Note: Descriptions are shown in the official language in which they were submitted.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-1-
CBP86, A Sperm Specific Protein
This application claims priority under 35 U.S.C. ~119(e) to provisional
patent application no. 60/176,887, filed January 19, 2000.
US Government Rights
This invention was made with United States Government support
under Grant No. HD U54 29099, awarded by the National Institutes of Health.
The
United States Government has certain rights in the invention.
Field of the Invention
The present invention is directed to acidic (pI 4.0) 86 kDa isoforms of
a novel, polymorphic, testis-specific protein, designated calcium binding
protein 86
(CBP86). This protein is tyrosine phosphorylated during i~ vitro capacitation
and
binds calcium after being phosphorylated.
Background of the Invention
Fertilization capacity is acquired. by spermatozoa only after residence
in the distinct microenvironments of the uterus o~ oviduct (depending on the
species)
for a finite period of time. The necessary series ~/of changes, termed
capacitation, was
first described independently by Chang and Austin in the early to mid 1950s.
Capacitation involves molecular changes in both the sperm head and tail which
allow
defined physiological endpoints to occur such as motility hyperactivation, a
whiplash-
like sperm tail motion, and regulated acrosomal exocytosis. Hyperactivation is
observed when sperm reach the oocyte and increase their flagellax bend
amplitude and
beat asymmetry which are thought to enhance the ability of sperm to penetrate
the egg
vestments by increasing forward progression and lateral flagellar thrust.
Our understanding of the molecular mechanisms underlying
capacitation and hyperactivation is rudimentary at present but there is
evidence that
Ca2+, cAMP and protein tyrosine phosphorylation are involved. Capacitation can
be
accomplished ih vitro using cauda epididymal or ejaculated sperm incubated in
defined media containing a protein source such as albumin, NaHC03, Caz+ and
energy
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-2-
substrates such as glucose, pyruvate or lactate. Conditions conducive to i~c
vitro
capacitation lead to increased tyrosine phosphorylation of a subset of
proteins in both
mouse and human sperm. The removal of albumin, NaHC03, or Ca2+ from
capacitation media prevents the occurrence of both tyrosine phosphorylation
and
capacitation. Two protein substrates for this capacitation-related
phosphorylation are
members of the A kinase anchoring protein family, AKAP 4 (originally called
AKAP82 or Fscl in mouse) and AKAP3 (originally called AI~AP95T, FSP95 or
AKAP 110), which are components of the fibrous sheath of the sperm tail.
Although little is known about the kinetics of intracellular calcium
during capacitation, a massive influx of Ca2+ occurs during the acrosome
reaction, and
extracellular calcium is required for sperm hyperactivation. If hyperactivated
sperm
are transferred to calcium free media for 30-60 min, none are hyperactive, but
hyperactivation can be restored by addition of 2 mM calcium. Calcium is also
known
to increase flagellar bend amplitude in demembranated sperm. Intracellular
calcium
[Ca2+;"] is increased in hyperactivated sperm in both the head and tail, and
Caz+;n
oscillates with each flagellar bend, indicating a direct relationship between
intracellular calcium and hyperactivation.
The cytosolic level of CAMP increases during capacitation, and
pharmacological stimulants which elevate intracellular cAMP such as the
phosphodiesterase inhibitors, caffeine and pentoxifylline enhance sperm
hyperactivated motility, enhance penetration of cervical mucus, increase tight
binding
to homologous zona pellucida, and increase fertilization. Calcium/calmodulin
is an
activator of both mammalian sperm adenylate cyclase (AC) and cyclic nucleotide
phosphodiesterase, and sperm AC is stimulated by HC03' anions. A soluble
testicular
adenylate cyclase has recently been cloned and shown to be sensitive to
bicarbonate.
Sperm protein tyrosine phosphorylation is accelerated by cAMP agonists, while
anatagonists of PKA inhibit tyrosine phosphorylation and capacitation. These
and
other observations suggest that sperm protein tyrosine phosphorylation and
capacitation are under the regulation of a cAMP/PKA pathway, which is
activated by
elevated cytosolic levels of calcium and HC03' anions. Mammalian sperm contain
all
three subtypes of the guanine nucleotide-binding regulatory proteins G; , and
G
proteins have been localized in particular to the sperm tail where protein
kinase A and
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-3-
C have also been reported. A cyclic nucleotide gated Ca2+ channel in mammalian
sperm plasma membranes has been reported, and N- and R-type Ca2+ channels have
been defined in mouse sperm.
The present invention is directed to targets at the intersection between
the calcium and protein tyrosine kinase signal transduction pathways in human
spermatozoa. In particular, the present invention describes the isolation and
characterization of a sperm calcium binding protein that is also
phosphorylated by
tyrosine kinases.
Definitions
In describing and claiming the invention, the following terminology
will be used in accordance with the definitions set forth below.
As used herein, "nucleic acid," "DNA," and similar terms also include
nucleic acid analogs, i.e. analogs having other than a phosphodiester
backbone. For
example, the so-called "peptide nucleic acids," which are known in the art and
have
peptide bonds instead of phosphodiester bonds in the backbone are considered
within
the scope of the present invention.
The term "peptide" encompasses a sequence of 3 or more amino acids
wherein the amino acids are naturally occurring or synthetic (non-naturally
occurring)
amino acids. Peptide mimetics include peptides having one or more of the
following
modifications:
1. peptides wherein one or more of the peptidyl --C(O)NR-- linkages (bonds)
have been replaced by a non-peptidyl linkage such as a --CH2_carbamate linkage
(--CH20C(O)NR--), a phosphonate linkage, a -CH2_sulfonamide (-CH 2__S(O)2NR--)
linkage, a urea (--NHC(O)NH--) linkage, a --CH2 -secondary amine linkage, or
with an
alkylated peptidyl linkage (--C(O)NR--) wherein R is C1-Cq. alkyl;
2. peptides wherein the N-terminus is derivatized to a --NRRl group, to a
-- NRC(O)R group, to a --NRC(O)OR group, to a --NRS(O)2R group, to a
--NHC(O)NHR group where R and Rl are hydrogen or C 1 _C4 alkyl with the
proviso that
R and Rl axe not both hydrogen;
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
_4-
3. peptides wherein the C terminus is derivatized to --C(O)R2 where R 2 is
selected from the group consisting of C 1 _C4 alkoxy, and --NR3R4 where R3 and
R4 are
independently selected from the group consisting of hydrogen and C 1 _C4
alkyl.
Naturally occurnng amino acid residues in peptides are abbreviated as
recommended by the IUPAC-IUB Biochemical Nomenclature Commission as follows:
Phenylalanine is Phe or F; Leucine is Leu or L; Isoleucine is Ile or I;
Methionine is Met
or M; Norleucine is Nle; Valine is Vat or V; Serine is Ser or S; Proline is
Pro or P;
Threonine is Thr or T; Alanine is Ala or A; Tyrosine is Tyr or Y; Histidine is
His or H;
Glutamine is Gln or Q; Asparagine is Asn or N; Lysine is Lys or K; Aspartic
Acid is Asp
or D; Glutamic Acid is Glu or E; Cysteine is Cys or C; Tryptophan is Trp or W;
Arginine
is Arg or R; Glycine is Gly or G, and X is any amino acid. Other naturally
occurring
amino acids include, by way of example, 4-hydroxyproline, 5-hydroxylysine, and
the
like.
Synthetic or non-naturally occurnng amino acids refer to amino acids
which do not naturally occur ih vivo but which, nevertheless, can be
incorporated into the
peptide structures described herein. The resulting "synthetic peptide" contain
amino
acids other than the 20 naturally occurring, genetically encoded amino acids
at one, two,
or more positions of the peptides. For instance, naphthylalanine can be
substituted for
trytophan to facilitate synthesis. Other synthetic amino acids that can be
substituted into
peptides include L-hydroxypropyl, L-3,4-dihydroxyphenylalanyl, alpha-amino
acids such
as L-alpha-hydroxylysyl and D-alpha-methylalanyl, L-alpha.-methylalanyl, beta.-
amino
acids, and isoquinolyl. D amino acids and non-naturally occurring synthetic
amino acids
can also be incorporated into the peptides. Other derivatives include
replacement of the
naturally occurring side chains of the 20 genetically encoded amino acids (or
any L or
D amino acid) with other side chains.
As used herein, the term "conservative amino acid substitution" are
defined herein as exchanges within one of the following five groups:
I. Small aliphatic, nonpolax or slightly polar residues:
Ala, Ser, Thr, Pro, Gly;
II. Polar, negatively charged residues and their amides:
Asp, Asn, Glu, Gln;
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-5-
III. Polar, positively charged residues:
His, Arg, Lys;
IV. Large, aliphatic, nonpolar residues:
Met Leu, Ile, Val, Cys
V. Large, aromatic residues:
Phe, Tyr, Trp
As used herein, the term "purified" and like terms relate to the isolation
of a molecule or compound in a farm that is substantially free of contaminants
normally associated with the molecule or compound in a native or natural
environment.
As used herein, the term "CBP86 polypeptide" and like terms refers to
polypeptides comprising SEQ ID NO: 2 and biologically active fragments
thereof.
As used herein, the term "biologically active fragments" or "bioactive
fragment" of an CBP86 polypeptide encompasses natural or synthetic portions of
SEQ
ID NO: 2 that are capable of specific binding to at least one of the natural
ligands of
the native CBP86 polypeptide.
"Operably linked" refers to a juxtaposition wherein the components are
configured so as to perform their usual function. Thus, control sequences or
promoters
operably linked to a coding sequence are capable of effecting the expression
of the
coding sequence.
As used herein, the term "pharmaceutically acceptable carrier"
encompasses any of the standard pharmaceutical carriers, such as a phosphate
buffered saline solution, water and emulsions such as an oil/water or
water/oil
emulsion, and various types of wetting agents.
Summary of the Invention
The present invention is directed to the isolation and characterization
of a novel testis and sperm-specific, calcium binding protein, CBP86, that is
expressed post-meiotically and localized in the sperm flagellum. This protein
exhibits
increased tyrosine phosphorylation during ih vitro capacitation and increased
calcium
binding isoforms during capacitation.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-6-
Brief Description of the Drawings
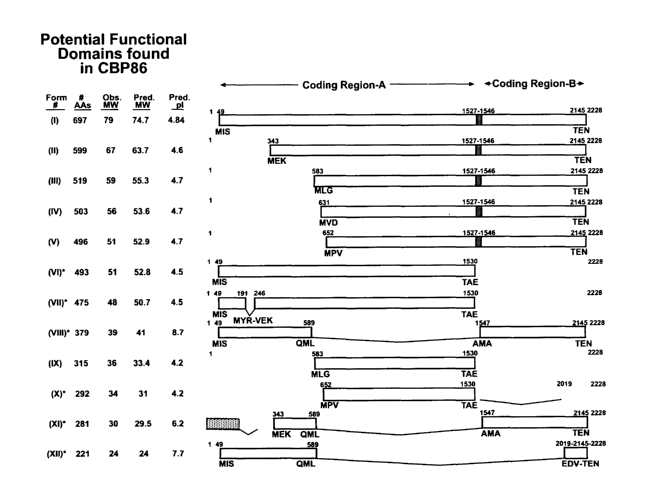
Fig. 1 is schematic representation of the potential translational variants
of CBP86. Twelve predicted CBP86 forms are indicated by Roman numerals (I
through XII, respectively). Forms I-V are predicted through alternative start
sites and
readthrough between amino acids 493 and 499. Splice variants VII, VIII and X-
XII,
indicated by asterisks, were cloned and sequenced from cDNA libraries. Clone
VI
was initially amplified from human testicular adaptor-ligated cDNA and was
verified
by cDNA library cloning. The predicted number of amino acids, pI's and
molecular
weights (MW) as well as the observed MW calculated from reduced and
carboxymethylated sperm peptides, are noted for each form. The coding regions
of
each of the CBP86 variants are shown as blocked regions. The stippled region
of
variant XI indicates a sequence not found in any other CBP86 cDNA sequence.
The
crosshatched region of variants I-V represents the readthrough region. Splice
junctions found in each variant axe numbered and the contiguous amino acid
sequences at the beginning and end of the splice sites are noted below each
junction.
Fig. 2A shows a multiple tissue Northern Blot, wherein CBP86 cDNA
corresponding to CR-A was radiolabeled with P3z and hybridized to 2 ug poly-
(A)+
mRNAs, revealing 2.4 and 1.4 Kb messages only in testicular RNA. Size of
molecular weight markers is indicated at left, lanes 1-8 contain poly-(A)+
mRNA
isolated from spleen, thymus, prostate, testis, ovary, small intestine, colon
and
leucocyte, respectively. The lower panel of Fig. 2A shows the identical blot
probed
with (3-actin cDNA as a positive control.
Fig. 2B shows a dot-blot tissue-mRNA Northern probed with P3z-
labeled CBP86 cDNA revealed hybridization only in testis (Dl). The normalized
(100-500 ng) poly-(A)+ mRNAs present on the grid were isolated from various
tissue
sources: A 1-8 represents whole brain, amygdala, caudate nucleus, cerebellum,
cerebral cortex, frontal lobe, hippocampus, medulla oblongata, respectively; B
1-7
represents occipitallobe, putamen, substantia nigra, temporal lobe, thalamus,
subthalmic nucleus, spinal chord, respectively; C 1-8 represents heart, aorta,
skeletal
muscle, colon, bladder, uterus, prostate, stomach, respectively; D 1-8
represents testis,
ovary, pancreas, pituitary gland, adrenal gland, thyroid gland, salivary
gland,
mammary gland, respectively; E 1-8 represents kidney, liver, small intestine,
spleen,
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-7-
thymus, peripheral leukocyte, lymph node, bone marrow, respectively; F 1-4
represents
appendix, lung, trachea, placenta, respectively; G 1-7 represents (All Fetal)
brain,
heart, kidney, liver, spleen, thymus, lung, respectively; and H 1-8 represents
100 ng
total yeast RNA, 100 ng yeast tRNA, 100 ng E. coli rRNA, 100 ng E. coli DNA,
100
ng poly r(A), 100 ng Cot 1 human DNA, 100 ng human DNA, 500 ng human DNA,
respectively.
Detailed Description of the Invention
Almost 50 years have elapsed since the independent discoveries of
capacitation by Chang and Austin, but molecular mechanisms to explain this
process
are not yet fully understood. Studies of CBP86 have now provided an added
dimension to the understanding of capacitation related molecular events in the
flagellum. The observations that a new calcium binding protein (CBP86) exists
in
the sperm tail throughout the entire length of the principle piece in
association with
the fibrous sheath adds another possible molecular component to the calcium
signaling pathway active during hyperactivation. CBP86 may be involved in
calcium
sequestration and episodic release and thus may play a direct role in
flagellar motility.
As reported herein the 86 kDa ~SCa binding isoforms of CBP86 are
composed of subunits. These isoforms increase during ih vitro capacitation,
and
dephosphorylation abolishes both calcium binding capacity and assembly of the
86
kDa isoforms. These observations point to a role for capacitation dependent
phosphorylation in calcium signaling. Although the time course for
capacitation ifz
vitro differs from species to species a median time for ih vitro capacitation
of human
sperm is three hours. This time course is similar to that observed for CBP86
phosphorylation and assembly, leading to the hypothesis that oligomerization
of
CBP86 into its calcium binding form is a capacitation related event requiring
tyrosine
phosphorylation and that the time required for this process may underlie the
temporal
requirements for capacitation and hyperactivation.
Furthermore, Northern and dot blot analysis of an extensive panel of
tissues place CBP86 in the category of a sperm and testis-specific protein.
Immunohistochemical analysis of human testis indicated that the CBP86 gene
first
becomes translated following meiosis and that the protein is present only in
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
_g_
spermatids, moving to the flagellum during the final stages of
spermatogenesis. In
contrast to calmodulin, which is considered to sequester sperm Ca2+ but is
present in
many somatic cell types, the tissue specificity of CBP86 may provide a unique
opportunity to target calcium sequestration and signaling in sperm. In
addition, the
post-meiotic pattern of protein localization and the tissue specificity of
gene
expression indicate that CBP86 should be given consideration as a candidate
for
targeted male contraception because of the possibility that antagonists of
CBP86
might act selectively during spermiogenesis.
Accordingly, the present invention is directed to therapeutic and
diagnostic methods and compositions based on CBP86 proteins and nucleic acids.
Antagonists of CBP86 function can be used to interfere with the capacitation
of
vertebrate sperm, and thus used as contraceptive agents. Furthermore,
antibodies
against the CBP86 protein can be used for the diagnosis of conditions or
diseases
characterized by expression or overexpression of CBP86, or in assays to
monitor
patients being treated with CBP86 agonists, antagonists or inhibitors.
In one embodiment, the present invention is directed to a purified
polypeptide comprising the amino acid sequence of SEQ ID NO: 2, or an amino
acid
sequence that differs from SEQ ID NO: 2 by one or more conservative amino acid
substitutions. More preferably, the purified polypeptide comprises an amino
acid
sequence that differs from SEQ ID NO: 2 by 20 or less conservative amino acid
substitutions, and more preferably by 10 or less conservative amino acid
substitutions.
Alternatively, the polypeptide may comprise an amino acid sequence that
differs from
SEQ ID NO: 2 by 1 to 5 alterations, wherein the alterations are independently
selected
from a single amino acid deletion, insertion or substitution.
Another embodiment of the present invention encompasses truncated
versions of the polypeptide of SEQ ID NO: 2, wherein the polypeptide is
translated
from one of several alternative start codons located downstream from the first
start
codon, at positions 343, 583, 631 and 652, respectively. For example, the
polypeptide
may comprise the sequence of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ
ID NO: 5 or SEQ ID NO: 6, or an amino acid sequence that differs from SEQ ID
NO:
2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5 or SEQ ID NO: 6 by one or more
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-9-
conservative amino acid substitutions, more preferably, by 10 or less
conservative
amino acid substitutions.
The present invention also comprises the various alternative spliced
forms of the CBP86 proteins as shown in Fig. 1. In particular, the present
invention is
directed to a polypeptide comprising the sequence of SEQ ID NO: 15 or an amino
acid sequence that differs from SEQ ID NO: 15 by one or more conservative
amino
acid substitutions. In another embodiment, the polypeptide comprises the
sequence of
SEQ ID NO: 16 or an amino acid sequence that differs from SEQ ID NO: 16 by one
or more conservative amino acid substitutions.
The CBP86 proteins also contain a number of binding motifs. Three of
the 6 known motifs of catapase, which are the signatures for the P-type ATPase
canon
transport superfamily are found in CR-A of CBP86:
LKTLLEGISR (SEQ ID NO: 7)
VSDNTGQEESGENSV (SEQ ID NO: 8)
SGTSVKSSSGP (SEQ ID NO: 9)
The N-terminus of CR-A contains 3 of 4 possible motifs that
constitute SH3 domains:
NQFAAAYFQEL (SEQ ID NO: 10)
VEKWSEGTTP (SEQ ID NO: 11)
KTTQFPSVYAVPG (SEQ ID NO: 12)
Further computer analysis found two (5 and 6) of the possible eight
progesterone receptor motifs:
PSSPPPTAVSPEFAYVP (SEQ ID NO: 13)
AEATALLSDTSLKGQPE (SEQ ID NO: 14)
In one embodiment, the present invention provides methods of
screening for agents, small molecules, or proteins that interact with CBP86.
The
invention encompasses both in vivo and i~. vat~o assays to screen small
molecules,
compounds, recombinant proteins, peptides, nucleic acids, antibodies etc.
which bind
to or modulate the activity of CBP86 and are thus useful as therapeutics or
diagnostic
markers for fertility.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-10-
In one embodiment the CBP86 polypeptide, or bioactive fragments
thereof, is used to isolate ligands that bind to the CBP86 polypeptide under
physiological conditions. The method comprises the steps of contacting the
CBP86
polypeptide with a mixture of compounds under physiological conditions,
removing
unbound and non-specifically bound material, and isolating the compounds that
remain bound to the CBP86 polypeptides. Typically, the CBP86 polypeptide will
be
bound to a solid support using standard techniques to allow rapid screening
compounds. The solid support can be selected from any surface that has been
used to
immobilize biological compounds and includes but is not limited to
polystyrene,
agarose, silica or nitrocellulose. In one embodiment the solid surface
comprises
functionalized silica or agarose beads. Screening for such compounds can be
accomplished using libraries of pharmaceutical agents and standard techniques
known
to the skilled practitioner.
The present invention also encompasses nucleic acid sequences that
encode the CBP86 polypeptide, and bioactive fragments and derivatives thereof.
In
particular the present invention is directed to nucleic acid sequences
comprising the
sequence of SEQ ID NO: 1 or fragments thereof. In one embodiment, purified
nucleic
acids comprising at least 8 contiguous nucleotides (i.e., a hybridizable
portion) that
are identical to any 8 contiguous nucleotides of SEQ ID NO: 1 are provided. In
other
embodiments, the nucleic acids comprises at least 25 (contiguous) nucleotides,
50
nucleotides, 100 nucleotides, 200 nucleotides, or 500 nucleotides of SEQ ID
NO: 1.
In one embodiment the nucleic acid sequence comprises a 350 by nucleic acid
sequence that is identical to a contiguous 350 by sequence of SEQ ID NO: 1. In
another embodiment the nucleic acid sequence comprises the sequence of SEQ ID
NO: 25 or SEQ ID NO: 26.
The present invention also includes nucleic acids that hybridize (under
conditions defined herein) to all or a portion of the nucleotide sequence
represented by
SEQ ID NO:1 or its complement. The hybridizing portion of the hybridizing
nucleic
acids is typically at least 15 (e.g., 20, 25, 30, or 50) nucleotides in
length. Hybridizing
nucleic acids of the type described herein can be used, for example, as a
cloning
probe, a primer (e.g., a PCR primer), or a diagnostic probe. It is anticipated
that the
DNA sequence of SEQ ID NO: 1, or fragments thereof can be used as probes to
detect
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-11-
additional members of the CBP86 families and to detect homologous genes from
other vertebrate species.
Nucleic acid duplex or hybrid stability is expressed as the melting
temperature or Tm, which is the temperature at which a nucleic acid duplex
dissociates into its component single stranded DNAs. This melting temperature
is
used to define the required stringency conditions. Typically a 1 % mismatch
results in
a 1 °C decrease in the Tm, and the temperature of the final wash in the
hybridization
reaction is reduced accordingly (for example, if two sequences having > 95%
identity,
the final wash temperature is decreased from the Tm by 5°C). In
practice, the change
in Tm can be between 0.5°C and 1.5°C per 1% mismatch.
The present invention is directed to the nucleic acid sequence of SEQ
ID NO: 1 and nucleic acid sequences that hybridize to that sequence (or
fragments
thereof) under stringent or highly stringent conditions. In accordance with
the present
invention highly stringent conditions are defined as conducting the
hybridization and
wash conditions at no lower than -5°C Tm. Stringent conditions are
defined as
involve hybridizing at 68°C in Sx SSC/Sx Denhardt's solution/1.0% SDS,
and washing
in 0.2x SSC/0.1% SDS at 68°C . Moderately stringent conditions include
hybridizing
at 68°C in Sx SSC/Sx Denhardt's solution/1.0% SDS and washing in 3x
SSC/0.1%
SDS at 42°C. Additional guidance regarding such conditions is readily
available in
the art, for example, by Sambrook et al., 1989, Molecular Cloning, A
Laboratory
Manual, Cold Spring Harbor Press, N.Y.; and Ausubel et al. (eds.), 1995,
Current
Protocols in Molecular Biology, (John Wiley & Sons, N.Y.) at Unit 2.10.
In another embodiment of the present invention, nucleic acid sequences
encoding the CBP86 receptor can be inserted into expression vectors and used
to
transfect cells to enhance the expression of those receptors on the target
cells. In
accordance with one embodiment, nucleic acid sequences encoding CBP86, or a
fragment or a derivative thereof, are inserted into a eukaryotic expression
vector in a
manner that operably links the gene sequences to the appropriate regulatory
sequences, and CBP86 is expressed in a eukaryotic host cell. Suitable
eukaryotic host
cells and vectors are known to those skilled in the art. In particular,
nucleic acid
sequences encoding CBP86 may be added to a cell or cells i~c vitro or in vivo
using
delivery mechanisms such as liposomes, viral based vectors, or microinjection.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-12-
Accordingly, one aspect of the present invention is directed to transgenic
cell lines
that contain recombinant genes that express CBP86.
Another embodiment of the present invention comprises antibodies
that are generated against CBP86. These antibodies can be formulated with
standard
carriers and optionally labeled to prepare therapeutic or diagnostic
compositions.
Antibodies to CBP86 may be generated using methods that are well known in the
art.
Such antibodies may include, but are not limited to, polyclonal, monoclonal,
chimeric
(i.e "humanized" antibodies), single chain (recombinant), Fab fragments, and
fragments produced by a Fab expression library. These antibodies can be used
as
diagnostic agents for the diagnosis of conditions or diseases characterized by
expression or overexpression of CBP86, or in assays to monitor patients being
treated
with CBP86 receptor agonists, antagonists or inhibitors. The antibodies useful
for
diagnostic purposes may be prepared in the same manner as those described
above for
therapeutics. The antibodies may be used with or without modification, and may
be
labeled by joining them, either covalently or non-covalently, with a reporter
molecule.
In accordance with one embodiment an antibody is provided that
specifically binds to the protein of SEQ ID NO: 2. More~particularly, the
antibody
binds to the amino acid sequence of SEQ ID NO: 15. Alternatively, the antibody
specifically binds to the amino acid sequence of SEQ ID NO: 16. In one
preferred
embodiment the antibody is a monoclonal antibody.
The invention also encompasses antibodies, including anti-idiotypic
antibodies, antagonists and agonists, as well as compounds or nucleotide
constructs
that inhibit expression of the CBP86 gene (transcription factor inhibitors,
antisense
and ribozyme molecules, or gene or regulatory sequence replacement
constructs), or
promote expression of CBP86 (e.g., expression constructs in which CBP86 coding
sequences are operatively associated with expression control elements such as
promoters, promoter/enhancers, etc. ).
The present invention also encompasses compositions that can be
placed in contact with sperm cells to inhibit the function of the CBP86
protein (i.e.
either by inhibiting the expression of the CBP86 protein or by interfering
with the
protein's function). In particular the compositions may comprise peptide
fragments of
CBP86, or analogs thereof that are taken up by the sperm cells and compete for
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-13-
binding with CBP86's natural ligands. Such inhibitory peptides can be modified
to
include fatty acid side chains to assist the peptides in penetrating the sperm
cell
membrane. Compositions comprising a CBP86 inhibitory agent can be used to
modulate fertility of an individual, and in one embodiment, the inhibitory
agents
function as a male contraceptive pharmaceutical. In accordance with one
embodiment
a composition is provided that comprises an eight to fifteen amino acid
sequence that
is identical to an eight to fifteen consecutive amino acid sequence of SEQ ID
NO: 2
and a pharmaceutically acceptable carrier.
The CBP86 protein contains a number of protein binding domains,
including three SH3 domains located at the 5' end of CR-A of the CBP86 gene.
In
addition, the 3' end of CR-A and the 5' end of the CR-B are relatively proline
rich.
Both the SH3 domains and the proline-rich stretches, referred to herein as the
putative
dimerization domains, provide CBP86 with potential sites for interaction with
other
flagellar proteins (such as the AI~APs). In accordance with one embodiment of
the
present invention the CBP86 polypeptide is used in an assay to screen for
compounds
that interfere with CBP's ability to bind to AKAPS. The assay comprises
combining
CRP with an AKAP in the presence of one or more potential inhibitors to
monitor the
ability of the potential inhibitor to prevent AKAP binding to the CBP
polypeptide
and/or the ability of the potential inhibitor to disrupt AKAP/CBP complexes.
Inhibitor of such binding interactions have utility as contraceptive agents
due to their
ability to prevent capacitation of sperm cells.
The CBP86 polypeptide and its splice derivatives can also be used in
accordance with the present invention as a marker for determining the extent
of
capacitation of sperm cells present in a sperm sample. The assay is based on
the
premise that phosphorylation and formation of the 86 kDa isoform of CBP86 is
correlated with capacitation of the sperm cells. Therefore measuring the
phosphorylation or oligomerization of CBP86 serves as a marker of
capacitation.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-14-
Example 1
Isolation of the CBP86 Protein
Materials and Methods
Solubilization and electrophoresis of human spermatozoal proteins
Preparation of semen specimens and solubilization of sperm proteins were
performed as previously described (Naaby-Hansen et al, 1997a.) For analytical
two-
dimensional electrophoresis the detergent/urea extracted proteins were
separated by
isoelectric focusing (IEF) in acrylamide tube gels prior to second dimensional
gel
electrophoresis (SDS-PAGE), which was performed in a Protean II xi Multi-Cell
apparatus (Bio-Rad, Richmond, CA) or on large format (23 x 23 cm) gels
(Investigator 2-D Electrophoresis System, ESA) which were also employed for
preparative 2D gel electrophoresis. Electrotransfer to nitrocellulose
membranes and
subsequent visualizing of the proteins by gold staining was accomplished as
previously described (Naaby-Hansen et al, 1997) while electrotransfer to PVDF
membranes (0.2 mrn pore size, Pierce) was carried out as described by Henzel
et al.
(1993) using the transfer buffer composition of Matsudaira (1987) (10 mM 3-
[cyclohexylamino]-1-propanesulfonic acid, 10% methanol, pH 11). The
immobilized
proteins were visualized by staining in a solution containing 0.1% Commassie
8250,
40% methanol and 0.1 % acetic acid for one minute, followed by destaining in a
solution of 10% acetic acid and 50% methanol for 3 x 3 minutes.
In vitro capacitatioh
Motile sperm were harvested by the swim up method of Bronson and Fusi
(1990). A control sample was removed and snap frozen (-70 C), while the
remaining
sperm were resuspended in one of the following media: Dulbecco's PBS, BWW,
BWW plus 3 mM b-cyclodextran (Sigma), BWW plus b-cyclodextran and 100 ~,~M
progesterone, human tubal fluid [HTF] (Irvine) plus HSA (30 mg/ml), HTF plus
HSA plus 2, 20 or 100 ~,~.M progesterone, HTF plus HSA plus 100 ~,~M
progesterone plus either 100, 200 or 400 ~,~,M of genestein or daidzein.
(Akiyama et
al., 1987). Capacitation was achieved by incubating the samples at 37°C
in 5% COZ
with sperm removed at various timepoints and isolated by centrifugation.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-15-
Detection of calcium binding proteins
Calcium binding proteins were demonstrated using a 45Ca overlay assay
modified from that described by Maruyama et al. (1984). The experiment was
replicated 4 times. In brief, the 2-D gel separated proteins were transferred
to PVDF
membranes (Jethmalani et a1.,1994), and the membranes were washed 3 x 20 min
in a
washing buffer (10 mM imidazole HCI, 60 mM KCl and 5 mM MgCl2, pH 6.8) and
incubated with 2 mCi/ml of 45CaC12 in washing buffer for 30 min at room
temperature. The membranes were subsequently rinsed for 2 min in distilled HZO
followed by 30 sec rinsing in 50% ethanol and were air dried on filterpaper
for 15-20
min. The membranes were then dried by hot air from a hairdryer and exposed on
phospho-imaging screens (Molecular Dynamics) for 10 days. The use of PVDF,
shortening of the final wash steps, and employment of phospho-imaging
detection
increased the signal to noise ratio compared to that achieved with the
procedure
originally proposed by Maruyama et al (1984). Some of the PVDF membranes were
subsequently stained with Commassie to localize the calcium binding proteins
within
the total 2-D protein pattern, while other membranes were used for western
blot
analysis as described below. Computerized pattern analysis and densitometry of
the
autoradiograms and the stained membranes were performed employing 2D Analyzer
software (BioImage 2000).
Generation of antiserum against gel purified CBP86
The 86 kDa Coomassie-stained protein spot was cored from three 1.5 mm
thick 2-D SDS-PAGE gels of human sperm extracts. The gel cylinders were minced
into a slurry in 1 ml of PBS and emulsified with an equal volume of complete
Freunds
adjuvant. Six hundred u1 of this emulsion was intradermally injected into a
New
Zealand white rabbit, followed by two monthly subcutaneous booster injections
of
similarly-prepared antigen with incomplete Freunds adjuvant. Serum was
collected 10
days after each booster injection. '
Dephosphorylation of sperm proteins
To examine the relationship between phosphorylation and calcium binding
capacity of the 86 kDa CBP86 form, sperm from 4 individuals were capacitated
for 5
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-16-
hr in HTF plus albumin, and the sperm were extracted in NP40/urea and the
extracts
pooled. The lysate was divided and one aliquot was treated with 2U/ml calf
intestinal
alkaline phosphatase (Boehringer Manheim) for %z hour at 37°C while the
other
aliquot remained untreated.
Microsequencing of the 86 kDa calcium binding tyrosine phosphorylated protein
The 86 kDa Coomassie stained protein spot was cored from a 1.5 mm thick 2D
SDS-polyacrylamide gel and fragmented into smaller pieces. The protein was
destained in methanol, reduced in 10 mM dithiothreitol and alkylated in 50 mM
iodoacetamide in 0.1 M ammonium bicarbonate. After removing the reagents, the
gel
pieces were incubated with 12.5 ng/ml trypsin in 50 mM ammonium bicarbonate
overnight at 37 °C. Peptides were extracted from the gel pieces in 50 %
acetonitrile
in 5% formic acid and microsequenced by tandem mass spectrometry and by Edman
degradation at the Biomolecular Research Facility of the University of
Virginia. Five
peptide sequences were obtained by mass spectrometry:
LVVPYGLK (SEQ ID NO: 17)
TLLEGISR (SEQ ID NO: 18)
TNPSNINQFAAAYFQELTMYR (SEQ ID NO: 19)
KYSSVYMEAEATALLSDTSL (SEQ ID NO: 20)
GQPEVPAQLLDAEGAI (SEQ ID NO: 21)
Differentiation of leucine and isoleucine in the sequences were determined by
Edman
sequencing of HPLC isolated peptides.
Cloning, sequencing and analysis of cDNAs
A degenerate deoxyinosine containing sense primer (5'- GGI-CAG-CCI-
GAG-GTI-CCI-GCI- CAA/G-C/TT - 3') (SEQ ID NO: 22) was designed from
peptide number 5 (GQPEVPAQL; SEQ ID NO: 23) and obtained from GIBCO BRL
(Life Technologies, CA). Using this forward primer and an adapter primer (AP 1
), a
3'-RACE (rapid amplification of cDNA ends) PCR was performed with 0.25 ng of
human testicular Marathon ready cDNA (CLONTECH, CA) in a 25 ~,1 assay system
for 40 cycles. Thermal cycling was done in a MJ Research (Watertown, MA)
thermal
cycler (PTC-200 DNA engine) using a program of one 3 min.cycle at 94 °C
followed
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-17-
by 40 cycles of denaturation, annealing and elongation at 94 °C for 30
sec, 60 °C for 1
min and 68 °C for 2 min. PCR products were separated on a 1.7% NuSieve
(FMC,
ME) agarose gel and a unique 1.0 kb DNA fragment was reamplified, cloned into
the
pCR 2.1-TOPO vector (Invitrogen, CA), and sequenced on a Perkin-Elmer Applied
Biosystems DNA sequences using BigDyeO fluoresence dye terminator chemistry
with Taq DNA polymerase (Perkin-Elmer, NJ). The 3'clone contained 1001 by
including a portion of CR-A and all of CR-B. The 5' end of the cDNA was also
amplified by 5' RACE PCR from the same template using an adapter primer (AP 1
)
and an antisense 3' gene-specific primer (5'- TTA-TTC-AGC-TGT-TGA- TTC-CCC-
TTC-TGG-TTC-AAT-TTC-TGG -3') (SEQ ID NO: 24) which was 263 by
downstream from the 5' end of the 1.0 kb 3' clone. A product of 1530 by was
obtained and cloned into the pCR 2.1-TOPO vector. The 5'clone revealed a 48 by
untranslated region and an open reading frame of 1479 bp. The cDNA clones were
sequenced in both directions using vector-derived and insert-specific primers.
The
nucleotide and amino acid sequence data were assembled.
Cloning of alternatively spliced forms of the transcript was performed by
probing a 5'-
Stretch A2~DR2 human testis cDNA library (Clontech, CA) according to
manufacturers instructions with the full-length 32P-labeled cDNA obtained
through the
RACE protocol. Purified tertiary plaques were converted to their plasmid
forms,
plated, grown in LB broth and the plasmid DNA isolated by Qiagen Mini-Kit
columns
before sequencing with both plasmid and gene-specific primers.
Northern and dot blot analyses
A Northern blot containing 2 mg of poly(A)''- RNA from eight selected human
tissues and a normalized RNA dot blot containing 89 to 514 ng of mRNA from 50
different human tissues were obtained from Clontech. The Northern blot was
probed
with a 32P-labeled 1479 by DNA corresponding to by 49-1527 of CR-A. Probes
were
prepared by random oligonucleotide prime labeling (Feinberg and Vogelstein,
1983).
Hybridization was performed in ExpressHyb solution (Clontech) at 68 °C
for 1 h
followed by three washes in 2x SSC, 0.05% SDS at room temperature and two
washes
in O.lx SSC, 0.1% SDS for 20 min at 50 °C. The blot was exposed to X-
ray film at -
70 °C for 60 h with two intensifying screens. The dot blot was probed
with the same
3zP-labeled cDNA corresponding to coding region A. The blot was hybridized in
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-18-
ExpressHyb solution (Clontech) containing salmon sperm DNA and human placental
Cot-1 DNA overnight at 65 °C. The blot was then washed three times in
2x SSC, 1%
SDS at 65 °C followed by two additional washes in O.lx SSC, 0.5% SDS
at 55 °C
before exposing the filter to X-Ray film for 18 h at -70 °C with two
intensifying
screens.
Reduction and carboxymethylation of human sperm proteins.
Washed sperm samples (Naaby-Hansen et al, 1997) were extracted in 8 M urea
in 0.36 M Tris-HCI, pH 8.6 containing 2% NP40 for 1h at 4 °C. The
supernatant was
precipitated, washed twice in 80% ethanol (final) and reconstituted in the
urea buffer
with no NP40. An aliquot of 1.5 mg protein was incubated in ~ M urea, 0.2%
EDTA,
119 mM mercaptoethanol in 0.36 M Tris-HCI, pH ~.6 at room temp for 4h under
nitrag~en in screw-cap tuhes (Crestfield et al, 1963). The mix was then
treated with a
freshly prepared solution of iodoacetic acid (0.111 M final concentration) in
1 N
NaOH for 15 min at room temp in the dark. After the reaction, the
carboxymethylated
proteins were washed in ethanol and used for Western analyses.
Expression and purification of the recombinant protein and antibody production
The cDNA encoding CR-A of CBP86 was amplified by polyinerase chain
reaction from human testicular Marathon ready cDNA (Clontech). Primers were
designed to create a NcoI site at the 5' end and a Not I site at the 3' end of
the
polymerase chain reaction product. The amplified cDNA was cloned into the NcoI
-
Not I sites of the pET28b expression vector (Novagen) and Escherichia coli
strain
NovaBlue(DE3) was transformed with the plasmid construct. The resulting
construct
appended six residues of histidine tag on the C-terminus of the protein. The
expression plasmid construct was sequenced at the 5'and 3' ends to verify the
reading
frame of the construct.
A single positive colony was inoculated in 1 liter of LB broth with 30 mglml
kanamycin and grown at 37 °C until the A6oo reached 0.6. Then
recombinant protein
expression was induced by addition of 1.0 mM IPTG (isopropyl-1-thio-b-D-
galactopyranoside), and growth was continued for another 3.0 h. The cells were
pelleted, resuspended in lx binding buffer (20 mM Tris-HCI, pH 7.9, 0.5 M
NaCI, 5
mM immidazole) containing 0.1% NP40 (Sigma) and 0.1 mg/ml lysozyme on ice for
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-19-
30 min, and sonicated briefly. The insoluble pellet resulting from
centrifugation at
15000 x g for 15 min was dissolved in 6 M urea in lx binding buffer for 1 h on
ice.
After recentrifugation at 15000 x g for 15 min the urea soluble fraction was
loaded
onto a Niz+-activated His-Binding resin column (Novagen) following
manufacturers
protocol, and the recombinant protein was eluted with 300 mM immidazole in lx
binding buffer containing 6 M urea. The affinity purified recombinant protein
was
used for immunization of female Lewis rats (200 ug/rat) in Freunds complete
adjuvant. Animals were boosted twice at an interval of 14 days with 200 ~.g of
recombinant protein in incomplete Freunds adjuvant and serum was collected 7
days
after each boost.
Immuno-blotting
Western blotting was performed employing a 1 : 3500 dilution of the rabbit
antiserum raised against gel purified CBP86 antigen and a 1 : 2500 dilution of
the rat
antiserum to rCBP86. Sperm proteins phosphorylated on tyrosine residues were
identified by immunoblotting with horseradish peroxidase-conjugated anti-
phosphotyrosine monoclonal antibody RC-20 (Transduction Laboratories) at a
1:2500
dilution in 10 mM Tris (pH 7.5), 0.1 M NaCI, and 0.05% Tween 20 for 20 min at
37°C (Ruff Jamisson et al, 1993)
Diagonal Gels
Human sperm cells, purified by swim-up, were solubilized for 20 min at
22°C
in Laemmli sample buffer (600 x 106 cells/ml), lacking beta-mercaptoethanol
and
containing 2 mM PMSF and 5 mM EDTA to inhibit protease activity. The
supernatant was heated and 50 plane were loaded on SDS-PAGE gradient gels (5-
12%) with a 5% stacking gel. Afterwards the gel was cut into strips (lanes)
and some
strips were incubated for 45 min at 37°C in reducing buffer (0.5% (w/v)
DTT, 0.1%
(w/v) SDS, 125 mM Tris, pH 6.8). Reduced and unreduced Gel-strips were then
laid
horizontally on top of 7.5% SDS-PAGE gels and proteins were run out. Proteins
were
transferred to nitrocellulose membranes and probed with anti-rec-CBP86 as
above.
Localization of CBP86 in the seminiferous epithelium of human testis
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-20-
Testes were obtained from three patients undergoing elective orchiectomies.
Testes were sliced once with a razor blade and immersed in neutral buffered
formalin
(4%) solution (Sigma) for one hour. The tissue was then minced and placed into
fresh
fixative overnight. The tissue was dehydrated in a graded series of ethanols,
cleared
in xylene, and embedded in paraffin. 2.5 ~,m thick sections were cut, mounted
onto
slides, de-paraffinized, rehydrated and permeabilized with 100% methanol.
Sections
were incubated in blocking solution containing 10% NGS in PBS, incubated with
anti-rCBP86 antiserum or pre-immune serum (1:200) in PBS containing 1% NGS
(PBS-NGS), washed, incubated with FITC-labeled goat anti-rat IgG (1:400;
Jackson
~10 Immunoresearch) in PBS-NGS, washed, and mounted with Slow Fade (Molecular
Probes, Eugene, OR) containing DAPI II counterstain (Vysis, Downers Grove,
IL).
Sections were observed by epifluorescence microscopy using a Zeiss microscope.
Individual blue and green fluorescent images were obtained using a digital
camera
(Hamamatsu) and compiled using Openlab software (Improvision Inc., Boston,
MA).
Indirect Immunofluorescence of Human Sperm
For immunofluorescence studies fresh human sperm were harvested over a
discontinuous 55%/80% Percoll gradient and subsequently washed 3 x with Hams F-
10 media. The sperm were counted using a hemocytometer and diluted to a
concentration of 1 x 106 sperm/ml. A 20 ~,1 aliquot of the sperm suspension
was
added per well (2 x 105 sperm) onto poly-L-lysine coated slides. The slides
were
dried at 40°C and then methanol fixed for 10 min. In some experiments
no fixation
was performed and the sperm were simply air dried onto the slide. After
washing 3 x
5 min in PBS, the slides were frozen at -70°C for 1 week. All
subsequent incubations
were done in a humid chamber. The preparations were blocked in 10% normal goat
serum (NGS) in PBS with 0.05% Tween-20 (PBS-tw) for 30 min. The primary
antiserum, either rabbit anti CBP86 antiserum or rat anti recombinant CBP86
and
their pre-immune controls, was diluted 400-fold with 10% NGS in PBS-tw and
were
incubated with the specimen overnight at 4°C. The slides were then
washed 3 x 5 min
in PBS-tw, and the secondary antibody, goat anti-rabbit IgG FITC conjugated
(Jackson ImmunoResearch) or goat anti-rat IgG FITC conjugated (Jackson
ImmunoResearch), were applied at 1:200 dilutions in 10% NGS in PBS-tw for 1
hour
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-21-
at 37°C. The slides were washed 3 x 5 min in PBS-tw, and Slow Fade-
Light Antifade
Kit (Molecular Probes, Inc.) was used to reduce the fading rate of the
fluorescein.
Electron Microscopic Localization
Sperm from four donors were pooled and washed twice by centrifugation at
550 x g in wash buffer, (Ham's F10 Nutrient Mixture (GibcoBRL) with 3%
sucrose).
The washed sperm were resuspended in fixative consisting of 4%
paraformaldehyde
and 0.2% glutaraldehyde in wash buffer for 15 minutes at room temperature.
After
removing fixative by centrifugation and washing 3X With wash buffer, the sperm
were
dehydrated through a graded series of ethanols from 40% to 100%. The cells
were
infiltrated with and embedded in Lowicryl K4M (Electron Microscopy Sciences,
Ft.
Washington, PA) according to the manufacturer's recommendations. The blocks
were
polymerized with UV light for 72 hrs at -20°C and ultrathin sections of
100 nm
thickness were cut.
Non-specific sperm-antibody interactions were blocked by incubating the
sections in undiluted normal goat serum for 15 minutes at room temperature and
washing once with wash buffer. Rat antiserum to rCBP86 and pre-immune serum
were diluted 1:50 in wash buffer with 1 % normal goat serum, 1 % bovine serum
albumin and 0.05% Tween 20. Lowicryl sections were incubated with diluted anti-
rCBP86 or wash buffer alone at 4°C for 16 hours. After washing four
times in wash
buffer, they were incubated for 1.5 hours at room temperature with 5 nm gold-
conjugated secondary antibody, goat anti-rat IgG (Goldmark Biologicals,
Phillipsburg
NJ) diluted 1:35 in wash buffer. The sections were washed with distilled water
and
stained with uranyl acetate before examination with a JEOL 100CX electron
microscope.
In vitro phosphorylation of recombinant CSP86 with c-Src
Baculovirus expressed c-Src was purchased from Upstate Biotechnology, Inc.
(Lake Placid, NY). Recombinant CBP86 was phosphorylated by c-Src in an in
vitro
kinase assay in which 0, 0.8, 0.16, or 0.03 ~.~.g of CBP86 was incubated in
the
presence or absence of 1 unit of c-Src in a 50 ~,~.1 reaction containing 50 mM
HEPES,
pH 7.4, 5 mM MnCl2, 70 nM ATP, 10 Ci [3zP]ATP (6000 Ci/mmol) for 10 min. The
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-22-
reaction was terminated with Laemlli SDS sample buffer and subjected to SDS-
PAGE
and autoradiography.
Results
Identification and characterization of calcium binding proteins (CBPs) in
human spermatozoa.
The 45Ca overlay technique of Maruyama et al (1983) was employed on 2-D
blots of human sperm proteins to identify more than 20 calcium binding protein
spots
(CBPs) in the range of 12.5 kDa to 115 kDa and pIs of 3.8 to 5.3. The relative
intensity of each spot, indicative of the concentration of the binding protein
and/or its
calcium binding capacity, was determined by computer densitometry. More than
90%
of the 45Ca was bound by eleven major CBPs migrating at MWs (kDa)/pI of
86/4.0,
80.4/4.3, 60.5/4.2, 55/4.9, 55/5.25, 26.5/5.2, 25/4.6, 24.7/4.75, 16.5/3.9,
15.8/4.7 and
14.5/3.95 in four replications of the experiment. The 45Ca overlay procedure,
which
was conducted at pH 6.8, did not detect human sperm CBP's in he neutral and
basic
areas (pH 6.2-8.5) of the IEF/PAGE gels. The protein which bound the majority
(60%) of the 45Ca was identified as calmodulin (CaM) based on its
electrophoretic
migration at 16.5 kDa and pI of 3.9.
Three prominent calcium binding proteins migrating at 86(84-88) kDa/4.0
(3.9-4.1), 60.5 kDa/4.2 and 26.5 kDa/5.2 were excised and microsequenced by
CAD
Mass Spectrometry (MS). Five internal peptide sequences and 15 N-terminal
amino
acids were obtained from the 60.5 kDa CBP, which identified the protein as
calreticulin (CRT). A 26.5 kDa CBP was previously identified as a human sperm
surface protein by vectorial labeling with'ZSI and was also detected in human
seminal
fluid. Six peptide sequences obtained by MS and 22 N-terminal amino acids
obtained
by Edman degradation identified the 26.5 kDa CBP as serum amyloid P-component
precursor (SAP). Calcium binding to CaM and SAP resides within EF-hand motifs,
while CRT's calcium binding occurs in repeated, polyacidic C-terminal domains.
The
ability of these proteins to bind 45Ca validated the sensitivity and
specificity of the
~SCa overlay procedure on 2-D gels.
The 86 kDa region of the gel which contained a train of protein spots which
readily bound 45Ca, was designated calcium binding protein 86 [CBP86].
Densitometry of the 45 Ca overlays indicated CBP86 was the second most intense
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-23-
staining region on the 2-D image after calinodulin. MS microsequence data from
5
peptides obtained after tryptic digestion of the excised 86 kDa spot (SEQ ID
NOS:
17-21) did not match any known peptide sequences in any protein or gene
database.
Silver staining showed several isoforms of CBP86 varying slightly in mass and
charge. The acidic isoforms of CBP86 bound more calcium than the more basic
isoforms even though the two differentially charged groups of CBP86 showed
similar
staining with silver nitrate. The acidic CBP86 isoforms appeared to be more
readily
soluble than the basic isoforms because they appeared after only 20 seconds of
solubilization in non-ionic detergent/urea when little if any of the basic 86
kDa
isoforms were solubilized.
CBP86 variants showed shifts in pI after dephosphorylation.
The central, dense portion of the 86 kDa protein cluster was excised from
several preparative 2-D gels and a rabbit antiserum was raised to the gel
purified
proteins. On 2-D immunoblots this antiserum recognized the 86kDa immunogen (
as
well as prominent clusters of protein spots at 27-38, 38-42, 50-56, and 63-72,
each of
which showed charge heterogeneity. Western blots of sperm proteins that had
been
solubilized in the presence of calf intestinal alkaline phosphatase resulted
in the
virtual disappearance of the more acidic 86 kDa immunoreactive isoforms
although
the more basic isofonns remained. In addition, isofonns in the 38-42 and 50-56
kDa
clusters shifted to more basic pIs after phosphatase treatment, indicating
that the
charge heterogeneity of these CBP86 forms is in part due to phosphorylation.
Cloning of CBP86 and its alternatively spliced vaunts.
A degenerate inosine-containing forward primer designed from peptide
number 5, GQPEVPAQL (SEQ ID NO: 23), was employed to amplify a 1.0 kb region
of cDNA by 3'-RACE PCR from human testicular Marathon-Ready cDNA (Clontech,
CA). A 1530 by 5'-cDNA fragment, including a 48 by untranslated region; was
similarly amplified and cloned using standard 5'-RACE PCR with an antisense 3'
reverse primer generated to a sequence 263 by downstream from the 5'-end of
the 1.0
kb 3'-clone. A nucleotide sequence for a composit 2228 by CBP86 cDNA (SEQ ID
NO: 1) was obtained by sequencing the two PCR fragments in both directions.
This
2228 by cDNA was the longest CBP86 cDNA obtained. This cDNA was P3z labeled
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-24-
and employed to screen a human testicular A?~DR2 5'-Stretch cDNA library
(Clontech, CA). Phage isolates were digested with restriction endonuclease,
grouped
according to restriction fragment sizes, and sequenced to yield several cDNAs
also of
2228 by as well as five alternative splice variants, which were submitted to
Genbank
under accession numbers AF295037, AF29038, AF295039, AF329634 and
AF007205.
The five splice variants and the 2228 by CBP86 cDNA are noted by asterisk in
Figure 1 (forms VI-VII and X-XII). Analysis of these sequences led to the
initial
conclusion that the CBP86 sequence was divided into two coding regions, CR-A
and
CR-B. CR-A begins at by 49 and ends at by 1527 (codons 1-494) with a stop
codon
TAA at by 1528-30 serving as an authentic termination codon for CR-A. CR-A
encodes a predicted protein of 493 amino acids with a mass of 52.8 kDa and pI
of 4.5.
Eighteen in frame nucleotides [1531-1546] then separate CR-A from the ATG
start
codon [1547-1550] of CR-B. CR-B [nucleotides 1547-2145] encodes a peptide that
serves as the carboxy terminus on several CBP86 variants. Splice variants were
sequenced containing alternative start codons at bps 49-51 [clones VI, VII,
VII and
XII], bps 343-345 [clone XI], bps 583-585 [clone IX] or bps 652-654 [clone X].
Assuming the stop codon at by 1528-30 was functional, these splice variants
contained deletions of all bf CR-B [clones VI, VII], a small N-terminal region
of CR-
A [clone VII], major portion of CR-A [clones VIII and XI], and a large domain
spanning CR-A and B [clone XII].
To determine if the splice variants resulted in translated products human
sperm
proteins were reduced and carboxymethylated, separated on 1-D gels, and
western
blotted with an antisera raised to recombinant CR-A. Twelve immunoreactive
CBP86
peptides ranging in apparent mass from 79 to 24 were identified. Isoforms at
67, 59
and 51 kDa were most immunoreactive. Importantly, CBP86 proteins were detected
with masses higher than those predicted from coding region A or from any of
the
variants, including deletions of coding regions A or B or variants with splice
junctions
into coding region B. This observation, coupled to the fact that the
intervening
nucleotides between CR-A and CR-B were in-frame, led to the conclusion that a
translational readthrough of the UAA translation terminating signal at the end
of CR-
A occurs in some instances. This translation readthrough accounts for the 12
translated peptides observed ih vivo from the six variant cDNAs.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-25-
CBP86 transcripts are testis specific
A 3aP-labeled cDNA probe corresponding to CR-A was employed for Northern
analysis of mRNA from several tissues (Figure 2A) and a dot blot (Figure 2B)
containing mRNAs from 50 distinct human tissues (Clontech, CA). Interestingly,
two
broad bands of approximately 2.4 and 1.4 Kb were noted in the testicular mRNA
(Figure 2A, lane 4), indicating that several CBP86 messages of different sizes
were
expressed in the human testis, a finding in concert with the cloning and
sequencing of
six cDNAs, including five splice variants noted above. The 2.4 kb transcript
(Fig.
2A) detected in pooled human testicular mRNAs may be accounted for by the
splice
variants of forms I-VI and IX (cDNAs of 2228 bp) or form VII (2173 bp)
assuming
approximately 200 by of untranslated region, while the 1.4 kb transcript may
be
accounted for by forms VIII (1270bp) or X (1088bp). A mRNA of approximately
0.9
to 1.0 kb is predicted for clone XII. Only a faint message of this size was
detected on
overexposed Northern blots, indicating that clone XII mRNAs as well as the 24
l~Da
protein are present in relatively lower abundance than other CBP86 mRNAs and
proteins. Importantly, CBP86 transcripts were expressed in testis (Fig. 2A,
lane 4 and
Fig. 2B, spot Dl) but not in other human tissues.
Motif analysis of splice variants revealed MAP4, RII dimerization, and
extensin domains
Analysis of the amino acid sequences deduced from the six CBP86 variants
sequenced to date, assuming translation readthrough of the stop codon
terminating
CR-A, yields 12 predicted proteins ranging in mass from 24 to 74.7 kDa.
(Figure 1).
Two of the predicted proteins [forms V and VI, Fig 4] are nearly identical in
mass
(52.8 and 52.9 kDa). The masses for the 12 deduced proteins are several kDa
less
than the masses observed for the 12 reduced and blocked CBP86 translated
proteins,
indicating some post-translational modifications) are occurring. Assuming
several
kDa of mass due to post-translational modification, the number and the pattern
of the
apparent masses of CBP86 proteins detected in reduced and carboxymethylated
sperm
protein extracts corresponds to both the number and the masses of the proteins
predicted from the six variants.
All five of the tryptic peptides microsequenced by MS from the original 86
kDa spot excised from the 2-D gel were recovered in the predicted amino acid
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-26-
sequence of CR-A. This finding validated that cDNAs corresponding to the 86
kDa
protein spot originally identified as a Ca2+ binding protein and cored from
preparative
2D gels had been cloned.
Computer analyses to ascertain functional domains of CBP86 revealed that
amino acids 94-493 bore a 25% identity with amino acids 308-717 of human
microtubule associated protein 4 (MAP4). However, the homologous region did
not
involve the microtubule binding domain of MAP4, nor were the 18-mer repeats
characteristic of the microtubule binding domain of MAPS present in CR-A or B.
A
98 amino acid stretch at the N-terminus of CBP86 (residues 10 to 108) bore 30%
identity to the testis-specific sperm protein SP17. Importantly, embedded
within this
domain, sequence' similarity to the regulatory subunit of type II cAMP-
dependent
protein kinase was noted. In particular, Val'°-Leu44 bore a 40%
identity and 57%
similarity to amino acids 7-41 of RIIaa (Newton et al, 1999).
This amino terminal region of RII contains both the RII dimerization domain
and the AKAP binding domain. This region also includes one domain with
similarity
to catatpase and one SH3 motif. Three of the 6 known motifs of catapase, which
are
the signatures for the P-type ATPase canon transport superfamily, were noted
in CR-
A of CBP86. A sub-family of this superfamily are Ca~2-pump ATPases which, like
CBP86, have Ca''-2-binding activity.
Motif analysis was employed to screen a list of proteins with weak overall
homology to CBP86 for those proteins having a known interaction with calcium.
Analysed in this way, the C-terminal third of CR-A revealed similarities with
cation
transporters in overlapping but distinct segments (e.g. a 98 residue region,
G1n36'-
G1y,46s showed 25% identity and 45% conserved homology with the beta-3
regulatory
subunit from the L-type voltage dependent calcium channel [Fugu Yubripes];
while a
67 residue region, Ser42$-G1u,493 revealed 34% identity and 42% conserved
homology
with the Na-Ca+K exchanger [Bos taurus]; and a 57 residue region, G1u33'-
Leu38'
revealed 19% identity and 50% conserved homology to the central domain of the
ion-
channel forming colocin lA toxin [E. coli].
The N-terminus of CR-A contained 3 of 4 possible motifs that constitute SH3
domains. Such Src homology-3 domains serve as sites for intermolecular protein
binding, interacting with proline-rich sequences on a range of signalling and
cytoskeletal proteins. Three PXXP consensus motifs, the cognate sites for SH3
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-27-
interaction, are present in CR-A [aa 396-399, 471-474, and 473-476) and three
were
present in CR-B (aa 211-214, 214-217, 326-329). No extended helical domains or
transmembrane domains were apparent within CR-A or B. However, in view of the
fact that CBP86 undergoes oligomerization (see below), it is noteworthy that
four
elements, each 22 amino acids in length, with similarity to the 7 element
fingerprint
for G-protein-coupled receptors were noted in CR-A at positions 185-206, 204-
225,
295-316, and 455-476. Oligomerization of CBP86 may confer function on these
elements.
Six potential phosphorylation sites for PKC, two phosphorylation sites for
~ CKII as well as four tyrosine residues were present in the C- terminus of
the CBP86
CR-A, suggesting that this region may be regulated by phosphorylation.
Interestingly,
this C-terminal domain, including two catatpase sites, was deleted in clones
VIII, XI
and XII suggesting that full length CBP86 differs in function from these
splice
variants.
Further computer analysis found two (5 and 6) of the possible eight
progesterone receptor motifs. A region covering residues 17 to 102 shared a
20%
identity with helix domains 9, 10, 11 and 12 of the progesterone receptor
binding
domain. 63% of the residues in this region were either identical or
conservative
replacements for the progesterone receptor binding domain. Potential N-linked
glycosylation sites (residues 50, 109 and 237) and two potential O-
glycosylation sites
(residues 258-261; 467-468) were also detected within CR-A of the CBP86
sequence.
The 5' region of CR-B is proline rich and contains two proline triplets, while
overall,
CR-B contains three cysteine residues.
A BLAST search revealed the highest alignment score to be a 40% similarity
(25% identity) between as 225-329 of ORF-B and the proline-rich extensin
glycoprotein found in plant cell walls (Keller and Lamb, 1989). Extensins are
members of the hydroxyproline-rich glycoprotein family (HRGPs) and contain a
characteristic pentapeptide repeat Ser-Pro4 (Chen and Varner, 1985) which in
CR-B
may be represented by a modified Ser-Pro3 domain at as 212-215. Interestingly,
a
similar Ser-Pro3 motif is present in CR-A at position 155-158.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-28_
Western Analyses with Antiserum to Recombinant CBP86 Indicate
Protein Polymorphism and Oligomerization
The cDNA sequence encoding the CBP86 ORF-A was cloned into the
bacterial expression vector pET28b and introduced into NovaBlue(DE3) cells.
The
recombinant protein was purified by immobilized metal affinity chromatography
using Ni2+- Sephaxose. Antiserum against purified rCBP86 was subsequently
raised
in female rats. Like the rabbit antisera to gel purified CBP86 this
monospecific rat
antiserum to rCBP86 also recognized multiple protein spots on 2D western blots
of
human sperm proteins. Immunoreactive species migrated in five major groups
based
on size: 1) 27-38 kDa; 2) 38-42 kDa; 3) 50-56 kDa; 4) 63-72 kDa; 5).81-87 kDa.
The
finding of similar patterns of CBP86 isofonns on 2D gels probed with antisera
to both
the gel purified and recombinant CBP86 confirmed that alternative splice
variants
identified as cDNAs during cloning were expressed at the protein level
resulting in
considerable CBP86 heterogeneity. As a further proof of the specificity of the
rat and
rabbit antisera to CBP86, immunoblots of purified recombinant were probed with
the
two antisera. Both antisera recognized identical MW forms of the recombinant
protein, including high molecular weight complexes >140 kDa, suggestive of
oligomerization of the recombinant proteins.
Relationships between the CBP86 isoforms were revealed on 1-D Western
blots of SDS extracts of human sperm electrophoresed under reducing conditions
where three major immunoreactive forms of CBP86 at 31, 43, and 72 kDa were
noted
along with several less abundant antigenic bands at 51 and 90-102 kDa. Western
blots of non-reduced samples revealed the same abundant 31, 43 and 72 kDa
species
observed on reduced gels along with prominent immunoreactive bands at 64 and
86
kDa as well as less immunoreactive 34 kDa , 45 kDa, 76 kDa and several higher
molecular weight forms. The finding of additional CBP86 forms on nonreduced
gels
indicated the presence of complexes composed of lower molecular weight forms
stabilized by S-S bridges or heterodimerization between LMW CBP86 forms and
unknown partner proteins-interactions which had not been fully dissociated by
the
relatively mild lysis procedure employed for the 2-D gel electrophoresis.
Further
evidence for CBP86 oligomerization was noted when only one major high
molecular
weight [HMW] complex was detected on immunoblots obtained from non-reduced
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-29-
native 1-D PAGE gels of human sperm proteins solubilized in 0.2% DOC and 1
NP40 in the absence of reducing agents.
Immunoblotting diagonal gels, in which Laemmli extracts of sperm were
analysed by 1D SDS-PAGE in a non-reduced first dimension and then reduced in
the
second dimension, revealed disaggregation of several high molecular weight
CBP86
species. The protein running at 86 kDa on nonreducing gels was shown in the
reducing dimension to be comprised of 43 kDa monomers. Similarly, a 76 kDa
protein (migrating above the prominent 72 kDa protein on nonreducing gels)
appeared
to be comprised of 43 kDa and 31 kDa monomers, while the 64 kDa protein was
comprised of 31 kDa monomers. The 43 kDa and 31 kDa subunits did not
dissociate
in the reducing dimension and migrated at the same mass in both reduced and
non-
reduced 1-D gels. From these immunoblots of diagonal gels it may be concluded
that
the two major CBP86 forms running on reduced gels at 31 and 43 kDa participate
in
HMW complexes by both homodimerization and heterodimerization.
The 86 kDa form of CBP86 increases with capacitation.
A comparison of extracts from freshly ejaculated human sperm to sperm
capacitated i~ vitro for 5 hours, revealed a substantial increase in the
amount of the 86
kDa CBP86 isoforms visible following capacitation. In addition, acidic
proteins from
groups 2 and 3 of the CBP86 forms (approximate MW 38-42 and 50-56 kDa) were
also more prominent in capacitated sperm, including the phosphorylated forms
of
group 2 previously noted.
Localization of CBP86 in the seminiferous epithelium
Immunofluorescent localization of CBP86 in the human testes using the
antibody to recombinant CBP86 showed staining of round and elongating
spermatids
in the seminiferous epithelium and testicular spermatozoa within the lumen of
the
tubules, indicative of a post-meiotic pattern of expression of the CBP86 gene.
The
staining patterns suggested a gradual migration of the CBP86 protein from a
diffuse
cytoplasmic localization in round spermatids to the posterior pole of early
spermatids
and then to the flagellum as the tail formed. Testes from three patients
showed
identical localization patterns.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-3 0-
Localization of CBP86 to the principal piece of the mature human sperm
flagellum by immunofluorescence and immuno-electron microscopy
Antibodies raised against rCBP86 recognized the entire length of the principal
piece of ejaculated methanol fixed spermatozoa with an intense signal by
indirect
immunofluorescence microscopy, while both the midpiece and the endpiece
exhibited
much fainter staining patterns. No CBP86 immunofluorescence was noted in the
human sperm head in these non-capacitated sperm. Importantly, no
immunofluorescence staining was observed on live motile sperm, indicating that
CBP86 epitopes were not accessible on the plasma membrane. A similar staining
pattern was achieved with the antiserum raised against gel excised CBP86.
When the distribution of CBP86 in freshly ejaculated human sperm was
examined by electron microscopic immunocytochemical staining, gold particles
were
distributed over the fibrous sheath compartment including the surface of the
longitudinal columns and ribs. Smaller numbers of gold partricles were present
in the
periaxonemal space. CBP86 was not detected in the annular ring or
mitochondrial
sheath and there was no evidence for CBP86 localization within the axoneme in
either
the principal piece or distal to the termination of the outer dense fibers.
CBP86 is tyrosine phosphorylated during in vitro capacitation
Proteins phosphorylated on tyrosine residues during capacitation were
identified on 2-D immuno-blots of freshly ejaculated sperm or from sperm
capacitated
for 3 or 6 hr by staining with the monoclonal anti-phosphotyrosine antibody RC-
20.
After 3 and 6 hours of ih vitro capacitation a significant increase was
observed in
tyrosine phosphorylation of several sperm proteins including AKAP 3 (fibrous
sheath
protein 95) and the 64 and 86 kDa forms of CBP86. Following 3 h capacitation
the
major acidic tyrosine phosphorylated component was a 64 kDa protein. However,
after 6 hrs of capacitation the intensity of the 64 kDa protein had diminished
and the
dominant tyrosine phosphoprotein in the region was the 86 kDa form of CBP86.
In
addition, a 53 kDa protein showed weak tyrosine phosphorylation after 3 hours
of
capacitation, while a further increase in phosphorylation of this CBP86 group
was
observed during the °subsequent 3 hours.
Tyrosine phosphorylation of the 86 kDa form of CBP86 varied with the
composition of the capacitation medium. Tyrosine phosphorylation of the 86kDa
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-31-
form of CBP86 in human tubal fluid plus albumin was higher than that observed
in
Dulbecco's PBS. Interestingly, addition of 100 microM progesterone to the HTF
+
albumin containing capacitation media further enhanced the phosphorylation of
the
most acidic of the CBP86 isoforms. Capacitation-induced tyrosine
phosphorylation
of the 86 kDa CBP86 isoforms was inhibited in a concentration dependent manner
by
treatment with the tyrosine kinase inhibitor, genistein, while similar
concentrations of
the analogue, daidzein had an inhibitory effect on phosphorylation of CBP86
but not
FSP 95 (AKAP 3). As a further proof that CBP86 can serve as a substrate for
tyrosine
kinase, recombinant CBP86 was phosphorylated using an ih vitro kinase assay
which
employed purified baculovirus-expressed c-Src. An increase in tyrosine
phosphorylation of CBP86 was noted as its concentration increased in the
presence of
a constant amount of c-Src, which autophosphorylated as expected. The data
confirmed that the acidic tyrosine phosphorylated 86 kDa forms of CBP86 are
immunoreactive with anti-rCBP86 and at high resolution the increases in the
acidic
immunoreactive forms of CBP86 were evident after capacitation in the presence
of
human tubal fluid and progesterone.
Comparison of 45Ca-binding to normal and alkaline phosphatase treated sperm
proteins revealed that 45Ca readily bound to the acidic (phosphorylated)
isoforms of
CBP86 in untreated sperm extracts, but in extracts treated with alkaline
phosphatase
priox to electrophoresis, the calcium binding capacity of the 86 kDa form of
CBP86
was abolished, while calcium binding to calreticulin was unaffected.
Comparison of
the immunoreactive forms of CBP86 present before and after treatment with
alkaline
phosphatase indicates that alkaline phosphatase reduces the presence of the
acidic
86kDa isoforms. Taken together, these observations indicate that both the 45Ca
binding capacity and the assembly of the 86 kDa form of CBP86 is dependent on
phosphorylation.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-32-
Discussion
CBP86 is a highly polymorphic protein derived from alternatively spliced
messages and possibly translational readthrough
Six cDNAs for CBP86 have been cloned and sequenced including five
alternative splice variants deleting domains of coding regions A or B. On
Northern
blots broad bands of 2.4 and 1.4 by were noted indicating polymorphic CBP86
mRNAs. Immunoblotting of reduced and carboxymethylated sperm protein extracts
on 1-D gels with antisera to recombinant CBP86 revealed 12 translated forms of
CBP86 from 24 to 79 kDa. Isoforms at 67, 59 and 51 kDa were most
immunoreactive. Since four CBP86 peptides were found to be larger than the
predicted molecular weight (~53 kDa) from CR-A or splice variants including
portions of CR-B (forms VI-VIII and X-XII in Fig. 1) a question is raised
about the
efficiency of the UAA translation termination signal at the end of CR-A.
The efficiency of translation termination is sensitive to both the 5' and 3'
sequences adjacent to the stop codon. A strong bias in the relative
frequencies of the
3'- flanking bases is found in highly expressed E. coli genes. For UAA the 3'-
flanking base preference is U»G>A>C while for UGA it is U»A>G>C (Brown et
al., 1990). The affinity of release factor for a stop codon is probably
enhanced by a
specific interaction with the 3' base (Pedersen et al., 199I). Furthermore,
although
, the last 5 amino acids in proteins are generally not important for protein
function, the
last 2 amino acids have a cooperative major influence on translation
termination
(Bjornsson et al., 1996). For the -2 amino acid residue, its acidic/basic as
well as
hydrophobic/hydrophilic properties are important for termination efficiency.
For
UAA, the preferred -1 amino acid is a basic residue, lysine in E. coli genes
(Brown et
al., 1990). In UGA, neutral -2 amino acids provide efficient termination if
they have
a hydrophilic side chain, and inefficient termination if the side chain is
hydrophobic.
Examination of the 3' and 5' sequences adjacent to UAA in CBP86 revealed that
the
3' adjacent nucleotide (which is G instead of a preferred U), and the 5' -2
amino acid
(which is a hydrophobic residue alanine, instead of a hydrophilic residue) and
the 5' -1
amino acid (which is a acidic residue, glutamic acid instead of a preferred
basic
residue like lysine) may not favor efficient termination of translation in CR-
A of
CBP86.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-33-
Accordingly, one explanation for the presence of CBP86 peptides larger than
those predicted from CR-A or the splice variants (e.g., forms. VI, VII, VIII,
X, XI &
XII) posits translational readthrough of the UAA termination signal encoded by
nucleotides 1528-30. Although stop codon suppression in mammalian cells has
been
well documented in the translation of different sized proteins from viral
genomes such
as HIV and MLV (Hatfield et al, 1992) and in the Drosophila kelch gene
(Robinson
and Cooley, 1997), CBP86 appears to be the first example of translational
readthrough of a normal human gene. Previously, translational readthrough was
noted
in fibroblasts of aspartylglucosaminuria patients with a mutation that creates
a
premature stop codon in the second exon of the aspartylglucosaminidase [AGA]
gene.
In this case limited amounts of normally sized AGA was detected.
Identification of a
normal, endogenous human gene that exhibits translational readthrough may lead
to
better understanding of both spermiogenesis and HIV susceptibility.
Relationships between different forms of CBP86
Immunoblots of gels run under various conditions revealed oligomerization of
CBP86 and relationships between low and high molecular weight forms. When
native
gels were run and immunoblotted CBP86 migrated as one major high molecular
weight complex. On blots of unreduced 1-D SDS-PAGE gels more than 10 distinct
immunoreactive CBP86 bands were resolved including prominent bands at 86, 72,
64,
43 and 31 kDa. Reduced but unblocked sperm extracts on 1-D SDS-PAGE gels
showed CBP86 immunoreactive forms at 72, 51, 43, and 31 kDa with the 86kDa and
64 kDa forms present on non-reduced gels no longer apparent. This indicated
that the
86 kDa calcium binding form as well as the 64 kDa form were comprised of
mercaptoethanol- sensitive subunits. Immunoblot analysis of diagonal gels run
in
unreduced and then reduced dimensions revealed subunit relationships. One 43
kDa
immunoreactive subunit was observed when the 86 kDa form of CBP86 was reduced,
suggesting the 86 kDa form consists of homodimers of the protein migrating at
43
kDa or heterodimers between this CBP86 form and an unknown partner(s).
Similarly,
a 31 kDa immunoreactive form of CBP86 was resolved from reduction of the 64
kDa
form suggesting additional homodimerization.
On western blots of 2-D IEF-PAGE gels, five major immunoreactive groups
were noted: 1) 27-38 kDa; 2) 38-42 kDa; 3) 50-56 kDa; 4) 63-72 kDa; and 5) 81-
87
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-34-
with each group showing considerable charge heterogeneity ranging overall from
pI
4-6. Because the masses deduced from the CBP86 cDNAs differ from the apparent
masses of immunoreactive CBP86 forms on gels of reduced and carboxymethylated
sperm proteins, post-translational modifications of the proteins encoded by
CBP86
splice variants are likely occurring which affect both charge and mass.
Antibodies
produced to the gel purified 86KDa spot as well as to rCBP86 show similar
immunoreactive groups of proteins on 2-D gels, confirming the heterogeneous
nature
of the related isoforms and indicating that the alternatively spliced
testicular mRNAs
which were cloned and sequenced result in translated CBP86 proteins.
Calcium binds to the acidic 86 kDa isoforms of CBP86 which increase
after in vitro capacitation
Of the human sperm proteins which bound 45calcium only calmodulin gave a
stronger signal than the 86 kDa phosphorylated isoforms of CBP86. Although
other
immunoreactive forms of CBP86 evidence a range of masses and charges on 2-D
immunoblots, calcium binding appears to be a property unique to the
phosphorylated,
oligomerized 86 kDa isoforms with pI of approximately 3.8 to 4Ø Computer
analysis did not reveal typical EF-hand motifs or typical poly-acidic
stretches in the
CBP86 sequence and the calcium binding domain of CBP86 remains unmapped. The
oligomerization of the 43 kDa subunit demonstrated by diagonal gel analysis
likely
relates to the acquisition of calcium binding capacity of the 86 kDa complex.
Increased amounts of the 86 kDa acidic isoforms of CBP86 as well as
phosphorylated members of other forms of CBP86 (groups 2 and 3) were observed
after in vitro capacitation. Since the 86 kDa CBP86 isoforms consist of 43 kDa
immunoreactive subunits, this increase in the 86 kDa isoforms provides the
first
demonstration, to our knowledge, of a human sperm protein that undergoes
oligomerization during capacitation.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-3 5-
Tyrosine phosphorylation of the acidic 86 kDa forms of CSP86 occurs
during isZ vitro. capacitation
Increased tyrosine phosphorylation of the acidic 86 kDa and 64 kDa isoforms
of CBP86 occurred during capacitation. The 64 kDa forms of CBP86 were
prominently phosphorylated after three hours of capacitation. Six hours of
capacitation were required for the 86 kDa isoforms to be prominently
phosphorylated.
The acidic isoforms of CBP86 (pI of 4.0) are the forms that bound calcium45
and no
calcium binding was observed in the area when the sample was dephosphorylated
prior to electrophoretic separation. It was notable that calreticulin served
as a positive
control in this experiment since it did not lose calcium binding capability
after the
sperm extract was treated with alkaline phosphatase prior to electrophoretic
separation. Interestingly, treatment of PVDF membranes with alkaline
phosphatase
after proteins had been transferred did not abolish the calcium binding
properties of
the 86 kDa CBP86 complex. This suggests that phosphorylation is not essential
for
calcium binding once the 86 kDa complex has formed.
Together these observations point to a model involving sequential
phosphorylation, assembly and acquisition of the calcium binding capacity of
the 86
kDa form of CBP86 during the capacitation period. CBP86 represents the first
example of calcium binding to a human sperm protein which is tyrosine
phosphorylated during capacitation. The characterization of CBP86 adds a third
protein substrate to AKAP 4 [originally called AKAP82 or Fscl in mouse] and
AKAP3 (originally called AKAP95T, FSP95 or AKAP110], which are protein
components of the fibrous sheath and were previously shown to be
phosphorylated
during in vitro capacitation.
Motifs in CSP86
CBP86 possesses putative motifs for self assembly and for progesterone and
AKA.P binding, and it has an extensin homology. Three of four possible motifs
of a
SH3 fingerprint axe present in the N-terminus of CBP86 and these three SH3
domains
are conserved in forms I, VI, VII, VIII, and XII while clones II and XI retain
one SH3
domain. Thus, eight of the predicted CBP86 forms have at least one SH3 domain.
SH3 domains occur in many proteins and axe thought to act as protein binding
structures and may be involved in linking signals transmitted from the cell
surface by
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-36-
protein tyrosine kinases. The crystal structures of several SH3 domains have
been
determined, and the view has emerged that SH3 domains facilitate binding to
partner
proteins by interaction with cognate proline-rich regions containing the
consensus
sequence Pro-X-X-Pro (PXXP). The three SH3 domains are located at the 5' end
of
CR-A of the CBP86 gene, while the 3' end of CR-A and the 5' end of the CR-B
are
relatively proline rich. Three PXXP consensus motifs are present in CR-A [aa
396-
399, 471-474, and 473-476) and three are present in CR-B (aa 529-532, 532-535,
643-
646), providing structural modules to account for CBP86 self assembly. Both
the
SH3 domains and the proline-rich stretches, which we term here the putative
dimerization domains, also provide CBP86 with potential sites for interaction
with
other flagellar proteins (such as the AKAPs).
The domain in CR-A of CBP86 which possesses similarity to the sperm
protein SP17 (Richardson et al. 1994) includes amino acids ZO-108. Embedded
within this SP17 domain at amino acids 10-44 is a region of 40% identity and
57%
similarity to the type II regulatory chain of cAMP dependent protein kinase.
This
homologous region of the mouse and human RII subunits lies in the N-terminus
between amino acids 7-41 and contains both the dimerization domain and the
AKAP
binding region (Newton et al, 1999). The dimerization domain is the site
responsible
for the interaction between individual RII subunits while the AKAP binding
region,
which requires RII dimerization for assembly, mediates binding to A-Kinase
Anchor
Proteins (AKAPs). The region of RII which is required for high affinity
binding to
AKAPs is contained within a X-type four-helix bundle dimerization motif with
an
extended hydrophobic face, a dimerization motif present in another class of
signalling
molecules, the S 100 proteins S 1 OOB and calcyclin. By anchoring PK-A to
specific
regions within a cell, AK.APs direct and specify PK-A action. The RII subunit
of
PK-A has been demonstrated to bind to two components of the fibrous sheath,
AI~AP
3 and AK.AP 4. AKAP3 localizes to the ribs of the fibrous sheath in human
sperm
and mouse AKAP 4 has been localized to the ribs and longitudinal columns. The
presence of the RII regulatory chain dimerization domain in CBP86 suggests
this
region of CBP86 may interact with AKAP3 and/or AKAP4 contributing to the
supermolecular structure of the fibrous sheath. It may also indicate that
CBP86 is
phosphorylated by PK-A.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-3 7-
CR-B contains an interesting alignment with the extensin family of
hydroxyproline -rich glycoproteins (HRGP's) which are expressed in specific
populations of cells in plant roots where they are thought to reinforce the
cell wall
against mechanical pressure. CBP86 contains two Ser-Pro3 repeats which
resemble
the Ser-Pro4 repeats that characterize the extensins. In view of its
structural similarity
to the extensins, CBP86's localization throughout the entire principal piece
suggests a
functional role in strengthening the cytoskeletal framework of the fibrous
sheath to
resist the mechanical forces of microtubule sliding.
Analysis of the CBP86 sequence detected two motifs of the possible eight of
the progesterone receptor. Residues 17 to 102 shared a 20.0% identity with
helix
domains 9, 10, 11 and 12 of the progesterone receptor binding domain, and a
local
alignment revealed that 63% of the residues in this region are either
identical or
conservative replacements. Helix domains 11 and 12 are directly involved with
ligand binding. This observation is noteworthy in light of the oligomerization
of
CBP86 and the possibility that the CBP86 progesterone domains may assemble.
Due
to CBP86's location in the sperm tail, any progesterone receptor associated
with it
may be similar to isoform C which lacks the highly conserved DNA binding
domain,
so a complete match with known progesterone receptor fingerprints is not
expected. It
is noteworthy that the two progesterone receptor motifs lie within domains of
CR-A
that undergo alternative splicing, so forms with and without this motif are
possible.
Progesterone induces the capacitation-related events of hyperactivation and
acrosome
reaction in human sperm. By stimulating a poorly characterized receptor which
appears to differ from nuclear progesterone receptors progesterone triggers a
rapid
influx of extracellular calcium which results in increased levels of free
intracellular
calcium followed by phosphatidylinositol 4,5-biphosphate hydrolysis in human
sperm. Progesterone induced calcium waves in human sperm are characterized by
an
initial transient peak followed by a sustained plateau phase lasting for
several minutes.
Both of these effects depend on extracellular calcium since they do not occur
in
calcium free medium. Progeterone has also been shown to promote tyrosine
phosphorylation of human sperm proteins. It is noteworthy that addition of
progesterone to the ih vitro capacitation medium leads to phosphorylation of
tyrosine
residues on the acidic CBP86 isoforms. Progesterone receptors appear as
heterodimers
(Nishikawa et al, 1995) similar to the hetero polymer complexes formed by
CBP86.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-38-
Example 2
Isolation of the Mouse CBP86 Homolog
A mouse testis ~,TriplEx cDNA library was screened using human CBP85
cDNA as a probe. A positive clone was obtained (named clone 1-7), a S'
truncated
alternatively spliced product which had about 80% similarity to human CBP86-2
and
the same splice site. This cDNA clone was then used to screen a mouse Lambda
FIX
II genomic library.
The mCBP86 genomic insert was about l2kb in size. The mouse full-length
I O cDNA and genomic sequences of CBP86 had remained unknown so far. For
further
analysis of mCBP86 genomic clone, it was necessary to get the full-length and
alternatively spliced cDNA sequences of mCBP86. The mouse ~,TriplEx cDNA
library was rescreened with a clone 1-7 cDNA probe. 10 positive clones were
obtained. The restriction map and DNA sequencing analysis of these clones
proved
that no full-length cDNA was obtained. The comparison of the cDNA sequences of
mCB,P86 clone 1- 7 and hCBP86 revealed that there was about 80% similarity
between them.
PCR was conducted using a Marathon mouse testis cDNA template and the
designed forward primers and 1-7R3, 1-7R2' reverse primers, and the expected
size
bands were obtained. Two bands appeared at the PCR product lane amplified with
the
forward primers and 1- 7R2 ' reverse primer on the agarose gel. The higher
band
conformed to that of the expected in size, and the lower band was similar in
size to
the band amplified with the forward primer and 1- 7R3 reverse primer. It
should be
reasonable to suppose that this band is the PCR product of alternatively
spliced
sequence similar to hCBP86-5.
The sequencing analysis of the PCR products above revealed that at least the
four alternatively spliced products of mCBP86 existed in mouse and they had
the
same splice sites as the corresponding hCBP86 variants. Comparison of cDNA and
amino acid sequences between human and mouse CBP86:
1. The cDNA sequences of mouse CBP86 had 78% (CBP86-4) to 8I
(CBP86-1 and CBP86-2) similarity and identity with that of human CBP86. For
the
amino acid sequences, mCBP86-2 had the highest similarity (80% ) and identity
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-39-
(76% ) with hCBP86; the next to it was mCBP86-4 (similarity 78% and identity
72%). The mCBP86-1 had 72% similarity and 66% identity with hCBP86.
2. The deleted region in mCBP86-2 cDNA sequence from 669bp (human
590bp) to 1493bp (human 1456bp) of mCBP86-1 had many large gaps compared with
the sequence of human CBP86. The sequences before the splice site-668bp (human
589bp) and after the splice site-1494bp (human 1457bp) were more conserved
than
the middle region from 669bp (human 590) to 1493bp (human 1546bp) of CBP86.
3. According to the results of the Northern blot, library screening and PCR,
it
is possible that mCBP86-2 was the most abundant variant of the alternatively
spliced
products of mCBP86.
4. The mCBP86-1 had calculated MW 48.28kD and PI 4.48 (hCBP86-1 MW
52.75, PI4.51); the mCBP86-2, MW 41.251cD and PI 6.55 ( hCBP86-2 MW 4lkD, PI
8.65); the mCBP86-3, MW 23.93, P16.45 (hCBP86-3 MW 23.96, PI 7.68).
5. The comparison of the sequences of the motifSH3, progesterone receptor
and CATATPASE displayed that there was high similaxity between mCBP86 and
hCBP86 except CATATPASE 4 from 6laa to 7laa of hCBP86 which was located on
the long gap region of mCBP86.
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-1-
SEQUENCE LISTING
<110> Naaby-Hansen, Soren
Wolltowicz, Michael
Mandal, Arabinda
Buer, Sen
Herr, John C.
1~ <120> CBP86, A Sperm Specific Protein
<130> 00492-02
<140>
<141>
<150> 60/176,887
<151> 2000-O1-19
<160> 26
<170> PatentIn Ver. 2.1
<210> 1
<211> 2228
<212> DNA
<213> Homo sapiens
<400> 1
cttaagagcg cggccggaaa gcagttgagt tacagacatc ctgccaaaat gatttcttca 60
aagcccagac ttgtcgtacc ctatggcctc aagactctgc tcgagggaat tagcagagct 120
gttctcaaaa ccaacccatc aaacatcaac cagtttgcag cagcttattt tcaagaactt 180
actatgtata gagggaatac tactatggat ataaaagatc tggttaaaca atttcatcag 240
attaaagtag agaaatggtc agaaggaacg acaccacaga agaaattaga atgtttaaaa 300
gaaccaggaa aaacatctgt agaatctaaa gtacctaccc agatggaaaa atctacagac 360
acagacgagg acaatgtaac cagaacagaa tatagtgaca aaaccaccca gtttccatca 420
gtttatgctg tgccaggcac tgagcaaacg gaagcagttg gtggtctttc ttccaaacca 480
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
S
gccaccccta agactactac cccaccctca tcaccacctc caacagctgt ctcaccagag 540
tttgcctacg tcccagctga cccagctcag cttgctgctc agatgttagg taaagtttca 600
tctattcatt ctgatcaatc tgatgtgtta atggtggatg tggcaaccag tatgcctgtt 660
gttatcaagg aggtgccaag ctcagaggct gctgaagatg tcatggtggc tgctcctctt 720
gtgtgttctg gaaaggtgct agaagtgcag gttgtgaacc aaacatctgt ccatgtagat 780
1S
ttgggttctc aacctaaaga aaatgaggct gaaccatcaa cggcttcctc agtccccttg 840
caggatgaac aagaacctcc tgcttatgat caagctcctg aggtcacttt gcaggctgat 900
attgaggtta tgtcaactgt tcatatatca tctgtctata acgatgtgcc tgtgactgaa 960
ggagttgttt atatcgagca actgccagaa caaatagtta tcccttttac tgatcaagtt 1020
gcttgtctta aagaaaatga gcagtcaaaa gaaaatgagc agtcaccacg agttagtccc 1080
2S
aaatctgtag tagaaaagac cacctctggc atgtctaaaa aatctgtaga gtccgtaaaa 1140
cttgcacagt tggaggagaa tgcaaaatat tcctcagtat atatggaggc agaagcaaca 1200
gctctgctct ctgacacatc tttgaaaggt cagcctgagg tacctgcaca actcctggat 1260
gcagaaggtg ctatcaaaat aggctctgaa aaatctctgc accttgaagt ggaggtcact 1320
tcaatagtct ctgacaatac tgggcaggag gagtctgggg aaaactctgt accccaggag 1380
atggaaggca gacctgtgct ctctggggaa gctgcagaag cagtgcactc aggtacatct 1440
gtaaagtcat ctagtggccc cttccctcct gctccagaag gccttactgc accagaaatt 1500
gaaccagaag gggaatcaac agctgaataa ggtttgatga agccagcaat ggcaacaagt 1560
gaacgaggac aaccaccacc atgttctaac atgtggaccc tttattgtct aactgataag 1620
aatcaacaag gtcacccatc accgccacct gcacctgggc cttttcccca agcaaccctc 1680
tatttaccta atcctaagga tccacagttt cagcagcatc caccaaaagt cacttttcca 1740
acttatgtga tgggcgacac caagaagacc agtgccccac cttttatctt agtaggctca 1800
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-3-
aatgttcagg aagcacaggg atggaaacct cttcccggac atgctgtcgt ttcacagtca 1860
gatgtcttga gatatgttgc aatgcaagtg cccattgctg ttcctgcaga tgagaaatac 1920
S cagaaacata ccctaagtcc ccagaatgct aatcctccaa gtggacaaga tgtccccagg 1980
ccaaaaagcc ctgttttcct ttctgttgct ttcccagtag aagatgtagc taaaaaaagt 2040
tcaggatctg gtgacaaatg tgctcccttt ggaagttacg gtattgctgg ggaggtaacc 2100
gtgactactg ctcacaaacg tcgcaaagca gaaactgaaa actgatccag aaatgacgct 2160
gtctgggtca acatttcagg gaggagtctg ccaccagtgt aatgtatcaa taaacttcat 2220
IS gcaagctt 2228
<210> 2
<211> 697
<212> PRT
<213> Homo Sapiens
<400> 2
25 Met Ile Ser Ser Lys Pro Arg Leu Val Val Pro Tyr Gly Leu Lys Thr
1 5 10 15
Leu Leu Glu Gly Ile Ser Arg Ala Val Leu Lys Thr Asn Pro Ser Asn
20 25 30
Ile Asn Gln Phe Ala Ala Ala Tyr Phe Gln Glu Leu Thr Met Tyr Arg
40 45
Gly Asn Thr Thr Met Asp Ile Lys Asp Leu Val Lys Gln Phe His Gln
35 50 55 60
Ile Lys Val Glu Lys Trp Ser Glu Gly Thr Thr Pro Gln Lys Lys Leu
65 70 75 80
Glu Cys Leu Lys Glu Pro Gly Lys Thr Ser Val Glu Ser Lys Val Pro
85 90 95
Thr Gln Met Glu Lys Ser Thr Asp Thr Asp Glu Asp Asn Val Thr Arg
100 105 110
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-4_
Thr Glu Tyr Ser Asp Lys Thr Thr Gln Phe Pro Ser Val Tyr Ala Val
115 120 125
Pro Gly Thr Glu Gln Thr Glu Ala Val Gly Gly Leu Ser Ser Lys Pro
130 135 140
Ala Thr Pro Lys Thr Thr Thr Pro Pro Ser Ser Pro Pro Pro Thr Ala
145 150 155 160
Val Ser Pro Glu Phe Ala Tyr Val Pro Ala Asp Pro Ala Gln Leu Ala
165 170 175
Ala Gln Met Leu Gly Lys Val Ser Ser Ile His Ser Asp Gln Ser Asp
180 185 190
Val Leu Met Val Asp Val Ala Thr Ser Met Pro Val Val Ile Lys Glu
195 200 205
Val Pro Ser Ser Glu Ala Ala Glu Asp Val Met Val Ala Ala Pro Leu
210 215 220
Val Cys Ser Gly Lys Val Leu Glu Val Gln Val Val Asn Gln Thr Ser
225 230 235 240
Val His Val Asp Leu Gly Ser Gln Pro Lys Glu Asn Glu Ala Glu Pro
245 250 255
Ser Thr Ala Ser Ser Val Pro Leu Gln Asp Glu Gln Glu Pro Pro Ala
260 265 270
Tyr Asp Gln Ala Pro Glu Val Thr Leu Gln Ala Asp Ile Glu Val Met
275 280 285
Ser Thr Val His Ile Ser Ser Val Tyr Asn Asp Val Pro Val Thr Glu
290 295 300
Gly Val Val Tyr Ile Glu Gln Leu Pro Glu Gln Ile Val Ile Pro Phe
305 310 315 320
Thr Asp Gln Val Ala Cys Leu Lys Glu Asn Glu Gln Ser Lys Glu Asn
325 330 335
Glu Gln Ser Pro Arg Val Ser Pro Lys Ser Val Val Glu Lys Thr Thr
340 345 350
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-5-
Ser Gly Met Ser Lys Lys Ser Val Glu Ser Val Lys Leu Ala Gln Leu
355 360 365
Glu Glu Asn Ala Lys Tyr Ser Ser Val Tyr Met Glu Ala Glu Ala Thr
370 375 380
Ala Leu Leu Ser Asp Thr Ser Leu Lys Gly Gln Pro Glu Val Pro Ala
385 390 395 400
1~ Gln Leu Leu Asp Ala Glu Gly Ala Ile Lys Ile Gly Ser Glu Lys Ser
405 410 415
Leu His Leu Glu Val Glu Val Thr Ser Ile Val Ser Asp Asn Thr Gly
420 425 430
Gln Glu Glu Ser Gly Glu Asn Ser Val Pro Gln Glu Met Glu Gly Arg
435 440 445
Pro Val Leu Ser Gly Glu Ala Ala Glu Ala Val His Ser Gly Thr Ser
450 455 460
Val Lys Ser Ser Ser Gly Pro Phe Pro Pro Ala Pro Glu Gly Leu Thr
465 470 475 480
Ala Pro Glu Ile Glu Pro Glu Gly Glu Ser Thr Ala Glu Gly Leu Met
485 490 495
Lys Pro Ala Met Ala Thr Ser Glu Arg Gly G1n Pro Pro Pro Cys Ser
500 505 510
Asn Met Trp Thr Leu Tyr Cys Leu Thr Asp Lys Asn Gln Gln Gly His
515 520 525
Pro Ser Pro Pro Pro Ala Pro Gly Pro Phe Pro Gln Ala Thr Leu Tyr
3$ 530 535 540
Leu Pro Asn Pro Lys Asp Pro Gln Phe Gln Gln His Pro Pro Lys Val
545 550 555 560
Thr Phe Pro Thr Tyr Val Met Gly Asp Thr Lys Lys Thr Ser Ala Pro
565 570 575
Pro Phe Ile Leu Val Gly Ser Asn Val Gln Glu Ala Gln Gly Trp Lys
580 585 590
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-6-
Pro Leu Pro Gly His Ala Val Val Ser Gln Ser Asp Val Leu Arg Tyr
595 600 605
Val Ala Met Gln Val Pro Ile Ala Val Pro Ala Asp Glu Lys Tyr Gln
610 615 620
Lys His Thr Leu Ser Pro Gln Asn Ala Asn Pro Pro Ser Gly Gln Asp
625 630 635 640
Val Pro Arg Pro Lys Ser Pro Val Phe Leu Ser Val Ala Phe Pro Val
645 650 655
Glu Asp Val Ala Lys Lys Ser Ser Asp Ser Gly Asp Lys Cys Ala Pro
660 ~ 665 670
1$
Phe Gly Ser Tyr Gly Ile Ala Gly Glu Val Thr Val Thr Thr Ala His
675 680 685
Lys Arg Arg Lys Ala Glu Thr Glu Asn
690 695
<210> 3
<211> 599
<212> PRT
<213> Homo Sapiens
<400> 3
Met Glu Lys Ser Thr Asp Thr Asp Glu Asp Asn Val Thr Arg Thr Glu
1 5 10 15
Tyr Ser Asp Lys Thr Thr Gln Phe Pro Ser Val Tyr Ala Val Pro Gly
20 25 30
Thr Glu Gln Thr Glu Ala Val Gly Gly Leu Ser Ser Lys Pro Ala Thr
35 40 45
Pro Lys Thr Thr Thr Pro Pro Ser Ser Pro Pro Pro Thr Ala Val Ser
50 55 60
Pro Glu Phe Ala Tyr Val Pro Ala Asp Pro Ala Gln Leu Ala Ala Gln
65 70 75 80
Met Leu Gly Lys Val Ser Ser Ile His Ser Asp Gln Ser Asp Val Leu
85 90 95
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
_7_
Met Val Asp Val Ala Thr Ser Met Pro Val Val Ile Lys Glu Val Pro
100 105 110
Ser Ser Glu Ala Ala Glu Asp Val Met Val Ala Ala Pro Leu Val Cys
115 120 125
Ser Gly Lys Val Leu Glu Val Gln Val Val Asn Gln Thr Sex Val His
130 135 140
Val Asp Leu Gly Ser Gln Pro Lys Glu Asn Glu Ala Glu Pro Ser Thr
145 150 155 160
Ala Ser Ser Val Pro Leu Gln Asp Glu Gln Glu Pro Pro Ala Tyr Asp
1$ 165 170 175
y
Gln Ala Pro Glu Val Thr Leu Gln Ala Asp Ile Glu Val Met Ser Thr
180 185 190
Val His Ile Ser Ser Val Tyr Asn Asp Val Pro Val Thr Glu Gly Val
195 200 205
Val Tyr Ile Glu Gln Leu Pro Glu Gln Ile Val Ile Pro Phe Thr Asp
210 215 220
Gln Val Ala Cys Leu Lys Glu Asn Glu Gln Ser Lys Glu Asn Glu Gln
225 230 235 240
Ser Pro Arg Va1 Ser Pro Lys Ser Val Val Glu Lys Thr Thr Ser Gly
245 250 255
Met Ser Lys Lys Ser Val Glu Ser Val Lys Leu Ala Gln Leu Glu Glu
260 265 270
Asn Ala Lys Tyr Ser Ser Val Tyr Met Glu Ala Glu Ala Thr Ala Leu
275 280 285
Leu Ser Asp Thr Ser Leu Lys Gly Gln Pro Glu Val Pro Ala Gln Leu
290 295 300
Leu Asp Ala Glu Gly Ala Ile Lys Ile Gly Ser Glu Lys Ser Leu His
305 310 315 320
Leu Glu Val Glu Val Thr Ser Ile Val Ser Asp Asn Thr Gly Gln Glu
325 330 335
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
_g_
Glu Ser Gly Glu Asn Ser Val Pro Gln Glu Met Glu Gly Arg Pro Val
340 345 350
Leu Ser Gly Glu Ala Ala Glu Ala Val His Ser Gly Thr Ser Val Lys
355 360 365
5er Ser Ser Gly Pro Phe Pro Pro Ala Pro Glu GIy Leu Thr Ala Pro
370 375 380
Glu Ile Glu Pro Glu Gly Glu Ser Thr Ala Glu Gly Leu Met Lys Pro
385 390 395 400
Ala Met Ala Thr Ser Glu Arg Gly Gln Pro Pro Pro Cys Ser Asn Met
405 410 41S
Trp Thr Leu Tyr Cys Leu Thr Asp Lys Asn Gln Gln Gly His Pro Ser
420 425 430
Pro Pro Pro Ala Pro Gly Pro Phe Pro Gln Ala Thr Leu Tyr Leu Pro
435 440 445
Asn Pro Lys Asp Pro Gln Phe Gln Gln His Pro Pro Lys Val Thr Phe
450 455 460
Pro Thr Tyr Val Met Gly Asp Thr Lys Lys Thr Ser AIa Pro Pro Phe
465 470 475 480
Ile Leu Val Gly Ser Asn Val Gln Glu Ala Gln Gly Trp Lys Pro Leu
485 490 495
Pro Gly His Ala Val Val Ser Gln Ser Asp Val Leu Arg Tyr Val Ala
500 505 510
Met Gln Val Pro IIe Ala Val Pro Ala Asp Glu Lys Tyr Gln Lys His
515 520 525
Thr Leu Ser Pro Gln Asn Ala Asn Pro Pro Ser Gly Gln Asp Val Pro
530 535 540
Arg Pro Lys Ser Pro Val Phe Leu Ser Val Ala Phe Pro Val Glu Asp
545 550 555 560
Val Ala Lys Lys Ser Ser Asp Ser Gly Asp Lys Cys Ala Pro Phe Gly
565 570 575
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-9-
Ser Tyr Gly Ile Ala Gly Glu Val Thr Val Thr Thr Ala His Lys Arg
580 585 590
Arg Lys Ala Glu Thr Glu Asn
595
<210> 4
<211> 519
<212 > P12T
<213> Homo sapiens
<400> 4
IS Met Leu Gly Lys Val Ser Ser Ile His Ser Asp Gln Ser Asp Val Leu
5 10 15
Met Val Asp Val Ala Thr Ser Met Pro Val Val Ile Lys Glu Val Pro
25 30
Ser Ser Glu Ala Ala Glu Asp Val Met Val Ala Ala Pro Leu Val Cys
3S 40 45
Ser Gly Lys Val Leu Glu Val Gln Val Val Asn Gln Thr Ser Val His
50 55 60
Val Asp Leu Gly Ser Gln Pro Lys Glu Asn Glu Ala Glu Pro Ser Thr
65 70 75 80
Ala Ser Ser Val Pro Leu Gln Asp Glu Gln Glu Pro Pro Ala Tyr Asp
85 90 95
Gln Ala Pro Glu Val Thr Leu Gln Ala Asp Lle Glu Val Met Ser Thr
100 105 110
Val His Ile Ser Ser Val Tyr Asn Asp Val Pro Val Thr Glu Gly Val
115 120 125
Val Tyr Ile Glu Gln Leu Pro Glu Gln Ile Val Tle Pro Phe Thr Asp
130 135 140
GIn Val,Ala Cys Leu Lys Glu Asn Glu Gln Ser Lys Glu Asn Glu Gln
145 150 155 160
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-10-
Ser Pro Arg Val Ser Pro Lys Ser Val Val Glu Lys Thr Thr Ser Gly
165 170 175
Met Ser Lys Lys Ser Val Glu Ser Val Lys Leu Ala Gln Leu Glu Glu
S 180 185 190
Asn Ala Lys Tyr Ser Ser Val Tyr Met Glu Ala Glu Ala Thr Ala Leu
195 200 205
Leu Ser Asp Thr Ser Leu Lys Gly Gln Pro Glu Val Pro Ala Gln Leu
210 215 220
Leu Asp Ala Glu Gly Ala Ile Lys Ile Gly Ser Glu Lys Ser Leu His
225 230 235 240
1S
Leu Glu Val Glu Val Thr Ser Ile Val Ser Asp Asn Thr Gly GIn Glu
245 250 255
Glu Ser Gly Glu Asn Ser Val Pro Gln Glu Met Glu Gly Arg Pro Val
260 265 270
Leu Ser GIy Glu Ala Ala Glu AIa Val His Ser Gly Thr Ser Val Lys
27S 280 285
2S Ser Ser Ser Gly Pro Phe Pro Pro Ala Pro Glu Gly Leu Thr Ala Pro
290 295 300
G1u Ile Glu Pro Glu Gly Glu Ser Thr Ala Glu Gly Leu Met Lys Pro
305 310 315 320
Ala Met Ala Thr Ser Glu Arg Gly Gln Pro Pro Pro Cys Ser Asn Met
325 330 335
Trp Thr Leu Tyr Cys Leu Thr Asp Lys Asn Gln Gln Gly His Pro Ser
3S 340 34S 350
Pro Pro Pro Ala Pro Gly Pro Phe Pro Gln Ala Thr Leu Tyr Leu Pro
355 360 365
Asn Pro Lys Asp Pro Gln Phe Gln Gln His Pro Pro Lys Val Thr Phe
370 375 380
Pro Thr Tyr Val Met Gly Asp Thr Lys Lys Thr Ser Ala Pro Pro Phe
385 390 395 400
4S
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-11-
Ile Leu Val Gly Ser Asn Val Gln Glu Ala Gln Gly Trp Lys Pro Leu
405 410 415
Pro Gly His Ala Val Val Ser Gln Ser Asp Val Leu Arg Tyr Val Ala
420 425 430
Met Gln Val Pro Ile Ala Val Pro Ala Asp Glu Lys Tyr Gln Lys His
435 440 445
Thr Leu Ser Pro Gln Asn Ala Asn Pro Pro Ser Gly Gln Asp Val Pro
450 455 460
Arg Pro Lys Ser Pro Val Phe Leu Ser Val Ala Phe Pro Val Glu Asp
465 470 475 480
Val Ala Lys Lys Ser Ser Asp Ser Gly Asp Lys Cys Ala Pro Phe Gly
485 490 495
Ser Tyr Gly Ile Ala Gly Glu Val Thr Val Thr Thr Ala His Lys Arg
500 505 510
Arg Lys Ala Glu Thr Glu Asn
515
<210> 5
<211> 503
<212> PRT
<213> Homo Sapiens
<400> 5
Met Val Asp Val Ala Thr Ser Met Pro Val Val Ile Lys Glu Val Pro
Z 5 10 15
Ser Ser Glu Ala Ala Glu Asp Val Met Val Ala Ala Pro Leu Val Cys
20 25 30
Ser Gly Lys Val Leu Glu Val Gln Val Val Asn Gln Thr Ser Val His
35 40 45
Val Asp Leu Gly Ser Gln Pro Lys Glu Asn Glu Ala Glu Pro Ser Thr
55 ~60
Ala Ser Ser Val Pro Leu Gln Asp Glu Gln Glu Pro Pro Ala Tyr Asp
45 65 70 75 80
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-12-
Gln Ala Pro Glu Val Thr Leu Gln Ala Asp Ile Glu Val Met Ser Thr
85 90 95
$ Val His Ile Ser Ser Val Tyr Asn Asp Val Pro Val Thr Glu Gly Val
100 105 ' 110
Val Tyr Ile Glu Gln Leu Pro Glu Gln Ile Val Ile Pro Phe Thr Asp
115 120 125
Gln Val Ala Cys Leu Lys Glu Asn Glu Gln Ser Lys Glu Asn Glu Gln
130 135 140
Ser Pro Arg Val Ser Pro Lys Ser Val Val Glu Lys Thr Thr Ser Gly
145 150 155 160
Met Ser Lys Lys Ser Val Glu Ser Val Lys Leu Ala Gln Leu Glu Glu
165 170 175
Asn Ala Lys Tyr Ser Ser Val Tyr Met Glu Ala Glu Ala Thr Ala Leu
180 185 190
Leu Ser Asp Thr Ser Leu Lys Gly Gln Pro Glu Val Pro Ala Gln Leu
195 200 205
Leu Asp Ala Glu Gly Ala Ile Lys Ile Gly Ser Glu Lys Ser Leu His
210 215 220
Leu Glu Val Glu Val Thr Ser Ile Val Ser Asp Asn Thr Gly Gln Glu
225 230 235 240
Glu Ser Gly Glu Asn Ser Val Pro Gln Glu Met Glu Gly Arg Pro Val
245 250 255
Leu Ser Gly Glu Ala Ala Glu Ala Val His Ser Gly Thr Ser Val Ly's
260 265 270
Ser Ser Ser Gly Pro Phe Pro Pro Ala Pro Glu Gly Leu Thr Ala Pro
275 280 28S
Glu Ile Glu Pro Glu Gly Glu Ser Thr Ala Glu Gly Leu Met Lys Pro
290 295 300
Ala Met Ala Thr Ser Glu Arg Gly Gln Pro Pro Pro Cys Ser Asn Met
305 310 315 320
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-13-
Trp Thr Leu Tyr Cys Leu Thr Asp Lys Asn Gln Gln Gly His Pro Ser
325 330 335
$ Pro Pro Pro Ala Pro Gly Pro Phe Pro Gln Ala Thr Leu Tyr Leu Pro
340 345 350
Asn Pro Lys Asp Pro Gln Phe Gln Gln His Pro Pro Lys Val Thr Phe
355 360 365
Pro Thr Tyr Val Met Gly Asp Thr Lys Lys Thr Ser Ala Pro Pro Phe
370 375 380
Ile Leu Val Gly Ser Asn Val Gln Glu Ala Gln Gly Trp Lys Pro Leu
1$ 385 390 395 400
Pro Gly His Ala Val Val Ser Gln Ser Asp Val Leu Arg Tyr Val Ala
405 410 415
Met Gln Val Pro Ile Ala Val Pro Ala Asp Glu Lys Tyr Gln Lys His
420 425 430
Thr Leu Ser Pro Gln Asn Ala Asn Pro Pro Ser Gly Gln Asp Val Pro
435 440 445
Arg Pro Lys Ser Pro Val Phe Leu Ser Val Ala Phe Pro Val Glu Asp
450 455 460
Val Ala Lys Lys Ser Ser Asp Ser Gly Asp Lys Cys Ala Pro Phe Gly
465 470 475 480
Ser Tyr Gly Ile Ala Gly Glu Val Thr Val Thr Thr Ala His Lys Arg
485 490 495
Arg Lys Ala Glu Thr Glu Asn
500
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-14-
<210> 6
<211> 496
<212> PRT
<213> Homo Sapiens
<400> 6
Met Pro Val Val Ile Lys Glu Val Pro Ser Ser Glu Ala Ala Glu Asp
1 5 10 15
Val Met Val Ala Ala Pro Leu Val Cys Ser Gly Lys Val Leu Glu Val
25 30
Gln VaI VaI Asn Gln Thr Ser Val His Val Asp Leu Gly Ser Gln Pro
35 ' 40 45
Lys Glu Asn Glu Ala Glu Pro Ser Thr Ala Ser Ser Val Pro Leu Gln
50 55 60
Asp Glu Gln Glu Pro Pro Ala Tyr Asp Gln Ala Pro Glu Val Thr Leu
65 70 75 80
Gln Ala Asp Ile Glu Val Met Ser Thr Val His Ile Ser Ser Val Tyr
85 90 95
Asn Asp VaI Pro Val Thr Glu Gly Val Val Tyr Ile Glu Gln Leu Pro
100 105 110
Glu Gln Ile Val Ile Pro Phe Thr Asp Gln Val Ala Cys Leu Lys Glu
115 120 125
Asn Glu Gln Ser Lys Glu Asn Glu Gln Ser Pro Arg Val Ser Pro Lys
130 l35 140
Ser Val Val Glu Lys Thr Thr Ser Gly Met Ser Lys Lys Ser Val Glu
145 150 155 160
Ser Val Lys Leu Ala Gln Leu Glu GIu Asn Ala Lys Tyr Ser Ser Val
165 170 175
Tyr Met Glu Ala Glu Ala Thr Ala Leu Leu Ser Asp Thr Ser Leu Lys
180 185 190
Gly Gln Pro Glu Val Pro Ala Gln Leu Leu Asp Ala Glu Gly Ala Ile
195 200 205
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-15-
Lys Ile Gly Ser Glu Lys Ser Leu His Leu Glu Val Glu Val Thr Ser
210 2l5 220
Ile Val Ser Asp Asn Thr Gly Gln Glu Glu Ser Gly Glu Asn Ser Val
5. 225 230 235 240
Pro Gln Glu Met Glu Gly Arg Pro Val Leu Ser Gly Glu Ala Ala Glu
245 250 255
Ala Val His Ser Gly Thr Ser Val Lys Ser Ser Ser Gly Pro Phe Pro
260 265 270
Pro Ala Pro Glu Gly Leu Thr Ala Pro Glu Ile G1u Pro Glu Gly Glu
275 280 285
Ser Thr Ala Glu Gly Leu Met Lys Pro Ala Met Ala Thr Ser Glu Arg
290 295 300
Gly Gln Pro Pro Pro Cys Ser Asn Met Trp Thr Leu Tyr Cys Leu Thr
305 310 315 320
Asp Lys Asn Gln Gln Gly His Pro Ser Pro Pro Pro Ala Pro Gly Pro
325 330 335
Phe Pro Gln Ala Thr Leu Tyr Leu Pro Asn Pro Lys Asp Pro Gln Phe
340 345 350
Gln Gln His Pro Pro Lys Val Thr Phe Pro Thr Tyr Val Met Gly Asp
355 360 365
Thr Lys Lys Thr Ser Ala Pro Pro Phe Ile Leu Val Gly Ser Asn Val
370 375 380
Gln Glu Ala Gln Gly Trp Lys Pro Leu Pro Gly His Ala Val Val Ser
385 390 395 400
Gln Ser Asp Val Leu Arg Tyr Val Ala Met Gln Val Pro Ile Ala Val
405 410 415
Pro Ala Asp Glu Lys Tyr Gln Lys His Thr Leu Ser Pro Gln Asn Ala
420 425 430
Asn Pro Pro Ser Gly Gln Asp Val Pro Arg Pro Lys Ser Pro Val Phe
435 440 445
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-16-
Leu Ser Val Ala Phe Pro Val Glu Asp Val Ala Lys Lys Ser Ser Asp
450 455 460
Ser Gly Asp Lys Cys Ala Pro Phe Gly Ser Tyr Gly Ile Ala Gly Glu
465 470 475 480
Val Thr Val Thr Thr Ala His Lys Arg Arg Lys Ala Glu Thr Glu Asn
485 490 ~ 495
<210> 7
<211> 14
<212> PRT
1$ <213> Homo Sapiens
<400> 7
Leu Lys Thr Leu Leu Glu Gly Ile Ser Arg Ala Val Leu Lys
1 5 10
<z1o> s
<211> 15
<212> PRT
<213> Homo Sapiens
<400> 8
Val Ser Asp Asn Thr Gly Gln Glu Glu Ser Gly Glu Asn Ser Val
1 5 10 15
<210> 9
<211> 11
<212> PRT
3$ <213> Homo Sapiens
<400> 9
Ser Gly Thr Ser Val Lys Ser Ser Ser Gly Arg
1 5 10
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-17-
<210> 10
<211> 11
<212> PRT
<213> Homo Sapiens
<400> 10
Asn Gln Phe Ala Ala Ala Tyr Phe Gln Glu Leu
1 5 10
<210> 11
<211> 10
<212> PRT
<213> Homo Sapiens
<400> 11
Val Glu Lys Trp Ser Glu Gly Thr Thr Pro
1 5 10
<210>12
<211>13
<212>PRT
<213>Homo Sapiens
<400> 12
Lys Thr Thr Gln Phe Pro Ser Val Tyr Ala Val Pro Gly
1 5 10
<210> 13
<211> 17
<212> PRT
<213> Homo Sapiens
<400> 13
Pro Ser Ser Pro Pro Pro Thr Ala Val Sex Pro Glu Phe Ala Tyr Val
1 5 10 15
Pro
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
_1g_
<210>14
<211>17
<212>PRT
<213>Homo sapiens
<400> 14
Ala Glu Ala Thr Ala Leu Leu Ser Asp Thr Ser Leu Lys Gly Gln Pro
1 5 10 15
Glu
<210> 15
<211> 291
<212> PRT
<213> Homo Sapiens
<400> 15
Met Pro Val Val Ile Lys Glu Val Pro Ser Ser Glu Ala Ala Glu Asp
1 5 10 15
Val Met Val Ala Ala Pro Leu Val Cys Sex Gly Lys Val Leu Glu Val
25 30
Gln Val Val Asn Gln Thr Ser Val His Val Asp Leu Gly Ser Gln Pro
40 45
Lys Glu Asn Glu Ala Glu Pro Ser Thr Ala Ser Ser Val Pro Leu Gln
30 50 55 60
Asp Glu Gln Glu Pro Pro Ala Tyr Asp Gln Ala Pro Glu Val Thr Leu
65 70 75 80
3S Gln Ala Asp Ile Glu Val Met Ser Thr Val His Ile Ser Ser Val Tyr
85 90 95
Asn Asp Val Pro Val Thr Glu Gly Val Val Tyr Ile Glu Gln Leu Pro
100 105 110
Glu Gln Ile Va1 Ile Pro Phe Thr Asp Gln Val Ala Cys Leu Lys Glu
115 120 125
Asn Glu Gln Ser Lys Glu Asn Glu Gln Ser Pro Arg Val Ser Pro Lys
130 135 140
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-19-
Ser Val Val Glu Lys Thr Thr Ser Gly Met Ser Lys Lys Ser Val Glu
145 150 155 160
Ser Val Lys Leu Ala Gln Leu GIu Glu Asn AIa Lys Tyr Ser Ser Val
165 170 175
Tyr Met Glu Ala Glu Ala Thr Ala Leu Leu Ser Asp Thr Ser Leu Lys
180 185 190
to
Gly Gln Pro Glu Val Pro Ala Gln Leu Leu Asp Ala Glu Gly Ala Ile
195 200 205
Lys Ile Gly Ser Glu Lys Ser Leu His Leu Glu Val Glu Val Thr Ser
210 215 220
Ile Val Ser Asp Asn Thr Gly Gln Glu Glu Ser Gly Glu Asn Ser Val
225 230 235 240
2~ Pro Gln Glu Met Glu Gly Arg Pro Val Leu Ser Gly Glu Ala Ala Glu
245 250 255
Ala Val His Ser Gly Thr Ser Val Lys Ser Ser Ser Gly Pro Phe Pro
260 265 270
Pro Ala Pro Glu Gly Leu Thr Ala Pro Glu Ile Glu Pro Glu Gly Glu
275 280 285
Ser Thr Ala Glu
290
<210> 16
<211> 204
<212> PRT
<213> Homo sapiens
<400> 16
Gly Leu~Met Lys Pro Ala Met Ala Thr Ser Glu Arg Gly Gln Pro Pro
4~ 1 5 10 15
Pro Cys Ser Asn Met Trp Thr Leu Tyr Cys Leu Thr Asp Lys Asn Gln
20 25 30
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-20-
Gln Gly His Pro Ser Pro Pro Pro Ala Pro Gly Pro Phe Pro Gln Ala
35 40 45
Thr Leu Tyr Leu Pro Asn Pro Lys Asp Pro Gln Phe Gln Gln His Pro
50 55 60
Pro Lys Val Thr Phe Pro Thr Tyr Val Met Gly Asp Thr Lys Lys Thr
65 70 75 80
Ser Ala Pro Pro Phe Ile Leu Val Gly Ser Asri Val Gln Glu Ala Gln
85 90 95
Gly Trp ~Lys Pro Leu Pro Gly His Ala Val Val Ser Gln Ser Asp Val
100 105 110
Leu Arg Tyr Val Ala Met Gln Val Pro Ile Ala Val Pro Ala Asp Glu
115 120 125
Lys Tyr Gln Lys His Thr Leu Ser Pro Gln Asn Ala Asn Pro Pro Ser
130 135 140
Gly Gln Asp Val Pro Arg Pro Lys Ser Pro Val Phe Leu Ser Val Ala
145 150 155 160
Phe Pro Val Glu Asp Val Ala Lys Lys Ser Ser Asp Ser Gly Asp Lys
165 170 ~ 175
Cys Ala Pro Phe Gly Ser Tyr Gly Tle Ala Gly Glu Val Thr Val Thr
180 185 190
Thr Ala His Lys Arg Arg Lys Ala Glu Thr Glu Asn
195 200
<210> 17
<211> 8
<212> PRT
<213> Homo sapiens
<400> 17
Leu Val Val Pro Tyr Gly Leu Lys
1 5
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-21 _
<210>18
<211>8
<212>PRT
<213>Homo sapiens
<400> 18
Thr Leu Leu Glu Gly Ile Ser Arg
1 ° 5
1~
<210> 19
<2l1> 21
<212> PRT
<213> Homo Sapiens
<400> 19
Thr Asn Pro Ser Asn Ile Asn Gln Phe Ala Ala Ala Tyr Phe Gln Glu
1 5 10 15
Leu Thr Met Tyr Arg
<210> 20
<211> 20
<212> PRT
<213> Homo Sapiens
<400> 20
3~ Lys Tyr Ser Ser Val Tyr Met Glu Ala Glu Ala Thr Ala Leu Leu Ser
1 5 10 15
Asp Thr Ser Leu
35
<210> 21
<211> 16
<212> PRT
4~ <213> Homo Sapiens
<400> 21
Gly Gln Pro Glu Val Pro Ala Gln Leu Leu Asp Ala Glu Gly Ala Ile
1 5 10 15
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-22-
<210> 22
<211> 26
<212> DNA
$ <213> Artificial Sequence
<220>
<221> primer bind
<222> (1) . . (26)
<223> n is inosine
<220>
<223> Description of Artificial Sequence: PCR degenerate
deoxyinosine sense primer
<400> 22
ggncagccng aggtnccngc ncaryt 26
<210> 23
<211> 9
<212> PRT
<213> Homo Sapiens
<400> 23 ,
Gly Gln Pro Glu Val Pro Ala Gln Leu
1 5
<210> 24
<211> 39
<212> DNA
<213> Artificial Sequence
<z2o>
<221> primer bind
<222> (1) . . (39)
<220>
<223> Description of Artificial Sequence: PCR antisense
primer
<400> 24
4S ttattcagct gttgattccc cttctggttc aatttctgg 39
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-23-
<210> 25
$ <211> 878
<212> DNA
<213> Homo Sapiens
<400> 25
atgcctgttg ttatcaagga ggtgccaagc tcagaggctg ctgaagatgt catggtggct 60
1$
gctcctcttg tgtgttctgg aaaggtgcta gaagtgcagg ttgtgaacca aacatctgtc 120
catgtagatt tgggttctca acctaaagaa aatgaggctg aaccatcaac ggcttcctca 180
gtccccttgc aggatgaaca agaacctcct gcttatgatc aagctcctga ggtcactttg 240
caggctgata ttgaggttat gtcaactgtt catatatcat ctgtctataa cgatgtgcct 300
gtgactgaag gagttgttta tatcgagcaa ctgccagaac aaatagttat cccttttact 360
2$
gatcaagttg cttgtcttaa agaaaatgag cagtcaaaag aaaatgagca gtcaccacga 420
gttagtccca aatctgtagt agaaaagaec acetctggca tgtctaaaaa atctgtagag 480
tccgtaaaao ttgcacagtt ggaggagaat gcaaaatatt cctcagtata tatggaggca 540
gaagcaacag ctctgctctc tgacacatct ttgaaaggtc agcctgaggt acctgcacaa 600
ctcctggatg cagaaggtgc tatcaaaata ggctctgaaa aatctctgca ccttgaagtg 660
3$
gaggtcactt caatagtctc tgacaatact gggcaggagg agtctgggga aaactctgta 720
ccccaggaga tggaaggcag acctgtgctc tctggggaag ctgcagaagc agtgcactca 780
ggtacatctg taaagtcatc tagtggcccc ttccctcctg ctccagaagg ccttactgca 840
ccagaaattg aaccagaagg ggaatcaaca gctgaataa 879
CA 02397901 2002-07-17
WO 01/53338 PCT/USO1/01715
-24-
1~
<210> 26
<211> 600
<212> DNA
<213> Homo sapiens
<400> 26
gcaatggcaa caagtgaacg aggacaacca ccaccatgtt ctaacatgtg gaccctttat 60
tgtctaactg ataagaatca acaaggtcac ccatcaccgc cacctgcacc tgggcctttt 120
ccccaagcaa ccctctattt acctaatcct aaggatccac agtttcagca gcatccacca 180
aaagtcactt ttccaactta tgtgatgggc gacaccaaga agaccagtgc cccacctttt 240
IS atcttagtag gctcaaatgt tcaggaagca cagggatgga aacctcttcc cggacatgct 300
gtcgtttcac agtcagatgt cttgagatat gttgcaatgc aagtgcccat tgctgttcct 360
gcagatgaga aataccagaa acatacccta agtccccaga atgctaatcc tccaagtgga 420
caagatgtcc ccaggccaaa aagccctgtt,ttcctttctg ttgctttccc agtagaagat 480
gtagctaaaa aaagttcagg atctggtgac aaatgtgctc cctttggaag ttacggtatt 540
gctggggagg taaccgtgac tactgctcac aaacgtcgca aagcagaaac tgaaaactga 600