Note: Descriptions are shown in the official language in which they were submitted.
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
Binding Compounds and Methods for Identifying Binding Compounds
Related Applications
This application claims the benefit of U.S.S.N. 60/190,946, U.S.S.N.
60/190,996 and
U.S.S.N. 60/191,299, the disclosure of each of which is incorporated by
reference herein.
Reference is also made to the applications filed on March 20, 2001 identified
by attorney docket
nos. CNS-005 and CNS-006, the disclosure of each of which is incorporated by
reference herein.
Field of the Invention
The invention generally relates to Cysteine-X-Cysteine Chemolcine Receptor 4
("CXCR4" or "CXC chemokine receptor 4"), and more particularly, to binding
compounds for
CXC chemokine receptor 4. Methods of the invention are useful for the
treatment of disease by
to identifying and preparing binding compounds for CXC chemokine receptor 4.
Background of the Invention
Chemokines (chemoattractant cytol~ines) comprise a family of structurally
related
secreted proteins of about 70-110 amino acids that share the ability to induce
migration and
activation of specific types of blood cells. See Proost P., et al. (1996) Int.
J. Clin. Lab. Rse. 26:
15 211-223; Premack, et al. (1996) Nature Medicine 2: 1174-1178; Yoshie, et
al. (1997) J.
Leukocyte Biol. 62: 634-644. Over 30 different human chemol~ines have been
described to date.
While they are primarily responsible for the activation and recruitment of
leulcocytes, they vary
in their specificities for different leulcocyte types (neutrophils, monocytes,
eosinophils,
basophils, lymphocytes, dendritic cells, etc.), and in the types of cells and
tissues where the
2o chemolcines are synthesized. Further analysis of this family of proteins
has shown that it can be
divided up into two fiuther subfamilies of proteins. These have been termed
CXC or a-
chemolcines, and the CC or J3-chemokines based on the spacings of two
conserved cysteine
residues near the amino terminus of the proteins.
Chemokines are typically produced at sites of tissue injury or stress where
they promote
25 the infiltration of leukocytes into tissues and facilitate an inflammatory
response. Some
chemolcines act selectively on immune system cells such as subsets of T-cells
or B lymphocytes
or antigen presenting cells, and may thereby promote immune responses to
antigens. In addition,
some chemokines have the ability to regulate the growth or migration of
hematopoietic
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-2-
progenitor and stem cells that normally differentiate into specific leukocyte
types, thereby
regulating leukocyte numbers in the blood.
The activities of chemolcines axe mediated by cell surface receptors that are
members of a
family of seven transmembrane ("7TM"), G-protein coupled receptors ("GPCR").
At least
twelve different human chemokine receptors are known, including CCRl, CCR2,
CCR3, CCR4,
CCRS, CCR6, CCR7, CCRB, CXCRl, CXCR2, CXCR3, and CXCR4. These receptors vary
in
their specificities for specific chemolcines. Some receptors bind to a single
known chemolcine,
while others bind to multiple chemokines. Binding of a chemokine to its
receptor typically
induces intracellular signaling responses such as a transient rise in
cytosolic calcium
to concentration, followed by cellular biological responses such as
chemotaxis. In addition, some
chemolcine receptors, such as CXCR4, serve as co-receptors for Human
Immunodeficiency Virus
(HIV), such that they interact with HIV and with the cellular CD4 receptor to
facilitate viral
entry into cells. '
Chemolcines are important in medicine because they regulate the movement and
~ biological activities of leukocytes in many disease situations, including,
but not limited to:
allergic disorders, autoinunune diseases, ischemia/reperfusion injury,
development of
atherosclerotic plaques, cancer (including mobilization of hematopoietic stem
cells for use in
chemotherapy or myeloprotection during chemotherapy), chronic inflammatory
disorders,
chronic rejection of transplanted organs or tissue grafts, chronic myelogenous
leukemia, and
2o infection by HIV and other pathogens Furthermore, CXCR4, in particular, has
been implicated
in diseases such as glioblastoma multiforme tumor, hepatocellular carcinoma,
colon cancer,
esophageal cancer, gastric cancers, breast cancer metastasis, pancreatic
cancer and in renal
allograft rejection. See e.g., Sehgal A, et. al., J. Surg. Oncol. 69(2)99-104
(1998); Begum NA, et.
al., Int. J. Ohcol. 14(5)927-934 (1999); Mitra P, et. al., I~t. J. Oncol.
14(5):917-25 (1999);
Muller A, et. al., Nature 410(6824)50-6 (2001); Koshiba T, et. al., Clip.
Cancer Res. 6(9):3530-
5 (2000); and Eitrler F, et. al., Transplantation 66(11):1551-7 (1998).
Antagonists of chemokine receptors may be of benefit in many of these diseases
by
reducing excessive inflammation and immune system responses. In the case of
HIV infection,
chemokines and antagonists that bind to HIV co-receptors may have utility in
inhibiting viral
3o entry into cells. HIV causes Acquired Immune Deficiency Syndrome ("AIDS"),
which is one of
the leading causes of death in the United States and throughout the world.
According to the
Center for Disease Control, at least 30.6 million people world-wide have been
infected with
HIV. HIV attacks the immune system and leaves the body vulnerable to a variety
of life-
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-3-
threatening illnesses. Common bacteria, yeast, and viruses that would not
cause disease in
people with a fully functional immune system often cause these illnesses in
people affected with
HIV.
Not all patients infected with HIV have AIDS. Typically, a patient who has
been
infected with HIV will slowly develop AIDS as HIV damages his immune system.
The severity
of the immune system damage is measured by an absolute CD4+ lymphocyte count;
a patient
having a count of less than 200 cells/~.1 is considered to have AIDS. The CD4
protein is a
glycoprotein of approximately 60,000 molecular weight and is expressed on the
cell membrane
of mature, thymus-derived (T) lymphocytes, and to a lesser extent on cells of
the
to monocyte/macrophage lineage. Typically, CD4 cells appear to function by
providing an
activating signal to B cells, by inducing T lymphocytes bearing the reciprocal
CD8 marker to
become cytotoxic/suppressor cells, and/or by interacting with targets bearing
major
histocompatibility complex (MHC) class II molecules.
The search for a preventative or therapeutic agent for HIV and AIDS has been
especially
15 intense as this epidemic has proliferated world-wide. Research has
discovered that the ability of
HIV to enter cells requires the binding of the HIV envelope glycoproteins
encoded by the env
gene to the CD4 receptor. These glycoproteins are encoded by the env gene and
translated as a
precursor, gp160, which is subsequently cleaved into gp 120 and gp41. Gp 120
binds to the CD4
protein present on the surface of susceptible target cells, resulting in the
fusion of virus with the
2o cell membranes, and facilitating virus entry into the host. The eventual
expression of env on the
surface of the HIV-infected host cell enables this cell to fuse with
uninfected, CD4+ cells,
thereby spreading the virus. However, in response to infection with HIV, the
host immune
system will produce antibodies targeted against various antigenic sites, or
determinants, of
gp120. Some of those antibodies will have a neutralizing effect and will
inhibit HIV infectivity.
25 It is believed that this neutralizing effect is due to the antibodies'
ability to interfere with HIV's
cellular attachment. It is also believed that this effect may explain in part,
the rather long latency
period between the initial seroconversion and the onset of clinical symptoms.
Recent studies have shown that the HIV fusion process occurs with a wide range
of
human cell types that either express human CD4 endogenously or have been
engineered to
3o express human CD4. The fusion process, however, does not occur with
nonhuman cell types
engineered to express human CD4. Although such nonhuman cells can still bind
env, membrane
fusion does not follow. The disparity between human and nonhuman cell types
exists apparently
because membrane fusion requires the co-expression of human CD4 and a co-
receptor specific to
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-4-
human cell types. Because they lack this co-receptor accessory factor,
nonhuman cell types
engineered to express only human CD4 are incapable of membrane fusion, and are
thus
nonpermissive for HIV infection. Furthermore, expression of CD4 in some human
cell lines was
insufficient to confer resistance to HIV-1 infection. In addition, some HIV-1
strains were T cell
tropic (T-tropic) while others were macrophage tropic (M-tropic), though both
cells possessed
the CD4 antigen. Further research has shown that certain chemolcines could
blocl~ the infectivity
of M-tropic but not T-tropic HIV strains. Thereafter, it was shown that an
orphan receptor,
CXCR4 was required for the activity of T-tropic strains. See e.g., Horulc R.,
Immunol Today
20(2):89-94 (1999); Doms RW, Peiper SC., Virology 235(2):179-90 (1997); Ward
SG, Bacon
to K, Westwiclc J., Immunity 9(1):1-11 (1998); Berson JF, Doms RW., Semin
linmtmol 10(3):237-
48 (1998).
While it has been demonstrated that HIV uses the CXCR4 as a co-receptor for
cellular
entry, it has been difficult for researchers to obtain high resolution X-ray
crystallographic
structures of a CXCR4 because of difficulties in crystallizing such a 7TM
protein which requires
complex interactions with lipids for its native conformation. The requirement
of the interaction
with lipids also malces difficult the preparation of biologically active forms
of such GPCRs,
because, in the absence of those lipids, they readily form denatured
aggregates with minimal to
no ability to specifically bind ligands unless great care is taken to preserve
the biologically active
conformation during solubilization. In the absence of an X-ray structure, a
variety of approaches
2o have been used to define the regions of CXCR4 that are involved in gp120
binding and viral
uptalce. These approaches generally involve comparing results with non-human
homologues,
chimeric receptors, and point mutants to study the structural requirements for
the co-receptor
activity of CXCR4. CXCR4, is a 3 S2 amino acid protein, has seven putative
transmembrane
("TM") segments (TM1=residues 40-64, TM2=77-99, TM3 = 111-131, TM4 =177-197,
TMS =
204-223, TM6 = 241-261, and TM7 = 283-307, putatively), and extracellular N-
terminus, three
extracellular loops and three intracellular loops connecting the transmembrane
segments, and an
intracellular C-terminus. The second extracellular loop is the region most
required for the entry
of HIV into the cell; however, the N-terminus and the third extracellular loop
are also involved.
Several charged residues on the extracellular side of the receptor have been
implicated in binding
(Asp-I 1, Asp-265, Glu-275, Glu-278, and Arg-280) using mutagenesis studies.
See e.g., Zhou
H, et. al., Arch. Biochem. Biophys. 373(1):211-7 (2000). In addition, the N-
terminus of the
ligand SDF-1 has been implicated as important for binding to the receptor. See
e.g., Crump MP,
et. al., EMBO J. 16(23):6996-7007 (1997). Furthermore, synthetic peptides have
been used to
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-5-
study the effect of high positive charge in peptides on the interaction with
CXCR4. See e.g., Luo
Z, et. al., Biochem. Biophys. Res. Comm. 263:691-5 (1999).
Several inhibitors of CXCR4 have been reported. These include positively
charged
peptides such as T22 and T140 and small molecule inhibitors such as ALX40-4C
and T134.
AMD 3100, a heterocyclic bicyclarn (one positive charge on each of two rings)
has been
reported as well. See e.g., Tamamura H, et. al., Bioorg Med Chem 6(7):1033-41
(I998);
Tamamura H, et. al., Biochem. Biophys. Rs. Comm. 252:877-82 (1998); Doranz BJ,
et. al., JExp
Med I86(8):1395-400 (1997); Aralcalci R, et. al., J. yi~ol. 73(2):1719-23
(1999); and Donzella
GA, et. al., Nat Med 4(1):72-7 (1998). AMD 3100, however, caused the
conversion from T- to
1o M-tropic viruses in Peripheral Blood Monocytes ("PBMCs"). See e.g., De
Clercq E., Mol.
Phaf°rrzacol. 57:833-839 (2000). In addition, bicyclams may interfere
with other CXCR4-lilce
receptors. See e.g., Schols D, et. al., J. Exp. Med 186(8):1383-1388 (1997).
Furthermore, T22
inhibits calcium mobilization, therefore interfering with CXCR4's natural
required signaling.
See e.g., Muralcami T, et. al., J. Exp. Med. 186(8):1389-1393 (1997).
15 As a result of the limitations of prior inhibitors of CXCR4 binding, a need
still remains
for effective HIV preventative and therapeutic agents, and methods for
identifying candidates
thereof. It has been demonstrated that HIV uses the CXCR4 as a co-receptor for
cellular entry
that can be blocked by its natural ligands and this malces a high affinity
ligand for CXCR4 an
important therapeutic target. GPCRs in general, and CXCR4 in particular, are
very difficult to
2o solubilize and purify because they normally need to fold and be maintained
in the presence of the
native lipids of the cell membrane. Simple expression and precipitation with
antibodies result
routinely in denatured aggregates with little or no ability to specifically
bind native ligands.
Accordingly, there is a need in the art for methods of identifying CXCR4
binding compounds
and identification of CXCR4 binding therapeutics with which to prevent or
treat diseases such as
25 AIDS. Such therapeutics may comprise peptides, peptidomimetics, or small
molecules that can
inhibit natural ligand binding to CXCR4. Such methods and compositions are
provided herein.
Summary of the Invention
The present invention provides binding compounds for CXCR4 and methods for
identifying those binding compounds. In one embodiment, screening methods are
provided to
3o identify binding motifs for CXCR4, as well as ligands capable of binding to
CXCR4. In another
embodiment, the invention comprises the design and identification of
therapeutic peptides,
peptidomimetics, or small molecules suitable for use in the prevention or
treatment of HIV and
AIDS.
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-6-
In one embodiment, methods of the invention provide for the synthesis and
purification
of linear and cyclic peptide libraries useful for screening and identifying a
binding motif for
CXCR4, as well as screening for potential ligands thereof. Methods of the
invention provide for
the incorporation of unnatural amino acids and amino acids of the D
configuration into linear or
cyclic peptides for use in such libraries. Libraries comprising peptides
having such amino acids
demonstrate enhanced binding affinity and duration of action i~ vivo resulting
from resistance to
proteolysis.
In a preferred embodiment, the invention provides fox the use of highly
diverse libraries
of peptide (linear and cyclic, natural and unnatural amino acids),
peptidomimetic, and small
to molecule compounds for the lead ligand identification step. Such Iigands
may be directly or
' indirectly agonistic or antagonistic to CXCR4 binding activity.
In a preferred embodiment, the invention provides for the use of phage display
methods
for the identification of preliminary motif information, followed by
additional rounds of affinity
purification with purified receptor preparations of the invention and highly
diverse libraries. In a
1s particularly preferred embodiment, phage display technology is combined
with the use of cyclic
peptide and/or peptidomimetic Libraries.
In another embodiment of the invention, computer-aided design technology is
used to
virtually screen, identify, design, or validate lead compounds for agonistic
or antagonistic
potential with regard to CXCR4 activity. Such technology uses computer-
generated, three-
2o dimensional images based upon molecular and structural information of both
the CXCR4 and the
potential binding partners by virtually aligung the protein with the binding
partners. In the case
of a library designed for computer-aided screening, a great deal of the
information necessary for
lead optimization is obtained directly from the library design. In one
embodiment, potential
leads are identified by prior screening of an actual Library or through some
other means. One
25 embodiment of the invention involves the screening of biologically
appropriate drugs that relies
on structure based rational drug design. In such cases, a three dimensional
structure of the
protein (or similar family member), peptide or molecule is determined and
potential agonists
and/or antagonists are designed with the aid of computer modeling. In a
preferred embodiment
of the invention, after an appropriate drug is identified, the drug is
contacted with CXCR4,
3o whereby a binding complex is formed between the potential drug and CXCR4.
Methods of
contacting the drug to CXCR4 are generally understood by anyone having slcill
in the art of drug
development.
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
In another embodiment, the present invention provides for the use of partially
purified
CXCR4 receptor protein as the agent for carrying out the selection,
identification, and
improvement of tight binding ligands in identifying therapeutically useful
compounds. In a
preferred embodiment, the invention comprises the use of tagging methods to
generate a
modified CXCR4 receptor protein that functions to facilitate purification and
identification steps
involved in the screening methods. In another embodiment, the invention
comprises a nucleic
acid sequence corresponding to the receptor CXCR4 fused to tag sequences
(i.e., GST, FLAG,
6xHis, dual tagged with FLAG-GST, C-MYC, MBP, V5, Xpress, CBP, HA) with
appropriate
specific protease sites engineered into the vector.
l0 In a particularly preferred embodiment, methods of the invention provide
for
solubilization and immobilization of CXCR4 to facilitate ligand selection
methods provided
herein. CXCR4 may be derived from any source, including without limitation:
inactive,
precipitated protein preparations; cell membrane preparations; and, whole cell
preparations. In
one embodiment, the invention provides for a method of screening combinatorial
libraries
directly for general aff nity determination using membranes from baculovirus
expression
systems or any other appropriate expression system. In one embodiment of the
invention,
partially purified CXCR4 is used in carrying out the selection,
identification, and improvement
of tight binding ligands. In a preferred embodiment, partially purified,
tagged CXCR4 is used in
a sequestered form to screen diverse libraries (focused or highly diverse) for
the affinity
2o purification of a tight binding ligand. In a highly preferred embodiment of
the invention, the
conditions for solubilization and immobilization of the appropriate ligand
provide for the use of
low salt, such as, for example, low magnesium or calcium conocentrations; and
no sodium
chloride ("NaCI") (O.OnM NaCI).
In another embodiment, the invention comprises the step of eluting bound
components of
the libraries from the immobilized protein with specific N-terminally blocked
peptides or other
non-sequencable analogs. In yet another embodiment, the invention comprises
the step of
binding combinatorial libraries to a resin-immobilized protein. In another
embodiment, the
invention comprises a purified polypeptide with tag sequences, which may be
immobilized onto
an appropriate affinity resin for assay. A further embodiment comprises the
step of releasing or
3o eluting tagged protein with its bound library with specific N-terminally
blocked peptides or other
non-sequencable analogs. In yet another embodiment, a method of the invention
comprises the
step of cleaving a tag from a protein of interest using a specific protease
(as designed into the
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
_g_
protein/vector) after immobilization onto an affinity resin and after the
combinatorial library is
bound to release the complex.
In yet another embodiment, the target ligand is selected from a linear peptide
library, a
peptidomimetic library, a cyclic peptide library, or a focused library
developed using an initial
motif identified by phage display techniques or a library combining any of the
foregoing. In
another embodiment, a target Iigand is eluted from the receptor preparation
using a peptide or
other ligand, or by using pH change or chaotropic agents, such as urea or
guanidine
hydrochloride, that can disrupt the hydrogen bonding structure of water and
denature proteins in
concentrated solutions by reducing the hydrophobic effect. Also contemplated
by the invention
1o are ligands for GXCR4 identified using the methods disclosed herein. In yet
another
embodiment of the invention, protein sequencing techniques are used for the
determination of
the structure of the ligand identified by the affinity purification step.
In another embodiment, the invention comprises therapeutic agents, such as,
for example,
a small molecule antagonist of CXCR4 binding that are identified using methods
of the invention
appropriate for the treatment of a disease or disorder, such as, for example,
HIV infection or
AIDS. In another embodiment, a patient infected with HIV is treated with a
therapeutic agent
comprising a compound identified using methods of the invention, or a small
molecule
antagonist of CXCR4 binding. In another embodiment, a patient infected with
HIV is treated
through the use of combinations of therapeutics that include, for example,
CXCR4 inhibitors and
2o reverse transcriptase and protease inhibitors.
A detailed description of certain preferred embodiments of the invention is
provided
below. Other embodiments of the invention are apparent upon review of the
detailed description
that follows.
Description of the Drawings
Figure 1 shows a peptide library with a fixed, non-degenerate lysine or
arginine and eight
degenerate positions consisting of eighteen amino acids in approximately equal
proportion.
Figure 2 shows a peptide library screening using binding domains.
Figure 2a shows a SDS-PAGE of cell Iysate containing CXCR4 and of purified
CXCR4
stained with coomassie.
3o Figure 3 shows an isolated human CXCR4 cDNA sequence.
Figure 4 shows a baculovirus transfer vector for CXCR4-HIS.
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-9-
Figure 5 shows a baculovirus transfer vector for CXCR4-FLAG.
Figure 6 shows a baculovirus transfer vector for CXCR4-GST.
Figure 7a is a chart showing the radioligand saturation binding studies using
membrane
preparations of GST-CXCR4 (High Five).
Figure 7b is a chart showing the displacement curves fox the radioligand
binding studies
using membrane preparations of GST-CXCR4 (High Five).
Figure 8 shows the immobilization of GPCRs for affinity purification from
libraries.
Figure 9 is a chart showing the displacement curves for CXCR4.
Figure 10 is a chart showing the high-throughput ligand-binding inhibition for
CXCR4.
to Figure 11 is a chart showing the ICSO's for CXCR4 analogs.
Figure 12a and Figure 12b are charts showing the inhibition of HIV infection
by CXCR4
peptide inhibitors.
Detailed Description of the Invention
Generally, methods of the invention provide for the determination of a binding
motif for
15 CXCR4. Further, methods of the invention provide for the identification of
agonists or
antagonists of the interaction of CXCR4 with its natural ligand, thereby
providing for the
identification of therapeutic lead compounds. Methods for library design and
synthesis, and
library screening that are particularly useful in the invention are described
in the following patent
and patent applications, the disclosure of each of which is incorporated by
reference herein:
2o Cantley et al., U.S. Patent No. 5,532,167; Cantley, et al., U.S.S.N.
08/369,643, filed December
17, 1998; Cantley, et al., U.S.S.N. 08/438,673, filed November 12, 1999; Hung-
Sen, et al.,
U.S.S.N. 091086,371, filed May 28, 1998; Hung-Sen, et al., U.S.S.N.
08/864,392, filed June 24,
1999; and Lai, et al., U.S.S.N. 09/387,590, filed August 31, 1999.
According to the methods of the invention, CXCR4 is cloned and expressed, and
tested
25 for activity. The CXCR4 may be tagged on the C-terminus or on the N-
terminus to facilitate the
determination of the character of the CXCR4's ligand-binding properties.
Exemplary tags
include, without limitation, 6xHis, FLAG, GST, V5, Xpress, c-myc, HA, CBD, and
MBP. The
tagged CXCR4 is used in screening of libraries comprising, for example, linear
and/or cyclic
peptides having natural and/or unnatural amino acids, peptidomimetics and/or
small molecules.
3o Such peptidomimetics and small molecules may comprise any natural or
synthetic compound,
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-10-
composition, chemical, protein, or any combination or modification of any of
the foregoing that
is used to screen for binding compounds of CXCR4.
In one aspect, an oriented degenerate peptide library method useful in methods
of the
invention uses soluble peptide libraries consisting of one or more amino acids
in non-degenerate
positions, known or suspected to be important for ligand binding, and eighteen
amino acids in
approximately equal proportions in degenerate positions. Cysteine and
tryptophan may be
omitted to avoid certain analytical difficulties on sequencing. Such a library
is shown in Figure
1, where X represents a degenerate position consisting of any of eighteen
amino acids and a
lysine or arginine is fixed at a non-degenerate position. Furthermore, the
selection of arginine or
to lysine as an orienting residue is based on the fact that basic residues of
gp120 are important
determinants in binding to CXCR4. Another aspect of the invention involves the
selection of
any amino acid as an orienting residue. Additional residues can be added to
the N-terminal of
the sequence shown in Figure 1 because there are often interfering substances
present in the first
and second sequencing cycles. Additional residues can be added at the C-
terminal end to
15 provide amino acids to better anchor the peptide to the filter in the
sequencer cartridge.
Another aspect of the invention provides for the use of highly diverse
libraries of peptide
(linear and cyclic, natural and unnatural amino acids), peptidomimetic, and
small molecule
compounds for the lead identification step. For example, these ligands can be
agonistic or
antagonistic in their function on the receptor. Generally, the invention uses
partially purified
2o CXCR4 as the agent for carrying out the selection, identification, and
improvement of tight
binding Iigands as a route to therapeutically useful compounds. In addition,
the invention
provides for the development and use of solubilization and immobilization
procedures that
facilitate efficient ligand selection methods provided herein. Specifically,
the optimal conditions
for the solubilization and immobilization for efficient ligand selection
comprise the use of low
25 salt, such as, for example, low or no magnesium or calcium concentrations,
and no NaCI
concentrations (O.OnM NaCI). Ligand selection methods using, for example,
inactive,
precipitated protein, cell membrane preparations, and whole cell preparations
are further
provided herein.
In one aspect of the invention, the screening step may comprise phage display
3o technology. Such phage display systems have been used to screen peptide
libraries for binding
to selected target molecules and to display functional proteins with the
potential of screening
these proteins for desired properties. More recent improvements of the display
approach have
made it possible to express enzymes as well as antibody fragments on the
bacteriophage surface
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-11-
thus allowing for selection of specific properties by selecting with specific
ligands. See e.g.,
Smith SF, et. al., Methods Enzym. 217:228-257 (1993). Phage display methods
may be used fox
the identification of preliminary motif information, and followed by
additional rounds of aff nity
purification with purified receptor preparations of the invention and highly
diverse libraries,
especially cyclic peptide and peptidomimetic libraries. The phage display
methods allow the
identif cation of motifs of natural amino acids. Information derived from
phage display can be
taken into affinity purification methods using, for example, synthetic
libraries containing novel
amino acid analogs or cyclic peptides to select ligands that have enhanced
pharmaceutical
characteristics. The use of initial, secondary and tertiary libraries allows a
more complete
to definition of the specificity of the binding site. Secondary libraries may
be sequenced
incorporating information from the initial library. With the first library,
some degenerate
positions may yield high preferences for specific amino acids and these may
become non-
degenerate positions consisting of the preferred amino acid in a second
library. See e.g., Wu R, J
Biol Chem 271(27):15934-41 (1996).
Alternatively, or in addition, computer-aided design technology may be used in
the
screening and/or designing of peptides, peptidomimetics, and small molecules.
Together with
information such as, fox example, the crystal structure of rhodopsin (see
e.g., Palczewslci, et al.,
Science 289(5480):739-745 (2000)) along with the sequence of CCRS,
transmembrane
predictions, and any structural information obtained from mutagenesis studies,
computer aided
2o design teclmology may virtually screen, identify, design and validate
potential compounds with
regards to their CXCR4 activity. Computer programs that may be used to aid in
the design of
appropriate peptides, peptidomimetics and small molecules include, for
example, Dock, Frodo
and Insight. An example of a method for screening of biologically appropriate
drugs relies on
structure based rational drug design. In such cases, a three dimensional
structure of the protein,
peptide or molecule is determined (or modeled after a close family member) and
potential
agonists and/or antagonists are designed with the aid of computer modeling.
See e.g., Butt et al.,
Scientific American, December 92-98 (1993); West et al., TIPS, 16:67-74
(1995); Dunbraclc et
al., Folding & Design, 2:27-42 (1997). After an appropriate drug is
identified, the drug is
contacted with CXCR4, wherein a binding complex forms between the potential
drug and
3o CXCR4. Methods of contacting the drug to CXCR4 are generally understood by
anyone having
skill in the art of drug development.
The screening step rnay be performed in solution phase, or with the CXCR4
immobilized
on affinity columns. In addition to the immobilization of tagged CXCR4 using
an affinity resin,
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
- 12-
other forms of sequestration can be used to perform the affinity purification
of select ligands
from libraries. These include, but are not limited to the following examples.
The receptor and
bound library components can be separated from non-bound library components
using
equilibrium dialysis. The tagged receptor can be bound to specific affinity
membranes, which
are in the form of plates or are separate. The libraries can then be incubated
with the membrane
and easily washed to remove non-specific binding components. Size exclusion
methodology can
be used to separate a purified receptor bound library complex from unbound
components after
pre-incubating the receptor with the library. Additionally, a micellar complex
containing the
receptor (which may or may not incorporate lipids as well as detergent) can be
separated after
to binding select affinity components from a library by differential
centrifugation. Generally, the
high affinity ligand can be released using low pH or high salt conditions and
the structure
identified by sequencing as described herein.
In order to determine those ligands that had the highest affinity to the
target receptor,
generally, over 200 peptide libraries were screened to determine each
library's respective
inhibition binding. In general, a greater than 10% inhibition at 100 ~M was
significant for
continued evaluation of the sequence via affinity purification. In additional
aspects of the
invention, once preferred amino acid residues are identified due to high
preference values by
CXCR4 at the degenerate positions of the library, specific peptides are
synthesized by the same
methods as employed for library synthesis. In one embodiment of the invention,
a high
2o preference value is greater than 1. The value is determined by subtracting
the control value from
the sample value and dividing by the reference value. In a preferred
embodiment of the
invention, the preference value is greater than 1.2. In a highly preferred
embodiment of the
invention, the preference value is greater than 2. After synthesis of the
identified peptide
sequence, the peptide is purified by, for example, High Performance Liquid
Chromatography
("HPLC") and compositions are confirmed by Matrix-Assisted Laser Desorption
Ionization-
Time of Flight Mass Spectrometer ("MALDI-TOF MS") and Edman Sequencing.
Generally,
relative affinities may be measured by modifying the radiolabel binding assay
used in receptor
purification.
To enhance the specificity of the motif obtained from the affinity purified
peptides, other
3o methods can be used. The bound components of the libraries can be eluted
from the
immobilized protein with specific N-terminally blocked peptides or other non-
sequencable
analogs. To avoid the release of minor contaminants from the affinity resin
after binding of the
library, the release/elution of the tagged CXCR4 with its bound libraxy can be
accomplished
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-13-
using specific N-terminally blocked peptides or other non-sequencable analogs.
This can be
done using acetylated FLAG peptide to elute CXCR4-FLAG receptor from the
resin.
Alternatively, the tag from CXCR4 may be cleaved using a specific protease (as
designed into
the protein/vector; either enterokinase or thrombin) after immobilization onto
the affinity resin
and after the combinatorial library is bound to release the complex. Finally,
libraries can be
prescreened for their ability to bind to the receptor (using significantly
less protein) by a binding
assay using CXCR4-containing membranes from, for example, Sft7 (Spodopte~a
f~ugiperda) or
High Five (Ti~ichoplusia hi) cells (both obtained from Invitrogen) in a single
assay or in an array
assay. This screening may be performed using CXCR4 and a number of linear and
cyclic
libraries to determine their effectiveness in inhibiting the natural ligand to
bind.
Methods of the invention further comprise the design of therapeutic agents
comprising
peptides, peptidomimetics, and/or small molecules that are antagonistic to
CXCR4 activity
appropriate fox the treatment of patients with a disease, such as AIDS.
Binding compounds for
CXCR4 and the identification of optimal synthesis and purification thereof
provides for an
effective treatment of AIDS and HIV infection. For example, the small peptide
ligand binding
compounds of the invention, both cyclic and linear peptide ligands,
demonstrate enhanced
binding affinity and action, and are resistant to proteolysis as identified,
for example, in Table 1.
Amino acids and peptides are abbreviated and designated following the rules of
the IUPAC-IUB
Commission of Biochemical Nomenclature in J. Biol. Chem. 247, 977-983 (1972).
Amino acid
2o symbols denote the L-configuration unless indicated otherwise,
In general, amino acids from the residues of gp 120 that are crucial for viral
uptake have
been used to specify fixed, or non-degenerate positions in the peptide
libraries that have been
designed for use in the oriented peptide library method described below and in
U.S. Patent
5,532,167, the disclosure of which is incorporated by reference herein. See
e.g., Rizzuto CD, et.
al., Science 280(5371):1949-53 (1998).
Table 1: The Sequences for the CXCR4-binding Peptides Discovered and Analogs
Thereof
CPI Pe ide
t Se
uence
1221 M AR S L I W RP A K A K K K
1312 A
1310 A
1301 A
1306 A
1295 A
1305 A
1304 A
1308 A
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
- 14-
13_07 A
1328 A
A
1251 Ac M A R S L I W R P A K A K K K
1334 G P
_1302 Tic
1303 Aib
1293 w
1309 2-Nal
1291 1-Nal
1296 F
1331 K K K A R S L I W R P A K A K K K
1300 F H E F R S L I W R P A K A K K K
1299 Y H E F R S L I W R P A K A K K K
1247 R S L I W R P A K A K K K
1248 f S L I W R P A K A K K K
1250 R S L I W R P A K
1249 M A R S L I W R P A K K K
1289 M A R S L I W R P A K A R R R
1297 A R S L I W R P A R R R R R
1365 A R S L I 2-NalR A A R R 2-NalR R
1366 AcA R S L I 2-NalR A A R R 2-NalR R
1330 F A R S L I E R A A R R W R R
1372 M A R S L I W E P A R R W R R
1373 M A R S L I W R P A E R W R R
1374 M A E S L I W R P A R R W R R
1377 M A R S L I W R P A R E W R R
1381 A R S L I 2-NalR L A R R 2-NalR R
1382 A R S I W R L A R R W R R
1384 A R S L I CI-FR L A R R CI-F R R
1389 F A R S L I 2-NalE A A R R 2-NalR R
1390 F A R S L I 2-NalA A R R 2-NalR R
1379 Aca r s I I 2-Nalr a a r r 2-Nalr r
1410 F A R S L I 2-NalR L A R R 2-NalR R
~
1411 Y A R S L I 2-NalR L A R R 2-NalR R
1457 F R S L I 2-NalR L A R R 2-Na!R R
1456 R R A R S L I 2-NalR A A R R 2-NalR R
1458 A R S L I 2-NalR TicA R R 2-NalR R
1448 AcA R S L I 2-NalR A A R R 2-NalR R
1443 A R S L I 2-NalR H A R R 2-NalR R
1424 K K K A R S L I 2-NalR L A R R 2-NalR R
1425 A R S L I W R L A R R W R R
1426 r r 2-nalr r a I r 2-nalI I s r a
1292 A R S L I W R P A K A K K K
1298 M A R S T I W R P A K A K K K
1329 M A A S L I W R P A K A K K K
1332 M A R S L I W R P A R R R R R
1629 A R S L I F4FR L A R R 2-NalR R
1630 A R H L f 2-NalR H A R R 2-NalR R
1631 H R S L I 2-NalR H A R R 2-NalR R
1632 2-NalR H A R R 2-NalR R
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-15-
1633 A R S L I 2-NalR L A R R F4F R R
1634 A R S L I 2-NalR ticA R R 2-NalR R
1641 2-Nal2-NalR H A R R 2-NalR R
1701 A R S L I 2-NalR P A R R 2-NalR R
Certain bodiments escribed
em of in
the the
invention following
are examples,
d which
are
not meant to be limiting.
EXAMPLES
EXAMPLE 1 : Preparation of Tagged CXCR4; Screening Of Linear Peptide Libraries
Various CXCR4 vectors were prepared for the baculovirus expression system
containing
epitope tags using standard techniques known by those skilled in the art that
allowed for easier
purification of the receptor. Tags may be incorporated at the N- or C-terminus
of proteins. For
CXCR4, tags were incorporated at the C-terminus of the receptor to determine
the character of
the receptor's ligand-binding properties that are in the N-terminal region of
the molecule to
to allow easier purification of the receptor. For CXCR4, tags were
incorporated at the C-terminus
of the receptor to determine the receptor's ligand-binding properties at the N-
terminal region of
the molecule to allow easier purification of the receptor. Tags were placed at
the N-terminus of
proteins. For CXCR4, tags may be incorporated either at the N- or C-termini of
the receptor.
There were no commercially available baculovirus transfer vectors with C-
terminal tags.
15 The construction of C terminal 6xHis tagged and C-terminal FLAG constructs
are provided
below as examples. Alternative tags may include, for example, GST, V5, Xpress,
c-myc, HA,
CBD, and MBP. These constructs were made by analogous procedures using
standard
techniques known by those skilled in the art.
The 6xHis tag enables a one-step purification using nickel chelation. The cDNA
for
2o CXCR4 was isolated from a spleen cDNA library using Polymerase Chain
Reaction ("PCR")
and primers for the 3' and 5' ends of CXCR4, as well as to the middle of the
gene. To create a C-
terminal 6xHis tag, CXCR4 was subcloned into an E. coli vector, pET30a, with a
C-terminal
6xHis tag. The newly created CXCR4-6xHis was then excised and ligated into
pBlueBac, a
baculovirus transfer vector (Invitrogen, Carlsbad, CA). The construct was
analyzed using both
25 restriction digest and sequencing, and transfected into Sf9 insect cells
(Pharmingen, San Diego,
CA) for expression as typically done by those skilled in the art of protein
expression.
A C-terminal bacterial FLAG construct was available from Sigma (pFLAG-CTC). A
similar strategy using standard techniques was employed for the construction
of this vector. The
CXCR4 was subcloned into the pFLAG-CTC plasmid, excised with the C-terminal
FLAG tag
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-16-
and then ligated into the digested pBlueBac vector. The construct was analyzed
using both
restriction digest and sequencing, and transfected into S~ or High Five insect
cells for
expression.
To express the CXCR4 gene in S~ or High Five cells, the pBlueBac vector
containing
the CXCR4 insert was cotransfected with Bac-N-Blue DNA using cationic liposome
mediated
transfection using standard techniques. The CXCR4 was inserted into the
baculovirus genome
by homologous recombination. Cells were monitored from 24 hours
posttransfection to 4-5
days. After about 72 hours, the transfection supernatant was assayed for
recombinant plaques
using a standard plaque assay. Cells which have the recombinant virus produce
blue plaques
to when grown in the presence of X-gal (5-bromo-4-chloro-3-indoyh-(3-D-
galactoside). These
plaques were purified and the isolate was verified by PCR for correctness of
recombination using
standard techniques. From this, a high-titer stock was generated and infection
performed from
this stoclc for expression worlc using standard techniques. Controls for
transfection include cells
only and transfer vector.
Sf7 or High Five cells were maintained both as adherent and suspension
cultures using
standard techniques known to those skilled in the art. The adherent cells were
grown to
confluence and passaged using the sloughing technique at a ratio of 1:5.
Suspension cells were
maintained in spinner flasks with 0.1 % pluronic F-68 (to minimize shearing)
for 2-3 months by
sub-culturing to a density of 1 x 106 cells/ml.
2o A time course after infection with recombinant virus was used to define
optimal growth
conditions for expression using standard techniques. Aliquots of cells from
spinner flasks were
taken fox this time course, centrifuged at 800 x g for 10 minutes at
4°C and both supernatant and
pellet assayed by SDS-PAGE/Western blot analysis. Figure 2a shows the SDS-
PAGE/Western
Blots of cell lysate containing CXCR4 and purified CXCR4 stained with
coornassie. The
CXCR4 was expected to be in the membrane fraction (pellet). All viable systems
were assayed
in this fashion for levels of expression. The systems with the best expression
levels was assayed
for activity using a standard binding assay on a membrane preparation using
SDF-1 (Chemicon)
and [l2sl~_SDF-1 (New Enghand Nuclear, "NEN", Boston, MA).
The membrane fraction was isolated by first pelleting the whole S~ cells (800
x g for 10
3o minutes at 4°C), then resuspending the pelhet in a lysis buffer with
homogenization. Typical
hysis buffer is around neutral pH and contains a cocktail of protease
inhibitors, all of which are
standard techniques for those skilled in the art. Membranes were pelleted.
Solubilization was
also conducted using varying NaCh concentrations. Despite conventional
thinking, the step of
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-17-
solubilization using low salt, for example, low calcium and magnesium
concentrations
substantially in the absence of NaCI provided unexpected optimal conditions
for solubilization
when compared for quantity and activity. Having O.OnM NaCI, although counter-
intuitive,
provided the best conditions when solubilizing and immobilizing candidates
with the binding
property of CXCR4. The solubilization of the receptor by different detergents
(such as, but not
limited to, (3-dodecylmaltoside, n-octyl-glucoside, CHAPS, deoxycholate, NP-
40, Triton X-100,
Tween-20, digitonin, Zwittergents, CYMAL, lauroylsarcosine, etc.) was compared
for quantity
and activity. A candidate for isolation was carried through for purification
as described below.
After determining an appropriate detergent for solubilization and activity,
such as, for
l0 example, Np-40, CXCR4 was purified from the membrane fraction. The exact
purification
scheme will depend on the construct chosen, which is subject to activity and
ease of
solubilization. For purification of the 6xHis-tagged CXCR4, the membrane
fraction was loaded
onto a Ni-NTA column (Qiagen, Valencia, CA) in the presence of detergent,
washed extensively,
and eluted with imidazole. Purification of the FLAG-tagged CXCR4 was performed
using the
anti-FLAG M2 affinity matrix (Sigma, St. Louis, MO) in the presence of NP-40
and eluted with
glycine. The purification was performed in the presence of NP-40 in the
experiment described
above. Activity of the purified receptor was assessed using a standard
binding/displacement
assay using SDF-1 and [laSl]_SDF-1.
Peptides were assembled on Riu~ amide resin (NovaBiochem, substitution level
0/.54
2o mmol/g) using an Applied Biosystems 433A synthesizer via 9-
fluorenylmethyloxycarbonyl/te~t.-
butyl ("Fmoc"/"tBu") based methods. tBu was used for the protection of side-
chains of Asp,
Glu, Ser, Thr, and Tyr, tert.-butyloxycarbonyl ("Boc") for Lys and Trp,
2,2,4,6,7-
pentamethyldihydrobenzofuran-5-sulfonyl ("Pbf') for Arg, and triphenylmethyl
("trityl", "Tut")
for Cys, His, Asn and Gln. The scale of the synthesis was 0.20 mmol. The resin
was initially
washed with N methylpyrrolidinone ("NMP") followed by a 1 x 3 minutes and 1 x
7.6 minutes
treatment of piperidine:NMP (1:4) for 1Va-Fmoc removal. All Fmoc-amino acids
were coupled
with N [(1H benzotriazol-1-yl)(dimethylamino)methylene]-N methylmethanaminium
hexafluorophosphate N oxide ("HBTU") according to the manufacturer's protocol:
(a) 1.0 mmol
of derivatized amino acid was dissolved in 2.1 g of NMP; (b) 0.9 mmol of 0.5 M
HBTU in N,N
3o dimethylformamide ("DMF") was added to the amino acid cartridge and the
solution was mixed
for 6 minutes; (c) 1.0 mL of 2.0 M N,N diisopropylethylamine ("DIEA") in NMP
was added to
the cartridge; (d) the HBTU solution was transferred to the resin and reacted
for 40 minutes at
ambient temperature while mixing. The resin was filtered and rinsed six times
with a total of 90
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-18-
ml of NMP and the cycle was repeated. In the one pot method to construct the
highly degenerate
oriented peptide libraries, a batch of resin was allowed to react with
mixtures of the
combinatorial amino acids without any partitioning of the resin.
Adjusting the concentrations of the amino acids in the starting mixture
controls the
relative coupling rates, thereby ensuring equal incorporation of the amino
acids in the library.
The optimization of a mixture of natural Boc and Fmoc protected amino acids
for the one pot
synthesis has been previously described (see e.g., U.S. Patent No. 5,225,533;
Ivanetich, et. Al.,
Corrabihato~ial Chemistry, vo1267, Academic Press, San Diego, CA USA, p 247-
260 (1996);
Buettner, et al., Innovations and Perspectives in Solid Phase Synthesis:
Peptides, Proteins, and
to Nucleic Acids, Mayflower Worldwide Ltd., Birmingham, UK, p 169-174 (1994);
Ostresh, et al.,
Biopoly~ae~s 34:1681-9 (1994); Songyang, et. al., Methods ih Mol Biol 87:87-98
(1998); and
Herman, et al., Molecular Diversity 2:147-155 (1996). Cleavage reactions were
performed by
stirring the peptidyl-resin in trifluoroacetic acid ("TFA"):H20:anisole:
triisopropylsilane
("iPr3SiH") (87.5:5:5:2.5, ~ 6 mL) for 3 hours at 25°C (see e.g.,
Herman et al., 1996). The
15 filtrates were collected and the resin was further washed with TFA. Cold (-
78°C) diethyl ether
was added to the combined extracts and the solution was cooled to -
78°C. After removing the
supernatant, the obtained precipitate was washed several times with cold
ether, dissolved in
glacial acetic acid and lyophilized.
For cyclic peptide libraries, Fmoc-Asp(OH)-ODmab (Dmab, 4-[N (1-(4,4-dimethyl-
2,6-
2o dioxoxcyclohexylidene)-3-methylbutyl)axnino]-benzyl) was side-chain
anchored to Rink amide
resin followed by chain elongation as described above. Following linear
assembly, removal of
the Dmab and Fmoc group was accomplished by treatments with hydrazine:DMF
(1:49) for 7
minutes and piperidine:NMP (1:4) for 6 x 3 minutes, respectively. The resin
was transferred to a
syringe containing a polypropylene frit for manual cyclization. On-resin head-
to-tail cyclization
25 was performed using 7-azabenzotriazol-1-yloxy)-tris(pyrrolidino)phosphonium
hexafluorophosphate ("PyAOP"):DIEA (1:2, 4 equiv) in a solution containing 1%
Trition X in
NMP:DMF:dichloromethane, methylene chloride, DCM) (1:1:1) for 2 hours at
55°C. The
unreacted linear precursor was treated with Fmoc-Nva-OH/PyAOP/DIEA ("Nva",
"norvaline")(1:1:2, 4 equiv) in DMF for 1 x 18 hours and 1 x 3 hours.
Subsequent cleavage and
3o side-chain deprotection as described above yielded a mixture containing a
cyclic peptide library
and the corresponding linear (uncyclized) sequences. The desired cyclic
peptide library was
purified to remove the linear contaminants by reversed-phase high performance
liquid
chromatography ("RP-HPLC").
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-19-
Peptides and peptide libraries were characterized by HPLC, MALDI-TOF MS
(Louisiana
State University), and Edman degradation. MALDI-TOF MS analysis is capable of
detecting the
presence of high-molecular weight impurities due to incomplete deprotection,
debloclcing, or re-
all~ylation. Edman degradation provides quantitative information about the
amount of each
amino acid in each degenerate position in a library.
The initial libraries synthesized had single, non-degenerate orienting amino
acids (i.e.,
M-X-X-X-X-R-X-X-X-X-A, where X is a degenerate equimolar mixture of all amino
acids
except cysteine). Cyclic libraries (head-to-tail) were also prepared with
single, non-degenerate
orienting amino acids. Through the use of these initial libraries, the optimal
residues at some
1o degenerate positions become defined and secondary libraries were made
fixing these positions.
For example, the head to tail cyclized library cyclo(M-X-X-X-X-R-X-X-X-X-N)
indicated that
the -4 position (from the fixed R) should be lysine, the -2 position should be
aspartate, the -1
position should be histidine, and the +3 position should be lysine so the
secondary library was
cyclo(M-K-X-D-H-R-X-X-K-N).
15 An oriented linear peptide library was applied to a column containing
immobilized
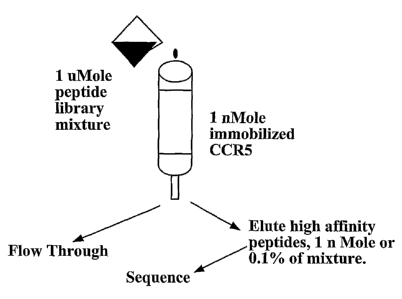
CXCR4 and a small fraction of isolated high affinity peptides. A schematic
diagram showing
the peptide library using binding domains can be seen in Figure 2. After
washing, bound
peptides were eluted from the column. Next, bound peptides and the entire
library applied to the
column were submitted individually to Edman degradation, to determine the
distribution of
2o amino acids as a function of position. Finally, the preferences of amino
acids at the degenerate
positions was determined. For example, if serine was 5% of the amino acids at
position +1 in
starting library but 15% of the amino acids in position +1 in the high
affinity peptides, there
would be a selection for serine at the +1 position. A preference value of 3 at
that position would
be obtained. Table 2 provides a selective review of the use of the peptide
library method with
25 binding domains.
Table 2: Use
Of Oriented
Linear Peptide
Libraries
To Determine
Preferred
Amino Acids
For Binding
Domains
(residue used
for orienting
sequence is
shown with
underline
(p = phospho-)
-- "pX"
Binding DomainPreferred PeptideKd (nM)
PDZ KKKKETDV 42
Src EPQ~YEEIPIYLK 80
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-20-
14-3-3 RLSH~SLP 55.7
~
SH2 (src) PYEEIY 100
SH3 PXRPXR
(axnphiphysin)
SHC NPXpY
Lim GPHydGPHydY/F
Example 2: Preparation of GST Tagged CXCR4; Screening Using Same
Libraries were synthesized on an ABI 433A (Applied Biosystems, Forster City,
CA) with
9-fluorenyhnethoxycarbonyl (Fmoc) protecting groups using a Rinlc Amide MBHA
resin
(substitution: 0.54 mmol/gm). To obtain approximately equal coupling of amino
acids for
degenerate positions, the amounts of amino acids are adjusted empirically
after considering
literature values. See e.g., Ostresh JM, et. al., Biopolyme~s 34(12):1681-9
(1994). The coupling
reagent was HBTU/HOBT/DIEA, 1 equivalent per equivalent of peptide. Cleavage
was effected
l0 by a cocktail (82% TFA, 5% phenol, 5% thioanisol, 2.5% 1,2-ethanedithiol,
5% water).
Peptides were precipitated from methyl tertiary butyl ether. Libraries were
characterized by
MALDI-TOF MS (Louisiana State University) and by amino acid sequencing.
The initial library used a single, non-degenerate basic amino acid (i.e., M-A-
X-X-X-X-R-
X-X-X-X-K-K-K). Secondary libraries were made fixing optimal residues found at
some
15 degenerate positions. For example, M-A-X-X-X-X-W-X-X-X-X-A-K-K-K may
indicate that the
-4 position should be arginine, -1 should be isoleucine, and +1 should be
arginine so the
secondary library would be M-A-R-X-X-I-W-R-X-X-X-A-K-K-K.
In the case of the 6xHis tagged CXCR4, the receptor was exposed to the
library, and
separation of free and bound peptides was accomplished by pelleting the
membranes by
2o centrifugation. The 6xHis-tagged CXCR4 purified receptor was incubated with
a peptide
library, about 1 p,mole of peptide and about 1 nmole of binding sites. After
incubation, receptor
with bound peptide was separated from unbound peptides by centrifugation
(receptor~peptide
complex in the pellet, unbound peptide in the supernatant). Nonspecifically
bound peptides were
removed by exhaustive washing, and resuspension of the pellet in low pH (<_
2.5) was used to
25 remove the bound peptide. This peptide was sequenced to determine the
consensus sequence.
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-21 -
When a FLAG-tagged CXCR4 was used for peptide library work, the receptor was
immobilized on an anti-FLAG M2 affinity matrix (St. Louis, MO). An additional
purification
approach used the CXCR4-GST construct and immobilized glutathione (Pierce,
Roclcford, IL).
Both the bound peptide mixture and the starting peptide library were sequenced
using
standard techniques. The amounts of each amino acid, as a function of
position, were
determined. Preference values for each amino acid at each position were
calculated by
comparing the amounts of amino acids present in the starting library and bound
fraction of
peptides. These procedures were used to generate preferred sequences of
peptides interacting
with many binding domains and have been described in Table 2.
to Also, secondary libraries were sequenced incorporating information from the
initial
library. For the first round of characterization, phage display technology is
also used to identify
preliminary binding motifs. The phage display method provides for the
identification of motifs
of natural amino acids. Phage display technology involves the insertion of DNA
sequences into
a gene coding for one of the phage coat proteins. The gene is inserted in a
particular location so
that the expressed protein insert can interact with other molecules. As a
result, the encoded
peptide or protein sequence will be presented on the surface of the phage and
exposed for
binding. By inserting degenerate nucleotides, each phage can express a
different peptide
sequence ("a phage library"). Incubation of this phage library with the
immobilized receptor can
be used to identify sequences which specifically bind to the receptor. Even
weak signals can
2o detected because they can be amplified by growing the isolated phage.
Information derived from
phage display is applicable to affinity purification methods using synthetic
libraries containing
novel amino acid analogs or cyclic peptides to select ligands that have
enhanced pharmaceutical
characteristics. The use of initial, secondary and tertiary libraries provided
a complete definition
of the specificity of the binding site.
Once preferred amino acids residues were identified using high preference
values by
CXCR4 at the degenerate positions of the library, specific peptides were
synthesized by methods
as employed for library synthesis. Peptides were then purified by HPLC and
compositions
confirmed by MALDI-TOF MS.
Relative affinities were measured by modifying the radiolabel binding assay
used in
3o receptor purification. Therefore, the ability of these peptides to displace
[l2sl]-SDF-1 from
purified CXCR4 membranes was measured.
Example 3: Preparation and Analysis of Tagged CXCR4; Screening Using Same
A. Cloning and Expression
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-22-
CXCR4 was isolated from a spleen cDNA library in two halves and spliced
together.
These two fragments were isolated using PCR technology and primers to the 3'
and 5' ends and
the middle of the CXCR4 gene. A full-length clone was not isolated with the 3'
and 5' primers;
however, two halves were isolated and ligated together using a unique BamHI
site in the gene.
s The identity of the construct was confirmed by sequencing. The sequence of
the isolate is in
Figure 3. An alternate splice shorter form was also isolated, which is called
CXCR4s. Tags
were added to the C-terminus of the receptor for use in immobilizing them for
affinity
purification assays using standard techniques. The following are specific
examples from
experiments using the tagging method.
l0 Const~~uction of CXCR4 with C-Terminal Histidine Tag (Insect Select
Expression System)
A previous construct containing the gene for GnRHR (gonadotropin releasing
hormone receptor)
was used to make the first CXCR4 construct. The gene for GnRIiR was spliced
out and replaced
by the isolated cDNA for CXCR4. This vector was originally the pet30a vector
with the 6xHis
tag at the C-terminus.
15 Cohst~uction of CXCR4 cohst~uct with C-te~mihal FLAG tag: PCR was performed
using
the primers 5' BspEl CXCR4 and 3' Bgl CXCR4 engineered with unique sites for
ligation of
CXCR4 in frame with the FLAG tag of pFLAG-CTC (a bacterial expression vector)
from Sigma.
This construct is called CXCR4-FLAG-CTC. CXCR4-FLAG was then removed by
digestion
and filled in with Klenow fragment. The fragment containing CXCR4-FLAG was
ligated into
2o pBluebac 4.5 that was first digested then blunted with Klenow. This final
construct is called
CXCR4-FLAG. The construct was confirmed by restriction digestion and
sequencing using
standard techniques. This construct has been used for expression and has been
determined to be
expressed sufficiently and in active form for use in affinity purification
screening.
Co~cstructioh of CXCR4 Construct with C-terminal GST tag: The newly
constructed
25 CXCR4-FLAG cDNA was removed from the CTC vector and subcloned into another
construct,
CCRS-GST, in place of the CCRS (using Bgl and BspEl ). This created the vector
for CXCR4-
GST using one step. The construct was confirmed by restriction digestion and
sequencing using
standard techniques. This construct has been used for expression and has been
determined to be
expressed sufficiently and in active form for use in affinity purification
screening.
3o Construction of CXCR4 with N termi~aal 6xHis tag: This construct was
prepared by
subcloning the CXCR4 into the commercially available vector, pBluebacHis2b
(Invitrogen).
The construct was confirmed by restriction digestion and sequencing using
standard techniques.
Plasmid maps for these vectors are found in Figures 4-6.
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
- 23 -
The vectors for the three new constructs (for CXCR4-FLAG, CXCR4-GST, and CXCR4-
HTS) were used to co-transfect S~ cells for the production of a viral stock of
each. These viral
stocks were purified using a standard plaque assay and then used in
experiments to infect for the
optimization of expression of CXCR4 with its various C-terminal tags. High
Five cells
(Invitrogen) were also transfected with these CXCR4 tagged constructs and
tested for expression
of CXCR4. All constructs were determined to express the appropriately tagged
receptor.
Expression levels after 72 hours were as much as 5 times greater in High Five
cells than those
for S~ cells. All of the above described experiments were done using standard
techniques
lcnovm to those skilled in the art.
1o A fourth construct for the expression of CXGR4 was made from the starting
vector
pBhueBac 4.5 (Invitrogen) to remove the thrombin and enterolcinase cleavage
sites in the
previously described vectors. The GST tag was added into the multiple cloning
site by using
PCR to generate the GST tag, then ligating into the digested vector
(SmaI/EcoRI) using standard
procedures known to those slcilled in the art. Next, the vector was made
compatible with the
Gateway technology from Lifetech for ease of manipulation. This was done by
ligating into the
SmaI site the cassette containing the recombination sites required for this
technology (from
Lifetech). CXCR4 was amplified using PCR with primers to extend the gene to
contain the
attachment sites for recombination. Then, the PCR product was incorporated
into the
baculovirus vector using BP clonase (the enzyme required for homologous
recombination) to
2o make a vector for baculovirus expression containing CXCR4 with a C-terminal
GST tag without
the enterohcinase or thrombin cleavage sites. This vector was cotransfected
into S~ cells for
preparation of the virus stock necessary for expression. The virus was plaque
purified, and a
PCR and sequence checked clone was used for expression of CXCR4. A time course
with this
construct showed that less proteolysis of the protein was observed and less
time was necessary to
obtain maximal expression of the receptor.
B. Act- ivity
Each of the tagged CXCR4 genes (CXCR4-FLAG, CXCR4-GST, and CXCR4-HIS)
were used to co-transfect S~ and High Five cells, as described in Example 1.
Whole celhs from
Sf9 and High five cell lines were lysed using hypotonic buffers (10 rnM Tris,
pH 7.4, 5 mM
3o EDTA), and membrane preparations were made by homogenization and
centrifugation using
standard techniques known to those slcilled in the art. Membrane preparations
for CXCR4-GST,
CXCR4-FLAG, and CXCR4-HIS were assayed using a standard radioligand binding
assay. The
radiohigand (lasT]_SDF-la was incubated with membranes (0.5 ~,g) in binding
buffer at 27°C for
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-24-
1 hour with and without unlabelled SDF-1. For filtration and washing, the
reaction was
transferred to Millipore Multiscreen filter plates (HATF 0.45~,m; pre-blocked
with 10% BSA),
filtered using vacuum, washed 4-5 times with 200 ~,L ice-cold buffer, and
radioactive counts
bound were detected using scintillation counting. All points were done in
triplicate. Uninfected
cells were used as a control for this experiment. To determine the KD,
saturation binding was
measured using increasing concentrations of [l2sl]_SDF-la (from 0.5 nM to 10
nM).
Nonspecific binding was measured in the presence of 400 nM unlabelled SDF-la.
Competitive
binding assays were performed by incubating CXCR4-containing membranes with
0.5 nM [125I]-
SDF-Ia and serial dilutions of unlabelled SDF-la or peptide Iigand. Analyses
of KD and ICso
to were performed using non-linear curve fitting in Kaleidagraph. Figure 7
generally shows charts
exemplifying radioligand binding studies using membrane preparations of GST-
CXCR4 (High
Five). Saturation binding demonstrated a KD of 3.27 nM as seen in Figure 7a.
Competitive
binding assays yielded an ICso of 12.5 nM and a Hill coefficient of 0.93 as
seen by the
displacement ciuves in Figure 7b. The assays analyzed for the charts in Figure
7 were performed
by methods provided herein, and were performed in triplicate. In a preferred
embodiment,
methods of the invention yield at least 20% active protein.
C. Solubilization
Both lysed whole cells and membrane preparations have been used for
solubilization.
Solubilization of the tagged versions of CXCR4 (CXCR4-FLAG, CXCR4-GST, and
CXCR4-
2o HIS) have been performed using many different combinations of detergents
(NP-40, Triton X-
I00, ~i-D-maltoside, n-octylglucoside, CYMAL, Zwittergents, Tween-20,
lysophosphatidyl
choline, CHAPS, etc.), salts (NaCl, CaCl2, MgCl2, MnClz, KCI, etc.), buffers
(Tris, Hepes,
Hepps, Pipes, Mes, Mops, acetate, phosphate, imidazole, etc.), and various
pH's (range 6.8-8.2).
Conditions for optimal solubilization were found using Zwittergent 3-14 and
low salt, e.g. low
magnesium and calcium, but no NaCI (O.OnM NaCI) and buffered at pH 8.1. In a
preferred
embodiment, at least 20% of the solubilized, immobilized protein is active. In
highly preferred
embodiments, at least 30%, 40%, 50% and 75% of the solubilized, immobilized
protein is active.
D. Immobilization
After solubilization, CXCR4-GST were immobilized onto affinity columns for
3o purification and as an active protein ready for use in screening of peptide
libraries. A schematic
diagram showing the immobilization of GPCRs for affinity purification from
libraries is shown
in Figure 8. CXCR4-GST was bound and immobilized onto glutathione-agarose
(Pierce) and
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
- 25 -
glutathione-sepharose (Amersham Pharmacia Biotech). The immobilization of the
functional
protein was accomplished by first solubilizing the receptor in 0.3% NP-40, 10
mM Hepes (or
Pipes), pH 7.5, then binding it to the glutathione-sepharose resin in 0.3% NP-
40, 10 mM Hepes
(or Pipes), pH 7.5, 3 mM CaCl2, 15 mM MgCl2. The activity of the immobilized
receptor was
s determined by incubation for 1 hour at 4°C with the radiolabeled SDF-
la, (as with the membrane
assay above) and competition with cold SDF-1 a. Uninfected cells were used as
controls for this
activity, as well as the column alone. These experiments demonstrated the
ability to immobilize
microgram quantities of the receptor in pure form (sufficient for affinity
purification screening;
see Figure 8) onto resin in active form. Figure 9 is a chart showing the
displacement curves for
CXCR4. Binding displacement experiments were performed both on CXCR4-
containing
membranes and on the immobilized receptor with radiolabeled SDF-1 displaced by
increasing
concentrations of cold SDF-1.
E. Peptide Library Synthesis
Libraries were synthesized on an ABI 433A (Applied Biosystems, Forster City,
CA) with
9-fluorenylmethoxycarbonyl (Fmoc) protecting groups using a Rink Amide MBHA
resin
(substitution: 0.54 mmol/gm). When a mixture of amino acids was used for
degenerate
positions, the approximately equal coupling of amino acids was obtained by
adjusting the
amounts of amino acids empirically after considering literature values. See.
e.g., Ivanetich et.
al., Combinatorial Chemistry, vol 267, Academic Press, San Diego, CA USA, p
247-260 (1996).
2o The coupling reagent was HBTU/HOBT/DIEA, 1 equivalent per equivalent of
peptide.
Cleavage was effected by a cocktail (82% TFA, 5% phenol, 5% thioanisol, 2.5%
1,2-
ethanedithiol, 5% water). Peptides were precipitated from methyl tertiary
butyl ether. Libraries
were characterized by MALDI-TOF MS (Louisiana State University) and by amino
acid
sequencing.
The initial libraries used a single, non-degenerate basic amino acid (i.e., M-
X-X-X-X-
W-X-X-X-X-A-K-K-K). Through the use of these initial libraries, the optimal
residues at some
or all degenerate positions became defined. Secondary libraries were made if
not all of the
positions were defined, fixing the defined positions.
F. Screening of Peptide Libraries Using Immobilized CXCR4
3o With active, large quantities of protein (1 nmol) immobilized to the
specific resin (for
example, CXCR4-GST to glutathione-sepharose), screening of billions of
compound can take
place by incubating them together and allowing the natural preferences and
binding affinities to
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-26-
purify the ligands which are preferred by CXCR4. These experiments have been
performed with
eleven libraries and may be performed with other libraries.
To identify which peptide libraries to screen, a membrane binding assay was
developed
to use with the peptide libraries. Each peptide library (100 ~,M final
concentration, average
molecular weight) was incubated with the receptor (in membranes) and the
ability of the peptides
in the library to inhibit (Iasl]-SDFla binding was determined. Figure 10 is a
chart showing the
high-throughput ligand-binding inhibition for CXCR4. CXCR4-containing
membranes were
incubated in the presence of 100 ~M library and radiolabeled SDF-1. Percent
inhibition was
calculated to determine the effect of each library. Peptide libraries with the
highest percent
1o inhibition were assayed first.
Approximately 500 mL of I x I06 cell/mL of High Five cells expressing CXCR4-
GST
were used per affinity purification. Immobilized CXCR4-GST (as described
herein) was
incubated with 1 mg of the peptide library CPI-10064 (M-A-X-X-X-X-W-X-X-X-X-A-
K-K-K)
for 20 minutes at room temperature. Unbound peptides were removed by washing.
Bound
peptides were eluted with 30% acetic acid. Eluted peptide was filtered using a
Centricon-10 to
remove any protein that might have co-eluted with acetic acid. The filtrate
was dried under
vacuum, dissolved in water and subjected to peptide sequencing.
The sequence for the consensus motif for the specific ligand was identified
from this
screening to be M-A-R-S-L-I-W-R-P-A-K-A-K-K-K. The affinity for the receptor
was
2o determined using standard radioligand displacement methodology as performed
by those slcilled
in the art. This ligand was determined to have an ICSO of ~60 ~,M. Figure 11,
for example, is a
chart showing the ICSO for CXCR4 analogs. Selective analogs increased binding
inhibition. CPI-
1221, a 15-mer, was the original peptide. Analogs of this peptide were made to
increase
specificity of binding as demonstrated by the curves in Figure 11.
Rational analoging of this peptide was performed. Minimal length of the
peptide
sequence was determined by deleting from both the N- and C-termini. Approaches
to protect
against metabolism were conducted using synthetic analogs. Insertion of a
rigid segment of the
peptide was accomplished with cyclic amino acids such as azetidine-2-
carboxylic acid,
tetrahydroisoquinoline (Tic), pipecolic acid (Pip), thiazolidine-4-carboxylic
acid (Thz), 1-amino-
1-cyclopentane-carboxylic acid and 1-amino-1-cyclohexanecarboxylic acid
(Sawyer, 1995) as
well as a,a-dialkyl residues such as aminoisobutyric acid (Aib) and
diethylglycine (Deg). For
refinement of activity and bioavailabilty, unnatural amino acids were
substituted into a sequence.
Analogs of Tyr, Phe (aromatic substitution), Arg, Lys (1VG, IVG-dimethyl-
arginine, 2,3-
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
_27-
diamW opropanoic acid), Ala, Gly, Val, Leu (oc-allylalanine, 2,3-
diphenylglycine, phenylglycine,
4,4,4-trifluorovaline, 5,5,5-trifluoroleucine, (3-dimethylaminoleucine) as
well as analogs for the
other proteogenic amino acids were examined. Hydrophobic unnatural amino acid
such as
naphthylalanine and cyclohexylalanine substitutions were used successfully.
Table 3 is a
summary of the substitutions made at the various positions in the peptide
ligand which were
synthesized and screened for activity using inhibition of [l2sl]_SDFla binding
and toxicity on
Jurlcat and CEM cells. In addition, all positions were individually
substituted with Ala, several
positions were substituted with D-amino acids, the N-terminus of several
peptides were
acetylated, deletions of the N- and C-termini were made, and several retro-
inverso peptides were
to synthesized. All of these peptides and peptide analogs all were discovered
to bind to CXCR4
and inhibit SDF1 a binding with varying degrees of activity. Examples of the
binding inhibition
curves for the original peptide and several analogs can be seen in Figure 11.
These curves
demonstrate the reduction in potency obtained by the analoging. Several of
these peptides have
also been screened for activity in prevention of HIV activity and determined
to be effective in
specifically blocking the entrance of HIV into the cells with ECSO's in the nM
range. For
example, the charts in Figure 12 shows the inhibition of HIV infection by
CXCR4 peptide
inhibitors. Specifically, Figure 12 (a) shows the infection by HIVaib was
completely inhibited by
CPI-1500 with an ECSO of 280 nM. CPI-1500 (ARSLI(2-Nal)R(Tic)ARR(2-Nal)RR)
also
demonstrated specificity for the X4 virus IITb over the RS virus 9881. Figure
12 (b) shows that
other CXCR4 peptide inhibitors demonstrated inhibition of the X4 virus, with
specificity, as
well.
Table 3: Peptides and Peptide analogs for CXCR4. Standard one-letter
abbreviations are
used for the 20 natural amino acids. Other abbreviations: 2-Nal (2-
naphthalalanine); 1-Nal (1-
naphthalalanine); Cl-F (chloro-phenylalanine); F-4-F (4-phenyl-phenyalanine);
Aib
(aminoisobutyric acid); Tic (tetrahydroisoquinoline); hArg(R2) (homo-arginine
where R = lower
alkyl substitution especially ethyl); Hyp (hydroxyproline); Orn (ornithine);
Pya (3-pyridyl-
alanine); Phg (phenylglycine); Dap (2,3~-diaminoproprionic acid); and Cha (0-
cyclohexyl-
alanine).
3o A'-B'-C'-D'-E'-E'-F'-C'-G'-F'-C'-B'/C'-F'/C'-C'-C'
Where: A'=M, K, hArg(R2), Orn, Dap
B'=A,L,I, Cha
C'= R,K,hArg(R2), Orn, Dap
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
_2g_
D'=S,A,T
E'=L, I, V, F, Pya, Phg
F'= W, I-Nal, 2-Nal, 4Cl-Phe, 4F-Phe, Pya
G'= P, H, Tic, A, Hyp, azetidine-2-carboxylic acid
G. Phase Display
As an alternative or additional method useful in screening, immobilized
functional
CXCR4 was used to isolate phase which bind to CXCR4 using standard techniques
known to
those skilled in the art. Subtraction of the baclcground from the glutathione-
sepharose beads,
to BSA, and SDFI a used in the assay was performed by incubation of the phase
library with the
mixture of these components. After incubating subtracted phase libraries
(i.e., NEB PhD C7C)
with the receptor, bound phase were eluted both with the natural CXCR4 ligand
(SDFla) and
with glycine, pH 2.2. PhD C7C is a particular phase library with 7 random
amino acids between
disulfides, and may be obtained from New England Biolabs ("NEB"). Multiple
rounds of
15 screening were performed. Both conditions have provided specific sequences
(see Table 4)
which bind to CXCR4 and inhibit ligand binding.
Table 4: Phase sequences isolated from CXCR4 screening. Standard one-letter
abbreviations
are used for the 20 natural amino acids.
P A H Y P M~ L
Q Y A T P N I~
Q Q R S T A F
P F R A T T E
T D I~ L L L D
H T Q H V R T
L G V I~ A P S
D L Q A R Y S
S L T E P S L
S T W P L A
R T T S D A L
Example 4: Prevention or Treatment of HIV Infection or AIDS
Presently, certain complications, however, are encountered during the
production,
formulation and use of therapeutic peptides, peptidomimetic, or small molecule
antagonists or
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-29-
agonists of CXCR4 binding used for the prevention and treatment of AIDS and
HIV infection.
Biologically appropriate antagonists or agonists that minimize the cost and
technical difficulty of
commercial production of therapeutic binding compounds of CXCR4 are further
contemplated
by the present invention. In addition, biologically appropriate antagonists or
agonists of CXCR4
binding that do not confer an immunilogical response to the antagonist or
agonist such that it
interferes with the effectiveness thereof are contemplated by the invention.
Moreover,
appropriate formulations that confer commercially reasonable shelf life of the
produced
antagonist or agonist of CXCR4 binding, without significant loss of biological
efficacy are
contemplated in the present invention. Furthermore, useful dosages for
administration to an
to individual are contemplated in the present invention appropriate for the
prevention and treatment
of AIDS and HIV infection.
The identification of appropriate candidates that, alone or admixed with other
suitable
molecules, that are competent in inhibiting CXCR4 binding are contemplated by
the invention.
Such candidates further contemplate the production of commercially significant
quantities of the
aforementioned identified candidates that are biologically appropriate for the
prevention and
treatment of AIDS and HIV infection. Moreover, the invention provides for the
production of
therapeutic grade commercially significant quantities of CXCR4 binding
antagonists, agonists or
derivatives in which any undesirable properties of the initially identified
analog, such as in vivo
toxicity or a tendency to degrade upon storage, are mitigated.
2o Methods of preventing and treating AIDS and HIV infection also, after the
identification
and design of a peptide, peptidomimetic, or small molecule antagonist of CXCR4
binding
activity, comprise the step of administering a composition comprising such a
compound capable
of inhibiting CXCR4 binding as described herein. Administration may be by any
compatible
route. Thus, as appropriate, administration may include oral or parenteral,
including intravenous
and intraperitoneal routes of administration. A particularly preferred method
is by controlled-
release injection of a suitable formulation. In addition, administration may
be by periodic
injections of a bolus of a composition, or may be made more continuous by
intravenous or
intraperitoneal administration from a reservoir that is external (e.g., an
intravenous bag) or
internal (e.g., a bioerodable implant).
3o Therapeutic compositions contemplated by the present invention may be
provided to an
individual by any suitable means, directly (e.g., locally, as by injection,
implantation or topical
administration to a tissue locus) or systemically (e.g., parenterally or
orally). Where the
composition is to be provided parenterally, such as by intravenous,
subcutaneous, intramolecular,
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-30-
ophthalmic, intraperitoneal, intramuscular, buccal, rectal, vaginal,
intraorbital, intracerebral,
intracranial, intraspinal, intraventricular, intrathecal, intracisternal,
intracapsular, intranasal or by
aerosol administration, the composition may comprise part of an aqueous or
physiologically
compatible fluid suspension or solution. Thus, the carrier or vehicle is
physiologically
s acceptable so that in addition to delivery of the desired composition to the
patient, it does not
otherwise adversely affect the patient's electrolyte and/or volume balance.
Useful solutions for parenteral administration may be prepared by any of the
methods
well known in the pharmaceutical art, described, for example, in REM1NGTON'S
PHARMACEUTICAL SCIENCES (Gennaro, A., ed.), Maclc Pub., 1990. Formulations of
the
1o therapeutic agents of the invention may include, for example, polyallcylene
glycols such as
polyethylene glycol, oils of vegetable. origin, hydrogenated naphthalenes, and
the lilce.
Formulations for direct administration, in particular, may include glycerol
and other
compositions of high viscosity to help maintain the agent at the desired
locus. Biocompatible,
preferably bioresorbable, polymers, including, for example, hyaluronic acid,
collagen, tricalcium
15 phosphate, polybutyrate, lactide, and glycolide polymers and
lactide/glycolide copolymers, may
be useful excipients to control the release of the agent in vivo. The concept
of a controlled
release injectable formulation for peptide drugs is well-accepted and offers
several advantages.
First, for example, bioavailabilities are high. Second, treatment regimens can
consist of once per
month or per three months (like Abbott's Leupron0), or once per year (e.g.
Alza's Viadur~).
2o Third, controlled release injectable formulations substantially reduces the
doses that can be used
(the Leupron injection dose is 1 mg/day but the 90 day formulation uses is
11.25 mg total).
Also, increased efficacy can be achieved if the therapeutic is present
continuously to prevent
infectivity. This consideration is particularly important in view of the need
to approach a cure
for this disease by preventing the reformation of slow-to-clear deposits of
infection such as the
25 memory T cell compartment. See e.g., Lee, V., ed. Peptide and P~oteisz Drug
Delivef y. Marcel
Deldcer, Inc., NY (1991).
Other potentially useful parenteral delivery systems for these agents include
ethylene-vinyl acetate copolymer particles, osmotic pumps, implantable
infusion systems, and
liposomes. Formulations for inhalation administration contain as excipients,
for example,
30 lactose, or may be aqueous solutions containing, for example,
polyoxyethylene-9-lauryl ether,
glycocholate and deoxycholate, or oily solutions for administration in the
form of nasal drops, or
as a gel to be applied intranasally. Formulations for parenteral
administration may also include
CA 02403784 2002-09-20
WO 01/70768 PCT/USO1/09160
-31 -
glycocholate for buccal administration, methoxysalicylate for rectal
administration, or cutric acid
for vaginal administration.
Additional aspects and embodiments of the invention are apparent to the
skilled artisan.