA, and the peptide
sequences
SVSESDTIRSISIAS (SEQ ID NO: 668; corresponding to a.a. 125-139 of SEQ ID NO:
525) and ISIASQCPTAGNSCL (SEQ ID NO: 667; corresponding to a.a. 135-149 of
SEQ ID NO: 525) recognized by the T cell line II-4C may be naturally processed
epitopes of the P703P protein.
EXAMPLE 11
EXPRESSION OF A BREAST TUMOR-DERIVED ANTIGEN
2O IN PROSTATE
Isolation of the antigen B305D from breast tumor by differential display
is described in US Patent Application No. 081700,014, filed August 20, 1996.
Several
different splice forms of this antigen were isolated. The determined cDNA
sequences
for these splice forms are provided in SEQ ID NO: 366-375, with the predicted
amino
acid sequences corresponding to the sequences of SEQ ID NO: 292, 298 and 301-
303
being provided in SEQ ID NO: 299-306, respectively. In further studies, a
splice
variant of the cDNA sequence of SEQ ID NO: 366 was isolated which was found to
contain an additional guanine residue at position 884 (SEQ ID NO: 530),
leading to a
frameshift in the open reading frame. The determined DNA sequence of this ORF
is
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
153
provided in SEQ ID NO: 531. This frameshift generates a protein sequence
(provided
in SEQ ID NO: 532) of 293 amino acids that contains the C-terminal domain
common
to the other isoforms of B305D but that differs in the N-terminal region.
The expression levels of B305D in a variety of tumor and normal tissues
were examined by real time PCR and by Northern analysis. The results indicated
that
B305D is highly expressed in breast tumor, prostate tumor, normal prostate and
normal
testes, with expression being low or undetectable in all other tissues
examined (colon
tumor, lung tumor, ovary tumor, and normal bone marrow, colon, kidney, liver,
lung,
ovary, skin, small intestine, stomach). Using real-time PCR on a panel of
prostate
tumors, expression of B305D in prostate tumors was shown to increase with
increasing
Gleason grade, demonstrating that expression of B305D increases as prostate
cancer
progresses.
EXAMPLE 12
1 S GENERATION OF HUMAN CTL IN VITRO USING WHOLE GENE PRIMING AND STIMULATION
TECHNIQUES WITH THE PROSTATE-SPECIFIC ANTIGEN PSO1 S
Using in vitro whole-gene priming with PSO1S-vaccinia infected DC
(see, for example, Yee et al, The Journal of Immunology, 157(9):4079-86,
1996),
human CTL lines were derived that specifically recognize autologous
fibroblasts
transduced with PSO1S (also known as L1-12), as determined by interferon y
ELISPOT
analysis as described above. Using a panel of HLA-mismatched B-LCL lines
transduced with P501 S, these CTL lines were shown to be likely restricted to
HLAB
class I allele. Specifically, dendritic cells (DC) were differentiated from
monocyte
cultures derived from PBMC of normal human donors by growing for five days in
RPMI medium containing 10% human serum, 50 nglml human GM-CSF and 30 ng/ml
human IL-4. Following culture, DC were infected overnight with recombinant
P501 S
vaccinia virus at a multiplicity of infection (M.O.I) of five, and matured
overnight by
the addition of 3 ~.g/ml CD40 ligand. Virus was inactivated by UV irradiation.
CD8+
T cells were isolated using a magnetic bead system, and priming cultures were
initiated
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
154
using standard culture techniques. Cultures were restimulated every 7-10 days
using
autologous primary fibroblasts retrovirally transduced with PSO1S and CD80.
Following four stimulation cycles, CD8+ T cell lines were identified that
specifically
produced interferon-y when stimulated with P501 S and CD80-transduced
autologous
fibroblasts. A panel of HLA-mismatched B-LCL lines transduced with P501 S were
generated to define the restriction allele of the response. By measuring
interferon y in
an ELISPOT assay, the P501 S specific response was shown to be likely
restricted by
HLA B alleles. These results demonstrate that a CD8+ CTL response to P501 S
can be
elicited.
To identify the epitope(s) recognized, cDNA encoding P501 S was
fragmented by various restriction digests, and sub-cloned into the retroviral
expression
vector pBIB-KS. Retroviral supernatants were generated by transfection of the
helper
packaging line Phoenix-Ampho. Supernatants were then used to transduce
Jurkat/A2Kb cells for CTL screening. CTL were screened in IFN-gamma ELISPOT
assays against these A2Kb targets transduced with the "library" of P501 S
fragments.
Initial positive fragments P501 S/H3 and P501 S/F2 were sequenced and found to
encode
amino acids 106-553 and amino acids 136-547, respectively, of SEQ ID NO: 113.
A
truncation of H3 was made to encode amino acid residues 106-351 of SEQ ID NO:
113,
which was unable to stimulate the CTL, thus localizing the epitope to amino
acid
residues 351-547. Additional fragments encoding amino acids 1-472 (Fragment A)
and
amino acids 1-351 (Fragment B) were also constructed. Fragment A but not
Fragment
B stimulated the CTL thus localizing the epitope to amino acid residues 351-
472.
Overlapping 20-mer and 18-mer peptides representing this region were tested by
pulsing
Jurkat/A2Kb cells versus CTL in an IFN-gamma assay. Only peptides P501 S-
369(20)
and PSO1S-369(18) stimulated the CTL. Nine-mer and 10-mer peptides
representing
this region were synthesized and similarly tested. Peptide P501 S-370 (SEQ ID
NO:
539) was the minimal 9-mer giving a strong response. Peptide P501 S-376 (SEQ
ID NO:
540) also gave a weak response, suggesting that it might represent a cross-
reactive
epitope.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
155
In subsequent studies, ~ the ability of primary human B cells transduced
with PSOlS to prime MHC class I-restricted, PSOIS-specific, autologous CD8 T
cells
was examined. Primary B cells were derived from PBMC of a homozygous HLA-A2
donor by culture in CD40 ligand and IL-4, transduced at high frequency with
recombinant P501 S in the vector pBIB, and selected with blastocidin-S. For in
vitro
priming, purified CD8+ T cells were cultured with autologous CD40 ligand + IL-
4
derived, P501 S-transduced B cells in a 96-well microculture format. These CTL
microcultures were re-stimulated with P501 S-transduced B cells and then
assayed for
specificity. Following this initial screen, microcultures with significant
signal above
background were cloned on autologous EBV-transformed B cells (BLCL), also
transduced with P501 S. Using IFN-gamma ELISPOT for detection, several of
these
CD8 T cell clones were found to be specific for P501 S, as demonstrated by
reactivity to
BLCL/P501 S but not BLCL transduced with control antigen. It was further
demonstrated that the anti-P501 S CD8 T cell specif city is HLA-A2-restricted.
First,
antibody blocking experiments with anti-HLA-A,B,C monoclonal antibody (W6.32),
anti-HLA-B,C monoclonal antibody (B 1.23.2) and a control monoclonal antibody
showed that only the anti-HLA-A,B,C antibody blocked recognition of P501 S
expressing autologous BLCL. Secondly, the anti-P501 S CTL also recognized an
HLA
A2 matched, heterologous BLCL transduced with P501 S, but not the
corresponding
EGFP transduced control BLCL.
A naturally processed, CD8, class I-restricted peptide epitope of P501 S
was identified as follows. Dendritic Cells (DC) were isolated by Percol
gradient
followed by differential adherence, and cultured for 5 days in the presence of
RPMI
medium containing 1% human serum, SOng/ml GM-CSF and 34ng/ml IL-4. Following
culture, DC were infected for 24 hours with P501 S-expressing adenovirus at an
MOI of
10 and matured for an additional 24 hours by the addition of 2ug/ml CD40
ligand. CD8
cells were enriched for by the subtraction of CD4+, CD14+ and CD16+
populations
from PBMC with magnetic beads. Priming cultures containing 10,000 P501 S-
expressing DC and 100,000 CD8+ T cells per well were set up in 96-well V-
bottom
plates with RPMI containing 10% human serum, Sng/ml IL-12 and lOng/ml IL-6.
Cultures were stimulated every 7 days using autologous fibroblasts
retrovirally
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
156
transduced to express P501S and CD80, and were treated with IFN-gamma for 48-
72
hours to upregulate MHC Class I expression. l0u/ml IL-2 was added at the time
of
stimulation and on days 2 and 5 following stimulation. Following 4 stimulation
cycles,
one PSO1S-specific CD8+ T cell line (refereed to as 2A2) was identified that
produced
IFN-gamma in response to IFN-gamma-treated P501 S/CD80 expressing autologous
fibroblasts, but not in response to IFN-gamma-treated P703P/CD80 expressing
autologous fibroblasts in a y-IFN Elispot assay. Line 2A2 was cloned in 96-
well plates
with 0.5 cell/well or 2 cells/well in the presence of 75,000 PBMC/well, 10,000
B
LCL/well, 30ng/ml OKT3 and SOu/ml IL-2. Twelve clones were isolated that
showed
strong P501 S specificity in response to transduced fibroblasts.
Fluorescence activated cell sorting (FACS) analysis was performed on
P501 S-specific clones using CD3-, CD4- and CD8-specific antibodies conjugated
to
PercP, FITC and PE respectively. Consistent with the use of CD8 enriched T
cells in
the priming cultures, P5401 S-specific clones were determined to be CD3+, CD8+
and
CD4-.
To identify the relevant P501 S epitope recognized by P501 S specific
CTL, pools of 18-20 mer or 30-mer peptides that spanned the majority of the
amino
acid sequence of PSOI S were loaded onto autologous B-LCL and tested in y-IFN
Elispot
assays for the ability to stimulate two P501 S-specific CTL clones, referred
to as 4E5
and 4E7. One pool, composed of five I 8-20 mer peptides that spanned amino
acids 411-
486 of P50I S (SEQ ID NO: 113), was found to be recognized by both P501 S-
specific
clones. To identify the specific 18-20 mer peptide recognized by the clones,
each of the
18-20 mer peptides that comprised the positive pool were tested individually
in y-IFN
Elispot assays for the ability to stimulate the two P501 S-specific CTL
clones, 4E5 and
4E7. Both 4E5 and 4E7 specifically recognized one 20-mer peptide (SEQ ID NO:
710;
cDNA sequence provided in SEQ ID NO: 711) that spanned amino acids 453-472 of
P501 S. Since the minimal epitope recognized by CD8+ T cells is almost always
either
a 9 or 10-mer peptide sequence, 10-mer peptides that spanned the entire
sequence of
SEQ ID NO: 710 were synthesized that differed by 1 amino acid. Each of these
10-mer
peptides was tested for the ability to stimulate two P501 S-specific clones,
(referred to as
1D5 and 1E12). One 10-mer peptide (SEQ ID NO: 712; cDNA sequence provided in
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
157
SEQ ID NO: 713) was identified that specifically stimulated the P501 S-specif
c clones.
This epitope spans amino acids 463-472 of P501 S. This sequence defines a
minimal 10
mer epitope from PSO1S that can be naturally processed and to which CTL
responses
can be identified in normal PBMC. Thus, this epitope is a candidate for use as
a vaccine
moiety, and as a therapeutic and/or diagnostic reagent for prostate cancer.
To identify the class I restriction element for the P501 S-derived sequence
of SEQ ID NO: 712, HLA blocking and mismatch analyses were performed. In y-IFN
Elispot assays, the specific response of clones 4A7 and 4E5 to P501 S-
transduced
autologous fibroblasts was blocked by pre-incubation with 25ug/ml W6/32 (pan-
Class I
blocking antibody) and B 1.23.2 (HLA-B/C blocking antibody). These results
demonstrate that the SEQ ID NO: 712-specific response is restricted to an HLA-
B or
HLA-C allele.
For the HLA mismatch analysis, autologous B-LCL (HLA-
A1,A2,B8,B51, Cwl, Cw7) and heterologous B-LCL (HLA-
A2,A3,B18,BS1,Cw5,Cw14) that share the HLAB51 allele were pulsed for one hour
with 20ug/ml of peptide of SEQ ID NO: 712, washed, and tested in y-IFN Elispot
assays
for the ability to stimulate clones 4A7 and 4E5. Antibody blocking assays with
the
B 1.23.2 (HLA-B/C blocking antibody) were also performed. SEQ ID NO: 712-
specific
response was detected using both the autologous (D326) and heterologous (D107)
B-
LCL, and furthermore the responses were blocked by pre-incubation with 25ug/ml
of
B 1.23.2 HLA-B/C blocking antibody. Together these results demonstrate that
the
P501 S-specific response to the peptide of SEQ ID NO: 712 is restricted to the
HLA-
B51 class I allele. Molecular cloning and sequence analysis of the HLA-B51
allele from
D3326 revealed that the HLA-B51 subtype of D326 is HLA-B51011.
Based on the 10-mer P501 S-derived epitope of SEQ ID NO: 712, two 9-
mers with the sequences of SEQ ID NO: 714 and 715 were synthesized and tested
in
Elispot assays for the ability to stimulate two P501 S-specific CTL clones
derived from
line 2A2. The 10-mer peptide of SEQ ID NO: 712, as well as the 9-mer peptide
of SEQ
ID NO: 715, but not the 9-mer peptide of SEQ ID NO: 714, were capable of
stimulating
the P501 S-specific CTL to produce IFN-gamma. These results demonstrate that
the
peptide of SEQ ID NO: 715 is a 9-mer P501 S-derived epitope recognized by P501
S-
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
158
specif c CTL. The DNA sequence encoding the epitope of SEQ ID NO: 715 is
provided
in SEQ ID NO: 716.
To identify the class I restricting,allele for the P501S-derived peptide of
SEQ ID NO: 712 and 715 specific response, each of the HLA B and C alleles were
cloned from the donor used in the in vitro priming experiment. Sequence
analysis
indicated that the relevant alleles were HLA-B8, HLA-B51, HLA-Cw01 and HLA-
Cw07. Each of these alleles were subcloned into an expression vector and co-
transfected together with the P501 S gene into VA-13 cells. Transfected VA-13
cells
were then tested for the ability to specifically stimulate the P501 S-specific
CTL in
ELISPOT assays. VA-13 cells transfected with P501 S and HLA-B51 were capable
of
stimulating the P501 S-specific CTL to secrete gamma-IFN. VA-13 cells
transfected
with HLA-B51 alone or PSO1S + the other HLA-alleles were not capable of
stimulating
the P501 S-specific CTL. These results demonstrate that the restricting allele
for the
P501 S-specific response is the HLABS I allele. Sequence analysis revealed
that the
subtype of the relevant restricting allele is HLA-B51011.
To determine if the P501 S-specific CTL could recognize prostate tumor
cells that express P501 S, the P501 S-positive lines LnCAP and C1ZL,2422 (both
expressing "moderate" amounts of P501 S mRNA and protein), and PC-3
(expressing
low amounts of P501 S mRNA and protein), plus the P501 S-negative cell line DU-
145
were retrovirally transduced with the HLA-B51011 allele that was cloned from
the
donor used 'to generate the P501 S-specific CTL. HLA-B51 O l 1- or EGFP-
transduced
and selected tumor cells were treated with gamma-interferon and androgen (to
upregulate stimulatory functions and P501 S, respectively) and used in gamma
interferon Elispot assays with the P501 S-specific CTL clones 4E5 and 4E7.
Untreated
cells were used as a control.
Both 4E5 and 4E7 efficiently and specifically recognized LnCAP and
CRL2422 cells that were transduced with the HLA-B51011 allele, but not the
same cell
lines transduced with EGFP. Additionally, both CTL clones specifically
recognized
PC-3 cells transduced with HLA-B51011, but not the P501 S-negative tumor cell
line
DU-145. Treatment with gamma-interferon or androgen did not enhance the
ability of
CTL to recognize tumor cells. These results demonstrate that P501 S-specific
CTL,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
159
generated by in vitro whole gene priming, specifically and efficiently
recognize prostate
tumor cell lines that express P501 S.
A naturally processed CD4 epitope of P501 S was identified as follows.
CD4 cells specific for P501 S were prepared as described above. A series
of 16 overlapping peptides were synthesized that spanned approximately 50% of
the
amino terminal portion of the P501 S gene (amino acids 1- 325 of SEQ ID NO:
113).
For priming, peptides were combined into pools of 4 peptides, pulsed at 4
~glml onto
dendritic cells (DC) for 24 hours, with TNF-alpha. DC were then washed and
mixed
with negatively selected CD4+ T cells in 96 well U-bottom plates. Cultures
were re-
stimulated weekly on fresh DC loaded with peptide pools. Following a total of
4
stimulation cycles, cells were rested for an additional week and tested for
specificity to
APC pulsed with peptide pools using y-IFN ELISA and proliferation assays. For
these
assays, adherent monocytes loaded with either the relevant peptide pool at
4ug/ml or an
irrelevant peptide at ~,g/ml were used as APC. T cell lines that demonstrated
either
specific cytokine secretion or proliferation were then tested for recognition
of individual
peptides that were present in the pool. T cell lines could be identified from
pools A and
B that recognized individual peptides from these pools.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
160
From pool A, lines AD9 and AE10 specifically recognized peptide 1
(SEQ ID NO: 719), and line AF5 recognized peptide 39 (SEQ ID NO: 718). From
pool
B, line BC6 could be identified that recognized peptide 58 (SEQ ID NO: 717).
Each of
these lines were stimulated on the specific peptide and tested for specific
recognition of
the peptide in a titration assay as well as cell lysates generated by
infection of HEK 293
cells with adenovirus expressing either P501 S or an irrelevant antigen. For
these assays,
APC-adherent monocytes were pulsed with either 10, 1, or 0.1 ~g/ml individual
P501S
peptides, and DC were pulsed overnight with a 1:5 dilution of adenovirally
infected cell
lysates. Lines AD9, AE10 and AF5 retained significant recognition of the
relevant
P501 S-derived peptides even at 0.1 mg/ml. Furthermore, line AD9 demonstrated
significant (8.1 fold stimulation index) specific activity for lysates from
adenovirus-
P501 S infected cells. These results demonstrate that high affinity CD4 T cell
lines can
be generated toward P501 S-derived epitopes, and that at least a subset of
these T cells
specific for the P501 S derived sequence of SEQ ID NO: 719 are specific for an
epitope
that is naturally processed by human cells. The DNA sequences encoding the
amino
acid sequences of SEQ ID NO: 717-719 are provided in SEQ ID NO: 720-722,
respectively.
To further characterize the P501 S-specific activity of AD9, the line was
cloned using anti-CD3. Three clones, referred to as 1A1, 1A9 and IFS, were
identified
that were specific for the P501 S-1 peptide (SEQ ID NO: 719). To determine the
HLA
restriction allele for the P501 S-specific response, each of these clones was
tested in
class II antibody blocking and HLA mismatch assays using proliferation and
gamma-
interferon assays. In antibody blocking assays and measuring gamma-interferon
production using ELISA assays, the ability of all three clones to recognize
peptide
pulsed APC was specifically blocked by co-incubation with either a pan-class
II
blocking antibody or a HLA-DR blocking antibody, but not with a HLA-DQ or an
irrelevant antibody. Proliferation assays performed simultaneously with the
same cells
confirmed these results. These data indicate that the P501 S-specific response
of the
clones is restricted by an HLA-DR allele. Further studies demonstrated that
the
restricting allele for the PSOlS-specific response is HLA-DRB1501.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
161
EXAMPLE 13
IDENTIFICATION OF PROSTATE-SPECIFIC ANTIGENS
BY MICROARRAY ANALYSIS
This Example describes the isolation of certain prostate-specific
polypeptides from a prostate tumor cDNA library.
A human prostate tumor cDNA expression library as described above
was screened using microarray analysis to identify clones that display at
least a three
fold over-expression in prostate tumor and/or normal prostate tissue, as
compared to
non-prostate normal tissues (not including testis). 372 clones were
identified, and 319
were successfully sequenced. Table I presents a summary of these clones, which
are
shown in SEQ ID NOs:385-400. Of these sequences SEQ ID NOs:386, 389, 390 and
392 correspond to novel genes, and SEQ ID NOs: 393 and 396 correspond to
previously
identified sequences. The others (SEQ ID NOs:385, 387, 388, 391, 394, 395 and
397-
I 5 400) correspond to known sequences, as shown in Table I.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
162
Table I
Summary of Prostate Tumor Antigens
Known Genes Previously IdentifiedNovel Genes
Genes
T-cell gamma chain P504S 23379 (SEQ ID
N0:389)
Kallikrein PI OOOC 23399 (SEQ ID
N0:392)
Vector P501 S 23320 (SEQ 1D
N0:386)
CGI-82 protein mRNA (23319; P503S 23381 (SEQ ID
SEQ ID N0:385) N0:390)
PSA PS10S
Ald. 6 Dehyd. P784P
L-iditol-2 dehydrogenase (23376;P502S
SEQ ID N0:388)
Ets transcription factor PDEFP706P
(22672; SEQ ID
N0:398)
hTGR (22678; SEQ ID NO:399) 19142.2, bangur.seq
(22621;
SEQ ID N0:396)
KIAA0295(22685; SEQ ID N0:400)5566.1 Wang (23404;
SEQ ID N0:393)
Prostatic Acid Phosphatase(22655;P712P
SEQ ID
N0:397)
transglutaminase (22611; SEQ P778P
ID N0:395)
HDLBP (23508; SEQ ID N0:394)
CGI-69 Protein(23367; SEQ
ID N0:387)
KIAA0122(23383; SEQ ID N0:391)
TEEG
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
163
CGI-82 showed 4.06 fold over-expression in prostate tissues as
compared to other normal tissues tested. It was over-expressed in 43% of
prostate
tumors, 25% normal prostate, not detected in other normal tissues tested. L-
iditol-2
dehydrogenase showed 4.94 fold over-expression in prostate tissues as compared
to
other normal tissues tested. It was over-expressed in 90% of prostate tumors,
100% of
normal prostate, and not detected in other normal tissues tested. Ets
transcription factor
PDEF showed 5.55 fold over-expression in prostate tissues as compared to other
normal
tissues tested. It was over-expressed in 47% prostate tumors, 25% normal
prostate and
not detected in other normal tissues tested. hTGRl showed 9.11 fold over-
expression in
prostate tissues as compared to other normal tissues tested. It was over-
expressed in
63% of prostate tumors and is not detected in normal tissues tested including
normal
prostate. I~IAA0295 showed 5.59 fold over-expression in prostate tissues as
compared
to other normal tissues tested. It was over-expressed in 47% of prostate
tumors, low to
undetectable in normal tissues tested including normal prostate tissues.
Prostatic acid
phosphatase showed 9.14 fold over-expression in prostate tissues as compared
to other
normal tissues tested. It was over-expressed in 67% of prostate tumors, 50% of
normal
prostate, and not detected in other normal tissues tested. Transglutaminase
showed
14.84 fold over-expression in prostate tissues as compared to other normal
tissues
tested. It was over-expressed in 30% of prostate tumors, 50% of normal
prostate, and is
not detected in other normal tissues tested. High density lipoprotein binding
protein
(HDLBP) showed 28.06 fold over-expression in prostate tissues as compared to
other
normal tissues tested. It was over-expressed in 97% of prostate tumors, 75% of
normal
prostate, and is undetectable in all other normal tissues tested. CGI-69
showed 3.56
fold over-expression in prostate tissues as compared to other normal tissues
tested. It is
a low abundant gene, detected in more than 90% of prostate tumors, and in 75%
normal
prostate tissues. The expression of this gene in normal tissues was very low.
KIAA0122 showed 4.24 fold over-expression in prostate tissues as compared to
other
normal tissues tested. It was over-expressed in 57% of prostate tumors, it was
undetectable in all normal tissues tested including normal prostate tissues.
19142.2
bangur showed 23.25 fold over-expression in prostate tissues as compared to
other
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
164
normal tissues tested. It was over-expressed in 97% of prostate tumors and
100% of
normal prostate. It was undetectable in other normal tissues tested. .5566..1
Wang
showed 3.31 fold over-expression in prostate tissues as compared to other
normal
tissues tested. It was over-expressed in 97% of prostate tumors, 75% normal
prostate
and was also over-expressed in noiTnal bone marrow, pancreas, and activated
PBMC.
Novel clone 23379 (also referred to as P553S) showed 4.86 fold over-expression
in
prostate tissues as compared to other normal tissues tested. It was detectable
in 97% of
prostate tumors and 75% normal prostate and is undetectable in all other
normal tissues
tested. Novel clone 23399 showed 4.09 fold over-expression in prostate tissues
as
compared to other normal tissues tested. It was over-expressed in 27% of
prostate
tumors and was undetectable in all normal tissues tested including normal
prostate
tissues. Novel clone 23320 showed 3.15 fold over-expression in prostate
tissues as
compared to other normal tissues tested. It was detectable in all prostate
tumors and
50% of normal prostate tissues. It was also expressed in normal colon and
trachea.
Other normal tissues do not express this gene at high level.
Subsequent full-length cloning studies on P553S, using standard
techniques, revealed that this clone is an incomplete spliced form of PSO1S.
The
determined, cDNA sequences for four splice variants of P553S are provided in
SEQ ID
NO: 623-626. An amino acid sequence encoded by SEQ ID NO: 626 is provided in
SEQ ID NO: 627. The cDNA sequence of SEQ ID NO: 623 was found to contain two
open reading frames (ORFs). The amino acid sequences encoded by these two ORFs
are provided in SEQ ID NO: 628 and 629.
EXAMPLE 14
IDENTIFICATION OF PROSTATE-SPECIFIC ANTIGENS
BY ELECTRONIC SUBTRACTION
This Example describes the use of an electronic subtraction technique to
identify prostate-specific antigens. .
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
I65
Potential prostate-specific genes present in the GenBank human EST
database were identified by electronic subtraction (similar to that described
by
Vasmatizis et al., Proc. Natl. Acad. Sci. LISA 95:300-304, 1998). The
sequences of EST
clones (43,482) derived from various prostate libraries were obtained from the
GenBank
public human EST database. Each prostate EST sequence was used as a query
sequence
in a BLASTN (National Center for Biotechnology Information) search against the
human EST database. All matches considered identical (length of matching
sequence
>100 base pairs, density of identical matches over this region > 70%) were
grouped
(aligned) together in a cluster. Clusters containing more than 200 ESTs were
discarded
IO since they probably represented repetitive elements or highly expressed
genes such as
those for ribosomal proteins. If two or more clusters shared common ESTs,
those
clusters were grouped together into a "supercluster," resulting in 4,345
prostate
superclusters.
Records for the 479 human cDNA libraries represented in the GenBank
release were downloaded to create a database of these cDNA library records.
These 479
cDNA libraries were grouped into three groups: Plus (normal prostate and
prostate
tumor libraries, and breast cell line libraries, in which expression was
desired), Minus
(libraries from other normal adult tissues, in which expression was not
desirable), and
Other (libraries from fetal tissue, infant tissue, tissues found only in
women, non-
prostate tumors and cell lines other than prostate cell lines, in which
expression was
considered to be irrelevant). A summary of these library groups is presented
in Table II.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
166
Table II
Prostate cDNA Libraries and ESTs
Library # of Libraries# of ESTs
Plus 25 43,482
Normal 11 18,875
Tumor 11 21,769
Cell lines 3 2,838
Minus 166
Other 287
Each supercluster was analyzed in terms of the ESTs within the
supercluster. The tissue source of each EST clone was noted and used to
classify the
superclusters into four groups: Type 1- EST clones found in the Plus group
libraries
only; no expression detected in Minus or Other group libraries; Type 2- EST
clones
derived from the Plus and Other group libraries only; no expression detected
in the
Minus group; Type 3- EST clones derived from the Plus, Minus and Other group
libraries, but the number of ESTs derived from the Plus group is higher than
in either
the Minus or Other groups; and Type 4- EST clones derived from Plus, Minus and
Other group libraries, but the number derived from the Plus group is higher
than the
number derived from the Minus group. This analysis identified 4,345 breast
clusters
(see Table III). From these clusters, 3,172 EST clones were ordered from
Research
Genetics, Inc., and were received as frozen glycerol stocks in 96-well plates.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
167
Table III
Prostate Cluster Summary
# of # of ESTs
Type SuperclustersOrdered
1 688 677
2 2899 2484
3 85 11
4 673 0
Total 4345 3172
The EST clone inserts were PCR-amplified using amino-linked PCR
primers for Synteni microarray analysis. When more than one PCR product was
obtained for a particular clone, that PCR product was not used for expression
analysis.
In total, 2,528 clones from the electronic subtraction method were analyzed by
microarray analysis to identify electronic subtraction breast clones that had
high levels
of tumor vs. normal tissue mRNA. Such screens were performed using a Synteni
(Palo
Alto, CA) microarray, according to the manufacturer's instructions (and
essentially as
described by Schena et al., P~oc. Natl. Acad. Sci. USA 93:10614-10619, 1996
and
Heller et al., Proc. Natl. Acad. Sci. USA 94:2150-2155, 1997). Within these
analyses,
the clones were arrayed on the chip, which was then probed with fluorescent
probes
1 S generated from normal and tumor prostate cDNA, as well as various other
normal
tissues. The slides were scanned and the fluorescence intensity was measured.
Clones with an expression ratio greater than 3 (i. e., the level in prostate
tumor and normal prostate mRNA was at least three times the level in other
normal
tissue mRNA) were identified as prostate tumor-specific sequences (Table IV).
The
sequences of these clones are provided in SEQ ID NO: 401-453, with certain
novel
sequences shown in SEQ ID NO: 407, 413, 416-419, 422, 426, 427 and 450.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
168
Table IV
Prostate-tumor Specific Clones
SEQ ID NO. Sequence Comments
Designation
401 22545 reviously identified P1000C
402 22547 reviously identified P704P
403 22548 known
404 22550 known
405 22551 PSA
406 22552 rostate secretory rotein 94
40 2 novel
7 2553
_ _ reviously identified P509S
40 _
8 22558
_ 22562 landular kallikrein
409
410 22565 reviously identified P1000C
411 22567 PAP
412 22568 B 10060 (breast tumor antigen)
413 22570 novel
414 22571 PSA
415 22572 reviously identified P706P
416 22573 novel
417 22574 novel
418 22575 novel
419 22580 novel
420 22581 PAP
42I 22582 rostatic secretory rotein 94
422 22583 novel
423 22584 rostatic secretory rotein 94
424 22585 prostatic secretory rotein
94
425 22586 known
426 22587 novel
427 22588 novel
428 22589 PAP
429 22590 known
430 22591 PSA
431 22592 known
432 22593 Previously identified P777P
433 22594 T cell rece for gamma chain
434 22595 Previously identified P705P
435 22596 Previously identified P707P
436 22847 PAP
43 7 22848 known
438 22849 prostatic secretory protein
57
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
169
439 22851 PAP
440 22852 PAP
441 22853 PAP
442 22854 reviously identified P509S
443 22855 reviously identified P705P
444 22856 reviously identified P774P
445 22857 PSA
446 23601 reviously identified P777P
447 23602 PSA
448 23605 PSA
449 23606 PSA
450 23612 novel
451 23614 PSA
452 23618 reviously identified P1000C
453 23622 previously identified P705P
Further studies on the clone of SEQ ID NO: 407 (also referred to as
P1020C) led to the isolation of an extended cDNA sequence provided in SEQ ID
NO:
591. This extended cDNA sequence was found to contain an open reading frame
that
encodes the predicted amino acid sequence of SEQ ID NO: 592. The P1020C cDNA
and amino acid sequences were found to show some similarity to the human
endogenous retroviral HERV-I~ pol gene and protein.
EXAMPLE 15
1 O FURTHER IDENTIFICATION OF PROSTATE-SPECIFIC ANTIGENS BY MICROARRAY
ANALYSIS
This Example describes the isolation of additional prostate-specific
polypeptides from a prostate tumor cDNA library.
A human prostate tumor cDNA expression library as described above
was screened using microarray analysis to identify clones that display at
least a three
fold over-expression in prostate tumor and/or normal prostate tissue, as
compared to
non-prostate normal tissues (not including testis). 142 clones were identified
and
sequenced. Certain of these clones are shown in SEQ ID NO: 454-467. Of these
sequences, SEQ ID NO: 459-460 represent novel genes. The others (SEQ ID NO:
454-
458 and 461-467) correspond to known sequences. Comparison of the determined
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
170
cDNA sequence of SEQ ID NO: 461 with sequences in the Genbank database using
the
BLAST program revealed homology to the previously identified transmembrane
protease serine 2 (TMPRSS2). The full-length cDNA sequence for this clone is
provided in SEQ ID NO: 751, with the corresponding amino acid sequence being
provided in SEQ ID NO: 752. The cDNA sequence encoding the first 209 amino
acids
of TMPRSS2 is provided in SEQ ID NO: 753, with the first 209 amino acids being
provided in SEQ ID NO: 754.
The sequence of SEQ ID NO: 462 (referred to as P835P) was found to
correspond to the previously identified clone FLJ13518 (Accession AK023643;
SEQ ID
NO: 774), which had no associated open reading frame (ORF). This clone was
used to
search the Geneseq DNA database and matched a clone previously identified as a
G
protein-coupled receptor protein (DNA Geneseq Accession A09351; amino acid
Geneseq Accession Y92365), that is characterized by the presence of seven
transmembrane domains. The sequences of fragments between these domains are
provided in SEQ ID NO: 778-785, with SEQ ID NO: 778, 780, 782 and 784
representing extracellular domains and SEQ ID NO: 779, 781, 783 and 785
representing
intracellular domains. SEQ ID NO: 778-785 represent amino acids 1-28, 53-61,
83-
103, 124-143, 165-201, 226-238, 263-272 and 297-381, respectively, of P835P.
The
full-length cDNA sequence for P835P is provided in SEQ ID NO: 773. The cDNA
sequence of the open reading frame for P835P, including stop codon, is
provided in
SEQ ID NO: 775, with the open reading frame without stop codan being provided
in
SEQ ID NO: 776 and the corresponding amino acid sequence being provided in SEQ
ID
NO: 777.
EXAMPLE 16
FURTHER CHARACTERIZATION OF PROSTATE-SPECIFIC ANTIGEN P7I OP
This Example describes the full length cloning of P71 OP.
The prostate cDNA library described above was screened with the P710P
fragment described above. One million colonies were plated on LB/Ampicillin
plates.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
I71
Nylon membrane filters were used to lift these colonies, and the cDNAs picked
up by
these filters were then denatured and cross-linked to the filters by UV light.
The P710P
fragment was radiolabeled and used to hybridize with the filters. Positive
cDNA clones
were selected and their cDNAs recovered and sequenced by an automatic Perkin
Elmer/Applied Biosystems Division Sequencer. Four sequences were obtained, and
are
presented in SEQ ID NO: 468-471. These sequences appear to represent different
splice
variants of the P71 OP gene. Subsequent comparison of the cDNA sequences of
P710P
with those in Genbank revealed homology to the DD3 gene (Genbank accession
numbers AF103907 & AF103908). The cDNA sequence of DD3 is provided in SEQ ID
NO: 618.
EXAMPLE 17
PROTEIN EXPRESSION OF PROSTATE-SPECIFIC ANTIGENS
This example describes the expression and purification of prostate-
specific antigens in E. coli, baculovirus, mammalian and yeast cells.
a) Expression of P501 S in E, coli
Expression of the full-length form of P501 S was attempted by first
cloning P501 S without the leader sequence (amino acids 36-553 of SEQ ID NO:
113)
downstream of the first 30 amino acids of the M. tuberculosis antigen Ral2
(SEQ ID
NO: 484) in pETl7b. Specifically, PSOlS DNA was used to perform PCR using the
primers AW025 (SEQ ID NO: 485) and AW003 (SEQ ID NO: 486). AW025 is a sense
cloning primer that contains a HindIIT site. AW003 is an antisense cloning
primer that
contains an EcoRI site. DNA amplification was performed using 5 ~l lOX Pfu
buffer, 1
x.120 mM dNTPs, 1 ~1 each of the PCR primers at 10 ~.M concentration, 40 ~1
water, 1
~1 Pfu DNA polymerase (Stratagene, La Jolla, CA) and 1 ~1 DNA at 100 ng/~1.
Denaturation at 95°C was performed for 30 sec, followed by 10 cycles of
95°C for 30
sec, 60°C for 1 min and by 72°C for 3 min. 20 cycles of
95°C for 30 sec, 65°C for 1 min
and by 72°C for 3 min, and lastly by 1 cycle of 72°C for 10 min.
The PCR product was
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
172
cloned to Ral2m/pETl7b using HindIII and EcoRI. The sequence of the resulting
fusion construct (referred to as Ral2-P501 S-F) was cbnfirmed by DNA
sequencing.
The fusion construct was transformed into BL21 (DE3)pLysE, pLysS and
CodonPlus E, coli (Stratagene) and grown overnight in LB broth with kanamycin.
The
resulting culture was induced with IPTG. Protein was transferred to PVDF
membrane
and blocked with 5% non-fat milk (in PBS-Tween buffer), washed three times and
incubated with mouse anti-His tag antibody (Clontech) for 1 hour. The membrane
was
washed 3 times and probed with HRP-Protein A (Zymed) for 30 min. Finally, the
membrane was washed 3 times and developed with ECL (Amersham). No expression
was detected by Western blot. Similarly, no expression was detected by Western
blot
when the Ral2-P501 S-F fusion was used for expression in ,BL21 CodonPlus by
CE6
phage (Irivitrogen).
An N-terminal fragment of P501 S (amino acids 36-325 of SEQ ID NO:
113) was cloned down-stream of the first 30 amino acids of the M. tuberculosis
antigen
RaI2 in pETl7b as follows. PSOlS DNA was used to perform PCR using the primers
AW025 (SEQ ID NO: 485) and AW027 (SEQ ID NO: 487). AW027 is an antisense
cloning primer that contains an EcoRI site and a stop codon. DNA amplification
was
performed essentially as described above. The resulting PCR product was cloned
to
Ral2 in pETl7b at the HindIII and EcoRI sites. The fusion construct (referred
to as
Ral2-P501 S-N) was confirmed by DNA sequencing.
The Ral2-P501 S-N fusion construct was used for expression in
BL21 (DE3)pLysE, pLysS and CodonPlus, essentially as described above. Using
Western blot analysis, protein bands were observed at the expected molecular
weight of
36 kDa. Some high molecular weight bands were also observed, probably due to
aggregation of the recombinant protein. No expression was detected by Western
blot
when the Ral2-PSO1S-F fusion was used for expression in BL2lCodonPlus by CE6
phage.
A fusion construct comprising a C-terminal portion of P501 S (amino
acids 257-553 of SEQ ID NO: 113) located down-stream of the first 30 amino
acids of
the M. tubey~culosis antigen Ral2 (SEQ ID NO: 484) was prepared as follows.
P501 S
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
173
DNA was used to perform PCR using the primers AW026 (SEQ ID NO: 488) and
AW003 (SEQ ID NO: 486). AW026 is a sense cloning primer that contains a
HindIII
site. DNA amplification was performed essentially as described above. The
resulting
PCR product was cloned to Ral2 in pETl7b at the HindIII and EcoRI sites. The
sequence for the fusion construct (referred to as Ral2-PSOI S-C) was
confirmed.
The Ral2-PSOI S-C fusion construct was used for expression in
BL21(DE3)pLysE, pLysS and CodonPlus, as described above. A small amount of
protein was detected by Western blot, with some molecular weight aggregates
also
being observed. Expression was also detected by Western blot when the Ral2-
P501 S-C
fusion was used for expression in BL21 CodonPlus induced by CE6 phage.
A fusion construct comprising a fragment of P501 S (amino acids 36-298
of SEQ ID NO: 113) located down-stream of the M. tuberculosis antigen Ral2
(SEQ ID
NO: 705) was prepared as follows. P501 S DNA was used to perform PCR using the
primers AW042 (SEQ ID NO: 706) and AW053 (SEQ ID NO: 707). AW042 is a sense
I S cloning primer that contains a EcoRI site. AW053 is an antisense primer
with stop and
Xho I sites. DNA amplification was performed essentially as described above.
The
resulting PCR product was cloned to RaI2 in pETl7b at the EcoRI and Xho I
sites. The
resulting fusion construct (referred to as Ral2-P501 S-E2) was expressed in
B834 (DE3)
pLys S E. coli host cells in TB media for 2 h at room temperature. Expressed
protein
was purified by washing the inclusion bodies and running on a Ni-NTA column.
The
purified protein stayed soluble in buffer containing 20 mM Tris-HCl (pH 8),
100 mM
NaCI, 10 mM (3-Me and 5% glycerol. The determined cDNA and amino acid
sequences
for the expressed fusion protein are provided in SEQ ID NO: 708 and 709,
respectfully.
bL Expression of P501 S in Baculovirus
The Bac-to-Bac baculovirus expression system (BRL Life Technologies,
Inc.) was used to express P501 S protein in insect cells. Full-length P501 S
(SEQ ID
NO: 1 I3) was amplified by PCR and cloned into the Xbal site of the donor
plasmid
pFastBacI. The recombinant bacmid and baculovirus were prepared according to
the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
174
manufacturer's instructions. The recombinant baculovirus was amplified in Sf~
cells
and the high titer viral stocks were utilized to infect High Five cells
(Invitrogen) to
make the recombinant protein. The identity of the full-length protein was
confirmed by
N-terminal sequencing of the recombinant protein and by Western blot analysis
(Figure
7). Specifically, 0.6 million High Five cells in 6-well plates were infected
with either
the unrelated control virus BV/ECD PD (lane 2), with recombinant baculovirus
for
P501 S at different amounts or MOIs (lanes 4-8), or were uninfected (lane 3).
Cell
lysates were run on SDS-PAGE under reducing conditions and analyzed by Western
blot with the anti-P501 S monoclonal antibody P501 S-10E3-G4D3 (prepared as
described below). Lane 1 is the biotinylated protein molecular weight marker
(BioLabs).
The localization of recombinant P501 S in the insect cells was
investigated as follows. The insect cells overeXpressing PSOI S were
fractionated into
fractions of nucleus, mitochondria, membrane and cytosol. Equal amounts of
protein
from each fraction were analyzed by Western blot with a monoclonal antibody
against
P501 S. Due to the scheme of fractionation, both nucleus and mitochondria
fractions
contain some plasma membrane components. However, the membrane fraction is
basically free from mitochondria and nucleus. P501 S was found to be present
in all
fractions that contain the membrane component, suggesting that P501 S may be
associated with plasma membrane of the insect cells expressing the recombinant
protein.
c) Expression of P501 S in. Mammalian Cells
Full-length P501 S (553 amino acids; SEQ ID NO: 113) was cloned into
various mammalian expression vectors, including pCEP4 (Invitrogen), pVR1012
(Vical, San Diego, CA) and a modified form of the retroviral vector pBMN,
referred to
as pBIB. Transfection of P501 S/pCEP4 and P501 S/pVR1012 into HEK293
fibroblasts
was carried out using the Fugene transfection reagent (Boehringer Mannheim).
Briefly,
2 u1 of Fugene reagent was diluted into 100 u1 of serum-free media and
incubated at
room temperature for 5-10 min. This mixture was added to 1 ug of PSO1S plasmid
DNA, mixed briefly and incubated for 30 minutes at room temperature. The
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
175
Fugene/DNA mixture was added to cells and incubated for 24-48 hours.
Expression of
recombinant PSOlS in transfected HEK293 fibroblasts was detected by means of
Western blot employing a monoclonal antibody to P501 S.
Transfection of p501 S/pCEP4 into CHO-K cells (American Type
Culture Collection, Rockville, MD) was carried out using GenePorter
transfection
reagent (Gene Therapy Systems, San Diego, CA). Briefly, 15 ~I of GenePorter
was
diluted in 500 ~,l of serum-free media and incubated at room temperature for
10 min.
The GenePorter/media mixture was added to 2 ~.g of plasmid DNA that was
diluted in
500 ~.l of serum-free media, mixed briefly and incubated for 30 min at room
temperature. CHO-K cells were rinsed in PBS to remove serum proteins, and the
GenePorter/DNA mix was added and incubated for 5 hours. The transfected cells
were
then fed an equal volume of 2x media and incubated for 24-48 hours.
FACS analysis of PSO1S transiently infected CHO-K cells, demonstrated
surface expression of P501 S. Expression was detected using rabbit polyclonal
antisera
xaised against a P501 S peptide, as described below. Flow cytometric analysis
was
performed using a FaCScan (Becton Dickinson), and the data were analyzed using
the
Cell Quest program.
dl Expression of P501 S in S. cep°evisiae
P501 S was expressed in yeast, directed in membranes, using the yeast a
prepro signal sequence. The natural signal sequence and first lumenal domain
of PSOlS
was deleted in order to conserve the natural positioning of the expressed P501
S protein.
Specifically, the a prepro signal sequence of S. cerevisiae linked to
amino acids 55-553 of SEQ ID NO: 113 with a His tag tail was cloned into the
plasmid
pRIT15068 with the CUPl promoter and transfected into S. ce~evisiae strain
Y1790.
The Y1790 strain is Leu+ and His-. Expression of protein was induced by
addition of
either 500 p,M or 250 ~,M of CuS04 at 30 °C in minimal medium
supplemented with
histidine. Cells were harvested 24 hours after induction. Extracts were
prepared by
growing cells to a concentration of OD600 5.0 in 50 mM citrate phosphate
buffer (pH
4.0) plus 130 mM NaCI supplemented with protease inhibitors. Cells were
disrupted
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
176
using glass beads and centrifuged for 20 min at 15,000 g. The recombinant
protein was
found to be 100% pellet associated.
Expression of the recombinant protein (molecular weight 63 kD) was
demonstrated by Western blot analysis, using the anti-P501 S monoclonal
antibody 10E-
D4-G3 described below. The amino acid sequence of the expressed protein is
provided
in SEQ ID NO: 792.
Fermentation processes for the production of the a, prepro-P501 S-His tag
recombinant protein in S cerevisiae (strain Y1790 - CUPI inducible promoter)
were
evaluated as follows. One hundred ~,l of a master seed containing 2.5 x 108
cells/mI of
transformed S ce~evisiae YI790 were spread on FSC004AA solid medium. The
composition of the FSC004AA medium is as follows: glucose 10 g/1; Na2Mo04.2H20
0.0002 g/1; folic acid 0.000064 g/1; KHZP04 1 g/1; MnSO4.H20 0.0004 g/1;
Inositol
0.064 g/1; MgS04.7H20 0.5 g/1; H3B03 0.0005 g/1; Pyridoxine 0.008 g/1;
CaC12.2H20
0.1 g/1; KI 0.0001 g/1; Thiamine 0.008 g/1; NaCI 0.1 g/1; CoC12.6H20 0.00009
g/1;
Niacin 0.000032 g/1; FeC13.6H20 0.0002 g/1; Riboflavin 0.000016 g/1;
Panthotenate Ca
0.008 g/1; CuS04.5H2O 0.00004 g/1; Biotin 0.000064 g/1; para-aminobenzoic acid
0.000016 g/1; ZnSO4.7H20 0.0004 g/1; (NH4)2SO4 5 g/1; agar 18 g/1; Histidine
0.1 g/1.
Two plates were incubated for 26 h at 30 °C. These solid pre-
cultures
were harvested in 5 ml of liquid medium FSC007AA and 0.5 rnl (or 9.3 x 107
cells) of
this suspension was used to inoculate 2 liquid pre-cultures.
The composition of the FSC007AA medium is as follows: Glucose 10
g/1; Na2Mo0~.2H20 0.0002 g/1; folic acid 0.000064 g/1; KH2PO4 1 g/1; MnS04.H20
0.0004 g11; Inositol 0.064 g/1; MgS04.7H20 0.5 g/1; H3BO3 0.0005 g/1;
Pyridoxine 0.008
g11; CaC12.2H20 0.1 g/1; KI 0.0001 g/1; Thiamine 0.008 g/1; NaCI 0.1 g/1;
CoC12.6H20
0.00009 g11; Niacine 0.000032 g/1; FeC13.6H20 0.0002 g/1; Riboflavin 0.000016
g/1;
Panthotenate Ca 0.008 g/1; CuS04.5H20 0.00004 g/1; Biotin 0.000064 g/1; para-
aminobenzoic acid 0.000016 g/1; ZnS04.7Ha0 0.0004 g/1; (NHQ)2SO4 5 g/1;
Histidine
0.1 g/I.
These pre-cultures were run for 20 hours in 2L flasks containing 400 ml
of medium FSC007AA in order to obtain an OD of I.8. The other characteristics
of
these pre-cultures are as follows: pH 2.8; .glucose 2.3 .~/L; ethanol 3.4 g~.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
177
The best timing for liquid pre-cultures for strain YI790 was determined
in preliminary experiments. Liquid pre-cultures containing 400 ml of medium
and
inoculated with various volumes of Master Seed (0.25, 0.5, 1 or 2 ml) were
monitored
in order to identify the best inoculum size and timing. Glucose, ethanol, pH,
OD and
cell number (determined by flow cytometry) were followed between 16 and 23
hours of
culture. Glucose exhaustion and maximal biomass were obtained after 20 hour
incubation with 0.5 inoculum. These conditions were adopted for transferring
the pre-
culture into fermentation.
In total, 800m1 of pre-culture were used to inoculate a 20 L fermenter
containing SL of medium FSC002AA. Three ml of irradiated antifoam were added
before inoculation. The composition of the FSC002AA medium is as follows:
(NH4)2SO4 6.4 g/1; Na2Mo04.2H20 2.05 mg/1; folic acid 0.54 mg/1; I~H2PO4 8.25
g/1;
MnS04.H20 4.1 mg/1; inositol S40 mg/; MgS04.7H20 4.69 g/1; H3B03 5.17 m/1;
pyridoxine 68 mg/l; CaC12.2H20 0.92 g/1; KI 1.03 mg/1; thiamine 68 mg/l; NaCI
0.06g11;
IS CoC12.6H20 0.92 mg/l; Niacine 0.27 mg/l; HCl 1 m1/1; FeC13.6H20 9.92 mg/1;
Riboflavin 0.13 mg/l; CuS04.5H2O 0.41 mg/l; Glucose 0.14 g/1; Panthotenate Ca
68
mg/1; ZnS04.7H20 4.1 mg/1; Biotin 0.54 mg/l; para-aminobenzoic acid O.I3 mg/l;
Histidine 0.3 g/1
The carbon source (glucose) was supplemented by a continuous feeding
of FFB004AA medium. The composition of the FFB004AA medium is as follows:
glucose 350 g/1; Na2Mo04.2H20 5.15 mg/l; folic acid 1.36 mgll; KH2PO4 20.6
g/1;
MnS04.H20 10.3 mg/1inositol 1350 mg/1; MgS04.7H20 11.7 g/1; H3B03 12.9 m/1;
pyridoxine 170 mg/1; CaC12.2H20 2.35 g/1; KI 2.6 mg/1; thiamine 170 g!1; NaCI
0.15 g/1;
CoC12.6H20 2.3 mg/1; niacine 0.67 mg/1; HCl 2.5 m1/1; FeC13.6H20 24.8 mg/1;
riboflavin; 0.33 mg/l; CuS04.5H20 I.03 mg/1; biotin 1.36 mg/1; panthotenate Ca
170
mg/l; ZnS04.7H20 10.3 mg/l; para-aminobenzoic acid: 0.33 mg/l; histidine 5.35
g/1.
The residual glucose concentration was maintained very low (~ 50 mg/L)
in order to minimize ethanol production by fermentation. This was achieved by
limiting
the development of the microorganism using a limited glucose feed rate. The
Standard
biomass content (OD 80-90) was reached in fermentation after 44 hour growth
phase.
CUPl promoter was then induced by adding SOO~CM CuS04 in order to
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
178
produce P501 S antigen. CuS04 addition was followed by ethanol accumulation
(up to 6
g/L), and the glucose feeding rate was then reduced in order to consume the
ethanol.
The copper available for the microorganism was monitored by testing Cu ion
concentration in the broth supernatant using a spectrophotometric copper assay
(DETC
method). The fermentation was then supplemented by CuS04 throughout the
induction
phase in order to maintain its concentration between 150 and 2S0 ~.M in the
supernatant. The biomass reached an OD of 100 at the end of induction. Cells
were
harvested after 8 hours of induction.
Cell homogenate was prepared and analysed by SDS-PAGE and Western
Blot using standard protocols. A major protein band with the expected
molecular weight
of 62KD was detected by Western blot using anti-P501 S monoclonal antibodies.
Western blot analysis also showed that the major 62KD band was progressively
produced from 30 minutes of induction on, and reached a maximum after 3 hours.
No
more antigen seemed to be produced between 3 and 12 hours of induction.
The number of passages through a French Press necessary to extract all
the antigen from the cells was evaluated. One, three and five passages were
tested and
total cell lysates, supernatants and pellets of cell lysates were analysed by
Western blot.
Three passages through a French Press were sufficient to completely extract
the antigen.
The antigen was present in the insoluble fraction.
expression of P703P in Baculovirus
The cDNA for full-length P703P-DES (SEQ ID NO: 326), together with
several flanking restriction sites, was obtained by digesting the plasmid
pCDNA703
with restriction endonucleases Xba I and Hind III. The resulting restriction
fragment
(approx. 800 base pairs) was ligated into the transfer plasmid pFastBacI which
was
digested with the same restriction enzymes. The sequence of the insert was
confirmed
by DNA sequencing. The recombinant transfer plasmid pFBP703 was used to make
recombinant bacmid DNA and baculovirus using the Bac-To-Bac Baculovirus
expression system (BRL Life Technologies). High Five cells were infected with
the
recombinant virus BVP703, as described above, to obtain recombinant P703P
protein.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
179
e) Expression of P788P in E. Coli
A truncated, N-terminal portion, of P788P (residues 1-644 of SEQ ID
NO: 777; referred to as P788P-N) fused with a C-terminal 6xHis Tag was
expressed in
E. coli as follows. P788P cDNA was amplified using the primers AW080 and AW081
(SEQ ID NO: 672 and 673). AW080 is a sense cloning primer with an Ndel site.
AW081 is an antisense cloning primer with a XhoI site. The PCR-amplified
P788P, as
well as the vector pCRXl, were digested with NdeI and XhoI. Vector and insert
were
ligated and transformed into NovaBIue cells. Colonies were randomly screened
for
insert and then sequenced. P788P-N clone #6 was confirmed to be identical to
the
designed construct. The expression construct P788P-N #6/pCRXl was transformed
into E. coli BL21 CodonPlus-RIL competent cells. After induction, most of the
cells
grew well, achieving OD600 of greater than 2.0 after 3 hr. Coomassie stained
SDS-
PAGE showed an over-expressed band at about 75 kD. Western blot analysis using
a
6xHisTag antibody confirmed the band was P788P-N. The determined cDNA sequence
for P788P-N is provided in SEQ ID NO: 674, with the corresponding amino acid
sequence being provided in SEQ ID NO: 675.
fl Expression of PS l OS in E. Coli
The PS l OS protein has 9 potential transmembrane domains and is
predicted to be located at the plasma membrane. The C-terminal protein of this
protein, as well as the predicted third extracellular domain of PS l OS were
expressed in
E. coli as follows.
The expression construct referred to as Ral2-P501 S-C was designed to
have a 6 HisTag at the N-terminal enc, followed by the M. tuberculosis antigen
Ral2
(SEQ ID NO: 676) and then the C-terminal portion of PS l OS (amino residues
1176-
1261 of SEQ ID NO: 538). Full-length PS l OS was used to amplify the PS l OS-C
fragment by PCR using the primers AW056 and AW057 (SEQ ID NO: 677 and 678,
respectively). AW056 is a sense cloning primer with an EcoRI site. AW057 is an
antisense primer with stop and XhoI sites. The amplified PSOIS fragment and
Ral2/pCRXl were digested with EcoRI and XhoI and then purified. The insert and
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
180
vector were ligated together and transformed into NovaBlue. Colonies were
randomly
screened for insert and sequences. For protein expression, the expression
construct was
transformed into E. coli BL21 (DE3) CodonPlus-RIL competent cells. A mini-
induction screen was performed to optimize the expression conditions. After
induction
the cells grew well, achieving OD 600 nm greater than 2.0 after 3 hours.
Coomassie
stain SDS-PAGE showed a highly over-expressed band at approx. 30 kD. Though
this
is higher than the expected molecular weight, western blot analysis was
positive,
showing this band to be the His tag-containing protein. The optimized culture
conditions are as follows. Dilute overnight culture/daytime culture (LB +
kanamycin +
chloramphenicol) into 2xYT (with kanamycin and chloramphenicol) at a ratio of
25 ml
culture to 1 liter 2xYT. Allow to grow at 37 °C until OD600 = 0.6. Take
an aliquot out
as TO sample. Add 1 mM IPTG and allow to grow at 30 °C for 3 hours.
Take out a T3
sample, spin down cells and store at -80 °C. The determined cDNA and
amino acid
sequences for the Ral2-PS l OS-C construct are provided in SEQ ID NO: 679 and
682,
respectively.
The expression construct PS l OS-C was designed to have a 5' added start codon
and a glycine (GGA) codon and then the PS l OS C terminal fragment followed by
the in
frame 6x histidine tag and stop codon from the pET28b vector. The cloning
strategy is
similar to that used for RaI2-PS10S-C, except that the PCR primers employed
were
those shown in SEQ ID NO: 685 and 686, respectively and the NcoI/XhoI cut in
pET28b was used. The primer of SEQ ID NO: 685 created a 5' NcoI site and added
a
start codon. The antisense primer of SEQ ID NO: 686 creates a XhoI site on PS
l OS C
terminal fragment. Clones were confirmed by sequencing. For protein
expression, the
expression construct was transformed into E. coli BL21 (DE3) CodonPlus-RIL
competent cells. An OD600 of greater than 2.0 was obtained 30 hours after
induction.
Coomassie stained SDS-PAGE showed an over-expressed band at about 11 kD.
Western blot analysis confirmed that the band was PS l OS-C, as did N-terminal
protein
sequencing. The optimized culture conditions are as follows: dilute overnight
culture/daytime culture (LB + kanamycin + chloramphenicol) into 2x YT (+
kanamycin
and chloramphenicol) at a ratio of 25 mL culture to 1 liter 2x YT, and allow
to grow at
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
181
37 °C until an OD 600 of about O.S is reached. Take out an aliquot as
TO sample. Add
1 mM IPTG and allow to grow at 30 °C for 3 hours. Spin down the cells
and store at -
80 °C until purification. The determined cDNA and amino acid sequences
for the
PS l OS-C construct are shown in SEQ ID NO: 680 and 683, respectively.
S The predicted third extracellular domain of PS I OS (PS l OS-E3; residues
328-676 of SEQ ID NO: S38) was expressed in E. coli as follows. The PSIOS
fragment
was amplified by PCR using the primers shown in SEQ ID NO: 687 and 688. The
primer of SEQ ID NO: 687 is a sense primer with an NdeI site for use in
ligating into
pPDM. The primer of SEQ ID NO: 688 is an antisense primer with an added XhoI
site
for use in ligating into pPDM. The resulting fragment was cloned to pPDM at
the NdeI
and XhoI sites. Clones were confirmed by sequencing. For protein expression,
the
clone ws transformed into E. coli BL21 (DE3) CodonPlus-RIL competent cells.
After
induction, an OD600 of greater than 2.0 Was achieved after 3 hours. Coomassie
stained
SDS-PAGE showed an over-expressed band at about 39 kD, and N-terminal
sequencing
I S confirmed the N-terminal to be that of PS l OS-E3. Optimized culture
conditions are as
follows: dilute overnight culture/daytime culture (LB + kanamycin +
chloramphenicol)
info Zx YT (kanamycin and chloramphenicol) at a ratio of 2S ml culture to 1
liter 2x
YT. Allow to grow at 37 °C until OD 600 equals 0.6. Take out an
aliquot as TO
sample. Add 1 mM IPTG and allow to grow at 30 °C for 3 hours. Take out
a T3
.sample, spin down the cells and store at -80 °C until purif cation.
The determined
cDNA and amino acid sequences for the PSOlS-E3 construct are provided in SEQ
ID
NO: 681 and 684, respectively.
g) Expression of P77SS in E. Coli
The antigen P77SP contains multiple open reading frames (ORF). The
2S third ORF, encoding the protein of SEQ ID NO: 483, has the best emotif
score. An
expression fusion construct containing the M. tuberculosis antigen Ral2 (SEQ
ID NO:
676) and P77SP-ORF3 with an N-terminal 6x HisTag was prepared as follows.
P77SP-
ORF3 was amplified using the sense PCR primers of SEQ ID NO: 689 and the anti-
sense PCR primer of SEQ ID NO: 690. The PCR amplified fragment of P77SP and
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
182
Ral2/pCRXI were digested with the restriction enzymes EcoRI and XhoI. Vector
and
insert were ligated and then transformed into NovaBlue cells. Colonies were
randomly
screened for insert and then sequenced. A 'clone having the desired sequence
was
transformed into E. coli BL21 (DE3) CodonPlus-RIL competent cells. Two hours
after
induction, the cell density peaked at OD600 of approximately 1.8. Coomassie
stained
SDS-PAGE showed an over-expressed band at about 31 kD. Western blot using 6x
HisTag antibody confirmed that the band was Ral2-P775P-ORF3. The determined
cDNA and amino acid sequences for the fusion construct are provided in SEQ ID
NO:
691 and 692, respectively.
H) EXPRESSION OF A P703P HIS TAG FUSION PROTEIN IN E. COLI
The cDNA for the coding region of P703P was prepared by PCR using
the primers of SEQ ID NO: 693 and 694. The PCR product was digested with EcoRI
restriction enzyme, gel purified and cloned into a modified pET28 vector with
a His tag
in frame, which had been digested with Eco72I and EcoRI restriction enzymes.
The
correct construct was confrmed by DNA sequence analysis and then transformed
into
E. coli BL21 (DE3) pLys S expression host cells. The determined amino acid and
cDNA sequences fox the expressed recombinant P703P are provided in SEQ ID NO:
695 and 696, respectively.
I) EXPRESSION OF A P705P HIS TAG FUSION PROTEIN IN E. COLI
The cDNA for the coding region of P705P was prepared by PCR using
the primers of SEQ ID NO: 697 and 698. The PCR product was digested with EcoRI
restriction enzyme, gel purified and cloned into a modified pET28 vector with
a His tag
in frame, which had been digested with Eco72I and EcoRI restriction enzymes.
The
correct construct was conf rmed by DNA sequence analysis and then transformed
into
E. coli BL21 (DE3) pLys S and BL21 (DE3) CodonPlus expression host cells. The
determined amino acid and cDNA sequences for the expressed recombinant P705P
are
provided in SEQ ID NO: 699 and 700, respectively.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
183
EXPRESSION OF A P711P HIS TAG FUSION PROTEIN IN E. COLI
The cDNA for the coding region of P711P was prepared by PCR using
the primers of SEQ ID NO: 701 and 702. The PCR product was digested with EcoRI
restriction enzyme, gel purified and cloned into a modified pET28 vector with
a His tag
in frame, which had been digested with Eco72I and EcoRI restriction enzymes.
The
correct construct was confirmed by DNA sequence analysis and then transformed
into
E. coli BL21 (DE3) pLys S and BL21 (DE3) CodonPlus expression host cells. The
determined amino acid and cDNA sequences for the expressed recombinant P711P
are
provided in SEQ ID NO: 703 and 704, respectively.
EXAMPLE 18
PREPARATION AND CHARACTERIZATION OF ANTIBODIES
AGAINST PROSTATE-SPECIFIC POLYPEPTIDES
a) Preparation and Characterization of Polyclonal Antibodies against P703P,
P504S and P509S
Polyclonal antibodies against P703P, P504S and P509S were prepared as
follows.
Each prostate tumor antigen expressed in an E. coli recombinant
expression system was grown overnight in LB broth with the appropriate
antibiotics at
37°C in a shaking incubator. The next morning, 10 ml of the overnight
culture was
added to 500 ml to 2x YT plus appropriate antibiotics in a 2L-baffled
Erlenmeyer flask.
When the Optical Density (at 560 nm) of the culture reached 0.4-0.6, the cells
were
induced with IPTG (1 mM). Four hours after induction with IPTG, the cells were
harvested by centrifugation. The cells were then washed with phosphate
buffered saline
and centrifuged again. The supernatant was discarded and the cells were either
frozen
for future use or immediately processed. Twenty ml of lysis buffer was added
to the
cell pellets and vortexed. To break open the E. coli cells, this mixture was
then run
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
184
through the French Press at a pressure of 16,000 psi. The cells were then
centrifuged
again and the supernatant and pellet were checked by SDS-PAGE for the
partitioning of
the recombinant protein. For proteins that localized to the cell pellet, the
pellet was
resuspended in IO mM Tris pH 8.0, 1% CHAPS and the inclusion body pellet was
washed and centrifuged again. This procedure was repeated twice more. The
washed
inclusion body pellet was solubilized with either 8 M urea or 6 M guanidine
HCl
containing 10 mM Tris pH 8.0 plus 10 mM imidazole. The solubilized protein was
added to 5 ml of nickel-chelate resin (Qiagen) and incubated for 45 min to 1
hour at
room temperature with continuous agitation. After incubation, the resin and
protein
mixture were poured through a disposable column and the flow through was
collected.
The column was then washed with 10-20 column volumes of the solubilization
buffer.
The antigen was then eluted from the column using 8M urea, 10 mM Tris pH 8.0
and
300 mM imidazole and collected in 3 ml fractions. A SDS-PAGE gel was run to
determine which fractions to pool for further purification.
As a final purification step, a strong anion exchange resin such as
HiPrepQ (Biorad) was equilibrated with the appropriate buffer and the pooled
fractions
from above were loaded onto the column. Each antigen was eluted off the column
with
a increasing salt gradient. Fractions were collected as the column was run and
another
SDS-PAGE gel was run to determine which fractions from the column to pool. The
pooled fractions were dialyzed against 10 mM Tris pH 8Ø The proteins were
then
vialed after filtration through a 0.22 micron filter and the antigens were
frozen until
needed for immunization.
Four hundred micrograms of each prostate antigen was combined with
100 micrograms of muramyldipeptide (MDP). Every four weeks rabbits were
boosted
with 100 micrograms mixed with an equal volume of Incomplete Freund's Adjuvant
(IFA). Seven days, following each boost, the animal was bled. Sera was
generated by
incubating the blood at 4°C for 12-4 hours followed by centrifugation.
Ninety-six well plates were coated with antigen by incubating with 50
microliters (typically 1 microgram) of recombinant protein at 4 °C for
20 hours. 250
microliters of BSA blocking buffer was added to the wells and incubated at
room
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
18S
temperature for 2 hours. Plates were washed 6 times with PBS/0.01% Tween.
Rabbit
sera was diluted in PBS. Fifty microliters of diluted sera was added to each
well and
incubated at room temperature for 30 min. Plates were washed as described
above
before SO microlifers of goat anti-rabbit horse radish peroxidase (HRP) at a
1:10000
S dilution was added and incubated at room temperature for 30 min. Plates were
again
washed as described above and 100 microliters of TMB microwell peroxidase
substrate
was added to each well. Following a 1 S min incubation in the dark at room
temperature, the colorimetric reaction was stopped with 100 microliters of 1N
H2S04
and read immediately at 4S0 nm. All polyclonal antibodies showed
immunoreactivity
to the appropriate antigen.
b) Preparation and Characterization of Antibodies against PS01 S
A marine monoclonal antibody directed against the carboxy-terminus of
the prostate-specific antigen PSO1 S was prepared as follows.
A truncated fragment of PSO1S (amino acids 3SS-S26 of SEQ ID NO:
IS 113) was generated and cloned into the pET28b vector (Novagen) and
expressed in E.
coli as a thioredoxin fusion protein with a histidine tag. The trx-PSO 1 S
fusion protein
was purified by nickel chromatography, digested with thrombin to remove the
trx
fragment and further purified by an acid precipitation procedure followed by
reverse
phase HPLC.
Mice were immunized with truncated P501 S protein. Serum bleeds from
mice that potentially contained anti-PSO1 S polyclonal sera were tested for
PSO1 S-
specific reactivity using ELISA assays with purified PS01 S and trx-PSO1 S
proteins.
Serum bleeds that appeared to react specifically with PSOI S were then
screened for
PSOl S reactivity by Western analysis. Mice that contained a PSO1 S-specific
antibody
2S component were sacrificed and spleen cells were used to generate anti-PSOlS
antibody
producing hybridomas using standard techniques. Hybridoma supernatants were
tested
for PSO1 S-specific reactivity initially by ELISA, and subsequently by FACS
analysis of
reactivity with PSO1S transduced cells. Based on these results, a monoclonal
hybridoma
referred to as 10E3 was chosen for further subcloning. A number of subclones
were
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
186
generated, tested for specific reactivity to P501 S using ELISA and typed for
IgG
isotype. The results of this analysis are shown below in Table V. Of the 16
subclones
tested, the monoclonal antibody 10E3-G4-D3 was selected for further study.
Table V
Isotype analysis of murine anti-P501 S monoclonal antibodies
Hybridoma clone Isotype Estimated [Ig] in su ernatant
( g/ml)
4D11 I Gl 14.6
1 G1 IgGl 0.6
4F6 IgGl 72
4H5 IgGl 13.8
4H5-E12 I Gl 10.7
4H5-EH2 I G1 9.2
4H5-H2-A10 I G1 10
4H5-H2-A3 I G 1 12.8
4H5-H2-A10-G6 I Gl 13.6
4H5-H2-B 1 I IgGl 12.3
1 OE3 IgG2a 3.4
1 OE3-D4 IgG2a 3.8
10E3-D4-G3 IgG2a 9.5
1 OE3-D4-G6 IgG2a 10.4
l0E3-E7 IgG2a 6.5
8H12 ~ IgG2a 0.6
The specificity of 10E3-G4-D3 for P501 S was examined by FACS
analysis. Specifically, cells were fixed (2% formaldehyde, 10 minutes),
permeabilized
(0.1 % saponin, 10 minutes) and stained with 1 OE3-G4-D3 at 0.5 - 1 ~g/ml,
followed by
incubation with a secondary, FITC-conjugated goat anti-mouse Ig antibody
(Pharmingen, San Diego, CA). Cells were then analyzed for FITC fluorescence
using
an Excalibur fluorescence activated cell sorter. For FACS analysis of
transduced cells,
B-LCL were retrovirally transduced with P501 S. For analysis of infected
cells, B-LCL
were infected with a vaccinia vector that expresses P501 S. To demonstrate
specificity
in these assays, B-LCL transduced with a different antigen (P703P) and
uninfected B-
LCL vectors were utilized. 10E3-G4-D3 was shown to bind with P501 S-transduced
B-
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
187
LCL and also with P501 S-infected B-LCL, but not with either uninfected cells
or
P703P-transduced cells.
To determine whether the epitope recognized by 10E3-G4-D3 was found
on the surface or in an intracellular compartment of cells, B-LCL were
transduced with
P501 S or HLA-B8 'as a control antigen and either fixed and permeabilized as
described
above or directly stained with 1 OE3-G4-D3 and analyzed as above. Specific
recognition
of P501 S by 10E3-G4-D3 was found to require permeabilization, suggesting that
the
epitope recognized by this anfibody is intracellular.
The reactivity of 10E3-G4-D3 with the three prostate tumor cell lines
Lncap, PC-3 and DU-145, which are known to express high, medium and very low
levels of P501 S, respectively, was examined by permeabilizing the cells and
treating
them as described above. Higher reactivity of 10E3-G4-D3 was seen with Lncap
than
with PC-3, which in turn showed higher reactivity that DU-145. These results
are in
agreement with the real time PCR and demonstrate that the antibody
specifically
recognizes P501 S in these tumor cell lines and that the epitope recognized in
prostate
tumor cell lines is also intracellular.
Specificity of 10E3-G4-D3 for P501 S was also demonstrated by Western
blot analysis. Lysates from the prostate tumor cell lines Lncap, DU-145 and PC-
3, from
P50I S-transiently transfected HEK293 cells, and from non-transfected HEK293
cells
were generated. Western blot analysis of these lysates with 10E3-G4-D3
revealed a 46
kDa immunoreactive band in Lncap, PC-3 and P501 S-transfected HEK cells, but
not in
DU-145 cells or non-transfected HEK293 cells. P501 S mRNA expression is
consistent
with these results since semi-quantitative PCR analysis revealed that P501 S
mRNA is
expressed in Lncap, to a lesser but detectable level in PC-3 and not at all in
DU-145
cells. Bacterially expressed and purified recombinant P50I S (referred to as
P501 SStr2)
was recognized by 10E3-G4-D3 (24 kDa), as was full-length P501 S that was
transiently
expressed in HEK293 cells using either the expression vector VRI012 or pCEP4.
Although the predicted molecular weight of P501 S is 60.5 kDa, both
transfected and
"native" P501 S run at a slightly lower mobility due to its hydrophobic
nature.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
188
Immunohistochemical analysis was performed on prostate tumor and a
panel of normal tissue sections (prostate, adrenal, breast, cervix, colon,
duodenum, gall
bladder, ileum, kidney, ovary, pancreas, parotid gland, skeletal muscle,
spleen and
testis). Tissue samples were fixed in formalin solution for 24 hours and
embedded in
paraffin before being sliced into 10 micron sections. Tissue sections were
permeabilized and incubated with 10E3-G4-D3 antibody for 1 hr. HRP-labeled
anti-
mouse followed by incubation with DAB chromogen was used to visualize P501 S
immunoreactivity. P501 S was found to be highly expressed in both normal
prostate and
prostate tumor tissue but was not detected in any of the other tissues tested.
To identify the epitope recognized by 10E3-G4-D3, an epitope mapping
approach was pursued. A series of 13 overlapping 20-21 mers (5 amino acid
overlap;
SEQ ID NO: 489-501) was synthesized that spanned the fragment of PSO1S used to
generate 10E3-G4-D3. Flat bottom 96 well microtiter plates were coated with
either the
peptides or the P501 S fragment used to immunize mice, at 1 microgram/ml for 2
hours
at 37 °C. Wells were then aspirated and blocked with phosphate buffered
saline
containing 1% (w/v) BSA for 2 hours at room temperature, and subsequently
washed in
PBS containing 0.1% Tween 20 (PBST). Purified antibody 10E3-G4-D3 was added at
2 fold dilutions (1000 ng - 16 ng) in PBST and incubated fox 30 minutes at
room
temperature. This was followed by washing 6 times with PBST and subsequently
incubating with HRP-conjugated donkey anti-mouse IgG (H+L)Affinipure F(ab')
fragment (Jackson Immunoresearch, West Grove, PA) at 1:20000 for 30 minutes.
Plates were then washed and incubated for 15 minutes in tetramethyl benzidine.
Reactions were stopped by the addition of 1N sulfuric acid and plates were
read at 450
nm using an ELISA plate reader. As shown in Fig. 8, reactivity was seen with
the
peptide of SEQ ID NO: 496 (corresponding to amino acids 439-459 of PSO1S) and
with
the P501 S fragment but not with the remaining peptides, demonstrating that
the epitope
recognized by 10E3-G4-D3 is localized to amino acids 439-459 of SEQ ID NO:
113.
In order to further evaluate the tissue specificity of P501 S, mufti-array
immunohistochemical analysis was performed on approximately 4700 different
human
tissues encompassing all the major normal organs as well as neoplasias derived
from
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
189
these tissues. Sixty-five of these human tissue samples were of prostate
origin. Tissue
sections 0.6 mm in diameter were formalin-fixed and paraffin embedded. Samples
were
pretreated with HIER using 10 mM citrate buffer pH 6.0 and boiling for 10 min.
Sections were stained with 10E3-G4-D3 and P501 S immunoreactivity was
visualized
with HRP. All the 65 prostate tissues samples (5 normal, 55 untreated prostate
tumors,
5 hormone refractory prostate tumors) were positive, showing distinct
perinuclear
staining. All other tissues examined wexe negative for P501 S expression.
c, Preparation and Characterization of Antibodies against P503S
A fragment of P503S (amino acids 113-241 of SEQ ID NO: 114) was
expressed and purified from bacteria essentially as described above for P501 S
and used
to immunize both rabbits and mice. Mouse monoclonal antibodies were isolated
using
standard hybridoma technology as described above. Rabbit monoclonal antibodies
were
isolated using Selected Lymphocyte Antibody Method (SLAM) technology at
Immgenics Pharmaceuticals (Vancouver, BC, Canada). Table VI, below, lists the
monoclonal antibodies that were developed against P503S.
Table VI
Antibody S ecies
20D4 Rabbit
JA1 Rabbit
1 A4 Mouse
1 C3 Mouse
1 C9 Mouse
1 D I 2 Mouse
2A1 1 Mouse
2H9 Mouse
4H7 Mouse
8A8 Mouse
8D 10 Mouse
9C 12 Mouse
6D 12 Mouse
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
190
The DNA sequences encoding the complementarity determining regions
(CDRs) for the rabbit monoclonal antibodies 20D4 and JAl were determined and
are
provided in SEQ ID NO: 502 and 503, respectively.
In order to better define the ~ epitope binding region of each of the
antibodies, a series of overlapping peptides were generated that span amino
acids 109-
213 of SEQ ID NO: 114. These peptides were used to epitope map the anti-P503S
monoclonal antibodies by ELISA as follows. The recombinant fragment of P503S
that
was employed as the immunogen was used as a positive control. Ninety-six well
microtiter plates were coated with either peptide or recombinant antigen at 20
ng/well
overnight at 4 °C. Plates were aspirated and blocked with phosphate
buffered saline
containing 1% (w/v) BSA for 2 hours at room temperature then washed in PBS
containing 0.1% Tween 20 (PBST). Purified rabbit monoclonal antibodies diluted
in
PBST were added to the wells and incubated for 30 min at room temperature.
This was
followed by washing 6 times with PBST and incubation with Protein-A HRP
conjugate
at a 1:2000 dilution for a further 30 min. Plates were washed six times in
PBST and
incubated with tetramethylbenzidine (TMB) substrate for a further 15 min. The
reaction
was stopped by the addition of 1N sulfuric acid and plates were read at 450 nm
using at
ELISA plate reader. ELISA with the mouse monoclonal antibodies was performed
with
supernatants from tissue culture run neat in the assay.
All of the antibodies bound to the recombinant P503S fragment, with the
exception of the negative control SP2 supernatant. 20D4, JA1 and 1 D 12 bound
strictly
to peptide #2101 (SEQ ID NO: 504), which corresponds to amino acids 151-169 of
SEQ ID NO: 114. 1 C3 bound to peptide #2102 (SEQ ID NO: 505), which
corresponds
to amino acids 165-184 of SEQ ID NO: 114. 9C 12 bound to peptide #2099 (SEQ ID
NO: 522), which corresponds to amino acids 120-139 of SEQ ID NO: 114. The
other
antibodies bind to regions that were not examined in these studies.
Subsequent to epitope mapping, the antibodies were tested by FACS
analysis on a cell line that stably expressed P503S to confirm that the
antibodies bind to
cell surface epitopes. Cells stably transfected with a control plasmid were
employed as
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
191
a negative control. Cells were stained live with no fixative. 0.S ug of anti-
PS03S
monoclonal antibody was added and cells were incubated on ice for 30 min
before being
washed twice and incubated with a FITC-labelled goat anti-rabbit or mouse
secondary
antibody for 20 min. After being washed twice, cells were analyzed with an
Excalibur
S fluorescent activated cell sorter. The monoclonal antibodies 1 C3, 1 D 12,
9C 12, 20D4
and JA1, but not 8D3, were found to bind to a cell surface epitope of PS03S.
In order to determine which tissues express PS03S,
immunohistochemical analysis was performed, essentially as described above, on
a
panel of normal tissues (prostate, adrenal, breast, cervix, colon, duodenum,
gall bladder,
ileum, kidney, ovary, pancreas, parotid gland, skeletal muscle, spleen and
testis). HRP
labeled anti-mouse or anti-rabbit antibody followed by incubation with TMB was
used
to visualize P503S immunoreactivity. PS03S was found to be highly expressed in
prostate tissue, with lower levels of expression being observed in cervix,
colon, ileum
and kidney, and no expression being observed in adrenal, breast, duodenum,
gall
1 S bladder, ovary, pancreas, parotid gland, skeletal muscle, spleen and
testis.
Western blot analysis was used to characterize anti-PS03S monoclonal
antibody specificity. SDS-PAGE was performed on recombinant (rec) PS03S
expressed
in and purified from bacteria and on lysates from HEK293 cells transfected
with full
length PS03S. Protein was transferred to nitrocellulose and then Western
blotted with
each of the anti-PS03S monoclonal antibodies (20D4, JA1, 1D12, 6D12 and 9C12)
at
an antibody concentration of 1 ug/ml. Protein was detected using horse radish
peroxidase (HRP) conjugated to either a goat anti-mouse monoclonal antibody or
to
protein A-sepharose. The monoclonal antibody 20D4 detected the appropriate
molecular weight 14 kDa recombinant PS03S (amino acids l I3-241) and the 23.5
kDa
2S species in the HEK293 cell lysates transfected with full length PS03S.
Other anti-
PS03S monoclonal antibodies displayed similax specificity by Western blot.
d) Preparation and Characterization of Antibodies against P703P
Rabbits were immunized with either a truncated (P703Ptrl; SEQ ID NO:
172) or full-length mature form (P703Pfl; SEQ ID NO: S23) of recombinant P703P
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
192
protein was expressed in and purif ed from bacteria as described above.
Affinity
purified polyclonal antibody was generated using immunogen P703Pfl or P703Ptr1
attached to a solid support. Rabbit monoclonal antibodies were isolated using
SLAM
technology at Immgenics Pharmaceuticals. Table VII below lists both the
polyclonal
and monoclonal antibodies that were generated against P703P.
Table VII
Antibody Immuno en S ecies/ty a
Aff. Purif. P703P (truncated);P703Ptrl Rabbit olyclonal
#2594
Aff. Purif. P703P (full P703Pf1 Rabbit olyclonal
len th); #9245
2D4 P703Ptr1 Rabbit monoclonal
8H2 P703Ptr1 Rabbit monoclonal
7H8 P703Ptr1 Rabbit monoclonal
The DNA sequences encoding the complementarily determining regions
(CDRs) for the rabbit monoclonal antibodies 8H2, 7H8 and 2D4 were determined
and
are provided in SEQ ID NO: 506-508, respectively.
Epitope mapping studies were performed as described above.
Monoclonal antibodies 2D4 and 7H8 were found to specifically bind to the
peptides of
SEQ ID NO: 509 (corresponding to amino acids 145-159 of SEQ ID NO: 172) and
SEQ
ID NO: 510 (corresponding to amino acids 11-25 of SEQ ID NO: 172),
respectively.
The polyclonal antibody 2594 was found to bind to the peptides of SEQ ID NO:
511-
514, with the polyclonal antibody 9427 binding to the peptides of SEQ ID NO:
515-517.
The specificity of the anti-P703P antibodies was determined by Western
blot analysis as follows. SDS-PAGE was performed on (1) bacterially expressed
recombinant antigen; (2) lysates of HEK293 cells and Ltk-/- cells either
untransfected or
txansfected with, a plasmid expressing full length P703P; and (3) supernatant
isolated
from these cell cultures. Protein was transferred to nitrocellulose and then
Western
blotted using the anti-P703P polyclonal antibody #2594 at an antibody
concentration of
I ug/ml. Protein was detected using horse radish peroxidase (HRP) conjugated
to an
anti-rabbit antibody. A 35 kDa immunoreactive band could be observed with
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
I93
recombinant P703P. Recombinant P703P runs at a slightly higher molecular
weight
since it is epitope tagged. In lysates and supernatants from cells transfected
with full
length P703P, a 30 kDa band corresponding to P703P was observed. To assure
specificity, lysates from HEK293 cells stably transfected with a control
plasmid were
also tested and were negative for P703P expression. Other anti-P703P
antibodies
showed similar results.
Immunohistochemical studies were performed as described above, using
anti-P703P monoclonal antibody. P703P was found to be expressed at high levels
in
normal prostate and prostate tumor tissue but was not detectable in all other
tissues
tested (breast tumor, lung tumor and normal kidney).
e) Preparation and Characterization of Antibodies against P504S
Full-length P504S (SEQ ID NO: 108) was expressed and purified from
bacteria essentially as described above for P501 S and employed to raise
rabbit
monoclonal antibodies using Selected Lymphocyte Antibody Method (SLAM)
technology at Immgenics Pharmaceuticals (Vancouver, BC, Canada). The anti-
P504S
monoclonal antibody 13H4 was shown by Western blot to bind to both expressed
recombinant P504S and to native P504S in tumor cells.
Immunohistochemical studies using I3H4 to assess P504S expression in
various prostate tissues were performed as described above. A total of 104
cases,
including 65 cases of radical prostatectomies with prostate cancer (PC), 26
cases of
prostate biopsies and 13 cases of benign prostate hyperplasia (BPH), were
stained with
the anti-P504S monoclonal antibody I3H4. P504S showed strongly cytoplasmic
granular staining in 64/65 (98.5%) of PCs in prostatectomies and 26/26 (100% )
of PCs
in prostatic biopsies. P504S was stained strongly and diffusely in carcinomas
(4+ in
91.2% of cases of PC; 3+ in 5.5%; 2+ in 2.2% and 1+ in .1.1 %) and high grade
prostatic
intraepithelial neoplasia (4+ in all cases). The expression of P504S did not
vary with
Gleason score. Only 17/91 (18.7%) of cases ofNP/BPH around PC and 2/13 (15.4%)
of
BPH cases were focally (1+, no 2+ to 4+ in aII cases) and weakly positive for
P504S in
large glands. Expression of P504S was not found in small atrophic glands,
postatrophic
hyperplasia, basal cell hyperplasia and transitional cell metaplasia in either
biopsies or
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
194
prostatectomies. P504S was thus found to be over-expressed in all Gleason
scoxes of
prostate cancer (98.5 to 100% of sensitivity) and exhibited only focal
positivities in
large normal glands in 19/104 of cases (82.3% of specificity). These findings
indicate
that P504S may be usefully employed for the diagnosis of prostate cancex.
EXAMPLE 19
CHARACTERIZATION OF CELL SURFACE EXPRESSION AND
CHROMOSOME LOCALIZATION OF THE PROSTATE-SPECIFIC ANTIGEN P501 S
This example describes studies demonstrating that the prostate-specific
antigen P501 S is expressed on the surface of cells, together with studies to
determine
the probable chromosomal location of P501 S.
The protein P501 S (SEQ ID NO: 113) is predicted to have 11
transmembrane domains. Based on the discovery that the epitope recognized by
the anti-
P501 S monoclonal antibody 1 OE3-G4-D3 (described above in Example 17) is
intracellular, it was predicted that following transmembrane determinants
would allow
the prediction of extracellular domains of P501 S. Fig. 9 is a schematic
representation
of the P501 S protein showing the predicted location of the transmembrane
domains and
the intracellular epitope described in Example 17. Underlined sequence
represents the
predicted transmembrane domains, bold sequence represents the predicted
extracellular
domains, and italicized sequence represents the predicted intracellular
domains.
Sequence that is both bold and underlined represents sequence employed to
generate
polyclonal rabbit serum. The location of the transmembrane domains was
predicted
using HHMTOP as described by Tusnady and Simon (Principles Governing Amino
Acid Composition of Integral Membrane Proteins: Applications to Topology
Prediction,
J. Mol. Biol. 283:489-506, 1998).
Based on Fig. 9, the P501 S domain flanked by the transmembrane
domains corresponding to amino acids 274-295 and 323-342 is predicted to be
extracellular. The peptide of SEQ ID NO: 518 corresponds to amino acids 306-
320 of
P501 S and lies in the predicted extracellular domain. The peptide of SEQ ID
NO: 519,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
195
which is identical to the peptide of SEQ ID NO: 518 with the exception of the
substitution of the histidine with an asparginine, was synthesized as
described above. A
Cys-Gly was added to the C-terminus of the peptide to facilitate conjugation
to the
carrier protein. Cleavage of the peptide from the solid support was carried
out using the
S following cleavage mixture: trifluoroacetic
acid:ethanediolahioanisol:water:phenol
(40:1:2:2:3). After cleaving for two hours, the peptide was precipitated in
cold ether.
The peptide pellet was then dissolved in 10% v/v acetic acid and lyophilized
prior to
purification by C 18 reverse phase hplc. A gradient of 5-60% acetonitrile
(containing
0.05% TFA) in water (containing 0.05% TFA) was used to elute the peptide. The
purity
of the peptide was verified by hplc and mass spectrometry, and was determined
to be
>95%. The purified peptide was used to generate rabbit polyclonal antisera as
described
above.
Surface expression of P501 S was examined by FACS analysis. Cells
were stained with the polyclonal anti-P501 S peptide serum at 10 ~g/ml,
washed,
incubated with a secondary FITC-conjugated goat anti-rabbit Ig antibody (ICN),
washed
and analyzed for FITC fluorescence using an Excalibur fluorescence activated
cell
sorter. For FACS analysis of transduced cells, B-LCL were retrovirally
transduced with
P501 S. To demonstrate specificity in these assays, B-LCL transduced with an
irrelevant
antigen (P703P) or nontransduced were stained in parallel. For FACS analysis
of
prostate tumor cell lines, Lncap, PC-3 and DU-145 were utilized. Prostate
tumor cell
lines were dissociated from tissue culture plates using cell dissociation
medium and
stained as above. All samples were treated with propidium iodide (PI) prior to
FACS
analysis, and data was obtained from PI-excluding (i. e., intact and non-
permeabilized)
cells. The rabbit polyclonal serum generated against the peptide of SEQ ID NO:
519
was shown to specifically recognize the surface of cells transduced to express
P501 S,
demonstrating that the epitope recognized by the polyclonal serum is
extracellular:
To determine biochemically if P501S is expressed on the cell surface,
peripheral membranes from Lncap cells were isolated and subjected to Western.
blot
analysis. Specifically, Lncap cells were lysed using a dounce homogenizes. in
5 rnl of
homogenization buffer (250 mM sucrose, 10 mM HEPES, 1mM EDTA, pH 8.0, 1
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
196
complete protease inhibitor tablet (Boehringer Mannheim)). Lysate samples were
spun
at 1000 g for S min at 4 °C. The supernatant was then spun at 8000g for
10 min at 4 °C.
Supernatant from the 8000g spin was recovered and subjected to a 100,000g spin
for 30
min at ~4 °C to recover peripheral membrane. Samples were then
separated by SDS-
S PAGE and Western blotted with the mouse monoclonal antibody 10E3-G4-D3
(described above in Example 17) using conditions described above. Recombinant
purified PSOl S, as well as HEK293 cells transfected with and over-expressing
PSOl S
were included as positive controls for PSO1 S detection. LCL cell lysate was
included as
a negative control. PSOl S could be detected in Lncap total cell lysate, the
8000g
(internal membrane) fraction and also in the IOO,OOOg (plasma membrane)
fraction. '
These results indicate that PSO1 S is expressed at, and localizes to, the
peripheral
membrane.
To demonstrate that the rabbit polyclonal antiserum generated to the
peptide of SEQ ID NO: S 19 specifically recognizes this peptide as well as the
1 S corresponding native peptide of SEQ ID NO: S 18, ELISA analyses were
performed.
For these analyses, flat-bottomed 96 well microtiter plates were coated with
either the
peptide of SEQ ID NO: S 19, the longer peptide of SEQ ID NO: S20 that spans
the entire
predicted extracellular domain, the peptide of SEQ ID NO: S21 which represents
the
epitope recognized by the PSOlS-specific antibody 10E3-G4-D3, or a PSO1S
fragment
(corresponding to amino acids 3SS-S26 of SEQ ID NO: 113) that does not include
the
immunizing peptide sequence, at I ~g/mI for 2 hours at 37 °C. Wells
were aspirated,
blocked with phosphate buffered saline containing 1% (w/v) BSA for 2 hours at
room
temperature and subsequently washed in PBS containing 0.1% Tween 20 (PBST).
Purified anti-PSO1 S polyclonal rabbit serum was added at 2 fold dilutions
(1000 ng -
12S ng) in PBST and incubated for 30 min at room temperature. This was
followed by
washing 6 times with PBST and incubating with HRP-conjugated goat anti-rabbit
IgG
(H+L) Affinipure F(ab') fragment at 1:20000 for 30 min. Plates were then
washed and
incubated for 1 S min in tetramethyl benzidine. Reactions were stopped by the
addition
of 1N sulfuric acid and plates were read at 4S0 nm using an ELISA plate
reader. As
shown in Fig. 11, the anti-PSO1 S polyclonal rabbit serum specifically
recognized the
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
197
peptide of SEQ ID NO: 519 used in the immunization as well as the longer
peptide of
SEQ ID NO: 520, but did not recognize the irrelevant P501 S-derived peptides
and
fragments.
In further studies, rabbits were immunized with peptides derived from
the P501 S sequence and predicted to be either extracellular or intracellular,
as shown in
Fig. 9. Polyclonal rabbit sera were isolated and polyclonal antibodies in the
serum were
purif ed, as described above. To determine specific reactivity with P501 S,
FACS
analysis was employed, utilizing either B-LCL transduced with P501 S or the
irrelevant
antigen P703P, of B-LCL infected with vaccinia virus-expressing P501 S. For
surface
expression, dead and non-intact cells were excluded from the analysis as
described
above. For intracellular staining, cells were fixed and permeabilized as
described
above. Rabbit polyclonal serum generated against the peptide of SEQ ID NO:
548,
which corresponds to amino acids 181-198 of PSO1S, was found to recognize a
surface
epitope of P501 S. Rabbit polyclonal serum generated against the peptide SEQ
ID NO:
551, which corresponds to amino acids 543-553 of PSOlS, was found to recognize
an
epitope that was either potentially extracellular or intracellular since in
different
experiments intact or permeabilized cells were recognized by the polyclonal
sera.
Based on similar deductive .reasoning, the sequences of SEQ ID NO: 541-547,
549 and
550, which correspond to amino acids 109-122, 539-553, 509-520, 37-54, 342-
359,
295-323, 217-274, 143-160 and 75-88, respectively, of P501 S, can be
considered to be
potential surface epitopes of P501 S recognized by antibodies.
In further studies, mouse monoclonal antibodies were raised against
amino acids 296 to 322 to P501 S, which are predicted to be in an
extracellular domain.
A/J mice were immunized with PSOI Sladenovirus, followed by subsequent boosts
with
an E. coli recombinant protein, referred to as PSOlN, that contains amino
acids 296 to
322 of P501 S, and with peptide 296-322 (SEQ ID NO: 755) coupled with I~LLH.
The
mice were subsequently used for splenic B cell fusions to generate anti-
peptide
hybridomas. The resulting 3 clones, referred to as 4F4 (IgGl,kappa), 4G5
(IgG2a,kappa) and 9B9 (IgGl,kappa), were grown for antibody production. The
4G5
mAb was purified by passing the supernatant over a Protein A-sepharose column,
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
198
followed by antibody elution using 0.2M glycine, pH 2.3. Purified antibody was
neutralized by the addition of 1M Tris, pH 8, and buffer exchanged into PBS.
For ELISA analysis, 96 well plates were coated with P501 S peptide 296-
322 (referred to as P501-long), an irrelevant P775 peptide, P501 S-N, P501
TR2, P501 S-
long-KLH, PSO1S peptide 306-319 (referred to as P501-short)-KL,H, or the
irrelevant
peptide 2073-KLH, all at a concentration of 2 ug/ml and allowed to incubate
for 60
minutes at 37 °C. After coating, plates were washed SX with PBS + 0.1%
Tween and
then blocked with PBS, 0.5% BSA, 0.4% Tween20 for 2 hours at room temperature.
Following the addition of supernatants ox purified mAb, the plates were
incubated for
60 minutes at room temperature. Plates were washed as above and donkey anti-
mouse
IgHRP-linked secondary antibody was added and incubated for 30 minutes at room
temperature, followed by a final washing as above. TMB peroxidase substrate
was
added and incubated 15 minutes at room temperature in the dark. The reaction
was
stopped by the addition of 1N H2S04 and the OD was read at 450 nM. All three
hybrid
clones secreted mAb that recognized peptide 296-322 and the recombinant
protein
PSO1N.
For FACS analysis, HEI~293 cells were transiently transfected with a
P501 S/VR1012 expression constructs using Fugene 6 reagent. After 2 days of
culture,
cells were harvested and washed, then incubated with purified 4G5 mAb for 30
minutes
on ice. After several washes in PBS, 0.5% BSA, 0.01% azide, goat anti-mouse Ig-
FITC
was added to the cells and incubated for 30 minutes on ice. Cells were washed
and
resuspended in wash buffer including 1% propidium iodide and subjected to FACS
analysis. The FACS analysis confirmed that amino acids 296-322 of PSO1S are in
an
extracellular domain and axe cell surface expressed.
The chromosomal location of P501 S was determined using the
GeneBridge 4 Radiation Hybrid panel (Research Genetics). The PCR primers of
SEQ
ID NO: 528 and 529 were employed in PCR with DNA pools from the hybrid panel
according to the manufacturer's directions. After 38 cycles of amplification,
the
reaction products were separated on a 1.2% agarose gel, and the results were
analyzed
through the Whitehead Institute/MIT Center for Genome Research web server
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
199
(http://www-genome.wi.mit.edu/cgi-bin/contig/rhmapper.pl) to determine the
probable
chromosomal location. Using this approach, PSOl S was mapped to the long arm
of
chromosome 1 at WI-9641 between q32 and q42. This region of chromosome 1 has
been linked to prostate cancer susceptibility in hereditary prostate cancer
(Smith et al.
S Science 274:1371-1374, 1996 and Berthon et al. Ayn. J. Huyn. Genet. 62:1416-
1424,
1998). These results suggest that PSOl S may play a role in prostate cancer
malignancy.
EXAMPLE 20
REGULATION OF EXPRESSION OF THE PROSTATE-SPECIFIC ANTIGEN PSOl S
Steroid (androgen) hormone modulation is a common treatment modality
in prostate cancer. The expression of a number of prostate tissue-specific
antigens have
previously been demonstrated to respond to androgen. The responsiveness of the
prostate-specific antigen PSO1 S to androgen treatment was examined in a
tissue culture
1 S system as follows.
Cells from the prostate tumor cell line LNCaP were plated at 1.S x 106
cells/T7S flask (for RNA isolation) or 3 x 105 cells/well of a 6-well plate
(for FACS
analysis) and grown overnight in RPMI 1640 media containing 10% charcoal-
stripped
fetal calf serum (BRL Life Technologies, Gaithersburg, MD). Cell culture was
continued for an additional 72 hours in RPMI 1640 media containing 10%
charcoal-
stripped fetal calf serum, with 1 nM of the synthetic androgen
Methyltrienolone
(R1881; New England Nuclear) added at various time points. Cells were then
harvested
for RNA isolation and FACS analysis at 0, 1, 2, 4, 8, 16, 24, 28 and 72-hours
post
androgen addition. FACS analysis was performed using the anti-PSOlS antibody
10E3-
2S G4-D3 and permeabilized cells.
For Northern analysis, S-10 micrograms of total RNA was run on a
formaldehyde denaturing gel, transferred to Hybond-N nylon membrane (Amersham
Pharmacia Biotech, Piscataway, NJ), cross-linked and stained with rnethylene
blue. The
filter was then prehybridized with Church's Buffer (2S0 mM Na2HP04, 70 mM
H3P04,
1 mM EDTA, 1% SDS, 1% BSA in pH 7.2) at 6S °C for 1 hour. PSOlS DNA was
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
200
labeled with 32P using High Prime random-primed DNA labeling kit (Boehringer
Mannheim). Unincorporated label was removed using MicroSpin 5300-HR columns
(Amersham Pharmacia Biotech). The RNA filter was then hybridized with fresh
Church's Buffer containing labeled cDNA overnight, washed with 1X SCP (0.1 M
NaCI, 0.03 M Na2HP04.7Ha0, 0.001 M Na2EDTA), 1% sarkosyl (n-lauroylsarcosine)
and exposed to X-ray film.
Using both FACS and Northern analysis, P501 S message and protein
levels were found in increase in response to androgen treatment.
EXAMPLE 20
PREPARATION OF FUSION PROTEINS OF PROSTATE-SPECIFIC ANTIGENS
The example describes the preparation of a fusion protein of the prostate-
specific antigen P703P and a truncated form of the known prostate antigen PSA.
The
truncated form of PSA has a 21 amino acid deletion around the active serine
site. The
expression construct for the fusion protein also has a restriction site at 3'
end,
immediately prior to the termination codon, to aid in adding cDNA for
additional
antigens.
The full-length cDNA for PSA was obtained by RT-PCR from a pool of
RNA from human prostate tumor tissues using the primers of SEQ ID NO: 607 and
608,
and cloned in the vector pCR-Blunt II-TOPO. The resulting cDNA was employed as
a
template to make two different fragments of PSA by PCR with two sets of
primers
(SEQ ID NO: 609 and 610; and SEQ ID NO: 611 and 612). The PCR products having
the expected size were used as templates to make truncated forms of PSA by PCR
with
the primers of SEQ ID NO: 611 and 613, which generated PSA (delta 208-218. in
amino
acids). The cDNA for the mature form of P703P with a 6X histidine tag at the
5' end,
was prepared by PCR with P703P and the primers of SEQ ID NO: 614 and 615. The
cDNA for the fusion of P703P with the truncated form of PSA (referred to as
FOPP)
was then obtained by PCR using the modified P703P cDNA and the truncated form
of
PSA cDNA as templates and the primers of SEQ ID NO: 614 and 615. The FOPP
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
201
cDNA was cloned into the NdeI site and XhoI site of the expression vector
pCRXl, and
confirmed by DNA sequencing. The determined cDNA sequence for the fusion
construct FOPP is provided in SEQ ID NO: 616, with the amino acid sequence
being
provided in SEQ ID NO: 617.
The fusion FOPP was expressed as a single recombinant protein in E.
coli as follows. The expression plasmid pCRXIFOPP was transformed into the E.
coli
strain BL21-CodonPlus RIL. The transformant was shown to express FOPP protein
upon induction with 1 mM IPTG. The culture of the corresponding expression
clone
was inoculated into 25 ml LB broth containing 50 ug/ml kanamycin and 34 ug/ml
IO chloramphenicol, grown at 37 °C to OD600 of about 1, and stored at 4
°C overnight.
The culture was diluted into 1 liter of TB LB containing 50 ug/ml kanamycin
and 34
ug/ml chloramphenicol, and grown at 37 °C to OD600 of 0.4. IPTG was
added to a
final concentration of 1 mM, and the culture was incubated at 30 °C for
3 hours. The
cells were pelleted by centrifugation at 5,000 RPM for 8 min. To purify the
protein, the
cell pellet was suspended in 25 ml of 10 mM Tris-Cl pH 8.0, 2mM PMSF, complete
protease inhibitor and 15 ug lysozyme. The cells were lysed at 4 °C for
30 minutes,
sonicated several times and the lysate centrifuged for 30 minutes at 10,000 x
g. The
precipitate, which contained the inclusion body, was washed twice with 10 mM
Tris-Cl
pH 8.0 and 1 % CHAPS. The inclusion body was dissolved in 40 ml of 10 mM Tris-
Cl
pH 8.0, 100 mM sodium phosphate and 8 M urea. The solution was bound to 8 ml
Ni-
NTA (Qiagen) for one hour at room temperature. The mixture was poured into a
25 ml
column and washed with 50 ml of 10 mM Tris-Cl pH 6.3, 100 mM sodium phosphate,
0.5% DOC and 8M urea. The bound protein was eluted with 350 mM imidazole, 10
mM Tris-Cl pH 8.0, 100 mM sodium phosphate and 8 M urea. The fractions
containing
FOPP proteins were combined and dialyzed extensively against 10 mM Tris-Cl pH
4.6,
aliquoted and stored at - 70 °C.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
202
EXAMPLE 21
REAL-TIME PCR CHARACTERIZATION OF THE PROSTATE-SPECIFIC ANTIGEN PSO1 S IN
PERIPHERAL BLOOD OF PROSTATE CANCER PATIENTS
Circulating epithelial cells were isolated from fresh blood of normal
individuals and metastatic prostate cancer patients, mRNA isolated and cDNA
prepared
using real-time PCR procedures. Real-time PCR was performed with the TaqmanTM
procedure using both gene specific primers and probes to determine the levels
of gene
expression.
Epithelial cells were enriched from blood samples using an
immunomagnetic bead separation method (Dynal A.S., Oslo, Norway). Isolated
cells
were lysed and the magnetic beads removed. The lysate was then processed for
poly A+
mRNA isolation using magnetic beads coated with Oligo(dT)25. After washing the
beads in buffer, bead/poly A+ RNA samples were suspended in 10 mM Tris HCl pH
8.0
and subjected to reversed transcription. The resulting cDNA was subjected to
real-time
PCR using gene specific primers. Beta-actin content was also determined and
used for
normalization. Samples with P501 S copies greater than the mean of the normal
samples + 3 standard deviations were considered positive. Real time PCR on
blood
samples was performed using the TaqmanTM procedure but extending to 50 cycles
using
forward and reverse primers and probes specific for P501 S. Of the eight
samples tested,
6 were positive for P501 S and /3-actin signal. The remaining 2 samples had no
detectable (3-actin or P501 S. No P501 S signal was observed in the four
normal blood
samples tested.
EXAMPLE 22
EXPRESSION OF THE PROSTATE-SPECIFIC ANTIGENS P7O3P AND PSO1 S IN
SLID MOUSE-PASSAGED PROSTATE TUMORS
When considering the effectiveness of antigens in the treatment of
prostate cancer, the continued presence of the antigens in tumors during
androgen
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
203
ablation therapy is important. The presence of the prostate-specific antigens
P703P and
P501 S in prostate ,tumor samples grown in SCID mice in the presence of
testosterone
was evaluated as follows.
Two prostate tumors that had metastasized to the bone were removed
from patients, implanted into SCID mice and grown in the presence of
testosterone.
Tumors were evaluated for mRNA expression of P703P, P501 S and PSA using
quantitative real time PCR with the SYBR green assay method. Expression of
P703P
and PSOI S in a prostate tumor was used as a positive control and the absence
in normal
intestine and normal heart as negative controls. In both cases, the specific
mRNA was
present in late passage tumors. Since the bone metastases were grown in the
presence
of testosterone, this implies that the presence of these genes would not be
lost during
androgen ablation therapy.
EXAMPLE 23
I S ANTI-P503S MONOCLONAL ANTIBODY INHIBITS TUMOR GROWTH IN VIVO
The ability of the anti-P503S monoclonal antibody 20D4 to suppress
tumor formation in mice was examined as follows.
Ten SCID mice were injected subcutaneously with HEK293 cells that
expressed P503S. Five mice received 150 micrograms of 20D4 intravenously at
day 0
(time of tumor cell injection), day 5 and day 9. Tumor size was measured for
50 days.
Of the five animals that received no 20D4, three formed detectable tumors
after about 2
weeks which continued to enlarge throughout the study. In contrast, none of
the five
mice that received 20D4 formed tumors. These results demonstrate that the anti-
P503S
Mab 20D4 displays potent anti-tumor activity in vivo.
EXAMPLE 24
CHARACTERIZATION OF A T CELL RECEPTOR CLONE FROM A
P501 S-SPECIFIC T CELL CLONE
T cells have a limited lifespan. However, cloning of T cell receptor
(TCR) chains and subsequent transfer essentially enables infinite propagation
of the T
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
204
cell specificity. Cloning of tumor-antigen TCR chains allows the transfer of
the
specificity into T cells isolated from patients that share the TCR MHC-
restricting allele.
Such T cells could then be expanded and used in adoptive transfer settings to
introduce
the tumor antigen specificity into patients carrying tumors that express the
antigen. T
S cell receptor alpha and beta chains from a CD8 T cell ~ clone specific for
the prostate-
specific antigen PSO1 S were isolated and sequenced as follows.
Total rnRNA from 2 x 106 cells from CTL clone 4ES (described above in
Example 12) was isolated using Trizol reagent and cDNA was synthesized. To
determine Va and Vb sequences in this clone, a panel of Va and Vb subtype-
specific
primers was synthesized and used in RT-PCR reactions with cDNA generated from
each of the clones. The RT-PCR reactions demonstrated that each of the clones
expressed a common Vb sequence that corresponded to the Vb7 subfamily.
Futhermore, using cDNA generated from the clone, the Va sequence expressed was
determined to be Va6. To clone the full TCR alpha and beta chains from clone
4ES,
1 S primers were designed that spanned the initiator and terminator-coding TCR
nucleotides. The primers were as follows: TCR Valpha-6 S'(sense): GGATCC---
GCCGCCACC-ATGTCACTTTCTAGCCTGCT (SEQ ID NO: 7S6) BamHI site
Kozak TCR alpha sequence TCR alpha 3' (antisense): GTCGAC---
TCAGCTGGACCACAGCCGCAG (SEQ ID NO: 7S7) SaII site TCR alpha constant
sequence TCR Vbeta-7. S'(sense): GGATCC---GCCGCCACC--
ATGGGCTGCAGGCTGCTCT (SEQ ID NO: 7S8) BamHI site, Kozak TCR alpha
sequence TCR beta 3' (antisense): GTCGAC---TCAGAAATCCTTTCTCTTGAC (SEQ
ID NO: 7S9) SaII site TCR beta constant sequence. Standard 3S cycle RT-PCR
reactions were established using cDNA synthesized from the CTL clone and the
above
primers, employing the proofreading thermostable polymerase PWO (Roche,
Nutley,
NJ).
The resultant specific bands (approx. 8S0 by for alpha and approx. 9S0
for beta) were ligated into the PCR blunt vector (Invitrogen) and transformed
into E.
coli. E . coli transformed with plasmids containing full-length alpha and beta
chains
were identified, and large scale preparations of the corresponding plasmids
were
generated. Plasmids containing full-length TCR alpha and beta chains were
submitted
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
205
for sequencing. The sequencing reactions demonstrated the cloning of full-
length TCR
alpha and beta chains with the determined cDNA sequences for the Vb and Va
chains
being shown in SEQ ID NO: 760 and 761, respectively. The corresponding amino
acid
sequences are shown in SEQ ID NO: 762 and 763, respectively. The Va sequence
was
shown by nucleotide sequence alignment to be 99% identical (347/348) to Va6.2,
and
the Vb to be 99% identical to Vb7 (336/338).
From the foregoing it will be appreciated that, although specific
embodiments of the invention have been described herein for purposes of
illustration,
various modifications may be made without deviating from the spirit and scope
of the
invention. Accordingly, the invention is not limited except as by the appended
claims.
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
1
SEQUENCE LISTING
<110> Corixa Corporation
Smithkline Beechan Biologicals S.A.
Xu, Jiangchun
Dillon, Davin C.
Mitcham, Jennifer L.
Harlocker, Susan L.
Jiang, Yuqui
Reed, Steven G.
Kalos, Michael D.
Fanger, Gary R.
Retter, Marc W.
Stolk, John A.
Day, Craig H.
Skeiky, Yasir A.W.
Wang, Aijun
Meagher, Medeleine Joy
Vanderbrugge, Didier
Dewerchin, Marianne
Dehottay, Ph.
de Rop, Philippe
<120> COMPOSITIONS AND METHODS FOR THE THERAPY AND
DIAGNOSIS OF PROSTATE CANCER
<130> 210121.42722PC
<140> PCT
<141> 2001-01-16
<160> 792
<170> FastSEQ for Windows Version 3.0
<210> 1
<211> 814
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(814)
<223> n = A,T,C or G
<400>
1
tttttttttttttttcacagtataacagctctttatttctgtgagttctactaggaaatc 60
atcaaatctgagggttgtctggaggacttcaatacacctccccccatagtgaatcagctt 120
ccagggggtccagtccctctccttacttcatccccatcccatgccaaaggaagaccctcc 180
ctccttggctcacagccttctctaggcttcccagtgcctccaggacagagtgggttatgt 240
tttcagctccatccttgctgtgagtgtctggtgcgttgtgcctccagcttctgctcagtg 300
cttcatggacagtgtccagcacatgtcactctccactctctcagtgtggatccactagtt 360
ctagagcggccgccaccgcggtggagctccagcttttgttccctttagtgagggttaatt 420
gcgcgcttggcgtaatcatggtcataactgtttcctgtgtgaaattgttatccgctcaca 480
attccacacaacatacgagccggaagcataaagtgtaaagcctggggtgcctaatgagtg 540
anctaactcacattaattgcgttgcgctcactgnccgctttccagtcnggaaaactgtcg 600
tgccagctgcattaatgaatcggccaacgcncggggaaaagcggtttgcgttttgggggc 660
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
2
tcttccgctt ctcgctcact nantcctgcg ctcggtcntt cggctgcggg gaacggtatc 720
actcctcaaa ggnggtatta cggttatccn naaatcnggg gatacccngg aaaaaanttt 780
aacaaaaggg cancaaaggg cngaaacgta aaaa 814
<210> 2
<211> 816
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(816)
<223> n = A,T,C or G
<400> 2
acagaaatgttggatggtggagcacctttctatacgacttacaggacagcagatggggaa 60
ttcatggctgttggagcaatagaaccccagttctacgagctgctgatcaaaggacttgga 120
ctaaagtctgatgaacttcccaatcagatgagcatggatgattggccagaaatgaagaag 180
aagtttgcagatgtatttgcaaagaagacgaaggcagagtggtgtcaaatctttgacggc 240
acagatgcctgtgtgactccggttctgacttttgaggaggttgttcatcatgatcacaac 300
aaggaacggggctcgtttatcaccagtgaggagcaggacgtgagcccccgccctgcacct 360
ctgctgttaaacaccccagccatcccttctttcaaaagggatccactagttctagaagcg 420
gccgccaccgcggtggagctccagcttttgttccctttagtgagggttaattgcgcgctt 480
ggcgtaatcatggtcatagctgtttcctgtgtgaaattgttatccgctcacaattccccc 540
aacatacgagccggaacataaagtgttaagcctggggtgcctaatgantgagctaactcn 600
cattaattgcgttgcgctcactgcccgctttccagtcgggaaaactgtcgtgccactgcn 660
ttantgaatcngccaccccccgggaaaaggcggttgcnttttgggcctcttccgctttcc 720
tcgctcattgatcctngcncccggtcttcggctgcggngaacggttcactcctcaaaggc 780
ggtntnccggttatccccaaacnggggatacccnga 816
<210> 3
<211> 773
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(773)
<223> n = A,T,C or G
<400> 3
cttttgaaagaagggatggctggggtgtttaacagcagaggtgcagggcgggggctcacg 60
tcctgctcctcactggtgataaacgagccccgttccttgttgtgatcatgatgaacaacc 120
tcctcaaaagtcagaaccggagtcacacaggcatctgtgccgtcaaagatttgacaccac 180
tctgccttcgtcttctttgcaaatacatctgcaaacttcttcttcatttctggccaatca 240
tccatgctcatctgattgggaagttcatcagactttagtccanntcctttgatcagcagc 300
tcgtagaactggggttctattgctccaacagccatgaattccccatctgctgtcctgtaa 360
gtcgtatagaaaggtgctccaccatccaacatgttctgtcctcgagggggggcccggtac 420
ccaattcgccctatantgagtcgtattacgcgcgctcactggccgtcgttttacaacgtc 480
gtgactgggaaaaccctgggcgttaccaacttaatcgccttgcagcacatccccctttcg 540
ccagctgggcgtaatancgaaaaggcccgcaccgatcgcccttccaacagttgcgcacct 600
gaatgggnaaatgggacccccctgttaccgcgeattnaacccccgcngggtttngttgtt 660
acccccacntnnaccgcttacactttgccagcgccttancgcccgctccctttcnccttt 720
cttcccttcctttcncnccnctttcccccggggtttcccccntcaaaccccna 773
<210> 4
<211> 828
<212> DNA
_r~ .,.,.., , .._ _.r.~J~~_b;k.~ :~F 5.,,;.~ ~'SSia..~._x~-...~ t,..:t~v
'~'a~..~~_ ......~ .~..:.. ~:.t,...~irz.,,.sr.~x.~,.zr.~. -w;.,~.., t_ ~~'-
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
3
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(828)
<223> n = A,T,C or G
<400>
4
cctcctgagtcctactgacctgtgctttctggtgtggagtccagggctgctaggaaaagg 60
aatgggcagacacaggtgtatgccaatgtttctgaaatgggtataatttcgtcctctcct 120
tcggaacactggctgtctctgaagacttctcgctcagtttcagtgaggacacacacaaag 180
acgtgggtgaccatgttgtttgtggggtgcagagatgggaggggtggggcccaccctgga 240
agagtggacagtgacacaaggtggacactctctacagatcactgaggataagctggagcc 300
acaatgcatgaggcacacacacagcaaggatgacnctgtaaacatagcccacgctgtcct 360
gngggcactgggaagcctanatnaggccgtgagcanaaagaaggggaggatccactagtt 420
ctanagcggccgccaccgcggtgganctccancttttgttccctttagtgagggttaatt 480
gcgcgcttggcntaatcatggtcatanctntttcctgtgtgaaattgttatccgctcaca 590
attccacacaacatacganccggaaacataaantgtaaacctggggtgcctaatgantga 600
ctaactcacattaattgcgttgcgctcactgcccgctttccaatcnggaaacctgtcttg 660
ccncttgcattnatgaatcngccaacccccggggaaaagcgtttgcgttttgggcgctct 720
tccgcttcctcnctcanttantccctncnctcggtcattcoggctgcngcaaaccggttc 780
accncctccaaagggggtattccggtttccccnaatccgggganancc 828
<210> 5
<211> 834
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(834)
<223> n = A,T,C or G
<400>
tttttttttttttttactgatagatggaatttattaagcttttcacatgtgatagcacat 60
agttttaattgcatccaaagtactaacaaaaactctagcaatcaagaatggcagcatgtt 120
attttataacaatcaacacctgtggcttttaaaatttggttttcataagataatttatac 180
tgaagtaaatctagccatgcttttaaaaaatgctttaggtcactccaagcttggcagtta 240
acatttggcataaacaataataaaacaatcacaatttaataaataacaaatacaacattg 300
taggccataatcatatacagtataaggaaaaggtggtagtgttgagtaagcagttattag 360
aatagaataccttggcctctatgcaaatatgtctagacactttgattcactcagccctga 420
cattcagttttcaaagtaggagacaggttctacagtatcattttacagtttccaacacat 480
tgaaaacaagtagaaaatgatgagttgatttttattaatgcattacatcctcaagagtta 540
tcaccaacccctcagttataaaaaattttcaagttatattagtcatataacttggtgtgc 600
ttattttaaattagtgctaaatggattaagtgaagacaacaatggtcccctaatgtgatt 660
gatattggtcatttttaccagcttctaaatctnaactttcaggcttttgaactggaacat 720
tgnatnacagtgttccanagttncaacctactggaacattacagtgtgcttgattcaaaa 780
tgttattttgttaaaaattaaattttaacctggtggaaaaataatttgaaatna 834
<210> 6
<211> 818
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(818)
<223> n = A, T, C or G
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
4
<400>
6
ttttttttttttttttttttaagaccctcatcaatagatggagacatacagaaatagtca 60
aaccacatctacaaaatgccagtatcaggcggcggcttcgaagccaaagtgatgtttgga 120
tgtaaagtgaaatattagttggcggatgaagcagatagtgaggaaagttgagccaataat 180
gacgtgaagtccgtggaagcctgtggctacaaaaaatgttgagccgtagatgccgtcgga 240
aatggtgaagggagactcgaagtactctgaggcttgtaggagggtaaaatagagacccag 300
taaaattgtaataagcagtgcttgaattatttggtttcggttgttttctattagactatg 360
gtgagctcaggtgattgatactcctgatgcgagtaatacggatgtgtttaggagtgggac 420
ttctaggggatttagcggggtgatgcctgttgggggccagtgccctcctagttggggggt 480
aggggctaggctggagtggtaaaaggctcagaaaaatcctgcgaagaaaaaaacttctga 540
ggtaataaataggattatcccgtatcgaaggcctttttggacaggtggtgtgtggtggcc 600
ttggtatgtgctttctcgtgttacatcgcgccatcattggtatatggttagtgtgttggg 660
ttantanggcctantatgaagaacttttggantggaattaaatcaatngcttggccggaa 720
gtcattanganggctnaaaaggccctgttangggtctgggctnggttttacccnacccat 780
ggaatncnccccccggacnantgnatccctattcttaa 818
<210> 7
<211> 817
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (l)...(817)
<223> n = A,T,C or G
<400>
7
tttttttttttttttttttttggctctagagggggtagagggggtgctatagggtaaata 60
cgggccctatttcaaagatttttaggggaattaattctaggacgatgggtatgaaactgt 120
ggtttgctccacagatttcagagcattgaccgtagtatacccccggtcgtgtagcggtga l80
aagtggtttggtttagacgtccgggaattgcatctgtttttaagcctaatgtggggacag 240
ctcatgagtgcaagacgtcttgtgatgtaattattatacnaatgggggcttcaatcggga 300
gtactactcgattgtcaacgtcaaggagtcgcaggtcgcctggttctaggaataatgggg 360
gaagtatgtaggaattgaagattaatccgccgtagtcggtgttctcctaggttcaatacc 420
attggtggccaattgatttgatggtaaggggagggatcgttgaactcgtctgttatgtaa 480
aggatnccttngggatgggaaggcnatnaaggactanggatnaatggcgggcangatatt 540
tcaaacngtctctanttcctgaaacgtctgaaatgttaataanaattaantttngttatt 600
gaatnttnnggaaaagggcttacaggactagaaaccaaatangaaaantaatnntaangg 660
cnttatcntnaaaggtnataaccnctcctatnatcccacccaatngnattccccacncnn 720
acnattggatnccccanttccanaaanggccnccccccggtgnannccnccttttgttcc 780
cttnantganggttattcncccctngcnttatcancc 817
<210> 8
<211> 799
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(799)
<223> n = A,T,C or G
<400> 8
catttccggg tttactttct aaggaaagcc gagcggaagc tgctaacgtg ggaatcggtg 60
cataaggaga actttctgct ggcacgcgct agggacaagc gggagagcga ctccgagcgt 120
ctgaagcgca cgtcccagaa ggtggacttg gcactgaaac agctgggaca catccgcgag 180
tacgaacagc gcctgaaagt gctggagcgg gaggtccagc agtgtagccg cgtcctgggg 240
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
tgggtggccgangcctganccgctctgccttgctgcccccangtgggccgccaccccctg 300
acctgccta,.ggtccaaacactgagccctgctggcggacttcaagganaacccccacangg 360
ggattttgctcctanantaaggctcatctgggcctcggcccccccacctggttggccttg 420
tctttgangtgagccccatgtccatctgggccactgtcnggaccacctttngggagtgtt 480
ctccttacaaccacannatgcccggctcctcccggaaaccantcccancctgngaaggat 540
caagncctgnatccactnntnctanaaccggccnccnccgcngtggaacccnccttntgt 600
tccttttcnttnagggttaatnncgccttggccttnccanngtcctncncnttttccnnt 660
gttnaaattgttangcncccnccnntcccncnncnncnancccgacccnnannttnnann 720
ncctgggggtnccnncngattgacccnnccnccctntanttgcnttngggnncnntgccc 780
ctttccctctnggganncg 799
<210> 9
<211> 801
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(801)
<223> n = A,T,C or G
<400>
9
acgccttgatcctcccaggctgggactggttctgggaggagccgggcatgctgtggtttg 60
taangatgacactcccaaaggtggtcctgacagtggcccagatggacatggggctcacct 120
caaggacaaggccaccaggtgcgggggccgaagcccacatgatccttactctatgagcaa 180
aatcccctgtgggggcttctccttgaagtccgccancagggctcagtctttggacccang 240
caggtcatggggttgtngnccaactgggggccncaacgcaaaanggcncagggcctcngn 300
cacccatcccangacgcggctacactnctggacctcccnctccaccactttcatgcgctg 360
ttcntacccgcgnatntgtcccanctgtttcngtgccnactccancttctnggacgtgcg 420
ctacatacgcccggantcncnctcccgctttgtccctatccacgtnccancaacaaattt 480
cnccntantgcaccnattcccacntttnncagntttccncnncgngcttccttntaaaag 540
ggttganccccggaaaatnccccaaagggggggggccnggtacccaactnccccctnata 600
gctgaantccecatnaccnngnctcnatgganccntccnttttaannacnttctnaactt 660
gggaananccctcgnccntncccccnttaatcccnccttgcnangnncntcccccnntcc 720
ncccnnntnggcntntnanncnaaaaaggcccnnnancaatctcctnncncctcanttcg 780
ccanccctcgaaatcggccnc 801
<210> 10
<211> 789
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(789)
<223> n = A,T,C or G
<400>
cagtctatntggccagtgtggcagctttccctgtggctgccggtgccacatgcctgtccc 60
acagtgtggccgtggtgacagcttcagccgccctcaccgggttcaccttctcagccctgc 120
agatcctgccctacacactggcctccctctaccaccgggagaagcaggtgttcctgccca 180
aataccgaggggacactggaggtgctagcagtgaggacagcctgatgaccagcttcctgc 240
caggccctaagcctggagctcccttccctaatggacacgtgggtgctggaggcagtggcc 300
tgctcccacctccacocgcgctctgcggggcctctgcctgtgatgtctccgtacgtgtgg 360
tggtgggtgagcccaccgangccagggtggttccgggccggggcatctgcctggacctcg 420
ccatcctggatagtgcttcctgctgtcccangtggccccatccctgtttatgggctccat 480
tgtccagctcagccagtctgtcactgcctatatggtgtctgccgcaggcctgggtctggt 540
cccatttactttgctacacaggtantatttgacaagaacganttggccaaatactcagcg 600
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
6
ttaaaaaatt ccagcaacat tgggggtgga aggcctgcct cactgggtcc aactccccgc 660
tcctgttaac cccatggggc tgccggcttg gocgccaatt tctgttgctg ccaaantnat 720
gtggctctct gctgccacct gttgctggct gaagtgcnta cngcncanct nggggggtng 780
ggngttccc 789
<210> 11
<211> 772
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(772)
<223> n = A,T,C or G
<400> 11
cccaccctacccaaatattagacaccaacacagaaaagctagcaatggattcccttctac 60
tttgttaaataaataagttaaatatttaaatgcctgtgtctctgtgatggcaacagaagg 120
accaacaggccacatcctgataaaaggtaagaggggggtggatcagcaaaaagacagtgc 180
tgtgggctgaggggacctggttcttgtgtgttgcccctcaggactcttcccctacaaata 240
actttcatatgttcaaatcccatggaggagtgtttcatcctagaaactcccatgcaagag 300
ctacattaaacgaagctgcaggttaaggggcttanagatgggaaaccaggtgactgagtt 360
tattcagctcccaaaaacccttctctaggtgtgtctcaactaggaggctagctgttaacc 420
ctgagcctgggtaatccacctgcagagtccccgcattccagtgcatggaacccttctggc 480
ctccctgtataagtccagac~tgaaacccccttggaaggnctccagtcaggcagccctana 540
aactggggaaaaaagaaaaggacgccccancccccagctgtgcanctacgcacctcaaca 600
gcacagggtggcagcaaaaaaaccactttactttggcacaaacaaaaactngggggggca 660
accccggcaccccnangggggttaacaggaancngggnaacntggaacccaattnaggca 720
ggcccnccaccccnaatnttgctgggaaatttttcctcccctaaattntttc 772
<210> 12
<211> 751
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(751)
<223> n = A,T,C or G
<400>
12
gccccaattccagctgccacaccacccacggtgactgcattagttcggatgtcatacaaa 60
agctgattgaagcaaccctctactttttggtcgtgagccttttgcttggtgcaggtttca 120
ttggctgtgttggtgacgttgtcattgcaacagaatgggggaaaggcactgttctctttg 180
aagtanggtgagtcctcaaaatccgtatagttggtgaagccacagcacttgagccctttc 240
atggtggtgttccacacttgagtgaagtcttcctgggaaccataatctttcttgatggca 300
ggcactaccagcaacgtcagggaagtgctcagccattgtggtgtacaccaaggcgaccac 360
agcagctgcnacctcagcaatgaagatgangaggangatgaagaagaacgtcncgagggc 420
acacttgctctcagtcttancaccatancagcccntgaaaaccaanancaaagaccacna 480
cnccggctgcgatgaagaaatnaccccncgttgacaaacttgcatggcactggganccac 540
agtggcccnaaaaatcttcaaaaaggatgccccatcnattgaccccccaaatgcccactg 600
ccaacaggggctgccccacncncnnaacgatganccnattgnacaagatctncntggtct 660
tnatnaacntgaaccctgcntngtggctcctgttcaggnccnnggcctgacttctnaann 720
aangaactcngaagnccccacnggananncg 751
<210> 13
<21I> 729
<212> DNA
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
7
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(729)
<223> n = A,T,C or G
<400> 13
gagccaggcgtccctctgcctgcccactcagtggcaacacccgggagctgttttgtcctt 60
tgtggancctcagcagtnccctctttcagaactcantgccaaganccctgaacaggagcc 120
accatgcagtgcttcagcttcattaagaccatgatgatcctcttcaatttgctcatcttt 180
ctgtgtggtgcagccctgttggcagtgggcatctgggtgtcaatcgatggggcatccttt 240
ctgaagatcttcgggccactgtcgtccagtgccatgcagtttgtcaacgtgggctacttc 300
ctcatcgcagccggcgttgtggtcttagctctaggtttcctgggctgctatggtgctaag 360
actgagagcaagtgtgccctcgtgacgttcttcttcatcctcctcctcatcttcattgct 420
gaggttgcaatgctgtggtcgccttggtgtacaccacaatggetgagcacttcctgacgt 480
tgctggtaatgcctgccatcaanaaaagattatgggttcccaggaanacttcactcaagt 540
gttggaacaccaccatgaaagggctcaagtgctgtggcttcnnccaactatacggatttt 600
gaagantcacctacttcaaagaaaanagtgcctttcccccatttctgttgcaattgacaa 660
acgtccccaacacagccaattgaaaacctgcacccaacccaaangggtccccaaccanaa 720
attnaaggg 729
<210> 14
<211> 816
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(8l6)
<223> n = A,T,C or G
<400> 14
tgctcttcctcaaagttgttcttgttgccataacaaccaccataggtaaagcgggcgcag 60
tgttcgctgaaggggttgtagtaccagcgcgggatgctctccttgcagagtcctgtgtct 120
ggcaggtccacgcagtgccctttgtcactggggaaatggatgcgctggagctcgtcaaag 180
ccactcgtgtatttttcacaggcagcctcgtccgacgcgtcggggcagttgggggtgtct 240
tcacactccaggaaactgtcnatgcagcagccattgctgcagcggaactgggtgggctga 300
cangtgccagagcacactggatggcgcctttccatgnnangggccctgngggaaagtccc 360
tganecccananctgcctctcaaangccccaccttgcacaccccgacaggctagaatgga 420
atcttcttcccgaaaggtagttnttcttgttgcccaanccanccccntaaacaaactctt 480
gcanatctgctccgngggggtcntantaccancgtgggaaaagaaccccaggcngcgaac 540
caancttgtttggatncgaagcnataatctnctnttctgcttggtggacagcaccantna 600
ctgtnnanctttagnccntggtcctcntgggttgnncttgaacctaatcnccnntcaact 660
gggacaaggtaantngccntcctttnaattcccnancntnccccctggtttggggttttn 720
cncnctcctaccccagaaannccgtgttcccccccaactaggggccnaaaccnnttnttc 780
cacaaccctnccccacccacgggttcngntggttng 816
<210> 15
<211> 783
<212> DNA
<213> Homo sapien
<220>
<22l> misc_feature
<222> (1)...(783)
<223> n = A,T,C or G
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
g
<400> 15
ccaaggcctgggcaggcatanacttgaaggtacaaccccaggaaoccctggtgctgaagg 60
atgtggaaaacacagattggcgcctactgcggggtgacacggatgtcagggtagagagga 120
aagacccaaaccaggtggaactgtggggactcaaggaangcacctacctgttccagctga 180
cagtgactagctcagaccacccagaggacacggecaacgtcacagtcactgtgctgtcca 240
ocaagcagacagaagactactgcctcgcatccaacaangtgggtcgctgccggggctctt 300
tcccacgctggtactatgaccccacggagcagatctgcaagagtttcgtttatggaggct 360
gcttgggcaacaagaacaactaccttcgggaagaagagtgcattctancctgtcngggtg 420
tgcaaggtgggcctttganangcanctctggggctcangcgactttcccccagggcccct 480
ccatggaaaggcgccatccantgttctctggcacctgtcagcccacccagttccgctgca 540
ncaatggctgctgcatcnacantttcctngaattgtgacaa'cacccccoantgcccccaa 600
ccctcccaacaaagcttccctgttnaaaaatacnccanttggcttttnacaaacncccgg 660
cncctccnttttccccnntnaacaaagggcnctngcntttgaactgcccnaacccnggaa 720
tctnccnnggaaaaantnccccccctggttcctnnaancocctccncnaaanctnccccc 780
ccc 783
<210> 16
<211> 801
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)..,(801)
<223> n = A,T,C or G
<400>
16
gccccaattccagctgccacaccacccacggtgactgcattagttcggatgtcatacaaa 60
agctgattgaagcaaccctctactttttggtcgtgagccttttgcttggtgcaggtttca 120
ttggctgtgttggtgacgttgtcattgcaacagaatgggggaaaggcactgttctctttg 180
aagtagggtgagtcctcaaaatccgtatagttggtgaagccacagcacttgagccctttc 240
atggtggtgttccacacttgagtgaagtcttcctgggaaccataatctttcttgatggca 300
ggcactaccagcaacgtcaggaagtgctcagccattgtggtgtacaccaaggcgaccaca 360
gcagctgcaacctcagcaatgaagatgaggaggaggatgaagaagaacgtcncgagggca 420
cacttgctctccgtcttagcaccatagcagcccangaaaccaagagcaaagaccacaacg 480
ccngctgcgaatgaaagaaantacccacgttgacaaactgcatggccactggacgacagt 540
tggcccgaanatcttcagaaaagggatgccccatcgattgaacacccanatgcccactgc 600
cnacagggctgcnccncncngaaagaatgagccattgaagaaggatcntcntggtcttaa 660
tgaactgaaaccntgcatggtggcccctgttcagggctcttggcagtgaattctganaaa 720
aaggaacngcntnagcccccccaaanganaaaacacccccgggtgttgccctgaattggc 780
ggccaagganccctgccccng 801
<210> 17
<211> 740
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(740)
<223> n = A,T,C or G
<400> 17
gtgagagccaggcgtccctctgcctgcccactcagtggcaacacccgggagctgttttgt 60
cctttgtggagcctcagcagttccctctttcagaactcactgccaagagccctgaacagg 120
agccaccatgcagtgcttcagcttcattaagaccatgatgatcctcttcaatttgctcat 180
ctttctgtgtggtgcagccctgttggcagtgggcatctgggtgtcaategatggggcatc 240
ctttctgaagatcttcgggccactgtcgtccagtgccatgcagtttgtcaacgtgggcta 300
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
9
cttcctcatcgcagccggcgttgtggtctttgctcttggtttcctgggctgctatggtgc 360
taagacggagagcaagtgtgccctcgtgacgttcttcttcatcctcctcctcatcttcat 420
tgctgaagttgcagctgctgtggtcgccttggtgtacaccacaatggctgaaccattcct 480
gacgttgctggtantgcctgccatcaanaaagattatgggttcccaggaaaaattcactc 540
aantntggaacaccnccatgaaaagggctccaatttctgntggcttccccaactataccg 600
gaattttgaaagantcnccctacttccaaaaaaaaananttgcctttncccccnttctgt 660
tgcaatgaaaacntcccaanacngccaatnaaaacctgcccnnncaaaaaggntcncaaa 720
caaaaaaantnnaagggttn 740
<210> 18
<211> 802
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(802)
<223> n = A,T,C or G
<400>
18
ccgctggttgcgctggtccagngnagccacgaagcacgtcagcatacacagcctcaatca 60
caaggtcttccagctgccgcacattacgcagggcaagagcctccagcaacactgcatatg 120
ggatacactttactttagcagccagggtgacaactgagaggtgtcgaagcttattcttct '180
gagcctctgttagtggaggaagattccgggcttcagctaagtagtcagcgtatgtcccat 240
aagcaaacactgtgagcagccggaaggtagaggcaaagtcactctcagccagctctctaa 300
cattgggcatgtccagcagttctccaaacacgtagacaccagnggcctccagcacctgat 36,0
ggatgagtgtggccagcgctgcccccttggccgacttggctaggagcagaaattgctcct 420
ggttctgccctgtcaccttcacttccgcactcatcactgcactgagtgtgggggacttgg 480
gctcaggatgtccagagacgtggttccgccccctcncttaatgacaccgnccanncaaoc 540
gtcggctcccgccgantgngttcgtcgtncctgggtcagggtctgctggccnctacttgc 600
aancttcgtcnggcccatggaattcaccncaccggaactngtangatccactnnttctat 660
aaccggncgccaccgcnnntggaactccactcttnttncctttacttgagggttaaggtc 720
acccttnncgttaccttggtccaaaccntnccntgtgtcganatngtnaatcnggnccna 780
tnccanccncatangaagccng 802
<210> 19
<211> 731
<212> DNA
<213> Homo sapien
<220>
<22l> misc_feature
<222> (1)...(731)
<223> n = A,T,C or G
<400> 19
cnaagcttccaggtnacgggccgcnaancctgacccnaggtancanaangcagncngcgg 60
gagcccaccgtcacgnggnggngtctttatnggagggggcggagccacatcnctggacnt 120
cntgaccccaactccccnccncncantgcagtgatgagtgcagaactgaaggtnacgtgg 180
caggaaccaagancaaannctgctccnntccaagtcggcnnagggggcggggctggccac 240
gcncatccntcnagtgctgnaaagccccnncctgtctacttgtttggagaacngcnnnga 300
catgcccagngttanataacnggcngagagtnantttgcctctcccttccggctgcgcan 360
cgngtntgcttagnggacataacctgactacttaactgaacccnngaatctnccncccct 420
ccactaagctcagaacaaaaaacttcgacaccactcanttgtcacctgnctgctcaagta 480
aagtgtaccccatncccaatgtntgctngangctctgncctgcnttangttcggtcctgg 540
gaagacctatcaattnaagctatgtttctgactgcctcttgctccctgnaacaancnacc 600
cnncnntccaagggggggncggcccccaatccccccaaccntnaattnantttanccccn 660
cccccnggcccggccttttacnancntcnnnnacngggnaaaaccnnngctttncccaac 720
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
1~
nnaatccncc t 731
<210> 20
<211> 754
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(754)
<223> n = A,T,C or G
<400> 20
tttttttttttttttttttttaaaaaccccctccattnaatgnaaacttccgaaattgtc 60
caaccccctcntccaaatnnccntttccgggngggggttccaaacccaanttanntttgg 120
annttaaattaaatnttnnttggnggnnnaanccnaatgtnangaaagttnaacccanta 180
tnancttnaatncctggaaaccngtngnttccaaaaatntttaacccttaantccctccg 240
aaatngttnanggaaaacccaanttctcntaaggttgtttgaaggntnaatnaaaanccc 300
nnccaattgtttttngccacgcctgaattaattggnttccgntgttttccnttaaaanaa 360
ggnnanccccggttantnaatccccccnnccccaattataccgantttttttngaattgg 420
gancccncgggaattaacggggnnnntccctnttggggggcnggnnccccccccntcggg 480
ggttngggncaggncnnaattgtttaagggtccgaaaaatccctccnagaaaaaaanctc 540
ccaggntgagnntngggtttnccccccccccanggcccctctcgnanagttggggtttgg 600
ggggcctgggattttntttcccctnttncctcccccccccccnggganagaggttngngt 660
tttgntcnncggccccnccnaaganctttnccganttnanttaaatccntgcctnggcga 720
agtccnttgnagggntaaanggccccctnncggg 754
<210> 21
<211> 755
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(755)
<223> n = A,T,C or G
<400>
21
atcancccatgaccccnaacnngggaccnctcanccggncnnncnaccnccggccnatca 60
nngtnagnncactncnnttnnatcacnccccnccnactacgcccncnanccnacgcncta 120
nncanatnccactganngcgcgangtnganngagaaanctnataccanagncaccanacn 180
ccagctgtccnanaangcctnnnatacnggnnnatccaatntgnancctccnaagtattn 240
nncnncanatgattttcctnanccgattacecntnccccctancccctcccccccaacna 300
cgaaggcnctggnccnaaggnngcgncnccccgctagntccccnncaagtcncncnccta 360
aactcanccnnattacncgcttcntgagtatcactccccgaatctcaccctactcaactc 420
aaaaanatcngatacaaaataatncaagcctgnttatnacactntgactgggtctctatt 480
ttagnggtccntnaancntcctaatacttccagtctnccttcnccaatttccnaanggct 540
ctttcngacagcatnttttggttcccnnttgggttcttanngaattgcccttcntngaac 600
gggctcntcttttccttcggttancctggnttcnnccggccagttattatttcccntttt 660
aaattcntnccntttanttttggcnttcnaaacccccggccttgaaaacggccccctggt 720
aaaaggttgttttganaaaatttttgttttgttcc 755
<210> 22
<2l1> 849
<212> DNA
<213> Homo sapien
<220>
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
11
<221> misc_feature
<222> (1)...(849)
<223> n = A,T,C or G
<400> 22
tttttttttttttttangtgtngtcgtgcaggtagaggcttactacaantgtgaanacgt 60
acgctnggantaangcgacccganttctagganncnccctaaaatcanactgtgaagatn l20
atcctgnnnacggaanggtcaccggnngatnntgctagggtgnccnctcccannncnttn l80
cataactcngnggccctgcccaccaccttcggcggcccngngnocgggcccgggtcattn 240
gnnttaaccncactnngcnancggtttccnnccccnncngacccnggcgatccggggtnc 300
tctgtcttcccctgnagncnanaaantgggccncggncccctttacccctnnacaagcca 360
cngccntctanccncngccccccctccantnngggggactgccnanngctccgttnctng 420
nnacaccnnngggtncctcggttgtcgantcnaccgnangccanggattccnaaggaagg 480
tgcgttnttggcccctacccttcgctncggnncacccttcccgacnanganccgctcccg 540
cncnncgnngcctcncctcgcaacacccgcnctcntcngtncggnnncccccccacccgc 600
nccctcncncngncgnancnctccnccnccgtctcanncaccaccccgccccgccaggcc 660
ntcanccacnggnngacnngnagcncnntcgcnccgcgcngcgncnccctcgccncngaa 720
ctncntcnggccantnncgctcaanccnnacnaaacgccgctgcgcggcccgnagcgncc 780
ncctccncgagtcctcccgncttccnacccangnnttccncgaggacacnnnaccccgcc 840
nncangcgg 849
<210> 23
<211> 872
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(872)
<223> n = A,T,C or G
<400> 23
gcgcaaactatacttcgctcgnactcgtgcgcctcgctnctcttttcctccgcaaccatg 60
tctgacnancccgattnggcngatatcnanaagntcgancagtccaaactgantaacaca 120
cacacncnanaganaaatccnctgccttccanagtanacnattgaacnngagaaccangc 180
nggcgaatcgtaatnaggcgtgcgccgccaatntgtcnccgtttattntnccagcntcnc 240
ctnccnaccctacntcttcnnagctgtcnnacccctngtncgnaccccccnaggtcggga 300
tcgggtttnnnntgaccgngcnncccctccccccntccatnacganccncccgcaccacc 360
nanngcncgcnccccgnnctcttcgccnccctgtcctntncccctgtngcctggcncngn 420
accgcattgaccctcgccnnctncnngaaancgnanacgtccgggttgnnannancgctg 480
tgggnnngcgtctgcnccgcgttccttccnncnncttccaccatcttcnttacngggtct 540
ccncgccntctcnnncacnccctgggacgctntcctntgccccccttnactccccccctt 600
cgncgtgneccgnccccaccntcatttncanacgntcttcacaannncctggntnnctcc 660
cnancngncngtcanccnagggaagggnggggnnccnntgnttgacgttgnggngangtc 720
cgaanantcctcnccntcancnctacccctcgggcgnnctctcngttnccaacttancaa 780
ntctcccccgngngcncntctcagcctcncccnccccnctctctgcantgtnctctgctc 840
tnaccnntacgantnttcgncnccctctttcc 872
<210> 24
<211> 815
<212> DNA
<2l3> Homo sapien
<220>
<221> misc_feature
<222> (1)...(815)
<223> n = A,T,C or G
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
12
<400> 24
gcatgcaagcttgagtattctatagngtcacctaaatancttggcntaatcatggtcnta 60
nctgncttcctgtgtcaaatgtatacnaantanatatgaatctnatntgacaaganngta 120
tcntncattagtaacaantgtnntgtccatcctgtcngancanattcccatnnattncgn 180
cgcattcncngcncantatntaatngggaantcnnntnnnncaccnncatctatcntncc 240
gcnccctgactggnagagatggatnanttctnntntgaccnacatgttcatcttggattn 300
aananccccccgcngnccaccggttngnngcnagccnntcccaagacctcctgtggaggt 360
aacctgegtcaganncatcaaacntgggaaacccgcnnccangtnnaagtngnnncanan 420
gatcccgtccaggnttnaccatcccttcncagcgccccctttngtgccttanagngnagc 480
gtgtccnanccnctcaacatganacgcgccagnccanccgcaattnggcacaatgtcgnc 540
gaaccccctagggggantnatncaaanccccaggattgtccncncangaaatcccncanc 600
cccnccctacccnnctttgggacngtgaccaantcccggagtnccagtccggccngnctc 660
ccccaccggtnnccntgggggggtgaanctcngnntcanccngncgaggnntcgnaagga 720
accggncctnggncgaanngancnntcngaagngccncntcgtataaccccccctcncca 780
nccnacngntagntcccccccngggtncggaangg 8l5
<210> 25
<211> 775
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(775)
<223> n = A,T,C or G
<400> 25
ccgagatgtctcgctccgtggccttagctgtgctcgcgctactctctctttctggcctgg 60
aggctatccagcgtactccaaagattcaggtttactcacgtcatccagcagagaatggaa l20
agtcaaatttcctgaattgctatgtgtctgggtttcatccatccgacattgaanttgact 180
tactgaagaatgganagagaattgaaaaagtggagcattcagacttgtctttcagcaagg 240
actggtctttctatctcntgtactacactgaattcacccccactgaaaaagatgagtatg 300
cctgccgtgtgaaccatgtgactttgtcacagcccaagatagttaagtgggatcgagaca 360
tgtaagcagncnncatggaagtttgaagatgccgcatttggattggatgaattccaaatt 420
ctgcttgcttgcnttttaatantgatatgcntatacaccctaccctttatgnccccaaat 480
tgtaggggttacatnantgttcncntnggacatgatcttcctttataantccnccnttcg 540
aattgcccgtcncccngttnngaatgtttccnnaaccacggttggctcccccaggtcncc 600
tcttacggaagggcctgggccnctttncaaggttgggggaaccnaaaatttcncttntgc 660
ccncccnccacnntcttgngnncncantttggaacccttccnattccccttggcctcnna 720
nccttnnctaanaaaacttnaaancgtngcnaaanntttnacttccccccttacc 775
<210> 26
<211> 820
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(820)
<223> n = A,T,C or G
<400>
26
anattantacagtgtaatcttttcccagaggtgtgtanagggaacggggcctagaggcat 60
cccanagatancttatancaacagtgctttgaccaagagctgctgggcacatttcctgca 120
gaaaaggtggcggtccccatcactcctcctctcccatagccatcccagaggggtgagtag 180
ccatcangccttcggtgggagggagtcanggaaacaacanaccacagagcanacagacca 240
ntgatgaccatgggcgggagcgagcctcttccctgnaccggggtggcananganagccta 300
nctgaggggtcacactataaacgttaacgaccnagatnancacctgcttcaagtgcaccc 360
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
13
ttcctacctg acnaccagng accnnnaact gcngcctggg gacagcnctg ggancagcta 420
acnnagcactcacctgcccccccatggccgtncgcntccctggtcctgncaagggaagct 480
ccctgttggaattncgggganaccaaggganccccctcctccanctgtgaaggaaaaann 540
gatggaattttncccttccggccnntcccctcttcctttacacgccccctnntactcntc 600
tccctctnttntcctgncncacttttnaccccnnnatttcccttnattgatcggannctn 660
ganattccactnncgcctnccntcnatcngnaanacnaaanactntctnacccnggggat 720
gggnncctcgntcatectctctttttcnctaccnccnnttctttgcctctccttngatca 780
tccaaccntcgntggccntncccccccnnntcctttnccc ~ 820
<210> 27
<211> 818
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(818)
<223> n = A,T,C or G
<400>
27
tctgggtgatggcctcttcctcctcagggacctctgactgctctgggccaaagaatctct 60
tgtttcttctccgagccccaggcagcggtgattcagccctgcccaacctgattctgatga 120
ctgcggatgctgtgacggacccaaggggcaaatagggtcccagggtccagggaggggcgc 180
ctgctgagcacttccgcccctcaccctgcccagcccctgccatgagctctgggctgggtc 240
tccgcctccagggttctgctcttccangcangccancaagtggcgctgggccacactggc 300
ttcttcctgccccntccctggctctgantctctgtcttcctgtcctgtgcangcnccttg 360
gatctcagtttccctcnctcanngaactctgtttctganntcttcanttaactntgantt 420
tatnaccnantggnctgtnctgtcnnactttaatgggccngaccggctaatccctccctc 480
nctcccttccanttcnnnnaaccngcttnccntcntctccccntancccgccngggaanc 540
ctcctttgccctnaccangggccnnnaccgcccntnnctnggggggcnnggtnnctncnc 600
ctgntnnccccnctcncnnttncctcgtcccnncnncgcnnngcannttcncngtcccnn 660
tnnctcttcnngtntcgnaangntcncntntnnnnngncnngntnntncntccctctcnc 720
cnnntgnangtnnttnnnncncngnnccccnnnncnnnnnnggnnntnnntctncncngc 780
cccnncccccngnattaaggcctccnntctccggccnc 818
<210> 28
<211> 731
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(731)
<223> n = A,T,C or G
<400> 28
aggaagggcggagggatattgtangggattgagggataggagnataangggggaggtgtg 60
tcccaacatganggtgnngttctcttttgaangagggttgngtttttannccnggtgggt 120
gattnaaccccattgtatggagnnaaaggntttnagggatttttcggctcttatcagtat 180
ntanattcctgtnaatcggaaaatnatntttcnncnggaaaatnttgctcccatccgnaa 240
attnctcccgggtagtgcatnttngggggncngccangtttcccaggctgctanaatcgt 300
actaaagnttnaagtgggantncaaatgaaaacctnncacagagnatccntacccgactg 360
tnnnttnccttcgccctntgactctgcnngagcccaatacccnngngnatgtcncccngn 420
nnngcgncnctgaaannnnctcgnggctnngancatcanggggtttcgcatcaaaagcnn 480
cgtttcncatnaaggcactttngcctcatccaaccnctngccctcnnccatttngccgtc 540
nggttcncctacgctnntngcncctnnntnganattttncccgcctngggnaancctcct 600
gnaatgggtagggncttntcttttnaccnngnggtntactaatcnnctncacgcntnctt 660
tctcnaceccccccctttttcaatcccancggcnaatggggtctccccnncgangggggg 720
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
14
nnncccannc c 731
<210> 29
<211> 822
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(822)
<223> n = A,T,C or G
<400>
29
actagtccagtgtggtggaattccattgtgttggggncncttctatgantantnttagat 60
cgctcanacctcacancctoccnacnangcctataangaanannaataganctgtncnnt 120
atntntacnctcatanncctcnnnacccactccctcttaacccntactgtgcctatngcn 180
tnnctantctntgccgcctncnanccaccngtgggccnaccncnngnattctcnatctcc 240
tcnccatntngcctanantangtncataccctatacctacnccaatgctannnctaancn 300
tccatnanttannntaactaccactgacntngactttcncatnanctcctaatttgaatc 360
tactctgactcccacngcctannnattagcancntcccccnacnatntctcaaccaaatc 420
ntcaacaacctatctanctgttcnccaaccnttncctccgatccccnnacaacccccctc 480
ccaaatacccnccacctgacncctaacccncaccatcccggcaagccnanggncatttan 540
ccactggaatcacnatngganaaaaaaaacccnaactctctancncnnatctccctaana 600
aatnctcctnnaatttactnncantnccatcaancccacntgaaacnnaacccctgtttt 660
tanatcccttctttcgaaaaccnaccctttannncccaacctttngggcccccccnctnc 720
ccnaatgaaggncncccaatcnangaaacgnccntgaaaaancnaggcnaanannntccg 780
canatcctatcccttanttnggggncccttncccngggcccc 822
<210> 30
<211> 787
<2l2> DNA
<213> Homo sapien
<220>
<22l> misc_feature
<222> (1)...(787)
<223> n = A,T,C or G
<400>
30
cggccgcctgctctggcacatgcctcctgaatggcatcaaaagtgatggactgcccattg 60
ctagagaagaccttctctcctactgtcattatggagccctgcagactgagggctcccctt 120
gtctgcaggatttgatgtctgaagtcgtggagtgtggcttggagctcctcatctacatna 180
gctggaagccctggagggcctctctcgccagcctcccccttctctccacgctctccangg 240
acaccaggggctccaggcagcccattattcccagnangacatggtgtttctccacgcgga 300
cccatggggcctgnaaggccagggtctcctttgacaccatctctcccgtcctgcctggca 360
ggccgtgggatccactanttctanaacggncgccaccncggtgggagctccagcttttgt 420
tcccnttaatgaaggttaattgcncgcttggcgtaatcatnggtcanaactntttcctgt 480
gtgaaattgtttntcccctcncnattccncncnacatacnaacccggaancataaagtgt 540
taaagcctgggggtngcctnnngaatnaactnaactcaattaattgcgttggctcatggc 600
ccgctttccnttcnggaaaactgtcntcccctgcnttnntgaatcggccaccccccnggg 660
aaaagcggtttgcnttttngggggntccttccncttcccccctcnctaanccctncgcct 720
cggtcgttncnggtngcggggaangggnatnnnctcccncnaagggggngagnnngntat 780
cccoaaa 787
<210> 31
<21l> 799
<212> DNA
<213> Homo sapien
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
IS
<220>
<221> misc_feature
<222> (1)...(799)
<223> n = A,T,C or G
<400> 31
tttttttttttttttttggcgatgctactgtttaattgcaggaggtgggggtgtgtgtac 60
catgtaccagggctattagaagcaagaaggaaggagggagggcagagcgccctgctgagc 120
aacaaaggactcctgcagccttctctgtctgtctcttggcgcaggcacatggggaggcct l80
cccgcagggtgggggccaccagtccaggggtgggagcactacanggggtgggagtgggtg 240
gtggctggtncnaatggcctgncacanatccctacgattcttgacacctggatttcacca 300
ggggaccttctgttctcccanggnaacttcntnnatctcnaaagaacacaactgtttctt 360
cngcanttctggctgttcatggaaagcacaggtgtccnatttnggctgggacttggtaca 420
tatggttccggcccacctctcccntcnaanaagtaattcacccccccccnccntctnttg 480
cctgggcccttaantacccacaccggaactcanttanttattcatcttnggntgggcttg 540
ntnatcnccncctgaangcgccaagttgaaaggccacgccgtncccnctccccatagnan 600
nttttnncntcanctaatgcccccccnggcaacnatccaatccccccccntgggggcccc 660
agcccanggcccccgnctcgggnnnccngncncgnantccccaggntctcccantcngnc 720
ccnnngcncccccgcacgcagaacanaaggntngagccnccgcannnnnnnggtnncnac 780
ctcgccccccccnncgnng 799
<210> 32
<211> 789
<212> DNA
<2l3> Homo sapien
<220>
<221> misc_feature
<222> (1)...(789)
<223> n = A,T,C or G
<400>~
32
tttttttttttttttttttttttttttttttttttttttttttttttttttttttttttt 60
ttttnccnagggcaggtttattgacaacctcncgggacacaancaggctggggacaggac 120
ggcaacaggctccggcggcggcggcggcggccctacctgcggtaccaaatntgcagcctc 180
cgctcccgcttgatnttcctctgcagctgcaggatgccntaaaacagggcctcggccntn 240
ggtgggcaccctgggatttnaatttccacgggcacaatgcggtcgcancccctcaccacc 300
nattaggaatagtggtnttacccnccnccgttggcncactccccntggaaaccacttntc 360
gcggctccggcatctggtcttaaaccttgcaaacnctggggccctctttttggttantnt 420
nccngccacaatcatnactcagactggcncgggctggccccaaaaaancnccccaaaacc 480
ggnccatgtcttnncggggttgctgcnatntncatcacctcccgggcncancaggncaac 540
ccaaaagttcttgnggcccncaaaaaanctccggggggncccagtttcaacaaagtcatc 600
ccccttggcccccaaatcctccccccgnttnctgggtttgggaacccacgcctctnnctt 660
tggnnggcaagntggntcccccttcgggcccccggtgggcccnnctctaangaaaacncc 720
ntcctnnncaccatccccccnngnnacgnctancaangnatccctttttttanaaacggg 780
ccccccncg 789
<210> 33
<211> 793
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(793)
<223> n = A,T,C or G
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
16
<400>
33
gacagaacatgttggatggtggagcacctttctatacgacttacaggacagcagatgggg 60
aattcatggctgttggagcaatanaaccccagttctacgagctgctgatcaaaggacttg 120
gactaaagtctgatgaacttcccaatcagatgagcatggatgattggccagaaatgaana 180
agaagtttgcagatgtatttgcaaagaagacgaaggcagagtggtgtcaaatctttgacg 240
gcacagatgcctgtgtgactccggttctgacttttgaggaggttgttcatcatgatcaca 300
acaangaacggggctcgtttatcaccantgaggagcaggacgtgagcccccgccctgcac 360
ctctgctgttaaacaccccagccatcccttctttcaaaagggatccactacttctagagc 420
ggncgccaccgcggtggagctccagcttttgttccctttagtgagggttaattgcgcgct 480
tggcgtaatcatggtcatanctgtttcctgtgtgaaattgttatccgctcacaattccac 540
acaacatacganccggaagcatnaaattttaaagcctggnggtngcctaatgantgaact 600
nactcacattaattggctttgcgctcactgcccgctttccagtccggaaaacctgtcctt 660
gccagctgccnttaatgaatcnggccaccccccggggaaaaggcngtttgcttnttgggg 720
cgcncttcccgctttctcgcttcctgaantccttccccccggtctttcggcttgcggcna 780
acggtatcnacct 793
<210> 34
<211> 756
<212> DNA
<213> Homo sapien
<220>
<221> miso_feature
<222> (1)...(756)
<223> n = A, T, C or G
<400> 34
gccgcgaccggcatgtacgagcaactcaagggcgagtggaaccgtaaaagccccaatctt 60
ancaagtgcggggaanagctgggtcgactcaagctagttcttctggagctcaacttcttg 120
ccaaccacagggaccaagctgaccaaacagcagctaattctggcccgtgacatactggag 180
atcggggcccaatggagcatcctacgcaangacatcccctccttcgagcgctacatggcc 240
cagctcaaatgctactactttgattacaangagcagctccccgagtcagcctatatgcac 300
cagctcttgggcctcaacetcctcttcctgctgtcccagaaccgggtggctgantnccac 360
acgganttgg.ancggctgcctgcccaangacatacanaccaatgtctacatcnaccacca 420
gtgtcctggagcaatactgatgganggcagctaccncaaagtnttcctggccnagggtaa 480
catcccccgccgagagctacaccttcttcattgacatcctgctcgacactatcagggatg 540
aaaatcgcngggttgctccagaaaggctncaanaanatccttttcnctgaaggcccccgg 600
atncnctagtnctagaatcggcccgccatcgcggtggancctccaacctttcgttnccct 660
ttactgagggttnattgccgcccttggcgttatcatggtcacnccngttncctgtgttga 720
aattnttaaccccccacaattccacgccnacattng 756
<210> 35
<211> 834
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(834)
<223> n = A,T,C or G
<400> 35
ggggatctctanatcnacctgnatgcatggttgtcggtgtggtcgctgtcgatgaanatg 60
aacaggatcttgcccttgaagctctcggctgctgtntttaagttgctcagtctgccgtca 120
tagtcagacacnctcttgggcaaaaaacancaggatntgagtcttgatttcacctccaat 180
aatcttcngggctgtctgctcggtgaactcgatgacnangggcagctggttgtgtntgat 240
aaantccancangttctccttggtgacctccccttcaaagttgttccggccttcatcaaa 300
cttctnnaanangannancccanctttgtcgagctggnatttgganaacacgtcactgtt 360
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
17
ggaaactgatcccaaatggtatgtcatccatcgcctctgctgcctgcaaaaaacttgctt 420
ggcncaaatccgactccccntccttgaaagaagccnatcacacccccctccctggactcc 480
nncaangactctnccgctnccccntccnngcagggttggtggcannccgggcccntgcgc 540
ttcttcagccagttcacnatnttcatcagcccctctgccagctgttntattccttggggg 600
ggaanccgtctctcccttcctgaannaactttgaccgtnggaatagccgcgcntcnccnt 660
aontnetgggccgggttcaaantccctccnttgnonntcncctcgggccattctggattt 720
nccnaactttttccttcccccnccccncggngtttggntttttcatngggccccaactct 780
gctnttggccantcccctgggggcntntancnccccctntggtcccntngggcc 834
<210> 36
<211> 814
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(814)
<223> n = A,T,C or G
<400>
36
cggncgctttccngccgcgccccgtttccatgacnaaggctcccttcangttaaatacnn 60
cctagnaaacattaatgggttgctctactaatacatcatacnaaccagtaagcctgccca 120
naacgccaactcaggccattcctaccaaaggaagaaaggctggtctctccaccccctgta 180
ggaaaggcctgccttgtaagacaccacaatncggctgaatctnaagtcttgtgttttact 240
aatggaaaaaaaaaataaacaanaggttttgttctcatggctgcccaccgcagcctggca 300
ctaaaacancccagcgctcacttctgcttgganaaatattctttgctcttttggacatca 360
ggcttgatggtatcactgccacntttccacccagctgggcncccttcccccatntttgtc 420
antganctggaaggcctgaancttagtctccaaaagtctcngcccacaagaccggccacc 480
aggggangtcntttncagtggatctgccaaanantacccntatcatcnntgaataaaaag 540
gcccctgaacganatgcttccancancctttaagacccataatcctngaaccatggtgcc 600
cttccggtctgatccnaaaggaatgttcctgggtcccantccctcctttgttncttacgt 660
tgtnttggacccntgctngnatnacccaantganatccccngaagcaccctncccctggc 720
atttgantttcntaaattctctgccctacnnctgaaagcacnattccctnggcnccnaan 780
ggngaactcaagaaggtctnngaaaaaccacncn 814
<210> 37
<211> 760
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(760)
<223> n = A,T,C or G
<400>
37
gcatgctgctcttcctcaaagttgttcttgttgccataacaaccaccataggtaaagcgg 60
gcgcagtgttcgctgaaggggttgtagtaccagcgcgggatgctctecttgcagagtcct 120
gtgtctggcaggtccacgcaatgccctttgtcactggggaaatggatgcgctggagctcg 180
tcnaanccactcgtgtatttttcacangcagcctcctccgaagcntccgggcagttgggg 240
gtgtcgtcacactccactaaactgtcgatncancagcccattgctgcagcggaactgggt 300
gggctgacaggtgccagaacacactggatnggcctttccatggaagggcctgggggaaat 360
cncctnancccaaactgcctctcaaaggccaccttgcacaccccgacaggctagaaatgc 420
actcttcttcccaaaggtagttgttcttgttgcccaagcancctccancaaaccaaaanc 480
ttgcaaaatctgctccgtgggggtcatnnntaccanggttggggaaanaaacccggcngn 540
ganccnccttgtttgaatgcnaaggnaataatcctcctgtcttgcttgggtggaanagca 600
caattgaactgttaacnttgggccgngttccnctngggtggtctgaaactaatcaccgtc 660
actggaaaaaggtangtgccttccttgaattcccaaanttcccctngntttgggtnnttt 720
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
Ig
ctcctctncc ctaaaaatcg tnttcccccc ccntanggcg 760
<210> 38
<211> 724
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(724)
<223> n = A,T,C or G
<400> 38
tttttttttttttttttttttttttttttttttttaaaaaccccctccattgaatgaaaa 60
cttccnaaattgtccaaccecctcnnccaaatnnccatttccgggggggggttccaaacc l20
caaattaattttggantttaaattaaatnttnattnggggaanaanccaaatgtnaagaa 180
aatttaacccattatnaacttaaatncctngaaacccntggnttccaaaaatttttaacc 240
cttaaatccctccgaaattgntaanggaaaaccaaattcncctaaggctntttgaaggtt 300
ngatttaaacccccttnanttnttttnacccnngnctnaantatttngnttccggtgttt 360
tcctnttaancntnggtaactcccgntaatgaannnccctaanccaattaaaccgaattt 420
tttttgaattggaaattccnngggaattnaccggggtttttcccntttgggggccatncc 480
cccnctttcggggtttgggnntaggttgaatttttnnangncccaaaaaancccccaana 540
aaaaaactcccaagnnttaattngaatntcccccttcccaggccttttgggaaaggnggg 600
tttntgggggccnggganttcnttcccccnttnccnccccccccccnggtaaanggttat 660
ngnntttggtttttgggccccttnanggaccttccggatngaaattaaatccccgggncg 720
gccg 724
<210> 39
<211> 751
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(751)
<223> n = A,T,C or G
<400>
39
tttttttttttttttctttgctcacatttaatttttattttgattttttttaatgctgca 60
caacacaatatttatttcatttgtttcttttatttcattttatttgtttgctgctgctgt 120
tttatttatttttactgaaagtgagagggaacttttgtggccttttttcctttttctgta 180
ggccgccttaagctttctaaatttggaacatctaagcaagctgaanggaaaagggggttt 240
cgcaaaatcactcgggggaanggaaaggttgctttgttaatcatgccctatggtgggtga 300
ttaactgcttgtacaattacntttcacttttaattaattgtgctnaangctttaattana 360
cttgggggttccctccccanaccaaccccnctgacaaaaagtgccngccctcaaatnatg 420
tcccggcnntcnttgaaacacacngcngaangttctcattntccccncnccaggtnaaaa 480
tgaagggttaccatntttaacnccacctccacntggcnnngcctgaatcctcnaaaancn 540
ccctcaancnaattnctnngccccggtcncgcntnngtcccncccgggctccgggaantn 600
cacccccngaanncnntnncnaacnaaattccgaaaatattcccnntcnctcaattcccc 660
cnnagactntcctcnncnancncaattttcttttnntcacgaacncgnnccnnaaaatgn 720
nnnncncctccnctngtccnnaatcnccanc 751
<210> 40
<211> 753
<212> DNA
<213> Homo sapien
<220>
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
19
<221> misc_feature
<222> (1)...(753)
<223> n = A,T,C or G
<400>
40
gtggtattttctgtaagatcaggtgttcctccctcgtaggtttagaggaaacaccctcat 60
agatgaaaacccccccgagacagcagcactgcaactgccaagcagccggggtaggagggg 120
cgccctatgcacagctgggcccttgagacagcagggcttcgatgtcaggctcgatgtcaa 180
tggtctggaagcggcggctgtacctgcgtaggggcacaccgtcagggcccaccaggaact 240
tctcaaagttccaggcaacntcgttgcgacacaccggagaccaggtgatnagcttggggt 300
cggtcataancgcggtggcgtcgtcgctgggagctggcagggcctcccgcaggaaggcna 360
ataaaaggtgcgcccccgcaccgttcanctcgcacttctcnaanaccatgangttgggct 420
cnaacccaccaccannccggacttccttganggaattcccaaatctcttcgntcttgggc 480
ttctnctgatgccctanctggttgcccngnatgccaancanccccaanccccggggtcct 540
aaancacccncctcctcntttcatctgggttnttntccccggaccntggttcctctcaag 600
ggancccatatctcnaccantactcaccntncccccccntgnnacccanccttctanngn 660
ttcccncccgncctctggcccntcaaanangcttncacnacctgggtctgccttcccccc 720
tnccctatctgnaccccncntttgtctcantnt 753
<210> 41
<211> 341
<212> DNA
<213> Homo sapien
<400> 41
actatatccatcacaacagacatgcttcatcccatagacttcttgacatagcttcaaatg 60
agtgaacccatccttgatttatatacatatatgttctcagtattttgggagcctttccac 120
ttctttaaaccttgttcattatgaacactgaaaataggaatttgtgaagagttaaaaagt 180
tatagcttgtttacgtagtaagtttttgaagtctacattcaatccagacacttagttgag 240
tgttaaactgtgatttttaaaaaatatcatttgagaatattctttcagaggtattttcat 300
ttttactttttgattaattgtgttttatatattagggtagt 341
<210> 42
<211> 101
<212> DNA
<213> Homo sapien
<400> 42
acttactgaa tttagttctg tgctcttcct tatttagtgt tgtatcataa atactttgat 60
gtttcaaaca ttctaaataa ataattttca gtggcttcat a 101
<210> 43
<211> 305
<2l2> DNA
<213> Homo sapien
<400> 43
acatctttgt tacagtctaa gatgtgttct taaatcacca ttccttcctg gtcctcaccc 60
tccagggtgg tctcacactg taattagagc tattgaggag tctttacagc aaattaagat 120
tcagatgcct tgctaagtct agagttctag agttatgttt cagaaagtct aagaaaccca 180
cctcttgaga ggtcagtaaa~gaggacttaa tatttcatat ctacaaaatg accacaggat 240
tggatacaga acgagagtta tcctggataa ctcagagctg agtacctgcc cgggggccgc 300
tcgaa 305
<210> 44
<211> 852
<212> DNA
<213> Homo sapien
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<220>
<221> misc_feature
<222> (1)...(852)
<223> n = A,T,C or G
<400>
44
acataaatatcagagaaaagtagtctttgaaatatttacgtccaggagttctttgtttct 60
gattatttggtgtgtgttttggtttgtgtccaaagtattggcagcttcagttttcatttt 120
ctctccatcctcgggcattcttcccaaatttatataccagtcttcgtccatccacacgct 180
ccagaatttctcttttgtagtaatatctcatagctcggctgagcttttcataggtcatgc 240
tgctgttgttcttctttttaccccatagctgagccactgcctctgatttcaagaacctga 300
agacgccctcagatcggtcttcccattttattaatcctgggttcttgtctgggttcaaga 360
ggatgtcgcggatgaattcccataagtgagtccctctcgggttgtgctttttggtgtggc 420
acttggcaggggggtcttgctcctttttcatatcaggtgactctgcaacaggaaggtgac 480
tggtggttgtcatggagatctgagcccggcagaaagttttgctgtccaacaaatctactg 540
tgctaccatagttggtgtcatataaatagttctngtctttccaggtgttcatgatggaag 600
gctcagtttgttcagtcttgacaatgacattgtgtgtggactggaacaggtcactactgc 660
actggcegttccacttcagatgctgcaagttgctgtagaggagntgccccgccgtccctg 720
ccgcccgggtgaactcctgcaaactcatgctgcaaaggtgctcgccgttgatgtcgaact 780
cntggaaagggatacaattggcatccagctggttggtgtccaggaggtgatggagccact 840
cccacacctggt 852
<210> 45
<211> 234
<212> DNA
<2l3> Homo sapien
<400> 45
acaacagacc cttgctcgct aacgacctca tgctcatcaa gttggacgaa tccgtgtccg 60
agtctgacac catccggagc atcagcattg cttcgcagtg ccctaccgcg gggaactctt 120
gcctcgtttc tggctggggt ctgctggcga acggcagaat gcctaccgtg ctgcagtgcg 180
tgaacgtgtc ggtggtgtct gaggaggtct gcagtaagct ctatgacccg ctgt 234
<210> 46
<211> 590
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(590)
<223> n = A,T,C or G
<400>
46
actttttatttaaatgtttataaggcagatctatgagaatgatagaaaacatggtgtgta 60
atttgatagcaatattttggagattacagagttttagtaattaccaattacacagttaaa 120
aagaagataatatattccaagcanatacaaaatatctaatgaaagatcaaggcaggaaaa 180
tgantataactaattgacaatggaaaatcaattttaatgtgaattgcacattatccttta 240
aaagctttcaaaanaaanaattattgcagtctanttaattcaaacagtgttaaatggtat 300
caggataaanaactgaagggcanaaagaattaattttcacttcatgtaacncacccanat 360
ttacaatggcttaaatgcanggaaaaagcagtggaagtagggaagtantcaaggtctttc 420
tggtctctaatctgccttactctttgggtgtggctttgatcctctggagacagctgccag 480
ggctcctgttatatccacaatcccagcagcaagatgaagggatgaaaaaggacacatgct 540
gccttcctttgaggagacttcatctcactggccaacactcagtcacatgt 590
<210> 47
<211> 774
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
21
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(774)
<223> n = A,T,C or G
<400>
47
acaagggggcataatgaaggagtgggganagattttaaagaaggaaaaaaaacgaggccc 60
tgaacagaattttcctgnacaacggggcttcaaaataattttcttggggaggttcaagac l20
gcttcactgcttgaaaottaaatggatgtgggacanaattttctgtaatgaccctgaggg 180
cattacagacgggactctgggaggaaggataaacagaaaggggacaaaggctaatcccaa 240
aacatcaaagaaaggaaggtggcgtcatacctcccagcctacacagttctccagggctct 300
cctcatccctggaggacgacagtggaggaacaactgaccatgtccccaggctcctgtgtg 360
ctggctcctggtcttcagcccccagctctggaagcccaccctctgctgatcctgcgtggc 420
ccacactccttgaacacacatccccaggttatattcctggacatggctgaacctcctatt 480
cctacttccgagatgccttgctccctgcagcctgtcaaaatcccactcaccctccaaacc 540
acggcatgggaagcctttctgacttgcctgattactccagcatcttggaacaatccctga 600
ttccccactccttagaggcaagatagggtggttaagagtagggctggaccacttggagcc 660
aggctgctggcttcaaattntggctcatttacgagctatgggaccttgggcaagtnatct 720
tcacttctatgggcntcattttgttctacctgcaaaatgggggataataatagt 774
<210> 48
<211> 124
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(124)
<223> n = A,T,C or G
<400> 48
canaaattga aattttataa aaaggcattt ttctcttata tecataaaat gatataattt 60
ttgcaantat anaaatgtgt cataaattat aatgttcctt aattacagct caacgcaact 120
tggt 124
<210> 49
<211> 147
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(147)
<223> n = A,T,C or G
<400> 49
gccgatgcta ctattttatt gcaggaggtg ggggtgtttt tattattctc tcaacagctt 60
tgtggctaca ggtggtgtct gactgcatna aaaanttttt tacgggtgat tgcaaaaatt 120
ttagggcacc catatcccaa gcantgt 147
<210> 50
<211> 107
<212> DNA
<213> Homo sapien
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
22
<400> 50
acattaaatt aataaaagga ctgttggggt tctgctaaaa cacatggctt gatatattgc 60
atggtttgag gttaggagga gttaggcata tgttttggga gaggggt ~ 107
<210> 51
<211> 204
<212> DNA
<223>.Homo sapien
<400> 51
gtcctaggaa gtctagggga cacacgactc tggggtcacg gggccgacac acttgcacgg 60
cgggaaggaa aggcagagaa gtgacaccgt cagggggaaa tgacagaaag gaaaatcaag 120
gccttgcaag gtcagaaagg ggactcaggg cttccaccac agccctgccc cacttggcca 180
cctccctttt gggaccagca atgt 204
<210> 52
<211> 491
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(491)
<223> n = A,T,C or G
<400>
52
acaaagataacatttatcttataacaaaaatttgatagttttaaaggttagtattgtgta 60
gggtattttccaaaagactaaagagataactcaggtaaaaagttagaaatgtataaaaca 120
ccatcagacaggtttttaaaaaacaacatattacaaaattagacaatcatccttaaaaaa 180
aaaacttcttgtatcaatttcttttgttcaaaatgactgacttaantatttttaaatatt 240
tcanaaacacttcctcaaaaattttcaanatggtagctttcanatgtnccctcagtccca 300
atgttgctcagataaataaatctcgtgagaacttaccacccaccacaagctttctggggc 360
atgcaaeagtgtcttttctttnctttttctttttttttttttacaggcacagaaactcat 420
caattttatttggataacaaagggtctccaaattatattgaaaaataaatccaagttaat 480
atcactcttgt 491
<210> 53
<211> 484
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(484)
<223> n = A, T, C or G
<400> 53
acataatttagcagggctaattaccataagatgctatttattaanaggtntatgatctga 60
gtattaacagttgctgaagtttggtatttttatgcagcattttctttttgctttgataac 120
actacagaacccttaaggacactgaaaattagtaagtaaagttcagaaacattagctgct 180
caatcaaatctctacataacactatagtaattaaaacgttaaaaaaaagtgttgaaatct 240
gcactagtatanaccgctcctgtcaggataanactgctttggaacagaaagggaaaaanc 300
agctttgantttctttgtgctgatangaggaaaggctgaattaccttgttgcctctccct 360
aatgattggcaggtcnggtaaatnccaaaacatattccaactcaacacttcttttccncg 420
tancttgantctgtgtattccaggancaggcggatggaatgggccagcccncggatgttc 480
cant 484
<210> 54
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
23
<211> 151
<212> DNA
<213> Homo sapien
<400> 54
actaaacctc gtgcttgtga actccataca gaaaacggtg ccatccctga acacggctgg 60
ccactgggta tactgctgac aaccgcaaca acaaaaacac aaatccttgg cactggctag 120
tctatgtcct ctcaagtgcc tttttgtttg t 151
<210> 55
<211> 91
<212> DNA
<213> Homo sapien
<400> 55
acctggcttg tctccgggtg gttcccggcg ccccccacgg tccccagaac ggacactttc 60
gccctccagt ggatactcga gccaaagtgg t 91
<210> 56
<212> 133
<212> DNA
<213> Homo sapien
<400> 56
ggcggatgtg cgttggttat atacaaatat gtcattttat gtaagggact tgagtatact 60
tggatttttg gtatctgtgg gttgggggga cggtccagga accaataccc catggatacc l20
aagggacaac tgt 133
<220> 57
<211> 147
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(147)
<223> n = A,T,C or G
<400> 57
actctggaga acctgagccg ctgctccgcc tctgggatga ggtgatgcan gcngtggcgc 60
gactgggagc tgagcccttc cctttgcgcc tgcctcagag gattgttgcc gacntgcana 120
tctcantggg ctggatncat gcagggt 147
<210> 58
<211> 198
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> ( 1 ) . . . 098 )
<223> n = A,T,C or G
<400> 58
acagggatat aggtttnaag ttattgtnat tgtaaaatac attgaatttt ctgtatactc 60
tgattacata catttatcct ttaaaaaaga tgtaaatctt aatttttatg ccatctatta 120
atttaccaat gagttacctt gtaaatgaga agtcatgata gcactgaatt ttaactagtt 180
ttgacttcta agtttggt 198
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
24
<210> 59
<211> 330
<212> DNA
<213> Homo sapien
<400> 59
acaacaaatg ggttgtgagg aagtcttatc agcaaaactg gtgatggcta ctgaaaagat 60
ccattgaaaa ttatcattaa tgattttaaa tgacaagtta tcaaaaactc actcaatttt 120
cacctgtgct agcttgctaa aatgggagtt aactctagag caaatatagt atcttctgaa 180
taoagtcaat aaatgacaaa gccagggcct acaggtggtt tccagacttt ccagacccag 240
cagaaggaat ctattttatc acatggatct ccgtctgtgc tcaaaatacc taatgatatt 300
tttcgtcttt attggacttc tttgaagagt 330
<210> 60
<211> 175
<212> DNA
<213> Homo sapien
<400> 60
accgtgggtg ccttctacat tcctgacggc tccttcacca acatctggtt ctacttcggc 60
gtcgtgggct ccttcctctt catcctcatc cagctggtgc tgctcatcga ctttgcgcac 120
tcctggaacc agcggtggct gggcaaggcc gaggagtgcg attcccgtgc ctggt 175
<210> 61
<211> 154
<212> DNA
<213> Homo sapien
<400> 61
accccacttt tcctcctgtg agcagtctgg acttctcact gctacatgat gagggtgagt 60
ggttgttgct cttcaacagt atcctcccct ttccggatct gctgagccgg acagcagtgc 120
tggactgcac agccccgggg ctccacattg ctgt 154
<210> 62
<211> 30
<212> DNA
<213> Homo sapien
<400> 62
cgctcgagcc ctatagtgag tcgtattaga 30
<210> 63
<211> 89
<212> DNA
<213> Homo sapien
<400> 63
acaagtcatt tcagcaccct ttgctcttca aaactgacca tcttttatat ttaatgcttc 60
ctgtatgaat aaaaatggtt atgtcaagt 89
<210> 64
<211> 97
<212> DNA
<213> Homo sapien
<400> 64
accggagtaa ctgagtcggg acgctgaatc tgaatccacc aataaataaa ggttctgcag 60
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
2S
aatcagtgca tccaggattg gtccttggat ctggggt 97.
<210> 65
<211> 377
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(377)
<223> n = A,T,C or G
<400>
65
acaacaanaantcccttctttaggccactgatggaaacctggaacccccttttgatggca 60
gcatggcgtcctaggccttgacacagcggctggggtttgggctntcccaaaccgcacacc 120
ccaaccctggtctacccacanttctggctatgggctgtctctgccactgaacatcagggt 180
tcggtcataanatgaaatcccaanggggacagaggtcagtagaggaagctcaatgagaaa 240
ggtgctgtttgctcagccagaaaacagctgcctggcattcgccgctgaactatgaacccg 300
tgggggtgaactacccccangaggaatcatgcctgggcgatgcaanggtgccaacaggag 360
gggcgggaggagcatgt 377
<210> 66
<211> 305
<212> DNA
<213> Homo sapien
<400> 66
acgcctttccctcagaattcagggaagagactgtcgcctgccttcctccgttgttgcgtg 60
agaacccgtgtgccccttcccaccatatccaccctcgctccatctttgaactcaaacacg 120
aggaactaactgcaccctggtcctctccccagtccccagttcaccctccatccctcacct 180
tcctccactctaagggatatcaacactgcccagcacaggggccctgaatttatgtggttt 240
ttatatattttttaataagatgcactttatgtcattttttaataaagtctgaagaattac 300
tgttt 305
<210> 67
<211> 385
<212> DNA
<213> Homo sapien
<400>
67
actacacacactccacttgcccttgtgagacactttgtcccagcactttaggaatgctga 60
ggtcggaccagccacatctcatgtgcaagattgcccagcagacatcaggtctgagagttc 120
cccttttaaaaaaggggacttgcttaaaaaagaagtctagccacgattgtgtagagcagc 180
tgtgctgtgctggagattcacttttgagagagttctcctctgagacctgatctttagagg 240
ctgggcagtcttgcacatgagatggggctggtctgatctcagcactccttagtctgcttg 300
cctctcccagggccccagcctggccacacctgcttacagggcactctcagatgcccatac 360
catagtttctgtgctagtggaccgt 385
<210> 68
<211> 73
<212> DNA
<213> Homo sapien
<400> 68
aattaaccag atatattttt accccagatg gggatattct ttgtaaaaaa tgaaaataaa 60
gtttttttaa tgg
73
<210> 69
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
26
<211> 536
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(536)
<223> n = A,T,C or G
<400>
69
actagtccagtgtggtggaattccattgtgttgggggctctcaccctcctctcctgcagc 60
tccagctttgtgctctgcctctgaggagaccatggcccagcatctgagtaccctgctgct 120
cctgctggccaccctagctgtggccctggcctggagccccaaggaggaggataggataat 180
cccgggtggcatctataacgcagacctcaatgatgagtgggtacagcgtgcccttcactt 240
cgccatcagcgagtataacaaggccaccaaagatgactactacagacgtccgctgcgggt 300
actaagagccaggcaacagaccgttgggggggtgaattacttcttcgacgtagaggtggg 360
ccgaaccatatgtaccaagtcccagcccaacttggacacctgtgccttccatgaacagcc 420
agaactgcagaagaaacagttgtgctctttcgagatctacgaagttccctggggagaaca 480
gaangtccctgggtgaaatccaggtgtcaagaaatcctanggatctgttgccaggc 536
<210> 70
<211> 477
<212> DNA
<213> Homo sapien
<400> 70
atgacccctaacaggggccctctcagccctcctaatgacctccggcctagccatgtgatt 60
tcacttccactccataacgctcctcatactaggcctactaaccaacacactaaccatata 120
ccaatgatggcgcgatgtaacacgagaaagcacataccaaggccaccacacaccacctgt 180
ccaaaaaggccttcgatacgggataatcctatttattacctcagaagtttttttcttcgc 240
agggatttttctgagccttttaccactccagcctagcccctaccccccaactaggagggc 300
actggcccccaacaggcatcaccccgctaaatcccctagaagtcccactcctaaacacat 360
ccgtattactcgcatcaggagtatcaatcacctgagctcaccatagtctaatagaaaaca 420
accgaaaccaaattattcaaagcactgcttattacaattttactgggtctctatttt 477
<210> 71
<211> 533
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1).,.(533)
<223> n = A,T,C or G
<400> 71
agagctataggtacagtgtgatctcagctttgcaaacacattttctacatagatagtact 60
aggtattaatagatatgtaaagaaagaaatcacaccattaataatggtaagattggttta 120
tgtgattttagtggtatttttggcacccttatatatgttttccaaactttcagcagtgat 180
attatttccataacttaaaaagtgagtttgaaaaagaaaatctccagcaagcatctcatt 240
taaataaaggtttgtcatctttaaaaatacagcaatatgtgactttttaaaaaagctgtc 300
aaataggtgtgaccctactaataattattagaaatacatttaaaaacatcgagtacctca 360
agtcagtttgccttgaaaaatatcaaatataactcttagagaaatgtacataaaagaatg 420
cttcgtaattttggagtangaggttccctcctcaattttgtatttttaaaaagtacatgg 480
taaaaaaaaaaattcacaacagtatataaggctgtaaaatgaagaattctgcc 533
<210> 72
<21I> 511
ccaaccctggtctacccacanttctggctatgggctgtctctgccactgaacatcagggt 180
tcggtcataanatgaaatcccaanggggacagaggtcagt
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(511)
<223> n = A,T,C or G
<400> 72
tattacggaaaaacacaccacataattcaactancaaagaanactgcttcagggcgtgta 60
aaatgaaaggcttccaggcagttatctgattaaagaacactaaaagagggacaaggctaa 120
aagccgcaggatgtctacactatancaggcgctatttgggttggctggaggagctgtgga 180
aaacatgganagattggtgctgganatcgccgtggctattcctcattgttattaoanagt 240
gaggttctctgtgtgcccactggtttgaaaaccgttctncaataatgatagaatagtaca 300
cacatgagaactgaaatggcccaaacccagaaagaaagcccaactagatcctcagaanac 360
gcttctagggacaataaccgatgaagaaaagatggcctccttgtgcccccgtctgttatg 420
atttctctccattgcagcnanaaacccgttcttctaagcaaacncaggtgatgatggcna 480
aaatacaccccctcttgaagnaccnggagga 511
<210> 73
<211> 499
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(499)
<223> n = A,T,C or G
<400>
73
cagtgccagcactggtgecagtaccagtaccaataacagtgccagtgccagtgccagcac 60
cagtggtggcttcagtgctggtgccagcctgaccgccactctcacatttgggctcttcgc 120
tggccttggtggagctggtgccagcaccagtggcagctctggtgcctgtggtttctccta 180
caagtgagattttagatattgttaatcctgccagtctttctcttcaagccagggtgcatc 240
ctcagaaacctactcaacacagcactctaggcagccactatcaatcaattgaagttgaca 300
ctctgcattaaatctatttgccatttctgaaaaaaaaaaaaaaaaaagggcggccgctcg 360
antctagagggcccgtttaaacccgctgatcagcctcgactgtgccttctanttgccagc 420
catctgttgtttgcccctcccccgntgccttccttgaccctggaaagtgccactcccact 480
gtcctttcctaantaaaat 499
<210> 74
<211> 537
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(537)
<223> n = A,T,C or G
<400> 74
tttcatagga gaacacactg aggagatact tgaagaattt ggattcagcc gcgaagagat 60
ttatcagctt aactcagata aaatcattga aagtaataag gtaaaagcta gtctctaact 120
tccaggccca cggctcaagt gaatttgaat actgcattta cagtgtagag taacacataa 180
cattgtatgc atggaaacat ggaggaacag tattacagtg tcctaccact ctaatcaaga 240
aaagaattac agactctgat tctacagtga tgattgaatt ctaaaaatgg taatcattag 300
ggcttttgat ttataanact ttgggtactt atactaaatt atggtagtta tactgccttc 360
cagtttgctt gatatatttg ttgatattaa gattcttgac ttatattttg aatgggttct 420
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
28
actgaaaaan gaatgatata ttcttgaaga catcgatata catttattta cactcttgat 480
tctacaatgt agaaaatgaa ggaaatgccc caaattgtat ggtgataaaa gtcccgt 537
<210> 75
<211> 467
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(467)
<223> n = A,T,C or G
<400> 75
caaanacaattgttcaaaagatgcaaatgatacactactgctgcagctcacaaacacctc 60
tgcatattacacgtacctcctcctgctcctcaagtagtgtggtctattttgccatcatca 120
cctgctgtctgcttagaagaacggctttctgctgcaanggagagaaatcataacagacgg 180
tggcacaaggaggccatcttttcctcatcggttattgtccctagaagcgtcttctgagga 240
tctagttgggctttctttctgggtttgggccatttcanttctcatgtgtgtactattcta 300
tcattattgtataacggttttcaaaccngtgggcacncagagaacctcactctgtaataa 360
caatgaggaatagccacggtgatctccagcaccaaatctctccatgttnttccagagctc 420
ctccagccaacccaaatagccgctgctatngtgtagaacatccctgn 467
<2l0> 76
<211> 400
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(400)
<223> n = A,T,C or G
<400> 76
aagctgacagcattcgggccgagatgtctcgctccgtggccttagctgtgctcgcgctac 60
tctctctttctggcctggaggctatccagcgtactccaaagattcaggtttactcacgtc l20
atccagcagagaatggaaagtcaaatttcctgaattgctatgtgtctgggtttcatccat 180
ccgacattgaagttgacttactgaagaatggagagagaattgaaaaagtggagcattcag 240
acttgtctttcagcaaggactggtctttctatctcttgtaotacactgaattoaccccca 300
ctgaaaaagatgagtatgcctgccgtgtgaaccatgtgactttgtcacagcccaagatng 360
ttnagtgggatcganacatgtaagcagcancatgggaggt 400
<210> 77
<211> 248
<212> DNA
<213> Homo sapien
<400>
77
ctggagtgccttggtgtttcaagcccctgcaggaagcagaatgcaccttctgaggcacct 60
ccagctgccccggcgggggatgcgaggctcggagcacccttgcccggctgtgattgctgc 120
caggcactgttcatctcagcttttctgtccctttgctcccggcaagcgcttctgctgaaa 180
gttcatatctggagcctgatgtcttaacgaataaaggtcccatgctccacccgaaaaaaa 240
aaaaaaaa 248
<210> 78
<211> 201
<212> DNA
<213> Homo sapien
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
29
<400> 78
actagtccag tgtggtggaa ttccattgtg ttgggcccaa cacaatggct acctttaaca 60
tcacccagac cccgccctgc ccgtgcccca cgctgctgct aacgacagta tgatgcttac 120
tctgctactc ggaaactatt tttatgtaat taatgtatgc tttcttgttt ataaatgcct 180
gatttaaaaa aaaaaaaaaa a 201
<210> 79
<211> 552
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(552)
<223> n = A,T,C or G
<400>
79
tccttttgttaggtttttgagacaaccctagacctaaactgtgtcacagacttctgaatg 60
tttaggcagtgctagtaatttcctcgtaatgattctgttattactttcctattctttatt 120
cctctttcttctgaagattaatgaagttgaaaattgaggtggataaatacaaaaaggtag 180
tgtgatagtataagtatctaagtgcagatgaaagtgtgttatatatatccattcaaaatt 240
atgcaagttagtaattactcagggttaactaaattactttaatatgctgttgaacctact 300
ctgttccttggctagaaaaaattataaacaggactttgttagtttgggaagccaaattga 360
taatattctatgttctaaaagttgggctatacataaantatnaagaaatatggaatttta 420
ttcccaggaatatggggttcatttatgaatantacccggganagaagttttgantnaaac 480
cngttttggttaatacgttaatatgtcctnaatnaacaaggcntgacttatttccaaaaa 540
aaaaaaaaaaas 552
<210> 80
<211> 476
<212> DNA
. <213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(476)
<223> n = A, T, C or G
<400> 80
acagggatttgagatgctaaggccccagagatcgtttgatccaaccctcttattttcaga 60
ggggaaaatggggcctagaagttacagagcatctagctggtgcgctggcacccctggcct 120
cacacagactcccgagtagctgggactacaggcacacagtcactgaagcaggccctgttt 180
gcaattcacgttgccacctccaacttaaacattcttcatatgtgatgtccttagtcacta 240
aggttaaactttcccacccagaaaaggcaacttagataaaatcttagagtactttcatac 300
tcttctaagtcctcttccagcctcactttgagtcctccttgggggttgataggaantntc 360
tcttggctttctcaataaaatctctatccatctcatgtttaatttggtacgcntaaaaat 420
gctgaaaaaattaaaatgttctggtttcnctttaaaaaaaaaaaaaaaaaaaaaaa 476
<210> 81
<211> 232
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(232)
<223> n = A, T, C or G
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<400> 81
tttttttttg tatgccntcn ctgtggngtt attgttgctg ccaccctgga ggagcccagt 60
ttcttctgta tctttctttt ctgggggatc ttcctggctc tgcccctcca ttcccagcct 120
ctcatcccca tcttgcactt ttgctagggt tggaggcgct ttcctggtag cccctcagag 180
actcagtcag cgggaataag tcctaggggt ggggggtgtg gcaagccggc ct 232
<210> 82
<211> 383
<212> DNA
<213> Homo sapien
<220>
<221> m.isc_feature
<222> (1). .(383)
<223> n = A,T,C or G
<400> 82
aggcgggagcagaagctaaagccaaagcccaagaagagtggcagtgccagcactggtgcc 60
agtaccagtaccaataacatgccagtgccagtgccagcaccagtggtggcttcagtgctg 120
gtgccagcctgaccgccactctcacatttgggctcttcgctggccttggtggagctggtg 180
ccagcaccagtggcagctctggtgcctgtggtttctcctacaagtgagattttagatatt 240
gttaatcctgccagtctttctcttcaagccagggtgcatcctcagaaacctactcaacac 300
agcactctnggcagccactatcaatcaattgaagttgacactctgcattaaatctatttg 360
ccatttcaaaaaaaaaaaaaaaa 383
<210> 83
<211> 494
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(494)
<223> n = A,T,C or G
<400> 83
accgaattgggaccgctggcttataagcgatcatgtcctccagtattacctcaacgagca 60
gggagatcgagtctatacgctgaagaaatttgacccgatgggacaacagacctgctcagc 120
ccatcctgctcggttctccccagatgacaaatactctcgacaccgaatcaccatcaagaa 180
acgcttcaaggtgctcatgacccagcaaccgcgccctgtcctctgagggtccttaaactg 240
atgtcttttctgccacctgttacccctcggagactccgtaaccaaactcttcggactgtg 300
agccctgatgcctttttgccagccatactctttggcntccagtctctcgtggcgattgat 360
tatgcttgtgtgaggcaatcatggtggcatcacccatnaagggaacacatttganttttt 420
tttcncatattttaaattacnaccagaatanttcagaataaatgaattgaaaaactctta 480
aaaaaaaaaaaaaa 494
<210> 84
<211> 380
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(380)
<223> n = A,T,C or G
<400> 84
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
31
gctggtagcctatggcgtggccacggangggctcctgaggcacgggacagtgacttccca 60
agtatcctgcgccgcgtcttctaccgtccctacctgcagatcttcgggcagattccccag 120
gaggacatggacgtggccctcatggagcacagcaactgctcgtcggagcccggcttctgg 180
gcacaccctcctggggcccaggcgggcacctgcgtctcccagtatgccaactggctggtg 240
gtgctgctcctcgtcatcttcctgctcgtggccaacatcctgctggtcacttgctcattg 300
ccatgttcagttacacattcggcaaagtacagggcaacagcnatctctactgggaaggcc 360
agcgttnccgcctcatccgg 380
<210> 85
<21l> 481
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(481)
<223> n = A,T,C or G
<400> 85
gagttagctcctccacaaccttgatgaggtcgtctgcagtggcctctcgcttcataccgc 60
tnccatcgtcatactgtaggtttgccaccacctcctgcatcttggggcggctaatatcca 120
ggaaactctcaatcaagtcaccgtcnatnaaacctgtggctggttctgtcttccgctcgg 180
tgtgaaaggatctccagaaggagtgctcgatcttccccacacttttgatgactttattga 240
gtcgattctgcatgtccagcaggaggttgtaccagctctctgacagtgaggtcaccagcc 300
ctatcatgccnttgaacgtgccgaagaacaccgagccttgtgtggggggtgnagtctcac 360
ccagattctgcattaccaganagccgtggcaaaaganattgacaactcgcccaggnngaa 420
aaagaacacctcctggaagtgctngccgctcctcgtccnttggtggnngcgcntnccttt 480
t 481
<210> 86
<211> 472
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(472)
<223> n = A,T,C or G
<400>
86
aacatcttcctgtataatgctgtgtaatatcgatccgatnttgtctgctgagaattcatt 60
acttggaaaagcaacttnaagcctggacactggtattaaaattcacaatatgcaacactt 120
taaacagtgtgtcaatctgctcccttactttgtcatcaccagtctgggaataagggtatg l80
ccctattcacacctgttaaaagggcgctaagcatttttgattcaacatctttttttttga 240
cacaagtccgaaaaaagcaaaagtaaacagttnttaatttgttagccaattcactttctt 300
catgggacagagccatttgatttaaaaagcaaattgcataatattgagctttgggagctg 360
atatntgagcggaagantagectttctacttcaccagacacaactcctttcatattggga 420
tgttnacnaaagttatgtctcttacagatgggatgcttttgtggcaattctg 472
<210> 87
<211> 413
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(413)
<223> n = A,T,C or G
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
32
<400> 87
agaaaccagtatctctnaaaacaacctctcataccttgtggacctaattttgtgtgcgtg 60
tgtgtgtgcgcgcatattatatagacaggcacatcttttttacttttgtaaaagcttatg 120
cctctttggtatctatatctgtgaaagttttaatgatctgccataatgtcttggggacct 180
ttgtcttctgtgtaaatggtactagagaaaacacctatnttatgagtcaatctagttngt 240
tttattcgacatgaaggaaatttccagatnacaacactnacaaactctcccttgactagg 300
ggggacaaagaaaagcanaactgaacatnagaaacaattncctggtgagaaattncataa 360
acagaaattgggtngtatattgaaananngcatcattnaaacgtttttttttt 413
<210> 88
<211> 448
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(448)
<223> n = A,T,C or G
<400>
88
cgcagcgggtcctctctatctagctccagcctctcgcctgccccactccccgcgtcccgc 60
gtcctagccnaccatggccgggcccctgcgcgccccgctgctcctgctggccatcctggc 120
egtggccctggccgtgagccccgeggccggctccagtcecggcaagcegccgcgcctggt 180
gggaggcccatggaccccgcgtggaagaagaaggtgtgcggcgtgcactggactttgccg 240
tcggcnantacaacaaacccgcaacnacttttaccnagcncgcgctgcaggttgtgccgc 300
cccaancaaattgttactnggggtaantaattcttggaagttgaacctgggccaaacnng 360
tttaccagaaccnagccaattngaacaattncccctccataacagccccttttaaaaagg 420
gaancantcctgntcttttccaaatttt 448
<210> 89
<211> 463
<212> DNA
<213> Homo sapien
<220>
<22l> misc_feature
<222> (1)...(463)
<223> n = A,T,C or G
<400> 89
gaattttgtg cactggccac tgtgatggaa ccattgggcc aggatgcttt gagtttatca 60
gtagtgattc tgccaaagtt ggtgttgtaa catgagtatg taaaatgtca aaaaattagc 120
agaggtctag gtctgcatat cagcagacag tttgtccgtg tattttgtag ccttgaagtt 180
ctcagtgaca agttnnttct gatgcgaagt tctnattcca gtgttttagt cctttgcatc 240
tttnatgttn agacttgcct ctntnaaatt gcttttgtnt tctgcaggta ctatctgtgg 300
tttaacaaaa tagaannact tctctgcttn gaanatttga atatcttaca tctnaaaatn 360
aattctctcc ccatannaaa acccangccc ttggganaat ttgaaaaang gntccttcnn 420
aattcnnana anttcagntn tcatacaaca naacngganc ccc 463
<210> 90
<211> 400
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(400)
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
33
<223> n = A,T,C or G
<400> 90
agggattgaa ggtctnttnt actgtcggac tgttcancca ccaactctac aagttgctgt 60
cttccactca ctgtctgtaa gcntnttaac ccagactgta tcttcataaa tagaacaaat 120
tcttcaccag tcacatcttc taggaccttt ttggattcag ttagtataag ctcttccact 180
tectttgtta agacttcatc tggtaaagtc ttaagttttg tagaaaggaa tttaattgct 240
cgttctctaa caatgtcctc tccttgaagt atttggctga acaacccacc tnaagtccct 300
ttgtgcatcc attttaaata tacttaatag ggcattggtn cactaggtta aattctgcaa 360
gagtcatctg tctgcaaaag ttgcgttagt atatctgcca 400
<210> 91
<2l1> 480
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(480)
<223> n = A,T,C or G
<400>
91
gagctcggatccaataatctttgtctgagggcagcacacatatncagtgccatggnaact 60
ggtctaccccacatgggagcagcatgccgtagntatataaggtcattccctgagtcagac 120
atgcctctttgactaccgtgtgccagtgctggtgattctcacacacctccnnccgctctt 180
tgtggaaaaactggcacttgnctggaactagcaagacatcacttacaaattcacccacga 240
gacacttgaaaggtgtaacaaagcgactcttgcattgctttttgtccctccggcaccagt 300
tgtcaatactaacccgctggtttgcctccatcacatttgtgatctgtagctctggataca 360
tctcctgacagtactgaagaacttcttcttttgtttcaaaagcaactcttggtgcctgtt 420
ngatcaggttcccatttcccagtccgaatgttcacatggcatatnttacttcccacaaaa 480
<210> 92
<211> 477
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(477)
<223> n = A,T,C or G
<400> 92
atacagcccanatcccaccacgaagatgcgcttgttgactgagaacctgatgcggtcact 60
ggtcccgctgtagccccagcgactctccacctgctggaagcggttgatgctgcactcctt 120
cccacgcaggcagcagcggggccggtcaatgaactccactcgtggcttggggttgacggt 180
taantgcaggaagaggctgaccacctcgcggtccaccaggatgcccgactgtgcgggacc 240
tgcagcgaaactcctcgatggtcatgagcgggaagcgaatgangcccagggccttgccca 300
gaaccttccgcctgttctctggcgtcacctgcagctgctgccgctnacactcggcctcgg 360
accagcggacaaacggcgttgaacagccgcacctcacggatgcccantgtgtcgcgctcc 420
aggaacggcnccagcgtgtccaggtcaatgtcggtgaancctccgcgggtaatggeg 477
<2l0> 93
<211> 377
<212> DNA
<213> Homo sapien
<220>
<221> misc feature
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
34
<222> (1)...(377)
<223> n = A,T,C or G
<400> 93
gaacggctggaccttgcctcgcattgtgctgctggcaggaataccttggcaagcagctcc 60
agtccgagcagccccagaccgctgccgcccgaagctaagcctgcctctggccttcccctc 120
cgcctcaatgcagaaccantagtgggagcactgtgtttagagttaagagtgaacactgtn 180
tgattttacttgggaatttcctctgttatatagcttttcccaatgctaatttccaaacaa 240
caacaacaaaataacatgtttgcctgttnagttgtataaaagtangtgattctgtatnta 300
aagaaaatattactgttacatatactgcttgcaanttctgtatttattggtnctctggaa 360
ataaatatattattaaa 377
<210> 94
<211> 495
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(495)
<223> n = A,T,C or G
<400> 94
ccctttgaggggttagggtccagttcccagtggaagaaacaggccaggagaantgcgtgc 60
cgagctgangcagatttcccacagtgaccccagagccctgggetatagtctctgacccct 120
ccaaggaaagaccaccttctggggacatgggctggagggcaggacctagaggoaccaagg 180
gaaggccccattccggggctgttccccgaggaggaagggaaggggctctgtgtgcccccc 240
acgaggaanaggccctgantcctgggatcanacaccccttcacgtgtatccccacacaaa 300
tgcaagctcaccaaggtcccctctcagtcccttccctacaccctgaacggncactggccc 360
acacccaccc-agancanccacccgccatggggaatgtnctcaaggaatcgcngggcaacg 420
tggactctngtcccnnaagggggcagaatctccaatagangganngaacccttgctnana 480
aaaaaaaanaaaaaa 495
<210> 95
<211> 472
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(472)
<223> n = A,T,C or G
<400>
95
ggttacttggtttcattgccaccacttagtggatgtcatttagaaccattttgtctgctc 60
cctctggaagccttgcgcagagcggactttgtaat'tgttggagaataactgctgaatttt 120
tagctgttttgagttgattcgcaccactgcaccacaactcaatatgaaaactatttnact 180
tatttattatcttgtgaaaagtatacaatgaaaattttgttcatactgtatttatcaagt 240
atgatgaaaagcaatagatatatattcttttattatgttnaattatgattgccattatta 300
atcggcaaaatgtggagtgtatgttcttttcacagtaatatatgccttttgtaacttcac 360
ttggttattttattgtaaatgaattacaaaattcttaatttaagaaaatggtangttata 420
tttanttcantaatttctttccttgtttacgttaattttgaaaagaatgcat 472
<210> 96
<211> 476
<212> DNA
<213> Homo sapien
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<220>
<221> misc_feature
<222> (1)...(476)
<223> n = A, T, C or G
<400>
96
ctgaagcatttcttcaaacttntctacttttgtcattgatacctgtagtaagttgacaat 60
gtggtgaaatttcaaaattatatgtaacttctactagttttactttctcccccaagtctt 120
ttttaactcatgatttttacacacacaatccagaacttattatatagcctctaagtcttt 180
attcttcacagtagatgatgaaagagtcctccagtgtcttgngcanaatgttctagntat 240
agctggatacatacngtgggagttctataaactcatacctcagtgggactnaaccaaaat 300
tgtgttagtctcaattcctaccacactgagggagcctcccaaatcactatattcttatct 360
gcaggtactcctccagaaaaacngacagggcaggcttgcatgaaaaagtnacatctgcgt 420
tacaaagtctatcttcctcanangtctgtnaaggaacaatttaatcttctagcttt 476
<210> 97
<211> 479
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(479)
<223> n = A,T,C or G
<400>
97
actctttctaatgctgatatgatcttgagtataagaatgcatatgtcactagaatggata 60
aaataatgctgcaaacttaatgttcttatgcaaaatggaacgctaatgaaacacagctta 120
caatcgcaaatcaaaactcacaagtgctcatctgttgtagatttagtgtaataagactta 180
gattgtgctccttcggatatgattgtttctcanatcttgggcaatnttccttagtcaaat 240
caggctactagaattctgttattggatatntgagagcatgaaatttttaanaatacactt 300
gtgattatnaaattaatcacaaatttcacttatacctgctatcagcagctagaaaaacat 360
ntnntttttanatcaaagtattttgtgtttggaantgtnnaaatgaaatctgaatgtggg 420
ttcnatcttattttttcccngacnactanttncttttttagggnctattctganccatc 479
<210> 98
<211> 461
<212> DNA
<213> Homo sapien
<400>
98
agtgacttgtcctccaacaaaaccccttgatcaagtttgtggcactgacaatcagaccta 60
tgctagttectgtcatctattcgctactaaatgcagactggaggggaccaaaaaggggca 120
tcaactccagctggattattttggagcctgcaaatctattcctacttgtacggactttga 180
agtgattcagtttcctctacggatgagagactggctcaagaatatcctcatgcagcttta 240
tgaagccactctgaacacgctggttatctagatgagaacagagaaataaagtcagaaaat 300
ttacctggagaaaagaggctttggctggggaccatcccattgaaccttctcttaaggact 360
ttaagaaaaactaccacatgttgtgtatcctggtgccggccgtttatgaactgaccaccc 420
tttggaataatcttgacgctcctgaacttgctcctctgcga 461
<210> 99
<211> 171
<212> DNA
<213> Homo sapien
<400> 99
gtggccgcgc gcaggtgttt cctcgtaccg cagggecccc tcccttcccc aggcgtccct 60
cggcgcctct gcgggcccga ggaggagcgg ctggcgggtg gggggagtgt gacccaccct 120
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
36
cggtgagaaa agccttctct agcgatctga gaggcgtgcc ttgggggtac c 171
<210> 100
<211> 269
<212> DNA
<213> Homo sapien
<400> 100
cggccgcaagtgcaactccagctggggccgtgcggacgaagattctgccagcagttggtc 60
cgactgcgacgacggcggcggcgacagtcgcaggtgcagcgcgggcgcctggggtcttgc 120
aaggctgagctgacgccgcagaggtcgtgtcacgtcccacgaccttgacgccgtcgggga 180
cagccggaacagagcccggtgaagcgggaggcctcggggagcccctcgggaagggcggcc 240
cgagagatacgcaggtgcaggtggccgcc 269
<210> 101
<211> 405
<212> DNA
<213> Homo sapien
<400> 101
tttttttttt ttttggaatc tactgcgagc acagcaggtc agcaacaagt ttattttgca 60
gctagcaagg taacagggta gggcatggtt acatgttcag gtcaacttcc tttgtcgtgg 120
ttgattggtt tgtctttatg ggggcggggt ggggtagggg aaacgaagca aataacatgg 180
agtgggtgca ccctccctgt agaacctggt tacaaagctt ggggcagttc acctggtctg 240
tgaccgtcat tttcttgaca tcaatgttat tagaagtcag gatatctttt agagagtcca 300
ctgttctgga gggagattag ggtttcttgc caaatccaac aaaatccact gaaaaagttg 360
gatgatcagt acgaataccg aggcatattc tcatatcggt ggcca 405
<210> 102
<211> 470
<212> DNA
<213> Homo sapien
<400>
102
tttttttttttttttttttttttttttttttttttttttttttttttttttttttttttt 60
ggcacttaatccatttttatttcaaaatgtctacaaatttaatcccattatacggtattt 120
tcaaaatctaaattattcaaattagccaaatccttaccaaataatacccaaaaatcaaaa 180
atatacttctttcagcaaacttgttacataaattaaaaaaatatatacggctggtgtttt 240
caaagtacaattatcttaacactgcaaacattttaaggaactaaaataaaaaaaaacact 300
ccgcaaaggttaaagggaacaacaaattcttttacaacaccattataaaaatcatatctc 360
aaatcttaggggaatatatacttcacacgggatcttaacttttactcactttgtttattt 420
ttttaaaccattgtttgggcccaacacaatggaatcccccctggactagt 470
<210> 103
<211> 581
<212> DNA
<213> Homo sapien
<400>
103
ttttttttttttttttttgacccccctcttataaaaaacaagttaccattttattttact 60
tacacatatttattttataattggtattagatattcaaaaggcagcttttaaaatcaaac 120
taaatggaaactgccttagatacataattcttaggaattagcttaaaatctgcctaaagt 180
gaaaatcttctctagctcttttgactgtaaatttttgactcttgtaaaacatccaaattc 240
atttttcttgtctttaaaattatctaatctttccattttttccctattccaagtcaattt 300
gcttctctagcctcatttcctagctcttatctactattagtaagtggcttttttcctaaa 360
agggaaaacaggaagagaaatggcacacaaaacaaacattttatattcatatttctacct 420
acgttaataaaatagcattttgtgaagccagctcaaaagaaggcttagatccttttatgt 480
ccattttagtcactaaacgatatcaaagtgccagaatgcaaaaggtttgtgaacatttat 540
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
37
tcaaaagcta atataagata tttcacatac tcatctttct g 581
<210> 104
<211> 578
<212> DNA
<213> Homo sapien
<400>
104
tttttttttttttttttttttttttctcttctttttttttgaaatgaggatcgagttttt 60
cactctctagatagggcatgaagaaaactcatctttcoagctttaaaataacaatcaaat 120
ctcttatgctatatcatattttaagttaaactaatgagtcactggcttatcttctcctga 180
aggaaatctgttcattcttctcattcatatagttatatcaagtactaccttgcatattga 240
gaggtttttcttctctatttacacatatatttccatgtgaatttgtatcaaacctttatt 300
ttcatgcaaactagaaaataatgtttcttttgcataagagaagagaacaatatagcatta 360
caaaactgctcaaattgtttgttaagttatccattataattagttggaaggagctaatac 420
aaatoacatttacgacagcaataataaaactgaagtaccagttaaatatccaaaataatt 480
aaaggaacatttttagcctgggtataattagctaattcactttacaagcatttattagaa 540
tgaattcacatgttattattcctagcocaacacaatgg 578
<210> l05
<211> 538
<212> DNA
<213> Homo sapien
<400> 105
tttttttttttttttcagtaataatcagaacaatatttatttttatatttaaaattoata 60
gaaaagtgccttacatttaataaaagtttgtttctcaaagtgatcagaggaattagatat 120
gtcttgaacaccaatattaatttgaggaaaatacaccaaaatacattaagtaaattattt 180
aagatcatagagcttgtaagtgaaaagataaaatttgacctcagaaactctgagcattaa 240
aaatccactattagcaaataaattactatggacttcttgctttaattttgtgatgaatat 300
ggggtgtcactggtaaaccaacacattctgaaggatacattacttagtgatagattctta 360
tgtactttgctaatacgtggatatgagttgacaagtttctotttcttcaatcttttaagg 420
ggcgagaaatgaggaagaaaagaaaaggattacgcatactgttctttctatggaaggatt 480
agatatgtttcctttgccaatattaaaaaaataataatgtttactactagtgaaacoc 538
<210> 106
<211> 473
<212> DNA
<213> Homo sapien
<400>
106
ttttttttttttttttagtcaagtttctatttttattataattaaagtcttggtcatttc 60
atttattagctctgcaacttacatatttaaattaaagaaacgttttagacaactgtacaa l20
tttataaatgtaaggtgccattattgagtaatatattcctccaagagtggatgtgtccct 180
~
tctcccaccaactaatgaacagcaacattagtttaattttattagtagatatacactgct 240
gcaaacgetaattctcttctccatccccatgtgatattgtgtatatgtgtgagttggtag 300
aatgcatcacaatctacaatcaacagcaagatgaagctaggctgggctttcggtgaaaat 360
agactgtgtctgtctgaatcaaatgatctgacctatcctcggtggcaagaactcttcgaa 420
ccgcttcctcaaaggcgctgccacatttgtggctctttgcacttgtttcaaaa 473
<210> 107
<211> 1621
<212> DNA
<213> Homo sapien
<400> 107
cgccatggca ctgcagggca tctcggtcat ggagctgtcc ggoctggccc cgggcccgtt 60
ctgtgctatg gtcctggctg acttcggggc gcgtgtggta cgcgtggacc ggcccggctc 120
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
38
ccgctacgacgtgagccgcttgggccggggcaagcgctcgctagtgctggacctgaagca180
gccgcggggagccgccgtgctgcggcgtctgtgcaagcggtcggatgtgctgctggagcc240
cttccgccgcggtgtcatggagaaactccagctgggcccagagattctgcagcgggaaaa300
tccaaggcttatttatgccaggctgagtggatttggccagtcaggaagcttctgceggtt360
agctggccacgatatcaactatttggctttgtcaggtgttctctcaaaaattggcagaag420
tggtgagaatccgtatgccccgctgaatctcctggctgactttgctggtggtggccttat480
gtgtgcactgggcattataatggctctttttgaccgcacacgcactgacaagggtcaggt540
cattgatgcaaatatggtggaaggaacagcatatttaagttcttttctgtggaaaactca600
gaaatcgagtctgtgggaagcacctcgaggacagaacatgttggatggtggagcaccttt660
ctatacgacttacaggacagcagatggggaattcatggctgttggagcaatagaacccca720
gttctacgagctgctgatcaaaggacttggactaaagtctgatgaacttcccaatcagat780
gagcatggatgattggccagaaatgaagaagaagtttgcagatgtatttgcaaagaagac840
gaaggcagagtggtgtcaaatctttgacggcacagatgcctgtgtgactccggttctgac900
ttttgaggaggttgttcatcatgatcacaacaaggaacggggctcgtttatcaccagtga960
ggagcaggacgtgagcccccgccctgcacctctgctgttaaacaccccagccatcccttc1020
tttcaaaagggatcctttcataggagaacacactgaggagatacttgaagaatttggatt1080
cagccgcgaagagatttatcagcttaactcagataaaatcattgaaagtaataaggtaaa1140
agctagtctctaacttccaggcccacggctcaagtgaatttgaatactgcatttacagtg1200
tagagtaacacataacattgtatgcatggaaacatggaggaacagtattacagtgtccta1260
ccactctaatcaagaaaagaattacagactctgattctacagtgatgattgaattctaaa1320
aatggttatcattagggcttttgatttataaaactttgggtacttatactaaattatggt1380
agttattctgccttccagtttgcttgatatatttgttgatattaagattcttgacttata1440
ttttgaatgggttctagtgaaaaaggaatgatatattcttgaagacatcgatatacattt1500
atttacactcttgattctacaatgtagaaaatgaggaaatgccacaaattgtatggtgat1560
aaaagtcacgtgaaacaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa1620
a 1621
<210> 1os
<211> 382
<212> PRT
<213> Homo sapien
<400> 108
Met Ala Leu Gln Gly Ile Ser Val Met Glu Leu Ser Gly Leu Ala Pro
1 5 10 15
Gly Pro Phe Cys Ala Met Val Leu Ala Asp Phe Gly Ala Arg Val Val
20 25 30
Arg Val Asp Arg Pro Gly Ser Arg Tyr Asp Val Ser Arg Leu Gly Arg
35 40 45
Gly Lys Arg Ser Leu Val Leu Asp Leu Lys Gln Pro Arg Gly Ala Ala
50 55 60
Val Leu Arg Arg Leu Cys Lys Arg Ser Asp Val Leu Leu G1u Pro Phe
65 70 75 80
Arg Arg Gly Val Met Glu Lys Leu Gln Leu Gly Pro Glu Ile Leu Gln
85 90 95
Arg Glu Asn Pro Arg Leu Ile Tyr Ala Arg Leu Ser Gly Phe Gly Gln
100 105 110
Ser Gly Ser Phe Cys Arg Leu Ala Gly His Asp Ile Asn Tyr Leu Ala
115 120 125
Leu Ser Gly Val Leu Ser Lys Tle Gly Arg Ser Gly Glu Asn Pro Tyr
130 135 140
Ala Pro Leu Asn Leu Leu Ala Asp Phe Ala Gly Gly Gly Leu Met Cys
145 150 155 160
Ala Leu Gly Ile Ile Met Ala Leu Phe Asp Arg Thr Arg Thr Asp Lys
165 170 175
Gly Gln Val Ile Asp Ala~Asn Met Val Glu Gly Thr Ala Tyr Leu Ser
180 185 190
Ser Phe Leu Trp Lys Thr Gln Lys Ser Ser Leu Trp Glu Ala Pro Arg
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
39
195 200 205
Gly Gln Asn Met Leu Asp Gly Gly Ala Pro Phe Tyr Thr Thr Tyr Arg
210 215 220
Thr A1a Asp Gly Glu Phe Met Ala Val Gly Ala Ile Glu Pro Gln Phe
225 230 235 240
Tyr Glu Leu Leu Ile Lys Gly Leu Gly Leu Lys Ser Asp Glu Leu Pro
245 250 255
Asn Gln Met Ser Met Asp Asp Trp Pro Glu Met Lys Lys Lys Phe Ala
260 265 270
Asp Val Phe Ala Lys Lys Thr Lys Ala Glu Trp Cys Gln Ile Phe Asp
275 280 285
Gly Thr Asp Ala Cys Val Thr Pro Val Leu Thr Phe Glu Glu Val Val
290 295 300
His His Asp His Asn Lys Glu Arg Gly Ser Phe Ile Thr Ser Glu Glu
305 310 315 320
Gln Asp Val Ser Pro Arg Pro Ala Pro Leu Leu Leu Asn Thr Pro Ala
325 330 335
Ile Pro Ser Phe Lys Arg Asp Pro Phe Ile Gly Glu His Thr Glu Glu
340 345 350
T1e Leu Glu Glu Phe Gly Phe Ser Arg Glu Glu Ile Tyr Gln Leu Asn
355 360 365
Ser Asp Lys Ile Ile Glu Ser Asn Lys Val Lys Ala Ser Leu
370 375 380
<210> 109
<211> 1524
<212> DNA
<213> Homo sapien z
<400>
109
ggcacgaggctgcgccagggcctgagcggaggcgggggcagcctcgccagcgggggcccc 60
gggcctggccatgcctcactgagccagcgcctgcgcctctacctcgccgacagctggaac 120
cagtgcgacctagtggctctcacctgcttcctcctgggcgtgggctgccggctgaccccg 180
ggtttgtaccacctgggccgcactgtcctctgcatcgacttcatggttttcacggtgcgg 240
ctgcttcacatcttcacggtcaacaaacagctggggcccaagatcgtcatcgtgagcaag 300
atgatgaagg acgtgttcttcttcctcttcttcctcggcgtgtggctggtagcctatggc 360
gtggccacggaggggctcctgaggccacgggacagtgacttcccaagtatcctgcgccgc 420
gtcttctaccgtccctacctgcagatcttcgggcagattccccaggaggacatggacgtg 480
gccctcatggagcacagcaactgctcgtcggagcccggcttctgggcacaccctcctggg 540
gcccaggcgggcacctgcgtctcccagtatgccaactggctggtggtgctgctcctcgtc 600
atcttcctgctcgtggccaacatcctgctggtcaacttgctcattgccatgttcagttac 660
acattcggcaaagtacagggcaacagcgatctctactggaaggcgcagcgttaccgcctc 720
atCCgggaattccactctcggcccgcgctggccccgccctttatcgtcatctcccacttg 780
cgcctcctgctcaggcaattgtgcaggcgaccccggagcccccagccgtcctccccggcc 840
ctcgagcatttccgggtttacctttctaaggaagccgagcggaagctgctaacgtgggaa 900
tcggtgcataaggagaactttctgctggcacgcgctagggacaagcgggagagcgactcc 960
gagcgtctgaagcgcacgtcccagaaggtggacttggcactgaaacagctgggacacatc 1020
cgcgagtacgaacagcgcctgaaagtgctggagcgggaggtccagcagtgtagccgcgtc 1080
ctggggtgggtggccgaggccctgagccgctctgccttgctgcccccaggtgggccgcca 1140
ccccctgacctgcctgggtccaaagactgagccctgctggcggacttcaaggagaagccc 1200
ccacaggggattttgctcctagagtaaggctcatctgggcctcggccccagcacctggtg 1260
gccttgtccttgaggtgagccccatgtccatctgggccactgtcaggaccacctttggga 1320
gtgtcatccttacaaaccacagcatgcccggctcctcccagaaccagtcccagcctggga 1380
ggatcaaggcctggatcccgggccgttatccatctggaggctgcagggtccttggggtaa 1440
cagggaccacagacccctcaccactcacagattcctcacactggggaaataaagccattt 1500
cagaggaaaaaaaaaaaaaaaaaa 1524
<210> 110
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<211> 3410
<212> DNA
<213> Homo sapien
<400>
1l0
gggaaccagcctgcacgcgctggctccgggtgacagccgcgcgcctcggccaggatctga60
gtgatgagacgtgtccccactgaggtgccccacagcagcaggtgttgagcatgggctgag120
aagctggaccggcaccaaagggctggcagaaatgggcgcctggetgattcctaggcagtt180
ggcggcagcaaggaggagaggccgcagcttctggagcagagccgagacgaagcagttctg240
gagtgcctgaacggccccctgagccctacccgcctggcccactatggtccagaggctgtg300
ggtgagccgcctgctgcggcaccggaaagcccagctcttgctggtcaacctgctaacctt360
tggcctggaggtgtgtttggccgcaggcatcacctatgtgccgcctctgctgctggaagt420
gggggtagaggagaagttcatgaccatggtgctgggcattggtccagtgctgggcctggt480
ctgtgtcccgctcctaggctcagccagtgaccactggcgtggacgctatggccgccgccg540
gcccttcatctgggcactgtccttgggcatcctgctgagcctctttctcatcccaagggc600
cggctggctagcagggctgctgtgcccggatcccaggcccctggagctggcactgctcat660
cctgggcgtggggctgctggacttctgtggccaggtgtgcttcactccactggaggccct720
gctctctgacctcttccgggacccggaccactgtcgccaggcctactctgtctatgcctt780
catgatcagtcttgggggctgcctgggctacctcctgcctgccattgactgggacaccag840
tgccctggccccctacctgggcacccaggaggagtgcctctttggcctgctcaccctcat900
cttcctcacctgcgtagcagccacactgctggtggctgaggaggcagcgctgggccccac960
cgagccagcagaagggctgtcggccccctccttgtcgccccactgctgtccatgccgggc1020
ccgcttggctttccggaacctgggcgccctgcttccccggctgcaccagctgtgctgccg1080
catgccccgcaccctgcgccggctcttcgtggctgagctgtgcagctggatggcactcat1140
gaccttcacgctgttttacacggatttcgtgggcgaggggctgt,accagggcgtgcccag1200
agctgagccgggcaccgaggcccggagacactatgatgaaggcgttcggatgggcagcct1260
ggggctgttcctgcagtgcgccatctccctggtcttctctctggtcatggaccggctggt1320
gcagcgattcggcactcgagcagtctatttggccagtgtggcagctttccctgtggctgc1380
cggtgccacatgcctgtcccacagtgtggccgtggtgacagcttcagccgccctcaccgg1440
gttcaccttctcagccctgcagatcctgccctacacactggcctccctctaccaccggga1500
gaagcaggtgttcctgcccaaataccgaggggacactggaggtgctagcagtgaggacag1560
cctgatgaccagcttcctgccaggccctaagcctggagctcccttccctaatggacacgt1620
gggtgctggaggcagtggcctgctcccacctccacccgcgctctgcggggcctctgcetg1680
tgatgtctccgtacgtgtggtggtgggtgagcccaccgaggccagggtggttccgggccg1740
gggcatctgcctggacctcgccatcctggatagtgccttcctgctgtcccaggtggcccc1800
atccctgtttatgggctccattgtccagctcagccagtctgtcactgcctatatggtgtc1860
tgccgcaggcctgggtctggtcgccatttactttgctacacaggtagtatttgacaagag1920
cgacttggccaaatactcagcgtagaaaacttccagcacattggggtggagggcctgcct1980
cactgggtcccagctccccgctcctgttagccccatggggctgccgggctggccgccagt2040
ttctgttgctgccaaagtaatgtggctctctgctgccaccctgtgctgctgaggtgcgta2100
gctgcacagctgggggctggggcgtccctctcctctctccccagtctctagggctgcctg2160
actggaggccttccaagggggtttcagtctggacttatacagggaggccagaagggctcc2220
atgcactggaatgcggggactctgcaggtggattacccaggctcagggttaacagctagc2280
ctcctagttgagacacacctagagaagggtttttgggagctgaataaactcagtcacctg2340
gtttcccatctctaagccccttaacctgcagcttcgtttaatgtagctcttgcatgggag2400
tttctaggatgaaacactcctccatgggatttgaacatatgacttatttgtaggggaaga2460
gtcctgaggggcaacacacaagaaccaggtcccctcagcccacagcactgtctttttgct2520
gatccacccccctcttaccttttatcaggatgtggcctgttggtccttctgttgccatca2580
cagagacacaggcatttaaatatttaacttatttatttaacaaagtagaagggaatccat2640
tgctagcttttctgtgttggtgtctaatatttgggtagggtgggggatccccaacaatca2700
ggtcccctgagatagctggtcattgggctgatcattgccagaatcttcttctcctggggt2760
ctggccccccaaaatgcctaacccaggaccttggaaattctactcatcccaaatgataat2820
tccaaatgctgttacccaaggttagggtgttgaaggaaggtagagggtggggcttcaggt2880
ctcaacggcttccctaaccacccctcttctcttggcccagcctggttccccccacttcca2940
ctcccctctactctctctaggactgggctgatgaaggcactgcccaaaatttcccctacc3000
cccaactttcccctacccccaactttccccaccagctccacaaccctgtttggagctact3060
gcaggaccagaagcacaaagtgcggtttcccaagcctttgtccatctcagcccccagagt3120
atatctgtgcttggggaatctcacacagaaactcaggagcaccccctgcctgagctaagg3180
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
41
gaggtcttat ctctcagggg gggtttaagt gccgtttgca ataatgtcgt cttatttatt 3240
tagcggggtg aatattttat actgtaagtg agcaatcaga gtataatgtt tatggtgaca 3300
aaattaaagg ctttcttata tgtttaaaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa 3360
aaaaaaaara aaaaaaaaaa aaaaaaaaaa aaaaaaataa aaaaaaaaaa 3410
<210> 111
<211> 1289
<212> DNA
<213> Homo sapien
<400>
111
agccaggcgtccctctgcctgcccactcagtggcaacacccgggagctgttttgtccttt60
gtggagcctcagcagttccctctttcagaactcactgccaagagccctgaacaggagcca120
ccatgcagtgcttcagcttcattaagaccatgatgatcctcttcaatttgctcatctttc180
tgtgtggtgcagccctgttggcagtgggcatctgggtgtcaatcgatggggcatcctttc240
tgaagatcttcgggccactgtcgtccagtgccatgcagtttgtcaacgtgggctacttcc300
tcatcgcagccggcgttgtggtctttgctcttggtttcctgggctgctatggtgctaaga360
ctgagagcaagtgtgccctcgtgacgttcttcttcatcctcctcctcatcttcattgctg420
aggttgcagctgctgtggtcgccttggtgtacaccacaatggctgagcacttcctgacgt480
tgctggtagtgcctgccatcaagaaagattatggttcccaggaagacttcactcaagtgt540
ggaacaccaccatgaaagggctcaagtgctgtggcttcaccaactatacggattttgagg600
actcaccctacttcaaagagaacagtgcctttcccccattctgttgcaatgacaacgtca660
ccaacacagccaatgaaacctgcaccaagcaaaaggctcacgaccaaaaagtagagggtt720
gcttcaatcagcttttgtatgacatccgaactaatgcagtcaccgtgggtggtgtggcag780
ctggaattgggggcctcgagctggctgccatgattgtgtccatgtatctgtactgcaatc840
tacaataagtccacttctgcctctgccactactgctgccacatgggaactgtgaagaggc900
accctggcaagcagcagtgattgggggaggggacaggatctaacaatgtcacttgggcca960
gaatggacctgccctttctgctccagacttggggctagatagggaccactccttttagcg1020
atgcctgactttccttccattggtgggtggatgggtggggggcattccagagcctctaag1080
gtagccagttctgttgcccattcccccagtctattaaacccttgatatgccccctaggcc1140
tagtggtgatcccagtgctctactgggggatgagagaaaggcattttatagcctgggcat1200
aagtgaaatcagcagagcctctgggtggatgtgtagaaggcacttcaaaatgcataaacc1260
tgttacaatgttaaaaaaaaaaaaaaaaa 1289
<2l0> 112
<211> 315
<212> PRT
<213> Homo sapien
<400> 112
Met Val Phe Thr Val Arg Leu Leu His Tle Phe Thr Val Asn Lys Gln
1 5 10 15
Leu Gly Pro Lys Ile Val Ile Val Ser Lys Met Met Lys Asp Val Phe
20 25 30
Phe Phe Leu Phe Phe Leu Gly Val Trp Leu Val Ala Tyr Gly Val Ala
35 40 45
Thr Glu Gly Leu Leu Arg Pro Arg Asp Ser Asp Phe Pro Ser Ile Leu
50 55 60
Arg Arg Val Phe Tyr Arg Pro Tyr Leu Gln Ile Phe Gly Gln Ile Pro
65 70 75 80
Gln Glu Asp Met Asp Val Ala Leu Met Glu His Ser Asn Cys Ser Ser
85 90 95
Glu Pro G1y Phe Trp Ala His Pro Pro Gly Ala Gln Ala Gly Thr Cys
l00 l05 110
Val Ser Gln Tyr Ala Asn Trp Leu Val Val Leu Leu Leu Val I1e Phe
1l5 120 125
Leu Leu Val Ala Asn Ile Leu Leu Val Asn Leu Leu Ile Ala Met Phe
130 135 140
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
42
Sex Tyr Thr Phe Gly Lys Val Gln Gly Asn Ser Asp Leu Tyr Trp Lys
145 150 155 160
Ala Gln Arg Tyr Arg Leu Ile Arg Glu Phe His Ser Arg Pro Ala Leu
165 170 175
Ala Pro Pro Phe IIe Val Ile Ser His Leu Arg Leu Leu Leu Arg Gln
180 185 190
Leu Cys Arg Arg Pro Arg Ser Pro Gln Pro Ser Ser Pro Ala Leu Glu
195 200 205
His Phe Arg Val Tyr Leu Ser Lys Glu Ala Glu Arg Lys Leu Leu Thr
210 215 220
Trp Glu Ser Val His Lys Glu Asn Phe Leu Leu Ala Arg Ala Arg Asp
225 230 235 240
Lys Arg Glu Ser Asp Ser Glu Arg Leu Lys Arg Thr Ser Gln Lys Val
245 250 255
Asp Leu Ala Leu Lys Gln Leu Gly His Ile Arg Glu Tyr Glu Gln Arg
260 265 270
Leu Lys Val Leu Glu Arg Glu Val Gln Gln Cys Ser Arg Val Leu Gly
275 280 285
Trp Val Ala Glu Ala Leu Ser Arg Ser Ala Leu Leu Pro Pro Gly Gly
290 295 300
Pro Pro Pro Pro Asp Leu Pro Gly Ser Lys Asp
305 310 315
<210> 113
<211> 553
<212> PRT
<213> Homo sapien
<400> 113
Met Val Gln Arg Leu Trp Val Ser Arg Leu Leu Arg His Arg Lys Ala
1 5 10 15
Gln Leu Leu Leu Val Asn Leu Leu Thr Phe Gly Leu Glu Val Cys Leu
20 25 30
Ala Ala Gly Ile Thr Tyr Val Pro Pro Leu Leu Leu Glu Val Gly Val
35 40 45
Glu Glu Lys Phe Met Thr Met Val Leu Gly Ile Gly Pro Val Leu Gly
50 55 60
Leu Val Cys Val Pro Leu Leu Gly Ser Ala Ser Asp His Trp Arg Gly
65 70 75 80
Arg Tyr Gly Arg Arg Arg Pro Phe Ile Trp Ala Leu Ser Leu Gly Ile
85 90 95
Leu Leu Ser Leu Phe Leu Ile Pro Arg Ala Gly Trp Leu Ala Gly Leu
100 105 110
Leu Cys Pro Asp Pro Arg Pro Leu Glu Leu Ala Leu Leu Ile Leu Gly
115 120 125
Val Gly Leu Leu Asp Phe Cys Gly Gln Val Cys Phe Thr Pro Leu Glu
230 135 140
Ala Leu Leu Ser Asp Leu Phe Arg Asp Pro Asp His Cys Arg Gln Ala
145 150 155 160
Tyr Ser Val Tyr Ala Phe Met Tle Ser Leu Gly Gly Cys Leu Gly Tyr
165 170 175
Leu Leu Pro Ala Ile Asp Trp Asp Thr Ser Ala Leu Ala Pro Tyr Leu
180 185 190
Gly Thr Gln Glu Glu Cys Leu Phe Gly Leu Leu Thr Leu Ile Phe Leu
195 200 205
Thr Cys Val Ala Ala Thr Leu Leu Val Ala Glu Glu Ala Ala Leu Gly
210 215 220
Pro Thr Glu Pro Ala G1u Gly Leu Ser Ala Pro Ser Leu Ser Pro His
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
43
225 230 235 240
Cys Cys Pro Cys Arg Ala Arg Leu Ala Phe Arg Asn Leu Gly Ala Leu
245 250 255
Leu Pro Arg Leu His Gln Leu Cys Cys Arg Met Pro Arg Thr Leu Arg
260 265 270
Arg Leu Phe Va1 Ala Glu Leu Cys Ser Trp Met Ala Leu Met Thr Phe
275 280 285
Thr Leu Phe Tyr Thr Asp Phe Val Gly Glu Gly Leu Tyr Gln Gly Val
290 295 300
Pro Arg Ala Glu Pro Gly Thr Glu Ala Arg Arg His Tyr Asp Glu Gly
305 310 315 320
Val Arg Met Gly Ser Leu Gly Leu Phe Leu Gln Cys A1a Ile Ser Leu
325 330 335
Val Phe Ser Leu Val Met Asp Arg Leu Val Gln Arg Phe Gly Thr Arg
340 345 350
Ala Val Tyr Leu Ala Ser Val Ala Ala Phe Pro Val Ala Ala Gly Ala
355 360 365
Thr Cys Leu Ser His Ser Val Ala Val Val Thr Ala Ser Ala Ala Leu
370 375 380
Thr Gly Phe Thr Phe Ser Ala Leu Gln Ile Leu Pro Tyr Thr Leu Ala
385 390 395 400
Ser Leu Tyr His Arg Glu Lys Gln Val Phe Leu Pro Lys Tyr Arg Gly
405 410 415
Asp Thr Gly Gly Ala Ser Ser Glu Asp Ser Leu Met Thr Ser Phe Leu
420 425 430
Pro Gly Pro Lys Pro Gly Ala Pro Phe Pro Asn Gly His Val Gly Ala
435 440 445
Gly Gly Ser Gly Leu Leu Pro Pro Pro Pro Ala Leu Cys G1y Ala Ser
450 455 460
Ala Cys Asp Val Ser Val Arg Val Val Val Gly Glu Pro Thr Glu Ala
465 470 475 480
Arg Val Val Pro Gly Arg Gly Ile Cys Leu Asp Leu Ala Ile Leu Asp
485 490 495
Ser Ala Phe Leu Leu Ser Gln Val Ala Pro Ser Leu Phe Met Gly Ser
500 505 510
Ile Val Gln Leu Ser Gln Ser Va1 Thr Ala Tyr Met Val Ser Ala Ala
515 520 525
Gly Leu Gly Leu Val Ala Ile Tyr Phe Ala Thr Gln Va1 Val Phe Asp
530 535 540
Lys Ser Asp Leu Ala Lys Tyr Ser Ala
545 550
<210> 114
<211> 241
<212> PRT
<223> Homo sapien
<400> 114
Met Gln Cys Phe Ser Phe Ile Lys Thr Met Met Ile Leu Phe Asn Leu
1 5 10 15
Leu Ile Phe Leu Cys Gly Ala Ala Leu Leu Ala Val Gly Ile Trp Val
20 25 30
Ser Ile Asp Gly Ala Ser Phe Leu Lys Ile Phe Gly Pro Leu Ser Ser
35 40 45
Ser A1a Met Gln Phe Val Asn Val Gly Tyr Phe Leu Ile Ala Ala Gly
50 55 60
Val Val Val Phe Ala Leu Gly Phe Leu Gly Cys Tyr Gly Ala Lys Thr
65 70 75 80
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
44
Glu Ser Lys Cys Ala Leu Val Thr Phe Phe Phe Ile Leu Leu Leu Ile
85 90 95
Phe Ile Ala Glu Val Ala Ala Ala Val Val Ala Leu Val Tyr Thr Thr
100 105 110
Met Ala Glu His Phe Leu Thr Leu Leu Val Val Pro Ala Ile Lys Lys
115 120 125
Asp Tyr Gly Ser Gln Glu Asp Phe Thr Gln Val Trp Asn Thr Thr Met
130 135 140
Lys Gly Leu Lys Cys Cys Gly Phe Thr Asn Tyr Thr Asp Phe Glu Asp
145 150 155 160
Ser Pro Tyr Phe Lys Glu Asn Ser Ala Phe Pro Pro Phe Cys Cys Asn
165 170 175
Asp Asn Val Thr Asn Thr Ala Asn Glu Thr Cys Thr Lys Gln Lys Ala
180 185 190
His Asp Gln Lys Val Glu Gly Cys Phe Asn Gln Leu Leu Tyr Asp Ile
195 200 205
Arg Thr Asn Ala Val Thr Val Gly Gly Val Ala Ala Gly Ile Gly Gly
210 215 220
Leu Glu Leu Ala A1a Met Ile Val Ser Met Tyr Leu Tyr Cys Asn Leu
225 230 235 240
Gln
<210> 1l5
<211> 366
<212> DNA
<213> Homo.sapien
<400> 1l5
gctctttctctcccctcctctgaatttaattctttcaacttgcaatttgcaaggattaca 60
catttcactgtgatgtatattgtgttgcaaaaaaaaaaaagtgtctttgtttaaaattac 120
ttggtttgtgaatccatcttgctttttccccattggaactagtcattaacccatctctga 180
actggtagaaaaacatctgaagagctagtctatcagcatctgacaggtgaattggatggt 240
tctcagaaccatttcacccagacagcctgtttctatcctgtttaataaattagtttgggt 300
tctctacatgcataacaaaccctgctccaatctgtcacataaaagtctgtgacttgaagt 360
ttagtc 366
<210> 116
<211> 282
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(282)
<223> n = A,T,C or G
<400> 116
acaaagatga accatttcct atattatagc aaaattaaaa ~tctacccgta ttctaatatt 60
gagaaatgag atnaaacaca atnttataaa gtctacttag agaagatcaa gtgacctcaa 120
agactttact attttcatat tttaagacac atgatttatc ctattttagt aacctggttc 180
atacgttaaa caaaggataa tgtgaacagc agagaggatt tgttggcaga aaatctatgt 240
tcaatctnga actatctana tcacagacat ttctattcct tt 282
<210> 117
<211> 305
<212> DNA
<213> Homo sapien
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<220>
<221> misc_feature
<222> (1)...(305)
<223> n = A,T,C or G
<400> 1l7
acacatgtcg cttcactgcc ttcttagatg cttctggtca acatanagga acagggacca 60
tatttatcct ccctcctgaa acaattgcaa aataanacaa aatatatgaa acaattgcaa l20
aataaggcaa aatatatgaa acaacaggtc tcgagatatt ggaaatcagt caatgaagga l80
tactgatccc tgatcactgt cctaatgcag gatgtgggaa acagatgagg tcacctctgt 240
gactgcccca gcttactgcc tgtagagagt ttctangctg cagttcagac agggagaaat 300
tgggt 305
<210> 118
<211> 71
<212> DNA
<213> Homo sapien
<220>
<22l> misc_feature
<222> (1)...(71)
<223> n = A,T,C or G
<400> 118
accaaggtgt ntgaatctct gacgtgggga tctctgattc ccgcacaatc tgagtggaaa 60
aantcctggg t 71
<210> l19
<211> 212
<212> DNA
<2l3> Homo sapien
<220>
<221> misc_feature
<222> (1)...(212)
<223> n = A,T,C or G
<400> 119
actccggttg gtgtcagcag cacgtggcat tgaacatngc aatgtggagc ccaaaccaca 60
gaaaatgggg tgaaattggc caactttcta tnaacttatg ttggcaantt tgccaccaac 120
agtaagctgg cccttctaat aaaagaaaat tgaaaggttt ctcactaanc ggaattaant 180
aatggantca aganactccc aggcctcagc gt 212
<210> 120
<211> 90
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1) .. . (90)
<223> n = A,T,C or G
<400> 120
actcgttgca natcaggggc cccccagagt caccgttgca ggagtccttc tggtcttgcc 60
ctccgccggc gcagaacatg ctggggtggt 90
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
46
<210> 121
<211> 218
<212> DNA
<213> Homo sapien
<220>
<22l> misc_feature
<222> (I)...(218)
<223> n = A,T,C or G
<400> 121
tgtancgtga anacgacaga nagggttgtc aaaaatggag aanccttgaa gtcattttga 60
gaataagatt tgctaaaaga tttggggcta aaacatggtt attgggagac atttctgaag 120
atatncangt aaattangga atgaattcat ggttcttttg ggaattcctt tacgatngcc 180
agcatanact tcatgtgggg atancagcta cccttgta 218
<210> 122
<211> 171
<212> DNA
<2l3> Homo sapien
<400> 122
taggggtgta tgcaactgta aggacaaaaa ttgagactca actggcttaa ccaataaagg 60
catttgttag ctcatggaac aggaagtcgg atggtggggc atcttcagtg ctgcatgagt 120
caccaccccg gcggggtcat ctgtgccaca ggtccctgtt gacagtgcgg t 171
<210> 123
<211> 76
<212> DNA
<213> Homo sapien
<220>
<22l> misc_feature
<222> (1)...(76)
<223> n = A,T,C or G
<400> 123
tgtagcgtga agacnacaga atggtgtgtg ctgtgctatc caggaacaca tttattatca 60
ttatcaanta ttgtgt 76
<210> 124
<211> 131
<212> DNA
<213> Homo sapien
<400> 124
acctttoccc aaggccaatg tcctgtgtgc taactggccg gctgcaggac agctgcaatt 60
caatgtgctg ggtcatatgg aggggaggag actctaaaat agccaatttt attctcttgg 120
ttaagatttg t 131
<210> 125
<211> 432
<212> DNA
<213> Homo sapien
<400> 125
actttatcta ctggctatga aatagatggt ggaaaattgc gttaccaact ataccactgg 60
cttgaaaaag aggtgatagc tcttcagagg acttgtgact tttgctcaga tgctgaagaa 120
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
47
ctaoagtctgcatttggcagaaatgaagatgaatttggattaaatgaggatgctgaagat 180
ttgcctcaccaaacaaaagtgaaacaactgagagaaaattttcaggaaaaaagacagtgg 240
ctcttgaagtatcagtcacttttgagaatgtttcttagttactgcatacttcatggatcc 300
catggtgggggtcttgcatctgtaagaatggaattgattttgcttttgcaagaatctcag 360
caggaaacatcagaaccactattttctagccctctgtcagagcaaacctcagtgcctctc 420
ctctttgcttgt 432
<210> 126
<211> 112
<212> DNA
<213> Homo sapien
<400> 226
acacaacttg aatagtaaaa tagaaactga gctgaaattt ctaattcact ttctaaccat 60
agtaagaatg atatttcccc ccagggatca ccaaatattt ataaaaattt gt 112
<210> 127
<211> 54
<212> DNA
<213> Homo sapien
<400> 127
accacgaaac cacaaacaag atggaagcat caatccactt gccaagcaca gcag 54
<210> 128
<211> 323
<212> DNA
<213> Homo sapien
<400> 128
acctcattagtaattgttttgttgtttcatttttttctaatgtctcccctctaccagctc 60
acctgagataacagaatgaaaatggaaggacagccagatttctcctttgctctctgctca 120
ttctctctgaagtctaggttacccattttggggacccattataggcaataaacacagttc 180
ccaaagcatttggacagtttcttgttgtgttttagaatggttttcctttttcttagcctt 240
ttcctgcaaaaggctcactcagtcccttgcttgctcagtggactgggctccccagggcct 300
aggctgccttcttttccatgtcc 323
<210> 129
<211> 192
<212> DNA
<2l3> Homo sapien
<220>
<221> misc_feature
<222> (1)...(192)
<223> n = A,T,C or G
<400> 129
acatacatgt gtgtatattt ttaaatatca cttttgtatc actctgactt tttagcatac 60
tgaaaacaca ctaacataat ttntgtgaac catgatcaga tacaacccaa atcattcatc 120
tagcacattc atctgtgata naaagatagg tgagtttcat ttccttcacg ttggccaatg 180
gataaacaaa gt 192
<210> 130
<21l> 362
<212> DNA
<213> Homo sapien
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
48
<220>
<221> misc_feature
<222> (1)...(362)
<223> n = A,T,C or G
<400> 130
cccttttttatggaatgagtagactgtatgtttgaanatttanccacaacctctttgaca 60
tataatgacgcaacaaaaaggtgctgtttagtcctatggttcagtttatgcccctgacaa 120
gtttccattgtgttttgccgatcttctggctaatcgtggtatcctccatgttattagtaa 180
ttctgtattccattttgttaacgcctggtagatgtaacctgctangaggctaactttata 240
cttatttaaaagctcttattttgtggtcattaaaatggcaatttatgtgcagcactttat 300
tgcagcaggaagcacgtgtgggttggttgtaaagctctttgctaatcttaaaaagtaatg 360
gg 362
<210> 131
<21l> 332
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1) . . (332)
<223> n = A,T,C or G
<400>
131
ctttttgaaagatcgtgtccactcctgtggacatcttgttttaatggagtttcccatgca 60
gtangactggtatggttgcagctgtccagataaaaacatttgaagagctccaaaatgaga 120
gttctcccaggttcgccctgctgctccaagtctcagcagcagcctcttttaggaggcatc 180
ttctgaactagattaaggcagcttgtaaatctgatgtgatttggtttattatccaactaa 240
cttccatctgttatcactggagaaagcccagactccccangacnggtacggattgtgggc 300
atanaaggattgggtgaagctggcgttgtggt 332
<210> 132
<211> 322
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(322)
<223> n = A,T,C or G
<400>
132
acttttgccattttgtatatataaacaatcttgggacattctcctgaaaactaggtgtcc 60
agtggctaagagaactcgatttcaagcaattctgaaaggaaaaccagcatgacacagaat 120
ctcaaattcccaaacaggggctctgtgggaaaaatgagggaggacctttgtatctcgggt 180
tttagcaagttaaaatgaanatgacaggaaaggcttatttatcaacaaagagaagagttg 240
ggatgcttctaaaaaaaactttggtagagaaaataggaatgctnaatcctagggaagcct 300
gtaacaatctacaattggtcca 322
<210> 133
<211> 278
<212> DNA
<2l3> Homo sapien
<220>
<221> misc_feature
<222> (1)...(278)
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
49
<223> n = A,T,C or G
<400>
133
acaagccttcacaagtttaactaaattgggattaatctttctgtanttatctgcataatt 60
cttgtttttctttccatctggctcctgggttgacaatttgtggaaacaactctattgcta l20
ctatttaaaaaaaatcacaaatctttccctttaagctatgttnaattcaaactattcctg 180
ctattcctgttttgtcaaagaaattatatttttcaaaatatgtntatttgtttgatgggt 240
cccacgaaacactaataaaaaccacagagaccagcctg 278
<210> 134
<211> 12l
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(121)
<223> n = A,T,C or G
<400> 134
gtttanaaaa cttgtttagc tccatagagg aaagaatgtt aaactttgta ttttaaaaca 60
tgattctctg aggttaaact tggttttcaa atgttatttt tacttgtatt ttgcttttgg 120
t 121
<210> 135
<2l1> 350
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(350)
<223> n = A,T,C or G
<400> 135
acttanaacc atgcctagca catcagaatc cctcaaagaa catcagtata atcctatacc 60
atancaagtg gtgactggtt aagcgtgcga caaaggtcag ctggcacatt acttgtgtgc 120
aaacttgata cttttgttct aagtaggaac tagtatacag tncctaggan tggtactcca 180
gggtgccccc caactcctgc agccgctcct ctgtgccagn occtgnaagg aactttcgct 240
ccacctcaat caagccctgg gccatgctac ctgcaattgg ctgaacaaac gtttgctgag 300
ttcccaagga tgcaaagcct ggtgctcaac tcctggggcg tcaactcagt 350
<210> l36
<211> 399
<212> DNA
<2l3> Homo sapien
<220>
<221> misc_feature
<222> (1)...(399)
<223> n = A,T,C or G
<400> 136
tgtaccgtgaagacgacagaagttgcatggcagggacagggcagggccgaggccagggtt 60
gctgtgattgtatccgaatantcctcgtgagaaaagataatgagatgacgtgagcagcct 120
gcagacttgtgtctgccttcaanaagccagacaggaaggccctgcctgccttggctctga l80
cctggcggccagccagccagccacaggtgggcttcttccttttgtggtgacaacnccaag 240
aaaactgcagaggcccagggtcaggtgtnagtgggtangtgaccataaaacaccaggtgc 300
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
S~
tcccaggaac ccgggcaaag gccatcccca cctacagcca gcatgcccac tggcgtgatg 360
ggtgcagang gatgaagcag coagntgttc tgctgt ggt 399
<210> 137
<211> 165
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(165)
<223> n = A,T,C or G
<400> 137
actggtgtgg tngggggtga tgctggtggt anaagttgan gtgacttcan gatggtgtgt 60
ggaggaagtg tgtgaacgta gggatgtaga ngttttggcc gtgctaaatg agcttcggga 120
ttggctggtc ccactggtgg tcactgtcat tggtggggtt cctgt 165
<210> 138
<211> 338
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(338)
<223> n = A,T,C or G
<400>
138
actcactggaatgccacattcacaacagaatcagaggtctgtgaaaacattaatggctcc 60
ttaacttctccagtaagaatcagggacttgaaatggaaacgttaacagccacatgcccaa 120
tgctgggcagtctcccatgccttccacagtgaaagggcttgagaaaaatcacatccaatg 180
tcatgtgtttccagccacaccaaaaggtgcttggggtggagggctgggggcatananggt 240
cangcctcaggaagcctcaagttccattcagctttgccactgtacattccccatntttaa 300
aaaaactgatgccttttttttttttttttgtaaaattc 338
<210> 139
<211> 382
<212> DNA
<213> Homo sapien
<400>
139
gggaatcttggtttttggcatctggtttgcctatagccgaggccactttgacagaacaaa 60
gaaagggacttcgagtaagaaggtgatttacagccagcctagtgcccgaagtgaaggaga 120
attcaaacagacctcgtcattcctggtgtgagcctggtcggctcaccgcctatcatctgc l80
atttgccttactcaggtgctaccggactctggcccctgatgtctgtagtttcacaggatg 240
ccttatttgtcttctacaccccacagggccccctacttcttcggatgtgtttttaataat 300
gtcagctatgtgccccatcctccttcatgccctccctccctttcctaccactgctgagtg 360
gcctggaacttgtttaaagtgt 382
<210> 140
<211> 200
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(200)
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
51
<223> n = A,T,C or G
<400> 140
accaaanctt ctttctgttg tgttngattt tactataggg gtttngcttn ttctaaanat 60
acttttcatt taacancttt tgttaagtgt caggctgcac tttgctccat anaattattg 120
ttttcacatt tcaacttgta tgtgtttgtc tcttanagca ttggtgaaat cacatatttt 180
atattcagca taaaggagaa 200
<210> 141
<211> 335
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(335)
<223> n = A,T,C or G
<400>
241
actttattttcaaaacactcatatgttgcaaaaaacacatagaaaaataaagtttggtgg 60
gggtgctgactaaacttcaagtcacagacttttatgtgacagattggagcagggtttgtt 120
atgcatgtagagaacccaaactaatttattaaacaggatagaaacaggctgtctgggtga 180
aatggttctgagaaccatccaattcacctgtcagatgctgatanactagctcttcagatg 240
tttttctaccagttcagagatnggttaatgactanttccaatggggaaaaagcaagatgg 300
attcacaaaccaagtaattttaaacaaagacactt 335
<210> 142
<211> 459
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(459)
<223> n = A,T,C or G
<400> l42
accaggttaatattgccacatatatcctttccaattgcgggctaaacagacgtgtattta 60
gggttgtttaaagacaacccagcttaatatcaagagaaattgtgacctttcatggagtat 120
ctgatggagaaaacactgagttttgacaaatcttattttattcagatagcagtctgatca 180
cacatggtccaacaacactcaaataataaatcaaatatnatcagatgttaaagattggtc 240
ttcaaacatcatagccaatgatgccccgcttgcctataatctctccgacataaaaccaca 300
tcaacacctcagtggccaccaaaccattcagcacagcttccttaactgtgagctgtttga 360
agctaccagtctgagcactattgactatntttttcangctctgaatagctctagggatct 420
cagcangggtgggaggaaccagctcaaccttggcgtant 459
<210> 143
<211> 140
<212> DNA
<213> Homo sapien
<400> 143
acatttcctt ccaccaagtc aggactcctg gcttctgtgg gagttcttat cacctgaggg 60
aaatccaaac agtctctcct agaaaggaat agtgtcacca accccaccca tctccctgag 120
accatccgac ttccctgtgt 140
<2l0> 144
<211> 164
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
52
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1). .(164)
<223> n = A,T,C or G
<400> 144
acttcagtaa caacatacaa taacaacatt aagtgtatat tgccatcttt gtcattttct 60
atctatacca ctctcccttc tgaaaacaan aatcactanc caatcactta tacaaatttg 120
aggcaattaa tccatatttg ttttcaataa ggaaaaaaag atgt 164
<2l0> l45
<211> 303
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(303)
<223> n = A, T, C or G
<400>
145
acgtagaccatccaactttgtatttgtaatggcaaacatccagnagcaattcctaaacaa 60
actggagggtatttatacccaattatcccattcattaacatgccctcctcctcaggctat 120
gcaggacagctatcataagtcggcccaggcatccagatactaccatttgtataaacttca 180
gtaggggagtccatccaagtgacaggtctaatcaaaggaggaaatggaacataagcccag 240
tagtaaaatnttgcttagctgaaacagccacaaaagacttaccgccgtggtgattaccat 300
caa 303
<210> 146
<211> 327
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(327j
<223> n = A,T,C or G
<400> 146
actgcagctc aattagaagt ggtctctgac tttcatcanc ttctccctgg gctccatgac 60
actggcctgg agtgactcat tgctctggtt ggttgagaga gctcctttgc caacaggcct 120
ccaagtcagg gctgggattt gtttcctttc cacattctag caacaatatg ctggccactt 180
cctgaacagg gagggtggga ggagccagca tggaacaagc tgccactttc taaagtagcc 240
agacttgcec ctgggcctgt cacacctact gatgaccttc tgtgcctgca ggatggaatg 300
taggggtgag ctgtgtgact ctatggt 327
<210> 147
<211> 173
<212> DNA
<2l3> Homo sapien
<220>
<221> misc_feature
<222> (1)...(173)
<223> n = A,T,C or G
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
53
<400> 147
acattgtttt tttgagataa agcattgana gagctctcct taacgtgaca caatggaagg 60
actggaacac atacccacat ctttgttctg agggataatt ttctgataaa gtcttgctgt 120
atattcaagc acatatgtta tatattattc agttccatgt ttatagccta gtt l73
<210> 148
<211> 477
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)....(477)
<223> n = A,T,C or G
<400>
148
acaaccactttatctcatcgaatttttaacccaaactcactcactgtgcctttctatcct 60
atgggatatattatttgatgctccatttcatcacacatatatgaataatacactcatact 120
gccctactacctgctgcaataatcacattcccttcctgtcctgaccctgaagccattggg 180
gtggtcctagtggccatcagtccangcctgcaccttgagcccttgagctccattgctcac 240
nccancccacctcaccgaccccatcctcttacacagctacctccttgctctctaacccca 300
tagattatntccaaattcagtcaattaagttactattaacactctacccgacatgtccag 360
caccactggtaagccttctccagccaacacacacacacacacacncacacacacacatat 420
ccaggcacaggctacctcatcttcacaatcacccctttaattaccatgctatggtgg 477
<210> 149
<211> 207
<212> DNA
<213> Homo sapien
<400> 149
acagttgtat tataatatca agaaataaac ttgcaatgag agcatttaag agggaagaac 60
taacgtattt tagagagcca aggaaggttt ctgtggggag tgggatgtaa ggtggggcct 120
gatgataaat aagagtcagc caggtaagtg ggtggtgtgg tatgggcaca gtgaagaaca 180
tttcaggcag agggaacagc agtgaaa 207
<210> l50
<211> 111
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(111)
<223> n = A, T, C or G
<400> 150
accttgattt cattgctgct ctgatggaaa cccaactatc taatttagct aaaacatggg 60
cacttaaatg tggtcagtgt ttggacttgt taactantgg catctttggg t 111
<210> 151
<211> 196
<212> DNA
<213> Homo sapien
<400> 151
agcgcggcag gtcatattga acattccaga tacctatcat tactcgatgc tgttgataac 60
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
54
agcaagatgg ctttgaactc agggtcacca ccagctattg gaccttacta tgaaaaccat 120
ggataccaac cggaaaaccc ctatcccgca cagcccactg tggtccccac tgtctacgag 180
gtgcatccgg ctcagt 196
<210> 152
<211> 132
<212> DNA
<213> Homo sapien
<400> 152
acagcacttt cacatgtaag aagggagaaa ttcctaaatg taggagaaag ataacagaac 60
cttccccttt tcatctagtg gtggaaacct gatgctttat gttgacagga atagaaccag 120
gagggagttt gt 132
<2l0> 153
<211'> 285
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(285)
<223> n = A,T,C or G
<400> 153
acaanaccca nganaggcca ctggccgtgg tgtcatggcc tccaaacatg aaagtgtcag 60
cttctgctct tatgtcctca tctgacaact ctttaccatt tttatcctcg ctcagcagga 120
gcacatcaat aaagtccaaa gtcttggact tggccttggc ttggaggaag tcatcaacac 180
cctggctagt gagggtgcgg cgccgctcct ggatgacggc atctgtgaag tcgtgcacca 240
gtctgcaggc cctgtggaag cgccgtccac acggagtnag gaatt 285
<2l0> 154
<211> 333
<212> DNA
<213> Homo sapien
<400> 154
accacagtcctgttgggccagggcttcatgaccctttctgtgaaaagccatattatcacc 60
accccaaatttttccttaaatatctttaactgaaggggtcagcctcttgactgcaaagac 120
cctaagccggttacacagctaactcccactggccctgatttgtgaaattgctgctgcctg 180
attggcacaggagtcgaaggtgttcagctcccctcctccgtggaacgagactctgatttg 240
agtttcacaaattctcgggccacctcgtcattgctcctctgaaataaaatccggagaatg 300
gtcaggcctgtctcatccatatggatcttccgg 333
<210> 155
<211> 308
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(308)
<223> n = A,T,C or G
<400> 155
actggaaata ataaaaccca catcacagtg ttgtgtcaaa gatcatcagg gcatggatgg 60
gaaagtgctt tgggaactgt aaagtgccta acacatgatc gatgattttt gttataatat 120
ttgaatcacg gtgcatacaa actctcctgc ctgctcctcc tgggccccag ccccagcccc 180
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
atcacagctc actgctctgt tcatccaggc ccagcatgta gtggctgatt cttcttggct 240
gcttttagcc tccanaagtt tctctgaagc caaccaaacc tctangtgta aggcatgctg 300
gccctggt 308
<210> 156
<211> 295
<212> DNA
<213> Homo sapien
<400> 156
accttgctcggtgcttggaacatattaggaactcaaaatatgagatgataacagtgccta 60
ttattgattactgagagaactgttagacatttagttgaagattttctacacaggaactga 120
gaataggagattatgtttggccctcatattctctcctatcctccttgcctcattctatgt 180
ctaatatattctcaatcaaataaggttagcataatcaggaaatcgaccaaataccaatat 240
aaaaccagatgtctatccttaagattttcaaatagaaaacaaattaacagactat 295
<210> 157
<211> 126
<212> DNA
<213> Homo sapien
<400> 157
acaagtttaa atagtgctgt cactgtgcat gtgctgaaat gtgaaatcca ccacatttct 60
gaagagcaaa acaaattctg tcatgtaatc tctatcttgg gtcgtgggta tatctgtccc l20
cttagt 126
<210> 158
<211> 442
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(442)
<223> n = A,T,C or G
<400>
158
acccactggtettggaaacacccatccttaatacgatgatttttctgtcgtgtgaaaatg 60
aanccagcaggctgcccctagtcagtccttccttccagagaaaaagagatttgagaaagt 120
gcctgggtaattcaccattaatttcctcccccaaactctctgagtcttcccttaatattt 180
ctggtggttctgaccaaagcaggtcatggtttgttgagcatttgggatcccagtgaagta 240
natgtttgtagccttgcatacttagcccttcccacgcacaaacggagtggcagagtggtg 300
ccaaccctgttttcccagtccacgtagacagattcacagtgcggaattctggaagctgga 360
nacagacgggctctttgcagagccgggactctgaganggacatgagggcctctgcctctg 420
tgttcattctctgatgtcctgt 442
<210> 159
<211> 498
<212> DNA
<213> Homo sapien
<220>
<221> miso_feature
<222> (1)...(498)
<223> n = A,T,C or G
<400> 159
acttccaggt aacgttgttg tttccgttga gcctgaactg atgggtgacg ttgtaggttc 60
att
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
56
tccaacaagaactgaggttgcagagcgggtagggaagagtgctgttccagttgcacctgg 120
gctgctgtggactgttgttgattcctcactacggcccaaggttgtggaactggcanaaag 180
gtgtgttgttgganttgagctcgggcggctgtggtaggttgtgggctcttcaacaggggc 240
tgctgtggtgccgggangtgaangtgttgtgtcacttgagcttggccagctctggaaagt 300
antanattcttcctgaaggccagcgcttgtggagctggcangggtcantgttgtgtgtaa 360
cgaaccagtgctgctgtgggtgggtgtanatcctccacaaagcctgaagttatggtgtcn 420
tcaggtaanaatgtggtttcagtgtccctgggcngctgtggaaggttgtanattgtcacc 480
aagggaataagctgtggt 498
<210> 160
<211> 380
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> {1)...{380)
<223> n = A, T, C or G
<400> 160
acctgcatccagcttccctgccaaactcacaaggagacatcaacctctagacagggaaac 60
agcttcaggatacttccaggagacagagccaccagcagcaaaacaaatattcccatgcct 120
ggagcatggcatagaggaagctganaaatgtggggtctgaggaagccatttgagtctggc 180
cactagacatctcatcagccacttgtgtgaagagatgccccatgaccccagatgcctctc 240
ccacccttacctccatctcacacacttgagctttccactctgtataattctaacatcctg 300
gagaaaaatggcagtttgaccgaacctgttcacaacggtagaggctgatttctaacgaaa 360
cttgtagaatgaagcctgga 380
<210> 161
<211> 114
<212> DNA
<213> Homo sapien
<400> 161
actccacatc ccctctgagc aggcggttgt cgttcaaggt gtatttggcc ttgcctgtca 60
cactgtccac tggcccctta tccacttggt gcttaatccc tcgaaagagc atgt 114
<210> 162
<211> 177
<212> DNA
<213> Homo sapien
<400> 162
actttctgaa tcgaatcaaa tgatacttag tgtagtttta atatcctcat atatatcaaa 60
gttttactac tctgataatt ttgtaaacca ggtaaceaga acatccagtc atacagcttt 120
tggtgatata taacttggca ataacccagt ctggtgatac ataaaactac tcactgt 177
<210> 163
<211> 137
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(137)
<223> n = A,T,C or G
<400> 163
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
catttataca gacaggcgtg aagacattca cgacaaaaac gcgaaattct atcccgtgac 60
canagaaggc agctacggct actcctacat cctggcgtgg gtggccttcg cctgcacctt 120
catcagcggc atgatgt 137
<210> l64
<211> 469
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(469)
<223> n = A,T,C or G
<400> 164
cttatcacaatgaatgttctcctgggcagcgttgtgatctttgccaccttcgtgaottta 60
tgcaatgcatcatgctatttcatacctaatgagggagttccaggagattcaaccaggaaa 120
tgcatggatctcaaaggaaacaaacacccaataaactcggagtggcagactgacaactgt 180
gagacatgcacttgctacgaaacagaaatttcatgttgcacccttgtttctacacctgtg 240
ggttatgacaaagacaactgccaaagaatcttcaagaaggaggactgcaagtatatcgtg 300
gtggagaagaaggacccaaaaaagacctgttctgtcagtgaatggataatctaatgtgct 360
tctagtaggcacagggctcccaggccaggcctcattctcctctggcctctaatagtcaat 420
gattgtgtagccatgcctatcagtaaaaagatntttgagcaaacacttt 469
<2l0> 165
<211> 195
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1) . . (195)
<223> n = A,T,C or G
<400> 165
acagtttttt atanatatcg acattgccgg cacttgtgtt cagtttcata aagctggtgg 60
atccgctgtc atccactatt ccttggctag agtaaaaatt attcttatag cccatgtccc 120
tgcaggccgc ccgcccgtag ttctcgttcc agtcgtcttg gcacacaggg tgccaggact 180
tcctctgaga tgagt 195
<210> 166
<211> 383
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1).,.(383)
<223> n = A,T,C or G
<400>
166
acatcttagtagtgtggcacatcagggggccatcagggtcacagtcactcatagcctcgc60
cgaggtcggagtccacaccaccggtgtaggtgtgctcaatcttgggcttggcgcccacct120
ttggagaagggatatgctgcacacacatgtccacaaagcctgtgaactcgccaaagaattl80
tttgcagaccagcctgagcaaggggcggatgttcagcttcagctcctccttcgtcaggtg240
gatgccaacctcgtctanggtccgtgggaagctggtgtccacntcacctacaacctgggc300
gangatcttataaagaggctccnagataaactccacgaaacttctctgggagctgctagt360
nggggcctttttggtgaactttc 383
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
S8
<210> 167
<211> 247
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(247)
<223> n = A,T,C or G
<400>
167
acagagccagaccttggccataaatgaancagagattaagactaaaccccaagtcganat60
tggagcagaaactggagcaagaagtgggcctggggctgaagtagagaccaaggccactgc120
tatanccatacacagagccaactctcaggccaaggcnatggttggggcaganccagagac180
tcaatctgantccaaagtggtggctggaacactggtcatgacanaggcagtgactctgac240
tgangtc 247
<210> l68
<211> 273
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(273)
<223> n = A,T,C or G
<400>
168
acttctaagttttctagaagtggaaggattgtantcatcctgaaaatgggtttacttcaa60
aatccctcanccttgttcttcacnactgtctatactganagtgtcatgtttccacaaagg120
gctgacacctgagcctgnattttcactcatccctgagaagccctttccagtagggtgggc180
aattcccaacttccttgccacaagcttcccaggctttctcccctggaaaactccagcttg240
agtcccagatacactcatgggctgccctgggca 273
<210> 169
<211> 431
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(431)
<223> n = A, T, C or G
<400> 169
acagccttgg cttccccaaa ctccacagtc tcagtgcaga aagatcatct tccagcagtc 60
agctcagacc agggtcaaag gatgtgacat caacagtttc tggtttcaga acaggttcta 120
ctactgtcaa atgacccccc atacttcctc aaaggctgtg gtaagttttg cacaggtgag 180
ggcagcagaa agggggtant tactgatgga caccatcttc tctgtatact ccacactgac 240
cttgccatgg gcaaaggccc ctaccacaaa aacaatagga tcactgctgg gcaccagctc 300
acgcacatca ctgacaaccg ggatggaaaa agaantgcca actttcatac atccaactgg 360
aaagtgatct gatactggat tcttaattac cttcaaaagc ttctgggggc catcagctgc 420
tcgaacactg a 431
<210> 170
<211> 266
<212> DNA
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
59
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(266)
<223> n = A,T,C or G
<400> 170
acctgtgggctgggctgttatgcctgtgccggctgctgaaagggagttcagaggtggagc 60
tcaaggagctctgcaggcattttgccaancctctccanagcanagggagcaacctacact 120
ccccgctagaaagacaccagattggagtcctgggagggggagttggggtgggcatttgat 180
gtatacttgtcacctgaatgaangagccagagaggaangagacgaanatganattggcct 240
tcaaagctaggggtctggcaggtgga 266
<210> 171
<211> 1248
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(1248)
<223> n = A,T,C or G
<400>
171
ggcagccaaatcataaacggcgaggactgcagcccgcactcgcagccctggcaggcggca 60
ctggtcatggaaaacgaattgttctgctcgggcgtcctggtgcatccgcagtgggtgctg 120
tcagccgcacactgtttccagaagtgagtgcagagctcctacaccatcgggctgggcctg 180
cacagtcttgaggccgaccaagagccagggagccagatggtggaggccagcctctccgta 240
cggcacccagagtacaacagacccttgctcgctaacgacctcatgctcatcaagttggac 300
gaatccgtgtccgagtctgacaccatccggagcatcagcattgcttcgcagtgccctacc 360
gcggggaactctt~gcctcgtttctggctggggtctgctggcgaacggcagaatgcctacc 420
gtgctgcagtgcgtgaacgtgtcggtggtgtctgaggaggtctgcagtaagctctatgac 480
ccgctgtaccaccccagcatgttctgcgccggcggagggcaagaccagaaggactcctgc 540
aacggtgactctggggggcccctgatctgcaacgggtacttgcagggccttgtgtctttc 600
ggaaaagccccgtgtggccaagttggcgtgccaggtgtctacaccaacctctgcaaattc 660
actgagtggatagagaaaaccgtccaggccagttaactctggggactgggaacccatgaa 720
attgacccccaaatacatcctgcggaaggaattcaggaatatctgttcccagcccctcct 780
ccctcaggcccaggagtccaggcccccagcccctcctccctcaaaccaagggtacagatc 840
cccagcccctcctccctcagacccaggagtccagaccccccagcccctcctccctcagac 900
ccaggagtccagcccctcctccctcagacccaggagtccagaccccccagcccctcctcc 960
ctcagacccaggggtccaggcccccaacccctcctccctcagactcagaggtccaagccc 1020
ccaacccntcattccccagacccagaggtccaggtcceagcccctcntccctcagaccca 1080
gcggtccaatgccacctagactntccctgtacacagtgcccccttgtggcacgttgaccc 1140
aaccttaccagttggtttttcatttttngtccctttcccctagatccagaaataaagttt 1200
aagagaagngcaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa 1248
<210> 172
<211> 159
<212> PRT
<213> Homo sapien
<220>
<221> VARIANT
<222> (1)...(159)
<223> Xaa = Any Amino Acid
<400> 172
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
Met Val Glu Ala Ser Leu Ser Val Arg His Pro Glu Tyr Asn Arg Pro
1 5 10 15
Leu Leu Ala Asn Asp Leu Met Leu Ile Lys Leu Asp Glu Ser Val Ser
20 25 30
Glu Ser Asp Thr Ile Arg Ser Ile Ser Tle Ala Ser Gln Cys Pro Thr
35 40 45
Ala Gly Asn Ser Cys Leu Val Ser Gly Trp Gly Leu Leu Ala Asn Gly
50 55 60
Arg Met Pro Thr Val Leu Gln Cys Val Asn Val Ser Val Val Ser Glu
70 75 80
G1u Val Cys Ser Lys Leu Tyr Asp Pro Leu Tyr His Pro Ser Met Phe
85 90 95
Cys Ala Gly Gly Gly Gln Xaa Gln Xaa Asp Ser Cys Asn Gly Asp Ser
100 105 110
Gly Gly Pro Leu I1e Cys Asn Gly Tyr Leu Gln Gly Leu Val Ser Phe
115 120 125
G1y Lys Ala Pro Cys Gly Gln Val Gly Val Pro Gly Val Tyr Thr Asn
130 135 140
Leu Cys Lys Phe Thr Glu Trp Tle Glu Lys Thr Val Gln Ala Ser
145 150 155
<210> 173
<211> 1265
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(1265)
<223> n = A,T,C or G
<400>
173
ggcagcccgcactcgcagccctggcaggcggcactggtcatggaaaacgaattgttctgc 60
tcgggcgtcctggtgcatccgcagtgggtgctgtcagccgcacactgtttccagaactcc 120
tacaccatcgggctgggcctgcacagtcttgaggccgaccaagagccagggagccagatg 180
gtggaggccagcctctccgtacggcacccagagtacaacagacccttgctcgctaacgac 240
ctcatgctcatcaagttggacgaatccgtgtccgagtctgacaccatccggagcatcagc 300
attgcttcgcagtgccctaccgcggggaactcttgcctcgtttctggctggggtctgctg 360
gcgaacggtgagctcacgggtgtgtgtctgccctcttcaaggaggtcctctgcccagtcg 420
cgggggctgacccagagctctgcgtcccaggcagaatgcctaccgtgctgcagtgcgtga 480
acgtgtcggtggtgtctgaggaggtctgcagtaagctctatgacccgctgtaccacccca 540
gcatgttctgcgccggcggagggcaagaccagaaggactcctgcaacggtgactctgggg 600
ggcccctgatctgcaacgggtacttgcagggccttgtgtctttcggaaaagccccgtgtg 660
gccaagttggcgtgccaggtgtctacaccaacctctgcaaattcactgagtggatagaga 720
aaaccgtccaggccagttaactctggggactgggaacccatgaaattgacccccaaatac 780
atcctgcggaaggaattcaggaatatctgttcccagcccctcctccctcaggcccaggag 840
tccaggcccccagcccctcctccctcaaaccaagggtacagatccccagcccctcctccc 900
tcagacccaggagtccagaccccccagcccctcctccctcagacccaggagtccagcccc 960
tcctccntcagacccaggagtccagaccccccagcccctcctccctcagacccaggggtt 1020
gaggcccccaacccctcctccttcagagtcagaggtccaagcccccaacccctcgttccc 1080
cagacccagaggtnnaggtcccagcccctcttccntcagacccagnggtccaatgccacc 1140
tagattttccctgnacacagtgcccccttgtggnangttgacccaaccttaccagttggt 1200
ttttcatttttngtccctttcccctagatccagaaataaagtttaagagangngcaaaaa 1260
aaaaa 1265
<210> 174
<211> 1459
<212> DNA
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
61
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(1459)
<223> n = A,T,C or G
<400> 174
ggtcagccgcacactgtttccagaagtgagtgcagagctcctacaccatcgggctgggcc60
tgcacagtcttgaggccgaccaagagccagggagccagatggtggaggccagcctctccg120
tacggcacccagagtacaacagacccttgctcgctaacgacctcatgctcatcaagttgg180
acgaatccgtgtccgagtctgacaccatccggagcatcagcattgcttcgcagtgcccta240
ccgcggggaactcttgcctcgtttctggctggggtctgctggcgaacggtgagctcacgg300
gtgtgtgtctgccctcttcaaggaggtcctctgcccagtcgcgggggctgacccagagct360
ctgcgtcccaggcagaatgcctaccgtgctgcagtgcgtgaacgtgtcggtggtgtctga420
ngaggtctgcantaagctctatgacccgctgtaccaccccancatgttctgcgccggcgg480
agggcaagaccagaaggactcctgcaacgtgagagaggggaaaggggagggcaggcgact540
cagggaagggtggagaagggggagacagagacacacagggccgcatggcgagatgcagag600
atggagagacacacagggagacagtgacaactagagagagaaactgagagaaacagagaa660
ataaacacaggaataaagagaagcaaaggaagagagaaacagaaacagacatggggaggc720
agaaacacacacacatagaaatgcagttgaccttccaacagcatggggcctgagggcggt780
gacctccacccaatagaaaatcctcttataacttttgactccccaaaaacctgactagaa840
atagcctactgttgacggggagccttaccaataacataaatagtcgatttatgcatacgt900
tttatgcattcatgatatacctttgttggaattttttgatatttctaagctacacagttc960
gtctgtgaatttttttaaattgttgcaactctcctaaaatttttctgatgtgtttattga1020
aaaaatccaagtataagtggacttgtgcattcaaaccagggttgttcaagggtcaactgt1080
gtacccagagggaaacagtgacacagattcatagaggtgaaacacgaagagaaacaggaa1140
aaatcaagactctacaaagaggctgggcagggtggctcatgcctgtaatcccagcacttt1200
gggaggcgaggcaggcagatcacttgaggtaaggagttcaagaccagcctggccaaaatg1260
gtgaaatcctgtctgtactaaaaatacaaaagttagctggatatggtggcaggcgcctgt1320
aatcccagctacttgggaggctgaggcaggagaattgcttgaatatgggaggcagaggtt1380
gaagtgagttgagatcacaccactatactccagctggggcaacagagtaagactctgtct1440
caaaaaaaaaaaaaaaaaa 1459
<210> 175
<211> 1167
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(1167)
<223> n = A,T,C or G
<400> 175
gcgcagccctggcaggcggcactggtcatggaaaacgaattgttctgctcgggcgtcctg60
gtgcatccgcagtgggtgctgtcagccgcacactgtttccagaactcctacaccatcgggl20
ctgggcctgcacagtcttgaggccgaccaagagccagggagccagatggtggaggccagc180
ctctccgtacggcacccagagtacaacagactcttgctcgctaacgacctcatgctcatc240
aagttggacgaatccgtgtccgagtctgacaccatccggagcatcagcattgcttcgcag300
tgccctaccgcggggaactcttgcctcgtntctggctggggtctgctggcgaacggcaga360
atgcctaccgtgctgcactgcgtgaacgtgtcggtggtgtctgaggangtctgcagtaag420
ctctatgacccgctgtaccaccccagcatgttctgcgccggcggagggcaagaccagaag480
gactcctgcaacggtgactctggggggcccctgatctgcaacgggtacttgcagggcctt540
gtgtctttcggaaaagccccgtgtggccaacttggcgtgccaggtgtctacaccaacctc600
tgcaaattcactgagtggatagagaaaaccgtccagnccagttaactctggggactggga660
acccatgaaattgacccccaaatacatcctgcggaangaattcaggaatatctgttccca720
gCCCC'tCCtCCC'tCaggCCCaggagtCCaggCCCCCagCCCC'tCCtCCCtcaaaccaagg780
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
62
gtacagatccccagcccctcctccctcagacccaggagtccagaccccccagcccctont 840
ccntcagacccaggagtccagcccctcctccntcagacgcaggagtccagaccccccagc 900
ccntcntccgtcagacccaggggtgcaggcccccaacccctcntccntcagagtcagagg 960
tccaagcccccaacccctcgttccccagacccagaggtncaggtcccagcccctcctccc 1020
tcagacccagcggtccaatgccacctagantntccctgtacacagtgcccccttgtggca 1080
ngttgacccaaccttaccagttggtttttcattttttgtccctttcccctagatccagaa 1140
ataaagtntaagagaagcgcaaaaaaa 1167
<210> 176
<211> 205
<212> PRT
<213> Homo sapien
<220>
<221> VARIANT
<222> (1)...(205)
<223> Xaa = Any Amino Acid
<400> 176
Met Glu Asn Glu Leu Phe Cys Ser Gly Val Leu Val His Pro Gln Trp
1 5 10 15
Val Leu Ser Ala Ala His Cys Phe Gln Asn Ser Tyr Thr Ile Gly Leu
20 25 30
Gly Leu His Ser Leu Glu Ala Asp Gln Glu Pro Gly Ser Gln Met Val
35 40 45
Glu A1a Ser Leu Ser Val Arg His Pro Glu Tyr Asn Arg Leu Leu Leu
50 55 60
Ala Asn Asp Leu Met Leu Ile Lys Leu Asp Glu Ser Val Ser Glu Ser
65 70 75 80
Asp Thr Ile Arg Ser Ile Ser Ile Ala Ser Gln Cys Pro Thr Ala Gly
85 90 95
Asn Ser Cys Leu Val Ser Gly Trp Gly Leu Leu Ala Asn Gly Arg Met
100 105 110
Pro Thr Val Leu His Cys Val Asn Val Ser Val Val Ser Glu Xaa Val
1l5 120 125
Cys Ser Lys Leu Tyr Asp Pro Leu Tyr His Pro Ser Met Phe Cys Ala
130 135 140
Gly Gly Gly Gln Asp Gln Lys Asp Ser Cys Asn Gly Asp Ser Gly Gly
145 150 155 160
Pro Leu Ile Cys Asn Gly Tyr Leu Gln Gly Leu Val Ser Phe Gly Lys
165 170 175
Ala Pro Cys Gly Gln Leu Gly Val Pro Gly Val Tyr Thr Asn Leu Cys
180 185 190
Lys Phe Thr Glu Trp Ile Glu Lys Thr Val Gln Xaa Ser
195 200 205
<210> 177
<211> 1119
<212> DNA
<2l3> Homo sapien
<400> 177
gcgcactcgcagccctggcaggcggcactggtcatggaaaacgaattgttctgctcgggc 60
gtcctggtgcatccgcagtgggtgctgtcagccgcacactgtttccagaactcctacacc 120
atcgggctgggcctgcacagtcttgaggccgaccaagagccagggagccagatggtggag 180
gccagcctctccgtacggcacccagagtacaacagacccttgctcgctaacgacctcatg 240
ctcatcaagttggacgaatccgtgtccgagtctgacaccatccggagcatcagcattgct 300
tcgcagtgccctaccgcggggaactcttgcctcgtttctggctggggtctgctggcgaac 360
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
63
gatgctgtgattgccatccagtcccagactgtgggaggctgggagtgtgagaagctttcc 420
caaccctggcagggttgtaccatttcggcaacttccagtgcaaggacgtcctgctgcatc 480
ctcactgggtgctcactactgctcactgcatcacccggaacactgtgatcaactagccag 540
caccatagttctccgaagtcagactatcatgattactgtgttgactgtgctgtctattgt 600
actaaccatgccgatgtttaggtgaaattagcgtcacttggcctcaaccatcttggtatc 660
cagttatcctcactgaattgagatttcctgcttcagtgtcagccattcccacataatttc 720
tgacctacagaggtgagggatcatatagctcttcaaggatgctggtactcccctcacaaa 780
ttcatttctcctgttgtagtgaaaggtgcgccctctggagcctcccagggtgggtgtgca 840
ggtcacaatgatgaatgtatgatcgtgttcccattacccaaagcctttaaatccctcatg 900
ctcagtacaccagggcaggtctagcatttcttcatttagtgtatgctgtccattcatgca 960
accacctcaggactcctggattctctgcctagttgagctcctgcatgctgcctccttggg 1020
gaggtgagggagagggcccatggttcaatgggatctgtgcagttgtaacacattaggtgc 1080
ttaataaacagaagctgtgatgttaaaaaaaaaaaaaaa 1119
<210> 178
<212> 164
<212> PRT
<213> Homo sapien
<220>
<221> VARIANT
<222> (1)...(164)
<223> Xaa = Any Amino Acid
<400> 178
Met Glu Asn Glu Leu Phe Cys Ser Gly Val Leu Val His Pro Gln Trp
1 5 10 15
Val Leu Ser Ala Ala His Cys Phe Gln Asn Ser Tyr Thr Ile Gly Leu
20 25 30
Gly Leu His Ser Leu Glu Ala Asp Gln Glu Pro Gly Ser Gln Met Val
35 40 45
Glu Ala Ser Leu Ser Val Arg His Pro Glu Tyr Asn Arg Pro Leu Leu
50 55 60
Ala Asn Asp Leu Met Leu Tle Lys Leu Asp Glu Ser Val Ser Glu Ser
65 70 75 80
Asp Thr Ile Arg Ser Ile Ser Ile Ala Ser Gln Cys Pro Thr Ala Gly
85 90 95
Asn Ser Cys Leu Val Ser Gly Trp Gly Leu Leu Ala Asn Asp Ala Val
100 105 110
Ile Ala Ile Gln Ser Xaa Thr Val Gly Gly Trp Glu Cys Glu Lys Leu
115 120 125
Ser Gln Pro Trp Gln Gly Cys Thr Ile Ser Ala Thr Ser Ser Ala Arg
130 135 140
Thr Ser Cys Cys Ile Leu Thr Gly Cys Ser Leu Leu Leu Thr Ala Ser
145 150 155 160
Pro Gly Thr Leu
<210> 179
<21l> 250
<212> DNA
<213> Homo sapien
<400> 179
ctggagtgcc ttggtgtttc aagcccctgc aggaagcaga atgcaccttc tgaggcacct 60
ccagctgccc ccggccgggg gatgcgaggc tcggagcacc cttgcccggc tgtgattgct 120
gccaggcact gttcatctca gcttttctgt ccctttgctc ccggcaagcg cttctgctga 180
aagttcatat ctggagcctg atgtcttaac gaataaaggt cccatgctcc acccgaaaaa 240
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
64
aaaaaaaaaa 250
<210> 180
<211> 202
<212> DNA
<213> Homo sapien
<400> 180
actagtccag tgtggtggaa ttccattgtg ttgggcccaa cacaatggct acctttaaca 60
tcacccagac cccgcccctg cccgtgcccc acgctgctgc taacgacagt atgatgctta 120
ctctgctact cggaaactat ttttatgtaa ttaatgtatg ctttcttgtt tataaatgcc 180
tgatttaaaa aaaaaaaaaa as 202
<210> 181
<211> 558
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> {1)...(558)
<223> n = A,T,C or G
<400> 18l
tccytttgktnaggtttkkgagacamccckagacctwaanctgtgtcacagacttcyngg 60
aatgtttaggcagtgctagtaatttcytcgtaatgattctgttattactttcctnattct 120
ttattcctctttcttctgaagattaatgaagttgaaaattgaggtggataaatacaaaaa 180
ggtagtgtgatagtataagtatctaagtgcagatgaaagtgtgttatatatatccattca 240
aaattatgcaagttagtaattactcagggttaactaaattactttaatatgctgttgaac 300
ctactctgttccttggctagaaaaaattataaacaggactttgttagtttgggaagccaa 360
attgataatattctatgttctaaaagttgggctatacataaattattaagaaatatggaw 420
ttttattcccaggaatatggkgttcattttatgaatattacscrggatagawgtwtgagt 480
aaaaycagttttggtwaataygtwaatatgtcmtaaataaacaakgctttgacttatttc 540
caaaaaaaaaaaaaaaaa 558
<210> 182
<211> 479
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(479)
<223> n = A,T,C or G
<400>
182
acagggwttkgrggatgctaagsccccrgarwtygtttgatccaaccctggcttwttttc 60
agaggggaaaatggggcctagaagttacagmscatytagytggtgcgmtggcacccctgg 120
cstcacacagastcccgagtagctgggactacaggcacacagtcactgaagcaggccctg 180
ttwgcaattcacgttgccacctccaacttaaacattcttcatatgtgatgtccttagtca 240
ctaaggttaaactttcccacccagaaaaggcaacttagataaaatcttagagtactttca 300
tactmttctaagtcctcttccagcctcactkkgagtcctmcytgggggttgataggaant 360
ntctcttggctttctcaataaartctctatycatctcatgtttaatttggtacgcatara 420
awtgstgaraaaattaaaatgttctggttymactttaaaaaraaaaaaaaaaaaaaaaa 479
<210> 183
<211> 384
<212> DNA
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<213> Homo sapien
<400>
183
aggcgggagcagaagctaaagccaaagcccaagaagagtggcagtgccagcactggtgcc 60
agtaccagtaccaataacagtgccagtgccagtgccagcaccagtggtggcttcagtgct 120
ggtgccagcctgaccgccactctcacatttgggctcttcgctggccttggtggagctggt 180
gccagcaccagtggcagctctggtgcctgtggtttctcctacaagtgagattttagatat 240
tgttaatcctgccagtctttctcttcaagccagggtgcatcctcagaaacctactcaaca 300
cagcactctaggcagccactatcaatcaattgaagttgacactctgcattaratctattt 360
gccatttcaaaaaaaaaaaaaaaa 384
<210> l84
<211> 496
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1j...(496)
<223> n = A,T,C or G
<400>
184
accgaattgggaccgctggcttataagcgatcatgtyyntccrgtatkacctcaacgagc 60
agggagatcgagtctatacgctgaagaaatttgacccgatgggacaacagacctgctcag l20
cccatcctgctcggttctccccagatgacaaatactctsgacaccgaatcaccatcaaga 180
aacgcttcaaggtgctcatgacccagcaaccgcgccctgtcctctgagggtcccttaaac 240
tgatgtcttttctgccacctgttaeccctcggagactccgtaaccaaactcttcggactg 300
tgagccctgatgcctttttgccagccatactctttggcatccagtctctcgtggcgattg 360
attatgcttgtgtgaggcaatcatggtggcatcacccataaagggaacacatttgacttt 420
tttttctcatattttaaattactacmagawtattwmagawwaaatgawttgaaaaactst 480
taaaaaaaaaaaaaaa 496
<210> 185
<211> 384
<212> DNA
<213> Homo sapien
<400> I85
gctggtagcctatggegkggcccacggaggggctcctgaggccacggracagtgacttcc 60
caagtatcytgcgcsgcgtcttctaccgtccctacctgcagatcttcgggcagattcccc 120
aggaggacatggacgtggccctcatggagcacagcaactgytcgtcggagcccggcttct 180
gggcacaccctcctggggcccaggcgggcacctgcgtctcccagtatgccaactggctgg 240
tggtgctgctcctcgtcatcttcctgctcgtggccaacatcctgctggtcaacttgctca 300
ttgccatgttcagttacacattcggcaaagtacagggcaacagcgatctctactgggaag 360
gcgcagcgttaccgcctcatccgg 384
<210> 186
<211> 577
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1j. .(577)
<223> n = A,T,C or G
<400> 7.86
gagttagctc ctccacaacc ttgatgaggt cgtctgcagt ggcctctcgc ttcataccgc 60
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
66
tnccatcgtcatactgtaggtttgccaccacytcctggcatcttggggcggcntaatatt 120
ccaggaaactctcaatcaagtcaccgtcgatgaaacctgtgggctggttctgtcttccgc 180
tcggtgtgaaaggatctcccagaaggagtgctcgatcttccccacacttttgatgacttt 240
attgagtcgattctgcatgtccagcaggaggttgtaccagctctctgacagtgaggtcac 300
cagccctatcatgccgttgamcgtgccgaagarcaccgagccttgtgtgggggkkgaagt 360
ctcacccagattctgcattaccagagagccgtggcaaaagacattgacaaactcgcccag 420
gtggaaaaagamcamctcctggargtgctngccgctcctcgtcmgttggtggcagcgctw 480
tccttttgacacacaaacaagttaaaggcattttcagcccccagaaanttgtcatcatcc 540
aagatntcgcacagcactnatccagttgggattaaat 577
<210> 187
<211> 534
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(534)
<223> n = A,T,C or G
<400>
187
aacatcttcctgtataatgctgtgtaatatcgatccgatnttgtctgstgagaatycatw 60
actkggaaaagmaacattaaagcctggacactggtattaaaattcacaatatgcaacact 120
ttaaacagtgtgtcaatctgctcccyynactttgtcatcaccagtctgggaakaagggta 180
tgccctattcacacctgttaaaagggcgctaagcatttttgattcaacatcttttttttt 240
gacacaagtccgaaaaaagcaaaagtaaacagttatyaatttgttagccaattcactttc 300
ttcatgggacagagccatytgatttaaaaagcaaattgcataatattgagcttygggagc 360
tgatatttgagcggaagagtagcctttctacttcaccagacacaactccctttcatattg 420
ggatgttnacnaaagtwatgtctctwacagatgggatgcttttgtggcaattctgttctg 480
aggatctcccagtttatttaccacttgcacaagaaggcgttttcttcctcaggc 534
<210> 188
<211> 761
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(761)
<223> n = A,T,C or G
<400>
188
agaaaccagtatctctnaaaacaacctctcataccttgtggacctaattttgtgtgcgtg 60
tgtgtgtgcgcgcatattatatagacaggcacatcttttttacttttgtaaaagcttatg 120
cctctttggtatctatatctgtgaaagttttaatgatctgccataatgtcttggggacct 180
ttgtcttctgtgtaaatggtactagagaaaacacctatnttatgagtcaatctagttngt 240
tttattcgacatgaaggaaatttccagatnacaacactnacaaactctccctkgackarg 300
ggggacaaagaaaagcaaaactgamcataaraaacaatwacctggtgagaarttgcataa 360
acagaaatwrggtagtatattgaarnacagcatcattaaarmgttwtkttwttctccctt 420
gcaaaaaacatgtacngacttcccgttgagtaatgccaagttgttttttttatnataaaa 480
cttgcccttcattacatgtttnaaagtggtgtggtgggccaaaatattgaaatgatggaa 540
ctgactgataaagctgtacaaataagcagtgtgcctaacaagcaacacagtaatgttgac 600
atgcttaattcacaaatgctaatttcattataaatgtttgctaaaatacactttgaacta 660
tttttctgtnttcccagagctgagatnttagattttatgtagtatnaagtgaaaaantac 720
gaaaataataacattgaagaaaaananaaaaaanaaaaaaa 761
<210> 189
<211> 482
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
67
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(482)
<223> n = A, T, C or G
<400>
189
tttttttttttttgccgatnctactattttattgcaggangtgggggtgtatgcaccgca 60
caccggggctatnagaagcaagaaggaaggagggagggcacagccccttgctgagcaaca 120
aagccgcctgctgccttctctgtctgtctcctggtgcaggcacatggggagaccttcccc 180
aaggcaggggccaccagtccaggggtgggaatacagggggtgggangtgtgcataagaag 240
tgataggcacaggccacccggtacagacccctcggctcctgacaggtngatttcgaccag 300
gtcattgtgccctgcccaggcacagcgtanatctggaaaagacagaatgctttccttttc 360
aaatttggctngtcatngaangggcanttttccaanttnggctnggtcttggtacncttg 420
gttcggcccagctccncgtccaaaaantattcacccnnctccnaattgcttgcnggnccc 480
cc 482
<210> 190
<211> 471
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(471)
<223> n = A,T,C or G
<400> 190
ttttttttttttttaaaacagtttttcacaacaaaatttattagaagaatagtggttttg 60
aaaactctcgcatccagtgagaactaccatacaccacattacagctnggaatgtnctcca 120
aatgtctggtcaaatgatacaatggaaccattcaatcttacacatgcacgaaagaacaag 180
cgcttttgacatacaatgcacaaaaaaaaaaggggggggggaccacatggattaaaattt 240
taagtactcatcacatacattaagacacagttctagtccagtcnaaaatcagaactgcnt 300
tgaaaaatttcatgtatgcaatccaaccaaagaacttnattggtgatcatgantnctcta 360
ctacatcnaccttgatcattgccaggaacnaaaagttnaaancacncngtacaaaaanaa 420
tctgtaattnanttcaacctccgtacngaaaaatnttnnttatacactccc 471
<210> 191
<211> 402
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(402)
<223> n = A, T, C or G
<400>
191
gagggattgaaggtctgttctastgtcggmctgttcagccaccaactctaacaagttgct 60
gtcttccactcactgtctgtaagctttttaacccagacwgtatcttcataaatagaacaa 120
attcttcaccagtcacatcttctaggacctttttggattcagttagtataagctcttcca 180
cttcctttgttaagacttcatctggtaaagtcttaagttttgtagaaaggaattyaattg 240
ctcgttctctaacaatgtcctctccttgaagtatttggctgaacaacccacctaaagtcc 300
ctttgtgcatccattttaaatatacttaatagggcattgktncactaggttaaattctgc 360
aagagtcatctgtctgcaaaagttgcgttagtatatctgcca 402
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
68
<210> 192
<211> 601
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(601)
<223> n = A,T,C or G
<400>
192
gagctcggatccaataatctttgtctgagggcagcacacatatncagtgccatggnaact 60
ggtctaccccacatgggagcagcatgccgtagntatataaggtcattccetgagtcagac 120
atgcytytttgaytaccgtgtgccaagtgctggtgattctyaacacacytccatcccgyt 180
cttttgtggaaaaactggcacttktctggaactagcargacatcacttacaaattcaccc 240
acgagacacttgaaaggtgtaacaaagcgaytcttgcattgctttttgtccctccggcac 300
cagttgtcaatactaacccgctggtttgcctccatcacatttgtgatctgtagctctgga 360
tacatctcctgacagtactgaagaacttcttcttttgtttcaaaagcarctcttggtgcc 420
tgttggatcaggttcccatttcccagtcygaatgttcacatggcatatttwacttcccac 480
aaaacattgcgatttgaggctcagcaacagcaaatcctgttccggcattggctgcaagag 540
cctcgatgtagccggccagcgccaaggcaggcgccgtgagccccaccagcagcagaagca 600
g 601
<210> 193
<211> 608
<212> DNA
<2l3> Homo sapien
<220>
<22l> misc_feature
<222> (1)...(608)
<223> n = A,T,C or G
<400> 193
atacagcccanatcccaccacgaagatgcgcttgttgactgagaacctgatgcggtcact 60
ggtcccgctgtagccccagcgactctccacctgctggaagcggttgatgctgcactcytt 120
cccaacgcaggcagmagcgggsccggtcaatgaactccaytcgtggcttggggtkgacgg 180
tkaagtgcaggaagaggctgaccacctcgcggtccaccaggatgcccgactgtgcgggac 240
ctgcagcgaaactcctcgatggtcatgagcgggaagcgaatgaggcccagggccttgccc 300
agaaccttccgcctgttctctggcgtcacctgcagctgctgccgctgacactcggcctcg 360
gaccagcggacaaacggerttgaaeagccgcacctcacggatgcccagtgtgtcgcgctc 420
caggammgscaccagcgtgtccaggtcaatgtcggtgaagccctccgcgggtratggcgt 480
ctgcagtgtttttgtcgatgttctccaggcacaggctggccagctgcggttcatcgaaga 540
gtcgagcctgcgtgagcagcatgaaggcgttgtcggctcgcagttcttcttcaggaactc 600
cacgcaat 608
<210> 194
<211> 392
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1) . . . (392)
<223> n = A, T, C or G
<400> 194
gaacggctgg accttgcctc gcattgtgct tgctggcagg gaataccttg gcaagcagyt 60
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
69
ccagtccgagcagccccagaccgctgcogcccgaagctaagcctgcctctggccttcccc120
tccgcctcaatgcagaaccagtagtgggagcactgtgtttagagttaagagtgaacactg180
tttgattttacttgggaatttcctctgttatatagcttttcccaatgctaatttccaaac240
aacaacaacaaaataacatgtttgcctgttaagttgtataaaagtaggtgattctgtatt300
taaagaaaatattactgttacatatactgcttgcaatttctgtatttattgktnctstgg360
aaataaatatagttattaaaggttgtcantcc 392
<220> 195
<211> 502
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(502)
<223> n = A,T,C or G
<400>
195
ccsttkgaggggtkaggkyccagttyccgagtggaagaaacaggccaggagaagtgcgtg 60
ccgagctgaggcagatgttcccacagtgacccccagagccstgggstatagtytctgacc 120
cctcncaaggaaagaccacsttctggggacatgggctggagggcaggacctagaggcacc 180
aagggaaggccccattccggggstgttccccgaggaggaagggaaggggctctgtgtgcc 240
ccccasgaggaagaggccctgagtcctgggatcagacaccccttcacgtgtatccccaca 300
caaatgcaagctcaccaaggtcccctctcagtccccttccstacaccctgamcggccact 360
gscscacacccacccagagcacgccacccgccatggggartgtgctcaaggartcgcngg 420
gcarcgtggacatctngtoccagaagggggcagaatctccaataganggactgarcmstt 480
gctnanaaaaaaaaanaaaaas 502
<210> 196
<211> 665
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(665)
<223> n = A,T,C or G
<400>
196
ggttacttggtttcattgccaccacttagtggatgtcatttagaaccattttgtctgctc 60
cctctggaagccttgcgcagagcggactttgtaattgttggagaataactgctgaatttt 120
wagctgtttkgagttgattsgcaccactgcacccacaacttcaatatgaaaacyawttga 180
actwatttattatcttgtgaaaagtataacaatgaaaattttgttcatactgtattkatc 240
aagtatgatgaaaagcaawagatatatattcttttattatgttaaattatgattgccatt 300
attaatcggcaaaatgtggagtgtatgttcttttcacagtaatatatgccttttgtaaot 360
tcacttggttattttattgtaaatgarttacaaaattcttaatttaagaraatggtatgt 420
watatttatttcattaatttctttcctkgtttacgtwaattttgaaaagawtgcatgatt 480
tcttgacagaaatcgatcttgatgctgtggaagtagtttgacccacatccctatgagttt 540
ttcttagaatgtataaaggttgtagcccatcnaacttcaaagaaaaaaatgaccacatac 600
tttgcaatcaggctgaaatgtggoatgctnttctaattccaactttataaactagcaaan 660
aagtg
665
<210> 197
<211> 492
<212> DNA
<213> Homo sapien
<220>
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<221> misc_feature
<222> (1)...(492)
<223> n = A,T,C or G
<400>
197
ttttntttttttttttttgcaggaaggattccatttattgtggatgcattttcacaatat 60
atgtttattggagcgatccattatcagtgaaaagtatcaagtgtttataanatttttagg 120
aaggcagattcacagaacatgctngtcngcttgcagttttacctcgtanagatnacagag 180
aattatagtcnaaccagtaaacnaggaatttacttttcaaaagattaaatccaaactgaa 240
caaaattctaccctgaaacttactccatccaaatattggaataanagtcagcagtgatac 300
attctcttctgaactttagattttctagaaaaatatgtaatagtgatcaggaagagctct 360
tgttcaaaagtacaacnaagcaatgttcccttaccataggccttaattcaaactttgatc 420
catttcactcccatcacgggagtcaatgctacctgggacacttgtattttgttcatnctg 480
ancntggcttas 492
<210> 198
<211> 478
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(478)
<223> n = A,T,C or G
<400>
198
tttnttttgnatttcantctgtannaantattttcattatgtttattanaaaaatatnaa 60
tgtntccacnacaaatcatnttacntnagtaagaggccanctacattgtacaacatacac 120
tgagtatattttgaaaaggacaagtttaaagtanacncatattgccgancatancacatt 180
tatacatggcttgattgatatttagcacagcanaaactgagtgagttaccagaaanaaat 240
natatatgtcaatcngatttaagatacaaaacagatcctatggtacatancatcntgtag 300
gagttgtggctttatgtttactgaaagtcaatgcagttcctgtacaaagagatggccgta 360
agcattctagtacctctactccatggttaagaatcgtacacttatgtttacatatgtnca 420
gggtaagaattgtgttaagtnaanttatggagaggtccangagaaaaatttgatncaa 478
<2l0> 199
<211> 482
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(482)
<223> n = A,T,C or G
<400>
199
agtgacttgtcctccaacaaaaccccttgatcaagtttgtggcactgacaatcagaccta 60
tgctagttcctgtcatctattcgctactaaatgcagactggaggggaccaaaaaggggca 120
tcaactccagctggattattttggagcctgcaaatctattcctacttgtacggactttga 180
agtgattcagtttcctctacggatgagagactggctcaagaatatcctcatgcagcttta 240
tgaagccnactctgaacacgctggttatctnagatgagaancagagaaataaagtcnaga 300
aaatttacctggangaaaagaggctttnggctggggaccatcccattgaaccttctctta 360
anggactttaagaanaaactaccacatgtntgtngtatcctggtgccnggccgtttantg 420'
aacntngacnncacccttntggaatanantcttgacngcntcctgaacttgctcctctgc 480
ga 482
<210> 200
<211> 270
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
71
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(270)
<223> n = A,T,C or G
<400> 200
cggccgcaagtgcaactccagctggggccgtgcggacgaagattctgccagcagttggtc 60
cgactgcgacgacggcggcggcgacagtcgcaggtgcagcgcgggcgcctggggtcttgc 120
aaggctgagctgacgccgcagaggtcgtgtcacgtcccacgaccttgacgccgtcgggga 180
cagccggaacagagcccggtgaangcgggaggcctcggggagcccctcgggaagggcggc 240
ccgagagatacgcaggtgcaggtggccgcc 270
<210> 201
<21l> 419
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (l)...(419)
<223> ri = A, T, C or G
<400>
201
ttttttttttttttggaatctactgcgagcacagcaggtcagcaacaagtttattttgca 60
gctagcaaggtaacagggtagggcatggttacatgttcaggtcaacttcctttgtcgtgg 120
ttgattggtttgtctttatgggggcggggtggggtaggggaaancgaagcanaantaaca 180
tggagtgggtgcaccctccctgtagaacctggttacnaaagcttggggcagttcacctgg 240
tctgtgaccgtcattttcttgacatcaatgttattagaagtcaggatatcttttagagag 300
tccactgtntctggagggagattagggtttcttgccaanatccaancaaaatccacntga 360
aaaagttggatgatncangtacngaataccganggcatanttctcatantcggtggcca 419
<210> 202
<211> 509
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(509)
<223> n = A,T,C or G
<400>
202
tttntttttttttttttttttttttttttttttttttttttttttttttttttttttttt 60
tggcacttaatccatttttatttcaaaatgtctacaaantttnaatncnccattatacng 120
gtnattttncaaaatctaaannttattcaaatntnagccaaantccttacncaaatnnaa 180
tacncncaaaaatcaaaaatatacntntctttcagcaaacttngttacataaattaaaaa 240
aatatatacggctggtgttttcaaagtacaattatcttaacactgcaaacatntttnnaa 300
ggaactaaaataaaaaaaaacactnccgcaaaggttaaagggaacaacaaattcntttta 360
caacancnncnattataaaaatcatatctcaaatcttaggggaatatatacttcacacng 420
ggatcttaacttttactncactttgtttatttttttanaaccattgtnttgggcccaaca 480
caatggnaatnccnccncnctggactagt 509
<210> 203
<2l1> 583
<212> DNA
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
72
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(583)
<223> n = A,T,C or G
<400>
203
ttttttttttttttttttgacccccctcttataaaaaacaagttaccattttattttact 60
tacacatatttattttataattggtattagatattcaaaaggcagcttttaaaatcaaac 120
taaatggaaactgccttagatacataattcttaggaattagcttaaaatctgcctaaagt 180
gaaaatcttctctagctcttttgactgtaaatttttgactcttgtaaaacatccaaattc 240
atttttcttgtctttaaaattatctaatctttccattttttccctattccaagtcaattt 300
gcttctctagcctcatttcctagctcttatctactattagtaagtggcttttttcctaaa 360
agggaaaacaggaagaganaatggcacacaaaacaaacattttatattcatatttctacc 420
tacgttaataaaatagcattttgtgaagccagctcaaaagaaggcttagatccttttatg 480
tccattttagtcactaaacgatatcnaaagtgccagaatgcaaaaggtttgtgaacattt 540
attcaaaagctaatataagatatttcacatactcatctttctg 583
<210> 204
<211> 589
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(589)
<223> n = A,T,C or G
<400>
204
ttttttttntttttttttttttttttnctcttctttttttttganaatgaggatcgagtt 60
tttcactctctagatagggcatgaagaaaactcatctttccagctttaaaataacaatca 120
aatctcttatgctatatcatattttaagttaaactaatgagtcactggcttatcttctcc 180
tgaaggaaatctgttcattcttctcattcatatagttatatcaagtactaccttgcatat 240
tgagaggtttttcttctctatttacacatatatttccatgtgaatttgtatcaaaccttt 300
attttcatgcaaactagaaaataatgtnttcttttgcataagagaagagaacaatatnag 360
cattacaaaactgctcaaattgtttgttaagnttatccattataattagttnggcaggag 420
ctaatacaaatcacatttacngacnagcaataataaaactgaagtaccagttaaatatcc 480
aaaataattaaaggaacatttttagcctgggtataattagctaattcactttacaagcat 540
ttattnagaatgaattcacatgttattattccntagcccaacacaatgg 589
<210> 205
<211> 545
<212> DNA
<2l3> Homo sapien
<220>
<221> misc_feature
<222> (1)...(545)
<223> n = A,T,C or G
<400>
205
tttttnttttttttttcagtaataatcagaacaatatttatttttatatttaaaattcat 60
agaaaagtgccttacatttaataaaagtttgtttctcaaagtgatcagaggaattagata 120
tngtcttgaacaccaatattaatttgaggaaaatacaccaaaatacattaagtaaattat 180
ttaagatcatagagcttgtaagtgaaaagataaaatttgacctcagaaactctgagcatt 240
aaaaatccactattagcaaataaattactatggacttcttgctttaattttgtgatgaat 300
atggggtgtcactggtaaaccaacacattctgaaggatacattacttagtgatagattct 360
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
73
tatgtacttt gctanatnac gtggatatga gttgacaagt ttctctttct tcaatctttt 420
aaggggcnga ngaaatgagg aagaaaagaa aaggattacg catactgttc tttctatngg 480
aaggattaga tatgtttcct ttgccaatat taaaaaaata ataatgttta ctactagtga 540
aaccc 545
<210> 206
<211> 487
<212> DNA
<213> Homo sapien
<220>
<22l> misc_feature
<222>. (1) . .. (487)
<223> n = A,T,C or G
<400>
206
ttttttttttttttttagtcaagtttctnatttttattataattaaagtcttggtcattt 60
catttattagctctgcaacttacatatttaaattaaagaaacgttnttagacaactgtna 120
caatttataaatgtaaggtgccattattgagtanatatattcctccaagagtggatgtgt 180
cccttctcccaccaactaatgaancagcaacattagtttaattttattagtagatnatac 240
actgctgcaaacgctaattctcttctccatccccatgtngatattgtgtatatgtgtgag 300
ttggtnagaatgcatcancaatctnacaatcaacagcaagatgaagctaggcntgggctt 360
tcggtgaaaatagactgtgtctgtctgaatcaaatgatctgacctatcctcggtggcaag 420
aactcttcgaaccgcttcctcaaaggcngctgccacatttgtggcntctnttgcacttgt 480
ttcaaaa 487
<210> 207
<211> 332
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(332)
<223> n = A,T,C or G
<400> 207
tgaattggct aaaagactgc atttttanaa ctagcaactc ttatttcttt cctttaaaaa 60
tacatagcat taaatcccaa atcctattta aagacctgac agcttgagaa ggtcactact 120
gcatttatag gaccttetgg tggttctgct gttacntttg aantctgaca atecttgana 180
atctttgcat gcagaggagg taaaaggtat tggattttca cagaggaana acacagcgca 240
gaaatgaagg ggccaggctt actgagcttg tccactggag ggctcatggg tgggacatgg 300
aaaagaaggc agcctaggcc ctggggagcc ca 332
<210> 208
<211> 524
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(524)
<223> n = A,T,C or G
<400> 208
agggcgtggt gcggagggcg ttactgtttt gtctcagtaa caataaatac aaaaagactg 60
gttgtgttcc ggccccatcc aaccacgaag ttgatttctc ttgtgtgcag agtgactgat 120
tttaaaggac atggagcttg tcacaatgtc acaatgtcac agtgtgaagg gcacactcac 180
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
74
tcccgcgtgattcacatttagcaaccaacaatagctcatgagtccatacttgtaaatact 240
tttggcagaatacttnttgaaacttgcagatgataactaagatccaaga.tatttcccaaa 300
gtaaatagaagtgggtcataatattaattacctgttcacatcagcttccatttacaagtc 360
atgagcccagacactgacatcaaactaagcccacttagactcctcaccaccagtctgtcc 420
tgtcatcagacaggaggctgtcaccttgaccaaattctcaccagtcaatcatctatccaa 480
aaaccattacctgatccacttccggtaatgcaccaccttggtga 524
<2l0> 209
<211> 159
<212> DNA
<213> Homo sapien
<400> 209
gggtgaggaa atccagagtt gccatggaga aaattccagt gtcagcattc ttgctccttg 60
tggccctctc ctacactctg gccagagata ccacagtcaa acctggagcc aaaaaggaca 120
caaaggactc tcgacccaaa ctgccccaga ccctctcca 159
<2l0> 210
<2I1> 256
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(256)
<223> n = A,T,C or G
<400> 210
actccctggc agacaaaggc agaggagaga gctctgttag ttctgtgttg ttgaactgcc 60
actgaatttc tttccacttg gactattaca tgccanttga gggactaatg gaaaaacgta 120
tggggagatt ttanccaatt tangtntgta aatggggaga ctggggcagg cgggagagat 180
ttgcagggtg naaatgggan ggctggtttg ttanatgaac agggacatag gaggtaggca 240
ccaggatgct aaatca ' 256
<210> 211
<211> 264
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(264)
<223> n = A,T,C or G
<400>
211
acattgtttttttgagataaagcattgagagagctctccttaacgtgacacaatggaagg 60
actggaacacatacccacatctttgttctgagggataattttctgataaagtcttgctgt 120
atattcaagcacatatgttatatattattcagttccatgtttatagcctagttaaggaga 180
ggggagatacattcngaaagaggactgaaagaaatactcaagtnggaaaacagaaaaaga 240
aaaaaaggagcaaatgagaagcct ! 264
<210> 212
<211> 328
<212> DNA
<2l3> Homo sapien
<220>
<221> mist feature
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
<222> (1)...(328)
<223> n = A,T,C or G
<400> 212
acccaaaaatccaatgctgaatatttggcttcattattcccanattctttgattgtcaaa 60
ggatttaatgttgtctcagcttgggcacttcagttaggacctaaggatgccagccggcag 120
gtttatatatgcagcaacaatattcaagcgcgacaacaggttattgaacttgcccgccag 180
ttnaatttcattcccattgacttgggatccttatcatcagccagagagattgaaaattta 240
cccctacnactctttactctctgganagggccagtggtggtagctataagcttggccaca 300
tttttttttcctttattcctttgtcaga 328
<210> 213
<211> 250
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(250)
<223> n = A,T,C or G
<400>
213
acttatgagcagagcgacatatccnagtgtagactgaataaaactgaattctctccagtt 60
taaagcattgctcactgaagggatagaagtgactgccaggagggaaagtaagccaaggct 120
cattatgccaaagganatatacatttcaattctccaaacttcttcctcattccaagagtt 180
ttcaatatttgcatgaacctgctgataanccatgttaanaaacaaatatctctctnacct 240
tctcatcggt 250
<210> 214
<2l1> 444
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(444)
<223> n = A,T,C or G
<400>
214
acccagaatccaatgctgaatatttggcttcattattcccagattctttgattgtcaaag 60
gatttaatgttgtctcagcttgggcacttcagttaggacctaaggatgccagccggcagg 120
tttatatatgcagcaacaatattcaagcgcgacaacaggttattgaacttgcccgccagt 180
tgaatttcattcccattgacttgggatccttatcatcagccanagagattgaaaatttac 240
ccctacgactctttactctctggagagggccagtggtggtagctataagcttggccacat 300
ttttttttcctttattcctttgtcagagatgcgattcatccatatgctanaaaccaacag 360
agtgacttttacaaaattcctataganattgtgaataaaaccttacctatagttgccatt 420
actttgctctccctaatatacctc 444
<210> 215
<211> 366
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (l)...(366)
<223> n = A,T,C or G
CA 02397741 2002-07-15
WO 01/51633 PCT/USO1/01574
76
<400> 215
acttatgagcagagcgacatatccaagtgtanactgaataaaactgaattctctccagtt 60
taaagcattgctcactgaagggatagaagtgactgccaggagggaaagtaagccaaggct 120
cattatgccaaagganatatacatttcaattctccaaacttcttcctcattccaagagtt 180
ttcaatatttgcatgaacctgctgataagccatgttgagaaacaaatatctctctgacct 240
tctcatcggtaagcagaggctgtaggcaacatggaccatagcgaanaaaaaacttagtaa 300
tccaagctgttttctacactgtaaccaggtttccaaccaaggtggaaatctcctatactt 360
ggtgcc
366
<210> 216
<211> 260
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (l)...(260)
<223> n = A,T,C or G
<400>
216
ctgtataaacagaactccactgcangagggagggccgggccaggagaatctccgcttgtc 60
caagacaggggcctaaggagggtctccacactgctnntaagggctnttncatttttttat 120
taataaaaagtnnaaaaggcctcttctcaacttttttcccttnggctggaaaatttaaaa 180
atcaaaaatttcctnaagttntcaagctatcatatatactntatcctgaaaaagcaacat 24f
aattcttccttccctccttt 260
<210> 217
<21l> 262
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(262)
<223> n = A,T,C or G
<400>
217
acctacgtgggtaagtttanaaatgttataatttcaggaanaggaacgcatataattgta 60
tcttgcctataattttctattttaataaggaaatagcaaattggggtggggggaatgtag 120
ggcattctacagtttgagcaaaatgcaattaaatgtggaaggacagcactgaaaaatttt 180
atgaataatctgtatgattatatgtctctagagtagatttataattagccacttacccta 240
atatccttcatgcttgtaaagt 262
<210> 218
<211> 205
<212> DNA
<213> Homo sapien
<220>
<221> misc_feature
<222> (1)...(205)
<223> n = A, T, C or G
<400> 218
accaaggtgg tgcattaccg gaantggatc aangacacca tcgtggccaa cccctgagca 60
cccc'tatcaa ctcccttttg tagtaaactt ggaaccttgg aaatgaccag gccaagactc 120
aggcctcccc agttctactg acctttgtcc ttangtntna ngtccagggt tgctaggaaa 180
anaaatcagc agacacaggt gtaaa 205
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
~~ TTENANT LES PAGES 1 A 281
NOTE : Pour les tomes additionels, veuillez contacter 1e Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 2
CONTAINING PAGES 1 TO 281
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME
NOTE POUR LE TOME / VOLUME NOTE: