Note: Descriptions are shown in the official language in which they were submitted.
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
1
COMPOSITIONS AND METHODS FOR EPITOPE MAPPING
BACKGROUND OF THE INVENTION
The present invention relates generally to drug
development and diagnostics and more specifically to
immunological assays for determining epitope expression.
Greater than 300,000 different proteins are
estimated to be present in humans. Of these proteins,
there are about 15,000 potential molecular therapeutic
targets. To date, less than 1000 have been identified
and exploited for pharmaceuticals. In an attempt to
identify which of the remaining 299,000 proteins are
viable pharmacological targets, various genomic tools
have been developed to analyze anomalies in the genetic
code or mRNA levels.
Genomics has been developed over the last
decade in part to identify new targets and has led to the
development of new diagnostic methods. Leads identified
by changes in mRNA levels have fueled the high throughput
screening groups of the major pharmaceutical companies,
many of which screen as many as 100 targets per year.
The genomics approach is, however, limited in that a
disease is manifested at the protein level. Therefore,
the changes in mRNA levels that form the cornerstone of
genomics is a poor approximation for biochemical changes
in a diseased tissue. Biological function, or aberrant
function, is the result of changes in protein levels or
processing. The correlation between mRNA levels and
protein expression is less than a 0.5 (Anderson and
Seilhamer, Electrophoresis 18:533-537 (1997)). With the
measurement of changes in mRNA using the tools of
genomics, the actual biologically active species, the
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
2
proteins, are not assessed. In addition, genomic
analysis has no way of identifying changes in post
translational modification, such as glycosylation or
phosphorylation. It is only by direct analysis of the
proteins that changes indicative of a disease will become
evident.
Consequently, the actual success rate for
genomic leads consequently is very low. Following
identification of a lead from genomics analysis, the
protein must be expressed in a variety of cell or animal
models in order to attribute functionality or some
correlative property between the protein and a disease.
The direct measurement of protein levels or processing
within a diseased tissue would greatly enhance the
success rate of target identification and eliminate some
of the intermediate steps necessary for validating a
target.
In part due to the limitations of genomic
analysis and in part due to the need to functionally
characterize genomic leads, the field of proteomics was
developed. In spite of its acknowledged advantages over
genomics for identifying biologically significant changes
in protein levels as the result of a disease state,
proteomics has lagged in its incorporation into the
biotechnology sector and drug discovery efforts. This
shortcoming is the result of reliance on the adaptation
of old techniques to proteomics studies, particularly
mass spectroscopy and 2-D electrophoresis. While these
techniques have been available for over thirty years,
automation, reproducibility, quantification, and rapid
throughput have proven to be formidable hurdles blocking
the incorporation of proteomics into the discovery stream
of biotechnology.
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
3
The traditional techniques for proteomics,
2D-electrophoresis and mass spectroscopy, are technically
limiting in that only about 200 of the proteins loaded on
a 2D-electrophoresis gel are visible, and of those, only
the proteins with masses ranging between 10 kDa and 100
kDa are readily separated. Relevant expression
differences are difficult to assign and validate since
multiple gels are difficult to prepare in a reproducible
manner. As a result of these technical hurdles, the
study of proteomics has not found its place in the drug
discovery pipeline.
Thus, there exists a need for convenient and
efficient methods to analyze proteins and modifications
thereof for drug discovery and diagnostic purposes. The
present invention satisfies this need and provides
related advantages as well.
SUMMARY OF THE INVENTION
The invention provides a composition comprising
a diverse population of reagent ligands attached to a
solid support and a diverse population of antibodies
specifically bound to the reagent ligands. The ligands
can be peptides, oligosaccharides, oligonucleotides, or
organic molecules. The invention additionally provides
methods of determining an epitope in a sample by
contacting a composition comprising a diverse population
of reagent ligands attached to a solid support and a
diverse population of antibodies specifically bound to
the reagent ligands with a sample; and detecting the
antibodies bound to the diverse population of reagent
ligands. The invention further provides methods of
diagnosing a disease, identifying a potential therapeutic
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
4
agent, and mapping accessible epitopes of a polypeptide
using invention compositions.
BRIEF DESCRIPTION OF THE DRAWINGS
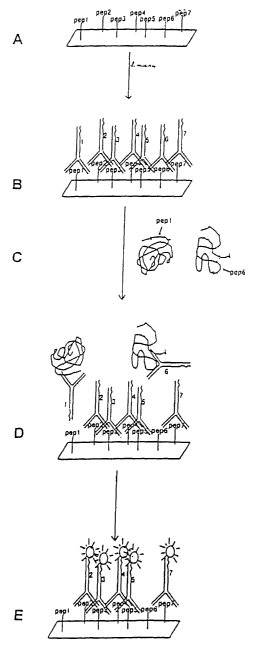
Figure 1 shows an outline for determining
epitope expression. In step A, a combinatorial peptide
library is synthesized on a solid support. In step B,
antibodies are specifically bound to the peptides to form
a ProtoChip. In step C, a sample is applied to the
. ProtoChip. In step D, epitopes expressed in the sample
competitively bind to the antibodies. In step E,
antibodies remaining bound to the peptides are
visualized.
Figure 2 shows the construction of a peptide
library.
Figure 3 shows the ScFv plasmid for expression
of a recombinant antibody library.
DETAINED DESCRIPTION OF THE INVENTION
The present invention provides a composition
comprising a plurality of reagent ligands attached to a
solid support and a plurality of reagent antibodies
specifically bound to the ligands, which is termed a
ProtoChip. The ligands can be peptides,
oligonucleotides, oligosaccharides or other organic
molecules. The invention also provides methods of
determining epitope expression in a sample using a
ProtoChip. The present invention draws from the fields
of molecular biology, immunology, combinatorial
chemistry, and high throughput screening. The present
invention can be advantageously used to overcome the
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
difficulties associated with traditional proteomics
techniques such as mass spectroscopy and 2-D
electrophoresis.
The present invention provides an advancement
5 of useful proteomics techniques that uses aspects of the
competitive immunoassay and is readily automatable for
the mapping of the epitome, an analysis of epitopes
expressed in a cell. The present invention provides a
method that is rapid, reproducible, quantifiable, and
provides an accurate snapshot of the proteome. Among
many applications, the present invention can be applied
to drug target discovery, diagnostics, drug development,
pharmacoproteomics, agricultural biotechnology, and
structural bioinformatics.
The invention ProtoChip has advantages over
current proteomics methodology. Essentially all possible
epitopes can be quantified using the invention ProtoChip,
with no size restriction for proteins or peptides. All
proteins that can be solubilized, even membrane bound
proteins that are difficult to analyze by traditional
proteomics techniques such as 2D electrophoresis, can be
quantified with the invention ProtoChip. The invention
allows for highly reproducible results, which can be
readily compared from experiment-to-experiment. The
invention allows detection of proteins 2 to 3 orders of
magnitude lower in concentration than by electrophoresis.
Known proteins can be easily quantified using methods of
the invention.
The invention can be used in diagnostic
applications and provides advantages similar to those
observed with nucleic acid based diagnostics. These
advantages include product standardization,
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
6
miniaturization, automation, and information management.
The invention provides advantages over other
immunochemistry based assays, including improved
sensitivity and specificity and allowing simultaneous
analysis of multiple epitopes. The invention is also
advantageous in that automation of all steps of sample
processing can be readily achieved.
As used herein, a "ligand" refers to a molecule
that can specifically bind to an antibody. The term
specifically means that the binding interaction is
detectable over non-specific interactions by a
quantifiable assay. A ligand can be essentially any type
of molecule such as a peptide or polypeptide, nucleic
acid or oligonucleotide, carbohydrate such as
oligosaccharides, or any organic derived compound.
As used herein, a "reagent ligand" refers to a
ligand used as a reagent for analysis of a sample, that
is, a non-analyte ligand. Although a reagent ligand can
be derived from a natural source or chemically
synthesized, it is understood that a reagent ligand
specifically excludes ligands in a sample to be analyzed.
As used herein, the term reagent ligand specifically
excludes antibodies, that is, the reagent ligand is a
non-antibody ligand.
As used herein, the term "polypeptide" refers
to a peptide, polypeptide or protein of two or more amino
acids. A polypeptide can also be modified by naturally
occurring modifications such as post-translational
modifications or synthetic modifications, including
phosphorylation, lipidation, prenylation, sulfatio.n,
hydroxylation, acetylation, addition of carbohydrate,
addition of prosthetic groups or cofactors, formation of
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
7
disulfide bonds, proteolysis, assembly into
macromolecular complexes, and the like.
A modification of a peptide can also include
non-naturally occurring derivatives, analogues and
functional mimetics thereof generated by chemical
synthesis. Derivatives can include chemical
modifications of the polypeptide such as alkylation,
acylation, carbamylation, iodination, or any modification
that derivatizes the polypeptide. Such derivatized
molecules include, for example, those molecules in which
free amino groups have been derivatized to form amine
hydrochlorides, p-toluene sulfonyl groups, carbobenzoxy
groups, t-butyloxycarbonyl groups, .chloroacetyl groups or
formyl groups. Free carboxyl groups can be derivatized
to form salts, methyl and ethyl esters or other types of
esters or hydrazides. Free hydroxyl groups can be
derivatized to form 0-acyl or 0-alkyl derivatives. The
imidazole nitrogen of histidine can be derivatized to
form N-im-benzylhistidine. Also included as derivatives
or analogues are those polypeptides which contain one or
more naturally occurring amino acid derivatives of the
twenty standard amino acids, for example,
4-hydroxyproline, 5-hydroxylysine, 3-methylhistidine,
homoserine, ornithine or carboxyglutamate, and can
include amino acids that are not linked by peptide bonds.
As used herein, the term "nucleic acid" or
"oligonucleotide" means a polynucleotide such as
deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
As used herein, the term "oligosaccharide" refers to
polymers of monosaccharides that can be linear or
branched. Oligosaccharides include modifications of
monosaccharides. As used herein, the term "organic
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
8
molecule" refers to organic molecules that are chemically
synthesized or are natural products.
As used herein, the term "antibody" is used in
its broadest sense to include polyclonal and monoclonal
antibodies, as well as antigen binding fragments of such
antibodies. An antibody useful in the invention, or
antigen binding fragment of such an antibody, is
characterized by having specific binding activity for a
ligand or sample epitope of at least about 1 x 105 M'1.
Thus, Fab, F(ab')2, Fd, Fv, single chain Fv (scFv)
fragments of an antibody and the like, which retain
specific binding activity for a ligand, are included
within the definition of an antibody. Specific binding
activity of an antibody for a ligand can be readily
determined by one skilled in the art, for example, by
comparing the binding activity of an antibody to a
particular ligand versus a control ligand that differs
from the particular ligand. Specific binding can
similarly be determined for a binding molecule for the
ligand that is not an antibody. Methods of preparing
polyclonal or monoclonal antibodies are well known to
those skilled in the art (see, for example, Harlow and
Lane, Antibodies: A Laboratory Manual, Cold Spring Harbor
Laboratory Press (1988)).
In addition, the term "antibody" as used herein
includes naturally occurring antibodies as well as
non-naturally occurring antibodies, including, for
example, single chain antibodies, chimeric, bifunctional
and humanized antibodies, as well as antigen-binding
fragments thereof. Such non-naturally occurring
antibodies can be constructed using solid phase peptide
synthesis, can be produced recombinantly or can be
obtained, for example, by screening combinatorial
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
9
libraries consisting of variable heavy chains and
variable light chains as described by Huse et al.
(Science 246:1275-1281 (1989)). These and other methods
of making functional antibodies are well known to those
skilled in the art (Winter and Harris, Immunol. Today
14:243-246 (1993): Ward et al., Nature 341:544-546 (1989)
Harlow and Zane, supra, 1988); Hilyard et al., Protein
Enaineerina: A practical approach (IRZ Press 1992);
Borrabeck, Antibody Enaineerinq, 2d ed. (Oxford
University Press 1995)).
A particularly useful method for generating
antibodies is based on using combinatorial libraries
consisting of variable heavy chains. and variable light
chains (Kang et al., Proc. Natl. Acad. Sci. USA, 88:4363-
4366 (1991), Huse et al., Science 246:1275-1281 (1989)).
The advantage of using such a combinatorial antibody
library is that antibodies do not have to be individually
generated for each ligand of the ProtoChip. No prior
knowledge of the exact characteristics of the ligands on
the ProtoChip is required when using a combinatorial
antibody library.
As used herein, a "reagent antibody" refers to
an antibody used as a reagent for analysis of a sample,
that is, a non-analyte antibody. Although a reagent
antibody can be derived from a natural source, chemically
synthesized, or expressed recombinantly, it is understood
that a reagent antibody specifically excludes antibodies
in a sample to be analyzed. Similarly, a "reagent
binding molecule" such as a reagent receptor, polypeptide
or enzyme, as disclosed herein, is a binding molecule
used as a reagent for analysis of a sample, that is, a
non-analyte binding molecule.
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
As used herein, the term "population" is
intended to refer to a group of two or more different
molecules. Populations can range from two to tens to
hundreds to thousands, or even millions or billions or
5 more molecules. For example, a population can contain
about 3 or more, about 5 or more, about 7 or more, about
10 or more, about 15 or more, about 20 or more, about 30
or more, about 40 or more, about 50 or more, about 75 or
more, about 100 or more, about 200 or more, about 500 or
10 more, or even about 1000 or more molecules. A population
can also contain about 104 or more, about 105 or more,
about 106 or more, about 10' or more, about 10$ or more or
about 109 or more molecules, about 101° or more molecules,
about 1011 or more molecules, about 1012 or more molecules,
or even greater numbers of molecules. As used herein, a
"subset" when used in reference to a population refers to
group of molecules that is less than all of the
population.
As used herein, a molecule in a sample can be
essentially any type of molecule such as a polypeptide,
nucleic acid, carbohydrate, lipid, or any organic derived
compound. Moreover, derivatives and analogues are also
intended to be included within the definition of this
term. For example, polypeptides can be modified by
postranslational modifications or synthetic
modifications, including phosphorylation, lipidation,
prenylation, sulfation, hydroxylation, acetylation,
addition of carbohydrate, addition of prosthetic groups
or cofactors, formation of disulfide bonds, proteolysis,
assembly into macromolecular complexes, and the like.
The invention provides a composition comprising
a diverse population of reagent ligands attached to a
solid support and a diverse population of antibodies
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
11
specifically bound to the reagent ligands. Such a
composition is also termed a ProtoChip. The ligands can
be peptides, oligosaccharides, oligonucleotides, or
organic molecules.
The present invention provides compositions and
methods useful for determining the expressed epitopes of
a molecule in a sample from an individual. The methods
of the invention are particularly useful for mapping
epitopes on polypeptides expressed in a sample. Epitope
mapping has been described as a means to identify the
specific site to which an antibody binds on the surface
of a polypeptide. Traditionally, epitope mapping has
been done by synthesizing all the 5 to 15 amino acid
stretches of a known protein with a known sequence, where
the peptides are offset from each other by 3 to 10 amino
acids. The peptide epitope is identified as the one that
complexes with the antibody.
The present invention provides methods allowing
epitopes present and accessible on essentially any
polypeptide or molecule in a sample to be determined.
The invention is advantageous in that no prior knowledge
of the sample polypeptide or sequence is required, and
the analysis of samples containing unknown protein
mixtures becomes feasible.
The invention provides a ProtoChip, which is a
diverse population of reagent ligands attached to a solid
support and a diverse population of reagent antibodies
specifically bound to the ligands. In one embodiment,
the ligands are peptides attached to a solid support and
are essentially an immobilized combinatorial epitope
peptide library made up of combinations of amino acids
(Figure 1, step A). The ligands, which have binding
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
12
activity for antibodies, can be complexed with antibodies
to form a ProtoChip (Figure 1, step B). The antibodies
can be, for example, antibodies expressed as recombinant
ScFv.
The ProtoChip functions to detect the presence
of epitopes in a sample. If a sample is exposed to a
ProtoChip, those epitopes present in the sample and
accessible to antibody binding compete for binding of
antibodies to ligands (Figure 1, steps C and D). Thus,
antibodies having binding activity for epitopes present
in the sample, for example, epitopes on the surface of
polypeptides, are displaced from their specific ligand
epitope when exposed to competing epitopes in the sample.
The invention thus also provides a composition
comprising a diverse population of reagent ligands
attached to a solid support and a diverse population of
reagent antibodies specifically bound to a subset of the
reagent ligands, wherein an unbound ligand has binding
activity for an antibody having specificity for a
molecule in a sample (Figure 1).
The antibodies remaining bound to the subset of
ligands can be detected (Figure 1, step E). By
identifying the ligands which are unbound by antibody,
that is, ligands having binding activity for the
displaced antibodies specific for a molecule in a sample,
epitope expression in the sample can be determined.
Thus, a map of epitopes is generated that provides a
proteome fingerprint for a sample such as a biological
fluid. The present invention provides methods that are
accurate, reproducible, and fast. The methods can be
applied to pharmaceutical target identification, drug
discovery, diagnostics, pharmacoproteomics, structural
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
13
bioinformatics, agricultural biotechnology, drug
development, and species identification.
The invention additionally provides a method of
determining an epitope in a sample. The method includes
the steps of contacting a composition comprising a
diverse population of reagent ligands attached to a solid
support and a diverse population of antibodies
specifically bound to the reagent ligands with a sample;
and detecting the antibodies bound to the diverse
population of reagent ligands. The method can further
include the step of identifying which of the reagent
ligands is unbound by antibody. In the method, a reagent
ligand unbound by reagent antibody has binding activity
fox an antibody having specificity for a molecule in the
sample.
The compositions of the invention for
determining an epitope using antibodies or binding
activity using binding molecules contain a diverse
population of reagent ligands attached to a solid
support. The reagent ligands are bound by a diverse
population of reagent antibodies or reagent binding
molecules. If desired, each of the ligands can be bound
by antibody or binding molecules. This can be
accomplished by removing any ligands from the solid
support for which a corresponding binding antibody or
binding molecule is not found. Alternatively, prior to
addition of the sample, less than all of the reagent
ligands can have bound molecules, for example, to use as
a control or because corresponding binding molecules are
not found. In such a case, the ligands having unbound
antibodies or binding molecules can be tested prior to
addition of sample and discarded or used as a control, as
desired.
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
14
Thus, the invention provides a solid support
comprising a diverse population of reagent ligands and a
diverse population of reagent antibodies or reagent
binding molecules specifically bound to the ligands,
where all of the ligands are bound, about 99o of the
ligands are bound, about 980 of the ligands are bound,
about 950 of the ligands are bound about 900 of the
ligands are bound, about 850 of the ligands are bound,
about 800 of the ligands are bound, about 75o of the
ligands are bound, about 700 of the ligands are bound,
about 600 of the ligands are bound, about 500 of the
ligands are bound, about 400 of the ligands are bound,
about 300 of the ligands are bound, about 200 of the
ligands are bound, about 100 of the ligands are bound,
about 50 of the ligands are bound, or even less, if
desired.
Proteins are formed by a series of amino acids
linked together in long chains which fold into a
3-dimensional structure. Exposed on the surface of this
structure are short peptide segments that are
recognizable to antibodies. These antigenic peptides are
called epitopes. Other epitopes include any antigenic
determinant that can specifically bind to an antibody.
By analogy to the terms genome and proteome, the epitome
would be the entire collection of antigenic epitopes
present in an organism.
The epitome is unique to an organism, a
disease, or an individual. A map of the epitome would
therefore provide convenient, quantitative information
useful for identifying changes in the protein levels of
diseased tissues and identifying different organisms by
mapping all the antigenic surface peptides of the
proteome. The epitome would also contain small molecule
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
components and other antigenic biomolecules like
oligosaccharides and oligonucleotides.
A diverse population of peptide ligands can be
generated by methods well known to those skilled in the
5 art. For example, the peptides can be synthesized by
well known combinatorial methods (see, for example,
Eichler et al., Med. Res. Rev. 15:481-496 (1995); Wilson
and Czarnik, eds., Combinatorial Chemistry: Synthesis and
Application, John Wiley & Sons, New York (1997); U.S.
10 Patent Nos. 5,264,563 and 5,405,783; Haridason et al.,
Proc. Indian Natl. Sci. Acad. Part A' 53:717-728 (1987;
Furka et al., Int. J. Peptide Protein Res. 37:487-493
(1991)). Methods of synthesizing nucleic acids or
oligonucleotides ligands, oligosaccharide ligands, and
15 organic molecule ligands are well known to those skilled
in the art (see, for example, Ausubel et al., Current
Protocols in Molecular Bioloay (Supplement 47), John
Wiley & Sons, New York (1999); Sofia, Mol. Divers. 3:75-
94 (1998); Eichler et al., Med. Res. Rev. 15:481-496
(1995); Gordon et al., J. Med. Chem. 37: 1233-1251
(1994) Gordon et al., J. Med. Chem. 37: 1385-1401
(1994): Gordon et al., Acc. Chem. Res. 29:144-154 (1996);
Wilson and Czarnik, eds., Combinatorial Chemistry:
Synthesis and Application, John Wiley & Sons, New York
(1997)).
The epitome can be approximated in a
combinatorial fashion by synthetically building ligand
libraries, for example, peptide libraries, on a solid
support in such a way that the peptide sequence is known
based on its location on a ProtoChip. For example, a
5-mer peptide synthesized from 6 amino acids would result
in 65 (7776) possible combinations. A peptide library 5
amino acids long synthesized from the 20 naturally
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
17
to a single antibody that can be displaced by an epitope
in a sample.
A diverse population of antibodies can also be
synthesized by methods well known to those skilled in the
5~ art, as described above (see, for example, Huse et al.,
Science 246:1275-1281 (1989)). Variability in antibody
recognition is afforded by six
complementarity-determining regions (CDRs) on the heavy
and light chains of the antibody. By synthesizing the
cDNA stretches that encode the
complementarity-determining regions in a mixed pool
random fashion and presenting them on various mouse
antibody framework regions, a soluble antibody library
can be prepared containing at least 10'-6 different
antibodies (Breitling and Diibel, Recombinant Antibodies
John Wiley, New York (1998)). This antibody library
would present sufficient diversity to provide specific
tight-binding antibodies for each of the combinatorial
peptide epitope analogs or other ligand epitope analogs.
Depending on the nature and complexity of the sample to
be analyzed, the antibody library can be a naturally
occurring library of antibodies expressed in an organism,
in particular a mammal such as a human, primate, mouse,
rabbit, goat, and the like, as disclosed herein (see
Example I).
The form of the antibody used in the invention
can be any of the well known forms described herein. A
particularly useful form can be the ScFv form. The ScFv
form of an antibody can be conveniently generated as a
diverse population of antibodies for use in the invention
(Figure 3). The hypervariable regions in the heavy and
light chain variable regions can be synthesized with a
random DNA library that generates a diverse population of
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
18
ScFv antibodies. Such an antibody library will have
diverse binding affinities and specificities that can
bind to the diverse population of ligands. Commercial
systems are available for the expression of a recombinant
antibody library (see, for example, Amersham Pharmacia
Biotech; Piscataway NJ).
Panning the antibody library over a high
density peptide library such as a peptide chip, ligands
immobilized on microwell plates, or other ligand
libraries, allows the antibodies with the highest
affinity to associate with a specific peptide or other
ligand to generate the invention ProtoChip. Non-binding
antibodies are removed by washing. Challenging the
antibody bound ligands with a sample biological extract
causes competing sample molecules that contain the same
epitopes as the immobilized ligands to displace the
antibody from the surface of the chip. Following
washing, the remaining associated antibodies can be
visualized using a variety of methods, as disclosed
herein. For a 5 amino acid peptide library, the
generated library would amount to 3.2 million individual,
simultaneous immunoassays. As such, each of the epitopes
would be both identified and quantified. The epitopes
present generate a map of the protein extract.
The antibodies remaining bound to the diverse
population of ligands attached to the solid support can
be detected using well known methods. For example, an
antibody can be directly modified or a secondary agent
can be generated or modified to include a detectable
moiety, for example, a radiolabel, a fluorochrome, a
chromogen, a ferromagnetic substance, a luminescent tag,
a detectable binding agent such as biotin, an enzyme such
as horse radish peroxidase (HRP), alkaline phosphatase,
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
19
glucose oxidase, and the like, or other detectable
moieties known in the art that are detectable by
analytical methods. A particularly useful detectable
label is a fluorescent label. Methods suitable for
detecting such moieties include, for example,
fluorescence spectroscopy, autoradiography or
phosphorimaging, colorimetric detection, light detection,
or surface plasmon resonance.
As used herein, a label refers to single atoms
and molecules that are either directly or indirectly
involved in the production of a detectable signal. Any
label can be linked to an antibody or secondary agent.
These detectable atoms or molecules can be.used alone or
in conjunction with additional reagents. Such additional
reagents are well-known in clinical diagnostic chemistry.
The linking of a label to an antibody or secondary agent
is well known in the art. Antibodies can be labeled by
conjugating detectable labels, including enzymes, using
cross linking agents or, if the antibodies are expressed
recombinantly, for example, using antibody libraries, the
antibodies can be labeled by expressing the antibodies as
a fusion with a detectable peptide tag, for example, the
E tag or similar peptide tags (see Figure 3).
A secondary agent, which can specifically bind
to an antibody, can also be directly labeled or be
detectable by another reagent that is detectable. Thus,
an antibody directly labeled or bound to a secondary
agent that is labeled or detectable by another reagent
can be detected using well known immunological detection
methods (Harlow and Lane, supra, 1988; Harlow and Lane,
Using Antibodies: A Laboratory Manual, Cold Spring Harbor
Press (1999) ) .
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
The use of detectable labels is also convenient
far quantitating the amount of epitope in a sample. A
particularly useful detectable label for quantitation of
the amount of epitope is a fluorescent label.
5 Quantitative determinations can be made using well known
methods for describing binding interactions. The
relative concentration of an epitope can be related to
fluorescence intensity. Specific epitopes can be
quantified using a standard solution of the purified
10 epitope and generating a calibration curve.
Alternatively, the relative concentration for an unknown
epitope can be determined in relation to its dissociation
constant.
Although the methods of the invention are most
15 conveniently used with a detectable label of either the
antibodies or secondary agent, the binding of antibody
can also be detected using mass spectroscopy, for
example, matrix-assisted laser desorption-time of flight
(MALDI-TOF) mass spectroscopy, if desired. Detection by
20 MALDI-TOF analysis can also be used to determine partial
sequences of the antibodies, for example, by determining
the sequence of variable regions~or CDRs of the detected
antibodies.
The reagent ligands of the invention ProtoChip
,are conveniently attached to a solid support. The solid
support can be a membrane such as a nylon or
nitrocellulose membrane, glass, derivatized glass,
silicon, plastic or other substrates. The ligands can be
bound to a flat surface such as a membrane or plate or
can be bound to spheres or beads. In one embodiment, the
solid support can be in the form of a compact disc (CD).
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
21
A convenient format for the ligands can be an
array containing a plurality of ligands. As used herein,
an array refers to a format for presenting ligands where
the ligands are stably bound to a solid support and
arranged such that the binding to an antibody on the
array can be detected. An array format is particularly
convenient when the diverse population of ligands is a
large population and is useful as a high density
screening format.
For example, the format of the ProtoChip can
take the form of a CD in which the ligand library is
synthesized in discrete locations on the surface of the
CD. In addition to encoded data, instructions and
protocols using standard CD formatting, the ligand
library such as a peptide library can be synthesized
along the CD groove in discrete micron sized pits. The
standard sized CD contains sufficient space to
conservatively snoods 310 million different peptides.
Audio CDs measure the reflection of an infrared
photodiode laser's light from the surface of the CD. By
decreasing the wavelength to 340 nm using commercially
available photodiode laser and measuring the emitted
light from fluorescently tagged antibodies or secondary
agents, a table top confocal flourimeter can be
constructed. Increased sensitivity arises from having
the fluorophore immobilized on a solid support, which
effectively reduces the sample volume to a range that
would allow single molecule detection (Lu et al.,
Science, 282:1877-1882 (1998)). If desired, the methods
of the invention using ProtoChip technology can be
conveniently automated. Thus, coupled with a CD
processing unit, the ProtoChip of the invention can be
conveniently read using a desktop instrument in a doctors
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
22
office or diagnostic laboratory. The ligands can also be
attached in a multiwell format, if desired.
The ligands can be stably bound to a solid
support via covalent interactions or non-covalent
interactions so long as the ligands remain bound to the
solid support during incubation or wash steps required
for binding of antibodies and/or contacting with a
sample. Generally, ligands are attached to a solid
support, for example, through covalent bonds such as
chemical crosslinks, A ligand can also be modified with
an affinity tag that facilitates binding and or
crosslinking of the ligand to the solid support.
The sample is contacted with the ProtoChip
under conditions that allow specific binding of the
sample molecules to the antibodies such that the
antibodies are displaced from the ProtoChip. As used
herein, specific binding means binding that is measurably
different from a non-specific interaction. Specific
binding can be measured, for example, by determining
binding of a molecule compared to binding of a control
molecule, which generally is a molecule of similar
structure that does not have binding activity, for
example, a peptide of similar size that lacks binding
activity. Specificity of binding also can be determined,
for example, by competition with a control molecule, for
example, competition with an excess of the same molecule.
In this case, specific binding is indicated if the
binding of a molecule is competitively inhibited by
itself. Thus, specific binding between an antibody and
antigen is measurably different from a non-specific
interaction and occurs via the antigen binding site of
the antibody. An antigen such as a peptide has binding
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
23
activity for the antibody if the antibody specifically
binds to the peptide.
As used herein, selective binding refers to a
binding interaction that is both specific and
discriminating between molecules, for example, an
antibody that binds to a single molecule or closely
related molecules. For example, an antibody can exhibit
specificity for an antigen that can be both specific and
selective for the antigen if the epitope is unique to a
molecule. Thus, a molecule having selective binding can
differentiate between molecules, as exemplified by an
antibody having specificity for an epitope unique to one
molecule or closely related molecules. Alternatively, an
antibody can have specificity for an epitope that is
common to many molecules, for example, a carbohydrate
that is expressed on a number of molecules. Such an
antibody has specific binding but is not selective for
one molecule or closely related molecules.
As used herein, the term "sample" is intended
to mean any biological fluid, body fluid, cell, tissue,
organ or portion thereof, that includes one or more
different molecules that can function as antigens for
antibodies bound to ligands on the ProtoChip or for
binding molecules bound to ligands on the ProtoChip. The
molecules in the sample are potential analyte molecules.
The term includes samples obtained or derived from the
individual. For example, a sample can be a fluid sample
such as body fluid, including blood, plasma, urine,
saliva or sputum. A sample can also be a tissue section
obtained by biopsy, cells that are placed in or adapted
to tissue culture, or fractions or components purified or
extracted from a biological fluid, tissue or cell. When
using a cell or tissue sample, the sample can be
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
24
processed to generate an extract that can be conveniently
contacted with a ProtoChip using methods well known to
those skilled in the art (Harlow and Lane, supra, 1988;
Harlow and Lane, supra, 1999).
If desired, the sample can be prepared with
denaturants, including detergents such as sodium dodecyl
sulfate (SDS). In the absence of denaturants, the
epitopes accessible for binding to antibodies are the
epitopes expressed on the surface of molecules, for
example, the surface peptides of a folded protein. In
the presence of denaturants, essentially all of the
epitopes can become accessible, for example, due to
unfolding of a protein and exposure of buried amino acid
residues. Thus, conditions for treating the sample can
be chosen to determine either epitopes accessible to
antibody binding under native conditions or epitopes
accessible under denaturing conditions.
The identity of the proteins or other molecules
associated with increases or decreases in a given epitope
can be obtained by comparing the epitope sequence to a
sequence database such as that being generated by the
human genome project. Alternatively, the protein of
interest can be isolated using immunoaffinity techniques
with the antibody specific for that epitope and sequenced
using standard biochemical techniques. Mass spectroscopy
can also be used to identify the antibody. In addition,
the corresponding gene can be amplified from a cDNA
library by polymerase chain reaction (PCR) using a
degenerate primer corresponding to the epitope peptide.
Methods of amplifying sequences by PCR are well known to
those skilled in the art (Dieffenbach and Dveksler, PCR
Primer: A Laboratory Manual, Cold Spring Harbor Press
(1995); Ausubel et al., Current Protocols in Molecular
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
Bioloay (Supplement 47), John Wiley & Sons, New York
(1999) ) .
The invention further provides a method of
diagnosing a disease. The method includes the steps of
5 contacting a composition comprising a diverse population
of reagent ligands attached to a solid support and a
diverse population of reagent antibodies specifically
bound to the reagent ligands with a sample from an
individual; detecting the reagent antibodies bound to the
10 diverse population of reagent ligands; and identifying
which of the reagent ligands is unbound by reagent
antibody, wherein a reagent ligand unbound by reagent
antibody has binding activity for an antibody having
specificity for a molecule associated with the disease.
15 The methods of the invention can be applied to
generate a database of epitope maps for a variety of
tissues, causative proteins or those affected by a
disease, which can be readily identified and quantified.
Since the methods of the invention are used to measure
20 epitopes as opposed to whole protein sequences, changes
in post translational modification and proteolytic
processing can also be directly identified. The methods
of the invention can be used to determine if an
individual has a particular disease such as cancer,
25 Alzheimer's disease, cardiovascular diseases,
cerebrovascular diseases, congenital anomalies,
infectious diseases, parasitic diseases, endocrine
related diseases, nutritional diseases, metabolic
diseases, metabolic disorders, diabetes, blood diseases,
mental disorders, diseases of the nervous system,
circulatory diseases, respiratory diseases, digestive
diseases, genitourinary diseases, skin diseases,
perinatal conditions, inflammatory diseases, arthritis,
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
26
erectile or fertility disorders, renal diseases, liver
diseases, and gastrointestinal diseases.
The methods of the invention are therefore
useful for diagnostic applications. The. high specificity
of antibodies make them invaluable diagnostic tools. To
date, the development of antibody based diagnostics has
required a prior knowledge of the antigen. The
identification of these antigens in many cases is the
result of years of academic and industrial research.
Subsequently, specific epitopes on the antigen must be
identified, analogs synthesized, and injected into mice
in order to generate monoclonal antibodies which are
frequently nonspecific o:r have poor binding
characteristics. Since the present invention is directed
to measuring the epitome, analysis of biological fluids
can immediately generate a panel of specific tight
binding antibodies for disease related proteins without
requiring any prior knowledge of the antigen.
If desired, specific antibodies can be
recreated by immunization of mice with the identified
epitope to generate monoclonal antibodies. In addition,
specific antibodies can be generated by analyzing
biological fluids using a phage display antibody library
or by panning an antibody library over the ligand
followed by isolation and sequence analysis of the
recombinant antibody. However, identification of
specific antibodies is not required since a disease
specific ProtoChip can be produced based on disease
specific epitopes identified by methods of the invention.
In another embodiment, a diagnostic ProtoChip can be
produced that holds the epitopes that are diagnostic for
a wide variety of diseases or medical conditions.
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
27
The method of the invention can be used to
identify therapeutically useful antibodies. For example,
the identification of a tumor specific epitope using a
ProtoChip of the invention also provides the tumor
specific antibody associated with that epitope. This
antibody can useful therapeutically for the treatment of
cancer. Examples of antibody therapeutics for the
treatment of cancer include Herceptin and Rituxan. Due
to the specificity of the identified antibodies for the
tumor, the antibody can be used to target a tumor for
therapeutic or diagnostic purposes, or other disease
targets, as desired.
The methods of the invention can also be used
to identify antigens useful in the development of
vaccines. Screening an infectious agent using methods of
the invention using, for example, a ProtoChip, allows
identification of epitopes associated with the infectious
agent. The epitope can be used for preparation of a
vaccine, for example, by coupling the epitope to a
suitable carrier, and administered to an individual in a
pharmaceutical composition suitable for stimulating an
immune response. Such compositions suitable for
stimulating an immune response are well known to those
skilled in the art and can include, for example, a
physiologically acceptable carrier and/or an adjuvant
suitable for stimulating an immune response, as desired.
The methods of the invention can be
conveniently automated, if desired. Following automatic
washing and reagent additions within a ProtoChip
analyzer, the ProtoChip can be quantified, for example,
using fluorescence to detect bound antibodies. By
applying a droplet of body fluid on a diagnostic
ProtoChip and placing the chip into a processor and
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
28
reader, immediate in-office diagnostics can be applied to
a panel of disorders. The generated epitope fingerprint
is compared to a database of values that results in an
easily interpreted readout of the diagnosis. Among the
many foreseeable diagnostic applications, ProtoChips
specific for vascular diseases, neurological disorders,
metabolic diseases, or infectious diseases can be
produced in addition to an all purpose panel useful for
annual checkups.
An advantage of the present invention using
ProtoChip based diagnostics is that panels of antibodies
can be generated without any prior knowledge or
prejudices of the disease. Additionally, with the
appropriate fluids, specific diagnostics can be generated
in a matter of days or weeks as opposed to the current
standard of months or years. The present invention
provides more specificity as a result of multiple epitope
probes, more flexibility as a result of the ability to
multiplex different diagnostics on the same chip, and, as
a result of the ease of discovery, a shorter product
development time than other immunoassay diagnostics.
The methods of the invention are also useful
for drug development and pharmacoproteomic applications.
The unachieved goal of genetically characterising patient
populations in order to more efficiently target drugs to
those who would respond has been termed pharmacogenomics.
Three markets have been suggested for this proposed
application of genomic techniques: 1) assisting in drug
development at the clinical trial stage by targeting
patient populations who will most benefit, 2) reanalyses
of approved drugs that show disappointing efficacy in
order to reposition the patient population to those who
are most likely to improve, 3) reviving failed drug
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
29
candidates by weeding out patients prone to side effects
or non-response. As yet, pharmacogenomics has not become
a reality in part due to the poor correlation between
mRNA levels and biological response. Unlike genomic
approaches, the present invention allows for the
quantification of protein levels. As such, clinical
trials have a greater chance of success if the epitome of
the patient population is mapped to a homogenous group of
responders, the efficacy of marketed drugs can be
optimized to prescription practice as they change
resulting from analysis using the invention ProtoChip,
and failed drugs can be revived as the result of
uncovering the patient requirements through ProtoChip
mapping. The goals set forth for pharmacogenomics can be
realized using invention ProtoChip technology by
analyzing the epitome of the patient population.
The methods of the invention thus can be used
to provide information useful in drug development. For
example, if insulin were to be tested against a random
population of diabetics, it would likely show no
significant effect on the lowering of glucose levels. It
is only after selecting a group of subjects based upon
age of onset of symptoms that the therapeutic value of
insulin is realized for juvenile onset diabetes. In the
design of clinical trials, the selection of the wrong
patient subpopulation for the study or the lack of
selection criteria can lead to the failure of a
potentially valuable drug. By prescreening trial
candidates using methods of the invention, a near
homogeneous group of patients can be enlisted in order to
ensure the greatest chances for success.
Alternatively, ProtoChip analysis of patients
from an unbiased trial population can uncover specific
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
markers suggestive of the potential outcome of treatment.
Accordingly, without stratifying the patients prior to
the trial, it is possible that those subjects with a
given amount of a specific epitope show a greater chance
5 for responding to the drug. This observation can be
taken forward to the design of epitome based parameters
for the prescription of drugs. While this strategy can
serve to reduce the patient population to only those who
respond to a drug, the improved accuracy of prescriptions
10 can generate new markets for drugs that previously showed
limited efficacy or by reviving drugs that failed to
prove sufficient efficacy during clinical trials. Thus,
methods of the invention can be used in new clinical
trials for drugs that failed to show statistically
15 significant efficacy in previous clinical trials.
The methods of the invention can be used to
determine the epitome map and generate databases
describing the epitome for a variety of organisms. These
databases can include various pathogenic species, healthy
20 and diseased tissues from humans and economically
valuable animal species, drug efficacy profiles, plants,
insects, and other organisms such as bacteria, yeasts and
immortalized cell lines. Thus, the methods of the
invention are useful for identifying a species of
25 organism such as a species animal, plant or bacteria.
For example, a particular bacterium or strain
of bacterium can be identified using methods of the
invention. The methods can be used to identify various
bacteria such as pathogenic bacteria. For example, a
30 pathogenic strain such as a methacillin-resistant
Staphylococcus aureus strain can be identified using
methods of the invention. The precise identification of
a bacterial strain in a sample can be used to select an
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
31
appropriate antibiotic effective against the particular
organism.
Specific proteins can be mapped using protein
standards or proteins purified using the identified
antibody can be sequenced such that changes in the
epitome are correlated to a specific protein or group of
proteins. The databases identified by methods of the
invention are useful for the discovery of new
pharmacological targets, new agricultural traits,
insecticides and the development of diagnostic tools.
The methods of the invention can be used in diagnostic
applications such that a physician can place biological
samples into a ProtoChip reader and immediately be.
provided with the identity of infectious bacteria or
viruses and the recommended treatment guidelines based
upon that specific organism and its resistance profile.
The invention can also be used without a
combinatorial antibody library bound to the ligands.
Instead, a protein of interest can be applied to the
immobilized ligands. Evaluation of bound protein can be
used to identify ligands for the protein. These ligands
can then be used as leads for drug optimization, target
validation tools for pharmacology models, or for the
development of high throughput screening assays. This
method eliminates the need for any prior knowledge of
protein function or activity and allows a single assay
protocol to be used for high throughput screens.
The invention further provides a method of
mapping accessible epitopes of a polypeptide. The method
includes the steps of contacting a composition comprising
a diverse population of reagent ligands attached to a
solid support and a diverse population of reagent
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
32
antibodies specifically bound to the reagent ligands with
a polypeptide; detecting the reagent antibodies bound to
the diverse population of reagent ligands; and
identifying which of the reagent ligands is unbound by
reagent antibody, wherein a reagent ligand unbound by
reagent antibody has binding activity for an antibody
having specificity for a polypeptide epitope accessible
to the antibody. Such a method is particularly useful
when the ligands are peptides.
The methods of the invention can also be used
for protein structural determinations. The value of
genome sequence information is only realized upon
determination of the functional significance of the
encoded proteins. This function is imparted not through
the primary structure of the sequence itself but through
the tertiary structure, the three dimensional shape of
the protein. Structure determination methods have had
limited success in accurately predicting the structure of
a protein based solely on its sequence. The experimental
determination of a protein structure is slow and tedious.
Since the epitope map identifies surface peptides of a
protein, the methods of the invention using a specific
protein in place of the biological fluid sample provide
experimental structural information that can be coupled
with sequence information to predict the tertiary protein
structure. These predictions can be refined by structure
or sequence comparison to proteins with known structure
and function. The methods of the invention can thus be
used for the rapid functional analysis of genomic and
proteomic leads without the need to express and isolate
large amounts of protein and without the investment of
large amounts of time as is required using traditional
structural methods.
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
33
The identification of surface epitopes can be
combined with computational protein structure prediction
algorithms, including ab initio folding algorithms such
as the strings method (Moult, Curr. Opion. Biotechnol.
10:583-588 (1999) Selbig et al., Bioinformatics 15:1039-
1046 (1999); Osguthorpe, Curr. Opin. Struct. Biol.
10:146-152 (2000); Jonassen et al., Proteins 34:206-219
(1999)). Computational protein structure algorithms are
well known to those skilled in the art. The combination
of the identification of surface epitopes and folding
algorithms allows a more accurate prediction of tertiary
protein structure than with computational methods alone.
Competition with the ProtoChip and a purified protein
allows identification of the surface epitopes of the
protein. Under the constraint of having these epitopes
on the surface of the protein, there are fewer degrees of
freedom, for example, fewer low energy states, accessible
to the computational calculation. Thus, the combination
of the methods of the invention directed to identifying
surface epitopes of a protein with computational protein
structure prediction algorithms can be used to greatly
improve the accuracy and structure prediction of
polypeptides.
The determination of the three dimensional
structure of a protein has become a key component of drug
discovery. Currently this is accomplished through X-ray
crystallography or by NMR. Both of these methods are
limited by the physical properties of the protein, its
solubility, and its ability to crystallize. Frequently,
the determination of the three dimensional structure
takes a year or more. With the identification of
hundreds of potential targets from genomic and proteomic
studies, a method to calculate the three dimensional
structure based upon the protein sequence would
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
34
accelerate the drug discovery process. The epitope map
generated using the invention ProtoChip for a given
protein provides a low resolution map of the protein
that, when used in conjunction with computational
methods, can yield accurate representations of the
protein.
Immunoaffinity purification of proteins is
hampered by the difficulty in identifying appropriate
antibodies for the protein of interest. The protein must
first be purified in sufficient quantities to immunize
rabbits for the production of polyclonal antibodies or
mice for monoclonal antibodies. If a satisfactory immune
response is obtained, then the-antibodies can be
immobilized on a solid support to make an immunoaffinity
column. As a result of the high affinity of
traditionally prepared monoclonal or polyclonal
antibodies, elution of the studied protein from an
immunoaffinity column frequently results in the
denaturation of the protein. Therefore, the antibodies
raised against the protein are often not satisfactory for
use in purification columns. Using the ProtoChip of the
present invention, an antibody for any protein, without
prior purification or even characterization of that
protein, can be generated having a predefined
dissociation constant selected for binding
characteristics based on wash conditions in the ProtoChip
analysis. Exemplary variable wash conditions include
changing the pH, changing ionic strength, changing
temperature, changing wash time, or any combination
thereof. For example, using higher stringency wash
conditions such as increasing ionic strength and/or
varying other buffer components and conditions can be
used to select for antibodies having tighter binding
activity for the ligand. Therefore, the invention
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
ProtoChip can be used to develop specific immunoaffinity
columns for any protein.
While the invention ProtoChip and related
methods are useful in human health applications, the
5 methods of the invention can similarly be applied to
animal health and agricultural uses. The epitope map can
be determined using methods of the invention and used in
quality control, for example, of meat processing, animal
breeding programs, and disease screening. The ability to
10 quickly establish specific epitope maps can be used to
boost the success of captive breeding programs by
maximizing phenotypic rather than genotypic diversity.
Agricultural applications of the invention
methods can be extended to plants with the
15 characterization and identification of proteins that
impart beneficial effects such as insect resistance or
improved growth characteristics of a crop plant. Plant
epitome characterization can also be used in the
identification and classification of different plants.
20 Plant characterization can be useful in the development
of novel pharmaceuticals. For example, taxol was
discovered in the bark of the rare slow growing Taxus
brevifolia. Due to the scarcity of this plant,
production of this valuable drug was economically
25 limited. Tedious analysis of other plants in the Taxus
family showed that the common fast growing Taxus baccata
produced a chemically similar compound in its leaves that
is easily converted to the biologically active drug
taxol. Other examples of plant-derived drugs include the
30 lymphoma drug vinblastine, which is derived from the
Madagascar rosy periwinkle, and the muscle relaxant
curare, which is derived from the South American curare
vine. Similar botanical findings using methods of the
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
36
invention can prove useful in drug discovery while
preserving ecologically susceptible species.
The methods of the invention can also be used
to identify drug targets and are therefore useful in drug
discovery. Comparison of the epitope map of a biological
fluid from a healthy individual to the epitope map of a
biological fluid from a diseased individual can be used
to reveal epitopes specific for the disease state. By
identifying the protein associated with these disease-
associated epitopes, potential therapeutic targets can be
determined.
The invention additionally provides a method of
identifying a potential therapeutic target. The method
includes the steps of contacting a composition comprising
a diverse population of reagent ligands attached to a
solid support and a diverse population of reagent
antibodies specifically bound to the reagent ligands with
a sample from an individual having a disease; detecting
reagent antibody binding to the diverse population of
reagent ligands; comparing the reagent antibody binding
to the diverse population of reagent ligands to the
reagent antibody binding of a normal sample contacted
with the composition; and determining which of the
reagent ligands differs in reagent antibody binding
between the sample from the individual having a disease
and the normal sample, wherein a reagent ligand differing
in reagent antibody binding between the samples is a
potential therapeutic target.
Comparison of antibody binding in a sample from
a diseased individual to a normal sample, that is, a
sample from an individual not having the disease, can be
used to determine epitopes related to the disease based
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
37
on differences in antibody binding. A ligand of the
invention composition that differs in binding between
these samples is a potential therapeutic target. If
desired, a group of diseased individuals can be analyzed
and compared to a group of normal individuals, that is,
individuals not having the disease. A statistically
significant number of individuals can be selected for the
groups and used for comparison to determine which ligands
differ in antibody binding. For example, 50 individuals
can be selected for a group. The ligands that differ in
antibody binding between the samples can be further
characterized by the methods disclosed herein and used as
a potential therapeutic target to screen for drug
candidates useful in treating the disease.
The compositions and methods disclosed above
use antibodies bound to ligands. However, it is
understood that other binding molecules can be used to
bind to ligands for detecting the presence of a
corresponding binding activity in a sample using the
methods disclosed herein using antibodies. Other binding
molecules can include polypeptides, receptors, enzymes,
carbohydrates, lipids, and the like, so long as the
binding molecule can bind to the reagent ligand and has
the ability to potentially bind to a corresponding sample
molecule, such that displacement of the binding molecule
can be used to detect the presence of a molecule in the
sample, as disclosed herein.
When using antibodies attached to ligands, the
binding activity in the sample identified by methods of
the invention is referred to as an epitope. In the case
of using binding molecules other than antibodies, the
binding activity of the sample molecules is determined.
Accordingly, a sample that displaces a binding molecule
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
38
from a ligand has a binding activity for that binding
molecule, analogous to an epitope when an antibody is
used.
Thus, the invention provides a method of
determining a binding activity in a sample. The method
includes the steps of contacting a composition comprising
a diverse population of reagent ligands attached to a
solid support and a diverse population of reagent binding
molecules specifically bound to the reagent ligands with
a sample; and detecting the reagent binding molecules
bound to the diverse population of reagent ligands. The
method can further comprise the step of identifying which
of the reagent ligands is unbound by reagent binding
molecule. The reagent ligand unbound lay reagent molecule
has binding activity for a binding molecule having
specificity for a molecule in the sample.
It is understood that modifications which do
not substantially affect the activity of the various
embodiments of this invention are also included within
the definition of the invention provided herein.
Accordingly, the following examples are intended to
illustrate but not limit the present invention.
EXAMPLE I
Epitope Mapping of Plasmodium falciparum Merozoite
Surface Protein 1
This example describes mapping of epitopes of
the l9kDa C-terminal region of merozoite surface protein
1 (MSP1-19) from Plasmodium falciparum.
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
39
A natural human IgG antibody library was tested
for its ability to bind to peptides associated with the
l9kDa C-terminal region of merozoite surface protein 1
(MSP1-19) from Plasmodium falciparum (Kaslow et al., Mol.
Biochem. Parasitoloay 63:283-289 (1994)). The 89 amino
acid sequence from MSP1-19 was used for the epitope
mapping experiment (see Table 1).
Briefly, a library of pentamer peptides was
synthesized on polypropylene pins following the
procedures described by Geysen et al., Proc. Natl. Acad.
Sci. USA 81:3998-4002 (1984). These peptides represented
all five-amino-acid stretches of MSP1-19 offset by one
residue (Table 1). Peptide pins were precoated in
phosphate buffered saline (PBS), pH 7.2, containing 20
BSA and 0.1o TWEEN 20 for one hour at room temperature.
Five successive washes of the pins were carried out for
five minutes each with agitation in PBS. Human IgG
(Calbiochem; San Diego CA) was complexed to the peptides
by incubating 0.1 mg/mL IgG in PBS at 4°C for 30 hours.
Unbound antibody was removed by washing as described
above in 10 mM TRIS, pH 7.4 buffer containing 150 mM NaCl
(TBS) using new microtiter plates for transferring the
pins for each of the five washings. Anti-human IgG
(goat) alkaline phosphatase conjugate was diluted to 0.1
mg specific antibody/mL in TBS and incubated with the
peptide/antibody complex for one hour at room temperature
before washing five times with TBS, as described above.
The pins were then incubated in assay solution containing
10 mM TRIS, pH 8.0, 150 mM NaCl, 0.5 mM MgCl2, and 0.1 mM
4-methylumbelliferyl-phosphate for 30 minutes at room
temperature in the dark. Following incubation, the
fluorescence intensity of the assay solution was measured
in a Spectromax Gemini plate reader (Molecular Devices;
Sunnyvale CA)(ex 358nm/em 450 nm).
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
The peptide/antibody complexes on the pins were
washed five times in TBS, as described above, and
incubated with 50mg/mh MSP1-19 diluted in TBS for one
hour at room temperature. The pins were washed in TBS as
5 before and incubated in assay mixture for 30 minutes in
the dark prior to measurement of the fluorescent
intensity. Change in binding was determined using the
equation:
= C FMSP F b,MSP~ ~ C F 100.MSP F b,MSP~ - C FO F b.OJ ~ ~ F100, 0 F b.0~
10 where FMSP and Fo are the fluorescence intensities of
peptide containing pins after and before exposure to
MSP1-19, respectively. F b~MSP and F b,o are the
fluorescence intensities of pins with no peptide after
and before exposure to MSP1-19, respectively. F IOO,MSP and
15 Floo,o are the fluorescence intensities of pins containing
a control peptide, GZAQG (SEQ ID N0:90), after and prior
to exposure to MSP1-19.
Human IgG complexed with all peptide-containing
pins, with an average relative fluorescent intensity of
20 46744 (arbitrary units) while control pins without
peptides had an average relative fluorescence of 244.
The large fluorescence relative to the blank indicates
human IgG bound to the peptides, while non-specific
binding was not observed to pins lacking bound peptides.
25 The pins were pre-exposed to BSA. If significant amounts
of BSA were to bind to the pins, it would be expected
that the IgG would associate with BSA on the surface of
the pins as a result of IgG affinity for BSA. The
absence of IgG on the control pins indicates that BSA
30 does not associate in a non-specific fashion with the
pins under the assay conditions. The range of relative
fluorescence for the peptide pins was 25560 to 57880,
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
41
suggesting a gradient of binding affinities and
population density of the specific peptide-binding
antibodies. Exposure of the antibody/peptide pins to
MSP1-19 caused a decrease in fluorescence of greater than
10o in eleven peptides associated with two regions
corresponding to the sequences C49-D57 and N70-D88
(Table 1). There was no significant decrease in
fluorescence of control peptides upon exposure to
MSP1-19. Therefore, the antibodies bound to the peptides
dissociated from the pins as the result of competition by
equivalent epitopes on MSP1-19.
Table 1: Epitope Map of MSP1-19
Pep- Sequ- F Pep- Sequ- F Pep- Sequ- F
tide# ence (o) tide# ence (o) ide ence (o)
#
1 NISQH <5 31 LLNYK <5 61 KCTEE
2 ISQHQ <5 32 LNYKQ <5 62 CTEED <5
3 SQHQC <5 33 NYKQE <5 63 TEEDS <5
4 QHQCV <5 34 YKQEG <5 64 EEDSG 7
5 HQCVK <5 35 KQEGD <5 65 EDSGS 7
6 QCVKK <5 36 QEGDK <5 66 DSGSN 9
7 CVKKQ <5 37 EGDKC <5 67 SGSNG <5
8 VKKQC <5 38 GDKCV <5 68 GSNGK <5
9 KKQCP <5 39 DKCVE <5 69 SNGKK <5
10 KQCPQ <5 40 KCVEN <5 70 NGKKI 12
11 QCPQN <5 41 CVENP <5 71 GKKIT 6
12 CPQNS <5 42 VENPN <5 72 KKITC 9
13 PQNSG <5 43 ENPNP <5 73 KITCE 7
14 QNSGC <5 44 NPNPT <5 74 ITCEC 5
15 NSGCF <5 45 PNPTC <5 75 TCECT 12
16 SGCFR <5 46 NPTCN <5 76 CECTK <5
17 GCFRH <5 47 PTCNE <5 77 ECTKP 13
18 CFRHL <5 48 TCNEN <5 78 CTKPD 10
19 FRHLD <5 49 CNENN 16 79 TKPDS 11
20 RHLDE <5 50 NENNG 5 80 KPDSY 11
21 HLDER <5 51 ENNGG <5 81 PDSYP 9
22 LDERE <5 52 NNGGC 6 82 DSYPL 9
23 DEREE <5 53 NGGCD 10 83 SYPLF 13
24 EREEC <5 54 GGCDA <5 84 YPLFD 12
25 REECK <5 55 GCDAD 7 85 PLFDG 16
26 EECKC <5 56 CDADA <5 86 LFDGI 11
27 ECKCL <5 57 DADAK <5 87 FDGIF 9
28 CKCLL <5 58 ADAKC <5 88 DGIFC 7
29 KCLLN <5 59 DAKCT <5 89 GIFCS 11
30 CLLNY <5 60 AKCTE 5
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
42
Peptides 1-89 correspond to SEQ ID NOS:1-89,
respectively.
The most commonly identified serum anitibody
response for Kenyan malaria immune positive donors to
MSP1-19 peptides corresponded to C78-G91 (Egan et al.,
Infection and Immunity 65:3024-3031 (1997)). This
sequence overlaps with the N70-D88 epitope region
identified by epitope mapping in this study. The study
by Egan et al. showed that the region corresponding to
the C49-D57 epitope was observed at a lower frequency as
a serum antibody response, while other infrequently
observed MSP1-19 epitopes were also~:identified.
The maximum amount of antibody dissociated from
the peptide as the result of exposure to MSPl-19 was 16%.
The x-ray structure of MSP1-19 shows that the majority of
the amino acid residues in this protein are solvent
exposed and would be expected to have the potential to
bind antibodies. Epitope mapping, however identified
only two regions with significant IgG binding affinity.
C49-D57, corresponding to a short (3-sheet on the protein
surface, and N70-D88, a long strand of a ~i-sheet exposed
to the surface, were identified as epitopes, while the
inaccessible antiparallel strand was not identified as an
epitope.
These results demonstrate that antibody/peptide
arrays can be formed by the combination of peptide
libraries and antibody libraries. Furthermore, these
results demonstrate that antibodies from a peptide
library will associate with an antibody library, and
those antibodies can be dissociated upon exposure to a
competing protein or peptide.
CA 02408835 2002-11-12
WO 01/88538 PCT/USO1/15450
43
Throughout this application various
publications have been referenced. The disclosures of
these publications in their entireties are hereby
incorporated by reference in this application in order to
more fully describe the state of the art to which this
invention pertains. Although the invention has been
described with reference to the examples provided above,
it should be understood that various modifications can be
made without departing from the spirit of the invention.